QTL Analysis - binary [ICI vs PBS] (conditioning on eoi vs. ici peaks)
Belinda Cornes
2022-07-12
Last updated: 2022-07-12
Checks: 5 2
Knit directory: Serreze-T1D_Workflow/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R
Markdown file created these results, you’ll want to first commit it to
the Git repo. If you’re still working on the analysis, you can ignore
this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20220210) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /Users/corneb/Documents/MyJax/CS/Projects/Serreze/qc/workflowr/Serreze-T1D_Workflow | . |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version a8c2d1a. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: analysis/.DS_Store
Ignored: data/.DS_Store
Untracked files:
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_0.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_52.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_mis_0.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_mis_52.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_mis_vo.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_vo.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_ici-early.vs.pbs.ici-late_5.batches.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_ici-early.vs.pbs.ici-late_5.batches_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs.ici-late_5.batches.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs.ici-late_5.batches_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs.ici-late_snpsqc_5.batches.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs.ici-late_snpsqc_5.batches_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs.ici-late_snpsqc_dis_no-x_updated_5.batches.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs.ici-late_snpsqc_dis_no-x_updated_5.batches_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.eoi_snpsqc_dis_no-x_updated_5.batches_11.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.eoi_snpsqc_dis_no-x_updated_5.batches_mis_11.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_conditional.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_conditional_1-peak.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_conditional_1-peak.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_conditional_1-peak_ahb.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_conditional_1-peak_test.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_conditional_2-peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_conditional_2-peaks.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_conditional_2-peaks_ahb.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_conditional_3-peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_conditional_3-peaks.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_conditional_3-peaks_ahb.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_mis_conditional.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_mis_conditional_1-peak.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_mis_conditional_1-peak.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_mis_conditional_1-peak_ahb.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_mis_conditional_2-peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_mis_conditional_2-peaks.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_mis_conditional_2-peaks_ahb.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_mis_conditional_3-peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_mis_conditional_3-peaks.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_5.batches_mis_conditional_3-peaks_ahb.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_1.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_conditional.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_conditional_1-peak.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_conditional_1-peak.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_conditional_1-peak_ahb.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_conditional_2-peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_conditional_2-peaks.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_conditional_2-peaks_ahb.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_conditional_3-peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_conditional_3-peaks.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_conditional_3-peaks_ahb.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_mis_1.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_mis_conditional.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_mis_conditional_1-peak.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_mis_conditional_1-peak.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_mis_conditional_1-peak_ahb.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_mis_conditional_2-peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_mis_conditional_2-peaks.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_mis_conditional_2-peaks_ahb.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_mis_conditional_3-peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_mis_conditional_3-peaks.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_mis_conditional_3-peaks_ahb.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_1.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_11.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_conditional.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_conditional_1-peak.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_conditional_1-peak.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_conditional_1-peak_ahb.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_conditional_2-peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_conditional_2-peaks.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_conditional_2-peaks_ahb.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_conditional_3-peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_conditional_3-peaks.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_conditional_3-peaks_ahb.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_11.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_conditional.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_conditional_1-peak.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_conditional_1-peak.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_conditional_1-peak_ahb.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_conditional_1.peak.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_conditional_2-peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_conditional_2-peaks.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_conditional_2-peaks_ahb.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_conditional_3-peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_conditional_3-peaks.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_conditional_3-peaks_ahb.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs.ici-late_pheno.corrected.cleaned_5.batches.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs.ici-late_pheno.corrected.cleaned_5.batches_mis.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs.ici-late_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs.ici-late_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs.ici-late_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs.ici-late_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_changed.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_changed.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_scanone_5.batches.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_scanone_5.batches_mis.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_oops.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_v2.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.4.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.4_miss.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.5.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.5_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_pheno.corrected.cleaned_5.batches_conditional.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_pheno.corrected.cleaned_5.batches_mis_conditional.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_conditional.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_mis_conditional.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_conditional.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_conditional.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs.ici-late_pheno.corrected.cleaned_5.batches.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs.ici-late_pheno.corrected.cleaned_5.batches_mis.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs.ici-late_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs.ici-late_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs.ici-late_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs.ici-late_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_vo.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_vo1.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_vo.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_vo1.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_pheno.corrected.cleaned_5.batches_conditional.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_pheno.corrected.cleaned_5.batches_mis_conditional.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_conditional.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_mis_conditional.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_conditional.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_conditional.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_conditional_1-peak.Rmd.R
Untracked: analysis/genotype.frequencies_ici-early.vs.pbs.ici-late_5.batches.Rmd
Untracked: analysis/genotype.frequencies_ici-early.vs.pbs.ici-late_5.batches_mis.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.eoi_5.batches_0.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.eoi_5.batches_52.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.eoi_5.batches_mis_0.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.eoi_5.batches_mis_52.Rmd
Untracked: analysis/index_5.batches_additional_vo.Rmd
Untracked: analysis/power.analysis_ici.vs.eoi_5.batches.Rmd
Untracked: analysis/power.analysis_ici.vs.eoi_5.batches.Rmd.R
Untracked: analysis/power.analysis_ici.vs.eoi_5.batches_mis.Rmd
Untracked: analysis/power.analysis_ici.vs.eoi_5.batches_mis.Rmd.R
Untracked: analysis/power.analysis_ici.vs.pbs_5.batches.Rmd
Untracked: analysis/power.analysis_ici.vs.pbs_5.batches.Rmd.R
Untracked: analysis/power.analysis_ici.vs.pbs_5.batches_mis.Rmd
Untracked: analysis/power.analysis_ici.vs.pbs_5.batches_mis.Rmd.R
Untracked: data/GM_covar.csv
Untracked: data/GM_covar_BC312.csv
Untracked: data/bad_markers_all_4.batches.RData
Untracked: data/bad_markers_all_5.batches.RData
Untracked: data/blup_sub_chr10_lod.drop-1.5_5.batches.csv
Untracked: data/blup_sub_chr3_lod.drop-1.5.csv
Untracked: data/blup_sub_chr3_lod.drop-1.5_5.batches.csv
Untracked: data/blup_sub_chr4_lod.drop-1.5.csv
Untracked: data/blup_sub_chr4_lod.drop-1.5_5.batches.csv
Untracked: data/covar_cleaned_ici.vs.eoi.csv
Untracked: data/covar_cleaned_ici.vs.pbs.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs.ici-late_5.batches.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs.ici-late_5.batches_mis.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_0.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_52.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_mis.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_mis_0.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_mis_52.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi1.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_0.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_52.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_mis.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_mis_0.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_mis_52.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs1.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_0.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_52.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_mis.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_mis_0.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_mis_52.csv
Untracked: data/covar_corrected_ici-early.vs.pbs.ici-late_5.batches.csv
Untracked: data/covar_corrected_ici-early.vs.pbs.ici-late_5.batches_mis.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_0.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_52.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_mis.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_mis_0.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_mis_52.csv
Untracked: data/covar_corrected_ici.vs.eoi.csv
Untracked: data/covar_corrected_ici.vs.eoi1.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_0.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_52.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_mis.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_mis_0.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_mis_52.csv
Untracked: data/covar_corrected_ici.vs.pbs.csv
Untracked: data/covar_corrected_ici.vs.pbs1.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_0.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_52.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_mis.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_mis_0.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_mis_52.csv
Untracked: data/e.RData
Untracked: data/e_BC312.RData
Untracked: data/e_snpg_samqc_4.batches.RData
Untracked: data/e_snpg_samqc_4.batches_bc.RData
Untracked: data/e_snpg_samqc_5.batches.RData
Untracked: data/errors_ind_4.batches.RData
Untracked: data/errors_ind_4.batches_bc.RData
Untracked: data/errors_ind_5.batches.RData
Untracked: data/files.to.sync.txt
Untracked: data/fitqtl_chr3.peak_chr4.peak_additive.txt
Untracked: data/fitqtl_chr3.peak_chr4.peak_interacting.txt
Untracked: data/fitqtl_chr3.peak_chr4.peak_sex_additive.txt
Untracked: data/fitqtl_chr3.peak_chr4.peak_sex_interacting.txt
Untracked: data/g2blup_effects.csv
Untracked: data/g2blup_effects.xlsx
Untracked: data/genes_chr10_lod.drop-1.5_5.batches.csv
Untracked: data/genes_chr3_lod.drop-1.5.csv
Untracked: data/genes_chr3_lod.drop-1.5_5.batches.csv
Untracked: data/genes_chr4_lod.drop-1.5.csv
Untracked: data/genetic_map.csv
Untracked: data/genetic_map_BC312.csv
Untracked: data/genotype_errors_marker_4.batches.RData
Untracked: data/genotype_errors_marker_5.batches.RData
Untracked: data/genotype_freq_marker_4.batches.RData
Untracked: data/genotype_freq_marker_5.batches.RData
Untracked: data/gm_allqc_4.batches.RData
Untracked: data/gm_allqc_5.batches.RData
Untracked: data/gm_allqc_5.batches_mis.RData
Untracked: data/gm_samqc_3.batches.RData
Untracked: data/gm_samqc_4.batches.RData
Untracked: data/gm_samqc_4.batches_bc.RData
Untracked: data/gm_samqc_5.batches.RData
Untracked: data/gm_serreze.192.RData
Untracked: data/gm_serreze.BC312.RData
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-additive.covariates_blup_sub_chr-6_peak.marker-UNC11006703_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-additive.covariates_blup_sub_chr-6_peak.marker-UNC11006703_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-additive.covariates_blup_sub_chr-6_peak.marker-cr27snv203_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-additive.covariates_genes_chr-6_peak.marker-UNC11006703_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-additive.covariates_genes_chr-6_peak.marker-UNC11006703_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-additive.covariates_genes_chr-6_peak.marker-cr27snv203_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-interactive.covariate_blup_sub_chr-1_peak.marker-ICR134_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-interactive.covariate_blup_sub_chr-1_peak.marker-ICR4119_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-interactive.covariate_blup_sub_chr-1_peak.marker-JAX00262446_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-interactive.covariate_blup_sub_chr-1_peak.marker-UNCHS001938_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-interactive.covariate_blup_sub_chr-6_peak.marker-UNC11008761_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-interactive.covariate_blup_sub_chr-6_peak.marker-UNCHS016990_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-interactive.covariate_genes_chr-1_peak.marker-ICR134_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-interactive.covariate_genes_chr-1_peak.marker-ICR4119_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-interactive.covariate_genes_chr-1_peak.marker-JAX00262446_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-interactive.covariate_genes_chr-1_peak.marker-UNCHS001938_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-interactive.covariate_genes_chr-6_peak.marker-UNC11008761_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-interactive.covariate_genes_chr-6_peak.marker-UNCHS016990_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNC28655293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_blup_sub_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_blup_sub_chr-5_peak.marker-UNC8675939_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNC28655293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_genes_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_genes_chr-5_peak.marker-UNC8675939_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_genes_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-10_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-10_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-10_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-11_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-11_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-11_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-12_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-12_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-12_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-13_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-13_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-13_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-14_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-14_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-14_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-15_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-15_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-15_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-16_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-16_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-16_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-17_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-17_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-17_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-18_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-18_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-18_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-19_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-19_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-19_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-1_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-1_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-1_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-2_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-2_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-2_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-3_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-3_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-3_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-4_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-4_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-4_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-5_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-6_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-6_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-6_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-7_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-7_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-7_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-8_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-8_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-8_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-9_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-9_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-9_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-X_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-X_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-X_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-10_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-10_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-10_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-11_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-11_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-11_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-12_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-12_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-12_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-13_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-13_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-13_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-14_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-14_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-14_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-15_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-15_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-15_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-16_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-16_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-16_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-17_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-17_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-17_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-18_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-18_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-18_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-19_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-19_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-19_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-1_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-1_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-1_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-2_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-2_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-2_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-3_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-3_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-3_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-4_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-4_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-4_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-5_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-6_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-6_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-6_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-7_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-7_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-7_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-8_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-8_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-8_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-9_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-9_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-9_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-X_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-X_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-X_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_gm_qtl_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_gm_qtl_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_gm_qtl_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_gm_qtl_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_gm_qtl_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_marker.freq_low.geno.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_marker.freq_low.probs.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs.ici-late_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs.ici-late_scanone_5.batches.Rdata
Untracked: data/ici-early.vs.pbs.ici-late_scanone_5.batches_mis.Rdata
Untracked: data/ici-early.vs.pbs.ici-late_scanone_snpsqc_5.batches.Rdata
Untracked: data/ici-early.vs.pbs.ici-late_scanone_snpsqc_5.batches_mis.Rdata
Untracked: data/ici-early.vs.pbs.ici-late_scanone_snpsqc_dis_no-x_updated_5.batches.Rdata
Untracked: data/ici-early.vs.pbs.ici-late_scanone_snpsqc_dis_no-x_updated_5.batches_mis.Rdata
Untracked: data/ici-early.vs.pbs_age.of.onset-additive.covariates_blup_sub_chr-7_peak.marker-UNC13388811_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-additive.covariates_genes_chr-7_peak.marker-UNC13388811_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-15_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-15_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-15_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-17_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-17_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-17_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_5.batches.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_scanone_5.batches.Rdata
Untracked: data/ici-early.vs.pbs_scanone_5.batches_mis.Rdata
Untracked: data/ici-early.vs.pbs_scanone_snpsqc_5.batches.Rdata
Untracked: data/ici-early.vs.pbs_scanone_snpsqc_5.batches_mis.Rdata
Untracked: data/ici-early.vs.pbs_scanone_snpsqc_dis_no-x_updated_5.batches.Rdata
Untracked: data/ici-early.vs.pbs_scanone_snpsqc_dis_no-x_updated_5.batches_mis.Rdata
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00020646_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00292499_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00294019_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18216614_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18240977_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18343181_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18363544_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18363544_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18363544_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-11_peak.marker-UNC20090524_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-2_peak.marker-UNCHS008007_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-2_peak.marker-UNCHS008007_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR5263_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR5263_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00105649_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00105649_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00106210_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00107193_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00516812_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00516812_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00522922_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00524205_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00535764_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4901083_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4901083_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4909519_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4909519_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4915446_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4915446_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4938565_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4938565_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4959266_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4959266_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4960612_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4960612_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4961542_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4961542_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5046545_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5055259_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5062524_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5068215_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5076935_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5082757_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5173293_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5193970_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5257268_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5264696_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5267813_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5270225_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5277061_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5278229_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5278229_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5282259_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5287977_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5301460_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5309136_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5321559_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5322455_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5329171_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5360055_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5778977_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5916997_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008229_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008229_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008234_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008234_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008242_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008260_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008260_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008281_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008281_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008338_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008338_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008363_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008363_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008409_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008432_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008609_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008614_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008627_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008659_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008725_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009066_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009067_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009068_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009112_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009693_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009821_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001276_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-sanger2496q_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-sanger2496q_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-4_peak.marker-UNC8250659_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-4_peak.marker-UNC8439633_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-4_peak.marker-UNC8439633_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-4_peak.marker-UNCHS012955_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-6_peak.marker-UNC11108920_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-6_peak.marker-UNC11108920_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00020646_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00292499_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00294019_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18216614_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18240977_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18343181_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18363544_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18363544_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18363544_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-11_peak.marker-UNC20090524_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-2_peak.marker-UNCHS008007_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-2_peak.marker-UNCHS008007_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR5263_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR5263_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00105649_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00105649_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00106210_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00107193_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00516812_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00516812_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00522922_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00524205_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00535764_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4901083_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4901083_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4909519_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4909519_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4915446_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4915446_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4938565_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4938565_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4959266_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4959266_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4960612_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4960612_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4961542_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4961542_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5046545_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5055259_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5062524_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5068215_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5076935_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5082757_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5173293_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5193970_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5257268_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5264696_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5267813_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5270225_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5277061_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5278229_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5278229_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5282259_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5287977_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5301460_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5309136_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5321559_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5322455_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5329171_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5360055_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5778977_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5916997_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008229_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008229_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008234_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008234_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008242_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008260_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008260_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008281_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008281_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008338_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008338_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008363_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008363_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008409_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008432_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008609_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008614_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008627_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008659_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008725_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009066_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009067_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009068_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009112_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009693_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009821_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001276_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-sanger2496q_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-sanger2496q_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-4_peak.marker-UNC8250659_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-4_peak.marker-UNC8439633_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-4_peak.marker-UNC8439633_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-4_peak.marker-UNCHS012955_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-6_peak.marker-UNC11108920_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-6_peak.marker-UNC11108920_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_blup.full_chr-10_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-11_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-12_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-13_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-14_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-15_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-16_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-17_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-18_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-19_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-1_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-2_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-3_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-4_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-6_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-7_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-8_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-9_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-X_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-ICR4295_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-JAX00292499_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18154330_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18195765_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18216614_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18240977_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18856953_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNCJPD004316_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNCJPD009532_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNC19155926_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNC19481366_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNCJPD004792_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-12_peak.marker-ICR4497_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-13_peak.marker-UNCHS036964_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-18_peak.marker-UNCHS045344_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-19_peak.marker-UNCJPD007087_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-1_peak.marker-ICR010_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-1_peak.marker-ICR010_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-2_peak.marker-UNC4609660_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-B6_rs31740628_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR1269_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR5325_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR5347_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR5348_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00104971_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00105059_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00105203_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00107930_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00107980_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00108068_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00108114_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00109750_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00189245_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00517218_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00517906_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00518418_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00518684_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00520122_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00524717_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00524828_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00525380_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00525709_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00527291_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00527587_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00529707_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00531601_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00532122_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4673471_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4681411_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4793677_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4799475_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4909830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4914130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4964062_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4985064_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4991565_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5001723_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5042361_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5055259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5076935_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5082757_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5173293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5182782_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5193970_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5240484_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5256922_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5266664_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5266711_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5267813_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5274554_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5277061_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5282259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5287977_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5301460_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5321559_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5322455_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5328596_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5350384_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5354724_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5375208_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5384072_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5400314_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5433945_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5435874_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5466669_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5517972_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5522257_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5523265_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5532424_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5553882_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5572051_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5582753_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5589260_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5595555_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5623239_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5639830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5680234_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5682314_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5725356_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5727351_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5734420_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5743257_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5751567_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5752623_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5776974_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5778977_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5788507_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5792468_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5794598_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5795561_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5802298_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5806628_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5817478_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5836999_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5841501_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5847927_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5966417_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008129_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008173_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008231_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008235_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008242_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008253_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008256_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008311_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008317_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008372_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008410_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008415_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008431_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008609_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008614_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008649_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008651_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008723_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008802_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008803_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008808_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008834_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008838_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008928_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008948_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008959_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008973_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008987_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009001_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009025_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009034_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009087_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009109_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009142_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009207_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009216_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009306_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009316_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009364_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009377_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009382_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009387_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009395_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009413_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009428_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009463_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009503_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009532_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009541_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009560_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009574_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001276_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001315_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001355_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001373_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001382_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC6851205_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8250659_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8299912_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8300499_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8381981_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8436434_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8439633_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8530763_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS012948_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS012976_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013139_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013304_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-5_peak.marker-JAX00590770_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-ICR1830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-UNC13170794_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-UNCHS019480_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-cr31snv87_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-UNC15430711_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-9_peak.marker-UNC17203329_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-9_peak.marker-UNCHS024593_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-X_peak.marker-UNC30627130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-X_peak.marker-UNCHS048313_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_snpsqc_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_snpsqc_dis_no-x_updated_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_snpsqc_dis_no-x_updated_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_snpsqc_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_snpsqc_dis_no-x_updated_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_snpsqc_dis_no-x_updated_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_snpsqc_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_snpsqc_dis_no-x_updated_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_snpsqc_dis_no-x_updated_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_snpsqc_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_snpsqc_dis_no-x_updated_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_snpsqc_dis_no-x_updated_5.batches_mis.mis.txt
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-ICR4295_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-JAX00292499_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18154330_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18195765_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18216614_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18240977_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18856953_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNCJPD004316_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNCJPD009532_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNC19155926_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNC19481366_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNCJPD004792_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-12_peak.marker-ICR4497_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-13_peak.marker-UNCHS036964_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-18_peak.marker-UNCHS045344_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-19_peak.marker-UNCJPD007087_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-1_peak.marker-ICR010_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-1_peak.marker-ICR010_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-2_peak.marker-UNC4609660_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-B6_rs31740628_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR1269_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR5325_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR5347_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR5348_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00104971_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00105059_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00105203_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00107930_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00107980_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00108068_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00108114_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00109750_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00189245_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00517218_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00517906_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00518418_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00518684_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00520122_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00524717_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00524828_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00525380_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00525709_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00527291_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00527587_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00529707_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00531601_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00532122_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4673471_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4681411_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4793677_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4799475_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4909830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4914130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4964062_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4985064_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4991565_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5001723_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5042361_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5055259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5076935_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5082757_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5173293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5182782_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5193970_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5240484_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5256922_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5266664_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5266711_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5267813_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5274554_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5277061_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5282259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5287977_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5301460_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5321559_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5322455_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5328596_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5350384_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5354724_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5375208_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5384072_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5400314_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5433945_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5435874_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5466669_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5517972_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5522257_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5523265_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5532424_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5553882_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5572051_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5582753_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5589260_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5595555_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5623239_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5639830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5680234_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5682314_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5725356_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5727351_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5734420_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5743257_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5751567_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5752623_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5776974_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5778977_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5788507_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5792468_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5794598_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5795561_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5802298_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5806628_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5817478_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5836999_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5841501_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5847927_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5966417_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008129_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008173_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008231_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008235_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008242_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008253_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008256_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008311_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008317_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008372_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008410_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008415_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008431_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008609_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008614_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008649_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008651_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008723_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008802_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008803_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008808_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008834_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008838_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008928_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008948_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008959_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008973_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008987_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009001_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009025_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009034_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009087_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009109_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009142_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009207_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009216_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009306_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009316_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009364_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009377_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009382_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009387_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009395_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009413_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009428_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009463_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009503_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009532_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009541_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009560_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009574_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001276_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001315_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001355_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001373_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001382_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC6851205_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8250659_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8299912_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8300499_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8381981_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8436434_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8439633_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8530763_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS012948_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS012976_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013139_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013304_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-5_peak.marker-JAX00590770_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-ICR1830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-UNC13170794_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-UNCHS019480_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-cr31snv87_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-UNC15430711_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-9_peak.marker-UNC17203329_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-9_peak.marker-UNCHS024593_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-X_peak.marker-UNC30627130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-X_peak.marker-UNCHS048313_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_gm_qtl_5.batches.csv
Untracked: data/ici.vs.eoi_gm_qtl_5.batches_mis.csv
Untracked: data/ici.vs.eoi_gm_qtl_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_gm_qtl_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_gm_qtl_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18343181_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18376338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18382395_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNCHS029024_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-1_peak.marker-UNC228500_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-1_peak.marker-UNC228500_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-ICR2047_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00112499_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00219554_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00529707_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4837069_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5006948_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5266664_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5300878_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5360055_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5679155_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5945286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC6227106_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008595_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009067_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009693_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001341_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-sanger2623_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-4_peak.marker-UNCHS013125_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-4_peak.marker-UNCHS013125_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-6_peak.marker-UNCHS018170_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-6_peak.marker-UNCHS018231_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18343181_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18376338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18382395_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNCHS029024_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-1_peak.marker-UNC228500_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-1_peak.marker-UNC228500_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-ICR2047_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00112499_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00219554_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00529707_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4837069_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5006948_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5266664_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5300878_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5360055_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5679155_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5945286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC6227106_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008595_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS009067_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS009693_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCJPD001246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCJPD001341_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-sanger2623_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-4_peak.marker-UNCHS013125_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-4_peak.marker-UNCHS013125_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-6_peak.marker-UNCHS018170_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-6_peak.marker-UNCHS018231_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.geno.freq.removed.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.geno.freq.removed_5.batches.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.geno.freq.removed_5.batches_mis.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_5.batches.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_5.batches_mis.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.probs.freq.removed.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.probs.freq.removed_5.batches.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.probs.freq.removed_5.batches_mis.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_5.batches.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_5.batches_mis.csv
Untracked: data/ici.vs.eoi_scanone_5.batches.Rdata
Untracked: data/ici.vs.eoi_scanone_5.batches_mis.Rdata
Untracked: data/ici.vs.eoi_scanone_snpsqc_5.batches.Rdata
Untracked: data/ici.vs.eoi_scanone_snpsqc_5.batches_mis.Rdata
Untracked: data/ici.vs.eoi_scanone_snpsqc_dis_no-x_updated_5.batches.Rdata
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNC28655293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-5_peak.marker-UNC8675939_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNC28655293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-5_peak.marker-UNC8675939_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_bad.markers_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-15_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-15_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-15_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-17_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-17_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-17_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-13_peak.marker-UNC22384241_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_conditional_1-peak-chr10.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_conditional_1-peak-chr3.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_conditional_1-peak-chr4.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_conditional_2-peaks-chr3-10.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_conditional_2-peaks-chr3-4.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_conditional_2-peaks-chr4-10.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_conditional_3-peaks-chr3-4-10.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-19_peak.marker-UNC29893228_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-19_peak.marker-UNC29990033_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-19_peak.marker-UNCHS047192_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-4_peak.marker-UNCHS012304_lod.drop-1.5_5.batches_mis_conditional_2-peaks-chr4-10.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-5_peak.marker-UNCJPD008862_lod.drop-1.5_5.batches_mis_conditional_1-peak-chr10.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-5_peak.marker-UNCJPD008862_lod.drop-1.5_5.batches_mis_conditional_2-peaks-chr4-10.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-8_peak.marker-UNCHS023353_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-8_peak.marker-UNCHS023353_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-8_peak.marker-UNCHS023364_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_fitqtl_peaks_additive_snpsqc_dis_no-x_updated_5.batches_mis_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_additive_snpsqc_dis_no-x_updated_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_additive_snpsqc_snpsqc_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_interacting_snpsqc_dis_no-x_updated_5.batches_mis_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_interacting_snpsqc_dis_no-x_updated_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_interacting_snpsqc_snpsqc_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_additive_snpsqc_dis_no-x_updated_5.batches_mis_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_additive_snpsqc_dis_no-x_updated_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_additive_snpsqc_snpsqc_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_interacting_snpsqc_dis_no-x_updated_5.batches_mis_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_interacting_snpsqc_dis_no-x_updated_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_interacting_snpsqc_snpsqc_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_genes_chr-13_peak.marker-UNC22384241_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_conditional_1-peak-chr10.csv
Untracked: data/ici.vs.pbs_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_conditional_1-peak-chr3.csv
Untracked: data/ici.vs.pbs_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_conditional_1-peak-chr4.csv
Untracked: data/ici.vs.pbs_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_conditional_2-peaks-chr3-10.csv
Untracked: data/ici.vs.pbs_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_conditional_2-peaks-chr3-4.csv
Untracked: data/ici.vs.pbs_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_conditional_2-peaks-chr4-10.csv
Untracked: data/ici.vs.pbs_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_conditional_3-peaks-chr3-4-10.csv
Untracked: data/ici.vs.pbs_genes_chr-19_peak.marker-UNC29893228_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-19_peak.marker-UNC29990033_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-19_peak.marker-UNCHS047192_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-4_peak.marker-UNCHS012304_lod.drop-1.5_5.batches_mis_conditional_2-peaks-chr4-10.csv
Untracked: data/ici.vs.pbs_genes_chr-5_peak.marker-UNCJPD008862_lod.drop-1.5_5.batches_mis_conditional_1-peak-chr10.csv
Untracked: data/ici.vs.pbs_genes_chr-5_peak.marker-UNCJPD008862_lod.drop-1.5_5.batches_mis_conditional_2-peaks-chr4-10.csv
Untracked: data/ici.vs.pbs_genes_chr-8_peak.marker-UNCHS023353_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-8_peak.marker-UNCHS023353_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-8_peak.marker-UNCHS023364_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_conditional_1-peak-chr10.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_conditional_1-peak-chr3.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_conditional_1-peak-chr4.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_conditional_2-peaks-chr3-10.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_conditional_2-peaks-chr3-4.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_conditional_2-peaks-chr4-10.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_conditional_3-peaks-chr3-4-10.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_mis.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_mis_conditional_1-peak-chr10.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_mis_conditional_1-peak-chr3.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_mis_conditional_1-peak-chr4.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_mis_conditional_2-peaks-chr3-10.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_mis_conditional_2-peaks-chr3-4.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_mis_conditional_2-peaks-chr4-10.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_mis_conditional_3-peaks-chr3-4-10.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_conditional_2-peaks-chr3-10.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_conditional_2-peaks-chr3-4.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_conditional_2-peaks-chr4-10.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_conditional_3-peaks-chr3-4-10.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_mis_conditional_1-peak-chr10.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_mis_conditional_1-peak-chr3.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_mis_conditional_1-peak-chr4.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_mis_conditional_2-peaks-chr3-10.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_mis_conditional_2-peaks-chr3-4.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_mis_conditional_2-peaks-chr4-10.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_mis_conditional_3-peaks-chr3-4-10.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_conditional_1-peak-chr10.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_conditional_1-peak-chr3.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_conditional_1-peak-chr4.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_conditional_2-peaks-chr3-10.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_conditional_2-peaks-chr3-4.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_conditional_2-peaks-chr4-10.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_conditional_3-peaks-chr3-4-10.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis_conditional_1-peak-chr10.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis_conditional_1-peak-chr3.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis_conditional_1-peak-chr4.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis_conditional_2-peaks-chr3-10.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis_conditional_2-peaks-chr3-4.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis_conditional_2-peaks-chr4-10.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis_conditional_3-peaks-chr3-4-10.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratiov_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratiov_5.batches_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-JAX00153527_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-JAX00153527_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNCHS020443_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNCHS020443_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNCHS020486_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-JAX00153527_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-JAX00153527_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNCHS020443_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNCHS020443_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNCHS020486_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_5.batches.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_5.batches_mis.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_5.batches.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_5.batches_mis.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed_5.batches.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed_5.batches_mis.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_5.batches.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_5.batches_mis.csv
Untracked: data/ici.vs.pbs_scanone_5.batches.Rdata
Untracked: data/ici.vs.pbs_scanone_5.batches_mis.Rdata
Untracked: data/ici.vs.pbs_scanone_snpsqc_5.batches.Rdata
Untracked: data/ici.vs.pbs_scanone_snpsqc_5.batches_mis.Rdata
Untracked: data/ici.vs.pbs_scanone_snpsqc_dis_no-x_updated_5.batches.Rdata
Untracked: data/ici.vs.pbs_scanone_snpsqc_dis_no-x_updated_5.batches_mis.Rdata
Untracked: data/mean.differences_age.of.onset_ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_5.batches.csv
Untracked: data/mean.differences_age.of.onset_ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_5.batches_mis.csv
Untracked: data/mean.differences_age.of.onset_ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_5.batches.csv
Untracked: data/mean.differences_age.of.onset_ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_5.batches_mis.csv
Untracked: data/mean.differences_group_ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_5.batches.csv
Untracked: data/mean.differences_group_ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_5.batches_mis.csv
Untracked: data/mean.differences_group_ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_5.batches.csv
Untracked: data/mean.differences_group_ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_5.batches_mis.csv
Untracked: data/percent_missing_id_3.batches.RData
Untracked: data/percent_missing_id_4.batches.RData
Untracked: data/percent_missing_id_4.batches_bc.RData
Untracked: data/percent_missing_id_5.batches.RData
Untracked: data/percent_missing_marker_4.batches.RData
Untracked: data/percent_missing_marker_5.batches.RData
Untracked: data/pheno.csv
Untracked: data/pheno_BC312.csv
Untracked: data/physical_map.csv
Untracked: data/physical_map_BC312.csv
Untracked: data/qc_info_bad_sample_3.batches.RData
Untracked: data/qc_info_bad_sample_4.batches.RData
Untracked: data/qc_info_bad_sample_4.batches_bc.RData
Untracked: data/qc_info_bad_sample_5.batches.RData
Untracked: data/remaining.markers_geno.freq.xlsx
Untracked: data/sample_geno.csv
Untracked: data/sample_geno_AHB_BC312.csv
Untracked: data/sample_geno_bc.csv
Untracked: data/sample_geno_bc_BC312.csv
Untracked: data/serreze_probs.rds
Untracked: data/serreze_probs_BC312.rds
Untracked: data/serreze_probs_allqc.rds
Untracked: data/serreze_probs_allqc_5.batches.rds
Untracked: data/serreze_probs_allqc_5.batches_mis.rds
Untracked: data/summary.cg_3.batches.RData
Untracked: data/summary.cg_4.batches.RData
Untracked: data/summary.cg_4.batches_bc.RData
Untracked: data/summary.cg_5.batches.RData
Untracked: output/Percent_missing_genotype_data_5.batches.pdf
Untracked: output/Percent_missing_genotype_data_per_marker_5.batches.pdf
Untracked: output/Proportion_matching_genotypes_before_removal_of_bad_samples_5.batches.pdf
Untracked: output/genotype_error_marker_5.batches.pdf
Untracked: output/genotype_frequency_marker_5.batches.pdf
Unstaged changes:
Modified: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs_5.batches.Rmd
Modified: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs_5.batches_mis.Rmd
Modified: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs_snpsqc_5.batches.Rmd
Modified: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs_snpsqc_5.batches_mis.Rmd
Modified: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs_snpsqc_dis_no-x_updated_5.batches.Rmd
Modified: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis.Rmd
Modified: analysis/4.1.1_qtl.analysis_binary_ici.vs.eoi_snpsqc_dis_no-x_updated.Rmd
Modified: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/genotype.frequencies_ici.vs.eoi_5.batches.Rmd
Modified: analysis/genotype.frequencies_ici.vs.eoi_5.batches_mis.Rmd
Modified: analysis/genotype.frequencies_ici.vs.pbs_5.batches.Rmd
Modified: analysis/genotype.frequencies_ici.vs.pbs_5.batches_mis.Rmd
Modified: analysis/index_5.batches.Rmd
Modified: analysis/index_5.batches_additional.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with
wflow_publish() to start tracking its development.
Loading Data
We will load the data and subset indivials out that are in the groups of interest. We will create a binary phenotype from this (PBS ==0, ICI == 1).
load("data/gm_allqc_5.batches_mis.RData")
#gm_allqc
gm=gm_allqc
gmObject of class cross2 (crosstype "bc")
Total individuals 308
No. genotyped individuals 308
No. phenotyped individuals 308
No. with both geno & pheno 308
No. phenotypes 1
No. covariates 6
No. phenotype covariates 0
No. chromosomes 20
Total markers 131356
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
9956 9987 7848 7586 7609 7736 7399 6458 6713 6385 7143 6110 6082 5966 5346 5015
17 18 19 X
5080 4605 3562 4770 #pr <- readRDS("data/serreze_probs_allqc.rds")
#pr <- readRDS("data/serreze_probs.rds")
##extracting animals with ici and pbs group status
miceinfo <- gm$covar[gm$covar$group == "PBS" | gm$covar$group == "ICI",]
table(miceinfo$group)
ICI PBS
104 34 mice.ids <- rownames(miceinfo)
gm <- gm[mice.ids]
gmObject of class cross2 (crosstype "bc")
Total individuals 138
No. genotyped individuals 138
No. phenotyped individuals 138
No. with both geno & pheno 138
No. phenotypes 1
No. covariates 6
No. phenotype covariates 0
No. chromosomes 20
Total markers 131356
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
9956 9987 7848 7586 7609 7736 7399 6458 6713 6385 7143 6110 6082 5966 5346 5015
17 18 19 X
5080 4605 3562 4770 #pr.qc <- pr
#for (i in 1:20){pr.qc[[i]] = pr.qc[[i]][mice.ids,,]}
#bin_pheno <- NULL
#bin_pheno$PBS <- ifelse(gm$covar$group == "PBS", 1, 0)
#bin_pheno$ICI <- ifelse(gm$covar$group == "ICI", 1, 0)
#bin_pheno <- as.data.frame(bin_pheno)
#rownames(bin_pheno) <- rownames(gm$covar)
gm$covar$ICI.vs.PBS <- ifelse(gm$covar$group == "PBS", 0, 1)
gm.full <- gm
##adding peaks as covariates: UNCHS008487, UNC8250659, UNC18240977
g.covar <- read.csv("/Users/corneb/Documents/MyJax/CS/Projects/Serreze/haplotype.reconstruction/output_5.batches/sample_geno_AHB.csv")
colnames(g.covar) <- gsub("\\.","-", colnames(g.covar))
rownames(g.covar) <- g.covar$marker
#g.covar <- g.covar[-1]
mar.covar <- g.covar[g.covar$marker == "UNCHS008487"| g.covar$marker == "UNC8250659"| g.covar$marker == "UNC18240977",]
mar.covar <- mar.covar[-1]
mar.covar[mar.covar=='A'] <- 0
mar.covar[mar.covar=='H'] <- 1
mar.covar[mar.covar=='B'] <- 2
mar.covar[mar.covar=='-'] <- ''
#mar.covar <- pull_markers(gm, c("UNCHS008487", "UNC8250659", "UNC18240977"))
mar.covar.g <- t(mar.covar)
mar.covar.g <- as.data.frame(mar.covar.g)
covars <- merge(gm$covar, mar.covar.g, by='row.names', sort=F)
table(covars$group)
ICI PBS
104 34 rownames(covars) <- covars$Row.names
covars <- covars[-1]
##removing problmetic marker
gm <- drop_markers(gm, "UNCHS013106")
##dropping monomorphic markers within the dataset
g <- do.call("cbind", gm$geno)
gf_mar <- t(apply(g, 2, function(a) table(factor(a, 1:2))/sum(a != 0)))
#gn_mar <- t(apply(g, 2, function(a) table(factor(a, 1:2))))
gf_mar <- gf_mar[gf_mar[,2] != "NaN",]
count <- rowSums(gf_mar <=0.05)
low_freq_df <- merge(as.data.frame(gf_mar),as.data.frame(count), by="row.names",all=T)
low_freq_df[is.na(low_freq_df)] <- ''
low_freq_df <- low_freq_df[low_freq_df$count == 1,]
rownames(low_freq_df) <- low_freq_df$Row.names
low_freq <- find_markerpos(gm, rownames(low_freq_df))
low_freq$id <- rownames(low_freq)
nrow(low_freq)[1] 98665low_freq_bad <- merge(low_freq,low_freq_df, by="row.names",all=T)
names(low_freq_bad)[1] <- c("marker")
gf_mar <- gf_mar[gf_mar[,2] != "NaN",]
MAF <- apply(gf_mar, 1, function(x) min(x))
MAF <- as.data.frame(MAF)
MAF$index <- 1:nrow(gf_mar)
gf_mar_maf <- merge(gf_mar,as.data.frame(MAF), by="row.names")
gf_mar_maf <- gf_mar_maf[order(gf_mar_maf$index),]
gfmar <- NULL
gfmar$gfmar_mar_0 <- sum(gf_mar_maf$MAF==0)
gfmar$gfmar_mar_1 <- sum(gf_mar_maf$MAF< 0.01)
gfmar$gfmar_mar_5 <- sum(gf_mar_maf$MAF< 0.05)
gfmar$gfmar_mar_10 <- sum(gf_mar_maf$MAF< 0.10)
gfmar$gfmar_mar_15 <- sum(gf_mar_maf$MAF< 0.15)
gfmar$gfmar_mar_25 <- sum(gf_mar_maf$MAF< 0.25)
gfmar$gfmar_mar_50 <- sum(gf_mar_maf$MAF< 0.50)
gfmar$total_snps <- nrow(as.data.frame(gf_mar_maf))
gfmar <- t(as.data.frame(gfmar))
gfmar <- as.data.frame(gfmar)
gfmar$count <- gfmar$V1
gfmar[c(2)] %>%
kable(escape = F,align = c("ccccccccc"),linesep ="\\hline") %>%
kable_styling(full_width = F) %>%
kable_styling("striped", full_width = F) %>%
row_spec(8 ,bold=T,color= "white",background = "black")| count | |
|---|---|
| gfmar_mar_0 | 88924 |
| gfmar_mar_1 | 92460 |
| gfmar_mar_5 | 98665 |
| gfmar_mar_10 | 99264 |
| gfmar_mar_15 | 99368 |
| gfmar_mar_25 | 100303 |
| gfmar_mar_50 | 130381 |
| total_snps | 131355 |
gm_qc <- drop_markers(gm, low_freq_bad$marker)
gm_qc <- drop_nullmarkers(gm_qc)
gm = gm_qc
gmObject of class cross2 (crosstype "bc")
Total individuals 138
No. genotyped individuals 138
No. phenotyped individuals 138
No. with both geno & pheno 138
No. phenotypes 1
No. covariates 7
No. phenotype covariates 0
No. chromosomes 20
Total markers 32690
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2952 2874 2058 2071 1955 2053 1862 1700 2009 1229 2069 1396 1628 1692 1064 940
17 18 19 X
401 1095 1067 575 markers <- marker_names(gm)
gmapdf <- read.csv("/Users/corneb/Documents/MyJax/CS/Projects/Serreze/haplotype.reconstruction/output_5.batches/genetic_map.csv")
pmapdf <- read.csv("/Users/corneb/Documents/MyJax/CS/Projects/Serreze/haplotype.reconstruction/output_5.batches/physical_map.csv")
#mapdf <- merge(gmapdf,pmapdf, by=c("marker","chr"), all=T)
#rownames(mapdf) <- mapdf$marker
#mapdf <- mapdf[markers,]
#names(mapdf) <- c('marker','chr','gmapdf','pmapdf')
#mapdfnd <- mapdf[!duplicated(mapdf[c(2:3)]),]
pr.qc <- calc_genoprob(gm)Conditionaing on eoi vs. ici chr 3 peak (UNCHS008487) & chr 4 peak (UNC8250659)
Genome-wide scan
#gm
Xcovar <- get_x_covar(gm)
#addcovar = model.matrix(~Sex, data = covars)[,-1]
addcovar = model.matrix(~UNCHS008487+UNC8250659, data = covars)[,-1]
#K <- calc_kinship(pr.qc, type = "loco")
#heatmap(K[[1]])
#K.overall <- calc_kinship(pr.qc, type = "overall")
#heatmap(K.overall)
kinship <- calc_kinship(pr.qc)
heatmap(kinship)
#operm <- scan1perm(pr.qc, gm$covar$phenos, Xcovar=Xcovar, n_perm=2000)
#operm <- scan1perm(pr.qc, gm$covar$phenos, addcovar = addcovar, n_perm=2000)
#operm <- scan1perm(pr.qc, gm$covar$phenos, n_perm=2000)
operm <- scan1perm(pr.qc, gm$covar["ICI.vs.PBS"], model="binary", n_perm=10, perm_Xsp=TRUE, chr_lengths=chr_lengths(gm$gmap), addcovar = addcovar)
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
names(summary_table) <- c("autosomes","X")
summary_table$significance.level <- rownames(summary_table)
rownames(summary_table) <- NULL
summary_table[c(3,1:2)] %>%
kable(escape = F,align = c("ccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| significance.level | autosomes | X |
|---|---|---|
| 0.01 | 3.090048 | 3.657299 |
| 0.05 | 3.029934 | 3.124699 |
| 0.1 | 2.954606 | 2.428212 |
The figures below show QTL maps for each phenotype
out <- scan1(pr.qc, gm$covar["ICI.vs.PBS"], Xcovar=Xcovar, model="binary", addcovar = addcovar)
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
plot_lod<-function(out,map){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- 11 # overall maximum LOD score
plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " [positions in cM] \n(using same scale as eoi vs ici for easier comparison)"))
add_threshold(map, summary(operm, alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
plot_lod(out,gm$gmap)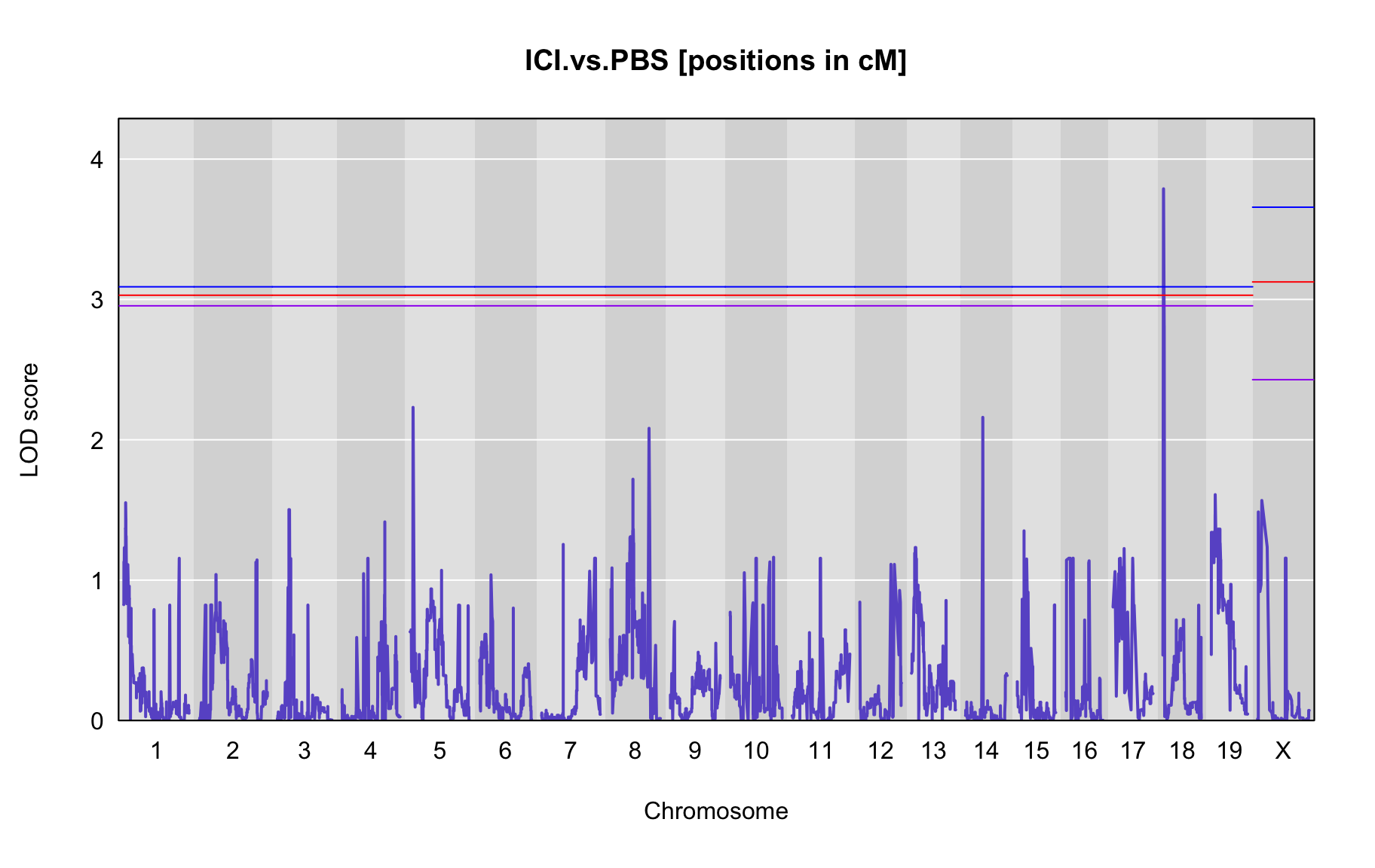
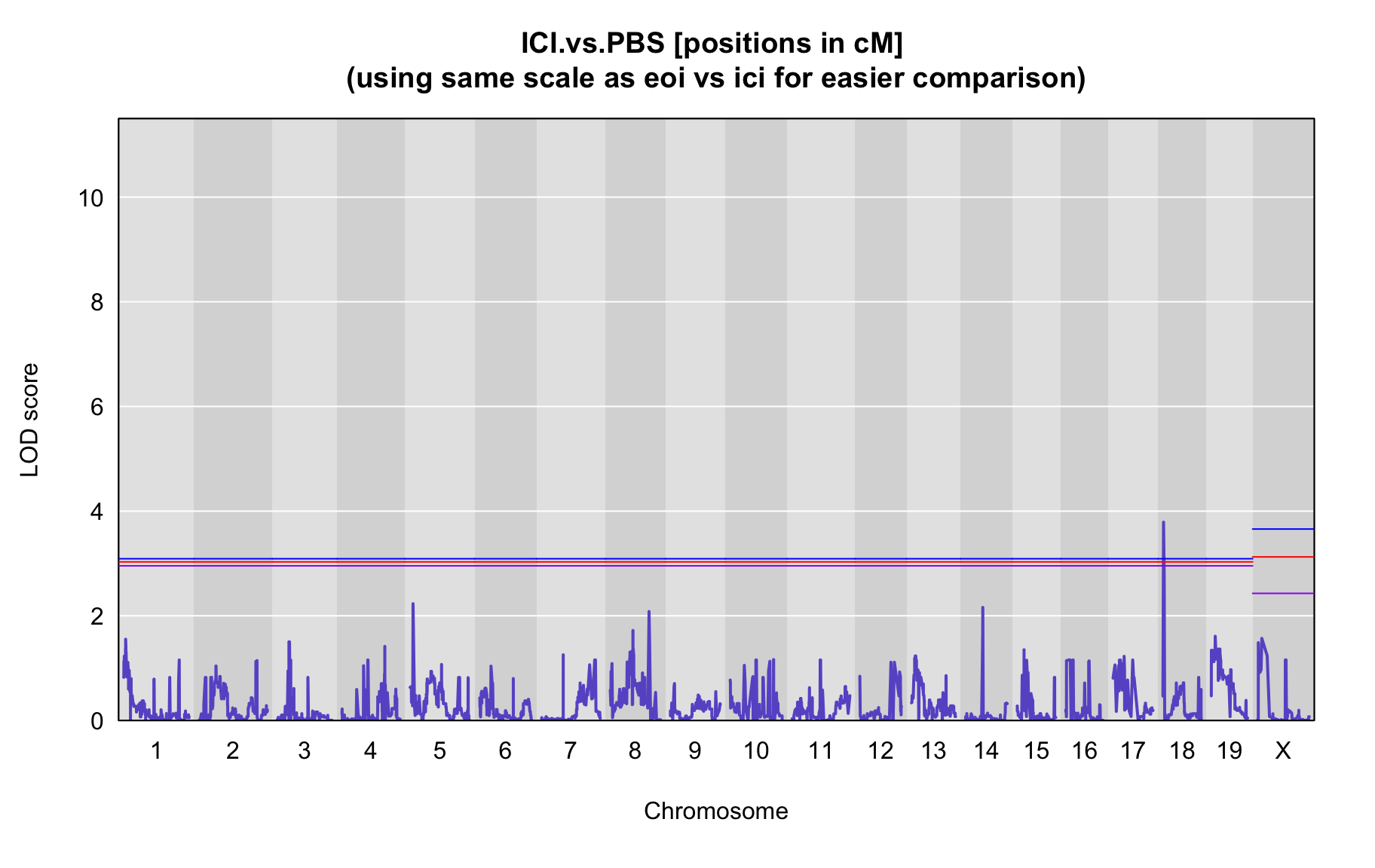
LOD peaks
The table below shows QTL peaks associated with the phenotype. We use the 95% threshold from the permutations to find peaks.
Centimorgan (cM)
peaks <- find_peaks(out, gm$gmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) >0){
peaks$marker <- find_marker(gm$gmap, chr=peaks$chr,pos=peaks$pos)
names(peaks)[2] <- c("phenotype")
peaks <- peaks[-1]
rownames(peaks) <- NULL
#print(kable(peaks, "html")
# %>% kable_styling("striped", full_width = T) %>%
# column_spec(1, bold=TRUE)
# )
print(kable(peaks, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
#peaks[] %>%
# kable(escape = F,align = c("cccccccc")) %>%
# kable_styling("striped", full_width = T) %>%
# column_spec(1, bold=TRUE)
#plot only peak chromosomes
plot_lod_chr<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
ymx <- 11
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in cM]\n(using same scale as eoi vs. ici for easier comparison)"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
for(i in unique(peaks$chr)){
#for (i in 1:nrow(peaks)){
#plot_lod_chr(out,gm$gmap, peaks$chr[i])
plot_lod_chr(out,gm$gmap, i)
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}| phenotype | chr | pos | lod | ci_lo | ci_hi | marker |
|---|---|---|---|---|---|---|
| ICI.vs.PBS | 18 | 2.934 | 3.788369 | 1.972 | 4.621 | UNCHS045343 |
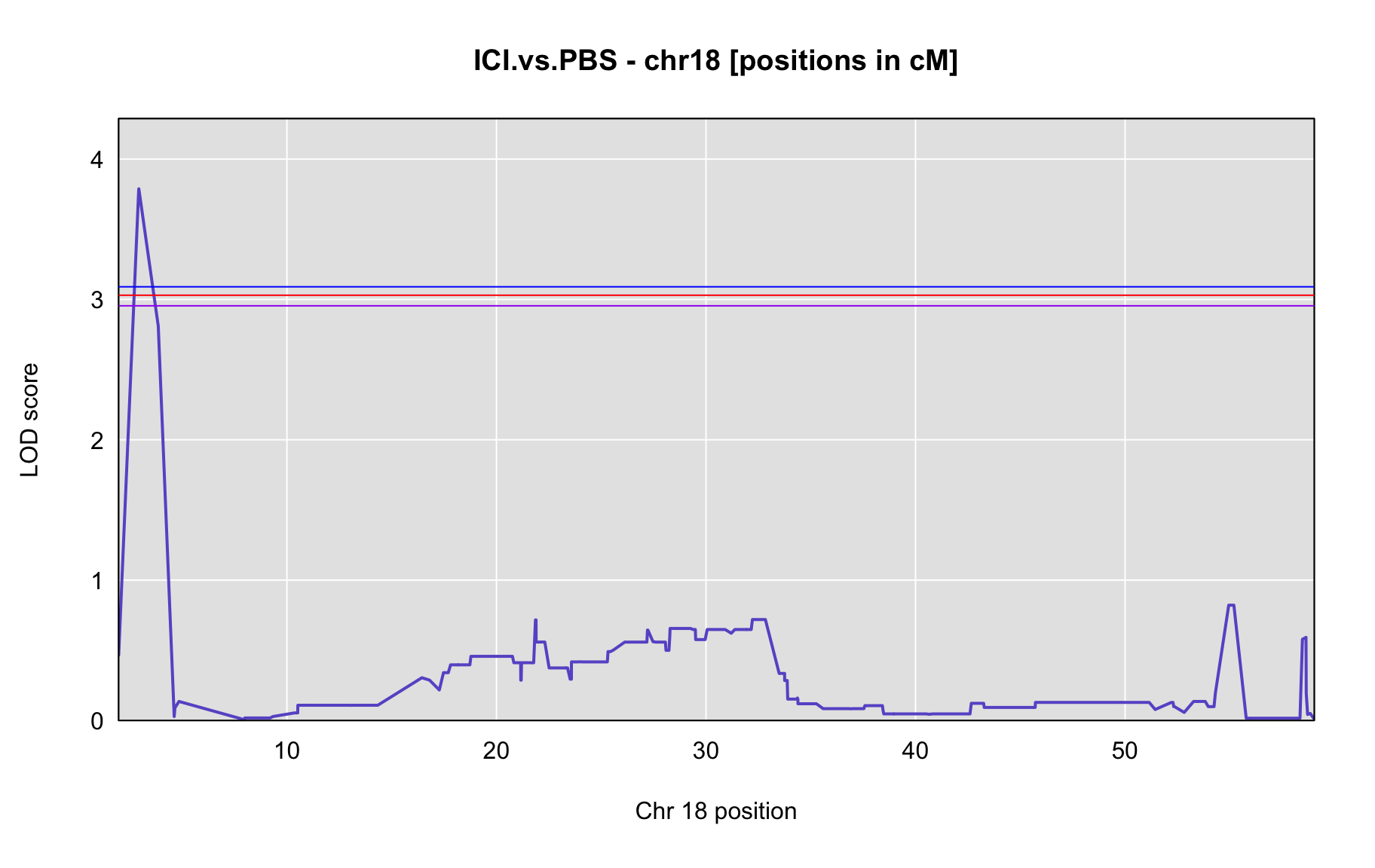
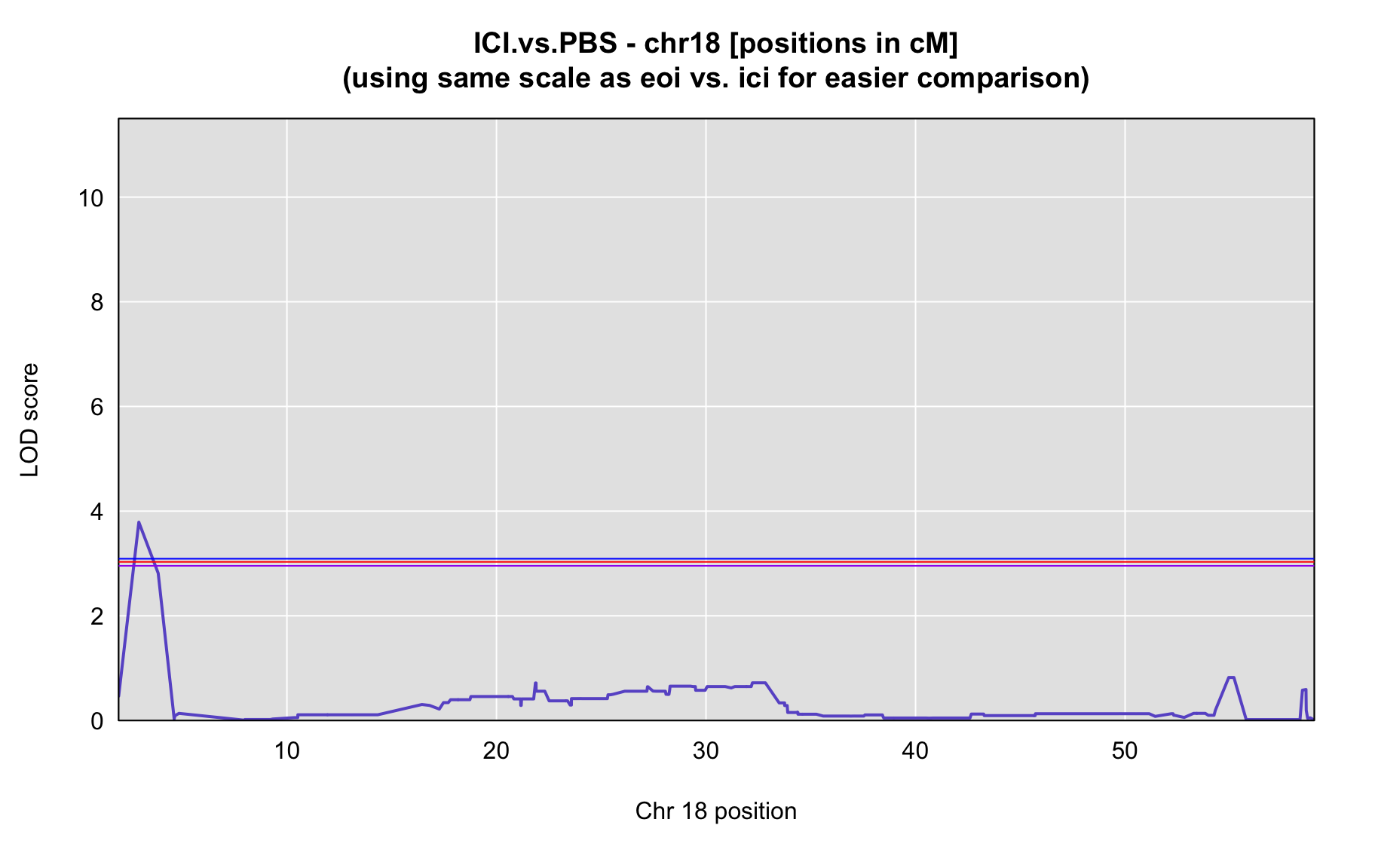
Megabase (MB)
print("peaks in MB positions")[1] “peaks in MB positions”
peaks_mba <- find_peaks(out, gm$pmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) >0){
peaks_mba$marker <- find_marker(gm$pmap, chr=peaks_mba$chr,pos=peaks_mba$pos)
names(peaks_mba)[2] <- c("phenotype")
peaks_mba <- peaks_mba[-1]
#peaks_mbl <- list()
##corresponding info in Mb
#for(i in 1:nrow(peaks)){
# #lodindex <- peaks$lodindex[i]
# phenotype <- peaks$phenotype[i]
# chr <- as.character(peaks$chr[i])
# lod <- peaks$lod[i]
# mark <- peaks$marker[i]
# pos <- mapdf[mapdf$marker==mark,]$pmapdf
# ci_lo <- mapdfnd$pmapdf[which(mapdfnd$gmapdf == peaks$ci_lo[i] & mapdfnd$chr == peaks$chr[i])]
# ci_hi <- mapdfnd$pmapdf[which(mapdfnd$gmapdf == peaks$ci_hi[i] & mapdfnd$chr == peaks$chr[i])]
# peaks_mb=as.data.frame(cbind(phenotype, chr, pos, lod, ci_lo, ci_hi, mark))
# names(peaks_mb)[7] <- c("marker")
# peaks_mbl[[i]] <- peaks_mb
#}
#peaks_mba2 <- do.call(rbind, peaks_mbl)
#peaks_mba2 <- as.data.frame(peaks_mba)
#peaks_mba[,c("chr", "pos", "lod", "ci_lo", "ci_hi")] <- sapply(peaks_mba[,c("chr", "pos", "lod", "ci_lo", "ci_hi")], as.numeric)
rownames(peaks_mba) <- NULL
#print(kable(peaks_mba, "html")
# %>% kable_styling("striped", full_width = T) %>%
# column_spec(1, bold=TRUE)
# )
print(kable(peaks_mba, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
#peaks_mba[] %>%
# kable(escape = F,align = c("cccccccc")) %>%
# kable_styling("striped", full_width = T) %>%
# column_spec(1, bold=TRUE)
plot_lod_chr_mb<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in MB]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
ymx <- 11
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in MB]\n(using same scale as eoi vs. ici for easier comparison)"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
for(i in unique(peaks_mba$chr)){
#for (i in 1:nrow(peaks_mba)){
#plot_lod_chr_mb(out,gm$pmap, peaks_mba$chr[i])
plot_lod_chr_mb(out,gm$pmap,i)
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}| phenotype | chr | pos | lod | ci_lo | ci_hi | marker |
|---|---|---|---|---|---|---|
| ICI.vs.PBS | 18 | 4.861199 | 3.788369 | 3.01212 | 7.97679 | UNCHS045343 |
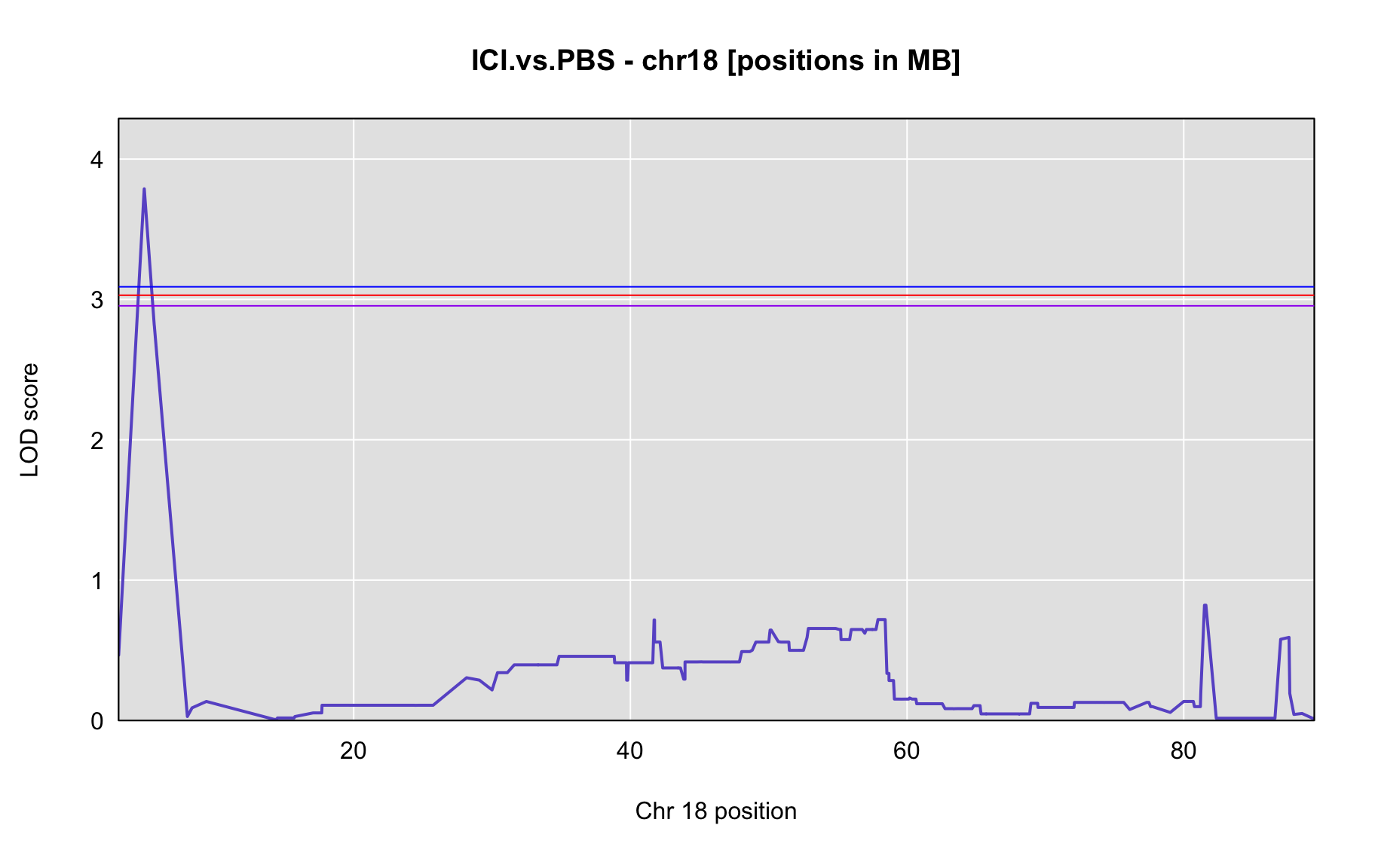
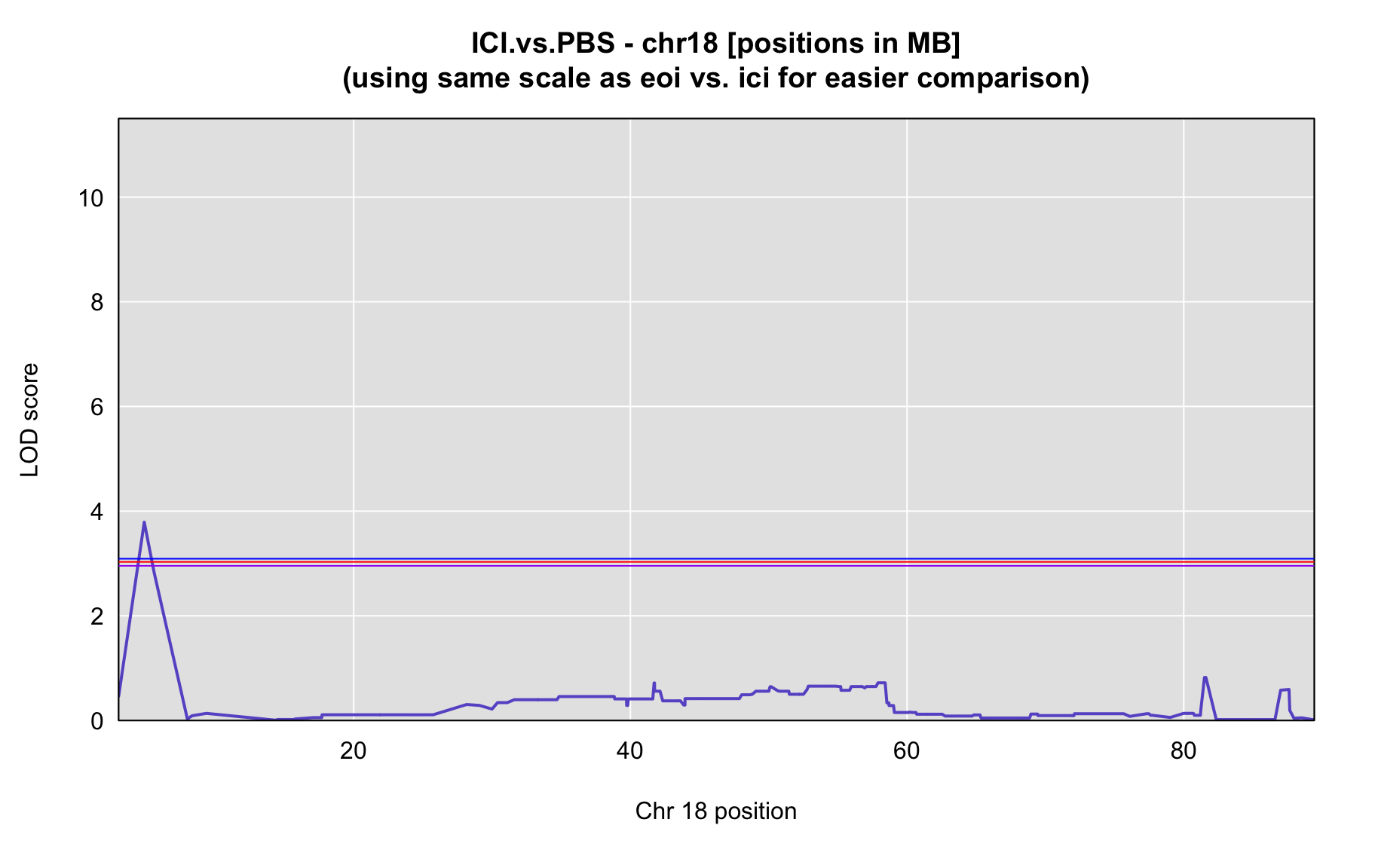
QTL effects
For each peak LOD location we give a list of gene
query_variants <- create_variant_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/cc_variants.sqlite")
query_genes <- create_gene_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/mouse_genes_mgi.sqlite")
if(nrow(peaks) >0){
for (i in 1:nrow(peaks)){
#for (i in 1:1){
#Plot 1
#marker = find_marker(gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i])
#gp <- g[,marker]
#gp[gp==1] <- "AA"
#gp[gp==2] <- "AB"
#gp[gp==0] <- NA
#plot_pxg(gp, gm$covar[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
#title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
###dev.off()
g <- maxmarg(pr.qc, gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i], return_char=TRUE)
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/","qtl_effect_", i, ".png"))
#par(mar=c(4.1, 4.1, 1.5, 0.6))
plot_pxg(g, gm$covar[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
##dev.off()
chr = peaks$chr[i]
# Plot 2
pr_sub <- pull_genoprobint(pr.qc, gm$gmap, chr, c(peaks$ci_lo[i], peaks$ci_hi[i]))
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar)
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], Xcovar=Xcovar)
#coeff <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
#coeff_sub <- scan1coef(pr_sub[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
blup <- scan1blup(pr.qc[,chr], gm$covar[peaks$phenotype[i]], addcovar = addcovar)
blup_sub <- scan1blup(pr_sub[,chr], gm$covar[peaks$phenotype[i]], addcovar = addcovar)
write.csv(as.data.frame(blup_sub), paste0("data/ici.vs.pbs_blup_sub_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_snpsqc_5.batches_mis_conditional_2-peaks-chr3-4.csv"), quote=F)
#plot_coef(coeff,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i]," [scan1coeff; positions in cM])")
# )
#plot_coef(coeff_sub,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i],"; 1.5 LOD drop interval [scan1coeff; positions in cM] ) ")
# )
plot_coef(blup,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," [scan1blup; positions in cM])")
)
plot_coef(blup_sub,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i],"; 1.5 LOD drop interval [scan1blup; positions in cM])")
)
# Plot 3
#c2effB <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary", contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
#c2effBb <- scan1blup(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]],Xcovar=Xcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
#plot(c2effB, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
#plot(c2effBb, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
##last_coef <- unclass(c2effB)[nrow(c2effB),2:3] # last two coefficients
##for(t in seq(along=last_coef))
## axis(side=4, at=last_coef[t], names(last_coef)[t], tick=FALSE)
#Table 1
chr = peaks_mba$chr[i]
start=as.numeric(peaks_mba$ci_lo[i])
end=as.numeric(peaks_mba$ci_hi[i])
genesgss = query_genes(chr, start, end)
write.csv(genesgss, file=paste0("data/ici.vs.pbs_genes_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_snpsqc_5.batches_mis_conditional_2-peaks-chr3-4.csv"), quote=F)
rownames(genesgss) <- NULL
genesgss$strand_old = genesgss$strand
genesgss$strand[genesgss$strand=="+"] <- "positive"
genesgss$strand[genesgss$strand=="-"] <- "negative"
#genesgss <-
#table <-
#genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")] %>%
#kable(escape = F,align = c("ccccccccccc")) %>%
#kable_styling("striped", full_width = T) #%>%
#cat #%>%
#column_spec(1, bold=TRUE)
#
#print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], escape = F,align = c("ccccccccccc")))
print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], "html") %>% kable_styling("striped", full_width = T))
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}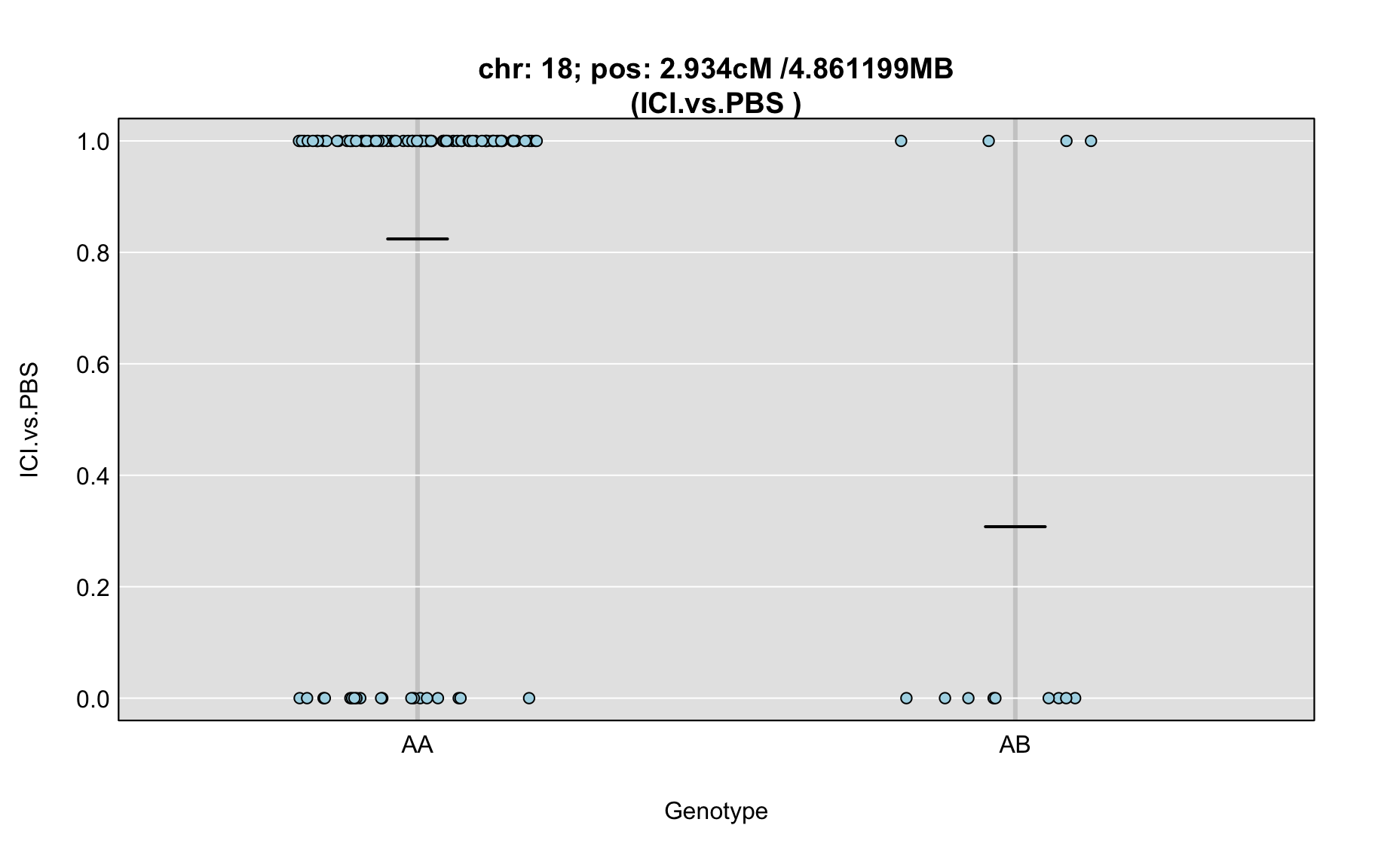
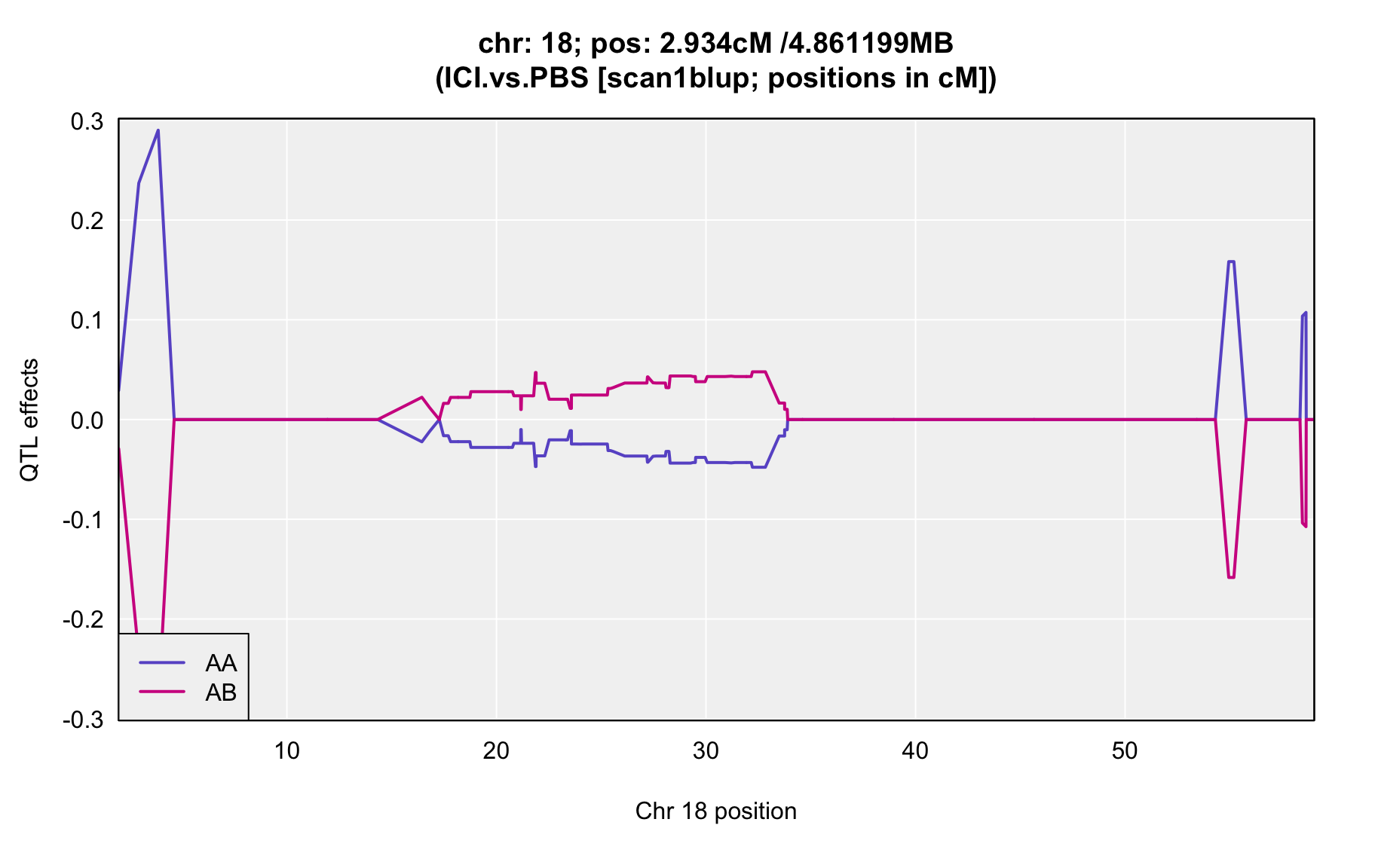
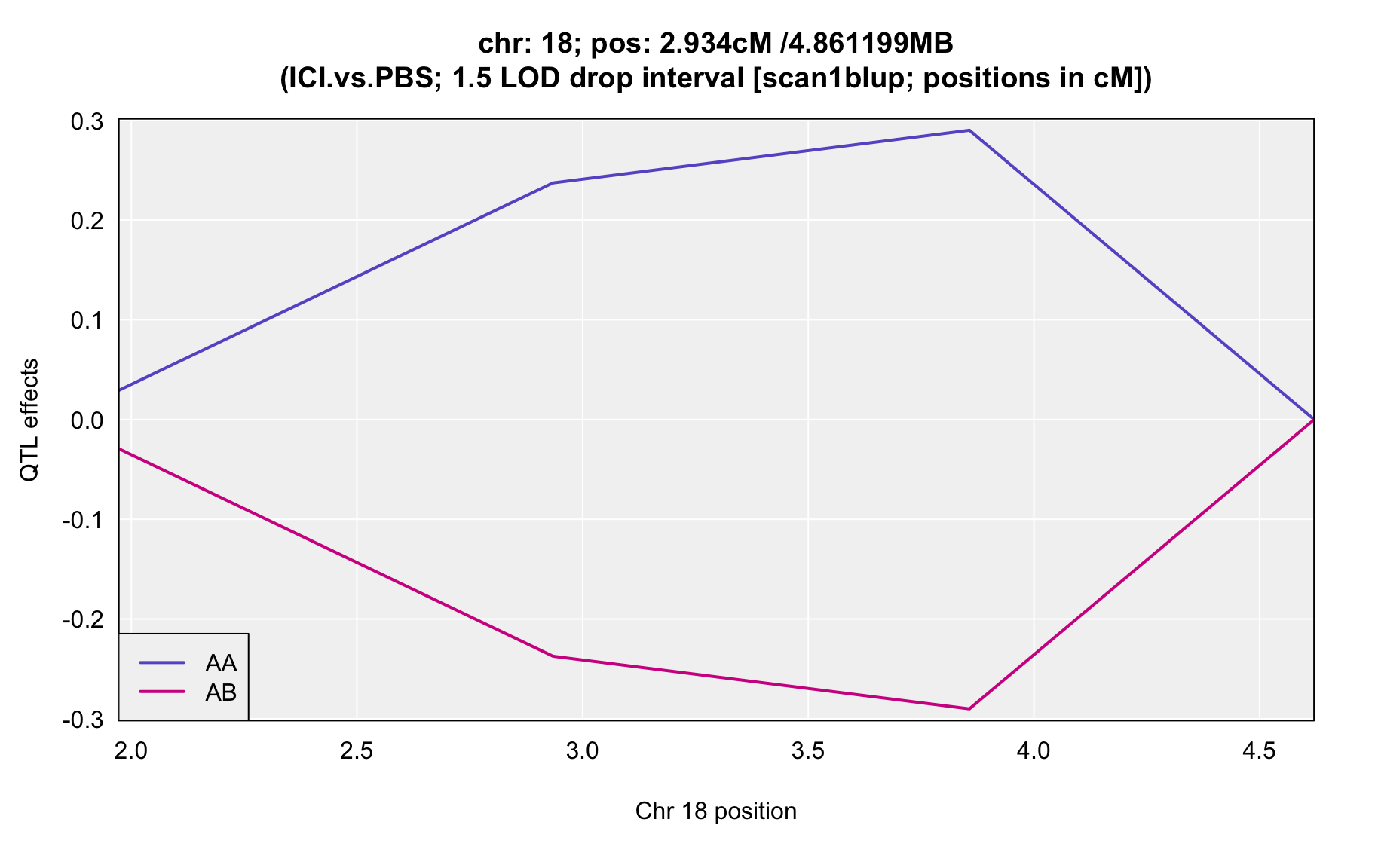
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 18 | pseudogene | 3.015908 | 3.016159 | positive | MGI_C57BL6J_6275399 | Gm50072 | ENSEMBL:ENSMUSG00000117547 | MGI:6275399 | pseudogene | predicted gene, 50072 |
| 18 | pseudogene | 3.026901 | 3.027882 | negative | MGI_C57BL6J_3852490 | Vmn1r-ps151 | NCBI_Gene:100312519,ENSEMBL:ENSMUSG00000093774 | MGI:3852490 | pseudogene | vomeronasal 1 receptor, pseudogene 151 |
| 18 | pseudogene | 3.080778 | 3.081476 | negative | MGI_C57BL6J_3852491 | Vmn1r-ps152 | NCBI_Gene:100312520,ENSEMBL:ENSMUSG00000093444 | MGI:3852491 | pseudogene | vomeronasal 1 receptor, pseudogene 152 |
| 18 | pseudogene | 3.105167 | 3.105396 | negative | MGI_C57BL6J_6275400 | Gm50073 | ENSEMBL:ENSMUSG00000117531 | MGI:6275400 | pseudogene | predicted gene, 50073 |
| 18 | gene | 3.122492 | 3.130012 | negative | MGI_C57BL6J_3852494 | Vmn1r238 | NCBI_Gene:100312476,ENSEMBL:ENSMUSG00000091539 | MGI:3852494 | protein coding gene | vomeronasal 1 receptor, 238 |
| 18 | pseudogene | 3.139032 | 3.139305 | negative | MGI_C57BL6J_6275401 | Gm50074 | ENSEMBL:ENSMUSG00000117353 | MGI:6275401 | pseudogene | predicted gene, 50074 |
| 18 | pseudogene | 3.171447 | 3.171932 | negative | MGI_C57BL6J_6275402 | Gm50075 | ENSEMBL:ENSMUSG00000117464 | MGI:6275402 | pseudogene | predicted gene, 50075 |
| 18 | gene | 3.266048 | 3.338176 | negative | MGI_C57BL6J_88495 | Crem | NCBI_Gene:12916,ENSEMBL:ENSMUSG00000063889 | MGI:88495 | protein coding gene | cAMP responsive element modulator |
| 18 | gene | 3.336416 | 3.366863 | positive | MGI_C57BL6J_3643252 | Gm6225 | NCBI_Gene:633947,ENSEMBL:ENSMUSG00000097746 | MGI:3643252 | lncRNA gene | predicted gene 6225 |
| 18 | gene | 3.382970 | 3.436700 | positive | MGI_C57BL6J_1918995 | Cul2 | NCBI_Gene:71745,ENSEMBL:ENSMUSG00000024231 | MGI:1918995 | protein coding gene | cullin 2 |
| 18 | pseudogene | 3.446654 | 3.447503 | positive | MGI_C57BL6J_3648491 | Gm6235 | NCBI_Gene:621501,ENSEMBL:ENSMUSG00000117370 | MGI:3648491 | pseudogene | predicted gene 6235 |
| 18 | pseudogene | 3.466371 | 3.466502 | negative | MGI_C57BL6J_6275423 | Gm50088 | ENSEMBL:ENSMUSG00000117389 | MGI:6275423 | pseudogene | predicted gene, 50088 |
| 18 | gene | 3.471630 | 3.474315 | positive | MGI_C57BL6J_3645649 | G430049J08Rik | ENSEMBL:ENSMUSG00000096528 | MGI:3645649 | unclassified gene | RIKEN cDNA G430049J08 gene |
| 18 | pseudogene | 3.484218 | 3.485193 | positive | MGI_C57BL6J_6275424 | Gm50089 | ENSEMBL:ENSMUSG00000117469 | MGI:6275424 | pseudogene | predicted gene, 50089 |
| 18 | gene | 3.507923 | 3.516404 | positive | MGI_C57BL6J_1915260 | Bambi | NCBI_Gene:68010,ENSEMBL:ENSMUSG00000024232 | MGI:1915260 | protein coding gene | BMP and activin membrane-bound inhibitor |
| 18 | pseudogene | 3.558675 | 3.560037 | positive | MGI_C57BL6J_6275425 | Gm50090 | ENSEMBL:ENSMUSG00000117454 | MGI:6275425 | pseudogene | predicted gene, 50090 |
| 18 | gene | 3.616281 | 3.618242 | positive | MGI_C57BL6J_6275426 | Gm50091 | ENSEMBL:ENSMUSG00000117506 | MGI:6275426 | unclassified gene | predicted gene, 50091 |
| 18 | gene | 3.770741 | 3.770804 | negative | MGI_C57BL6J_5453420 | Gm23643 | ENSEMBL:ENSMUSG00000088480 | MGI:5453420 | snRNA gene | predicted gene, 23643 |
| 18 | pseudogene | 3.857730 | 3.858585 | negative | MGI_C57BL6J_3647465 | Rpl7a-ps6 | NCBI_Gene:435549,ENSEMBL:ENSMUSG00000117463 | MGI:3647465 | pseudogene | ribosomal protein L7A, pseudogene 6 |
| 18 | gene | 3.860108 | 3.860430 | positive | MGI_C57BL6J_5454409 | Gm24632 | ENSEMBL:ENSMUSG00000084719 | MGI:5454409 | unclassified non-coding RNA gene | predicted gene, 24632 |
| 18 | pseudogene | 4.010143 | 4.011335 | positive | MGI_C57BL6J_3646622 | Gm7378 | NCBI_Gene:664867,ENSEMBL:ENSMUSG00000117395 | MGI:3646622 | pseudogene | predicted gene 7378 |
| 18 | pseudogene | 4.052351 | 4.052771 | negative | MGI_C57BL6J_3644899 | Gm6248 | NCBI_Gene:621666,ENSEMBL:ENSMUSG00000117565 | MGI:3644899 | pseudogene | predicted gene 6248 |
| 18 | gene | 4.165832 | 4.182236 | positive | MGI_C57BL6J_1914578 | Lyzl1 | NCBI_Gene:67328,ENSEMBL:ENSMUSG00000024233 | MGI:1914578 | protein coding gene | lysozyme-like 1 |
| 18 | pseudogene | 4.198320 | 4.199135 | negative | MGI_C57BL6J_3708638 | Gm10557 | NCBI_Gene:383374,ENSEMBL:ENSMUSG00000073647 | MGI:3708638 | pseudogene | predicted gene 10557 |
| 18 | gene | 4.244121 | 4.250779 | negative | MGI_C57BL6J_5624542 | Gm41657 | NCBI_Gene:105246358 | MGI:5624542 | lncRNA gene | predicted gene, 41657 |
| 18 | pseudogene | 4.293097 | 4.294538 | negative | MGI_C57BL6J_3647488 | Gm7400 | NCBI_Gene:664909,ENSEMBL:ENSMUSG00000117482 | MGI:3647488 | pseudogene | predicted gene 7400 |
| 18 | gene | 4.331325 | 4.353412 | negative | MGI_C57BL6J_1346878 | Map3k8 | NCBI_Gene:26410,ENSEMBL:ENSMUSG00000024235 | MGI:1346878 | protein coding gene | mitogen-activated protein kinase kinase kinase 8 |
| 18 | gene | 4.353547 | 4.368945 | positive | MGI_C57BL6J_1921165 | 4833419F23Rik | NCBI_Gene:73915,ENSEMBL:ENSMUSG00000117401 | MGI:1921165 | lncRNA gene | RIKEN cDNA 4833419F23 gene |
| 18 | gene | 4.353644 | 4.380723 | positive | MGI_C57BL6J_6275434 | Gm50096 | ENSEMBL:ENSMUSG00000117594 | MGI:6275434 | lncRNA gene | predicted gene, 50096 |
| 18 | gene | 4.373669 | 4.379985 | negative | MGI_C57BL6J_5477359 | Gm26865 | ENSEMBL:ENSMUSG00000097641 | MGI:5477359 | lncRNA gene | predicted gene, 26865 |
| 18 | gene | 4.375592 | 4.398798 | positive | MGI_C57BL6J_1914690 | Mtpap | NCBI_Gene:67440,ENSEMBL:ENSMUSG00000024234 | MGI:1914690 | protein coding gene | mitochondrial poly(A) polymerase |
| 18 | gene | 4.399328 | 4.400910 | positive | MGI_C57BL6J_6275321 | Gm50023 | ENSEMBL:ENSMUSG00000117579 | MGI:6275321 | lncRNA gene | predicted gene, 50023 |
| 18 | pseudogene | 4.484469 | 4.485501 | positive | MGI_C57BL6J_3644066 | Gm7411 | NCBI_Gene:664931,ENSEMBL:ENSMUSG00000117640 | MGI:3644066 | pseudogene | predicted gene 7411 |
| 18 | gene | 4.541612 | 4.562599 | negative | MGI_C57BL6J_5624543 | Gm41658 | NCBI_Gene:105246359 | MGI:5624543 | lncRNA gene | predicted gene, 41658 |
| 18 | gene | 4.590763 | 4.616064 | positive | MGI_C57BL6J_6275323 | Gm50024 | ENSEMBL:ENSMUSG00000117521 | MGI:6275323 | lncRNA gene | predicted gene, 50024 |
| 18 | gene | 4.590792 | 4.682869 | positive | MGI_C57BL6J_2685174 | Jcad | NCBI_Gene:240185,ENSEMBL:ENSMUSG00000033960 | MGI:2685174 | protein coding gene | junctional cadherin 5 associated |
| 18 | pseudogene | 4.743061 | 4.743876 | positive | MGI_C57BL6J_3645969 | Gm5047 | NCBI_Gene:268988,ENSEMBL:ENSMUSG00000117585 | MGI:3645969 | pseudogene | predicted gene 5047 |
| 18 | gene | 4.812486 | 4.850959 | positive | MGI_C57BL6J_3642809 | Gm10556 | NCBI_Gene:100038439,ENSEMBL:ENSMUSG00000097043 | MGI:3642809 | lncRNA gene | predicted gene 10556 |
| 18 | gene | 4.820303 | 4.826659 | negative | MGI_C57BL6J_5593428 | Gm34269 | NCBI_Gene:102637466 | MGI:5593428 | lncRNA gene | predicted gene, 34269 |
| 18 | gene | 4.869777 | 4.912263 | positive | MGI_C57BL6J_5624544 | Gm41659 | NCBI_Gene:105246360 | MGI:5624544 | lncRNA gene | predicted gene, 41659 |
| 18 | gene | 4.920245 | 5.119299 | positive | MGI_C57BL6J_2147319 | Svil | NCBI_Gene:225115,ENSEMBL:ENSMUSG00000024236 | MGI:2147319 | protein coding gene | supervillin |
| 18 | gene | 4.959636 | 4.960300 | positive | MGI_C57BL6J_3642287 | BC025933 | NA | NA | protein coding gene | cDNA sequence BC025933 |
| 18 | gene | 5.042818 | 5.048867 | negative | MGI_C57BL6J_5593485 | Gm34326 | NCBI_Gene:102637544 | MGI:5593485 | lncRNA gene | predicted gene, 34326 |
| 18 | gene | 5.139349 | 5.142068 | positive | MGI_C57BL6J_5593700 | Gm34541 | NCBI_Gene:102637831 | MGI:5593700 | lncRNA gene | predicted gene, 34541 |
| 18 | gene | 5.141762 | 5.165731 | negative | MGI_C57BL6J_5477176 | Gm26682 | NCBI_Gene:102637682,ENSEMBL:ENSMUSG00000097888 | MGI:5477176 | lncRNA gene | predicted gene, 26682 |
| 18 | gene | 5.210027 | 5.334963 | negative | MGI_C57BL6J_2444919 | Zfp438 | NCBI_Gene:240186,ENSEMBL:ENSMUSG00000050945 | MGI:2444919 | protein coding gene | zinc finger protein 438 |
| 18 | gene | 5.348258 | 5.361861 | negative | MGI_C57BL6J_6275385 | Gm50064 | ENSEMBL:ENSMUSG00000117520 | MGI:6275385 | lncRNA gene | predicted gene, 50064 |
| 18 | gene | 5.368541 | 5.370482 | negative | MGI_C57BL6J_6275387 | Gm50065 | ENSEMBL:ENSMUSG00000117617 | MGI:6275387 | lncRNA gene | predicted gene, 50065 |
| 18 | gene | 5.389802 | 5.402870 | negative | MGI_C57BL6J_5593789 | Gm34630 | NCBI_Gene:102637944 | MGI:5593789 | lncRNA gene | predicted gene, 34630 |
| 18 | gene | 5.491501 | 5.593505 | negative | MGI_C57BL6J_3642044 | Gm10125 | NCBI_Gene:791318,ENSEMBL:ENSMUSG00000063087 | MGI:3642044 | lncRNA gene | predicted gene 10125 |
| 18 | gene | 5.572824 | 5.575055 | negative | MGI_C57BL6J_5477069 | Gm26575 | ENSEMBL:ENSMUSG00000097926 | MGI:5477069 | lncRNA gene | predicted gene, 26575 |
| 18 | gene | 5.588043 | 5.591711 | negative | MGI_C57BL6J_3780408 | Gm2238 | ENSEMBL:ENSMUSG00000103138 | MGI:3780408 | unclassified gene | predicted gene 2238 |
| 18 | gene | 5.591860 | 5.775468 | positive | MGI_C57BL6J_1344313 | Zeb1 | NCBI_Gene:21417,ENSEMBL:ENSMUSG00000024238 | MGI:1344313 | protein coding gene | zinc finger E-box binding homeobox 1 |
| 18 | pseudogene | 5.611558 | 5.611930 | negative | MGI_C57BL6J_3801802 | Gm16147 | ENSEMBL:ENSMUSG00000090078 | MGI:3801802 | pseudogene | predicted gene 16147 |
| 18 | gene | 5.614473 | 5.614583 | positive | MGI_C57BL6J_5452486 | Gm22709 | ENSEMBL:ENSMUSG00000080543 | MGI:5452486 | miRNA gene | predicted gene, 22709 |
| 18 | gene | 5.885791 | 5.889876 | negative | MGI_C57BL6J_5593899 | Gm34740 | NCBI_Gene:102638095 | MGI:5593899 | lncRNA gene | predicted gene, 34740 |
| 18 | gene | 5.945183 | 5.966908 | positive | MGI_C57BL6J_5593963 | Gm34804 | NCBI_Gene:102638178,ENSEMBL:ENSMUSG00000117508 | MGI:5593963 | lncRNA gene | predicted gene, 34804 |
| 18 | gene | 6.004578 | 6.015278 | positive | MGI_C57BL6J_5624548 | Gm41663 | NCBI_Gene:105246364 | MGI:5624548 | lncRNA gene | predicted gene, 41663 |
| 18 | gene | 6.014253 | 6.024013 | negative | MGI_C57BL6J_5624547 | Gm41662 | NCBI_Gene:105246363,ENSEMBL:ENSMUSG00000117548 | MGI:5624547 | lncRNA gene | predicted gene, 41662 |
| 18 | gene | 6.024427 | 6.136098 | negative | MGI_C57BL6J_1922665 | Arhgap12 | NCBI_Gene:75415,ENSEMBL:ENSMUSG00000041225 | MGI:1922665 | protein coding gene | Rho GTPase activating protein 12 |
| 18 | pseudogene | 6.029179 | 6.030456 | negative | MGI_C57BL6J_5011425 | Gm19240 | NCBI_Gene:100418482 | MGI:5011425 | pseudogene | predicted gene, 19240 |
| 18 | gene | 6.201002 | 6.242174 | negative | MGI_C57BL6J_1098268 | Kif5b | NCBI_Gene:16573,ENSEMBL:ENSMUSG00000006740 | MGI:1098268 | protein coding gene | kinesin family member 5B |
| 18 | gene | 6.213913 | 6.227324 | positive | MGI_C57BL6J_4937863 | Gm17036 | ENSEMBL:ENSMUSG00000091165 | MGI:4937863 | lncRNA gene | predicted gene 17036 |
| 18 | gene | 6.234853 | 6.326693 | positive | MGI_C57BL6J_5624549 | Gm41664 | NCBI_Gene:105246365,ENSEMBL:ENSMUSG00000117519 | MGI:5624549 | lncRNA gene | predicted gene, 41664 |
| 18 | pseudogene | 6.332591 | 6.333082 | positive | MGI_C57BL6J_3646174 | Rpl27-ps3 | NCBI_Gene:621100,ENSEMBL:ENSMUSG00000073640 | MGI:3646174 | pseudogene | ribosomal protein L27, pseudogene 3 |
| 18 | pseudogene | 6.368360 | 6.368797 | positive | MGI_C57BL6J_6275267 | Gm49986 | ENSEMBL:ENSMUSG00000117387 | MGI:6275267 | pseudogene | predicted gene, 49986 |
| 18 | pseudogene | 6.371352 | 6.371660 | negative | MGI_C57BL6J_3648803 | Gm6291 | NCBI_Gene:102637200,ENSEMBL:ENSMUSG00000095614 | MGI:3648803 | pseudogene | predicted gene 6291 |
| 18 | gene | 6.435951 | 6.516108 | negative | MGI_C57BL6J_1278322 | Epc1 | NCBI_Gene:13831,ENSEMBL:ENSMUSG00000024240 | MGI:1278322 | protein coding gene | enhancer of polycomb homolog 1 |
| 18 | gene | 6.490143 | 6.496962 | positive | MGI_C57BL6J_5579235 | Gm28529 | ENSEMBL:ENSMUSG00000101320 | MGI:5579235 | lncRNA gene | predicted gene 28529 |
| 18 | gene | 6.490564 | 6.490646 | negative | MGI_C57BL6J_3811424 | Mir1893 | miRBase:MI0008327,NCBI_Gene:100316773,ENSEMBL:ENSMUSG00000084514 | MGI:3811424 | miRNA gene | microRNA 1893 |
| 18 | pseudogene | 6.557130 | 6.557796 | positive | MGI_C57BL6J_3645335 | Gm7464 | NCBI_Gene:665047,ENSEMBL:ENSMUSG00000117425 | MGI:3645335 | pseudogene | predicted gene 7464 |
| 18 | gene | 6.603629 | 6.640435 | positive | MGI_C57BL6J_1918151 | 4921524L21Rik | NCBI_Gene:70901,ENSEMBL:ENSMUSG00000039540 | MGI:1918151 | protein coding gene | RIKEN cDNA 4921524L21 gene |
| 18 | pseudogene | 6.687843 | 6.691317 | negative | MGI_C57BL6J_6275405 | Gm50077 | ENSEMBL:ENSMUSG00000117620 | MGI:6275405 | pseudogene | predicted gene, 50077 |
| 18 | gene | 6.733905 | 6.794429 | positive | MGI_C57BL6J_102790 | Rab18 | NCBI_Gene:19330,ENSEMBL:ENSMUSG00000073639 | MGI:102790 | protein coding gene | RAB18, member RAS oncogene family |
| 18 | pseudogene | 6.760073 | 6.760959 | negative | MGI_C57BL6J_3648356 | Gm7466 | NCBI_Gene:665053,ENSEMBL:ENSMUSG00000117359 | MGI:3648356 | pseudogene | predicted gene 7466 |
| 18 | gene | 6.788768 | 6.792137 | positive | MGI_C57BL6J_6275412 | Gm50081 | ENSEMBL:ENSMUSG00000117461 | MGI:6275412 | lncRNA gene | predicted gene, 50081 |
| 18 | gene | 6.910459 | 6.936621 | negative | MGI_C57BL6J_3826781 | Gm2350 | ENSEMBL:ENSMUSG00000117652 | MGI:3826781 | lncRNA gene | predicted gene 2350 |
| 18 | gene | 6.934518 | 7.004780 | negative | MGI_C57BL6J_2687286 | Mkx | NCBI_Gene:210719,ENSEMBL:ENSMUSG00000061013 | MGI:2687286 | protein coding gene | mohawk homeobox |
| 18 | gene | 7.004963 | 7.066322 | positive | MGI_C57BL6J_5826270 | Gm46633 | NCBI_Gene:108168391,ENSEMBL:ENSMUSG00000117356 | MGI:5826270 | lncRNA gene | predicted gene, 46633 |
| 18 | pseudogene | 7.041939 | 7.044195 | negative | MGI_C57BL6J_3642728 | Gm10350 | NCBI_Gene:664778,ENSEMBL:ENSMUSG00000117381 | MGI:3642728 | pseudogene | predicted gene 10350 |
| 18 | gene | 7.088209 | 7.298304 | negative | MGI_C57BL6J_1922184 | Armc4 | NCBI_Gene:74934,ENSEMBL:ENSMUSG00000061802 | MGI:1922184 | protein coding gene | armadillo repeat containing 4 |
| 18 | pseudogene | 7.107181 | 7.108183 | negative | MGI_C57BL6J_5010766 | Gm18581 | NCBI_Gene:100417382,ENSEMBL:ENSMUSG00000117360 | MGI:5010766 | pseudogene | predicted gene, 18581 |
| 18 | gene | 7.150344 | 7.151777 | negative | MGI_C57BL6J_1915234 | 4930415O11Rik | ENSEMBL:ENSMUSG00000117383 | MGI:1915234 | unclassified gene | RIKEN cDNA 4930415O11 gene |
| 18 | gene | 7.332445 | 7.346299 | negative | MGI_C57BL6J_5594138 | Gm34979 | NCBI_Gene:102638402 | MGI:5594138 | lncRNA gene | predicted gene, 34979 |
| 18 | gene | 7.347959 | 7.626893 | negative | MGI_C57BL6J_1922989 | Mpp7 | NCBI_Gene:75739,ENSEMBL:ENSMUSG00000057440 | MGI:1922989 | protein coding gene | membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| 18 | pseudogene | 7.548932 | 7.549781 | positive | MGI_C57BL6J_5011133 | Gm18948 | NCBI_Gene:100418013,ENSEMBL:ENSMUSG00000117556 | MGI:5011133 | pseudogene | predicted gene, 18948 |
| 18 | pseudogene | 7.581482 | 7.582130 | positive | MGI_C57BL6J_3644282 | Fabp5l2 | NCBI_Gene:622384,ENSEMBL:ENSMUSG00000094334 | MGI:3644282 | pseudogene | fatty acid binding protein 5-like 2 |
| 18 | pseudogene | 7.684076 | 7.693411 | negative | MGI_C57BL6J_5010948 | Gm18763 | NCBI_Gene:100417683,ENSEMBL:ENSMUSG00000117542 | MGI:5010948 | pseudogene | predicted gene, 18763 |
| 18 | pseudogene | 7.798784 | 7.799900 | negative | MGI_C57BL6J_3780506 | Gm2336 | NCBI_Gene:100039606 | MGI:3780506 | pseudogene | predicted gene 2336 |
| 18 | gene | 7.835177 | 7.835283 | positive | MGI_C57BL6J_5454069 | Gm24292 | ENSEMBL:ENSMUSG00000064822 | MGI:5454069 | snRNA gene | predicted gene, 24292 |
| 18 | pseudogene | 7.844684 | 7.844935 | negative | MGI_C57BL6J_6275329 | Gm50027 | ENSEMBL:ENSMUSG00000117462 | MGI:6275329 | pseudogene | predicted gene, 50027 |
| 18 | gene | 7.868018 | 7.869113 | negative | MGI_C57BL6J_3641793 | Gm9993 | ENSEMBL:ENSMUSG00000117505 | MGI:3641793 | lncRNA gene | predicted gene 9993 |
| 18 | gene | 7.868832 | 7.929028 | positive | MGI_C57BL6J_2387357 | Wac | NCBI_Gene:225131,ENSEMBL:ENSMUSG00000024283 | MGI:2387357 | protein coding gene | WW domain containing adaptor with coiled-coil |
| 18 | gene | 7.894300 | 7.895792 | positive | MGI_C57BL6J_1924412 | A430102J17Rik | NA | NA | unclassified gene | RIKEN cDNA A430102J17 gene |
| 18 | pseudogene | 7.932728 | 7.933556 | positive | MGI_C57BL6J_3643245 | Gm7502 | NCBI_Gene:665121,ENSEMBL:ENSMUSG00000091019 | MGI:3643245 | pseudogene | predicted gene 7502 |
R/qtl
scanone
gmObject of class cross2 (crosstype "bc")
Total individuals 138
No. genotyped individuals 138
No. phenotyped individuals 138
No. with both geno & pheno 138
No. phenotypes 1
No. covariates 7
No. phenotype covariates 0
No. chromosomes 20
Total markers 32690
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2952 2874 2058 2071 1955 2053 1862 1700 2009 1229 2069 1396 1628 1692 1064 940
17 18 19 X
401 1095 1067 575 #detach("package:qtl2", unload=TRUE)
#library(qtl)
cross <- qtl::read.cross("csv", file = "data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_mis_conditional_2-peaks-chr3-4.csv",alleles=c("A","B")) --Read the following data:
138 individuals
32690 markers
5 phenotypes
--Cross type: bc cross <- qtl::jittermap(cross)
summary(cross) Backcross
No. individuals: 138
No. phenotypes: 5
Percent phenotyped: 100 100 100 99.3 98.6
No. chromosomes: 20
Autosomes: 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19
X chr: X
Total markers: 32690
No. markers: 2952 2874 2058 2071 1955 2053 1862 1700 2009 1229 2069
1396 1628 1692 1064 940 401 1095 1067 575
Percent genotyped: 99.5
Genotypes (%):
Autosomes: AA:54.3 AB:45.7
X chromosome: AA:52.7 AB:47.3 cross.probs <- qtl::calc.genoprob(cross)
print("method == hk")[1] "method == hk"add.covars = qtl::pull.pheno(cross.probs, c("UNCHS008487","UNC8250659"))
scanone.hk <-qtl::scanone(cross.probs, pheno.col="ICI.vs.PBS" , model="binary", method="hk", addcovar = add.covars)
operm.hk <- qtl::scanone(cross.probs, method = "hk", pheno.col="ICI.vs.PBS", n.perm = 10, perm.Xsp = TRUE, model="binary", verbose=FALSE, addcovar = add.covars)
plot(operm.hk)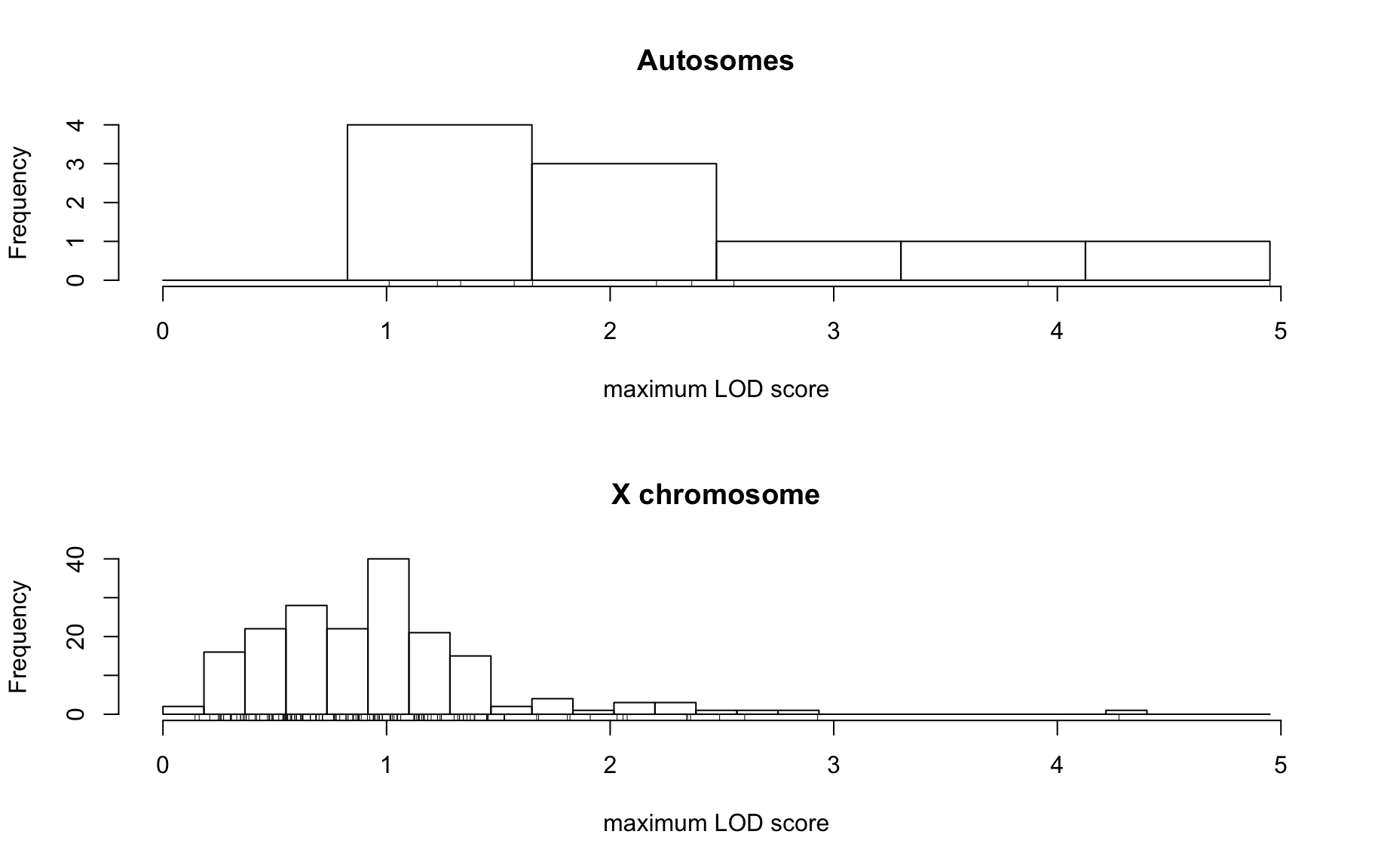
print(summary(operm.hk, alpha=c(0.01, 0.05, 0.1)))Autosome LOD thresholds (10 permutations)
lod
1% 4.86
5% 4.49
10% 4.03
X chromosome LOD thresholds (183 permutations)
lod
1% 4.15
5% 3.62
10% 2.94#plot(scanone.hk, bandcol = "grey90",lty=1, cex=1, col = "steelblue")
#qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.01, col = 'blue')
#qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.05, col = 'red')
#qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.1, col = 'purple')
ymx <- maxlod(out) # overall maximum LOD score
plot(scanone.hk, bandcol = "grey90",lty=1, cex=1, col = "slateblue", ylim=c(0, ymx+0.5))
title(main = paste0(colnames(out), " [positions in cM]"))
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.01, col = 'blue')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.05, col = 'red')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.1, col = 'purple')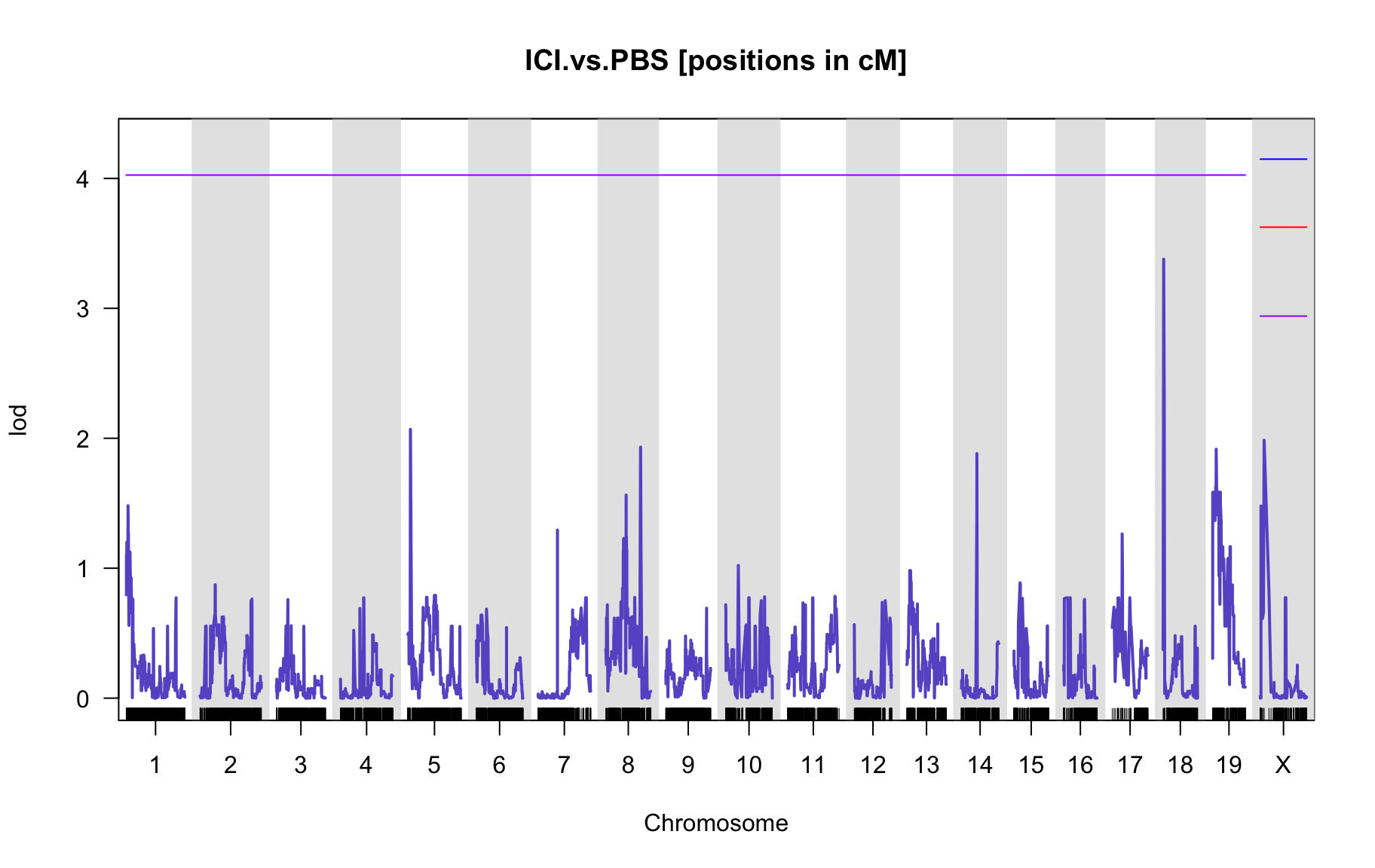
ymx <- 11
plot(scanone.hk, bandcol = "grey90",lty=1, cex=1, col = "slateblue", ylim=c(0, ymx+0.5))
title(main = paste0(colnames(out), " [positions in cM]\n(using same scale as eoi vs ici for easier comparison)"))
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.01, col = 'blue')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.05, col = 'red')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.1, col = 'purple')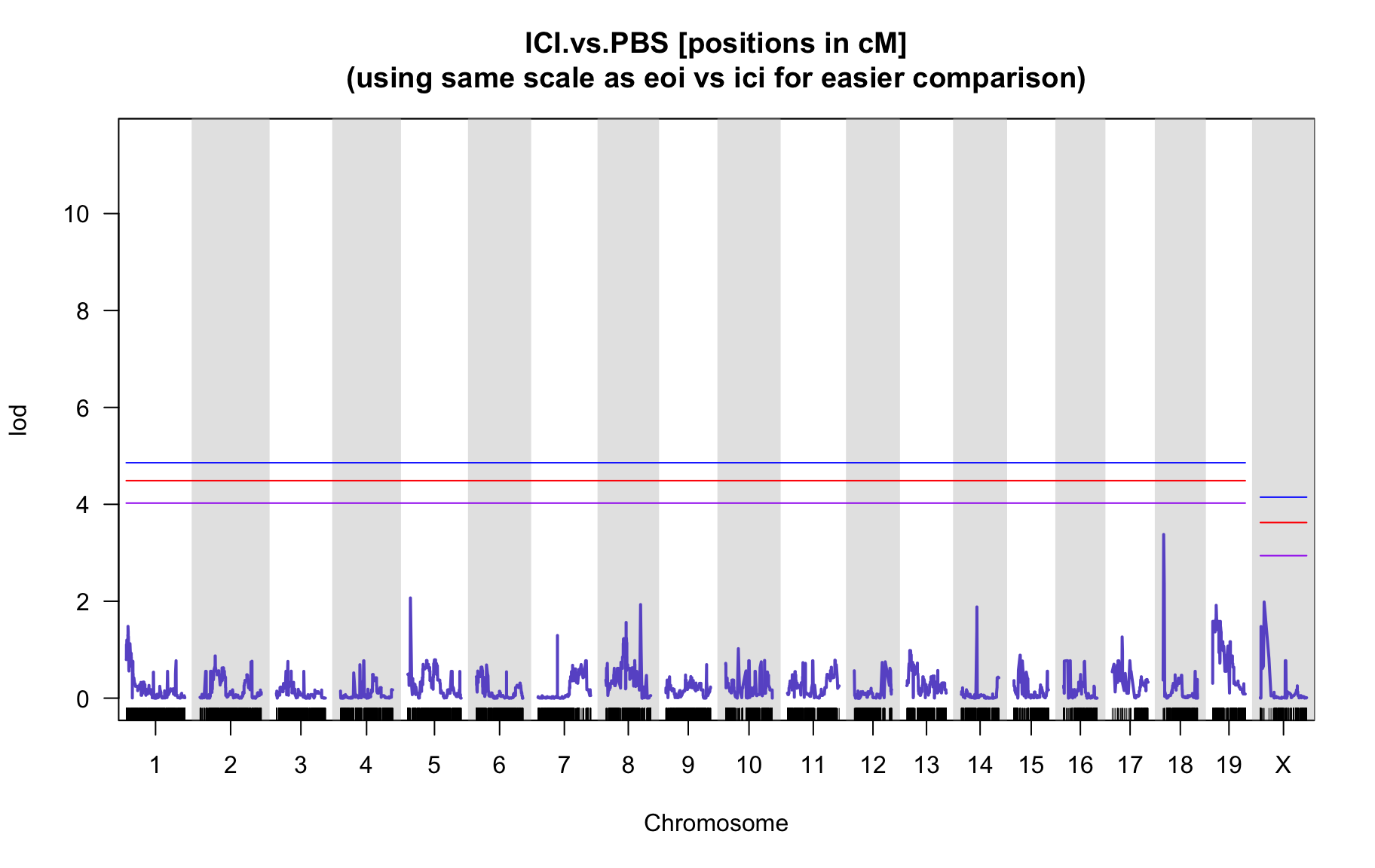
print(as.data.frame(summary(scanone.hk, perms=operm.hk, pvalues=TRUE, format="allpeaks"))) chr pos lod pval
UNCHS000141 1 4.439109 1.4810382 0.7191035
UNC2981117 2 27.613431 0.8724495 1.0000000
JAX00521745 3 20.613360 0.7586221 1.0000000
JAX00556948 4 41.038754 0.7726880 1.0000000
UNC8675939 5 6.193174 2.0684019 0.5185858
UNC10923091 6 18.321306 0.6856206 1.0000000
UNCHS020066 7 34.113657 1.2936681 0.8168395
UNC15524531 8 59.462590 1.9315437 0.5185858
UNC17203597 9 68.527929 0.6928610 1.0000000
UNCHS028001 10 21.783362 1.0219015 0.9118239
UNC20465557 11 80.904972 0.7841081 1.0000000
UNC21787084 12 53.288332 0.7505664 1.0000000
JAX00351755 13 7.044200 0.9809396 1.0000000
UNC24056202 14 30.156730 1.8824837 0.5185858
UNC25275535 15 11.967146 0.8880487 1.0000000
UNCHS042071 16 13.176096 0.7730967 1.0000000
UNCrs47191360 17 18.433013 1.2636005 0.8168395
UNCHS045343 18 2.934001 3.3781249 0.2096965
UNC29920552 19 8.810092 1.9159935 0.5185858
UNCHS048314 X 8.974041 1.9849011 0.6619190print("all peaks with a p-value less or equal to 0.05 (suggestive)")[1] "all peaks with a p-value less or equal to 0.05 (suggestive)"print(as.data.frame(summary(scanone.hk, perms=operm.hk, alpha=0.05, pvalues=TRUE, format="allpeaks")))[1] chr pos lod
<0 rows> (or 0-length row.names)#print("method == ehk")
#scanone.ehk <-qtl::scanone(cross.probs, pheno.col="ICI.vs.PBS" , model="binary", method="ehk")
#operm.ehk <- qtl::scanone(cross.probs, method = "ehk", pheno.col="ICI.vs.PBS", n.perm = 10, perm.Xsp = TRUE, model="binary", verbose=FALSE)
#plot(operm.ehk)
#print(summary(operm.ehk, alpha=c(0.01, 0.05, 0.1)))
#plot(scanone.ehk, bandcol = "grey90",lty=1, cex=1, col = "steelblue")
#qtl::add.threshold(scanone.ehk, perms= operm.ehk, alpha=0.01, col = 'blue')
#qtl::add.threshold(scanone.ehk, perms= operm.ehk, alpha=0.05, col = 'red')
#qtl::add.threshold(scanone.ehk, perms= operm.ehk, alpha=0.1, col = 'purple')
#print(as.data.frame(summary(scanone.ehk)))
#print(as.data.frame(summary(scanone.ehk, perms=operm.ehk, alpha=0.05, pvalues=TRUE, format="allpeaks")))Conditionaing on eoi vs. ici chr 3 peak (UNCHS008487) & chr 10 peak (UNC18240977)
Genome-wide scan
#gm
Xcovar <- get_x_covar(gm)
#addcovar = model.matrix(~Sex, data = covars)[,-1]
addcovar = model.matrix(~UNCHS008487+UNC18240977, data = covars)[,-1]
#K <- calc_kinship(pr.qc, type = "loco")
#heatmap(K[[1]])
#K.overall <- calc_kinship(pr.qc, type = "overall")
#heatmap(K.overall)
kinship <- calc_kinship(pr.qc)
heatmap(kinship)
#operm <- scan1perm(pr.qc, gm$covar$phenos, Xcovar=Xcovar, n_perm=2000)
#operm <- scan1perm(pr.qc, gm$covar$phenos, addcovar = addcovar, n_perm=2000)
#operm <- scan1perm(pr.qc, gm$covar$phenos, n_perm=2000)
operm <- scan1perm(pr.qc, gm$covar["ICI.vs.PBS"], model="binary", n_perm=10, perm_Xsp=TRUE, chr_lengths=chr_lengths(gm$gmap), addcovar = addcovar)
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
names(summary_table) <- c("autosomes","X")
summary_table$significance.level <- rownames(summary_table)
rownames(summary_table) <- NULL
summary_table[c(3,1:2)] %>%
kable(escape = F,align = c("ccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| significance.level | autosomes | X |
|---|---|---|
| 0.01 | 2.738061 | 2.910347 |
| 0.05 | 2.671339 | 2.756903 |
| 0.1 | 2.587731 | 2.556243 |
The figures below show QTL maps for each phenotype
out <- scan1(pr.qc, gm$covar["ICI.vs.PBS"], Xcovar=Xcovar, model="binary", addcovar = addcovar)
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
plot_lod<-function(out,map){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- 11 # overall maximum LOD score
plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " [positions in cM] \n(using same scale as eoi vs ici for easier comparison)"))
add_threshold(map, summary(operm, alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
plot_lod(out,gm$gmap)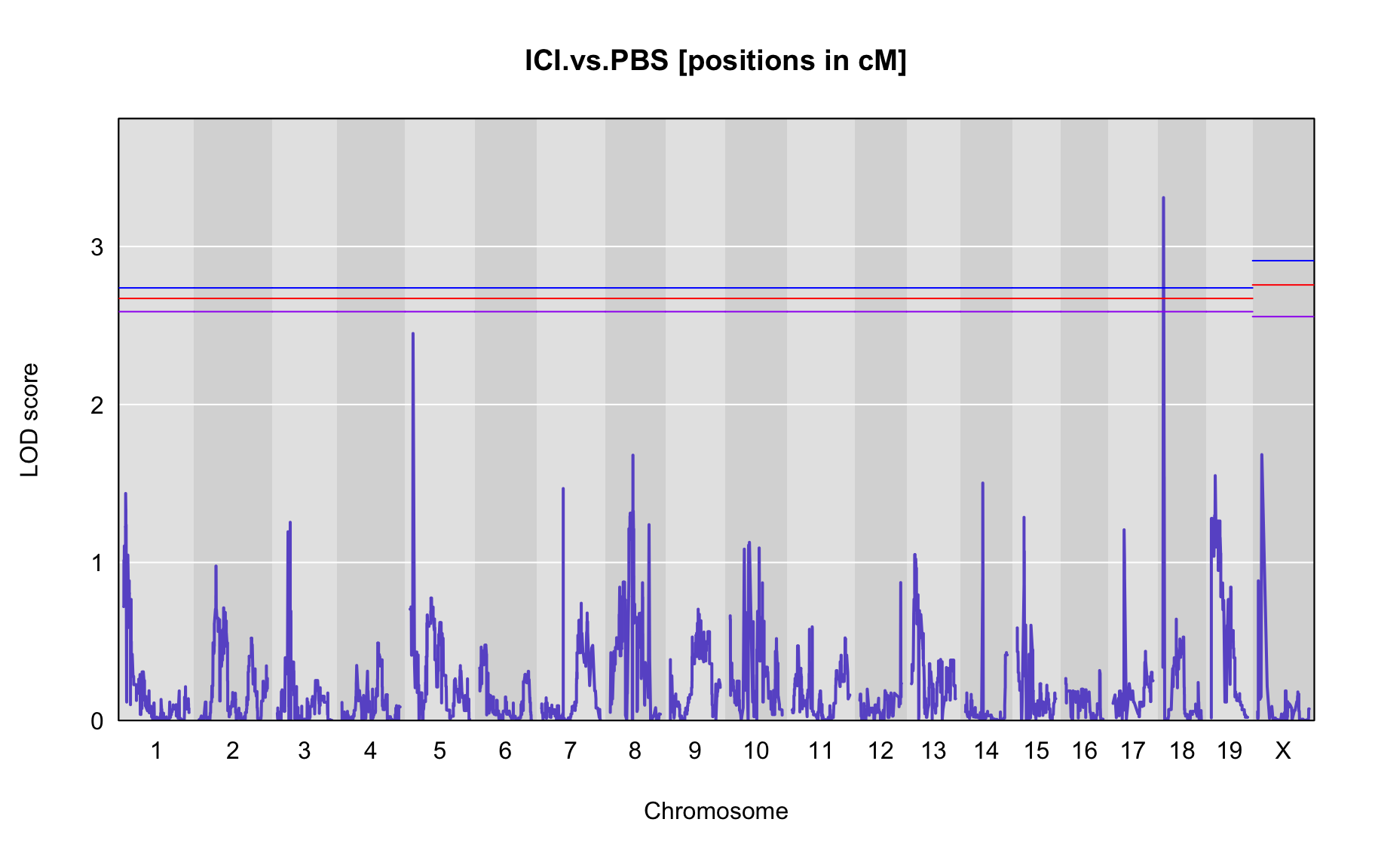
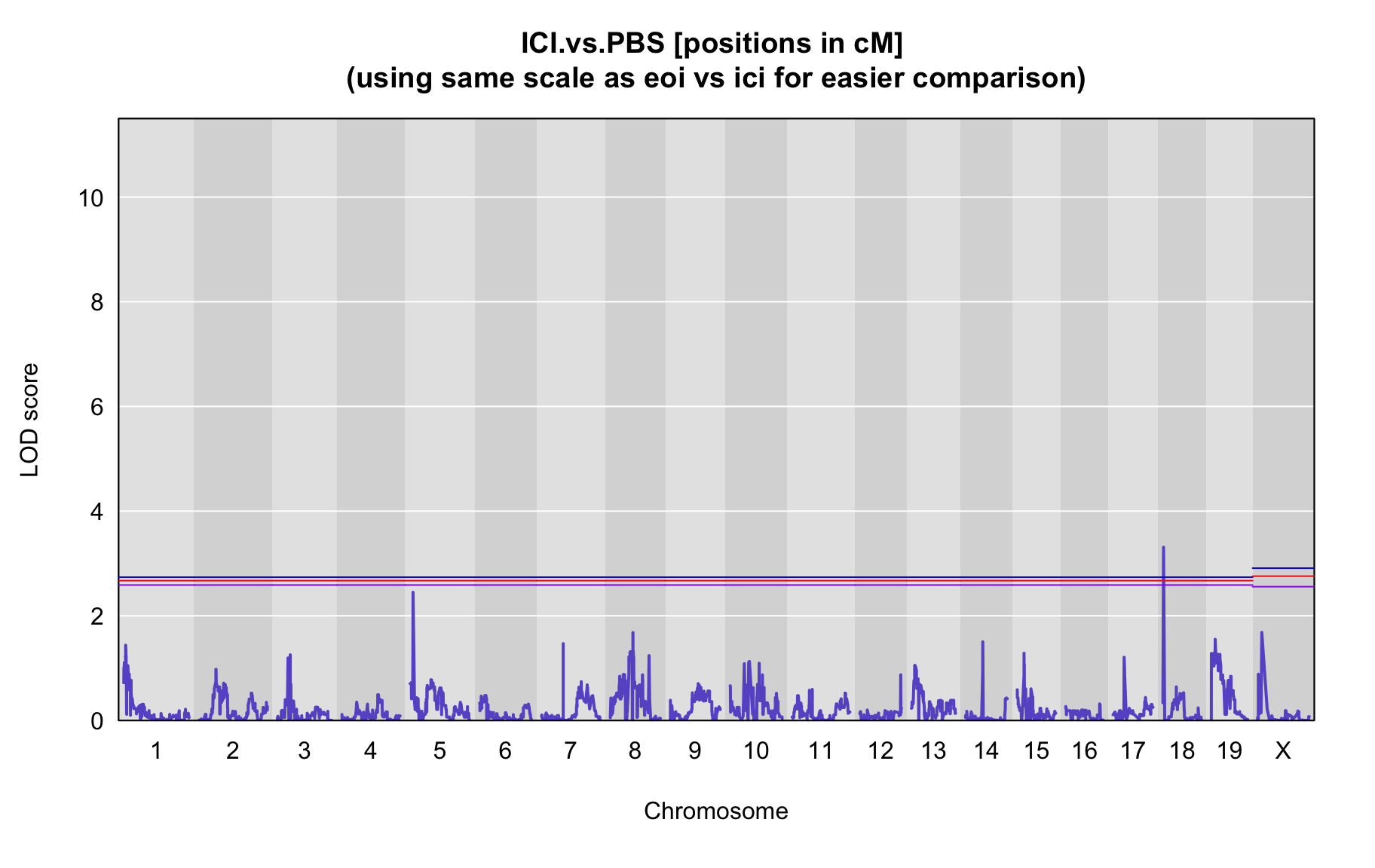
LOD peaks
The table below shows QTL peaks associated with the phenotype. We use the 95% threshold from the permutations to find peaks.
Centimorgan (cM)
peaks <- find_peaks(out, gm$gmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) >0){
peaks$marker <- find_marker(gm$gmap, chr=peaks$chr,pos=peaks$pos)
names(peaks)[2] <- c("phenotype")
peaks <- peaks[-1]
rownames(peaks) <- NULL
#print(kable(peaks, "html")
# %>% kable_styling("striped", full_width = T) %>%
# column_spec(1, bold=TRUE)
# )
print(kable(peaks, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
#peaks[] %>%
# kable(escape = F,align = c("cccccccc")) %>%
# kable_styling("striped", full_width = T) %>%
# column_spec(1, bold=TRUE)
#plot only peak chromosomes
plot_lod_chr<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
ymx <- 11
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in cM]\n(using same scale as eoi vs. ici for easier comparison)"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
for(i in unique(peaks$chr)){
#for (i in 1:nrow(peaks)){
#plot_lod_chr(out,gm$gmap, peaks$chr[i])
plot_lod_chr(out,gm$gmap, i)
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}| phenotype | chr | pos | lod | ci_lo | ci_hi | marker |
|---|---|---|---|---|---|---|
| ICI.vs.PBS | 18 | 2.934 | 3.309497 | 1.972 | 3.857 | UNCHS045343 |
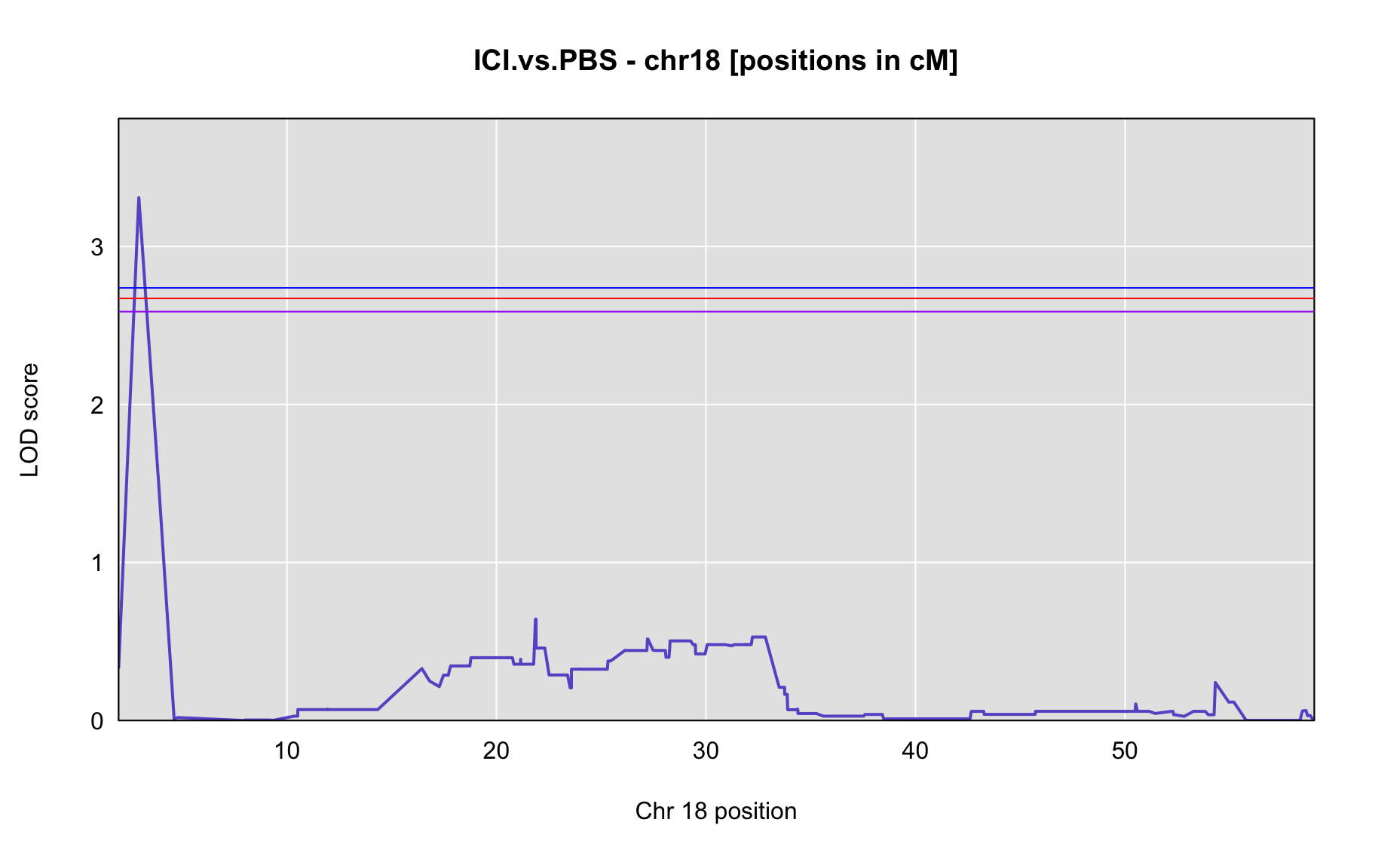
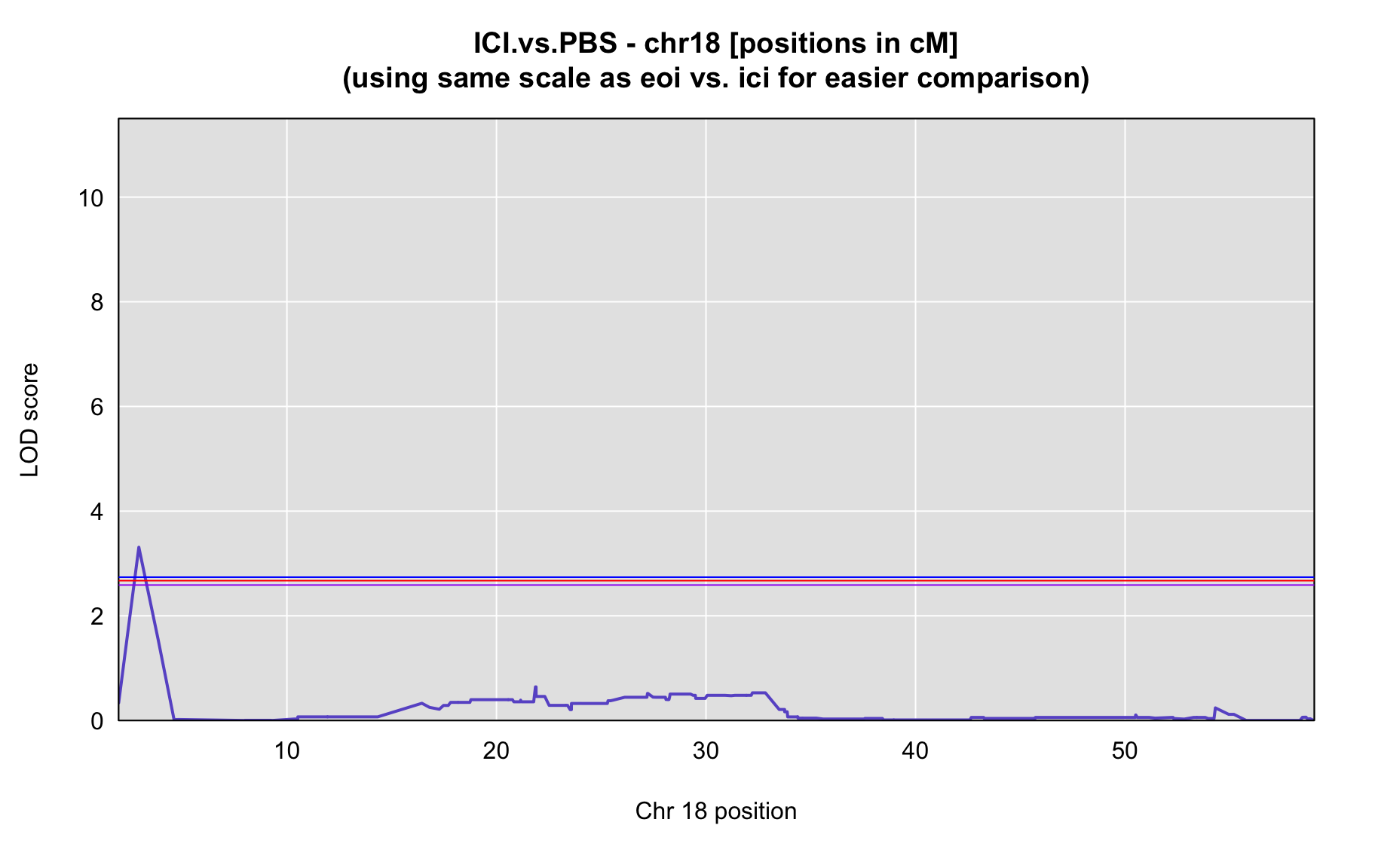
Megabase (MB)
print("peaks in MB positions")[1] “peaks in MB positions”
peaks_mba <- find_peaks(out, gm$pmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) >0){
peaks_mba$marker <- find_marker(gm$pmap, chr=peaks_mba$chr,pos=peaks_mba$pos)
names(peaks_mba)[2] <- c("phenotype")
peaks_mba <- peaks_mba[-1]
#peaks_mbl <- list()
##corresponding info in Mb
#for(i in 1:nrow(peaks)){
# #lodindex <- peaks$lodindex[i]
# phenotype <- peaks$phenotype[i]
# chr <- as.character(peaks$chr[i])
# lod <- peaks$lod[i]
# mark <- peaks$marker[i]
# pos <- mapdf[mapdf$marker==mark,]$pmapdf
# ci_lo <- mapdfnd$pmapdf[which(mapdfnd$gmapdf == peaks$ci_lo[i] & mapdfnd$chr == peaks$chr[i])]
# ci_hi <- mapdfnd$pmapdf[which(mapdfnd$gmapdf == peaks$ci_hi[i] & mapdfnd$chr == peaks$chr[i])]
# peaks_mb=as.data.frame(cbind(phenotype, chr, pos, lod, ci_lo, ci_hi, mark))
# names(peaks_mb)[7] <- c("marker")
# peaks_mbl[[i]] <- peaks_mb
#}
#peaks_mba2 <- do.call(rbind, peaks_mbl)
#peaks_mba2 <- as.data.frame(peaks_mba)
#peaks_mba[,c("chr", "pos", "lod", "ci_lo", "ci_hi")] <- sapply(peaks_mba[,c("chr", "pos", "lod", "ci_lo", "ci_hi")], as.numeric)
rownames(peaks_mba) <- NULL
#print(kable(peaks_mba, "html")
# %>% kable_styling("striped", full_width = T) %>%
# column_spec(1, bold=TRUE)
# )
print(kable(peaks_mba, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
#peaks_mba[] %>%
# kable(escape = F,align = c("cccccccc")) %>%
# kable_styling("striped", full_width = T) %>%
# column_spec(1, bold=TRUE)
plot_lod_chr_mb<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in MB]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
ymx <- 11
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in MB]\n(using same scale as eoi vs. ici for easier comparison)"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
for(i in unique(peaks_mba$chr)){
#for (i in 1:nrow(peaks_mba)){
#plot_lod_chr_mb(out,gm$pmap, peaks_mba$chr[i])
plot_lod_chr_mb(out,gm$pmap,i)
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}| phenotype | chr | pos | lod | ci_lo | ci_hi | marker |
|---|---|---|---|---|---|---|
| ICI.vs.PBS | 18 | 4.861199 | 3.309497 | 3.01212 | 5.584328 | UNCHS045343 |
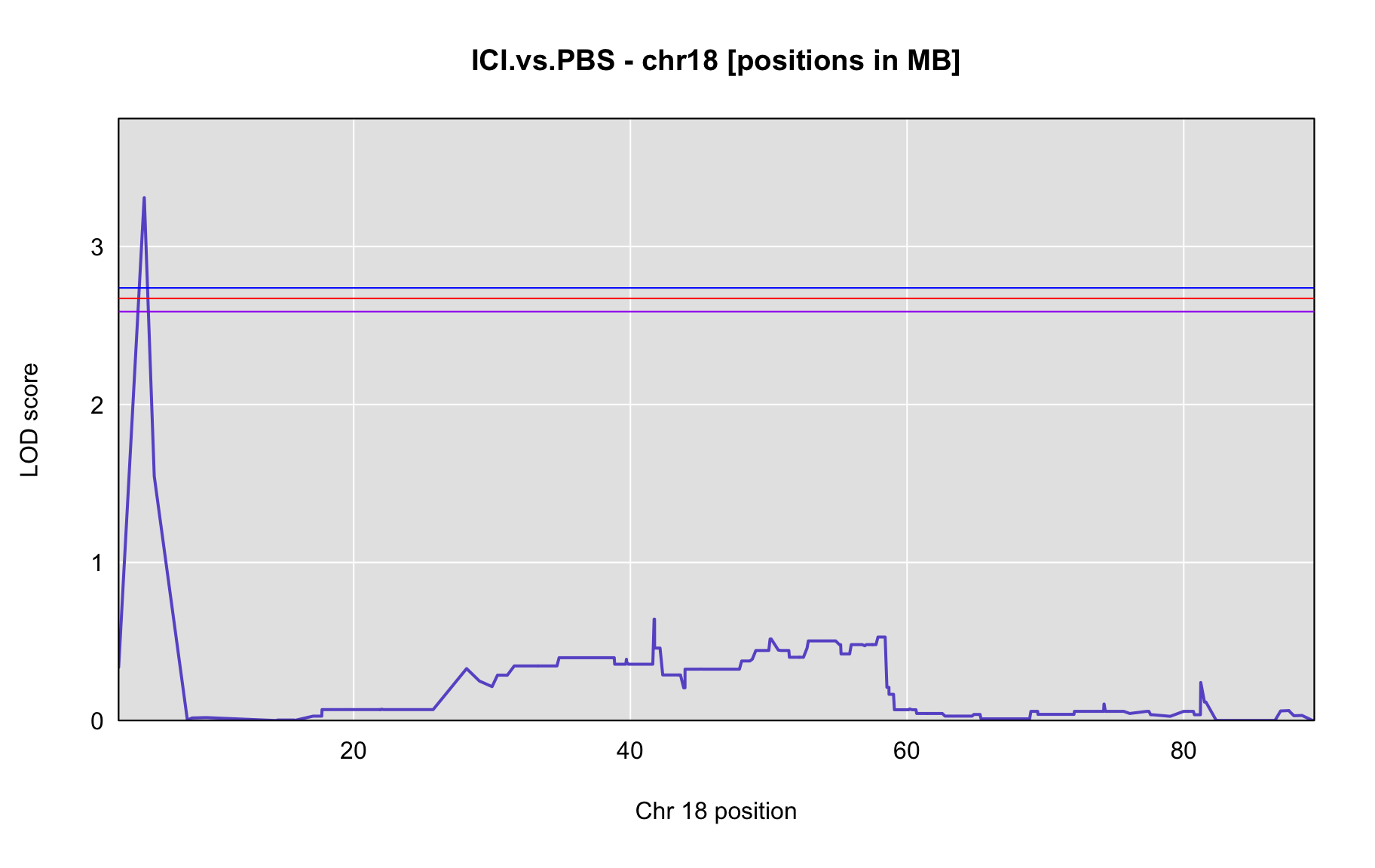
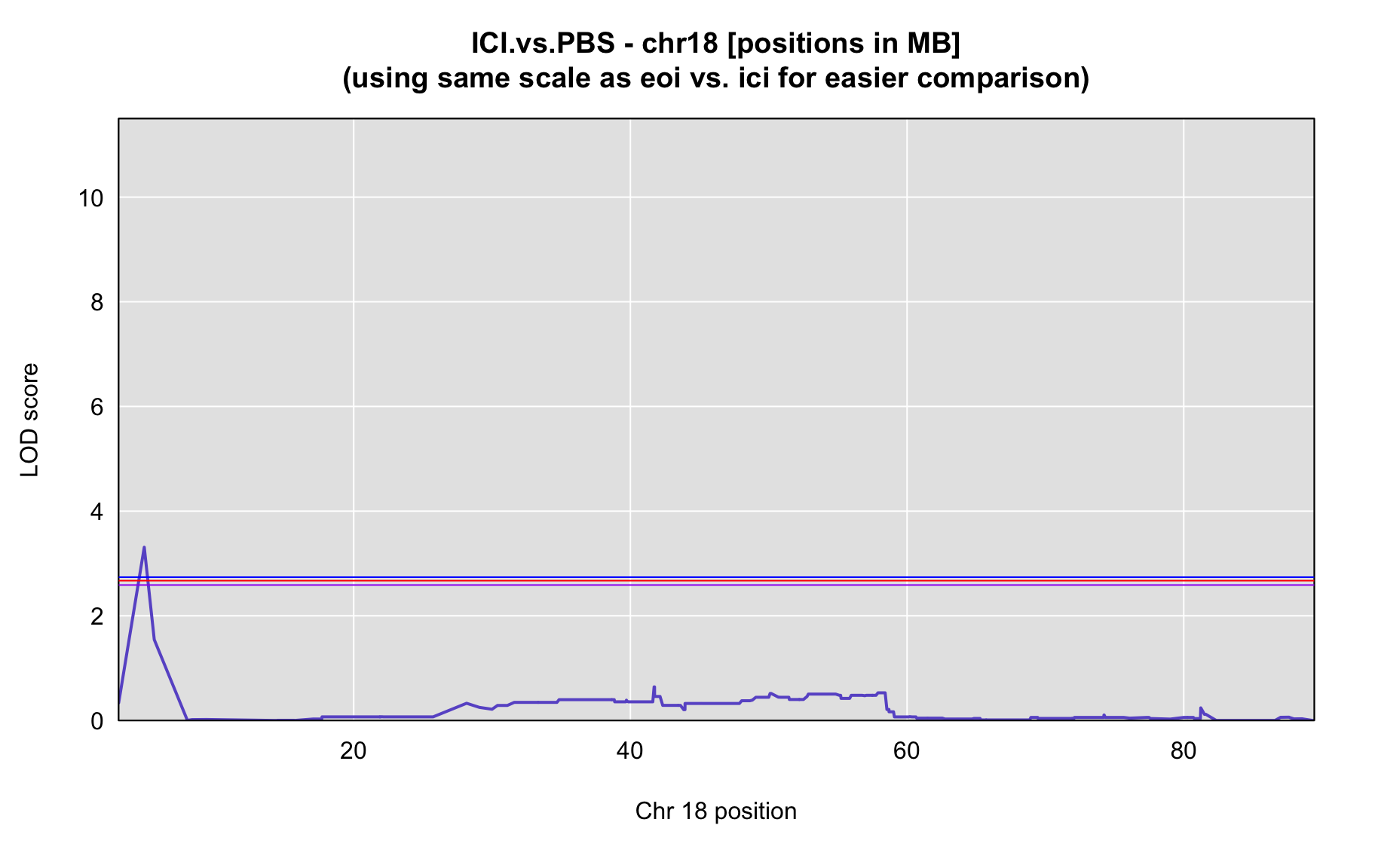
QTL effects
For each peak LOD location we give a list of gene
query_variants <- create_variant_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/cc_variants.sqlite")
query_genes <- create_gene_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/mouse_genes_mgi.sqlite")
if(nrow(peaks) >0){
for (i in 1:nrow(peaks)){
#for (i in 1:1){
#Plot 1
#marker = find_marker(gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i])
#gp <- g[,marker]
#gp[gp==1] <- "AA"
#gp[gp==2] <- "AB"
#gp[gp==0] <- NA
#plot_pxg(gp, gm$covar[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
#title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
###dev.off()
g <- maxmarg(pr.qc, gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i], return_char=TRUE)
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/","qtl_effect_", i, ".png"))
#par(mar=c(4.1, 4.1, 1.5, 0.6))
plot_pxg(g, gm$covar[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
##dev.off()
chr = peaks$chr[i]
# Plot 2
pr_sub <- pull_genoprobint(pr.qc, gm$gmap, chr, c(peaks$ci_lo[i], peaks$ci_hi[i]))
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar)
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], Xcovar=Xcovar)
#coeff <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
#coeff_sub <- scan1coef(pr_sub[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
blup <- scan1blup(pr.qc[,chr], gm$covar[peaks$phenotype[i]], addcovar = addcovar)
blup_sub <- scan1blup(pr_sub[,chr], gm$covar[peaks$phenotype[i]], addcovar = addcovar)
write.csv(as.data.frame(blup_sub), paste0("data/ici.vs.pbs_blup_sub_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_snpsqc_5.batches_mis_conditional_2-peaks-chr3-10.csv"), quote=F)
#plot_coef(coeff,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i]," [scan1coeff; positions in cM])")
# )
#plot_coef(coeff_sub,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i],"; 1.5 LOD drop interval [scan1coeff; positions in cM] ) ")
# )
plot_coef(blup,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," [scan1blup; positions in cM])")
)
plot_coef(blup_sub,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i],"; 1.5 LOD drop interval [scan1blup; positions in cM])")
)
# Plot 3
#c2effB <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary", contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
#c2effBb <- scan1blup(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]],Xcovar=Xcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
#plot(c2effB, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
#plot(c2effBb, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
##last_coef <- unclass(c2effB)[nrow(c2effB),2:3] # last two coefficients
##for(t in seq(along=last_coef))
## axis(side=4, at=last_coef[t], names(last_coef)[t], tick=FALSE)
#Table 1
chr = peaks_mba$chr[i]
start=as.numeric(peaks_mba$ci_lo[i])
end=as.numeric(peaks_mba$ci_hi[i])
genesgss = query_genes(chr, start, end)
write.csv(genesgss, file=paste0("data/ici.vs.pbs_genes_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_snpsqc_5.batches_mis_conditional_2-peaks-chr3-10.csv"), quote=F)
rownames(genesgss) <- NULL
genesgss$strand_old = genesgss$strand
genesgss$strand[genesgss$strand=="+"] <- "positive"
genesgss$strand[genesgss$strand=="-"] <- "negative"
#genesgss <-
#table <-
#genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")] %>%
#kable(escape = F,align = c("ccccccccccc")) %>%
#kable_styling("striped", full_width = T) #%>%
#cat #%>%
#column_spec(1, bold=TRUE)
#
#print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], escape = F,align = c("ccccccccccc")))
print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], "html") %>% kable_styling("striped", full_width = T))
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}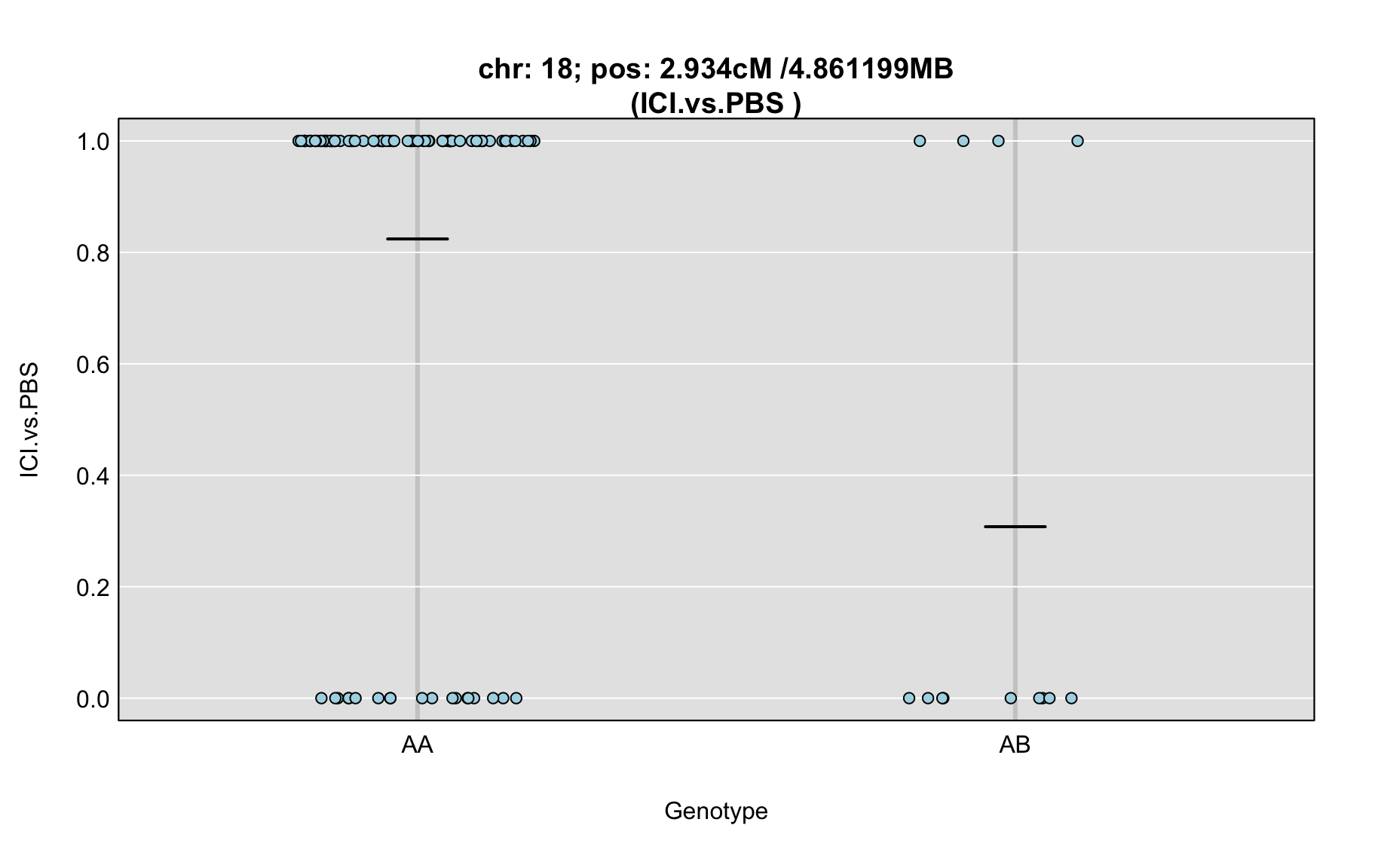
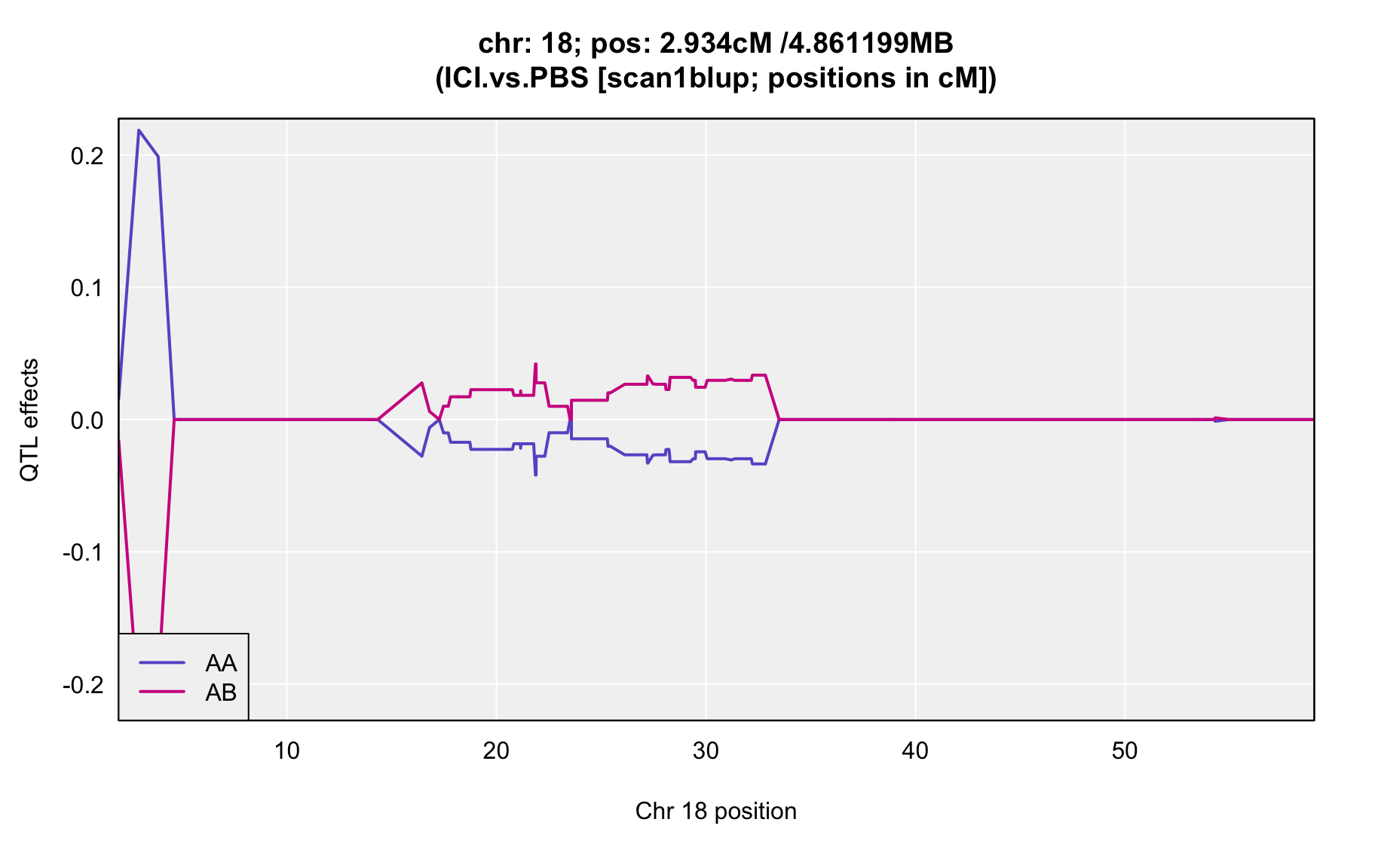
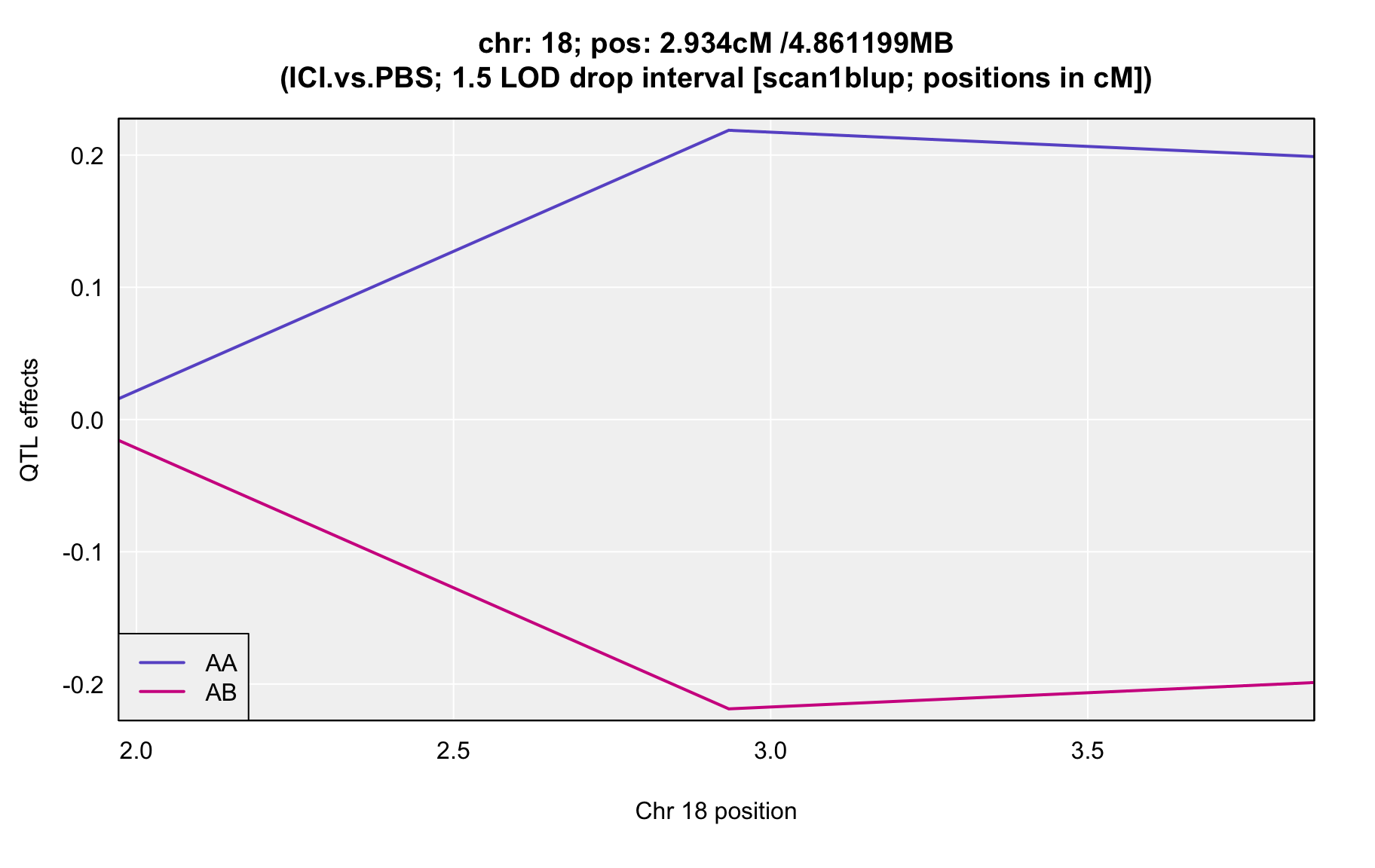
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 18 | pseudogene | 3.015908 | 3.016159 | positive | MGI_C57BL6J_6275399 | Gm50072 | ENSEMBL:ENSMUSG00000117547 | MGI:6275399 | pseudogene | predicted gene, 50072 |
| 18 | pseudogene | 3.026901 | 3.027882 | negative | MGI_C57BL6J_3852490 | Vmn1r-ps151 | NCBI_Gene:100312519,ENSEMBL:ENSMUSG00000093774 | MGI:3852490 | pseudogene | vomeronasal 1 receptor, pseudogene 151 |
| 18 | pseudogene | 3.080778 | 3.081476 | negative | MGI_C57BL6J_3852491 | Vmn1r-ps152 | NCBI_Gene:100312520,ENSEMBL:ENSMUSG00000093444 | MGI:3852491 | pseudogene | vomeronasal 1 receptor, pseudogene 152 |
| 18 | pseudogene | 3.105167 | 3.105396 | negative | MGI_C57BL6J_6275400 | Gm50073 | ENSEMBL:ENSMUSG00000117531 | MGI:6275400 | pseudogene | predicted gene, 50073 |
| 18 | gene | 3.122492 | 3.130012 | negative | MGI_C57BL6J_3852494 | Vmn1r238 | NCBI_Gene:100312476,ENSEMBL:ENSMUSG00000091539 | MGI:3852494 | protein coding gene | vomeronasal 1 receptor, 238 |
| 18 | pseudogene | 3.139032 | 3.139305 | negative | MGI_C57BL6J_6275401 | Gm50074 | ENSEMBL:ENSMUSG00000117353 | MGI:6275401 | pseudogene | predicted gene, 50074 |
| 18 | pseudogene | 3.171447 | 3.171932 | negative | MGI_C57BL6J_6275402 | Gm50075 | ENSEMBL:ENSMUSG00000117464 | MGI:6275402 | pseudogene | predicted gene, 50075 |
| 18 | gene | 3.266048 | 3.338176 | negative | MGI_C57BL6J_88495 | Crem | NCBI_Gene:12916,ENSEMBL:ENSMUSG00000063889 | MGI:88495 | protein coding gene | cAMP responsive element modulator |
| 18 | gene | 3.336416 | 3.366863 | positive | MGI_C57BL6J_3643252 | Gm6225 | NCBI_Gene:633947,ENSEMBL:ENSMUSG00000097746 | MGI:3643252 | lncRNA gene | predicted gene 6225 |
| 18 | gene | 3.382970 | 3.436700 | positive | MGI_C57BL6J_1918995 | Cul2 | NCBI_Gene:71745,ENSEMBL:ENSMUSG00000024231 | MGI:1918995 | protein coding gene | cullin 2 |
| 18 | pseudogene | 3.446654 | 3.447503 | positive | MGI_C57BL6J_3648491 | Gm6235 | NCBI_Gene:621501,ENSEMBL:ENSMUSG00000117370 | MGI:3648491 | pseudogene | predicted gene 6235 |
| 18 | pseudogene | 3.466371 | 3.466502 | negative | MGI_C57BL6J_6275423 | Gm50088 | ENSEMBL:ENSMUSG00000117389 | MGI:6275423 | pseudogene | predicted gene, 50088 |
| 18 | gene | 3.471630 | 3.474315 | positive | MGI_C57BL6J_3645649 | G430049J08Rik | ENSEMBL:ENSMUSG00000096528 | MGI:3645649 | unclassified gene | RIKEN cDNA G430049J08 gene |
| 18 | pseudogene | 3.484218 | 3.485193 | positive | MGI_C57BL6J_6275424 | Gm50089 | ENSEMBL:ENSMUSG00000117469 | MGI:6275424 | pseudogene | predicted gene, 50089 |
| 18 | gene | 3.507923 | 3.516404 | positive | MGI_C57BL6J_1915260 | Bambi | NCBI_Gene:68010,ENSEMBL:ENSMUSG00000024232 | MGI:1915260 | protein coding gene | BMP and activin membrane-bound inhibitor |
| 18 | pseudogene | 3.558675 | 3.560037 | positive | MGI_C57BL6J_6275425 | Gm50090 | ENSEMBL:ENSMUSG00000117454 | MGI:6275425 | pseudogene | predicted gene, 50090 |
| 18 | gene | 3.616281 | 3.618242 | positive | MGI_C57BL6J_6275426 | Gm50091 | ENSEMBL:ENSMUSG00000117506 | MGI:6275426 | unclassified gene | predicted gene, 50091 |
| 18 | gene | 3.770741 | 3.770804 | negative | MGI_C57BL6J_5453420 | Gm23643 | ENSEMBL:ENSMUSG00000088480 | MGI:5453420 | snRNA gene | predicted gene, 23643 |
| 18 | pseudogene | 3.857730 | 3.858585 | negative | MGI_C57BL6J_3647465 | Rpl7a-ps6 | NCBI_Gene:435549,ENSEMBL:ENSMUSG00000117463 | MGI:3647465 | pseudogene | ribosomal protein L7A, pseudogene 6 |
| 18 | gene | 3.860108 | 3.860430 | positive | MGI_C57BL6J_5454409 | Gm24632 | ENSEMBL:ENSMUSG00000084719 | MGI:5454409 | unclassified non-coding RNA gene | predicted gene, 24632 |
| 18 | pseudogene | 4.010143 | 4.011335 | positive | MGI_C57BL6J_3646622 | Gm7378 | NCBI_Gene:664867,ENSEMBL:ENSMUSG00000117395 | MGI:3646622 | pseudogene | predicted gene 7378 |
| 18 | pseudogene | 4.052351 | 4.052771 | negative | MGI_C57BL6J_3644899 | Gm6248 | NCBI_Gene:621666,ENSEMBL:ENSMUSG00000117565 | MGI:3644899 | pseudogene | predicted gene 6248 |
| 18 | gene | 4.165832 | 4.182236 | positive | MGI_C57BL6J_1914578 | Lyzl1 | NCBI_Gene:67328,ENSEMBL:ENSMUSG00000024233 | MGI:1914578 | protein coding gene | lysozyme-like 1 |
| 18 | pseudogene | 4.198320 | 4.199135 | negative | MGI_C57BL6J_3708638 | Gm10557 | NCBI_Gene:383374,ENSEMBL:ENSMUSG00000073647 | MGI:3708638 | pseudogene | predicted gene 10557 |
| 18 | gene | 4.244121 | 4.250779 | negative | MGI_C57BL6J_5624542 | Gm41657 | NCBI_Gene:105246358 | MGI:5624542 | lncRNA gene | predicted gene, 41657 |
| 18 | pseudogene | 4.293097 | 4.294538 | negative | MGI_C57BL6J_3647488 | Gm7400 | NCBI_Gene:664909,ENSEMBL:ENSMUSG00000117482 | MGI:3647488 | pseudogene | predicted gene 7400 |
| 18 | gene | 4.331325 | 4.353412 | negative | MGI_C57BL6J_1346878 | Map3k8 | NCBI_Gene:26410,ENSEMBL:ENSMUSG00000024235 | MGI:1346878 | protein coding gene | mitogen-activated protein kinase kinase kinase 8 |
| 18 | gene | 4.353547 | 4.368945 | positive | MGI_C57BL6J_1921165 | 4833419F23Rik | NCBI_Gene:73915,ENSEMBL:ENSMUSG00000117401 | MGI:1921165 | lncRNA gene | RIKEN cDNA 4833419F23 gene |
| 18 | gene | 4.353644 | 4.380723 | positive | MGI_C57BL6J_6275434 | Gm50096 | ENSEMBL:ENSMUSG00000117594 | MGI:6275434 | lncRNA gene | predicted gene, 50096 |
| 18 | gene | 4.373669 | 4.379985 | negative | MGI_C57BL6J_5477359 | Gm26865 | ENSEMBL:ENSMUSG00000097641 | MGI:5477359 | lncRNA gene | predicted gene, 26865 |
| 18 | gene | 4.375592 | 4.398798 | positive | MGI_C57BL6J_1914690 | Mtpap | NCBI_Gene:67440,ENSEMBL:ENSMUSG00000024234 | MGI:1914690 | protein coding gene | mitochondrial poly(A) polymerase |
| 18 | gene | 4.399328 | 4.400910 | positive | MGI_C57BL6J_6275321 | Gm50023 | ENSEMBL:ENSMUSG00000117579 | MGI:6275321 | lncRNA gene | predicted gene, 50023 |
| 18 | pseudogene | 4.484469 | 4.485501 | positive | MGI_C57BL6J_3644066 | Gm7411 | NCBI_Gene:664931,ENSEMBL:ENSMUSG00000117640 | MGI:3644066 | pseudogene | predicted gene 7411 |
| 18 | gene | 4.541612 | 4.562599 | negative | MGI_C57BL6J_5624543 | Gm41658 | NCBI_Gene:105246359 | MGI:5624543 | lncRNA gene | predicted gene, 41658 |
| 18 | gene | 4.590763 | 4.616064 | positive | MGI_C57BL6J_6275323 | Gm50024 | ENSEMBL:ENSMUSG00000117521 | MGI:6275323 | lncRNA gene | predicted gene, 50024 |
| 18 | gene | 4.590792 | 4.682869 | positive | MGI_C57BL6J_2685174 | Jcad | NCBI_Gene:240185,ENSEMBL:ENSMUSG00000033960 | MGI:2685174 | protein coding gene | junctional cadherin 5 associated |
| 18 | pseudogene | 4.743061 | 4.743876 | positive | MGI_C57BL6J_3645969 | Gm5047 | NCBI_Gene:268988,ENSEMBL:ENSMUSG00000117585 | MGI:3645969 | pseudogene | predicted gene 5047 |
| 18 | gene | 4.812486 | 4.850959 | positive | MGI_C57BL6J_3642809 | Gm10556 | NCBI_Gene:100038439,ENSEMBL:ENSMUSG00000097043 | MGI:3642809 | lncRNA gene | predicted gene 10556 |
| 18 | gene | 4.820303 | 4.826659 | negative | MGI_C57BL6J_5593428 | Gm34269 | NCBI_Gene:102637466 | MGI:5593428 | lncRNA gene | predicted gene, 34269 |
| 18 | gene | 4.869777 | 4.912263 | positive | MGI_C57BL6J_5624544 | Gm41659 | NCBI_Gene:105246360 | MGI:5624544 | lncRNA gene | predicted gene, 41659 |
| 18 | gene | 4.920245 | 5.119299 | positive | MGI_C57BL6J_2147319 | Svil | NCBI_Gene:225115,ENSEMBL:ENSMUSG00000024236 | MGI:2147319 | protein coding gene | supervillin |
| 18 | gene | 4.959636 | 4.960300 | positive | MGI_C57BL6J_3642287 | BC025933 | NA | NA | protein coding gene | cDNA sequence BC025933 |
| 18 | gene | 5.042818 | 5.048867 | negative | MGI_C57BL6J_5593485 | Gm34326 | NCBI_Gene:102637544 | MGI:5593485 | lncRNA gene | predicted gene, 34326 |
| 18 | gene | 5.139349 | 5.142068 | positive | MGI_C57BL6J_5593700 | Gm34541 | NCBI_Gene:102637831 | MGI:5593700 | lncRNA gene | predicted gene, 34541 |
| 18 | gene | 5.141762 | 5.165731 | negative | MGI_C57BL6J_5477176 | Gm26682 | NCBI_Gene:102637682,ENSEMBL:ENSMUSG00000097888 | MGI:5477176 | lncRNA gene | predicted gene, 26682 |
| 18 | gene | 5.210027 | 5.334963 | negative | MGI_C57BL6J_2444919 | Zfp438 | NCBI_Gene:240186,ENSEMBL:ENSMUSG00000050945 | MGI:2444919 | protein coding gene | zinc finger protein 438 |
| 18 | gene | 5.348258 | 5.361861 | negative | MGI_C57BL6J_6275385 | Gm50064 | ENSEMBL:ENSMUSG00000117520 | MGI:6275385 | lncRNA gene | predicted gene, 50064 |
| 18 | gene | 5.368541 | 5.370482 | negative | MGI_C57BL6J_6275387 | Gm50065 | ENSEMBL:ENSMUSG00000117617 | MGI:6275387 | lncRNA gene | predicted gene, 50065 |
| 18 | gene | 5.389802 | 5.402870 | negative | MGI_C57BL6J_5593789 | Gm34630 | NCBI_Gene:102637944 | MGI:5593789 | lncRNA gene | predicted gene, 34630 |
| 18 | gene | 5.491501 | 5.593505 | negative | MGI_C57BL6J_3642044 | Gm10125 | NCBI_Gene:791318,ENSEMBL:ENSMUSG00000063087 | MGI:3642044 | lncRNA gene | predicted gene 10125 |
| 18 | gene | 5.572824 | 5.575055 | negative | MGI_C57BL6J_5477069 | Gm26575 | ENSEMBL:ENSMUSG00000097926 | MGI:5477069 | lncRNA gene | predicted gene, 26575 |
R/qtl
scanone
gmObject of class cross2 (crosstype "bc")
Total individuals 138
No. genotyped individuals 138
No. phenotyped individuals 138
No. with both geno & pheno 138
No. phenotypes 1
No. covariates 7
No. phenotype covariates 0
No. chromosomes 20
Total markers 32690
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2952 2874 2058 2071 1955 2053 1862 1700 2009 1229 2069 1396 1628 1692 1064 940
17 18 19 X
401 1095 1067 575 #detach("package:qtl2", unload=TRUE)
#library(qtl)
cross <- qtl::read.cross("csv", file = "data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_mis_conditional_2-peaks-chr3-10.csv",alleles=c("A","B")) --Read the following data:
138 individuals
32690 markers
5 phenotypes
--Cross type: bc cross <- qtl::jittermap(cross)
summary(cross) Backcross
No. individuals: 138
No. phenotypes: 5
Percent phenotyped: 100 100 100 99.3 98.6
No. chromosomes: 20
Autosomes: 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19
X chr: X
Total markers: 32690
No. markers: 2952 2874 2058 2071 1955 2053 1862 1700 2009 1229 2069
1396 1628 1692 1064 940 401 1095 1067 575
Percent genotyped: 99.5
Genotypes (%):
Autosomes: AA:54.3 AB:45.7
X chromosome: AA:52.7 AB:47.3 cross.probs <- qtl::calc.genoprob(cross)
print("method == hk")[1] "method == hk"add.covars = qtl::pull.pheno(cross.probs, c("UNCHS008487","UNC18240977"))
scanone.hk <-qtl::scanone(cross.probs, pheno.col="ICI.vs.PBS" , model="binary", method="hk", addcovar = add.covars)
operm.hk <- qtl::scanone(cross.probs, method = "hk", pheno.col="ICI.vs.PBS", n.perm = 10, perm.Xsp = TRUE, model="binary", verbose=FALSE, addcovar = add.covars)
plot(operm.hk)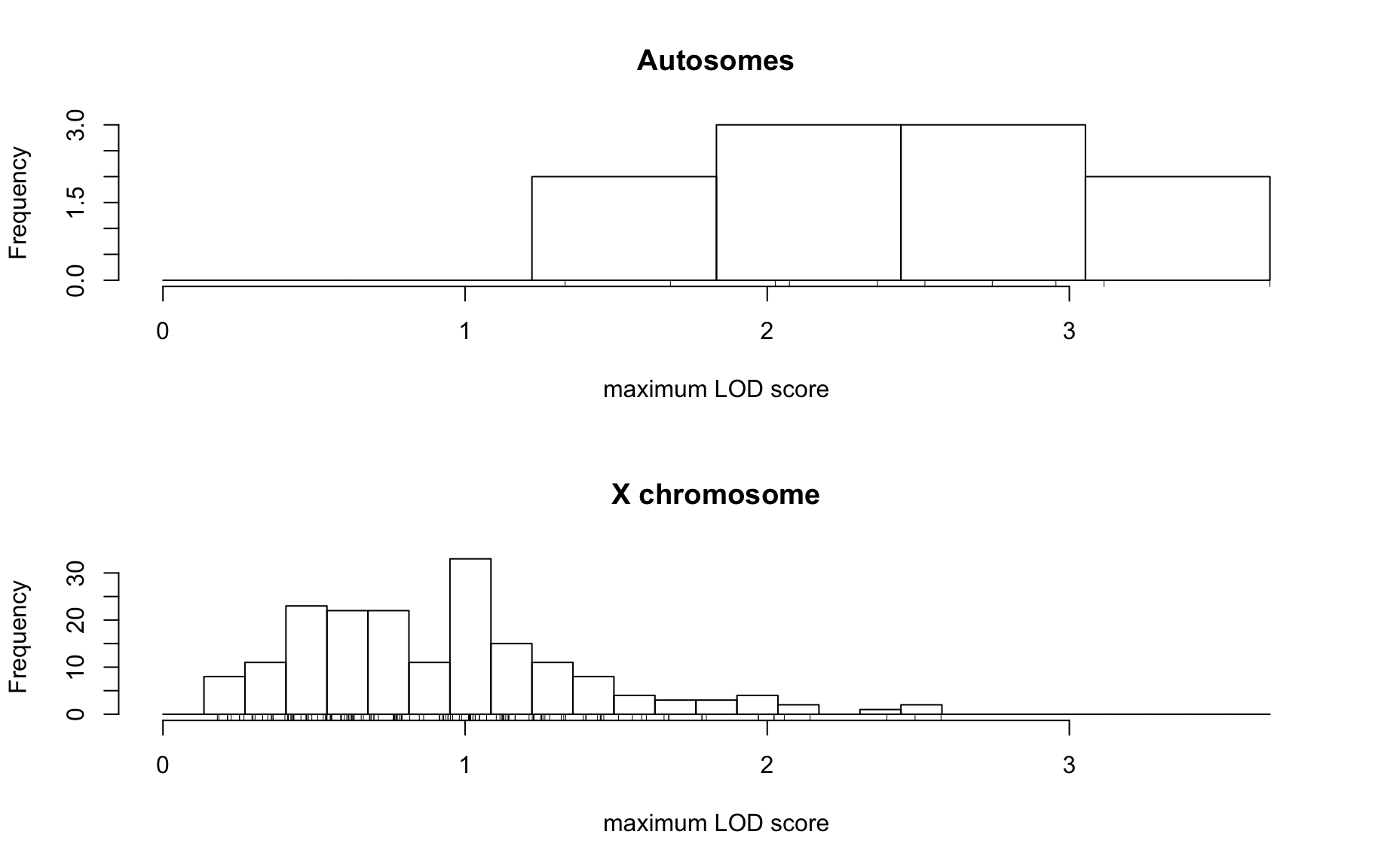
print(summary(operm.hk, alpha=c(0.01, 0.05, 0.1)))Autosome LOD thresholds (10 permutations)
lod
1% 3.62
5% 3.43
10% 3.19
X chromosome LOD thresholds (183 permutations)
lod
1% 2.57
5% 2.53
10% 2.49#plot(scanone.hk, bandcol = "grey90",lty=1, cex=1, col = "steelblue")
#qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.01, col = 'blue')
#qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.05, col = 'red')
#qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.1, col = 'purple')
ymx <- maxlod(out) # overall maximum LOD score
plot(scanone.hk, bandcol = "grey90",lty=1, cex=1, col = "slateblue", ylim=c(0, ymx+0.5))
title(main = paste0(colnames(out), " [positions in cM]"))
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.01, col = 'blue')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.05, col = 'red')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.1, col = 'purple')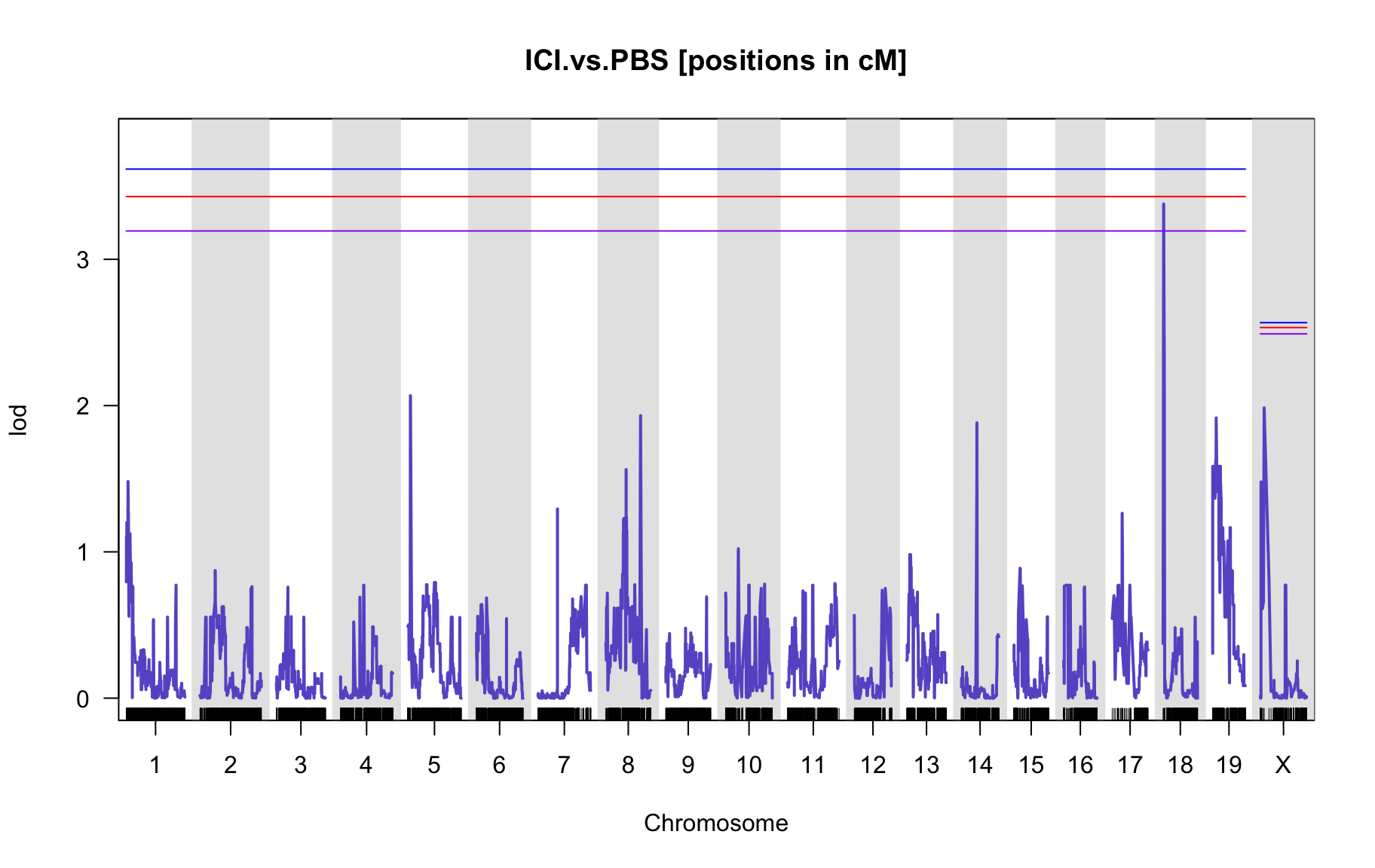
ymx <- 11
plot(scanone.hk, bandcol = "grey90",lty=1, cex=1, col = "slateblue", ylim=c(0, ymx+0.5))
title(main = paste0(colnames(out), " [positions in cM]\n(using same scale as eoi vs ici for easier comparison)"))
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.01, col = 'blue')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.05, col = 'red')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.1, col = 'purple')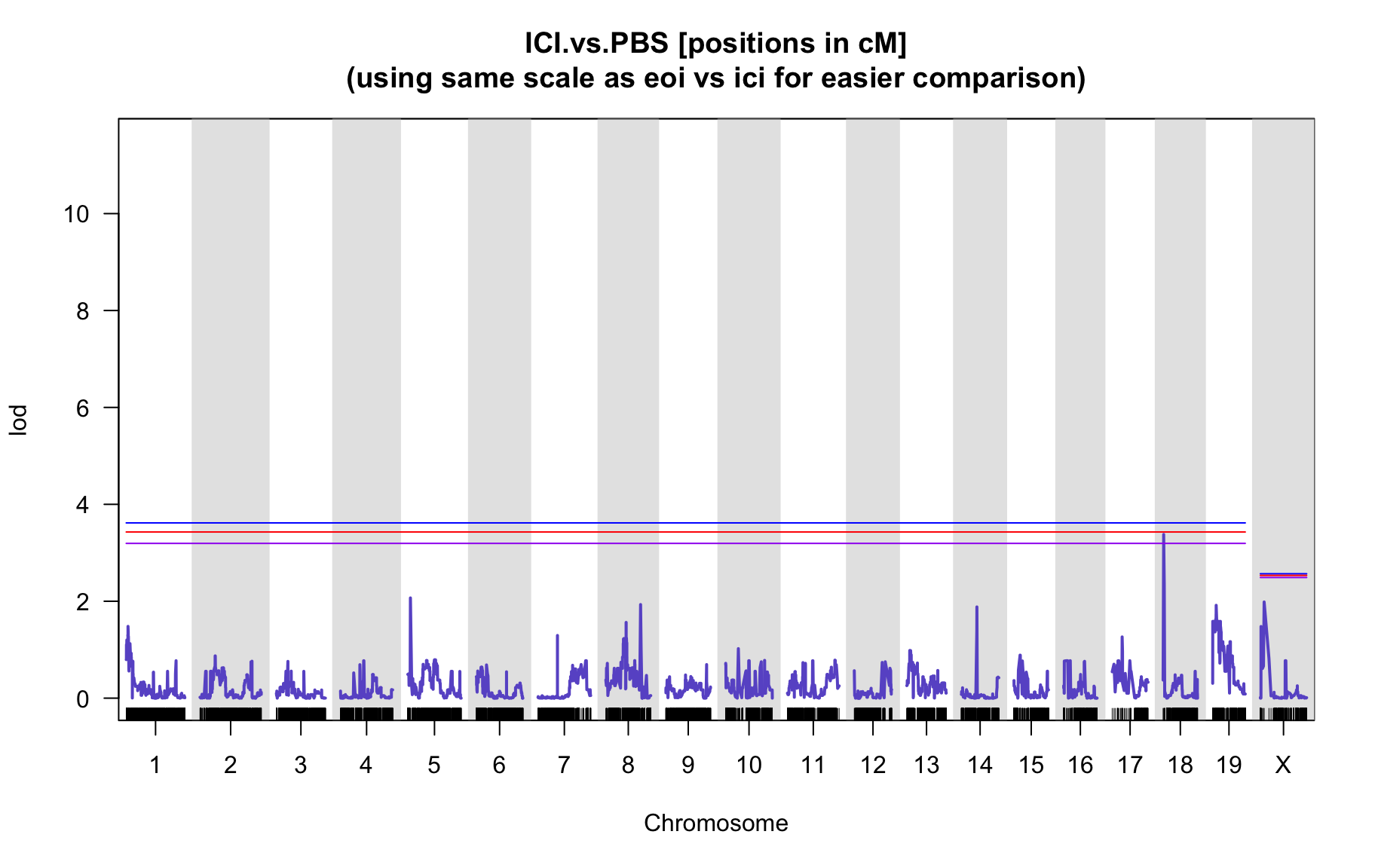
print(as.data.frame(summary(scanone.hk, perms=operm.hk, pvalues=TRUE, format="allpeaks"))) chr pos lod pval
UNCHS000141 1 4.439109 1.4810382 0.9118239
UNC2981117 2 27.613431 0.8724495 1.0000000
JAX00521745 3 20.613360 0.7586221 1.0000000
JAX00556948 4 41.038754 0.7726880 1.0000000
UNC8675939 5 6.193174 2.0684019 0.7191035
UNC10923091 6 18.321306 0.6856206 1.0000000
UNCHS020066 7 34.113657 1.2936681 1.0000000
UNC15524531 8 59.462590 1.9315437 0.8168395
UNC17203597 9 68.527929 0.6928610 1.0000000
UNCHS028001 10 21.783362 1.0219015 1.0000000
UNC20465557 11 80.904972 0.7841081 1.0000000
UNC21787084 12 53.288332 0.7505664 1.0000000
JAX00351755 13 7.044200 0.9809396 1.0000000
UNC24056202 14 30.156730 1.8824837 0.8168395
UNC25275535 15 11.967146 0.8880487 1.0000000
UNCHS042071 16 13.176096 0.7730967 1.0000000
UNCrs47191360 17 18.433013 1.2636005 1.0000000
UNCHS045343 18 2.934001 3.3781249 0.1051672
UNC29920552 19 8.810092 1.9159935 0.8168395
UNCHS048314 X 8.974041 1.9849011 0.5288989print("all peaks with a p-value less or equal to 0.05 (suggestive)")[1] "all peaks with a p-value less or equal to 0.05 (suggestive)"print(as.data.frame(summary(scanone.hk, perms=operm.hk, alpha=0.05, pvalues=TRUE, format="allpeaks")))[1] chr pos lod
<0 rows> (or 0-length row.names)#print("method == ehk")
#scanone.ehk <-qtl::scanone(cross.probs, pheno.col="ICI.vs.PBS" , model="binary", method="ehk")
#operm.ehk <- qtl::scanone(cross.probs, method = "ehk", pheno.col="ICI.vs.PBS", n.perm = 10, perm.Xsp = TRUE, model="binary", verbose=FALSE)
#plot(operm.ehk)
#print(summary(operm.ehk, alpha=c(0.01, 0.05, 0.1)))
#plot(scanone.ehk, bandcol = "grey90",lty=1, cex=1, col = "steelblue")
#qtl::add.threshold(scanone.ehk, perms= operm.ehk, alpha=0.01, col = 'blue')
#qtl::add.threshold(scanone.ehk, perms= operm.ehk, alpha=0.05, col = 'red')
#qtl::add.threshold(scanone.ehk, perms= operm.ehk, alpha=0.1, col = 'purple')
#print(as.data.frame(summary(scanone.ehk)))
#print(as.data.frame(summary(scanone.ehk, perms=operm.ehk, alpha=0.05, pvalues=TRUE, format="allpeaks")))Conditionaing on eoi vs. ici chr 4 peak (UNCHS008487) & chr 10 peak (UNC18240977)
Genome-wide scan
#gm
Xcovar <- get_x_covar(gm)
#addcovar = model.matrix(~Sex, data = covars)[,-1]
addcovar = model.matrix(~UNC8250659+UNC18240977, data = covars)[,-1]
#K <- calc_kinship(pr.qc, type = "loco")
#heatmap(K[[1]])
#K.overall <- calc_kinship(pr.qc, type = "overall")
#heatmap(K.overall)
kinship <- calc_kinship(pr.qc)
heatmap(kinship)
#operm <- scan1perm(pr.qc, gm$covar$phenos, Xcovar=Xcovar, n_perm=2000)
#operm <- scan1perm(pr.qc, gm$covar$phenos, addcovar = addcovar, n_perm=2000)
#operm <- scan1perm(pr.qc, gm$covar$phenos, n_perm=2000)
operm <- scan1perm(pr.qc, gm$covar["ICI.vs.PBS"], model="binary", n_perm=10, perm_Xsp=TRUE, chr_lengths=chr_lengths(gm$gmap), addcovar = addcovar)
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
names(summary_table) <- c("autosomes","X")
summary_table$significance.level <- rownames(summary_table)
rownames(summary_table) <- NULL
summary_table[c(3,1:2)] %>%
kable(escape = F,align = c("ccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| significance.level | autosomes | X |
|---|---|---|
| 0.01 | 3.572011 | 3.009770 |
| 0.05 | 3.301449 | 2.940770 |
| 0.1 | 2.962412 | 2.850537 |
The figures below show QTL maps for each phenotype
out <- scan1(pr.qc, gm$covar["ICI.vs.PBS"], Xcovar=Xcovar, model="binary", addcovar = addcovar)
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
plot_lod<-function(out,map){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- 11 # overall maximum LOD score
plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " [positions in cM] \n(using same scale as eoi vs ici for easier comparison)"))
add_threshold(map, summary(operm, alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
plot_lod(out,gm$gmap)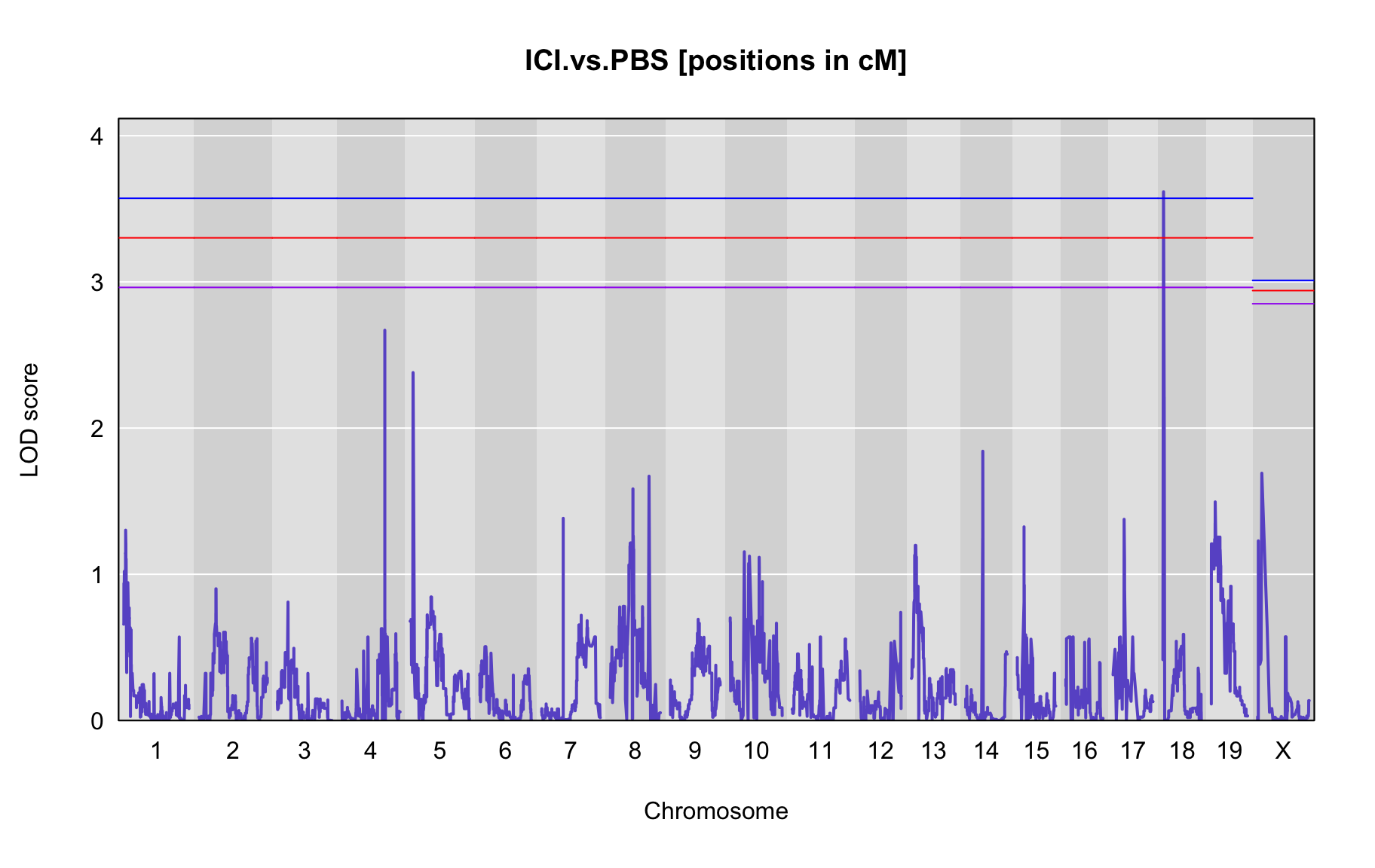
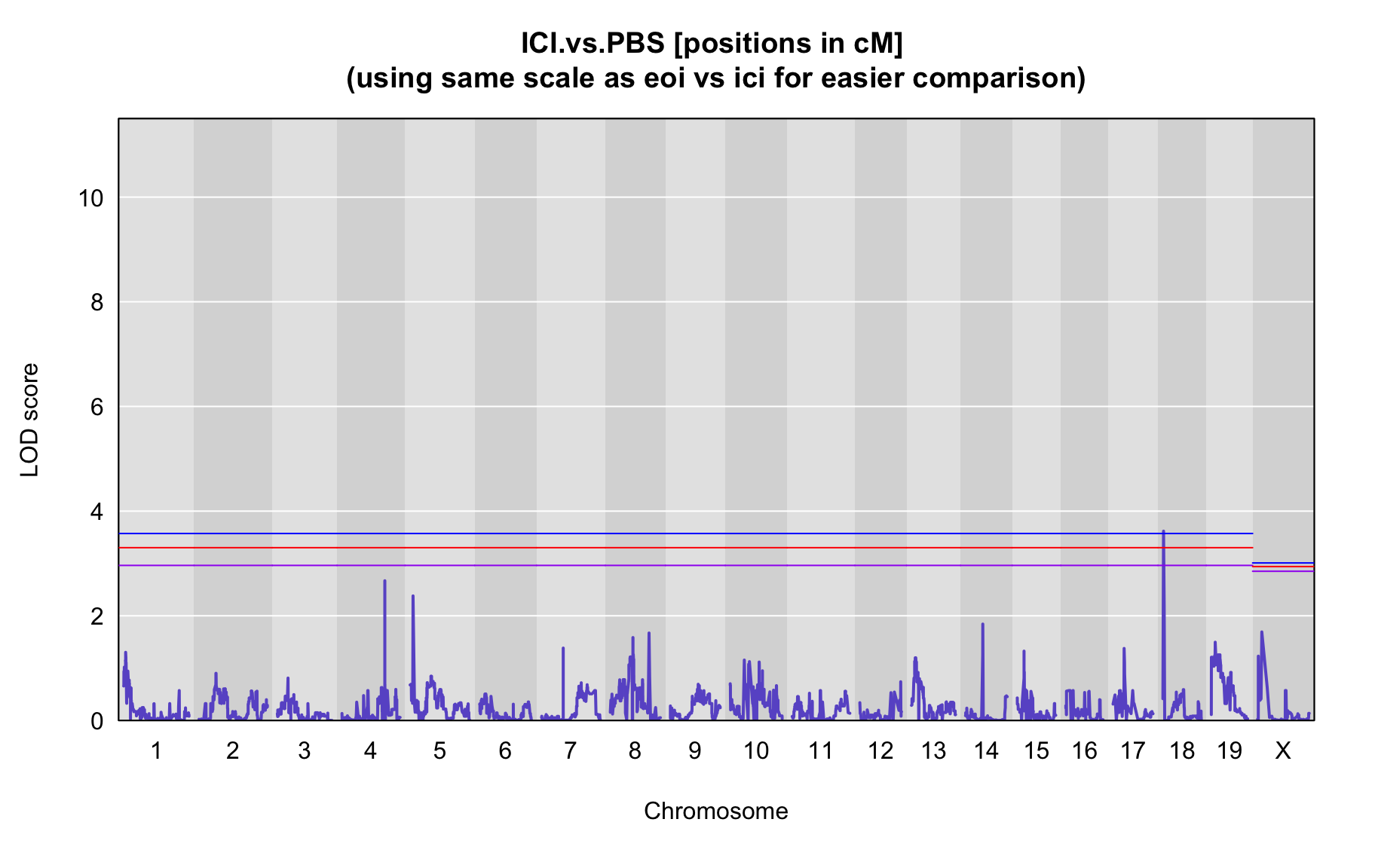
LOD peaks
The table below shows QTL peaks associated with the phenotype. We use the 95% threshold from the permutations to find peaks.
Centimorgan (cM)
peaks <- find_peaks(out, gm$gmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) >0){
peaks$marker <- find_marker(gm$gmap, chr=peaks$chr,pos=peaks$pos)
names(peaks)[2] <- c("phenotype")
peaks <- peaks[-1]
rownames(peaks) <- NULL
#print(kable(peaks, "html")
# %>% kable_styling("striped", full_width = T) %>%
# column_spec(1, bold=TRUE)
# )
print(kable(peaks, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
#peaks[] %>%
# kable(escape = F,align = c("cccccccc")) %>%
# kable_styling("striped", full_width = T) %>%
# column_spec(1, bold=TRUE)
#plot only peak chromosomes
plot_lod_chr<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
ymx <- 11
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in cM]\n(using same scale as eoi vs. ici for easier comparison)"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
for(i in unique(peaks$chr)){
#for (i in 1:nrow(peaks)){
#plot_lod_chr(out,gm$gmap, peaks$chr[i])
plot_lod_chr(out,gm$gmap, i)
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}| phenotype | chr | pos | lod | ci_lo | ci_hi | marker |
|---|---|---|---|---|---|---|
| ICI.vs.PBS | 18 | 2.934 | 3.616886 | 1.972 | 4.621 | UNCHS045343 |
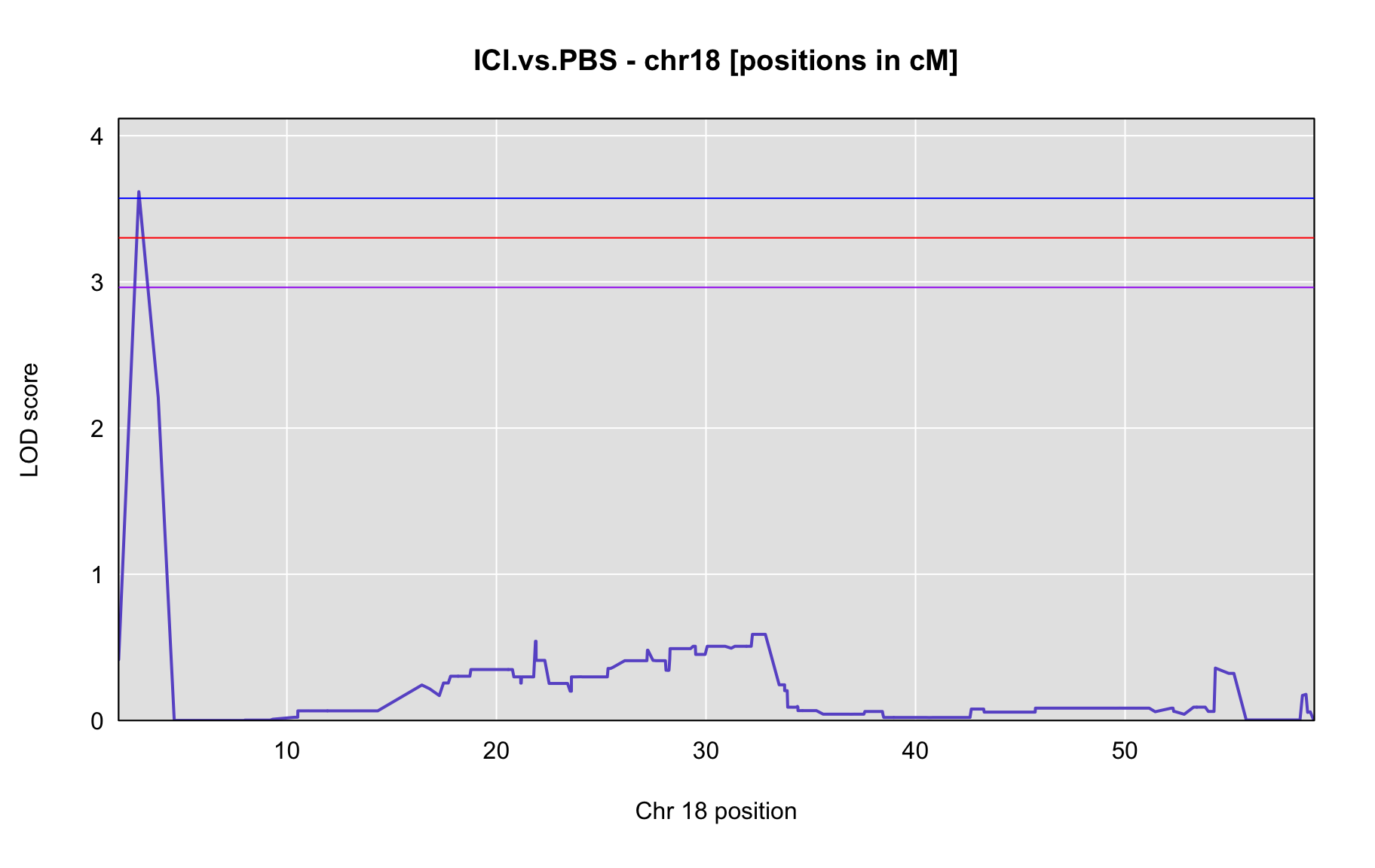
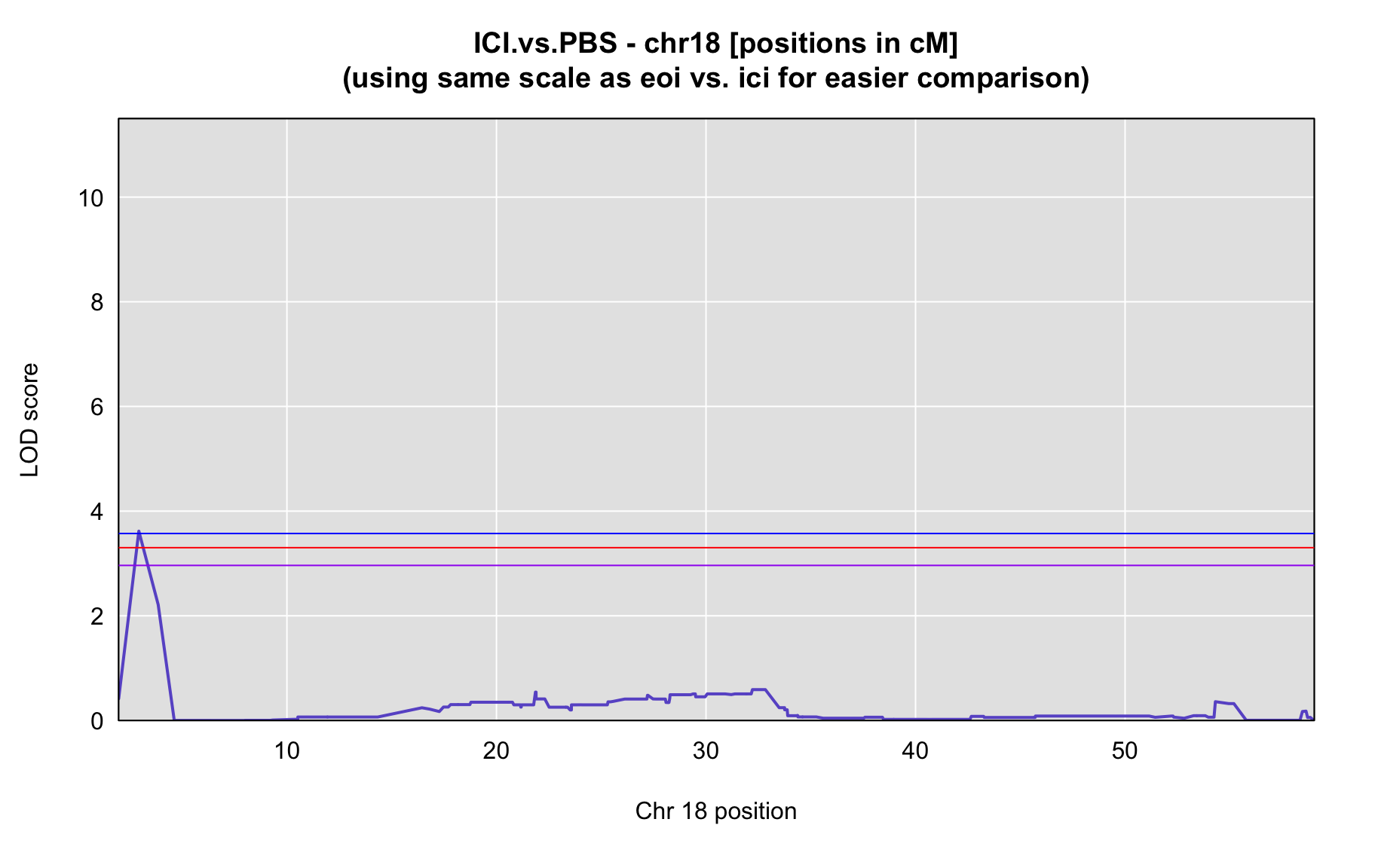
Megabase (MB)
print("peaks in MB positions")[1] “peaks in MB positions”
peaks_mba <- find_peaks(out, gm$pmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) >0){
peaks_mba$marker <- find_marker(gm$pmap, chr=peaks_mba$chr,pos=peaks_mba$pos)
names(peaks_mba)[2] <- c("phenotype")
peaks_mba <- peaks_mba[-1]
#peaks_mbl <- list()
##corresponding info in Mb
#for(i in 1:nrow(peaks)){
# #lodindex <- peaks$lodindex[i]
# phenotype <- peaks$phenotype[i]
# chr <- as.character(peaks$chr[i])
# lod <- peaks$lod[i]
# mark <- peaks$marker[i]
# pos <- mapdf[mapdf$marker==mark,]$pmapdf
# ci_lo <- mapdfnd$pmapdf[which(mapdfnd$gmapdf == peaks$ci_lo[i] & mapdfnd$chr == peaks$chr[i])]
# ci_hi <- mapdfnd$pmapdf[which(mapdfnd$gmapdf == peaks$ci_hi[i] & mapdfnd$chr == peaks$chr[i])]
# peaks_mb=as.data.frame(cbind(phenotype, chr, pos, lod, ci_lo, ci_hi, mark))
# names(peaks_mb)[7] <- c("marker")
# peaks_mbl[[i]] <- peaks_mb
#}
#peaks_mba2 <- do.call(rbind, peaks_mbl)
#peaks_mba2 <- as.data.frame(peaks_mba)
#peaks_mba[,c("chr", "pos", "lod", "ci_lo", "ci_hi")] <- sapply(peaks_mba[,c("chr", "pos", "lod", "ci_lo", "ci_hi")], as.numeric)
rownames(peaks_mba) <- NULL
#print(kable(peaks_mba, "html")
# %>% kable_styling("striped", full_width = T) %>%
# column_spec(1, bold=TRUE)
# )
print(kable(peaks_mba, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
#peaks_mba[] %>%
# kable(escape = F,align = c("cccccccc")) %>%
# kable_styling("striped", full_width = T) %>%
# column_spec(1, bold=TRUE)
plot_lod_chr_mb<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in MB]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
ymx <- 11
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in MB]\n(using same scale as eoi vs. ici for easier comparison)"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
for(i in unique(peaks_mba$chr)){
#for (i in 1:nrow(peaks_mba)){
#plot_lod_chr_mb(out,gm$pmap, peaks_mba$chr[i])
plot_lod_chr_mb(out,gm$pmap,i)
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}| phenotype | chr | pos | lod | ci_lo | ci_hi | marker |
|---|---|---|---|---|---|---|
| ICI.vs.PBS | 18 | 4.861199 | 3.616886 | 3.01212 | 7.97679 | UNCHS045343 |
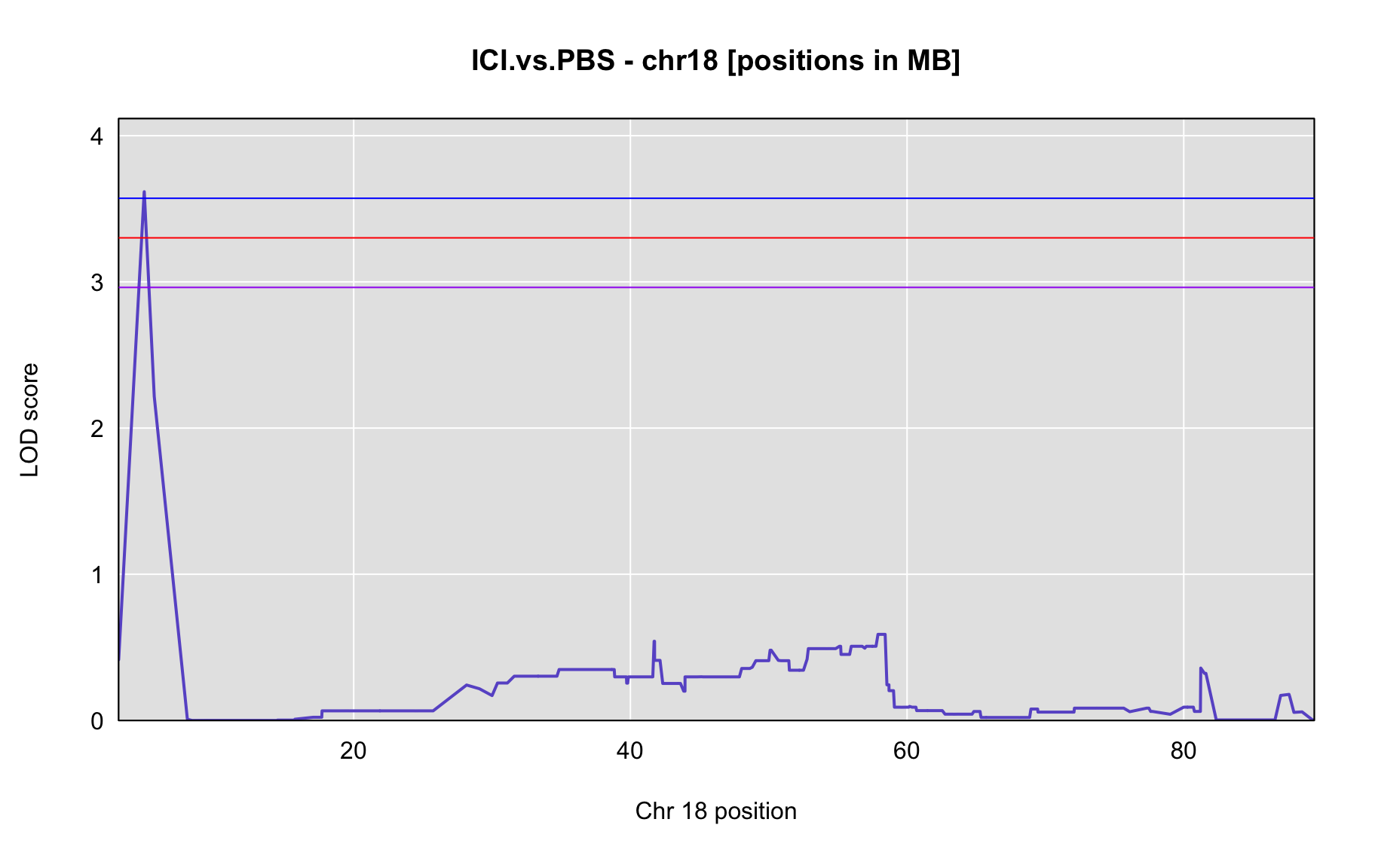
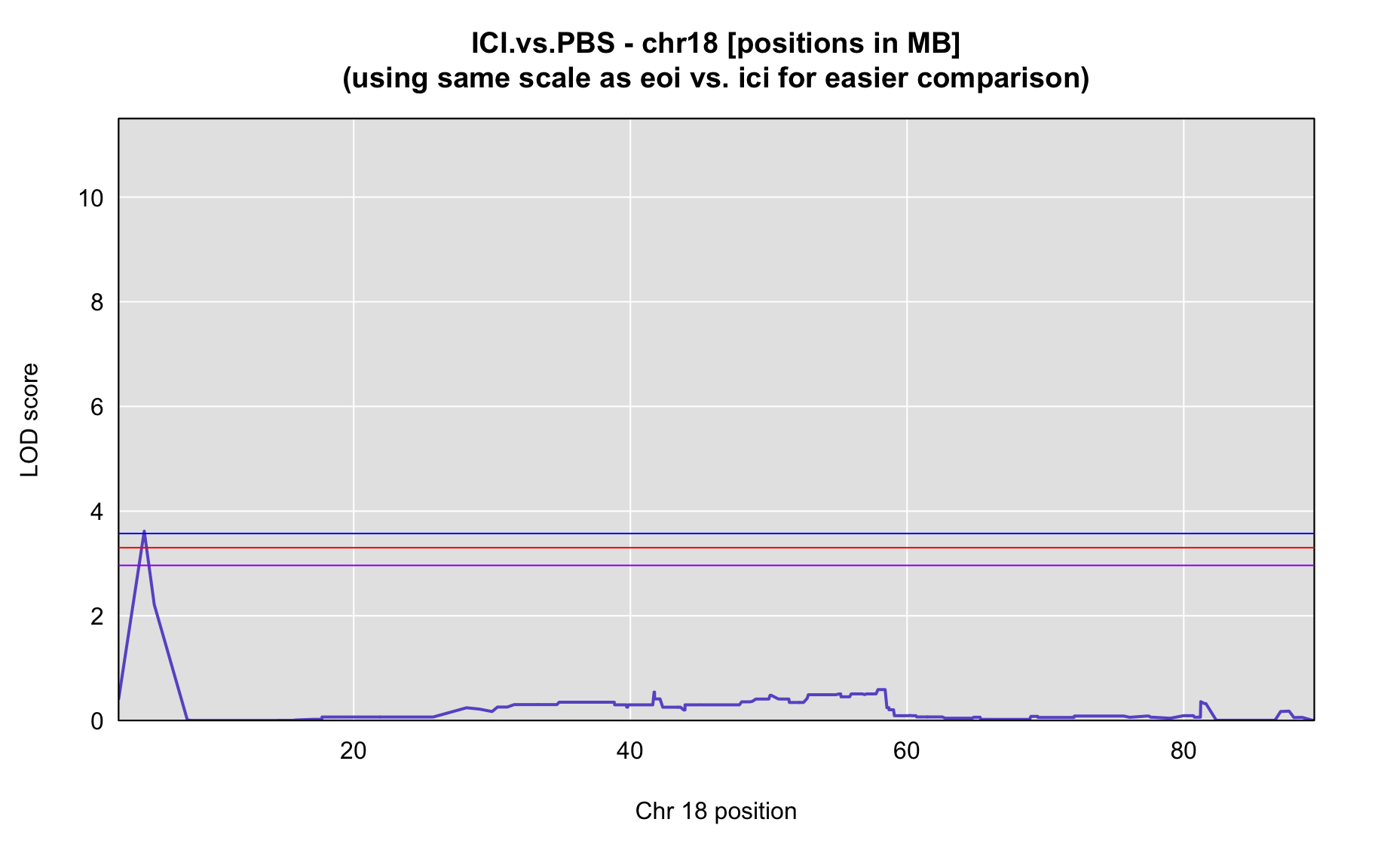
QTL effects
For each peak LOD location we give a list of gene
query_variants <- create_variant_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/cc_variants.sqlite")
query_genes <- create_gene_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/mouse_genes_mgi.sqlite")
if(nrow(peaks) >0){
for (i in 1:nrow(peaks)){
#for (i in 1:1){
#Plot 1
#marker = find_marker(gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i])
#gp <- g[,marker]
#gp[gp==1] <- "AA"
#gp[gp==2] <- "AB"
#gp[gp==0] <- NA
#plot_pxg(gp, gm$covar[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
#title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
###dev.off()
g <- maxmarg(pr.qc, gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i], return_char=TRUE)
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/","qtl_effect_", i, ".png"))
#par(mar=c(4.1, 4.1, 1.5, 0.6))
plot_pxg(g, gm$covar[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
##dev.off()
chr = peaks$chr[i]
# Plot 2
pr_sub <- pull_genoprobint(pr.qc, gm$gmap, chr, c(peaks$ci_lo[i], peaks$ci_hi[i]))
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar)
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], Xcovar=Xcovar)
#coeff <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
#coeff_sub <- scan1coef(pr_sub[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
blup <- scan1blup(pr.qc[,chr], gm$covar[peaks$phenotype[i]], addcovar = addcovar)
blup_sub <- scan1blup(pr_sub[,chr], gm$covar[peaks$phenotype[i]], addcovar = addcovar)
write.csv(as.data.frame(blup_sub), paste0("data/ici.vs.pbs_blup_sub_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_snpsqc_5.batches_mis_conditional_2-peaks-chr4-10.csv"), quote=F)
#plot_coef(coeff,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i]," [scan1coeff; positions in cM])")
# )
#plot_coef(coeff_sub,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i],"; 1.5 LOD drop interval [scan1coeff; positions in cM] ) ")
# )
plot_coef(blup,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," [scan1blup; positions in cM])")
)
plot_coef(blup_sub,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i],"; 1.5 LOD drop interval [scan1blup; positions in cM])")
)
# Plot 3
#c2effB <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary", contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
#c2effBb <- scan1blup(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]],Xcovar=Xcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
#plot(c2effB, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
#plot(c2effBb, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
##last_coef <- unclass(c2effB)[nrow(c2effB),2:3] # last two coefficients
##for(t in seq(along=last_coef))
## axis(side=4, at=last_coef[t], names(last_coef)[t], tick=FALSE)
#Table 1
chr = peaks_mba$chr[i]
start=as.numeric(peaks_mba$ci_lo[i])
end=as.numeric(peaks_mba$ci_hi[i])
genesgss = query_genes(chr, start, end)
write.csv(genesgss, file=paste0("data/ici.vs.pbs_genes_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_snpsqc_5.batches_mis_conditional_2-peaks-chr4-10.csv"), quote=F)
rownames(genesgss) <- NULL
genesgss$strand_old = genesgss$strand
genesgss$strand[genesgss$strand=="+"] <- "positive"
genesgss$strand[genesgss$strand=="-"] <- "negative"
#genesgss <-
#table <-
#genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")] %>%
#kable(escape = F,align = c("ccccccccccc")) %>%
#kable_styling("striped", full_width = T) #%>%
#cat #%>%
#column_spec(1, bold=TRUE)
#
#print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], escape = F,align = c("ccccccccccc")))
print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], "html") %>% kable_styling("striped", full_width = T))
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}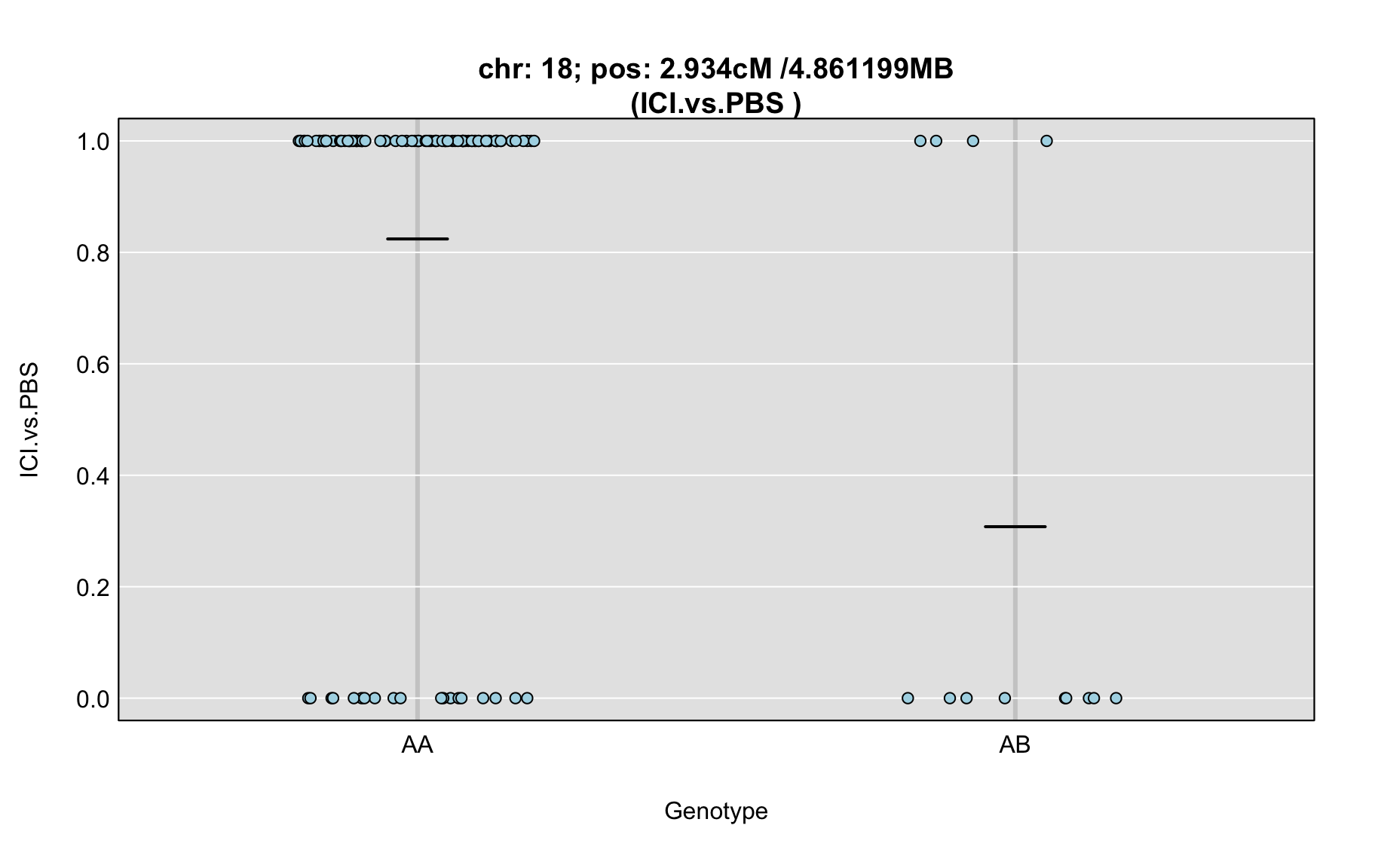
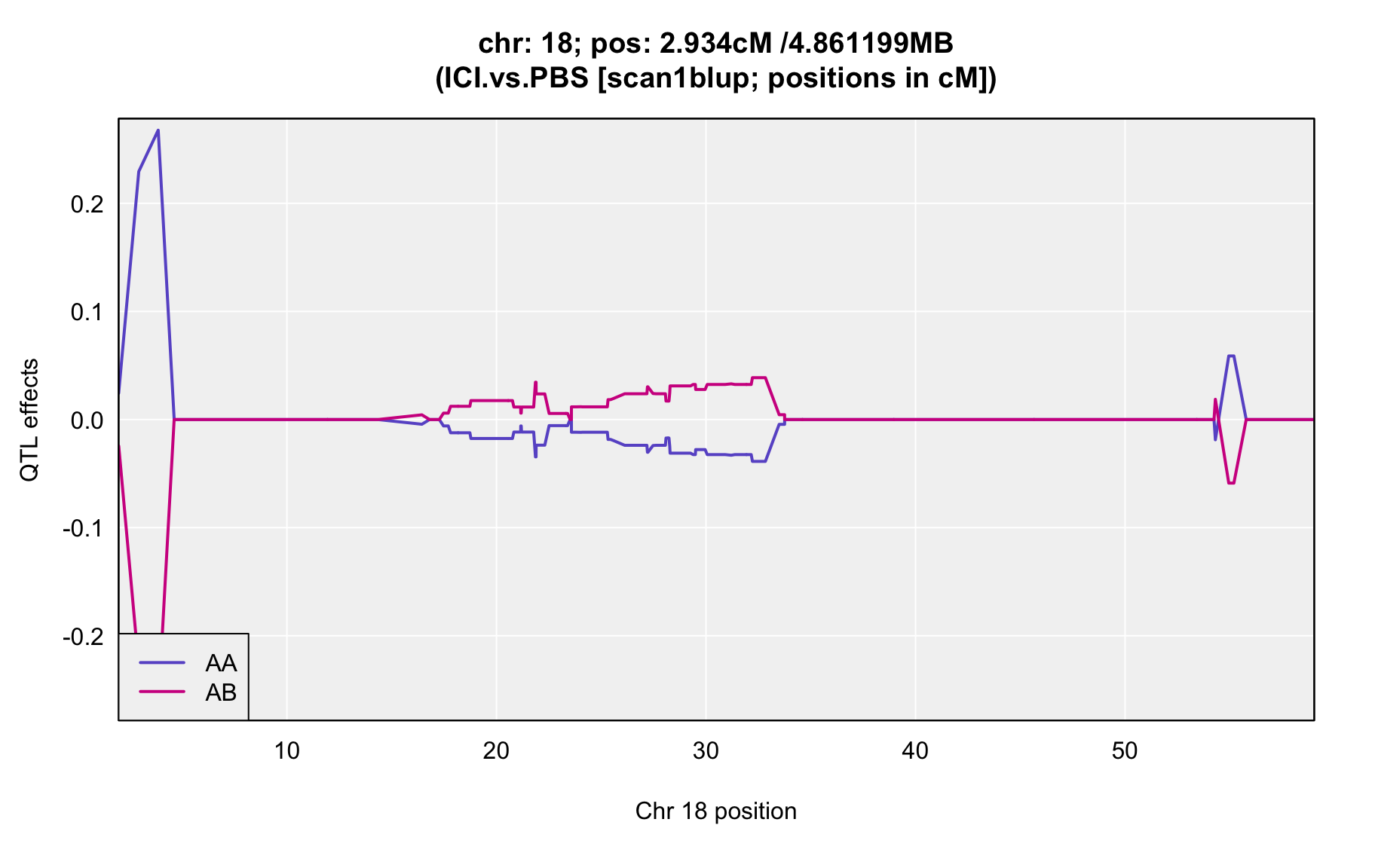
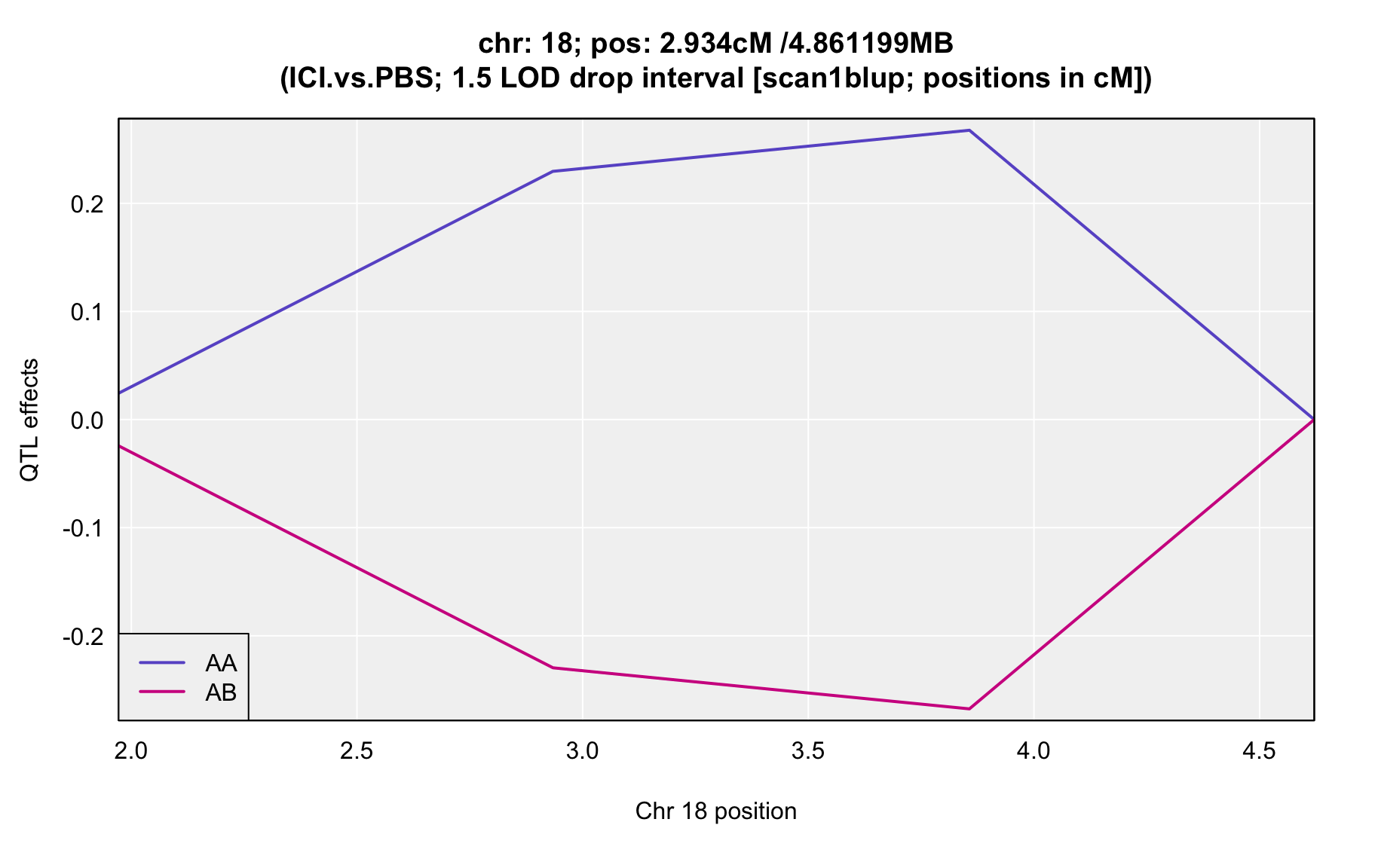
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 18 | pseudogene | 3.015908 | 3.016159 | positive | MGI_C57BL6J_6275399 | Gm50072 | ENSEMBL:ENSMUSG00000117547 | MGI:6275399 | pseudogene | predicted gene, 50072 |
| 18 | pseudogene | 3.026901 | 3.027882 | negative | MGI_C57BL6J_3852490 | Vmn1r-ps151 | NCBI_Gene:100312519,ENSEMBL:ENSMUSG00000093774 | MGI:3852490 | pseudogene | vomeronasal 1 receptor, pseudogene 151 |
| 18 | pseudogene | 3.080778 | 3.081476 | negative | MGI_C57BL6J_3852491 | Vmn1r-ps152 | NCBI_Gene:100312520,ENSEMBL:ENSMUSG00000093444 | MGI:3852491 | pseudogene | vomeronasal 1 receptor, pseudogene 152 |
| 18 | pseudogene | 3.105167 | 3.105396 | negative | MGI_C57BL6J_6275400 | Gm50073 | ENSEMBL:ENSMUSG00000117531 | MGI:6275400 | pseudogene | predicted gene, 50073 |
| 18 | gene | 3.122492 | 3.130012 | negative | MGI_C57BL6J_3852494 | Vmn1r238 | NCBI_Gene:100312476,ENSEMBL:ENSMUSG00000091539 | MGI:3852494 | protein coding gene | vomeronasal 1 receptor, 238 |
| 18 | pseudogene | 3.139032 | 3.139305 | negative | MGI_C57BL6J_6275401 | Gm50074 | ENSEMBL:ENSMUSG00000117353 | MGI:6275401 | pseudogene | predicted gene, 50074 |
| 18 | pseudogene | 3.171447 | 3.171932 | negative | MGI_C57BL6J_6275402 | Gm50075 | ENSEMBL:ENSMUSG00000117464 | MGI:6275402 | pseudogene | predicted gene, 50075 |
| 18 | gene | 3.266048 | 3.338176 | negative | MGI_C57BL6J_88495 | Crem | NCBI_Gene:12916,ENSEMBL:ENSMUSG00000063889 | MGI:88495 | protein coding gene | cAMP responsive element modulator |
| 18 | gene | 3.336416 | 3.366863 | positive | MGI_C57BL6J_3643252 | Gm6225 | NCBI_Gene:633947,ENSEMBL:ENSMUSG00000097746 | MGI:3643252 | lncRNA gene | predicted gene 6225 |
| 18 | gene | 3.382970 | 3.436700 | positive | MGI_C57BL6J_1918995 | Cul2 | NCBI_Gene:71745,ENSEMBL:ENSMUSG00000024231 | MGI:1918995 | protein coding gene | cullin 2 |
| 18 | pseudogene | 3.446654 | 3.447503 | positive | MGI_C57BL6J_3648491 | Gm6235 | NCBI_Gene:621501,ENSEMBL:ENSMUSG00000117370 | MGI:3648491 | pseudogene | predicted gene 6235 |
| 18 | pseudogene | 3.466371 | 3.466502 | negative | MGI_C57BL6J_6275423 | Gm50088 | ENSEMBL:ENSMUSG00000117389 | MGI:6275423 | pseudogene | predicted gene, 50088 |
| 18 | gene | 3.471630 | 3.474315 | positive | MGI_C57BL6J_3645649 | G430049J08Rik | ENSEMBL:ENSMUSG00000096528 | MGI:3645649 | unclassified gene | RIKEN cDNA G430049J08 gene |
| 18 | pseudogene | 3.484218 | 3.485193 | positive | MGI_C57BL6J_6275424 | Gm50089 | ENSEMBL:ENSMUSG00000117469 | MGI:6275424 | pseudogene | predicted gene, 50089 |
| 18 | gene | 3.507923 | 3.516404 | positive | MGI_C57BL6J_1915260 | Bambi | NCBI_Gene:68010,ENSEMBL:ENSMUSG00000024232 | MGI:1915260 | protein coding gene | BMP and activin membrane-bound inhibitor |
| 18 | pseudogene | 3.558675 | 3.560037 | positive | MGI_C57BL6J_6275425 | Gm50090 | ENSEMBL:ENSMUSG00000117454 | MGI:6275425 | pseudogene | predicted gene, 50090 |
| 18 | gene | 3.616281 | 3.618242 | positive | MGI_C57BL6J_6275426 | Gm50091 | ENSEMBL:ENSMUSG00000117506 | MGI:6275426 | unclassified gene | predicted gene, 50091 |
| 18 | gene | 3.770741 | 3.770804 | negative | MGI_C57BL6J_5453420 | Gm23643 | ENSEMBL:ENSMUSG00000088480 | MGI:5453420 | snRNA gene | predicted gene, 23643 |
| 18 | pseudogene | 3.857730 | 3.858585 | negative | MGI_C57BL6J_3647465 | Rpl7a-ps6 | NCBI_Gene:435549,ENSEMBL:ENSMUSG00000117463 | MGI:3647465 | pseudogene | ribosomal protein L7A, pseudogene 6 |
| 18 | gene | 3.860108 | 3.860430 | positive | MGI_C57BL6J_5454409 | Gm24632 | ENSEMBL:ENSMUSG00000084719 | MGI:5454409 | unclassified non-coding RNA gene | predicted gene, 24632 |
| 18 | pseudogene | 4.010143 | 4.011335 | positive | MGI_C57BL6J_3646622 | Gm7378 | NCBI_Gene:664867,ENSEMBL:ENSMUSG00000117395 | MGI:3646622 | pseudogene | predicted gene 7378 |
| 18 | pseudogene | 4.052351 | 4.052771 | negative | MGI_C57BL6J_3644899 | Gm6248 | NCBI_Gene:621666,ENSEMBL:ENSMUSG00000117565 | MGI:3644899 | pseudogene | predicted gene 6248 |
| 18 | gene | 4.165832 | 4.182236 | positive | MGI_C57BL6J_1914578 | Lyzl1 | NCBI_Gene:67328,ENSEMBL:ENSMUSG00000024233 | MGI:1914578 | protein coding gene | lysozyme-like 1 |
| 18 | pseudogene | 4.198320 | 4.199135 | negative | MGI_C57BL6J_3708638 | Gm10557 | NCBI_Gene:383374,ENSEMBL:ENSMUSG00000073647 | MGI:3708638 | pseudogene | predicted gene 10557 |
| 18 | gene | 4.244121 | 4.250779 | negative | MGI_C57BL6J_5624542 | Gm41657 | NCBI_Gene:105246358 | MGI:5624542 | lncRNA gene | predicted gene, 41657 |
| 18 | pseudogene | 4.293097 | 4.294538 | negative | MGI_C57BL6J_3647488 | Gm7400 | NCBI_Gene:664909,ENSEMBL:ENSMUSG00000117482 | MGI:3647488 | pseudogene | predicted gene 7400 |
| 18 | gene | 4.331325 | 4.353412 | negative | MGI_C57BL6J_1346878 | Map3k8 | NCBI_Gene:26410,ENSEMBL:ENSMUSG00000024235 | MGI:1346878 | protein coding gene | mitogen-activated protein kinase kinase kinase 8 |
| 18 | gene | 4.353547 | 4.368945 | positive | MGI_C57BL6J_1921165 | 4833419F23Rik | NCBI_Gene:73915,ENSEMBL:ENSMUSG00000117401 | MGI:1921165 | lncRNA gene | RIKEN cDNA 4833419F23 gene |
| 18 | gene | 4.353644 | 4.380723 | positive | MGI_C57BL6J_6275434 | Gm50096 | ENSEMBL:ENSMUSG00000117594 | MGI:6275434 | lncRNA gene | predicted gene, 50096 |
| 18 | gene | 4.373669 | 4.379985 | negative | MGI_C57BL6J_5477359 | Gm26865 | ENSEMBL:ENSMUSG00000097641 | MGI:5477359 | lncRNA gene | predicted gene, 26865 |
| 18 | gene | 4.375592 | 4.398798 | positive | MGI_C57BL6J_1914690 | Mtpap | NCBI_Gene:67440,ENSEMBL:ENSMUSG00000024234 | MGI:1914690 | protein coding gene | mitochondrial poly(A) polymerase |
| 18 | gene | 4.399328 | 4.400910 | positive | MGI_C57BL6J_6275321 | Gm50023 | ENSEMBL:ENSMUSG00000117579 | MGI:6275321 | lncRNA gene | predicted gene, 50023 |
| 18 | pseudogene | 4.484469 | 4.485501 | positive | MGI_C57BL6J_3644066 | Gm7411 | NCBI_Gene:664931,ENSEMBL:ENSMUSG00000117640 | MGI:3644066 | pseudogene | predicted gene 7411 |
| 18 | gene | 4.541612 | 4.562599 | negative | MGI_C57BL6J_5624543 | Gm41658 | NCBI_Gene:105246359 | MGI:5624543 | lncRNA gene | predicted gene, 41658 |
| 18 | gene | 4.590763 | 4.616064 | positive | MGI_C57BL6J_6275323 | Gm50024 | ENSEMBL:ENSMUSG00000117521 | MGI:6275323 | lncRNA gene | predicted gene, 50024 |
| 18 | gene | 4.590792 | 4.682869 | positive | MGI_C57BL6J_2685174 | Jcad | NCBI_Gene:240185,ENSEMBL:ENSMUSG00000033960 | MGI:2685174 | protein coding gene | junctional cadherin 5 associated |
| 18 | pseudogene | 4.743061 | 4.743876 | positive | MGI_C57BL6J_3645969 | Gm5047 | NCBI_Gene:268988,ENSEMBL:ENSMUSG00000117585 | MGI:3645969 | pseudogene | predicted gene 5047 |
| 18 | gene | 4.812486 | 4.850959 | positive | MGI_C57BL6J_3642809 | Gm10556 | NCBI_Gene:100038439,ENSEMBL:ENSMUSG00000097043 | MGI:3642809 | lncRNA gene | predicted gene 10556 |
| 18 | gene | 4.820303 | 4.826659 | negative | MGI_C57BL6J_5593428 | Gm34269 | NCBI_Gene:102637466 | MGI:5593428 | lncRNA gene | predicted gene, 34269 |
| 18 | gene | 4.869777 | 4.912263 | positive | MGI_C57BL6J_5624544 | Gm41659 | NCBI_Gene:105246360 | MGI:5624544 | lncRNA gene | predicted gene, 41659 |
| 18 | gene | 4.920245 | 5.119299 | positive | MGI_C57BL6J_2147319 | Svil | NCBI_Gene:225115,ENSEMBL:ENSMUSG00000024236 | MGI:2147319 | protein coding gene | supervillin |
| 18 | gene | 4.959636 | 4.960300 | positive | MGI_C57BL6J_3642287 | BC025933 | NA | NA | protein coding gene | cDNA sequence BC025933 |
| 18 | gene | 5.042818 | 5.048867 | negative | MGI_C57BL6J_5593485 | Gm34326 | NCBI_Gene:102637544 | MGI:5593485 | lncRNA gene | predicted gene, 34326 |
| 18 | gene | 5.139349 | 5.142068 | positive | MGI_C57BL6J_5593700 | Gm34541 | NCBI_Gene:102637831 | MGI:5593700 | lncRNA gene | predicted gene, 34541 |
| 18 | gene | 5.141762 | 5.165731 | negative | MGI_C57BL6J_5477176 | Gm26682 | NCBI_Gene:102637682,ENSEMBL:ENSMUSG00000097888 | MGI:5477176 | lncRNA gene | predicted gene, 26682 |
| 18 | gene | 5.210027 | 5.334963 | negative | MGI_C57BL6J_2444919 | Zfp438 | NCBI_Gene:240186,ENSEMBL:ENSMUSG00000050945 | MGI:2444919 | protein coding gene | zinc finger protein 438 |
| 18 | gene | 5.348258 | 5.361861 | negative | MGI_C57BL6J_6275385 | Gm50064 | ENSEMBL:ENSMUSG00000117520 | MGI:6275385 | lncRNA gene | predicted gene, 50064 |
| 18 | gene | 5.368541 | 5.370482 | negative | MGI_C57BL6J_6275387 | Gm50065 | ENSEMBL:ENSMUSG00000117617 | MGI:6275387 | lncRNA gene | predicted gene, 50065 |
| 18 | gene | 5.389802 | 5.402870 | negative | MGI_C57BL6J_5593789 | Gm34630 | NCBI_Gene:102637944 | MGI:5593789 | lncRNA gene | predicted gene, 34630 |
| 18 | gene | 5.491501 | 5.593505 | negative | MGI_C57BL6J_3642044 | Gm10125 | NCBI_Gene:791318,ENSEMBL:ENSMUSG00000063087 | MGI:3642044 | lncRNA gene | predicted gene 10125 |
| 18 | gene | 5.572824 | 5.575055 | negative | MGI_C57BL6J_5477069 | Gm26575 | ENSEMBL:ENSMUSG00000097926 | MGI:5477069 | lncRNA gene | predicted gene, 26575 |
| 18 | gene | 5.588043 | 5.591711 | negative | MGI_C57BL6J_3780408 | Gm2238 | ENSEMBL:ENSMUSG00000103138 | MGI:3780408 | unclassified gene | predicted gene 2238 |
| 18 | gene | 5.591860 | 5.775468 | positive | MGI_C57BL6J_1344313 | Zeb1 | NCBI_Gene:21417,ENSEMBL:ENSMUSG00000024238 | MGI:1344313 | protein coding gene | zinc finger E-box binding homeobox 1 |
| 18 | pseudogene | 5.611558 | 5.611930 | negative | MGI_C57BL6J_3801802 | Gm16147 | ENSEMBL:ENSMUSG00000090078 | MGI:3801802 | pseudogene | predicted gene 16147 |
| 18 | gene | 5.614473 | 5.614583 | positive | MGI_C57BL6J_5452486 | Gm22709 | ENSEMBL:ENSMUSG00000080543 | MGI:5452486 | miRNA gene | predicted gene, 22709 |
| 18 | gene | 5.885791 | 5.889876 | negative | MGI_C57BL6J_5593899 | Gm34740 | NCBI_Gene:102638095 | MGI:5593899 | lncRNA gene | predicted gene, 34740 |
| 18 | gene | 5.945183 | 5.966908 | positive | MGI_C57BL6J_5593963 | Gm34804 | NCBI_Gene:102638178,ENSEMBL:ENSMUSG00000117508 | MGI:5593963 | lncRNA gene | predicted gene, 34804 |
| 18 | gene | 6.004578 | 6.015278 | positive | MGI_C57BL6J_5624548 | Gm41663 | NCBI_Gene:105246364 | MGI:5624548 | lncRNA gene | predicted gene, 41663 |
| 18 | gene | 6.014253 | 6.024013 | negative | MGI_C57BL6J_5624547 | Gm41662 | NCBI_Gene:105246363,ENSEMBL:ENSMUSG00000117548 | MGI:5624547 | lncRNA gene | predicted gene, 41662 |
| 18 | gene | 6.024427 | 6.136098 | negative | MGI_C57BL6J_1922665 | Arhgap12 | NCBI_Gene:75415,ENSEMBL:ENSMUSG00000041225 | MGI:1922665 | protein coding gene | Rho GTPase activating protein 12 |
| 18 | pseudogene | 6.029179 | 6.030456 | negative | MGI_C57BL6J_5011425 | Gm19240 | NCBI_Gene:100418482 | MGI:5011425 | pseudogene | predicted gene, 19240 |
| 18 | gene | 6.201002 | 6.242174 | negative | MGI_C57BL6J_1098268 | Kif5b | NCBI_Gene:16573,ENSEMBL:ENSMUSG00000006740 | MGI:1098268 | protein coding gene | kinesin family member 5B |
| 18 | gene | 6.213913 | 6.227324 | positive | MGI_C57BL6J_4937863 | Gm17036 | ENSEMBL:ENSMUSG00000091165 | MGI:4937863 | lncRNA gene | predicted gene 17036 |
| 18 | gene | 6.234853 | 6.326693 | positive | MGI_C57BL6J_5624549 | Gm41664 | NCBI_Gene:105246365,ENSEMBL:ENSMUSG00000117519 | MGI:5624549 | lncRNA gene | predicted gene, 41664 |
| 18 | pseudogene | 6.332591 | 6.333082 | positive | MGI_C57BL6J_3646174 | Rpl27-ps3 | NCBI_Gene:621100,ENSEMBL:ENSMUSG00000073640 | MGI:3646174 | pseudogene | ribosomal protein L27, pseudogene 3 |
| 18 | pseudogene | 6.368360 | 6.368797 | positive | MGI_C57BL6J_6275267 | Gm49986 | ENSEMBL:ENSMUSG00000117387 | MGI:6275267 | pseudogene | predicted gene, 49986 |
| 18 | pseudogene | 6.371352 | 6.371660 | negative | MGI_C57BL6J_3648803 | Gm6291 | NCBI_Gene:102637200,ENSEMBL:ENSMUSG00000095614 | MGI:3648803 | pseudogene | predicted gene 6291 |
| 18 | gene | 6.435951 | 6.516108 | negative | MGI_C57BL6J_1278322 | Epc1 | NCBI_Gene:13831,ENSEMBL:ENSMUSG00000024240 | MGI:1278322 | protein coding gene | enhancer of polycomb homolog 1 |
| 18 | gene | 6.490143 | 6.496962 | positive | MGI_C57BL6J_5579235 | Gm28529 | ENSEMBL:ENSMUSG00000101320 | MGI:5579235 | lncRNA gene | predicted gene 28529 |
| 18 | gene | 6.490564 | 6.490646 | negative | MGI_C57BL6J_3811424 | Mir1893 | miRBase:MI0008327,NCBI_Gene:100316773,ENSEMBL:ENSMUSG00000084514 | MGI:3811424 | miRNA gene | microRNA 1893 |
| 18 | pseudogene | 6.557130 | 6.557796 | positive | MGI_C57BL6J_3645335 | Gm7464 | NCBI_Gene:665047,ENSEMBL:ENSMUSG00000117425 | MGI:3645335 | pseudogene | predicted gene 7464 |
| 18 | gene | 6.603629 | 6.640435 | positive | MGI_C57BL6J_1918151 | 4921524L21Rik | NCBI_Gene:70901,ENSEMBL:ENSMUSG00000039540 | MGI:1918151 | protein coding gene | RIKEN cDNA 4921524L21 gene |
| 18 | pseudogene | 6.687843 | 6.691317 | negative | MGI_C57BL6J_6275405 | Gm50077 | ENSEMBL:ENSMUSG00000117620 | MGI:6275405 | pseudogene | predicted gene, 50077 |
| 18 | gene | 6.733905 | 6.794429 | positive | MGI_C57BL6J_102790 | Rab18 | NCBI_Gene:19330,ENSEMBL:ENSMUSG00000073639 | MGI:102790 | protein coding gene | RAB18, member RAS oncogene family |
| 18 | pseudogene | 6.760073 | 6.760959 | negative | MGI_C57BL6J_3648356 | Gm7466 | NCBI_Gene:665053,ENSEMBL:ENSMUSG00000117359 | MGI:3648356 | pseudogene | predicted gene 7466 |
| 18 | gene | 6.788768 | 6.792137 | positive | MGI_C57BL6J_6275412 | Gm50081 | ENSEMBL:ENSMUSG00000117461 | MGI:6275412 | lncRNA gene | predicted gene, 50081 |
| 18 | gene | 6.910459 | 6.936621 | negative | MGI_C57BL6J_3826781 | Gm2350 | ENSEMBL:ENSMUSG00000117652 | MGI:3826781 | lncRNA gene | predicted gene 2350 |
| 18 | gene | 6.934518 | 7.004780 | negative | MGI_C57BL6J_2687286 | Mkx | NCBI_Gene:210719,ENSEMBL:ENSMUSG00000061013 | MGI:2687286 | protein coding gene | mohawk homeobox |
| 18 | gene | 7.004963 | 7.066322 | positive | MGI_C57BL6J_5826270 | Gm46633 | NCBI_Gene:108168391,ENSEMBL:ENSMUSG00000117356 | MGI:5826270 | lncRNA gene | predicted gene, 46633 |
| 18 | pseudogene | 7.041939 | 7.044195 | negative | MGI_C57BL6J_3642728 | Gm10350 | NCBI_Gene:664778,ENSEMBL:ENSMUSG00000117381 | MGI:3642728 | pseudogene | predicted gene 10350 |
| 18 | gene | 7.088209 | 7.298304 | negative | MGI_C57BL6J_1922184 | Armc4 | NCBI_Gene:74934,ENSEMBL:ENSMUSG00000061802 | MGI:1922184 | protein coding gene | armadillo repeat containing 4 |
| 18 | pseudogene | 7.107181 | 7.108183 | negative | MGI_C57BL6J_5010766 | Gm18581 | NCBI_Gene:100417382,ENSEMBL:ENSMUSG00000117360 | MGI:5010766 | pseudogene | predicted gene, 18581 |
| 18 | gene | 7.150344 | 7.151777 | negative | MGI_C57BL6J_1915234 | 4930415O11Rik | ENSEMBL:ENSMUSG00000117383 | MGI:1915234 | unclassified gene | RIKEN cDNA 4930415O11 gene |
| 18 | gene | 7.332445 | 7.346299 | negative | MGI_C57BL6J_5594138 | Gm34979 | NCBI_Gene:102638402 | MGI:5594138 | lncRNA gene | predicted gene, 34979 |
| 18 | gene | 7.347959 | 7.626893 | negative | MGI_C57BL6J_1922989 | Mpp7 | NCBI_Gene:75739,ENSEMBL:ENSMUSG00000057440 | MGI:1922989 | protein coding gene | membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| 18 | pseudogene | 7.548932 | 7.549781 | positive | MGI_C57BL6J_5011133 | Gm18948 | NCBI_Gene:100418013,ENSEMBL:ENSMUSG00000117556 | MGI:5011133 | pseudogene | predicted gene, 18948 |
| 18 | pseudogene | 7.581482 | 7.582130 | positive | MGI_C57BL6J_3644282 | Fabp5l2 | NCBI_Gene:622384,ENSEMBL:ENSMUSG00000094334 | MGI:3644282 | pseudogene | fatty acid binding protein 5-like 2 |
| 18 | pseudogene | 7.684076 | 7.693411 | negative | MGI_C57BL6J_5010948 | Gm18763 | NCBI_Gene:100417683,ENSEMBL:ENSMUSG00000117542 | MGI:5010948 | pseudogene | predicted gene, 18763 |
| 18 | pseudogene | 7.798784 | 7.799900 | negative | MGI_C57BL6J_3780506 | Gm2336 | NCBI_Gene:100039606 | MGI:3780506 | pseudogene | predicted gene 2336 |
| 18 | gene | 7.835177 | 7.835283 | positive | MGI_C57BL6J_5454069 | Gm24292 | ENSEMBL:ENSMUSG00000064822 | MGI:5454069 | snRNA gene | predicted gene, 24292 |
| 18 | pseudogene | 7.844684 | 7.844935 | negative | MGI_C57BL6J_6275329 | Gm50027 | ENSEMBL:ENSMUSG00000117462 | MGI:6275329 | pseudogene | predicted gene, 50027 |
| 18 | gene | 7.868018 | 7.869113 | negative | MGI_C57BL6J_3641793 | Gm9993 | ENSEMBL:ENSMUSG00000117505 | MGI:3641793 | lncRNA gene | predicted gene 9993 |
| 18 | gene | 7.868832 | 7.929028 | positive | MGI_C57BL6J_2387357 | Wac | NCBI_Gene:225131,ENSEMBL:ENSMUSG00000024283 | MGI:2387357 | protein coding gene | WW domain containing adaptor with coiled-coil |
| 18 | gene | 7.894300 | 7.895792 | positive | MGI_C57BL6J_1924412 | A430102J17Rik | NA | NA | unclassified gene | RIKEN cDNA A430102J17 gene |
| 18 | pseudogene | 7.932728 | 7.933556 | positive | MGI_C57BL6J_3643245 | Gm7502 | NCBI_Gene:665121,ENSEMBL:ENSMUSG00000091019 | MGI:3643245 | pseudogene | predicted gene 7502 |
R/qtl
scanone
gmObject of class cross2 (crosstype "bc")
Total individuals 138
No. genotyped individuals 138
No. phenotyped individuals 138
No. with both geno & pheno 138
No. phenotypes 1
No. covariates 7
No. phenotype covariates 0
No. chromosomes 20
Total markers 32690
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2952 2874 2058 2071 1955 2053 1862 1700 2009 1229 2069 1396 1628 1692 1064 940
17 18 19 X
401 1095 1067 575 #detach("package:qtl2", unload=TRUE)
#library(qtl)
cross <- qtl::read.cross("csv", file = "data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_mis_conditional_2-peaks-chr4-10.csv",alleles=c("A","B")) --Read the following data:
138 individuals
32690 markers
5 phenotypes
--Cross type: bc cross <- qtl::jittermap(cross)
summary(cross) Backcross
No. individuals: 138
No. phenotypes: 5
Percent phenotyped: 100 100 100 98.6 98.6
No. chromosomes: 20
Autosomes: 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19
X chr: X
Total markers: 32690
No. markers: 2952 2874 2058 2071 1955 2053 1862 1700 2009 1229 2069
1396 1628 1692 1064 940 401 1095 1067 575
Percent genotyped: 99.5
Genotypes (%):
Autosomes: AA:54.3 AB:45.7
X chromosome: AA:52.7 AB:47.3 cross.probs <- qtl::calc.genoprob(cross)
print("method == hk")[1] "method == hk"add.covars = qtl::pull.pheno(cross.probs, c("UNC8250659","UNC18240977"))
scanone.hk <-qtl::scanone(cross.probs, pheno.col="ICI.vs.PBS" , model="binary", method="hk", addcovar = add.covars)
operm.hk <- qtl::scanone(cross.probs, method = "hk", pheno.col="ICI.vs.PBS", n.perm = 10, perm.Xsp = TRUE, model="binary", verbose=FALSE, addcovar = add.covars)
plot(operm.hk)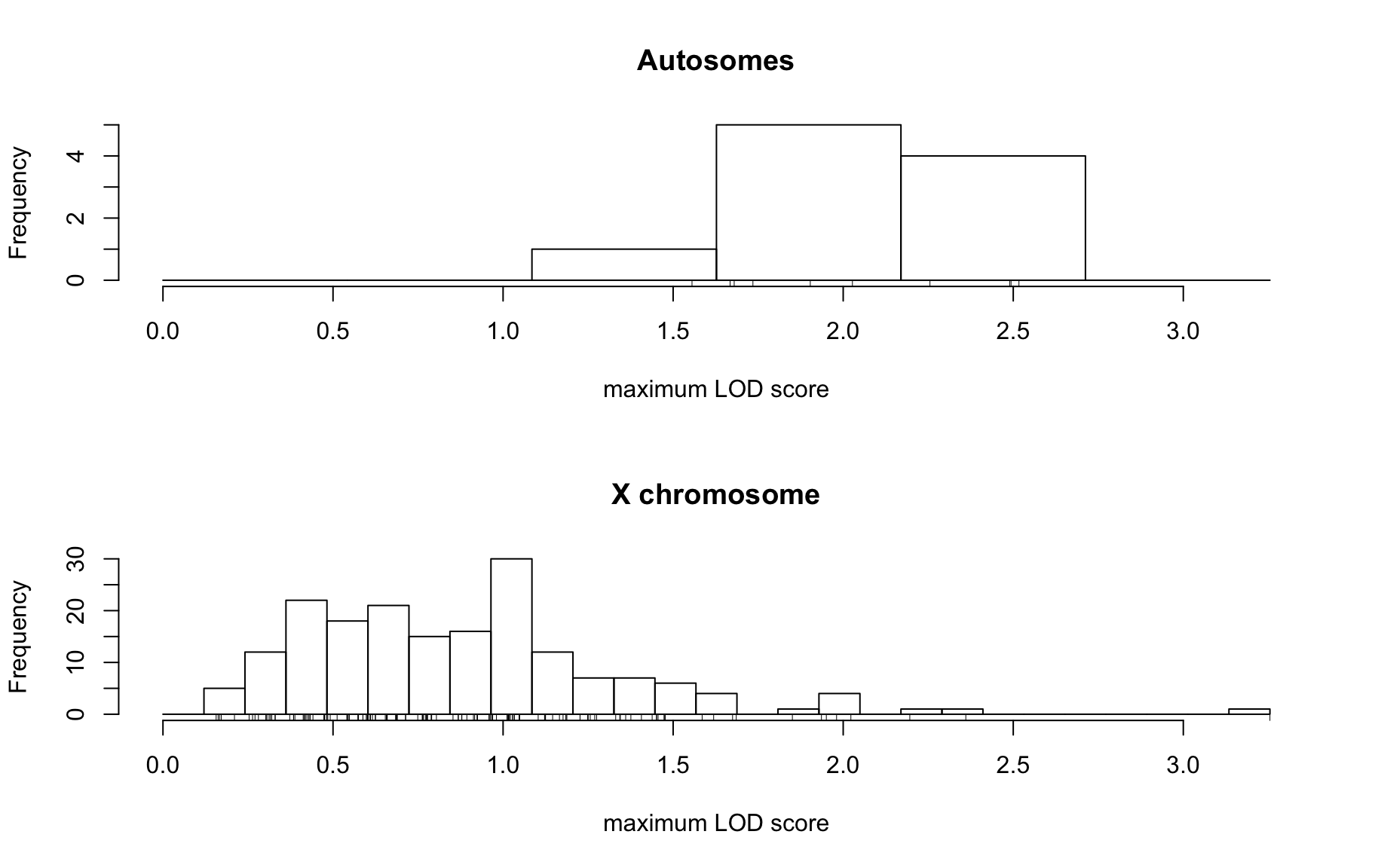
print(summary(operm.hk, alpha=c(0.01, 0.05, 0.1)))Autosome LOD thresholds (10 permutations)
lod
1% 2.51
5% 2.51
10% 2.50
X chromosome LOD thresholds (183 permutations)
lod
1% 3.17
5% 2.82
10% 2.37#plot(scanone.hk, bandcol = "grey90",lty=1, cex=1, col = "steelblue")
#qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.01, col = 'blue')
#qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.05, col = 'red')
#qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.1, col = 'purple')
ymx <- maxlod(out) # overall maximum LOD score
plot(scanone.hk, bandcol = "grey90",lty=1, cex=1, col = "slateblue", ylim=c(0, ymx+0.5))
title(main = paste0(colnames(out), " [positions in cM]"))
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.01, col = 'blue')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.05, col = 'red')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.1, col = 'purple')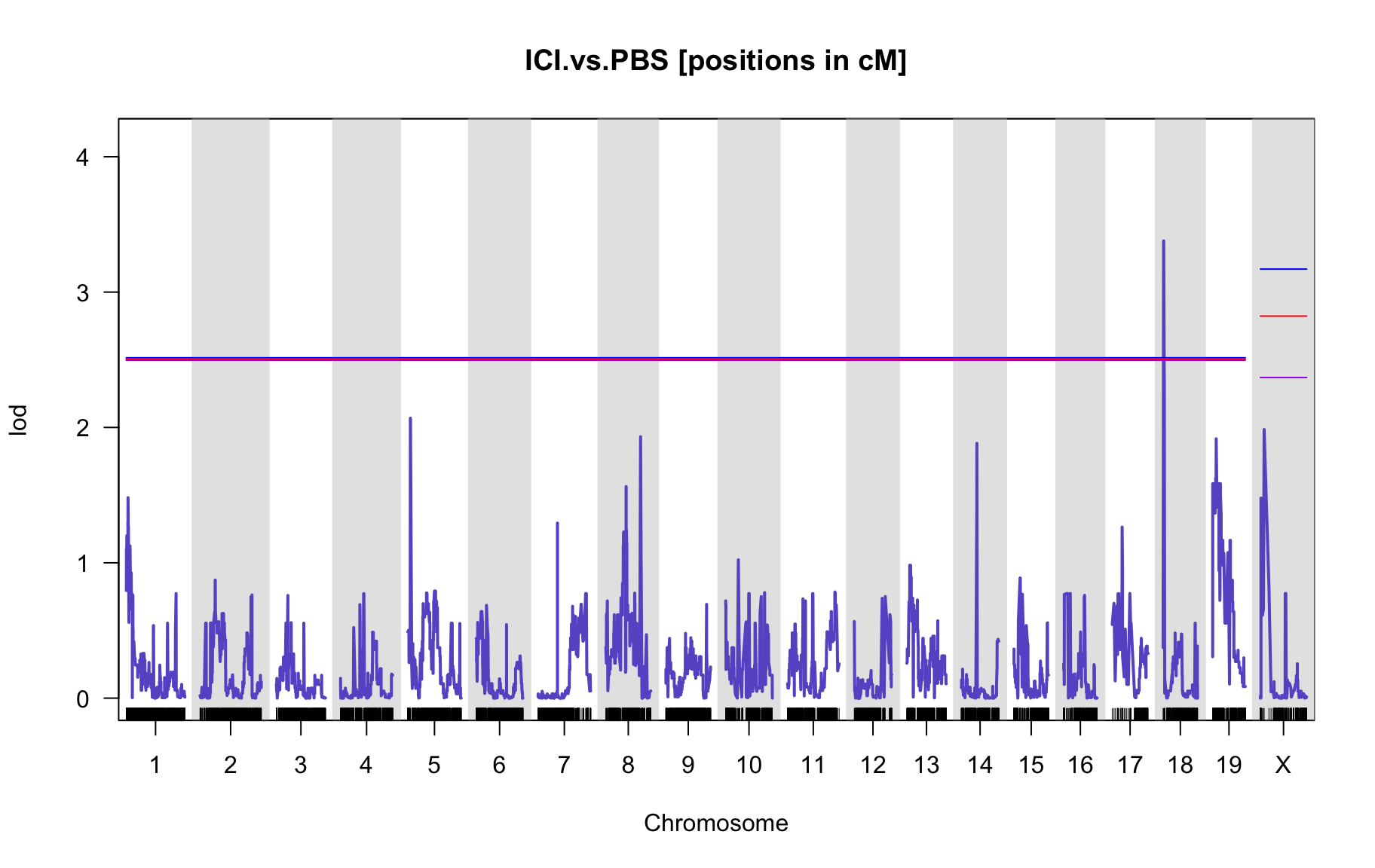
ymx <- 11
plot(scanone.hk, bandcol = "grey90",lty=1, cex=1, col = "slateblue", ylim=c(0, ymx+0.5))
title(main = paste0(colnames(out), " [positions in cM]\n(using same scale as eoi vs ici for easier comparison)"))
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.01, col = 'blue')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.05, col = 'red')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.1, col = 'purple')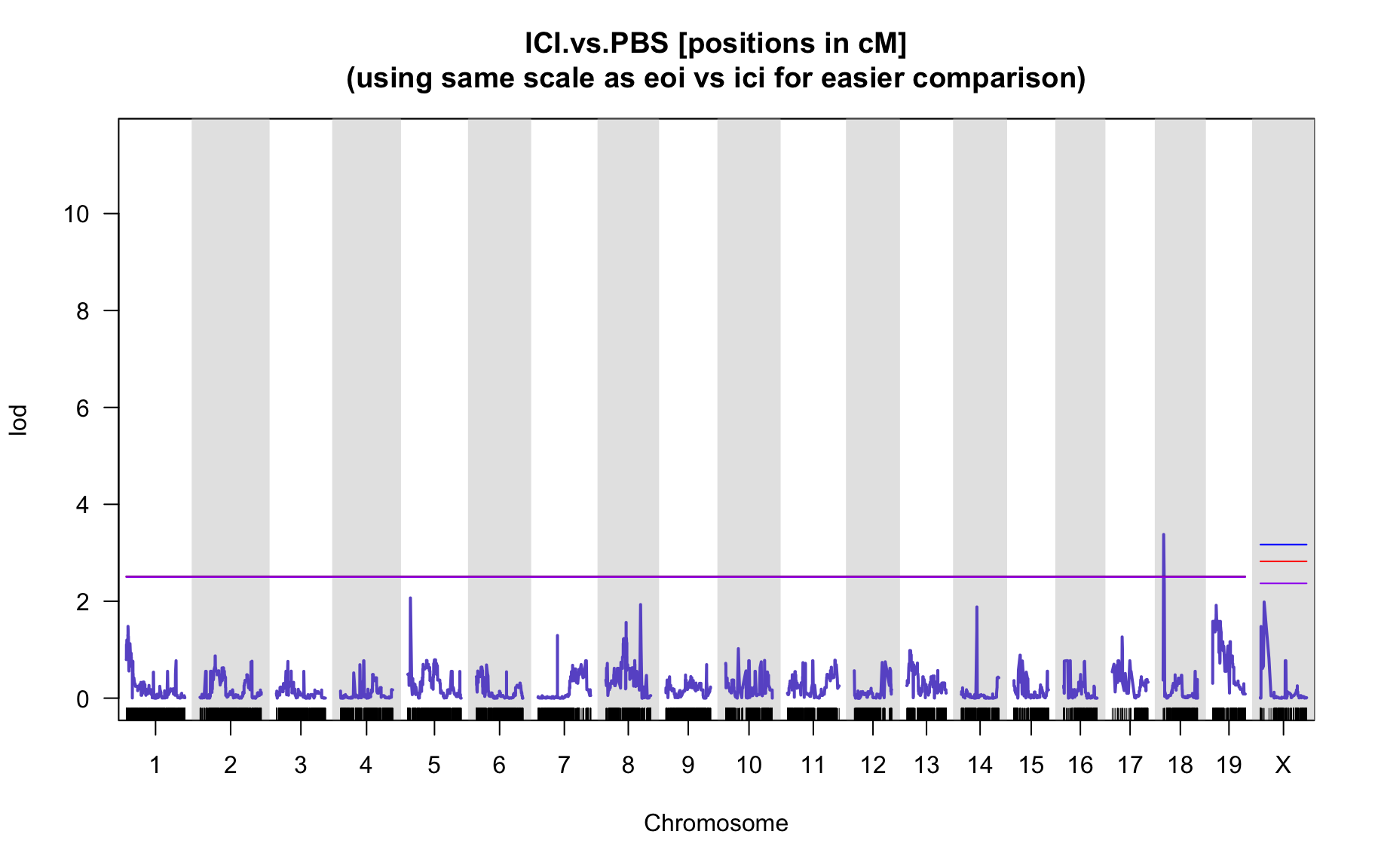
print(as.data.frame(summary(scanone.hk, perms=operm.hk, pvalues=TRUE, format="allpeaks"))) chr pos lod pval
UNCHS000141 1 4.439109 1.4810382 1.0000000
UNC2981117 2 27.613431 0.8724495 1.0000000
JAX00521745 3 20.613360 0.7586221 1.0000000
JAX00556948 4 41.038754 0.7726880 1.0000000
UNC8675939 5 6.193174 2.0684019 0.4165181
UNC10923091 6 18.321306 0.6856206 1.0000000
UNCHS020066 7 34.113657 1.2936681 1.0000000
UNC15524531 8 59.462590 1.9315437 0.5185858
UNC17203597 9 68.527929 0.6928610 1.0000000
UNCHS028001 10 21.783362 1.0219015 1.0000000
UNC20465557 11 80.904972 0.7841081 1.0000000
UNC21787084 12 53.288332 0.7505664 1.0000000
JAX00351755 13 7.044200 0.9809396 1.0000000
UNC24056202 14 30.156730 1.8824837 0.6195366
UNC25275535 15 11.967146 0.8880487 1.0000000
UNCHS042071 16 13.176096 0.7730967 1.0000000
UNCrs47191360 17 18.433013 1.2636005 1.0000000
UNCHS045343 18 2.934001 3.3781249 0.0000000
UNC29920552 19 8.810092 1.9159935 0.5185858
UNCHS048314 X 8.974041 1.9849011 0.3472122print("all peaks with a p-value less or equal to 0.05 (suggestive)")[1] "all peaks with a p-value less or equal to 0.05 (suggestive)"print(as.data.frame(summary(scanone.hk, perms=operm.hk, alpha=0.05, pvalues=TRUE, format="allpeaks"))) chr pos lod pval
UNCHS045343 18 2.934001 3.378125 0#print("method == ehk")
#scanone.ehk <-qtl::scanone(cross.probs, pheno.col="ICI.vs.PBS" , model="binary", method="ehk")
#operm.ehk <- qtl::scanone(cross.probs, method = "ehk", pheno.col="ICI.vs.PBS", n.perm = 10, perm.Xsp = TRUE, model="binary", verbose=FALSE)
#plot(operm.ehk)
#print(summary(operm.ehk, alpha=c(0.01, 0.05, 0.1)))
#plot(scanone.ehk, bandcol = "grey90",lty=1, cex=1, col = "steelblue")
#qtl::add.threshold(scanone.ehk, perms= operm.ehk, alpha=0.01, col = 'blue')
#qtl::add.threshold(scanone.ehk, perms= operm.ehk, alpha=0.05, col = 'red')
#qtl::add.threshold(scanone.ehk, perms= operm.ehk, alpha=0.1, col = 'purple')
#print(as.data.frame(summary(scanone.ehk)))
#print(as.data.frame(summary(scanone.ehk, perms=operm.ehk, alpha=0.05, pvalues=TRUE, format="allpeaks")))
R version 3.5.1 (2018-07-02)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS 10.15.7
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_AU.UTF-8/en_AU.UTF-8/en_AU.UTF-8/C/en_AU.UTF-8/en_AU.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] abind_1.4-5 qtl2_0.22 reshape2_1.4.3 ggplot2_3.3.6
[5] tibble_3.1.7 psych_2.2.5 readxl_1.4.0 cluster_2.0.7-1
[9] dplyr_1.0.9 optparse_1.6.6 rhdf5_2.26.2 mclust_5.4.5
[13] tidyr_1.2.0 data.table_1.14.2 knitr_1.29 kableExtra_1.3.4
[17] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] httr_1.4.3 bit64_0.9-7 viridisLite_0.3.0 assertthat_0.2.1
[5] highr_0.8 blob_1.2.1 cellranger_1.1.0 yaml_2.2.1
[9] gdtools_0.2.1 pillar_1.7.0 RSQLite_2.2.0 backports_1.1.5
[13] lattice_0.20-35 glue_1.6.2 digest_0.6.25 promises_1.1.0
[17] rvest_1.0.2 colorspace_1.4-1 htmltools_0.5.2 httpuv_1.5.2
[21] plyr_1.8.6 pkgconfig_2.0.3 purrr_0.3.4 scales_1.1.1
[25] webshot_0.5.3 svglite_1.2.3 qtl_1.46-2 getopt_1.20.3
[29] later_1.0.0 git2r_0.26.1 generics_0.0.2 ellipsis_0.3.2
[33] withr_2.5.0 cli_3.3.0 mnormt_1.5-6 magrittr_2.0.3
[37] crayon_1.5.1 memoise_1.1.0 evaluate_0.14 fs_1.3.2
[41] fansi_0.4.1 nlme_3.1-137 xml2_1.3.2 tools_3.5.1
[45] lifecycle_1.0.1 stringr_1.4.0 Rhdf5lib_1.4.3 munsell_0.5.0
[49] compiler_3.5.1 systemfonts_0.1.1 rlang_1.0.3 grid_3.5.1
[53] rstudioapi_0.13 rmarkdown_2.3 gtable_0.3.0 DBI_1.1.0
[57] R6_2.4.1 fastmap_1.1.0 bit_1.1-15.2 utf8_1.1.4
[61] rprojroot_1.3-2 stringi_1.4.6 parallel_3.5.1 Rcpp_1.0.4.6
[65] vctrs_0.4.1 tidyselect_1.1.2 xfun_0.15