QTL Analysis - binary [ICI vs PBS] (conditioning on eoi vs. ici peaks)
Belinda Cornes
2022-07-02
Last updated: 2022-07-02
Checks: 5 2
Knit directory: Serreze-T1D_Workflow/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R
Markdown file created these results, you’ll want to first commit it to
the Git repo. If you’re still working on the analysis, you can ignore
this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20220210) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /Users/corneb/Documents/MyJax/CS/Projects/Serreze/qc/workflowr/Serreze-T1D_Workflow | . |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version a8c2d1a. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: analysis/.DS_Store
Ignored: data/.DS_Store
Untracked files:
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_0.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_52.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_mis_0.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_mis_52.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_mis_vo.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_vo.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_ici-early.vs.pbs.ici-late_5.batches.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_ici-early.vs.pbs.ici-late_5.batches_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs.ici-late_5.batches.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs.ici-late_5.batches_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs.ici-late_snpsqc_5.batches.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs.ici-late_snpsqc_5.batches_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs.ici-late_snpsqc_dis_no-x_updated_5.batches.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs.ici-late_snpsqc_dis_no-x_updated_5.batches_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.eoi_snpsqc_dis_no-x_updated_5.batches_11.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.eoi_snpsqc_dis_no-x_updated_5.batches_mis_11.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_conditional.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_conditional_1-peak.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_conditional_1-peak.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_conditional_2-peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_conditional_2-peaks.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_conditional_3-peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_conditional_3-peaks.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_mis_conditional.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_mis_conditional_1-peak.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_mis_conditional_1-peak.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_mis_conditional_2-peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_mis_conditional_2-peaks.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_mis_conditional_3-peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_mis_conditional_3-peaks.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_1.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_conditional.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_conditional_1-peak.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_conditional_1-peak.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_conditional_2-peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_conditional_2-peaks.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_conditional_3-peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_conditional_3-peaks.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_mis_1.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_mis_conditional.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_mis_conditional_1-peak.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_mis_conditional_1-peak.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_mis_conditional_2-peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_mis_conditional_2-peaks.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_mis_conditional_3-peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_mis_conditional_3-peaks.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_1.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_11.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_conditional.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_conditional_1-peak.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_conditional_1-peak.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_conditional_2-peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_conditional_2-peaks.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_conditional_3-peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_conditional_3-peaks.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_11.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_conditional.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_conditional_1-peak.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_conditional_1-peak.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_conditional_1.peak.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_conditional_2-peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_conditional_2-peaks.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_conditional_3-peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_conditional_3-peaks.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs.ici-late_pheno.corrected.cleaned_5.batches.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs.ici-late_pheno.corrected.cleaned_5.batches_mis.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs.ici-late_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs.ici-late_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs.ici-late_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs.ici-late_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_changed.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_changed.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_scanone_5.batches.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_scanone_5.batches_mis.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_oops.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_v2.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.4.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.4_miss.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.5.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.5_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_pheno.corrected.cleaned_5.batches_conditional.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_pheno.corrected.cleaned_5.batches_mis_conditional.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_conditional.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_mis_conditional.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_conditional.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_conditional.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs.ici-late_pheno.corrected.cleaned_5.batches.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs.ici-late_pheno.corrected.cleaned_5.batches_mis.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs.ici-late_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs.ici-late_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs.ici-late_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs.ici-late_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_vo.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_vo1.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_vo.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_vo1.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_pheno.corrected.cleaned_5.batches_conditional.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_pheno.corrected.cleaned_5.batches_mis_conditional.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_conditional.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_mis_conditional.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_conditional.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_conditional.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_conditional_1-peak.Rmd.R
Untracked: analysis/genotype.frequencies_ici-early.vs.pbs.ici-late_5.batches.Rmd
Untracked: analysis/genotype.frequencies_ici-early.vs.pbs.ici-late_5.batches_mis.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.eoi_5.batches_0.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.eoi_5.batches_52.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.eoi_5.batches_mis_0.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.eoi_5.batches_mis_52.Rmd
Untracked: analysis/index_5.batches_additional_vo.Rmd
Untracked: analysis/power.analysis_ici.vs.eoi_5.batches.Rmd
Untracked: analysis/power.analysis_ici.vs.eoi_5.batches.Rmd.R
Untracked: analysis/power.analysis_ici.vs.eoi_5.batches_mis.Rmd
Untracked: analysis/power.analysis_ici.vs.eoi_5.batches_mis.Rmd.R
Untracked: analysis/power.analysis_ici.vs.pbs_5.batches.Rmd
Untracked: analysis/power.analysis_ici.vs.pbs_5.batches.Rmd.R
Untracked: analysis/power.analysis_ici.vs.pbs_5.batches_mis.Rmd
Untracked: analysis/power.analysis_ici.vs.pbs_5.batches_mis.Rmd.R
Untracked: data/GM_covar.csv
Untracked: data/GM_covar_BC312.csv
Untracked: data/bad_markers_all_4.batches.RData
Untracked: data/bad_markers_all_5.batches.RData
Untracked: data/blup_sub_chr10_lod.drop-1.5_5.batches.csv
Untracked: data/blup_sub_chr3_lod.drop-1.5.csv
Untracked: data/blup_sub_chr3_lod.drop-1.5_5.batches.csv
Untracked: data/blup_sub_chr4_lod.drop-1.5.csv
Untracked: data/blup_sub_chr4_lod.drop-1.5_5.batches.csv
Untracked: data/covar_cleaned_ici.vs.eoi.csv
Untracked: data/covar_cleaned_ici.vs.pbs.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs.ici-late_5.batches.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs.ici-late_5.batches_mis.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_0.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_52.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_mis.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_mis_0.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_mis_52.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi1.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_0.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_52.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_mis.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_mis_0.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_mis_52.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs1.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_0.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_52.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_mis.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_mis_0.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_mis_52.csv
Untracked: data/covar_corrected_ici-early.vs.pbs.ici-late_5.batches.csv
Untracked: data/covar_corrected_ici-early.vs.pbs.ici-late_5.batches_mis.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_0.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_52.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_mis.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_mis_0.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_mis_52.csv
Untracked: data/covar_corrected_ici.vs.eoi.csv
Untracked: data/covar_corrected_ici.vs.eoi1.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_0.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_52.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_mis.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_mis_0.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_mis_52.csv
Untracked: data/covar_corrected_ici.vs.pbs.csv
Untracked: data/covar_corrected_ici.vs.pbs1.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_0.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_52.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_mis.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_mis_0.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_mis_52.csv
Untracked: data/e.RData
Untracked: data/e_BC312.RData
Untracked: data/e_snpg_samqc_4.batches.RData
Untracked: data/e_snpg_samqc_4.batches_bc.RData
Untracked: data/e_snpg_samqc_5.batches.RData
Untracked: data/errors_ind_4.batches.RData
Untracked: data/errors_ind_4.batches_bc.RData
Untracked: data/errors_ind_5.batches.RData
Untracked: data/files.to.sync.txt
Untracked: data/fitqtl_chr3.peak_chr4.peak_additive.txt
Untracked: data/fitqtl_chr3.peak_chr4.peak_interacting.txt
Untracked: data/fitqtl_chr3.peak_chr4.peak_sex_additive.txt
Untracked: data/fitqtl_chr3.peak_chr4.peak_sex_interacting.txt
Untracked: data/g2blup_effects.csv
Untracked: data/g2blup_effects.xlsx
Untracked: data/genes_chr10_lod.drop-1.5_5.batches.csv
Untracked: data/genes_chr3_lod.drop-1.5.csv
Untracked: data/genes_chr3_lod.drop-1.5_5.batches.csv
Untracked: data/genes_chr4_lod.drop-1.5.csv
Untracked: data/genetic_map.csv
Untracked: data/genetic_map_BC312.csv
Untracked: data/genotype_errors_marker_4.batches.RData
Untracked: data/genotype_errors_marker_5.batches.RData
Untracked: data/genotype_freq_marker_4.batches.RData
Untracked: data/genotype_freq_marker_5.batches.RData
Untracked: data/gm_allqc_4.batches.RData
Untracked: data/gm_allqc_5.batches.RData
Untracked: data/gm_allqc_5.batches_mis.RData
Untracked: data/gm_samqc_3.batches.RData
Untracked: data/gm_samqc_4.batches.RData
Untracked: data/gm_samqc_4.batches_bc.RData
Untracked: data/gm_samqc_5.batches.RData
Untracked: data/gm_serreze.192.RData
Untracked: data/gm_serreze.BC312.RData
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-additive.covariates_blup_sub_chr-6_peak.marker-UNC11006703_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-additive.covariates_blup_sub_chr-6_peak.marker-UNC11006703_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-additive.covariates_blup_sub_chr-6_peak.marker-cr27snv203_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-additive.covariates_genes_chr-6_peak.marker-UNC11006703_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-additive.covariates_genes_chr-6_peak.marker-UNC11006703_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-additive.covariates_genes_chr-6_peak.marker-cr27snv203_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-interactive.covariate_blup_sub_chr-1_peak.marker-ICR134_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-interactive.covariate_blup_sub_chr-1_peak.marker-ICR4119_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-interactive.covariate_blup_sub_chr-1_peak.marker-JAX00262446_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-interactive.covariate_blup_sub_chr-1_peak.marker-UNCHS001938_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-interactive.covariate_blup_sub_chr-6_peak.marker-UNC11008761_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-interactive.covariate_blup_sub_chr-6_peak.marker-UNCHS016990_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-interactive.covariate_genes_chr-1_peak.marker-ICR134_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-interactive.covariate_genes_chr-1_peak.marker-ICR4119_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-interactive.covariate_genes_chr-1_peak.marker-JAX00262446_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-interactive.covariate_genes_chr-1_peak.marker-UNCHS001938_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-interactive.covariate_genes_chr-6_peak.marker-UNC11008761_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-interactive.covariate_genes_chr-6_peak.marker-UNCHS016990_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNC28655293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_blup_sub_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_blup_sub_chr-5_peak.marker-UNC8675939_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNC28655293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_genes_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_genes_chr-5_peak.marker-UNC8675939_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_genes_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-10_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-10_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-10_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-11_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-11_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-11_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-12_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-12_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-12_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-13_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-13_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-13_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-14_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-14_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-14_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-15_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-15_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-15_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-16_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-16_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-16_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-17_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-17_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-17_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-18_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-18_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-18_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-19_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-19_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-19_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-1_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-1_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-1_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-2_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-2_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-2_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-3_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-3_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-3_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-4_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-4_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-4_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-5_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-6_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-6_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-6_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-7_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-7_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-7_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-8_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-8_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-8_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-9_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-9_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-9_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-X_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-X_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-X_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-10_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-10_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-10_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-11_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-11_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-11_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-12_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-12_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-12_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-13_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-13_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-13_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-14_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-14_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-14_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-15_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-15_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-15_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-16_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-16_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-16_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-17_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-17_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-17_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-18_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-18_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-18_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-19_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-19_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-19_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-1_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-1_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-1_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-2_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-2_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-2_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-3_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-3_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-3_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-4_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-4_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-4_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-5_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-6_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-6_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-6_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-7_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-7_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-7_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-8_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-8_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-8_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-9_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-9_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-9_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-X_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-X_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-X_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_gm_qtl_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_gm_qtl_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_gm_qtl_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_gm_qtl_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_gm_qtl_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_marker.freq_low.geno.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_marker.freq_low.probs.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_scanone_5.batches.Rdata
Untracked: data/ici-early.vs.pbs.ici-late_scanone_5.batches_mis.Rdata
Untracked: data/ici-early.vs.pbs.ici-late_scanone_snpsqc_5.batches.Rdata
Untracked: data/ici-early.vs.pbs.ici-late_scanone_snpsqc_5.batches_mis.Rdata
Untracked: data/ici-early.vs.pbs.ici-late_scanone_snpsqc_dis_no-x_updated_5.batches.Rdata
Untracked: data/ici-early.vs.pbs.ici-late_scanone_snpsqc_dis_no-x_updated_5.batches_mis.Rdata
Untracked: data/ici-early.vs.pbs_age.of.onset-additive.covariates_blup_sub_chr-7_peak.marker-UNC13388811_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-additive.covariates_genes_chr-7_peak.marker-UNC13388811_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-15_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-15_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-15_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-17_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-17_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-17_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_5.batches.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_scanone_5.batches.Rdata
Untracked: data/ici-early.vs.pbs_scanone_5.batches_mis.Rdata
Untracked: data/ici-early.vs.pbs_scanone_snpsqc_5.batches.Rdata
Untracked: data/ici-early.vs.pbs_scanone_snpsqc_5.batches_mis.Rdata
Untracked: data/ici-early.vs.pbs_scanone_snpsqc_dis_no-x_updated_5.batches.Rdata
Untracked: data/ici-early.vs.pbs_scanone_snpsqc_dis_no-x_updated_5.batches_mis.Rdata
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00020646_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00292499_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00294019_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18216614_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18240977_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18343181_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18363544_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18363544_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18363544_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-11_peak.marker-UNC20090524_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-2_peak.marker-UNCHS008007_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-2_peak.marker-UNCHS008007_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR5263_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR5263_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00105649_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00105649_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00106210_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00107193_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00516812_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00516812_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00522922_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00524205_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00535764_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4901083_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4901083_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4909519_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4909519_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4915446_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4915446_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4938565_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4938565_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4959266_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4959266_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4960612_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4960612_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4961542_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4961542_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5046545_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5055259_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5062524_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5068215_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5076935_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5082757_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5173293_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5193970_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5257268_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5264696_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5267813_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5270225_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5277061_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5278229_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5278229_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5282259_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5287977_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5301460_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5309136_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5321559_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5322455_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5329171_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5360055_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5778977_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5916997_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008229_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008229_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008234_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008234_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008242_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008260_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008260_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008281_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008281_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008338_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008338_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008363_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008363_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008409_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008432_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008609_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008614_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008627_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008659_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008725_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009066_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009067_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009068_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009112_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009693_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009821_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001276_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-sanger2496q_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-sanger2496q_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-4_peak.marker-UNC8250659_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-4_peak.marker-UNC8439633_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-4_peak.marker-UNC8439633_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-4_peak.marker-UNCHS012955_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-6_peak.marker-UNC11108920_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-6_peak.marker-UNC11108920_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00020646_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00292499_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00294019_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18216614_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18240977_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18343181_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18363544_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18363544_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18363544_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-11_peak.marker-UNC20090524_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-2_peak.marker-UNCHS008007_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-2_peak.marker-UNCHS008007_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR5263_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR5263_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00105649_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00105649_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00106210_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00107193_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00516812_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00516812_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00522922_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00524205_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00535764_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4901083_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4901083_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4909519_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4909519_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4915446_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4915446_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4938565_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4938565_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4959266_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4959266_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4960612_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4960612_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4961542_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4961542_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5046545_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5055259_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5062524_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5068215_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5076935_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5082757_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5173293_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5193970_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5257268_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5264696_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5267813_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5270225_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5277061_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5278229_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5278229_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5282259_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5287977_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5301460_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5309136_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5321559_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5322455_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5329171_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5360055_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5778977_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5916997_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008229_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008229_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008234_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008234_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008242_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008260_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008260_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008281_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008281_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008338_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008338_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008363_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008363_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008409_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008432_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008609_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008614_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008627_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008659_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008725_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009066_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009067_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009068_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009112_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009693_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009821_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001276_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-sanger2496q_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-sanger2496q_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-4_peak.marker-UNC8250659_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-4_peak.marker-UNC8439633_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-4_peak.marker-UNC8439633_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-4_peak.marker-UNCHS012955_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-6_peak.marker-UNC11108920_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-6_peak.marker-UNC11108920_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_blup.full_chr-10_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-11_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-12_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-13_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-14_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-15_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-16_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-17_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-18_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-19_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-1_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-2_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-3_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-4_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-6_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-7_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-8_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-9_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-X_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-ICR4295_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-JAX00292499_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18154330_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18195765_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18216614_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18240977_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18856953_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNCJPD004316_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNCJPD009532_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNC19155926_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNC19481366_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNCJPD004792_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-12_peak.marker-ICR4497_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-13_peak.marker-UNCHS036964_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-18_peak.marker-UNCHS045344_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-19_peak.marker-UNCJPD007087_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-1_peak.marker-ICR010_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-1_peak.marker-ICR010_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-2_peak.marker-UNC4609660_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-B6_rs31740628_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR1269_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR5325_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR5347_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR5348_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00104971_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00105059_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00105203_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00107930_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00107980_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00108068_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00108114_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00109750_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00189245_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00517218_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00517906_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00518418_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00518684_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00520122_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00524717_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00524828_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00525380_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00525709_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00527291_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00527587_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00529707_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00531601_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00532122_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4673471_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4681411_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4793677_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4799475_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4909830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4914130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4964062_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4985064_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4991565_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5001723_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5042361_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5055259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5076935_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5082757_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5173293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5182782_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5193970_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5240484_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5256922_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5266664_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5266711_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5267813_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5274554_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5277061_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5282259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5287977_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5301460_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5321559_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5322455_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5328596_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5350384_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5354724_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5375208_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5384072_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5400314_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5433945_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5435874_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5466669_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5517972_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5522257_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5523265_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5532424_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5553882_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5572051_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5582753_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5589260_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5595555_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5623239_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5639830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5680234_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5682314_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5725356_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5727351_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5734420_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5743257_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5751567_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5752623_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5776974_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5778977_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5788507_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5792468_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5794598_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5795561_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5802298_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5806628_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5817478_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5836999_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5841501_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5847927_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5966417_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008129_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008173_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008231_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008235_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008242_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008253_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008256_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008311_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008317_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008372_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008410_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008415_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008431_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008609_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008614_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008649_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008651_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008723_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008802_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008803_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008808_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008834_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008838_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008928_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008948_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008959_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008973_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008987_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009001_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009025_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009034_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009087_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009109_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009142_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009207_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009216_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009306_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009316_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009364_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009377_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009382_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009387_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009395_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009413_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009428_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009463_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009503_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009532_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009541_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009560_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009574_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001276_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001315_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001355_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001373_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001382_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC6851205_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8250659_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8299912_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8300499_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8381981_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8436434_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8439633_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8530763_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS012948_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS012976_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013139_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013304_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-5_peak.marker-JAX00590770_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-ICR1830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-UNC13170794_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-UNCHS019480_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-cr31snv87_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-UNC15430711_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-9_peak.marker-UNC17203329_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-9_peak.marker-UNCHS024593_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-X_peak.marker-UNC30627130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-X_peak.marker-UNCHS048313_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_snpsqc_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_snpsqc_dis_no-x_updated_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_snpsqc_dis_no-x_updated_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_snpsqc_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_snpsqc_dis_no-x_updated_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_snpsqc_dis_no-x_updated_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_snpsqc_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_snpsqc_dis_no-x_updated_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_snpsqc_dis_no-x_updated_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_snpsqc_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_snpsqc_dis_no-x_updated_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_snpsqc_dis_no-x_updated_5.batches_mis.mis.txt
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-ICR4295_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-JAX00292499_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18154330_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18195765_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18216614_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18240977_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18856953_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNCJPD004316_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNCJPD009532_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNC19155926_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNC19481366_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNCJPD004792_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-12_peak.marker-ICR4497_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-13_peak.marker-UNCHS036964_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-18_peak.marker-UNCHS045344_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-19_peak.marker-UNCJPD007087_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-1_peak.marker-ICR010_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-1_peak.marker-ICR010_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-2_peak.marker-UNC4609660_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-B6_rs31740628_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR1269_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR5325_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR5347_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR5348_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00104971_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00105059_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00105203_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00107930_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00107980_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00108068_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00108114_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00109750_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00189245_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00517218_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00517906_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00518418_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00518684_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00520122_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00524717_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00524828_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00525380_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00525709_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00527291_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00527587_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00529707_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00531601_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00532122_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4673471_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4681411_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4793677_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4799475_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4909830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4914130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4964062_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4985064_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4991565_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5001723_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5042361_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5055259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5076935_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5082757_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5173293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5182782_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5193970_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5240484_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5256922_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5266664_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5266711_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5267813_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5274554_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5277061_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5282259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5287977_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5301460_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5321559_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5322455_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5328596_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5350384_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5354724_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5375208_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5384072_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5400314_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5433945_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5435874_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5466669_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5517972_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5522257_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5523265_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5532424_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5553882_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5572051_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5582753_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5589260_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5595555_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5623239_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5639830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5680234_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5682314_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5725356_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5727351_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5734420_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5743257_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5751567_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5752623_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5776974_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5778977_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5788507_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5792468_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5794598_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5795561_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5802298_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5806628_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5817478_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5836999_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5841501_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5847927_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5966417_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008129_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008173_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008231_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008235_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008242_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008253_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008256_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008311_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008317_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008372_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008410_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008415_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008431_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008609_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008614_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008649_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008651_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008723_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008802_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008803_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008808_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008834_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008838_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008928_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008948_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008959_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008973_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008987_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009001_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009025_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009034_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009087_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009109_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009142_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009207_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009216_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009306_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009316_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009364_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009377_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009382_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009387_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009395_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009413_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009428_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009463_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009503_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009532_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009541_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009560_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009574_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001276_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001315_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001355_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001373_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001382_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC6851205_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8250659_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8299912_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8300499_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8381981_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8436434_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8439633_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8530763_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS012948_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS012976_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013139_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013304_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-5_peak.marker-JAX00590770_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-ICR1830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-UNC13170794_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-UNCHS019480_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-cr31snv87_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-UNC15430711_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-9_peak.marker-UNC17203329_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-9_peak.marker-UNCHS024593_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-X_peak.marker-UNC30627130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-X_peak.marker-UNCHS048313_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_gm_qtl_5.batches.csv
Untracked: data/ici.vs.eoi_gm_qtl_5.batches_mis.csv
Untracked: data/ici.vs.eoi_gm_qtl_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_gm_qtl_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_gm_qtl_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18343181_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18376338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18382395_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNCHS029024_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-1_peak.marker-UNC228500_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-1_peak.marker-UNC228500_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-ICR2047_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00112499_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00219554_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00529707_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4837069_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5006948_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5266664_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5300878_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5360055_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5679155_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5945286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC6227106_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008595_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009067_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009693_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001341_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-sanger2623_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-4_peak.marker-UNCHS013125_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-4_peak.marker-UNCHS013125_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-6_peak.marker-UNCHS018170_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-6_peak.marker-UNCHS018231_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18343181_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18376338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18382395_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNCHS029024_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-1_peak.marker-UNC228500_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-1_peak.marker-UNC228500_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-ICR2047_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00112499_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00219554_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00529707_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4837069_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5006948_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5266664_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5300878_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5360055_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5679155_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5945286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC6227106_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008595_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS009067_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS009693_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCJPD001246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCJPD001341_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-sanger2623_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-4_peak.marker-UNCHS013125_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-4_peak.marker-UNCHS013125_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-6_peak.marker-UNCHS018170_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-6_peak.marker-UNCHS018231_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.geno.freq.removed.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.geno.freq.removed_5.batches.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.geno.freq.removed_5.batches_mis.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_5.batches.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_5.batches_mis.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.probs.freq.removed.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.probs.freq.removed_5.batches.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.probs.freq.removed_5.batches_mis.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_5.batches.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_5.batches_mis.csv
Untracked: data/ici.vs.eoi_scanone_5.batches.Rdata
Untracked: data/ici.vs.eoi_scanone_5.batches_mis.Rdata
Untracked: data/ici.vs.eoi_scanone_snpsqc_5.batches.Rdata
Untracked: data/ici.vs.eoi_scanone_snpsqc_5.batches_mis.Rdata
Untracked: data/ici.vs.eoi_scanone_snpsqc_dis_no-x_updated_5.batches.Rdata
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNC28655293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-5_peak.marker-UNC8675939_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNC28655293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-5_peak.marker-UNC8675939_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_bad.markers_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-15_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-15_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-15_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-17_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-17_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-17_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-13_peak.marker-UNC22384241_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_conditional_1-peak-chr10.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_conditional_1-peak-chr3.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_conditional_1-peak-chr4.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-19_peak.marker-UNC29893228_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-19_peak.marker-UNC29990033_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-19_peak.marker-UNCHS047192_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-8_peak.marker-UNCHS023353_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-8_peak.marker-UNCHS023353_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-8_peak.marker-UNCHS023364_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_fitqtl_peaks_additive_snpsqc_dis_no-x_updated_5.batches_mis_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_additive_snpsqc_dis_no-x_updated_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_additive_snpsqc_snpsqc_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_interacting_snpsqc_dis_no-x_updated_5.batches_mis_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_interacting_snpsqc_dis_no-x_updated_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_interacting_snpsqc_snpsqc_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_additive_snpsqc_dis_no-x_updated_5.batches_mis_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_additive_snpsqc_dis_no-x_updated_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_additive_snpsqc_snpsqc_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_interacting_snpsqc_dis_no-x_updated_5.batches_mis_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_interacting_snpsqc_dis_no-x_updated_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_interacting_snpsqc_snpsqc_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_genes_chr-13_peak.marker-UNC22384241_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_conditional_1-peak-chr10.csv
Untracked: data/ici.vs.pbs_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_conditional_1-peak-chr3.csv
Untracked: data/ici.vs.pbs_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_conditional_1-peak-chr4.csv
Untracked: data/ici.vs.pbs_genes_chr-19_peak.marker-UNC29893228_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-19_peak.marker-UNC29990033_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-19_peak.marker-UNCHS047192_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-8_peak.marker-UNCHS023353_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-8_peak.marker-UNCHS023353_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-8_peak.marker-UNCHS023364_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_conditional_1-peak-chr10.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_conditional_1-peak-chr3.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_conditional_1-peak-chr4.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_conditional_2-peaks-chr3-10.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_conditional_2-peaks-chr3-4.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_conditional_2-peaks-chr4-10.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_mis.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_mis_conditional_1-peak-chr10.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_mis_conditional_1-peak-chr3.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_mis_conditional_1-peak-chr4.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_mis_conditional_1-peak-chr10.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_mis_conditional_1-peak-chr3.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_mis_conditional_1-peak-chr4.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis_conditional_1-peak-chr3.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis_conditional_3-peaks-chr3-4-10.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratiov_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratiov_5.batches_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-JAX00153527_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-JAX00153527_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNCHS020443_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNCHS020443_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNCHS020486_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-JAX00153527_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-JAX00153527_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNCHS020443_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNCHS020443_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNCHS020486_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_5.batches.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_5.batches_mis.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_5.batches.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_5.batches_mis.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed_5.batches.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed_5.batches_mis.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_5.batches.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_5.batches_mis.csv
Untracked: data/ici.vs.pbs_scanone_5.batches.Rdata
Untracked: data/ici.vs.pbs_scanone_5.batches_mis.Rdata
Untracked: data/ici.vs.pbs_scanone_snpsqc_5.batches.Rdata
Untracked: data/ici.vs.pbs_scanone_snpsqc_5.batches_mis.Rdata
Untracked: data/ici.vs.pbs_scanone_snpsqc_dis_no-x_updated_5.batches.Rdata
Untracked: data/ici.vs.pbs_scanone_snpsqc_dis_no-x_updated_5.batches_mis.Rdata
Untracked: data/mean.differences_age.of.onset_ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_5.batches.csv
Untracked: data/mean.differences_age.of.onset_ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_5.batches_mis.csv
Untracked: data/mean.differences_age.of.onset_ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_5.batches.csv
Untracked: data/mean.differences_age.of.onset_ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_5.batches_mis.csv
Untracked: data/mean.differences_group_ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_5.batches.csv
Untracked: data/mean.differences_group_ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_5.batches_mis.csv
Untracked: data/mean.differences_group_ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_5.batches.csv
Untracked: data/mean.differences_group_ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_5.batches_mis.csv
Untracked: data/percent_missing_id_3.batches.RData
Untracked: data/percent_missing_id_4.batches.RData
Untracked: data/percent_missing_id_4.batches_bc.RData
Untracked: data/percent_missing_id_5.batches.RData
Untracked: data/percent_missing_marker_4.batches.RData
Untracked: data/percent_missing_marker_5.batches.RData
Untracked: data/pheno.csv
Untracked: data/pheno_BC312.csv
Untracked: data/physical_map.csv
Untracked: data/physical_map_BC312.csv
Untracked: data/qc_info_bad_sample_3.batches.RData
Untracked: data/qc_info_bad_sample_4.batches.RData
Untracked: data/qc_info_bad_sample_4.batches_bc.RData
Untracked: data/qc_info_bad_sample_5.batches.RData
Untracked: data/remaining.markers_geno.freq.xlsx
Untracked: data/sample_geno.csv
Untracked: data/sample_geno_AHB_BC312.csv
Untracked: data/sample_geno_bc.csv
Untracked: data/sample_geno_bc_BC312.csv
Untracked: data/serreze_probs.rds
Untracked: data/serreze_probs_BC312.rds
Untracked: data/serreze_probs_allqc.rds
Untracked: data/serreze_probs_allqc_5.batches.rds
Untracked: data/serreze_probs_allqc_5.batches_mis.rds
Untracked: data/summary.cg_3.batches.RData
Untracked: data/summary.cg_4.batches.RData
Untracked: data/summary.cg_4.batches_bc.RData
Untracked: data/summary.cg_5.batches.RData
Untracked: output/Percent_missing_genotype_data_5.batches.pdf
Untracked: output/Percent_missing_genotype_data_per_marker_5.batches.pdf
Untracked: output/Proportion_matching_genotypes_before_removal_of_bad_samples_5.batches.pdf
Untracked: output/genotype_error_marker_5.batches.pdf
Untracked: output/genotype_frequency_marker_5.batches.pdf
Unstaged changes:
Modified: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs_5.batches.Rmd
Modified: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs_5.batches_mis.Rmd
Modified: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs_snpsqc_5.batches.Rmd
Modified: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs_snpsqc_5.batches_mis.Rmd
Modified: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs_snpsqc_dis_no-x_updated_5.batches.Rmd
Modified: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis.Rmd
Modified: analysis/4.1.1_qtl.analysis_binary_ici.vs.eoi_snpsqc_dis_no-x_updated.Rmd
Modified: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/genotype.frequencies_ici.vs.eoi_5.batches.Rmd
Modified: analysis/genotype.frequencies_ici.vs.eoi_5.batches_mis.Rmd
Modified: analysis/genotype.frequencies_ici.vs.pbs_5.batches.Rmd
Modified: analysis/genotype.frequencies_ici.vs.pbs_5.batches_mis.Rmd
Modified: analysis/index_5.batches.Rmd
Modified: analysis/index_5.batches_additional.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with
wflow_publish() to start tracking its development.
Loading Data
We will load the data and subset indivials out that are in the groups of interest. We will create a binary phenotype from this (PBS ==0, ICI == 1).
load("data/gm_allqc_5.batches_mis.RData")
#gm_allqc
gm=gm_allqc
gmObject of class cross2 (crosstype "bc")
Total individuals 308
No. genotyped individuals 308
No. phenotyped individuals 308
No. with both geno & pheno 308
No. phenotypes 1
No. covariates 6
No. phenotype covariates 0
No. chromosomes 20
Total markers 131356
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
9956 9987 7848 7586 7609 7736 7399 6458 6713 6385 7143 6110 6082 5966 5346 5015
17 18 19 X
5080 4605 3562 4770 #pr <- readRDS("data/serreze_probs_allqc_5.batches_mis.rds")
#pr <- readRDS("data/serreze_probs.rds")
##extracting animals with ici and pbs group status
miceinfo <- gm$covar[gm$covar$group == "PBS" | gm$covar$group == "ICI",]
table(miceinfo$group)
ICI PBS
104 34 mice.ids <- rownames(miceinfo)
gm <- gm[mice.ids]
gmObject of class cross2 (crosstype "bc")
Total individuals 138
No. genotyped individuals 138
No. phenotyped individuals 138
No. with both geno & pheno 138
No. phenotypes 1
No. covariates 6
No. phenotype covariates 0
No. chromosomes 20
Total markers 131356
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
9956 9987 7848 7586 7609 7736 7399 6458 6713 6385 7143 6110 6082 5966 5346 5015
17 18 19 X
5080 4605 3562 4770 #pr.qc <- pr
#for (i in 1:20){pr.qc[[i]] = pr.qc[[i]][mice.ids,,]}
#bin_pheno <- NULL
#bin_pheno$PBS <- ifelse(gm$covar$group == "PBS", 1, 0)
#bin_pheno$ICI <- ifelse(gm$covar$group == "ICI", 1, 0)
#bin_pheno <- as.data.frame(bin_pheno)
#rownames(bin_pheno) <- rownames(gm$covar)
gm$covar$ICI.vs.PBS <- ifelse(gm$covar$group == "PBS", 0, 1)
gm.full <- gm
mar.covar <- pull_markers(gm, c("UNCHS008487", "UNC8250659", "UNC18240977"))
mar.covar.g <- do.call("cbind", mar.covar$geno)
covars <- merge(gm$covar, mar.covar.g, by='row.names', sort=F)
table(covars$group)
ICI PBS
104 34 rownames(covars) <- covars$Row.names
covars <- covars[-1]
##removing problmetic marker
gm <- drop_markers(gm, "UNCHS013106")
markers <- marker_names(gm)
gmapdf <- read.csv("/Users/corneb/Documents/MyJax/CS/Projects/Serreze/haplotype.reconstruction/output_5.batches/genetic_map.csv")
pmapdf <- read.csv("/Users/corneb/Documents/MyJax/CS/Projects/Serreze/haplotype.reconstruction/output_5.batches/physical_map.csv")
#mapdf <- merge(gmapdf,pmapdf, by=c("marker","chr"), all=T)
#rownames(mapdf) <- mapdf$marker
#mapdf <- mapdf[markers,]
#names(mapdf) <- c('marker','chr','gmapdf','pmapdf')
#mapdfnd <- mapdf[!duplicated(mapdf[c(2:3)]),]
pr.qc <- calc_genoprob(gm)
gmObject of class cross2 (crosstype "bc")
Total individuals 138
No. genotyped individuals 138
No. phenotyped individuals 138
No. with both geno & pheno 138
No. phenotypes 1
No. covariates 7
No. phenotype covariates 0
No. chromosomes 20
Total markers 131355
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
9956 9987 7848 7585 7609 7736 7399 6458 6713 6385 7143 6110 6082 5966 5346 5015
17 18 19 X
5080 4605 3562 4770 Conditionaing on eoi vs. ici chr 3 peak (UNCHS008487) & chr 4 peak (UNC8250659)
Genome-wide scan
Genome-wide scan
Xcovar <- get_x_covar(gm)
addcovar = model.matrix(~UNCHS008487+UNC8250659, data = covars)[,-1]
#K <- calc_kinship(pr.qc, type = "loco")
#heatmap(K[[1]])
#K.overall <- calc_kinship(pr.qc, type = "overall")
#heatmap(K.overall)
kinship <- calc_kinship(pr.qc)
heatmap(kinship)
#operm <- scan1perm(pr.qc, gm$covar$phenos, Xcovar=Xcovar, n_perm=2000)
#operm <- scan1perm(pr.qc, gm$covar$phenos, addcovar = addcovar, n_perm=2000)
#operm <- scan1perm(pr.qc, gm$covar$phenos, n_perm=2000)
operm <- scan1perm(pr.qc, gm$covar["ICI.vs.PBS"], model="binary", n_perm=10, perm_Xsp=TRUE, chr_lengths=chr_lengths(gm$gmap), addcovar = addcovar)
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
names(summary_table) <- c("autosomes","X")
summary_table$significance.level <- rownames(summary_table)
rownames(summary_table) <- NULL
summary_table[c(3,1:2)] %>%
kable(escape = F,align = c("ccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| significance.level | autosomes | X |
|---|---|---|
| 0.01 | 4.538200 | 5.289082 |
| 0.05 | 4.034056 | 4.476800 |
| 0.1 | 3.402310 | 3.414586 |
The figures below show QTL maps for each phenotype
out <- scan1(pr.qc, gm$covar["ICI.vs.PBS"], Xcovar=Xcovar, model="binary", addcovar = addcovar)
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
plot_lod<-function(out,map){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
##legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- 14 # overall maximum LOD score
plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
##legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " [positions in cM] \n(using same scale as eoi vs. ici for easier comparison)"))
add_threshold(map, summary(operm, alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
plot_lod(out,gm$gmap)
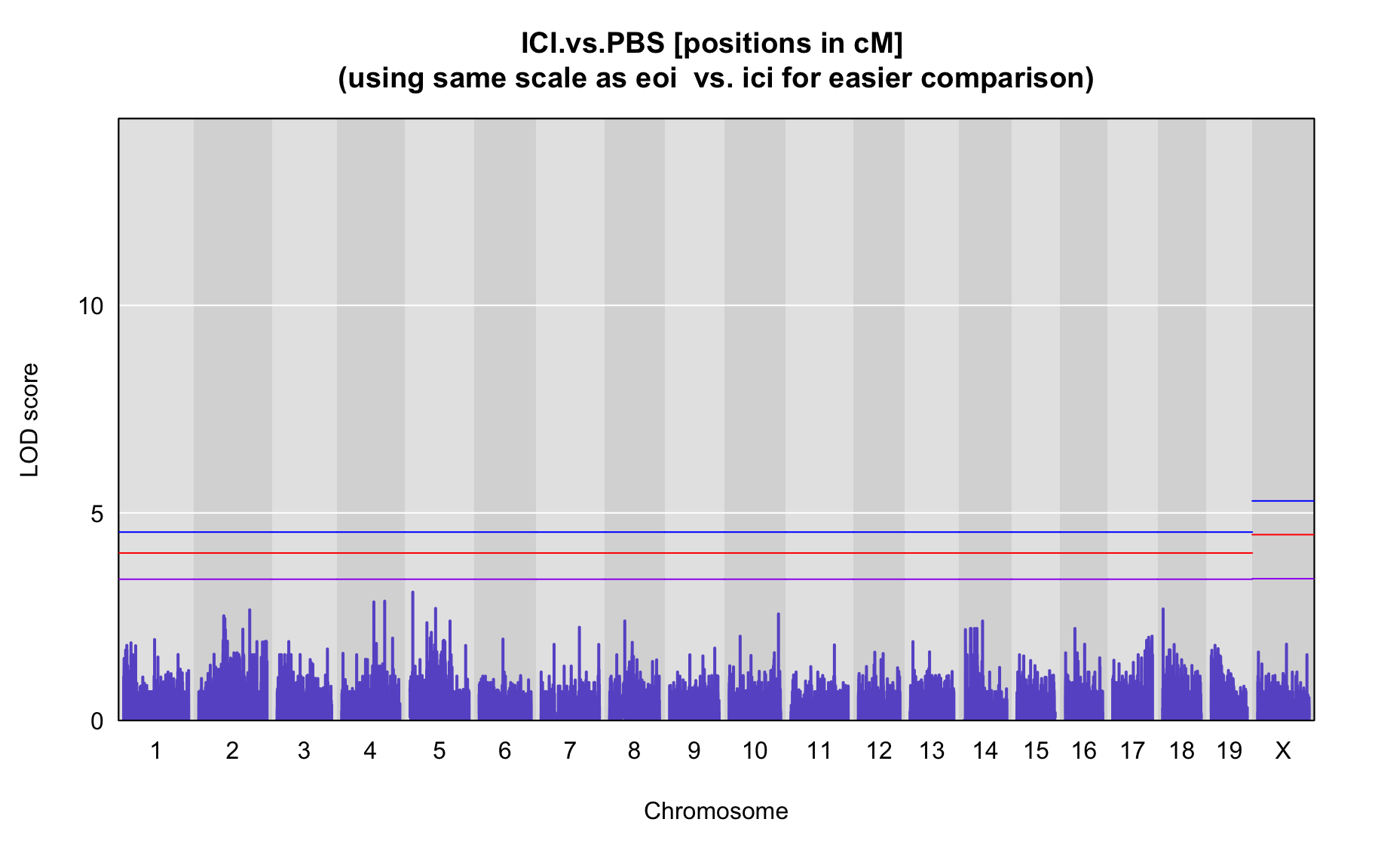
LOD peaks
The table below shows QTL peaks associated with the phenotype. We use the 95% threshold from the permutations to find peaks.
Centimorgan (cM)
peaks <- find_peaks(out, gm$gmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) >0){
peaks$marker <- find_marker(gm$gmap, chr=peaks$chr,pos=peaks$pos)
names(peaks)[2] <- c("phenotype")
peaks <- peaks[-1]
rownames(peaks) <- NULL
print(kable(peaks, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
#plot only peak chromosomes
plot_lod_chr<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
ymx <- 14
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in cM]\n(using same scale as eoi vs. ici for easier comparison)"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
for(i in unique(peaks$chr)){
#for (i in 1:nrow(peaks)){
#plot_lod_chr(out,gm$gmap, peaks$chr[i])
plot_lod_chr(out,gm$gmap, i)
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are no peaks that have a LOD that reaches suggestive (p<0.05) level of 4.03405558593242 [autosomes]/4.47679966592167 [x-chromosome]”
Megabase (MB)
print("peaks in MB positions")[1] “peaks in MB positions”
peaks_mba <- find_peaks(out, gm$pmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) >0){
peaks_mba$marker <- find_marker(gm$pmap, chr=peaks_mba$chr,pos=peaks_mba$pos)
names(peaks_mba)[2] <- c("phenotype")
peaks_mba <- peaks_mba[-1]
#peaks_mbl <- list()
##corresponding info in Mb
#for(i in 1:nrow(peaks)){
# #lodindex <- peaks$lodindex[i]
# phenotype <- peaks$phenotype[i]
# chr <- as.character(peaks$chr[i])
# lod <- peaks$lod[i]
# mark <- peaks$marker[i]
# pos <- mapdf[mapdf$marker==mark,]$pmapdf
# ci_lo <- mapdfnd$pmapdf[which(mapdfnd$gmapdf == peaks$ci_lo[i] & mapdfnd$chr == peaks$chr[i])]
# ci_hi <- mapdfnd$pmapdf[which(mapdfnd$gmapdf == peaks$ci_hi[i] & mapdfnd$chr == peaks$chr[i])]
# peaks_mb=as.data.frame(cbind(phenotype, chr, pos, lod, ci_lo, ci_hi, mark))
# names(peaks_mb)[7] <- c("marker")
# peaks_mbl[[i]] <- peaks_mb
#}
#peaks_mba2 <- do.call(rbind, peaks_mbl)
#peaks_mba2 <- as.data.frame(peaks_mba)
#peaks_mba[,c("chr", "pos", "lod", "ci_lo", "ci_hi")] <- sapply(peaks_mba[,c("chr", "pos", "lod", "ci_lo", "ci_hi")], as.numeric)
rownames(peaks_mba) <- NULL
print(kable(peaks_mba, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
plot_lod_chr_mb<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in MB]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
ymx <- 14
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in MB]\n(using same scale as eoi vs. ici for easier comparison)"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
for(i in unique(peaks_mba$chr)){
#for (i in 1:nrow(peaks_mba)){
#plot_lod_chr_mb(out,gm$pmap, peaks_mba$chr[i])
plot_lod_chr_mb(out,gm$pmap,i)
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are no peaks that have a LOD that reaches suggestive (p<0.05) level of 4.03405558593242 [autosomes]/4.47679966592167 [x-chromosome]”
QTL effects
For each peak LOD location we give a list of gene
query_variants <- create_variant_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/cc_variants.sqlite")
query_genes <- create_gene_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/mouse_genes_mgi.sqlite")
if(nrow(peaks) >0){
for (i in 1:nrow(peaks)){
#for (i in 1:1){
#Plot 1
g <- maxmarg(pr.qc, gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i], return_char=TRUE)
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/","qtl_effect_", i, ".png"))
#par(mar=c(4.1, 4.1, 1.5, 0.6))
plot_pxg(g, gm$covar[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
##dev.off()
chr = peaks$chr[i]
# Plot 2
pr_sub <- pull_genoprobint(pr.qc, gm$gmap, chr, c(peaks$ci_lo[i], peaks$ci_hi[i]))
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar)
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], Xcovar=Xcovar)
#coeff <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
#coeff_sub <- scan1coef(pr_sub[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
blup <- scan1blup(pr.qc[,chr], gm$covar[peaks$phenotype[i]], addcovar = addcovar)
blup_sub <- scan1blup(pr_sub[,chr], gm$covar[peaks$phenotype[i]], addcovar = addcovar)
write.csv(as.data.frame(blup_sub), paste0("data/ici.vs.pbs_blup_sub_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_5.batches_mis_conditional_2-peaks-chr3-4.csv"), quote=F)
#plot_coef(coeff,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i]," [scan1coeff; positions in cM])")
# )
#plot_coef(coeff_sub,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i],"; 1.5 LOD drop interval [scan1coeff; positions in cM] ) ")
# )
plot_coef(blup,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i], "; pos: ", peaks$pos[i], "cM / ",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," [scan1blup; positions in cM])")
)
plot_coef(blup_sub,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i],"; 1.5 LOD drop interval [scan1blup; positions in cM])")
)
# Plot 3
#c2effB <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary", contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
#c2effBb <- scan1blup(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]],Xcovar=Xcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
#plot(c2effB, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
#plot(c2effBb, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
##last_coef <- unclass(c2effB)[nrow(c2effB),2:3] # last two coefficients
##for(t in seq(along=last_coef))
## axis(side=4, at=last_coef[t], names(last_coef)[t], tick=FALSE)
#Table 1
chr = peaks_mba$chr[i]
start=as.numeric(peaks_mba$ci_lo[i])
end=as.numeric(peaks_mba$ci_hi[i])
genesgss = query_genes(chr, start, end)
write.csv(genesgss, file=paste0("data/ici.vs.pbs_genes_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_5.batches_mis_conditional_2-peaks-chr3-4.csv"), quote=F)
rownames(genesgss) <- NULL
genesgss$strand_old = genesgss$strand
genesgss$strand[genesgss$strand=="+"] <- "positive"
genesgss$strand[genesgss$strand=="-"] <- "negative"
#genesgss <-
#table <-
#genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")] %>%
#kable(escape = F,align = c("ccccccccccc")) %>%
#kable_styling("striped", full_width = T) #%>%
#cat #%>%
#column_spec(1, bold=TRUE)
#
#print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], escape = F,align = c("ccccccccccc")))
print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], "html") %>% kable_styling("striped", full_width = T))
#table
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are no peaks that have a LOD that reaches suggestive (p<0.05) level of 4.03405558593242 [autosomes]/4.47679966592167 [x-chromosome]”
R/qtl
scanone
gmObject of class cross2 (crosstype "bc")
Total individuals 138
No. genotyped individuals 138
No. phenotyped individuals 138
No. with both geno & pheno 138
No. phenotypes 1
No. covariates 7
No. phenotype covariates 0
No. chromosomes 20
Total markers 131355
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
9956 9987 7848 7585 7609 7736 7399 6458 6713 6385 7143 6110 6082 5966 5346 5015
17 18 19 X
5080 4605 3562 4770 #detach("package:qtl2", unload=TRUE)
#library(qtl)
cross <- qtl::read.cross("csv", file = "data/ici.vs.pbs_gm_qtl_5.batches_mis_conditional_2-peaks-chr3-4.csv",alleles=c("A","B")) --Read the following data:
138 individuals
131355 markers
5 phenotypes
--Cross type: bc cross <- qtl::jittermap(cross)
summary(cross) Backcross
No. individuals: 138
No. phenotypes: 5
Percent phenotyped: 100 100 100 100 100
No. chromosomes: 20
Autosomes: 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19
X chr: X
Total markers: 131355
No. markers: 9956 9987 7848 7585 7609 7736 7399 6458 6713 6385 7143
6110 6082 5966 5346 5015 5080 4605 3562 4770
Percent genotyped: 99.5
Genotypes (%):
Autosomes: AA:88.1 AB:11.9
X chromosome: AA:93.9 AB:6.1 cross.probs <- qtl::calc.genoprob(cross)
print("method == hk")[1] "method == hk"add.covars = qtl::pull.pheno(cross.probs, c("UNCHS008487","UNC8250659"))
scanone.hk <-qtl::scanone(cross.probs, pheno.col="ICI.vs.PBS" , model="binary", method="hk", addcovar = add.covars)
operm.hk <- qtl::scanone(cross.probs, method = "hk", pheno.col="ICI.vs.PBS", n.perm = 10, perm.Xsp = TRUE, model="binary", verbose=FALSE, addcovar = add.covars)
plot(operm.hk)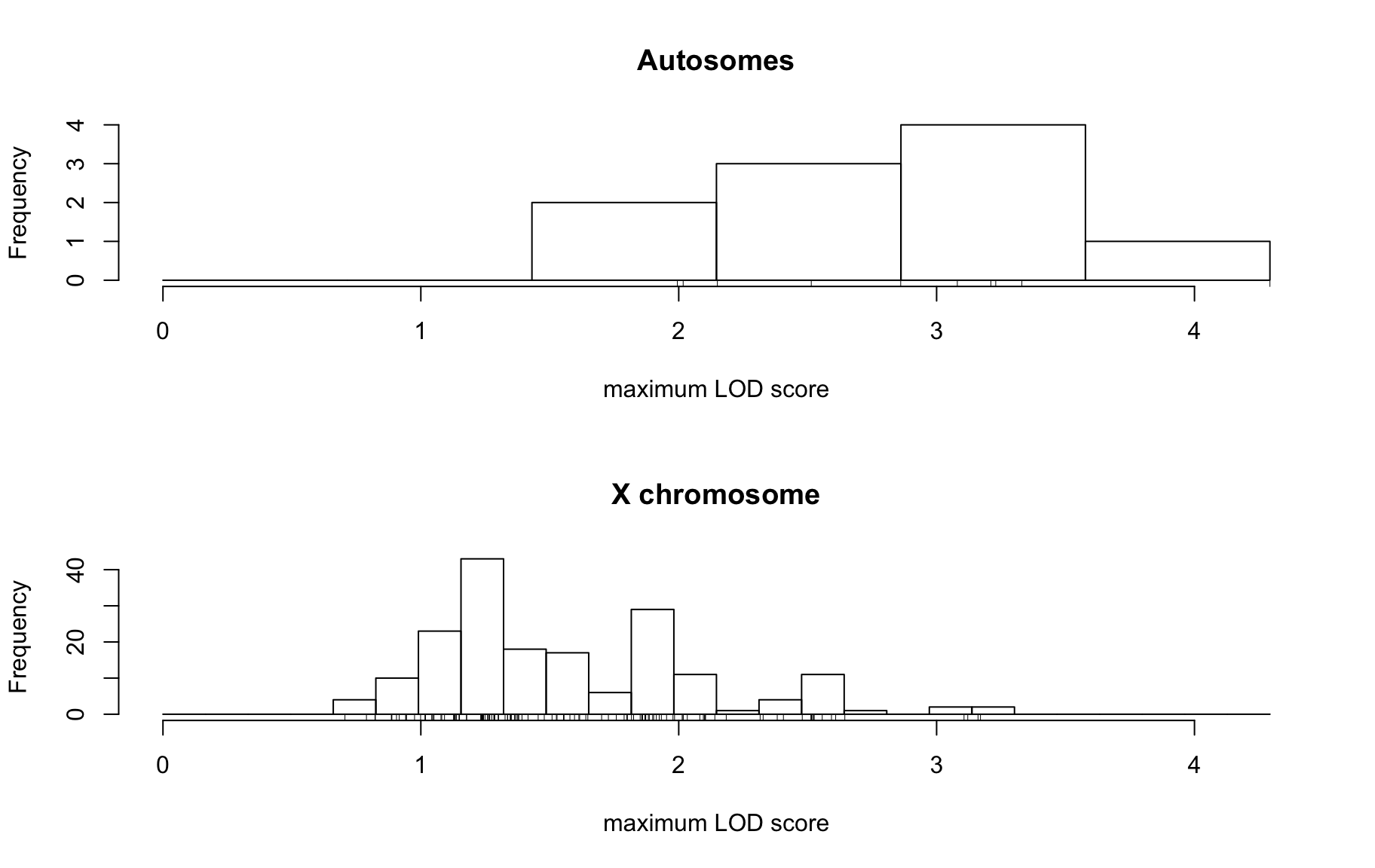
print(summary(operm.hk, alpha=c(0.01, 0.05, 0.1)))Autosome LOD thresholds (10 permutations)
lod
1% 4.21
5% 3.88
10% 3.47
X chromosome LOD thresholds (182 permutations)
lod
1% 3.17
5% 3.17
10% 3.16#plot(scanone.hk, bandcol = "grey90",lty=1, cex=1, col = "steelblue")
#qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.01, col = 'blue')
#qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.05, col = 'red')
#qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.1, col = 'purple')
ymx <- maxlod(out) # overall maximum LOD score
plot(scanone.hk, bandcol = "grey90",lty=1, cex=1, col = "slateblue", ylim=c(0, ymx+0.5))
title(main = paste0(colnames(out), " [positions in cM]"))
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.01, col = 'blue')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.05, col = 'red')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.1, col = 'purple')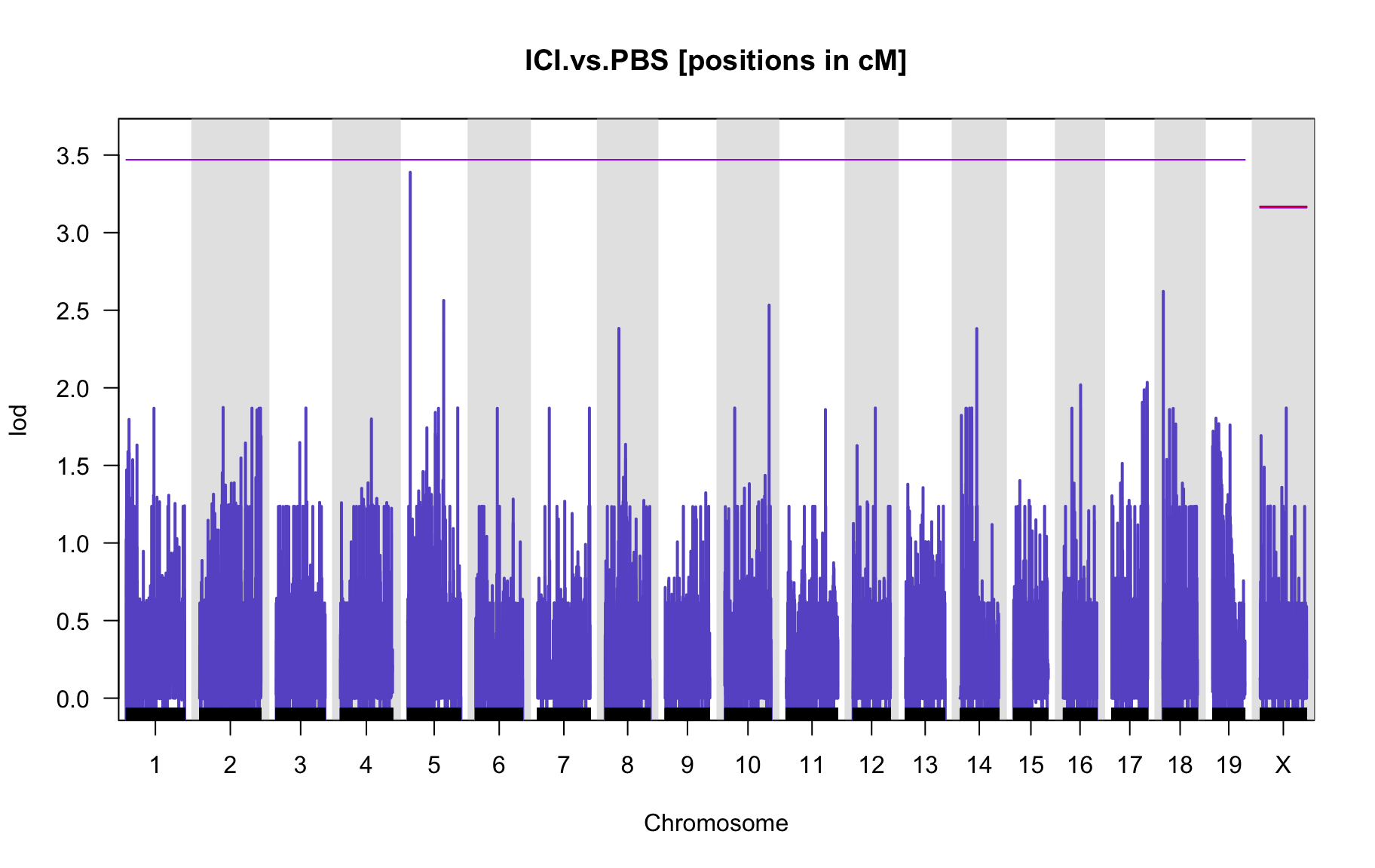
ymx <- 14
plot(scanone.hk, bandcol = "grey90",lty=1, cex=1, col = "slateblue", ylim=c(0, ymx+0.5))
title(main = paste0(colnames(out), " [positions in cM]\n(using same scale as ici vs. eoi for easier comparison)"))
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.01, col = 'blue')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.05, col = 'red')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.1, col = 'purple')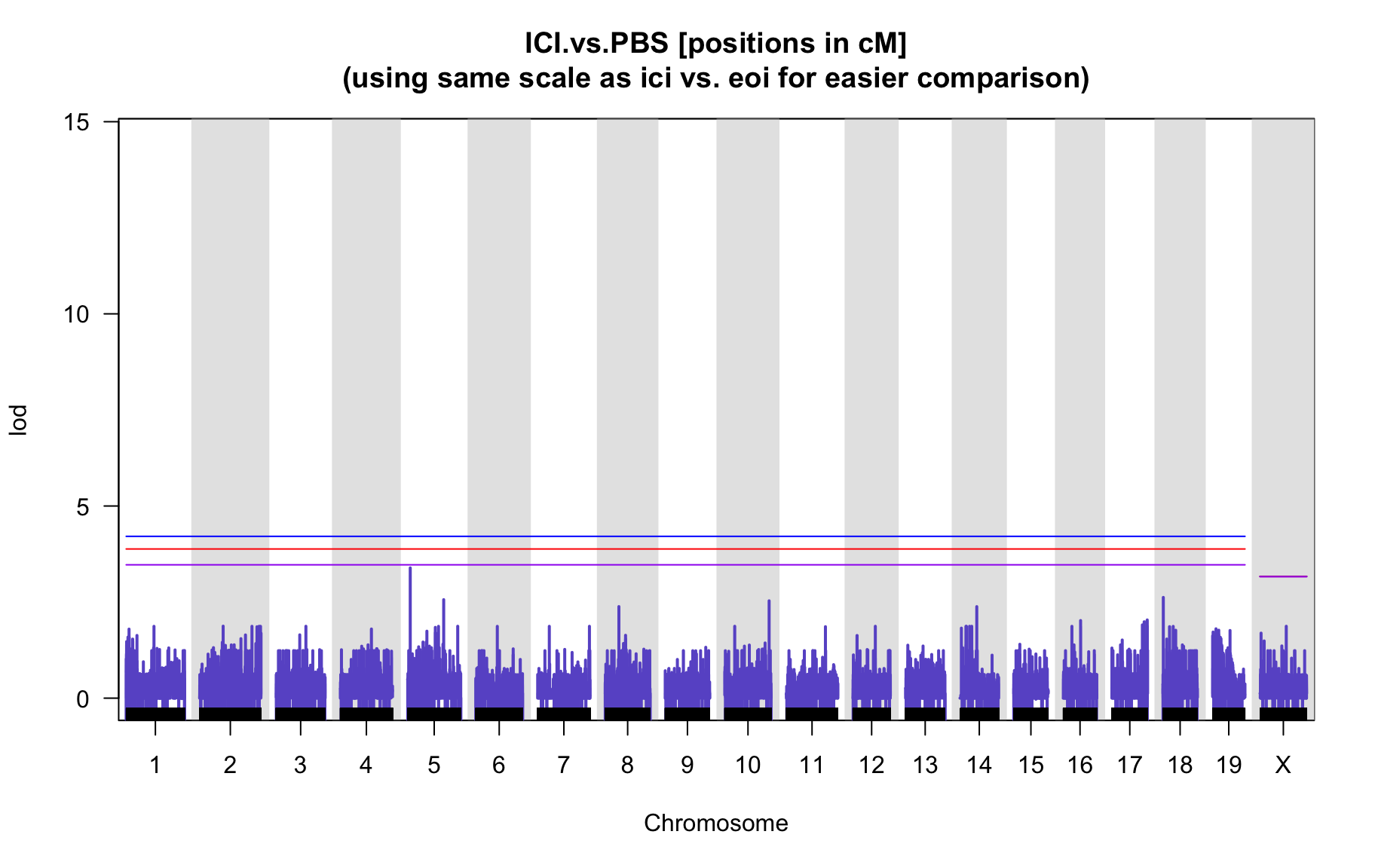
print(as.data.frame(summary(scanone.hk, perms=operm.hk, pvalues=TRUE, format="allpeaks"))) chr pos lod pval
UNC1197721 1 47.673577 1.868502 1.0000000
UNCHS005266 2 40.908610 1.873368 1.0000000
UNC6045859 3 50.620447 1.870152 1.0000000
JAX00123711 4 53.542073 1.799165 1.0000000
JAX00574407 5 6.213413 3.388862 0.1052020
UNCHS017906 6 37.001766 1.868162 1.0000000
JAX00233659 7 87.998157 1.870072 1.0000000
JAX00667121 8 24.443849 2.382541 0.7192282
UNC17203329 9 68.519175 1.323467 1.0000000
JAX00302150 10 74.547218 2.532706 0.6196652
UNC20281687 11 67.380801 1.859331 1.0000000
UNCHS034188 12 38.836978 1.870151 1.0000000
JAX00350973 13 5.743505 1.378528 1.0000000
JAX00380911 14 30.154517 2.381660 0.7192282
UNC25275535 15 11.968141 1.402063 1.0000000
UNC26790882 16 30.832641 2.019242 0.8169482
UNCHS045288 17 60.699043 2.034920 0.8169482
JAX00450853 18 2.913076 2.621475 0.6196652
UNCHS047190 19 8.712408 1.804318 1.0000000
XiB2 X 45.699951 1.870087 0.9986399print("all peaks with a p-value less or equal to 0.05 (suggestive)")[1] "all peaks with a p-value less or equal to 0.05 (suggestive)"print(as.data.frame(summary(scanone.hk, perms=operm.hk, alpha=0.05, pvalues=TRUE, format="allpeaks")))[1] chr pos lod
<0 rows> (or 0-length row.names)#print("method == ehk")
#scanone.ehk <-qtl::scanone(cross.probs, pheno.col="ICI.vs.PBS" , model="binary", method="ehk")
#operm.ehk <- qtl::scanone(cross.probs, method = "ehk", pheno.col="ICI.vs.PBS", n.perm = 10, perm.Xsp = TRUE, model="binary", verbose=FALSE)
#plot(operm.ehk)
#print(summary(operm.ehk, alpha=c(0.01, 0.05, 0.1)))
#plot(scanone.ehk, bandcol = "grey90",lty=1, cex=1, col = "steelblue")
#qtl::add.threshold(scanone.ehk, perms= operm.ehk, alpha=0.01, col = 'blue')
#qtl::add.threshold(scanone.ehk, perms= operm.ehk, alpha=0.05, col = 'red')
#qtl::add.threshold(scanone.ehk, perms= operm.ehk, alpha=0.1, col = 'purple')
#print(as.data.frame(summary(scanone.ehk)))
#print(as.data.frame(summary(scanone.ehk, perms=operm.ehk, alpha=0.05, pvalues=TRUE, format="allpeaks")))Conditionaing on eoi vs. ici chr 3 peak (UNCHS008487) & chr 10 peak (UNC18240977)
Genome-wide scan
Xcovar <- get_x_covar(gm)
addcovar = model.matrix(~UNCHS008487+UNC18240977, data = covars)[,-1]
#K <- calc_kinship(pr.qc, type = "loco")
#heatmap(K[[1]])
#K.overall <- calc_kinship(pr.qc, type = "overall")
#heatmap(K.overall)
kinship <- calc_kinship(pr.qc)
heatmap(kinship)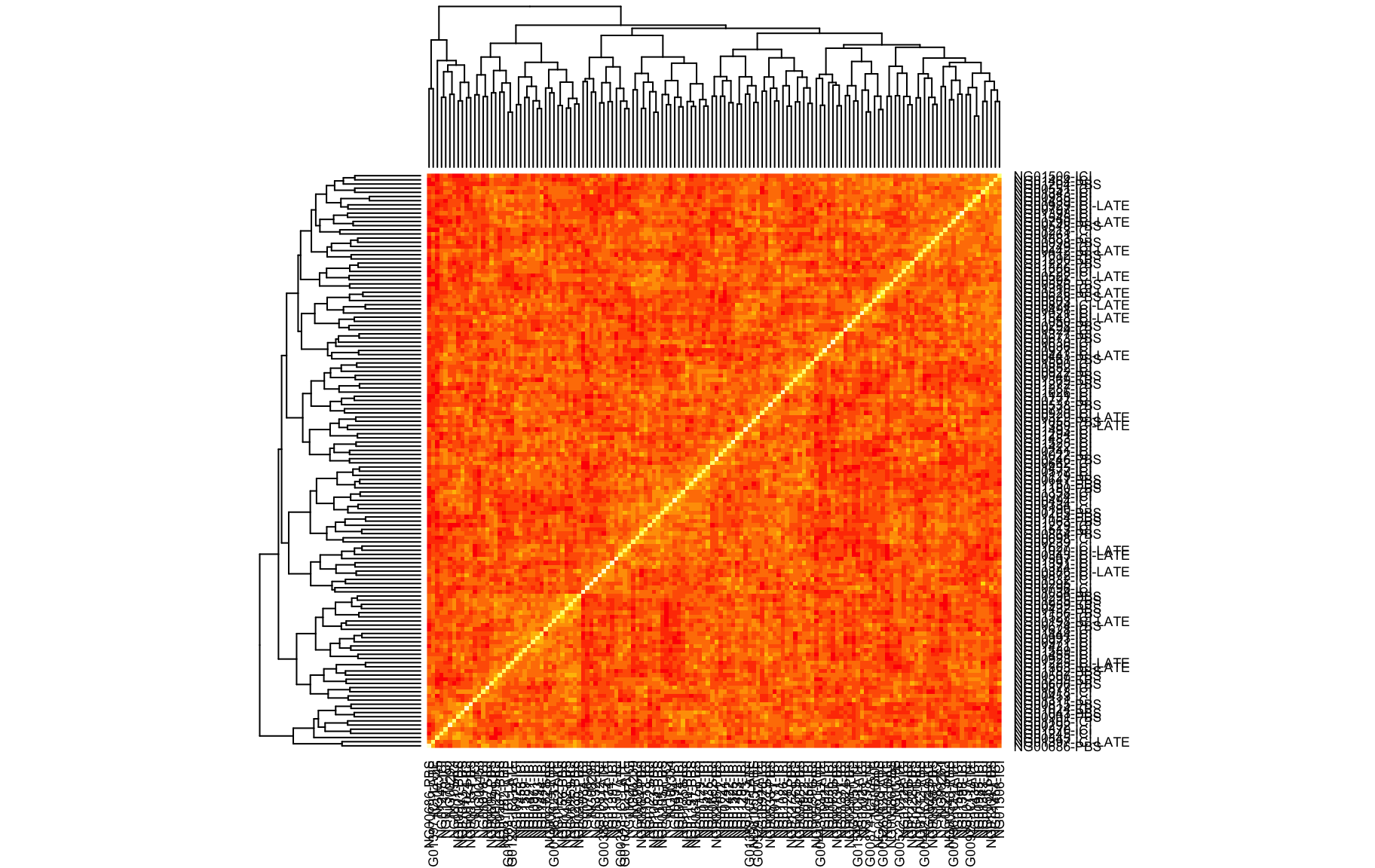
#operm <- scan1perm(pr.qc, gm$covar$phenos, Xcovar=Xcovar, n_perm=2000)
#operm <- scan1perm(pr.qc, gm$covar$phenos, addcovar = addcovar, n_perm=2000)
#operm <- scan1perm(pr.qc, gm$covar$phenos, n_perm=2000)
operm <- scan1perm(pr.qc, gm$covar["ICI.vs.PBS"], model="binary", n_perm=10, perm_Xsp=TRUE, chr_lengths=chr_lengths(gm$gmap), addcovar = addcovar)
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
names(summary_table) <- c("autosomes","X")
summary_table$significance.level <- rownames(summary_table)
rownames(summary_table) <- NULL
summary_table[c(3,1:2)] %>%
kable(escape = F,align = c("ccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| significance.level | autosomes | X |
|---|---|---|
| 0.01 | 3.764568 | 3.519636 |
| 0.05 | 3.758664 | 3.355931 |
| 0.1 | 3.751265 | 3.141856 |
The figures below show QTL maps for each phenotype
out <- scan1(pr.qc, gm$covar["ICI.vs.PBS"], Xcovar=Xcovar, model="binary", addcovar = addcovar)
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
plot_lod<-function(out,map){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
##legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- 14 # overall maximum LOD score
plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
##legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " [positions in cM] \n(using same scale as eoi vs. ici for easier comparison)"))
add_threshold(map, summary(operm, alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
plot_lod(out,gm$gmap)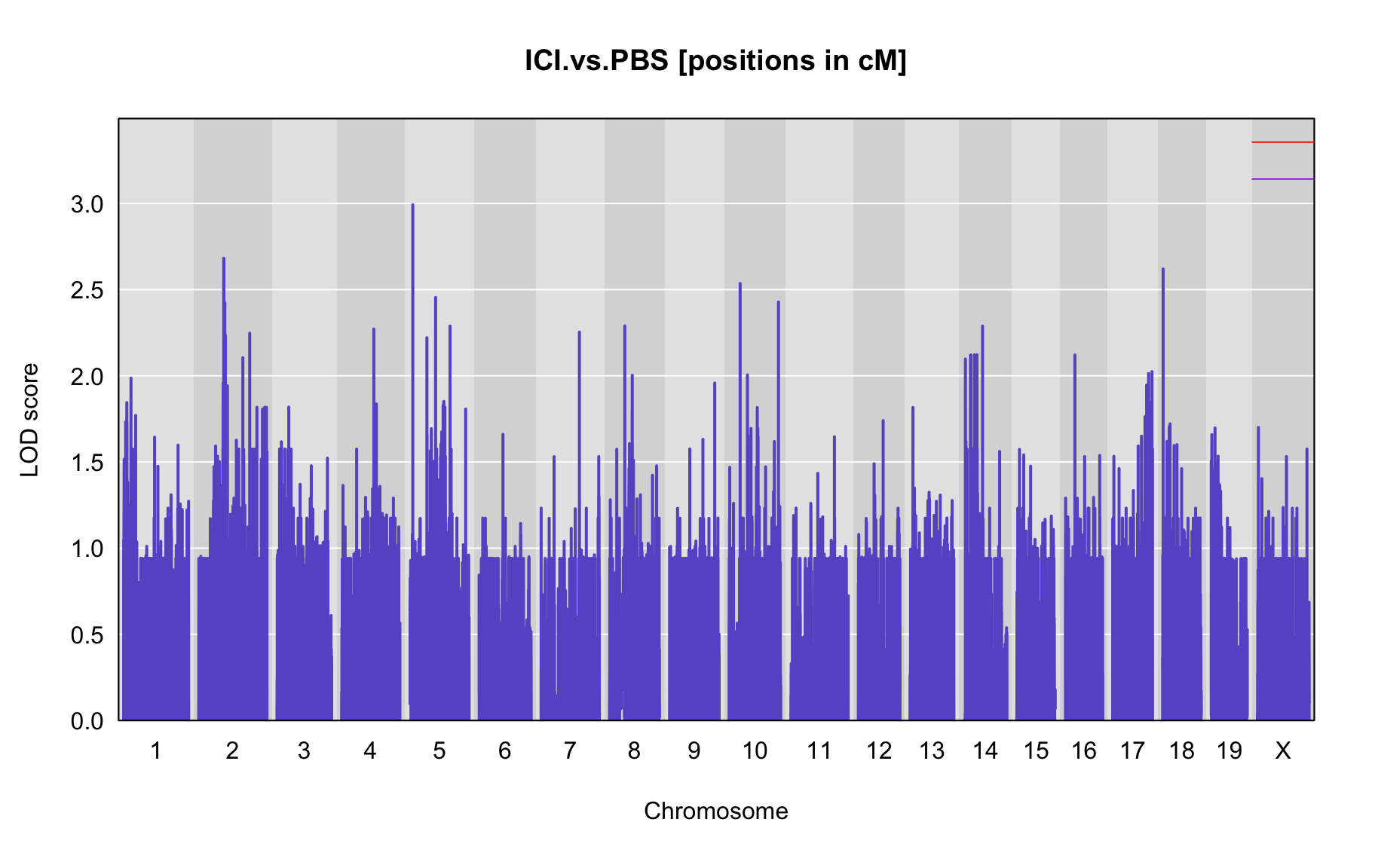
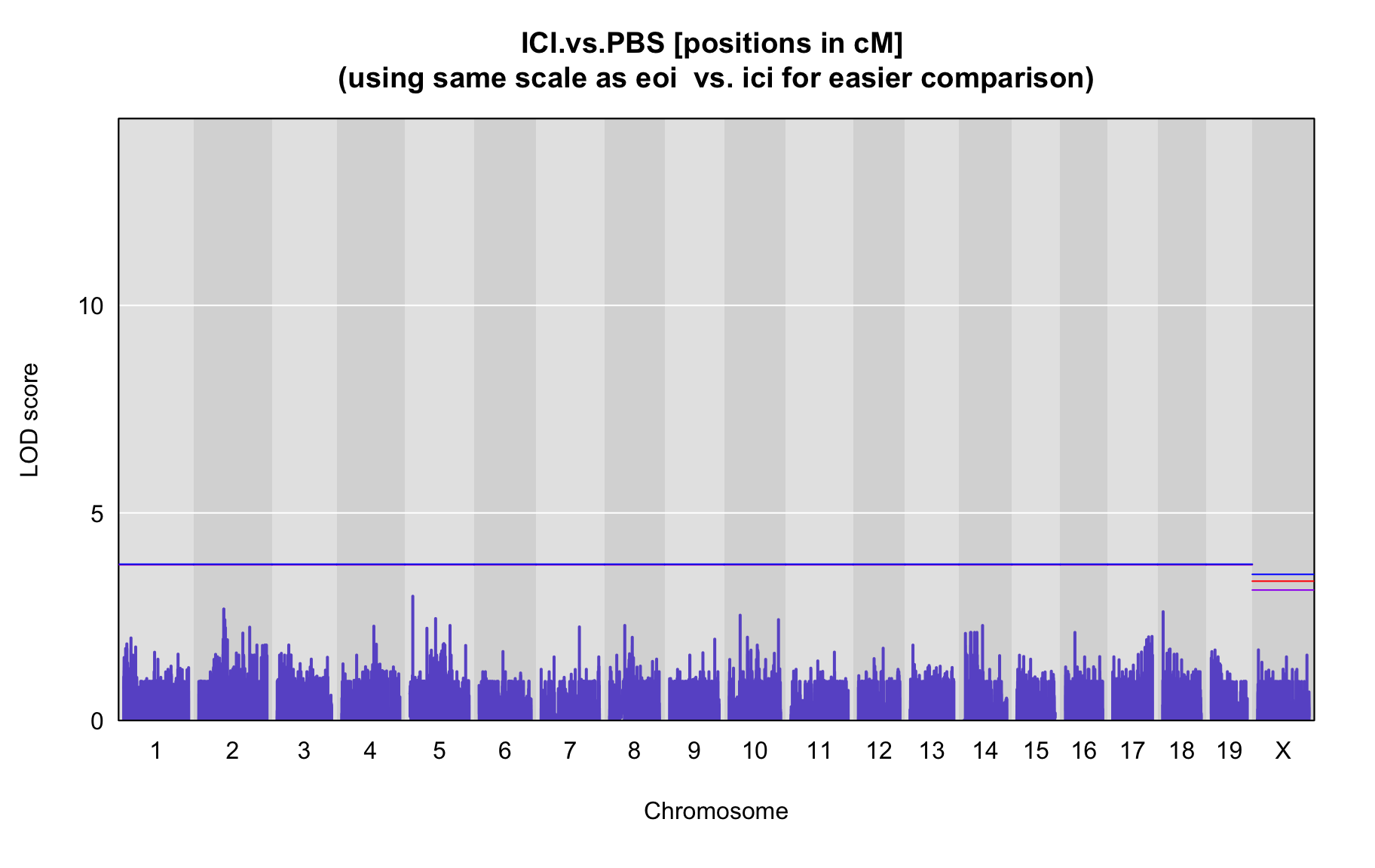
LOD peaks
The table below shows QTL peaks associated with the phenotype. We use the 95% threshold from the permutations to find peaks.
Centimorgan (cM)
peaks <- find_peaks(out, gm$gmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) >0){
peaks$marker <- find_marker(gm$gmap, chr=peaks$chr,pos=peaks$pos)
names(peaks)[2] <- c("phenotype")
peaks <- peaks[-1]
rownames(peaks) <- NULL
print(kable(peaks, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
#plot only peak chromosomes
plot_lod_chr<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
ymx <- 14
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in cM]\n(using same scale as eoi vs. ici for easier comparison)"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
for(i in unique(peaks$chr)){
#for (i in 1:nrow(peaks)){
#plot_lod_chr(out,gm$gmap, peaks$chr[i])
plot_lod_chr(out,gm$gmap, i)
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are no peaks that have a LOD that reaches suggestive (p<0.05) level of 3.75866362627187 [autosomes]/3.3559311891303 [x-chromosome]”
Megabase (MB)
print("peaks in MB positions")[1] “peaks in MB positions”
peaks_mba <- find_peaks(out, gm$pmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) >0){
peaks_mba$marker <- find_marker(gm$pmap, chr=peaks_mba$chr,pos=peaks_mba$pos)
names(peaks_mba)[2] <- c("phenotype")
peaks_mba <- peaks_mba[-1]
#peaks_mbl <- list()
##corresponding info in Mb
#for(i in 1:nrow(peaks)){
# #lodindex <- peaks$lodindex[i]
# phenotype <- peaks$phenotype[i]
# chr <- as.character(peaks$chr[i])
# lod <- peaks$lod[i]
# mark <- peaks$marker[i]
# pos <- mapdf[mapdf$marker==mark,]$pmapdf
# ci_lo <- mapdfnd$pmapdf[which(mapdfnd$gmapdf == peaks$ci_lo[i] & mapdfnd$chr == peaks$chr[i])]
# ci_hi <- mapdfnd$pmapdf[which(mapdfnd$gmapdf == peaks$ci_hi[i] & mapdfnd$chr == peaks$chr[i])]
# peaks_mb=as.data.frame(cbind(phenotype, chr, pos, lod, ci_lo, ci_hi, mark))
# names(peaks_mb)[7] <- c("marker")
# peaks_mbl[[i]] <- peaks_mb
#}
#peaks_mba2 <- do.call(rbind, peaks_mbl)
#peaks_mba2 <- as.data.frame(peaks_mba)
#peaks_mba[,c("chr", "pos", "lod", "ci_lo", "ci_hi")] <- sapply(peaks_mba[,c("chr", "pos", "lod", "ci_lo", "ci_hi")], as.numeric)
rownames(peaks_mba) <- NULL
print(kable(peaks_mba, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
plot_lod_chr_mb<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in MB]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
ymx <- 14
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in MB]\n(using same scale as eoi vs. ici for easier comparison)"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
for(i in unique(peaks_mba$chr)){
#for (i in 1:nrow(peaks_mba)){
#plot_lod_chr_mb(out,gm$pmap, peaks_mba$chr[i])
plot_lod_chr_mb(out,gm$pmap,i)
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are no peaks that have a LOD that reaches suggestive (p<0.05) level of 3.75866362627187 [autosomes]/3.3559311891303 [x-chromosome]”
QTL effects
For each peak LOD location we give a list of gene
query_variants <- create_variant_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/cc_variants.sqlite")
query_genes <- create_gene_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/mouse_genes_mgi.sqlite")
if(nrow(peaks) >0){
for (i in 1:nrow(peaks)){
#for (i in 1:1){
#Plot 1
g <- maxmarg(pr.qc, gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i], return_char=TRUE)
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/","qtl_effect_", i, ".png"))
#par(mar=c(4.1, 4.1, 1.5, 0.6))
plot_pxg(g, gm$covar[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
##dev.off()
chr = peaks$chr[i]
# Plot 2
pr_sub <- pull_genoprobint(pr.qc, gm$gmap, chr, c(peaks$ci_lo[i], peaks$ci_hi[i]))
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar)
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], Xcovar=Xcovar)
#coeff <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
#coeff_sub <- scan1coef(pr_sub[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
blup <- scan1blup(pr.qc[,chr], gm$covar[peaks$phenotype[i]], addcovar = addcovar)
blup_sub <- scan1blup(pr_sub[,chr], gm$covar[peaks$phenotype[i]], addcovar = addcovar)
write.csv(as.data.frame(blup_sub), paste0("data/ici.vs.pbs_blup_sub_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_5.batches_mis_conditional_2-peaks-chr3-10.csv"), quote=F)
#plot_coef(coeff,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i]," [scan1coeff; positions in cM])")
# )
#plot_coef(coeff_sub,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i],"; 1.5 LOD drop interval [scan1coeff; positions in cM] ) ")
# )
plot_coef(blup,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i], "; pos: ", peaks$pos[i], "cM / ",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," [scan1blup; positions in cM])")
)
plot_coef(blup_sub,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i],"; 1.5 LOD drop interval [scan1blup; positions in cM])")
)
# Plot 3
#c2effB <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary", contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
#c2effBb <- scan1blup(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]],Xcovar=Xcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
#plot(c2effB, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
#plot(c2effBb, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
##last_coef <- unclass(c2effB)[nrow(c2effB),2:3] # last two coefficients
##for(t in seq(along=last_coef))
## axis(side=4, at=last_coef[t], names(last_coef)[t], tick=FALSE)
#Table 1
chr = peaks_mba$chr[i]
start=as.numeric(peaks_mba$ci_lo[i])
end=as.numeric(peaks_mba$ci_hi[i])
genesgss = query_genes(chr, start, end)
write.csv(genesgss, file=paste0("data/ici.vs.pbs_genes_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_5.batches_mis_conditional_2-peaks-chr3-10.csv"), quote=F)
rownames(genesgss) <- NULL
genesgss$strand_old = genesgss$strand
genesgss$strand[genesgss$strand=="+"] <- "positive"
genesgss$strand[genesgss$strand=="-"] <- "negative"
#genesgss <-
#table <-
#genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")] %>%
#kable(escape = F,align = c("ccccccccccc")) %>%
#kable_styling("striped", full_width = T) #%>%
#cat #%>%
#column_spec(1, bold=TRUE)
#
#print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], escape = F,align = c("ccccccccccc")))
print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], "html") %>% kable_styling("striped", full_width = T))
#table
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are no peaks that have a LOD that reaches suggestive (p<0.05) level of 3.75866362627187 [autosomes]/3.3559311891303 [x-chromosome]”
R/qtl
scanone
gmObject of class cross2 (crosstype "bc")
Total individuals 138
No. genotyped individuals 138
No. phenotyped individuals 138
No. with both geno & pheno 138
No. phenotypes 1
No. covariates 7
No. phenotype covariates 0
No. chromosomes 20
Total markers 131355
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
9956 9987 7848 7585 7609 7736 7399 6458 6713 6385 7143 6110 6082 5966 5346 5015
17 18 19 X
5080 4605 3562 4770 #detach("package:qtl2", unload=TRUE)
#library(qtl)
cross <- qtl::read.cross("csv", file = "data/ici.vs.pbs_gm_qtl_5.batches_mis_conditional_2-peaks-chr3-10.csv",alleles=c("A","B")) --Read the following data:
138 individuals
131355 markers
5 phenotypes
--Cross type: bc cross <- qtl::jittermap(cross)
summary(cross) Backcross
No. individuals: 138
No. phenotypes: 5
Percent phenotyped: 100 100 100 100 100
No. chromosomes: 20
Autosomes: 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19
X chr: X
Total markers: 131355
No. markers: 9956 9987 7848 7585 7609 7736 7399 6458 6713 6385 7143
6110 6082 5966 5346 5015 5080 4605 3562 4770
Percent genotyped: 99.5
Genotypes (%):
Autosomes: AA:88.1 AB:11.9
X chromosome: AA:93.9 AB:6.1 cross.probs <- qtl::calc.genoprob(cross)
print("method == hk")[1] "method == hk"add.covars = qtl::pull.pheno(cross.probs, c("UNCHS008487","UNC18240977"))
scanone.hk <-qtl::scanone(cross.probs, pheno.col="ICI.vs.PBS" , model="binary", method="hk", addcovar = add.covars)
operm.hk <- qtl::scanone(cross.probs, method = "hk", pheno.col="ICI.vs.PBS", n.perm = 10, perm.Xsp = TRUE, model="binary", verbose=FALSE, addcovar = add.covars)
plot(operm.hk)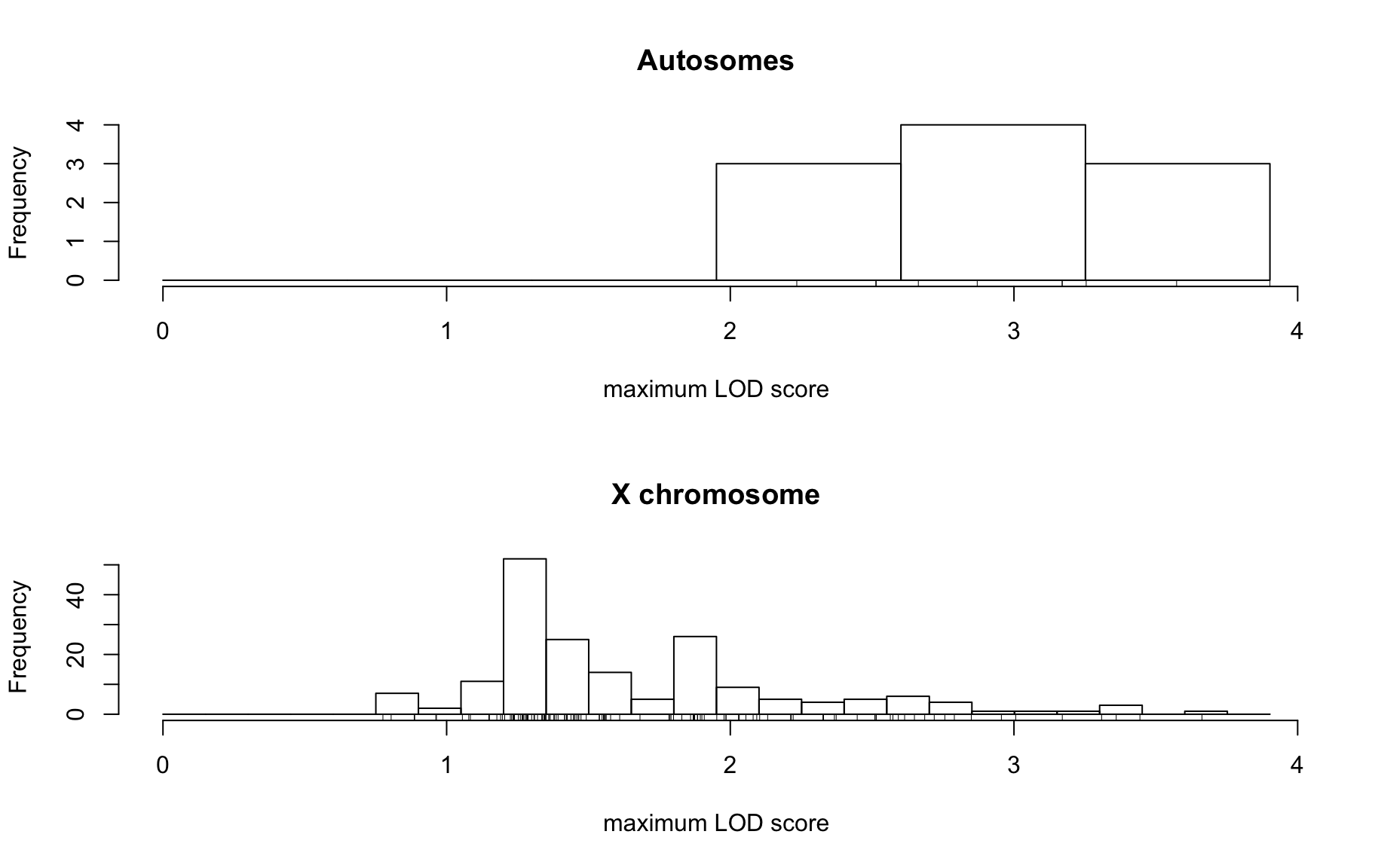
print(summary(operm.hk, alpha=c(0.01, 0.05, 0.1)))Autosome LOD thresholds (10 permutations)
lod
1% 3.87
5% 3.76
10% 3.62
X chromosome LOD thresholds (182 permutations)
lod
1% 3.64
5% 3.56
10% 3.45#plot(scanone.hk, bandcol = "grey90",lty=1, cex=1, col = "steelblue")
#qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.01, col = 'blue')
#qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.05, col = 'red')
#qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.1, col = 'purple')
ymx <- maxlod(out) # overall maximum LOD score
plot(scanone.hk, bandcol = "grey90",lty=1, cex=1, col = "slateblue", ylim=c(0, ymx+0.5))
title(main = paste0(colnames(out), " [positions in cM]"))
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.01, col = 'blue')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.05, col = 'red')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.1, col = 'purple')
ymx <- 14
plot(scanone.hk, bandcol = "grey90",lty=1, cex=1, col = "slateblue", ylim=c(0, ymx+0.5))
title(main = paste0(colnames(out), " [positions in cM]\n(using same scale as ici vs. eoi for easier comparison)"))
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.01, col = 'blue')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.05, col = 'red')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.1, col = 'purple')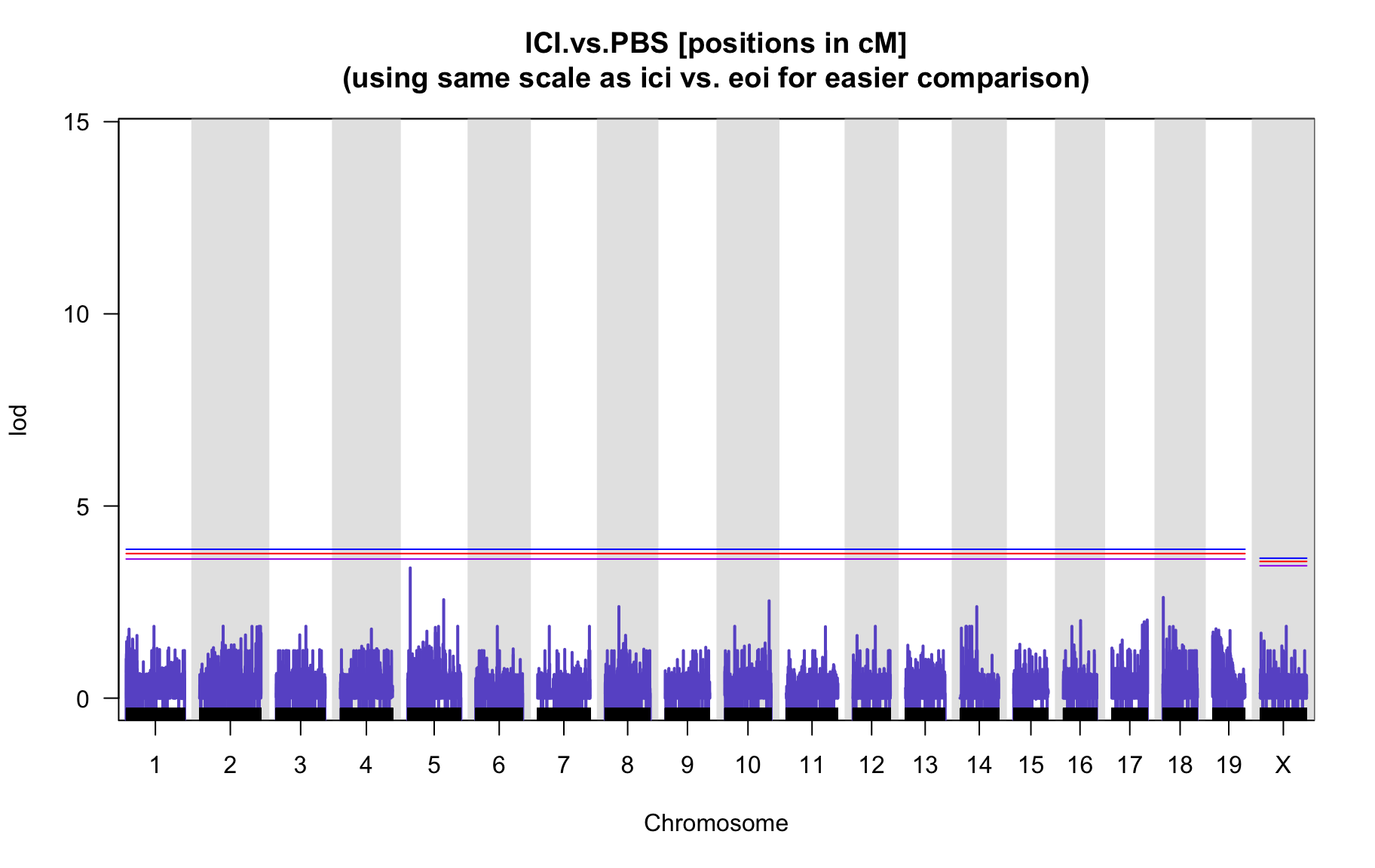
print(as.data.frame(summary(scanone.hk, perms=operm.hk, pvalues=TRUE, format="allpeaks"))) chr pos lod pval
UNC1197721 1 47.673577 1.868502 1.0000000
UNCHS005266 2 40.908610 1.873368 1.0000000
UNC6045859 3 50.620447 1.870152 1.0000000
JAX00123711 4 53.542073 1.799165 1.0000000
JAX00574407 5 6.213413 3.388862 0.2097615
UNCHS017906 6 37.001766 1.868162 1.0000000
JAX00233659 7 87.998157 1.870072 1.0000000
JAX00667121 8 24.443849 2.382541 0.9118988
UNC17203329 9 68.519175 1.323467 1.0000000
JAX00302150 10 74.547218 2.532706 0.7192282
UNC20281687 11 67.380801 1.859331 1.0000000
UNCHS034188 12 38.836978 1.870151 1.0000000
JAX00350973 13 5.743505 1.378528 1.0000000
JAX00380911 14 30.154517 2.381660 0.9118988
UNC25275535 15 11.968141 1.402063 1.0000000
UNC26790882 16 30.832641 2.019242 1.0000000
UNCHS045288 17 60.699043 2.034920 1.0000000
JAX00450853 18 2.913076 2.621475 0.7192282
UNCHS047190 19 8.712408 1.804318 1.0000000
XiB2 X 45.699951 1.870087 0.9992566print("all peaks with a p-value less or equal to 0.05 (suggestive)")[1] "all peaks with a p-value less or equal to 0.05 (suggestive)"print(as.data.frame(summary(scanone.hk, perms=operm.hk, alpha=0.05, pvalues=TRUE, format="allpeaks")))[1] chr pos lod
<0 rows> (or 0-length row.names)#print("method == ehk")
#scanone.ehk <-qtl::scanone(cross.probs, pheno.col="ICI.vs.PBS" , model="binary", method="ehk")
#operm.ehk <- qtl::scanone(cross.probs, method = "ehk", pheno.col="ICI.vs.PBS", n.perm = 10, perm.Xsp = TRUE, model="binary", verbose=FALSE)
#plot(operm.ehk)
#print(summary(operm.ehk, alpha=c(0.01, 0.05, 0.1)))
#plot(scanone.ehk, bandcol = "grey90",lty=1, cex=1, col = "steelblue")
#qtl::add.threshold(scanone.ehk, perms= operm.ehk, alpha=0.01, col = 'blue')
#qtl::add.threshold(scanone.ehk, perms= operm.ehk, alpha=0.05, col = 'red')
#qtl::add.threshold(scanone.ehk, perms= operm.ehk, alpha=0.1, col = 'purple')
#print(as.data.frame(summary(scanone.ehk)))
#print(as.data.frame(summary(scanone.ehk, perms=operm.ehk, alpha=0.05, pvalues=TRUE, format="allpeaks")))Conditionaing on eoi vs. ici chr 4 peak (UNCHS008487) & chr 10 peak (UNC18240977)
Genome-wide scan
Xcovar <- get_x_covar(gm)
addcovar = model.matrix(~UNC8250659+UNC18240977, data = covars)[,-1]
#K <- calc_kinship(pr.qc, type = "loco")
#heatmap(K[[1]])
#K.overall <- calc_kinship(pr.qc, type = "overall")
#heatmap(K.overall)
kinship <- calc_kinship(pr.qc)
heatmap(kinship)
#operm <- scan1perm(pr.qc, gm$covar$phenos, Xcovar=Xcovar, n_perm=2000)
#operm <- scan1perm(pr.qc, gm$covar$phenos, addcovar = addcovar, n_perm=2000)
#operm <- scan1perm(pr.qc, gm$covar$phenos, n_perm=2000)
operm <- scan1perm(pr.qc, gm$covar["ICI.vs.PBS"], model="binary", n_perm=10, perm_Xsp=TRUE, chr_lengths=chr_lengths(gm$gmap), addcovar = addcovar)
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
names(summary_table) <- c("autosomes","X")
summary_table$significance.level <- rownames(summary_table)
rownames(summary_table) <- NULL
summary_table[c(3,1:2)] %>%
kable(escape = F,align = c("ccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| significance.level | autosomes | X |
|---|---|---|
| 0.01 | 3.086790 | 4.197360 |
| 0.05 | 3.058033 | 3.823126 |
| 0.1 | 3.021996 | 3.333744 |
The figures below show QTL maps for each phenotype
out <- scan1(pr.qc, gm$covar["ICI.vs.PBS"], Xcovar=Xcovar, model="binary", addcovar = addcovar)
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
plot_lod<-function(out,map){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
##legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- 14 # overall maximum LOD score
plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
##legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " [positions in cM] \n(using same scale as eoi vs. ici for easier comparison)"))
add_threshold(map, summary(operm, alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
plot_lod(out,gm$gmap)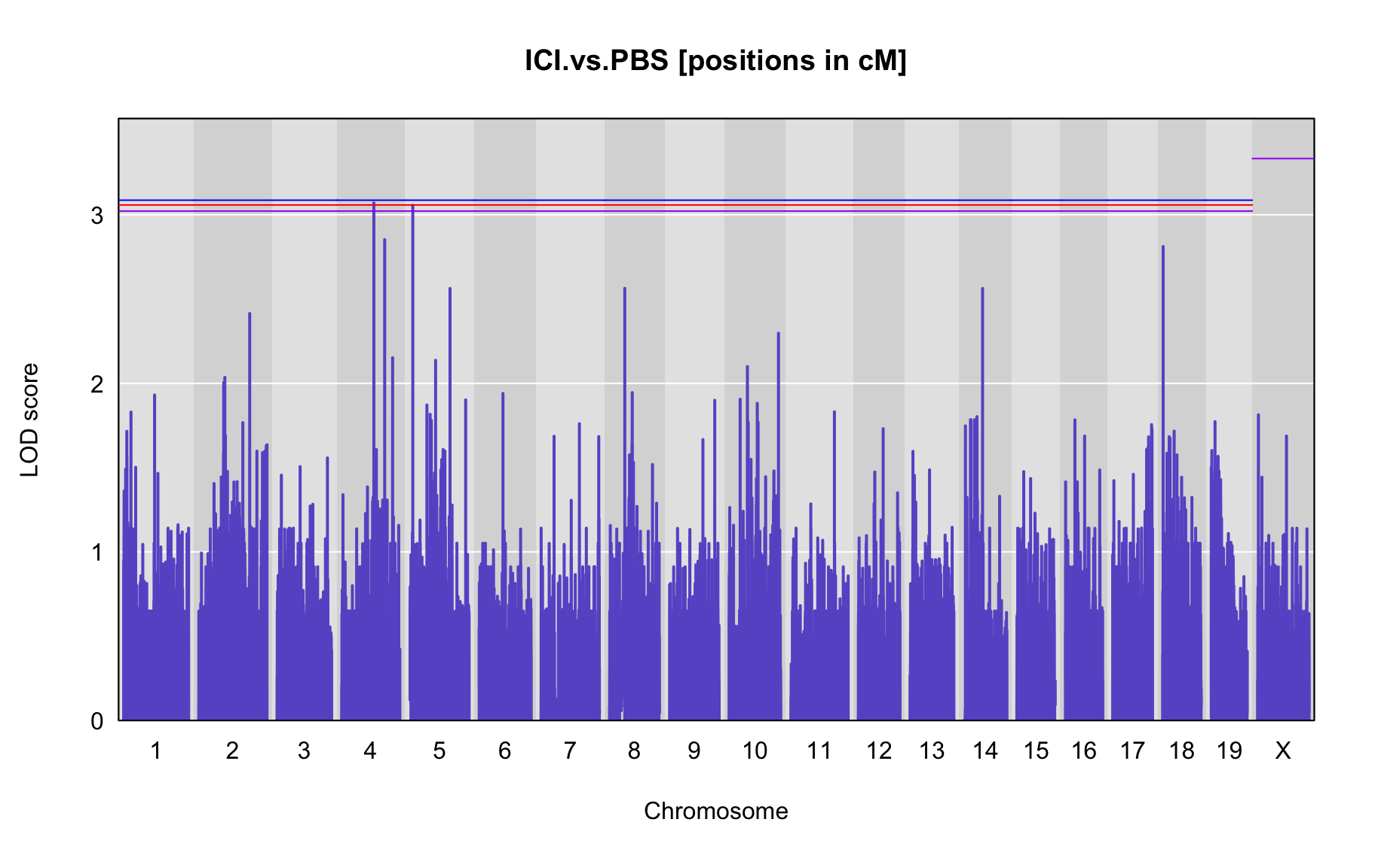
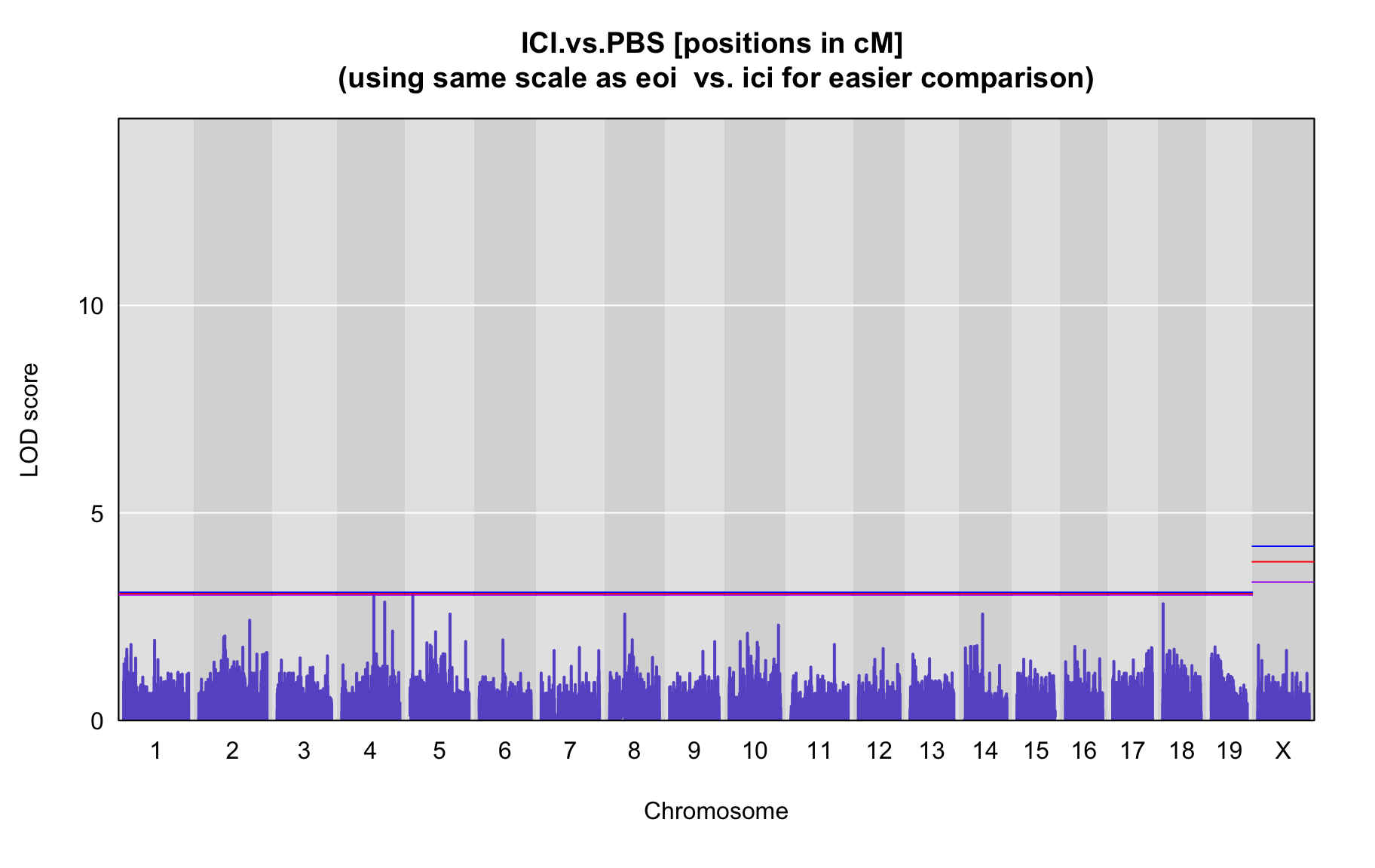
LOD peaks
The table below shows QTL peaks associated with the phenotype. We use the 95% threshold from the permutations to find peaks.
Centimorgan (cM)
peaks <- find_peaks(out, gm$gmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) >0){
peaks$marker <- find_marker(gm$gmap, chr=peaks$chr,pos=peaks$pos)
names(peaks)[2] <- c("phenotype")
peaks <- peaks[-1]
rownames(peaks) <- NULL
print(kable(peaks, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
#plot only peak chromosomes
plot_lod_chr<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
ymx <- 14
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in cM]\n(using same scale as eoi vs. ici for easier comparison)"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
for(i in unique(peaks$chr)){
#for (i in 1:nrow(peaks)){
#plot_lod_chr(out,gm$gmap, peaks$chr[i])
plot_lod_chr(out,gm$gmap, i)
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}| phenotype | chr | pos | lod | ci_lo | ci_hi | marker |
|---|---|---|---|---|---|---|
| ICI.vs.PBS | 4 | 49.777 | 3.070548 | 49.776 | 77.787 | UNCHS012304 |
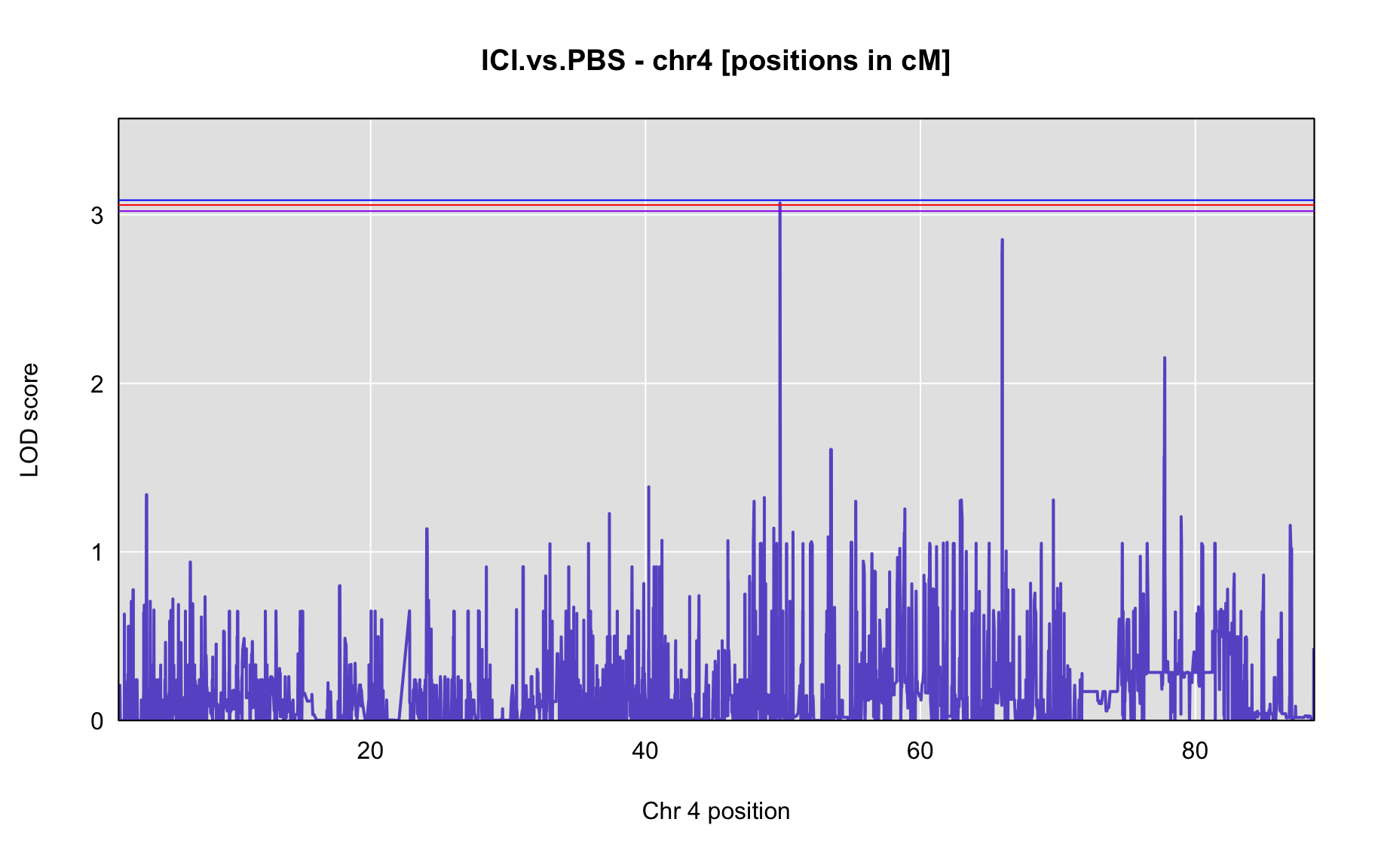
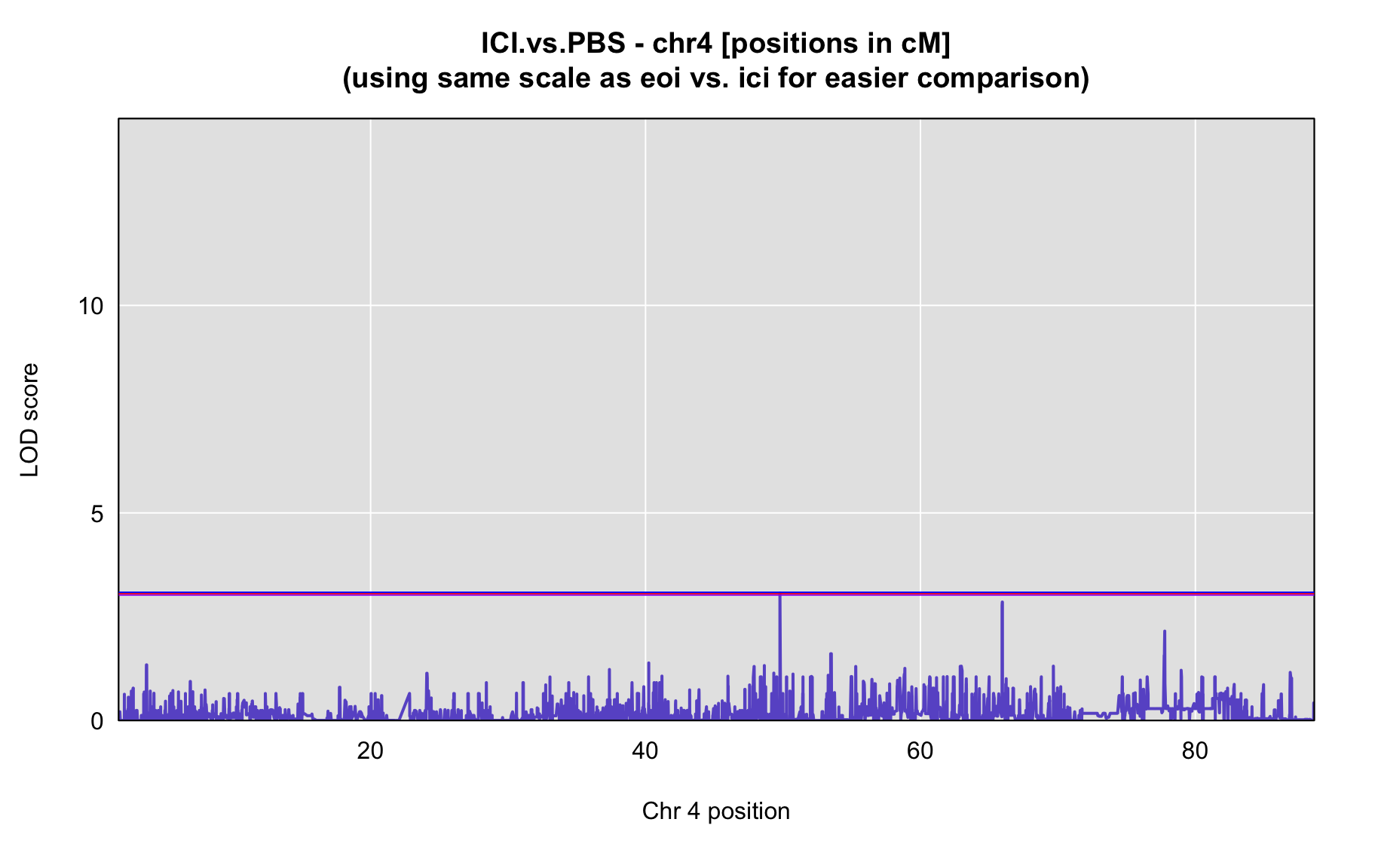
Megabase (MB)
print("peaks in MB positions")[1] “peaks in MB positions”
peaks_mba <- find_peaks(out, gm$pmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) >0){
peaks_mba$marker <- find_marker(gm$pmap, chr=peaks_mba$chr,pos=peaks_mba$pos)
names(peaks_mba)[2] <- c("phenotype")
peaks_mba <- peaks_mba[-1]
#peaks_mbl <- list()
##corresponding info in Mb
#for(i in 1:nrow(peaks)){
# #lodindex <- peaks$lodindex[i]
# phenotype <- peaks$phenotype[i]
# chr <- as.character(peaks$chr[i])
# lod <- peaks$lod[i]
# mark <- peaks$marker[i]
# pos <- mapdf[mapdf$marker==mark,]$pmapdf
# ci_lo <- mapdfnd$pmapdf[which(mapdfnd$gmapdf == peaks$ci_lo[i] & mapdfnd$chr == peaks$chr[i])]
# ci_hi <- mapdfnd$pmapdf[which(mapdfnd$gmapdf == peaks$ci_hi[i] & mapdfnd$chr == peaks$chr[i])]
# peaks_mb=as.data.frame(cbind(phenotype, chr, pos, lod, ci_lo, ci_hi, mark))
# names(peaks_mb)[7] <- c("marker")
# peaks_mbl[[i]] <- peaks_mb
#}
#peaks_mba2 <- do.call(rbind, peaks_mbl)
#peaks_mba2 <- as.data.frame(peaks_mba)
#peaks_mba[,c("chr", "pos", "lod", "ci_lo", "ci_hi")] <- sapply(peaks_mba[,c("chr", "pos", "lod", "ci_lo", "ci_hi")], as.numeric)
rownames(peaks_mba) <- NULL
print(kable(peaks_mba, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
plot_lod_chr_mb<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in MB]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
ymx <- 14
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in MB]\n(using same scale as eoi vs. ici for easier comparison)"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
for(i in unique(peaks_mba$chr)){
#for (i in 1:nrow(peaks_mba)){
#plot_lod_chr_mb(out,gm$pmap, peaks_mba$chr[i])
plot_lod_chr_mb(out,gm$pmap,i)
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}| phenotype | chr | pos | lod | ci_lo | ci_hi | marker |
|---|---|---|---|---|---|---|
| ICI.vs.PBS | 4 | 106.7137 | 3.070548 | 106.7113 | 143.751 | UNCHS012304 |
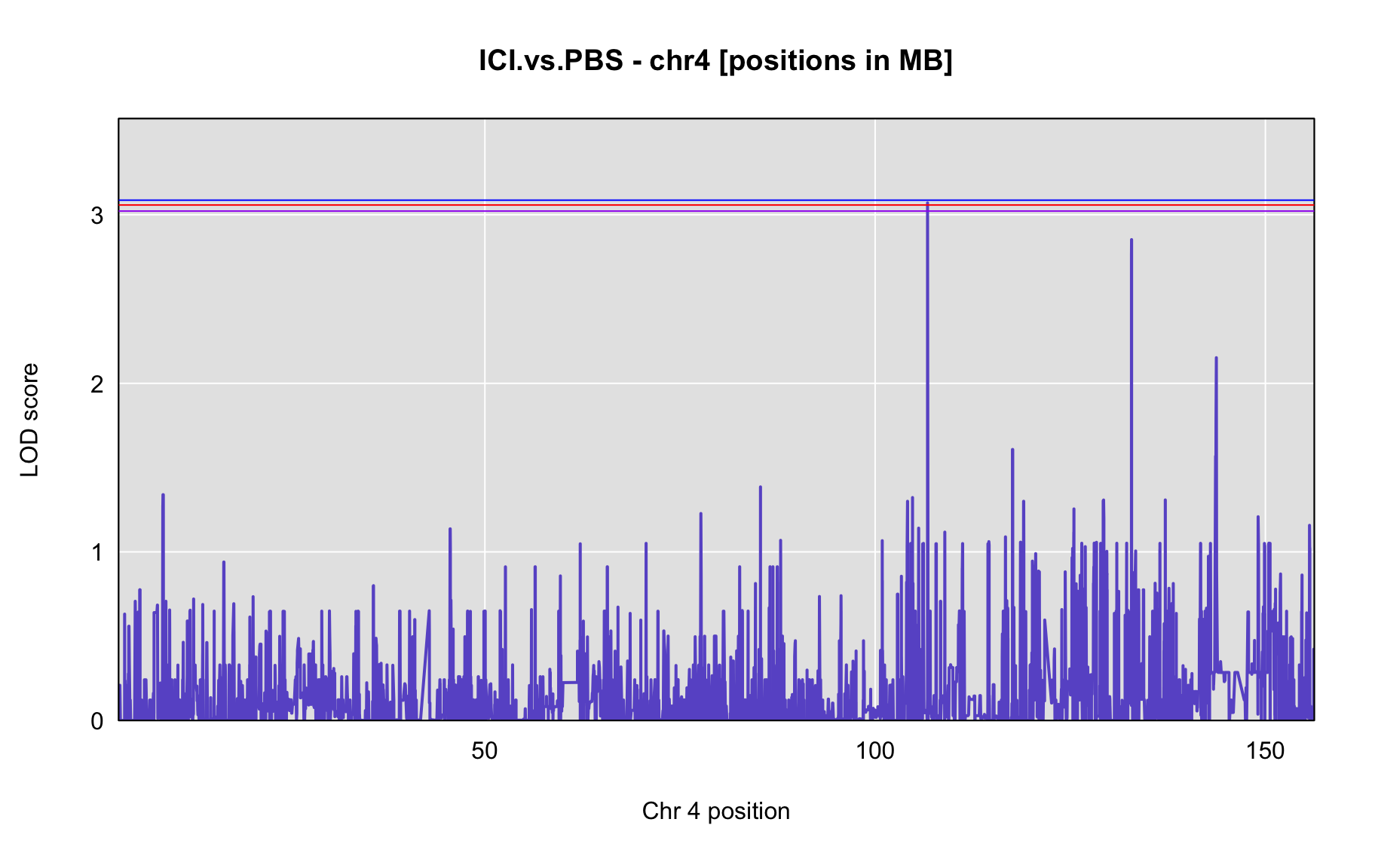
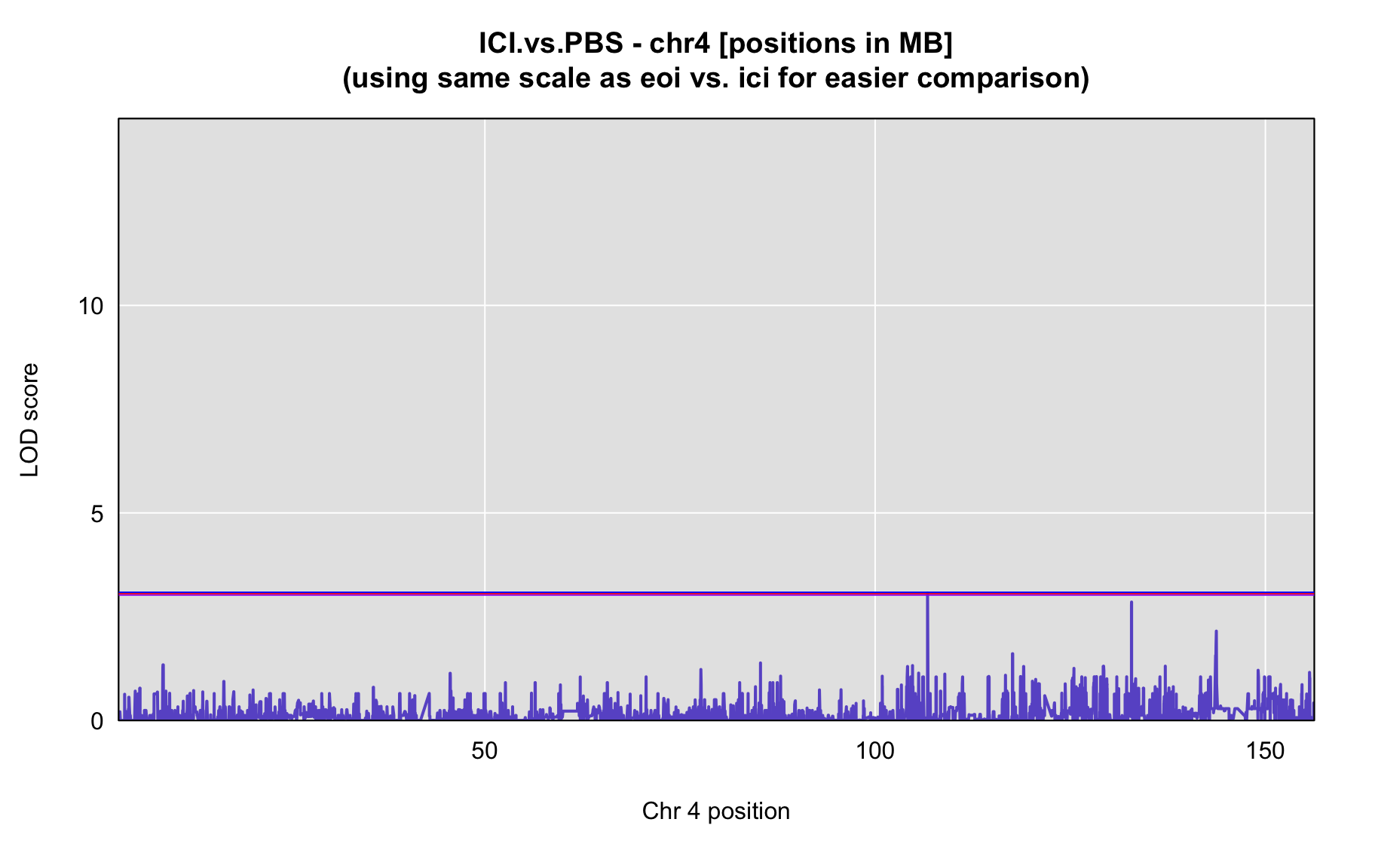
QTL effects
For each peak LOD location we give a list of gene
query_variants <- create_variant_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/cc_variants.sqlite")
query_genes <- create_gene_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/mouse_genes_mgi.sqlite")
if(nrow(peaks) >0){
for (i in 1:nrow(peaks)){
#for (i in 1:1){
#Plot 1
g <- maxmarg(pr.qc, gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i], return_char=TRUE)
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/","qtl_effect_", i, ".png"))
#par(mar=c(4.1, 4.1, 1.5, 0.6))
plot_pxg(g, gm$covar[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
##dev.off()
chr = peaks$chr[i]
# Plot 2
pr_sub <- pull_genoprobint(pr.qc, gm$gmap, chr, c(peaks$ci_lo[i], peaks$ci_hi[i]))
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar)
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], Xcovar=Xcovar)
#coeff <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
#coeff_sub <- scan1coef(pr_sub[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
blup <- scan1blup(pr.qc[,chr], gm$covar[peaks$phenotype[i]], addcovar = addcovar)
blup_sub <- scan1blup(pr_sub[,chr], gm$covar[peaks$phenotype[i]], addcovar = addcovar)
write.csv(as.data.frame(blup_sub), paste0("data/ici.vs.pbs_blup_sub_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_5.batches_mis_conditional_2-peaks-chr4-10.csv"), quote=F)
#plot_coef(coeff,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i]," [scan1coeff; positions in cM])")
# )
#plot_coef(coeff_sub,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i],"; 1.5 LOD drop interval [scan1coeff; positions in cM] ) ")
# )
plot_coef(blup,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i], "; pos: ", peaks$pos[i], "cM / ",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," [scan1blup; positions in cM])")
)
plot_coef(blup_sub,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i],"; 1.5 LOD drop interval [scan1blup; positions in cM])")
)
# Plot 3
#c2effB <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary", contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
#c2effBb <- scan1blup(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]],Xcovar=Xcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
#plot(c2effB, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
#plot(c2effBb, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
##last_coef <- unclass(c2effB)[nrow(c2effB),2:3] # last two coefficients
##for(t in seq(along=last_coef))
## axis(side=4, at=last_coef[t], names(last_coef)[t], tick=FALSE)
#Table 1
chr = peaks_mba$chr[i]
start=as.numeric(peaks_mba$ci_lo[i])
end=as.numeric(peaks_mba$ci_hi[i])
genesgss = query_genes(chr, start, end)
write.csv(genesgss, file=paste0("data/ici.vs.pbs_genes_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_5.batches_mis_conditional_2-peaks-chr4-10.csv"), quote=F)
rownames(genesgss) <- NULL
genesgss$strand_old = genesgss$strand
genesgss$strand[genesgss$strand=="+"] <- "positive"
genesgss$strand[genesgss$strand=="-"] <- "negative"
#genesgss <-
#table <-
#genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")] %>%
#kable(escape = F,align = c("ccccccccccc")) %>%
#kable_styling("striped", full_width = T) #%>%
#cat #%>%
#column_spec(1, bold=TRUE)
#
#print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], escape = F,align = c("ccccccccccc")))
print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], "html") %>% kable_styling("striped", full_width = T))
#table
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}
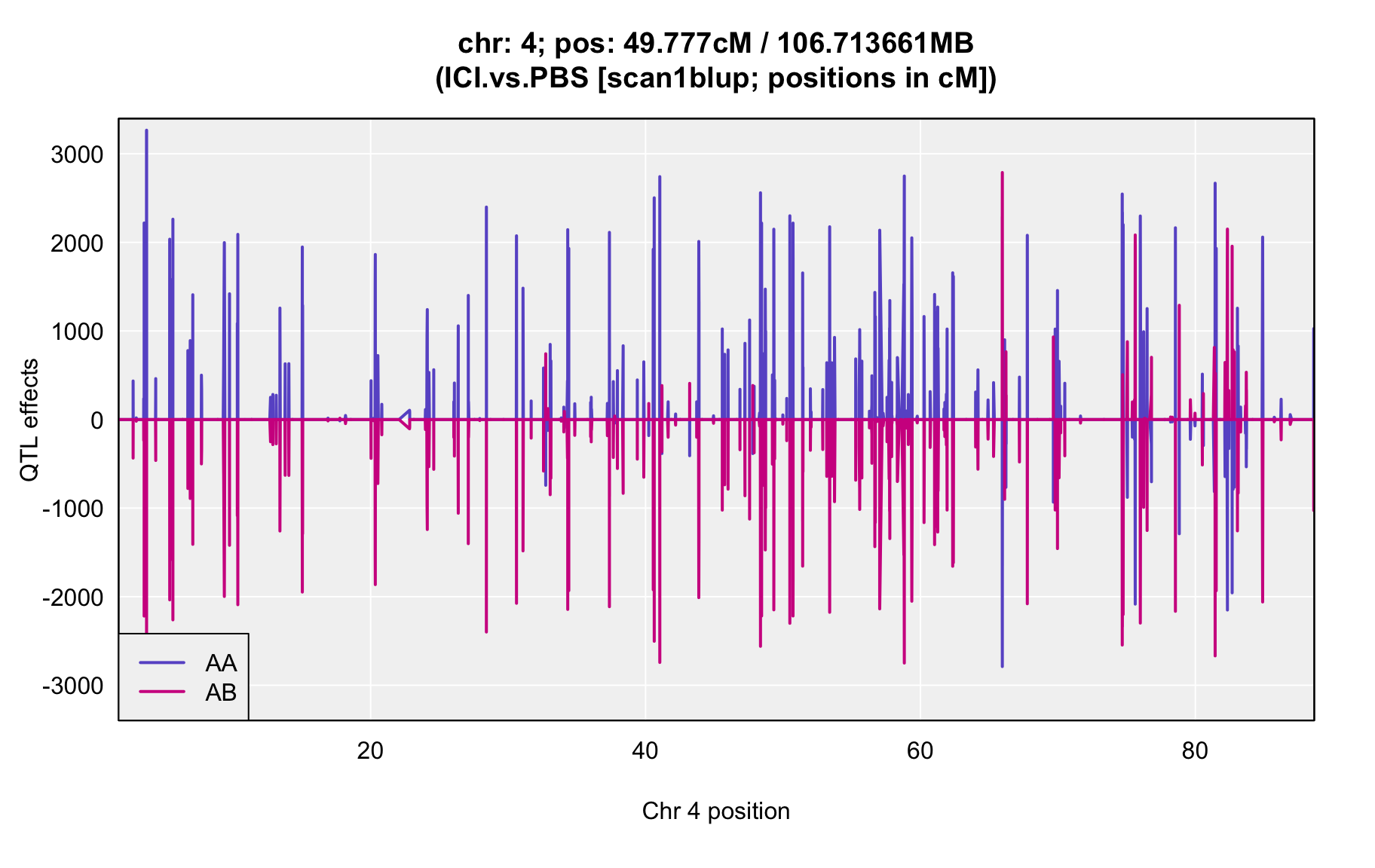
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 4 | gene | 106.6804 | 106.7366 | negative | MGI_C57BL6J_2685873 | Mroh7 | NCBI_Gene:381538,ENSEMBL:ENSMUSG00000047502 | MGI:2685873 | protein coding gene | maestro heat-like repeat family member 7 |
| 4 | gene | 106.7339 | 106.7483 | positive | MGI_C57BL6J_2657115 | Fam151a | NCBI_Gene:230579,ENSEMBL:ENSMUSG00000034871 | MGI:2657115 | protein coding gene | family with sequence simliarity 151, member A |
| 4 | gene | 106.7446 | 106.8061 | negative | MGI_C57BL6J_1913736 | Acot11 | NCBI_Gene:329910,ENSEMBL:ENSMUSG00000034853 | MGI:1913736 | protein coding gene | acyl-CoA thioesterase 11 |
| 4 | pseudogene | 106.7748 | 106.7750 | positive | MGI_C57BL6J_3651452 | Gm12745 | ENSEMBL:ENSMUSG00000081110 | MGI:3651452 | pseudogene | predicted gene 12745 |
| 4 | gene | 106.8149 | 106.8283 | negative | MGI_C57BL6J_5590224 | Gm31065 | NCBI_Gene:102633170 | MGI:5590224 | lncRNA gene | predicted gene, 31065 |
| 4 | gene | 106.8393 | 106.8402 | negative | MGI_C57BL6J_5590284 | Gm31125 | NCBI_Gene:102633257 | MGI:5590284 | lncRNA gene | predicted gene, 31125 |
| 4 | gene | 106.8418 | 106.8479 | negative | MGI_C57BL6J_3652135 | Gm12746 | ENSEMBL:ENSMUSG00000086940 | MGI:3652135 | lncRNA gene | predicted gene 12746 |
| 4 | gene | 106.9002 | 106.9129 | negative | MGI_C57BL6J_5826502 | Gm46865 | NCBI_Gene:108168970 | MGI:5826502 | lncRNA gene | predicted gene, 46865 |
| 4 | gene | 106.9107 | 107.0497 | positive | MGI_C57BL6J_1919725 | Ssbp3 | NCBI_Gene:72475,ENSEMBL:ENSMUSG00000061887 | MGI:1919725 | protein coding gene | single-stranded DNA binding protein 3 |
| 4 | gene | 107.0179 | 107.0225 | negative | MGI_C57BL6J_3650114 | Gm12786 | ENSEMBL:ENSMUSG00000085581 | MGI:3650114 | lncRNA gene | predicted gene 12786 |
| 4 | gene | 107.0559 | 107.0669 | negative | MGI_C57BL6J_1926268 | Mrpl37 | NCBI_Gene:56280,ENSEMBL:ENSMUSG00000028622 | MGI:1926268 | protein coding gene | mitochondrial ribosomal protein L37 |
| 4 | gene | 107.0669 | 107.0883 | positive | MGI_C57BL6J_1919657 | Cyb5rl | NCBI_Gene:230582,ENSEMBL:ENSMUSG00000028621 | MGI:1919657 | protein coding gene | cytochrome b5 reductase-like |
| 4 | gene | 107.0969 | 107.1131 | positive | MGI_C57BL6J_3045328 | Cdcp2 | NCBI_Gene:242603,ENSEMBL:ENSMUSG00000047636 | MGI:3045328 | protein coding gene | CUB domain containing protein 2 |
| 4 | gene | 107.1266 | 107.1363 | positive | MGI_C57BL6J_3649683 | Gm12802 | ENSEMBL:ENSMUSG00000085304 | MGI:3649683 | lncRNA gene | predicted gene 12802 |
| 4 | gene | 107.1342 | 107.1791 | negative | MGI_C57BL6J_1913776 | Tceanc2 | NCBI_Gene:66526,ENSEMBL:ENSMUSG00000028619 | MGI:1913776 | protein coding gene | transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| 4 | gene | 107.1784 | 107.2010 | positive | MGI_C57BL6J_1929278 | Tmem59 | NCBI_Gene:56374,ENSEMBL:ENSMUSG00000028618 | MGI:1929278 | protein coding gene | transmembrane protein 59 |
| 4 | gene | 107.2091 | 107.2211 | positive | MGI_C57BL6J_3652166 | Ldlrad1 | NCBI_Gene:546840,ENSEMBL:ENSMUSG00000070877 | MGI:3652166 | protein coding gene | low density lipoprotein receptor class A domain containing 1 |
| 4 | gene | 107.2335 | 107.2535 | negative | MGI_C57BL6J_1925059 | Lrrc42 | NCBI_Gene:77809,ENSEMBL:ENSMUSG00000028617 | MGI:1925059 | protein coding gene | leucine rich repeat containing 42 |
| 4 | gene | 107.2536 | 107.2845 | positive | MGI_C57BL6J_1920188 | Hspb11 | NCBI_Gene:72938,ENSEMBL:ENSMUSG00000063172 | MGI:1920188 | protein coding gene | heat shock protein family B (small), member 11 |
| 4 | pseudogene | 107.2704 | 107.2709 | positive | MGI_C57BL6J_3651240 | Gm12850 | ENSEMBL:ENSMUSG00000082941 | MGI:3651240 | pseudogene | predicted gene 12850 |
| 4 | gene | 107.2915 | 107.3072 | negative | MGI_C57BL6J_94896 | Dio1 | NCBI_Gene:13370,ENSEMBL:ENSMUSG00000034785 | MGI:94896 | protein coding gene | deiodinase, iodothyronine, type I |
| 4 | gene | 107.3142 | 107.3598 | positive | MGI_C57BL6J_1915532 | Yipf1 | NCBI_Gene:230584,ENSEMBL:ENSMUSG00000057375 | MGI:1915532 | protein coding gene | Yip1 domain family, member 1 |
| 4 | pseudogene | 107.3268 | 107.3285 | negative | MGI_C57BL6J_3649408 | Gm12870 | NCBI_Gene:652920,ENSEMBL:ENSMUSG00000082279 | MGI:3649408 | pseudogene | predicted gene 12870 |
| 4 | gene | 107.3490 | 107.3514 | negative | MGI_C57BL6J_3649409 | Gm12869 | ENSEMBL:ENSMUSG00000086041 | MGI:3649409 | lncRNA gene | predicted gene 12869 |
| 4 | gene | 107.3678 | 107.4163 | positive | MGI_C57BL6J_1920037 | Ndc1 | NCBI_Gene:72787,ENSEMBL:ENSMUSG00000028614 | MGI:1920037 | protein coding gene | NDC1 transmembrane nucleoporin |
| 4 | gene | 107.4266 | 107.6351 | positive | MGI_C57BL6J_2386723 | Glis1 | NCBI_Gene:230587,ENSEMBL:ENSMUSG00000034762 | MGI:2386723 | protein coding gene | GLIS family zinc finger 1 |
| 4 | gene | 107.4338 | 107.4345 | negative | MGI_C57BL6J_5826503 | Gm46866 | NCBI_Gene:108168971 | MGI:5826503 | lncRNA gene | predicted gene, 46866 |
| 4 | gene | 107.4346 | 107.4347 | positive | MGI_C57BL6J_5454581 | Gm24804 | ENSEMBL:ENSMUSG00000092723 | MGI:5454581 | miRNA gene | predicted gene, 24804 |
| 4 | gene | 107.5918 | 107.6013 | negative | MGI_C57BL6J_1925456 | 4930552P06Rik | NCBI_Gene:102633643 | MGI:1925456 | lncRNA gene | RIKEN cDNA 4930552P06 gene |
| 4 | gene | 107.6763 | 107.6843 | negative | MGI_C57BL6J_1927125 | Dmrtb1 | NCBI_Gene:56296,ENSEMBL:ENSMUSG00000028610 | MGI:1927125 | protein coding gene | DMRT-like family B with proline-rich C-terminal, 1 |
| 4 | pseudogene | 107.6881 | 107.6887 | positive | MGI_C57BL6J_3650162 | Gm12899 | NCBI_Gene:100417388,ENSEMBL:ENSMUSG00000081916 | MGI:3650162 | pseudogene | predicted gene 12899 |
| 4 | gene | 107.6896 | 107.7590 | positive | MGI_C57BL6J_1920570 | 1700047F07Rik | NCBI_Gene:102633724,ENSEMBL:ENSMUSG00000085549 | MGI:1920570 | lncRNA gene | RIKEN cDNA 1700047F07 gene |
| 4 | gene | 107.6978 | 107.6980 | negative | MGI_C57BL6J_5454900 | Gm25123 | ENSEMBL:ENSMUSG00000094267 | MGI:5454900 | miRNA gene | predicted gene, 25123 |
| 4 | gene | 107.6982 | 107.7365 | negative | MGI_C57BL6J_5590841 | Gm31682 | NCBI_Gene:102633989 | MGI:5590841 | lncRNA gene | predicted gene, 31682 |
| 4 | pseudogene | 107.7594 | 107.7605 | negative | MGI_C57BL6J_3650884 | Gm12906 | NCBI_Gene:433743,ENSEMBL:ENSMUSG00000084874 | MGI:3650884 | pseudogene | predicted gene 12906 |
| 4 | gene | 107.7595 | 107.7710 | positive | MGI_C57BL6J_3650882 | Gm12907 | NCBI_Gene:102634068,ENSEMBL:ENSMUSG00000087195 | MGI:3650882 | lncRNA gene | predicted gene 12907 |
| 4 | gene | 107.7837 | 107.7899 | positive | MGI_C57BL6J_1914798 | 4933424M12Rik | NCBI_Gene:67548,ENSEMBL:ENSMUSG00000087289 | MGI:1914798 | lncRNA gene | RIKEN cDNA 4933424M12 gene |
| 4 | gene | 107.7931 | 107.8022 | negative | MGI_C57BL6J_3651133 | Lrp8os3 | NCBI_Gene:105244644,ENSEMBL:ENSMUSG00000087200 | MGI:3651133 | bidirectional promoter lncRNA gene | low density lipoprotein receptor-related protein 8, apolipoprotein e receptor, opposite strand 3 |
| 4 | gene | 107.8019 | 107.8768 | positive | MGI_C57BL6J_1340044 | Lrp8 | NCBI_Gene:16975,ENSEMBL:ENSMUSG00000028613 | MGI:1340044 | protein coding gene | low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| 4 | gene | 107.8044 | 107.8232 | negative | MGI_C57BL6J_3588233 | Lrp8os2 | NCBI_Gene:619295,ENSEMBL:ENSMUSG00000073779 | MGI:3588233 | antisense lncRNA gene | low density lipoprotein receptor-related protein 8, apolipoprotein e receptor, opposite strand 2 |
| 4 | gene | 107.8279 | 107.8308 | negative | MGI_C57BL6J_1925359 | Lrp8os1 | ENSEMBL:ENSMUSG00000028612 | MGI:1925359 | antisense lncRNA gene | low density lipoprotein receptor-related protein 8, apolipoprotein e receptor, opposite strand 1 |
| 4 | gene | 107.8798 | 107.8874 | positive | MGI_C57BL6J_1330312 | Magoh | NCBI_Gene:17149,ENSEMBL:ENSMUSG00000028609 | MGI:1330312 | protein coding gene | mago homolog, exon junction complex core component |
| 4 | gene | 107.8898 | 107.8994 | positive | MGI_C57BL6J_1921348 | Czib | NCBI_Gene:74098,ENSEMBL:ENSMUSG00000028608 | MGI:1921348 | protein coding gene | CXXC motif containing zinc binding protein |
| 4 | gene | 107.9040 | 107.9236 | negative | MGI_C57BL6J_109176 | Cpt2 | NCBI_Gene:12896,ENSEMBL:ENSMUSG00000028607 | MGI:109176 | protein coding gene | carnitine palmitoyltransferase 2 |
| 4 | gene | 107.9375 | 107.9474 | positive | MGI_C57BL6J_5623106 | Gm40221 | NCBI_Gene:105244646 | MGI:5623106 | lncRNA gene | predicted gene, 40221 |
| 4 | gene | 107.9539 | 107.9607 | positive | MGI_C57BL6J_1924473 | 4930407G08Rik | NCBI_Gene:77223,ENSEMBL:ENSMUSG00000086713 | MGI:1924473 | lncRNA gene | RIKEN cDNA 4930407G08 gene |
| 4 | gene | 107.9683 | 108.0135 | positive | MGI_C57BL6J_2444087 | Slc1a7 | NCBI_Gene:242607,ENSEMBL:ENSMUSG00000008932 | MGI:2444087 | protein coding gene | solute carrier family 1 (glutamate transporter), member 7 |
| 4 | gene | 108.0148 | 108.0964 | negative | MGI_C57BL6J_2674939 | Podn | NCBI_Gene:242608,ENSEMBL:ENSMUSG00000028600 | MGI:2674939 | protein coding gene | podocan |
| 4 | gene | 108.0417 | 108.0418 | negative | MGI_C57BL6J_5453131 | Gm23354 | ENSEMBL:ENSMUSG00000089051 | MGI:5453131 | snRNA gene | predicted gene, 23354 |
| 4 | gene | 108.0438 | 108.1450 | negative | MGI_C57BL6J_98254 | Scp2 | NCBI_Gene:20280,ENSEMBL:ENSMUSG00000028603 | MGI:98254 | protein coding gene | sterol carrier protein 2, liver |
| 4 | gene | 108.0874 | 108.0874 | positive | MGI_C57BL6J_5531386 | Mir6397 | miRBase:MI0021933,NCBI_Gene:102465213,ENSEMBL:ENSMUSG00000098980 | MGI:5531386 | miRNA gene | microRNA 6397 |
| 4 | gene | 108.1654 | 108.1793 | positive | MGI_C57BL6J_1289238 | Echdc2 | NCBI_Gene:52430,ENSEMBL:ENSMUSG00000028601 | MGI:1289238 | protein coding gene | enoyl Coenzyme A hydratase domain containing 2 |
| 4 | gene | 108.1802 | 108.2182 | negative | MGI_C57BL6J_2446208 | Zyg11a | NCBI_Gene:230590,ENSEMBL:ENSMUSG00000034645 | MGI:2446208 | protein coding gene | zyg-11 family member A, cell cycle regulator |
| 4 | gene | 108.2180 | 108.2205 | positive | MGI_C57BL6J_5623107 | Gm40222 | NCBI_Gene:105244647 | MGI:5623107 | lncRNA gene | predicted gene, 40222 |
| 4 | gene | 108.2278 | 108.3011 | negative | MGI_C57BL6J_2685277 | Zyg11b | NCBI_Gene:414872,ENSEMBL:ENSMUSG00000034636 | MGI:2685277 | protein coding gene | zyg-ll family member B, cell cycle regulator |
| 4 | pseudogene | 108.3104 | 108.3112 | negative | MGI_C57BL6J_3651646 | Gm12742 | ENSEMBL:ENSMUSG00000083957 | MGI:3651646 | pseudogene | predicted gene 12742 |
| 4 | pseudogene | 108.3223 | 108.3226 | positive | MGI_C57BL6J_3651641 | Gm12740 | ENSEMBL:ENSMUSG00000081734 | MGI:3651641 | pseudogene | predicted gene 12740 |
| 4 | gene | 108.3281 | 108.3415 | positive | MGI_C57BL6J_1917143 | Coa7 | NCBI_Gene:69893,ENSEMBL:ENSMUSG00000048351 | MGI:1917143 | protein coding gene | cytochrome c oxidase assembly factor 7 |
| 4 | gene | 108.3455 | 108.3512 | negative | MGI_C57BL6J_5826526 | Gm46889 | NCBI_Gene:108169005 | MGI:5826526 | lncRNA gene | predicted gene, 46889 |
| 4 | gene | 108.3678 | 108.3858 | negative | MGI_C57BL6J_3651644 | Shisal2a | NCBI_Gene:545667,ENSEMBL:ENSMUSG00000059816 | MGI:3651644 | protein coding gene | shisa like 2A |
| 4 | gene | 108.3833 | 108.4066 | positive | MGI_C57BL6J_5591456 | Gm32297 | NCBI_Gene:105244648 | MGI:5591456 | lncRNA gene | predicted gene, 32297 |
| 4 | gene | 108.4002 | 108.4070 | negative | MGI_C57BL6J_1914555 | Gpx7 | NCBI_Gene:67305,ENSEMBL:ENSMUSG00000028597 | MGI:1914555 | protein coding gene | glutathione peroxidase 7 |
| 4 | gene | 108.4306 | 108.4542 | positive | MGI_C57BL6J_5623108 | Gm40223 | NCBI_Gene:105244650 | MGI:5623108 | lncRNA gene | predicted gene, 40223 |
| 4 | pseudogene | 108.4507 | 108.4514 | negative | MGI_C57BL6J_3651217 | Gm12738 | NCBI_Gene:665728,ENSEMBL:ENSMUSG00000083255 | MGI:3651217 | pseudogene | predicted gene 12738 |
| 4 | gene | 108.4594 | 108.5607 | positive | MGI_C57BL6J_2445126 | Tut4 | NCBI_Gene:230594,ENSEMBL:ENSMUSG00000034610 | MGI:2445126 | protein coding gene | terminal uridylyl transferase 4 |
| 4 | gene | 108.4669 | 108.4671 | negative | MGI_C57BL6J_5454744 | Gm24967 | ENSEMBL:ENSMUSG00000095361 | MGI:5454744 | snRNA gene | predicted gene, 24967 |
| 4 | pseudogene | 108.4841 | 108.4849 | negative | MGI_C57BL6J_3651018 | Gm12739 | NCBI_Gene:105244651,ENSEMBL:ENSMUSG00000083783 | MGI:3651018 | pseudogene | predicted gene 12739 |
| 4 | gene | 108.4980 | 108.4981 | positive | MGI_C57BL6J_5452915 | Gm23138 | ENSEMBL:ENSMUSG00000065299 | MGI:5452915 | snRNA gene | predicted gene, 23138 |
| 4 | gene | 108.5138 | 108.5168 | positive | MGI_C57BL6J_3651021 | Gm12737 | ENSEMBL:ENSMUSG00000085093 | MGI:3651021 | lncRNA gene | predicted gene 12737 |
| 4 | gene | 108.5219 | 108.5220 | negative | MGI_C57BL6J_5455604 | Gm25827 | ENSEMBL:ENSMUSG00000087714 | MGI:5455604 | snRNA gene | predicted gene, 25827 |
| 4 | gene | 108.5375 | 108.5376 | negative | MGI_C57BL6J_5452122 | Gm22345 | ENSEMBL:ENSMUSG00000084593 | MGI:5452122 | snRNA gene | predicted gene, 22345 |
| 4 | gene | 108.5632 | 108.5793 | negative | MGI_C57BL6J_1916962 | Prpf38a | NCBI_Gene:230596,ENSEMBL:ENSMUSG00000063800 | MGI:1916962 | protein coding gene | PRP38 pre-mRNA processing factor 38 (yeast) domain containing A |
| 4 | gene | 108.5756 | 108.6160 | positive | MGI_C57BL6J_1328337 | Orc1 | NCBI_Gene:18392,ENSEMBL:ENSMUSG00000028587 | MGI:1328337 | protein coding gene | origin recognition complex, subunit 1 |
| 4 | gene | 108.5924 | 108.6198 | negative | MGI_C57BL6J_3651449 | Gm12743 | ENSEMBL:ENSMUSG00000084968 | MGI:3651449 | lncRNA gene | predicted gene 12743 |
| 4 | gene | 108.6198 | 108.6355 | positive | MGI_C57BL6J_2443076 | Cc2d1b | NCBI_Gene:319965,ENSEMBL:ENSMUSG00000028582 | MGI:2443076 | protein coding gene | coiled-coil and C2 domain containing 1B |
| 4 | gene | 108.6229 | 108.6252 | negative | MGI_C57BL6J_5313040 | Gm20731 | ENSEMBL:ENSMUSG00000102912 | MGI:5313040 | protein coding gene | predicted gene, 20731 |
| 4 | gene | 108.6338 | 108.6374 | negative | MGI_C57BL6J_4936988 | Gm17354 | ENSEMBL:ENSMUSG00000091985 | MGI:4936988 | lncRNA gene | predicted gene, 17354 |
| 4 | gene | 108.6375 | 108.7822 | negative | MGI_C57BL6J_2652838 | Zfyve9 | NCBI_Gene:230597,ENSEMBL:ENSMUSG00000034557 | MGI:2652838 | protein coding gene | zinc finger, FYVE domain containing 9 |
| 4 | gene | 108.7196 | 108.7819 | positive | MGI_C57BL6J_1920383 | 3110021N24Rik | ENSEMBL:ENSMUSG00000094958 | MGI:1920383 | protein coding gene | RIKEN cDNA 3110021N24 gene |
| 4 | pseudogene | 108.7317 | 108.7324 | positive | MGI_C57BL6J_3651642 | Gm12741 | NCBI_Gene:100040326,ENSEMBL:ENSMUSG00000082374 | MGI:3651642 | pseudogene | predicted gene 12741 |
| 4 | gene | 108.8143 | 108.8343 | negative | MGI_C57BL6J_1915312 | Btf3l4 | NCBI_Gene:70533,ENSEMBL:ENSMUSG00000028568 | MGI:1915312 | protein coding gene | basic transcription factor 3-like 4 |
| 4 | gene | 108.8346 | 108.8621 | positive | MGI_C57BL6J_1913323 | Txndc12 | NCBI_Gene:66073,ENSEMBL:ENSMUSG00000028567 | MGI:1913323 | protein coding gene | thioredoxin domain containing 12 (endoplasmic reticulum) |
| 4 | gene | 108.8474 | 108.8486 | negative | MGI_C57BL6J_3704235 | A730015C16Rik | NA | NA | lncRNA gene | RIKEN cDNA A730015C16 gene |
| 4 | gene | 108.8478 | 108.8494 | positive | MGI_C57BL6J_1923547 | Kti12 | NCBI_Gene:100087,ENSEMBL:ENSMUSG00000073775 | MGI:1923547 | protein coding gene | KTI12 homolog, chromatin associated |
| 4 | gene | 108.8509 | 108.8520 | positive | MGI_C57BL6J_1924896 | C030043A13Rik | NA | NA | unclassified gene | RIKEN cDNA C030043A13 gene |
| 4 | gene | 108.8791 | 108.9433 | positive | MGI_C57BL6J_1917158 | Rab3b | NCBI_Gene:69908,ENSEMBL:ENSMUSG00000003411 | MGI:1917158 | protein coding gene | RAB3B, member RAS oncogene family |
| 4 | pseudogene | 108.9336 | 108.9338 | negative | MGI_C57BL6J_3650723 | Gm12736 | ENSEMBL:ENSMUSG00000082699 | MGI:3650723 | pseudogene | predicted gene 12736 |
| 4 | gene | 108.9428 | 108.9721 | negative | MGI_C57BL6J_1924423 | 8030443G20Rik | NCBI_Gene:77173,ENSEMBL:ENSMUSG00000086483 | MGI:1924423 | lncRNA gene | RIKEN cDNA 8030443G20 gene |
| 4 | gene | 108.9986 | 108.9987 | negative | MGI_C57BL6J_5453366 | Gm23589 | ENSEMBL:ENSMUSG00000088285 | MGI:5453366 | snRNA gene | predicted gene, 23589 |
| 4 | gene | 109.0006 | 109.0618 | positive | MGI_C57BL6J_1201386 | Nrd1 | NCBI_Gene:230598,ENSEMBL:ENSMUSG00000053510 | MGI:1201386 | protein coding gene | nardilysin, N-arginine dibasic convertase, NRD convertase 1 |
| 4 | gene | 109.0177 | 109.0177 | positive | MGI_C57BL6J_3691609 | Mir761 | miRBase:MI0004306,NCBI_Gene:791075,ENSEMBL:ENSMUSG00000076444 | MGI:3691609 | miRNA gene | microRNA 761 |
| 4 | gene | 109.0611 | 109.2023 | negative | MGI_C57BL6J_1923784 | Osbpl9 | NCBI_Gene:100273,ENSEMBL:ENSMUSG00000028559 | MGI:1923784 | protein coding gene | oxysterol binding protein-like 9 |
| 4 | gene | 109.0698 | 109.0702 | negative | MGI_C57BL6J_1925486 | 5330417P21Rik | NA | NA | unclassified gene | RIKEN cDNA 5330417P21 gene |
| 4 | gene | 109.0785 | 109.0812 | positive | MGI_C57BL6J_5591740 | Gm32581 | NCBI_Gene:102635178 | MGI:5591740 | lncRNA gene | predicted gene, 32581 |
| 4 | gene | 109.1030 | 109.1035 | negative | MGI_C57BL6J_1924670 | C030032G21Rik | NA | NA | unclassified gene | RIKEN cDNA C030032G21 gene |
| 4 | gene | 109.1609 | 109.1612 | negative | MGI_C57BL6J_5453520 | Gm23743 | ENSEMBL:ENSMUSG00000088014 | MGI:5453520 | unclassified non-coding RNA gene | predicted gene, 23743 |
| 4 | gene | 109.2022 | 109.2085 | positive | MGI_C57BL6J_5826504 | Gm46867 | NCBI_Gene:108168972 | MGI:5826504 | lncRNA gene | predicted gene, 46867 |
| 4 | gene | 109.2184 | 109.2551 | positive | MGI_C57BL6J_2140435 | Calr4 | NCBI_Gene:108802,ENSEMBL:ENSMUSG00000028558 | MGI:2140435 | protein coding gene | calreticulin 4 |
| 4 | gene | 109.2241 | 109.2801 | negative | MGI_C57BL6J_5623110 | Gm40225 | NCBI_Gene:105244653 | MGI:5623110 | lncRNA gene | predicted gene, 40225 |
| 4 | gene | 109.2803 | 109.3878 | positive | MGI_C57BL6J_104583 | Eps15 | NCBI_Gene:13858,ENSEMBL:ENSMUSG00000028552 | MGI:104583 | protein coding gene | epidermal growth factor receptor pathway substrate 15 |
| 4 | gene | 109.3554 | 109.3600 | negative | MGI_C57BL6J_5623109 | Gm40224 | NCBI_Gene:105244652 | MGI:5623109 | lncRNA gene | predicted gene, 40224 |
| 4 | pseudogene | 109.3737 | 109.3749 | negative | MGI_C57BL6J_3652196 | Gm12749 | NCBI_Gene:666144,ENSEMBL:ENSMUSG00000083512 | MGI:3652196 | pseudogene | predicted gene 12749 |
| 4 | gene | 109.3941 | 109.4447 | positive | MGI_C57BL6J_2444350 | Ttc39a | NCBI_Gene:230603,ENSEMBL:ENSMUSG00000028555 | MGI:2444350 | protein coding gene | tetratricopeptide repeat domain 39A |
| 4 | gene | 109.4023 | 109.4063 | negative | MGI_C57BL6J_3651956 | Ttc39aos1 | NCBI_Gene:102635290,ENSEMBL:ENSMUSG00000085873 | MGI:3651956 | antisense lncRNA gene | Ttc39a opposite strand RNA 1 |
| 4 | gene | 109.4511 | 109.4847 | negative | MGI_C57BL6J_1352759 | Rnf11 | NCBI_Gene:29864,ENSEMBL:ENSMUSG00000028557 | MGI:1352759 | protein coding gene | ring finger protein 11 |
| 4 | gene | 109.5053 | 109.5313 | negative | MGI_C57BL6J_1914896 | 4930522H14Rik | NCBI_Gene:67646,ENSEMBL:ENSMUSG00000060491 | MGI:1914896 | protein coding gene | RIKEN cDNA 4930522H14 gene |
| 4 | gene | 109.5370 | 109.5372 | positive | MGI_C57BL6J_5452274 | Gm22497 | ENSEMBL:ENSMUSG00000077687 | MGI:5452274 | snoRNA gene | predicted gene, 22497 |
| 4 | gene | 109.5510 | 109.5540 | positive | MGI_C57BL6J_5623111 | Gm40226 | NCBI_Gene:105244654 | MGI:5623111 | lncRNA gene | predicted gene, 40226 |
| 4 | gene | 109.5692 | 109.6568 | positive | MGI_C57BL6J_3650795 | Gm12811 | ENSEMBL:ENSMUSG00000087641 | MGI:3650795 | lncRNA gene | predicted gene 12811 |
| 4 | pseudogene | 109.5994 | 109.6000 | negative | MGI_C57BL6J_5011297 | Gm19112 | NCBI_Gene:100418274 | MGI:5011297 | pseudogene | predicted gene, 19112 |
| 4 | pseudogene | 109.6296 | 109.6309 | positive | MGI_C57BL6J_3651634 | Gm12803 | NCBI_Gene:100418450,ENSEMBL:ENSMUSG00000082771 | MGI:3651634 | pseudogene | predicted gene 12803 |
| 4 | gene | 109.6309 | 109.6491 | negative | MGI_C57BL6J_5623112 | Gm40227 | NCBI_Gene:105244655 | MGI:5623112 | lncRNA gene | predicted gene, 40227 |
| 4 | gene | 109.6515 | 109.6579 | negative | MGI_C57BL6J_2442649 | 9630013D21Rik | NCBI_Gene:319743,ENSEMBL:ENSMUSG00000059027 | MGI:2442649 | lncRNA gene | RIKEN cDNA 9630013D21 gene |
| 4 | gene | 109.6609 | 109.6672 | negative | MGI_C57BL6J_105388 | Cdkn2c | NCBI_Gene:12580,ENSEMBL:ENSMUSG00000028551 | MGI:105388 | protein coding gene | cyclin dependent kinase inhibitor 2C |
| 4 | gene | 109.6669 | 109.6682 | negative | MGI_C57BL6J_3826683 | Gm2703 | NA | NA | protein coding gene | predicted gene 2703 |
| 4 | gene | 109.6766 | 109.9640 | positive | MGI_C57BL6J_109419 | Faf1 | NCBI_Gene:14084,ENSEMBL:ENSMUSG00000010517 | MGI:109419 | protein coding gene | Fas-associated factor 1 |
| 4 | pseudogene | 109.7544 | 109.7548 | negative | MGI_C57BL6J_3650597 | Gm12808 | ENSEMBL:ENSMUSG00000083995 | MGI:3650597 | pseudogene | predicted gene 12808 |
| 4 | gene | 109.9732 | 109.9778 | negative | MGI_C57BL6J_1924703 | Dmrta2os | NCBI_Gene:77453,ENSEMBL:ENSMUSG00000085256 | MGI:1924703 | antisense lncRNA gene | doublesex and mab-3 related transcription factor like family A2, opposite strand |
| 4 | gene | 109.9778 | 109.9837 | positive | MGI_C57BL6J_2653629 | Dmrta2 | NCBI_Gene:242620,ENSEMBL:ENSMUSG00000047143 | MGI:2653629 | protein coding gene | doublesex and mab-3 related transcription factor like family A2 |
| 4 | gene | 110.1527 | 110.1567 | positive | MGI_C57BL6J_5592229 | Gm33070 | NCBI_Gene:102635827 | MGI:5592229 | lncRNA gene | predicted gene, 33070 |
| 4 | gene | 110.2037 | 110.3519 | negative | MGI_C57BL6J_107427 | Elavl4 | NCBI_Gene:15572,ENSEMBL:ENSMUSG00000028546 | MGI:107427 | protein coding gene | ELAV like RNA binding protein 4 |
| 4 | gene | 110.3976 | 111.6643 | positive | MGI_C57BL6J_1918244 | Agbl4 | NCBI_Gene:78933,ENSEMBL:ENSMUSG00000061298 | MGI:1918244 | protein coding gene | ATP/GTP binding protein-like 4 |
| 4 | gene | 110.4348 | 110.4360 | positive | MGI_C57BL6J_1924569 | C030007D22Rik | NA | NA | unclassified gene | RIKEN cDNA C030007D22 gene |
| 4 | pseudogene | 110.5100 | 110.5103 | positive | MGI_C57BL6J_3651080 | Gm12806 | ENSEMBL:ENSMUSG00000081813 | MGI:3651080 | pseudogene | predicted gene 12806 |
| 4 | pseudogene | 110.5128 | 110.5143 | negative | MGI_C57BL6J_3650599 | Gm12807 | NCBI_Gene:102635892,ENSEMBL:ENSMUSG00000082348 | MGI:3650599 | pseudogene | predicted gene 12807 |
| 4 | pseudogene | 111.2901 | 111.2911 | positive | MGI_C57BL6J_3651639 | Gm12804 | NCBI_Gene:100417659,ENSEMBL:ENSMUSG00000081068 | MGI:3651639 | pseudogene | predicted gene 12804 |
| 4 | pseudogene | 111.3195 | 111.3206 | negative | MGI_C57BL6J_3650124 | Gm12805 | NCBI_Gene:100417245,ENSEMBL:ENSMUSG00000083904 | MGI:3650124 | pseudogene | predicted gene 12805 |
| 4 | gene | 111.4138 | 111.4603 | positive | MGI_C57BL6J_1914871 | Bend5 | NCBI_Gene:67621,ENSEMBL:ENSMUSG00000028545 | MGI:1914871 | protein coding gene | BEN domain containing 5 |
| 4 | gene | 111.4278 | 111.4279 | negative | MGI_C57BL6J_4422056 | n-R5s191 | ENSEMBL:ENSMUSG00000084673 | MGI:4422056 | rRNA gene | nuclear encoded rRNA 5S 191 |
| 4 | gene | 111.6956 | 111.7240 | negative | MGI_C57BL6J_5592351 | Gm33192 | NCBI_Gene:102635996 | MGI:5592351 | lncRNA gene | predicted gene, 33192 |
| 4 | gene | 111.7199 | 111.8292 | positive | MGI_C57BL6J_1915196 | Spata6 | NCBI_Gene:67946,ENSEMBL:ENSMUSG00000034401 | MGI:1915196 | protein coding gene | spermatogenesis associated 6 |
| 4 | pseudogene | 111.7906 | 111.7917 | negative | MGI_C57BL6J_3650787 | Gm12961 | NCBI_Gene:100417901,ENSEMBL:ENSMUSG00000082686 | MGI:3650787 | pseudogene | predicted gene 12961 |
| 4 | pseudogene | 111.8070 | 111.8085 | negative | MGI_C57BL6J_3651525 | Gm12960 | NCBI_Gene:105244657,ENSEMBL:ENSMUSG00000100514 | MGI:3651525 | pseudogene | predicted gene 12960 |
| 4 | gene | 111.8754 | 111.9029 | negative | MGI_C57BL6J_2140201 | Slc5a9 | NCBI_Gene:230612,ENSEMBL:ENSMUSG00000028544 | MGI:2140201 | protein coding gene | solute carrier family 5 (sodium/glucose cotransporter), member 9 |
| 4 | gene | 111.9194 | 111.9504 | positive | MGI_C57BL6J_3651523 | Skint8 | NCBI_Gene:639774,ENSEMBL:ENSMUSG00000078599 | MGI:3651523 | protein coding gene | selection and upkeep of intraepithelial T cells 8 |
| 4 | gene | 111.9729 | 111.9883 | positive | MGI_C57BL6J_3041190 | Skint7 | NCBI_Gene:328505,ENSEMBL:ENSMUSG00000049214 | MGI:3041190 | protein coding gene | selection and upkeep of intraepithelial T cells 7 |
| 4 | gene | 111.9887 | 111.9890 | negative | MGI_C57BL6J_5455621 | Gm25844 | ENSEMBL:ENSMUSG00000084536 | MGI:5455621 | unclassified non-coding RNA gene | predicted gene, 25844 |
| 4 | gene | 112.0063 | 112.0295 | positive | MGI_C57BL6J_3649627 | Skint1 | NCBI_Gene:639781,ENSEMBL:ENSMUSG00000089773 | MGI:3649627 | protein coding gene | selection and upkeep of intraepithelial T cells 1 |
| 4 | gene | 112.0720 | 112.1681 | positive | MGI_C57BL6J_2444425 | Skint4 | NCBI_Gene:320640,ENSEMBL:ENSMUSG00000055960 | MGI:2444425 | protein coding gene | selection and upkeep of intraepithelial T cells 4 |
| 4 | pseudogene | 112.1210 | 112.1213 | negative | MGI_C57BL6J_3652018 | Gm12820 | ENSEMBL:ENSMUSG00000084326 | MGI:3652018 | pseudogene | predicted gene 12820 |
| 4 | pseudogene | 112.1461 | 112.1462 | negative | MGI_C57BL6J_3652016 | Gm12815 | ENSEMBL:ENSMUSG00000082638 | MGI:3652016 | pseudogene | predicted gene 12815 |
| 4 | gene | 112.2322 | 112.3005 | positive | MGI_C57BL6J_3045331 | Skint3 | NCBI_Gene:195564,ENSEMBL:ENSMUSG00000070868 | MGI:3045331 | protein coding gene | selection and upkeep of intraepithelial T cells 3 |
| 4 | gene | 112.3860 | 112.4340 | negative | MGI_C57BL6J_3045341 | Skint9 | NCBI_Gene:329918,ENSEMBL:ENSMUSG00000049972 | MGI:3045341 | protein coding gene | selection and upkeep of intraepithelial T cells 9 |
| 4 | pseudogene | 112.4738 | 112.5215 | positive | MGI_C57BL6J_3652017 | Gm12819 | NCBI_Gene:100504555,ENSEMBL:ENSMUSG00000081166 | MGI:3652017 | pseudogene | predicted gene 12819 |
| 4 | pseudogene | 112.5371 | 112.5377 | positive | MGI_C57BL6J_3649625 | Gm12821 | NCBI_Gene:102636060,ENSEMBL:ENSMUSG00000083599 | MGI:3649625 | pseudogene | predicted gene 12821 |
| 4 | gene | 112.5572 | 112.6522 | positive | MGI_C57BL6J_3649629 | Skint2 | NCBI_Gene:329919,ENSEMBL:ENSMUSG00000034359 | MGI:3649629 | protein coding gene | selection and upkeep of intraepithelial T cells 2 |
| 4 | pseudogene | 112.5597 | 112.5933 | positive | MGI_C57BL6J_3651208 | Gm12814 | NCBI_Gene:630887,ENSEMBL:ENSMUSG00000083753 | MGI:3651208 | pseudogene | predicted gene 12814 |
| 4 | gene | 112.5908 | 112.5909 | negative | MGI_C57BL6J_5453876 | Gm24099 | ENSEMBL:ENSMUSG00000096564 | MGI:5453876 | snRNA gene | predicted gene, 24099 |
| 4 | pseudogene | 112.7051 | 112.7062 | positive | MGI_C57BL6J_3649633 | Gm12817 | NCBI_Gene:100040581,ENSEMBL:ENSMUSG00000081066 | MGI:3649633 | pseudogene | predicted gene 12817 |
| 4 | gene | 112.7108 | 112.7749 | negative | MGI_C57BL6J_2685416 | Skint10 | NCBI_Gene:230613,ENSEMBL:ENSMUSG00000048766 | MGI:2685416 | protein coding gene | selection and upkeep of intraepithelial T cells 10 |
| 4 | pseudogene | 112.7721 | 112.7733 | positive | MGI_C57BL6J_5010585 | Gm18400 | NCBI_Gene:100417108 | MGI:5010585 | pseudogene | predicted gene, 18400 |
| 4 | gene | 112.8045 | 113.2870 | negative | MGI_C57BL6J_3649262 | Skint6 | NCBI_Gene:230622,ENSEMBL:ENSMUSG00000087194 | MGI:3649262 | protein coding gene | selection and upkeep of intraepithelial T cells 6 |
| 4 | gene | 112.9237 | 112.9453 | positive | MGI_C57BL6J_3651665 | Gm12823 | ENSEMBL:ENSMUSG00000085878 | MGI:3651665 | lncRNA gene | predicted gene 12823 |
| 4 | pseudogene | 112.9550 | 112.9563 | negative | MGI_C57BL6J_3649628 | Gm12816 | NCBI_Gene:433745,ENSEMBL:ENSMUSG00000083011 | MGI:3649628 | pseudogene | predicted gene 12816 |
| 4 | pseudogene | 112.9785 | 112.9798 | negative | MGI_C57BL6J_3649634 | Gm12818 | NCBI_Gene:100534383,ENSEMBL:ENSMUSG00000082678 | MGI:3649634 | pseudogene | predicted gene 12818 |
| 4 | pseudogene | 113.3682 | 113.3945 | positive | MGI_C57BL6J_3650999 | Gm12813 | NCBI_Gene:100040607,ENSEMBL:ENSMUSG00000083384 | MGI:3650999 | pseudogene | predicted gene 12813 |
| 4 | gene | 113.4779 | 113.9995 | negative | MGI_C57BL6J_3650151 | Skint5 | NCBI_Gene:242627,ENSEMBL:ENSMUSG00000078598 | MGI:3650151 | protein coding gene | selection and upkeep of intraepithelial T cells 5 |
| 4 | pseudogene | 114.0963 | 114.0974 | negative | MGI_C57BL6J_3650153 | Gm12825 | NCBI_Gene:100418129,ENSEMBL:ENSMUSG00000080696 | MGI:3650153 | pseudogene | predicted gene 12825 |
| 4 | gene | 114.1634 | 114.2450 | positive | MGI_C57BL6J_2685415 | Skint11 | NCBI_Gene:230623,ENSEMBL:ENSMUSG00000057977 | MGI:2685415 | protein coding gene | selection and upkeep of intraepithelial T cells 11 |
| 4 | pseudogene | 114.1651 | 114.1659 | negative | MGI_C57BL6J_3783183 | Gm15741 | NCBI_Gene:100417107,ENSEMBL:ENSMUSG00000082825 | MGI:3783183 | pseudogene | predicted gene 15741 |
| 4 | gene | 114.3299 | 114.3466 | negative | MGI_C57BL6J_5623113 | Gm40228 | NCBI_Gene:105244658 | MGI:5623113 | lncRNA gene | predicted gene, 40228 |
| 4 | gene | 114.4062 | 114.6151 | positive | MGI_C57BL6J_3650152 | Trabd2b | NCBI_Gene:666048,ENSEMBL:ENSMUSG00000070867 | MGI:3650152 | protein coding gene | TraB domain containing 2B |
| 4 | gene | 114.4140 | 114.4149 | negative | MGI_C57BL6J_5592869 | Gm33710 | NCBI_Gene:102636714 | MGI:5592869 | lncRNA gene | predicted gene, 33710 |
| 4 | gene | 114.5769 | 114.5922 | negative | MGI_C57BL6J_5592814 | Gm33655 | NCBI_Gene:102636647 | MGI:5592814 | lncRNA gene | predicted gene, 33655 |
| 4 | gene | 114.6727 | 114.6733 | negative | MGI_C57BL6J_5579570 | Gm28864 | ENSEMBL:ENSMUSG00000100998 | MGI:5579570 | lncRNA gene | predicted gene 28864 |
| 4 | gene | 114.6803 | 114.6876 | positive | MGI_C57BL6J_3650003 | Gm12829 | NCBI_Gene:102636500,ENSEMBL:ENSMUSG00000085870 | MGI:3650003 | lncRNA gene | predicted gene 12829 |
| 4 | gene | 114.8161 | 114.9029 | negative | MGI_C57BL6J_1918862 | 9130410C08Rik | NCBI_Gene:105244659,ENSEMBL:ENSMUSG00000085323 | MGI:1918862 | lncRNA gene | RIKEN cDNA 9130410C08 gene |
| 4 | gene | 114.8217 | 114.8562 | positive | MGI_C57BL6J_3649998 | Gm12830 | NCBI_Gene:433746,ENSEMBL:ENSMUSG00000055198 | MGI:3649998 | protein coding gene | predicted gene 12830 |
| 4 | gene | 114.8450 | 114.8451 | negative | MGI_C57BL6J_5453007 | Gm23230 | ENSEMBL:ENSMUSG00000087923 | MGI:5453007 | snRNA gene | predicted gene, 23230 |
| 4 | gene | 114.9063 | 114.9089 | negative | MGI_C57BL6J_1347471 | Foxd2 | NCBI_Gene:17301,ENSEMBL:ENSMUSG00000055210 | MGI:1347471 | protein coding gene | forkhead box D2 |
| 4 | gene | 114.9093 | 114.9220 | positive | MGI_C57BL6J_2444065 | Foxd2os | NCBI_Gene:100040736,ENSEMBL:ENSMUSG00000085399 | MGI:2444065 | lncRNA gene | forkhead box D2, opposite strand |
| 4 | gene | 114.9251 | 114.9260 | negative | MGI_C57BL6J_1353569 | Foxe3 | NCBI_Gene:30923,ENSEMBL:ENSMUSG00000044518 | MGI:1353569 | protein coding gene | forkhead box E3 |
| 4 | gene | 114.9468 | 114.9508 | negative | MGI_C57BL6J_5623114 | Gm40229 | NCBI_Gene:105244660 | MGI:5623114 | lncRNA gene | predicted gene, 40229 |
| 4 | gene | 114.9593 | 114.9872 | negative | MGI_C57BL6J_1913838 | Cmpk1 | NCBI_Gene:66588,ENSEMBL:ENSMUSG00000028719 | MGI:1913838 | protein coding gene | cytidine monophosphate (UMP-CMP) kinase 1 |
| 4 | gene | 114.9880 | 114.9881 | positive | MGI_C57BL6J_5453822 | Gm24045 | ENSEMBL:ENSMUSG00000065047 | MGI:5453822 | unclassified non-coding RNA gene | predicted gene, 24045 |
| 4 | gene | 115.0001 | 115.0432 | positive | MGI_C57BL6J_107477 | Stil | NCBI_Gene:20460,ENSEMBL:ENSMUSG00000028718 | MGI:107477 | protein coding gene | Scl/Tal1 interrupting locus |
| 4 | gene | 115.0564 | 115.0718 | positive | MGI_C57BL6J_98480 | Tal1 | NCBI_Gene:21349,ENSEMBL:ENSMUSG00000028717 | MGI:98480 | protein coding gene | T cell acute lymphocytic leukemia 1 |
| 4 | gene | 115.0576 | 115.0582 | negative | MGI_C57BL6J_5592945 | Gm33786 | NCBI_Gene:102636820 | MGI:5592945 | lncRNA gene | predicted gene, 33786 |
| 4 | gene | 115.0887 | 115.0939 | positive | MGI_C57BL6J_1914432 | Pdzk1ip1 | NCBI_Gene:67182,ENSEMBL:ENSMUSG00000028716 | MGI:1914432 | protein coding gene | PDZK1 interacting protein 1 |
| 4 | gene | 115.1063 | 115.1343 | negative | MGI_C57BL6J_1932403 | Cyp4x1 | NCBI_Gene:81906,ENSEMBL:ENSMUSG00000047155 | MGI:1932403 | protein coding gene | cytochrome P450, family 4, subfamily x, polypeptide 1 |
| 4 | gene | 115.1338 | 115.1379 | positive | MGI_C57BL6J_3649427 | Cyp4x1os | NCBI_Gene:105244662,ENSEMBL:ENSMUSG00000086896 | MGI:3649427 | antisense lncRNA gene | cytochrome P450, family 4, subfamily x, polypeptide 1, opposite strand |
| 4 | pseudogene | 115.1386 | 115.1565 | negative | MGI_C57BL6J_1932405 | Cyp4a28-ps | NCBI_Gene:547220,ENSEMBL:ENSMUSG00000081823 | MGI:1932405 | pseudogene | cytochrome P450, family 4, subfamily a, polypeptide 28, pseudogene |
| 4 | pseudogene | 115.1758 | 115.1768 | positive | MGI_C57BL6J_3651920 | Cyp4a29-ps1 | ENSEMBL:ENSMUSG00000082889 | MGI:3651920 | pseudogene | cytochrome P450, family 4, subfamily a, polypeptide 29, pseudogene 1 |
| 4 | pseudogene | 115.2388 | 115.2416 | negative | MGI_C57BL6J_3649428 | Gm12834 | NCBI_Gene:384042,ENSEMBL:ENSMUSG00000081138 | MGI:3649428 | pseudogene | predicted gene 12834 |
| 4 | gene | 115.2421 | 115.2546 | positive | MGI_C57BL6J_3717143 | Cyp4a29 | NCBI_Gene:230639,ENSEMBL:ENSMUSG00000083138 | MGI:3717143 | protein coding gene | cytochrome P450, family 4, subfamily a, polypeptide 29 |
| 4 | gene | 115.2990 | 115.3328 | positive | MGI_C57BL6J_88612 | Cyp4a12a | NCBI_Gene:277753,ENSEMBL:ENSMUSG00000066071 | MGI:88612 | protein coding gene | cytochrome P450, family 4, subfamily a, polypeptide 12a |
| 4 | pseudogene | 115.3438 | 115.3631 | positive | MGI_C57BL6J_3649426 | Cyp4a30-ps | NCBI_Gene:546842,ENSEMBL:ENSMUSG00000083190 | MGI:3649426 | pseudogene | cytochrome P450, family 4, subfamily a, member 30, pseudogene |
| 4 | gene | 115.4116 | 115.4390 | positive | MGI_C57BL6J_3611747 | Cyp4a12b | NCBI_Gene:13118,ENSEMBL:ENSMUSG00000078597 | MGI:3611747 | protein coding gene | cytochrome P450, family 4, subfamily a, polypeptide 12B |
| 4 | pseudogene | 115.4161 | 115.4165 | negative | MGI_C57BL6J_3649425 | Gm12837 | ENSEMBL:ENSMUSG00000084415 | MGI:3649425 | pseudogene | predicted gene 12837 |
| 4 | gene | 115.4525 | 115.4714 | positive | MGI_C57BL6J_3717145 | Cyp4a30b | NCBI_Gene:435802,ENSEMBL:ENSMUSG00000084346 | MGI:3717145 | protein coding gene | cytochrome P450, family 4, subfamily a, polypeptide 30b |
| 4 | pseudogene | 115.4538 | 115.4541 | negative | MGI_C57BL6J_3649431 | Gm12838 | ENSEMBL:ENSMUSG00000080142 | MGI:3649431 | pseudogene | predicted gene 12838 |
| 4 | pseudogene | 115.4795 | 115.4813 | positive | MGI_C57BL6J_3651919 | Gm12833 | NCBI_Gene:230641,ENSEMBL:ENSMUSG00000086724 | MGI:3651919 | pseudogene | predicted gene 12833 |
| 4 | gene | 115.4862 | 115.4962 | negative | MGI_C57BL6J_1096550 | Cyp4a14 | NCBI_Gene:13119,ENSEMBL:ENSMUSG00000028715 | MGI:1096550 | protein coding gene | cytochrome P450, family 4, subfamily a, polypeptide 14 |
| 4 | gene | 115.5183 | 115.5336 | positive | MGI_C57BL6J_88611 | Cyp4a10 | NCBI_Gene:13117,ENSEMBL:ENSMUSG00000066072 | MGI:88611 | protein coding gene | cytochrome P450, family 4, subfamily a, polypeptide 10 |
| 4 | pseudogene | 115.5434 | 115.5451 | negative | MGI_C57BL6J_3651921 | Cyp4b1-ps1 | NCBI_Gene:100418216,ENSEMBL:ENSMUSG00000081006 | MGI:3651921 | pseudogene | cytochrome P450, family 4, subfamily b, polypeptide 1, pseudogene 1 |
| 4 | gene | 115.5636 | 115.5790 | positive | MGI_C57BL6J_3028580 | Cyp4a31 | NCBI_Gene:666168,ENSEMBL:ENSMUSG00000028712 | MGI:3028580 | protein coding gene | cytochrome P450, family 4, subfamily a, polypeptide 31 |
| 4 | pseudogene | 115.5820 | 115.5831 | negative | MGI_C57BL6J_3649236 | Cyp4b1-ps2 | NCBI_Gene:631037,ENSEMBL:ENSMUSG00000083457 | MGI:3649236 | pseudogene | cytochrome P450, family 4, subfamily b, polypeptide 1, pseudogene 2 |
| 4 | gene | 115.6009 | 115.6225 | positive | MGI_C57BL6J_3717148 | Cyp4a32 | NCBI_Gene:100040843,ENSEMBL:ENSMUSG00000063929 | MGI:3717148 | protein coding gene | cytochrome P450, family 4, subfamily a, polypeptide 32 |
| 4 | gene | 115.6247 | 115.6477 | negative | MGI_C57BL6J_103225 | Cyp4b1 | NCBI_Gene:13120,ENSEMBL:ENSMUSG00000028713 | MGI:103225 | protein coding gene | cytochrome P450, family 4, subfamily b, polypeptide 1 |
| 4 | pseudogene | 115.6524 | 115.6535 | positive | MGI_C57BL6J_3650820 | Gm12848 | NCBI_Gene:546843,ENSEMBL:ENSMUSG00000082043 | MGI:3650820 | pseudogene | predicted gene 12848 |
| 4 | pseudogene | 115.7073 | 115.7083 | positive | MGI_C57BL6J_3650623 | Gm12849 | ENSEMBL:ENSMUSG00000081172 | MGI:3650623 | pseudogene | predicted gene 12849 |
| 4 | gene | 115.7377 | 115.7773 | positive | MGI_C57BL6J_2442397 | Efcab14 | NCBI_Gene:230648,ENSEMBL:ENSMUSG00000034210 | MGI:2442397 | protein coding gene | EF-hand calcium binding domain 14 |
| 4 | gene | 115.7798 | 115.7827 | negative | MGI_C57BL6J_1922423 | Tex38 | NCBI_Gene:75173,ENSEMBL:ENSMUSG00000044556 | MGI:1922423 | protein coding gene | testis expressed 38 |
| 4 | gene | 115.7848 | 115.8188 | positive | MGI_C57BL6J_2180560 | Atpaf1 | NCBI_Gene:230649,ENSEMBL:ENSMUSG00000028710 | MGI:2180560 | protein coding gene | ATP synthase mitochondrial F1 complex assembly factor 1 |
| 4 | gene | 115.8189 | 115.8227 | positive | MGI_C57BL6J_3650822 | Gm12847 | ENSEMBL:ENSMUSG00000085249 | MGI:3650822 | lncRNA gene | predicted gene 12847 |
| 4 | gene | 115.8279 | 115.8362 | positive | MGI_C57BL6J_2140623 | Mob3c | NCBI_Gene:100465,ENSEMBL:ENSMUSG00000028709 | MGI:2140623 | protein coding gene | MOB kinase activator 3C |
| 4 | gene | 115.8390 | 115.8793 | positive | MGI_C57BL6J_894316 | Mknk1 | NCBI_Gene:17346,ENSEMBL:ENSMUSG00000028708 | MGI:894316 | protein coding gene | MAP kinase-interacting serine/threonine kinase 1 |
| 4 | gene | 115.8783 | 115.8786 | negative | MGI_C57BL6J_5580140 | Gm29434 | ENSEMBL:ENSMUSG00000100081 | MGI:5580140 | lncRNA gene | predicted gene 29434 |
| 4 | gene | 115.8844 | 115.8895 | positive | MGI_C57BL6J_3614952 | Kncn | NCBI_Gene:654462,ENSEMBL:ENSMUSG00000073774 | MGI:3614952 | protein coding gene | kinocilin |
| 4 | gene | 115.8907 | 115.8992 | positive | MGI_C57BL6J_1923487 | 6430628N08Rik | NCBI_Gene:76237,ENSEMBL:ENSMUSG00000034185 | MGI:1923487 | protein coding gene | RIKEN cDNA 6430628N08 gene |
| 4 | gene | 115.9151 | 115.9406 | negative | MGI_C57BL6J_2153518 | Dmbx1 | NCBI_Gene:140477,ENSEMBL:ENSMUSG00000028707 | MGI:2153518 | protein coding gene | diencephalon/mesencephalon homeobox 1 |
| 4 | gene | 115.9671 | 115.9709 | negative | MGI_C57BL6J_5621377 | Gm38492 | NCBI_Gene:102637048 | MGI:5621377 | lncRNA gene | predicted gene, 38492 |
| 4 | gene | 115.9671 | 116.0187 | negative | MGI_C57BL6J_109609 | Faah | NCBI_Gene:14073,ENSEMBL:ENSMUSG00000034171 | MGI:109609 | protein coding gene | fatty acid amide hydrolase |
| 4 | gene | 116.0318 | 116.0539 | negative | MGI_C57BL6J_1919431 | Nsun4 | NCBI_Gene:72181,ENSEMBL:ENSMUSG00000028706 | MGI:1919431 | protein coding gene | NOL1/NOP2/Sun domain family, member 4 |
| 4 | gene | 116.0670 | 116.0751 | negative | MGI_C57BL6J_1913826 | Uqcrh | NCBI_Gene:66576,ENSEMBL:ENSMUSG00000063882 | MGI:1913826 | protein coding gene | ubiquinol-cytochrome c reductase hinge protein |
| 4 | pseudogene | 116.0672 | 116.0677 | positive | MGI_C57BL6J_3650410 | Gm12854 | NCBI_Gene:102637129,ENSEMBL:ENSMUSG00000078238 | MGI:3650410 | pseudogene | predicted gene 12854 |
| 4 | gene | 116.0753 | 116.0971 | positive | MGI_C57BL6J_2441984 | Lrrc41 | NCBI_Gene:230654,ENSEMBL:ENSMUSG00000028703 | MGI:2441984 | protein coding gene | leucine rich repeat containing 41 |
| 4 | gene | 116.0943 | 116.1237 | negative | MGI_C57BL6J_894697 | Rad54l | NCBI_Gene:19366,ENSEMBL:ENSMUSG00000028702 | MGI:894697 | protein coding gene | RAD54 like (S. cerevisiae) |
| 4 | gene | 116.1238 | 116.1388 | positive | MGI_C57BL6J_1924036 | 2510003B16Rik | NCBI_Gene:76786 | MGI:1924036 | lncRNA gene | RIKEN cDNA 2510003B16 gene |
| 4 | gene | 116.1238 | 116.1598 | positive | MGI_C57BL6J_1915523 | Pomgnt1 | NCBI_Gene:68273,ENSEMBL:ENSMUSG00000028700 | MGI:1915523 | protein coding gene | protein O-linked mannose beta 1,2-N-acetylglucosaminyltransferase |
| 4 | gene | 116.1324 | 116.1513 | negative | MGI_C57BL6J_1915325 | Lurap1 | NCBI_Gene:68075,ENSEMBL:ENSMUSG00000028701 | MGI:1915325 | protein coding gene | leucine rich adaptor protein 1 |
| 4 | gene | 116.1619 | 116.1676 | negative | MGI_C57BL6J_1914055 | Tspan1 | NCBI_Gene:66805,ENSEMBL:ENSMUSG00000028699 | MGI:1914055 | protein coding gene | tetraspanin 1 |
| 4 | gene | 116.1734 | 116.1743 | positive | MGI_C57BL6J_1914573 | 1700042G07Rik | NCBI_Gene:67323,ENSEMBL:ENSMUSG00000078593 | MGI:1914573 | protein coding gene | RIKEN cDNA 1700042G07 gene |
| 4 | gene | 116.1734 | 116.2998 | positive | MGI_C57BL6J_6121526 | Gm49337 | ENSEMBL:ENSMUSG00000111410 | MGI:6121526 | protein coding gene | predicted gene, 49337 |
| 4 | gene | 116.1762 | 116.1777 | positive | MGI_C57BL6J_3649505 | Gm12951 | ENSEMBL:ENSMUSG00000085920 | MGI:3649505 | lncRNA gene | predicted gene 12951 |
| 4 | pseudogene | 116.2004 | 116.2010 | negative | MGI_C57BL6J_3649506 | Llph-ps1 | NCBI_Gene:433748,ENSEMBL:ENSMUSG00000081459 | MGI:3649506 | pseudogene | LLP homolog, pseudogene 1 |
| 4 | gene | 116.2214 | 116.3031 | positive | MGI_C57BL6J_109277 | Pik3r3 | NCBI_Gene:18710,ENSEMBL:ENSMUSG00000028698 | MGI:109277 | protein coding gene | phosphoinositide-3-kinase regulatory subunit 3 |
| 4 | pseudogene | 116.2500 | 116.2511 | negative | MGI_C57BL6J_5010720 | Gm18535 | NCBI_Gene:100417330 | MGI:5010720 | pseudogene | predicted gene, 18535 |
| 4 | gene | 116.3068 | 116.4643 | negative | MGI_C57BL6J_894676 | Mast2 | NCBI_Gene:17776,ENSEMBL:ENSMUSG00000003810 | MGI:894676 | protein coding gene | microtubule associated serine/threonine kinase 2 |
| 4 | gene | 116.3823 | 116.3857 | negative | MGI_C57BL6J_2444793 | A630078A22Rik | NA | NA | unclassified gene | RIKEN cDNA A630078A22 gene |
| 4 | pseudogene | 116.4488 | 116.4492 | positive | MGI_C57BL6J_3649504 | Gm12950 | ENSEMBL:ENSMUSG00000082592 | MGI:3649504 | pseudogene | predicted gene 12950 |
| 4 | gene | 116.5075 | 116.5426 | positive | MGI_C57BL6J_96581 | Ipp | NCBI_Gene:16351,ENSEMBL:ENSMUSG00000028696 | MGI:96581 | protein coding gene | IAP promoted placental gene |
| 4 | gene | 116.5515 | 116.5570 | negative | MGI_C57BL6J_3045357 | Tmem69 | NCBI_Gene:230657,ENSEMBL:ENSMUSG00000055900 | MGI:3045357 | protein coding gene | transmembrane protein 69 |
| 4 | gene | 116.5517 | 116.5526 | positive | MGI_C57BL6J_3649497 | Gm12953 | ENSEMBL:ENSMUSG00000087299 | MGI:3649497 | lncRNA gene | predicted gene 12953 |
| 4 | gene | 116.5572 | 116.5939 | positive | MGI_C57BL6J_1924360 | Gpbp1l1 | NCBI_Gene:77110,ENSEMBL:ENSMUSG00000034042 | MGI:1924360 | protein coding gene | GC-rich promoter binding protein 1-like 1 |
| 4 | gene | 116.5897 | 116.5976 | negative | MGI_C57BL6J_3612454 | C530005A16Rik | NCBI_Gene:654318,ENSEMBL:ENSMUSG00000085408 | MGI:3612454 | lncRNA gene | RIKEN cDNA C530005A16 gene |
| 4 | gene | 116.5967 | 116.6003 | positive | MGI_C57BL6J_1915667 | Ccdc17 | NCBI_Gene:622665,ENSEMBL:ENSMUSG00000034035 | MGI:1915667 | protein coding gene | coiled-coil domain containing 17 |
| 4 | gene | 116.6011 | 116.6280 | negative | MGI_C57BL6J_1355328 | Nasp | NCBI_Gene:50927,ENSEMBL:ENSMUSG00000028693 | MGI:1355328 | protein coding gene | nuclear autoantigenic sperm protein (histone-binding) |
| 4 | gene | 116.6268 | 116.6287 | positive | MGI_C57BL6J_2441802 | A430091L06Rik | NA | NA | unclassified gene | RIKEN cDNA A430091L06 gene |
| 4 | gene | 116.6365 | 116.6517 | negative | MGI_C57BL6J_1929955 | Akr1a1 | NCBI_Gene:58810,ENSEMBL:ENSMUSG00000028692 | MGI:1929955 | protein coding gene | aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| 4 | gene | 116.6742 | 116.6743 | positive | MGI_C57BL6J_5454914 | Gm25137 | ENSEMBL:ENSMUSG00000065185 | MGI:5454914 | snRNA gene | predicted gene, 25137 |
| 4 | gene | 116.6855 | 116.7008 | positive | MGI_C57BL6J_99523 | Prdx1 | NCBI_Gene:18477,ENSEMBL:ENSMUSG00000028691 | MGI:99523 | protein coding gene | peroxiredoxin 1 |
| 4 | gene | 116.6971 | 116.6975 | positive | MGI_C57BL6J_1924686 | 9530001N24Rik | NA | NA | unclassified gene | RIKEN cDNA 9530001N24 gene |
| 4 | gene | 116.7023 | 116.7084 | negative | MGI_C57BL6J_1914346 | Mmachc | NCBI_Gene:67096,ENSEMBL:ENSMUSG00000028690 | MGI:1914346 | protein coding gene | methylmalonic aciduria cblC type, with homocystinuria |
| 4 | gene | 116.7081 | 116.7084 | positive | MGI_C57BL6J_1925287 | 4930565M07Rik | NA | NA | unclassified gene | RIKEN cDNA 4930565M07 gene |
| 4 | gene | 116.7085 | 116.7512 | positive | MGI_C57BL6J_1915644 | Ccdc163 | NCBI_Gene:68394,ENSEMBL:ENSMUSG00000028689 | MGI:1915644 | protein coding gene | coiled-coil domain containing 163 |
| 4 | gene | 116.7209 | 116.8060 | positive | MGI_C57BL6J_2385204 | Tesk2 | NCBI_Gene:230661,ENSEMBL:ENSMUSG00000033985 | MGI:2385204 | protein coding gene | testis-specific kinase 2 |
| 4 | gene | 116.7259 | 116.7260 | positive | MGI_C57BL6J_5531070 | Gm27688 | ENSEMBL:ENSMUSG00000098491 | MGI:5531070 | unclassified non-coding RNA gene | predicted gene, 27688 |
| 4 | gene | 116.7943 | 116.8076 | negative | MGI_C57BL6J_1915526 | Toe1 | NCBI_Gene:68276,ENSEMBL:ENSMUSG00000028688 | MGI:1915526 | protein coding gene | target of EGR1, member 1 (nuclear) |
| 4 | gene | 116.8077 | 116.8194 | positive | MGI_C57BL6J_1917853 | Mutyh | NCBI_Gene:70603,ENSEMBL:ENSMUSG00000028687 | MGI:1917853 | protein coding gene | mutY DNA glycosylase |
| 4 | gene | 116.8199 | 116.8217 | negative | MGI_C57BL6J_2444646 | Hpdl | NCBI_Gene:242642,ENSEMBL:ENSMUSG00000043155 | MGI:2444646 | protein coding gene | 4-hydroxyphenylpyruvate dioxygenase-like |
| 4 | gene | 116.8243 | 116.8306 | positive | MGI_C57BL6J_3650481 | Gm12996 | NCBI_Gene:100502973,ENSEMBL:ENSMUSG00000086417 | MGI:3650481 | lncRNA gene | predicted gene 12996 |
| 4 | pseudogene | 116.8399 | 116.8402 | negative | MGI_C57BL6J_3650480 | Rpl36-ps8 | NCBI_Gene:625336,ENSEMBL:ENSMUSG00000075390 | MGI:3650480 | pseudogene | ribosomal protein L36, pseudogene 8 |
| 4 | pseudogene | 116.8527 | 116.8531 | positive | MGI_C57BL6J_3650479 | Gm12994 | ENSEMBL:ENSMUSG00000082841 | MGI:3650479 | pseudogene | predicted gene 12994 |
| 4 | pseudogene | 116.8746 | 116.8755 | negative | MGI_C57BL6J_3650477 | Gm12993 | ENSEMBL:ENSMUSG00000082063 | MGI:3650477 | pseudogene | predicted gene 12993 |
| 4 | gene | 116.8774 | 116.9893 | positive | MGI_C57BL6J_1921714 | Zswim5 | NCBI_Gene:74464,ENSEMBL:ENSMUSG00000033948 | MGI:1921714 | protein coding gene | zinc finger SWIM-type containing 5 |
| 4 | gene | 116.9046 | 116.9047 | positive | MGI_C57BL6J_5456132 | Gm26355 | ENSEMBL:ENSMUSG00000088845 | MGI:5456132 | snRNA gene | predicted gene, 26355 |
| 4 | gene | 116.9152 | 116.9155 | positive | MGI_C57BL6J_5455212 | Gm25435 | ENSEMBL:ENSMUSG00000088902 | MGI:5455212 | unclassified non-coding RNA gene | predicted gene, 25435 |
| 4 | gene | 116.9224 | 116.9225 | negative | MGI_C57BL6J_5451869 | Gm22092 | ENSEMBL:ENSMUSG00000095805 | MGI:5451869 | snRNA gene | predicted gene, 22092 |
| 4 | pseudogene | 116.9457 | 116.9465 | negative | MGI_C57BL6J_3651168 | Gm12998 | NCBI_Gene:100417429,ENSEMBL:ENSMUSG00000082207 | MGI:3651168 | pseudogene | predicted gene 12998 |
| 4 | gene | 116.9900 | 116.9944 | negative | MGI_C57BL6J_98916 | Urod | NCBI_Gene:22275,ENSEMBL:ENSMUSG00000028684 | MGI:98916 | protein coding gene | uroporphyrinogen decarboxylase |
| 4 | gene | 116.9953 | 117.0053 | positive | MGI_C57BL6J_1923858 | Hectd3 | NCBI_Gene:76608,ENSEMBL:ENSMUSG00000046861 | MGI:1923858 | protein coding gene | HECT domain E3 ubiquitin protein ligase 3 |
| 4 | gene | 117.0022 | 117.0030 | negative | MGI_C57BL6J_1919485 | 1700021J08Rik | ENSEMBL:ENSMUSG00000086890 | MGI:1919485 | lncRNA gene | RIKEN cDNA 1700021J08 gene |
| 4 | pseudogene | 117.0151 | 117.0158 | negative | MGI_C57BL6J_3650026 | Gm12997 | NCBI_Gene:100416056,ENSEMBL:ENSMUSG00000082249 | MGI:3650026 | pseudogene | predicted gene 12997 |
| 4 | gene | 117.0194 | 117.0873 | positive | MGI_C57BL6J_1313286 | Eif2b3 | NCBI_Gene:108067,ENSEMBL:ENSMUSG00000028683 | MGI:1313286 | protein coding gene | eukaryotic translation initiation factor 2B, subunit 3 |
| 4 | gene | 117.0272 | 117.0273 | positive | MGI_C57BL6J_5452112 | Gm22335 | ENSEMBL:ENSMUSG00000088349 | MGI:5452112 | snRNA gene | predicted gene, 22335 |
| 4 | gene | 117.0958 | 117.1161 | positive | MGI_C57BL6J_1095405 | Ptch2 | NCBI_Gene:19207,ENSEMBL:ENSMUSG00000028681 | MGI:1095405 | protein coding gene | patched 2 |
| 4 | gene | 117.1192 | 117.1257 | negative | MGI_C57BL6J_1925861 | Btbd19 | NCBI_Gene:78611,ENSEMBL:ENSMUSG00000073771 | MGI:1925861 | protein coding gene | BTB (POZ) domain containing 19 |
| 4 | gene | 117.1245 | 117.1287 | positive | MGI_C57BL6J_3045358 | Tctex1d4 | NCBI_Gene:242646,ENSEMBL:ENSMUSG00000047671 | MGI:3045358 | protein coding gene | Tctex1 domain containing 4 |
| 4 | gene | 117.1287 | 117.1340 | negative | MGI_C57BL6J_109604 | Plk3 | NCBI_Gene:12795,ENSEMBL:ENSMUSG00000028680 | MGI:109604 | protein coding gene | polo like kinase 3 |
| 4 | pseudogene | 117.1453 | 117.1486 | positive | MGI_C57BL6J_3663793 | Best4-ps | NCBI_Gene:230664,ENSEMBL:ENSMUSG00000033872 | MGI:3663793 | pseudogene | bestrophin 4, pseudogene |
| 4 | pseudogene | 117.1489 | 117.1493 | positive | MGI_C57BL6J_3651767 | Gm13015 | NCBI_Gene:625405,ENSEMBL:ENSMUSG00000070834 | MGI:3651767 | pseudogene | predicted gene 13015 |
| 4 | gene | 117.1538 | 117.1562 | negative | MGI_C57BL6J_98166 | Rps8 | NCBI_Gene:20116,ENSEMBL:ENSMUSG00000047675 | MGI:98166 | protein coding gene | ribosomal protein S8 |
| 4 | gene | 117.1541 | 117.1542 | negative | MGI_C57BL6J_5452757 | Gm22980 | ENSEMBL:ENSMUSG00000064542 | MGI:5452757 | snoRNA gene | predicted gene, 22980 |
| 4 | gene | 117.1545 | 117.1546 | negative | MGI_C57BL6J_3819531 | Snord38a | NCBI_Gene:100217424,ENSEMBL:ENSMUSG00000065680 | MGI:3819531 | snoRNA gene | small nucleolar RNA, C/D box 38A |
| 4 | gene | 117.1552 | 117.1553 | negative | MGI_C57BL6J_5456107 | Gm26330 | ENSEMBL:ENSMUSG00000064751 | MGI:5456107 | snoRNA gene | predicted gene, 26330 |
| 4 | gene | 117.1558 | 117.1558 | negative | MGI_C57BL6J_3819543 | Snord55 | NCBI_Gene:100216533,ENSEMBL:ENSMUSG00000092680 | MGI:3819543 | snoRNA gene | small nucleolar RNA, C/D box 55 |
| 4 | gene | 117.1583 | 117.1584 | negative | MGI_C57BL6J_5452175 | Gm22398 | ENSEMBL:ENSMUSG00000095209 | MGI:5452175 | miRNA gene | predicted gene, 22398 |
| 4 | gene | 117.1596 | 117.1826 | negative | MGI_C57BL6J_1921054 | Kif2c | NCBI_Gene:73804,ENSEMBL:ENSMUSG00000028678 | MGI:1921054 | protein coding gene | kinesin family member 2C |
| 4 | gene | 117.1899 | 117.1901 | positive | MGI_C57BL6J_5454876 | Gm25099 | ENSEMBL:ENSMUSG00000095676 | MGI:5454876 | snRNA gene | predicted gene, 25099 |
| 4 | gene | 117.2025 | 117.2026 | positive | MGI_C57BL6J_5452920 | Gm23143 | ENSEMBL:ENSMUSG00000094405 | MGI:5452920 | snRNA gene | predicted gene, 23143 |
| 4 | gene | 117.2050 | 117.2054 | positive | MGI_C57BL6J_3629729 | Gt(pU21)107Imeg | NA | NA | unclassified gene | gene trap 107%2c Institute of Molecular Embryology and Genetics |
| 4 | gene | 117.2108 | 117.2110 | positive | MGI_C57BL6J_5453064 | Gm23287 | ENSEMBL:ENSMUSG00000096280 | MGI:5453064 | snRNA gene | predicted gene, 23287 |
| 4 | gene | 117.2133 | 117.2521 | negative | MGI_C57BL6J_2686507 | Armh1 | NCBI_Gene:381544,ENSEMBL:ENSMUSG00000060268 | MGI:2686507 | protein coding gene | armadillo-like helical domain containing 1 |
| 4 | gene | 117.2133 | 117.2138 | negative | MGI_C57BL6J_1923635 | 1700012C08Rik | ENSEMBL:ENSMUSG00000086482 | MGI:1923635 | lncRNA gene | RIKEN cDNA 1700012C08 gene |
| 4 | gene | 117.2520 | 117.2686 | positive | MGI_C57BL6J_1916027 | Tmem53 | NCBI_Gene:68777,ENSEMBL:ENSMUSG00000048772 | MGI:1916027 | protein coding gene | transmembrane protein 53 |
| 4 | gene | 117.2715 | 117.4971 | negative | MGI_C57BL6J_1913993 | Rnf220 | NCBI_Gene:66743,ENSEMBL:ENSMUSG00000028677 | MGI:1913993 | protein coding gene | ring finger protein 220 |
| 4 | gene | 117.2894 | 117.3054 | positive | MGI_C57BL6J_3651051 | Gm12843 | NCBI_Gene:102637763,ENSEMBL:ENSMUSG00000085749 | MGI:3651051 | lncRNA gene | predicted gene 12843 |
| 4 | gene | 117.3117 | 117.3194 | negative | MGI_C57BL6J_5623116 | Gm40231 | NCBI_Gene:105244664 | MGI:5623116 | lncRNA gene | predicted gene, 40231 |
| 4 | gene | 117.3281 | 117.3290 | negative | MGI_C57BL6J_1924582 | C030012D19Rik | NA | NA | unclassified gene | RIKEN cDNA C030012D19 gene |
| 4 | gene | 117.4417 | 117.4550 | positive | MGI_C57BL6J_3650145 | Gm12828 | NCBI_Gene:108168973,ENSEMBL:ENSMUSG00000086876 | MGI:3650145 | lncRNA gene | predicted gene 12828 |
| 4 | gene | 117.4959 | 117.5093 | positive | MGI_C57BL6J_3651358 | Gm12827 | NCBI_Gene:102637578,ENSEMBL:ENSMUSG00000087228 | MGI:3651358 | lncRNA gene | predicted gene 12827 |
| 4 | gene | 117.5094 | 117.5196 | positive | MGI_C57BL6J_5593828 | Gm34669 | NCBI_Gene:102637993 | MGI:5593828 | lncRNA gene | predicted gene, 34669 |
| 4 | pseudogene | 117.5138 | 117.5142 | negative | MGI_C57BL6J_3650150 | Gm12826 | ENSEMBL:ENSMUSG00000083446 | MGI:3650150 | pseudogene | predicted gene 12826 |
| 4 | gene | 117.5503 | 117.6743 | positive | MGI_C57BL6J_2153887 | Eri3 | NCBI_Gene:140546,ENSEMBL:ENSMUSG00000033423 | MGI:2153887 | protein coding gene | exoribonuclease 3 |
| 4 | gene | 117.5701 | 117.5702 | positive | MGI_C57BL6J_5455336 | Gm25559 | ENSEMBL:ENSMUSG00000087807 | MGI:5455336 | miRNA gene | predicted gene, 25559 |
| 4 | pseudogene | 117.5988 | 117.5994 | positive | MGI_C57BL6J_5011377 | Gm19192 | NCBI_Gene:100418411 | MGI:5011377 | pseudogene | predicted gene, 19192 |
| 4 | gene | 117.6747 | 117.6823 | negative | MGI_C57BL6J_1913483 | Dmap1 | NCBI_Gene:66233,ENSEMBL:ENSMUSG00000009640 | MGI:1913483 | protein coding gene | DNA methyltransferase 1-associated protein 1 |
| 4 | gene | 117.6991 | 117.7040 | positive | MGI_C57BL6J_5593905 | Gm34746 | NCBI_Gene:102638104 | MGI:5593905 | lncRNA gene | predicted gene, 34746 |
| 4 | gene | 117.7002 | 117.7009 | positive | MGI_C57BL6J_3650917 | Gm12840 | ENSEMBL:ENSMUSG00000086320 | MGI:3650917 | lncRNA gene | predicted gene 12840 |
| 4 | gene | 117.7244 | 117.7269 | positive | MGI_C57BL6J_3651666 | Gm12845 | NCBI_Gene:100416740,ENSEMBL:ENSMUSG00000108127 | MGI:3651666 | protein coding gene | predicted gene 12845 |
| 4 | pseudogene | 117.7281 | 117.7296 | negative | MGI_C57BL6J_3651053 | Gm12844 | NCBI_Gene:433749,ENSEMBL:ENSMUSG00000084260 | MGI:3651053 | pseudogene | predicted gene 12844 |
| 4 | gene | 117.7567 | 117.7657 | negative | MGI_C57BL6J_2181068 | Klf17 | NCBI_Gene:75753,ENSEMBL:ENSMUSG00000048626 | MGI:2181068 | protein coding gene | Kruppel-like factor 17 |
| 4 | gene | 117.7817 | 117.7871 | negative | MGI_C57BL6J_5623117 | Gm40232 | NCBI_Gene:105244666 | MGI:5623117 | lncRNA gene | predicted gene, 40232 |
| 4 | pseudogene | 117.8250 | 117.8257 | negative | MGI_C57BL6J_3651050 | Gm12842 | NCBI_Gene:606540,ENSEMBL:ENSMUSG00000083507 | MGI:3651050 | pseudogene | predicted gene 12842 |
| 4 | gene | 117.8345 | 117.8752 | positive | MGI_C57BL6J_95760 | Slc6a9 | NCBI_Gene:14664,ENSEMBL:ENSMUSG00000028542 | MGI:95760 | protein coding gene | solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| 4 | gene | 117.8353 | 117.8686 | negative | MGI_C57BL6J_4937941 | Gm17114 | ENSEMBL:ENSMUSG00000091237 | MGI:4937941 | lncRNA gene | predicted gene 17114 |
| 4 | gene | 117.8665 | 117.8727 | negative | MGI_C57BL6J_2685874 | Ccdc24 | NCBI_Gene:381546,ENSEMBL:ENSMUSG00000078588 | MGI:2685874 | protein coding gene | coiled-coil domain containing 24 |
| 4 | gene | 117.8693 | 117.8835 | negative | MGI_C57BL6J_1858493 | B4galt2 | NCBI_Gene:53418,ENSEMBL:ENSMUSG00000028541 | MGI:1858493 | protein coding gene | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| 4 | gene | 117.8745 | 117.8793 | positive | MGI_C57BL6J_5623118 | Gm40233 | NCBI_Gene:105244667 | MGI:5623118 | lncRNA gene | predicted gene, 40233 |
| 4 | gene | 117.8836 | 117.8862 | positive | MGI_C57BL6J_3651048 | Gm12841 | NCBI_Gene:108168974,ENSEMBL:ENSMUSG00000085278 | MGI:3651048 | lncRNA gene | predicted gene 12841 |
| 4 | gene | 117.8843 | 117.8873 | negative | MGI_C57BL6J_1890510 | Atp6v0b | NCBI_Gene:114143,ENSEMBL:ENSMUSG00000033379 | MGI:1890510 | protein coding gene | ATPase, H+ transporting, lysosomal V0 subunit B |
| 4 | gene | 117.8886 | 117.8920 | negative | MGI_C57BL6J_1914978 | Dph2 | NCBI_Gene:67728,ENSEMBL:ENSMUSG00000028540 | MGI:1914978 | protein coding gene | DPH2 homolog |
| 4 | gene | 117.8945 | 117.9152 | negative | MGI_C57BL6J_2385205 | Ipo13 | NCBI_Gene:230673,ENSEMBL:ENSMUSG00000033365 | MGI:2385205 | protein coding gene | importin 13 |
| 4 | gene | 117.9259 | 117.9308 | negative | MGI_C57BL6J_1333791 | Artn | NCBI_Gene:11876,ENSEMBL:ENSMUSG00000028539 | MGI:1333791 | protein coding gene | artemin |
| 4 | gene | 117.9322 | 118.1349 | negative | MGI_C57BL6J_1316659 | St3gal3 | NCBI_Gene:20441,ENSEMBL:ENSMUSG00000028538 | MGI:1316659 | protein coding gene | ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
| 4 | gene | 117.9724 | 117.9953 | positive | MGI_C57BL6J_1925865 | 9530034E10Rik | NCBI_Gene:78615,ENSEMBL:ENSMUSG00000086554 | MGI:1925865 | lncRNA gene | RIKEN cDNA 9530034E10 gene |
| 4 | gene | 118.1370 | 118.1800 | negative | MGI_C57BL6J_2446210 | Kdm4a | NCBI_Gene:230674,ENSEMBL:ENSMUSG00000033326 | MGI:2446210 | protein coding gene | lysine (K)-specific demethylase 4A |
| 4 | gene | 118.1641 | 118.1642 | positive | MGI_C57BL6J_5453159 | Gm23382 | ENSEMBL:ENSMUSG00000088303 | MGI:5453159 | snoRNA gene | predicted gene, 23382 |
| 4 | gene | 118.2082 | 118.2914 | negative | MGI_C57BL6J_102695 | Ptprf | NCBI_Gene:19268,ENSEMBL:ENSMUSG00000033295 | MGI:102695 | protein coding gene | protein tyrosine phosphatase, receptor type, F |
| 4 | gene | 118.2105 | 118.2106 | negative | MGI_C57BL6J_5531012 | Mir7226 | miRBase:MI0023721,NCBI_Gene:102466820,ENSEMBL:ENSMUSG00000098596 | MGI:5531012 | miRNA gene | microRNA 7226 |
| 4 | gene | 118.2523 | 118.2575 | negative | MGI_C57BL6J_5623120 | Gm40235 | NCBI_Gene:105244669 | MGI:5623120 | lncRNA gene | predicted gene, 40235 |
| 4 | pseudogene | 118.3600 | 118.3627 | positive | MGI_C57BL6J_1915430 | Hyi | NCBI_Gene:68180,ENSEMBL:ENSMUSG00000006395 | MGI:1915430 | polymorphic pseudogene | hydroxypyruvate isomerase (putative) |
| 4 | gene | 118.3627 | 118.4093 | negative | MGI_C57BL6J_3033336 | Szt2 | NCBI_Gene:230676,ENSEMBL:ENSMUSG00000033253 | MGI:3033336 | protein coding gene | SZT2 subunit of KICSTOR complex |
| 4 | gene | 118.4093 | 118.4158 | positive | MGI_C57BL6J_1915269 | Med8 | NCBI_Gene:80509,ENSEMBL:ENSMUSG00000006392 | MGI:1915269 | protein coding gene | mediator complex subunit 8 |
| 4 | gene | 118.4281 | 118.4330 | positive | MGI_C57BL6J_1858959 | Elovl1 | NCBI_Gene:54325,ENSEMBL:ENSMUSG00000006390 | MGI:1858959 | protein coding gene | elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
| 4 | gene | 118.4329 | 118.4374 | negative | MGI_C57BL6J_1859866 | Cdc20 | NCBI_Gene:107995,ENSEMBL:ENSMUSG00000006398 | MGI:1859866 | protein coding gene | cell division cycle 20 |
| 4 | pseudogene | 118.4410 | 118.4412 | negative | MGI_C57BL6J_3651259 | Gm12858 | ENSEMBL:ENSMUSG00000081921 | MGI:3651259 | pseudogene | predicted gene 12858 |
| 4 | gene | 118.4424 | 118.4575 | negative | MGI_C57BL6J_97076 | Mpl | NCBI_Gene:17480,ENSEMBL:ENSMUSG00000006389 | MGI:97076 | protein coding gene | myeloproliferative leukemia virus oncogene |
| 4 | gene | 118.4712 | 118.4901 | negative | MGI_C57BL6J_99906 | Tie1 | NCBI_Gene:21846,ENSEMBL:ENSMUSG00000033191 | MGI:99906 | protein coding gene | tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| 4 | pseudogene | 118.4907 | 118.4976 | negative | MGI_C57BL6J_3651649 | Gm12859 | NCBI_Gene:545675,ENSEMBL:ENSMUSG00000080857 | MGI:3651649 | pseudogene | predicted gene 12859 |
| 4 | pseudogene | 118.5220 | 118.5227 | positive | MGI_C57BL6J_3652230 | Gm12857 | NCBI_Gene:666596,ENSEMBL:ENSMUSG00000081233 | MGI:3652230 | pseudogene | predicted gene 12857 |
| 4 | gene | 118.5270 | 118.5302 | positive | MGI_C57BL6J_1913701 | 2610528J11Rik | NCBI_Gene:66451,ENSEMBL:ENSMUSG00000028536 | MGI:1913701 | protein coding gene | RIKEN cDNA 2610528J11 gene |
| 4 | gene | 118.5409 | 118.5450 | negative | MGI_C57BL6J_1923409 | Tmem125 | NCBI_Gene:230678,ENSEMBL:ENSMUSG00000050854 | MGI:1923409 | protein coding gene | transmembrane protein 125 |
| 4 | gene | 118.5471 | 118.5483 | positive | MGI_C57BL6J_5623122 | Gm40237 | NCBI_Gene:105244671 | MGI:5623122 | lncRNA gene | predicted gene, 40237 |
| 4 | gene | 118.5546 | 118.6208 | negative | MGI_C57BL6J_2686209 | Cfap57 | NCBI_Gene:68625,ENSEMBL:ENSMUSG00000028730 | MGI:2686209 | protein coding gene | cilia and flagella associated protein 57 |
| 4 | gene | 118.6208 | 118.6278 | positive | MGI_C57BL6J_1916322 | Ebna1bp2 | NCBI_Gene:69072,ENSEMBL:ENSMUSG00000028729 | MGI:1916322 | protein coding gene | EBNA1 binding protein 2 |
| 4 | gene | 118.6264 | 118.6319 | positive | MGI_C57BL6J_1277206 | D4Ertd617e | NCBI_Gene:100041290 | MGI:1277206 | lncRNA gene | DNA segment, Chr 4, ERATO Doi 617, expressed |
| 4 | gene | 118.6310 | 118.6378 | negative | MGI_C57BL6J_3650185 | Gm12853 | NCBI_Gene:102638941,ENSEMBL:ENSMUSG00000085292 | MGI:3650185 | lncRNA gene | predicted gene 12853 |
| 4 | gene | 118.6590 | 118.6680 | positive | MGI_C57BL6J_1333887 | Olfr62 | NCBI_Gene:18363,ENSEMBL:ENSMUSG00000043698 | MGI:1333887 | protein coding gene | olfactory receptor 62 |
| 4 | pseudogene | 118.6726 | 118.6735 | negative | MGI_C57BL6J_3031177 | Olfr1343-ps1 | NCBI_Gene:258707,ENSEMBL:ENSMUSG00000086521 | MGI:3031177 | pseudogene | olfactory receptor 1343, pseudogene 1 |
| 4 | gene | 118.6875 | 118.6930 | negative | MGI_C57BL6J_3031176 | Olfr1342 | NCBI_Gene:258708,ENSEMBL:ENSMUSG00000043383 | MGI:3031176 | protein coding gene | olfactory receptor 1342 |
| 4 | gene | 118.7035 | 118.7134 | positive | MGI_C57BL6J_3031175 | Olfr1341 | NCBI_Gene:258852,ENSEMBL:ENSMUSG00000046790 | MGI:3031175 | protein coding gene | olfactory receptor 1341 |
| 4 | gene | 118.7201 | 118.7294 | positive | MGI_C57BL6J_3031174 | Olfr1340 | NCBI_Gene:258301,ENSEMBL:ENSMUSG00000070821 | MGI:3031174 | protein coding gene | olfactory receptor 1340 |
| 4 | pseudogene | 118.7228 | 118.7255 | negative | MGI_C57BL6J_5439393 | Gm21942 | NCBI_Gene:257848 | MGI:5439393 | pseudogene | predicted gene, 21942 |
| 4 | gene | 118.7322 | 118.7369 | positive | MGI_C57BL6J_3031173 | Olfr1339 | NCBI_Gene:258851,ENSEMBL:ENSMUSG00000070820 | MGI:3031173 | protein coding gene | olfactory receptor 1339 |
| 4 | gene | 118.7520 | 118.7587 | negative | MGI_C57BL6J_3031172 | Olfr1338 | NCBI_Gene:258259,ENSEMBL:ENSMUSG00000095218 | MGI:3031172 | protein coding gene | olfactory receptor 1338 |
| 4 | gene | 118.7816 | 118.7826 | negative | MGI_C57BL6J_3031171 | Olfr1337 | NCBI_Gene:258306,ENSEMBL:ENSMUSG00000111159 | MGI:3031171 | protein coding gene | olfactory receptor 1337 |
| 4 | pseudogene | 118.7910 | 118.7919 | negative | MGI_C57BL6J_3649662 | Gm12855 | NCBI_Gene:257847,ENSEMBL:ENSMUSG00000081474 | MGI:3649662 | pseudogene | predicted gene 12855 |
| 4 | gene | 118.8088 | 118.8099 | negative | MGI_C57BL6J_3031168 | Olfr1335 | NCBI_Gene:435804,ENSEMBL:ENSMUSG00000066061 | MGI:3031168 | protein coding gene | olfactory receptor 1335 |
| 4 | gene | 118.8277 | 118.8363 | negative | MGI_C57BL6J_3031167 | Olfr1333 | NCBI_Gene:258265,ENSEMBL:ENSMUSG00000110947 | MGI:3031167 | protein coding gene | olfactory receptor 1333 |
| 4 | pseudogene | 118.8541 | 118.8550 | positive | MGI_C57BL6J_3031166 | Olfr1332-ps1 | NCBI_Gene:257952,ENSEMBL:ENSMUSG00000083336 | MGI:3031166 | pseudogene | olfactory receptor 1332, pseudogene 1 |
| 4 | gene | 118.8646 | 118.8717 | positive | MGI_C57BL6J_3031165 | Olfr1331 | NCBI_Gene:258159,ENSEMBL:ENSMUSG00000073769 | MGI:3031165 | protein coding gene | olfactory receptor 1331 |
| 4 | gene | 118.8886 | 118.8996 | positive | MGI_C57BL6J_3031164 | Olfr1330 | NCBI_Gene:258331,ENSEMBL:ENSMUSG00000073768 | MGI:3031164 | protein coding gene | olfactory receptor 1330 |
| 4 | gene | 118.8980 | 118.9974 | negative | MGI_C57BL6J_3651214 | Gm12865 | NCBI_Gene:105244672,ENSEMBL:ENSMUSG00000087238 | MGI:3651214 | lncRNA gene | predicted gene 12865 |
| 4 | gene | 118.9165 | 118.9175 | negative | MGI_C57BL6J_3031163 | Olfr1329 | NCBI_Gene:258214,ENSEMBL:ENSMUSG00000096705 | MGI:3031163 | protein coding gene | olfactory receptor 1329 |
| 4 | gene | 118.9301 | 118.9386 | negative | MGI_C57BL6J_3031162 | Olfr1328 | NCBI_Gene:258394,ENSEMBL:ENSMUSG00000111259 | MGI:3031162 | protein coding gene | olfactory receptor 1328 |
| 4 | pseudogene | 118.9421 | 118.9434 | negative | MGI_C57BL6J_3649664 | Gm12856 | NCBI_Gene:100415963,ENSEMBL:ENSMUSG00000084133 | MGI:3649664 | pseudogene | predicted gene 12856 |
| 4 | gene | 118.9543 | 118.9549 | negative | MGI_C57BL6J_3651212 | Gm12863 | NCBI_Gene:100038571,ENSEMBL:ENSMUSG00000073765 | MGI:3651212 | lncRNA gene | predicted gene 12863 |
| 4 | gene | 118.9616 | 118.9689 | positive | MGI_C57BL6J_2140628 | Lao1 | NCBI_Gene:100470,ENSEMBL:ENSMUSG00000024903 | MGI:2140628 | protein coding gene | L-amino acid oxidase 1 |
| 4 | gene | 118.9713 | 118.9995 | positive | MGI_C57BL6J_3651891 | Gm12861 | ENSEMBL:ENSMUSG00000085895 | MGI:3651891 | lncRNA gene | predicted gene 12861 |
| 4 | gene | 118.9803 | 118.9804 | positive | MGI_C57BL6J_5454129 | Gm24352 | ENSEMBL:ENSMUSG00000089591 | MGI:5454129 | snoRNA gene | predicted gene, 24352 |
| 4 | pseudogene | 118.9870 | 118.9882 | positive | MGI_C57BL6J_3649273 | Gm12864 | NCBI_Gene:666703,ENSEMBL:ENSMUSG00000082143 | MGI:3649273 | pseudogene | predicted gene 12864 |
| 4 | gene | 118.9997 | 119.0005 | negative | MGI_C57BL6J_3651209 | Gm12862 | ENSEMBL:ENSMUSG00000087464 | MGI:3651209 | lncRNA gene | predicted gene 12862 |
| 4 | gene | 119.0507 | 119.0620 | positive | MGI_C57BL6J_5428673 | Lincred2 | NCBI_Gene:100996932 | MGI:5428673 | lncRNA gene | long intergenic non-protein coding RNA of erythroid differentiation 2 |
| 4 | gene | 119.0548 | 119.0691 | negative | MGI_C57BL6J_3649462 | Gm12866 | NCBI_Gene:433751,ENSEMBL:ENSMUSG00000066060 | MGI:3649462 | lncRNA gene | predicted gene 12866 |
| 4 | gene | 119.0730 | 119.0844 | positive | MGI_C57BL6J_5594669 | Gm35510 | NCBI_Gene:102639122 | MGI:5594669 | lncRNA gene | predicted gene, 35510 |
| 4 | gene | 119.1087 | 119.1380 | positive | MGI_C57BL6J_95755 | Slc2a1 | NCBI_Gene:20525,ENSEMBL:ENSMUSG00000028645 | MGI:95755 | protein coding gene | solute carrier family 2 (facilitated glucose transporter), member 1 |
| 4 | gene | 119.1298 | 119.1298 | positive | MGI_C57BL6J_3837121 | Mir1957a | miRBase:MI0009954,ENSEMBL:ENSMUSG00000088552 | MGI:3837121 | miRNA gene | microRNA 1957a |
| 4 | gene | 119.1303 | 119.1310 | negative | MGI_C57BL6J_3651003 | Gm12867 | ENSEMBL:ENSMUSG00000086683 | MGI:3651003 | lncRNA gene | predicted gene 12867 |
| 4 | gene | 119.1337 | 119.1388 | negative | MGI_C57BL6J_3651004 | Gm12868 | ENSEMBL:ENSMUSG00000085505 | MGI:3651004 | lncRNA gene | predicted gene 12868 |
| 4 | gene | 119.1695 | 119.1742 | negative | MGI_C57BL6J_3041163 | Zfp691 | NCBI_Gene:195522,ENSEMBL:ENSMUSG00000045268 | MGI:3041163 | protein coding gene | zinc finger protein 691 |
| 4 | gene | 119.1755 | 119.1900 | negative | MGI_C57BL6J_1349816 | Ermap | NCBI_Gene:27028,ENSEMBL:ENSMUSG00000028644 | MGI:1349816 | protein coding gene | erythroblast membrane-associated protein |
| 4 | gene | 119.1953 | 119.2013 | positive | MGI_C57BL6J_1916466 | Svbp | NCBI_Gene:69216,ENSEMBL:ENSMUSG00000028643 | MGI:1916466 | protein coding gene | small vasohibin binding protein |
| 4 | gene | 119.2036 | 119.2328 | negative | MGI_C57BL6J_2140466 | AU022252 | NCBI_Gene:230696,ENSEMBL:ENSMUSG00000078584 | MGI:2140466 | protein coding gene | expressed sequence AU022252 |
| 4 | gene | 119.2050 | 119.2186 | negative | MGI_C57BL6J_1922430 | Tmem269 | NCBI_Gene:75180,ENSEMBL:ENSMUSG00000028642 | MGI:1922430 | protein coding gene | transmembrane protein 269 |
| 4 | gene | 119.2232 | 119.2248 | positive | MGI_C57BL6J_3650325 | Gm12898 | ENSEMBL:ENSMUSG00000085626 | MGI:3650325 | lncRNA gene | predicted gene 12898 |
| 4 | gene | 119.2329 | 119.2490 | positive | MGI_C57BL6J_1888921 | P3h1 | NCBI_Gene:56401,ENSEMBL:ENSMUSG00000028641 | MGI:1888921 | protein coding gene | prolyl 3-hydroxylase 1 |
| 4 | gene | 119.2525 | 119.2570 | negative | MGI_C57BL6J_3651776 | Gm12927 | NCBI_Gene:102639193,ENSEMBL:ENSMUSG00000085203 | MGI:3651776 | lncRNA gene | predicted gene 12927 |
| 4 | gene | 119.2554 | 119.2624 | positive | MGI_C57BL6J_3033992 | Cldn19 | NCBI_Gene:242653,ENSEMBL:ENSMUSG00000066058 | MGI:3033992 | protein coding gene | claudin 19 |
| 4 | gene | 119.2773 | 119.2947 | negative | MGI_C57BL6J_99146 | Ybx1 | NCBI_Gene:22608,ENSEMBL:ENSMUSG00000028639 | MGI:99146 | protein coding gene | Y box protein 1 |
| 4 | gene | 119.2941 | 119.2951 | positive | MGI_C57BL6J_1920791 | 1700102N10Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 1700102N10 gene |
| 4 | gene | 119.2950 | 119.2955 | negative | MGI_C57BL6J_5611045 | Gm37817 | ENSEMBL:ENSMUSG00000102819 | MGI:5611045 | unclassified gene | predicted gene, 37817 |
| 4 | gene | 119.3000 | 119.3205 | negative | MGI_C57BL6J_106499 | Ppih | NCBI_Gene:66101,ENSEMBL:ENSMUSG00000060288 | MGI:106499 | protein coding gene | peptidyl prolyl isomerase H |
| 4 | gene | 119.3225 | 119.4155 | negative | MGI_C57BL6J_1920582 | Ccdc30 | NCBI_Gene:73332,ENSEMBL:ENSMUSG00000028637 | MGI:1920582 | protein coding gene | coiled-coil domain containing 30 |
| 4 | pseudogene | 119.3339 | 119.3355 | negative | MGI_C57BL6J_96761 | Ldha-ps2 | NCBI_Gene:16830,ENSEMBL:ENSMUSG00000083836 | MGI:96761 | pseudogene | lactate dehydrogenase A, pseudogene 2 |
| 4 | gene | 119.3935 | 119.3999 | positive | MGI_C57BL6J_6324757 | Gm50484 | ENSEMBL:ENSMUSG00000118403 | MGI:6324757 | lncRNA gene | predicted gene, 50484 |
| 4 | gene | 119.4185 | 119.4224 | negative | MGI_C57BL6J_1915237 | Ppcs | NCBI_Gene:106564,ENSEMBL:ENSMUSG00000028636 | MGI:1915237 | protein coding gene | phosphopantothenoylcysteine synthetase |
| 4 | gene | 119.4227 | 119.4674 | positive | MGI_C57BL6J_2140259 | Zmynd12 | NCBI_Gene:332934,ENSEMBL:ENSMUSG00000070806 | MGI:2140259 | protein coding gene | zinc finger, MYND domain containing 12 |
| 4 | gene | 119.4653 | 119.4926 | negative | MGI_C57BL6J_3040686 | Rimkla | NCBI_Gene:194237,ENSEMBL:ENSMUSG00000048899 | MGI:3040686 | protein coding gene | ribosomal modification protein rimK-like family member A |
| 4 | pseudogene | 119.5019 | 119.5025 | negative | MGI_C57BL6J_3652229 | Gm12954 | ENSEMBL:ENSMUSG00000082009 | MGI:3652229 | pseudogene | predicted gene 12954 |
| 4 | pseudogene | 119.5096 | 119.5177 | negative | MGI_C57BL6J_3652227 | Gm12956 | NCBI_Gene:102640085,ENSEMBL:ENSMUSG00000082814 | MGI:3652227 | pseudogene | predicted gene 12956 |
| 4 | pseudogene | 119.5191 | 119.5201 | negative | MGI_C57BL6J_3652228 | Gm12955 | NCBI_Gene:666766,ENSEMBL:ENSMUSG00000080773 | MGI:3652228 | pseudogene | predicted gene 12955 |
| 4 | pseudogene | 119.5209 | 119.5215 | positive | MGI_C57BL6J_3649304 | Gm12957 | NCBI_Gene:384052,ENSEMBL:ENSMUSG00000084379 | MGI:3649304 | pseudogene | predicted gene 12957 |
| 4 | pseudogene | 119.5279 | 119.5282 | positive | MGI_C57BL6J_97991 | Rnu6-ps2 | NCBI_Gene:19864,ENSEMBL:ENSMUSG00000094494 | MGI:97991 | pseudogene | U6 small nuclear RNA, pseudogene 2 |
| 4 | gene | 119.5303 | 119.5395 | negative | MGI_C57BL6J_3035485 | Frg2f1 | NCBI_Gene:433752,ENSEMBL:ENSMUSG00000087385 | MGI:3035485 | protein coding gene | FSHD region gene 2 family member 1 |
| 4 | gene | 119.5370 | 119.6291 | positive | MGI_C57BL6J_2443432 | Foxj3 | NCBI_Gene:230700,ENSEMBL:ENSMUSG00000032998 | MGI:2443432 | protein coding gene | forkhead box J3 |
| 4 | pseudogene | 119.5537 | 119.5541 | negative | MGI_C57BL6J_3649444 | Gm12959 | ENSEMBL:ENSMUSG00000081678 | MGI:3649444 | pseudogene | predicted gene 12959 |
| 4 | gene | 119.6377 | 119.6395 | positive | MGI_C57BL6J_102738 | Guca2a | NCBI_Gene:14915,ENSEMBL:ENSMUSG00000023247 | MGI:102738 | protein coding gene | guanylate cyclase activator 2a (guanylin) |
| 4 | gene | 119.6566 | 119.6590 | negative | MGI_C57BL6J_1270851 | Guca2b | NCBI_Gene:14916,ENSEMBL:ENSMUSG00000032978 | MGI:1270851 | protein coding gene | guanylate cyclase activator 2b (retina) |
| 4 | gene | 119.6590 | 119.6638 | positive | MGI_C57BL6J_5594920 | Gm35761 | NCBI_Gene:102639449 | MGI:5594920 | lncRNA gene | predicted gene, 35761 |
| 4 | gene | 119.6957 | 119.7241 | negative | MGI_C57BL6J_5595116 | Gm35957 | NCBI_Gene:102639707 | MGI:5595116 | lncRNA gene | predicted gene, 35957 |
| 4 | gene | 119.7338 | 120.1380 | positive | MGI_C57BL6J_106589 | Hivep3 | NCBI_Gene:16656,ENSEMBL:ENSMUSG00000028634 | MGI:106589 | protein coding gene | human immunodeficiency virus type I enhancer binding protein 3 |
| 4 | gene | 120.0046 | 120.0495 | negative | MGI_C57BL6J_5621517 | Gm38632 | NCBI_Gene:102642778 | MGI:5621517 | lncRNA gene | predicted gene, 38632 |
| 4 | gene | 120.0658 | 120.0747 | negative | MGI_C57BL6J_5595374 | Gm36215 | NCBI_Gene:102640050 | MGI:5595374 | lncRNA gene | predicted gene, 36215 |
| 4 | gene | 120.1542 | 120.2622 | negative | MGI_C57BL6J_5595293 | Gm36134 | NCBI_Gene:102639939 | MGI:5595293 | lncRNA gene | predicted gene, 36134 |
| 4 | gene | 120.1612 | 120.1674 | positive | MGI_C57BL6J_95284 | Edn2 | NCBI_Gene:13615,ENSEMBL:ENSMUSG00000028635 | MGI:95284 | protein coding gene | endothelin 2 |
| 4 | gene | 120.1762 | 120.1813 | positive | MGI_C57BL6J_5595232 | Gm36073 | NCBI_Gene:102639859 | MGI:5595232 | lncRNA gene | predicted gene, 36073 |
| 4 | gene | 120.1908 | 120.2003 | positive | MGI_C57BL6J_5595172 | Gm36013 | NCBI_Gene:102639782 | MGI:5595172 | lncRNA gene | predicted gene, 36013 |
| 4 | gene | 120.2671 | 120.2873 | negative | MGI_C57BL6J_2676586 | Foxo6 | NCBI_Gene:329934,ENSEMBL:ENSMUSG00000052135 | MGI:2676586 | protein coding gene | forkhead box O6 |
| 4 | gene | 120.2911 | 120.3039 | positive | MGI_C57BL6J_3028036 | Foxo6os | NCBI_Gene:402730,ENSEMBL:ENSMUSG00000084929 | MGI:3028036 | antisense lncRNA gene | forkhead box O6, opposite strand |
| 4 | gene | 120.3097 | 120.3129 | negative | MGI_C57BL6J_5595658 | Gm36499 | NCBI_Gene:102640438 | MGI:5595658 | lncRNA gene | predicted gene, 36499 |
| 4 | gene | 120.3191 | 120.3972 | negative | MGI_C57BL6J_5595733 | Gm36574 | NCBI_Gene:102640537 | MGI:5595733 | lncRNA gene | predicted gene, 36574 |
| 4 | gene | 120.4053 | 120.5302 | positive | MGI_C57BL6J_1352762 | Scmh1 | NCBI_Gene:29871,ENSEMBL:ENSMUSG00000000085 | MGI:1352762 | protein coding gene | sex comb on midleg homolog 1 |
| 4 | gene | 120.5321 | 120.5367 | positive | MGI_C57BL6J_3045330 | Slfnl1 | NCBI_Gene:194219,ENSEMBL:ENSMUSG00000047518 | MGI:3045330 | protein coding gene | schlafen like 1 |
| 4 | gene | 120.5399 | 120.5703 | negative | MGI_C57BL6J_1858304 | Ctps | NCBI_Gene:51797,ENSEMBL:ENSMUSG00000028633 | MGI:1858304 | protein coding gene | cytidine 5’-triphosphate synthase |
| 4 | gene | 120.5887 | 120.6097 | positive | MGI_C57BL6J_3648581 | Gm8439 | NCBI_Gene:667063,ENSEMBL:ENSMUSG00000091297 | MGI:3648581 | protein coding gene | predicted gene 8439 |
| 4 | gene | 120.6158 | 120.6227 | positive | MGI_C57BL6J_3651651 | Gm12860 | ENSEMBL:ENSMUSG00000086187 | MGI:3651651 | lncRNA gene | predicted gene 12860 |
| 4 | gene | 120.6442 | 120.6481 | negative | MGI_C57BL6J_5791415 | Gm45579 | ENSEMBL:ENSMUSG00000110389 | MGI:5791415 | lncRNA gene | predicted gene 45579 |
| 4 | gene | 120.6596 | 120.6633 | positive | MGI_C57BL6J_5589050 | Gm29891 | NCBI_Gene:102631594 | MGI:5589050 | lncRNA gene | predicted gene, 29891 |
| 4 | gene | 120.6666 | 120.6678 | positive | MGI_C57BL6J_1861694 | Cited4 | NCBI_Gene:56222,ENSEMBL:ENSMUSG00000070803 | MGI:1861694 | protein coding gene | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| 4 | gene | 120.6961 | 120.7486 | negative | MGI_C57BL6J_1926803 | Kcnq4 | NCBI_Gene:60613,ENSEMBL:ENSMUSG00000028631 | MGI:1926803 | protein coding gene | potassium voltage-gated channel, subfamily Q, member 4 |
| 4 | gene | 120.7574 | 120.8316 | negative | MGI_C57BL6J_107901 | Nfyc | NCBI_Gene:18046,ENSEMBL:ENSMUSG00000032897 | MGI:107901 | protein coding gene | nuclear transcription factor-Y gamma |
| 4 | gene | 120.7695 | 120.7696 | negative | MGI_C57BL6J_2676909 | Mir30c-1 | miRBase:MI0000547,NCBI_Gene:387227,ENSEMBL:ENSMUSG00000065490 | MGI:2676909 | miRNA gene | microRNA 30c-1 |
| 4 | gene | 120.7726 | 120.7727 | positive | MGI_C57BL6J_5562761 | Mir30f | miRBase:MI0021961,NCBI_Gene:102466650,ENSEMBL:ENSMUSG00000105585 | MGI:5562761 | miRNA gene | microRNA 30f |
| 4 | gene | 120.7726 | 120.7727 | negative | MGI_C57BL6J_3619329 | Mir30e | miRBase:MI0000259,NCBI_Gene:723836,ENSEMBL:ENSMUSG00000065409 | MGI:3619329 | miRNA gene | microRNA 30e |
| 4 | gene | 120.8375 | 120.8376 | positive | MGI_C57BL6J_5453713 | Gm23936 | ENSEMBL:ENSMUSG00000096876 | MGI:5453713 | miRNA gene | predicted gene, 23936 |
| 4 | gene | 120.8548 | 120.8966 | positive | MGI_C57BL6J_2443331 | Rims3 | NCBI_Gene:242662,ENSEMBL:ENSMUSG00000032890 | MGI:2443331 | protein coding gene | regulating synaptic membrane exocytosis 3 |
| 4 | pseudogene | 120.9011 | 120.9028 | positive | MGI_C57BL6J_3649219 | Gm12871 | NCBI_Gene:102631670,ENSEMBL:ENSMUSG00000082200 | MGI:3649219 | pseudogene | predicted gene 12871 |
| 4 | gene | 120.9035 | 120.9036 | positive | MGI_C57BL6J_5454455 | Gm24678 | ENSEMBL:ENSMUSG00000089381 | MGI:5454455 | snRNA gene | predicted gene, 24678 |
| 4 | gene | 120.9212 | 120.9250 | negative | MGI_C57BL6J_1920422 | Exo5 | NCBI_Gene:73172,ENSEMBL:ENSMUSG00000028629 | MGI:1920422 | protein coding gene | exonuclease 5 |
| 4 | gene | 120.9301 | 120.9534 | negative | MGI_C57BL6J_107794 | Zfp69 | NCBI_Gene:381549,ENSEMBL:ENSMUSG00000064141 | MGI:107794 | protein coding gene | zinc finger protein 69 |
| 4 | gene | 120.9683 | 121.0173 | negative | MGI_C57BL6J_1917030 | Smap2 | NCBI_Gene:69780,ENSEMBL:ENSMUSG00000032870 | MGI:1917030 | protein coding gene | small ArfGAP 2 |
| 4 | gene | 120.9733 | 120.9733 | negative | MGI_C57BL6J_5562762 | Mir7015 | miRBase:MI0022864,NCBI_Gene:102465614,ENSEMBL:ENSMUSG00000106436 | MGI:5562762 | miRNA gene | microRNA 7015 |
| 4 | gene | 121.0394 | 121.0553 | positive | MGI_C57BL6J_88466 | Col9a2 | NCBI_Gene:12840,ENSEMBL:ENSMUSG00000028626 | MGI:88466 | protein coding gene | collagen, type IX, alpha 2 |
| 4 | gene | 121.0592 | 121.0982 | negative | MGI_C57BL6J_1890508 | Zmpste24 | NCBI_Gene:230709,ENSEMBL:ENSMUSG00000043207 | MGI:1890508 | protein coding gene | zinc metallopeptidase, STE24 |
| 4 | gene | 121.0758 | 121.0759 | negative | MGI_C57BL6J_5455963 | Gm26186 | ENSEMBL:ENSMUSG00000088314 | MGI:5455963 | snoRNA gene | predicted gene, 26186 |
| 4 | gene | 121.0840 | 121.0841 | negative | MGI_C57BL6J_5453317 | Gm23540 | ENSEMBL:ENSMUSG00000088168 | MGI:5453317 | snRNA gene | predicted gene, 23540 |
| 4 | gene | 121.1057 | 121.1092 | negative | MGI_C57BL6J_1916719 | Tmco2 | NCBI_Gene:69469,ENSEMBL:ENSMUSG00000078577 | MGI:1916719 | protein coding gene | transmembrane and coiled-coil domains 2 |
| 4 | pseudogene | 121.1148 | 121.1153 | negative | MGI_C57BL6J_3651897 | Gm12882 | NCBI_Gene:100042174,ENSEMBL:ENSMUSG00000081423 | MGI:3651897 | pseudogene | predicted gene 12882 |
| 4 | pseudogene | 121.1209 | 121.1302 | negative | MGI_C57BL6J_3651892 | Gm12881 | NCBI_Gene:102640296,ENSEMBL:ENSMUSG00000082891 | MGI:3651892 | pseudogene | predicted gene 12881 |
| 4 | pseudogene | 121.1301 | 121.1313 | positive | MGI_C57BL6J_3649632 | Gm12879 | ENSEMBL:ENSMUSG00000081305 | MGI:3649632 | pseudogene | predicted gene 12879 |
| 4 | gene | 121.1347 | 121.1350 | positive | MGI_C57BL6J_5452759 | Gm22982 | ENSEMBL:ENSMUSG00000064547 | MGI:5452759 | unclassified non-coding RNA gene | predicted gene, 22982 |
| 4 | gene | 121.1454 | 121.2151 | negative | MGI_C57BL6J_1924705 | Rlf | NCBI_Gene:109263,ENSEMBL:ENSMUSG00000049878 | MGI:1924705 | protein coding gene | rearranged L-myc fusion sequence |
| 4 | pseudogene | 121.2271 | 121.2275 | negative | MGI_C57BL6J_3652134 | Gm12890 | ENSEMBL:ENSMUSG00000080706 | MGI:3652134 | pseudogene | predicted gene 12890 |
| 4 | gene | 121.2309 | 121.2392 | positive | MGI_C57BL6J_5589261 | Gm30102 | NCBI_Gene:102631880 | MGI:5589261 | lncRNA gene | predicted gene, 30102 |
| 4 | gene | 121.2357 | 121.2415 | negative | MGI_C57BL6J_5623124 | Gm40239 | NCBI_Gene:105244675 | MGI:5623124 | lncRNA gene | predicted gene, 40239 |
| 4 | gene | 121.3163 | 121.3249 | negative | MGI_C57BL6J_3652130 | Gm12888 | NCBI_Gene:545677,ENSEMBL:ENSMUSG00000073764 | MGI:3652130 | protein coding gene | predicted gene 12888 |
| 4 | pseudogene | 121.3798 | 121.3800 | positive | MGI_C57BL6J_3652132 | Gm12889 | ENSEMBL:ENSMUSG00000083941 | MGI:3652132 | pseudogene | predicted gene 12889 |
| 4 | pseudogene | 121.3806 | 121.3809 | positive | MGI_C57BL6J_3651896 | Gm12885 | ENSEMBL:ENSMUSG00000083901 | MGI:3651896 | pseudogene | predicted gene 12885 |
| 4 | pseudogene | 121.3814 | 121.3818 | positive | MGI_C57BL6J_3651895 | Gm12884 | ENSEMBL:ENSMUSG00000081465 | MGI:3651895 | pseudogene | predicted gene 12884 |
| 4 | pseudogene | 121.3930 | 121.3966 | positive | MGI_C57BL6J_3651898 | Gm12883 | ENSEMBL:ENSMUSG00000081667 | MGI:3651898 | pseudogene | predicted gene 12883 |
| 4 | gene | 121.3942 | 121.3966 | negative | MGI_C57BL6J_3648197 | Gm8359 | NCBI_Gene:666914 | MGI:3648197 | protein coding gene | predicted gene 8359 |
| 4 | gene | 121.4147 | 121.4231 | negative | MGI_C57BL6J_3651888 | Gm12886 | NCBI_Gene:666921,ENSEMBL:ENSMUSG00000078576 | MGI:3651888 | protein coding gene | predicted gene 12886 |
| 4 | pseudogene | 121.4768 | 121.5828 | negative | MGI_C57BL6J_3651893 | Gm12880 | NCBI_Gene:100504652,ENSEMBL:ENSMUSG00000082821 | MGI:3651893 | pseudogene | predicted gene 12880 |
| 4 | gene | 121.5095 | 121.5096 | positive | MGI_C57BL6J_5455939 | Gm26162 | ENSEMBL:ENSMUSG00000088418 | MGI:5455939 | snoRNA gene | predicted gene, 26162 |
| 4 | gene | 121.6140 | 121.6221 | negative | MGI_C57BL6J_3652131 | Gm12887 | NCBI_Gene:666927,ENSEMBL:ENSMUSG00000078575 | MGI:3652131 | protein coding gene | predicted gene 12887 |
| 4 | pseudogene | 121.6414 | 121.6418 | positive | MGI_C57BL6J_3649631 | Gm12878 | ENSEMBL:ENSMUSG00000083615 | MGI:3649631 | pseudogene | predicted gene 12878 |
| 4 | gene | 121.9079 | 121.9208 | negative | MGI_C57BL6J_5434759 | Gm21404 | NCBI_Gene:100862019 | MGI:5434759 | protein coding gene | predicted gene, 21404 |
| 4 | pseudogene | 122.0081 | 122.0096 | positive | MGI_C57BL6J_3649649 | Gm12892 | NCBI_Gene:666937,ENSEMBL:ENSMUSG00000083679 | MGI:3649649 | pseudogene | predicted gene 12892 |
| 4 | pseudogene | 122.1228 | 122.1231 | positive | MGI_C57BL6J_3650312 | Gm12896 | ENSEMBL:ENSMUSG00000083558 | MGI:3650312 | pseudogene | predicted gene 12896 |
| 4 | pseudogene | 122.1877 | 122.2067 | positive | MGI_C57BL6J_3649906 | Gm12897 | NCBI_Gene:102632107,ENSEMBL:ENSMUSG00000083949 | MGI:3649906 | pseudogene | predicted gene 12897 |
| 4 | pseudogene | 122.2350 | 122.2359 | positive | MGI_C57BL6J_3652102 | Gm12893 | ENSEMBL:ENSMUSG00000083867 | MGI:3652102 | pseudogene | predicted gene 12893 |
| 4 | gene | 122.3009 | 122.3138 | negative | MGI_C57BL6J_3649909 | Gm10573 | NCBI_Gene:666931 | MGI:3649909 | protein coding gene | predicted gene 10573 |
| 4 | pseudogene | 122.4552 | 122.4554 | positive | MGI_C57BL6J_3649907 | Gm12895 | ENSEMBL:ENSMUSG00000083365 | MGI:3649907 | pseudogene | predicted gene 12895 |
| 4 | gene | 122.5202 | 122.5392 | positive | MGI_C57BL6J_5589564 | Gm30405 | NCBI_Gene:102632293 | MGI:5589564 | lncRNA gene | predicted gene, 30405 |
| 4 | pseudogene | 122.5827 | 122.6049 | negative | MGI_C57BL6J_3650567 | Gm12877 | ENSEMBL:ENSMUSG00000081377 | MGI:3650567 | pseudogene | predicted gene 12877 |
| 4 | pseudogene | 122.5930 | 122.5941 | positive | MGI_C57BL6J_3651901 | Gm12874 | NCBI_Gene:100504420,ENSEMBL:ENSMUSG00000084196 | MGI:3651901 | pseudogene | predicted gene 12874 |
| 4 | pseudogene | 122.6403 | 122.6483 | negative | MGI_C57BL6J_3649744 | Gm12875 | ENSEMBL:ENSMUSG00000080871 | MGI:3649744 | pseudogene | predicted gene 12875 |
| 4 | pseudogene | 122.6609 | 122.6773 | negative | MGI_C57BL6J_3651904 | Gm12876 | ENSEMBL:ENSMUSG00000083109 | MGI:3651904 | pseudogene | predicted gene 12876 |
| 4 | gene | 122.6893 | 122.7051 | positive | MGI_C57BL6J_1924682 | 9530002B09Rik | NCBI_Gene:77432,ENSEMBL:ENSMUSG00000023263 | MGI:1924682 | protein coding gene | RIKEN cDNA 9530002B09 gene |
| 4 | gene | 122.7265 | 122.7395 | negative | MGI_C57BL6J_5589707 | Gm30548 | NCBI_Gene:102632489 | MGI:5589707 | lncRNA gene | predicted gene, 30548 |
| 4 | pseudogene | 122.7303 | 122.7347 | negative | MGI_C57BL6J_3649389 | Gm12872 | ENSEMBL:ENSMUSG00000084082 | MGI:3649389 | pseudogene | predicted gene 12872 |
| 4 | pseudogene | 122.7715 | 122.7938 | negative | MGI_C57BL6J_3651900 | Gm12873 | NCBI_Gene:102632555,ENSEMBL:ENSMUSG00000083494 | MGI:3651900 | pseudogene | predicted gene 12873 |
| 4 | gene | 122.8361 | 122.8592 | positive | MGI_C57BL6J_1298204 | Ppt1 | NCBI_Gene:19063,ENSEMBL:ENSMUSG00000028657 | MGI:1298204 | protein coding gene | palmitoyl-protein thioesterase 1 |
| 4 | gene | 122.8590 | 122.8861 | negative | MGI_C57BL6J_88262 | Cap1 | NCBI_Gene:12331,ENSEMBL:ENSMUSG00000028656 | MGI:88262 | protein coding gene | CAP, adenylate cyclase-associated protein 1 (yeast) |
| 4 | pseudogene | 122.9258 | 122.9262 | positive | MGI_C57BL6J_3649646 | Gm12891 | NCBI_Gene:102632907,ENSEMBL:ENSMUSG00000080929 | MGI:3649646 | pseudogene | predicted gene 12891 |
| 4 | gene | 122.9468 | 122.9612 | negative | MGI_C57BL6J_1923824 | Mfsd2a | NCBI_Gene:76574,ENSEMBL:ENSMUSG00000028655 | MGI:1923824 | protein coding gene | major facilitator superfamily domain containing 2A |
| 4 | gene | 122.9613 | 122.9635 | positive | MGI_C57BL6J_1918347 | 4933421A08Rik | ENSEMBL:ENSMUSG00000086443 | MGI:1918347 | lncRNA gene | RIKEN cDNA 4933421A08 gene |
| 4 | gene | 122.9956 | 123.0025 | positive | MGI_C57BL6J_96799 | Mycl | NCBI_Gene:16918,ENSEMBL:ENSMUSG00000028654 | MGI:96799 | protein coding gene | v-myc avian myelocytomatosis viral oncogene lung carcinoma derived |
| 4 | gene | 123.0165 | 123.0549 | positive | MGI_C57BL6J_1914216 | Trit1 | NCBI_Gene:66966,ENSEMBL:ENSMUSG00000028653 | MGI:1914216 | protein coding gene | tRNA isopentenyltransferase 1 |
| 4 | gene | 123.0812 | 123.0894 | positive | MGI_C57BL6J_5826505 | Gm46868 | NCBI_Gene:108168975 | MGI:5826505 | lncRNA gene | predicted gene, 46868 |
| 4 | gene | 123.0812 | 123.0940 | negative | MGI_C57BL6J_3651539 | Gm12923 | NCBI_Gene:105244676,ENSEMBL:ENSMUSG00000086153 | MGI:3651539 | lncRNA gene | predicted gene 12923 |
| 4 | gene | 123.1043 | 123.1321 | positive | MGI_C57BL6J_107335 | Bmp8b | NCBI_Gene:12164,ENSEMBL:ENSMUSG00000002384 | MGI:107335 | protein coding gene | bone morphogenetic protein 8b |
| 4 | gene | 123.1133 | 123.1168 | negative | MGI_C57BL6J_4936878 | Gm17244 | ENSEMBL:ENSMUSG00000102997 | MGI:4936878 | unclassified gene | predicted gene, 17244 |
| 4 | gene | 123.1162 | 123.1180 | positive | MGI_C57BL6J_2664115 | Oxct2b | NCBI_Gene:353371,ENSEMBL:ENSMUSG00000076438 | MGI:2664115 | protein coding gene | 3-oxoacid CoA transferase 2B |
| 4 | gene | 123.1271 | 123.1400 | negative | MGI_C57BL6J_1917118 | Ppie | NCBI_Gene:56031,ENSEMBL:ENSMUSG00000028651 | MGI:1917118 | protein coding gene | peptidylprolyl isomerase E (cyclophilin E) |
| 4 | gene | 123.1832 | 123.1947 | positive | MGI_C57BL6J_2157521 | Hpcal4 | NCBI_Gene:170638,ENSEMBL:ENSMUSG00000046093 | MGI:2157521 | protein coding gene | hippocalcin-like 4 |
| 4 | gene | 123.2012 | 123.2253 | positive | MGI_C57BL6J_2155700 | Nt5c1a | NCBI_Gene:230718,ENSEMBL:ENSMUSG00000054958 | MGI:2155700 | protein coding gene | 5’-nucleotidase, cytosolic IA |
| 4 | gene | 123.2336 | 123.2499 | positive | MGI_C57BL6J_1860511 | Heyl | NCBI_Gene:56198,ENSEMBL:ENSMUSG00000032744 | MGI:1860511 | protein coding gene | hairy/enhancer-of-split related with YRPW motif-like |
| 4 | pseudogene | 123.2580 | 123.2589 | positive | MGI_C57BL6J_3650176 | Gm12901 | NCBI_Gene:194197,ENSEMBL:ENSMUSG00000083720 | MGI:3650176 | pseudogene | predicted gene 12901 |
| 4 | gene | 123.2624 | 123.2989 | positive | MGI_C57BL6J_2385206 | Pabpc4 | NCBI_Gene:230721,ENSEMBL:ENSMUSG00000011257 | MGI:2385206 | protein coding gene | poly(A) binding protein, cytoplasmic 4 |
| 4 | pseudogene | 123.2749 | 123.2755 | negative | MGI_C57BL6J_3650387 | Gm12900 | NCBI_Gene:622334,ENSEMBL:ENSMUSG00000082634 | MGI:3650387 | pseudogene | predicted gene 12900 |
| 4 | gene | 123.2913 | 123.2915 | positive | MGI_C57BL6J_5455565 | Gm25788 | ENSEMBL:ENSMUSG00000064655 | MGI:5455565 | snoRNA gene | predicted gene, 25788 |
| 4 | gene | 123.2932 | 123.2933 | positive | MGI_C57BL6J_5451931 | Gm22154 | ENSEMBL:ENSMUSG00000065778 | MGI:5451931 | snoRNA gene | predicted gene, 22154 |
| 4 | gene | 123.3126 | 123.3433 | negative | MGI_C57BL6J_104515 | Bmp8a | NCBI_Gene:12163,ENSEMBL:ENSMUSG00000032726 | MGI:104515 | protein coding gene | bone morphogenetic protein 8a |
| 4 | gene | 123.3219 | 123.3237 | negative | MGI_C57BL6J_1891061 | Oxct2a | NCBI_Gene:64059,ENSEMBL:ENSMUSG00000076436 | MGI:1891061 | protein coding gene | 3-oxoacid CoA transferase 2A |
| 4 | gene | 123.3496 | 123.6844 | negative | MGI_C57BL6J_108559 | Macf1 | NCBI_Gene:11426,ENSEMBL:ENSMUSG00000028649 | MGI:108559 | protein coding gene | microtubule-actin crosslinking factor 1 |
| 4 | gene | 123.4036 | 123.4119 | negative | MGI_C57BL6J_3045367 | D830031N03Rik | NCBI_Gene:442834 | MGI:3045367 | protein coding gene | RIKEN cDNA D830031N03 gene |
| 4 | pseudogene | 123.4352 | 123.4364 | positive | MGI_C57BL6J_3651538 | Gm12924 | NCBI_Gene:102633376,ENSEMBL:ENSMUSG00000081528 | MGI:3651538 | pseudogene | predicted gene 12924 |
| 4 | gene | 123.4902 | 123.4903 | positive | MGI_C57BL6J_5453325 | Gm23548 | ENSEMBL:ENSMUSG00000089588 | MGI:5453325 | unclassified non-coding RNA gene | predicted gene, 23548 |
| 4 | gene | 123.5504 | 123.5570 | positive | MGI_C57BL6J_5590315 | Gm31156 | NCBI_Gene:102633296 | MGI:5590315 | lncRNA gene | predicted gene, 31156 |
| 4 | gene | 123.6164 | 123.6165 | positive | MGI_C57BL6J_5531039 | Mir6398 | miRBase:MI0021934,NCBI_Gene:102465214,ENSEMBL:ENSMUSG00000099005 | MGI:5531039 | miRNA gene | microRNA 6398 |
| 4 | pseudogene | 123.6303 | 123.6307 | positive | MGI_C57BL6J_3651535 | Gm12926 | ENSEMBL:ENSMUSG00000081585 | MGI:3651535 | pseudogene | predicted gene 12926 |
| 4 | pseudogene | 123.6446 | 123.6479 | negative | MGI_C57BL6J_5594400 | Gm35241 | NCBI_Gene:102638752 | MGI:5594400 | pseudogene | predicted gene, 35241 |
| 4 | gene | 123.6659 | 123.6712 | positive | MGI_C57BL6J_3651536 | Gm12925 | NCBI_Gene:102633214,ENSEMBL:ENSMUSG00000087307 | MGI:3651536 | lncRNA gene | predicted gene 12925 |
| 4 | pseudogene | 123.7092 | 123.7101 | negative | MGI_C57BL6J_3651537 | Gm12922 | NCBI_Gene:100039110,ENSEMBL:ENSMUSG00000081170 | MGI:3651537 | pseudogene | predicted gene 12922 |
| 4 | gene | 123.7127 | 123.7182 | negative | MGI_C57BL6J_1890889 | Ndufs5 | NCBI_Gene:595136,ENSEMBL:ENSMUSG00000028648 | MGI:1890889 | protein coding gene | NADH:ubiquinone oxidoreductase core subunit S5 |
| 4 | gene | 123.7346 | 123.7503 | negative | MGI_C57BL6J_1915300 | Akirin1 | NCBI_Gene:68050,ENSEMBL:ENSMUSG00000023075 | MGI:1915300 | protein coding gene | akirin 1 |
| 4 | gene | 123.7879 | 123.8302 | positive | MGI_C57BL6J_3608413 | Rhbdl2 | NCBI_Gene:230726,ENSEMBL:ENSMUSG00000043333 | MGI:3608413 | protein coding gene | rhomboid like 2 |
| 4 | gene | 123.8336 | 123.8595 | negative | MGI_C57BL6J_1925891 | 1700121C08Rik | NCBI_Gene:105244677,ENSEMBL:ENSMUSG00000086275 | MGI:1925891 | lncRNA gene | RIKEN cDNA 1700121C08 gene |
| 4 | pseudogene | 123.8381 | 123.8395 | positive | MGI_C57BL6J_3650403 | Gm12904 | NCBI_Gene:667064,ENSEMBL:ENSMUSG00000082310 | MGI:3650403 | pseudogene | predicted gene 12904 |
| 4 | pseudogene | 123.8460 | 123.8495 | negative | MGI_C57BL6J_5826497 | Gm46860 | NCBI_Gene:108168958 | MGI:5826497 | pseudogene | predicted gene, 46860 |
| 4 | gene | 123.8597 | 123.8632 | positive | MGI_C57BL6J_3613760 | 4933427I04Rik | ENSEMBL:ENSMUSG00000073761 | MGI:3613760 | protein coding gene | Riken cDNA 4933427I04 gene |
| 4 | gene | 123.8772 | 123.8806 | positive | MGI_C57BL6J_5590468 | Gm31309 | NCBI_Gene:102633498 | MGI:5590468 | protein coding gene | predicted gene, 31309 |
| 4 | gene | 123.8953 | 123.8966 | negative | MGI_C57BL6J_3642540 | Gm10572 | NA | NA | unclassified gene | predicted gene 10572 |
| 4 | gene | 123.8979 | 123.9012 | negative | MGI_C57BL6J_5623125 | Gm40240 | NCBI_Gene:105244678 | MGI:5623125 | lncRNA gene | predicted gene, 40240 |
| 4 | gene | 123.8986 | 123.9048 | negative | MGI_C57BL6J_5477100 | Gm26606 | ENSEMBL:ENSMUSG00000097337 | MGI:5477100 | lncRNA gene | predicted gene, 26606 |
| 4 | pseudogene | 123.9021 | 123.9050 | negative | MGI_C57BL6J_3045257 | D130007C19Rik | NCBI_Gene:442805,ENSEMBL:ENSMUSG00000054304 | MGI:3045257 | pseudogene | RIKEN cDNA D130007C19 gene |
| 4 | pseudogene | 123.9024 | 123.9030 | positive | MGI_C57BL6J_3650402 | Gm12903 | ENSEMBL:ENSMUSG00000080993 | MGI:3650402 | pseudogene | predicted gene 12903 |
| 4 | gene | 123.9048 | 123.9123 | positive | MGI_C57BL6J_1891750 | Mycbp | NCBI_Gene:56309,ENSEMBL:ENSMUSG00000028647 | MGI:1891750 | protein coding gene | MYC binding protein |
| 4 | gene | 123.9051 | 123.9180 | negative | MGI_C57BL6J_3702581 | Gm12905 | ENSEMBL:ENSMUSG00000085875 | MGI:3702581 | lncRNA gene | predicted gene 12905 |
| 4 | gene | 123.9153 | 123.9180 | negative | MGI_C57BL6J_1922470 | 4930535I16Rik | NA | NA | protein coding gene | RIKEN cDNA 4930535I16 gene |
| 4 | gene | 123.9174 | 123.9370 | positive | MGI_C57BL6J_1858751 | Rragc | NCBI_Gene:54170,ENSEMBL:ENSMUSG00000028646 | MGI:1858751 | protein coding gene | Ras-related GTP binding C |
| 4 | pseudogene | 123.9262 | 123.9266 | positive | MGI_C57BL6J_3650399 | Gm12902 | ENSEMBL:ENSMUSG00000082515 | MGI:3650399 | pseudogene | predicted gene 12902 |
| 4 | gene | 124.0540 | 124.0701 | positive | MGI_C57BL6J_5623126 | Gm40241 | NCBI_Gene:105244679 | MGI:5623126 | lncRNA gene | predicted gene, 40241 |
| 4 | gene | 124.2714 | 124.2724 | negative | MGI_C57BL6J_5623127 | Gm40242 | NCBI_Gene:105244680 | MGI:5623127 | lncRNA gene | predicted gene, 40242 |
| 4 | gene | 124.2778 | 124.2792 | negative | MGI_C57BL6J_5610895 | Gm37667 | ENSEMBL:ENSMUSG00000103541 | MGI:5610895 | unclassified gene | predicted gene, 37667 |
| 4 | gene | 124.2931 | 124.2999 | negative | MGI_C57BL6J_3649992 | Gm12916 | NCBI_Gene:102633918,ENSEMBL:ENSMUSG00000086087 | MGI:3649992 | lncRNA gene | predicted gene 12916 |
| 4 | gene | 124.3028 | 124.3327 | negative | MGI_C57BL6J_1923660 | 1700021L23Rik | NCBI_Gene:76410,ENSEMBL:ENSMUSG00000086955 | MGI:1923660 | lncRNA gene | RIKEN cDNA 1700021L23 gene |
| 4 | gene | 124.3263 | 124.3325 | negative | MGI_C57BL6J_1918570 | 4933435F18Rik | ENSEMBL:ENSMUSG00000086978 | MGI:1918570 | lncRNA gene | RIKEN cDNA 4933435F18 gene |
| 4 | gene | 124.3356 | 124.3357 | negative | MGI_C57BL6J_5452760 | Gm22983 | ENSEMBL:ENSMUSG00000088933 | MGI:5452760 | snRNA gene | predicted gene, 22983 |
| 4 | gene | 124.4144 | 124.4860 | negative | MGI_C57BL6J_1925710 | 1700057H15Rik | NCBI_Gene:78460,ENSEMBL:ENSMUSG00000084757 | MGI:1925710 | lncRNA gene | RIKEN cDNA 1700057H15 gene |
| 4 | gene | 124.4514 | 124.4564 | positive | MGI_C57BL6J_5590989 | Gm31830 | NCBI_Gene:102634185 | MGI:5590989 | lncRNA gene | predicted gene, 31830 |
| 4 | gene | 124.4653 | 124.4681 | negative | MGI_C57BL6J_5590928 | Gm31769 | NCBI_Gene:102634107 | MGI:5590928 | lncRNA gene | predicted gene, 31769 |
| 4 | gene | 124.5229 | 124.6575 | negative | MGI_C57BL6J_3780334 | Gm2164 | NCBI_Gene:100039332,ENSEMBL:ENSMUSG00000098760 | MGI:3780334 | lncRNA gene | predicted gene 2164 |
| 4 | gene | 124.5692 | 124.5755 | positive | MGI_C57BL6J_1918352 | 4933407E24Rik | NCBI_Gene:108809,ENSEMBL:ENSMUSG00000073759 | MGI:1918352 | lncRNA gene | RIKEN cDNA 4933407E24 gene |
| 4 | gene | 124.6099 | 124.6108 | negative | MGI_C57BL6J_1917499 | 3100002H09Rik | NA | NA | lncRNA gene | RIKEN cDNA 3100002H09 gene |
| 4 | gene | 124.6399 | 124.6408 | positive | MGI_C57BL6J_1915732 | 1110007E10Rik | NA | NA | unclassified gene | RIKEN cDNA 1110007E10 gene |
| 4 | gene | 124.6568 | 124.6607 | positive | MGI_C57BL6J_101896 | Pou3f1 | NCBI_Gene:18991,ENSEMBL:ENSMUSG00000090125 | MGI:101896 | protein coding gene | POU domain, class 3, transcription factor 1 |
| 4 | gene | 124.6643 | 124.6687 | negative | MGI_C57BL6J_5621430 | Gm38545 | NCBI_Gene:102641270 | MGI:5621430 | lncRNA gene | predicted gene, 38545 |
| 4 | gene | 124.6782 | 124.6936 | negative | MGI_C57BL6J_1914455 | Utp11 | NCBI_Gene:67205,ENSEMBL:ENSMUSG00000028907 | MGI:1914455 | protein coding gene | UTP11 small subunit processome component |
| 4 | gene | 124.6960 | 124.7086 | positive | MGI_C57BL6J_1341092 | Fhl3 | NCBI_Gene:14201,ENSEMBL:ENSMUSG00000032643 | MGI:1341092 | protein coding gene | four and a half LIM domains 3 |
| 4 | gene | 124.6975 | 124.6976 | negative | MGI_C57BL6J_5454257 | Gm24480 | ENSEMBL:ENSMUSG00000088067 | MGI:5454257 | snRNA gene | predicted gene, 24480 |
| 4 | gene | 124.7148 | 124.7325 | positive | MGI_C57BL6J_1922312 | Sf3a3 | NCBI_Gene:75062,ENSEMBL:ENSMUSG00000028902 | MGI:1922312 | protein coding gene | splicing factor 3a, subunit 3 |
| 4 | pseudogene | 124.7190 | 124.7196 | positive | MGI_C57BL6J_3652006 | Gm12917 | NCBI_Gene:664991,ENSEMBL:ENSMUSG00000083104 | MGI:3652006 | pseudogene | predicted gene 12917 |
| 4 | gene | 124.7317 | 124.7318 | positive | MGI_C57BL6J_3629652 | Mir697 | miRBase:MI0004681,NCBI_Gene:735277,ENSEMBL:ENSMUSG00000076271 | MGI:3629652 | miRNA gene | microRNA 697 |
| 4 | gene | 124.7353 | 124.7514 | negative | MGI_C57BL6J_3650273 | Gm12915 | NCBI_Gene:102634646,ENSEMBL:ENSMUSG00000086589 | MGI:3650273 | lncRNA gene | predicted gene 12915 |
| 4 | gene | 124.7358 | 124.7359 | positive | MGI_C57BL6J_5454498 | Gm24721 | ENSEMBL:ENSMUSG00000096411 | MGI:5454498 | miRNA gene | predicted gene, 24721 |
| 4 | gene | 124.7374 | 124.7401 | positive | MGI_C57BL6J_5591276 | Gm32117 | NCBI_Gene:102634569 | MGI:5591276 | lncRNA gene | predicted gene, 32117 |
| 4 | gene | 124.7418 | 124.8015 | positive | MGI_C57BL6J_103257 | Inpp5b | NCBI_Gene:16330,ENSEMBL:ENSMUSG00000028894 | MGI:103257 | protein coding gene | inositol polyphosphate-5-phosphatase B |
| 4 | gene | 124.7438 | 124.7439 | positive | MGI_C57BL6J_3629653 | Mir698 | miRBase:MI0004682,NCBI_Gene:735263,ENSEMBL:ENSMUSG00000076119 | MGI:3629653 | miRNA gene | microRNA 698 |
| 4 | gene | 124.8021 | 124.8498 | positive | MGI_C57BL6J_101786 | Mtf1 | NCBI_Gene:17764,ENSEMBL:ENSMUSG00000028890 | MGI:101786 | protein coding gene | metal response element binding transcription factor 1 |
| 4 | gene | 124.8208 | 124.8209 | positive | MGI_C57BL6J_4422057 | n-R5s192 | ENSEMBL:ENSMUSG00000084526 | MGI:4422057 | rRNA gene | nuclear encoded rRNA 5S 192 |
| 4 | gene | 124.8491 | 124.8512 | negative | MGI_C57BL6J_1916170 | 1110065P20Rik | NCBI_Gene:68920,ENSEMBL:ENSMUSG00000078570 | MGI:1916170 | protein coding gene | RIKEN cDNA 1110065P20 gene |
| 4 | gene | 124.8507 | 124.8552 | positive | MGI_C57BL6J_2387201 | Yrdc | NCBI_Gene:230734,ENSEMBL:ENSMUSG00000028889 | MGI:2387201 | protein coding gene | yrdC domain containing (E.coli) |
| 4 | gene | 124.8552 | 124.8809 | negative | MGI_C57BL6J_2684896 | Maneal | NCBI_Gene:215090,ENSEMBL:ENSMUSG00000042763 | MGI:2684896 | protein coding gene | mannosidase, endo-alpha-like |
| 4 | gene | 124.8809 | 124.9178 | positive | MGI_C57BL6J_3586824 | Epha10 | NCBI_Gene:230735,ENSEMBL:ENSMUSG00000028876 | MGI:3586824 | protein coding gene | Eph receptor A10 |
| 4 | gene | 124.8909 | 124.8948 | negative | MGI_C57BL6J_1920878 | 1700125G02Rik | NCBI_Gene:73628,ENSEMBL:ENSMUSG00000085263 | MGI:1920878 | lncRNA gene | RIKEN cDNA 1700125G02 gene |
| 4 | gene | 124.9185 | 124.9393 | negative | MGI_C57BL6J_1196274 | Cdca8 | NCBI_Gene:52276,ENSEMBL:ENSMUSG00000028873 | MGI:1196274 | protein coding gene | cell division cycle associated 8 |
| 4 | gene | 124.9370 | 124.9447 | positive | MGI_C57BL6J_3041172 | 9930104L06Rik | NCBI_Gene:194268,ENSEMBL:ENSMUSG00000044730 | MGI:3041172 | protein coding gene | RIKEN cDNA 9930104L06 gene |
| 4 | gene | 124.9457 | 124.9628 | positive | MGI_C57BL6J_5623128 | Gm40243 | NCBI_Gene:105244681 | MGI:5623128 | lncRNA gene | predicted gene, 40243 |
| 4 | gene | 124.9576 | 124.9870 | negative | MGI_C57BL6J_3650529 | Gm12930 | ENSEMBL:ENSMUSG00000087011 | MGI:3650529 | lncRNA gene | predicted gene 12930 |
| 4 | gene | 124.9676 | 124.9746 | negative | MGI_C57BL6J_5591530 | Gm32371 | NCBI_Gene:102634896 | MGI:5591530 | lncRNA gene | predicted gene, 32371 |
| 4 | gene | 124.9864 | 125.0091 | positive | MGI_C57BL6J_2183426 | Rspo1 | NCBI_Gene:192199,ENSEMBL:ENSMUSG00000028871 | MGI:2183426 | protein coding gene | R-spondin 1 |
| 4 | gene | 125.0166 | 125.0554 | positive | MGI_C57BL6J_2385207 | Gnl2 | NCBI_Gene:230737,ENSEMBL:ENSMUSG00000028869 | MGI:2385207 | protein coding gene | guanine nucleotide binding protein-like 2 (nucleolar) |
| 4 | gene | 125.0177 | 125.0210 | positive | MGI_C57BL6J_5621357 | Gm38472 | NCBI_Gene:102635036 | MGI:5621357 | lncRNA gene | predicted gene, 38472 |
| 4 | pseudogene | 125.0195 | 125.0198 | positive | MGI_C57BL6J_3650713 | Gm12931 | ENSEMBL:ENSMUSG00000080934 | MGI:3650713 | pseudogene | predicted gene 12931 |
| 4 | gene | 125.0553 | 125.0657 | negative | MGI_C57BL6J_1922813 | Dnali1 | NCBI_Gene:75563,ENSEMBL:ENSMUSG00000042707 | MGI:1922813 | protein coding gene | dynein, axonemal, light intermediate polypeptide 1 |
| 4 | gene | 125.0667 | 125.0741 | positive | MGI_C57BL6J_2156003 | Snip1 | NCBI_Gene:76793,ENSEMBL:ENSMUSG00000050213 | MGI:2156003 | protein coding gene | Smad nuclear interacting protein 1 |
| 4 | pseudogene | 125.0755 | 125.0765 | positive | MGI_C57BL6J_5010665 | Gm18480 | NCBI_Gene:100417248 | MGI:5010665 | pseudogene | predicted gene, 18480 |
| 4 | gene | 125.0829 | 125.0849 | negative | MGI_C57BL6J_3651151 | Gm12932 | ENSEMBL:ENSMUSG00000086342 | MGI:3651151 | lncRNA gene | predicted gene 12932 |
| 4 | gene | 125.0851 | 125.1132 | positive | MGI_C57BL6J_1917338 | Meaf6 | NCBI_Gene:70088,ENSEMBL:ENSMUSG00000028863 | MGI:1917338 | protein coding gene | MYST/Esa1-associated factor 6 |
| 4 | gene | 125.1184 | 125.1279 | negative | MGI_C57BL6J_2385891 | Zc3h12a | NCBI_Gene:230738,ENSEMBL:ENSMUSG00000042677 | MGI:2385891 | protein coding gene | zinc finger CCCH type containing 12A |
| 4 | gene | 125.1375 | 125.1477 | negative | MGI_C57BL6J_5623131 | Gm40246 | NCBI_Gene:105244684 | MGI:5623131 | lncRNA gene | predicted gene, 40246 |
| 4 | gene | 125.1452 | 125.2904 | positive | MGI_C57BL6J_5591770 | Gm32611 | NCBI_Gene:102635216 | MGI:5591770 | protein coding gene | predicted gene, 32611 |
| 4 | gene | 125.1500 | 125.1871 | positive | MGI_C57BL6J_5623130 | Gm40245 | NCBI_Gene:105244683 | MGI:5623130 | lncRNA gene | predicted gene, 40245 |
| 4 | gene | 125.2184 | 125.2199 | positive | MGI_C57BL6J_5623129 | Gm40244 | NCBI_Gene:105244682 | MGI:5623129 | lncRNA gene | predicted gene, 40244 |
| 4 | gene | 125.2286 | 125.2421 | positive | MGI_C57BL6J_1920510 | 1700041M05Rik | ENSEMBL:ENSMUSG00000087010 | MGI:1920510 | lncRNA gene | RIKEN cDNA 1700041M05 gene |
| 4 | gene | 125.2905 | 125.3015 | positive | MGI_C57BL6J_5826506 | Gm46869 | NCBI_Gene:108168977 | MGI:5826506 | lncRNA gene | predicted gene, 46869 |
| 4 | gene | 125.3123 | 125.3135 | negative | MGI_C57BL6J_5591855 | Gm32696 | NCBI_Gene:102635326 | MGI:5591855 | lncRNA gene | predicted gene, 32696 |
| 4 | gene | 125.3817 | 125.3945 | positive | MGI_C57BL6J_5623132 | Gm40247 | NCBI_Gene:105244685 | MGI:5623132 | lncRNA gene | predicted gene, 40247 |
| 4 | gene | 125.4511 | 125.4772 | positive | MGI_C57BL6J_5591915 | Gm32756 | NCBI_Gene:102635406 | MGI:5591915 | lncRNA gene | predicted gene, 32756 |
| 4 | gene | 125.4902 | 125.7142 | positive | MGI_C57BL6J_95816 | Grik3 | NCBI_Gene:14807,ENSEMBL:ENSMUSG00000001985 | MGI:95816 | protein coding gene | glutamate receptor, ionotropic, kainate 3 |
| 4 | pseudogene | 125.5041 | 125.5050 | positive | MGI_C57BL6J_95590 | Ftl2-ps | NCBI_Gene:14337,ENSEMBL:ENSMUSG00000082062 | MGI:95590 | pseudogene | ferritin light polypeptide 2, pseudogene |
| 4 | gene | 125.5047 | 125.5049 | positive | MGI_C57BL6J_3629654 | Mir692-2 | NCBI_Gene:735288,ENSEMBL:ENSMUSG00000094403 | MGI:3629654 | miRNA gene | microRNA 692-2 |
| 4 | gene | 125.5502 | 125.5561 | negative | MGI_C57BL6J_5591966 | Gm32807 | NCBI_Gene:102635479 | MGI:5591966 | lncRNA gene | predicted gene, 32807 |
| 4 | gene | 125.8297 | 125.8368 | negative | MGI_C57BL6J_5623134 | Gm40249 | NCBI_Gene:105244687 | MGI:5623134 | lncRNA gene | predicted gene, 40249 |
| 4 | gene | 125.8879 | 125.9224 | negative | MGI_C57BL6J_1919645 | 2610028E06Rik | NCBI_Gene:72395,ENSEMBL:ENSMUSG00000085562 | MGI:1919645 | lncRNA gene | RIKEN cDNA 2610028E06 gene |
| 4 | gene | 126.0112 | 126.0245 | negative | MGI_C57BL6J_5592067 | Gm32908 | NCBI_Gene:102635619 | MGI:5592067 | lncRNA gene | predicted gene, 32908 |
| 4 | gene | 126.0234 | 126.0450 | positive | MGI_C57BL6J_1339755 | Csf3r | NCBI_Gene:12986,ENSEMBL:ENSMUSG00000028859 | MGI:1339755 | protein coding gene | colony stimulating factor 3 receptor (granulocyte) |
| 4 | gene | 126.0469 | 126.0555 | positive | MGI_C57BL6J_1913657 | Mrps15 | NCBI_Gene:66407,ENSEMBL:ENSMUSG00000028861 | MGI:1913657 | protein coding gene | mitochondrial ribosomal protein S15 |
| 4 | gene | 126.0560 | 126.0922 | positive | MGI_C57BL6J_1916308 | Oscp1 | NCBI_Gene:230751,ENSEMBL:ENSMUSG00000042616 | MGI:1916308 | protein coding gene | organic solute carrier partner 1 |
| 4 | pseudogene | 126.0698 | 126.0703 | negative | MGI_C57BL6J_3650282 | Rpl28-ps3 | NCBI_Gene:100039645,ENSEMBL:ENSMUSG00000079942 | MGI:3650282 | pseudogene | ribosomal protein L28, pseudogene 3 |
| 4 | gene | 126.0966 | 126.0986 | positive | MGI_C57BL6J_2151045 | Lsm10 | NCBI_Gene:116748,ENSEMBL:ENSMUSG00000050188 | MGI:2151045 | protein coding gene | U7 snRNP-specific Sm-like protein LSM10 |
| 4 | gene | 126.1005 | 126.1047 | negative | MGI_C57BL6J_1924911 | 1520401O13Rik | NA | NA | unclassified gene | RIKEN cDNA 1520401O13 gene |
| 4 | gene | 126.1039 | 126.1410 | positive | MGI_C57BL6J_1921428 | Stk40 | NCBI_Gene:74178,ENSEMBL:ENSMUSG00000042608 | MGI:1921428 | protein coding gene | serine/threonine kinase 40 |
| 4 | gene | 126.1457 | 126.1499 | positive | MGI_C57BL6J_1922063 | Eva1b | NCBI_Gene:230752,ENSEMBL:ENSMUSG00000050212 | MGI:1922063 | protein coding gene | eva-1 homolog B (C. elegans) |
| 4 | gene | 126.1460 | 126.1477 | positive | MGI_C57BL6J_3650946 | Gm12946 | ENSEMBL:ENSMUSG00000086961 | MGI:3650946 | lncRNA gene | predicted gene 12946 |
| 4 | gene | 126.1506 | 126.1640 | negative | MGI_C57BL6J_1914188 | Sh3d21 | NCBI_Gene:66938,ENSEMBL:ENSMUSG00000073758 | MGI:1914188 | protein coding gene | SH3 domain containing 21 |
| 4 | gene | 126.1641 | 126.2028 | negative | MGI_C57BL6J_2442637 | Thrap3 | NCBI_Gene:230753,ENSEMBL:ENSMUSG00000043962 | MGI:2442637 | protein coding gene | thyroid hormone receptor associated protein 3 |
| 4 | gene | 126.2012 | 126.2013 | negative | MGI_C57BL6J_5455100 | Gm25323 | ENSEMBL:ENSMUSG00000084504 | MGI:5455100 | snRNA gene | predicted gene, 25323 |
| 4 | gene | 126.2029 | 126.2039 | positive | MGI_C57BL6J_1925109 | D730050C22Rik | NA | NA | unclassified gene | RIKEN cDNA D730050C22 gene |
| 4 | gene | 126.2322 | 126.2565 | negative | MGI_C57BL6J_2384297 | Map7d1 | NCBI_Gene:245877,ENSEMBL:ENSMUSG00000028849 | MGI:2384297 | protein coding gene | MAP7 domain containing 1 |
| 4 | gene | 126.2623 | 126.2759 | positive | MGI_C57BL6J_1351486 | Trappc3 | NCBI_Gene:27096,ENSEMBL:ENSMUSG00000028847 | MGI:1351486 | protein coding gene | trafficking protein particle complex 3 |
| 4 | gene | 126.2868 | 126.3143 | positive | MGI_C57BL6J_88464 | Col8a2 | NCBI_Gene:329941,ENSEMBL:ENSMUSG00000056174 | MGI:88464 | protein coding gene | collagen, type VIII, alpha 2 |
| 4 | gene | 126.3160 | 126.3217 | negative | MGI_C57BL6J_2140364 | Adprhl2 | NCBI_Gene:100206,ENSEMBL:ENSMUSG00000042558 | MGI:2140364 | protein coding gene | ADP-ribosylhydrolase like 2 |
| 4 | gene | 126.3221 | 126.3258 | negative | MGI_C57BL6J_1346335 | Tekt2 | NCBI_Gene:24084,ENSEMBL:ENSMUSG00000028845 | MGI:1346335 | protein coding gene | tektin 2 |
| 4 | gene | 126.3236 | 126.3252 | positive | MGI_C57BL6J_3704237 | Gm12945 | ENSEMBL:ENSMUSG00000087394 | MGI:3704237 | lncRNA gene | predicted gene 12945 |
| 4 | gene | 126.3270 | 126.4297 | negative | MGI_C57BL6J_2446634 | Ago3 | NCBI_Gene:214150,ENSEMBL:ENSMUSG00000028842 | MGI:2446634 | protein coding gene | argonaute RISC catalytic subunit 3 |
| 4 | gene | 126.4350 | 126.4687 | negative | MGI_C57BL6J_2446630 | Ago1 | NCBI_Gene:236511,ENSEMBL:ENSMUSG00000041530 | MGI:2446630 | protein coding gene | argonaute RISC catalytic subunit 1 |
| 4 | pseudogene | 126.4763 | 126.4767 | negative | MGI_C57BL6J_3652021 | Gm12929 | ENSEMBL:ENSMUSG00000084099 | MGI:3652021 | pseudogene | predicted gene 12929 |
| 4 | pseudogene | 126.4767 | 126.4773 | negative | MGI_C57BL6J_3652022 | Gm12928 | ENSEMBL:ENSMUSG00000082381 | MGI:3652022 | pseudogene | predicted gene 12928 |
| 4 | gene | 126.4895 | 126.5336 | negative | MGI_C57BL6J_1924100 | Ago4 | NCBI_Gene:76850,ENSEMBL:ENSMUSG00000042500 | MGI:1924100 | protein coding gene | argonaute RISC catalytic subunit 4 |
| 4 | pseudogene | 126.5217 | 126.5221 | positive | MGI_C57BL6J_3651149 | Gm12935 | ENSEMBL:ENSMUSG00000083461 | MGI:3651149 | pseudogene | predicted gene 12935 |
| 4 | pseudogene | 126.5243 | 126.5247 | negative | MGI_C57BL6J_3651150 | Gm12934 | ENSEMBL:ENSMUSG00000082888 | MGI:3651150 | pseudogene | predicted gene 12934 |
| 4 | gene | 126.5569 | 126.5945 | positive | MGI_C57BL6J_2445153 | Clspn | NCBI_Gene:269582,ENSEMBL:ENSMUSG00000042489 | MGI:2445153 | protein coding gene | claspin |
| 4 | gene | 126.5599 | 126.5599 | positive | MGI_C57BL6J_5530942 | Mir7119 | miRBase:MI0022970,NCBI_Gene:102465673,ENSEMBL:ENSMUSG00000098831 | MGI:5530942 | miRNA gene | microRNA 7119 |
| 4 | pseudogene | 126.5844 | 126.5851 | negative | MGI_C57BL6J_3651145 | Gm12936 | NCBI_Gene:102635893,ENSEMBL:ENSMUSG00000081319 | MGI:3651145 | pseudogene | predicted gene 12936 |
| 4 | gene | 126.6098 | 126.6144 | positive | MGI_C57BL6J_3609248 | 5730409E04Rik | NCBI_Gene:230757,ENSEMBL:ENSMUSG00000073755 | MGI:3609248 | protein coding gene | RIKEN cDNA 5730409E04Rik gene |
| 4 | gene | 126.6144 | 126.6148 | negative | MGI_C57BL6J_5623135 | Gm40250 | NCBI_Gene:105244688 | MGI:5623135 | lncRNA gene | predicted gene, 40250 |
| 4 | gene | 126.6164 | 126.6203 | negative | MGI_C57BL6J_5826507 | Gm46870 | NCBI_Gene:108168978 | MGI:5826507 | lncRNA gene | predicted gene, 46870 |
| 4 | pseudogene | 126.6652 | 126.6662 | positive | MGI_C57BL6J_3651147 | Gm12933 | NCBI_Gene:620877,ENSEMBL:ENSMUSG00000081210 | MGI:3651147 | pseudogene | predicted gene 12933 |
| 4 | gene | 126.6776 | 126.7097 | positive | MGI_C57BL6J_1347045 | Psmb2 | NCBI_Gene:26445,ENSEMBL:ENSMUSG00000028837 | MGI:1347045 | protein coding gene | proteasome (prosome, macropain) subunit, beta type 2 |
| 4 | gene | 126.6995 | 126.7040 | positive | MGI_C57BL6J_2442195 | E330021A06Rik | NA | NA | unclassified gene | RIKEN cDNA E330021A06 gene |
| 4 | gene | 126.7160 | 126.7363 | negative | MGI_C57BL6J_2679630 | Tfap2e | NCBI_Gene:332937,ENSEMBL:ENSMUSG00000042477 | MGI:2679630 | protein coding gene | transcription factor AP-2, epsilon |
| 4 | gene | 126.7438 | 126.7536 | negative | MGI_C57BL6J_1347351 | Ncdn | NCBI_Gene:26562,ENSEMBL:ENSMUSG00000028833 | MGI:1347351 | protein coding gene | neurochondrin |
| 4 | gene | 126.7535 | 126.8701 | positive | MGI_C57BL6J_2140475 | AU040320 | NCBI_Gene:100317,ENSEMBL:ENSMUSG00000028830 | MGI:2140475 | protein coding gene | expressed sequence AU040320 |
| 4 | pseudogene | 126.7955 | 126.7964 | negative | MGI_C57BL6J_3650044 | Gm12937 | ENSEMBL:ENSMUSG00000082498 | MGI:3650044 | pseudogene | predicted gene 12937 |
| 4 | gene | 126.8180 | 126.8181 | negative | MGI_C57BL6J_5453938 | Gm24161 | ENSEMBL:ENSMUSG00000077780 | MGI:5453938 | snoRNA gene | predicted gene, 24161 |
| 4 | gene | 126.8610 | 126.8611 | negative | MGI_C57BL6J_5455381 | Gm25604 | ENSEMBL:ENSMUSG00000088984 | MGI:5455381 | snoRNA gene | predicted gene, 25604 |
| 4 | gene | 126.8618 | 126.9688 | negative | MGI_C57BL6J_1915035 | Zmym4 | NCBI_Gene:67785,ENSEMBL:ENSMUSG00000042446 | MGI:1915035 | protein coding gene | zinc finger, MYM-type 4 |
| 4 | gene | 126.9681 | 126.9694 | positive | MGI_C57BL6J_1918435 | 4933424C08Rik | NA | NA | unclassified gene | RIKEN cDNA 4933424C08 gene |
| 4 | pseudogene | 126.9834 | 126.9836 | positive | MGI_C57BL6J_3652092 | Gm12939 | ENSEMBL:ENSMUSG00000081609 | MGI:3652092 | pseudogene | predicted gene 12939 |
| 4 | gene | 127.0206 | 127.0511 | positive | MGI_C57BL6J_1918764 | Sfpq | NCBI_Gene:71514,ENSEMBL:ENSMUSG00000028820 | MGI:1918764 | protein coding gene | splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) |
| 4 | gene | 127.0337 | 127.0494 | positive | MGI_C57BL6J_3702626 | Gm12940 | ENSEMBL:ENSMUSG00000085334 | MGI:3702626 | lncRNA gene | predicted gene 12940 |
| 4 | pseudogene | 127.0464 | 127.0466 | positive | MGI_C57BL6J_3650501 | Gm12941 | ENSEMBL:ENSMUSG00000084088 | MGI:3650501 | pseudogene | predicted gene 12941 |
| 4 | gene | 127.0471 | 127.0612 | negative | MGI_C57BL6J_1915560 | Zmym1 | NCBI_Gene:68310,ENSEMBL:ENSMUSG00000043872 | MGI:1915560 | protein coding gene | zinc finger, MYM domain containing 1 |
| 4 | pseudogene | 127.0559 | 127.0567 | positive | MGI_C57BL6J_3650498 | Gm12944 | NCBI_Gene:102635997,ENSEMBL:ENSMUSG00000084149 | MGI:3650498 | pseudogene | predicted gene 12944 |
| 4 | gene | 127.0774 | 127.1244 | positive | MGI_C57BL6J_106505 | Zmym6 | NCBI_Gene:100177,ENSEMBL:ENSMUSG00000042408 | MGI:106505 | protein coding gene | zinc finger, MYM-type 6 |
| 4 | gene | 127.1085 | 127.1086 | negative | MGI_C57BL6J_5455377 | Gm25600 | ENSEMBL:ENSMUSG00000088982 | MGI:5455377 | snRNA gene | predicted gene, 25600 |
| 4 | gene | 127.1237 | 127.1297 | positive | MGI_C57BL6J_3758095 | Tmem35b | NCBI_Gene:100039968,ENSEMBL:ENSMUSG00000070737 | MGI:3758095 | protein coding gene | transmembrane protein 35B |
| 4 | pseudogene | 127.1412 | 127.1415 | negative | MGI_C57BL6J_3650497 | Gm12943 | NCBI_Gene:621588,ENSEMBL:ENSMUSG00000082220 | MGI:3650497 | pseudogene | predicted gene 12943 |
| 4 | gene | 127.1685 | 127.2370 | positive | MGI_C57BL6J_3039563 | Dlgap3 | NCBI_Gene:242667,ENSEMBL:ENSMUSG00000042388 | MGI:3039563 | protein coding gene | DLG associated protein 3 |
| 4 | gene | 127.2438 | 127.2478 | positive | MGI_C57BL6J_1933141 | Smim12 | NCBI_Gene:80284,ENSEMBL:ENSMUSG00000042380 | MGI:1933141 | protein coding gene | small integral membrane protein 12 |
| 4 | gene | 127.2487 | 127.3134 | positive | MGI_C57BL6J_5592428 | Gm33269 | NCBI_Gene:102636104 | MGI:5592428 | lncRNA gene | predicted gene, 33269 |
| 4 | gene | 127.2847 | 127.2848 | negative | MGI_C57BL6J_5451998 | Gm22221 | ENSEMBL:ENSMUSG00000087987 | MGI:5451998 | snRNA gene | predicted gene, 22221 |
| 4 | pseudogene | 127.2968 | 127.2975 | negative | MGI_C57BL6J_3649530 | Gm12948 | NCBI_Gene:100417246,ENSEMBL:ENSMUSG00000082047 | MGI:3649530 | pseudogene | predicted gene 12948 |
| 4 | gene | 127.3114 | 127.3140 | negative | MGI_C57BL6J_95715 | Gja4 | NCBI_Gene:14612,ENSEMBL:ENSMUSG00000050234 | MGI:95715 | protein coding gene | gap junction protein, alpha 4 |
| 4 | gene | 127.3252 | 127.3308 | negative | MGI_C57BL6J_95721 | Gjb3 | NCBI_Gene:14620,ENSEMBL:ENSMUSG00000042367 | MGI:95721 | protein coding gene | gap junction protein, beta 3 |
| 4 | gene | 127.3292 | 127.3433 | positive | MGI_C57BL6J_5623136 | Gm40251 | NCBI_Gene:105244689 | MGI:5623136 | lncRNA gene | predicted gene, 40251 |
| 4 | gene | 127.3410 | 127.3541 | negative | MGI_C57BL6J_95722 | Gjb4 | NCBI_Gene:14621,ENSEMBL:ENSMUSG00000046623 | MGI:95722 | protein coding gene | gap junction protein, beta 4 |
| 4 | gene | 127.3548 | 127.3582 | negative | MGI_C57BL6J_95723 | Gjb5 | NCBI_Gene:14622,ENSEMBL:ENSMUSG00000042357 | MGI:95723 | protein coding gene | gap junction protein, beta 5 |
| 4 | pseudogene | 127.4083 | 127.4097 | negative | MGI_C57BL6J_3649527 | Gm12947 | NCBI_Gene:667296,ENSEMBL:ENSMUSG00000079998 | MGI:3649527 | pseudogene | predicted gene 12947 |
| 4 | gene | 127.4171 | 127.4248 | positive | MGI_C57BL6J_5623137 | Gm40252 | NCBI_Gene:105244690 | MGI:5623137 | lncRNA gene | predicted gene, 40252 |
| 4 | gene | 127.4290 | 127.4299 | positive | MGI_C57BL6J_5623138 | Gm40253 | NA | NA | lncRNA gene | predicted gene%2c 40253 |
| 4 | gene | 127.4748 | 127.5329 | positive | MGI_C57BL6J_1920738 | 1700080G11Rik | NCBI_Gene:73488,ENSEMBL:ENSMUSG00000086771 | MGI:1920738 | lncRNA gene | RIKEN cDNA 1700080G11 gene |
| 4 | gene | 127.4748 | 127.4768 | positive | MGI_C57BL6J_1925335 | 4930471C06Rik | ENSEMBL:ENSMUSG00000086138 | MGI:1925335 | lncRNA gene | RIKEN cDNA 4930471C06 gene |
| 4 | gene | 127.5451 | 127.5705 | positive | MGI_C57BL6J_2444470 | A630031M04Rik | ENSEMBL:ENSMUSG00000087470 | MGI:2444470 | lncRNA gene | RIKEN cDNA A630031M04 gene |
| 4 | gene | 127.6187 | 127.6199 | positive | MGI_C57BL6J_5623140 | Gm40255 | NCBI_Gene:105244693 | MGI:5623140 | lncRNA gene | predicted gene, 40255 |
| 4 | gene | 127.8106 | 127.8122 | negative | MGI_C57BL6J_1920846 | 1700112K13Rik | ENSEMBL:ENSMUSG00000087510 | MGI:1920846 | lncRNA gene | RIKEN cDNA 1700112K13 gene |
| 4 | gene | 127.9139 | 127.9164 | positive | MGI_C57BL6J_5623141 | Gm40256 | NCBI_Gene:105244694 | MGI:5623141 | lncRNA gene | predicted gene, 40256 |
| 4 | gene | 127.9276 | 127.9881 | negative | MGI_C57BL6J_3616080 | CK137956 | NCBI_Gene:635169,ENSEMBL:ENSMUSG00000028813 | MGI:3616080 | protein coding gene | cDNA sequence CK137956 |
| 4 | gene | 127.9716 | 127.9724 | positive | MGI_C57BL6J_5621456 | Gm38571 | NCBI_Gene:102641725 | MGI:5621456 | lncRNA gene | predicted gene, 38571 |
| 4 | gene | 127.9878 | 128.5677 | positive | MGI_C57BL6J_2386401 | Csmd2 | NCBI_Gene:329942,ENSEMBL:ENSMUSG00000028804 | MGI:2386401 | protein coding gene | CUB and Sushi multiple domains 2 |
| 4 | gene | 128.1322 | 128.1670 | negative | MGI_C57BL6J_1922239 | Csmd2os | NCBI_Gene:74989,ENSEMBL:ENSMUSG00000087459 | MGI:1922239 | antisense lncRNA gene | CUB and Sushi multiple domains 2, opposite strand |
| 4 | gene | 128.2602 | 128.2610 | negative | MGI_C57BL6J_1916567 | Hmgb4 | NCBI_Gene:69317,ENSEMBL:ENSMUSG00000048686 | MGI:1916567 | protein coding gene | high-mobility group box 4 |
| 4 | gene | 128.2610 | 128.2630 | positive | MGI_C57BL6J_1922492 | Hmgb4os | NCBI_Gene:75242,ENSEMBL:ENSMUSG00000085717 | MGI:1922492 | antisense lncRNA gene | high-mobility group box 4, opposite strand |
| 4 | gene | 128.5062 | 128.5334 | negative | MGI_C57BL6J_5592815 | Gm33656 | NCBI_Gene:102636648 | MGI:5592815 | lncRNA gene | predicted gene, 33656 |
| 4 | gene | 128.5787 | 128.6102 | negative | MGI_C57BL6J_2679268 | Zscan20 | NCBI_Gene:269585,ENSEMBL:ENSMUSG00000061894 | MGI:2679268 | protein coding gene | zinc finger and SCAN domains 20 |
| 4 | gene | 128.6154 | 128.6186 | negative | MGI_C57BL6J_3045221 | Tlr12 | NCBI_Gene:384059,ENSEMBL:ENSMUSG00000062545 | MGI:3045221 | protein coding gene | toll-like receptor 12 |
| 4 | gene | 128.6372 | 128.6392 | positive | MGI_C57BL6J_3652007 | Gm12958 | NCBI_Gene:105244695,ENSEMBL:ENSMUSG00000084891 | MGI:3652007 | lncRNA gene | predicted gene 12958 |
| 4 | gene | 128.6547 | 128.7529 | positive | MGI_C57BL6J_1860454 | Phc2 | NCBI_Gene:54383,ENSEMBL:ENSMUSG00000028796 | MGI:1860454 | protein coding gene | polyhomeotic 2 |
| 4 | gene | 128.7213 | 128.7275 | negative | MGI_C57BL6J_5592871 | Gm33712 | NA | NA | unclassified non-coding RNA gene | predicted gene%2c 33712 |
| 4 | gene | 128.7554 | 128.7702 | positive | MGI_C57BL6J_2685279 | A3galt2 | NCBI_Gene:215493,ENSEMBL:ENSMUSG00000028794 | MGI:2685279 | protein coding gene | alpha 1,3-galactosyltransferase 2 (isoglobotriaosylceramide synthase) |
| 4 | gene | 128.7731 | 128.8061 | negative | MGI_C57BL6J_2652839 | Zfp362 | NCBI_Gene:230761,ENSEMBL:ENSMUSG00000028799 | MGI:2652839 | protein coding gene | zinc finger protein 362 |
| 4 | gene | 128.8462 | 128.8737 | positive | MGI_C57BL6J_3649345 | Gm12968 | ENSEMBL:ENSMUSG00000085701 | MGI:3649345 | lncRNA gene | predicted gene 12968 |
| 4 | pseudogene | 128.8680 | 128.8694 | negative | MGI_C57BL6J_3650134 | Gm12969 | NCBI_Gene:667354,ENSEMBL:ENSMUSG00000084011 | MGI:3650134 | pseudogene | predicted gene 12969 |
| 4 | gene | 128.8720 | 128.8774 | negative | MGI_C57BL6J_5592973 | Gm33814 | NCBI_Gene:102636861 | MGI:5592973 | lncRNA gene | predicted gene, 33814 |
| 4 | gene | 128.8776 | 128.8823 | positive | MGI_C57BL6J_5592919 | Gm33760 | NA | NA | unclassified non-coding RNA gene | predicted gene%2c 33760 |
| 4 | gene | 128.8817 | 128.9113 | positive | MGI_C57BL6J_1914775 | Trim62 | NCBI_Gene:67525,ENSEMBL:ENSMUSG00000041000 | MGI:1914775 | protein coding gene | tripartite motif-containing 62 |
| 4 | gene | 128.9173 | 128.9223 | positive | MGI_C57BL6J_5593060 | Gm33901 | NCBI_Gene:102636978 | MGI:5593060 | lncRNA gene | predicted gene, 33901 |
| 4 | gene | 128.9302 | 128.9625 | negative | MGI_C57BL6J_2442093 | Azin2 | NCBI_Gene:242669,ENSEMBL:ENSMUSG00000028789 | MGI:2442093 | protein coding gene | antizyme inhibitor 2 |
| 4 | gene | 128.9360 | 128.9402 | positive | MGI_C57BL6J_3801803 | Gm15904 | NCBI_Gene:105244696,ENSEMBL:ENSMUSG00000085961 | MGI:3801803 | lncRNA gene | predicted gene 15904 |
| 4 | gene | 128.9920 | 129.0115 | positive | MGI_C57BL6J_87978 | Ak2 | NCBI_Gene:11637,ENSEMBL:ENSMUSG00000028792 | MGI:87978 | protein coding gene | adenylate kinase 2 |
| 4 | gene | 129.0501 | 129.0581 | negative | MGI_C57BL6J_1914599 | 1700086P04Rik | NCBI_Gene:67349,ENSEMBL:ENSMUSG00000085085 | MGI:1914599 | lncRNA gene | RIKEN cDNA 1700086P04 gene |
| 4 | gene | 129.0581 | 129.0859 | positive | MGI_C57BL6J_1922484 | Rnf19b | NCBI_Gene:75234,ENSEMBL:ENSMUSG00000028793 | MGI:1922484 | protein coding gene | ring finger protein 19B |
| 4 | gene | 129.1055 | 129.1116 | positive | MGI_C57BL6J_1913510 | Tmem54 | NCBI_Gene:66260,ENSEMBL:ENSMUSG00000028786 | MGI:1913510 | protein coding gene | transmembrane protein 54 |
| 4 | gene | 129.1116 | 129.1221 | negative | MGI_C57BL6J_1336200 | Hpca | NCBI_Gene:15444,ENSEMBL:ENSMUSG00000028785 | MGI:1336200 | protein coding gene | hippocalcin |
| 4 | gene | 129.1351 | 129.1369 | positive | MGI_C57BL6J_3801906 | Gm15906 | ENSEMBL:ENSMUSG00000087416 | MGI:3801906 | lncRNA gene | predicted gene 15906 |
| 4 | gene | 129.1369 | 129.1446 | positive | MGI_C57BL6J_1917614 | Fndc5 | NCBI_Gene:384061,ENSEMBL:ENSMUSG00000001334 | MGI:1917614 | protein coding gene | fibronectin type III domain containing 5 |
| 4 | gene | 129.1480 | 129.1897 | negative | MGI_C57BL6J_1921898 | S100pbp | NCBI_Gene:74648,ENSEMBL:ENSMUSG00000040928 | MGI:1921898 | protein coding gene | S100P binding protein |
| 4 | gene | 129.1898 | 129.2196 | positive | MGI_C57BL6J_2147627 | Yars | NCBI_Gene:107271,ENSEMBL:ENSMUSG00000028811 | MGI:2147627 | protein coding gene | tyrosyl-tRNA synthetase |
| 4 | gene | 129.2196 | 129.2614 | negative | MGI_C57BL6J_2140651 | C77080 | NCBI_Gene:97130,ENSEMBL:ENSMUSG00000050390 | MGI:2140651 | protein coding gene | expressed sequence C77080 |
| 4 | gene | 129.2394 | 129.2868 | positive | MGI_C57BL6J_3650499 | Gm12976 | NCBI_Gene:102637131,ENSEMBL:ENSMUSG00000087575 | MGI:3650499 | lncRNA gene | predicted gene 12976 |
| 4 | gene | 129.2829 | 129.2829 | negative | MGI_C57BL6J_5452771 | Gm22994 | ENSEMBL:ENSMUSG00000093163 | MGI:5452771 | miRNA gene | predicted gene, 22994 |
| 4 | gene | 129.2875 | 129.3094 | positive | MGI_C57BL6J_1916078 | Sync | NCBI_Gene:68828,ENSEMBL:ENSMUSG00000001333 | MGI:1916078 | protein coding gene | syncoilin |
| 4 | gene | 129.3071 | 129.3354 | negative | MGI_C57BL6J_1194912 | Rbbp4 | NCBI_Gene:19646,ENSEMBL:ENSMUSG00000057236 | MGI:1194912 | protein coding gene | retinoblastoma binding protein 4, chromatin remodeling factor |
| 4 | gene | 129.3091 | 129.3184 | positive | MGI_C57BL6J_5477216 | Gm26722 | ENSEMBL:ENSMUSG00000096944 | MGI:5477216 | lncRNA gene | predicted gene, 26722 |
| 4 | gene | 129.3356 | 129.3545 | positive | MGI_C57BL6J_1914356 | Zbtb8os | NCBI_Gene:67106,ENSEMBL:ENSMUSG00000057572 | MGI:1914356 | protein coding gene | zinc finger and BTB domain containing 8 opposite strand |
| 4 | gene | 129.3536 | 129.3781 | negative | MGI_C57BL6J_1920930 | Zbtb8a | NCBI_Gene:73680,ENSEMBL:ENSMUSG00000028807 | MGI:1920930 | protein coding gene | zinc finger and BTB domain containing 8a |
| 4 | pseudogene | 129.3798 | 129.3802 | positive | MGI_C57BL6J_3651320 | Gm12980 | ENSEMBL:ENSMUSG00000082468 | MGI:3651320 | pseudogene | predicted gene 12980 |
| 4 | gene | 129.3825 | 129.4075 | positive | MGI_C57BL6J_2444986 | C330020E22Rik | NCBI_Gene:320896 | MGI:2444986 | lncRNA gene | RIKEN cDNA C330020E22 gene |
| 4 | gene | 129.4258 | 129.4468 | negative | MGI_C57BL6J_2387181 | Zbtb8b | NCBI_Gene:215627,ENSEMBL:ENSMUSG00000048485 | MGI:2387181 | protein coding gene | zinc finger and BTB domain containing 8b |
| 4 | gene | 129.4584 | 129.4585 | positive | MGI_C57BL6J_5454398 | Gm24621 | ENSEMBL:ENSMUSG00000064613 | MGI:5454398 | snRNA gene | predicted gene, 24621 |
| 4 | gene | 129.4602 | 129.4615 | negative | MGI_C57BL6J_1918074 | 4921504P20Rik | NA | NA | unclassified gene | RIKEN cDNA 4921504P20 gene |
| 4 | gene | 129.4616 | 129.4885 | positive | MGI_C57BL6J_1913466 | Bsdc1 | NCBI_Gene:100383,ENSEMBL:ENSMUSG00000040859 | MGI:1913466 | protein coding gene | BSD domain containing 1 |
| 4 | gene | 129.4890 | 129.4910 | negative | MGI_C57BL6J_1929914 | Tssk3 | NCBI_Gene:58864,ENSEMBL:ENSMUSG00000000411 | MGI:1929914 | protein coding gene | testis-specific serine kinase 3 |
| 4 | gene | 129.4912 | 129.4920 | positive | MGI_C57BL6J_1915483 | Fam229a | NCBI_Gene:68233,ENSEMBL:ENSMUSG00000078554 | MGI:1915483 | protein coding gene | family with sequence similarity 229, member A |
| 4 | gene | 129.4920 | 129.5097 | negative | MGI_C57BL6J_3651303 | Gm12979 | ENSEMBL:ENSMUSG00000086336 | MGI:3651303 | lncRNA gene | predicted gene 12979 |
| 4 | gene | 129.5136 | 129.5160 | positive | MGI_C57BL6J_97143 | Marcksl1 | NCBI_Gene:17357,ENSEMBL:ENSMUSG00000047945 | MGI:97143 | protein coding gene | MARCKS-like 1 |
| 4 | gene | 129.5161 | 129.5427 | negative | MGI_C57BL6J_108086 | Hdac1 | NCBI_Gene:433759,ENSEMBL:ENSMUSG00000028800 | MGI:108086 | protein coding gene | histone deacetylase 1 |
| 4 | gene | 129.5483 | 129.5736 | negative | MGI_C57BL6J_96756 | Lck | NCBI_Gene:16818,ENSEMBL:ENSMUSG00000000409 | MGI:96756 | protein coding gene | lymphocyte protein tyrosine kinase |
| 4 | gene | 129.5577 | 129.5578 | negative | MGI_C57BL6J_5530949 | Mir8119 | miRBase:MI0026051,NCBI_Gene:102465997,ENSEMBL:ENSMUSG00000099070 | MGI:5530949 | miRNA gene | microRNA 8119 |
| 4 | gene | 129.5768 | 129.5786 | negative | MGI_C57BL6J_2668032 | Fam167b | NCBI_Gene:230766,ENSEMBL:ENSMUSG00000050493 | MGI:2668032 | protein coding gene | family with sequence similarity 167, member B |
| 4 | gene | 129.5920 | 129.6006 | negative | MGI_C57BL6J_1860763 | Eif3i | NCBI_Gene:54709,ENSEMBL:ENSMUSG00000028798 | MGI:1860763 | protein coding gene | eukaryotic translation initiation factor 3, subunit I |
| 4 | gene | 129.6007 | 129.6241 | positive | MGI_C57BL6J_1924049 | Tmem234 | NCBI_Gene:76799,ENSEMBL:ENSMUSG00000028797 | MGI:1924049 | protein coding gene | transmembrane protein 234 |
| 4 | gene | 129.6083 | 129.6145 | negative | MGI_C57BL6J_2686212 | Dcdc2b | NCBI_Gene:100504491,ENSEMBL:ENSMUSG00000078552 | MGI:2686212 | protein coding gene | doublecortin domain containing 2b |
| 4 | gene | 129.6151 | 129.6191 | negative | MGI_C57BL6J_2446212 | Iqcc | NCBI_Gene:230767,ENSEMBL:ENSMUSG00000040795 | MGI:2446212 | protein coding gene | IQ motif containing C |
| 4 | gene | 129.6193 | 129.6239 | negative | MGI_C57BL6J_1913514 | Ccdc28b | NCBI_Gene:66264,ENSEMBL:ENSMUSG00000028795 | MGI:1913514 | protein coding gene | coiled coil domain containing 28B |
| 4 | gene | 129.6231 | 129.6241 | positive | MGI_C57BL6J_5593464 | Gm34305 | NCBI_Gene:102637517 | MGI:5593464 | lncRNA gene | predicted gene, 34305 |
| 4 | gene | 129.6243 | 129.6251 | negative | MGI_C57BL6J_1920034 | 2810452K05Rik | NA | NA | unclassified gene | RIKEN cDNA 2810452K05 gene |
| 4 | gene | 129.6261 | 129.6411 | negative | MGI_C57BL6J_105968 | Txlna | NCBI_Gene:109658,ENSEMBL:ENSMUSG00000053841 | MGI:105968 | protein coding gene | taxilin alpha |
| 4 | gene | 129.6440 | 129.6728 | negative | MGI_C57BL6J_1100836 | Kpna6 | NCBI_Gene:16650,ENSEMBL:ENSMUSG00000003731 | MGI:1100836 | protein coding gene | karyopherin (importin) alpha 6 |
| 4 | gene | 129.6764 | 129.6968 | negative | MGI_C57BL6J_2682939 | Tmem39b | NCBI_Gene:230770,ENSEMBL:ENSMUSG00000053730 | MGI:2682939 | protein coding gene | transmembrane protein 39b |
| 4 | gene | 129.6845 | 129.6846 | negative | MGI_C57BL6J_5531376 | Mir7016 | miRBase:MI0022865,NCBI_Gene:102465615,ENSEMBL:ENSMUSG00000098422 | MGI:5531376 | miRNA gene | microRNA 7016 |
| 4 | gene | 129.7024 | 129.7428 | negative | MGI_C57BL6J_893579 | Khdrbs1 | NCBI_Gene:20218,ENSEMBL:ENSMUSG00000028790 | MGI:893579 | protein coding gene | KH domain containing, RNA binding, signal transduction associated 1 |
| 4 | pseudogene | 129.7616 | 129.7619 | negative | MGI_C57BL6J_3652042 | Gm12967 | NCBI_Gene:100040532,ENSEMBL:ENSMUSG00000080944 | MGI:3652042 | pseudogene | predicted gene 12967 |
| 4 | gene | 129.7623 | 129.7624 | negative | MGI_C57BL6J_5531399 | Gm28017 | ENSEMBL:ENSMUSG00000099322 | MGI:5531399 | miRNA gene | predicted gene, 28017 |
| 4 | gene | 129.8112 | 129.8500 | positive | MGI_C57BL6J_1277117 | Ptp4a2 | NCBI_Gene:19244,ENSEMBL:ENSMUSG00000028788 | MGI:1277117 | protein coding gene | protein tyrosine phosphatase 4a2 |
| 4 | gene | 129.8207 | 129.8221 | positive | MGI_C57BL6J_1923343 | 5830469G19Rik | NA | NA | unclassified gene | RIKEN cDNA 5830469G19 gene |
| 4 | gene | 129.8525 | 129.8537 | negative | MGI_C57BL6J_1921548 | 1700108I11Rik | NCBI_Gene:105244698 | MGI:1921548 | lncRNA gene | RIKEN cDNA 1700108I11 gene |
| 4 | pseudogene | 129.8551 | 129.8567 | positive | MGI_C57BL6J_3708701 | Gm10571 | ENSEMBL:ENSMUSG00000073753 | MGI:3708701 | pseudogene | predicted gene 10571 |
| 4 | gene | 129.8956 | 129.8957 | positive | MGI_C57BL6J_5453622 | Gm23845 | ENSEMBL:ENSMUSG00000065768 | MGI:5453622 | snRNA gene | predicted gene, 23845 |
| 4 | pseudogene | 129.8989 | 129.9004 | positive | MGI_C57BL6J_3652043 | Gm12966 | NCBI_Gene:622469,ENSEMBL:ENSMUSG00000070729 | MGI:3652043 | pseudogene | predicted gene 12966 |
| 4 | gene | 129.9062 | 129.9198 | positive | MGI_C57BL6J_2442924 | E330017L17Rik | NCBI_Gene:319894,ENSEMBL:ENSMUSG00000086369 | MGI:2442924 | lncRNA gene | RIKEN cDNA E330017L17 gene |
| 4 | gene | 129.9288 | 129.9571 | positive | MGI_C57BL6J_3652045 | Spocd1 | NCBI_Gene:622480,ENSEMBL:ENSMUSG00000028784 | MGI:3652045 | protein coding gene | SPOC domain containing 1 |
| 4 | gene | 129.9604 | 129.9651 | positive | MGI_C57BL6J_1919475 | 1700003M07Rik | NCBI_Gene:72225,ENSEMBL:ENSMUSG00000085389 | MGI:1919475 | lncRNA gene | RIKEN cDNA 1700003M07 gene |
| 4 | gene | 129.9849 | 130.0226 | positive | MGI_C57BL6J_2451244 | Adgrb2 | NCBI_Gene:230775,ENSEMBL:ENSMUSG00000028782 | MGI:2451244 | protein coding gene | adhesion G protein-coupled receptor B2 |
| 4 | gene | 130.0230 | 130.0497 | negative | MGI_C57BL6J_3651569 | Gm12963 | NCBI_Gene:102640839,ENSEMBL:ENSMUSG00000085517 | MGI:3651569 | lncRNA gene | predicted gene 12963 |
| 4 | gene | 130.0474 | 130.0993 | positive | MGI_C57BL6J_1095396 | Col16a1 | NCBI_Gene:107581,ENSEMBL:ENSMUSG00000040690 | MGI:1095396 | protein coding gene | collagen, type XVI, alpha 1 |
| 4 | gene | 130.0762 | 130.0795 | negative | MGI_C57BL6J_5826508 | Gm46871 | NCBI_Gene:108168980 | MGI:5826508 | lncRNA gene | predicted gene, 46871 |
| 4 | gene | 130.1026 | 130.1281 | positive | MGI_C57BL6J_1915148 | Pef1 | NCBI_Gene:67898,ENSEMBL:ENSMUSG00000028779 | MGI:1915148 | protein coding gene | penta-EF hand domain containing 1 |
| 4 | gene | 130.1055 | 130.1083 | negative | MGI_C57BL6J_5623143 | Gm40258 | NCBI_Gene:105244699 | MGI:5623143 | lncRNA gene | predicted gene, 40258 |
| 4 | gene | 130.1302 | 130.1397 | negative | MGI_C57BL6J_2385650 | Hcrtr1 | NCBI_Gene:230777,ENSEMBL:ENSMUSG00000028778 | MGI:2385650 | protein coding gene | hypocretin (orexin) receptor 1 |
| 4 | gene | 130.1440 | 130.1489 | positive | MGI_C57BL6J_5623144 | Gm40259 | NCBI_Gene:105244700 | MGI:5623144 | lncRNA gene | predicted gene, 40259 |
| 4 | gene | 130.1645 | 130.1751 | negative | MGI_C57BL6J_2137617 | Tinagl1 | NCBI_Gene:94242,ENSEMBL:ENSMUSG00000028776 | MGI:2137617 | protein coding gene | tubulointerstitial nephritis antigen-like 1 |
| 4 | gene | 130.2090 | 130.2224 | negative | MGI_C57BL6J_2685699 | Ldc1 | NCBI_Gene:332942,ENSEMBL:ENSMUSG00000023120 | MGI:2685699 | protein coding gene | leucine decarboxylase 1 |
| 4 | gene | 130.2366 | 130.2491 | positive | MGI_C57BL6J_5826361 | Gm46724 | NCBI_Gene:108168712 | MGI:5826361 | lncRNA gene | predicted gene, 46724 |
| 4 | gene | 130.2535 | 130.2792 | negative | MGI_C57BL6J_1919132 | Serinc2 | NCBI_Gene:230779,ENSEMBL:ENSMUSG00000023232 | MGI:1919132 | protein coding gene | serine incorporator 2 |
| 4 | gene | 130.2757 | 130.2809 | positive | MGI_C57BL6J_5594116 | Gm34957 | NCBI_Gene:102638376 | MGI:5594116 | lncRNA gene | predicted gene, 34957 |
| 4 | gene | 130.2912 | 130.3087 | negative | MGI_C57BL6J_3642427 | Gm10570 | NCBI_Gene:102638454,ENSEMBL:ENSMUSG00000073752 | MGI:3642427 | lncRNA gene | predicted gene 10570 |
| 4 | gene | 130.3086 | 130.3155 | positive | MGI_C57BL6J_95476 | Fabp3 | NCBI_Gene:14077,ENSEMBL:ENSMUSG00000028773 | MGI:95476 | protein coding gene | fatty acid binding protein 3, muscle and heart |
| 4 | gene | 130.3154 | 130.3600 | negative | MGI_C57BL6J_1919955 | Zcchc17 | NCBI_Gene:619605,ENSEMBL:ENSMUSG00000028772 | MGI:1919955 | protein coding gene | zinc finger, CCHC domain containing 17 |
| 4 | gene | 130.3177 | 130.3333 | positive | MGI_C57BL6J_5594233 | Gm35074 | NCBI_Gene:102638533 | MGI:5594233 | lncRNA gene | predicted gene, 35074 |
| 4 | gene | 130.3601 | 130.3900 | positive | MGI_C57BL6J_1913835 | Snrnp40 | NCBI_Gene:66585,ENSEMBL:ENSMUSG00000074088 | MGI:1913835 | protein coding gene | small nuclear ribonucleoprotein 40 (U5) |
| 4 | gene | 130.3710 | 130.3720 | negative | MGI_C57BL6J_1922417 | 4930527F18Rik | NA | NA | unclassified gene | RIKEN cDNA 4930527F18 gene |
| 4 | gene | 130.3799 | 130.3808 | positive | MGI_C57BL6J_1920110 | 2900017G11Rik | NA | NA | unclassified gene | RIKEN cDNA 2900017G11 gene |
| 4 | gene | 130.5301 | 130.5742 | negative | MGI_C57BL6J_1914399 | Nkain1 | NCBI_Gene:67149,ENSEMBL:ENSMUSG00000078532 | MGI:1914399 | protein coding gene | Na+/K+ transporting ATPase interacting 1 |
| 4 | gene | 130.5723 | 130.5731 | negative | MGI_C57BL6J_1916206 | 1500006G06Rik | NA | NA | unclassified gene | RIKEN cDNA 1500006G06 gene |
| 4 | gene | 130.6633 | 130.7816 | positive | MGI_C57BL6J_1931749 | Pum1 | NCBI_Gene:80912,ENSEMBL:ENSMUSG00000028580 | MGI:1931749 | protein coding gene | pumilio RNA-binding family member 1 |
| 4 | gene | 130.7097 | 130.7105 | positive | MGI_C57BL6J_1919888 | 2810017D21Rik | NA | NA | unclassified gene | RIKEN cDNA 2810017D21 gene |
| 4 | gene | 130.7496 | 130.7497 | positive | MGI_C57BL6J_3819558 | Snord85 | NCBI_Gene:100217460,ENSEMBL:ENSMUSG00000065196 | MGI:3819558 | snoRNA gene | small nucleolar RNA, C/D box 85 |
| 4 | gene | 130.7586 | 130.7624 | positive | MGI_C57BL6J_3650041 | Gm12971 | ENSEMBL:ENSMUSG00000087449 | MGI:3650041 | lncRNA gene | predicted gene 12971 |
| 4 | gene | 130.7663 | 130.7664 | positive | MGI_C57BL6J_5453868 | Gm24091 | ENSEMBL:ENSMUSG00000093793 | MGI:5453868 | snoRNA gene | predicted gene, 24091 |
| 4 | gene | 130.7748 | 130.7792 | negative | MGI_C57BL6J_5477010 | Gm26516 | ENSEMBL:ENSMUSG00000097158 | MGI:5477010 | lncRNA gene | predicted gene, 26516 |
| 4 | gene | 130.7757 | 130.7758 | positive | MGI_C57BL6J_5453488 | Gm23711 | ENSEMBL:ENSMUSG00000095119 | MGI:5453488 | snoRNA gene | predicted gene, 23711 |
| 4 | gene | 130.7845 | 130.7880 | positive | MGI_C57BL6J_3650043 | Gm12972 | NCBI_Gene:108168710,ENSEMBL:ENSMUSG00000086548 | MGI:3650043 | lncRNA gene | predicted gene 12972 |
| 4 | gene | 130.7925 | 130.8263 | positive | MGI_C57BL6J_1349163 | Sdc3 | NCBI_Gene:20970,ENSEMBL:ENSMUSG00000025743 | MGI:1349163 | protein coding gene | syndecan 3 |
| 4 | gene | 130.8116 | 130.8123 | negative | MGI_C57BL6J_3650040 | Gm12970 | ENSEMBL:ENSMUSG00000087451 | MGI:3650040 | lncRNA gene | predicted gene 12970 |
| 4 | gene | 130.8244 | 130.8263 | negative | MGI_C57BL6J_5477210 | Gm26716 | ENSEMBL:ENSMUSG00000097224 | MGI:5477210 | lncRNA gene | predicted gene, 26716 |
| 4 | gene | 130.8278 | 130.8342 | negative | MGI_C57BL6J_5625201 | Gm42316 | NCBI_Gene:105247171 | MGI:5625201 | lncRNA gene | predicted gene, 42316 |
| 4 | gene | 130.8578 | 130.8579 | negative | MGI_C57BL6J_5452437 | Gm22660 | ENSEMBL:ENSMUSG00000077381 | MGI:5452437 | snoRNA gene | predicted gene, 22660 |
| 4 | gene | 130.8710 | 130.8729 | positive | MGI_C57BL6J_5594453 | Gm35294 | NCBI_Gene:102638819 | MGI:5594453 | lncRNA gene | predicted gene, 35294 |
| 4 | gene | 130.9061 | 130.9146 | negative | MGI_C57BL6J_5621484 | Gm38599 | NCBI_Gene:102642196 | MGI:5621484 | lncRNA gene | predicted gene, 38599 |
| 4 | gene | 130.9129 | 130.9361 | positive | MGI_C57BL6J_108046 | Laptm5 | NCBI_Gene:16792,ENSEMBL:ENSMUSG00000028581 | MGI:108046 | protein coding gene | lysosomal-associated protein transmembrane 5 |
| 4 | gene | 130.9444 | 130.9555 | positive | MGI_C57BL6J_106591 | Matn1 | NCBI_Gene:17180,ENSEMBL:ENSMUSG00000040533 | MGI:106591 | protein coding gene | matrilin 1, cartilage matrix protein |
| 4 | gene | 130.9747 | 131.0013 | positive | MGI_C57BL6J_3650042 | Gm12973 | NCBI_Gene:108168713,ENSEMBL:ENSMUSG00000085804 | MGI:3650042 | lncRNA gene | predicted gene 12973 |
| 4 | gene | 131.0008 | 131.0105 | negative | MGI_C57BL6J_1925085 | A930031H19Rik | NCBI_Gene:77835,ENSEMBL:ENSMUSG00000086262 | MGI:1925085 | lncRNA gene | RIKEN cDNA A930031H19 gene |
| 4 | gene | 131.0639 | 131.0656 | negative | MGI_C57BL6J_5625202 | Gm42317 | NCBI_Gene:105247172 | MGI:5625202 | lncRNA gene | predicted gene, 42317 |
| 4 | gene | 131.0757 | 131.0803 | negative | MGI_C57BL6J_5594641 | Gm35482 | NCBI_Gene:102639082 | MGI:5594641 | lncRNA gene | predicted gene, 35482 |
| 4 | gene | 131.1563 | 131.1627 | positive | MGI_C57BL6J_5625203 | Gm42318 | NCBI_Gene:105247173 | MGI:5625203 | lncRNA gene | predicted gene, 42318 |
| 4 | gene | 131.2393 | 131.2394 | positive | MGI_C57BL6J_5455038 | Gm25261 | ENSEMBL:ENSMUSG00000088041 | MGI:5455038 | snoRNA gene | predicted gene, 25261 |
| 4 | gene | 131.4091 | 131.4283 | positive | MGI_C57BL6J_5594793 | Gm35634 | NCBI_Gene:102639287 | MGI:5594793 | lncRNA gene | predicted gene, 35634 |
| 4 | gene | 131.4563 | 131.4826 | positive | MGI_C57BL6J_5826360 | Gm46723 | NCBI_Gene:108168711 | MGI:5826360 | lncRNA gene | predicted gene, 46723 |
| 4 | gene | 131.4584 | 131.4855 | negative | MGI_C57BL6J_5594938 | Gm35779 | NCBI_Gene:102639475 | MGI:5594938 | lncRNA gene | predicted gene, 35779 |
| 4 | gene | 131.4872 | 131.5055 | positive | MGI_C57BL6J_2685677 | Gm831 | NCBI_Gene:329950,ENSEMBL:ENSMUSG00000078531 | MGI:2685677 | lncRNA gene | predicted gene 831 |
| 4 | gene | 131.6174 | 131.6351 | positive | MGI_C57BL6J_5625205 | Gm42320 | NCBI_Gene:105247175 | MGI:5625205 | lncRNA gene | predicted gene, 42320 |
| 4 | gene | 131.6470 | 131.6778 | negative | MGI_C57BL6J_3650901 | Gm12962 | ENSEMBL:ENSMUSG00000087171 | MGI:3650901 | lncRNA gene | predicted gene 12962 |
| 4 | gene | 131.6471 | 131.7105 | negative | MGI_C57BL6J_5625206 | Gm42321 | NCBI_Gene:105247176 | MGI:5625206 | lncRNA gene | predicted gene, 42321 |
| 4 | gene | 131.6946 | 131.7105 | negative | MGI_C57BL6J_3802056 | Gm16080 | ENSEMBL:ENSMUSG00000086856 | MGI:3802056 | lncRNA gene | predicted gene 16080 |
| 4 | gene | 131.7685 | 131.8383 | negative | MGI_C57BL6J_1321151 | Ptpru | NCBI_Gene:19273,ENSEMBL:ENSMUSG00000028909 | MGI:1321151 | protein coding gene | protein tyrosine phosphatase, receptor type, U |
| 4 | gene | 131.8434 | 131.8678 | positive | MGI_C57BL6J_1349441 | Mecr | NCBI_Gene:26922,ENSEMBL:ENSMUSG00000028910 | MGI:1349441 | protein coding gene | mitochondrial trans-2-enoyl-CoA reductase |
| 4 | gene | 131.8736 | 131.9017 | positive | MGI_C57BL6J_1890577 | Srsf4 | NCBI_Gene:57317,ENSEMBL:ENSMUSG00000028911 | MGI:1890577 | protein coding gene | serine/arginine-rich splicing factor 4 |
| 4 | gene | 131.8786 | 131.8832 | positive | MGI_C57BL6J_2442442 | A930004J17Rik | ENSEMBL:ENSMUSG00000102602 | MGI:2442442 | unclassified gene | RIKEN cDNA A930004J17 gene |
| 4 | gene | 131.8879 | 131.8894 | positive | MGI_C57BL6J_1918083 | 4733401A01Rik | NA | NA | unclassified gene | RIKEN cDNA 4733401A01 gene |
| 4 | gene | 131.8995 | 131.9200 | negative | MGI_C57BL6J_3702644 | Gm12992 | NCBI_Gene:545681,ENSEMBL:ENSMUSG00000085667 | MGI:3702644 | lncRNA gene | predicted gene 12992 |
| 4 | pseudogene | 131.9027 | 131.9033 | negative | MGI_C57BL6J_3705382 | Gm12978 | NCBI_Gene:100418326,ENSEMBL:ENSMUSG00000083013 | MGI:3705382 | pseudogene | predicted gene 12978 |
| 4 | gene | 131.9200 | 131.9240 | positive | MGI_C57BL6J_3646343 | Tmem200b | NCBI_Gene:623230,ENSEMBL:ENSMUSG00000070720 | MGI:3646343 | protein coding gene | transmembrane protein 200B |
| 4 | gene | 131.9208 | 131.9215 | negative | MGI_C57BL6J_6324745 | Gm50475 | ENSEMBL:ENSMUSG00000118394 | MGI:6324745 | lncRNA gene | predicted gene, 50475 |
| 4 | gene | 131.9234 | 132.0756 | negative | MGI_C57BL6J_95401 | Epb41 | NCBI_Gene:269587,ENSEMBL:ENSMUSG00000028906 | MGI:95401 | protein coding gene | erythrocyte membrane protein band 4.1 |
| 4 | gene | 131.9922 | 131.9923 | negative | MGI_C57BL6J_5530827 | Gm27445 | ENSEMBL:ENSMUSG00000098577 | MGI:5530827 | miRNA gene | predicted gene, 27445 |
| 4 | gene | 132.0482 | 132.0490 | positive | MGI_C57BL6J_3649474 | Gm13063 | ENSEMBL:ENSMUSG00000084869 | MGI:3649474 | lncRNA gene | predicted gene 13063 |
| 4 | gene | 132.0495 | 132.0498 | positive | MGI_C57BL6J_5595316 | Gm36157 | NCBI_Gene:102639968 | MGI:5595316 | lncRNA gene | predicted gene, 36157 |
| 4 | gene | 132.0747 | 132.0770 | positive | MGI_C57BL6J_3641670 | Gm10300 | ENSEMBL:ENSMUSG00000070717 | MGI:3641670 | protein coding gene | predicted gene 10300 |
| 4 | pseudogene | 132.0939 | 132.0943 | positive | MGI_C57BL6J_3650812 | Gm13214 | ENSEMBL:ENSMUSG00000083368 | MGI:3650812 | pseudogene | predicted gene 13214 |
| 4 | gene | 132.1048 | 132.1094 | negative | MGI_C57BL6J_5610939 | Gm37711 | ENSEMBL:ENSMUSG00000102796 | MGI:5610939 | unclassified gene | predicted gene, 37711 |
| 4 | gene | 132.1077 | 132.1445 | negative | MGI_C57BL6J_97438 | Oprd1 | NCBI_Gene:18386,ENSEMBL:ENSMUSG00000050511 | MGI:97438 | protein coding gene | opioid receptor, delta 1 |
| 4 | gene | 132.1253 | 132.1253 | positive | MGI_C57BL6J_4413695 | n-TAcgc3 | NCBI_Gene:102467236 | MGI:4413695 | tRNA gene | nuclear encoded tRNA alanine 3 (anticodon CGC) |
| 4 | pseudogene | 132.1484 | 132.1489 | negative | MGI_C57BL6J_3650813 | Gm13215 | NCBI_Gene:664894,ENSEMBL:ENSMUSG00000081394 | MGI:3650813 | pseudogene | predicted gene 13215 |
| 4 | gene | 132.1849 | 132.2123 | negative | MGI_C57BL6J_2444233 | Ythdf2 | NCBI_Gene:213541,ENSEMBL:ENSMUSG00000040025 | MGI:2444233 | protein coding gene | YTH N6-methyladenosine RNA binding protein 2 |
| 4 | gene | 132.2184 | 132.2185 | negative | MGI_C57BL6J_5451810 | Gm22033 | ENSEMBL:ENSMUSG00000080597 | MGI:5451810 | miRNA gene | predicted gene, 22033 |
| 4 | pseudogene | 132.2199 | 132.2206 | negative | MGI_C57BL6J_3652187 | Rps15a-ps4 | NCBI_Gene:664903,ENSEMBL:ENSMUSG00000083757 | MGI:3652187 | pseudogene | ribosomal protein S15A, pseudogene 4 |
| 4 | gene | 132.2210 | 132.2616 | negative | MGI_C57BL6J_2135604 | Gmeb1 | NCBI_Gene:56809,ENSEMBL:ENSMUSG00000028901 | MGI:2135604 | protein coding gene | glucocorticoid modulatory element binding protein 1 |
| 4 | pseudogene | 132.2447 | 132.2460 | negative | MGI_C57BL6J_3651953 | Gm13252 | NCBI_Gene:105247177,ENSEMBL:ENSMUSG00000084105 | MGI:3651953 | pseudogene | predicted gene 13252 |
| 4 | gene | 132.2605 | 132.2688 | positive | MGI_C57BL6J_5579580 | Gm28874 | NCBI_Gene:102640216,ENSEMBL:ENSMUSG00000100813 | MGI:5579580 | lncRNA gene | predicted gene 28874 |
| 4 | gene | 132.2701 | 132.2702 | positive | MGI_C57BL6J_5579578 | Gm28872 | ENSEMBL:ENSMUSG00000099937 | MGI:5579578 | lncRNA gene | predicted gene 28872 |
| 4 | gene | 132.2701 | 132.2702 | negative | MGI_C57BL6J_2148804 | Rnu11 | NCBI_Gene:353373,ENSEMBL:ENSMUSG00000077323 | MGI:2148804 | snRNA gene | U11 small nuclear RNA |
| 4 | gene | 132.2741 | 132.2743 | negative | MGI_C57BL6J_3629726 | Gt(pU21)115Imeg | NA | NA | unclassified gene | gene trap 115%2c Institute of Molecular Embryology and Genetics |
| 4 | gene | 132.2744 | 132.2958 | positive | MGI_C57BL6J_1913714 | Taf12 | NCBI_Gene:66464,ENSEMBL:ENSMUSG00000028899 | MGI:1913714 | protein coding gene | TATA-box binding protein associated factor 12 |
| 4 | gene | 132.3004 | 132.3034 | negative | MGI_C57BL6J_2441753 | Rab42 | NCBI_Gene:242681,ENSEMBL:ENSMUSG00000089687 | MGI:2441753 | protein coding gene | RAB42, member RAS oncogene family |
| 4 | gene | 132.3086 | 132.3110 | positive | MGI_C57BL6J_1916721 | Snhg12 | NCBI_Gene:100039864,ENSEMBL:ENSMUSG00000086290 | MGI:1916721 | lncRNA gene | small nucleolar RNA host gene 12 |
| 4 | gene | 132.3095 | 132.3096 | positive | MGI_C57BL6J_4361141 | Snora16a | NCBI_Gene:100310813,ENSEMBL:ENSMUSG00000065097,ENSEMBL:ENSMUSG00000105911 | MGI:4361141 | snoRNA gene | small nucleolar RNA, H/ACA box 16A |
| 4 | gene | 132.3099 | 132.3101 | positive | MGI_C57BL6J_3819506 | Snora44 | NCBI_Gene:100217418,ENSEMBL:ENSMUSG00000064604 | MGI:3819506 | snoRNA gene | small nucleolar RNA, H/ACA box 44 |
| 4 | gene | 132.3102 | 132.3104 | positive | MGI_C57BL6J_3819510 | Snora61 | NCBI_Gene:100217440,ENSEMBL:ENSMUSG00000064949 | MGI:3819510 | snoRNA gene | small nucleolar RNA, H/ACA box 61 |
| 4 | gene | 132.3107 | 132.3108 | positive | MGI_C57BL6J_3819570 | Snord99 | NCBI_Gene:100217437,ENSEMBL:ENSMUSG00000080615 | MGI:3819570 | snoRNA gene | small nucleolar RNA, C/D box 99 |
| 4 | gene | 132.3118 | 132.3295 | negative | MGI_C57BL6J_1919037 | Trnau1ap | NCBI_Gene:71787,ENSEMBL:ENSMUSG00000028898 | MGI:1919037 | protein coding gene | tRNA selenocysteine 1 associated protein 1 |
| 4 | gene | 132.3319 | 132.3536 | negative | MGI_C57BL6J_1913989 | Rcc1 | NCBI_Gene:100088,ENSEMBL:ENSMUSG00000028896 | MGI:1913989 | protein coding gene | regulator of chromosome condensation 1 |
| 4 | gene | 132.3480 | 132.3537 | negative | MGI_C57BL6J_2684817 | Snhg3 | NCBI_Gene:399101,ENSEMBL:ENSMUSG00000085241 | MGI:2684817 | lncRNA gene | small nucleolar RNA host gene 3 |
| 4 | gene | 132.3518 | 132.3534 | positive | MGI_C57BL6J_4936934 | Gm17300 | ENSEMBL:ENSMUSG00000091021 | MGI:4936934 | lncRNA gene | predicted gene, 17300 |
| 4 | gene | 132.3523 | 132.3525 | negative | MGI_C57BL6J_4360049 | Snora73b | NCBI_Gene:100306945,ENSEMBL:ENSMUSG00000065353 | MGI:4360049 | snoRNA gene | small nucleolar RNA, H/ACA box 73b |
| 4 | gene | 132.3528 | 132.3530 | negative | MGI_C57BL6J_4360046 | Snora73a | NCBI_Gene:100306944,ENSEMBL:ENSMUSG00000064387 | MGI:4360046 | snoRNA gene | small nucleolar RNA, H/ACA box 73a |
| 4 | gene | 132.3559 | 132.4225 | negative | MGI_C57BL6J_2140327 | Phactr4 | NCBI_Gene:100169,ENSEMBL:ENSMUSG00000066043 | MGI:2140327 | protein coding gene | phosphatase and actin regulator 4 |
| 4 | gene | 132.4083 | 132.4084 | positive | MGI_C57BL6J_5455484 | Gm25707 | ENSEMBL:ENSMUSG00000096350 | MGI:5455484 | miRNA gene | predicted gene, 25707 |
| 4 | gene | 132.4420 | 132.4421 | positive | MGI_C57BL6J_5452832 | Gm23055 | ENSEMBL:ENSMUSG00000064739 | MGI:5452832 | snRNA gene | predicted gene, 23055 |
| 4 | gene | 132.4587 | 132.4639 | negative | MGI_C57BL6J_1914469 | Med18 | NCBI_Gene:67219,ENSEMBL:ENSMUSG00000066042 | MGI:1914469 | protein coding gene | mediator complex subunit 18 |
| 4 | gene | 132.4920 | 132.5105 | negative | MGI_C57BL6J_2651874 | Sesn2 | NCBI_Gene:230784,ENSEMBL:ENSMUSG00000028893 | MGI:2651874 | protein coding gene | sestrin 2 |
| 4 | gene | 132.5036 | 132.5096 | negative | MGI_C57BL6J_3651528 | Gm12981 | ENSEMBL:ENSMUSG00000086997 | MGI:3651528 | lncRNA gene | predicted gene 12981 |
| 4 | gene | 132.5160 | 132.5161 | positive | MGI_C57BL6J_4413681 | n-TAagc8 | NCBI_Gene:102467230 | MGI:4413681 | tRNA gene | nuclear encoded tRNA alanine 8 (anticodon AGC) |
| 4 | gene | 132.5306 | 132.5354 | negative | MGI_C57BL6J_1196457 | Atpif1 | NCBI_Gene:11983,ENSEMBL:ENSMUSG00000054428 | MGI:1196457 | protein coding gene | ATPase inhibitory factor 1 |
| 4 | gene | 132.5327 | 132.5336 | positive | MGI_C57BL6J_3651167 | Gm12999 | ENSEMBL:ENSMUSG00000087352 | MGI:3651167 | lncRNA gene | predicted gene 12999 |
| 4 | gene | 132.5356 | 132.5537 | positive | MGI_C57BL6J_1915848 | Dnajc8 | NCBI_Gene:68598,ENSEMBL:ENSMUSG00000054405 | MGI:1915848 | protein coding gene | DnaJ heat shock protein family (Hsp40) member C8 |
| 4 | gene | 132.5471 | 132.5472 | negative | MGI_C57BL6J_5454539 | Gm24762 | ENSEMBL:ENSMUSG00000065623 | MGI:5454539 | snRNA gene | predicted gene, 24762 |
| 4 | gene | 132.5641 | 132.5827 | positive | MGI_C57BL6J_106066 | Ptafr | NCBI_Gene:19204,ENSEMBL:ENSMUSG00000056529 | MGI:106066 | protein coding gene | platelet-activating factor receptor |
| 4 | gene | 132.6390 | 132.7248 | positive | MGI_C57BL6J_109339 | Eya3 | NCBI_Gene:14050,ENSEMBL:ENSMUSG00000028886 | MGI:109339 | protein coding gene | EYA transcriptional coactivator and phosphatase 3 |
| 4 | gene | 132.6391 | 132.6403 | positive | MGI_C57BL6J_1923270 | 5830409B07Rik | NA | NA | unclassified gene | RIKEN cDNA 5830409B07 gene |
| 4 | gene | 132.7249 | 132.7326 | negative | MGI_C57BL6J_2685877 | Xkr8 | NCBI_Gene:381560,ENSEMBL:ENSMUSG00000037752 | MGI:2685877 | protein coding gene | X-linked Kx blood group related 8 |
| 4 | gene | 132.7330 | 132.7573 | negative | MGI_C57BL6J_1916022 | Smpdl3b | NCBI_Gene:100340,ENSEMBL:ENSMUSG00000028885 | MGI:1916022 | protein coding gene | sphingomyelin phosphodiesterase, acid-like 3B |
| 4 | gene | 132.7683 | 132.7788 | positive | MGI_C57BL6J_1339939 | Rpa2 | NCBI_Gene:19891,ENSEMBL:ENSMUSG00000028884 | MGI:1339939 | protein coding gene | replication protein A2 |
| 4 | gene | 132.7818 | 132.7964 | negative | MGI_C57BL6J_2446213 | Themis2 | NCBI_Gene:230787,ENSEMBL:ENSMUSG00000037731 | MGI:2446213 | protein coding gene | thymocyte selection associated family member 2 |
| 4 | pseudogene | 132.8020 | 132.8023 | negative | MGI_C57BL6J_3651775 | Gm13022 | ENSEMBL:ENSMUSG00000081647 | MGI:3651775 | pseudogene | predicted gene 13022 |
| 4 | gene | 132.8269 | 132.8432 | negative | MGI_C57BL6J_2140494 | Ppp1r8 | NCBI_Gene:100336,ENSEMBL:ENSMUSG00000028882 | MGI:2140494 | protein coding gene | protein phosphatase 1, regulatory subunit 8 |
| 4 | gene | 132.8384 | 132.8385 | negative | MGI_C57BL6J_5452544 | Gm22767 | ENSEMBL:ENSMUSG00000088990 | MGI:5452544 | snoRNA gene | predicted gene, 22767 |
| 4 | gene | 132.8535 | 132.8845 | negative | MGI_C57BL6J_1931027 | Stx12 | NCBI_Gene:100226,ENSEMBL:ENSMUSG00000028879 | MGI:1931027 | protein coding gene | syntaxin 12 |
| 4 | gene | 132.8822 | 132.8833 | negative | MGI_C57BL6J_1924875 | C530007A02Rik | NA | NA | unclassified gene | RIKEN cDNA C530007A02 gene |
| 4 | pseudogene | 132.8848 | 132.8866 | positive | MGI_C57BL6J_3649305 | Gm13033 | NCBI_Gene:100038703,ENSEMBL:ENSMUSG00000083816 | MGI:3649305 | pseudogene | predicted gene 13033 |
| 4 | gene | 132.8992 | 132.9231 | negative | MGI_C57BL6J_2385211 | Fam76a | NCBI_Gene:230789,ENSEMBL:ENSMUSG00000028878 | MGI:2385211 | protein coding gene | family with sequence similarity 76, member A |
| 4 | gene | 132.9312 | 132.9313 | negative | MGI_C57BL6J_5454690 | Gm24913 | ENSEMBL:ENSMUSG00000087911 | MGI:5454690 | snRNA gene | predicted gene, 24913 |
| 4 | gene | 132.9484 | 132.9607 | negative | MGI_C57BL6J_5595677 | Gm36518 | NCBI_Gene:102640462 | MGI:5595677 | lncRNA gene | predicted gene, 36518 |
| 4 | gene | 132.9741 | 133.0019 | positive | MGI_C57BL6J_95527 | Fgr | NCBI_Gene:14191,ENSEMBL:ENSMUSG00000028874 | MGI:95527 | protein coding gene | FGR proto-oncogene, Src family tyrosine kinase |
| 4 | gene | 133.0027 | 133.0082 | negative | MGI_C57BL6J_5595758 | Gm36599 | NCBI_Gene:102640565 | MGI:5595758 | lncRNA gene | predicted gene, 36599 |
| 4 | gene | 133.0113 | 133.0781 | positive | MGI_C57BL6J_2444218 | Ahdc1 | NCBI_Gene:230793,ENSEMBL:ENSMUSG00000037692 | MGI:2444218 | protein coding gene | AT hook, DNA binding motif, containing 1 |
| 4 | gene | 133.0834 | 133.0891 | positive | MGI_C57BL6J_5477109 | Gm26615 | NCBI_Gene:102640635,ENSEMBL:ENSMUSG00000097088 | MGI:5477109 | lncRNA gene | predicted gene, 26615 |
| 4 | gene | 133.1145 | 133.1146 | negative | MGI_C57BL6J_4413870 | n-TGgcc1 | NCBI_Gene:102467429 | MGI:4413870 | tRNA gene | nuclear encoded tRNA glycine 1 (anticodon GCC) |
| 4 | gene | 133.1305 | 133.1998 | positive | MGI_C57BL6J_1098641 | Wasf2 | NCBI_Gene:242687,ENSEMBL:ENSMUSG00000028868 | MGI:1098641 | protein coding gene | WAS protein family, member 2 |
| 4 | gene | 133.1714 | 133.1717 | negative | MGI_C57BL6J_5454413 | Gm24636 | ENSEMBL:ENSMUSG00000089298 | MGI:5454413 | unclassified non-coding RNA gene | predicted gene, 24636 |
| 4 | gene | 133.1957 | 133.1957 | positive | MGI_C57BL6J_5531327 | Mir7017 | miRBase:MI0022866,NCBI_Gene:102466215,ENSEMBL:ENSMUSG00000099202 | MGI:5531327 | miRNA gene | microRNA 7017 |
| 4 | gene | 133.2093 | 133.2125 | negative | MGI_C57BL6J_101908 | Gpr3 | NCBI_Gene:14748,ENSEMBL:ENSMUSG00000049649 | MGI:101908 | protein coding gene | G-protein coupled receptor 3 |
| 4 | gene | 133.2121 | 133.2246 | positive | MGI_C57BL6J_1916905 | Cd164l2 | NCBI_Gene:69655,ENSEMBL:ENSMUSG00000028865 | MGI:1916905 | protein coding gene | CD164 sialomucin-like 2 |
| 4 | pseudogene | 133.2315 | 133.2393 | positive | MGI_C57BL6J_3510254 | Fcnc-ps | NCBI_Gene:493918 | MGI:3510254 | pseudogene | ficolin C pseudogene |
| 4 | gene | 133.2397 | 133.2529 | positive | MGI_C57BL6J_1855691 | Map3k6 | NCBI_Gene:53608,ENSEMBL:ENSMUSG00000028862 | MGI:1855691 | protein coding gene | mitogen-activated protein kinase kinase kinase 6 |
| 4 | gene | 133.2531 | 133.2631 | negative | MGI_C57BL6J_1933365 | Sytl1 | NCBI_Gene:269589,ENSEMBL:ENSMUSG00000028860 | MGI:1933365 | protein coding gene | synaptotagmin-like 1 |
| 4 | gene | 133.2660 | 133.2778 | negative | MGI_C57BL6J_1098568 | Tmem222 | NCBI_Gene:52174,ENSEMBL:ENSMUSG00000028857 | MGI:1098568 | protein coding gene | transmembrane protein 222 |
| 4 | pseudogene | 133.2809 | 133.2813 | negative | MGI_C57BL6J_3700962 | Gm13259 | ENSEMBL:ENSMUSG00000083241 | MGI:3700962 | pseudogene | predicted gene 13259 |
| 4 | gene | 133.2925 | 133.3535 | negative | MGI_C57BL6J_2685541 | Wdtc1 | NCBI_Gene:230796,ENSEMBL:ENSMUSG00000037622 | MGI:2685541 | protein coding gene | WD and tetratricopeptide repeats 1 |
| 4 | gene | 133.2925 | 133.2928 | positive | MGI_C57BL6J_3781803 | Gm3627 | NA | NA | protein coding gene | predicted gene 3627 |
| 4 | gene | 133.3697 | 133.4237 | positive | MGI_C57BL6J_102462 | Slc9a1 | NCBI_Gene:20544,ENSEMBL:ENSMUSG00000028854 | MGI:102462 | protein coding gene | solute carrier family 9 (sodium/hydrogen exchanger), member 1 |
| 4 | gene | 133.3698 | 133.3699 | positive | MGI_C57BL6J_4950447 | Mir5122 | miRBase:MI0018031,NCBI_Gene:100628620,ENSEMBL:ENSMUSG00000092745 | MGI:4950447 | miRNA gene | microRNA 5122 |
| 4 | gene | 133.4327 | 133.4349 | negative | MGI_C57BL6J_1920802 | 1700091J24Rik | NCBI_Gene:105247178 | MGI:1920802 | lncRNA gene | RIKEN cDNA 1700091J24 gene |
| 4 | gene | 133.4371 | 133.4404 | negative | MGI_C57BL6J_5625207 | Gm42322 | NCBI_Gene:105247179 | MGI:5625207 | lncRNA gene | predicted gene, 42322 |
| 4 | gene | 133.4794 | 133.4852 | negative | MGI_C57BL6J_3651057 | Gm13257 | NCBI_Gene:102640829,ENSEMBL:ENSMUSG00000086850 | MGI:3651057 | lncRNA gene | predicted gene 13257 |
| 4 | gene | 133.4801 | 133.4879 | positive | MGI_C57BL6J_2140500 | Tent5b | NCBI_Gene:100342,ENSEMBL:ENSMUSG00000046694 | MGI:2140500 | protein coding gene | terminal nucleotidyltransferase 5B |
| 4 | gene | 133.4911 | 133.4992 | negative | MGI_C57BL6J_1916789 | Trnp1 | NCBI_Gene:69539,ENSEMBL:ENSMUSG00000056596 | MGI:1916789 | protein coding gene | TMF1-regulated nuclear protein 1 |
| 4 | gene | 133.5129 | 133.5223 | negative | MGI_C57BL6J_5625208 | Gm42323 | NCBI_Gene:105247180 | MGI:5625208 | protein coding gene | predicted gene, 42323 |
| 4 | gene | 133.5189 | 133.5308 | positive | MGI_C57BL6J_1916323 | Kdf1 | NCBI_Gene:69073,ENSEMBL:ENSMUSG00000037600 | MGI:1916323 | protein coding gene | keratinocyte differentiation factor 1 |
| 4 | gene | 133.5325 | 133.5460 | negative | MGI_C57BL6J_106014 | Nudc | NCBI_Gene:18221,ENSEMBL:ENSMUSG00000028851 | MGI:106014 | protein coding gene | nudC nuclear distribution protein |
| 4 | gene | 133.5534 | 133.5567 | positive | MGI_C57BL6J_1346344 | Nr0b2 | NCBI_Gene:23957,ENSEMBL:ENSMUSG00000037583 | MGI:1346344 | protein coding gene | nuclear receptor subfamily 0, group B, member 2 |
| 4 | gene | 133.5588 | 133.5589 | negative | MGI_C57BL6J_5452935 | Gm23158 | ENSEMBL:ENSMUSG00000093886 | MGI:5452935 | miRNA gene | predicted gene, 23158 |
| 4 | gene | 133.5747 | 133.5842 | positive | MGI_C57BL6J_2442492 | Gpatch3 | NCBI_Gene:242691,ENSEMBL:ENSMUSG00000028850 | MGI:2442492 | protein coding gene | G patch domain containing 3 |
| 4 | gene | 133.5844 | 133.5917 | positive | MGI_C57BL6J_2140368 | Gpn2 | NCBI_Gene:100210,ENSEMBL:ENSMUSG00000028848 | MGI:2140368 | protein coding gene | GPN-loop GTPase 2 |
| 4 | gene | 133.5949 | 133.5960 | negative | MGI_C57BL6J_5625209 | Gm42324 | NCBI_Gene:105247181 | MGI:5625209 | lncRNA gene | predicted gene, 42324 |
| 4 | gene | 133.6006 | 133.6022 | negative | MGI_C57BL6J_1891831 | Sfn | NCBI_Gene:55948,ENSEMBL:ENSMUSG00000047281 | MGI:1891831 | protein coding gene | stratifin |
| 4 | gene | 133.6047 | 133.6502 | negative | MGI_C57BL6J_3527792 | Zdhhc18 | NCBI_Gene:503610,ENSEMBL:ENSMUSG00000037553 | MGI:3527792 | protein coding gene | zinc finger, DHHC domain containing 18 |
| 4 | pseudogene | 133.6381 | 133.6506 | negative | MGI_C57BL6J_3651606 | Gm13213 | NCBI_Gene:545683,ENSEMBL:ENSMUSG00000083885 | MGI:3651606 | pseudogene | predicted gene 13213 |
| 4 | gene | 133.6540 | 133.6543 | positive | MGI_C57BL6J_5452439 | Gm22662 | ENSEMBL:ENSMUSG00000093031 | MGI:5452439 | unclassified non-coding RNA gene | predicted gene, 22662 |
| 4 | gene | 133.6599 | 133.6732 | negative | MGI_C57BL6J_2442480 | Pigv | NCBI_Gene:230801,ENSEMBL:ENSMUSG00000043257 | MGI:2442480 | protein coding gene | phosphatidylinositol glycan anchor biosynthesis, class V |
| 4 | gene | 133.6727 | 133.6732 | positive | MGI_C57BL6J_1920565 | 1700041L08Rik | NA | NA | unclassified gene | RIKEN cDNA 1700041L08 gene |
| 4 | gene | 133.6790 | 133.7568 | negative | MGI_C57BL6J_1935147 | Arid1a | NCBI_Gene:93760,ENSEMBL:ENSMUSG00000007880 | MGI:1935147 | protein coding gene | AT rich interactive domain 1A (SWI-like) |
| 4 | gene | 133.7171 | 133.7171 | negative | MGI_C57BL6J_5562763 | Mir7227 | miRBase:MI0023722,NCBI_Gene:102465707,ENSEMBL:ENSMUSG00000104566 | MGI:5562763 | miRNA gene | microRNA 7227 |
| 4 | gene | 133.7681 | 133.7754 | negative | MGI_C57BL6J_5589178 | Gm30019 | NCBI_Gene:102631762 | MGI:5589178 | lncRNA gene | predicted gene, 30019 |
| 4 | gene | 133.7750 | 133.7813 | positive | MGI_C57BL6J_5589101 | Gm29942 | NCBI_Gene:102631659 | MGI:5589101 | lncRNA gene | predicted gene, 29942 |
| 4 | gene | 133.7920 | 133.8215 | negative | MGI_C57BL6J_5593455 | Gm34296 | NCBI_Gene:102637505 | MGI:5593455 | lncRNA gene | predicted gene, 34296 |
| 4 | gene | 133.7976 | 133.7977 | negative | MGI_C57BL6J_5455047 | Gm25270 | ENSEMBL:ENSMUSG00000084555 | MGI:5455047 | snRNA gene | predicted gene, 25270 |
| 4 | gene | 133.8152 | 133.8216 | positive | MGI_C57BL6J_3650037 | Gm12974 | NCBI_Gene:105247182,ENSEMBL:ENSMUSG00000084789 | MGI:3650037 | lncRNA gene | predicted gene 12974 |
| 4 | gene | 133.8473 | 133.8878 | negative | MGI_C57BL6J_104558 | Rps6ka1 | NCBI_Gene:20111,ENSEMBL:ENSMUSG00000003644 | MGI:104558 | protein coding gene | ribosomal protein S6 kinase polypeptide 1 |
| 4 | gene | 133.8716 | 133.8772 | positive | MGI_C57BL6J_3650699 | Gm12977 | NCBI_Gene:102631939,ENSEMBL:ENSMUSG00000085009 | MGI:3650699 | lncRNA gene | predicted gene 12977 |
| 4 | pseudogene | 133.8829 | 133.8831 | negative | MGI_C57BL6J_3651383 | Gm12985 | ENSEMBL:ENSMUSG00000080946 | MGI:3651383 | pseudogene | predicted gene 12985 |
| 4 | gene | 133.8834 | 133.8835 | negative | MGI_C57BL6J_5452300 | Gm22523 | ENSEMBL:ENSMUSG00000089321 | MGI:5452300 | snRNA gene | predicted gene, 22523 |
| 4 | gene | 133.9110 | 133.9114 | negative | MGI_C57BL6J_1921148 | 4930429E23Rik | NA | NA | unclassified gene | RIKEN cDNA 4930429E23 gene |
| 4 | gene | 133.9647 | 133.9686 | negative | MGI_C57BL6J_96136 | Hmgn2 | NCBI_Gene:15331,ENSEMBL:ENSMUSG00000003038 | MGI:96136 | protein coding gene | high mobility group nucleosomal binding domain 2 |
| 4 | gene | 133.9690 | 134.0009 | negative | MGI_C57BL6J_1914672 | Dhdds | NCBI_Gene:67422,ENSEMBL:ENSMUSG00000012117 | MGI:1914672 | protein coding gene | dehydrodolichyl diphosphate synthase |
| 4 | gene | 134.0033 | 134.0192 | negative | MGI_C57BL6J_1890546 | Lin28a | NCBI_Gene:83557,ENSEMBL:ENSMUSG00000050966 | MGI:1890546 | protein coding gene | lin-28 homolog A (C. elegans) |
| 4 | gene | 134.0034 | 134.0049 | positive | MGI_C57BL6J_3710625 | Gm10299 | NA | NA | unclassified gene | predicted gene 10299 |
| 4 | gene | 134.0181 | 134.0199 | positive | MGI_C57BL6J_3650259 | Gm13061 | ENSEMBL:ENSMUSG00000087288 | MGI:3650259 | lncRNA gene | predicted gene 13061 |
| 4 | gene | 134.0512 | 134.0590 | positive | MGI_C57BL6J_3650254 | Zfp683 | NCBI_Gene:100503878,ENSEMBL:ENSMUSG00000049410 | MGI:3650254 | protein coding gene | zinc finger protein 683 |
| 4 | gene | 134.0607 | 134.0925 | positive | MGI_C57BL6J_1334463 | Crybg2 | NCBI_Gene:230806,ENSEMBL:ENSMUSG00000012123 | MGI:1334463 | protein coding gene | crystallin beta-gamma domain containing 2 |
| 4 | gene | 134.0824 | 134.0951 | negative | MGI_C57BL6J_1346088 | Cd52 | NCBI_Gene:23833,ENSEMBL:ENSMUSG00000000682 | MGI:1346088 | protein coding gene | CD52 antigen |
| 4 | gene | 134.1025 | 134.1276 | positive | MGI_C57BL6J_1914836 | Ubxn11 | NCBI_Gene:67586,ENSEMBL:ENSMUSG00000012126 | MGI:1914836 | protein coding gene | UBX domain protein 11 |
| 4 | gene | 134.1274 | 134.1288 | negative | MGI_C57BL6J_1920973 | Sh3bgrl3 | NCBI_Gene:73723,ENSEMBL:ENSMUSG00000028843 | MGI:1920973 | protein coding gene | SH3 domain binding glutamic acid-rich protein-like 3 |
| 4 | gene | 134.1299 | 134.1871 | negative | MGI_C57BL6J_1917262 | Cep85 | NCBI_Gene:70012,ENSEMBL:ENSMUSG00000037443 | MGI:1917262 | protein coding gene | centrosomal protein 85 |
| 4 | gene | 134.1600 | 134.1641 | positive | MGI_C57BL6J_5826531 | Gm46894 | NCBI_Gene:108169013 | MGI:5826531 | protein coding gene | predicted gene, 46894 |
| 4 | gene | 134.1678 | 134.1679 | negative | MGI_C57BL6J_5531001 | Gm27619 | ENSEMBL:ENSMUSG00000098693 | MGI:5531001 | snoRNA gene | predicted gene, 27619 |
| 4 | gene | 134.1908 | 134.2030 | negative | MGI_C57BL6J_3702974 | Gm7534 | NCBI_Gene:665186,ENSEMBL:ENSMUSG00000073747 | MGI:3702974 | protein coding gene | predicted gene 7534 |
| 4 | pseudogene | 134.1998 | 134.2002 | negative | MGI_C57BL6J_3651062 | Gm13131 | ENSEMBL:ENSMUSG00000081772 | MGI:3651062 | pseudogene | predicted gene 13131 |
| 4 | gene | 134.2120 | 134.2274 | negative | MGI_C57BL6J_3043288 | Catsper4 | NCBI_Gene:329954,ENSEMBL:ENSMUSG00000048003 | MGI:3043288 | protein coding gene | cation channel, sperm associated 4 |
| 4 | gene | 134.2280 | 134.2384 | negative | MGI_C57BL6J_2670958 | Cnksr1 | NCBI_Gene:194231,ENSEMBL:ENSMUSG00000028841 | MGI:2670958 | protein coding gene | connector enhancer of kinase suppressor of Ras 1 |
| 4 | gene | 134.2433 | 134.2456 | negative | MGI_C57BL6J_1915290 | Zfp593 | NCBI_Gene:68040,ENSEMBL:ENSMUSG00000028840 | MGI:1915290 | protein coding gene | zinc finger protein 593 |
| 4 | gene | 134.2437 | 134.2459 | positive | MGI_C57BL6J_3528958 | E130218I03Rik | NCBI_Gene:77490,ENSEMBL:ENSMUSG00000086322 | MGI:3528958 | lncRNA gene | RIKEN cDNA E130218I03 gene |
| 4 | gene | 134.2469 | 134.2500 | positive | MGI_C57BL6J_5589350 | Gm30191 | ENSEMBL:ENSMUSG00000108398 | MGI:5589350 | protein coding gene | predicted gene, 30191 |
| 4 | gene | 134.2511 | 134.2662 | negative | MGI_C57BL6J_1919940 | Grrp1 | NCBI_Gene:72690,ENSEMBL:ENSMUSG00000050105 | MGI:1919940 | protein coding gene | glycine/arginine rich protein 1 |
| 4 | gene | 134.2558 | 134.2657 | positive | MGI_C57BL6J_5625210 | Gm42325 | NCBI_Gene:105247183 | MGI:5625210 | lncRNA gene | predicted gene, 42325 |
| 4 | gene | 134.2750 | 134.2879 | negative | MGI_C57BL6J_2385213 | Pdik1l | NCBI_Gene:230809,ENSEMBL:ENSMUSG00000050890 | MGI:2385213 | protein coding gene | PDLIM1 interacting kinase 1 like |
| 4 | gene | 134.3151 | 134.3296 | positive | MGI_C57BL6J_2447992 | Trim63 | NCBI_Gene:433766,ENSEMBL:ENSMUSG00000028834 | MGI:2447992 | protein coding gene | tripartite motif-containing 63 |
| 4 | pseudogene | 134.3355 | 134.3358 | negative | MGI_C57BL6J_3652269 | Gm13195 | ENSEMBL:ENSMUSG00000083065 | MGI:3652269 | pseudogene | predicted gene 13195 |
| 4 | gene | 134.3430 | 134.3545 | positive | MGI_C57BL6J_106637 | Slc30a2 | NCBI_Gene:230810,ENSEMBL:ENSMUSG00000028836 | MGI:106637 | protein coding gene | solute carrier family 30 (zinc transporter), member 2 |
| 4 | gene | 134.3564 | 134.3838 | negative | MGI_C57BL6J_1888742 | Extl1 | NCBI_Gene:56219,ENSEMBL:ENSMUSG00000028838 | MGI:1888742 | protein coding gene | exostoses (multiple)-like 1 |
| 4 | gene | 134.3849 | 134.3870 | positive | MGI_C57BL6J_3779501 | Gm5589 | NA | NA | unclassified gene | predicted gene 5589 |
| 4 | gene | 134.3963 | 134.4274 | positive | MGI_C57BL6J_2140321 | Pafah2 | NCBI_Gene:100163,ENSEMBL:ENSMUSG00000037366 | MGI:2140321 | protein coding gene | platelet-activating factor acetylhydrolase 2 |
| 4 | gene | 134.4337 | 134.4483 | negative | MGI_C57BL6J_5589612 | Gm30453 | NCBI_Gene:102632361 | MGI:5589612 | lncRNA gene | predicted gene, 30453 |
| 4 | gene | 134.4488 | 134.4502 | negative | MGI_C57BL6J_1916659 | 1700021N21Rik | NCBI_Gene:69409,ENSEMBL:ENSMUSG00000087343 | MGI:1916659 | lncRNA gene | RIKEN cDNA 1700021N21 gene |
| 4 | gene | 134.4683 | 134.4738 | positive | MGI_C57BL6J_96739 | Stmn1 | NCBI_Gene:16765,ENSEMBL:ENSMUSG00000028832 | MGI:96739 | protein coding gene | stathmin 1 |
| 4 | gene | 134.4684 | 134.4685 | positive | MGI_C57BL6J_5453411 | Gm23634 | ENSEMBL:ENSMUSG00000092812 | MGI:5453411 | miRNA gene | predicted gene, 23634 |
| 4 | pseudogene | 134.4932 | 134.4940 | positive | MGI_C57BL6J_3650425 | Gm13250 | NCBI_Gene:100416260,ENSEMBL:ENSMUSG00000083001 | MGI:3650425 | pseudogene | predicted gene 13250 |
| 4 | gene | 134.4967 | 134.5102 | positive | MGI_C57BL6J_1919154 | Paqr7 | NCBI_Gene:71904,ENSEMBL:ENSMUSG00000037348 | MGI:1919154 | protein coding gene | progestin and adipoQ receptor family member VII |
| 4 | gene | 134.5110 | 134.5239 | positive | MGI_C57BL6J_1917135 | Aunip | NCBI_Gene:69885,ENSEMBL:ENSMUSG00000078521 | MGI:1917135 | protein coding gene | aurora kinase A and ninein interacting protein |
| 4 | gene | 134.5256 | 134.5354 | negative | MGI_C57BL6J_1924074 | Mtfr1l | NCBI_Gene:76824,ENSEMBL:ENSMUSG00000046671 | MGI:1924074 | protein coding gene | mitochondrial fission regulator 1-like |
| 4 | gene | 134.5379 | 134.5522 | negative | MGI_C57BL6J_2151208 | Selenon | NCBI_Gene:74777,ENSEMBL:ENSMUSG00000050989 | MGI:2151208 | protein coding gene | selenoprotein N |
| 4 | gene | 134.5617 | 134.7043 | negative | MGI_C57BL6J_2446214 | Man1c1 | NCBI_Gene:230815,ENSEMBL:ENSMUSG00000037306 | MGI:2446214 | protein coding gene | mannosidase, alpha, class 1C, member 1 |
| 4 | gene | 134.5677 | 134.5679 | negative | MGI_C57BL6J_5531374 | Mir6403 | miRBase:MI0021939,NCBI_Gene:102465216,ENSEMBL:ENSMUSG00000099289 | MGI:5531374 | miRNA gene | microRNA 6403 |
| 4 | gene | 134.5914 | 134.5950 | positive | MGI_C57BL6J_5625211 | Gm42326 | NCBI_Gene:105247184 | MGI:5625211 | lncRNA gene | predicted gene, 42326 |
| 4 | gene | 134.7416 | 134.7680 | negative | MGI_C57BL6J_2140175 | Ldlrap1 | NCBI_Gene:100017,ENSEMBL:ENSMUSG00000037295 | MGI:2140175 | protein coding gene | low density lipoprotein receptor adaptor protein 1 |
| 4 | gene | 134.8028 | 134.8536 | negative | MGI_C57BL6J_1913396 | Maco1 | NCBI_Gene:66146,ENSEMBL:ENSMUSG00000028826 | MGI:1913396 | protein coding gene | macoilin 1 |
| 4 | gene | 134.8645 | 134.8962 | positive | MGI_C57BL6J_1202882 | Rhd | NCBI_Gene:19746,ENSEMBL:ENSMUSG00000028825 | MGI:1202882 | protein coding gene | Rh blood group, D antigen |
| 4 | gene | 134.8978 | 134.9150 | negative | MGI_C57BL6J_1919067 | Tmem50a | NCBI_Gene:71817,ENSEMBL:ENSMUSG00000028822 | MGI:1919067 | protein coding gene | transmembrane protein 50A |
| 4 | gene | 134.9236 | 134.9277 | positive | MGI_C57BL6J_106498 | Rsrp1 | NCBI_Gene:27981,ENSEMBL:ENSMUSG00000037266 | MGI:106498 | protein coding gene | arginine/serine rich protein 1 |
| 4 | gene | 134.9309 | 134.9375 | positive | MGI_C57BL6J_1915842 | Syf2 | NCBI_Gene:68592,ENSEMBL:ENSMUSG00000028821 | MGI:1915842 | protein coding gene | SYF2 homolog, RNA splicing factor (S. cerevisiae) |
| 4 | gene | 135.0437 | 135.0514 | negative | MGI_C57BL6J_5625212 | Gm42327 | NCBI_Gene:105247185 | MGI:5625212 | lncRNA gene | predicted gene, 42327 |
| 4 | gene | 135.1182 | 135.1204 | negative | MGI_C57BL6J_3801903 | Gm16225 | ENSEMBL:ENSMUSG00000086884 | MGI:3801903 | lncRNA gene | predicted gene 16225 |
| 4 | gene | 135.1206 | 135.1780 | positive | MGI_C57BL6J_102672 | Runx3 | NCBI_Gene:12399,ENSEMBL:ENSMUSG00000070691 | MGI:102672 | protein coding gene | runt related transcription factor 3 |
| 4 | gene | 135.1434 | 135.1443 | negative | MGI_C57BL6J_3801902 | Gm16224 | ENSEMBL:ENSMUSG00000084986 | MGI:3801902 | lncRNA gene | predicted gene 16224 |
| 4 | gene | 135.2140 | 135.2728 | negative | MGI_C57BL6J_1352754 | Clic4 | NCBI_Gene:29876,ENSEMBL:ENSMUSG00000037242 | MGI:1352754 | protein coding gene | chloride intracellular channel 4 (mitochondrial) |
| 4 | gene | 135.2831 | 135.2891 | positive | MGI_C57BL6J_5589926 | Gm30767 | NCBI_Gene:102632783 | MGI:5589926 | lncRNA gene | predicted gene, 30767 |
| 4 | pseudogene | 135.2903 | 135.2907 | negative | MGI_C57BL6J_3651378 | Gm12983 | NCBI_Gene:676072,ENSEMBL:ENSMUSG00000087270 | MGI:3651378 | pseudogene | predicted gene 12983 |
| 4 | gene | 135.2966 | 135.2994 | negative | MGI_C57BL6J_5589860 | Gm30701 | NCBI_Gene:102632698 | MGI:5589860 | lncRNA gene | predicted gene, 30701 |
| 4 | gene | 135.3073 | 135.3095 | positive | MGI_C57BL6J_3651384 | Gm12984 | ENSEMBL:ENSMUSG00000085002 | MGI:3651384 | lncRNA gene | predicted gene 12984 |
| 4 | pseudogene | 135.3078 | 135.3080 | negative | MGI_C57BL6J_3651381 | Gm12982 | ENSEMBL:ENSMUSG00000082833 | MGI:3651381 | pseudogene | predicted gene 12982 |
| 4 | gene | 135.3205 | 135.3533 | negative | MGI_C57BL6J_1858303 | Srrm1 | NCBI_Gene:51796,ENSEMBL:ENSMUSG00000028809 | MGI:1858303 | protein coding gene | serine/arginine repetitive matrix 1 |
| 4 | gene | 135.3696 | 135.3983 | negative | MGI_C57BL6J_2444888 | Ncmap | NCBI_Gene:230822,ENSEMBL:ENSMUSG00000043924 | MGI:2444888 | protein coding gene | noncompact myelin associated protein |
| 4 | pseudogene | 135.3772 | 135.3772 | positive | MGI_C57BL6J_3651750 | Gm12991 | ENSEMBL:ENSMUSG00000084032 | MGI:3651750 | pseudogene | predicted gene 12991 |
| 4 | pseudogene | 135.4032 | 135.4037 | negative | MGI_C57BL6J_3651107 | Gm12990 | ENSEMBL:ENSMUSG00000081136 | MGI:3651107 | pseudogene | predicted gene 12990 |
| 4 | gene | 135.4123 | 135.4339 | negative | MGI_C57BL6J_1858220 | Rcan3 | NCBI_Gene:53902,ENSEMBL:ENSMUSG00000059713 | MGI:1858220 | protein coding gene | regulator of calcineurin 3 |
| 4 | gene | 135.4166 | 135.4166 | negative | MGI_C57BL6J_3629655 | Mir700 | miRBase:MI0004684,NCBI_Gene:735285,ENSEMBL:ENSMUSG00000076123 | MGI:3629655 | miRNA gene | microRNA 700 |
| 4 | gene | 135.4207 | 135.4278 | positive | MGI_C57BL6J_5589981 | Gm30822 | NCBI_Gene:102632859 | MGI:5589981 | lncRNA gene | predicted gene, 30822 |
| 4 | gene | 135.4301 | 135.4308 | negative | MGI_C57BL6J_3801882 | Gm15979 | ENSEMBL:ENSMUSG00000087016 | MGI:3801882 | lncRNA gene | predicted gene 15979 |
| 4 | gene | 135.4454 | 135.4950 | negative | MGI_C57BL6J_1921802 | Nipal3 | NCBI_Gene:74552,ENSEMBL:ENSMUSG00000028803 | MGI:1921802 | protein coding gene | NIPA-like domain containing 3 |
| 4 | gene | 135.4509 | 135.4512 | positive | MGI_C57BL6J_5610498 | Gm37270 | ENSEMBL:ENSMUSG00000103955 | MGI:5610498 | unclassified gene | predicted gene, 37270 |
| 4 | gene | 135.4948 | 135.5378 | positive | MGI_C57BL6J_1926056 | Stpg1 | NCBI_Gene:78806,ENSEMBL:ENSMUSG00000028801 | MGI:1926056 | protein coding gene | sperm tail PG rich repeat containing 1 |
| 4 | gene | 135.5124 | 135.5125 | negative | MGI_C57BL6J_5455094 | Gm25317 | ENSEMBL:ENSMUSG00000080474 | MGI:5455094 | miRNA gene | predicted gene, 25317 |
| 4 | gene | 135.5419 | 135.5736 | negative | MGI_C57BL6J_2655333 | Grhl3 | NCBI_Gene:230824,ENSEMBL:ENSMUSG00000037188 | MGI:2655333 | protein coding gene | grainyhead like transcription factor 3 |
| 4 | gene | 135.5736 | 135.5802 | positive | MGI_C57BL6J_5590047 | Gm30888 | NCBI_Gene:102632942 | MGI:5590047 | lncRNA gene | predicted gene, 30888 |
| 4 | gene | 135.5837 | 135.5838 | positive | MGI_C57BL6J_5452883 | Gm23106 | ENSEMBL:ENSMUSG00000088179 | MGI:5452883 | snRNA gene | predicted gene, 23106 |
| 4 | gene | 135.6267 | 135.6302 | positive | MGI_C57BL6J_1913597 | 1700029M20Rik | NCBI_Gene:73937,ENSEMBL:ENSMUSG00000086788 | MGI:1913597 | lncRNA gene | RIKEN cDNA 1700029M20 gene |
| 4 | gene | 135.6503 | 135.6540 | negative | MGI_C57BL6J_5590100 | Gm30941 | NCBI_Gene:102633011 | MGI:5590100 | lncRNA gene | predicted gene, 30941 |
| 4 | gene | 135.6863 | 135.7082 | positive | MGI_C57BL6J_2429859 | Ifnlr1 | NCBI_Gene:242700,ENSEMBL:ENSMUSG00000062157 | MGI:2429859 | protein coding gene | interferon lambda receptor 1 |
| 4 | pseudogene | 135.7157 | 135.7167 | negative | MGI_C57BL6J_3652245 | Gm12989 | NCBI_Gene:100418217,ENSEMBL:ENSMUSG00000083678 | MGI:3652245 | pseudogene | predicted gene 12989 |
| 4 | gene | 135.7282 | 135.7554 | positive | MGI_C57BL6J_2663588 | Il22ra1 | NCBI_Gene:230828,ENSEMBL:ENSMUSG00000037157 | MGI:2663588 | protein coding gene | interleukin 22 receptor, alpha 1 |
| 4 | pseudogene | 135.7556 | 135.7568 | negative | MGI_C57BL6J_3650876 | Gm12988 | NCBI_Gene:665334,ENSEMBL:ENSMUSG00000082743 | MGI:3650876 | pseudogene | predicted gene 12988 |
| 4 | gene | 135.7588 | 135.8156 | positive | MGI_C57BL6J_2685280 | Myom3 | NCBI_Gene:242702,ENSEMBL:ENSMUSG00000037139 | MGI:2685280 | protein coding gene | myomesin family, member 3 |
| 4 | pseudogene | 135.8537 | 135.8542 | positive | MGI_C57BL6J_3650940 | Gm13000 | ENSEMBL:ENSMUSG00000081672 | MGI:3650940 | pseudogene | predicted gene 13000 |
| 4 | gene | 135.8557 | 135.8699 | positive | MGI_C57BL6J_1333805 | Srsf10 | NCBI_Gene:14105,ENSEMBL:ENSMUSG00000028676 | MGI:1333805 | protein coding gene | serine/arginine-rich splicing factor 10 |
| 4 | gene | 135.8709 | 135.8739 | negative | MGI_C57BL6J_106512 | Pnrc2 | NCBI_Gene:52830,ENSEMBL:ENSMUSG00000028675 | MGI:106512 | protein coding gene | proline-rich nuclear receptor coactivator 2 |
| 4 | pseudogene | 135.8913 | 135.8915 | negative | MGI_C57BL6J_3651355 | Gm13006 | ENSEMBL:ENSMUSG00000083430 | MGI:3651355 | pseudogene | predicted gene 13006 |
| 4 | gene | 135.8952 | 135.9202 | positive | MGI_C57BL6J_104650 | Cnr2 | NCBI_Gene:12802,ENSEMBL:ENSMUSG00000062585 | MGI:104650 | protein coding gene | cannabinoid receptor 2 (macrophage) |
| 4 | gene | 135.9207 | 135.9403 | positive | MGI_C57BL6J_95593 | Fuca1 | NCBI_Gene:71665,ENSEMBL:ENSMUSG00000028673 | MGI:95593 | protein coding gene | fucosidase, alpha-L- 1, tissue |
| 4 | gene | 135.9464 | 135.9626 | positive | MGI_C57BL6J_96158 | Hmgcl | NCBI_Gene:15356,ENSEMBL:ENSMUSG00000028672 | MGI:96158 | protein coding gene | 3-hydroxy-3-methylglutaryl-Coenzyme A lyase |
| 4 | gene | 135.9637 | 135.9682 | positive | MGI_C57BL6J_1921496 | Gale | NCBI_Gene:74246,ENSEMBL:ENSMUSG00000028671 | MGI:1921496 | protein coding gene | galactose-4-epimerase, UDP |
| 4 | gene | 135.9682 | 135.9726 | negative | MGI_C57BL6J_1347000 | Lypla2 | NCBI_Gene:26394,ENSEMBL:ENSMUSG00000028670 | MGI:1347000 | protein coding gene | lysophospholipase 2 |
| 4 | gene | 135.9755 | 135.9873 | negative | MGI_C57BL6J_1913443 | Pithd1 | NCBI_Gene:66193,ENSEMBL:ENSMUSG00000028669 | MGI:1913443 | protein coding gene | PITH (C-terminal proteasome-interacting domain of thioredoxin-like) domain containing 1 |
| 4 | pseudogene | 135.9975 | 135.9979 | positive | MGI_C57BL6J_3649722 | Gm13007 | NCBI_Gene:669445,ENSEMBL:ENSMUSG00000083852 | MGI:3649722 | pseudogene | predicted gene 13007 |
| 4 | gene | 136.0034 | 136.0218 | negative | MGI_C57BL6J_1351315 | Eloa | NCBI_Gene:27224,ENSEMBL:ENSMUSG00000028668 | MGI:1351315 | protein coding gene | elongin A |
| 4 | gene | 136.0036 | 136.0109 | positive | MGI_C57BL6J_3702670 | Gm13008 | ENSEMBL:ENSMUSG00000085714 | MGI:3702670 | lncRNA gene | predicted gene 13008 |
| 4 | gene | 136.0181 | 136.0185 | negative | MGI_C57BL6J_5454139 | Gm24362 | ENSEMBL:ENSMUSG00000097995 | MGI:5454139 | lncRNA gene | predicted gene, 24362 |
| 4 | gene | 136.0283 | 136.0534 | negative | MGI_C57BL6J_1914275 | Rpl11 | NCBI_Gene:67025,ENSEMBL:ENSMUSG00000059291 | MGI:1914275 | protein coding gene | ribosomal protein L11 |
| 4 | pseudogene | 136.0745 | 136.0750 | negative | MGI_C57BL6J_3651086 | Gm13009 | ENSEMBL:ENSMUSG00000082877 | MGI:3651086 | pseudogene | predicted gene 13009 |
| 4 | gene | 136.1375 | 136.1376 | positive | MGI_C57BL6J_5455778 | Gm26001 | ENSEMBL:ENSMUSG00000088614 | MGI:5455778 | snRNA gene | predicted gene, 26001 |
| 4 | gene | 136.1386 | 136.1434 | negative | MGI_C57BL6J_5625214 | Gm42329 | NCBI_Gene:105247187 | MGI:5625214 | lncRNA gene | predicted gene, 42329 |
| 4 | gene | 136.1435 | 136.1458 | positive | MGI_C57BL6J_96398 | Id3 | NCBI_Gene:15903,ENSEMBL:ENSMUSG00000007872 | MGI:96398 | protein coding gene | inhibitor of DNA binding 3 |
| 4 | gene | 136.1723 | 136.1961 | positive | MGI_C57BL6J_1096341 | E2f2 | NCBI_Gene:242705,ENSEMBL:ENSMUSG00000018983 | MGI:1096341 | protein coding gene | E2F transcription factor 2 |
| 4 | gene | 136.2063 | 136.2466 | positive | MGI_C57BL6J_2684986 | Asap3 | NCBI_Gene:230837,ENSEMBL:ENSMUSG00000036995 | MGI:2684986 | protein coding gene | ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| 4 | gene | 136.2145 | 136.2166 | positive | MGI_C57BL6J_2441962 | D230019A03Rik | NA | NA | unclassified gene | RIKEN cDNA D230019A03 gene |
| 4 | gene | 136.2477 | 136.2749 | positive | MGI_C57BL6J_1196908 | Tcea3 | NCBI_Gene:21401,ENSEMBL:ENSMUSG00000001604 | MGI:1196908 | protein coding gene | transcription elongation factor A (SII), 3 |
| 4 | gene | 136.2842 | 136.2939 | positive | MGI_C57BL6J_99192 | Zfp46 | NCBI_Gene:22704,ENSEMBL:ENSMUSG00000051351 | MGI:99192 | protein coding gene | zinc finger protein 46 |
| 4 | pseudogene | 136.3074 | 136.3079 | negative | MGI_C57BL6J_3650319 | Gm13013 | ENSEMBL:ENSMUSG00000081330 | MGI:3650319 | pseudogene | predicted gene 13013 |
| 4 | gene | 136.3109 | 136.3594 | positive | MGI_C57BL6J_1891692 | Hnrnpr | NCBI_Gene:74326,ENSEMBL:ENSMUSG00000066037 | MGI:1891692 | protein coding gene | heterogeneous nuclear ribonucleoprotein R |
| 4 | gene | 136.3478 | 136.3494 | negative | MGI_C57BL6J_5504137 | Gm27022 | ENSEMBL:ENSMUSG00000098155 | MGI:5504137 | lncRNA gene | predicted gene, 27022 |
| 4 | gene | 136.3503 | 136.3594 | positive | MGI_C57BL6J_1918848 | 9130020K20Rik | NCBI_Gene:105247188 | MGI:1918848 | lncRNA gene | RIKEN cDNA 9130020K20 gene |
| 4 | gene | 136.3558 | 136.3594 | positive | MGI_C57BL6J_4937022 | Gm17388 | NA | NA | lncRNA gene | predicted gene%2c 17388 |
| 4 | gene | 136.3875 | 136.3890 | positive | MGI_C57BL6J_5590454 | Gm31295 | NCBI_Gene:102633481 | MGI:5590454 | lncRNA gene | predicted gene, 31295 |
| 4 | gene | 136.4235 | 136.4444 | positive | MGI_C57BL6J_96276 | Htr1d | NCBI_Gene:15552,ENSEMBL:ENSMUSG00000070687 | MGI:96276 | protein coding gene | 5-hydroxytryptamine (serotonin) receptor 1D |
| 4 | gene | 136.4623 | 136.4640 | positive | MGI_C57BL6J_2443447 | 6030445D17Rik | ENSEMBL:ENSMUSG00000066178 | MGI:2443447 | protein coding gene | RIKEN cDNA 6030445D17 gene |
| 4 | gene | 136.4697 | 136.5548 | positive | MGI_C57BL6J_107629 | Luzp1 | NCBI_Gene:269593,ENSEMBL:ENSMUSG00000001089 | MGI:107629 | protein coding gene | leucine zipper protein 1 |
| 4 | gene | 136.5505 | 136.6028 | negative | MGI_C57BL6J_1196256 | Kdm1a | NCBI_Gene:99982,ENSEMBL:ENSMUSG00000036940 | MGI:1196256 | protein coding gene | lysine (K)-specific demethylase 1A |
| 4 | pseudogene | 136.5842 | 136.5847 | negative | MGI_C57BL6J_3649751 | Gm12987 | ENSEMBL:ENSMUSG00000082711 | MGI:3649751 | pseudogene | predicted gene 12987 |
| 4 | gene | 136.6069 | 136.6133 | positive | MGI_C57BL6J_1914913 | Tex46 | NCBI_Gene:67663,ENSEMBL:ENSMUSG00000036921 | MGI:1914913 | protein coding gene | testis expressed 46 |
| 4 | pseudogene | 136.6088 | 136.6099 | negative | MGI_C57BL6J_3651584 | Gm12986 | NCBI_Gene:433770,ENSEMBL:ENSMUSG00000083054 | MGI:3651584 | pseudogene | predicted gene 12986 |
| 4 | gene | 136.6098 | 136.6099 | negative | MGI_C57BL6J_5451957 | Gm22180 | ENSEMBL:ENSMUSG00000092747 | MGI:5451957 | miRNA gene | predicted gene, 22180 |
| 4 | gene | 136.6226 | 136.6398 | positive | MGI_C57BL6J_2448566 | Lactbl1 | NCBI_Gene:242707,ENSEMBL:ENSMUSG00000070683 | MGI:2448566 | protein coding gene | lactamase, beta-like 1 |
| 4 | gene | 136.6396 | 136.6421 | positive | MGI_C57BL6J_5610811 | Gm37583 | ENSEMBL:ENSMUSG00000103119 | MGI:5610811 | unclassified gene | predicted gene, 37583 |
| 4 | gene | 136.6475 | 136.8360 | negative | MGI_C57BL6J_99611 | Ephb2 | NCBI_Gene:13844,ENSEMBL:ENSMUSG00000028664 | MGI:99611 | protein coding gene | Eph receptor B2 |
| 4 | gene | 136.8360 | 136.8421 | positive | MGI_C57BL6J_5590695 | Gm31536 | NCBI_Gene:102633796 | MGI:5590695 | lncRNA gene | predicted gene, 31536 |
| 4 | gene | 136.8801 | 136.8862 | negative | MGI_C57BL6J_88224 | C1qb | NCBI_Gene:12260,ENSEMBL:ENSMUSG00000036905 | MGI:88224 | protein coding gene | complement component 1, q subcomponent, beta polypeptide |
| 4 | gene | 136.8898 | 136.8931 | negative | MGI_C57BL6J_88225 | C1qc | NCBI_Gene:12262,ENSEMBL:ENSMUSG00000036896 | MGI:88225 | protein coding gene | complement component 1, q subcomponent, C chain |
| 4 | gene | 136.8959 | 136.8988 | negative | MGI_C57BL6J_88223 | C1qa | NCBI_Gene:12259,ENSEMBL:ENSMUSG00000036887 | MGI:88223 | protein coding gene | complement component 1, q subcomponent, alpha polypeptide |
| 4 | gene | 136.9294 | 136.9568 | negative | MGI_C57BL6J_109378 | Epha8 | NCBI_Gene:13842,ENSEMBL:ENSMUSG00000028661 | MGI:109378 | protein coding gene | Eph receptor A8 |
| 4 | gene | 136.9797 | 137.0494 | negative | MGI_C57BL6J_2682254 | Zbtb40 | NCBI_Gene:230848,ENSEMBL:ENSMUSG00000060862 | MGI:2682254 | protein coding gene | zinc finger and BTB domain containing 40 |
| 4 | gene | 137.0257 | 137.0258 | positive | MGI_C57BL6J_5453611 | Gm23834 | ENSEMBL:ENSMUSG00000077624 | MGI:5453611 | snoRNA gene | predicted gene, 23834 |
| 4 | gene | 137.0766 | 137.0951 | positive | MGI_C57BL6J_5590807 | Gm31648 | NCBI_Gene:102633949 | MGI:5590807 | lncRNA gene | predicted gene, 31648 |
| 4 | gene | 137.1167 | 137.1183 | negative | MGI_C57BL6J_3702655 | Gm13001 | NCBI_Gene:102634021,ENSEMBL:ENSMUSG00000087417 | MGI:3702655 | lncRNA gene | predicted gene 13001 |
| 4 | gene | 137.1167 | 137.1176 | negative | MGI_C57BL6J_1921513 | 1700037C06Rik | ENSEMBL:ENSMUSG00000085542 | MGI:1921513 | lncRNA gene | RIKEN cDNA 1700037C06 gene |
| 4 | gene | 137.1395 | 137.1425 | negative | MGI_C57BL6J_5590921 | Gm31762 | NCBI_Gene:102634099 | MGI:5590921 | lncRNA gene | predicted gene, 31762 |
| 4 | gene | 137.1591 | 137.1651 | negative | MGI_C57BL6J_3650099 | Gm13003 | NCBI_Gene:100503595,ENSEMBL:ENSMUSG00000085833 | MGI:3650099 | lncRNA gene | predicted gene 13003 |
| 4 | gene | 137.1713 | 137.1781 | negative | MGI_C57BL6J_5826509 | Gm46872 | NCBI_Gene:108168981 | MGI:5826509 | lncRNA gene | predicted gene, 46872 |
| 4 | gene | 137.1840 | 137.1841 | positive | MGI_C57BL6J_5455176 | Gm25399 | ENSEMBL:ENSMUSG00000093294 | MGI:5455176 | miRNA gene | predicted gene, 25399 |
| 4 | gene | 137.1841 | 137.1841 | positive | MGI_C57BL6J_5453040 | Gm23263 | ENSEMBL:ENSMUSG00000092848 | MGI:5453040 | miRNA gene | predicted gene, 23263 |
| 4 | gene | 137.1841 | 137.2528 | negative | MGI_C57BL6J_5590979 | Gm31820 | NCBI_Gene:102634171 | MGI:5590979 | lncRNA gene | predicted gene, 31820 |
| 4 | pseudogene | 137.2190 | 137.2204 | negative | MGI_C57BL6J_3650102 | Gm13005 | NCBI_Gene:100047000,ENSEMBL:ENSMUSG00000080806 | MGI:3650102 | pseudogene | predicted gene 13005 |
| 4 | pseudogene | 137.2226 | 137.2233 | negative | MGI_C57BL6J_3650101 | Gm13002 | ENSEMBL:ENSMUSG00000082149 | MGI:3650101 | pseudogene | predicted gene 13002 |
| 4 | gene | 137.2775 | 137.2997 | positive | MGI_C57BL6J_98957 | Wnt4 | NCBI_Gene:22417,ENSEMBL:ENSMUSG00000036856 | MGI:98957 | protein coding gene | wingless-type MMTV integration site family, member 4 |
| 4 | gene | 137.3197 | 137.3578 | negative | MGI_C57BL6J_106211 | Cdc42 | NCBI_Gene:12540,ENSEMBL:ENSMUSG00000006699 | MGI:106211 | protein coding gene | cell division cycle 42 |
| 4 | gene | 137.3256 | 137.3256 | positive | MGI_C57BL6J_5455549 | Gm25772 | ENSEMBL:ENSMUSG00000089199 | MGI:5455549 | snRNA gene | predicted gene, 25772 |
| 4 | gene | 137.3364 | 137.3401 | negative | MGI_C57BL6J_2441797 | A430061O12Rik | NA | NA | unclassified gene | RIKEN cDNA A430061O12 gene |
| 4 | gene | 137.3873 | 137.3884 | negative | MGI_C57BL6J_1917225 | 2810405F17Rik | ENSEMBL:ENSMUSG00000101493 | MGI:1917225 | lncRNA gene | RIKEN cDNA 2810405F17 gene |
| 4 | pseudogene | 137.3953 | 137.3957 | positive | MGI_C57BL6J_3650100 | Rpl31-ps10 | NCBI_Gene:665533,ENSEMBL:ENSMUSG00000070667 | MGI:3650100 | pseudogene | ribosomal protein L31, pseudogene 10 |
| 4 | gene | 137.4016 | 137.4098 | negative | MGI_C57BL6J_3651647 | Cela3a | NCBI_Gene:242711,ENSEMBL:ENSMUSG00000078520 | MGI:3651647 | protein coding gene | chymotrypsin-like elastase family, member 3A |
| 4 | gene | 137.4210 | 137.4305 | negative | MGI_C57BL6J_1915118 | Cela3b | NCBI_Gene:67868,ENSEMBL:ENSMUSG00000023433 | MGI:1915118 | protein coding gene | chymotrypsin-like elastase family, member 3B |
| 4 | gene | 137.4366 | 137.4671 | negative | MGI_C57BL6J_3651180 | Gm13010 | NCBI_Gene:105247191,ENSEMBL:ENSMUSG00000086845 | MGI:3651180 | lncRNA gene | predicted gene 13010 |
| 4 | gene | 137.4533 | 137.4555 | positive | MGI_C57BL6J_1916630 | 1700013G24Rik | NCBI_Gene:69380,ENSEMBL:ENSMUSG00000041399 | MGI:1916630 | protein coding gene | RIKEN cDNA 1700013G24 gene |
| 4 | gene | 137.4688 | 137.5706 | positive | MGI_C57BL6J_96257 | Hspg2 | NCBI_Gene:15530,ENSEMBL:ENSMUSG00000028763 | MGI:96257 | protein coding gene | perlecan (heparan sulfate proteoglycan 2) |
| 4 | gene | 137.5383 | 137.5384 | positive | MGI_C57BL6J_5530988 | Mir7018 | miRBase:MI0022867,NCBI_Gene:102465616,ENSEMBL:ENSMUSG00000099134 | MGI:5530988 | miRNA gene | microRNA 7018 |
| 4 | gene | 137.5707 | 137.5824 | negative | MGI_C57BL6J_3588210 | Ldlrad2 | NCBI_Gene:435811,ENSEMBL:ENSMUSG00000094035 | MGI:3588210 | protein coding gene | low density lipoprotein receptor class A domain containing 2 |
| 4 | gene | 137.5938 | 137.6585 | positive | MGI_C57BL6J_2158502 | Usp48 | NCBI_Gene:170707,ENSEMBL:ENSMUSG00000043411 | MGI:2158502 | protein coding gene | ubiquitin specific peptidase 48 |
| 4 | gene | 137.6647 | 137.7299 | positive | MGI_C57BL6J_109338 | Rap1gap | NCBI_Gene:110351,ENSEMBL:ENSMUSG00000041351 | MGI:109338 | protein coding gene | Rap1 GTPase-activating protein |
| 4 | gene | 137.6970 | 137.7074 | negative | MGI_C57BL6J_1917058 | Rap1gapos | NCBI_Gene:69808,ENSEMBL:ENSMUSG00000087476 | MGI:1917058 | antisense lncRNA gene | RAP1 GTPase activating protein, opposite strand |
| 4 | gene | 137.7417 | 137.7964 | negative | MGI_C57BL6J_87983 | Alpl | NCBI_Gene:11647,ENSEMBL:ENSMUSG00000028766 | MGI:87983 | protein coding gene | alkaline phosphatase, liver/bone/kidney |
| 4 | gene | 137.8063 | 137.8120 | negative | MGI_C57BL6J_5591238 | Gm32079 | NCBI_Gene:102634516 | MGI:5591238 | lncRNA gene | predicted gene, 32079 |
| 4 | gene | 137.8622 | 137.9652 | positive | MGI_C57BL6J_1101357 | Ece1 | NCBI_Gene:230857,ENSEMBL:ENSMUSG00000057530 | MGI:1101357 | protein coding gene | endothelin converting enzyme 1 |
| 4 | gene | 137.8820 | 137.8839 | negative | MGI_C57BL6J_3652270 | Gm13012 | NCBI_Gene:102634683,ENSEMBL:ENSMUSG00000085823 | MGI:3652270 | lncRNA gene | predicted gene 13012 |
| 4 | gene | 137.9154 | 137.9155 | negative | MGI_C57BL6J_1921862 | 4833410I11Rik | NA | NA | unclassified gene | RIKEN cDNA 4833410I11 gene |
| 4 | gene | 137.9918 | 137.9923 | positive | MGI_C57BL6J_3035105 | BE691133 | NA | NA | unclassified gene | expressed sequence BE691133 |
| 4 | gene | 137.9930 | 138.2085 | positive | MGI_C57BL6J_1923935 | Eif4g3 | NCBI_Gene:230861,ENSEMBL:ENSMUSG00000028760 | MGI:1923935 | protein coding gene | eukaryotic translation initiation factor 4 gamma, 3 |
| 4 | gene | 137.9942 | 138.0279 | positive | MGI_C57BL6J_5591553 | Gm32394 | NCBI_Gene:102634927 | MGI:5591553 | lncRNA gene | predicted gene, 32394 |
| 4 | gene | 138.0140 | 138.0184 | positive | MGI_C57BL6J_3611235 | 8030494B02Rik | NA | NA | unclassified non-coding RNA gene | Riken cDNA 8030494B02 gene |
| 4 | gene | 138.1376 | 138.1385 | negative | MGI_C57BL6J_1920813 | 1700095J12Rik | NCBI_Gene:73563,ENSEMBL:ENSMUSG00000084804 | MGI:1920813 | lncRNA gene | RIKEN cDNA 1700095J12 gene |
| 4 | gene | 138.1632 | 138.1633 | positive | MGI_C57BL6J_5531049 | Mir6399 | miRBase:MI0021935,NCBI_Gene:102466907,ENSEMBL:ENSMUSG00000098513 | MGI:5531049 | miRNA gene | microRNA 6399 |
| 4 | gene | 138.2157 | 138.2162 | positive | MGI_C57BL6J_1916844 | 2310026L22Rik | ENSEMBL:ENSMUSG00000078519 | MGI:1916844 | lncRNA gene | RIKEN cDNA 2310026L22 gene |
| 4 | gene | 138.2163 | 138.2447 | positive | MGI_C57BL6J_109369 | Hp1bp3 | NCBI_Gene:15441,ENSEMBL:ENSMUSG00000028759 | MGI:109369 | protein coding gene | heterochromatin protein 1, binding protein 3 |
| 4 | gene | 138.2504 | 138.2613 | positive | MGI_C57BL6J_2446215 | Sh2d5 | NCBI_Gene:230863,ENSEMBL:ENSMUSG00000045349 | MGI:2446215 | protein coding gene | SH2 domain containing 5 |
| 4 | gene | 138.2504 | 138.3020 | positive | MGI_C57BL6J_1098229 | Kif17 | NCBI_Gene:16559,ENSEMBL:ENSMUSG00000028758 | MGI:1098229 | protein coding gene | kinesin family member 17 |
| 4 | gene | 138.3047 | 138.3126 | positive | MGI_C57BL6J_1194508 | Ddost | NCBI_Gene:13200,ENSEMBL:ENSMUSG00000028757 | MGI:1194508 | protein coding gene | dolichyl-di-phosphooligosaccharide-protein glycotransferase |
| 4 | gene | 138.3134 | 138.3263 | negative | MGI_C57BL6J_1916193 | Pink1 | NCBI_Gene:68943,ENSEMBL:ENSMUSG00000028756 | MGI:1916193 | protein coding gene | PTEN induced putative kinase 1 |
| 4 | gene | 138.3161 | 138.3162 | negative | MGI_C57BL6J_5531329 | Mir7019 | miRBase:MI0022868,NCBI_Gene:102466788,ENSEMBL:ENSMUSG00000099200 | MGI:5531329 | miRNA gene | microRNA 7019 |
| 4 | gene | 138.3260 | 138.3261 | negative | MGI_C57BL6J_5531028 | Gm27646 | ENSEMBL:ENSMUSG00000098364 | MGI:5531028 | miRNA gene | predicted gene, 27646 |
| 4 | gene | 138.3384 | 138.3683 | negative | MGI_C57BL6J_1919519 | Cda | NCBI_Gene:72269,ENSEMBL:ENSMUSG00000028755 | MGI:1919519 | protein coding gene | cytidine deaminase |
| 4 | gene | 138.3941 | 138.3965 | negative | MGI_C57BL6J_3651622 | Fam43b | NCBI_Gene:625638,ENSEMBL:ENSMUSG00000078235 | MGI:3651622 | protein coding gene | family with sequence similarity 43, member B |
| 4 | gene | 138.3952 | 138.3977 | positive | MGI_C57BL6J_1931024 | AB041806 | ENSEMBL:ENSMUSG00000046109 | MGI:1931024 | lncRNA gene | hypothetical protein, MNCb-2457 |
| 4 | pseudogene | 138.4257 | 138.4260 | positive | MGI_C57BL6J_3649580 | Rpl38-ps1 | NCBI_Gene:625646,ENSEMBL:ENSMUSG00000083326 | MGI:3649580 | pseudogene | ribosomal protein L38, pseudogene 1 |
| 4 | gene | 138.4347 | 138.4423 | positive | MGI_C57BL6J_1915600 | Mul1 | NCBI_Gene:68350,ENSEMBL:ENSMUSG00000041241 | MGI:1915600 | protein coding gene | mitochondrial ubiquitin ligase activator of NFKB 1 |
| 4 | gene | 138.4543 | 138.4601 | positive | MGI_C57BL6J_1913509 | Camk2n1 | NCBI_Gene:66259,ENSEMBL:ENSMUSG00000046447 | MGI:1913509 | protein coding gene | calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| 4 | gene | 138.4976 | 138.5285 | positive | MGI_C57BL6J_5591708 | Gm32549 | NCBI_Gene:102635131 | MGI:5591708 | lncRNA gene | predicted gene, 32549 |
| 4 | gene | 138.5642 | 138.6359 | negative | MGI_C57BL6J_1922968 | Vwa5b1 | NCBI_Gene:75718,ENSEMBL:ENSMUSG00000028753 | MGI:1922968 | protein coding gene | von Willebrand factor A domain containing 5B1 |
| 4 | gene | 138.5876 | 138.5877 | negative | MGI_C57BL6J_5452738 | Gm22961 | ENSEMBL:ENSMUSG00000088589 | MGI:5452738 | snRNA gene | predicted gene, 22961 |
| 4 | gene | 138.6390 | 138.6576 | positive | MGI_C57BL6J_5591821 | Gm32662 | NCBI_Gene:102635281 | MGI:5591821 | lncRNA gene | predicted gene, 32662 |
| 4 | gene | 138.6775 | 138.7117 | positive | MGI_C57BL6J_5591929 | Gm32770 | NCBI_Gene:102635431 | MGI:5591929 | lncRNA gene | predicted gene, 32770 |
| 4 | gene | 138.6778 | 138.6782 | positive | MGI_C57BL6J_1914908 | 4930563F15Rik | NA | NA | unclassified gene | RIKEN cDNA 4930563F15 gene |
| 4 | gene | 138.7098 | 138.7384 | negative | MGI_C57BL6J_2443123 | Ubxn10 | NCBI_Gene:212190,ENSEMBL:ENSMUSG00000043621 | MGI:2443123 | protein coding gene | UBX domain protein 10 |
| 4 | gene | 138.7248 | 138.7461 | positive | MGI_C57BL6J_106638 | Pla2g2c | NCBI_Gene:18781,ENSEMBL:ENSMUSG00000028750 | MGI:106638 | protein coding gene | phospholipase A2, group IIC |
| 4 | gene | 138.7505 | 138.7576 | negative | MGI_C57BL6J_1349661 | Pla2g2f | NCBI_Gene:26971,ENSEMBL:ENSMUSG00000028749 | MGI:1349661 | protein coding gene | phospholipase A2, group IIF |
| 4 | gene | 138.7614 | 138.7697 | negative | MGI_C57BL6J_5625217 | Gm42332 | NCBI_Gene:105247193 | MGI:5625217 | lncRNA gene | predicted gene, 42332 |
| 4 | gene | 138.7725 | 138.7802 | negative | MGI_C57BL6J_5592030 | Gm32871 | NCBI_Gene:102635577 | MGI:5592030 | lncRNA gene | predicted gene, 32871 |
| 4 | gene | 138.7757 | 138.7821 | positive | MGI_C57BL6J_1341796 | Pla2g2d | NCBI_Gene:18782,ENSEMBL:ENSMUSG00000041202 | MGI:1341796 | protein coding gene | phospholipase A2, group IID |
| 4 | gene | 138.7992 | 138.8635 | negative | MGI_C57BL6J_101899 | Pla2g5 | NCBI_Gene:18784,ENSEMBL:ENSMUSG00000041193 | MGI:101899 | protein coding gene | phospholipase A2, group V |
| 4 | gene | 138.8274 | 138.8275 | negative | MGI_C57BL6J_5455057 | Gm25280 | ENSEMBL:ENSMUSG00000087894 | MGI:5455057 | snRNA gene | predicted gene, 25280 |
| 4 | pseudogene | 138.8319 | 138.8352 | positive | MGI_C57BL6J_104642 | Pla2g2a | NCBI_Gene:18780,ENSEMBL:ENSMUSG00000058908 | MGI:104642 | polymorphic pseudogene | phospholipase A2, group IIA (platelets, synovial fluid) |
| 4 | gene | 138.8635 | 138.8686 | positive | MGI_C57BL6J_5592089 | Gm32930 | NCBI_Gene:102635643 | MGI:5592089 | lncRNA gene | predicted gene, 32930 |
| 4 | gene | 138.8710 | 138.8740 | negative | MGI_C57BL6J_3651765 | Gm13030 | NCBI_Gene:105734733,ENSEMBL:ENSMUSG00000078518 | MGI:3651765 | protein coding gene | predicted gene 13030 |
| 4 | gene | 138.8779 | 138.8828 | positive | MGI_C57BL6J_1349660 | Pla2g2e | NCBI_Gene:26970,ENSEMBL:ENSMUSG00000028751 | MGI:1349660 | protein coding gene | phospholipase A2, group IIE |
| 4 | gene | 138.8911 | 138.9140 | negative | MGI_C57BL6J_1920412 | Otud3 | NCBI_Gene:73162,ENSEMBL:ENSMUSG00000041161 | MGI:1920412 | protein coding gene | OTU domain containing 3 |
| 4 | pseudogene | 138.9352 | 138.9357 | negative | MGI_C57BL6J_5010282 | Gm18097 | NCBI_Gene:100416403 | MGI:5010282 | pseudogene | predicted gene, 18097 |
| 4 | gene | 138.9364 | 138.9386 | negative | MGI_C57BL6J_5592170 | Gm33011 | NCBI_Gene:102635752 | MGI:5592170 | lncRNA gene | predicted gene, 33011 |
| 4 | gene | 138.9619 | 138.9647 | negative | MGI_C57BL6J_5625218 | Gm42333 | NCBI_Gene:105247194 | MGI:5625218 | lncRNA gene | predicted gene, 42333 |
| 4 | gene | 138.9671 | 138.9684 | positive | MGI_C57BL6J_1914075 | Rnf186 | NCBI_Gene:66825,ENSEMBL:ENSMUSG00000070661 | MGI:1914075 | protein coding gene | ring finger protein 186 |
| 4 | gene | 138.9721 | 139.0592 | positive | MGI_C57BL6J_1924306 | Tmco4 | NCBI_Gene:77056,ENSEMBL:ENSMUSG00000041143 | MGI:1924306 | protein coding gene | transmembrane and coiled-coil domains 4 |
| 4 | gene | 139.0032 | 139.0196 | negative | MGI_C57BL6J_5592373 | Gm33214 | NCBI_Gene:102636025 | MGI:5592373 | lncRNA gene | predicted gene, 33214 |
| 4 | gene | 139.0597 | 139.0756 | negative | MGI_C57BL6J_1196627 | Htr6 | NCBI_Gene:15565,ENSEMBL:ENSMUSG00000028747 | MGI:1196627 | protein coding gene | 5-hydroxytryptamine (serotonin) receptor 6 |
| 4 | gene | 139.0749 | 139.0772 | positive | MGI_C57BL6J_5791369 | Gm45533 | NCBI_Gene:108168960,ENSEMBL:ENSMUSG00000110296 | MGI:5791369 | lncRNA gene | predicted gene 45533 |
| 4 | gene | 139.0823 | 139.0930 | negative | MGI_C57BL6J_104591 | Nbl1 | NCBI_Gene:17965,ENSEMBL:ENSMUSG00000041120 | MGI:104591 | protein coding gene | NBL1, DAN family BMP antagonist |
| 4 | gene | 139.1018 | 139.1311 | negative | MGI_C57BL6J_1913628 | Micos10 | NCBI_Gene:433771,ENSEMBL:ENSMUSG00000050608 | MGI:1913628 | protein coding gene | mitochondrial contact site and cristae organizing system subunit 10 |
| 4 | gene | 139.1341 | 139.1653 | negative | MGI_C57BL6J_5592463 | Gm33304 | NCBI_Gene:102636159 | MGI:5592463 | lncRNA gene | predicted gene, 33304 |
| 4 | pseudogene | 139.1502 | 139.1508 | positive | MGI_C57BL6J_1935164 | Hspe1-ps4 | NCBI_Gene:93751,ENSEMBL:ENSMUSG00000083238 | MGI:1935164 | pseudogene | heat shock protein 1 (chaperonin 10), pseudogene 4 |
| 4 | gene | 139.1754 | 139.1807 | negative | MGI_C57BL6J_3650322 | Gm16287 | NCBI_Gene:100038595,ENSEMBL:ENSMUSG00000073739 | MGI:3650322 | lncRNA gene | predicted gene 16287 |
| 4 | gene | 139.1922 | 139.1927 | negative | MGI_C57BL6J_1925704 | 9530077C14Rik | NA | NA | unclassified gene | RIKEN cDNA 9530077C14 gene |
| 4 | gene | 139.1929 | 139.2918 | positive | MGI_C57BL6J_104652 | Capzb | NCBI_Gene:12345,ENSEMBL:ENSMUSG00000028745 | MGI:104652 | protein coding gene | capping protein (actin filament) muscle Z-line, beta |
| 4 | gene | 139.2940 | 139.3107 | negative | MGI_C57BL6J_2384837 | Pqlc2 | NCBI_Gene:212555,ENSEMBL:ENSMUSG00000028744 | MGI:2384837 | protein coding gene | PQ loop repeat containing 2 |
| 4 | gene | 139.3107 | 139.3188 | positive | MGI_C57BL6J_107796 | Akr7a5 | NCBI_Gene:110198,ENSEMBL:ENSMUSG00000028743 | MGI:107796 | protein coding gene | aldo-keto reductase family 7, member A5 (aflatoxin aldehyde reductase) |
| 4 | gene | 139.3382 | 139.3383 | positive | MGI_C57BL6J_5455509 | Gm25732 | ENSEMBL:ENSMUSG00000092909 | MGI:5455509 | miRNA gene | predicted gene, 25732 |
| 4 | gene | 139.3383 | 139.3384 | positive | MGI_C57BL6J_5454022 | Gm24245 | ENSEMBL:ENSMUSG00000087943 | MGI:5454022 | miRNA gene | predicted gene, 24245 |
| 4 | pseudogene | 139.3422 | 139.3433 | positive | MGI_C57BL6J_3652310 | Gm13019 | ENSEMBL:ENSMUSG00000083404 | MGI:3652310 | pseudogene | predicted gene 13019 |
| 4 | pseudogene | 139.3444 | 139.3463 | negative | MGI_C57BL6J_1100505 | Ccnd3-ps | NCBI_Gene:626000,ENSEMBL:ENSMUSG00000080708 | MGI:1100505 | pseudogene | cyclin D3, pseudogene |
| 4 | gene | 139.3474 | 139.3526 | negative | MGI_C57BL6J_1917152 | Mrto4 | NCBI_Gene:69902,ENSEMBL:ENSMUSG00000028741 | MGI:1917152 | protein coding gene | mRNA turnover 4, ribosome maturation factor |
| 4 | gene | 139.3526 | 139.3787 | positive | MGI_C57BL6J_2443696 | Emc1 | NCBI_Gene:230866,ENSEMBL:ENSMUSG00000078517 | MGI:2443696 | protein coding gene | ER membrane protein complex subunit 1 |
| 4 | gene | 139.3526 | 139.4896 | positive | MGI_C57BL6J_1916366 | Ubr4 | NCBI_Gene:69116,ENSEMBL:ENSMUSG00000066036 | MGI:1916366 | protein coding gene | ubiquitin protein ligase E3 component n-recognin 4 |
| 4 | gene | 139.3765 | 139.3804 | negative | MGI_C57BL6J_1919807 | 2700016F22Rik | ENSEMBL:ENSMUSG00000097731 | MGI:1919807 | lncRNA gene | RIKEN cDNA 2700016F22 gene |
| 4 | gene | 139.4305 | 139.4306 | negative | MGI_C57BL6J_4414084 | n-TVaac1 | NCBI_Gene:102467443 | MGI:4414084 | tRNA gene | nuclear encoded tRNA valine 1 (anticodon AAC) |
| 4 | gene | 139.5305 | 139.6204 | positive | MGI_C57BL6J_2140675 | Iffo2 | NCBI_Gene:212632,ENSEMBL:ENSMUSG00000041025 | MGI:2140675 | protein coding gene | intermediate filament family orphan 2 |
| 4 | gene | 139.5889 | 139.5890 | negative | MGI_C57BL6J_5455348 | Gm25571 | ENSEMBL:ENSMUSG00000094359 | MGI:5455348 | miRNA gene | predicted gene, 25571 |
| 4 | gene | 139.6032 | 139.6627 | positive | MGI_C57BL6J_5439438 | Gm21969 | ENSEMBL:ENSMUSG00000094439 | MGI:5439438 | protein coding gene | predicted gene 21969 |
| 4 | gene | 139.6229 | 139.6497 | positive | MGI_C57BL6J_2443883 | Aldh4a1 | NCBI_Gene:212647,ENSEMBL:ENSMUSG00000028737 | MGI:2443883 | protein coding gene | aldehyde dehydrogenase 4 family, member A1 |
| 4 | gene | 139.6440 | 139.6441 | positive | MGI_C57BL6J_5531141 | Mir7020 | miRBase:MI0022869,NCBI_Gene:102465617,ENSEMBL:ENSMUSG00000098569 | MGI:5531141 | miRNA gene | microRNA 7020 |
| 4 | gene | 139.6535 | 139.6703 | positive | MGI_C57BL6J_1933546 | Tas1r2 | NCBI_Gene:83770,ENSEMBL:ENSMUSG00000028738 | MGI:1933546 | protein coding gene | taste receptor, type 1, member 2 |
| 4 | gene | 139.7350 | 139.7357 | positive | MGI_C57BL6J_5625219 | Gm42334 | NCBI_Gene:105247195 | MGI:5625219 | lncRNA gene | predicted gene, 42334 |
| 4 | gene | 139.7371 | 139.8335 | negative | MGI_C57BL6J_97491 | Pax7 | NCBI_Gene:18509,ENSEMBL:ENSMUSG00000028736 | MGI:97491 | protein coding gene | paired box 7 |
| 4 | gene | 139.8331 | 139.8529 | positive | MGI_C57BL6J_5826510 | Gm46873 | NCBI_Gene:108168982 | MGI:5826510 | lncRNA gene | predicted gene, 46873 |
| 4 | gene | 139.9105 | 139.9344 | positive | MGI_C57BL6J_2686513 | Gm1667 | NCBI_Gene:102636380,ENSEMBL:ENSMUSG00000087646 | MGI:2686513 | lncRNA gene | predicted gene 1667 |
| 4 | gene | 139.9233 | 139.9332 | positive | MGI_C57BL6J_3651504 | Gm13028 | ENSEMBL:ENSMUSG00000087628 | MGI:3651504 | lncRNA gene | predicted gene 13028 |
| 4 | gene | 139.9261 | 139.9435 | negative | MGI_C57BL6J_1918485 | 4933427I22Rik | NCBI_Gene:71235,ENSEMBL:ENSMUSG00000085928 | MGI:1918485 | lncRNA gene | RIKEN cDNA 4933427I22 gene |
| 4 | gene | 139.9602 | 139.9680 | negative | MGI_C57BL6J_2444612 | Klhdc7a | NCBI_Gene:242721,ENSEMBL:ENSMUSG00000078234 | MGI:2444612 | protein coding gene | kelch domain containing 7A |
| 4 | gene | 139.9675 | 139.9676 | negative | MGI_C57BL6J_4358926 | Mir2139 | miRBase:MI0010752,NCBI_Gene:100316727,ENSEMBL:ENSMUSG00000089294 | MGI:4358926 | miRNA gene | microRNA 2139 |
| 4 | gene | 140.0268 | 140.2469 | negative | MGI_C57BL6J_2681842 | Igsf21 | NCBI_Gene:230868,ENSEMBL:ENSMUSG00000040972 | MGI:2681842 | protein coding gene | immunoglobulin superfamily, member 21 |
| 4 | gene | 140.1086 | 140.1450 | positive | MGI_C57BL6J_3651764 | Gm13029 | NCBI_Gene:102636673,ENSEMBL:ENSMUSG00000086164 | MGI:3651764 | lncRNA gene | predicted gene 13029 |
| 4 | gene | 140.1319 | 140.1334 | positive | MGI_C57BL6J_3651912 | Gm13027 | ENSEMBL:ENSMUSG00000085968 | MGI:3651912 | lncRNA gene | predicted gene 13027 |
| 4 | gene | 140.2185 | 140.2225 | positive | MGI_C57BL6J_5592701 | Gm33542 | NCBI_Gene:102636490 | MGI:5592701 | lncRNA gene | predicted gene, 33542 |
| 4 | gene | 140.3207 | 140.3233 | negative | MGI_C57BL6J_3642724 | Gm9867 | ENSEMBL:ENSMUSG00000103107 | MGI:3642724 | lncRNA gene | predicted gene 9867 |
| 4 | gene | 140.3234 | 140.3305 | positive | MGI_C57BL6J_5593054 | Gm33895 | NCBI_Gene:102636971 | MGI:5593054 | lncRNA gene | predicted gene, 33895 |
| 4 | gene | 140.3554 | 140.3619 | positive | MGI_C57BL6J_5593233 | Gm34074 | NCBI_Gene:102637201 | MGI:5593233 | lncRNA gene | predicted gene, 34074 |
| 4 | gene | 140.3805 | 140.3864 | positive | MGI_C57BL6J_3650527 | Gm13016 | NCBI_Gene:329970,ENSEMBL:ENSMUSG00000087084 | MGI:3650527 | lncRNA gene | predicted gene 13016 |
| 4 | gene | 140.3904 | 140.3969 | positive | MGI_C57BL6J_3650528 | Gm13017 | NCBI_Gene:102637278,ENSEMBL:ENSMUSG00000086960 | MGI:3650528 | lncRNA gene | predicted gene 13017 |
| 4 | gene | 140.4225 | 140.4319 | positive | MGI_C57BL6J_3649637 | Gm13021 | NCBI_Gene:102637389,ENSEMBL:ENSMUSG00000086437 | MGI:3649637 | lncRNA gene | predicted gene 13021 |
| 4 | gene | 140.4380 | 140.4613 | positive | MGI_C57BL6J_3650881 | Gm13026 | ENSEMBL:ENSMUSG00000085047 | MGI:3650881 | lncRNA gene | predicted gene 13026 |
| 4 | pseudogene | 140.4458 | 140.4462 | negative | MGI_C57BL6J_6324716 | Gm50453 | ENSEMBL:ENSMUSG00000118416 | MGI:6324716 | pseudogene | predicted gene, 50453 |
| 4 | gene | 140.5145 | 140.6660 | negative | MGI_C57BL6J_1920004 | Arhgef10l | NCBI_Gene:72754,ENSEMBL:ENSMUSG00000040964 | MGI:1920004 | protein coding gene | Rho guanine nucleotide exchange factor (GEF) 10-like |
| 4 | gene | 140.6051 | 140.6066 | positive | MGI_C57BL6J_5625220 | Gm42335 | NCBI_Gene:105247196 | MGI:5625220 | lncRNA gene | predicted gene, 42335 |
| 4 | gene | 140.6797 | 140.6863 | positive | MGI_C57BL6J_3650883 | Gm13025 | NCBI_Gene:102637546,ENSEMBL:ENSMUSG00000086159 | MGI:3650883 | lncRNA gene | predicted gene 13025 |
| 4 | gene | 140.6927 | 140.6928 | positive | MGI_C57BL6J_5452822 | Gm23045 | ENSEMBL:ENSMUSG00000095878 | MGI:5452822 | miRNA gene | predicted gene, 23045 |
| 4 | gene | 140.7005 | 140.7232 | positive | MGI_C57BL6J_1919784 | Rcc2 | NCBI_Gene:108911,ENSEMBL:ENSMUSG00000040945 | MGI:1919784 | protein coding gene | regulator of chromosome condensation 2 |
| 4 | gene | 140.7170 | 140.7171 | positive | MGI_C57BL6J_5455728 | Gm25951 | ENSEMBL:ENSMUSG00000093071 | MGI:5455728 | miRNA gene | predicted gene, 25951 |
| 4 | gene | 140.7274 | 140.7426 | negative | MGI_C57BL6J_2655198 | Padi6 | NCBI_Gene:242726,ENSEMBL:ENSMUSG00000040935 | MGI:2655198 | protein coding gene | peptidyl arginine deiminase, type VI |
| 4 | gene | 140.7455 | 140.7742 | negative | MGI_C57BL6J_1338898 | Padi4 | NCBI_Gene:18602,ENSEMBL:ENSMUSG00000025330 | MGI:1338898 | protein coding gene | peptidyl arginine deiminase, type IV |
| 4 | gene | 140.7854 | 140.8106 | negative | MGI_C57BL6J_1338891 | Padi3 | NCBI_Gene:18601,ENSEMBL:ENSMUSG00000025328 | MGI:1338891 | protein coding gene | peptidyl arginine deiminase, type III |
| 4 | gene | 140.8110 | 140.8175 | positive | MGI_C57BL6J_3702685 | Gm13032 | NCBI_Gene:100049161,ENSEMBL:ENSMUSG00000085918 | MGI:3702685 | lncRNA gene | predicted gene 13032 |
| 4 | gene | 140.8130 | 140.8458 | negative | MGI_C57BL6J_1338893 | Padi1 | NCBI_Gene:18599,ENSEMBL:ENSMUSG00000025329 | MGI:1338893 | protein coding gene | peptidyl arginine deiminase, type I |
| 4 | gene | 140.8727 | 140.8781 | positive | MGI_C57BL6J_1922378 | 4930515B02Rik | NCBI_Gene:75128,ENSEMBL:ENSMUSG00000087045 | MGI:1922378 | lncRNA gene | RIKEN cDNA 4930515B02 gene |
| 4 | gene | 140.9063 | 140.9526 | positive | MGI_C57BL6J_1338892 | Padi2 | NCBI_Gene:18600,ENSEMBL:ENSMUSG00000028927 | MGI:1338892 | protein coding gene | peptidyl arginine deiminase, type II |
| 4 | gene | 140.9090 | 140.9103 | negative | MGI_C57BL6J_5826356 | Gm46719 | NCBI_Gene:108168688 | MGI:5826356 | lncRNA gene | predicted gene, 46719 |
| 4 | gene | 140.9187 | 140.9188 | positive | MGI_C57BL6J_5456003 | Gm26226 | ENSEMBL:ENSMUSG00000064443 | MGI:5456003 | snRNA gene | predicted gene, 26226 |
| 4 | gene | 140.9272 | 140.9321 | negative | MGI_C57BL6J_5593534 | Gm34375 | NCBI_Gene:102637611 | MGI:5593534 | lncRNA gene | predicted gene, 34375 |
| 4 | gene | 140.9477 | 140.9579 | negative | MGI_C57BL6J_3702680 | Gm13031 | NCBI_Gene:100126227,ENSEMBL:ENSMUSG00000087698 | MGI:3702680 | lncRNA gene | predicted gene 13031 |
| 4 | gene | 140.9604 | 140.9613 | negative | MGI_C57BL6J_5593590 | Gm34431 | NCBI_Gene:102637683 | MGI:5593590 | lncRNA gene | predicted gene, 34431 |
| 4 | gene | 140.9612 | 140.9792 | positive | MGI_C57BL6J_1914930 | Sdhb | NCBI_Gene:67680,ENSEMBL:ENSMUSG00000009863 | MGI:1914930 | protein coding gene | succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| 4 | gene | 140.9869 | 141.0077 | positive | MGI_C57BL6J_1922022 | Atp13a2 | NCBI_Gene:74772,ENSEMBL:ENSMUSG00000036622 | MGI:1922022 | protein coding gene | ATPase type 13A2 |
| 4 | gene | 141.0104 | 141.0160 | positive | MGI_C57BL6J_99559 | Mfap2 | NCBI_Gene:17150,ENSEMBL:ENSMUSG00000060572 | MGI:99559 | protein coding gene | microfibrillar-associated protein 2 |
| 4 | gene | 141.0166 | 141.0606 | negative | MGI_C57BL6J_3529431 | Crocc | NCBI_Gene:230872,ENSEMBL:ENSMUSG00000040860 | MGI:3529431 | protein coding gene | ciliary rootlet coiled-coil, rootletin |
| 4 | gene | 141.0665 | 141.0784 | negative | MGI_C57BL6J_1913397 | Necap2 | NCBI_Gene:66147,ENSEMBL:ENSMUSG00000028923 | MGI:1913397 | protein coding gene | NECAP endocytosis associated 2 |
| 4 | pseudogene | 141.0848 | 141.0857 | positive | MGI_C57BL6J_3651114 | Gm13047 | NCBI_Gene:638991,ENSEMBL:ENSMUSG00000082246 | MGI:3651114 | pseudogene | predicted gene 13047 |
| 4 | gene | 141.0883 | 141.1128 | positive | MGI_C57BL6J_3607787 | Spata21 | NCBI_Gene:329972,ENSEMBL:ENSMUSG00000045004 | MGI:3607787 | protein coding gene | spermatogenesis associated 21 |
| 4 | gene | 141.1130 | 141.1398 | negative | MGI_C57BL6J_1098672 | Szrd1 | NCBI_Gene:213491,ENSEMBL:ENSMUSG00000040842 | MGI:1098672 | protein coding gene | SUZ RNA binding domain containing 1 |
| 4 | gene | 141.1157 | 141.1181 | positive | MGI_C57BL6J_3704238 | 4921514A10Rik | ENSEMBL:ENSMUSG00000097620 | MGI:3704238 | lncRNA gene | RIKEN cDNA 4921514A10 gene |
| 4 | gene | 141.1479 | 141.2041 | positive | MGI_C57BL6J_1924992 | Fbxo42 | NCBI_Gene:213499,ENSEMBL:ENSMUSG00000028920 | MGI:1924992 | protein coding gene | F-box protein 42 |
| 4 | gene | 141.2137 | 141.2268 | positive | MGI_C57BL6J_1923416 | Cplane2 | NCBI_Gene:76166,ENSEMBL:ENSMUSG00000073733 | MGI:1923416 | protein coding gene | ciliogenesis and planar polarity effector 2 |
| 4 | gene | 141.2382 | 141.2395 | negative | MGI_C57BL6J_3641643 | C630004L07Rik | ENSEMBL:ENSMUSG00000103366 | MGI:3641643 | unclassified gene | RIKEN cDNA C630004L07 gene |
| 4 | gene | 141.2395 | 141.2579 | positive | MGI_C57BL6J_1925912 | Arhgef19 | NCBI_Gene:213649,ENSEMBL:ENSMUSG00000028919 | MGI:1925912 | protein coding gene | Rho guanine nucleotide exchange factor (GEF) 19 |
| 4 | gene | 141.2618 | 141.2660 | negative | MGI_C57BL6J_3649999 | Gm13055 | ENSEMBL:ENSMUSG00000085774 | MGI:3649999 | lncRNA gene | predicted gene 13055 |
| 4 | gene | 141.2784 | 141.2798 | positive | MGI_C57BL6J_3650725 | Gm13056 | NCBI_Gene:100503810,ENSEMBL:ENSMUSG00000085395 | MGI:3650725 | lncRNA gene | predicted gene 13056 |
| 4 | gene | 141.3012 | 141.3294 | positive | MGI_C57BL6J_95278 | Epha2 | NCBI_Gene:13836,ENSEMBL:ENSMUSG00000006445 | MGI:95278 | protein coding gene | Eph receptor A2 |
| 4 | gene | 141.3462 | 141.3462 | positive | MGI_C57BL6J_5455467 | Gm25690 | ENSEMBL:ENSMUSG00000093180 | MGI:5455467 | miRNA gene | predicted gene, 25690 |
| 4 | gene | 141.3478 | 141.3509 | positive | MGI_C57BL6J_3651959 | Gm13074 | ENSEMBL:ENSMUSG00000085836 | MGI:3651959 | lncRNA gene | predicted gene 13074 |
| 4 | pseudogene | 141.3569 | 141.3571 | negative | MGI_C57BL6J_3651957 | Gm13076 | NCBI_Gene:102637915,ENSEMBL:ENSMUSG00000084238 | MGI:3651957 | pseudogene | predicted gene 13076 |
| 4 | gene | 141.3682 | 141.3842 | positive | MGI_C57BL6J_2685539 | Fam131c | NCBI_Gene:277743,ENSEMBL:ENSMUSG00000006218 | MGI:2685539 | protein coding gene | family with sequence similarity 131, member C |
| 4 | gene | 141.3846 | 141.3987 | negative | MGI_C57BL6J_1329026 | Clcnka | NCBI_Gene:12733,ENSEMBL:ENSMUSG00000033770 | MGI:1329026 | protein coding gene | chloride channel, voltage-sensitive Ka |
| 4 | gene | 141.3988 | 141.4112 | positive | MGI_C57BL6J_3651958 | Gm13075 | ENSEMBL:ENSMUSG00000086061 | MGI:3651958 | lncRNA gene | predicted gene 13075 |
| 4 | gene | 141.3998 | 141.4006 | negative | MGI_C57BL6J_5593852 | Gm34693 | NCBI_Gene:102638025 | MGI:5593852 | lncRNA gene | predicted gene, 34693 |
| 4 | gene | 141.4044 | 141.4160 | negative | MGI_C57BL6J_1930643 | Clcnkb | NCBI_Gene:56365,ENSEMBL:ENSMUSG00000006216 | MGI:1930643 | protein coding gene | chloride channel, voltage-sensitive Kb |
| 4 | gene | 141.4208 | 141.4253 | positive | MGI_C57BL6J_1352494 | Hspb7 | NCBI_Gene:29818,ENSEMBL:ENSMUSG00000006221 | MGI:1352494 | protein coding gene | heat shock protein family, member 7 (cardiovascular) |
| 4 | gene | 141.4327 | 141.4361 | negative | MGI_C57BL6J_2685540 | Srarp | NCBI_Gene:277744,ENSEMBL:ENSMUSG00000070637 | MGI:2685540 | protein coding gene | steroid receptor associated and regulated protein |
| 4 | gene | 141.4440 | 141.4447 | negative | MGI_C57BL6J_5625222 | Gm42337 | NCBI_Gene:105247198 | MGI:5625222 | lncRNA gene | predicted gene, 42337 |
| 4 | gene | 141.4447 | 141.4679 | positive | MGI_C57BL6J_107410 | Zbtb17 | NCBI_Gene:22642,ENSEMBL:ENSMUSG00000006215 | MGI:107410 | protein coding gene | zinc finger and BTB domain containing 17 |
| 4 | gene | 141.4679 | 141.5386 | negative | MGI_C57BL6J_1891706 | Spen | NCBI_Gene:56381,ENSEMBL:ENSMUSG00000040761 | MGI:1891706 | protein coding gene | spen family transcription repressor |
| 4 | gene | 141.5355 | 141.5374 | positive | MGI_C57BL6J_3782299 | Gm4123 | NCBI_Gene:100042951 | MGI:3782299 | unclassified gene | predicted gene 4123 |
| 4 | gene | 141.5392 | 141.5436 | positive | MGI_C57BL6J_2443748 | B830004H01Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA B830004H01 gene |
| 4 | gene | 141.5462 | 141.5483 | positive | MGI_C57BL6J_2444063 | B330016D10Rik | NCBI_Gene:320456,ENSEMBL:ENSMUSG00000048406 | MGI:2444063 | lncRNA gene | RIKEN cDNA B330016D10 gene |
| 4 | gene | 141.5761 | 141.6061 | negative | MGI_C57BL6J_1921452 | Fblim1 | NCBI_Gene:74202,ENSEMBL:ENSMUSG00000006219 | MGI:1921452 | protein coding gene | filamin binding LIM protein 1 |
| 4 | gene | 141.6134 | 141.6186 | negative | MGI_C57BL6J_2384869 | Tmem82 | NCBI_Gene:213989,ENSEMBL:ENSMUSG00000043085 | MGI:2384869 | protein coding gene | transmembrane protein 82 |
| 4 | gene | 141.6140 | 141.6156 | positive | MGI_C57BL6J_2140323 | AI507597 | NCBI_Gene:100165,ENSEMBL:ENSMUSG00000073731 | MGI:2140323 | lncRNA gene | expressed sequence AI507597 |
| 4 | gene | 141.6188 | 141.6240 | negative | MGI_C57BL6J_2686215 | Slc25a34 | NCBI_Gene:384071,ENSEMBL:ENSMUSG00000040740 | MGI:2686215 | protein coding gene | solute carrier family 25, member 34 |
| 4 | gene | 141.6257 | 141.6649 | negative | MGI_C57BL6J_1916832 | Plekhm2 | NCBI_Gene:69582,ENSEMBL:ENSMUSG00000028917 | MGI:1916832 | protein coding gene | pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| 4 | gene | 141.6521 | 141.6549 | positive | MGI_C57BL6J_1921207 | 4930451G21Rik | NA | NA | unclassified gene | RIKEN cDNA 4930451G21 gene |
| 4 | gene | 141.6775 | 141.7234 | negative | MGI_C57BL6J_1917244 | Ddi2 | NCBI_Gene:68817,ENSEMBL:ENSMUSG00000078515 | MGI:1917244 | protein coding gene | DNA-damage inducible protein 2 |
| 4 | gene | 141.6836 | 141.6857 | negative | MGI_C57BL6J_3526447 | Rsc1a1 | NCBI_Gene:69994,ENSEMBL:ENSMUSG00000040715 | MGI:3526447 | protein coding gene | regulatory solute carrier protein, family 1, member 1 |
| 4 | gene | 141.7295 | 141.7438 | positive | MGI_C57BL6J_5594022 | Gm34863 | NCBI_Gene:102638257 | MGI:5594022 | lncRNA gene | predicted gene, 34863 |
| 4 | gene | 141.7467 | 141.7593 | positive | MGI_C57BL6J_1923236 | Agmat | NCBI_Gene:75986,ENSEMBL:ENSMUSG00000040706 | MGI:1923236 | protein coding gene | agmatine ureohydrolase (agmatinase) |
| 4 | gene | 141.7602 | 141.7909 | negative | MGI_C57BL6J_2442146 | Dnajc16 | NCBI_Gene:214063,ENSEMBL:ENSMUSG00000040697 | MGI:2442146 | protein coding gene | DnaJ heat shock protein family (Hsp40) member C16 |
| 4 | gene | 141.7936 | 141.8160 | positive | MGI_C57BL6J_1277950 | Casp9 | NCBI_Gene:12371,ENSEMBL:ENSMUSG00000028914 | MGI:1277950 | protein coding gene | caspase 9 |
| 4 | gene | 141.8150 | 141.8262 | negative | MGI_C57BL6J_95316 | Cela2a | NCBI_Gene:13706,ENSEMBL:ENSMUSG00000058579 | MGI:95316 | protein coding gene | chymotrypsin-like elastase family, member 2A |
| 4 | gene | 141.8150 | 141.8464 | negative | MGI_C57BL6J_1923951 | Ctrc | NCBI_Gene:76701,ENSEMBL:ENSMUSG00000062478 | MGI:1923951 | protein coding gene | chymotrypsin C (caldecrin) |
| 4 | gene | 141.8443 | 141.8470 | positive | MGI_C57BL6J_1916389 | Ctrcos | NCBI_Gene:69139,ENSEMBL:ENSMUSG00000086818 | MGI:1916389 | antisense lncRNA gene | chymotrypsin C (caldecrin), opposite strand |
| 4 | gene | 141.8483 | 141.8585 | positive | MGI_C57BL6J_5594193 | Gm35034 | NCBI_Gene:108168984 | MGI:5594193 | lncRNA gene | predicted gene, 35034 |
| 4 | gene | 141.8581 | 141.8749 | negative | MGI_C57BL6J_106504 | Efhd2 | NCBI_Gene:27984,ENSEMBL:ENSMUSG00000040659 | MGI:106504 | protein coding gene | EF hand domain containing 2 |
| 4 | gene | 141.8757 | 141.8770 | positive | MGI_C57BL6J_5580073 | Gm29367 | ENSEMBL:ENSMUSG00000100396 | MGI:5580073 | lncRNA gene | predicted gene 29367 |
| 4 | gene | 141.8797 | 141.8856 | negative | MGI_C57BL6J_5826511 | Gm46874 | NCBI_Gene:108168983 | MGI:5826511 | lncRNA gene | predicted gene, 46874 |
| 4 | gene | 141.8813 | 141.9003 | positive | MGI_C57BL6J_3651870 | Fhad1os2 | NCBI_Gene:102638933,ENSEMBL:ENSMUSG00000085922 | MGI:3651870 | antisense lncRNA gene | forkhead-associated (FHA) phosphopeptide binding domain 1, opposite strand 2 |
| 4 | gene | 141.8904 | 142.0151 | negative | MGI_C57BL6J_1920323 | Fhad1 | NCBI_Gene:329977,ENSEMBL:ENSMUSG00000051435 | MGI:1920323 | protein coding gene | forkhead-associated (FHA) phosphopeptide binding domain 1 |
| 4 | gene | 141.9099 | 141.9159 | positive | MGI_C57BL6J_5625223 | Gm42338 | NCBI_Gene:105247199 | MGI:5625223 | lncRNA gene | predicted gene, 42338 |
| 4 | gene | 141.9209 | 141.9328 | positive | MGI_C57BL6J_5594454 | Gm35295 | NCBI_Gene:102638820 | MGI:5594454 | lncRNA gene | predicted gene, 35295 |
| 4 | pseudogene | 141.9768 | 141.9773 | positive | MGI_C57BL6J_3650255 | Gm13059 | ENSEMBL:ENSMUSG00000081079 | MGI:3650255 | pseudogene | predicted gene 13059 |
| 4 | gene | 141.9830 | 141.9868 | positive | MGI_C57BL6J_3649995 | Fhad1os1 | NCBI_Gene:545691,ENSEMBL:ENSMUSG00000085424 | MGI:3649995 | antisense lncRNA gene | forkhead-associated (FHA) phosphopeptide binding domain 1, opposite strand 1 |
| 4 | gene | 142.0179 | 142.0290 | positive | MGI_C57BL6J_1926167 | 4930455G09Rik | NCBI_Gene:78917,ENSEMBL:ENSMUSG00000085761 | MGI:1926167 | lncRNA gene | RIKEN cDNA 4930455G09 gene |
| 4 | gene | 142.0310 | 142.0846 | negative | MGI_C57BL6J_2384874 | Tmem51 | NCBI_Gene:214359,ENSEMBL:ENSMUSG00000040616 | MGI:2384874 | protein coding gene | transmembrane protein 51 |
| 4 | gene | 142.0429 | 142.0486 | negative | MGI_C57BL6J_3651989 | Gm13053 | ENSEMBL:ENSMUSG00000085574 | MGI:3651989 | lncRNA gene | predicted gene 13053 |
| 4 | gene | 142.0840 | 142.0881 | positive | MGI_C57BL6J_3642733 | Tmem51os1 | NCBI_Gene:100038693,ENSEMBL:ENSMUSG00000073728 | MGI:3642733 | antisense lncRNA gene | Tmem51 opposite strand 1 |
| 4 | gene | 142.1024 | 142.2394 | negative | MGI_C57BL6J_1918779 | Kazn | NCBI_Gene:71529,ENSEMBL:ENSMUSG00000040606 | MGI:1918779 | protein coding gene | kazrin, periplakin interacting protein |
| 4 | gene | 142.1267 | 142.1282 | positive | MGI_C57BL6J_5594740 | Gm35581 | NCBI_Gene:102639222 | MGI:5594740 | lncRNA gene | predicted gene, 35581 |
| 4 | gene | 142.1806 | 142.1841 | positive | MGI_C57BL6J_3649476 | Gm13062 | ENSEMBL:ENSMUSG00000087525 | MGI:3649476 | lncRNA gene | predicted gene 13062 |
| 4 | gene | 142.2394 | 142.2426 | positive | MGI_C57BL6J_5594690 | Gm35531 | NCBI_Gene:102639151 | MGI:5594690 | lncRNA gene | predicted gene, 35531 |
| 4 | gene | 142.2557 | 142.2561 | negative | MGI_C57BL6J_1920742 | 1700085G17Rik | NA | NA | unclassified gene | RIKEN cDNA 1700085G17 gene |
| 4 | gene | 142.3078 | 142.3465 | negative | MGI_C57BL6J_3651990 | Gm13052 | ENSEMBL:ENSMUSG00000085816 | MGI:3651990 | lncRNA gene | predicted gene 13052 |
| 4 | gene | 142.3487 | 142.3610 | positive | MGI_C57BL6J_5826512 | Gm46875 | NCBI_Gene:108168985 | MGI:5826512 | lncRNA gene | predicted gene, 46875 |
| 4 | gene | 142.4071 | 142.4143 | negative | MGI_C57BL6J_5625224 | Gm42339 | NCBI_Gene:105247200 | MGI:5625224 | lncRNA gene | predicted gene, 42339 |
| 4 | gene | 142.4217 | 142.4230 | negative | MGI_C57BL6J_3665284 | C030029H13Rik | NA | NA | unclassified gene | Riken cDNA C030029H13 gene |
| 4 | gene | 142.4956 | 142.4996 | positive | MGI_C57BL6J_5594911 | Gm35752 | NCBI_Gene:102639438 | MGI:5594911 | lncRNA gene | predicted gene, 35752 |
| 4 | gene | 142.5984 | 143.0748 | negative | MGI_C57BL6J_1917975 | 6330411D24Rik | ENSEMBL:ENSMUSG00000110067 | MGI:1917975 | lncRNA gene | RIKEN cDNA 6330411D24 gene |
| 4 | gene | 142.6317 | 142.6354 | positive | MGI_C57BL6J_5625227 | Gm42342 | NCBI_Gene:105247203 | MGI:5625227 | lncRNA gene | predicted gene, 42342 |
| 4 | gene | 142.6418 | 142.6631 | positive | MGI_C57BL6J_5625225 | Gm42340 | NCBI_Gene:105247201 | MGI:5625225 | lncRNA gene | predicted gene, 42340 |
| 4 | gene | 142.6434 | 142.6476 | negative | MGI_C57BL6J_5625226 | Gm42341 | NCBI_Gene:105247202 | MGI:5625226 | lncRNA gene | predicted gene, 42341 |
| 4 | gene | 142.7908 | 142.7910 | positive | MGI_C57BL6J_5610852 | Gm37624 | ENSEMBL:ENSMUSG00000103678 | MGI:5610852 | unclassified gene | predicted gene, 37624 |
| 4 | gene | 143.1027 | 143.1063 | negative | MGI_C57BL6J_1926077 | 5830426C09Rik | NA | NA | unclassified gene | RIKEN cDNA 5830426C09 gene |
| 4 | gene | 143.1074 | 143.2130 | negative | MGI_C57BL6J_107628 | Prdm2 | NCBI_Gene:110593,ENSEMBL:ENSMUSG00000057637 | MGI:107628 | protein coding gene | PR domain containing 2, with ZNF domain |
| 4 | gene | 143.1361 | 143.1362 | negative | MGI_C57BL6J_5562764 | Mir7021 | miRBase:MI0022870,NCBI_Gene:102465618,ENSEMBL:ENSMUSG00000106597 | MGI:5562764 | miRNA gene | microRNA 7021 |
| 4 | pseudogene | 143.1526 | 143.1530 | positive | MGI_C57BL6J_3650684 | Gm13038 | ENSEMBL:ENSMUSG00000081838 | MGI:3650684 | pseudogene | predicted gene 13038 |
| 4 | pseudogene | 143.1666 | 143.1674 | positive | MGI_C57BL6J_3649997 | Gm13039 | NCBI_Gene:100384872,ENSEMBL:ENSMUSG00000083826 | MGI:3649997 | pseudogene | predicted gene 13039 |
| 4 | gene | 143.2041 | 143.2042 | positive | MGI_C57BL6J_5451816 | Gm22039 | ENSEMBL:ENSMUSG00000087903 | MGI:5451816 | snoRNA gene | predicted gene, 22039 |
| 4 | gene | 143.2120 | 143.2150 | positive | MGI_C57BL6J_3646960 | Gm6687 | NA | NA | protein coding gene | predicted gene 6687 |
| 4 | pseudogene | 143.2420 | 143.2434 | negative | MGI_C57BL6J_3650664 | Eef2-ps2 | NCBI_Gene:433776,ENSEMBL:ENSMUSG00000082575 | MGI:3650664 | pseudogene | eukaryotic translation elongation factor 2, pseudogene 2 |
| 4 | gene | 143.2674 | 143.2996 | negative | MGI_C57BL6J_103098 | Pdpn | NCBI_Gene:14726,ENSEMBL:ENSMUSG00000028583 | MGI:103098 | protein coding gene | podoplanin |
| 4 | gene | 143.3404 | 143.3407 | positive | MGI_C57BL6J_5456090 | Gm26313 | ENSEMBL:ENSMUSG00000088395 | MGI:5456090 | unclassified non-coding RNA gene | predicted gene, 26313 |
| 4 | gene | 143.3465 | 143.3710 | positive | MGI_C57BL6J_2442845 | Lrrc38 | NCBI_Gene:242735,ENSEMBL:ENSMUSG00000028584 | MGI:2442845 | protein coding gene | leucine rich repeat containing 38 |
| 4 | pseudogene | 143.3895 | 143.3905 | positive | MGI_C57BL6J_3651262 | Anp32b-ps1 | NCBI_Gene:621961,ENSEMBL:ENSMUSG00000081792 | MGI:3651262 | pseudogene | Bacidic (leucine-rich) nuclear phosphoprotein 32 family, member B, pseudogene 1 |
| 4 | gene | 143.3944 | 143.4002 | positive | MGI_C57BL6J_1890541 | Pramel1 | NCBI_Gene:83491,ENSEMBL:ENSMUSG00000041805 | MGI:1890541 | protein coding gene | preferentially expressed antigen in melanoma-like 1 |
| 4 | pseudogene | 143.4023 | 143.4034 | positive | MGI_C57BL6J_3649502 | Gm13045 | ENSEMBL:ENSMUSG00000082407 | MGI:3649502 | pseudogene | predicted gene 13045 |
| 4 | gene | 143.4123 | 143.4211 | positive | MGI_C57BL6J_2140473 | Pramef8 | NCBI_Gene:242736,ENSEMBL:ENSMUSG00000046862 | MGI:2140473 | protein coding gene | PRAME family member 8 |
| 4 | gene | 143.4372 | 143.4503 | negative | MGI_C57BL6J_2684051 | Oog4 | NCBI_Gene:242737,ENSEMBL:ENSMUSG00000047799 | MGI:2684051 | protein coding gene | oogenesin 4 |
| 4 | pseudogene | 143.4568 | 143.4606 | negative | MGI_C57BL6J_3651155 | Gm13042 | NCBI_Gene:666015,ENSEMBL:ENSMUSG00000083930 | MGI:3651155 | pseudogene | predicted gene 13042 |
| 4 | pseudogene | 143.4643 | 143.4648 | negative | MGI_C57BL6J_3649503 | Gm13046 | ENSEMBL:ENSMUSG00000084059 | MGI:3649503 | pseudogene | predicted gene 13046 |
| 4 | pseudogene | 143.4994 | 143.5025 | positive | MGI_C57BL6J_3649499 | Gm13041 | NCBI_Gene:626700,ENSEMBL:ENSMUSG00000083403 | MGI:3649499 | pseudogene | predicted gene 13041 |
| 4 | gene | 143.5113 | 143.5178 | positive | MGI_C57BL6J_3649500 | Gm13043 | NCBI_Gene:545693,ENSEMBL:ENSMUSG00000095409 | MGI:3649500 | protein coding gene | predicted gene 13043 |
| 4 | gene | 143.5360 | 143.5425 | positive | MGI_C57BL6J_3649498 | Gm13040 | NCBI_Gene:100040854,ENSEMBL:ENSMUSG00000070616 | MGI:3649498 | protein coding gene | predicted gene 13040 |
| 4 | gene | 143.5517 | 143.5582 | positive | MGI_C57BL6J_3649690 | Gm13057 | NCBI_Gene:100040861,ENSEMBL:ENSMUSG00000096154 | MGI:3649690 | protein coding gene | predicted gene 13057 |
| 4 | gene | 143.5517 | 143.5738 | positive | MGI_C57BL6J_3525148 | BC080695 | NCBI_Gene:329986,ENSEMBL:ENSMUSG00000070618 | MGI:3525148 | protein coding gene | cDNA sequence BC080695 |
| 4 | pseudogene | 143.5950 | 143.6010 | negative | MGI_C57BL6J_3650002 | Gm13058 | NCBI_Gene:666049,ENSEMBL:ENSMUSG00000081523 | MGI:3650002 | pseudogene | predicted gene 13058 |
| 4 | pseudogene | 143.6116 | 143.6122 | negative | MGI_C57BL6J_3650235 | Gm13080 | NCBI_Gene:100044633,ENSEMBL:ENSMUSG00000044377 | MGI:3650235 | pseudogene | predicted gene 13080 |
| 4 | gene | 143.6150 | 143.6187 | positive | MGI_C57BL6J_3650232 | Gm13083 | NCBI_Gene:279185,ENSEMBL:ENSMUSG00000066688 | MGI:3650232 | protein coding gene | predicted gene 13083 |
| 4 | gene | 143.6538 | 143.6606 | negative | MGI_C57BL6J_3649971 | Gm13088 | NCBI_Gene:277668,ENSEMBL:ENSMUSG00000078513 | MGI:3649971 | protein coding gene | predicted gene 13088 |
| 4 | pseudogene | 143.6631 | 143.6638 | negative | MGI_C57BL6J_3650236 | Gm13085 | ENSEMBL:ENSMUSG00000084351 | MGI:3650236 | pseudogene | predicted gene 13085 |
| 4 | pseudogene | 143.6711 | 143.6736 | positive | MGI_C57BL6J_3650233 | Gm13082 | NCBI_Gene:100040924,ENSEMBL:ENSMUSG00000081227 | MGI:3650233 | pseudogene | predicted gene 13082 |
| 4 | pseudogene | 143.6819 | 143.6853 | positive | MGI_C57BL6J_5826498 | Gm46861 | NCBI_Gene:108168961 | MGI:5826498 | pseudogene | predicted gene, 46861 |
| 4 | gene | 143.6965 | 143.7027 | negative | MGI_C57BL6J_3649972 | Gm13089 | NCBI_Gene:277667,ENSEMBL:ENSMUSG00000070617 | MGI:3649972 | protein coding gene | predicted gene 13089 |
| 4 | gene | 143.7195 | 143.7292 | positive | MGI_C57BL6J_3650231 | Gm13078 | NCBI_Gene:277666,ENSEMBL:ENSMUSG00000046435 | MGI:3650231 | protein coding gene | predicted gene 13078 |
| 4 | pseudogene | 143.7353 | 143.7356 | negative | MGI_C57BL6J_3649970 | Gm13087 | ENSEMBL:ENSMUSG00000082717 | MGI:3649970 | pseudogene | predicted gene 13087 |
| 4 | pseudogene | 143.7458 | 143.7514 | positive | MGI_C57BL6J_3650234 | Gm13081 | ENSEMBL:ENSMUSG00000082030 | MGI:3650234 | pseudogene | predicted gene 13081 |
R/qtl
scanone
gmObject of class cross2 (crosstype "bc")
Total individuals 138
No. genotyped individuals 138
No. phenotyped individuals 138
No. with both geno & pheno 138
No. phenotypes 1
No. covariates 7
No. phenotype covariates 0
No. chromosomes 20
Total markers 131355
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
9956 9987 7848 7585 7609 7736 7399 6458 6713 6385 7143 6110 6082 5966 5346 5015
17 18 19 X
5080 4605 3562 4770 #detach("package:qtl2", unload=TRUE)
#library(qtl)
cross <- qtl::read.cross("csv", file = "data/ici.vs.pbs_gm_qtl_5.batches_mis_conditional_2-peaks-chr4-10.csv",alleles=c("A","B")) --Read the following data:
138 individuals
131355 markers
5 phenotypes
--Cross type: bc cross <- qtl::jittermap(cross)
summary(cross) Backcross
No. individuals: 138
No. phenotypes: 5
Percent phenotyped: 100 100 100 100 100
No. chromosomes: 20
Autosomes: 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19
X chr: X
Total markers: 131355
No. markers: 9956 9987 7848 7585 7609 7736 7399 6458 6713 6385 7143
6110 6082 5966 5346 5015 5080 4605 3562 4770
Percent genotyped: 99.5
Genotypes (%):
Autosomes: AA:88.1 AB:11.9
X chromosome: AA:93.9 AB:6.1 cross.probs <- qtl::calc.genoprob(cross)
print("method == hk")[1] "method == hk"add.covars = qtl::pull.pheno(cross.probs, c("UNC8250659","UNC18240977"))
scanone.hk <-qtl::scanone(cross.probs, pheno.col="ICI.vs.PBS" , model="binary", method="hk", addcovar = add.covars)
operm.hk <- qtl::scanone(cross.probs, method = "hk", pheno.col="ICI.vs.PBS", n.perm = 10, perm.Xsp = TRUE, model="binary", verbose=FALSE, addcovar = add.covars)
plot(operm.hk)
print(summary(operm.hk, alpha=c(0.01, 0.05, 0.1)))Autosome LOD thresholds (10 permutations)
lod
1% 3.33
5% 3.28
10% 3.21
X chromosome LOD thresholds (182 permutations)
lod
1% 3.39
5% 3.29
10% 3.17#plot(scanone.hk, bandcol = "grey90",lty=1, cex=1, col = "steelblue")
#qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.01, col = 'blue')
#qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.05, col = 'red')
#qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.1, col = 'purple')
ymx <- maxlod(out) # overall maximum LOD score
plot(scanone.hk, bandcol = "grey90",lty=1, cex=1, col = "slateblue", ylim=c(0, ymx+0.5))
title(main = paste0(colnames(out), " [positions in cM]"))
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.01, col = 'blue')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.05, col = 'red')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.1, col = 'purple')
ymx <- 14
plot(scanone.hk, bandcol = "grey90",lty=1, cex=1, col = "slateblue", ylim=c(0, ymx+0.5))
title(main = paste0(colnames(out), " [positions in cM]\n(using same scale as ici vs. eoi for easier comparison)"))
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.01, col = 'blue')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.05, col = 'red')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.1, col = 'purple')
print(as.data.frame(summary(scanone.hk, perms=operm.hk, pvalues=TRUE, format="allpeaks"))) chr pos lod pval
UNC1197721 1 47.673577 1.868502 1.0000000
UNCHS005266 2 40.908610 1.873368 1.0000000
UNC6045859 3 50.620447 1.870152 1.0000000
JAX00123711 4 53.542073 1.799165 1.0000000
JAX00574407 5 6.213413 3.388862 0.0000000
UNCHS017906 6 37.001766 1.868162 1.0000000
JAX00233659 7 87.998157 1.870072 1.0000000
JAX00667121 8 24.443849 2.382541 0.6196652
UNC17203329 9 68.519175 1.323467 1.0000000
JAX00302150 10 74.547218 2.532706 0.5187088
UNC20281687 11 67.380801 1.859331 1.0000000
UNCHS034188 12 38.836978 1.870151 1.0000000
JAX00350973 13 5.743505 1.378528 1.0000000
JAX00380911 14 30.154517 2.381660 0.6196652
UNC25275535 15 11.968141 1.402063 1.0000000
UNC26790882 16 30.832641 2.019242 0.9118988
UNCHS045288 17 60.699043 2.034920 0.9118988
JAX00450853 18 2.913076 2.621475 0.5187088
UNCHS047190 19 8.712408 1.804318 1.0000000
XiB2 X 45.699951 1.870087 0.9996602print("all peaks with a p-value less or equal to 0.05 (suggestive)")[1] "all peaks with a p-value less or equal to 0.05 (suggestive)"print(as.data.frame(summary(scanone.hk, perms=operm.hk, alpha=0.05, pvalues=TRUE, format="allpeaks"))) chr pos lod pval
JAX00574407 5 6.213413 3.388862 0#print("method == ehk")
#scanone.ehk <-qtl::scanone(cross.probs, pheno.col="ICI.vs.PBS" , model="binary", method="ehk")
#operm.ehk <- qtl::scanone(cross.probs, method = "ehk", pheno.col="ICI.vs.PBS", n.perm = 10, perm.Xsp = TRUE, model="binary", verbose=FALSE)
#plot(operm.ehk)
#print(summary(operm.ehk, alpha=c(0.01, 0.05, 0.1)))
#plot(scanone.ehk, bandcol = "grey90",lty=1, cex=1, col = "steelblue")
#qtl::add.threshold(scanone.ehk, perms= operm.ehk, alpha=0.01, col = 'blue')
#qtl::add.threshold(scanone.ehk, perms= operm.ehk, alpha=0.05, col = 'red')
#qtl::add.threshold(scanone.ehk, perms= operm.ehk, alpha=0.1, col = 'purple')
#print(as.data.frame(summary(scanone.ehk)))
#print(as.data.frame(summary(scanone.ehk, perms=operm.ehk, alpha=0.05, pvalues=TRUE, format="allpeaks")))
R version 3.5.1 (2018-07-02)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS 10.15.7
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_AU.UTF-8/en_AU.UTF-8/en_AU.UTF-8/C/en_AU.UTF-8/en_AU.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] abind_1.4-5 qtl2_0.22 reshape2_1.4.3 ggplot2_3.3.6
[5] tibble_3.1.7 psych_2.2.5 readxl_1.4.0 cluster_2.0.7-1
[9] dplyr_1.0.9 optparse_1.6.6 rhdf5_2.26.2 mclust_5.4.5
[13] tidyr_1.2.0 data.table_1.14.2 knitr_1.29 kableExtra_1.3.4
[17] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] httr_1.4.3 bit64_0.9-7 viridisLite_0.3.0 assertthat_0.2.1
[5] highr_0.8 blob_1.2.1 cellranger_1.1.0 yaml_2.2.1
[9] gdtools_0.2.1 pillar_1.7.0 RSQLite_2.2.0 backports_1.1.5
[13] lattice_0.20-35 glue_1.6.2 digest_0.6.25 promises_1.1.0
[17] rvest_1.0.2 colorspace_1.4-1 htmltools_0.5.2 httpuv_1.5.2
[21] plyr_1.8.6 pkgconfig_2.0.3 purrr_0.3.4 scales_1.1.1
[25] webshot_0.5.3 svglite_1.2.3 qtl_1.46-2 getopt_1.20.3
[29] later_1.0.0 git2r_0.26.1 generics_0.0.2 ellipsis_0.3.2
[33] withr_2.5.0 cli_3.3.0 mnormt_1.5-6 magrittr_2.0.3
[37] crayon_1.5.1 memoise_1.1.0 evaluate_0.14 fs_1.3.2
[41] fansi_0.4.1 nlme_3.1-137 xml2_1.3.2 tools_3.5.1
[45] lifecycle_1.0.1 stringr_1.4.0 Rhdf5lib_1.4.3 munsell_0.5.0
[49] compiler_3.5.1 systemfonts_0.1.1 rlang_1.0.3 grid_3.5.1
[53] rstudioapi_0.13 rmarkdown_2.3 gtable_0.3.0 DBI_1.1.0
[57] R6_2.4.1 fastmap_1.1.0 bit_1.1-15.2 utf8_1.1.4
[61] rprojroot_1.3-2 stringi_1.4.6 parallel_3.5.1 Rcpp_1.0.4.6
[65] vctrs_0.4.1 tidyselect_1.1.2 xfun_0.15