diagnosis_qc_gigamuga_nine_batches
Hao He
2019-09-23
- Genotype diagnostics for diversity outbred mice
- library
- Generate json file for all batches
- Missing data per sample
- Sexes
- Sample duplicates
- Array intensities and Genotype frequencies
- Crossover counts and Genotyping error LOD scores
- Missing data in Markers and Genotype frequencies Markers
- Remove bad samples
Last updated: 2019-09-23
Checks: 7 0
Knit directory: csna_workflow/
This reproducible R Markdown analysis was created with workflowr (version 1.4.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190918) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Untracked files:
Untracked: analysis/01_geneseek2qtl2.R
Untracked: analysis/01_geneseek2qtl2.Rout
Untracked: analysis/01_geneseek2qtl2.script
Untracked: analysis/01_geneseek2qtl2.stderr
Untracked: analysis/01_geneseek2qtl2.stdout
Untracked: analysis/02_geneseek2intensity.R
Untracked: analysis/02_geneseek2intensity.Rout
Untracked: analysis/02_geneseek2intensity.script
Untracked: analysis/02_geneseek2intensity.stderr
Untracked: analysis/02_geneseek2intensity.stdout
Untracked: analysis/03_firstgm2genoprobs.R
Untracked: analysis/03_firstgm2genoprobs.Rout
Untracked: analysis/03_firstgm2genoprobs.script
Untracked: analysis/03_firstgm2genoprobs.stderr
Untracked: analysis/03_firstgm2genoprobs.stdout
Untracked: analysis/04_diagnosis_qc_gigamuga_nine_batches.R
Untracked: analysis/04_diagnosis_qc_gigamuga_nine_batches.Rout
Untracked: analysis/04_diagnosis_qc_gigamuga_nine_batches.script
Untracked: analysis/04_diagnosis_qc_gigamuga_nine_batches.stderr
Untracked: analysis/04_diagnosis_qc_gigamuga_nine_batches.stdout
Untracked: analysis/05_after_diagnosis_qc_gigamuga_nine_batches.R
Untracked: analysis/05_after_diagnosis_qc_gigamuga_nine_batches.Rout
Untracked: analysis/05_after_diagnosis_qc_gigamuga_nine_batches.script
Untracked: analysis/05_after_diagnosis_qc_gigamuga_nine_batches.stderr
Untracked: analysis/05_after_diagnosis_qc_gigamuga_nine_batches.stdout
Untracked: analysis/06_final_pr_apr_69K.R
Untracked: analysis/06_final_pr_apr_69K.Rout
Untracked: analysis/06_final_pr_apr_69K.script
Untracked: analysis/06_final_pr_apr_69K.stderr
Untracked: analysis/06_final_pr_apr_69K.stdout
Untracked: analysis/07.1_html_founder_prop.R
Untracked: analysis/07.1_html_founder_prop.Rout
Untracked: analysis/07.1_html_founder_prop.script
Untracked: analysis/07.1_html_founder_prop.stderr
Untracked: analysis/07.1_html_founder_prop.stdout
Untracked: analysis/07_recomb_size_founder_prop.R
Untracked: analysis/07_recomb_size_founder_prop.Rmd
Untracked: analysis/07_recomb_size_founder_prop.Rout
Untracked: analysis/07_recomb_size_founder_prop.script
Untracked: analysis/07_recomb_size_founder_prop.stderr
Untracked: analysis/07_recomb_size_founder_prop.stdout
Untracked: analysis/08_gcta_herit.R
Untracked: analysis/09_qtlmapping.R
Untracked: analysis/Novelty_resids_datarelease_07302918.gcta.Rout
Untracked: analysis/Novelty_resids_datarelease_07302918.gcta.script
Untracked: analysis/Novelty_resids_datarelease_07302918.gcta.stderr
Untracked: analysis/Novelty_resids_datarelease_07302918.gcta.stdout
Untracked: analysis/Novelty_resids_datarelease_07302918_m1.Rout
Untracked: analysis/Novelty_resids_datarelease_07302918_m1.script
Untracked: analysis/Novelty_resids_datarelease_07302918_m1.stderr
Untracked: analysis/Novelty_resids_datarelease_07302918_m1.stdout
Untracked: analysis/Novelty_resids_datarelease_07302918_m2.Rout
Untracked: analysis/Novelty_resids_datarelease_07302918_m2.script
Untracked: analysis/Novelty_resids_datarelease_07302918_m2.stderr
Untracked: analysis/Novelty_resids_datarelease_07302918_m2.stdout
Untracked: analysis/Novelty_resids_datarelease_07302918_m3.Rout
Untracked: analysis/Novelty_resids_datarelease_07302918_m3.script
Untracked: analysis/Novelty_resids_datarelease_07302918_m3.stderr
Untracked: analysis/Novelty_resids_datarelease_07302918_m3.stdout
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918.gcta.Rout
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918.gcta.script
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918.gcta.stderr
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918.gcta.stdout
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918_m1.Rout
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918_m1.script
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918_m1.stderr
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918_m1.stdout
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918_m2.Rout
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918_m2.script
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918_m2.stderr
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918_m2.stdout
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918_m3.Rout
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918_m3.script
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918_m3.stderr
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918_m3.stdout
Untracked: analysis/run_01_geneseek2qtl2.R
Untracked: analysis/run_02_geneseek2intensity.R
Untracked: analysis/run_03_firstgm2genoprobs.R
Untracked: analysis/run_04_diagnosis_qc_gigamuga_nine_batches.R
Untracked: analysis/run_05_after_diagnosis_qc_gigamuga_nine_batches.R
Untracked: analysis/run_06_final_pr_apr_69K.R
Untracked: analysis/run_07.1_html_founder_prop.R
Untracked: analysis/run_07_recomb_size_founder_prop.R
Untracked: analysis/run_08_gcta_herit.R
Untracked: analysis/run_09_qtlmapping.R
Untracked: code/reconst_utils.R
Untracked: data/FinalReport/
Untracked: data/GCTA/
Untracked: data/GM/
Untracked: data/Jackson_Lab_Bubier_MURGIGV01/
Untracked: data/cc_variants.sqlite
Untracked: data/marker_grid_0.02cM_plus.txt
Untracked: data/mouse_genes_mgi.sqlite
Untracked: data/pheno/
Untracked: docs/figure/
Untracked: output/AfterQC_Percent_missing_genotype_data.pdf
Untracked: output/AfterQC_Proportion_matching_genotypes_after_removal_of_bad_samples.pdf
Untracked: output/AfterQC_Proportion_matching_genotypes_before_removal_of_bad_samples.pdf
Untracked: output/AfterQC_number_crossover.pdf
Untracked: output/Percent_genotype_errors_obs_vs_predicted.pdf
Untracked: output/Percent_missing_genotype_data.pdf
Untracked: output/Percent_missing_genotype_data_per_marker.pdf
Untracked: output/Proportion_matching_genotypes_after_removal_of_bad_samples.pdf
Untracked: output/Proportion_matching_genotypes_before_removal_of_bad_samples.pdf
Untracked: output/genotype_error_marker.pdf
Untracked: output/genotype_frequency_marker.pdf
Untracked: output/m1.Novelty_resids_datarelease_07302918.RData
Untracked: output/m1.Novelty_residuals_RankNormal_datarelease_07302918.RData
Untracked: output/m2.Novelty_resids_datarelease_07302918.RData
Untracked: output/m2.Novelty_residuals_RankNormal_datarelease_07302918.RData
Untracked: output/num.geno.pheno.in.Novelty_resids_datarelease_07302918.csv
Untracked: output/num.geno.pheno.in.Novelty_residuals_RankNormal_datarelease_07302918.csv
Untracked: output/number_crossover.pdf
Untracked: output/prop_across_generation_chr_p.RData
Unstaged changes:
Modified: README.md
Modified: _workflowr.yml
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 680f68c | xhyuo | 2019-09-23 | First build |
Genotype diagnostics for diversity outbred mice
We first load the R/qtl2 package and the data. We’ll also load the R/broman package for some utilities and plotting functions, and R/qtlcharts for interactive graphs.
library
library(broman)
library(qtl2)
library(qtlcharts)
library(ggplot2)
library(ggrepel)
library(DOQTL)
library(mclust)
source("code/reconst_utils.R")Generate json file for all batches
#total sample id 2514
#load json file for the nine batches
gm <- get(load("data/Jackson_Lab_Bubier_MURGIGV01/gm_2514.RData"))
gmObject of class cross2 (crosstype "do")
Total individuals 2514
No. genotyped individuals 2514
No. phenotyped individuals 2514
No. with both geno & pheno 2514
No. phenotypes 1
No. covariates 2
No. phenotype covariates 0
No. chromosomes 20
Total markers 112729
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15
8555 8666 6420 6615 6571 6444 6294 5677 5870 5447 6352 5167 5274 5039 4555
16 17 18 19 X
4369 4330 4002 3108 3974 Missing data per sample
gmObject of class cross2 (crosstype "do")
Total individuals 2514
No. genotyped individuals 2514
No. phenotyped individuals 2514
No. with both geno & pheno 2514
No. phenotypes 1
No. covariates 2
No. phenotype covariates 0
No. chromosomes 20
Total markers 112729
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15
8555 8666 6420 6615 6571 6444 6294 5677 5870 5447 6352 5167 5274 5039 4555
16 17 18 19 X
4369 4330 4002 3108 3974 percent_missing <- n_missing(gm, "ind", "prop")*100
setScreenSize(height=100, width=300)Set screen size to height=100 x width=300labels <- paste0(names(percent_missing), " (", round(percent_missing,2), "%)")
iplot(seq_along(percent_missing), percent_missing, indID=labels,
chartOpts=list(xlab="Mouse", ylab="Percent missing genotype data",
ylim=c(0, 60)))#save into pdf
pdf(file = "output/Percent_missing_genotype_data.pdf", width = 20, height = 20)
labels <- as.character(do.call(rbind.data.frame, strsplit(ind_ids(gm), "V01_"))[,2])
labels[percent_missing < 5] = ""
# Change point shapes and colors
p <- ggplot(data = data.frame(Mouse=seq_along(percent_missing),
Percent_missing_genotype_data = percent_missing,
batch = factor(as.character(do.call(rbind.data.frame, strsplit(ind_ids(gm), "_"))[,5]))),
aes(x=Mouse, y=Percent_missing_genotype_data, color = batch)) +
geom_point() +
geom_hline(yintercept=5, linetype="solid", color = "red") +
geom_text_repel(aes(label=labels), vjust = 0, nudge_y = 0.01, show.legend = FALSE, size=3) +
theme(text = element_text(size = 20))
p
dev.off()png
2 p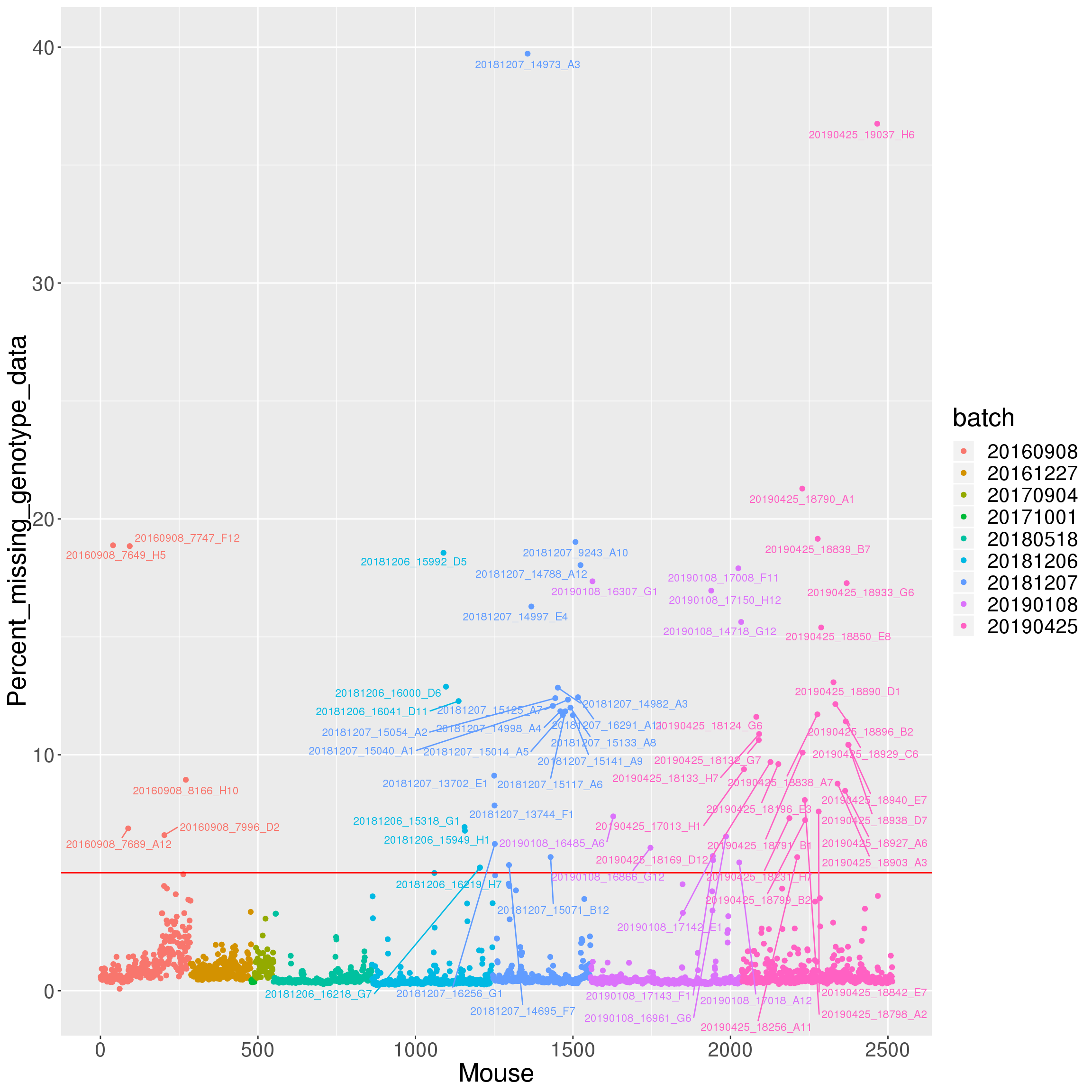
save(percent_missing,
file = "data/Jackson_Lab_Bubier_MURGIGV01/percent_missing_id.RData")Sexes
xint <- read_csv_numer("data/Jackson_Lab_Bubier_MURGIGV01/Jackson_Lab_Bubier_MURGIGV01_qtl2_chrXint.csv", transpose=TRUE)
yint <- read_csv_numer("data/Jackson_Lab_Bubier_MURGIGV01/Jackson_Lab_Bubier_MURGIGV01_qtl2_chrYint.csv", transpose=TRUE)
# Gigamuga marker annotation file from UNC.
gm_marker_file = "http://csbio.unc.edu/MUGA/snps.gigamuga.Rdata" #FIXED
# Read in the UNC GigaMUGA SNPs and clusters.
load(url(gm_marker_file))
#subset down to gm
snps$marker = as.character(snps$marker)
#snp <- snps[snps$marker %in% marker_names(gm),]
#load the intensities.fst.RData
load("data/Jackson_Lab_Bubier_MURGIGV01/intensities.fst.RData")
#X and Y channel
X <- result[result$channel == "X",]
rownames(X) <- X$snp
X <- X[,c(-1,-2)]
Y <- result[result$channel == "Y",]
rownames(Y) <- Y$snp
Y <- Y[,c(-1,-2)]
#determine sex
sex = determine_sex_chry_m(x = X, y = Y, markers = snps)$sex
gm$covar <- merge(data.frame(id = names(sex),
predict.sex = sex,stringsAsFactors = F),
gm$covar,
by.x = "id",
by.y = "row.names")
rownames(gm$covar) <- gm$covar$id
#sex order
sex <- gm$covar[rownames(xint),"predict.sex"]
x_pval <- apply(xint, 2, function(a) t.test(a ~ sex)$p.value)
y_pval <- apply(yint, 2, function(a) t.test(a ~ sex)$p.value)
xint_ave <- rowMeans(xint[, x_pval < 0.05/length(x_pval)], na.rm=TRUE)
yint_ave <- rowMeans(yint[, y_pval < 0.05/length(y_pval)], na.rm=TRUE)
point_colors <- as.character( brocolors("web")[c("green", "purple")] )
labels <- paste0(names(xint_ave))
iplot(xint_ave, yint_ave, group=sex, indID=labels,
chartOpts=list(pointcolor=point_colors, pointsize=4,
xlab="Average X chr intensity", ylab="Average Y chr intensity"))phetX <- rowSums(gm$geno$X == 2)/rowSums(gm$geno$X != 0)
phetX <- phetX[names(phetX) %in% names(xint_ave)]
iplot(xint_ave, phetX, group=sex, indID=labels,
chartOpts=list(pointcolor=point_colors, pointsize=4,
xlab="Average X chr intensity", ylab="Proportion het on X chr"))Sample duplicates
cg <- compare_geno(gm, cores=10)
summary.cg <- summary(cg)
summary.cg ind1
Jackson_Lab_Bubier_MURGIGV01_20190108_16305_E1
Jackson_Lab_Bubier_MURGIGV01_20190425_18160_C11
Jackson_Lab_Bubier_MURGIGV01_20170904_8673_B4
Jackson_Lab_Bubier_MURGIGIV01_20171001_9373_H7
Jackson_Lab_Bubier_MURGIGIV01_20171001_9375_E7
Jackson_Lab_Bubier_MURGIGIV01_20171001_9255_H11
Jackson_Lab_Bubier_MURGIGIV01_20171001_9371_G8
Jackson_Lab_Bubier_MURGIGV01_20180518_13885_B9
Jackson_Lab_Bubier_MURGIGIV01_20171001_9258_F8
Jackson_Lab_Bubier_MURGIGIV01_20171001_9254_F11
Jackson_Lab_Bubier_MURGIGV01_20170904_8616_C4
Jackson_Lab_Bubier_MURGIGV01_20170904_8618_E3
Jackson_Lab_Bubier_MURGIGV01_20170904_8617_A4
Jackson_Lab_Bubier_MURGIGV01_20161227_8664_E10
Jackson_Lab_Bubier_MURGIGV01_20170904_9378_C8
Jackson_Lab_Bubier_MURGIGV01_20161227_8585_B8
Jackson_Lab_Bubier_MURGIGV01_20170904_8674_F3
Jackson_Lab_Bubier_MURGIGV01_20160908_8184_E10
Jackson_Lab_Bubier_MURGIGV01_20161227_8166_H3
Jackson_Lab_Bubier_MURGIGV01_20160908_8167_F10
Jackson_Lab_Bubier_MURGIGV01_20160908_8168_D10
Jackson_Lab_Bubier_MURGIGV01_20160908_8183_G10
Jackson_Lab_Bubier_MURGIGV01_20160908_8185_C10
Jackson_Lab_Bubier_MURGIGV01_20181207_14998_A4
Jackson_Lab_Bubier_MURGIGV01_20181207_15054_A2
Jackson_Lab_Bubier_MURGIGV01_20181207_15054_A2
Jackson_Lab_Bubier_MURGIGV01_20181207_15014_A5
Jackson_Lab_Bubier_MURGIGV01_20181207_15014_A5
Jackson_Lab_Bubier_MURGIGV01_20181207_14982_A3
Jackson_Lab_Bubier_MURGIGV01_20181207_14998_A4
Jackson_Lab_Bubier_MURGIGV01_20181207_14998_A4
Jackson_Lab_Bubier_MURGIGV01_20181207_15014_A5
Jackson_Lab_Bubier_MURGIGV01_20181207_15054_A2
Jackson_Lab_Bubier_MURGIGV01_20181207_14998_A4
Jackson_Lab_Bubier_MURGIGV01_20181207_15014_A5
Jackson_Lab_Bubier_MURGIGV01_20181207_15040_A1
Jackson_Lab_Bubier_MURGIGV01_20181207_15014_A5
Jackson_Lab_Bubier_MURGIGV01_20181207_15054_A2
Jackson_Lab_Bubier_MURGIGV01_20181207_14982_A3
Jackson_Lab_Bubier_MURGIGV01_20181207_15054_A2
Jackson_Lab_Bubier_MURGIGV01_20181207_15054_A2
Jackson_Lab_Bubier_MURGIGV01_20181207_15040_A1
Jackson_Lab_Bubier_MURGIGV01_20181207_14998_A4
Jackson_Lab_Bubier_MURGIGV01_20181207_15040_A1
Jackson_Lab_Bubier_MURGIGV01_20181207_15133_A8
Jackson_Lab_Bubier_MURGIGV01_20181207_14998_A4
Jackson_Lab_Bubier_MURGIGV01_20181207_15054_A2
Jackson_Lab_Bubier_MURGIGV01_20181207_14982_A3
Jackson_Lab_Bubier_MURGIGV01_20181207_15117_A6
Jackson_Lab_Bubier_MURGIGV01_20181207_15117_A6
Jackson_Lab_Bubier_MURGIGV01_20181207_15054_A2
Jackson_Lab_Bubier_MURGIGV01_20181207_14982_A3
Jackson_Lab_Bubier_MURGIGV01_20181207_15040_A1
Jackson_Lab_Bubier_MURGIGV01_20181207_14982_A3
Jackson_Lab_Bubier_MURGIGV01_20181207_15141_A9
Jackson_Lab_Bubier_MURGIGV01_20181207_15040_A1
Jackson_Lab_Bubier_MURGIGV01_20181207_15133_A8
Jackson_Lab_Bubier_MURGIGV01_20181207_15040_A1
Jackson_Lab_Bubier_MURGIGV01_20181207_15125_A7
Jackson_Lab_Bubier_MURGIGV01_20181207_15040_A1
Jackson_Lab_Bubier_MURGIGV01_20160908_7996_D2
Jackson_Lab_Bubier_MURGIGV01_20181207_15125_A7
Jackson_Lab_Bubier_MURGIGV01_20181207_14982_A3
Jackson_Lab_Bubier_MURGIGV01_20181207_14982_A3
Jackson_Lab_Bubier_MURGIGV01_20181206_16041_D11
Jackson_Lab_Bubier_MURGIGV01_20181207_15117_A6
Jackson_Lab_Bubier_MURGIGV01_20181207_15117_A6
Jackson_Lab_Bubier_MURGIGV01_20181206_16000_D6
Jackson_Lab_Bubier_MURGIGV01_20181206_16041_D11
Jackson_Lab_Bubier_MURGIGV01_20181207_15040_A1
Jackson_Lab_Bubier_MURGIGV01_20181207_15040_A1
Jackson_Lab_Bubier_MURGIGV01_20181207_15125_A7
Jackson_Lab_Bubier_MURGIGV01_20181206_16041_D11
Jackson_Lab_Bubier_MURGIGV01_20181206_16000_D6
Jackson_Lab_Bubier_MURGIGV01_20181206_16000_D6
Jackson_Lab_Bubier_MURGIGV01_20181206_16041_D11
Jackson_Lab_Bubier_MURGIGV01_20190425_18229_F7
Jackson_Lab_Bubier_MURGIGV01_20181206_16000_D6
Jackson_Lab_Bubier_MURGIGV01_20181206_16041_D11
Jackson_Lab_Bubier_MURGIGV01_20181206_16041_D11
Jackson_Lab_Bubier_MURGIGV01_20181206_16000_D6
Jackson_Lab_Bubier_MURGIGV01_20181206_16041_D11
Jackson_Lab_Bubier_MURGIGV01_20181206_16000_D6
Jackson_Lab_Bubier_MURGIGV01_20181206_16000_D6
Jackson_Lab_Bubier_MURGIGV01_20181206_16041_D11
Jackson_Lab_Bubier_MURGIGV01_20181206_16000_D6
Jackson_Lab_Bubier_MURGIGV01_20181206_16041_D11
Jackson_Lab_Bubier_MURGIGV01_20181206_16000_D6
Jackson_Lab_Bubier_MURGIGV01_20181206_16041_D11
Jackson_Lab_Bubier_MURGIGV01_20181206_16000_D6
Jackson_Lab_Bubier_MURGIGV01_20181206_16000_D6
Jackson_Lab_Bubier_MURGIGV01_20160908_8166_H10
Jackson_Lab_Bubier_MURGIGV01_20160908_8166_H10
ind2 prop_match n_mismatch
Jackson_Lab_Bubier_MURGIGV01_20190108_16487_C10 1.00 30
Jackson_Lab_Bubier_MURGIGV01_20190425_18162_E11 1.00 40
Jackson_Lab_Bubier_MURGIGV01_20170904_8669_D5 1.00 46
Jackson_Lab_Bubier_MURGIGV01_20181207_9373_G10 1.00 52
Jackson_Lab_Bubier_MURGIGV01_20181207_9375_B10 1.00 57
Jackson_Lab_Bubier_MURGIGV01_20181207_9255_D10 1.00 61
Jackson_Lab_Bubier_MURGIGV01_20181207_9371_F10 1.00 62
Jackson_Lab_Bubier_MURGIGV01_20181206_13885_A8 1.00 67
Jackson_Lab_Bubier_MURGIGV01_20170904_9386_E9 1.00 79
Jackson_Lab_Bubier_MURGIGV01_20181207_9254_C10 1.00 81
Jackson_Lab_Bubier_MURGIGV01_20170904_8597_E5 1.00 85
Jackson_Lab_Bubier_MURGIGV01_20170904_8614_A5 1.00 86
Jackson_Lab_Bubier_MURGIGV01_20170904_8613_C5 1.00 94
Jackson_Lab_Bubier_MURGIGV01_20170904_8664_C1 1.00 111
Jackson_Lab_Bubier_MURGIGV01_20181207_9378_E10 1.00 168
Jackson_Lab_Bubier_MURGIGV01_20170904_8585_B1 1.00 172
Jackson_Lab_Bubier_MURGIGV01_20170904_8670_B5 1.00 246
Jackson_Lab_Bubier_MURGIGV01_20161227_8184_F3 1.00 288
Jackson_Lab_Bubier_MURGIGV01_20161227_8166_C1 1.00 294
Jackson_Lab_Bubier_MURGIGV01_20161227_8167_F4 1.00 383
Jackson_Lab_Bubier_MURGIGV01_20161227_8168_E3 0.99 666
Jackson_Lab_Bubier_MURGIGV01_20161227_8183_G3 0.99 677
Jackson_Lab_Bubier_MURGIGV01_20161227_8185_B4 0.99 1253
Jackson_Lab_Bubier_MURGIGV01_20181207_15014_A5 0.98 1999
Jackson_Lab_Bubier_MURGIGV01_20181207_15014_A5 0.98 2029
Jackson_Lab_Bubier_MURGIGV01_20181207_14998_A4 0.98 2090
Jackson_Lab_Bubier_MURGIGV01_20181207_15141_A9 0.98 2136
Jackson_Lab_Bubier_MURGIGV01_20181207_15133_A8 0.98 2161
Jackson_Lab_Bubier_MURGIGV01_20181207_15014_A5 0.98 2172
Jackson_Lab_Bubier_MURGIGV01_20181207_15141_A9 0.98 2231
Jackson_Lab_Bubier_MURGIGV01_20181207_15133_A8 0.98 2239
Jackson_Lab_Bubier_MURGIGV01_20181207_15117_A6 0.98 2257
Jackson_Lab_Bubier_MURGIGV01_20181207_15141_A9 0.98 2256
Jackson_Lab_Bubier_MURGIGV01_20181207_15117_A6 0.98 2274
Jackson_Lab_Bubier_MURGIGV01_20181207_16291_A11 0.98 2261
Jackson_Lab_Bubier_MURGIGV01_20181207_15014_A5 0.98 2272
Jackson_Lab_Bubier_MURGIGV01_20181207_15125_A7 0.97 2279
Jackson_Lab_Bubier_MURGIGV01_20181207_15133_A8 0.97 2280
Jackson_Lab_Bubier_MURGIGV01_20181207_14998_A4 0.97 2286
Jackson_Lab_Bubier_MURGIGV01_20181207_14982_A3 0.97 2300
Jackson_Lab_Bubier_MURGIGV01_20181207_15117_A6 0.97 2341
Jackson_Lab_Bubier_MURGIGV01_20181207_14998_A4 0.97 2355
Jackson_Lab_Bubier_MURGIGV01_20181207_16291_A11 0.97 2358
Jackson_Lab_Bubier_MURGIGV01_20181207_15054_A2 0.97 2360
Jackson_Lab_Bubier_MURGIGV01_20181207_15141_A9 0.97 2405
Jackson_Lab_Bubier_MURGIGV01_20181207_15125_A7 0.97 2402
Jackson_Lab_Bubier_MURGIGV01_20181207_16291_A11 0.97 2384
Jackson_Lab_Bubier_MURGIGV01_20181207_15133_A8 0.97 2406
Jackson_Lab_Bubier_MURGIGV01_20181207_15133_A8 0.97 2444
Jackson_Lab_Bubier_MURGIGV01_20181207_15141_A9 0.97 2462
Jackson_Lab_Bubier_MURGIGV01_20181207_15125_A7 0.97 2439
Jackson_Lab_Bubier_MURGIGV01_20181207_15141_A9 0.97 2460
Jackson_Lab_Bubier_MURGIGV01_20181207_15141_A9 0.97 2502
Jackson_Lab_Bubier_MURGIGV01_20181207_15117_A6 0.97 2498
Jackson_Lab_Bubier_MURGIGV01_20181207_16291_A11 0.97 2528
Jackson_Lab_Bubier_MURGIGV01_20181207_14982_A3 0.97 2510
Jackson_Lab_Bubier_MURGIGV01_20181207_16291_A11 0.97 2528
Jackson_Lab_Bubier_MURGIGV01_20181207_15133_A8 0.97 2543
Jackson_Lab_Bubier_MURGIGV01_20181207_15141_A9 0.97 2551
Jackson_Lab_Bubier_MURGIGV01_20181207_15117_A6 0.97 2560
Jackson_Lab_Bubier_MURGIGV01_20161227_7996_B1 0.97 2959
Jackson_Lab_Bubier_MURGIGV01_20181207_15133_A8 0.97 2575
Jackson_Lab_Bubier_MURGIGV01_20181207_16291_A11 0.97 2552
Jackson_Lab_Bubier_MURGIGV01_20181207_15125_A7 0.97 2556
Jackson_Lab_Bubier_MURGIGV01_20181207_15014_A5 0.97 2592
Jackson_Lab_Bubier_MURGIGV01_20181207_16291_A11 0.97 2602
Jackson_Lab_Bubier_MURGIGV01_20181207_15125_A7 0.97 2620
Jackson_Lab_Bubier_MURGIGV01_20181207_15014_A5 0.97 2609
Jackson_Lab_Bubier_MURGIGV01_20181207_14998_A4 0.97 2624
Jackson_Lab_Bubier_MURGIGV01_20181207_16291_A11 0.97 2612
Jackson_Lab_Bubier_MURGIGV01_20181207_15125_A7 0.97 2637
Jackson_Lab_Bubier_MURGIGV01_20181207_16291_A11 0.97 2655
Jackson_Lab_Bubier_MURGIGV01_20181207_15054_A2 0.97 2670
Jackson_Lab_Bubier_MURGIGV01_20181207_14998_A4 0.97 2711
Jackson_Lab_Bubier_MURGIGV01_20181207_15054_A2 0.97 2707
Jackson_Lab_Bubier_MURGIGV01_20181207_15141_A9 0.97 2767
Jackson_Lab_Bubier_MURGIGV01_20190425_18231_H7 0.97 3197
Jackson_Lab_Bubier_MURGIGV01_20181207_15141_A9 0.97 2827
Jackson_Lab_Bubier_MURGIGV01_20181207_15117_A6 0.97 2872
Jackson_Lab_Bubier_MURGIGV01_20181207_15133_A8 0.97 2872
Jackson_Lab_Bubier_MURGIGV01_20181207_15133_A8 0.97 2855
Jackson_Lab_Bubier_MURGIGV01_20181207_14982_A3 0.97 2867
Jackson_Lab_Bubier_MURGIGV01_20181207_14982_A3 0.97 2861
Jackson_Lab_Bubier_MURGIGV01_20181207_15040_A1 0.97 2890
Jackson_Lab_Bubier_MURGIGV01_20181207_15040_A1 0.97 2940
Jackson_Lab_Bubier_MURGIGV01_20181207_16291_A11 0.97 2935
Jackson_Lab_Bubier_MURGIGV01_20181207_16291_A11 0.97 2960
Jackson_Lab_Bubier_MURGIGV01_20181207_15117_A6 0.97 2972
Jackson_Lab_Bubier_MURGIGV01_20181207_15125_A7 0.97 2981
Jackson_Lab_Bubier_MURGIGV01_20181207_15125_A7 0.97 2987
Jackson_Lab_Bubier_MURGIGV01_20181206_16041_D11 0.96 3283
Jackson_Lab_Bubier_MURGIGV01_20161227_8166_C1 0.95 5134
Jackson_Lab_Bubier_MURGIGV01_20161227_8166_H3 0.95 5480
n_typed n_match index1 index2
112143 112113 1560 1630
112012 111972 2118 2120
111435 111389 509 517
112133 112081 485 1514
111952 111895 482 1509
112077 112016 481 1511
112153 112091 488 1513
112005 111938 711 1206
111431 111352 487 542
111927 111846 479 1510
110987 110902 510 518
110927 110841 506 514
110546 110452 508 516
111653 111542 459 492
111837 111669 534 1512
111428 111256 440 491
109186 108940 507 515
110967 110679 268 308
109711 109417 310 385
110540 110157 269 316
109746 109080 267 307
109152 108475 270 309
108741 107488 266 312
91336 89337 1460 1468
90922 88893 1444 1468
90690 88600 1444 1460
91521 89385 1468 1500
91299 89138 1468 1492
90443 88271 1452 1468
91320 89089 1460 1500
91033 88794 1460 1492
91459 89202 1468 1476
90796 88540 1444 1500
91243 88969 1460 1476
90653 88392 1468 1516
91068 88796 1436 1468
90846 88567 1468 1484
90537 88257 1444 1492
90241 87955 1452 1460
89711 87411 1444 1452
90689 88348 1444 1476
90856 88501 1436 1460
90478 88120 1460 1516
90309 87949 1436 1444
91116 88711 1492 1500
90592 88190 1460 1484
89846 87462 1444 1516
90057 87651 1452 1492
91035 88591 1476 1492
91329 88867 1476 1500
90122 87683 1444 1484
90276 87816 1452 1500
91009 88507 1436 1500
90197 87699 1452 1476
90564 88036 1500 1516
89898 87388 1436 1452
90274 87746 1492 1516
90723 88180 1436 1492
90781 88230 1484 1500
90882 88322 1436 1476
104715 101756 203 384
90506 87931 1484 1492
89470 86918 1452 1516
89608 87052 1452 1484
90854 88262 1137 1468
90487 87885 1476 1516
90661 88041 1476 1484
90220 87611 1097 1468
90599 87975 1137 1460
90095 87483 1436 1516
90322 87685 1436 1484
89799 87144 1484 1516
90056 87386 1137 1444
90055 87344 1097 1460
89568 86861 1097 1444
90715 87948 1137 1500
104191 100994 2185 2187
90169 87342 1097 1500
90585 87713 1137 1476
90454 87582 1137 1492
89841 86986 1097 1492
89627 86760 1137 1452
89078 86217 1097 1452
89756 86866 1097 1436
90250 87310 1137 1436
89297 86362 1097 1516
89885 86925 1137 1516
90108 87136 1097 1476
89937 86956 1137 1484
89421 86434 1097 1484
89510 86227 1097 1137
101152 96018 271 385
101684 96204 271 310summary.cg$Name.ind1 <- as.character(do.call(rbind.data.frame, strsplit(as.character(summary.cg$ind1), "_"))[,6])
summary.cg$Name.ind2 <- as.character(do.call(rbind.data.frame, strsplit(as.character(summary.cg$ind2), "_"))[,6])
summary.cg$miss.ind1 <- percent_missing[match(summary.cg$ind1, names(percent_missing))]
summary.cg$miss.ind2 <- percent_missing[match(summary.cg$ind2, names(percent_missing))]
summary.cg$remove.id <- ifelse(summary.cg$miss.ind1 > summary.cg$miss.ind2, summary.cg$ind1, summary.cg$ind2)
summary.cg$remove.id [1] "Jackson_Lab_Bubier_MURGIGV01_20190108_16305_E1"
[2] "Jackson_Lab_Bubier_MURGIGV01_20190425_18162_E11"
[3] "Jackson_Lab_Bubier_MURGIGV01_20170904_8669_D5"
[4] "Jackson_Lab_Bubier_MURGIGIV01_20171001_9373_H7"
[5] "Jackson_Lab_Bubier_MURGIGIV01_20171001_9375_E7"
[6] "Jackson_Lab_Bubier_MURGIGV01_20181207_9255_D10"
[7] "Jackson_Lab_Bubier_MURGIGIV01_20171001_9371_G8"
[8] "Jackson_Lab_Bubier_MURGIGV01_20181206_13885_A8"
[9] "Jackson_Lab_Bubier_MURGIGV01_20170904_9386_E9"
[10] "Jackson_Lab_Bubier_MURGIGV01_20181207_9254_C10"
[11] "Jackson_Lab_Bubier_MURGIGV01_20170904_8597_E5"
[12] "Jackson_Lab_Bubier_MURGIGV01_20170904_8618_E3"
[13] "Jackson_Lab_Bubier_MURGIGV01_20170904_8613_C5"
[14] "Jackson_Lab_Bubier_MURGIGV01_20170904_8664_C1"
[15] "Jackson_Lab_Bubier_MURGIGV01_20170904_9378_C8"
[16] "Jackson_Lab_Bubier_MURGIGV01_20161227_8585_B8"
[17] "Jackson_Lab_Bubier_MURGIGV01_20170904_8670_B5"
[18] "Jackson_Lab_Bubier_MURGIGV01_20160908_8184_E10"
[19] "Jackson_Lab_Bubier_MURGIGV01_20161227_8166_C1"
[20] "Jackson_Lab_Bubier_MURGIGV01_20160908_8167_F10"
[21] "Jackson_Lab_Bubier_MURGIGV01_20160908_8168_D10"
[22] "Jackson_Lab_Bubier_MURGIGV01_20160908_8183_G10"
[23] "Jackson_Lab_Bubier_MURGIGV01_20160908_8185_C10"
[24] "Jackson_Lab_Bubier_MURGIGV01_20181207_14998_A4"
[25] "Jackson_Lab_Bubier_MURGIGV01_20181207_15054_A2"
[26] "Jackson_Lab_Bubier_MURGIGV01_20181207_15054_A2"
[27] "Jackson_Lab_Bubier_MURGIGV01_20181207_15014_A5"
[28] "Jackson_Lab_Bubier_MURGIGV01_20181207_15133_A8"
[29] "Jackson_Lab_Bubier_MURGIGV01_20181207_14982_A3"
[30] "Jackson_Lab_Bubier_MURGIGV01_20181207_14998_A4"
[31] "Jackson_Lab_Bubier_MURGIGV01_20181207_15133_A8"
[32] "Jackson_Lab_Bubier_MURGIGV01_20181207_15117_A6"
[33] "Jackson_Lab_Bubier_MURGIGV01_20181207_15054_A2"
[34] "Jackson_Lab_Bubier_MURGIGV01_20181207_14998_A4"
[35] "Jackson_Lab_Bubier_MURGIGV01_20181207_16291_A11"
[36] "Jackson_Lab_Bubier_MURGIGV01_20181207_15040_A1"
[37] "Jackson_Lab_Bubier_MURGIGV01_20181207_15125_A7"
[38] "Jackson_Lab_Bubier_MURGIGV01_20181207_15054_A2"
[39] "Jackson_Lab_Bubier_MURGIGV01_20181207_14982_A3"
[40] "Jackson_Lab_Bubier_MURGIGV01_20181207_14982_A3"
[41] "Jackson_Lab_Bubier_MURGIGV01_20181207_15054_A2"
[42] "Jackson_Lab_Bubier_MURGIGV01_20181207_15040_A1"
[43] "Jackson_Lab_Bubier_MURGIGV01_20181207_16291_A11"
[44] "Jackson_Lab_Bubier_MURGIGV01_20181207_15054_A2"
[45] "Jackson_Lab_Bubier_MURGIGV01_20181207_15133_A8"
[46] "Jackson_Lab_Bubier_MURGIGV01_20181207_15125_A7"
[47] "Jackson_Lab_Bubier_MURGIGV01_20181207_16291_A11"
[48] "Jackson_Lab_Bubier_MURGIGV01_20181207_14982_A3"
[49] "Jackson_Lab_Bubier_MURGIGV01_20181207_15133_A8"
[50] "Jackson_Lab_Bubier_MURGIGV01_20181207_15117_A6"
[51] "Jackson_Lab_Bubier_MURGIGV01_20181207_15054_A2"
[52] "Jackson_Lab_Bubier_MURGIGV01_20181207_14982_A3"
[53] "Jackson_Lab_Bubier_MURGIGV01_20181207_15040_A1"
[54] "Jackson_Lab_Bubier_MURGIGV01_20181207_14982_A3"
[55] "Jackson_Lab_Bubier_MURGIGV01_20181207_16291_A11"
[56] "Jackson_Lab_Bubier_MURGIGV01_20181207_14982_A3"
[57] "Jackson_Lab_Bubier_MURGIGV01_20181207_16291_A11"
[58] "Jackson_Lab_Bubier_MURGIGV01_20181207_15040_A1"
[59] "Jackson_Lab_Bubier_MURGIGV01_20181207_15125_A7"
[60] "Jackson_Lab_Bubier_MURGIGV01_20181207_15040_A1"
[61] "Jackson_Lab_Bubier_MURGIGV01_20160908_7996_D2"
[62] "Jackson_Lab_Bubier_MURGIGV01_20181207_15125_A7"
[63] "Jackson_Lab_Bubier_MURGIGV01_20181207_14982_A3"
[64] "Jackson_Lab_Bubier_MURGIGV01_20181207_14982_A3"
[65] "Jackson_Lab_Bubier_MURGIGV01_20181206_16041_D11"
[66] "Jackson_Lab_Bubier_MURGIGV01_20181207_16291_A11"
[67] "Jackson_Lab_Bubier_MURGIGV01_20181207_15125_A7"
[68] "Jackson_Lab_Bubier_MURGIGV01_20181206_16000_D6"
[69] "Jackson_Lab_Bubier_MURGIGV01_20181206_16041_D11"
[70] "Jackson_Lab_Bubier_MURGIGV01_20181207_16291_A11"
[71] "Jackson_Lab_Bubier_MURGIGV01_20181207_15125_A7"
[72] "Jackson_Lab_Bubier_MURGIGV01_20181207_16291_A11"
[73] "Jackson_Lab_Bubier_MURGIGV01_20181207_15054_A2"
[74] "Jackson_Lab_Bubier_MURGIGV01_20181206_16000_D6"
[75] "Jackson_Lab_Bubier_MURGIGV01_20181206_16000_D6"
[76] "Jackson_Lab_Bubier_MURGIGV01_20181206_16041_D11"
[77] "Jackson_Lab_Bubier_MURGIGV01_20190425_18231_H7"
[78] "Jackson_Lab_Bubier_MURGIGV01_20181206_16000_D6"
[79] "Jackson_Lab_Bubier_MURGIGV01_20181206_16041_D11"
[80] "Jackson_Lab_Bubier_MURGIGV01_20181206_16041_D11"
[81] "Jackson_Lab_Bubier_MURGIGV01_20181206_16000_D6"
[82] "Jackson_Lab_Bubier_MURGIGV01_20181207_14982_A3"
[83] "Jackson_Lab_Bubier_MURGIGV01_20181206_16000_D6"
[84] "Jackson_Lab_Bubier_MURGIGV01_20181206_16000_D6"
[85] "Jackson_Lab_Bubier_MURGIGV01_20181206_16041_D11"
[86] "Jackson_Lab_Bubier_MURGIGV01_20181206_16000_D6"
[87] "Jackson_Lab_Bubier_MURGIGV01_20181207_16291_A11"
[88] "Jackson_Lab_Bubier_MURGIGV01_20181206_16000_D6"
[89] "Jackson_Lab_Bubier_MURGIGV01_20181207_15125_A7"
[90] "Jackson_Lab_Bubier_MURGIGV01_20181206_16000_D6"
[91] "Jackson_Lab_Bubier_MURGIGV01_20181206_16000_D6"
[92] "Jackson_Lab_Bubier_MURGIGV01_20160908_8166_H10"
[93] "Jackson_Lab_Bubier_MURGIGV01_20160908_8166_H10" save(summary.cg,
file = "data/Jackson_Lab_Bubier_MURGIGV01/summary.cg.RData")
pdf(file = "output/Proportion_matching_genotypes_before_removal_of_bad_samples.pdf", width = 20, height = 20)
par(mar=c(5.1,0.6,0.6, 0.6))
hist(cg[upper.tri(cg)], breaks=seq(0, 1, length=201),
main="", yaxt="n", ylab="", xlab="Proportion matching genotypes")
rug(cg[upper.tri(cg)])
dev.off()png
2 par(mar=c(5.1,0.6,0.6, 0.6))
hist(cg[upper.tri(cg)], breaks=seq(0, 1, length=201),
main="", yaxt="n", ylab="", xlab="Proportion matching genotypes")
rug(cg[upper.tri(cg)])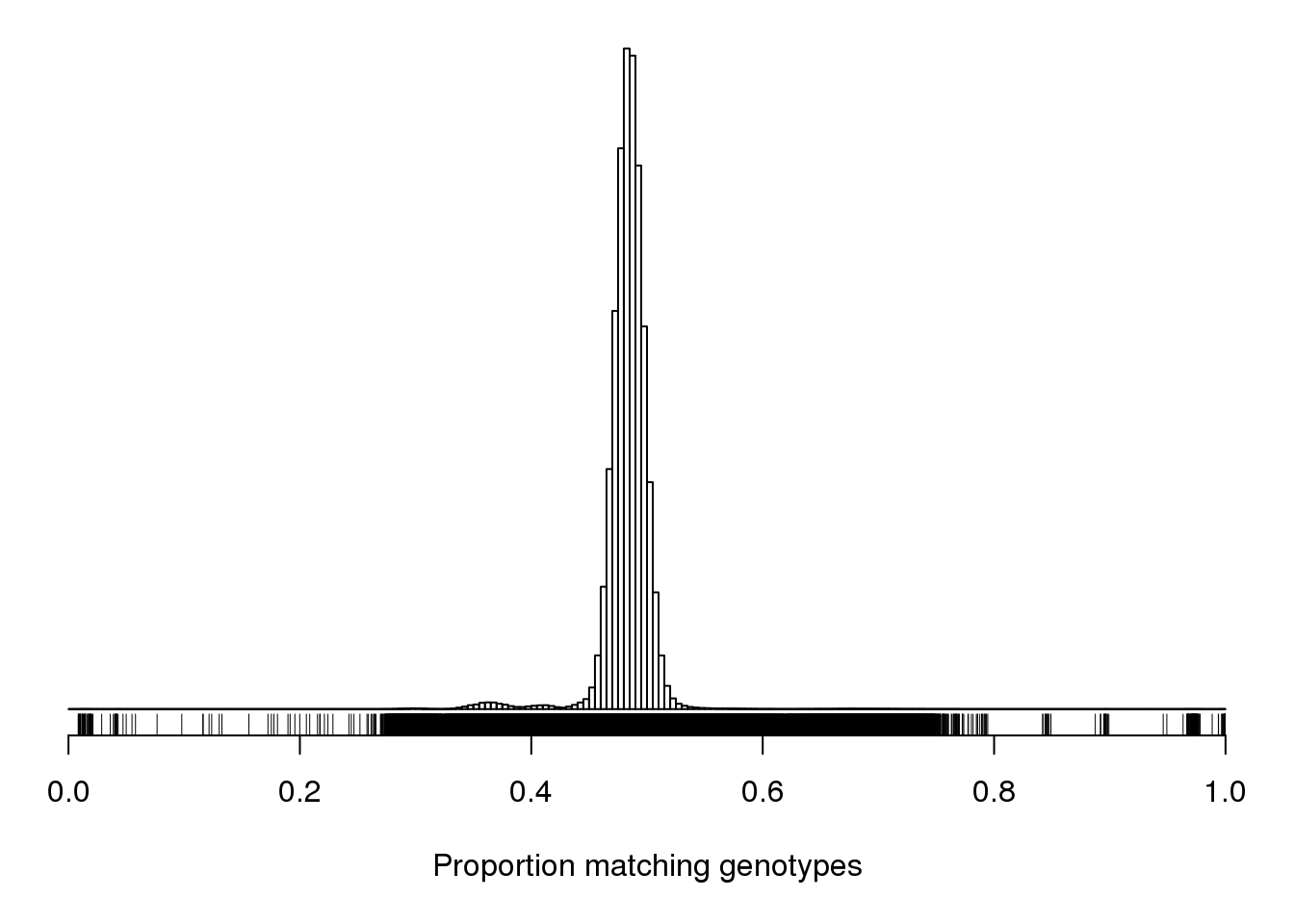
pdf(file = "output/Proportion_matching_genotypes_after_removal_of_bad_samples.pdf",width = 20, height = 20)
cgsub <- cg[percent_missing < 5, percent_missing < 5]
par(mar=c(5.1,0.6,0.6, 0.6))
hist(cgsub[upper.tri(cgsub)], breaks=seq(0, 1, length=201),
main="", yaxt="n", ylab="", xlab="Proportion matching genotypes")
rug(cgsub[upper.tri(cgsub)])
dev.off()png
2 cgsub <- cg[percent_missing < 5, percent_missing < 5]
par(mar=c(5.1,0.6,0.6, 0.6))
hist(cgsub[upper.tri(cgsub)], breaks=seq(0, 1, length=201),
main="", yaxt="n", ylab="", xlab="Proportion matching genotypes")
rug(cgsub[upper.tri(cgsub)])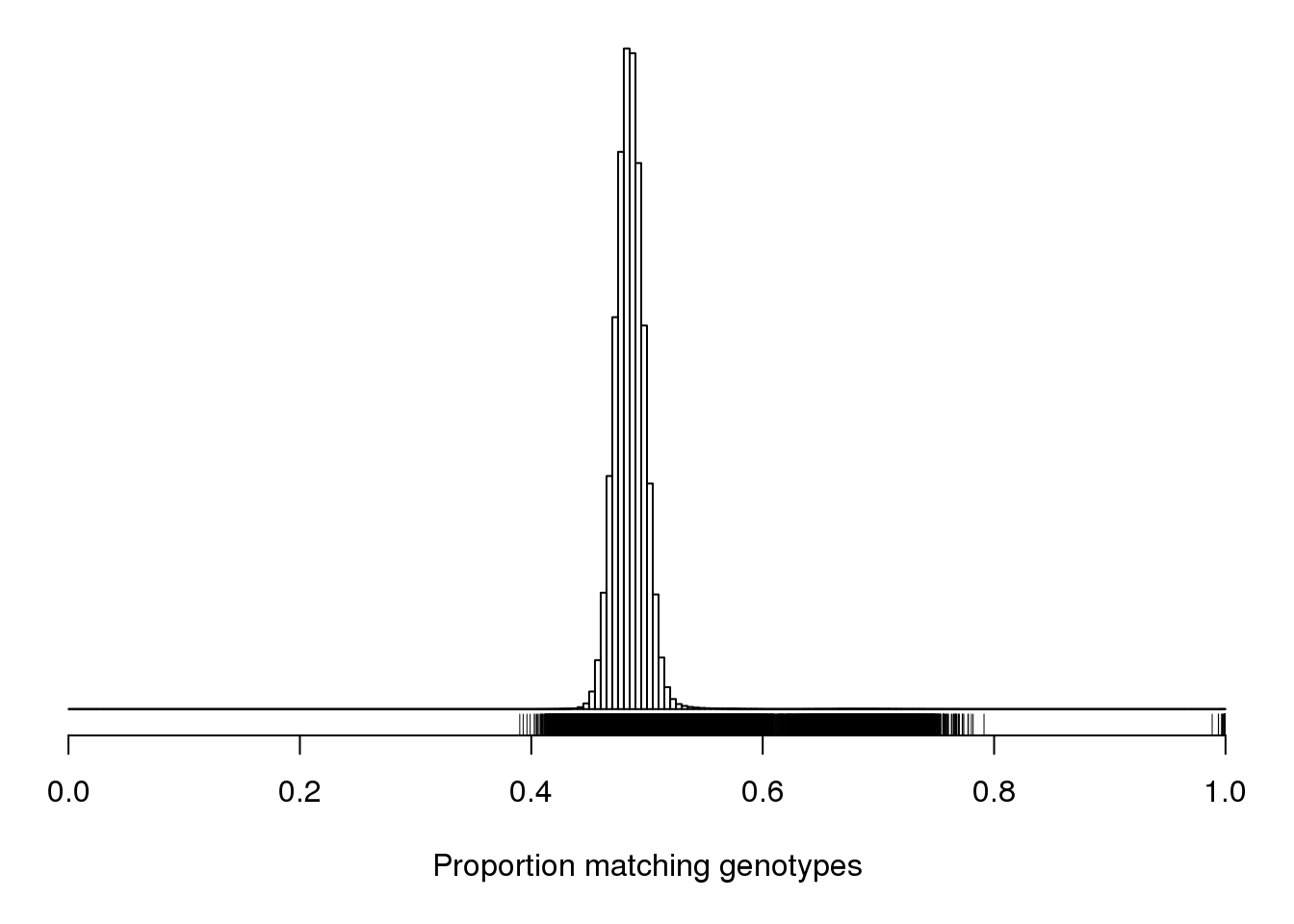
#show top 20 samples with missing genotypes
percent_missing <- n_missing(gm, "ind", "prop")*100
round(sort(percent_missing, decreasing=TRUE)[1:20], 1) Jackson_Lab_Bubier_MURGIGV01_20181207_14973_A3
39.7
Jackson_Lab_Bubier_MURGIGV01_20190425_19037_H6
36.8
Jackson_Lab_Bubier_MURGIGV01_20190425_18790_A1
21.3
Jackson_Lab_Bubier_MURGIGV01_20190425_18839_B7
19.2
Jackson_Lab_Bubier_MURGIGV01_20181207_9243_A10
19.0
Jackson_Lab_Bubier_MURGIGV01_20160908_7649_H5
18.9
Jackson_Lab_Bubier_MURGIGV01_20160908_7747_F12
18.8
Jackson_Lab_Bubier_MURGIGV01_20181206_15992_D5
18.6
Jackson_Lab_Bubier_MURGIGV01_20181207_14788_A12
18.0
Jackson_Lab_Bubier_MURGIGV01_20190108_17008_F11
17.9
Jackson_Lab_Bubier_MURGIGV01_20190108_16307_G1
17.4
Jackson_Lab_Bubier_MURGIGV01_20190425_18933_G6
17.3
Jackson_Lab_Bubier_MURGIGV01_20190108_17150_H12
17.0
Jackson_Lab_Bubier_MURGIGV01_20181207_14997_E4
16.3
Jackson_Lab_Bubier_MURGIGV01_20190108_14718_G12
15.6
Jackson_Lab_Bubier_MURGIGV01_20190425_18850_E8
15.4
Jackson_Lab_Bubier_MURGIGV01_20190425_18890_D1
13.1
Jackson_Lab_Bubier_MURGIGV01_20181206_16000_D6
12.9
Jackson_Lab_Bubier_MURGIGV01_20181207_14982_A3
12.8
Jackson_Lab_Bubier_MURGIGV01_20181207_16291_A11
12.4 Array intensities and Genotype frequencies
int <- result
rm(result)
int <- int[seq(1, nrow(int), by=2),-(1:2)] + int[-seq(1, nrow(int), by=2),-(1:2)]
int <- int[,intersect(ind_ids(gm), colnames(int))]
n <- names(sort(percent_missing[intersect(ind_ids(gm), colnames(int))], decreasing=TRUE))
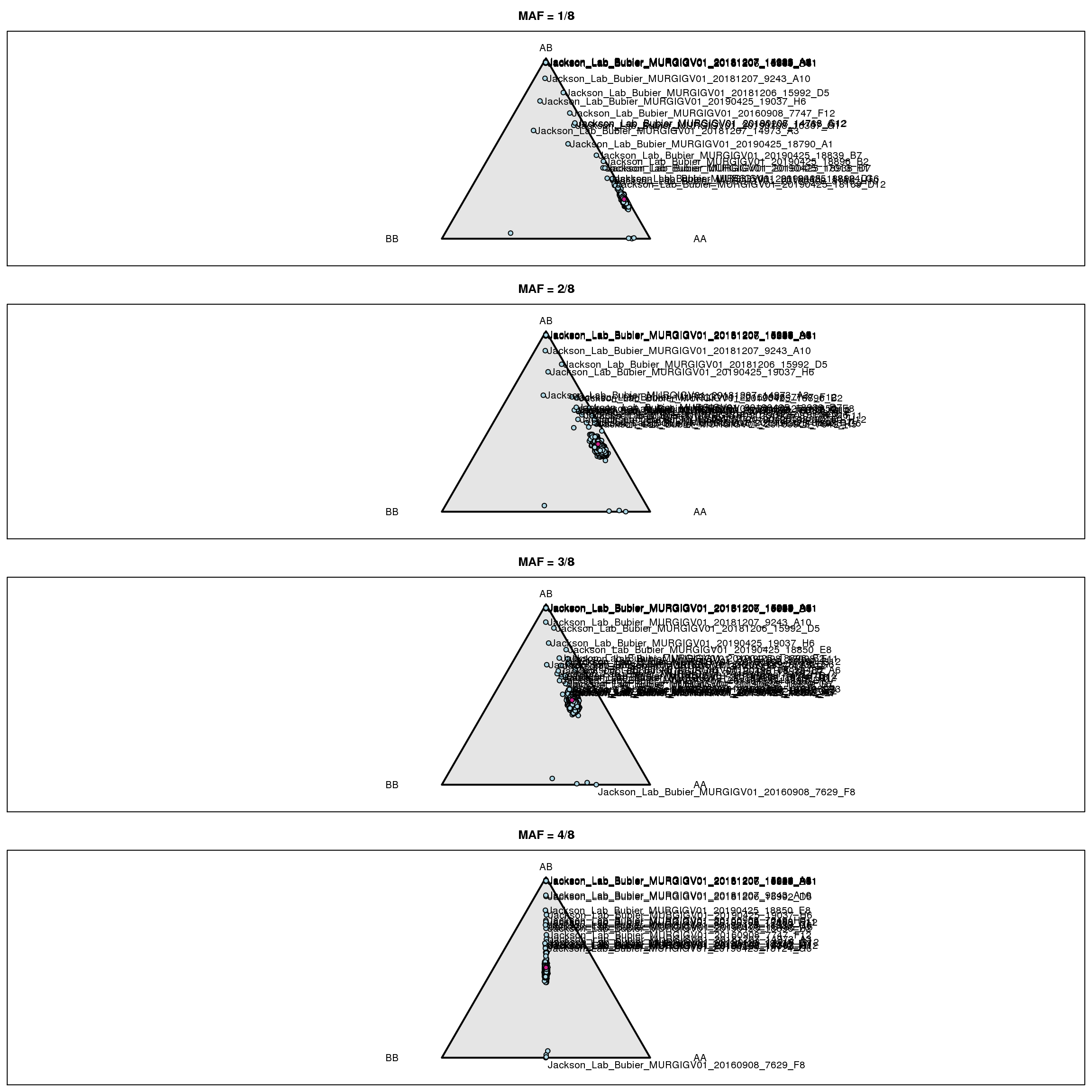
iboxplot(log10(t(int[,n])+1), orderByMedian=FALSE, chartOpts=list(ylab="log10(SNP intensity + 1)"))# Genotype frequencies
g <- do.call("cbind", gm$geno[1:19])
fg <- do.call("cbind", gm$founder_geno[1:19])
g <- g[,colSums(fg==0)==0]
fg <- fg[,colSums(fg==0)==0]
fgn <- colSums(fg==3)
gf_ind <- vector("list", 4)
for(i in 1:4) {
gf_ind[[i]] <- t(apply(g[,fgn==i], 1, function(a) table(factor(a, 1:3))/sum(a != 0)))
}
par(mfrow=c(4,1), mar=c(0.6, 0.6, 2.6, 0.6))
for(i in 1:4) {
triplot(c("AA", "AB", "BB"), main=paste0("MAF = ", i, "/8"))
tripoints(gf_ind[[i]], pch=21, bg="lightblue")
tripoints(c((1-i/8)^2, 2*i/8*(1-i/8), (i/8)^2), pch=21, bg="violetred")
if(i>=3) { # label mouse with lowest het
wh <- which(gf_ind[[i]][,2] == min(gf_ind[[i]][,2]))
tritext(gf_ind[[i]][wh,,drop=FALSE] + c(0.02, -0.02, 0),
names(wh), adj=c(0, 1))
}
# label other mice
if(i==1) {
lab <- rownames(gf_ind[[i]])[gf_ind[[i]][,2]>0.3]
}
else if(i==2) {
lab <- rownames(gf_ind[[i]])[gf_ind[[i]][,2]>0.48]
}
else if(i==3) {
lab <- rownames(gf_ind[[i]])[gf_ind[[i]][,2]>0.51]
}
else if(i==4) {
lab <- rownames(gf_ind[[i]])[gf_ind[[i]][,2]>0.6]
}
for(ind in lab) {
if(grepl("^F", ind) && i != 3) {
tritext(gf_ind[[i]][ind,,drop=FALSE] + c(-0.01, 0, +0.01), ind, adj=c(1,0.5))
} else {
tritext(gf_ind[[i]][ind,,drop=FALSE] + c(0.01, 0, -0.01), ind, adj=c(0,0.5))
}
}
}
Crossover counts and Genotyping error LOD scores
#load pre-caluated results
load("data/Jackson_Lab_Bubier_MURGIGV01/pr.RData")
load("data/Jackson_Lab_Bubier_MURGIGV01/m.RData")
load("data/Jackson_Lab_Bubier_MURGIGV01/nxo.RData")
#crossover
totxo <- rowSums(nxo)[rownames(gm$covar)]
iplot(seq_along(totxo),
totxo,
group=gm$covar$ngen,
chartOpts=list(xlab="Mouse", ylab="Number of crossovers",
margin=list(left=80,top=40,right=40,bottom=40,inner=5),
axispos=list(xtitle=25,ytitle=50,xlabel=5,ylabel=5)))#save crossover into pdf
pdf(file = "output/number_crossover.pdf")
cross_over <- data.frame(Mouse = seq_along(totxo), Number_crossovers = totxo, generation = gm$covar$ngen)
names(totxo) <- as.character(do.call(rbind.data.frame, strsplit(names(totxo), "V01_"))[,2])
names(totxo)[totxo <= 800] = ""
# Change point shapes and colors
p <-ggplot(cross_over, aes(x=Mouse, y=Number_crossovers, fill = generation, color=generation)) +
geom_point() +
geom_text_repel(aes(label=names(totxo),hjust=0,vjust=0), show.legend = FALSE)
p
dev.off()png
2 p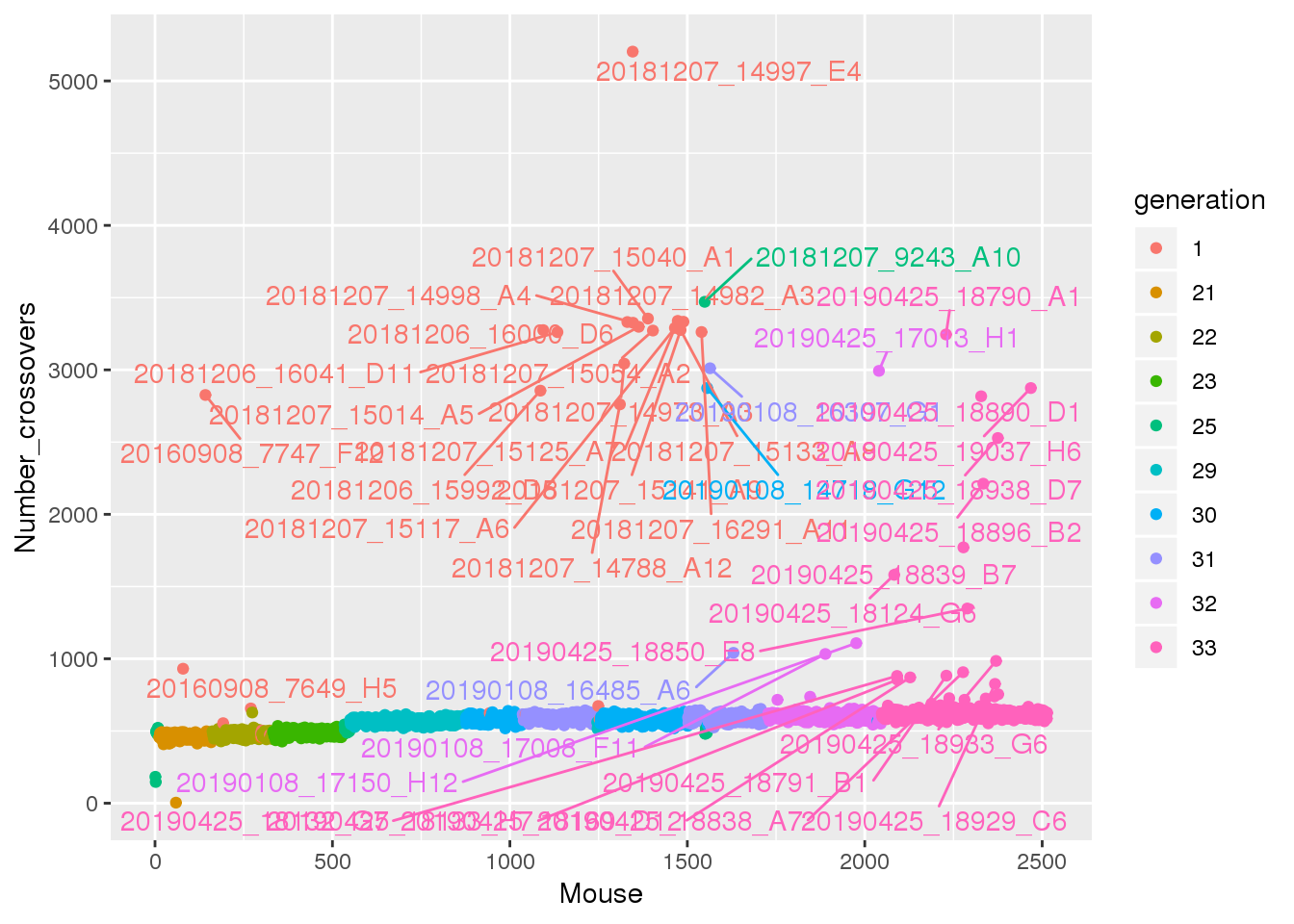
#Here are the crossover counts for those mice:
tmp <- cbind(percent_missing=round(percent_missing,2), total_xo=totxo)[percent_missing >= 5,]
tmp[order(tmp[,1]),] percent_missing total_xo
Jackson_Lab_Bubier_MURGIGV01_20181206_16218_G7 5.21 574
Jackson_Lab_Bubier_MURGIGV01_20181206_16219_H7 5.23 590
Jackson_Lab_Bubier_MURGIGV01_20181207_14695_F7 5.33 558
Jackson_Lab_Bubier_MURGIGV01_20190108_17018_A12 5.44 587
Jackson_Lab_Bubier_MURGIGV01_20190108_17143_F1 5.53 582
Jackson_Lab_Bubier_MURGIGV01_20190425_18256_A11 5.66 564
Jackson_Lab_Bubier_MURGIGV01_20181207_15071_B12 5.67 599
Jackson_Lab_Bubier_MURGIGV01_20190108_17142_E1 5.71 592
Jackson_Lab_Bubier_MURGIGV01_20190108_16866_G12 6.06 617
Jackson_Lab_Bubier_MURGIGV01_20181207_16256_G1 6.22 558
Jackson_Lab_Bubier_MURGIGV01_20190108_16961_G6 6.54 605
Jackson_Lab_Bubier_MURGIGV01_20160908_7996_D2 6.60 474
Jackson_Lab_Bubier_MURGIGV01_20181206_15949_H1 6.78 602
Jackson_Lab_Bubier_MURGIGV01_20160908_7689_A12 6.88 439
Jackson_Lab_Bubier_MURGIGV01_20181206_15318_G1 6.94 585
Jackson_Lab_Bubier_MURGIGV01_20190425_18799_B2 7.23 694
Jackson_Lab_Bubier_MURGIGV01_20190425_18231_H7 7.32 620
Jackson_Lab_Bubier_MURGIGV01_20190108_16485_A6 7.39 599
Jackson_Lab_Bubier_MURGIGV01_20190425_18842_E7 7.60 649
Jackson_Lab_Bubier_MURGIGV01_20181207_13744_F1 7.85 551
Jackson_Lab_Bubier_MURGIGV01_20190425_18798_A2 8.08 633
Jackson_Lab_Bubier_MURGIGV01_20190425_18927_A6 8.47 580
Jackson_Lab_Bubier_MURGIGV01_20190425_18903_A3 8.78 614
Jackson_Lab_Bubier_MURGIGV01_20160908_8166_H10 8.94 448
Jackson_Lab_Bubier_MURGIGV01_20181207_13702_E1 9.12 556
Jackson_Lab_Bubier_MURGIGV01_20190425_17013_H1 9.39 601
Jackson_Lab_Bubier_MURGIGV01_20190425_18196_E3 9.61 607
Jackson_Lab_Bubier_MURGIGV01_20190425_18169_D12 9.70 592
Jackson_Lab_Bubier_MURGIGV01_20190425_18791_B1 10.10 3245
Jackson_Lab_Bubier_MURGIGV01_20190425_18940_E7 10.42 2528
Jackson_Lab_Bubier_MURGIGV01_20190425_18938_D7 10.43 600
Jackson_Lab_Bubier_MURGIGV01_20190425_18132_G7 10.63 648
Jackson_Lab_Bubier_MURGIGV01_20190425_18133_H7 10.88 881
Jackson_Lab_Bubier_MURGIGV01_20190425_18929_C6 11.41 607
Jackson_Lab_Bubier_MURGIGV01_20190425_18124_G6 11.61 650
Jackson_Lab_Bubier_MURGIGV01_20181207_15141_A9 11.68 538
Jackson_Lab_Bubier_MURGIGV01_20181207_15014_A5 11.69 562
Jackson_Lab_Bubier_MURGIGV01_20190425_18838_A7 11.72 648
Jackson_Lab_Bubier_MURGIGV01_20181207_14998_A4 11.85 578
Jackson_Lab_Bubier_MURGIGV01_20181207_15117_A6 11.85 566
Jackson_Lab_Bubier_MURGIGV01_20181207_15133_A8 12.00 584
Jackson_Lab_Bubier_MURGIGV01_20181207_15040_A1 12.07 549
Jackson_Lab_Bubier_MURGIGV01_20190425_18896_B2 12.15 622
Jackson_Lab_Bubier_MURGIGV01_20181206_16041_D11 12.28 565
Jackson_Lab_Bubier_MURGIGV01_20181207_15125_A7 12.34 567
Jackson_Lab_Bubier_MURGIGV01_20181207_15054_A2 12.40 560
Jackson_Lab_Bubier_MURGIGV01_20181207_16291_A11 12.44 593
Jackson_Lab_Bubier_MURGIGV01_20181207_14982_A3 12.85 620
Jackson_Lab_Bubier_MURGIGV01_20181206_16000_D6 12.89 601
Jackson_Lab_Bubier_MURGIGV01_20190425_18890_D1 13.08 654
Jackson_Lab_Bubier_MURGIGV01_20190425_18850_E8 15.40 638
Jackson_Lab_Bubier_MURGIGV01_20190108_14718_G12 15.63 593
Jackson_Lab_Bubier_MURGIGV01_20181207_14997_E4 16.29 596
Jackson_Lab_Bubier_MURGIGV01_20190108_17150_H12 16.96 621
Jackson_Lab_Bubier_MURGIGV01_20190425_18933_G6 17.28 607
Jackson_Lab_Bubier_MURGIGV01_20190108_16307_G1 17.36 570
Jackson_Lab_Bubier_MURGIGV01_20190108_17008_F11 17.91 623
Jackson_Lab_Bubier_MURGIGV01_20181207_14788_A12 18.04 591
Jackson_Lab_Bubier_MURGIGV01_20181206_15992_D5 18.56 567
Jackson_Lab_Bubier_MURGIGV01_20160908_7747_F12 18.85 447
Jackson_Lab_Bubier_MURGIGV01_20160908_7649_H5 18.88 412
Jackson_Lab_Bubier_MURGIGV01_20181207_9243_A10 19.02 611
Jackson_Lab_Bubier_MURGIGV01_20190425_18839_B7 19.16 907
Jackson_Lab_Bubier_MURGIGV01_20190425_18790_A1 21.29 602
Jackson_Lab_Bubier_MURGIGV01_20190425_19037_H6 36.75 634
Jackson_Lab_Bubier_MURGIGV01_20181207_14973_A3 39.72 619# Genotyping error LOD scores
load("data/Jackson_Lab_Bubier_MURGIGV01/e.RData")
errors_ind <- rowSums(e>2)/n_typed(gm)*100
lab <- paste0(names(errors_ind), " (", myround(percent_missing,1), "%)")
iplot(seq_along(errors_ind), errors_ind, indID=lab,
chartOpts=list(xlab="Mouse", ylab="Percent genotyping errors", ylim=c(0, 8),
axispos=list(xtitle=25, ytitle=50, xlabel=5, ylabel=5)))save(errors_ind, file = "data/Jackson_Lab_Bubier_MURGIGV01/errors_ind.RData")
# Apparent genotyping errors
load("data/Jackson_Lab_Bubier_MURGIGV01/snpg.RData")
gobs <- do.call("cbind", gm$geno)
gobs[gobs==0] <- NA
par(pty="s")
err_direct <- rowMeans(snpg != gobs, na.rm=TRUE)*100
errors_ind_0 <- rowSums(e > 0)/n_typed(gm)*100
par(mar=c(4.1,4.1,0.6, 0.6))
grayplot(errors_ind_0, err_direct,
xlab="Percent errors (error LOD > 0)",
ylab="Percent errors (obs vs predicted)",
xlim=c(0, 2), ylim=c(0, 2))
abline(0,1,lty=2, col="gray60")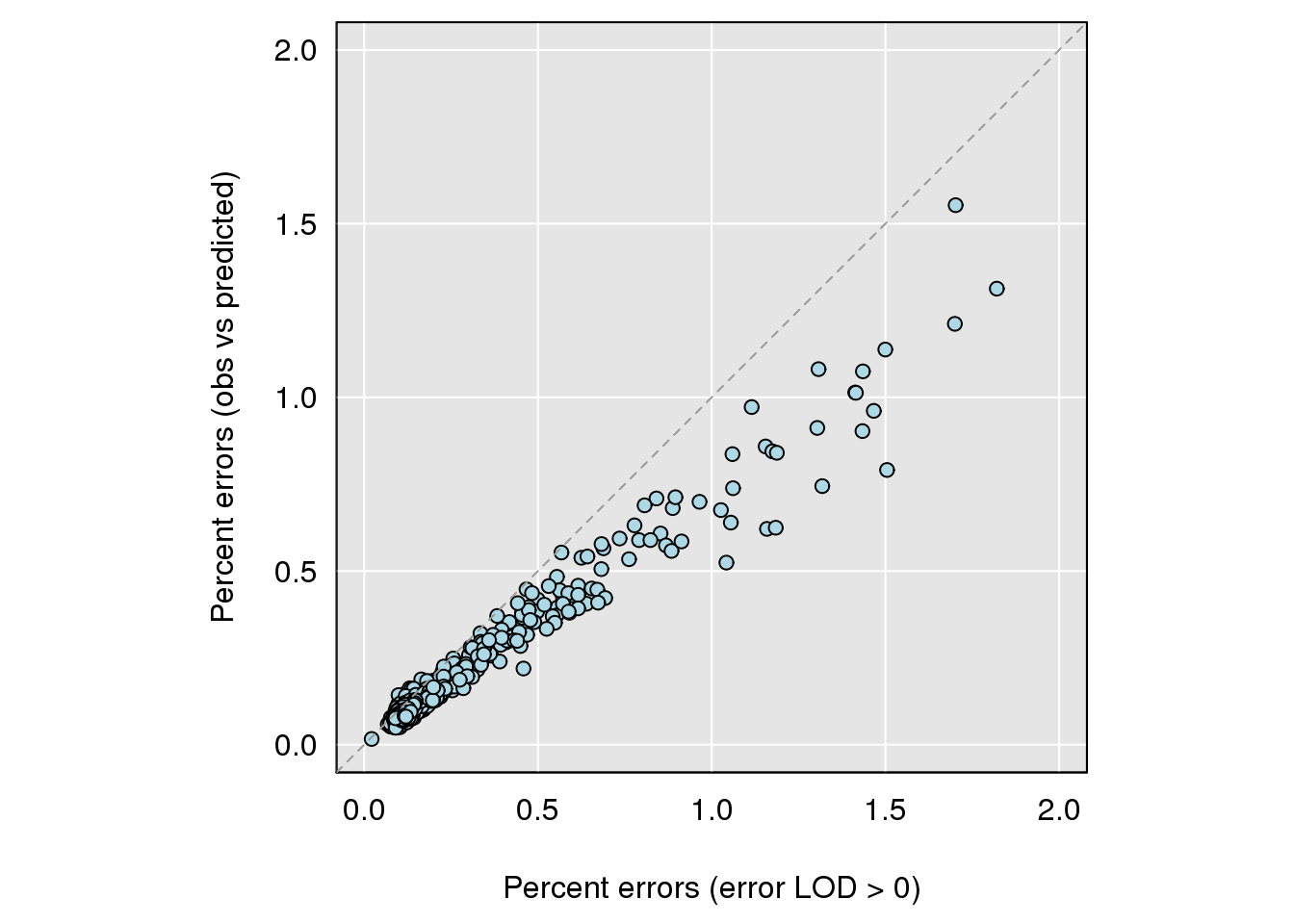
pdf(file = "output/Percent_genotype_errors_obs_vs_predicted.pdf",width = 20, height = 20)
par(pty="s")
err_direct <- rowMeans(snpg != gobs, na.rm=TRUE)*100
errors_ind_0 <- rowSums(e > 0)/n_typed(gm)*100
par(mar=c(4.1,4.1,0.6, 0.6))
grayplot(errors_ind_0, err_direct,
xlab="Percent errors (error LOD > 0)",
ylab="Percent errors (obs vs predicted)",
xlim=c(0, 2), ylim=c(0, 2))
abline(0,1,lty=2, col="gray60")
dev.off()png
2 Missing data in Markers and Genotype frequencies Markers
#It can also be useful to look at the proportion of missing genotypes by marker.
#Markers with a lot of missing data were likely difficult to call, and so the genotypes that were called may contain a lot of errors.
pmis_mar <- n_missing(gm, "marker", "proportion")*100
par(mar=c(5.1,0.6,0.6, 0.6))
hist(pmis_mar, breaks=seq(0, 100, length=201),
main="", yaxt="n", ylab="", xlab="Percent missing genotypes")
rug(pmis_mar)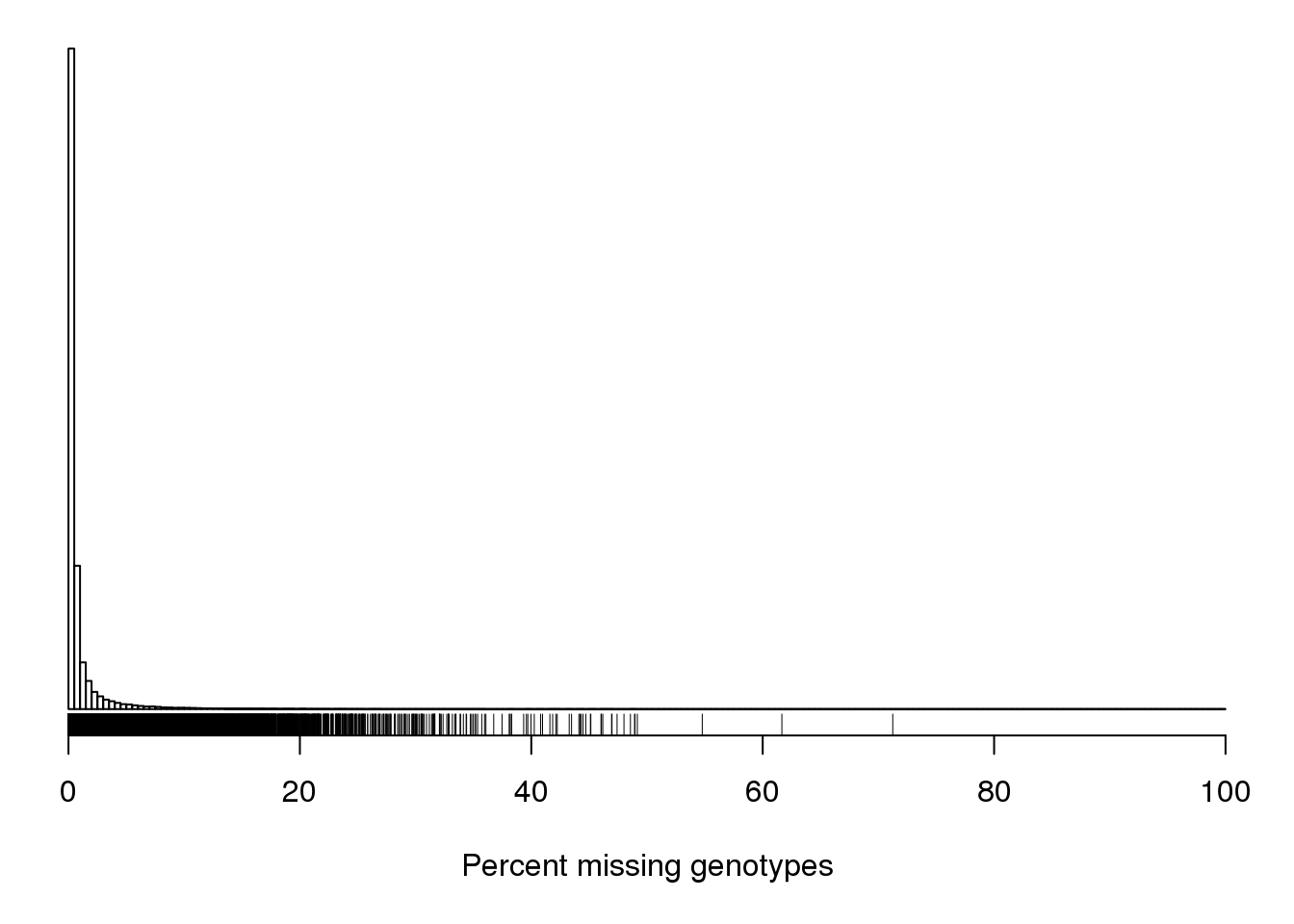
pdf(file = "output/Percent_missing_genotype_data_per_marker.pdf")
par(mar=c(5.1,0.6,0.6, 0.6))
hist(pmis_mar, breaks=seq(0, 100, length=201),
main="", yaxt="n", ylab="", xlab="Percent missing genotypes")
rug(pmis_mar)
dev.off()png
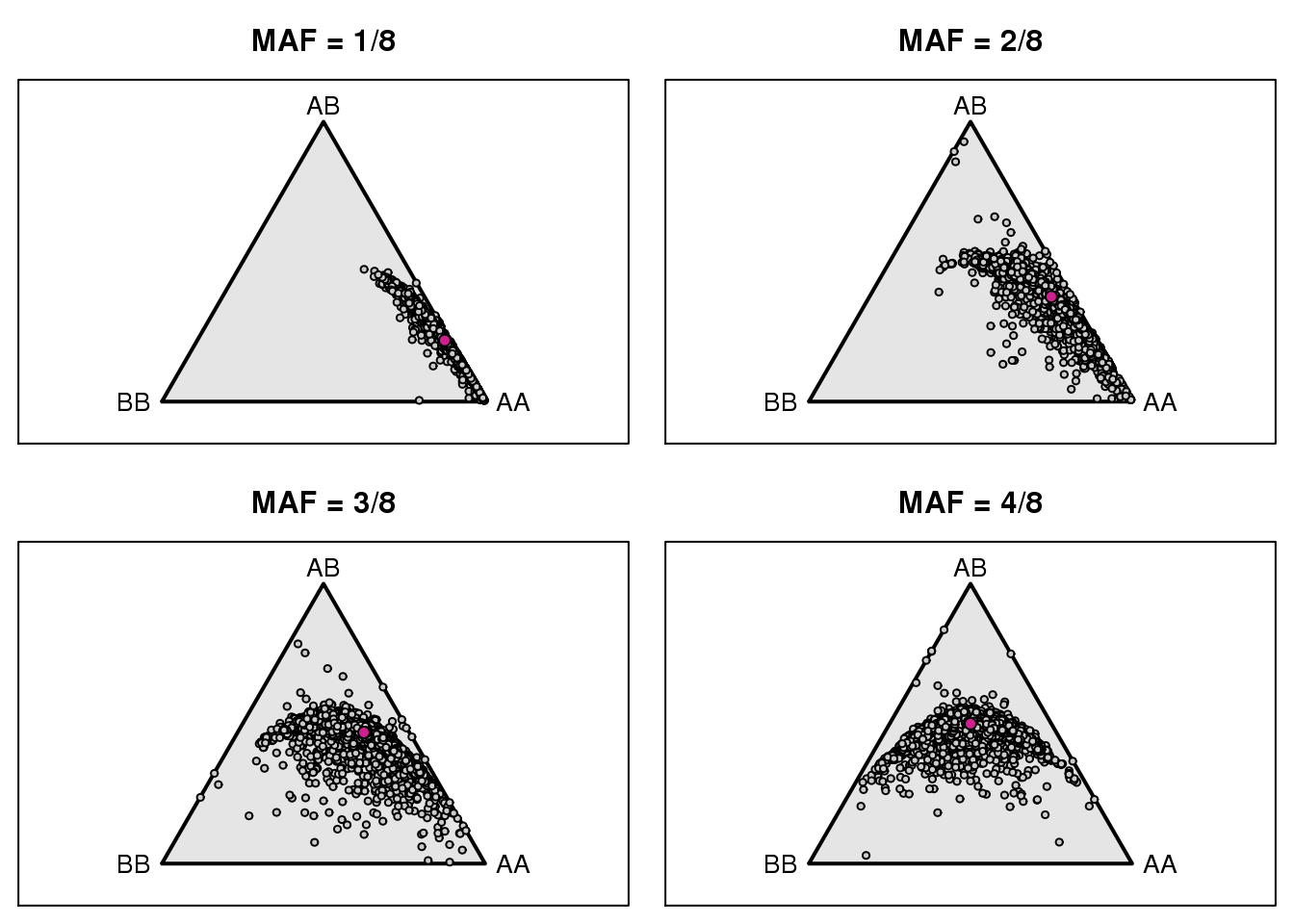
2 # Genotype frequencies Markers
gf_mar <- t(apply(g, 2, function(a) table(factor(a, 1:3))/sum(a != 0)))
gn_mar <- t(apply(g, 2, function(a) table(factor(a, 1:3))))
pdf(file = "output/genotype_frequency_marker.pdf")
par(mfrow=c(2,2), mar=c(0.6, 0.6, 2.6, 0.6))
for(i in 1:4) {
triplot(c("AA", "AB", "BB"), main=paste0("MAF = ", i, "/8"))
z <- gf_mar[fgn==i,]
z <- z[rowSums(is.na(z)) < 3,]
tripoints(z, pch=21, bg="gray80", cex=0.6)
tripoints(c((1-i/8)^2, 2*i/8*(1-i/8), (i/8)^2), pch=21, bg="violetred")
}
dev.off()png
2 par(mfrow=c(2,2), mar=c(0.6, 0.6, 2.6, 0.6))
for(i in 1:4) {
triplot(c("AA", "AB", "BB"), main=paste0("MAF = ", i, "/8"))
z <- gf_mar[fgn==i,]
z <- z[rowSums(is.na(z)) < 3,]
tripoints(z, pch=21, bg="gray80", cex=0.6)
tripoints(c((1-i/8)^2, 2*i/8*(1-i/8), (i/8)^2), pch=21, bg="violetred")
}
# Genotype errors Markers
errors_mar <- colSums(e>2)/n_typed(gm, "marker")*100
grayplot(pmis_mar, errors_mar,
xlab="Proportion missing", ylab="Proportion genotyping errors")
pdf(file = "output/genotype_error_marker.pdf")
grayplot(pmis_mar, errors_mar,
xlab="Proportion missing", ylab="Proportion genotyping errors")
dev.off()png
2 Remove bad samples
#percent missing
qc_info <- merge(gm$covar,
data.frame(id = names(percent_missing),
percent_missing = percent_missing,stringsAsFactors = F),by = "id")
#cross_over
qc_info <- merge(qc_info,
data.frame(id = rownames(cross_over),
Number_crossovers = cross_over$Number_crossovers,stringsAsFactors = F),by = "id")
#missing sex
qc_info$sex.match <- ifelse(qc_info$predict.sex == qc_info$sex, TRUE, FALSE)
#genotype errors
qc_info <- merge(qc_info,
data.frame(id = names(errors_ind),
genotype_erros = errors_ind,stringsAsFactors = F),by = "id")
#duplicated id to be remove
qc_info$remove.id.duplicated <- ifelse(qc_info$id %in% summary.cg$remove.id, TRUE,FALSE)
bad.sample <- qc_info[qc_info$ngen ==1 | qc_info$Number_crossovers <= 200 | qc_info$Number_crossovers >=1000 | qc_info$percent_missing >= 10 | qc_info$genotype_erros >= 1 | qc_info$remove.id.duplicated == TRUE,]
save(qc_info, bad.sample, file = "data/Jackson_Lab_Bubier_MURGIGV01/qc_info_bad_sample.RData")
#remove 77 bad samples
gm.no.bad <- gm[paste0("-",as.character(bad.sample$id)),]
#fix sex mismatch
gm.no.bad$covar[gm.no.bad$covar$id %in% names(yint_ave[!names(yint_ave) %in% bad.sample$id][yint_ave[!names(yint_ave) %in% bad.sample$id] <= 0.1]),"predict.sex"] <- "F"
gm.no.badObject of class cross2 (crosstype "do")
Total individuals 2437
No. genotyped individuals 2437
No. phenotyped individuals 2437
No. with both geno & pheno 2437
No. phenotypes 1
No. covariates 4
No. phenotype covariates 0
No. chromosomes 20
Total markers 112729
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15
8555 8666 6420 6615 6571 6444 6294 5677 5870 5447 6352 5167 5274 5039 4555
16 17 18 19 X
4369 4330 4002 3108 3974 #2437 subjects
# update other stuff
e <- e[ind_ids(gm.no.bad),]
g <- g[ind_ids(gm.no.bad),]
snpg <- snpg[ind_ids(gm.no.bad),]
length(errors_mar[errors_mar > 5])[1] 273# omit the 273 markers with error rates >5%.
bad_markers <- find_markerpos(gm.no.bad, names(errors_mar[errors_mar > 5]))
save(bad_markers, file = "data/Jackson_Lab_Bubier_MURGIGV01/bad_markers.RData")
#drop bad markers
gm_DO2437_qc <- drop_markers(gm.no.bad, names(errors_mar)[errors_mar > 5])
gm_DO2437_qcObject of class cross2 (crosstype "do")
Total individuals 2437
No. genotyped individuals 2437
No. phenotyped individuals 2437
No. with both geno & pheno 2437
No. phenotypes 1
No. covariates 4
No. phenotype covariates 0
No. chromosomes 20
Total markers 112456
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15
8533 8646 6402 6599 6558 6427 6281 5660 5856 5438 6338 5152 5256 5026 4547
16 17 18 19 X
4356 4321 3987 3102 3971 save(gm_DO2437_qc, file = "data/Jackson_Lab_Bubier_MURGIGV01/gm_DO2437_qc.RData")
save(e,g,snpg, file = "data/Jackson_Lab_Bubier_MURGIGV01/e_g_snpg_qc.RData")
sessionInfo()R version 3.3.2 (2016-10-31)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS release 6.5 (Final)
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats4 parallel methods stats graphics grDevices utils
[8] datasets base
other attached packages:
[1] mclust_5.2.1 DOQTL_1.10.0
[3] VariantAnnotation_1.20.3 Rsamtools_1.26.2
[5] SummarizedExperiment_1.4.0 Biobase_2.34.0
[7] BSgenome.Mmusculus.UCSC.mm10_1.4.0 BSgenome_1.42.0
[9] rtracklayer_1.34.2 Biostrings_2.42.1
[11] XVector_0.14.1 GenomicRanges_1.26.4
[13] GenomeInfoDb_1.10.3 IRanges_2.8.2
[15] S4Vectors_0.12.2 BiocGenerics_0.20.0
[17] ggrepel_0.8.1 ggplot2_3.1.0
[19] qtlcharts_0.9-6 qtl2_0.18
[21] broman_0.68-2
loaded via a namespace (and not attached):
[1] bitops_1.0-6 fs_1.2.6
[3] bit64_0.9-7 doParallel_1.0.10
[5] rprojroot_1.3-2 prabclus_2.2-6
[7] regress_1.3-15 tools_3.3.2
[9] backports_1.1.2 R6_2.4.0
[11] DBI_1.0.0 lazyeval_0.2.1
[13] colorspace_1.4-0 trimcluster_0.1-2
[15] annotationTools_1.48.0 nnet_7.3-12
[17] withr_2.1.2 tidyselect_0.2.5
[19] bit_1.1-14 git2r_0.23.0
[21] labeling_0.3 diptest_0.75-7
[23] scales_1.0.0 QTLRel_1.0
[25] DEoptimR_1.0-8 mvtnorm_1.0-5
[27] robustbase_0.92-7 stringr_1.3.1
[29] digest_0.6.18 rmarkdown_1.11
[31] pkgconfig_2.0.1 htmltools_0.3.6
[33] htmlwidgets_1.3 rlang_0.4.0
[35] RSQLite_2.1.1 jsonlite_1.6
[37] hwriter_1.3.2 gtools_3.5.0
[39] BiocParallel_1.8.2 dplyr_0.8.3
[41] RCurl_1.95-4.12 magrittr_1.5
[43] modeltools_0.2-21 qtl_1.41-6
[45] Matrix_1.2-14 Rcpp_1.0.2
[47] munsell_0.5.0 stringi_1.2.4
[49] whisker_0.3-2 yaml_2.2.0
[51] MASS_7.3-50 zlibbioc_1.20.0
[53] rhdf5_2.18.0 flexmix_2.3-13
[55] plyr_1.8.4 grid_3.3.2
[57] blob_1.1.1 gdata_2.18.0
[59] crayon_1.3.4 lattice_0.20-35
[61] GenomicFeatures_1.26.2 annotate_1.52.1
[63] knitr_1.20 pillar_1.3.1
[65] RUnit_0.4.31 fpc_2.1-10
[67] corpcor_1.6.9 codetools_0.2-15
[69] biomaRt_2.30.0 XML_3.98-1.16
[71] glue_1.3.1 evaluate_0.10
[73] data.table_1.11.4 foreach_1.4.4
[75] gtable_0.2.0 purrr_0.3.2
[77] kernlab_0.9-25 assertthat_0.2.1
[79] xtable_1.8-2 class_7.3-14
[81] tibble_2.1.3 iterators_1.0.10
[83] GenomicAlignments_1.10.1 AnnotationDbi_1.36.2
[85] memoise_1.1.0 workflowr_1.4.0
[87] cluster_2.0.7-1