Prj01_RL_pheno_qtl2_DO_11092020_69k
Hao He
Last updated: 2022-09-19
Checks: 6 1
Knit directory: csna_workflow/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190922) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /projects/csna/csna_workflow/ | . |
| /projects/csna/csna_workflow/data/69k_grid_pgmap.RData | data/69k_grid_pgmap.RData |
| /projects/csna/csna_workflow/data/cc_variants.sqlite | data/cc_variants.sqlite |
| /projects/csna/csna_workflow/data/mouse_genes_mgi.sqlite | data/mouse_genes_mgi.sqlite |
| /projects/csna/csna_workflow/data/GCTA/12_batches.fam | data/GCTA/12_batches.fam |
| /projects/csna/csna_workflow/data/GCTA/Prj01_RL_pheno_qttl2_DO_11092020/update_id.txt | data/GCTA/Prj01_RL_pheno_qttl2_DO_11092020/update_id.txt |
| /projects/csna/csna_workflow/data/GCTA/Prj01_RL_pheno_qttl2_DO_11092020/update_sex.txt | data/GCTA/Prj01_RL_pheno_qttl2_DO_11092020/update_sex.txt |
| /projects/csna/csna_workflow/data/GCTA/Prj01_RL_pheno_qttl2_DO_11092020/update_covar.txt | data/GCTA/Prj01_RL_pheno_qttl2_DO_11092020/update_covar.txt |
| /projects/csna/csna_workflow/data/GCTA/Prj01_RL_pheno_qttl2_DO_11092020/update.pheno.txt | data/GCTA/Prj01_RL_pheno_qttl2_DO_11092020/update.pheno.txt |
| /projects/csna/csna_workflow/data/GCTA/ | data/GCTA |
| /projects/csna/csna_workflow/data/GCTA/Prj01_RL_pheno_qttl2_DO_11092020/ | data/GCTA/Prj01_RL_pheno_qttl2_DO_11092020 |
| /projects/csna/csna_workflow/data/GCTA/Prj01_RL_pheno_qttl2_DO_11092020/h2.csv | data/GCTA/Prj01_RL_pheno_qttl2_DO_11092020/h2.csv |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version e1ea743. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: output/qtlmapping/
Untracked files:
Untracked: analysis/.nfs0000000135a08b9b00002448
Untracked: analysis/.nfs0000000135a08ba50000244a
Untracked: analysis/Prj01_RL_pheno_qtl2_DO_11092020_69k.sh
Untracked: analysis/Prj01_RL_pheno_qtl2_DO_11092020_69k.stderr
Untracked: analysis/Prj01_RL_pheno_qtl2_DO_11092020_69k.stdout
Untracked: analysis/RL_h2.R
Untracked: analysis/csna_image.sif
Untracked: analysis/draft.R
Untracked: analysis/for_johnathan.R
Untracked: analysis/index0.Rmd
Untracked: analysis/meta_robyn.R
Untracked: analysis/meta_robyn_Jentsch2.R
Untracked: analysis/meta_robyn_Price.R
Untracked: analysis/meta_robyn_cc.R
Untracked: analysis/ped.R
Untracked: analysis/pylmm_hmdp.R
Untracked: analysis/qtlmapping_results.Rmd
Untracked: analysis/rstudio.4.0.3.simg
Untracked: analysis/sample_num_qtl.R
Untracked: analysis/samplesize/
Untracked: analysis/scripts/
Untracked: analysis/snp_UCLA1_mm10_mm39.R
Untracked: analysis/snp_gene_eqtl_coeff.txt
Untracked: analysis/thecube_cc_rix.R
Untracked: analysis/thecube_impute_dogenome.R
Untracked: analysis/thecube_impute_dogenome.err
Untracked: analysis/thecube_impute_dogenome.out
Untracked: analysis/thecube_impute_dogenome.sh
Untracked: analysis/workflow_proc.R
Untracked: analysis/workflow_proc.sh
Untracked: analysis/workflow_proc.stderr
Untracked: analysis/workflow_proc.stdout
Untracked: archived/
Untracked: code/FinalReport2Plink.sh
Untracked: code/FinalReport2cross2.R
Untracked: code/csna_qtlmapping_plot.R
Untracked: code/csna_qtlmapping_workflow.R
Untracked: code/csna_snp_gwas.R
Untracked: code/do2sanger.helper.R
Untracked: code/herit_boot.R
Untracked: code/impute_dogenome.R
Untracked: code/plink-1.07-x86_64.zip
Untracked: code/plink-1.07-x86_64/
Untracked: code/plink2
Untracked: code/plink2_linux_x86_64.zip
Untracked: code/predict_sanger_snp_geno.R
Untracked: code/pylmm/
Untracked: code/pylmm0/
Untracked: code/qtlsnp_eqtl_sv.R
Untracked: code/reconst_utils.R
Untracked: code/sample.size.num.qtl.R
Untracked: code/test_qtlsnp_eqtl_sv.R
Untracked: data/69k_grid_pgmap.RData
Untracked: data/CSNA03_animaldata.csv
Untracked: data/DO_DrugNaive_Striatum/
Untracked: data/DO_Striatum.rdata
Untracked: data/Dickson2.csv
Untracked: data/Dickson2.csv.1
Untracked: data/Dickson2.csv.2
Untracked: data/Dickson2.csv.3
Untracked: data/Dickson2.csv.4
Untracked: data/FinalReport/
Untracked: data/GCTA/
Untracked: data/GM/
Untracked: data/Jackson_Lab_11_batches/
Untracked: data/Jackson_Lab_12_batches/
Untracked: data/Jackson_Lab_Bubier_MURGIGV01/
Untracked: data/Jackson_Lab_Gagnon/
Untracked: data/RTG/
Untracked: data/cc.apr.RData
Untracked: data/cc_do_founders_key.rds
Untracked: data/cc_variants.sqlite
Untracked: data/founder_SVs_modified.bed
Untracked: data/gene.anno.cis.RData
Untracked: data/hmdp/
Untracked: data/marker_grid_0.02cM_plus.txt
Untracked: data/meta/
Untracked: data/mm10ToMm39.over.chain
Untracked: data/mouse_genes_mgi.sqlite
Untracked: data/pheno/
Untracked: data/rlb/
Untracked: data/snp_UCLA1_mm10_mm39.csv
Untracked: data/thecube/
Untracked: launch_rstudio.sbatch
Untracked: output/DO.allrawpheno.corr.fullmat.RData
Untracked: output/DO_IVSA_pheno.csv
Untracked: output/DO_csna03_novelty_pheno.csv
Untracked: output/Prj01_RL_11092020_cutoff6.pheno.herit.csv
Untracked: output/Prj01_RL_11092020_cutoff6.qtlout.69k.RData
Untracked: output/Prj01_RL_pheno.herit.csv
Untracked: output/RL_prj/Prj01_RL_11092020_cutoff6.pheno.herit.csv
Untracked: output/RL_prj/apr.RDS
Untracked: output/archived_output/
Untracked: output/archived_qtlmapping/
Untracked: output/circle_chromosome.pdf
Untracked: output/corr.cc.fd.fullmat.RData
Untracked: output/corr.cc.fullmat.RData
Untracked: output/corr.fd.fullmat.RData
Untracked: output/csna.allqtl.69k.peaks.RData
Untracked: output/csna.allqtl.69k.peaks.csv
Untracked: output/csna.allqtl.array.peaks.RData
Untracked: output/csna.allqtl.array.peaks.csv
Untracked: output/csna.allqtl.peaks.RData
Untracked: output/csna.allqtl.peaks.csv
Untracked: output/gm.RData
Untracked: output/hmdp/
Untracked: output/hmdp_ped/
Untracked: output/lod.69k.RData
Untracked: output/meta/
Untracked: output/pdf/
Untracked: output/peak.69k.RData
Untracked: output/qtl.out.sample.1000.RData
Untracked: output/qtl.out.sample.1200.RData
Untracked: output/qtl.out.sample.1500.RData
Untracked: output/qtl.out.sample.2000.RData
Untracked: output/qtl.out.sample.300.RData
Untracked: output/qtl.out.sample.3000.RData
Untracked: output/qtl.out.sample.400.RData
Untracked: output/qtl.out.sample.500.RData
Untracked: output/qtl.out.sample.600.RData
Untracked: output/qtlmapping.heatmap.pdf
Untracked: output/qtlmapping.heatmap.sens.pdf
Untracked: output/qtlmapping_plot/
Untracked: output/rlb/
Untracked: output/sample.szie.qtl.HB.Repeat.Entries.ranknorm.RData
Untracked: output/sample.szie.qtl.LD.pct.time.in.light.ranknorm.RData
Untracked: output/sample.szie.qtl.NPP.NoveltyPreference.ZoneTime_GreyWhiteBlack_Total.ranknorm.RData
Untracked: output/sample.szie.qtl.OFA.pct.time.corner.ranknorm.RData
Untracked: output/sample.szie.qtl.OFA.total.distance.traveled.ranknorm.RData
Untracked: output/sample.szie.vs.num.qtls.pdf
Untracked: output/snp_gwas/
Untracked: output/test_gemma/
Untracked: output/thecube/
Untracked: rstudio.4.0.3_v1.2.simg
Unstaged changes:
Modified: .Rprofile
Modified: .gitignore
Modified: README.md
Modified: _workflowr.yml
Modified: analysis/00_Process_all_phenotypes_in_founder_CC_DO.Rmd
Deleted: analysis/01_geneseek2qtl2.R
Deleted: analysis/02_geneseek2intensity.R
Deleted: analysis/03_firstgm2genoprobs.R
Deleted: analysis/04_diagnosis_qc_gigamuga_11_batches.Rmd
Modified: analysis/04_diagnosis_qc_gigamuga_12_batches.Rmd
Deleted: analysis/04_diagnosis_qc_gigamuga_nine_batches.R
Deleted: analysis/04_diagnosis_qc_gigamuga_nine_batches.Rmd
Deleted: analysis/05_after_diagnosis_qc_gigamuga_11_batches.Rmd
Modified: analysis/05_after_diagnosis_qc_gigamuga_12_batches.Rmd
Deleted: analysis/05_after_diagnosis_qc_gigamuga_nine_batches.R
Deleted: analysis/05_after_diagnosis_qc_gigamuga_nine_batches.Rmd
Deleted: analysis/06_final_pr_apr_69K.R
Deleted: analysis/07.1_html_founder_prop.R
Modified: analysis/07_do_diversity_report.Rmd
Deleted: analysis/07_recomb_size_founder_prop.R
Deleted: analysis/07_recomb_size_founder_prop.Rmd
Deleted: analysis/08_gcta_herit.R
Deleted: analysis/09_qtlmapping.R
Deleted: analysis/10_qtl_permu.R
Deleted: analysis/11_qtl_blup.R
Deleted: analysis/12_plot_69k_qtl_mapping_2.Rmd
Deleted: analysis/12_plot_qtl_mapping_1.Rmd
Deleted: analysis/12_plot_qtl_mapping_2.Rmd
Deleted: analysis/13_plot_69k_conditional_m2_qtlmapping.Rmd
Deleted: analysis/13_plot_conditional_m2_qtlmapping.Rmd
Deleted: analysis/16_diagnosis_qc_gigamuga_gagnon.Rmd
Deleted: analysis/16_diagnosis_qc_gigamuga_gagnon2.Rmd
Deleted: analysis/17_plot_qtl_mapping.Rmd
Modified: analysis/_site.yml
Deleted: analysis/run_01_geneseek2qtl2.R
Deleted: analysis/run_02_geneseek2intensity.R
Deleted: analysis/run_03_firstgm2genoprobs.R
Deleted: analysis/run_04_diagnosis_qc_gigamuga_nine_batches.R
Deleted: analysis/run_05_after_diagnosis_qc_gigamuga_nine_batches.R
Deleted: analysis/run_06_final_pr_apr_69K.R
Deleted: analysis/run_07.1_html_founder_prop.R
Deleted: analysis/run_07_recomb_size_founder_prop.R
Deleted: analysis/run_08_gcta_herit.R
Deleted: analysis/run_09_qtlmapping.R
Deleted: analysis/run_10_qtl_permu.R
Deleted: analysis/run_11_qtl_blup.R
Modified: code/README.md
Modified: csna_workflow.Rproj
Modified: data/README.md
Modified: output/README.md
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_AcqAnticipatoryCorrectResponses_coefplot.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_AcqAnticipatoryCorrectResponses_coefplot_blup.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_AcqAnticipatoryCorrectResponsesnooutlier_coefplot.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_AcqAnticipatoryCorrectResponsesnooutlier_coefplot_blup.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_AcqAnticipatoryCorrectResponsesnooutlierrz_coefplot.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_AcqAnticipatoryCorrectResponsesnooutlierrz_coefplot_blup.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_AcqAnticipatoryCorrectResponsesrz_coefplot.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_AcqAnticipatoryCorrectResponsesrz_coefplot_blup.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_AcqTotalTrials_coefplot.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_AcqTotalTrials_coefplot_blup.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_AcqTotalTrialsrz_coefplot.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_AcqTotalTrialsrz_coefplot_blup.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_DiffTotalTrials_coefplot.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_DiffTotalTrials_coefplot_blup.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_DiffTotalTrialsrz_coefplot.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_DiffTotalTrialsrz_coefplot_blup.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_RevAnticipatoryIncorrectResponses_coefplot.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_RevAnticipatoryIncorrectResponses_coefplot_blup.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_RevAnticipatoryIncorrectResponsesboxcoxtrans_coefplot.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_RevAnticipatoryIncorrectResponsesboxcoxtrans_coefplot_blup.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_RevAnticipatoryIncorrectResponsesnooutlier_coefplot.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_RevAnticipatoryIncorrectResponsesnooutlier_coefplot_blup.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_RevAnticipatoryIncorrectResponsesnooutlierrz_coefplot.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_RevAnticipatoryIncorrectResponsesnooutlierrz_coefplot_blup.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_RevAnticipatoryIncorrectResponsesrz_coefplot.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_RevAnticipatoryIncorrectResponsesrz_coefplot_blup.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_RevTotalTrials_coefplot.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_RevTotalTrials_coefplot_blup.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_RevTotalTrialsrz_coefplot.pdf
Modified: output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_RevTotalTrialsrz_coefplot_blup.pdf
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/Prj01_RL_pheno_qtl2_DO_11092020_69k.Rmd) and HTML (docs/Prj01_RL_pheno_qtl2_DO_11092020_69k.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | e1ea743 | xhyuo | 2022-09-16 | Build site. |
| Rmd | b8db415 | xhyuo | 2022-09-16 | bootstrap |
| html | 0395735 | xhyuo | 2022-09-07 | Build site. |
| Rmd | bebe974 | xhyuo | 2022-09-07 | section |
| html | 9baa624 | xhyuo | 2022-08-29 | Build site. |
| Rmd | 8541f24 | xhyuo | 2022-08-29 | Prj01_RL_pheno_qtl2_DO_11092020_69k |
Last update: 2022-09-19
loading libraries
library(ggplot2)
library(gridExtra)
library(GGally)
library(parallel)
library(qtl2)
library(parallel)
library(survival)
library(regress)
library(abind)
library(openxlsx)
library(tidyverse)
library(SimDesign)
library(MASS)
library(EnvStats)
rz.transform <- function(y) {
rankY=rank(y, ties.method="average", na.last="keep")
rzT=qnorm(rankY/(length(na.exclude(rankY))+1))
return(rzT)
}
# heritability
# function to simulate data to estimate heritability intervals
h2_sim <- function(trait, K, covar, nsim=100){
# trait, K and covar are matrices with row.names in qtl2-style
# covar adjustment applied to data but not in simulated data
# reduce mice to common set with kinship and trait values
# mice.names <- intersect(row.names(K), row.names(trait))
# K <- K[mice.names, mice.names]
# trait <- as.matrix(trait[mice.names,],ncol=1)
N <- dim(trait)[1]
# estimate heritability
h2 <- as.numeric(est_herit(trait, K, covar))
# simulate from kinship matrix and re-estimate
h2_sim <- rep(0,nsim)
for(i in 1:nsim){
trait_sim <- t(rmvnorm(n = 1, mean = rep(0,N), sigma = h2*2*K + (1-h2)*diag(1,N)))
rownames(trait_sim) <- rownames(trait)
h2_sim[i] <- as.numeric(est_herit(trait_sim, K))
}
# return estimated h2 and simulations
list(N=N, h2=h2, sim=h2_sim)
}
setwd("/projects/csna/csna_workflow/")Read phenotype data
#Prj01_RL_pheno_qttl2_DO_11092020
pheno <- readr::read_csv("data/pheno/Prj01_RL_pheno_qttl2_DO_11092020.csv", na = c("N/A", "NA"))
pheno <- pheno %>%
filter(apply(pheno, 1, function(x) sum(is.na(x))) < 5) %>% # remove rows with 5 NAs
mutate(
Acq.Anticipatory.Correct.Responses.nooutlier = case_when(
(Acq.Anticipatory.Correct.Responses > 5 | is.na(Acq.Anticipatory.Correct.Responses)) ~ NA_real_,
TRUE ~ Acq.Anticipatory.Correct.Responses),
Rev.Anticipatory.Incorrect.Responses.nooutlier = case_when(
(Rev.Anticipatory.Incorrect.Responses > 6) ~ NA_real_,
TRUE ~ Rev.Anticipatory.Incorrect.Responses
)) %>% #boxcoxtransformation
mutate(across(c(Rev.Anticipatory.Incorrect.Responses), ~ EnvStats::boxcoxTransform(.x, lambda = EnvStats::boxcox(.x, optimize = TRUE)$lambda),
.names ="{.col}.boxcox.trans")) %>% #omit values >6 for Acq.Anticipatory.Correct.Responses and Rev.Anticipatory.Incorrect.Responses variables.
mutate(across(Acq.Total.Trials:Rev.Anticipatory.Incorrect.Responses.nooutlier, ~ rz.transform(.x),
.names ="{.col}.rz"))
#load gm data
load("data/Jackson_Lab_12_batches/gm_DO3173_qc_newid.RData")
#subset
overlap.id <- intersect(pheno$mouseID, ind_ids(gm_after_qc))
load("data/Jackson_Lab_12_batches/qc_info.RData")
#merge pheno with covar
pheno <- pheno %>%
left_join(qc_info) %>%
filter(mouseID %in% overlap.id) %>%
filter(bad.sample == FALSE) %>%
mutate(sex = case_when(
sex == "M" ~ 1,
sex == "F" ~ 0
)) %>%
mutate_at(vars(sex,
ngen),
list(factor))
#pgmap
load("/projects/csna/csna_workflow/data/69k_grid_pgmap.RData")
query_variants <- create_variant_query_func("/projects/csna/csna_workflow/data/cc_variants.sqlite")
query_genes <- create_gene_query_func("/projects/csna/csna_workflow/data/mouse_genes_mgi.sqlite")Plot qtl mapping on 69k
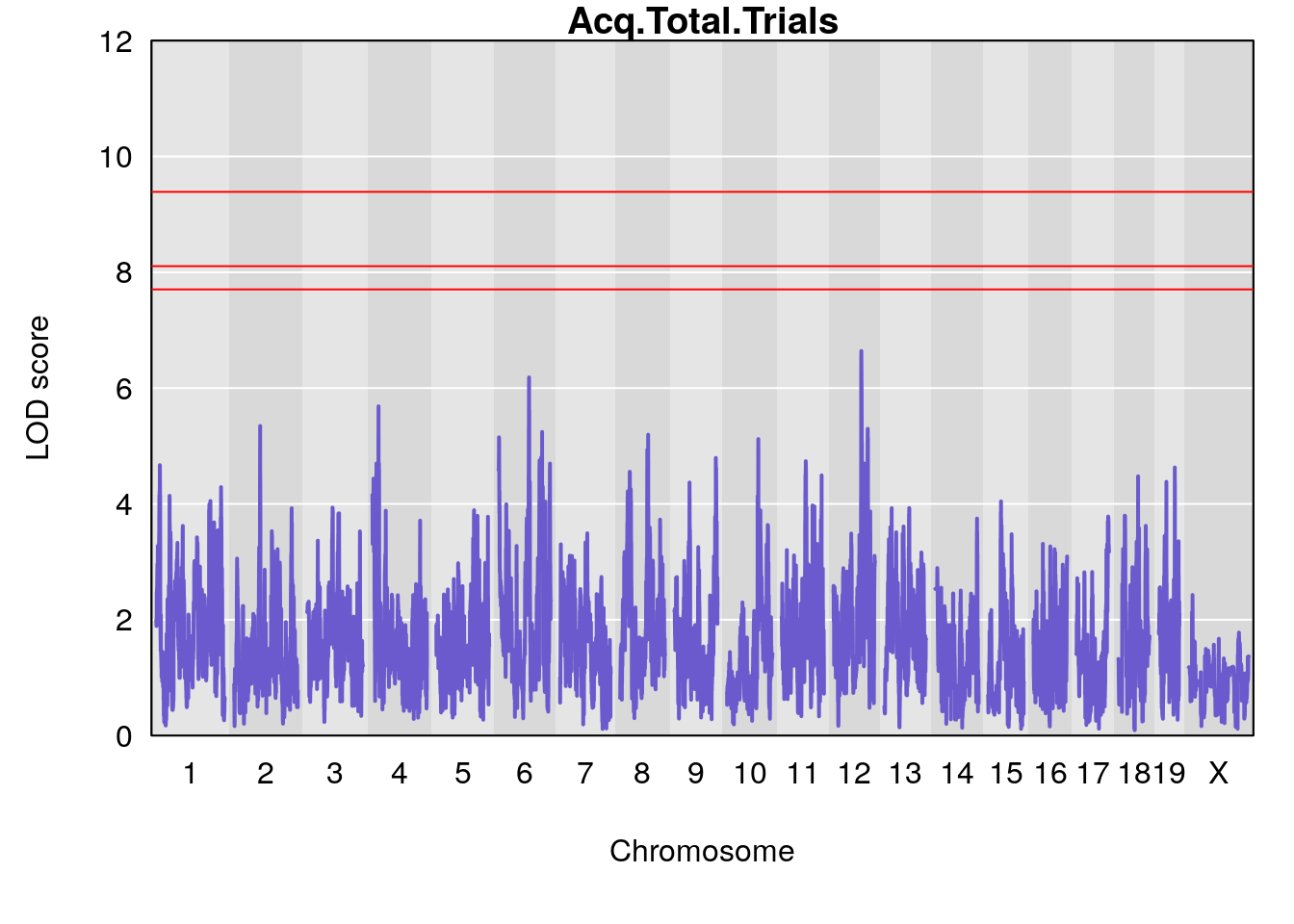
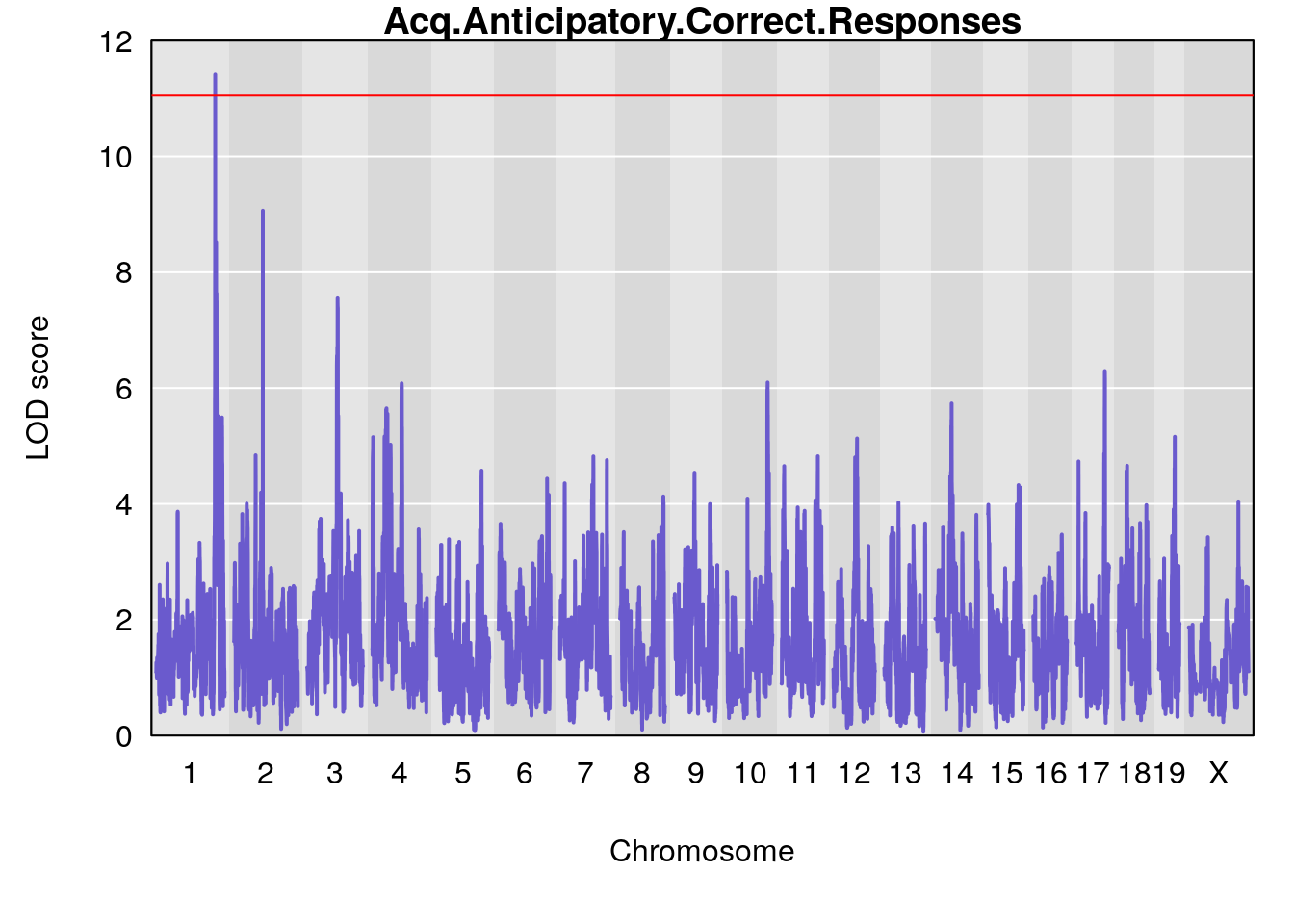
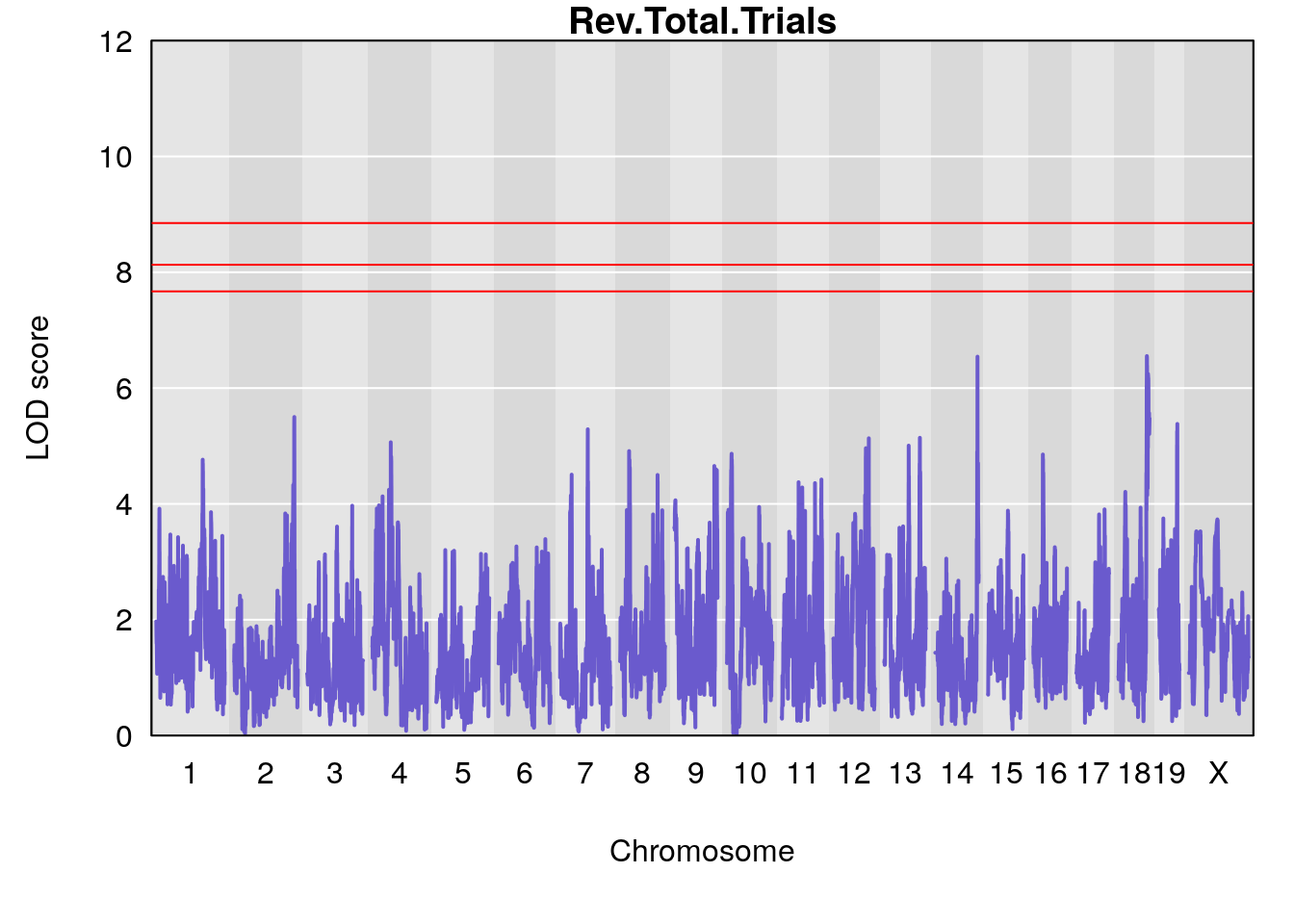
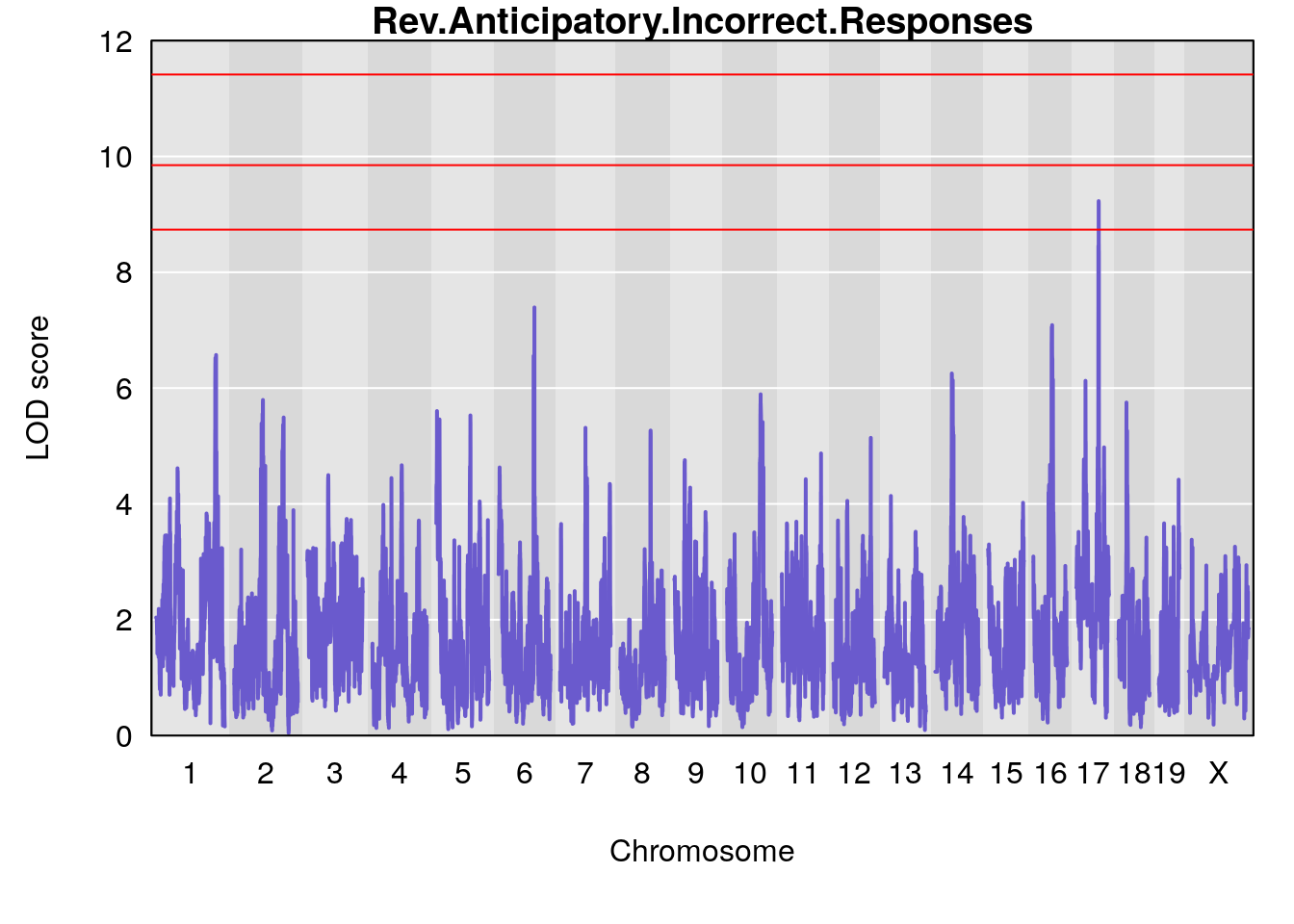
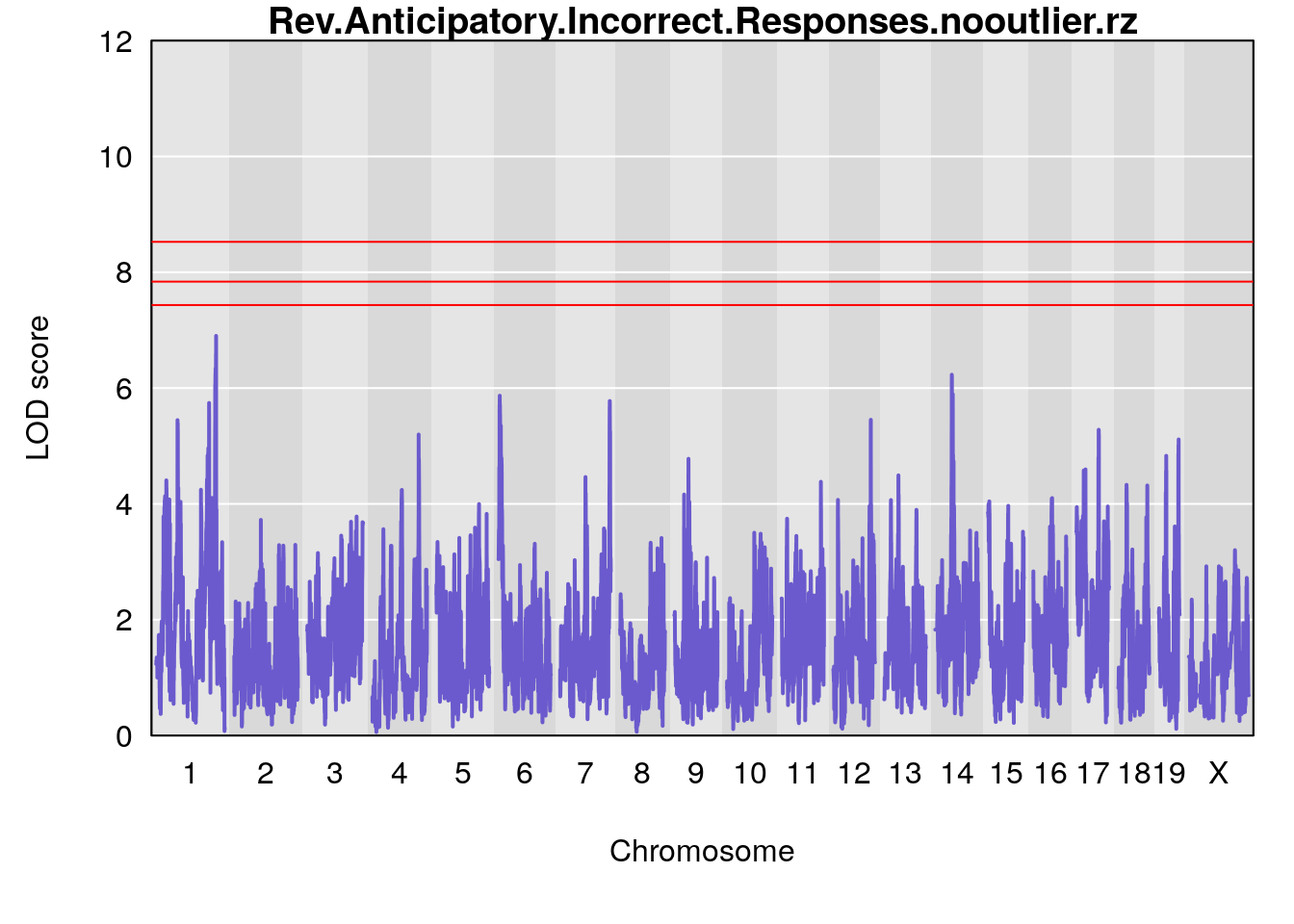
Plot by using its output Prj01_RL_11092020_cutoff6.qtlout.69k.RData
load("output/Prj01_RL_11092020_cutoff6.qtlout.69k.RData")
#genome-wide plot
for(i in names(m2.qtl.out)){
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(m2.qtl.out[[i]]) # overall maximum LOD score
plot(m2.qtl.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, 12))
abline(h=summary(m2.permu[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(m2.permu[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(m2.permu[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0(i))
}
| Version | Author | Date |
|---|---|---|
| 9baa624 | xhyuo | 2022-08-29 |
| Version | Author | Date |
|---|---|---|
| 9baa624 | xhyuo | 2022-08-29 |
| Version | Author | Date |
|---|---|---|
| 9baa624 | xhyuo | 2022-08-29 |
| Version | Author | Date |
|---|---|---|
| 9baa624 | xhyuo | 2022-08-29 |
| Version | Author | Date |
|---|---|---|
| 9baa624 | xhyuo | 2022-08-29 |
| Version | Author | Date |
|---|---|---|
| 9baa624 | xhyuo | 2022-08-29 |

| Version | Author | Date |
|---|---|---|
| 9baa624 | xhyuo | 2022-08-29 |
| Version | Author | Date |
|---|---|---|
| 9baa624 | xhyuo | 2022-08-29 |
| Version | Author | Date |
|---|---|---|
| 9baa624 | xhyuo | 2022-08-29 |
| Version | Author | Date |
|---|---|---|
| 9baa624 | xhyuo | 2022-08-29 |
| Version | Author | Date |
|---|---|---|
| 9baa624 | xhyuo | 2022-08-29 |

| Version | Author | Date |
|---|---|---|
| 9baa624 | xhyuo | 2022-08-29 |
| Version | Author | Date |
|---|---|---|
| 9baa624 | xhyuo | 2022-08-29 |
| Version | Author | Date |
|---|---|---|
| 9baa624 | xhyuo | 2022-08-29 |
| Version | Author | Date |
|---|---|---|
| 9baa624 | xhyuo | 2022-08-29 |
pdf(file = paste0("output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout.pdf"), width = 16, height =8)
#save genome-wide plo
for(i in names(m2.qtl.out)){
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(m2.qtl.out[[i]]) # overall maximum LOD score
plot(m2.qtl.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, 10))
abline(h=summary(m2.permu[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(m2.permu[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(m2.permu[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0(i))
}
dev.off()
# png
# 2Coeff plot of qtl mapping
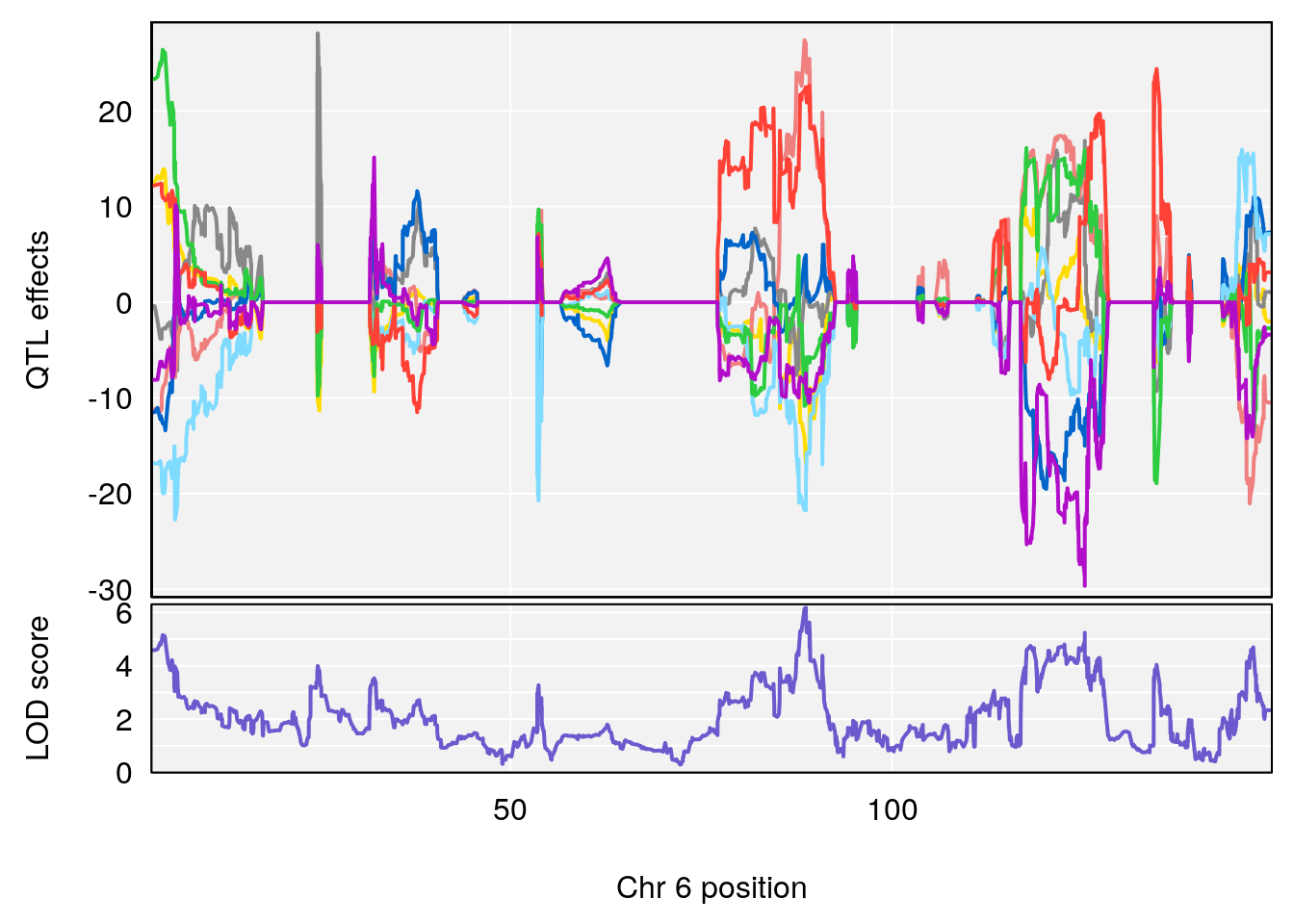
#peaks coeff plot
for(i in names(m2.qtl.out)){
print(i)
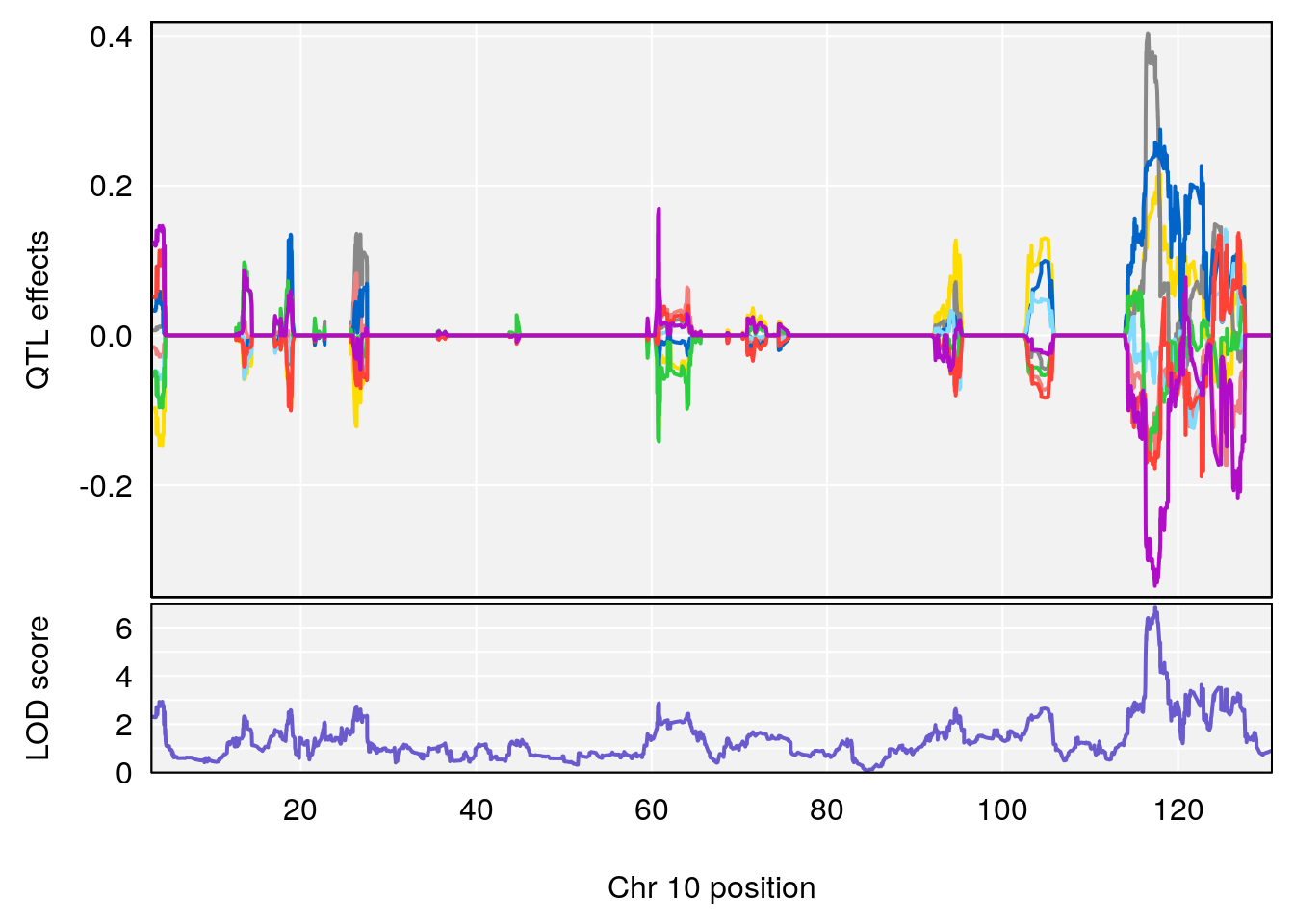
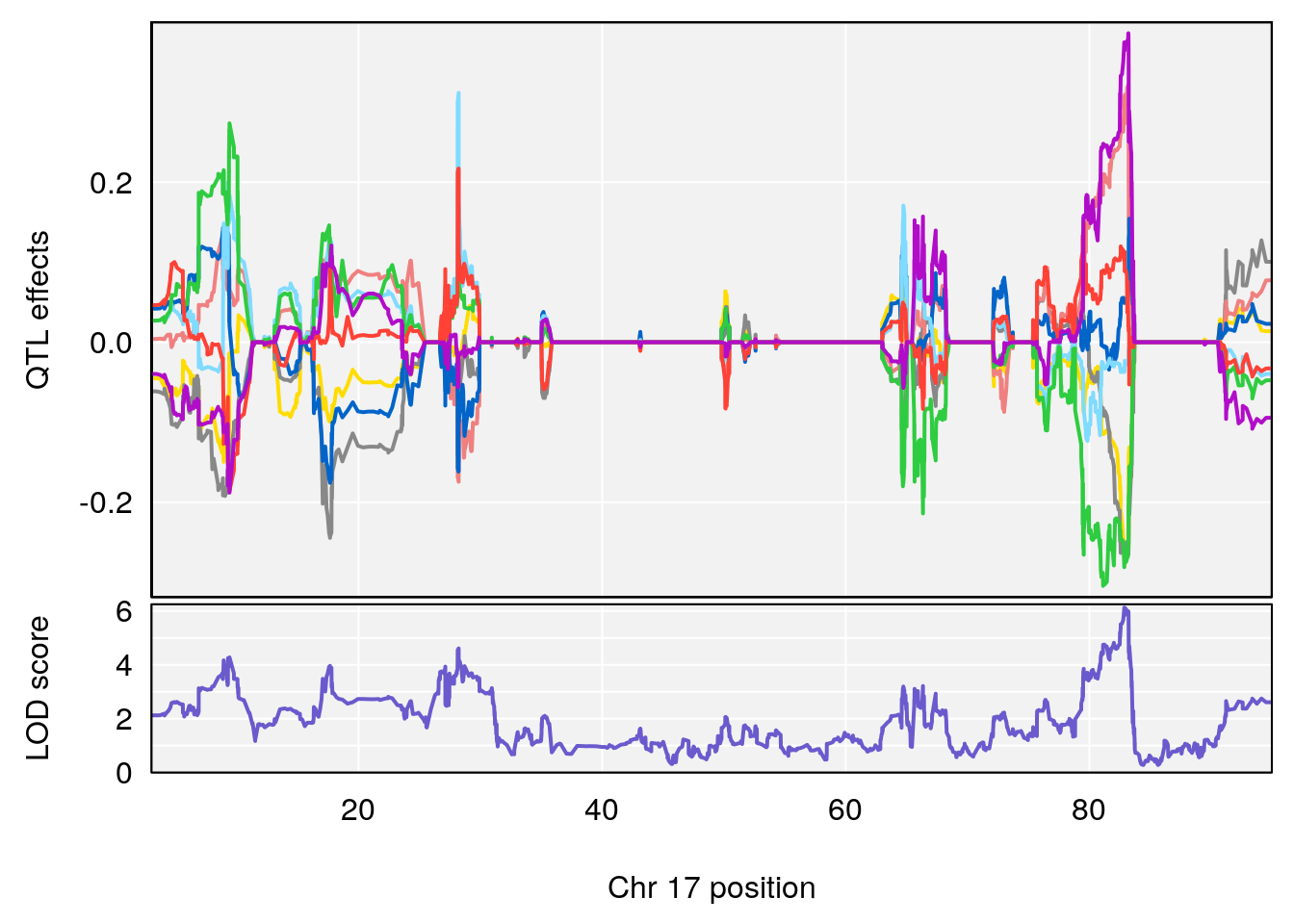
peaks <- find_peaks(m2.qtl.out[[i]], map=pmap, threshold=6, drop=1.5)
if(nrow(peaks) > 0) {
print(peaks)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
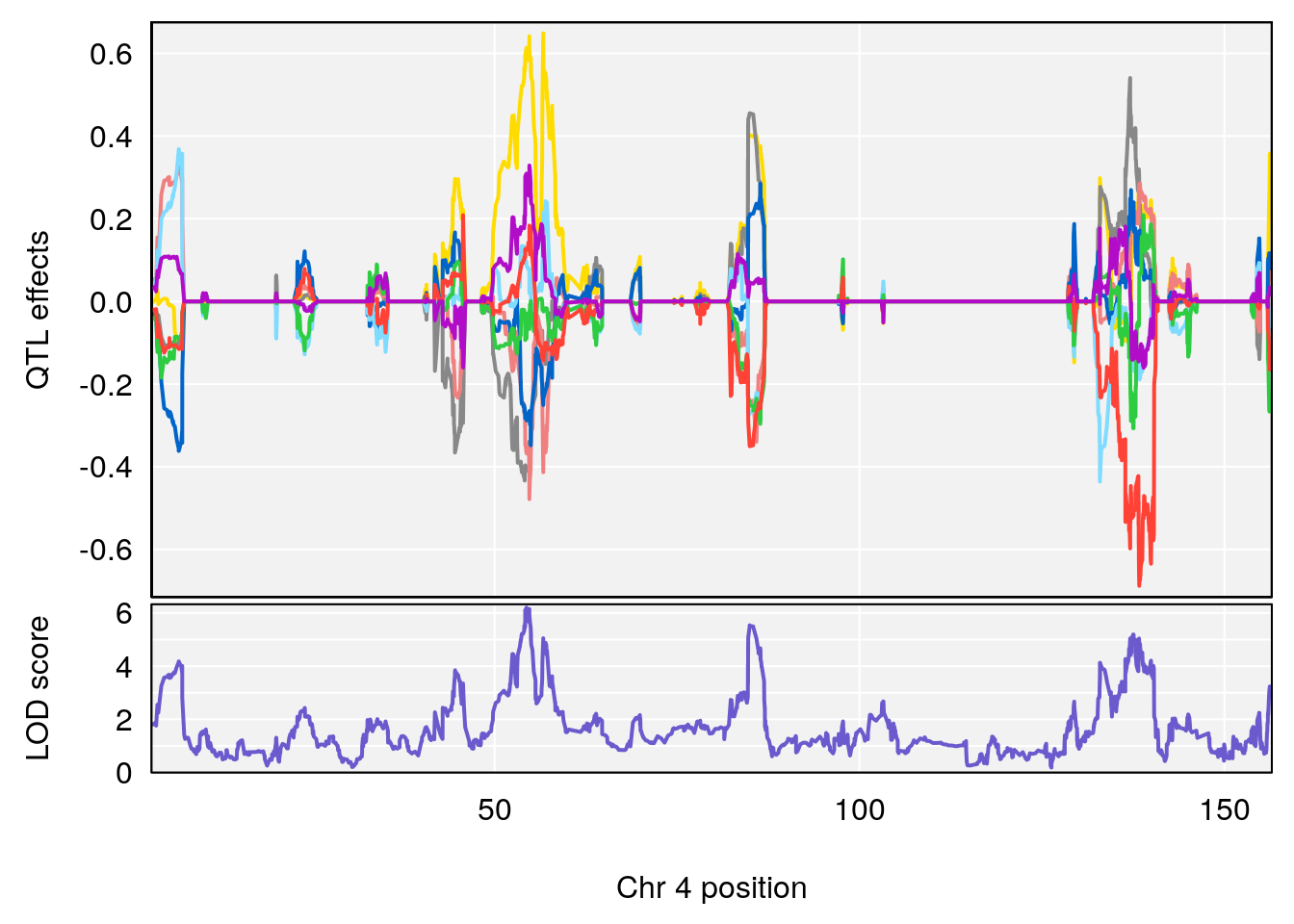
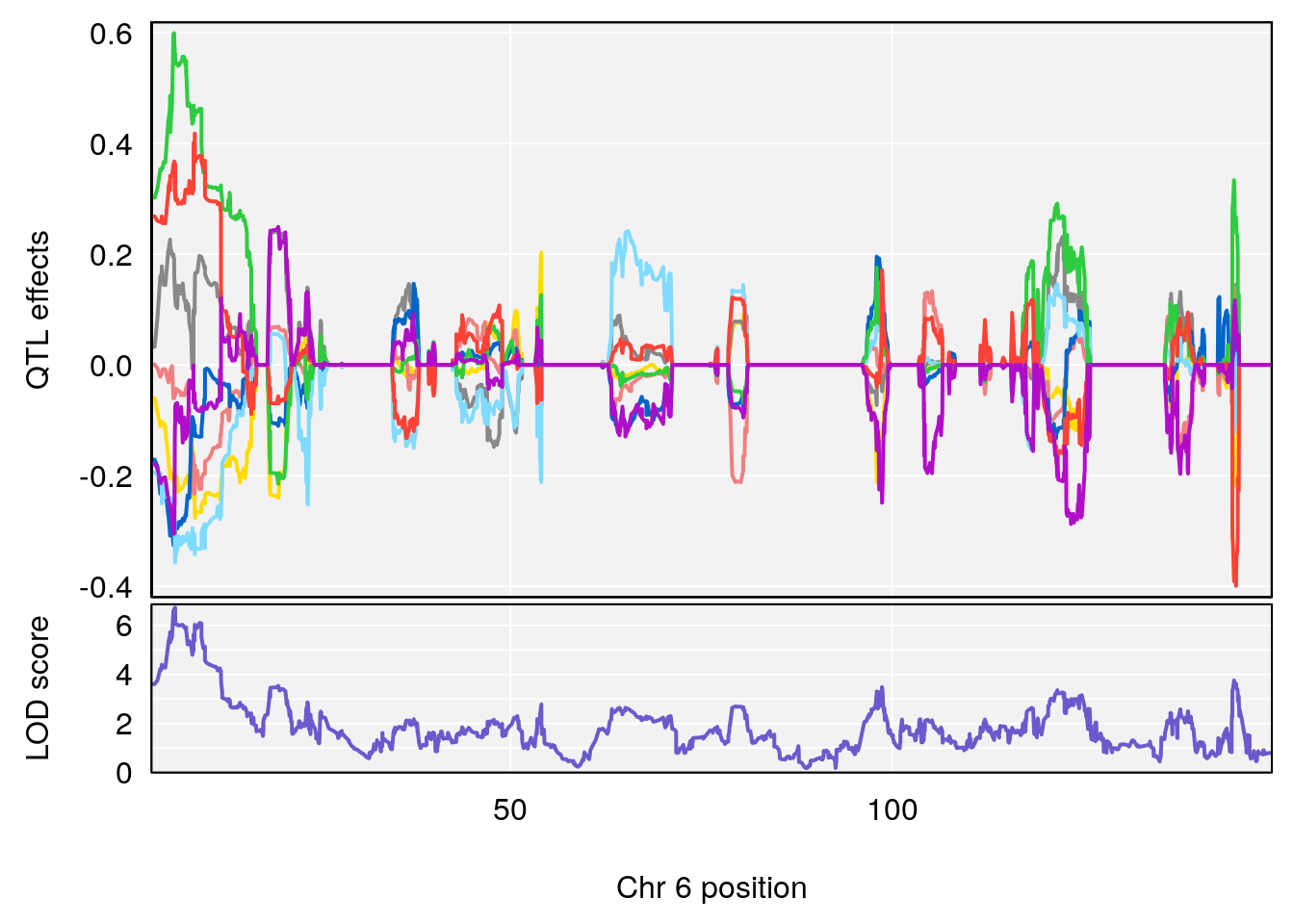
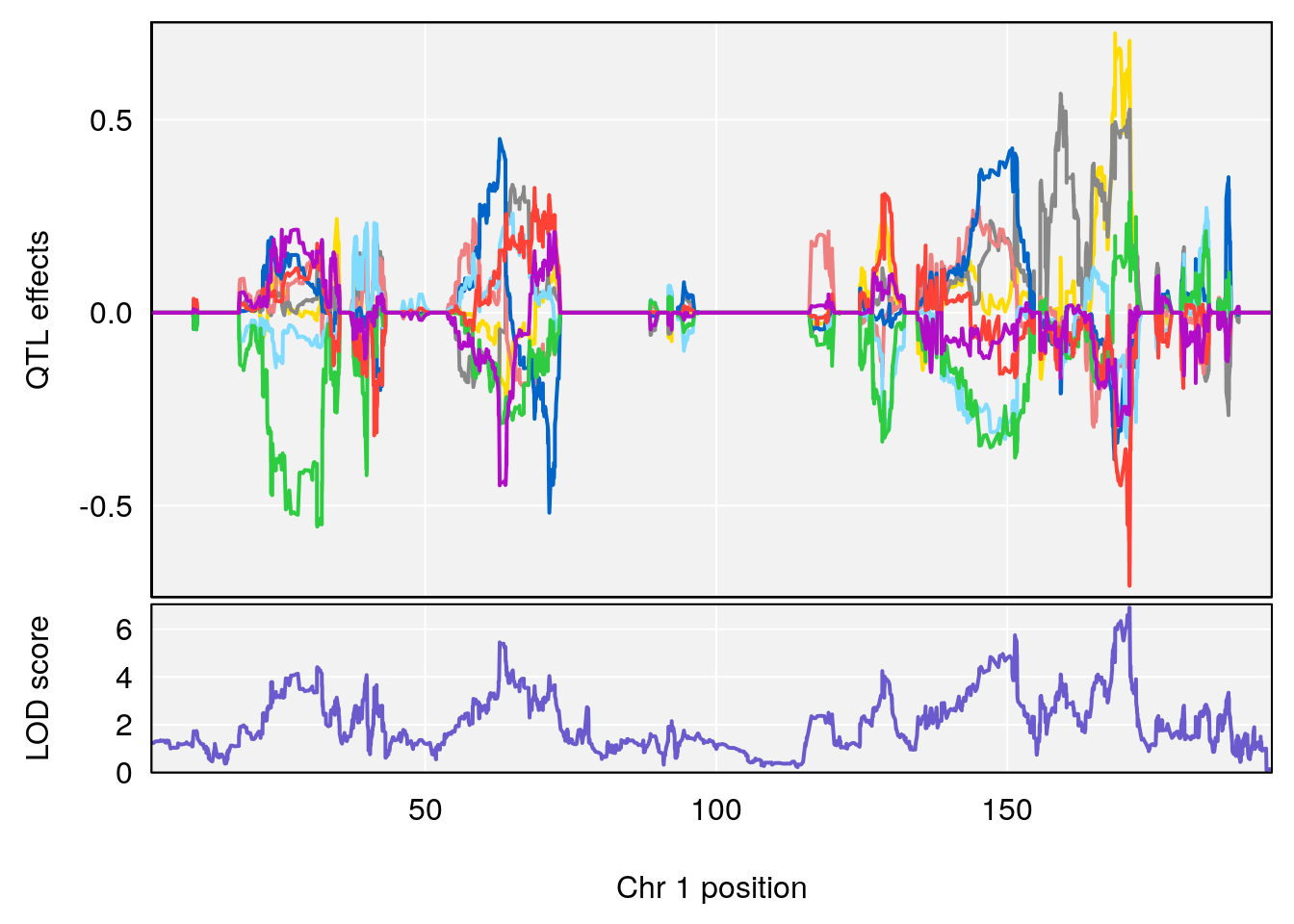
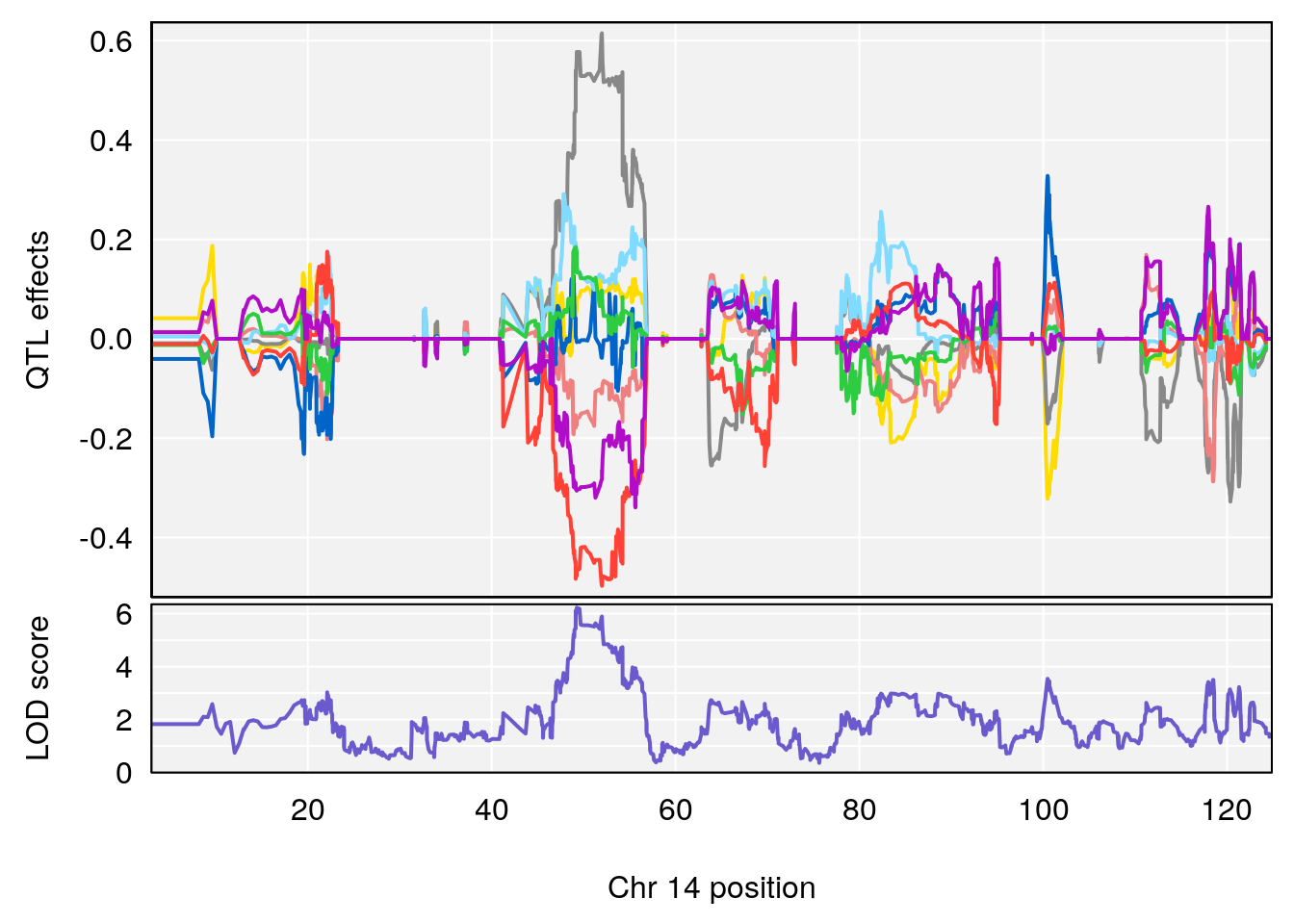
plot_coefCC(m2.sigqtl.coef[[i]][[p]], pmap[chr], scan1_output=m2.qtl.out[[i]], bgcolor="gray95", legend=NULL)
plot_coefCC(m2.sigqtl.blup[[i]][[p]], pmap[chr], scan1_output=m2.qtl.out[[i]], bgcolor="gray95", legend=NULL)
#plot(m2.sigqtl.snps[[i]][[p]]$lod, m2.sigqtl.snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=m2.sigqtl.genes[[i]][[p]])
}
}
}
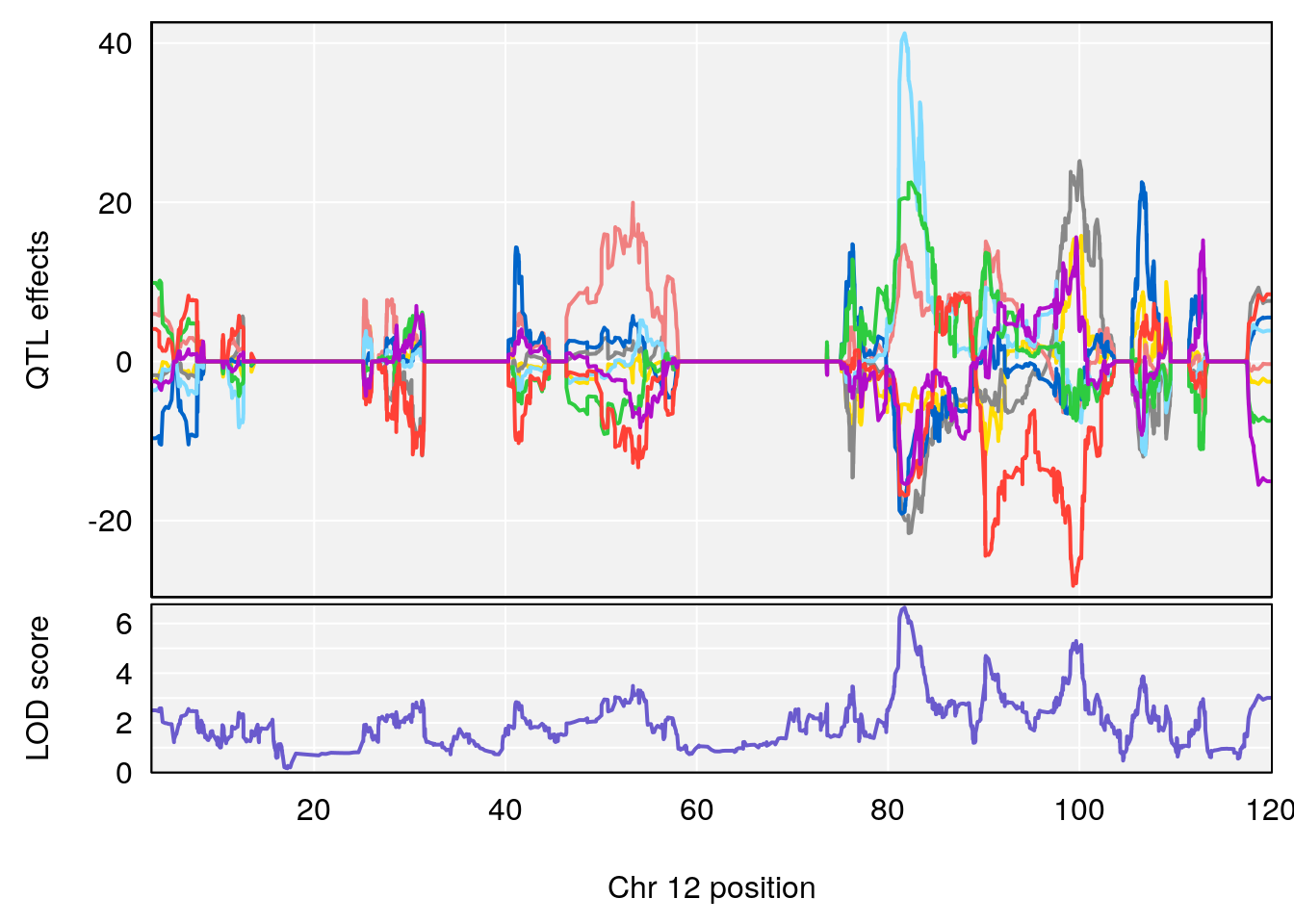
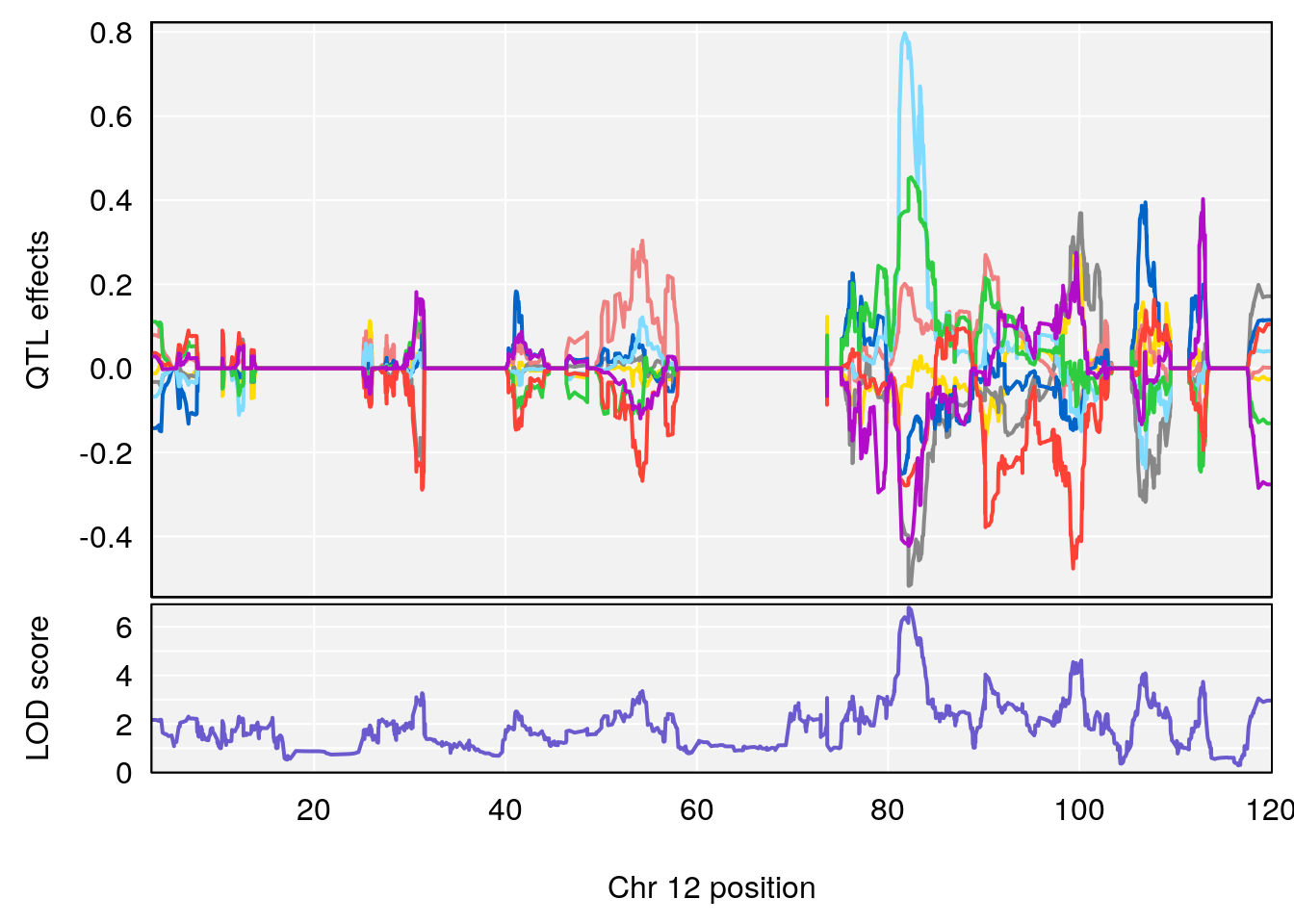
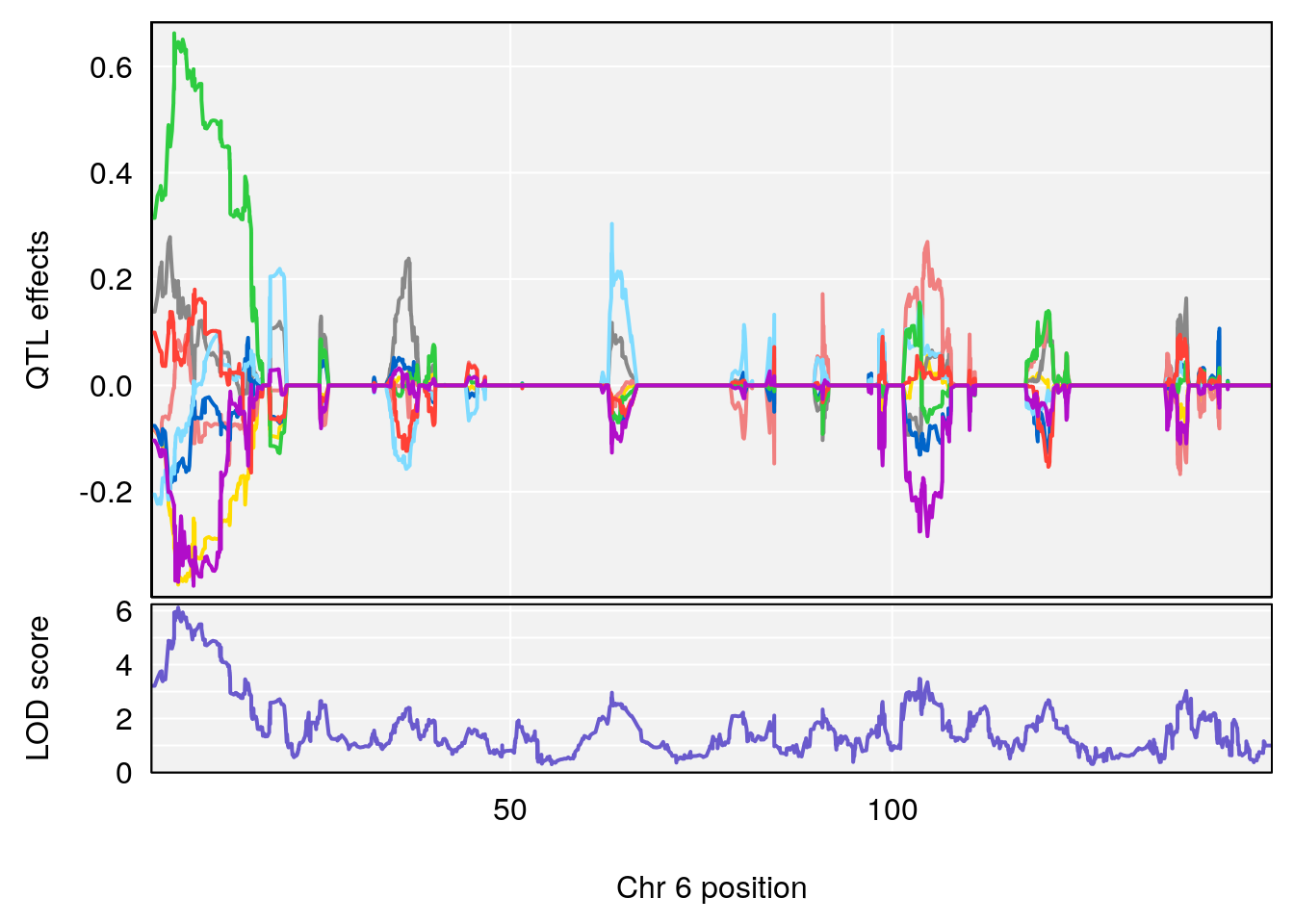
# [1] "Acq.Total.Trials"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
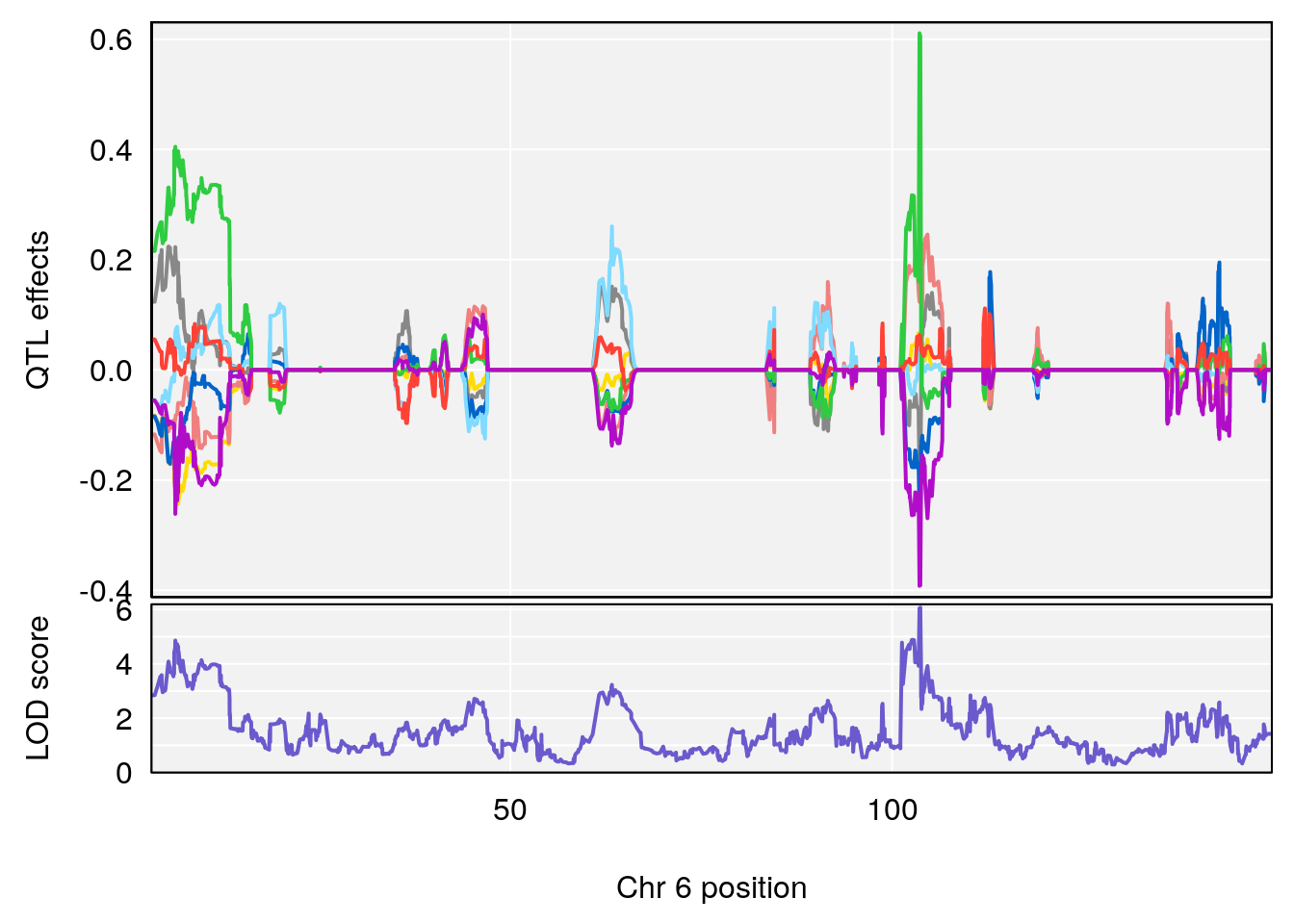
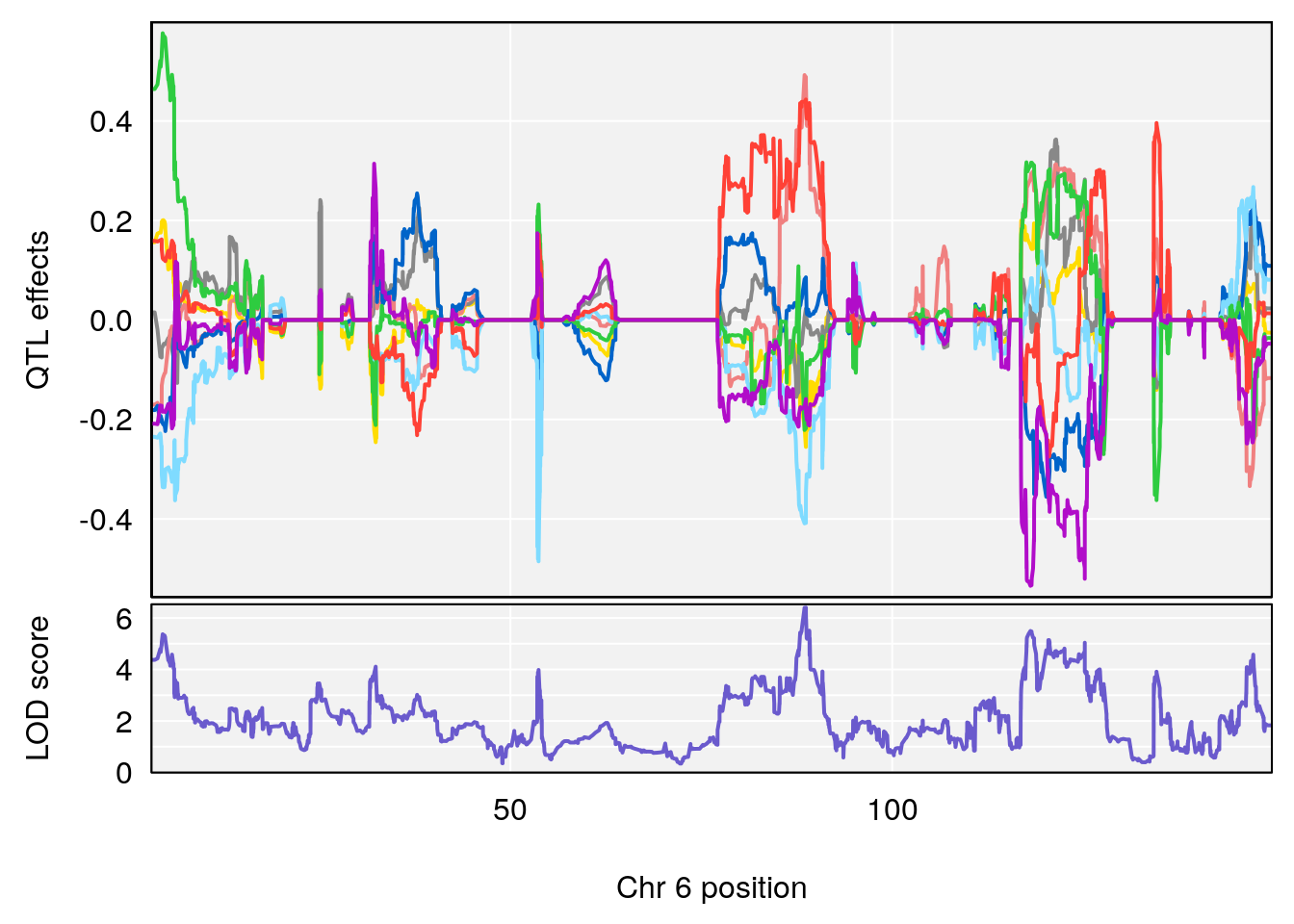
# 1 1 pheno1 6 88.69893 6.184173 3.77429 147.36991
# 2 1 pheno1 12 81.73145 6.640822 81.11268 99.75717
# [1] 1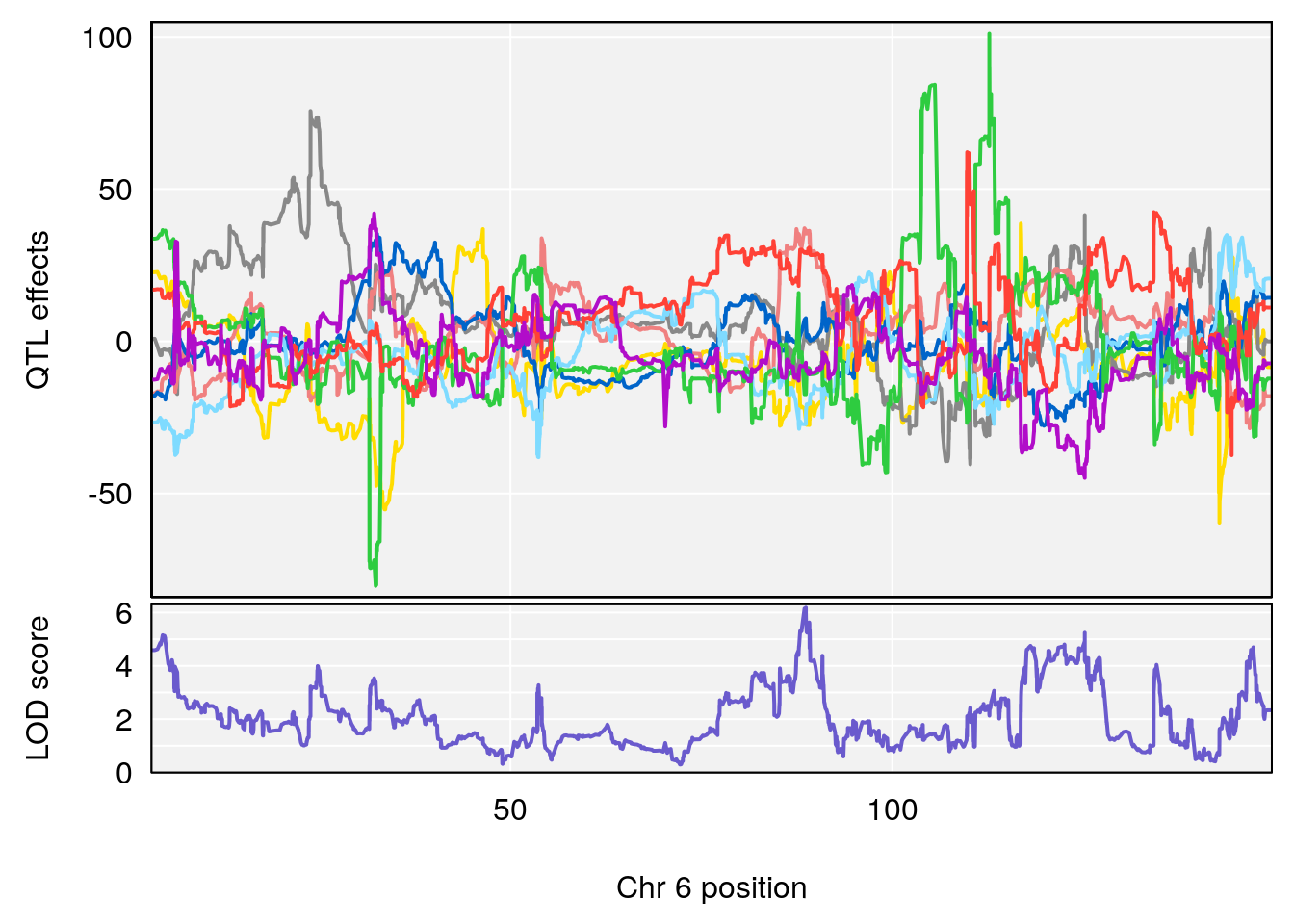
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 2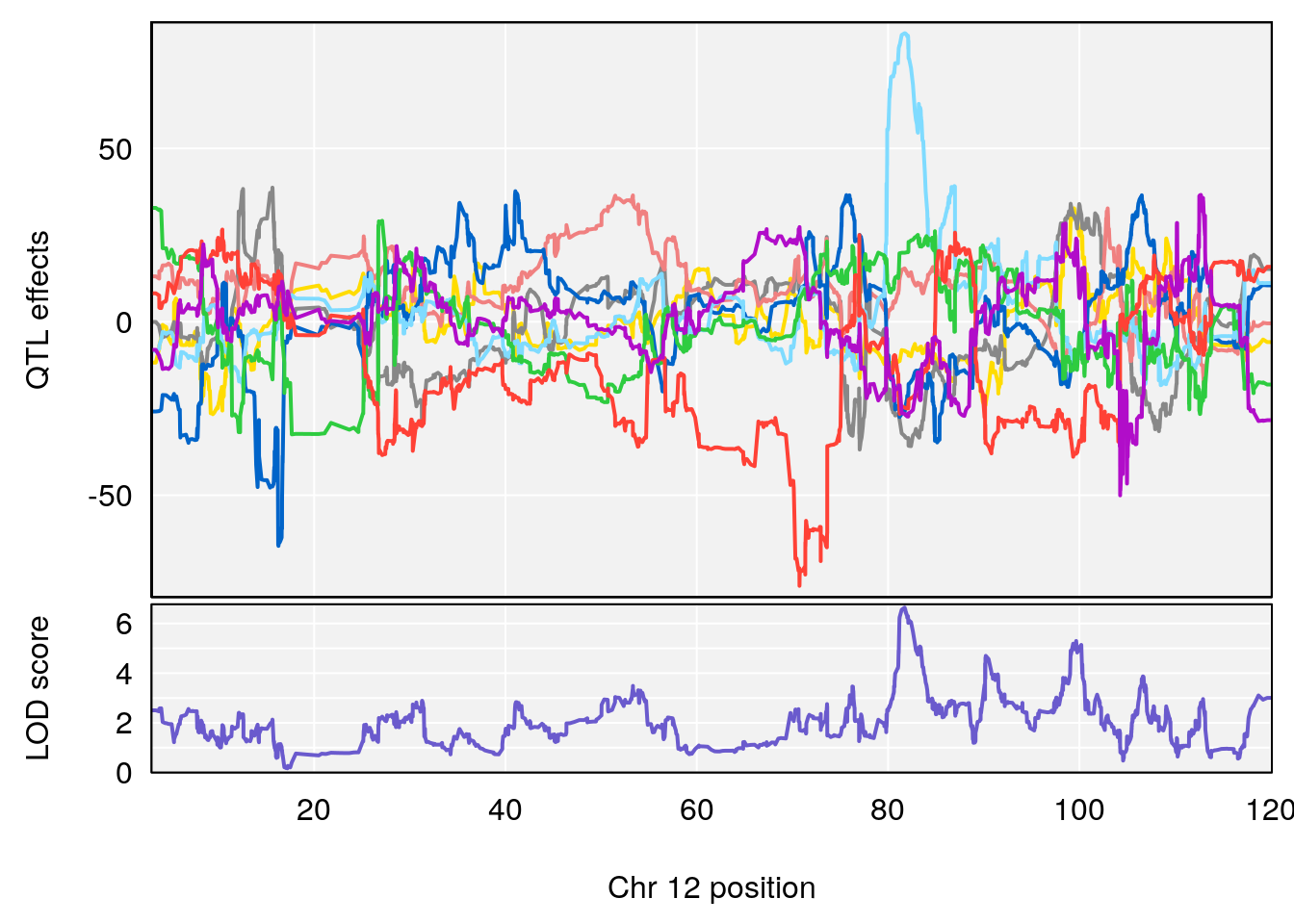
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
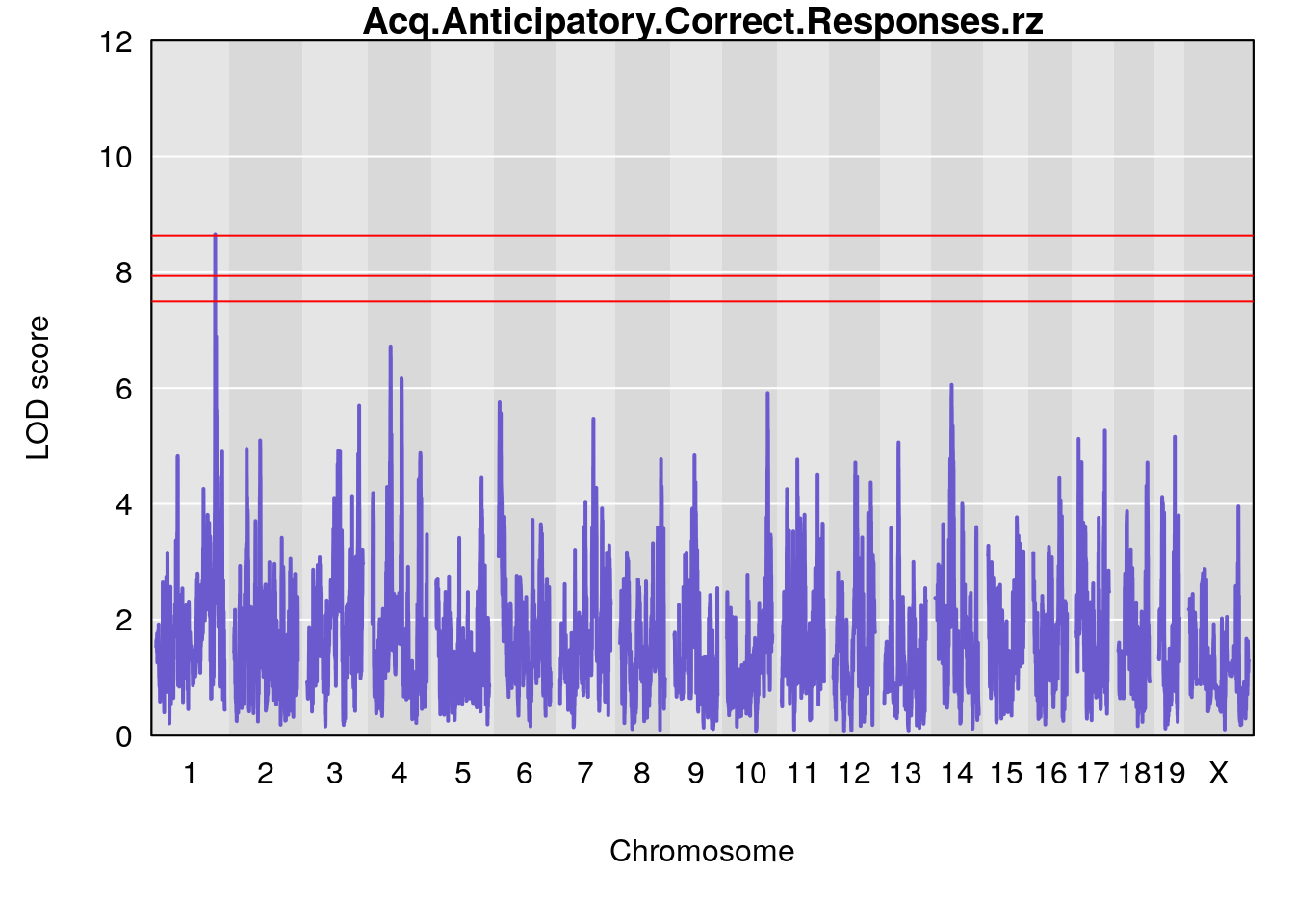
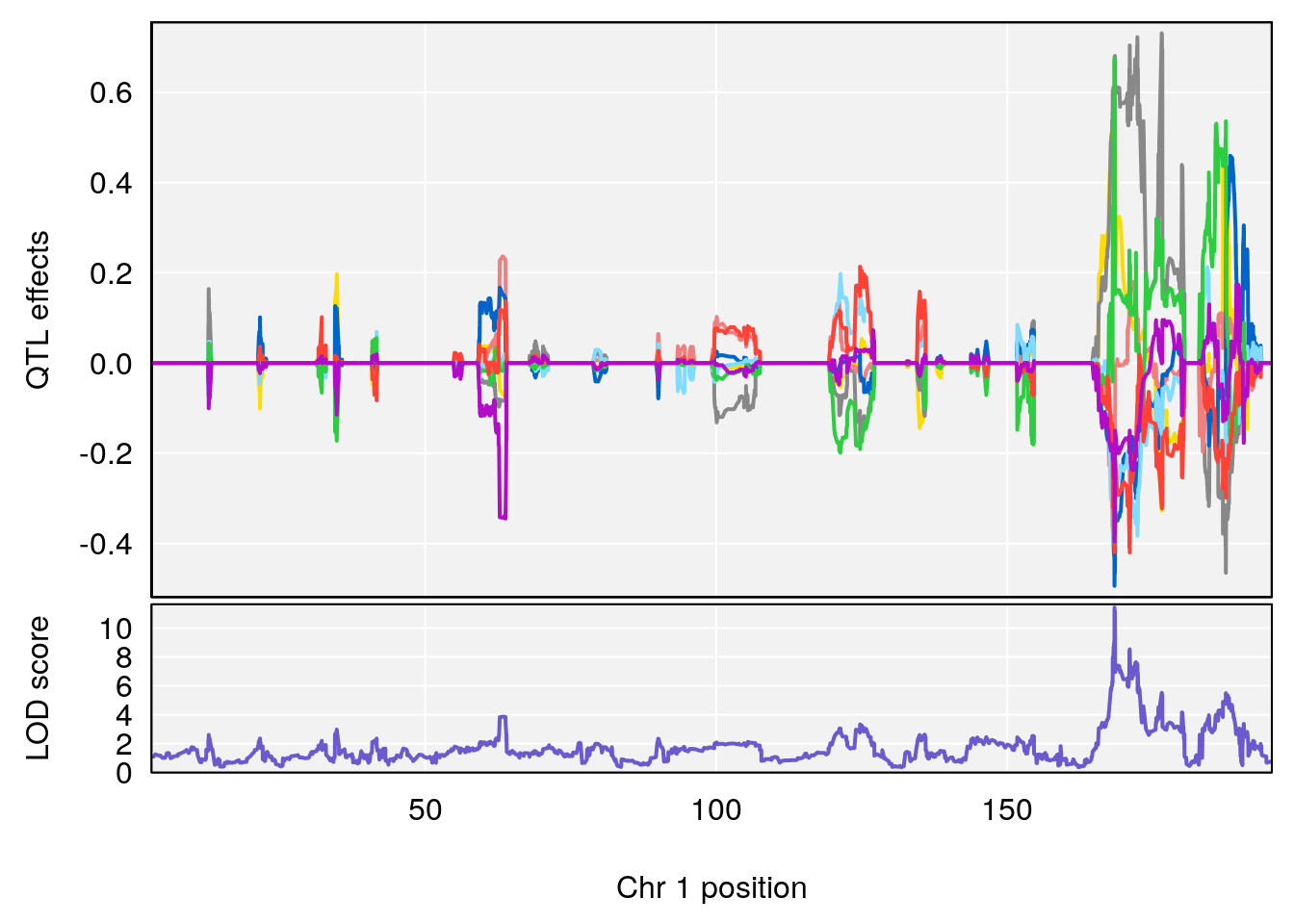
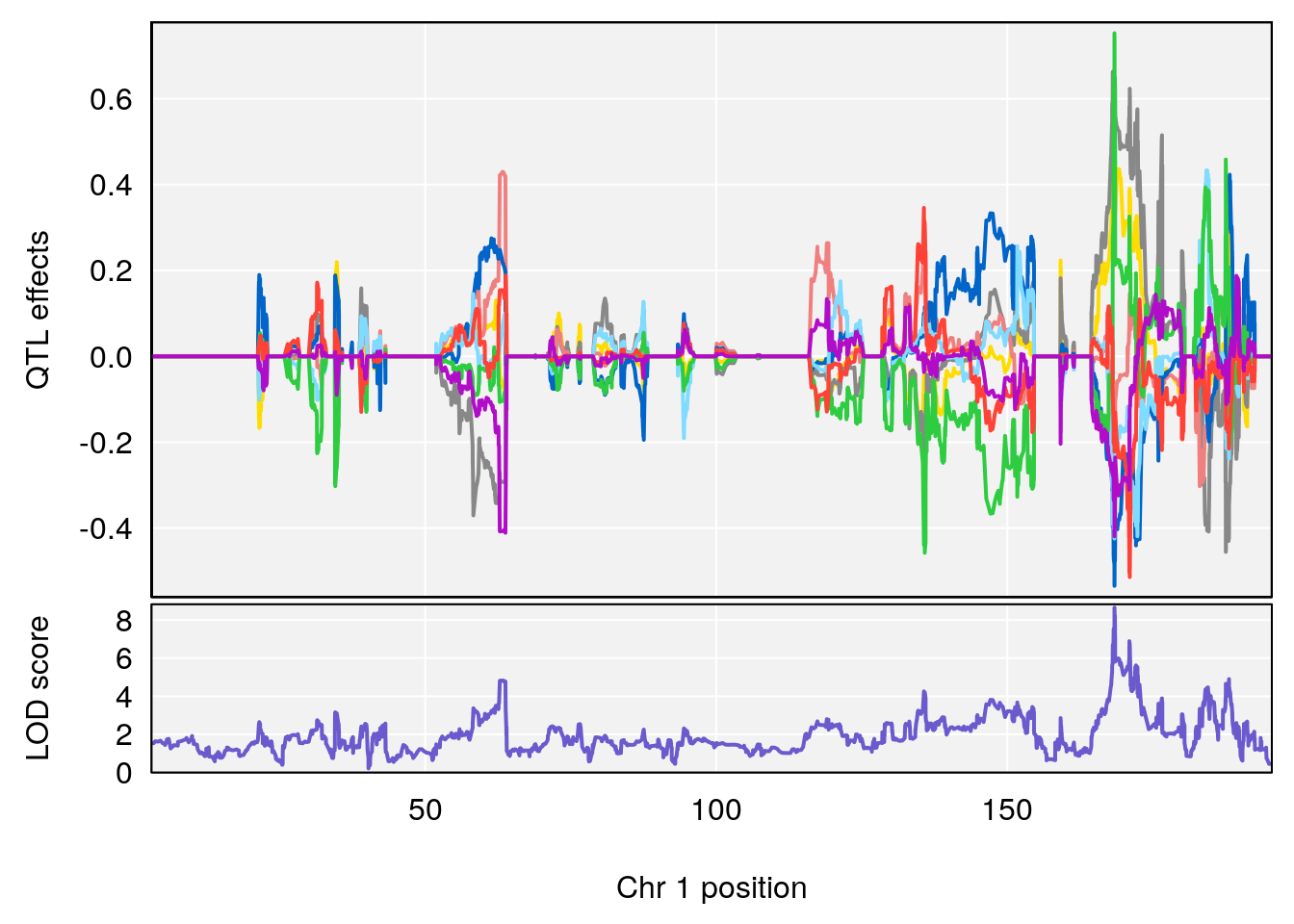
# [1] "Acq.Anticipatory.Correct.Responses"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 168.41339 11.416644 168.398842 168.52005
# 2 1 pheno1 2 83.81222 9.064175 83.529734 83.82600
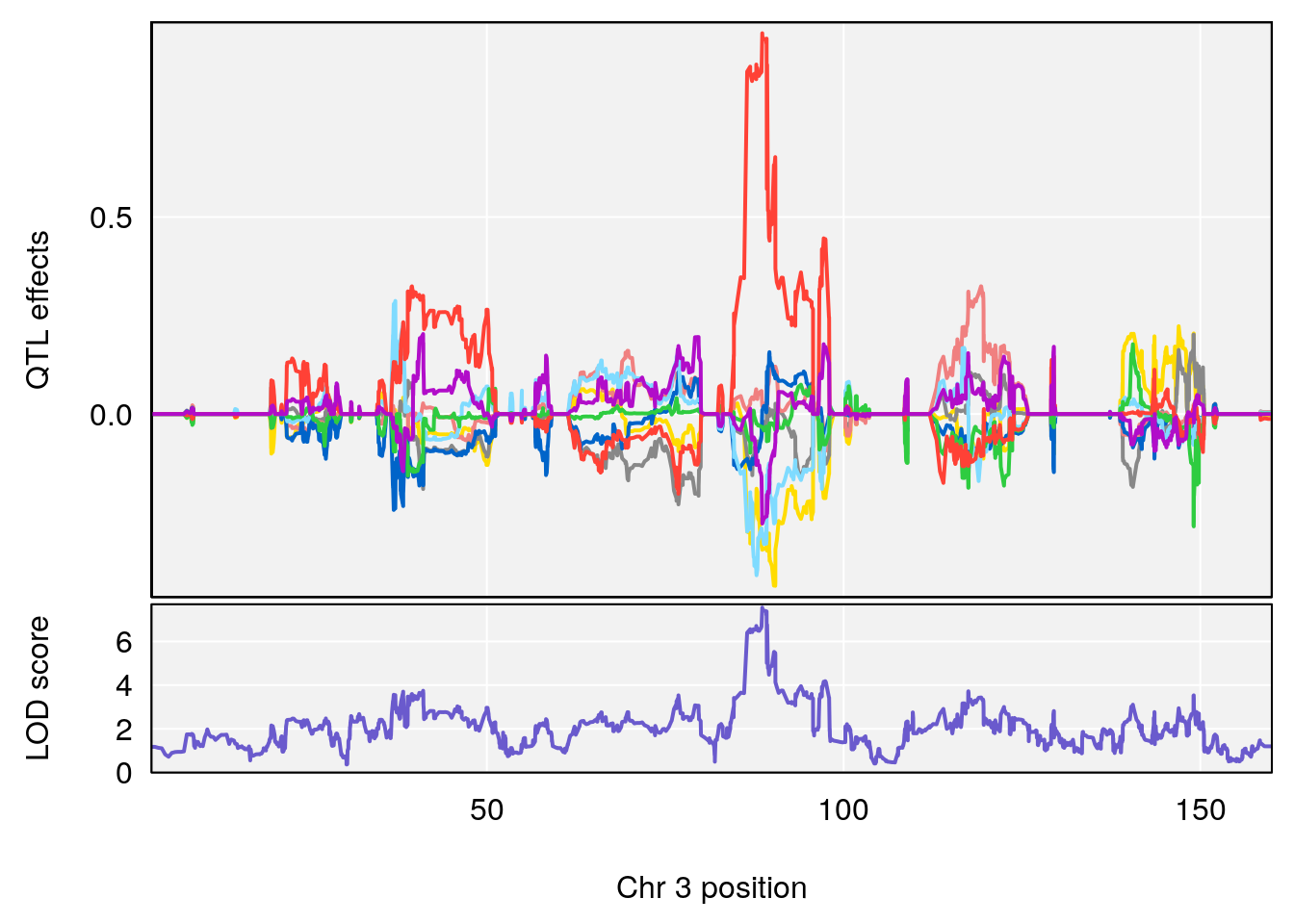
# 3 1 pheno1 3 88.59530 7.550793 86.050147 89.39324
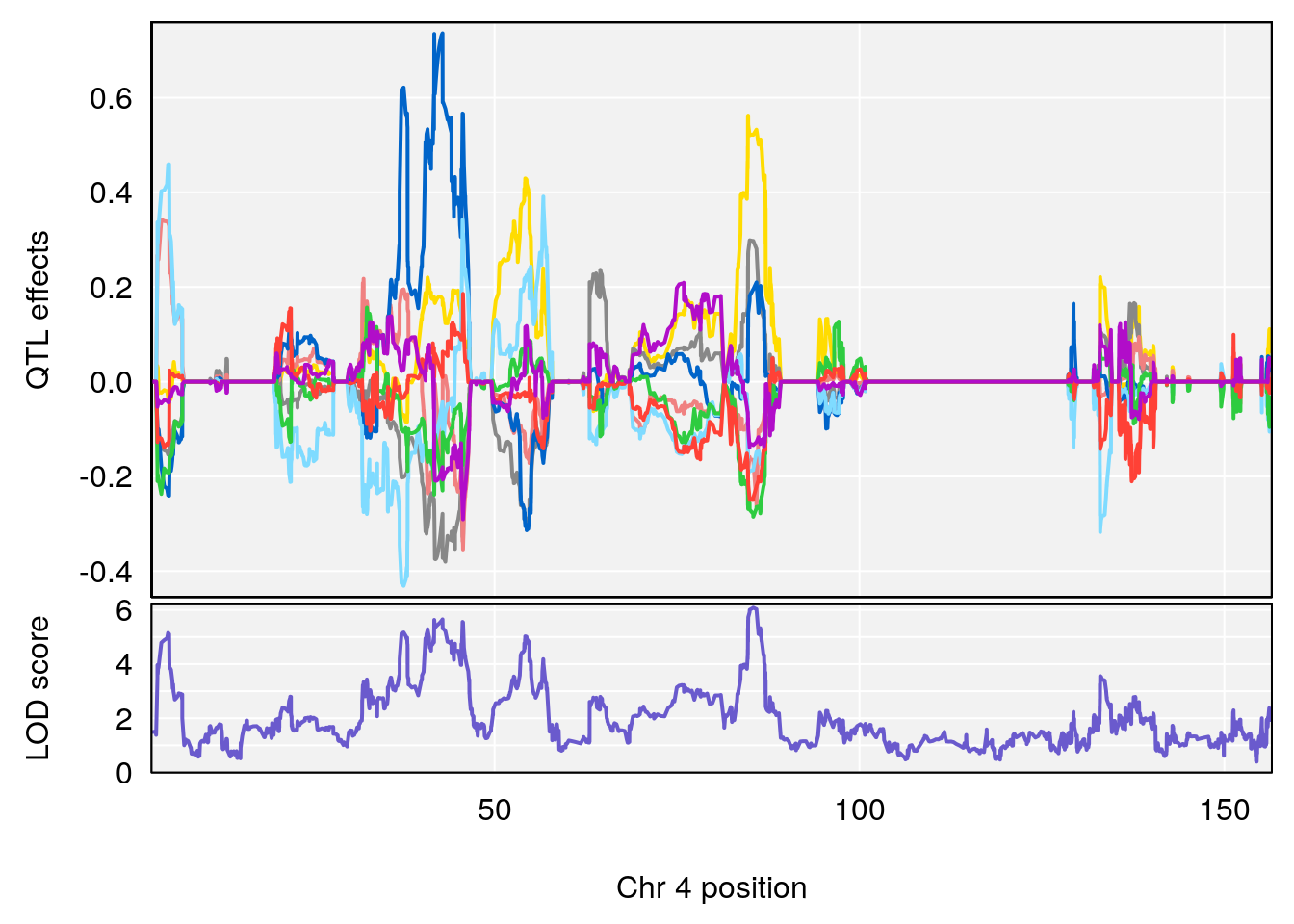
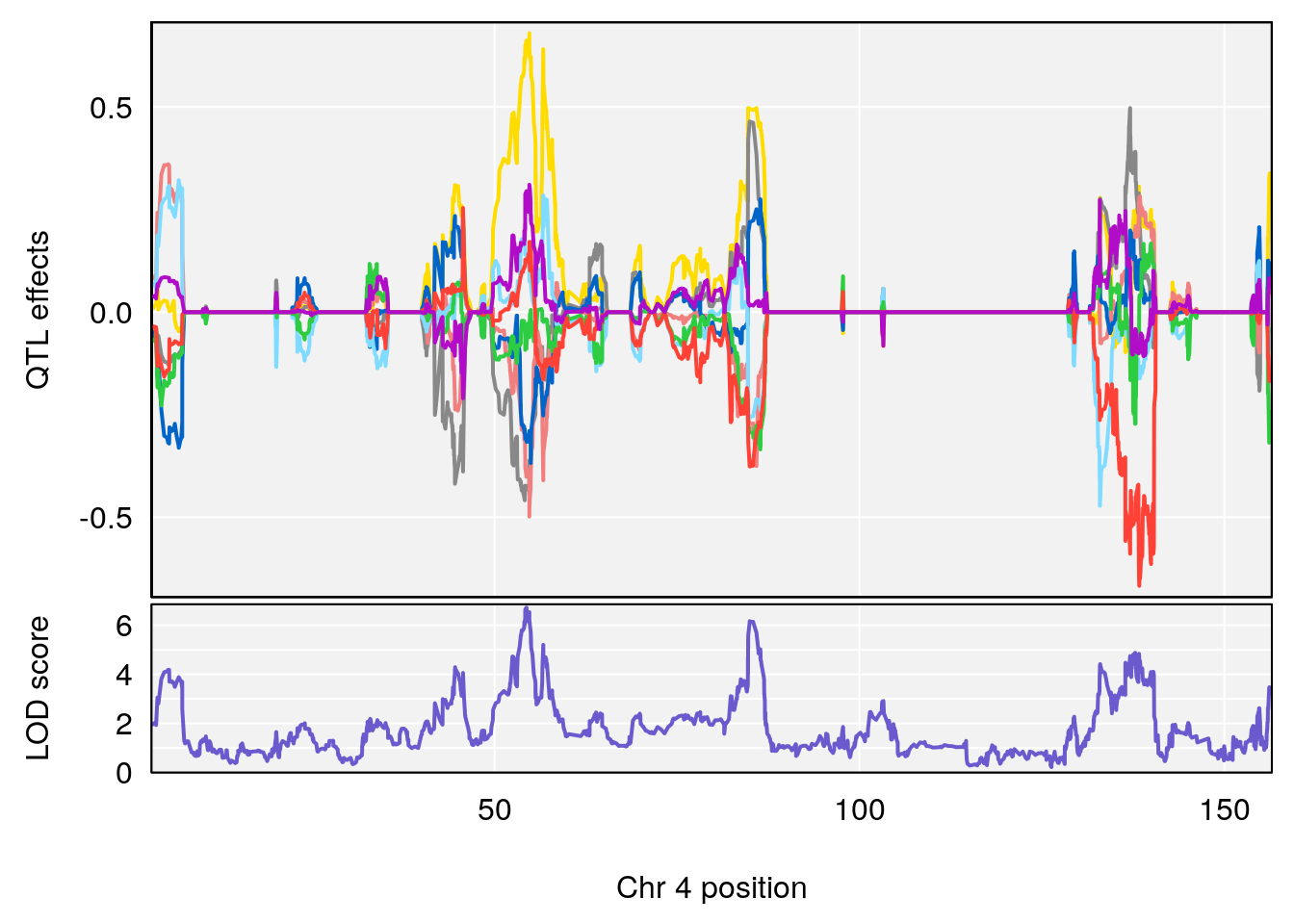
# 4 1 pheno1 4 85.44520 6.082516 3.945062 86.84903
# 5 1 pheno1 10 117.29331 6.097238 116.264165 118.73253
# 6 1 pheno1 17 82.91709 6.294987 81.121859 83.40925
# [1] 1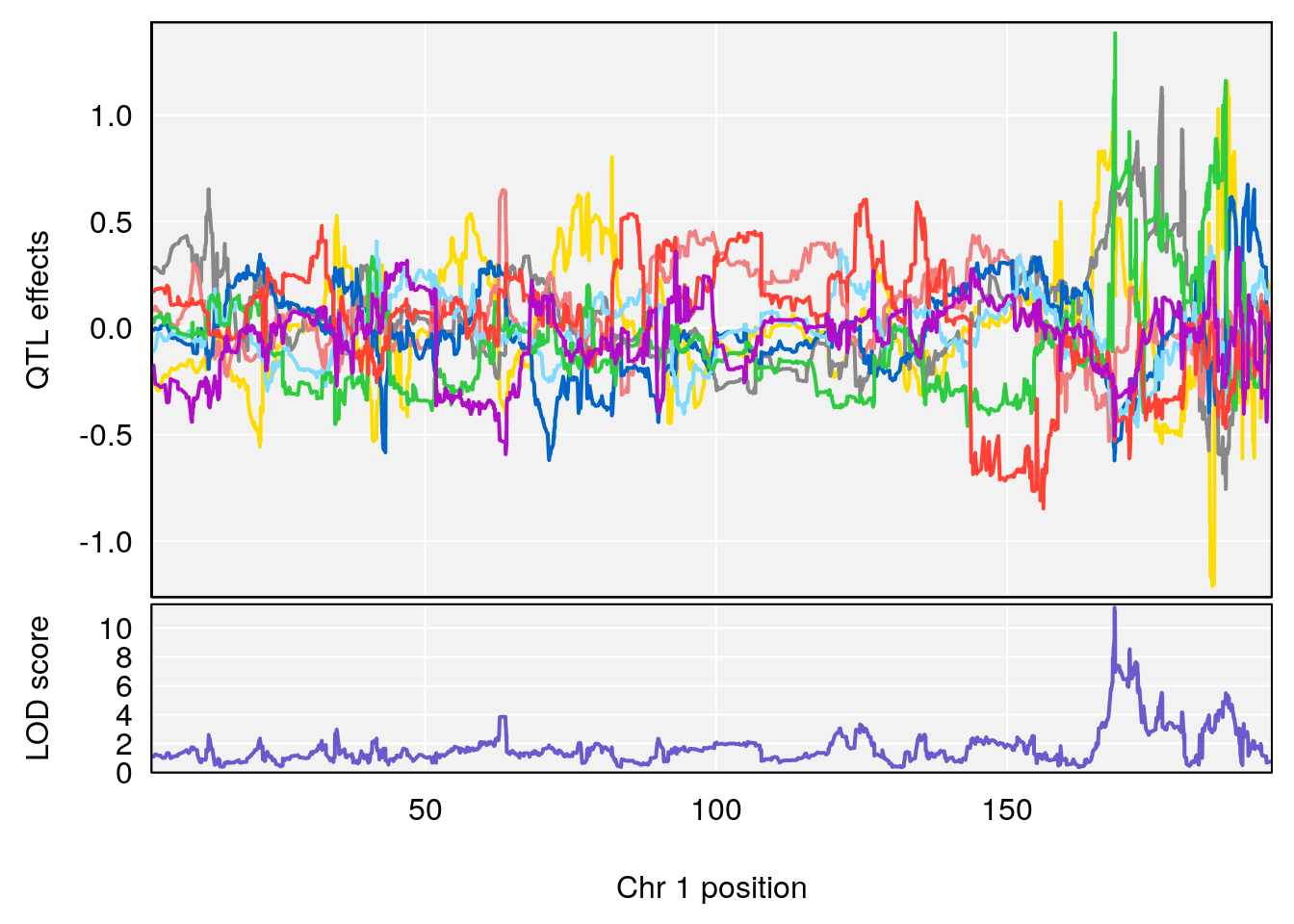
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 2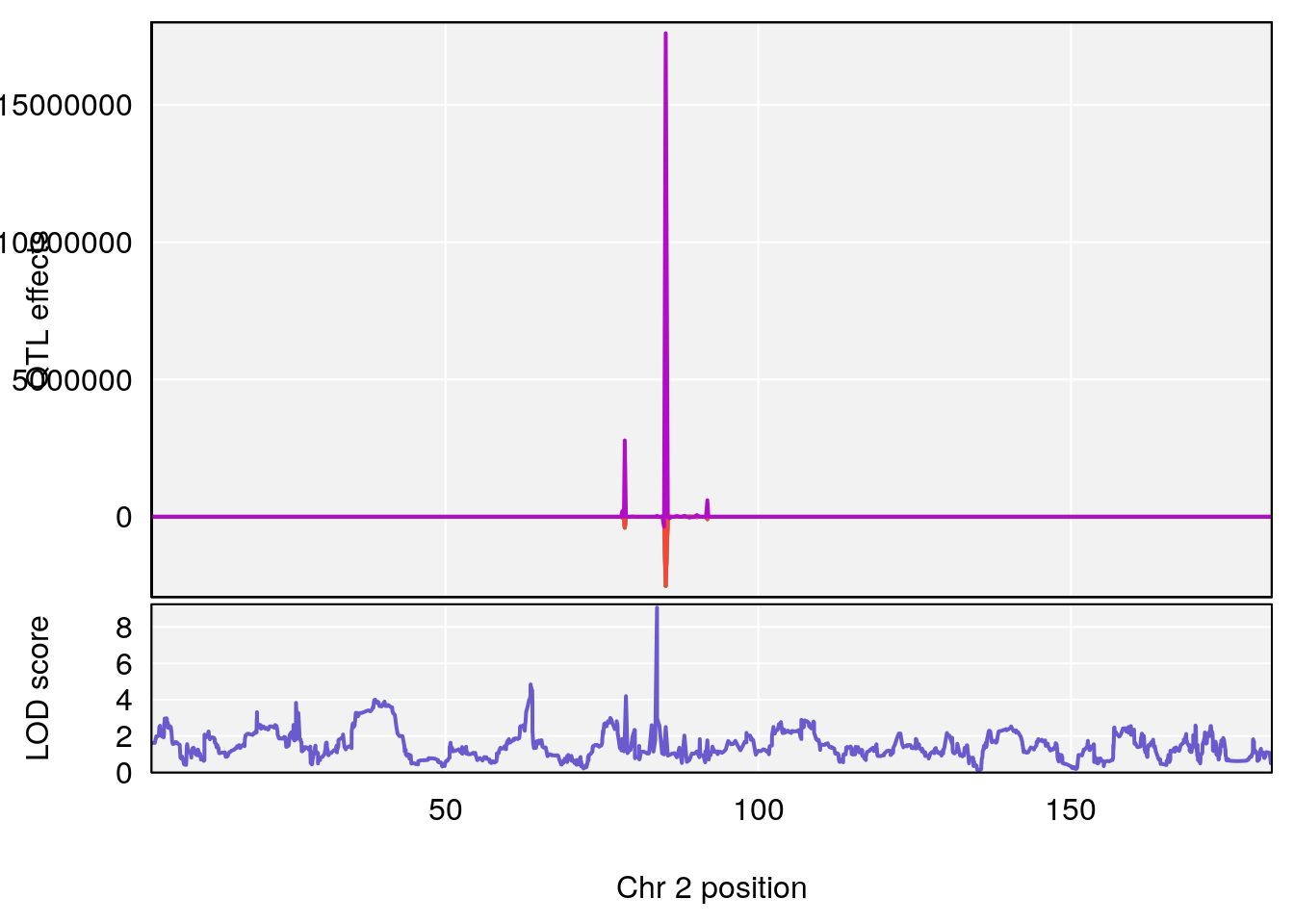
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
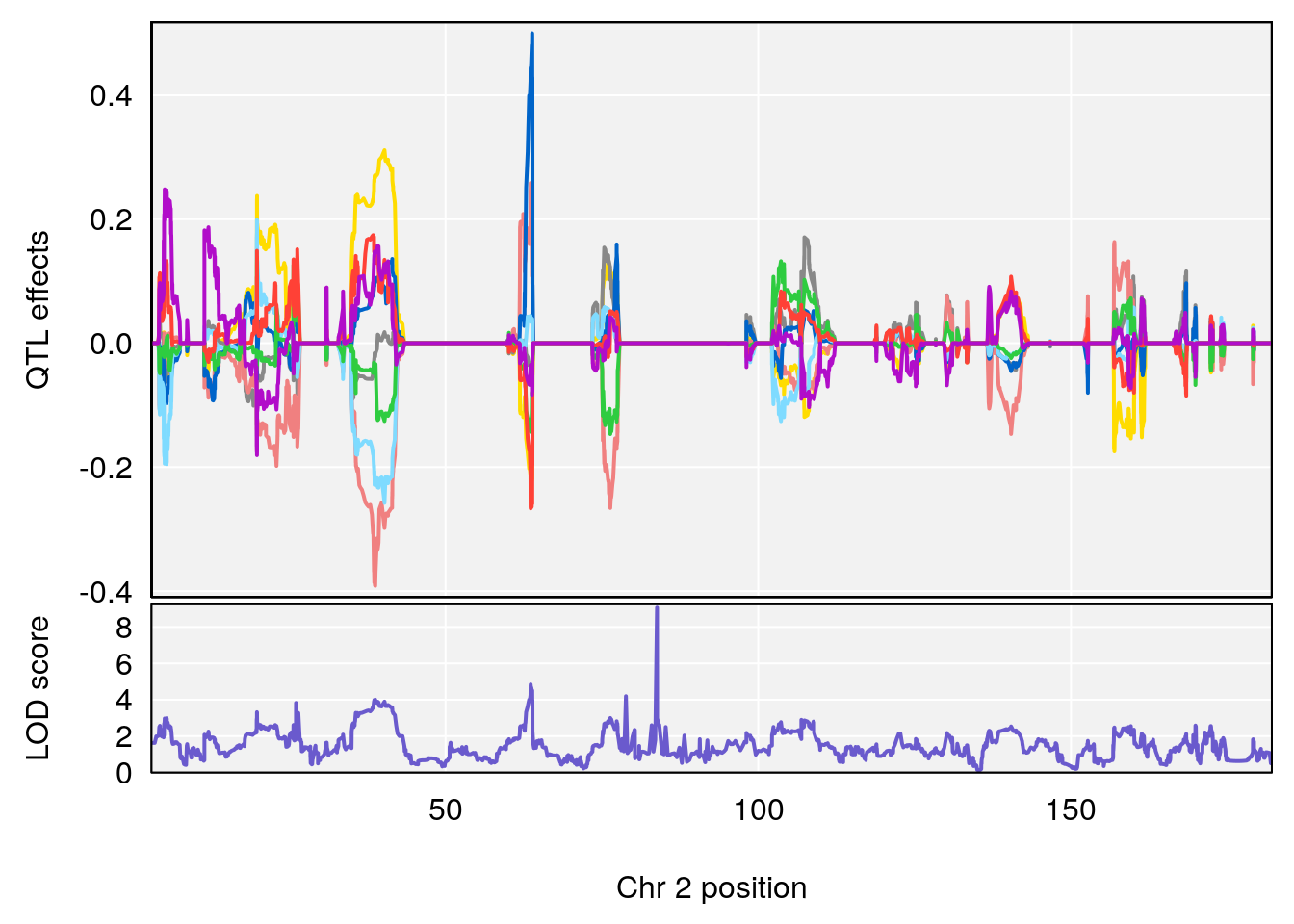
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 3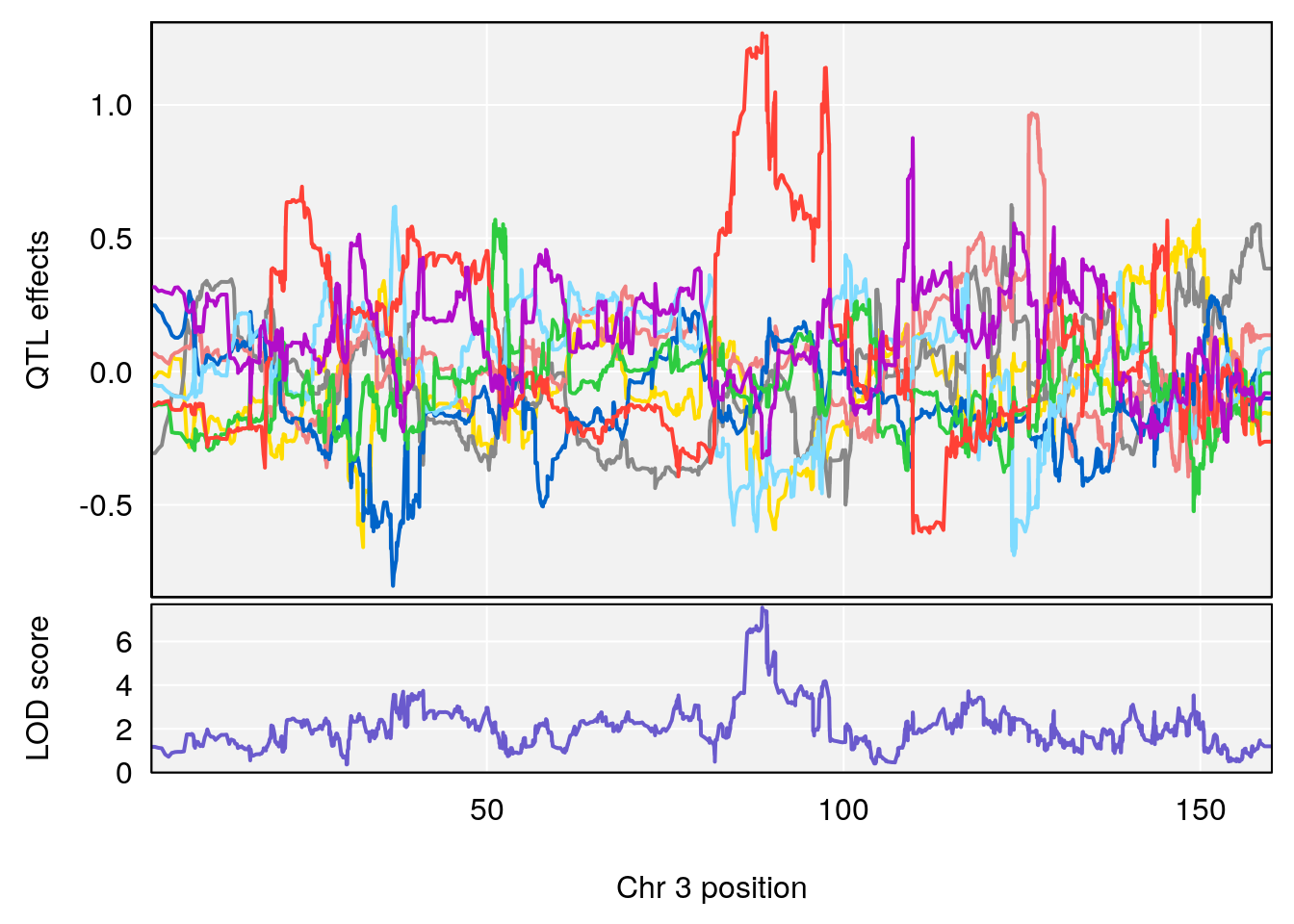
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 4
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 5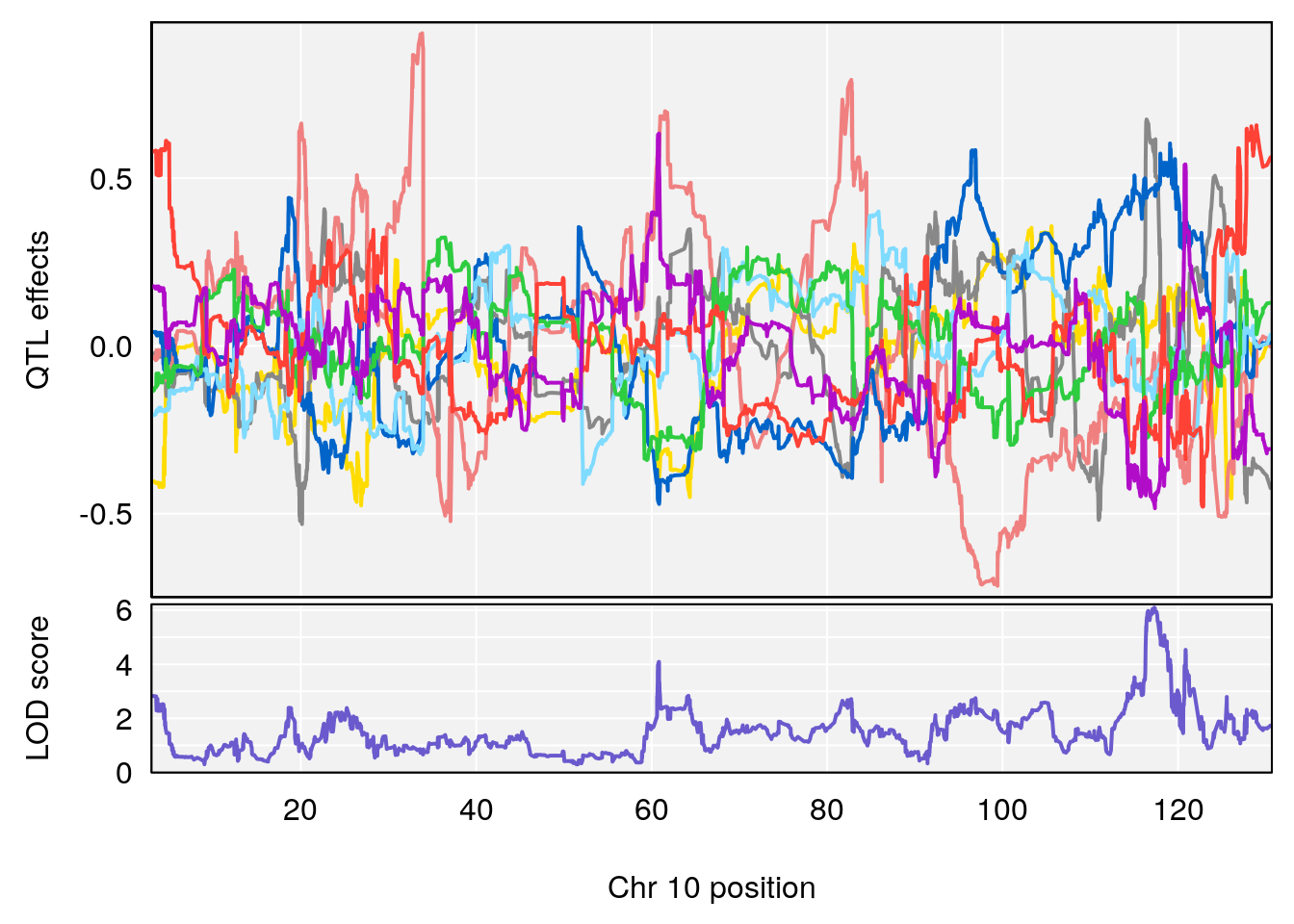
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
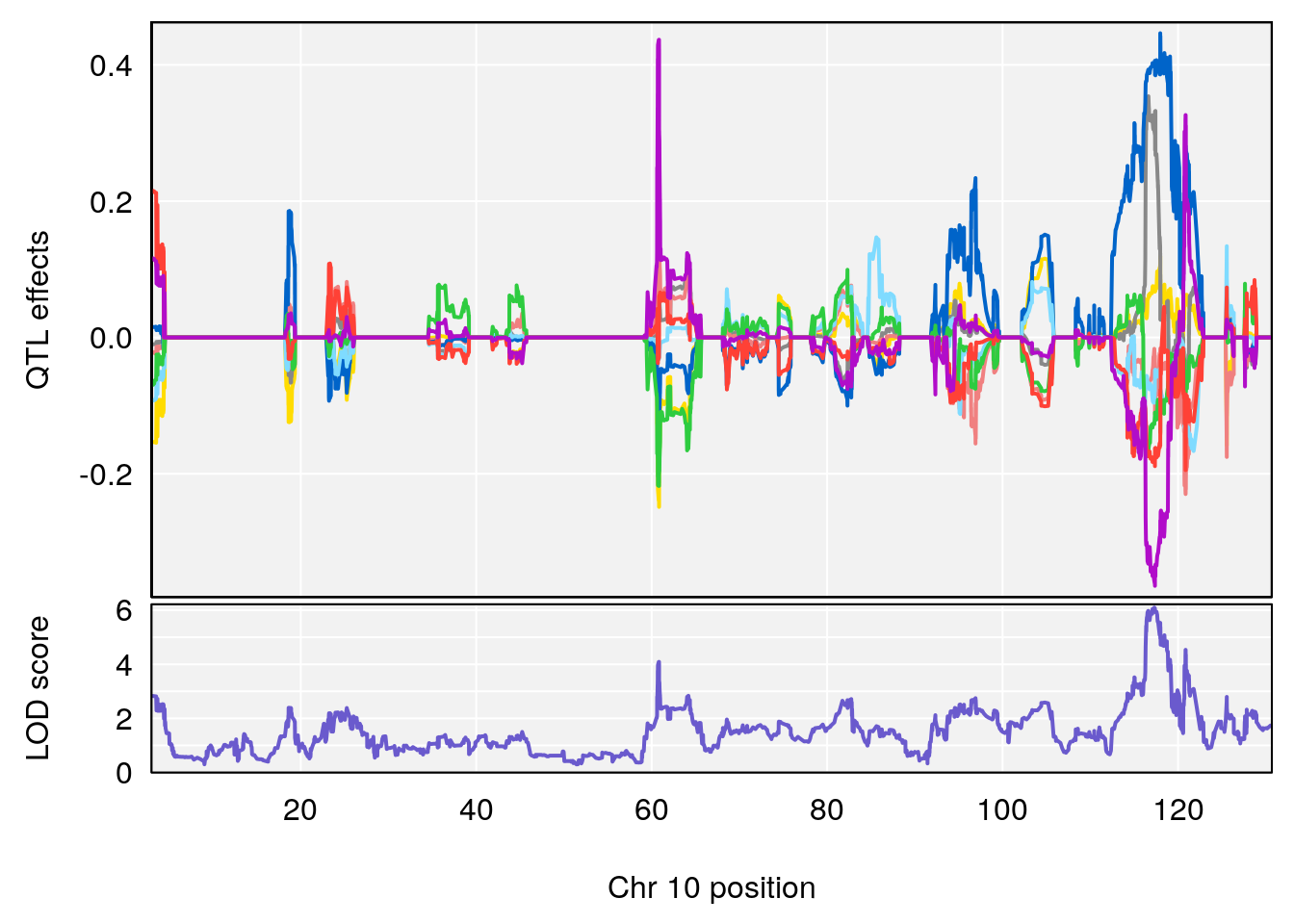
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 6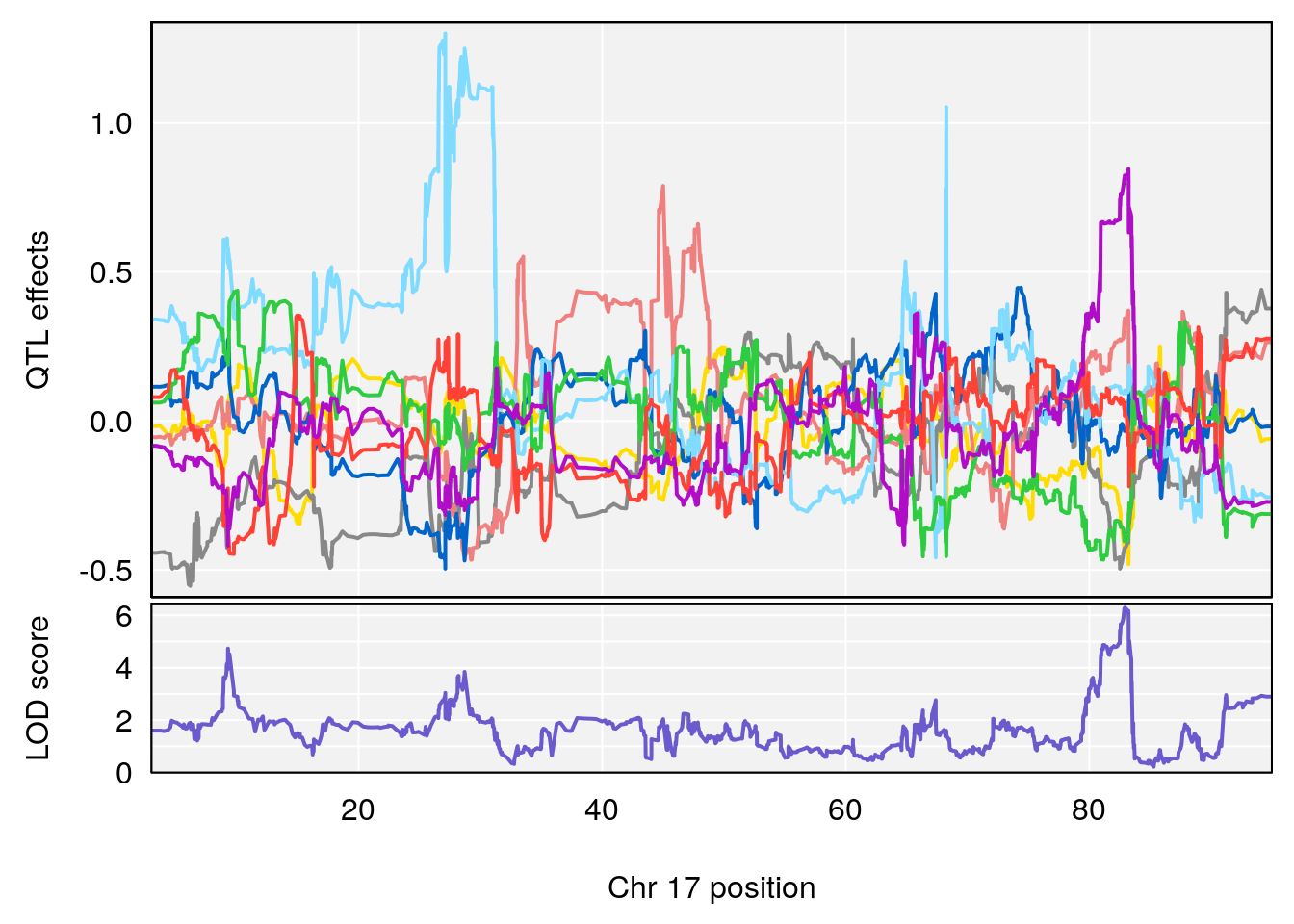
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
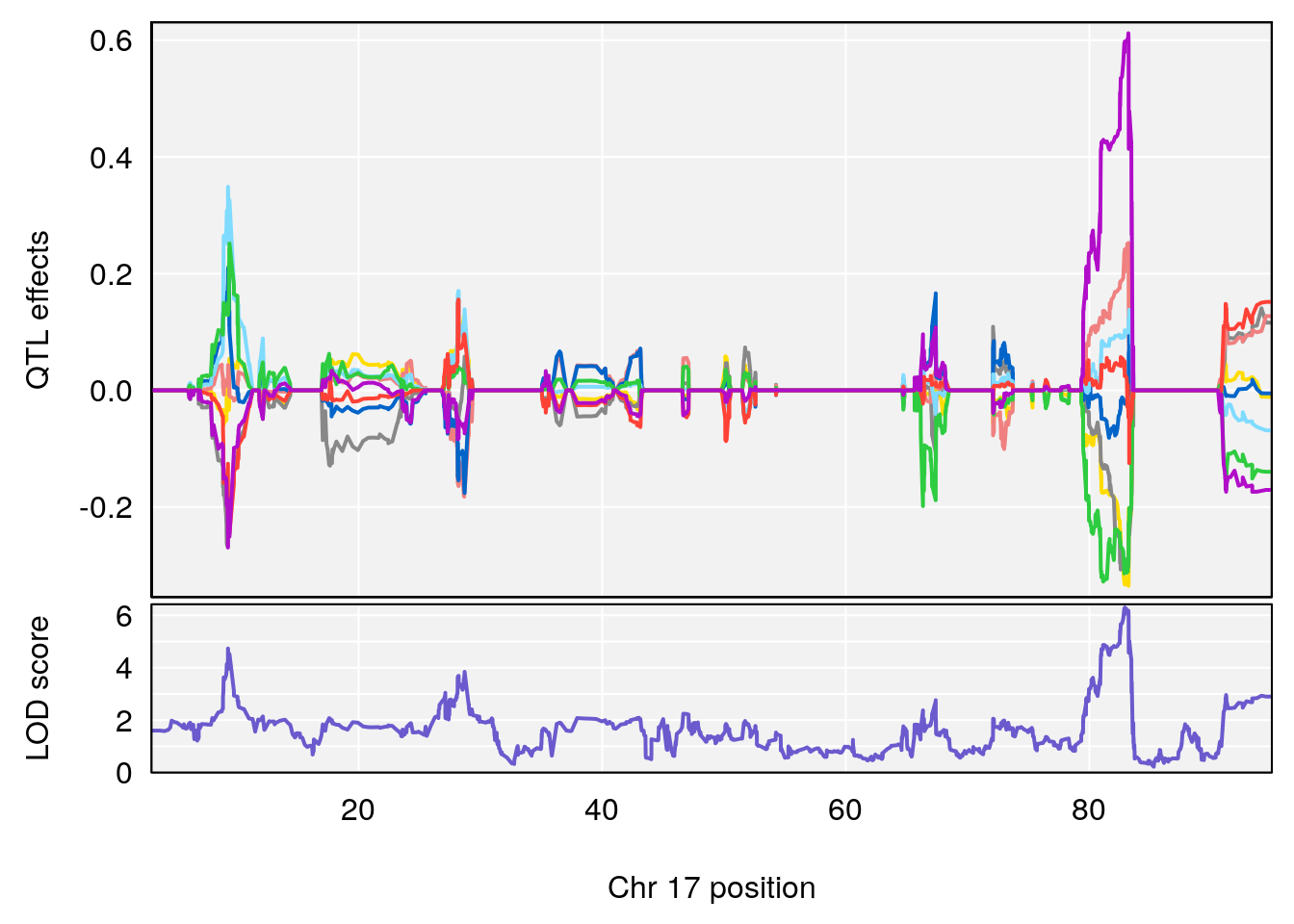
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
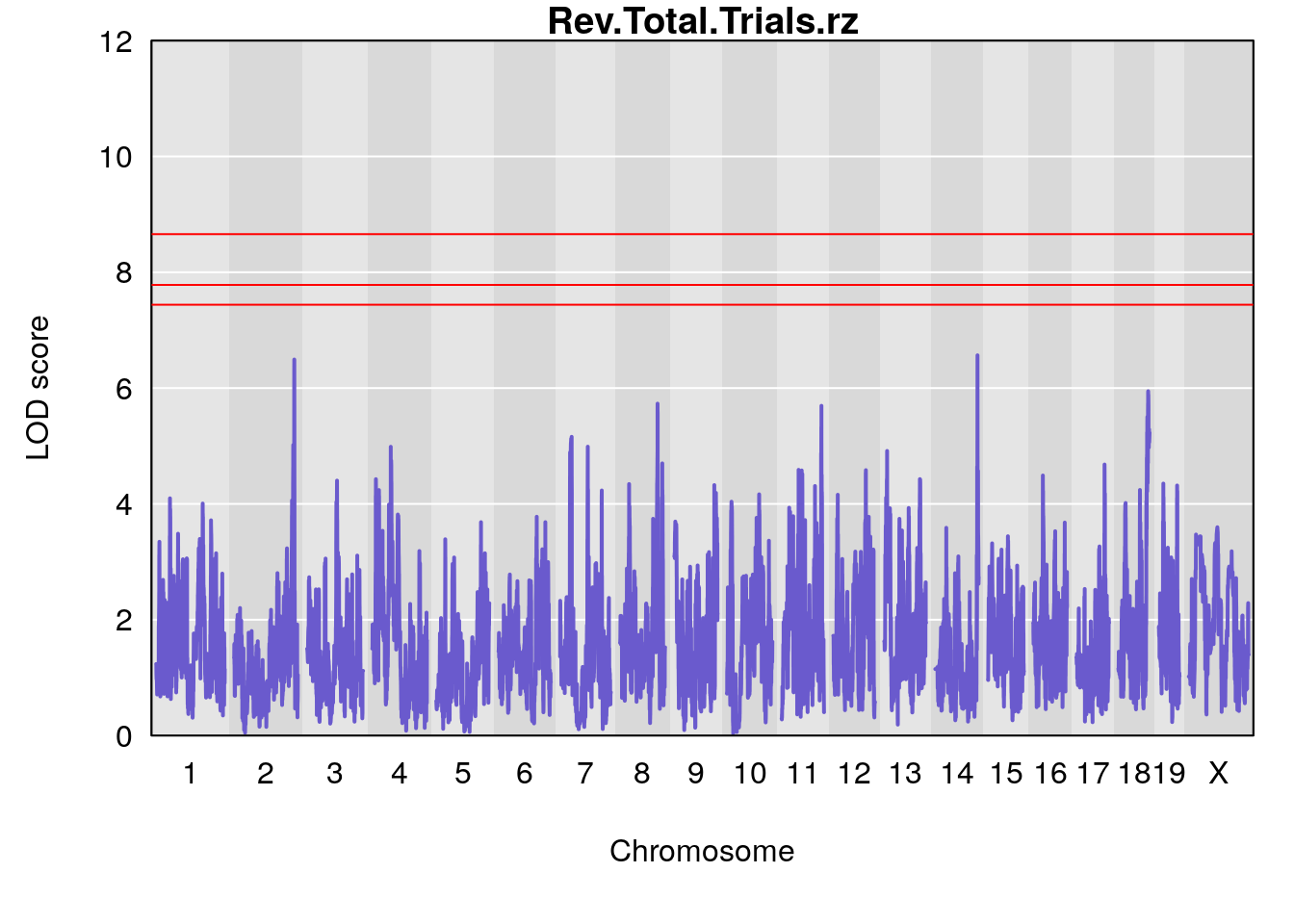
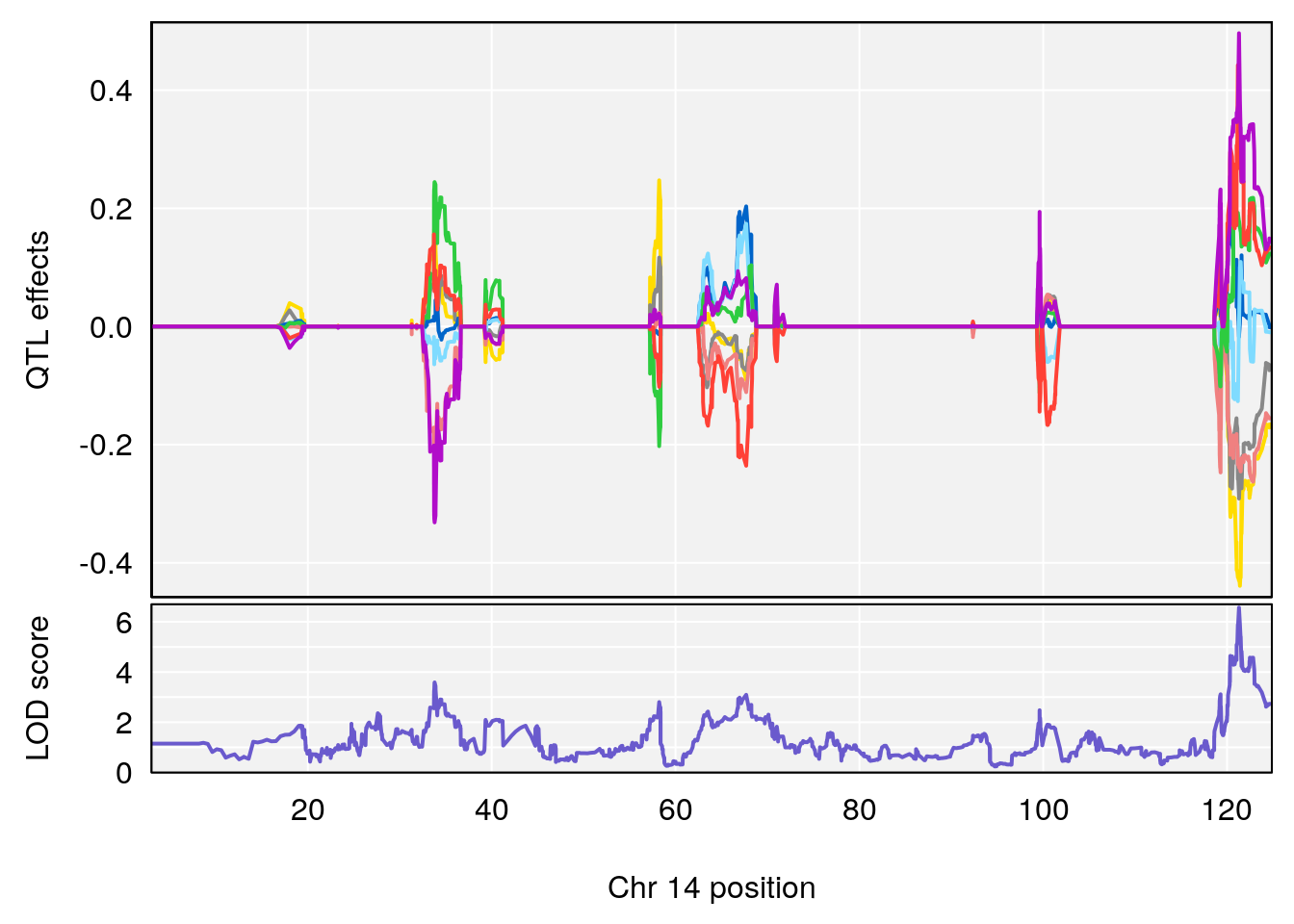
# [1] "Rev.Total.Trials"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 14 121.29650 6.542708 121.11874 121.5717
# 2 1 pheno1 18 83.08088 6.553229 82.31526 90.6726
# [1] 1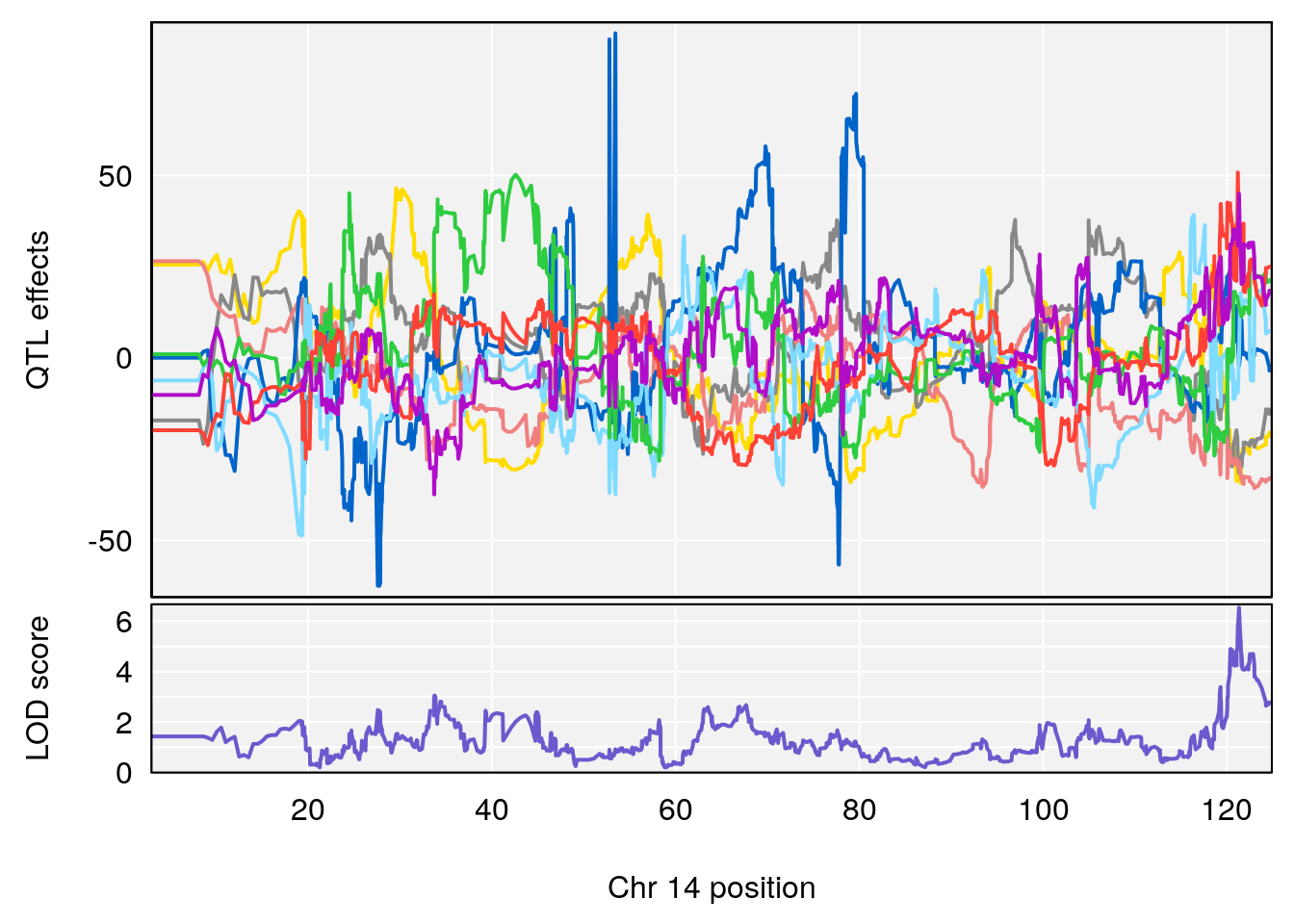
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
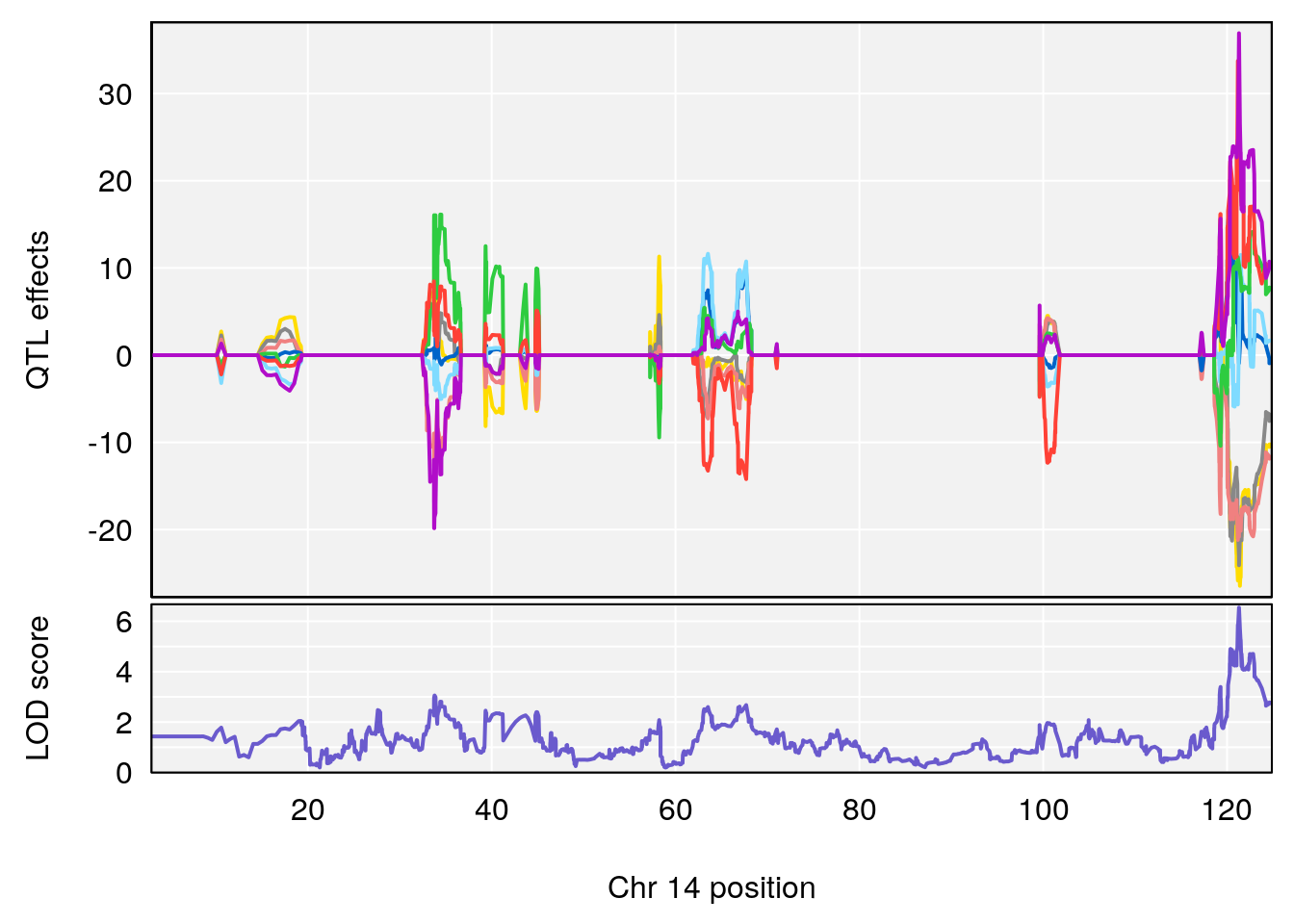
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 2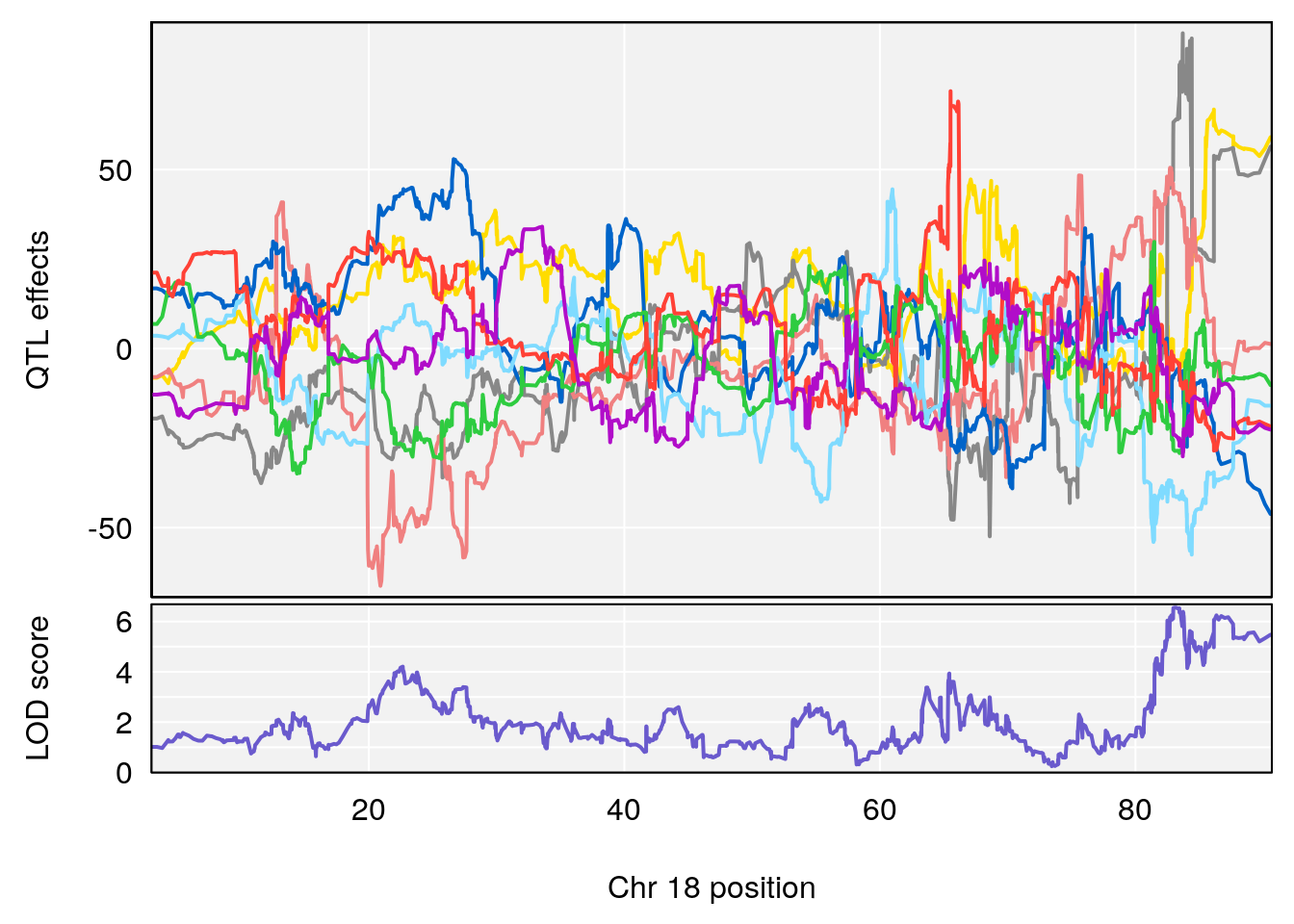
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
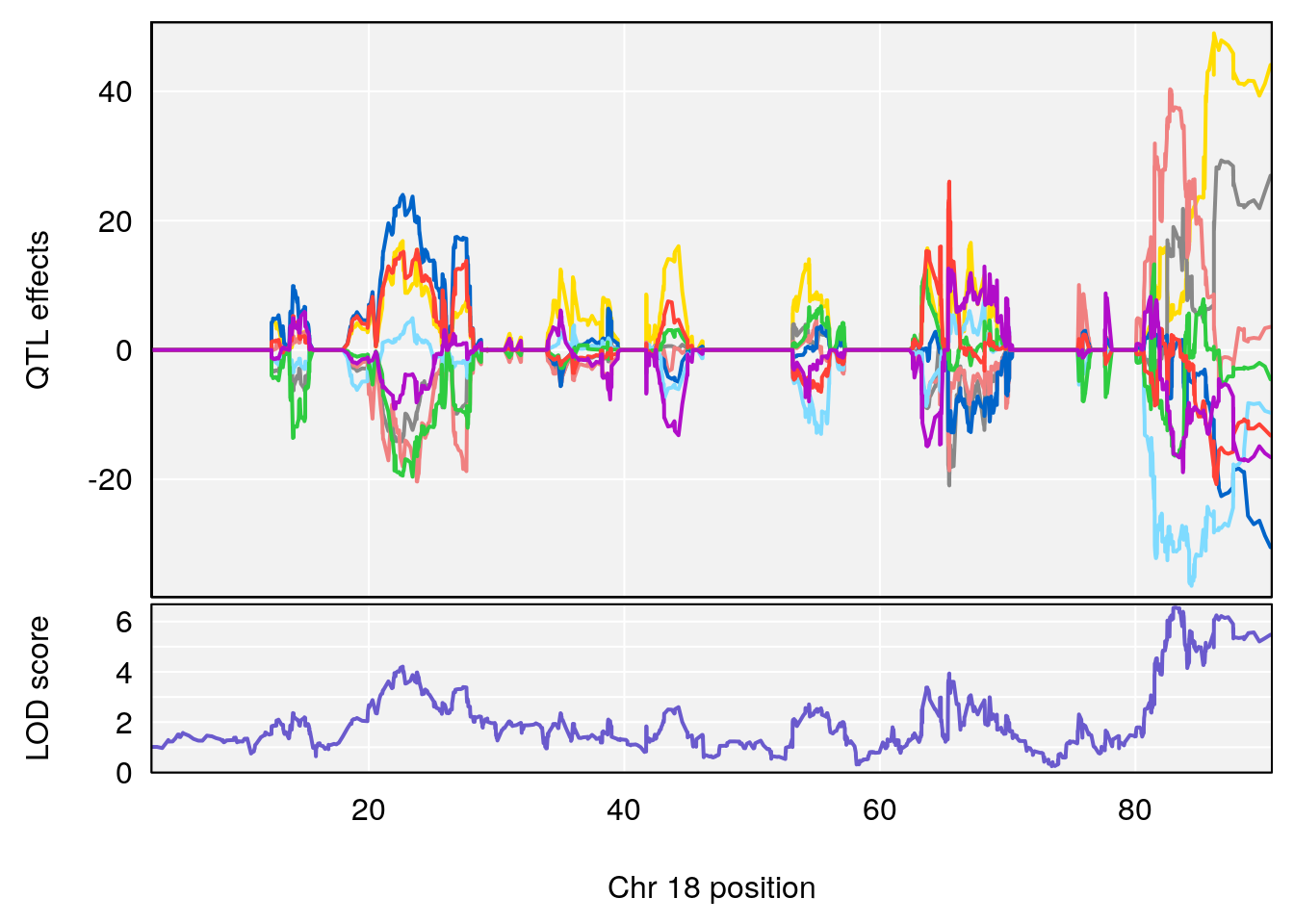
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
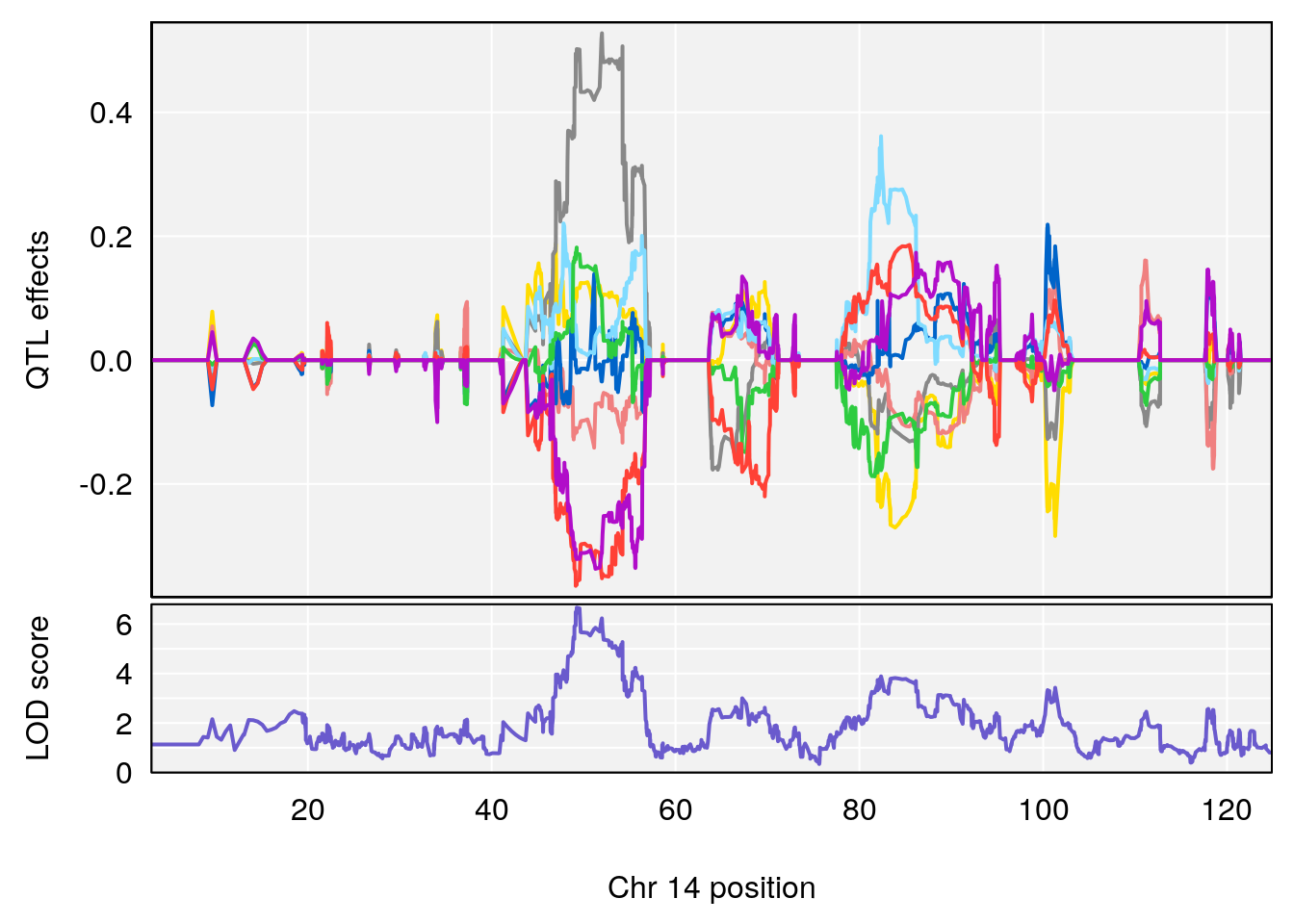
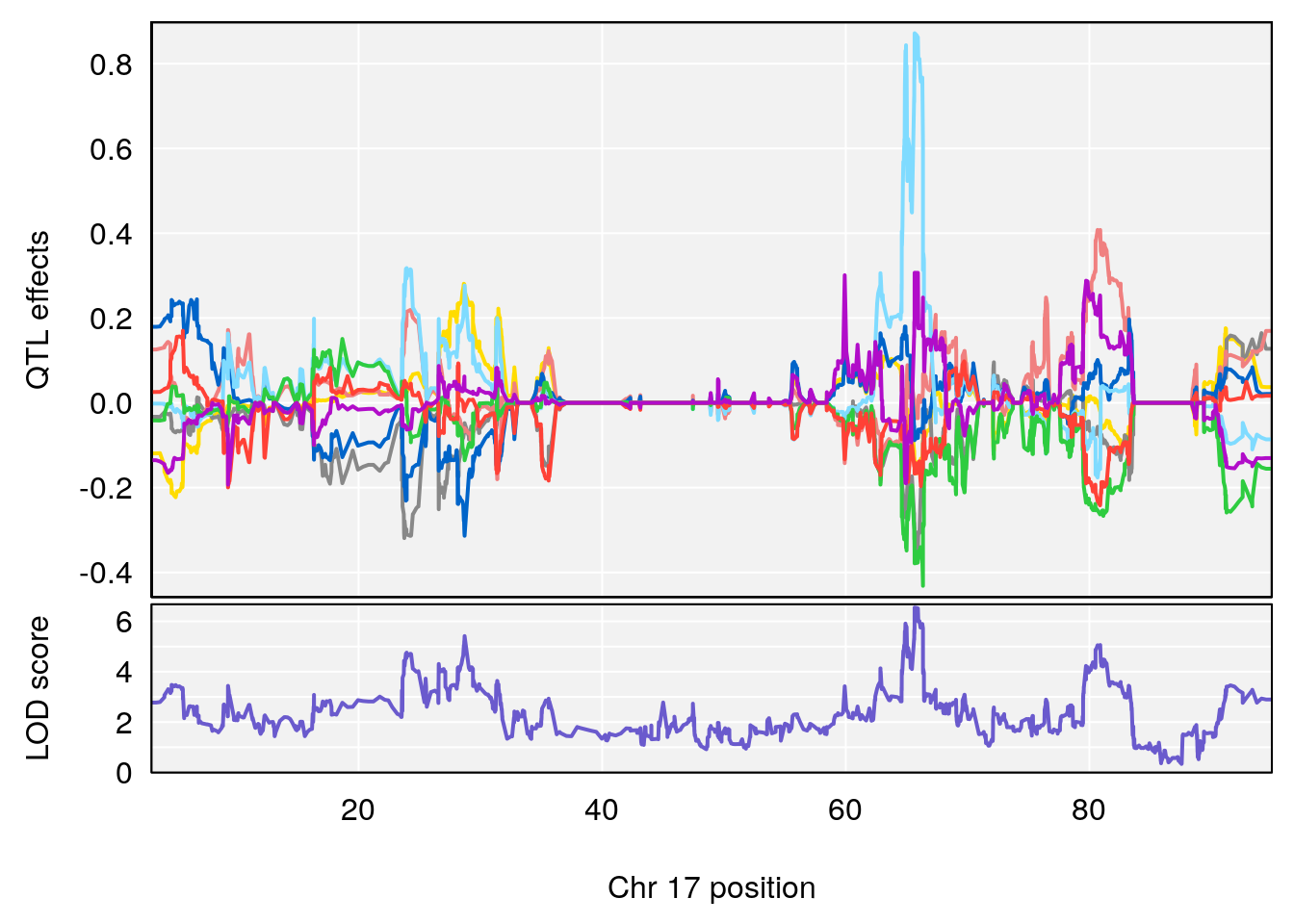
# [1] "Rev.Anticipatory.Incorrect.Responses"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 170.96387 6.571598 168.25531 171.06618
# 2 1 pheno1 6 103.68554 7.392701 101.08006 103.77874
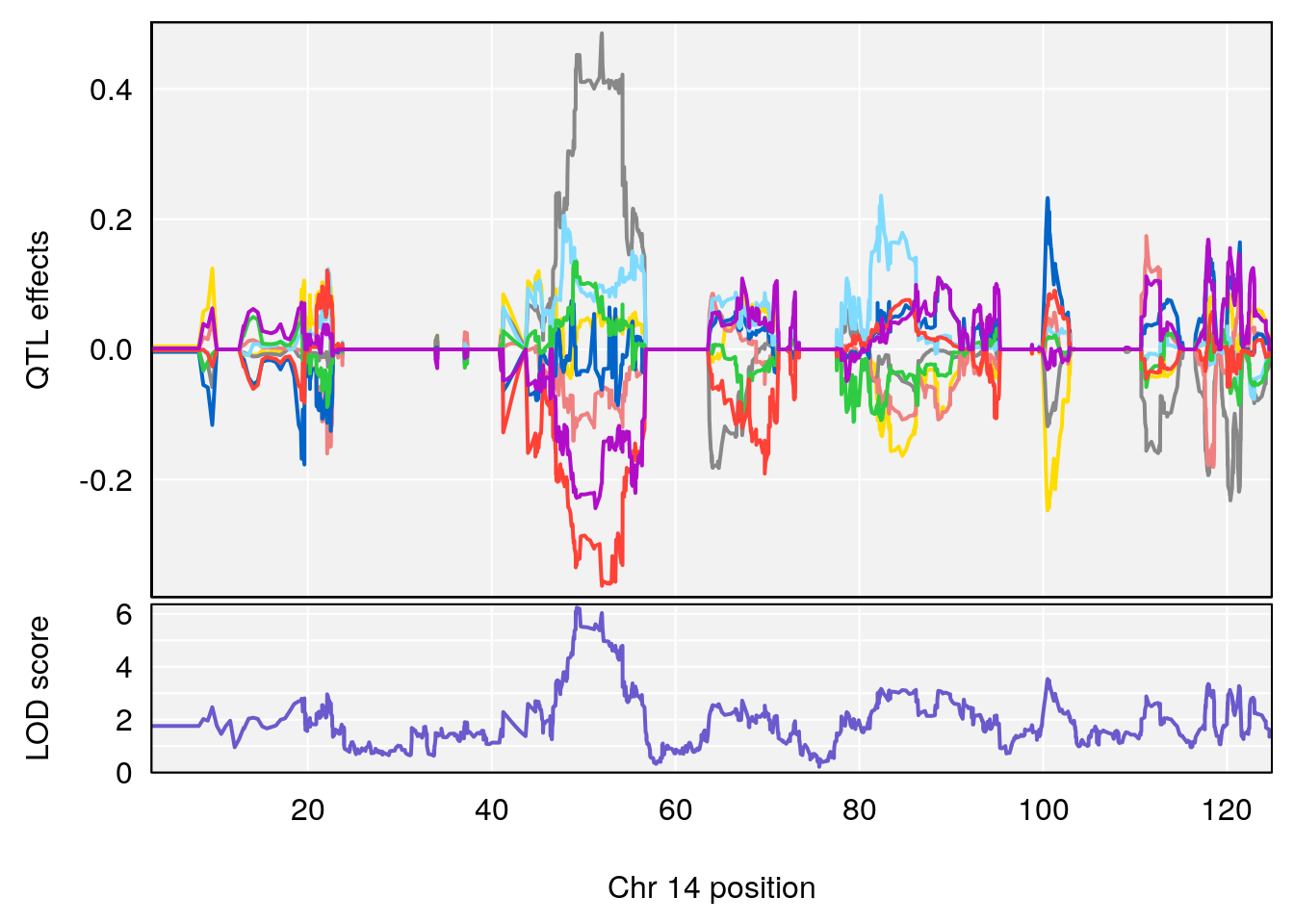
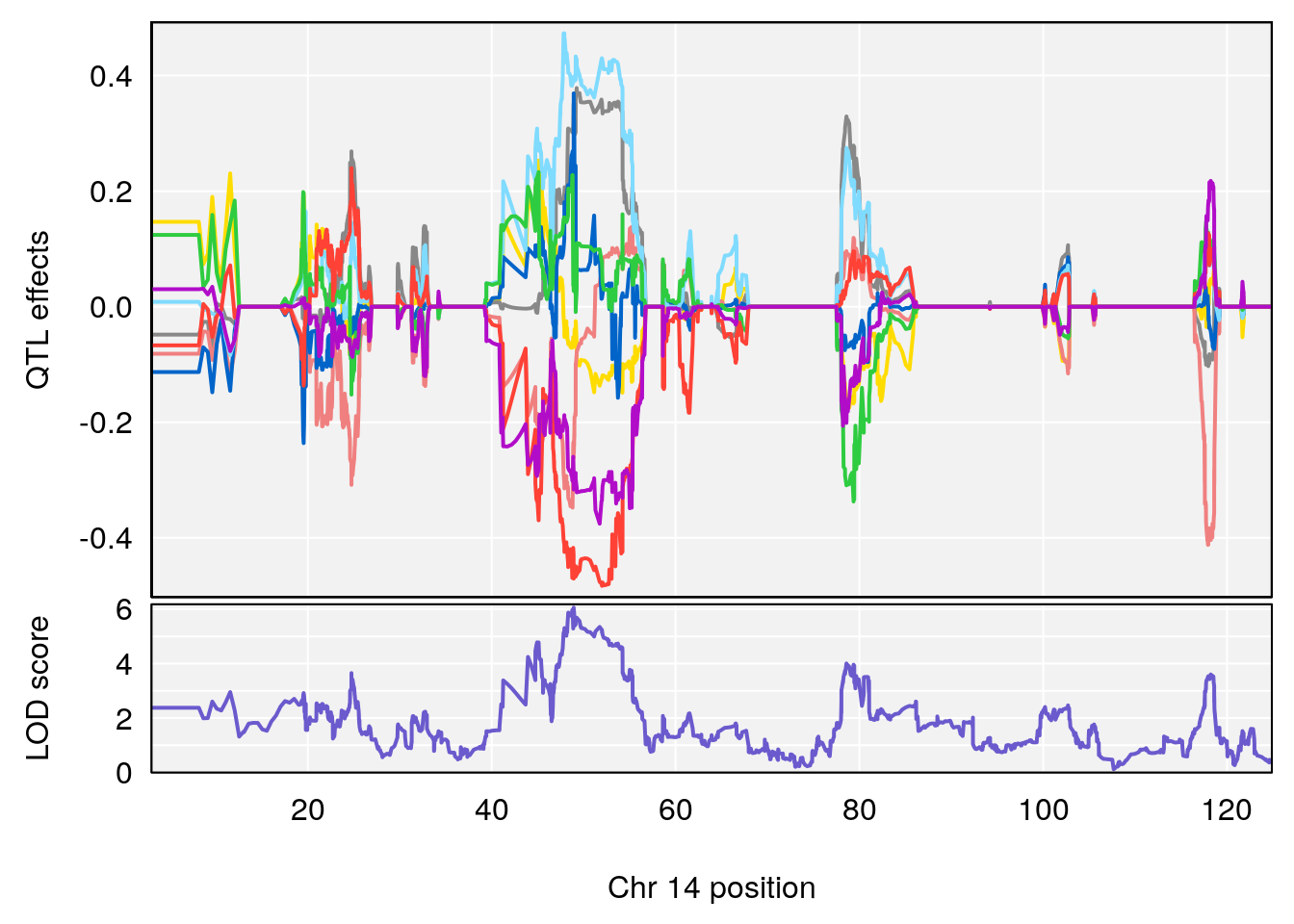
# 3 1 pheno1 14 49.25670 6.252322 48.80084 54.24423
# 4 1 pheno1 16 56.17223 7.087466 54.37418 58.59104
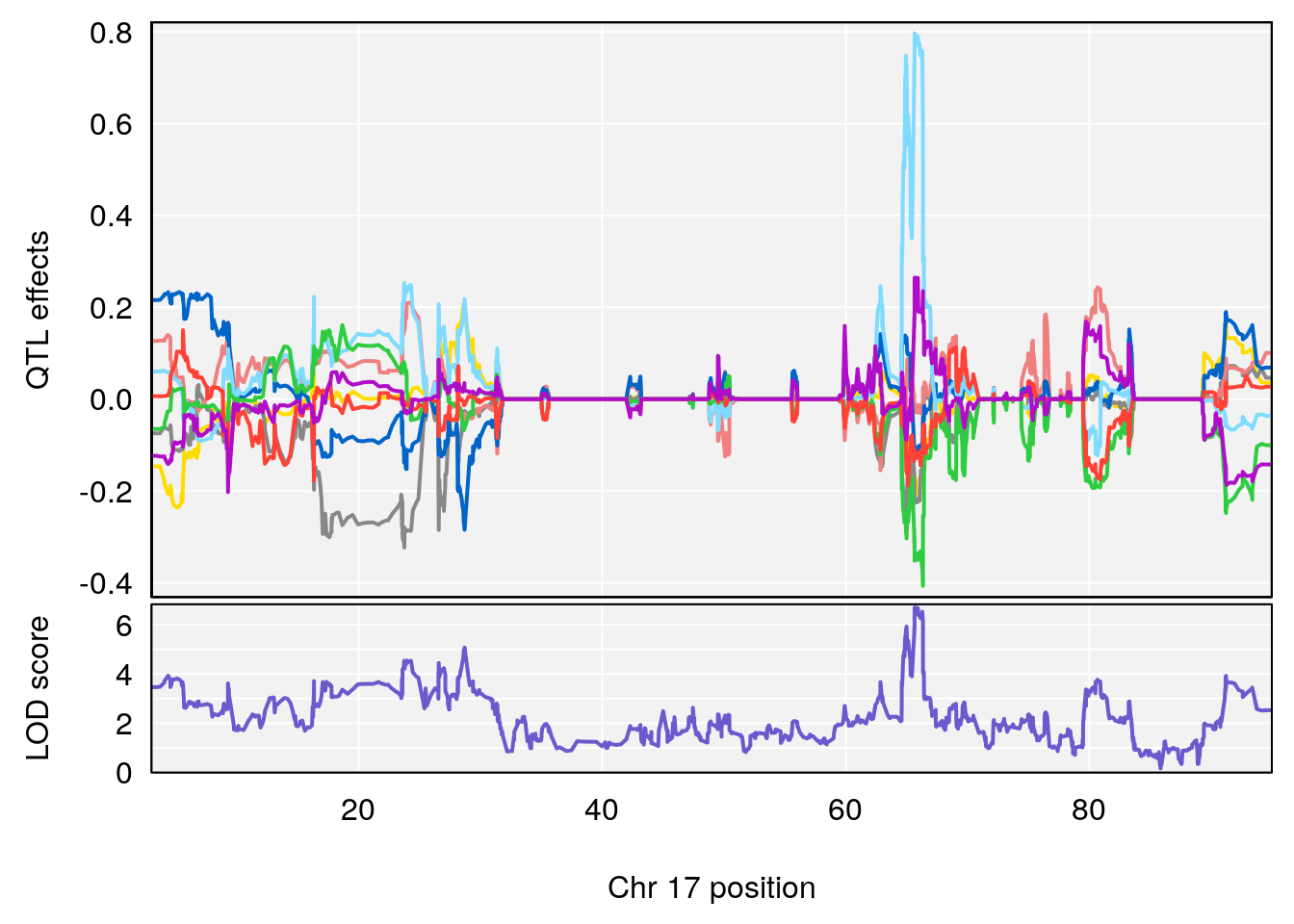
# 5 1 pheno1 17 65.65263 9.228873 64.83160 66.34240
# [1] 1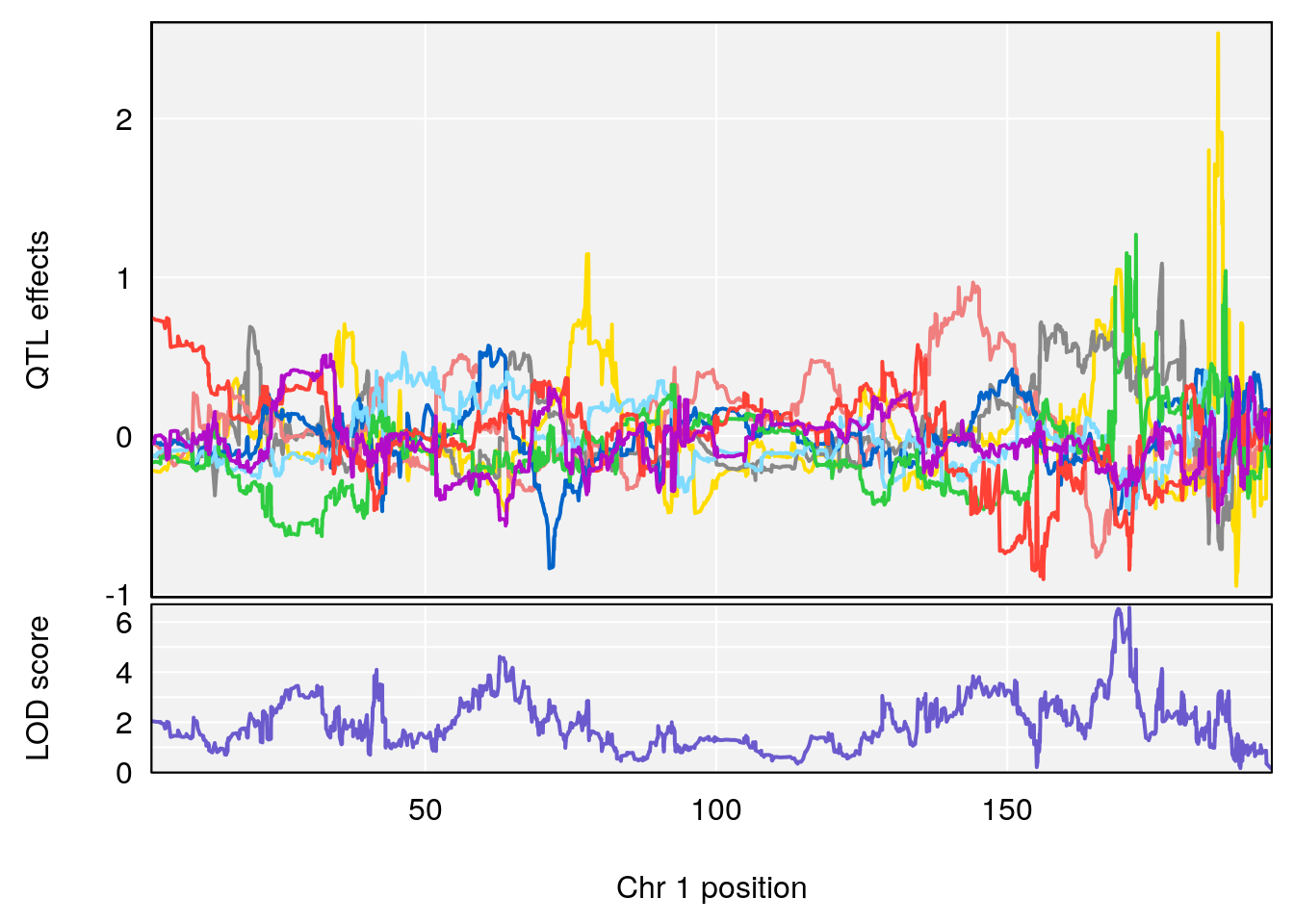
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
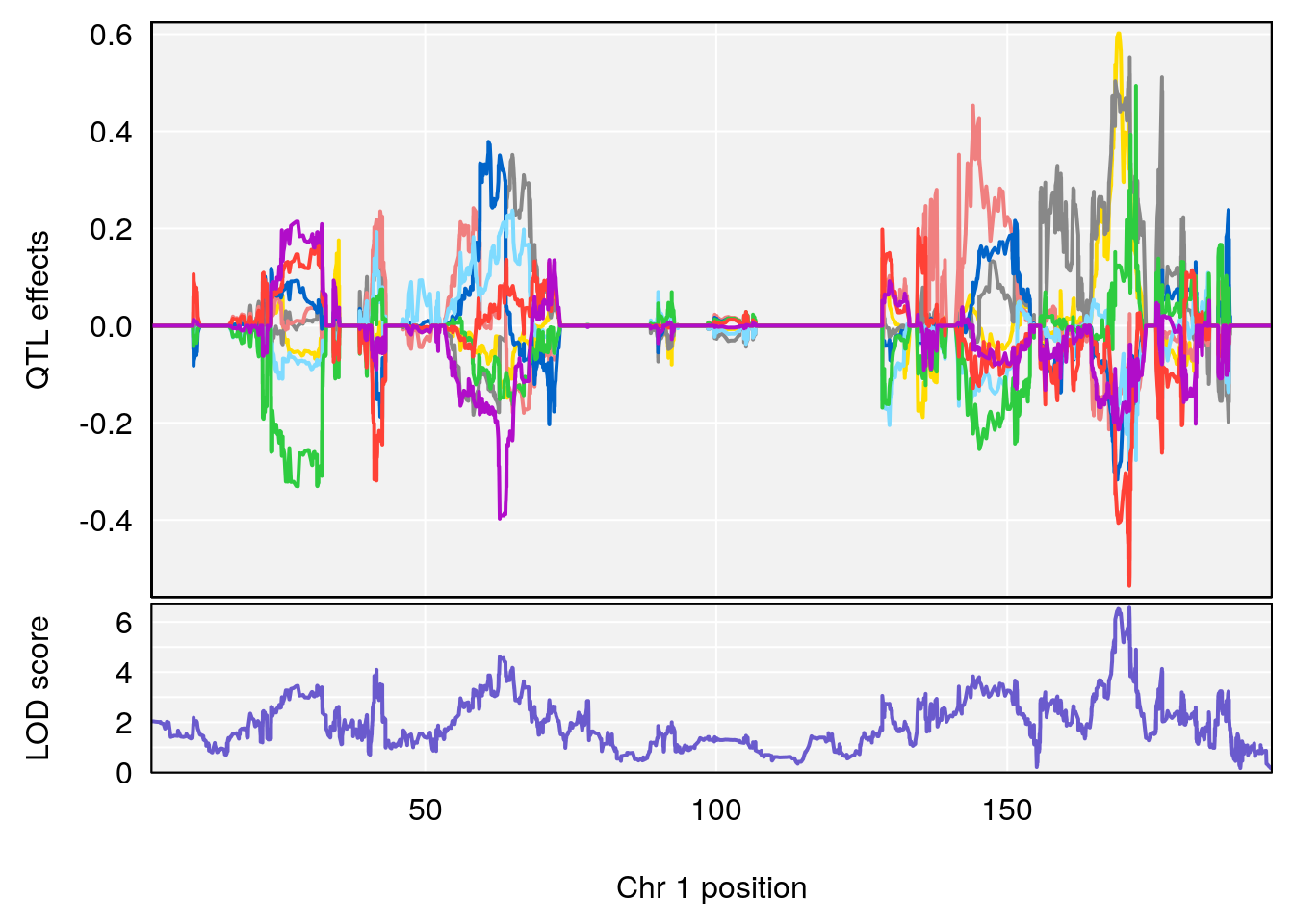
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 2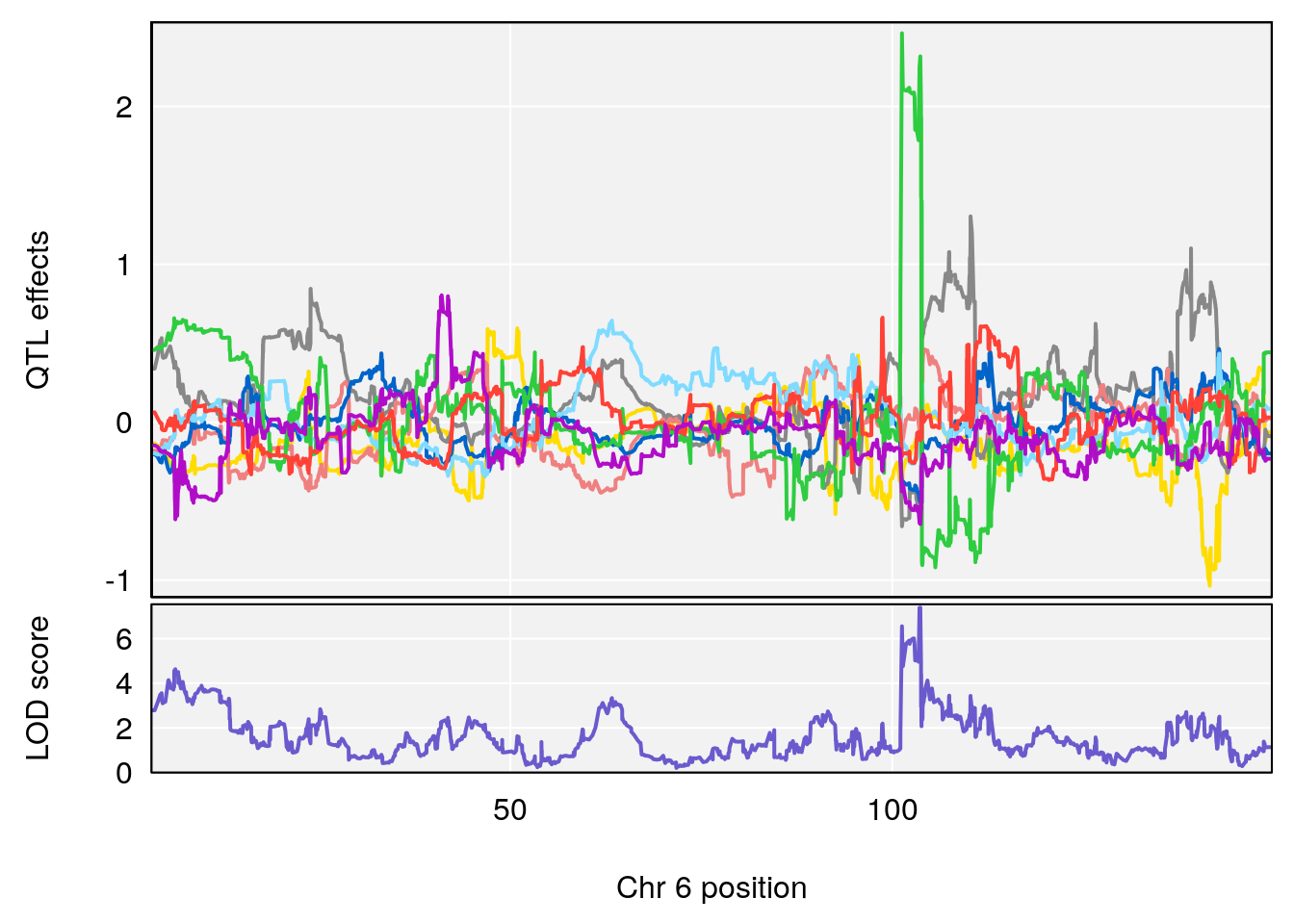
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
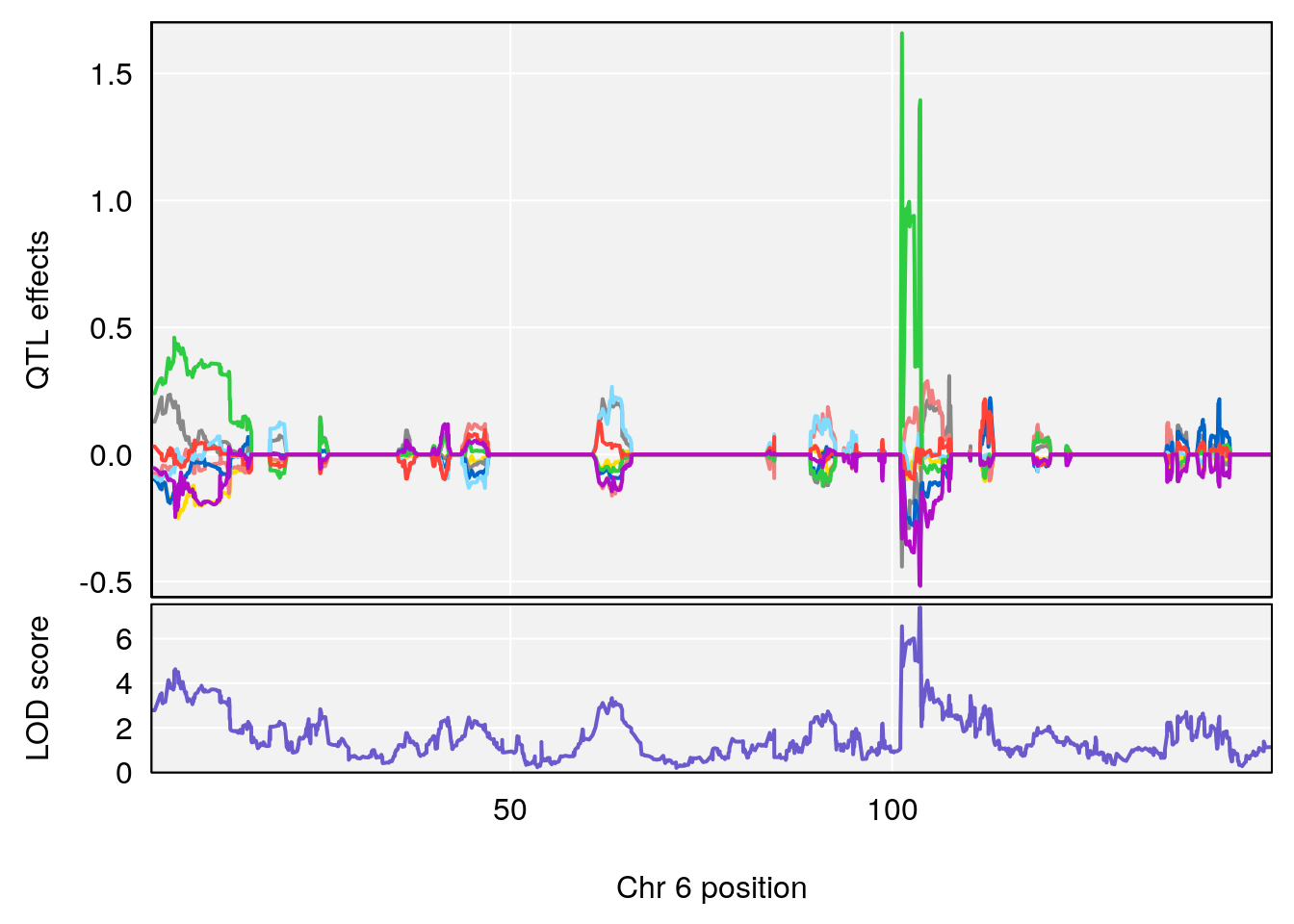
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 3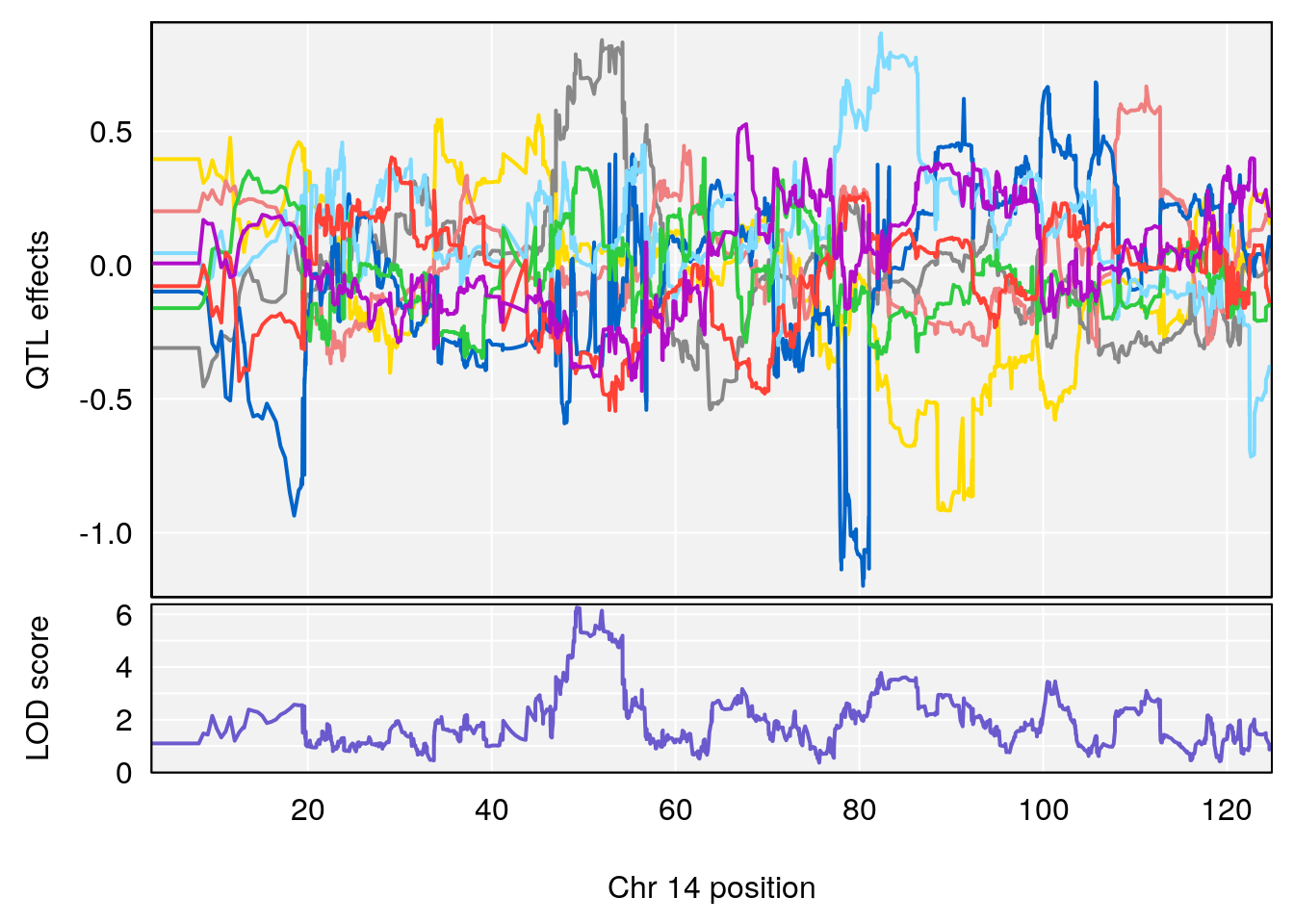
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
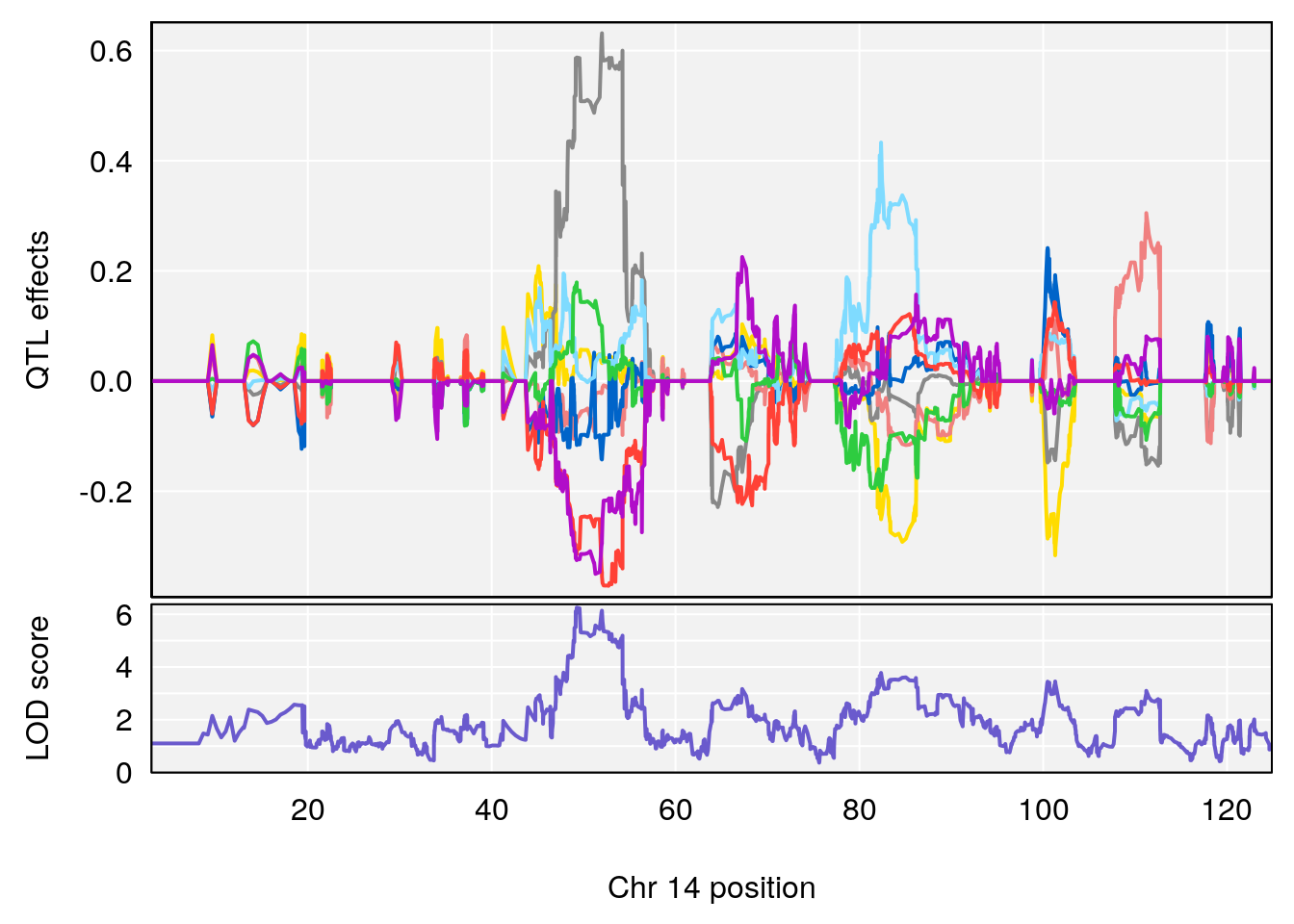
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 4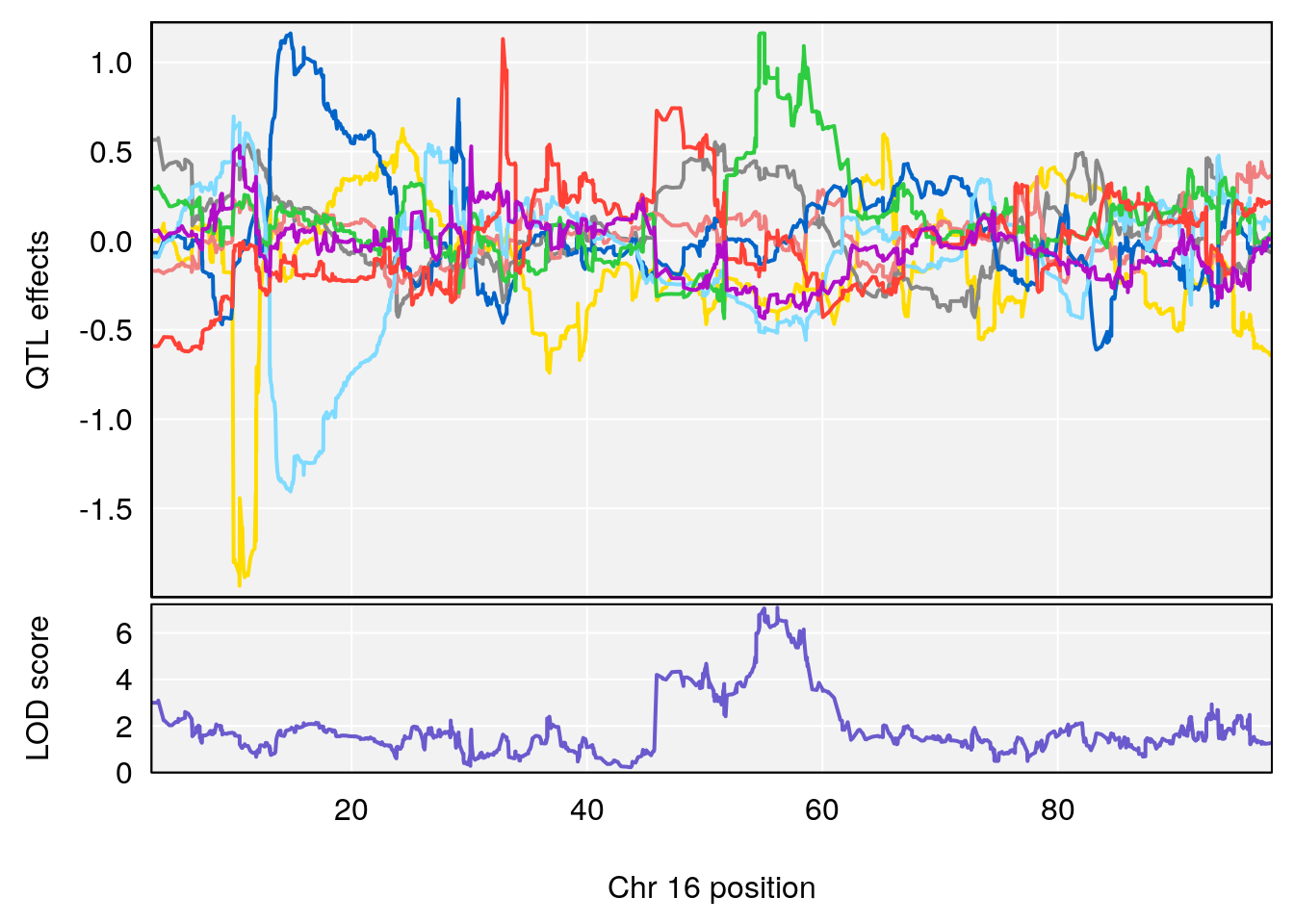
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
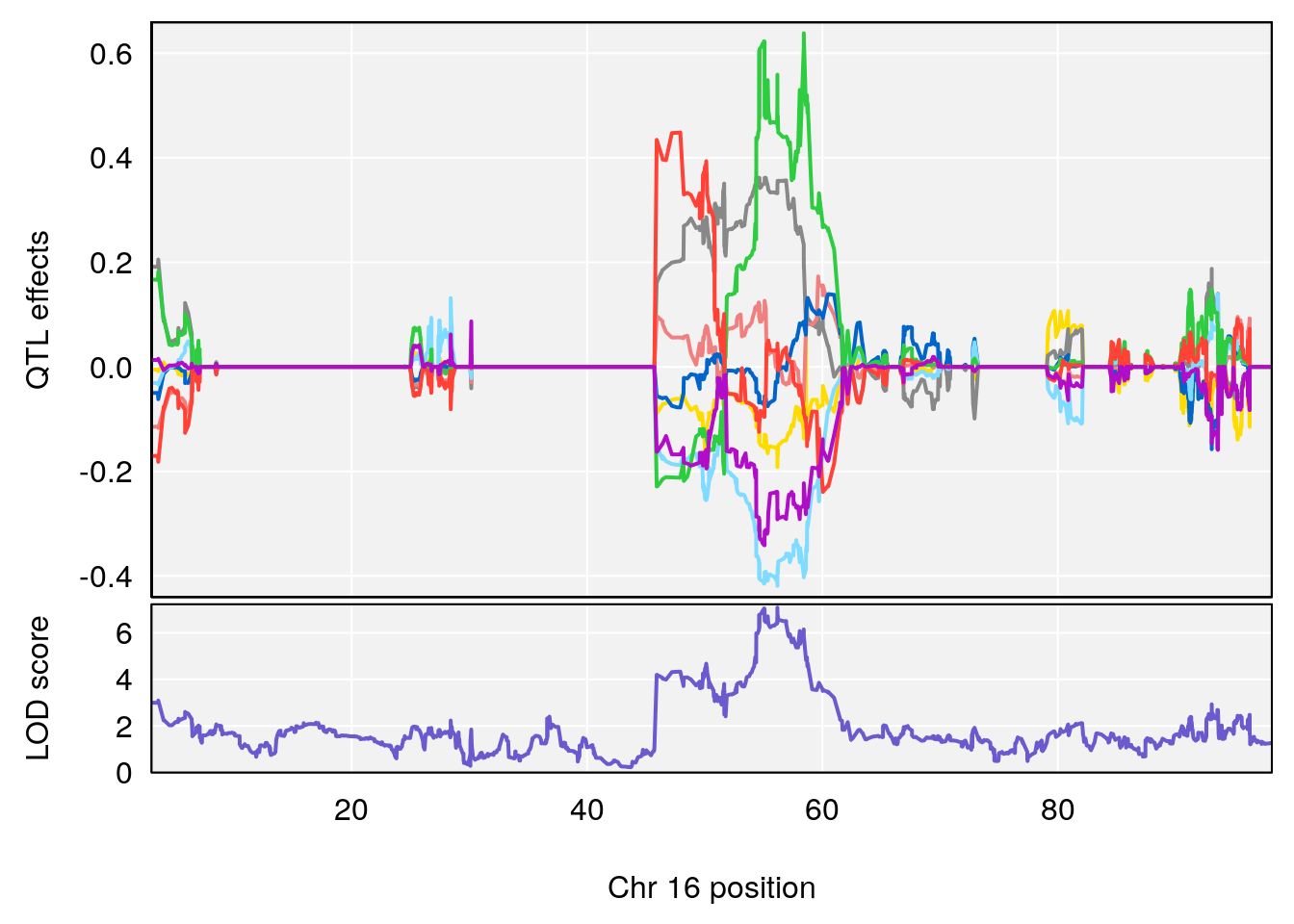
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 5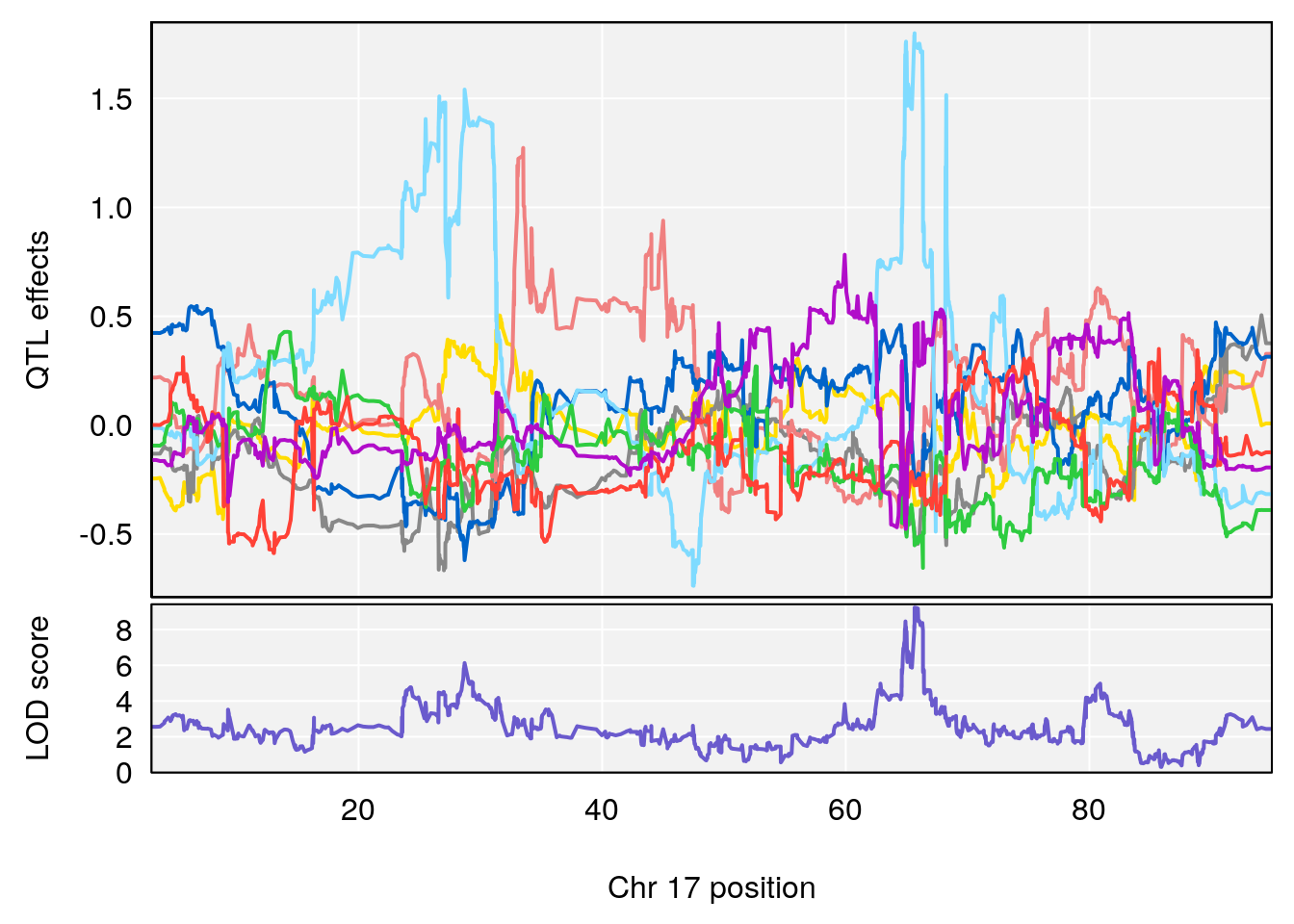
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
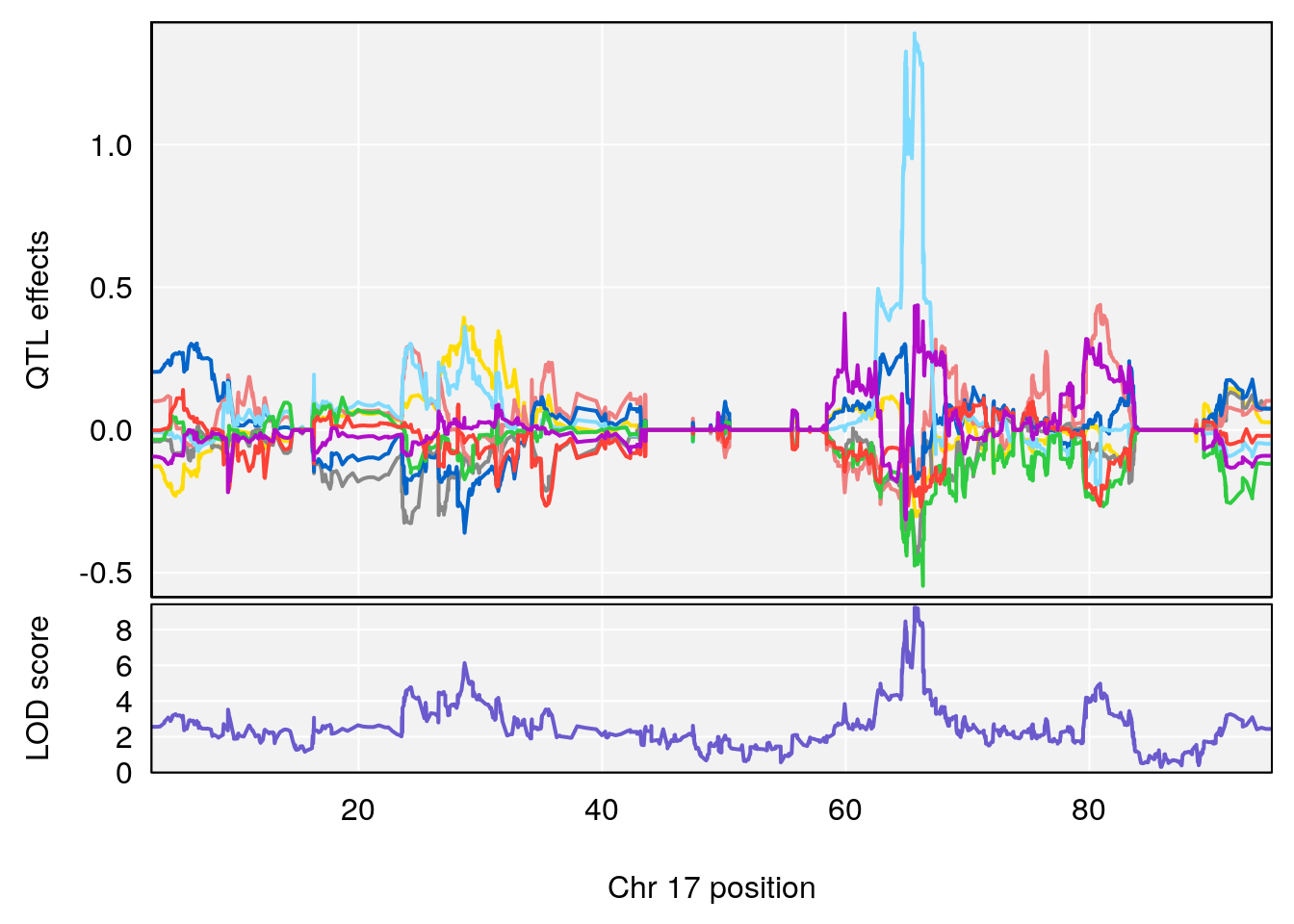
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
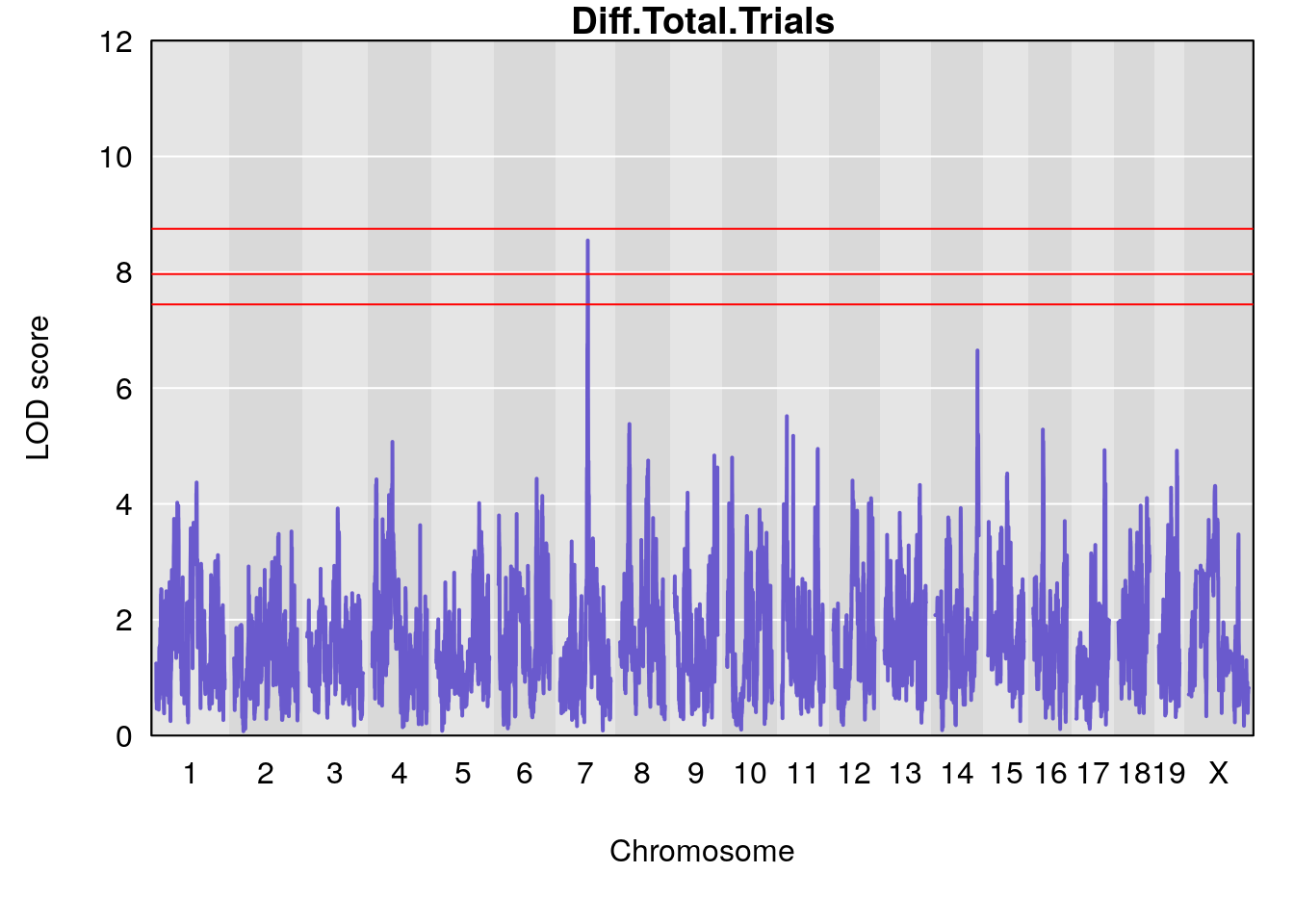
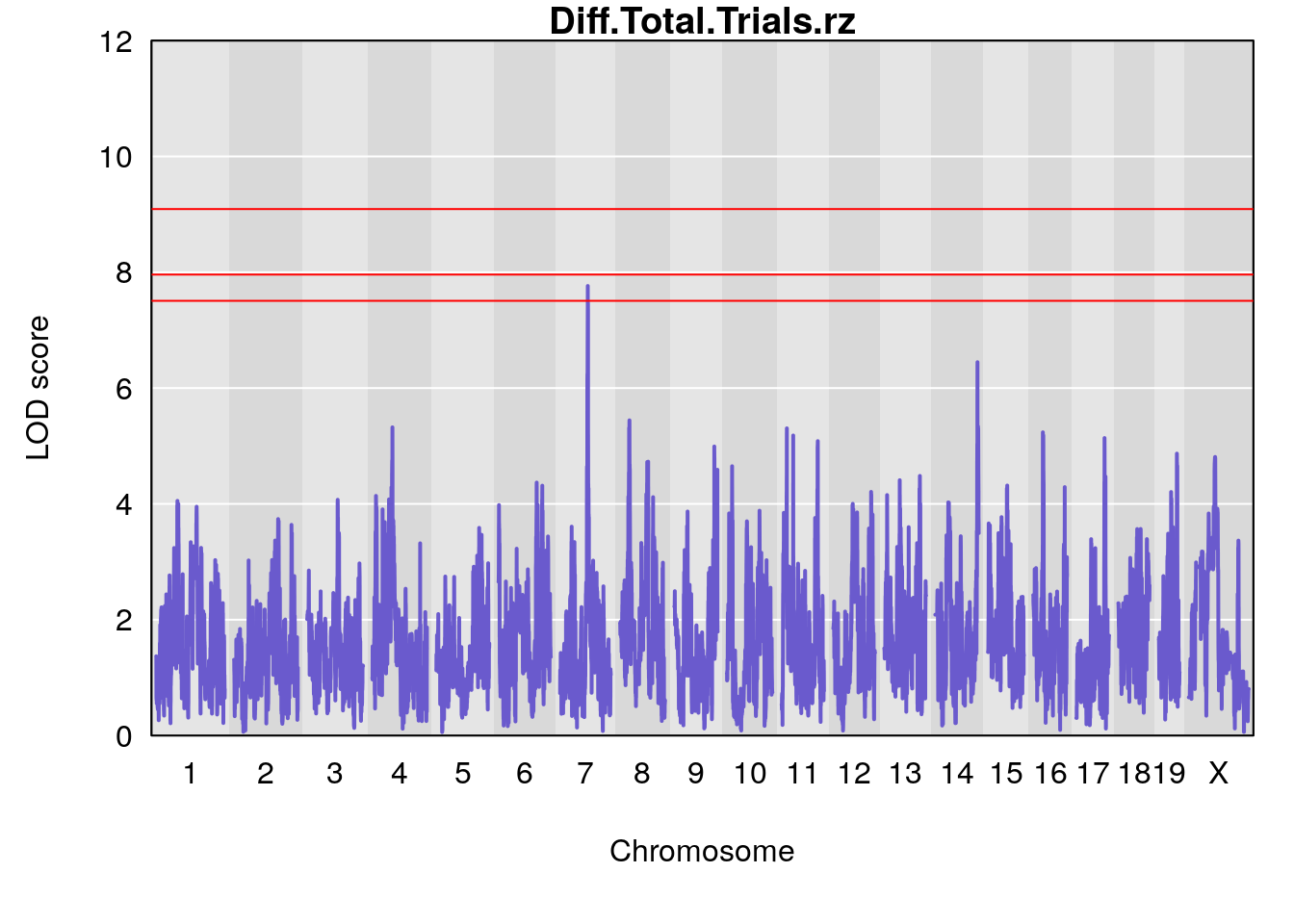
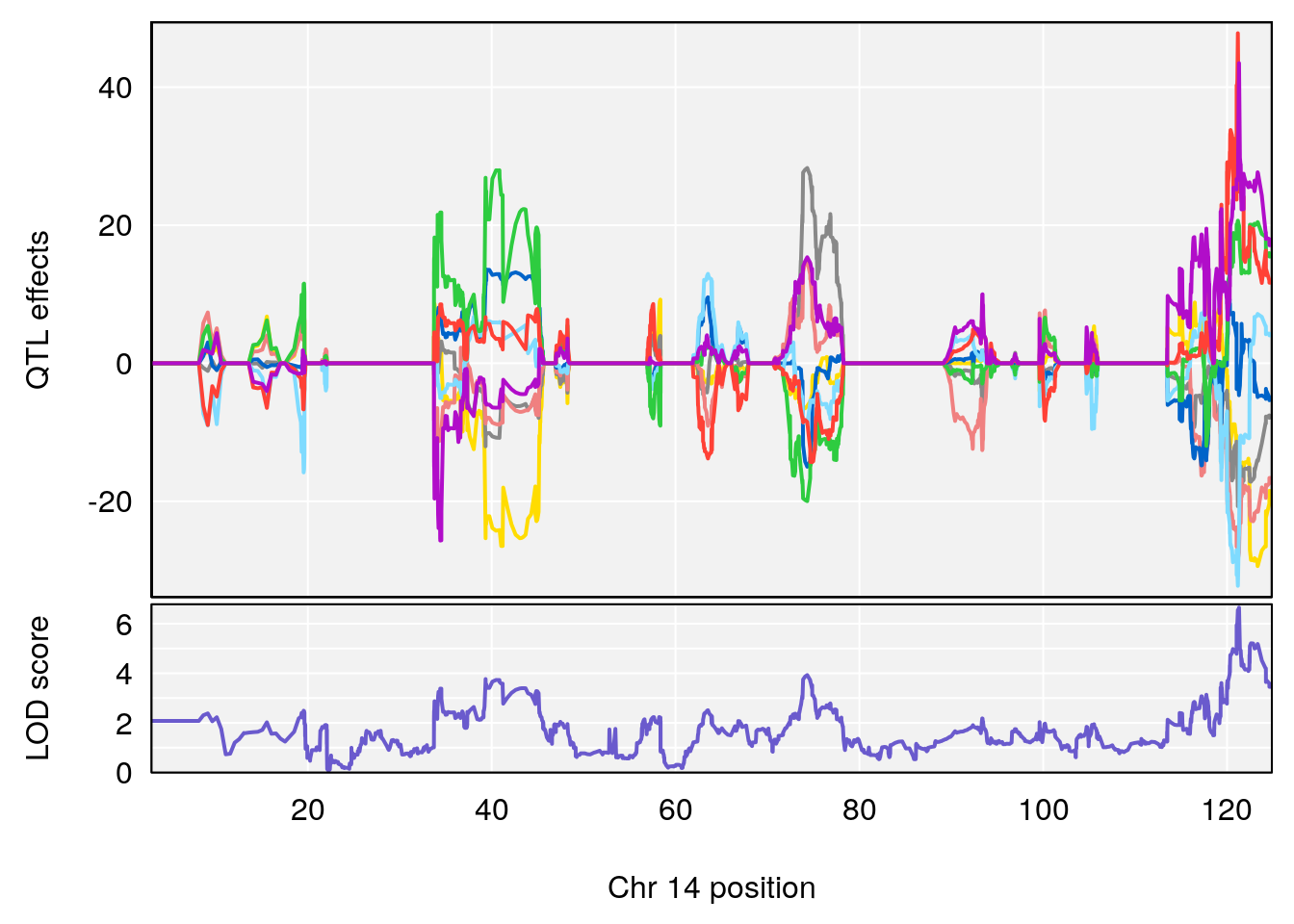
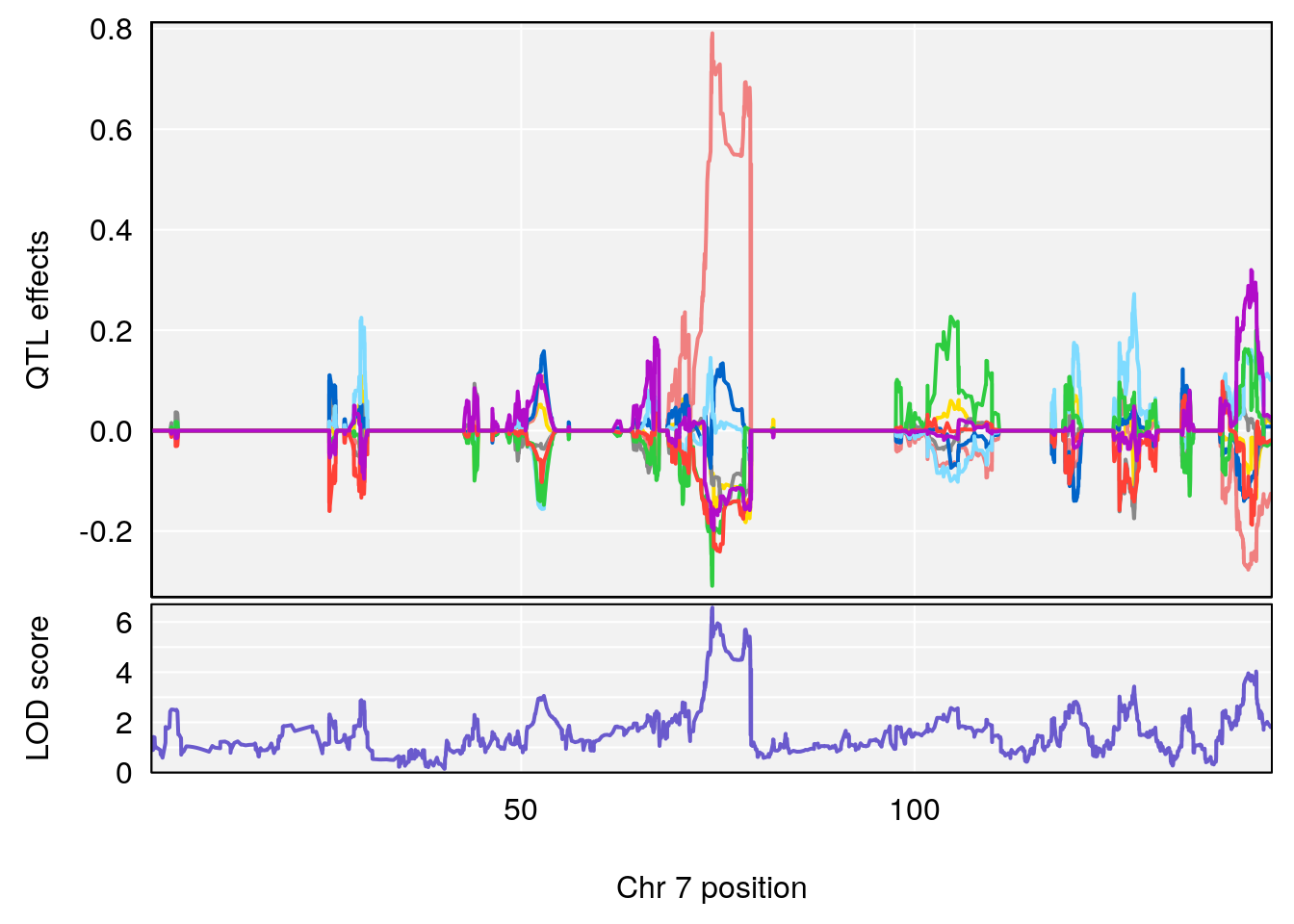
# [1] "Diff.Total.Trials"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 7 80.80292 8.549071 80.09433 81.52958
# 2 1 pheno1 14 121.29944 6.649824 121.03534 123.78109
# [1] 1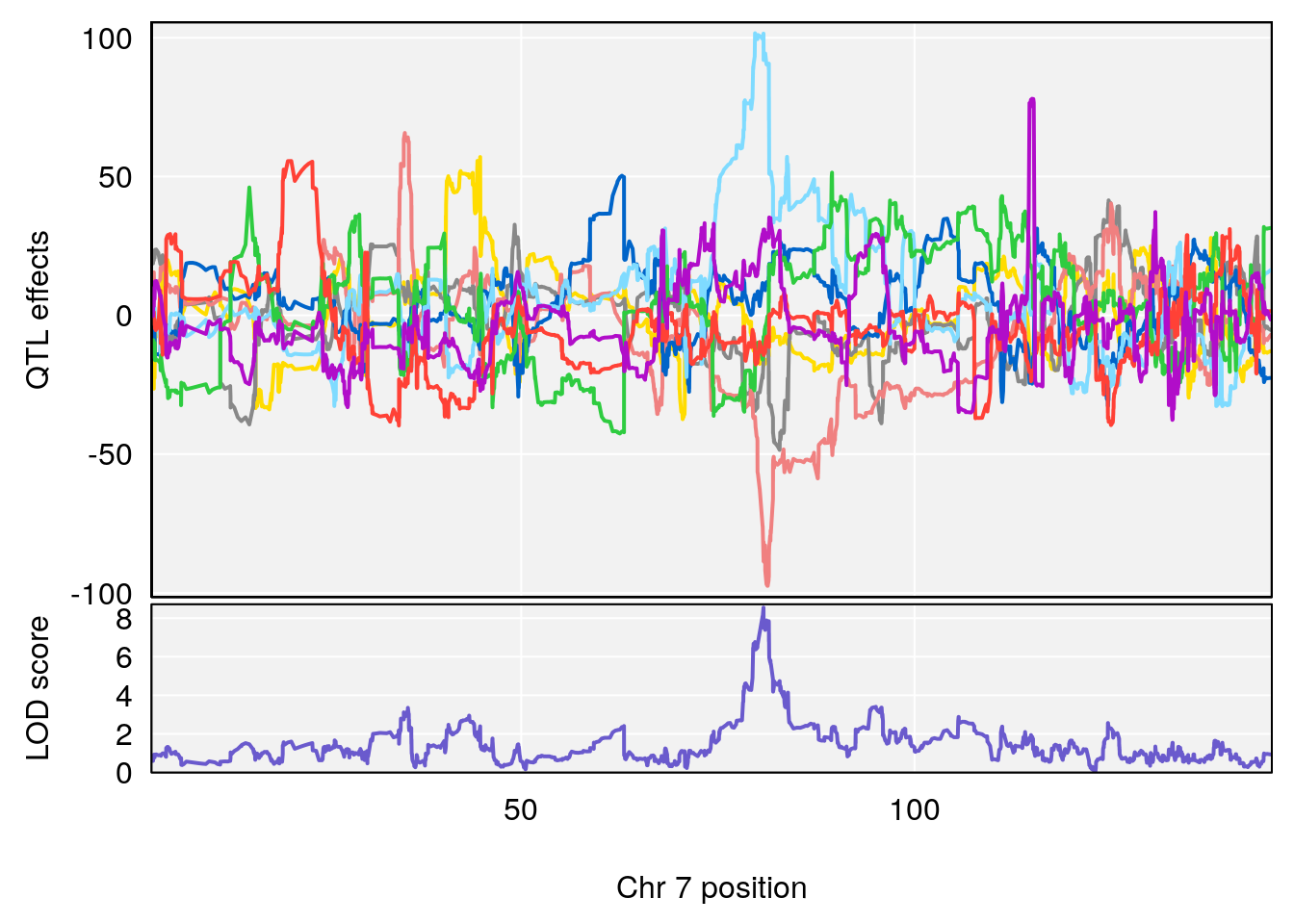
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
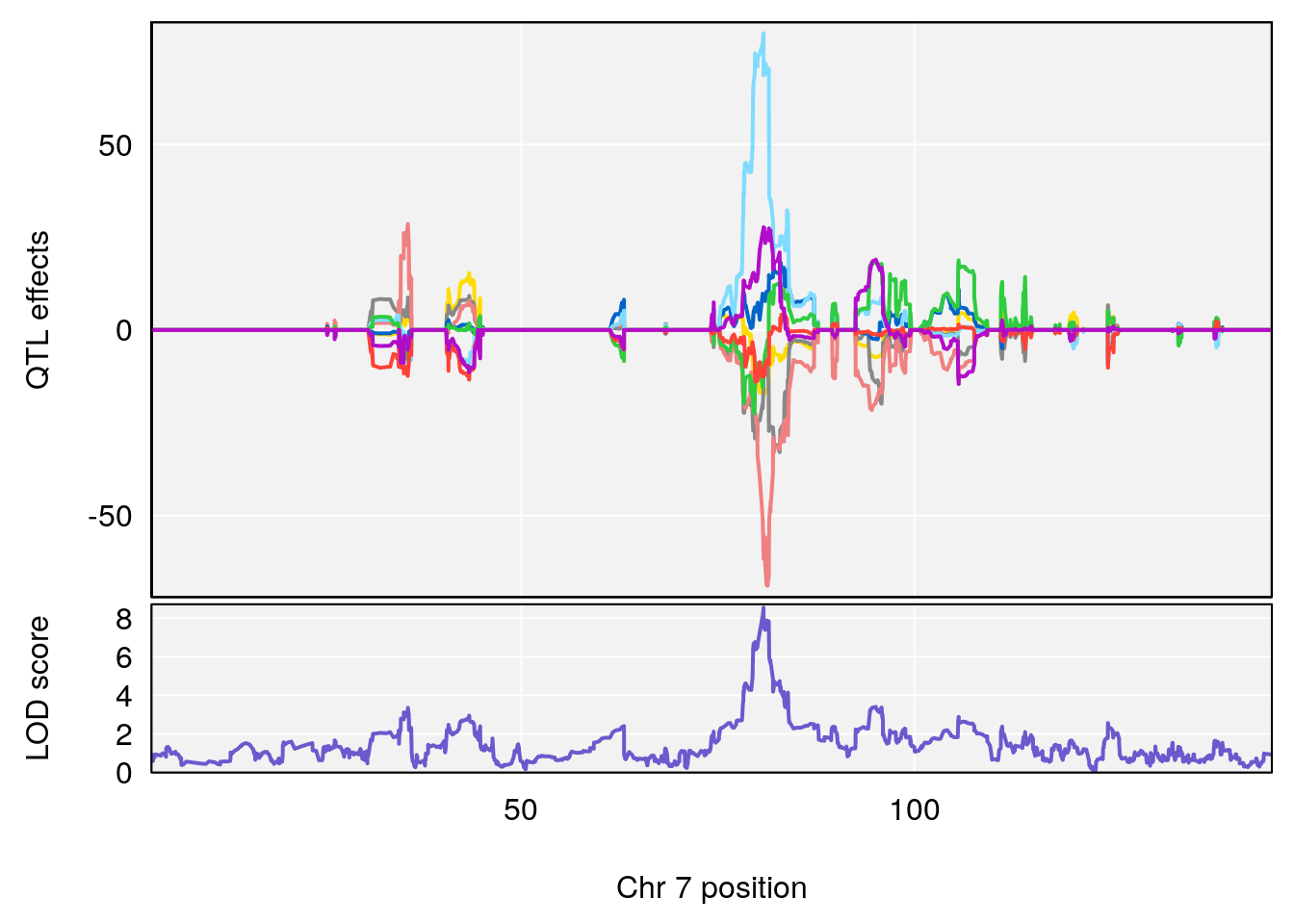
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 2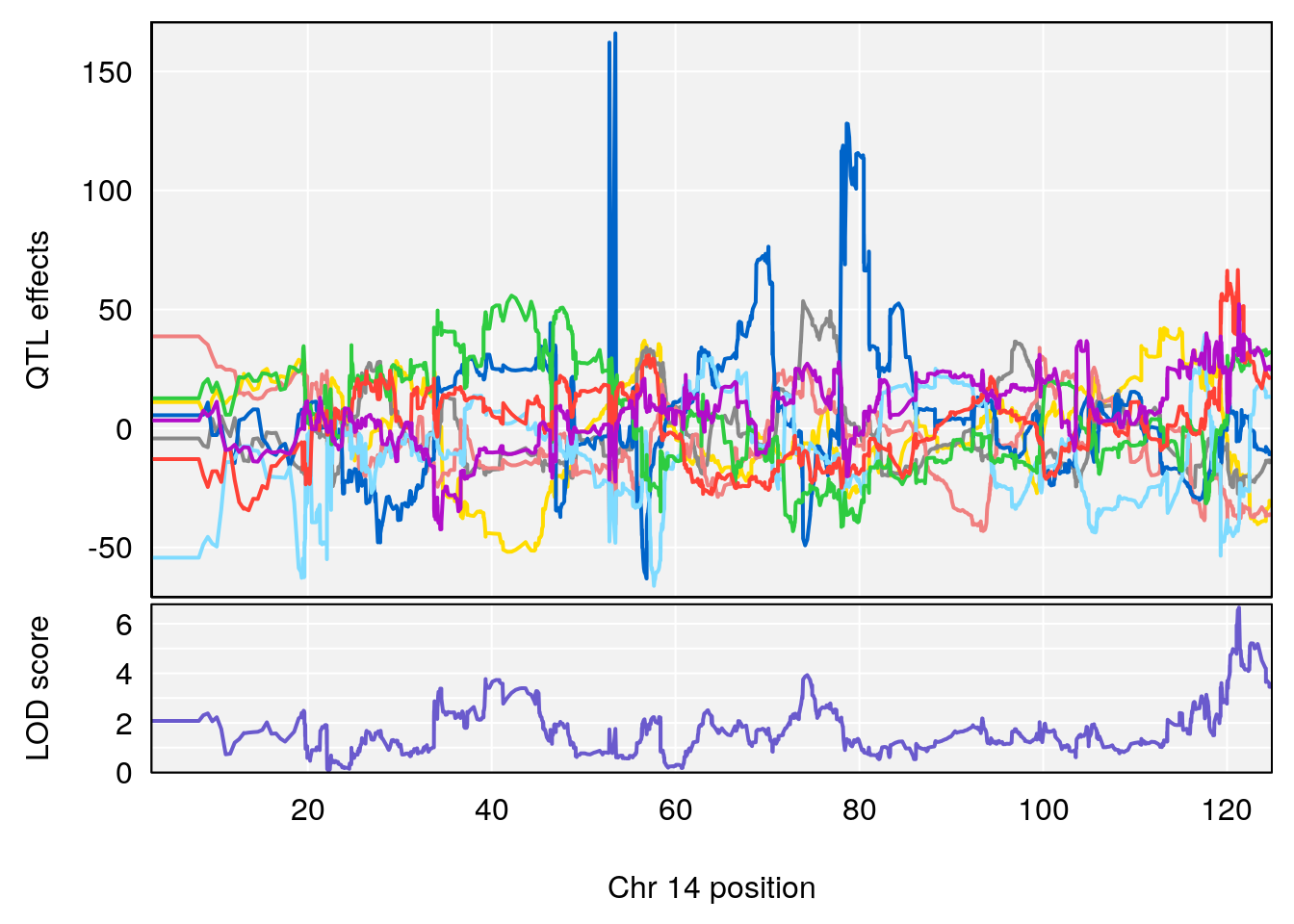
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
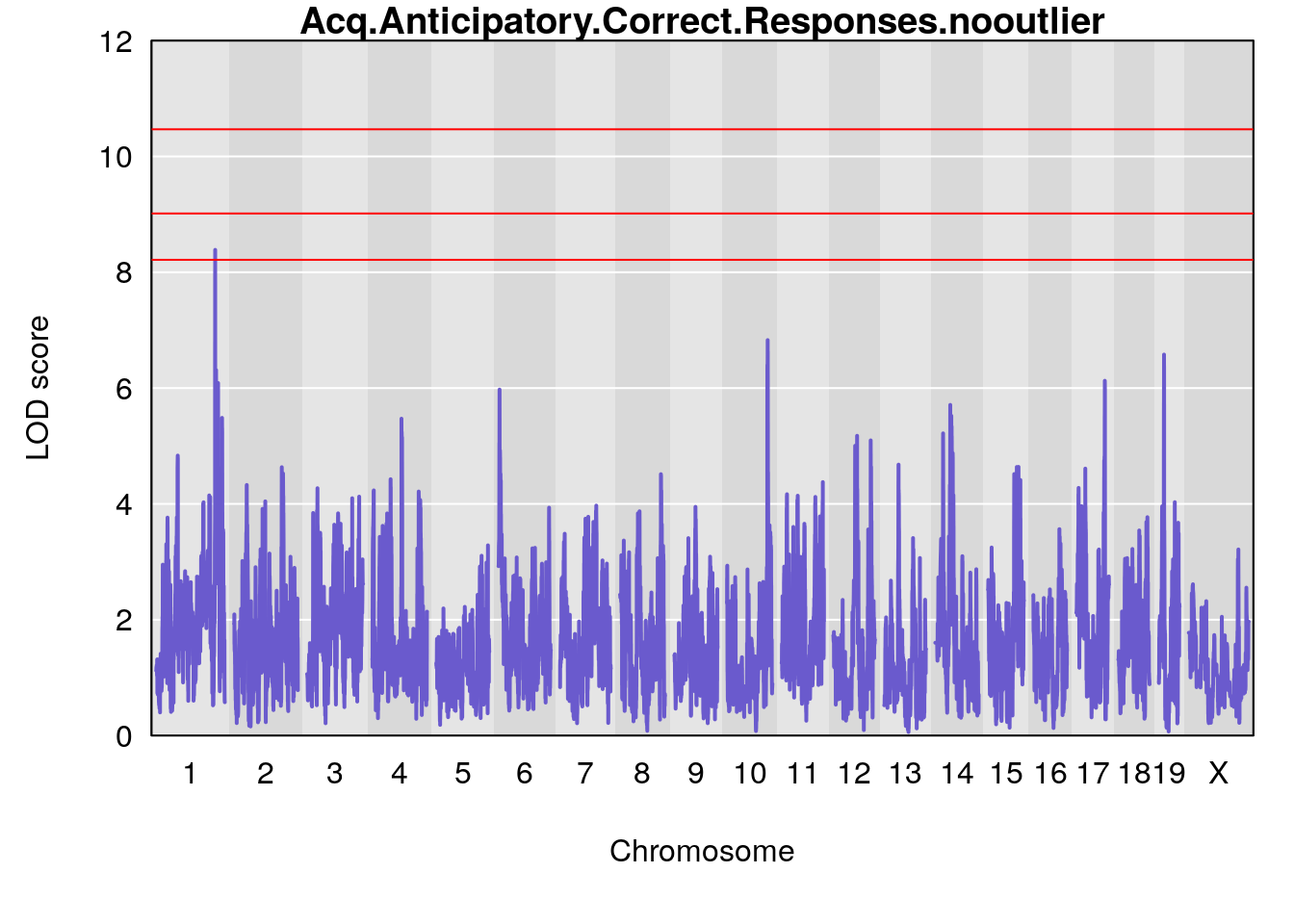
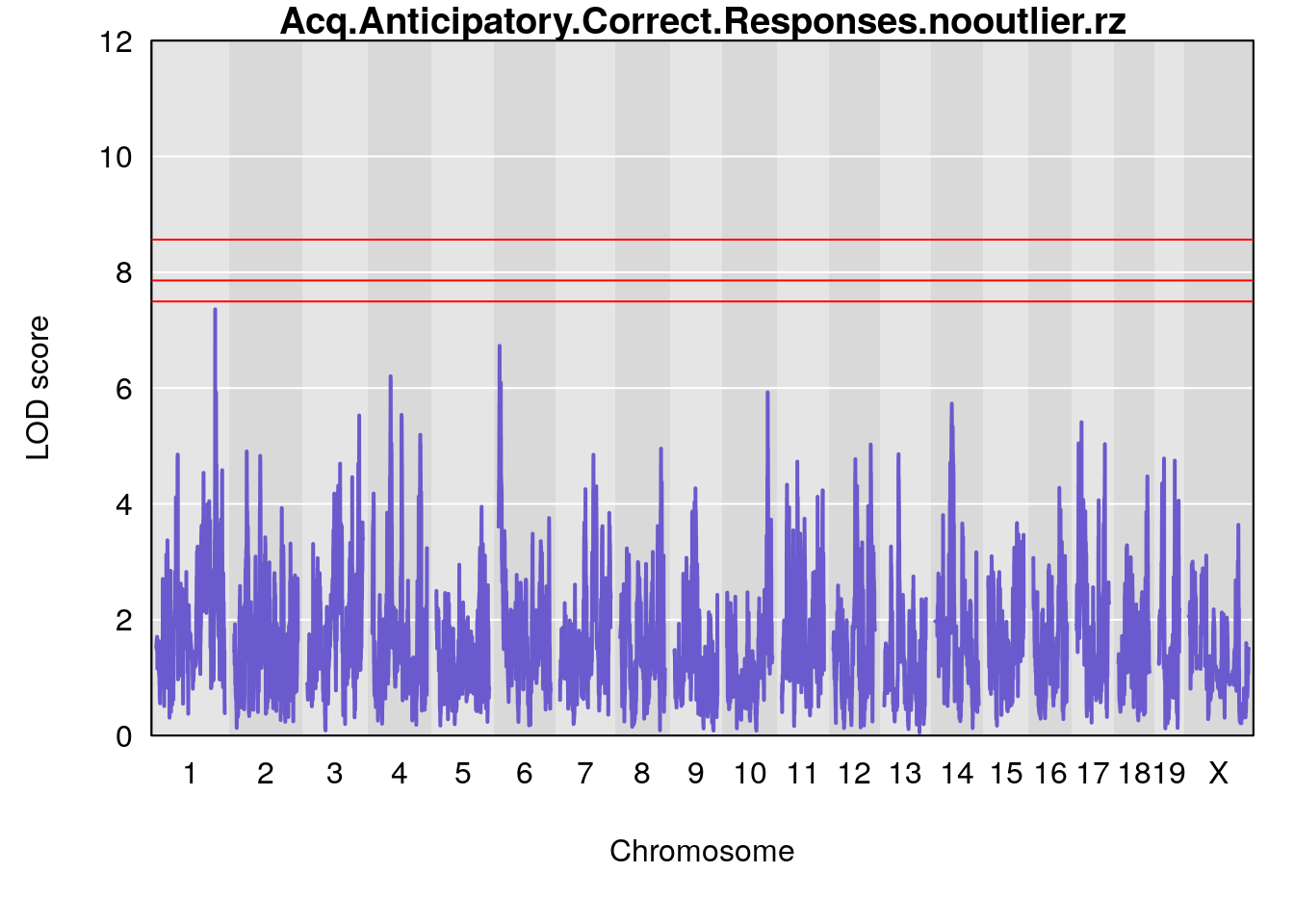
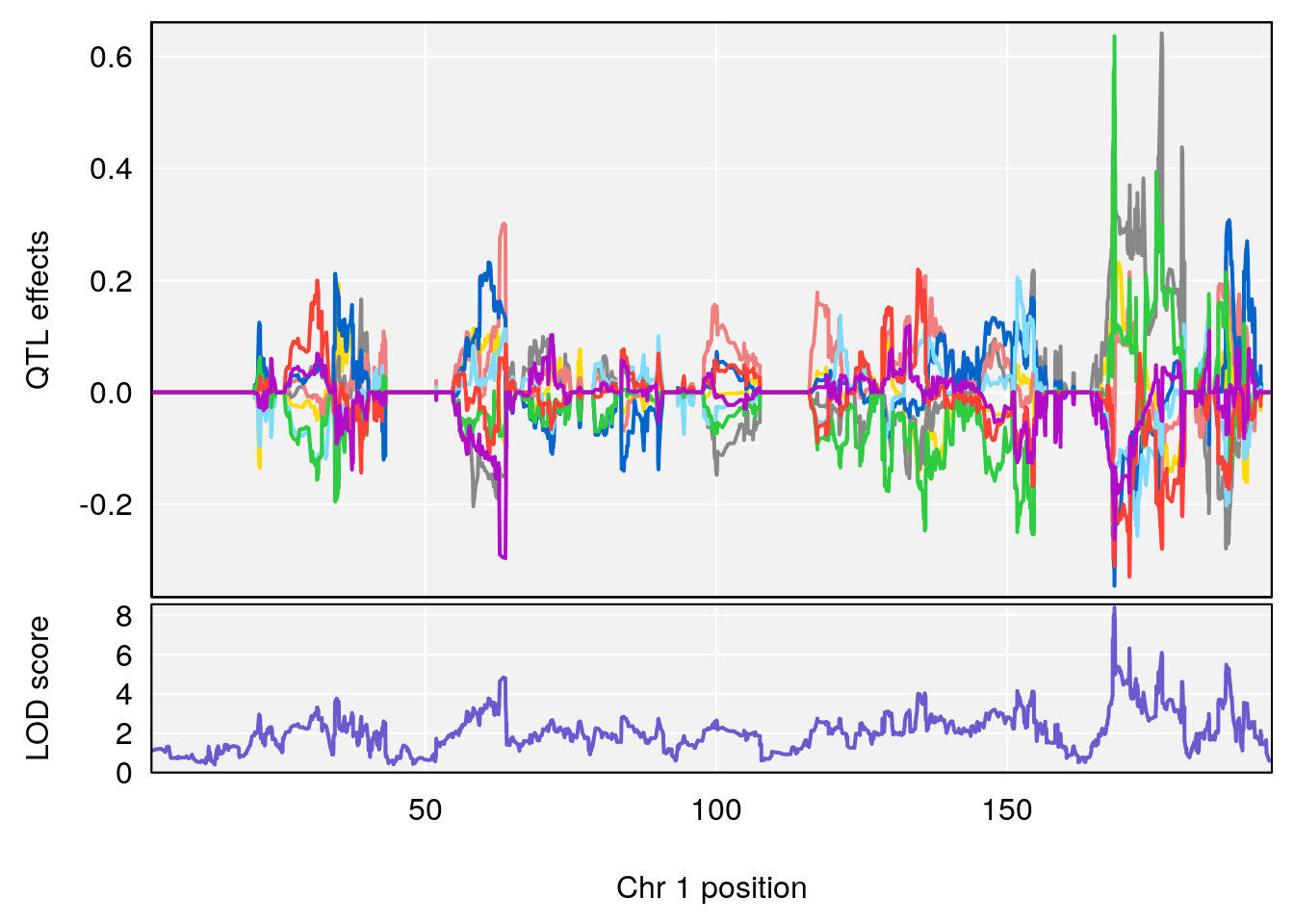
# [1] "Acq.Anticipatory.Correct.Responses.nooutlier"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 168.40300 8.387058 168.22298 168.52005
# 2 1 pheno1 10 117.42104 6.828398 116.30325 117.97397
# 3 1 pheno1 17 82.87091 6.126227 81.12186 83.40925
# 4 1 pheno1 19 17.71985 6.578499 17.23709 18.07340
# [1] 1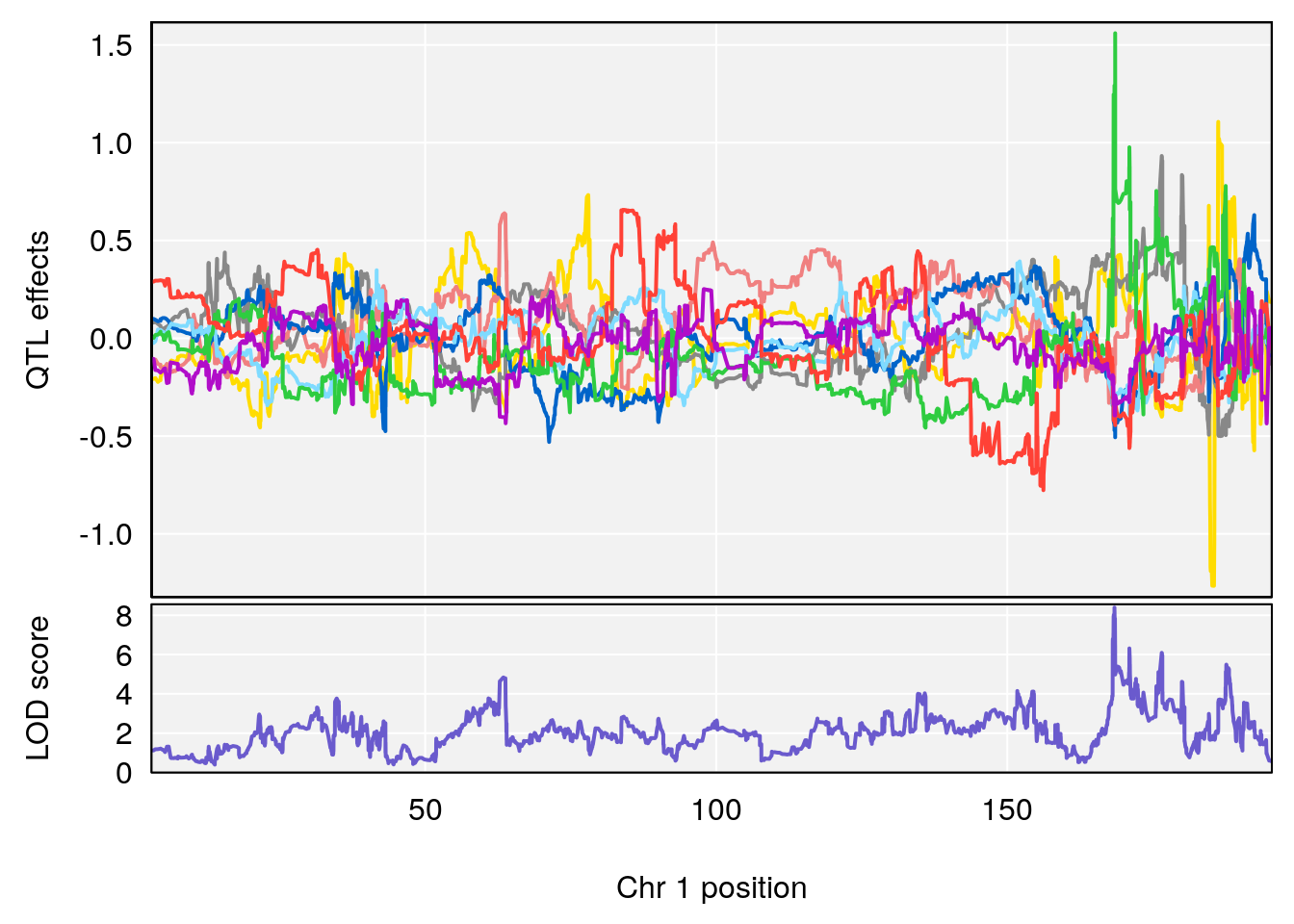
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 2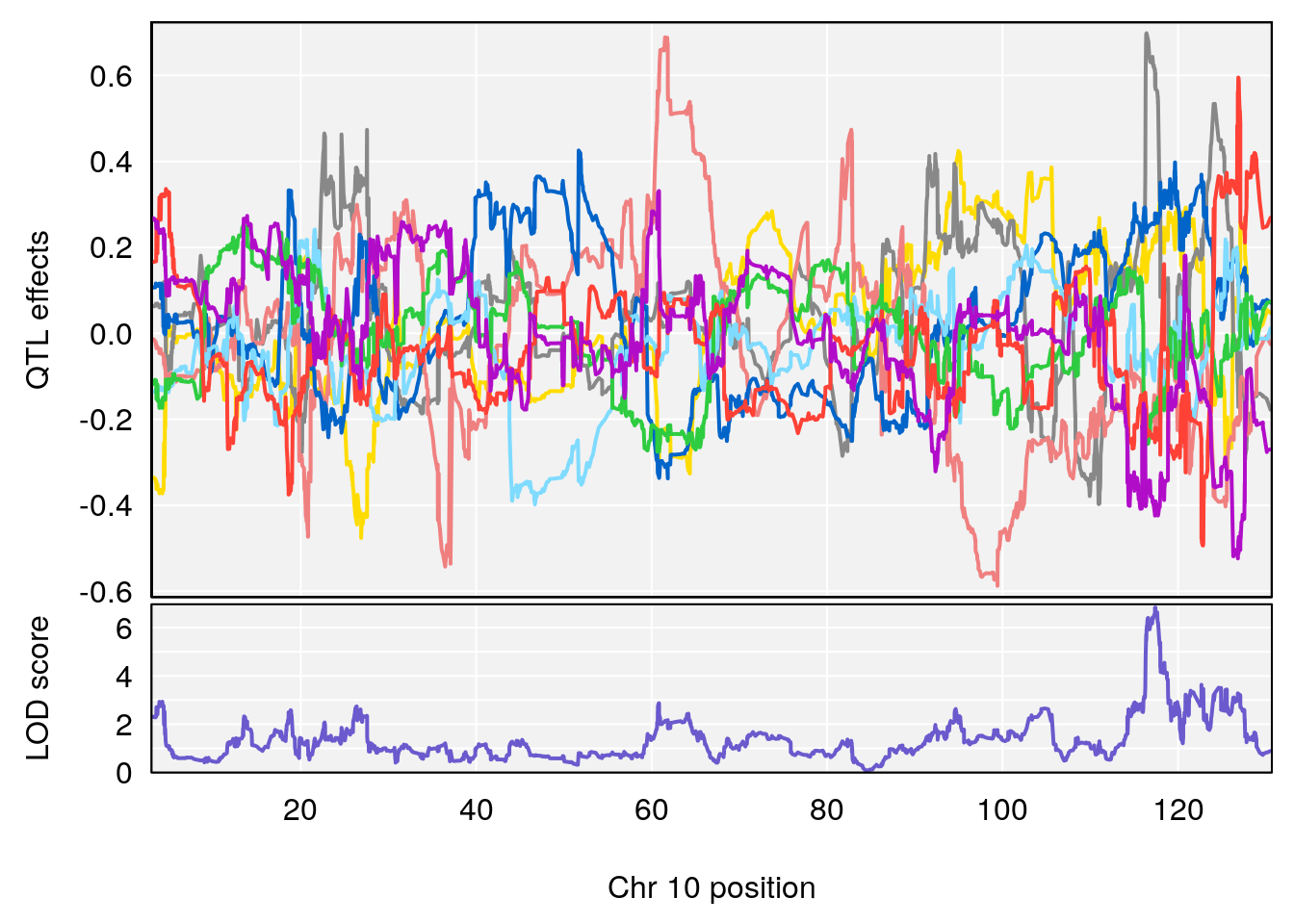
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 3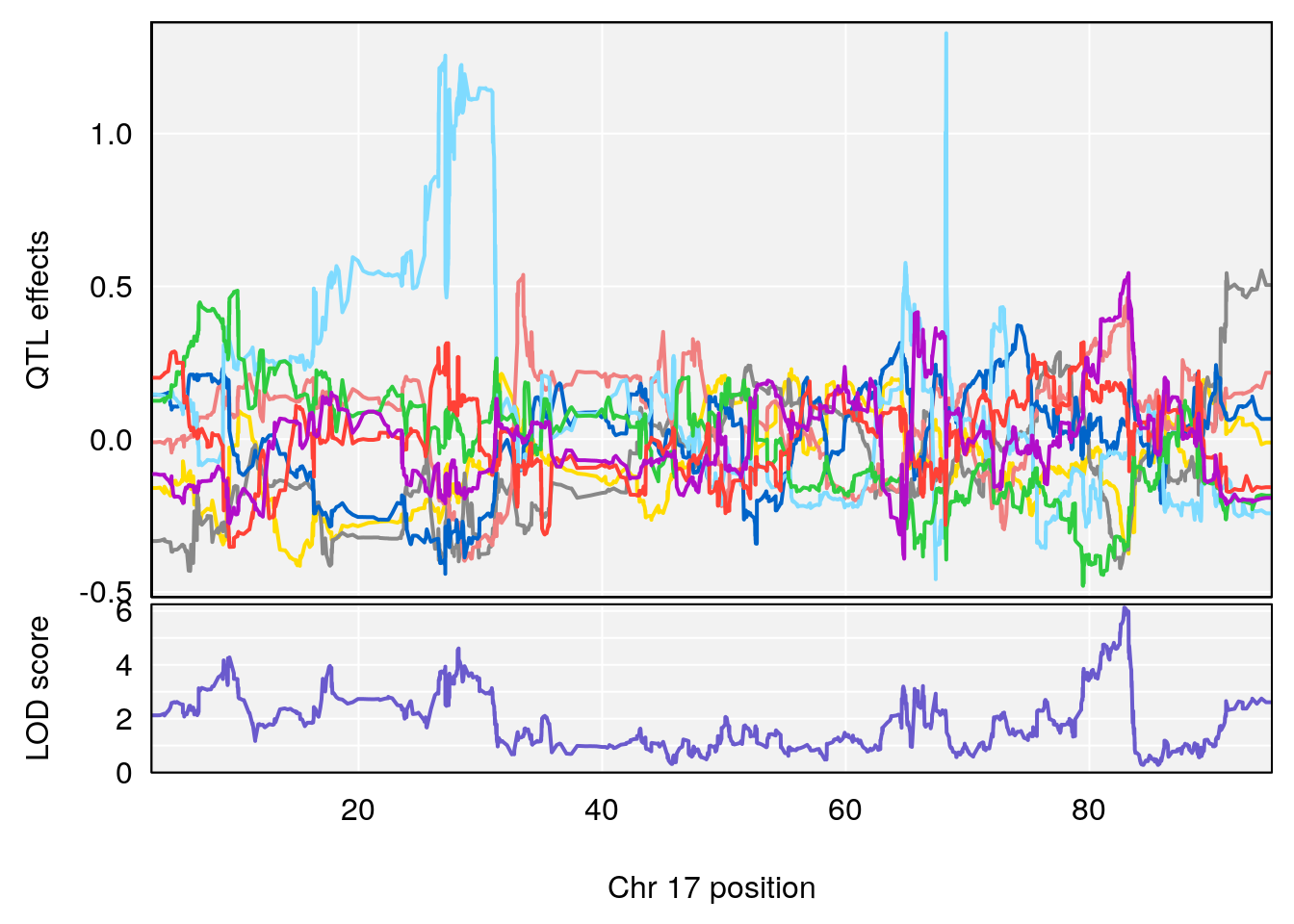
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 4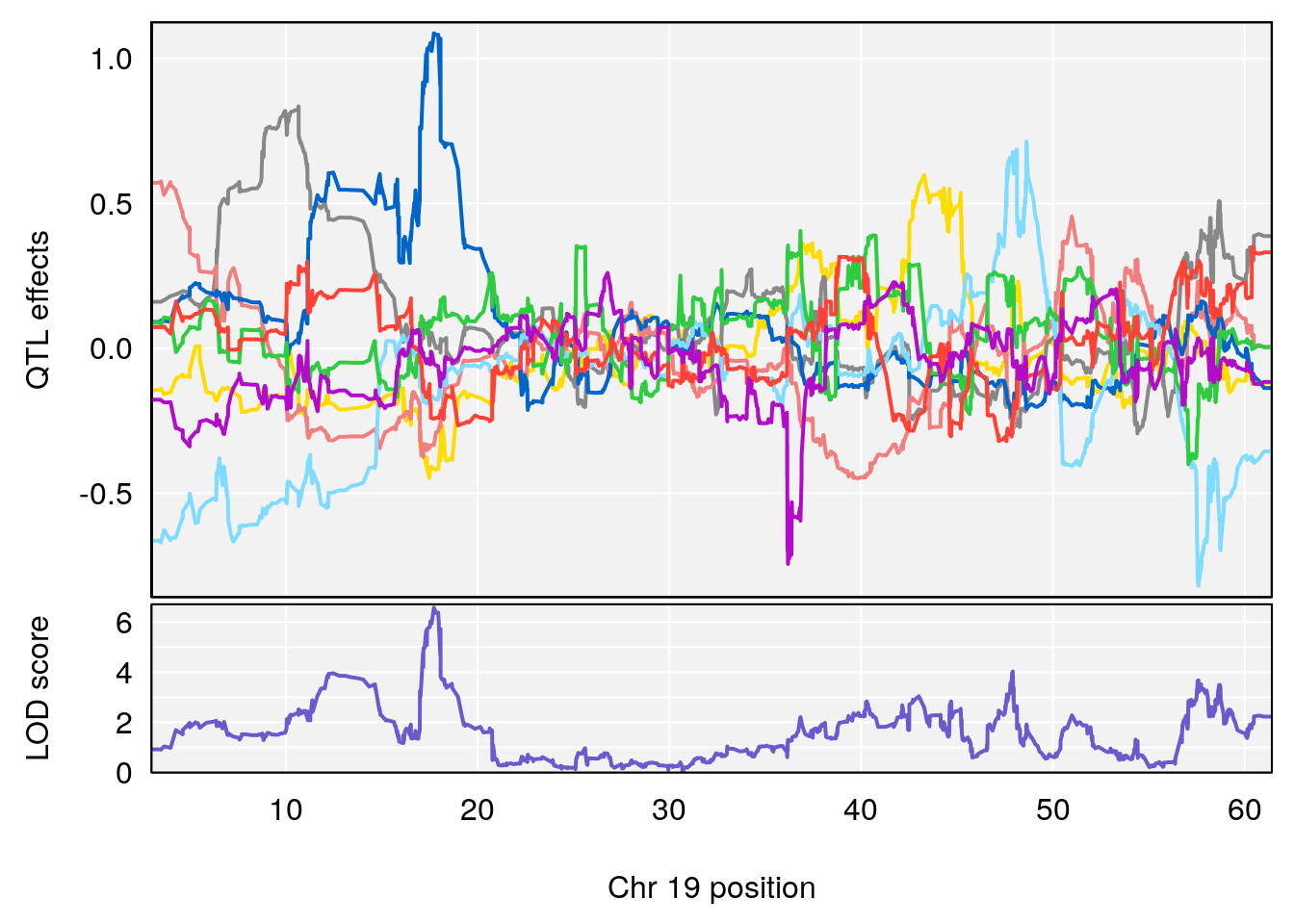
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
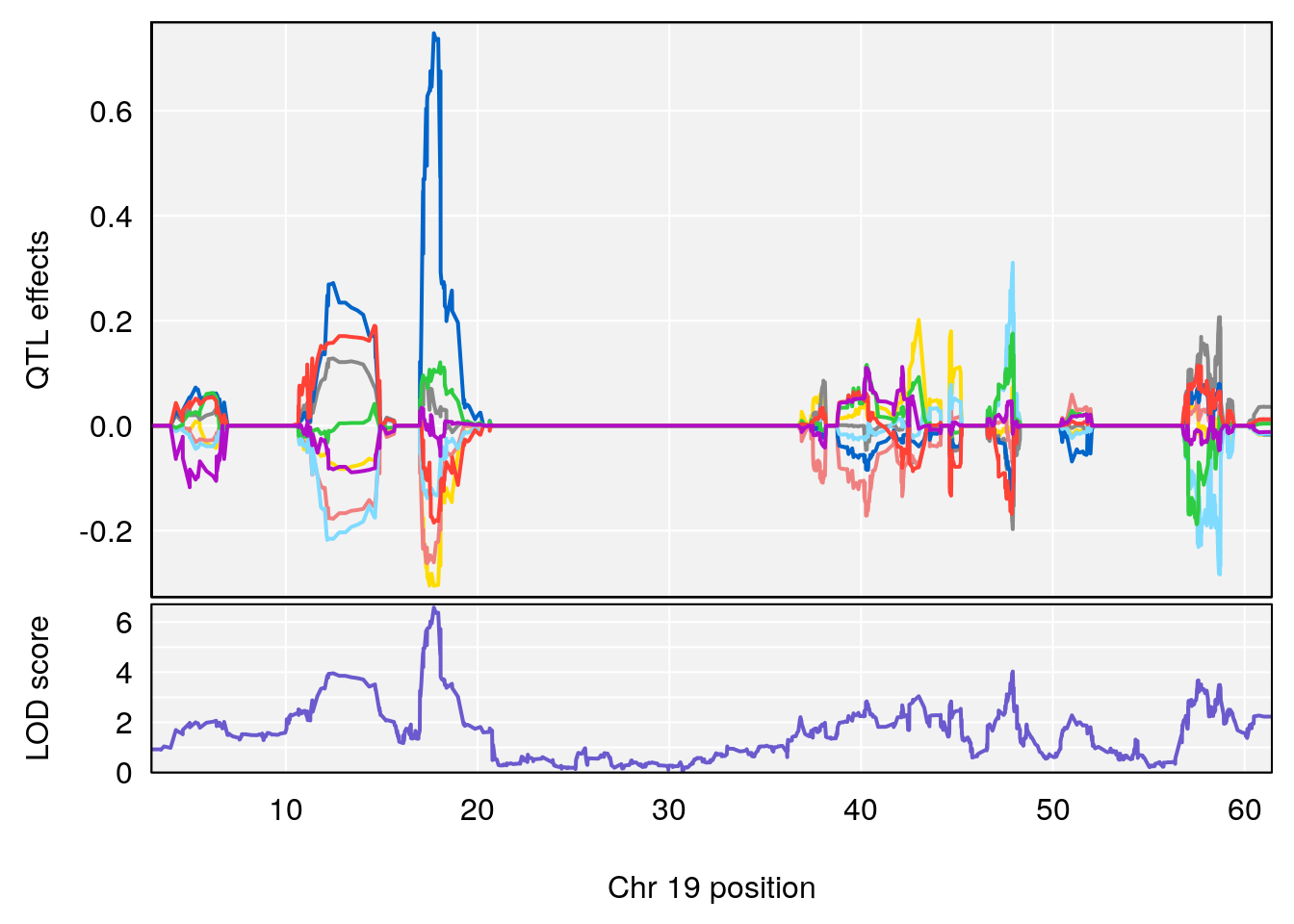
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] "Rev.Anticipatory.Incorrect.Responses.nooutlier"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 170.96387 6.013441 41.45143 171.18444
# 2 1 pheno1 6 103.68554 6.071478 6.05109 103.77874
# 3 1 pheno1 7 74.30691 6.576426 74.07750 79.11953
# 4 1 pheno1 14 49.25670 6.665675 48.80084 54.24423
# 5 1 pheno1 16 56.17223 6.112808 54.36765 58.59104
# 6 1 pheno1 17 65.65263 6.545085 28.65423 80.94284
# [1] 1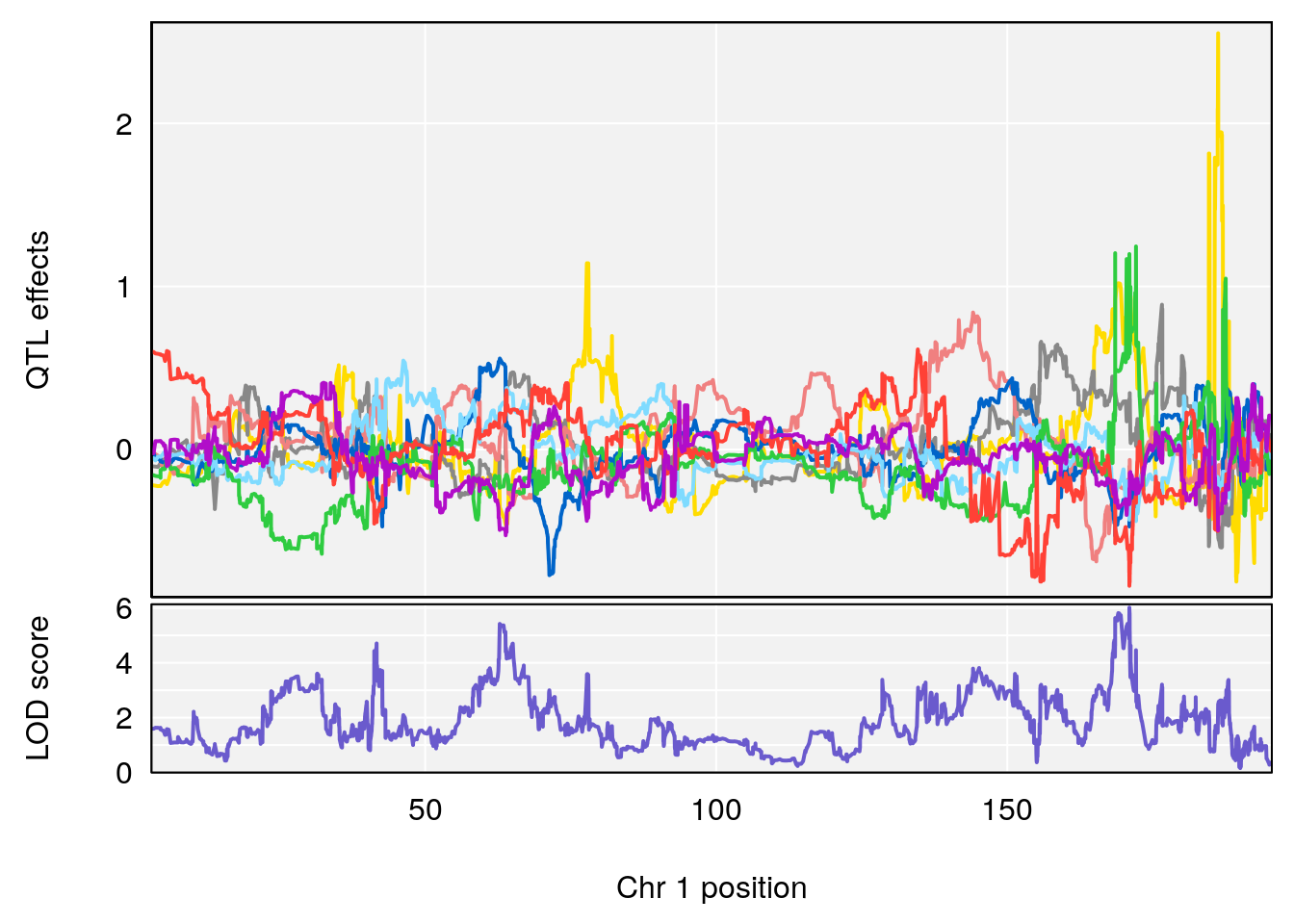
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
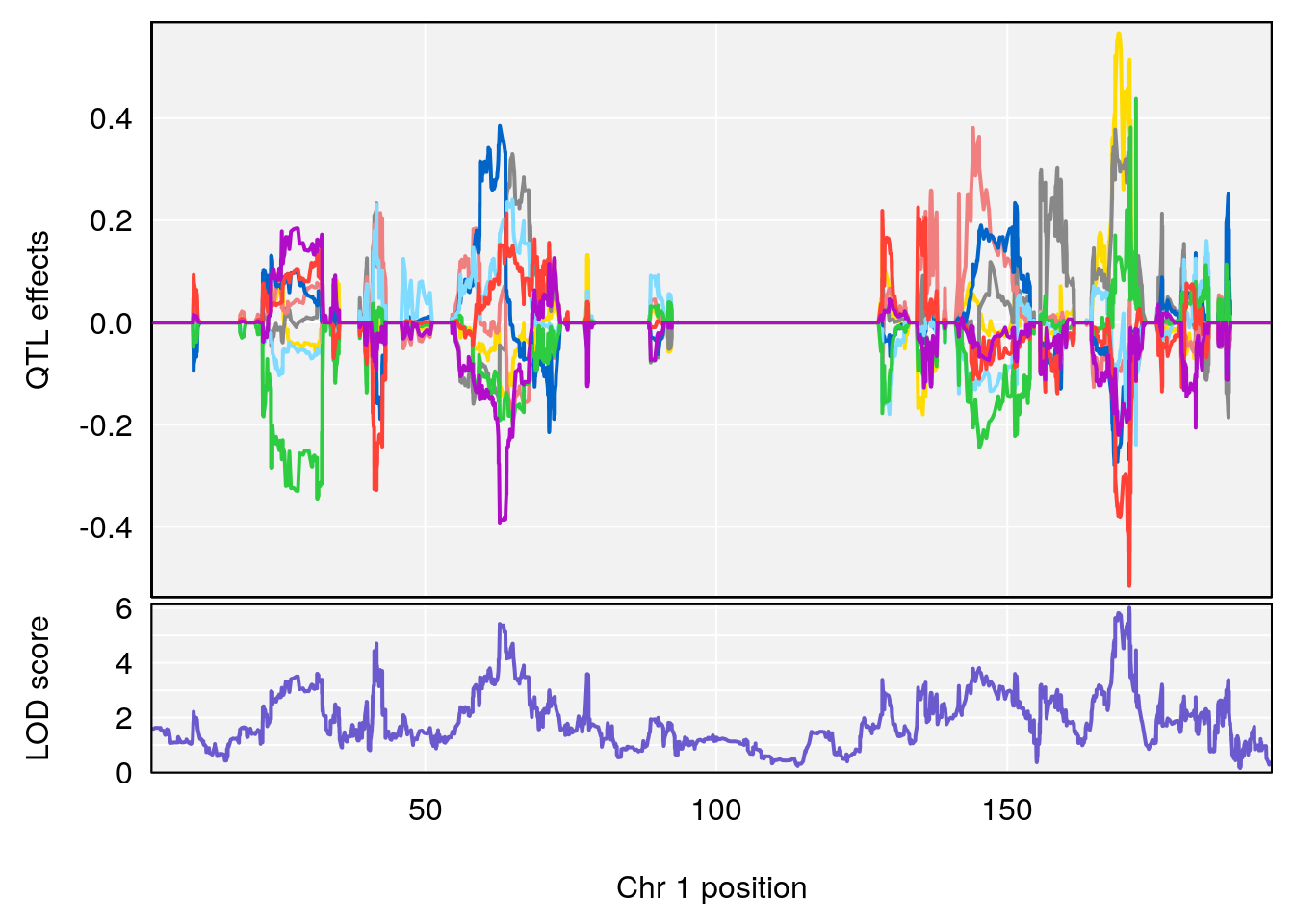
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 2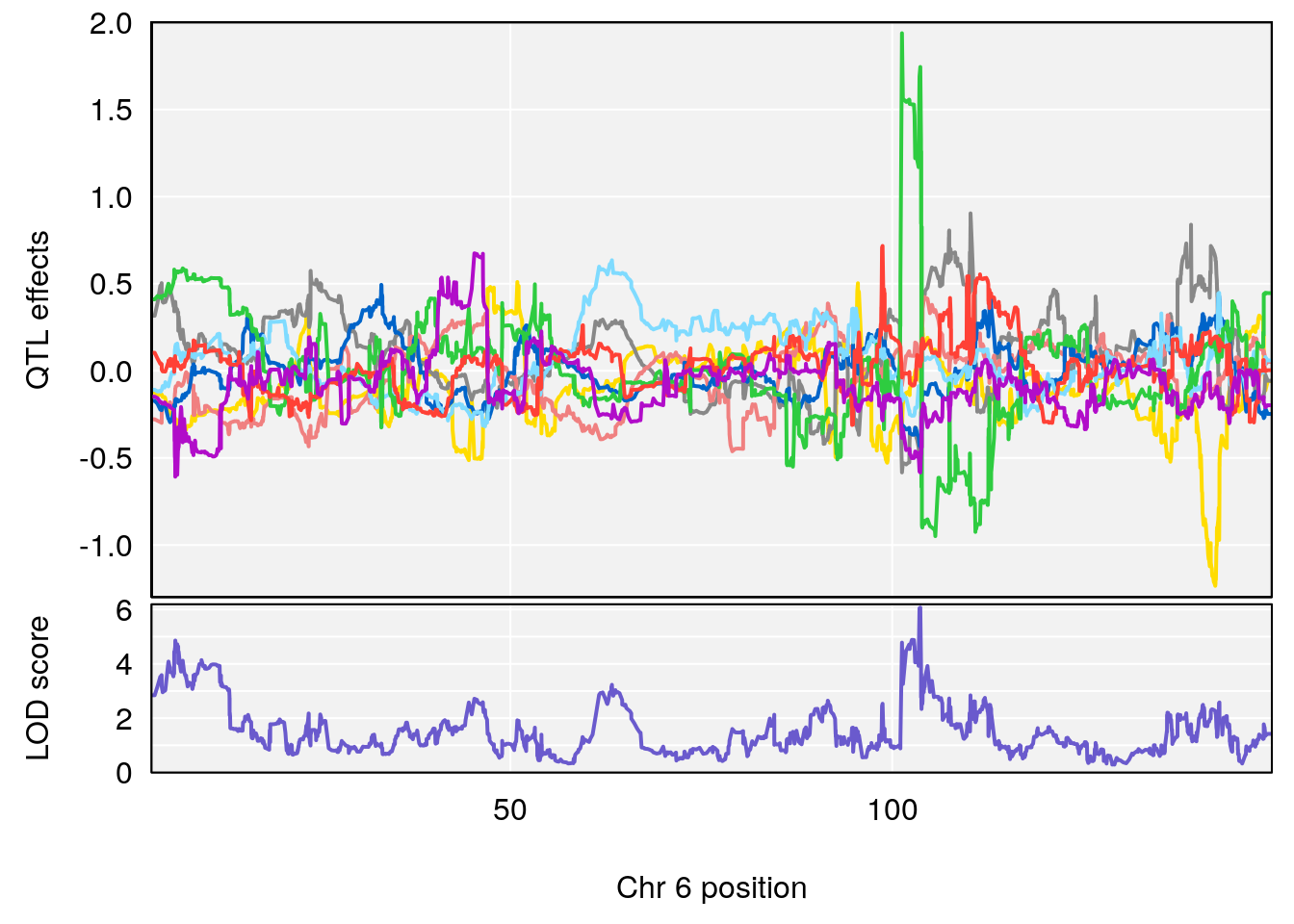
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 3
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 4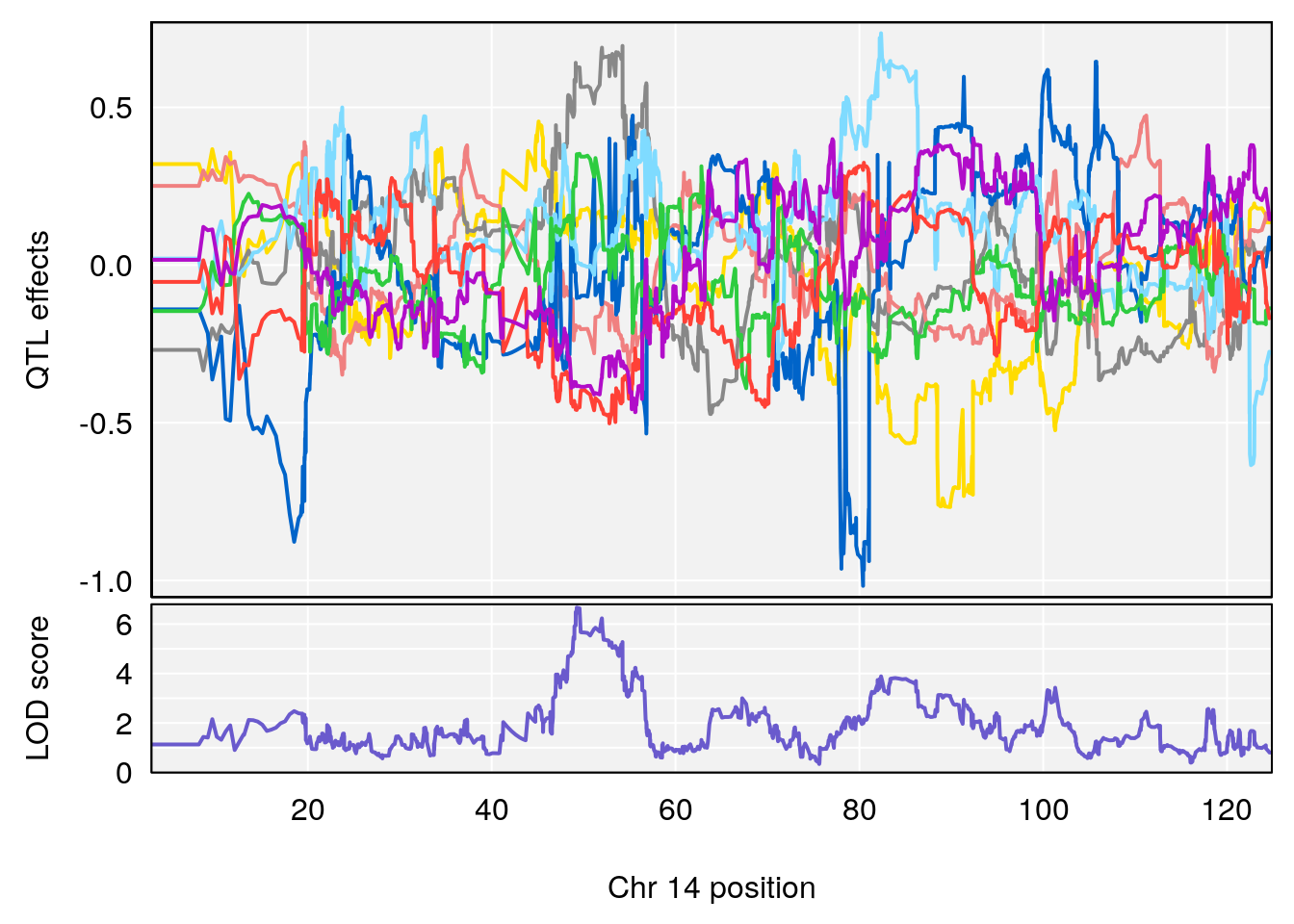
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 5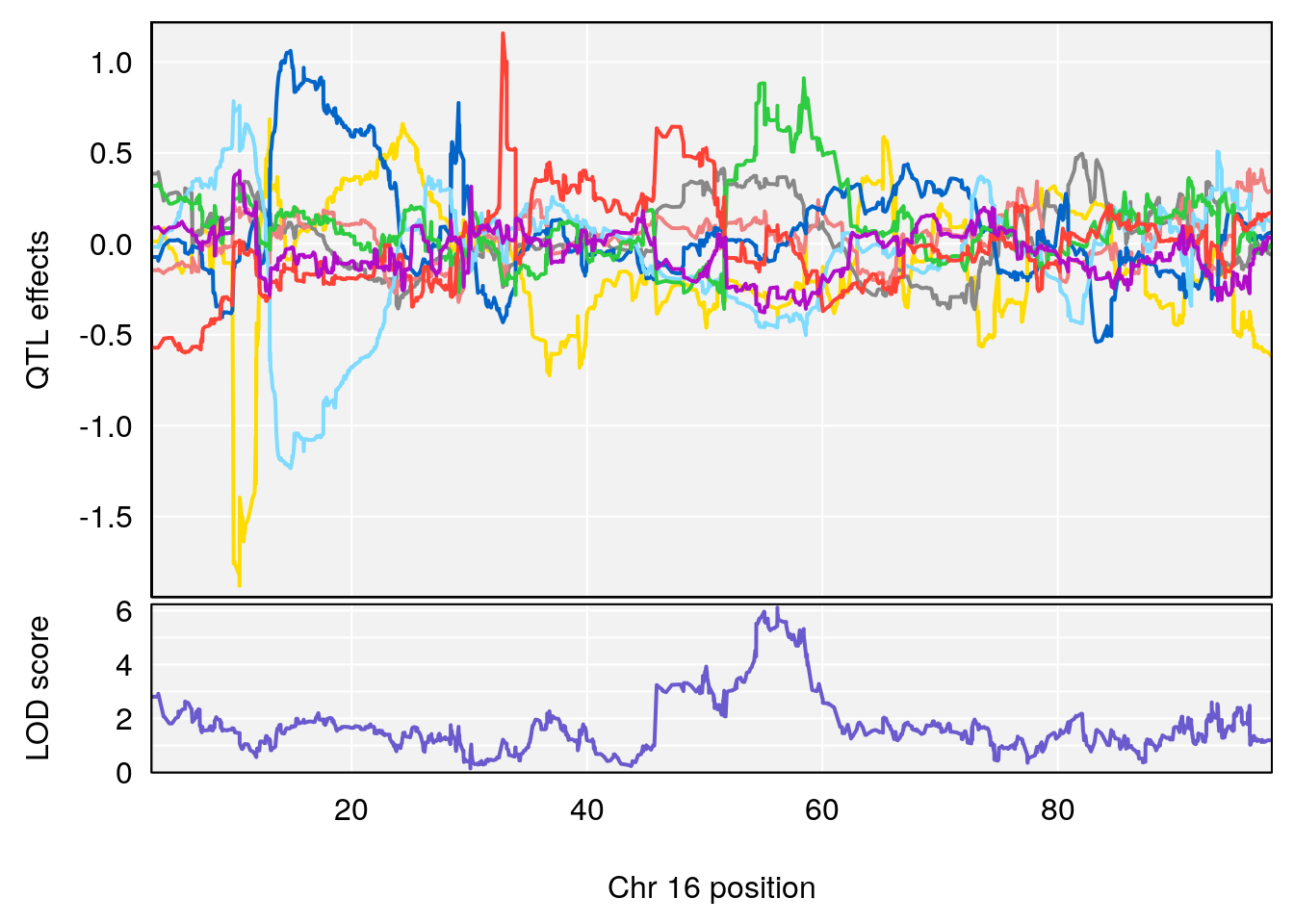
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |

| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 6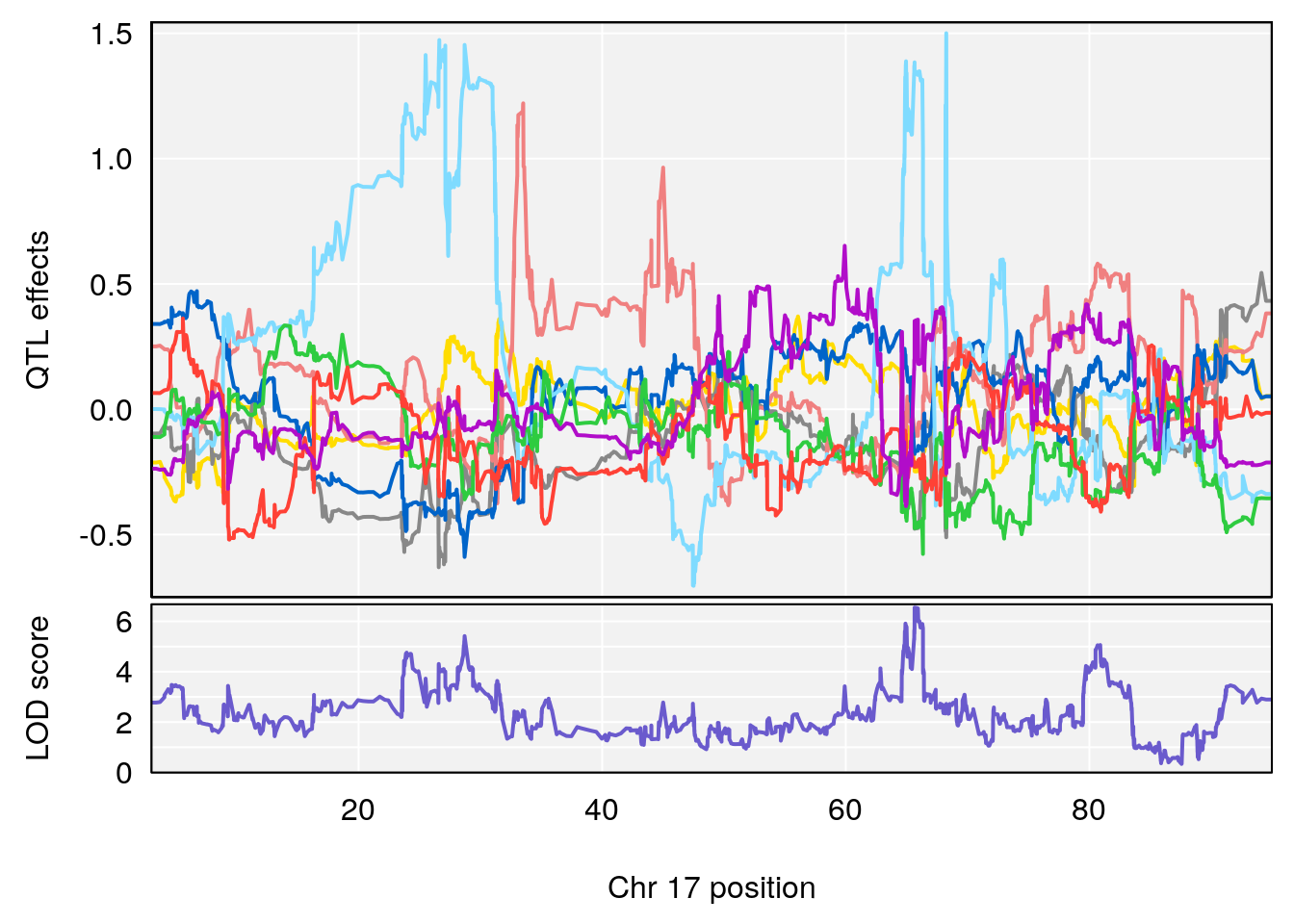
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
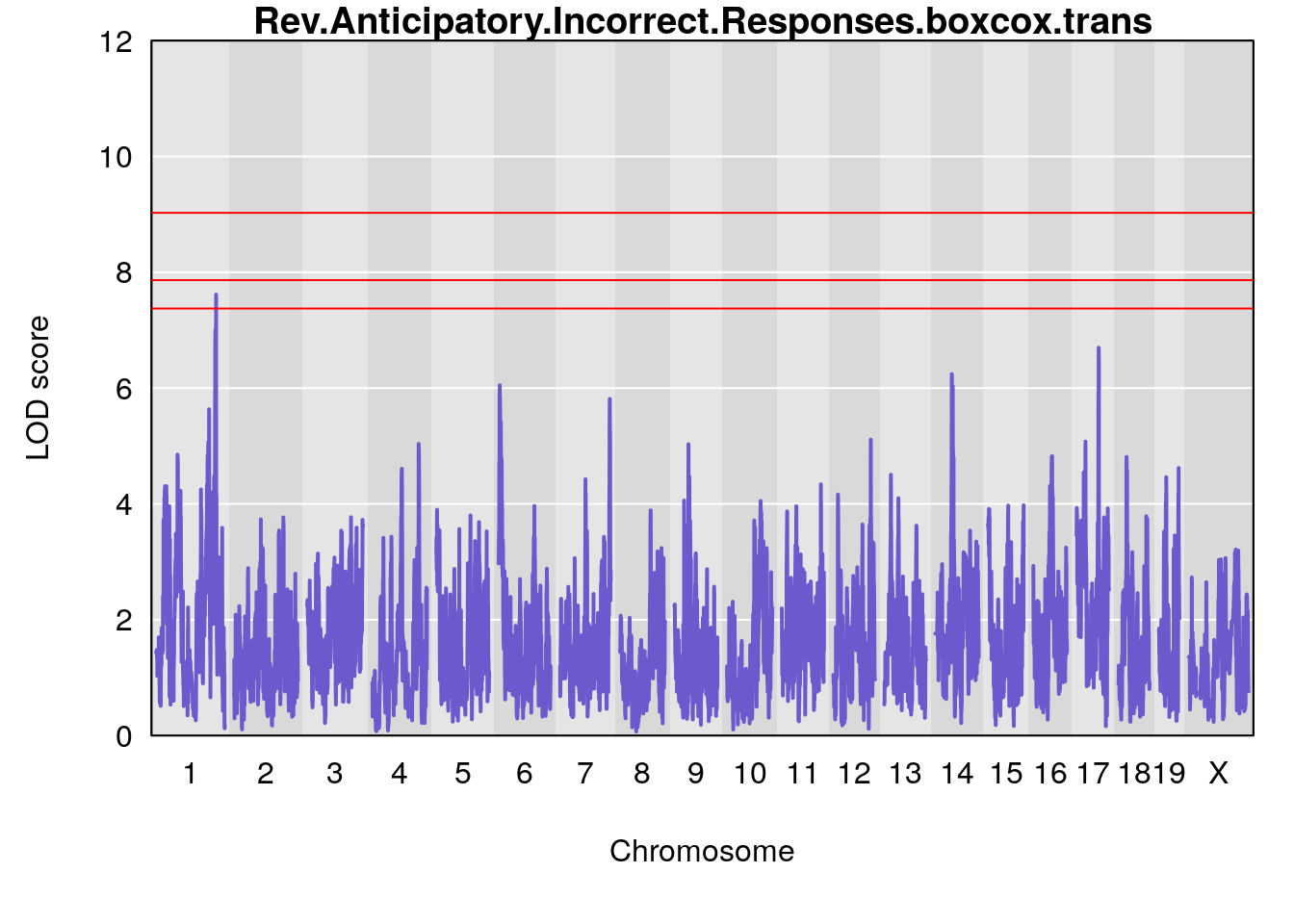
# [1] "Rev.Anticipatory.Incorrect.Responses.boxcox.trans"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 170.963873 7.614860 168.522985 171.06618
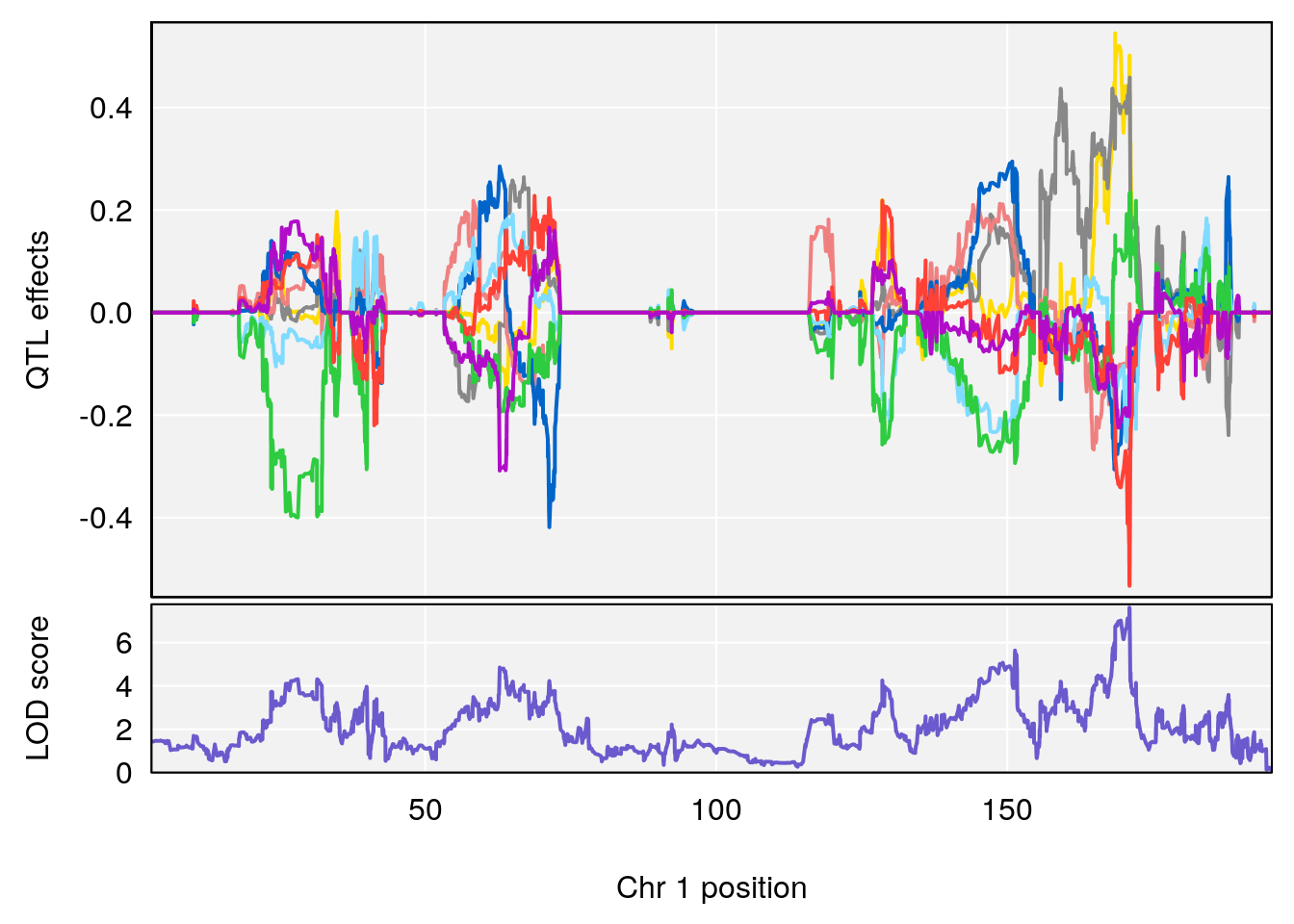
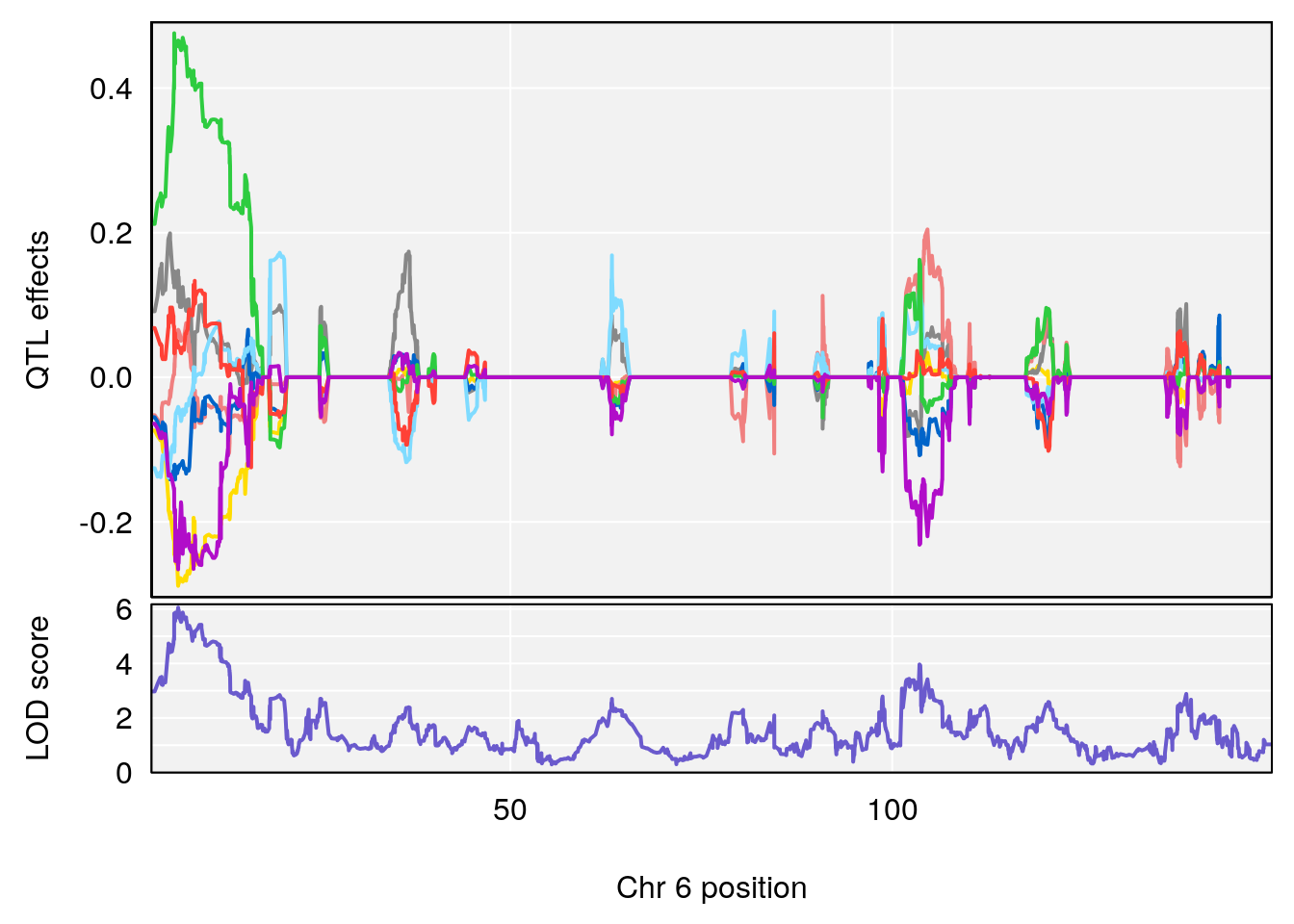
# 2 1 pheno1 6 6.483562 6.049903 4.801035 12.08985
# 3 1 pheno1 14 49.256704 6.241500 48.800840 54.24423
# 4 1 pheno1 17 65.652629 6.697057 64.831601 66.34240
# [1] 1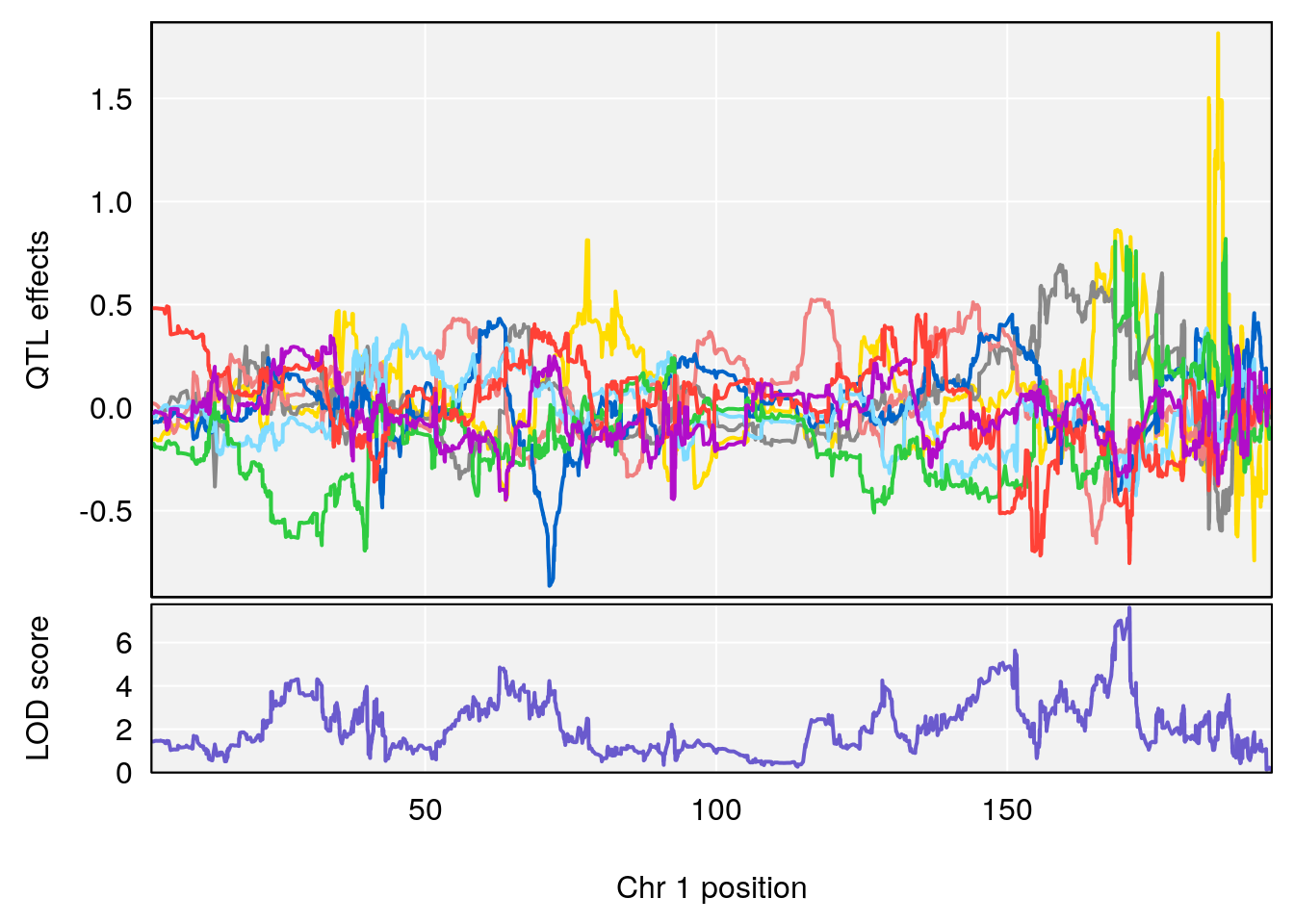
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 2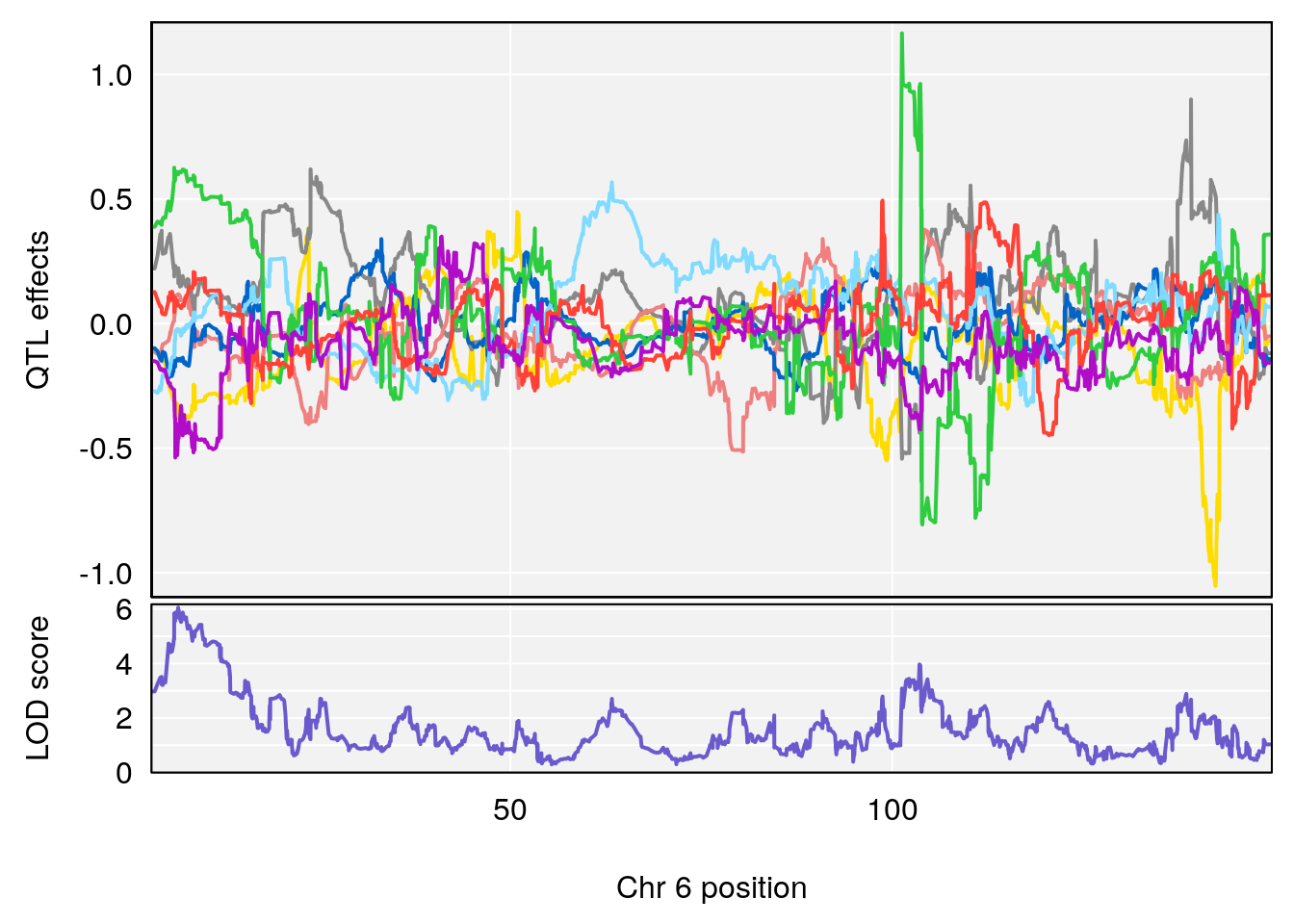
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 3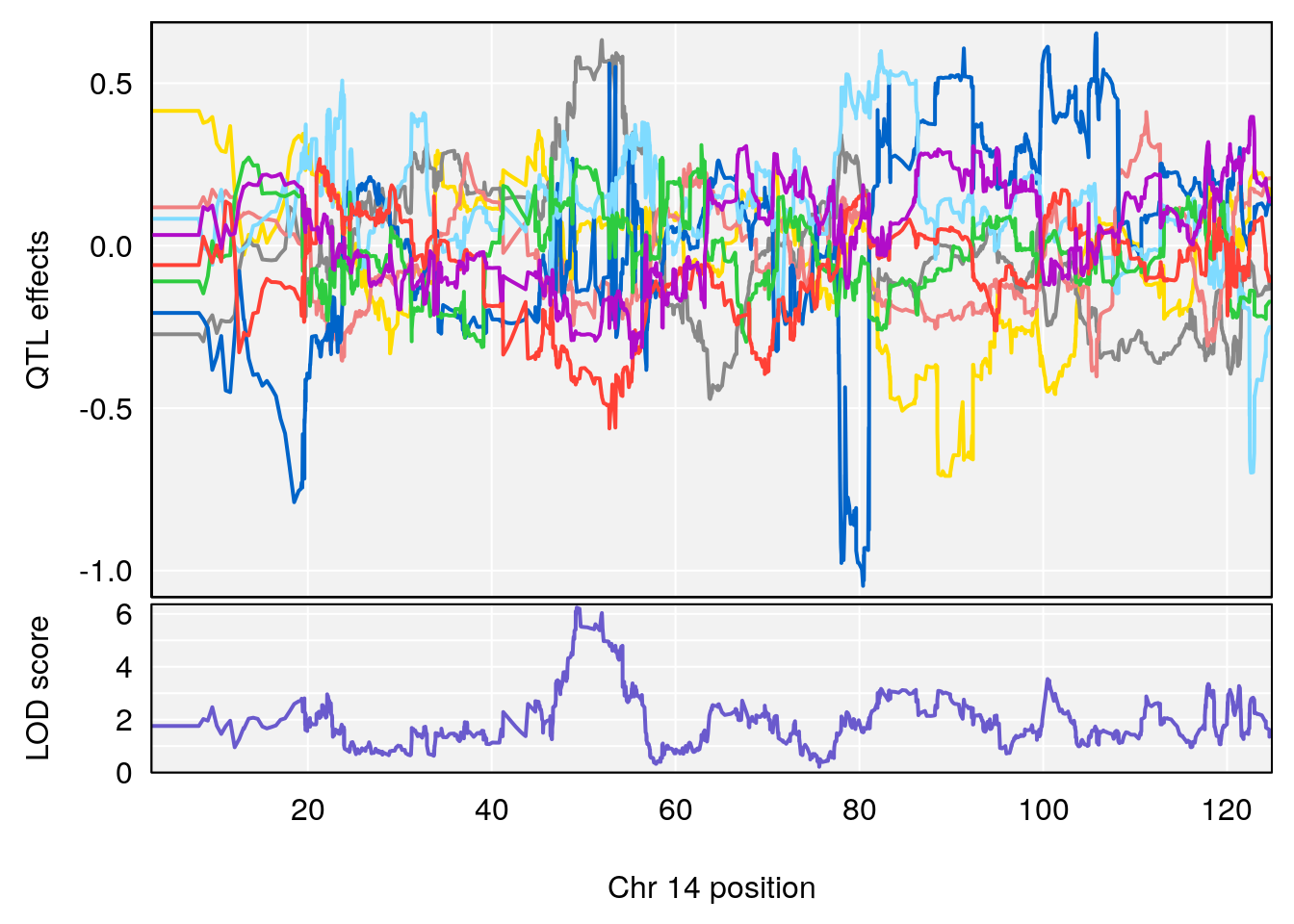
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 4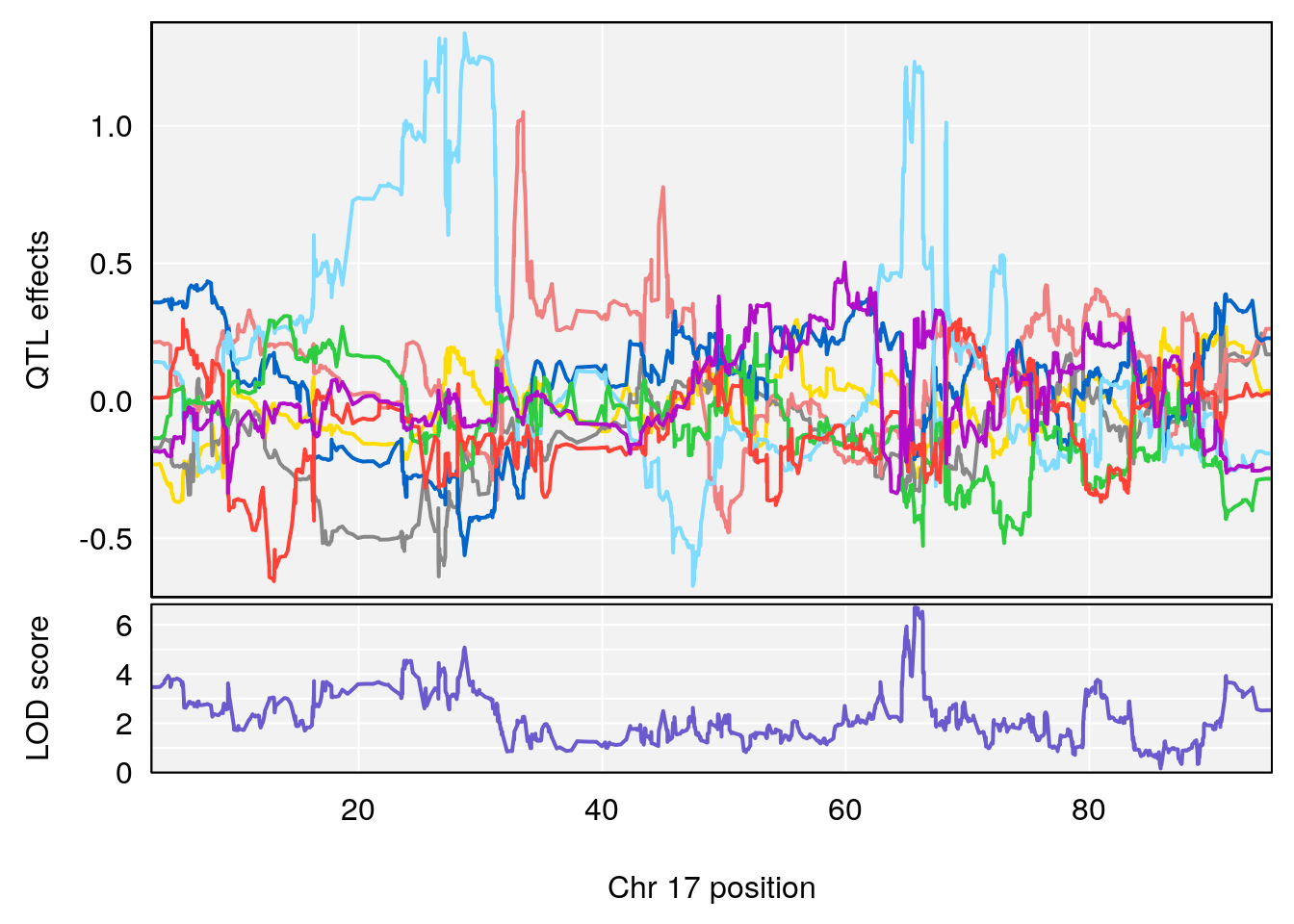
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
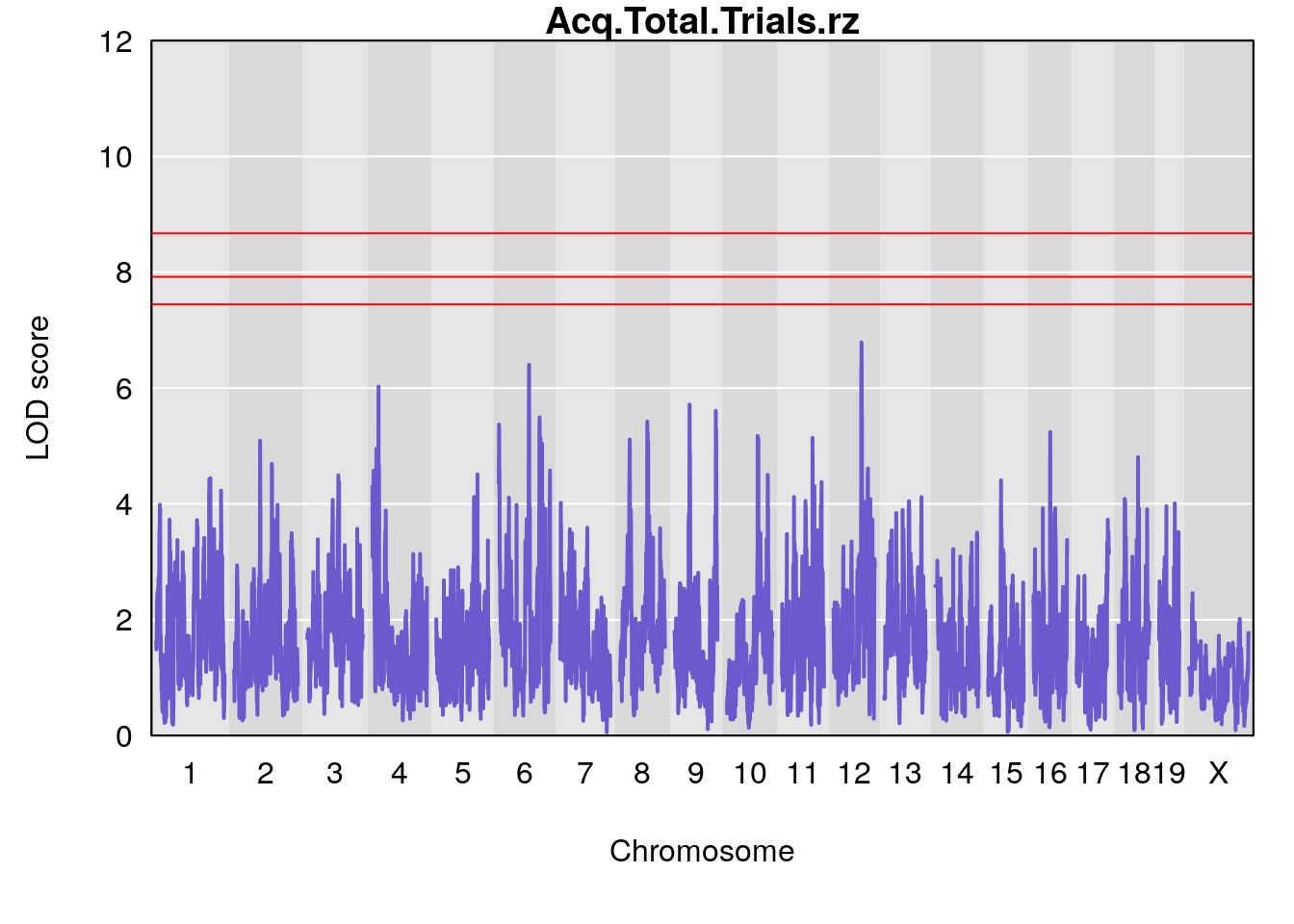
# [1] "Acq.Total.Trials.rz"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 4 20.49269 6.027420 6.185280 21.84250
# 2 1 pheno1 6 88.69893 6.403379 4.194618 125.22781
# 3 1 pheno1 12 82.15231 6.789289 81.112677 83.46144
# [1] 1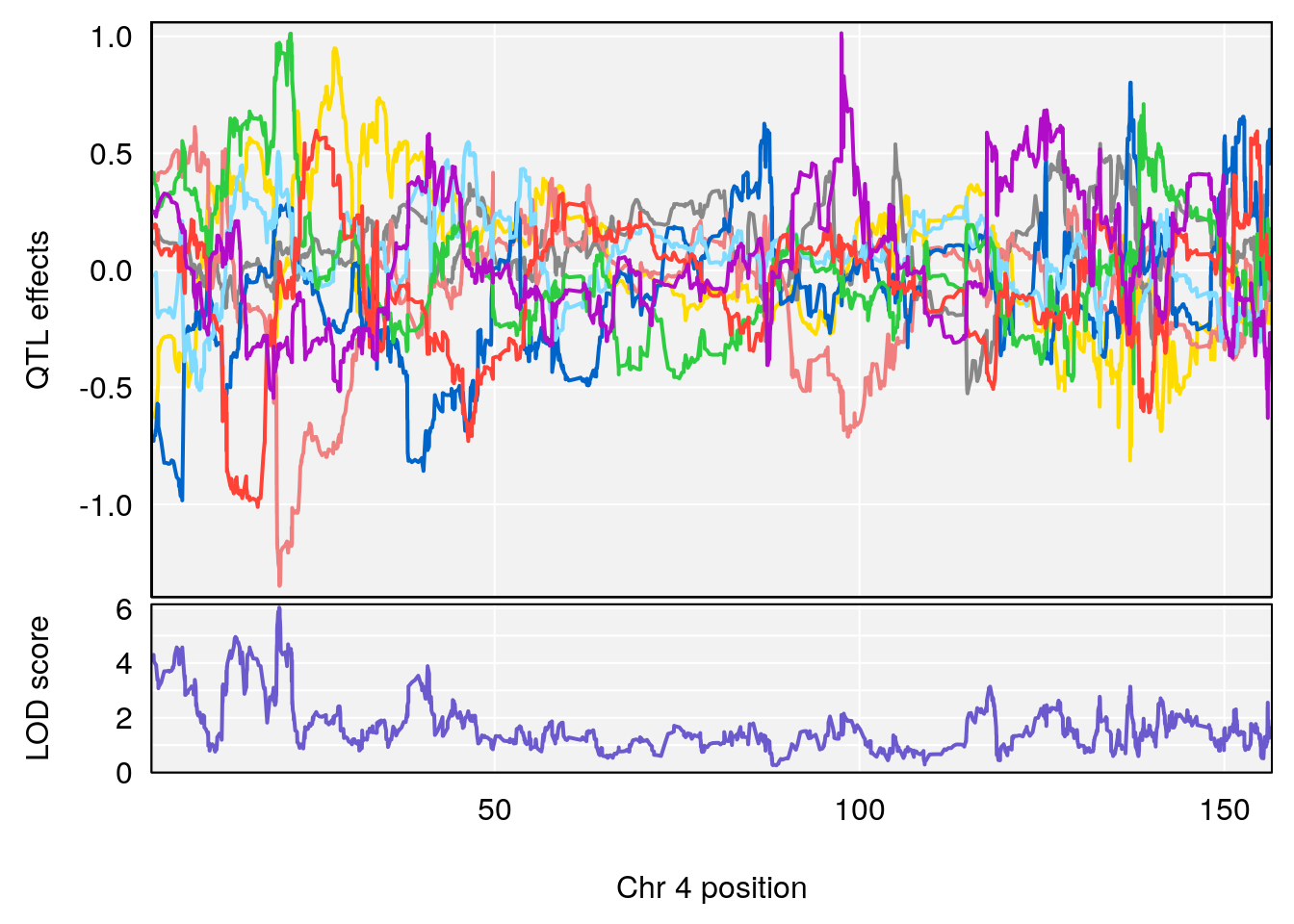
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
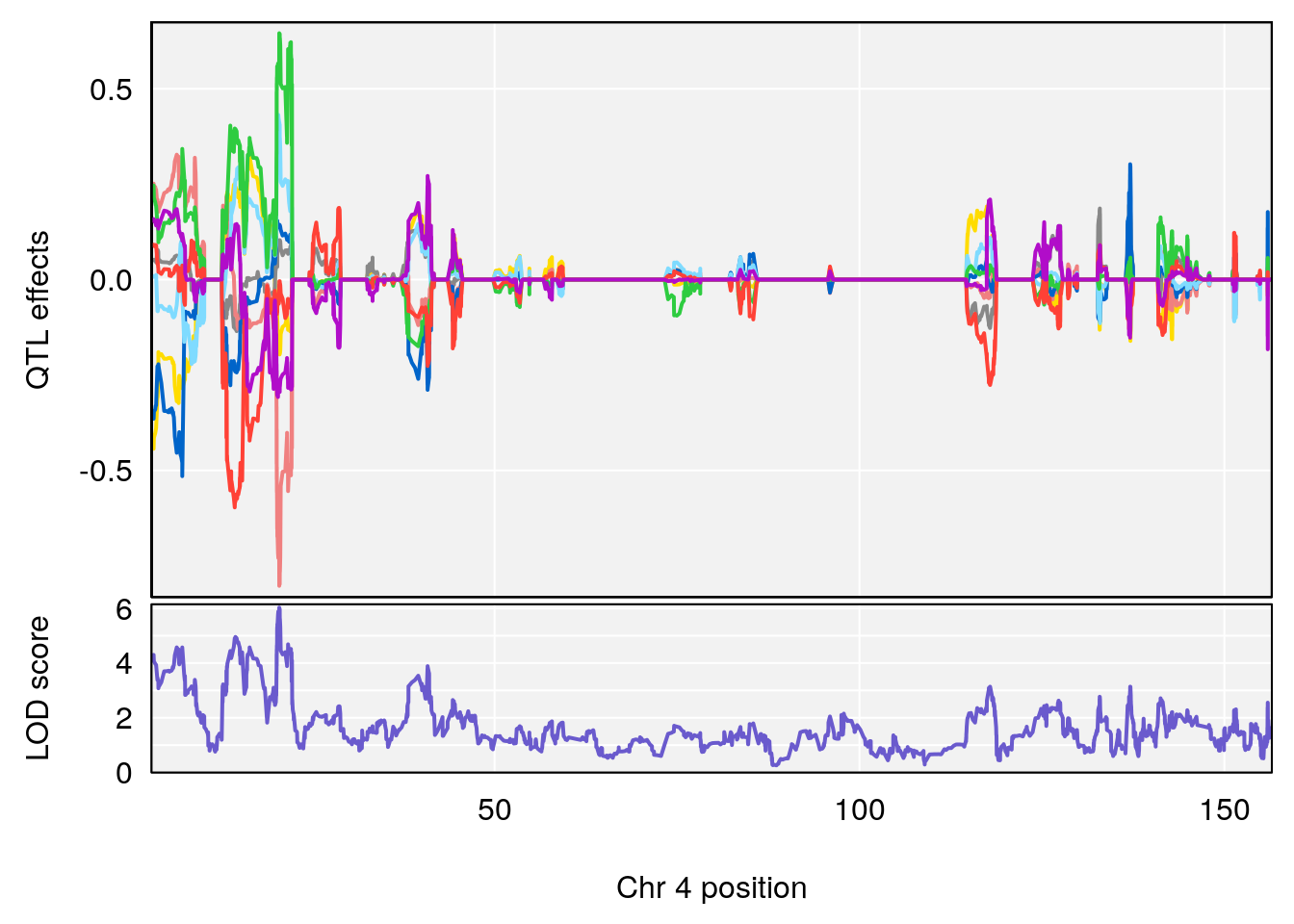
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 2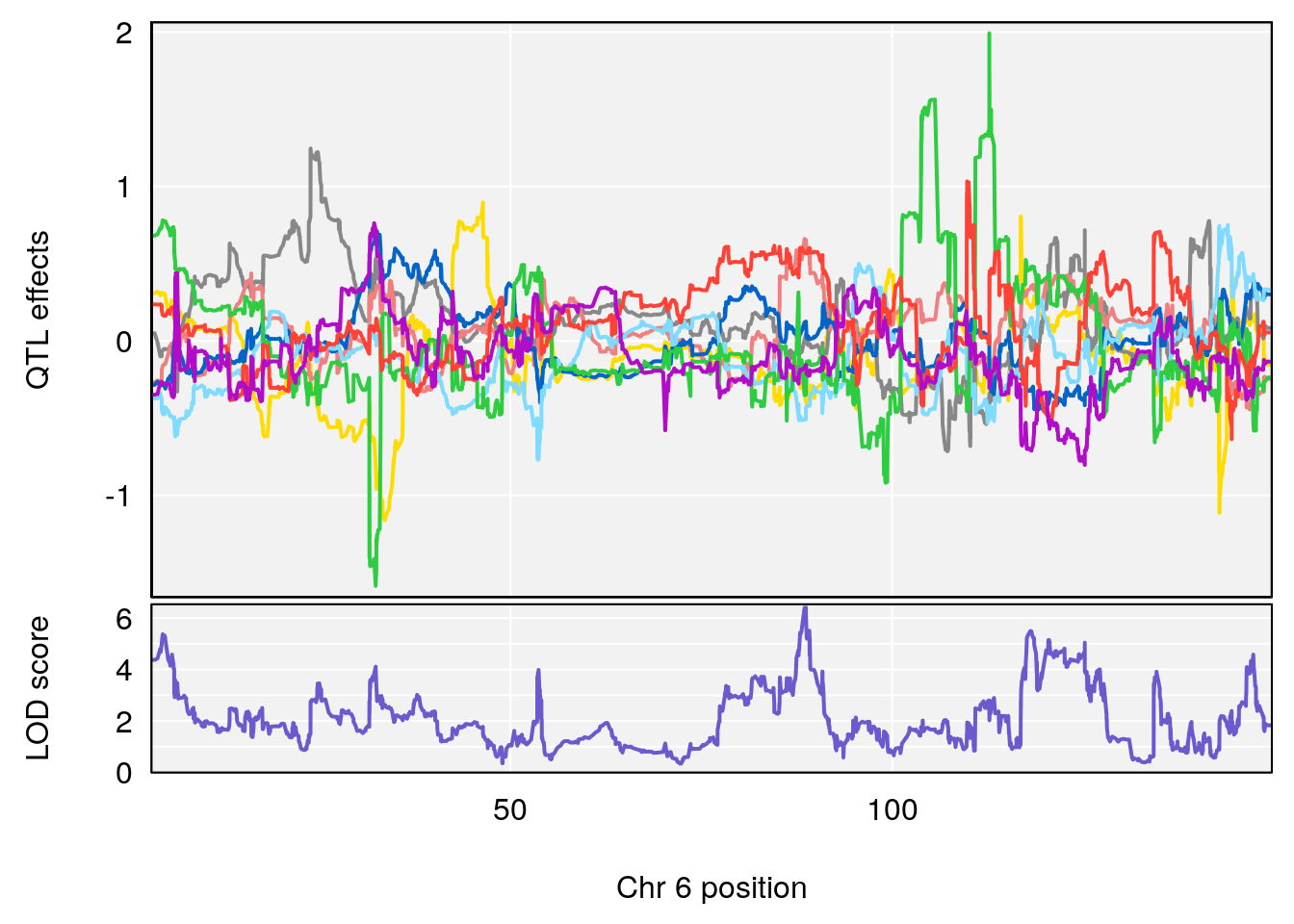
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 3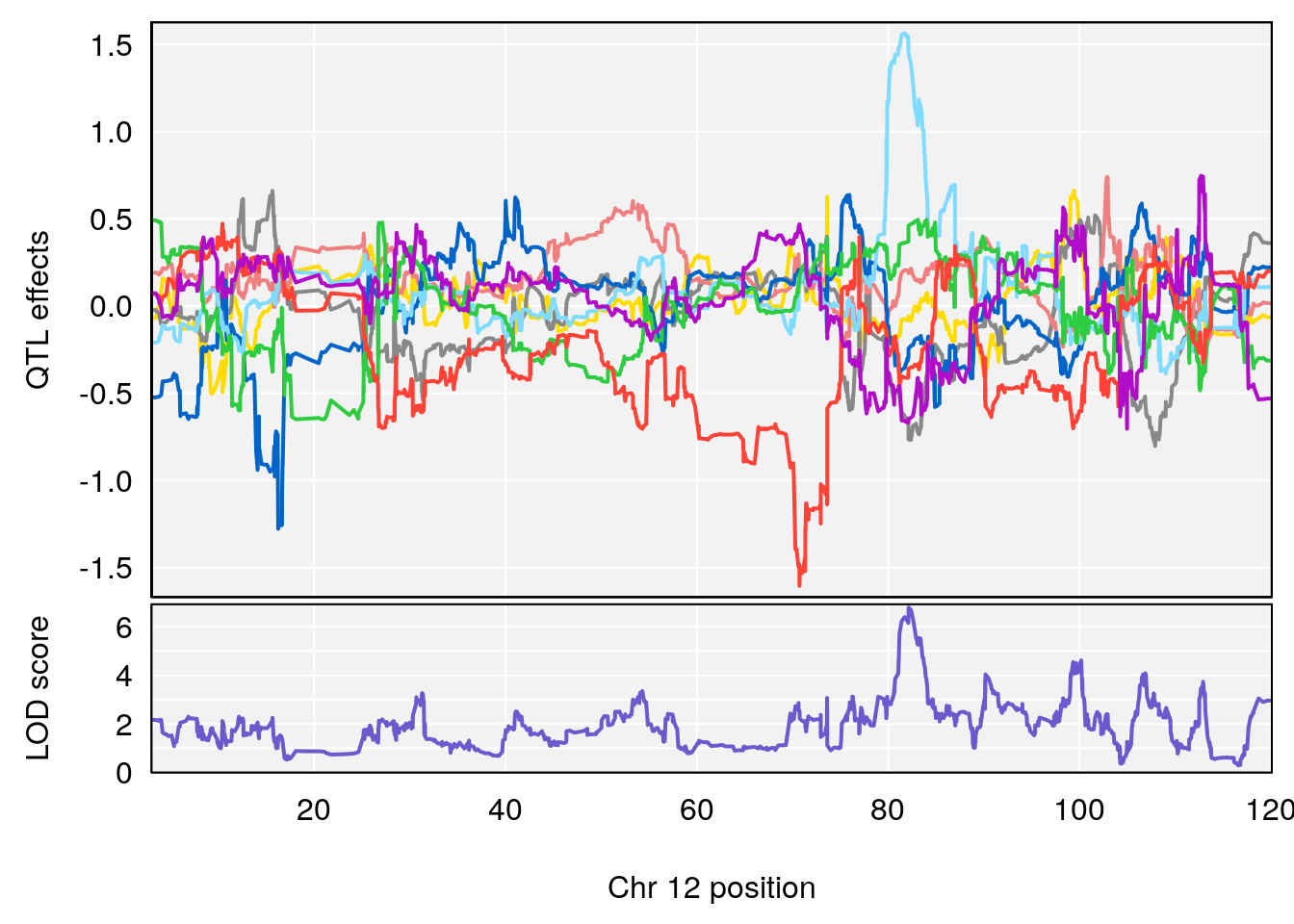
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] "Acq.Anticipatory.Correct.Responses.rz"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 168.41339 8.655344 168.23921 168.52005
# 2 1 pheno1 4 54.37504 6.720086 53.45208 86.15855
# 3 1 pheno1 14 48.92034 6.057568 44.75345 54.22123
# [1] 1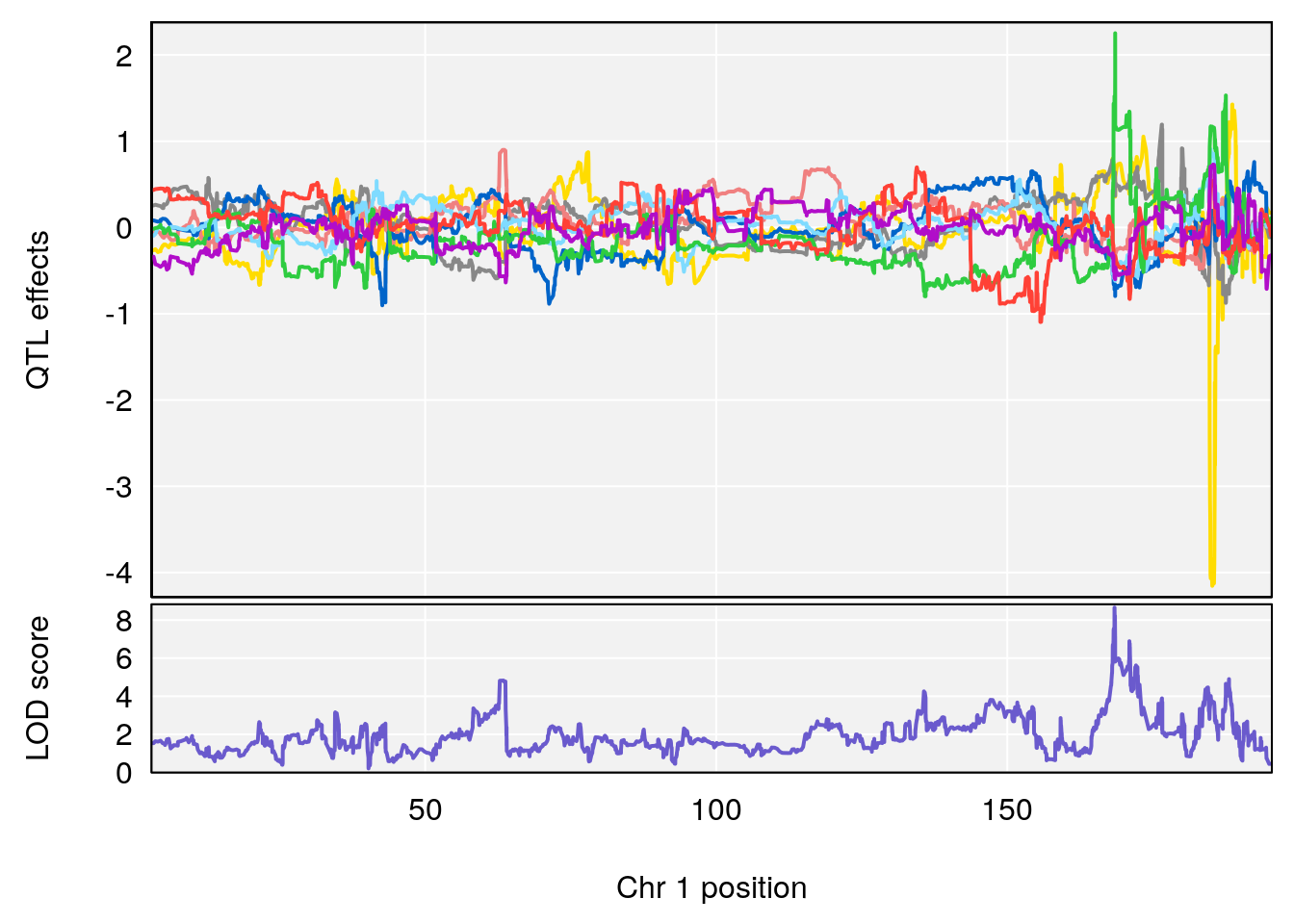
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 2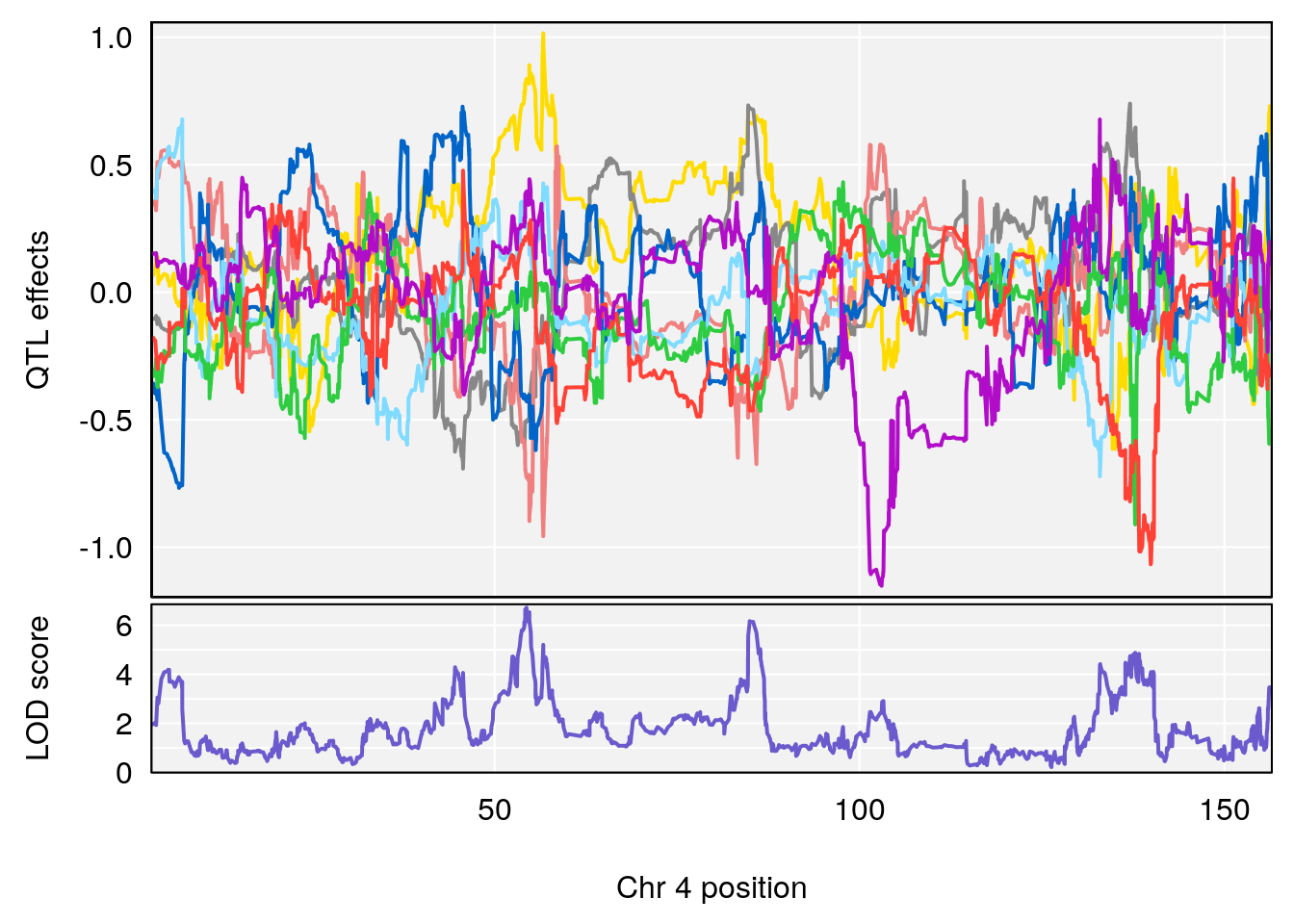
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 3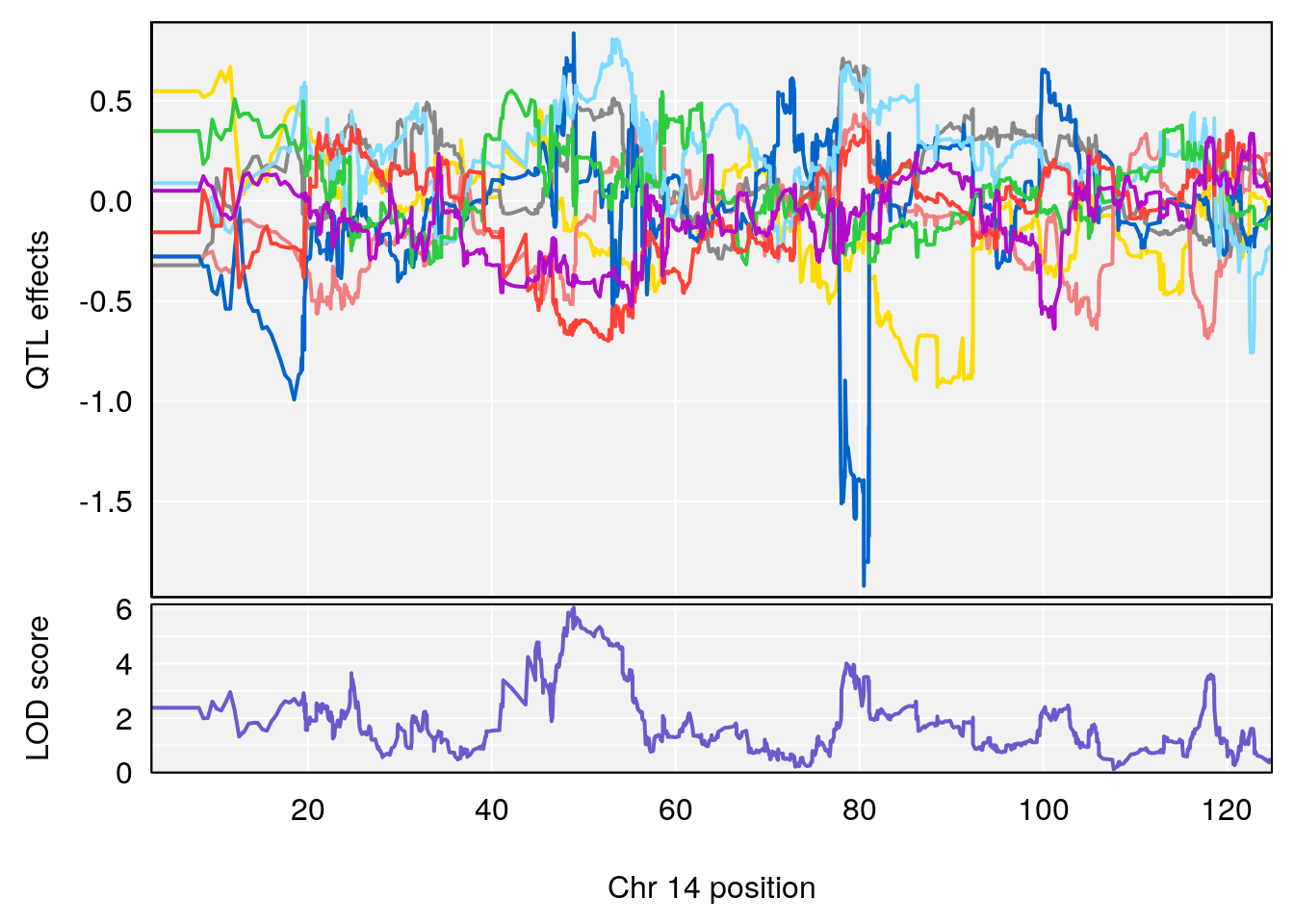
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] "Rev.Total.Trials.rz"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 2 171.9407 6.492934 168.9687 172.3610
# 2 1 pheno1 14 121.2965 6.565981 121.0353 121.5717
# [1] 1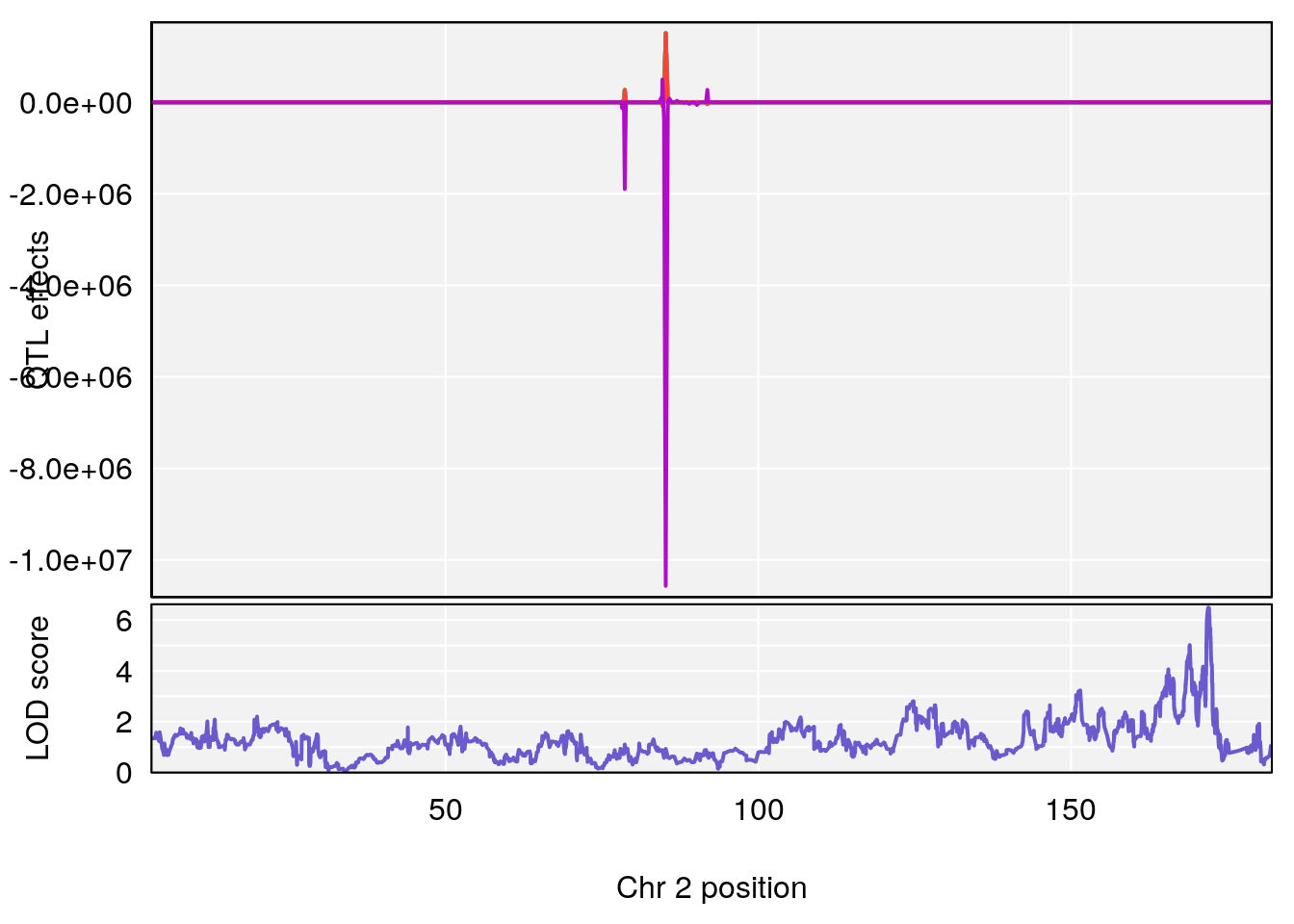
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
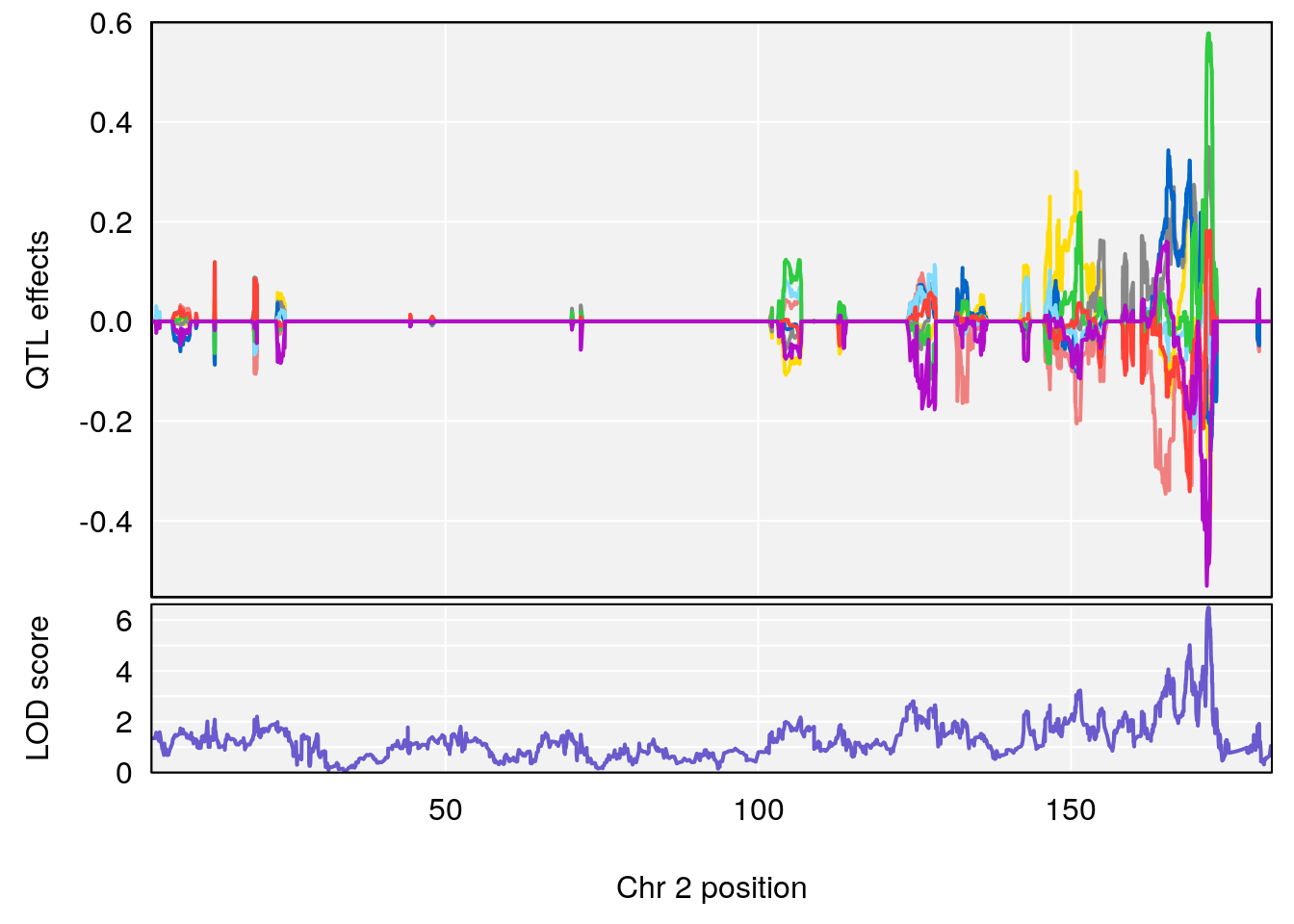
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 2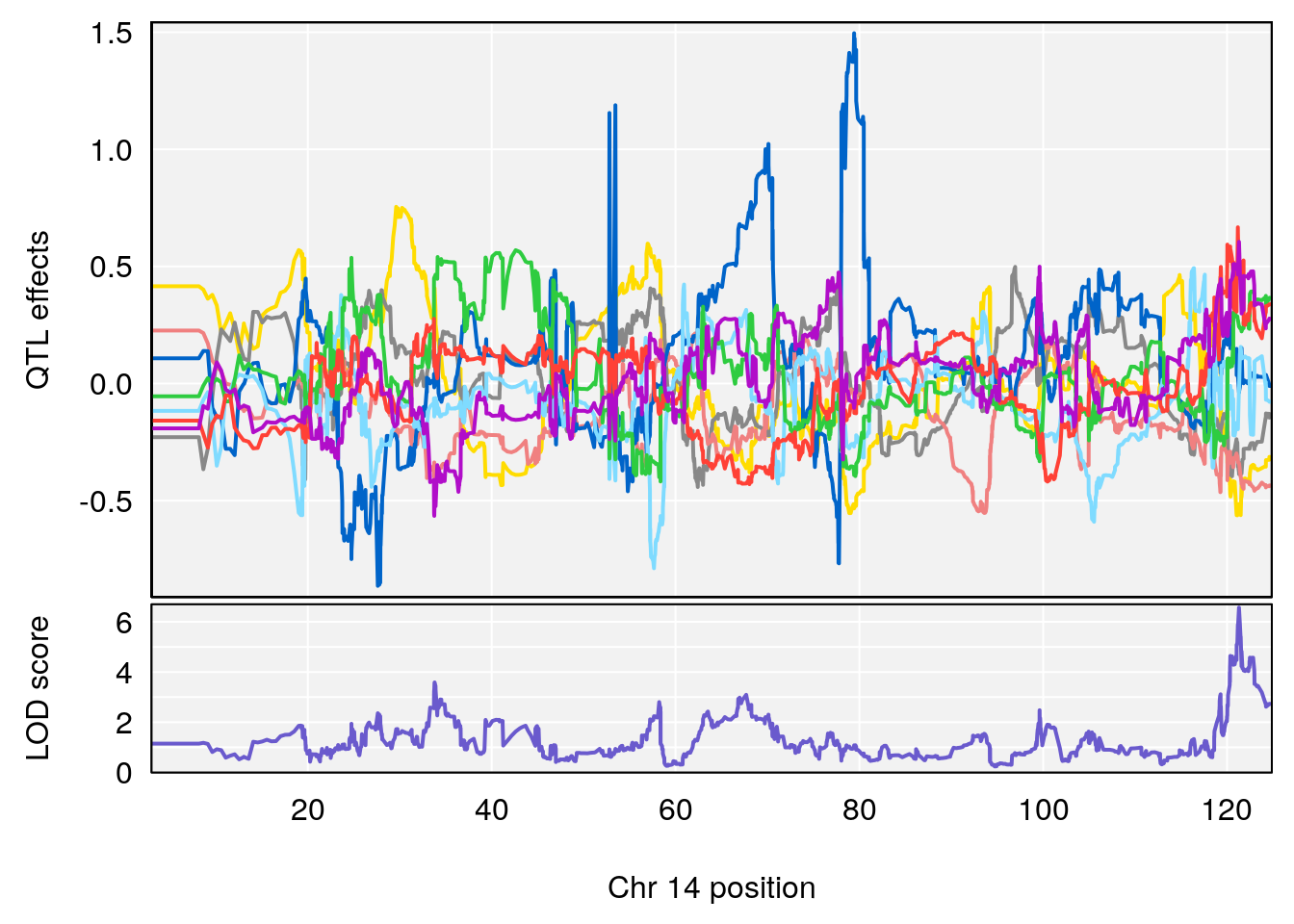
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] "Rev.Anticipatory.Incorrect.Responses.rz"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 170.963873 7.424719 168.522985 171.06618
# 2 1 pheno1 6 6.483562 6.114980 4.801035 12.08985
# 3 1 pheno1 14 49.256704 6.106932 48.800840 54.24423
# 4 1 pheno1 17 65.686785 6.125969 28.589293 66.34240
# [1] 1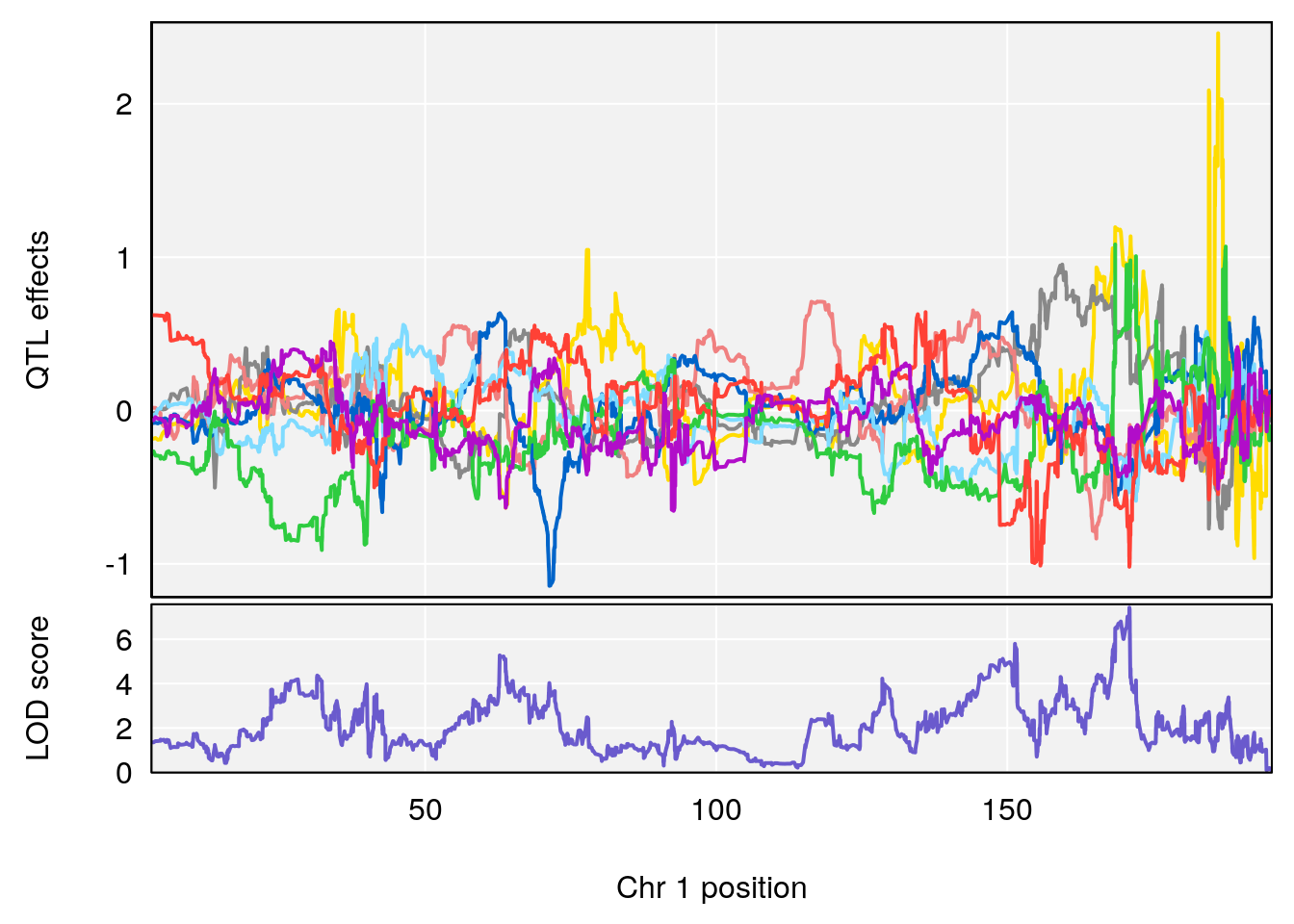
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
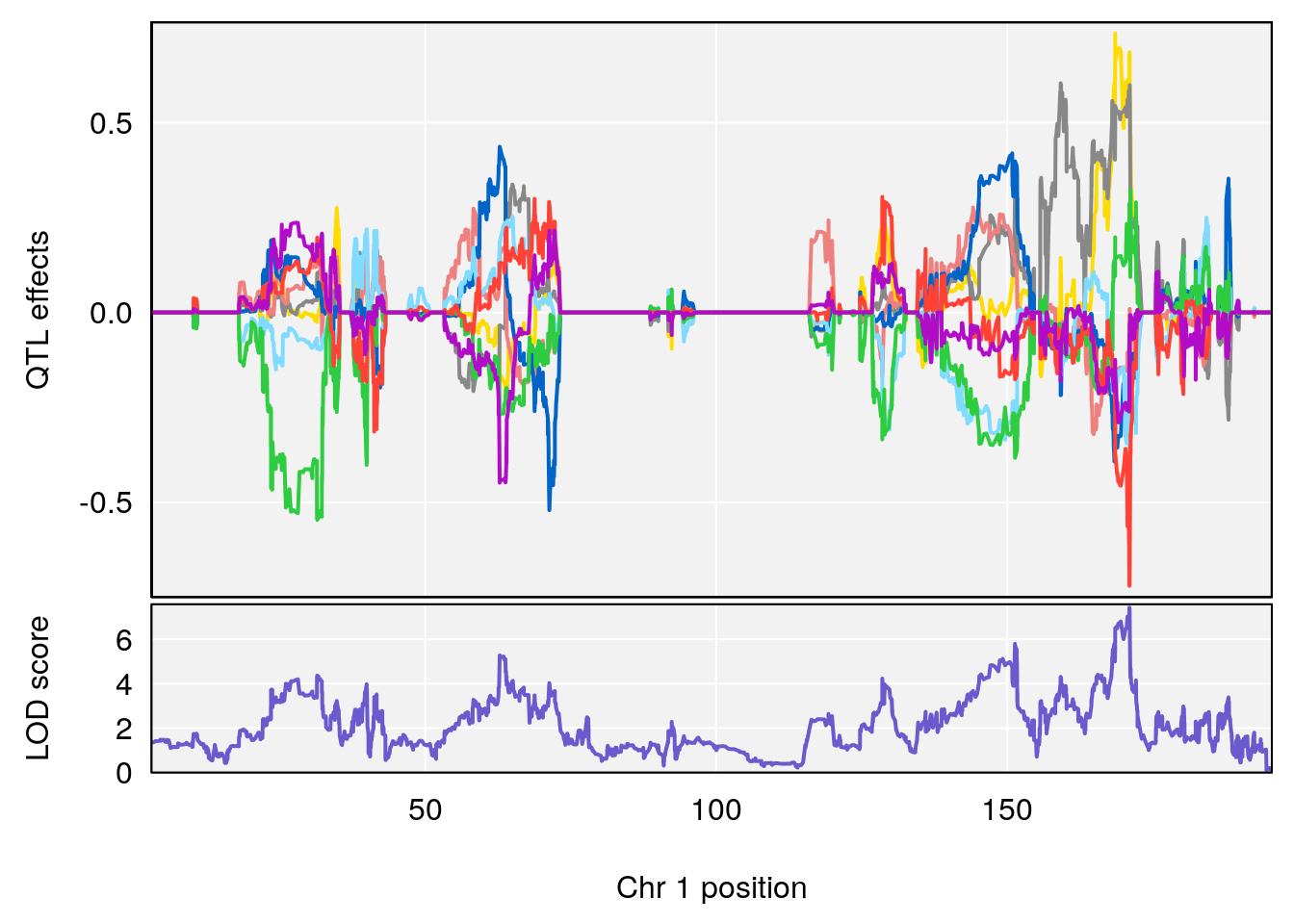
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 2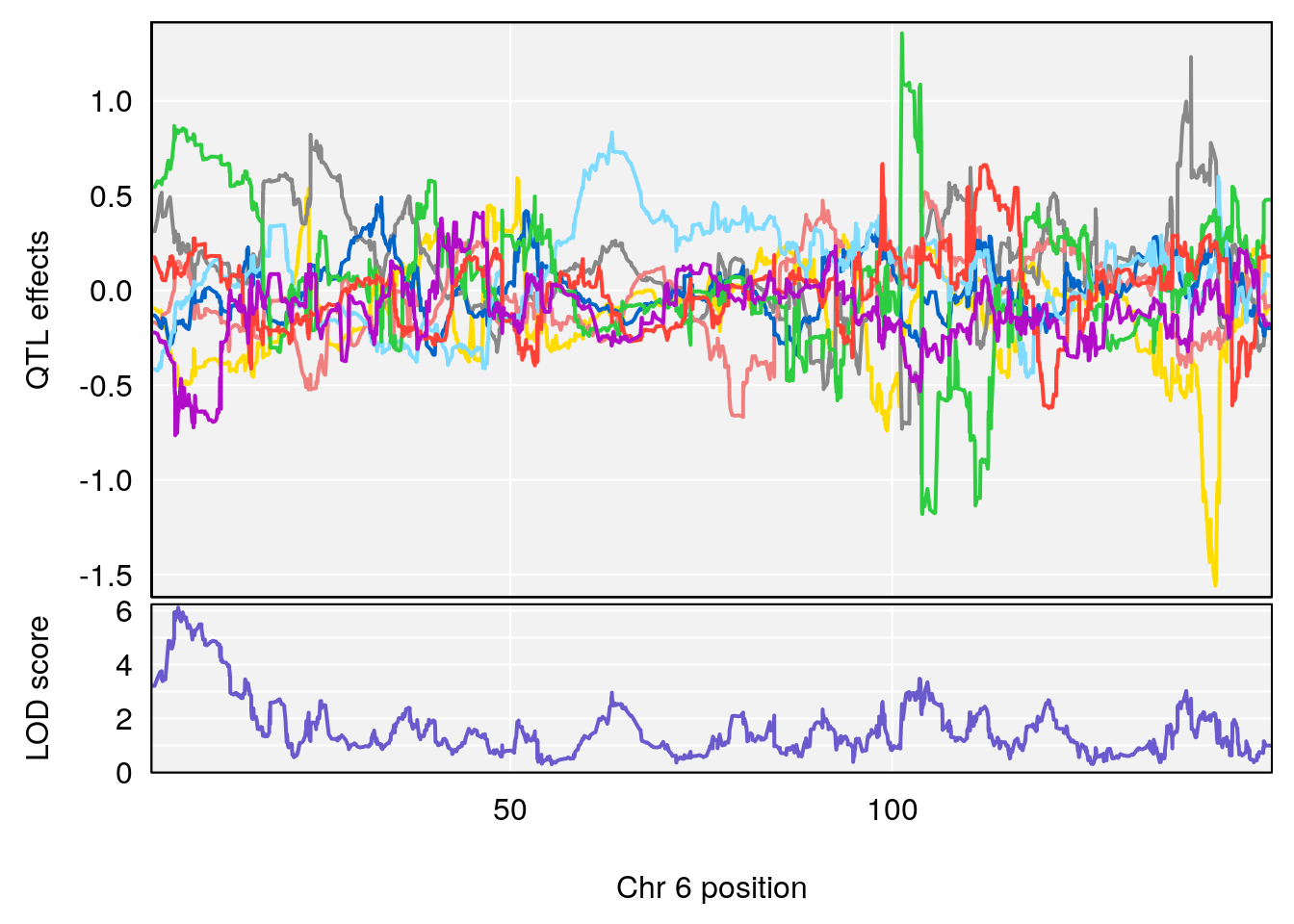
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 3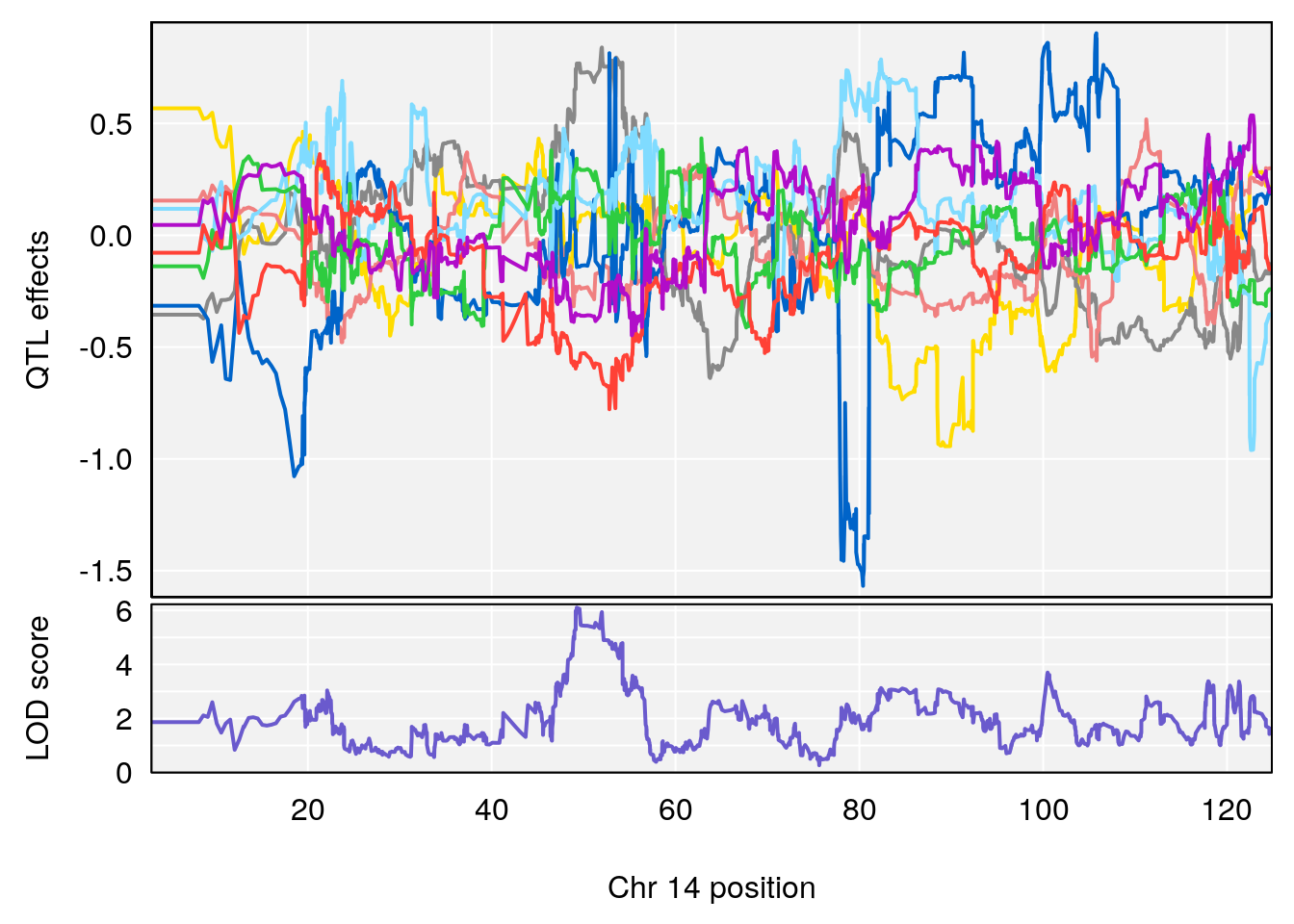
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
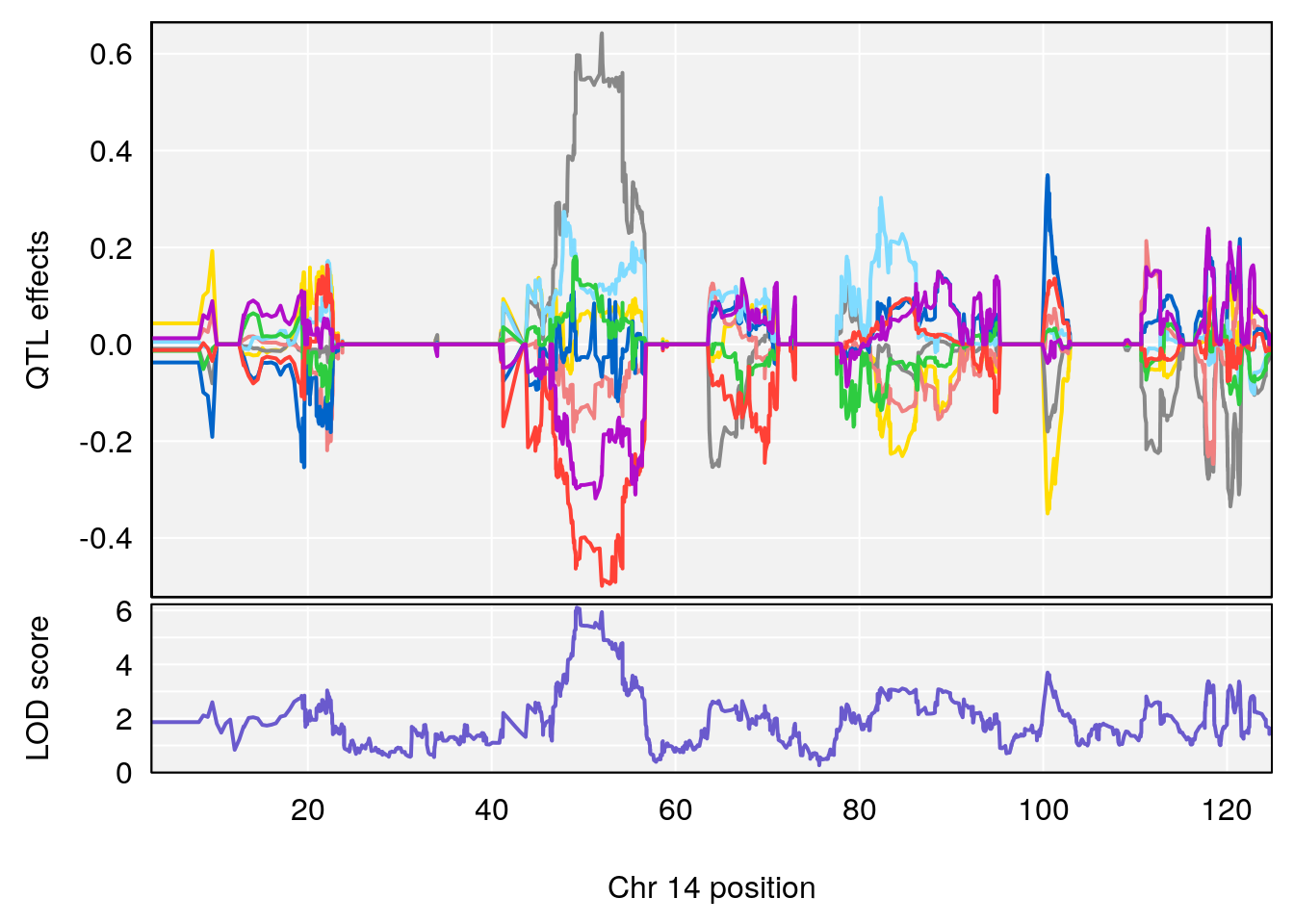
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 4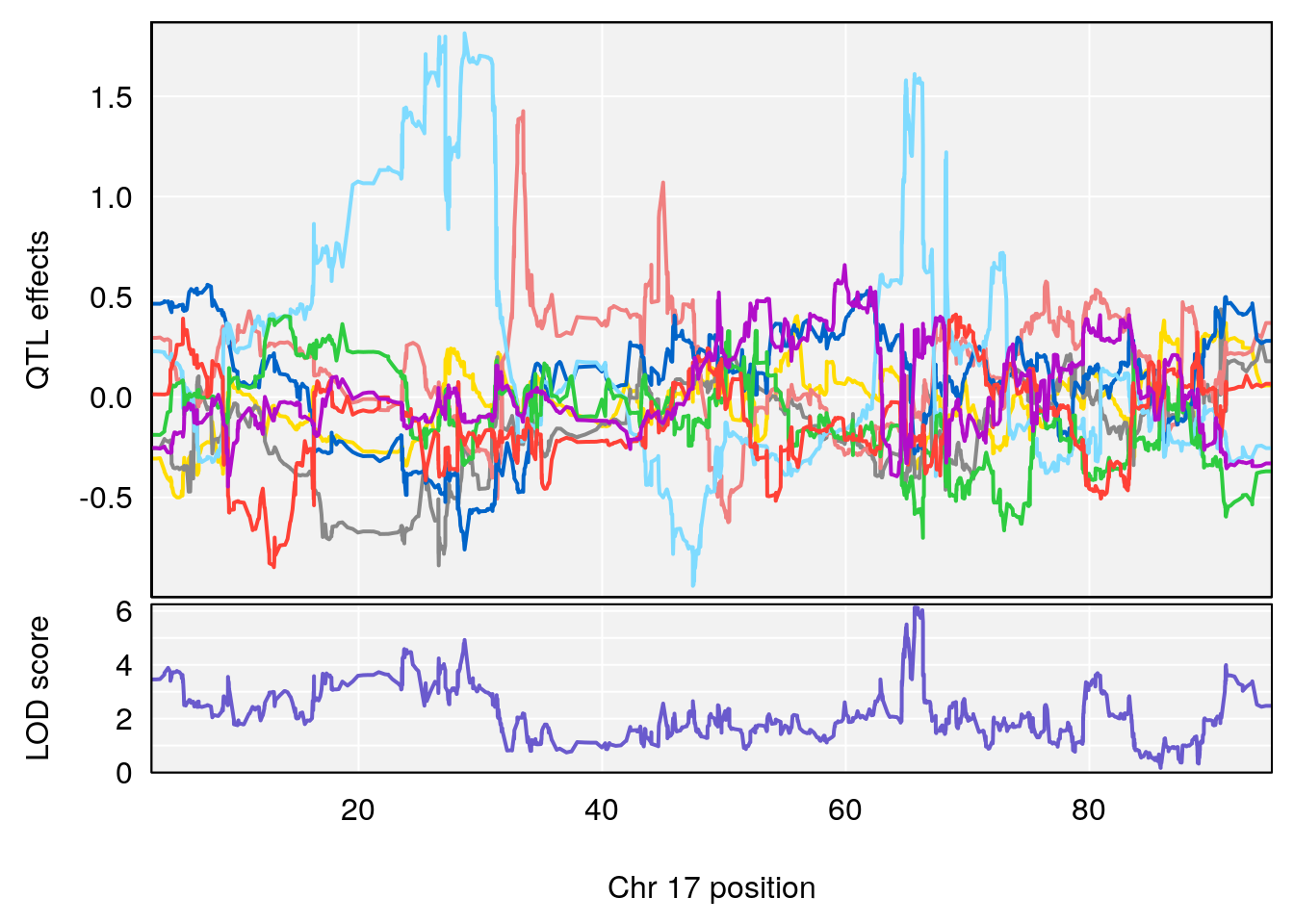
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
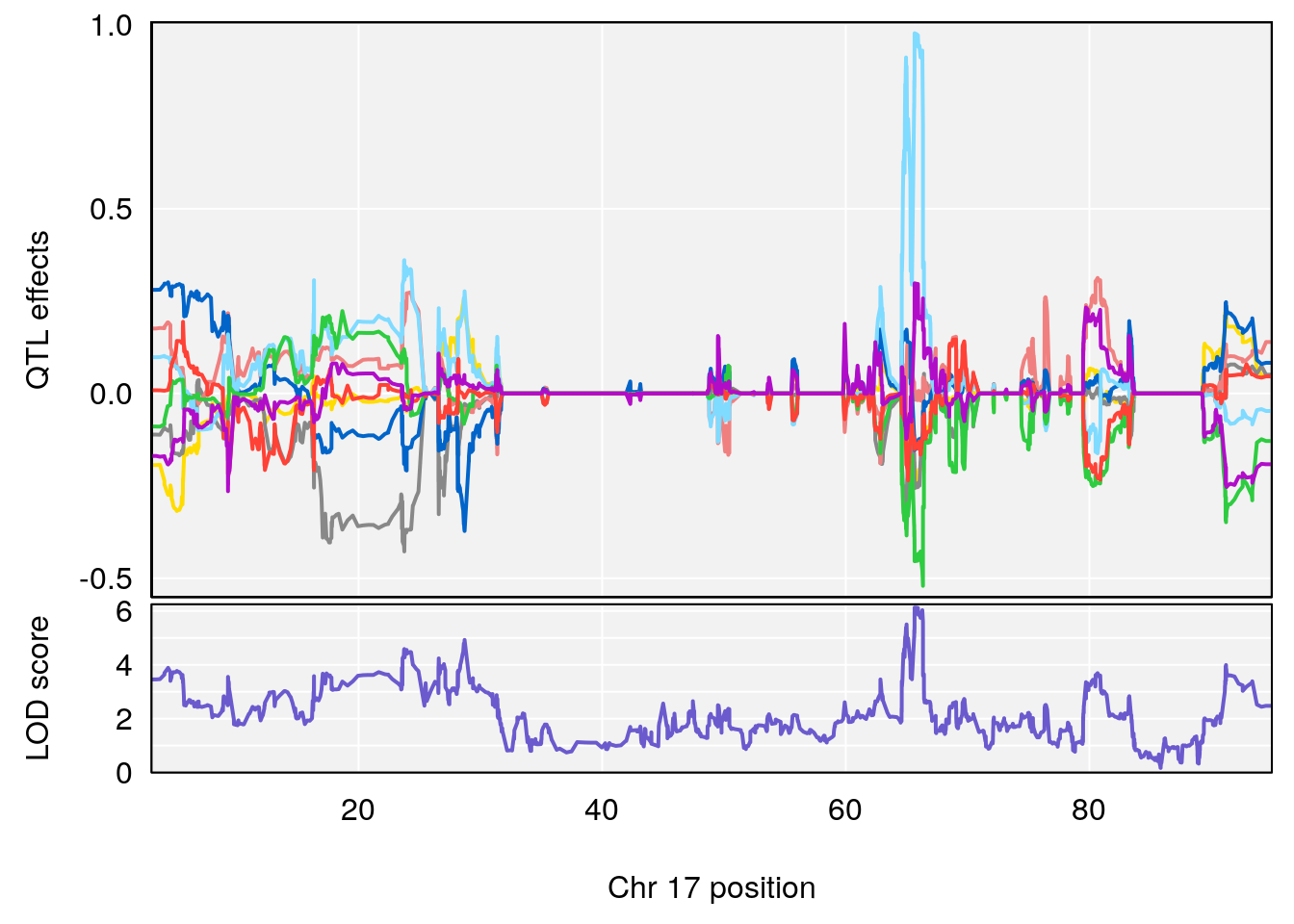
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] "Diff.Total.Trials.rz"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 7 80.80292 7.763503 80.09433 81.52958
# 2 1 pheno1 14 121.29944 6.447166 120.54323 123.78109
# [1] 1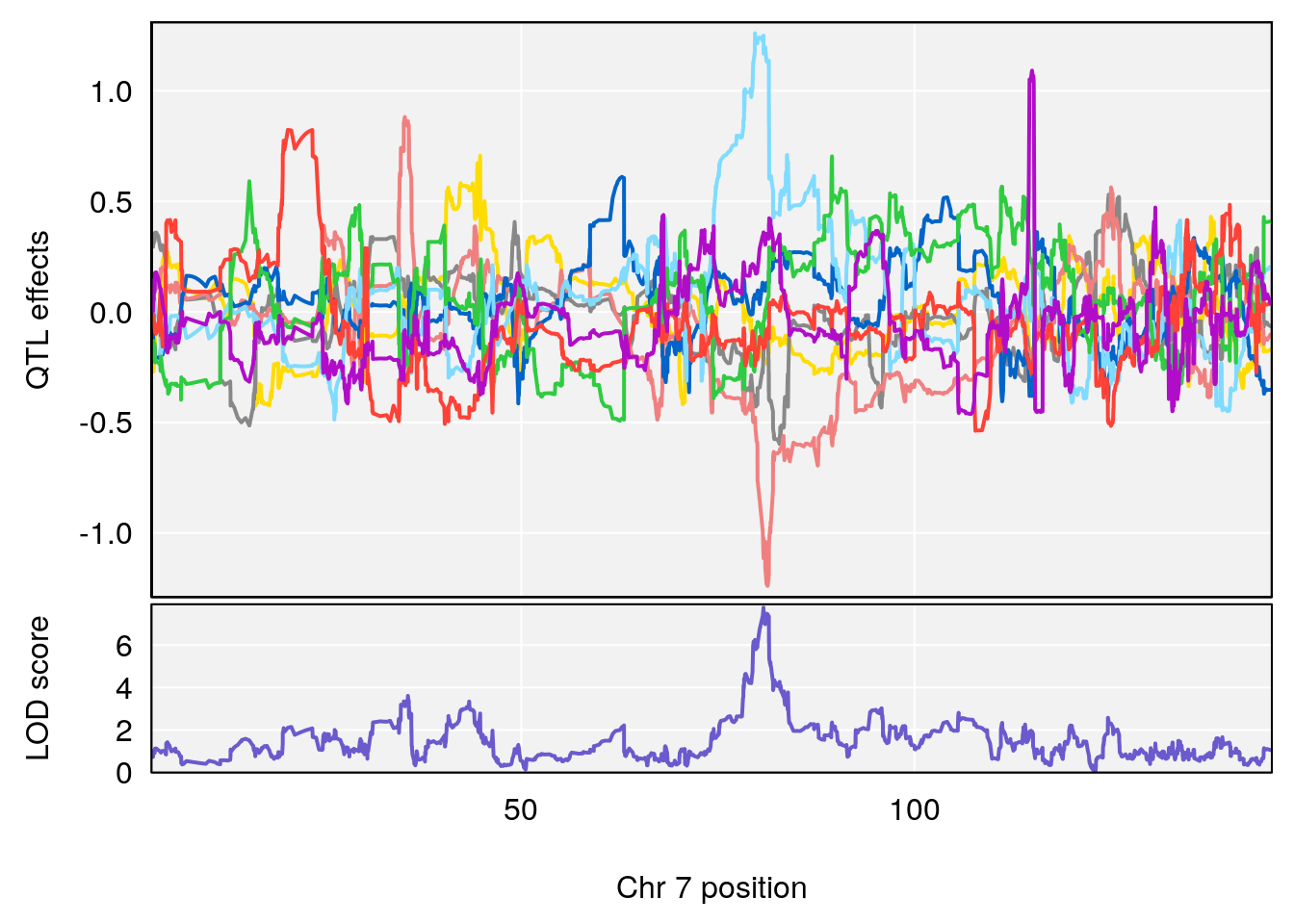
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
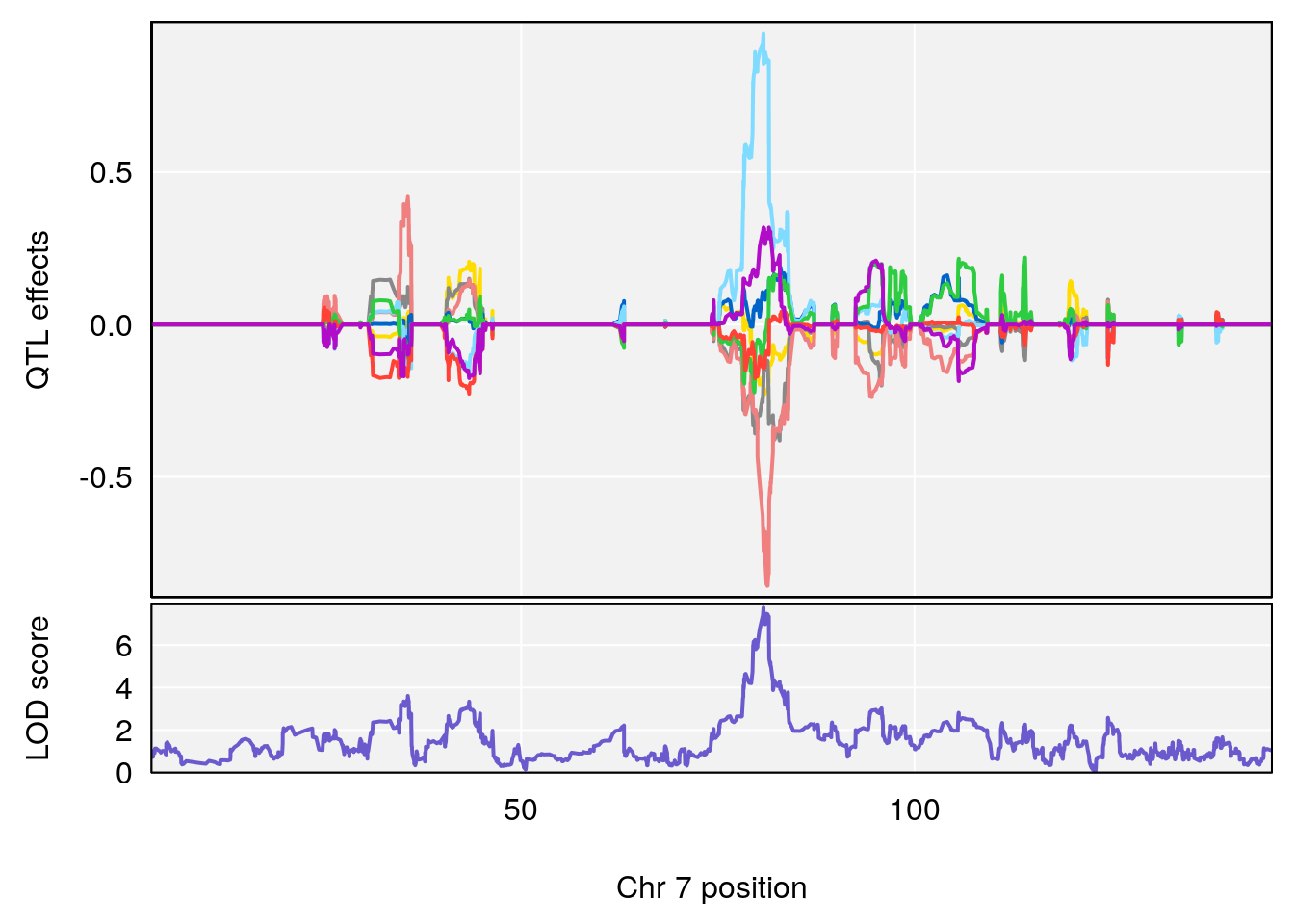
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 2
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |

| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] "Acq.Anticipatory.Correct.Responses.nooutlier.rz"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 168.400920 7.359211 168.222977 170.984487
# 2 1 pheno1 4 54.375041 6.204213 53.176714 138.505716
# 3 1 pheno1 6 6.072045 6.729573 4.801035 9.764711
# [1] 1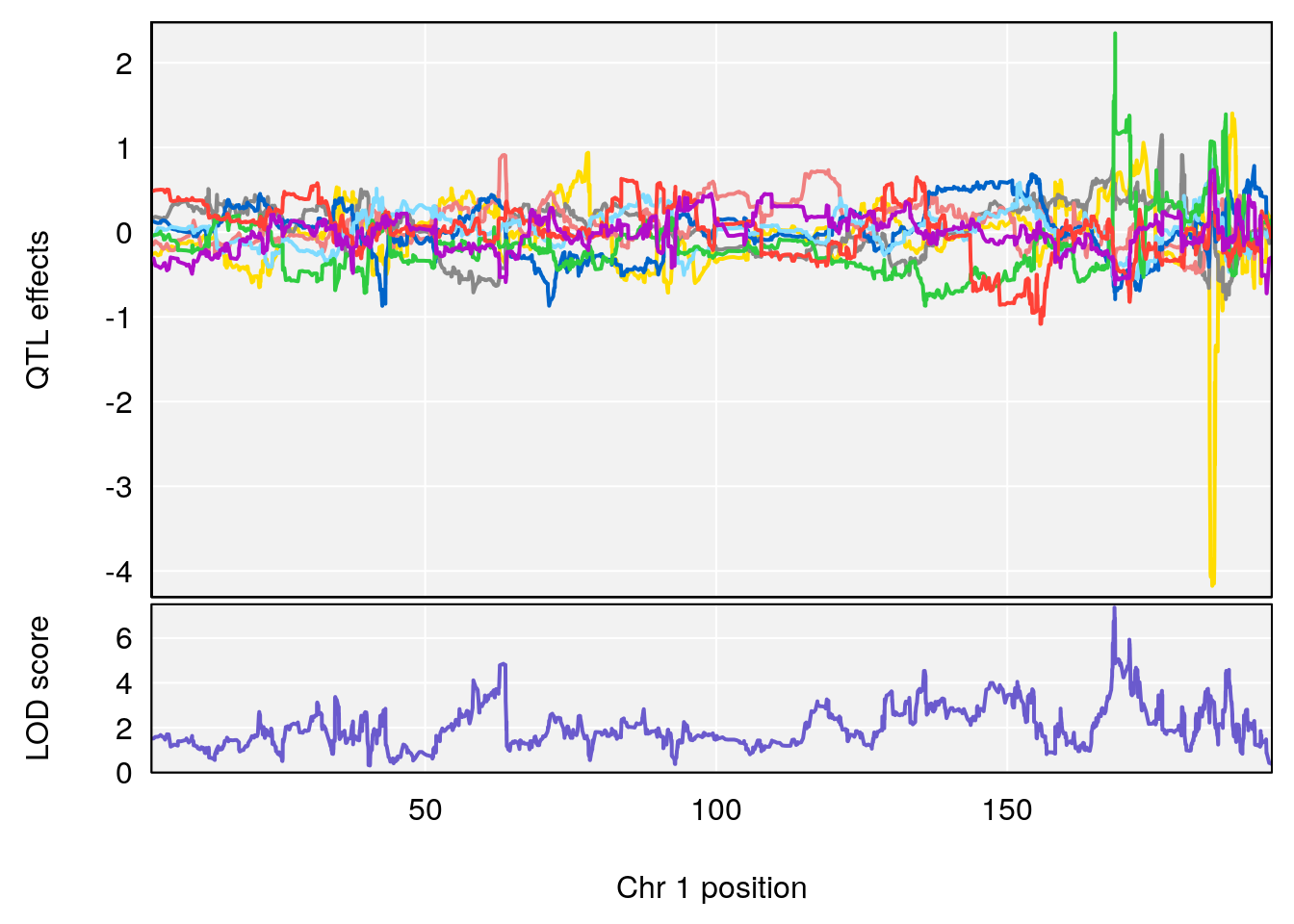
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
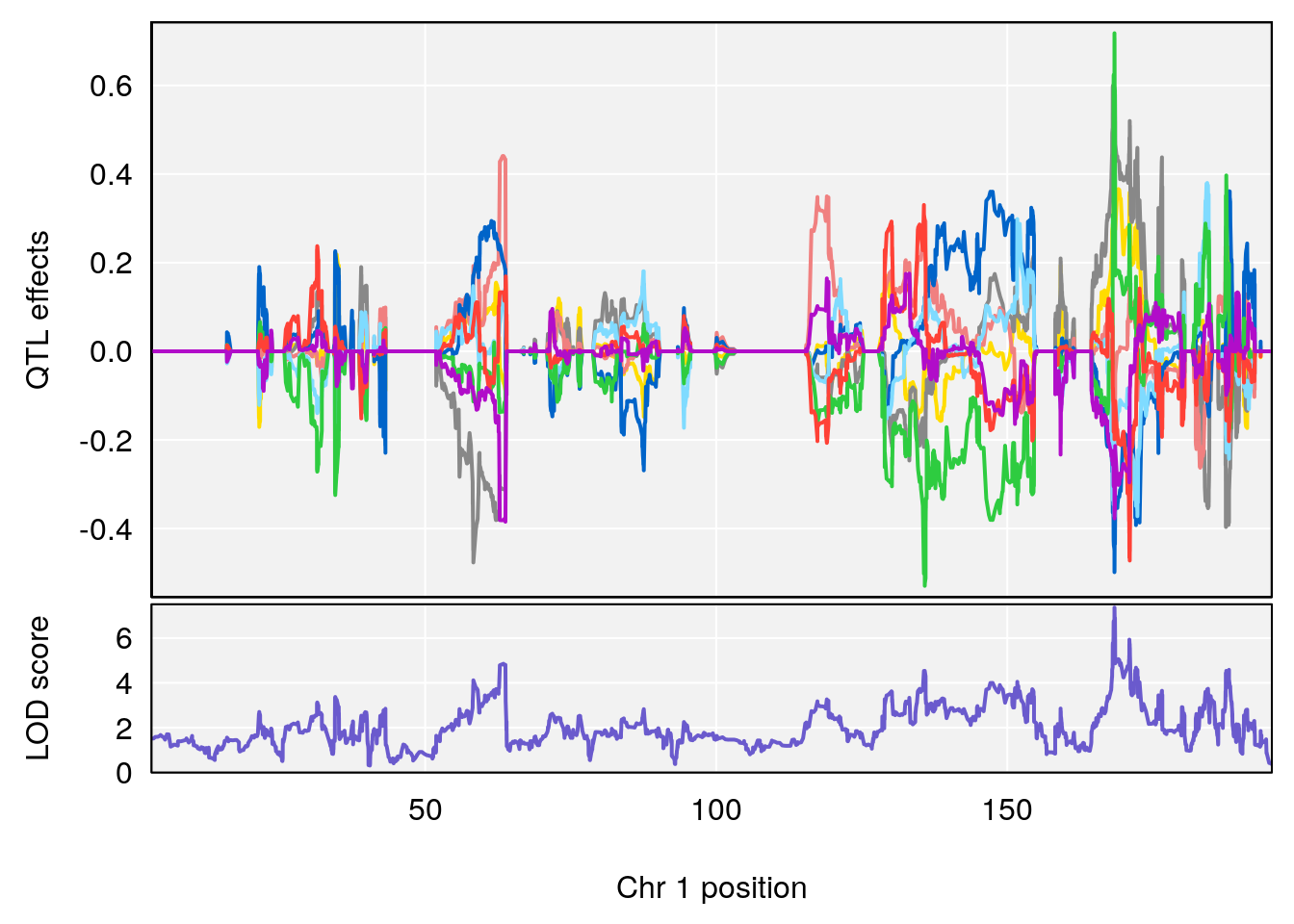
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 2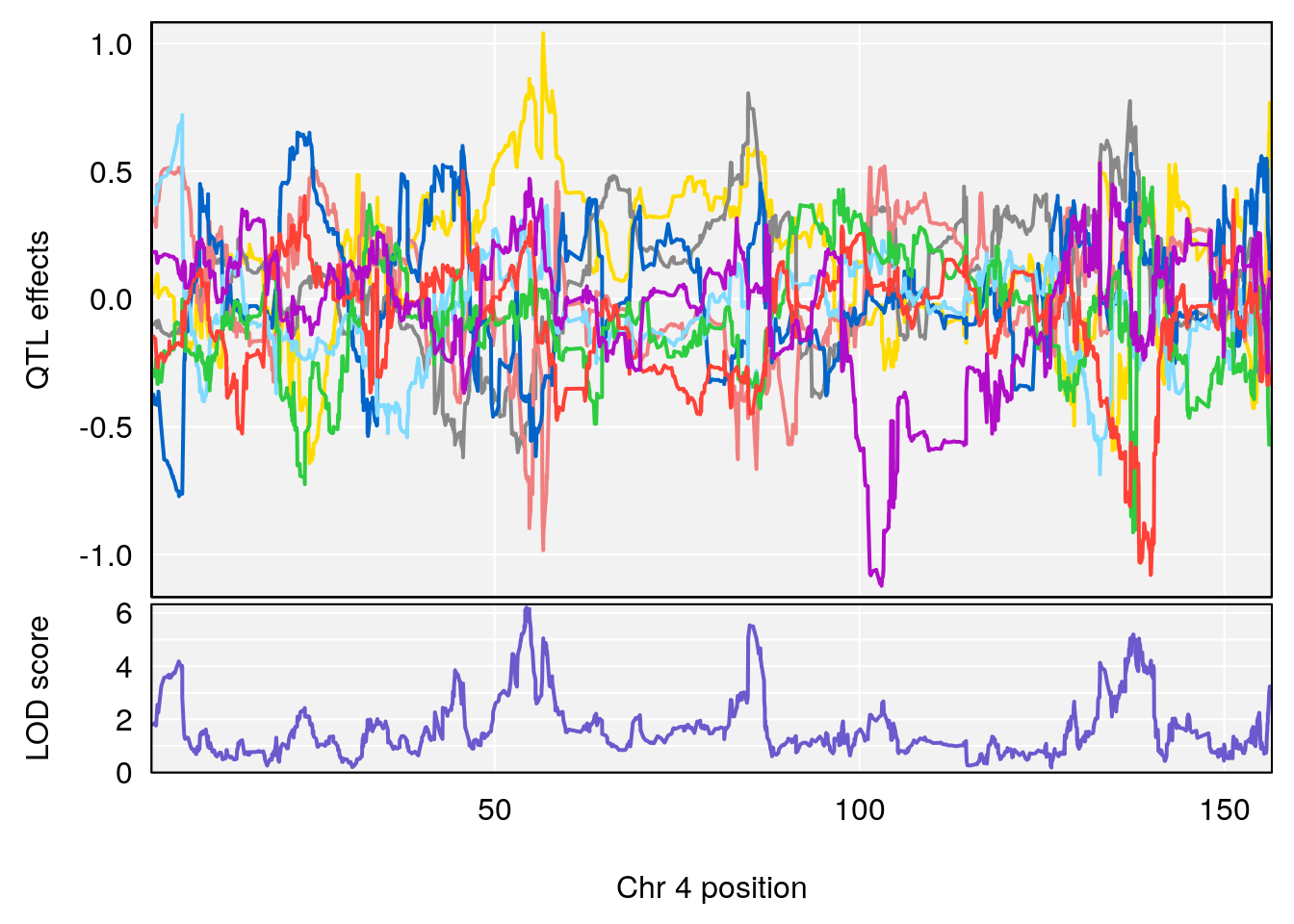
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 3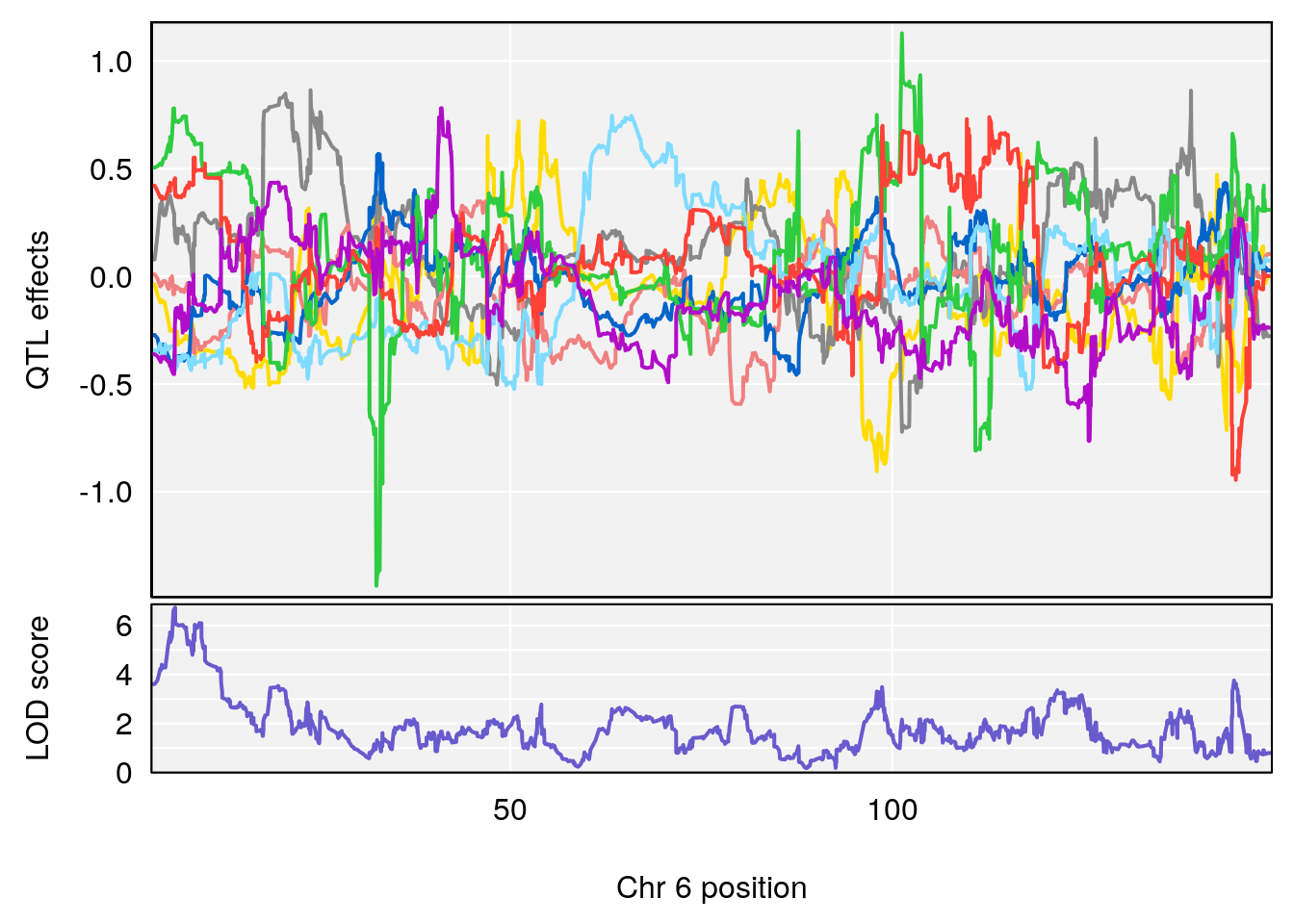
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] "Rev.Anticipatory.Incorrect.Responses.nooutlier.rz"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 170.9639 6.901609 62.68856 171.0662
# 2 1 pheno1 14 49.2567 6.230687 48.80084 54.2312
# [1] 1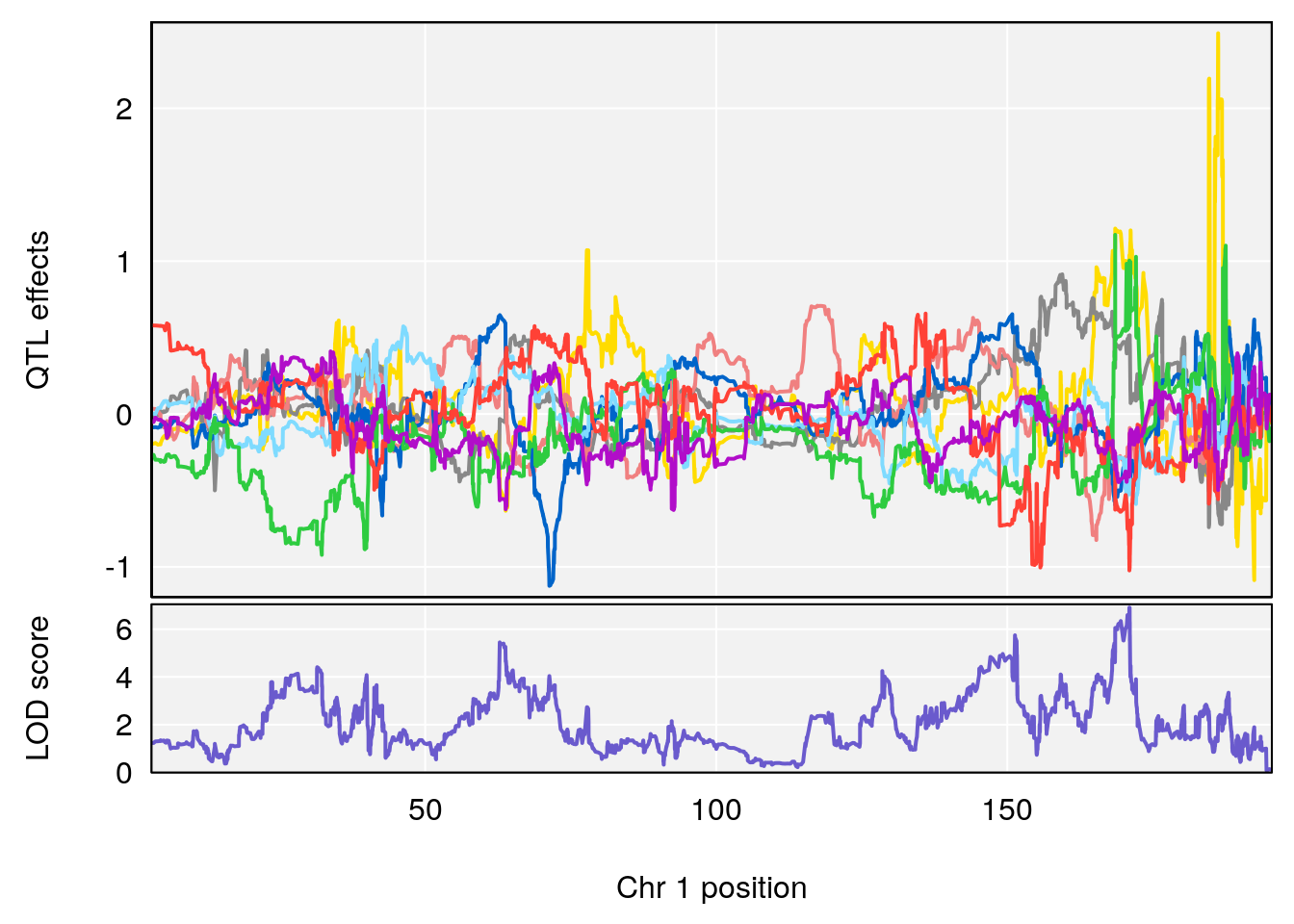
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
# [1] 2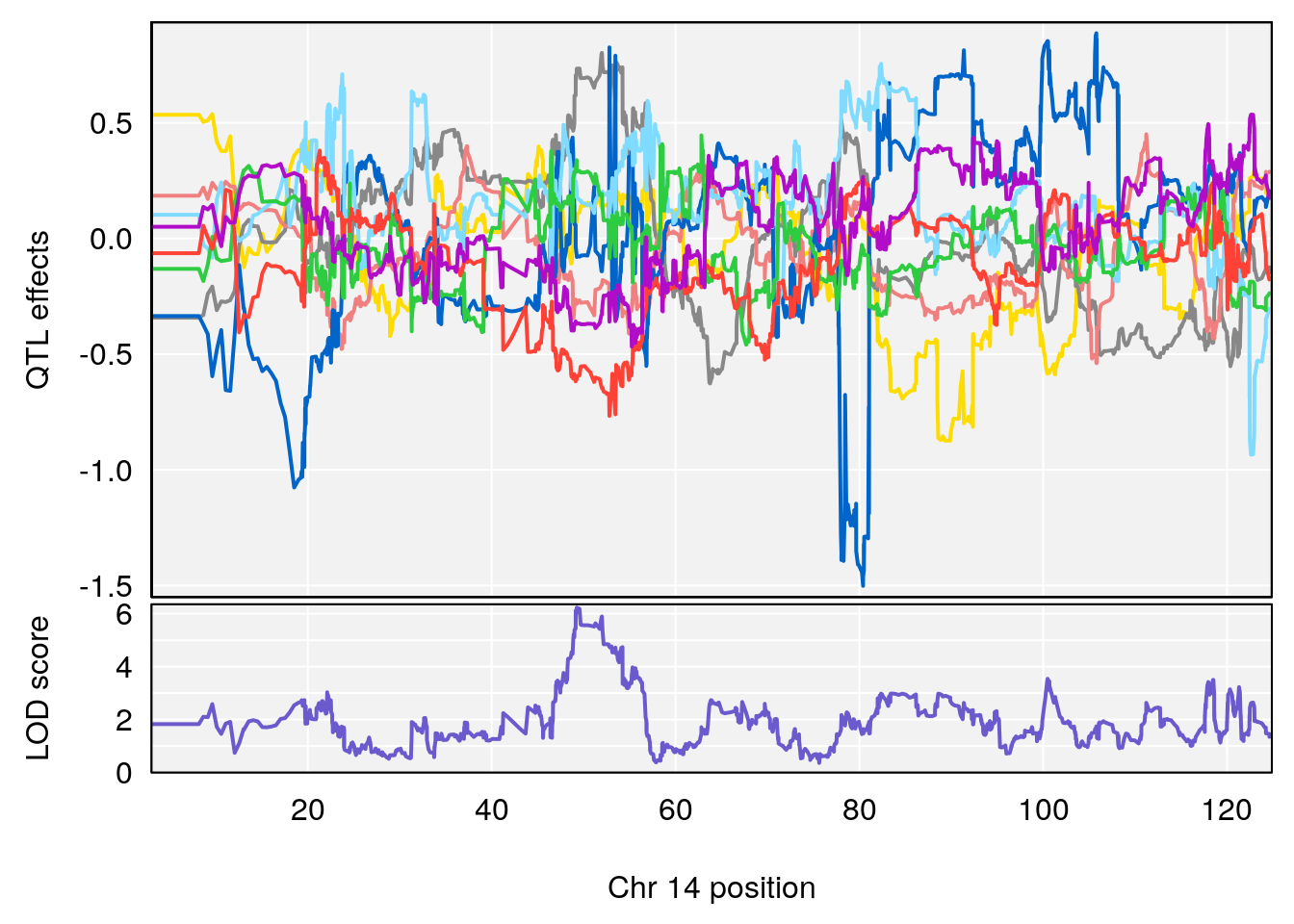
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
| Version | Author | Date |
|---|---|---|
| 0395735 | xhyuo | 2022-09-07 |
#save peaks coeff plot
for(i in names(m2.qtl.out)){
print(i)
peaks <- find_peaks(m2.qtl.out[[i]], map=pmap, threshold=6, drop=1.5)
if(nrow(peaks) > 0){
fname <- paste("output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_", str_replace_all(i, "[[:punct:]]", "") ,"_coefplot.pdf",sep="")
pdf(file = fname, width = 16, height =8)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(m2.sigqtl.coef[[i]][[p]], pmap[chr], scan1_output=m2.qtl.out[[i]], bgcolor="gray95", legend=NULL)
plot_coefCC(m2.sigqtl.blup[[i]][[p]], pmap[chr], scan1_output=m2.qtl.out[[i]], bgcolor="gray95", legend=NULL)
#plot(m2.sigqtl.snps[[i]][[p]]$lod, m2.sigqtl.snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=m2.sigqtl.genes[[i]][[p]])
}
dev.off()
}
}
# [1] "Acq.Total.Trials"
# [1] 1
# [1] 2
# [1] "Acq.Anticipatory.Correct.Responses"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] 5
# [1] 6
# [1] "Rev.Total.Trials"
# [1] 1
# [1] 2
# [1] "Rev.Anticipatory.Incorrect.Responses"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] 5
# [1] "Diff.Total.Trials"
# [1] 1
# [1] 2
# [1] "Acq.Anticipatory.Correct.Responses.nooutlier"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] "Rev.Anticipatory.Incorrect.Responses.nooutlier"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] 5
# [1] 6
# [1] "Rev.Anticipatory.Incorrect.Responses.boxcox.trans"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] "Acq.Total.Trials.rz"
# [1] 1
# [1] 2
# [1] 3
# [1] "Acq.Anticipatory.Correct.Responses.rz"
# [1] 1
# [1] 2
# [1] 3
# [1] "Rev.Total.Trials.rz"
# [1] 1
# [1] 2
# [1] "Rev.Anticipatory.Incorrect.Responses.rz"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] "Diff.Total.Trials.rz"
# [1] 1
# [1] 2
# [1] "Acq.Anticipatory.Correct.Responses.nooutlier.rz"
# [1] 1
# [1] 2
# [1] 3
# [1] "Rev.Anticipatory.Incorrect.Responses.nooutlier.rz"
# [1] 1
# [1] 2
#save peaks coeff blup plot
for(i in names(m2.qtl.out)){
print(i)
peaks <- find_peaks(m2.qtl.out[[i]], map=pmap, threshold=6, drop=1.5)
if(nrow(peaks) > 0) {
fname <- paste("output/RL_prj/Prj01_RL_11092020_cutoff6.qtlout_", str_replace_all(i, "[[:punct:]]", "") ,"_coefplot_blup.pdf",sep="")
pdf(file = fname, width = 16, height =8)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(m2.sigqtl.blup[[i]][[p]], pmap[chr], scan1_output=m2.qtl.out[[i]], bgcolor="gray95", legend=NULL)
#plot(m2.sigqtl.snps[[i]][[p]]$lod, m2.sigqtl.snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=m2.sigqtl.genes[[i]][[p]])
}
dev.off()
}
}
# [1] "Acq.Total.Trials"
# [1] 1
# [1] 2
# [1] "Acq.Anticipatory.Correct.Responses"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] 5
# [1] 6
# [1] "Rev.Total.Trials"
# [1] 1
# [1] 2
# [1] "Rev.Anticipatory.Incorrect.Responses"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] 5
# [1] "Diff.Total.Trials"
# [1] 1
# [1] 2
# [1] "Acq.Anticipatory.Correct.Responses.nooutlier"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] "Rev.Anticipatory.Incorrect.Responses.nooutlier"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] 5
# [1] 6
# [1] "Rev.Anticipatory.Incorrect.Responses.boxcox.trans"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] "Acq.Total.Trials.rz"
# [1] 1
# [1] 2
# [1] 3
# [1] "Acq.Anticipatory.Correct.Responses.rz"
# [1] 1
# [1] 2
# [1] 3
# [1] "Rev.Total.Trials.rz"
# [1] 1
# [1] 2
# [1] "Rev.Anticipatory.Incorrect.Responses.rz"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] "Diff.Total.Trials.rz"
# [1] 1
# [1] 2
# [1] "Acq.Anticipatory.Correct.Responses.nooutlier.rz"
# [1] 1
# [1] 2
# [1] 3
# [1] "Rev.Anticipatory.Incorrect.Responses.nooutlier.rz"
# [1] 1
# [1] 2heritability by GCTA
#gcta id
gcta.id <- read.table("/projects/csna/csna_workflow/data/GCTA/12_batches.fam", header = F, sep = " ")
#overlap
pheno.overlap <- pheno[pheno$mouseID %in% intersect(gcta.id$V1, pheno$mouseID),]
pheno.overlap$sex <- as.numeric(pheno.overlap$sex)
pheno.overlap$sex[pheno.overlap$sex == 0] <- 2 #female
dim(pheno.overlap)
#subset id
idlist <- cbind(pheno.overlap$mouseID, pheno.overlap$mouseID)
write.table(idlist, file = "/projects/csna/csna_workflow/data/GCTA/Prj01_RL_pheno_qttl2_DO_11092020/update_id.txt", sep = "\t", col.names = FALSE, row.names = FALSE, quote = FALSE)
#update sex
sex <- data.frame(id1 = pheno.overlap$mouseID,
id2 = pheno.overlap$mouseID,
sex = as.character(pheno.overlap$sex))
write.table(sex, file = "/projects/csna/csna_workflow/data/GCTA/Prj01_RL_pheno_qttl2_DO_11092020/update_sex.txt",sep = "\t", col.names = FALSE, row.names = FALSE, quote = FALSE)
#updated covariate
covar <- data.frame(id1 = pheno.overlap$mouseID,
id2 = pheno.overlap$mouseID,
sex = as.character(pheno.overlap$sex)
)
write.table(covar, file = "/projects/csna/csna_workflow/data/GCTA/Prj01_RL_pheno_qttl2_DO_11092020/update_covar.txt",sep = "\t", col.names = FALSE, row.names = FALSE, quote = FALSE)
#update pheno
update.pheno <- cbind(data.frame(FID = pheno.overlap$mouseID,
IID = pheno.overlap$mouseID),
pheno.overlap[,4:17]
)
write.table(update.pheno, file = "/projects/csna/csna_workflow/data/GCTA/Prj01_RL_pheno_qttl2_DO_11092020/update.pheno.txt",sep = "\t", col.names = FALSE, row.names = FALSE, quote = FALSE)
#subset id and update sex
system(paste0("cd /projects/csna/csna_workflow/data/GCTA/Prj01_RL_pheno_qttl2_DO_11092020/; /projects/csna/csna_workflow/data/GCTA/plink -bfile ",
"/projects/csna/csna_workflow/data/GCTA/","12_batches ",
"--keep update_id.txt ",
"--update-sex update_sex.txt ",
"--make-bed --out ",
"Prj01_RL_pheno_qttl2_DO_11092020"))
# Estimate the GRM
system(paste0("cd /projects/csna/csna_workflow/data/GCTA/Prj01_RL_pheno_qttl2_DO_11092020/; /projects/csna/csna_workflow/data/GCTA/gcta64 --bfile ",
"Prj01_RL_pheno_qttl2_DO_11092020", " --make-grm --out ",
"Prj01_RL_pheno_qttl2_DO_11092020"))
for(i in 1:length(colnames(pheno.overlap)[4:17])){
system(
paste0("cd /projects/csna/csna_workflow/data/GCTA/Prj01_RL_pheno_qttl2_DO_11092020/; /projects/csna/csna_workflow/data/GCTA/gcta64 --reml --grm ",
"Prj01_RL_pheno_qttl2_DO_11092020", " --pheno update.pheno.txt --covar update_covar.txt --mpheno ",
i," --out ",colnames(pheno.overlap)[4:17][i],".out")
)
}
#plot heritability by GCTA
hsq.gcta <- list()
phe.name <- colnames(pheno.overlap)[4:17]
for (i in phe.name){
hsq.gcta[[i]] <- read.table(file = paste0("/projects/csna/csna_workflow/data/GCTA/Prj01_RL_pheno_qttl2_DO_11092020/",i,".out.hsq"),sep = "\t", header = FALSE,fill = TRUE, stringsAsFactors = FALSE)
}
h <- data.frame(
Phenotype = phe.name,
Heritability = as.numeric(as.vector(unlist(lapply(hsq.gcta, FUN = function(x){x[5,2]})))),
SE = as.numeric(as.vector(unlist(lapply(hsq.gcta, FUN = function(x){x[5,3]})))),
Sample_size = as.numeric(as.vector(unlist(lapply(hsq.gcta, FUN = function(x){x[11,2]})))),
# Domain = sub("\\..*", "", phe.name),
stringsAsFactors = FALSE)
write.csv(h, file = "/projects/csna/csna_workflow/data/GCTA/Prj01_RL_pheno_qttl2_DO_11092020/h2.csv", row.names = F, quote = F)
#histgram
h$Heritability <- round(h$Heritability,2)
pdf(file = paste0("/projects/csna/csna_workflow/data/GCTA/Prj01_RL_pheno_qttl2_DO_11092020/","Prj01_RL_pheno_qttl2_DO_11092020__heritability_by_GCTA.pdf"), height = 10, width = 10)
p<-ggplot(data=h, aes(x=Phenotype, y=Heritability)) + #, fill=Domain, color = Domain)) +
geom_bar(stat="identity", fill = "blue", color = "blue", show.legend = FALSE) +
scale_y_continuous(breaks=seq(0.0, 1.0, 0.1)) +
geom_text(aes(label = Heritability, y = Heritability + 0.005), position = position_dodge(0.9),vjust = 0) +
theme(axis.text.x = element_text(angle = 90, hjust = 1))
p
dev.off()
#plot heritability by qtl2 array --------------
herit <- data.frame(Phenotype = names(unlist(qtl.hsq)),
Heritability = round(unlist(qtl.hsq),3))
herit <- herit %>%
arrange(desc(Heritability))
herit$Phenotype <- factor(herit$Phenotype, levels = herit$Phenotype)
#histgram
p2 <- ggplot(data=herit, aes(x=Phenotype, y=Heritability)) + #, fill=Domain, color = Domain)) +
geom_bar(stat="identity", fill = "blue", color = "blue", show.legend = FALSE) +
scale_y_continuous(breaks=seq(0.0, 1.0, 0.1)) +
geom_text(aes(label = Heritability, y = Heritability + 0.005), position = position_dodge(0.9),vjust = 0) +
theme(axis.text.x = element_text(angle = 90, hjust = 1)) +
ggtitle("Heritability by qtl2 array")
p2
heritability by qtl2 and the bootstrap estimates
apr <- readRDS("output/RL_prj/apr.RDS")
#kinship
k_overall = calc_kinship(probs = apr, type = "overall", use_allele_probs = TRUE, cores = 20)
# #heritability -----------------------------------------------------------
# loop through traits
#pheno list
pheno <- as.data.frame(pheno)
pheno.names <- colnames(pheno)[4:18]
pheno.herit <- data.frame(Name = pheno.names, h2=0, N=0, ll=0, ul=0)
covar = model.matrix(~sex, data = pheno)[,-1, drop=F]
rownames(covar) <- pheno$mouseID
colnames(covar)[1] = "sex"
#loop
for(i in 1:length(pheno.names)){
j = pheno.names[[i]]
print(j)
phe = pheno[,j,drop=F]
rownames(phe) = pheno$mouseID
tmp <- h2_sim(phe, k_overall, covar, 100)
pheno.herit$N[i] <- tmp$N
pheno.herit$h2[i] <- tmp$h2
pheno.herit$ll[i] <- quantile(tmp$sim, 0.1)
pheno.herit$ul[i] <- quantile(tmp$sim, 0.9)
}
# [1] "Acq.Total.Trials"
# [1] "Acq.Anticipatory.Correct.Responses"
# [1] "Rev.Total.Trials"
# [1] "Rev.Anticipatory.Incorrect.Responses"
# [1] "Diff.Total.Trials"
# [1] "Acq.Anticipatory.Correct.Responses.nooutlier"
# [1] "Rev.Anticipatory.Incorrect.Responses.nooutlier"
# [1] "Rev.Anticipatory.Incorrect.Responses.boxcox.trans"
# [1] "Acq.Total.Trials.rz"
# [1] "Acq.Anticipatory.Correct.Responses.rz"
# [1] "Rev.Total.Trials.rz"
# [1] "Rev.Anticipatory.Incorrect.Responses.rz"
# [1] "Diff.Total.Trials.rz"
# [1] "Acq.Anticipatory.Correct.Responses.nooutlier.rz"
# [1] "Rev.Anticipatory.Incorrect.Responses.nooutlier.rz"
write.csv(pheno.herit, file = "output/RL_prj/Prj01_RL_11092020_cutoff6.pheno.herit.csv",
row.names = FALSE, quote = F)
sessionInfo()
# R version 4.0.3 (2020-10-10)
# Platform: x86_64-pc-linux-gnu (64-bit)
# Running under: Ubuntu 20.04.2 LTS
#
# Matrix products: default
# BLAS/LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.8.so
#
# locale:
# [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
# [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
# [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=C
# [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
# [9] LC_ADDRESS=C LC_TELEPHONE=C
# [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
#
# attached base packages:
# [1] parallel stats graphics grDevices utils datasets methods
# [8] base
#
# other attached packages:
# [1] EnvStats_2.4.0 MASS_7.3-53.1 SimDesign_2.2 forcats_0.5.1
# [5] stringr_1.4.0 dplyr_1.0.4 purrr_0.3.4 readr_1.4.0
# [9] tidyr_1.1.2 tibble_3.0.6 tidyverse_1.3.0 openxlsx_4.2.3
# [13] abind_1.4-5 regress_1.3-21 survival_3.2-7 qtl2_0.24
# [17] GGally_2.1.0 gridExtra_2.3 ggplot2_3.3.3 workflowr_1.6.2
#
# loaded via a namespace (and not attached):
# [1] fs_1.5.0 lubridate_1.7.9.2 bit64_4.0.5 RColorBrewer_1.1-2
# [5] httr_1.4.2 rprojroot_2.0.2 tools_4.0.3 backports_1.2.1
# [9] R6_2.5.0 DBI_1.1.1 colorspace_2.0-0 withr_2.4.1
# [13] tidyselect_1.1.0 bit_4.0.4 compiler_4.0.3 git2r_0.28.0
# [17] cli_2.3.0 rvest_0.3.6 xml2_1.3.2 scales_1.1.1
# [21] pbapply_1.4-3 digest_0.6.27 rmarkdown_2.6 pkgconfig_2.0.3
# [25] htmltools_0.5.1.1 dbplyr_2.1.0 fastmap_1.1.0 highr_0.8
# [29] rlang_1.0.2 readxl_1.3.1 rstudioapi_0.13 RSQLite_2.2.3
# [33] generics_0.1.0 jsonlite_1.7.2 zip_2.1.1 magrittr_2.0.1
# [37] Matrix_1.3-2 Rcpp_1.0.6 munsell_0.5.0 lifecycle_1.0.0
# [41] stringi_1.5.3 whisker_0.4 yaml_2.2.1 plyr_1.8.6
# [45] grid_4.0.3 blob_1.2.1 promises_1.2.0.1 crayon_1.4.1
# [49] lattice_0.20-41 haven_2.3.1 splines_4.0.3 hms_1.0.0
# [53] knitr_1.31 pillar_1.4.7 codetools_0.2-18 reprex_1.0.0
# [57] glue_1.4.2 evaluate_0.14 data.table_1.13.6 modelr_0.1.8
# [61] vctrs_0.3.6 httpuv_1.5.5 foreach_1.5.1 cellranger_1.1.0
# [65] gtable_0.3.0 reshape_0.8.8 assertthat_0.2.1 cachem_1.0.4
# [69] xfun_0.21 broom_0.7.4 later_1.1.0.1 iterators_1.0.13
# [73] memoise_2.0.0 ellipsis_0.3.1