Step 1: Sample QC [4.batches_myo]
Belinda Cornes
2023-01-11
Last updated: 2023-01-11
Checks: 5 2
Knit directory: Serreze-T1D_Workflow/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R
Markdown file created these results, you’ll want to first commit it to
the Git repo. If you’re still working on the analysis, you can ignore
this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20220210) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /Users/corneb/Documents/MyJax/CS/Projects/Serreze/qc/workflowr/Serreze-T1D_Workflow | . |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version c9fc66b. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: analysis/.DS_Store
Untracked files:
Untracked: analysis/0.1.1_preparing.data_bqc_4.batches_myo.Rmd
Untracked: analysis/0.1.1_preparing.data_bqc_4.batches_myo.Rmd.R
Untracked: analysis/0.1_samples_batch_20220729.Rmd
Untracked: analysis/0.1_samples_batch_20220729.Rmd.R
Untracked: analysis/0.1_samples_batch_20220826.Rmd
Untracked: analysis/0.1_samples_batch_20220826.Rmd.R
Untracked: analysis/0.1_samples_batch_20221006.Rmd
Untracked: analysis/0.1_samples_batch_20221006.Rmd.R
Untracked: analysis/0.1_samples_batch_20221116.Rmd
Untracked: analysis/0.1_samples_batch_20221116.Rmd.R
Untracked: analysis/0.2_haplotype_comparison_bqc_4.batches_myo_minprob.Rmd
Untracked: analysis/0.2_haplotype_comparison_bqc_4.batches_myo_minprob.Rmd.R
Untracked: analysis/2.1_sample_bqc_4.batches_myo.Rmd
Untracked: analysis/2.1_sample_bqc_4.batches_myo.Rmd.R
Untracked: analysis/2.2.1_snp_qc_4.batches_myo.Rmd
Untracked: analysis/2.2.1_snp_qc_4.batches_myo.Rmd.R
Untracked: analysis/2.2.1_snp_qc_4.batches_myo_mis.Rmd
Untracked: analysis/2.2.1_snp_qc_4.batches_myo_mis.Rmd.R
Untracked: analysis/2.4_preparing.data_aqc_4.batches_myo.Rmd
Untracked: analysis/2.4_preparing.data_aqc_4.batches_myo.Rmd.R
Untracked: analysis/2.4_preparing.data_aqc_4.batches_myo_mis.Rmd
Untracked: analysis/2.4_preparing.data_aqc_4.batches_myo_mis.Rmd.R
Untracked: analysis/3.1_phenotype.qc_corrected_4.batches_myo.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_4.batches_myo.Rmd.R
Untracked: analysis/3.1_phenotype.qc_corrected_4.batches_myo_mis.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_4.batches_myo_mis.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici-myo-yes.vs.het-ici-myo-no_snpsqc_dis_no-x_updated_4.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici-myo-yes.vs.het-ici-myo-no_snpsqc_dis_no-x_updated_4.batches_myo.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici.vs.het-pbs_snpsqc_dis_no-x_updated_4.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici.vs.het-pbs_snpsqc_dis_no-x_updated_4.batches_myo.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-myo-yes.vs.ici-myo-no_snpsqc_dis_no-x_updated_4.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-myo-yes.vs.ici-myo-no_snpsqc_dis_no-x_updated_4.batches_myo.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-sick.vs.ici-eoi_snpsqc_dis_no-x_updated_4.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-sick.vs.ici-eoi_snpsqc_dis_no-x_updated_4.batches_myo.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_4.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_4.batches_myo.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_4.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_4.batches_myo_mis.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_myo-yes.vs.myo-no_snpsqc_dis_no-x_updated_4.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_myo-yes.vs.myo-no_snpsqc_dis_no-x_updated_4.batches_myo.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_pbs-myo-yes.vs.pbs-myo-no_snpsqc_dis_no-x_updated_4.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_pbs-myo-yes.vs.pbs-myo-no_snpsqc_dis_no-x_updated_4.batches_myo.Rmd.R
Untracked: analysis/genotype.frequencies_het-ici-myo-yes.vs.het-ici-myo-no_4.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_het-ici-myo-yes.vs.het-ici-myo-no_4.batches_myo.Rmd.R
Untracked: analysis/genotype.frequencies_het-ici-myo-yes.vs.het-ici-myo-no_4.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_het-ici-myo-yes.vs.het-ici-myo-no_4.batches_myo_mis.Rmd.R
Untracked: analysis/genotype.frequencies_het-ici.vs.het-pbs_4.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_het-ici.vs.het-pbs_4.batches_myo.Rmd.R
Untracked: analysis/genotype.frequencies_het-ici.vs.het-pbs_4.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_het-ici.vs.het-pbs_4.batches_myo_mis.Rmd.R
Untracked: analysis/genotype.frequencies_ici-myo-yes.vs.ici-myo-no_4.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_ici-myo-yes.vs.ici-myo-no_4.batches_myo.Rmd.R
Untracked: analysis/genotype.frequencies_ici-myo-yes.vs.ici-myo-no_4.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_ici-myo-yes.vs.ici-myo-no_4.batches_myo_mis.Rmd.R
Untracked: analysis/genotype.frequencies_ici-sick.vs.ici-eoi_4.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_ici-sick.vs.ici-eoi_4.batches_myo.Rmd.R
Untracked: analysis/genotype.frequencies_ici-sick.vs.ici-eoi_4.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_ici-sick.vs.ici-eoi_4.batches_myo_mis.Rmd.R
Untracked: analysis/genotype.frequencies_ici.vs.pbs_4.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.pbs_4.batches_myo.Rmd.R
Untracked: analysis/genotype.frequencies_ici.vs.pbs_4.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.pbs_4.batches_myo_mis.Rmd.R
Untracked: analysis/genotype.frequencies_myo-yes.vs.myo-no_4.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_myo-yes.vs.myo-no_4.batches_myo.Rmd.R
Untracked: analysis/genotype.frequencies_myo-yes.vs.myo-no_4.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_myo-yes.vs.myo-no_4.batches_myo_mis.Rmd.R
Untracked: analysis/genotype.frequencies_pbs-myo-yes.vs.pbs-myo-no_4.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_pbs-myo-yes.vs.pbs-myo-no_4.batches_myo.Rmd.R
Untracked: analysis/genotype.frequencies_pbs-myo-yes.vs.pbs-myo-no_4.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_pbs-myo-yes.vs.pbs-myo-no_4.batches_myo_mis.Rmd.R
Untracked: analysis/gwas_csq_OTU_OTU_18_unclassified_Lachnospiraceae.log.txt
Untracked: analysis/index_4.batches_myo.Rmd
Untracked: analysis/index_4.batches_myo.Rmd.R
Untracked: data/GM_covar_4.batches_myo.csv
Untracked: data/bad_markers_all_4.batches_myo.RData
Untracked: data/covar_corrected.cleaned_het-ici-myo-yes.vs.het-ici-myo-no_4.batches_myo.csv
Untracked: data/covar_corrected.cleaned_het-ici-myo-yes.vs.het-ici-myo-no_4.batches_myo_mis.csv
Untracked: data/covar_corrected.cleaned_het-ici.vs.het-pbs_4.batches_myo.csv
Untracked: data/covar_corrected.cleaned_het-ici.vs.het-pbs_4.batches_myo_mis.csv
Untracked: data/covar_corrected.cleaned_ici-myo-yes.vs.ici-myo-no_4.batches_myo.csv
Untracked: data/covar_corrected.cleaned_ici-myo-yes.vs.ici-myo-no_4.batches_myo_mis.csv
Untracked: data/covar_corrected.cleaned_ici-sick.vs.ici-eoi_4.batches_myo.csv
Untracked: data/covar_corrected.cleaned_ici-sick.vs.ici-eoi_4.batches_myo_mis.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_4.batches_myo.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_4.batches_myo_mis.csv
Untracked: data/covar_corrected.cleaned_myo-yes.vs.myo-no_4.batches_myo.csv
Untracked: data/covar_corrected.cleaned_myo-yes.vs.myo-no_4.batches_myo_mis.csv
Untracked: data/covar_corrected.cleaned_pbs-myo-yes.vs.pbs-myo-no_4.batches_myo.csv
Untracked: data/covar_corrected.cleaned_pbs-myo-yes.vs.pbs-myo-no_4.batches_myo_mis.csv
Untracked: data/covar_corrected_het-ici-myo-yes.vs.het-ici-myo-no_4.batches_myo.csv
Untracked: data/covar_corrected_het-ici-myo-yes.vs.het-ici-myo-no_4.batches_myo_mis.csv
Untracked: data/covar_corrected_het-ici.vs.het-pbs_4.batches_myo.csv
Untracked: data/covar_corrected_het-ici.vs.het-pbs_4.batches_myo_mis.csv
Untracked: data/covar_corrected_ici-myo-yes.vs.ici-myo-no_4.batches_myo.csv
Untracked: data/covar_corrected_ici-myo-yes.vs.ici-myo-no_4.batches_myo_mis.csv
Untracked: data/covar_corrected_ici-sick.vs.ici-eoi_4.batches_myo.csv
Untracked: data/covar_corrected_ici-sick.vs.ici-eoi_4.batches_myo_mis.csv
Untracked: data/covar_corrected_ici.vs.pbs_4.batches_myo.csv
Untracked: data/covar_corrected_ici.vs.pbs_4.batches_myo_mis.csv
Untracked: data/covar_corrected_myo-yes.vs.myo-no_4.batches_myo.csv
Untracked: data/covar_corrected_myo-yes.vs.myo-no_4.batches_myo_mis.csv
Untracked: data/covar_corrected_pbs-myo-yes.vs.pbs-myo-no_4.batches_myo.csv
Untracked: data/covar_corrected_pbs-myo-yes.vs.pbs-myo-no_4.batches_myo_mis.csv
Untracked: data/e_4.batches_myo.RData
Untracked: data/e_snpg_samqc_4.batches_myo.RData
Untracked: data/errors_ind_4.batches_myo.RData
Untracked: data/genetic_map_4.batches_myo.csv
Untracked: data/genotype_errors_marker_4.batches_myo.RData
Untracked: data/genotype_freq_marker_4.batches_myo.RData
Untracked: data/gm_allqc_4.batches_myo.RData
Untracked: data/gm_allqc_4.batches_myo_mis.RData
Untracked: data/gm_samqc_4.batches_myo.RData
Untracked: data/gm_serreze.BC312.RData
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_marker.freq_low.geno.freq.removed_geno.ratio_4.batches_myo.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_marker.freq_low.geno.freq.removed_geno.ratio_4.batches_myo_mis.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratiov_4.batches_myo.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratiov_4.batches_myo_mis.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_marker.freq_low.probs.freq.removed_geno.ratio_4.batches_myo.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_marker.freq_low.probs.freq.removed_geno.ratio_4.batches_myo_mis.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_4.batches_myo.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_4.batches_myo_mis.csv
Untracked: data/het-ici.vs.het-pbs_marker.freq_low.geno.freq.removed_geno.ratio_4.batches_myo.csv
Untracked: data/het-ici.vs.het-pbs_marker.freq_low.geno.freq.removed_geno.ratio_4.batches_myo_mis.csv
Untracked: data/het-ici.vs.het-pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratiov_4.batches_myo.csv
Untracked: data/het-ici.vs.het-pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratiov_4.batches_myo_mis.csv
Untracked: data/het-ici.vs.het-pbs_marker.freq_low.probs.freq.removed_geno.ratio_4.batches_myo.csv
Untracked: data/het-ici.vs.het-pbs_marker.freq_low.probs.freq.removed_geno.ratio_4.batches_myo_mis.csv
Untracked: data/het-ici.vs.het-pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_4.batches_myo.csv
Untracked: data/het-ici.vs.het-pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_4.batches_myo_mis.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_marker.freq_low.geno.freq.removed_geno.ratio_4.batches_myo.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_marker.freq_low.geno.freq.removed_geno.ratio_4.batches_myo_mis.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratiov_4.batches_myo.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratiov_4.batches_myo_mis.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_marker.freq_low.probs.freq.removed_geno.ratio_4.batches_myo.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_marker.freq_low.probs.freq.removed_geno.ratio_4.batches_myo_mis.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_4.batches_myo.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_4.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-10_peak.marker-UNC18805053_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-10_peak.marker-UNCHS029427_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-11_peak.marker-UNCHS031753_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-11_peak.marker-UNCHS031802_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-12_peak.marker-JAX00326005_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-12_peak.marker-UNC21995304_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-13_peak.marker-JAX00370189_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-13_peak.marker-UNCHS035661_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-14_peak.marker-UNC24597582_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-14_peak.marker-UNCHS039096_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-15_peak.marker-UNC25489755_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-15_peak.marker-UNCHS040614_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-16_peak.marker-UNCHS042686_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-17_peak.marker-UNCHS043777_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-17_peak.marker-UNCHS043880_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-18_peak.marker-UNC29296831_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-18_peak.marker-UNC29297751_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-19_peak.marker-UNC30069852_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-19_peak.marker-UNC30386742_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-1_peak.marker-UNCHS001121_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-1_peak.marker-UNCHS002308_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-2_peak.marker-UNC3990359_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-2_peak.marker-UNCHS006135_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-3_peak.marker-JAX00105915_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-3_peak.marker-UNC6020011_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-4_peak.marker-UNC8099452_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-4_peak.marker-UNC8161950_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-5_peak.marker-UNC9678100_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-5_peak.marker-UNC9678931_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-6_peak.marker-UNC12162881_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-6_peak.marker-backupUNC060363218_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-7_peak.marker-UNC12719038_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-7_peak.marker-UNCHS022024_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-8_peak.marker-UNC14948439_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-8_peak.marker-UNCHS023592_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-9_peak.marker-UNC16009822_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-9_peak.marker-UNC17271730_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-X_peak.marker-UNC31358512_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-X_peak.marker-UNCHS049472_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-10_peak.marker-UNC18805053_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-10_peak.marker-UNCHS029427_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-11_peak.marker-UNCHS031753_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-11_peak.marker-UNCHS031802_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-12_peak.marker-JAX00326005_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-12_peak.marker-UNC21995304_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-13_peak.marker-JAX00370189_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-13_peak.marker-UNCHS035661_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-14_peak.marker-UNC24597582_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-14_peak.marker-UNCHS039096_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-15_peak.marker-UNC25489755_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-15_peak.marker-UNCHS040614_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-16_peak.marker-UNCHS042686_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-17_peak.marker-UNCHS043777_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-17_peak.marker-UNCHS043880_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-18_peak.marker-UNC29296831_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-18_peak.marker-UNC29297751_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-19_peak.marker-UNC30069852_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-19_peak.marker-UNC30386742_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-1_peak.marker-UNCHS001121_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-1_peak.marker-UNCHS002308_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-2_peak.marker-UNC3990359_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-2_peak.marker-UNCHS006135_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-3_peak.marker-JAX00105915_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-3_peak.marker-UNC6020011_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-4_peak.marker-UNC8099452_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-4_peak.marker-UNC8161950_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-5_peak.marker-UNC9678100_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-5_peak.marker-UNC9678931_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-6_peak.marker-UNC12162881_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-6_peak.marker-backupUNC060363218_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-7_peak.marker-UNC12719038_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-7_peak.marker-UNCHS022024_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-8_peak.marker-UNC14948439_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-8_peak.marker-UNCHS023592_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-9_peak.marker-UNC16009822_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-9_peak.marker-UNC17271730_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-X_peak.marker-UNC31358512_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-X_peak.marker-UNCHS049472_lod.drop-1.5_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_gm_qtl_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_marker.freq_low.geno.freq.removed_geno.ratio_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_marker.freq_low.geno.freq.removed_geno.ratio_4.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratiov_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratiov_4.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_marker.freq_low.probs.freq.removed_geno.ratio_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_marker.freq_low.probs.freq.removed_geno.ratio_4.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_4.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_4.batches_myo_mis.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_4.batches_myo.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_4.batches_myo.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_4.batches_myo_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratiov_4.batches_myo.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratiov_4.batches_myo_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_4.batches_myo.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_4.batches_myo_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_4.batches_myo.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_4.batches_myo_mis.csv
Untracked: data/myo-yes.vs.myo-no_marker.freq_low.geno.freq.removed_geno.ratio_4.batches_myo.csv
Untracked: data/myo-yes.vs.myo-no_marker.freq_low.geno.freq.removed_geno.ratio_4.batches_myo_mis.csv
Untracked: data/myo-yes.vs.myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratiov_4.batches_myo.csv
Untracked: data/myo-yes.vs.myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratiov_4.batches_myo_mis.csv
Untracked: data/myo-yes.vs.myo-no_marker.freq_low.probs.freq.removed_geno.ratio_4.batches_myo.csv
Untracked: data/myo-yes.vs.myo-no_marker.freq_low.probs.freq.removed_geno.ratio_4.batches_myo_mis.csv
Untracked: data/myo-yes.vs.myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_4.batches_myo.csv
Untracked: data/myo-yes.vs.myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_4.batches_myo_mis.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.geno.freq.removed_geno.ratio_4.batches_myo.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.geno.freq.removed_geno.ratio_4.batches_myo_mis.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratiov_4.batches_myo.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratiov_4.batches_myo_mis.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.probs.freq.removed_geno.ratio_4.batches_myo.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.probs.freq.removed_geno.ratio_4.batches_myo_mis.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_4.batches_myo.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_4.batches_myo_mis.csv
Untracked: data/percent_missing_id_4.batches_myo.RData
Untracked: data/percent_missing_marker_4.batches_myo.RData
Untracked: data/pheno_4.batches_myo.csv
Untracked: data/physical_map_4.batches_myo.csv
Untracked: data/qc_info_bad_sample_4.batches_myo.RData
Untracked: data/sample_geno_AHB_4.batches_myo.csv
Untracked: data/sample_geno_bc_4.batches_myo.csv
Untracked: data/serreze_probs_4.batches_myo.rds
Untracked: data/serreze_probs_allqc_4.batches_myo.rds
Untracked: data/serreze_probs_allqc_4.batches_myo_mis.rds
Untracked: data/summary.cg_4.batches_myo.RData
Untracked: output/Percent_missing_genotype_data_4.batches_myo.pdf
Untracked: output/Percent_missing_genotype_data_per_marker_4.batches_myo.pdf
Untracked: output/Proportion_matching_genotypes_before_removal_of_bad_samples_4.batches_myo.pdf
Untracked: output/genotype_error_marker_4.batches_myo.pdf
Untracked: output/genotype_frequency_marker_4.batches_myo.pdf
Unstaged changes:
Modified: analysis/index_5.batches.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with
wflow_publish() to start tracking its development.
This script is running genotype QC on raw data (with some outcomes already seen in the project at a glace). Here, we first load the R/qtl2 package and the data. We’ll also load the R/broman package for some utilities and plotting functions, and R/qtlcharts for interactive graphs.
We will follow the steps by Karl Broman found here
Loading Project
gm <- get(load(paste0(filepaths,"/gm_serreze_bc_4.batches_myo_BC217.RData")))
gmWarning in check_cross2(object): 1249 invalid genotypes in crossObject of class cross2 (crosstype "bc")
Total individuals 217
No. genotyped individuals 217
No. phenotyped individuals 217
No. with both geno & pheno 217
No. phenotypes 1
No. covariates 11
No. phenotype covariates 0
No. chromosomes 20
Total markers 133716
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13
10159 10172 7987 7736 7778 7911 7548 6561 6823 6471 7276 6226 6177
14 15 16 17 18 19 X
6082 5421 5075 5162 4682 3612 4857 sample_file <- dir(path = filepaths, pattern = "^DODB_*", full.names = TRUE)
samples <- read.csv(sample_file)
all.equal(as.character(ind_ids(gm)), as.character(samples$Original.Mouse.ID))[1] TRUEMissing Data
percent_missing <- n_missing(gm, "ind", "prop")*100
#labels <- paste0(as.character(do.call(rbind.data.frame, strsplit(names(percent_missing), "_"))[,7]), " (", round(percent_missing,2), "%)")
labels <- paste0(names(percent_missing), " (", round(percent_missing,2), "%)")
iplot(seq_along(percent_missing), percent_missing, indID=labels,
chartOpts=list(xlab="Mouse", ylab="Percent missing genotype data",
ylim=c(0, 70)))Set screen size to height=700 x width=1000#save into pdf
pdf(file = "output/Percent_missing_genotype_data_4.batches_myo.pdf", width = 20, height = 20)
#labels <- as.character(do.call(rbind.data.frame, strsplit(names(totxo), "V01_"))[,2])
#labels <- as.character(do.call(rbind.data.frame, strsplit(ind_ids(gm), "_"))[,7])
#labels <- paste0(names(percent_missing), " (", round(percent_missing,2), "%)")
labels <- ind_ids(gm)
labels[percent_missing < 10] = ""
# Change point shapes and colors
p <- ggplot(data = data.frame(Mouse=seq_along(percent_missing),
Percent_missing_genotype_data = percent_missing,
batch = factor(as.character(do.call(rbind.data.frame, strsplit(as.character(samples$Unique.Sample.ID), "_"))[,6]))
#batch = factor(as.character(do.call(rbind.data.frame, strsplit(as.character(samples$Directory), "_"))[,5]))
),
aes(x=Mouse, y=Percent_missing_genotype_data, color = batch)) +
geom_point() +
geom_hline(yintercept=10, linetype="solid", color = "red") +
geom_text_repel(aes(label=labels), vjust = 0, nudge_y = 0.01, show.legend = FALSE, size=3) +
theme(text = element_text(size = 10))
p
dev.off()quartz_off_screen
2 p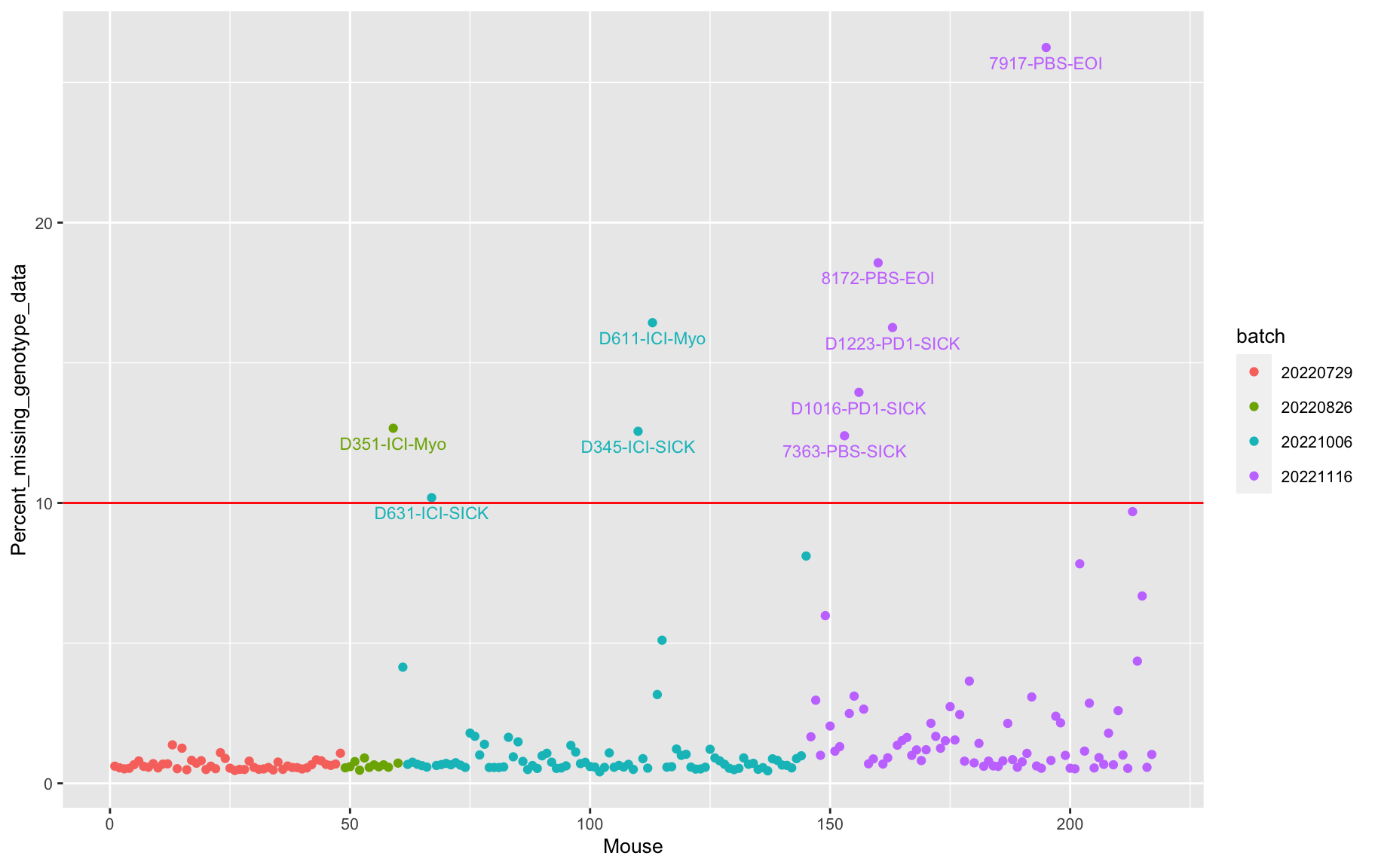
save(percent_missing,file = "data/percent_missing_id_4.batches_myo.RData")
gm.covar = data.frame(id=rownames(gm$covar),gm$covar)
qc_info_cr <- merge(gm.covar,
data.frame(id = names(percent_missing),percent_missing = percent_missing,stringsAsFactors = F),by = "id")
bad.sample.cr <- qc_info_cr[qc_info_cr$percent_missing >= 10,]| Sample_ID | percent_missing |
|---|---|
| 7363-PBS-SICK | 12.3971701217506 |
| 7917-PBS-EOI | 26.244428490233 |
| 8172-PBS-EOI | 18.566214963056 |
| D1016-PD1-SICK | 13.9452271979419 |
| D1223-PD1-SICK | 16.256842860989 |
| D345-ICI-SICK | 12.5557150976697 |
| D351-ICI-Myo | 12.6649017320291 |
| D611-ICI-Myo | 16.431092763768 |
| D631-ICI-SICK | 10.1880104101229 |
Sex
hdf5_filename <- dir(path = filepaths, pattern = "^hdf5_*", full.names = TRUE)
snps_file <- "/Users/corneb/Documents/MyJax/CS/Projects/support.files/MUGAarrays/UWisc/gm_uwisc_v1.csv"
snps <- read.csv(snps_file)
snps <- snps[snps$unique == TRUE, ]
#snps <- snps[snps$chr %in% c(1:19, "X"), ]
snps$chr <- sub("^chr", "", snps$chr) ###remove prefix "chr"
colnames(snps)[colnames(snps)=="bp_mm10"] <- "pos"
colnames(snps)[colnames(snps)=="cM_cox"] <- "cM"
snps <- snps %>% drop_na(chr, marker)
snps$pos <- snps$pos * 1e-6
rownames(snps) <- snps$marker
colnames(snps)[1:4] <- c("marker", "chr", "pos", "pos")
# g <- h5read(hdf5_filename, "G")
# g <- do.call(cbind, g)
x <- h5read(hdf5_filename, "X") # X channel intensities
x <- do.call(cbind, x)
y <- h5read(hdf5_filename, "Y") # Y channel intensities
y <- do.call(cbind, y)
rn <- h5read(hdf5_filename, "rownames")[[1]] # markers
cn <- h5read(hdf5_filename, "colnames") # samples
cn <- do.call(c, cn)
# dimnames(g) <- list(rn, cn)
dimnames(x) <- list(rn, cn)
dimnames(y) <- list(rn, cn)
#cr <- colMeans(g != "--") # Call rate for each sample avg 0.95
# sex <- determine_sex(x = x, y = y, markers = snps)$se
markers <- snps
chrx <- markers$marker[which(markers$chr == "X")]
chry <- markers$marker[which(markers$chr == "Y")]
#x[chrx,ind_ids(gm)]
chrx_int <- colMeans(x[chrx,as.character(ind_ids(gm))] + y[chrx,as.character(ind_ids(gm))], na.rm = T)
chry_int <- colMeans(x[chry,as.character(ind_ids(gm))] + y[chry,as.character(ind_ids(gm))], na.rm = T)
all.equal(as.character(ind_ids(gm)), as.character(samples$Original.Mouse.ID))[1] TRUE#sex order
#samples$Sex <- 'F'
sex <- samples$Sex
point_colors <- as.character( brocolors("web")[c("green", "purple")] )
percent_missing <- n_missing(gm, summary="proportion")*100
labels <- paste0(names(chrx_int), " (", round(percent_missing), "%)")
iplot( chrx_int, chry_int, group=sex, indID=labels,
chartOpts=list(pointcolor=point_colors, pointsize=4,
xlab="Average X chr intensity", ylab="Average Y chr intensity"))For figures above and below, those labelled as female in metadata
given, are coloured green, with those labelled as male are
coloured as purple. The above is an interactive scatterplot
of the average SNP intensity on the Y chromosome versus the average SNP
intensity on the X chromosome.
phetX <- rowSums(gm$geno$X == 2)/rowSums(gm$geno$X != 0)
phetX <- phetX[as.character(ind_ids(gm)) %in% names(chrx_int)]
names(phetX) <- as.character(ind_ids(gm))
iplot(chrx_int, phetX, group=sex, indID=labels,
chartOpts=list(pointcolor=point_colors, pointsize=4,
xlab="Average X chr intensity", ylab="Proportion het on X chr"))In the above scatterplot, we show the proportion of hets vs the average intensity for the X chromosome SNPs. In calculating the proportion of heterozygous genotypes for the individuals, we look at X chromosome genotypes equal to 2 which corresponds to the heterozygote) relative to not being 0 (which is used to encode missing genotypes). The genotypes are arranged with rows being individuals and columns being markers.
The following are the mice that have had sex incorrectly assigned:
| Neogen_Sample_ID | Sample_ID | Sex | Inferred.Sex |
|---|---|---|---|
| D63-ICI-Myo | The_Jackson_Lab_Serreze_MURGIGV01_20220729_D63-ICI-Myo_B7 | M | F |
| D38-ICI-Myo | The_Jackson_Lab_Serreze_MURGIGV01_20220826_D38-ICI-Myo_B9 | F | M |
| D351-ICI-Myo | The_Jackson_Lab_Serreze_MURGIGV01_20220826_D351-ICI-Myo_C10 | F | M |
| D320-ICI-SICK | The_Jackson_Lab_Serreze_MURGIGV01_20221006_D320-ICI-SICK_H3 | F | M |
| D611-ICI-Myo | The_Jackson_Lab_Serreze_MURGIGV01_20221006_D611-ICI-Myo_D6 | F | M |
| 7363-PBS-SICK | The_Jackson_Lab_Serreze_MURGIGV01_20221116_7363-PBS-SICK_C11 | F | M |
| 8172-PBS-EOI | The_Jackson_Lab_Serreze_MURGIGV01_20221116_8172-PBS-EOI_B12 | F | M |
| D1223-PD1-SICK | The_Jackson_Lab_Serreze_MURGIGV01_20221116_D1223-PD1-SICK_E12 | F | M |
| 7917-PBS-EOI | The_Jackson_Lab_Serreze_MURGIGV01_20221116_7917-PBS-EOI_E5 | F | M |
| 8144-PBS-EOI | The_Jackson_Lab_Serreze_MURGIGV01_20221116_8144-PBS-EOI_E8 | F | M |
Sample Duplicates
cg <- compare_geno(gm, cores=10)
summary.cg <- summary(cg)Here is a histogram of the proportion of matching genotypes. The tick marks below the histogram indicate individual pairs.
save(summary.cg,file = "data/summary.cg_4.batches_myo.RData")
pdf(file = "output/Proportion_matching_genotypes_before_removal_of_bad_samples_4.batches_myo.pdf", width = 20, height = 20)
par(mar=c(5.1,0.6,0.6, 0.6))
hist(cg[upper.tri(cg)], breaks=seq(0, 1, length=201),
main="", yaxt="n", ylab="", xlab="Proportion matching genotypes")
rug(cg[upper.tri(cg)])
dev.off()quartz_off_screen
2 par(mar=c(5.1,0.6,0.6, 0.6))
hist(cg[upper.tri(cg)], breaks=seq(0, 1, length=201),
main="", yaxt="n", ylab="", xlab="Proportion matching genotypes")
rug(cg[upper.tri(cg)])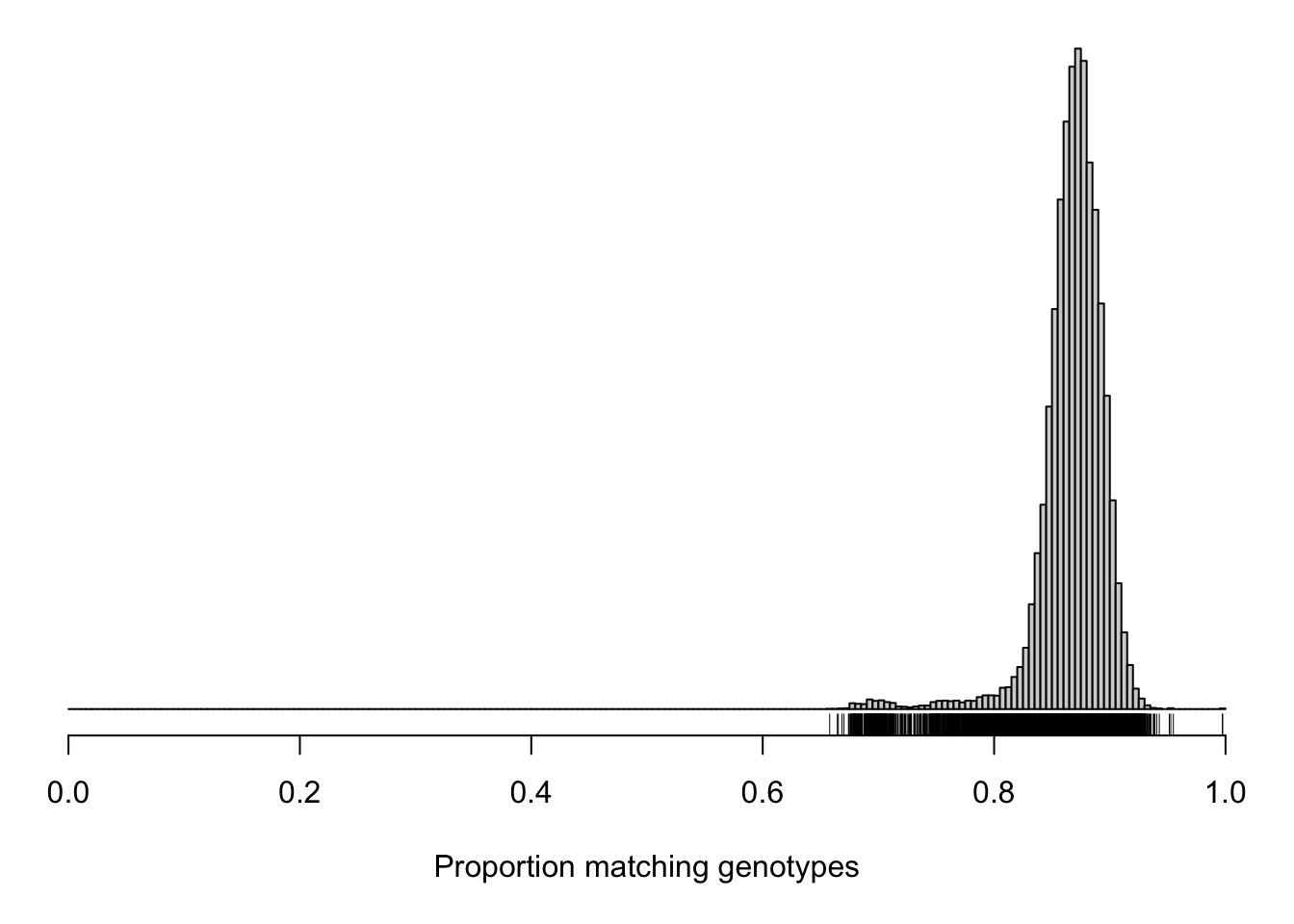
cgsub <- cg[percent_missing < 10, percent_missing < 10]
par(mar=c(5.1,0.6,0.6, 0.6))
hist(cgsub[upper.tri(cgsub)], breaks=seq(0, 1, length=201),
main="", yaxt="n", ylab="", xlab="Proportion matching genotypes [percent missing < 10%]")
rug(cgsub[upper.tri(cgsub)])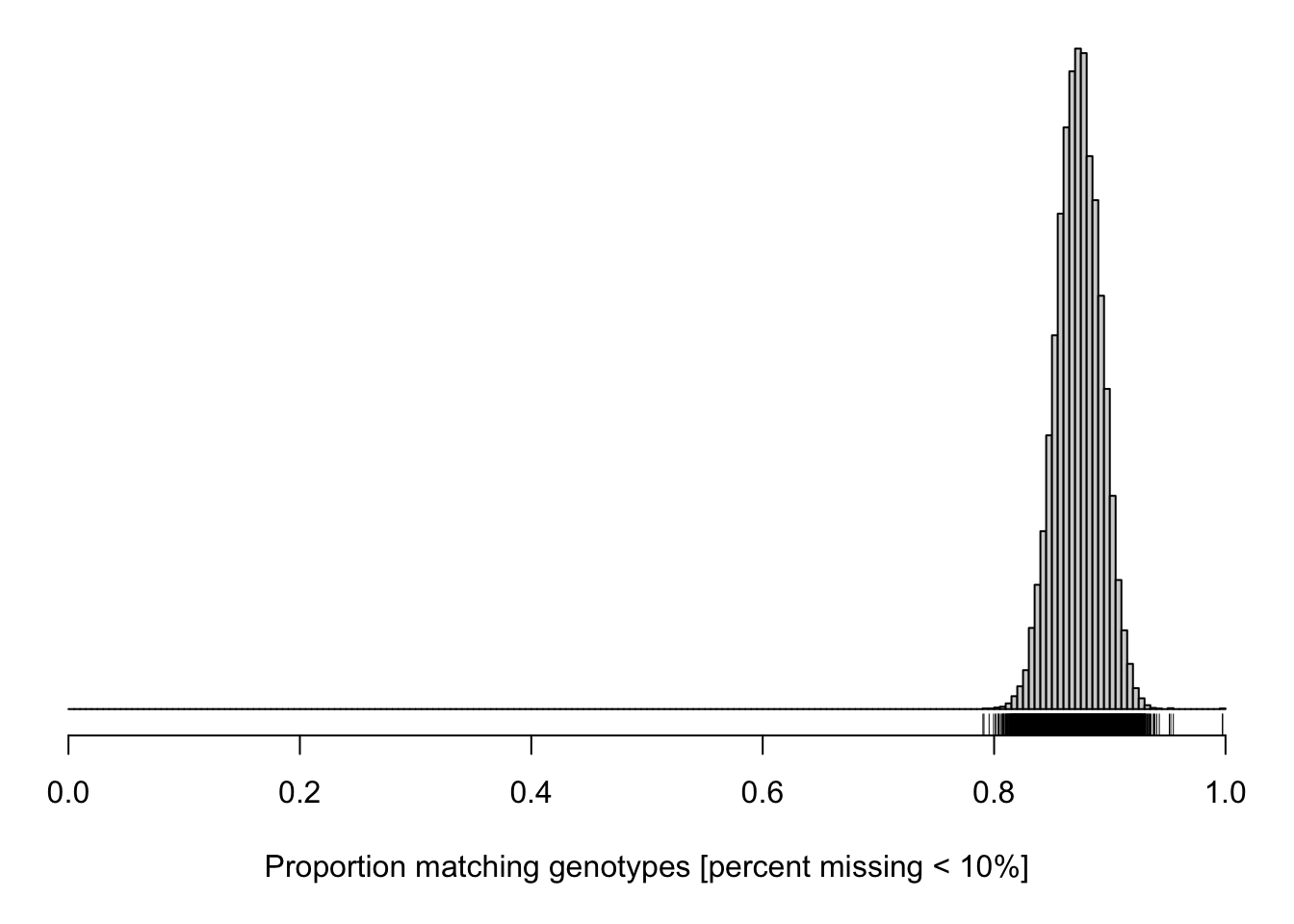
Array Intensities
#load the intensities.fst_4.batches_myo.RData
#load("data/intensities.fst_4.batches_myo.RData")
xn <- x[,as.character(ind_ids(gm))]
xn <- xn[snps$marker,]
xnm <- rownames(xn)
yn <- y[,as.character(ind_ids(gm))]
yn <- yn[snps$marker,]
# bring together in one matrix
result <- cbind(snp=rep(snps$marker, 2),
channel=rep(c("x", "y"), each=length(snps$marker)),
as.data.frame(rbind(xn, yn)))
rownames(result) <- 1:nrow(result)
# bring SNP rows together
result <- result[as.numeric(t(cbind(seq_along(snps$marker), seq_along(snps$marker)+length(snps$marker)))),]
rownames(result) <- 1:nrow(result)
#load the intensities.fst_4.batches_myo.RData
#load("data/heh/intensities.fst_4.batches_myo.RData")
#X and Y channel
X <- result[result$channel == "x",]
rownames(X) <- X$snp
X <- X[,c(-1,-2)]
Y <- result[result$channel == "y",]
rownames(Y) <- Y$snp
Y <- Y[,c(-1,-2)]
int <- result
#int <- result
#rm(result)
int <- int[seq(1, nrow(int), by=2),-(1:2)] + int[-seq(1, nrow(int), by=2),-(1:2)]
int <- int[,intersect(as.character(ind_ids(gm)), colnames(int))]
names(percent_missing) <- as.character(names(percent_missing))
n <- names(sort(percent_missing[intersect(as.character(ind_ids(gm)), colnames(int))], decreasing=TRUE))
iboxplot(log10(t(int[,n])+1), orderByMedian=FALSE, chartOpts=list(ylab="log10(SNP intensity + 1)"))In the above plot, distributions of array intensities (after a log10(x+1) transformation) are displayed.
The arrays are sorted by the proportion of missing genotype data for the sample, and the curves connect various quantiles of the intensities.
qu <- apply(int, 2, quantile, c(0.01, 0.99), na.rm=TRUE)
group <- (percent_missing >= 19.97) + (percent_missing > 5) + (percent_missing > 2) + 1
labels <- paste0(colnames(qu), " (", round(percent_missing), "%)")
iplot(qu[1,], qu[2,], indID=labels, group=group,
chartOpts=list(xlab="1 %ile of array intensities",
ylab="99 %ile of array intensities",
pointcolor=c("#ccc", "slateblue", "Orchid", "#ff851b")))For this particular set of arrays, a plot of the 1 %ile vs the 99 %ile is quite revealing. In the following, the orange points are those with > 20% missing genotypes, the pink points are the samples with 5-20% missing genotypes, and the blue points are the samples with 2-5% missing genotypes.
Genotyping Error LOD Scores
load("/Users/corneb/Documents/MyJax/CS/Projects/Serreze/haplotype.reconstruction/output_4.batches_myo_corrected/e_bc_4.batches_myo_BC217.RData")
errors_ind <- rowSums(e>2)[rownames(gm$covar)]/n_typed(gm)*100
lab <- paste0(as.character(names(errors_ind)), " (", myround(percent_missing[as.character(rownames(gm$covar))],1), "%)")
iplot(seq_along(errors_ind), errors_ind, indID=lab,
chartOpts=list(xlab="Mouse", ylab="Percent genotyping errors", ylim=c(0, 15),
axispos=list(xtitle=25, ytitle=50, xlabel=5, ylabel=5)))save(errors_ind, file = "data/errors_ind_4.batches_myo.RData")Removing Samples
##percent missing
gm.covar = data.frame(id=as.character(rownames(gm$covar)),gm$covar)
qc_info <- merge(gm.covar,
data.frame(id = names(percent_missing),percent_missing = percent_missing,stringsAsFactors = F),by = "id")
#missing sex
#qc_info$sex.match <- ifelse(qc_info$sexp == qc_info$sex, TRUE, FALSE)
rownames(samples) <- as.character(samples$Original.Mouse.ID)
samples <- samples[as.character(qc_info$id),]
#samples$Unique.Sample.ID <- as.character(samples$Unique.Sample.ID)
all.equal(as.character(qc_info$id), as.character(samples$Original.Mouse.ID))[1] TRUEqc_info$sex.match <- ifelse((samples$Inferred.Sex == samples$Sex), TRUE, FALSE)
#genotype errors
qc_info <- merge(qc_info,
data.frame(id = as.character(names(errors_ind)),
genotype_erros = errors_ind,stringsAsFactors = F),by = "id")
##duplicated id to be remove
qc_info$duplicate.id <- ifelse(qc_info$id %in% as.character(summary.cg$remove.id), TRUE,FALSE)
#bad.sample <- qc_info[qc_info$generation ==1 | qc_info$Number_crossovers <= 200 | qc_info$Number_crossovers >=1000 | qc_info$percent_missing >= 10 | qc_info$genotype_erros >= 1 | qc_info$remove.id.duplicated == TRUE,]
bad.sample <- qc_info[qc_info$percent_missing >= 10 | qc_info$genotype_erros >= 8,]
save(qc_info, bad.sample, file = "data/qc_info_bad_sample_4.batches_myo.RData")
gm_samqc <- gm[paste0("-",as.character(bad.sample$id.1)),]
gm_samqcWarning in check_cross2(object): 1249 invalid genotypes in crossObject of class cross2 (crosstype "bc")
Total individuals 208
No. genotyped individuals 208
No. phenotyped individuals 208
No. with both geno & pheno 208
No. phenotypes 1
No. covariates 11
No. phenotype covariates 0
No. chromosomes 20
Total markers 133716
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13
10159 10172 7987 7736 7778 7911 7548 6561 6823 6471 7276 6226 6177
14 15 16 17 18 19 X
6082 5421 5075 5162 4682 3612 4857 save(gm_samqc, file = "data/gm_samqc_4.batches_myo.RData")
# update other stuff
e <- e[ind_ids(gm_samqc),]
#g <- g[ind_ids(gm_samqc),]
#snpg <- snpg[ind_ids(gm_samqc),]
#save(e,g,snpg, file = "data/e_g_snpg_samqc_4.batches_myo.RData")
save(e, file = "data/e_snpg_samqc_4.batches_myo.RData")Here is the list of samples that were removed:
| Sample_ID |
|---|
| 7363-PBS-SICK |
| 7917-PBS-EOI |
| 8172-PBS-EOI |
| D1016-PD1-SICK |
| D1223-PD1-SICK |
| D345-ICI-SICK |
| D351-ICI-Myo |
| D611-ICI-Myo |
| D631-ICI-SICK |
Below is a table summarising the problematic samples found throughout QC. These include the following:
- no_pheno == sample does not have a phenotype (if applicable)
- high_miss == sample has higher than 10% missing genotypes
- diff_sex == sample has sex mismatch
- high_geno_errors == Sample has geno errors above 10%
- dublicate_ID == sample was duplicated (or highly concordant)
NB: For duplcate pairs, the one that was chosen to be removed was the one that had a higher missing rate
| Sample_ID | high_miss | diff_sex | high_geno.errors | highly_concordant |
|---|---|---|---|---|
| 6404-PBS-EOI | XX | |||
| 6411-PBS-Myo | XX | |||
| 6603-PBS-EOI | XX | |||
| 6614-PBS-EOI | XX | |||
| 6704-PBS-EOI | XX | |||
| 6705-PBS-EOI | XX | |||
| 6707-PBS-EOI | XX | |||
| 6708-PBS-EOI | XX | |||
| 6863-PBS-Myo | XX | |||
| 6872-PBS-Myo | XX | |||
| 6874-PBS-Myo | XX | |||
| 6890-PBS-EOI | XX | |||
| 6892-PBS-EOI | XX | |||
| 6894-PBS-EOI | XX | |||
| 6964-PBS-EOI | XX | |||
| 7168-ICI-Myo | XX | |||
| 7169-ICI-Myo | XX | |||
| 7175-PD1-EOI | XX | |||
| 7176-ICI-Myo | XX | |||
| 7179-ICI-Myo | XX | |||
| 7181-PBS-EOI | XX | |||
| 7182-PBS-Myo | XX | |||
| 7269-PBS-Myo | XX | |||
| 7276-PBS-EOI | XX | |||
| 7329-ICI-Myo | XX | |||
| 7333-ICI-EOI | XX | |||
| 7338-PD1-EOI | XX | |||
| 7340-PD1-EOI | XX | |||
| 7346-PBS-Myo | XX | |||
| 7348-PBS-EOI | XX | |||
| 7351-PBS-EOI | XX | |||
| 7357-PBS-EOI | XX | |||
| 7358-PBS-EOI | XX | |||
| 7359-PBS-Myo | XX | |||
| 7360-PBS-SICK | XX | |||
| 7363-PBS-SICK | XX | XX | ||
| 7364-PBS-EOI | XX | |||
| 7782-ICI-Myo | XX | |||
| 7786-ICI-Myo | XX | |||
| 7788-ICI-Myo | XX | |||
| 7789-ICI-Myo | XX | |||
| 7792-ICI-Myo | XX | |||
| 7805-PBS-EOI | XX | |||
| 7904-PBS-EOI | XX | |||
| 7911-PBS-EOI | XX | |||
| 7912-PBS-EOI | XX | |||
| 7915-PBS-EOI | XX | |||
| 7917-PBS-EOI | XX | XX | XX | |
| 7921-PBS-EOI | XX | |||
| 7924-ICI-EOI | XX | |||
| 7927-ICI-Myo | XX | |||
| 7937-ICI-EOI | XX | |||
| 8144-PBS-EOI | XX | XX | ||
| 8149-PBS-EOI | XX | |||
| 8153-PBS-EOI | XX | |||
| 8169-PBS-EOI | XX | |||
| 8172-PBS-EOI | XX | XX | ||
| 8604-PBS-Myo | XX | |||
| 8608-PBS-EOI | XX | |||
| 8615-PBS-EOI | XX | |||
| 8626-PBS-EOI | XX | |||
| 8829-PBS-EOI | XX | |||
| 8839-PBS-Myo | XX | |||
| 8840-PBS-EOI | XX | |||
| 8851-PBS-SICK | XX | |||
| D1-PBS-Myo | XX | |||
| D100-ICI-EOI | XX | |||
| D1014-PD1-SICK | XX | |||
| D1016-PD1-SICK | XX | |||
| D1062-PBS-SICK | XX | |||
| D1086-PD1-SICK | XX | |||
| D1088-PD1-SICK | XX | |||
| D110-PBS-EOI | XX | |||
| D1104-PD1-SICK | XX | |||
| D1108-PD1-SICK | XX | |||
| D1109-PD1-SICK | XX | |||
| D1110-PD1-SICK | XX | |||
| D113-PBS-EOI | XX | |||
| D114-PBS-EOI | XX | |||
| D1142-PD1-SICK | XX | |||
| D115-ICI-EOI | XX | |||
| D1154-PD1-SICK | XX | |||
| D118-ICI-EOI | XX | |||
| D12-ICI-EOI | XX | |||
| D1206-PD1-SICK | XX | |||
| D1208-PD1-SICK | XX | |||
| D121-ICI-Myo | XX | |||
| D1223-PD1-SICK | XX | XX | ||
| D1262-PD1-SICK | XX | |||
| D127-PBS-EOI | XX | |||
| D128-PBS-Myo | XX | |||
| D1280-PD1-SICK | XX | |||
| D1281-PD1-SICK | XX | |||
| D1283-PD1-SICK | XX | |||
| D1285-PD1-SICK | XX | |||
| D1290-PD1-SICK | XX | |||
| D13-ICI-EOI | XX | |||
| D132-PBS-EOI | XX | |||
| D137-PBS-EOI | XX | |||
| D1422-PD1-SICK | XX | |||
| D1452-PD1-SICK | XX | |||
| D1454-PD1-SICK | XX | |||
| D158-PBS-EOI | XX | |||
| D159-PBS-EOI | XX | |||
| D162-PBS-EOI | XX | |||
| D183-PBS-EOI | XX | |||
| D19-PBS-EOI | XX | |||
| D2-PBS-EOI | XX | |||
| D20-PBS-EOI | XX | |||
| D206-PBS-EOI | XX | |||
| D21-PBS-EOI | XX | |||
| D24-ICI-EOI | XX | |||
| D250-PBS-EOI | XX | |||
| D251-PBS-Myo | XX | |||
| D30-ICI-Myo | XX | |||
| D300-ICI-SICK | XX | |||
| D304-ICI-Myo | XX | |||
| D309-ICI-Myo | XX | |||
| D31-PBS-Myo | XX | |||
| D313-ICI-Myo | XX | |||
| D315-PD1-SICK | XX | |||
| D318-ICI-Myo | XX | |||
| D320-ICI-SICK | XX | XX | ||
| D329-PBS-SICK | XX | |||
| D343-ICI-SICK | XX | |||
| D344-ICI-Myo | XX | |||
| D345-ICI-SICK | XX | |||
| D35-PBS-EOI | XX | |||
| D351-ICI-Myo | XX | XX | ||
| D357-PD1-SICK | XX | |||
| D362-ICI-Myo | XX | |||
| D37-PBS-EOI | XX | |||
| D373-ICI-Myo | XX | |||
| D374-ICI-Myo | XX | |||
| D38-ICI-Myo | XX | XX | ||
| D386-ICI-Myo | XX | |||
| D398-ICI-Myo | XX | |||
| D4-PBS-EOI | XX | |||
| D40-ICI-Myo | XX | |||
| D402-ICI-Myo | XX | |||
| D408-ICI-Myo | XX | |||
| D410-ICI-Myo | XX | |||
| D436-PBS-Myo | XX | |||
| D463-ICI-SICK | XX | |||
| D467-ICI-Myo | XX | |||
| D491-PBS-SICK | XX | |||
| D50-PBS-EOI | XX | |||
| D503-ICI-Myo | XX | |||
| D506-ICI-Myo | XX | |||
| D509-ICI-Myo | XX | |||
| D517-ICI-Myo | XX | |||
| D529-ICI-Myo | XX | |||
| D532-ICI-SICK | XX | |||
| D538-PD1-SICK | XX | |||
| D548-ICI-Myo | XX | |||
| D551-ICI-SICK | XX | |||
| D552-ICI-Myo | XX | |||
| D558-ICI-SICK | XX | |||
| D563-ICI-Myo | XX | |||
| D6-PBS-Myo | XX | |||
| D60-PBS-EOI | XX | |||
| D61-ICI-EOI | XX | |||
| D611-ICI-Myo | XX | XX | XX | |
| D62-ICI-Myo | XX | |||
| D629-PD1-SICK | XX | |||
| D63-ICI-Myo | XX | XX | ||
| D631-ICI-SICK | XX | |||
| D66-ICI-Myo | XX | |||
| D667-ICI-Myo | XX | |||
| D678-ICI-Myo | XX | |||
| D679-ICI-SICK | XX | |||
| D682-ICI-Myo | XX | |||
| D69-PBS-EOI | XX | |||
| D699-ICI-Myo | XX | |||
| D70-PBS-EOI | XX | |||
| D704-PD1-SICK | XX | |||
| D727-PBS-Myo | XX | |||
| D74-ICI-EOI | XX | |||
| D743-ICI-Myo | XX | |||
| D75-ICI-EOI | XX | |||
| D752-PD1-SICK | XX | |||
| D755-ICI-SICK | XX | |||
| D761-PD1-SICK | XX | |||
| D767-ICI-Myo | XX | |||
| D8-PBS-Myo | XX | |||
| D82-PBS-EOI | XX | |||
| D821-ICI-Myo | XX | |||
| D83-PBS-EOI | XX | |||
| D857-PD1-SICK | XX | |||
| D86-ICI-Myo | XX | |||
| D868-ICI-Myo | XX | |||
| D870-ICI-Myo | XX | |||
| D879-ICI-Myo | XX | |||
| D882-ICI-Myo | XX | |||
| D89-ICI-Myo | XX | |||
| D90-ICI-EOI | XX | |||
| D927-ICI-Myo | XX | |||
| D931-ICI-Myo | XX | |||
| D934-ICI-Myo | XX | |||
| D964-PD1-SICK | XX | |||
| D969-PD1-SICK | XX | |||
| D975-PD1-SICK | XX | |||
| D976-PD1-SICK | XX | |||
| D981-PD1-SICK | XX | |||
| D985-ICI-Myo | XX | |||
| D993-PBS-Myo | XX | |||
| NM00155-ICI-Myo | XX | |||
| NM00161-ICI-EOI | XX | |||
| NM00170-PBS-EOI | XX | |||
| NM00174-PBS-Myo | XX | |||
| NM00175-ICI-EOI | XX | |||
| NM00180-PBS-Myo | XX | |||
| NM00181-PBS-Myo | XX | |||
| NM00183-PBS-Myo | XX | |||
| NM00184-ICI-Myo | XX | |||
| NM00185-ICI-Myo | XX |
sessionInfo()R version 4.2.2 (2022-10-31)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Big Sur ... 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] readxl_1.4.1 cluster_2.1.4 dplyr_1.0.10 optparse_1.7.3
[5] rhdf5_2.40.0 tidyr_1.2.1 data.table_1.14.6 fst_0.9.8
[9] knitr_1.41 kableExtra_1.3.4 mclust_6.0.0 ggrepel_0.9.2
[13] ggplot2_3.4.0 qtlcharts_0.16 qtl2_0.30 broman_0.80
[17] workflowr_1.7.0
loaded via a namespace (and not attached):
[1] httr_1.4.4 sass_0.4.4 bit64_4.0.5 jsonlite_1.8.4
[5] viridisLite_0.4.1 bslib_0.4.1 assertthat_0.2.1 getPass_0.2-2
[9] highr_0.9 blob_1.2.3 cellranger_1.1.0 yaml_2.3.6
[13] pillar_1.8.1 RSQLite_2.2.19 glue_1.6.2 digest_0.6.30
[17] promises_1.2.0.1 rvest_1.0.3 colorspace_2.0-3 htmltools_0.5.3
[21] httpuv_1.6.6 pkgconfig_2.0.3 purrr_0.3.5 scales_1.2.1
[25] webshot_0.5.4 processx_3.8.0 svglite_2.1.0 qtl_1.54
[29] whisker_0.4.1 getopt_1.20.3 later_1.3.0 git2r_0.30.1
[33] tibble_3.1.8 farver_2.1.1 generics_0.1.3 ellipsis_0.3.2
[37] cachem_1.0.6 withr_2.5.0 cli_3.4.1 magrittr_2.0.3
[41] memoise_2.0.1 evaluate_0.18 ps_1.7.2 fs_1.5.2
[45] fansi_1.0.3 xml2_1.3.3 tools_4.2.2 lifecycle_1.0.3
[49] stringr_1.5.0 Rhdf5lib_1.18.2 munsell_0.5.0 callr_3.7.3
[53] compiler_4.2.2 jquerylib_0.1.4 systemfonts_1.0.4 rlang_1.0.6
[57] grid_4.2.2 fstcore_0.9.12 rhdf5filters_1.8.0 rstudioapi_0.14
[61] htmlwidgets_1.5.4 labeling_0.4.2 rmarkdown_2.18 gtable_0.3.1
[65] DBI_1.1.3 R6_2.5.1 fastmap_1.1.0 bit_4.0.5
[69] utf8_1.2.2 rprojroot_2.0.3 stringi_1.7.8 parallel_4.2.2
[73] Rcpp_1.0.9 vctrs_0.5.1 tidyselect_1.2.0 xfun_0.35