QTL Analysis - binary [ICI vs PBS]
Belinda Cornes
2022-04-12
Last updated: 2022-04-12
Checks: 5 2
Knit directory: Serreze-T1D_Workflow/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20220210) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /Users/corneb/Documents/MyJax/CS/Projects/Serreze/qc/workflowr/Serreze-T1D_Workflow | . |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version d0c63e8. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: analysis/.DS_Store
Ignored: analysis/figure/
Ignored: data/.DS_Store
Untracked files:
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_mis_vo.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_vo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs_5.batches.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs_5.batches_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs_snpsqc_5.batches.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs_snpsqc_5.batches_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs_snpsqc_dis_no-x_updated_5.batches.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-early.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.eoi_snpsqc_dis_no-x_updated_5.batches_11.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.eoi_snpsqc_dis_no-x_updated_5.batches_mis_11.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_1.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_mis_1.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_peaks.Rmd.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_1.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_11.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_11.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_peaks.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_peaks.Rmd
Untracked: analysis/genotype.frequencies_ici-early.vs.pbs_5.batches.Rmd
Untracked: analysis/genotype.frequencies_ici-early.vs.pbs_5.batches_mis.Rmd
Untracked: analysis/index_5.batches_additional.Rmd.R
Untracked: data/GM_covar.csv
Untracked: data/GM_covar_BC312.csv
Untracked: data/bad_markers_all_4.batches.RData
Untracked: data/bad_markers_all_5.batches.RData
Untracked: data/blup_sub_chr10_lod.drop-1.5_5.batches.csv
Untracked: data/blup_sub_chr3_lod.drop-1.5.csv
Untracked: data/blup_sub_chr3_lod.drop-1.5_5.batches.csv
Untracked: data/blup_sub_chr4_lod.drop-1.5.csv
Untracked: data/blup_sub_chr4_lod.drop-1.5_5.batches.csv
Untracked: data/covar_cleaned_ici.vs.eoi.csv
Untracked: data/covar_cleaned_ici.vs.pbs.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_mis.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi1.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_mis.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs1.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_mis.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_mis.csv
Untracked: data/covar_corrected_ici.vs.eoi.csv
Untracked: data/covar_corrected_ici.vs.eoi1.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_mis.csv
Untracked: data/covar_corrected_ici.vs.pbs.csv
Untracked: data/covar_corrected_ici.vs.pbs1.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_mis.csv
Untracked: data/e.RData
Untracked: data/e_BC312.RData
Untracked: data/e_snpg_samqc_4.batches.RData
Untracked: data/e_snpg_samqc_4.batches_bc.RData
Untracked: data/e_snpg_samqc_5.batches.RData
Untracked: data/errors_ind_4.batches.RData
Untracked: data/errors_ind_4.batches_bc.RData
Untracked: data/errors_ind_5.batches.RData
Untracked: data/fitqtl_chr3.peak_chr4.peak_additive.txt
Untracked: data/fitqtl_chr3.peak_chr4.peak_interacting.txt
Untracked: data/fitqtl_chr3.peak_chr4.peak_sex_additive.txt
Untracked: data/fitqtl_chr3.peak_chr4.peak_sex_interacting.txt
Untracked: data/g2blup_effects.csv
Untracked: data/g2blup_effects.xlsx
Untracked: data/genes_chr10_lod.drop-1.5_5.batches.csv
Untracked: data/genes_chr3_lod.drop-1.5.csv
Untracked: data/genes_chr3_lod.drop-1.5_5.batches.csv
Untracked: data/genes_chr4_lod.drop-1.5.csv
Untracked: data/genetic_map.csv
Untracked: data/genetic_map_BC312.csv
Untracked: data/genotype_errors_marker_4.batches.RData
Untracked: data/genotype_errors_marker_5.batches.RData
Untracked: data/genotype_freq_marker_4.batches.RData
Untracked: data/genotype_freq_marker_5.batches.RData
Untracked: data/gm_allqc_4.batches.RData
Untracked: data/gm_allqc_5.batches.RData
Untracked: data/gm_allqc_5.batches_mis.RData
Untracked: data/gm_samqc_3.batches.RData
Untracked: data/gm_samqc_4.batches.RData
Untracked: data/gm_samqc_4.batches_bc.RData
Untracked: data/gm_samqc_5.batches.RData
Untracked: data/gm_serreze.192.RData
Untracked: data/gm_serreze.BC312.RData
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-15_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-15_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-15_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-17_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-17_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-17_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_5.batches.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratiov_5.batches.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratiov_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_scanone_5.batches.Rdata
Untracked: data/ici-early.vs.pbs_scanone_5.batches_mis.Rdata
Untracked: data/ici-early.vs.pbs_scanone_snpsqc_5.batches.Rdata
Untracked: data/ici-early.vs.pbs_scanone_snpsqc_5.batches_mis.Rdata
Untracked: data/ici-early.vs.pbs_scanone_snpsqc_dis_no-x_updated_5.batches.Rdata
Untracked: data/ici-early.vs.pbs_scanone_snpsqc_dis_no-x_updated_5.batches_mis.Rdata
Untracked: data/ici.vs.eoi_blup.full_chr-10_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-11_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-12_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-13_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-14_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-15_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-16_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-17_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-18_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-19_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-1_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-2_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-3_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-4_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-6_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-7_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-8_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-9_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-X_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-ICR4295_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-JAX00292499_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18154330_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18195765_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18216614_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18240977_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18856953_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNCJPD004316_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNCJPD009532_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNC19155926_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNC19481366_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNCJPD004792_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-12_peak.marker-ICR4497_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-13_peak.marker-UNCHS036964_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-18_peak.marker-UNCHS045344_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-19_peak.marker-UNCJPD007087_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-1_peak.marker-ICR010_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-1_peak.marker-ICR010_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-2_peak.marker-UNC4609660_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-B6_rs31740628_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR1269_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR5325_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR5347_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR5348_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00104971_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00105059_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00105203_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00107930_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00107980_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00108068_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00108114_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00109750_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00189245_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00517218_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00517906_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00518418_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00518684_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00520122_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00524717_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00524828_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00525380_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00525709_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00527291_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00527587_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00529707_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00531601_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00532122_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4673471_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4681411_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4793677_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4799475_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4909830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4914130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4964062_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4985064_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4991565_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5001723_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5042361_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5055259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5076935_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5082757_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5173293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5182782_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5193970_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5240484_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5256922_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5266664_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5266711_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5267813_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5274554_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5277061_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5282259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5287977_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5301460_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5321559_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5322455_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5328596_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5350384_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5354724_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5375208_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5384072_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5400314_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5433945_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5435874_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5466669_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5517972_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5522257_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5523265_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5532424_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5553882_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5572051_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5582753_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5589260_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5595555_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5623239_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5639830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5680234_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5682314_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5725356_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5727351_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5734420_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5743257_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5751567_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5752623_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5776974_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5778977_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5788507_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5792468_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5794598_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5795561_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5802298_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5806628_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5817478_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5836999_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5841501_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5847927_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5966417_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008129_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008173_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008231_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008235_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008242_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008253_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008256_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008311_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008317_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008372_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008410_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008415_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008431_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008609_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008614_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008649_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008651_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008723_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008802_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008803_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008808_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008834_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008838_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008928_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008948_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008959_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008973_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008987_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009001_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009025_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009034_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009087_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009109_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009142_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009207_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009216_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009306_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009316_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009364_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009377_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009382_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009387_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009395_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009413_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009428_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009463_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009503_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009532_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009541_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009560_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009574_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001276_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001315_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001355_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001373_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001382_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC6851205_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8250659_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8299912_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8300499_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8381981_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8436434_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8439633_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8530763_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS012948_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS012976_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013139_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013304_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-5_peak.marker-JAX00590770_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-ICR1830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-UNC13170794_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-UNCHS019480_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-cr31snv87_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-UNC15430711_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-9_peak.marker-UNC17203329_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-9_peak.marker-UNCHS024593_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-X_peak.marker-UNC30627130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-X_peak.marker-UNCHS048313_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_snpsqc_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_snpsqc_dis_no-x_updated_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_snpsqc_dis_no-x_updated_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_snpsqc_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_snpsqc_dis_no-x_updated_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_snpsqc_dis_no-x_updated_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_snpsqc_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_snpsqc_dis_no-x_updated_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_snpsqc_dis_no-x_updated_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_snpsqc_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_snpsqc_dis_no-x_updated_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_snpsqc_dis_no-x_updated_5.batches_mis.mis.txt
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-ICR4295_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-JAX00292499_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18154330_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18195765_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18216614_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18240977_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18856953_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNCJPD004316_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNCJPD009532_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNC19155926_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNC19481366_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNCJPD004792_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-12_peak.marker-ICR4497_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-13_peak.marker-UNCHS036964_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-18_peak.marker-UNCHS045344_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-19_peak.marker-UNCJPD007087_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-1_peak.marker-ICR010_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-1_peak.marker-ICR010_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-2_peak.marker-UNC4609660_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-B6_rs31740628_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR1269_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR5325_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR5347_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR5348_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00104971_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00105059_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00105203_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00107930_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00107980_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00108068_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00108114_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00109750_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00189245_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00517218_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00517906_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00518418_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00518684_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00520122_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00524717_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00524828_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00525380_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00525709_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00527291_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00527587_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00529707_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00531601_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00532122_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4673471_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4681411_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4793677_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4799475_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4909830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4914130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4964062_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4985064_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4991565_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5001723_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5042361_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5055259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5076935_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5082757_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5173293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5182782_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5193970_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5240484_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5256922_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5266664_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5266711_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5267813_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5274554_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5277061_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5282259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5287977_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5301460_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5321559_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5322455_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5328596_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5350384_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5354724_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5375208_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5384072_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5400314_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5433945_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5435874_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5466669_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5517972_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5522257_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5523265_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5532424_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5553882_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5572051_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5582753_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5589260_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5595555_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5623239_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5639830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5680234_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5682314_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5725356_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5727351_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5734420_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5743257_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5751567_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5752623_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5776974_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5778977_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5788507_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5792468_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5794598_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5795561_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5802298_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5806628_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5817478_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5836999_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5841501_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5847927_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5966417_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008129_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008173_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008231_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008235_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008242_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008253_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008256_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008311_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008317_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008372_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008410_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008415_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008431_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008609_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008614_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008649_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008651_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008723_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008802_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008803_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008808_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008834_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008838_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008928_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008948_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008959_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008973_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008987_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009001_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009025_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009034_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009087_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009109_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009142_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009207_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009216_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009306_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009316_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009364_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009377_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009382_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009387_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009395_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009413_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009428_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009463_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009503_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009532_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009541_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009560_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009574_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001276_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001315_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001355_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001373_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001382_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC6851205_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8250659_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8299912_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8300499_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8381981_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8436434_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8439633_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8530763_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS012948_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS012976_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013139_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013304_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-5_peak.marker-JAX00590770_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-ICR1830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-UNC13170794_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-UNCHS019480_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-cr31snv87_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-UNC15430711_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-9_peak.marker-UNC17203329_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-9_peak.marker-UNCHS024593_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-X_peak.marker-UNC30627130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-X_peak.marker-UNCHS048313_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_gm_qtl_5.batches.csv
Untracked: data/ici.vs.eoi_gm_qtl_5.batches_mis.csv
Untracked: data/ici.vs.eoi_gm_qtl_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_gm_qtl_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_gm_qtl_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.geno.freq.removed.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.probs.freq.removed.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.eoi_scanone_5.batches.Rdata
Untracked: data/ici.vs.eoi_scanone_5.batches_mis.Rdata
Untracked: data/ici.vs.eoi_scanone_snpsqc_5.batches.Rdata
Untracked: data/ici.vs.eoi_scanone_snpsqc_5.batches_mis.Rdata
Untracked: data/ici.vs.eoi_scanone_snpsqc_dis_no-x_updated_5.batches.Rdata
Untracked: data/ici.vs.pbs_bad.markers_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-15_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-15_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-15_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-17_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-17_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-17_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-13_peak.marker-UNC22384241_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-19_peak.marker-UNC29893228_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-19_peak.marker-UNC29990033_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-19_peak.marker-UNCHS047192_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-8_peak.marker-UNCHS023353_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-8_peak.marker-UNCHS023353_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-8_peak.marker-UNCHS023364_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_fitqtl_peaks_additive_snpsqc_dis_no-x_updated_5.batches_mis_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_additive_snpsqc_dis_no-x_updated_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_interacting_snpsqc_dis_no-x_updated_5.batches_mis_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_interacting_snpsqc_dis_no-x_updated_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_additive_snpsqc_dis_no-x_updated_5.batches_mis_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_additive_snpsqc_dis_no-x_updated_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_interacting_snpsqc_dis_no-x_updated_5.batches_mis_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_interacting_snpsqc_dis_no-x_updated_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_genes_chr-13_peak.marker-UNC22384241_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_genes_chr-19_peak.marker-UNC29893228_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-19_peak.marker-UNC29990033_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-19_peak.marker-UNCHS047192_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-8_peak.marker-UNCHS023353_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-8_peak.marker-UNCHS023353_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-8_peak.marker-UNCHS023364_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_mis.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratiov_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratiov_5.batches_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.pbs_scanone_5.batches.Rdata
Untracked: data/ici.vs.pbs_scanone_5.batches_mis.Rdata
Untracked: data/ici.vs.pbs_scanone_snpsqc_5.batches.Rdata
Untracked: data/ici.vs.pbs_scanone_snpsqc_5.batches_mis.Rdata
Untracked: data/ici.vs.pbs_scanone_snpsqc_dis_no-x_updated_5.batches.Rdata
Untracked: data/ici.vs.pbs_scanone_snpsqc_dis_no-x_updated_5.batches_mis.Rdata
Untracked: data/percent_missing_id_3.batches.RData
Untracked: data/percent_missing_id_4.batches.RData
Untracked: data/percent_missing_id_4.batches_bc.RData
Untracked: data/percent_missing_id_5.batches.RData
Untracked: data/percent_missing_marker_4.batches.RData
Untracked: data/percent_missing_marker_5.batches.RData
Untracked: data/pheno.csv
Untracked: data/pheno_BC312.csv
Untracked: data/physical_map.csv
Untracked: data/physical_map_BC312.csv
Untracked: data/qc_info_bad_sample_3.batches.RData
Untracked: data/qc_info_bad_sample_4.batches.RData
Untracked: data/qc_info_bad_sample_4.batches_bc.RData
Untracked: data/qc_info_bad_sample_5.batches.RData
Untracked: data/remaining.markers_geno.freq.xlsx
Untracked: data/sample_geno.csv
Untracked: data/sample_geno_AHB_BC312.csv
Untracked: data/sample_geno_bc.csv
Untracked: data/sample_geno_bc_BC312.csv
Untracked: data/serreze_probs.rds
Untracked: data/serreze_probs_BC312.rds
Untracked: data/serreze_probs_allqc.rds
Untracked: data/serreze_probs_allqc_5.batches.rds
Untracked: data/serreze_probs_allqc_5.batches_mis.rds
Untracked: data/summary.cg_3.batches.RData
Untracked: data/summary.cg_4.batches.RData
Untracked: data/summary.cg_4.batches_bc.RData
Untracked: data/summary.cg_5.batches.RData
Untracked: output/Percent_missing_genotype_data_5.batches.pdf
Untracked: output/Percent_missing_genotype_data_per_marker_5.batches.pdf
Untracked: output/Proportion_matching_genotypes_before_removal_of_bad_samples_5.batches.pdf
Untracked: output/genotype_error_marker_5.batches.pdf
Untracked: output/genotype_frequency_marker_5.batches.pdf
Unstaged changes:
Modified: analysis/3.1_phenotype.qc_corrected_5.batches.Rmd
Modified: analysis/3.1_phenotype.qc_corrected_5.batches_mis.Rmd
Modified: analysis/4.1.1_qtl.analysis_binary_ici.vs.eoi_snpsqc_dis_no-x_updated.Rmd
Modified: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated.Rmd
Modified: analysis/index_5.batches.Rmd
Modified: analysis/index_5.batches_additional.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with wflow_publish() to start tracking its development.
Loading Data
We will load the data and subset indivials out that are in the groups of interest. We will create a binary phenotype from this (PBS ==0, ICI == 1).
load("data/gm_allqc_5.batches.RData")
#gm_allqc
gm=gm_allqc
gmObject of class cross2 (crosstype "bc")
Total individuals 308
No. genotyped individuals 308
No. phenotyped individuals 308
No. with both geno & pheno 308
No. phenotypes 1
No. covariates 6
No. phenotype covariates 0
No. chromosomes 20
Total markers 34537
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2643 2629 1857 1890 1774 1941 1672 1627 1878 1176 1871 1300 1549 1578 1257 935
17 18 19 X
501 913 1014 4532 #pr <- readRDS("data/serreze_probs_allqc.rds")
#pr <- readRDS("data/serreze_probs.rds")
##extracting animals with ici and pbs group status
miceinfo <- gm$covar[gm$covar$group == "PBS" | gm$covar$group == "ICI",]
table(miceinfo$group)
ICI PBS
104 34 mice.ids <- rownames(miceinfo)
gm <- gm[mice.ids]
gmObject of class cross2 (crosstype "bc")
Total individuals 138
No. genotyped individuals 138
No. phenotyped individuals 138
No. with both geno & pheno 138
No. phenotypes 1
No. covariates 6
No. phenotype covariates 0
No. chromosomes 20
Total markers 34537
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2643 2629 1857 1890 1774 1941 1672 1627 1878 1176 1871 1300 1549 1578 1257 935
17 18 19 X
501 913 1014 4532 #pr.qc <- pr
#for (i in 1:20){pr.qc[[i]] = pr.qc[[i]][mice.ids,,]}
#bin_pheno <- NULL
#bin_pheno$PBS <- ifelse(gm$covar$group == "PBS", 1, 0)
#bin_pheno$ICI <- ifelse(gm$covar$group == "ICI", 1, 0)
#bin_pheno <- as.data.frame(bin_pheno)
#rownames(bin_pheno) <- rownames(gm$covar)
gm$covar$ICI.vs.PBS <- ifelse(gm$covar$group == "PBS", 0, 1)
gm.full <- gm
##removing problmetic marker
gm <- drop_markers(gm, "UNCHS013106")
##dropping monomorphic markers within the dataset
g <- do.call("cbind", gm$geno)
gf_mar <- t(apply(g, 2, function(a) table(factor(a, 1:2))/sum(a != 0)))
#gn_mar <- t(apply(g, 2, function(a) table(factor(a, 1:2))))
gf_mar <- gf_mar[gf_mar[,2] != "NaN",]
count <- rowSums(gf_mar <=0.05)
low_freq_df <- merge(as.data.frame(gf_mar),as.data.frame(count), by="row.names",all=T)
low_freq_df[is.na(low_freq_df)] <- ''
low_freq_df <- low_freq_df[low_freq_df$count == 1,]
rownames(low_freq_df) <- low_freq_df$Row.names
low_freq <- find_markerpos(gm, rownames(low_freq_df))
low_freq$id <- rownames(low_freq)
nrow(low_freq)[1] 7989low_freq_bad <- merge(low_freq,low_freq_df, by="row.names",all=T)
names(low_freq_bad)[1] <- c("marker")
gf_mar <- gf_mar[gf_mar[,2] != "NaN",]
MAF <- apply(gf_mar, 1, function(x) min(x))
MAF <- as.data.frame(MAF)
MAF$index <- 1:nrow(gf_mar)
gf_mar_maf <- merge(gf_mar,as.data.frame(MAF), by="row.names")
gf_mar_maf <- gf_mar_maf[order(gf_mar_maf$index),]
gfmar <- NULL
gfmar$gfmar_mar_0 <- sum(gf_mar_maf$MAF==0)
gfmar$gfmar_mar_1 <- sum(gf_mar_maf$MAF< 0.01)
gfmar$gfmar_mar_5 <- sum(gf_mar_maf$MAF< 0.05)
gfmar$gfmar_mar_10 <- sum(gf_mar_maf$MAF< 0.10)
gfmar$gfmar_mar_15 <- sum(gf_mar_maf$MAF< 0.15)
gfmar$gfmar_mar_25 <- sum(gf_mar_maf$MAF< 0.25)
gfmar$gfmar_mar_50 <- sum(gf_mar_maf$MAF< 0.50)
gfmar$total_snps <- nrow(as.data.frame(gf_mar_maf))
gfmar <- t(as.data.frame(gfmar))
gfmar <- as.data.frame(gfmar)
gfmar$count <- gfmar$V1
gfmar[c(2)] %>%
kable(escape = F,align = c("ccccccccc"),linesep ="\\hline") %>%
kable_styling(full_width = F) %>%
kable_styling("striped", full_width = F) %>%
row_spec(8 ,bold=T,color= "white",background = "black")| count | |
|---|---|
| gfmar_mar_0 | 3814 |
| gfmar_mar_1 | 4068 |
| gfmar_mar_5 | 7989 |
| gfmar_mar_10 | 8527 |
| gfmar_mar_15 | 8598 |
| gfmar_mar_25 | 9313 |
| gfmar_mar_50 | 33753 |
| total_snps | 34537 |
gm_qc <- drop_markers(gm, low_freq_bad$marker)
gm_qc <- drop_nullmarkers(gm_qc)
gm = gm_qc
gmObject of class cross2 (crosstype "bc")
Total individuals 138
No. genotyped individuals 138
No. phenotyped individuals 138
No. with both geno & pheno 138
No. phenotypes 1
No. covariates 7
No. phenotype covariates 0
No. chromosomes 20
Total markers 26548
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2374 2374 1623 1691 1568 1703 1464 1468 1685 974 1671 1134 1377 1411 876 751
17 18 19 X
330 770 920 384 markers <- marker_names(gm)
gmapdf <- read.csv("/Users/corneb/Documents/MyJax/CS/Projects/Serreze/haplotype.reconstruction/output_5.batches/genetic_map.csv")
pmapdf <- read.csv("/Users/corneb/Documents/MyJax/CS/Projects/Serreze/haplotype.reconstruction/output_5.batches/physical_map.csv")
#mapdf <- merge(gmapdf,pmapdf, by=c("marker","chr"), all=T)
#rownames(mapdf) <- mapdf$marker
#mapdf <- mapdf[markers,]
#names(mapdf) <- c('marker','chr','gmapdf','pmapdf')
#mapdfnd <- mapdf[!duplicated(mapdf[c(2:3)]),]
pr.qc <- calc_genoprob(gm)Genome-wide scan
For each of the phenotype analyzed, permutations were used for each model to obtain genome-wide LOD significance threshold for p < 0.01, p < 0.05, p < 0.10, respectively, separately for X and automsomes (A).
The table shows the estimated significance thresholds from permutation test.
We also looked at the kinship to see how correlated each sample is. Kinship values between pairs of samples range between 0 (no relationship) and 1.0 (completely identical). The darker the colour the more indentical the pairs are.
Xcovar <- get_x_covar(gm)
#addcovar = model.matrix(~Sex, data = covars)[,-1]
#K <- calc_kinship(pr.qc, type = "loco")
#heatmap(K[[1]])
#K.overall <- calc_kinship(pr.qc, type = "overall")
#heatmap(K.overall)
kinship <- calc_kinship(pr.qc)
heatmap(kinship)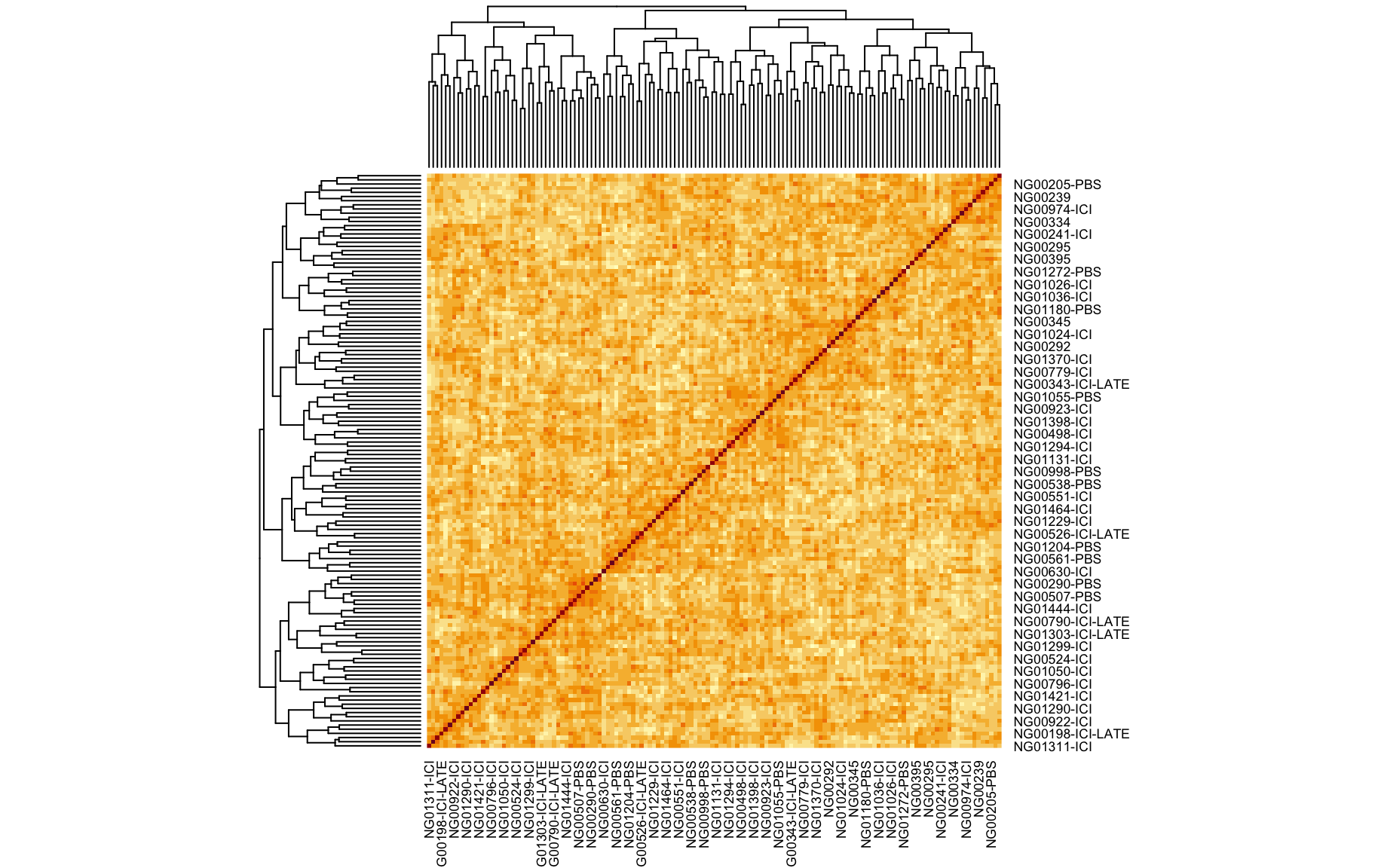
#operm <- scan1perm(pr.qc, gm$covar$phenos, Xcovar=Xcovar, n_perm=2000)
#operm <- scan1perm(pr.qc, gm$covar$phenos, addcovar = addcovar, n_perm=2000)
#operm <- scan1perm(pr.qc, gm$covar$phenos, n_perm=2000)
operm <- scan1perm(pr.qc, gm$covar["ICI.vs.PBS"], model="binary", n_perm=10, perm_Xsp=TRUE, chr_lengths=chr_lengths(gm$gmap))
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
names(summary_table) <- c("autosomes","X")
summary_table$significance.level <- rownames(summary_table)
rownames(summary_table) <- NULL
summary_table[c(3,1:2)] %>%
kable(escape = F,align = c("ccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| significance.level | autosomes | X |
|---|---|---|
| 0.01 | 2.771651 | 3.882077 |
| 0.05 | 2.742675 | 3.306148 |
| 0.1 | 2.706367 | 2.552978 |
The figures below show QTL maps for each phenotype
out <- scan1(pr.qc, gm$covar["ICI.vs.PBS"], Xcovar=Xcovar, model="binary")
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
plot_lod<-function(out,map){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- 11 # overall maximum LOD score
plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " [positions in cM] \n(using same scale as eoi vs ici for easier comparison)"))
add_threshold(map, summary(operm, alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
plot_lod(out,gm$gmap)

LOD peaks
The table below shows QTL peaks associated with the phenotype. We use the 95% threshold from the permutations to find peaks.
Centimorgan (cM)
#peaks <- find_peaks(out, gm$gmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
peaks <- find_peaks(out, gm$gmap, threshold=1.5, drop=1.5)
if(nrow(peaks) >0){
peaks$marker <- find_marker(gm$gmap, chr=peaks$chr,pos=peaks$pos)
names(peaks)[2] <- c("phenotype")
peaks <- peaks[-1]
rownames(peaks) <- NULL
print(kable(peaks, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
#plot only peak chromosomes
plot_lod_chr<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
ymx <- 11
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in cM]\n(using same scale as eoi vs. ici for easier comparison)"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
for(i in unique(peaks$chr)){
#for (i in 1:nrow(peaks)){
#plot_lod_chr(out,gm$gmap, peaks$chr[i])
plot_lod_chr(out,gm$gmap, i)
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}| phenotype | chr | pos | lod | ci_lo | ci_hi | marker |
|---|---|---|---|---|---|---|
| ICI.vs.PBS | 8 | 35.540 | 1.563047 | 1.952 | 70.682 | UNCHS023364 |
| ICI.vs.PBS | 13 | 13.831 | 1.603866 | 2.989 | 67.317 | UNC22384241 |
| ICI.vs.PBS | 19 | 8.029 | 1.629859 | 3.029 | 54.895 | UNC29893228 |

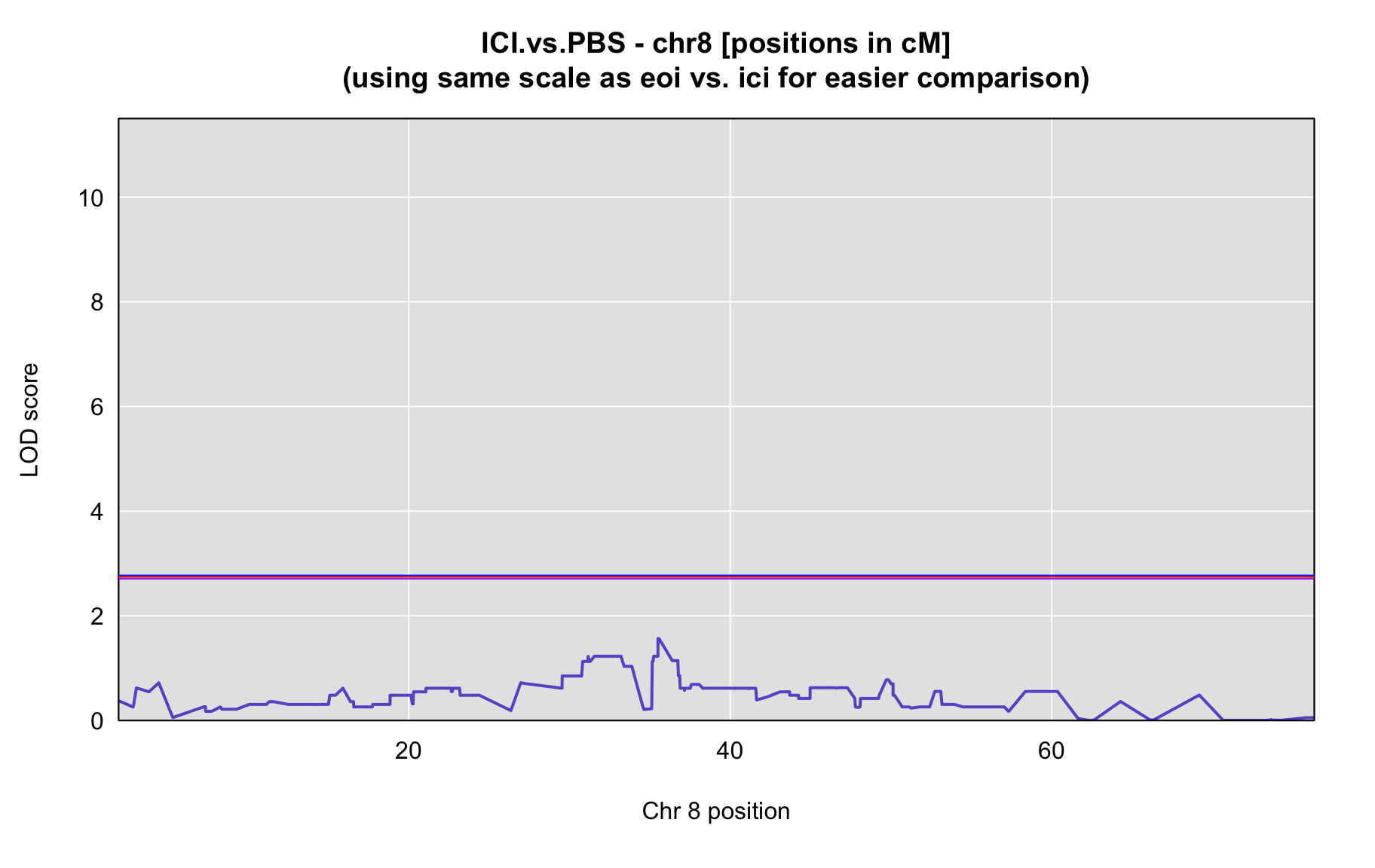
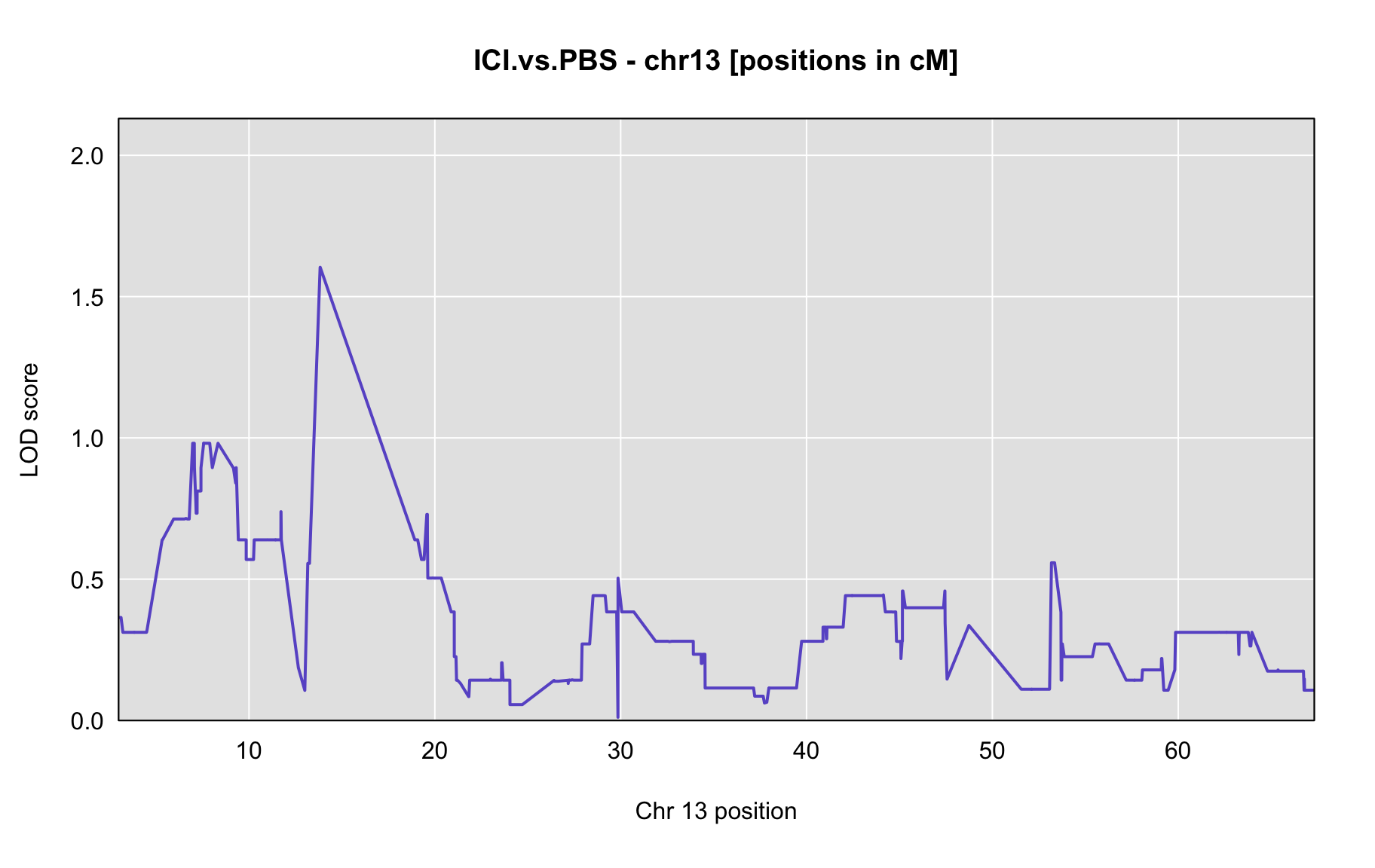
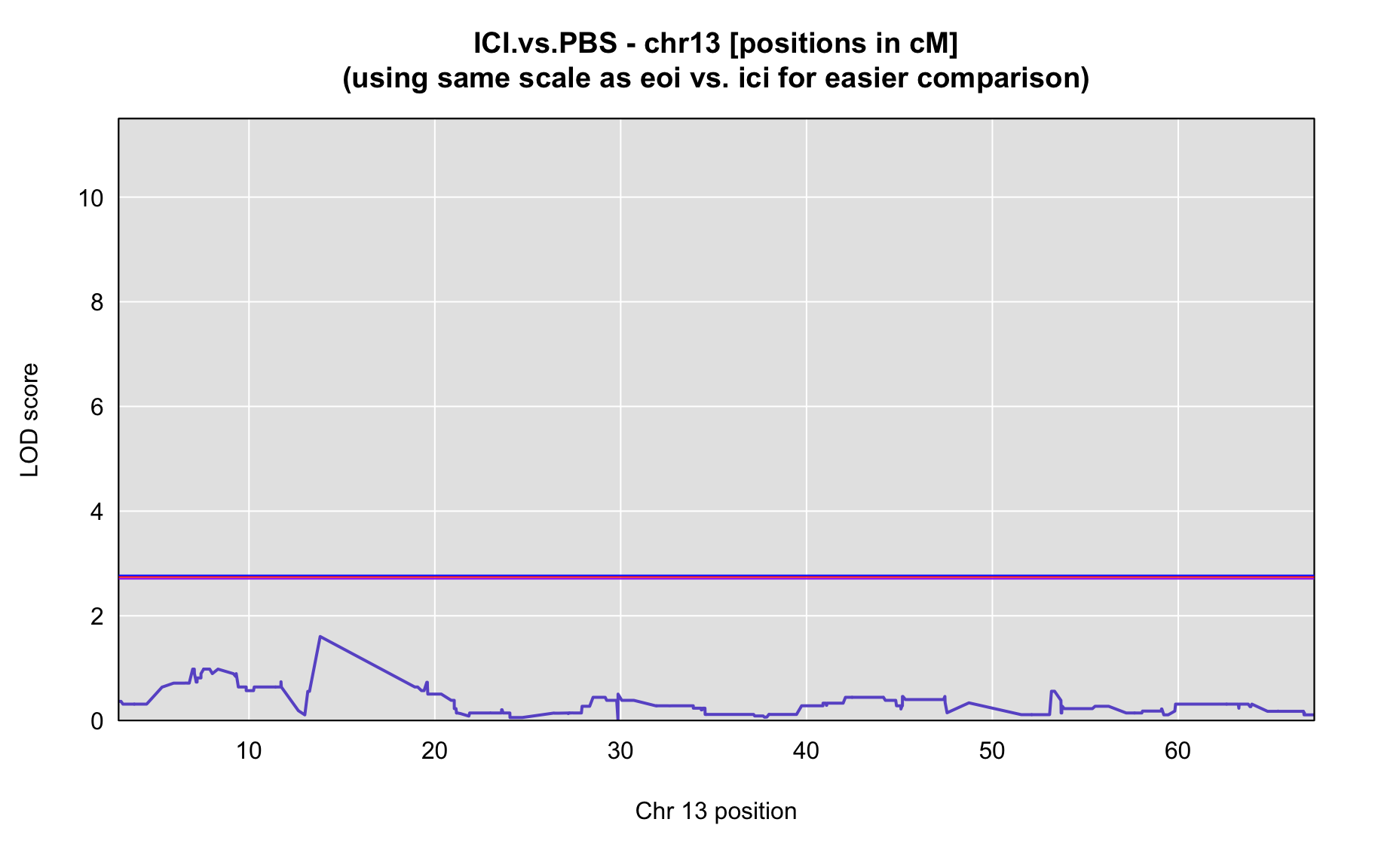
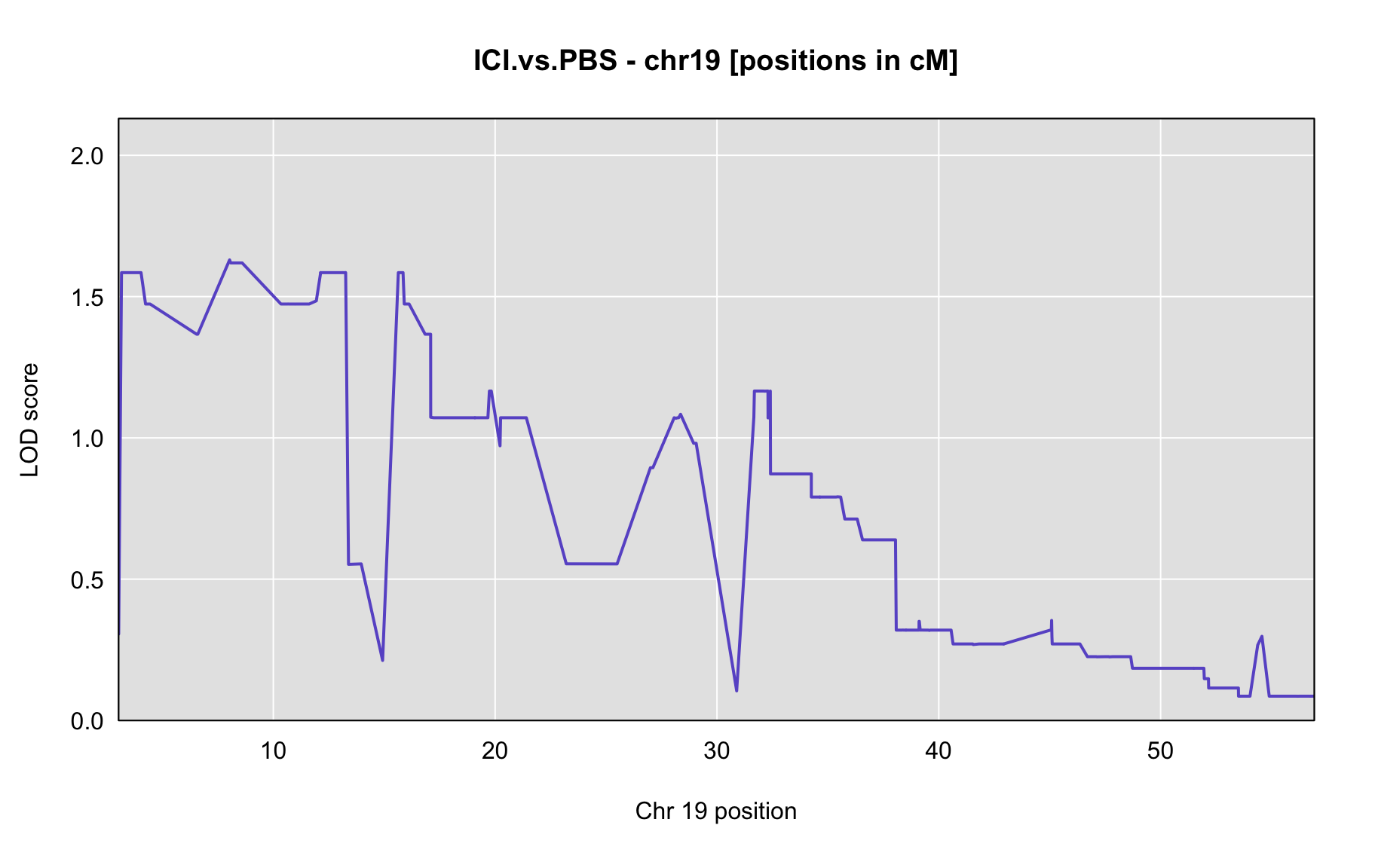
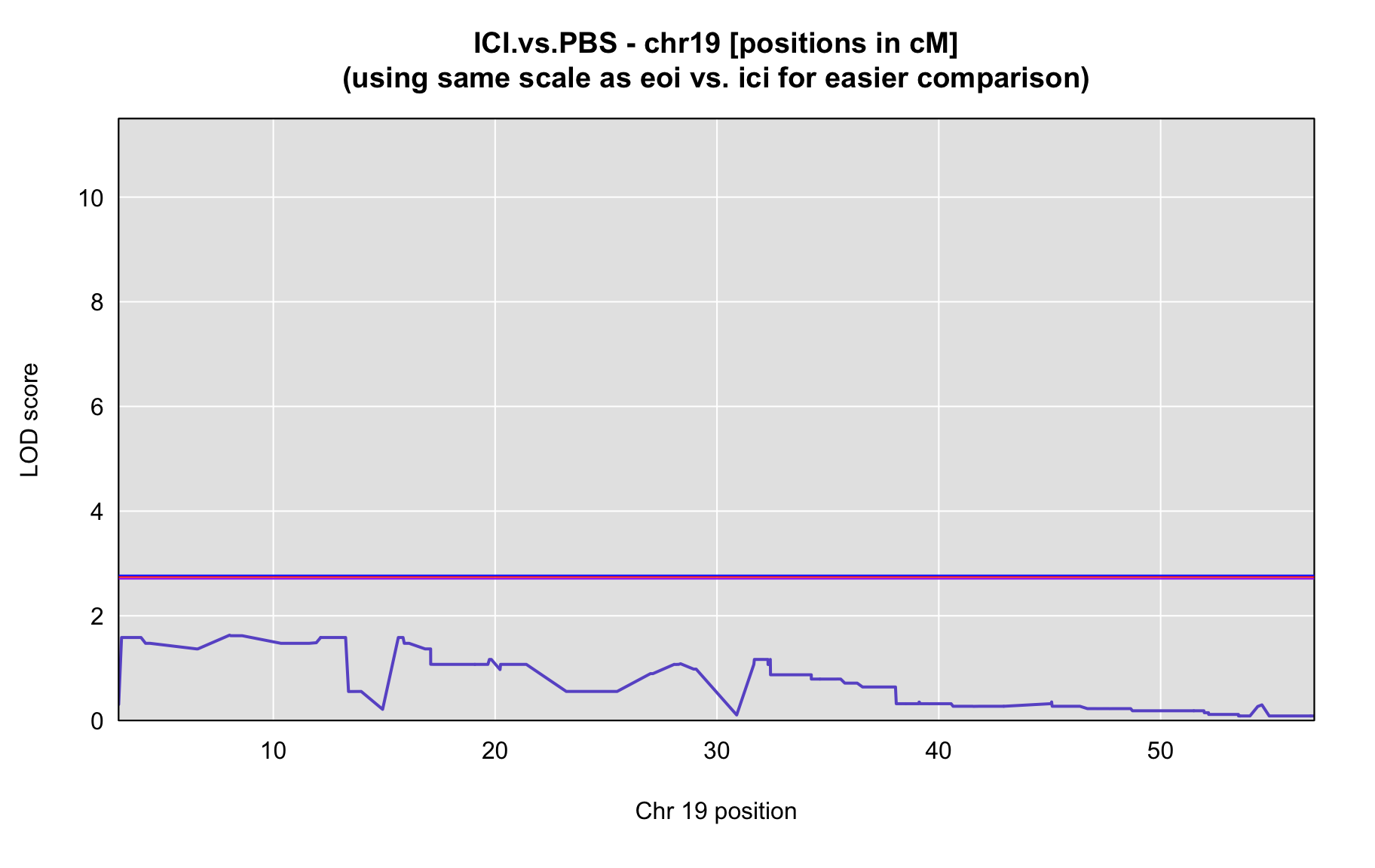
Megabase (MB)
print("peaks in MB positions")[1] “peaks in MB positions”
#peaks_mba <- find_peaks(out, gm$pmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
peaks_mba <- find_peaks(out, gm$pmap, threshold=1.5, drop=1.5)
if(nrow(peaks) >0){
peaks_mba$marker <- find_marker(gm$pmap, chr=peaks_mba$chr,pos=peaks_mba$pos)
names(peaks_mba)[2] <- c("phenotype")
peaks_mba <- peaks_mba[-1]
#peaks_mbl <- list()
##corresponding info in Mb
#for(i in 1:nrow(peaks)){
# #lodindex <- peaks$lodindex[i]
# phenotype <- peaks$phenotype[i]
# chr <- as.character(peaks$chr[i])
# lod <- peaks$lod[i]
# mark <- peaks$marker[i]
# pos <- mapdf[mapdf$marker==mark,]$pmapdf
# ci_lo <- mapdfnd$pmapdf[which(mapdfnd$gmapdf == peaks$ci_lo[i] & mapdfnd$chr == peaks$chr[i])]
# ci_hi <- mapdfnd$pmapdf[which(mapdfnd$gmapdf == peaks$ci_hi[i] & mapdfnd$chr == peaks$chr[i])]
# peaks_mb=as.data.frame(cbind(phenotype, chr, pos, lod, ci_lo, ci_hi, mark))
# names(peaks_mb)[7] <- c("marker")
# peaks_mbl[[i]] <- peaks_mb
#}
#peaks_mba2 <- do.call(rbind, peaks_mbl)
#peaks_mba2 <- as.data.frame(peaks_mba)
#peaks_mba[,c("chr", "pos", "lod", "ci_lo", "ci_hi")] <- sapply(peaks_mba[,c("chr", "pos", "lod", "ci_lo", "ci_hi")], as.numeric)
rownames(peaks_mba) <- NULL
print(kable(peaks_mba, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
plot_lod_chr_mb<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in MB]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
ymx <- 11
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in MB]\n(using same scale as eoi vs. ici for easier comparison)"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
for(i in unique(peaks_mba$chr)){
#for (i in 1:nrow(peaks_mba)){
#plot_lod_chr_mb(out,gm$pmap, peaks_mba$chr[i])
plot_lod_chr_mb(out,gm$pmap,i)
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}| phenotype | chr | pos | lod | ci_lo | ci_hi | marker |
|---|---|---|---|---|---|---|
| ICI.vs.PBS | 8 | 75.15958 | 1.563047 | 3.964084 | 121.65538 | UNCHS023364 |
| ICI.vs.PBS | 13 | 33.67626 | 1.603866 | 5.935749 | 119.26793 | UNC22384241 |
| ICI.vs.PBS | 19 | 11.25598 | 1.629859 | 3.263752 | 59.37816 | UNC29893228 |


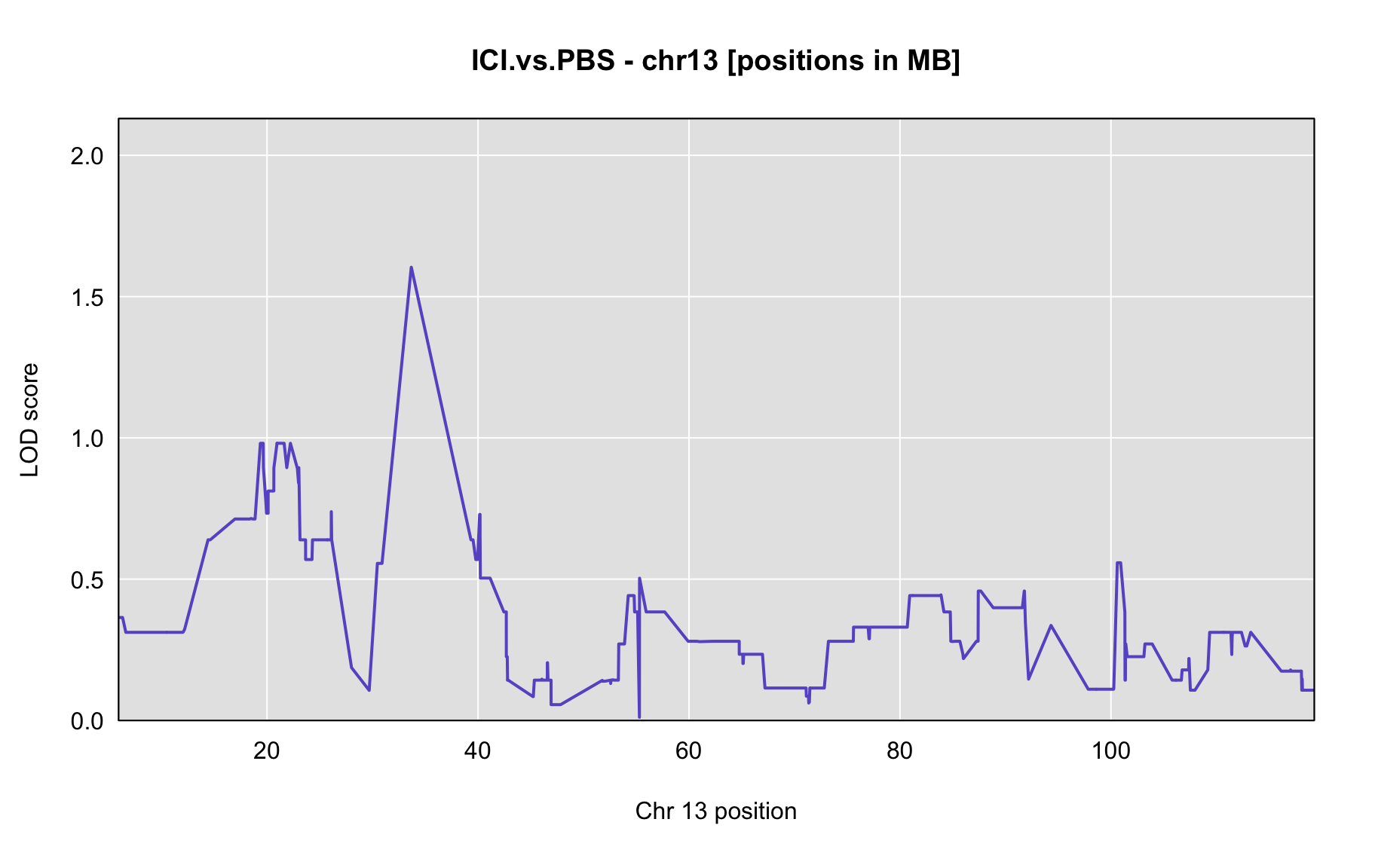
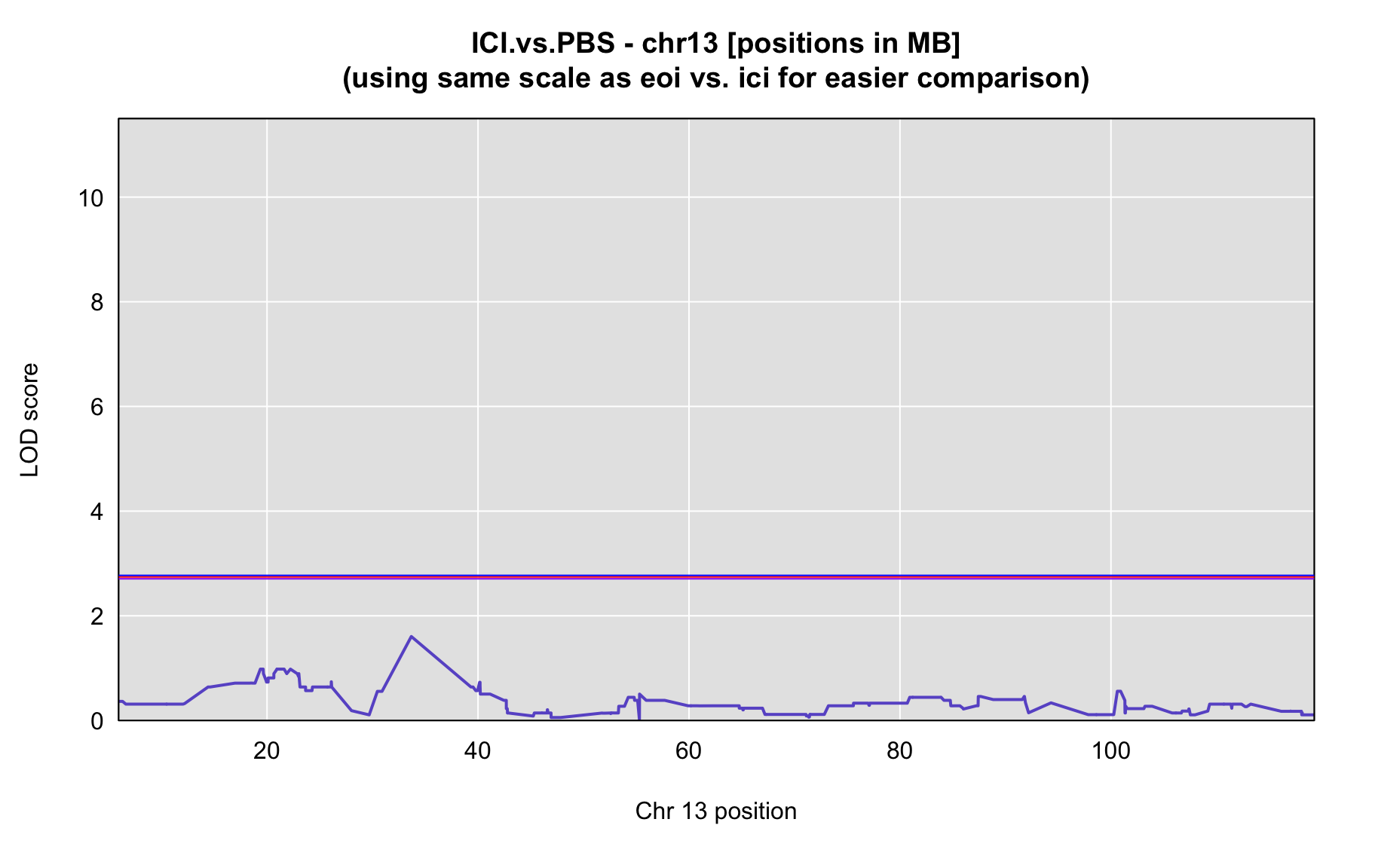
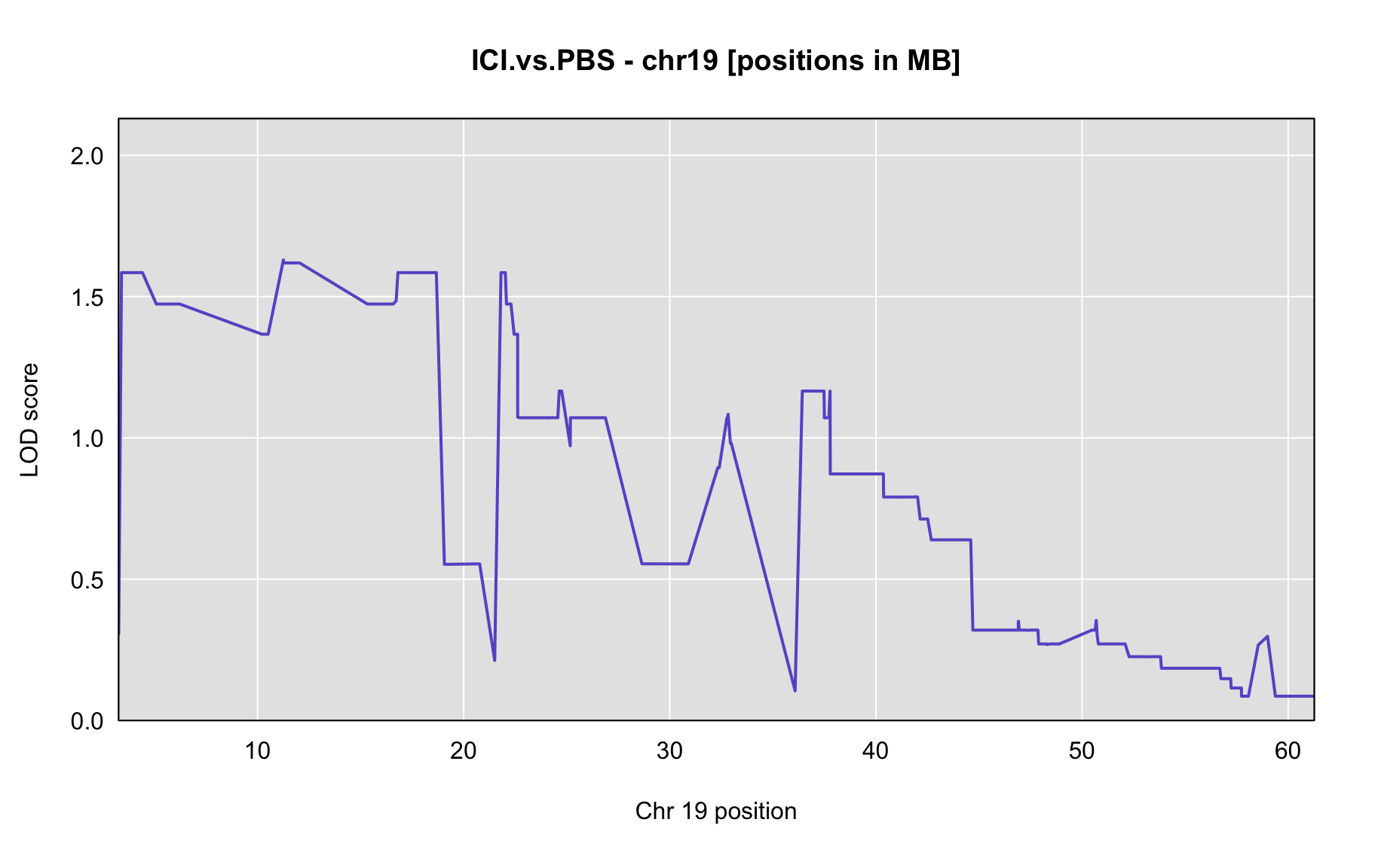

QTL effects
For each peak LOD location we give a list of gene
Chromsome 8
query_variants <- create_variant_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/cc_variants.sqlite")
query_genes <- create_gene_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/mouse_genes_mgi.sqlite")
if(nrow(peaks) >0){
for (i in 1){
#for (i in 1:1){
#Plot 1
#marker = find_marker(gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i])
#gp <- g[,marker]
#gp[gp==1] <- "AA"
#gp[gp==2] <- "AB"
#gp[gp==0] <- NA
#plot_pxg(gp, gm$covar[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
#title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
###dev.off()
g <- maxmarg(pr.qc, gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i], return_char=TRUE)
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/","qtl_effect_", i, ".png"))
#par(mar=c(4.1, 4.1, 1.5, 0.6))
plot_pxg(g, gm$covar[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
##dev.off()
chr = peaks$chr[i]
# Plot 2
pr_sub <- pull_genoprobint(pr.qc, gm$gmap, chr, c(peaks$ci_lo[i], peaks$ci_hi[i]))
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar)
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], Xcovar=Xcovar)
#coeff <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
#coeff_sub <- scan1coef(pr_sub[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
blup <- scan1blup(pr.qc[,chr], gm$covar[peaks$phenotype[i]])
blup_sub <- scan1blup(pr_sub[,chr], gm$covar[peaks$phenotype[i]])
write.csv(as.data.frame(blup_sub), paste0("data/ici.vs.pbs_blup_sub_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv"), quote=F)
#plot_coef(coeff,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i]," [scan1coeff; positions in cM])")
# )
#plot_coef(coeff_sub,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i],"; 1.5 LOD drop interval [scan1coeff; positions in cM] ) ")
# )
plot_coef(blup,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," [scan1blup; positions in cM])")
)
plot_coef(blup_sub,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i],"; 1.5 LOD drop interval [scan1blup; positions in cM])")
)
# Plot 3
#c2effB <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary", contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
#c2effBb <- scan1blup(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]],Xcovar=Xcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
#plot(c2effB, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
#plot(c2effBb, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
##last_coef <- unclass(c2effB)[nrow(c2effB),2:3] # last two coefficients
##for(t in seq(along=last_coef))
## axis(side=4, at=last_coef[t], names(last_coef)[t], tick=FALSE)
#Table 1
chr = peaks_mba$chr[i]
start=as.numeric(peaks_mba$ci_lo[i])
end=as.numeric(peaks_mba$ci_hi[i])
genesgss = query_genes(chr, start, end)
write.csv(genesgss, file=paste0("data/ici.vs.pbs_genes_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv"), quote=F)
rownames(genesgss) <- NULL
genesgss$strand_old = genesgss$strand
genesgss$strand[genesgss$strand=="+"] <- "positive"
genesgss$strand[genesgss$strand=="-"] <- "negative"
#genesgss <-
#table <-
#genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")] %>%
#kable(escape = F,align = c("ccccccccccc")) %>%
#kable_styling("striped", full_width = T) #%>%
#cat #%>%
#column_spec(1, bold=TRUE)
#
#print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], escape = F,align = c("ccccccccccc")))
print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], "html") %>% kable_styling("striped", full_width = T))
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}
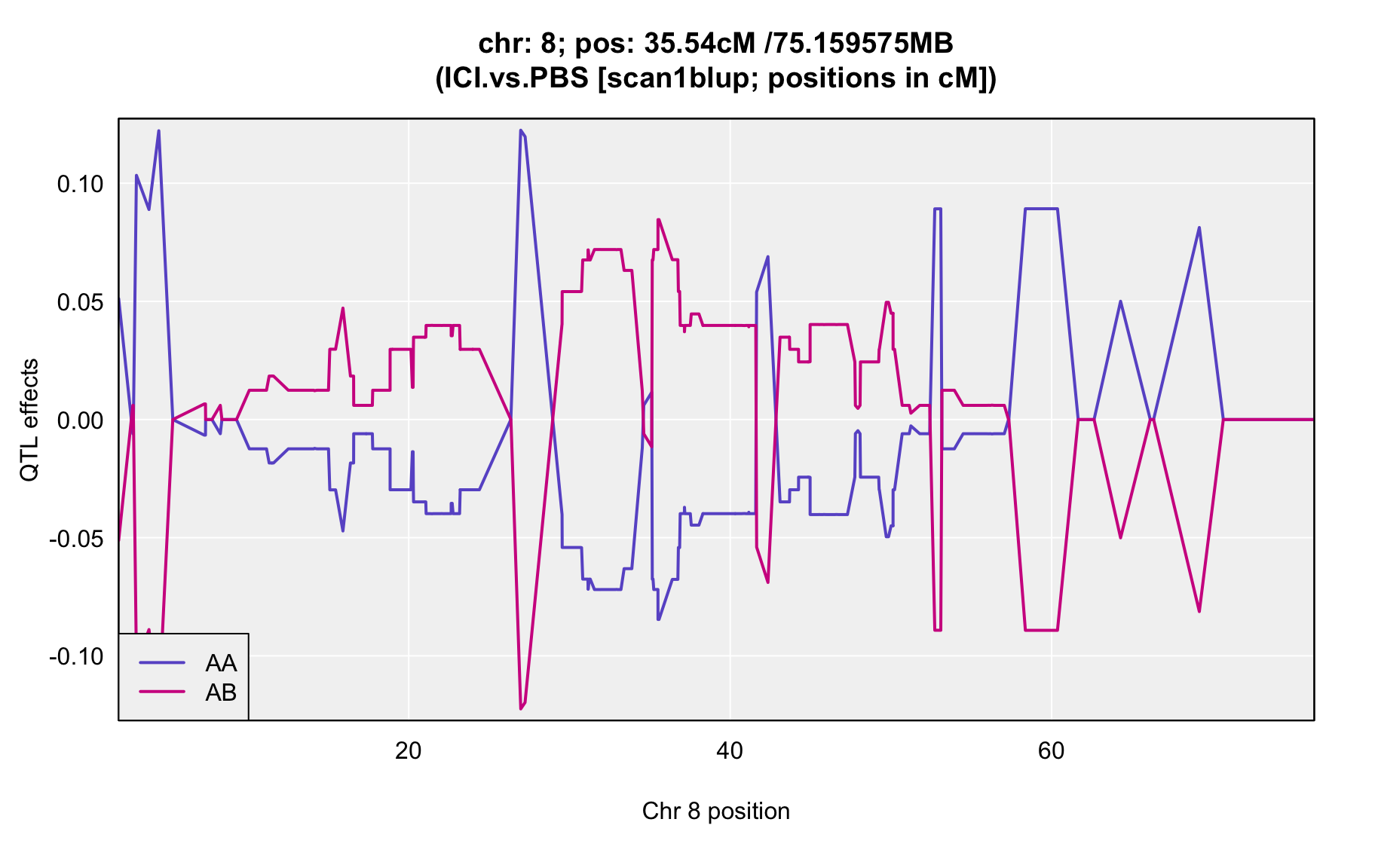
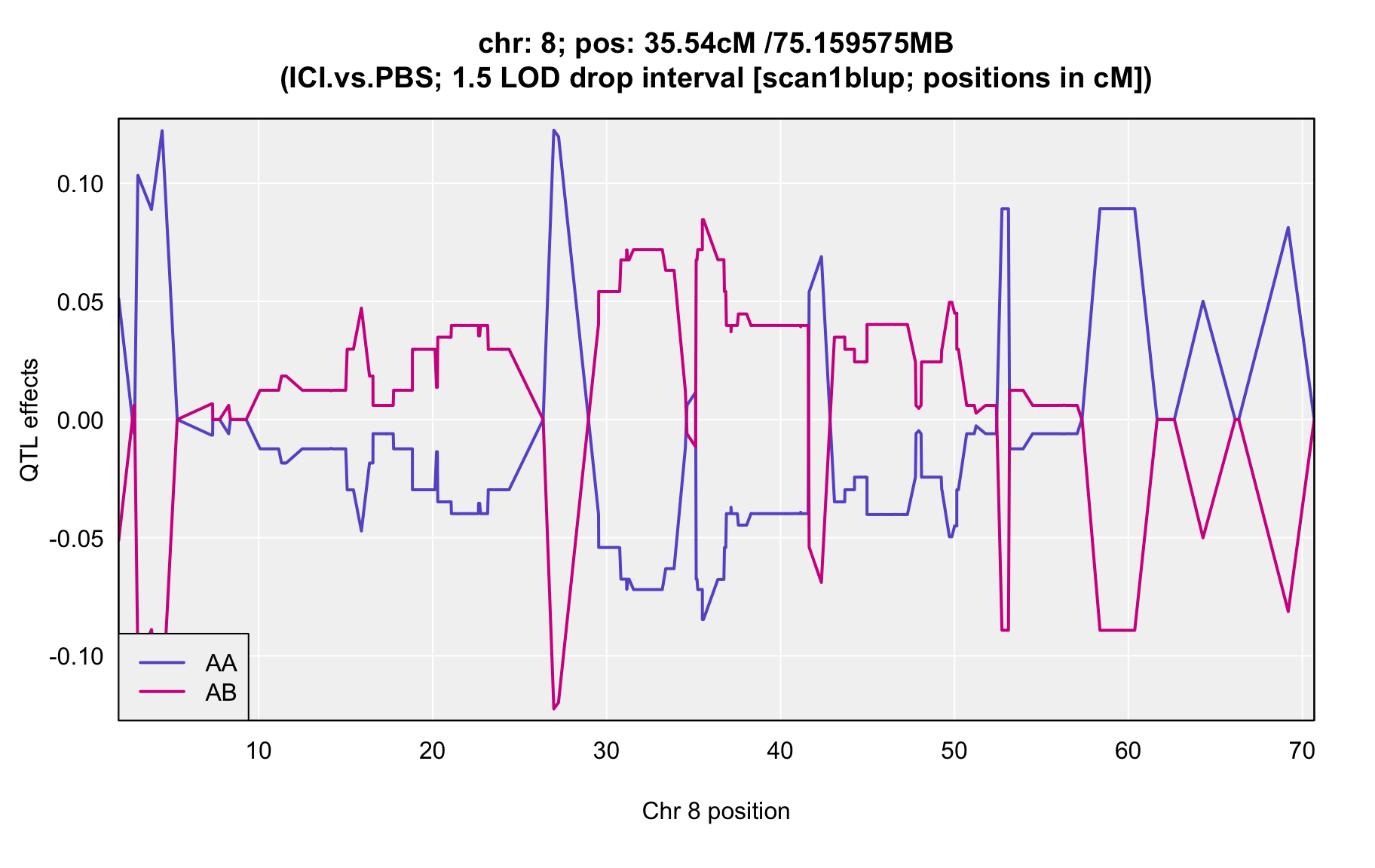
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 8 | gene | 3.965712 | 3.969482 | negative | MGI_C57BL6J_5622007 | Gm39122 | NCBI_Gene:105243110 | MGI:5622007 | lncRNA gene | predicted gene, 39122 |
| 8 | pseudogene | 3.994626 | 3.997509 | positive | MGI_C57BL6J_3645153 | Gm7389 | NCBI_Gene:664888,ENSEMBL:ENSMUSG00000097657 | MGI:3645153 | pseudogene | predicted gene 7389 |
| 8 | pseudogene | 4.016454 | 4.016882 | negative | MGI_C57BL6J_5753624 | Gm45048 | ENSEMBL:ENSMUSG00000109267 | MGI:5753624 | pseudogene | predicted gene 45048 |
| 8 | pseudogene | 4.018973 | 4.021489 | positive | MGI_C57BL6J_3647493 | Gm7397 | NCBI_Gene:664905,ENSEMBL:ENSMUSG00000109377 | MGI:3647493 | pseudogene | predicted gene 7397 |
| 8 | pseudogene | 4.034157 | 4.034747 | negative | MGI_C57BL6J_5753625 | Gm45049 | ENSEMBL:ENSMUSG00000109512 | MGI:5753625 | pseudogene | predicted gene 45049 |
| 8 | pseudogene | 4.039854 | 4.040421 | negative | MGI_C57BL6J_3647246 | Gm7407 | NCBI_Gene:664922,ENSEMBL:ENSMUSG00000109255 | MGI:3647246 | pseudogene | predicted gene 7407 |
| 8 | pseudogene | 4.087219 | 4.089901 | positive | MGI_C57BL6J_4418561 | Gm16589 | NCBI_Gene:631071,ENSEMBL:ENSMUSG00000074482 | MGI:4418561 | pseudogene | predicted gene 16589 |
| 8 | gene | 4.088149 | 4.089166 | negative | MGI_C57BL6J_5477244 | Gm26750 | ENSEMBL:ENSMUSG00000097544 | MGI:5477244 | lncRNA gene | predicted gene, 26750 |
| 8 | gene | 4.102784 | 4.105838 | negative | MGI_C57BL6J_1916392 | Cd209f | NCBI_Gene:69142,ENSEMBL:ENSMUSG00000051906 | MGI:1916392 | protein coding gene | CD209f antigen |
| 8 | gene | 4.115513 | 4.115765 | negative | MGI_C57BL6J_5453665 | Gm23888 | ENSEMBL:ENSMUSG00000084639 | MGI:5453665 | unclassified non-coding RNA gene | predicted gene, 23888 |
| 8 | gene | 4.134098 | 4.137707 | positive | MGI_C57BL6J_1917442 | Cd209g | NCBI_Gene:70192,ENSEMBL:ENSMUSG00000079168 | MGI:1917442 | protein coding gene | CD209g antigen |
| 8 | gene | 4.162372 | 4.206882 | negative | MGI_C57BL6J_1926149 | 4932443L11Rik | NCBI_Gene:78899,ENSEMBL:ENSMUSG00000086805 | MGI:1926149 | lncRNA gene | RIKEN cDNA 4932443L11 gene |
| 8 | gene | 4.166538 | 4.211257 | positive | MGI_C57BL6J_2442167 | Evi5l | NCBI_Gene:213027,ENSEMBL:ENSMUSG00000011832 | MGI:2442167 | protein coding gene | ecotropic viral integration site 5 like |
| 8 | gene | 4.209478 | 4.217535 | negative | MGI_C57BL6J_3605626 | Prr36 | NCBI_Gene:73072,ENSEMBL:ENSMUSG00000064125 | MGI:3605626 | protein coding gene | proline rich 36 |
| 8 | gene | 4.224269 | 4.237470 | positive | MGI_C57BL6J_1919517 | Lrrc8e | NCBI_Gene:72267,ENSEMBL:ENSMUSG00000046589 | MGI:1919517 | protein coding gene | leucine rich repeat containing 8 family, member E |
| 8 | gene | 4.238740 | 4.247897 | positive | MGI_C57BL6J_1346871 | Map2k7 | NCBI_Gene:26400,ENSEMBL:ENSMUSG00000002948 | MGI:1346871 | protein coding gene | mitogen-activated protein kinase kinase 7 |
| 8 | gene | 4.238865 | 4.251423 | positive | MGI_C57BL6J_6121499 | Gm49320 | ENSEMBL:ENSMUSG00000109061 | MGI:6121499 | protein coding gene | predicted gene, 49320 |
| 8 | gene | 4.248214 | 4.251423 | positive | MGI_C57BL6J_3833469 | Tgfbr3l | NCBI_Gene:100044509,ENSEMBL:ENSMUSG00000089736 | MGI:3833469 | protein coding gene | transforming growth factor, beta receptor III-like |
| 8 | gene | 4.253080 | 4.256220 | positive | MGI_C57BL6J_1914861 | Snapc2 | NCBI_Gene:102209,ENSEMBL:ENSMUSG00000011837 | MGI:1914861 | protein coding gene | small nuclear RNA activating complex, polypeptide 2 |
| 8 | gene | 4.257646 | 4.259274 | negative | MGI_C57BL6J_88566 | Ctxn1 | NCBI_Gene:330695,ENSEMBL:ENSMUSG00000048644 | MGI:88566 | protein coding gene | cortexin 1 |
| 8 | gene | 4.259731 | 4.275913 | negative | MGI_C57BL6J_1343262 | Timm44 | NCBI_Gene:21856,ENSEMBL:ENSMUSG00000002949 | MGI:1343262 | protein coding gene | translocase of inner mitochondrial membrane 44 |
| 8 | gene | 4.274853 | 4.275136 | positive | MGI_C57BL6J_5455879 | Gm26102 | ENSEMBL:ENSMUSG00000087876 | MGI:5455879 | unclassified non-coding RNA gene | predicted gene, 26102 |
| 8 | gene | 4.284781 | 4.325413 | negative | MGI_C57BL6J_1100851 | Elavl1 | NCBI_Gene:15568,ENSEMBL:ENSMUSG00000040028 | MGI:1100851 | protein coding gene | ELAV (embryonic lethal, abnormal vision)-like 1 (Hu antigen R) |
| 8 | gene | 4.325160 | 4.360020 | positive | MGI_C57BL6J_1099448 | Ccl25 | NCBI_Gene:20300,ENSEMBL:ENSMUSG00000023235 | MGI:1099448 | protein coding gene | chemokine (C-C motif) ligand 25 |
| 8 | pseudogene | 4.362298 | 4.362654 | negative | MGI_C57BL6J_5753445 | Gm44869 | ENSEMBL:ENSMUSG00000108818 | MGI:5753445 | pseudogene | predicted gene 44869 |
| 8 | pseudogene | 4.368596 | 4.402845 | negative | MGI_C57BL6J_5753694 | Gm45118 | ENSEMBL:ENSMUSG00000109356 | MGI:5753694 | pseudogene | predicted gene 45118 |
| 8 | pseudogene | 4.368797 | 4.392170 | positive | MGI_C57BL6J_3644530 | Gm7434 | NCBI_Gene:664977,ENSEMBL:ENSMUSG00000109241 | MGI:3644530 | pseudogene | predicted gene 7434 |
| 8 | gene | 4.399238 | 4.401011 | positive | MGI_C57BL6J_5753693 | Gm45117 | ENSEMBL:ENSMUSG00000109397 | MGI:5753693 | unclassified gene | predicted gene 45117 |
| 8 | pseudogene | 4.465542 | 4.466206 | positive | MGI_C57BL6J_3648421 | Gm6070 | NCBI_Gene:606543,ENSEMBL:ENSMUSG00000108860 | MGI:3648421 | pseudogene | predicted gene 6070 |
| 8 | gene | 4.492979 | 4.531680 | positive | MGI_C57BL6J_1914510 | Cers4 | NCBI_Gene:67260,ENSEMBL:ENSMUSG00000008206 | MGI:1914510 | protein coding gene | ceramide synthase 4 |
| 8 | gene | 4.521912 | 4.522125 | negative | MGI_C57BL6J_5610184 | Gm36956 | ENSEMBL:ENSMUSG00000102201 | MGI:5610184 | unclassified gene | predicted gene, 36956 |
| 8 | gene | 4.527036 | 4.529608 | positive | MGI_C57BL6J_2442727 | A830058I18Rik | NA | NA | unclassified gene | RIKEN cDNA A830058I18 gene |
| 8 | pseudogene | 4.529615 | 4.538190 | negative | MGI_C57BL6J_3757941 | Vmn2r-ps89 | NCBI_Gene:674360,ENSEMBL:ENSMUSG00000108829 | MGI:3757941 | pseudogene | vomeronasal 2, receptor, pseudogene 89 |
| 8 | gene | 4.613167 | 4.632327 | positive | MGI_C57BL6J_2385298 | Zfp958 | NCBI_Gene:233987,ENSEMBL:ENSMUSG00000058748 | MGI:2385298 | protein coding gene | zinc finger protein 958 |
| 8 | pseudogene | 4.641372 | 4.643729 | positive | MGI_C57BL6J_3645148 | Gm7451 | NCBI_Gene:665021,ENSEMBL:ENSMUSG00000109489 | MGI:3645148 | pseudogene | predicted gene 7451 |
| 8 | pseudogene | 4.651995 | 4.653615 | positive | MGI_C57BL6J_5589661 | Gm30502 | NCBI_Gene:102632427 | MGI:5589661 | pseudogene | predicted gene, 30502 |
| 8 | pseudogene | 4.652064 | 4.652477 | positive | MGI_C57BL6J_5753800 | Gm45224 | ENSEMBL:ENSMUSG00000109448 | MGI:5753800 | pseudogene | predicted gene 45224 |
| 8 | gene | 4.656156 | 4.665350 | negative | MGI_C57BL6J_5622008 | Gm39123 | NCBI_Gene:105243112 | MGI:5622008 | lncRNA gene | predicted gene, 39123 |
| 8 | gene | 4.677526 | 4.688740 | positive | MGI_C57BL6J_3643419 | Gm6410 | NCBI_Gene:623223,ENSEMBL:ENSMUSG00000097308 | MGI:3643419 | lncRNA gene | predicted gene 6410 |
| 8 | gene | 4.677539 | 4.678365 | negative | MGI_C57BL6J_3645338 | Gm7461 | ENSEMBL:ENSMUSG00000048484 | MGI:3645338 | unclassified non-coding RNA gene | predicted gene 7461 |
| 8 | pseudogene | 4.713104 | 4.715351 | positive | MGI_C57BL6J_3780845 | Gm2676 | NCBI_Gene:100040234,ENSEMBL:ENSMUSG00000109055 | MGI:3780845 | pseudogene | predicted gene 2676 |
| 8 | pseudogene | 4.722606 | 4.722836 | negative | MGI_C57BL6J_5753535 | Gm44959 | ENSEMBL:ENSMUSG00000108962 | MGI:5753535 | pseudogene | predicted gene 44959 |
| 8 | gene | 4.724636 | 4.728105 | negative | MGI_C57BL6J_5622009 | Gm39124 | NCBI_Gene:105243113 | MGI:5622009 | lncRNA gene | predicted gene, 39124 |
| 8 | gene | 4.735976 | 4.779567 | negative | MGI_C57BL6J_1338802 | Shcbp1 | NCBI_Gene:20419,ENSEMBL:ENSMUSG00000022322 | MGI:1338802 | protein coding gene | Shc SH2-domain binding protein 1 |
| 8 | pseudogene | 4.777802 | 4.778940 | positive | MGI_C57BL6J_3647382 | Gm6334 | NCBI_Gene:622568,ENSEMBL:ENSMUSG00000109462 | MGI:3647382 | pseudogene | predicted gene 6334 |
| 8 | pseudogene | 4.812530 | 4.812670 | positive | MGI_C57BL6J_5753536 | Gm44960 | ENSEMBL:ENSMUSG00000109034 | MGI:5753536 | pseudogene | predicted gene 44960 |
| 8 | pseudogene | 4.812817 | 4.822699 | negative | MGI_C57BL6J_3779867 | Gm9457 | NCBI_Gene:669393,ENSEMBL:ENSMUSG00000095600 | MGI:3779867 | pseudogene | predicted gene 9457 |
| 8 | pseudogene | 4.821035 | 4.821230 | negative | MGI_C57BL6J_5753537 | Gm44961 | ENSEMBL:ENSMUSG00000109063 | MGI:5753537 | pseudogene | predicted gene 44961 |
| 8 | pseudogene | 4.861124 | 4.861718 | positive | MGI_C57BL6J_3648110 | Rpl21-ps14 | NCBI_Gene:631359,ENSEMBL:ENSMUSG00000062152 | MGI:3648110 | pseudogene | ribosomal protein L21, pseudogene 14 |
| 8 | gene | 5.069259 | 5.069377 | negative | MGI_C57BL6J_4421941 | n-R5s93 | ENSEMBL:ENSMUSG00000096219 | MGI:4421941 | rRNA gene | nuclear encoded rRNA 5S 93 |
| 8 | pseudogene | 5.079294 | 5.080996 | positive | MGI_C57BL6J_3645094 | Gm5739 | NCBI_Gene:436026,ENSEMBL:ENSMUSG00000109295 | MGI:3645094 | pseudogene | predicted gene 5739 |
| 8 | gene | 5.083219 | 5.105351 | negative | MGI_C57BL6J_1201406 | Slc10a2 | NCBI_Gene:20494,ENSEMBL:ENSMUSG00000023073 | MGI:1201406 | protein coding gene | solute carrier family 10, member 2 |
| 8 | pseudogene | 5.117281 | 5.118859 | positive | MGI_C57BL6J_3647902 | Gm5605 | NCBI_Gene:100417643,ENSEMBL:ENSMUSG00000109181 | MGI:3647902 | pseudogene | predicted gene 5605 |
| 8 | gene | 5.332995 | 5.333113 | negative | MGI_C57BL6J_4421942 | n-R5s94 | ENSEMBL:ENSMUSG00000064507 | MGI:4421942 | rRNA gene | nuclear encoded rRNA 5S 94 |
| 8 | gene | 5.436984 | 5.440990 | negative | MGI_C57BL6J_5753269 | Gm44693 | ENSEMBL:ENSMUSG00000109313 | MGI:5753269 | lncRNA gene | predicted gene 44693 |
| 8 | pseudogene | 5.527689 | 5.528542 | positive | MGI_C57BL6J_3646017 | Gm6426 | NCBI_Gene:623409,ENSEMBL:ENSMUSG00000109266 | MGI:3646017 | pseudogene | predicted gene 6426 |
| 8 | pseudogene | 5.639580 | 5.641011 | positive | MGI_C57BL6J_3037698 | Gm1840 | NCBI_Gene:669429,ENSEMBL:ENSMUSG00000043192 | MGI:3037698 | pseudogene | predicted gene 1840 |
| 8 | gene | 5.705427 | 5.705495 | positive | MGI_C57BL6J_5452342 | Gm22565 | ENSEMBL:ENSMUSG00000093281 | MGI:5452342 | miRNA gene | predicted gene, 22565 |
| 8 | pseudogene | 5.993981 | 5.994723 | negative | MGI_C57BL6J_3780903 | Gm2734 | NCBI_Gene:100040360,ENSEMBL:ENSMUSG00000109069 | MGI:3780903 | pseudogene | predicted gene 2734 |
| 8 | gene | 6.034867 | 6.036262 | positive | MGI_C57BL6J_5753419 | Gm44843 | ENSEMBL:ENSMUSG00000109518 | MGI:5753419 | unclassified gene | predicted gene 44843 |
| 8 | gene | 6.218668 | 6.222039 | positive | MGI_C57BL6J_5753418 | Gm44842 | ENSEMBL:ENSMUSG00000109534 | MGI:5753418 | unclassified gene | predicted gene 44842 |
| 8 | gene | 6.223819 | 6.223962 | negative | MGI_C57BL6J_5454065 | Gm24288 | ENSEMBL:ENSMUSG00000088255 | MGI:5454065 | snoRNA gene | predicted gene, 24288 |
| 8 | pseudogene | 6.437744 | 6.438079 | positive | MGI_C57BL6J_5753420 | Gm44844 | ENSEMBL:ENSMUSG00000109543 | MGI:5753420 | pseudogene | predicted gene 44844 |
| 8 | pseudogene | 6.830814 | 6.830978 | negative | MGI_C57BL6J_5753485 | Gm44909 | ENSEMBL:ENSMUSG00000109457 | MGI:5753485 | pseudogene | predicted gene 44909 |
| 8 | pseudogene | 7.001878 | 7.002803 | negative | MGI_C57BL6J_3643243 | Gm7506 | NCBI_Gene:665127,ENSEMBL:ENSMUSG00000109463 | MGI:3643243 | pseudogene | predicted gene 7506 |
| 8 | pseudogene | 7.040157 | 7.041218 | positive | MGI_C57BL6J_5753487 | Gm44911 | ENSEMBL:ENSMUSG00000108852 | MGI:5753487 | pseudogene | predicted gene 44911 |
| 8 | pseudogene | 7.132569 | 7.132964 | negative | MGI_C57BL6J_5753486 | Gm44910 | ENSEMBL:ENSMUSG00000109538 | MGI:5753486 | pseudogene | predicted gene 44910 |
| 8 | pseudogene | 7.231234 | 7.231499 | positive | MGI_C57BL6J_5753484 | Gm44908 | ENSEMBL:ENSMUSG00000108914 | MGI:5753484 | pseudogene | predicted gene 44908 |
| 8 | gene | 7.380948 | 7.428572 | positive | MGI_C57BL6J_4938042 | Gm17215 | NCBI_Gene:102632928,ENSEMBL:ENSMUSG00000092092 | MGI:4938042 | lncRNA gene | predicted gene 17215 |
| 8 | gene | 7.420552 | 7.580231 | negative | MGI_C57BL6J_5590113 | Gm30954 | NCBI_Gene:102633030,ENSEMBL:ENSMUSG00000109260 | MGI:5590113 | lncRNA gene | predicted gene, 30954 |
| 8 | gene | 7.428222 | 7.428328 | negative | MGI_C57BL6J_5456204 | Gm26427 | ENSEMBL:ENSMUSG00000093992 | MGI:5456204 | snRNA gene | predicted gene, 26427 |
| 8 | gene | 7.521636 | 7.521834 | positive | MGI_C57BL6J_5530967 | Gm27585 | ENSEMBL:ENSMUSG00000098667 | MGI:5530967 | unclassified non-coding RNA gene | predicted gene, 27585 |
| 8 | gene | 7.525256 | 7.525457 | positive | MGI_C57BL6J_5530927 | Gm27545 | ENSEMBL:ENSMUSG00000098978 | MGI:5530927 | unclassified non-coding RNA gene | predicted gene, 27545 |
| 8 | gene | 7.638764 | 7.638892 | positive | MGI_C57BL6J_5454946 | Gm25169 | ENSEMBL:ENSMUSG00000077378 | MGI:5454946 | snoRNA gene | predicted gene, 25169 |
| 8 | gene | 7.855847 | 7.863406 | positive | MGI_C57BL6J_5578968 | Gm28262 | ENSEMBL:ENSMUSG00000099890 | MGI:5578968 | lncRNA gene | predicted gene 28262 |
| 8 | gene | 7.985368 | 7.988466 | positive | MGI_C57BL6J_5753736 | Gm45160 | ENSEMBL:ENSMUSG00000109316 | MGI:5753736 | unclassified gene | predicted gene 45160 |
| 8 | gene | 8.044577 | 8.484730 | negative | MGI_C57BL6J_5590294 | Gm31135 | NCBI_Gene:102633271,ENSEMBL:ENSMUSG00000109127 | MGI:5590294 | lncRNA gene | predicted gene, 31135 |
| 8 | gene | 8.066666 | 8.093355 | positive | MGI_C57BL6J_5590560 | Gm31401 | NCBI_Gene:102633617,ENSEMBL:ENSMUSG00000109566 | MGI:5590560 | lncRNA gene | predicted gene, 31401 |
| 8 | gene | 8.334081 | 8.336213 | negative | MGI_C57BL6J_5753652 | Gm45076 | ENSEMBL:ENSMUSG00000109134 | MGI:5753652 | lncRNA gene | predicted gene 45076 |
| 8 | gene | 8.335077 | 8.342428 | positive | MGI_C57BL6J_2443964 | A630009H07Rik | ENSEMBL:ENSMUSG00000109056 | MGI:2443964 | lncRNA gene | RIKEN cDNA A630009H07 gene |
| 8 | gene | 8.377090 | 8.391702 | positive | MGI_C57BL6J_5590502 | Gm31343 | ENSEMBL:ENSMUSG00000109515 | MGI:5590502 | lncRNA gene | predicted gene, 31343 |
| 8 | pseudogene | 8.454832 | 8.455441 | negative | MGI_C57BL6J_3780615 | Gm2448 | NCBI_Gene:100416079,ENSEMBL:ENSMUSG00000108913 | MGI:3780615 | pseudogene | predicted gene 2448 |
| 8 | gene | 8.488302 | 8.495535 | positive | MGI_C57BL6J_1925967 | D530014G21Rik | ENSEMBL:ENSMUSG00000108941 | MGI:1925967 | lncRNA gene | RIKEN cDNA D530014G21 gene |
| 8 | gene | 8.576522 | 8.580342 | positive | MGI_C57BL6J_5590622 | Gm31463 | NCBI_Gene:102633702,ENSEMBL:ENSMUSG00000108932 | MGI:5590622 | lncRNA gene | predicted gene, 31463 |
| 8 | gene | 8.612264 | 8.615862 | positive | MGI_C57BL6J_5753651 | Gm45075 | ENSEMBL:ENSMUSG00000109436 | MGI:5753651 | lncRNA gene | predicted gene 45075 |
| 8 | gene | 8.617235 | 8.661260 | negative | MGI_C57BL6J_105097 | Efnb2 | NCBI_Gene:13642,ENSEMBL:ENSMUSG00000001300 | MGI:105097 | protein coding gene | ephrin B2 |
| 8 | gene | 8.629175 | 8.635073 | negative | MGI_C57BL6J_2442650 | E230020D15Rik | ENSEMBL:ENSMUSG00000108912 | MGI:2442650 | unclassified gene | RIKEN cDNA E230020D15 gene |
| 8 | gene | 8.637966 | 8.639205 | negative | MGI_C57BL6J_5753650 | Gm45074 | ENSEMBL:ENSMUSG00000109568 | MGI:5753650 | unclassified gene | predicted gene 45074 |
| 8 | gene | 8.642766 | 8.647012 | negative | MGI_C57BL6J_5753649 | Gm45073 | ENSEMBL:ENSMUSG00000109438 | MGI:5753649 | unclassified gene | predicted gene 45073 |
| 8 | gene | 8.661801 | 8.664728 | positive | MGI_C57BL6J_1913969 | 4921522P10Rik | ENSEMBL:ENSMUSG00000040467 | MGI:1913969 | lncRNA gene | RIKEN cDNA 4921522P10 gene |
| 8 | gene | 8.665075 | 8.690549 | negative | MGI_C57BL6J_2442985 | Arglu1 | NCBI_Gene:234023,ENSEMBL:ENSMUSG00000040459 | MGI:2442985 | protein coding gene | arginine and glutamate rich 1 |
| 8 | gene | 8.689665 | 8.691821 | positive | MGI_C57BL6J_3642584 | B930078G14Rik | NA | NA | protein coding gene | RIKEN cDNA B930078G14 gene |
| 8 | gene | 8.690093 | 8.690162 | negative | MGI_C57BL6J_5531130 | Mir7654 | miRBase:MI0024994,NCBI_Gene:102465762,ENSEMBL:ENSMUSG00000098917 | MGI:5531130 | miRNA gene | microRNA 7654 |
| 8 | pseudogene | 8.711484 | 8.712614 | positive | MGI_C57BL6J_5010578 | Gm18393 | NCBI_Gene:100417089,ENSEMBL:ENSMUSG00000109117 | MGI:5010578 | pseudogene | predicted gene, 18393 |
| 8 | gene | 8.716924 | 8.719622 | positive | MGI_C57BL6J_2142723 | AU045717 | NA | NA | unclassified gene | expressed sequence AU045717 |
| 8 | gene | 8.716924 | 8.721308 | negative | MGI_C57BL6J_5753197 | Gm44621 | ENSEMBL:ENSMUSG00000109190 | MGI:5753197 | lncRNA gene | predicted gene 44621 |
| 8 | gene | 8.722957 | 8.755164 | positive | MGI_C57BL6J_5622012 | Gm39127 | NCBI_Gene:105243116 | MGI:5622012 | lncRNA gene | predicted gene, 39127 |
| 8 | gene | 8.727111 | 8.853647 | positive | MGI_C57BL6J_5753202 | Gm44626 | ENSEMBL:ENSMUSG00000109371 | MGI:5753202 | lncRNA gene | predicted gene 44626 |
| 8 | gene | 8.735970 | 8.755337 | positive | MGI_C57BL6J_5753198 | Gm44622 | ENSEMBL:ENSMUSG00000109062 | MGI:5753198 | lncRNA gene | predicted gene 44622 |
| 8 | gene | 8.831447 | 8.831907 | negative | MGI_C57BL6J_1922402 | 4930540A11Rik | NA | NA | unclassified gene | RIKEN cDNA 4930540A11 gene |
| 8 | gene | 8.920937 | 8.932699 | negative | MGI_C57BL6J_5753091 | Gm44515 | ENSEMBL:ENSMUSG00000109346 | MGI:5753091 | lncRNA gene | predicted gene 44515 |
| 8 | gene | 9.125412 | 9.132291 | negative | MGI_C57BL6J_1918499 | 4933430N04Rik | NCBI_Gene:71249,ENSEMBL:ENSMUSG00000108848 | MGI:1918499 | lncRNA gene | RIKEN cDNA 4933430N04 gene |
| 8 | gene | 9.141110 | 9.144570 | negative | MGI_C57BL6J_5590969 | Gm31810 | NCBI_Gene:102634157 | MGI:5590969 | lncRNA gene | predicted gene, 31810 |
| 8 | gene | 9.144669 | 9.149050 | positive | MGI_C57BL6J_1921938 | 4930453L07Rik | NCBI_Gene:74688,ENSEMBL:ENSMUSG00000109215 | MGI:1921938 | lncRNA gene | RIKEN cDNA 4930453L07 gene |
| 8 | gene | 9.148011 | 9.152138 | negative | MGI_C57BL6J_5590885 | Gm31726 | NCBI_Gene:102634045 | MGI:5590885 | lncRNA gene | predicted gene, 31726 |
| 8 | gene | 9.152600 | 9.157221 | negative | MGI_C57BL6J_5753092 | Gm44516 | ENSEMBL:ENSMUSG00000109124 | MGI:5753092 | lncRNA gene | predicted gene 44516 |
| 8 | gene | 9.167517 | 9.174793 | positive | MGI_C57BL6J_5622013 | Gm39128 | NCBI_Gene:105243117,ENSEMBL:ENSMUSG00000109064 | MGI:5622013 | lncRNA gene | predicted gene, 39128 |
| 8 | gene | 9.205902 | 9.773322 | negative | MGI_C57BL6J_2142765 | Fam155a | NCBI_Gene:270028,ENSEMBL:ENSMUSG00000079157 | MGI:2142765 | protein coding gene | family with sequence similarity 155, member A |
| 8 | gene | 9.213097 | 9.217410 | negative | MGI_C57BL6J_5753118 | Gm44542 | ENSEMBL:ENSMUSG00000109040 | MGI:5753118 | unclassified gene | predicted gene 44542 |
| 8 | gene | 9.342220 | 9.345126 | negative | MGI_C57BL6J_5753117 | Gm44541 | ENSEMBL:ENSMUSG00000109286 | MGI:5753117 | unclassified gene | predicted gene 44541 |
| 8 | gene | 9.370957 | 9.375326 | negative | MGI_C57BL6J_5753094 | Gm44518 | ENSEMBL:ENSMUSG00000109385 | MGI:5753094 | unclassified gene | predicted gene 44518 |
| 8 | gene | 9.459092 | 9.461218 | positive | MGI_C57BL6J_1923047 | 4930435N07Rik | ENSEMBL:ENSMUSG00000109551 | MGI:1923047 | unclassified gene | RIKEN cDNA 4930435N07 gene |
| 8 | gene | 9.568132 | 9.568249 | negative | MGI_C57BL6J_4421943 | n-R5s95 | ENSEMBL:ENSMUSG00000064618 | MGI:4421943 | rRNA gene | nuclear encoded rRNA 5S 95 |
| 8 | gene | 9.595109 | 9.596945 | negative | MGI_C57BL6J_5753373 | Gm44797 | ENSEMBL:ENSMUSG00000109359 | MGI:5753373 | unclassified gene | predicted gene 44797 |
| 8 | gene | 9.598799 | 9.600653 | negative | MGI_C57BL6J_5753376 | Gm44800 | ENSEMBL:ENSMUSG00000109418 | MGI:5753376 | unclassified gene | predicted gene 44800 |
| 8 | gene | 9.603823 | 9.605882 | negative | MGI_C57BL6J_5753377 | Gm44801 | ENSEMBL:ENSMUSG00000109529 | MGI:5753377 | unclassified gene | predicted gene 44801 |
| 8 | gene | 9.618153 | 9.620562 | negative | MGI_C57BL6J_5753374 | Gm44798 | ENSEMBL:ENSMUSG00000109139 | MGI:5753374 | unclassified gene | predicted gene 44798 |
| 8 | gene | 9.658248 | 9.659449 | negative | MGI_C57BL6J_5753375 | Gm44799 | ENSEMBL:ENSMUSG00000109095 | MGI:5753375 | unclassified gene | predicted gene 44799 |
| 8 | gene | 9.685829 | 9.690276 | negative | MGI_C57BL6J_1920130 | 2900027M19Rik | ENSEMBL:ENSMUSG00000109162 | MGI:1920130 | unclassified non-coding RNA gene | RIKEN cDNA 2900027M19 gene |
| 8 | gene | 9.709171 | 9.711086 | negative | MGI_C57BL6J_5753378 | Gm44802 | ENSEMBL:ENSMUSG00000108918 | MGI:5753378 | unclassified gene | predicted gene 44802 |
| 8 | gene | 9.770728 | 9.770880 | negative | MGI_C57BL6J_6096189 | Gm47312 | ENSEMBL:ENSMUSG00000105098 | MGI:6096189 | miRNA gene | predicted gene, 47312 |
| 8 | gene | 9.800194 | 9.811974 | positive | MGI_C57BL6J_5753337 | Gm44761 | ENSEMBL:ENSMUSG00000109183 | MGI:5753337 | lncRNA gene | predicted gene 44761 |
| 8 | gene | 9.864251 | 9.870315 | positive | MGI_C57BL6J_3642099 | Gm10217 | ENSEMBL:ENSMUSG00000067627 | MGI:3642099 | protein coding gene | predicted gene 10217 |
| 8 | gene | 9.875530 | 9.919463 | negative | MGI_C57BL6J_3708768 | Gm10067 | ENSEMBL:ENSMUSG00000095279 | MGI:3708768 | lncRNA gene | predicted gene 10067 |
| 8 | gene | 9.969049 | 9.977686 | negative | MGI_C57BL6J_1335098 | Lig4 | NCBI_Gene:319583,ENSEMBL:ENSMUSG00000049717 | MGI:1335098 | protein coding gene | ligase IV, DNA, ATP-dependent |
| 8 | gene | 9.977707 | 9.992155 | positive | MGI_C57BL6J_1916154 | Abhd13 | NCBI_Gene:68904,ENSEMBL:ENSMUSG00000040396 | MGI:1916154 | protein coding gene | abhydrolase domain containing 13 |
| 8 | gene | 10.003513 | 10.007539 | negative | MGI_C57BL6J_3642447 | Gm10703 | NA | NA | protein coding gene | predicted gene 10703 |
| 8 | gene | 10.006203 | 10.039072 | positive | MGI_C57BL6J_1344376 | Tnfsf13b | NCBI_Gene:24099,ENSEMBL:ENSMUSG00000031497 | MGI:1344376 | protein coding gene | tumor necrosis factor (ligand) superfamily, member 13b |
| 8 | gene | 10.016427 | 10.019527 | negative | MGI_C57BL6J_5753365 | Gm44789 | ENSEMBL:ENSMUSG00000109054 | MGI:5753365 | unclassified gene | predicted gene 44789 |
| 8 | gene | 10.065054 | 10.119632 | positive | MGI_C57BL6J_5753364 | Gm44788 | ENSEMBL:ENSMUSG00000109545 | MGI:5753364 | lncRNA gene | predicted gene 44788 |
| 8 | gene | 10.153911 | 10.634742 | positive | MGI_C57BL6J_2685951 | Myo16 | NCBI_Gene:244281,ENSEMBL:ENSMUSG00000039057 | MGI:2685951 | protein coding gene | myosin XVI |
| 8 | gene | 10.673361 | 10.676646 | positive | MGI_C57BL6J_5591489 | Gm32330 | NCBI_Gene:102634842 | MGI:5591489 | lncRNA gene | predicted gene, 32330 |
| 8 | gene | 10.675348 | 10.676913 | negative | MGI_C57BL6J_3588224 | 5330413D20Rik | ENSEMBL:ENSMUSG00000109258 | MGI:3588224 | lncRNA gene | RIKEN cDNA 5330413D20 gene |
| 8 | gene | 10.685351 | 10.687390 | positive | MGI_C57BL6J_5753362 | Gm44786 | ENSEMBL:ENSMUSG00000109503 | MGI:5753362 | lncRNA gene | predicted gene 44786 |
| 8 | gene | 10.687948 | 10.691706 | positive | MGI_C57BL6J_5753361 | Gm44785 | ENSEMBL:ENSMUSG00000109432 | MGI:5753361 | lncRNA gene | predicted gene 44785 |
| 8 | pseudogene | 10.716707 | 10.738674 | negative | MGI_C57BL6J_5589825 | Gm30666 | NCBI_Gene:102632644 | MGI:5589825 | pseudogene | predicted gene, 30666 |
| 8 | gene | 10.723486 | 10.725161 | positive | MGI_C57BL6J_5753360 | Gm44784 | ENSEMBL:ENSMUSG00000109067 | MGI:5753360 | lncRNA gene | predicted gene 44784 |
| 8 | gene | 10.732676 | 10.736533 | positive | MGI_C57BL6J_5753621 | Gm45045 | ENSEMBL:ENSMUSG00000108997 | MGI:5753621 | lncRNA gene | predicted gene 45045 |
| 8 | gene | 10.738737 | 10.770849 | positive | MGI_C57BL6J_5622014 | Gm39129 | NCBI_Gene:105243118,ENSEMBL:ENSMUSG00000109271 | MGI:5622014 | lncRNA gene | predicted gene, 39129 |
| 8 | gene | 10.740250 | 10.747285 | negative | MGI_C57BL6J_5591588 | Gm32429 | NCBI_Gene:102634980 | MGI:5591588 | lncRNA gene | predicted gene, 32429 |
| 8 | pseudogene | 10.808655 | 10.809182 | negative | MGI_C57BL6J_3647039 | Rps16-ps3 | NCBI_Gene:384782,ENSEMBL:ENSMUSG00000096258 | MGI:3647039 | pseudogene | ribosomal protein S16, pseudogene 3 |
| 8 | gene | 10.817012 | 10.817721 | negative | MGI_C57BL6J_5591644 | Gm32485 | NCBI_Gene:102635050 | MGI:5591644 | lncRNA gene | predicted gene, 32485 |
| 8 | gene | 10.847429 | 10.855147 | positive | MGI_C57BL6J_5622015 | Gm39130 | NCBI_Gene:105243119 | MGI:5622015 | lncRNA gene | predicted gene, 39130 |
| 8 | gene | 10.855721 | 10.866277 | negative | MGI_C57BL6J_5591699 | Gm32540 | NCBI_Gene:102635120,ENSEMBL:ENSMUSG00000108961 | MGI:5591699 | lncRNA gene | predicted gene, 32540 |
| 8 | gene | 10.870422 | 10.882454 | negative | MGI_C57BL6J_2443166 | B930025P03Rik | NCBI_Gene:320014,ENSEMBL:ENSMUSG00000109473 | MGI:2443166 | lncRNA gene | RIKEN cDNA B930025P03 gene |
| 8 | gene | 10.884453 | 10.891591 | negative | MGI_C57BL6J_5753620 | Gm45044 | ENSEMBL:ENSMUSG00000108969 | MGI:5753620 | lncRNA gene | predicted gene 45044 |
| 8 | gene | 10.897493 | 10.900558 | negative | MGI_C57BL6J_3780984 | Gm2814 | NCBI_Gene:100040507,ENSEMBL:ENSMUSG00000109291 | MGI:3780984 | lncRNA gene | predicted gene 2814 |
| 8 | gene | 10.899922 | 10.903893 | positive | MGI_C57BL6J_1918874 | 4833411C07Rik | NCBI_Gene:71624,ENSEMBL:ENSMUSG00000109089 | MGI:1918874 | lncRNA gene | RIKEN cDNA 4833411C07 gene |
| 8 | gene | 10.919619 | 10.921084 | negative | MGI_C57BL6J_5591753 | Gm32594 | NCBI_Gene:102635194 | MGI:5591753 | lncRNA gene | predicted gene, 32594 |
| 8 | gene | 10.924427 | 10.928696 | negative | MGI_C57BL6J_1921277 | 3930402G23Rik | NCBI_Gene:665306,ENSEMBL:ENSMUSG00000038917 | MGI:1921277 | lncRNA gene | RIKEN cDNA 3930402G23 gene |
| 8 | pseudogene | 10.929887 | 10.930097 | positive | MGI_C57BL6J_5753618 | Gm45042 | ENSEMBL:ENSMUSG00000109480 | MGI:5753618 | pseudogene | predicted gene 45042 |
| 8 | gene | 10.943326 | 10.947890 | negative | MGI_C57BL6J_5753531 | Gm44955 | ENSEMBL:ENSMUSG00000109084 | MGI:5753531 | lncRNA gene | predicted gene 44955 |
| 8 | gene | 10.962957 | 10.970814 | negative | MGI_C57BL6J_5753532 | Gm44956 | ENSEMBL:ENSMUSG00000109508 | MGI:5753532 | lncRNA gene | predicted gene 44956 |
| 8 | gene | 10.984681 | 11.008458 | negative | MGI_C57BL6J_109334 | Irs2 | NCBI_Gene:384783,ENSEMBL:ENSMUSG00000038894 | MGI:109334 | protein coding gene | insulin receptor substrate 2 |
| 8 | gene | 11.007850 | 11.054541 | positive | MGI_C57BL6J_3588277 | 9530052E02Rik | NCBI_Gene:619321,ENSEMBL:ENSMUSG00000096938 | MGI:3588277 | lncRNA gene | RIKEN cDNA 9530052E02 gene |
| 8 | gene | 11.071046 | 11.076881 | positive | MGI_C57BL6J_1925942 | B830042I05Rik | ENSEMBL:ENSMUSG00000108847 | MGI:1925942 | lncRNA gene | RIKEN cDNA B830042I05 gene |
| 8 | gene | 11.116555 | 11.117373 | positive | MGI_C57BL6J_5753290 | Gm44714 | ENSEMBL:ENSMUSG00000109151 | MGI:5753290 | lncRNA gene | predicted gene 44714 |
| 8 | gene | 11.135641 | 11.149246 | negative | MGI_C57BL6J_5753293 | Gm44717 | ENSEMBL:ENSMUSG00000109029 | MGI:5753293 | lncRNA gene | predicted gene 44717 |
| 8 | gene | 11.187755 | 11.204503 | positive | MGI_C57BL6J_3705275 | Gm15418 | ENSEMBL:ENSMUSG00000085573 | MGI:3705275 | lncRNA gene | predicted gene 15418 |
| 8 | gene | 11.198423 | 11.312826 | negative | MGI_C57BL6J_88454 | Col4a1 | NCBI_Gene:12826,ENSEMBL:ENSMUSG00000031502 | MGI:88454 | protein coding gene | collagen, type IV, alpha 1 |
| 8 | gene | 11.258088 | 11.285958 | positive | MGI_C57BL6J_5591868 | Gm32709 | NCBI_Gene:102635346 | MGI:5591868 | lncRNA gene | predicted gene, 32709 |
| 8 | gene | 11.290558 | 11.291267 | negative | MGI_C57BL6J_5753292 | Gm44716 | ENSEMBL:ENSMUSG00000109358 | MGI:5753292 | unclassified gene | predicted gene 44716 |
| 8 | gene | 11.312805 | 11.449287 | positive | MGI_C57BL6J_88455 | Col4a2 | NCBI_Gene:12827,ENSEMBL:ENSMUSG00000031503 | MGI:88455 | protein coding gene | collagen, type IV, alpha 2 |
| 8 | gene | 11.321303 | 11.351695 | negative | MGI_C57BL6J_5825667 | Gm46030 | NCBI_Gene:108167519 | MGI:5825667 | lncRNA gene | predicted gene, 46030 |
| 8 | gene | 11.399186 | 11.417892 | negative | MGI_C57BL6J_3705320 | Gm15419 | ENSEMBL:ENSMUSG00000084783 | MGI:3705320 | lncRNA gene | predicted gene 15419 |
| 8 | gene | 11.453513 | 11.478710 | negative | MGI_C57BL6J_102789 | Rab20 | NCBI_Gene:19332,ENSEMBL:ENSMUSG00000031504 | MGI:102789 | protein coding gene | RAB20, member RAS oncogene family |
| 8 | gene | 11.457471 | 11.459742 | negative | MGI_C57BL6J_5012285 | Gm20100 | ENSEMBL:ENSMUSG00000110245 | MGI:5012285 | unclassified gene | predicted gene, 20100 |
| 8 | gene | 11.477929 | 11.480241 | positive | MGI_C57BL6J_2142601 | E230013L22Rik | NCBI_Gene:102132,ENSEMBL:ENSMUSG00000096957 | MGI:2142601 | lncRNA gene | RIKEN cDNA E230013L22 gene |
| 8 | gene | 11.497506 | 11.514960 | positive | MGI_C57BL6J_1913353 | Naxd | NCBI_Gene:69225,ENSEMBL:ENSMUSG00000031505 | MGI:1913353 | protein coding gene | NAD(P)HX dehydratase |
| 8 | gene | 11.513977 | 11.550783 | negative | MGI_C57BL6J_1919191 | Cars2 | NCBI_Gene:71941,ENSEMBL:ENSMUSG00000056228 | MGI:1919191 | protein coding gene | cysteinyl-tRNA synthetase 2 (mitochondrial)(putative) |
| 8 | gene | 11.555571 | 11.563251 | positive | MGI_C57BL6J_1349481 | Ing1 | NCBI_Gene:26356,ENSEMBL:ENSMUSG00000045969 | MGI:1349481 | protein coding gene | inhibitor of growth family, member 1 |
| 8 | gene | 11.611581 | 11.635877 | negative | MGI_C57BL6J_1921840 | Ankrd10 | NCBI_Gene:102334,ENSEMBL:ENSMUSG00000031508 | MGI:1921840 | protein coding gene | ankyrin repeat domain 10 |
| 8 | gene | 11.622373 | 11.623241 | negative | MGI_C57BL6J_1919755 | 2610319H10Rik | NA | NA | unclassified gene | RIKEN cDNA 2610319H10 gene |
| 8 | gene | 11.635694 | 11.635736 | negative | MGI_C57BL6J_5562781 | Mir7238 | miRBase:MI0023733,NCBI_Gene:102465714,ENSEMBL:ENSMUSG00000104958 | MGI:5562781 | miRNA gene | microRNA 7238 |
| 8 | gene | 11.637282 | 11.638324 | positive | MGI_C57BL6J_5791428 | Gm45592 | ENSEMBL:ENSMUSG00000109810 | MGI:5791428 | unclassified gene | predicted gene 45592 |
| 8 | gene | 11.654921 | 11.701063 | negative | MGI_C57BL6J_1923663 | 1700016D06Rik | NCBI_Gene:76413,ENSEMBL:ENSMUSG00000031509 | MGI:1923663 | protein coding gene | RIKEN cDNA 1700016D06 gene |
| 8 | gene | 11.674574 | 11.677139 | positive | MGI_C57BL6J_3645249 | Gm7562 | NCBI_Gene:665271,ENSEMBL:ENSMUSG00000109623 | MGI:3645249 | unclassified gene | predicted gene 7562 |
| 8 | gene | 11.678808 | 11.682211 | positive | MGI_C57BL6J_1925260 | 4930465I24Rik | ENSEMBL:ENSMUSG00000109986 | MGI:1925260 | lncRNA gene | RIKEN cDNA 4930465I24 gene |
| 8 | pseudogene | 11.683394 | 11.684260 | negative | MGI_C57BL6J_3647502 | Gm7600 | NCBI_Gene:665363,ENSEMBL:ENSMUSG00000109933 | MGI:3647502 | pseudogene | predicted gene 7600 |
| 8 | pseudogene | 11.692982 | 11.694514 | negative | MGI_C57BL6J_3648756 | Gm6524 | NCBI_Gene:624710,ENSEMBL:ENSMUSG00000110177 | MGI:3648756 | pseudogene | predicted gene 6524 |
| 8 | gene | 11.696615 | 11.696858 | negative | MGI_C57BL6J_5531236 | Gm27854 | ENSEMBL:ENSMUSG00000093319 | MGI:5531236 | unclassified non-coding RNA gene | predicted gene, 27854 |
| 8 | gene | 11.713260 | 11.722620 | positive | MGI_C57BL6J_1920868 | 1700128E19Rik | NCBI_Gene:73618,ENSEMBL:ENSMUSG00000031510 | MGI:1920868 | lncRNA gene | RIKEN cDNA 1700128E19 gene |
| 8 | pseudogene | 11.714755 | 11.715239 | negative | MGI_C57BL6J_5791256 | Gm45420 | NCBI_Gene:108167520,ENSEMBL:ENSMUSG00000110313 | MGI:5791256 | pseudogene | predicted gene 45420 |
| 8 | gene | 11.726065 | 11.727375 | negative | MGI_C57BL6J_1918390 | 4933413I22Rik | NA | NA | unclassified gene | RIKEN cDNA 4933413I22 gene |
| 8 | gene | 11.727721 | 11.835219 | positive | MGI_C57BL6J_1860493 | Arhgef7 | NCBI_Gene:54126,ENSEMBL:ENSMUSG00000031511 | MGI:1860493 | protein coding gene | Rho guanine nucleotide exchange factor (GEF7) |
| 8 | pseudogene | 11.738621 | 11.739366 | positive | MGI_C57BL6J_5011176 | Gm18991 | NCBI_Gene:100418085,ENSEMBL:ENSMUSG00000110394 | MGI:5011176 | pseudogene | predicted gene, 18991 |
| 8 | gene | 11.750648 | 11.772803 | negative | MGI_C57BL6J_5825668 | Gm46031 | NCBI_Gene:108167521 | MGI:5825668 | lncRNA gene | predicted gene, 46031 |
| 8 | gene | 11.838978 | 11.840400 | negative | MGI_C57BL6J_3801964 | Gm15875 | ENSEMBL:ENSMUSG00000086500 | MGI:3801964 | lncRNA gene | predicted gene 15875 |
| 8 | gene | 11.840474 | 11.855761 | positive | MGI_C57BL6J_1922778 | Tex29 | NCBI_Gene:75528,ENSEMBL:ENSMUSG00000031512 | MGI:1922778 | protein coding gene | testis expressed 29 |
| 8 | gene | 11.874159 | 11.896690 | positive | MGI_C57BL6J_5622017 | Gm39132 | NCBI_Gene:105243121,ENSEMBL:ENSMUSG00000109772 | MGI:5622017 | lncRNA gene | predicted gene, 39132 |
| 8 | gene | 11.945465 | 11.948297 | negative | MGI_C57BL6J_5791516 | Gm45680 | ENSEMBL:ENSMUSG00000110002 | MGI:5791516 | lncRNA gene | predicted gene 45680 |
| 8 | gene | 11.948339 | 12.000216 | positive | MGI_C57BL6J_3588231 | B020031H02Rik | ENSEMBL:ENSMUSG00000109995 | MGI:3588231 | lncRNA gene | RIKEN cDNA B020031H02 gene |
| 8 | pseudogene | 11.960969 | 11.961515 | negative | MGI_C57BL6J_5010503 | Gm18318 | NCBI_Gene:100416915 | MGI:5010503 | pseudogene | predicted gene, 18318 |
| 8 | gene | 12.234676 | 12.239916 | negative | MGI_C57BL6J_5592188 | Gm33029 | NCBI_Gene:102635778 | MGI:5592188 | lncRNA gene | predicted gene, 33029 |
| 8 | gene | 12.277338 | 12.283147 | positive | MGI_C57BL6J_3588221 | A230072I06Rik | NCBI_Gene:619290,ENSEMBL:ENSMUSG00000074473 | MGI:3588221 | protein coding gene | RIKEN cDNA A230072I06 gene |
| 8 | gene | 12.282211 | 12.282612 | positive | MGI_C57BL6J_5579881 | Gm29175 | ENSEMBL:ENSMUSG00000101899 | MGI:5579881 | lncRNA gene | predicted gene 29175 |
| 8 | gene | 12.314026 | 12.319896 | negative | MGI_C57BL6J_5592334 | Gm33175 | NCBI_Gene:102635976,ENSEMBL:ENSMUSG00000110412 | MGI:5592334 | lncRNA gene | predicted gene, 33175 |
| 8 | gene | 12.356153 | 12.356469 | positive | MGI_C57BL6J_5451984 | Gm22207 | ENSEMBL:ENSMUSG00000089234 | MGI:5451984 | unclassified non-coding RNA gene | predicted gene, 22207 |
| 8 | gene | 12.385771 | 12.436768 | positive | MGI_C57BL6J_3645522 | Sox1ot | NCBI_Gene:434280,ENSEMBL:ENSMUSG00000047935 | MGI:3645522 | sense overlapping lncRNA gene | Sox1 overlapping transcript |
| 8 | gene | 12.390168 | 12.391639 | positive | MGI_C57BL6J_5791396 | Gm45560 | ENSEMBL:ENSMUSG00000110160 | MGI:5791396 | unclassified gene | predicted gene 45560 |
| 8 | gene | 12.390168 | 12.391639 | negative | MGI_C57BL6J_3647910 | Gm5606 | NA | NA | unclassified gene | predicted gene 5606 |
| 8 | gene | 12.395295 | 12.400126 | positive | MGI_C57BL6J_98357 | Sox1 | NCBI_Gene:20664,ENSEMBL:ENSMUSG00000096014 | MGI:98357 | protein coding gene | SRY (sex determining region Y)-box 1 |
| 8 | gene | 12.396403 | 12.396488 | positive | MGI_C57BL6J_5455016 | Gm25239 | ENSEMBL:ENSMUSG00000092656 | MGI:5455016 | miRNA gene | predicted gene, 25239 |
| 8 | gene | 12.458858 | 12.466964 | negative | MGI_C57BL6J_5592485 | Gm33326 | NCBI_Gene:102636189,ENSEMBL:ENSMUSG00000109921 | MGI:5592485 | lncRNA gene | predicted gene, 33326 |
| 8 | gene | 12.472134 | 12.474458 | negative | MGI_C57BL6J_5592691 | Gm33532 | NCBI_Gene:102636475 | MGI:5592691 | lncRNA gene | predicted gene, 33532 |
| 8 | gene | 12.502899 | 12.505341 | positive | MGI_C57BL6J_1925389 | 4933439N14Rik | ENSEMBL:ENSMUSG00000109582 | MGI:1925389 | lncRNA gene | RIKEN cDNA 4933439N14 gene |
| 8 | gene | 12.554272 | 12.573320 | negative | MGI_C57BL6J_3642132 | D630011A20Rik | ENSEMBL:ENSMUSG00000097421 | MGI:3642132 | lncRNA gene | RIKEN cDNA D630011A20 gene |
| 8 | gene | 12.573000 | 12.600744 | positive | MGI_C57BL6J_1925884 | Spaca7 | NCBI_Gene:78634,ENSEMBL:ENSMUSG00000010435 | MGI:1925884 | protein coding gene | sperm acrosome associated 7 |
| 8 | gene | 12.614277 | 12.672248 | negative | MGI_C57BL6J_2183752 | Tubgcp3 | NCBI_Gene:259279,ENSEMBL:ENSMUSG00000000759 | MGI:2183752 | protein coding gene | tubulin, gamma complex associated protein 3 |
| 8 | gene | 12.666095 | 12.666198 | positive | MGI_C57BL6J_5456260 | Gm26483 | ENSEMBL:ENSMUSG00000088641 | MGI:5456260 | miRNA gene | predicted gene, 26483 |
| 8 | gene | 12.672186 | 12.679238 | positive | MGI_C57BL6J_1918216 | 4931415C17Rik | NCBI_Gene:70966,ENSEMBL:ENSMUSG00000054910 | MGI:1918216 | lncRNA gene | RIKEN cDNA 4931415C17 gene |
| 8 | gene | 12.706944 | 12.719127 | negative | MGI_C57BL6J_3642533 | Gm15348 | NCBI_Gene:100038554,ENSEMBL:ENSMUSG00000074469 | MGI:3642533 | lncRNA gene | predicted gene 15348 |
| 8 | gene | 12.749201 | 12.751308 | positive | MGI_C57BL6J_5622018 | Gm39133 | NCBI_Gene:105243123 | MGI:5622018 | lncRNA gene | predicted gene, 39133 |
| 8 | gene | 12.753823 | 12.756988 | negative | MGI_C57BL6J_5592746 | Gm33587 | NCBI_Gene:102636552 | MGI:5592746 | lncRNA gene | predicted gene, 33587 |
| 8 | gene | 12.757014 | 12.868728 | positive | MGI_C57BL6J_1354735 | Atp11a | NCBI_Gene:50770,ENSEMBL:ENSMUSG00000031441 | MGI:1354735 | protein coding gene | ATPase, class VI, type 11A |
| 8 | gene | 12.828335 | 12.833865 | negative | MGI_C57BL6J_3705181 | Gm15350 | NCBI_Gene:100504249,ENSEMBL:ENSMUSG00000084780 | MGI:3705181 | lncRNA gene | predicted gene 15350 |
| 8 | gene | 12.860358 | 12.877344 | negative | MGI_C57BL6J_3705310 | Gm15347 | ENSEMBL:ENSMUSG00000085465 | MGI:3705310 | lncRNA gene | predicted gene 15347 |
| 8 | gene | 12.872344 | 12.873542 | negative | MGI_C57BL6J_5592796 | Gm33637 | NCBI_Gene:102636626 | MGI:5592796 | lncRNA gene | predicted gene, 33637 |
| 8 | gene | 12.873544 | 13.020905 | positive | MGI_C57BL6J_103263 | Mcf2l | NCBI_Gene:17207,ENSEMBL:ENSMUSG00000031442 | MGI:103263 | protein coding gene | mcf.2 transforming sequence-like |
| 8 | gene | 12.879878 | 12.885542 | negative | MGI_C57BL6J_3705219 | Gm15353 | NCBI_Gene:105243125,ENSEMBL:ENSMUSG00000079146 | MGI:3705219 | lncRNA gene | predicted gene 15353 |
| 8 | gene | 12.899146 | 12.915758 | negative | MGI_C57BL6J_3705299 | Gm15351 | NCBI_Gene:105243124,ENSEMBL:ENSMUSG00000086770 | MGI:3705299 | lncRNA gene | predicted gene 15351 |
| 8 | gene | 12.947941 | 12.948070 | positive | MGI_C57BL6J_5621556 | Gm38671 | NCBI_Gene:104797311,ENSEMBL:ENSMUSG00000105030 | MGI:5621556 | miRNA gene | predicted gene, 38671 |
| 8 | pseudogene | 12.971826 | 12.972029 | negative | MGI_C57BL6J_3705665 | Gm15349 | ENSEMBL:ENSMUSG00000082682 | MGI:3705665 | pseudogene | predicted gene 15349 |
| 8 | pseudogene | 13.021868 | 13.022856 | positive | MGI_C57BL6J_3641922 | Gm15352 | NCBI_Gene:791304,ENSEMBL:ENSMUSG00000081951 | MGI:3641922 | pseudogene | predicted gene 15352 |
| 8 | gene | 13.025506 | 13.035809 | positive | MGI_C57BL6J_109325 | F7 | NCBI_Gene:14068,ENSEMBL:ENSMUSG00000031443 | MGI:109325 | protein coding gene | coagulation factor VII |
| 8 | gene | 13.037308 | 13.056676 | positive | MGI_C57BL6J_103107 | F10 | NCBI_Gene:14058,ENSEMBL:ENSMUSG00000031444 | MGI:103107 | protein coding gene | coagulation factor X |
| 8 | gene | 13.060581 | 13.076026 | positive | MGI_C57BL6J_1860488 | Proz | NCBI_Gene:66901,ENSEMBL:ENSMUSG00000031445 | MGI:1860488 | protein coding gene | protein Z, vitamin K-dependent plasma glycoprotein |
| 8 | gene | 13.068774 | 13.071080 | positive | MGI_C57BL6J_5592850 | Gm33691 | NCBI_Gene:102636690 | MGI:5592850 | protein coding gene | predicted gene, 33691 |
| 8 | gene | 13.075476 | 13.105459 | negative | MGI_C57BL6J_2443003 | Pcid2 | NCBI_Gene:234069,ENSEMBL:ENSMUSG00000038542 | MGI:2443003 | protein coding gene | PCI domain containing 2 |
| 8 | gene | 13.076065 | 13.082740 | positive | MGI_C57BL6J_4937850 | Gm17023 | NCBI_Gene:108167522,ENSEMBL:ENSMUSG00000091443 | MGI:4937850 | lncRNA gene | predicted gene 17023 |
| 8 | pseudogene | 13.086509 | 13.087430 | positive | MGI_C57BL6J_4937849 | Gm17022 | ENSEMBL:ENSMUSG00000091784 | MGI:4937849 | pseudogene | predicted gene 17022 |
| 8 | gene | 13.105520 | 13.147940 | positive | MGI_C57BL6J_1914487 | Cul4a | NCBI_Gene:99375,ENSEMBL:ENSMUSG00000031446 | MGI:1914487 | protein coding gene | cullin 4A |
| 8 | gene | 13.120355 | 13.121331 | positive | MGI_C57BL6J_1921336 | 4933411D12Rik | NA | NA | unclassified gene | RIKEN cDNA 4933411D12 gene |
| 8 | gene | 13.136072 | 13.136132 | positive | MGI_C57BL6J_5531253 | Mir7065 | miRBase:MI0022914,NCBI_Gene:102465645,ENSEMBL:ENSMUSG00000098532 | MGI:5531253 | miRNA gene | microRNA 7065 |
| 8 | gene | 13.159161 | 13.175338 | positive | MGI_C57BL6J_96745 | Lamp1 | NCBI_Gene:16783,ENSEMBL:ENSMUSG00000031447 | MGI:96745 | protein coding gene | lysosomal-associated membrane protein 1 |
| 8 | gene | 13.172022 | 13.200652 | negative | MGI_C57BL6J_1914040 | Grtp1 | NCBI_Gene:66790,ENSEMBL:ENSMUSG00000038515 | MGI:1914040 | protein coding gene | GH regulated TBC protein 1 |
| 8 | gene | 13.189031 | 13.189098 | negative | MGI_C57BL6J_3837215 | Mir1968 | miRBase:MI0009965,NCBI_Gene:100316713,ENSEMBL:ENSMUSG00000088054 | MGI:3837215 | miRNA gene | microRNA 1968 |
| 8 | gene | 13.200720 | 13.207714 | positive | MGI_C57BL6J_1925585 | 2810030D12Rik | NCBI_Gene:78335,ENSEMBL:ENSMUSG00000099338 | MGI:1925585 | lncRNA gene | RIKEN cDNA 2810030D12 gene |
| 8 | gene | 13.201168 | 13.214598 | negative | MGI_C57BL6J_5593664 | Gm34505 | NCBI_Gene:102637779 | MGI:5593664 | lncRNA gene | predicted gene, 34505 |
| 8 | gene | 13.218782 | 13.254632 | negative | MGI_C57BL6J_2442168 | Adprhl1 | NCBI_Gene:234072,ENSEMBL:ENSMUSG00000031448 | MGI:2442168 | protein coding gene | ADP-ribosylhydrolase like 1 |
| 8 | gene | 13.255963 | 13.288131 | negative | MGI_C57BL6J_2142792 | Dcun1d2 | NCBI_Gene:102323,ENSEMBL:ENSMUSG00000038506 | MGI:2142792 | protein coding gene | DCN1, defective in cullin neddylation 1, domain containing 2 (S. cerevisiae) |
| 8 | gene | 13.264102 | 13.265187 | negative | MGI_C57BL6J_2142689 | E330037G11Rik | ENSEMBL:ENSMUSG00000107531 | MGI:2142689 | unclassified gene | RIKEN cDNA E330037G11 gene |
| 8 | gene | 13.283115 | 13.283217 | negative | MGI_C57BL6J_5454791 | Gm25014 | ENSEMBL:ENSMUSG00000089026 | MGI:5454791 | snRNA gene | predicted gene, 25014 |
| 8 | gene | 13.285352 | 13.287466 | negative | MGI_C57BL6J_2443889 | 9530085L11Rik | ENSEMBL:ENSMUSG00000107742 | MGI:2443889 | unclassified gene | RIKEN cDNA 9530085L11 gene |
| 8 | gene | 13.287913 | 13.322924 | positive | MGI_C57BL6J_2444946 | Tmco3 | NCBI_Gene:234076,ENSEMBL:ENSMUSG00000038497 | MGI:2444946 | protein coding gene | transmembrane and coiled-coil domains 3 |
| 8 | gene | 13.338726 | 13.378448 | positive | MGI_C57BL6J_101934 | Tfdp1 | NCBI_Gene:21781,ENSEMBL:ENSMUSG00000038482 | MGI:101934 | protein coding gene | transcription factor Dp 1 |
| 8 | gene | 13.386205 | 13.396825 | negative | MGI_C57BL6J_88114 | Atp4b | NCBI_Gene:11945,ENSEMBL:ENSMUSG00000031449 | MGI:88114 | protein coding gene | ATPase, H+/K+ exchanging, beta polypeptide |
| 8 | gene | 13.391106 | 13.392198 | positive | MGI_C57BL6J_5791246 | Gm45410 | ENSEMBL:ENSMUSG00000110311 | MGI:5791246 | unclassified gene | predicted gene 45410 |
| 8 | gene | 13.405081 | 13.421951 | positive | MGI_C57BL6J_1345146 | Grk1 | NCBI_Gene:24013,ENSEMBL:ENSMUSG00000031450 | MGI:1345146 | protein coding gene | G protein-coupled receptor kinase 1 |
| 8 | gene | 13.435164 | 13.483437 | positive | MGI_C57BL6J_2685533 | Tmem255b | NCBI_Gene:272465,ENSEMBL:ENSMUSG00000038457 | MGI:2685533 | protein coding gene | transmembrane protein 255B |
| 8 | gene | 13.465374 | 13.494535 | negative | MGI_C57BL6J_95660 | Gas6 | NCBI_Gene:14456,ENSEMBL:ENSMUSG00000031451 | MGI:95660 | protein coding gene | growth arrest specific 6 |
| 8 | gene | 13.537678 | 13.547028 | negative | MGI_C57BL6J_3779504 | Gm5608 | ENSEMBL:ENSMUSG00000110278 | MGI:3779504 | lncRNA gene | predicted gene 5608 |
| 8 | gene | 13.550722 | 13.562554 | negative | MGI_C57BL6J_1913751 | 1700029H14Rik | NCBI_Gene:66501,ENSEMBL:ENSMUSG00000031452 | MGI:1913751 | protein coding gene | RIKEN cDNA 1700029H14 gene |
| 8 | gene | 13.566948 | 13.677603 | negative | MGI_C57BL6J_1197013 | Rasa3 | NCBI_Gene:19414,ENSEMBL:ENSMUSG00000031453 | MGI:1197013 | protein coding gene | RAS p21 protein activator 3 |
| 8 | gene | 13.590313 | 13.591997 | positive | MGI_C57BL6J_5593954 | Gm34795 | NCBI_Gene:102638168 | MGI:5593954 | lncRNA gene | predicted gene, 34795 |
| 8 | gene | 13.672926 | 13.676869 | negative | MGI_C57BL6J_2443836 | 4930442M18Rik | NA | NA | unclassified gene | RIKEN cDNA 4930442M18 gene |
| 8 | gene | 13.682868 | 13.785106 | negative | MGI_C57BL6J_5825669 | Gm46032 | NCBI_Gene:108167523 | MGI:5825669 | lncRNA gene | predicted gene, 46032 |
| 8 | gene | 13.705449 | 13.744599 | positive | MGI_C57BL6J_2685952 | Cfap97d2 | NCBI_Gene:403185,ENSEMBL:ENSMUSG00000090336 | MGI:2685952 | protein coding gene | CFAP97 domain containing 2 |
| 8 | pseudogene | 13.745742 | 13.747572 | negative | MGI_C57BL6J_3648911 | Gm7669 | NCBI_Gene:665516,ENSEMBL:ENSMUSG00000109617 | MGI:3648911 | pseudogene | predicted gene 7669 |
| 8 | gene | 13.757649 | 13.781938 | positive | MGI_C57BL6J_1917207 | Cdc16 | NCBI_Gene:69957,ENSEMBL:ENSMUSG00000038416 | MGI:1917207 | protein coding gene | CDC16 cell division cycle 16 |
| 8 | gene | 13.785233 | 13.799193 | positive | MGI_C57BL6J_1914281 | Upf3a | NCBI_Gene:67031,ENSEMBL:ENSMUSG00000038398 | MGI:1914281 | protein coding gene | UPF3 regulator of nonsense transcripts homolog A (yeast) |
| 8 | gene | 13.835229 | 13.838389 | negative | MGI_C57BL6J_2667157 | AF366264 | NCBI_Gene:231201,ENSEMBL:ENSMUSG00000057116 | MGI:2667157 | protein coding gene | cDNA sequence AF366264 |
| 8 | gene | 13.869641 | 13.881639 | positive | MGI_C57BL6J_1196398 | Champ1 | NCBI_Gene:101994,ENSEMBL:ENSMUSG00000047710 | MGI:1196398 | protein coding gene | chromosome alignment maintaining phosphoprotein 1 |
| 8 | gene | 13.876091 | 13.876184 | positive | MGI_C57BL6J_6096119 | Gm47277 | ENSEMBL:ENSMUSG00000104767 | MGI:6096119 | miRNA gene | predicted gene, 47277 |
| 8 | gene | 13.876097 | 13.876226 | positive | MGI_C57BL6J_5454475 | Gm24698 | ENSEMBL:ENSMUSG00000065817 | MGI:5454475 | snoRNA gene | predicted gene, 24698 |
| 8 | gene | 13.884787 | 13.890327 | negative | MGI_C57BL6J_1913673 | Coprs | NCBI_Gene:66423,ENSEMBL:ENSMUSG00000031458 | MGI:1913673 | protein coding gene | coordinator of PRMT5, differentiation stimulator |
| 8 | pseudogene | 13.896155 | 13.896818 | positive | MGI_C57BL6J_3644053 | Gm7676 | NCBI_Gene:665536,ENSEMBL:ENSMUSG00000068631 | MGI:3644053 | pseudogene | predicted gene 7676 |
| 8 | gene | 13.907742 | 13.940544 | positive | MGI_C57BL6J_1914072 | Fbxo25 | NCBI_Gene:66822,ENSEMBL:ENSMUSG00000038365 | MGI:1914072 | protein coding gene | F-box protein 25 |
| 8 | gene | 13.952008 | 13.975453 | negative | MGI_C57BL6J_1919398 | Tdrp | NCBI_Gene:72148,ENSEMBL:ENSMUSG00000050052 | MGI:1919398 | protein coding gene | testis development related protein |
| 8 | gene | 13.986090 | 14.003561 | positive | MGI_C57BL6J_3643437 | Gm5907 | NCBI_Gene:546036,ENSEMBL:ENSMUSG00000110058 | MGI:3643437 | lncRNA gene | predicted gene 5907 |
| 8 | gene | 13.996369 | 14.018303 | negative | MGI_C57BL6J_5594125 | Gm34966 | NCBI_Gene:105243127 | MGI:5594125 | lncRNA gene | predicted gene, 34966 |
| 8 | gene | 14.022030 | 14.025440 | negative | MGI_C57BL6J_5622019 | Gm39134 | NCBI_Gene:105243128 | MGI:5622019 | lncRNA gene | predicted gene, 39134 |
| 8 | gene | 14.025882 | 14.090338 | negative | MGI_C57BL6J_3588201 | Erich1 | NCBI_Gene:234086,ENSEMBL:ENSMUSG00000051978 | MGI:3588201 | protein coding gene | glutamate rich 1 |
| 8 | gene | 14.065788 | 14.067818 | negative | MGI_C57BL6J_5791356 | Gm45520 | ENSEMBL:ENSMUSG00000109982 | MGI:5791356 | unclassified gene | predicted gene 45520 |
| 8 | gene | 14.090461 | 14.092590 | positive | MGI_C57BL6J_3642530 | Gm10699 | ENSEMBL:ENSMUSG00000074461 | MGI:3642530 | protein coding gene | predicted gene 10699 |
| 8 | gene | 14.094614 | 14.848626 | positive | MGI_C57BL6J_2443181 | Dlgap2 | NCBI_Gene:244310,ENSEMBL:ENSMUSG00000047495 | MGI:2443181 | protein coding gene | DLG associated protein 2 |
| 8 | pseudogene | 14.136151 | 14.137412 | positive | MGI_C57BL6J_3781339 | Gm3160 | NCBI_Gene:100041140,ENSEMBL:ENSMUSG00000085371 | MGI:3781339 | pseudogene | predicted pseudogene 3160 |
| 8 | gene | 14.273181 | 14.273311 | negative | MGI_C57BL6J_5455961 | Gm26184 | ENSEMBL:ENSMUSG00000088311 | MGI:5455961 | snoRNA gene | predicted gene, 26184 |
| 8 | gene | 14.350178 | 14.350285 | positive | MGI_C57BL6J_5690739 | Gm44347 | ENSEMBL:ENSMUSG00000106670 | MGI:5690739 | miRNA gene | predicted gene, 44347 |
| 8 | gene | 14.381732 | 14.507710 | positive | MGI_C57BL6J_1924327 | 9030205G03Rik | NA | NA | unclassified gene | RIKEN cDNA 9030205G03 gene |
| 8 | gene | 14.745329 | 14.767361 | negative | MGI_C57BL6J_1924725 | C030037F17Rik | NCBI_Gene:77475,ENSEMBL:ENSMUSG00000084945 | MGI:1924725 | lncRNA gene | RIKEN cDNA C030037F17 gene |
| 8 | gene | 14.881335 | 14.901720 | positive | MGI_C57BL6J_1349447 | Cln8 | NCBI_Gene:26889,ENSEMBL:ENSMUSG00000026317 | MGI:1349447 | protein coding gene | ceroid-lipofuscinosis, neuronal 8 |
| 8 | gene | 14.908494 | 15.001085 | positive | MGI_C57BL6J_2444453 | Arhgef10 | NCBI_Gene:234094,ENSEMBL:ENSMUSG00000071176 | MGI:2444453 | protein coding gene | Rho guanine nucleotide exchange factor (GEF) 10 |
| 8 | gene | 14.910590 | 14.911492 | negative | MGI_C57BL6J_1922318 | 4930513N24Rik | NA | NA | unclassified gene | RIKEN cDNA 4930513N24 gene |
| 8 | gene | 14.947271 | 14.953177 | negative | MGI_C57BL6J_5594444 | Gm35285 | NCBI_Gene:102638808 | MGI:5594444 | lncRNA gene | predicted gene, 35285 |
| 8 | gene | 14.957193 | 14.961586 | negative | MGI_C57BL6J_3840133 | Gm16350 | NCBI_Gene:102638701,ENSEMBL:ENSMUSG00000090024 | MGI:3840133 | lncRNA gene | predicted gene 16350 |
| 8 | gene | 14.994134 | 14.996912 | negative | MGI_C57BL6J_5594308 | Gm35149 | NCBI_Gene:102638629 | MGI:5594308 | lncRNA gene | predicted gene, 35149 |
| 8 | gene | 15.011005 | 15.033333 | positive | MGI_C57BL6J_1922151 | Kbtbd11 | NCBI_Gene:74901,ENSEMBL:ENSMUSG00000055675 | MGI:1922151 | protein coding gene | kelch repeat and BTB (POZ) domain containing 11 |
| 8 | gene | 15.015092 | 15.018907 | negative | MGI_C57BL6J_1923332 | 5830468F06Rik | ENSEMBL:ENSMUSG00000098322 | MGI:1923332 | lncRNA gene | RIKEN cDNA 5830468F06 gene |
| 8 | gene | 15.024326 | 15.028422 | negative | MGI_C57BL6J_5611072 | Gm37844 | ENSEMBL:ENSMUSG00000102615 | MGI:5611072 | unclassified gene | predicted gene, 37844 |
| 8 | gene | 15.038550 | 15.040422 | positive | MGI_C57BL6J_5594499 | Gm35340 | NCBI_Gene:102638883 | MGI:5594499 | lncRNA gene | predicted gene, 35340 |
| 8 | gene | 15.041444 | 15.046078 | negative | MGI_C57BL6J_2142823 | BB014433 | NCBI_Gene:434285,ENSEMBL:ENSMUSG00000049008 | MGI:2142823 | protein coding gene | expressed sequence BB014433 |
| 8 | gene | 15.044563 | 15.045648 | positive | MGI_C57BL6J_5791112 | Gm45276 | ENSEMBL:ENSMUSG00000109871 | MGI:5791112 | unclassified gene | predicted gene 45276 |
| 8 | gene | 15.046212 | 15.050770 | positive | MGI_C57BL6J_5791110 | Gm45274 | ENSEMBL:ENSMUSG00000109965 | MGI:5791110 | lncRNA gene | predicted gene 45274 |
| 8 | gene | 15.057653 | 15.133541 | positive | MGI_C57BL6J_1328358 | Myom2 | NCBI_Gene:17930,ENSEMBL:ENSMUSG00000031461 | MGI:1328358 | protein coding gene | myomesin 2 |
| 8 | pseudogene | 15.100329 | 15.101350 | negative | MGI_C57BL6J_3783120 | Gm15678 | ENSEMBL:ENSMUSG00000083000 | MGI:3783120 | pseudogene | predicted gene 15678 |
| 8 | gene | 15.141044 | 15.371982 | positive | MGI_C57BL6J_3643599 | Gm7706 | ENSEMBL:ENSMUSG00000110286 | MGI:3643599 | lncRNA gene | predicted gene 7706 |
| 8 | gene | 15.182089 | 15.189612 | negative | MGI_C57BL6J_5791107 | Gm45271 | ENSEMBL:ENSMUSG00000109886 | MGI:5791107 | lncRNA gene | predicted gene 45271 |
| 8 | gene | 15.227405 | 15.227537 | negative | MGI_C57BL6J_5454008 | Gm24231 | ENSEMBL:ENSMUSG00000077503 | MGI:5454008 | snoRNA gene | predicted gene, 24231 |
| 8 | gene | 15.375639 | 15.382429 | positive | MGI_C57BL6J_5594551 | Gm35392 | NCBI_Gene:102638957,ENSEMBL:ENSMUSG00000109748 | MGI:5594551 | lncRNA gene | predicted gene, 35392 |
| 8 | gene | 15.412324 | 15.412416 | positive | MGI_C57BL6J_5453999 | Gm24222 | ENSEMBL:ENSMUSG00000089459 | MGI:5453999 | rRNA gene | predicted gene, 24222 |
| 8 | gene | 15.413980 | 15.414089 | positive | MGI_C57BL6J_5455648 | Gm25871 | ENSEMBL:ENSMUSG00000094608 | MGI:5455648 | miRNA gene | predicted gene, 25871 |
| 8 | gene | 15.418536 | 15.418645 | positive | MGI_C57BL6J_5455032 | Gm25255 | ENSEMBL:ENSMUSG00000094971 | MGI:5455032 | miRNA gene | predicted gene, 25255 |
| 8 | pseudogene | 15.437866 | 15.438160 | positive | MGI_C57BL6J_5791514 | Gm45678 | ENSEMBL:ENSMUSG00000110288 | MGI:5791514 | pseudogene | predicted gene 45678 |
| 8 | gene | 15.530401 | 15.579574 | positive | MGI_C57BL6J_5825670 | Gm46033 | NCBI_Gene:108167524 | MGI:5825670 | lncRNA gene | predicted gene, 46033 |
| 8 | gene | 15.537582 | 15.538922 | positive | MGI_C57BL6J_5791513 | Gm45677 | ENSEMBL:ENSMUSG00000109839 | MGI:5791513 | unclassified gene | predicted gene 45677 |
| 8 | gene | 15.580279 | 15.583962 | negative | MGI_C57BL6J_5594612 | Gm35453 | ENSEMBL:ENSMUSG00000110356 | MGI:5594612 | lncRNA gene | predicted gene, 35453 |
| 8 | gene | 15.581943 | 15.584719 | positive | MGI_C57BL6J_5791512 | Gm45676 | ENSEMBL:ENSMUSG00000110392 | MGI:5791512 | lncRNA gene | predicted gene 45676 |
| 8 | gene | 15.584723 | 15.670425 | positive | MGI_C57BL6J_5791244 | Gm45408 | ENSEMBL:ENSMUSG00000110217 | MGI:5791244 | lncRNA gene | predicted gene 45408 |
| 8 | gene | 15.629981 | 15.633409 | positive | MGI_C57BL6J_5791511 | Gm45675 | ENSEMBL:ENSMUSG00000110201 | MGI:5791511 | unclassified gene | predicted gene 45675 |
| 8 | pseudogene | 15.877660 | 15.878209 | positive | MGI_C57BL6J_3779675 | Gm7128 | NCBI_Gene:634008,ENSEMBL:ENSMUSG00000109757 | MGI:3779675 | pseudogene | predicted gene 7128 |
| 8 | gene | 15.892537 | 17.535586 | negative | MGI_C57BL6J_2137383 | Csmd1 | NCBI_Gene:94109,ENSEMBL:ENSMUSG00000060924 | MGI:2137383 | protein coding gene | CUB and Sushi multiple domains 1 |
| 8 | gene | 15.933416 | 15.934667 | positive | MGI_C57BL6J_5825710 | Gm46073 | NCBI_Gene:108167587 | MGI:5825710 | lncRNA gene | predicted gene, 46073 |
| 8 | gene | 16.168757 | 16.168838 | negative | MGI_C57BL6J_4834227 | Mir3106 | miRBase:MI0004486,NCBI_Gene:100526464,ENSEMBL:ENSMUSG00000093142 | MGI:4834227 | miRNA gene | microRNA 3106 |
| 8 | gene | 16.226003 | 16.232305 | positive | MGI_C57BL6J_3840154 | Gm16346 | ENSEMBL:ENSMUSG00000085150 | MGI:3840154 | lncRNA gene | predicted gene 16346 |
| 8 | gene | 16.431315 | 16.468209 | positive | MGI_C57BL6J_3840141 | Gm16347 | NCBI_Gene:102639135,ENSEMBL:ENSMUSG00000087363 | MGI:3840141 | lncRNA gene | predicted gene 16347 |
| 8 | gene | 16.547242 | 16.549872 | positive | MGI_C57BL6J_5791229 | Gm45393 | ENSEMBL:ENSMUSG00000109616 | MGI:5791229 | unclassified gene | predicted gene 45393 |
| 8 | pseudogene | 16.581824 | 16.582722 | positive | MGI_C57BL6J_5010504 | Gm18319 | NCBI_Gene:100416916 | MGI:5010504 | pseudogene | predicted gene, 18319 |
| 8 | gene | 16.751319 | 16.752493 | positive | MGI_C57BL6J_5791230 | Gm45394 | ENSEMBL:ENSMUSG00000110132 | MGI:5791230 | unclassified gene | predicted gene 45394 |
| 8 | gene | 16.852168 | 16.852287 | positive | MGI_C57BL6J_5455055 | Gm25278 | ENSEMBL:ENSMUSG00000087892 | MGI:5455055 | snoRNA gene | predicted gene, 25278 |
| 8 | gene | 16.912706 | 16.916182 | negative | MGI_C57BL6J_1920469 | 3110080E11Rik | ENSEMBL:ENSMUSG00000109625 | MGI:1920469 | unclassified gene | RIKEN cDNA 3110080E11 gene |
| 8 | gene | 16.959941 | 16.964159 | negative | MGI_C57BL6J_2442083 | C030026K05Rik | NA | NA | unclassified gene | RIKEN cDNA C030026K05 gene |
| 8 | gene | 17.286531 | 17.293135 | positive | MGI_C57BL6J_5622021 | Gm39136 | NCBI_Gene:105243132 | MGI:5622021 | lncRNA gene | predicted gene, 39136 |
| 8 | gene | 17.438812 | 17.439238 | positive | MGI_C57BL6J_1925706 | C030002A05Rik | NA | NA | unclassified gene | RIKEN cDNA C030002A05 gene |
| 8 | gene | 17.562287 | 17.574415 | positive | MGI_C57BL6J_5622022 | Gm39137 | NCBI_Gene:105243134 | MGI:5622022 | lncRNA gene | predicted gene, 39137 |
| 8 | pseudogene | 18.052917 | 18.053666 | negative | MGI_C57BL6J_3644412 | Gm7724 | NCBI_Gene:665638 | MGI:3644412 | pseudogene | predicted gene 7724 |
| 8 | gene | 18.274712 | 18.274821 | negative | MGI_C57BL6J_5455442 | Gm25665 | ENSEMBL:ENSMUSG00000088320 | MGI:5455442 | snoRNA gene | predicted gene, 25665 |
| 8 | gene | 18.312472 | 18.334338 | negative | MGI_C57BL6J_1924173 | 1700014L14Rik | NCBI_Gene:108167578,ENSEMBL:ENSMUSG00000110056 | MGI:1924173 | lncRNA gene | RIKEN cDNA 1700014L14 gene |
| 8 | gene | 18.497497 | 18.522065 | negative | MGI_C57BL6J_5623348 | Gm40463 | NCBI_Gene:105244941 | MGI:5623348 | lncRNA gene | predicted gene, 40463 |
| 8 | gene | 18.512515 | 18.512645 | negative | MGI_C57BL6J_5454246 | Gm24469 | ENSEMBL:ENSMUSG00000077675 | MGI:5454246 | snoRNA gene | predicted gene, 24469 |
| 8 | gene | 18.541975 | 18.545256 | negative | MGI_C57BL6J_5594899 | Gm35740 | NCBI_Gene:102639420 | MGI:5594899 | lncRNA gene | predicted gene, 35740 |
| 8 | gene | 18.595131 | 18.805912 | positive | MGI_C57BL6J_2443308 | Mcph1 | NCBI_Gene:244329,ENSEMBL:ENSMUSG00000039842 | MGI:2443308 | protein coding gene | microcephaly, primary autosomal recessive 1 |
| 8 | gene | 18.613981 | 18.614109 | negative | MGI_C57BL6J_5455540 | Gm25763 | ENSEMBL:ENSMUSG00000077287 | MGI:5455540 | snoRNA gene | predicted gene, 25763 |
| 8 | pseudogene | 18.648689 | 18.657074 | negative | MGI_C57BL6J_4937976 | Gm17149 | NCBI_Gene:108167565,ENSEMBL:ENSMUSG00000090334 | MGI:4937976 | pseudogene | predicted gene 17149 |
| 8 | gene | 18.652801 | 18.678056 | negative | MGI_C57BL6J_4439649 | Gm16725 | ENSEMBL:ENSMUSG00000087300 | MGI:4439649 | lncRNA gene | predicted gene, 16725 |
| 8 | gene | 18.690263 | 18.741562 | negative | MGI_C57BL6J_1202890 | Angpt2 | NCBI_Gene:11601,ENSEMBL:ENSMUSG00000031465 | MGI:1202890 | protein coding gene | angiopoietin 2 |
| 8 | gene | 18.844181 | 18.846776 | negative | MGI_C57BL6J_3698177 | 9630029G12Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 9630029G12 gene |
| 8 | gene | 18.846277 | 18.891361 | positive | MGI_C57BL6J_1196345 | Agpat5 | NCBI_Gene:52123,ENSEMBL:ENSMUSG00000031467 | MGI:1196345 | protein coding gene | 1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) |
| 8 | gene | 18.862196 | 18.866143 | positive | MGI_C57BL6J_5791329 | Gm45493 | ENSEMBL:ENSMUSG00000109784 | MGI:5791329 | unclassified gene | predicted gene 45493 |
| 8 | gene | 18.889210 | 18.902316 | negative | MGI_C57BL6J_5595093 | Gm35934 | NCBI_Gene:102639679,ENSEMBL:ENSMUSG00000110530 | MGI:5595093 | lncRNA gene | predicted gene, 35934 |
| 8 | pseudogene | 18.905439 | 18.906533 | negative | MGI_C57BL6J_5804885 | Gm45770 | ENSEMBL:ENSMUSG00000110651 | MGI:5804885 | pseudogene | predicted gene 45770 |
| 8 | gene | 18.924152 | 18.952366 | negative | MGI_C57BL6J_2442327 | Xkr5 | NCBI_Gene:319581,ENSEMBL:ENSMUSG00000039814 | MGI:2442327 | protein coding gene | X-linked Kx blood group related 5 |
| 8 | gene | 18.932989 | 18.933091 | positive | MGI_C57BL6J_5452328 | Gm22551 | ENSEMBL:ENSMUSG00000088420 | MGI:5452328 | snoRNA gene | predicted gene, 22551 |
| 8 | gene | 18.950289 | 18.970684 | positive | MGI_C57BL6J_5623349 | Gm40464 | NCBI_Gene:105244942 | MGI:5623349 | lncRNA gene | predicted gene, 40464 |
| 8 | gene | 18.974940 | 18.978256 | negative | MGI_C57BL6J_2672976 | Defb40 | NCBI_Gene:360217,ENSEMBL:ENSMUSG00000053678 | MGI:2672976 | protein coding gene | defensin beta 40 |
| 8 | gene | 18.986233 | 18.991055 | negative | MGI_C57BL6J_2672966 | Defb37 | NCBI_Gene:353320,ENSEMBL:ENSMUSG00000053695 | MGI:2672966 | protein coding gene | defensin beta 37 |
| 8 | gene | 18.989004 | 18.998649 | positive | MGI_C57BL6J_5622023 | Gm39138 | NCBI_Gene:105243135 | MGI:5622023 | lncRNA gene | predicted gene, 39138 |
| 8 | gene | 19.023407 | 19.026566 | negative | MGI_C57BL6J_2672972 | Defb38 | NCBI_Gene:360212,ENSEMBL:ENSMUSG00000053790 | MGI:2672972 | protein coding gene | defensin beta 38 |
| 8 | pseudogene | 19.035221 | 19.045486 | negative | MGI_C57BL6J_3709611 | Gm15366 | NCBI_Gene:100504390,ENSEMBL:ENSMUSG00000080726 | MGI:3709611 | pseudogene | predicted gene 15366 |
| 8 | gene | 19.052826 | 19.064810 | negative | MGI_C57BL6J_2672974 | Defb39 | NCBI_Gene:360214,ENSEMBL:ENSMUSG00000061847 | MGI:2672974 | protein coding gene | defensin beta 39 |
| 8 | gene | 19.111931 | 19.114839 | negative | MGI_C57BL6J_1924924 | Defb12 | NCBI_Gene:77674,ENSEMBL:ENSMUSG00000043787 | MGI:1924924 | protein coding gene | defensin beta 12 |
| 8 | gene | 19.123728 | 19.126540 | positive | MGI_C57BL6J_2672979 | Defb34 | NCBI_Gene:360211,ENSEMBL:ENSMUSG00000052554 | MGI:2672979 | protein coding gene | defensin beta 34 |
| 8 | gene | 19.140759 | 19.143010 | positive | MGI_C57BL6J_3647173 | Spag11b | NCBI_Gene:546038,ENSEMBL:ENSMUSG00000059463 | MGI:3647173 | protein coding gene | sperm associated antigen 11B |
| 8 | gene | 19.140778 | 19.159440 | positive | MGI_C57BL6J_5804941 | Gm45826 | ENSEMBL:ENSMUSG00000110641 | MGI:5804941 | protein coding gene | predicted gene 45826 |
| 8 | gene | 19.157213 | 19.159920 | positive | MGI_C57BL6J_1925378 | Spag11a | NCBI_Gene:78128,ENSEMBL:ENSMUSG00000079842 | MGI:1925378 | protein coding gene | sperm associated antigen 11A |
| 8 | gene | 19.160244 | 19.162222 | positive | MGI_C57BL6J_5825671 | Gm46034 | NCBI_Gene:108167525 | MGI:5825671 | lncRNA gene | predicted gene, 46034 |
| 8 | gene | 19.163156 | 19.195309 | positive | MGI_C57BL6J_2675345 | Defb14 | NCBI_Gene:244332,ENSEMBL:ENSMUSG00000046354 | MGI:2675345 | protein coding gene | defensin beta 14 |
| 8 | gene | 19.163520 | 19.188065 | negative | MGI_C57BL6J_5622024 | Gm39139 | NCBI_Gene:105243136,ENSEMBL:ENSMUSG00000110542 | MGI:5622024 | lncRNA gene | predicted gene, 39139 |
| 8 | gene | 19.198704 | 19.201547 | positive | MGI_C57BL6J_1927667 | Defb4 | NCBI_Gene:56519,ENSEMBL:ENSMUSG00000059230 | MGI:1927667 | protein coding gene | defensin beta 4 |
| 8 | pseudogene | 19.210462 | 19.212767 | positive | MGI_C57BL6J_3647175 | Defa-ps12 | NCBI_Gene:654452,ENSEMBL:ENSMUSG00000081163 | MGI:3647175 | pseudogene | defensin, alpha, pseudogene 12 |
| 8 | gene | 19.225478 | 19.228209 | positive | MGI_C57BL6J_2151044 | Defb6 | NCBI_Gene:116746,ENSEMBL:ENSMUSG00000050756 | MGI:2151044 | protein coding gene | defensin beta 6 |
| 8 | gene | 19.239916 | 19.242143 | positive | MGI_C57BL6J_3646525 | Defb46 | NCBI_Gene:574081,ENSEMBL:ENSMUSG00000071169 | MGI:3646525 | protein coding gene | defensin beta 46 |
| 8 | gene | 19.247580 | 19.250828 | positive | MGI_C57BL6J_1933153 | Defb5 | NCBI_Gene:81007,ENSEMBL:ENSMUSG00000039785 | MGI:1933153 | protein coding gene | defensin beta 5 |
| 8 | pseudogene | 19.279009 | 19.279846 | negative | MGI_C57BL6J_3705466 | Gm15357 | ENSEMBL:ENSMUSG00000081901 | MGI:3705466 | pseudogene | predicted gene 15357 |
| 8 | gene | 19.293360 | 19.295339 | positive | MGI_C57BL6J_1351612 | Defb3 | NCBI_Gene:27358,ENSEMBL:ENSMUSG00000039775 | MGI:1351612 | protein coding gene | defensin beta 3 |
| 8 | pseudogene | 19.300677 | 19.304794 | positive | MGI_C57BL6J_3705864 | Defa-ps13 | NCBI_Gene:654456,ENSEMBL:ENSMUSG00000081154 | MGI:3705864 | pseudogene | defensin, alpha, pseudogene 13 |
| 8 | pseudogene | 19.422506 | 19.423444 | positive | MGI_C57BL6J_3705817 | Defa-ps14 | NCBI_Gene:100040514,ENSEMBL:ENSMUSG00000082710 | MGI:3705817 | pseudogene | defensin, alpha, pseudogene 14 |
| 8 | gene | 19.445769 | 19.447606 | negative | MGI_C57BL6J_2654206 | Defb8 | NCBI_Gene:244334,ENSEMBL:ENSMUSG00000031471 | MGI:2654206 | protein coding gene | defensin beta 8 |
| 8 | pseudogene | 19.472893 | 19.474732 | negative | MGI_C57BL6J_3705808 | Defa-ps15 | NCBI_Gene:100040528,ENSEMBL:ENSMUSG00000080894 | MGI:3705808 | pseudogene | defensin, alpha, pseudogene 15 |
| 8 | pseudogene | 19.492906 | 19.493609 | positive | MGI_C57BL6J_3642193 | Rpl19-ps11 | NCBI_Gene:100041511,ENSEMBL:ENSMUSG00000081094 | MGI:3642193 | pseudogene | ribosomal protein L19, pseudogene 11 |
| 8 | gene | 19.493189 | 19.493268 | positive | MGI_C57BL6J_5530670 | Gm27288 | ENSEMBL:ENSMUSG00000098798 | MGI:5530670 | miRNA gene | predicted gene, 27288 |
| 8 | gene | 19.495097 | 19.497775 | positive | MGI_C57BL6J_2179200 | Defb7 | NCBI_Gene:246080,ENSEMBL:ENSMUSG00000037790 | MGI:2179200 | protein coding gene | defensin beta 7 |
| 8 | pseudogene | 19.513853 | 19.517012 | negative | MGI_C57BL6J_3705720 | Gm15358 | NCBI_Gene:102633232,ENSEMBL:ENSMUSG00000082650 | MGI:3705720 | pseudogene | predicted gene 15358 |
| 8 | gene | 19.535319 | 19.535408 | negative | MGI_C57BL6J_5531234 | Gm27852 | ENSEMBL:ENSMUSG00000098476 | MGI:5531234 | miRNA gene | predicted gene, 27852 |
| 8 | pseudogene | 19.535331 | 19.541206 | positive | MGI_C57BL6J_3643338 | Gm16425 | NCBI_Gene:670326,ENSEMBL:ENSMUSG00000083218 | MGI:3643338 | pseudogene | predicted gene 16425 |
| 8 | pseudogene | 19.579893 | 19.581049 | positive | MGI_C57BL6J_3705375 | Selenot-ps | NCBI_Gene:100040561,ENSEMBL:ENSMUSG00000081920 | MGI:3705375 | pseudogene | selenoprotein T, pseudogene |
| 8 | gene | 19.580900 | 19.665322 | negative | MGI_C57BL6J_5595157 | Gm35998 | NCBI_Gene:102639764,ENSEMBL:ENSMUSG00000110463 | MGI:5595157 | lncRNA gene | predicted gene, 35998 |
| 8 | pseudogene | 19.632898 | 19.635441 | positive | MGI_C57BL6J_5588948 | Gm29789 | NCBI_Gene:101056171 | MGI:5588948 | pseudogene | predicted gene, 29789 |
| 8 | pseudogene | 19.682266 | 19.697736 | positive | MGI_C57BL6J_3644574 | Gm6483 | NCBI_Gene:624198,ENSEMBL:ENSMUSG00000087153 | MGI:3644574 | pseudogene | predicted gene 6483 |
| 8 | gene | 19.704311 | 19.753603 | positive | MGI_C57BL6J_1923056 | 4930467E23Rik | NCBI_Gene:626415,ENSEMBL:ENSMUSG00000096265 | MGI:1923056 | protein coding gene | RIKEN cDNA 4930467E23 gene |
| 8 | pseudogene | 19.711094 | 19.711748 | negative | MGI_C57BL6J_3643584 | Gm7760 | NCBI_Gene:665730,ENSEMBL:ENSMUSG00000110531 | MGI:3643584 | pseudogene | predicted gene 7760 |
| 8 | pseudogene | 19.784746 | 19.784856 | positive | MGI_C57BL6J_3710638 | Gm10060 | NCBI_Gene:100041567,ENSEMBL:ENSMUSG00000110669 | MGI:3710638 | pseudogene | predicted gene 10060 |
| 8 | gene | 19.788050 | 19.935330 | positive | MGI_C57BL6J_5590530 | Gm31371 | NCBI_Gene:78672,ENSEMBL:ENSMUSG00000110591 | MGI:5590530 | protein coding gene | predicted gene, 31371 |
| 8 | gene | 19.789798 | 19.798548 | positive | MGI_C57BL6J_5477347 | Gm26853 | ENSEMBL:ENSMUSG00000097230 | MGI:5477347 | lncRNA gene | predicted gene, 26853 |
| 8 | pseudogene | 19.942830 | 19.943265 | negative | MGI_C57BL6J_5434142 | Gm20786 | NCBI_Gene:673754,ENSEMBL:ENSMUSG00000110460 | MGI:5434142 | pseudogene | predicted gene, 20786 |
| 8 | gene | 19.973994 | 19.979937 | negative | MGI_C57BL6J_5825712 | Gm46075 | NCBI_Gene:108167590 | MGI:5825712 | lncRNA gene | predicted gene, 46075 |
| 8 | gene | 19.979994 | 20.012604 | positive | MGI_C57BL6J_5623351 | Gm40466 | NCBI_Gene:105244944,ENSEMBL:ENSMUSG00000110649 | MGI:5623351 | lncRNA gene | predicted gene, 40466 |
| 8 | gene | 20.013842 | 20.016268 | positive | MGI_C57BL6J_5595325 | Gm36166 | NCBI_Gene:102639983 | MGI:5595325 | lncRNA gene | predicted gene, 36166 |
| 8 | pseudogene | 20.019133 | 20.019411 | negative | MGI_C57BL6J_5804869 | Gm45754 | ENSEMBL:ENSMUSG00000110589 | MGI:5804869 | pseudogene | predicted gene 45754 |
| 8 | pseudogene | 20.034379 | 20.066067 | positive | MGI_C57BL6J_5434134 | Gm20778 | NCBI_Gene:665756,ENSEMBL:ENSMUSG00000094421 | MGI:5434134 | pseudogene | predicted gene, 20778 |
| 8 | pseudogene | 20.122251 | 20.151665 | positive | MGI_C57BL6J_5433975 | Gm21811 | NCBI_Gene:101056094,ENSEMBL:ENSMUSG00000096401 | MGI:5433975 | pseudogene | predicted gene, 21811 |
| 8 | pseudogene | 20.122384 | 20.122494 | positive | MGI_C57BL6J_5433933 | Gm21769 | ENSEMBL:ENSMUSG00000110458 | MGI:5433933 | pseudogene | predicted gene, 21769 |
| 8 | gene | 20.257557 | 20.261199 | negative | MGI_C57BL6J_5623353 | Gm40468 | NCBI_Gene:105244946 | MGI:5623353 | lncRNA gene | predicted gene, 40468 |
| 8 | pseudogene | 20.262613 | 20.297432 | negative | MGI_C57BL6J_3694236 | 6820431F20Rik | NCBI_Gene:547150,ENSEMBL:ENSMUSG00000071796 | MGI:3694236 | pseudogene | RIKEN cDNA 6820431F20 gene |
| 8 | gene | 20.292477 | 20.420781 | positive | MGI_C57BL6J_5477298 | Gm26804 | ENSEMBL:ENSMUSG00000097263 | MGI:5477298 | lncRNA gene | predicted gene, 26804 |
| 8 | pseudogene | 20.297324 | 20.297428 | negative | MGI_C57BL6J_5295689 | Gm20583 | NCBI_Gene:100384904 | MGI:5295689 | pseudogene | predicted gene, 20583 |
| 8 | gene | 20.326254 | 20.331188 | negative | MGI_C57BL6J_1925922 | 9530057J20Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 9530057J20 gene |
| 8 | gene | 20.331461 | 20.363760 | negative | MGI_C57BL6J_3796981 | Gm15319 | NCBI_Gene:100040599,ENSEMBL:ENSMUSG00000074449 | MGI:3796981 | protein coding gene | predicted gene 15319 |
| 8 | pseudogene | 20.381027 | 20.381692 | positive | MGI_C57BL6J_3643361 | Gm7807 | NCBI_Gene:665822,ENSEMBL:ENSMUSG00000110637 | MGI:3643361 | pseudogene | predicted gene 7807 |
| 8 | pseudogene | 20.385782 | 20.424814 | negative | MGI_C57BL6J_1914283 | 2610005L07Rik | NCBI_Gene:381598,ENSEMBL:ENSMUSG00000071793 | MGI:1914283 | pseudogene | RIKEN cDNA 2610005L07 gene |
| 8 | pseudogene | 20.424592 | 20.424696 | negative | MGI_C57BL6J_5295690 | Gm20584 | NCBI_Gene:100384905 | MGI:5295690 | pseudogene | predicted gene, 20584 |
| 8 | gene | 20.498274 | 20.512049 | positive | MGI_C57BL6J_5623352 | Gm40467 | NCBI_Gene:105244945 | MGI:5623352 | lncRNA gene | predicted gene, 40467 |
| 8 | gene | 20.549350 | 20.550331 | negative | MGI_C57BL6J_5623354 | Gm40469 | NA | NA | protein coding gene | predicted gene%2c 40469 |
| 8 | pseudogene | 20.556693 | 20.557486 | negative | MGI_C57BL6J_5434537 | Gm21182 | ENSEMBL:ENSMUSG00000095766 | MGI:5434537 | pseudogene | predicted gene, 21182 |
| 8 | pseudogene | 20.567424 | 20.582797 | positive | MGI_C57BL6J_5434447 | Gm21092 | NCBI_Gene:100861634,ENSEMBL:ENSMUSG00000095403 | MGI:5434447 | pseudogene | predicted gene, 21092 |
| 8 | pseudogene | 20.596266 | 20.597041 | negative | MGI_C57BL6J_5434467 | Gm21112 | NCBI_Gene:108167588,ENSEMBL:ENSMUSG00000110574 | MGI:5434467 | pseudogene | predicted gene, 21112 |
| 8 | gene | 20.614406 | 20.640703 | positive | MGI_C57BL6J_5434474 | Gm21119 | NCBI_Gene:100861668,ENSEMBL:ENSMUSG00000095294 | MGI:5434474 | protein coding gene | predicted gene, 21119 |
| 8 | gene | 20.649947 | 20.652334 | positive | MGI_C57BL6J_5825711 | Gm46074 | NCBI_Gene:108167589 | MGI:5825711 | lncRNA gene | predicted gene, 46074 |
| 8 | pseudogene | 20.828015 | 20.835070 | positive | MGI_C57BL6J_5434301 | Gm20946 | NCBI_Gene:100415900,ENSEMBL:ENSMUSG00000110699 | MGI:5434301 | pseudogene | predicted gene, 20946 |
| 8 | pseudogene | 20.841352 | 20.842350 | negative | MGI_C57BL6J_5434153 | Gm20797 | NCBI_Gene:100039061,ENSEMBL:ENSMUSG00000096890 | MGI:5434153 | pseudogene | predicted gene, 20797 |
| 8 | pseudogene | 20.869332 | 20.870658 | negative | MGI_C57BL6J_5434300 | Gm20945 | NCBI_Gene:100415898,ENSEMBL:ENSMUSG00000094534 | MGI:5434300 | pseudogene | predicted gene, 20945 |
| 8 | pseudogene | 20.876499 | 20.889113 | positive | MGI_C57BL6J_5439395 | Gm21944 | NCBI_Gene:654455 | MGI:5439395 | pseudogene | predicted gene, 21944 |
| 8 | pseudogene | 20.879130 | 20.888483 | negative | MGI_C57BL6J_5434152 | Gm20796 | NCBI_Gene:100039037,ENSEMBL:ENSMUSG00000093800 | MGI:5434152 | pseudogene | predicted gene, 20796 |
| 8 | gene | 20.892667 | 20.897723 | positive | MGI_C57BL6J_3647176 | Defb33 | NCBI_Gene:654453,ENSEMBL:ENSMUSG00000074454 | MGI:3647176 | protein coding gene | defensin beta 33 |
| 8 | gene | 20.900606 | 20.902627 | negative | MGI_C57BL6J_3705859 | Gm15056 | NCBI_Gene:100504014,ENSEMBL:ENSMUSG00000082976 | MGI:3705859 | protein coding gene | predicted gene 15056 |
| 8 | gene | 20.901960 | 20.922102 | negative | MGI_C57BL6J_3646527 | Gm6040 | NCBI_Gene:100503992,ENSEMBL:ENSMUSG00000071165 | MGI:3646527 | protein coding gene | predicted gene 6040 |
| 8 | gene | 20.943694 | 20.944748 | negative | MGI_C57BL6J_3630303 | AY761185 | NCBI_Gene:100503970,ENSEMBL:ENSMUSG00000079120 | MGI:3630303 | protein coding gene | cDNA sequence AY761185 |
| 8 | gene | 21.025545 | 21.026517 | positive | MGI_C57BL6J_1913548 | Defa21 | NCBI_Gene:66298,ENSEMBL:ENSMUSG00000074447 | MGI:1913548 | protein coding gene | defensin, alpha, 21 |
| 8 | gene | 21.040256 | 21.040360 | negative | MGI_C57BL6J_5452919 | Gm23142 | ENSEMBL:ENSMUSG00000094406 | MGI:5452919 | snRNA gene | predicted gene, 23142 |
| 8 | gene | 21.055040 | 21.056016 | positive | MGI_C57BL6J_3630381 | Defa23 | NCBI_Gene:497114,ENSEMBL:ENSMUSG00000074446 | MGI:3630381 | protein coding gene | defensin, alpha, 23 |
| 8 | gene | 21.065038 | 21.066001 | positive | MGI_C57BL6J_3711900 | Defa35 | NCBI_Gene:100041688,ENSEMBL:ENSMUSG00000061845 | MGI:3711900 | protein coding gene | defensin, alpha, 35 |
| 8 | pseudogene | 21.078141 | 21.078846 | positive | MGI_C57BL6J_3705791 | Defa-ps3 | ENSEMBL:ENSMUSG00000087663 | MGI:3705791 | pseudogene | defensin, alpha, pseudogene 3 |
| 8 | gene | 21.084442 | 21.085285 | positive | MGI_C57BL6J_3630385 | Defa25 | NCBI_Gene:13236,ENSEMBL:ENSMUSG00000094687 | MGI:3630385 | protein coding gene | defensin, alpha, 25 |
| 8 | pseudogene | 21.091333 | 21.091883 | positive | MGI_C57BL6J_3645828 | Gm6689 | NCBI_Gene:626605,ENSEMBL:ENSMUSG00000085898 | MGI:3645828 | pseudogene | predicted gene 6689 |
| 8 | gene | 21.094971 | 21.096050 | negative | MGI_C57BL6J_3709605 | Defa38 | NCBI_Gene:634825,ENSEMBL:ENSMUSG00000061958 | MGI:3709605 | protein coding gene | defensin, alpha, 38 |
| 8 | gene | 21.095624 | 21.095715 | negative | MGI_C57BL6J_5454345 | Gm24568 | ENSEMBL:ENSMUSG00000094516 | MGI:5454345 | snoRNA gene | predicted gene, 24568 |
| 8 | pseudogene | 21.100487 | 21.103271 | positive | MGI_C57BL6J_3781046 | Gm2869 | NCBI_Gene:100040619,ENSEMBL:ENSMUSG00000085173 | MGI:3781046 | pseudogene | predicted gene 2869 |
| 8 | pseudogene | 21.119166 | 21.119348 | positive | MGI_C57BL6J_3705782 | Defa-ps4 | ENSEMBL:ENSMUSG00000083509 | MGI:3705782 | pseudogene | defensin, alpha, pseudogene 4 |
| 8 | gene | 21.134642 | 21.135598 | positive | MGI_C57BL6J_3808881 | Defa30 | NCBI_Gene:100038927,ENSEMBL:ENSMUSG00000074444 | MGI:3808881 | protein coding gene | defensin, alpha, 30 |
| 8 | gene | 21.162277 | 21.163249 | positive | MGI_C57BL6J_3639039 | Defa22 | NCBI_Gene:382059,ENSEMBL:ENSMUSG00000074443 | MGI:3639039 | protein coding gene | defensin, alpha, 22 |
| 8 | gene | 21.176768 | 21.176872 | negative | MGI_C57BL6J_5456017 | Gm26240 | ENSEMBL:ENSMUSG00000095431 | MGI:5456017 | snRNA gene | predicted gene, 26240 |
| 8 | gene | 21.191573 | 21.192550 | positive | MGI_C57BL6J_102509 | Defa31 | NCBI_Gene:13226,ENSEMBL:ENSMUSG00000074442 | MGI:102509 | protein coding gene | defensin, alpha, 31 |
| 8 | gene | 21.201561 | 21.202539 | positive | MGI_C57BL6J_3705230 | Defa41 | NCBI_Gene:100041759,ENSEMBL:ENSMUSG00000079116 | MGI:3705230 | protein coding gene | defensin, alpha, 41 |
| 8 | pseudogene | 21.218984 | 21.219763 | positive | MGI_C57BL6J_3705778 | Defa-ps5 | ENSEMBL:ENSMUSG00000094321 | MGI:3705778 | pseudogene | defensin, alpha, pseudogene 5 |
| 8 | pseudogene | 21.225406 | 21.226247 | positive | MGI_C57BL6J_3705855 | Defa-ps6 | NCBI_Gene:672740,ENSEMBL:ENSMUSG00000095328 | MGI:3705855 | pseudogene | defensin, alpha, pseudogene 6 |
| 8 | pseudogene | 21.232066 | 21.232233 | negative | MGI_C57BL6J_3705773 | Defa-ps7 | ENSEMBL:ENSMUSG00000080722 | MGI:3705773 | pseudogene | defensin, alpha, pseudogene 7 |
| 8 | pseudogene | 21.236510 | 21.239297 | positive | MGI_C57BL6J_3705378 | Gm15303 | NCBI_Gene:665969,ENSEMBL:ENSMUSG00000082957 | MGI:3705378 | pseudogene | predicted gene 15303 |
| 8 | gene | 21.249754 | 21.250556 | positive | MGI_C57BL6J_3708769 | Defa40 | NCBI_Gene:100041787,ENSEMBL:ENSMUSG00000074441 | MGI:3708769 | protein coding gene | defensin, alpha, 40 |
| 8 | pseudogene | 21.270693 | 21.271997 | positive | MGI_C57BL6J_3832672 | Defa-ps8 | ENSEMBL:ENSMUSG00000094180 | MGI:3832672 | pseudogene | defensin, alpha, pseudogene 8 |
| 8 | gene | 21.287409 | 21.288377 | positive | MGI_C57BL6J_94883 | Defa3 | NCBI_Gene:13237,ENSEMBL:ENSMUSG00000074440 | MGI:94883 | protein coding gene | defensin, alpha, 3 |
| 8 | gene | 21.297394 | 21.298375 | positive | MGI_C57BL6J_99583 | Defa5 | NCBI_Gene:13239,ENSEMBL:ENSMUSG00000074439 | MGI:99583 | protein coding gene | defensin, alpha, 5 |
| 8 | pseudogene | 21.309162 | 21.309942 | positive | MGI_C57BL6J_3705799 | Gm15300 | ENSEMBL:ENSMUSG00000083251 | MGI:3705799 | pseudogene | predicted gene 15300 |
| 8 | gene | 21.315546 | 21.316481 | positive | MGI_C57BL6J_3642780 | Defa27 | NCBI_Gene:100041811,ENSEMBL:ENSMUSG00000082211 | MGI:3642780 | protein coding gene | defensin, alpha, 27 |
| 8 | gene | 21.325887 | 21.327020 | negative | MGI_C57BL6J_94881 | Defa29 | NCBI_Gene:13218,ENSEMBL:ENSMUSG00000074437 | MGI:94881 | protein coding gene | defensin, alpha, 29 |
| 8 | gene | 21.326548 | 21.326639 | negative | MGI_C57BL6J_5453338 | Gm23561 | ENSEMBL:ENSMUSG00000095221 | MGI:5453338 | snoRNA gene | predicted gene, 23561 |
| 8 | pseudogene | 21.330783 | 21.333576 | positive | MGI_C57BL6J_3705638 | Gm15302 | NCBI_Gene:100415769,ENSEMBL:ENSMUSG00000084377 | MGI:3705638 | pseudogene | predicted gene 15302 |
| 8 | pseudogene | 21.342586 | 21.342757 | positive | MGI_C57BL6J_3705774 | Defa-ps16 | NCBI_Gene:100040668,ENSEMBL:ENSMUSG00000082918 | MGI:3705774 | pseudogene | defensin, alpha, pseudogene 16 |
| 8 | gene | 21.378515 | 21.379495 | positive | MGI_C57BL6J_94882 | Defa2 | NCBI_Gene:100294660,ENSEMBL:ENSMUSG00000096295 | MGI:94882 | protein coding gene | defensin, alpha, 2 |
| 8 | gene | 21.391811 | 21.392792 | positive | MGI_C57BL6J_5434357 | Defa33 | NCBI_Gene:100505096,ENSEMBL:ENSMUSG00000094362 | MGI:5434357 | protein coding gene | defensin, alpha, 33 |
| 8 | gene | 21.427411 | 21.428391 | positive | MGI_C57BL6J_5434853 | Defa36 | NCBI_Gene:100862126,ENSEMBL:ENSMUSG00000094662 | MGI:5434853 | protein coding gene | defensin, alpha, 36 |
| 8 | pseudogene | 21.438675 | 21.439467 | positive | MGI_C57BL6J_3705798 | Gm15305 | ENSEMBL:ENSMUSG00000084344 | MGI:3705798 | pseudogene | predicted gene 15305 |
| 8 | pseudogene | 21.445069 | 21.446004 | positive | MGI_C57BL6J_3642785 | Defa-ps18 | NCBI_Gene:100041843,ENSEMBL:ENSMUSG00000074436 | MGI:3642785 | pseudogene | defensin, alpha, pseudogene 18 |
| 8 | pseudogene | 21.454293 | 21.454762 | positive | MGI_C57BL6J_3705458 | Gm15309 | NCBI_Gene:100040678,ENSEMBL:ENSMUSG00000083106 | MGI:3705458 | pseudogene | predicted gene 15309 |
| 8 | gene | 21.455427 | 21.456560 | negative | MGI_C57BL6J_3645033 | Defa42 | NCBI_Gene:665927,ENSEMBL:ENSMUSG00000079114 | MGI:3645033 | protein coding gene | defensin, alpha, 42 |
| 8 | gene | 21.456090 | 21.456181 | negative | MGI_C57BL6J_5453345 | Gm23568 | ENSEMBL:ENSMUSG00000096558 | MGI:5453345 | snoRNA gene | predicted gene, 23568 |
| 8 | pseudogene | 21.460794 | 21.463597 | positive | MGI_C57BL6J_3705536 | Gm15304 | NCBI_Gene:100415768,ENSEMBL:ENSMUSG00000081321 | MGI:3705536 | pseudogene | predicted gene 15304 |
| 8 | pseudogene | 21.473151 | 21.473522 | positive | MGI_C57BL6J_3705788 | Defa-ps17 | NCBI_Gene:100040693,ENSEMBL:ENSMUSG00000095925 | MGI:3705788 | pseudogene | defensin, alpha, pseudogene 17 |
| 8 | gene | 21.509258 | 21.510237 | positive | MGI_C57BL6J_1915259 | Defa20 | NCBI_Gene:68009,ENSEMBL:ENSMUSG00000095066 | MGI:1915259 | protein coding gene | defensin, alpha, 20 |
| 8 | gene | 21.529030 | 21.530012 | positive | MGI_C57BL6J_3709042 | Defa32 | NCBI_Gene:100041890,ENSEMBL:ENSMUSG00000094818 | MGI:3709042 | protein coding gene | defensin, alpha, 32 |
| 8 | gene | 21.548060 | 21.548165 | negative | MGI_C57BL6J_5454200 | Gm24423 | ENSEMBL:ENSMUSG00000075934 | MGI:5454200 | snRNA gene | predicted gene, 24423 |
| 8 | gene | 21.565386 | 21.566350 | positive | MGI_C57BL6J_3705236 | Defa37 | NCBI_Gene:100041895,ENSEMBL:ENSMUSG00000065956 | MGI:3705236 | protein coding gene | defensin, alpha, 37 |
| 8 | pseudogene | 21.576640 | 21.577426 | positive | MGI_C57BL6J_3705786 | Gm15311 | ENSEMBL:ENSMUSG00000083987 | MGI:3705786 | pseudogene | predicted gene 15311 |
| 8 | gene | 21.582960 | 21.583961 | positive | MGI_C57BL6J_3646688 | Defa28 | NCBI_Gene:626682,ENSEMBL:ENSMUSG00000074434 | MGI:3646688 | protein coding gene | defensin, alpha, 28 |
| 8 | pseudogene | 21.592341 | 21.592611 | positive | MGI_C57BL6J_3705692 | Gm15312 | NCBI_Gene:100040706,ENSEMBL:ENSMUSG00000084041 | MGI:3705692 | pseudogene | predicted gene 15312 |
| 8 | gene | 21.593375 | 21.594508 | negative | MGI_C57BL6J_3648003 | Defa43 | NCBI_Gene:665956,ENSEMBL:ENSMUSG00000079113 | MGI:3648003 | protein coding gene | defensin, alpha, 43 |
| 8 | gene | 21.594038 | 21.594129 | negative | MGI_C57BL6J_5454606 | Gm24829 | ENSEMBL:ENSMUSG00000095703 | MGI:5454606 | snoRNA gene | predicted gene, 24829 |
| 8 | pseudogene | 21.598741 | 21.601526 | positive | MGI_C57BL6J_3705655 | Gm15314 | NCBI_Gene:635093,ENSEMBL:ENSMUSG00000082323 | MGI:3705655 | pseudogene | predicted gene 15314 |
| 8 | gene | 21.618168 | 21.618972 | positive | MGI_C57BL6J_3630390 | Defa26 | NCBI_Gene:626708,ENSEMBL:ENSMUSG00000060070 | MGI:3630390 | protein coding gene | defensin, alpha, 26 |
| 8 | pseudogene | 21.639071 | 21.640378 | positive | MGI_C57BL6J_3705784 | Gm15313 | ENSEMBL:ENSMUSG00000084366 | MGI:3705784 | pseudogene | predicted gene 15313 |
| 8 | gene | 21.655767 | 21.656736 | positive | MGI_C57BL6J_1345152 | Defa17 | NCBI_Gene:23855,ENSEMBL:ENSMUSG00000060208 | MGI:1345152 | protein coding gene | defensin, alpha, 17 |
| 8 | gene | 21.665797 | 21.666733 | positive | MGI_C57BL6J_3709048 | Defa34 | NCBI_Gene:100041952,ENSEMBL:ENSMUSG00000063206 | MGI:3709048 | protein coding gene | defensin, alpha, 34 |
| 8 | pseudogene | 21.688648 | 21.689428 | positive | MGI_C57BL6J_3705785 | Defa-ps9 | ENSEMBL:ENSMUSG00000082859 | MGI:3705785 | pseudogene | defensin, alpha, pseudogene 9 |
| 8 | pseudogene | 21.695012 | 21.695853 | positive | MGI_C57BL6J_3630392 | Defa-ps1 | NCBI_Gene:727720,ENSEMBL:ENSMUSG00000095813 | MGI:3630392 | pseudogene | defensin, alpha, pseudogene 1 |
| 8 | gene | 21.702519 | 21.703647 | negative | MGI_C57BL6J_3611585 | Defa39 | NCBI_Gene:382000,ENSEMBL:ENSMUSG00000058618 | MGI:3611585 | protein coding gene | defensin, alpha, 39 |
| 8 | gene | 21.703175 | 21.703266 | negative | MGI_C57BL6J_5452224 | Gm22447 | ENSEMBL:ENSMUSG00000095507 | MGI:5452224 | snoRNA gene | predicted gene, 22447 |
| 8 | pseudogene | 21.707866 | 21.710605 | positive | MGI_C57BL6J_3644630 | Gm7869 | NCBI_Gene:100415770,ENSEMBL:ENSMUSG00000081668 | MGI:3644630 | pseudogene | predicted gene 7869 |
| 8 | pseudogene | 21.722057 | 21.722241 | positive | MGI_C57BL6J_3705783 | Defa-ps10 | NCBI_Gene:100040729,ENSEMBL:ENSMUSG00000096340 | MGI:3705783 | pseudogene | defensin, alpha, pseudogene 10 |
| 8 | gene | 21.733883 | 21.735386 | positive | MGI_C57BL6J_99582 | Defa6 | NA | NA | protein coding gene | defensin%2c alpha%2c 6 |
| 8 | gene | 21.734494 | 21.735471 | positive | MGI_C57BL6J_3630383 | Defa24 | NCBI_Gene:503491,ENSEMBL:ENSMUSG00000064213 | MGI:3630383 | protein coding gene | defensin, alpha, 24 |
| 8 | gene | 21.735268 | 21.735386 | positive | MGI_C57BL6J_99589 | Defa12 | NA | NA | protein coding gene | defensin%2c alpha%2c 12 |
| 8 | pseudogene | 21.749175 | 21.750000 | positive | MGI_C57BL6J_3705879 | Defa-ps11 | NCBI_Gene:100040739,ENSEMBL:ENSMUSG00000084254 | MGI:3705879 | pseudogene | defensin, alpha, pseudogene 11 |
| 8 | gene | 21.776555 | 21.795185 | positive | MGI_C57BL6J_1096878 | Defb1 | NCBI_Gene:13214,ENSEMBL:ENSMUSG00000044748 | MGI:1096878 | protein coding gene | defensin beta 1 |
| 8 | pseudogene | 21.816964 | 21.817384 | negative | MGI_C57BL6J_3705494 | Gm15316 | ENSEMBL:ENSMUSG00000082698 | MGI:3705494 | pseudogene | predicted gene 15316 |
| 8 | gene | 21.823539 | 21.831296 | positive | MGI_C57BL6J_3055870 | Defb50 | NCBI_Gene:387334,ENSEMBL:ENSMUSG00000058568 | MGI:3055870 | protein coding gene | defensin beta 50 |
| 8 | gene | 21.839926 | 21.843482 | positive | MGI_C57BL6J_1338754 | Defb2 | NCBI_Gene:13215,ENSEMBL:ENSMUSG00000006570 | MGI:1338754 | protein coding gene | defensin beta 2 |
| 8 | gene | 21.858901 | 21.862011 | positive | MGI_C57BL6J_2179205 | Defb10 | NCBI_Gene:246085,ENSEMBL:ENSMUSG00000044743 | MGI:2179205 | protein coding gene | defensin beta 10 |
| 8 | gene | 21.881713 | 21.885434 | negative | MGI_C57BL6J_2179198 | Defb9 | NCBI_Gene:246079,ENSEMBL:ENSMUSG00000047390 | MGI:2179198 | protein coding gene | defensin beta 9 |
| 8 | gene | 21.905374 | 21.906432 | negative | MGI_C57BL6J_2179197 | Defb11 | NCBI_Gene:246081,ENSEMBL:ENSMUSG00000045337 | MGI:2179197 | protein coding gene | defensin beta 11 |
| 8 | gene | 21.906664 | 21.934313 | positive | MGI_C57BL6J_5596038 | Gm36879 | ENSEMBL:ENSMUSG00000110616 | MGI:5596038 | lncRNA gene | predicted gene, 36879 |
| 8 | gene | 21.929799 | 21.932710 | negative | MGI_C57BL6J_2179202 | Defb15 | NCBI_Gene:246082,ENSEMBL:ENSMUSG00000048500 | MGI:2179202 | protein coding gene | defensin beta 15 |
| 8 | pseudogene | 21.935351 | 21.935824 | positive | MGI_C57BL6J_5804911 | Gm45796 | ENSEMBL:ENSMUSG00000110681 | MGI:5804911 | pseudogene | predicted gene 45796 |
| 8 | gene | 21.938352 | 21.940878 | positive | MGI_C57BL6J_2179204 | Defb35 | NCBI_Gene:246084,ENSEMBL:ENSMUSG00000058052 | MGI:2179204 | protein coding gene | defensin beta 35 |
| 8 | gene | 21.946762 | 21.948850 | positive | MGI_C57BL6J_2179203 | Defb13 | NCBI_Gene:246083,ENSEMBL:ENSMUSG00000044222 | MGI:2179203 | protein coding gene | defensin beta 13 |
| 8 | gene | 21.947674 | 21.969608 | negative | MGI_C57BL6J_5588980 | Gm29821 | NCBI_Gene:102631498 | MGI:5588980 | lncRNA gene | predicted gene, 29821 |
| 8 | gene | 21.958714 | 21.964303 | positive | MGI_C57BL6J_2142877 | Fam90a1a | NCBI_Gene:97476,ENSEMBL:ENSMUSG00000079112 | MGI:2142877 | protein coding gene | family with sequence similarity 90, member A1A |
| 8 | gene | 21.969741 | 21.974054 | positive | MGI_C57BL6J_1915179 | Ccdc70 | NCBI_Gene:67929,ENSEMBL:ENSMUSG00000017049 | MGI:1915179 | protein coding gene | coiled-coil domain containing 70 |
| 8 | gene | 21.992783 | 22.060305 | negative | MGI_C57BL6J_103297 | Atp7b | NCBI_Gene:11979,ENSEMBL:ENSMUSG00000006567 | MGI:103297 | protein coding gene | ATPase, Cu++ transporting, beta polypeptide |
| 8 | gene | 22.060721 | 22.071627 | positive | MGI_C57BL6J_2142632 | Alg11 | NCBI_Gene:207958,ENSEMBL:ENSMUSG00000063362 | MGI:2142632 | protein coding gene | asparagine-linked glycosylation 11 (alpha-1,2-mannosyltransferase) |
| 8 | gene | 22.073615 | 22.125418 | negative | MGI_C57BL6J_2142824 | Nek5 | NCBI_Gene:330721,ENSEMBL:ENSMUSG00000037738 | MGI:2142824 | protein coding gene | NIMA (never in mitosis gene a)-related expressed kinase 5 |
| 8 | gene | 22.128283 | 22.167002 | negative | MGI_C57BL6J_1344371 | Nek3 | NCBI_Gene:23954,ENSEMBL:ENSMUSG00000031478 | MGI:1344371 | protein coding gene | NIMA (never in mitosis gene a)-related expressed kinase 3 |
| 8 | gene | 22.168149 | 22.185819 | negative | MGI_C57BL6J_1931797 | Ckap2 | NCBI_Gene:80986,ENSEMBL:ENSMUSG00000037725 | MGI:1931797 | protein coding gene | cytoskeleton associated protein 2 |
| 8 | gene | 22.185915 | 22.187152 | positive | MGI_C57BL6J_5589037 | Gm29878 | NCBI_Gene:102631575 | MGI:5589037 | lncRNA gene | predicted gene, 29878 |
| 8 | gene | 22.192809 | 22.220843 | positive | MGI_C57BL6J_1917410 | Vps36 | NCBI_Gene:70160,ENSEMBL:ENSMUSG00000031479 | MGI:1917410 | protein coding gene | vacuolar protein sorting 36 |
| 8 | gene | 22.225862 | 22.261334 | positive | MGI_C57BL6J_1929096 | Thsd1 | NCBI_Gene:56229,ENSEMBL:ENSMUSG00000031480 | MGI:1929096 | protein coding gene | thrombospondin, type I, domain 1 |
| 8 | pseudogene | 22.230847 | 22.231320 | negative | MGI_C57BL6J_1321400 | Rpl30-ps2 | NCBI_Gene:19948,ENSEMBL:ENSMUSG00000090007 | MGI:1321400 | pseudogene | ribosomal protein L30, pseudogene 2 |
| 8 | gene | 22.283315 | 22.371419 | positive | MGI_C57BL6J_2446460 | Tpte | NCBI_Gene:234129,ENSEMBL:ENSMUSG00000031481 | MGI:2446460 | protein coding gene | transmembrane phosphatase with tensin homology |
| 8 | pseudogene | 22.307086 | 22.307416 | positive | MGI_C57BL6J_5825672 | Gm46035 | NCBI_Gene:108167526 | MGI:5825672 | pseudogene | predicted gene, 46035 |
| 8 | gene | 22.375550 | 22.398621 | negative | MGI_C57BL6J_1342274 | Slc25a15 | NCBI_Gene:18408,ENSEMBL:ENSMUSG00000031482 | MGI:1342274 | protein coding gene | solute carrier family 25 (mitochondrial carrier ornithine transporter), member 15 |
| 8 | gene | 22.398721 | 22.399596 | positive | MGI_C57BL6J_1916358 | 1810012K16Rik | NCBI_Gene:69108,ENSEMBL:ENSMUSG00000099825 | MGI:1916358 | lncRNA gene | RIKEN cDNA 1810012K16 gene |
| 8 | pseudogene | 22.402576 | 22.402752 | negative | MGI_C57BL6J_5791288 | Gm45452 | ENSEMBL:ENSMUSG00000110193 | MGI:5791288 | pseudogene | predicted gene 45452 |
| 8 | pseudogene | 22.407204 | 22.409382 | positive | MGI_C57BL6J_3647295 | Gm6009 | NCBI_Gene:102631749,ENSEMBL:ENSMUSG00000109938 | MGI:3647295 | pseudogene | predicted gene 6009 |
| 8 | gene | 22.410457 | 22.411309 | positive | MGI_C57BL6J_5791289 | Gm45453 | ENSEMBL:ENSMUSG00000110129 | MGI:5791289 | unclassified gene | predicted gene 45453 |
| 8 | gene | 22.411181 | 22.429669 | positive | MGI_C57BL6J_1913153 | Mrps31 | NCBI_Gene:57312,ENSEMBL:ENSMUSG00000031533 | MGI:1913153 | protein coding gene | mitochondrial ribosomal protein S31 |
| 8 | gene | 22.453132 | 22.454753 | negative | MGI_C57BL6J_5622025 | Gm39140 | NCBI_Gene:105243137 | MGI:5622025 | lncRNA gene | predicted gene, 39140 |
| 8 | gene | 22.462614 | 22.476882 | negative | MGI_C57BL6J_2142501 | Smim19 | NCBI_Gene:102032,ENSEMBL:ENSMUSG00000031534 | MGI:2142501 | protein coding gene | small integral membrane protein 19 |
| 8 | gene | 22.476700 | 22.569616 | positive | MGI_C57BL6J_97851 | Slc20a2 | NCBI_Gene:20516,ENSEMBL:ENSMUSG00000037656 | MGI:97851 | protein coding gene | solute carrier family 20, member 2 |
| 8 | gene | 22.480291 | 22.483332 | positive | MGI_C57BL6J_4937125 | Gm17491 | ENSEMBL:ENSMUSG00000097042 | MGI:4937125 | lncRNA gene | predicted gene, 17491 |
| 8 | gene | 22.577075 | 22.593813 | negative | MGI_C57BL6J_106922 | Vdac3 | NCBI_Gene:22335,ENSEMBL:ENSMUSG00000008892 | MGI:106922 | protein coding gene | voltage-dependent anion channel 3 |
| 8 | gene | 22.594063 | 22.611124 | positive | MGI_C57BL6J_5589245 | Gm30086 | NCBI_Gene:102631860 | MGI:5589245 | lncRNA gene | predicted gene, 30086 |
| 8 | gene | 22.609081 | 22.609423 | positive | MGI_C57BL6J_1920678 | 1700058M10Rik | NA | NA | unclassified gene | RIKEN cDNA 1700058M10 gene |
| 8 | gene | 22.614848 | 22.619586 | positive | MGI_C57BL6J_5589294 | Gm30135 | NCBI_Gene:102631927 | MGI:5589294 | lncRNA gene | predicted gene, 30135 |
| 8 | gene | 22.623685 | 22.627547 | positive | MGI_C57BL6J_2385299 | Dkk4 | NCBI_Gene:234130,ENSEMBL:ENSMUSG00000031535 | MGI:2385299 | protein coding gene | dickkopf WNT signaling pathway inhibitor 4 |
| 8 | gene | 22.628119 | 22.653466 | negative | MGI_C57BL6J_97740 | Polb | NCBI_Gene:18970,ENSEMBL:ENSMUSG00000031536 | MGI:97740 | protein coding gene | polymerase (DNA directed), beta |
| 8 | gene | 22.634371 | 22.636809 | negative | MGI_C57BL6J_1915324 | A930013F10Rik | NCBI_Gene:68074 | MGI:1915324 | lncRNA gene | RIKEN cDNA A930013F10 gene |
| 8 | gene | 22.644861 | 22.646286 | positive | MGI_C57BL6J_5477403 | Gm26909 | ENSEMBL:ENSMUSG00000097839 | MGI:5477403 | lncRNA gene | predicted gene, 26909 |
| 8 | gene | 22.659205 | 22.706591 | negative | MGI_C57BL6J_1338071 | Ikbkb | NCBI_Gene:16150,ENSEMBL:ENSMUSG00000031537 | MGI:1338071 | protein coding gene | inhibitor of kappaB kinase beta |
| 8 | gene | 22.686265 | 22.695646 | positive | MGI_C57BL6J_3705174 | Gm15346 | ENSEMBL:ENSMUSG00000085199 | MGI:3705174 | lncRNA gene | predicted gene 15346 |
| 8 | gene | 22.757722 | 22.782848 | positive | MGI_C57BL6J_97610 | Plat | NCBI_Gene:18791,ENSEMBL:ENSMUSG00000031538 | MGI:97610 | protein coding gene | plasminogen activator, tissue |
| 8 | gene | 22.787354 | 22.805654 | negative | MGI_C57BL6J_1929214 | Ap3m2 | NCBI_Gene:64933,ENSEMBL:ENSMUSG00000031539 | MGI:1929214 | protein coding gene | adaptor-related protein complex 3, mu 2 subunit |
| 8 | gene | 22.805766 | 22.809537 | positive | MGI_C57BL6J_1920549 | 1700041G16Rik | ENSEMBL:ENSMUSG00000054822 | MGI:1920549 | lncRNA gene | RIKEN cDNA 1700041G16 gene |
| 8 | gene | 22.811025 | 22.861510 | negative | MGI_C57BL6J_5825673 | Gm46036 | NCBI_Gene:108167527 | MGI:5825673 | lncRNA gene | predicted gene, 46036 |
| 8 | gene | 22.822428 | 22.822545 | negative | MGI_C57BL6J_5454512 | Gm24735 | ENSEMBL:ENSMUSG00000089484 | MGI:5454512 | snRNA gene | predicted gene, 24735 |
| 8 | gene | 22.859516 | 22.943262 | positive | MGI_C57BL6J_2442415 | Kat6a | NCBI_Gene:244349,ENSEMBL:ENSMUSG00000031540 | MGI:2442415 | protein coding gene | K(lysine) acetyltransferase 6A |
| 8 | pseudogene | 22.871588 | 22.872171 | negative | MGI_C57BL6J_5010860 | Gm18675 | NCBI_Gene:100417541 | MGI:5010860 | pseudogene | predicted gene, 18675 |
| 8 | gene | 22.873589 | 22.876190 | positive | MGI_C57BL6J_5791391 | Gm45555 | ENSEMBL:ENSMUSG00000109652 | MGI:5791391 | unclassified gene | predicted gene 45555 |
| 8 | pseudogene | 22.958030 | 22.958627 | negative | MGI_C57BL6J_3641648 | Gm10043 | NCBI_Gene:100042253,ENSEMBL:ENSMUSG00000109793 | MGI:3641648 | pseudogene | predicted pseudogene 10043 |
| 8 | gene | 22.974836 | 23.150501 | positive | MGI_C57BL6J_88024 | Ank1 | NCBI_Gene:11733,ENSEMBL:ENSMUSG00000031543 | MGI:88024 | protein coding gene | ankyrin 1, erythroid |
| 8 | gene | 23.032881 | 23.034691 | negative | MGI_C57BL6J_5589341 | Gm30182 | NCBI_Gene:105243139 | MGI:5589341 | lncRNA gene | predicted gene, 30182 |
| 8 | gene | 23.101972 | 23.102096 | negative | MGI_C57BL6J_5690757 | Gm44365 | ENSEMBL:ENSMUSG00000106605 | MGI:5690757 | miRNA gene | predicted gene, 44365 |
| 8 | pseudogene | 23.127456 | 23.127710 | positive | MGI_C57BL6J_3801793 | Gm15815 | ENSEMBL:ENSMUSG00000083285 | MGI:3801793 | pseudogene | predicted gene 15815 |
| 8 | gene | 23.138785 | 23.149585 | negative | MGI_C57BL6J_3802114 | Gm15816 | NCBI_Gene:101056408,ENSEMBL:ENSMUSG00000086741 | MGI:3802114 | lncRNA gene | predicted gene 15816 |
| 8 | gene | 23.142555 | 23.142682 | positive | MGI_C57BL6J_3619423 | Mir486 | miRBase:MI0003493,NCBI_Gene:723876,ENSEMBL:ENSMUSG00000070084 | MGI:3619423 | miRNA gene | microRNA 486 |
| 8 | gene | 23.142573 | 23.142662 | negative | MGI_C57BL6J_4834325 | Mir3107 | miRBase:MI0014103,NCBI_Gene:100526510,ENSEMBL:ENSMUSG00000106471 | MGI:4834325 | miRNA gene | microRNA 3107 |
| 8 | gene | 23.153269 | 23.158948 | positive | MGI_C57BL6J_1921811 | Nkx6-3 | NCBI_Gene:74561,ENSEMBL:ENSMUSG00000063672 | MGI:1921811 | protein coding gene | NK6 homeobox 3 |
| 8 | gene | 23.164726 | 23.169554 | negative | MGI_C57BL6J_5589465 | Gm30306 | NCBI_Gene:102632159 | MGI:5589465 | lncRNA gene | predicted gene, 30306 |
| 8 | gene | 23.167760 | 23.171183 | positive | MGI_C57BL6J_5589406 | Gm30247 | NCBI_Gene:102632080 | MGI:5589406 | lncRNA gene | predicted gene, 30247 |
| 8 | gene | 23.171265 | 23.208453 | negative | MGI_C57BL6J_2142716 | Gpat4 | NCBI_Gene:102247,ENSEMBL:ENSMUSG00000031545 | MGI:2142716 | protein coding gene | glycerol-3-phosphate acyltransferase 4 |
| 8 | gene | 23.208512 | 23.209362 | positive | MGI_C57BL6J_1925092 | B230112G18Rik | ENSEMBL:ENSMUSG00000103925 | MGI:1925092 | unclassified gene | RIKEN cDNA B230112G18 gene |
| 8 | gene | 23.226610 | 23.237668 | negative | MGI_C57BL6J_1923847 | Gins4 | NCBI_Gene:109145,ENSEMBL:ENSMUSG00000031546 | MGI:1923847 | protein coding gene | GINS complex subunit 4 (Sld5 homolog) |
| 8 | gene | 23.241326 | 23.257434 | negative | MGI_C57BL6J_1931029 | Golga7 | NCBI_Gene:57437,ENSEMBL:ENSMUSG00000015341 | MGI:1931029 | protein coding gene | golgi autoantigen, golgin subfamily a, 7 |
| 8 | gene | 23.250409 | 23.252984 | positive | MGI_C57BL6J_5622026 | Gm39141 | NCBI_Gene:105243140 | MGI:5622026 | lncRNA gene | predicted gene, 39141 |
| 8 | gene | 23.256635 | 23.258909 | positive | MGI_C57BL6J_1918905 | 4930518F22Rik | NA | NA | unclassified gene | RIKEN cDNA 4930518F22 gene |
| 8 | gene | 23.256635 | 23.258909 | positive | MGI_C57BL6J_5791248 | Gm45412 | ENSEMBL:ENSMUSG00000110256 | MGI:5791248 | unclassified gene | predicted gene 45412 |
| 8 | gene | 23.290033 | 23.290164 | negative | MGI_C57BL6J_5454112 | Gm24335 | ENSEMBL:ENSMUSG00000077399 | MGI:5454112 | snoRNA gene | predicted gene, 24335 |
| 8 | gene | 23.387121 | 23.404313 | negative | MGI_C57BL6J_5589529 | Gm30370 | NCBI_Gene:102632237 | MGI:5589529 | lncRNA gene | predicted gene, 30370 |
| 8 | gene | 23.411382 | 23.449632 | positive | MGI_C57BL6J_892014 | Sfrp1 | NCBI_Gene:20377,ENSEMBL:ENSMUSG00000031548 | MGI:892014 | protein coding gene | secreted frizzled-related protein 1 |
| 8 | gene | 23.534472 | 23.535739 | positive | MGI_C57BL6J_1918892 | 4930413E20Rik | NCBI_Gene:102632305 | MGI:1918892 | lncRNA gene | RIKEN cDNA 4930413E20 gene |
| 8 | gene | 23.635080 | 23.648505 | negative | MGI_C57BL6J_5589630 | Gm30471 | NCBI_Gene:102632384 | MGI:5589630 | lncRNA gene | predicted gene, 30471 |
| 8 | gene | 23.636019 | 24.156585 | positive | MGI_C57BL6J_2443497 | Zmat4 | NCBI_Gene:320158,ENSEMBL:ENSMUSG00000037492 | MGI:2443497 | protein coding gene | zinc finger, matrin type 4 |
| 8 | gene | 23.696005 | 23.697635 | positive | MGI_C57BL6J_5753741 | Gm45165 | ENSEMBL:ENSMUSG00000109413 | MGI:5753741 | unclassified gene | predicted gene 45165 |
| 8 | gene | 23.752083 | 23.752765 | negative | MGI_C57BL6J_5753742 | Gm45166 | ENSEMBL:ENSMUSG00000109158 | MGI:5753742 | unclassified gene | predicted gene 45166 |
| 8 | gene | 24.173904 | 24.185568 | positive | MGI_C57BL6J_2685430 | A730045E13Rik | ENSEMBL:ENSMUSG00000053979 | MGI:2685430 | lncRNA gene | RIKEN cDNA A730045E13 gene |
| 8 | gene | 24.220510 | 24.230146 | positive | MGI_C57BL6J_5753739 | Gm45163 | ENSEMBL:ENSMUSG00000109419 | MGI:5753739 | lncRNA gene | predicted gene 45163 |
| 8 | gene | 24.353944 | 24.359436 | negative | MGI_C57BL6J_5753740 | Gm45164 | ENSEMBL:ENSMUSG00000109299 | MGI:5753740 | lncRNA gene | predicted gene 45164 |
| 8 | gene | 24.382131 | 24.391067 | positive | MGI_C57BL6J_5589792 | Gm30633 | NCBI_Gene:102632603 | MGI:5589792 | lncRNA gene | predicted gene, 30633 |
| 8 | gene | 24.412179 | 24.416937 | positive | MGI_C57BL6J_5589851 | Gm30692 | NCBI_Gene:102632684,ENSEMBL:ENSMUSG00000109141 | MGI:5589851 | lncRNA gene | predicted gene, 30692 |
| 8 | gene | 24.422404 | 24.440008 | positive | MGI_C57BL6J_5477208 | Gm26714 | ENSEMBL:ENSMUSG00000097654 | MGI:5477208 | lncRNA gene | predicted gene, 26714 |
| 8 | gene | 24.429438 | 24.431124 | negative | MGI_C57BL6J_5589916 | Gm30757 | NCBI_Gene:102632769 | MGI:5589916 | lncRNA gene | predicted gene, 30757 |
| 8 | gene | 24.437180 | 24.438984 | negative | MGI_C57BL6J_1916318 | Tcim | NCBI_Gene:69068,ENSEMBL:ENSMUSG00000056313 | MGI:1916318 | protein coding gene | transcriptional and immune response regulator |
| 8 | gene | 24.440880 | 24.447864 | negative | MGI_C57BL6J_5590004 | Gm30845 | NCBI_Gene:102632886 | MGI:5590004 | lncRNA gene | predicted gene, 30845 |
| 8 | gene | 24.447097 | 24.449983 | positive | MGI_C57BL6J_5753196 | Gm44620 | ENSEMBL:ENSMUSG00000108820 | MGI:5753196 | unclassified gene | predicted gene 44620 |
| 8 | gene | 24.502201 | 24.512539 | positive | MGI_C57BL6J_1924373 | 9130214F15Rik | NCBI_Gene:102632957,ENSEMBL:ENSMUSG00000109237 | MGI:1924373 | lncRNA gene | RIKEN cDNA 9130214F15 gene |
| 8 | gene | 24.531892 | 24.576408 | negative | MGI_C57BL6J_2142489 | Ido2 | NCBI_Gene:209176,ENSEMBL:ENSMUSG00000031549 | MGI:2142489 | protein coding gene | indoleamine 2,3-dioxygenase 2 |
| 8 | gene | 24.584133 | 24.597009 | negative | MGI_C57BL6J_96416 | Ido1 | NCBI_Gene:15930,ENSEMBL:ENSMUSG00000031551 | MGI:96416 | protein coding gene | indoleamine 2,3-dioxygenase 1 |
| 8 | gene | 24.602246 | 24.674755 | negative | MGI_C57BL6J_105986 | Adam18 | NCBI_Gene:13524,ENSEMBL:ENSMUSG00000031552 | MGI:105986 | protein coding gene | a disintegrin and metallopeptidase domain 18 |
| 8 | gene | 24.677225 | 24.726039 | negative | MGI_C57BL6J_102518 | Adam3 | NCBI_Gene:11497,ENSEMBL:ENSMUSG00000031553 | MGI:102518 | protein coding gene | a disintegrin and metallopeptidase domain 3 (cyritestin) |
| 8 | gene | 24.680814 | 24.680937 | positive | MGI_C57BL6J_5455985 | Gm26208 | ENSEMBL:ENSMUSG00000089302 | MGI:5455985 | snoRNA gene | predicted gene, 26208 |
| 8 | gene | 24.711406 | 24.714550 | positive | MGI_C57BL6J_5791364 | Gm45528 | ENSEMBL:ENSMUSG00000110225 | MGI:5791364 | unclassified gene | predicted gene 45528 |
| 8 | gene | 24.727093 | 24.824369 | negative | MGI_C57BL6J_104730 | Adam5 | NCBI_Gene:11499,ENSEMBL:ENSMUSG00000031554 | MGI:104730 | protein coding gene | a disintegrin and metallopeptidase domain 5 |
| 8 | gene | 24.836140 | 24.948918 | negative | MGI_C57BL6J_2653822 | Adam32 | NCBI_Gene:353188,ENSEMBL:ENSMUSG00000037437 | MGI:2653822 | protein coding gene | a disintegrin and metallopeptidase domain 32 |
| 8 | gene | 24.938300 | 24.938402 | positive | MGI_C57BL6J_5453675 | Gm23898 | ENSEMBL:ENSMUSG00000088736 | MGI:5453675 | snRNA gene | predicted gene, 23898 |
| 8 | gene | 24.949611 | 25.016934 | negative | MGI_C57BL6J_105376 | Adam9 | NCBI_Gene:11502,ENSEMBL:ENSMUSG00000031555 | MGI:105376 | protein coding gene | a disintegrin and metallopeptidase domain 9 (meltrin gamma) |
| 8 | gene | 25.017211 | 25.023260 | positive | MGI_C57BL6J_1916992 | Tm2d2 | NCBI_Gene:69742,ENSEMBL:ENSMUSG00000031556 | MGI:1916992 | protein coding gene | TM2 domain containing 2 |
| 8 | gene | 25.024928 | 25.038962 | negative | MGI_C57BL6J_3036260 | Htra4 | NCBI_Gene:330723,ENSEMBL:ENSMUSG00000037406 | MGI:3036260 | protein coding gene | HtrA serine peptidase 4 |
| 8 | gene | 25.037052 | 25.041892 | positive | MGI_C57BL6J_4439857 | Gm16933 | ENSEMBL:ENSMUSG00000086289 | MGI:4439857 | lncRNA gene | predicted gene, 16933 |
| 8 | gene | 25.039144 | 25.104929 | negative | MGI_C57BL6J_1928144 | Plekha2 | NCBI_Gene:83436,ENSEMBL:ENSMUSG00000031557 | MGI:1928144 | protein coding gene | pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| 8 | gene | 25.101216 | 25.109986 | positive | MGI_C57BL6J_1921994 | 5830408C22Rik | ENSEMBL:ENSMUSG00000110148 | MGI:1921994 | lncRNA gene | RIKEN cDNA 5830408C22 gene |
| 8 | gene | 25.103489 | 25.104668 | negative | MGI_C57BL6J_5804860 | Gm45745 | ENSEMBL:ENSMUSG00000109684 | MGI:5804860 | unclassified gene | predicted gene 45745 |
| 8 | gene | 25.129572 | 25.133208 | positive | MGI_C57BL6J_5622028 | Gm39143 | NCBI_Gene:105243143 | MGI:5622028 | lncRNA gene | predicted gene, 39143 |
| 8 | gene | 25.154552 | 25.256588 | negative | MGI_C57BL6J_2443510 | Tacc1 | NCBI_Gene:320165,ENSEMBL:ENSMUSG00000065954 | MGI:2443510 | protein coding gene | transforming, acidic coiled-coil containing protein 1 |
| 8 | gene | 25.159125 | 25.159233 | negative | MGI_C57BL6J_5530866 | Mir8108 | miRBase:MI0026039,NCBI_Gene:102467006,ENSEMBL:ENSMUSG00000099142 | MGI:5530866 | miRNA gene | microRNA 8108 |
| 8 | gene | 25.256481 | 25.258509 | positive | MGI_C57BL6J_5622029 | Gm39144 | NCBI_Gene:105243144 | MGI:5622029 | lncRNA gene | predicted gene, 39144 |
| 8 | gene | 25.271046 | 25.279558 | positive | MGI_C57BL6J_5622030 | Gm39145 | NCBI_Gene:105243145 | MGI:5622030 | lncRNA gene | predicted gene, 39145 |
| 8 | gene | 25.302650 | 25.306270 | negative | MGI_C57BL6J_1921739 | 5430421F17Rik | NCBI_Gene:74489,ENSEMBL:ENSMUSG00000110140 | MGI:1921739 | lncRNA gene | RIKEN cDNA 5430421F17 gene |
| 8 | gene | 25.353360 | 25.389823 | negative | MGI_C57BL6J_5622031 | Gm39146 | NCBI_Gene:105243146 | MGI:5622031 | lncRNA gene | predicted gene, 39146 |
| 8 | pseudogene | 25.399020 | 25.427436 | positive | MGI_C57BL6J_5622032 | Gm39147 | NCBI_Gene:105243147,ENSEMBL:ENSMUSG00000110172 | MGI:5622032 | pseudogene | predicted gene, 39147 |
| 8 | gene | 25.415119 | 25.454212 | negative | MGI_C57BL6J_5590137 | Gm30978 | NCBI_Gene:102633061,ENSEMBL:ENSMUSG00000110110 | MGI:5590137 | lncRNA gene | predicted gene, 30978 |
| 8 | gene | 25.440618 | 25.448762 | positive | MGI_C57BL6J_5791073 | Gm45237 | ENSEMBL:ENSMUSG00000110017 | MGI:5791073 | lncRNA gene | predicted gene 45237 |
| 8 | gene | 25.443668 | 25.454107 | negative | MGI_C57BL6J_5791074 | Gm45238 | ENSEMBL:ENSMUSG00000110399 | MGI:5791074 | lncRNA gene | predicted gene 45238 |
| 8 | pseudogene | 25.454244 | 25.454751 | positive | MGI_C57BL6J_3642613 | Gm10689 | NCBI_Gene:100113396,ENSEMBL:ENSMUSG00000074412 | MGI:3642613 | pseudogene | predicted gene 10689 |
| 8 | gene | 25.463677 | 25.487585 | negative | MGI_C57BL6J_5590204 | Gm31045 | NCBI_Gene:102633143,ENSEMBL:ENSMUSG00000110063 | MGI:5590204 | lncRNA gene | predicted gene, 31045 |
| 8 | gene | 25.513623 | 25.575718 | positive | MGI_C57BL6J_95522 | Fgfr1 | NCBI_Gene:14182,ENSEMBL:ENSMUSG00000031565 | MGI:95522 | protein coding gene | fibroblast growth factor receptor 1 |
| 8 | gene | 25.520786 | 25.543249 | negative | MGI_C57BL6J_5590447 | Gm31288 | NCBI_Gene:102633471 | MGI:5590447 | lncRNA gene | predicted gene, 31288 |
| 8 | gene | 25.548661 | 25.556083 | negative | MGI_C57BL6J_3801949 | Gm16159 | NCBI_Gene:102633315,ENSEMBL:ENSMUSG00000086134 | MGI:3801949 | lncRNA gene | predicted gene 16159 |
| 8 | gene | 25.578482 | 25.597984 | negative | MGI_C57BL6J_2444979 | Letm2 | NCBI_Gene:270035,ENSEMBL:ENSMUSG00000037363 | MGI:2444979 | protein coding gene | leucine zipper-EF-hand containing transmembrane protein 2 |
| 8 | gene | 25.594454 | 25.595106 | negative | MGI_C57BL6J_5791406 | Gm45570 | ENSEMBL:ENSMUSG00000110038 | MGI:5791406 | lncRNA gene | predicted gene 45570 |
| 8 | gene | 25.597012 | 25.599207 | positive | MGI_C57BL6J_3698180 | D830025C05Rik | ENSEMBL:ENSMUSG00000043794 | MGI:3698180 | lncRNA gene | RIKEN cDNA D830025C05 gene |
| 8 | gene | 25.601508 | 25.719667 | positive | MGI_C57BL6J_2142581 | Nsd3 | NCBI_Gene:234135,ENSEMBL:ENSMUSG00000054823 | MGI:2142581 | protein coding gene | nuclear receptor binding SET domain protein 3 |
| 8 | gene | 25.610428 | 25.610558 | negative | MGI_C57BL6J_5452961 | Gm23184 | ENSEMBL:ENSMUSG00000084543 | MGI:5452961 | snoRNA gene | predicted gene, 23184 |
| 8 | gene | 25.631950 | 25.632079 | positive | MGI_C57BL6J_5454769 | Gm24992 | ENSEMBL:ENSMUSG00000088037 | MGI:5454769 | snoRNA gene | predicted gene, 24992 |
| 8 | gene | 25.678950 | 25.679271 | positive | MGI_C57BL6J_5452871 | Gm23094 | ENSEMBL:ENSMUSG00000088176 | MGI:5452871 | unclassified non-coding RNA gene | predicted gene, 23094 |
| 8 | gene | 25.687369 | 25.688196 | positive | MGI_C57BL6J_5791179 | Gm45343 | ENSEMBL:ENSMUSG00000110088 | MGI:5791179 | unclassified gene | predicted gene 45343 |
| 8 | gene | 25.692146 | 25.693508 | positive | MGI_C57BL6J_5791178 | Gm45342 | ENSEMBL:ENSMUSG00000109679 | MGI:5791178 | unclassified gene | predicted gene 45342 |
| 8 | gene | 25.719779 | 25.724887 | positive | MGI_C57BL6J_1919160 | Plpp5 | NCBI_Gene:71910,ENSEMBL:ENSMUSG00000031570 | MGI:1919160 | protein coding gene | phospholipid phosphatase 5 |
| 8 | gene | 25.725324 | 25.754596 | negative | MGI_C57BL6J_1919358 | Ddhd2 | NCBI_Gene:72108,ENSEMBL:ENSMUSG00000061313 | MGI:1919358 | protein coding gene | DDHD domain containing 2 |
| 8 | gene | 25.744390 | 25.745580 | negative | MGI_C57BL6J_1923464 | 6430710M23Rik | ENSEMBL:ENSMUSG00000110127 | MGI:1923464 | unclassified gene | RIKEN cDNA 6430710M23 gene |
| 8 | gene | 25.754480 | 25.769704 | positive | MGI_C57BL6J_4937118 | Gm17484 | ENSEMBL:ENSMUSG00000091514 | MGI:4937118 | lncRNA gene | predicted gene, 17484 |
| 8 | gene | 25.764538 | 25.785287 | negative | MGI_C57BL6J_1914634 | Bag4 | NCBI_Gene:67384,ENSEMBL:ENSMUSG00000037316 | MGI:1914634 | protein coding gene | BCL2-associated athanogene 4 |
| 8 | pseudogene | 25.775836 | 25.776513 | negative | MGI_C57BL6J_5011260 | Gm19075 | NCBI_Gene:100418210 | MGI:5011260 | pseudogene | predicted gene, 19075 |
| 8 | gene | 25.785288 | 25.803975 | positive | MGI_C57BL6J_1914457 | Lsm1 | NCBI_Gene:67207,ENSEMBL:ENSMUSG00000037296 | MGI:1914457 | protein coding gene | LSM1 homolog, mRNA degradation associated |
| 8 | pseudogene | 25.788823 | 25.789353 | negative | MGI_C57BL6J_5825674 | Gm46037 | NCBI_Gene:108167528 | MGI:5825674 | pseudogene | predicted gene, 46037 |
| 8 | gene | 25.806555 | 25.815982 | positive | MGI_C57BL6J_102760 | Star | NCBI_Gene:20845,ENSEMBL:ENSMUSG00000031574 | MGI:102760 | protein coding gene | steroidogenic acute regulatory protein |
| 8 | gene | 25.815996 | 25.847694 | negative | MGI_C57BL6J_1344416 | Ash2l | NCBI_Gene:23808,ENSEMBL:ENSMUSG00000031575 | MGI:1344416 | protein coding gene | ASH2 like histone lysine methyltransferase complex subunit |
| 8 | gene | 25.848627 | 25.937943 | positive | MGI_C57BL6J_1202300 | Kcnu1 | NCBI_Gene:16532,ENSEMBL:ENSMUSG00000031576 | MGI:1202300 | protein coding gene | potassium channel, subfamily U, member 1 |
| 8 | gene | 25.944453 | 25.976753 | negative | MGI_C57BL6J_1196297 | Hgsnat | NCBI_Gene:52120,ENSEMBL:ENSMUSG00000037260 | MGI:1196297 | protein coding gene | heparan-alpha-glucosaminide N-acetyltransferase |
| 8 | gene | 25.980604 | 25.994133 | negative | MGI_C57BL6J_1921903 | Pomk | NCBI_Gene:74653,ENSEMBL:ENSMUSG00000037251 | MGI:1921903 | protein coding gene | protein-O-mannose kinase |
| 8 | gene | 25.992654 | 25.998229 | positive | MGI_C57BL6J_5590561 | Gm31402 | NCBI_Gene:102633618 | MGI:5590561 | lncRNA gene | predicted gene, 31402 |
| 8 | gene | 25.998722 | 26.015650 | negative | MGI_C57BL6J_104683 | Fnta | NCBI_Gene:14272,ENSEMBL:ENSMUSG00000015994 | MGI:104683 | protein coding gene | farnesyltransferase, CAAX box, alpha |
| 8 | gene | 26.021421 | 26.119433 | negative | MGI_C57BL6J_2443554 | Hook3 | NCBI_Gene:320191,ENSEMBL:ENSMUSG00000037234 | MGI:2443554 | protein coding gene | hook microtubule tethering protein 3 |
| 8 | gene | 26.023410 | 26.032253 | positive | MGI_C57BL6J_5791247 | Gm45411 | ENSEMBL:ENSMUSG00000110183 | MGI:5791247 | lncRNA gene | predicted gene 45411 |
| 8 | gene | 26.051607 | 26.052546 | negative | MGI_C57BL6J_1924339 | 5730564C23Rik | NA | NA | unclassified gene | RIKEN cDNA 5730564C23 gene |
| 8 | gene | 26.062515 | 26.066052 | negative | MGI_C57BL6J_5791249 | Gm45413 | ENSEMBL:ENSMUSG00000109640 | MGI:5791249 | unclassified gene | predicted gene 45413 |
| 8 | gene | 26.082609 | 26.096522 | positive | MGI_C57BL6J_1923854 | 1700047A11Rik | NCBI_Gene:76604,ENSEMBL:ENSMUSG00000085437 | MGI:1923854 | lncRNA gene | RIKEN cDNA 1700047A11 gene |
| 8 | pseudogene | 26.103616 | 26.104228 | positive | MGI_C57BL6J_5010549 | Gm18364 | NCBI_Gene:100417018 | MGI:5010549 | pseudogene | predicted gene, 18364 |
| 8 | gene | 26.119365 | 26.151790 | positive | MGI_C57BL6J_1924983 | Rnf170 | NCBI_Gene:77733,ENSEMBL:ENSMUSG00000013878 | MGI:1924983 | protein coding gene | ring finger protein 170 |
| 8 | gene | 26.119534 | 26.160819 | positive | MGI_C57BL6J_5804807 | Gm45692 | ENSEMBL:ENSMUSG00000109850 | MGI:5804807 | protein coding gene | predicted gene 45692 |
| 8 | gene | 26.131178 | 26.132642 | negative | MGI_C57BL6J_5791087 | Gm45251 | ENSEMBL:ENSMUSG00000110120 | MGI:5791087 | unclassified gene | predicted gene 45251 |
| 8 | gene | 26.151888 | 26.155479 | negative | MGI_C57BL6J_5791086 | Gm45250 | ENSEMBL:ENSMUSG00000110317 | MGI:5791086 | lncRNA gene | predicted gene 45250 |
| 8 | gene | 26.158141 | 26.164151 | positive | MGI_C57BL6J_1921004 | Thap1 | NCBI_Gene:73754,ENSEMBL:ENSMUSG00000037214 | MGI:1921004 | protein coding gene | THAP domain containing, apoptosis associated protein 1 |
| 8 | gene | 26.219461 | 26.219549 | positive | MGI_C57BL6J_5690691 | Gm44299 | ENSEMBL:ENSMUSG00000105292 | MGI:5690691 | miRNA gene | predicted gene, 44299 |
| 8 | pseudogene | 26.237222 | 26.237407 | positive | MGI_C57BL6J_6324752 | Gm50479 | ENSEMBL:ENSMUSG00000118460 | MGI:6324752 | pseudogene | predicted gene, 50479 |
| 8 | gene | 26.240150 | 26.240408 | negative | MGI_C57BL6J_5452004 | Gm22227 | ENSEMBL:ENSMUSG00000087981 | MGI:5452004 | unclassified non-coding RNA gene | predicted gene, 22227 |
| 8 | gene | 26.242724 | 26.244051 | positive | MGI_C57BL6J_5791141 | Gm45305 | ENSEMBL:ENSMUSG00000110075 | MGI:5791141 | lncRNA gene | predicted gene 45305 |
| 8 | gene | 26.266878 | 26.289106 | positive | MGI_C57BL6J_5590886 | Gm31727 | NCBI_Gene:102634047,ENSEMBL:ENSMUSG00000110417 | MGI:5590886 | lncRNA gene | predicted gene, 31727 |
| 8 | gene | 26.298209 | 26.312463 | negative | MGI_C57BL6J_5590943 | Gm31784 | NCBI_Gene:102634123,ENSEMBL:ENSMUSG00000110229 | MGI:5590943 | lncRNA gene | predicted gene, 31784 |
| 8 | gene | 26.298634 | 26.354163 | positive | MGI_C57BL6J_5791143 | Gm45307 | ENSEMBL:ENSMUSG00000109682 | MGI:5791143 | lncRNA gene | predicted gene 45307 |
| 8 | gene | 26.381686 | 26.389138 | positive | MGI_C57BL6J_5591002 | Gm31843 | NCBI_Gene:102634202 | MGI:5591002 | lncRNA gene | predicted gene, 31843 |
| 8 | gene | 26.386110 | 26.389066 | negative | MGI_C57BL6J_5622033 | Gm39148 | NCBI_Gene:105243149 | MGI:5622033 | lncRNA gene | predicted gene, 39148 |
| 8 | gene | 26.427319 | 26.439263 | negative | MGI_C57BL6J_5591057 | Gm31898 | NCBI_Gene:102634274,ENSEMBL:ENSMUSG00000109996 | MGI:5591057 | lncRNA gene | predicted gene, 31898 |
| 8 | gene | 26.438829 | 26.440492 | positive | MGI_C57BL6J_5791416 | Gm45580 | ENSEMBL:ENSMUSG00000110383 | MGI:5791416 | lncRNA gene | predicted gene 45580 |
| 8 | gene | 26.450460 | 26.484652 | negative | MGI_C57BL6J_5591142 | Gm31983 | NCBI_Gene:102634391,ENSEMBL:ENSMUSG00000109961 | MGI:5591142 | lncRNA gene | predicted gene, 31983 |
| 8 | gene | 26.559750 | 26.580222 | negative | MGI_C57BL6J_5622034 | Gm39149 | NCBI_Gene:105243150,ENSEMBL:ENSMUSG00000110222 | MGI:5622034 | lncRNA gene | predicted gene, 39149 |
| 8 | gene | 26.626741 | 26.629395 | negative | MGI_C57BL6J_5591209 | Gm32050 | NCBI_Gene:102634474,ENSEMBL:ENSMUSG00000110351 | MGI:5591209 | lncRNA gene | predicted gene, 32050 |
| 8 | gene | 26.637675 | 26.639952 | positive | MGI_C57BL6J_5791420 | Gm45584 | ENSEMBL:ENSMUSG00000110338 | MGI:5791420 | lncRNA gene | predicted gene 45584 |
| 8 | gene | 26.663301 | 26.698634 | negative | MGI_C57BL6J_5591257 | Gm32098 | NCBI_Gene:102634544,ENSEMBL:ENSMUSG00000109680 | MGI:5591257 | lncRNA gene | predicted gene, 32098 |
| 8 | gene | 26.745929 | 26.760561 | negative | MGI_C57BL6J_5791482 | Gm45646 | ENSEMBL:ENSMUSG00000109689 | MGI:5791482 | lncRNA gene | predicted gene 45646 |
| 8 | gene | 26.768747 | 26.770539 | negative | MGI_C57BL6J_5825704 | Gm46067 | NCBI_Gene:108167579 | MGI:5825704 | lncRNA gene | predicted gene, 46067 |
| 8 | pseudogene | 26.780315 | 26.780509 | positive | MGI_C57BL6J_5791483 | Gm45647 | ENSEMBL:ENSMUSG00000110297 | MGI:5791483 | pseudogene | predicted gene 45647 |
| 8 | gene | 26.814593 | 26.884380 | negative | MGI_C57BL6J_1919566 | 2310008N11Rik | NCBI_Gene:72316,ENSEMBL:ENSMUSG00000100335 | MGI:1919566 | lncRNA gene | RIKEN cDNA 2310008N11 gene |
| 8 | pseudogene | 26.823247 | 26.823772 | positive | MGI_C57BL6J_5588887 | Gm29728 | NCBI_Gene:101055839 | MGI:5588887 | pseudogene | predicted gene, 29728 |
| 8 | gene | 26.960786 | 26.977077 | negative | MGI_C57BL6J_5622035 | Gm39150 | NCBI_Gene:105243151 | MGI:5622035 | lncRNA gene | predicted gene, 39150 |
| 8 | gene | 26.966434 | 26.967474 | positive | MGI_C57BL6J_5791408 | Gm45572 | ENSEMBL:ENSMUSG00000110345 | MGI:5791408 | lncRNA gene | predicted gene 45572 |
| 8 | gene | 26.977325 | 26.981462 | positive | MGI_C57BL6J_2662729 | Zfp703 | NCBI_Gene:353310,ENSEMBL:ENSMUSG00000085795 | MGI:2662729 | protein coding gene | zinc finger protein 703 |
| 8 | gene | 26.977391 | 26.980001 | negative | MGI_C57BL6J_5791207 | Gm45371 | ENSEMBL:ENSMUSG00000109954 | MGI:5791207 | lncRNA gene | predicted gene 45371 |
| 8 | gene | 26.996250 | 26.998954 | positive | MGI_C57BL6J_5791206 | Gm45370 | ENSEMBL:ENSMUSG00000109621 | MGI:5791206 | lncRNA gene | predicted gene 45370 |
| 8 | gene | 27.007567 | 27.012903 | positive | MGI_C57BL6J_5591548 | Gm32389 | NCBI_Gene:102634919,ENSEMBL:ENSMUSG00000109603 | MGI:5591548 | lncRNA gene | predicted gene, 32389 |
| 8 | pseudogene | 27.019454 | 27.020093 | negative | MGI_C57BL6J_5011349 | Gm19164 | NCBI_Gene:100418365,ENSEMBL:ENSMUSG00000110081 | MGI:5011349 | pseudogene | predicted gene, 19164 |
| 8 | gene | 27.023261 | 27.040328 | positive | MGI_C57BL6J_2387215 | Erlin2 | NCBI_Gene:244373,ENSEMBL:ENSMUSG00000031483 | MGI:2387215 | protein coding gene | ER lipid raft associated 2 |
| 8 | gene | 27.042377 | 27.043042 | negative | MGI_C57BL6J_3704307 | Proscos | ENSEMBL:ENSMUSG00000091154 | MGI:3704307 | antisense lncRNA gene | proline synthetase co-transcribed, opposite strand |
| 8 | gene | 27.042384 | 27.056132 | positive | MGI_C57BL6J_1891207 | Plpbp | NCBI_Gene:114863,ENSEMBL:ENSMUSG00000031485 | MGI:1891207 | protein coding gene | pyridoxal phosphate binding protein |
| 8 | gene | 27.085491 | 27.123436 | positive | MGI_C57BL6J_1925810 | Adgra2 | NCBI_Gene:78560,ENSEMBL:ENSMUSG00000031486 | MGI:1925810 | protein coding gene | adhesion G protein-coupled receptor A2 |
| 8 | gene | 27.086269 | 27.107449 | negative | MGI_C57BL6J_5622036 | Gm39151 | NCBI_Gene:105243152 | MGI:5622036 | lncRNA gene | predicted gene, 39151 |
| 8 | gene | 27.123339 | 27.128699 | negative | MGI_C57BL6J_1913903 | Brf2 | NCBI_Gene:66653,ENSEMBL:ENSMUSG00000031487 | MGI:1913903 | protein coding gene | BRF2, RNA polymerase III transcription initiation factor 50kDa subunit |
| 8 | gene | 27.138773 | 27.174646 | negative | MGI_C57BL6J_1923017 | Rab11fip1 | NCBI_Gene:75767,ENSEMBL:ENSMUSG00000031488 | MGI:1923017 | protein coding gene | RAB11 family interacting protein 1 (class I) |
| 8 | gene | 27.142910 | 27.149706 | positive | MGI_C57BL6J_1918332 | 4933416M07Rik | NCBI_Gene:71082,ENSEMBL:ENSMUSG00000097262 | MGI:1918332 | lncRNA gene | RIKEN cDNA 4933416M07 gene |
| 8 | gene | 27.159828 | 27.170691 | positive | MGI_C57BL6J_5791103 | Gm45267 | ENSEMBL:ENSMUSG00000109589 | MGI:5791103 | lncRNA gene | predicted gene 45267 |
| 8 | gene | 27.168840 | 27.171012 | negative | MGI_C57BL6J_5791102 | Gm45266 | ENSEMBL:ENSMUSG00000109719 | MGI:5791102 | unclassified gene | predicted gene 45266 |
| 8 | gene | 27.197459 | 27.223844 | negative | MGI_C57BL6J_1923865 | Got1l1 | NCBI_Gene:76615,ENSEMBL:ENSMUSG00000039720 | MGI:1923865 | protein coding gene | glutamic-oxaloacetic transaminase 1-like 1 |
| 8 | gene | 27.225776 | 27.250616 | negative | MGI_C57BL6J_87939 | Adrb3 | NCBI_Gene:11556,ENSEMBL:ENSMUSG00000031489 | MGI:87939 | protein coding gene | adrenergic receptor, beta 3 |
| 8 | gene | 27.228686 | 27.240332 | positive | MGI_C57BL6J_5791306 | Gm45470 | NCBI_Gene:108167529,ENSEMBL:ENSMUSG00000109674 | MGI:5791306 | lncRNA gene | predicted gene 45470 |
| 8 | gene | 27.260327 | 27.276674 | positive | MGI_C57BL6J_103267 | Eif4ebp1 | NCBI_Gene:13685,ENSEMBL:ENSMUSG00000031490 | MGI:103267 | protein coding gene | eukaryotic translation initiation factor 4E binding protein 1 |
| 8 | gene | 27.279441 | 27.281070 | positive | MGI_C57BL6J_1918674 | 5430430B14Rik | ENSEMBL:ENSMUSG00000109953 | MGI:1918674 | unclassified gene | RIKEN cDNA 5430430B14 gene |
| 8 | pseudogene | 27.294677 | 27.295815 | positive | MGI_C57BL6J_5010451 | Gm18266 | NCBI_Gene:100416805 | MGI:5010451 | pseudogene | predicted gene, 18266 |
| 8 | pseudogene | 27.296045 | 27.297200 | positive | MGI_C57BL6J_3780135 | Gm9731 | NCBI_Gene:732482,ENSEMBL:ENSMUSG00000110154 | MGI:3780135 | pseudogene | predicted gene 9731 |
| 8 | gene | 27.344323 | 27.349430 | positive | MGI_C57BL6J_1921539 | Tex24 | NCBI_Gene:541463,ENSEMBL:ENSMUSG00000071138 | MGI:1921539 | protein coding gene | testis expressed gene 24 |
| 8 | gene | 27.368711 | 27.399730 | positive | MGI_C57BL6J_106212 | Chrnb3 | NCBI_Gene:108043,ENSEMBL:ENSMUSG00000031492 | MGI:106212 | protein coding gene | cholinergic receptor, nicotinic, beta polypeptide 3 |
| 8 | gene | 27.403212 | 27.413944 | negative | MGI_C57BL6J_106213 | Chrna6 | NCBI_Gene:11440,ENSEMBL:ENSMUSG00000031491 | MGI:106213 | protein coding gene | cholinergic receptor, nicotinic, alpha polypeptide 6 |
| 8 | gene | 27.421324 | 27.421430 | negative | MGI_C57BL6J_5454187 | Gm24410 | ENSEMBL:ENSMUSG00000065367 | MGI:5454187 | snRNA gene | predicted gene, 24410 |
| 8 | gene | 27.447100 | 27.620917 | positive | MGI_C57BL6J_5804976 | Gm45861 | ENSEMBL:ENSMUSG00000110333 | MGI:5804976 | protein coding gene | predicted gene 45861 |
| 8 | gene | 27.447527 | 27.582717 | positive | MGI_C57BL6J_1918202 | Poteg | NCBI_Gene:70952,ENSEMBL:ENSMUSG00000063932 | MGI:1918202 | protein coding gene | POTE ankyrin domain family, member G |
| 8 | gene | 27.582692 | 27.620453 | positive | MGI_C57BL6J_2686544 | Gm1698 | NCBI_Gene:382003,ENSEMBL:ENSMUSG00000074404 | MGI:2686544 | lncRNA gene | predicted gene 1698 |
| 8 | gene | 27.615817 | 27.615904 | positive | MGI_C57BL6J_5690885 | Gm44493 | ENSEMBL:ENSMUSG00000104743 | MGI:5690885 | miRNA gene | predicted gene, 44493 |
| 8 | gene | 27.761009 | 27.761111 | negative | MGI_C57BL6J_5453979 | Gm24202 | ENSEMBL:ENSMUSG00000064882 | MGI:5453979 | snRNA gene | predicted gene, 24202 |
| 8 | gene | 27.893861 | 27.896154 | negative | MGI_C57BL6J_5622038 | Gm39153 | NCBI_Gene:105243154 | MGI:5622038 | lncRNA gene | predicted gene, 39153 |
| 8 | gene | 28.082694 | 28.084266 | negative | MGI_C57BL6J_5804866 | Gm45751 | ENSEMBL:ENSMUSG00000110343 | MGI:5804866 | unclassified gene | predicted gene 45751 |
| 8 | pseudogene | 28.083194 | 28.084920 | negative | MGI_C57BL6J_3645752 | Gm8096 | NCBI_Gene:666422,ENSEMBL:ENSMUSG00000110372 | MGI:3645752 | pseudogene | predicted gene 8096 |
| 8 | gene | 28.084828 | 28.085596 | positive | MGI_C57BL6J_1922692 | 1700008N11Rik | ENSEMBL:ENSMUSG00000109657 | MGI:1922692 | lncRNA gene | RIKEN cDNA 1700008N11 gene |
| 8 | pseudogene | 28.185822 | 28.187040 | positive | MGI_C57BL6J_3649005 | Gm8100 | NCBI_Gene:666430,ENSEMBL:ENSMUSG00000098056 | MGI:3649005 | pseudogene | predicted gene 8100 |
| 8 | pseudogene | 28.520676 | 28.529237 | positive | MGI_C57BL6J_3645532 | Gm8110 | NCBI_Gene:666446,ENSEMBL:ENSMUSG00000110322 | MGI:3645532 | pseudogene | predicted gene 8110 |
| 8 | gene | 28.527826 | 28.529717 | negative | MGI_C57BL6J_5791407 | Gm45571 | ENSEMBL:ENSMUSG00000109794 | MGI:5791407 | lncRNA gene | predicted gene 45571 |
| 8 | gene | 28.593505 | 28.595599 | positive | MGI_C57BL6J_5477289 | Gm26795 | NCBI_Gene:105243155,ENSEMBL:ENSMUSG00000097776 | MGI:5477289 | lncRNA gene | predicted gene, 26795 |
| 8 | gene | 28.646717 | 29.219681 | negative | MGI_C57BL6J_2389364 | Unc5d | NCBI_Gene:210801,ENSEMBL:ENSMUSG00000063626 | MGI:2389364 | protein coding gene | unc-5 netrin receptor D |
| 8 | gene | 28.696941 | 28.708641 | positive | MGI_C57BL6J_5591664 | Gm32505 | NCBI_Gene:102635079 | MGI:5591664 | lncRNA gene | predicted gene, 32505 |
| 8 | gene | 29.084685 | 29.087486 | negative | MGI_C57BL6J_3708772 | Gm9911 | ENSEMBL:ENSMUSG00000109874 | MGI:3708772 | unclassified gene | predicted gene 9911 |
| 8 | gene | 29.088073 | 29.091903 | negative | MGI_C57BL6J_5791488 | Gm45652 | ENSEMBL:ENSMUSG00000110263 | MGI:5791488 | unclassified gene | predicted gene 45652 |
| 8 | gene | 29.108837 | 29.112776 | negative | MGI_C57BL6J_2385655 | Svet1 | ENSEMBL:ENSMUSG00000109648 | MGI:2385655 | unclassified gene | subventricular expressed transcript 1 |
| 8 | gene | 29.228598 | 29.235975 | negative | MGI_C57BL6J_5591726 | Gm32567 | NCBI_Gene:102635158,ENSEMBL:ENSMUSG00000110180 | MGI:5591726 | lncRNA gene | predicted gene, 32567 |
| 8 | pseudogene | 29.286223 | 29.286981 | negative | MGI_C57BL6J_2142657 | Nudc-ps1 | NCBI_Gene:100041237,ENSEMBL:ENSMUSG00000110331 | MGI:2142657 | pseudogene | nuclear distribution gene C homolog (Aspergillus), pseudogene 1 |
| 8 | pseudogene | 29.417354 | 29.417856 | negative | MGI_C57BL6J_5010861 | Gm18676 | NCBI_Gene:100417542 | MGI:5010861 | pseudogene | predicted gene, 18676 |
| 8 | gene | 29.530349 | 29.532136 | positive | MGI_C57BL6J_5591780 | Gm32621 | NCBI_Gene:102635230 | MGI:5591780 | lncRNA gene | predicted gene, 32621 |
| 8 | pseudogene | 29.533059 | 29.533405 | negative | MGI_C57BL6J_5588975 | Gm29816 | NCBI_Gene:102238521 | MGI:5588975 | pseudogene | predicted gene, 29816 |
| 8 | gene | 29.610770 | 29.614259 | negative | MGI_C57BL6J_5591837 | Gm32678 | NCBI_Gene:102635303 | MGI:5591837 | lncRNA gene | predicted gene, 32678 |
| 8 | pseudogene | 30.081673 | 30.081960 | negative | MGI_C57BL6J_5791328 | Gm45492 | ENSEMBL:ENSMUSG00000109597 | MGI:5791328 | pseudogene | predicted gene 45492 |
| 8 | gene | 30.111969 | 30.143385 | negative | MGI_C57BL6J_5622039 | Gm39154 | NCBI_Gene:105243156,ENSEMBL:ENSMUSG00000109666 | MGI:5622039 | lncRNA gene | predicted gene, 39154 |
| 8 | pseudogene | 30.177932 | 30.183860 | negative | MGI_C57BL6J_3644291 | Gm6853 | NCBI_Gene:628189,ENSEMBL:ENSMUSG00000109586 | MGI:3644291 | pseudogene | predicted gene 6853 |
| 8 | gene | 30.439747 | 30.442224 | negative | MGI_C57BL6J_5791326 | Gm45490 | ENSEMBL:ENSMUSG00000110006 | MGI:5791326 | lncRNA gene | predicted gene 45490 |
| 8 | gene | 30.482983 | 30.513811 | negative | MGI_C57BL6J_5791325 | Gm45489 | ENSEMBL:ENSMUSG00000110223 | MGI:5791325 | lncRNA gene | predicted gene 45489 |
| 8 | gene | 30.568393 | 30.575573 | positive | MGI_C57BL6J_1918564 | 4933433F19Rik | NCBI_Gene:71314,ENSEMBL:ENSMUSG00000097044 | MGI:1918564 | lncRNA gene | RIKEN cDNA 4933433F19 gene |
| 8 | gene | 30.915147 | 30.916548 | positive | MGI_C57BL6J_5591894 | Gm32735 | NCBI_Gene:102635378 | MGI:5591894 | lncRNA gene | predicted gene, 32735 |
| 8 | gene | 30.942695 | 30.946606 | negative | MGI_C57BL6J_2441976 | D430032J08Rik | NA | NA | unclassified gene | RIKEN cDNA D430032J08 gene |
| 8 | gene | 30.942701 | 30.959699 | negative | MGI_C57BL6J_5591950 | Gm32791 | NCBI_Gene:102635459 | MGI:5591950 | lncRNA gene | predicted gene, 32791 |
| 8 | gene | 31.001395 | 31.009494 | negative | MGI_C57BL6J_5592000 | Gm32841 | NCBI_Gene:102635531 | MGI:5592000 | lncRNA gene | predicted gene, 32841 |
| 8 | pseudogene | 31.003908 | 31.004304 | positive | MGI_C57BL6J_5791088 | Gm45252 | ENSEMBL:ENSMUSG00000109614 | MGI:5791088 | pseudogene | predicted gene 45252 |
| 8 | gene | 31.089471 | 31.097047 | positive | MGI_C57BL6J_1914209 | Dusp26 | NCBI_Gene:66959,ENSEMBL:ENSMUSG00000039661 | MGI:1914209 | protein coding gene | dual specificity phosphatase 26 (putative) |
| 8 | gene | 31.106629 | 31.109225 | positive | MGI_C57BL6J_5623369 | Gm40484 | NCBI_Gene:105244966 | MGI:5623369 | lncRNA gene | predicted gene, 40484 |
| 8 | gene | 31.111670 | 31.132071 | positive | MGI_C57BL6J_1916117 | Rnf122 | NCBI_Gene:68867,ENSEMBL:ENSMUSG00000039328 | MGI:1916117 | protein coding gene | ring finger protein 122 |
| 8 | gene | 31.149861 | 31.149967 | negative | MGI_C57BL6J_3819521 | Snord13 | NCBI_Gene:100217422,ENSEMBL:ENSMUSG00000088252 | MGI:3819521 | snoRNA gene | small nucleolar RNA, C/D box 13 |
| 8 | gene | 31.149862 | 31.149968 | negative | MGI_C57BL6J_1923944 | 1810029E06Rik | NA | NA | unclassified gene | RIKEN cDNA 1810029E06 gene |
| 8 | gene | 31.150315 | 31.164703 | positive | MGI_C57BL6J_2384576 | Tti2 | NCBI_Gene:234138,ENSEMBL:ENSMUSG00000031577 | MGI:2384576 | protein coding gene | TELO2 interacting protein 2 |
| 8 | gene | 31.159463 | 31.168764 | negative | MGI_C57BL6J_1915170 | Mak16 | NCBI_Gene:67920,ENSEMBL:ENSMUSG00000031578 | MGI:1915170 | protein coding gene | MAK16 homolog |
| 8 | gene | 31.187331 | 31.265284 | positive | MGI_C57BL6J_2384748 | Fut10 | NCBI_Gene:171167,ENSEMBL:ENSMUSG00000046152 | MGI:2384748 | protein coding gene | fucosyltransferase 10 |
| 8 | pseudogene | 31.213221 | 31.215425 | negative | MGI_C57BL6J_3647834 | Gm8051 | NCBI_Gene:102635732,ENSEMBL:ENSMUSG00000083588 | MGI:3647834 | pseudogene | predicted gene 8051 |
| 8 | pseudogene | 31.279699 | 31.280865 | negative | MGI_C57BL6J_3648130 | Gm8168 | NCBI_Gene:666568,ENSEMBL:ENSMUSG00000110133 | MGI:3648130 | pseudogene | predicted gene 8168 |
| 8 | gene | 31.367749 | 31.382439 | positive | MGI_C57BL6J_1921767 | 7420700N18Rik | NCBI_Gene:74517,ENSEMBL:ENSMUSG00000109915 | MGI:1921767 | lncRNA gene | RIKEN cDNA 7420700N18 gene |
| 8 | pseudogene | 31.379254 | 31.380551 | negative | MGI_C57BL6J_5595272 | Gm36113 | NCBI_Gene:102639913 | MGI:5595272 | pseudogene | predicted gene, 36113 |
| 8 | pseudogene | 31.380551 | 31.381316 | negative | MGI_C57BL6J_5011281 | Gm19096 | NCBI_Gene:100418243,ENSEMBL:ENSMUSG00000109705 | MGI:5011281 | pseudogene | predicted gene, 19096 |
| 8 | gene | 31.491976 | 31.500090 | positive | MGI_C57BL6J_5791139 | Gm45303 | ENSEMBL:ENSMUSG00000109605 | MGI:5791139 | lncRNA gene | predicted gene 45303 |
| 8 | gene | 31.544463 | 31.544686 | negative | MGI_C57BL6J_5453316 | Gm23539 | ENSEMBL:ENSMUSG00000088162 | MGI:5453316 | unclassified non-coding RNA gene | predicted gene, 23539 |
| 8 | gene | 31.598979 | 31.663824 | negative | MGI_C57BL6J_5621457 | Gm38572 | NA | NA | lncRNA gene | predicted gene%2c 38572 |
| 8 | gene | 31.598979 | 31.663828 | negative | MGI_C57BL6J_5791140 | Gm45304 | ENSEMBL:ENSMUSG00000110329 | MGI:5791140 | lncRNA gene | predicted gene 45304 |
| 8 | gene | 31.661331 | 31.661431 | negative | MGI_C57BL6J_5453961 | Gm24184 | ENSEMBL:ENSMUSG00000084524 | MGI:5453961 | unclassified non-coding RNA gene | predicted gene, 24184 |
| 8 | pseudogene | 31.737208 | 31.739760 | negative | MGI_C57BL6J_3648041 | Gm5117 | NCBI_Gene:330731,ENSEMBL:ENSMUSG00000093862 | MGI:3648041 | pseudogene | predicted gene 5117 |
| 8 | gene | 31.809465 | 32.884820 | negative | MGI_C57BL6J_96083 | Nrg1 | NCBI_Gene:211323,ENSEMBL:ENSMUSG00000062991 | MGI:96083 | protein coding gene | neuregulin 1 |
| 8 | gene | 31.919554 | 31.926205 | positive | MGI_C57BL6J_5592382 | Gm33223 | NCBI_Gene:102636040 | MGI:5592382 | lncRNA gene | predicted gene, 33223 |
| 8 | gene | 32.008504 | 32.018405 | positive | MGI_C57BL6J_5477072 | Gm26578 | NCBI_Gene:102641860,ENSEMBL:ENSMUSG00000097928 | MGI:5477072 | lncRNA gene | predicted gene, 26578 |
| 8 | gene | 32.151895 | 32.181847 | positive | MGI_C57BL6J_3646474 | Gm8179 | NCBI_Gene:666589 | MGI:3646474 | lncRNA gene | predicted gene 8179 |
| 8 | gene | 32.186129 | 32.186248 | positive | MGI_C57BL6J_5451917 | Gm22140 | ENSEMBL:ENSMUSG00000089370 | MGI:5451917 | snRNA gene | predicted gene, 22140 |
| 8 | pseudogene | 32.471078 | 32.477346 | negative | MGI_C57BL6J_5825713 | Gm46076 | NCBI_Gene:108167591 | MGI:5825713 | pseudogene | predicted gene, 46076 |
| 8 | gene | 32.751734 | 32.853374 | positive | MGI_C57BL6J_5622040 | Gm39155 | NCBI_Gene:105243158 | MGI:5622040 | lncRNA gene | predicted gene, 39155 |
| 8 | gene | 32.888505 | 32.950026 | negative | MGI_C57BL6J_3782158 | Gm3985 | NCBI_Gene:100042715,ENSEMBL:ENSMUSG00000079070 | MGI:3782158 | lncRNA gene | predicted gene 3985 |
| 8 | gene | 32.890742 | 32.892657 | positive | MGI_C57BL6J_5592241 | Gm33082 | NCBI_Gene:108167592 | MGI:5592241 | lncRNA gene | predicted gene, 33082 |
| 8 | gene | 32.972831 | 33.056091 | negative | MGI_C57BL6J_5791423 | Gm45587 | ENSEMBL:ENSMUSG00000110214 | MGI:5791423 | lncRNA gene | predicted gene 45587 |
| 8 | gene | 33.071052 | 33.095369 | negative | MGI_C57BL6J_5622041 | Gm39156 | NCBI_Gene:105243160 | MGI:5622041 | lncRNA gene | predicted gene, 39156 |
| 8 | gene | 33.116803 | 33.149512 | negative | MGI_C57BL6J_5592440 | Gm33281 | NCBI_Gene:102636119 | MGI:5592440 | lncRNA gene | predicted gene, 33281 |
| 8 | pseudogene | 33.189364 | 33.192332 | negative | MGI_C57BL6J_3644703 | Gm6877 | NCBI_Gene:628412,ENSEMBL:ENSMUSG00000083557 | MGI:3644703 | pseudogene | predicted pseudogene 6877 |
| 8 | gene | 33.234372 | 33.385542 | negative | MGI_C57BL6J_109635 | Wrn | NCBI_Gene:22427,ENSEMBL:ENSMUSG00000031583 | MGI:109635 | protein coding gene | Werner syndrome RecQ like helicase |
| 8 | gene | 33.385614 | 33.417472 | positive | MGI_C57BL6J_1922279 | Purg | NCBI_Gene:75029,ENSEMBL:ENSMUSG00000049184 | MGI:1922279 | protein coding gene | purine-rich element binding protein G |
| 8 | gene | 33.428705 | 33.432014 | positive | MGI_C57BL6J_2442538 | 5930422O12Rik | NA | NA | lncRNA gene | RIKEN cDNA 5930422O12 gene |
| 8 | pseudogene | 33.434510 | 33.435307 | negative | MGI_C57BL6J_3646495 | Gm8201 | NCBI_Gene:666630 | MGI:3646495 | pseudogene | predicted gene 8201 |
| 8 | pseudogene | 33.484196 | 33.485757 | positive | MGI_C57BL6J_104764 | Hmgb1-rs17 | NCBI_Gene:628431,ENSEMBL:ENSMUSG00000109750 | MGI:104764 | pseudogene | high mobility group box 1, related sequence 17 |
| 8 | gene | 33.516684 | 33.585585 | positive | MGI_C57BL6J_1934816 | Tex15 | NCBI_Gene:104271,ENSEMBL:ENSMUSG00000009628 | MGI:1934816 | protein coding gene | testis expressed gene 15 |
| 8 | gene | 33.599621 | 33.619804 | positive | MGI_C57BL6J_1321161 | Ppp2cb | NCBI_Gene:19053,ENSEMBL:ENSMUSG00000009630 | MGI:1321161 | protein coding gene | protein phosphatase 2 (formerly 2A), catalytic subunit, beta isoform |
| 8 | gene | 33.619586 | 33.645451 | negative | MGI_C57BL6J_1337129 | Ubxn8 | NCBI_Gene:108159,ENSEMBL:ENSMUSG00000052906 | MGI:1337129 | protein coding gene | UBX domain protein 8 |
| 8 | gene | 33.645537 | 33.652876 | positive | MGI_C57BL6J_5592592 | Gm33433 | NCBI_Gene:102636335 | MGI:5592592 | lncRNA gene | predicted gene, 33433 |
| 8 | gene | 33.652523 | 33.698163 | positive | MGI_C57BL6J_95804 | Gsr | NCBI_Gene:14782,ENSEMBL:ENSMUSG00000031584 | MGI:95804 | protein coding gene | glutathione reductase |
| 8 | gene | 33.691801 | 33.691944 | positive | MGI_C57BL6J_5454504 | Gm24727 | ENSEMBL:ENSMUSG00000088856 | MGI:5454504 | snoRNA gene | predicted gene, 24727 |
| 8 | pseudogene | 33.692018 | 33.693056 | positive | MGI_C57BL6J_5009989 | Gm17803 | NCBI_Gene:100286834 | MGI:5009989 | pseudogene | predicted gene, 17803 |
| 8 | gene | 33.701775 | 33.709143 | negative | MGI_C57BL6J_5623370 | Gm40485 | NCBI_Gene:105244967 | MGI:5623370 | lncRNA gene | predicted gene, 40485 |
| 8 | gene | 33.730534 | 33.731819 | negative | MGI_C57BL6J_1925886 | 1700104B16Rik | ENSEMBL:ENSMUSG00000049476 | MGI:1925886 | lncRNA gene | RIKEN cDNA 1700104B16 gene |
| 8 | gene | 33.731833 | 33.777173 | positive | MGI_C57BL6J_1915403 | Gtf2e2 | NCBI_Gene:68153,ENSEMBL:ENSMUSG00000031585 | MGI:1915403 | protein coding gene | general transcription factor II E, polypeptide 2 (beta subunit) |
| 8 | gene | 33.742112 | 33.748020 | negative | MGI_C57BL6J_1919882 | Smim18 | NCBI_Gene:72632,ENSEMBL:ENSMUSG00000094500 | MGI:1919882 | protein coding gene | small integral membrane protein 18 |
| 8 | gene | 33.782643 | 33.929896 | negative | MGI_C57BL6J_1334446 | Rbpms | NCBI_Gene:19663,ENSEMBL:ENSMUSG00000031586 | MGI:1334446 | protein coding gene | RNA binding protein gene with multiple splicing |
| 8 | gene | 33.813203 | 33.833094 | positive | MGI_C57BL6J_5477126 | Gm26632 | ENSEMBL:ENSMUSG00000097442 | MGI:5477126 | lncRNA gene | predicted gene, 26632 |
| 8 | gene | 33.884774 | 33.885747 | negative | MGI_C57BL6J_5504093 | Gm26978 | ENSEMBL:ENSMUSG00000098166 | MGI:5504093 | lncRNA gene | predicted gene, 26978 |
| 8 | gene | 33.928167 | 33.928769 | negative | MGI_C57BL6J_1924643 | 9530006C21Rik | NA | NA | unclassified gene | RIKEN cDNA 9530006C21 gene |
| 8 | gene | 33.964213 | 33.974504 | positive | MGI_C57BL6J_5592747 | Gm33588 | NCBI_Gene:102636553 | MGI:5592747 | lncRNA gene | predicted gene, 33588 |
| 8 | gene | 33.985444 | 33.987200 | negative | MGI_C57BL6J_5804932 | Gm45817 | ENSEMBL:ENSMUSG00000110614 | MGI:5804932 | unclassified gene | predicted gene 45817 |
| 8 | gene | 33.985914 | 33.990524 | positive | MGI_C57BL6J_5622042 | Gm39157 | NCBI_Gene:105243162,ENSEMBL:ENSMUSG00000110663 | MGI:5622042 | lncRNA gene | predicted gene, 39157 |
| 8 | gene | 34.045172 | 34.064659 | positive | MGI_C57BL6J_5623372 | Gm40487 | NCBI_Gene:105244969 | MGI:5623372 | lncRNA gene | predicted gene, 40487 |
| 8 | gene | 34.049262 | 34.057046 | positive | MGI_C57BL6J_3641827 | Gm9951 | ENSEMBL:ENSMUSG00000054618 | MGI:3641827 | lncRNA gene | predicted gene 9951 |
| 8 | gene | 34.052489 | 34.055877 | negative | MGI_C57BL6J_5623371 | Gm40486 | NCBI_Gene:105244968 | MGI:5623371 | lncRNA gene | predicted gene, 40486 |
| 8 | gene | 34.090420 | 34.108796 | negative | MGI_C57BL6J_1343154 | Dctn6 | NCBI_Gene:22428,ENSEMBL:ENSMUSG00000031516 | MGI:1343154 | protein coding gene | dynactin 6 |
| 8 | gene | 34.094548 | 34.096149 | positive | MGI_C57BL6J_5477262 | Gm26768 | ENSEMBL:ENSMUSG00000097257 | MGI:5477262 | lncRNA gene | predicted gene, 26768 |
| 8 | gene | 34.108139 | 34.125185 | positive | MGI_C57BL6J_2685017 | Mboat4 | NCBI_Gene:234155,ENSEMBL:ENSMUSG00000071113 | MGI:2685017 | protein coding gene | membrane bound O-acyltransferase domain containing 4 |
| 8 | pseudogene | 34.110991 | 34.111737 | negative | MGI_C57BL6J_3704308 | Gm10131 | NCBI_Gene:100042777,ENSEMBL:ENSMUSG00000063412 | MGI:3704308 | pseudogene | predicted pseudogene 10131 |
| 8 | gene | 34.119011 | 34.120208 | negative | MGI_C57BL6J_5622043 | Gm39158 | NCBI_Gene:105243163,ENSEMBL:ENSMUSG00000109869 | MGI:5622043 | lncRNA gene | predicted gene, 39158 |
| 8 | gene | 34.135572 | 34.147033 | negative | MGI_C57BL6J_1915442 | Leprotl1 | NCBI_Gene:68192,ENSEMBL:ENSMUSG00000031513 | MGI:1915442 | protein coding gene | leptin receptor overlapping transcript-like 1 |
| 8 | pseudogene | 34.152690 | 34.152971 | positive | MGI_C57BL6J_5791105 | Gm45269 | ENSEMBL:ENSMUSG00000110247 | MGI:5791105 | pseudogene | predicted gene 45269 |
| 8 | gene | 34.154563 | 34.170847 | positive | MGI_C57BL6J_1915137 | Saraf | NCBI_Gene:67887,ENSEMBL:ENSMUSG00000031532 | MGI:1915137 | protein coding gene | store-operated calcium entry-associated regulatory factor |
| 8 | gene | 34.166622 | 34.166712 | negative | MGI_C57BL6J_5530943 | Mir6395 | miRBase:MI0021930,NCBI_Gene:102465211,ENSEMBL:ENSMUSG00000098785 | MGI:5530943 | miRNA gene | microRNA 6395 |
| 8 | gene | 34.171547 | 34.173638 | negative | MGI_C57BL6J_5592797 | Gm33638 | NCBI_Gene:102636627 | MGI:5592797 | lncRNA gene | predicted gene, 33638 |
| 8 | gene | 34.216772 | 34.226145 | positive | MGI_C57BL6J_5579949 | Gm29243 | NCBI_Gene:102636691,ENSEMBL:ENSMUSG00000100605 | MGI:5579949 | lncRNA gene | predicted gene 29243 |
| 8 | pseudogene | 34.269032 | 34.328465 | positive | MGI_C57BL6J_3647233 | Gm4889 | NCBI_Gene:108167567,ENSEMBL:ENSMUSG00000056797 | MGI:3647233 | pseudogene | predicted gene 4889 |
| 8 | gene | 34.295856 | 34.295976 | negative | MGI_C57BL6J_5452954 | Gm23177 | ENSEMBL:ENSMUSG00000080434 | MGI:5452954 | miRNA gene | predicted gene, 23177 |
| 8 | pseudogene | 34.307074 | 34.307719 | negative | MGI_C57BL6J_3644987 | Gm6100 | NCBI_Gene:619780,ENSEMBL:ENSMUSG00000095202 | MGI:3644987 | pseudogene | predicted gene 6100 |
| 8 | pseudogene | 34.321656 | 34.322842 | negative | MGI_C57BL6J_3647610 | Gm8254 | NCBI_Gene:666718,ENSEMBL:ENSMUSG00000098046 | MGI:3647610 | pseudogene | predicted gene 8254 |
| 8 | gene | 34.344696 | 34.362842 | negative | MGI_C57BL6J_5791186 | Gm45350 | ENSEMBL:ENSMUSG00000109964 | MGI:5791186 | lncRNA gene | predicted gene 45350 |
| 8 | gene | 34.377076 | 34.386762 | negative | MGI_C57BL6J_5592990 | Gm33831 | NCBI_Gene:102636881,ENSEMBL:ENSMUSG00000110178 | MGI:5592990 | lncRNA gene | predicted gene, 33831 |
| 8 | gene | 34.419204 | 34.437622 | negative | MGI_C57BL6J_5593069 | Gm33910 | NCBI_Gene:102636992 | MGI:5593069 | lncRNA gene | predicted gene, 33910 |
| 8 | gene | 34.419237 | 34.472995 | negative | MGI_C57BL6J_5791185 | Gm45349 | ENSEMBL:ENSMUSG00000110369 | MGI:5791185 | lncRNA gene | predicted gene 45349 |
| 8 | gene | 34.440726 | 34.455836 | positive | MGI_C57BL6J_5622044 | Gm39159 | NCBI_Gene:105243165 | MGI:5622044 | lncRNA gene | predicted gene, 39159 |
| 8 | gene | 34.456918 | 34.488105 | positive | MGI_C57BL6J_5825675 | Gm46038 | NCBI_Gene:108167530 | MGI:5825675 | lncRNA gene | predicted gene, 46038 |
| 8 | gene | 34.462682 | 34.472999 | negative | MGI_C57BL6J_5622045 | Gm39160 | NCBI_Gene:105243166 | MGI:5622045 | lncRNA gene | predicted gene, 39160 |
| 8 | gene | 34.488965 | 34.492650 | positive | MGI_C57BL6J_5623374 | Gm40489 | NCBI_Gene:105244971 | MGI:5623374 | lncRNA gene | predicted gene, 40489 |
| 8 | pseudogene | 34.501561 | 34.507707 | positive | MGI_C57BL6J_3648998 | Gm8268 | NCBI_Gene:666746,ENSEMBL:ENSMUSG00000109580 | MGI:3648998 | pseudogene | predicted gene 8268 |
| 8 | gene | 34.520468 | 34.526628 | positive | MGI_C57BL6J_5593127 | Gm33968 | NCBI_Gene:102637063,ENSEMBL:ENSMUSG00000110015 | MGI:5593127 | lncRNA gene | predicted gene, 33968 |
| 8 | gene | 34.583025 | 34.584493 | negative | MGI_C57BL6J_1921960 | 4930507A01Rik | NA | NA | unclassified gene | RIKEN cDNA 4930507A01 gene |
| 8 | gene | 34.583456 | 34.585026 | negative | MGI_C57BL6J_5825705 | Gm46068 | NCBI_Gene:108167580 | MGI:5825705 | lncRNA gene | predicted gene, 46068 |
| 8 | gene | 34.589420 | 34.640316 | negative | MGI_C57BL6J_5439417 | B930018H19Rik | NCBI_Gene:330734,ENSEMBL:ENSMUSG00000109819 | MGI:5439417 | lncRNA gene | RIKEN cDNA B930018H19 gene |
| 8 | gene | 34.606700 | 34.619519 | positive | MGI_C57BL6J_5593255 | Gm34096 | NCBI_Gene:102637227,ENSEMBL:ENSMUSG00000109880 | MGI:5593255 | lncRNA gene | predicted gene, 34096 |
| 8 | gene | 34.658106 | 34.659369 | negative | MGI_C57BL6J_5791463 | Gm45627 | ENSEMBL:ENSMUSG00000109846 | MGI:5791463 | lncRNA gene | predicted gene 45627 |
| 8 | gene | 34.688679 | 34.690159 | negative | MGI_C57BL6J_5825706 | Gm46069 | NCBI_Gene:108167581 | MGI:5825706 | lncRNA gene | predicted gene, 46069 |
| 8 | gene | 34.807297 | 34.819894 | positive | MGI_C57BL6J_2442191 | Dusp4 | NCBI_Gene:319520,ENSEMBL:ENSMUSG00000031530 | MGI:2442191 | protein coding gene | dual specificity phosphatase 4 |
| 8 | gene | 34.809536 | 34.810409 | negative | MGI_C57BL6J_3780055 | Gm9648 | NA | NA | unclassified gene | predicted gene 9648 |
| 8 | gene | 34.826460 | 34.965690 | negative | MGI_C57BL6J_1341087 | Tnks | NCBI_Gene:21951,ENSEMBL:ENSMUSG00000031529 | MGI:1341087 | protein coding gene | tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| 8 | gene | 34.965052 | 34.974147 | positive | MGI_C57BL6J_3708775 | Gm9939 | NCBI_Gene:105243167,ENSEMBL:ENSMUSG00000054247 | MGI:3708775 | lncRNA gene | predicted gene 9939 |
| 8 | gene | 34.967274 | 34.979874 | negative | MGI_C57BL6J_5593527 | Gm34368 | ENSEMBL:ENSMUSG00000109717 | MGI:5593527 | lncRNA gene | predicted gene, 34368 |
| 8 | pseudogene | 35.052122 | 35.053454 | negative | MGI_C57BL6J_3648459 | Gm8291 | NCBI_Gene:666792,ENSEMBL:ENSMUSG00000110361 | MGI:3648459 | pseudogene | predicted gene 8291 |
| 8 | gene | 35.094539 | 35.095242 | negative | MGI_C57BL6J_1920879 | 1700123E06Rik | NA | NA | unclassified gene | RIKEN cDNA 1700123E06 gene |
| 8 | gene | 35.094545 | 35.113490 | positive | MGI_C57BL6J_5593578 | Gm34419 | NCBI_Gene:102637665,ENSEMBL:ENSMUSG00000110130 | MGI:5593578 | lncRNA gene | predicted gene, 34419 |
| 8 | pseudogene | 35.132995 | 35.133532 | negative | MGI_C57BL6J_3648456 | Rpl31-ps23 | NCBI_Gene:666797,ENSEMBL:ENSMUSG00000097106 | MGI:3648456 | pseudogene | ribosomal protein L31, pseudogene 23 |
| 8 | pseudogene | 35.150086 | 35.150446 | negative | MGI_C57BL6J_5791462 | Gm45626 | ENSEMBL:ENSMUSG00000109678 | MGI:5791462 | pseudogene | predicted gene 45626 |
| 8 | gene | 35.179103 | 35.183886 | negative | MGI_C57BL6J_5791461 | Gm45625 | ENSEMBL:ENSMUSG00000110314 | MGI:5791461 | lncRNA gene | predicted gene 45625 |
| 8 | gene | 35.218638 | 35.222071 | positive | MGI_C57BL6J_5593633 | Gm34474 | NCBI_Gene:102637740,ENSEMBL:ENSMUSG00000109773 | MGI:5593633 | lncRNA gene | predicted gene, 34474 |
| 8 | gene | 35.222941 | 35.226103 | positive | MGI_C57BL6J_5593692 | Gm34533 | NCBI_Gene:102637819,ENSEMBL:ENSMUSG00000109607 | MGI:5593692 | lncRNA gene | predicted gene, 34533 |
| 8 | gene | 35.232953 | 35.245479 | negative | MGI_C57BL6J_5593756 | Gm34597 | NCBI_Gene:102637901,ENSEMBL:ENSMUSG00000109688 | MGI:5593756 | lncRNA gene | predicted gene, 34597 |
| 8 | gene | 35.249761 | 35.261735 | negative | MGI_C57BL6J_5593806 | Gm34647 | NCBI_Gene:102637967 | MGI:5593806 | lncRNA gene | predicted gene, 34647 |
| 8 | gene | 35.348589 | 35.367831 | negative | MGI_C57BL6J_5593864 | Gm34705 | NCBI_Gene:102638044 | MGI:5593864 | lncRNA gene | predicted gene, 34705 |
| 8 | gene | 35.367750 | 35.376392 | negative | MGI_C57BL6J_5791305 | Gm45469 | ENSEMBL:ENSMUSG00000109698 | MGI:5791305 | lncRNA gene | predicted gene 45469 |
| 8 | gene | 35.367857 | 35.373295 | negative | MGI_C57BL6J_5622046 | Gm39161 | NCBI_Gene:105243169 | MGI:5622046 | lncRNA gene | predicted gene, 39161 |
| 8 | gene | 35.375667 | 35.388139 | positive | MGI_C57BL6J_2177268 | Ppp1r3b | NCBI_Gene:244416,ENSEMBL:ENSMUSG00000046794 | MGI:2177268 | protein coding gene | protein phosphatase 1, regulatory subunit 3B |
| 8 | gene | 35.407471 | 35.409506 | negative | MGI_C57BL6J_5791137 | Gm45301 | ENSEMBL:ENSMUSG00000110384 | MGI:5791137 | lncRNA gene | predicted gene 45301 |
| 8 | gene | 35.410285 | 35.415504 | negative | MGI_C57BL6J_5594070 | Gm34911 | NCBI_Gene:102638319 | MGI:5594070 | lncRNA gene | predicted gene, 34911 |
| 8 | gene | 35.436574 | 35.444961 | positive | MGI_C57BL6J_5594012 | Gm34853 | NCBI_Gene:102641483,ENSEMBL:ENSMUSG00000110367 | MGI:5594012 | lncRNA gene | predicted gene, 34853 |
| 8 | gene | 35.465253 | 35.496196 | negative | MGI_C57BL6J_1914526 | Eri1 | NCBI_Gene:67276,ENSEMBL:ENSMUSG00000031527 | MGI:1914526 | protein coding gene | exoribonuclease 1 |
| 8 | gene | 35.518396 | 35.535312 | negative | MGI_C57BL6J_5622047 | Gm39162 | NCBI_Gene:105243170 | MGI:5622047 | lncRNA gene | predicted gene, 39162 |
| 8 | gene | 35.578977 | 35.580074 | negative | MGI_C57BL6J_1924565 | C030008P14Rik | NA | NA | unclassified gene | RIKEN cDNA C030008P14 gene |
| 8 | gene | 35.582503 | 35.589020 | negative | MGI_C57BL6J_4439717 | Gm16793 | NCBI_Gene:100504714,ENSEMBL:ENSMUSG00000097357 | MGI:4439717 | lncRNA gene | predicted gene, 16793 |
| 8 | gene | 35.587798 | 35.679449 | positive | MGI_C57BL6J_1098644 | Mfhas1 | NCBI_Gene:52065,ENSEMBL:ENSMUSG00000070056 | MGI:1098644 | protein coding gene | malignant fibrous histiocytoma amplified sequence 1 |
| 8 | gene | 35.707660 | 35.719949 | negative | MGI_C57BL6J_1922784 | 1700015I17Rik | NCBI_Gene:102638388,ENSEMBL:ENSMUSG00000110335 | MGI:1922784 | lncRNA gene | RIKEN cDNA 1700015I17 gene |
| 8 | gene | 35.723356 | 35.764170 | positive | MGI_C57BL6J_5594180 | Gm35021 | NCBI_Gene:102638463,ENSEMBL:ENSMUSG00000109714 | MGI:5594180 | lncRNA gene | predicted gene, 35021 |
| 8 | gene | 35.727683 | 35.729206 | negative | MGI_C57BL6J_5623377 | Gm40492 | NCBI_Gene:105244974 | MGI:5623377 | lncRNA gene | predicted gene, 40492 |
| 8 | gene | 35.747449 | 35.747562 | positive | MGI_C57BL6J_5451993 | Gm22216 | ENSEMBL:ENSMUSG00000088182 | MGI:5451993 | rRNA gene | predicted gene, 22216 |
| 8 | gene | 35.763930 | 35.764056 | negative | MGI_C57BL6J_5690866 | Gm44474 | ENSEMBL:ENSMUSG00000104820 | MGI:5690866 | miRNA gene | predicted gene, 44474 |
| 8 | gene | 35.765661 | 35.818048 | positive | MGI_C57BL6J_5011595 | Gm19410 | NCBI_Gene:100502846,ENSEMBL:ENSMUSG00000109372 | MGI:5011595 | protein coding gene | predicted gene, 19410 |
| 8 | pseudogene | 35.800686 | 35.801853 | negative | MGI_C57BL6J_5791402 | Gm45566 | ENSEMBL:ENSMUSG00000110359 | MGI:5791402 | pseudogene | predicted gene 45566 |
| 8 | gene | 35.824709 | 35.826559 | negative | MGI_C57BL6J_1919158 | Cldn23 | NCBI_Gene:71908,ENSEMBL:ENSMUSG00000055976 | MGI:1919158 | protein coding gene | claudin 23 |
| 8 | gene | 35.864085 | 35.882957 | negative | MGI_C57BL6J_5439418 | 5430403N17Rik | NCBI_Gene:330737,ENSEMBL:ENSMUSG00000109761 | MGI:5439418 | lncRNA gene | RIKEN cDNA 5430403N17 gene |
| 8 | gene | 35.977516 | 35.977578 | negative | MGI_C57BL6J_5451807 | Gm22030 | ENSEMBL:ENSMUSG00000080592 | MGI:5451807 | miRNA gene | predicted gene, 22030 |
| 8 | pseudogene | 36.025677 | 36.026148 | positive | MGI_C57BL6J_3642518 | Rps12-ps24 | NCBI_Gene:671641,ENSEMBL:ENSMUSG00000098181 | MGI:3642518 | pseudogene | ribosomal protein S12, pseudogene 24 |
| 8 | gene | 36.066346 | 36.073607 | positive | MGI_C57BL6J_5622048 | Gm39163 | NCBI_Gene:105243171 | MGI:5622048 | lncRNA gene | predicted gene, 39163 |
| 8 | gene | 36.094625 | 36.147787 | positive | MGI_C57BL6J_1196223 | Prag1 | NCBI_Gene:244418,ENSEMBL:ENSMUSG00000050271 | MGI:1196223 | protein coding gene | PEAK1 related kinase activating pseudokinase 1 |
| 8 | gene | 36.096192 | 36.096987 | positive | MGI_C57BL6J_1924338 | 3010031K01Rik | NA | NA | unclassified gene | RIKEN cDNA 3010031K01 gene |
| 8 | gene | 36.156010 | 36.161588 | positive | MGI_C57BL6J_5621299 | Gm38414 | NCBI_Gene:330738,ENSEMBL:ENSMUSG00000109704 | MGI:5621299 | lncRNA gene | predicted gene, 38414 |
| 8 | gene | 36.174140 | 36.176075 | negative | MGI_C57BL6J_5622049 | Gm39164 | NCBI_Gene:105243172,ENSEMBL:ENSMUSG00000110321 | MGI:5622049 | lncRNA gene | predicted gene, 39164 |
| 8 | gene | 36.176141 | 36.178423 | positive | MGI_C57BL6J_1918528 | 4933430A20Rik | ENSEMBL:ENSMUSG00000110116 | MGI:1918528 | lncRNA gene | RIKEN cDNA 4933430A20 gene |
| 8 | gene | 36.188372 | 36.202847 | positive | MGI_C57BL6J_5594679 | Gm35520 | NCBI_Gene:102639136,ENSEMBL:ENSMUSG00000110176 | MGI:5594679 | lncRNA gene | predicted gene, 35520 |
| 8 | gene | 36.216064 | 36.249516 | negative | MGI_C57BL6J_3609241 | Lonrf1 | NCBI_Gene:244421,ENSEMBL:ENSMUSG00000039633 | MGI:3609241 | protein coding gene | LON peptidase N-terminal domain and ring finger 1 |
| 8 | gene | 36.226421 | 36.228514 | positive | MGI_C57BL6J_5622050 | Gm39165 | NCBI_Gene:105243173 | MGI:5622050 | lncRNA gene | predicted gene, 39165 |
| 8 | gene | 36.247998 | 36.315793 | positive | MGI_C57BL6J_5594871 | Gm35712 | NCBI_Gene:102639386 | MGI:5594871 | lncRNA gene | predicted gene, 35712 |
| 8 | pseudogene | 36.372396 | 36.373651 | negative | MGI_C57BL6J_3811197 | Gm5787 | NCBI_Gene:544878,ENSEMBL:ENSMUSG00000098154 | MGI:3811197 | pseudogene | predicted gene 5787 |
| 8 | gene | 36.406094 | 36.406244 | negative | MGI_C57BL6J_5455927 | Gm26150 | ENSEMBL:ENSMUSG00000088051 | MGI:5455927 | snRNA gene | predicted gene, 26150 |
| 8 | pseudogene | 36.450232 | 36.450800 | positive | MGI_C57BL6J_5011325 | Gm19140 | NCBI_Gene:100418322,ENSEMBL:ENSMUSG00000109977 | MGI:5011325 | pseudogene | predicted gene, 19140 |
| 8 | gene | 36.457286 | 36.516570 | positive | MGI_C57BL6J_2442328 | Trmt9b | NCBI_Gene:319582,ENSEMBL:ENSMUSG00000039620 | MGI:2442328 | protein coding gene | tRNA methyltransferase 9B |
| 8 | gene | 36.567739 | 36.953170 | negative | MGI_C57BL6J_1354949 | Dlc1 | NCBI_Gene:50768,ENSEMBL:ENSMUSG00000031523 | MGI:1354949 | protein coding gene | deleted in liver cancer 1 |
| 8 | gene | 36.646994 | 36.647965 | negative | MGI_C57BL6J_1924613 | 9530004P13Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 9530004P13 gene |
| 8 | gene | 36.758821 | 36.761679 | negative | MGI_C57BL6J_2142813 | 9430047G12Rik | NA | NA | unclassified gene | RIKEN cDNA 9430047G12 gene |
| 8 | gene | 36.876508 | 36.878373 | negative | MGI_C57BL6J_5011459 | Gm19274 | NA | NA | unclassified gene | predicted gene%2c 19274 |
| 8 | gene | 36.954740 | 36.959348 | positive | MGI_C57BL6J_5622051 | Gm39166 | NCBI_Gene:105243174 | MGI:5622051 | lncRNA gene | predicted gene, 39166 |
| 8 | gene | 36.985573 | 37.024238 | negative | MGI_C57BL6J_3041241 | G630064G18Rik | NCBI_Gene:414128,ENSEMBL:ENSMUSG00000110242 | MGI:3041241 | lncRNA gene | RIKEN cDNA G630064G18 gene |
| 8 | gene | 36.993575 | 36.995533 | positive | MGI_C57BL6J_2142538 | AI429214 | NCBI_Gene:621080,ENSEMBL:ENSMUSG00000074384 | MGI:2142538 | protein coding gene | expressed sequence AI429214 |
| 8 | pseudogene | 37.027093 | 37.027338 | positive | MGI_C57BL6J_5791526 | Gm45690 | ENSEMBL:ENSMUSG00000110158 | MGI:5791526 | pseudogene | predicted gene 45690 |
| 8 | gene | 37.088936 | 37.090309 | negative | MGI_C57BL6J_3641650 | Gm10683 | ENSEMBL:ENSMUSG00000109817 | MGI:3641650 | protein coding gene | predicted gene 10683 |
| 8 | pseudogene | 37.093318 | 37.094468 | positive | MGI_C57BL6J_5010452 | Gm18267 | NCBI_Gene:100416806 | MGI:5010452 | pseudogene | predicted gene, 18267 |
| 8 | pseudogene | 37.094695 | 37.095854 | positive | MGI_C57BL6J_3643490 | Gm8342 | NCBI_Gene:666877,ENSEMBL:ENSMUSG00000109668 | MGI:3643490 | pseudogene | predicted gene 8342 |
| 8 | gene | 37.097018 | 37.188061 | positive | MGI_C57BL6J_5791360 | Gm45524 | ENSEMBL:ENSMUSG00000110285 | MGI:5791360 | lncRNA gene | predicted gene 45524 |
| 8 | pseudogene | 37.103795 | 37.106002 | positive | MGI_C57BL6J_4361947 | Gm16530 | NCBI_Gene:76418 | MGI:4361947 | pseudogene | predicted gene, 16530 |
| 8 | gene | 37.103796 | 37.105508 | negative | MGI_C57BL6J_1923668 | 1700016D18Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 1700016D18 gene |
| 8 | pseudogene | 37.103813 | 37.110133 | positive | MGI_C57BL6J_3781611 | Gm3433 | NCBI_Gene:100041616,ENSEMBL:ENSMUSG00000110337 | MGI:3781611 | pseudogene | predicted gene 3433 |
| 8 | pseudogene | 37.149628 | 37.159699 | positive | MGI_C57BL6J_3648719 | Gm7319 | NCBI_Gene:641121,ENSEMBL:ENSMUSG00000109932 | MGI:3648719 | pseudogene | predicted gene 7319 |
| 8 | pseudogene | 37.183218 | 37.184232 | negative | MGI_C57BL6J_5791359 | Gm45523 | ENSEMBL:ENSMUSG00000109646 | MGI:5791359 | pseudogene | predicted gene 45523 |
| 8 | pseudogene | 37.250821 | 37.250986 | positive | MGI_C57BL6J_5791358 | Gm45522 | ENSEMBL:ENSMUSG00000109795 | MGI:5791358 | pseudogene | predicted gene 45522 |
| 8 | gene | 37.317477 | 37.317578 | negative | MGI_C57BL6J_5454903 | Gm25126 | ENSEMBL:ENSMUSG00000065277 | MGI:5454903 | snRNA gene | predicted gene, 25126 |
| 8 | gene | 37.322385 | 37.350753 | negative | MGI_C57BL6J_5791204 | Gm45368 | ENSEMBL:ENSMUSG00000110312 | MGI:5791204 | lncRNA gene | predicted gene 45368 |
| 8 | pseudogene | 37.371728 | 37.373277 | negative | MGI_C57BL6J_3645773 | Gm6203 | NCBI_Gene:621149,ENSEMBL:ENSMUSG00000110415 | MGI:3645773 | pseudogene | predicted gene 6203 |
| 8 | pseudogene | 37.400898 | 37.401020 | negative | MGI_C57BL6J_5791490 | Gm45654 | ENSEMBL:ENSMUSG00000109820 | MGI:5791490 | pseudogene | predicted gene 45654 |
| 8 | gene | 37.517840 | 38.662638 | negative | MGI_C57BL6J_2388820 | Sgcz | NCBI_Gene:244431,ENSEMBL:ENSMUSG00000039539 | MGI:2388820 | protein coding gene | sarcoglycan zeta |
| 8 | pseudogene | 37.787664 | 37.788940 | negative | MGI_C57BL6J_3801883 | Gm16205 | NCBI_Gene:436033,ENSEMBL:ENSMUSG00000081316 | MGI:3801883 | pseudogene | predicted gene 16205 |
| 8 | pseudogene | 38.076714 | 38.079639 | positive | MGI_C57BL6J_3801788 | Gm16204 | NCBI_Gene:100417841,ENSEMBL:ENSMUSG00000081517 | MGI:3801788 | pseudogene | predicted gene 16204 |
| 8 | gene | 38.213201 | 38.213283 | positive | MGI_C57BL6J_4413932 | n-TLcag2 | NCBI_Gene:102467334 | MGI:4413932 | tRNA gene | nuclear encoded tRNA leucine 2 (anticodon CAG) |
| 8 | gene | 38.252133 | 38.252202 | negative | MGI_C57BL6J_3619393 | Mir383 | miRBase:MI0000800,NCBI_Gene:723860,ENSEMBL:ENSMUSG00000065457 | MGI:3619393 | miRNA gene | microRNA 383 |
| 8 | gene | 38.513400 | 38.517639 | negative | MGI_C57BL6J_2444454 | 9330187G09Rik | NA | NA | unclassified gene | RIKEN cDNA 9330187G09 gene |
| 8 | gene | 38.659693 | 38.671308 | positive | MGI_C57BL6J_5623378 | Gm40493 | NCBI_Gene:105244975,ENSEMBL:ENSMUSG00000110236 | MGI:5623378 | lncRNA gene | predicted gene, 40493 |
| 8 | gene | 38.982903 | 38.983035 | negative | MGI_C57BL6J_5452905 | Gm23128 | ENSEMBL:ENSMUSG00000106007,ENSEMBL:ENSMUSG00000077445 | MGI:5452905 | snoRNA gene | predicted gene, 23128 |
| 8 | gene | 39.005468 | 39.165640 | positive | MGI_C57BL6J_1933134 | Tusc3 | NCBI_Gene:80286,ENSEMBL:ENSMUSG00000039530 | MGI:1933134 | protein coding gene | tumor suppressor candidate 3 |
| 8 | gene | 39.171656 | 39.174192 | positive | MGI_C57BL6J_1924133 | 6430500C12Rik | NA | NA | unclassified gene | RIKEN cDNA 6430500C12 gene |
| 8 | pseudogene | 39.200936 | 39.201506 | positive | MGI_C57BL6J_5434303 | Gm20948 | NCBI_Gene:100417019,ENSEMBL:ENSMUSG00000109922 | MGI:5434303 | pseudogene | predicted gene, 20948 |
| 8 | pseudogene | 39.204018 | 39.204462 | positive | MGI_C57BL6J_5009930 | Gm17766 | NCBI_Gene:100044748,ENSEMBL:ENSMUSG00000110153 | MGI:5009930 | pseudogene | predicted gene, 17766 |
| 8 | pseudogene | 39.206468 | 39.206632 | positive | MGI_C57BL6J_5791234 | Gm45398 | ENSEMBL:ENSMUSG00000110128 | MGI:5791234 | pseudogene | predicted gene 45398 |
| 8 | pseudogene | 39.282243 | 39.285152 | positive | MGI_C57BL6J_1332224 | Zfp148-ps1 | NCBI_Gene:632066,ENSEMBL:ENSMUSG00000109601 | MGI:1332224 | pseudogene | zinc finger protein 148, pseudogene 1 |
| 8 | gene | 39.297980 | 39.368268 | positive | MGI_C57BL6J_3646754 | Gm6213 | NCBI_Gene:621335,ENSEMBL:ENSMUSG00000109827 | MGI:3646754 | lncRNA gene | predicted gene 6213 |
| 8 | pseudogene | 39.310452 | 39.311695 | negative | MGI_C57BL6J_5010829 | Gm18644 | NCBI_Gene:100417492,ENSEMBL:ENSMUSG00000109955 | MGI:5010829 | pseudogene | predicted gene, 18644 |
| 8 | pseudogene | 39.455754 | 39.458691 | positive | MGI_C57BL6J_3643259 | Gm5609 | NCBI_Gene:434304,ENSEMBL:ENSMUSG00000110118 | MGI:3643259 | pseudogene | predicted gene 5609 |
| 8 | gene | 39.543232 | 39.642750 | negative | MGI_C57BL6J_98257 | Msr1 | NCBI_Gene:20288,ENSEMBL:ENSMUSG00000025044 | MGI:98257 | protein coding gene | macrophage scavenger receptor 1 |
| 8 | gene | 39.636247 | 39.708088 | positive | MGI_C57BL6J_5595900 | Gm36741 | NCBI_Gene:102640745 | MGI:5595900 | lncRNA gene | predicted gene, 36741 |
| 8 | pseudogene | 39.718624 | 39.767145 | negative | MGI_C57BL6J_3780202 | Gm2033 | NCBI_Gene:100039072,ENSEMBL:ENSMUSG00000109789 | MGI:3780202 | pseudogene | predicted gene 2033 |
| 8 | gene | 39.872396 | 39.928157 | negative | MGI_C57BL6J_5595954 | Gm36795 | NCBI_Gene:102640817 | MGI:5595954 | lncRNA gene | predicted gene, 36795 |
| 8 | gene | 39.930986 | 39.933257 | negative | MGI_C57BL6J_3641844 | Gm10682 | NA | NA | protein coding gene | predicted gene 10682 |
| 8 | pseudogene | 39.940061 | 39.940665 | positive | MGI_C57BL6J_3647783 | Gm8381 | NCBI_Gene:666959 | MGI:3647783 | pseudogene | predicted gene 8381 |
| 8 | pseudogene | 40.016464 | 40.018292 | positive | MGI_C57BL6J_3647792 | Gm7082 | NCBI_Gene:632166,ENSEMBL:ENSMUSG00000109815 | MGI:3647792 | pseudogene | predicted gene 7082 |
| 8 | gene | 40.279166 | 40.308331 | negative | MGI_C57BL6J_1891346 | Fgf20 | NCBI_Gene:80857,ENSEMBL:ENSMUSG00000031603 | MGI:1891346 | protein coding gene | fibroblast growth factor 20 |
| 8 | gene | 40.307435 | 40.308397 | negative | MGI_C57BL6J_1917015 | 1500004F05Rik | NA | NA | unclassified gene | RIKEN cDNA 1500004F05 gene |
| 8 | gene | 40.307458 | 40.388782 | positive | MGI_C57BL6J_1925756 | Micu3 | NCBI_Gene:78506,ENSEMBL:ENSMUSG00000039478 | MGI:1925756 | protein coding gene | mitochondrial calcium uptake family, member 3 |
| 8 | gene | 40.307875 | 40.307929 | negative | MGI_C57BL6J_5531297 | Mir7666 | miRBase:MI0025006,NCBI_Gene:102466237,ENSEMBL:ENSMUSG00000098431 | MGI:5531297 | miRNA gene | microRNA 7666 |
| 8 | gene | 40.363808 | 40.363843 | negative | MGI_C57BL6J_1917101 | 2010007E15Rik | NA | NA | unclassified gene | RIKEN cDNA 2010007E15 gene |
| 8 | gene | 40.423002 | 40.510268 | positive | MGI_C57BL6J_1923452 | Zdhhc2 | NCBI_Gene:70546,ENSEMBL:ENSMUSG00000039470 | MGI:1923452 | protein coding gene | zinc finger, DHHC domain containing 2 |
| 8 | gene | 40.492538 | 40.515847 | negative | MGI_C57BL6J_1298230 | Cnot7 | NCBI_Gene:18983,ENSEMBL:ENSMUSG00000031601 | MGI:1298230 | protein coding gene | CCR4-NOT transcription complex, subunit 7 |
| 8 | gene | 40.511783 | 40.551134 | positive | MGI_C57BL6J_1261835 | Vps37a | NCBI_Gene:52348,ENSEMBL:ENSMUSG00000031600 | MGI:1261835 | protein coding gene | vacuolar protein sorting 37A |
| 8 | gene | 40.550143 | 40.634923 | negative | MGI_C57BL6J_1891693 | Mtmr7 | NCBI_Gene:54384,ENSEMBL:ENSMUSG00000039431 | MGI:1891693 | protein coding gene | myotubularin related protein 7 |
| 8 | gene | 40.650598 | 40.660294 | positive | MGI_C57BL6J_3780252 | Gm2085 | NA | NA | unclassified non-coding RNA gene | predicted gene 2085 |
| 8 | gene | 40.675077 | 40.682199 | positive | MGI_C57BL6J_105984 | Adam24 | NCBI_Gene:13526,ENSEMBL:ENSMUSG00000046723 | MGI:105984 | protein coding gene | a disintegrin and metallopeptidase domain 24 (testase 1) |
| 8 | pseudogene | 40.683497 | 40.683647 | negative | MGI_C57BL6J_5791496 | Gm45660 | ENSEMBL:ENSMUSG00000109658 | MGI:5791496 | pseudogene | predicted gene 45660 |
| 8 | pseudogene | 40.712681 | 40.713287 | negative | MGI_C57BL6J_3645600 | Gm8423 | NCBI_Gene:667019,ENSEMBL:ENSMUSG00000110315 | MGI:3645600 | pseudogene | predicted gene 8423 |
| 8 | pseudogene | 40.724832 | 40.726407 | positive | MGI_C57BL6J_3780247 | Gm2080 | NCBI_Gene:100039170,ENSEMBL:ENSMUSG00000110228 | MGI:3780247 | pseudogene | predicted gene 2080 |
| 8 | pseudogene | 40.747890 | 40.749180 | negative | MGI_C57BL6J_3647304 | Gm5749 | NCBI_Gene:436137,ENSEMBL:ENSMUSG00000110370 | MGI:3647304 | pseudogene | predicted gene 5749 |
| 8 | gene | 40.752208 | 40.756187 | positive | MGI_C57BL6J_1345157 | Adam25 | NCBI_Gene:23793,ENSEMBL:ENSMUSG00000071937 | MGI:1345157 | protein coding gene | a disintegrin and metallopeptidase domain 25 (testase 2) |
| 8 | pseudogene | 40.765276 | 40.766734 | positive | MGI_C57BL6J_3643054 | Gm8433 | NCBI_Gene:667041 | MGI:3643054 | pseudogene | predicted gene 8433 |
| 8 | pseudogene | 40.789057 | 40.790167 | negative | MGI_C57BL6J_3642972 | Gm8436 | NCBI_Gene:667052,ENSEMBL:ENSMUSG00000110032 | MGI:3642972 | pseudogene | predicted gene 8436 |
| 8 | gene | 40.793273 | 40.797342 | positive | MGI_C57BL6J_2152342 | Adam20 | NCBI_Gene:384806,ENSEMBL:ENSMUSG00000046282 | MGI:2152342 | protein coding gene | a disintegrin and metallopeptidase domain 20 |
| 8 | gene | 40.822990 | 40.827037 | positive | MGI_C57BL6J_3045694 | Adam39 | NCBI_Gene:546055,ENSEMBL:ENSMUSG00000054033 | MGI:3045694 | protein coding gene | a disintegrin and metallopeptidase domain 39 |
| 8 | pseudogene | 40.853625 | 40.855045 | negative | MGI_C57BL6J_3644320 | Gm5345 | NCBI_Gene:384808,ENSEMBL:ENSMUSG00000098050 | MGI:3644320 | pseudogene | predicted gene 5345 |
| 8 | pseudogene | 40.855231 | 40.856397 | positive | MGI_C57BL6J_3647926 | Gm8445 | NCBI_Gene:667077 | MGI:3647926 | pseudogene | predicted gene 8445 |
| 8 | gene | 40.862367 | 40.922308 | positive | MGI_C57BL6J_99828 | Slc7a2 | NCBI_Gene:11988,ENSEMBL:ENSMUSG00000031596 | MGI:99828 | protein coding gene | solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| 8 | gene | 40.862395 | 40.865647 | negative | MGI_C57BL6J_5477078 | Gm26584 | ENSEMBL:ENSMUSG00000097474 | MGI:5477078 | lncRNA gene | predicted gene, 26584 |
| 8 | gene | 40.926212 | 40.990785 | positive | MGI_C57BL6J_1916047 | Pdgfrl | NCBI_Gene:68797,ENSEMBL:ENSMUSG00000031595 | MGI:1916047 | protein coding gene | platelet-derived growth factor receptor-like |
| 8 | gene | 40.988597 | 40.991065 | negative | MGI_C57BL6J_3704309 | Gm9868 | NA | NA | unclassified gene | predicted gene 9868 |
| 8 | gene | 40.990912 | 41.134350 | negative | MGI_C57BL6J_2142572 | Mtus1 | NCBI_Gene:102103,ENSEMBL:ENSMUSG00000045636 | MGI:2142572 | protein coding gene | mitochondrial tumor suppressor 1 |
| 8 | gene | 41.017468 | 41.024216 | positive | MGI_C57BL6J_1926099 | B430010I23Rik | NCBI_Gene:78849,ENSEMBL:ENSMUSG00000084960 | MGI:1926099 | lncRNA gene | RIKEN cDNA B430010I23 gene |
| 8 | gene | 41.039756 | 41.042053 | positive | MGI_C57BL6J_3802024 | Gm16192 | ENSEMBL:ENSMUSG00000086773 | MGI:3802024 | lncRNA gene | predicted gene 16192 |
| 8 | gene | 41.053110 | 41.064706 | positive | MGI_C57BL6J_3802025 | Gm16193 | ENSEMBL:ENSMUSG00000085672 | MGI:3802025 | lncRNA gene | predicted gene 16193 |
| 8 | gene | 41.154052 | 41.172984 | positive | MGI_C57BL6J_5622052 | Gm39167 | NCBI_Gene:105243175 | MGI:5622052 | lncRNA gene | predicted gene, 39167 |
| 8 | gene | 41.191434 | 41.217978 | negative | MGI_C57BL6J_102795 | Fgl1 | NCBI_Gene:234199,ENSEMBL:ENSMUSG00000031594 | MGI:102795 | protein coding gene | fibrinogen-like protein 1 |
| 8 | gene | 41.218131 | 41.221100 | positive | MGI_C57BL6J_3840113 | Gm16348 | NCBI_Gene:102640962,ENSEMBL:ENSMUSG00000085445 | MGI:3840113 | lncRNA gene | predicted gene 16348 |
| 8 | gene | 41.239641 | 41.334087 | positive | MGI_C57BL6J_1277958 | Pcm1 | NCBI_Gene:18536,ENSEMBL:ENSMUSG00000031592 | MGI:1277958 | protein coding gene | pericentriolar material 1 |
| 8 | gene | 41.340197 | 41.397644 | negative | MGI_C57BL6J_1277124 | Asah1 | NCBI_Gene:11886,ENSEMBL:ENSMUSG00000031591 | MGI:1277124 | protein coding gene | N-acylsphingosine amidohydrolase 1 |
| 8 | gene | 41.389181 | 41.417121 | negative | MGI_C57BL6J_893597 | Frg1 | NCBI_Gene:14300,ENSEMBL:ENSMUSG00000031590 | MGI:893597 | protein coding gene | FSHD region gene 1 |
| 8 | pseudogene | 41.608470 | 41.609687 | negative | MGI_C57BL6J_3780363 | Gapdh-ps14 | NCBI_Gene:100039376,ENSEMBL:ENSMUSG00000098149 | MGI:3780363 | pseudogene | glyceraldehyde-3-phosphate dehydrogenase, pseudogene 14 |
| 8 | pseudogene | 41.657514 | 41.658670 | negative | MGI_C57BL6J_3037673 | Smarce1-ps1 | NCBI_Gene:211895,ENSEMBL:ENSMUSG00000094568 | MGI:3037673 | pseudogene | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1, pseodogene 1 |
| 8 | gene | 41.827267 | 42.046202 | positive | MGI_C57BL6J_1917216 | 2810404M03Rik | NCBI_Gene:69966,ENSEMBL:ENSMUSG00000102046 | MGI:1917216 | lncRNA gene | RIKEN cDNA 2810404M03 gene |
| 8 | pseudogene | 41.923520 | 41.927130 | positive | MGI_C57BL6J_3779956 | Hspd1-ps1 | NCBI_Gene:671994,ENSEMBL:ENSMUSG00000108069 | MGI:3779956 | pseudogene | heat shock protein 1 (chaperonin), pseudogene 1 |
| 8 | pseudogene | 42.081329 | 42.083312 | negative | MGI_C57BL6J_2387419 | Zfp353-ps | NCBI_Gene:234203,ENSEMBL:ENSMUSG00000095057 | MGI:2387419 | pseudogene | zinc finger protein 353, pseudogene |
| 8 | pseudogene | 42.246836 | 42.247795 | positive | MGI_C57BL6J_3643972 | Gm6180 | NCBI_Gene:620772,ENSEMBL:ENSMUSG00000053038 | MGI:3643972 | pseudogene | predicted pseudogene 6180 |
| 8 | gene | 42.522136 | 42.522203 | negative | MGI_C57BL6J_5453308 | Gm23531 | ENSEMBL:ENSMUSG00000077755 | MGI:5453308 | snoRNA gene | predicted gene, 23531 |
| 8 | gene | 42.742807 | 42.742912 | positive | MGI_C57BL6J_5455958 | Gm26181 | ENSEMBL:ENSMUSG00000088312 | MGI:5455958 | snRNA gene | predicted gene, 26181 |
| 8 | pseudogene | 42.769434 | 42.770276 | positive | MGI_C57BL6J_5010599 | Gm18414 | NCBI_Gene:100417132,ENSEMBL:ENSMUSG00000109987 | MGI:5010599 | pseudogene | predicted gene, 18414 |
| 8 | pseudogene | 43.085587 | 43.085848 | positive | MGI_C57BL6J_5791426 | Gm45590 | ENSEMBL:ENSMUSG00000109627 | MGI:5791426 | pseudogene | predicted gene 45590 |
| 8 | gene | 43.129806 | 43.180489 | negative | MGI_C57BL6J_2687279 | Triml1 | NCBI_Gene:244448,ENSEMBL:ENSMUSG00000031651 | MGI:2687279 | protein coding gene | tripartite motif family-like 1 |
| 8 | pseudogene | 43.158754 | 43.163946 | positive | MGI_C57BL6J_3643970 | Gm6284 | NCBI_Gene:622082,ENSEMBL:ENSMUSG00000109991 | MGI:3643970 | pseudogene | predicted gene 6284 |
| 8 | gene | 43.180491 | 43.193884 | positive | MGI_C57BL6J_3642989 | Triml2 | NCBI_Gene:622117,ENSEMBL:ENSMUSG00000091490 | MGI:3642989 | protein coding gene | tripartite motif family-like 2 |
| 8 | pseudogene | 43.199967 | 43.200543 | positive | MGI_C57BL6J_3780391 | Gm2221 | NCBI_Gene:100039417,ENSEMBL:ENSMUSG00000109974 | MGI:3780391 | pseudogene | predicted gene 2221 |
| 8 | gene | 43.295063 | 43.307009 | negative | MGI_C57BL6J_99187 | Zfp42 | NCBI_Gene:22702,ENSEMBL:ENSMUSG00000051176 | MGI:99187 | protein coding gene | zinc finger protein 42 |
| 8 | gene | 43.312626 | 43.316640 | negative | MGI_C57BL6J_5623383 | Gm40498 | NCBI_Gene:105244980 | MGI:5623383 | protein coding gene | predicted gene, 40498 |
| 8 | gene | 43.377975 | 43.378104 | positive | MGI_C57BL6J_5452763 | Gm22986 | ENSEMBL:ENSMUSG00000084702 | MGI:5452763 | snoRNA gene | predicted gene, 22986 |
| 8 | gene | 43.519762 | 43.528137 | negative | MGI_C57BL6J_3588304 | Adam26b | NCBI_Gene:382007,ENSEMBL:ENSMUSG00000063900 | MGI:3588304 | protein coding gene | a disintegrin and metallopeptidase domain 26B |
| 8 | gene | 43.568273 | 43.576707 | negative | MGI_C57BL6J_105985 | Adam26a | NCBI_Gene:13525,ENSEMBL:ENSMUSG00000048516 | MGI:105985 | protein coding gene | a disintegrin and metallopeptidase domain 26A (testase 3) |
| 8 | pseudogene | 43.617078 | 43.618771 | negative | MGI_C57BL6J_5012023 | Gm19838 | NCBI_Gene:100503694 | MGI:5012023 | pseudogene | predicted gene, 19838 |
| 8 | gene | 43.624951 | 43.703316 | negative | MGI_C57BL6J_3647273 | Adam34l | NCBI_Gene:384813,ENSEMBL:ENSMUSG00000050190 | MGI:3647273 | protein coding gene | a disintegrin and metallopeptidase domain 34 like |
| 8 | gene | 43.650309 | 43.710049 | negative | MGI_C57BL6J_2181992 | Adam34 | NCBI_Gene:252866,ENSEMBL:ENSMUSG00000079058 | MGI:2181992 | protein coding gene | a disintegrin and metallopeptidase domain 34 |
| 8 | gene | 43.673658 | 43.699500 | negative | MGI_C57BL6J_3647274 | Gm5347 | NCBI_Gene:384814,ENSEMBL:ENSMUSG00000109723 | MGI:3647274 | protein coding gene | predicted gene 5347 |
| 8 | pseudogene | 43.699258 | 43.701498 | negative | MGI_C57BL6J_5594788 | Gm35629 | NCBI_Gene:102639281,ENSEMBL:ENSMUSG00000110155 | MGI:5594788 | pseudogene | predicted gene, 35629 |
| 8 | gene | 43.703479 | 43.754163 | positive | MGI_C57BL6J_5791324 | Gm45488 | ENSEMBL:ENSMUSG00000110168 | MGI:5791324 | lncRNA gene | predicted gene 45488 |
| 8 | pseudogene | 43.722269 | 43.724065 | negative | MGI_C57BL6J_3644337 | Gm6011 | NCBI_Gene:547011,ENSEMBL:ENSMUSG00000110224 | MGI:3644337 | pseudogene | predicted gene 6011 |
| 8 | pseudogene | 43.729219 | 43.729967 | negative | MGI_C57BL6J_3647931 | Gm6303 | NCBI_Gene:622263,ENSEMBL:ENSMUSG00000110036 | MGI:3647931 | pseudogene | predicted gene 6303 |
| 8 | pseudogene | 43.749052 | 43.750400 | negative | MGI_C57BL6J_5791493 | Gm45657 | ENSEMBL:ENSMUSG00000110347 | MGI:5791493 | pseudogene | predicted gene 45657 |
| 8 | pseudogene | 43.804844 | 43.805423 | positive | MGI_C57BL6J_5791494 | Gm45658 | ENSEMBL:ENSMUSG00000109826 | MGI:5791494 | pseudogene | predicted gene 45658 |
| 8 | gene | 43.908656 | 43.914604 | negative | MGI_C57BL6J_5791495 | Gm45659 | ENSEMBL:ENSMUSG00000109993 | MGI:5791495 | lncRNA gene | predicted gene 45659 |
| 8 | gene | 44.039250 | 44.039361 | positive | MGI_C57BL6J_5454368 | Gm24591 | ENSEMBL:ENSMUSG00000080348 | MGI:5454368 | miRNA gene | predicted gene, 24591 |
| 8 | pseudogene | 44.048106 | 44.048369 | negative | MGI_C57BL6J_5012040 | Gm19855 | NCBI_Gene:102639731,ENSEMBL:ENSMUSG00000109856 | MGI:5012040 | pseudogene | predicted gene, 19855 |
| 8 | gene | 44.227718 | 44.228545 | negative | MGI_C57BL6J_5611200 | Gm37972 | ENSEMBL:ENSMUSG00000102378 | MGI:5611200 | unclassified gene | predicted gene, 37972 |
| 8 | gene | 44.547974 | 44.548074 | negative | MGI_C57BL6J_5456018 | Gm26241 | ENSEMBL:ENSMUSG00000092685 | MGI:5456018 | miRNA gene | predicted gene, 26241 |
| 8 | gene | 44.601833 | 44.601939 | positive | MGI_C57BL6J_5455866 | Gm26089 | ENSEMBL:ENSMUSG00000088523 | MGI:5455866 | rRNA gene | predicted gene, 26089 |
| 8 | gene | 44.747479 | 44.766239 | negative | MGI_C57BL6J_5589556 | Gm30397 | NCBI_Gene:102632276 | MGI:5589556 | lncRNA gene | predicted gene, 30397 |
| 8 | gene | 44.934712 | 44.935159 | negative | MGI_C57BL6J_3708779 | Gm9908 | ENSEMBL:ENSMUSG00000110409 | MGI:3708779 | lncRNA gene | predicted gene 9908 |
| 8 | gene | 44.935046 | 45.052257 | positive | MGI_C57BL6J_109168 | Fat1 | NCBI_Gene:14107,ENSEMBL:ENSMUSG00000070047 | MGI:109168 | protein coding gene | FAT atypical cadherin 1 |
| 8 | gene | 45.017216 | 45.022385 | negative | MGI_C57BL6J_3780534 | Gm2366 | ENSEMBL:ENSMUSG00000097491 | MGI:3780534 | lncRNA gene | predicted gene 2366 |
| 8 | gene | 45.060702 | 45.065418 | negative | MGI_C57BL6J_3525069 | AY512931 | NCBI_Gene:434310,ENSEMBL:ENSMUSG00000066158 | MGI:3525069 | lncRNA gene | cDNA sequence AY512931 |
| 8 | gene | 45.069137 | 45.089233 | positive | MGI_C57BL6J_102967 | Mtnr1a | NCBI_Gene:17773,ENSEMBL:ENSMUSG00000054764 | MGI:102967 | protein coding gene | melatonin receptor 1A |
| 8 | gene | 45.110481 | 45.344439 | positive | MGI_C57BL6J_5589663 | Gm30504 | NCBI_Gene:102632429,ENSEMBL:ENSMUSG00000110260 | MGI:5589663 | lncRNA gene | predicted gene, 30504 |
| 8 | gene | 45.160494 | 45.165219 | negative | MGI_C57BL6J_3646898 | Gm6329 | NCBI_Gene:622523,ENSEMBL:ENSMUSG00000110231 | MGI:3646898 | lncRNA gene | predicted gene 6329 |
| 8 | gene | 45.240659 | 45.262050 | negative | MGI_C57BL6J_99481 | F11 | NCBI_Gene:109821,ENSEMBL:ENSMUSG00000031645 | MGI:99481 | protein coding gene | coagulation factor XI |
| 8 | gene | 45.266688 | 45.294859 | negative | MGI_C57BL6J_102849 | Klkb1 | NCBI_Gene:16621,ENSEMBL:ENSMUSG00000109764 | MGI:102849 | protein coding gene | kallikrein B, plasma 1 |
| 8 | gene | 45.269671 | 45.308466 | negative | MGI_C57BL6J_5804868 | Gm45753 | ENSEMBL:ENSMUSG00000031640 | MGI:5804868 | protein coding gene | predicted gene 45753 |
| 8 | gene | 45.304944 | 45.333216 | negative | MGI_C57BL6J_2142763 | Cyp4v3 | NCBI_Gene:102294,ENSEMBL:ENSMUSG00000079057 | MGI:2142763 | protein coding gene | cytochrome P450, family 4, subfamily v, polypeptide 3 |
| 8 | gene | 45.336715 | 45.382291 | negative | MGI_C57BL6J_2387177 | Fam149a | NCBI_Gene:212326,ENSEMBL:ENSMUSG00000070044 | MGI:2387177 | protein coding gene | family with sequence similarity 149, member A |
| 8 | gene | 45.395665 | 45.411949 | negative | MGI_C57BL6J_2156367 | Tlr3 | NCBI_Gene:142980,ENSEMBL:ENSMUSG00000031639 | MGI:2156367 | protein coding gene | toll-like receptor 3 |
| 8 | gene | 45.410063 | 45.410760 | positive | MGI_C57BL6J_5791378 | Gm45542 | ENSEMBL:ENSMUSG00000109785 | MGI:5791378 | lncRNA gene | predicted gene 45542 |
| 8 | gene | 45.507788 | 45.827906 | positive | MGI_C57BL6J_1924574 | Sorbs2 | NCBI_Gene:234214,ENSEMBL:ENSMUSG00000031626 | MGI:1924574 | protein coding gene | sorbin and SH3 domain containing 2 |
| 8 | gene | 45.526389 | 45.551328 | negative | MGI_C57BL6J_5589918 | Gm30759 | NCBI_Gene:102632772 | MGI:5589918 | lncRNA gene | predicted gene, 30759 |
| 8 | gene | 45.539594 | 45.540115 | negative | MGI_C57BL6J_5791294 | Gm45458 | ENSEMBL:ENSMUSG00000109665 | MGI:5791294 | unclassified gene | predicted gene 45458 |
| 8 | gene | 45.547377 | 45.605140 | negative | MGI_C57BL6J_3840153 | Gm16351 | NCBI_Gene:102632686,ENSEMBL:ENSMUSG00000084954 | MGI:3840153 | lncRNA gene | predicted gene 16351 |
| 8 | gene | 45.581615 | 45.581751 | negative | MGI_C57BL6J_5453381 | Gm23604 | ENSEMBL:ENSMUSG00000105754,ENSEMBL:ENSMUSG00000077499 | MGI:5453381 | snoRNA gene | predicted gene, 23604 |
| 8 | gene | 45.649946 | 45.665137 | negative | MGI_C57BL6J_5589794 | Gm30635 | NCBI_Gene:102632605 | MGI:5589794 | lncRNA gene | predicted gene, 30635 |
| 8 | gene | 45.723325 | 45.819297 | negative | MGI_C57BL6J_2443013 | Sorbs2os | NCBI_Gene:319940,ENSEMBL:ENSMUSG00000085440 | MGI:2443013 | antisense lncRNA gene | sorbin and SH3 domain containing 2, opposite strand |
| 8 | gene | 45.779228 | 45.782316 | positive | MGI_C57BL6J_5791293 | Gm45457 | ENSEMBL:ENSMUSG00000110411 | MGI:5791293 | unclassified gene | predicted gene 45457 |
| 8 | gene | 45.864478 | 45.864603 | negative | MGI_C57BL6J_5455086 | Gm25309 | ENSEMBL:ENSMUSG00000080378 | MGI:5455086 | snRNA gene | predicted gene, 25309 |
| 8 | gene | 45.885452 | 45.919548 | positive | MGI_C57BL6J_1859274 | Pdlim3 | NCBI_Gene:53318,ENSEMBL:ENSMUSG00000031636 | MGI:1859274 | protein coding gene | PDZ and LIM domain 3 |
| 8 | gene | 45.920872 | 45.922424 | positive | MGI_C57BL6J_5791348 | Gm45512 | ENSEMBL:ENSMUSG00000110071 | MGI:5791348 | unclassified gene | predicted gene 45512 |
| 8 | gene | 45.934619 | 45.945230 | positive | MGI_C57BL6J_2685018 | Ccdc110 | NCBI_Gene:212392,ENSEMBL:ENSMUSG00000071104 | MGI:2685018 | protein coding gene | coiled-coil domain containing 110 |
| 8 | gene | 45.939366 | 45.975456 | negative | MGI_C57BL6J_1916729 | 1700029J07Rik | NCBI_Gene:69479,ENSEMBL:ENSMUSG00000071103 | MGI:1916729 | protein coding gene | RIKEN cDNA 1700029J07 gene |
| 8 | pseudogene | 45.949612 | 45.949917 | positive | MGI_C57BL6J_3801745 | Gm15878 | ENSEMBL:ENSMUSG00000087542 | MGI:3801745 | pseudogene | predicted gene 15878 |
| 8 | gene | 45.975528 | 45.996958 | positive | MGI_C57BL6J_1913679 | Ufsp2 | NCBI_Gene:192169,ENSEMBL:ENSMUSG00000031634 | MGI:1913679 | protein coding gene | UFM1-specific peptidase 2 |
| 8 | gene | 45.996902 | 46.005707 | negative | MGI_C57BL6J_3603344 | Ankrd37 | NCBI_Gene:654824,ENSEMBL:ENSMUSG00000050914 | MGI:3603344 | protein coding gene | ankyrin repeat domain 37 |
| 8 | gene | 45.999303 | 46.029478 | positive | MGI_C57BL6J_1914870 | Lrp2bp | NCBI_Gene:67620,ENSEMBL:ENSMUSG00000031637 | MGI:1914870 | protein coding gene | Lrp2 binding protein |
| 8 | gene | 46.033261 | 46.152206 | negative | MGI_C57BL6J_2142610 | Snx25 | NCBI_Gene:102141,ENSEMBL:ENSMUSG00000038291 | MGI:2142610 | protein coding gene | sorting nexin 25 |
| 8 | gene | 46.107259 | 46.107531 | positive | MGI_C57BL6J_5452807 | Gm23030 | ENSEMBL:ENSMUSG00000088629 | MGI:5452807 | unclassified non-coding RNA gene | predicted gene, 23030 |
| 8 | gene | 46.151771 | 46.195590 | positive | MGI_C57BL6J_1914006 | Cfap97 | NCBI_Gene:66756,ENSEMBL:ENSMUSG00000031631 | MGI:1914006 | protein coding gene | cilia and flagella associated protein 97 |
| 8 | gene | 46.152610 | 46.156100 | positive | MGI_C57BL6J_3783078 | Gm15634 | ENSEMBL:ENSMUSG00000086279 | MGI:3783078 | lncRNA gene | predicted gene 15634 |
| 8 | gene | 46.162090 | 46.163246 | positive | MGI_C57BL6J_5804808 | Gm45693 | ENSEMBL:ENSMUSG00000109902 | MGI:5804808 | unclassified gene | predicted gene 45693 |
| 8 | gene | 46.206797 | 46.211284 | negative | MGI_C57BL6J_1353495 | Slc25a4 | NCBI_Gene:11739,ENSEMBL:ENSMUSG00000031633 | MGI:1353495 | protein coding gene | solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4 |
| 8 | pseudogene | 46.228994 | 46.230128 | positive | MGI_C57BL6J_87907 | Actg-ps1 | NCBI_Gene:621269,ENSEMBL:ENSMUSG00000074978 | MGI:87907 | pseudogene | actin, gamma, pseudogene 1 |
| 8 | gene | 46.229794 | 46.229877 | positive | MGI_C57BL6J_5453589 | Gm23812 | ENSEMBL:ENSMUSG00000092897 | MGI:5453589 | miRNA gene | predicted gene, 23812 |
| 8 | pseudogene | 46.254486 | 46.255696 | negative | MGI_C57BL6J_3642852 | Gm10313 | NCBI_Gene:100039875,ENSEMBL:ENSMUSG00000071102 | MGI:3642852 | pseudogene | predicted pseudogene 10313 |
| 8 | gene | 46.272051 | 46.272147 | positive | MGI_C57BL6J_5454446 | Gm24669 | ENSEMBL:ENSMUSG00000089561 | MGI:5454446 | snRNA gene | predicted gene, 24669 |
| 8 | pseudogene | 46.279608 | 46.280195 | positive | MGI_C57BL6J_3643885 | Gm8575 | NCBI_Gene:667325,ENSEMBL:ENSMUSG00000109692 | MGI:3643885 | pseudogene | predicted gene 8575 |
| 8 | gene | 46.281945 | 46.284669 | negative | MGI_C57BL6J_5590039 | Gm30880 | NCBI_Gene:102632930 | MGI:5590039 | lncRNA gene | predicted gene, 30880 |
| 8 | gene | 46.292039 | 46.294671 | negative | MGI_C57BL6J_3040955 | Helt | NCBI_Gene:234219,ENSEMBL:ENSMUSG00000047171 | MGI:3040955 | protein coding gene | helt bHLH transcription factor |
| 8 | gene | 46.299131 | 46.325729 | positive | MGI_C57BL6J_5622053 | Gm39168 | NCBI_Gene:105243177,ENSEMBL:ENSMUSG00000110179 | MGI:5622053 | lncRNA gene | predicted gene, 39168 |
| 8 | gene | 46.313801 | 46.314611 | positive | MGI_C57BL6J_5791091 | Gm45255 | ENSEMBL:ENSMUSG00000109743 | MGI:5791091 | unclassified gene | predicted gene 45255 |
| 8 | gene | 46.363251 | 46.367212 | positive | MGI_C57BL6J_5590090 | Gm30931 | NCBI_Gene:102632998,ENSEMBL:ENSMUSG00000110035 | MGI:5590090 | lncRNA gene | predicted gene, 30931 |
| 8 | gene | 46.372106 | 46.385888 | negative | MGI_C57BL6J_5791089 | Gm45253 | ENSEMBL:ENSMUSG00000109612 | MGI:5791089 | lncRNA gene | predicted gene 45253 |
| 8 | pseudogene | 46.451929 | 46.452304 | negative | MGI_C57BL6J_5791081 | Gm45245 | ENSEMBL:ENSMUSG00000109912 | MGI:5791081 | pseudogene | predicted gene 45245 |
| 8 | gene | 46.469991 | 46.470951 | negative | MGI_C57BL6J_5590209 | Gm31050 | NCBI_Gene:102633148 | MGI:5590209 | lncRNA gene | predicted gene, 31050 |
| 8 | gene | 46.471037 | 46.536051 | positive | MGI_C57BL6J_102797 | Acsl1 | NCBI_Gene:14081,ENSEMBL:ENSMUSG00000018796 | MGI:102797 | protein coding gene | acyl-CoA synthetase long-chain family member 1 |
| 8 | gene | 46.471391 | 46.473590 | negative | MGI_C57BL6J_5791080 | Gm45244 | ENSEMBL:ENSMUSG00000109807 | MGI:5791080 | unclassified gene | predicted gene 45244 |
| 8 | gene | 46.552019 | 46.581144 | positive | MGI_C57BL6J_1919126 | Cenpu | NCBI_Gene:71876,ENSEMBL:ENSMUSG00000031629 | MGI:1919126 | protein coding gene | centromere protein U |
| 8 | gene | 46.575579 | 46.617234 | negative | MGI_C57BL6J_3603756 | Primpol | NCBI_Gene:408022,ENSEMBL:ENSMUSG00000038225 | MGI:3603756 | protein coding gene | primase and polymerase (DNA-directed) |
| 8 | gene | 46.604749 | 46.605963 | positive | MGI_C57BL6J_5791443 | Gm45607 | ENSEMBL:ENSMUSG00000110270 | MGI:5791443 | lncRNA gene | predicted gene 45607 |
| 8 | gene | 46.617291 | 46.639698 | positive | MGI_C57BL6J_107739 | Casp3 | NCBI_Gene:12367,ENSEMBL:ENSMUSG00000031628 | MGI:107739 | protein coding gene | caspase 3 |
| 8 | gene | 46.668808 | 46.739515 | negative | MGI_C57BL6J_4439599 | Gm16675 | NCBI_Gene:100503498,ENSEMBL:ENSMUSG00000097534 | MGI:4439599 | lncRNA gene | predicted gene, 16675 |
| 8 | gene | 46.668942 | 46.678998 | negative | MGI_C57BL6J_5590270 | Gm31111 | NCBI_Gene:102633233 | MGI:5590270 | lncRNA gene | predicted gene, 31111 |
| 8 | gene | 46.739732 | 46.847458 | positive | MGI_C57BL6J_96591 | Irf2 | NCBI_Gene:16363,ENSEMBL:ENSMUSG00000031627 | MGI:96591 | protein coding gene | interferon regulatory factor 2 |
| 8 | gene | 46.763251 | 46.764973 | negative | MGI_C57BL6J_5791315 | Gm45479 | ENSEMBL:ENSMUSG00000109751 | MGI:5791315 | lncRNA gene | predicted gene 45479 |
| 8 | gene | 46.770547 | 46.773358 | negative | MGI_C57BL6J_5590331 | Gm31172 | NCBI_Gene:102633317,ENSEMBL:ENSMUSG00000110374 | MGI:5590331 | lncRNA gene | predicted gene, 31172 |
| 8 | gene | 46.826701 | 46.829108 | negative | MGI_C57BL6J_5622055 | Gm39170 | NCBI_Gene:105243179 | MGI:5622055 | lncRNA gene | predicted gene, 39170 |
| 8 | gene | 46.833036 | 46.833912 | negative | MGI_C57BL6J_5791317 | Gm45481 | ENSEMBL:ENSMUSG00000110414 | MGI:5791317 | unclassified gene | predicted gene 45481 |
| 8 | gene | 46.939197 | 47.096762 | positive | MGI_C57BL6J_2445171 | Enpp6 | NCBI_Gene:320981,ENSEMBL:ENSMUSG00000038173 | MGI:2445171 | protein coding gene | ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| 8 | gene | 46.946795 | 46.948799 | negative | MGI_C57BL6J_1916535 | 1700011L03Rik | ENSEMBL:ENSMUSG00000109759 | MGI:1916535 | lncRNA gene | RIKEN cDNA 1700011L03 gene |
| 8 | gene | 46.994795 | 47.014177 | negative | MGI_C57BL6J_5791436 | Gm45600 | ENSEMBL:ENSMUSG00000109888 | MGI:5791436 | lncRNA gene | predicted gene 45600 |
| 8 | gene | 47.026805 | 47.038177 | negative | MGI_C57BL6J_1923153 | 4930579M01Rik | ENSEMBL:ENSMUSG00000086190 | MGI:1923153 | lncRNA gene | RIKEN cDNA 4930579M01 gene |
| 8 | gene | 47.075018 | 47.082406 | negative | MGI_C57BL6J_5590390 | Gm31231 | NCBI_Gene:102633396 | MGI:5590390 | lncRNA gene | predicted gene, 31231 |
| 8 | gene | 47.135651 | 47.163704 | positive | MGI_C57BL6J_3041231 | E330018M18Rik | ENSEMBL:ENSMUSG00000110264 | MGI:3041231 | lncRNA gene | RIKEN cDNA E330018M18 gene |
| 8 | gene | 47.180048 | 47.446362 | negative | MGI_C57BL6J_1918319 | Stox2 | NCBI_Gene:71069,ENSEMBL:ENSMUSG00000038143 | MGI:1918319 | protein coding gene | storkhead box 2 |
| 8 | gene | 47.224459 | 47.227166 | negative | MGI_C57BL6J_5791373 | Gm45537 | ENSEMBL:ENSMUSG00000109606 | MGI:5791373 | unclassified gene | predicted gene 45537 |
| 8 | gene | 47.294272 | 47.446362 | negative | MGI_C57BL6J_1926155 | 4930448N21Rik | NA | NA | unclassified gene | RIKEN cDNA 4930448N21 gene |
| 8 | gene | 47.323514 | 47.324515 | positive | MGI_C57BL6J_5622056 | Gm39171 | NCBI_Gene:105243181 | MGI:5622056 | lncRNA gene | predicted gene, 39171 |
| 8 | gene | 47.412408 | 47.414164 | positive | MGI_C57BL6J_3642418 | Gm10083 | NCBI_Gene:102639847 | MGI:3642418 | protein coding gene | predicted gene 10083 |
| 8 | gene | 47.440418 | 47.440551 | positive | MGI_C57BL6J_5452634 | Gm22857 | ENSEMBL:ENSMUSG00000092710 | MGI:5452634 | snoRNA gene | predicted gene, 22857 |
| 8 | gene | 47.490115 | 47.533470 | negative | MGI_C57BL6J_2444585 | Trappc11 | NCBI_Gene:320714,ENSEMBL:ENSMUSG00000038102 | MGI:2444585 | protein coding gene | trafficking protein particle complex 11 |
| 8 | gene | 47.533645 | 47.552955 | positive | MGI_C57BL6J_2681000 | Rwdd4a | NCBI_Gene:192174,ENSEMBL:ENSMUSG00000031568 | MGI:2681000 | protein coding gene | RWD domain containing 4A |
| 8 | gene | 47.557529 | 47.567896 | positive | MGI_C57BL6J_5791412 | Gm45576 | ENSEMBL:ENSMUSG00000110363 | MGI:5791412 | lncRNA gene | predicted gene 45576 |
| 8 | gene | 47.591668 | 47.591747 | positive | MGI_C57BL6J_5530791 | Gm27409 | ENSEMBL:ENSMUSG00000098402 | MGI:5530791 | miRNA gene | predicted gene, 27409 |
| 8 | gene | 47.594821 | 47.595550 | positive | MGI_C57BL6J_5622057 | Gm39172 | NCBI_Gene:105243182 | MGI:5622057 | lncRNA gene | predicted gene, 39172 |
| 8 | pseudogene | 47.627553 | 47.628100 | negative | MGI_C57BL6J_5791411 | Gm45575 | ENSEMBL:ENSMUSG00000109960 | MGI:5791411 | pseudogene | predicted gene 45575 |
| 8 | pseudogene | 47.632276 | 47.632779 | negative | MGI_C57BL6J_3644689 | Gm8623 | NCBI_Gene:667422,ENSEMBL:ENSMUSG00000062458 | MGI:3644689 | pseudogene | predicted gene 8623 |
| 8 | gene | 47.667178 | 47.675556 | negative | MGI_C57BL6J_1916510 | Ing2 | NCBI_Gene:69260,ENSEMBL:ENSMUSG00000063049 | MGI:1916510 | protein coding gene | inhibitor of growth family, member 2 |
| 8 | gene | 47.675391 | 47.685239 | positive | MGI_C57BL6J_3034595 | AA386476 | ENSEMBL:ENSMUSG00000074357 | MGI:3034595 | lncRNA gene | expressed sequence AA386476 |
| 8 | gene | 47.681281 | 47.695396 | negative | MGI_C57BL6J_5590851 | Gm31692 | NCBI_Gene:102634002 | MGI:5590851 | lncRNA gene | predicted gene, 31692 |
| 8 | gene | 47.685665 | 47.700416 | positive | MGI_C57BL6J_5590797 | Gm31638 | NCBI_Gene:102633936 | MGI:5590797 | lncRNA gene | predicted gene, 31638 |
| 8 | gene | 47.709344 | 47.714854 | negative | MGI_C57BL6J_1918175 | Cdkn2aip | NCBI_Gene:70925,ENSEMBL:ENSMUSG00000038069 | MGI:1918175 | protein coding gene | CDKN2A interacting protein |
| 8 | gene | 47.713266 | 47.715021 | positive | MGI_C57BL6J_3704310 | E030037K01Rik | ENSEMBL:ENSMUSG00000097706 | MGI:3704310 | lncRNA gene | RIKEN cDNA E030037K01 gene |
| 8 | gene | 47.719246 | 47.719369 | positive | MGI_C57BL6J_4421944 | n-R5s96 | ENSEMBL:ENSMUSG00000064508 | MGI:4421944 | rRNA gene | nuclear encoded rRNA 5S 96 |
| 8 | gene | 47.743977 | 47.750352 | negative | MGI_C57BL6J_5590945 | Gm31786 | NCBI_Gene:102634125,ENSEMBL:ENSMUSG00000110665 | MGI:5590945 | lncRNA gene | predicted gene, 31786 |
| 8 | gene | 47.764054 | 47.766125 | negative | MGI_C57BL6J_5804939 | Gm45824 | ENSEMBL:ENSMUSG00000110604 | MGI:5804939 | lncRNA gene | predicted gene 45824 |
| 8 | gene | 47.773555 | 47.777802 | positive | MGI_C57BL6J_5804979 | Gm45864 | ENSEMBL:ENSMUSG00000110539 | MGI:5804979 | lncRNA gene | predicted gene 45864 |
| 8 | gene | 47.811723 | 47.818294 | negative | MGI_C57BL6J_1920682 | 1700061N14Rik | NCBI_Gene:105243184,ENSEMBL:ENSMUSG00000110464 | MGI:1920682 | lncRNA gene | RIKEN cDNA 1700061N14 gene |
| 8 | gene | 47.822143 | 47.822805 | positive | MGI_C57BL6J_3712484 | Cldn24 | NCBI_Gene:100039801,ENSEMBL:ENSMUSG00000061974 | MGI:3712484 | protein coding gene | claudin 24 |
| 8 | gene | 47.823959 | 47.990924 | negative | MGI_C57BL6J_1261872 | Wwc2 | NCBI_Gene:52357,ENSEMBL:ENSMUSG00000031563 | MGI:1261872 | protein coding gene | WW, C2 and coiled-coil domain containing 2 |
| 8 | gene | 47.824482 | 47.825476 | positive | MGI_C57BL6J_1922927 | Cldn22 | NCBI_Gene:75677,ENSEMBL:ENSMUSG00000038064 | MGI:1922927 | protein coding gene | claudin 22 |
| 8 | gene | 47.899182 | 47.899294 | positive | MGI_C57BL6J_4421945 | n-R5s97 | ENSEMBL:ENSMUSG00000064519 | MGI:4421945 | rRNA gene | nuclear encoded rRNA 5S 97 |
| 8 | gene | 47.943096 | 47.952674 | negative | MGI_C57BL6J_5622058 | Gm39173 | NCBI_Gene:105243183 | MGI:5622058 | lncRNA gene | predicted gene, 39173 |
| 8 | gene | 47.971635 | 47.975581 | negative | MGI_C57BL6J_5591143 | Gm31984 | NCBI_Gene:102634392 | MGI:5591143 | lncRNA gene | predicted gene, 31984 |
| 8 | gene | 48.002210 | 48.009157 | negative | MGI_C57BL6J_5591211 | Gm32052 | NCBI_Gene:102634476,ENSEMBL:ENSMUSG00000110624 | MGI:5591211 | lncRNA gene | predicted gene, 32052 |
| 8 | pseudogene | 48.070002 | 48.070628 | negative | MGI_C57BL6J_3780775 | Gm2607 | ENSEMBL:ENSMUSG00000110544 | MGI:3780775 | pseudogene | predicted gene 2607 |
| 8 | gene | 48.099092 | 48.153233 | positive | MGI_C57BL6J_2444529 | Dctd | NCBI_Gene:320685,ENSEMBL:ENSMUSG00000031562 | MGI:2444529 | protein coding gene | dCMP deaminase |
| 8 | gene | 48.144215 | 48.154388 | positive | MGI_C57BL6J_5622059 | Gm39174 | NCBI_Gene:105243185 | MGI:5622059 | lncRNA gene | predicted gene, 39174 |
| 8 | gene | 48.206587 | 48.218650 | positive | MGI_C57BL6J_5591353 | Gm32194 | NCBI_Gene:102634662 | MGI:5591353 | lncRNA gene | predicted gene, 32194 |
| 8 | gene | 48.220031 | 48.225571 | positive | MGI_C57BL6J_5592082 | Gm32923 | NCBI_Gene:102635635 | MGI:5592082 | lncRNA gene | predicted gene, 32923 |
| 8 | gene | 48.225665 | 48.935162 | negative | MGI_C57BL6J_1345183 | Tenm3 | NCBI_Gene:23965,ENSEMBL:ENSMUSG00000031561 | MGI:1345183 | protein coding gene | teneurin transmembrane protein 3 |
| 8 | pseudogene | 48.270248 | 48.270985 | negative | MGI_C57BL6J_5592001 | Gm32842 | ENSEMBL:ENSMUSG00000110480 | MGI:5592001 | pseudogene | predicted gene, 32842 |
| 8 | gene | 48.378507 | 48.379685 | negative | MGI_C57BL6J_1924529 | 9430019C24Rik | NA | NA | unclassified gene | RIKEN cDNA 9430019C24 gene |
| 8 | gene | 48.485270 | 48.488748 | negative | MGI_C57BL6J_2443853 | D930044I17Rik | NA | NA | unclassified gene | RIKEN cDNA D930044I17 gene |
| 8 | pseudogene | 48.630930 | 48.631169 | negative | MGI_C57BL6J_5804887 | Gm45772 | ENSEMBL:ENSMUSG00000110509 | MGI:5804887 | pseudogene | predicted gene 45772 |
| 8 | gene | 48.660798 | 48.671892 | positive | MGI_C57BL6J_5622063 | Gm39178 | NCBI_Gene:105243189 | MGI:5622063 | lncRNA gene | predicted gene, 39178 |
| 8 | gene | 48.841995 | 48.843142 | negative | MGI_C57BL6J_1920270 | 2900073C17Rik | NA | NA | unclassified gene | RIKEN cDNA 2900073C17 gene |
| 8 | gene | 48.843665 | 48.870497 | positive | MGI_C57BL6J_5011929 | Gm19744 | NCBI_Gene:100503519,ENSEMBL:ENSMUSG00000100706 | MGI:5011929 | lncRNA gene | predicted gene, 19744 |
| 8 | gene | 48.958350 | 48.958481 | negative | MGI_C57BL6J_5455607 | Gm25830 | ENSEMBL:ENSMUSG00000089548 | MGI:5455607 | snoRNA gene | predicted gene, 25830 |
| 8 | gene | 49.044280 | 49.046017 | positive | MGI_C57BL6J_5804945 | Gm45830 | ENSEMBL:ENSMUSG00000110503 | MGI:5804945 | lncRNA gene | predicted gene 45830 |
| 8 | gene | 49.063879 | 49.071859 | positive | MGI_C57BL6J_5804944 | Gm45829 | ENSEMBL:ENSMUSG00000110606 | MGI:5804944 | lncRNA gene | predicted gene 45829 |
| 8 | gene | 49.184533 | 49.186666 | positive | MGI_C57BL6J_96220 | Hpvc1 | NA | NA | protein coding gene | human papillomavirus 18 E5 central sequence motif gene 1 |
| 8 | gene | 49.230527 | 49.231969 | negative | MGI_C57BL6J_5804947 | Gm45832 | ENSEMBL:ENSMUSG00000110465 | MGI:5804947 | unclassified gene | predicted gene 45832 |
| 8 | gene | 49.369290 | 49.549013 | positive | MGI_C57BL6J_1922609 | 4930555F03Rik | NCBI_Gene:75359,ENSEMBL:ENSMUSG00000031559 | MGI:1922609 | lncRNA gene | RIKEN cDNA 4930555F03 gene |
| 8 | gene | 49.462453 | 49.464739 | negative | MGI_C57BL6J_5791413 | Gm45577 | ENSEMBL:ENSMUSG00000110137 | MGI:5791413 | unclassified gene | predicted gene 45577 |
| 8 | gene | 49.485770 | 49.488645 | negative | MGI_C57BL6J_5804812 | Gm45697 | ENSEMBL:ENSMUSG00000110564 | MGI:5804812 | unclassified gene | predicted gene 45697 |
| 8 | gene | 49.521424 | 49.522408 | positive | MGI_C57BL6J_5804811 | Gm45696 | ENSEMBL:ENSMUSG00000110688 | MGI:5804811 | unclassified gene | predicted gene 45696 |
| 8 | gene | 49.738480 | 49.739896 | negative | MGI_C57BL6J_1924135 | 6430514K02Rik | NA | NA | unclassified gene | RIKEN cDNA 6430514K02 gene |
| 8 | gene | 49.818937 | 49.949006 | positive | MGI_C57BL6J_5791157 | Gm45321 | ENSEMBL:ENSMUSG00000109907 | MGI:5791157 | lncRNA gene | predicted gene 45321 |
| 8 | gene | 49.838068 | 49.839428 | positive | MGI_C57BL6J_5791158 | Gm45322 | ENSEMBL:ENSMUSG00000110051 | MGI:5791158 | lncRNA gene | predicted gene 45322 |
| 8 | gene | 49.959483 | 50.013384 | negative | MGI_C57BL6J_5791159 | Gm45323 | ENSEMBL:ENSMUSG00000110375 | MGI:5791159 | lncRNA gene | predicted gene 45323 |
| 8 | gene | 49.989907 | 49.995544 | positive | MGI_C57BL6J_5791160 | Gm45324 | ENSEMBL:ENSMUSG00000110147 | MGI:5791160 | unclassified gene | predicted gene 45324 |
| 8 | gene | 50.117745 | 50.517889 | negative | MGI_C57BL6J_5791177 | Gm45341 | ENSEMBL:ENSMUSG00000109838 | MGI:5791177 | lncRNA gene | predicted gene 45341 |
| 8 | gene | 50.201613 | 50.517680 | negative | MGI_C57BL6J_5791291 | Gm45455 | ENSEMBL:ENSMUSG00000109741 | MGI:5791291 | lncRNA gene | predicted gene 45455 |
| 8 | gene | 50.368112 | 50.416089 | positive | MGI_C57BL6J_3780683 | Gm2516 | NCBI_Gene:100039954,ENSEMBL:ENSMUSG00000110281 | MGI:3780683 | lncRNA gene | predicted gene 2516 |
| 8 | gene | 50.389092 | 50.390097 | positive | MGI_C57BL6J_5791095 | Gm45259 | ENSEMBL:ENSMUSG00000110050 | MGI:5791095 | unclassified gene | predicted gene 45259 |
| 8 | gene | 50.563241 | 50.563303 | positive | MGI_C57BL6J_5453763 | Gm23986 | ENSEMBL:ENSMUSG00000087769 | MGI:5453763 | snRNA gene | predicted gene, 23986 |
| 8 | gene | 50.743291 | 50.916997 | negative | MGI_C57BL6J_2442759 | C130073E24Rik | ENSEMBL:ENSMUSG00000110246 | MGI:2442759 | lncRNA gene | RIKEN cDNA C130073E24 gene |
| 8 | gene | 50.764515 | 50.918746 | negative | MGI_C57BL6J_1922799 | 1700019L22Rik | NCBI_Gene:75549 | MGI:1922799 | lncRNA gene | RIKEN cDNA 1700019L22 gene |
| 8 | pseudogene | 51.137630 | 51.137847 | positive | MGI_C57BL6J_5791222 | Gm45386 | ENSEMBL:ENSMUSG00000109840 | MGI:5791222 | pseudogene | predicted gene 45386 |
| 8 | gene | 51.177683 | 51.180608 | positive | MGI_C57BL6J_5592134 | Gm32975 | ENSEMBL:ENSMUSG00000109944 | MGI:5592134 | lncRNA gene | predicted gene, 32975 |
| 8 | pseudogene | 51.275890 | 51.276925 | positive | MGI_C57BL6J_3646571 | Gm8679 | NCBI_Gene:667517,ENSEMBL:ENSMUSG00000109691 | MGI:3646571 | pseudogene | predicted gene 8679 |
| 8 | pseudogene | 51.453395 | 51.454271 | positive | MGI_C57BL6J_5010096 | Gm17911 | NCBI_Gene:100416081,ENSEMBL:ENSMUSG00000110085 | MGI:5010096 | pseudogene | predicted gene, 17911 |
| 8 | pseudogene | 51.794303 | 51.794893 | negative | MGI_C57BL6J_5010178 | Gm17993 | NCBI_Gene:100416238 | MGI:5010178 | pseudogene | predicted gene, 17993 |
| 8 | pseudogene | 51.878446 | 51.880771 | positive | MGI_C57BL6J_3645274 | Gm6458 | NCBI_Gene:623872,ENSEMBL:ENSMUSG00000109897 | MGI:3645274 | pseudogene | predicted gene 6458 |
| 8 | gene | 52.024987 | 52.031271 | negative | MGI_C57BL6J_5791225 | Gm45389 | ENSEMBL:ENSMUSG00000110163 | MGI:5791225 | lncRNA gene | predicted gene 45389 |
| 8 | gene | 52.041424 | 52.041568 | negative | MGI_C57BL6J_5690779 | Gm44387 | ENSEMBL:ENSMUSG00000106349 | MGI:5690779 | miRNA gene | predicted gene, 44387 |
| 8 | gene | 52.078604 | 52.078742 | positive | MGI_C57BL6J_5454706 | Gm24929 | ENSEMBL:ENSMUSG00000065337 | MGI:5454706 | snRNA gene | predicted gene, 24929 |
| 8 | gene | 52.080163 | 52.099750 | positive | MGI_C57BL6J_5791224 | Gm45388 | ENSEMBL:ENSMUSG00000109823 | MGI:5791224 | lncRNA gene | predicted gene 45388 |
| 8 | pseudogene | 52.195768 | 52.197187 | negative | MGI_C57BL6J_3701610 | Gm9892 | NCBI_Gene:100040220,ENSEMBL:ENSMUSG00000052825 | MGI:3701610 | pseudogene | predicted gene 9892 |
| 8 | pseudogene | 52.278313 | 52.279487 | negative | MGI_C57BL6J_3644114 | Gm6463 | NCBI_Gene:623935,ENSEMBL:ENSMUSG00000098231 | MGI:3644114 | pseudogene | predicted gene 6463 |
| 8 | gene | 52.336915 | 52.337105 | negative | MGI_C57BL6J_5455590 | Gm25813 | ENSEMBL:ENSMUSG00000064682 | MGI:5455590 | snRNA gene | predicted gene, 25813 |
| 8 | gene | 52.356611 | 52.403339 | positive | MGI_C57BL6J_3036227 | 1190028D05Rik | NCBI_Gene:330767,ENSEMBL:ENSMUSG00000110013 | MGI:3036227 | lncRNA gene | RIKEN cDNA 1190028D05 gene |
| 8 | pseudogene | 52.554357 | 52.554714 | negative | MGI_C57BL6J_5791509 | Gm45673 | ENSEMBL:ENSMUSG00000109919 | MGI:5791509 | pseudogene | predicted gene 45673 |
| 8 | pseudogene | 52.745403 | 52.774710 | positive | MGI_C57BL6J_3780093 | Gm9685 | NCBI_Gene:676688,ENSEMBL:ENSMUSG00000109983 | MGI:3780093 | pseudogene | predicted gene 9685 |
| 8 | gene | 53.006408 | 53.006523 | positive | MGI_C57BL6J_5453443 | Gm23666 | ENSEMBL:ENSMUSG00000088678 | MGI:5453443 | snoRNA gene | predicted gene, 23666 |
| 8 | gene | 53.436100 | 53.480650 | positive | MGI_C57BL6J_5791390 | Gm45554 | ENSEMBL:ENSMUSG00000110362 | MGI:5791390 | lncRNA gene | predicted gene 45554 |
| 8 | gene | 53.436263 | 53.446491 | positive | MGI_C57BL6J_5622064 | Gm39179 | NCBI_Gene:105243190 | MGI:5622064 | lncRNA gene | predicted gene, 39179 |
| 8 | gene | 53.478190 | 53.480388 | positive | MGI_C57BL6J_5622065 | Gm39180 | NCBI_Gene:105243191 | MGI:5622065 | lncRNA gene | predicted gene, 39180 |
| 8 | gene | 53.493763 | 53.493824 | positive | MGI_C57BL6J_5531346 | Gm27964 | ENSEMBL:ENSMUSG00000098878 | MGI:5531346 | snRNA gene | predicted gene, 27964 |
| 8 | gene | 53.493853 | 53.493914 | positive | MGI_C57BL6J_5452618 | Gm22841 | ENSEMBL:ENSMUSG00000089068 | MGI:5452618 | snRNA gene | predicted gene, 22841 |
| 8 | gene | 53.511686 | 53.523422 | positive | MGI_C57BL6J_104873 | Aga | NCBI_Gene:11593,ENSEMBL:ENSMUSG00000031521 | MGI:104873 | protein coding gene | aspartylglucosaminidase |
| 8 | gene | 53.565595 | 53.583539 | negative | MGI_C57BL6J_5622066 | Gm39181 | NCBI_Gene:105243192 | MGI:5622066 | lncRNA gene | predicted gene, 39181 |
| 8 | gene | 53.586867 | 53.639065 | negative | MGI_C57BL6J_2384588 | Neil3 | NCBI_Gene:234258,ENSEMBL:ENSMUSG00000039396 | MGI:2384588 | protein coding gene | nei like 3 (E. coli) |
| 8 | pseudogene | 53.911739 | 53.912656 | positive | MGI_C57BL6J_5012106 | Gm19921 | NCBI_Gene:100503841,ENSEMBL:ENSMUSG00000110334 | MGI:5012106 | pseudogene | predicted gene, 19921 |
| 8 | gene | 54.076070 | 54.077773 | negative | MGI_C57BL6J_5592312 | Gm33153 | NCBI_Gene:102635944 | MGI:5592312 | protein coding gene | predicted gene, 33153 |
| 8 | gene | 54.077532 | 54.187096 | positive | MGI_C57BL6J_109124 | Vegfc | NCBI_Gene:22341,ENSEMBL:ENSMUSG00000031520 | MGI:109124 | protein coding gene | vascular endothelial growth factor C |
| 8 | gene | 54.513644 | 54.514150 | negative | MGI_C57BL6J_5791389 | Gm45553 | ENSEMBL:ENSMUSG00000110403 | MGI:5791389 | unclassified gene | predicted gene 45553 |
| 8 | gene | 54.520433 | 54.530751 | negative | MGI_C57BL6J_1923937 | Spcs3 | NCBI_Gene:76687,ENSEMBL:ENSMUSG00000054408 | MGI:1923937 | protein coding gene | signal peptidase complex subunit 3 homolog (S. cerevisiae) |
| 8 | gene | 54.529580 | 54.587842 | positive | MGI_C57BL6J_1923544 | Asb5 | NCBI_Gene:76294,ENSEMBL:ENSMUSG00000031519 | MGI:1923544 | protein coding gene | ankyrin repeat and SOCs box-containing 5 |
| 8 | gene | 54.568550 | 54.600987 | negative | MGI_C57BL6J_1924353 | 5330439A09Rik | NCBI_Gene:77103,ENSEMBL:ENSMUSG00000089708 | MGI:1924353 | lncRNA gene | RIKEN cDNA 5330439A09 gene |
| 8 | gene | 54.596123 | 54.599395 | positive | MGI_C57BL6J_5825676 | Gm46039 | NCBI_Gene:108167531 | MGI:5825676 | lncRNA gene | predicted gene, 46039 |
| 8 | gene | 54.600781 | 54.610098 | positive | MGI_C57BL6J_1916531 | Spata4 | NCBI_Gene:69281,ENSEMBL:ENSMUSG00000031518 | MGI:1916531 | protein coding gene | spermatogenesis associated 4 |
| 8 | gene | 54.629055 | 54.887184 | negative | MGI_C57BL6J_1924662 | Wdr17 | NCBI_Gene:244484,ENSEMBL:ENSMUSG00000039375 | MGI:1924662 | protein coding gene | WD repeat domain 17 |
| 8 | gene | 54.708987 | 54.710381 | positive | MGI_C57BL6J_3783201 | Gm15758 | ENSEMBL:ENSMUSG00000089719 | MGI:3783201 | lncRNA gene | predicted gene 15758 |
| 8 | gene | 54.779434 | 55.060878 | positive | MGI_C57BL6J_107671 | Gpm6a | NCBI_Gene:234267,ENSEMBL:ENSMUSG00000031517 | MGI:107671 | protein coding gene | glycoprotein m6a |
| 8 | gene | 54.866261 | 54.866379 | positive | MGI_C57BL6J_4421946 | n-R5s98 | ENSEMBL:ENSMUSG00000084498 | MGI:4421946 | rRNA gene | nuclear encoded rRNA 5S 98 |
| 8 | gene | 54.959819 | 54.961193 | positive | MGI_C57BL6J_5791099 | Gm45263 | ENSEMBL:ENSMUSG00000110342 | MGI:5791099 | unclassified gene | predicted gene 45263 |
| 8 | gene | 55.005285 | 55.007781 | positive | MGI_C57BL6J_5791101 | Gm45265 | ENSEMBL:ENSMUSG00000109725 | MGI:5791101 | unclassified gene | predicted gene 45265 |
| 8 | gene | 55.028401 | 55.032395 | positive | MGI_C57BL6J_5791100 | Gm45264 | ENSEMBL:ENSMUSG00000109927 | MGI:5791100 | unclassified gene | predicted gene 45264 |
| 8 | gene | 55.043059 | 55.044521 | positive | MGI_C57BL6J_5791098 | Gm45262 | ENSEMBL:ENSMUSG00000109942 | MGI:5791098 | unclassified gene | predicted gene 45262 |
| 8 | gene | 55.055874 | 55.057729 | positive | MGI_C57BL6J_3045287 | A230085B16Rik | ENSEMBL:ENSMUSG00000110047 | MGI:3045287 | unclassified non-coding RNA gene | RIKEN cDNA A230085B16 gene |
| 8 | gene | 55.073249 | 55.077134 | positive | MGI_C57BL6J_2443625 | 9330121K16Rik | ENSEMBL:ENSMUSG00000110249 | MGI:2443625 | unclassified gene | RIKEN cDNA 9330121K16 gene |
| 8 | pseudogene | 55.079907 | 55.082008 | negative | MGI_C57BL6J_3647934 | Gm8734 | NCBI_Gene:667624,ENSEMBL:ENSMUSG00000109736 | MGI:3647934 | pseudogene | predicted gene 8734 |
| 8 | gene | 55.348649 | 55.351792 | positive | MGI_C57BL6J_5825700 | Gm46063 | NCBI_Gene:108167568 | MGI:5825700 | protein coding gene | predicted gene, 46063 |
| 8 | pseudogene | 55.423876 | 55.424191 | positive | MGI_C57BL6J_5595546 | Gm36387 | NCBI_Gene:102640276 | MGI:5595546 | pseudogene | predicted gene, 36387 |
| 8 | pseudogene | 55.532284 | 55.532837 | positive | MGI_C57BL6J_5295692 | Gm20586 | NCBI_Gene:100417493,ENSEMBL:ENSMUSG00000110025 | MGI:5295692 | pseudogene | predicted gene, 20586 |
| 8 | pseudogene | 55.552813 | 55.553128 | positive | MGI_C57BL6J_5595612 | Gm36453 | NCBI_Gene:102640382 | MGI:5595612 | pseudogene | predicted gene, 36453 |
| 8 | pseudogene | 55.659353 | 55.659947 | positive | MGI_C57BL6J_5010505 | Gm18320 | NCBI_Gene:100416917,ENSEMBL:ENSMUSG00000110064 | MGI:5010505 | pseudogene | predicted gene, 18320 |
| 8 | pseudogene | 55.777199 | 55.777982 | positive | MGI_C57BL6J_5791446 | Gm45610 | ENSEMBL:ENSMUSG00000109943 | MGI:5791446 | pseudogene | predicted gene 45610 |
| 8 | gene | 55.870912 | 55.906964 | negative | MGI_C57BL6J_2676326 | Adam29 | NCBI_Gene:244486,ENSEMBL:ENSMUSG00000046258 | MGI:2676326 | protein coding gene | a disintegrin and metallopeptidase domain 29 |
| 8 | gene | 55.940411 | 56.130070 | positive | MGI_C57BL6J_95749 | Glra3 | NCBI_Gene:110304,ENSEMBL:ENSMUSG00000038257 | MGI:95749 | protein coding gene | glycine receptor, alpha 3 subunit |
| 8 | pseudogene | 55.973640 | 55.974219 | negative | MGI_C57BL6J_3643013 | Gm7351 | NCBI_Gene:664812,ENSEMBL:ENSMUSG00000110164 | MGI:3643013 | pseudogene | predicted gene 7351 |
| 8 | pseudogene | 56.016717 | 56.017100 | negative | MGI_C57BL6J_5791202 | Gm45366 | ENSEMBL:ENSMUSG00000109662 | MGI:5791202 | pseudogene | predicted gene 45366 |
| 8 | gene | 56.129641 | 56.130066 | positive | MGI_C57BL6J_1924799 | 9330109K16Rik | NA | NA | unclassified gene | RIKEN cDNA 9330109K16 gene |
| 8 | gene | 56.294552 | 56.321046 | positive | MGI_C57BL6J_108085 | Hpgd | NCBI_Gene:15446,ENSEMBL:ENSMUSG00000031613 | MGI:108085 | protein coding gene | hydroxyprostaglandin dehydrogenase 15 (NAD) |
| 8 | gene | 56.311304 | 56.314909 | positive | MGI_C57BL6J_5791376 | Gm45540 | ENSEMBL:ENSMUSG00000110397 | MGI:5791376 | unclassified gene | predicted gene 45540 |
| 8 | pseudogene | 56.453515 | 56.454439 | positive | MGI_C57BL6J_3779743 | Gm7419 | NCBI_Gene:664948,ENSEMBL:ENSMUSG00000110309 | MGI:3779743 | pseudogene | predicted gene 7419 |
| 8 | gene | 56.465622 | 56.484968 | negative | MGI_C57BL6J_1922373 | 4930518J21Rik | NCBI_Gene:75123,ENSEMBL:ENSMUSG00000110310 | MGI:1922373 | lncRNA gene | RIKEN cDNA 4930518J21 gene |
| 8 | gene | 56.521372 | 56.521503 | negative | MGI_C57BL6J_5454152 | Gm24375 | ENSEMBL:ENSMUSG00000077585 | MGI:5454152 | snoRNA gene | predicted gene, 24375 |
| 8 | gene | 56.531516 | 56.551047 | negative | MGI_C57BL6J_3525111 | Cep44 | NCBI_Gene:382010,ENSEMBL:ENSMUSG00000038215 | MGI:3525111 | protein coding gene | centrosomal protein 44 |
| 8 | gene | 56.551090 | 56.593939 | positive | MGI_C57BL6J_1354696 | Fbxo8 | NCBI_Gene:50753,ENSEMBL:ENSMUSG00000038206 | MGI:1354696 | protein coding gene | F-box protein 8 |
| 8 | pseudogene | 56.560914 | 56.561544 | positive | MGI_C57BL6J_3780936 | Gm2767 | NCBI_Gene:100040431,ENSEMBL:ENSMUSG00000110318 | MGI:3780936 | pseudogene | predicted gene 2767 |
| 8 | gene | 56.561301 | 56.561409 | positive | MGI_C57BL6J_5455769 | Gm25992 | ENSEMBL:ENSMUSG00000076392 | MGI:5455769 | miRNA gene | predicted gene, 25992 |
| 8 | pseudogene | 56.713049 | 56.715273 | positive | MGI_C57BL6J_3644902 | Gm6503 | NCBI_Gene:624433,ENSEMBL:ENSMUSG00000109760 | MGI:3644902 | pseudogene | predicted gene 6503 |
| 8 | gene | 56.860568 | 56.888893 | negative | MGI_C57BL6J_5592660 | Gm33501 | NCBI_Gene:102636438,ENSEMBL:ENSMUSG00000110000 | MGI:5592660 | lncRNA gene | predicted gene, 33501 |
| 8 | pseudogene | 56.952728 | 56.955271 | positive | MGI_C57BL6J_3644705 | Gm7328 | NCBI_Gene:654355,ENSEMBL:ENSMUSG00000110043 | MGI:3644705 | pseudogene | predicted gene 7328 |
| 8 | gene | 57.281117 | 57.324233 | negative | MGI_C57BL6J_5578769 | Hand2os1 | NCBI_Gene:102636514,ENSEMBL:ENSMUSG00000100510 | MGI:5578769 | antisense lncRNA gene | Hand2, opposite strand 1 |
| 8 | pseudogene | 57.291520 | 57.292052 | negative | MGI_C57BL6J_5010179 | Gm17994 | NCBI_Gene:100416239,ENSEMBL:ENSMUSG00000110327 | MGI:5010179 | pseudogene | predicted gene, 17994 |
| 8 | gene | 57.320983 | 57.324633 | positive | MGI_C57BL6J_103580 | Hand2 | NCBI_Gene:15111,ENSEMBL:ENSMUSG00000038193 | MGI:103580 | protein coding gene | heart and neural crest derivatives expressed 2 |
| 8 | gene | 57.331691 | 57.338863 | negative | MGI_C57BL6J_5593189 | Gm34030 | NCBI_Gene:102637146,ENSEMBL:ENSMUSG00000109697 | MGI:5593189 | lncRNA gene | predicted gene, 34030 |
| 8 | gene | 57.332118 | 57.354580 | positive | MGI_C57BL6J_1923246 | 5033428I22Rik | NCBI_Gene:75996,ENSEMBL:ENSMUSG00000097910 | MGI:1923246 | lncRNA gene | RIKEN cDNA 5033428I22 gene |
| 8 | gene | 57.403104 | 57.415052 | positive | MGI_C57BL6J_5593258 | Gm34099 | NCBI_Gene:102637230,ENSEMBL:ENSMUSG00000109629 | MGI:5593258 | lncRNA gene | predicted gene, 34099 |
| 8 | pseudogene | 57.446581 | 57.447101 | negative | MGI_C57BL6J_5010321 | Gm18136 | NCBI_Gene:100416508 | MGI:5010321 | pseudogene | predicted gene, 18136 |
| 8 | gene | 57.455880 | 57.479360 | positive | MGI_C57BL6J_1328308 | Scrg1 | NCBI_Gene:20284,ENSEMBL:ENSMUSG00000031610 | MGI:1328308 | protein coding gene | scrapie responsive gene 1 |
| 8 | gene | 57.482702 | 57.487860 | negative | MGI_C57BL6J_1929129 | Sap30 | NCBI_Gene:60406,ENSEMBL:ENSMUSG00000031609 | MGI:1929129 | protein coding gene | sin3 associated polypeptide |
| 8 | gene | 57.487963 | 57.510262 | positive | MGI_C57BL6J_1925603 | 2500002B13Rik | NCBI_Gene:78353,ENSEMBL:ENSMUSG00000096917 | MGI:1925603 | lncRNA gene | RIKEN cDNA 2500002B13 gene |
| 8 | gene | 57.511843 | 57.515999 | positive | MGI_C57BL6J_96157 | Hmgb2 | NCBI_Gene:97165,ENSEMBL:ENSMUSG00000054717 | MGI:96157 | protein coding gene | high mobility group box 2 |
| 8 | pseudogene | 57.519635 | 57.522350 | negative | MGI_C57BL6J_3644785 | Gm6012 | NCBI_Gene:547012,ENSEMBL:ENSMUSG00000109899 | MGI:3644785 | pseudogene | predicted gene 6012 |
| 8 | gene | 57.523825 | 57.653055 | negative | MGI_C57BL6J_1349449 | Galnt7 | NCBI_Gene:108150,ENSEMBL:ENSMUSG00000031608 | MGI:1349449 | protein coding gene | polypeptide N-acetylgalactosaminyltransferase 7 |
| 8 | gene | 57.549775 | 57.549888 | positive | MGI_C57BL6J_5455156 | Gm25379 | ENSEMBL:ENSMUSG00000077526 | MGI:5455156 | snoRNA gene | predicted gene, 25379 |
| 8 | pseudogene | 57.550284 | 57.551484 | positive | MGI_C57BL6J_3704311 | Gm10284 | NCBI_Gene:100040634,ENSEMBL:ENSMUSG00000070025 | MGI:3704311 | pseudogene | predicted pseudogene 10284 |
| 8 | gene | 57.604818 | 57.607156 | negative | MGI_C57BL6J_5791370 | Gm45534 | ENSEMBL:ENSMUSG00000110405 | MGI:5791370 | unclassified gene | predicted gene 45534 |
| 8 | gene | 57.626083 | 57.627487 | negative | MGI_C57BL6J_5791196 | Gm45360 | ENSEMBL:ENSMUSG00000109852 | MGI:5791196 | unclassified gene | predicted gene 45360 |
| 8 | gene | 57.639330 | 57.640133 | negative | MGI_C57BL6J_5011454 | Gm19269 | ENSEMBL:ENSMUSG00000110238 | MGI:5011454 | unclassified gene | predicted gene, 19269 |
| 8 | gene | 57.641518 | 57.643822 | negative | MGI_C57BL6J_5791194 | Gm45358 | ENSEMBL:ENSMUSG00000109585 | MGI:5791194 | unclassified gene | predicted gene 45358 |
| 8 | gene | 57.650889 | 57.652593 | negative | MGI_C57BL6J_1922099 | 4930412F12Rik | NCBI_Gene:74849,ENSEMBL:ENSMUSG00000110289 | MGI:1922099 | unclassified non-coding RNA gene | RIKEN cDNA 4930412F12 gene |
| 8 | gene | 57.651753 | 57.663430 | positive | MGI_C57BL6J_2142745 | AW046200 | NCBI_Gene:100502619,ENSEMBL:ENSMUSG00000110419 | MGI:2142745 | lncRNA gene | expressed sequence AW046200 |
| 8 | gene | 57.658262 | 57.658688 | positive | MGI_C57BL6J_5791195 | Gm45359 | ENSEMBL:ENSMUSG00000109728 | MGI:5791195 | lncRNA gene | predicted gene 45359 |
| 8 | gene | 57.696723 | 57.699462 | positive | MGI_C57BL6J_5791480 | Gm45644 | ENSEMBL:ENSMUSG00000109636 | MGI:5791480 | unclassified gene | predicted gene 45644 |
| 8 | gene | 57.699774 | 57.727344 | positive | MGI_C57BL6J_5622067 | Gm39182 | NA | NA | lncRNA gene | predicted gene%2c 39182 |
| 8 | gene | 57.699774 | 57.727344 | positive | MGI_C57BL6J_5791192 | Gm45356 | ENSEMBL:ENSMUSG00000110162 | MGI:5791192 | lncRNA gene | predicted gene 45356 |
| 8 | gene | 57.772489 | 58.912640 | negative | MGI_C57BL6J_1913581 | Galntl6 | NCBI_Gene:270049,ENSEMBL:ENSMUSG00000096914 | MGI:1913581 | protein coding gene | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
| 8 | gene | 57.799505 | 57.802906 | positive | MGI_C57BL6J_3781099 | Gm2921 | NCBI_Gene:105243194 | MGI:3781099 | lncRNA gene | predicted gene 2921 |
| 8 | gene | 57.891743 | 57.894995 | positive | MGI_C57BL6J_5791469 | Gm45633 | ENSEMBL:ENSMUSG00000109753 | MGI:5791469 | unclassified gene | predicted gene 45633 |
| 8 | gene | 58.171435 | 58.298866 | positive | MGI_C57BL6J_5477399 | Gm26905 | ENSEMBL:ENSMUSG00000097695 | MGI:5477399 | lncRNA gene | predicted gene, 26905 |
| 8 | gene | 58.246587 | 58.248720 | positive | MGI_C57BL6J_5791471 | Gm45635 | ENSEMBL:ENSMUSG00000109963 | MGI:5791471 | unclassified gene | predicted gene 45635 |
| 8 | gene | 58.523294 | 58.524326 | positive | MGI_C57BL6J_3801770 | Gm15882 | ENSEMBL:ENSMUSG00000090003 | MGI:3801770 | lncRNA gene | predicted gene 15882 |
| 8 | gene | 58.602605 | 58.604594 | negative | MGI_C57BL6J_5791470 | Gm45634 | ENSEMBL:ENSMUSG00000110227 | MGI:5791470 | unclassified gene | predicted gene 45634 |
| 8 | gene | 58.662429 | 58.663492 | negative | MGI_C57BL6J_5791472 | Gm45636 | ENSEMBL:ENSMUSG00000109791 | MGI:5791472 | unclassified gene | predicted gene 45636 |
| 8 | pseudogene | 58.698849 | 58.700796 | negative | MGI_C57BL6J_3801769 | Gm15881 | NCBI_Gene:100271928,ENSEMBL:ENSMUSG00000085254 | MGI:3801769 | pseudogene | predicted gene 15881 |
| 8 | gene | 58.911755 | 58.914298 | positive | MGI_C57BL6J_2447770 | BC030500 | NCBI_Gene:234290,ENSEMBL:ENSMUSG00000049946 | MGI:2447770 | protein coding gene | cDNA sequence BC030500 |
| 8 | pseudogene | 59.092611 | 59.093428 | negative | MGI_C57BL6J_3646612 | Gm7499 | NCBI_Gene:665116,ENSEMBL:ENSMUSG00000109746 | MGI:3646612 | pseudogene | predicted gene 7499 |
| 8 | pseudogene | 59.219235 | 59.221300 | positive | MGI_C57BL6J_3645689 | Gm5740 | NCBI_Gene:436042,ENSEMBL:ENSMUSG00000110401 | MGI:3645689 | pseudogene | predicted gene 5740 |
| 8 | gene | 59.419306 | 59.451513 | positive | MGI_C57BL6J_5593529 | Gm34370 | NCBI_Gene:102637603,ENSEMBL:ENSMUSG00000110381 | MGI:5593529 | lncRNA gene | predicted gene, 34370 |
| 8 | gene | 59.458540 | 59.478458 | negative | MGI_C57BL6J_5593579 | Gm34420 | NCBI_Gene:102637666 | MGI:5593579 | lncRNA gene | predicted gene, 34420 |
| 8 | gene | 59.559315 | 59.559422 | positive | MGI_C57BL6J_5454624 | Gm24847 | ENSEMBL:ENSMUSG00000088657 | MGI:5454624 | snRNA gene | predicted gene, 24847 |
| 8 | gene | 59.825649 | 59.844723 | positive | MGI_C57BL6J_1922267 | 4930470O06Rik | NCBI_Gene:75017,ENSEMBL:ENSMUSG00000110406 | MGI:1922267 | lncRNA gene | RIKEN cDNA 4930470O06 gene |
| 8 | gene | 60.079590 | 60.079693 | positive | MGI_C57BL6J_5455941 | Gm26164 | ENSEMBL:ENSMUSG00000065727 | MGI:5455941 | snRNA gene | predicted gene, 26164 |
| 8 | gene | 60.195802 | 60.280351 | positive | MGI_C57BL6J_5593782 | Gm34623 | NCBI_Gene:102637934,ENSEMBL:ENSMUSG00000109966 | MGI:5593782 | lncRNA gene | predicted gene, 34623 |
| 8 | pseudogene | 60.262262 | 60.265885 | negative | MGI_C57BL6J_5011415 | Gm19230 | NCBI_Gene:100418468 | MGI:5011415 | pseudogene | predicted gene, 19230 |
| 8 | gene | 60.430024 | 60.503198 | negative | MGI_C57BL6J_3647628 | Gm10283 | ENSEMBL:ENSMUSG00000070023 | MGI:3647628 | protein coding gene | predicted gene 10283 |
| 8 | gene | 60.505868 | 60.545677 | positive | MGI_C57BL6J_1345167 | Aadat | NCBI_Gene:23923,ENSEMBL:ENSMUSG00000057228 | MGI:1345167 | protein coding gene | aminoadipate aminotransferase |
| 8 | gene | 60.616416 | 60.632731 | negative | MGI_C57BL6J_5593889 | Gm34730 | NCBI_Gene:102638082,ENSEMBL:ENSMUSG00000109930 | MGI:5593889 | lncRNA gene | predicted gene, 34730 |
| 8 | gene | 60.632825 | 60.676731 | positive | MGI_C57BL6J_1918556 | Mfap3l | NCBI_Gene:71306,ENSEMBL:ENSMUSG00000031647 | MGI:1918556 | protein coding gene | microfibrillar-associated protein 3-like |
| 8 | gene | 60.719291 | 60.725806 | positive | MGI_C57BL6J_5791486 | Gm45650 | ENSEMBL:ENSMUSG00000109992 | MGI:5791486 | lncRNA gene | predicted gene 45650 |
| 8 | pseudogene | 60.756236 | 60.757453 | positive | MGI_C57BL6J_5791487 | Gm45651 | NCBI_Gene:108167569,ENSEMBL:ENSMUSG00000109988 | MGI:5791487 | pseudogene | predicted gene 45651 |
| 8 | pseudogene | 60.792221 | 60.793697 | positive | MGI_C57BL6J_5010183 | Stambp-ps2 | NCBI_Gene:100416243,ENSEMBL:ENSMUSG00000109797 | MGI:5010183 | pseudogene | STAM binding protein, pseudogene 2 |
| 8 | gene | 60.807739 | 60.807846 | negative | MGI_C57BL6J_5453106 | Gm23329 | ENSEMBL:ENSMUSG00000088040 | MGI:5453106 | snoRNA gene | predicted gene, 23329 |
| 8 | pseudogene | 60.846312 | 60.846668 | positive | MGI_C57BL6J_5791321 | Gm45485 | ENSEMBL:ENSMUSG00000110261 | MGI:5791321 | pseudogene | predicted gene 45485 |
| 8 | gene | 60.890394 | 60.908671 | positive | MGI_C57BL6J_1919862 | Hpf1 | NCBI_Gene:72612,ENSEMBL:ENSMUSG00000038005 | MGI:1919862 | protein coding gene | histone PARylation factor 1 |
| 8 | gene | 60.910389 | 60.983311 | negative | MGI_C57BL6J_103555 | Clcn3 | NCBI_Gene:12725,ENSEMBL:ENSMUSG00000004319 | MGI:103555 | protein coding gene | chloride channel, voltage-sensitive 3 |
| 8 | pseudogene | 60.935642 | 60.936200 | positive | MGI_C57BL6J_5010579 | Gm18394 | NCBI_Gene:100417090 | MGI:5010579 | pseudogene | predicted gene, 18394 |
| 8 | gene | 60.963039 | 60.965022 | negative | MGI_C57BL6J_1921797 | 9030622M22Rik | NA | NA | unclassified gene | RIKEN cDNA 9030622M22 gene |
| 8 | gene | 60.983304 | 60.990502 | positive | MGI_C57BL6J_2443920 | B230317F23Rik | NCBI_Gene:320383,ENSEMBL:ENSMUSG00000045746 | MGI:2443920 | lncRNA gene | RIKEN cDNA B230317F23 gene |
| 8 | gene | 60.993193 | 61.131349 | positive | MGI_C57BL6J_97303 | Nek1 | NCBI_Gene:18004,ENSEMBL:ENSMUSG00000031644 | MGI:97303 | protein coding gene | NIMA (never in mitosis gene a)-related expressed kinase 1 |
| 8 | gene | 61.024131 | 61.025271 | positive | MGI_C57BL6J_5791320 | Gm45484 | ENSEMBL:ENSMUSG00000110046 | MGI:5791320 | unclassified gene | predicted gene 45484 |
| 8 | gene | 61.050049 | 61.053942 | positive | MGI_C57BL6J_5804924 | Gm45809 | ENSEMBL:ENSMUSG00000109708 | MGI:5804924 | unclassified gene | predicted gene 45809 |
| 8 | gene | 61.075365 | 61.077046 | positive | MGI_C57BL6J_5791319 | Gm45483 | ENSEMBL:ENSMUSG00000109693 | MGI:5791319 | unclassified gene | predicted gene 45483 |
| 8 | pseudogene | 61.112130 | 61.112502 | negative | MGI_C57BL6J_3801988 | Gm16179 | ENSEMBL:ENSMUSG00000080956 | MGI:3801988 | pseudogene | predicted gene 16179 |
| 8 | gene | 61.117605 | 61.194648 | negative | MGI_C57BL6J_3801783 | Gm16178 | ENSEMBL:ENSMUSG00000086681 | MGI:3801783 | lncRNA gene | predicted gene 16178 |
| 8 | gene | 61.223872 | 61.464778 | positive | MGI_C57BL6J_1913066 | Sh3rf1 | NCBI_Gene:59009,ENSEMBL:ENSMUSG00000031642 | MGI:1913066 | protein coding gene | SH3 domain containing ring finger 1 |
| 8 | gene | 61.281522 | 61.288872 | negative | MGI_C57BL6J_1916529 | 1700001D01Rik | NCBI_Gene:69279,ENSEMBL:ENSMUSG00000099419 | MGI:1916529 | lncRNA gene | RIKEN cDNA 1700001D01 gene |
| 8 | gene | 61.307663 | 61.313426 | positive | MGI_C57BL6J_5623384 | Gm40499 | NCBI_Gene:105244981 | MGI:5623384 | lncRNA gene | predicted gene, 40499 |
| 8 | pseudogene | 61.363827 | 61.364498 | negative | MGI_C57BL6J_3809114 | Gm10015 | NCBI_Gene:100502680,ENSEMBL:ENSMUSG00000056943 | MGI:3809114 | pseudogene | predicted gene 10015 |
| 8 | pseudogene | 61.380465 | 61.381419 | negative | MGI_C57BL6J_3645713 | Gm7432 | NCBI_Gene:664975,ENSEMBL:ENSMUSG00000109610 | MGI:3645713 | pseudogene | predicted gene 7432 |
| 8 | gene | 61.384937 | 61.429030 | negative | MGI_C57BL6J_1917732 | 5730405A17Rik | NCBI_Gene:102638389 | MGI:1917732 | lncRNA gene | RIKEN cDNA 5730405A17 gene |
| 8 | gene | 61.487734 | 61.506694 | positive | MGI_C57BL6J_2384567 | Cbr4 | NCBI_Gene:234309,ENSEMBL:ENSMUSG00000031641 | MGI:2384567 | protein coding gene | carbonyl reductase 4 |
| 8 | gene | 61.511429 | 61.902700 | negative | MGI_C57BL6J_1919583 | Palld | NCBI_Gene:72333,ENSEMBL:ENSMUSG00000058056 | MGI:1919583 | protein coding gene | palladin, cytoskeletal associated protein |
| 8 | gene | 61.627808 | 61.631325 | positive | MGI_C57BL6J_5594413 | Gm35254 | NCBI_Gene:102638770 | MGI:5594413 | lncRNA gene | predicted gene, 35254 |
| 8 | gene | 61.662003 | 61.663845 | negative | MGI_C57BL6J_3781192 | Gm3014 | NA | NA | unclassified gene | predicted gene 3014 |
| 8 | pseudogene | 61.819562 | 61.821525 | negative | MGI_C57BL6J_5010251 | Gm18066 | NCBI_Gene:100416355,ENSEMBL:ENSMUSG00000110131 | MGI:5010251 | pseudogene | predicted gene, 18066 |
| 8 | gene | 61.849006 | 61.924768 | positive | MGI_C57BL6J_1922361 | 4930512H18Rik | NCBI_Gene:75111,ENSEMBL:ENSMUSG00000086866 | MGI:1922361 | lncRNA gene | RIKEN cDNA 4930512H18 gene |
| 8 | gene | 61.921746 | 61.928028 | negative | MGI_C57BL6J_5791254 | Gm45418 | ENSEMBL:ENSMUSG00000109729 | MGI:5791254 | lncRNA gene | predicted gene 45418 |
| 8 | gene | 61.928064 | 62.038244 | positive | MGI_C57BL6J_2384570 | Ddx60 | NCBI_Gene:234311,ENSEMBL:ENSMUSG00000037921 | MGI:2384570 | protein coding gene | DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| 8 | gene | 62.057042 | 62.123193 | negative | MGI_C57BL6J_1347090 | Anxa10 | NCBI_Gene:26359,ENSEMBL:ENSMUSG00000031635 | MGI:1347090 | protein coding gene | annexin A10 |
| 8 | pseudogene | 62.193439 | 62.194246 | negative | MGI_C57BL6J_3781139 | Gm2961 | NCBI_Gene:100040775,ENSEMBL:ENSMUSG00000110252 | MGI:3781139 | pseudogene | predicted gene 2961 |
| 8 | pseudogene | 62.486395 | 62.488549 | negative | MGI_C57BL6J_3647040 | Hspd1-ps2 | NCBI_Gene:384830,ENSEMBL:ENSMUSG00000109687 | MGI:3647040 | pseudogene | heat shock protein 1 (chaperonin), pseudogene 2 |
| 8 | pseudogene | 62.539590 | 62.540469 | positive | MGI_C57BL6J_3781153 | Gm2975 | NCBI_Gene:100040804,ENSEMBL:ENSMUSG00000109895 | MGI:3781153 | pseudogene | predicted gene 2975 |
| 8 | pseudogene | 62.546946 | 62.547864 | negative | MGI_C57BL6J_3648480 | Gm7561 | NCBI_Gene:665267,ENSEMBL:ENSMUSG00000109740 | MGI:3648480 | pseudogene | predicted gene 7561 |
| 8 | gene | 62.616317 | 62.634645 | negative | MGI_C57BL6J_5791296 | Gm45460 | ENSEMBL:ENSMUSG00000109767 | MGI:5791296 | lncRNA gene | predicted gene 45460 |
| 8 | gene | 62.646335 | 62.855382 | negative | MGI_C57BL6J_5791295 | Gm45459 | ENSEMBL:ENSMUSG00000109994 | MGI:5791295 | lncRNA gene | predicted gene 45459 |
| 8 | gene | 62.749686 | 62.761858 | positive | MGI_C57BL6J_5791298 | Gm45462 | ENSEMBL:ENSMUSG00000110364 | MGI:5791298 | lncRNA gene | predicted gene 45462 |
| 8 | gene | 62.783612 | 62.783725 | positive | MGI_C57BL6J_5452101 | Gm22324 | ENSEMBL:ENSMUSG00000089528 | MGI:5452101 | snoRNA gene | predicted gene, 22324 |
| 8 | gene | 62.951009 | 63.357103 | positive | MGI_C57BL6J_1920152 | Spock3 | NCBI_Gene:72902,ENSEMBL:ENSMUSG00000054162 | MGI:1920152 | protein coding gene | sparc/osteonectin, cwcv and kazal-like domains proteoglycan 3 |
| 8 | gene | 62.979221 | 62.983755 | positive | MGI_C57BL6J_5791297 | Gm45461 | ENSEMBL:ENSMUSG00000109744 | MGI:5791297 | unclassified gene | predicted gene 45461 |
| 8 | gene | 63.020866 | 63.025047 | positive | MGI_C57BL6J_5791352 | Gm45516 | ENSEMBL:ENSMUSG00000110308 | MGI:5791352 | unclassified gene | predicted gene 45516 |
| 8 | gene | 63.108674 | 63.108807 | negative | MGI_C57BL6J_5454013 | Gm24236 | ENSEMBL:ENSMUSG00000088419 | MGI:5454013 | snoRNA gene | predicted gene, 24236 |
| 8 | gene | 63.198972 | 63.201439 | positive | MGI_C57BL6J_5791353 | Gm45517 | ENSEMBL:ENSMUSG00000109622 | MGI:5791353 | unclassified gene | predicted gene 45517 |
| 8 | pseudogene | 63.522920 | 63.525890 | positive | MGI_C57BL6J_5010830 | Gm18645 | NCBI_Gene:100417494,ENSEMBL:ENSMUSG00000110300,ENSEMBL:ENSMUSG00000110678 | MGI:5010830 | pseudogene | predicted gene, 18645 |
| 8 | gene | 63.545943 | 63.552805 | positive | MGI_C57BL6J_5594527 | Gm35368 | ENSEMBL:ENSMUSG00000109999 | MGI:5594527 | lncRNA gene | predicted gene, 35368 |
| 8 | gene | 63.700863 | 63.739577 | negative | MGI_C57BL6J_5791425 | Gm45589 | ENSEMBL:ENSMUSG00000109825 | MGI:5791425 | lncRNA gene | predicted gene 45589 |
| 8 | gene | 63.748212 | 63.777476 | negative | MGI_C57BL6J_5594583 | Gm35424 | NCBI_Gene:102639000 | MGI:5594583 | lncRNA gene | predicted gene, 35424 |
| 8 | pseudogene | 63.778033 | 63.779251 | positive | MGI_C57BL6J_3647041 | Gm5350 | NCBI_Gene:384831,ENSEMBL:ENSMUSG00000098106 | MGI:3647041 | pseudogene | predicted gene 5350 |
| 8 | gene | 63.878196 | 63.878326 | positive | MGI_C57BL6J_5454357 | Gm24580 | ENSEMBL:ENSMUSG00000077153 | MGI:5454357 | snoRNA gene | predicted gene, 24580 |
| 8 | gene | 63.924694 | 63.952248 | negative | MGI_C57BL6J_3644562 | Sgo2b | NCBI_Gene:244495,ENSEMBL:ENSMUSG00000094443 | MGI:3644562 | protein coding gene | shugoshin 2B |
| 8 | pseudogene | 63.932151 | 63.933363 | positive | MGI_C57BL6J_5594872 | Gm35713 | NCBI_Gene:102639387,ENSEMBL:ENSMUSG00000109818 | MGI:5594872 | pseudogene | predicted gene, 35713 |
| 8 | pseudogene | 63.999252 | 63.999627 | negative | MGI_C57BL6J_5791349 | Gm45513 | ENSEMBL:ENSMUSG00000110305 | MGI:5791349 | pseudogene | predicted gene 45513 |
| 8 | gene | 64.013098 | 64.206271 | negative | MGI_C57BL6J_106923 | Tll1 | NCBI_Gene:21892,ENSEMBL:ENSMUSG00000053626 | MGI:106923 | protein coding gene | tolloid-like |
| 8 | gene | 64.161405 | 64.161526 | positive | MGI_C57BL6J_5452363 | Gm22586 | ENSEMBL:ENSMUSG00000088132 | MGI:5452363 | rRNA gene | predicted gene, 22586 |
| 8 | gene | 64.236378 | 64.237497 | positive | MGI_C57BL6J_5594631 | Gm35472 | NCBI_Gene:102639070 | MGI:5594631 | lncRNA gene | predicted gene, 35472 |
| 8 | gene | 64.408643 | 64.417100 | negative | MGI_C57BL6J_5594680 | Gm35521 | NCBI_Gene:102639138,ENSEMBL:ENSMUSG00000109579 | MGI:5594680 | lncRNA gene | predicted gene, 35521 |
| 8 | gene | 64.514332 | 64.514441 | negative | MGI_C57BL6J_3629713 | Mir710 | miRBase:MI0004694,NCBI_Gene:735270,ENSEMBL:ENSMUSG00000076042 | MGI:3629713 | miRNA gene | microRNA 710 |
| 8 | gene | 64.592542 | 64.693109 | negative | MGI_C57BL6J_101932 | Cpe | NCBI_Gene:12876,ENSEMBL:ENSMUSG00000037852 | MGI:101932 | protein coding gene | carboxypeptidase E |
| 8 | gene | 64.654527 | 64.659525 | negative | MGI_C57BL6J_2445218 | 9330197I21Rik | NA | NA | unclassified gene | RIKEN cDNA 9330197I21 gene |
| 8 | gene | 64.701107 | 64.711076 | negative | MGI_C57BL6J_5791354 | Gm45518 | ENSEMBL:ENSMUSG00000110368 | MGI:5791354 | lncRNA gene | predicted gene 45518 |
| 8 | gene | 64.718139 | 64.733792 | negative | MGI_C57BL6J_1913484 | Msmo1 | NCBI_Gene:66234,ENSEMBL:ENSMUSG00000031604 | MGI:1913484 | protein coding gene | methylsterol monoxygenase 1 |
| 8 | gene | 64.739673 | 64.850029 | negative | MGI_C57BL6J_1924363 | Klhl2 | NCBI_Gene:77113,ENSEMBL:ENSMUSG00000031605 | MGI:1924363 | protein coding gene | kelch-like 2, Mayven |
| 8 | gene | 64.775415 | 64.811680 | positive | MGI_C57BL6J_5825677 | Gm46040 | NCBI_Gene:108167532 | MGI:5825677 | lncRNA gene | predicted gene, 46040 |
| 8 | gene | 64.807639 | 64.809138 | negative | MGI_C57BL6J_3779213 | Gm10997 | ENSEMBL:ENSMUSG00000109879 | MGI:3779213 | unclassified gene | predicted gene 10997 |
| 8 | gene | 64.838234 | 64.839390 | positive | MGI_C57BL6J_5791181 | Gm45345 | ENSEMBL:ENSMUSG00000109854 | MGI:5791181 | lncRNA gene | predicted gene 45345 |
| 8 | gene | 64.876997 | 64.881068 | negative | MGI_C57BL6J_5623386 | Gm40501 | NCBI_Gene:105244984 | MGI:5623386 | lncRNA gene | predicted gene, 40501 |
| 8 | pseudogene | 64.910964 | 64.912003 | negative | MGI_C57BL6J_5791241 | Gm45405 | NCBI_Gene:108167582,ENSEMBL:ENSMUSG00000109613 | MGI:5791241 | pseudogene | predicted gene 45405 |
| 8 | gene | 64.931972 | 64.932062 | positive | MGI_C57BL6J_5455987 | Gm26210 | ENSEMBL:ENSMUSG00000089305 | MGI:5455987 | unclassified non-coding RNA gene | predicted gene, 26210 |
| 8 | gene | 64.937399 | 64.944735 | positive | MGI_C57BL6J_5591666 | Gm32507 | NCBI_Gene:102635081,ENSEMBL:ENSMUSG00000110157 | MGI:5591666 | lncRNA gene | predicted gene, 32507 |
| 8 | gene | 64.945590 | 64.947718 | negative | MGI_C57BL6J_5791078 | Gm45242 | ENSEMBL:ENSMUSG00000110378 | MGI:5791078 | unclassified gene | predicted gene 45242 |
| 8 | gene | 64.947178 | 64.969036 | positive | MGI_C57BL6J_1920317 | Tmem192 | NCBI_Gene:73067,ENSEMBL:ENSMUSG00000025521 | MGI:1920317 | protein coding gene | transmembrane protein 192 |
| 8 | gene | 64.980884 | 64.987831 | negative | MGI_C57BL6J_2685640 | Trim75 | NCBI_Gene:333307,ENSEMBL:ENSMUSG00000071089 | MGI:2685640 | protein coding gene | tripartite motif-containing 75 |
| 8 | gene | 64.998448 | 65.018688 | negative | MGI_C57BL6J_2387430 | Trim60 | NCBI_Gene:234329,ENSEMBL:ENSMUSG00000053490 | MGI:2387430 | protein coding gene | tripartite motif-containing 60 |
| 8 | gene | 65.012976 | 65.018688 | negative | MGI_C57BL6J_2387432 | Trim61 | NCBI_Gene:260296,ENSEMBL:ENSMUSG00000109718 | MGI:2387432 | protein coding gene | tripartite motif-containing 61 |
| 8 | gene | 65.014856 | 65.037096 | negative | MGI_C57BL6J_5805001 | Gm45886 | ENSEMBL:ENSMUSG00000109805 | MGI:5805001 | lncRNA gene | predicted gene 45886 |
| 8 | pseudogene | 65.015592 | 65.015800 | positive | MGI_C57BL6J_5791284 | Gm45448 | ENSEMBL:ENSMUSG00000109650 | MGI:5791284 | pseudogene | predicted gene 45448 |
| 8 | gene | 65.028417 | 65.037336 | negative | MGI_C57BL6J_3642370 | Apela | NCBI_Gene:100038489,ENSEMBL:ENSMUSG00000079042 | MGI:3642370 | protein coding gene | apelin receptor early endogenous ligand |
| 8 | gene | 65.040966 | 65.079475 | positive | MGI_C57BL6J_5622068 | Gm39183 | NCBI_Gene:105243196 | MGI:5622068 | lncRNA gene | predicted gene, 39183 |
| 8 | gene | 65.075033 | 65.077480 | positive | MGI_C57BL6J_3642419 | Gm10663 | ENSEMBL:ENSMUSG00000074302 | MGI:3642419 | protein coding gene | predicted gene 10663 |
| 8 | gene | 65.085613 | 65.129563 | negative | MGI_C57BL6J_3039572 | Smim31 | NCBI_Gene:407795,ENSEMBL:ENSMUSG00000074300 | MGI:3039572 | protein coding gene | small integral membrane protein 31 |
| 8 | pseudogene | 65.092154 | 65.093163 | negative | MGI_C57BL6J_5791281 | Gm45445 | NCBI_Gene:108167570,ENSEMBL:ENSMUSG00000109711 | MGI:5791281 | pseudogene | predicted gene 45445 |
| 8 | pseudogene | 65.092959 | 65.096043 | negative | MGI_C57BL6J_6098523 | Gm48810 | NCBI_Gene:102639877,ENSEMBL:ENSMUSG00000109620 | MGI:6098523 | pseudogene | predicted gene, 48810 |
| 8 | gene | 65.093831 | 65.093914 | negative | MGI_C57BL6J_5454110 | Gm24333 | ENSEMBL:ENSMUSG00000092842 | MGI:5454110 | miRNA gene | predicted gene, 24333 |
| 8 | gene | 65.104115 | 65.109866 | positive | MGI_C57BL6J_5622069 | Gm39184 | NCBI_Gene:105243197 | MGI:5622069 | lncRNA gene | predicted gene, 39184 |
| 8 | pseudogene | 65.130889 | 65.132350 | negative | MGI_C57BL6J_3644741 | Gm7619 | NCBI_Gene:665396,ENSEMBL:ENSMUSG00000109935 | MGI:3644741 | pseudogene | predicted gene 7619 |
| 8 | pseudogene | 65.184244 | 65.184591 | positive | MGI_C57BL6J_5791085 | Gm45249 | ENSEMBL:ENSMUSG00000110283 | MGI:5791085 | pseudogene | predicted gene 45249 |
| 8 | pseudogene | 65.278747 | 65.279232 | negative | MGI_C57BL6J_3647111 | Gm7624 | NCBI_Gene:665405,ENSEMBL:ENSMUSG00000110226 | MGI:3647111 | pseudogene | predicted gene 7624 |
| 8 | pseudogene | 65.367300 | 65.367442 | positive | MGI_C57BL6J_2673991 | Vaultrc-ps1 | NCBI_Gene:378473 | MGI:2673991 | pseudogene | vault RNA component pseudogene 1 |
| 8 | gene | 65.367301 | 65.367443 | positive | MGI_C57BL6J_5455180 | Gm25403 | ENSEMBL:ENSMUSG00000064905 | MGI:5455180 | unclassified non-coding RNA gene | predicted gene, 25403 |
| 8 | gene | 65.617900 | 66.471637 | positive | MGI_C57BL6J_1920175 | March1 | NCBI_Gene:72925,ENSEMBL:ENSMUSG00000036469 | MGI:1920175 | protein coding gene | membrane-associated ring finger (C3HC4) 1 |
| 8 | gene | 65.667595 | 65.667701 | negative | MGI_C57BL6J_5453292 | Gm23515 | ENSEMBL:ENSMUSG00000064763 | MGI:5453292 | snRNA gene | predicted gene, 23515 |
| 8 | pseudogene | 65.816697 | 65.872502 | negative | MGI_C57BL6J_3642792 | Gm15355 | NCBI_Gene:100040986,ENSEMBL:ENSMUSG00000083905 | MGI:3642792 | pseudogene | predicted gene 15355 |
| 8 | gene | 65.902057 | 65.902191 | positive | MGI_C57BL6J_5455900 | Gm26123 | ENSEMBL:ENSMUSG00000065297 | MGI:5455900 | snoRNA gene | predicted gene, 26123 |
| 8 | gene | 66.002346 | 66.002452 | negative | MGI_C57BL6J_5453982 | Gm24205 | ENSEMBL:ENSMUSG00000064887 | MGI:5453982 | snRNA gene | predicted gene, 24205 |
| 8 | gene | 66.139016 | 66.140712 | positive | MGI_C57BL6J_3028044 | B230308H17Rik | NA | NA | unclassified gene | RIKEN cDNA B230308H17 gene |
| 8 | gene | 66.384623 | 66.387535 | negative | MGI_C57BL6J_3705093 | Gm15354 | ENSEMBL:ENSMUSG00000086156 | MGI:3705093 | lncRNA gene | predicted gene 15354 |
| 8 | gene | 66.439455 | 66.442809 | negative | MGI_C57BL6J_3705102 | Gm15356 | ENSEMBL:ENSMUSG00000085109 | MGI:3705102 | lncRNA gene | predicted gene 15356 |
| 8 | gene | 66.473118 | 66.486549 | negative | MGI_C57BL6J_1913532 | Tma16 | NCBI_Gene:66282,ENSEMBL:ENSMUSG00000025591 | MGI:1913532 | protein coding gene | translation machinery associated 16 |
| 8 | gene | 66.495059 | 66.496946 | positive | MGI_C57BL6J_5591727 | Gm32568 | ENSEMBL:ENSMUSG00000110500 | MGI:5591727 | lncRNA gene | predicted gene, 32568 |
| 8 | gene | 66.511740 | 66.519199 | positive | MGI_C57BL6J_1921669 | Tktl2 | NCBI_Gene:74419,ENSEMBL:ENSMUSG00000025519 | MGI:1921669 | protein coding gene | transketolase-like 2 |
| 8 | pseudogene | 66.588712 | 66.589008 | negative | MGI_C57BL6J_3840144 | Gm16330 | ENSEMBL:ENSMUSG00000085082 | MGI:3840144 | pseudogene | predicted gene 16330 |
| 8 | gene | 66.679965 | 66.688576 | negative | MGI_C57BL6J_108082 | Npy5r | NCBI_Gene:18168,ENSEMBL:ENSMUSG00000044014 | MGI:108082 | protein coding gene | neuropeptide Y receptor Y5 |
| 8 | gene | 66.697116 | 66.706800 | positive | MGI_C57BL6J_104963 | Npy1r | NCBI_Gene:18166,ENSEMBL:ENSMUSG00000036437 | MGI:104963 | protein coding gene | neuropeptide Y receptor Y1 |
| 8 | gene | 66.780080 | 66.795561 | positive | MGI_C57BL6J_5805026 | Gm45911 | ENSEMBL:ENSMUSG00000110454 | MGI:5805026 | lncRNA gene | predicted gene 45911 |
| 8 | pseudogene | 66.845797 | 66.847510 | positive | MGI_C57BL6J_3646609 | LOC665443 | NCBI_Gene:665443,ENSEMBL:ENSMUSG00000110533 | MGI:3646609 | pseudogene | RNA binding motif protein 7 pseudogene |
| 8 | gene | 66.859957 | 66.890564 | positive | MGI_C57BL6J_2682306 | Naf1 | NCBI_Gene:234344,ENSEMBL:ENSMUSG00000014907 | MGI:2682306 | protein coding gene | nuclear assembly factor 1 ribonucleoprotein |
| 8 | gene | 66.905997 | 66.938808 | positive | MGI_C57BL6J_5622070 | Gm39185 | NCBI_Gene:105243198,ENSEMBL:ENSMUSG00000110468 | MGI:5622070 | lncRNA gene | predicted gene, 39185 |
| 8 | gene | 66.918650 | 66.918778 | negative | MGI_C57BL6J_5455052 | Gm25275 | ENSEMBL:ENSMUSG00000077485 | MGI:5455052 | snoRNA gene | predicted gene, 25275 |
| 8 | pseudogene | 67.053622 | 67.054138 | negative | MGI_C57BL6J_3645906 | Gm7639 | NCBI_Gene:665453,ENSEMBL:ENSMUSG00000110583 | MGI:3645906 | pseudogene | predicted gene 7639 |
| 8 | gene | 67.176771 | 67.177787 | positive | MGI_C57BL6J_5804942 | Gm45827 | ENSEMBL:ENSMUSG00000110512 | MGI:5804942 | lncRNA gene | predicted gene 45827 |
| 8 | gene | 67.198946 | 67.235307 | negative | MGI_C57BL6J_5591782 | Gm32623 | NCBI_Gene:102635232 | MGI:5591782 | lncRNA gene | predicted gene, 32623 |
| 8 | gene | 67.213296 | 67.213419 | negative | MGI_C57BL6J_5454051 | Gm24274 | ENSEMBL:ENSMUSG00000080437 | MGI:5454051 | snRNA gene | predicted gene, 24274 |
| 8 | gene | 67.279082 | 67.279231 | negative | MGI_C57BL6J_5454454 | Gm24677 | ENSEMBL:ENSMUSG00000089380 | MGI:5454454 | snoRNA gene | predicted gene, 24677 |
| 8 | pseudogene | 67.310687 | 67.311539 | positive | MGI_C57BL6J_5804995 | Gm45880 | ENSEMBL:ENSMUSG00000110632 | MGI:5804995 | pseudogene | predicted gene 45880 |
| 8 | gene | 67.352104 | 67.365004 | negative | MGI_C57BL6J_3642761 | Gm10660 | ENSEMBL:ENSMUSG00000074292 | MGI:3642761 | lncRNA gene | predicted gene 10660 |
| 8 | gene | 67.480921 | 67.492582 | positive | MGI_C57BL6J_97279 | Nat1 | NCBI_Gene:17960,ENSEMBL:ENSMUSG00000025588 | MGI:97279 | protein coding gene | N-acetyl transferase 1 |
| 8 | gene | 67.494858 | 67.502644 | positive | MGI_C57BL6J_109201 | Nat2 | NCBI_Gene:17961,ENSEMBL:ENSMUSG00000051147 | MGI:109201 | protein coding gene | N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| 8 | pseudogene | 67.514000 | 67.515658 | negative | MGI_C57BL6J_3642279 | Gm9755 | NCBI_Gene:100041265,ENSEMBL:ENSMUSG00000030735 | MGI:3642279 | pseudogene | predicted pseudogene 9755 |
| 8 | gene | 67.523854 | 67.548627 | positive | MGI_C57BL6J_102537 | Nat3 | NCBI_Gene:17962,ENSEMBL:ENSMUSG00000056426 | MGI:102537 | protein coding gene | N-acetyltransferase 3 |
| 8 | pseudogene | 67.584903 | 67.585334 | negative | MGI_C57BL6J_5595406 | Gm36247 | NCBI_Gene:102640094,ENSEMBL:ENSMUSG00000110452 | MGI:5595406 | pseudogene | predicted gene, 36247 |
| 8 | gene | 67.629014 | 67.629106 | negative | MGI_C57BL6J_5690783 | Gm44391 | ENSEMBL:ENSMUSG00000104757 | MGI:5690783 | miRNA gene | predicted gene, 44391 |
| 8 | gene | 67.689082 | 68.212027 | negative | MGI_C57BL6J_1918215 | Psd3 | NCBI_Gene:234353,ENSEMBL:ENSMUSG00000030465 | MGI:1918215 | protein coding gene | pleckstrin and Sec7 domain containing 3 |
| 8 | gene | 67.862928 | 67.863826 | negative | MGI_C57BL6J_5592047 | Gm32888 | NCBI_Gene:102635597 | MGI:5592047 | lncRNA gene | predicted gene, 32888 |
| 8 | gene | 67.949793 | 68.077600 | positive | MGI_C57BL6J_3801813 | Gm15991 | NCBI_Gene:102635305,ENSEMBL:ENSMUSG00000087266 | MGI:3801813 | lncRNA gene | predicted gene 15991 |
| 8 | gene | 68.010340 | 68.010470 | positive | MGI_C57BL6J_5451795 | Gm22018 | ENSEMBL:ENSMUSG00000077545 | MGI:5451795 | snoRNA gene | predicted gene, 22018 |
| 8 | gene | 68.218173 | 68.228413 | negative | MGI_C57BL6J_5477153 | Gm26659 | ENSEMBL:ENSMUSG00000097609 | MGI:5477153 | lncRNA gene | predicted gene, 26659 |
| 8 | gene | 68.276528 | 68.347704 | positive | MGI_C57BL6J_1919531 | Sh2d4a | NCBI_Gene:72281,ENSEMBL:ENSMUSG00000053886 | MGI:1919531 | protein coding gene | SH2 domain containing 4A |
| 8 | pseudogene | 68.299069 | 68.299456 | positive | MGI_C57BL6J_5804854 | Gm45739 | ENSEMBL:ENSMUSG00000110472 | MGI:5804854 | pseudogene | predicted gene 45739 |
| 8 | pseudogene | 68.316973 | 68.318087 | negative | MGI_C57BL6J_5011355 | Gm19170 | NCBI_Gene:100418379,ENSEMBL:ENSMUSG00000110650 | MGI:5011355 | pseudogene | predicted gene, 19170 |
| 8 | gene | 68.356781 | 68.735675 | negative | MGI_C57BL6J_2442354 | Csgalnact1 | NCBI_Gene:234356,ENSEMBL:ENSMUSG00000036356 | MGI:2442354 | protein coding gene | chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| 8 | gene | 68.368434 | 68.375710 | positive | MGI_C57BL6J_1920859 | 1700125H03Rik | NCBI_Gene:73609,ENSEMBL:ENSMUSG00000085197 | MGI:1920859 | lncRNA gene | RIKEN cDNA 1700125H03 gene |
| 8 | gene | 68.457148 | 68.458681 | positive | MGI_C57BL6J_3783099 | Gm15656 | ENSEMBL:ENSMUSG00000087489 | MGI:3783099 | lncRNA gene | predicted gene 15656 |
| 8 | gene | 68.536035 | 68.540261 | negative | MGI_C57BL6J_2445212 | 9530019H20Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 9530019H20 gene |
| 8 | pseudogene | 68.620138 | 68.620437 | positive | MGI_C57BL6J_3783098 | Gm15654 | ENSEMBL:ENSMUSG00000085487 | MGI:3783098 | pseudogene | predicted gene 15654 |
| 8 | gene | 68.627265 | 68.630145 | negative | MGI_C57BL6J_2443038 | A130009I22Rik | NA | NA | unclassified gene | RIKEN cDNA A130009I22 gene |
| 8 | gene | 68.793928 | 68.831667 | positive | MGI_C57BL6J_1918135 | Ints10 | NCBI_Gene:70885,ENSEMBL:ENSMUSG00000031864 | MGI:1918135 | protein coding gene | integrator complex subunit 10 |
| 8 | pseudogene | 68.798866 | 68.799573 | positive | MGI_C57BL6J_3782998 | Gm15549 | NCBI_Gene:100416244,ENSEMBL:ENSMUSG00000081745 | MGI:3782998 | pseudogene | predicted gene 15549 |
| 8 | gene | 68.880491 | 68.907448 | positive | MGI_C57BL6J_96820 | Lpl | NCBI_Gene:16956,ENSEMBL:ENSMUSG00000015568 | MGI:96820 | protein coding gene | lipoprotein lipase |
| 8 | pseudogene | 68.950321 | 68.950544 | positive | MGI_C57BL6J_5804895 | Gm45780 | ENSEMBL:ENSMUSG00000110449 | MGI:5804895 | pseudogene | predicted gene 45780 |
| 8 | gene | 68.954003 | 68.956183 | positive | MGI_C57BL6J_5592156 | Gm32997 | NCBI_Gene:102635735 | MGI:5592156 | lncRNA gene | predicted gene, 32997 |
| 8 | gene | 68.970449 | 69.003612 | negative | MGI_C57BL6J_5592262 | Gm33103 | NCBI_Gene:102635877,ENSEMBL:ENSMUSG00000110594 | MGI:5592262 | lncRNA gene | predicted gene, 33103 |
| 8 | gene | 68.993387 | 68.999665 | positive | MGI_C57BL6J_5592211 | Gm33052 | NCBI_Gene:102635805 | MGI:5592211 | lncRNA gene | predicted gene, 33052 |
| 8 | gene | 69.037708 | 69.090633 | negative | MGI_C57BL6J_106684 | Slc18a1 | NCBI_Gene:110877,ENSEMBL:ENSMUSG00000036330 | MGI:106684 | protein coding gene | solute carrier family 18 (vesicular monoamine), member 1 |
| 8 | gene | 69.044754 | 69.063422 | positive | MGI_C57BL6J_3783158 | Gm15716 | ENSEMBL:ENSMUSG00000089995 | MGI:3783158 | lncRNA gene | predicted gene 15716 |
| 8 | gene | 69.088646 | 69.113718 | positive | MGI_C57BL6J_109618 | Atp6v1b2 | NCBI_Gene:11966,ENSEMBL:ENSMUSG00000006273 | MGI:109618 | protein coding gene | ATPase, H+ transporting, lysosomal V1 subunit B2 |
| 8 | gene | 69.132669 | 69.133349 | positive | MGI_C57BL6J_2142622 | C230098O21Rik | NA | NA | unclassified gene | RIKEN cDNA C230098O21 gene |
| 8 | gene | 69.132669 | 69.184225 | negative | MGI_C57BL6J_2684762 | Lzts1 | NCBI_Gene:211134,ENSEMBL:ENSMUSG00000036306 | MGI:2684762 | protein coding gene | leucine zipper, putative tumor suppressor 1 |
| 8 | gene | 69.157186 | 69.159724 | positive | MGI_C57BL6J_5622072 | Gm39187 | NCBI_Gene:105243202 | MGI:5622072 | lncRNA gene | predicted gene, 39187 |
| 8 | gene | 69.206376 | 69.208981 | negative | MGI_C57BL6J_5622073 | Gm39188 | NCBI_Gene:105243203 | MGI:5622073 | lncRNA gene | predicted gene, 39188 |
| 8 | gene | 69.209038 | 69.230539 | positive | MGI_C57BL6J_2675306 | Zfp930 | NCBI_Gene:234358,ENSEMBL:ENSMUSG00000059897 | MGI:2675306 | protein coding gene | zinc finger protein 930 |
| 8 | pseudogene | 69.225450 | 69.225807 | negative | MGI_C57BL6J_5592337 | Gm33178 | NCBI_Gene:102635979,ENSEMBL:ENSMUSG00000110538 | MGI:5592337 | pseudogene | predicted gene, 33178 |
| 8 | pseudogene | 69.238882 | 69.240095 | negative | MGI_C57BL6J_3644097 | Gm7684 | NCBI_Gene:665553,ENSEMBL:ENSMUSG00000110657 | MGI:3644097 | pseudogene | predicted gene 7684 |
| 8 | gene | 69.271053 | 69.314207 | positive | MGI_C57BL6J_2444324 | D130040H23Rik | NCBI_Gene:211135,ENSEMBL:ENSMUSG00000079038 | MGI:2444324 | protein coding gene | RIKEN cDNA D130040H23 gene |
| 8 | pseudogene | 69.329073 | 69.329717 | positive | MGI_C57BL6J_3645865 | Gm7688 | NCBI_Gene:665561,ENSEMBL:ENSMUSG00000110514 | MGI:3645865 | pseudogene | predicted gene 7688 |
| 8 | gene | 69.366510 | 69.369566 | negative | MGI_C57BL6J_2443005 | E230024E03Rik | NA | NA | unclassified gene | RIKEN cDNA E230024E03 gene |
| 8 | pseudogene | 69.366510 | 69.395544 | negative | MGI_C57BL6J_3642803 | Gm10033 | NCBI_Gene:378466,ENSEMBL:ENSMUSG00000110444 | MGI:3642803 | pseudogene | predicted gene 10033 |
| 8 | gene | 69.399725 | 69.400020 | positive | MGI_C57BL6J_5455438 | Gm25661 | ENSEMBL:ENSMUSG00000088329 | MGI:5455438 | ribozyme gene | predicted gene, 25661 |
| 8 | gene | 69.419497 | 69.425233 | positive | MGI_C57BL6J_5804963 | Gm45848 | ENSEMBL:ENSMUSG00000110474 | MGI:5804963 | lncRNA gene | predicted gene 45848 |
| 8 | gene | 69.420690 | 69.454048 | negative | MGI_C57BL6J_3779905 | Gm9495 | NCBI_Gene:670593,ENSEMBL:ENSMUSG00000107280 | MGI:3779905 | protein coding gene | predicted gene 9495 |
| 8 | pseudogene | 69.470069 | 69.470406 | positive | MGI_C57BL6J_5804965 | Gm45850 | ENSEMBL:ENSMUSG00000110536 | MGI:5804965 | pseudogene | predicted gene 45850 |
| 8 | gene | 69.473492 | 69.477662 | negative | MGI_C57BL6J_5663400 | Gm43263 | NCBI_Gene:108167571,ENSEMBL:ENSMUSG00000106683 | MGI:5663400 | lncRNA gene | predicted gene 43263 |
| 8 | gene | 69.481346 | 69.522976 | negative | MGI_C57BL6J_3649141 | Gm7697 | NCBI_Gene:665578,ENSEMBL:ENSMUSG00000106824 | MGI:3649141 | protein coding gene | predicted gene 7697 |
| 8 | pseudogene | 69.540903 | 69.541414 | positive | MGI_C57BL6J_3779585 | Rps7-ps1 | NCBI_Gene:622336,ENSEMBL:ENSMUSG00000110434 | MGI:3779585 | pseudogene | ribosomal protein S7, pseudogene 1 |
| 8 | pseudogene | 69.554777 | 69.585162 | negative | MGI_C57BL6J_3781543 | Gm3365 | NCBI_Gene:100041482,ENSEMBL:ENSMUSG00000111594 | MGI:3781543 | pseudogene | predicted gene 3365 |
| 8 | gene | 69.610654 | 69.625559 | negative | MGI_C57BL6J_2142546 | Zfp868 | NCBI_Gene:234362,ENSEMBL:ENSMUSG00000060427 | MGI:2142546 | protein coding gene | zinc finger protein 868 |
| 8 | gene | 69.654463 | 69.667132 | positive | MGI_C57BL6J_3646490 | Zfp964 | NCBI_Gene:636741,ENSEMBL:ENSMUSG00000091764 | MGI:3646490 | protein coding gene | zinc finger protein 964 |
| 8 | gene | 69.702656 | 69.716990 | negative | MGI_C57BL6J_1914119 | Zfp869 | NCBI_Gene:66869,ENSEMBL:ENSMUSG00000054648 | MGI:1914119 | protein coding gene | zinc finger protein 869 |
| 8 | gene | 69.741639 | 69.749970 | negative | MGI_C57BL6J_4867078 | Zfp963 | NCBI_Gene:620419,ENSEMBL:ENSMUSG00000092260 | MGI:4867078 | protein coding gene | zinc finger protein 963 |
| 8 | gene | 69.742862 | 69.774886 | negative | MGI_C57BL6J_5141887 | Gm20422 | ENSEMBL:ENSMUSG00000092544 | MGI:5141887 | protein coding gene | predicted gene 20422 |
| 8 | gene | 69.749693 | 69.750626 | positive | MGI_C57BL6J_1923873 | 1700093C20Rik | NA | NA | unclassified gene | RIKEN cDNA 1700093C20 gene |
| 8 | pseudogene | 69.755790 | 69.756396 | positive | MGI_C57BL6J_5012078 | Gm19893 | NCBI_Gene:100503784 | MGI:5012078 | pseudogene | predicted gene, 19893 |
| 8 | gene | 69.761324 | 69.791178 | negative | MGI_C57BL6J_3584369 | Zfp866 | NCBI_Gene:330788,ENSEMBL:ENSMUSG00000043090 | MGI:3584369 | protein coding gene | zinc finger protein 866 |
| 8 | gene | 69.787010 | 69.791199 | negative | MGI_C57BL6J_5622074 | Gm39189 | NCBI_Gene:105243205 | MGI:5622074 | lncRNA gene | predicted gene, 39189 |
| 8 | gene | 69.791163 | 69.807749 | positive | MGI_C57BL6J_2180801 | Atp13a1 | NCBI_Gene:170759,ENSEMBL:ENSMUSG00000031862 | MGI:2180801 | protein coding gene | ATPase type 13A1 |
| 8 | gene | 69.808249 | 69.821870 | positive | MGI_C57BL6J_1926066 | Gmip | NCBI_Gene:78816,ENSEMBL:ENSMUSG00000036246 | MGI:1926066 | protein coding gene | Gem-interacting protein |
| 8 | gene | 69.813310 | 69.814081 | negative | MGI_C57BL6J_5592854 | Gm33695 | NCBI_Gene:102636695 | MGI:5592854 | lncRNA gene | predicted gene, 33695 |
| 8 | gene | 69.822429 | 69.831102 | positive | MGI_C57BL6J_1858422 | Lpar2 | NCBI_Gene:53978,ENSEMBL:ENSMUSG00000031861 | MGI:1858422 | protein coding gene | lysophosphatidic acid receptor 2 |
| 8 | gene | 69.832475 | 69.872292 | positive | MGI_C57BL6J_1931321 | Pbx4 | NCBI_Gene:80720,ENSEMBL:ENSMUSG00000031860 | MGI:1931321 | protein coding gene | pre B cell leukemia homeobox 4 |
| 8 | gene | 69.880366 | 69.887687 | negative | MGI_C57BL6J_1915959 | Cilp2 | NCBI_Gene:68709,ENSEMBL:ENSMUSG00000044006 | MGI:1915959 | protein coding gene | cartilage intermediate layer protein 2 |
| 8 | gene | 69.882980 | 69.888384 | positive | MGI_C57BL6J_5592905 | Gm33746 | NCBI_Gene:102636765 | MGI:5592905 | lncRNA gene | predicted gene, 33746 |
| 8 | gene | 69.887788 | 69.902558 | negative | MGI_C57BL6J_1914434 | Ndufa13 | NCBI_Gene:67184,ENSEMBL:ENSMUSG00000036199 | MGI:1914434 | protein coding gene | NADH:ubiquinone oxidoreductase subunit A13 |
| 8 | gene | 69.887788 | 69.902712 | negative | MGI_C57BL6J_2681845 | Yjefn3 | ENSEMBL:ENSMUSG00000048967 | MGI:2681845 | protein coding gene | YjeF N-terminal domain containing 3 |
| 8 | gene | 69.902184 | 69.903518 | positive | MGI_C57BL6J_2148775 | Tssk6 | NCBI_Gene:83984,ENSEMBL:ENSMUSG00000047654 | MGI:2148775 | protein coding gene | testis-specific serine kinase 6 |
| 8 | gene | 69.907069 | 69.996384 | negative | MGI_C57BL6J_2384585 | Gatad2a | NCBI_Gene:234366,ENSEMBL:ENSMUSG00000036180 | MGI:2384585 | protein coding gene | GATA zinc finger domain containing 2A |
| 8 | pseudogene | 70.010549 | 70.011002 | negative | MGI_C57BL6J_3647355 | Gm7730 | ENSEMBL:ENSMUSG00000110545 | MGI:3647355 | pseudogene | predicted gene 7730 |
| 8 | gene | 70.016123 | 70.042734 | negative | MGI_C57BL6J_1921799 | Mau2 | NCBI_Gene:74549,ENSEMBL:ENSMUSG00000031858 | MGI:1921799 | protein coding gene | MAU2 sister chromatid cohesion factor |
| 8 | gene | 70.042785 | 70.072347 | positive | MGI_C57BL6J_1917866 | Sugp1 | NCBI_Gene:70616,ENSEMBL:ENSMUSG00000011306 | MGI:1917866 | protein coding gene | SURP and G patch domain containing 1 |
| 8 | gene | 70.072924 | 70.080066 | positive | MGI_C57BL6J_1933210 | Tm6sf2 | NCBI_Gene:107770,ENSEMBL:ENSMUSG00000036151 | MGI:1933210 | protein coding gene | transmembrane 6 superfamily member 2 |
| 8 | gene | 70.083457 | 70.090862 | positive | MGI_C57BL6J_2679531 | Hapln4 | NCBI_Gene:330790,ENSEMBL:ENSMUSG00000007594 | MGI:2679531 | protein coding gene | hyaluronan and proteoglycan link protein 4 |
| 8 | gene | 70.093085 | 70.120873 | negative | MGI_C57BL6J_104694 | Ncan | NCBI_Gene:13004,ENSEMBL:ENSMUSG00000002341 | MGI:104694 | protein coding gene | neurocan |
| 8 | gene | 70.102524 | 70.102586 | negative | MGI_C57BL6J_5530672 | Mir7066 | miRBase:MI0022915,NCBI_Gene:102465646,ENSEMBL:ENSMUSG00000099042 | MGI:5530672 | miRNA gene | microRNA 7066 |
| 8 | gene | 70.123028 | 70.123153 | negative | MGI_C57BL6J_5455683 | Gm25906 | ENSEMBL:ENSMUSG00000089114 | MGI:5455683 | snoRNA gene | predicted gene, 25906 |
| 8 | gene | 70.130794 | 70.139197 | negative | MGI_C57BL6J_1333865 | Rfxank | NCBI_Gene:19727,ENSEMBL:ENSMUSG00000036120 | MGI:1333865 | protein coding gene | regulatory factor X-associated ankyrin-containing protein |
| 8 | gene | 70.131327 | 70.133752 | positive | MGI_C57BL6J_1922942 | Nr2c2ap | NCBI_Gene:75692,ENSEMBL:ENSMUSG00000071078 | MGI:1922942 | protein coding gene | nuclear receptor 2C2-associated protein |
| 8 | gene | 70.139499 | 70.148635 | positive | MGI_C57BL6J_1919618 | Borcs8 | NCBI_Gene:72368,ENSEMBL:ENSMUSG00000002345 | MGI:1919618 | protein coding gene | BLOC-1 related complex subunit 8 |
| 8 | gene | 70.139499 | 70.167488 | positive | MGI_C57BL6J_5825566 | Gm45929 | NCBI_Gene:105980076 | MGI:5825566 | protein coding gene | predicted gene, 45929 |
| 8 | gene | 70.139711 | 70.167488 | positive | MGI_C57BL6J_104526 | Mef2b | NCBI_Gene:17259,ENSEMBL:ENSMUSG00000079033 | MGI:104526 | protein coding gene | myocyte enhancer factor 2B |
| 8 | gene | 70.172329 | 70.183681 | positive | MGI_C57BL6J_2384577 | Tmem161a | NCBI_Gene:234371,ENSEMBL:ENSMUSG00000002342 | MGI:2384577 | protein coding gene | transmembrane protein 161A |
| 8 | gene | 70.184340 | 70.212305 | negative | MGI_C57BL6J_1920345 | Slc25a42 | NCBI_Gene:73095,ENSEMBL:ENSMUSG00000002346 | MGI:1920345 | protein coding gene | solute carrier family 25, member 42 |
| 8 | gene | 70.220172 | 70.234466 | negative | MGI_C57BL6J_1924063 | Armc6 | NCBI_Gene:76813,ENSEMBL:ENSMUSG00000002343 | MGI:1924063 | protein coding gene | armadillo repeat containing 6 |
| 8 | gene | 70.234209 | 70.279915 | positive | MGI_C57BL6J_2678085 | Sugp2 | NCBI_Gene:234373,ENSEMBL:ENSMUSG00000036054 | MGI:2678085 | protein coding gene | SURP and G patch domain containing 2 |
| 8 | gene | 70.272349 | 70.279916 | positive | MGI_C57BL6J_5621312 | Gm38427 | NCBI_Gene:100503831 | MGI:5621312 | unclassified gene | predicted gene, 38427 |
| 8 | gene | 70.282827 | 70.294361 | positive | MGI_C57BL6J_1347359 | Homer3 | NCBI_Gene:26558,ENSEMBL:ENSMUSG00000003573 | MGI:1347359 | protein coding gene | homer scaffolding protein 3 |
| 8 | gene | 70.292866 | 70.302511 | negative | MGI_C57BL6J_2136689 | Ddx49 | NCBI_Gene:234374,ENSEMBL:ENSMUSG00000057788 | MGI:2136689 | protein coding gene | DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| 8 | gene | 70.302518 | 70.312993 | positive | MGI_C57BL6J_1891702 | Cope | NCBI_Gene:59042,ENSEMBL:ENSMUSG00000055681 | MGI:1891702 | protein coding gene | coatomer protein complex, subunit epsilon |
| 8 | gene | 70.315775 | 70.331592 | positive | MGI_C57BL6J_2136690 | Cers1 | NCBI_Gene:93898,ENSEMBL:ENSMUSG00000087408 | MGI:2136690 | protein coding gene | ceramide synthase 1 |
| 8 | gene | 70.315775 | 70.331592 | positive | MGI_C57BL6J_95683 | Gdf1 | NCBI_Gene:14559,ENSEMBL:ENSMUSG00000109523 | MGI:95683 | protein coding gene | growth differentiation factor 1 |
| 8 | gene | 70.331522 | 70.353422 | negative | MGI_C57BL6J_107995 | Upf1 | NCBI_Gene:19704,ENSEMBL:ENSMUSG00000058301 | MGI:107995 | protein coding gene | UPF1 regulator of nonsense transcripts homolog (yeast) |
| 8 | gene | 70.373548 | 70.382066 | positive | MGI_C57BL6J_88469 | Comp | NCBI_Gene:12845,ENSEMBL:ENSMUSG00000031849 | MGI:88469 | protein coding gene | cartilage oligomeric matrix protein |
| 8 | gene | 70.382352 | 70.439636 | negative | MGI_C57BL6J_2142523 | Crtc1 | NCBI_Gene:382056,ENSEMBL:ENSMUSG00000003575 | MGI:2142523 | protein coding gene | CREB regulated transcription coactivator 1 |
| 8 | gene | 70.450219 | 70.476976 | negative | MGI_C57BL6J_2443079 | Klhl26 | NCBI_Gene:234378,ENSEMBL:ENSMUSG00000055707 | MGI:2443079 | protein coding gene | kelch-like 26 |
| 8 | gene | 70.483863 | 70.487471 | negative | MGI_C57BL6J_1915187 | Tmem59l | NCBI_Gene:67937,ENSEMBL:ENSMUSG00000035964 | MGI:1915187 | protein coding gene | transmembrane protein 59-like |
| 8 | gene | 70.488181 | 70.490691 | positive | MGI_C57BL6J_1921485 | 1700020G03Rik | NCBI_Gene:105243207,ENSEMBL:ENSMUSG00000110052 | MGI:1921485 | lncRNA gene | RIKEN cDNA 1700020G03 gene |
| 8 | gene | 70.493107 | 70.504081 | positive | MGI_C57BL6J_1340030 | Crlf1 | NCBI_Gene:12931,ENSEMBL:ENSMUSG00000007888 | MGI:1340030 | protein coding gene | cytokine receptor-like factor 1 |
| 8 | gene | 70.504292 | 70.506773 | negative | MGI_C57BL6J_1913712 | Rex1bd | NCBI_Gene:66462,ENSEMBL:ENSMUSG00000058833 | MGI:1913712 | protein coding gene | required for excision 1-B domain containing |
| 8 | gene | 70.508263 | 70.510801 | negative | MGI_C57BL6J_98887 | Uba52 | NCBI_Gene:22186,ENSEMBL:ENSMUSG00000090137 | MGI:98887 | protein coding gene | ubiquitin A-52 residue ribosomal protein fusion product 1 |
| 8 | gene | 70.508272 | 70.527956 | negative | MGI_C57BL6J_1922870 | Kxd1 | NCBI_Gene:75620,ENSEMBL:ENSMUSG00000055553 | MGI:1922870 | protein coding gene | KxDL motif containing 1 |
| 8 | gene | 70.510474 | 70.512652 | positive | MGI_C57BL6J_1925433 | 4930522P08Rik | NCBI_Gene:78183 | MGI:1925433 | unclassified non-coding RNA gene | RIKEN cDNA 4930522P08 gene |
| 8 | gene | 70.527714 | 70.535328 | positive | MGI_C57BL6J_1341070 | Fkbp8 | NCBI_Gene:14232,ENSEMBL:ENSMUSG00000019428 | MGI:1341070 | protein coding gene | FK506 binding protein 8 |
| 8 | gene | 70.539457 | 70.592858 | positive | MGI_C57BL6J_109377 | Ell | NCBI_Gene:13716,ENSEMBL:ENSMUSG00000070002 | MGI:109377 | protein coding gene | elongation factor RNA polymerase II |
| 8 | gene | 70.594373 | 70.597290 | positive | MGI_C57BL6J_1919030 | Isyna1 | NCBI_Gene:71780,ENSEMBL:ENSMUSG00000019139 | MGI:1919030 | protein coding gene | myo-inositol 1-phosphate synthase A1 |
| 8 | gene | 70.597485 | 70.608872 | negative | MGI_C57BL6J_1924150 | Ssbp4 | NCBI_Gene:76900,ENSEMBL:ENSMUSG00000070003 | MGI:1924150 | protein coding gene | single stranded DNA binding protein 4 |
| 8 | gene | 70.610425 | 70.612818 | positive | MGI_C57BL6J_5791382 | Gm45546 | ENSEMBL:ENSMUSG00000110030 | MGI:5791382 | unclassified gene | predicted gene 45546 |
| 8 | gene | 70.616155 | 70.621483 | positive | MGI_C57BL6J_2445284 | Lrrc25 | NCBI_Gene:211228,ENSEMBL:ENSMUSG00000049988 | MGI:2445284 | protein coding gene | leucine rich repeat containing 25 |
| 8 | gene | 70.629393 | 70.632470 | negative | MGI_C57BL6J_1346047 | Gdf15 | NCBI_Gene:23886,ENSEMBL:ENSMUSG00000038508 | MGI:1346047 | protein coding gene | growth differentiation factor 15 |
| 8 | gene | 70.632518 | 70.637910 | positive | MGI_C57BL6J_5593313 | Gm34154 | NCBI_Gene:102637309 | MGI:5593313 | lncRNA gene | predicted gene, 34154 |
| 8 | gene | 70.646435 | 70.660388 | negative | MGI_C57BL6J_1913772 | Pgpep1 | NCBI_Gene:66522,ENSEMBL:ENSMUSG00000056204 | MGI:1913772 | protein coding gene | pyroglutamyl-peptidase I |
| 8 | gene | 70.673231 | 70.678752 | positive | MGI_C57BL6J_1354692 | Lsm4 | NCBI_Gene:50783,ENSEMBL:ENSMUSG00000031848 | MGI:1354692 | protein coding gene | LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
| 8 | gene | 70.683309 | 70.692591 | negative | MGI_C57BL6J_5622076 | Gm39191 | NCBI_Gene:105243209 | MGI:5622076 | lncRNA gene | predicted gene, 39191 |
| 8 | gene | 70.697739 | 70.700616 | positive | MGI_C57BL6J_96648 | Jund | NCBI_Gene:16478,ENSEMBL:ENSMUSG00000071076 | MGI:96648 | protein coding gene | jun D proto-oncogene |
| 8 | gene | 70.699162 | 70.700080 | negative | MGI_C57BL6J_3809055 | Gm11175 | ENSEMBL:ENSMUSG00000080058 | MGI:3809055 | lncRNA gene | predicted gene 11175 |
| 8 | gene | 70.702025 | 70.717650 | positive | MGI_C57BL6J_3708784 | Iqcn | NCBI_Gene:637079,ENSEMBL:ENSMUSG00000110622 | MGI:3708784 | protein coding gene | IQ motif containing N |
| 8 | gene | 70.708458 | 70.724507 | negative | MGI_C57BL6J_5593414 | Gm34255 | NCBI_Gene:102637450 | MGI:5593414 | lncRNA gene | predicted gene, 34255 |
| 8 | gene | 70.718345 | 70.722635 | positive | MGI_C57BL6J_3781514 | Gm3336 | NCBI_Gene:100502950,ENSEMBL:ENSMUSG00000095026 | MGI:3781514 | protein coding gene | predicted gene 3336 |
| 8 | gene | 70.723720 | 70.751186 | positive | MGI_C57BL6J_99556 | Pde4c | NCBI_Gene:110385,ENSEMBL:ENSMUSG00000031842 | MGI:99556 | protein coding gene | phosphodiesterase 4C, cAMP specific |
| 8 | gene | 70.754679 | 70.758686 | positive | MGI_C57BL6J_97843 | Rab3a | NCBI_Gene:19339,ENSEMBL:ENSMUSG00000031840 | MGI:97843 | protein coding gene | RAB3A, member RAS oncogene family |
| 8 | gene | 70.758649 | 70.760946 | negative | MGI_C57BL6J_2681846 | Mpv17l2 | NCBI_Gene:234384,ENSEMBL:ENSMUSG00000035559 | MGI:2681846 | protein coding gene | MPV17 mitochondrial membrane protein-like 2 |
| 8 | gene | 70.762773 | 70.766663 | negative | MGI_C57BL6J_2137648 | Ifi30 | NCBI_Gene:65972,ENSEMBL:ENSMUSG00000031838 | MGI:2137648 | protein coding gene | interferon gamma inducible protein 30 |
| 8 | gene | 70.766579 | 70.768220 | positive | MGI_C57BL6J_1923413 | 6330537M06Rik | ENSEMBL:ENSMUSG00000110575 | MGI:1923413 | unclassified gene | RIKEN cDNA 6330537M06 gene |
| 8 | gene | 70.768176 | 70.776715 | negative | MGI_C57BL6J_1098772 | Pik3r2 | NCBI_Gene:18709,ENSEMBL:ENSMUSG00000031834 | MGI:1098772 | protein coding gene | phosphoinositide-3-kinase regulatory subunit 2 |
| 8 | gene | 70.773797 | 70.774424 | positive | MGI_C57BL6J_3781775 | Gm3598 | NA | NA | protein coding gene | predicted gene 3598 |
| 8 | gene | 70.776815 | 70.777823 | positive | MGI_C57BL6J_1919343 | 2010320M18Rik | NCBI_Gene:72093,ENSEMBL:ENSMUSG00000100691 | MGI:1919343 | lncRNA gene | RIKEN cDNA 2010320M18 gene |
| 8 | gene | 70.778117 | 70.805068 | negative | MGI_C57BL6J_2683541 | Mast3 | NCBI_Gene:546071,ENSEMBL:ENSMUSG00000031833 | MGI:2683541 | protein coding gene | microtubule associated serine/threonine kinase 3 |
| 8 | gene | 70.798135 | 70.798192 | positive | MGI_C57BL6J_5530869 | Mir7240 | miRBase:MI0023735,NCBI_Gene:102466823,ENSEMBL:ENSMUSG00000099031 | MGI:5530869 | miRNA gene | microRNA 7240 |
| 8 | gene | 70.808359 | 70.821424 | positive | MGI_C57BL6J_104579 | Il12rb1 | NCBI_Gene:16161,ENSEMBL:ENSMUSG00000000791 | MGI:104579 | protein coding gene | interleukin 12 receptor, beta 1 |
| 8 | gene | 70.835129 | 70.839720 | negative | MGI_C57BL6J_1918057 | Arrdc2 | NCBI_Gene:70807,ENSEMBL:ENSMUSG00000002910 | MGI:1918057 | protein coding gene | arrestin domain containing 2 |
| 8 | gene | 70.842049 | 70.863258 | negative | MGI_C57BL6J_1933993 | Kcnn1 | NCBI_Gene:84036,ENSEMBL:ENSMUSG00000002908 | MGI:1933993 | protein coding gene | potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
| 8 | gene | 70.863041 | 70.865431 | positive | MGI_C57BL6J_3045239 | A230052G05Rik | NCBI_Gene:442796,ENSEMBL:ENSMUSG00000020887 | MGI:3045239 | unclassified gene | RIKEN cDNA A230052G05 gene |
| 8 | gene | 70.868226 | 70.873935 | negative | MGI_C57BL6J_1916403 | Ccdc124 | NCBI_Gene:234388,ENSEMBL:ENSMUSG00000007721 | MGI:1916403 | protein coding gene | coiled-coil domain containing 124 |
| 8 | gene | 70.882885 | 70.893052 | negative | MGI_C57BL6J_2149330 | Slc5a5 | NCBI_Gene:114479,ENSEMBL:ENSMUSG00000000792 | MGI:2149330 | protein coding gene | solute carrier family 5 (sodium iodide symporter), member 5 |
| 8 | gene | 70.894722 | 70.897443 | negative | MGI_C57BL6J_1924058 | Rpl18a | NCBI_Gene:76808,ENSEMBL:ENSMUSG00000045128 | MGI:1924058 | protein coding gene | ribosomal protein L18A |
| 8 | gene | 70.895508 | 70.895570 | negative | MGI_C57BL6J_5531413 | Mir7067 | miRBase:MI0022916,NCBI_Gene:102465984,ENSEMBL:ENSMUSG00000098710 | MGI:5531413 | miRNA gene | microRNA 7067 |
| 8 | gene | 70.895759 | 70.895889 | negative | MGI_C57BL6J_2148181 | Snora68 | NCBI_Gene:104370,ENSEMBL:ENSMUSG00000077563 | MGI:2148181 | snoRNA gene | small nucleolar RNA, H/ACA box 68 |
| 8 | gene | 70.905932 | 70.917529 | positive | MGI_C57BL6J_2443304 | Map1s | NCBI_Gene:270058,ENSEMBL:ENSMUSG00000019261 | MGI:2443304 | protein coding gene | microtubule-associated protein 1S |
| 8 | gene | 70.925525 | 70.925618 | positive | MGI_C57BL6J_3837216 | Mir1969 | miRBase:MI0009966,NCBI_Gene:100316777,ENSEMBL:ENSMUSG00000088248 | MGI:3837216 | miRNA gene | microRNA 1969 |
| 8 | gene | 70.931315 | 70.933053 | positive | MGI_C57BL6J_3643366 | Gm10654 | NCBI_Gene:665828,ENSEMBL:ENSMUSG00000074252 | MGI:3643366 | protein coding gene | predicted gene 10654 |
| 8 | gene | 70.948246 | 71.085758 | negative | MGI_C57BL6J_1916669 | 1700026F02Rik | NCBI_Gene:69419,ENSEMBL:ENSMUSG00000101854 | MGI:1916669 | lncRNA gene | RIKEN cDNA 1700026F02 gene |
| 8 | gene | 71.026792 | 71.126576 | positive | MGI_C57BL6J_5594415 | Gm35256 | ENSEMBL:ENSMUSG00000110554 | MGI:5594415 | lncRNA gene | predicted gene, 35256 |
| 8 | pseudogene | 71.040837 | 71.041758 | negative | MGI_C57BL6J_5010394 | Gm18209 | NCBI_Gene:100416709 | MGI:5010394 | pseudogene | predicted gene, 18209 |
| 8 | gene | 71.042295 | 71.085766 | negative | MGI_C57BL6J_4937210 | Gm17576 | NCBI_Gene:105243210 | MGI:4937210 | lncRNA gene | predicted gene, 17576 |
| 8 | pseudogene | 71.059326 | 71.074910 | negative | MGI_C57BL6J_3761337 | Vmn2r-ps90 | NCBI_Gene:100124571 | MGI:3761337 | pseudogene | vomeronasal 2, receptor, pseudogene 90 |
| 8 | gene | 71.086005 | 71.108291 | positive | MGI_C57BL6J_3781819 | Gm3643 | NCBI_Gene:100042057 | MGI:3781819 | unclassified gene | predicted gene 3643 |
| 8 | gene | 71.091701 | 71.171456 | positive | MGI_C57BL6J_3646513 | Gm7827 | NCBI_Gene:665858 | MGI:3646513 | protein coding gene | predicted gene 7827 |
| 8 | gene | 71.119342 | 71.119448 | positive | MGI_C57BL6J_5531010 | Gm27628 | ENSEMBL:ENSMUSG00000098594 | MGI:5531010 | miRNA gene | predicted gene, 27628 |
| 8 | gene | 71.166044 | 71.206948 | negative | MGI_C57BL6J_5621300 | Gm38415 | NCBI_Gene:330796 | MGI:5621300 | lncRNA gene | predicted gene, 38415 |
| 8 | pseudogene | 71.214298 | 71.216456 | negative | MGI_C57BL6J_5588860 | Gm29701 | NCBI_Gene:101055629 | MGI:5588860 | pseudogene | predicted gene, 29701 |
| 8 | pseudogene | 71.223915 | 71.224453 | positive | MGI_C57BL6J_88062 | Aprt-ps | NCBI_Gene:621377,ENSEMBL:ENSMUSG00000110664 | MGI:88062 | pseudogene | adenine phosphoribosyl transferase, pseudogene |
| 8 | gene | 71.224493 | 71.225480 | positive | MGI_C57BL6J_3648141 | B930053N02Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA B930053N02 gene |
| 8 | gene | 71.248561 | 71.272934 | negative | MGI_C57BL6J_1923728 | Haus8 | NCBI_Gene:76478,ENSEMBL:ENSMUSG00000035439 | MGI:1923728 | protein coding gene | 4HAUS augmin-like complex, subunit 8 |
| 8 | gene | 71.272619 | 71.360713 | positive | MGI_C57BL6J_106624 | Myo9b | NCBI_Gene:17925,ENSEMBL:ENSMUSG00000004677 | MGI:106624 | protein coding gene | myosin IXb |
| 8 | gene | 71.366848 | 71.369732 | positive | MGI_C57BL6J_1914273 | Use1 | NCBI_Gene:67023,ENSEMBL:ENSMUSG00000002395 | MGI:1914273 | protein coding gene | unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| 8 | gene | 71.371298 | 71.379361 | positive | MGI_C57BL6J_1924340 | Ocel1 | NCBI_Gene:77090,ENSEMBL:ENSMUSG00000002396 | MGI:1924340 | protein coding gene | occludin/ELL domain containing 1 |
| 8 | gene | 71.374118 | 71.381960 | negative | MGI_C57BL6J_1352453 | Nr2f6 | NCBI_Gene:13864,ENSEMBL:ENSMUSG00000002393 | MGI:1352453 | protein coding gene | nuclear receptor subfamily 2, group F, member 6 |
| 8 | gene | 71.384272 | 71.395832 | negative | MGI_C57BL6J_1922920 | Ushbp1 | NCBI_Gene:234395,ENSEMBL:ENSMUSG00000034911 | MGI:1922920 | protein coding gene | USH1 protein network component harmonin binding protein 1 |
| 8 | gene | 71.389122 | 71.389673 | positive | MGI_C57BL6J_2441824 | A730052K04Rik | NA | NA | unclassified gene | RIKEN cDNA A730052K04 gene |
| 8 | gene | 71.396161 | 71.396775 | negative | MGI_C57BL6J_1922931 | 2310041K03Rik | NA | NA | unclassified gene | RIKEN cDNA 2310041K03 gene |
| 8 | gene | 71.396420 | 71.404772 | positive | MGI_C57BL6J_1915501 | Babam1 | NCBI_Gene:68251,ENSEMBL:ENSMUSG00000031820 | MGI:1915501 | protein coding gene | BRISC and BRCA1 A complex member 1 |
| 8 | gene | 71.405348 | 71.405947 | negative | MGI_C57BL6J_5622078 | Gm39193 | NCBI_Gene:105243212 | MGI:5622078 | lncRNA gene | predicted gene, 39193 |
| 8 | gene | 71.406002 | 71.410547 | positive | MGI_C57BL6J_1918775 | Ankle1 | NCBI_Gene:234396,ENSEMBL:ENSMUSG00000046295 | MGI:1918775 | protein coding gene | ankyrin repeat and LEM domain containing 1 |
| 8 | gene | 71.411197 | 71.415831 | positive | MGI_C57BL6J_5622079 | Gm39194 | NCBI_Gene:105243213 | MGI:5622079 | lncRNA gene | predicted gene, 39194 |
| 8 | gene | 71.456700 | 71.463657 | negative | MGI_C57BL6J_1918946 | Abhd8 | NCBI_Gene:64296,ENSEMBL:ENSMUSG00000007950 | MGI:1918946 | protein coding gene | abhydrolase domain containing 8 |
| 8 | gene | 71.464926 | 71.465753 | positive | MGI_C57BL6J_2137227 | Mrpl34 | NCBI_Gene:94065,ENSEMBL:ENSMUSG00000034880 | MGI:2137227 | protein coding gene | mitochondrial ribosomal protein L34 |
| 8 | gene | 71.467111 | 71.467968 | negative | MGI_C57BL6J_1921183 | 4930412O06Rik | NA | NA | unclassified gene | RIKEN cDNA 4930412O06 gene |
| 8 | gene | 71.469171 | 71.476098 | positive | MGI_C57BL6J_1913748 | Dda1 | NCBI_Gene:66498,ENSEMBL:ENSMUSG00000074247 | MGI:1913748 | protein coding gene | DET1 and DDB1 associated 1 |
| 8 | gene | 71.476017 | 71.486067 | negative | MGI_C57BL6J_2687327 | Ano8 | NCBI_Gene:382014,ENSEMBL:ENSMUSG00000034863 | MGI:2687327 | protein coding gene | anoctamin 8 |
| 8 | gene | 71.487863 | 71.499583 | positive | MGI_C57BL6J_1917609 | Gtpbp3 | NCBI_Gene:70359,ENSEMBL:ENSMUSG00000007610 | MGI:1917609 | protein coding gene | GTP binding protein 3 |
| 8 | gene | 71.497753 | 71.511804 | negative | MGI_C57BL6J_1890497 | Plvap | NCBI_Gene:84094,ENSEMBL:ENSMUSG00000034845 | MGI:1890497 | protein coding gene | plasmalemma vesicle associated protein |
| 8 | gene | 71.502836 | 71.505696 | negative | MGI_C57BL6J_5825678 | Gm46041 | NCBI_Gene:108167533 | MGI:5825678 | protein coding gene | predicted gene, 46041 |
| 8 | gene | 71.520118 | 71.527858 | negative | MGI_C57BL6J_3588239 | Ccdc194 | NCBI_Gene:619297,ENSEMBL:ENSMUSG00000108900 | MGI:3588239 | protein coding gene | coiled-coil domain containing 194 |
| 8 | pseudogene | 71.523558 | 71.524326 | positive | MGI_C57BL6J_3645028 | Gm7850 | NCBI_Gene:101056163,ENSEMBL:ENSMUSG00000110619 | MGI:3645028 | pseudogene | predicted gene 7850 |
| 8 | gene | 71.534255 | 71.537456 | negative | MGI_C57BL6J_1916800 | Bst2 | NCBI_Gene:69550,ENSEMBL:ENSMUSG00000046718 | MGI:1916800 | protein coding gene | bone marrow stromal cell antigen 2 |
| 8 | gene | 71.542879 | 71.548085 | positive | MGI_C57BL6J_1920961 | Mvb12a | NCBI_Gene:73711,ENSEMBL:ENSMUSG00000031813 | MGI:1920961 | protein coding gene | multivesicular body subunit 12A |
| 8 | gene | 71.554238 | 71.559018 | negative | MGI_C57BL6J_3525074 | Tmem221 | NCBI_Gene:434325,ENSEMBL:ENSMUSG00000043664 | MGI:3525074 | protein coding gene | transmembrane protein 221 |
| 8 | gene | 71.560555 | 71.566649 | negative | MGI_C57BL6J_1924446 | Nxnl1 | NCBI_Gene:234404,ENSEMBL:ENSMUSG00000034829 | MGI:1924446 | protein coding gene | nucleoredoxin-like 1 |
| 8 | gene | 71.568873 | 71.587304 | positive | MGI_C57BL6J_1347098 | Slc27a1 | NCBI_Gene:26457,ENSEMBL:ENSMUSG00000031808 | MGI:1347098 | protein coding gene | solute carrier family 27 (fatty acid transporter), member 1 |
| 8 | gene | 71.592176 | 71.601092 | positive | MGI_C57BL6J_1913421 | Pgls | NCBI_Gene:66171,ENSEMBL:ENSMUSG00000031807 | MGI:1913421 | protein coding gene | 6-phosphogluconolactonase |
| 8 | gene | 71.596356 | 71.608149 | positive | MGI_C57BL6J_3686743 | Fam129c | NCBI_Gene:100037278,ENSEMBL:ENSMUSG00000043243 | MGI:3686743 | protein coding gene | family with sequence similarity 129, member C |
| 8 | gene | 71.610991 | 71.624911 | positive | MGI_C57BL6J_1924348 | Colgalt1 | NCBI_Gene:234407,ENSEMBL:ENSMUSG00000034807 | MGI:1924348 | protein coding gene | collagen beta(1-O)galactosyltransferase 1 |
| 8 | gene | 71.624417 | 71.671775 | negative | MGI_C57BL6J_3051532 | Unc13a | NCBI_Gene:382018,ENSEMBL:ENSMUSG00000034799 | MGI:3051532 | protein coding gene | unc-13 homolog A |
| 8 | gene | 71.629524 | 71.630910 | positive | MGI_C57BL6J_5622080 | Gm39195 | NCBI_Gene:105243215 | MGI:5622080 | lncRNA gene | predicted gene, 39195 |
| 8 | gene | 71.631047 | 71.631106 | negative | MGI_C57BL6J_5530928 | Mir6769b | miRBase:MI0022917,NCBI_Gene:102466799,ENSEMBL:ENSMUSG00000098976 | MGI:5530928 | miRNA gene | microRNA 6769b |
| 8 | gene | 71.674949 | 71.676677 | negative | MGI_C57BL6J_3641937 | Gm9933 | NA | NA | unclassified gene | predicted gene 9933 |
| 8 | gene | 71.676296 | 71.690577 | positive | MGI_C57BL6J_99928 | Jak3 | NCBI_Gene:16453,ENSEMBL:ENSMUSG00000031805 | MGI:99928 | protein coding gene | Janus kinase 3 |
| 8 | gene | 71.689214 | 71.690577 | positive | MGI_C57BL6J_108427 | Insl3 | NCBI_Gene:16336,ENSEMBL:ENSMUSG00000079019 | MGI:108427 | protein coding gene | insulin-like 3 |
| 8 | gene | 71.690756 | 71.702625 | negative | MGI_C57BL6J_2152535 | B3gnt3 | NCBI_Gene:72297,ENSEMBL:ENSMUSG00000031803 | MGI:2152535 | protein coding gene | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 |
| 8 | pseudogene | 71.695608 | 71.696014 | positive | MGI_C57BL6J_5804871 | Gm45756 | ENSEMBL:ENSMUSG00000110438 | MGI:5804871 | pseudogene | predicted gene 45756 |
| 8 | pseudogene | 71.704205 | 71.704968 | negative | MGI_C57BL6J_5011045 | Gm18860 | NCBI_Gene:100417840,ENSEMBL:ENSMUSG00000110569 | MGI:5011045 | pseudogene | predicted gene, 18860 |
| 8 | gene | 71.708387 | 71.725716 | negative | MGI_C57BL6J_1921265 | Fcho1 | NCBI_Gene:74015,ENSEMBL:ENSMUSG00000070000 | MGI:1921265 | protein coding gene | FCH domain only 1 |
| 8 | gene | 71.751873 | 71.781322 | negative | MGI_C57BL6J_5594731 | Gm35572 | NCBI_Gene:102639212,ENSEMBL:ENSMUSG00000110462 | MGI:5594731 | lncRNA gene | predicted gene, 35572 |
| 8 | pseudogene | 71.760118 | 71.780287 | positive | MGI_C57BL6J_5588864 | Gm29705 | NCBI_Gene:101055648,ENSEMBL:ENSMUSG00000110473 | MGI:5588864 | pseudogene | predicted gene, 29705 |
| 8 | gene | 71.815988 | 71.828713 | positive | MGI_C57BL6J_5623389 | Gm40504 | NCBI_Gene:105244987 | MGI:5623389 | lncRNA gene | predicted gene, 40504 |
| 8 | gene | 71.824641 | 71.880519 | negative | MGI_C57BL6J_3779485 | Gm5373 | NCBI_Gene:385024,ENSEMBL:ENSMUSG00000110685 | MGI:3779485 | protein coding gene | predicted gene 5373 |
| 8 | pseudogene | 71.877720 | 71.878584 | positive | MGI_C57BL6J_3761339 | Vmn2r-ps91 | NCBI_Gene:675190,ENSEMBL:ENSMUSG00000110490 | MGI:3761339 | pseudogene | vomeronasal 2, receptor, pseudogene 91 |
| 8 | gene | 71.882019 | 71.895727 | positive | MGI_C57BL6J_2384299 | Zfp709 | NCBI_Gene:236193,ENSEMBL:ENSMUSG00000056019 | MGI:2384299 | protein coding gene | zinc finger protein 709 |
| 8 | gene | 71.908606 | 71.918851 | positive | MGI_C57BL6J_3642748 | Zfp882 | NCBI_Gene:382019,ENSEMBL:ENSMUSG00000089857 | MGI:3642748 | protein coding gene | zinc finger protein 882 |
| 8 | gene | 71.908635 | 71.934630 | positive | MGI_C57BL6J_2684459 | Zfp617 | NCBI_Gene:170938,ENSEMBL:ENSMUSG00000066880 | MGI:2684459 | protein coding gene | zinc finger protein 617 |
| 8 | pseudogene | 71.916556 | 71.918851 | negative | MGI_C57BL6J_3779896 | Ccnb2-ps | NCBI_Gene:670453,ENSEMBL:ENSMUSG00000083718 | MGI:3779896 | pseudogene | cyclin B2, pseudogene |
| 8 | gene | 71.951038 | 71.993792 | positive | MGI_C57BL6J_3583954 | Zfp961 | NCBI_Gene:234413,ENSEMBL:ENSMUSG00000052446 | MGI:3583954 | protein coding gene | zinc finger protein 961 |
| 8 | gene | 71.983903 | 71.987153 | positive | MGI_C57BL6J_5804809 | Gm45694 | ENSEMBL:ENSMUSG00000110529 | MGI:5804809 | unclassified gene | predicted gene 45694 |
| 8 | gene | 71.988482 | 72.009660 | negative | MGI_C57BL6J_1919304 | Cyp4f18 | NCBI_Gene:72054,ENSEMBL:ENSMUSG00000003484 | MGI:1919304 | protein coding gene | cytochrome P450, family 4, subfamily f, polypeptide 18 |
| 8 | pseudogene | 72.035230 | 72.035969 | positive | MGI_C57BL6J_5804849 | Gm45734 | NCBI_Gene:108167535,ENSEMBL:ENSMUSG00000110546 | MGI:5804849 | pseudogene | predicted gene 45734 |
| 8 | pseudogene | 72.036067 | 72.036588 | negative | MGI_C57BL6J_5011109 | Gm18924 | NCBI_Gene:100417967,ENSEMBL:ENSMUSG00000110643 | MGI:5011109 | pseudogene | predicted gene, 18924 |
| 8 | gene | 72.037642 | 72.037799 | positive | MGI_C57BL6J_5454829 | Gm25052 | ENSEMBL:ENSMUSG00000084610 | MGI:5454829 | snRNA gene | predicted gene, 25052 |
| 8 | gene | 72.041218 | 72.060630 | positive | MGI_C57BL6J_3030206 | Olfr372 | NCBI_Gene:404315,ENSEMBL:ENSMUSG00000069998 | MGI:3030206 | protein coding gene | olfactory receptor 372 |
| 8 | pseudogene | 72.047428 | 72.047515 | positive | MGI_C57BL6J_5791380 | Gm45544 | ENSEMBL:ENSMUSG00000110096 | MGI:5791380 | pseudogene | predicted gene 45544 |
| 8 | pseudogene | 72.078774 | 72.079092 | negative | MGI_C57BL6J_5804850 | Gm45735 | ENSEMBL:ENSMUSG00000110522 | MGI:5804850 | pseudogene | predicted gene 45735 |
| 8 | gene | 72.083177 | 72.103084 | positive | MGI_C57BL6J_3030207 | Olfr373 | NCBI_Gene:258532,ENSEMBL:ENSMUSG00000061561 | MGI:3030207 | protein coding gene | olfactory receptor 373 |
| 8 | pseudogene | 72.089668 | 72.089755 | positive | MGI_C57BL6J_5791379 | Gm45543 | ENSEMBL:ENSMUSG00000109990 | MGI:5791379 | pseudogene | predicted gene 45543 |
| 8 | gene | 72.107040 | 72.118200 | positive | MGI_C57BL6J_3030208 | Olfr374 | NCBI_Gene:258335,ENSEMBL:ENSMUSG00000046881 | MGI:3030208 | protein coding gene | olfactory receptor 374 |
| 8 | gene | 72.111234 | 72.111320 | positive | MGI_C57BL6J_5454524 | Gm24747 | ENSEMBL:ENSMUSG00000088460 | MGI:5454524 | rRNA gene | predicted gene, 24747 |
| 8 | gene | 72.118451 | 72.125924 | negative | MGI_C57BL6J_5622081 | Gm39196 | NCBI_Gene:105243217 | MGI:5622081 | lncRNA gene | predicted gene, 39196 |
| 8 | gene | 72.130174 | 72.153142 | positive | MGI_C57BL6J_2449202 | Tpm4 | NCBI_Gene:326618,ENSEMBL:ENSMUSG00000031799 | MGI:2449202 | protein coding gene | tropomyosin 4 |
| 8 | gene | 72.137389 | 72.140820 | negative | MGI_C57BL6J_5477080 | Gm26586 | ENSEMBL:ENSMUSG00000097472 | MGI:5477080 | lncRNA gene | predicted gene, 26586 |
| 8 | gene | 72.147601 | 72.160855 | negative | MGI_C57BL6J_3802007 | Gm16091 | ENSEMBL:ENSMUSG00000087502 | MGI:3802007 | lncRNA gene | predicted gene 16091 |
| 8 | gene | 72.161200 | 72.183904 | positive | MGI_C57BL6J_96960 | Rab8a | NCBI_Gene:17274,ENSEMBL:ENSMUSG00000003037 | MGI:96960 | protein coding gene | RAB8A, member RAS oncogene family |
| 8 | gene | 72.189638 | 72.201527 | positive | MGI_C57BL6J_2676364 | Hsh2d | NCBI_Gene:209488,ENSEMBL:ENSMUSG00000062007 | MGI:2676364 | protein coding gene | hematopoietic SH2 domain containing |
| 8 | gene | 72.204335 | 72.213086 | negative | MGI_C57BL6J_2685953 | Cib3 | NCBI_Gene:234421,ENSEMBL:ENSMUSG00000074240 | MGI:2685953 | protein coding gene | calcium and integrin binding family member 3 |
| 8 | gene | 72.213494 | 72.213636 | negative | MGI_C57BL6J_3779257 | Gm11034 | ENSEMBL:ENSMUSG00000079016 | MGI:3779257 | protein coding gene | predicted gene 11034 |
| 8 | gene | 72.219730 | 72.224418 | positive | MGI_C57BL6J_1915172 | Fam32a | NCBI_Gene:67922,ENSEMBL:ENSMUSG00000003039 | MGI:1915172 | protein coding gene | family with sequence similarity 32, member A |
| 8 | gene | 72.240018 | 72.257385 | positive | MGI_C57BL6J_102776 | Ap1m1 | NCBI_Gene:11767,ENSEMBL:ENSMUSG00000003033 | MGI:102776 | protein coding gene | adaptor-related protein complex AP-1, mu subunit 1 |
| 8 | pseudogene | 72.283642 | 72.285883 | negative | MGI_C57BL6J_5622082 | Gm39197 | NCBI_Gene:105243218,ENSEMBL:ENSMUSG00000110443 | MGI:5622082 | pseudogene | predicted gene, 39197 |
| 8 | gene | 72.300466 | 72.302154 | positive | MGI_C57BL6J_5623390 | Gm40505 | NCBI_Gene:105244988 | MGI:5623390 | lncRNA gene | predicted gene, 40505 |
| 8 | gene | 72.304108 | 72.305310 | negative | MGI_C57BL6J_3704312 | Gm10282 | NCBI_Gene:100042405,ENSEMBL:ENSMUSG00000070713 | MGI:3704312 | protein coding gene | predicted pseudogene 10282 |
| 8 | gene | 72.319033 | 72.321656 | positive | MGI_C57BL6J_1342772 | Klf2 | NCBI_Gene:16598,ENSEMBL:ENSMUSG00000055148 | MGI:1342772 | protein coding gene | Kruppel-like factor 2 (lung) |
| 8 | gene | 72.340990 | 72.421541 | negative | MGI_C57BL6J_104582 | Eps15l1 | NCBI_Gene:13859,ENSEMBL:ENSMUSG00000006276 | MGI:104582 | protein coding gene | epidermal growth factor receptor pathway substrate 15-like 1 |
| 8 | gene | 72.417409 | 72.421472 | negative | MGI_C57BL6J_5804894 | Gm45779 | ENSEMBL:ENSMUSG00000110558 | MGI:5804894 | unclassified gene | predicted gene 45779 |
| 8 | gene | 72.421540 | 72.438567 | positive | MGI_C57BL6J_5825680 | Gm46043 | NCBI_Gene:108167537 | MGI:5825680 | lncRNA gene | predicted gene, 46043 |
| 8 | gene | 72.424176 | 72.443870 | negative | MGI_C57BL6J_1920566 | Calr3 | NCBI_Gene:73316,ENSEMBL:ENSMUSG00000019732 | MGI:1920566 | protein coding gene | calreticulin 3 |
| 8 | pseudogene | 72.429550 | 72.429952 | negative | MGI_C57BL6J_3782007 | Gm3835 | ENSEMBL:ENSMUSG00000083922 | MGI:3782007 | pseudogene | predicted pseudogene 3835 |
| 8 | gene | 72.443880 | 72.461001 | positive | MGI_C57BL6J_1919504 | 1700030K09Rik | NCBI_Gene:72254,ENSEMBL:ENSMUSG00000052794 | MGI:1919504 | protein coding gene | RIKEN cDNA 1700030K09 gene |
| 8 | gene | 72.460482 | 72.475682 | negative | MGI_C57BL6J_106417 | Cherp | NCBI_Gene:27967,ENSEMBL:ENSMUSG00000052488 | MGI:106417 | protein coding gene | calcium homeostasis endoplasmic reticulum protein |
| 8 | gene | 72.470016 | 72.470089 | negative | MGI_C57BL6J_5530952 | Mir7068 | miRBase:MI0022918,NCBI_Gene:102465647,ENSEMBL:ENSMUSG00000099079 | MGI:5530952 | miRNA gene | microRNA 7068 |
| 8 | gene | 72.477995 | 72.492614 | negative | MGI_C57BL6J_2142403 | Slc35e1 | NCBI_Gene:270066,ENSEMBL:ENSMUSG00000019731 | MGI:2142403 | protein coding gene | solute carrier family 35, member E1 |
| 8 | gene | 72.492708 | 72.506270 | positive | MGI_C57BL6J_4937069 | Gm17435 | NCBI_Gene:102639683,ENSEMBL:ENSMUSG00000097000 | MGI:4937069 | lncRNA gene | predicted gene, 17435 |
| 8 | gene | 72.494558 | 72.548310 | negative | MGI_C57BL6J_1917875 | Med26 | NCBI_Gene:70625,ENSEMBL:ENSMUSG00000045248 | MGI:1917875 | protein coding gene | mediator complex subunit 26 |
| 8 | pseudogene | 72.515213 | 72.516621 | positive | MGI_C57BL6J_5010831 | Gm18646 | NCBI_Gene:100417495,ENSEMBL:ENSMUSG00000110446 | MGI:5010831 | pseudogene | predicted gene, 18646 |
| 8 | pseudogene | 72.530933 | 72.531312 | positive | MGI_C57BL6J_5595016 | Gm35857 | NCBI_Gene:102639577,ENSEMBL:ENSMUSG00000110532 | MGI:5595016 | pseudogene | predicted gene, 35857 |
| 8 | gene | 72.541480 | 72.541580 | negative | MGI_C57BL6J_5531401 | Gm28019 | ENSEMBL:ENSMUSG00000094041 | MGI:5531401 | miRNA gene | predicted gene, 28019 |
| 8 | gene | 72.548406 | 72.559318 | positive | MGI_C57BL6J_5825679 | Gm46042 | NCBI_Gene:108167536 | MGI:5825679 | lncRNA gene | predicted gene, 46042 |
| 8 | gene | 72.548416 | 72.551683 | positive | MGI_C57BL6J_1920762 | 1700085D22Rik | NA | NA | unclassified gene | RIKEN cDNA 1700085D22 gene |
| 8 | gene | 72.565188 | 72.571088 | negative | MGI_C57BL6J_1914068 | Smim7 | NCBI_Gene:66818,ENSEMBL:ENSMUSG00000044600 | MGI:1914068 | protein coding gene | small integral membrane protein 7 |
| 8 | gene | 72.571823 | 72.587284 | positive | MGI_C57BL6J_1921416 | Tmem38a | NCBI_Gene:74166,ENSEMBL:ENSMUSG00000031791 | MGI:1921416 | protein coding gene | transmembrane protein 38A |
| 8 | gene | 72.646225 | 72.717876 | positive | MGI_C57BL6J_2442268 | Nwd1 | NCBI_Gene:319555,ENSEMBL:ENSMUSG00000048148 | MGI:2442268 | protein coding gene | NACHT and WD repeat domain containing 1 |
| 8 | gene | 72.684314 | 72.684438 | positive | MGI_C57BL6J_5456014 | Gm26237 | ENSEMBL:ENSMUSG00000093276 | MGI:5456014 | miRNA gene | predicted gene, 26237 |
| 8 | gene | 72.714927 | 72.717876 | positive | MGI_C57BL6J_2442193 | A730056I06Rik | NA | NA | unclassified gene | RIKEN cDNA A730056I06 gene |
| 8 | gene | 72.723270 | 72.758203 | positive | MGI_C57BL6J_107158 | Sin3b | NCBI_Gene:20467,ENSEMBL:ENSMUSG00000031622 | MGI:107158 | protein coding gene | transcriptional regulator, SIN3B (yeast) |
| 8 | gene | 72.760883 | 72.763898 | positive | MGI_C57BL6J_1298207 | F2rl3 | NCBI_Gene:14065,ENSEMBL:ENSMUSG00000050147 | MGI:1298207 | protein coding gene | coagulation factor II (thrombin) receptor-like 3 |
| 8 | gene | 72.765481 | 72.767614 | positive | MGI_C57BL6J_5804820 | Gm45705 | ENSEMBL:ENSMUSG00000110481 | MGI:5804820 | unclassified gene | predicted gene 45705 |
| 8 | gene | 72.765481 | 72.767616 | negative | MGI_C57BL6J_5804821 | Gm45706 | ENSEMBL:ENSMUSG00000110696 | MGI:5804821 | unclassified gene | predicted gene 45706 |
| 8 | gene | 72.766388 | 72.784995 | negative | MGI_C57BL6J_5622083 | Gm39198 | NCBI_Gene:105243219 | MGI:5622083 | lncRNA gene | predicted gene, 39198 |
| 8 | pseudogene | 72.771658 | 72.784333 | negative | MGI_C57BL6J_5804819 | Gm45704 | ENSEMBL:ENSMUSG00000110585 | MGI:5804819 | pseudogene | predicted gene 45704 |
| 8 | pseudogene | 72.795136 | 72.795487 | negative | MGI_C57BL6J_5804906 | Gm45791 | ENSEMBL:ENSMUSG00000110563 | MGI:5804906 | pseudogene | predicted gene 45791 |
| 8 | gene | 72.814598 | 73.353577 | negative | MGI_C57BL6J_1342270 | Large1 | NCBI_Gene:16795,ENSEMBL:ENSMUSG00000004383 | MGI:1342270 | protein coding gene | LARGE xylosyl- and glucuronyltransferase 1 |
| 8 | gene | 73.016512 | 73.016591 | positive | MGI_C57BL6J_4950385 | Mir28b | miRBase:MI0016979,NCBI_Gene:100628574,ENSEMBL:ENSMUSG00000093140 | MGI:4950385 | miRNA gene | microRNA 28b |
| 8 | gene | 73.087957 | 73.091176 | negative | MGI_C57BL6J_2442692 | B930093H17Rik | NA | NA | unclassified gene | RIKEN cDNA B930093H17 gene |
| 8 | gene | 73.175119 | 73.175258 | negative | MGI_C57BL6J_5452309 | Gm22532 | ENSEMBL:ENSMUSG00000089447 | MGI:5452309 | snoRNA gene | predicted gene, 22532 |
| 8 | gene | 73.199258 | 73.200674 | negative | MGI_C57BL6J_2142655 | A730070K21Rik | NA | NA | unclassified gene | RIKEN cDNA A730070K21 gene |
| 8 | pseudogene | 73.436519 | 73.437656 | negative | MGI_C57BL6J_3646604 | Gm7943 | NCBI_Gene:666134,ENSEMBL:ENSMUSG00000110450 | MGI:3646604 | pseudogene | predicted gene 7943 |
| 8 | gene | 73.641396 | 73.641505 | negative | MGI_C57BL6J_5453032 | Gm23255 | ENSEMBL:ENSMUSG00000076199 | MGI:5453032 | miRNA gene | predicted gene, 23255 |
| 8 | gene | 73.645314 | 73.645352 | negative | MGI_C57BL6J_1921232 | 4930448E06Rik | NA | NA | unclassified gene | RIKEN cDNA 4930448E06 gene |
| 8 | pseudogene | 73.749451 | 73.750803 | negative | MGI_C57BL6J_88514 | Crry-ps | NCBI_Gene:101056221,ENSEMBL:ENSMUSG00000047657 | MGI:88514 | pseudogene | complement receptor related protein, pseudogene |
| 8 | pseudogene | 73.871693 | 73.872612 | positive | MGI_C57BL6J_3643172 | Gm7948 | NCBI_Gene:666146,ENSEMBL:ENSMUSG00000110609 | MGI:3643172 | pseudogene | predicted gene 7948 |
| 8 | gene | 74.081645 | 74.093684 | negative | MGI_C57BL6J_5804936 | Gm45821 | ENSEMBL:ENSMUSG00000110703 | MGI:5804936 | lncRNA gene | predicted gene 45821 |
| 8 | gene | 74.270247 | 74.270436 | positive | MGI_C57BL6J_5531246 | Gm27864 | ENSEMBL:ENSMUSG00000098378 | MGI:5531246 | miRNA gene | predicted gene, 27864 |
| 8 | gene | 74.590987 | 74.833947 | negative | MGI_C57BL6J_3779256 | Gm11033 | ENSEMBL:ENSMUSG00000079011 | MGI:3779256 | lncRNA gene | predicted gene 11033 |
| 8 | gene | 74.873173 | 74.893506 | positive | MGI_C57BL6J_1918847 | Isx | NCBI_Gene:71597,ENSEMBL:ENSMUSG00000031621 | MGI:1918847 | protein coding gene | intestine specific homeobox |
| 8 | gene | 74.993356 | 75.031978 | positive | MGI_C57BL6J_1918073 | Hmgxb4 | NCBI_Gene:70823,ENSEMBL:ENSMUSG00000034518 | MGI:1918073 | protein coding gene | HMG box domain containing 4 |
| 8 | gene | 75.024097 | 75.033941 | negative | MGI_C57BL6J_5804937 | Gm45822 | ENSEMBL:ENSMUSG00000110593 | MGI:5804937 | lncRNA gene | predicted gene 45822 |
| 8 | gene | 75.033658 | 75.070121 | positive | MGI_C57BL6J_1338026 | Tom1 | NCBI_Gene:21968,ENSEMBL:ENSMUSG00000042870 | MGI:1338026 | protein coding gene | target of myb1 trafficking protein |
| 8 | gene | 75.037434 | 75.045246 | negative | MGI_C57BL6J_5804935 | Gm45820 | ENSEMBL:ENSMUSG00000110433 | MGI:5804935 | lncRNA gene | predicted gene 45820 |
| 8 | pseudogene | 75.079026 | 75.080152 | positive | MGI_C57BL6J_3780226 | Gm2059 | NCBI_Gene:100039134,ENSEMBL:ENSMUSG00000110429 | MGI:3780226 | pseudogene | predicted gene 2059 |
| 8 | gene | 75.093618 | 75.100593 | positive | MGI_C57BL6J_96163 | Hmox1 | NCBI_Gene:15368,ENSEMBL:ENSMUSG00000005413 | MGI:96163 | protein coding gene | heme oxygenase 1 |
| 8 | gene | 75.109508 | 75.128439 | positive | MGI_C57BL6J_103197 | Mcm5 | NCBI_Gene:17218,ENSEMBL:ENSMUSG00000005410 | MGI:103197 | protein coding gene | minichromosome maintenance complex component 5 |
| 8 | gene | 75.176458 | 75.177733 | positive | MGI_C57BL6J_5805014 | Gm45899 | ENSEMBL:ENSMUSG00000110693 | MGI:5805014 | lncRNA gene | predicted gene 45899 |
| 8 | gene | 75.201439 | 75.210042 | positive | MGI_C57BL6J_5805018 | Gm45903 | ENSEMBL:ENSMUSG00000110640 | MGI:5805018 | lncRNA gene | predicted gene 45903 |
| 8 | gene | 75.213944 | 75.224113 | positive | MGI_C57BL6J_1922391 | Rasd2 | NCBI_Gene:75141,ENSEMBL:ENSMUSG00000034472 | MGI:1922391 | protein coding gene | RASD family, member 2 |
| 8 | pseudogene | 75.274474 | 75.275716 | negative | MGI_C57BL6J_3780051 | Gm9644 | NCBI_Gene:675507,ENSEMBL:ENSMUSG00000110515 | MGI:3780051 | pseudogene | predicted gene 9644 |
| 8 | pseudogene | 75.294570 | 75.294762 | positive | MGI_C57BL6J_5805022 | Gm45907 | ENSEMBL:ENSMUSG00000110432 | MGI:5805022 | pseudogene | predicted gene 45907 |
| 8 | pseudogene | 75.383716 | 75.385670 | positive | MGI_C57BL6J_1278338 | Umpk-ps | NCBI_Gene:384845,ENSEMBL:ENSMUSG00000110543 | MGI:1278338 | pseudogene | uridine monophosphate kinase, pseudogene |
| 8 | pseudogene | 75.398508 | 75.399049 | positive | MGI_C57BL6J_3645839 | Gm7984 | NCBI_Gene:666211,ENSEMBL:ENSMUSG00000049891 | MGI:3645839 | pseudogene | predicted gene 7984 |
| 8 | pseudogene | 75.401968 | 75.402311 | positive | MGI_C57BL6J_5012070 | Gm19885 | NCBI_Gene:100503770,ENSEMBL:ENSMUSG00000110668 | MGI:5012070 | pseudogene | predicted gene, 19885 |
| 8 | gene | 75.405337 | 75.406589 | negative | MGI_C57BL6J_3708786 | Gm10358 | NCBI_Gene:100039229,ENSEMBL:ENSMUSG00000110469 | MGI:3708786 | protein coding gene | predicted gene 10358 |
| 8 | gene | 75.448694 | 75.984508 | positive | MGI_C57BL6J_1919081 | Iqcm | NCBI_Gene:71831,ENSEMBL:ENSMUSG00000031620 | MGI:1919081 | protein coding gene | IQ motif containing M |
| 8 | gene | 75.595779 | 75.596185 | negative | MGI_C57BL6J_5611498 | Gm38270 | ENSEMBL:ENSMUSG00000103054 | MGI:5611498 | unclassified gene | predicted gene, 38270 |
| 8 | pseudogene | 75.711462 | 75.713305 | negative | MGI_C57BL6J_3645047 | Gm7901 | NCBI_Gene:666036,ENSEMBL:ENSMUSG00000101431 | MGI:3645047 | pseudogene | predicted gene 7901 |
| 8 | gene | 75.992509 | 76.415040 | negative | MGI_C57BL6J_5804855 | Gm45740 | ENSEMBL:ENSMUSG00000110485 | MGI:5804855 | lncRNA gene | predicted gene 45740 |
| 8 | gene | 76.010026 | 76.011139 | positive | MGI_C57BL6J_5804856 | Gm45741 | ENSEMBL:ENSMUSG00000110603 | MGI:5804856 | lncRNA gene | predicted gene 45741 |
| 8 | gene | 76.011004 | 76.011147 | positive | MGI_C57BL6J_1923882 | 1700093P08Rik | NA | NA | unclassified gene | RIKEN cDNA 1700093P08 gene |
| 8 | gene | 76.021929 | 76.022055 | negative | MGI_C57BL6J_5452321 | Gm22544 | ENSEMBL:ENSMUSG00000077537 | MGI:5452321 | snoRNA gene | predicted gene, 22544 |
| 8 | pseudogene | 76.116595 | 76.117027 | negative | MGI_C57BL6J_5804857 | Gm45742 | ENSEMBL:ENSMUSG00000110553 | MGI:5804857 | pseudogene | predicted gene 45742 |
| 8 | gene | 76.274344 | 76.274450 | negative | MGI_C57BL6J_5452286 | Gm22509 | ENSEMBL:ENSMUSG00000064709 | MGI:5452286 | snRNA gene | predicted gene, 22509 |
| 8 | gene | 76.289516 | 76.296013 | positive | MGI_C57BL6J_5804913 | Gm45798 | ENSEMBL:ENSMUSG00000110655 | MGI:5804913 | lncRNA gene | predicted gene 45798 |
| 8 | pseudogene | 76.293955 | 76.295177 | positive | MGI_C57BL6J_3645735 | Gm7069 | NCBI_Gene:631594,ENSEMBL:ENSMUSG00000098215 | MGI:3645735 | pseudogene | predicted gene 7069 |
| 8 | pseudogene | 76.395132 | 76.395724 | negative | MGI_C57BL6J_3649098 | Gm7990 | NCBI_Gene:666226,ENSEMBL:ENSMUSG00000110595 | MGI:3649098 | pseudogene | predicted gene 7990 |
| 8 | gene | 76.437089 | 76.445882 | negative | MGI_C57BL6J_5622084 | Gm39199 | NCBI_Gene:105243220 | MGI:5622084 | lncRNA gene | predicted gene, 39199 |
| 8 | gene | 76.605226 | 76.900621 | negative | MGI_C57BL6J_3642042 | Gm10649 | NCBI_Gene:102640923,ENSEMBL:ENSMUSG00000074235 | MGI:3642042 | lncRNA gene | predicted gene 10649 |
| 8 | gene | 76.619155 | 76.619282 | negative | MGI_C57BL6J_5530737 | Gm27355 | ENSEMBL:ENSMUSG00000099012 | MGI:5530737 | rRNA gene | predicted gene, 27355 |
| 8 | pseudogene | 76.862210 | 76.863060 | negative | MGI_C57BL6J_3643866 | Gm5353 | NCBI_Gene:634222,ENSEMBL:ENSMUSG00000087321 | MGI:3643866 | pseudogene | predicted gene 5353 |
| 8 | gene | 76.899442 | 77.245012 | positive | MGI_C57BL6J_99459 | Nr3c2 | NCBI_Gene:110784,ENSEMBL:ENSMUSG00000031618 | MGI:99459 | protein coding gene | nuclear receptor subfamily 3, group C, member 2 |
| 8 | gene | 77.146351 | 77.146480 | negative | MGI_C57BL6J_5454615 | Gm24838 | ENSEMBL:ENSMUSG00000089309 | MGI:5454615 | snoRNA gene | predicted gene, 24838 |
| 8 | gene | 77.194695 | 77.194866 | positive | MGI_C57BL6J_5453037 | Gm23260 | ENSEMBL:ENSMUSG00000088942 | MGI:5453037 | snRNA gene | predicted gene, 23260 |
| 8 | gene | 77.250366 | 77.517953 | negative | MGI_C57BL6J_1925764 | Arhgap10 | NCBI_Gene:78514,ENSEMBL:ENSMUSG00000037148 | MGI:1925764 | protein coding gene | Rho GTPase activating protein 10 |
| 8 | gene | 77.282861 | 77.286207 | positive | MGI_C57BL6J_5791242 | Gm45406 | ENSEMBL:ENSMUSG00000110258 | MGI:5791242 | lncRNA gene | predicted gene 45406 |
| 8 | gene | 77.305533 | 77.313299 | positive | MGI_C57BL6J_1918415 | 4933421D24Rik | ENSEMBL:ENSMUSG00000110410 | MGI:1918415 | lncRNA gene | RIKEN cDNA 4933421D24 gene |
| 8 | gene | 77.331970 | 77.336922 | positive | MGI_C57BL6J_5791121 | Gm45285 | ENSEMBL:ENSMUSG00000109732 | MGI:5791121 | lncRNA gene | predicted gene 45285 |
| 8 | gene | 77.368841 | 77.372516 | negative | MGI_C57BL6J_5791243 | Gm45407 | ENSEMBL:ENSMUSG00000109873 | MGI:5791243 | unclassified gene | predicted gene 45407 |
| 8 | gene | 77.517056 | 77.523898 | positive | MGI_C57BL6J_1917595 | 0610038B21Rik | NCBI_Gene:70345,ENSEMBL:ENSMUSG00000097882 | MGI:1917595 | lncRNA gene | RIKEN cDNA 0610038B21 gene |
| 8 | gene | 77.518115 | 77.523898 | positive | MGI_C57BL6J_4361268 | Gm16529 | NA | NA | antisense lncRNA gene | predicted gene%2c 16529 |
| 8 | gene | 77.549387 | 77.581338 | positive | MGI_C57BL6J_2142651 | Prmt9 | NCBI_Gene:102182,ENSEMBL:ENSMUSG00000037134 | MGI:2142651 | protein coding gene | protein arginine methyltransferase 9 |
| 8 | gene | 77.569495 | 77.569602 | negative | MGI_C57BL6J_5453245 | Gm23468 | ENSEMBL:ENSMUSG00000087750 | MGI:5453245 | snRNA gene | predicted gene, 23468 |
| 8 | gene | 77.574219 | 77.575568 | positive | MGI_C57BL6J_5791122 | Gm45286 | ENSEMBL:ENSMUSG00000109787 | MGI:5791122 | unclassified gene | predicted gene 45286 |
| 8 | gene | 77.595978 | 77.610760 | negative | MGI_C57BL6J_2384562 | Tmem184c | NCBI_Gene:234463,ENSEMBL:ENSMUSG00000031617 | MGI:2384562 | protein coding gene | transmembrane protein 184C |
| 8 | gene | 77.610860 | 77.769682 | positive | MGI_C57BL6J_1921725 | 4933431K23Rik | NCBI_Gene:105243223,NCBI_Gene:74475,NCBI_Gene:108167319,ENSEMBL:ENSMUSG00000086451 | MGI:1921725 | lncRNA gene | RIKEN cDNA 4933431K23 gene |
| 8 | gene | 77.612090 | 77.621554 | positive | MGI_C57BL6J_5622085 | Gm39200 | NCBI_Gene:105243221 | MGI:5622085 | lncRNA gene | predicted gene, 39200 |
| 8 | gene | 77.624172 | 77.628957 | positive | MGI_C57BL6J_1914607 | 1700092C02Rik | NCBI_Gene:67357,ENSEMBL:ENSMUSG00000086781 | MGI:1914607 | lncRNA gene | RIKEN cDNA 1700092C02 gene |
| 8 | gene | 77.659691 | 77.662412 | positive | MGI_C57BL6J_5804829 | Gm45714 | ENSEMBL:ENSMUSG00000109866 | MGI:5804829 | unclassified gene | predicted gene 45714 |
| 8 | gene | 77.663029 | 77.724464 | negative | MGI_C57BL6J_105923 | Ednra | NCBI_Gene:13617,ENSEMBL:ENSMUSG00000031616 | MGI:105923 | protein coding gene | endothelin receptor type A |
| 8 | gene | 77.728853 | 77.735012 | negative | MGI_C57BL6J_5622086 | Gm39201 | NCBI_Gene:105243222 | MGI:5622086 | lncRNA gene | predicted gene, 39201 |
| 8 | gene | 77.759747 | 77.764880 | negative | MGI_C57BL6J_5589004 | Gm29845 | NCBI_Gene:102631532 | MGI:5589004 | lncRNA gene | predicted gene, 29845 |
| 8 | gene | 77.807535 | 77.807640 | positive | MGI_C57BL6J_5453758 | Gm23981 | ENSEMBL:ENSMUSG00000087761 | MGI:5453758 | snRNA gene | predicted gene, 23981 |
| 8 | gene | 78.044010 | 78.055473 | positive | MGI_C57BL6J_5589054 | Gm29895 | NCBI_Gene:102631598,ENSEMBL:ENSMUSG00000110382 | MGI:5589054 | lncRNA gene | predicted gene, 29895 |
| 8 | gene | 78.072451 | 78.073588 | positive | MGI_C57BL6J_3643019 | Gm6360 | NCBI_Gene:102631676 | MGI:3643019 | lncRNA gene | predicted gene 6360 |
| 8 | pseudogene | 78.107960 | 78.108110 | positive | MGI_C57BL6J_5791124 | Gm45288 | ENSEMBL:ENSMUSG00000109948 | MGI:5791124 | pseudogene | predicted gene 45288 |
| 8 | pseudogene | 78.197516 | 78.198157 | negative | MGI_C57BL6J_5791410 | Gm45574 | NCBI_Gene:108167573,ENSEMBL:ENSMUSG00000110328 | MGI:5791410 | pseudogene | predicted gene 45574 |
| 8 | gene | 78.204128 | 78.213186 | negative | MGI_C57BL6J_5622087 | Gm39202 | NCBI_Gene:105243224 | MGI:5622087 | lncRNA gene | predicted gene, 39202 |
| 8 | gene | 78.213297 | 78.394326 | positive | MGI_C57BL6J_1920551 | Ttc29 | NCBI_Gene:73301,ENSEMBL:ENSMUSG00000037101 | MGI:1920551 | protein coding gene | tetratricopeptide repeat domain 29 |
| 8 | gene | 78.433009 | 78.436652 | negative | MGI_C57BL6J_102524 | Pou4f2 | NCBI_Gene:18997,ENSEMBL:ENSMUSG00000031688 | MGI:102524 | protein coding gene | POU domain, class 4, transcription factor 2 |
| 8 | pseudogene | 78.485975 | 78.486464 | negative | MGI_C57BL6J_3780395 | Gm2225 | NCBI_Gene:100039422,ENSEMBL:ENSMUSG00000110057 | MGI:3780395 | pseudogene | predicted gene 2225 |
| 8 | gene | 78.505269 | 78.508928 | negative | MGI_C57BL6J_1343045 | Rbmxl1 | NCBI_Gene:19656,ENSEMBL:ENSMUSG00000037070 | MGI:1343045 | protein coding gene | RNA binding motif protein, X-linked like-1 |
| 8 | gene | 78.509328 | 78.734012 | positive | MGI_C57BL6J_1924025 | Slc10a7 | NCBI_Gene:76775,ENSEMBL:ENSMUSG00000031684 | MGI:1924025 | protein coding gene | solute carrier family 10 (sodium/bile acid cotransporter family), member 7 |
| 8 | pseudogene | 78.604291 | 78.605055 | positive | MGI_C57BL6J_3780423 | Gm2253 | NCBI_Gene:100039466,ENSEMBL:ENSMUSG00000110244 | MGI:3780423 | pseudogene | predicted gene 2253 |
| 8 | gene | 78.675558 | 78.678997 | positive | MGI_C57BL6J_5622088 | Gm39203 | NCBI_Gene:105243225 | MGI:5622088 | lncRNA gene | predicted gene, 39203 |
| 8 | pseudogene | 78.754916 | 78.774545 | negative | MGI_C57BL6J_6096013 | Gm47209 | ENSEMBL:ENSMUSG00000112190 | MGI:6096013 | pseudogene | predicted gene, 47209 |
| 8 | pseudogene | 78.778002 | 78.778217 | negative | MGI_C57BL6J_5791282 | Gm45446 | ENSEMBL:ENSMUSG00000109768 | MGI:5791282 | pseudogene | predicted gene 45446 |
| 8 | gene | 78.804865 | 78.821152 | negative | MGI_C57BL6J_1925901 | Lsm6 | NCBI_Gene:78651,ENSEMBL:ENSMUSG00000031683 | MGI:1925901 | protein coding gene | LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| 8 | pseudogene | 78.869386 | 78.869869 | negative | MGI_C57BL6J_3643693 | Gm8054 | NCBI_Gene:666340,ENSEMBL:ENSMUSG00000069986 | MGI:3643693 | pseudogene | predicted pseudogene 8054 |
| 8 | gene | 78.950851 | 78.953914 | negative | MGI_C57BL6J_5791162 | Gm45326 | ENSEMBL:ENSMUSG00000110094 | MGI:5791162 | lncRNA gene | predicted gene 45326 |
| 8 | gene | 78.953926 | 79.014119 | positive | MGI_C57BL6J_5622089 | Gm39204 | NCBI_Gene:105243226,ENSEMBL:ENSMUSG00000109583 | MGI:5622089 | lncRNA gene | predicted gene, 39204 |
| 8 | pseudogene | 78.986229 | 78.987093 | negative | MGI_C57BL6J_3643698 | Gm8056 | NCBI_Gene:666346 | MGI:3643698 | pseudogene | predicted gene 8056 |
| 8 | gene | 79.014364 | 79.014470 | negative | MGI_C57BL6J_5451821 | Gm22044 | ENSEMBL:ENSMUSG00000064926 | MGI:5451821 | snRNA gene | predicted gene, 22044 |
| 8 | gene | 79.027983 | 79.193768 | positive | MGI_C57BL6J_2444807 | Zfp827 | NCBI_Gene:622675,ENSEMBL:ENSMUSG00000071064 | MGI:2444807 | protein coding gene | zinc finger protein 827 |
| 8 | gene | 79.084867 | 79.085647 | negative | MGI_C57BL6J_2443468 | C030019G06Rik | NA | NA | unclassified gene | RIKEN cDNA C030019G06 gene |
| 8 | gene | 79.174185 | 79.177163 | positive | MGI_C57BL6J_5804843 | Gm45728 | ENSEMBL:ENSMUSG00000109782 | MGI:5804843 | unclassified gene | predicted gene 45728 |
| 8 | gene | 79.181407 | 79.203571 | negative | MGI_C57BL6J_5589430 | Gm30271 | NCBI_Gene:102632114 | MGI:5589430 | lncRNA gene | predicted gene, 30271 |
| 8 | gene | 79.210423 | 79.248897 | negative | MGI_C57BL6J_1914937 | 1700011L22Rik | NCBI_Gene:67687,ENSEMBL:ENSMUSG00000031682 | MGI:1914937 | protein coding gene | RIKEN cDNA 1700011L22 gene |
| 8 | gene | 79.225930 | 79.233224 | positive | MGI_C57BL6J_5589488 | Gm30329 | ENSEMBL:ENSMUSG00000110388 | MGI:5589488 | lncRNA gene | predicted gene, 30329 |
| 8 | gene | 79.263598 | 79.294980 | negative | MGI_C57BL6J_1923805 | Mmaa | NCBI_Gene:109136,ENSEMBL:ENSMUSG00000037022 | MGI:1923805 | protein coding gene | methylmalonic aciduria (cobalamin deficiency) type A |
| 8 | gene | 79.295078 | 79.308961 | positive | MGI_C57BL6J_3779445 | Gm4890 | NCBI_Gene:234479,ENSEMBL:ENSMUSG00000097174 | MGI:3779445 | lncRNA gene | predicted gene 4890 |
| 8 | gene | 79.338395 | 79.399518 | negative | MGI_C57BL6J_109452 | Smad1 | NCBI_Gene:17125,ENSEMBL:ENSMUSG00000031681 | MGI:109452 | protein coding gene | SMAD family member 1 |
| 8 | gene | 79.394726 | 79.404683 | positive | MGI_C57BL6J_5589548 | Gm30389 | NCBI_Gene:102632263 | MGI:5589548 | lncRNA gene | predicted gene, 30389 |
| 8 | gene | 79.404148 | 79.411216 | positive | MGI_C57BL6J_5791437 | Gm45601 | ENSEMBL:ENSMUSG00000109945 | MGI:5791437 | lncRNA gene | predicted gene 45601 |
| 8 | gene | 79.440341 | 79.448774 | positive | MGI_C57BL6J_5622090 | Gm39205 | NCBI_Gene:105243227 | MGI:5622090 | lncRNA gene | predicted gene, 39205 |
| 8 | pseudogene | 79.489741 | 79.490248 | negative | MGI_C57BL6J_5622173 | Gm39288 | NCBI_Gene:105243339,ENSEMBL:ENSMUSG00000109851 | MGI:5622173 | pseudogene | predicted gene, 39288 |
| 8 | gene | 79.491142 | 79.499737 | positive | MGI_C57BL6J_5791438 | Gm45602 | ENSEMBL:ENSMUSG00000109774 | MGI:5791438 | lncRNA gene | predicted gene 45602 |
| 8 | pseudogene | 79.497153 | 79.498217 | negative | MGI_C57BL6J_3643188 | Gm8077 | NCBI_Gene:666388,ENSEMBL:ENSMUSG00000109591 | MGI:3643188 | pseudogene | predicted gene 8077 |
| 8 | pseudogene | 79.549581 | 79.550764 | negative | MGI_C57BL6J_3646128 | Gm8079 | NCBI_Gene:666392,ENSEMBL:ENSMUSG00000097975 | MGI:3646128 | pseudogene | predicted gene 8079 |
| 8 | gene | 79.639618 | 79.677755 | positive | MGI_C57BL6J_1098801 | Otud4 | NCBI_Gene:73945,ENSEMBL:ENSMUSG00000036990 | MGI:1098801 | protein coding gene | OTU domain containing 4 |
| 8 | pseudogene | 79.678835 | 79.680916 | negative | MGI_C57BL6J_107737 | Tpd52-ps | NCBI_Gene:107730,ENSEMBL:ENSMUSG00000094594 | MGI:107737 | pseudogene | tumor protein D52, pseudogene |
| 8 | gene | 79.683442 | 79.711740 | negative | MGI_C57BL6J_1195458 | Abce1 | NCBI_Gene:24015,ENSEMBL:ENSMUSG00000058355 | MGI:1195458 | protein coding gene | ATP-binding cassette, sub-family E (OABP), member 1 |
| 8 | pseudogene | 79.705597 | 79.706539 | positive | MGI_C57BL6J_5504121 | Gm27006 | ENSEMBL:ENSMUSG00000098101 | MGI:5504121 | pseudogene | predicted gene, 27006 |
| 8 | gene | 79.711748 | 79.779630 | positive | MGI_C57BL6J_1916249 | Anapc10 | NCBI_Gene:68999,ENSEMBL:ENSMUSG00000036977 | MGI:1916249 | protein coding gene | anaphase promoting complex subunit 10 |
| 8 | gene | 79.747267 | 79.752413 | negative | MGI_C57BL6J_5791271 | Gm45435 | ENSEMBL:ENSMUSG00000110202 | MGI:5791271 | lncRNA gene | predicted gene 45435 |
| 8 | pseudogene | 79.772497 | 79.773516 | negative | MGI_C57BL6J_5791270 | Gm45434 | NCBI_Gene:108167538,ENSEMBL:ENSMUSG00000110044 | MGI:5791270 | pseudogene | predicted gene 45434 |
| 8 | pseudogene | 79.803365 | 79.803611 | negative | MGI_C57BL6J_5791269 | Gm45433 | ENSEMBL:ENSMUSG00000110048 | MGI:5791269 | pseudogene | predicted gene 45433 |
| 8 | gene | 79.836777 | 79.845820 | positive | MGI_C57BL6J_5589653 | Gm30494 | NCBI_Gene:102632418 | MGI:5589653 | lncRNA gene | predicted gene, 30494 |
| 8 | pseudogene | 79.863664 | 79.864079 | negative | MGI_C57BL6J_5791268 | Gm45432 | ENSEMBL:ENSMUSG00000110188 | MGI:5791268 | pseudogene | predicted gene 45432 |
| 8 | gene | 79.965850 | 80.058069 | negative | MGI_C57BL6J_1341847 | Hhip | NCBI_Gene:15245,ENSEMBL:ENSMUSG00000064325 | MGI:1341847 | protein coding gene | Hedgehog-interacting protein |
| 8 | gene | 79.970390 | 79.970564 | positive | MGI_C57BL6J_5531202 | Gm27820 | ENSEMBL:ENSMUSG00000098688 | MGI:5531202 | unclassified non-coding RNA gene | predicted gene, 27820 |
| 8 | gene | 80.002398 | 80.007412 | positive | MGI_C57BL6J_5622091 | Gm39206 | NCBI_Gene:105243228 | MGI:5622091 | lncRNA gene | predicted gene, 39206 |
| 8 | pseudogene | 80.086205 | 80.086697 | negative | MGI_C57BL6J_5592255 | Gm33096 | ENSEMBL:ENSMUSG00000109738 | MGI:5592255 | pseudogene | predicted gene, 33096 |
| 8 | pseudogene | 80.110417 | 80.112168 | negative | MGI_C57BL6J_3646686 | Gm5909 | NCBI_Gene:546075,ENSEMBL:ENSMUSG00000082491 | MGI:3646686 | pseudogene | predicted gene 5909 |
| 8 | gene | 80.128194 | 80.138667 | negative | MGI_C57BL6J_5791267 | Gm45431 | ENSEMBL:ENSMUSG00000109593 | MGI:5791267 | lncRNA gene | predicted gene 45431 |
| 8 | gene | 80.168697 | 80.177911 | positive | MGI_C57BL6J_5589711 | Gm30552 | NCBI_Gene:102632494 | MGI:5589711 | lncRNA gene | predicted gene, 30552 |
| 8 | gene | 80.195199 | 80.197656 | positive | MGI_C57BL6J_5791266 | Gm45430 | ENSEMBL:ENSMUSG00000110294 | MGI:5791266 | lncRNA gene | predicted gene 45430 |
| 8 | gene | 80.284843 | 80.303069 | positive | MGI_C57BL6J_5622093 | Gm39208 | NCBI_Gene:105243230 | MGI:5622093 | lncRNA gene | predicted gene, 39208 |
| 8 | gene | 80.312967 | 80.326758 | negative | MGI_C57BL6J_5589819 | Gm30660 | NCBI_Gene:102632634 | MGI:5589819 | lncRNA gene | predicted gene, 30660 |
| 8 | gene | 80.328624 | 80.395214 | negative | MGI_C57BL6J_5589764 | Gm30605 | NCBI_Gene:102632566 | MGI:5589764 | lncRNA gene | predicted gene, 30605 |
| 8 | gene | 80.493781 | 80.510785 | positive | MGI_C57BL6J_95880 | Gypa | NCBI_Gene:14934,ENSEMBL:ENSMUSG00000051839 | MGI:95880 | protein coding gene | glycophorin A |
| 8 | gene | 80.511842 | 80.514490 | negative | MGI_C57BL6J_1921959 | 4930505O20Rik | NCBI_Gene:74709,ENSEMBL:ENSMUSG00000086912 | MGI:1921959 | lncRNA gene | RIKEN cDNA 4930505O20 gene |
| 8 | gene | 80.512544 | 80.526165 | positive | MGI_C57BL6J_5622094 | Gm39209 | NCBI_Gene:105243231 | MGI:5622094 | lncRNA gene | predicted gene, 39209 |
| 8 | gene | 80.525705 | 80.592537 | negative | MGI_C57BL6J_5825681 | Gm46044 | NCBI_Gene:108167539 | MGI:5825681 | lncRNA gene | predicted gene, 46044 |
| 8 | gene | 80.611039 | 80.695557 | positive | MGI_C57BL6J_2685641 | Frem3 | NCBI_Gene:333315,ENSEMBL:ENSMUSG00000042353 | MGI:2685641 | protein coding gene | Fras1 related extracellular matrix protein 3 |
| 8 | gene | 80.698507 | 80.739497 | negative | MGI_C57BL6J_1935129 | Smarca5 | NCBI_Gene:93762,ENSEMBL:ENSMUSG00000031715 | MGI:1935129 | protein coding gene | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| 8 | gene | 80.727554 | 80.729137 | negative | MGI_C57BL6J_5804987 | Gm45872 | ENSEMBL:ENSMUSG00000110142 | MGI:5804987 | unclassified gene | predicted gene 45872 |
| 8 | gene | 80.764431 | 80.880519 | negative | MGI_C57BL6J_108088 | Gab1 | NCBI_Gene:14388,ENSEMBL:ENSMUSG00000031714 | MGI:108088 | protein coding gene | growth factor receptor bound protein 2-associated protein 1 |
| 8 | pseudogene | 80.770596 | 80.772176 | positive | MGI_C57BL6J_3646045 | Gm7997 | NCBI_Gene:666237,ENSEMBL:ENSMUSG00000109584 | MGI:3646045 | pseudogene | predicted gene 7997 |
| 8 | gene | 80.797958 | 80.799803 | negative | MGI_C57BL6J_5012495 | Gm20310 | NA | NA | unclassified gene | predicted gene%2c 20310 |
| 8 | gene | 80.859386 | 80.860004 | positive | MGI_C57BL6J_1919462 | 1700019H22Rik | NA | NA | unclassified gene | RIKEN cDNA 1700019H22 gene |
| 8 | gene | 80.879938 | 80.881864 | positive | MGI_C57BL6J_5590264 | Gm31105 | NCBI_Gene:102633223,ENSEMBL:ENSMUSG00000109587 | MGI:5590264 | lncRNA gene | predicted gene, 31105 |
| 8 | gene | 80.933534 | 80.938667 | negative | MGI_C57BL6J_5504163 | Gm27048 | ENSEMBL:ENSMUSG00000098001 | MGI:5504163 | lncRNA gene | predicted gene, 27048 |
| 8 | pseudogene | 80.933659 | 80.934774 | positive | MGI_C57BL6J_3645744 | Gm4891 | NCBI_Gene:234501,ENSEMBL:ENSMUSG00000098223 | MGI:3645744 | pseudogene | predicted gene 4891 |
| 8 | gene | 80.948963 | 80.984052 | positive | MGI_C57BL6J_5590382 | Gm31223 | NCBI_Gene:102633388 | MGI:5590382 | lncRNA gene | predicted gene, 31223 |
| 8 | gene | 80.980724 | 81.015031 | negative | MGI_C57BL6J_1922091 | Usp38 | NCBI_Gene:74841,ENSEMBL:ENSMUSG00000038250 | MGI:1922091 | protein coding gene | ubiquitin specific peptidase 38 |
| 8 | gene | 81.013576 | 81.061276 | positive | MGI_C57BL6J_3646552 | Gm9725 | NCBI_Gene:105243233,ENSEMBL:ENSMUSG00000037982 | MGI:3646552 | lncRNA gene | predicted gene 9725 |
| 8 | gene | 81.061334 | 81.069803 | positive | MGI_C57BL6J_5791374 | Gm45538 | ENSEMBL:ENSMUSG00000109980 | MGI:5791374 | lncRNA gene | predicted gene 45538 |
| 8 | pseudogene | 81.196740 | 81.198375 | negative | MGI_C57BL6J_3644090 | Gm4899 | NCBI_Gene:236010,ENSEMBL:ENSMUSG00000094615 | MGI:3644090 | pseudogene | predicted gene 4899 |
| 8 | pseudogene | 81.294636 | 81.295063 | negative | MGI_C57BL6J_5791375 | Gm45539 | ENSEMBL:ENSMUSG00000109928 | MGI:5791375 | pseudogene | predicted gene 45539 |
| 8 | pseudogene | 81.307978 | 81.309167 | negative | MGI_C57BL6J_3646767 | Gm5354 | NCBI_Gene:384852,ENSEMBL:ENSMUSG00000097949 | MGI:3646767 | pseudogene | predicted gene 5354 |
| 8 | gene | 81.342342 | 82.134316 | positive | MGI_C57BL6J_2158925 | Inpp4b | NCBI_Gene:234515,ENSEMBL:ENSMUSG00000037940 | MGI:2158925 | protein coding gene | inositol polyphosphate-4-phosphatase, type II |
| 8 | gene | 81.517924 | 81.519168 | positive | MGI_C57BL6J_1923355 | 5830456J23Rik | NA | NA | unclassified gene | RIKEN cDNA 5830456J23 gene |
| 8 | gene | 81.665876 | 81.667118 | negative | MGI_C57BL6J_1923126 | 4930579O11Rik | ENSEMBL:ENSMUSG00000090732 | MGI:1923126 | lncRNA gene | RIKEN cDNA 4930579O11 gene |
| 8 | pseudogene | 81.790558 | 81.790908 | negative | MGI_C57BL6J_4937996 | Gm17169 | ENSEMBL:ENSMUSG00000090584 | MGI:4937996 | pseudogene | predicted gene 17169 |
| 8 | pseudogene | 81.827676 | 81.828356 | positive | MGI_C57BL6J_4937899 | Gm17072 | NCBI_Gene:100417496,ENSEMBL:ENSMUSG00000091998 | MGI:4937899 | pseudogene | predicted gene 17072 |
| 8 | gene | 82.152720 | 82.152782 | negative | MGI_C57BL6J_5455044 | Gm25267 | ENSEMBL:ENSMUSG00000089147 | MGI:5455044 | snRNA gene | predicted gene, 25267 |
| 8 | pseudogene | 82.160609 | 82.165639 | positive | MGI_C57BL6J_3779929 | Gm9520 | NCBI_Gene:671182,ENSEMBL:ENSMUSG00000109664 | MGI:3779929 | pseudogene | predicted gene 9520 |
| 8 | gene | 82.184799 | 82.207535 | negative | MGI_C57BL6J_5621475 | Gm38590 | NCBI_Gene:102641995,ENSEMBL:ENSMUSG00000110045 | MGI:5621475 | lncRNA gene | predicted gene, 38590 |
| 8 | gene | 82.331624 | 82.403272 | negative | MGI_C57BL6J_103014 | Il15 | NCBI_Gene:16168,ENSEMBL:ENSMUSG00000031712 | MGI:103014 | protein coding gene | interleukin 15 |
| 8 | gene | 82.403316 | 82.403719 | positive | MGI_C57BL6J_1924620 | 9530004M14Rik | NA | NA | lncRNA gene | RIKEN cDNA 9530004M14 gene |
| 8 | gene | 82.409091 | 82.409226 | positive | MGI_C57BL6J_5454915 | Gm25138 | ENSEMBL:ENSMUSG00000065182 | MGI:5454915 | snoRNA gene | predicted gene, 25138 |
| 8 | pseudogene | 82.577749 | 82.578902 | positive | MGI_C57BL6J_3644250 | Gm8167 | NCBI_Gene:666564,ENSEMBL:ENSMUSG00000097984 | MGI:3644250 | pseudogene | predicted gene 8167 |
| 8 | pseudogene | 82.747616 | 82.757987 | positive | MGI_C57BL6J_3780063 | Gm9655 | NCBI_Gene:675800,ENSEMBL:ENSMUSG00000109893 | MGI:3780063 | pseudogene | predicted gene 9655 |
| 8 | gene | 82.748281 | 82.755052 | negative | MGI_C57BL6J_3647995 | Gm8025 | NA | NA | unclassified gene | predicted gene 8025 |
| 8 | gene | 82.763607 | 82.774160 | negative | MGI_C57BL6J_1353574 | Zfp330 | NCBI_Gene:30932,ENSEMBL:ENSMUSG00000031711 | MGI:1353574 | protein coding gene | zinc finger protein 330 |
| 8 | gene | 82.796114 | 82.837534 | negative | MGI_C57BL6J_5590704 | Gm31545 | NCBI_Gene:102633811,ENSEMBL:ENSMUSG00000109643 | MGI:5590704 | lncRNA gene | predicted gene, 31545 |
| 8 | gene | 82.863013 | 83.091271 | positive | MGI_C57BL6J_2443860 | Rnf150 | NCBI_Gene:330812,ENSEMBL:ENSMUSG00000047747 | MGI:2443860 | protein coding gene | ring finger protein 150 |
| 8 | pseudogene | 82.896061 | 82.897291 | positive | MGI_C57BL6J_5825682 | Gm46045 | NCBI_Gene:108167540 | MGI:5825682 | pseudogene | predicted gene, 46045 |
| 8 | pseudogene | 82.952220 | 82.953412 | positive | MGI_C57BL6J_5010397 | Gm18212 | NCBI_Gene:100416715,ENSEMBL:ENSMUSG00000109600 | MGI:5010397 | pseudogene | predicted gene, 18212 |
| 8 | gene | 83.082258 | 83.086957 | negative | MGI_C57BL6J_5791285 | Gm45449 | ENSEMBL:ENSMUSG00000109876 | MGI:5791285 | lncRNA gene | predicted gene 45449 |
| 8 | gene | 83.165081 | 83.166170 | negative | MGI_C57BL6J_3704313 | Gm10645 | ENSEMBL:ENSMUSG00000074228 | MGI:3704313 | protein coding gene | predicted gene 10645 |
| 8 | gene | 83.165352 | 83.272940 | positive | MGI_C57BL6J_1918560 | Tbc1d9 | NCBI_Gene:71310,ENSEMBL:ENSMUSG00000031709 | MGI:1918560 | protein coding gene | TBC1 domain family, member 9 |
| 8 | gene | 83.222909 | 83.229996 | positive | MGI_C57BL6J_5624912 | Gm42027 | NCBI_Gene:105246802 | MGI:5624912 | lncRNA gene | predicted gene, 42027 |
| 8 | pseudogene | 83.267059 | 83.267628 | positive | MGI_C57BL6J_5010184 | Gm17999 | NCBI_Gene:100416245,ENSEMBL:ENSMUSG00000110422 | MGI:5010184 | pseudogene | predicted gene, 17999 |
| 8 | gene | 83.290348 | 83.298456 | positive | MGI_C57BL6J_98894 | Ucp1 | NCBI_Gene:22227,ENSEMBL:ENSMUSG00000031710 | MGI:98894 | protein coding gene | uncoupling protein 1 (mitochondrial, proton carrier) |
| 8 | gene | 83.312632 | 83.332486 | negative | MGI_C57BL6J_2445165 | Elmod2 | NCBI_Gene:244548,ENSEMBL:ENSMUSG00000035151 | MGI:2445165 | protein coding gene | ELMO/CED-12 domain containing 2 |
| 8 | gene | 83.332569 | 83.338952 | positive | MGI_C57BL6J_5591486 | Gm32327 | NCBI_Gene:102634836 | MGI:5591486 | lncRNA gene | predicted gene, 32327 |
| 8 | gene | 83.348471 | 83.382320 | positive | MGI_C57BL6J_1914805 | Mgat4d | NCBI_Gene:67555,ENSEMBL:ENSMUSG00000035057 | MGI:1914805 | protein coding gene | MGAT4 family, member C |
| 8 | pseudogene | 83.352556 | 83.353215 | positive | MGI_C57BL6J_3783102 | Gm15660 | NCBI_Gene:100416356,ENSEMBL:ENSMUSG00000082136 | MGI:3783102 | pseudogene | predicted gene 15660 |
| 8 | gene | 83.381469 | 83.389473 | negative | MGI_C57BL6J_5591536 | Gm32377 | NCBI_Gene:102634903 | MGI:5591536 | lncRNA gene | predicted gene, 32377 |
| 8 | gene | 83.389823 | 83.428552 | positive | MGI_C57BL6J_107472 | Clgn | NCBI_Gene:12745,ENSEMBL:ENSMUSG00000002190 | MGI:107472 | protein coding gene | calmegin |
| 8 | gene | 83.430084 | 83.458439 | negative | MGI_C57BL6J_1927654 | Scoc | NCBI_Gene:56367,ENSEMBL:ENSMUSG00000063253 | MGI:1927654 | protein coding gene | short coiled-coil protein |
| 8 | pseudogene | 83.477471 | 83.479216 | negative | MGI_C57BL6J_3646687 | Gm5910 | NCBI_Gene:546077,ENSEMBL:ENSMUSG00000110617 | MGI:3646687 | pseudogene | predicted gene 5910 |
| 8 | pseudogene | 83.489743 | 83.490829 | negative | MGI_C57BL6J_5592401 | Gm33242 | NCBI_Gene:102636069,ENSEMBL:ENSMUSG00000110448 | MGI:5592401 | pseudogene | predicted gene, 33242 |
| 8 | gene | 83.535780 | 83.545227 | positive | MGI_C57BL6J_3030204 | Olfr370 | NCBI_Gene:258267,ENSEMBL:ENSMUSG00000047286 | MGI:3030204 | protein coding gene | olfactory receptor 370 |
| 8 | pseudogene | 83.546440 | 83.548267 | positive | MGI_C57BL6J_5010185 | Gm18000 | NCBI_Gene:100416246,ENSEMBL:ENSMUSG00000110478 | MGI:5010185 | pseudogene | predicted gene, 18000 |
| 8 | gene | 83.566671 | 83.571626 | positive | MGI_C57BL6J_1914166 | Ndufb7 | NCBI_Gene:66916,ENSEMBL:ENSMUSG00000033938 | MGI:1914166 | protein coding gene | NADH:ubiquinone oxidoreductase subunit B7 |
| 8 | gene | 83.571698 | 83.607971 | negative | MGI_C57BL6J_1915408 | Tecr | NCBI_Gene:106529,ENSEMBL:ENSMUSG00000031708 | MGI:1915408 | protein coding gene | trans-2,3-enoyl-CoA reductase |
| 8 | gene | 83.576044 | 83.591856 | positive | MGI_C57BL6J_2442966 | D830024N08Rik | NCBI_Gene:105246804,ENSEMBL:ENSMUSG00000110580 | MGI:2442966 | lncRNA gene | RIKEN cDNA D830024N08 gene |
| 8 | gene | 83.584087 | 83.585525 | negative | MGI_C57BL6J_5804938 | Gm45823 | ENSEMBL:ENSMUSG00000110471 | MGI:5804938 | lncRNA gene | predicted gene 45823 |
| 8 | gene | 83.599835 | 83.608094 | negative | MGI_C57BL6J_5624914 | Gm42029 | NCBI_Gene:105246805 | MGI:5624914 | lncRNA gene | predicted gene, 42029 |
| 8 | pseudogene | 83.599941 | 83.600784 | positive | MGI_C57BL6J_5010186 | Gm18001 | NCBI_Gene:100416247,ENSEMBL:ENSMUSG00000110690 | MGI:5010186 | pseudogene | predicted gene, 18001 |
| 8 | gene | 83.608175 | 83.612653 | positive | MGI_C57BL6J_1931874 | Dnajb1 | NCBI_Gene:81489,ENSEMBL:ENSMUSG00000005483 | MGI:1931874 | protein coding gene | DnaJ heat shock protein family (Hsp40) member B1 |
| 8 | gene | 83.652677 | 83.664789 | positive | MGI_C57BL6J_1926252 | Gipc1 | NCBI_Gene:67903,ENSEMBL:ENSMUSG00000019433 | MGI:1926252 | protein coding gene | GIPC PDZ domain containing family, member 1 |
| 8 | gene | 83.666533 | 83.699179 | negative | MGI_C57BL6J_108022 | Pkn1 | NCBI_Gene:320795,ENSEMBL:ENSMUSG00000057672 | MGI:108022 | protein coding gene | protein kinase N1 |
| 8 | gene | 83.666640 | 83.672756 | positive | MGI_C57BL6J_97793 | Ptger1 | NCBI_Gene:19216,ENSEMBL:ENSMUSG00000019464 | MGI:97793 | protein coding gene | prostaglandin E receptor 1 (subtype EP1) |
| 8 | gene | 83.715177 | 83.726892 | positive | MGI_C57BL6J_1915528 | Ddx39 | NCBI_Gene:68278,ENSEMBL:ENSMUSG00000005481 | MGI:1915528 | protein coding gene | DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 |
| 8 | gene | 83.723251 | 83.741326 | negative | MGI_C57BL6J_1347095 | Adgre5 | NCBI_Gene:26364,ENSEMBL:ENSMUSG00000002885 | MGI:1347095 | protein coding gene | adhesion G protein-coupled receptor E5 |
| 8 | gene | 83.723878 | 83.723984 | negative | MGI_C57BL6J_5530972 | Mir1668 | miRBase:MI0026040,NCBI_Gene:102465897,ENSEMBL:ENSMUSG00000098465 | MGI:5530972 | miRNA gene | microRNA 1668 |
| 8 | gene | 83.741283 | 83.745645 | positive | MGI_C57BL6J_5825683 | Gm46046 | NCBI_Gene:108167541 | MGI:5825683 | lncRNA gene | predicted gene, 46046 |
| 8 | pseudogene | 83.749504 | 83.749656 | positive | MGI_C57BL6J_5804893 | Gm45778 | ENSEMBL:ENSMUSG00000110570 | MGI:5804893 | pseudogene | predicted gene 45778 |
| 8 | gene | 83.752918 | 83.753204 | positive | MGI_C57BL6J_5451801 | Gm22024 | ENSEMBL:ENSMUSG00000084682 | MGI:5451801 | unclassified non-coding RNA gene | predicted gene, 22024 |
| 8 | gene | 83.762019 | 83.762941 | negative | MGI_C57BL6J_5624915 | Gm42030 | NCBI_Gene:105246806 | MGI:5624915 | lncRNA gene | predicted gene, 42030 |
| 8 | gene | 83.887079 | 83.899771 | negative | MGI_C57BL6J_5477215 | Gm26721 | NCBI_Gene:105243235,ENSEMBL:ENSMUSG00000096943 | MGI:5477215 | lncRNA gene | predicted gene, 26721 |
| 8 | pseudogene | 83.891772 | 83.892305 | positive | MGI_C57BL6J_3646765 | Gm5355 | NCBI_Gene:384857 | MGI:3646765 | pseudogene | predicted gene 5355 |
| 8 | gene | 83.900098 | 83.941954 | positive | MGI_C57BL6J_1929461 | Adgrl1 | NCBI_Gene:330814,ENSEMBL:ENSMUSG00000013033 | MGI:1929461 | protein coding gene | adhesion G protein-coupled receptor L1 |
| 8 | gene | 83.929443 | 83.929556 | negative | MGI_C57BL6J_5690842 | Gm44450 | ENSEMBL:ENSMUSG00000105569 | MGI:5690842 | miRNA gene | predicted gene, 44450 |
| 8 | gene | 83.929688 | 83.929830 | negative | MGI_C57BL6J_5690722 | Gm44330 | ENSEMBL:ENSMUSG00000105878 | MGI:5690722 | miRNA gene | predicted gene, 44330 |
| 8 | gene | 83.932329 | 83.955428 | negative | MGI_C57BL6J_3704314 | Gm10644 | NCBI_Gene:100126034,ENSEMBL:ENSMUSG00000074219 | MGI:3704314 | protein coding gene | predicted gene 10644 |
| 8 | gene | 83.955507 | 83.970197 | positive | MGI_C57BL6J_1914179 | Asf1b | NCBI_Gene:66929,ENSEMBL:ENSMUSG00000005470 | MGI:1914179 | protein coding gene | anti-silencing function 1B histone chaperone |
| 8 | gene | 83.972978 | 83.996444 | positive | MGI_C57BL6J_97592 | Prkaca | NCBI_Gene:18747,ENSEMBL:ENSMUSG00000005469 | MGI:97592 | protein coding gene | protein kinase, cAMP dependent, catalytic, alpha |
| 8 | gene | 83.997672 | 84.000409 | positive | MGI_C57BL6J_2142433 | Samd1 | NCBI_Gene:666704,ENSEMBL:ENSMUSG00000079003 | MGI:2142433 | protein coding gene | sterile alpha motif domain containing 1 |
| 8 | gene | 84.000516 | 84.004770 | positive | MGI_C57BL6J_1920703 | 1700067K01Rik | NCBI_Gene:73453,ENSEMBL:ENSMUSG00000046408 | MGI:1920703 | protein coding gene | RIKEN cDNA 1700067K01 gene |
| 8 | gene | 84.005628 | 84.005764 | positive | MGI_C57BL6J_5531205 | Mir8111 | miRBase:MI0026043,NCBI_Gene:102466886,ENSEMBL:ENSMUSG00000098823 | MGI:5531205 | miRNA gene | microRNA 8111 |
| 8 | gene | 84.010222 | 84.012171 | negative | MGI_C57BL6J_1917384 | Misp3 | NCBI_Gene:70134,ENSEMBL:ENSMUSG00000074217 | MGI:1917384 | protein coding gene | MISP family member 3 |
| 8 | gene | 84.011515 | 84.011633 | negative | MGI_C57BL6J_3783372 | Mir1199 | miRBase:MI0006307,NCBI_Gene:100316677,ENSEMBL:ENSMUSG00000080665 | MGI:3783372 | miRNA gene | microRNA 1199 |
| 8 | gene | 84.021045 | 84.030295 | positive | MGI_C57BL6J_1921587 | Palm3 | NCBI_Gene:74337,ENSEMBL:ENSMUSG00000047986 | MGI:1921587 | protein coding gene | paralemmin 3 |
| 8 | gene | 84.030286 | 84.042629 | negative | MGI_C57BL6J_1355318 | Il27ra | NCBI_Gene:50931,ENSEMBL:ENSMUSG00000005465 | MGI:1355318 | protein coding gene | interleukin 27 receptor, alpha |
| 8 | gene | 84.042929 | 84.045000 | negative | MGI_C57BL6J_2158015 | Rln3 | NCBI_Gene:212108,ENSEMBL:ENSMUSG00000045232 | MGI:2158015 | protein coding gene | relaxin 3 |
| 8 | gene | 84.063561 | 84.065137 | negative | MGI_C57BL6J_3642507 | Gm10643 | ENSEMBL:ENSMUSG00000074215 | MGI:3642507 | protein coding gene | predicted gene 10643 |
| 8 | gene | 84.065236 | 84.067287 | negative | MGI_C57BL6J_3045248 | C330011M18Rik | ENSEMBL:ENSMUSG00000056753 | MGI:3045248 | lncRNA gene | RIKEN cDNA C330011M18 gene |
| 8 | gene | 84.066834 | 84.096995 | positive | MGI_C57BL6J_105982 | Rfx1 | NCBI_Gene:19724,ENSEMBL:ENSMUSG00000031706 | MGI:105982 | protein coding gene | regulatory factor X, 1 (influences HLA class II expression) |
| 8 | gene | 84.086099 | 84.086186 | positive | MGI_C57BL6J_3629717 | Mir709 | miRBase:MI0004693,NCBI_Gene:735271,ENSEMBL:ENSMUSG00000076144 | MGI:3629717 | miRNA gene | microRNA 709 |
| 8 | gene | 84.097072 | 84.104792 | negative | MGI_C57BL6J_2684420 | Dcaf15 | NCBI_Gene:212123,ENSEMBL:ENSMUSG00000037103 | MGI:2684420 | protein coding gene | DDB1 and CUL4 associated factor 15 |
| 8 | gene | 84.099436 | 84.100525 | positive | MGI_C57BL6J_5622095 | Gm39210 | NCBI_Gene:105243236 | MGI:5622095 | lncRNA gene | predicted gene, 39210 |
| 8 | gene | 84.125927 | 84.143017 | positive | MGI_C57BL6J_2685352 | Podnl1 | NCBI_Gene:244550,ENSEMBL:ENSMUSG00000012889 | MGI:2685352 | protein coding gene | podocan-like 1 |
| 8 | gene | 84.132828 | 84.147936 | negative | MGI_C57BL6J_2384831 | Cc2d1a | NCBI_Gene:212139,ENSEMBL:ENSMUSG00000036686 | MGI:2384831 | protein coding gene | coiled-coil and C2 domain containing 1A |
| 8 | gene | 84.147947 | 84.172597 | positive | MGI_C57BL6J_1921916 | 4930432K21Rik | NCBI_Gene:74666,ENSEMBL:ENSMUSG00000008129 | MGI:1921916 | protein coding gene | RIKEN cDNA 4930432K21 gene |
| 8 | gene | 84.156757 | 84.156877 | positive | MGI_C57BL6J_5454887 | Gm25110 | ENSEMBL:ENSMUSG00000080334 | MGI:5454887 | miRNA gene | predicted gene, 25110 |
| 8 | gene | 84.173730 | 84.185217 | negative | MGI_C57BL6J_2675387 | Nanos3 | NCBI_Gene:244551,ENSEMBL:ENSMUSG00000056155 | MGI:2675387 | protein coding gene | nanos C2HC-type zinc finger 3 |
| 8 | gene | 84.178716 | 84.178787 | negative | MGI_C57BL6J_3709847 | Mir181d | miRBase:MI0005450,NCBI_Gene:100049549,ENSEMBL:ENSMUSG00000076338 | MGI:3709847 | miRNA gene | microRNA 181d |
| 8 | gene | 84.178873 | 84.178961 | negative | MGI_C57BL6J_3618737 | Mir181c | miRBase:MI0000724,NCBI_Gene:723819,ENSEMBL:ENSMUSG00000065483 | MGI:3618737 | miRNA gene | microRNA 181c |
| 8 | gene | 84.192774 | 84.197667 | negative | MGI_C57BL6J_5477381 | Gm26887 | ENSEMBL:ENSMUSG00000097120 | MGI:5477381 | lncRNA gene | predicted gene, 26887 |
| 8 | gene | 84.200869 | 84.203049 | positive | MGI_C57BL6J_5610580 | Gm37352 | ENSEMBL:ENSMUSG00000103593 | MGI:5610580 | unclassified gene | predicted gene, 37352 |
| 8 | gene | 84.208518 | 84.208592 | positive | MGI_C57BL6J_2676897 | Mir23a | miRBase:MI0000571,NCBI_Gene:387216,ENSEMBL:ENSMUSG00000065611 | MGI:2676897 | miRNA gene | microRNA 23a |
| 8 | gene | 84.208672 | 84.208758 | positive | MGI_C57BL6J_2676902 | Mir27a | miRBase:MI0000578,NCBI_Gene:387220,ENSEMBL:ENSMUSG00000065444 | MGI:2676902 | miRNA gene | microRNA 27a |
| 8 | gene | 84.208815 | 84.208921 | positive | MGI_C57BL6J_3618755 | Mir24-2 | miRBase:MI0000572,NCBI_Gene:723960,ENSEMBL:ENSMUSG00000065541 | MGI:3618755 | miRNA gene | microRNA 24-2 |
| 8 | gene | 84.208825 | 84.208907 | negative | MGI_C57BL6J_4834326 | Mir3074-2 | miRBase:MI0014104,NCBI_Gene:100526528,ENSEMBL:ENSMUSG00000105458 | MGI:4834326 | miRNA gene | microRNA 3074-2 |
| 8 | gene | 84.208920 | 84.209609 | positive | MGI_C57BL6J_5477026 | Gm26532 | ENSEMBL:ENSMUSG00000097296 | MGI:5477026 | lncRNA gene | predicted gene, 26532 |
| 8 | gene | 84.210678 | 84.237055 | negative | MGI_C57BL6J_2443726 | Zswim4 | NCBI_Gene:212168,ENSEMBL:ENSMUSG00000035671 | MGI:2443726 | protein coding gene | zinc finger SWIM-type containing 4 |
| 8 | gene | 84.235112 | 84.238340 | positive | MGI_C57BL6J_5591919 | Gm32760 | NCBI_Gene:102635413 | MGI:5591919 | lncRNA gene | predicted gene, 32760 |
| 8 | gene | 84.246233 | 84.251711 | negative | MGI_C57BL6J_1289231 | D8Ertd738e | NCBI_Gene:101966,ENSEMBL:ENSMUSG00000019362 | MGI:1289231 | protein coding gene | DNA segment, Chr 8, ERATO Doi 738, expressed |
| 8 | gene | 84.249906 | 84.257326 | negative | MGI_C57BL6J_1915123 | Mri1 | NCBI_Gene:67873,ENSEMBL:ENSMUSG00000004996 | MGI:1915123 | protein coding gene | methylthioribose-1-phosphate isomerase 1 |
| 8 | gene | 84.257767 | 84.270458 | negative | MGI_C57BL6J_1914986 | Ccdc130 | NCBI_Gene:67736,ENSEMBL:ENSMUSG00000004994 | MGI:1914986 | protein coding gene | coiled-coil domain containing 130 |
| 8 | pseudogene | 84.280876 | 84.281604 | negative | MGI_C57BL6J_5010862 | Gm18677 | NCBI_Gene:100417543 | MGI:5010862 | pseudogene | predicted gene, 18677 |
| 8 | pseudogene | 84.302963 | 84.303733 | negative | MGI_C57BL6J_5791444 | Gm45608 | ENSEMBL:ENSMUSG00000109653 | MGI:5791444 | pseudogene | predicted gene 45608 |
| 8 | gene | 84.338630 | 84.640251 | positive | MGI_C57BL6J_109482 | Cacna1a | NCBI_Gene:12286,ENSEMBL:ENSMUSG00000034656 | MGI:109482 | protein coding gene | calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| 8 | gene | 84.380449 | 84.380581 | negative | MGI_C57BL6J_5453403 | Gm23626 | ENSEMBL:ENSMUSG00000064392 | MGI:5453403 | snoRNA gene | predicted gene, 23626 |
| 8 | gene | 84.583040 | 84.588733 | negative | MGI_C57BL6J_5592013 | Gm32854 | NCBI_Gene:102635550 | MGI:5592013 | lncRNA gene | predicted gene, 32854 |
| 8 | gene | 84.611202 | 84.617184 | negative | MGI_C57BL6J_3802174 | Gm16183 | ENSEMBL:ENSMUSG00000086067 | MGI:3802174 | lncRNA gene | predicted gene 16183 |
| 8 | gene | 84.624087 | 84.639255 | negative | MGI_C57BL6J_5622097 | Gm39212 | NCBI_Gene:105243238 | MGI:5622097 | lncRNA gene | predicted gene, 39212 |
| 8 | gene | 84.642119 | 84.643273 | positive | MGI_C57BL6J_5622096 | Gm39211 | NCBI_Gene:105243237 | MGI:5622096 | lncRNA gene | predicted gene, 39211 |
| 8 | gene | 84.661331 | 84.662854 | negative | MGI_C57BL6J_104815 | Ier2 | NCBI_Gene:15936,ENSEMBL:ENSMUSG00000053560 | MGI:104815 | protein coding gene | immediate early response 2 |
| 8 | gene | 84.663269 | 84.665026 | negative | MGI_C57BL6J_2148235 | 4930447I22Rik | NA | NA | unclassified gene | RIKEN cDNA 4930447I22 gene |
| 8 | gene | 84.665128 | 84.669406 | positive | MGI_C57BL6J_5477158 | Gm26664 | ENSEMBL:ENSMUSG00000097193 | MGI:5477158 | lncRNA gene | predicted gene, 26664 |
| 8 | pseudogene | 84.665445 | 84.668745 | positive | MGI_C57BL6J_6324731 | Gm50464 | ENSEMBL:ENSMUSG00000118487 | MGI:6324731 | pseudogene | predicted gene, 50464 |
| 8 | gene | 84.670477 | 84.687908 | negative | MGI_C57BL6J_1914080 | Nacc1 | NCBI_Gene:66830,ENSEMBL:ENSMUSG00000001910 | MGI:1914080 | protein coding gene | nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| 8 | gene | 84.686307 | 84.699808 | positive | MGI_C57BL6J_1289155 | Trmt1 | NCBI_Gene:212528,ENSEMBL:ENSMUSG00000001909 | MGI:1289155 | protein coding gene | tRNA methyltransferase 1 |
| 8 | gene | 84.699876 | 84.800344 | negative | MGI_C57BL6J_97311 | Nfix | NCBI_Gene:18032,ENSEMBL:ENSMUSG00000001911 | MGI:97311 | protein coding gene | nuclear factor I/X |
| 8 | gene | 84.701422 | 84.704940 | positive | MGI_C57BL6J_96891 | Lyl1 | NCBI_Gene:17095,ENSEMBL:ENSMUSG00000034041 | MGI:96891 | protein coding gene | lymphoblastomic leukemia 1 |
| 8 | gene | 84.721825 | 84.726844 | positive | MGI_C57BL6J_3588227 | G430095P16Rik | NCBI_Gene:619292,ENSEMBL:ENSMUSG00000074203 | MGI:3588227 | protein coding gene | RIKEN cDNA G430095P16 gene |
| 8 | gene | 84.815404 | 84.832409 | negative | MGI_C57BL6J_1344365 | Dand5 | NCBI_Gene:23863,ENSEMBL:ENSMUSG00000053226 | MGI:1344365 | protein coding gene | DAN domain family member 5, BMP antagonist |
| 8 | gene | 84.831522 | 84.835482 | positive | MGI_C57BL6J_1914947 | Gadd45gip1 | NCBI_Gene:102060,ENSEMBL:ENSMUSG00000033751 | MGI:1914947 | protein coding gene | growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| 8 | gene | 84.834019 | 84.840748 | negative | MGI_C57BL6J_105126 | Rad23a | NCBI_Gene:19358,ENSEMBL:ENSMUSG00000003813 | MGI:105126 | protein coding gene | RAD23 homolog A, nucleotide excision repair protein |
| 8 | gene | 84.841850 | 84.846934 | negative | MGI_C57BL6J_88252 | Calr | NCBI_Gene:12317,ENSEMBL:ENSMUSG00000003814 | MGI:88252 | protein coding gene | calreticulin |
| 8 | gene | 84.847143 | 84.856462 | positive | MGI_C57BL6J_1920869 | 1700122E12Rik | NCBI_Gene:73619,ENSEMBL:ENSMUSG00000087685 | MGI:1920869 | lncRNA gene | RIKEN cDNA 1700122E12 gene |
| 8 | pseudogene | 84.853959 | 84.854609 | negative | MGI_C57BL6J_3645263 | Gm5911 | NCBI_Gene:546080 | MGI:3645263 | pseudogene | predicted gene 5911 |
| 8 | gene | 84.856970 | 84.869257 | positive | MGI_C57BL6J_1913840 | Farsa | NCBI_Gene:66590,ENSEMBL:ENSMUSG00000003808 | MGI:1913840 | protein coding gene | phenylalanyl-tRNA synthetase, alpha subunit |
| 8 | gene | 84.867947 | 84.868008 | positive | MGI_C57BL6J_5531159 | Mir7069 | miRBase:MI0022919,NCBI_Gene:102466220,ENSEMBL:ENSMUSG00000098331 | MGI:5531159 | miRNA gene | microRNA 7069 |
| 8 | gene | 84.872111 | 84.888221 | positive | MGI_C57BL6J_1919096 | Syce2 | NCBI_Gene:71846,ENSEMBL:ENSMUSG00000003824 | MGI:1919096 | protein coding gene | synaptonemal complex central element protein 2 |
| 8 | gene | 84.886387 | 84.893921 | negative | MGI_C57BL6J_104541 | Gcdh | NCBI_Gene:270076,ENSEMBL:ENSMUSG00000003809 | MGI:104541 | protein coding gene | glutaryl-Coenzyme A dehydrogenase |
| 8 | gene | 84.894018 | 84.900154 | positive | MGI_C57BL6J_5622098 | Gm39213 | NCBI_Gene:105243239 | MGI:5622098 | lncRNA gene | predicted gene, 39213 |
| 8 | gene | 84.901928 | 84.905295 | positive | MGI_C57BL6J_1342771 | Klf1 | NCBI_Gene:16596,ENSEMBL:ENSMUSG00000054191 | MGI:1342771 | protein coding gene | Kruppel-like factor 1 (erythroid) |
| 8 | gene | 84.908560 | 84.922915 | positive | MGI_C57BL6J_1329019 | Dnase2a | NCBI_Gene:13423,ENSEMBL:ENSMUSG00000003812 | MGI:1329019 | protein coding gene | deoxyribonuclease II alpha |
| 8 | gene | 84.911853 | 84.946131 | negative | MGI_C57BL6J_1861901 | Mast1 | NCBI_Gene:56527,ENSEMBL:ENSMUSG00000053693 | MGI:1861901 | protein coding gene | microtubule associated serine/threonine kinase 1 |
| 8 | gene | 84.913335 | 84.922915 | positive | MGI_C57BL6J_5621311 | Gm38426 | NCBI_Gene:100503676 | MGI:5621311 | lncRNA gene | predicted gene, 38426 |
| 8 | gene | 84.925525 | 84.925633 | negative | MGI_C57BL6J_5453974 | Gm24197 | ENSEMBL:ENSMUSG00000092826 | MGI:5453974 | miRNA gene | predicted gene, 24197 |
| 8 | gene | 84.946991 | 84.956606 | positive | MGI_C57BL6J_2443686 | Rtbdn | NCBI_Gene:234542,ENSEMBL:ENSMUSG00000048617 | MGI:2443686 | protein coding gene | retbindin |
| 8 | gene | 84.956610 | 84.969767 | negative | MGI_C57BL6J_1916974 | Rnaseh2a | NCBI_Gene:69724,ENSEMBL:ENSMUSG00000052926 | MGI:1916974 | protein coding gene | ribonuclease H2, large subunit |
| 8 | gene | 84.967678 | 84.969433 | negative | MGI_C57BL6J_6215100 | Gm49661 | ENSEMBL:ENSMUSG00000111184 | MGI:6215100 | protein coding gene | predicted gene, 49661 |
| 8 | gene | 84.969587 | 84.974834 | positive | MGI_C57BL6J_109486 | Prdx2 | NCBI_Gene:21672,ENSEMBL:ENSMUSG00000005161 | MGI:109486 | protein coding gene | peroxiredoxin 2 |
| 8 | gene | 84.974484 | 84.978748 | negative | MGI_C57BL6J_96647 | Junb | NCBI_Gene:16477,ENSEMBL:ENSMUSG00000052837 | MGI:96647 | protein coding gene | jun B proto-oncogene |
| 8 | gene | 84.990573 | 85.003364 | positive | MGI_C57BL6J_2181664 | Hook2 | NCBI_Gene:170833,ENSEMBL:ENSMUSG00000052566 | MGI:2181664 | protein coding gene | hook microtubule tethering protein 2 |
| 8 | gene | 85.007202 | 85.014531 | negative | MGI_C57BL6J_2387588 | Best2 | NCBI_Gene:212989,ENSEMBL:ENSMUSG00000052819 | MGI:2387588 | protein coding gene | bestrophin 2 |
| 8 | gene | 85.017931 | 85.025281 | negative | MGI_C57BL6J_1928379 | Asna1 | NCBI_Gene:56495,ENSEMBL:ENSMUSG00000052456 | MGI:1928379 | protein coding gene | arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) |
| 8 | gene | 85.026812 | 85.030286 | positive | MGI_C57BL6J_1922833 | Trir | NCBI_Gene:68544,ENSEMBL:ENSMUSG00000041203 | MGI:1922833 | protein coding gene | telomerase RNA component interacting RNase |
| 8 | gene | 85.036870 | 85.057585 | positive | MGI_C57BL6J_2384849 | Tnpo2 | NCBI_Gene:212999,ENSEMBL:ENSMUSG00000031691 | MGI:2384849 | protein coding gene | transportin 2 (importin 3, karyopherin beta 2b) |
| 8 | gene | 85.050207 | 85.050276 | positive | MGI_C57BL6J_5455283 | Gm25506 | ENSEMBL:ENSMUSG00000077527 | MGI:5455283 | snoRNA gene | predicted gene, 25506 |
| 8 | gene | 85.053160 | 85.060460 | negative | MGI_C57BL6J_2444082 | A230103J11Rik | NCBI_Gene:320466,ENSEMBL:ENSMUSG00000087026 | MGI:2444082 | lncRNA gene | RIKEN cDNA A230103J11 gene |
| 8 | gene | 85.060024 | 85.067124 | positive | MGI_C57BL6J_1915878 | Fbxw9 | NCBI_Gene:68628,ENSEMBL:ENSMUSG00000008167 | MGI:1915878 | protein coding gene | F-box and WD-40 domain protein 9 |
| 8 | gene | 85.062086 | 85.062171 | positive | MGI_C57BL6J_5530947 | Mir7070 | miRBase:MI0022920,NCBI_Gene:102465648,ENSEMBL:ENSMUSG00000099076 | MGI:5530947 | miRNA gene | microRNA 7070 |
| 8 | gene | 85.067568 | 85.067982 | negative | MGI_C57BL6J_3645690 | Gm5741 | NCBI_Gene:100503710,ENSEMBL:ENSMUSG00000095845 | MGI:3645690 | protein coding gene | predicted gene 5741 |
| 8 | gene | 85.069723 | 85.075162 | positive | MGI_C57BL6J_2683592 | Dhps | NCBI_Gene:330817,ENSEMBL:ENSMUSG00000060038 | MGI:2683592 | protein coding gene | deoxyhypusine synthase |
| 8 | gene | 85.075035 | 85.081306 | negative | MGI_C57BL6J_1915086 | Wdr83 | NCBI_Gene:67836,ENSEMBL:ENSMUSG00000005150 | MGI:1915086 | protein coding gene | WD repeat domain containing 83 |
| 8 | gene | 85.080740 | 85.082339 | positive | MGI_C57BL6J_3041257 | Wdr83os | NCBI_Gene:414077,ENSEMBL:ENSMUSG00000059355 | MGI:3041257 | protein coding gene | WD repeat domain 83 opposite strand |
| 8 | gene | 85.082729 | 85.098739 | positive | MGI_C57BL6J_107286 | Man2b1 | NCBI_Gene:17159,ENSEMBL:ENSMUSG00000005142 | MGI:107286 | protein coding gene | mannosidase 2, alpha B1 |
| 8 | gene | 85.107168 | 85.123095 | negative | MGI_C57BL6J_3648473 | Zfp791 | NCBI_Gene:244556,ENSEMBL:ENSMUSG00000074194 | MGI:3648473 | protein coding gene | zinc finger protein 791 |
| 8 | gene | 85.112012 | 85.112068 | negative | MGI_C57BL6J_5453165 | Gm23388 | ENSEMBL:ENSMUSG00000089213 | MGI:5453165 | miRNA gene | predicted gene, 23388 |
| 8 | pseudogene | 85.122728 | 85.122950 | positive | MGI_C57BL6J_5791153 | Gm45317 | ENSEMBL:ENSMUSG00000110219 | MGI:5791153 | pseudogene | predicted gene 45317 |
| 8 | gene | 85.125872 | 85.132289 | negative | MGI_C57BL6J_5624916 | Gm42031 | NCBI_Gene:105246807,ENSEMBL:ENSMUSG00000110386 | MGI:5624916 | lncRNA gene | predicted gene, 42031 |
| 8 | pseudogene | 85.135765 | 85.135926 | positive | MGI_C57BL6J_5791152 | Gm45316 | ENSEMBL:ENSMUSG00000110200 | MGI:5791152 | pseudogene | predicted gene 45316 |
| 8 | gene | 85.151924 | 85.173622 | positive | MGI_C57BL6J_3643620 | Cks1brt | NCBI_Gene:624855,ENSEMBL:ENSMUSG00000062687 | MGI:3643620 | protein coding gene | CDC28 protein kinase 1b, retrogene |
| 8 | gene | 85.152065 | 85.159145 | negative | MGI_C57BL6J_5592455 | Gm33296 | NCBI_Gene:102636144 | MGI:5592455 | lncRNA gene | predicted gene, 33296 |
| 8 | pseudogene | 85.154218 | 85.154923 | negative | MGI_C57BL6J_5010322 | Gm18137 | NCBI_Gene:100416509 | MGI:5010322 | pseudogene | predicted gene, 18137 |
| 8 | gene | 85.225988 | 85.231509 | positive | MGI_C57BL6J_3030205 | Olfr371 | NCBI_Gene:258858,ENSEMBL:ENSMUSG00000051952 | MGI:3030205 | protein coding gene | olfactory receptor 371 |
| 8 | gene | 85.260392 | 85.299802 | negative | MGI_C57BL6J_1890467 | Vps35 | NCBI_Gene:65114,ENSEMBL:ENSMUSG00000031696 | MGI:1890467 | protein coding gene | VPS35 retromer complex component |
| 8 | gene | 85.299632 | 85.308279 | positive | MGI_C57BL6J_1929285 | Orc6 | NCBI_Gene:56452,ENSEMBL:ENSMUSG00000031697 | MGI:1929285 | protein coding gene | origin recognition complex, subunit 6 |
| 8 | gene | 85.315140 | 85.315246 | positive | MGI_C57BL6J_5452113 | Gm22336 | ENSEMBL:ENSMUSG00000088344 | MGI:5452113 | snRNA gene | predicted gene, 22336 |
| 8 | gene | 85.324303 | 85.386345 | negative | MGI_C57BL6J_2443063 | Mylk3 | NCBI_Gene:213435,ENSEMBL:ENSMUSG00000031698 | MGI:2443063 | protein coding gene | myosin light chain kinase 3 |
| 8 | gene | 85.339343 | 85.340272 | positive | MGI_C57BL6J_2142438 | AA672651 | ENSEMBL:ENSMUSG00000084895 | MGI:2142438 | lncRNA gene | expressed sequence AA672651 |
| 8 | gene | 85.408759 | 85.432841 | negative | MGI_C57BL6J_1913964 | 4921524J17Rik | NCBI_Gene:66714,ENSEMBL:ENSMUSG00000036934 | MGI:1913964 | protein coding gene | RIKEN cDNA 4921524J17 gene |
| 8 | pseudogene | 85.414275 | 85.414515 | negative | MGI_C57BL6J_5791430 | Gm45594 | ENSEMBL:ENSMUSG00000109790 | MGI:5791430 | pseudogene | predicted gene 45594 |
| 8 | gene | 85.432686 | 85.433323 | positive | MGI_C57BL6J_3704315 | 1700051O22Rik | ENSEMBL:ENSMUSG00000103341 | MGI:3704315 | unclassified gene | RIKEN cDNA 1700051O22 Gene |
| 8 | gene | 85.492576 | 85.527560 | positive | MGI_C57BL6J_1915391 | Gpt2 | NCBI_Gene:108682,ENSEMBL:ENSMUSG00000031700 | MGI:1915391 | protein coding gene | glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| 8 | gene | 85.537633 | 85.555344 | negative | MGI_C57BL6J_1931882 | Dnaja2 | NCBI_Gene:56445,ENSEMBL:ENSMUSG00000031701 | MGI:1931882 | protein coding gene | DnaJ heat shock protein family (Hsp40) member A2 |
| 8 | gene | 85.599832 | 85.603131 | positive | MGI_C57BL6J_5791260 | Gm45424 | ENSEMBL:ENSMUSG00000110341 | MGI:5791260 | unclassified gene | predicted gene 45424 |
| 8 | gene | 85.602712 | 85.607270 | negative | MGI_C57BL6J_5592526 | Gm33367 | NCBI_Gene:102636248 | MGI:5592526 | lncRNA gene | predicted gene, 33367 |
| 8 | gene | 85.605462 | 85.610761 | positive | MGI_C57BL6J_5791261 | Gm45425 | ENSEMBL:ENSMUSG00000109731 | MGI:5791261 | lncRNA gene | predicted gene 45425 |
| 8 | gene | 85.636588 | 85.700924 | negative | MGI_C57BL6J_1921763 | Neto2 | NCBI_Gene:74513,ENSEMBL:ENSMUSG00000036902 | MGI:1921763 | protein coding gene | neuropilin (NRP) and tolloid (TLL)-like 2 |
| 8 | gene | 85.700905 | 85.701021 | positive | MGI_C57BL6J_5530877 | Mir8109 | miRBase:MI0026041,NCBI_Gene:102465898,ENSEMBL:ENSMUSG00000098388 | MGI:5530877 | miRNA gene | microRNA 8109 |
| 8 | gene | 85.701074 | 85.701826 | positive | MGI_C57BL6J_1916195 | 1500002J14Rik | NA | NA | unclassified gene | RIKEN cDNA 1500002J14 gene |
| 8 | gene | 85.717557 | 85.840949 | negative | MGI_C57BL6J_106419 | Itfg1 | NCBI_Gene:71927,ENSEMBL:ENSMUSG00000031703 | MGI:106419 | protein coding gene | integrin alpha FG-GAP repeat containing 1 |
| 8 | gene | 85.728374 | 85.765893 | positive | MGI_C57BL6J_5622099 | Gm39214 | NCBI_Gene:105243241,ENSEMBL:ENSMUSG00000109754 | MGI:5622099 | lncRNA gene | predicted gene, 39214 |
| 8 | pseudogene | 85.824313 | 85.824603 | positive | MGI_C57BL6J_107703 | Atp5k-ps1 | NCBI_Gene:101055750 | MGI:107703 | pseudogene | ATP synthase, H+ transporting, mitochondrial F1F0 complex, subunit E, pseudogene 1 |
| 8 | gene | 85.840939 | 86.064251 | positive | MGI_C57BL6J_97578 | Phkb | NCBI_Gene:102093,ENSEMBL:ENSMUSG00000036879 | MGI:97578 | protein coding gene | phosphorylase kinase beta |
| 8 | pseudogene | 85.853373 | 85.854437 | negative | MGI_C57BL6J_3802049 | Gm16167 | NCBI_Gene:100418211,ENSEMBL:ENSMUSG00000090043 | MGI:3802049 | pseudogene | predicted gene 16167 |
| 8 | pseudogene | 85.956060 | 85.956795 | negative | MGI_C57BL6J_5011356 | Gm19171 | NCBI_Gene:100418380 | MGI:5011356 | pseudogene | predicted gene, 19171 |
| 8 | gene | 85.994245 | 85.996642 | positive | MGI_C57BL6J_5440479 | Hm629797 | ENSEMBL:ENSMUSG00000098439 | MGI:5440479 | lncRNA gene | cDNA sequence HM629797 |
| 8 | gene | 86.063341 | 86.064250 | positive | MGI_C57BL6J_1917238 | 2010205J10Rik | NA | NA | unclassified gene | RIKEN cDNA 2010205J10 gene |
| 8 | pseudogene | 86.114535 | 86.118504 | negative | MGI_C57BL6J_5010659 | Gm18474 | NCBI_Gene:100417236 | MGI:5010659 | pseudogene | predicted gene, 18474 |
| 8 | pseudogene | 86.146153 | 86.147294 | negative | MGI_C57BL6J_5434385 | Gm21030 | NCBI_Gene:100534332 | MGI:5434385 | pseudogene | predicted gene, 21030 |
| 8 | gene | 86.158243 | 86.212023 | positive | MGI_C57BL6J_5592791 | Gm33632 | NCBI_Gene:102636621 | MGI:5592791 | lncRNA gene | predicted gene, 33632 |
| 8 | gene | 86.208021 | 86.211211 | negative | MGI_C57BL6J_5592736 | Gm33577 | NCBI_Gene:102636541 | MGI:5592736 | lncRNA gene | predicted gene, 33577 |
| 8 | gene | 86.233854 | 86.233977 | positive | MGI_C57BL6J_5453681 | Gm23904 | ENSEMBL:ENSMUSG00000088192 | MGI:5453681 | snoRNA gene | predicted gene, 23904 |
| 8 | pseudogene | 86.256555 | 86.258369 | negative | MGI_C57BL6J_3780809 | Gm2641 | NCBI_Gene:100040165 | MGI:3780809 | pseudogene | predicted gene 2641 |
| 8 | gene | 86.279797 | 86.284080 | positive | MGI_C57BL6J_5592846 | Gm33687 | NCBI_Gene:102636685 | MGI:5592846 | lncRNA gene | predicted gene, 33687 |
| 8 | gene | 86.292501 | 86.292598 | positive | MGI_C57BL6J_5454267 | Gm24490 | ENSEMBL:ENSMUSG00000065737 | MGI:5454267 | snRNA gene | predicted gene, 24490 |
| 8 | gene | 86.482260 | 86.580688 | negative | MGI_C57BL6J_2441679 | Abcc12 | NCBI_Gene:244562,ENSEMBL:ENSMUSG00000036872 | MGI:2441679 | protein coding gene | ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
| 8 | gene | 86.586962 | 86.587083 | positive | MGI_C57BL6J_5690740 | Gm44348 | ENSEMBL:ENSMUSG00000106513 | MGI:5690740 | miRNA gene | predicted gene, 44348 |
| 8 | gene | 86.624043 | 86.723873 | positive | MGI_C57BL6J_1914137 | Lonp2 | NCBI_Gene:66887,ENSEMBL:ENSMUSG00000047866 | MGI:1914137 | protein coding gene | lon peptidase 2, peroxisomal |
| 8 | pseudogene | 86.659318 | 86.660035 | negative | MGI_C57BL6J_3801751 | Gm16177 | ENSEMBL:ENSMUSG00000082484 | MGI:3801751 | pseudogene | predicted gene 16177 |
| 8 | gene | 86.671236 | 86.671343 | negative | MGI_C57BL6J_5452082 | Gm22305 | ENSEMBL:ENSMUSG00000064624 | MGI:5452082 | snRNA gene | predicted gene, 22305 |
| 8 | gene | 86.720454 | 86.746006 | negative | MGI_C57BL6J_108064 | Siah1a | NCBI_Gene:20437,ENSEMBL:ENSMUSG00000036840 | MGI:108064 | protein coding gene | siah E3 ubiquitin protein ligase 1A |
| 8 | gene | 86.745453 | 86.748655 | positive | MGI_C57BL6J_3704316 | Gm10638 | NCBI_Gene:666945,ENSEMBL:ENSMUSG00000074178 | MGI:3704316 | lncRNA gene | predicted gene 10638 |
| 8 | gene | 86.805454 | 86.826230 | negative | MGI_C57BL6J_5622100 | Gm39215 | NCBI_Gene:105243242 | MGI:5622100 | lncRNA gene | predicted gene, 39215 |
| 8 | gene | 86.808160 | 86.885258 | negative | MGI_C57BL6J_2136825 | N4bp1 | NCBI_Gene:80750,ENSEMBL:ENSMUSG00000031652 | MGI:2136825 | protein coding gene | NEDD4 binding protein 1 |
| 8 | gene | 86.849296 | 86.851618 | negative | MGI_C57BL6J_5593036 | Gm33877 | NCBI_Gene:102636944 | MGI:5593036 | lncRNA gene | predicted gene, 33877 |
| 8 | gene | 86.865500 | 86.869523 | negative | MGI_C57BL6J_2443611 | F830004M19Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA F830004M19 gene |
| 8 | gene | 86.885688 | 86.886566 | positive | MGI_C57BL6J_1923941 | 2010110E17Rik | ENSEMBL:ENSMUSG00000098434 | MGI:1923941 | lncRNA gene | RIKEN cDNA 2010110E17 gene |
| 8 | gene | 86.902453 | 86.919422 | positive | MGI_C57BL6J_5521010 | Gm27167 | ENSEMBL:ENSMUSG00000098534 | MGI:5521010 | lncRNA gene | predicted gene 27167 |
| 8 | pseudogene | 86.914783 | 86.915665 | positive | MGI_C57BL6J_104623 | Fth-ps3 | NCBI_Gene:14322,ENSEMBL:ENSMUSG00000109706 | MGI:104623 | pseudogene | ferritin heavy chain, pseudogene 3 |
| 8 | gene | 86.920477 | 86.921147 | negative | MGI_C57BL6J_5521011 | Gm27168 | ENSEMBL:ENSMUSG00000099162 | MGI:5521011 | lncRNA gene | predicted gene 27168 |
| 8 | gene | 86.922117 | 86.926374 | positive | MGI_C57BL6J_5592550 | Gm33391 | NCBI_Gene:102636279 | MGI:5592550 | lncRNA gene | predicted gene, 33391 |
| 8 | gene | 86.965503 | 86.965619 | negative | MGI_C57BL6J_5454558 | Gm24781 | ENSEMBL:ENSMUSG00000088261 | MGI:5454558 | snoRNA gene | predicted gene, 24781 |
| 8 | pseudogene | 87.047689 | 87.048226 | positive | MGI_C57BL6J_3647042 | Rps13-ps1 | NCBI_Gene:625298,ENSEMBL:ENSMUSG00000066362 | MGI:3647042 | pseudogene | ribosomal protein S13, pseudogene 1 |
| 8 | pseudogene | 87.051397 | 87.052248 | negative | MGI_C57BL6J_5010787 | Gm18602 | NCBI_Gene:100417420,ENSEMBL:ENSMUSG00000110349 | MGI:5010787 | pseudogene | predicted gene, 18602 |
| 8 | gene | 87.093307 | 87.135397 | positive | MGI_C57BL6J_3645880 | Gm5118 | NCBI_Gene:102637021,ENSEMBL:ENSMUSG00000098854 | MGI:3645880 | lncRNA gene | predicted gene 5118 |
| 8 | gene | 87.114645 | 87.198404 | positive | MGI_C57BL6J_5521089 | Gm27246 | NCBI_Gene:102637261,ENSEMBL:ENSMUSG00000098684 | MGI:5521089 | lncRNA gene | predicted gene 27246 |
| 8 | gene | 87.143147 | 87.146335 | negative | MGI_C57BL6J_5593184 | Gm34025 | NCBI_Gene:102637139 | MGI:5593184 | lncRNA gene | predicted gene, 34025 |
| 8 | gene | 87.169747 | 87.199850 | negative | MGI_C57BL6J_3641741 | Gm10637 | NCBI_Gene:100038706,ENSEMBL:ENSMUSG00000099207 | MGI:3641741 | lncRNA gene | predicted gene 10637 |
| 8 | gene | 87.196212 | 87.198404 | positive | MGI_C57BL6J_5622101 | Gm39216 | NCBI_Gene:105243243 | MGI:5622101 | lncRNA gene | predicted gene, 39216 |
| 8 | gene | 87.203026 | 87.293504 | positive | MGI_C57BL6J_5521012 | Gm27169 | ENSEMBL:ENSMUSG00000098257 | MGI:5521012 | lncRNA gene | predicted gene 27169 |
| 8 | pseudogene | 87.241585 | 87.242303 | negative | MGI_C57BL6J_5011160 | Gm18975 | NCBI_Gene:100418061 | MGI:5011160 | pseudogene | predicted gene, 18975 |
| 8 | gene | 87.354501 | 87.373075 | negative | MGI_C57BL6J_5521050 | Gm27207 | ENSEMBL:ENSMUSG00000098768 | MGI:5521050 | lncRNA gene | predicted gene 27207 |
| 8 | gene | 87.381059 | 87.387551 | negative | MGI_C57BL6J_5521051 | Gm27208 | ENSEMBL:ENSMUSG00000098391 | MGI:5521051 | lncRNA gene | predicted gene 27208 |
| 8 | gene | 87.458404 | 87.459437 | positive | MGI_C57BL6J_1922487 | 4930535O05Rik | NCBI_Gene:75237,ENSEMBL:ENSMUSG00000098360 | MGI:1922487 | lncRNA gene | RIKEN cDNA 4930535O05 gene |
| 8 | gene | 87.460469 | 87.463797 | positive | MGI_C57BL6J_5012057 | Gm19872 | NCBI_Gene:100503751,ENSEMBL:ENSMUSG00000098295 | MGI:5012057 | lncRNA gene | predicted gene, 19872 |
| 8 | gene | 87.460469 | 87.525554 | positive | MGI_C57BL6J_3780864 | Gm2694 | NCBI_Gene:100040294,ENSEMBL:ENSMUSG00000097248 | MGI:3780864 | lncRNA gene | predicted gene 2694 |
| 8 | gene | 87.468405 | 87.472609 | negative | MGI_C57BL6J_88281 | Cbln1 | NCBI_Gene:12404,ENSEMBL:ENSMUSG00000031654 | MGI:88281 | protein coding gene | cerebellin 1 precursor protein |
| 8 | gene | 87.476055 | 87.495462 | negative | MGI_C57BL6J_5504131 | Gm27016 | ENSEMBL:ENSMUSG00000097992 | MGI:5504131 | lncRNA gene | predicted gene, 27016 |
| 8 | gene | 87.500049 | 87.501231 | negative | MGI_C57BL6J_1923912 | 1700120C18Rik | ENSEMBL:ENSMUSG00000098086 | MGI:1923912 | lncRNA gene | RIKEN cDNA 1700120C18 gene |
| 8 | gene | 87.563853 | 87.589424 | positive | MGI_C57BL6J_3607717 | 4933402J07Rik | NCBI_Gene:330820,ENSEMBL:ENSMUSG00000069971 | MGI:3607717 | protein coding gene | RIKEN cDNA 4933402J07 gene |
| 8 | gene | 87.636098 | 87.642210 | negative | MGI_C57BL6J_1920807 | 1700096P03Rik | NCBI_Gene:73557 | MGI:1920807 | lncRNA gene | RIKEN cDNA 1700096P03 gene |
| 8 | gene | 87.636657 | 87.637013 | positive | MGI_C57BL6J_5454618 | Gm24841 | ENSEMBL:ENSMUSG00000089301 | MGI:5454618 | unclassified non-coding RNA gene | predicted gene, 24841 |
| 8 | gene | 87.661810 | 87.959595 | negative | MGI_C57BL6J_1891217 | Zfp423 | NCBI_Gene:94187,ENSEMBL:ENSMUSG00000045333 | MGI:1891217 | protein coding gene | zinc finger protein 423 |
| 8 | gene | 87.926875 | 87.929536 | positive | MGI_C57BL6J_5593571 | Gm34412 | NCBI_Gene:102637658 | MGI:5593571 | lncRNA gene | predicted gene, 34412 |
| 8 | gene | 87.968023 | 87.968913 | positive | MGI_C57BL6J_5624917 | Gm42032 | NCBI_Gene:105246808 | MGI:5624917 | lncRNA gene | predicted gene, 42032 |
| 8 | gene | 88.024687 | 88.034171 | positive | MGI_C57BL6J_5622102 | Gm39217 | NCBI_Gene:105243244 | MGI:5622102 | lncRNA gene | predicted gene, 39217 |
| 8 | pseudogene | 88.044898 | 88.118707 | negative | MGI_C57BL6J_3780885 | Gm2716 | NCBI_Gene:100040328,ENSEMBL:ENSMUSG00000110719 | MGI:3780885 | pseudogene | predicted gene 2716 |
| 8 | pseudogene | 88.074275 | 88.074804 | negative | MGI_C57BL6J_5010863 | Gm18678 | NCBI_Gene:100417544 | MGI:5010863 | pseudogene | predicted gene, 18678 |
| 8 | gene | 88.118735 | 88.135197 | positive | MGI_C57BL6J_1921981 | Cnep1r1 | NCBI_Gene:382030,ENSEMBL:ENSMUSG00000036810 | MGI:1921981 | protein coding gene | CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
| 8 | gene | 88.137855 | 88.172027 | positive | MGI_C57BL6J_2444491 | Heatr3 | NCBI_Gene:234549,ENSEMBL:ENSMUSG00000031657 | MGI:2444491 | protein coding gene | HEAT repeat containing 3 |
| 8 | pseudogene | 88.138079 | 88.138517 | negative | MGI_C57BL6J_3708787 | Gm9988 | ENSEMBL:ENSMUSG00000055968 | MGI:3708787 | pseudogene | predicted gene 9988 |
| 8 | gene | 88.138206 | 88.138274 | positive | MGI_C57BL6J_5531005 | Mir7071 | miRBase:MI0022921,NCBI_Gene:102465649,ENSEMBL:ENSMUSG00000099080 | MGI:5531005 | miRNA gene | microRNA 7071 |
| 8 | gene | 88.174560 | 88.175034 | positive | MGI_C57BL6J_1924515 | 9430002A10Rik | ENSEMBL:ENSMUSG00000062704 | MGI:1924515 | lncRNA gene | RIKEN cDNA 9430002A10 gene |
| 8 | gene | 88.199213 | 88.262997 | positive | MGI_C57BL6J_1917820 | Tent4b | NCBI_Gene:214627,ENSEMBL:ENSMUSG00000036779 | MGI:1917820 | protein coding gene | terminal nucleotidyltransferase 4B |
| 8 | gene | 88.272366 | 88.329965 | positive | MGI_C57BL6J_102891 | Adcy7 | NCBI_Gene:11513,ENSEMBL:ENSMUSG00000031659 | MGI:102891 | protein coding gene | adenylate cyclase 7 |
| 8 | gene | 88.330598 | 88.362251 | negative | MGI_C57BL6J_1349766 | Brd7 | NCBI_Gene:26992,ENSEMBL:ENSMUSG00000031660 | MGI:1349766 | protein coding gene | bromodomain containing 7 |
| 8 | gene | 88.381590 | 88.389037 | negative | MGI_C57BL6J_5593910 | Gm34751 | NCBI_Gene:102638112 | MGI:5593910 | lncRNA gene | predicted gene, 34751 |
| 8 | pseudogene | 88.391121 | 88.392228 | positive | MGI_C57BL6J_5791333 | Gm45497 | ENSEMBL:ENSMUSG00000110280 | MGI:5791333 | pseudogene | predicted gene 45497 |
| 8 | gene | 88.417460 | 88.428840 | positive | MGI_C57BL6J_5622103 | Gm39218 | NCBI_Gene:105243245 | MGI:5622103 | lncRNA gene | predicted gene, 39218 |
| 8 | gene | 88.480006 | 88.485050 | positive | MGI_C57BL6J_5594005 | Gm34846 | NCBI_Gene:102638237 | MGI:5594005 | lncRNA gene | predicted gene, 34846 |
| 8 | gene | 88.512558 | 88.519485 | positive | MGI_C57BL6J_5594235 | Gm35076 | NCBI_Gene:102638539 | MGI:5594235 | lncRNA gene | predicted gene, 35076 |
| 8 | gene | 88.519575 | 88.521220 | negative | MGI_C57BL6J_5594321 | Gm35162 | NCBI_Gene:102638649 | MGI:5594321 | lncRNA gene | predicted gene, 35162 |
| 8 | gene | 88.521344 | 88.594887 | positive | MGI_C57BL6J_2135954 | Nkd1 | NCBI_Gene:93960,ENSEMBL:ENSMUSG00000031661 | MGI:2135954 | protein coding gene | naked cuticle 1 |
| 8 | gene | 88.560033 | 88.562949 | positive | MGI_C57BL6J_5791332 | Gm45496 | ENSEMBL:ENSMUSG00000110187 | MGI:5791332 | unclassified gene | predicted gene 45496 |
| 8 | gene | 88.626561 | 88.636130 | negative | MGI_C57BL6J_1918857 | Snx20 | NCBI_Gene:71607,ENSEMBL:ENSMUSG00000031662 | MGI:1918857 | protein coding gene | sorting nexin 20 |
| 8 | gene | 88.647280 | 88.688474 | positive | MGI_C57BL6J_2429397 | Nod2 | NCBI_Gene:257632,ENSEMBL:ENSMUSG00000055994 | MGI:2429397 | protein coding gene | nucleotide-binding oligomerization domain containing 2 |
| 8 | gene | 88.697001 | 88.751946 | positive | MGI_C57BL6J_1921506 | Cyld | NCBI_Gene:74256,ENSEMBL:ENSMUSG00000036712 | MGI:1921506 | protein coding gene | CYLD lysine 63 deubiquitinase |
| 8 | pseudogene | 88.721076 | 88.721649 | negative | MGI_C57BL6J_5010180 | Gm17995 | NCBI_Gene:100416240,ENSEMBL:ENSMUSG00000109970 | MGI:5010180 | pseudogene | predicted gene, 17995 |
| 8 | pseudogene | 88.728203 | 88.729042 | negative | MGI_C57BL6J_5010095 | Gm17910 | NCBI_Gene:100416080,ENSEMBL:ENSMUSG00000110299 | MGI:5010095 | pseudogene | predicted gene, 17910 |
| 8 | pseudogene | 88.806360 | 88.807174 | positive | MGI_C57BL6J_3646827 | Rps6-ps2 | NCBI_Gene:675985,ENSEMBL:ENSMUSG00000056772 | MGI:3646827 | pseudogene | ribosomal protein S6, pseudogene 2 |
| 8 | gene | 88.840301 | 88.848306 | positive | MGI_C57BL6J_5624919 | Gm42034 | NCBI_Gene:105246810 | MGI:5624919 | lncRNA gene | predicted gene, 42034 |
| 8 | gene | 88.860968 | 88.863074 | negative | MGI_C57BL6J_5791340 | Gm45504 | ENSEMBL:ENSMUSG00000109778 | MGI:5791340 | unclassified gene | predicted gene 45504 |
| 8 | gene | 89.024735 | 89.024831 | positive | MGI_C57BL6J_5530953 | Mir8110 | miRBase:MI0026042,NCBI_Gene:102465899,ENSEMBL:ENSMUSG00000099078 | MGI:5530953 | miRNA gene | microRNA 8110 |
| 8 | gene | 89.027235 | 89.044162 | negative | MGI_C57BL6J_1889585 | Sall1 | NCBI_Gene:58198,ENSEMBL:ENSMUSG00000031665 | MGI:1889585 | protein coding gene | spalt like transcription factor 1 |
| 8 | gene | 89.039976 | 89.071021 | positive | MGI_C57BL6J_3781311 | Gm3134 | NCBI_Gene:100041089 | MGI:3781311 | lncRNA gene | predicted gene 3134 |
| 8 | gene | 89.045693 | 89.052979 | negative | MGI_C57BL6J_5825684 | Gm46047 | NCBI_Gene:108167542 | MGI:5825684 | lncRNA gene | predicted gene, 46047 |
| 8 | gene | 89.085418 | 89.130837 | negative | MGI_C57BL6J_5594517 | Gm35358 | NCBI_Gene:102638911 | MGI:5594517 | lncRNA gene | predicted gene, 35358 |
| 8 | gene | 89.137153 | 89.142424 | positive | MGI_C57BL6J_5594464 | Gm35305 | NCBI_Gene:102638836 | MGI:5594464 | lncRNA gene | predicted gene, 35305 |
| 8 | pseudogene | 89.146284 | 89.148109 | negative | MGI_C57BL6J_3647663 | Gm6625 | NCBI_Gene:625801,ENSEMBL:ENSMUSG00000094790 | MGI:3647663 | pseudogene | predicted gene 6625 |
| 8 | pseudogene | 89.186858 | 89.187589 | negative | MGI_C57BL6J_3643649 | Gm5356 | NCBI_Gene:384862,ENSEMBL:ENSMUSG00000069962 | MGI:3643649 | pseudogene | predicted pseudogene 5356 |
| 8 | gene | 89.219478 | 89.230325 | positive | MGI_C57BL6J_5594701 | Gm35542 | NCBI_Gene:102639167 | MGI:5594701 | lncRNA gene | predicted gene, 35542 |
| 8 | gene | 89.427227 | 89.462515 | positive | MGI_C57BL6J_5594752 | Gm35593 | NCBI_Gene:102639237 | MGI:5594752 | lncRNA gene | predicted gene, 35593 |
| 8 | gene | 89.589435 | 89.589541 | negative | MGI_C57BL6J_5456108 | Gm26331 | ENSEMBL:ENSMUSG00000064758 | MGI:5456108 | snRNA gene | predicted gene, 26331 |
| 8 | gene | 89.667173 | 89.667304 | negative | MGI_C57BL6J_5453989 | Gm24212 | ENSEMBL:ENSMUSG00000088883 | MGI:5453989 | snoRNA gene | predicted gene, 24212 |
| 8 | gene | 90.247040 | 90.349073 | negative | MGI_C57BL6J_3039593 | Tox3 | NCBI_Gene:244579,ENSEMBL:ENSMUSG00000043668 | MGI:3039593 | protein coding gene | TOX high mobility group box family member 3 |
| 8 | gene | 90.355284 | 90.359600 | negative | MGI_C57BL6J_5622104 | Gm39219 | NCBI_Gene:105243246 | MGI:5622104 | lncRNA gene | predicted gene, 39219 |
| 8 | gene | 90.359703 | 90.361755 | positive | MGI_C57BL6J_1925400 | 4930405E02Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 4930405E02 gene |
| 8 | gene | 90.417185 | 90.420255 | negative | MGI_C57BL6J_5825685 | Gm46048 | NCBI_Gene:108167543 | MGI:5825685 | lncRNA gene | predicted gene, 46048 |
| 8 | gene | 90.480943 | 90.491058 | negative | MGI_C57BL6J_5594956 | Gm35797 | NCBI_Gene:102639496 | MGI:5594956 | lncRNA gene | predicted gene, 35797 |
| 8 | gene | 90.520823 | 90.535881 | negative | MGI_C57BL6J_5622105 | Gm39220 | NCBI_Gene:105243247 | MGI:5622105 | lncRNA gene | predicted gene, 39220 |
| 8 | gene | 90.669805 | 90.674958 | negative | MGI_C57BL6J_5791475 | Gm45639 | ENSEMBL:ENSMUSG00000109672 | MGI:5791475 | lncRNA gene | predicted gene 45639 |
| 8 | pseudogene | 90.675240 | 90.676203 | negative | MGI_C57BL6J_5010181 | Gm17996 | NCBI_Gene:100416241 | MGI:5010181 | pseudogene | predicted gene, 17996 |
| 8 | gene | 90.685186 | 90.688063 | negative | MGI_C57BL6J_5791474 | Gm45638 | ENSEMBL:ENSMUSG00000109726 | MGI:5791474 | unclassified gene | predicted gene 45638 |
| 8 | gene | 90.709458 | 90.740775 | negative | MGI_C57BL6J_5595009 | Gm35850 | NCBI_Gene:102639570,ENSEMBL:ENSMUSG00000109870 | MGI:5595009 | lncRNA gene | predicted gene, 35850 |
| 8 | gene | 90.759213 | 90.762605 | negative | MGI_C57BL6J_5595122 | Gm35963 | NCBI_Gene:102639716,ENSEMBL:ENSMUSG00000110212 | MGI:5595122 | lncRNA gene | predicted gene, 35963 |
| 8 | gene | 90.790826 | 90.790947 | negative | MGI_C57BL6J_5690774 | Gm44382 | ENSEMBL:ENSMUSG00000105321 | MGI:5690774 | miRNA gene | predicted gene, 44382 |
| 8 | gene | 90.816145 | 90.817393 | negative | MGI_C57BL6J_5012120 | Gm19935 | NCBI_Gene:100503868,ENSEMBL:ENSMUSG00000110332 | MGI:5012120 | lncRNA gene | predicted gene, 19935 |
| 8 | gene | 90.828352 | 91.054516 | positive | MGI_C57BL6J_1924001 | Chd9 | NCBI_Gene:109151,ENSEMBL:ENSMUSG00000056608 | MGI:1924001 | protein coding gene | chromodomain helicase DNA binding protein 9 |
| 8 | gene | 90.848034 | 90.849846 | negative | MGI_C57BL6J_5791477 | Gm45641 | ENSEMBL:ENSMUSG00000110166 | MGI:5791477 | lncRNA gene | predicted gene 45641 |
| 8 | gene | 90.876581 | 90.878476 | positive | MGI_C57BL6J_5791476 | Gm45640 | ENSEMBL:ENSMUSG00000109812 | MGI:5791476 | unclassified gene | predicted gene 45640 |
| 8 | gene | 90.893155 | 90.894638 | positive | MGI_C57BL6J_5791479 | Gm45643 | ENSEMBL:ENSMUSG00000109863 | MGI:5791479 | unclassified gene | predicted gene 45643 |
| 8 | gene | 90.906532 | 90.908416 | negative | MGI_C57BL6J_3645470 | Gm6658 | ENSEMBL:ENSMUSG00000074171 | MGI:3645470 | protein coding gene | predicted gene 6658 |
| 8 | gene | 91.055264 | 91.056557 | positive | MGI_C57BL6J_5791199 | Gm45363 | ENSEMBL:ENSMUSG00000110377 | MGI:5791199 | unclassified gene | predicted gene 45363 |
| 8 | gene | 91.059351 | 91.061885 | positive | MGI_C57BL6J_5622106 | Gm39221 | NCBI_Gene:105243249 | MGI:5622106 | lncRNA gene | predicted gene, 39221 |
| 8 | gene | 91.061265 | 91.061396 | negative | MGI_C57BL6J_5452205 | Gm22428 | ENSEMBL:ENSMUSG00000089179 | MGI:5452205 | snoRNA gene | predicted gene, 22428 |
| 8 | gene | 91.070057 | 91.123844 | positive | MGI_C57BL6J_105085 | Rbl2 | NCBI_Gene:19651,ENSEMBL:ENSMUSG00000031666 | MGI:105085 | protein coding gene | RB transcriptional corepressor like 2 |
| 8 | gene | 91.107639 | 91.199976 | negative | MGI_C57BL6J_3693832 | Aktip | NCBI_Gene:14339,ENSEMBL:ENSMUSG00000031667 | MGI:3693832 | protein coding gene | thymoma viral proto-oncogene 1 interacting protein |
| 8 | gene | 91.113762 | 91.116671 | negative | MGI_C57BL6J_5791300 | Gm45464 | ENSEMBL:ENSMUSG00000109727 | MGI:5791300 | lncRNA gene | predicted gene 45464 |
| 8 | pseudogene | 91.152207 | 91.152374 | positive | MGI_C57BL6J_5791501 | Gm45665 | ENSEMBL:ENSMUSG00000110189 | MGI:5791501 | pseudogene | predicted gene 45665 |
| 8 | pseudogene | 91.157739 | 91.158890 | positive | MGI_C57BL6J_3648086 | Gm8489 | NCBI_Gene:667162,ENSEMBL:ENSMUSG00000109755 | MGI:3648086 | pseudogene | predicted gene 8489 |
| 8 | gene | 91.160516 | 91.171180 | positive | MGI_C57BL6J_5791502 | Gm45666 | ENSEMBL:ENSMUSG00000110106 | MGI:5791502 | lncRNA gene | predicted gene 45666 |
| 8 | gene | 91.184919 | 91.185183 | negative | MGI_C57BL6J_5453185 | Gm23408 | ENSEMBL:ENSMUSG00000089148 | MGI:5453185 | unclassified non-coding RNA gene | predicted gene, 23408 |
| 8 | gene | 91.210359 | 91.235269 | positive | MGI_C57BL6J_5595402 | Gm36243 | NCBI_Gene:102640089,ENSEMBL:ENSMUSG00000110089 | MGI:5595402 | lncRNA gene | predicted gene, 36243 |
| 8 | gene | 91.217030 | 91.313354 | negative | MGI_C57BL6J_1920563 | Rpgrip1l | NCBI_Gene:244585,ENSEMBL:ENSMUSG00000033282 | MGI:1920563 | protein coding gene | Rpgrip1-like |
| 8 | pseudogene | 91.249617 | 91.250142 | negative | MGI_C57BL6J_5010788 | Gm18603 | NCBI_Gene:100417421 | MGI:5010788 | pseudogene | predicted gene, 18603 |
| 8 | gene | 91.258438 | 91.260535 | negative | MGI_C57BL6J_5791498 | Gm45662 | ENSEMBL:ENSMUSG00000109828 | MGI:5791498 | unclassified gene | predicted gene 45662 |
| 8 | gene | 91.273747 | 91.300367 | positive | MGI_C57BL6J_5595322 | Gm36163 | NCBI_Gene:102639977 | MGI:5595322 | lncRNA gene | predicted gene, 36163 |
| 8 | gene | 91.305967 | 91.307755 | negative | MGI_C57BL6J_5791264 | Gm45428 | ENSEMBL:ENSMUSG00000109956 | MGI:5791264 | unclassified gene | predicted gene 45428 |
| 8 | gene | 91.313491 | 91.668439 | positive | MGI_C57BL6J_1347093 | Fto | NCBI_Gene:26383,ENSEMBL:ENSMUSG00000055932 | MGI:1347093 | protein coding gene | fat mass and obesity associated |
| 8 | gene | 91.365669 | 91.367266 | positive | MGI_C57BL6J_3026999 | B130011D17Rik | NA | NA | unclassified gene | RIKEN cDNA B130011D17 gene |
| 8 | gene | 91.390693 | 91.391945 | negative | MGI_C57BL6J_5791263 | Gm45427 | ENSEMBL:ENSMUSG00000110233 | MGI:5791263 | unclassified gene | predicted gene 45427 |
| 8 | gene | 91.449521 | 91.474598 | negative | MGI_C57BL6J_2444771 | 4831440D22Rik | NCBI_Gene:320798,ENSEMBL:ENSMUSG00000110138 | MGI:2444771 | lncRNA gene | RIKEN cDNA 4831440D22 gene |
| 8 | gene | 91.450306 | 91.452512 | positive | MGI_C57BL6J_5791128 | Gm45292 | ENSEMBL:ENSMUSG00000110079 | MGI:5791128 | unclassified gene | predicted gene 45292 |
| 8 | gene | 91.493019 | 91.495595 | positive | MGI_C57BL6J_5791125 | Gm45289 | ENSEMBL:ENSMUSG00000109642 | MGI:5791125 | unclassified gene | predicted gene 45289 |
| 8 | gene | 91.528835 | 91.532724 | negative | MGI_C57BL6J_5791126 | Gm45290 | ENSEMBL:ENSMUSG00000109709 | MGI:5791126 | unclassified gene | predicted gene 45290 |
| 8 | gene | 91.579288 | 91.581389 | positive | MGI_C57BL6J_5791130 | Gm45294 | ENSEMBL:ENSMUSG00000110068 | MGI:5791130 | unclassified gene | predicted gene 45294 |
| 8 | gene | 91.621448 | 91.629729 | negative | MGI_C57BL6J_3781413 | Gm3235 | ENSEMBL:ENSMUSG00000090778 | MGI:3781413 | lncRNA gene | predicted gene 3235 |
| 8 | gene | 91.633079 | 91.634890 | positive | MGI_C57BL6J_5791129 | Gm45293 | ENSEMBL:ENSMUSG00000110325 | MGI:5791129 | unclassified gene | predicted gene 45293 |
| 8 | gene | 91.670852 | 91.671897 | negative | MGI_C57BL6J_5791131 | Gm45295 | ENSEMBL:ENSMUSG00000110302 | MGI:5791131 | unclassified gene | predicted gene 45295 |
| 8 | pseudogene | 91.688172 | 91.689135 | negative | MGI_C57BL6J_3780027 | Gm9619 | NCBI_Gene:674324,ENSEMBL:ENSMUSG00000110371 | MGI:3780027 | pseudogene | predicted gene 9619 |
| 8 | pseudogene | 91.689701 | 91.690112 | positive | MGI_C57BL6J_3781052 | Gm2875 | NCBI_Gene:100040628,ENSEMBL:ENSMUSG00000110073 | MGI:3781052 | pseudogene | predicted gene 2875 |
| 8 | gene | 91.710233 | 91.715184 | negative | MGI_C57BL6J_5595484 | Gm36325 | NCBI_Gene:102640201,ENSEMBL:ENSMUSG00000110216 | MGI:5595484 | lncRNA gene | predicted gene, 36325 |
| 8 | gene | 91.788508 | 91.794248 | negative | MGI_C57BL6J_5595539 | Gm36380 | NCBI_Gene:102640269,ENSEMBL:ENSMUSG00000110304 | MGI:5595539 | lncRNA gene | predicted gene, 36380 |
| 8 | gene | 91.795574 | 91.797743 | negative | MGI_C57BL6J_5622107 | Gm39222 | NCBI_Gene:105243251 | MGI:5622107 | lncRNA gene | predicted gene, 39222 |
| 8 | gene | 91.798511 | 91.802067 | negative | MGI_C57BL6J_1197522 | Irx3 | NCBI_Gene:16373,ENSEMBL:ENSMUSG00000031734 | MGI:1197522 | protein coding gene | Iroquois related homeobox 3 |
| 8 | gene | 91.800415 | 91.807539 | positive | MGI_C57BL6J_2441953 | Irx3os | NCBI_Gene:319388,ENSEMBL:ENSMUSG00000046413 | MGI:2441953 | antisense lncRNA gene | iroquois homeobox 3, opposite strand |
| 8 | gene | 91.822584 | 91.830637 | positive | MGI_C57BL6J_5622108 | Gm39223 | NCBI_Gene:105243252 | MGI:5622108 | lncRNA gene | predicted gene, 39223 |
| 8 | gene | 91.864876 | 91.872995 | positive | MGI_C57BL6J_5579217 | Gm28511 | NCBI_Gene:108167544,ENSEMBL:ENSMUSG00000099664 | MGI:5579217 | lncRNA gene | predicted gene 28511 |
| 8 | gene | 91.907565 | 91.913066 | negative | MGI_C57BL6J_5579221 | Gm28515 | ENSEMBL:ENSMUSG00000099812 | MGI:5579221 | lncRNA gene | predicted gene 28515 |
| 8 | gene | 91.974095 | 91.991947 | negative | MGI_C57BL6J_5595829 | Gm36670 | NCBI_Gene:102640656,ENSEMBL:ENSMUSG00000109696 | MGI:5595829 | lncRNA gene | predicted gene, 36670 |
| 8 | gene | 91.997432 | 92.010860 | positive | MGI_C57BL6J_5596002 | Gm36843 | NCBI_Gene:102640885,ENSEMBL:ENSMUSG00000110274 | MGI:5596002 | lncRNA gene | predicted gene, 36843 |
| 8 | gene | 92.024442 | 92.038438 | positive | MGI_C57BL6J_5579222 | Gm28516 | NCBI_Gene:105243253,ENSEMBL:ENSMUSG00000101613 | MGI:5579222 | lncRNA gene | predicted gene 28516 |
| 8 | gene | 92.039917 | 92.040020 | positive | MGI_C57BL6J_5454031 | Gm24254 | ENSEMBL:ENSMUSG00000065636 | MGI:5454031 | snRNA gene | predicted gene, 24254 |
| 8 | gene | 92.152096 | 92.158266 | negative | MGI_C57BL6J_5791168 | Gm45332 | ENSEMBL:ENSMUSG00000110380 | MGI:5791168 | lncRNA gene | predicted gene 45332 |
| 8 | gene | 92.172587 | 92.189124 | positive | MGI_C57BL6J_5791171 | Gm45335 | ENSEMBL:ENSMUSG00000110198 | MGI:5791171 | lncRNA gene | predicted gene 45335 |
| 8 | gene | 92.233125 | 92.247959 | positive | MGI_C57BL6J_5791170 | Gm45334 | ENSEMBL:ENSMUSG00000110134 | MGI:5791170 | lncRNA gene | predicted gene 45334 |
| 8 | gene | 92.326031 | 92.356120 | negative | MGI_C57BL6J_1918546 | Crnde | NCBI_Gene:71296,ENSEMBL:ENSMUSG00000031736 | MGI:1918546 | lncRNA gene | colorectal neoplasia differentially expressed (non-protein coding) |
| 8 | gene | 92.336668 | 92.339250 | negative | MGI_C57BL6J_5433981 | Gm21817 | ENSEMBL:ENSMUSG00000095651 | MGI:5433981 | lncRNA gene | predicted gene, 21817 |
| 8 | gene | 92.357625 | 92.376286 | positive | MGI_C57BL6J_1859086 | Irx5 | NCBI_Gene:54352,ENSEMBL:ENSMUSG00000031737 | MGI:1859086 | protein coding gene | Iroquois homeobox 5 |
| 8 | gene | 92.465767 | 92.469665 | positive | MGI_C57BL6J_5791172 | Gm45336 | ENSEMBL:ENSMUSG00000110290 | MGI:5791172 | lncRNA gene | predicted gene 45336 |
| 8 | gene | 92.674288 | 92.680956 | positive | MGI_C57BL6J_1927642 | Irx6 | NCBI_Gene:64379,ENSEMBL:ENSMUSG00000031738 | MGI:1927642 | protein coding gene | Iroquois homeobox 6 |
| 8 | gene | 92.680172 | 92.682194 | negative | MGI_C57BL6J_5791341 | Gm45505 | ENSEMBL:ENSMUSG00000110023 | MGI:5791341 | lncRNA gene | predicted gene 45505 |
| 8 | pseudogene | 92.715746 | 92.716975 | negative | MGI_C57BL6J_3781450 | Gm3272 | NCBI_Gene:100041325,ENSEMBL:ENSMUSG00000078967 | MGI:3781450 | pseudogene | predicted pseudogene 3272 |
| 8 | gene | 92.827291 | 92.853421 | positive | MGI_C57BL6J_97009 | Mmp2 | NCBI_Gene:17390,ENSEMBL:ENSMUSG00000031740 | MGI:97009 | protein coding gene | matrix metallopeptidase 2 |
| 8 | gene | 92.853653 | 92.856334 | negative | MGI_C57BL6J_5791308 | Gm45472 | ENSEMBL:ENSMUSG00000109669 | MGI:5791308 | lncRNA gene | predicted gene 45472 |
| 8 | gene | 92.855329 | 92.919279 | positive | MGI_C57BL6J_3606214 | Lpcat2 | NCBI_Gene:270084,ENSEMBL:ENSMUSG00000033192 | MGI:3606214 | protein coding gene | lysophosphatidylcholine acyltransferase 2 |
| 8 | gene | 92.901395 | 92.902411 | positive | MGI_C57BL6J_1916793 | Capns2 | NCBI_Gene:69543,ENSEMBL:ENSMUSG00000078144 | MGI:1916793 | protein coding gene | calpain, small subunit 2 |
| 8 | gene | 92.913132 | 92.913189 | negative | MGI_C57BL6J_5451928 | Gm22151 | ENSEMBL:ENSMUSG00000080613 | MGI:5451928 | miRNA gene | predicted gene, 22151 |
| 8 | pseudogene | 92.922522 | 92.922870 | positive | MGI_C57BL6J_3781125 | Gm2947 | NCBI_Gene:100040752 | MGI:3781125 | pseudogene | predicted gene 2947 |
| 8 | gene | 92.960079 | 93.001667 | positive | MGI_C57BL6J_1270850 | Slc6a2 | NCBI_Gene:20538,ENSEMBL:ENSMUSG00000055368 | MGI:1270850 | protein coding gene | solute carrier family 6 (neurotransmitter transporter, noradrenalin), member 2 |
| 8 | gene | 93.011619 | 93.041516 | positive | MGI_C57BL6J_5791299 | Gm45463 | ENSEMBL:ENSMUSG00000109651 | MGI:5791299 | lncRNA gene | predicted gene 45463 |
| 8 | gene | 93.020214 | 93.048192 | negative | MGI_C57BL6J_3648919 | Ces1a | NCBI_Gene:244595,ENSEMBL:ENSMUSG00000071047 | MGI:3648919 | protein coding gene | carboxylesterase 1A |
| 8 | gene | 93.056725 | 93.080021 | negative | MGI_C57BL6J_3779470 | Ces1b | NCBI_Gene:382044,ENSEMBL:ENSMUSG00000078964 | MGI:3779470 | protein coding gene | carboxylesterase 1B |
| 8 | gene | 93.099015 | 93.131283 | negative | MGI_C57BL6J_95420 | Ces1c | NCBI_Gene:13884,ENSEMBL:ENSMUSG00000057400 | MGI:95420 | protein coding gene | carboxylesterase 1C |
| 8 | pseudogene | 93.141331 | 93.154437 | negative | MGI_C57BL6J_5805025 | Gm45910 | NCBI_Gene:108167545,ENSEMBL:ENSMUSG00000110714 | MGI:5805025 | pseudogene | predicted gene 45910 |
| 8 | gene | 93.166068 | 93.197838 | negative | MGI_C57BL6J_2148202 | Ces1d | NCBI_Gene:104158,ENSEMBL:ENSMUSG00000056973 | MGI:2148202 | protein coding gene | carboxylesterase 1D |
| 8 | gene | 93.190328 | 93.191358 | negative | MGI_C57BL6J_5805024 | Gm45909 | ENSEMBL:ENSMUSG00000110496 | MGI:5805024 | unclassified gene | predicted gene 45909 |
| 8 | gene | 93.201218 | 93.229631 | negative | MGI_C57BL6J_95432 | Ces1e | NCBI_Gene:13897,ENSEMBL:ENSMUSG00000061959 | MGI:95432 | protein coding gene | carboxylesterase 1E |
| 8 | pseudogene | 93.238653 | 93.251175 | negative | MGI_C57BL6J_5804842 | Gm45727 | ENSEMBL:ENSMUSG00000110659 | MGI:5804842 | pseudogene | predicted gene 45727 |
| 8 | gene | 93.256236 | 93.279766 | negative | MGI_C57BL6J_2142687 | Ces1f | NCBI_Gene:234564,ENSEMBL:ENSMUSG00000031725 | MGI:2142687 | protein coding gene | carboxylesterase 1F |
| 8 | gene | 93.302369 | 93.337308 | negative | MGI_C57BL6J_88378 | Ces1g | NCBI_Gene:12623,ENSEMBL:ENSMUSG00000057074 | MGI:88378 | protein coding gene | carboxylesterase 1G |
| 8 | gene | 93.351841 | 93.379818 | negative | MGI_C57BL6J_1922954 | Ces1h | NCBI_Gene:75704,ENSEMBL:ENSMUSG00000074156 | MGI:1922954 | protein coding gene | carboxylesterase 1H |
| 8 | gene | 93.363623 | 93.431049 | positive | MGI_C57BL6J_5804880 | Gm45765 | ENSEMBL:ENSMUSG00000110718 | MGI:5804880 | lncRNA gene | predicted gene 45765 |
| 8 | gene | 93.398905 | 93.403908 | positive | MGI_C57BL6J_5804879 | Gm45764 | ENSEMBL:ENSMUSG00000110687 | MGI:5804879 | lncRNA gene | predicted gene 45764 |
| 8 | gene | 93.415537 | 93.427654 | negative | MGI_C57BL6J_5589291 | Gm30132 | NCBI_Gene:102631922,ENSEMBL:ENSMUSG00000110670 | MGI:5589291 | lncRNA gene | predicted gene, 30132 |
| 8 | gene | 93.430475 | 93.437174 | negative | MGI_C57BL6J_5804881 | Gm45766 | ENSEMBL:ENSMUSG00000110695 | MGI:5804881 | lncRNA gene | predicted gene 45766 |
| 8 | gene | 93.499063 | 93.535879 | negative | MGI_C57BL6J_1915185 | Ces5a | NCBI_Gene:67935,ENSEMBL:ENSMUSG00000058019 | MGI:1915185 | protein coding gene | carboxylesterase 5A |
| 8 | gene | 93.565977 | 93.631795 | negative | MGI_C57BL6J_5804823 | Gm45708 | NCBI_Gene:108167364,ENSEMBL:ENSMUSG00000110534 | MGI:5804823 | lncRNA gene | predicted gene 45708 |
| 8 | gene | 93.644160 | 93.751706 | negative | MGI_C57BL6J_5477337 | Gm26843 | NCBI_Gene:102632635,ENSEMBL:ENSMUSG00000110559 | MGI:5477337 | lncRNA gene | predicted gene, 26843 |
| 8 | gene | 93.706771 | 93.719975 | negative | MGI_C57BL6J_5589765 | Gm30606 | NCBI_Gene:108167546,ENSEMBL:ENSMUSG00000110713 | MGI:5589765 | lncRNA gene | predicted gene, 30606 |
| 8 | gene | 93.731231 | 93.750223 | positive | MGI_C57BL6J_5622109 | Gm39224 | NCBI_Gene:105243254 | MGI:5622109 | lncRNA gene | predicted gene, 39224 |
| 8 | gene | 93.747575 | 93.749852 | negative | MGI_C57BL6J_5804901 | Gm45786 | ENSEMBL:ENSMUSG00000110672 | MGI:5804901 | unclassified gene | predicted gene 45786 |
| 8 | gene | 93.750147 | 93.813722 | negative | MGI_C57BL6J_1923059 | 4930488L21Rik | NCBI_Gene:75809,ENSEMBL:ENSMUSG00000097520 | MGI:1923059 | lncRNA gene | RIKEN cDNA 4930488L21 gene |
| 8 | gene | 93.809966 | 93.969388 | positive | MGI_C57BL6J_95775 | Gnao1 | NCBI_Gene:14681,ENSEMBL:ENSMUSG00000031748 | MGI:95775 | protein coding gene | guanine nucleotide binding protein, alpha O |
| 8 | gene | 93.933835 | 93.933936 | negative | MGI_C57BL6J_5453936 | Gm24159 | ENSEMBL:ENSMUSG00000093005 | MGI:5453936 | miRNA gene | predicted gene, 24159 |
| 8 | gene | 93.971588 | 94.012842 | negative | MGI_C57BL6J_1345634 | Amfr | NCBI_Gene:23802,ENSEMBL:ENSMUSG00000031751 | MGI:1345634 | protein coding gene | autocrine motility factor receptor |
| 8 | gene | 94.015496 | 94.037039 | negative | MGI_C57BL6J_1915469 | Nudt21 | NCBI_Gene:68219,ENSEMBL:ENSMUSG00000031754 | MGI:1915469 | protein coding gene | nudix (nucleoside diphosphate linked moiety X)-type motif 21 |
| 8 | gene | 94.037139 | 94.067922 | positive | MGI_C57BL6J_2442978 | Ogfod1 | NCBI_Gene:270086,ENSEMBL:ENSMUSG00000033009 | MGI:2442978 | protein coding gene | 2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| 8 | gene | 94.048781 | 94.050695 | positive | MGI_C57BL6J_5804988 | Gm45873 | ENSEMBL:ENSMUSG00000110499 | MGI:5804988 | unclassified gene | predicted gene 45873 |
| 8 | gene | 94.067954 | 94.099370 | negative | MGI_C57BL6J_2135267 | Bbs2 | NCBI_Gene:67378,ENSEMBL:ENSMUSG00000031755 | MGI:2135267 | protein coding gene | Bardet-Biedl syndrome 2 (human) |
| 8 | gene | 94.099542 | 94.100572 | positive | MGI_C57BL6J_5622111 | Gm39226 | NCBI_Gene:105243257 | MGI:5622111 | lncRNA gene | predicted gene, 39226 |
| 8 | gene | 94.137197 | 94.139031 | positive | MGI_C57BL6J_99692 | Mt4 | NCBI_Gene:17752,ENSEMBL:ENSMUSG00000031757 | MGI:99692 | protein coding gene | metallothionein 4 |
| 8 | gene | 94.152607 | 94.154148 | positive | MGI_C57BL6J_97173 | Mt3 | NCBI_Gene:17751,ENSEMBL:ENSMUSG00000031760 | MGI:97173 | protein coding gene | metallothionein 3 |
| 8 | gene | 94.166124 | 94.172524 | negative | MGI_C57BL6J_5622112 | Gm39227 | NCBI_Gene:105243258 | MGI:5622112 | lncRNA gene | predicted gene, 39227 |
| 8 | gene | 94.170748 | 94.173412 | negative | MGI_C57BL6J_5804889 | Gm45774 | ENSEMBL:ENSMUSG00000110588 | MGI:5804889 | lncRNA gene | predicted gene 45774 |
| 8 | gene | 94.172618 | 94.173568 | positive | MGI_C57BL6J_97172 | Mt2 | NCBI_Gene:17750,ENSEMBL:ENSMUSG00000031762 | MGI:97172 | protein coding gene | metallothionein 2 |
| 8 | gene | 94.179082 | 94.180327 | positive | MGI_C57BL6J_97171 | Mt1 | NCBI_Gene:17748,ENSEMBL:ENSMUSG00000031765 | MGI:97171 | protein coding gene | metallothionein 1 |
| 8 | gene | 94.182131 | 94.184982 | positive | MGI_C57BL6J_5622113 | Gm39228 | NCBI_Gene:105243259,ENSEMBL:ENSMUSG00000110524 | MGI:5622113 | lncRNA gene | predicted gene, 39228 |
| 8 | gene | 94.214540 | 94.317227 | positive | MGI_C57BL6J_1919055 | Nup93 | NCBI_Gene:71805,ENSEMBL:ENSMUSG00000032939 | MGI:1919055 | protein coding gene | nucleoporin 93 |
| 8 | pseudogene | 94.222952 | 94.223709 | positive | MGI_C57BL6J_3781583 | Rpl10-ps5 | NCBI_Gene:100041555,ENSEMBL:ENSMUSG00000110679 | MGI:3781583 | pseudogene | ribosomal protein L10, pseudogene 5 |
| 8 | gene | 94.273426 | 94.276860 | positive | MGI_C57BL6J_5804919 | Gm45804 | ENSEMBL:ENSMUSG00000110442 | MGI:5804919 | unclassified gene | predicted gene 45804 |
| 8 | gene | 94.322158 | 94.328100 | positive | MGI_C57BL6J_2142856 | C78859 | ENSEMBL:ENSMUSG00000110710 | MGI:2142856 | lncRNA gene | expressed sequence C78859 |
| 8 | gene | 94.324311 | 94.324381 | positive | MGI_C57BL6J_3618733 | Mir138-2 | miRBase:MI0000164,NCBI_Gene:723956,ENSEMBL:ENSMUSG00000065512 | MGI:3618733 | miRNA gene | microRNA 138-2 |
| 8 | gene | 94.329109 | 94.366222 | positive | MGI_C57BL6J_108114 | Slc12a3 | NCBI_Gene:20497,ENSEMBL:ENSMUSG00000031766 | MGI:108114 | protein coding gene | solute carrier family 12, member 3 |
| 8 | gene | 94.377872 | 94.380317 | positive | MGI_C57BL6J_3802159 | Gm15889 | ENSEMBL:ENSMUSG00000089712 | MGI:3802159 | lncRNA gene | predicted gene 15889 |
| 8 | gene | 94.379184 | 94.386972 | negative | MGI_C57BL6J_3802160 | Gm15890 | ENSEMBL:ENSMUSG00000090105 | MGI:3802160 | lncRNA gene | predicted gene 15890 |
| 8 | gene | 94.386138 | 94.395377 | positive | MGI_C57BL6J_1927406 | Herpud1 | NCBI_Gene:64209,ENSEMBL:ENSMUSG00000031770 | MGI:1927406 | protein coding gene | homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| 8 | pseudogene | 94.405111 | 94.406392 | positive | MGI_C57BL6J_1929217 | Ap3s1-ps2 | NCBI_Gene:434339,ENSEMBL:ENSMUSG00000094257 | MGI:1929217 | pseudogene | adaptor-related protein complex 3, sigma 1 subunit, pseudogene 2 |
| 8 | gene | 94.422898 | 94.435103 | negative | MGI_C57BL6J_2443913 | 9330175E14Rik | NCBI_Gene:320377,ENSEMBL:ENSMUSG00000097194 | MGI:2443913 | lncRNA gene | RIKEN cDNA 9330175E14 gene |
| 8 | gene | 94.434354 | 94.527272 | positive | MGI_C57BL6J_3612191 | Nlrc5 | NCBI_Gene:434341,ENSEMBL:ENSMUSG00000074151 | MGI:3612191 | protein coding gene | NLR family, CARD domain containing 5 |
| 8 | gene | 94.505046 | 94.505105 | positive | MGI_C57BL6J_5531047 | Mir7072 | miRBase:MI0022922,NCBI_Gene:102466800,ENSEMBL:ENSMUSG00000098511 | MGI:5531047 | miRNA gene | microRNA 7072 |
| 8 | gene | 94.532705 | 94.570531 | positive | MGI_C57BL6J_2387578 | Cpne2 | NCBI_Gene:234577,ENSEMBL:ENSMUSG00000034361 | MGI:2387578 | protein coding gene | copine II |
| 8 | gene | 94.550579 | 94.568584 | negative | MGI_C57BL6J_5621440 | Gm38555 | NCBI_Gene:102641396 | MGI:5621440 | lncRNA gene | predicted gene, 38555 |
| 8 | gene | 94.574138 | 94.601997 | negative | MGI_C57BL6J_1919637 | Fam192a | NCBI_Gene:102122,ENSEMBL:ENSMUSG00000031774 | MGI:1919637 | protein coding gene | family with sequence similarity 192, member A |
| 8 | gene | 94.601937 | 94.660276 | positive | MGI_C57BL6J_1914860 | Rspry1 | NCBI_Gene:67610,ENSEMBL:ENSMUSG00000050079 | MGI:1914860 | protein coding gene | ring finger and SPRY domain containing 1 |
| 8 | gene | 94.610777 | 94.612697 | positive | MGI_C57BL6J_5804912 | Gm45797 | ENSEMBL:ENSMUSG00000110504 | MGI:5804912 | unclassified gene | predicted gene 45797 |
| 8 | pseudogene | 94.640942 | 94.641524 | positive | MGI_C57BL6J_5010187 | Gm18002 | NCBI_Gene:100416248 | MGI:5010187 | pseudogene | predicted gene, 18002 |
| 8 | gene | 94.666600 | 94.674457 | positive | MGI_C57BL6J_1349429 | Arl2bp | NCBI_Gene:107566,ENSEMBL:ENSMUSG00000031776 | MGI:1349429 | protein coding gene | ADP-ribosylation factor-like 2 binding protein |
| 8 | gene | 94.674894 | 94.696278 | negative | MGI_C57BL6J_1915051 | Pllp | NCBI_Gene:67801,ENSEMBL:ENSMUSG00000031775 | MGI:1915051 | protein coding gene | plasma membrane proteolipid |
| 8 | gene | 94.675606 | 94.680166 | positive | MGI_C57BL6J_5804882 | Gm45767 | ENSEMBL:ENSMUSG00000110702 | MGI:5804882 | lncRNA gene | predicted gene 45767 |
| 8 | gene | 94.698016 | 94.698285 | negative | MGI_C57BL6J_5452623 | Gm22846 | ENSEMBL:ENSMUSG00000088331 | MGI:5452623 | unclassified non-coding RNA gene | predicted gene, 22846 |
| 8 | gene | 94.703631 | 94.703713 | positive | MGI_C57BL6J_4413935 | n-TLcag5 | NCBI_Gene:102467555 | MGI:4413935 | tRNA gene | nuclear encoded tRNA leucine 5 (anticodon CAG) |
| 8 | gene | 94.704004 | 94.704086 | negative | MGI_C57BL6J_4413934 | n-TLcag4 | NCBI_Gene:102467335 | MGI:4413934 | tRNA gene | nuclear encoded tRNA leucine 4 (anticodon CAG) |
| 8 | gene | 94.745590 | 94.751699 | positive | MGI_C57BL6J_1306779 | Ccl22 | NCBI_Gene:20299,ENSEMBL:ENSMUSG00000031779 | MGI:1306779 | protein coding gene | chemokine (C-C motif) ligand 22 |
| 8 | pseudogene | 94.760026 | 94.761784 | positive | MGI_C57BL6J_2668078 | Gnb1-ps3 | NCBI_Gene:360131,ENSEMBL:ENSMUSG00000110708 | MGI:2668078 | pseudogene | guanine nucleotide binding protein (G protein), beta 1, pseudogene 3 |
| 8 | gene | 94.772009 | 94.782427 | positive | MGI_C57BL6J_1097153 | Cx3cl1 | NCBI_Gene:20312,ENSEMBL:ENSMUSG00000031778 | MGI:1097153 | protein coding gene | chemokine (C-X3-C motif) ligand 1 |
| 8 | gene | 94.794169 | 94.802436 | negative | MGI_C57BL6J_1914614 | 1700121C10Rik | NA | NA | unclassified gene | RIKEN cDNA 1700121C10 gene |
| 8 | gene | 94.805250 | 94.812038 | positive | MGI_C57BL6J_1329039 | Ccl17 | NCBI_Gene:20295,ENSEMBL:ENSMUSG00000031780 | MGI:1329039 | protein coding gene | chemokine (C-C motif) ligand 17 |
| 8 | gene | 94.819804 | 94.838372 | negative | MGI_C57BL6J_1922083 | Ciapin1 | NCBI_Gene:109006,ENSEMBL:ENSMUSG00000031781 | MGI:1922083 | protein coding gene | cytokine induced apoptosis inhibitor 1 |
| 8 | pseudogene | 94.835698 | 94.836559 | positive | MGI_C57BL6J_3779742 | Gm7418 | NCBI_Gene:664947,ENSEMBL:ENSMUSG00000090188 | MGI:3779742 | pseudogene | predicted pseudogene 7418 |
| 8 | gene | 94.838321 | 94.854895 | positive | MGI_C57BL6J_1915164 | Coq9 | NCBI_Gene:67914,ENSEMBL:ENSMUSG00000031782 | MGI:1915164 | protein coding gene | coenzyme Q9 |
| 8 | gene | 94.857450 | 94.873535 | positive | MGI_C57BL6J_109299 | Polr2c | NCBI_Gene:20021,ENSEMBL:ENSMUSG00000031783 | MGI:109299 | protein coding gene | polymerase (RNA) II (DNA directed) polypeptide C |
| 8 | gene | 94.863828 | 94.876330 | negative | MGI_C57BL6J_2148865 | Dok4 | NCBI_Gene:114255,ENSEMBL:ENSMUSG00000040631 | MGI:2148865 | protein coding gene | docking protein 4 |
| 8 | gene | 94.902869 | 94.918098 | negative | MGI_C57BL6J_2686927 | Ccdc102a | NCBI_Gene:234582,ENSEMBL:ENSMUSG00000063605 | MGI:2686927 | protein coding gene | coiled-coil domain containing 102A |
| 8 | gene | 94.921811 | 94.943293 | positive | MGI_C57BL6J_2685955 | Adgrg5 | NCBI_Gene:382045,ENSEMBL:ENSMUSG00000061577 | MGI:2685955 | protein coding gene | adhesion G protein-coupled receptor G5 |
| 8 | gene | 94.964677 | 94.977041 | negative | MGI_C57BL6J_5590127 | Gm30968 | NCBI_Gene:102633049 | MGI:5590127 | lncRNA gene | predicted gene, 30968 |
| 8 | pseudogene | 94.964679 | 94.969756 | negative | MGI_C57BL6J_3642117 | Gm10286 | ENSEMBL:ENSMUSG00000070111 | MGI:3642117 | pseudogene | predicted gene 10286 |
| 8 | gene | 94.974751 | 95.014217 | positive | MGI_C57BL6J_1340051 | Adgrg1 | NCBI_Gene:14766,ENSEMBL:ENSMUSG00000031785 | MGI:1340051 | protein coding gene | adhesion G protein-coupled receptor G1 |
| 8 | gene | 95.017111 | 95.045250 | positive | MGI_C57BL6J_1859670 | Adgrg3 | NCBI_Gene:54672,ENSEMBL:ENSMUSG00000060470 | MGI:1859670 | protein coding gene | adhesion G protein-coupled receptor G3 |
| 8 | gene | 95.045981 | 95.047460 | positive | MGI_C57BL6J_5804872 | Gm45757 | ENSEMBL:ENSMUSG00000110426 | MGI:5804872 | unclassified gene | predicted gene 45757 |
| 8 | gene | 95.055103 | 95.078141 | positive | MGI_C57BL6J_2685616 | Drc7 | NCBI_Gene:330830,ENSEMBL:ENSMUSG00000031786 | MGI:2685616 | protein coding gene | dynein regulatory complex subunit 7 |
| 8 | gene | 95.081175 | 95.103149 | positive | MGI_C57BL6J_1921437 | Katnb1 | NCBI_Gene:74187,ENSEMBL:ENSMUSG00000031787 | MGI:1921437 | protein coding gene | katanin p80 (WD40-containing) subunit B 1 |
| 8 | gene | 95.089671 | 95.090676 | negative | MGI_C57BL6J_5804873 | Gm45758 | ENSEMBL:ENSMUSG00000110516 | MGI:5804873 | unclassified gene | predicted gene 45758 |
| 8 | gene | 95.099828 | 95.202857 | negative | MGI_C57BL6J_109202 | Kifc3 | NCBI_Gene:16582,ENSEMBL:ENSMUSG00000031788 | MGI:109202 | protein coding gene | kinesin family member C3 |
| 8 | gene | 95.113566 | 95.129609 | positive | MGI_C57BL6J_5590383 | Gm31224 | NCBI_Gene:102633389,ENSEMBL:ENSMUSG00000110517 | MGI:5590383 | lncRNA gene | predicted gene, 31224 |
| 8 | gene | 95.126666 | 95.134867 | positive | MGI_C57BL6J_5804927 | Gm45812 | ENSEMBL:ENSMUSG00000110428 | MGI:5804927 | lncRNA gene | predicted gene 45812 |
| 8 | gene | 95.138117 | 95.148583 | positive | MGI_C57BL6J_5590195 | Gm31036 | NCBI_Gene:102633132,ENSEMBL:ENSMUSG00000110475 | MGI:5590195 | lncRNA gene | predicted gene, 31036 |
| 8 | gene | 95.201090 | 95.205011 | positive | MGI_C57BL6J_5590436 | Gm31277 | NCBI_Gene:102633459 | MGI:5590436 | lncRNA gene | predicted gene, 31277 |
| 8 | gene | 95.239043 | 95.306585 | negative | MGI_C57BL6J_2664102 | Cngb1 | NCBI_Gene:333329,ENSEMBL:ENSMUSG00000031789 | MGI:2664102 | protein coding gene | cyclic nucleotide gated channel beta 1 |
| 8 | pseudogene | 95.307101 | 95.307800 | negative | MGI_C57BL6J_3643393 | Gm5359 | NCBI_Gene:384883 | MGI:3643393 | pseudogene | predicted gene 5359 |
| 8 | gene | 95.310566 | 95.326323 | positive | MGI_C57BL6J_1920657 | Tepp | NCBI_Gene:73407,ENSEMBL:ENSMUSG00000090206 | MGI:1920657 | protein coding gene | testis, prostate and placenta expressed |
| 8 | gene | 95.323011 | 95.331950 | negative | MGI_C57BL6J_1890618 | Zfp319 | NCBI_Gene:79233,ENSEMBL:ENSMUSG00000046556 | MGI:1890618 | protein coding gene | zinc finger protein 319 |
| 8 | gene | 95.331717 | 95.347513 | positive | MGI_C57BL6J_2142454 | Usb1 | NCBI_Gene:101985,ENSEMBL:ENSMUSG00000031792 | MGI:2142454 | protein coding gene | U6 snRNA biogenesis 1 |
| 8 | gene | 95.352268 | 95.375080 | positive | MGI_C57BL6J_109320 | Mmp15 | NCBI_Gene:17388,ENSEMBL:ENSMUSG00000031790 | MGI:109320 | protein coding gene | matrix metallopeptidase 15 |
| 8 | gene | 95.420249 | 95.434869 | negative | MGI_C57BL6J_107428 | Cfap20 | NCBI_Gene:14894,ENSEMBL:ENSMUSG00000031796 | MGI:107428 | protein coding gene | cilia and flagella associated protein 20 |
| 8 | gene | 95.446096 | 95.490039 | negative | MGI_C57BL6J_88547 | Csnk2a2 | NCBI_Gene:13000,ENSEMBL:ENSMUSG00000046707 | MGI:88547 | protein coding gene | casein kinase 2, alpha prime polypeptide |
| 8 | gene | 95.468646 | 95.472133 | positive | MGI_C57BL6J_5804969 | Gm45854 | ENSEMBL:ENSMUSG00000110598 | MGI:5804969 | unclassified gene | predicted gene 45854 |
| 8 | gene | 95.480453 | 95.483623 | negative | MGI_C57BL6J_5804968 | Gm45853 | ENSEMBL:ENSMUSG00000110635 | MGI:5804968 | unclassified gene | predicted gene 45853 |
| 8 | gene | 95.496430 | 95.503718 | positive | MGI_C57BL6J_1918290 | 4933406B17Rik | NCBI_Gene:71040,ENSEMBL:ENSMUSG00000110427 | MGI:1918290 | lncRNA gene | RIKEN cDNA 4933406B17 gene |
| 8 | gene | 95.534060 | 95.558890 | positive | MGI_C57BL6J_3606076 | Ccdc113 | NCBI_Gene:244608,ENSEMBL:ENSMUSG00000036598 | MGI:3606076 | protein coding gene | coiled-coil domain containing 113 |
| 8 | gene | 95.551793 | 95.552302 | positive | MGI_C57BL6J_1920844 | 1700112L15Rik | ENSEMBL:ENSMUSG00000110577 | MGI:1920844 | unclassified gene | RIKEN cDNA 1700112L15 gene |
| 8 | gene | 95.559066 | 95.576340 | negative | MGI_C57BL6J_1918243 | Prss54 | NCBI_Gene:70993,ENSEMBL:ENSMUSG00000048400 | MGI:1918243 | protein coding gene | protease, serine 54 |
| 8 | pseudogene | 95.577452 | 95.580952 | positive | MGI_C57BL6J_3645264 | Gm5912 | NCBI_Gene:546088,ENSEMBL:ENSMUSG00000081436 | MGI:3645264 | pseudogene | predicted gene 5912 |
| 8 | gene | 95.593422 | 95.621509 | positive | MGI_C57BL6J_5590677 | Gm31518 | NCBI_Gene:102633771,ENSEMBL:ENSMUSG00000110447 | MGI:5590677 | lncRNA gene | predicted gene, 31518 |
| 8 | gene | 95.629173 | 95.630391 | negative | MGI_C57BL6J_5804817 | Gm45702 | ENSEMBL:ENSMUSG00000110483 | MGI:5804817 | lncRNA gene | predicted gene 45702 |
| 8 | gene | 95.633453 | 95.645060 | positive | MGI_C57BL6J_1926083 | Gins3 | NCBI_Gene:78833,ENSEMBL:ENSMUSG00000031669 | MGI:1926083 | protein coding gene | GINS complex subunit 3 (Psf3 homolog) |
| 8 | gene | 95.653041 | 95.657255 | positive | MGI_C57BL6J_5622115 | Gm39230 | NCBI_Gene:105243261 | MGI:5622115 | lncRNA gene | predicted gene, 39230 |
| 8 | gene | 95.659425 | 95.662571 | negative | MGI_C57BL6J_5624922 | Gm42037 | NCBI_Gene:105246813 | MGI:5624922 | lncRNA gene | predicted gene, 42037 |
| 8 | pseudogene | 95.674693 | 95.675395 | negative | MGI_C57BL6J_3648988 | Gm6013 | NCBI_Gene:547022,ENSEMBL:ENSMUSG00000110701 | MGI:3648988 | pseudogene | predicted gene 6013 |
| 8 | gene | 95.676980 | 95.715119 | positive | MGI_C57BL6J_2384590 | Ndrg4 | NCBI_Gene:234593,ENSEMBL:ENSMUSG00000036564 | MGI:2384590 | protein coding gene | N-myc downstream regulated gene 4 |
| 8 | gene | 95.715113 | 95.719010 | positive | MGI_C57BL6J_1913333 | Setd6 | NCBI_Gene:66083,ENSEMBL:ENSMUSG00000031671 | MGI:1913333 | protein coding gene | SET domain containing 6 |
| 8 | gene | 95.719451 | 95.807505 | negative | MGI_C57BL6J_2442402 | Cnot1 | NCBI_Gene:234594,ENSEMBL:ENSMUSG00000036550 | MGI:2442402 | protein coding gene | CCR4-NOT transcription complex, subunit 1 |
| 8 | pseudogene | 95.721645 | 95.723031 | positive | MGI_C57BL6J_5825699 | Gm46062 | NCBI_Gene:108167566 | MGI:5825699 | pseudogene | predicted gene, 46062 |
| 8 | gene | 95.746060 | 95.746195 | negative | MGI_C57BL6J_5456270 | Gm26493 | ENSEMBL:ENSMUSG00000065089 | MGI:5456270 | snoRNA gene | predicted gene, 26493 |
| 8 | gene | 95.753151 | 95.753214 | negative | MGI_C57BL6J_5562782 | Mir7073 | miRBase:MI0022923,NCBI_Gene:102465650,ENSEMBL:ENSMUSG00000106145 | MGI:5562782 | miRNA gene | microRNA 7073 |
| 8 | gene | 95.756950 | 95.757084 | negative | MGI_C57BL6J_5456042 | Gm26265 | ENSEMBL:ENSMUSG00000065211,ENSEMBL:ENSMUSG00000105690 | MGI:5456042 | snoRNA gene | predicted gene, 26265 |
| 8 | gene | 95.775912 | 95.778530 | negative | MGI_C57BL6J_5804877 | Gm45762 | ENSEMBL:ENSMUSG00000110556 | MGI:5804877 | unclassified gene | predicted gene 45762 |
| 8 | gene | 95.806744 | 95.821728 | positive | MGI_C57BL6J_2443064 | 4930513N10Rik | NCBI_Gene:319960,ENSEMBL:ENSMUSG00000074136 | MGI:2443064 | lncRNA gene | RIKEN cDNA 4930513N10 gene |
| 8 | gene | 95.808552 | 95.818274 | negative | MGI_C57BL6J_5590818 | Gm31659 | ENSEMBL:ENSMUSG00000110489 | MGI:5590818 | lncRNA gene | predicted gene, 31659 |
| 8 | gene | 95.825346 | 95.826156 | positive | MGI_C57BL6J_3704317 | Sap18b | NCBI_Gene:100041953,ENSEMBL:ENSMUSG00000061104 | MGI:3704317 | protein coding gene | Sin3-associated polypeptide 18B |
| 8 | gene | 95.828598 | 95.834464 | negative | MGI_C57BL6J_5590964 | Gm31805 | NCBI_Gene:102634150,ENSEMBL:ENSMUSG00000110661 | MGI:5590964 | lncRNA gene | predicted gene, 31805 |
| 8 | gene | 95.835920 | 95.853577 | negative | MGI_C57BL6J_2679005 | Slc38a7 | NCBI_Gene:234595,ENSEMBL:ENSMUSG00000036534 | MGI:2679005 | protein coding gene | solute carrier family 38, member 7 |
| 8 | gene | 95.864134 | 95.888547 | negative | MGI_C57BL6J_95792 | Got2 | NCBI_Gene:14719,ENSEMBL:ENSMUSG00000031672 | MGI:95792 | protein coding gene | glutamatic-oxaloacetic transaminase 2, mitochondrial |
| 8 | gene | 95.870814 | 95.872595 | positive | MGI_C57BL6J_5804875 | Gm45760 | ENSEMBL:ENSMUSG00000110711 | MGI:5804875 | unclassified gene | predicted gene 45760 |
| 8 | gene | 95.910574 | 95.934042 | positive | MGI_C57BL6J_5805010 | Gm45895 | ENSEMBL:ENSMUSG00000110634 | MGI:5805010 | lncRNA gene | predicted gene 45895 |
| 8 | pseudogene | 95.921527 | 95.922613 | positive | MGI_C57BL6J_3645520 | Gm8637 | NCBI_Gene:667445,ENSEMBL:ENSMUSG00000110506 | MGI:3645520 | pseudogene | predicted gene 8637 |
| 8 | gene | 96.007041 | 96.010702 | positive | MGI_C57BL6J_5591051 | Gm31892 | NCBI_Gene:102634265 | MGI:5591051 | lncRNA gene | predicted gene, 31892 |
| 8 | gene | 96.309486 | 96.361833 | positive | MGI_C57BL6J_5591164 | Gm32005 | NCBI_Gene:102634420,ENSEMBL:ENSMUSG00000110623 | MGI:5591164 | lncRNA gene | predicted gene, 32005 |
| 8 | gene | 96.318487 | 96.318618 | negative | MGI_C57BL6J_5452557 | Gm22780 | ENSEMBL:ENSMUSG00000077272 | MGI:5452557 | snoRNA gene | predicted gene, 22780 |
| 8 | gene | 96.371707 | 96.404082 | negative | MGI_C57BL6J_5591281 | Gm32122 | NCBI_Gene:102634575,ENSEMBL:ENSMUSG00000110633 | MGI:5591281 | lncRNA gene | predicted gene, 32122 |
| 8 | gene | 96.512340 | 96.513979 | positive | MGI_C57BL6J_1920573 | 1700047G07Rik | ENSEMBL:ENSMUSG00000110717 | MGI:1920573 | lncRNA gene | RIKEN cDNA 1700047G07 gene |
| 8 | gene | 96.656129 | 96.656219 | positive | MGI_C57BL6J_5453909 | Gm24132 | ENSEMBL:ENSMUSG00000077938 | MGI:5453909 | miRNA gene | predicted gene, 24132 |
| 8 | gene | 97.149419 | 97.164356 | negative | MGI_C57BL6J_5804818 | Gm45703 | ENSEMBL:ENSMUSG00000110567 | MGI:5804818 | lncRNA gene | predicted gene 45703 |
| 8 | pseudogene | 97.163793 | 97.165030 | positive | MGI_C57BL6J_3644589 | Gm5131 | NCBI_Gene:333331,ENSEMBL:ENSMUSG00000110715 | MGI:3644589 | pseudogene | predicted gene 5131 |
| 8 | gene | 97.215971 | 97.218982 | positive | MGI_C57BL6J_5804903 | Gm45788 | ENSEMBL:ENSMUSG00000110552 | MGI:5804903 | lncRNA gene | predicted gene 45788 |
| 8 | gene | 97.222534 | 97.222635 | positive | MGI_C57BL6J_5454937 | Gm25160 | ENSEMBL:ENSMUSG00000089539 | MGI:5454937 | snRNA gene | predicted gene, 25160 |
| 8 | gene | 97.255841 | 97.255950 | positive | MGI_C57BL6J_5456176 | Gm26399 | ENSEMBL:ENSMUSG00000077125 | MGI:5456176 | miRNA gene | predicted gene, 26399 |
| 8 | pseudogene | 97.344676 | 97.344984 | positive | MGI_C57BL6J_5791083 | Gm45247 | ENSEMBL:ENSMUSG00000109937 | MGI:5791083 | pseudogene | predicted gene 45247 |
| 8 | pseudogene | 97.427712 | 97.429110 | negative | MGI_C57BL6J_3648292 | Gm5913 | NCBI_Gene:546090,ENSEMBL:ENSMUSG00000081111 | MGI:3648292 | pseudogene | predicted gene 5913 |
| 8 | pseudogene | 97.565400 | 97.566610 | negative | MGI_C57BL6J_3779693 | Gm7191 | NCBI_Gene:636859,ENSEMBL:ENSMUSG00000098116 | MGI:3779693 | pseudogene | predicted gene 7191 |
| 8 | gene | 97.840048 | 97.847267 | positive | MGI_C57BL6J_5622116 | Gm39231 | NCBI_Gene:105243262,ENSEMBL:ENSMUSG00000109615 | MGI:5622116 | lncRNA gene | predicted gene, 39231 |
| 8 | pseudogene | 97.876154 | 97.876520 | positive | MGI_C57BL6J_5791082 | Gm45246 | ENSEMBL:ENSMUSG00000109645 | MGI:5791082 | pseudogene | predicted gene 45246 |
| 8 | pseudogene | 98.196676 | 98.197515 | negative | MGI_C57BL6J_5010182 | Gm17997 | NCBI_Gene:100416242,ENSEMBL:ENSMUSG00000109681 | MGI:5010182 | pseudogene | predicted gene, 17997 |
| 8 | pseudogene | 98.223527 | 98.224663 | negative | MGI_C57BL6J_3704115 | Gm7192 | NCBI_Gene:636915,ENSEMBL:ENSMUSG00000110194 | MGI:3704115 | pseudogene | predicted gene 7192 |
| 8 | gene | 98.675077 | 98.675202 | negative | MGI_C57BL6J_5453271 | Gm23494 | ENSEMBL:ENSMUSG00000080480 | MGI:5453271 | snRNA gene | predicted gene, 23494 |
| 8 | pseudogene | 98.878491 | 98.878955 | negative | MGI_C57BL6J_5588976 | Gm29817 | NCBI_Gene:102238522,ENSEMBL:ENSMUSG00000110011 | MGI:5588976 | pseudogene | predicted gene, 29817 |
| 8 | gene | 99.007127 | 99.032740 | positive | MGI_C57BL6J_3783121 | Gm15679 | NCBI_Gene:100873092,ENSEMBL:ENSMUSG00000089819 | MGI:3783121 | lncRNA gene | predicted gene 15679 |
| 8 | gene | 99.024471 | 99.416486 | negative | MGI_C57BL6J_107434 | Cdh8 | NCBI_Gene:12564,ENSEMBL:ENSMUSG00000036510 | MGI:107434 | protein coding gene | cadherin 8 |
| 8 | gene | 99.045927 | 99.049409 | negative | MGI_C57BL6J_5791133 | Gm45297 | ENSEMBL:ENSMUSG00000109775 | MGI:5791133 | unclassified gene | predicted gene 45297 |
| 8 | gene | 99.072463 | 99.072569 | positive | MGI_C57BL6J_5454470 | Gm24693 | ENSEMBL:ENSMUSG00000065819 | MGI:5454470 | snRNA gene | predicted gene, 24693 |
| 8 | gene | 99.127368 | 99.148251 | positive | MGI_C57BL6J_3642106 | Gm10632 | NA | NA | unclassified gene | predicted gene 10632 |
| 8 | pseudogene | 99.144552 | 99.148150 | negative | MGI_C57BL6J_3783123 | Gm15681 | NCBI_Gene:100135655,ENSEMBL:ENSMUSG00000086842 | MGI:3783123 | pseudogene | predicted gene 15681 |
| 8 | gene | 99.397213 | 99.506770 | negative | MGI_C57BL6J_3783122 | Gm15680 | ENSEMBL:ENSMUSG00000090116 | MGI:3783122 | lncRNA gene | predicted gene 15680 |
| 8 | gene | 99.416642 | 99.733438 | positive | MGI_C57BL6J_2443215 | A330008L17Rik | NCBI_Gene:234624,ENSEMBL:ENSMUSG00000052479 | MGI:2443215 | lncRNA gene | RIKEN cDNA A330008L17 gene |
| 8 | pseudogene | 99.664019 | 99.665407 | positive | MGI_C57BL6J_3646820 | Gm8688 | ENSEMBL:ENSMUSG00000083586 | MGI:3646820 | pseudogene | predicted gene 8688 |
| 8 | pseudogene | 99.826882 | 99.827658 | negative | MGI_C57BL6J_5011417 | Gm19232 | NCBI_Gene:100418472,ENSEMBL:ENSMUSG00000109939 | MGI:5011417 | pseudogene | predicted gene, 19232 |
| 8 | pseudogene | 100.030493 | 100.032156 | positive | MGI_C57BL6J_3705743 | Impdh2-ps | NCBI_Gene:100042069,ENSEMBL:ENSMUSG00000071041 | MGI:3705743 | pseudogene | inosine monophosphate dehydrogenase 2, pseudogene |
| 8 | pseudogene | 100.060114 | 100.061064 | positive | MGI_C57BL6J_3648738 | Gm5742 | NCBI_Gene:436054,ENSEMBL:ENSMUSG00000081383 | MGI:3648738 | pseudogene | predicted gene 5742 |
| 8 | pseudogene | 100.438758 | 100.438962 | positive | MGI_C57BL6J_5791132 | Gm45296 | ENSEMBL:ENSMUSG00000110220 | MGI:5791132 | pseudogene | predicted gene 45296 |
| 8 | pseudogene | 100.636519 | 100.638016 | positive | MGI_C57BL6J_3646323 | Gm5361 | NCBI_Gene:384895,ENSEMBL:ENSMUSG00000110014 | MGI:3646323 | pseudogene | predicted gene 5361 |
| 8 | gene | 100.661909 | 100.726355 | negative | MGI_C57BL6J_5622117 | Gm39232 | NCBI_Gene:105243264,ENSEMBL:ENSMUSG00000109769 | MGI:5622117 | lncRNA gene | predicted gene, 39232 |
| 8 | gene | 101.539584 | 101.539708 | negative | MGI_C57BL6J_5452000 | Gm22223 | ENSEMBL:ENSMUSG00000087985 | MGI:5452000 | snoRNA gene | predicted gene, 22223 |
| 8 | gene | 101.590997 | 101.614593 | positive | MGI_C57BL6J_5591582 | Gm32423 | NCBI_Gene:102634971 | MGI:5591582 | lncRNA gene | predicted gene, 32423 |
| 8 | gene | 101.759270 | 101.766950 | negative | MGI_C57BL6J_5591636 | Gm32477 | NCBI_Gene:102635041 | MGI:5591636 | lncRNA gene | predicted gene, 32477 |
| 8 | gene | 101.782958 | 101.787866 | positive | MGI_C57BL6J_5825707 | Gm46070 | NCBI_Gene:108167583 | MGI:5825707 | lncRNA gene | predicted gene, 46070 |
| 8 | gene | 102.270509 | 102.312596 | negative | MGI_C57BL6J_5622118 | Gm39233 | NCBI_Gene:105243265 | MGI:5622118 | lncRNA gene | predicted gene, 39233 |
| 8 | gene | 102.523894 | 102.523956 | positive | MGI_C57BL6J_5454379 | Gm24602 | ENSEMBL:ENSMUSG00000088596 | MGI:5454379 | snRNA gene | predicted gene, 24602 |
| 8 | pseudogene | 102.534645 | 102.534890 | positive | MGI_C57BL6J_5791258 | Gm45422 | ENSEMBL:ENSMUSG00000110069 | MGI:5791258 | pseudogene | predicted gene 45422 |
| 8 | gene | 102.632095 | 102.785983 | negative | MGI_C57BL6J_99217 | Cdh11 | NCBI_Gene:12552,ENSEMBL:ENSMUSG00000031673 | MGI:99217 | protein coding gene | cadherin 11 |
| 8 | gene | 102.654191 | 102.658188 | negative | MGI_C57BL6J_5791093 | Gm45257 | ENSEMBL:ENSMUSG00000110084 | MGI:5791093 | unclassified gene | predicted gene 45257 |
| 8 | gene | 102.660815 | 102.663585 | positive | MGI_C57BL6J_5624923 | Gm42038 | NCBI_Gene:105246814 | MGI:5624923 | lncRNA gene | predicted gene, 42038 |
| 8 | gene | 102.706049 | 102.706790 | negative | MGI_C57BL6J_1924691 | 9430099M06Rik | ENSEMBL:ENSMUSG00000109997 | MGI:1924691 | unclassified gene | RIKEN cDNA 9430099M06 gene |
| 8 | gene | 102.732390 | 102.732484 | negative | MGI_C57BL6J_5456078 | Gm26301 | ENSEMBL:ENSMUSG00000078006 | MGI:5456078 | miRNA gene | predicted gene, 26301 |
| 8 | gene | 102.780190 | 102.781042 | negative | MGI_C57BL6J_5791094 | Gm45258 | ENSEMBL:ENSMUSG00000110269 | MGI:5791094 | unclassified gene | predicted gene 45258 |
| 8 | pseudogene | 102.864775 | 102.865872 | negative | MGI_C57BL6J_3644565 | Gm8730 | NCBI_Gene:667618,ENSEMBL:ENSMUSG00000063696 | MGI:3644565 | pseudogene | predicted pseudogene 8730 |
| 8 | gene | 103.130248 | 103.144946 | negative | MGI_C57BL6J_5791227 | Gm45391 | ENSEMBL:ENSMUSG00000109848 | MGI:5791227 | lncRNA gene | predicted gene 45391 |
| 8 | gene | 103.271152 | 103.340130 | positive | MGI_C57BL6J_5591690 | Gm32531 | NCBI_Gene:102635111,ENSEMBL:ENSMUSG00000109701 | MGI:5591690 | lncRNA gene | predicted gene, 32531 |
| 8 | gene | 103.296749 | 103.316593 | positive | MGI_C57BL6J_5622119 | Gm39234 | NCBI_Gene:105243266 | MGI:5622119 | lncRNA gene | predicted gene, 39234 |
| 8 | gene | 103.343878 | 103.347534 | negative | MGI_C57BL6J_1917044 | 1600027J07Rik | NCBI_Gene:69794,ENSEMBL:ENSMUSG00000110340 | MGI:1917044 | lncRNA gene | RIKEN cDNA 1600027J07 gene |
| 8 | gene | 103.488996 | 103.492625 | positive | MGI_C57BL6J_5591748 | Gm32589 | NCBI_Gene:102635187 | MGI:5591748 | lncRNA gene | predicted gene, 32589 |
| 8 | gene | 103.531645 | 103.563623 | negative | MGI_C57BL6J_5791113 | Gm45277 | ENSEMBL:ENSMUSG00000109699 | MGI:5791113 | lncRNA gene | predicted gene 45277 |
| 8 | gene | 103.568056 | 103.571656 | negative | MGI_C57BL6J_5591801 | Gm32642 | NCBI_Gene:102635256 | MGI:5591801 | lncRNA gene | predicted gene, 32642 |
| 8 | gene | 103.603782 | 103.680384 | positive | MGI_C57BL6J_1918308 | 4933400L20Rik | NCBI_Gene:71058,ENSEMBL:ENSMUSG00000110336 | MGI:1918308 | lncRNA gene | RIKEN cDNA 4933400L20 gene |
| 8 | gene | 103.722702 | 103.728600 | positive | MGI_C57BL6J_5622120 | Gm39235 | NCBI_Gene:105243267 | MGI:5622120 | lncRNA gene | predicted gene, 39235 |
| 8 | gene | 104.011068 | 104.036762 | negative | MGI_C57BL6J_5591887 | Gm32728 | NCBI_Gene:102635371 | MGI:5591887 | lncRNA gene | predicted gene, 32728 |
| 8 | gene | 104.059324 | 104.066136 | positive | MGI_C57BL6J_5622121 | Gm39236 | NCBI_Gene:105243268 | MGI:5622121 | lncRNA gene | predicted gene, 39236 |
| 8 | pseudogene | 104.071149 | 104.071735 | positive | MGI_C57BL6J_3644122 | Gm8748 | NCBI_Gene:667656,ENSEMBL:ENSMUSG00000110019 | MGI:3644122 | pseudogene | predicted gene 8748 |
| 8 | gene | 104.088916 | 104.105873 | negative | MGI_C57BL6J_5588841 | Gm29682 | NCBI_Gene:546096,ENSEMBL:ENSMUSG00000110053 | MGI:5588841 | lncRNA gene | predicted gene, 29682 |
| 8 | gene | 104.101613 | 104.144511 | positive | MGI_C57BL6J_105057 | Cdh5 | NCBI_Gene:12562,ENSEMBL:ENSMUSG00000031871 | MGI:105057 | protein coding gene | cadherin 5 |
| 8 | gene | 104.152706 | 104.154337 | positive | MGI_C57BL6J_5791499 | Gm45663 | ENSEMBL:ENSMUSG00000109909 | MGI:5791499 | unclassified gene | predicted gene 45663 |
| 8 | gene | 104.170372 | 104.219122 | positive | MGI_C57BL6J_1929597 | Bean1 | NCBI_Gene:65115,ENSEMBL:ENSMUSG00000031872 | MGI:1929597 | protein coding gene | brain expressed, associated with Nedd4, 1 |
| 8 | gene | 104.177795 | 104.184599 | negative | MGI_C57BL6J_3642022 | Gm10631 | ENSEMBL:ENSMUSG00000074128 | MGI:3642022 | lncRNA gene | predicted gene 10631 |
| 8 | gene | 104.217202 | 104.218299 | negative | MGI_C57BL6J_5805007 | Gm45892 | ENSEMBL:ENSMUSG00000110674 | MGI:5805007 | lncRNA gene | predicted gene 45892 |
| 8 | gene | 104.220612 | 104.223609 | positive | MGI_C57BL6J_5805002 | Gm45887 | ENSEMBL:ENSMUSG00000110638 | MGI:5805002 | unclassified gene | predicted gene 45887 |
| 8 | pseudogene | 104.223747 | 104.224100 | positive | MGI_C57BL6J_5804865 | Gm45750 | ENSEMBL:ENSMUSG00000110551 | MGI:5804865 | pseudogene | predicted gene 45750 |
| 8 | gene | 104.226685 | 104.248577 | negative | MGI_C57BL6J_1913266 | Tk2 | NCBI_Gene:57813,ENSEMBL:ENSMUSG00000035824 | MGI:1913266 | protein coding gene | thymidine kinase 2, mitochondrial |
| 8 | gene | 104.250861 | 104.264938 | positive | MGI_C57BL6J_1922708 | Cklf | NCBI_Gene:75458,ENSEMBL:ENSMUSG00000054400 | MGI:1922708 | protein coding gene | chemokine-like factor |
| 8 | gene | 104.281042 | 104.293181 | negative | MGI_C57BL6J_2447160 | Cmtm2a | NCBI_Gene:73381,ENSEMBL:ENSMUSG00000074127 | MGI:2447160 | protein coding gene | CKLF-like MARVEL transmembrane domain containing 2A |
| 8 | gene | 104.281347 | 104.310081 | negative | MGI_C57BL6J_5804826 | Gm45711 | ENSEMBL:ENSMUSG00000031876 | MGI:5804826 | protein coding gene | predicted gene 45711 |
| 8 | gene | 104.292622 | 104.310145 | negative | MGI_C57BL6J_2447159 | Cmtm1 | NCBI_Gene:100504164,ENSEMBL:ENSMUSG00000110430 | MGI:2447159 | protein coding gene | CKLF-like MARVEL transmembrane domain containing 1 |
| 8 | pseudogene | 104.310832 | 104.312769 | negative | MGI_C57BL6J_5804825 | Gm45710 | ENSEMBL:ENSMUSG00000110642 | MGI:5804825 | pseudogene | predicted gene 45710 |
| 8 | gene | 104.321567 | 104.321787 | positive | MGI_C57BL6J_3779240 | Gm11020 | ENSEMBL:ENSMUSG00000078953 | MGI:3779240 | lncRNA gene | predicted gene 11020 |
| 8 | gene | 104.322204 | 104.330764 | positive | MGI_C57BL6J_2447311 | Cmtm2b | NCBI_Gene:75502,ENSEMBL:ENSMUSG00000035785 | MGI:2447311 | protein coding gene | CKLF-like MARVEL transmembrane domain containing 2B |
| 8 | pseudogene | 104.324440 | 104.324680 | negative | MGI_C57BL6J_5804993 | Gm45878 | ENSEMBL:ENSMUSG00000110600 | MGI:5804993 | pseudogene | predicted gene 45878 |
| 8 | gene | 104.339383 | 104.347672 | positive | MGI_C57BL6J_2447162 | Cmtm3 | NCBI_Gene:68119,ENSEMBL:ENSMUSG00000031875 | MGI:2447162 | protein coding gene | CKLF-like MARVEL transmembrane domain containing 3 |
| 8 | gene | 104.348191 | 104.395807 | negative | MGI_C57BL6J_2142888 | Cmtm4 | NCBI_Gene:97487,ENSEMBL:ENSMUSG00000096188 | MGI:2142888 | protein coding gene | CKLF-like MARVEL transmembrane domain containing 4 |
| 8 | gene | 104.362602 | 104.363668 | negative | MGI_C57BL6J_5804992 | Gm45877 | ENSEMBL:ENSMUSG00000110486 | MGI:5804992 | lncRNA gene | predicted gene 45877 |
| 8 | gene | 104.417674 | 104.443051 | negative | MGI_C57BL6J_107738 | Dync1li2 | NCBI_Gene:234663,ENSEMBL:ENSMUSG00000035770 | MGI:107738 | protein coding gene | dynein, cytoplasmic 1 light intermediate chain 2 |
| 8 | gene | 104.438260 | 104.439462 | positive | MGI_C57BL6J_5804846 | Gm45731 | ENSEMBL:ENSMUSG00000110689 | MGI:5804846 | unclassified gene | predicted gene 45731 |
| 8 | gene | 104.442271 | 104.443786 | positive | MGI_C57BL6J_3642019 | K230015D01Rik | ENSEMBL:ENSMUSG00000110630 | MGI:3642019 | lncRNA gene | RIKEN cDNA K230015D01 gene |
| 8 | gene | 104.446718 | 104.509910 | negative | MGI_C57BL6J_2443187 | Terb1 | NCBI_Gene:320022,ENSEMBL:ENSMUSG00000052616 | MGI:2443187 | protein coding gene | telomere repeat binding bouquet formation protein 1 |
| 8 | gene | 104.507902 | 104.508008 | positive | MGI_C57BL6J_5451925 | Gm22148 | ENSEMBL:ENSMUSG00000077633 | MGI:5451925 | snRNA gene | predicted gene, 22148 |
| 8 | gene | 104.511028 | 104.534655 | negative | MGI_C57BL6J_2384561 | Nae1 | NCBI_Gene:234664,ENSEMBL:ENSMUSG00000031878 | MGI:2384561 | protein coding gene | NEDD8 activating enzyme E1 subunit 1 |
| 8 | gene | 104.534689 | 104.550347 | positive | MGI_C57BL6J_103100 | Car7 | NCBI_Gene:12354,ENSEMBL:ENSMUSG00000031883 | MGI:103100 | protein coding gene | carbonic anhydrase 7 |
| 8 | gene | 104.551907 | 104.556987 | negative | MGI_C57BL6J_5804958 | Gm45843 | ENSEMBL:ENSMUSG00000110607 | MGI:5804958 | lncRNA gene | predicted gene 45843 |
| 8 | pseudogene | 104.577591 | 104.577796 | negative | MGI_C57BL6J_3648165 | Gm6825 | NCBI_Gene:102635660 | MGI:3648165 | pseudogene | predicted gene 6825 |
| 8 | gene | 104.591451 | 104.599026 | positive | MGI_C57BL6J_1918878 | Pdp2 | NCBI_Gene:382051,ENSEMBL:ENSMUSG00000048371 | MGI:1918878 | protein coding gene | pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| 8 | gene | 104.601911 | 104.624396 | negative | MGI_C57BL6J_106671 | Cdh16 | NCBI_Gene:12556,ENSEMBL:ENSMUSG00000031881 | MGI:106671 | protein coding gene | cadherin 16 |
| 8 | gene | 104.628066 | 104.631367 | negative | MGI_C57BL6J_1930943 | Rrad | NCBI_Gene:56437,ENSEMBL:ENSMUSG00000031880 | MGI:1930943 | protein coding gene | Ras-related associated with diabetes |
| 8 | gene | 104.630832 | 104.636492 | positive | MGI_C57BL6J_5592182 | Gm33023 | NCBI_Gene:102635769,ENSEMBL:ENSMUSG00000110652 | MGI:5592182 | lncRNA gene | predicted gene, 33023 |
| 8 | gene | 104.639838 | 104.641965 | negative | MGI_C57BL6J_1915773 | Ciao2b | NCBI_Gene:68523,ENSEMBL:ENSMUSG00000031879 | MGI:1915773 | protein coding gene | cytosolic iron-sulfur assembly component 2B |
| 8 | gene | 104.641807 | 104.661492 | positive | MGI_C57BL6J_1920784 | 1700082M22Rik | NCBI_Gene:105243269,ENSEMBL:ENSMUSG00000023737 | MGI:1920784 | lncRNA gene | RIKEN cDNA 1700082M22 gene |
| 8 | gene | 104.661828 | 104.665082 | positive | MGI_C57BL6J_98767 | Tlm | NA | NA | unclassified gene | T lymphoma oncogene |
| 8 | pseudogene | 104.675318 | 104.682211 | negative | MGI_C57BL6J_3648500 | Gm7208 | NCBI_Gene:637550,ENSEMBL:ENSMUSG00000110568 | MGI:3648500 | pseudogene | predicted gene 7208 |
| 8 | pseudogene | 104.701705 | 104.720965 | negative | MGI_C57BL6J_3648109 | Gm5159 | NCBI_Gene:382052,ENSEMBL:ENSMUSG00000110557 | MGI:3648109 | pseudogene | predicted gene 5159 |
| 8 | gene | 104.709372 | 104.709446 | negative | MGI_C57BL6J_5690720 | Gm44328 | ENSEMBL:ENSMUSG00000105612 | MGI:5690720 | miRNA gene | predicted gene, 44328 |
| 8 | gene | 104.711249 | 104.718260 | positive | MGI_C57BL6J_2444007 | 9230110F11Rik | ENSEMBL:ENSMUSG00000110497 | MGI:2444007 | lncRNA gene | RIKEN cDNA 9230110F11 gene |
| 8 | gene | 104.734003 | 104.741634 | positive | MGI_C57BL6J_2142491 | Ces2a | NCBI_Gene:102022,ENSEMBL:ENSMUSG00000055730 | MGI:2142491 | protein coding gene | carboxylesterase 2A |
| 8 | pseudogene | 104.755716 | 104.787925 | positive | MGI_C57BL6J_3645552 | Gm6831 | NCBI_Gene:628050,ENSEMBL:ENSMUSG00000110639 | MGI:3645552 | pseudogene | predicted gene 6831 |
| 8 | pseudogene | 104.757654 | 104.758506 | negative | MGI_C57BL6J_5010660 | Gm18475 | NCBI_Gene:100417237 | MGI:5010660 | pseudogene | predicted gene, 18475 |
| 8 | pseudogene | 104.785828 | 104.787612 | positive | MGI_C57BL6J_5804897 | Gm45782 | ENSEMBL:ENSMUSG00000110694 | MGI:5804897 | pseudogene | predicted gene 45782 |
| 8 | gene | 104.794802 | 104.799052 | positive | MGI_C57BL6J_3045271 | 4932416K20Rik | NCBI_Gene:442809,ENSEMBL:ENSMUSG00000069925 | MGI:3045271 | lncRNA gene | RIKEN cDNA 4932416K20 gene |
| 8 | gene | 104.828494 | 104.840093 | positive | MGI_C57BL6J_2448547 | Ces2b | NCBI_Gene:234669,ENSEMBL:ENSMUSG00000050097 | MGI:2448547 | protein coding gene | carboxyesterase 2B |
| 8 | pseudogene | 104.844370 | 104.845927 | negative | MGI_C57BL6J_5592874 | Gm33715 | NCBI_Gene:102636722 | MGI:5592874 | pseudogene | predicted gene, 33715 |
| 8 | pseudogene | 104.844378 | 104.846879 | negative | MGI_C57BL6J_1298410 | Matr3-ps2 | NCBI_Gene:17186,ENSEMBL:ENSMUSG00000089748 | MGI:1298410 | pseudogene | matrin 3, pseudogene 2 |
| 8 | gene | 104.847068 | 104.854483 | positive | MGI_C57BL6J_2385905 | Ces2c | NCBI_Gene:234671,ENSEMBL:ENSMUSG00000061825 | MGI:2385905 | protein coding gene | carboxylesterase 2C |
| 8 | gene | 104.853178 | 104.868998 | negative | MGI_C57BL6J_5592377 | Gm33218 | NCBI_Gene:102636034 | MGI:5592377 | lncRNA gene | predicted gene, 33218 |
| 8 | pseudogene | 104.867424 | 104.874083 | positive | MGI_C57BL6J_3704319 | Ces2d-ps | NCBI_Gene:667754,ENSEMBL:ENSMUSG00000031884 | MGI:3704319 | pseudogene | carboxylesterase 2D, pseudogene |
| 8 | gene | 104.869099 | 104.888372 | negative | MGI_C57BL6J_5592300 | Gm33141 | NCBI_Gene:102635929 | MGI:5592300 | lncRNA gene | predicted gene, 33141 |
| 8 | pseudogene | 104.886798 | 104.902787 | positive | MGI_C57BL6J_3646429 | Gm8798 | NCBI_Gene:667760,ENSEMBL:ENSMUSG00000110597 | MGI:3646429 | pseudogene | predicted gene 8798 |
| 8 | gene | 104.926260 | 104.934672 | positive | MGI_C57BL6J_2443170 | Ces2e | NCBI_Gene:234673,ENSEMBL:ENSMUSG00000031886 | MGI:2443170 | protein coding gene | carboxylesterase 2E |
| 8 | gene | 104.947355 | 104.961130 | positive | MGI_C57BL6J_1919153 | Ces2f | NCBI_Gene:71903,ENSEMBL:ENSMUSG00000062826 | MGI:1919153 | protein coding gene | carboxylesterase 2F |
| 8 | gene | 104.961589 | 104.969537 | positive | MGI_C57BL6J_1919611 | Ces2g | NCBI_Gene:72361,ENSEMBL:ENSMUSG00000031877 | MGI:1919611 | protein coding gene | carboxylesterase 2G |
| 8 | pseudogene | 104.984464 | 104.996811 | positive | MGI_C57BL6J_3648739 | Gm5743 | NCBI_Gene:436058,ENSEMBL:ENSMUSG00000110479 | MGI:3648739 | pseudogene | predicted gene 5743 |
| 8 | gene | 105.000853 | 105.021178 | positive | MGI_C57BL6J_3648740 | Ces2h | NCBI_Gene:436059,ENSEMBL:ENSMUSG00000091813 | MGI:3648740 | protein coding gene | carboxylesterase 2H |
| 8 | pseudogene | 105.045166 | 105.045502 | positive | MGI_C57BL6J_5804840 | Gm45725 | ENSEMBL:ENSMUSG00000110549 | MGI:5804840 | pseudogene | predicted gene 45725 |
| 8 | gene | 105.048583 | 105.061928 | positive | MGI_C57BL6J_102773 | Ces3a | NCBI_Gene:382053,ENSEMBL:ENSMUSG00000069922 | MGI:102773 | protein coding gene | carboxylesterase 3A |
| 8 | gene | 105.051049 | 105.051468 | negative | MGI_C57BL6J_5804839 | Gm45724 | ENSEMBL:ENSMUSG00000110488 | MGI:5804839 | unclassified gene | predicted gene 45724 |
| 8 | gene | 105.083723 | 105.093929 | positive | MGI_C57BL6J_3644960 | Ces3b | NCBI_Gene:13909,ENSEMBL:ENSMUSG00000062181 | MGI:3644960 | protein coding gene | carboxylesterase 3B |
| 8 | pseudogene | 105.110622 | 105.111421 | positive | MGI_C57BL6J_3643513 | Gm8804 | NCBI_Gene:667774,ENSEMBL:ENSMUSG00000110502 | MGI:3643513 | pseudogene | predicted gene 8804 |
| 8 | gene | 105.131800 | 105.150109 | positive | MGI_C57BL6J_2384581 | Ces4a | NCBI_Gene:234677,ENSEMBL:ENSMUSG00000060560 | MGI:2384581 | protein coding gene | carboxylesterase 4A |
| 8 | gene | 105.164587 | 105.170367 | positive | MGI_C57BL6J_3782002 | Gm3830 | ENSEMBL:ENSMUSG00000099465 | MGI:3782002 | lncRNA gene | predicted gene 3830 |
| 8 | gene | 105.170674 | 105.217989 | positive | MGI_C57BL6J_99851 | Cbfb | NCBI_Gene:12400,ENSEMBL:ENSMUSG00000031885 | MGI:99851 | protein coding gene | core binding factor beta |
| 8 | gene | 105.172496 | 105.172602 | positive | MGI_C57BL6J_5451840 | Gm22063 | ENSEMBL:ENSMUSG00000064494 | MGI:5451840 | snRNA gene | predicted gene, 22063 |
| 8 | gene | 105.203993 | 105.204081 | positive | MGI_C57BL6J_5454406 | Gm24629 | ENSEMBL:ENSMUSG00000084711 | MGI:5454406 | unclassified non-coding RNA gene | predicted gene, 24629 |
| 8 | gene | 105.225145 | 105.253053 | positive | MGI_C57BL6J_2443049 | D230025D16Rik | NCBI_Gene:234678,ENSEMBL:ENSMUSG00000031889 | MGI:2443049 | protein coding gene | RIKEN cDNA D230025D16 gene |
| 8 | gene | 105.252638 | 105.255319 | negative | MGI_C57BL6J_2142841 | B3gnt9 | NCBI_Gene:97440,ENSEMBL:ENSMUSG00000069920 | MGI:2142841 | protein coding gene | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| 8 | gene | 105.258286 | 105.264609 | negative | MGI_C57BL6J_109200 | Tradd | NCBI_Gene:71609,ENSEMBL:ENSMUSG00000031887 | MGI:109200 | protein coding gene | TNFRSF1A-associated via death domain |
| 8 | gene | 105.264648 | 105.269326 | positive | MGI_C57BL6J_1354697 | Fbxl8 | NCBI_Gene:50788,ENSEMBL:ENSMUSG00000033313 | MGI:1354697 | protein coding gene | F-box and leucine-rich repeat protein 8 |
| 8 | gene | 105.269741 | 105.275845 | positive | MGI_C57BL6J_1347058 | Hsf4 | NCBI_Gene:26386,ENSEMBL:ENSMUSG00000033249 | MGI:1347058 | protein coding gene | heat shock transcription factor 4 |
| 8 | gene | 105.275948 | 105.281939 | positive | MGI_C57BL6J_1925938 | Nol3 | NCBI_Gene:78688,ENSEMBL:ENSMUSG00000014776 | MGI:1925938 | protein coding gene | nucleolar protein 3 (apoptosis repressor with CARD domain) |
| 8 | gene | 105.280409 | 105.289864 | negative | MGI_C57BL6J_1921606 | 4931428F04Rik | NCBI_Gene:74356,ENSEMBL:ENSMUSG00000014837 | MGI:1921606 | protein coding gene | RIKEN cDNA 4931428F04 gene |
| 8 | gene | 105.289678 | 105.291485 | positive | MGI_C57BL6J_5012348 | Gm20163 | NCBI_Gene:102636360,ENSEMBL:ENSMUSG00000110611 | MGI:5012348 | lncRNA gene | predicted gene, 20163 |
| 8 | gene | 105.289923 | 105.296964 | negative | MGI_C57BL6J_3041195 | Exoc3l | NCBI_Gene:277978,ENSEMBL:ENSMUSG00000043251 | MGI:3041195 | protein coding gene | exocyst complex component 3-like |
| 8 | gene | 105.297222 | 105.297618 | negative | MGI_C57BL6J_1925686 | A930018C05Rik | NA | NA | unclassified gene | RIKEN cDNA A930018C05 gene |
| 8 | gene | 105.297663 | 105.305370 | positive | MGI_C57BL6J_103012 | E2f4 | NCBI_Gene:104394,ENSEMBL:ENSMUSG00000014859 | MGI:103012 | protein coding gene | E2F transcription factor 4 |
| 8 | gene | 105.305550 | 105.310760 | positive | MGI_C57BL6J_2679007 | Elmo3 | NCBI_Gene:234683,ENSEMBL:ENSMUSG00000014791 | MGI:2679007 | protein coding gene | engulfment and cell motility 3 |
| 8 | gene | 105.307792 | 105.307900 | negative | MGI_C57BL6J_5530702 | Gm27320 | ENSEMBL:ENSMUSG00000099127 | MGI:5530702 | miRNA gene | predicted gene, 27320 |
| 8 | gene | 105.308364 | 105.308460 | negative | MGI_C57BL6J_3619340 | Mir328 | miRBase:MI0000603,NCBI_Gene:723841,ENSEMBL:ENSMUSG00000070130 | MGI:3619340 | miRNA gene | microRNA 328 |
| 8 | gene | 105.312265 | 105.326339 | negative | MGI_C57BL6J_2443262 | Lrrc29 | NCBI_Gene:234684,ENSEMBL:ENSMUSG00000041679 | MGI:2443262 | protein coding gene | leucine rich repeat containing 29 |
| 8 | gene | 105.326354 | 105.335233 | positive | MGI_C57BL6J_1913570 | Tmem208 | NCBI_Gene:66320,ENSEMBL:ENSMUSG00000014856 | MGI:1913570 | protein coding gene | transmembrane protein 208 |
| 8 | gene | 105.329160 | 105.347970 | negative | MGI_C57BL6J_2679008 | Fhod1 | NCBI_Gene:234686,ENSEMBL:ENSMUSG00000014778 | MGI:2679008 | protein coding gene | formin homology 2 domain containing 1 |
| 8 | gene | 105.344210 | 105.347645 | positive | MGI_C57BL6J_5825686 | Gm46049 | NCBI_Gene:108167547 | MGI:5825686 | lncRNA gene | predicted gene, 46049 |
| 8 | gene | 105.348118 | 105.369881 | positive | MGI_C57BL6J_2685542 | Slc9a5 | NCBI_Gene:277973,ENSEMBL:ENSMUSG00000014786 | MGI:2685542 | protein coding gene | solute carrier family 9 (sodium/hydrogen exchanger), member 5 |
| 8 | gene | 105.369639 | 105.382862 | positive | MGI_C57BL6J_2142544 | Plekhg4 | NCBI_Gene:102075,ENSEMBL:ENSMUSG00000014782 | MGI:2142544 | protein coding gene | pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| 8 | gene | 105.382807 | 105.413502 | negative | MGI_C57BL6J_3045294 | Kctd19 | NCBI_Gene:279499,ENSEMBL:ENSMUSG00000051648 | MGI:3045294 | protein coding gene | potassium channel tetramerisation domain containing 19 |
| 8 | gene | 105.413571 | 105.464086 | positive | MGI_C57BL6J_2448585 | Lrrc36 | NCBI_Gene:270091,ENSEMBL:ENSMUSG00000054320 | MGI:2448585 | protein coding gene | leucine rich repeat containing 36 |
| 8 | gene | 105.467492 | 105.471526 | negative | MGI_C57BL6J_1915221 | Tppp3 | NCBI_Gene:67971,ENSEMBL:ENSMUSG00000014846 | MGI:1915221 | protein coding gene | tubulin polymerization-promoting protein family member 3 |
| 8 | gene | 105.472419 | 105.497102 | negative | MGI_C57BL6J_1918046 | Zdhhc1 | NCBI_Gene:70796,ENSEMBL:ENSMUSG00000039199 | MGI:1918046 | protein coding gene | zinc finger, DHHC domain containing 1 |
| 8 | pseudogene | 105.503199 | 105.504084 | positive | MGI_C57BL6J_3643717 | Gm8838 | NCBI_Gene:667837,ENSEMBL:ENSMUSG00000110608 | MGI:3643717 | pseudogene | predicted gene 8838 |
| 8 | gene | 105.518746 | 105.523988 | positive | MGI_C57BL6J_104720 | Hsd11b2 | NCBI_Gene:15484,ENSEMBL:ENSMUSG00000031891 | MGI:104720 | protein coding gene | hydroxysteroid 11-beta dehydrogenase 2 |
| 8 | gene | 105.524465 | 105.566047 | negative | MGI_C57BL6J_1201778 | Atp6v0d1 | NCBI_Gene:11972,ENSEMBL:ENSMUSG00000013160 | MGI:1201778 | protein coding gene | ATPase, H+ transporting, lysosomal V0 subunit D1 |
| 8 | gene | 105.566182 | 105.582002 | positive | MGI_C57BL6J_5611478 | Gm38250 | NCBI_Gene:108167550,ENSEMBL:ENSMUSG00000104168 | MGI:5611478 | lncRNA gene | predicted gene, 38250 |
| 8 | gene | 105.566695 | 105.637393 | negative | MGI_C57BL6J_892013 | Agrp | NCBI_Gene:11604,ENSEMBL:ENSMUSG00000005705 | MGI:892013 | protein coding gene | agouti related neuropeptide |
| 8 | pseudogene | 105.585319 | 105.586884 | negative | MGI_C57BL6J_5804970 | Gm45855 | NCBI_Gene:108167548,ENSEMBL:ENSMUSG00000110686 | MGI:5804970 | pseudogene | predicted gene 45855 |
| 8 | gene | 105.594082 | 105.622219 | positive | MGI_C57BL6J_1922937 | Ripor1 | NCBI_Gene:75687,ENSEMBL:ENSMUSG00000038604 | MGI:1922937 | protein coding gene | RHO family interacting cell polarization regulator 1 |
| 8 | gene | 105.615466 | 105.615573 | positive | MGI_C57BL6J_3837211 | Mir1966 | miRBase:MI0009963,NCBI_Gene:100316712,ENSEMBL:ENSMUSG00000089432 | MGI:3837211 | miRNA gene | microRNA 1966 |
| 8 | gene | 105.624336 | 105.637485 | negative | MGI_C57BL6J_3779534 | Gm5914 | NCBI_Gene:546100,ENSEMBL:ENSMUSG00000098120 | MGI:3779534 | lncRNA gene | predicted gene 5914 |
| 8 | gene | 105.633750 | 105.634871 | positive | MGI_C57BL6J_5611612 | Gm38384 | ENSEMBL:ENSMUSG00000102593 | MGI:5611612 | unclassified gene | predicted gene, 38384 |
| 8 | gene | 105.636538 | 105.682922 | positive | MGI_C57BL6J_109447 | Ctcf | NCBI_Gene:13018,ENSEMBL:ENSMUSG00000005698 | MGI:109447 | protein coding gene | CCCTC-binding factor |
| 8 | pseudogene | 105.660167 | 105.663747 | negative | MGI_C57BL6J_3642938 | Gm5915 | NCBI_Gene:102635661,ENSEMBL:ENSMUSG00000080021 | MGI:3642938 | pseudogene | predicted pseudogene 5915 |
| 8 | gene | 105.682815 | 105.686220 | negative | MGI_C57BL6J_5592737 | Gm33578 | NCBI_Gene:102636542 | MGI:5592737 | lncRNA gene | predicted gene, 33578 |
| 8 | gene | 105.686010 | 105.698195 | positive | MGI_C57BL6J_2685431 | Carmil2 | NCBI_Gene:234695,ENSEMBL:ENSMUSG00000050357 | MGI:2685431 | protein coding gene | capping protein regulator and myosin 1 linker 2 |
| 8 | gene | 105.695860 | 105.701290 | negative | MGI_C57BL6J_87873 | Acd | NCBI_Gene:497652,ENSEMBL:ENSMUSG00000038000 | MGI:87873 | protein coding gene | adrenocortical dysplasia |
| 8 | gene | 105.701148 | 105.703496 | positive | MGI_C57BL6J_1927223 | Pard6a | NCBI_Gene:56513,ENSEMBL:ENSMUSG00000005699 | MGI:1927223 | protein coding gene | par-6 family cell polarity regulator alpha |
| 8 | gene | 105.703649 | 105.708221 | negative | MGI_C57BL6J_2142593 | Enkd1 | NCBI_Gene:102124,ENSEMBL:ENSMUSG00000013155 | MGI:2142593 | protein coding gene | enkurin domain containing 1 |
| 8 | gene | 105.707655 | 105.710246 | positive | MGI_C57BL6J_1918296 | 4933405L10Rik | NCBI_Gene:71046,ENSEMBL:ENSMUSG00000013158 | MGI:1918296 | protein coding gene | RIKEN cDNA 4933405L10 gene |
| 8 | gene | 105.713854 | 105.758665 | negative | MGI_C57BL6J_1917825 | Gfod2 | NCBI_Gene:70575,ENSEMBL:ENSMUSG00000013150 | MGI:1917825 | protein coding gene | glucose-fructose oxidoreductase domain containing 2 |
| 8 | gene | 105.768308 | 105.827350 | negative | MGI_C57BL6J_1921584 | Ranbp10 | NCBI_Gene:74334,ENSEMBL:ENSMUSG00000037415 | MGI:1921584 | protein coding gene | RAN binding protein 10 |
| 8 | gene | 105.827697 | 105.844680 | positive | MGI_C57BL6J_1919486 | Tsnaxip1 | NCBI_Gene:72236,ENSEMBL:ENSMUSG00000031893 | MGI:1919486 | protein coding gene | translin-associated factor X (Tsnax) interacting protein 1 |
| 8 | gene | 105.830549 | 105.833891 | negative | MGI_C57BL6J_5825687 | Gm46050 | NCBI_Gene:108167551 | MGI:5825687 | lncRNA gene | predicted gene, 46050 |
| 8 | gene | 105.844669 | 105.855038 | negative | MGI_C57BL6J_2443939 | Cenpt | NCBI_Gene:320394,ENSEMBL:ENSMUSG00000036672 | MGI:2443939 | protein coding gene | centromere protein T |
| 8 | gene | 105.855103 | 105.856950 | positive | MGI_C57BL6J_1930964 | Thap11 | NCBI_Gene:59016,ENSEMBL:ENSMUSG00000036442 | MGI:1930964 | protein coding gene | THAP domain containing 11 |
| 8 | gene | 105.859303 | 105.860422 | negative | MGI_C57BL6J_1923171 | 4930578M07Rik | ENSEMBL:ENSMUSG00000110586 | MGI:1923171 | lncRNA gene | RIKEN cDNA 4930578M07 gene |
| 8 | gene | 105.860580 | 105.880402 | positive | MGI_C57BL6J_1915301 | Nutf2 | NCBI_Gene:68051,ENSEMBL:ENSMUSG00000008450 | MGI:1915301 | protein coding gene | nuclear transport factor 2 |
| 8 | gene | 105.880873 | 105.894908 | positive | MGI_C57BL6J_2446249 | Edc4 | NCBI_Gene:234699,ENSEMBL:ENSMUSG00000036270 | MGI:2446249 | protein coding gene | enhancer of mRNA decapping 4 |
| 8 | gene | 105.893567 | 105.895023 | positive | MGI_C57BL6J_2443642 | Nrn1l | NCBI_Gene:234700,ENSEMBL:ENSMUSG00000044287 | MGI:2443642 | protein coding gene | neuritin 1-like |
| 8 | gene | 105.900441 | 105.931802 | positive | MGI_C57BL6J_3528383 | Pskh1 | NCBI_Gene:244631,ENSEMBL:ENSMUSG00000048310 | MGI:3528383 | protein coding gene | protein serine kinase H1 |
| 8 | gene | 105.931994 | 105.933862 | negative | MGI_C57BL6J_88558 | Ctrl | NCBI_Gene:109660,ENSEMBL:ENSMUSG00000031896 | MGI:88558 | protein coding gene | chymotrypsin-like |
| 8 | gene | 105.934333 | 105.945843 | positive | MGI_C57BL6J_3801720 | Gm16156 | ENSEMBL:ENSMUSG00000087436 | MGI:3801720 | lncRNA gene | predicted gene 16156 |
| 8 | gene | 105.935728 | 105.938454 | negative | MGI_C57BL6J_1096380 | Psmb10 | NCBI_Gene:19171,ENSEMBL:ENSMUSG00000031897 | MGI:1096380 | protein coding gene | proteasome (prosome, macropain) subunit, beta type 10 |
| 8 | gene | 105.939551 | 105.943402 | negative | MGI_C57BL6J_96755 | Lcat | NCBI_Gene:16816,ENSEMBL:ENSMUSG00000035237 | MGI:96755 | protein coding gene | lecithin cholesterol acyltransferase |
| 8 | gene | 105.943590 | 105.966149 | negative | MGI_C57BL6J_1309465 | Slc12a4 | NCBI_Gene:20498,ENSEMBL:ENSMUSG00000017765 | MGI:1309465 | protein coding gene | solute carrier family 12, member 4 |
| 8 | gene | 105.951637 | 105.951699 | negative | MGI_C57BL6J_5530985 | Mir7074 | miRBase:MI0022924,NCBI_Gene:102465651,ENSEMBL:ENSMUSG00000099131 | MGI:5530985 | miRNA gene | microRNA 7074 |
| 8 | gene | 105.973097 | 105.985879 | positive | MGI_C57BL6J_5804867 | Gm45752 | ENSEMBL:ENSMUSG00000110457 | MGI:5804867 | lncRNA gene | predicted gene 45752 |
| 8 | gene | 105.973513 | 105.979429 | negative | MGI_C57BL6J_1919104 | Dpep3 | NCBI_Gene:71854,ENSEMBL:ENSMUSG00000031898 | MGI:1919104 | protein coding gene | dipeptidase 3 |
| 8 | gene | 105.984944 | 106.006347 | negative | MGI_C57BL6J_2442042 | Dpep2 | NCBI_Gene:319446,ENSEMBL:ENSMUSG00000115067,ENSEMBL:ENSMUSG00000053687 | MGI:2442042 | protein coding gene | dipeptidase 2 |
| 8 | gene | 105.988501 | 105.988631 | negative | MGI_C57BL6J_5452749 | Gm22972 | ENSEMBL:ENSMUSG00000065231 | MGI:5452749 | snoRNA gene | predicted gene, 22972 |
| 8 | gene | 105.991337 | 106.053840 | positive | MGI_C57BL6J_1913619 | Dus2 | NCBI_Gene:66369,ENSEMBL:ENSMUSG00000031901 | MGI:1913619 | protein coding gene | dihydrouridine synthase 2 |
| 8 | gene | 105.995466 | 106.005975 | negative | MGI_C57BL6J_6121577 | Dpep2nb | ENSEMBL:ENSMUSG00000115768 | MGI:6121577 | protein coding gene | Dpep2 neighbor |
| 8 | gene | 106.009616 | 106.011882 | negative | MGI_C57BL6J_1919236 | Ddx28 | NCBI_Gene:71986,ENSEMBL:ENSMUSG00000045538 | MGI:1919236 | protein coding gene | DEAD (Asp-Glu-Ala-Asp) box polypeptide 28 |
| 8 | gene | 106.058840 | 106.130537 | positive | MGI_C57BL6J_103296 | Nfatc3 | NCBI_Gene:18021,ENSEMBL:ENSMUSG00000031902 | MGI:103296 | protein coding gene | nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
| 8 | gene | 106.071615 | 106.071874 | negative | MGI_C57BL6J_5531338 | Gm27956 | ENSEMBL:ENSMUSG00000093345 | MGI:5531338 | unclassified non-coding RNA gene | predicted gene, 27956 |
| 8 | gene | 106.130319 | 106.137006 | negative | MGI_C57BL6J_1924661 | Esrp2 | NCBI_Gene:77411,ENSEMBL:ENSMUSG00000084128 | MGI:1924661 | protein coding gene | epithelial splicing regulatory protein 2 |
| 8 | gene | 106.135400 | 106.138723 | positive | MGI_C57BL6J_1917021 | 1810019D21Rik | NCBI_Gene:69771,ENSEMBL:ENSMUSG00000086390 | MGI:1917021 | lncRNA gene | RIKEN cDNA 1810019D21 gene |
| 8 | gene | 106.150351 | 106.164715 | positive | MGI_C57BL6J_2178076 | Pla2g15 | NCBI_Gene:192654,ENSEMBL:ENSMUSG00000031903 | MGI:2178076 | protein coding gene | phospholipase A2, group XV |
| 8 | gene | 106.168836 | 106.198714 | positive | MGI_C57BL6J_2142598 | Slc7a6 | NCBI_Gene:330836,ENSEMBL:ENSMUSG00000031904 | MGI:2142598 | protein coding gene | solute carrier family 7 (cationic amino acid transporter, y+ system), member 6 |
| 8 | gene | 106.200438 | 106.210992 | negative | MGI_C57BL6J_1916951 | Slc7a6os | NCBI_Gene:66432,ENSEMBL:ENSMUSG00000033106 | MGI:1916951 | protein coding gene | solute carrier family 7, member 6 opposite strand |
| 8 | gene | 106.210936 | 106.252794 | positive | MGI_C57BL6J_2384879 | Prmt7 | NCBI_Gene:214572,ENSEMBL:ENSMUSG00000060098 | MGI:2384879 | protein coding gene | protein arginine N-methyltransferase 7 |
| 8 | gene | 106.237903 | 106.425728 | negative | MGI_C57BL6J_5477280 | Gm26786 | ENSEMBL:ENSMUSG00000097026 | MGI:5477280 | lncRNA gene | predicted gene, 26786 |
| 8 | gene | 106.252548 | 106.337988 | negative | MGI_C57BL6J_1927578 | Smpd3 | NCBI_Gene:58994,ENSEMBL:ENSMUSG00000031906 | MGI:1927578 | protein coding gene | sphingomyelin phosphodiesterase 3, neutral |
| 8 | gene | 106.282229 | 106.335754 | positive | MGI_C57BL6J_1922294 | 4930506A18Rik | NCBI_Gene:75044 | MGI:1922294 | lncRNA gene | RIKEN cDNA 4930506A18 gene |
| 8 | gene | 106.283307 | 106.300017 | positive | MGI_C57BL6J_3642307 | Gm10629 | ENSEMBL:ENSMUSG00000074113 | MGI:3642307 | lncRNA gene | predicted gene 10629 |
| 8 | gene | 106.338007 | 106.339454 | positive | MGI_C57BL6J_5825688 | Gm46051 | NCBI_Gene:108167552 | MGI:5825688 | lncRNA gene | predicted gene, 46051 |
| 8 | gene | 106.358116 | 106.371283 | positive | MGI_C57BL6J_5622123 | Gm39238 | NCBI_Gene:105243273 | MGI:5622123 | lncRNA gene | predicted gene, 39238 |
| 8 | gene | 106.400045 | 106.405736 | positive | MGI_C57BL6J_5593185 | Gm34026 | NCBI_Gene:102637140 | MGI:5593185 | lncRNA gene | predicted gene, 34026 |
| 8 | gene | 106.405620 | 106.406529 | negative | MGI_C57BL6J_1915413 | A930006D01Rik | NA | NA | unclassified gene | RIKEN cDNA A930006D01 gene |
| 8 | gene | 106.415283 | 106.426598 | positive | MGI_C57BL6J_104786 | Zfp90 | NCBI_Gene:22751,ENSEMBL:ENSMUSG00000031907 | MGI:104786 | protein coding gene | zinc finger protein 90 |
| 8 | gene | 106.500505 | 106.509656 | positive | MGI_C57BL6J_3045356 | 6030452D12Rik | ENSEMBL:ENSMUSG00000054069 | MGI:3045356 | protein coding gene | RIKEN cDNA 6030452D12 gene |
| 8 | gene | 106.510852 | 106.557297 | positive | MGI_C57BL6J_88356 | Cdh3 | NCBI_Gene:12560,ENSEMBL:ENSMUSG00000061048 | MGI:88356 | protein coding gene | cadherin 3 |
| 8 | pseudogene | 106.572965 | 106.573432 | negative | MGI_C57BL6J_3641908 | Gm10073 | NCBI_Gene:100042734,ENSEMBL:ENSMUSG00000060019 | MGI:3641908 | pseudogene | predicted pseudogene 10073 |
| 8 | gene | 106.587143 | 106.594820 | positive | MGI_C57BL6J_1915942 | 1110028F18Rik | NCBI_Gene:68692,ENSEMBL:ENSMUSG00000099139 | MGI:1915942 | lncRNA gene | RIKEN cDNA 1110028F18 gene |
| 8 | gene | 106.592673 | 106.595192 | negative | MGI_C57BL6J_5622124 | Gm39239 | NCBI_Gene:105243274 | MGI:5622124 | lncRNA gene | predicted gene, 39239 |
| 8 | gene | 106.603350 | 106.670247 | positive | MGI_C57BL6J_88354 | Cdh1 | NCBI_Gene:12550,ENSEMBL:ENSMUSG00000000303 | MGI:88354 | protein coding gene | cadherin 1 |
| 8 | gene | 106.683068 | 106.851439 | positive | MGI_C57BL6J_2142786 | Tango6 | NCBI_Gene:272538,ENSEMBL:ENSMUSG00000041949 | MGI:2142786 | protein coding gene | transport and golgi organization 6 |
| 8 | gene | 106.825491 | 106.846971 | negative | MGI_C57BL6J_5593356 | Gm34197 | NCBI_Gene:102637368 | MGI:5593356 | lncRNA gene | predicted gene, 34197 |
| 8 | gene | 106.870242 | 106.884566 | positive | MGI_C57BL6J_109599 | Has3 | NCBI_Gene:15118,ENSEMBL:ENSMUSG00000031910 | MGI:109599 | protein coding gene | hyaluronan synthase 3 |
| 8 | gene | 106.883863 | 106.893602 | negative | MGI_C57BL6J_2443370 | Chtf8 | NCBI_Gene:214987,ENSEMBL:ENSMUSG00000046691 | MGI:2443370 | protein coding gene | CTF8, chromosome transmission fidelity factor 8 |
| 8 | gene | 106.883890 | 106.893593 | negative | MGI_C57BL6J_6303050 | Gm50241 | ENSEMBL:ENSMUSG00000117748 | MGI:6303050 | protein coding gene | predicted gene, 50241 |
| 8 | gene | 106.893615 | 106.923094 | positive | MGI_C57BL6J_1096573 | Utp4 | NCBI_Gene:21771,ENSEMBL:ENSMUSG00000041438 | MGI:1096573 | protein coding gene | UTP4 small subunit processome component |
| 8 | pseudogene | 106.927701 | 106.928454 | negative | MGI_C57BL6J_3648679 | Rpl10-ps2 | NCBI_Gene:234703,ENSEMBL:ENSMUSG00000074108 | MGI:3648679 | pseudogene | ribosomal protein L10, pseudogene 2 |
| 8 | gene | 106.933985 | 106.935038 | negative | MGI_C57BL6J_1925916 | 9530067L11Rik | NA | NA | unclassified gene | RIKEN cDNA 9530067L11 gene |
| 8 | gene | 106.935750 | 107.019714 | positive | MGI_C57BL6J_101771 | Sntb2 | NCBI_Gene:20650,ENSEMBL:ENSMUSG00000041308 | MGI:101771 | protein coding gene | syntrophin, basic 2 |
| 8 | gene | 106.974833 | 106.976150 | positive | MGI_C57BL6J_1920237 | 2900057C01Rik | NA | NA | unclassified gene | RIKEN cDNA 2900057C01 gene |
| 8 | gene | 107.009502 | 107.009611 | negative | MGI_C57BL6J_5451862 | Gm22085 | ENSEMBL:ENSMUSG00000088969 | MGI:5451862 | snRNA gene | predicted gene, 22085 |
| 8 | gene | 107.029411 | 107.031192 | negative | MGI_C57BL6J_3802096 | Gm16208 | NCBI_Gene:102637442,ENSEMBL:ENSMUSG00000085464 | MGI:3802096 | lncRNA gene | predicted gene 16208 |
| 8 | gene | 107.031222 | 107.045756 | positive | MGI_C57BL6J_1890520 | Vps4a | NCBI_Gene:116733,ENSEMBL:ENSMUSG00000031913 | MGI:1890520 | protein coding gene | vacuolar protein sorting 4A |
| 8 | pseudogene | 107.037088 | 107.038236 | negative | MGI_C57BL6J_3802095 | Gm16209 | NCBI_Gene:100418381,ENSEMBL:ENSMUSG00000083734 | MGI:3802095 | pseudogene | predicted gene 16209 |
| 8 | gene | 107.044698 | 107.048614 | negative | MGI_C57BL6J_1915273 | NCBI_Gene:68023,ENSEMBL:ENSMUSG00000078931 | MGI:1915273 | protein coding gene | peptide deformylase (mitochondrial) | |
| 8 | gene | 107.046289 | 107.056737 | negative | MGI_C57BL6J_2142885 | Cog8 | NCBI_Gene:97484,ENSEMBL:ENSMUSG00000031916 | MGI:2142885 | protein coding gene | component of oligomeric golgi complex 8 |
| 8 | gene | 107.048244 | 107.050563 | positive | MGI_C57BL6J_3704321 | Gm10627 | NA | NA | unclassified gene | predicted gene 10627 |
| 8 | gene | 107.056877 | 107.060931 | positive | MGI_C57BL6J_1913414 | Nip7 | NCBI_Gene:66164,ENSEMBL:ENSMUSG00000031917 | MGI:1913414 | protein coding gene | NIP7, nucleolar pre-rRNA processing protein |
| 8 | gene | 107.061476 | 107.065644 | negative | MGI_C57BL6J_1913519 | Tmed6 | NCBI_Gene:66269,ENSEMBL:ENSMUSG00000031919 | MGI:1913519 | protein coding gene | transmembrane p24 trafficking protein 6 |
| 8 | gene | 107.069400 | 107.096645 | negative | MGI_C57BL6J_1195972 | Terf2 | NCBI_Gene:21750,ENSEMBL:ENSMUSG00000031921 | MGI:1195972 | protein coding gene | telomeric repeat binding factor 2 |
| 8 | gene | 107.096025 | 107.096106 | negative | MGI_C57BL6J_5531203 | Mir7075 | miRBase:MI0022925,NCBI_Gene:102465652,ENSEMBL:ENSMUSG00000098671 | MGI:5531203 | miRNA gene | microRNA 7075 |
| 8 | gene | 107.119110 | 107.119892 | positive | MGI_C57BL6J_1925821 | C630050I24Rik | ENSEMBL:ENSMUSG00000102271 | MGI:1925821 | unclassified non-coding RNA gene | RIKEN cDNA C630050I24 gene |
| 8 | gene | 107.124087 | 107.126761 | positive | MGI_C57BL6J_5622125 | Gm39240 | NCBI_Gene:105243276 | MGI:5622125 | lncRNA gene | predicted gene, 39240 |
| 8 | gene | 107.150640 | 107.187471 | positive | MGI_C57BL6J_1913677 | Cyb5b | NCBI_Gene:66427,ENSEMBL:ENSMUSG00000031924 | MGI:1913677 | protein coding gene | cytochrome b5 type B |
| 8 | gene | 107.205122 | 107.217659 | positive | MGI_C57BL6J_1914894 | 4930517G24Rik | NCBI_Gene:67644 | MGI:1914894 | lncRNA gene | RIKEN cDNA 4930517G24 gene |
| 8 | pseudogene | 107.262746 | 107.263269 | negative | MGI_C57BL6J_3642474 | Rps18-ps3 | NCBI_Gene:100042791,ENSEMBL:ENSMUSG00000057657 | MGI:3642474 | pseudogene | ribosomal protein S18, pseudogene 3 |
| 8 | gene | 107.293001 | 107.379517 | positive | MGI_C57BL6J_1859333 | Nfat5 | NCBI_Gene:54446,ENSEMBL:ENSMUSG00000003847 | MGI:1859333 | protein coding gene | nuclear factor of activated T cells 5 |
| 8 | gene | 107.309788 | 107.310804 | positive | MGI_C57BL6J_1917935 | 3830408D07Rik | NA | NA | unclassified gene | RIKEN cDNA 3830408D07 gene |
| 8 | gene | 107.312677 | 107.314037 | positive | MGI_C57BL6J_1915922 | 1110035E04Rik | NA | NA | unclassified gene | RIKEN cDNA 1110035E04 gene |
| 8 | gene | 107.335641 | 107.336471 | positive | MGI_C57BL6J_1924852 | C430050I21Rik | NA | NA | unclassified gene | RIKEN cDNA C430050I21 gene |
| 8 | gene | 107.388225 | 107.403206 | negative | MGI_C57BL6J_103187 | Nqo1 | NCBI_Gene:18104,ENSEMBL:ENSMUSG00000003849 | MGI:103187 | protein coding gene | NAD(P)H dehydrogenase, quinone 1 |
| 8 | gene | 107.412486 | 107.425051 | negative | MGI_C57BL6J_1914869 | Nob1 | NCBI_Gene:67619,ENSEMBL:ENSMUSG00000003848 | MGI:1914869 | protein coding gene | NIN1/RPN12 binding protein 1 homolog |
| 8 | gene | 107.436334 | 107.558597 | positive | MGI_C57BL6J_1914144 | Wwp2 | NCBI_Gene:66894,ENSEMBL:ENSMUSG00000031930 | MGI:1914144 | protein coding gene | WW domain containing E3 ubiquitin protein ligase 2 |
| 8 | gene | 107.436834 | 107.438478 | positive | MGI_C57BL6J_3708790 | A430107J10Rik | NA | NA | unclassified gene | RIKEN cDNA A430107J10 gene |
| 8 | pseudogene | 107.439131 | 107.439585 | negative | MGI_C57BL6J_3704322 | Rps26-ps1 | NCBI_Gene:100042812,ENSEMBL:ENSMUSG00000059775 | MGI:3704322 | pseudogene | ribosomal protein S26, pseudogene 1 |
| 8 | pseudogene | 107.444284 | 107.444692 | negative | MGI_C57BL6J_5805015 | Gm45900 | ENSEMBL:ENSMUSG00000110501 | MGI:5805015 | pseudogene | predicted gene 45900 |
| 8 | gene | 107.446962 | 107.450204 | negative | MGI_C57BL6J_5622126 | Gm39241 | NCBI_Gene:105243277 | MGI:5622126 | lncRNA gene | predicted gene, 39241 |
| 8 | gene | 107.455456 | 107.457325 | positive | MGI_C57BL6J_3028052 | E430022E14Rik | NA | NA | unclassified gene | RIKEN cDNA E430022E14 gene |
| 8 | gene | 107.551244 | 107.551313 | positive | MGI_C57BL6J_2676825 | Mir140 | miRBase:MI0000165,NCBI_Gene:387158,ENSEMBL:ENSMUSG00000065439 | MGI:2676825 | miRNA gene | microRNA 140 |
| 8 | gene | 107.580380 | 107.588482 | negative | MGI_C57BL6J_1351511 | Psmd7 | NCBI_Gene:17463,ENSEMBL:ENSMUSG00000039067 | MGI:1351511 | protein coding gene | proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 |
| 8 | pseudogene | 107.690626 | 107.690844 | negative | MGI_C57BL6J_3643403 | Gm8940 | ENSEMBL:ENSMUSG00000110508 | MGI:3643403 | pseudogene | predicted gene 8940 |
| 8 | gene | 107.779197 | 107.823290 | positive | MGI_C57BL6J_5624925 | Gm42040 | NCBI_Gene:105246816 | MGI:5624925 | lncRNA gene | predicted gene, 42040 |
| 8 | gene | 107.800429 | 107.834337 | negative | MGI_C57BL6J_5622127 | Gm39242 | NCBI_Gene:105243278 | MGI:5622127 | lncRNA gene | predicted gene, 39242 |
| 8 | gene | 107.840957 | 107.841057 | positive | MGI_C57BL6J_5453423 | Gm23646 | ENSEMBL:ENSMUSG00000095727 | MGI:5453423 | miRNA gene | predicted gene, 23646 |
| 8 | gene | 107.842451 | 107.842551 | positive | MGI_C57BL6J_5452602 | Gm22825 | ENSEMBL:ENSMUSG00000094718 | MGI:5452602 | miRNA gene | predicted gene, 22825 |
| 8 | gene | 107.869685 | 107.885011 | positive | MGI_C57BL6J_1923255 | 5033426E14Rik | ENSEMBL:ENSMUSG00000110625 | MGI:1923255 | lncRNA gene | RIKEN cDNA 5033426E14 gene |
| 8 | gene | 107.942644 | 108.961636 | positive | MGI_C57BL6J_99948 | Zfhx3 | NCBI_Gene:11906,ENSEMBL:ENSMUSG00000038872 | MGI:99948 | protein coding gene | zinc finger homeobox 3 |
| 8 | gene | 108.305826 | 108.310983 | negative | MGI_C57BL6J_5593834 | Gm34675 | NCBI_Gene:102638000 | MGI:5593834 | lncRNA gene | predicted gene, 34675 |
| 8 | gene | 108.452224 | 108.472312 | negative | MGI_C57BL6J_5622128 | Gm39243 | NCBI_Gene:105243279 | MGI:5622128 | lncRNA gene | predicted gene, 39243 |
| 8 | gene | 108.512291 | 108.524195 | negative | MGI_C57BL6J_5624926 | Gm42041 | NCBI_Gene:105246817 | MGI:5624926 | lncRNA gene | predicted gene, 42041 |
| 8 | gene | 108.526937 | 108.538488 | negative | MGI_C57BL6J_5622129 | Gm39244 | NCBI_Gene:105243280,ENSEMBL:ENSMUSG00000109868 | MGI:5622129 | lncRNA gene | predicted gene, 39244 |
| 8 | gene | 108.553252 | 108.584677 | negative | MGI_C57BL6J_1924437 | Lncbate1 | NCBI_Gene:109215,ENSEMBL:ENSMUSG00000110613 | MGI:1924437 | lncRNA gene | brown adipose tissue enriched long non-coding RNA 1 |
| 8 | gene | 108.637580 | 108.677384 | positive | MGI_C57BL6J_5624927 | Gm42042 | NCBI_Gene:105246818 | MGI:5624927 | lncRNA gene | predicted gene, 42042 |
| 8 | gene | 108.677589 | 108.695612 | negative | MGI_C57BL6J_5624928 | Gm42043 | NCBI_Gene:105246819 | MGI:5624928 | lncRNA gene | predicted gene, 42043 |
| 8 | gene | 108.702683 | 108.703088 | negative | MGI_C57BL6J_5592948 | Gm33789 | NA | NA | protein coding gene | predicted gene%2c 33789 |
| 8 | gene | 108.737593 | 108.742800 | positive | MGI_C57BL6J_5611270 | Gm38042 | ENSEMBL:ENSMUSG00000104292 | MGI:5611270 | unclassified gene | predicted gene, 38042 |
| 8 | gene | 108.867562 | 108.869267 | positive | MGI_C57BL6J_5611546 | Gm38318 | ENSEMBL:ENSMUSG00000102989 | MGI:5611546 | unclassified gene | predicted gene, 38318 |
| 8 | gene | 108.918264 | 108.921874 | negative | MGI_C57BL6J_5313133 | Gm20686 | ENSEMBL:ENSMUSG00000093697 | MGI:5313133 | lncRNA gene | predicted gene 20686 |
| 8 | gene | 108.936860 | 108.936938 | positive | MGI_C57BL6J_4834327 | Mir3108 | miRBase:MI0014105,NCBI_Gene:100526511,ENSEMBL:ENSMUSG00000093241 | MGI:4834327 | miRNA gene | microRNA 3108 |
| 8 | pseudogene | 109.039300 | 109.040438 | positive | MGI_C57BL6J_5011110 | Gm18925 | NCBI_Gene:100417971 | MGI:5011110 | pseudogene | predicted gene, 18925 |
| 8 | gene | 109.075222 | 109.273811 | positive | MGI_C57BL6J_3037424 | D030068K23Rik | NCBI_Gene:628686,ENSEMBL:ENSMUSG00000097393 | MGI:3037424 | lncRNA gene | RIKEN cDNA D030068K23 gene |
| 8 | gene | 109.145219 | 109.249862 | negative | MGI_C57BL6J_1918204 | 4922502B01Rik | NCBI_Gene:105243282 | MGI:1918204 | lncRNA gene | RIKEN cDNA 4922502B01 gene |
| 8 | gene | 109.180755 | 109.185634 | positive | MGI_C57BL6J_2142440 | A730093L10Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA A730093L10 gene |
| 8 | pseudogene | 109.339787 | 109.340908 | negative | MGI_C57BL6J_3037801 | Gm1943 | NCBI_Gene:384864,ENSEMBL:ENSMUSG00000110528 | MGI:3037801 | pseudogene | predicted gene 1943 |
| 8 | gene | 109.377815 | 109.542642 | positive | MGI_C57BL6J_1930136 | Pmfbp1 | NCBI_Gene:56523,ENSEMBL:ENSMUSG00000031727 | MGI:1930136 | protein coding gene | polyamine modulated factor 1 binding protein 1 |
| 8 | gene | 109.425644 | 109.425744 | negative | MGI_C57BL6J_5452940 | Gm23163 | ENSEMBL:ENSMUSG00000092821 | MGI:5452940 | miRNA gene | predicted gene, 23163 |
| 8 | gene | 109.447209 | 109.462969 | negative | MGI_C57BL6J_5594304 | Gm35145 | NCBI_Gene:102638622 | MGI:5594304 | lncRNA gene | predicted gene, 35145 |
| 8 | gene | 109.467860 | 109.493962 | negative | MGI_C57BL6J_5622131 | Gm39246 | NCBI_Gene:105243283 | MGI:5622131 | lncRNA gene | predicted gene, 39246 |
| 8 | gene | 109.545281 | 109.547262 | negative | MGI_C57BL6J_1921784 | 8430428J23Rik | NA | NA | unclassified gene | RIKEN cDNA 8430428J23 gene |
| 8 | gene | 109.548003 | 109.565861 | negative | MGI_C57BL6J_1927617 | Dhx38 | NCBI_Gene:64340,ENSEMBL:ENSMUSG00000037993 | MGI:1927617 | protein coding gene | DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| 8 | gene | 109.565892 | 109.574051 | positive | MGI_C57BL6J_2443724 | Txnl4b | NCBI_Gene:234723,ENSEMBL:ENSMUSG00000031723 | MGI:2443724 | protein coding gene | thioredoxin-like 4B |
| 8 | gene | 109.575128 | 109.579172 | negative | MGI_C57BL6J_96211 | Hp | NCBI_Gene:15439,ENSEMBL:ENSMUSG00000031722 | MGI:96211 | protein coding gene | haptoglobin |
| 8 | gene | 109.583210 | 109.584295 | negative | MGI_C57BL6J_5622132 | Gm39247 | NCBI_Gene:105243284 | MGI:5622132 | lncRNA gene | predicted gene, 39247 |
| 8 | gene | 109.591343 | 109.608703 | negative | MGI_C57BL6J_1928378 | Dhodh | NCBI_Gene:56749,ENSEMBL:ENSMUSG00000031730 | MGI:1928378 | protein coding gene | dihydroorotate dehydrogenase |
| 8 | gene | 109.613993 | 109.674386 | positive | MGI_C57BL6J_2664670 | Pkd1l3 | NCBI_Gene:244646,ENSEMBL:ENSMUSG00000048827 | MGI:2664670 | protein coding gene | polycystic kidney disease 1 like 3 |
| 8 | gene | 109.671321 | 109.693294 | negative | MGI_C57BL6J_1919205 | Ist1 | NCBI_Gene:71955,ENSEMBL:ENSMUSG00000031729 | MGI:1919205 | protein coding gene | increased sodium tolerance 1 homolog (yeast) |
| 8 | gene | 109.705494 | 109.724932 | positive | MGI_C57BL6J_1923121 | Zfp821 | NCBI_Gene:75871,ENSEMBL:ENSMUSG00000031728 | MGI:1923121 | protein coding gene | zinc finger protein 821 |
| 8 | gene | 109.710048 | 109.713503 | negative | MGI_C57BL6J_5622133 | Gm39248 | NCBI_Gene:105243285 | MGI:5622133 | lncRNA gene | predicted gene, 39248 |
| 8 | gene | 109.726451 | 109.737795 | negative | MGI_C57BL6J_3694797 | Atxn1l | NCBI_Gene:52335,ENSEMBL:ENSMUSG00000069895 | MGI:3694797 | protein coding gene | ataxin 1-like |
| 8 | pseudogene | 109.751037 | 109.751952 | negative | MGI_C57BL6J_3645294 | Gm8970 | NCBI_Gene:668089 | MGI:3645294 | pseudogene | predicted gene 8970 |
| 8 | gene | 109.778554 | 109.864210 | positive | MGI_C57BL6J_101919 | Ap1g1 | NCBI_Gene:11765,ENSEMBL:ENSMUSG00000031731 | MGI:101919 | protein coding gene | adaptor protein complex AP-1, gamma 1 subunit |
| 8 | gene | 109.839295 | 109.839379 | positive | MGI_C57BL6J_3819554 | Snord71 | NCBI_Gene:100217432,ENSEMBL:ENSMUSG00000077549 | MGI:3819554 | snoRNA gene | small nucleolar RNA, C/D box 71 |
| 8 | gene | 109.840588 | 109.840689 | negative | MGI_C57BL6J_5455098 | Gm25321 | ENSEMBL:ENSMUSG00000084507 | MGI:5455098 | snRNA gene | predicted gene, 25321 |
| 8 | gene | 109.845573 | 109.847164 | negative | MGI_C57BL6J_2442853 | A230107O07Rik | NA | NA | unclassified gene | RIKEN cDNA A230107O07 gene |
| 8 | gene | 109.859984 | 109.868455 | negative | MGI_C57BL6J_4936978 | Gm17344 | ENSEMBL:ENSMUSG00000078143 | MGI:4936978 | lncRNA gene | predicted gene, 17344 |
| 8 | gene | 109.868520 | 109.944671 | positive | MGI_C57BL6J_2444928 | Phlpp2 | NCBI_Gene:244650,ENSEMBL:ENSMUSG00000031732 | MGI:2444928 | protein coding gene | PH domain and leucine rich repeat protein phosphatase 2 |
| 8 | gene | 109.925388 | 109.933663 | positive | MGI_C57BL6J_3840114 | Gm16349 | ENSEMBL:ENSMUSG00000085678 | MGI:3840114 | lncRNA gene | predicted gene 16349 |
| 8 | gene | 109.947909 | 109.962205 | negative | MGI_C57BL6J_1920858 | Marveld3 | NCBI_Gene:73608,ENSEMBL:ENSMUSG00000001672 | MGI:1920858 | protein coding gene | MARVEL (membrane-associating) domain containing 3 |
| 8 | pseudogene | 109.973953 | 109.974352 | negative | MGI_C57BL6J_5804910 | Gm45795 | ENSEMBL:ENSMUSG00000110707 | MGI:5804910 | pseudogene | predicted gene 45795 |
| 8 | gene | 109.986052 | 109.986756 | positive | MGI_C57BL6J_1913800 | 2010109N18Rik | NA | NA | unclassified gene | RIKEN cDNA 2010109N18 gene |
| 8 | gene | 109.990436 | 109.999804 | positive | MGI_C57BL6J_98487 | Tat | NCBI_Gene:234724,ENSEMBL:ENSMUSG00000001670 | MGI:98487 | protein coding gene | tyrosine aminotransferase |
| 8 | gene | 110.006525 | 110.007526 | positive | MGI_C57BL6J_1920687 | 1700064F23Rik | NCBI_Gene:73437 | MGI:1920687 | lncRNA gene | RIKEN cDNA 1700064F23 gene |
| 8 | pseudogene | 110.011375 | 110.014324 | negative | MGI_C57BL6J_5805003 | Gm45888 | ENSEMBL:ENSMUSG00000110561 | MGI:5805003 | pseudogene | predicted gene 45888 |
| 8 | gene | 110.029075 | 110.039740 | negative | MGI_C57BL6J_1349479 | Chst4 | NCBI_Gene:26887,ENSEMBL:ENSMUSG00000035930 | MGI:1349479 | protein coding gene | carbohydrate sulfotransferase 4 |
| 8 | gene | 110.047432 | 110.057176 | positive | MGI_C57BL6J_1924421 | 8030455M16Rik | NCBI_Gene:77171,ENSEMBL:ENSMUSG00000110507 | MGI:1924421 | lncRNA gene | RIKEN cDNA 8030455M16 gene |
| 8 | gene | 110.079691 | 110.092753 | positive | MGI_C57BL6J_2443465 | Zfp612 | NCBI_Gene:234725,ENSEMBL:ENSMUSG00000044676 | MGI:2443465 | protein coding gene | zinc finger protein 612 |
| 8 | gene | 110.082803 | 110.102753 | negative | MGI_C57BL6J_4937354 | Gm17720 | ENSEMBL:ENSMUSG00000091041 | MGI:4937354 | protein coding gene | predicted gene, 17720 |
| 8 | gene | 110.101207 | 110.111961 | positive | MGI_C57BL6J_5439433 | Tle7 | NCBI_Gene:102638837,ENSEMBL:ENSMUSG00000095941 | MGI:5439433 | protein coding gene | TLE family member 7 |
| 8 | gene | 110.113773 | 110.132468 | positive | MGI_C57BL6J_5594702 | Gm35543 | NCBI_Gene:102639168 | MGI:5594702 | lncRNA gene | predicted gene, 35543 |
| 8 | gene | 110.137502 | 110.168210 | negative | MGI_C57BL6J_101914 | Calb2 | NCBI_Gene:12308,ENSEMBL:ENSMUSG00000003657 | MGI:101914 | protein coding gene | calbindin 2 |
| 8 | gene | 110.215347 | 110.224489 | positive | MGI_C57BL6J_2384580 | Cmtr2 | NCBI_Gene:234728,ENSEMBL:ENSMUSG00000046441 | MGI:2384580 | protein coding gene | cap methyltransferase 2 |
| 8 | gene | 110.255042 | 110.260549 | negative | MGI_C57BL6J_5622134 | Gm39249 | NCBI_Gene:105243286 | MGI:5622134 | lncRNA gene | predicted gene, 39249 |
| 8 | gene | 110.262146 | 110.267462 | negative | MGI_C57BL6J_5477326 | Gm26832 | NCBI_Gene:102639530,ENSEMBL:ENSMUSG00000097409 | MGI:5477326 | lncRNA gene | predicted gene, 26832 |
| 8 | gene | 110.266977 | 110.610253 | positive | MGI_C57BL6J_2389007 | Hydin | NCBI_Gene:244653,ENSEMBL:ENSMUSG00000059854 | MGI:2389007 | protein coding gene | HYDIN, axonemal central pair apparatus protein |
| 8 | gene | 110.421361 | 110.421467 | negative | MGI_C57BL6J_5453404 | Gm23627 | ENSEMBL:ENSMUSG00000064393 | MGI:5453404 | snRNA gene | predicted gene, 23627 |
| 8 | gene | 110.436824 | 110.444585 | negative | MGI_C57BL6J_5594835 | Gm35676 | NCBI_Gene:102639342 | MGI:5594835 | lncRNA gene | predicted gene, 35676 |
| 8 | gene | 110.495366 | 110.502356 | negative | MGI_C57BL6J_5434638 | Gm21283 | NCBI_Gene:100861869 | MGI:5434638 | lncRNA gene | predicted gene, 21283 |
| 8 | gene | 110.498852 | 110.499142 | positive | MGI_C57BL6J_5452478 | Gm22701 | ENSEMBL:ENSMUSG00000093112 | MGI:5452478 | unclassified non-coding RNA gene | predicted gene, 22701 |
| 8 | gene | 110.498985 | 110.499044 | positive | MGI_C57BL6J_6096177 | Gm47306 | ENSEMBL:ENSMUSG00000105131 | MGI:6096177 | miRNA gene | predicted gene, 47306 |
| 8 | gene | 110.556766 | 110.556862 | negative | MGI_C57BL6J_5690881 | Gm44489 | ENSEMBL:ENSMUSG00000105825 | MGI:5690881 | miRNA gene | predicted gene, 44489 |
| 8 | gene | 110.618566 | 110.720398 | positive | MGI_C57BL6J_2157980 | Vac14 | NCBI_Gene:234729,ENSEMBL:ENSMUSG00000010936 | MGI:2157980 | protein coding gene | Vac14 homolog (S. cerevisiae) |
| 8 | gene | 110.630553 | 110.630623 | negative | MGI_C57BL6J_4413880 | n-TGgcc11 | NCBI_Gene:102467533 | MGI:4413880 | tRNA gene | nuclear encoded tRNA glycine 11 (anticodon GCC) |
| 8 | gene | 110.631246 | 110.631316 | negative | MGI_C57BL6J_4413879 | n-TGgcc10 | NCBI_Gene:102467646 | MGI:4413879 | tRNA gene | nuclear encoded tRNA glycine 10 (anticodon GCC) |
| 8 | gene | 110.721283 | 110.741401 | positive | MGI_C57BL6J_3039591 | Mtss2 | NCBI_Gene:244654,ENSEMBL:ENSMUSG00000033763 | MGI:3039591 | protein coding gene | MTSS I-BAR domain containing 2 |
| 8 | gene | 110.741829 | 110.805945 | negative | MGI_C57BL6J_1923777 | Il34 | NCBI_Gene:76527,ENSEMBL:ENSMUSG00000031750 | MGI:1923777 | protein coding gene | interleukin 34 |
| 8 | gene | 110.754992 | 110.758972 | positive | MGI_C57BL6J_3801859 | Gm15894 | NCBI_Gene:102639717,ENSEMBL:ENSMUSG00000090039 | MGI:3801859 | lncRNA gene | predicted gene 15894 |
| 8 | gene | 110.760395 | 110.780955 | positive | MGI_C57BL6J_5622135 | Gm39250 | NCBI_Gene:105243287 | MGI:5622135 | lncRNA gene | predicted gene, 39250 |
| 8 | gene | 110.805638 | 110.807477 | positive | MGI_C57BL6J_3802071 | Gm15895 | ENSEMBL:ENSMUSG00000085068 | MGI:3802071 | lncRNA gene | predicted gene 15895 |
| 8 | gene | 110.810239 | 110.846806 | negative | MGI_C57BL6J_1289341 | Sf3b3 | NCBI_Gene:101943,ENSEMBL:ENSMUSG00000033732 | MGI:1289341 | protein coding gene | splicing factor 3b, subunit 3 |
| 8 | gene | 110.838528 | 110.838616 | negative | MGI_C57BL6J_3819518 | Snord111 | NCBI_Gene:100217465,ENSEMBL:ENSMUSG00000080396 | MGI:3819518 | snoRNA gene | small nucleolar RNA, C/D box 111 |
| 8 | gene | 110.842383 | 110.842458 | negative | MGI_C57BL6J_5451970 | Gm22193 | ENSEMBL:ENSMUSG00000080518 | MGI:5451970 | snoRNA gene | predicted gene, 22193 |
| 8 | gene | 110.846600 | 110.882234 | positive | MGI_C57BL6J_2142808 | Cog4 | NCBI_Gene:102339,ENSEMBL:ENSMUSG00000031753 | MGI:2142808 | protein coding gene | component of oligomeric golgi complex 4 |
| 8 | gene | 110.863229 | 110.940314 | negative | MGI_C57BL6J_5477310 | Gm26816 | ENSEMBL:ENSMUSG00000097380 | MGI:5477310 | lncRNA gene | predicted gene, 26816 |
| 8 | gene | 110.882456 | 110.902488 | negative | MGI_C57BL6J_1916071 | Fcsk | NCBI_Gene:234730,ENSEMBL:ENSMUSG00000033703 | MGI:1916071 | protein coding gene | fucose kinase |
| 8 | gene | 110.919833 | 110.972497 | positive | MGI_C57BL6J_99427 | St3gal2 | NCBI_Gene:20444,ENSEMBL:ENSMUSG00000031749 | MGI:99427 | protein coding gene | ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| 8 | gene | 110.923116 | 110.923263 | positive | MGI_C57BL6J_5455909 | Gm26132 | ENSEMBL:ENSMUSG00000065100 | MGI:5455909 | snoRNA gene | predicted gene, 26132 |
| 8 | gene | 110.936187 | 110.939366 | negative | MGI_C57BL6J_5825689 | Gm46052 | NCBI_Gene:108167554 | MGI:5825689 | lncRNA gene | predicted gene, 46052 |
| 8 | gene | 110.942300 | 110.949609 | negative | MGI_C57BL6J_5622136 | Gm39251 | NCBI_Gene:105243288 | MGI:5622136 | lncRNA gene | predicted gene, 39251 |
| 8 | gene | 110.974551 | 110.997823 | negative | MGI_C57BL6J_99526 | Ddx19a | NCBI_Gene:13680,ENSEMBL:ENSMUSG00000015023 | MGI:99526 | protein coding gene | DEAD (Asp-Glu-Ala-Asp) box polypeptide 19a |
| 8 | gene | 111.003186 | 111.031751 | negative | MGI_C57BL6J_2148251 | Ddx19b | NCBI_Gene:234733,ENSEMBL:ENSMUSG00000033658 | MGI:2148251 | protein coding gene | DEAD (Asp-Glu-Ala-Asp) box polypeptide 19b |
| 8 | gene | 111.016450 | 111.016530 | negative | MGI_C57BL6J_4950390 | Mir3473d | miRBase:MI0018033,NCBI_Gene:100628592,ENSEMBL:ENSMUSG00000093087 | MGI:4950390 | miRNA gene | microRNA 3473d |
| 8 | gene | 111.027827 | 111.057664 | positive | MGI_C57BL6J_2384560 | Aars | NCBI_Gene:234734,ENSEMBL:ENSMUSG00000031960 | MGI:2384560 | protein coding gene | alanyl-tRNA synthetase |
| 8 | gene | 111.041948 | 111.042024 | negative | MGI_C57BL6J_5453880 | Gm24103 | ENSEMBL:ENSMUSG00000092671 | MGI:5453880 | miRNA gene | predicted gene, 24103 |
| 8 | gene | 111.056339 | 111.057664 | positive | MGI_C57BL6J_1919794 | Exosc6 | NCBI_Gene:72544,ENSEMBL:ENSMUSG00000109941 | MGI:1919794 | protein coding gene | exosome component 6 |
| 8 | gene | 111.058697 | 111.065159 | negative | MGI_C57BL6J_5621408 | Gm38523 | NCBI_Gene:102639870 | MGI:5621408 | protein coding gene | predicted gene, 38523 |
| 8 | gene | 111.058920 | 111.092426 | negative | MGI_C57BL6J_2672935 | Clec18a | NCBI_Gene:353287,ENSEMBL:ENSMUSG00000033633 | MGI:2672935 | protein coding gene | C-type lectin domain family 18, member A |
| 8 | gene | 111.062561 | 111.062631 | positive | MGI_C57BL6J_4413871 | n-TGgcc2 | NCBI_Gene:102467530 | MGI:4413871 | tRNA gene | nuclear encoded tRNA glycine 2 (anticodon GCC) |
| 8 | gene | 111.094606 | 111.139781 | positive | MGI_C57BL6J_2442188 | Pdpr | NCBI_Gene:319518,ENSEMBL:ENSMUSG00000033624 | MGI:2442188 | protein coding gene | pyruvate dehydrogenase phosphatase regulatory subunit |
| 8 | gene | 111.127444 | 111.145498 | negative | MGI_C57BL6J_1924618 | 9430091E24Rik | NCBI_Gene:434350,ENSEMBL:ENSMUSG00000084808 | MGI:1924618 | lncRNA gene | RIKEN cDNA 9430091E24 gene |
| 8 | gene | 111.154421 | 111.259216 | negative | MGI_C57BL6J_104967 | Glg1 | NCBI_Gene:20340,ENSEMBL:ENSMUSG00000003316 | MGI:104967 | protein coding gene | golgi apparatus protein 1 |
| 8 | gene | 111.194964 | 111.195045 | positive | MGI_C57BL6J_5452970 | Gm23193 | ENSEMBL:ENSMUSG00000076382 | MGI:5452970 | miRNA gene | predicted gene, 23193 |
| 8 | gene | 111.270944 | 111.300222 | negative | MGI_C57BL6J_2384584 | Rfwd3 | NCBI_Gene:234736,ENSEMBL:ENSMUSG00000033596 | MGI:2384584 | protein coding gene | ring finger and WD repeat domain 3 |
| 8 | gene | 111.286612 | 111.287767 | negative | MGI_C57BL6J_1918757 | 8430437B07Rik | NA | NA | unclassified gene | RIKEN cDNA 8430437B07 gene |
| 8 | pseudogene | 111.309675 | 111.310962 | negative | MGI_C57BL6J_3648989 | Gm6014 | NCBI_Gene:105243289 | MGI:3648989 | pseudogene | predicted gene 6014 |
| 8 | gene | 111.311681 | 111.338456 | negative | MGI_C57BL6J_1921818 | Mlkl | NCBI_Gene:74568,ENSEMBL:ENSMUSG00000012519 | MGI:1921818 | protein coding gene | mixed lineage kinase domain-like |
| 8 | pseudogene | 111.318037 | 111.318430 | positive | MGI_C57BL6J_5804827 | Gm45712 | ENSEMBL:ENSMUSG00000110484 | MGI:5804827 | pseudogene | predicted gene 45712 |
| 8 | gene | 111.345135 | 111.393824 | negative | MGI_C57BL6J_2443327 | Fa2h | NCBI_Gene:338521,ENSEMBL:ENSMUSG00000033579 | MGI:2443327 | protein coding gene | fatty acid 2-hydroxylase |
| 8 | gene | 111.418732 | 111.418863 | positive | MGI_C57BL6J_5454570 | Gm24793 | ENSEMBL:ENSMUSG00000077477 | MGI:5454570 | snoRNA gene | predicted gene, 24793 |
| 8 | gene | 111.448781 | 111.522107 | negative | MGI_C57BL6J_2442115 | Wdr59 | NCBI_Gene:319481,ENSEMBL:ENSMUSG00000031959 | MGI:2442115 | protein coding gene | WD repeat domain 59 |
| 8 | gene | 111.450200 | 111.450322 | positive | MGI_C57BL6J_5531107 | Mir1957b | miRBase:MI0021929,ENSEMBL:ENSMUSG00000098410 | MGI:5531107 | miRNA gene | microRNA 1957b |
| 8 | gene | 111.536097 | 111.626030 | positive | MGI_C57BL6J_2177308 | Znrf1 | NCBI_Gene:170737,ENSEMBL:ENSMUSG00000033545 | MGI:2177308 | protein coding gene | zinc and ring finger 1 |
| 8 | gene | 111.623785 | 111.630374 | negative | MGI_C57BL6J_106428 | Ldhd | NCBI_Gene:52815,ENSEMBL:ENSMUSG00000031958 | MGI:106428 | protein coding gene | lactate dehydrogenase D |
| 8 | gene | 111.643401 | 111.671108 | positive | MGI_C57BL6J_99154 | Zfp1 | NCBI_Gene:22640,ENSEMBL:ENSMUSG00000055835 | MGI:99154 | protein coding gene | zinc finger protein 1 |
| 8 | gene | 111.686510 | 111.691010 | negative | MGI_C57BL6J_88559 | Ctrb1 | NCBI_Gene:66473,ENSEMBL:ENSMUSG00000031957 | MGI:88559 | protein coding gene | chymotrypsinogen B1 |
| 8 | gene | 111.689601 | 111.689674 | negative | MGI_C57BL6J_5530700 | Mir7076 | miRBase:MI0022926,NCBI_Gene:102466993,ENSEMBL:ENSMUSG00000099129 | MGI:5530700 | miRNA gene | microRNA 7076 |
| 8 | gene | 111.710474 | 111.743849 | negative | MGI_C57BL6J_108091 | Bcar1 | NCBI_Gene:12927,ENSEMBL:ENSMUSG00000031955 | MGI:108091 | protein coding gene | breast cancer anti-estrogen resistance 1 |
| 8 | gene | 111.748667 | 111.754711 | negative | MGI_C57BL6J_5622137 | Gm39252 | NCBI_Gene:105243290 | MGI:5622137 | lncRNA gene | predicted gene, 39252 |
| 8 | gene | 111.768473 | 111.854310 | negative | MGI_C57BL6J_1344403 | Cfdp1 | NCBI_Gene:23837,ENSEMBL:ENSMUSG00000031954 | MGI:1344403 | protein coding gene | craniofacial development protein 1 |
| 8 | gene | 111.863038 | 111.876751 | negative | MGI_C57BL6J_106426 | Tmem170 | NCBI_Gene:66817,ENSEMBL:ENSMUSG00000031953 | MGI:106426 | protein coding gene | transmembrane protein 170 |
| 8 | gene | 111.879304 | 111.879407 | positive | MGI_C57BL6J_5453630 | Gm23853 | ENSEMBL:ENSMUSG00000089000 | MGI:5453630 | snRNA gene | predicted gene, 23853 |
| 8 | gene | 111.889135 | 111.910447 | negative | MGI_C57BL6J_1931825 | Chst5 | NCBI_Gene:56773,ENSEMBL:ENSMUSG00000031952 | MGI:1931825 | protein coding gene | carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 5 |
| 8 | gene | 111.912011 | 111.933881 | negative | MGI_C57BL6J_2685024 | Tmem231 | NCBI_Gene:234740,ENSEMBL:ENSMUSG00000031951 | MGI:2685024 | protein coding gene | transmembrane protein 231 |
| 8 | gene | 111.914116 | 111.929092 | positive | MGI_C57BL6J_5595690 | Gm36531 | NCBI_Gene:102640482 | MGI:5595690 | lncRNA gene | predicted gene, 36531 |
| 8 | gene | 111.940703 | 111.953612 | positive | MGI_C57BL6J_1890602 | Gabarapl2 | NCBI_Gene:93739,ENSEMBL:ENSMUSG00000031950 | MGI:1890602 | protein coding gene | gamma-aminobutyric acid (GABA) A receptor-associated protein-like 2 |
| 8 | gene | 111.966897 | 111.992311 | negative | MGI_C57BL6J_1353631 | Adat1 | NCBI_Gene:30947,ENSEMBL:ENSMUSG00000031949 | MGI:1353631 | protein coding gene | adenosine deaminase, tRNA-specific 1 |
| 8 | gene | 111.977511 | 111.981933 | negative | MGI_C57BL6J_2142886 | C88045 | NA | NA | unclassified gene | expressed sequence C88045 |
| 8 | gene | 111.993439 | 112.011354 | negative | MGI_C57BL6J_1934754 | Kars | NCBI_Gene:85305,ENSEMBL:ENSMUSG00000031948 | MGI:1934754 | protein coding gene | lysyl-tRNA synthetase |
| 8 | gene | 112.011359 | 112.020528 | positive | MGI_C57BL6J_1929871 | Terf2ip | NCBI_Gene:57321,ENSEMBL:ENSMUSG00000033430 | MGI:1929871 | protein coding gene | telomeric repeat binding factor 2, interacting protein |
| 8 | pseudogene | 112.013433 | 112.015036 | negative | MGI_C57BL6J_3643578 | Gm6793 | NCBI_Gene:627828,ENSEMBL:ENSMUSG00000092086 | MGI:3643578 | pseudogene | predicted gene 6793 |
| 8 | gene | 112.024933 | 112.027351 | negative | MGI_C57BL6J_3779974 | Gm9564 | NCBI_Gene:672498 | MGI:3779974 | protein coding gene | predicted gene 9564 |
| 8 | gene | 112.061572 | 112.063676 | positive | MGI_C57BL6J_5595830 | Gm36671 | NCBI_Gene:102640657 | MGI:5595830 | lncRNA gene | predicted gene, 36671 |
| 8 | gene | 112.062505 | 112.062611 | positive | MGI_C57BL6J_5452604 | Gm22827 | ENSEMBL:ENSMUSG00000065700 | MGI:5452604 | snRNA gene | predicted gene, 22827 |
| 8 | pseudogene | 112.236332 | 112.237064 | negative | MGI_C57BL6J_3781811 | Gm3635 | NCBI_Gene:100042040,ENSEMBL:ENSMUSG00000110435 | MGI:3781811 | pseudogene | predicted gene 3635 |
| 8 | gene | 112.293848 | 112.297155 | negative | MGI_C57BL6J_5622138 | Gm39253 | NCBI_Gene:105243291 | MGI:5622138 | lncRNA gene | predicted gene, 39253 |
| 8 | gene | 112.472122 | 112.475506 | negative | MGI_C57BL6J_5622139 | Gm39254 | NCBI_Gene:105243292 | MGI:5622139 | lncRNA gene | predicted gene, 39254 |
| 8 | gene | 112.554617 | 112.569575 | negative | MGI_C57BL6J_5504109 | Gm26994 | NCBI_Gene:102640845,ENSEMBL:ENSMUSG00000098075 | MGI:5504109 | lncRNA gene | predicted gene, 26994 |
| 8 | gene | 112.569963 | 112.882717 | positive | MGI_C57BL6J_2183572 | Cntnap4 | NCBI_Gene:170571,ENSEMBL:ENSMUSG00000031772 | MGI:2183572 | protein coding gene | contactin associated protein-like 4 |
| 8 | gene | 112.581636 | 112.581740 | positive | MGI_C57BL6J_5452441 | Gm22664 | ENSEMBL:ENSMUSG00000089344 | MGI:5452441 | unclassified non-coding RNA gene | predicted gene, 22664 |
| 8 | gene | 112.880460 | 113.004997 | negative | MGI_C57BL6J_5805019 | Gm45904 | ENSEMBL:ENSMUSG00000110455 | MGI:5805019 | lncRNA gene | predicted gene 45904 |
| 8 | gene | 112.961448 | 112.962560 | positive | MGI_C57BL6J_1915879 | 1110013I04Rik | NA | NA | unclassified gene | RIKEN cDNA 1110013I04 gene |
| 8 | gene | 113.067262 | 113.093334 | positive | MGI_C57BL6J_3642651 | Gm10280 | NCBI_Gene:791378,ENSEMBL:ENSMUSG00000110541 | MGI:3642651 | lncRNA gene | predicted gene 10280 |
| 8 | gene | 113.134783 | 113.140159 | negative | MGI_C57BL6J_5589055 | Gm29896 | NCBI_Gene:102631599 | MGI:5589055 | lncRNA gene | predicted gene, 29896 |
| 8 | gene | 113.372042 | 113.425842 | negative | MGI_C57BL6J_5589114 | Gm29955 | NCBI_Gene:102631677 | MGI:5589114 | lncRNA gene | predicted gene, 29955 |
| 8 | gene | 113.517282 | 113.550808 | positive | MGI_C57BL6J_1925298 | 4930571O06Rik | NCBI_Gene:78048 | MGI:1925298 | lncRNA gene | RIKEN cDNA 4930571O06 gene |
| 8 | gene | 113.519476 | 113.519606 | negative | MGI_C57BL6J_5455543 | Gm25766 | ENSEMBL:ENSMUSG00000077289 | MGI:5455543 | snoRNA gene | predicted gene, 25766 |
| 8 | gene | 113.528510 | 113.528671 | negative | MGI_C57BL6J_5454068 | Gm24291 | ENSEMBL:ENSMUSG00000088258 | MGI:5454068 | snRNA gene | predicted gene, 24291 |
| 8 | pseudogene | 113.563320 | 113.573617 | negative | MGI_C57BL6J_5825701 | Gm46064 | NCBI_Gene:108167574 | MGI:5825701 | pseudogene | predicted gene, 46064 |
| 8 | pseudogene | 113.589042 | 113.591133 | positive | MGI_C57BL6J_5010580 | Gm18395 | NCBI_Gene:100417091 | MGI:5010580 | pseudogene | predicted gene, 18395 |
| 8 | gene | 113.633934 | 113.635780 | negative | MGI_C57BL6J_1918138 | 4921521C08Rik | NA | NA | unclassified gene | RIKEN cDNA 4921521C08 gene |
| 8 | gene | 113.635586 | 113.645192 | positive | MGI_C57BL6J_1923231 | Mon1b | NCBI_Gene:270096,ENSEMBL:ENSMUSG00000078908 | MGI:1923231 | protein coding gene | MON1 homolog B, secretory traffciking associated |
| 8 | gene | 113.643213 | 113.655533 | positive | MGI_C57BL6J_1922247 | Syce1l | NCBI_Gene:668110,ENSEMBL:ENSMUSG00000033409 | MGI:1922247 | protein coding gene | synaptonemal complex central element protein 1 like |
| 8 | pseudogene | 113.647900 | 113.648263 | negative | MGI_C57BL6J_5804904 | Gm45789 | ENSEMBL:ENSMUSG00000110581 | MGI:5804904 | pseudogene | predicted gene 45789 |
| 8 | gene | 113.667926 | 113.676458 | negative | MGI_C57BL6J_5589211 | Gm30052 | NCBI_Gene:102631810,ENSEMBL:ENSMUSG00000110582 | MGI:5589211 | lncRNA gene | predicted gene, 30052 |
| 8 | gene | 113.697123 | 113.850502 | negative | MGI_C57BL6J_2442600 | Adamts18 | NCBI_Gene:208936,ENSEMBL:ENSMUSG00000053399 | MGI:2442600 | protein coding gene | a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 18 |
| 8 | gene | 114.010658 | 114.081140 | negative | MGI_C57BL6J_5589266 | Gm30107 | NCBI_Gene:102631886 | MGI:5589266 | lncRNA gene | predicted gene, 30107 |
| 8 | gene | 114.133557 | 114.154739 | positive | MGI_C57BL6J_1914778 | Nudt7 | NCBI_Gene:67528,ENSEMBL:ENSMUSG00000031767 | MGI:1914778 | protein coding gene | nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| 8 | gene | 114.153407 | 114.154939 | positive | MGI_C57BL6J_2670975 | BC024137 | NA | NA | unclassified gene | cDNA sequence BC024137 |
| 8 | gene | 114.189017 | 114.205589 | negative | MGI_C57BL6J_5589431 | Gm30272 | NCBI_Gene:102632115 | MGI:5589431 | lncRNA gene | predicted gene, 30272 |
| 8 | gene | 114.205582 | 114.374074 | positive | MGI_C57BL6J_2142534 | Vat1l | NCBI_Gene:270097,ENSEMBL:ENSMUSG00000046844 | MGI:2142534 | protein coding gene | vesicle amine transport protein 1 like |
| 8 | gene | 114.341160 | 114.351479 | negative | MGI_C57BL6J_1918372 | 4933408N05Rik | NCBI_Gene:71122,ENSEMBL:ENSMUSG00000084848 | MGI:1918372 | lncRNA gene | RIKEN cDNA 4933408N05 gene |
| 8 | gene | 114.418034 | 114.428133 | positive | MGI_C57BL6J_2685642 | Clec3a | NCBI_Gene:403395,ENSEMBL:ENSMUSG00000008874 | MGI:2685642 | protein coding gene | C-type lectin domain family 3, member a |
| 8 | gene | 114.423020 | 114.435809 | negative | MGI_C57BL6J_5589489 | Gm30330 | NCBI_Gene:102632190 | MGI:5589489 | lncRNA gene | predicted gene, 30330 |
| 8 | gene | 114.439635 | 115.352712 | positive | MGI_C57BL6J_1931237 | Wwox | NCBI_Gene:80707,ENSEMBL:ENSMUSG00000004637 | MGI:1931237 | protein coding gene | WW domain-containing oxidoreductase |
| 8 | gene | 114.588688 | 114.601179 | negative | MGI_C57BL6J_3802040 | Gm16116 | NCBI_Gene:102632568,ENSEMBL:ENSMUSG00000090217 | MGI:3802040 | lncRNA gene | predicted gene 16116 |
| 8 | gene | 114.609330 | 114.620211 | negative | MGI_C57BL6J_5589713 | Gm30554 | NCBI_Gene:102632496 | MGI:5589713 | lncRNA gene | predicted gene, 30554 |
| 8 | gene | 114.639546 | 114.642114 | negative | MGI_C57BL6J_3801925 | Gm16117 | ENSEMBL:ENSMUSG00000089723 | MGI:3801925 | lncRNA gene | predicted gene 16117 |
| 8 | gene | 114.698439 | 114.711521 | negative | MGI_C57BL6J_3801926 | Gm16118 | NCBI_Gene:102632420,ENSEMBL:ENSMUSG00000089797 | MGI:3801926 | lncRNA gene | predicted gene 16118 |
| 8 | gene | 114.952104 | 114.962544 | negative | MGI_C57BL6J_5622142 | Gm39257 | NCBI_Gene:105243295 | MGI:5622142 | lncRNA gene | predicted gene, 39257 |
| 8 | gene | 114.976711 | 114.978180 | positive | MGI_C57BL6J_1920349 | 3100003L13Rik | NA | NA | unclassified gene | RIKEN cDNA 3100003L13 gene |
| 8 | gene | 115.052814 | 115.052913 | negative | MGI_C57BL6J_5452333 | Gm22556 | ENSEMBL:ENSMUSG00000088428 | MGI:5452333 | rRNA gene | predicted gene, 22556 |
| 8 | gene | 115.585392 | 115.595152 | positive | MGI_C57BL6J_1922282 | 4930488N15Rik | ENSEMBL:ENSMUSG00000110592 | MGI:1922282 | lncRNA gene | RIKEN cDNA 4930488N15 gene |
| 8 | gene | 115.591170 | 115.591273 | negative | MGI_C57BL6J_5453454 | Gm23677 | ENSEMBL:ENSMUSG00000065090 | MGI:5453454 | snRNA gene | predicted gene, 23677 |
| 8 | gene | 115.635165 | 115.644889 | negative | MGI_C57BL6J_5622141 | Gm39256 | NCBI_Gene:105243294 | MGI:5622141 | lncRNA gene | predicted gene, 39256 |
| 8 | gene | 115.682942 | 115.707794 | negative | MGI_C57BL6J_96909 | Maf | NCBI_Gene:17132,ENSEMBL:ENSMUSG00000055435 | MGI:96909 | protein coding gene | avian musculoaponeurotic fibrosarcoma oncogene homolog |
| 8 | gene | 115.687724 | 115.692320 | negative | MGI_C57BL6J_2444447 | 6030439D06Rik | NA | NA | unclassified gene | RIKEN cDNA 6030439D06 gene |
| 8 | gene | 115.705078 | 115.707233 | positive | MGI_C57BL6J_3805551 | Gm15655 | ENSEMBL:ENSMUSG00000086717 | MGI:3805551 | lncRNA gene | predicted gene 15655 |
| 8 | gene | 115.715872 | 115.730008 | positive | MGI_C57BL6J_5589871 | Gm30712 | NCBI_Gene:102632716 | MGI:5589871 | lncRNA gene | predicted gene, 30712 |
| 8 | gene | 115.770861 | 115.771963 | positive | MGI_C57BL6J_1922370 | 4930509E22Rik | NA | NA | unclassified gene | RIKEN cDNA 4930509E22 gene |
| 8 | gene | 115.830224 | 115.841969 | positive | MGI_C57BL6J_5590084 | Gm30925 | NCBI_Gene:102632989 | MGI:5590084 | lncRNA gene | predicted gene, 30925 |
| 8 | gene | 115.856253 | 115.869493 | positive | MGI_C57BL6J_5590196 | Gm31037 | NCBI_Gene:102633133 | MGI:5590196 | lncRNA gene | predicted gene, 31037 |
| 8 | gene | 115.872900 | 115.878136 | positive | MGI_C57BL6J_5590265 | Gm31106 | NCBI_Gene:102633224 | MGI:5590265 | lncRNA gene | predicted gene, 31106 |
| 8 | gene | 115.904142 | 115.906537 | negative | MGI_C57BL6J_5590323 | Gm31164 | NCBI_Gene:102633305 | MGI:5590323 | lncRNA gene | predicted gene, 31164 |
| 8 | gene | 115.971635 | 115.972071 | negative | MGI_C57BL6J_5804848 | Gm45733 | ENSEMBL:ENSMUSG00000110620 | MGI:5804848 | lncRNA gene | predicted gene 45733 |
| 8 | gene | 116.181451 | 116.187369 | negative | MGI_C57BL6J_1923031 | 4930422C21Rik | ENSEMBL:ENSMUSG00000110476 | MGI:1923031 | lncRNA gene | RIKEN cDNA 4930422C21 gene |
| 8 | gene | 116.190682 | 116.192533 | positive | MGI_C57BL6J_5590384 | Gm31225 | NCBI_Gene:102633390 | MGI:5590384 | lncRNA gene | predicted gene, 31225 |
| 8 | gene | 116.243515 | 116.352435 | negative | MGI_C57BL6J_5590437 | Gm31278 | NCBI_Gene:102633460 | MGI:5590437 | lncRNA gene | predicted gene, 31278 |
| 8 | gene | 116.357061 | 116.359550 | positive | MGI_C57BL6J_1920580 | 1700018P08Rik | ENSEMBL:ENSMUSG00000110709 | MGI:1920580 | lncRNA gene | RIKEN cDNA 1700018P08 gene |
| 8 | pseudogene | 116.489046 | 116.491218 | positive | MGI_C57BL6J_3643650 | Gm5357 | NCBI_Gene:384868,ENSEMBL:ENSMUSG00000110700 | MGI:3643650 | pseudogene | predicted gene 5357 |
| 8 | pseudogene | 116.501698 | 116.502572 | positive | MGI_C57BL6J_3643990 | Gm9058 | NCBI_Gene:668233 | MGI:3643990 | pseudogene | predicted gene 9058 |
| 8 | gene | 116.504974 | 116.515915 | positive | MGI_C57BL6J_1922715 | Dynlrb2 | NCBI_Gene:75465,ENSEMBL:ENSMUSG00000034467 | MGI:1922715 | protein coding gene | dynein light chain roadblock-type 2 |
| 8 | gene | 116.527885 | 116.529401 | negative | MGI_C57BL6J_5622143 | Gm39258 | NCBI_Gene:105243297 | MGI:5622143 | lncRNA gene | predicted gene, 39258 |
| 8 | gene | 116.568723 | 116.732991 | negative | MGI_C57BL6J_1923046 | Cdyl2 | NCBI_Gene:75796,ENSEMBL:ENSMUSG00000031758 | MGI:1923046 | protein coding gene | chromodomain protein, Y chromosome-like 2 |
| 8 | gene | 116.605918 | 116.610520 | positive | MGI_C57BL6J_5590526 | Gm31367 | NCBI_Gene:102633575 | MGI:5590526 | lncRNA gene | predicted gene, 31367 |
| 8 | pseudogene | 116.764144 | 116.765479 | negative | MGI_C57BL6J_3708792 | Gm10620 | ENSEMBL:ENSMUSG00000074072 | MGI:3708792 | pseudogene | predicted gene 10620 |
| 8 | gene | 116.781712 | 116.801412 | negative | MGI_C57BL6J_1925463 | 4930563M20Rik | NCBI_Gene:78213 | MGI:1925463 | lncRNA gene | RIKEN cDNA 4930563M20 gene |
| 8 | gene | 116.841972 | 116.853185 | negative | MGI_C57BL6J_5624930 | Gm42045 | NCBI_Gene:105246821 | MGI:5624930 | lncRNA gene | predicted gene, 42045 |
| 8 | gene | 116.888685 | 116.944209 | negative | MGI_C57BL6J_1913781 | Cmc2 | NCBI_Gene:66531,ENSEMBL:ENSMUSG00000014633 | MGI:1913781 | protein coding gene | COX assembly mitochondrial protein 2 |
| 8 | gene | 116.921667 | 116.941507 | positive | MGI_C57BL6J_1919405 | Cenpn | NCBI_Gene:72155,ENSEMBL:ENSMUSG00000031756 | MGI:1919405 | protein coding gene | centromere protein N |
| 8 | gene | 116.943388 | 116.960445 | positive | MGI_C57BL6J_2682328 | Atmin | NCBI_Gene:234776,ENSEMBL:ENSMUSG00000047388 | MGI:2682328 | protein coding gene | ATM interactor |
| 8 | gene | 116.969594 | 116.978959 | negative | MGI_C57BL6J_1916778 | 1700030J22Rik | NCBI_Gene:69528,ENSEMBL:ENSMUSG00000031847 | MGI:1916778 | protein coding gene | RIKEN cDNA 1700030J22 gene |
| 8 | gene | 116.981810 | 116.993537 | negative | MGI_C57BL6J_1915383 | Gcsh | NCBI_Gene:68133,ENSEMBL:ENSMUSG00000034424 | MGI:1915383 | protein coding gene | glycine cleavage system protein H (aminomethyl carrier) |
| 8 | gene | 116.995679 | 117.082449 | negative | MGI_C57BL6J_2664668 | Pkd1l2 | NCBI_Gene:76645,ENSEMBL:ENSMUSG00000034416 | MGI:2664668 | protein coding gene | polycystic kidney disease 1 like 2 |
| 8 | gene | 117.019043 | 117.021941 | positive | MGI_C57BL6J_5590678 | Gm31519 | NCBI_Gene:102633772 | MGI:5590678 | lncRNA gene | predicted gene, 31519 |
| 8 | gene | 117.032807 | 117.043639 | positive | MGI_C57BL6J_3705091 | Gm15395 | NCBI_Gene:102633692,ENSEMBL:ENSMUSG00000085688 | MGI:3705091 | lncRNA gene | predicted gene 15395 |
| 8 | gene | 117.095851 | 117.133720 | positive | MGI_C57BL6J_1926923 | Bco1 | NCBI_Gene:63857,ENSEMBL:ENSMUSG00000031845 | MGI:1926923 | protein coding gene | beta-carotene oxygenase 1 |
| 8 | pseudogene | 117.125335 | 117.125738 | positive | MGI_C57BL6J_5313141 | Gm20694 | ENSEMBL:ENSMUSG00000093446 | MGI:5313141 | pseudogene | predicted gene 20694 |
| 8 | gene | 117.158135 | 117.215997 | positive | MGI_C57BL6J_1890619 | Gan | NCBI_Gene:209239,ENSEMBL:ENSMUSG00000052557 | MGI:1890619 | protein coding gene | giant axonal neuropathy |
| 8 | gene | 117.211549 | 117.211579 | positive | MGI_C57BL6J_5012389 | Gm20204 | NA | NA | unclassified non-coding RNA gene | predicted gene%2c 20204 |
| 8 | gene | 117.257019 | 117.461506 | positive | MGI_C57BL6J_1921690 | Cmip | NCBI_Gene:74440,ENSEMBL:ENSMUSG00000034390 | MGI:1921690 | protein coding gene | c-Maf inducing protein |
| 8 | gene | 117.258079 | 117.260800 | positive | MGI_C57BL6J_2443594 | A130010C12Rik | NA | NA | unclassified gene | RIKEN cDNA A130010C12 gene |
| 8 | gene | 117.290669 | 117.292197 | positive | MGI_C57BL6J_1923167 | 4930572C08Rik | NA | NA | unclassified gene | RIKEN cDNA 4930572C08 gene |
| 8 | gene | 117.417466 | 117.423209 | negative | MGI_C57BL6J_5622145 | Gm39260 | NCBI_Gene:105243299 | MGI:5622145 | lncRNA gene | predicted gene, 39260 |
| 8 | gene | 117.453023 | 117.453079 | positive | MGI_C57BL6J_5531156 | Mir7077 | miRBase:MI0022927,NCBI_Gene:102466801,ENSEMBL:ENSMUSG00000099062 | MGI:5531156 | miRNA gene | microRNA 7077 |
| 8 | gene | 117.459265 | 117.459328 | positive | MGI_C57BL6J_5530768 | Mir7078 | miRBase:MI0022928,NCBI_Gene:102465653,ENSEMBL:ENSMUSG00000098483 | MGI:5530768 | miRNA gene | microRNA 7078 |
| 8 | gene | 117.498275 | 117.635142 | positive | MGI_C57BL6J_97616 | Plcg2 | NCBI_Gene:234779,ENSEMBL:ENSMUSG00000034330 | MGI:97616 | protein coding gene | phospholipase C, gamma 2 |
| 8 | gene | 117.546972 | 117.547026 | negative | MGI_C57BL6J_5530743 | Gm27361 | ENSEMBL:ENSMUSG00000092818 | MGI:5530743 | miRNA gene | predicted gene, 27361 |
| 8 | gene | 117.661399 | 117.673713 | negative | MGI_C57BL6J_1921282 | Sdr42e1 | NCBI_Gene:74032,ENSEMBL:ENSMUSG00000034308 | MGI:1921282 | protein coding gene | short chain dehydrogenase/reductase family 42E, member 1 |
| 8 | gene | 117.675815 | 117.680613 | negative | MGI_C57BL6J_5590876 | Gm31717 | NCBI_Gene:105243300,ENSEMBL:ENSMUSG00000110459 | MGI:5590876 | lncRNA gene | predicted gene, 31717 |
| 8 | gene | 117.681622 | 117.693198 | positive | MGI_C57BL6J_5621301 | Gm38416 | NCBI_Gene:330850,ENSEMBL:ENSMUSG00000110151 | MGI:5621301 | lncRNA gene | predicted gene, 38416 |
| 8 | gene | 117.700982 | 117.759030 | positive | MGI_C57BL6J_1096386 | Hsd17b2 | NCBI_Gene:15486,ENSEMBL:ENSMUSG00000031844 | MGI:1096386 | protein coding gene | hydroxysteroid (17-beta) dehydrogenase 2 |
| 8 | gene | 117.712570 | 117.714091 | positive | MGI_C57BL6J_5804891 | Gm45776 | ENSEMBL:ENSMUSG00000110520 | MGI:5804891 | unclassified gene | predicted gene 45776 |
| 8 | gene | 117.742499 | 117.748484 | negative | MGI_C57BL6J_5590933 | Gm31774 | NCBI_Gene:102634113,ENSEMBL:ENSMUSG00000110494 | MGI:5590933 | lncRNA gene | predicted gene, 31774 |
| 8 | gene | 117.761601 | 117.776462 | negative | MGI_C57BL6J_5622146 | Gm39261 | NCBI_Gene:105243301 | MGI:5622146 | lncRNA gene | predicted gene, 39261 |
| 8 | gene | 117.791642 | 117.801943 | negative | MGI_C57BL6J_1915783 | Mphosph6 | NCBI_Gene:68533,ENSEMBL:ENSMUSG00000031843 | MGI:1915783 | protein coding gene | M phase phosphoprotein 6 |
| 8 | pseudogene | 117.802062 | 117.803234 | positive | MGI_C57BL6J_3708793 | Gm10617 | ENSEMBL:ENSMUSG00000074065 | MGI:3708793 | pseudogene | predicted gene 10617 |
| 8 | gene | 117.944793 | 117.948430 | negative | MGI_C57BL6J_5622147 | Gm39262 | NCBI_Gene:105243302 | MGI:5622147 | lncRNA gene | predicted gene, 39262 |
| 8 | gene | 118.120954 | 118.121083 | positive | MGI_C57BL6J_5454977 | Gm25200 | ENSEMBL:ENSMUSG00000065028 | MGI:5454977 | snoRNA gene | predicted gene, 25200 |
| 8 | gene | 118.140017 | 119.324929 | positive | MGI_C57BL6J_99551 | Cdh13 | NCBI_Gene:12554,ENSEMBL:ENSMUSG00000031841 | MGI:99551 | protein coding gene | cadherin 13 |
| 8 | pseudogene | 118.381726 | 118.382191 | negative | MGI_C57BL6J_5804837 | Gm45722 | ENSEMBL:ENSMUSG00000110662 | MGI:5804837 | pseudogene | predicted gene 45722 |
| 8 | gene | 118.426493 | 118.426599 | negative | MGI_C57BL6J_5453812 | Gm24035 | ENSEMBL:ENSMUSG00000064801 | MGI:5453812 | snRNA gene | predicted gene, 24035 |
| 8 | gene | 118.607127 | 118.610499 | negative | MGI_C57BL6J_5804836 | Gm45721 | ENSEMBL:ENSMUSG00000110548 | MGI:5804836 | lncRNA gene | predicted gene 45721 |
| 8 | gene | 118.610989 | 118.615879 | negative | MGI_C57BL6J_5804835 | Gm45720 | ENSEMBL:ENSMUSG00000110667 | MGI:5804835 | lncRNA gene | predicted gene 45720 |
| 8 | gene | 118.726900 | 118.726987 | positive | MGI_C57BL6J_5452776 | Gm22999 | ENSEMBL:ENSMUSG00000087707 | MGI:5452776 | snoRNA gene | predicted gene, 22999 |
| 8 | gene | 119.052306 | 119.058195 | negative | MGI_C57BL6J_5591052 | Gm31893 | NCBI_Gene:102634266 | MGI:5591052 | lncRNA gene | predicted gene, 31893 |
| 8 | gene | 119.344488 | 119.348929 | positive | MGI_C57BL6J_1915446 | Hsbp1 | NCBI_Gene:68196,ENSEMBL:ENSMUSG00000031839 | MGI:1915446 | protein coding gene | heat shock factor binding protein 1 |
| 8 | gene | 119.378696 | 119.386537 | positive | MGI_C57BL6J_5804838 | Gm45723 | NCBI_Gene:108167584,ENSEMBL:ENSMUSG00000110470 | MGI:5804838 | lncRNA gene | predicted gene 45723 |
| 8 | gene | 119.394419 | 119.421940 | positive | MGI_C57BL6J_1928485 | Mlycd | NCBI_Gene:56690,ENSEMBL:ENSMUSG00000074064 | MGI:1928485 | protein coding gene | malonyl-CoA decarboxylase |
| 8 | gene | 119.406544 | 119.407549 | negative | MGI_C57BL6J_5825690 | Gm46053 | NCBI_Gene:108167555 | MGI:5825690 | lncRNA gene | predicted gene, 46053 |
| 8 | gene | 119.415104 | 119.429070 | negative | MGI_C57BL6J_5591165 | Gm32006 | NCBI_Gene:102634421 | MGI:5591165 | lncRNA gene | predicted gene, 32006 |
| 8 | gene | 119.434124 | 119.446256 | positive | MGI_C57BL6J_1919089 | Osgin1 | NCBI_Gene:71839,ENSEMBL:ENSMUSG00000074063 | MGI:1919089 | protein coding gene | oxidative stress induced growth inhibitor 1 |
| 8 | gene | 119.446719 | 119.472640 | positive | MGI_C57BL6J_2152211 | Necab2 | NCBI_Gene:117148,ENSEMBL:ENSMUSG00000031837 | MGI:2152211 | protein coding gene | N-terminal EF-hand calcium binding protein 2 |
| 8 | gene | 119.479574 | 119.501698 | negative | MGI_C57BL6J_2685433 | Slc38a8 | NCBI_Gene:234788,ENSEMBL:ENSMUSG00000034224 | MGI:2685433 | protein coding gene | solute carrier family 38, member 8 |
| 8 | gene | 119.508151 | 119.558825 | negative | MGI_C57BL6J_1927235 | Mbtps1 | NCBI_Gene:56453,ENSEMBL:ENSMUSG00000031835 | MGI:1927235 | protein coding gene | membrane-bound transcription factor peptidase, site 1 |
| 8 | gene | 119.561978 | 119.575200 | negative | MGI_C57BL6J_1919802 | Hsdl1 | NCBI_Gene:72552,ENSEMBL:ENSMUSG00000034189 | MGI:1919802 | protein coding gene | hydroxysteroid dehydrogenase like 1 |
| 8 | gene | 119.575235 | 119.598454 | positive | MGI_C57BL6J_1915520 | Dnaaf1 | NCBI_Gene:68270,ENSEMBL:ENSMUSG00000031831 | MGI:1915520 | protein coding gene | dynein, axonemal assembly factor 1 |
| 8 | gene | 119.597871 | 119.605240 | negative | MGI_C57BL6J_109576 | Taf1c | NCBI_Gene:21341,ENSEMBL:ENSMUSG00000031832 | MGI:109576 | protein coding gene | TATA-box binding protein associated factor, RNA polymerase I, C |
| 8 | pseudogene | 119.602120 | 119.602742 | negative | MGI_C57BL6J_3648822 | Rps13-ps4 | NCBI_Gene:628061,ENSEMBL:ENSMUSG00000081378 | MGI:3648822 | pseudogene | ribosomal protein S13, pseudogene 4 |
| 8 | gene | 119.612710 | 119.616926 | positive | MGI_C57BL6J_1923023 | Adad2 | NCBI_Gene:75773,ENSEMBL:ENSMUSG00000024266 | MGI:1923023 | protein coding gene | adenosine deaminase domain containing 2 |
| 8 | gene | 119.623854 | 119.635887 | negative | MGI_C57BL6J_1913983 | Kcng4 | NCBI_Gene:66733,ENSEMBL:ENSMUSG00000045246 | MGI:1913983 | protein coding gene | potassium voltage-gated channel, subfamily G, member 4 |
| 8 | gene | 119.631151 | 119.632876 | negative | MGI_C57BL6J_5825692 | Gm46055 | NCBI_Gene:108167557 | MGI:5825692 | lncRNA gene | predicted gene, 46055 |
| 8 | gene | 119.635818 | 119.646868 | positive | MGI_C57BL6J_5825691 | Gm46054 | NCBI_Gene:108167556 | MGI:5825691 | lncRNA gene | predicted gene, 46054 |
| 8 | gene | 119.658880 | 119.671411 | negative | MGI_C57BL6J_5591511 | Gm32352 | NCBI_Gene:102634871,ENSEMBL:ENSMUSG00000110453 | MGI:5591511 | lncRNA gene | predicted gene, 32352 |
| 8 | gene | 119.666365 | 119.688222 | positive | MGI_C57BL6J_1915116 | Wfdc1 | NCBI_Gene:67866,ENSEMBL:ENSMUSG00000023336 | MGI:1915116 | protein coding gene | WAP four-disulfide core domain 1 |
| 8 | gene | 119.700009 | 119.757718 | positive | MGI_C57BL6J_1916297 | Atp2c2 | NCBI_Gene:69047,ENSEMBL:ENSMUSG00000034112 | MGI:1916297 | protein coding gene | ATPase, Ca++ transporting, type 2C, member 2 |
| 8 | gene | 119.714454 | 119.715452 | positive | MGI_C57BL6J_5804862 | Gm45747 | ENSEMBL:ENSMUSG00000110445 | MGI:5804862 | unclassified gene | predicted gene 45747 |
| 8 | gene | 119.714454 | 119.715452 | negative | MGI_C57BL6J_1925480 | 5033405D04Rik | NA | NA | unclassified gene | RIKEN cDNA 5033405D04 gene |
| 8 | gene | 119.760076 | 119.778416 | negative | MGI_C57BL6J_1921597 | Meak7 | NCBI_Gene:74347,ENSEMBL:ENSMUSG00000034105 | MGI:1921597 | protein coding gene | MTOR associated protein , eak-7 homolog |
| 8 | gene | 119.809214 | 119.840579 | negative | MGI_C57BL6J_1919292 | Cotl1 | NCBI_Gene:72042,ENSEMBL:ENSMUSG00000031827 | MGI:1919292 | protein coding gene | coactosin-like 1 (Dictyostelium) |
| 8 | gene | 119.862231 | 119.876995 | positive | MGI_C57BL6J_2385305 | Klhl36 | NCBI_Gene:234796,ENSEMBL:ENSMUSG00000031828 | MGI:2385305 | protein coding gene | kelch-like 36 |
| 8 | gene | 119.910360 | 119.957560 | positive | MGI_C57BL6J_894652 | Usp10 | NCBI_Gene:22224,ENSEMBL:ENSMUSG00000031826 | MGI:894652 | protein coding gene | ubiquitin specific peptidase 10 |
| 8 | gene | 119.910841 | 124.345722 | positive | MGI_C57BL6J_5141853 | Gm20388 | ENSEMBL:ENSMUSG00000092329 | MGI:5141853 | protein coding gene | predicted gene 20388 |
| 8 | gene | 119.992438 | 120.052794 | positive | MGI_C57BL6J_1926142 | Crispld2 | NCBI_Gene:78892,ENSEMBL:ENSMUSG00000031825 | MGI:1926142 | protein coding gene | cysteine-rich secretory protein LCCL domain containing 2 |
| 8 | gene | 120.036194 | 120.045287 | negative | MGI_C57BL6J_3783126 | Gm15684 | ENSEMBL:ENSMUSG00000089776 | MGI:3783126 | lncRNA gene | predicted gene 15684 |
| 8 | gene | 120.080890 | 120.101482 | negative | MGI_C57BL6J_2142662 | Zdhhc7 | NCBI_Gene:102193,ENSEMBL:ENSMUSG00000031823 | MGI:2142662 | protein coding gene | zinc finger, DHHC domain containing 7 |
| 8 | gene | 120.095060 | 120.097736 | positive | MGI_C57BL6J_3802167 | Gm15898 | ENSEMBL:ENSMUSG00000089742 | MGI:3802167 | lncRNA gene | predicted gene 15898 |
| 8 | gene | 120.101582 | 120.103904 | positive | MGI_C57BL6J_3028043 | A130014A01Rik | ENSEMBL:ENSMUSG00000097944 | MGI:3028043 | lncRNA gene | RIKEN cDNA A130014A01 gene |
| 8 | gene | 120.109441 | 120.110965 | positive | MGI_C57BL6J_5611607 | Gm38379 | ENSEMBL:ENSMUSG00000102665 | MGI:5611607 | unclassified gene | predicted gene, 38379 |
| 8 | gene | 120.114152 | 120.165307 | positive | MGI_C57BL6J_2443793 | 6430548M08Rik | NCBI_Gene:234797,ENSEMBL:ENSMUSG00000031824 | MGI:2443793 | protein coding gene | RIKEN cDNA 6430548M08 gene |
| 8 | gene | 120.166397 | 120.177486 | negative | MGI_C57BL6J_3588213 | Fam92b | NCBI_Gene:436062,ENSEMBL:ENSMUSG00000042269 | MGI:3588213 | protein coding gene | family with sequence similarity 92, member B |
| 8 | gene | 120.204434 | 120.228230 | negative | MGI_C57BL6J_3045392 | A330074K22Rik | NCBI_Gene:434353,ENSEMBL:ENSMUSG00000097960 | MGI:3045392 | lncRNA gene | RIKEN cDNA A330074K22 gene |
| 8 | gene | 120.228456 | 120.581390 | positive | MGI_C57BL6J_1098275 | Gse1 | NCBI_Gene:382034,ENSEMBL:ENSMUSG00000031822 | MGI:1098275 | protein coding gene | genetic suppressor element 1, coiled-coil protein |
| 8 | gene | 120.327675 | 120.339024 | positive | MGI_C57BL6J_5592127 | Gm32968 | NCBI_Gene:102635696 | MGI:5592127 | lncRNA gene | predicted gene, 32968 |
| 8 | gene | 120.342211 | 120.345258 | negative | MGI_C57BL6J_5622149 | Gm39264 | NCBI_Gene:105243304 | MGI:5622149 | lncRNA gene | predicted gene, 39264 |
| 8 | gene | 120.443459 | 120.443549 | negative | MGI_C57BL6J_5452492 | Gm22715 | ENSEMBL:ENSMUSG00000080549 | MGI:5452492 | miRNA gene | predicted gene, 22715 |
| 8 | gene | 120.520615 | 120.521686 | positive | MGI_C57BL6J_5504086 | Gm26971 | ENSEMBL:ENSMUSG00000097983 | MGI:5504086 | lncRNA gene | predicted gene, 26971 |
| 8 | gene | 120.536892 | 120.537868 | negative | MGI_C57BL6J_1921477 | 1700016A09Rik | NCBI_Gene:74227,ENSEMBL:ENSMUSG00000102737 | MGI:1921477 | unclassified gene | RIKEN cDNA 1700016A09 gene |
| 8 | gene | 120.538696 | 120.538754 | positive | MGI_C57BL6J_5530955 | Mir7687 | miRBase:MI0025039,NCBI_Gene:102466849,ENSEMBL:ENSMUSG00000098280 | MGI:5530955 | miRNA gene | microRNA 7687 |
| 8 | gene | 120.578633 | 120.589304 | negative | MGI_C57BL6J_1921019 | Gins2 | NCBI_Gene:272551,ENSEMBL:ENSMUSG00000031821 | MGI:1921019 | protein coding gene | GINS complex subunit 2 (Psf2 homolog) |
| 8 | gene | 120.589531 | 120.599161 | positive | MGI_C57BL6J_3642104 | Gm10614 | NCBI_Gene:100038388,ENSEMBL:ENSMUSG00000097773 | MGI:3642104 | lncRNA gene | predicted gene 10614 |
| 8 | pseudogene | 120.597110 | 120.597563 | negative | MGI_C57BL6J_5804949 | Gm45834 | ENSEMBL:ENSMUSG00000110601 | MGI:5804949 | pseudogene | predicted gene 45834 |
| 8 | gene | 120.598552 | 120.599277 | positive | MGI_C57BL6J_5477233 | Gm26739 | ENSEMBL:ENSMUSG00000097702 | MGI:5477233 | lncRNA gene | predicted gene, 26739 |
| 8 | gene | 120.608600 | 120.668142 | negative | MGI_C57BL6J_5504136 | Gm27021 | NCBI_Gene:105734727,ENSEMBL:ENSMUSG00000097919 | MGI:5504136 | protein coding gene | predicted gene, 27021 |
| 8 | gene | 120.608602 | 120.647632 | negative | MGI_C57BL6J_1916168 | 1190005I06Rik | NCBI_Gene:68918,ENSEMBL:ENSMUSG00000043687 | MGI:1916168 | protein coding gene | RIKEN cDNA 1190005I06 gene |
| 8 | gene | 120.628524 | 120.632471 | positive | MGI_C57BL6J_5592234 | Gm33075 | NCBI_Gene:102635834 | MGI:5592234 | lncRNA gene | predicted gene, 33075 |
| 8 | gene | 120.643950 | 120.653687 | positive | MGI_C57BL6J_5477031 | Gm26537 | ENSEMBL:ENSMUSG00000097500 | MGI:5477031 | lncRNA gene | predicted gene, 26537 |
| 8 | gene | 120.653914 | 120.668573 | negative | MGI_C57BL6J_1343095 | Emc8 | NCBI_Gene:18117,ENSEMBL:ENSMUSG00000031819 | MGI:1343095 | protein coding gene | ER membrane protein complex subunit 8 |
| 8 | gene | 120.668222 | 120.674209 | positive | MGI_C57BL6J_88473 | Cox4i1 | NCBI_Gene:12857,ENSEMBL:ENSMUSG00000031818 | MGI:88473 | protein coding gene | cytochrome c oxidase subunit 4I1 |
| 8 | gene | 120.685399 | 120.686265 | positive | MGI_C57BL6J_5592301 | Gm33142 | ENSEMBL:ENSMUSG00000104484 | MGI:5592301 | unclassified non-coding RNA gene | predicted gene, 33142 |
| 8 | gene | 120.686054 | 120.701181 | negative | MGI_C57BL6J_5622150 | Gm39265 | NCBI_Gene:105243305 | MGI:5622150 | lncRNA gene | predicted gene, 39265 |
| 8 | gene | 120.717500 | 120.717754 | negative | MGI_C57BL6J_5454016 | Gm24239 | ENSEMBL:ENSMUSG00000088416 | MGI:5454016 | unclassified non-coding RNA gene | predicted gene, 24239 |
| 8 | pseudogene | 120.727211 | 120.728011 | positive | MGI_C57BL6J_5010894 | Gm18709 | NCBI_Gene:100417604,ENSEMBL:ENSMUSG00000097895 | MGI:5010894 | pseudogene | predicted gene, 18709 |
| 8 | gene | 120.736358 | 120.756694 | positive | MGI_C57BL6J_96395 | Irf8 | NCBI_Gene:15900,ENSEMBL:ENSMUSG00000041515 | MGI:96395 | protein coding gene | interferon regulatory factor 8 |
| 8 | gene | 120.767475 | 120.770229 | negative | MGI_C57BL6J_5622151 | Gm39266 | NCBI_Gene:105243306 | MGI:5622151 | lncRNA gene | predicted gene, 39266 |
| 8 | gene | 120.786929 | 120.795374 | positive | MGI_C57BL6J_5592358 | Gm33199 | NCBI_Gene:102636004 | MGI:5592358 | lncRNA gene | predicted gene, 33199 |
| 8 | gene | 120.851385 | 120.867621 | negative | MGI_C57BL6J_5622152 | Gm39267 | NCBI_Gene:105243307 | MGI:5622152 | lncRNA gene | predicted gene, 39267 |
| 8 | gene | 120.868990 | 120.874597 | negative | MGI_C57BL6J_5592527 | Gm33368 | NCBI_Gene:102636249 | MGI:5592527 | lncRNA gene | predicted gene, 33368 |
| 8 | gene | 120.879399 | 120.882481 | positive | MGI_C57BL6J_5477372 | Gm26878 | NCBI_Gene:102636330,ENSEMBL:ENSMUSG00000097087 | MGI:5477372 | lncRNA gene | predicted gene, 26878 |
| 8 | gene | 120.917008 | 120.919697 | positive | MGI_C57BL6J_5592630 | Gm33471 | NCBI_Gene:102636395 | MGI:5592630 | lncRNA gene | predicted gene, 33471 |
| 8 | gene | 121.002927 | 121.003190 | positive | MGI_C57BL6J_5579030 | Gm28324 | ENSEMBL:ENSMUSG00000101886 | MGI:5579030 | lncRNA gene | predicted gene 28324 |
| 8 | gene | 121.016513 | 121.023617 | negative | MGI_C57BL6J_5592738 | Gm33579 | NCBI_Gene:102636543 | MGI:5592738 | lncRNA gene | predicted gene, 33579 |
| 8 | gene | 121.035751 | 121.037957 | negative | MGI_C57BL6J_5825708 | Gm46071 | NCBI_Gene:108167585 | MGI:5825708 | lncRNA gene | predicted gene, 46071 |
| 8 | gene | 121.054882 | 121.083110 | negative | MGI_C57BL6J_1916040 | Fendrr | NCBI_Gene:68790,ENSEMBL:ENSMUSG00000097336 | MGI:1916040 | lncRNA gene | Foxf1 adjacent non-coding developmental regulatory RNA |
| 8 | gene | 121.084386 | 121.088154 | positive | MGI_C57BL6J_1347470 | Foxf1 | NCBI_Gene:15227,ENSEMBL:ENSMUSG00000042812 | MGI:1347470 | protein coding gene | forkhead box F1 |
| 8 | gene | 121.084609 | 121.084706 | negative | MGI_C57BL6J_5530912 | Gm27530 | ENSEMBL:ENSMUSG00000098560 | MGI:5530912 | miRNA gene | predicted gene, 27530 |
| 8 | gene | 121.091628 | 121.108415 | negative | MGI_C57BL6J_2679252 | Mthfsd | NCBI_Gene:234814,ENSEMBL:ENSMUSG00000031816 | MGI:2679252 | protein coding gene | methenyltetrahydrofolate synthetase domain containing |
| 8 | gene | 121.116171 | 121.118895 | positive | MGI_C57BL6J_1347481 | Foxc2 | NCBI_Gene:14234,ENSEMBL:ENSMUSG00000046714 | MGI:1347481 | protein coding gene | forkhead box C2 |
| 8 | gene | 121.127685 | 121.130644 | positive | MGI_C57BL6J_1347469 | Foxl1 | NCBI_Gene:14241,ENSEMBL:ENSMUSG00000097084 | MGI:1347469 | protein coding gene | forkhead box L1 |
| 8 | gene | 121.140943 | 121.141620 | positive | MGI_C57BL6J_5610837 | Gm37609 | ENSEMBL:ENSMUSG00000103830 | MGI:5610837 | unclassified gene | predicted gene, 37609 |
| 8 | gene | 121.166916 | 121.168363 | negative | MGI_C57BL6J_5592792 | Gm33633 | NCBI_Gene:102636622 | MGI:5592792 | lncRNA gene | predicted gene, 33633 |
| 8 | gene | 121.172324 | 121.183403 | positive | MGI_C57BL6J_5624931 | Gm42046 | NCBI_Gene:105246822 | MGI:5624931 | lncRNA gene | predicted gene, 42046 |
| 8 | gene | 121.198714 | 121.203602 | negative | MGI_C57BL6J_1923251 | 5033426O07Rik | NCBI_Gene:76001,ENSEMBL:ENSMUSG00000097097 | MGI:1923251 | lncRNA gene | RIKEN cDNA 5033426O07 gene |
| 8 | gene | 121.213807 | 121.222717 | positive | MGI_C57BL6J_5592898 | Gm33739 | NCBI_Gene:102636757 | MGI:5592898 | lncRNA gene | predicted gene, 33739 |
| 8 | gene | 121.256747 | 121.356836 | negative | MGI_C57BL6J_5477309 | Gm26815 | ENSEMBL:ENSMUSG00000097384 | MGI:5477309 | lncRNA gene | predicted gene, 26815 |
| 8 | gene | 121.261747 | 121.264665 | negative | MGI_C57BL6J_5592982 | Gm33823 | NCBI_Gene:102636872 | MGI:5592982 | lncRNA gene | predicted gene, 33823 |
| 8 | gene | 121.418627 | 121.429912 | positive | MGI_C57BL6J_5477241 | Gm26747 | NCBI_Gene:102636945,ENSEMBL:ENSMUSG00000097783 | MGI:5477241 | lncRNA gene | predicted gene, 26747 |
| 8 | gene | 121.434980 | 121.435389 | positive | MGI_C57BL6J_5477278 | Gm26784 | ENSEMBL:ENSMUSG00000097438 | MGI:5477278 | lncRNA gene | predicted gene, 26784 |
| 8 | gene | 121.530776 | 121.544360 | negative | MGI_C57BL6J_1923655 | 1700018B08Rik | NCBI_Gene:76405,ENSEMBL:ENSMUSG00000031809 | MGI:1923655 | protein coding gene | RIKEN cDNA 1700018B08 gene |
| 8 | gene | 121.542080 | 121.543481 | positive | MGI_C57BL6J_5504160 | Gm27045 | ENSEMBL:ENSMUSG00000098211 | MGI:5504160 | lncRNA gene | predicted gene, 27045 |
| 8 | gene | 121.544382 | 121.548342 | positive | MGI_C57BL6J_1919509 | 1700030M09Rik | NCBI_Gene:72259,ENSEMBL:ENSMUSG00000097424 | MGI:1919509 | lncRNA gene | RIKEN cDNA 1700030M09 gene |
| 8 | gene | 121.549440 | 121.578863 | negative | MGI_C57BL6J_1354708 | Fbxo31 | NCBI_Gene:76454,ENSEMBL:ENSMUSG00000052934 | MGI:1354708 | protein coding gene | F-box protein 31 |
| 8 | gene | 121.579229 | 121.579301 | negative | MGI_C57BL6J_4413994 | n-TMcat17 | NCBI_Gene:102467360 | MGI:4413994 | tRNA gene | nuclear encoded tRNA methionine 17 (anticodon CAT) |
| 8 | pseudogene | 121.581765 | 121.582193 | positive | MGI_C57BL6J_5009950 | Gm17786 | NCBI_Gene:100048204,ENSEMBL:ENSMUSG00000097987 | MGI:5009950 | pseudogene | predicted gene, 17786 |
| 8 | gene | 121.590361 | 121.598760 | positive | MGI_C57BL6J_1914693 | Map1lc3b | NCBI_Gene:67443,ENSEMBL:ENSMUSG00000031812 | MGI:1914693 | protein coding gene | microtubule-associated protein 1 light chain 3 beta |
| 8 | gene | 121.598697 | 121.652901 | negative | MGI_C57BL6J_2159407 | Zcchc14 | NCBI_Gene:142682,ENSEMBL:ENSMUSG00000061410 | MGI:2159407 | protein coding gene | zinc finger, CCHC domain containing 14 |
| 8 | gene | 121.616563 | 121.617908 | negative | MGI_C57BL6J_1919593 | 2600002B07Rik | NA | NA | unclassified gene | RIKEN cDNA 2600002B07 gene |
| 8 | gene | 121.654054 | 121.654517 | positive | MGI_C57BL6J_5504145 | Gm27030 | ENSEMBL:ENSMUSG00000098221 | MGI:5504145 | lncRNA gene | predicted gene, 27030 |
Chromsome 13
query_variants <- create_variant_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/cc_variants.sqlite")
query_genes <- create_gene_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/mouse_genes_mgi.sqlite")
if(nrow(peaks) >0){
for (i in 2){
#for (i in 1:1){
#Plot 1
#marker = find_marker(gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i])
#gp <- g[,marker]
#gp[gp==1] <- "AA"
#gp[gp==2] <- "AB"
#gp[gp==0] <- NA
#plot_pxg(gp, gm$covar[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
#title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
###dev.off()
g <- maxmarg(pr.qc, gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i], return_char=TRUE)
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/","qtl_effect_", i, ".png"))
#par(mar=c(4.1, 4.1, 1.5, 0.6))
plot_pxg(g, gm$covar[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
##dev.off()
chr = peaks$chr[i]
# Plot 2
pr_sub <- pull_genoprobint(pr.qc, gm$gmap, chr, c(peaks$ci_lo[i], peaks$ci_hi[i]))
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar)
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], Xcovar=Xcovar)
#coeff <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
#coeff_sub <- scan1coef(pr_sub[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
blup <- scan1blup(pr.qc[,chr], gm$covar[peaks$phenotype[i]])
blup_sub <- scan1blup(pr_sub[,chr], gm$covar[peaks$phenotype[i]])
write.csv(as.data.frame(blup_sub), paste0("data/ici.vs.pbs_blup_sub_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv"), quote=F)
#plot_coef(coeff,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i]," [scan1coeff; positions in cM])")
# )
#plot_coef(coeff_sub,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i],"; 1.5 LOD drop interval [scan1coeff; positions in cM] ) ")
# )
plot_coef(blup,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," [scan1blup; positions in cM])")
)
plot_coef(blup_sub,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i],"; 1.5 LOD drop interval [scan1blup; positions in cM])")
)
# Plot 3
#c2effB <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary", contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
#c2effBb <- scan1blup(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]],Xcovar=Xcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
#plot(c2effB, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
#plot(c2effBb, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
##last_coef <- unclass(c2effB)[nrow(c2effB),2:3] # last two coefficients
##for(t in seq(along=last_coef))
## axis(side=4, at=last_coef[t], names(last_coef)[t], tick=FALSE)
#Table 1
chr = peaks_mba$chr[i]
start=as.numeric(peaks_mba$ci_lo[i])
end=as.numeric(peaks_mba$ci_hi[i])
genesgss = query_genes(chr, start, end)
write.csv(genesgss, file=paste0("data/ici.vs.pbs_genes_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv"), quote=F)
rownames(genesgss) <- NULL
genesgss$strand_old = genesgss$strand
genesgss$strand[genesgss$strand=="+"] <- "positive"
genesgss$strand[genesgss$strand=="-"] <- "negative"
#genesgss <-
#table <-
#genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")] %>%
#kable(escape = F,align = c("ccccccccccc")) %>%
#kable_styling("striped", full_width = T) #%>%
#cat #%>%
#column_spec(1, bold=TRUE)
#
#print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], escape = F,align = c("ccccccccccc")))
print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], "html") %>% kable_styling("striped", full_width = T))
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}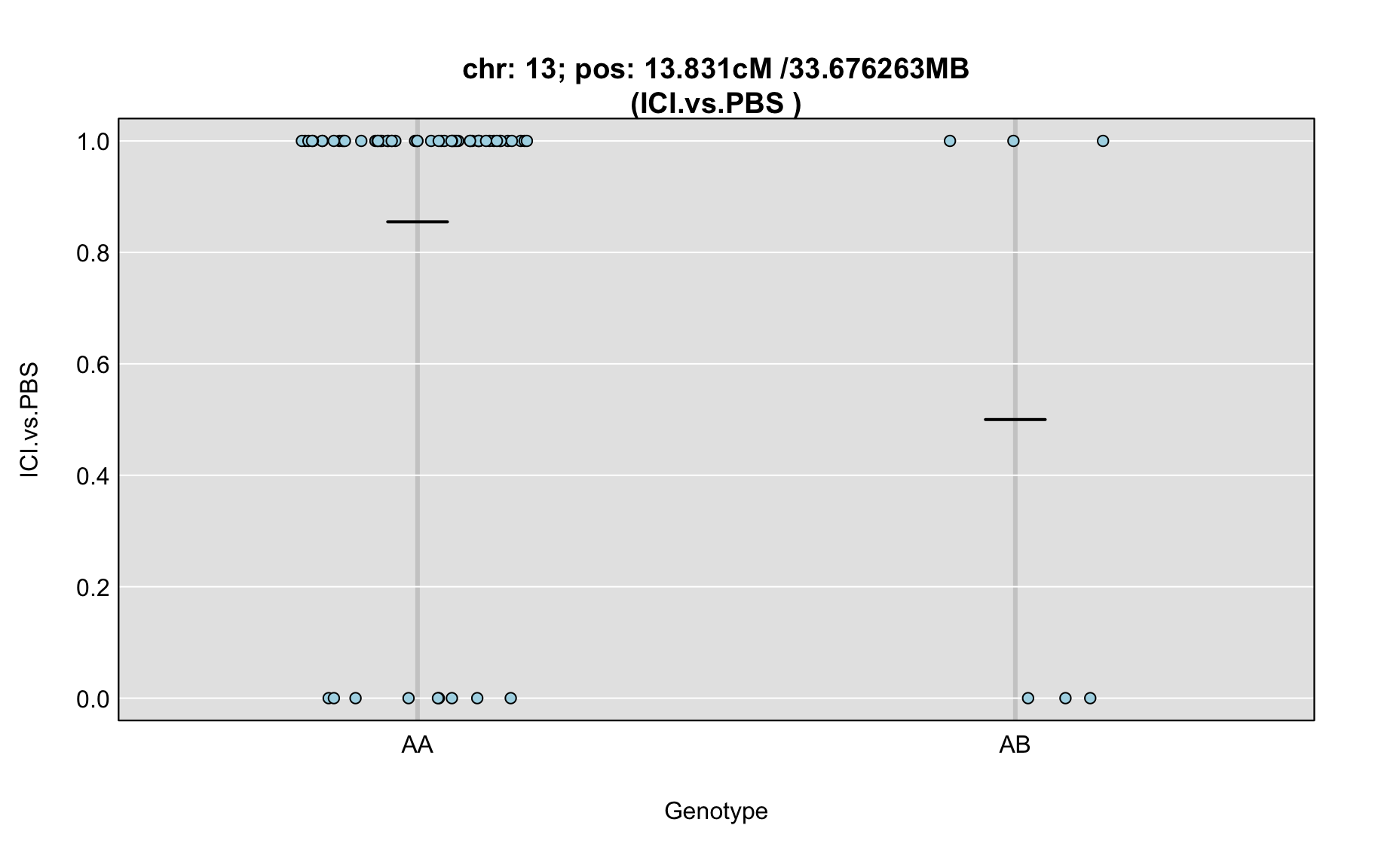
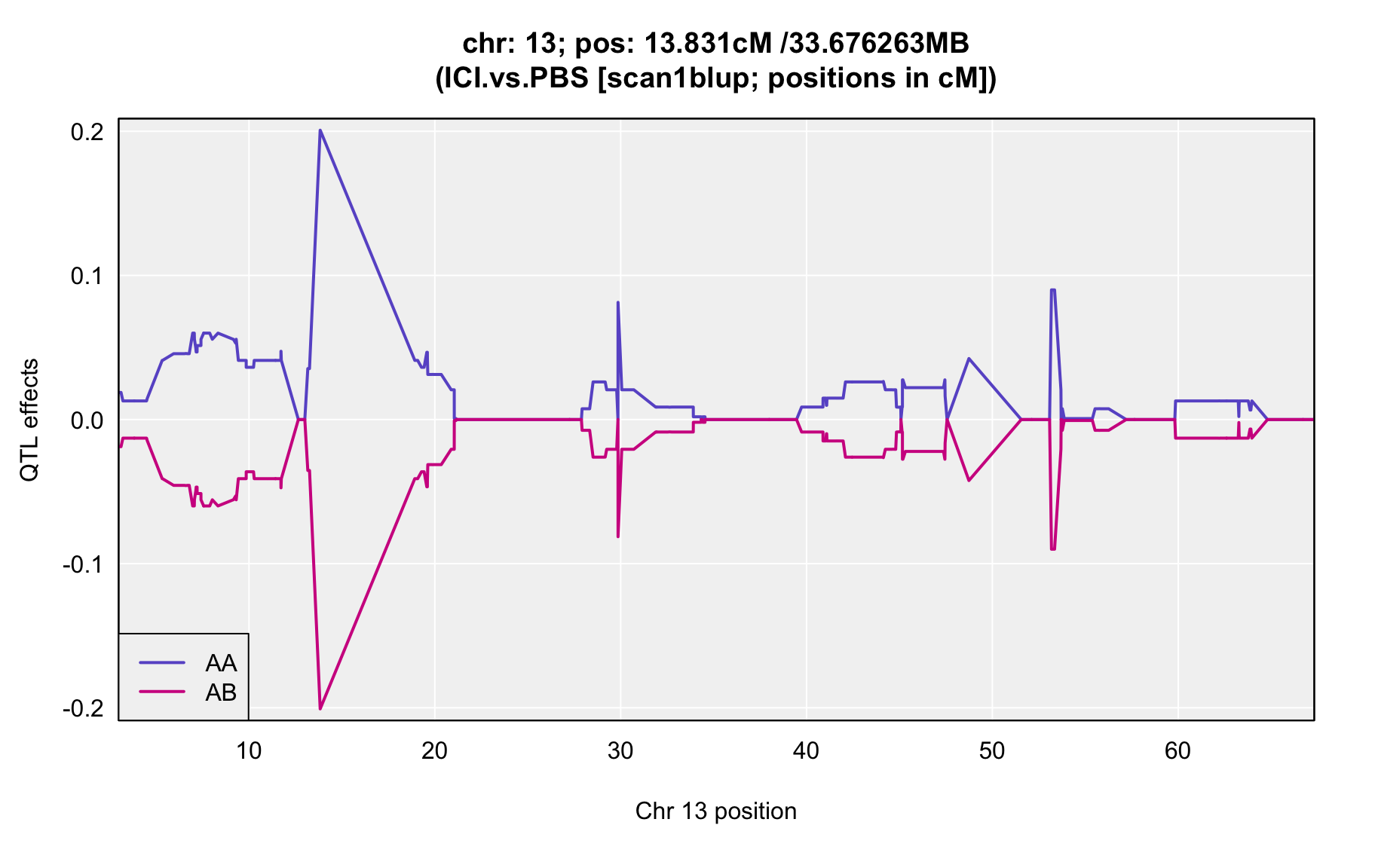
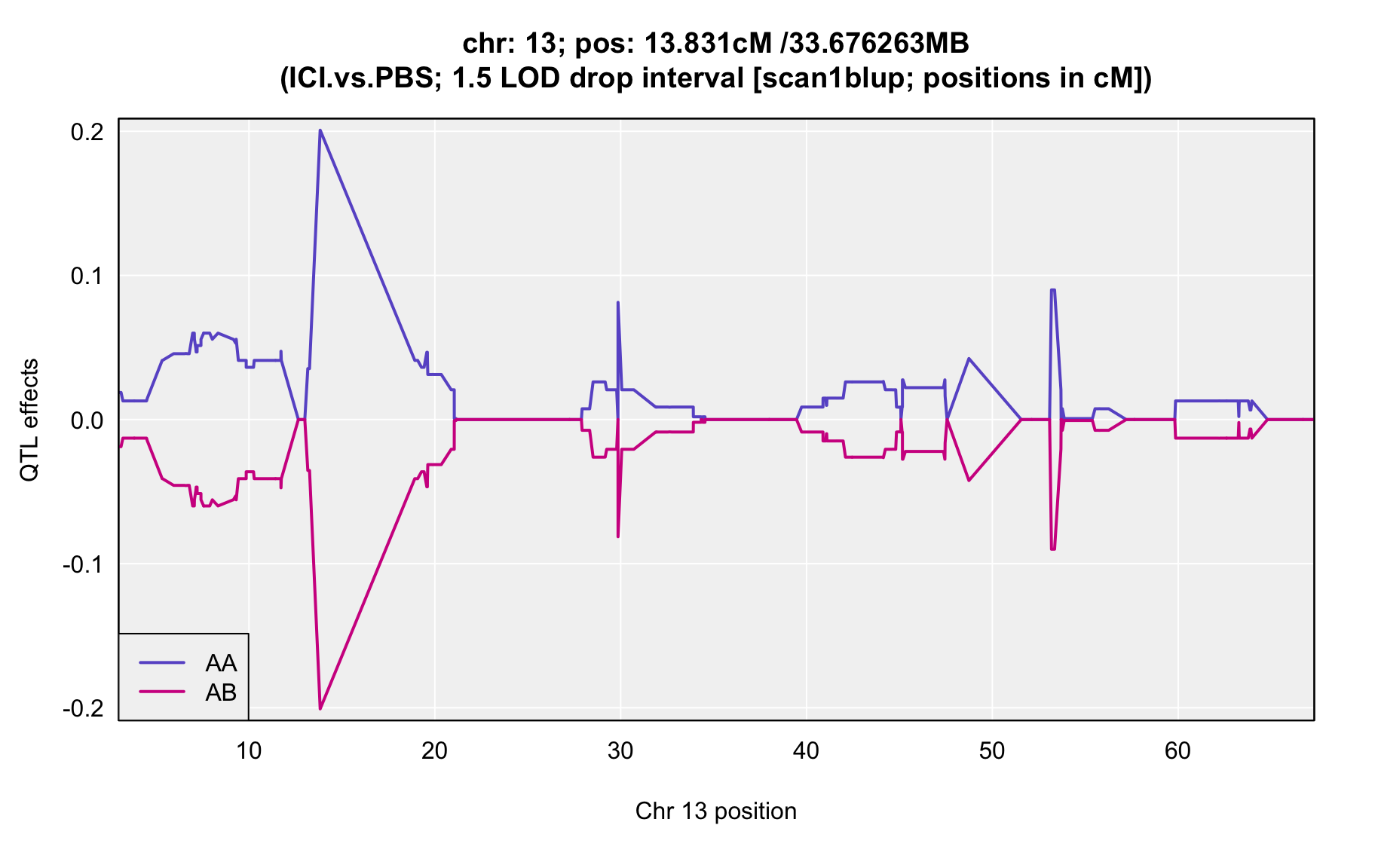
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 13 | gene | 5.915706 | 6.014285 | positive | MGI_C57BL6J_5826038 | Gm46401 | NCBI_Gene:108168063,ENSEMBL:ENSMUSG00000113017 | MGI:5826038 | lncRNA gene | predicted gene, 46401 |
| 13 | gene | 5.997455 | 5.997551 | negative | MGI_C57BL6J_5456036 | Gm26259 | ENSEMBL:ENSMUSG00000077971 | MGI:5456036 | miRNA gene | predicted gene, 26259 |
| 13 | gene | 6.024533 | 6.029004 | negative | MGI_C57BL6J_6096507 | Gm47514 | ENSEMBL:ENSMUSG00000113054 | MGI:6096507 | lncRNA gene | predicted gene, 47514 |
| 13 | gene | 6.073113 | 6.073782 | negative | MGI_C57BL6J_6096563 | Gm47548 | ENSEMBL:ENSMUSG00000113483 | MGI:6096563 | lncRNA gene | predicted gene, 47548 |
| 13 | gene | 6.269108 | 6.277026 | positive | MGI_C57BL6J_6096565 | Gm47549 | ENSEMBL:ENSMUSG00000113765 | MGI:6096565 | lncRNA gene | predicted gene, 47549 |
| 13 | gene | 6.280378 | 6.294714 | negative | MGI_C57BL6J_5594774 | Gm35615 | NCBI_Gene:102639262,ENSEMBL:ENSMUSG00000114097 | MGI:5594774 | lncRNA gene | predicted gene, 35615 |
| 13 | gene | 6.307568 | 6.320059 | positive | MGI_C57BL6J_5594830 | Gm35671 | NCBI_Gene:102639335 | MGI:5594830 | lncRNA gene | predicted gene, 35671 |
| 13 | gene | 6.395716 | 6.398740 | positive | MGI_C57BL6J_5579171 | Gm28465 | ENSEMBL:ENSMUSG00000101540 | MGI:5579171 | lncRNA gene | predicted gene 28465 |
| 13 | gene | 6.530595 | 6.531244 | positive | MGI_C57BL6J_1925651 | 2900042G08Rik | ENSEMBL:ENSMUSG00000114195 | MGI:1925651 | unclassified gene | RIKEN cDNA 2900042G08 gene |
| 13 | gene | 6.548122 | 6.580515 | positive | MGI_C57BL6J_1916867 | Pitrm1 | NCBI_Gene:69617,ENSEMBL:ENSMUSG00000021193 | MGI:1916867 | protein coding gene | pitrilysin metallepetidase 1 |
| 13 | gene | 6.579768 | 6.648792 | negative | MGI_C57BL6J_1891833 | Pfkp | NCBI_Gene:56421,ENSEMBL:ENSMUSG00000021196 | MGI:1891833 | protein coding gene | phosphofructokinase, platelet |
| 13 | gene | 6.631534 | 6.648563 | positive | MGI_C57BL6J_5594889 | Gm35730 | NCBI_Gene:102639406 | MGI:5594889 | lncRNA gene | predicted gene, 35730 |
| 13 | pseudogene | 6.662535 | 6.662971 | negative | MGI_C57BL6J_3641706 | Gm10029 | NCBI_Gene:100039532,ENSEMBL:ENSMUSG00000068324 | MGI:3641706 | pseudogene | predicted gene 10029 |
| 13 | pseudogene | 6.673139 | 6.673408 | positive | MGI_C57BL6J_6097648 | Gm48238 | ENSEMBL:ENSMUSG00000113545 | MGI:6097648 | pseudogene | predicted gene, 48238 |
| 13 | gene | 6.925283 | 6.925851 | negative | MGI_C57BL6J_1920105 | 2900024D18Rik | ENSEMBL:ENSMUSG00000113932 | MGI:1920105 | unclassified gene | RIKEN cDNA 2900024D18 gene |
| 13 | gene | 6.995654 | 7.076316 | negative | MGI_C57BL6J_5623547 | Gm40662 | NCBI_Gene:105245172,ENSEMBL:ENSMUSG00000114017 | MGI:5623547 | lncRNA gene | predicted gene, 40662 |
| 13 | gene | 7.120394 | 7.120503 | negative | MGI_C57BL6J_5452466 | Gm22689 | ENSEMBL:ENSMUSG00000076407 | MGI:5452466 | miRNA gene | predicted gene, 22689 |
| 13 | gene | 7.137850 | 7.161134 | negative | MGI_C57BL6J_5595084 | Gm35925 | NCBI_Gene:102639667 | MGI:5595084 | lncRNA gene | predicted gene, 35925 |
| 13 | gene | 7.139127 | 7.175925 | negative | MGI_C57BL6J_5623548 | Gm40663 | NCBI_Gene:105245173,ENSEMBL:ENSMUSG00000113241 | MGI:5623548 | lncRNA gene | predicted gene, 40663 |
| 13 | pseudogene | 7.307774 | 7.309215 | positive | MGI_C57BL6J_3644571 | Gm8725 | NCBI_Gene:667611,ENSEMBL:ENSMUSG00000113112 | MGI:3644571 | pseudogene | predicted gene 8725 |
| 13 | pseudogene | 7.387147 | 7.388238 | positive | MGI_C57BL6J_3647846 | Gm6437 | NCBI_Gene:623516 | MGI:3647846 | pseudogene | predicted gene 6437 |
| 13 | gene | 7.446509 | 7.476761 | positive | MGI_C57BL6J_5595233 | Gm36074 | NCBI_Gene:102639860,ENSEMBL:ENSMUSG00000113225 | MGI:5595233 | lncRNA gene | predicted gene, 36074 |
| 13 | gene | 7.507318 | 7.510231 | positive | MGI_C57BL6J_5595295 | Gm36136 | NCBI_Gene:102639941 | MGI:5595295 | lncRNA gene | predicted gene, 36136 |
| 13 | pseudogene | 7.633044 | 7.633191 | positive | MGI_C57BL6J_6097665 | Gm48251 | ENSEMBL:ENSMUSG00000112939 | MGI:6097665 | pseudogene | predicted gene, 48251 |
| 13 | pseudogene | 7.753170 | 7.753278 | negative | MGI_C57BL6J_6097666 | Gm48252 | ENSEMBL:ENSMUSG00000113828 | MGI:6097666 | pseudogene | predicted gene, 48252 |
| 13 | gene | 7.803436 | 7.804714 | negative | MGI_C57BL6J_6097667 | Gm48253 | ENSEMBL:ENSMUSG00000113316 | MGI:6097667 | unclassified gene | predicted gene, 48253 |
| 13 | pseudogene | 8.013696 | 8.038218 | negative | MGI_C57BL6J_3780137 | Gm9742 | NCBI_Gene:791083,ENSEMBL:ENSMUSG00000113508 | MGI:3780137 | pseudogene | predicted gene 9742 |
| 13 | pseudogene | 8.128050 | 8.130854 | positive | MGI_C57BL6J_3646226 | Gm5191 | NCBI_Gene:382722,ENSEMBL:ENSMUSG00000113412 | MGI:3646226 | pseudogene | predicted gene 5191 |
| 13 | gene | 8.199247 | 8.200494 | negative | MGI_C57BL6J_6097673 | Gm48257 | ENSEMBL:ENSMUSG00000114126 | MGI:6097673 | lncRNA gene | predicted gene, 48257 |
| 13 | gene | 8.202430 | 8.768762 | positive | MGI_C57BL6J_2151118 | Adarb2 | NCBI_Gene:94191,ENSEMBL:ENSMUSG00000052551 | MGI:2151118 | protein coding gene | adenosine deaminase, RNA-specific, B2 |
| 13 | gene | 8.278604 | 8.279289 | positive | MGI_C57BL6J_6097676 | Gm48259 | ENSEMBL:ENSMUSG00000113810 | MGI:6097676 | unclassified gene | predicted gene, 48259 |
| 13 | gene | 8.282248 | 8.286103 | positive | MGI_C57BL6J_6097679 | Gm48261 | ENSEMBL:ENSMUSG00000113073 | MGI:6097679 | unclassified gene | predicted gene, 48261 |
| 13 | pseudogene | 8.289850 | 8.290113 | negative | MGI_C57BL6J_3641865 | Gm10357 | ENSEMBL:ENSMUSG00000072421 | MGI:3641865 | pseudogene | predicted gene 10357 |
| 13 | pseudogene | 8.295315 | 8.299143 | negative | MGI_C57BL6J_6097678 | Gm48260 | ENSEMBL:ENSMUSG00000113978 | MGI:6097678 | pseudogene | predicted gene, 48260 |
| 13 | gene | 8.553020 | 8.557635 | positive | MGI_C57BL6J_6097681 | Gm48262 | ENSEMBL:ENSMUSG00000113742 | MGI:6097681 | unclassified gene | predicted gene, 48262 |
| 13 | gene | 8.797302 | 8.798402 | negative | MGI_C57BL6J_6097782 | Gm48327 | ENSEMBL:ENSMUSG00000113391 | MGI:6097782 | unclassified gene | predicted gene, 48327 |
| 13 | gene | 8.802966 | 8.872100 | negative | MGI_C57BL6J_1920393 | Wdr37 | NCBI_Gene:207615,ENSEMBL:ENSMUSG00000021147 | MGI:1920393 | protein coding gene | WD repeat domain 37 |
| 13 | gene | 8.833099 | 8.834372 | negative | MGI_C57BL6J_6097851 | Gm48375 | ENSEMBL:ENSMUSG00000114094 | MGI:6097851 | unclassified gene | predicted gene, 48375 |
| 13 | gene | 8.870341 | 8.872105 | positive | MGI_C57BL6J_6097849 | Gm48374 | ENSEMBL:ENSMUSG00000113589 | MGI:6097849 | unclassified gene | predicted gene, 48374 |
| 13 | gene | 8.884914 | 8.885823 | negative | MGI_C57BL6J_6098139 | Gm48574 | ENSEMBL:ENSMUSG00000113245 | MGI:6098139 | unclassified gene | predicted gene, 48574 |
| 13 | gene | 8.885501 | 8.892451 | positive | MGI_C57BL6J_2442264 | Idi1 | NCBI_Gene:319554,ENSEMBL:ENSMUSG00000058258 | MGI:2442264 | protein coding gene | isopentenyl-diphosphate delta isomerase |
| 13 | gene | 8.921336 | 8.921438 | negative | MGI_C57BL6J_5452527 | Gm22750 | ENSEMBL:ENSMUSG00000076030 | MGI:5452527 | miRNA gene | predicted gene, 22750 |
| 13 | pseudogene | 8.940301 | 8.940988 | negative | MGI_C57BL6J_3646227 | Rpl10a-ps2 | NCBI_Gene:382723,ENSEMBL:ENSMUSG00000061988 | MGI:3646227 | pseudogene | ribosomal protein L10A, pseudogene 2 |
| 13 | gene | 8.940986 | 8.948231 | negative | MGI_C57BL6J_3704398 | Gm9745 | NCBI_Gene:100039655,ENSEMBL:ENSMUSG00000021148 | MGI:3704398 | protein coding gene | predicted gene 9745 |
| 13 | gene | 8.945045 | 8.945179 | positive | MGI_C57BL6J_5455051 | Gm25274 | ENSEMBL:ENSMUSG00000077484 | MGI:5455051 | snoRNA gene | predicted gene, 25274 |
| 13 | gene | 8.949921 | 8.950053 | negative | MGI_C57BL6J_5451892 | Gm22115 | ENSEMBL:ENSMUSG00000096771 | MGI:5451892 | snoRNA gene | predicted gene, 22115 |
| 13 | gene | 8.952863 | 8.960945 | positive | MGI_C57BL6J_2444315 | Idi2 | NCBI_Gene:320581,ENSEMBL:ENSMUSG00000033520 | MGI:2444315 | protein coding gene | isopentenyl-diphosphate delta isomerase 2 |
| 13 | gene | 8.955932 | 8.956064 | negative | MGI_C57BL6J_5453700 | Gm23923 | ENSEMBL:ENSMUSG00000095414 | MGI:5453700 | snoRNA gene | predicted gene, 23923 |
| 13 | gene | 8.966331 | 8.996532 | negative | MGI_C57BL6J_1916487 | Gtpbp4 | NCBI_Gene:69237,ENSEMBL:ENSMUSG00000021149 | MGI:1916487 | protein coding gene | GTP binding protein 4 |
| 13 | pseudogene | 8.966705 | 8.968082 | positive | MGI_C57BL6J_6098258 | Gm48649 | ENSEMBL:ENSMUSG00000113466 | MGI:6098258 | pseudogene | predicted gene, 48649 |
| 13 | pseudogene | 8.993005 | 8.993238 | positive | MGI_C57BL6J_6098259 | Gm48650 | ENSEMBL:ENSMUSG00000113731 | MGI:6098259 | pseudogene | predicted gene, 48650 |
| 13 | gene | 9.001432 | 9.076451 | negative | MGI_C57BL6J_5595423 | Gm36264 | NCBI_Gene:102640119,ENSEMBL:ENSMUSG00000113543 | MGI:5595423 | lncRNA gene | predicted gene, 36264 |
| 13 | pseudogene | 9.025732 | 9.026545 | negative | MGI_C57BL6J_3642977 | Gm8782 | NCBI_Gene:667721 | MGI:3642977 | pseudogene | predicted gene 8782 |
| 13 | gene | 9.093814 | 9.174451 | positive | MGI_C57BL6J_106330 | Larp4b | NCBI_Gene:217980,ENSEMBL:ENSMUSG00000033499 | MGI:106330 | protein coding gene | La ribonucleoprotein domain family, member 4B |
| 13 | gene | 9.109305 | 9.109599 | positive | MGI_C57BL6J_5453330 | Gm23553 | ENSEMBL:ENSMUSG00000088442 | MGI:5453330 | unclassified non-coding RNA gene | predicted gene, 23553 |
| 13 | gene | 9.128239 | 9.130509 | positive | MGI_C57BL6J_3045272 | A130023E24Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA A130023E24 gene |
| 13 | pseudogene | 9.140731 | 9.140881 | negative | MGI_C57BL6J_5578861 | Gm28155 | ENSEMBL:ENSMUSG00000099428 | MGI:5578861 | pseudogene | predicted gene 28155 |
| 13 | pseudogene | 9.199486 | 9.224989 | positive | MGI_C57BL6J_3646031 | Gm8784 | NCBI_Gene:667727,ENSEMBL:ENSMUSG00000113936 | MGI:3646031 | pseudogene | predicted gene 8784 |
| 13 | gene | 9.268684 | 9.274143 | positive | MGI_C57BL6J_6098546 | Gm48825 | ENSEMBL:ENSMUSG00000113455 | MGI:6098546 | lncRNA gene | predicted gene, 48825 |
| 13 | pseudogene | 9.272686 | 9.273163 | negative | MGI_C57BL6J_5826028 | Gm46391 | NCBI_Gene:108168050,ENSEMBL:ENSMUSG00000113755 | MGI:5826028 | pseudogene | predicted gene, 46391 |
| 13 | gene | 9.276503 | 9.668928 | positive | MGI_C57BL6J_1920179 | Dip2c | NCBI_Gene:208440,ENSEMBL:ENSMUSG00000048264 | MGI:1920179 | protein coding gene | disco interacting protein 2 homolog C |
| 13 | gene | 9.321427 | 9.322979 | positive | MGI_C57BL6J_6098646 | Gm48887 | ENSEMBL:ENSMUSG00000113922 | MGI:6098646 | unclassified gene | predicted gene, 48887 |
| 13 | gene | 9.328630 | 9.330247 | positive | MGI_C57BL6J_6098652 | Gm48890 | ENSEMBL:ENSMUSG00000113984 | MGI:6098652 | unclassified gene | predicted gene, 48890 |
| 13 | gene | 9.344402 | 9.348425 | positive | MGI_C57BL6J_6098616 | Gm48870 | ENSEMBL:ENSMUSG00000113065 | MGI:6098616 | unclassified gene | predicted gene, 48870 |
| 13 | pseudogene | 9.352028 | 9.352689 | positive | MGI_C57BL6J_6098615 | Gm48869 | ENSEMBL:ENSMUSG00000113869 | MGI:6098615 | pseudogene | predicted gene, 48869 |
| 13 | pseudogene | 9.385729 | 9.387327 | negative | MGI_C57BL6J_5011376 | Gm19191 | NCBI_Gene:100418409 | MGI:5011376 | pseudogene | predicted gene, 19191 |
| 13 | gene | 9.387139 | 9.391741 | positive | MGI_C57BL6J_5477095 | Gm26601 | ENSEMBL:ENSMUSG00000097859 | MGI:5477095 | lncRNA gene | predicted gene, 26601 |
| 13 | gene | 9.412517 | 9.415160 | positive | MGI_C57BL6J_6098650 | Gm48889 | ENSEMBL:ENSMUSG00000112955 | MGI:6098650 | unclassified gene | predicted gene, 48889 |
| 13 | gene | 9.445900 | 9.448419 | positive | MGI_C57BL6J_6098618 | Gm48871 | ENSEMBL:ENSMUSG00000112948 | MGI:6098618 | unclassified gene | predicted gene, 48871 |
| 13 | gene | 9.557861 | 9.561989 | positive | MGI_C57BL6J_6098620 | Gm48872 | ENSEMBL:ENSMUSG00000113905 | MGI:6098620 | unclassified gene | predicted gene, 48872 |
| 13 | gene | 9.623380 | 9.623627 | negative | MGI_C57BL6J_5453273 | Gm23496 | ENSEMBL:ENSMUSG00000088366 | MGI:5453273 | unclassified non-coding RNA gene | predicted gene, 23496 |
| 13 | gene | 9.635329 | 9.637089 | negative | MGI_C57BL6J_6098648 | Gm48888 | ENSEMBL:ENSMUSG00000113355 | MGI:6098648 | unclassified gene | predicted gene, 48888 |
| 13 | gene | 9.674531 | 9.689544 | positive | MGI_C57BL6J_5595582 | Gm36423 | NCBI_Gene:102640331,ENSEMBL:ENSMUSG00000114138 | MGI:5595582 | lncRNA gene | predicted gene, 36423 |
| 13 | gene | 9.684833 | 9.766014 | negative | MGI_C57BL6J_1913755 | Zmynd11 | NCBI_Gene:66505,ENSEMBL:ENSMUSG00000021156 | MGI:1913755 | protein coding gene | zinc finger, MYND domain containing 11 |
| 13 | gene | 9.733897 | 9.738234 | positive | MGI_C57BL6J_6096981 | Gm47802 | ENSEMBL:ENSMUSG00000113885 | MGI:6096981 | lncRNA gene | predicted gene, 47802 |
| 13 | gene | 9.753741 | 9.756879 | negative | MGI_C57BL6J_6097020 | Gm47826 | ENSEMBL:ENSMUSG00000113650 | MGI:6097020 | unclassified gene | predicted gene, 47826 |
| 13 | gene | 9.796447 | 9.801823 | positive | MGI_C57BL6J_5623550 | Gm40665 | NCBI_Gene:105245175 | MGI:5623550 | lncRNA gene | predicted gene, 40665 |
| 13 | pseudogene | 9.820511 | 9.820733 | positive | MGI_C57BL6J_6096330 | Gm47401 | ENSEMBL:ENSMUSG00000113754 | MGI:6096330 | pseudogene | predicted gene, 47401 |
| 13 | gene | 9.834469 | 9.834525 | negative | MGI_C57BL6J_5453964 | Gm24187 | ENSEMBL:ENSMUSG00000088609 | MGI:5453964 | miRNA gene | predicted gene, 24187 |
| 13 | gene | 9.875486 | 10.360914 | negative | MGI_C57BL6J_88398 | Chrm3 | NCBI_Gene:12671,ENSEMBL:ENSMUSG00000046159 | MGI:88398 | protein coding gene | cholinergic receptor, muscarinic 3, cardiac |
| 13 | pseudogene | 9.892910 | 9.894050 | positive | MGI_C57BL6J_6096338 | Gm47406 | ENSEMBL:ENSMUSG00000113146 | MGI:6096338 | pseudogene | predicted gene, 47406 |
| 13 | gene | 10.048667 | 10.051848 | negative | MGI_C57BL6J_6096346 | Gm47411 | ENSEMBL:ENSMUSG00000113502 | MGI:6096346 | unclassified gene | predicted gene, 47411 |
| 13 | gene | 10.064758 | 10.070866 | negative | MGI_C57BL6J_6096336 | Gm47405 | ENSEMBL:ENSMUSG00000113680 | MGI:6096336 | lncRNA gene | predicted gene, 47405 |
| 13 | gene | 10.164744 | 10.165774 | negative | MGI_C57BL6J_1924453 | 9330159N22Rik | ENSEMBL:ENSMUSG00000113695 | MGI:1924453 | unclassified gene | RIKEN cDNA 9330159N22 gene |
| 13 | gene | 10.175784 | 10.180807 | negative | MGI_C57BL6J_6096334 | Gm47404 | ENSEMBL:ENSMUSG00000113261 | MGI:6096334 | lncRNA gene | predicted gene, 47404 |
| 13 | gene | 10.357595 | 10.797502 | positive | MGI_C57BL6J_5477355 | Gm26861 | NCBI_Gene:108168064,ENSEMBL:ENSMUSG00000097009 | MGI:5477355 | lncRNA gene | predicted gene, 26861 |
| 13 | gene | 10.517687 | 10.538811 | positive | MGI_C57BL6J_6096339 | Gm47407 | ENSEMBL:ENSMUSG00000113758 | MGI:6096339 | lncRNA gene | predicted gene, 47407 |
| 13 | gene | 10.539204 | 10.557739 | negative | MGI_C57BL6J_5595684 | Gm36525 | NCBI_Gene:102640472,ENSEMBL:ENSMUSG00000113243 | MGI:5595684 | lncRNA gene | predicted gene, 36525 |
| 13 | gene | 10.592484 | 10.592566 | negative | MGI_C57BL6J_5453192 | Gm23415 | ENSEMBL:ENSMUSG00000093260 | MGI:5453192 | miRNA gene | predicted gene, 23415 |
| 13 | gene | 10.674389 | 10.676726 | positive | MGI_C57BL6J_5623551 | Gm40666 | NCBI_Gene:105245176 | MGI:5623551 | lncRNA gene | predicted gene, 40666 |
| 13 | gene | 10.700161 | 10.830778 | positive | MGI_C57BL6J_6096341 | Gm47408 | ENSEMBL:ENSMUSG00000114134 | MGI:6096341 | lncRNA gene | predicted gene, 47408 |
| 13 | gene | 10.974423 | 10.979945 | positive | MGI_C57BL6J_6096464 | Gm47486 | ENSEMBL:ENSMUSG00000113673 | MGI:6096464 | lncRNA gene | predicted gene, 47486 |
| 13 | gene | 11.426779 | 11.426898 | negative | MGI_C57BL6J_5455273 | Gm25496 | ENSEMBL:ENSMUSG00000077618 | MGI:5455273 | snoRNA gene | predicted gene, 25496 |
| 13 | pseudogene | 11.522052 | 11.528852 | positive | MGI_C57BL6J_3690088 | Zp4-ps | NCBI_Gene:664793,ENSEMBL:ENSMUSG00000113593 | MGI:3690088 | pseudogene | zona pellucida glycoprotein 4, pseudogene |
| 13 | gene | 11.523096 | 11.523744 | negative | MGI_C57BL6J_6096466 | Gm47487 | ENSEMBL:ENSMUSG00000113799 | MGI:6096466 | unclassified gene | predicted gene, 47487 |
| 13 | gene | 11.553099 | 12.106945 | negative | MGI_C57BL6J_99685 | Ryr2 | NCBI_Gene:20191,ENSEMBL:ENSMUSG00000021313 | MGI:99685 | protein coding gene | ryanodine receptor 2, cardiac |
| 13 | gene | 11.818248 | 11.821734 | negative | MGI_C57BL6J_6096476 | Gm47494 | ENSEMBL:ENSMUSG00000113269 | MGI:6096476 | unclassified gene | predicted gene, 47494 |
| 13 | pseudogene | 11.939558 | 11.940543 | positive | MGI_C57BL6J_5504071 | Gm26956 | ENSEMBL:ENSMUSG00000098110 | MGI:5504071 | pseudogene | predicted gene, 26956 |
| 13 | gene | 12.047342 | 12.052055 | negative | MGI_C57BL6J_6096474 | Gm47493 | ENSEMBL:ENSMUSG00000112947 | MGI:6096474 | unclassified gene | predicted gene, 47493 |
| 13 | pseudogene | 12.160754 | 12.160964 | positive | MGI_C57BL6J_6096208 | Gm47323 | ENSEMBL:ENSMUSG00000113347 | MGI:6096208 | pseudogene | predicted gene, 47323 |
| 13 | pseudogene | 12.171504 | 12.172150 | negative | MGI_C57BL6J_3643012 | Gm16419 | NCBI_Gene:100039678,ENSEMBL:ENSMUSG00000113848 | MGI:3643012 | pseudogene | predicted gene 16419 |
| 13 | gene | 12.177458 | 12.179281 | negative | MGI_C57BL6J_6096206 | Gm47322 | ENSEMBL:ENSMUSG00000113789 | MGI:6096206 | unclassified gene | predicted gene, 47322 |
| 13 | gene | 12.182712 | 12.258193 | negative | MGI_C57BL6J_894292 | Mtr | NCBI_Gene:238505,ENSEMBL:ENSMUSG00000021311 | MGI:894292 | protein coding gene | 5-methyltetrahydrofolate-homocysteine methyltransferase |
| 13 | gene | 12.182717 | 12.186488 | negative | MGI_C57BL6J_3642355 | Gm10336 | NCBI_Gene:328186 | MGI:3642355 | lncRNA gene | predicted gene 10336 |
| 13 | pseudogene | 12.259793 | 12.260270 | negative | MGI_C57BL6J_6095708 | Gm47021 | ENSEMBL:ENSMUSG00000113036 | MGI:6095708 | pseudogene | predicted gene, 47021 |
| 13 | gene | 12.269426 | 12.340760 | negative | MGI_C57BL6J_109192 | Actn2 | NCBI_Gene:11472,ENSEMBL:ENSMUSG00000052374 | MGI:109192 | protein coding gene | actinin alpha 2 |
| 13 | gene | 12.309722 | 12.310844 | positive | MGI_C57BL6J_5589185 | Gm30026 | NCBI_Gene:102631774 | MGI:5589185 | lncRNA gene | predicted gene, 30026 |
| 13 | gene | 12.310900 | 12.323960 | positive | MGI_C57BL6J_5623552 | Gm40667 | NCBI_Gene:105245177 | MGI:5623552 | lncRNA gene | predicted gene, 40667 |
| 13 | pseudogene | 12.378158 | 12.378640 | positive | MGI_C57BL6J_3644181 | Gm5445 | NCBI_Gene:432725,ENSEMBL:ENSMUSG00000094185 | MGI:3644181 | pseudogene | predicted gene 5445 |
| 13 | gene | 12.395027 | 12.440289 | positive | MGI_C57BL6J_2442524 | Heatr1 | NCBI_Gene:217995,ENSEMBL:ENSMUSG00000050244 | MGI:2442524 | protein coding gene | HEAT repeat containing 1 |
| 13 | pseudogene | 12.424896 | 12.426147 | negative | MGI_C57BL6J_3646762 | Gm5928 | NCBI_Gene:102631952,ENSEMBL:ENSMUSG00000113931 | MGI:3646762 | pseudogene | predicted gene 5928 |
| 13 | gene | 12.439402 | 12.464977 | negative | MGI_C57BL6J_1928481 | Lgals8 | NCBI_Gene:56048,ENSEMBL:ENSMUSG00000057554 | MGI:1928481 | protein coding gene | lectin, galactose binding, soluble 8 |
| 13 | gene | 12.471209 | 12.520704 | negative | MGI_C57BL6J_1931001 | Edaradd | NCBI_Gene:171211,ENSEMBL:ENSMUSG00000095105 | MGI:1931001 | protein coding gene | EDAR (ectodysplasin-A receptor)-associated death domain |
| 13 | gene | 12.540143 | 12.562716 | positive | MGI_C57BL6J_5589398 | Gm30239 | ENSEMBL:ENSMUSG00000114115 | MGI:5589398 | lncRNA gene | predicted gene, 30239 |
| 13 | gene | 12.565808 | 12.611396 | positive | MGI_C57BL6J_1914725 | Ero1lb | NCBI_Gene:67475,ENSEMBL:ENSMUSG00000057069 | MGI:1914725 | protein coding gene | ERO1-like beta (S. cerevisiae) |
| 13 | gene | 12.589565 | 12.589791 | positive | MGI_C57BL6J_5455260 | Gm25483 | ENSEMBL:ENSMUSG00000065158 | MGI:5455260 | snoRNA gene | predicted gene, 25483 |
| 13 | pseudogene | 12.614065 | 12.650395 | negative | MGI_C57BL6J_3710533 | Gpr137b-ps | NCBI_Gene:664862,ENSEMBL:ENSMUSG00000097715 | MGI:3710533 | pseudogene | G protein-coupled receptor 137B, pseudogene |
| 13 | gene | 12.666051 | 12.666314 | positive | MGI_C57BL6J_5453842 | Gm24065 | ENSEMBL:ENSMUSG00000088112 | MGI:5453842 | unclassified non-coding RNA gene | predicted gene, 24065 |
| 13 | pseudogene | 12.675601 | 12.675957 | negative | MGI_C57BL6J_6097948 | Gm48440 | ENSEMBL:ENSMUSG00000113021 | MGI:6097948 | pseudogene | predicted gene, 48440 |
| 13 | pseudogene | 12.679747 | 12.679939 | negative | MGI_C57BL6J_6097956 | Gm48445 | ENSEMBL:ENSMUSG00000113308 | MGI:6097956 | pseudogene | predicted gene, 48445 |
| 13 | pseudogene | 12.679940 | 12.680359 | positive | MGI_C57BL6J_6097969 | Gm48455 | ENSEMBL:ENSMUSG00000113996 | MGI:6097969 | pseudogene | predicted gene, 48455 |
| 13 | pseudogene | 12.682289 | 12.683041 | positive | MGI_C57BL6J_6097970 | Gm48456 | ENSEMBL:ENSMUSG00000113148 | MGI:6097970 | pseudogene | predicted gene, 48456 |
| 13 | pseudogene | 12.683610 | 12.684013 | negative | MGI_C57BL6J_6097972 | Gm48458 | ENSEMBL:ENSMUSG00000113914 | MGI:6097972 | pseudogene | predicted gene, 48458 |
| 13 | gene | 12.701821 | 12.823297 | positive | MGI_C57BL6J_3780567 | Gm2399 | NCBI_Gene:105245453,ENSEMBL:ENSMUSG00000078141 | MGI:3780567 | protein coding gene | predicted gene 2399 |
| 13 | pseudogene | 12.734364 | 12.734785 | positive | MGI_C57BL6J_3643634 | Gm7042 | NCBI_Gene:108168065,ENSEMBL:ENSMUSG00000113448 | MGI:3643634 | pseudogene | predicted gene 7042 |
| 13 | gene | 12.790816 | 12.800093 | negative | MGI_C57BL6J_1341833 | Prl2c3 | NCBI_Gene:18812,ENSEMBL:ENSMUSG00000056457 | MGI:1341833 | protein coding gene | prolactin family 2, subfamily c, member 3 |
| 13 | pseudogene | 12.864124 | 12.866069 | positive | MGI_C57BL6J_5010447 | Gm18262 | NCBI_Gene:100416801,ENSEMBL:ENSMUSG00000113053 | MGI:5010447 | pseudogene | predicted gene, 18262 |
| 13 | pseudogene | 12.866445 | 12.874100 | negative | MGI_C57BL6J_3780712 | Gm2544 | NCBI_Gene:100039999,ENSEMBL:ENSMUSG00000113117 | MGI:3780712 | pseudogene | predicted gene 2544 |
| 13 | pseudogene | 12.879755 | 12.881253 | negative | MGI_C57BL6J_5011063 | Gm18878 | NCBI_Gene:100417883,ENSEMBL:ENSMUSG00000113353 | MGI:5011063 | pseudogene | predicted gene, 18878 |
| 13 | pseudogene | 12.917710 | 12.917965 | negative | MGI_C57BL6J_6098066 | Gm48527 | ENSEMBL:ENSMUSG00000113256 | MGI:6098066 | pseudogene | predicted gene, 48527 |
| 13 | gene | 12.925762 | 12.932245 | positive | MGI_C57BL6J_5623789 | Gm40904 | NCBI_Gene:105245452 | MGI:5623789 | lncRNA gene | predicted gene, 40904 |
| 13 | gene | 12.996120 | 13.005383 | negative | MGI_C57BL6J_97618 | Prl2c2 | NCBI_Gene:18811,ENSEMBL:ENSMUSG00000079092 | MGI:97618 | protein coding gene | prolactin family 2, subfamily c, member 2 |
| 13 | gene | 13.030789 | 13.033906 | positive | MGI_C57BL6J_5623788 | Gm40903 | NCBI_Gene:105245451 | MGI:5623788 | lncRNA gene | predicted gene, 40903 |
| 13 | gene | 13.034858 | 13.038936 | positive | MGI_C57BL6J_6098092 | Gm48544 | ENSEMBL:ENSMUSG00000113049 | MGI:6098092 | lncRNA gene | predicted gene, 48544 |
| 13 | pseudogene | 13.076118 | 13.078045 | positive | MGI_C57BL6J_5010448 | Gm18263 | NCBI_Gene:100416802,ENSEMBL:ENSMUSG00000113150 | MGI:5010448 | pseudogene | predicted gene, 18263 |
| 13 | pseudogene | 13.078699 | 13.081311 | negative | MGI_C57BL6J_5591459 | Gm32300 | NCBI_Gene:102634800,ENSEMBL:ENSMUSG00000113567 | MGI:5591459 | pseudogene | predicted gene, 32300 |
| 13 | pseudogene | 13.103874 | 13.105410 | positive | MGI_C57BL6J_5011064 | Gm18879 | NCBI_Gene:100417884,ENSEMBL:ENSMUSG00000113624 | MGI:5011064 | pseudogene | predicted gene, 18879 |
| 13 | pseudogene | 13.111476 | 13.118600 | positive | MGI_C57BL6J_3780036 | Gm9628 | NCBI_Gene:674723,ENSEMBL:ENSMUSG00000113319 | MGI:3780036 | pseudogene | predicted gene 9628 |
| 13 | pseudogene | 13.119254 | 13.121268 | negative | MGI_C57BL6J_5010444 | Gm18259 | NCBI_Gene:100416797,ENSEMBL:ENSMUSG00000113675 | MGI:5010444 | pseudogene | predicted gene, 18259 |
| 13 | gene | 13.168730 | 13.171177 | negative | MGI_C57BL6J_3642757 | Gm10811 | ENSEMBL:ENSMUSG00000113908 | MGI:3642757 | protein coding gene | predicted gene 10811 |
| 13 | gene | 13.182703 | 13.191930 | positive | MGI_C57BL6J_1858413 | Prl2c5 | NCBI_Gene:107849,ENSEMBL:ENSMUSG00000055360 | MGI:1858413 | protein coding gene | prolactin family 2, subfamily c, member 5 |
| 13 | pseudogene | 13.197379 | 13.198919 | positive | MGI_C57BL6J_5011065 | Gm18880 | NCBI_Gene:100417885,ENSEMBL:ENSMUSG00000113221 | MGI:5011065 | pseudogene | predicted gene, 18880 |
| 13 | pseudogene | 13.204876 | 13.207550 | positive | MGI_C57BL6J_6098160 | Gm48588 | ENSEMBL:ENSMUSG00000113371 | MGI:6098160 | pseudogene | predicted gene, 48588 |
| 13 | pseudogene | 13.224663 | 13.224925 | negative | MGI_C57BL6J_6098161 | Gm48589 | ENSEMBL:ENSMUSG00000113729 | MGI:6098161 | pseudogene | predicted gene, 48589 |
| 13 | pseudogene | 13.231493 | 13.232963 | negative | MGI_C57BL6J_3805957 | Gm2423 | NCBI_Gene:100039786,ENSEMBL:ENSMUSG00000061724 | MGI:3805957 | pseudogene | predicted gene 2423 |
| 13 | pseudogene | 13.275129 | 13.276085 | positive | MGI_C57BL6J_3779486 | Gm5374 | NCBI_Gene:385049,ENSEMBL:ENSMUSG00000114152 | MGI:3779486 | pseudogene | predicted gene 5374 |
| 13 | pseudogene | 13.283832 | 13.284247 | positive | MGI_C57BL6J_6098162 | Gm48590 | ENSEMBL:ENSMUSG00000112938 | MGI:6098162 | pseudogene | predicted gene, 48590 |
| 13 | gene | 13.298243 | 13.298342 | negative | MGI_C57BL6J_5455910 | Gm26133 | ENSEMBL:ENSMUSG00000096198 | MGI:5455910 | snRNA gene | predicted gene, 26133 |
| 13 | gene | 13.320830 | 13.323370 | negative | MGI_C57BL6J_3779654 | Gm7040 | NCBI_Gene:630663,ENSEMBL:ENSMUSG00000113027 | MGI:3779654 | protein coding gene | predicted gene 7040 |
| 13 | pseudogene | 13.323364 | 13.350427 | positive | MGI_C57BL6J_5010747 | Gm18562 | NCBI_Gene:100417363,ENSEMBL:ENSMUSG00000110123 | MGI:5010747 | pseudogene | predicted gene, 18562 |
| 13 | gene | 13.329484 | 13.329671 | positive | MGI_C57BL6J_5455581 | Gm25804 | ENSEMBL:ENSMUSG00000084734 | MGI:5455581 | snoRNA gene | predicted gene, 25804 |
| 13 | gene | 13.357620 | 13.394236 | negative | MGI_C57BL6J_1891463 | Gpr137b | NCBI_Gene:83924,ENSEMBL:ENSMUSG00000021306 | MGI:1891463 | protein coding gene | G protein-coupled receptor 137B |
| 13 | gene | 13.402836 | 13.403132 | positive | MGI_C57BL6J_5452858 | Gm23081 | ENSEMBL:ENSMUSG00000088470 | MGI:5452858 | unclassified non-coding RNA gene | predicted gene, 23081 |
| 13 | pseudogene | 13.412440 | 13.412794 | negative | MGI_C57BL6J_6098276 | Gm48661 | ENSEMBL:ENSMUSG00000113445 | MGI:6098276 | pseudogene | predicted gene, 48661 |
| 13 | pseudogene | 13.417147 | 13.418200 | positive | MGI_C57BL6J_6098291 | Gm48672 | ENSEMBL:ENSMUSG00000113444 | MGI:6098291 | pseudogene | predicted gene, 48672 |
| 13 | pseudogene | 13.418762 | 13.437551 | negative | MGI_C57BL6J_3647725 | Gm7426 | NCBI_Gene:664960,ENSEMBL:ENSMUSG00000094865 | MGI:3647725 | pseudogene | predicted gene 7426 |
| 13 | gene | 13.437551 | 13.512275 | positive | MGI_C57BL6J_97342 | Nid1 | NCBI_Gene:18073,ENSEMBL:ENSMUSG00000005397 | MGI:97342 | protein coding gene | nidogen 1 |
| 13 | gene | 13.474343 | 13.482405 | negative | MGI_C57BL6J_6098306 | Gm48682 | ENSEMBL:ENSMUSG00000113380 | MGI:6098306 | lncRNA gene | predicted gene, 48682 |
| 13 | gene | 13.514738 | 13.515817 | positive | MGI_C57BL6J_6098481 | Gm48783 | ENSEMBL:ENSMUSG00000114068 | MGI:6098481 | unclassified gene | predicted gene, 48783 |
| 13 | gene | 13.586046 | 13.591868 | negative | MGI_C57BL6J_5589995 | Gm30836 | NCBI_Gene:102632874 | MGI:5589995 | lncRNA gene | predicted gene, 30836 |
| 13 | gene | 13.590340 | 13.778803 | positive | MGI_C57BL6J_107448 | Lyst | NCBI_Gene:17101,ENSEMBL:ENSMUSG00000019726 | MGI:107448 | protein coding gene | lysosomal trafficking regulator |
| 13 | gene | 13.784059 | 13.827896 | positive | MGI_C57BL6J_102703 | Gng4 | NCBI_Gene:14706,ENSEMBL:ENSMUSG00000021303 | MGI:102703 | protein coding gene | guanine nucleotide binding protein (G protein), gamma 4 |
| 13 | gene | 13.789344 | 13.789633 | positive | MGI_C57BL6J_5455048 | Gm25271 | ENSEMBL:ENSMUSG00000087898 | MGI:5455048 | unclassified non-coding RNA gene | predicted gene, 25271 |
| 13 | pseudogene | 13.842146 | 13.844819 | positive | MGI_C57BL6J_3644941 | Gm7441 | NCBI_Gene:665000,ENSEMBL:ENSMUSG00000113493 | MGI:3644941 | pseudogene | predicted gene 7441 |
| 13 | gene | 13.862534 | 13.893979 | positive | MGI_C57BL6J_6097109 | Gm47882 | ENSEMBL:ENSMUSG00000113811 | MGI:6097109 | lncRNA gene | predicted gene, 47882 |
| 13 | pseudogene | 13.890760 | 13.891999 | negative | MGI_C57BL6J_3644947 | Gm7446 | NCBI_Gene:665008,ENSEMBL:ENSMUSG00000113258 | MGI:3644947 | pseudogene | predicted gene 7446 |
| 13 | pseudogene | 13.895807 | 13.910408 | negative | MGI_C57BL6J_3779656 | Gm7046 | NCBI_Gene:630894,ENSEMBL:ENSMUSG00000114087 | MGI:3779656 | pseudogene | predicted gene 7046 |
| 13 | gene | 13.953219 | 13.954338 | positive | MGI_C57BL6J_6097112 | Gm47884 | ENSEMBL:ENSMUSG00000113310 | MGI:6097112 | lncRNA gene | predicted gene, 47884 |
| 13 | gene | 13.954469 | 13.999103 | positive | MGI_C57BL6J_2145517 | B3galnt2 | NCBI_Gene:97884,ENSEMBL:ENSMUSG00000039242 | MGI:2145517 | protein coding gene | UDP-GalNAc:betaGlcNAc beta 1,3-galactosaminyltransferase, polypeptide 2 |
| 13 | pseudogene | 13.964475 | 13.971337 | negative | MGI_C57BL6J_5011041 | Gm18856 | NCBI_Gene:100417835,ENSEMBL:ENSMUSG00000096192 | MGI:5011041 | pseudogene | predicted gene, 18856 |
| 13 | gene | 13.997947 | 14.039639 | negative | MGI_C57BL6J_1917680 | Tbce | NCBI_Gene:70430,ENSEMBL:ENSMUSG00000039233 | MGI:1917680 | protein coding gene | tubulin-specific chaperone E |
| 13 | pseudogene | 14.041392 | 14.041599 | positive | MGI_C57BL6J_6097326 | Gm48017 | ENSEMBL:ENSMUSG00000113819 | MGI:6097326 | pseudogene | predicted gene, 48017 |
| 13 | gene | 14.049793 | 14.063488 | negative | MGI_C57BL6J_1341724 | Ggps1 | NCBI_Gene:14593,ENSEMBL:ENSMUSG00000021302 | MGI:1341724 | protein coding gene | geranylgeranyl diphosphate synthase 1 |
| 13 | gene | 14.063232 | 14.199603 | positive | MGI_C57BL6J_2137512 | Arid4b | NCBI_Gene:94246,ENSEMBL:ENSMUSG00000039219 | MGI:2137512 | protein coding gene | AT rich interactive domain 4B (RBP1-like) |
| 13 | gene | 14.065987 | 14.069060 | positive | MGI_C57BL6J_3041220 | D230040A04Rik | NA | NA | unclassified gene | RIKEN cDNA D230040A04 gene |
| 13 | gene | 14.087439 | 14.087545 | positive | MGI_C57BL6J_5455906 | Gm26129 | ENSEMBL:ENSMUSG00000064653 | MGI:5455906 | snRNA gene | predicted gene, 26129 |
| 13 | gene | 14.118743 | 14.118855 | negative | MGI_C57BL6J_4421897 | n-R5s52 | ENSEMBL:ENSMUSG00000077802 | MGI:4421897 | rRNA gene | nuclear encoded rRNA 5S 52 |
| 13 | gene | 14.171053 | 14.174488 | positive | MGI_C57BL6J_6097341 | Gm48027 | ENSEMBL:ENSMUSG00000113775 | MGI:6097341 | lncRNA gene | predicted gene, 48027 |
| 13 | pseudogene | 14.220545 | 14.221938 | negative | MGI_C57BL6J_3780659 | Gm2492 | NCBI_Gene:100039916,ENSEMBL:ENSMUSG00000113563 | MGI:3780659 | pseudogene | predicted gene 2492 |
| 13 | gene | 14.226438 | 14.523279 | negative | MGI_C57BL6J_2444115 | Hecw1 | NCBI_Gene:94253,ENSEMBL:ENSMUSG00000021301 | MGI:2444115 | protein coding gene | HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| 13 | gene | 14.377987 | 14.379573 | negative | MGI_C57BL6J_6095691 | Gm47011 | ENSEMBL:ENSMUSG00000113981 | MGI:6095691 | unclassified gene | predicted gene, 47011 |
| 13 | gene | 14.521486 | 14.525016 | positive | MGI_C57BL6J_5590052 | Gm30893 | NCBI_Gene:102632948,ENSEMBL:ENSMUSG00000113654 | MGI:5590052 | lncRNA gene | predicted gene, 30893 |
| 13 | pseudogene | 14.548554 | 14.549539 | positive | MGI_C57BL6J_5011043 | Gm18858 | NCBI_Gene:100417837 | MGI:5011043 | pseudogene | predicted gene, 18858 |
| 13 | pseudogene | 14.559118 | 14.559309 | positive | MGI_C57BL6J_6097444 | Gm48096 | ENSEMBL:ENSMUSG00000113345 | MGI:6097444 | pseudogene | predicted gene, 48096 |
| 13 | pseudogene | 14.594607 | 14.595687 | positive | MGI_C57BL6J_5591685 | Gm32526 | NCBI_Gene:102635105,ENSEMBL:ENSMUSG00000113627 | MGI:5591685 | pseudogene | predicted gene, 32526 |
| 13 | gene | 14.608184 | 14.613165 | negative | MGI_C57BL6J_2137226 | Mrpl32 | NCBI_Gene:75398,ENSEMBL:ENSMUSG00000015672 | MGI:2137226 | protein coding gene | mitochondrial ribosomal protein L32 |
| 13 | gene | 14.613240 | 14.674236 | positive | MGI_C57BL6J_104885 | Psma2 | NCBI_Gene:19166,ENSEMBL:ENSMUSG00000015671 | MGI:104885 | protein coding gene | proteasome (prosome, macropain) subunit, alpha type 2 |
| 13 | gene | 14.628619 | 14.628731 | negative | MGI_C57BL6J_5455221 | Gm25444 | ENSEMBL:ENSMUSG00000089624 | MGI:5455221 | snRNA gene | predicted gene, 25444 |
| 13 | gene | 14.630154 | 14.639096 | positive | MGI_C57BL6J_2145422 | AW209491 | NCBI_Gene:105351,ENSEMBL:ENSMUSG00000039182 | MGI:2145422 | protein coding gene | expressed sequence AW209491 |
| 13 | pseudogene | 14.862000 | 14.864137 | positive | MGI_C57BL6J_5011374 | Gm19189 | NCBI_Gene:100418406,ENSEMBL:ENSMUSG00000113686 | MGI:5011374 | pseudogene | predicted gene, 19189 |
| 13 | pseudogene | 14.943144 | 14.944695 | positive | MGI_C57BL6J_6097786 | Gm48329 | ENSEMBL:ENSMUSG00000114176 | MGI:6097786 | pseudogene | predicted gene, 48329 |
| 13 | gene | 14.996767 | 15.019558 | negative | MGI_C57BL6J_5590225 | Gm31066 | NCBI_Gene:102633171 | MGI:5590225 | lncRNA gene | predicted gene, 31066 |
| 13 | pseudogene | 15.104233 | 15.119249 | positive | MGI_C57BL6J_3779922 | Gm9512 | NCBI_Gene:670994,ENSEMBL:ENSMUSG00000113389 | MGI:3779922 | pseudogene | predicted gene 9512 |
| 13 | pseudogene | 15.118386 | 15.118631 | negative | MGI_C57BL6J_6097799 | Gm48338 | ENSEMBL:ENSMUSG00000113240 | MGI:6097799 | pseudogene | predicted gene, 48338 |
| 13 | gene | 15.375624 | 15.413100 | positive | MGI_C57BL6J_5012228 | Gm20043 | NCBI_Gene:100504070,ENSEMBL:ENSMUSG00000113410 | MGI:5012228 | lncRNA gene | predicted gene, 20043 |
| 13 | gene | 15.440302 | 15.464657 | negative | MGI_C57BL6J_3647197 | Gm6556 | NCBI_Gene:625121,ENSEMBL:ENSMUSG00000097806 | MGI:3647197 | lncRNA gene | predicted gene 6556 |
| 13 | gene | 15.459980 | 15.461295 | negative | MGI_C57BL6J_5477139 | Gm26645 | ENSEMBL:ENSMUSG00000097684 | MGI:5477139 | lncRNA gene | predicted gene, 26645 |
| 13 | gene | 15.463235 | 15.730026 | positive | MGI_C57BL6J_95729 | Gli3 | NCBI_Gene:14634,ENSEMBL:ENSMUSG00000021318 | MGI:95729 | protein coding gene | GLI-Kruppel family member GLI3 |
| 13 | pseudogene | 15.649935 | 15.650145 | positive | MGI_C57BL6J_6097806 | Gm48343 | ENSEMBL:ENSMUSG00000113191 | MGI:6097806 | pseudogene | predicted gene, 48343 |
| 13 | gene | 15.768990 | 15.770120 | negative | MGI_C57BL6J_6097898 | Gm48408 | ENSEMBL:ENSMUSG00000113309 | MGI:6097898 | unclassified gene | predicted gene, 48408 |
| 13 | gene | 15.781664 | 15.802630 | negative | MGI_C57BL6J_1918321 | 4933412O06Rik | NCBI_Gene:71071,ENSEMBL:ENSMUSG00000113960 | MGI:1918321 | lncRNA gene | RIKEN cDNA 4933412O06 gene |
| 13 | gene | 15.807243 | 15.827537 | positive | MGI_C57BL6J_5439406 | A530046M15Rik | NCBI_Gene:328190,ENSEMBL:ENSMUSG00000113338 | MGI:5439406 | lncRNA gene | RIKEN cDNA A530046M15 gene |
| 13 | pseudogene | 15.813548 | 16.023423 | negative | MGI_C57BL6J_3646895 | B230303A05Rik | NCBI_Gene:100503142,ENSEMBL:ENSMUSG00000113701 | MGI:3646895 | pseudogene | RIKEN cDNA B230303A05 gene |
| 13 | pseudogene | 15.814712 | 15.815429 | negative | MGI_C57BL6J_6117381 | Gm48907 | NCBI_Gene:100503142 | MGI:6117381 | pseudogene | predicted gene, 48907 |
| 13 | gene | 15.817316 | 15.820679 | positive | MGI_C57BL6J_2145514 | A530058O07Rik | ENSEMBL:ENSMUSG00000113290 | MGI:2145514 | unclassified gene | RIKEN cDNA A530058O07 gene |
| 13 | gene | 15.931054 | 15.945339 | positive | MGI_C57BL6J_5590609 | Gm31450 | NCBI_Gene:102633686,ENSEMBL:ENSMUSG00000114057 | MGI:5590609 | lncRNA gene | predicted gene, 31450 |
| 13 | pseudogene | 15.969864 | 15.970410 | negative | MGI_C57BL6J_6118242 | Gm48928 | ENSEMBL:ENSMUSG00000113080 | MGI:6118242 | pseudogene | predicted gene, 48928 |
| 13 | gene | 16.007743 | 16.031621 | positive | MGI_C57BL6J_96570 | Inhba | NCBI_Gene:16323,ENSEMBL:ENSMUSG00000041324 | MGI:96570 | protein coding gene | inhibin beta-A |
| 13 | gene | 16.025382 | 16.032275 | negative | MGI_C57BL6J_6097954 | Gm48444 | ENSEMBL:ENSMUSG00000113224 | MGI:6097954 | lncRNA gene | predicted gene, 48444 |
| 13 | gene | 16.041046 | 16.041152 | negative | MGI_C57BL6J_5452405 | Gm22628 | ENSEMBL:ENSMUSG00000095584 | MGI:5452405 | snRNA gene | predicted gene, 22628 |
| 13 | pseudogene | 16.045640 | 16.046119 | positive | MGI_C57BL6J_3643759 | Gm6562 | NCBI_Gene:625181,ENSEMBL:ENSMUSG00000113715 | MGI:3643759 | pseudogene | predicted gene 6562 |
| 13 | gene | 16.151561 | 16.261349 | negative | MGI_C57BL6J_5590870 | Gm31711 | NCBI_Gene:102634027,ENSEMBL:ENSMUSG00000112962 | MGI:5590870 | lncRNA gene | predicted gene, 31711 |
| 13 | gene | 16.273191 | 16.277838 | negative | MGI_C57BL6J_6098014 | Gm48491 | ENSEMBL:ENSMUSG00000113038 | MGI:6098014 | lncRNA gene | predicted gene, 48491 |
| 13 | gene | 16.368944 | 16.373180 | negative | MGI_C57BL6J_6098016 | Gm48492 | ENSEMBL:ENSMUSG00000113322 | MGI:6098016 | lncRNA gene | predicted gene, 48492 |
| 13 | pseudogene | 16.385919 | 16.389759 | negative | MGI_C57BL6J_6098018 | Gm48493 | ENSEMBL:ENSMUSG00000113924 | MGI:6098018 | pseudogene | predicted gene, 48493 |
| 13 | pseudogene | 16.390591 | 16.390854 | positive | MGI_C57BL6J_6098019 | Gm48494 | ENSEMBL:ENSMUSG00000113232 | MGI:6098019 | pseudogene | predicted gene, 48494 |
| 13 | pseudogene | 16.431525 | 16.431780 | negative | MGI_C57BL6J_6098022 | Gm48496 | ENSEMBL:ENSMUSG00000112969 | MGI:6098022 | pseudogene | predicted gene, 48496 |
| 13 | pseudogene | 16.533539 | 16.533821 | negative | MGI_C57BL6J_6098023 | Gm48497 | ENSEMBL:ENSMUSG00000113972 | MGI:6098023 | pseudogene | predicted gene, 48497 |
| 13 | pseudogene | 16.637881 | 16.639529 | negative | MGI_C57BL6J_3647620 | Gm7537 | NCBI_Gene:665191,ENSEMBL:ENSMUSG00000113862 | MGI:3647620 | pseudogene | predicted gene 7537 |
| 13 | gene | 16.857472 | 17.695553 | negative | MGI_C57BL6J_1923221 | Sugct | NCBI_Gene:192136,ENSEMBL:ENSMUSG00000055137 | MGI:1923221 | protein coding gene | succinyl-CoA glutarate-CoA transferase |
| 13 | gene | 16.859185 | 16.859472 | negative | MGI_C57BL6J_5455875 | Gm26098 | ENSEMBL:ENSMUSG00000087872 | MGI:5455875 | unclassified non-coding RNA gene | predicted gene, 26098 |
| 13 | gene | 16.937609 | 16.947045 | positive | MGI_C57BL6J_6098031 | Gm48503 | ENSEMBL:ENSMUSG00000113851 | MGI:6098031 | lncRNA gene | predicted gene, 48503 |
| 13 | pseudogene | 16.943846 | 16.945569 | positive | MGI_C57BL6J_6098025 | Gm48499 | ENSEMBL:ENSMUSG00000114114 | MGI:6098025 | pseudogene | predicted gene, 48499 |
| 13 | pseudogene | 17.050921 | 17.051878 | positive | MGI_C57BL6J_5504158 | Gm27043 | ENSEMBL:ENSMUSG00000098218 | MGI:5504158 | pseudogene | predicted gene, 27043 |
| 13 | gene | 17.206021 | 17.206127 | positive | MGI_C57BL6J_5454797 | Gm25020 | ENSEMBL:ENSMUSG00000094754 | MGI:5454797 | snRNA gene | predicted gene, 25020 |
| 13 | gene | 17.258771 | 17.261641 | positive | MGI_C57BL6J_6098029 | Gm48502 | ENSEMBL:ENSMUSG00000113024 | MGI:6098029 | unclassified gene | predicted gene, 48502 |
| 13 | pseudogene | 17.337557 | 17.338356 | positive | MGI_C57BL6J_5010075 | Gm17890 | NCBI_Gene:100416045,ENSEMBL:ENSMUSG00000113219 | MGI:5010075 | pseudogene | predicted gene, 17890 |
| 13 | pseudogene | 17.360294 | 17.364134 | positive | MGI_C57BL6J_6098028 | Gm48501 | ENSEMBL:ENSMUSG00000113889 | MGI:6098028 | pseudogene | predicted gene, 48501 |
| 13 | gene | 17.440790 | 17.440896 | positive | MGI_C57BL6J_5454650 | Gm24873 | ENSEMBL:ENSMUSG00000094665 | MGI:5454650 | snRNA gene | predicted gene, 24873 |
| 13 | gene | 17.682710 | 17.685778 | negative | MGI_C57BL6J_5623711 | Gm40826 | NCBI_Gene:105245356 | MGI:5623711 | lncRNA gene | predicted gene, 40826 |
| 13 | gene | 17.695192 | 17.699748 | positive | MGI_C57BL6J_1913558 | Mplkip | NCBI_Gene:66308,ENSEMBL:ENSMUSG00000012429 | MGI:1913558 | protein coding gene | M-phase specific PLK1 intereacting protein |
| 13 | gene | 17.708603 | 17.709864 | negative | MGI_C57BL6J_6098216 | Gm48621 | ENSEMBL:ENSMUSG00000113045 | MGI:6098216 | unclassified gene | predicted gene, 48621 |
| 13 | gene | 17.710315 | 17.805097 | negative | MGI_C57BL6J_1916812 | Cdk13 | NCBI_Gene:69562,ENSEMBL:ENSMUSG00000041297 | MGI:1916812 | protein coding gene | cyclin-dependent kinase 13 |
| 13 | gene | 17.735985 | 17.737054 | negative | MGI_C57BL6J_1918744 | 8430406P12Rik | ENSEMBL:ENSMUSG00000113432 | MGI:1918744 | unclassified gene | RIKEN cDNA 8430406P12 gene |
| 13 | gene | 17.747473 | 17.747593 | positive | MGI_C57BL6J_3783375 | Mir466i | miRBase:MI0006282,NCBI_Gene:100316665,ENSEMBL:ENSMUSG00000080574 | MGI:3783375 | miRNA gene | microRNA 466i |
| 13 | gene | 17.754516 | 17.757325 | negative | MGI_C57BL6J_6098222 | Gm48624 | ENSEMBL:ENSMUSG00000113140 | MGI:6098222 | unclassified gene | predicted gene, 48624 |
| 13 | gene | 17.809591 | 17.809697 | negative | MGI_C57BL6J_5453795 | Gm24018 | ENSEMBL:ENSMUSG00000095025 | MGI:5453795 | snRNA gene | predicted gene, 24018 |
| 13 | pseudogene | 17.820922 | 17.821774 | positive | MGI_C57BL6J_5011044 | Gm18859 | NCBI_Gene:100417838,ENSEMBL:ENSMUSG00000081285 | MGI:5011044 | pseudogene | predicted gene, 18859 |
| 13 | gene | 17.880571 | 17.944239 | negative | MGI_C57BL6J_1927243 | Rala | NCBI_Gene:56044,ENSEMBL:ENSMUSG00000008859 | MGI:1927243 | protein coding gene | v-ral simian leukemia viral oncogene A (ras related) |
| 13 | pseudogene | 17.894855 | 17.895383 | positive | MGI_C57BL6J_5010547 | Gm18362 | NCBI_Gene:100417014,ENSEMBL:ENSMUSG00000114177 | MGI:5010547 | pseudogene | predicted gene, 18362 |
| 13 | gene | 17.932386 | 17.938434 | positive | MGI_C57BL6J_6098432 | Gm48754 | ENSEMBL:ENSMUSG00000113100 | MGI:6098432 | lncRNA gene | predicted gene, 48754 |
| 13 | gene | 17.944291 | 17.946278 | positive | MGI_C57BL6J_5591858 | Gm32699 | ENSEMBL:ENSMUSG00000113528 | MGI:5591858 | protein coding gene | predicted gene, 32699 |
| 13 | pseudogene | 17.949901 | 17.951106 | positive | MGI_C57BL6J_3779506 | Gm5628 | NCBI_Gene:434585,ENSEMBL:ENSMUSG00000113750 | MGI:3779506 | pseudogene | predicted gene 5628 |
| 13 | gene | 17.951938 | 17.954912 | positive | MGI_C57BL6J_3026956 | A930027P06Rik | ENSEMBL:ENSMUSG00000113688 | MGI:3026956 | unclassified gene | RIKEN cDNA A930027P06 gene |
| 13 | pseudogene | 17.960852 | 17.961416 | negative | MGI_C57BL6J_6098471 | Gm48776 | ENSEMBL:ENSMUSG00000114077 | MGI:6098471 | pseudogene | predicted gene, 48776 |
| 13 | gene | 17.961150 | 17.992533 | negative | MGI_C57BL6J_6098473 | Gm48778 | ENSEMBL:ENSMUSG00000113030 | MGI:6098473 | lncRNA gene | predicted gene, 48778 |
| 13 | gene | 17.981330 | 17.993351 | negative | MGI_C57BL6J_1914258 | Yae1d1 | NCBI_Gene:67008,ENSEMBL:ENSMUSG00000075054 | MGI:1914258 | protein coding gene | Yae1 domain containing 1 |
| 13 | gene | 18.002310 | 18.008344 | negative | MGI_C57BL6J_6098505 | Gm48799 | ENSEMBL:ENSMUSG00000113847 | MGI:6098505 | lncRNA gene | predicted gene, 48799 |
| 13 | pseudogene | 18.030317 | 18.031629 | negative | MGI_C57BL6J_1270159 | Vdac3-ps1 | NCBI_Gene:22336,ENSEMBL:ENSMUSG00000075053 | MGI:1270159 | pseudogene | voltage-dependent anion channel 3, pseudogene 1 |
| 13 | pseudogene | 18.038561 | 18.039456 | positive | MGI_C57BL6J_3646645 | Gm6575 | NCBI_Gene:625310,ENSEMBL:ENSMUSG00000113496 | MGI:3646645 | pseudogene | predicted gene 6575 |
| 13 | gene | 18.084947 | 18.100202 | negative | MGI_C57BL6J_1921242 | 4930448F12Rik | NCBI_Gene:73992,ENSEMBL:ENSMUSG00000113470 | MGI:1921242 | lncRNA gene | RIKEN cDNA 4930448F12 gene |
| 13 | pseudogene | 18.094472 | 18.095954 | negative | MGI_C57BL6J_6098507 | Gm48800 | ENSEMBL:ENSMUSG00000113893 | MGI:6098507 | pseudogene | predicted gene, 48800 |
| 13 | gene | 18.121098 | 18.397686 | negative | MGI_C57BL6J_2443631 | Pou6f2 | NCBI_Gene:218030,ENSEMBL:ENSMUSG00000009734 | MGI:2443631 | protein coding gene | POU domain, class 6, transcription factor 2 |
| 13 | gene | 18.281144 | 18.284357 | positive | MGI_C57BL6J_5590991 | Gm31832 | NCBI_Gene:102634189 | MGI:5590991 | lncRNA gene | predicted gene, 31832 |
| 13 | gene | 18.340269 | 18.346639 | negative | MGI_C57BL6J_5826039 | Gm46402 | NCBI_Gene:108168066 | MGI:5826039 | lncRNA gene | predicted gene, 46402 |
| 13 | gene | 18.640257 | 18.645349 | negative | MGI_C57BL6J_6098551 | Gm48829 | ENSEMBL:ENSMUSG00000114118 | MGI:6098551 | lncRNA gene | predicted gene, 48829 |
| 13 | gene | 18.717286 | 18.866811 | positive | MGI_C57BL6J_1929215 | Vps41 | NCBI_Gene:218035,ENSEMBL:ENSMUSG00000041236 | MGI:1929215 | protein coding gene | VPS41 HOPS complex subunit |
| 13 | gene | 18.922577 | 18.922710 | positive | MGI_C57BL6J_5455949 | Gm26172 | ENSEMBL:ENSMUSG00000089046 | MGI:5455949 | snoRNA gene | predicted gene, 26172 |
| 13 | gene | 18.948137 | 19.154032 | positive | MGI_C57BL6J_103574 | Amph | NCBI_Gene:218038,ENSEMBL:ENSMUSG00000021314 | MGI:103574 | protein coding gene | amphiphysin |
| 13 | gene | 18.971702 | 18.974997 | negative | MGI_C57BL6J_6096744 | Gm47658 | ENSEMBL:ENSMUSG00000113576 | MGI:6096744 | lncRNA gene | predicted gene, 47658 |
| 13 | pseudogene | 18.973460 | 18.974141 | positive | MGI_C57BL6J_6096746 | Gm47659 | ENSEMBL:ENSMUSG00000113993 | MGI:6096746 | pseudogene | predicted gene, 47659 |
| 13 | pseudogene | 18.983852 | 18.984455 | positive | MGI_C57BL6J_5010250 | Gm18065 | NCBI_Gene:100416354,ENSEMBL:ENSMUSG00000113028 | MGI:5010250 | pseudogene | predicted gene, 18065 |
| 13 | gene | 19.029940 | 19.057896 | negative | MGI_C57BL6J_6096748 | Gm47661 | ENSEMBL:ENSMUSG00000114080 | MGI:6096748 | lncRNA gene | predicted gene, 47661 |
| 13 | pseudogene | 19.071197 | 19.071575 | negative | MGI_C57BL6J_6096747 | Gm47660 | ENSEMBL:ENSMUSG00000113378 | MGI:6096747 | pseudogene | predicted gene, 47660 |
| 13 | gene | 19.145775 | 19.193711 | negative | MGI_C57BL6J_5591046 | Gm31887 | NCBI_Gene:102634258,ENSEMBL:ENSMUSG00000105715 | MGI:5591046 | lncRNA gene | predicted gene, 31887 |
| 13 | gene | 19.178024 | 19.178492 | positive | MGI_C57BL6J_98637 | Trgv7 | NCBI_Gene:21641,ENSEMBL:ENSMUSG00000076744 | MGI:98637 | gene segment | T cell receptor gamma, variable 7 |
| 13 | gene | 19.184974 | 19.185506 | positive | MGI_C57BL6J_98634 | Trgv4 | NCBI_Gene:21638,ENSEMBL:ENSMUSG00000076745 | MGI:98634 | gene segment | T cell receptor gamma, variable 4 |
| 13 | gene | 19.190356 | 19.190929 | positive | MGI_C57BL6J_98636 | Trgv6 | NCBI_Gene:21640,ENSEMBL:ENSMUSG00000076746 | MGI:98636 | gene segment | T cell receptor gamma, variable 6 |
| 13 | gene | 19.192278 | 19.192725 | positive | MGI_C57BL6J_98635 | Trgv5 | NCBI_Gene:21639,ENSEMBL:ENSMUSG00000076747 | MGI:98635 | gene segment | T cell receptor gamma, variable 5 |
| 13 | gene | 19.210343 | 19.210402 | positive | MGI_C57BL6J_4440480 | Trgj1 | NCBI_Gene:100126431,ENSEMBL:ENSMUSG00000104962 | MGI:4440480 | gene segment | T cell receptor gamma joining 1 |
| 13 | gene | 19.214103 | 19.217024 | positive | MGI_C57BL6J_98625 | Trgc1 | NCBI_Gene:107573,ENSEMBL:ENSMUSG00000076749 | MGI:98625 | gene segment | T cell receptor gamma, constant 1 |
| 13 | gene | 19.242845 | 19.243301 | positive | MGI_C57BL6J_98633 | Trgv3 | NCBI_Gene:21637,ENSEMBL:ENSMUSG00000076750 | MGI:98633 | gene segment | T cell receptor gamma, variable 3 |
| 13 | gene | 19.257117 | 19.257176 | positive | MGI_C57BL6J_4439528 | Trgj3 | NCBI_Gene:100126432,ENSEMBL:ENSMUSG00000104630 | MGI:4439528 | gene segment | T cell receptor gamma joining 3 |
| 13 | gene | 19.260884 | 19.263528 | positive | MGI_C57BL6J_98627 | Trgc3 | NCBI_Gene:107575,ENSEMBL:ENSMUSG00000091682 | MGI:98627 | gene segment | T cell receptor gamma, constant 3 |
| 13 | gene | 19.304649 | 19.307551 | negative | MGI_C57BL6J_98626 | Trgc2 | NCBI_Gene:107574,ENSEMBL:ENSMUSG00000076752 | MGI:98626 | gene segment | T cell receptor gamma, constant 2 |
| 13 | gene | 19.311245 | 19.311304 | negative | MGI_C57BL6J_4439598 | Trgj2 | NCBI_Gene:100126433,ENSEMBL:ENSMUSG00000105830 | MGI:4439598 | gene segment | T cell receptor gamma joining 2 |
| 13 | gene | 19.336573 | 19.337167 | negative | MGI_C57BL6J_98632 | Trgv2 | NCBI_Gene:21636,ENSEMBL:ENSMUSG00000076754 | MGI:98632 | gene segment | T cell receptor gamma variable 2 |
| 13 | gene | 19.339989 | 19.340454 | positive | MGI_C57BL6J_98631 | Trgv1 | NCBI_Gene:21632,ENSEMBL:ENSMUSG00000076755 | MGI:98631 | gene segment | T cell receptor gamma, variable 1 |
| 13 | gene | 19.342152 | 19.342212 | positive | MGI_C57BL6J_4439597 | Trgj4 | NCBI_Gene:100126434,ENSEMBL:ENSMUSG00000098799 | MGI:4439597 | gene segment | T cell receptor gamma joining 4 |
| 13 | gene | 19.344558 | 19.352756 | positive | MGI_C57BL6J_98628 | Trgc4 | NCBI_Gene:107576,ENSEMBL:ENSMUSG00000076757 | MGI:98628 | gene segment | T cell receptor gamma, constant 4 |
| 13 | gene | 19.357676 | 19.395813 | negative | MGI_C57BL6J_1923455 | Stard3nl | NCBI_Gene:76205,ENSEMBL:ENSMUSG00000003062 | MGI:1923455 | protein coding gene | STARD3 N-terminal like |
| 13 | gene | 19.393126 | 19.395453 | negative | MGI_C57BL6J_5662820 | Gm42683 | ENSEMBL:ENSMUSG00000105848 | MGI:5662820 | unclassified gene | predicted gene 42683 |
| 13 | gene | 19.403876 | 19.425573 | positive | MGI_C57BL6J_5623712 | Gm40827 | NCBI_Gene:105245357 | MGI:5623712 | lncRNA gene | predicted gene, 40827 |
| 13 | pseudogene | 19.412800 | 19.413324 | positive | MGI_C57BL6J_5010316 | Gm18131 | NCBI_Gene:100416502,ENSEMBL:ENSMUSG00000104959 | MGI:5010316 | pseudogene | predicted gene, 18131 |
| 13 | gene | 19.462847 | 19.504367 | negative | MGI_C57BL6J_2442084 | 9330199G10Rik | NCBI_Gene:319466,ENSEMBL:ENSMUSG00000114189 | MGI:2442084 | lncRNA gene | RIKEN cDNA 9330199G10 gene |
| 13 | pseudogene | 19.525986 | 19.526716 | positive | MGI_C57BL6J_5010548 | Gm18363 | NCBI_Gene:100417015,ENSEMBL:ENSMUSG00000113251 | MGI:5010548 | pseudogene | predicted gene, 18363 |
| 13 | gene | 19.591706 | 19.619950 | negative | MGI_C57BL6J_2145369 | Epdr1 | NCBI_Gene:105298,ENSEMBL:ENSMUSG00000002808 | MGI:2145369 | protein coding gene | ependymin related protein 1 (zebrafish) |
| 13 | gene | 19.623104 | 19.632994 | positive | MGI_C57BL6J_892010 | Sfrp4 | NCBI_Gene:20379,ENSEMBL:ENSMUSG00000021319 | MGI:892010 | protein coding gene | secreted frizzled-related protein 4 |
| 13 | gene | 19.645061 | 19.697827 | negative | MGI_C57BL6J_1920662 | Nme8 | NCBI_Gene:73412,ENSEMBL:ENSMUSG00000041138 | MGI:1920662 | protein coding gene | NME/NM23 family member 8 |
| 13 | gene | 19.672382 | 19.673934 | negative | MGI_C57BL6J_6096663 | Gm47606 | ENSEMBL:ENSMUSG00000114009 | MGI:6096663 | unclassified gene | predicted gene, 47606 |
| 13 | gene | 19.727417 | 19.732955 | negative | MGI_C57BL6J_2441809 | Gpr141b | NCBI_Gene:319293,ENSEMBL:ENSMUSG00000047462 | MGI:2441809 | protein coding gene | G protein-coupled receptor 141B |
| 13 | gene | 19.749680 | 19.824336 | negative | MGI_C57BL6J_2672983 | Gpr141 | NCBI_Gene:353346,ENSEMBL:ENSMUSG00000053101 | MGI:2672983 | protein coding gene | G protein-coupled receptor 141 |
| 13 | pseudogene | 19.820739 | 19.821678 | positive | MGI_C57BL6J_5011339 | Gm19154 | NCBI_Gene:100418349,ENSEMBL:ENSMUSG00000113542 | MGI:5011339 | pseudogene | predicted gene, 19154 |
| 13 | pseudogene | 19.993651 | 19.995814 | negative | MGI_C57BL6J_3644854 | Gm5446 | NCBI_Gene:432730,ENSEMBL:ENSMUSG00000050243 | MGI:3644854 | pseudogene | predicted gene 5446 |
| 13 | gene | 20.090507 | 20.608353 | positive | MGI_C57BL6J_2153044 | Elmo1 | NCBI_Gene:140580,ENSEMBL:ENSMUSG00000041112 | MGI:2153044 | protein coding gene | engulfment and cell motility 1 |
| 13 | gene | 20.271905 | 20.274039 | positive | MGI_C57BL6J_6096756 | Gm47665 | ENSEMBL:ENSMUSG00000113649 | MGI:6096756 | unclassified gene | predicted gene, 47665 |
| 13 | gene | 20.445286 | 20.473490 | negative | MGI_C57BL6J_5591195 | Gm32036 | ENSEMBL:ENSMUSG00000113313 | MGI:5591195 | lncRNA gene | predicted gene, 32036 |
| 13 | gene | 20.499888 | 20.501227 | positive | MGI_C57BL6J_6096754 | Gm47664 | ENSEMBL:ENSMUSG00000113183 | MGI:6096754 | lncRNA gene | predicted gene, 47664 |
| 13 | gene | 20.504795 | 20.506598 | positive | MGI_C57BL6J_6096742 | Gm47657 | ENSEMBL:ENSMUSG00000113557 | MGI:6096742 | unclassified gene | predicted gene, 47657 |
| 13 | pseudogene | 20.645757 | 20.647164 | positive | MGI_C57BL6J_6096851 | Gm47721 | ENSEMBL:ENSMUSG00000113130 | MGI:6096851 | pseudogene | predicted gene, 47721 |
| 13 | pseudogene | 20.704383 | 20.705382 | negative | MGI_C57BL6J_3647727 | Gm7611 | ENSEMBL:ENSMUSG00000097951 | MGI:3647727 | pseudogene | predicted gene 7611 |
| 13 | pseudogene | 20.748465 | 20.748836 | positive | MGI_C57BL6J_3647732 | Gm7614 | NCBI_Gene:665386,ENSEMBL:ENSMUSG00000113168 | MGI:3647732 | pseudogene | predicted gene 7614 |
| 13 | gene | 20.794103 | 21.036617 | positive | MGI_C57BL6J_1350928 | Aoah | NCBI_Gene:27052,ENSEMBL:ENSMUSG00000021322 | MGI:1350928 | protein coding gene | acyloxyacyl hydrolase |
| 13 | pseudogene | 20.827836 | 20.828316 | positive | MGI_C57BL6J_6096859 | Gm47727 | ENSEMBL:ENSMUSG00000114070 | MGI:6096859 | pseudogene | predicted gene, 47727 |
| 13 | gene | 20.844754 | 20.844877 | positive | MGI_C57BL6J_5455382 | Gm25605 | ENSEMBL:ENSMUSG00000088989 | MGI:5455382 | snRNA gene | predicted gene, 25605 |
| 13 | gene | 20.936803 | 21.010596 | negative | MGI_C57BL6J_5591401 | Gm32242 | NCBI_Gene:102634724 | MGI:5591401 | lncRNA gene | predicted gene, 32242 |
| 13 | gene | 21.069334 | 21.074947 | negative | MGI_C57BL6J_3031204 | Olfr1370 | NCBI_Gene:258528,ENSEMBL:ENSMUSG00000042869 | MGI:3031204 | protein coding gene | olfactory receptor 1370 |
| 13 | pseudogene | 21.111483 | 21.119095 | positive | MGI_C57BL6J_3031203 | Olfr1369-ps1 | NCBI_Gene:404504,ENSEMBL:ENSMUSG00000060404 | MGI:3031203 | pseudogene | olfactory receptor 1369, pseudogene 1 |
| 13 | gene | 21.129639 | 21.136296 | positive | MGI_C57BL6J_3030097 | Olfr263 | NCBI_Gene:18341,ENSEMBL:ENSMUSG00000071522 | MGI:3030097 | protein coding gene | olfactory receptor 263 |
| 13 | gene | 21.141882 | 21.145867 | negative | MGI_C57BL6J_3031202 | Olfr1368 | NCBI_Gene:258527,ENSEMBL:ENSMUSG00000045474 | MGI:3031202 | protein coding gene | olfactory receptor 1368 |
| 13 | gene | 21.160785 | 21.160856 | negative | MGI_C57BL6J_4413851 | n-TEctc11 | NCBI_Gene:102467521 | MGI:4413851 | tRNA gene | nuclear encoded tRNA glutamic acid 11 (anticodon CTC) |
| 13 | gene | 21.161405 | 21.161477 | positive | MGI_C57BL6J_4413999 | n-TFgaa4 | NCBI_Gene:102467362 | MGI:4413999 | tRNA gene | nuclear encoded tRNA pheylalanine 4 (anticodon GAA) |
| 13 | gene | 21.166337 | 21.170774 | positive | MGI_C57BL6J_5591457 | Gm32298 | NCBI_Gene:102634798 | MGI:5591457 | lncRNA gene | predicted gene, 32298 |
| 13 | gene | 21.167604 | 21.167676 | positive | MGI_C57BL6J_4413991 | n-TMcat14 | NCBI_Gene:102467359 | MGI:4413991 | tRNA gene | nuclear encoded tRNA methionine 14 (anticodon CAT) |
| 13 | gene | 21.168075 | 21.168147 | negative | MGI_C57BL6J_4413975 | n-TKctt28 | NCBI_Gene:102467352 | MGI:4413975 | tRNA gene | nuclear encoded tRNA lysine 28 (anticodon TTT) |
| 13 | gene | 21.169687 | 21.169759 | negative | MGI_C57BL6J_4413992 | n-TMcat15 | NCBI_Gene:102467652 | MGI:4413992 | tRNA gene | nuclear encoded tRNA methionine 15 (anticodon CAT) |
| 13 | gene | 21.170036 | 21.170117 | positive | MGI_C57BL6J_4413925 | n-TLaag7 | NCBI_Gene:102467331 | MGI:4413925 | tRNA gene | nuclear encoded tRNA leucine 7 (anticodon AAG) |
| 13 | gene | 21.170206 | 21.170287 | positive | MGI_C57BL6J_4413923 | n-TLaag5 | NCBI_Gene:102467550 | MGI:4413923 | tRNA gene | nuclear encoded tRNA leucine 5 (anticodon AAG) |
| 13 | gene | 21.172351 | 21.172422 | positive | MGI_C57BL6J_4413831 | n-TQctg7 | NCBI_Gene:102467293 | MGI:4413831 | tRNA gene | nuclear encoded tRNA glutamine 7 (anticodon CTG) |
| 13 | gene | 21.172837 | 21.172942 | negative | MGI_C57BL6J_4413927 | n-TLcaa4 | NCBI_Gene:102467332 | MGI:4413927 | tRNA gene | nuclear encoded tRNA leucine 4 (anticodon CAA) |
| 13 | gene | 21.177187 | 21.187556 | negative | MGI_C57BL6J_6096988 | Gm47806 | ENSEMBL:ENSMUSG00000113104 | MGI:6096988 | lncRNA gene | predicted gene, 47806 |
| 13 | gene | 21.179445 | 21.194724 | positive | MGI_C57BL6J_97904 | Trim27 | NCBI_Gene:19720,ENSEMBL:ENSMUSG00000021326 | MGI:97904 | protein coding gene | tripartite motif-containing 27 |
| 13 | gene | 21.200204 | 21.200308 | positive | MGI_C57BL6J_4413928 | n-TLcaa1 | NCBI_Gene:102467552 | MGI:4413928 | tRNA gene | nuclear encoded tRNA leucine 1 (anticodon CAA) |
| 13 | gene | 21.214480 | 21.223827 | negative | MGI_C57BL6J_5591579 | Gm32420 | NCBI_Gene:102634966 | MGI:5591579 | lncRNA gene | predicted gene, 32420 |
| 13 | gene | 21.215458 | 21.215530 | negative | MGI_C57BL6J_4413722 | n-TRccg3 | NCBI_Gene:102467467 | MGI:4413722 | tRNA gene | nuclear encoded tRNA arginine 3 (anticodon CCG) |
| 13 | gene | 21.222586 | 21.222657 | positive | MGI_C57BL6J_4413689 | n-TAagc16 | NCBI_Gene:102467419 | MGI:4413689 | tRNA gene | nuclear encoded tRNA alanine 16 (anticodon AGC) |
| 13 | gene | 21.233557 | 21.254198 | negative | MGI_C57BL6J_5591684 | Gm32525 | NCBI_Gene:102635104 | MGI:5591684 | lncRNA gene | predicted gene, 32525 |
| 13 | gene | 21.234045 | 21.234116 | positive | MGI_C57BL6J_4413690 | n-TAagc17 | NCBI_Gene:102467234 | MGI:4413690 | tRNA gene | nuclear encoded tRNA alanine 17 (anticodon AGC) |
| 13 | gene | 21.240945 | 21.241016 | positive | MGI_C57BL6J_4413713 | n-TAtgc11 | NCBI_Gene:102467244 | MGI:4413713 | tRNA gene | nuclear encoded tRNA alanine 11 (anticodon TGC) |
| 13 | gene | 21.242570 | 21.242641 | positive | MGI_C57BL6J_4413691 | n-TAagc18 | NCBI_Gene:102467454 | MGI:4413691 | tRNA gene | nuclear encoded tRNA alanine 18 (anticodon AGC) |
| 13 | gene | 21.249194 | 21.249267 | negative | MGI_C57BL6J_4414050 | n-TTagt3 | NCBI_Gene:102467602 | MGI:4414050 | tRNA gene | nuclear encoded tRNA threonine 3 (anticodon AGT) |
| 13 | gene | 21.254220 | 21.254291 | positive | MGI_C57BL6J_4413701 | n-TAcgc9 | NCBI_Gene:102467458 | MGI:4413701 | tRNA gene | nuclear encoded tRNA alanine 9 (anticodon CGC) |
| 13 | gene | 21.259768 | 21.259839 | positive | MGI_C57BL6J_4413692 | n-TAagc19 | NCBI_Gene:102467444 | MGI:4413692 | tRNA gene | nuclear encoded tRNA alanine 19 (anticodon AGC) |
| 13 | gene | 21.264580 | 21.264653 | positive | MGI_C57BL6J_4414058 | n-TTcgt2 | NCBI_Gene:102467418 | MGI:4414058 | tRNA gene | nuclear encoded tRNA threonine 2 (anticodon CGT) |
| 13 | gene | 21.267643 | 21.267764 | negative | MGI_C57BL6J_1100841 | Targ1 | NA | NA | protein coding gene | transforming growth factor alpha regulated gene 1 |
| 13 | gene | 21.268469 | 21.268540 | negative | MGI_C57BL6J_4413837 | n-TQttg3 | NCBI_Gene:102467515 | MGI:4413837 | tRNA gene | nuclear encoded tRNA glutamine 3 (anticodon TTG) |
| 13 | gene | 21.275956 | 21.276029 | positive | MGI_C57BL6J_4414065 | n-TTtgt4 | NCBI_Gene:102467608 | MGI:4414065 | tRNA gene | nuclear encoded tRNA threonine 4 (anticodon TGT) |
| 13 | gene | 21.279696 | 21.279768 | positive | MGI_C57BL6J_4413729 | n-TRtcg2 | NCBI_Gene:102467470 | MGI:4413729 | tRNA gene | nuclear encoded tRNA arginine 2 (anticodon TCG) |
| 13 | gene | 21.286429 | 21.292731 | negative | MGI_C57BL6J_104886 | Gpx5 | NCBI_Gene:14780,ENSEMBL:ENSMUSG00000004344 | MGI:104886 | protein coding gene | glutathione peroxidase 5 |
| 13 | gene | 21.295465 | 21.319625 | positive | MGI_C57BL6J_1922762 | Gpx6 | NCBI_Gene:75512,ENSEMBL:ENSMUSG00000004341 | MGI:1922762 | protein coding gene | glutathione peroxidase 6 |
| 13 | pseudogene | 21.296138 | 21.296752 | negative | MGI_C57BL6J_3649599 | Gm11270 | NCBI_Gene:625752,ENSEMBL:ENSMUSG00000084414 | MGI:3649599 | pseudogene | predicted gene 11270 |
| 13 | gene | 21.322813 | 21.322886 | positive | MGI_C57BL6J_4414060 | n-TTcgt4 | NCBI_Gene:102467606 | MGI:4414060 | tRNA gene | nuclear encoded tRNA threonine 4 (anticodon CGT) |
| 13 | gene | 21.326717 | 21.326789 | negative | MGI_C57BL6J_4413990 | n-TMcat13 | NCBI_Gene:102467578 | MGI:4413990 | tRNA gene | nuclear encoded tRNA methionine 13 (anticodon CAT) |
| 13 | gene | 21.346910 | 21.347964 | positive | MGI_C57BL6J_3031201 | Olfr1367 | NCBI_Gene:258526,ENSEMBL:ENSMUSG00000045508 | MGI:3031201 | protein coding gene | olfactory receptor 1367 |
| 13 | gene | 21.362820 | 21.372302 | positive | MGI_C57BL6J_1099444 | Zscan12 | NCBI_Gene:22758,ENSEMBL:ENSMUSG00000036721 | MGI:1099444 | protein coding gene | zinc finger and SCAN domain containing 12 |
| 13 | pseudogene | 21.380691 | 21.381130 | negative | MGI_C57BL6J_3652214 | Gm11271 | ENSEMBL:ENSMUSG00000081191 | MGI:3652214 | pseudogene | predicted gene 11271 |
| 13 | gene | 21.387003 | 21.402755 | negative | MGI_C57BL6J_1919989 | Zkscan3 | NCBI_Gene:72739,ENSEMBL:ENSMUSG00000021327 | MGI:1919989 | protein coding gene | zinc finger with KRAB and SCAN domains 3 |
| 13 | gene | 21.396578 | 21.396638 | negative | MGI_C57BL6J_5530669 | Mir6942 | miRBase:MI0022789,NCBI_Gene:102465568,ENSEMBL:ENSMUSG00000098790 | MGI:5530669 | miRNA gene | microRNA 6942 |
| 13 | pseudogene | 21.412051 | 21.417906 | positive | MGI_C57BL6J_6324754 | Gm50481 | ENSEMBL:ENSMUSG00000118467 | MGI:6324754 | pseudogene | predicted gene, 50481 |
| 13 | gene | 21.421275 | 21.441058 | negative | MGI_C57BL6J_2441675 | Pgbd1 | NCBI_Gene:319207,ENSEMBL:ENSMUSG00000055313 | MGI:2441675 | protein coding gene | piggyBac transposable element derived 1 |
| 13 | gene | 21.440471 | 21.445934 | positive | MGI_C57BL6J_1922171 | 4930470G03Rik | NCBI_Gene:105245359,ENSEMBL:ENSMUSG00000085788 | MGI:1922171 | lncRNA gene | RIKEN cDNA 4930470G03 gene |
| 13 | gene | 21.442174 | 21.453765 | negative | MGI_C57BL6J_3531417 | Zscan26 | NCBI_Gene:432731,ENSEMBL:ENSMUSG00000022228 | MGI:3531417 | protein coding gene | zinc finger and SCAN domain containing 26 |
| 13 | gene | 21.445160 | 21.445240 | positive | MGI_C57BL6J_3811418 | Mir1896 | miRBase:MI0008322,NCBI_Gene:100316752,ENSEMBL:ENSMUSG00000084590 | MGI:3811418 | miRNA gene | microRNA 1896 |
| 13 | gene | 21.467047 | 21.468509 | negative | MGI_C57BL6J_1913957 | Nkapl | NCBI_Gene:66707,ENSEMBL:ENSMUSG00000059395 | MGI:1913957 | protein coding gene | NFKB activating protein-like |
| 13 | gene | 21.477615 | 21.485049 | negative | MGI_C57BL6J_5623713 | Gm40828 | NCBI_Gene:105245360,ENSEMBL:ENSMUSG00000114702 | MGI:5623713 | lncRNA gene | predicted gene, 40828 |
| 13 | gene | 21.477623 | 21.479443 | negative | MGI_C57BL6J_3642433 | Gm10065 | NA | NA | protein coding gene | predicted gene 10065 |
| 13 | gene | 21.478849 | 21.485507 | positive | MGI_C57BL6J_3649412 | Zkscan4 | NCBI_Gene:544922,ENSEMBL:ENSMUSG00000054931 | MGI:3649412 | protein coding gene | zinc finger with KRAB and SCAN domains 4 |
| 13 | gene | 21.486338 | 21.486401 | negative | MGI_C57BL6J_5453556 | Gm23779 | ENSEMBL:ENSMUSG00000088563 | MGI:5453556 | snRNA gene | predicted gene, 23779 |
| 13 | gene | 21.492845 | 21.492926 | negative | MGI_C57BL6J_4414036 | n-TSgct2 | NCBI_Gene:102467597 | MGI:4414036 | tRNA gene | nuclear encoded tRNA serine 2 (anticodon GCT) |
| 13 | gene | 21.495223 | 21.497146 | positive | MGI_C57BL6J_3574096 | AK157302 | NCBI_Gene:432732,ENSEMBL:ENSMUSG00000078139 | MGI:3574096 | protein coding gene | cDNA sequence AK157302 |
| 13 | gene | 21.500935 | 21.501452 | negative | MGI_C57BL6J_3649411 | Gm11273 | NCBI_Gene:100040340,ENSEMBL:ENSMUSG00000079941 | MGI:3649411 | protein coding gene | predicted gene 11273 |
| 13 | pseudogene | 21.504313 | 21.506533 | negative | MGI_C57BL6J_3641940 | Zfp389 | NCBI_Gene:100038371,ENSEMBL:ENSMUSG00000082396 | MGI:3641940 | pseudogene | zinc finger protein 389 |
| 13 | gene | 21.513221 | 21.531120 | negative | MGI_C57BL6J_1913815 | Zkscan8 | NCBI_Gene:93681,ENSEMBL:ENSMUSG00000063894 | MGI:1913815 | protein coding gene | zinc finger with KRAB and SCAN domains 8 |
| 13 | gene | 21.531069 | 21.558701 | positive | MGI_C57BL6J_5591883 | Gm32724 | NCBI_Gene:102635366 | MGI:5591883 | lncRNA gene | predicted gene, 32724 |
| 13 | gene | 21.535113 | 21.542808 | negative | MGI_C57BL6J_3031200 | Olfr1366 | NCBI_Gene:258280,ENSEMBL:ENSMUSG00000048996 | MGI:3031200 | protein coding gene | olfactory receptor 1366 |
| 13 | gene | 21.553894 | 21.559893 | negative | MGI_C57BL6J_3031369 | Olfr1535 | NCBI_Gene:404335,ENSEMBL:ENSMUSG00000054890 | MGI:3031369 | protein coding gene | olfactory receptor 1535 |
| 13 | gene | 21.571813 | 21.581759 | negative | MGI_C57BL6J_3031198 | Olfr1364 | NCBI_Gene:258533,ENSEMBL:ENSMUSG00000046016 | MGI:3031198 | protein coding gene | olfactory receptor 1364 |
| 13 | gene | 21.610978 | 21.614022 | negative | MGI_C57BL6J_3031196 | Olfr1362 | NCBI_Gene:258739,ENSEMBL:ENSMUSG00000051258 | MGI:3031196 | protein coding gene | olfactory receptor 1362 |
| 13 | gene | 21.636623 | 21.642518 | negative | MGI_C57BL6J_104715 | Olfr11 | NCBI_Gene:218066,ENSEMBL:ENSMUSG00000036658 | MGI:104715 | protein coding gene | olfactory receptor 11 |
| 13 | gene | 21.656880 | 21.664523 | negative | MGI_C57BL6J_3031195 | Olfr1361 | NCBI_Gene:258534,ENSEMBL:ENSMUSG00000049737 | MGI:3031195 | protein coding gene | olfactory receptor 1361 |
| 13 | gene | 21.673431 | 21.677405 | negative | MGI_C57BL6J_3031194 | Olfr1360 | NCBI_Gene:258536,ENSEMBL:ENSMUSG00000108534 | MGI:3031194 | protein coding gene | olfactory receptor 1360 |
| 13 | gene | 21.698111 | 21.706833 | positive | MGI_C57BL6J_3031193 | Olfr1359 | NCBI_Gene:258067,ENSEMBL:ENSMUSG00000108674 | MGI:3031193 | protein coding gene | olfactory receptor 1359 |
| 13 | gene | 21.710580 | 21.710650 | positive | MGI_C57BL6J_4413876 | n-TGgcc7 | NCBI_Gene:102467312 | MGI:4413876 | tRNA gene | nuclear encoded tRNA glycine 7 (anticodon GCC) |
| 13 | gene | 21.710994 | 21.711065 | positive | MGI_C57BL6J_4413984 | n-TMcat7 | NCBI_Gene:102467356 | MGI:4413984 | tRNA gene | nuclear encoded tRNA methionine 7 (anticodon CAT) |
| 13 | gene | 21.715713 | 21.716143 | negative | MGI_C57BL6J_2448403 | H2bc13 | NCBI_Gene:319185,ENSEMBL:ENSMUSG00000094338 | MGI:2448403 | protein coding gene | H2B clustered histone 13 |
| 13 | gene | 21.716412 | 21.716859 | positive | MGI_C57BL6J_2448457 | H2ac13 | NCBI_Gene:319191,ENSEMBL:ENSMUSG00000071516 | MGI:2448457 | protein coding gene | H2A clustered histone 13 |
| 13 | gene | 21.716667 | 21.716804 | negative | MGI_C57BL6J_5690759 | Gm44367 | ENSEMBL:ENSMUSG00000105014 | MGI:5690759 | miRNA gene | predicted gene, 44367 |
| 13 | gene | 21.717628 | 21.718115 | positive | MGI_C57BL6J_2448349 | H3c10 | NCBI_Gene:319152,ENSEMBL:ENSMUSG00000101355 | MGI:2448349 | protein coding gene | H3 clustered histone 10 |
| 13 | gene | 21.721433 | 21.721574 | positive | MGI_C57BL6J_5690749 | Gm44357 | ENSEMBL:ENSMUSG00000105238 | MGI:5690749 | miRNA gene | predicted gene, 44357 |
| 13 | pseudogene | 21.721442 | 21.721808 | negative | MGI_C57BL6J_2448312 | H2ac14 | NCBI_Gene:319174,ENSEMBL:ENSMUSG00000080076 | MGI:2448312 | pseudogene | H2A clustered histone 14 |
| 13 | gene | 21.722044 | 21.722567 | positive | MGI_C57BL6J_2448404 | H2bc14 | NCBI_Gene:319186,ENSEMBL:ENSMUSG00000114279 | MGI:2448404 | protein coding gene | H2B clustered histone 14 |
| 13 | gene | 21.735034 | 21.735837 | positive | MGI_C57BL6J_2448436 | H4c11 | NCBI_Gene:319159,ENSEMBL:ENSMUSG00000067455 | MGI:2448436 | protein coding gene | H4 clustered histone 11 |
| 13 | gene | 21.750145 | 21.750553 | negative | MGI_C57BL6J_2448439 | H4c12 | NCBI_Gene:319160,ENSEMBL:ENSMUSG00000064288 | MGI:2448439 | protein coding gene | H4 clustered histone 12 |
| 13 | gene | 21.753377 | 21.753912 | negative | MGI_C57BL6J_2448297 | H2ac15 | NCBI_Gene:319169,ENSEMBL:ENSMUSG00000063021 | MGI:2448297 | protein coding gene | H2A clustered histone 15 |
| 13 | gene | 21.753445 | 21.753582 | positive | MGI_C57BL6J_5690846 | Gm44454 | ENSEMBL:ENSMUSG00000105580 | MGI:5690846 | miRNA gene | predicted gene, 44454 |
| 13 | gene | 21.754123 | 21.754553 | positive | MGI_C57BL6J_2448407 | H2bc15 | NCBI_Gene:319187,ENSEMBL:ENSMUSG00000095217 | MGI:2448407 | protein coding gene | H2B clustered histone 15 |
| 13 | gene | 21.768129 | 21.771133 | positive | MGI_C57BL6J_3649225 | Gm11274 | NCBI_Gene:108168068,ENSEMBL:ENSMUSG00000085331 | MGI:3649225 | lncRNA gene | predicted gene 11274 |
| 13 | gene | 21.779832 | 21.780625 | negative | MGI_C57BL6J_1861461 | H1f5 | NCBI_Gene:56702,ENSEMBL:ENSMUSG00000058773 | MGI:1861461 | protein coding gene | H1.5 linker histone, cluster member |
| 13 | gene | 21.782915 | 21.783397 | negative | MGI_C57BL6J_2448350 | H3c11 | NCBI_Gene:319153,ENSEMBL:ENSMUSG00000101972 | MGI:2448350 | protein coding gene | H3 clustered histone 11 |
| 13 | gene | 21.786772 | 21.787218 | negative | MGI_C57BL6J_2448300 | Hist1h2an | NCBI_Gene:319170,ENSEMBL:ENSMUSG00000069309 | MGI:2448300 | protein coding gene | histone cluster 1, H2an |
| 13 | gene | 21.786875 | 21.787015 | positive | MGI_C57BL6J_5690738 | Gm44346 | ENSEMBL:ENSMUSG00000106281 | MGI:5690738 | miRNA gene | predicted gene, 44346 |
| 13 | gene | 21.787461 | 21.801561 | positive | MGI_C57BL6J_2448409 | Hist1h2bp | NCBI_Gene:319188,ENSEMBL:ENSMUSG00000069308 | MGI:2448409 | protein coding gene | histone cluster 1, H2bp |
| 13 | gene | 21.806412 | 21.810199 | negative | MGI_C57BL6J_3702051 | Hist1h2bq | NCBI_Gene:665596,ENSEMBL:ENSMUSG00000069307 | MGI:3702051 | protein coding gene | histone cluster 1, H2bq |
| 13 | gene | 21.810367 | 21.810944 | positive | MGI_C57BL6J_2448302 | Hist1h2ao | NCBI_Gene:665433,ENSEMBL:ENSMUSG00000094248 | MGI:2448302 | protein coding gene | histone cluster 1, H2ao |
| 13 | gene | 21.810712 | 21.810849 | negative | MGI_C57BL6J_5011843 | Gm19658 | ENSEMBL:ENSMUSG00000105256 | MGI:5011843 | miRNA gene | predicted gene, 19658 |
| 13 | gene | 21.811746 | 21.812150 | positive | MGI_C57BL6J_2448441 | Hist1h4m | NCBI_Gene:100041230,ENSEMBL:ENSMUSG00000069306 | MGI:2448441 | protein coding gene | histone cluster 1, H4m |
| 13 | gene | 21.813170 | 21.819385 | positive | MGI_C57BL6J_5826040 | Gm46403 | NCBI_Gene:108168067 | MGI:5826040 | protein coding gene | predicted gene, 46403 |
| 13 | gene | 21.831767 | 21.832196 | negative | MGI_C57BL6J_4843992 | Hist1h4n | NCBI_Gene:319161,ENSEMBL:ENSMUSG00000069305 | MGI:4843992 | protein coding gene | histone cluster 1, H4n |
| 13 | gene | 21.833024 | 21.833507 | negative | MGI_C57BL6J_3710573 | Hist1h2ap | NCBI_Gene:319171,ENSEMBL:ENSMUSG00000094777 | MGI:3710573 | protein coding gene | histone cluster 1, H2ap |
| 13 | gene | 21.833093 | 21.833230 | positive | MGI_C57BL6J_5690858 | Gm44466 | ENSEMBL:ENSMUSG00000106440 | MGI:5690858 | miRNA gene | predicted gene, 44466 |
| 13 | gene | 21.833743 | 21.837530 | positive | MGI_C57BL6J_3710645 | Hist1h2br | NCBI_Gene:665622,ENSEMBL:ENSMUSG00000069303 | MGI:3710645 | protein coding gene | histone cluster 1 H2br |
| 13 | gene | 21.848022 | 21.848104 | negative | MGI_C57BL6J_4413942 | n-TLtaa2 | NCBI_Gene:108168116 | MGI:4413942 | tRNA gene | nuclear encoded tRNA leucine 2 (anticodon TAA) |
| 13 | gene | 21.855320 | 21.855393 | negative | MGI_C57BL6J_4413912 | n-TIaat11 | NCBI_Gene:108168117 | MGI:4413912 | tRNA gene | nuclear encoded tRNA isoleucine 11 (anticodon AAT) |
| 13 | gene | 21.856021 | 21.856094 | positive | MGI_C57BL6J_4414051 | n-TTagt4 | NCBI_Gene:108168129 | MGI:4414051 | tRNA gene | nuclear encoded tRNA threonine 4 (anticodon AGT) |
| 13 | pseudogene | 21.875233 | 21.878024 | negative | MGI_C57BL6J_3651908 | Gm11278 | NCBI_Gene:102635439,ENSEMBL:ENSMUSG00000080770 | MGI:3651908 | pseudogene | predicted gene 11278 |
| 13 | gene | 21.882033 | 21.882106 | negative | MGI_C57BL6J_4413911 | n-TIaat10 | NCBI_Gene:108168118 | MGI:4413911 | tRNA gene | nuclear encoded tRNA isoleucine 10 (anticodon AAT) |
| 13 | gene | 21.882885 | 21.882957 | negative | MGI_C57BL6J_4413996 | n-TFgaa1 | NCBI_Gene:108168119 | MGI:4413996 | tRNA gene | nuclear encoded tRNA pheylalanine 1 (anticodon GAA) |
| 13 | gene | 21.896918 | 21.897049 | negative | MGI_C57BL6J_3837223 | Mir1983 | miRBase:MI0009990,NCBI_Gene:100316716,ENSEMBL:ENSMUSG00000088148 | MGI:3837223 | miRNA gene | microRNA 1983 |
| 13 | gene | 21.896938 | 21.897031 | negative | MGI_C57BL6J_4413915 | n-TItat14 | NCBI_Gene:108168120 | MGI:4413915 | tRNA gene | nuclear encoded tRNA isoleucine 14 (anticodon TAT) |
| 13 | gene | 21.897192 | 21.898593 | positive | MGI_C57BL6J_5623715 | Gm40830 | NA | NA | lncRNA gene | predicted gene%2c 40830 |
| 13 | pseudogene | 21.903156 | 21.904784 | positive | MGI_C57BL6J_3651909 | Gm11279 | NCBI_Gene:100418364,ENSEMBL:ENSMUSG00000081576 | MGI:3651909 | pseudogene | predicted gene 11279 |
| 13 | gene | 21.907111 | 21.907182 | positive | MGI_C57BL6J_4413979 | n-TMcat2 | NCBI_Gene:108168130 | MGI:4413979 | tRNA gene | nuclear encoded tRNA methionine 2 (anticodon CAT) |
| 13 | gene | 21.910872 | 21.912358 | positive | MGI_C57BL6J_5623716 | Gm40831 | NCBI_Gene:105245363 | MGI:5623716 | lncRNA gene | predicted gene, 40831 |
| 13 | gene | 21.911313 | 21.911384 | positive | MGI_C57BL6J_4413762 | n-TDgtc11 | NCBI_Gene:108168132 | MGI:4413762 | tRNA gene | nuclear encoded tRNA aspartic acid 11 (anticodon GTC) |
| 13 | gene | 21.914838 | 21.914943 | positive | MGI_C57BL6J_4413929 | n-TLcaa2 | NCBI_Gene:108168133 | MGI:4413929 | tRNA gene | nuclear encoded tRNA leucine 2 (anticodon CAA) |
| 13 | gene | 21.917162 | 21.917247 | negative | MGI_C57BL6J_4413736 | n-TRtct3 | NCBI_Gene:108168121 | MGI:4413736 | tRNA gene | nuclear encoded tRNA arginine 3 (anticodon TCT) |
| 13 | gene | 21.919215 | 21.919296 | positive | MGI_C57BL6J_4414025 | n-TSaga5 | NCBI_Gene:108168134 | MGI:4414025 | tRNA gene | nuclear encoded tRNA serine 5 (anticodon AGA) |
| 13 | gene | 21.920200 | 21.920271 | positive | MGI_C57BL6J_4413834 | n-TQctg10 | NCBI_Gene:108168135 | MGI:4413834 | tRNA gene | nuclear encoded tRNA glutamine 10 (anticodon CTG) |
| 13 | gene | 21.924928 | 21.925016 | negative | MGI_C57BL6J_5690848 | Gm44456 | NCBI_Gene:108168122,ENSEMBL:ENSMUSG00000105839 | MGI:5690848 | miRNA gene | predicted gene, 44456 |
| 13 | gene | 21.924935 | 21.925016 | negative | MGI_C57BL6J_4414045 | n-TStga2 | NCBI_Gene:108168122 | MGI:4414045 | tRNA gene | nuclear encoded tRNA serine 2 (anticodon TGA) |
| 13 | gene | 21.927857 | 21.927938 | positive | MGI_C57BL6J_4414026 | n-TSaga6 | NCBI_Gene:108168136 | MGI:4414026 | tRNA gene | nuclear encoded tRNA serine 6 (anticodon AGA) |
| 13 | gene | 21.931169 | 21.931240 | negative | MGI_C57BL6J_4413987 | n-TMcat10 | NCBI_Gene:108168123 | MGI:4413987 | tRNA gene | nuclear encoded tRNA methionine 10 (anticodon CAT) |
| 13 | gene | 21.933961 | 21.934032 | negative | MGI_C57BL6J_4413832 | n-TQctg8 | NCBI_Gene:108168124 | MGI:4413832 | tRNA gene | nuclear encoded tRNA glutamine 8 (anticodon CTG) |
| 13 | gene | 21.935497 | 21.935568 | negative | MGI_C57BL6J_4413764 | n-TDgtc13 | NCBI_Gene:108168125 | MGI:4413764 | tRNA gene | nuclear encoded tRNA aspartic acid 13 (anticodon GTC) |
| 13 | gene | 21.937094 | 21.937175 | negative | MGI_C57BL6J_4414030 | n-TSaga1 | NCBI_Gene:108168126 | MGI:4414030 | tRNA gene | nuclear encoded tRNA serine 1 (anticodon AGA) |
| 13 | gene | 21.940319 | 21.940390 | negative | MGI_C57BL6J_4413763 | n-TDgtc12 | NCBI_Gene:108168127 | MGI:4413763 | tRNA gene | nuclear encoded tRNA aspartic acid 12 (anticodon GTC) |
| 13 | gene | 21.940374 | 21.945197 | negative | MGI_C57BL6J_3705200 | Gm11280 | ENSEMBL:ENSMUSG00000085393 | MGI:3705200 | lncRNA gene | predicted gene 11280 |
| 13 | gene | 21.940914 | 21.940995 | negative | MGI_C57BL6J_4414028 | n-TSaga8 | NCBI_Gene:108168128 | MGI:4414028 | tRNA gene | nuclear encoded tRNA serine 8 (anticodon AGA) |
| 13 | gene | 21.944937 | 21.960779 | positive | MGI_C57BL6J_1922244 | Zfp184 | NCBI_Gene:193452,ENSEMBL:ENSMUSG00000006720 | MGI:1922244 | protein coding gene | zinc finger protein 184 (Kruppel-like) |
| 13 | pseudogene | 21.953053 | 21.953628 | negative | MGI_C57BL6J_3651906 | Gm11281 | ENSEMBL:ENSMUSG00000078917 | MGI:3651906 | pseudogene | predicted gene 11281 |
| 13 | pseudogene | 21.956708 | 21.958123 | negative | MGI_C57BL6J_3650570 | Gm11282 | NCBI_Gene:100418082,ENSEMBL:ENSMUSG00000081265 | MGI:3650570 | pseudogene | predicted gene 11282 |
| 13 | gene | 21.971788 | 21.971860 | positive | MGI_C57BL6J_4413969 | n-TKttt22 | NCBI_Gene:108168137 | MGI:4413969 | tRNA gene | nuclear encoded tRNA lysine 22 (anticodon TTT) |
| 13 | gene | 21.975032 | 21.975103 | positive | MGI_C57BL6J_4413985 | n-TMcat8 | NCBI_Gene:108168138 | MGI:4413985 | tRNA gene | nuclear encoded tRNA methionine 8 (anticodon CAT) |
| 13 | gene | 21.981181 | 21.988734 | positive | MGI_C57BL6J_2684870 | Pom121l2 | NCBI_Gene:195236,ENSEMBL:ENSMUSG00000016982 | MGI:2684870 | protein coding gene | POM121 membrane glycoprotein-like 2 (rat) |
| 13 | gene | 21.994190 | 21.994271 | negative | MGI_C57BL6J_4414042 | n-TSgct8 | NCBI_Gene:102467381 | MGI:4414042 | tRNA gene | nuclear encoded tRNA serine 8 (anticodon GCT) |
| 13 | gene | 21.996581 | 21.996652 | negative | MGI_C57BL6J_4413825 | n-TQctg1 | NCBI_Gene:102467510 | MGI:4413825 | tRNA gene | nuclear encoded tRNA glutamine 1 (anticodon CTG) |
| 13 | gene | 22.002173 | 22.009742 | negative | MGI_C57BL6J_1859181 | Prss16 | NCBI_Gene:54373,ENSEMBL:ENSMUSG00000006179 | MGI:1859181 | protein coding gene | protease, serine 16 (thymus) |
| 13 | gene | 22.014468 | 22.020738 | positive | MGI_C57BL6J_3651659 | Gm11290 | ENSEMBL:ENSMUSG00000085827 | MGI:3651659 | lncRNA gene | predicted gene 11290 |
| 13 | gene | 22.015791 | 22.015863 | negative | MGI_C57BL6J_4414086 | n-TVaac3 | NCBI_Gene:102467617 | MGI:4414086 | tRNA gene | nuclear encoded tRNA valine 3 (anticodon AAC) |
| 13 | gene | 22.021144 | 22.021216 | negative | MGI_C57BL6J_4413714 | n-TRacg1 | NCBI_Gene:102467628 | MGI:4413714 | tRNA gene | nuclear encoded tRNA arginine 1 (anticodon ACG) |
| 13 | gene | 22.022419 | 22.022500 | negative | MGI_C57BL6J_4414032 | n-TScga1 | NCBI_Gene:102467376 | MGI:4414032 | tRNA gene | nuclear encoded tRNA serine 1 (anticodon CGA) |
| 13 | gene | 22.026548 | 22.026620 | positive | MGI_C57BL6J_4414099 | n-TVcac8 | NCBI_Gene:102467406 | MGI:4414099 | tRNA gene | nuclear encoded tRNA valine 8 (anticodon CAC) |
| 13 | gene | 22.026699 | 22.032196 | positive | MGI_C57BL6J_3651660 | Gm11292 | ENSEMBL:ENSMUSG00000043145 | MGI:3651660 | lncRNA gene | predicted gene 11292 |
| 13 | gene | 22.029223 | 22.029296 | positive | MGI_C57BL6J_4413908 | n-TIaat7 | NCBI_Gene:102467544 | MGI:4413908 | tRNA gene | nuclear encoded tRNA isoleucine 7 (anticodon AAT) |
| 13 | gene | 22.030100 | 22.030173 | negative | MGI_C57BL6J_4414048 | n-TTagt1 | NCBI_Gene:102467601 | MGI:4414048 | tRNA gene | nuclear encoded tRNA threonine 1 (anticodon AGT) |
| 13 | gene | 22.035122 | 22.035643 | negative | MGI_C57BL6J_2448295 | H2ac12 | NCBI_Gene:319168,ENSEMBL:ENSMUSG00000069302 | MGI:2448295 | protein coding gene | H2A clustered histone 12 |
| 13 | gene | 22.035164 | 22.035307 | positive | MGI_C57BL6J_5690782 | Gm44390 | ENSEMBL:ENSMUSG00000104608 | MGI:5690782 | miRNA gene | predicted gene, 44390 |
| 13 | gene | 22.035821 | 22.036345 | positive | MGI_C57BL6J_2448399 | H2bc12 | NCBI_Gene:319184,ENSEMBL:ENSMUSG00000062727 | MGI:2448399 | protein coding gene | H2B clustered histone 12 |
| 13 | gene | 22.040636 | 22.041362 | negative | MGI_C57BL6J_2448432 | H4c9 | NCBI_Gene:319158,ENSEMBL:ENSMUSG00000060639 | MGI:2448432 | protein coding gene | H4 clustered histone 9 |
| 13 | gene | 22.042460 | 22.042949 | negative | MGI_C57BL6J_2448293 | H2ac11 | NCBI_Gene:319167,ENSEMBL:ENSMUSG00000069301 | MGI:2448293 | protein coding gene | H2A clustered histone 11 |
| 13 | gene | 22.042545 | 22.042682 | positive | MGI_C57BL6J_5690878 | Gm44486 | ENSEMBL:ENSMUSG00000105433 | MGI:5690878 | miRNA gene | predicted gene, 44486 |
| 13 | gene | 22.043214 | 22.043676 | positive | MGI_C57BL6J_2448388 | H2bc11 | NCBI_Gene:319183,ENSEMBL:ENSMUSG00000069300 | MGI:2448388 | protein coding gene | H2B clustered histone 11 |
| 13 | gene | 22.049514 | 22.051450 | positive | MGI_C57BL6J_6097822 | Gm48356 | ENSEMBL:ENSMUSG00000114817 | MGI:6097822 | unclassified gene | predicted gene, 48356 |
| 13 | gene | 22.050686 | 22.050767 | negative | MGI_C57BL6J_4414039 | n-TSgct5 | NCBI_Gene:102467438 | MGI:4414039 | tRNA gene | nuclear encoded tRNA serine 5 (anticodon GCT) |
| 13 | pseudogene | 22.057168 | 22.057912 | negative | MGI_C57BL6J_4439036 | Vmn1r-ps92 | NCBI_Gene:100312522,ENSEMBL:ENSMUSG00000083873 | MGI:4439036 | pseudogene | vomeronasal 1 receptor, pseudogene 92 |
| 13 | pseudogene | 22.078070 | 22.078684 | positive | MGI_C57BL6J_4439038 | Vmn1r-ps93 | NCBI_Gene:100312523,ENSEMBL:ENSMUSG00000082869 | MGI:4439038 | pseudogene | vomeronasal 1 receptor, pseudogene 93 |
| 13 | pseudogene | 22.083453 | 22.083787 | negative | MGI_C57BL6J_3651653 | Gm11285 | NCBI_Gene:665690,ENSEMBL:ENSMUSG00000084072 | MGI:3651653 | pseudogene | predicted gene 11285 |
| 13 | gene | 22.083541 | 22.083634 | negative | MGI_C57BL6J_5456077 | Gm26300 | ENSEMBL:ENSMUSG00000076237 | MGI:5456077 | miRNA gene | predicted gene, 26300 |
| 13 | gene | 22.084440 | 22.095916 | positive | MGI_C57BL6J_2182259 | Vmn1r188 | NCBI_Gene:252912,ENSEMBL:ENSMUSG00000069299 | MGI:2182259 | protein coding gene | vomeronasal 1 receptor 188 |
| 13 | pseudogene | 22.092119 | 22.092589 | positive | MGI_C57BL6J_4439040 | Vmn1r-ps94 | NCBI_Gene:100312524,ENSEMBL:ENSMUSG00000081334 | MGI:4439040 | pseudogene | vomeronasal 1 receptor, pseudogene 94 |
| 13 | gene | 22.099762 | 22.105824 | negative | MGI_C57BL6J_2182257 | Vmn1r189 | NCBI_Gene:252906,ENSEMBL:ENSMUSG00000099611 | MGI:2182257 | protein coding gene | vomeronasal 1 receptor 189 |
| 13 | pseudogene | 22.123289 | 22.124193 | positive | MGI_C57BL6J_4439042 | Vmn1r-ps95 | NCBI_Gene:100312525,ENSEMBL:ENSMUSG00000081469 | MGI:4439042 | pseudogene | vomeronasal 1 receptor, pseudogene 95 |
| 13 | pseudogene | 22.126715 | 22.127054 | positive | MGI_C57BL6J_3852506 | Vmn1r-ps96 | NCBI_Gene:100312526,ENSEMBL:ENSMUSG00000114620 | MGI:3852506 | pseudogene | vomeronasal 1 receptor, pseudogene 96 |
| 13 | pseudogene | 22.136424 | 22.137026 | negative | MGI_C57BL6J_4439044 | Vmn1r-ps97 | NCBI_Gene:100312527,ENSEMBL:ENSMUSG00000083167 | MGI:4439044 | pseudogene | vomeronasal 1 receptor, pseudogene 97 |
| 13 | pseudogene | 22.144441 | 22.145525 | negative | MGI_C57BL6J_2182261 | Vmn1r190-ps | NCBI_Gene:252908,ENSEMBL:ENSMUSG00000089973 | MGI:2182261 | pseudogene | vomeronasal 1 receptor 190, pseudogene |
| 13 | pseudogene | 22.152362 | 22.152727 | positive | MGI_C57BL6J_3852453 | Vmn1r-ps98 | NCBI_Gene:100312528 | MGI:3852453 | pseudogene | vomeronasal 1 receptor, pseudogene 98 |
| 13 | pseudogene | 22.163883 | 22.165807 | negative | MGI_C57BL6J_3651654 | Gm11284 | NCBI_Gene:665703,ENSEMBL:ENSMUSG00000083319 | MGI:3651654 | pseudogene | predicted gene 11284 |
| 13 | gene | 22.173220 | 22.183969 | negative | MGI_C57BL6J_2182258 | Vmn1r191 | NCBI_Gene:632534,ENSEMBL:ENSMUSG00000095916 | MGI:2182258 | protein coding gene | vomeronasal 1 receptor 191 |
| 13 | pseudogene | 22.174695 | 22.175595 | negative | MGI_C57BL6J_4439048 | Vmn1r-ps99 | NCBI_Gene:100312529,ENSEMBL:ENSMUSG00000083348 | MGI:4439048 | pseudogene | vomeronasal 1 receptor, pseudogene 97 |
| 13 | gene | 22.183997 | 22.191141 | negative | MGI_C57BL6J_2182260 | Vmn1r192 | NCBI_Gene:252907,ENSEMBL:ENSMUSG00000099787 | MGI:2182260 | protein coding gene | vomeronasal 1 receptor 192 |
| 13 | gene | 22.204575 | 22.204675 | negative | MGI_C57BL6J_5454861 | Gm25084 | ENSEMBL:ENSMUSG00000096817 | MGI:5454861 | miRNA gene | predicted gene, 25084 |
| 13 | gene | 22.212419 | 22.212519 | negative | MGI_C57BL6J_5453059 | Gm23282 | ENSEMBL:ENSMUSG00000095684 | MGI:5453059 | miRNA gene | predicted gene, 23282 |
| 13 | gene | 22.213672 | 22.223160 | negative | MGI_C57BL6J_2159695 | Vmn1r193 | NCBI_Gene:171259,ENSEMBL:ENSMUSG00000046932 | MGI:2159695 | protein coding gene | vomeronasal 1 receptor 193 |
| 13 | gene | 22.244215 | 22.245105 | positive | MGI_C57BL6J_3651596 | Vmn1r194 | NCBI_Gene:626299,ENSEMBL:ENSMUSG00000069297 | MGI:3651596 | protein coding gene | vomeronasal 1 receptor 194 |
| 13 | gene | 22.270522 | 22.281230 | positive | MGI_C57BL6J_2159692 | Vmn1r195 | NCBI_Gene:171257,ENSEMBL:ENSMUSG00000069296 | MGI:2159692 | protein coding gene | vomeronasal 1 receptor 195 |
| 13 | pseudogene | 22.282341 | 22.283548 | positive | MGI_C57BL6J_3651595 | Gm11295 | NCBI_Gene:665726,ENSEMBL:ENSMUSG00000084038 | MGI:3651595 | pseudogene | predicted gene 11295 |
| 13 | gene | 22.289920 | 22.298360 | positive | MGI_C57BL6J_4438447 | Vmn1r196 | NCBI_Gene:100312484,ENSEMBL:ENSMUSG00000069295 | MGI:4438447 | protein coding gene | vomeronasal 1 receptor 196 |
| 13 | pseudogene | 22.298612 | 22.299478 | negative | MGI_C57BL6J_4439051 | Vmn1r-ps100 | NCBI_Gene:100312532,ENSEMBL:ENSMUSG00000082785 | MGI:4439051 | pseudogene | vomeronasal 1 receptor, pseudogene 100 |
| 13 | gene | 22.322191 | 22.332866 | positive | MGI_C57BL6J_2159686 | Vmn1r197 | NCBI_Gene:171278,ENSEMBL:ENSMUSG00000069294 | MGI:2159686 | protein coding gene | vomeronasal 1 receptor 197 |
| 13 | pseudogene | 22.336315 | 22.336503 | positive | MGI_C57BL6J_6098012 | Gm48489 | ENSEMBL:ENSMUSG00000114499 | MGI:6098012 | pseudogene | predicted gene, 48489 |
| 13 | gene | 22.350364 | 22.360301 | positive | MGI_C57BL6J_2159689 | Vmn1r198 | NCBI_Gene:171254,ENSEMBL:ENSMUSG00000095125 | MGI:2159689 | protein coding gene | vomeronasal 1 receptor 198 |
| 13 | pseudogene | 22.370561 | 22.370832 | positive | MGI_C57BL6J_3852454 | Vmn1r-ps101 | NCBI_Gene:100312533,ENSEMBL:ENSMUSG00000114666 | MGI:3852454 | pseudogene | vomeronasal 1 receptor, pseudogene 101 |
| 13 | gene | 22.379818 | 22.388102 | positive | MGI_C57BL6J_2159662 | Vmn1r199 | NCBI_Gene:171247,ENSEMBL:ENSMUSG00000069292 | MGI:2159662 | protein coding gene | vomeronasal 1 receptor 199 |
| 13 | pseudogene | 22.385435 | 22.386534 | positive | MGI_C57BL6J_4439057 | Vmn1r-ps102 | NCBI_Gene:100312534,ENSEMBL:ENSMUSG00000081080 | MGI:4439057 | pseudogene | vomeronasal 1 receptor, pseudogene 102 |
| 13 | gene | 22.390373 | 22.400052 | positive | MGI_C57BL6J_2159660 | Vmn1r200 | NCBI_Gene:171246,ENSEMBL:ENSMUSG00000101073 | MGI:2159660 | protein coding gene | vomeronasal 1 receptor 200 |
| 13 | pseudogene | 22.441272 | 22.442243 | positive | MGI_C57BL6J_2159658 | Vmn1r-ps103 | NCBI_Gene:171245,ENSEMBL:ENSMUSG00000084924 | MGI:2159658 | pseudogene | vomeronasal 1 receptor, pseudogene 103 |
| 13 | pseudogene | 22.449641 | 22.450440 | positive | MGI_C57BL6J_4439058 | Vmn1r-ps104 | NCBI_Gene:100312535,ENSEMBL:ENSMUSG00000081283 | MGI:4439058 | pseudogene | vomeronasal 1 receptor, pseudogene 104 |
| 13 | pseudogene | 22.464296 | 22.467857 | positive | MGI_C57BL6J_3651396 | Gm11298 | NCBI_Gene:665749,ENSEMBL:ENSMUSG00000083940 | MGI:3651396 | pseudogene | predicted gene 11298 |
| 13 | gene | 22.471923 | 22.478701 | positive | MGI_C57BL6J_2159690 | Vmn1r201 | NCBI_Gene:171255,ENSEMBL:ENSMUSG00000094898 | MGI:2159690 | protein coding gene | vomeronasal 1 receptor 201 |
| 13 | pseudogene | 22.480631 | 22.483043 | positive | MGI_C57BL6J_4439328 | Vmn1r-ps106 | NCBI_Gene:100312577,ENSEMBL:ENSMUSG00000081717 | MGI:4439328 | pseudogene | vomeronasal 1 receptor, pseudogene 106 |
| 13 | pseudogene | 22.491605 | 22.492185 | positive | MGI_C57BL6J_4439059 | Vmn1r-ps107 | NCBI_Gene:100312536,ENSEMBL:ENSMUSG00000080697 | MGI:4439059 | pseudogene | vomeronasal 1 receptor, pseudogene 107 |
| 13 | pseudogene | 22.496034 | 22.496418 | positive | MGI_C57BL6J_4439060 | Vmn1r-ps108 | NCBI_Gene:100312537,ENSEMBL:ENSMUSG00000081495 | MGI:4439060 | pseudogene | vomeronasal 1 receptor, pseudogene 108 |
| 13 | gene | 22.497750 | 22.505381 | negative | MGI_C57BL6J_2159693 | Vmn1r202 | NCBI_Gene:171258,ENSEMBL:ENSMUSG00000094379 | MGI:2159693 | protein coding gene | vomeronasal 1 receptor 202 |
| 13 | gene | 22.518114 | 22.531992 | positive | MGI_C57BL6J_2159674 | Vmn1r203 | NCBI_Gene:171270,ENSEMBL:ENSMUSG00000069289 | MGI:2159674 | protein coding gene | vomeronasal 1 receptor 203 |
| 13 | gene | 22.523911 | 22.524744 | negative | MGI_C57BL6J_3651398 | Gm11300 | ENSEMBL:ENSMUSG00000087688 | MGI:3651398 | lncRNA gene | predicted gene 11300 |
| 13 | gene | 22.553198 | 22.558200 | positive | MGI_C57BL6J_3651399 | Vmn1r204 | NCBI_Gene:632793,ENSEMBL:ENSMUSG00000094637 | MGI:3651399 | protein coding gene | vomeronasal 1 receptor 204 |
| 13 | gene | 22.591900 | 22.593000 | negative | MGI_C57BL6J_2159667 | Vmn1r205 | NCBI_Gene:171251,ENSEMBL:ENSMUSG00000100296 | MGI:2159667 | protein coding gene | vomeronasal 1 receptor 205 |
| 13 | gene | 22.617456 | 22.624251 | negative | MGI_C57BL6J_2159665 | Vmn1r206 | NCBI_Gene:171250,ENSEMBL:ENSMUSG00000101578 | MGI:2159665 | protein coding gene | vomeronasal 1 receptor 206 |
| 13 | pseudogene | 22.625731 | 22.626482 | negative | MGI_C57BL6J_4439061 | Vmn1r-ps109 | NCBI_Gene:100312538,ENSEMBL:ENSMUSG00000081914 | MGI:4439061 | pseudogene | vomeronasal 1 receptor, pseudogene 109 |
| 13 | pseudogene | 22.628071 | 22.629164 | negative | MGI_C57BL6J_4439062 | Vmn1r-ps110 | NCBI_Gene:100312539,ENSEMBL:ENSMUSG00000083413 | MGI:4439062 | pseudogene | vomeronasal 1 receptor, pseudogene 110 |
| 13 | pseudogene | 22.675660 | 22.676969 | positive | MGI_C57BL6J_3650171 | Gm11307 | NCBI_Gene:382740,ENSEMBL:ENSMUSG00000082178 | MGI:3650171 | pseudogene | predicted gene 11307 |
| 13 | pseudogene | 22.690970 | 22.691966 | negative | MGI_C57BL6J_4439063 | Vmn1r-ps111 | NCBI_Gene:100312540,ENSEMBL:ENSMUSG00000083673 | MGI:4439063 | pseudogene | vomeronasal 1 receptor, pseudogene 111 |
| 13 | gene | 22.691299 | 22.714987 | positive | MGI_C57BL6J_3650169 | Gm11309 | ENSEMBL:ENSMUSG00000085221 | MGI:3650169 | lncRNA gene | predicted gene 11309 |
| 13 | pseudogene | 22.722717 | 22.729629 | negative | MGI_C57BL6J_3650396 | Vmn1r207-ps | NCBI_Gene:432735,ENSEMBL:ENSMUSG00000084136 | MGI:3650396 | pseudogene | vomeronasal 1 receptor 207, pseudogene |
| 13 | pseudogene | 22.733000 | 22.733923 | negative | MGI_C57BL6J_4439064 | Vmn1r-ps112 | NCBI_Gene:100312541,ENSEMBL:ENSMUSG00000082540 | MGI:4439064 | pseudogene | vomeronasal 1 receptor, pseudogene 112 |
| 13 | pseudogene | 22.736988 | 22.738093 | negative | MGI_C57BL6J_4439065 | Vmn1r-ps113 | NCBI_Gene:100312542,ENSEMBL:ENSMUSG00000081531 | MGI:4439065 | pseudogene | vomeronasal 1 receptor, pseudogene 113 |
| 13 | pseudogene | 22.758927 | 22.759358 | positive | MGI_C57BL6J_3650398 | Gm11313 | ENSEMBL:ENSMUSG00000080730 | MGI:3650398 | pseudogene | predicted gene 11313 |
| 13 | gene | 22.771474 | 22.777400 | negative | MGI_C57BL6J_2159668 | Vmn1r208 | NCBI_Gene:171252,ENSEMBL:ENSMUSG00000071493 | MGI:2159668 | protein coding gene | vomeronasal 1 receptor 208 |
| 13 | gene | 22.772656 | 22.773471 | positive | MGI_C57BL6J_3650397 | Gm11314 | ENSEMBL:ENSMUSG00000087643 | MGI:3650397 | lncRNA gene | predicted gene 11314 |
| 13 | gene | 22.800629 | 22.809682 | negative | MGI_C57BL6J_3650395 | Vmn1r209 | NCBI_Gene:432736,ENSEMBL:ENSMUSG00000071491 | MGI:3650395 | protein coding gene | vomeronasal 1 receptor 209 |
| 13 | gene | 22.825875 | 22.831348 | negative | MGI_C57BL6J_2159673 | Vmn1r210 | NCBI_Gene:171269,ENSEMBL:ENSMUSG00000061296 | MGI:2159673 | protein coding gene | vomeronasal 1 receptor 210 |
| 13 | pseudogene | 22.844994 | 22.846058 | negative | MGI_C57BL6J_4439066 | Vmn1r-ps114 | NCBI_Gene:100312543,ENSEMBL:ENSMUSG00000083297 | MGI:4439066 | pseudogene | vomeronasal 1 receptor, pseudogene 114 |
| 13 | gene | 22.847426 | 22.857561 | negative | MGI_C57BL6J_2159685 | Vmn1r211 | NCBI_Gene:171277,ENSEMBL:ENSMUSG00000063998 | MGI:2159685 | protein coding gene | vomeronasal 1 receptor 211 |
| 13 | pseudogene | 22.866640 | 22.866806 | positive | MGI_C57BL6J_3852455 | Vmn1r-ps115 | NCBI_Gene:100312544 | MGI:3852455 | pseudogene | vomeronasal 1 receptor, pseudogene 115 |
| 13 | pseudogene | 22.875102 | 22.875393 | negative | MGI_C57BL6J_4439067 | Vmn1r-ps116 | NCBI_Gene:100312545,ENSEMBL:ENSMUSG00000082037 | MGI:4439067 | pseudogene | vomeronasal 1 receptor, pseudogene 116 |
| 13 | gene | 22.879377 | 22.879507 | negative | MGI_C57BL6J_5453551 | Gm23774 | ENSEMBL:ENSMUSG00000089615 | MGI:5453551 | snoRNA gene | predicted gene, 23774 |
| 13 | gene | 22.883042 | 22.884324 | negative | MGI_C57BL6J_2159683 | Vmn1r212 | NCBI_Gene:171275,ENSEMBL:ENSMUSG00000071490 | MGI:2159683 | protein coding gene | vomeronasal 1 receptor 212 |
| 13 | pseudogene | 22.897719 | 22.898738 | negative | MGI_C57BL6J_3852456 | Vmn1r-ps117 | NCBI_Gene:100312546,ENSEMBL:ENSMUSG00000114527 | MGI:3852456 | pseudogene | vomeronasal 1 receptor, pseudogene 117 |
| 13 | pseudogene | 22.902338 | 22.902830 | negative | MGI_C57BL6J_3650584 | Gm11320 | NCBI_Gene:100312547,ENSEMBL:ENSMUSG00000094264 | MGI:3650584 | pseudogene | predicted gene 11320 |
| 13 | pseudogene | 22.931298 | 22.932199 | negative | MGI_C57BL6J_3651028 | Gm11321 | NCBI_Gene:100312548,ENSEMBL:ENSMUSG00000082340 | MGI:3651028 | pseudogene | predicted gene 11321 |
| 13 | pseudogene | 22.939199 | 22.940179 | negative | MGI_C57BL6J_4439068 | Vmn1r-ps120 | NCBI_Gene:100312549,ENSEMBL:ENSMUSG00000082268 | MGI:4439068 | pseudogene | vomeronasal 1 receptor, pseudogene 120 |
| 13 | pseudogene | 22.943812 | 22.944304 | negative | MGI_C57BL6J_3650586 | Gm11322 | NCBI_Gene:100312550,ENSEMBL:ENSMUSG00000094485 | MGI:3650586 | pseudogene | predicted gene 11322 |
| 13 | pseudogene | 22.960813 | 22.961773 | positive | MGI_C57BL6J_3651015 | Gm11318 | NCBI_Gene:100503070,ENSEMBL:ENSMUSG00000081060 | MGI:3651015 | pseudogene | predicted gene 11318 |
| 13 | pseudogene | 22.996143 | 22.996737 | negative | MGI_C57BL6J_4439075 | Vmn1r-ps123 | NCBI_Gene:100312552,ENSEMBL:ENSMUSG00000079064 | MGI:4439075 | pseudogene | vomeronasal 1 receptor, pseudogene 123 |
| 13 | gene | 23.007656 | 23.014482 | positive | MGI_C57BL6J_2159664 | Vmn1r213 | NCBI_Gene:171249,ENSEMBL:ENSMUSG00000060024 | MGI:2159664 | protein coding gene | vomeronasal 1 receptor 213 |
| 13 | pseudogene | 23.016210 | 23.016366 | negative | MGI_C57BL6J_3852458 | Vmn1r-ps124 | NCBI_Gene:100312553 | MGI:3852458 | pseudogene | vomeronasal 1 receptor, pseudogene 124 |
| 13 | pseudogene | 23.023251 | 23.023546 | positive | MGI_C57BL6J_4439076 | Vmn1r-ps125 | NCBI_Gene:100312554,ENSEMBL:ENSMUSG00000082656 | MGI:4439076 | pseudogene | vomeronasal 1 receptor, pseudogene 125 |
| 13 | gene | 23.030418 | 23.037957 | positive | MGI_C57BL6J_2159663 | Vmn1r214 | NCBI_Gene:171248,ENSEMBL:ENSMUSG00000061829 | MGI:2159663 | protein coding gene | vomeronasal 1 receptor 214 |
| 13 | pseudogene | 23.042413 | 23.042994 | negative | MGI_C57BL6J_5011338 | Gm19153 | NCBI_Gene:100418348,ENSEMBL:ENSMUSG00000114394 | MGI:5011338 | pseudogene | predicted gene, 19153 |
| 13 | pseudogene | 23.056683 | 23.057617 | positive | MGI_C57BL6J_4439077 | Vmn1r-ps126 | NCBI_Gene:100312555,ENSEMBL:ENSMUSG00000083124 | MGI:4439077 | pseudogene | vomeronasal 1 receptor, pseudogene 126 |
| 13 | gene | 23.068025 | 23.078006 | positive | MGI_C57BL6J_2159687 | Vmn1r215 | NCBI_Gene:171253,ENSEMBL:ENSMUSG00000099917 | MGI:2159687 | protein coding gene | vomeronasal 1 receptor 215 |
| 13 | gene | 23.096976 | 23.108732 | positive | MGI_C57BL6J_2159696 | Vmn1r216 | NCBI_Gene:171279,ENSEMBL:ENSMUSG00000116057,ENSEMBL:ENSMUSG00000115697 | MGI:2159696 | protein coding gene | vomeronasal 1 receptor 216 |
| 13 | pseudogene | 23.103454 | 23.103922 | positive | MGI_C57BL6J_3650589 | Gm11325 | ENSEMBL:ENSMUSG00000080861 | MGI:3650589 | pseudogene | predicted gene 11325 |
| 13 | pseudogene | 23.104296 | 23.104790 | positive | MGI_C57BL6J_4439078 | Vmn1r-ps127 | NCBI_Gene:100312556,ENSEMBL:ENSMUSG00000081912 | MGI:4439078 | pseudogene | vomeronasal 1 receptor, pseudogene 127 |
| 13 | gene | 23.105100 | 23.118226 | negative | MGI_C57BL6J_2159677 | Vmn1r217 | NCBI_Gene:171273,ENSEMBL:ENSMUSG00000115791 | MGI:2159677 | protein coding gene | vomeronasal 1 receptor 217 |
| 13 | pseudogene | 23.124489 | 23.126243 | positive | MGI_C57BL6J_3651039 | Vmn1r-ps128 | NCBI_Gene:100040639,ENSEMBL:ENSMUSG00000084315 | MGI:3651039 | pseudogene | vomeronasal 1 receptor, pseudogene 128 |
| 13 | gene | 23.133155 | 23.146112 | positive | MGI_C57BL6J_2159691 | Vmn1r218 | NCBI_Gene:171256,ENSEMBL:ENSMUSG00000115020 | MGI:2159691 | protein coding gene | vomeronasal 1 receptor 218 |
| 13 | pseudogene | 23.153310 | 23.153828 | positive | MGI_C57BL6J_3852461 | Vmn1r-ps129 | NCBI_Gene:100312557 | MGI:3852461 | pseudogene | vomeronasal 1 receptor, pseudogene 129 |
| 13 | gene | 23.159312 | 23.168154 | positive | MGI_C57BL6J_2159676 | Vmn1r219 | NCBI_Gene:171272,ENSEMBL:ENSMUSG00000061376 | MGI:2159676 | protein coding gene | vomeronasal 1 receptor 219 |
| 13 | pseudogene | 23.166611 | 23.167053 | positive | MGI_C57BL6J_3852462 | Vmn1r-ps130 | NCBI_Gene:100312558 | MGI:3852462 | pseudogene | vomeronasal 1 receptor, pseudogene 130 |
| 13 | gene | 23.178514 | 23.187975 | negative | MGI_C57BL6J_2159675 | Vmn1r220 | NCBI_Gene:171271,ENSEMBL:ENSMUSG00000096099 | MGI:2159675 | protein coding gene | vomeronasal 1 receptor 220 |
| 13 | pseudogene | 23.198292 | 23.198631 | negative | MGI_C57BL6J_3852468 | Vmn1r-ps131 | NCBI_Gene:100312559,ENSEMBL:ENSMUSG00000114699 | MGI:3852468 | pseudogene | vomeronasal 1 receptor, pseudogene 131 |
| 13 | pseudogene | 23.201161 | 23.202079 | negative | MGI_C57BL6J_4439079 | Vmn1r-ps132 | NCBI_Gene:100312560,ENSEMBL:ENSMUSG00000080794 | MGI:4439079 | pseudogene | vomeronasal 1 receptor, pseudogene 132 |
| 13 | gene | 23.214127 | 23.222913 | positive | MGI_C57BL6J_4438449 | Vmn1r221 | NCBI_Gene:100312485,ENSEMBL:ENSMUSG00000082316 | MGI:4438449 | protein coding gene | vomeronasal 1 receptor 221 |
| 13 | pseudogene | 23.228329 | 23.228773 | negative | MGI_C57BL6J_3852469 | Vmn1r-ps133 | NCBI_Gene:100312561,ENSEMBL:ENSMUSG00000114811 | MGI:3852469 | pseudogene | vomeronasal 1 receptor, pseudogene 133 |
| 13 | gene | 23.230836 | 23.235947 | negative | MGI_C57BL6J_2159679 | Vmn1r222 | NCBI_Gene:171274,ENSEMBL:ENSMUSG00000061022 | MGI:2159679 | protein coding gene | vomeronasal 1 receptor 222 |
| 13 | pseudogene | 23.241974 | 23.242365 | negative | MGI_C57BL6J_3852470 | Vmn1r-ps134 | NCBI_Gene:100312562 | MGI:3852470 | pseudogene | vomeronasal 1 receptor, pseudogene 134 |
| 13 | gene | 23.247134 | 23.253906 | positive | MGI_C57BL6J_3649245 | Vmn1r223 | NCBI_Gene:100036518,ENSEMBL:ENSMUSG00000069280 | MGI:3649245 | protein coding gene | vomeronasal 1 receptor 223 |
| 13 | gene | 23.251097 | 23.259019 | negative | MGI_C57BL6J_5592253 | Gm33094 | NCBI_Gene:102635862 | MGI:5592253 | lncRNA gene | predicted gene, 33094 |
| 13 | pseudogene | 23.266534 | 23.266821 | positive | MGI_C57BL6J_3649432 | Gm11331 | ENSEMBL:ENSMUSG00000082593 | MGI:3649432 | pseudogene | predicted gene 11331 |
| 13 | pseudogene | 23.268880 | 23.269501 | positive | MGI_C57BL6J_4439317 | Vmn1r-ps135 | NCBI_Gene:100312563,ENSEMBL:ENSMUSG00000084096 | MGI:4439317 | pseudogene | vomeronasal 1 receptor, pseudogene 135 |
| 13 | gene | 23.270421 | 23.270516 | negative | MGI_C57BL6J_4413916 | n-TIaat15 | NCBI_Gene:102467327 | MGI:4413916 | tRNA gene | nuclear encoded tRNA isoleucine 15 (anticodon TAT) |
| 13 | gene | 23.276204 | 23.313407 | negative | MGI_C57BL6J_1922572 | 4930557F10Rik | NCBI_Gene:75322,ENSEMBL:ENSMUSG00000085488 | MGI:1922572 | lncRNA gene | RIKEN cDNA 4930557F10 gene |
| 13 | gene | 23.278350 | 23.282267 | positive | MGI_C57BL6J_1921311 | 4933404K08Rik | NCBI_Gene:74061,ENSEMBL:ENSMUSG00000085122 | MGI:1921311 | lncRNA gene | RIKEN cDNA 4933404K08 gene |
| 13 | gene | 23.284323 | 23.286746 | negative | MGI_C57BL6J_5623717 | Gm40832 | NCBI_Gene:105245364 | MGI:5623717 | lncRNA gene | predicted gene, 40832 |
| 13 | gene | 23.285401 | 23.285473 | positive | MGI_C57BL6J_4414087 | n-TVaac4 | NCBI_Gene:102467400 | MGI:4414087 | tRNA gene | nuclear encoded tRNA valine 4 (anticodon AAC) |
| 13 | gene | 23.285649 | 23.285721 | positive | MGI_C57BL6J_4413683 | n-TAagc10 | NCBI_Gene:102467231 | MGI:4413683 | tRNA gene | nuclear encoded tRNA alanine 10 (anticodon AGC) |
| 13 | pseudogene | 23.298049 | 23.298196 | positive | MGI_C57BL6J_3649438 | Gm11333 | ENSEMBL:ENSMUSG00000080698 | MGI:3649438 | pseudogene | predicted gene 11333 |
| 13 | gene | 23.298781 | 23.298853 | positive | MGI_C57BL6J_4414089 | n-TVaac6 | NCBI_Gene:102467401 | MGI:4414089 | tRNA gene | nuclear encoded tRNA valine 6 (anticodon AAC) |
| 13 | gene | 23.299061 | 23.299133 | positive | MGI_C57BL6J_4413684 | n-TAagc11 | NCBI_Gene:102467451 | MGI:4413684 | tRNA gene | nuclear encoded tRNA alanine 11 (anticodon AGC) |
| 13 | gene | 23.300122 | 23.300194 | positive | MGI_C57BL6J_4414095 | n-TVcac4 | NCBI_Gene:102467404 | MGI:4414095 | tRNA gene | nuclear encoded tRNA valine 4 (anticodon CAC) |
| 13 | gene | 23.307629 | 23.307701 | positive | MGI_C57BL6J_4414102 | n-TVcac11 | NCBI_Gene:102467625 | MGI:4414102 | tRNA gene | nuclear encoded tRNA valine 11 (anticodon CAC) |
| 13 | gene | 23.309052 | 23.309125 | negative | MGI_C57BL6J_4413910 | n-TIaat9 | NCBI_Gene:102467431 | MGI:4413910 | tRNA gene | nuclear encoded tRNA isoleucine 9 (anticodon AAT) |
| 13 | gene | 23.309431 | 23.310169 | negative | MGI_C57BL6J_1922628 | 4930597G05Rik | NA | NA | unclassified gene | RIKEN cDNA 4930597G05 gene |
| 13 | gene | 23.313525 | 23.385799 | positive | MGI_C57BL6J_1923137 | 4930586N03Rik | NCBI_Gene:75887,ENSEMBL:ENSMUSG00000087097 | MGI:1923137 | lncRNA gene | RIKEN cDNA 4930586N03 gene |
| 13 | gene | 23.317466 | 23.317538 | positive | MGI_C57BL6J_4413989 | n-TMcat12 | NCBI_Gene:102467633 | MGI:4413989 | tRNA gene | nuclear encoded tRNA methionine 12 (anticodon CAT) |
| 13 | gene | 23.353103 | 23.369208 | negative | MGI_C57BL6J_2442566 | Zfp322a | NCBI_Gene:218100,ENSEMBL:ENSMUSG00000046351 | MGI:2442566 | protein coding gene | zinc finger protein 322A |
| 13 | pseudogene | 23.382297 | 23.382660 | positive | MGI_C57BL6J_3649250 | Gm11334 | NCBI_Gene:105245365,ENSEMBL:ENSMUSG00000083667 | MGI:3649250 | pseudogene | predicted gene 11334 |
| 13 | gene | 23.397060 | 23.397133 | positive | MGI_C57BL6J_4413904 | n-TIaat3 | NCBI_Gene:102467542 | MGI:4413904 | tRNA gene | nuclear encoded tRNA isoleucine 3 (anticodon AAT) |
| 13 | gene | 23.400795 | 23.400867 | negative | MGI_C57BL6J_4413685 | n-TAagc12 | NCBI_Gene:102467232 | MGI:4413685 | tRNA gene | nuclear encoded tRNA alanine 12 (anticodon AGC) |
| 13 | gene | 23.401073 | 23.401145 | negative | MGI_C57BL6J_4414085 | n-TVaac2 | NCBI_Gene:102467399 | MGI:4414085 | tRNA gene | nuclear encoded tRNA valine 2 (anticodon AAC) |
| 13 | gene | 23.412077 | 23.412149 | negative | MGI_C57BL6J_4414093 | n-TVcac2 | NCBI_Gene:102467403 | MGI:4414093 | tRNA gene | nuclear encoded tRNA valine 2 (anticodon CAC) |
| 13 | gene | 23.412941 | 23.413013 | negative | MGI_C57BL6J_4413680 | n-TAagc7 | NCBI_Gene:102467635 | MGI:4413680 | tRNA gene | nuclear encoded tRNA alanine 7 (anticodon AGC) |
| 13 | gene | 23.413248 | 23.413320 | negative | MGI_C57BL6J_4414088 | n-TVaac5 | NCBI_Gene:102467618 | MGI:4414088 | tRNA gene | nuclear encoded tRNA valine 5 (anticodon AAC) |
| 13 | gene | 23.418361 | 23.423866 | negative | MGI_C57BL6J_1353636 | Abt1 | NCBI_Gene:30946,ENSEMBL:ENSMUSG00000036376 | MGI:1353636 | protein coding gene | activator of basal transcription 1 |
| 13 | gene | 23.423985 | 23.425251 | positive | MGI_C57BL6J_1916940 | 2310057B04Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 2310057B04 gene |
| 13 | gene | 23.425646 | 23.425734 | negative | MGI_C57BL6J_4414076 | n-TYgta3 | NCBI_Gene:102467613 | MGI:4414076 | tRNA gene | nuclear encoded tRNA tyrosine 3 (anticodon GTA) |
| 13 | gene | 23.426505 | 23.426592 | positive | MGI_C57BL6J_4414077 | n-TYgta4 | NCBI_Gene:102467396 | MGI:4414077 | tRNA gene | nuclear encoded tRNA tyrosine 4 (anticodon GTA) |
| 13 | gene | 23.427095 | 23.427180 | negative | MGI_C57BL6J_4414078 | n-TYgta5 | NCBI_Gene:102467614 | MGI:4414078 | tRNA gene | nuclear encoded tRNA tyrosine 5 (anticodon GTA) |
| 13 | gene | 23.427975 | 23.431022 | negative | MGI_C57BL6J_2444850 | C230035I16Rik | NCBI_Gene:320842,ENSEMBL:ENSMUSG00000085024 | MGI:2444850 | lncRNA gene | RIKEN cDNA C230035I16 gene |
| 13 | gene | 23.428738 | 23.428826 | negative | MGI_C57BL6J_4414079 | n-TYgta6 | NCBI_Gene:102467397 | MGI:4414079 | tRNA gene | nuclear encoded tRNA tyrosine 6 (anticodon GTA) |
| 13 | gene | 23.431412 | 23.431484 | positive | MGI_C57BL6J_4413686 | n-TAagc13 | NCBI_Gene:102467452 | MGI:4413686 | tRNA gene | nuclear encoded tRNA alanine 13 (anticodon AGC) |
| 13 | gene | 23.432990 | 23.433077 | negative | MGI_C57BL6J_4414080 | n-TYgta7 | NCBI_Gene:102467446 | MGI:4414080 | tRNA gene | nuclear encoded tRNA tyrosine 7 (anticodon GTA) |
| 13 | gene | 23.433296 | 23.434672 | negative | MGI_C57BL6J_5623718 | Gm40833 | NCBI_Gene:105245366 | MGI:5623718 | lncRNA gene | predicted gene, 40833 |
| 13 | gene | 23.436340 | 23.436412 | positive | MGI_C57BL6J_4413958 | n-TKctt11 | NCBI_Gene:102467345 | MGI:4413958 | tRNA gene | nuclear encoded tRNA lysine 11 (anticodon CTT) |
| 13 | gene | 23.436994 | 23.437065 | negative | MGI_C57BL6J_4414005 | n-TPagg3 | NCBI_Gene:102467365 | MGI:4414005 | tRNA gene | nuclear encoded tRNA proline 3 (anticodon AGG) |
| 13 | gene | 23.438109 | 23.438182 | negative | MGI_C57BL6J_4413909 | n-TIaat8 | NCBI_Gene:102467324 | MGI:4413909 | tRNA gene | nuclear encoded tRNA isoleucine 8 (anticodon AAT) |
| 13 | gene | 23.438697 | 23.438768 | negative | MGI_C57BL6J_4413702 | n-TAcgc10 | NCBI_Gene:102467239 | MGI:4413702 | tRNA gene | nuclear encoded tRNA alanine 10 (anticodon CGC) |
| 13 | gene | 23.441820 | 23.445990 | negative | MGI_C57BL6J_5826041 | Gm46404 | NCBI_Gene:108168069,ENSEMBL:ENSMUSG00000113440 | MGI:5826041 | lncRNA gene | predicted gene, 46404 |
| 13 | gene | 23.446142 | 23.446214 | negative | MGI_C57BL6J_4414100 | n-TVcac9 | NCBI_Gene:102467624 | MGI:4414100 | tRNA gene | nuclear encoded tRNA valine 9 (anticodon CAC) |
| 13 | gene | 23.446792 | 23.446864 | negative | MGI_C57BL6J_4413718 | n-TRacg5 | NCBI_Gene:102467465 | MGI:4413718 | tRNA gene | nuclear encoded tRNA arginine 5 (anticodon ACG) |
| 13 | gene | 23.450622 | 23.450695 | positive | MGI_C57BL6J_4414053 | n-TTagt6 | NCBI_Gene:102467386 | MGI:4414053 | tRNA gene | nuclear encoded tRNA threonine 6 (anticodon AGT) |
| 13 | gene | 23.453432 | 23.453514 | negative | MGI_C57BL6J_4413940 | n-TLcag10 | NCBI_Gene:102467557 | MGI:4413940 | tRNA gene | nuclear encoded tRNA leucine 10 (anticodon CAG) |
| 13 | gene | 23.456984 | 23.465914 | negative | MGI_C57BL6J_103118 | Btn1a1 | NCBI_Gene:12231,ENSEMBL:ENSMUSG00000000706 | MGI:103118 | protein coding gene | butyrophilin, subfamily 1, member A1 |
| 13 | gene | 23.463535 | 23.463606 | positive | MGI_C57BL6J_4413858 | n-TEttc5 | NCBI_Gene:102467304 | MGI:4413858 | tRNA gene | nuclear encoded tRNA glutamic acid 5 (anticodon TTC) |
| 13 | gene | 23.467076 | 23.467165 | positive | MGI_C57BL6J_4414081 | n-TYgta8 | NCBI_Gene:102467615 | MGI:4414081 | tRNA gene | nuclear encoded tRNA tyrosine 8 (anticodon GTA) |
| 13 | gene | 23.477049 | 23.488858 | negative | MGI_C57BL6J_3606486 | Btn2a2 | NCBI_Gene:238555,ENSEMBL:ENSMUSG00000053216 | MGI:3606486 | protein coding gene | butyrophilin, subfamily 2, member A2 |
| 13 | gene | 23.498106 | 23.498177 | positive | MGI_C57BL6J_4414068 | n-TWcca3 | NCBI_Gene:102467392 | MGI:4414068 | tRNA gene | nuclear encoded tRNA trytophan 3 (anticodon CCA) |
| 13 | gene | 23.500080 | 23.500151 | positive | MGI_C57BL6J_4413982 | n-TMcat5 | NCBI_Gene:102467435 | MGI:4413982 | tRNA gene | nuclear encoded tRNA methionine 5 (anticodon CAT) |
| 13 | gene | 23.500522 | 23.500594 | negative | MGI_C57BL6J_4413717 | n-TRacg4 | NCBI_Gene:102467245 | MGI:4413717 | tRNA gene | nuclear encoded tRNA arginine 4 (anticodon ACG) |
| 13 | gene | 23.500957 | 23.501038 | negative | MGI_C57BL6J_4414027 | n-TSaga7 | NCBI_Gene:102467374 | MGI:4414027 | tRNA gene | nuclear encoded tRNA serine 7 (anticodon AGA) |
| 13 | gene | 23.503980 | 23.504052 | negative | MGI_C57BL6J_4413730 | n-TRtcg3 | NCBI_Gene:102467251 | MGI:4413730 | tRNA gene | nuclear encoded tRNA arginine 3 (anticodon TCG) |
| 13 | gene | 23.506519 | 23.506590 | positive | MGI_C57BL6J_4414069 | n-TWcca4 | NCBI_Gene:102467610 | MGI:4414069 | tRNA gene | nuclear encoded tRNA trytophan 4 (anticodon CCA) |
| 13 | gene | 23.517203 | 23.517273 | positive | MGI_C57BL6J_4413875 | n-TGgcc6 | NCBI_Gene:102467311 | MGI:4413875 | tRNA gene | nuclear encoded tRNA glycine 6 (anticodon GCC) |
| 13 | gene | 23.517692 | 23.517763 | positive | MGI_C57BL6J_4413983 | n-TMcat6 | NCBI_Gene:102467575 | MGI:4413983 | tRNA gene | nuclear encoded tRNA methionine 6 (anticodon CAT) |
| 13 | gene | 23.518142 | 23.518250 | positive | MGI_C57BL6J_4414044 | n-TStga1 | NCBI_Gene:102467382,ENSEMBL:ENSMUSG00000104750 | MGI:4414044 | tRNA gene | nuclear encoded tRNA serine 1 (anticodon TGA) |
| 13 | gene | 23.519050 | 23.519121 | positive | MGI_C57BL6J_4413838 | n-TQttg4 | NCBI_Gene:102467296 | MGI:4413838 | tRNA gene | nuclear encoded tRNA glutamine 4 (anticodon TTG) |
| 13 | gene | 23.519613 | 23.519684 | positive | MGI_C57BL6J_4413839 | n-TQttg5 | NCBI_Gene:102467516 | MGI:4413839 | tRNA gene | nuclear encoded tRNA glutamine 5 (anticodon TTG) |
| 13 | gene | 23.521870 | 23.521951 | positive | MGI_C57BL6J_4414038 | n-TSgct4 | NCBI_Gene:102467379 | MGI:4414038 | tRNA gene | nuclear encoded tRNA serine 4 (anticodon GCT) |
| 13 | gene | 23.526456 | 23.526528 | negative | MGI_C57BL6J_4413728 | n-TRtcg1 | NCBI_Gene:102467250 | MGI:4413728 | tRNA gene | nuclear encoded tRNA arginine 1 (anticodon TCG) |
| 13 | gene | 23.526997 | 23.530943 | negative | MGI_C57BL6J_3705194 | Gm11335 | NCBI_Gene:100034675,ENSEMBL:ENSMUSG00000085016 | MGI:3705194 | lncRNA gene | predicted gene 11335 |
| 13 | gene | 23.530348 | 23.530419 | negative | MGI_C57BL6J_4413986 | n-TMcat9 | NCBI_Gene:102467357 | MGI:4413986 | tRNA gene | nuclear encoded tRNA methionine 9 (anticodon CAT) |
| 13 | gene | 23.531044 | 23.531519 | positive | MGI_C57BL6J_2448427 | H2c8 | NCBI_Gene:69386,ENSEMBL:ENSMUSG00000060981 | MGI:2448427 | protein coding gene | H4 clustered histone 8 |
| 13 | gene | 23.533906 | 23.534378 | positive | MGI_C57BL6J_2448309 | H2ac10 | NCBI_Gene:319173,ENSEMBL:ENSMUSG00000061991 | MGI:2448309 | protein coding gene | H2A clustered histone 10 |
| 13 | gene | 23.534114 | 23.534254 | negative | MGI_C57BL6J_5690689 | Gm44297 | ENSEMBL:ENSMUSG00000105109 | MGI:5690689 | miRNA gene | predicted gene, 44297 |
| 13 | gene | 23.535418 | 23.535951 | positive | MGI_C57BL6J_2145541 | H3c8 | NCBI_Gene:97908,ENSEMBL:ENSMUSG00000099517 | MGI:2145541 | protein coding gene | H3 clustered histone 8 |
| 13 | gene | 23.541714 | 23.543444 | negative | MGI_C57BL6J_2448387 | H2bc9 | NCBI_Gene:319182,ENSEMBL:ENSMUSG00000114456 | MGI:2448387 | protein coding gene | H2B clustered histone 9 |
| 13 | gene | 23.544052 | 23.545312 | positive | MGI_C57BL6J_2448329 | H3c7 | NCBI_Gene:260423,ENSEMBL:ENSMUSG00000100210 | MGI:2448329 | protein coding gene | H3 clustered histone 7 |
| 13 | gene | 23.551258 | 23.551648 | negative | MGI_C57BL6J_2448425 | H4c6 | NCBI_Gene:319157,ENSEMBL:ENSMUSG00000069274 | MGI:2448425 | protein coding gene | H4 clustered histone 6 |
| 13 | gene | 23.553262 | 23.560686 | positive | MGI_C57BL6J_107502 | H1f3 | NCBI_Gene:14957,ENSEMBL:ENSMUSG00000052565 | MGI:107502 | protein coding gene | H1.3 linker histone, cluster member |
| 13 | gene | 23.561534 | 23.562369 | negative | MGI_C57BL6J_2448326 | H3c6 | NCBI_Gene:319151,ENSEMBL:ENSMUSG00000069273 | MGI:2448326 | protein coding gene | H3 clustered histone 6 |
| 13 | gene | 23.570517 | 23.571220 | negative | MGI_C57BL6J_2448290 | H2ac8 | NCBI_Gene:319166,ENSEMBL:ENSMUSG00000069272 | MGI:2448290 | protein coding gene | H2A clustered histone 8 |
| 13 | gene | 23.570726 | 23.570863 | positive | MGI_C57BL6J_5690786 | Gm44394 | ENSEMBL:ENSMUSG00000104771 | MGI:5690786 | miRNA gene | predicted gene, 44394 |
| 13 | gene | 23.571396 | 23.572013 | positive | MGI_C57BL6J_2448386 | H2bc8 | NCBI_Gene:319181,ENSEMBL:ENSMUSG00000058385 | MGI:2448386 | protein coding gene | H2B clustered histone 8 |
| 13 | gene | 23.573736 | 23.574196 | negative | MGI_C57BL6J_2448383 | H2bc7 | NCBI_Gene:319180,ENSEMBL:ENSMUSG00000069268 | MGI:2448383 | protein coding gene | H2B clustered histone 7 |
| 13 | gene | 23.574381 | 23.574952 | positive | MGI_C57BL6J_2448289 | H2ac7 | NCBI_Gene:319165,ENSEMBL:ENSMUSG00000071478 | MGI:2448289 | protein coding gene | H2A clustered histone 7 |
| 13 | gene | 23.574716 | 23.574853 | negative | MGI_C57BL6J_5690751 | Gm44359 | ENSEMBL:ENSMUSG00000105924 | MGI:5690751 | miRNA gene | predicted gene, 44359 |
| 13 | gene | 23.575763 | 23.577011 | positive | MGI_C57BL6J_2448322 | H3c4 | NCBI_Gene:319149,ENSEMBL:ENSMUSG00000099583 | MGI:2448322 | protein coding gene | H3 clustered histone 4 |
| 13 | gene | 23.581598 | 23.582735 | positive | MGI_C57BL6J_2448423 | H4c4 | NCBI_Gene:319156,ENSEMBL:ENSMUSG00000061482 | MGI:2448423 | protein coding gene | H4 clustered histone 4 |
| 13 | gene | 23.583670 | 23.621124 | negative | MGI_C57BL6J_2448380 | H2bc6 | NCBI_Gene:319179,ENSEMBL:ENSMUSG00000047246 | MGI:2448380 | protein coding gene | H2B clustered histone 6 |
| 13 | gene | 23.591186 | 23.606239 | positive | MGI_C57BL6J_5592546 | Gm33387 | NCBI_Gene:102636274 | MGI:5592546 | lncRNA gene | predicted gene, 33387 |
| 13 | gene | 23.620629 | 23.622558 | negative | MGI_C57BL6J_1931527 | H1f4 | NCBI_Gene:50709,ENSEMBL:ENSMUSG00000051627 | MGI:1931527 | protein coding gene | H1.4 linker histone, cluster member |
| 13 | pseudogene | 23.620730 | 23.621049 | negative | MGI_C57BL6J_3805547 | Gm11398 | ENSEMBL:ENSMUSG00000080741 | MGI:3805547 | pseudogene | predicted gene 11398 |
| 13 | gene | 23.663021 | 23.663124 | positive | MGI_C57BL6J_5452229 | Gm22452 | ENSEMBL:ENSMUSG00000084753 | MGI:5452229 | snRNA gene | predicted gene, 22452 |
| 13 | gene | 23.681467 | 23.683959 | negative | MGI_C57BL6J_2448287 | H2ac6 | NCBI_Gene:319164,ENSEMBL:ENSMUSG00000069270 | MGI:2448287 | protein coding gene | H2A clustered histone 6 |
| 13 | gene | 23.682545 | 23.682652 | negative | MGI_C57BL6J_5454768 | Gm24991 | ENSEMBL:ENSMUSG00000088030 | MGI:5454768 | rRNA gene | predicted gene, 24991 |
| 13 | gene | 23.683535 | 23.683672 | positive | MGI_C57BL6J_5690742 | Gm44350 | ENSEMBL:ENSMUSG00000104997 | MGI:5690742 | miRNA gene | predicted gene, 44350 |
| 13 | gene | 23.684199 | 23.692908 | positive | MGI_C57BL6J_1915274 | H2bc4 | NCBI_Gene:68024,ENSEMBL:ENSMUSG00000018102 | MGI:1915274 | protein coding gene | H2B clustered histone 4 |
| 13 | gene | 23.695811 | 23.696725 | positive | MGI_C57BL6J_1888530 | H1f6 | NCBI_Gene:107970,ENSEMBL:ENSMUSG00000036211 | MGI:1888530 | protein coding gene | H1.6 linker histone, cluster member |
| 13 | gene | 23.697635 | 23.698458 | negative | MGI_C57BL6J_2448421 | H4c3 | NCBI_Gene:319155,ENSEMBL:ENSMUSG00000060678 | MGI:2448421 | protein coding gene | H4 clustered histone 3 |
| 13 | gene | 23.702034 | 23.710986 | negative | MGI_C57BL6J_109191 | Hfe | NCBI_Gene:15216,ENSEMBL:ENSMUSG00000006611 | MGI:109191 | protein coding gene | homeostatic iron regulator |
| 13 | pseudogene | 23.728548 | 23.730106 | positive | MGI_C57BL6J_3651857 | Gm11338 | NCBI_Gene:100504088,ENSEMBL:ENSMUSG00000083206 | MGI:3651857 | pseudogene | predicted gene 11338 |
| 13 | gene | 23.738807 | 23.740367 | positive | MGI_C57BL6J_1931526 | H1f2 | NCBI_Gene:50708,ENSEMBL:ENSMUSG00000036181 | MGI:1931526 | protein coding gene | H1.2 linker histone, cluster member |
| 13 | gene | 23.744973 | 23.745602 | negative | MGI_C57BL6J_2448320 | H3c3 | NCBI_Gene:319148,ENSEMBL:ENSMUSG00000069310 | MGI:2448320 | protein coding gene | H3 clustered histone 3 |
| 13 | pseudogene | 23.746166 | 23.746499 | negative | MGI_C57BL6J_3651860 | H2ac5-ps | ENSEMBL:ENSMUSG00000080999 | MGI:3651860 | pseudogene | H2A clustered histone 5, pseudogene |
| 13 | gene | 23.746175 | 23.746312 | positive | MGI_C57BL6J_5690706 | Gm44314 | ENSEMBL:ENSMUSG00000106595 | MGI:5690706 | miRNA gene | predicted gene, 44314 |
| 13 | gene | 23.746734 | 23.747241 | positive | MGI_C57BL6J_2448377 | H2bc3 | NCBI_Gene:319178,ENSEMBL:ENSMUSG00000075031 | MGI:2448377 | protein coding gene | H2B clustered histone 3 |
| 13 | gene | 23.751088 | 23.751598 | positive | MGI_C57BL6J_2448306 | H2ac4 | NCBI_Gene:319172,ENSEMBL:ENSMUSG00000061615 | MGI:2448306 | protein coding gene | H2A clustered histone 4 |
| 13 | gene | 23.751378 | 23.751515 | negative | MGI_C57BL6J_5690876 | Gm44484 | ENSEMBL:ENSMUSG00000106203 | MGI:5690876 | miRNA gene | predicted gene, 44484 |
| 13 | gene | 23.752267 | 23.752886 | positive | MGI_C57BL6J_2448319 | H3c2 | NCBI_Gene:319150,ENSEMBL:ENSMUSG00000069267 | MGI:2448319 | protein coding gene | H3 clustered histone 2 |
| 13 | gene | 23.755822 | 23.757620 | negative | MGI_C57BL6J_3590517 | 4930558J22Rik | ENSEMBL:ENSMUSG00000075029 | MGI:3590517 | lncRNA gene | RIKEN cDNA 4930558J22 gene |
| 13 | gene | 23.756937 | 23.757409 | positive | MGI_C57BL6J_2448420 | H4c2 | NCBI_Gene:326620,ENSEMBL:ENSMUSG00000069266 | MGI:2448420 | protein coding gene | H4 clustered histone 2 |
| 13 | gene | 23.760692 | 23.761249 | negative | MGI_C57BL6J_2448419 | H4c1 | NCBI_Gene:326619,ENSEMBL:ENSMUSG00000060093 | MGI:2448419 | protein coding gene | H4 clustered histone 1 |
| 13 | gene | 23.761853 | 23.762386 | negative | MGI_C57BL6J_2668828 | H3c1 | NCBI_Gene:360198,ENSEMBL:ENSMUSG00000069265 | MGI:2668828 | protein coding gene | H3 clustered histone 1 |
| 13 | gene | 23.763666 | 23.764412 | positive | MGI_C57BL6J_1931523 | H1f1 | NCBI_Gene:80838,ENSEMBL:ENSMUSG00000049539 | MGI:1931523 | protein coding gene | H1.1 linker histone, cluster member |
| 13 | gene | 23.769913 | 23.791731 | positive | MGI_C57BL6J_2684869 | Trim38 | NCBI_Gene:214158,ENSEMBL:ENSMUSG00000064140 | MGI:2684869 | protein coding gene | tripartite motif-containing 38 |
| 13 | gene | 23.794402 | 23.804640 | negative | MGI_C57BL6J_3651858 | Gm11337 | NCBI_Gene:105245367,ENSEMBL:ENSMUSG00000087173 | MGI:3651858 | lncRNA gene | predicted gene 11337 |
| 13 | gene | 23.806940 | 23.825191 | positive | MGI_C57BL6J_2443098 | Slc17a2 | NCBI_Gene:218103,ENSEMBL:ENSMUSG00000036110 | MGI:2443098 | protein coding gene | solute carrier family 17 (sodium phosphate), member 2 |
| 13 | gene | 23.836917 | 23.860718 | positive | MGI_C57BL6J_2389216 | Slc17a3 | NCBI_Gene:105355,ENSEMBL:ENSMUSG00000036083 | MGI:2389216 | protein coding gene | solute carrier family 17 (sodium phosphate), member 3 |
| 13 | gene | 23.867723 | 23.895730 | positive | MGI_C57BL6J_103209 | Slc17a1 | NCBI_Gene:20504,ENSEMBL:ENSMUSG00000021335 | MGI:103209 | protein coding gene | solute carrier family 17 (sodium phosphate), member 1 |
| 13 | gene | 23.895890 | 23.915009 | negative | MGI_C57BL6J_2442850 | Slc17a4 | NCBI_Gene:319848,ENSEMBL:ENSMUSG00000021336 | MGI:2442850 | protein coding gene | solute carrier family 17 (sodium phosphate), member 4 |
| 13 | pseudogene | 23.923692 | 23.924164 | negative | MGI_C57BL6J_109487 | Snrp1c-ps2 | NCBI_Gene:20632,ENSEMBL:ENSMUSG00000082558 | MGI:109487 | pseudogene | U1 small nuclear ribonucleoprotein 1C, pseudogene 2 |
| 13 | gene | 23.933086 | 23.949115 | positive | MGI_C57BL6J_5592675 | Gm33516 | NCBI_Gene:102636457 | MGI:5592675 | lncRNA gene | predicted gene, 33516 |
| 13 | gene | 23.933725 | 23.934156 | negative | MGI_C57BL6J_2448375 | H2bc1 | NCBI_Gene:319177,ENSEMBL:ENSMUSG00000050799 | MGI:2448375 | protein coding gene | H2B clustered histone 1 |
| 13 | gene | 23.934462 | 23.934913 | positive | MGI_C57BL6J_2448285 | H2ac1 | NCBI_Gene:319163,ENSEMBL:ENSMUSG00000060081 | MGI:2448285 | protein coding gene | H2A clustered histone 1 |
| 13 | gene | 23.934707 | 23.934835 | negative | MGI_C57BL6J_5690736 | Gm44344 | ENSEMBL:ENSMUSG00000105874 | MGI:5690736 | miRNA gene | predicted gene, 44344 |
| 13 | gene | 23.953456 | 23.991330 | negative | MGI_C57BL6J_2384873 | Scgn | NCBI_Gene:214189,ENSEMBL:ENSMUSG00000021337 | MGI:2384873 | protein coding gene | secretagogin, EF-hand calcium binding protein |
| 13 | gene | 23.956809 | 23.966175 | positive | MGI_C57BL6J_3652101 | Gm11339 | ENSEMBL:ENSMUSG00000087603 | MGI:3652101 | lncRNA gene | predicted gene 11339 |
| 13 | gene | 24.012344 | 24.280795 | negative | MGI_C57BL6J_1915982 | Carmil1 | NCBI_Gene:68732,ENSEMBL:ENSMUSG00000021338 | MGI:1915982 | protein coding gene | capping protein regulator and myosin 1 linker 1 |
| 13 | gene | 24.059425 | 24.067678 | positive | MGI_C57BL6J_1918449 | 4933427I18Rik | NA | NA | protein coding gene | RIKEN cDNA 4933427I18 gene |
| 13 | gene | 24.178265 | 24.189252 | positive | MGI_C57BL6J_5592732 | Gm33573 | NCBI_Gene:102636534 | MGI:5592732 | lncRNA gene | predicted gene, 33573 |
| 13 | pseudogene | 24.195897 | 24.196518 | positive | MGI_C57BL6J_3650912 | Gm11343 | NCBI_Gene:100416503,ENSEMBL:ENSMUSG00000081707 | MGI:3650912 | pseudogene | predicted gene 11343 |
| 13 | pseudogene | 24.291394 | 24.291805 | negative | MGI_C57BL6J_3652096 | Gm11340 | ENSEMBL:ENSMUSG00000083409 | MGI:3652096 | pseudogene | predicted gene 11340 |
| 13 | gene | 24.316730 | 24.316852 | negative | MGI_C57BL6J_5453117 | Gm23340 | ENSEMBL:ENSMUSG00000089580 | MGI:5453117 | snoRNA gene | predicted gene, 23340 |
| 13 | pseudogene | 24.320218 | 24.320971 | positive | MGI_C57BL6J_3652095 | Gm11341 | NCBI_Gene:108168051,ENSEMBL:ENSMUSG00000081909 | MGI:3652095 | pseudogene | predicted gene 11341 |
| 13 | gene | 24.324108 | 24.414803 | negative | MGI_C57BL6J_3652098 | Gm11342 | NCBI_Gene:102636681,ENSEMBL:ENSMUSG00000084788 | MGI:3652098 | lncRNA gene | predicted gene 11342 |
| 13 | gene | 24.327404 | 24.477377 | positive | MGI_C57BL6J_103227 | Cmah | NCBI_Gene:12763,ENSEMBL:ENSMUSG00000016756 | MGI:103227 | protein coding gene | cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| 13 | pseudogene | 24.410106 | 24.410440 | negative | MGI_C57BL6J_3652103 | Gm11344 | ENSEMBL:ENSMUSG00000083165 | MGI:3652103 | pseudogene | predicted gene 11344 |
| 13 | gene | 24.486662 | 24.486764 | negative | MGI_C57BL6J_5451790 | Gm22013 | ENSEMBL:ENSMUSG00000089413 | MGI:5451790 | snRNA gene | predicted gene, 22013 |
| 13 | gene | 24.501541 | 24.733816 | positive | MGI_C57BL6J_2444879 | Ripor2 | NCBI_Gene:193385,ENSEMBL:ENSMUSG00000036006 | MGI:2444879 | protein coding gene | RHO family interacting cell polarization regulator 2 |
| 13 | pseudogene | 24.596347 | 24.604925 | negative | MGI_C57BL6J_1923274 | Gm11346 | NCBI_Gene:76024,ENSEMBL:ENSMUSG00000085603 | MGI:1923274 | pseudogene | predicted gene 11346 |
| 13 | gene | 24.631918 | 24.638567 | negative | MGI_C57BL6J_2444135 | C530050E15Rik | ENSEMBL:ENSMUSG00000097838 | MGI:2444135 | lncRNA gene | RIKEN cDNA C530050E15 gene |
| 13 | gene | 24.733875 | 24.746592 | positive | MGI_C57BL6J_1916676 | Armh2 | NCBI_Gene:666145,ENSEMBL:ENSMUSG00000085861 | MGI:1916676 | protein coding gene | armadillo-like helical domain containing 2 |
| 13 | gene | 24.751845 | 24.761937 | negative | MGI_C57BL6J_1927344 | Gmnn | NCBI_Gene:57441,ENSEMBL:ENSMUSG00000006715 | MGI:1927344 | protein coding gene | geminin |
| 13 | gene | 24.801657 | 24.816197 | positive | MGI_C57BL6J_2441726 | BC005537 | NCBI_Gene:79555,ENSEMBL:ENSMUSG00000019132 | MGI:2441726 | protein coding gene | cDNA sequence BC005537 |
| 13 | gene | 24.811447 | 24.811568 | negative | MGI_C57BL6J_5452135 | Gm22358 | ENSEMBL:ENSMUSG00000064811 | MGI:5452135 | snoRNA gene | predicted gene, 22358 |
| 13 | gene | 24.817948 | 24.831540 | negative | MGI_C57BL6J_1914084 | Acot13 | NCBI_Gene:66834,ENSEMBL:ENSMUSG00000006717 | MGI:1914084 | protein coding gene | acyl-CoA thioesterase 13 |
| 13 | gene | 24.831659 | 24.842153 | positive | MGI_C57BL6J_1860486 | Tdp2 | NCBI_Gene:56196,ENSEMBL:ENSMUSG00000035958 | MGI:1860486 | protein coding gene | tyrosyl-DNA phosphodiesterase 2 |
| 13 | gene | 24.845131 | 24.901439 | positive | MGI_C57BL6J_3036268 | D130043K22Rik | NCBI_Gene:210108,ENSEMBL:ENSMUSG00000006711 | MGI:3036268 | protein coding gene | RIKEN cDNA D130043K22 gene |
| 13 | gene | 24.905593 | 24.914272 | positive | MGI_C57BL6J_1921673 | 4932702P03Rik | NCBI_Gene:73326,ENSEMBL:ENSMUSG00000086796 | MGI:1921673 | lncRNA gene | RIKEN cDNA 4932702P03 gene |
| 13 | gene | 24.907579 | 24.937661 | negative | MGI_C57BL6J_2441982 | Aldh5a1 | NCBI_Gene:214579,ENSEMBL:ENSMUSG00000035936 | MGI:2441982 | protein coding gene | aldhehyde dehydrogenase family 5, subfamily A1 |
| 13 | gene | 24.937401 | 24.941417 | positive | MGI_C57BL6J_3604449 | 9330162012Rik | NCBI_Gene:493800,ENSEMBL:ENSMUSG00000086448 | MGI:3604449 | lncRNA gene | cDNA RIKEN 9330162012 gene |
| 13 | gene | 24.937785 | 24.992501 | positive | MGI_C57BL6J_106604 | Gpld1 | NCBI_Gene:14756,ENSEMBL:ENSMUSG00000021340 | MGI:106604 | protein coding gene | glycosylphosphatidylinositol specific phospholipase D1 |
| 13 | gene | 24.965108 | 24.968304 | negative | MGI_C57BL6J_5593178 | Gm34019 | NCBI_Gene:102637132 | MGI:5593178 | lncRNA gene | predicted gene, 34019 |
| 13 | gene | 24.987483 | 25.020379 | negative | MGI_C57BL6J_2685748 | Mrs2 | NCBI_Gene:380836,ENSEMBL:ENSMUSG00000021339 | MGI:2685748 | protein coding gene | MRS2 magnesium transporter |
| 13 | gene | 24.989319 | 24.989645 | negative | MGI_C57BL6J_3642849 | Gm9983 | NA | NA | protein coding gene | predicted gene 9983 |
| 13 | pseudogene | 25.035059 | 25.035328 | positive | MGI_C57BL6J_6096662 | Gm47605 | ENSEMBL:ENSMUSG00000114228 | MGI:6096662 | pseudogene | predicted gene, 47605 |
| 13 | gene | 25.055232 | 25.210706 | positive | MGI_C57BL6J_2652818 | Dcdc2a | NCBI_Gene:195208,ENSEMBL:ENSMUSG00000035910 | MGI:2652818 | protein coding gene | doublecortin domain containing 2a |
| 13 | pseudogene | 25.130747 | 25.131064 | negative | MGI_C57BL6J_3652295 | Gm11348 | ENSEMBL:ENSMUSG00000082667 | MGI:3652295 | pseudogene | predicted gene 11348 |
| 13 | gene | 25.170722 | 25.171831 | positive | MGI_C57BL6J_1922157 | 4930483C01Rik | NA | NA | unclassified gene | RIKEN cDNA 4930483C01 gene |
| 13 | gene | 25.252039 | 25.270502 | negative | MGI_C57BL6J_894662 | Nrsn1 | NCBI_Gene:22360,ENSEMBL:ENSMUSG00000048978 | MGI:894662 | protein coding gene | neurensin 1 |
| 13 | gene | 25.360511 | 25.371720 | positive | MGI_C57BL6J_5593281 | Gm34122 | NCBI_Gene:102637258 | MGI:5593281 | lncRNA gene | predicted gene, 34122 |
| 13 | gene | 25.428169 | 25.430127 | negative | MGI_C57BL6J_2442047 | F830002E08Rik | ENSEMBL:ENSMUSG00000114427 | MGI:2442047 | unclassified gene | RIKEN cDNA F830002E08 gene |
| 13 | pseudogene | 25.455710 | 25.456569 | positive | MGI_C57BL6J_98806 | Tpi-rs8 | NCBI_Gene:193438,ENSEMBL:ENSMUSG00000083919 | MGI:98806 | pseudogene | triosephosphate isomerase related sequence 8 |
| 13 | pseudogene | 25.582119 | 25.583465 | negative | MGI_C57BL6J_3652298 | Gm11349 | NCBI_Gene:214973,ENSEMBL:ENSMUSG00000080840 | MGI:3652298 | pseudogene | predicted gene 11349 |
| 13 | pseudogene | 25.680274 | 25.681146 | negative | MGI_C57BL6J_3652299 | Gm11350 | NCBI_Gene:100416798,ENSEMBL:ENSMUSG00000082161 | MGI:3652299 | pseudogene | predicted gene 11350 |
| 13 | gene | 25.921033 | 25.986293 | negative | MGI_C57BL6J_3649797 | Gm11351 | NCBI_Gene:100503530,ENSEMBL:ENSMUSG00000086531 | MGI:3649797 | lncRNA gene | predicted gene 11351 |
| 13 | pseudogene | 26.194676 | 26.195314 | positive | MGI_C57BL6J_3649796 | Gm11352 | NCBI_Gene:666221,ENSEMBL:ENSMUSG00000084358 | MGI:3649796 | pseudogene | predicted gene 11352 |
| 13 | gene | 26.283150 | 26.312406 | negative | MGI_C57BL6J_1920826 | 1700092E19Rik | NCBI_Gene:73576,ENSEMBL:ENSMUSG00000084811 | MGI:1920826 | lncRNA gene | RIKEN cDNA 1700092E19 gene |
| 13 | gene | 26.358954 | 26.360026 | negative | MGI_C57BL6J_6097107 | Gm47881 | ENSEMBL:ENSMUSG00000114660 | MGI:6097107 | unclassified gene | predicted gene, 47881 |
| 13 | pseudogene | 26.492502 | 26.493219 | positive | MGI_C57BL6J_3649794 | Gm11353 | NCBI_Gene:100042180,ENSEMBL:ENSMUSG00000060198 | MGI:3649794 | pseudogene | predicted gene 11353 |
| 13 | pseudogene | 26.680568 | 26.680693 | negative | MGI_C57BL6J_6097111 | Gm47883 | ENSEMBL:ENSMUSG00000114225 | MGI:6097111 | pseudogene | predicted gene, 47883 |
| 13 | gene | 26.768172 | 26.770165 | negative | MGI_C57BL6J_1194493 | Hdgfl1 | NCBI_Gene:15192,ENSEMBL:ENSMUSG00000045835 | MGI:1194493 | protein coding gene | HDGF like 1 |
| 13 | gene | 26.769393 | 26.772727 | positive | MGI_C57BL6J_1918166 | 4921520N01Rik | ENSEMBL:ENSMUSG00000114342 | MGI:1918166 | lncRNA gene | RIKEN cDNA 4921520N01 gene |
| 13 | pseudogene | 26.787312 | 26.787786 | negative | MGI_C57BL6J_3650603 | Gm11354 | NCBI_Gene:108168039,ENSEMBL:ENSMUSG00000081653 | MGI:3650603 | pseudogene | predicted gene 11354 |
| 13 | pseudogene | 26.899691 | 26.899996 | negative | MGI_C57BL6J_3649791 | Gm11355 | ENSEMBL:ENSMUSG00000082236 | MGI:3649791 | pseudogene | predicted gene 11355 |
| 13 | gene | 26.968491 | 27.058022 | negative | MGI_C57BL6J_6097118 | Gm47888 | ENSEMBL:ENSMUSG00000114646 | MGI:6097118 | lncRNA gene | predicted gene, 47888 |
| 13 | gene | 27.057570 | 27.065205 | positive | MGI_C57BL6J_97762 | Prl | NCBI_Gene:19109,ENSEMBL:ENSMUSG00000021342 | MGI:97762 | protein coding gene | prolactin |
| 13 | gene | 27.094189 | 27.100260 | positive | MGI_C57BL6J_97606 | Prl3d1 | NCBI_Gene:18775,ENSEMBL:ENSMUSG00000057170 | MGI:97606 | protein coding gene | prolactin family 3, subfamily d, member 1 |
| 13 | gene | 27.121632 | 27.127555 | positive | MGI_C57BL6J_2660935 | Prl3d2 | NCBI_Gene:215028,ENSEMBL:ENSMUSG00000062737 | MGI:2660935 | protein coding gene | prolactin family 3, subfamily d, member 1 |
| 13 | gene | 27.156769 | 27.162593 | positive | MGI_C57BL6J_2660938 | Prl3d3 | NCBI_Gene:215029,ENSEMBL:ENSMUSG00000062201 | MGI:2660938 | protein coding gene | prolactin family 3, subfamily d, member 3 |
| 13 | gene | 27.196659 | 27.203749 | positive | MGI_C57BL6J_1351649 | Prl3c1 | NCBI_Gene:27372,ENSEMBL:ENSMUSG00000017922 | MGI:1351649 | protein coding gene | prolactin family 3, subfamily c, member 1 |
| 13 | gene | 27.241847 | 27.249740 | positive | MGI_C57BL6J_97607 | Prl3b1 | NCBI_Gene:18776,ENSEMBL:ENSMUSG00000038891 | MGI:97607 | protein coding gene | prolactin family 3, subfamily b, member 1 |
| 13 | gene | 27.259436 | 27.276667 | positive | MGI_C57BL6J_1914250 | Prl3a1 | NCBI_Gene:67000,ENSEMBL:ENSMUSG00000038883 | MGI:1914250 | protein coding gene | prolactin family 3, subfamily a, member 1 |
| 13 | gene | 27.292616 | 27.306032 | positive | MGI_C57BL6J_1922371 | 4930511O05Rik | ENSEMBL:ENSMUSG00000086698 | MGI:1922371 | lncRNA gene | RIKEN cDNA 4930511O05 gene |
| 13 | pseudogene | 27.306712 | 27.306903 | positive | MGI_C57BL6J_3649562 | Gm11356 | ENSEMBL:ENSMUSG00000083096 | MGI:3649562 | pseudogene | predicted gene 11356 |
| 13 | gene | 27.312597 | 27.319254 | positive | MGI_C57BL6J_1206579 | Prl6a1 | NCBI_Gene:19111,ENSEMBL:ENSMUSG00000069259 | MGI:1206579 | protein coding gene | prolactin family 6, subfamily a, member 1 |
| 13 | pseudogene | 27.314084 | 27.315037 | positive | MGI_C57BL6J_3713473 | Gm11357 | NCBI_Gene:100417603,ENSEMBL:ENSMUSG00000089749 | MGI:3713473 | pseudogene | predicted gene 11357 |
| 13 | pseudogene | 27.331428 | 27.334404 | positive | MGI_C57BL6J_3649560 | Gm11358 | ENSEMBL:ENSMUSG00000082306 | MGI:3649560 | pseudogene | predicted gene 11358 |
| 13 | gene | 27.345673 | 27.354216 | positive | MGI_C57BL6J_894281 | Prl8a2 | NCBI_Gene:13529,ENSEMBL:ENSMUSG00000018259 | MGI:894281 | protein coding gene | prolactin family 8, subfamily a, member 2 |
| 13 | gene | 27.383339 | 27.390846 | negative | MGI_C57BL6J_1861444 | Prl2b1 | NCBI_Gene:66392,ENSEMBL:ENSMUSG00000069258 | MGI:1861444 | protein coding gene | prolactin family 2, subfamily b, member 1 |
| 13 | gene | 27.395407 | 27.432740 | positive | MGI_C57BL6J_5623721 | Gm40836 | NCBI_Gene:105245370 | MGI:5623721 | lncRNA gene | predicted gene, 40836 |
| 13 | gene | 27.432681 | 27.438688 | negative | MGI_C57BL6J_1332225 | Prl8a6 | NCBI_Gene:19112,ENSEMBL:ENSMUSG00000021345 | MGI:1332225 | protein coding gene | prolactin family 8, subfamily a, member 6 |
| 13 | gene | 27.506968 | 27.513217 | negative | MGI_C57BL6J_1921438 | Prl8a8 | NCBI_Gene:74188,ENSEMBL:ENSMUSG00000021346 | MGI:1921438 | protein coding gene | prolactin family 8, subfamily a, member 81 |
| 13 | gene | 27.557995 | 27.564606 | negative | MGI_C57BL6J_1914560 | Prl8a9 | NCBI_Gene:67310,ENSEMBL:ENSMUSG00000006490 | MGI:1914560 | protein coding gene | prolactin family8, subfamily a, member 9 |
| 13 | gene | 27.573922 | 27.582171 | negative | MGI_C57BL6J_1920494 | Prl8a1 | NCBI_Gene:73244,ENSEMBL:ENSMUSG00000019756 | MGI:1920494 | protein coding gene | prolactin family 8, subfamily a, member 1 |
| 13 | gene | 27.601812 | 27.610582 | negative | MGI_C57BL6J_1922846 | Prl7b1 | NCBI_Gene:75596,ENSEMBL:ENSMUSG00000021347 | MGI:1922846 | protein coding gene | prolactin family 7, subfamily b, member 1 |
| 13 | gene | 27.633364 | 27.642493 | negative | MGI_C57BL6J_1206572 | Prl7a1 | NCBI_Gene:19113,ENSEMBL:ENSMUSG00000006488 | MGI:1206572 | protein coding gene | prolactin family 7, subfamily a, member 1 |
| 13 | gene | 27.658584 | 27.668052 | negative | MGI_C57BL6J_1206571 | Prl7a2 | NCBI_Gene:19114,ENSEMBL:ENSMUSG00000046899 | MGI:1206571 | protein coding gene | prolactin family 7, subfamily a, member 2 |
| 13 | pseudogene | 27.683199 | 27.690869 | negative | MGI_C57BL6J_3651591 | Gm11359 | NCBI_Gene:494218,ENSEMBL:ENSMUSG00000083946 | MGI:3651591 | pseudogene | predicted gene 11359 |
| 13 | gene | 27.706337 | 27.716748 | negative | MGI_C57BL6J_97619 | Prl7d1 | NCBI_Gene:18814,ENSEMBL:ENSMUSG00000021348 | MGI:97619 | protein coding gene | prolactin family 7, subfamily d, member 1 |
| 13 | gene | 27.773495 | 27.780846 | negative | MGI_C57BL6J_1914755 | Prl7c1 | NCBI_Gene:67505,ENSEMBL:ENSMUSG00000060738 | MGI:1914755 | protein coding gene | prolactin family 7, subfamily c, member 1 |
| 13 | gene | 27.801654 | 27.808729 | positive | MGI_C57BL6J_1861446 | Prl2a1 | NCBI_Gene:56635,ENSEMBL:ENSMUSG00000022886 | MGI:1861446 | protein coding gene | prolactin family 2, subfamily a, member 1 |
| 13 | gene | 27.849327 | 27.857791 | positive | MGI_C57BL6J_3649927 | Prl2c1 | NCBI_Gene:666317,ENSEMBL:ENSMUSG00000062551 | MGI:3649927 | protein coding gene | Prolactin family 2, subfamily c, member 1 |
| 13 | gene | 27.874089 | 27.874188 | positive | MGI_C57BL6J_5452253 | Gm22476 | ENSEMBL:ENSMUSG00000095744 | MGI:5452253 | snRNA gene | predicted gene, 22476 |
| 13 | pseudogene | 27.928608 | 27.929601 | positive | MGI_C57BL6J_3643919 | Gm8045 | NCBI_Gene:666321 | MGI:3643919 | pseudogene | predicted gene 8045 |
| 13 | pseudogene | 27.929585 | 27.931698 | positive | MGI_C57BL6J_5593324 | Gm34165 | NCBI_Gene:102637325,ENSEMBL:ENSMUSG00000113672 | MGI:5593324 | pseudogene | predicted gene, 34165 |
| 13 | pseudogene | 27.956152 | 27.956415 | positive | MGI_C57BL6J_3649916 | Gm11360 | NCBI_Gene:100041036,ENSEMBL:ENSMUSG00000113281 | MGI:3649916 | pseudogene | predicted gene 11360 |
| 13 | pseudogene | 28.006508 | 28.008057 | positive | MGI_C57BL6J_5011066 | Gm18881 | NCBI_Gene:100417886,ENSEMBL:ENSMUSG00000113916 | MGI:5011066 | pseudogene | predicted gene, 18881 |
| 13 | gene | 28.016212 | 28.023559 | positive | MGI_C57BL6J_1206587 | Prl4a1 | NCBI_Gene:19110,ENSEMBL:ENSMUSG00000005891 | MGI:1206587 | protein coding gene | prolactin family 4, subfamily a, member 1 |
| 13 | pseudogene | 28.072061 | 28.090874 | positive | MGI_C57BL6J_6095705 | Gm47019 | ENSEMBL:ENSMUSG00000114182 | MGI:6095705 | pseudogene | predicted gene, 47019 |
| 13 | gene | 28.142484 | 28.151611 | positive | MGI_C57BL6J_106332 | Prl5a1 | NCBI_Gene:28078,ENSEMBL:ENSMUSG00000017064 | MGI:106332 | protein coding gene | prolactin family 5, subfamily a, member 1 |
| 13 | gene | 28.257528 | 28.258138 | positive | MGI_C57BL6J_3649931 | Rps18-ps5 | NCBI_Gene:100042388,ENSEMBL:ENSMUSG00000113061 | MGI:3649931 | protein coding gene | ribosomal protein S18, pseudogene 5 |
| 13 | pseudogene | 28.285569 | 28.309898 | positive | MGI_C57BL6J_6095953 | Gm47172 | ENSEMBL:ENSMUSG00000113418 | MGI:6095953 | pseudogene | predicted gene, 47172 |
| 13 | gene | 28.295761 | 28.297983 | positive | MGI_C57BL6J_5593544 | Gm34385 | NCBI_Gene:102637623 | MGI:5593544 | lncRNA gene | predicted gene, 34385 |
| 13 | pseudogene | 28.356621 | 28.364255 | negative | MGI_C57BL6J_3781993 | Gm3821 | NCBI_Gene:100042393 | MGI:3781993 | pseudogene | predicted gene 3821 |
| 13 | gene | 28.394453 | 28.420558 | negative | MGI_C57BL6J_5623726 | Gm40841 | NCBI_Gene:105245375,ENSEMBL:ENSMUSG00000113216 | MGI:5623726 | lncRNA gene | predicted gene, 40841 |
| 13 | gene | 28.398202 | 28.401625 | negative | MGI_C57BL6J_2444806 | A130022D17Rik | NA | NA | unclassified gene | RIKEN cDNA A130022D17 gene |
| 13 | gene | 28.409868 | 28.411311 | negative | MGI_C57BL6J_1925057 | A930025H08Rik | NA | NA | unclassified gene | RIKEN cDNA A930025H08 gene |
| 13 | gene | 28.460034 | 28.885620 | negative | MGI_C57BL6J_1919768 | 2610307P16Rik | NCBI_Gene:72518,ENSEMBL:ENSMUSG00000085936 | MGI:1919768 | lncRNA gene | RIKEN cDNA 2610307P16 gene |
| 13 | pseudogene | 28.510543 | 28.511232 | negative | MGI_C57BL6J_3645683 | Gm6081 | NCBI_Gene:606729,ENSEMBL:ENSMUSG00000086785 | MGI:3645683 | pseudogene | predicted gene 6081 |
| 13 | gene | 28.529805 | 28.533403 | negative | MGI_C57BL6J_3028028 | E130104P22Rik | NA | NA | unclassified gene | RIKEN cDNA E130104P22 gene |
| 13 | gene | 28.710781 | 28.710873 | negative | MGI_C57BL6J_5531182 | Mir6368 | miRBase:MI0021897,NCBI_Gene:102466635,ENSEMBL:ENSMUSG00000099158 | MGI:5531182 | miRNA gene | microRNA 6368 |
| 13 | gene | 28.725910 | 28.726348 | negative | MGI_C57BL6J_1917721 | 2610304O13Rik | NA | NA | unclassified gene | RIKEN cDNA 2610304O13 gene |
| 13 | gene | 28.769607 | 28.772199 | negative | MGI_C57BL6J_2443653 | 9430081I23Rik | NA | NA | unclassified gene | RIKEN cDNA 9430081I23 gene |
| 13 | pseudogene | 28.827123 | 28.827362 | positive | MGI_C57BL6J_4937162 | Gm17528 | ENSEMBL:ENSMUSG00000092093 | MGI:4937162 | pseudogene | predicted gene, 17528 |
| 13 | pseudogene | 28.836875 | 28.837724 | positive | MGI_C57BL6J_3651391 | Gm11362 | ENSEMBL:ENSMUSG00000082861 | MGI:3651391 | pseudogene | predicted gene 11362 |
| 13 | gene | 28.860705 | 28.863846 | negative | MGI_C57BL6J_2443407 | 5930430O07Rik | NA | NA | unclassified gene | RIKEN cDNA 5930430O07 gene |
| 13 | gene | 28.889458 | 28.898676 | negative | MGI_C57BL6J_5593798 | Gm34639 | NCBI_Gene:102637956,ENSEMBL:ENSMUSG00000113352 | MGI:5593798 | lncRNA gene | predicted gene, 34639 |
| 13 | gene | 28.905604 | 28.909600 | negative | MGI_C57BL6J_5593856 | Gm34697 | NCBI_Gene:102638031 | MGI:5593856 | lncRNA gene | predicted gene, 34697 |
| 13 | gene | 28.912736 | 28.916770 | negative | MGI_C57BL6J_5623743 | Gm40858 | NCBI_Gene:105245394 | MGI:5623743 | lncRNA gene | predicted gene, 40858 |
| 13 | gene | 28.943048 | 28.951671 | positive | MGI_C57BL6J_5477229 | Gm26735 | ENSEMBL:ENSMUSG00000097461 | MGI:5477229 | lncRNA gene | predicted gene, 26735 |
| 13 | gene | 28.945177 | 28.959261 | negative | MGI_C57BL6J_5826042 | Gm46405 | NCBI_Gene:108168071 | MGI:5826042 | lncRNA gene | predicted gene, 46405 |
| 13 | gene | 28.945388 | 28.948106 | positive | MGI_C57BL6J_5826043 | Gm46406 | NCBI_Gene:108168072 | MGI:5826043 | lncRNA gene | predicted gene, 46406 |
| 13 | gene | 28.948919 | 28.953713 | negative | MGI_C57BL6J_98366 | Sox4 | NCBI_Gene:20677,ENSEMBL:ENSMUSG00000076431 | MGI:98366 | protein coding gene | SRY (sex determining region Y)-box 4 |
| 13 | gene | 29.014399 | 29.040336 | positive | MGI_C57BL6J_1925170 | A330102I10Rik | NCBI_Gene:77920,ENSEMBL:ENSMUSG00000086363 | MGI:1925170 | lncRNA gene | RIKEN cDNA A330102I10 gene |
| 13 | gene | 29.015456 | 29.016121 | positive | MGI_C57BL6J_1924890 | C030039E19Rik | NA | NA | unclassified gene | RIKEN cDNA C030039E19 gene |
| 13 | pseudogene | 29.130211 | 29.130758 | negative | MGI_C57BL6J_3652072 | Gm11363 | ENSEMBL:ENSMUSG00000082730 | MGI:3652072 | pseudogene | predicted gene 11363 |
| 13 | gene | 29.191746 | 29.855684 | negative | MGI_C57BL6J_1921765 | Cdkal1 | NCBI_Gene:68916,ENSEMBL:ENSMUSG00000006191 | MGI:1921765 | protein coding gene | CDK5 regulatory subunit associated protein 1-like 1 |
| 13 | gene | 29.235777 | 29.236620 | negative | MGI_C57BL6J_3652071 | Gm11364 | ENSEMBL:ENSMUSG00000086423 | MGI:3652071 | lncRNA gene | predicted gene 11364 |
| 13 | gene | 29.906575 | 29.986119 | negative | MGI_C57BL6J_1096340 | E2f3 | NCBI_Gene:13557,ENSEMBL:ENSMUSG00000016477 | MGI:1096340 | protein coding gene | E2F transcription factor 3 |
| 13 | gene | 29.940283 | 29.942261 | positive | MGI_C57BL6J_3652070 | Gm11365 | ENSEMBL:ENSMUSG00000086893 | MGI:3652070 | lncRNA gene | predicted gene 11365 |
| 13 | gene | 30.003375 | 30.035446 | positive | MGI_C57BL6J_5623745 | Gm40860 | NCBI_Gene:105245398 | MGI:5623745 | lncRNA gene | predicted gene, 40860 |
| 13 | gene | 30.058677 | 30.062971 | positive | MGI_C57BL6J_3652073 | Gm11368 | ENSEMBL:ENSMUSG00000085188 | MGI:3652073 | lncRNA gene | predicted gene 11368 |
| 13 | gene | 30.098493 | 30.102042 | positive | MGI_C57BL6J_6096091 | Gm47259 | ENSEMBL:ENSMUSG00000113366 | MGI:6096091 | unclassified gene | predicted gene, 47259 |
| 13 | pseudogene | 30.112474 | 30.112593 | positive | MGI_C57BL6J_3652074 | Gm11367 | ENSEMBL:ENSMUSG00000083676 | MGI:3652074 | pseudogene | predicted gene 11367 |
| 13 | gene | 30.136449 | 30.246717 | positive | MGI_C57BL6J_2387184 | Mboat1 | NCBI_Gene:218121,ENSEMBL:ENSMUSG00000038732 | MGI:2387184 | protein coding gene | membrane bound O-acyltransferase domain containing 1 |
| 13 | gene | 30.148198 | 30.148381 | negative | MGI_C57BL6J_5452983 | Gm23206 | ENSEMBL:ENSMUSG00000084669 | MGI:5452983 | snRNA gene | predicted gene, 23206 |
| 13 | gene | 30.166206 | 30.177886 | positive | MGI_C57BL6J_5623754 | Gm40869 | NCBI_Gene:105245408 | MGI:5623754 | lncRNA gene | predicted gene, 40869 |
| 13 | pseudogene | 30.181690 | 30.182155 | positive | MGI_C57BL6J_3652075 | Gm11366 | NCBI_Gene:100846989,ENSEMBL:ENSMUSG00000082648 | MGI:3652075 | pseudogene | predicted gene 11366 |
| 13 | gene | 30.231667 | 30.235651 | negative | MGI_C57BL6J_5623748 | Gm40863 | NCBI_Gene:105245401 | MGI:5623748 | lncRNA gene | predicted gene, 40863 |
| 13 | gene | 30.336356 | 30.382867 | positive | MGI_C57BL6J_87964 | Agtr1a | NCBI_Gene:11607,ENSEMBL:ENSMUSG00000049115 | MGI:87964 | protein coding gene | angiotensin II receptor, type 1a |
| 13 | gene | 30.370359 | 30.377183 | positive | MGI_C57BL6J_6097531 | Gm48161 | ENSEMBL:ENSMUSG00000113424 | MGI:6097531 | lncRNA gene | predicted gene, 48161 |
| 13 | pseudogene | 30.399617 | 30.399934 | negative | MGI_C57BL6J_3652076 | Gm11369 | ENSEMBL:ENSMUSG00000083882 | MGI:3652076 | pseudogene | predicted gene 11369 |
| 13 | gene | 30.540308 | 30.545362 | negative | MGI_C57BL6J_1913944 | Uqcrfs1 | NCBI_Gene:66694,ENSEMBL:ENSMUSG00000038462 | MGI:1913944 | protein coding gene | ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| 13 | gene | 30.611041 | 30.614625 | negative | MGI_C57BL6J_5594268 | Gm35109 | NCBI_Gene:102638578 | MGI:5594268 | lncRNA gene | predicted gene, 35109 |
| 13 | gene | 30.615043 | 30.624259 | positive | MGI_C57BL6J_1922340 | 4930519D14Rik | NCBI_Gene:75090,ENSEMBL:ENSMUSG00000086263 | MGI:1922340 | lncRNA gene | RIKEN cDNA 4930519D14 gene |
| 13 | gene | 30.652759 | 30.659874 | negative | MGI_C57BL6J_5594317 | Gm35158 | NCBI_Gene:102638644 | MGI:5594317 | lncRNA gene | predicted gene, 35158 |
| 13 | gene | 30.659999 | 30.711232 | positive | MGI_C57BL6J_1915926 | Dusp22 | NCBI_Gene:105352,ENSEMBL:ENSMUSG00000069255 | MGI:1915926 | protein coding gene | dual specificity phosphatase 22 |
| 13 | pseudogene | 30.731777 | 30.732036 | positive | MGI_C57BL6J_3651832 | Gm11370 | ENSEMBL:ENSMUSG00000081905 | MGI:3651832 | pseudogene | predicted gene 11370 |
| 13 | gene | 30.749208 | 30.766982 | positive | MGI_C57BL6J_1096873 | Irf4 | NCBI_Gene:16364,ENSEMBL:ENSMUSG00000021356 | MGI:1096873 | protein coding gene | interferon regulatory factor 4 |
| 13 | gene | 30.784433 | 30.795226 | negative | MGI_C57BL6J_5588834 | Gm29675 | ENSEMBL:ENSMUSG00000113515 | MGI:5588834 | lncRNA gene | predicted gene, 29675 |
| 13 | gene | 30.789516 | 30.974273 | negative | MGI_C57BL6J_1913732 | Exoc2 | NCBI_Gene:66482,ENSEMBL:ENSMUSG00000021357 | MGI:1913732 | protein coding gene | exocyst complex component 2 |
| 13 | gene | 30.946573 | 30.947761 | negative | MGI_C57BL6J_2671003 | Hus1b | NCBI_Gene:210554,ENSEMBL:ENSMUSG00000076430 | MGI:2671003 | protein coding gene | HUS1 checkpoint clamp component B |
| 13 | gene | 30.974164 | 30.976248 | positive | MGI_C57BL6J_3644017 | Gm5447 | ENSEMBL:ENSMUSG00000056257 | MGI:3644017 | protein coding gene | predicted gene 5447 |
| 13 | pseudogene | 31.009177 | 31.010362 | positive | MGI_C57BL6J_3651829 | Gm11371 | ENSEMBL:ENSMUSG00000082014 | MGI:3651829 | pseudogene | predicted gene 11371 |
| 13 | gene | 31.072730 | 31.077845 | negative | MGI_C57BL6J_6098277 | Gm48662 | ENSEMBL:ENSMUSG00000114161 | MGI:6098277 | lncRNA gene | predicted gene, 48662 |
| 13 | gene | 31.080897 | 31.084403 | positive | MGI_C57BL6J_5594461 | Gm35302 | NCBI_Gene:102638831 | MGI:5594461 | lncRNA gene | predicted gene, 35302 |
| 13 | pseudogene | 31.199086 | 31.199655 | positive | MGI_C57BL6J_3651830 | Gm11372 | ENSEMBL:ENSMUSG00000084175 | MGI:3651830 | pseudogene | predicted gene 11372 |
| 13 | gene | 31.213410 | 31.241086 | negative | MGI_C57BL6J_1921174 | 4930401O12Rik | NCBI_Gene:73924,ENSEMBL:ENSMUSG00000087460 | MGI:1921174 | lncRNA gene | RIKEN cDNA 4930401O12 gene |
| 13 | gene | 31.321353 | 31.409096 | negative | MGI_C57BL6J_3041239 | G630018N14Rik | NCBI_Gene:414126,ENSEMBL:ENSMUSG00000085620 | MGI:3041239 | lncRNA gene | RIKEN cDNA G630018N14 gene |
| 13 | gene | 31.325934 | 31.330767 | negative | MGI_C57BL6J_3651828 | Gm11373 | ENSEMBL:ENSMUSG00000087662 | MGI:3651828 | lncRNA gene | predicted gene 11373 |
| 13 | gene | 31.352980 | 31.353094 | positive | MGI_C57BL6J_5452107 | Gm22330 | ENSEMBL:ENSMUSG00000088693 | MGI:5452107 | snoRNA gene | predicted gene, 22330 |
| 13 | gene | 31.392578 | 31.396579 | positive | MGI_C57BL6J_3650400 | Gm11376 | ENSEMBL:ENSMUSG00000087140 | MGI:3650400 | lncRNA gene | predicted gene 11376 |
| 13 | gene | 31.427255 | 31.427379 | positive | MGI_C57BL6J_5453128 | Gm23351 | ENSEMBL:ENSMUSG00000089052 | MGI:5453128 | snoRNA gene | predicted gene, 23351 |
| 13 | gene | 31.556134 | 31.559333 | negative | MGI_C57BL6J_3704402 | A530084C06Rik | ENSEMBL:ENSMUSG00000090863 | MGI:3704402 | protein coding gene | RIKEN cDNA A530084C06 gene |
| 13 | gene | 31.556134 | 31.560976 | positive | MGI_C57BL6J_1298228 | Foxq1 | NCBI_Gene:15220,ENSEMBL:ENSMUSG00000038415 | MGI:1298228 | protein coding gene | forkhead box Q1 |
| 13 | gene | 31.564749 | 31.573562 | positive | MGI_C57BL6J_3650186 | Gm11377 | ENSEMBL:ENSMUSG00000087220 | MGI:3650186 | lncRNA gene | predicted gene 11377 |
| 13 | gene | 31.565492 | 31.625792 | negative | MGI_C57BL6J_1916631 | 1700018A04Rik | NCBI_Gene:71307,ENSEMBL:ENSMUSG00000038408 | MGI:1916631 | lncRNA gene | RIKEN cDNA 1700018A04 gene |
| 13 | gene | 31.619168 | 31.620369 | positive | MGI_C57BL6J_3650784 | Gm11378 | ENSEMBL:ENSMUSG00000087276 | MGI:3650784 | lncRNA gene | predicted gene 11378 |
| 13 | gene | 31.625816 | 31.631406 | positive | MGI_C57BL6J_1347479 | Foxf2 | NCBI_Gene:14238,ENSEMBL:ENSMUSG00000038402 | MGI:1347479 | protein coding gene | forkhead box F2 |
| 13 | gene | 31.626446 | 31.626543 | negative | MGI_C57BL6J_5530898 | Gm27516 | ENSEMBL:ENSMUSG00000098266 | MGI:5530898 | miRNA gene | predicted gene, 27516 |
| 13 | gene | 31.737633 | 31.737740 | negative | MGI_C57BL6J_5454963 | Gm25186 | ENSEMBL:ENSMUSG00000065843 | MGI:5454963 | snRNA gene | predicted gene, 25186 |
| 13 | gene | 31.797539 | 31.800789 | positive | MGI_C57BL6J_3650783 | Gm11379 | ENSEMBL:ENSMUSG00000086144 | MGI:3650783 | lncRNA gene | predicted gene 11379 |
| 13 | gene | 31.806633 | 31.812476 | positive | MGI_C57BL6J_1347466 | Foxc1 | NCBI_Gene:17300,ENSEMBL:ENSMUSG00000050295 | MGI:1347466 | protein coding gene | forkhead box C1 |
| 13 | gene | 31.811162 | 31.812476 | positive | MGI_C57BL6J_1922550 | 4930548F15Rik | NA | NA | unclassified gene | RIKEN cDNA 4930548F15 gene |
| 13 | gene | 31.819579 | 32.338740 | negative | MGI_C57BL6J_1891112 | Gmds | NCBI_Gene:218138,ENSEMBL:ENSMUSG00000038372 | MGI:1891112 | protein coding gene | GDP-mannose 4, 6-dehydratase |
| 13 | gene | 31.832905 | 31.834736 | negative | MGI_C57BL6J_6098639 | Gm48883 | ENSEMBL:ENSMUSG00000113119 | MGI:6098639 | unclassified gene | predicted gene, 48883 |
| 13 | pseudogene | 31.882297 | 31.887358 | positive | MGI_C57BL6J_3650782 | Gm11380 | NCBI_Gene:108168073,ENSEMBL:ENSMUSG00000083149 | MGI:3650782 | pseudogene | predicted gene 11380 |
| 13 | gene | 31.968296 | 31.997837 | positive | MGI_C57BL6J_5594891 | Gm35732 | NCBI_Gene:102639408,ENSEMBL:ENSMUSG00000113935 | MGI:5594891 | lncRNA gene | predicted gene, 35732 |
| 13 | gene | 31.976648 | 31.979258 | negative | MGI_C57BL6J_2444742 | 9130221L21Rik | NA | NA | unclassified gene | RIKEN cDNA 9130221L21 gene |
| 13 | gene | 32.007295 | 32.092787 | positive | MGI_C57BL6J_5594831 | Gm35672 | NCBI_Gene:105245415 | MGI:5594831 | lncRNA gene | predicted gene, 35672 |
| 13 | gene | 32.095279 | 32.099175 | negative | MGI_C57BL6J_6098643 | Gm48885 | ENSEMBL:ENSMUSG00000112944 | MGI:6098643 | unclassified gene | predicted gene, 48885 |
| 13 | gene | 32.338819 | 32.342643 | positive | MGI_C57BL6J_2442493 | A730091E23Rik | ENSEMBL:ENSMUSG00000113965 | MGI:2442493 | unclassified gene | RIKEN cDNA A730091E23 gene |
| 13 | gene | 32.379907 | 32.383688 | negative | MGI_C57BL6J_3650781 | Gm11381 | ENSEMBL:ENSMUSG00000085770 | MGI:3650781 | lncRNA gene | predicted gene 11381 |
| 13 | gene | 32.465453 | 32.493376 | positive | MGI_C57BL6J_5623761 | Gm40876 | NCBI_Gene:105245418 | MGI:5623761 | lncRNA gene | predicted gene, 40876 |
| 13 | gene | 32.532919 | 32.536415 | negative | MGI_C57BL6J_6097405 | Gm48073 | ENSEMBL:ENSMUSG00000113449 | MGI:6097405 | lncRNA gene | predicted gene, 48073 |
| 13 | gene | 32.700827 | 32.806773 | negative | MGI_C57BL6J_3643758 | Mylk4 | NCBI_Gene:238564,ENSEMBL:ENSMUSG00000044951 | MGI:3643758 | protein coding gene | myosin light chain kinase family, member 4 |
| 13 | gene | 32.725851 | 32.726972 | negative | MGI_C57BL6J_1924005 | 2310045N14Rik | NA | NA | unclassified gene | RIKEN cDNA 2310045N14 gene |
| 13 | gene | 32.758202 | 32.758310 | positive | MGI_C57BL6J_5530724 | Gm27342 | ENSEMBL:ENSMUSG00000098788 | MGI:5530724 | snRNA gene | predicted gene, 27342 |
| 13 | gene | 32.800633 | 32.803091 | negative | MGI_C57BL6J_3686989 | D930007J09Rik | ENSEMBL:ENSMUSG00000042874 | MGI:3686989 | lncRNA gene | RIKEN cDNA D930007J09 gene |
| 13 | gene | 32.802030 | 32.822610 | positive | MGI_C57BL6J_1926153 | Wrnip1 | NCBI_Gene:78903,ENSEMBL:ENSMUSG00000021400 | MGI:1926153 | protein coding gene | Werner helicase interacting protein 1 |
| 13 | gene | 32.842092 | 32.851185 | negative | MGI_C57BL6J_1913472 | Serpinb1a | NCBI_Gene:66222,ENSEMBL:ENSMUSG00000044734 | MGI:1913472 | protein coding gene | serine (or cysteine) peptidase inhibitor, clade B, member 1a |
| 13 | pseudogene | 32.859948 | 32.863178 | negative | MGI_C57BL6J_2445366 | Serpinb1-ps1 | NCBI_Gene:282665,ENSEMBL:ENSMUSG00000081433 | MGI:2445366 | pseudogene | serine (or cysteine) peptidase inhibitor, clade B, member 1, pseudogene |
| 13 | gene | 32.865053 | 32.870912 | positive | MGI_C57BL6J_5595058 | Gm35899 | NCBI_Gene:102639635 | MGI:5595058 | lncRNA gene | predicted gene, 35899 |
| 13 | gene | 32.881397 | 32.940628 | negative | MGI_C57BL6J_2445363 | Serpinb1c | NCBI_Gene:380839,ENSEMBL:ENSMUSG00000079049 | MGI:2445363 | protein coding gene | serine (or cysteine) peptidase inhibitor, clade B, member 1c |
| 13 | pseudogene | 32.923221 | 32.924494 | negative | MGI_C57BL6J_5010496 | Gm18311 | NCBI_Gene:100416908 | MGI:5010496 | pseudogene | predicted gene, 18311 |
| 13 | pseudogene | 32.938513 | 32.938691 | negative | MGI_C57BL6J_3650578 | Gm11382 | ENSEMBL:ENSMUSG00000074973 | MGI:3650578 | pseudogene | predicted pseudogene 11382 |
| 13 | gene | 32.956811 | 32.979067 | positive | MGI_C57BL6J_894688 | Serpinb6b | NCBI_Gene:20708,ENSEMBL:ENSMUSG00000042842 | MGI:894688 | protein coding gene | serine (or cysteine) peptidase inhibitor, clade B, member 6b |
| 13 | gene | 32.997061 | 33.017957 | positive | MGI_C57BL6J_106603 | Serpinb9 | NCBI_Gene:20723,ENSEMBL:ENSMUSG00000045827 | MGI:106603 | protein coding gene | serine (or cysteine) peptidase inhibitor, clade B, member 9 |
| 13 | gene | 32.997062 | 33.001750 | positive | MGI_C57BL6J_6096750 | Gm47662 | ENSEMBL:ENSMUSG00000114185 | MGI:6096750 | lncRNA gene | predicted gene, 47662 |
| 13 | gene | 33.000998 | 33.006525 | negative | MGI_C57BL6J_5595340 | Gm36181 | NCBI_Gene:102640002 | MGI:5595340 | lncRNA gene | predicted gene, 36181 |
| 13 | gene | 33.027414 | 33.041884 | positive | MGI_C57BL6J_894668 | Serpinb9b | NCBI_Gene:20706,ENSEMBL:ENSMUSG00000021403 | MGI:894668 | protein coding gene | serine (or cysteine) peptidase inhibitor, clade B, member 9b |
| 13 | gene | 33.030407 | 33.034704 | negative | MGI_C57BL6J_5595258 | Gm36099 | NCBI_Gene:102639897,ENSEMBL:ENSMUSG00000113726 | MGI:5595258 | lncRNA gene | predicted gene, 36099 |
| 13 | pseudogene | 33.065295 | 33.066815 | positive | MGI_C57BL6J_5010501 | Gm18316 | NCBI_Gene:100416913 | MGI:5010501 | pseudogene | predicted gene, 18316 |
| 13 | gene | 33.078504 | 33.094380 | positive | MGI_C57BL6J_2445361 | Serpinb1b | NCBI_Gene:282663,ENSEMBL:ENSMUSG00000051029 | MGI:2445361 | protein coding gene | serine (or cysteine) peptidase inhibitor, clade B, member 1b |
| 13 | pseudogene | 33.105539 | 33.133493 | negative | MGI_C57BL6J_3651702 | Gm11384 | ENSEMBL:ENSMUSG00000083653 | MGI:3651702 | pseudogene | predicted gene 11384 |
| 13 | pseudogene | 33.120516 | 33.121322 | negative | MGI_C57BL6J_3649586 | Gm11383 | NCBI_Gene:628301,ENSEMBL:ENSMUSG00000082893 | MGI:3649586 | pseudogene | predicted gene 11383 |
| 13 | pseudogene | 33.137574 | 33.138240 | negative | MGI_C57BL6J_5010893 | Gm18708 | NCBI_Gene:100417602 | MGI:5010893 | pseudogene | predicted gene, 18708 |
| 13 | gene | 33.149275 | 33.159754 | negative | MGI_C57BL6J_894669 | Serpinb9c | NCBI_Gene:20707,ENSEMBL:ENSMUSG00000021404 | MGI:894669 | protein coding gene | serine (or cysteine) peptidase inhibitor, clade B, member 9c |
| 13 | gene | 33.192959 | 33.203980 | positive | MGI_C57BL6J_894667 | Serpinb9d | NCBI_Gene:20726,ENSEMBL:ENSMUSG00000054266 | MGI:894667 | protein coding gene | serine (or cysteine) peptidase inhibitor, clade B, member 9d |
| 13 | pseudogene | 33.217268 | 33.218151 | negative | MGI_C57BL6J_3705792 | Gm11385 | ENSEMBL:ENSMUSG00000081237 | MGI:3705792 | pseudogene | predicted gene 11385 |
| 13 | pseudogene | 33.237702 | 33.238305 | positive | MGI_C57BL6J_6096956 | Gm47788 | ENSEMBL:ENSMUSG00000113703 | MGI:6096956 | pseudogene | predicted gene, 47788 |
| 13 | gene | 33.249610 | 33.260850 | positive | MGI_C57BL6J_894672 | Serpinb9e | NCBI_Gene:20710,ENSEMBL:ENSMUSG00000062342 | MGI:894672 | protein coding gene | serine (or cysteine) peptidase inhibitor, clade B, member 9e |
| 13 | gene | 33.284223 | 33.293474 | positive | MGI_C57BL6J_5623790 | Gm40905 | NCBI_Gene:105245454 | MGI:5623790 | lncRNA gene | predicted gene, 40905 |
| 13 | gene | 33.324077 | 33.335370 | positive | MGI_C57BL6J_894671 | Serpinb9f | NCBI_Gene:20709,ENSEMBL:ENSMUSG00000038327 | MGI:894671 | protein coding gene | serine (or cysteine) peptidase inhibitor, clade B, member 9f |
| 13 | gene | 33.358862 | 33.368110 | positive | MGI_C57BL6J_5623791 | Gm40906 | NCBI_Gene:105245455 | MGI:5623791 | lncRNA gene | predicted gene, 40906 |
| 13 | gene | 33.395673 | 33.405285 | positive | MGI_C57BL6J_3709608 | Serpinb9h | NCBI_Gene:544923,ENSEMBL:ENSMUSG00000071452 | MGI:3709608 | protein coding gene | serine (or cysteine) peptidase inhibitor, clade B, member 9h |
| 13 | pseudogene | 33.409104 | 33.436782 | positive | MGI_C57BL6J_5011340 | Gm19155 | NCBI_Gene:100418350 | MGI:5011340 | pseudogene | predicted gene, 19155 |
| 13 | gene | 33.484790 | 33.496004 | positive | MGI_C57BL6J_1919260 | Serpinb9g | NCBI_Gene:93806,ENSEMBL:ENSMUSG00000057726 | MGI:1919260 | protein coding gene | serine (or cysteine) peptidase inhibitor, clade B, member 9g |
| 13 | pseudogene | 33.508127 | 33.526311 | positive | MGI_C57BL6J_3649582 | Gm11387 | NCBI_Gene:102635863,ENSEMBL:ENSMUSG00000081903 | MGI:3649582 | pseudogene | predicted gene 11387 |
| 13 | pseudogene | 33.531613 | 33.531959 | negative | MGI_C57BL6J_3649585 | Gm11386 | NCBI_Gene:100502609,ENSEMBL:ENSMUSG00000081288 | MGI:3649585 | pseudogene | predicted gene 11386 |
| 13 | pseudogene | 33.542999 | 33.550698 | positive | MGI_C57BL6J_3649834 | Gm11393 | ENSEMBL:ENSMUSG00000082722 | MGI:3649834 | pseudogene | predicted gene 11393 |
| 13 | gene | 33.562486 | 33.569317 | negative | MGI_C57BL6J_3648840 | Gm6093 | NCBI_Gene:619715,ENSEMBL:ENSMUSG00000112963 | MGI:3648840 | lncRNA gene | predicted gene 6093 |
| 13 | gene | 33.567794 | 33.568005 | positive | MGI_C57BL6J_5454820 | Gm25043 | ENSEMBL:ENSMUSG00000089229 | MGI:5454820 | ribozyme gene | predicted gene, 25043 |
| 13 | gene | 33.569021 | 33.572086 | positive | MGI_C57BL6J_6097166 | Gm47916 | ENSEMBL:ENSMUSG00000113016 | MGI:6097166 | lncRNA gene | predicted gene, 47916 |
| 13 | pseudogene | 33.595476 | 33.596953 | negative | MGI_C57BL6J_3645611 | Gm5194 | NCBI_Gene:382743,ENSEMBL:ENSMUSG00000113398 | MGI:3645611 | pseudogene | predicted gene 5194 |
| 13 | pseudogene | 33.641625 | 33.641783 | positive | MGI_C57BL6J_3649831 | Gm11388 | ENSEMBL:ENSMUSG00000083042 | MGI:3649831 | pseudogene | predicted gene 11388 |
| 13 | gene | 33.661405 | 33.671585 | positive | MGI_C57BL6J_2667783 | Serpinb6d | NCBI_Gene:238568,ENSEMBL:ENSMUSG00000047889 | MGI:2667783 | protein coding gene | serine (or cysteine) peptidase inhibitor, clade B, member 6d |
| 13 | pseudogene | 33.686457 | 33.689508 | positive | MGI_C57BL6J_3649830 | Gm11389 | ENSEMBL:ENSMUSG00000082986 | MGI:3649830 | pseudogene | predicted gene 11389 |
| 13 | pseudogene | 33.700473 | 33.701102 | negative | MGI_C57BL6J_5477195 | Gm26701 | ENSEMBL:ENSMUSG00000097202 | MGI:5477195 | pseudogene | predicted gene, 26701 |
| 13 | pseudogene | 33.718275 | 33.742136 | positive | MGI_C57BL6J_3649835 | Gm11392 | NCBI_Gene:193403,ENSEMBL:ENSMUSG00000097931 | MGI:3649835 | pseudogene | predicted gene 11392 |
| 13 | pseudogene | 33.729033 | 33.729861 | negative | MGI_C57BL6J_3649833 | Gm11394 | NCBI_Gene:100418363,ENSEMBL:ENSMUSG00000097190 | MGI:3649833 | pseudogene | predicted gene 11394 |
| 13 | pseudogene | 33.741240 | 33.742038 | positive | MGI_C57BL6J_3649832 | Gm11395 | ENSEMBL:ENSMUSG00000097054 | MGI:3649832 | pseudogene | predicted gene 11395 |
| 13 | pseudogene | 33.766920 | 33.811610 | positive | MGI_C57BL6J_3761346 | Vmn2r-ps94 | NCBI_Gene:619864,ENSEMBL:ENSMUSG00000080317 | MGI:3761346 | pseudogene | vomeronasal 2, receptor, pseudogene 94 |
| 13 | gene | 33.832292 | 33.843408 | negative | MGI_C57BL6J_2667778 | Serpinb6e | NCBI_Gene:435350,ENSEMBL:ENSMUSG00000069248 | MGI:2667778 | protein coding gene | serine (or cysteine) peptidase inhibitor, clade B, member 6e |
| 13 | gene | 33.879816 | 33.905708 | negative | MGI_C57BL6J_2145481 | Serpinb6c | NCBI_Gene:97848,ENSEMBL:ENSMUSG00000052180 | MGI:2145481 | protein coding gene | serine (or cysteine) peptidase inhibitor, clade B, member 6c |
| 13 | gene | 33.917918 | 34.002794 | negative | MGI_C57BL6J_103123 | Serpinb6a | NCBI_Gene:20719,ENSEMBL:ENSMUSG00000060147 | MGI:103123 | protein coding gene | serine (or cysteine) peptidase inhibitor, clade B, member 6a |
| 13 | gene | 33.935567 | 33.960181 | positive | MGI_C57BL6J_1916058 | 1110046J04Rik | NCBI_Gene:68808,ENSEMBL:ENSMUSG00000085457 | MGI:1916058 | lncRNA gene | RIKEN cDNA 1110046J04 gene |
| 13 | gene | 33.964466 | 33.988465 | positive | MGI_C57BL6J_104513 | Nqo2 | NCBI_Gene:18105,ENSEMBL:ENSMUSG00000046949 | MGI:104513 | protein coding gene | N-ribosyldihydronicotinamide quinone reductase 2 |
| 13 | gene | 34.002363 | 34.037147 | positive | MGI_C57BL6J_108212 | Ripk1 | NCBI_Gene:19766,ENSEMBL:ENSMUSG00000021408 | MGI:108212 | protein coding gene | receptor (TNFRSF)-interacting serine-threonine kinase 1 |
| 13 | pseudogene | 34.006966 | 34.008331 | negative | MGI_C57BL6J_4937910 | Gm17083 | ENSEMBL:ENSMUSG00000090628 | MGI:4937910 | pseudogene | predicted gene 17083 |
| 13 | pseudogene | 34.034526 | 34.035036 | positive | MGI_C57BL6J_4937911 | Gm17084 | ENSEMBL:ENSMUSG00000091462 | MGI:4937911 | pseudogene | predicted gene 17084 |
| 13 | gene | 34.037585 | 34.074074 | positive | MGI_C57BL6J_1915271 | Bphl | NCBI_Gene:68021,ENSEMBL:ENSMUSG00000038286 | MGI:1915271 | protein coding gene | biphenyl hydrolase-like (serine hydrolase, breast epithelial mucin-associated antigen) |
| 13 | gene | 34.074274 | 34.078008 | negative | MGI_C57BL6J_107861 | Tubb2a | NCBI_Gene:22151,ENSEMBL:ENSMUSG00000058672 | MGI:107861 | protein coding gene | tubulin, beta 2A class IIA |
| 13 | gene | 34.084155 | 34.113272 | negative | MGI_C57BL6J_1921231 | 4930447K03Rik | NCBI_Gene:432745,ENSEMBL:ENSMUSG00000113591 | MGI:1921231 | lncRNA gene | RIKEN cDNA 4930447K03 gene |
| 13 | pseudogene | 34.087137 | 34.089027 | negative | MGI_C57BL6J_6095778 | Gm47065 | ENSEMBL:ENSMUSG00000114067 | MGI:6095778 | pseudogene | predicted gene, 47065 |
| 13 | gene | 34.119701 | 34.122465 | positive | MGI_C57BL6J_6095797 | Gm47076 | ENSEMBL:ENSMUSG00000113645 | MGI:6095797 | lncRNA gene | predicted gene, 47076 |
| 13 | gene | 34.126748 | 34.130466 | negative | MGI_C57BL6J_1920960 | Tubb2b | NCBI_Gene:73710,ENSEMBL:ENSMUSG00000045136 | MGI:1920960 | protein coding gene | tubulin, beta 2B class IIB |
| 13 | gene | 34.130570 | 34.138468 | positive | MGI_C57BL6J_5595659 | Gm36500 | NCBI_Gene:102640439,ENSEMBL:ENSMUSG00000113883 | MGI:5595659 | lncRNA gene | predicted gene, 36500 |
| 13 | gene | 34.148432 | 34.151024 | negative | MGI_C57BL6J_5595708 | Gm36549 | NCBI_Gene:102640507 | MGI:5595708 | lncRNA gene | predicted gene, 36549 |
| 13 | gene | 34.162964 | 34.178173 | positive | MGI_C57BL6J_1916916 | Psmg4 | NCBI_Gene:69666,ENSEMBL:ENSMUSG00000071451 | MGI:1916916 | protein coding gene | proteasome (prosome, macropain) assembly chaperone 4 |
| 13 | gene | 34.179158 | 34.345959 | negative | MGI_C57BL6J_1920352 | Slc22a23 | NCBI_Gene:73102,ENSEMBL:ENSMUSG00000038267 | MGI:1920352 | protein coding gene | solute carrier family 22, member 23 |
| 13 | gene | 34.196633 | 34.196884 | negative | MGI_C57BL6J_1925659 | 2900079J23Rik | NA | NA | unclassified gene | RIKEN cDNA 2900079J23 gene |
| 13 | gene | 34.276965 | 34.277252 | negative | MGI_C57BL6J_5454307 | Gm24530 | ENSEMBL:ENSMUSG00000088573 | MGI:5454307 | unclassified non-coding RNA gene | predicted gene, 24530 |
| 13 | pseudogene | 34.318808 | 34.319217 | positive | MGI_C57BL6J_6095813 | Gm47086 | ENSEMBL:ENSMUSG00000113430 | MGI:6095813 | pseudogene | predicted gene, 47086 |
| 13 | gene | 34.342722 | 34.343456 | positive | MGI_C57BL6J_5826044 | Gm46407 | NCBI_Gene:108168074 | MGI:5826044 | lncRNA gene | predicted gene, 46407 |
| 13 | pseudogene | 34.393719 | 34.394539 | negative | MGI_C57BL6J_3645100 | Gm6129 | NCBI_Gene:620132,ENSEMBL:ENSMUSG00000113843 | MGI:3645100 | pseudogene | predicted gene 6129 |
| 13 | pseudogene | 34.426230 | 34.426431 | negative | MGI_C57BL6J_6095867 | Gm47120 | ENSEMBL:ENSMUSG00000114170 | MGI:6095867 | pseudogene | predicted gene, 47120 |
| 13 | gene | 34.459557 | 34.482181 | negative | MGI_C57BL6J_5623793 | Gm40908 | NCBI_Gene:105245458 | MGI:5623793 | lncRNA gene | predicted gene, 40908 |
| 13 | gene | 34.500122 | 34.506511 | negative | MGI_C57BL6J_5623794 | Gm40909 | NCBI_Gene:105245459,ENSEMBL:ENSMUSG00000113998 | MGI:5623794 | lncRNA gene | predicted gene, 40909 |
| 13 | pseudogene | 34.561176 | 34.563254 | positive | MGI_C57BL6J_5595917 | Gm36758 | NCBI_Gene:105245460,ENSEMBL:ENSMUSG00000113554 | MGI:5595917 | pseudogene | predicted gene, 36758 |
| 13 | gene | 34.610827 | 34.612246 | negative | MGI_C57BL6J_6095878 | Gm47127 | ENSEMBL:ENSMUSG00000113185 | MGI:6095878 | lncRNA gene | predicted gene, 47127 |
| 13 | gene | 34.627834 | 34.653486 | negative | MGI_C57BL6J_1914145 | Pxdc1 | NCBI_Gene:66895,ENSEMBL:ENSMUSG00000021411 | MGI:1914145 | protein coding gene | PX domain containing 1 |
| 13 | gene | 34.652923 | 34.666774 | positive | MGI_C57BL6J_3801931 | Gm15908 | NCBI_Gene:101056149,ENSEMBL:ENSMUSG00000086786 | MGI:3801931 | lncRNA gene | predicted gene 15908 |
| 13 | pseudogene | 34.680733 | 34.680921 | negative | MGI_C57BL6J_6095914 | Gm47149 | ENSEMBL:ENSMUSG00000113044 | MGI:6095914 | pseudogene | predicted gene, 47149 |
| 13 | gene | 34.711449 | 34.715030 | positive | MGI_C57BL6J_6095915 | Gm47150 | ENSEMBL:ENSMUSG00000113615 | MGI:6095915 | lncRNA gene | predicted gene, 47150 |
| 13 | gene | 34.718056 | 34.720751 | negative | MGI_C57BL6J_6095917 | Gm47151 | ENSEMBL:ENSMUSG00000113822 | MGI:6095917 | lncRNA gene | predicted gene, 47151 |
| 13 | gene | 34.720682 | 34.728661 | positive | MGI_C57BL6J_6095919 | Gm47152 | ENSEMBL:ENSMUSG00000113457 | MGI:6095919 | lncRNA gene | predicted gene, 47152 |
| 13 | gene | 34.734828 | 34.747623 | positive | MGI_C57BL6J_1351640 | Fam50b | NCBI_Gene:108161,ENSEMBL:ENSMUSG00000038246 | MGI:1351640 | protein coding gene | family with sequence similarity 50, member B |
| 13 | gene | 34.781585 | 34.814544 | negative | MGI_C57BL6J_6095928 | Gm47157 | ENSEMBL:ENSMUSG00000113484 | MGI:6095928 | lncRNA gene | predicted gene, 47157 |
| 13 | pseudogene | 34.865712 | 34.866610 | negative | MGI_C57BL6J_3646981 | Gm8231 | NCBI_Gene:666677,ENSEMBL:ENSMUSG00000114127 | MGI:3646981 | pseudogene | predicted gene 8231 |
| 13 | gene | 34.872512 | 34.875432 | negative | MGI_C57BL6J_5595998 | Gm36839 | NCBI_Gene:102640879,ENSEMBL:ENSMUSG00000113170 | MGI:5595998 | lncRNA gene | predicted gene, 36839 |
| 13 | gene | 34.875294 | 34.906064 | positive | MGI_C57BL6J_109584 | Prpf4b | NCBI_Gene:19134,ENSEMBL:ENSMUSG00000021413 | MGI:109584 | protein coding gene | pre-mRNA processing factor 4B |
| 13 | gene | 34.909959 | 34.924358 | negative | MGI_C57BL6J_1919114 | Fam217a | NCBI_Gene:71864,ENSEMBL:ENSMUSG00000021414 | MGI:1919114 | protein coding gene | family with sequence similarity 217, member A |
| 13 | gene | 34.916925 | 34.956218 | positive | MGI_C57BL6J_1914011 | 4933417A18Rik | NCBI_Gene:66761,ENSEMBL:ENSMUSG00000021415 | MGI:1914011 | protein coding gene | RIKEN cDNA 4933417A18 gene |
| 13 | pseudogene | 34.940632 | 34.941750 | positive | MGI_C57BL6J_3647220 | Gm8245 | NCBI_Gene:666697,ENSEMBL:ENSMUSG00000098209 | MGI:3647220 | pseudogene | predicted gene 8245 |
| 13 | gene | 34.946614 | 34.963938 | negative | MGI_C57BL6J_1916373 | Eci3 | NCBI_Gene:69123,ENSEMBL:ENSMUSG00000021416 | MGI:1916373 | protein coding gene | enoyl-Coenzyme A delta isomerase 3 |
| 13 | pseudogene | 34.962902 | 34.963060 | negative | MGI_C57BL6J_6096026 | Gm47217 | ENSEMBL:ENSMUSG00000113066 | MGI:6096026 | pseudogene | predicted gene, 47217 |
| 13 | gene | 34.963949 | 34.990452 | positive | MGI_C57BL6J_4439908 | Gm16984 | NCBI_Gene:102640991,ENSEMBL:ENSMUSG00000085962 | MGI:4439908 | lncRNA gene | predicted gene, 16984 |
| 13 | gene | 34.967435 | 34.968620 | positive | MGI_C57BL6J_1924675 | 9430081G06Rik | NA | NA | unclassified gene | RIKEN cDNA 9430081G06 gene |
| 13 | gene | 34.977748 | 35.027096 | negative | MGI_C57BL6J_1346064 | Eci2 | NCBI_Gene:23986,ENSEMBL:ENSMUSG00000021417 | MGI:1346064 | protein coding gene | enoyl-Coenzyme A delta isomerase 2 |
| 13 | pseudogene | 35.007680 | 35.011090 | negative | MGI_C57BL6J_5826045 | Gm46408 | NCBI_Gene:108168075 | MGI:5826045 | pseudogene | predicted gene, 46408 |
| 13 | gene | 35.027354 | 35.031946 | negative | MGI_C57BL6J_6098225 | Gm48626 | ENSEMBL:ENSMUSG00000114061 | MGI:6098225 | lncRNA gene | predicted gene, 48626 |
| 13 | gene | 35.052727 | 35.083396 | positive | MGI_C57BL6J_5623795 | Gm40910 | NCBI_Gene:105245461,ENSEMBL:ENSMUSG00000113738 | MGI:5623795 | lncRNA gene | predicted gene, 40910 |
| 13 | gene | 35.065461 | 35.067620 | positive | MGI_C57BL6J_6098230 | Gm48629 | ENSEMBL:ENSMUSG00000114233 | MGI:6098230 | lncRNA gene | predicted gene, 48629 |
| 13 | gene | 35.167146 | 35.172309 | negative | MGI_C57BL6J_1922447 | 4930529N20Rik | NCBI_Gene:75197,ENSEMBL:ENSMUSG00000113906 | MGI:1922447 | lncRNA gene | RIKEN cDNA 4930529N20 gene |
| 13 | gene | 35.167927 | 35.188017 | positive | MGI_C57BL6J_1922894 | 1700011B04Rik | NCBI_Gene:100503727,ENSEMBL:ENSMUSG00000102099 | MGI:1922894 | lncRNA gene | RIKEN cDNA 1700011B04 gene |
| 13 | gene | 35.184370 | 35.212048 | negative | MGI_C57BL6J_1922793 | 1700019C18Rik | NCBI_Gene:75543,ENSEMBL:ENSMUSG00000113770 | MGI:1922793 | lncRNA gene | RIKEN cDNA 1700019C18 gene |
| 13 | gene | 35.242971 | 35.256521 | negative | MGI_C57BL6J_6098325 | Gm48694 | ENSEMBL:ENSMUSG00000114237 | MGI:6098325 | lncRNA gene | predicted gene, 48694 |
| 13 | gene | 35.293147 | 35.293277 | negative | MGI_C57BL6J_5452451 | Gm22674 | ENSEMBL:ENSMUSG00000077601 | MGI:5452451 | snoRNA gene | predicted gene, 22674 |
| 13 | gene | 35.395383 | 35.455832 | positive | MGI_C57BL6J_6098342 | Gm48703 | ENSEMBL:ENSMUSG00000114838 | MGI:6098342 | lncRNA gene | predicted gene, 48703 |
| 13 | gene | 35.543902 | 35.545484 | positive | MGI_C57BL6J_6098344 | Gm48704 | ENSEMBL:ENSMUSG00000114337 | MGI:6098344 | lncRNA gene | predicted gene, 48704 |
| 13 | gene | 35.603030 | 35.660777 | negative | MGI_C57BL6J_6098349 | Gm48707 | ENSEMBL:ENSMUSG00000114661 | MGI:6098349 | lncRNA gene | predicted gene, 48707 |
| 13 | gene | 35.659810 | 35.874064 | positive | MGI_C57BL6J_1339956 | Cdyl | NCBI_Gene:12593,ENSEMBL:ENSMUSG00000059288 | MGI:1339956 | protein coding gene | chromodomain protein, Y chromosome-like |
| 13 | gene | 35.662135 | 35.662276 | positive | MGI_C57BL6J_5451903 | Gm22126 | ENSEMBL:ENSMUSG00000065178 | MGI:5451903 | snRNA gene | predicted gene, 22126 |
| 13 | gene | 35.773573 | 35.776896 | negative | MGI_C57BL6J_6098351 | Gm48708 | ENSEMBL:ENSMUSG00000114373 | MGI:6098351 | lncRNA gene | predicted gene, 48708 |
| 13 | gene | 35.798750 | 35.802136 | positive | MGI_C57BL6J_6098347 | Gm48706 | ENSEMBL:ENSMUSG00000114555 | MGI:6098347 | unclassified gene | predicted gene, 48706 |
| 13 | gene | 35.893472 | 35.911565 | negative | MGI_C57BL6J_1346084 | Rpp40 | NCBI_Gene:208366,ENSEMBL:ENSMUSG00000021418 | MGI:1346084 | protein coding gene | ribonuclease P 40 subunit |
| 13 | gene | 35.905457 | 35.921389 | positive | MGI_C57BL6J_6098418 | Gm48746 | ENSEMBL:ENSMUSG00000114270 | MGI:6098418 | lncRNA gene | predicted gene, 48746 |
| 13 | pseudogene | 35.937549 | 35.937833 | negative | MGI_C57BL6J_6098436 | Gm48756 | ENSEMBL:ENSMUSG00000114910 | MGI:6098436 | pseudogene | predicted gene, 48756 |
| 13 | gene | 35.957545 | 35.965008 | negative | MGI_C57BL6J_5589286 | Gm30127 | NCBI_Gene:102631915,ENSEMBL:ENSMUSG00000114647 | MGI:5589286 | lncRNA gene | predicted gene, 30127 |
| 13 | gene | 35.958799 | 35.970388 | positive | MGI_C57BL6J_1923737 | Ppp1r3g | NCBI_Gene:76487,ENSEMBL:ENSMUSG00000050423 | MGI:1923737 | protein coding gene | protein phosphatase 1, regulatory subunit 3G |
| 13 | gene | 35.978395 | 36.117526 | negative | MGI_C57BL6J_2683538 | Lyrm4 | NCBI_Gene:380840,ENSEMBL:ENSMUSG00000046573 | MGI:2683538 | protein coding gene | LYR motif containing 4 |
| 13 | pseudogene | 35.990257 | 35.995173 | negative | MGI_C57BL6J_3782198 | Tmed10-ps | NCBI_Gene:100042773,ENSEMBL:ENSMUSG00000114251 | MGI:3782198 | pseudogene | transmembrane p24 trafficking protein 10, pseudogene |
| 13 | pseudogene | 35.992132 | 35.993431 | positive | MGI_C57BL6J_5010317 | Gm18132 | NCBI_Gene:100416504,ENSEMBL:ENSMUSG00000114340 | MGI:5010317 | pseudogene | predicted gene, 18132 |
| 13 | gene | 35.995453 | 36.014464 | positive | MGI_C57BL6J_6098460 | Gm48770 | ENSEMBL:ENSMUSG00000114857 | MGI:6098460 | lncRNA gene | predicted gene, 48770 |
| 13 | gene | 36.006725 | 36.189723 | negative | MGI_C57BL6J_6098451 | Gm48765 | ENSEMBL:ENSMUSG00000114407 | MGI:6098451 | lncRNA gene | predicted gene, 48765 |
| 13 | gene | 36.110575 | 36.113825 | negative | MGI_C57BL6J_2442767 | 9530001P21Rik | NA | NA | unclassified gene | RIKEN cDNA 9530001P21 gene |
| 13 | gene | 36.117412 | 36.726280 | positive | MGI_C57BL6J_1917205 | Fars2 | NCBI_Gene:69955,ENSEMBL:ENSMUSG00000021420 | MGI:1917205 | protein coding gene | phenylalanine-tRNA synthetase 2 (mitochondrial) |
| 13 | pseudogene | 36.182712 | 36.183048 | negative | MGI_C57BL6J_6098459 | Gm48769 | ENSEMBL:ENSMUSG00000114371 | MGI:6098459 | pseudogene | predicted gene, 48769 |
| 13 | gene | 36.207753 | 36.228022 | positive | MGI_C57BL6J_5623797 | Gm40912 | NCBI_Gene:105245463 | MGI:5623797 | lncRNA gene | predicted gene, 40912 |
| 13 | gene | 36.252359 | 36.256920 | positive | MGI_C57BL6J_6098453 | Gm48766 | ENSEMBL:ENSMUSG00000114617 | MGI:6098453 | unclassified gene | predicted gene, 48766 |
| 13 | gene | 36.458447 | 36.459280 | negative | MGI_C57BL6J_6098447 | Gm48763 | ENSEMBL:ENSMUSG00000114714 | MGI:6098447 | unclassified gene | predicted gene, 48763 |
| 13 | pseudogene | 36.558329 | 36.559218 | negative | MGI_C57BL6J_3648013 | Gm8277 | NCBI_Gene:666758,ENSEMBL:ENSMUSG00000114581 | MGI:3648013 | pseudogene | predicted gene 8277 |
| 13 | gene | 36.569764 | 36.575934 | negative | MGI_C57BL6J_6098455 | Gm48767 | ENSEMBL:ENSMUSG00000114387 | MGI:6098455 | lncRNA gene | predicted gene, 48767 |
| 13 | gene | 36.582091 | 36.583526 | negative | MGI_C57BL6J_1918539 | 4933436N17Rik | ENSEMBL:ENSMUSG00000114746 | MGI:1918539 | unclassified gene | RIKEN cDNA 4933436N17 gene |
| 13 | gene | 36.612734 | 36.653073 | positive | MGI_C57BL6J_5826046 | Gm46409 | NCBI_Gene:108168076,ENSEMBL:ENSMUSG00000114851 | MGI:5826046 | lncRNA gene | predicted gene, 46409 |
| 13 | gene | 36.645120 | 36.653209 | positive | MGI_C57BL6J_5826047 | Gm46410 | NCBI_Gene:108168077 | MGI:5826047 | lncRNA gene | predicted gene, 46410 |
| 13 | gene | 36.654405 | 36.654536 | positive | MGI_C57BL6J_5456172 | Gm26395 | ENSEMBL:ENSMUSG00000077282 | MGI:5456172 | snoRNA gene | predicted gene, 26395 |
| 13 | gene | 36.717255 | 36.725176 | positive | MGI_C57BL6J_5621521 | Gm38636 | NCBI_Gene:102642800 | MGI:5621521 | lncRNA gene | predicted gene, 38636 |
| 13 | gene | 36.725361 | 36.735131 | negative | MGI_C57BL6J_1915654 | Nrn1 | NCBI_Gene:68404,ENSEMBL:ENSMUSG00000039114 | MGI:1915654 | protein coding gene | neuritin 1 |
| 13 | gene | 36.727258 | 36.729259 | positive | MGI_C57BL6J_5589336 | Gm30177 | NCBI_Gene:102631989,ENSEMBL:ENSMUSG00000114708 | MGI:5589336 | lncRNA gene | predicted gene, 30177 |
| 13 | gene | 36.823990 | 36.973329 | positive | MGI_C57BL6J_5589648 | Gm30489 | NCBI_Gene:102632411,ENSEMBL:ENSMUSG00000114329 | MGI:5589648 | lncRNA gene | predicted gene, 30489 |
| 13 | gene | 36.836366 | 36.885596 | positive | MGI_C57BL6J_6118556 | Gm49144 | ENSEMBL:ENSMUSG00000114483 | MGI:6118556 | lncRNA gene | predicted gene, 49144 |
| 13 | gene | 36.864016 | 36.864625 | negative | MGI_C57BL6J_6096772 | Gm47675 | ENSEMBL:ENSMUSG00000114884 | MGI:6096772 | lncRNA gene | predicted gene, 47675 |
| 13 | gene | 36.867178 | 37.050244 | negative | MGI_C57BL6J_1921395 | F13a1 | NCBI_Gene:74145,ENSEMBL:ENSMUSG00000039109 | MGI:1921395 | protein coding gene | coagulation factor XIII, A1 subunit |
| 13 | gene | 36.989093 | 36.997851 | positive | MGI_C57BL6J_5589399 | Gm30240 | NCBI_Gene:102632069 | MGI:5589399 | lncRNA gene | predicted gene, 30240 |
| 13 | gene | 37.070967 | 37.072531 | positive | MGI_C57BL6J_5589708 | Gm30549 | NCBI_Gene:102632490 | MGI:5589708 | lncRNA gene | predicted gene, 30549 |
| 13 | pseudogene | 37.092547 | 37.095675 | negative | MGI_C57BL6J_5010828 | Gm18643 | NCBI_Gene:100417491,ENSEMBL:ENSMUSG00000114336 | MGI:5010828 | pseudogene | predicted gene, 18643 |
| 13 | gene | 37.156018 | 37.157755 | positive | MGI_C57BL6J_6096825 | Gm47707 | ENSEMBL:ENSMUSG00000114428 | MGI:6096825 | lncRNA gene | predicted gene, 47707 |
| 13 | gene | 37.237649 | 37.249502 | negative | MGI_C57BL6J_5589759 | Gm30600 | NCBI_Gene:102632558,ENSEMBL:ENSMUSG00000114297 | MGI:5589759 | lncRNA gene | predicted gene, 30600 |
| 13 | gene | 37.248660 | 37.252165 | positive | MGI_C57BL6J_6096829 | Gm47709 | ENSEMBL:ENSMUSG00000114249 | MGI:6096829 | lncRNA gene | predicted gene, 47709 |
| 13 | gene | 37.326466 | 37.327047 | positive | MGI_C57BL6J_6096833 | Gm47711 | ENSEMBL:ENSMUSG00000114508 | MGI:6096833 | lncRNA gene | predicted gene, 47711 |
| 13 | gene | 37.333725 | 37.341997 | positive | MGI_C57BL6J_5623798 | Gm40913 | NCBI_Gene:105245464 | MGI:5623798 | lncRNA gene | predicted gene, 40913 |
| 13 | gene | 37.345208 | 37.419036 | positive | MGI_C57BL6J_1321404 | Ly86 | NCBI_Gene:17084,ENSEMBL:ENSMUSG00000021423 | MGI:1321404 | protein coding gene | lymphocyte antigen 86 |
| 13 | gene | 37.421833 | 37.435149 | negative | MGI_C57BL6J_3782209 | Gm4035 | NCBI_Gene:100042794,ENSEMBL:ENSMUSG00000100760 | MGI:3782209 | lncRNA gene | predicted gene 4035 |
| 13 | gene | 37.439593 | 37.440454 | negative | MGI_C57BL6J_6096864 | Gm47730 | ENSEMBL:ENSMUSG00000114791 | MGI:6096864 | lncRNA gene | predicted gene, 47730 |
| 13 | gene | 37.449400 | 37.451157 | negative | MGI_C57BL6J_1917395 | 2210022D18Rik | NA | NA | unclassified gene | RIKEN cDNA 2210022D18 gene |
| 13 | gene | 37.449400 | 37.451157 | negative | MGI_C57BL6J_5580165 | Gm29459 | ENSEMBL:ENSMUSG00000100932 | MGI:5580165 | lncRNA gene | predicted gene 29459 |
| 13 | gene | 37.452212 | 37.453208 | negative | MGI_C57BL6J_5580164 | Gm29458 | ENSEMBL:ENSMUSG00000099847 | MGI:5580164 | lncRNA gene | predicted gene 29458 |
| 13 | gene | 37.472038 | 37.479213 | positive | MGI_C57BL6J_6096866 | Gm47731 | ENSEMBL:ENSMUSG00000114701 | MGI:6096866 | lncRNA gene | predicted gene, 47731 |
| 13 | gene | 37.472038 | 37.479213 | negative | MGI_C57BL6J_2145343 | AU017834 | NA | NA | unclassified gene | expressed sequence AU017834 |
| 13 | gene | 37.477085 | 37.487980 | negative | MGI_C57BL6J_5623799 | Gm40914 | NCBI_Gene:105245465 | MGI:5623799 | lncRNA gene | predicted gene, 40914 |
| 13 | gene | 37.482385 | 37.483150 | positive | MGI_C57BL6J_6096868 | Gm47732 | ENSEMBL:ENSMUSG00000114573 | MGI:6096868 | lncRNA gene | predicted gene, 47732 |
| 13 | gene | 37.490045 | 37.532958 | negative | MGI_C57BL6J_5590022 | Gm30863 | NCBI_Gene:102632911 | MGI:5590022 | lncRNA gene | predicted gene, 30863 |
| 13 | gene | 37.504206 | 37.507752 | negative | MGI_C57BL6J_6096872 | Gm47734 | ENSEMBL:ENSMUSG00000114389 | MGI:6096872 | lncRNA gene | predicted gene, 47734 |
| 13 | gene | 37.508940 | 37.512421 | positive | MGI_C57BL6J_5580296 | Gm29590 | NCBI_Gene:102632831,ENSEMBL:ENSMUSG00000100022 | MGI:5580296 | lncRNA gene | predicted gene 29590 |
| 13 | gene | 37.540136 | 37.549725 | positive | MGI_C57BL6J_5590077 | Lncbate6 | NCBI_Gene:102632980,ENSEMBL:ENSMUSG00000114665 | MGI:5590077 | lncRNA gene | brown adipose tissue enriched long noncoding RNA 6 |
| 13 | gene | 37.561945 | 37.564972 | negative | MGI_C57BL6J_6096895 | Gm47751 | ENSEMBL:ENSMUSG00000114724 | MGI:6096895 | lncRNA gene | predicted gene, 47751 |
| 13 | gene | 37.581077 | 37.589313 | positive | MGI_C57BL6J_6096900 | Gm47754 | ENSEMBL:ENSMUSG00000114784 | MGI:6096900 | lncRNA gene | predicted gene, 47754 |
| 13 | pseudogene | 37.582271 | 37.582564 | positive | MGI_C57BL6J_6096899 | Gm47753 | ENSEMBL:ENSMUSG00000114718 | MGI:6096899 | pseudogene | predicted gene, 47753 |
| 13 | gene | 37.616600 | 37.627603 | negative | MGI_C57BL6J_5623800 | Gm40915 | NCBI_Gene:105245466,ENSEMBL:ENSMUSG00000114404 | MGI:5623800 | lncRNA gene | predicted gene, 40915 |
| 13 | gene | 37.633785 | 37.648236 | negative | MGI_C57BL6J_5623801 | Gm40916 | NCBI_Gene:105245467,ENSEMBL:ENSMUSG00000114355 | MGI:5623801 | lncRNA gene | predicted gene, 40916 |
| 13 | gene | 37.643905 | 37.648099 | negative | MGI_C57BL6J_5623802 | Gm40917 | NCBI_Gene:105245468 | MGI:5623802 | lncRNA gene | predicted gene, 40917 |
| 13 | gene | 37.657013 | 37.670077 | negative | MGI_C57BL6J_2145255 | AI463229 | NCBI_Gene:100042802,ENSEMBL:ENSMUSG00000114828 | MGI:2145255 | lncRNA gene | expressed sequence AI463229 |
| 13 | gene | 37.684925 | 37.686425 | negative | MGI_C57BL6J_6121551 | Gm49350 | ENSEMBL:ENSMUSG00000114546 | MGI:6121551 | lncRNA gene | predicted gene, 49350 |
| 13 | gene | 37.685631 | 37.717648 | negative | MGI_C57BL6J_5623803 | Gm40918 | NCBI_Gene:105245469,ENSEMBL:ENSMUSG00000114732 | MGI:5623803 | lncRNA gene | predicted gene, 40918 |
| 13 | pseudogene | 37.697306 | 37.698158 | negative | MGI_C57BL6J_6121553 | Gm49351 | ENSEMBL:ENSMUSG00000114259 | MGI:6121553 | pseudogene | predicted gene, 49351 |
| 13 | gene | 37.717382 | 37.734742 | negative | MGI_C57BL6J_5825573 | Gm45936 | NCBI_Gene:108167318 | MGI:5825573 | lncRNA gene | predicted gene, 45936 |
| 13 | gene | 37.761545 | 37.768147 | positive | MGI_C57BL6J_5590759 | Gm31600 | NCBI_Gene:102633878,ENSEMBL:ENSMUSG00000114800 | MGI:5590759 | lncRNA gene | predicted gene, 31600 |
| 13 | gene | 37.778380 | 37.952005 | positive | MGI_C57BL6J_2443664 | Rreb1 | NCBI_Gene:68750,ENSEMBL:ENSMUSG00000039087 | MGI:2443664 | protein coding gene | ras responsive element binding protein 1 |
| 13 | gene | 37.793382 | 37.800785 | negative | MGI_C57BL6J_5826048 | Gm46411 | NCBI_Gene:108168078,ENSEMBL:ENSMUSG00000114370 | MGI:5826048 | lncRNA gene | predicted gene, 46411 |
| 13 | gene | 37.966605 | 37.994236 | negative | MGI_C57BL6J_105082 | Ssr1 | NCBI_Gene:107513,ENSEMBL:ENSMUSG00000021427 | MGI:105082 | protein coding gene | signal sequence receptor, alpha |
| 13 | gene | 38.006052 | 38.037069 | negative | MGI_C57BL6J_1918463 | Cage1 | NCBI_Gene:71213,ENSEMBL:ENSMUSG00000044566 | MGI:1918463 | protein coding gene | cancer antigen 1 |
| 13 | gene | 38.036989 | 38.061433 | positive | MGI_C57BL6J_1918590 | Riok1 | NCBI_Gene:71340,ENSEMBL:ENSMUSG00000021428 | MGI:1918590 | protein coding gene | RIO kinase 1 |
| 13 | gene | 38.077059 | 38.080465 | negative | MGI_C57BL6J_5623805 | Gm40920 | NCBI_Gene:105245471 | MGI:5623805 | lncRNA gene | predicted gene, 40920 |
| 13 | gene | 38.086202 | 38.086317 | positive | MGI_C57BL6J_5530769 | Gm27387 | ENSEMBL:ENSMUSG00000098481 | MGI:5530769 | miRNA gene | predicted gene, 27387 |
| 13 | gene | 38.150477 | 38.151792 | negative | MGI_C57BL6J_3641890 | Gm10129 | ENSEMBL:ENSMUSG00000103789 | MGI:3641890 | protein coding gene | predicted gene 10129 |
| 13 | gene | 38.151082 | 38.198577 | positive | MGI_C57BL6J_109611 | Dsp | NCBI_Gene:109620,ENSEMBL:ENSMUSG00000054889 | MGI:109611 | protein coding gene | desmoplakin |
| 13 | gene | 38.204914 | 38.227665 | positive | MGI_C57BL6J_1915047 | Snrnp48 | NCBI_Gene:67797,ENSEMBL:ENSMUSG00000021431 | MGI:1915047 | protein coding gene | small nuclear ribonucleoprotein 48 (U11/U12) |
| 13 | gene | 38.258030 | 38.260264 | negative | MGI_C57BL6J_5623806 | Gm40921 | NCBI_Gene:105245472 | MGI:5623806 | lncRNA gene | predicted gene, 40921 |
| 13 | pseudogene | 38.270388 | 38.270609 | negative | MGI_C57BL6J_6097264 | Gm47978 | ENSEMBL:ENSMUSG00000114799 | MGI:6097264 | pseudogene | predicted gene, 47978 |
| 13 | gene | 38.320526 | 38.340721 | positive | MGI_C57BL6J_6097284 | Gm47990 | ENSEMBL:ENSMUSG00000114525 | MGI:6097284 | lncRNA gene | predicted gene, 47990 |
| 13 | pseudogene | 38.332572 | 38.332938 | positive | MGI_C57BL6J_6097286 | Gm47991 | ENSEMBL:ENSMUSG00000114861 | MGI:6097286 | pseudogene | predicted gene, 47991 |
| 13 | gene | 38.345107 | 38.500314 | positive | MGI_C57BL6J_88182 | Bmp6 | NCBI_Gene:12161,ENSEMBL:ENSMUSG00000039004 | MGI:88182 | protein coding gene | bone morphogenetic protein 6 |
| 13 | gene | 38.454149 | 38.461805 | negative | MGI_C57BL6J_1923104 | 4930579J19Rik | NCBI_Gene:75854,ENSEMBL:ENSMUSG00000114895 | MGI:1923104 | lncRNA gene | RIKEN cDNA 4930579J19 gene |
| 13 | gene | 38.500079 | 38.528824 | negative | MGI_C57BL6J_2145316 | Txndc5 | NCBI_Gene:105245,ENSEMBL:ENSMUSG00000038991 | MGI:2145316 | protein coding gene | thioredoxin domain containing 5 |
| 13 | pseudogene | 38.514096 | 38.514875 | positive | MGI_C57BL6J_6097290 | Gm47994 | ENSEMBL:ENSMUSG00000114399 | MGI:6097290 | pseudogene | predicted gene, 47994 |
| 13 | gene | 38.527403 | 38.532920 | positive | MGI_C57BL6J_5590993 | Gm31834 | NCBI_Gene:102634191,ENSEMBL:ENSMUSG00000114638 | MGI:5590993 | lncRNA gene | predicted gene, 31834 |
| 13 | gene | 38.535274 | 38.536680 | negative | MGI_C57BL6J_6097377 | Gm48056 | ENSEMBL:ENSMUSG00000114331 | MGI:6097377 | lncRNA gene | predicted gene, 48056 |
| 13 | gene | 38.554775 | 38.564815 | positive | MGI_C57BL6J_5623807 | Gm40922 | NCBI_Gene:105245473,ENSEMBL:ENSMUSG00000114866 | MGI:5623807 | lncRNA gene | predicted gene, 40922 |
| 13 | gene | 38.592842 | 38.635876 | negative | MGI_C57BL6J_2178598 | Bloc1s5 | NCBI_Gene:17828,ENSEMBL:ENSMUSG00000038982 | MGI:2178598 | protein coding gene | biogenesis of lysosomal organelles complex-1, subunit 5, muted |
| 13 | gene | 38.634971 | 38.637116 | positive | MGI_C57BL6J_6096328 | Gm47400 | ENSEMBL:ENSMUSG00000114830 | MGI:6096328 | lncRNA gene | predicted gene, 47400 |
| 13 | gene | 38.644207 | 38.659058 | negative | MGI_C57BL6J_1913393 | Eef1e1 | NCBI_Gene:66143,ENSEMBL:ENSMUSG00000001707 | MGI:1913393 | protein coding gene | eukaryotic translation elongation factor 1 epsilon 1 |
| 13 | pseudogene | 38.673878 | 38.674670 | negative | MGI_C57BL6J_3644054 | Gm6200 | NCBI_Gene:102634259,ENSEMBL:ENSMUSG00000114651 | MGI:3644054 | pseudogene | predicted gene 6200 |
| 13 | gene | 38.710054 | 38.908255 | positive | MGI_C57BL6J_5826029 | Gm46392 | NCBI_Gene:108168052,ENSEMBL:ENSMUSG00000114767 | MGI:5826029 | lncRNA gene | predicted gene, 46392 |
| 13 | gene | 38.723642 | 38.727222 | positive | MGI_C57BL6J_6096499 | Gm47510 | ENSEMBL:ENSMUSG00000114813 | MGI:6096499 | lncRNA gene | predicted gene, 47510 |
| 13 | gene | 38.775824 | 38.777191 | positive | MGI_C57BL6J_6096501 | Gm47511 | ENSEMBL:ENSMUSG00000114845 | MGI:6096501 | lncRNA gene | predicted gene, 47511 |
| 13 | gene | 38.782161 | 38.825755 | negative | MGI_C57BL6J_6096497 | Gm47509 | ENSEMBL:ENSMUSG00000114319 | MGI:6096497 | lncRNA gene | predicted gene, 47509 |
| 13 | gene | 38.898360 | 38.898587 | negative | MGI_C57BL6J_5451812 | Gm22035 | ENSEMBL:ENSMUSG00000087909 | MGI:5451812 | unclassified non-coding RNA gene | predicted gene, 22035 |
| 13 | gene | 38.917153 | 38.919219 | negative | MGI_C57BL6J_6096533 | Gm47530 | ENSEMBL:ENSMUSG00000114949 | MGI:6096533 | unclassified gene | predicted gene, 47530 |
| 13 | gene | 38.932136 | 38.960875 | negative | MGI_C57BL6J_1913978 | Slc35b3 | NCBI_Gene:108652,ENSEMBL:ENSMUSG00000021432 | MGI:1913978 | protein coding gene | solute carrier family 35, member B3 |
| 13 | gene | 38.945543 | 38.951313 | negative | MGI_C57BL6J_5477008 | Gm26514 | ENSEMBL:ENSMUSG00000097151 | MGI:5477008 | lncRNA gene | predicted gene, 26514 |
| 13 | gene | 38.960619 | 38.966605 | positive | MGI_C57BL6J_1923208 | 5033403F01Rik | NCBI_Gene:75958,ENSEMBL:ENSMUSG00000096932 | MGI:1923208 | lncRNA gene | RIKEN cDNA 5033403F01 gene |
| 13 | gene | 39.440565 | 39.449671 | positive | MGI_C57BL6J_6096246 | Gm47348 | ENSEMBL:ENSMUSG00000114512 | MGI:6096246 | lncRNA gene | predicted gene, 47348 |
| 13 | gene | 39.463227 | 39.467017 | negative | MGI_C57BL6J_3781686 | Gm3509 | ENSEMBL:ENSMUSG00000114821 | MGI:3781686 | lncRNA gene | predicted gene 3509 |
| 13 | gene | 39.467347 | 39.470134 | positive | MGI_C57BL6J_6096248 | Gm47349 | ENSEMBL:ENSMUSG00000114330 | MGI:6096248 | lncRNA gene | predicted gene, 47349 |
| 13 | gene | 39.530598 | 39.533103 | negative | MGI_C57BL6J_6096252 | Gm47351 | ENSEMBL:ENSMUSG00000114221 | MGI:6096252 | lncRNA gene | predicted gene, 47351 |
| 13 | gene | 39.571583 | 39.699499 | positive | MGI_C57BL6J_6096254 | Gm47352 | ENSEMBL:ENSMUSG00000114841 | MGI:6096254 | lncRNA gene | predicted gene, 47352 |
| 13 | gene | 39.617315 | 39.647352 | negative | MGI_C57BL6J_5591402 | Gm32243 | NCBI_Gene:102634725,ENSEMBL:ENSMUSG00000114300 | MGI:5591402 | lncRNA gene | predicted gene, 32243 |
| 13 | pseudogene | 39.757299 | 39.757967 | positive | MGI_C57BL6J_3645612 | Gm5195 | NCBI_Gene:382747,ENSEMBL:ENSMUSG00000114346 | MGI:3645612 | pseudogene | predicted gene 5195 |
| 13 | gene | 39.771790 | 39.791384 | negative | MGI_C57BL6J_5623808 | Gm40923 | NCBI_Gene:105245474,ENSEMBL:ENSMUSG00000114710 | MGI:5623808 | lncRNA gene | predicted gene, 40923 |
| 13 | gene | 39.791438 | 39.844840 | positive | MGI_C57BL6J_1925003 | A230103O09Rik | NCBI_Gene:77753,ENSEMBL:ENSMUSG00000114491 | MGI:1925003 | lncRNA gene | RIKEN cDNA A230103O09 gene |
| 13 | gene | 39.791448 | 39.794937 | positive | MGI_C57BL6J_5826030 | Gm46393 | NCBI_Gene:108168053 | MGI:5826030 | lncRNA gene | predicted gene, 46393 |
| 13 | pseudogene | 39.796321 | 39.796944 | positive | MGI_C57BL6J_6096270 | Gm47361 | ENSEMBL:ENSMUSG00000114381 | MGI:6096270 | pseudogene | predicted gene, 47361 |
| 13 | gene | 39.803393 | 39.806238 | negative | MGI_C57BL6J_5826031 | Gm46394 | NCBI_Gene:108168054 | MGI:5826031 | lncRNA gene | predicted gene, 46394 |
| 13 | gene | 39.858863 | 39.859027 | negative | MGI_C57BL6J_5453749 | Gm23972 | ENSEMBL:ENSMUSG00000065256 | MGI:5453749 | snRNA gene | predicted gene, 23972 |
| 13 | gene | 39.877035 | 39.884381 | negative | MGI_C57BL6J_6096197 | Gm47316 | ENSEMBL:ENSMUSG00000114957 | MGI:6096197 | lncRNA gene | predicted gene, 47316 |
| 13 | gene | 39.912866 | 39.923211 | negative | MGI_C57BL6J_5623809 | Gm40924 | NCBI_Gene:105245475 | MGI:5623809 | lncRNA gene | predicted gene, 40924 |
| 13 | gene | 40.001882 | 40.361450 | negative | MGI_C57BL6J_2658851 | Ofcc1 | NCBI_Gene:218165,ENSEMBL:ENSMUSG00000047094 | MGI:2658851 | protein coding gene | orofacial cleft 1 candidate 1 |
| 13 | pseudogene | 40.496188 | 40.497473 | positive | MGI_C57BL6J_3646048 | Gm8352 | NCBI_Gene:666894,ENSEMBL:ENSMUSG00000114248 | MGI:3646048 | pseudogene | predicted gene 8352 |
| 13 | pseudogene | 40.533570 | 40.533947 | positive | MGI_C57BL6J_6095738 | Gm47038 | ENSEMBL:ENSMUSG00000114215 | MGI:6095738 | pseudogene | predicted gene, 47038 |
| 13 | gene | 40.559320 | 40.606479 | negative | MGI_C57BL6J_6095739 | Gm47039 | ENSEMBL:ENSMUSG00000114198 | MGI:6095739 | lncRNA gene | predicted gene, 47039 |
| 13 | gene | 40.642607 | 40.644649 | negative | MGI_C57BL6J_6095743 | Gm47041 | ENSEMBL:ENSMUSG00000114322 | MGI:6095743 | unclassified gene | predicted gene, 47041 |
| 13 | gene | 40.704005 | 40.707949 | positive | MGI_C57BL6J_3642885 | Gm9979 | ENSEMBL:ENSMUSG00000055732 | MGI:3642885 | lncRNA gene | predicted gene 9979 |
| 13 | gene | 40.715302 | 40.738376 | negative | MGI_C57BL6J_104671 | Tfap2a | NCBI_Gene:21418,ENSEMBL:ENSMUSG00000021359 | MGI:104671 | protein coding gene | transcription factor AP-2, alpha |
| 13 | gene | 40.723110 | 40.725449 | positive | MGI_C57BL6J_6095771 | Gm47061 | ENSEMBL:ENSMUSG00000114936 | MGI:6095771 | lncRNA gene | predicted gene, 47061 |
| 13 | gene | 40.727808 | 40.735162 | positive | MGI_C57BL6J_5477182 | Gm26688 | NCBI_Gene:102633805,ENSEMBL:ENSMUSG00000097752 | MGI:5477182 | lncRNA gene | predicted gene, 26688 |
| 13 | gene | 40.737568 | 40.739021 | positive | MGI_C57BL6J_6095781 | Gm47067 | ENSEMBL:ENSMUSG00000114597 | MGI:6095781 | lncRNA gene | predicted gene, 47067 |
| 13 | gene | 40.754060 | 40.754996 | negative | MGI_C57BL6J_6097459 | Gm48107 | ENSEMBL:ENSMUSG00000114640 | MGI:6097459 | lncRNA gene | predicted gene, 48107 |
| 13 | pseudogene | 40.778040 | 40.778807 | positive | MGI_C57BL6J_3646756 | Gm6245 | NCBI_Gene:621629,ENSEMBL:ENSMUSG00000090475 | MGI:3646756 | pseudogene | predicted gene 6245 |
| 13 | pseudogene | 40.830771 | 40.838656 | negative | MGI_C57BL6J_3647123 | Gm7201 | NCBI_Gene:637157,ENSEMBL:ENSMUSG00000114263 | MGI:3647123 | pseudogene | predicted gene 7201 |
| 13 | gene | 40.846733 | 40.852426 | positive | MGI_C57BL6J_5594319 | Gm35160 | NCBI_Gene:102638646,ENSEMBL:ENSMUSG00000114560 | MGI:5594319 | protein coding gene | predicted gene, 35160 |
| 13 | gene | 40.859754 | 40.960892 | positive | MGI_C57BL6J_1100870 | Gcnt2 | NCBI_Gene:14538,ENSEMBL:ENSMUSG00000021360 | MGI:1100870 | protein coding gene | glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| 13 | gene | 40.865791 | 40.865862 | positive | MGI_C57BL6J_4950449 | Mir5124a | miRBase:MI0018035,NCBI_Gene:100628630,ENSEMBL:ENSMUSG00000093269 | MGI:4950449 | miRNA gene | microRNA 5124a |
| 13 | gene | 40.873498 | 40.874189 | negative | MGI_C57BL6J_6097574 | Gm48192 | ENSEMBL:ENSMUSG00000114672 | MGI:6097574 | unclassified gene | predicted gene, 48192 |
| 13 | gene | 40.897180 | 40.917895 | negative | MGI_C57BL6J_5590842 | Gm31683 | NCBI_Gene:102633993,ENSEMBL:ENSMUSG00000114662 | MGI:5590842 | lncRNA gene | predicted gene, 31683 |
| 13 | pseudogene | 40.911504 | 40.912207 | negative | MGI_C57BL6J_6097568 | Gm48188 | ENSEMBL:ENSMUSG00000114601 | MGI:6097568 | pseudogene | predicted gene, 48188 |
| 13 | gene | 40.927834 | 41.001151 | negative | MGI_C57BL6J_2441825 | A730081D07Rik | NCBI_Gene:102633880,ENSEMBL:ENSMUSG00000086693 | MGI:2441825 | lncRNA gene | RIKEN cDNA A730081D07 gene |
| 13 | gene | 40.948889 | 40.949631 | negative | MGI_C57BL6J_1918697 | 5530402G07Rik | ENSEMBL:ENSMUSG00000114723 | MGI:1918697 | unclassified gene | RIKEN cDNA 5530402G07 gene |
| 13 | gene | 41.001010 | 41.013033 | positive | MGI_C57BL6J_1915333 | Pak1ip1 | NCBI_Gene:68083,ENSEMBL:ENSMUSG00000038683 | MGI:1915333 | protein coding gene | PAK1 interacting protein 1 |
| 13 | gene | 41.016250 | 41.022586 | positive | MGI_C57BL6J_1913404 | Tmem14c | NCBI_Gene:66154,ENSEMBL:ENSMUSG00000021361 | MGI:1913404 | protein coding gene | transmembrane protein 14C |
| 13 | gene | 41.025007 | 41.079706 | negative | MGI_C57BL6J_96913 | Mak | NCBI_Gene:17152,ENSEMBL:ENSMUSG00000021363 | MGI:96913 | protein coding gene | male germ cell-associated kinase |
| 13 | pseudogene | 41.091430 | 41.091723 | positive | MGI_C57BL6J_6097807 | Gm48344 | ENSEMBL:ENSMUSG00000114385 | MGI:6097807 | pseudogene | predicted gene, 48344 |
| 13 | gene | 41.101427 | 41.111035 | negative | MGI_C57BL6J_1861438 | Gcm2 | NCBI_Gene:107889,ENSEMBL:ENSMUSG00000021362 | MGI:1861438 | protein coding gene | glial cells missing homolog 2 |
| 13 | gene | 41.114307 | 41.174351 | positive | MGI_C57BL6J_2685114 | Sycp2l | NCBI_Gene:637277,ENSEMBL:ENSMUSG00000038651 | MGI:2685114 | protein coding gene | synaptonemal complex protein 2-like |
| 13 | gene | 41.176888 | 41.179030 | positive | MGI_C57BL6J_6098042 | Gm48510 | ENSEMBL:ENSMUSG00000114388 | MGI:6098042 | lncRNA gene | predicted gene, 48510 |
| 13 | gene | 41.182381 | 41.220405 | negative | MGI_C57BL6J_1858960 | Elovl2 | NCBI_Gene:54326,ENSEMBL:ENSMUSG00000021364 | MGI:1858960 | protein coding gene | elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 |
| 13 | pseudogene | 41.234663 | 41.235100 | negative | MGI_C57BL6J_3647979 | Rps17-ps1 | NCBI_Gene:268650,ENSEMBL:ENSMUSG00000114238 | MGI:3647979 | pseudogene | ribosomal protein S17, pseudogene 1 |
| 13 | gene | 41.236046 | 41.242083 | positive | MGI_C57BL6J_5591222 | Gm32063 | NCBI_Gene:102634494,ENSEMBL:ENSMUSG00000114247 | MGI:5591222 | lncRNA gene | predicted gene, 32063 |
| 13 | gene | 41.249844 | 41.276577 | positive | MGI_C57BL6J_2652854 | Smim13 | NCBI_Gene:108934,ENSEMBL:ENSMUSG00000091264 | MGI:2652854 | protein coding gene | small integral membrane protein 13 |
| 13 | gene | 41.273803 | 41.273977 | negative | MGI_C57BL6J_4936998 | Gm17364 | ENSEMBL:ENSMUSG00000090853 | MGI:4936998 | protein coding gene | predicted gene, 17364 |
| 13 | gene | 41.309581 | 41.487362 | negative | MGI_C57BL6J_97302 | Nedd9 | NCBI_Gene:18003,ENSEMBL:ENSMUSG00000021365 | MGI:97302 | protein coding gene | neural precursor cell expressed, developmentally down-regulated gene 9 |
| 13 | gene | 41.359971 | 41.370908 | positive | MGI_C57BL6J_5477371 | Gm26877 | ENSEMBL:ENSMUSG00000097373 | MGI:5477371 | lncRNA gene | predicted gene, 26877 |
| 13 | gene | 41.379956 | 41.384725 | positive | MGI_C57BL6J_6098134 | Gm48571 | ENSEMBL:ENSMUSG00000114914 | MGI:6098134 | lncRNA gene | predicted gene, 48571 |
| 13 | gene | 41.388724 | 41.390797 | negative | MGI_C57BL6J_2445012 | A130026F07Rik | NA | NA | unclassified gene | RIKEN cDNA A130026F07 gene |
| 13 | gene | 41.402291 | 41.403353 | positive | MGI_C57BL6J_6098132 | Gm48570 | ENSEMBL:ENSMUSG00000114552 | MGI:6098132 | lncRNA gene | predicted gene, 48570 |
| 13 | gene | 41.407412 | 41.411210 | positive | MGI_C57BL6J_5591607 | Gm32448 | NCBI_Gene:102635003 | MGI:5591607 | lncRNA gene | predicted gene, 32448 |
| 13 | gene | 41.476526 | 41.477900 | negative | MGI_C57BL6J_6098130 | Gm48569 | ENSEMBL:ENSMUSG00000114537 | MGI:6098130 | unclassified gene | predicted gene, 48569 |
| 13 | gene | 41.486649 | 41.544567 | positive | MGI_C57BL6J_5591560 | Gm32401 | NCBI_Gene:102634940,ENSEMBL:ENSMUSG00000114585 | MGI:5591560 | lncRNA gene | predicted gene, 32401 |
| 13 | gene | 41.513893 | 41.605820 | negative | MGI_C57BL6J_5591343 | Gm32184 | NCBI_Gene:102634650,ENSEMBL:ENSMUSG00000114545 | MGI:5591343 | lncRNA gene | predicted gene, 32184 |
| 13 | gene | 41.577909 | 41.578074 | negative | MGI_C57BL6J_5454596 | Gm24819 | ENSEMBL:ENSMUSG00000092835 | MGI:5454596 | snRNA gene | predicted gene, 24819 |
| 13 | gene | 41.605907 | 41.641357 | positive | MGI_C57BL6J_3647046 | Tmem170b | NCBI_Gene:621976,ENSEMBL:ENSMUSG00000087370 | MGI:3647046 | protein coding gene | transmembrane protein 170B |
| 13 | gene | 41.649443 | 41.656777 | positive | MGI_C57BL6J_3644232 | Gm5082 | NCBI_Gene:328231,ENSEMBL:ENSMUSG00000054258 | MGI:3644232 | lncRNA gene | predicted gene 5082 |
| 13 | gene | 41.676858 | 41.683924 | positive | MGI_C57BL6J_5623810 | Gm40925 | NCBI_Gene:105245476 | MGI:5623810 | lncRNA gene | predicted gene, 40925 |
| 13 | gene | 41.763145 | 41.847684 | negative | MGI_C57BL6J_1924596 | Adtrp | NCBI_Gene:109254,ENSEMBL:ENSMUSG00000058022 | MGI:1924596 | protein coding gene | androgen dependent TFPI regulating protein |
| 13 | gene | 41.800783 | 41.801795 | positive | MGI_C57BL6J_1924241 | 1700061E18Rik | ENSEMBL:ENSMUSG00000085759 | MGI:1924241 | lncRNA gene | RIKEN cDNA 1700061E18 gene |
| 13 | gene | 41.843014 | 41.849768 | positive | MGI_C57BL6J_5579270 | Gm28564 | ENSEMBL:ENSMUSG00000099672 | MGI:5579270 | lncRNA gene | predicted gene 28564 |
| 13 | gene | 41.850718 | 41.851180 | positive | MGI_C57BL6J_5610926 | Gm37698 | ENSEMBL:ENSMUSG00000103479 | MGI:5610926 | unclassified gene | predicted gene, 37698 |
| 13 | gene | 41.865915 | 41.913092 | positive | MGI_C57BL6J_3641845 | Gm10790 | NCBI_Gene:100038577,ENSEMBL:ENSMUSG00000113856 | MGI:3641845 | lncRNA gene | predicted gene 10790 |
| 13 | gene | 41.901651 | 41.901754 | positive | MGI_C57BL6J_5451934 | Gm22157 | ENSEMBL:ENSMUSG00000089035 | MGI:5451934 | snRNA gene | predicted gene, 22157 |
| 13 | gene | 41.993919 | 42.021086 | negative | MGI_C57BL6J_5591714 | Gm32555 | NCBI_Gene:102635142 | MGI:5591714 | lncRNA gene | predicted gene, 32555 |
| 13 | gene | 42.046266 | 42.050567 | positive | MGI_C57BL6J_5579413 | Gm28707 | NCBI_Gene:108168079,ENSEMBL:ENSMUSG00000100071 | MGI:5579413 | lncRNA gene | predicted gene 28707 |
| 13 | gene | 42.051970 | 42.192537 | positive | MGI_C57BL6J_96100 | Hivep1 | NCBI_Gene:110521,ENSEMBL:ENSMUSG00000021366 | MGI:96100 | protein coding gene | human immunodeficiency virus type I enhancer binding protein 1 |
| 13 | gene | 42.186725 | 42.199473 | positive | MGI_C57BL6J_5591937 | Gm32778 | NCBI_Gene:102635441 | MGI:5591937 | lncRNA gene | predicted gene, 32778 |
| 13 | gene | 42.199733 | 42.203860 | positive | MGI_C57BL6J_2441672 | 4732499D12Rik | NA | NA | unclassified gene | RIKEN cDNA 4732499D12 gene |
| 13 | gene | 42.301476 | 42.307990 | positive | MGI_C57BL6J_95283 | Edn1 | NCBI_Gene:13614,ENSEMBL:ENSMUSG00000021367 | MGI:95283 | protein coding gene | endothelin 1 |
| 13 | gene | 42.302298 | 42.304991 | negative | MGI_C57BL6J_6095863 | Gm47118 | ENSEMBL:ENSMUSG00000113619 | MGI:6095863 | lncRNA gene | predicted gene, 47118 |
| 13 | gene | 42.389899 | 42.403389 | negative | MGI_C57BL6J_5591992 | Gm32833 | NCBI_Gene:102635520 | MGI:5591992 | lncRNA gene | predicted gene, 32833 |
| 13 | pseudogene | 42.472320 | 42.472777 | negative | MGI_C57BL6J_3645582 | Gm6049 | NCBI_Gene:606512,ENSEMBL:ENSMUSG00000113000 | MGI:3645582 | pseudogene | predicted gene 6049 |
| 13 | pseudogene | 42.569463 | 42.569886 | positive | MGI_C57BL6J_6095875 | Gm47125 | ENSEMBL:ENSMUSG00000113367 | MGI:6095875 | pseudogene | predicted gene, 47125 |
| 13 | gene | 42.624994 | 42.625084 | positive | MGI_C57BL6J_5452863 | Gm23086 | ENSEMBL:ENSMUSG00000095255 | MGI:5452863 | miRNA gene | predicted gene, 23086 |
| 13 | gene | 42.680571 | 43.138526 | positive | MGI_C57BL6J_2659021 | Phactr1 | NCBI_Gene:218194,ENSEMBL:ENSMUSG00000054728 | MGI:2659021 | protein coding gene | phosphatase and actin regulator 1 |
| 13 | gene | 42.782692 | 42.788258 | negative | MGI_C57BL6J_3802042 | Gm15810 | ENSEMBL:ENSMUSG00000089710 | MGI:3802042 | lncRNA gene | predicted gene 15810 |
| 13 | pseudogene | 42.868338 | 42.869602 | negative | MGI_C57BL6J_3801816 | Gm15812 | ENSEMBL:ENSMUSG00000085276 | MGI:3801816 | pseudogene | predicted gene 15812 |
| 13 | gene | 42.998511 | 43.006067 | positive | MGI_C57BL6J_3802046 | Gm15809 | ENSEMBL:ENSMUSG00000090135 | MGI:3802046 | lncRNA gene | predicted gene 15809 |
| 13 | gene | 43.052148 | 43.052410 | negative | MGI_C57BL6J_5455863 | Gm26086 | ENSEMBL:ENSMUSG00000088527 | MGI:5455863 | unclassified non-coding RNA gene | predicted gene, 26086 |
| 13 | gene | 43.117664 | 43.125717 | negative | MGI_C57BL6J_3801815 | Gm15813 | NCBI_Gene:105245478,ENSEMBL:ENSMUSG00000087202 | MGI:3801815 | lncRNA gene | predicted gene 15813 |
| 13 | gene | 43.151740 | 43.171501 | negative | MGI_C57BL6J_1914296 | Tbc1d7 | NCBI_Gene:67046,ENSEMBL:ENSMUSG00000021368 | MGI:1914296 | protein coding gene | TBC1 domain family, member 7 |
| 13 | gene | 43.171556 | 43.192505 | positive | MGI_C57BL6J_5623814 | Gm40929 | NCBI_Gene:105245481 | MGI:5623814 | lncRNA gene | predicted gene, 40929 |
| 13 | gene | 43.195240 | 43.304172 | negative | MGI_C57BL6J_2145304 | Gfod1 | NCBI_Gene:328232,ENSEMBL:ENSMUSG00000051335 | MGI:2145304 | protein coding gene | glucose-fructose oxidoreductase domain containing 1 |
| 13 | gene | 43.365439 | 43.395203 | positive | MGI_C57BL6J_1915596 | Sirt5 | NCBI_Gene:68346,ENSEMBL:ENSMUSG00000054021 | MGI:1915596 | protein coding gene | sirtuin 5 |
| 13 | gene | 43.398376 | 43.407097 | positive | MGI_C57BL6J_1917328 | Nol7 | NCBI_Gene:70078,ENSEMBL:ENSMUSG00000063200 | MGI:1917328 | protein coding gene | nucleolar protein 7 |
| 13 | gene | 43.402673 | 43.481283 | negative | MGI_C57BL6J_1928741 | Ranbp9 | NCBI_Gene:56705,ENSEMBL:ENSMUSG00000038546 | MGI:1928741 | protein coding gene | RAN binding protein 9 |
| 13 | pseudogene | 43.417534 | 43.417865 | positive | MGI_C57BL6J_6096648 | Gm47597 | ENSEMBL:ENSMUSG00000112942 | MGI:6096648 | pseudogene | predicted gene, 47597 |
| 13 | gene | 43.425742 | 43.425850 | negative | MGI_C57BL6J_5455841 | Gm26064 | ENSEMBL:ENSMUSG00000065315 | MGI:5455841 | snRNA gene | predicted gene, 26064 |
| 13 | gene | 43.489504 | 43.494756 | positive | MGI_C57BL6J_5592037 | Gm32878 | NCBI_Gene:102635584 | MGI:5592037 | lncRNA gene | predicted gene, 32878 |
| 13 | gene | 43.498897 | 43.500167 | negative | MGI_C57BL6J_6096785 | Gm47683 | ENSEMBL:ENSMUSG00000113891 | MGI:6096785 | lncRNA gene | predicted gene, 47683 |
| 13 | pseudogene | 43.508945 | 43.509242 | negative | MGI_C57BL6J_3779697 | Gm7214 | NCBI_Gene:102635720,ENSEMBL:ENSMUSG00000113177 | MGI:3779697 | pseudogene | predicted gene 7214 |
| 13 | gene | 43.516809 | 43.531785 | positive | MGI_C57BL6J_5592098 | Gm32939 | NCBI_Gene:102635654,ENSEMBL:ENSMUSG00000112983 | MGI:5592098 | lncRNA gene | predicted gene, 32939 |
| 13 | gene | 43.538393 | 43.560260 | negative | MGI_C57BL6J_1923387 | Mcur1 | NCBI_Gene:76137,ENSEMBL:ENSMUSG00000021371 | MGI:1923387 | protein coding gene | mitochondrial calcium uniporter regulator 1 |
| 13 | gene | 43.615162 | 43.670946 | positive | MGI_C57BL6J_3045355 | Rnf182 | NCBI_Gene:328234,ENSEMBL:ENSMUSG00000044164 | MGI:3045355 | protein coding gene | ring finger protein 182 |
| 13 | gene | 43.683120 | 43.701064 | positive | MGI_C57BL6J_6096860 | Gm47728 | ENSEMBL:ENSMUSG00000114121 | MGI:6096860 | lncRNA gene | predicted gene, 47728 |
| 13 | gene | 43.704435 | 43.705020 | positive | MGI_C57BL6J_1925787 | C030005H24Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA C030005H24 gene |
| 13 | gene | 43.716791 | 43.719038 | positive | MGI_C57BL6J_5434107 | Gm20751 | NCBI_Gene:622552,ENSEMBL:ENSMUSG00000110016 | MGI:5434107 | lncRNA gene | predicted gene, 20751 |
| 13 | gene | 43.784775 | 43.803133 | positive | MGI_C57BL6J_1328316 | Cd83 | NCBI_Gene:12522,ENSEMBL:ENSMUSG00000015396 | MGI:1328316 | protein coding gene | CD83 antigen |
| 13 | gene | 43.842438 | 43.846157 | positive | MGI_C57BL6J_5592274 | Gm33115 | NCBI_Gene:102635895,ENSEMBL:ENSMUSG00000114192 | MGI:5592274 | lncRNA gene | predicted gene, 33115 |
| 13 | gene | 43.863214 | 43.868589 | positive | MGI_C57BL6J_5592354 | Gm33195 | NCBI_Gene:102636000,ENSEMBL:ENSMUSG00000113780 | MGI:5592354 | lncRNA gene | predicted gene, 33195 |
| 13 | gene | 43.872802 | 43.872909 | positive | MGI_C57BL6J_5452894 | Gm23117 | ENSEMBL:ENSMUSG00000089107 | MGI:5452894 | snRNA gene | predicted gene, 23117 |
| 13 | gene | 43.948876 | 43.952699 | positive | MGI_C57BL6J_3780403 | Gm2233 | NCBI_Gene:100039432,ENSEMBL:ENSMUSG00000113671 | MGI:3780403 | lncRNA gene | predicted gene 2233 |
| 13 | gene | 43.990997 | 43.992795 | positive | MGI_C57BL6J_2442189 | A430062P19Rik | NA | NA | unclassified gene | RIKEN cDNA A430062P19 gene |
| 13 | gene | 44.010590 | 44.045135 | negative | MGI_C57BL6J_5592648 | Gm33489 | NCBI_Gene:102636424,ENSEMBL:ENSMUSG00000113252 | MGI:5592648 | lncRNA gene | predicted gene, 33489 |
| 13 | gene | 44.091968 | 44.099971 | negative | MGI_C57BL6J_5592789 | Gm33630 | NCBI_Gene:102636618,ENSEMBL:ENSMUSG00000113276 | MGI:5592789 | lncRNA gene | predicted gene, 33630 |
| 13 | gene | 44.121167 | 44.125179 | positive | MGI_C57BL6J_3779454 | Gm5083 | NCBI_Gene:328235,ENSEMBL:ENSMUSG00000089815 | MGI:3779454 | lncRNA gene | predicted gene 5083 |
| 13 | gene | 44.150588 | 44.152755 | positive | MGI_C57BL6J_5592843 | Gm33684 | NCBI_Gene:102636682,ENSEMBL:ENSMUSG00000113458 | MGI:5592843 | lncRNA gene | predicted gene, 33684 |
| 13 | gene | 44.185760 | 44.216487 | negative | MGI_C57BL6J_2443693 | A330076C08Rik | NCBI_Gene:100503592,ENSEMBL:ENSMUSG00000113186 | MGI:2443693 | lncRNA gene | RIKEN cDNA A330076C08 gene |
| 13 | gene | 44.232398 | 44.239606 | positive | MGI_C57BL6J_6096945 | Gm47781 | ENSEMBL:ENSMUSG00000113727 | MGI:6096945 | lncRNA gene | predicted gene, 47781 |
| 13 | gene | 44.249460 | 44.280449 | negative | MGI_C57BL6J_5588835 | Gm29676 | NCBI_Gene:432754,ENSEMBL:ENSMUSG00000113689 | MGI:5588835 | lncRNA gene | predicted gene, 29676 |
| 13 | gene | 44.363836 | 44.400161 | negative | MGI_C57BL6J_5593117 | Gm33958 | NCBI_Gene:102637052,ENSEMBL:ENSMUSG00000112936 | MGI:5593117 | lncRNA gene | predicted gene, 33958 |
| 13 | gene | 44.400436 | 44.455303 | negative | MGI_C57BL6J_5504122 | Gm27007 | ENSEMBL:ENSMUSG00000098107 | MGI:5504122 | lncRNA gene | predicted gene, 27007 |
| 13 | pseudogene | 44.438834 | 44.439813 | negative | MGI_C57BL6J_5504151 | Gm27036 | ENSEMBL:ENSMUSG00000098124 | MGI:5504151 | pseudogene | predicted gene, 27036 |
| 13 | gene | 44.439712 | 44.469030 | positive | MGI_C57BL6J_1916741 | 1700029N11Rik | NCBI_Gene:69491,ENSEMBL:ENSMUSG00000098144 | MGI:1916741 | lncRNA gene | RIKEN cDNA 1700029N11 gene |
| 13 | gene | 44.467020 | 44.470727 | negative | MGI_C57BL6J_5593179 | Gm34020 | NCBI_Gene:102637133 | MGI:5593179 | lncRNA gene | predicted gene, 34020 |
| 13 | gene | 44.472417 | 44.504548 | negative | MGI_C57BL6J_5593243 | Gm34084 | NCBI_Gene:102637213,ENSEMBL:ENSMUSG00000113010 | MGI:5593243 | lncRNA gene | predicted gene, 34084 |
| 13 | gene | 44.504628 | 44.508705 | positive | MGI_C57BL6J_5623815 | Gm40930 | NCBI_Gene:105245482 | MGI:5623815 | lncRNA gene | predicted gene, 40930 |
| 13 | gene | 44.509558 | 44.511037 | positive | MGI_C57BL6J_6096963 | Gm47792 | ENSEMBL:ENSMUSG00000113492 | MGI:6096963 | lncRNA gene | predicted gene, 47792 |
| 13 | gene | 44.533965 | 44.568668 | negative | MGI_C57BL6J_5593435 | Gm34276 | NCBI_Gene:102637478,ENSEMBL:ENSMUSG00000113977 | MGI:5593435 | lncRNA gene | predicted gene, 34276 |
| 13 | gene | 44.573582 | 44.637009 | negative | MGI_C57BL6J_5623816 | Gm40931 | NCBI_Gene:105245483 | MGI:5623816 | lncRNA gene | predicted gene, 40931 |
| 13 | gene | 44.622182 | 44.625217 | positive | MGI_C57BL6J_6096986 | Gm47805 | ENSEMBL:ENSMUSG00000113730 | MGI:6096986 | lncRNA gene | predicted gene, 47805 |
| 13 | gene | 44.729474 | 44.921643 | positive | MGI_C57BL6J_104813 | Jarid2 | NCBI_Gene:16468,ENSEMBL:ENSMUSG00000038518 | MGI:104813 | protein coding gene | jumonji, AT rich interactive domain 2 |
| 13 | gene | 44.812345 | 44.813923 | positive | MGI_C57BL6J_2442821 | A930104B17Rik | NA | NA | unclassified gene | RIKEN cDNA A930104B17 gene |
| 13 | gene | 44.838165 | 44.838283 | negative | MGI_C57BL6J_5451990 | Gm22213 | ENSEMBL:ENSMUSG00000088189 | MGI:5451990 | snoRNA gene | predicted gene, 22213 |
| 13 | gene | 44.922075 | 45.002147 | negative | MGI_C57BL6J_2137586 | Dtnbp1 | NCBI_Gene:94245,ENSEMBL:ENSMUSG00000057531 | MGI:2137586 | protein coding gene | dystrobrevin binding protein 1 |
| 13 | gene | 45.026252 | 45.026355 | negative | MGI_C57BL6J_5452881 | Gm23104 | ENSEMBL:ENSMUSG00000065656 | MGI:5452881 | snRNA gene | predicted gene, 23104 |
| 13 | gene | 45.075493 | 45.078269 | positive | MGI_C57BL6J_5593568 | Gm34409 | NCBI_Gene:102637654 | MGI:5593568 | lncRNA gene | predicted gene, 34409 |
| 13 | pseudogene | 45.078370 | 45.079184 | negative | MGI_C57BL6J_96241 | Hsp25-ps1 | NCBI_Gene:15508,ENSEMBL:ENSMUSG00000078915 | MGI:96241 | pseudogene | heat shock protein 25, pseudogene 1 |
| 13 | gene | 45.078692 | 45.081222 | positive | MGI_C57BL6J_3809102 | Gm9817 | ENSEMBL:ENSMUSG00000047061 | MGI:3809102 | protein coding gene | predicted gene 9817 |
| 13 | gene | 45.099957 | 45.117884 | positive | MGI_C57BL6J_5623817 | Gm40932 | NCBI_Gene:105245484,ENSEMBL:ENSMUSG00000113553 | MGI:5623817 | lncRNA gene | predicted gene, 40932 |
| 13 | pseudogene | 45.186332 | 45.187554 | negative | MGI_C57BL6J_3648246 | Gm8513 | NCBI_Gene:667204,ENSEMBL:ENSMUSG00000098136 | MGI:3648246 | pseudogene | predicted gene 8513 |
| 13 | gene | 45.235635 | 45.335588 | positive | MGI_C57BL6J_6096404 | Gm47448 | ENSEMBL:ENSMUSG00000113772 | MGI:6096404 | lncRNA gene | predicted gene, 47448 |
| 13 | gene | 45.246632 | 45.267772 | negative | MGI_C57BL6J_5593625 | Gm34466 | NCBI_Gene:102637730,ENSEMBL:ENSMUSG00000113919 | MGI:5593625 | lncRNA gene | predicted gene, 34466 |
| 13 | gene | 45.294654 | 45.294996 | negative | MGI_C57BL6J_5453532 | Gm23755 | ENSEMBL:ENSMUSG00000088203 | MGI:5453532 | unclassified non-coding RNA gene | predicted gene, 23755 |
| 13 | gene | 45.359100 | 45.371211 | positive | MGI_C57BL6J_5623818 | Gm40933 | NCBI_Gene:105245485 | MGI:5623818 | lncRNA gene | predicted gene, 40933 |
| 13 | gene | 45.389742 | 45.412022 | positive | MGI_C57BL6J_2388271 | Mylip | NCBI_Gene:218203,ENSEMBL:ENSMUSG00000038175 | MGI:2388271 | protein coding gene | myosin regulatory light chain interacting protein |
| 13 | pseudogene | 45.431002 | 45.431331 | positive | MGI_C57BL6J_6096415 | Gm47455 | ENSEMBL:ENSMUSG00000113446 | MGI:6096415 | pseudogene | predicted gene, 47455 |
| 13 | gene | 45.486224 | 45.486330 | negative | MGI_C57BL6J_5453164 | Gm23387 | ENSEMBL:ENSMUSG00000089211 | MGI:5453164 | snRNA gene | predicted gene, 23387 |
| 13 | gene | 45.507444 | 45.553800 | positive | MGI_C57BL6J_1913605 | Gmpr | NCBI_Gene:66355,ENSEMBL:ENSMUSG00000000253 | MGI:1913605 | protein coding gene | guanosine monophosphate reductase |
| 13 | gene | 45.521186 | 45.524440 | negative | MGI_C57BL6J_5593741 | Gm34582 | NCBI_Gene:102637886 | MGI:5593741 | lncRNA gene | predicted gene, 34582 |
| 13 | gene | 45.549755 | 45.965034 | negative | MGI_C57BL6J_104783 | Atxn1 | NCBI_Gene:20238,ENSEMBL:ENSMUSG00000046876 | MGI:104783 | protein coding gene | ataxin 1 |
| 13 | gene | 45.749572 | 45.755206 | positive | MGI_C57BL6J_6096423 | Gm47460 | ENSEMBL:ENSMUSG00000114131 | MGI:6096423 | lncRNA gene | predicted gene, 47460 |
| 13 | gene | 45.936040 | 45.936904 | positive | MGI_C57BL6J_1922143 | 4930453C13Rik | ENSEMBL:ENSMUSG00000113987 | MGI:1922143 | lncRNA gene | RIKEN cDNA 4930453C13 gene |
| 13 | gene | 45.939813 | 45.940698 | positive | MGI_C57BL6J_2444743 | 9930104M19Rik | NA | NA | unclassified gene | RIKEN cDNA 9930104M19 gene |
| 13 | gene | 45.965062 | 45.966508 | positive | MGI_C57BL6J_1923250 | 5033430I15Rik | NCBI_Gene:76000,ENSEMBL:ENSMUSG00000038152 | MGI:1923250 | lncRNA gene | RIKEN cDNA 5033430I15 gene |
| 13 | gene | 46.019856 | 46.037769 | positive | MGI_C57BL6J_5594150 | Gm34991 | NCBI_Gene:102638420 | MGI:5594150 | lncRNA gene | predicted gene, 34991 |
| 13 | gene | 46.060597 | 46.068765 | positive | MGI_C57BL6J_5825586 | Gm45949 | NCBI_Gene:108167338,ENSEMBL:ENSMUSG00000113860 | MGI:5825586 | lncRNA gene | predicted gene, 45949 |
| 13 | gene | 46.175428 | 46.191046 | negative | MGI_C57BL6J_3642714 | Gm10113 | ENSEMBL:ENSMUSG00000062282 | MGI:3642714 | lncRNA gene | predicted gene 10113 |
| 13 | gene | 46.273721 | 46.300115 | positive | MGI_C57BL6J_2686420 | Stmnd1 | NCBI_Gene:380842,ENSEMBL:ENSMUSG00000063529 | MGI:2686420 | protein coding gene | stathmin domain containing 1 |
| 13 | gene | 46.418176 | 46.431099 | positive | MGI_C57BL6J_3610364 | Rbm24 | NCBI_Gene:666794,ENSEMBL:ENSMUSG00000038132 | MGI:3610364 | protein coding gene | RNA binding motif protein 24 |
| 13 | gene | 46.472669 | 46.501984 | negative | MGI_C57BL6J_5594269 | Gm35110 | NCBI_Gene:102638580,ENSEMBL:ENSMUSG00000114752 | MGI:5594269 | lncRNA gene | predicted gene, 35110 |
| 13 | gene | 46.501848 | 46.650281 | positive | MGI_C57BL6J_1914502 | Cap2 | NCBI_Gene:67252,ENSEMBL:ENSMUSG00000021373 | MGI:1914502 | protein coding gene | CAP, adenylate cyclase-associated protein, 2 (yeast) |
| 13 | gene | 46.518684 | 46.518761 | positive | MGI_C57BL6J_5454692 | Gm24915 | ENSEMBL:ENSMUSG00000087913 | MGI:5454692 | rRNA gene | predicted gene, 24915 |
| 13 | gene | 46.605327 | 46.619780 | negative | MGI_C57BL6J_6096522 | Gm47523 | ENSEMBL:ENSMUSG00000114596 | MGI:6096522 | lncRNA gene | predicted gene, 47523 |
| 13 | gene | 46.669443 | 46.678064 | positive | MGI_C57BL6J_2145496 | Fam8a1 | NCBI_Gene:97863,ENSEMBL:ENSMUSG00000069237 | MGI:2145496 | protein coding gene | family with sequence similarity 8, member A1 |
| 13 | gene | 46.679902 | 46.727940 | negative | MGI_C57BL6J_2385621 | Nup153 | NCBI_Gene:218210,ENSEMBL:ENSMUSG00000021374 | MGI:2385621 | protein coding gene | nucleoporin 153 |
| 13 | gene | 46.720557 | 46.737102 | positive | MGI_C57BL6J_5826049 | Gm46412 | NCBI_Gene:108168080 | MGI:5826049 | lncRNA gene | predicted gene, 46412 |
| 13 | gene | 46.749087 | 46.931302 | negative | MGI_C57BL6J_1098264 | Kif13a | NCBI_Gene:16553,ENSEMBL:ENSMUSG00000021375 | MGI:1098264 | protein coding gene | kinesin family member 13A |
| 13 | gene | 46.767520 | 46.767649 | negative | MGI_C57BL6J_5454397 | Gm24620 | ENSEMBL:ENSMUSG00000064558 | MGI:5454397 | snoRNA gene | predicted gene, 24620 |
| 13 | gene | 46.804666 | 46.808331 | negative | MGI_C57BL6J_2444865 | D130012G24Rik | NA | NA | unclassified gene | RIKEN cDNA D130012G24 gene |
| 13 | gene | 46.881027 | 46.884898 | positive | MGI_C57BL6J_6097663 | Gm48250 | ENSEMBL:ENSMUSG00000114917 | MGI:6097663 | lncRNA gene | predicted gene, 48250 |
| 13 | gene | 46.902589 | 46.902692 | positive | MGI_C57BL6J_5452737 | Gm22960 | ENSEMBL:ENSMUSG00000084466 | MGI:5452737 | snRNA gene | predicted gene, 22960 |
| 13 | pseudogene | 46.945190 | 46.945693 | negative | MGI_C57BL6J_6097780 | Gm48325 | ENSEMBL:ENSMUSG00000114449 | MGI:6097780 | pseudogene | predicted gene, 48325 |
| 13 | gene | 46.979908 | 47.011195 | positive | MGI_C57BL6J_1917079 | 2010001K21Rik | ENSEMBL:ENSMUSG00000051606 | MGI:1917079 | lncRNA gene | RIKEN cDNA 2010001K21 gene |
| 13 | gene | 46.980720 | 46.980848 | positive | MGI_C57BL6J_5454546 | Gm24769 | ENSEMBL:ENSMUSG00000089131 | MGI:5454546 | snoRNA gene | predicted gene, 24769 |
| 13 | gene | 46.986447 | 47.008713 | positive | MGI_C57BL6J_2145354 | E030046B03Rik | NA | NA | lncRNA gene | RIKEN cDNA E030046B03 gene |
| 13 | pseudogene | 47.009346 | 47.011042 | negative | MGI_C57BL6J_3779518 | Gm5790 | NCBI_Gene:544928,ENSEMBL:ENSMUSG00000114858 | MGI:3779518 | pseudogene | predicted gene 5790 |
| 13 | gene | 47.012557 | 47.014850 | negative | MGI_C57BL6J_2145264 | Nhlrc1 | NCBI_Gene:105193,ENSEMBL:ENSMUSG00000044231 | MGI:2145264 | protein coding gene | NHL repeat containing 1 |
| 13 | gene | 47.021987 | 47.044737 | negative | MGI_C57BL6J_98812 | Tpmt | NCBI_Gene:22017,ENSEMBL:ENSMUSG00000021376 | MGI:98812 | protein coding gene | thiopurine methyltransferase |
| 13 | pseudogene | 47.024346 | 47.024986 | positive | MGI_C57BL6J_5010992 | Gm18807 | NCBI_Gene:100417752,ENSEMBL:ENSMUSG00000114376 | MGI:5010992 | pseudogene | predicted gene, 18807 |
| 13 | gene | 47.043467 | 47.085279 | positive | MGI_C57BL6J_2145261 | Kdm1b | NCBI_Gene:218214,ENSEMBL:ENSMUSG00000038080 | MGI:2145261 | protein coding gene | lysine (K)-specific demethylase 1B |
| 13 | gene | 47.084767 | 47.106227 | negative | MGI_C57BL6J_1926209 | Dek | NCBI_Gene:110052,ENSEMBL:ENSMUSG00000021377 | MGI:1926209 | protein coding gene | DEK oncogene (DNA binding) |
| 13 | gene | 47.106419 | 47.109519 | positive | MGI_C57BL6J_1919521 | 1700026N04Rik | ENSEMBL:ENSMUSG00000114681 | MGI:1919521 | lncRNA gene | RIKEN cDNA 1700026N04 gene |
| 13 | gene | 47.115684 | 47.132081 | negative | MGI_C57BL6J_5623820 | Gm40935 | NCBI_Gene:105245487 | MGI:5623820 | lncRNA gene | predicted gene, 40935 |
| 13 | gene | 47.122656 | 47.247991 | positive | MGI_C57BL6J_2384986 | Rnf144b | NCBI_Gene:218215,ENSEMBL:ENSMUSG00000038068 | MGI:2384986 | protein coding gene | ring finger protein 144B |
| 13 | gene | 47.143940 | 47.148670 | negative | MGI_C57BL6J_2442085 | A930002C04Rik | NCBI_Gene:319467,ENSEMBL:ENSMUSG00000114961 | MGI:2442085 | lncRNA gene | RIKEN cDNA A930002C04 gene |
| 13 | gene | 47.152357 | 47.155008 | negative | MGI_C57BL6J_6098148 | Gm48581 | ENSEMBL:ENSMUSG00000114882 | MGI:6098148 | unclassified gene | predicted gene, 48581 |
| 13 | gene | 47.219526 | 47.224544 | negative | MGI_C57BL6J_5623819 | Gm40934 | NCBI_Gene:105245486 | MGI:5623819 | lncRNA gene | predicted gene, 40934 |
| 13 | gene | 47.335251 | 47.340939 | negative | MGI_C57BL6J_6098200 | Gm48612 | ENSEMBL:ENSMUSG00000114958 | MGI:6098200 | lncRNA gene | predicted gene, 48612 |
| 13 | gene | 47.359429 | 47.359701 | positive | MGI_C57BL6J_2145166 | AA517650 | NA | NA | unclassified gene | expressed sequence AA517650 |
| 13 | gene | 47.361386 | 47.439554 | positive | MGI_C57BL6J_5594892 | Gm35733 | NCBI_Gene:102639410,ENSEMBL:ENSMUSG00000114843 | MGI:5594892 | lncRNA gene | predicted gene, 35733 |
| 13 | gene | 47.461264 | 47.464280 | positive | MGI_C57BL6J_5623821 | Gm40936 | NCBI_Gene:105245488 | MGI:5623821 | lncRNA gene | predicted gene, 40936 |
| 13 | gene | 47.667505 | 47.668393 | positive | MGI_C57BL6J_1922189 | 4930471G24Rik | NCBI_Gene:74939,ENSEMBL:ENSMUSG00000114509 | MGI:1922189 | lncRNA gene | RIKEN cDNA 4930471G24 gene |
| 13 | gene | 47.784231 | 47.789591 | negative | MGI_C57BL6J_5595087 | Gm35928 | NCBI_Gene:102639671 | MGI:5595087 | lncRNA gene | predicted gene, 35928 |
| 13 | gene | 47.843815 | 47.851943 | positive | MGI_C57BL6J_5595150 | Gm35991 | NCBI_Gene:102639755 | MGI:5595150 | lncRNA gene | predicted gene, 35991 |
| 13 | gene | 47.902558 | 47.948474 | negative | MGI_C57BL6J_5623823 | Gm40938 | NCBI_Gene:105245490 | MGI:5623823 | lncRNA gene | predicted gene, 40938 |
| 13 | gene | 47.910435 | 47.934301 | positive | MGI_C57BL6J_3588274 | G630093K05Rik | NCBI_Gene:619327,ENSEMBL:ENSMUSG00000114959 | MGI:3588274 | lncRNA gene | RIKEN cDNA G630093K05 gene |
| 13 | gene | 47.960330 | 48.018608 | negative | MGI_C57BL6J_1918221 | 4931429P17Rik | NCBI_Gene:70971,ENSEMBL:ENSMUSG00000047324 | MGI:1918221 | lncRNA gene | RIKEN cDNA 4931429P17 gene |
| 13 | gene | 48.028356 | 48.043795 | positive | MGI_C57BL6J_5595260 | Gm36101 | NCBI_Gene:102639899,ENSEMBL:ENSMUSG00000114810 | MGI:5595260 | lncRNA gene | predicted gene, 36101 |
| 13 | gene | 48.044231 | 48.262623 | negative | MGI_C57BL6J_2444369 | A330033J07Rik | NCBI_Gene:320614,ENSEMBL:ENSMUSG00000097622 | MGI:2444369 | lncRNA gene | RIKEN cDNA A330033J07 gene |
| 13 | gene | 48.107617 | 48.237251 | negative | MGI_C57BL6J_5595505 | Gm36346 | NCBI_Gene:102640229,ENSEMBL:ENSMUSG00000114457 | MGI:5595505 | lncRNA gene | predicted gene, 36346 |
| 13 | gene | 48.227702 | 48.241412 | positive | MGI_C57BL6J_5595342 | Gm36183 | NCBI_Gene:102640005 | MGI:5595342 | lncRNA gene | predicted gene, 36183 |
| 13 | gene | 48.239054 | 48.262591 | negative | MGI_C57BL6J_6118781 | Gm49291 | ENSEMBL:ENSMUSG00000114860 | MGI:6118781 | lncRNA gene | predicted gene, 49291 |
| 13 | gene | 48.242373 | 48.253779 | positive | MGI_C57BL6J_5623824 | Gm40939 | NCBI_Gene:105245491 | MGI:5623824 | lncRNA gene | predicted gene, 40939 |
| 13 | gene | 48.261228 | 48.266026 | positive | MGI_C57BL6J_99414 | Id4 | NCBI_Gene:15904,ENSEMBL:ENSMUSG00000021379 | MGI:99414 | protein coding gene | inhibitor of DNA binding 4 |
| 13 | gene | 48.272413 | 48.273884 | negative | MGI_C57BL6J_2443792 | A330048O09Rik | NCBI_Gene:320315,ENSEMBL:ENSMUSG00000097326 | MGI:2443792 | lncRNA gene | RIKEN cDNA A330048O09 gene |
| 13 | pseudogene | 48.312578 | 48.314384 | positive | MGI_C57BL6J_3646798 | Gm8588 | NCBI_Gene:667351,ENSEMBL:ENSMUSG00000114881 | MGI:3646798 | pseudogene | predicted gene 8588 |
| 13 | gene | 48.359459 | 48.408512 | negative | MGI_C57BL6J_6098256 | Gm48648 | ENSEMBL:ENSMUSG00000114695 | MGI:6098256 | lncRNA gene | predicted gene, 48648 |
| 13 | gene | 48.439665 | 48.446672 | negative | MGI_C57BL6J_5623825 | Gm40940 | NCBI_Gene:105245492 | MGI:5623825 | lncRNA gene | predicted gene, 40940 |
| 13 | gene | 48.465810 | 48.477602 | negative | MGI_C57BL6J_5623826 | Gm40941 | NCBI_Gene:105245493 | MGI:5623826 | lncRNA gene | predicted gene, 40941 |
| 13 | gene | 48.480763 | 48.513474 | negative | MGI_C57BL6J_1915161 | Zfp169 | NCBI_Gene:67911,ENSEMBL:ENSMUSG00000050954 | MGI:1915161 | protein coding gene | zinc finger protein 169 |
| 13 | gene | 48.491403 | 48.505499 | positive | MGI_C57BL6J_5595561 | Gm36402 | NCBI_Gene:102640299 | MGI:5595561 | lncRNA gene | predicted gene, 36402 |
| 13 | gene | 48.513540 | 48.515002 | positive | MGI_C57BL6J_3045249 | A830005F24Rik | NCBI_Gene:442803,ENSEMBL:ENSMUSG00000053181 | MGI:3045249 | protein coding gene | RIKEN cDNA A830005F24 gene |
| 13 | gene | 48.532905 | 48.546500 | negative | MGI_C57BL6J_1924398 | 6720427I07Rik | ENSEMBL:ENSMUSG00000097375 | MGI:1924398 | lncRNA gene | RIKEN cDNA 6720427I07 gene |
| 13 | gene | 48.536012 | 48.536114 | negative | MGI_C57BL6J_2676796 | Mirlet7d | miRBase:MI0000405,NCBI_Gene:387247,ENSEMBL:ENSMUSG00000065453 | MGI:2676796 | miRNA gene | microRNA let7d |
| 13 | gene | 48.537825 | 48.537921 | positive | MGI_C57BL6J_5453888 | Gm24111 | ENSEMBL:ENSMUSG00000093232 | MGI:5453888 | miRNA gene | predicted gene, 24111 |
| 13 | gene | 48.537829 | 48.537917 | negative | MGI_C57BL6J_2676798 | Mirlet7f-1 | miRBase:MI0000562,NCBI_Gene:387252,ENSEMBL:ENSMUSG00000105621 | MGI:2676798 | miRNA gene | microRNA let7f-1 |
| 13 | gene | 48.538179 | 48.538272 | negative | MGI_C57BL6J_2676793 | Mirlet7a-1 | miRBase:MI0000556,NCBI_Gene:387244,ENSEMBL:ENSMUSG00000065421 | MGI:2676793 | miRNA gene | microRNA let7a-1 |
| 13 | gene | 48.542644 | 48.545033 | negative | MGI_C57BL6J_5610466 | Gm37238 | ENSEMBL:ENSMUSG00000103373 | MGI:5610466 | unclassified gene | predicted gene, 37238 |
| 13 | gene | 48.572253 | 48.572384 | negative | MGI_C57BL6J_5455009 | Gm25232 | ENSEMBL:ENSMUSG00000095214 | MGI:5455009 | snoRNA gene | predicted gene, 25232 |
| 13 | gene | 48.577869 | 48.625672 | negative | MGI_C57BL6J_2145430 | Ptpdc1 | NCBI_Gene:218232,ENSEMBL:ENSMUSG00000038042 | MGI:2145430 | protein coding gene | protein tyrosine phosphatase domain containing 1 |
| 13 | gene | 48.593501 | 48.596017 | positive | MGI_C57BL6J_5595709 | Gm36550 | NCBI_Gene:108168081,ENSEMBL:ENSMUSG00000113037 | MGI:5595709 | lncRNA gene | predicted gene, 36550 |
| 13 | gene | 48.652722 | 48.662831 | negative | MGI_C57BL6J_5595765 | Gm36606 | NCBI_Gene:102640574 | MGI:5595765 | lncRNA gene | predicted gene, 36606 |
| 13 | gene | 48.662998 | 48.666507 | positive | MGI_C57BL6J_103124 | Barx1 | NCBI_Gene:12022,ENSEMBL:ENSMUSG00000021381 | MGI:103124 | protein coding gene | BarH-like homeobox 1 |
| 13 | gene | 48.708189 | 48.734084 | positive | MGI_C57BL6J_5595943 | Gm36784 | NCBI_Gene:102640802 | MGI:5595943 | lncRNA gene | predicted gene, 36784 |
| 13 | pseudogene | 48.763050 | 48.763610 | negative | MGI_C57BL6J_5010315 | Rpl17-ps6 | NCBI_Gene:100416501,ENSEMBL:ENSMUSG00000113297 | MGI:5010315 | pseudogene | ribosomal protein L17, pseudogene 6 |
| 13 | gene | 48.801750 | 48.871119 | negative | MGI_C57BL6J_1338034 | Phf2 | NCBI_Gene:18676,ENSEMBL:ENSMUSG00000038025 | MGI:1338034 | protein coding gene | PHD finger protein 2 |
| 13 | gene | 48.871037 | 48.878349 | positive | MGI_C57BL6J_3645592 | Phf2os1 | NCBI_Gene:102642832,ENSEMBL:ENSMUSG00000086889 | MGI:3645592 | antisense lncRNA gene | PHD finger protein 2, opposite strand 1 |
| 13 | gene | 48.879216 | 48.968017 | negative | MGI_C57BL6J_2446163 | Fam120a | NCBI_Gene:218236,ENSEMBL:ENSMUSG00000038014 | MGI:2446163 | protein coding gene | family with sequence similarity 120, member A |
| 13 | gene | 48.968112 | 48.972031 | positive | MGI_C57BL6J_1915378 | Fam120aos | NCBI_Gene:68128,ENSEMBL:ENSMUSG00000097059 | MGI:1915378 | lincRNA gene | family with sequence similarity 120A, opposite strand |
| 13 | pseudogene | 48.978708 | 48.981240 | negative | MGI_C57BL6J_5010497 | Gm18312 | NCBI_Gene:100416909 | MGI:5010497 | pseudogene | predicted gene, 18312 |
| 13 | gene | 49.036302 | 49.148574 | negative | MGI_C57BL6J_1922857 | Wnk2 | NCBI_Gene:75607,ENSEMBL:ENSMUSG00000037989 | MGI:1922857 | protein coding gene | WNK lysine deficient protein kinase 2 |
| 13 | gene | 49.161905 | 49.163639 | negative | MGI_C57BL6J_5623827 | Gm40942 | NCBI_Gene:105245494 | MGI:5623827 | lncRNA gene | predicted gene, 40942 |
| 13 | gene | 49.169808 | 49.183825 | negative | MGI_C57BL6J_5623828 | Gm40943 | NCBI_Gene:105245495 | MGI:5623828 | lncRNA gene | predicted gene, 40943 |
| 13 | gene | 49.187485 | 49.196251 | positive | MGI_C57BL6J_1196617 | Ninj1 | NCBI_Gene:18081,ENSEMBL:ENSMUSG00000037966 | MGI:1196617 | protein coding gene | ninjurin 1 |
| 13 | gene | 49.202950 | 49.216062 | negative | MGI_C57BL6J_1915730 | Card19 | NCBI_Gene:68480,ENSEMBL:ENSMUSG00000037960 | MGI:1915730 | protein coding gene | caspase recruitment domain family, member 19 |
| 13 | gene | 49.230689 | 49.248706 | negative | MGI_C57BL6J_1913579 | Susd3 | NCBI_Gene:66329,ENSEMBL:ENSMUSG00000021384 | MGI:1913579 | protein coding gene | sushi domain containing 3 |
| 13 | gene | 49.259044 | 49.261793 | positive | MGI_C57BL6J_4437722 | E030007A22Rik | NA | NA | lncRNA gene | RIKEN cDNA E030007A22 gene |
| 13 | gene | 49.261554 | 49.320469 | negative | MGI_C57BL6J_1353657 | Fgd3 | NCBI_Gene:30938,ENSEMBL:ENSMUSG00000037946 | MGI:1353657 | protein coding gene | FYVE, RhoGEF and PH domain containing 3 |
| 13 | gene | 49.340856 | 49.341376 | negative | MGI_C57BL6J_1925893 | 1700129O19Rik | NA | NA | unclassified gene | RIKEN cDNA 1700129O19 gene |
| 13 | gene | 49.341549 | 49.387026 | positive | MGI_C57BL6J_1924145 | Bicd2 | NCBI_Gene:76895,ENSEMBL:ENSMUSG00000037933 | MGI:1924145 | protein coding gene | BICD cargo adaptor 2 |
| 13 | gene | 49.391565 | 49.407347 | negative | MGI_C57BL6J_5826050 | Gm46413 | NCBI_Gene:108168082 | MGI:5826050 | lncRNA gene | predicted gene, 46413 |
| 13 | gene | 49.421190 | 49.464573 | positive | MGI_C57BL6J_1922928 | Ippk | NCBI_Gene:75678,ENSEMBL:ENSMUSG00000021385 | MGI:1922928 | protein coding gene | inositol 1,3,4,5,6-pentakisphosphate 2-kinase |
| 13 | gene | 49.464023 | 49.652897 | negative | MGI_C57BL6J_1913586 | Cenpp | NCBI_Gene:66336,ENSEMBL:ENSMUSG00000021391 | MGI:1913586 | protein coding gene | centromere protein P |
| 13 | pseudogene | 49.493318 | 49.496589 | positive | MGI_C57BL6J_3582925 | Tes3-ps | NCBI_Gene:54339,ENSEMBL:ENSMUSG00000113255 | MGI:3582925 | pseudogene | testis derived transcript 3, pseudogene |
| 13 | gene | 49.504810 | 49.532789 | positive | MGI_C57BL6J_3039578 | Ecm2 | NCBI_Gene:407800,ENSEMBL:ENSMUSG00000043631 | MGI:3039578 | protein coding gene | extracellular matrix protein 2, female organ and adipocyte specific |
| 13 | gene | 49.544443 | 49.567565 | positive | MGI_C57BL6J_1913945 | Aspn | NCBI_Gene:66695,ENSEMBL:ENSMUSG00000021388 | MGI:1913945 | protein coding gene | asporin |
| 13 | gene | 49.579754 | 49.592822 | positive | MGI_C57BL6J_1350918 | Omd | NCBI_Gene:27047,ENSEMBL:ENSMUSG00000048368 | MGI:1350918 | protein coding gene | osteomodulin |
| 13 | gene | 49.608046 | 49.624501 | positive | MGI_C57BL6J_109278 | Ogn | NCBI_Gene:18295,ENSEMBL:ENSMUSG00000021390 | MGI:109278 | protein coding gene | osteoglycin |
| 13 | gene | 49.653078 | 49.679016 | positive | MGI_C57BL6J_1918180 | Nol8 | NCBI_Gene:70930,ENSEMBL:ENSMUSG00000021392 | MGI:1918180 | protein coding gene | nucleolar protein 8 |
| 13 | gene | 49.682100 | 49.734267 | positive | MGI_C57BL6J_2145219 | Iars | NCBI_Gene:105148,ENSEMBL:ENSMUSG00000037851 | MGI:2145219 | protein coding gene | isoleucine-tRNA synthetase |
| 13 | gene | 49.684301 | 49.684433 | positive | MGI_C57BL6J_5455171 | Gm25394 | ENSEMBL:ENSMUSG00000087963 | MGI:5455171 | snoRNA gene | predicted gene, 25394 |
| 13 | gene | 49.714119 | 49.714240 | positive | MGI_C57BL6J_5453259 | Gm23482 | ENSEMBL:ENSMUSG00000077360 | MGI:5453259 | snoRNA gene | predicted gene, 23482 |
| 13 | gene | 49.750462 | 49.752616 | positive | MGI_C57BL6J_5011519 | Gm19334 | NCBI_Gene:100502710,ENSEMBL:ENSMUSG00000118398 | MGI:5011519 | protein coding gene | predicted gene, 19334 |
| 13 | gene | 49.764328 | 49.765303 | positive | MGI_C57BL6J_2685116 | Gm270 | ENSEMBL:ENSMUSG00000113823 | MGI:2685116 | protein coding gene | predicted gene 270 |
| 13 | gene | 49.784742 | 49.799001 | negative | MGI_C57BL6J_5589461 | Gm30302 | NCBI_Gene:102632152,ENSEMBL:ENSMUSG00000114073 | MGI:5589461 | protein coding gene | predicted gene, 30302 |
| 13 | gene | 49.784981 | 49.787292 | positive | MGI_C57BL6J_6097262 | Gm47977 | ENSEMBL:ENSMUSG00000112995 | MGI:6097262 | lncRNA gene | predicted gene, 47977 |
| 13 | gene | 49.787804 | 49.887777 | positive | MGI_C57BL6J_2685118 | Gm272 | NCBI_Gene:102631805,ENSEMBL:ENSMUSG00000113254 | MGI:2685118 | protein coding gene | predicted gene 272 |
| 13 | pseudogene | 49.804744 | 49.805956 | negative | MGI_C57BL6J_3646144 | Gm7251 | NCBI_Gene:638833,ENSEMBL:ENSMUSG00000069236 | MGI:3646144 | pseudogene | predicted pseudogene 7251 |
| 13 | gene | 49.858623 | 49.864688 | negative | MGI_C57BL6J_3643443 | Gm8672 | NCBI_Gene:667503,ENSEMBL:ENSMUSG00000113530 | MGI:3643443 | protein coding gene | predicted gene 8672 |
| 13 | pseudogene | 49.882956 | 49.883766 | positive | MGI_C57BL6J_5011196 | Gm19011 | NCBI_Gene:100418118,ENSEMBL:ENSMUSG00000113333 | MGI:5011196 | pseudogene | predicted gene, 19011 |
| 13 | pseudogene | 49.890423 | 49.892868 | negative | MGI_C57BL6J_6097315 | Gm48009 | ENSEMBL:ENSMUSG00000113782 | MGI:6097315 | pseudogene | predicted gene, 48009 |
| 13 | gene | 49.898842 | 49.904597 | negative | MGI_C57BL6J_3645762 | Gm8674 | NCBI_Gene:667507,ENSEMBL:ENSMUSG00000093976 | MGI:3645762 | protein coding gene | predicted gene 8674 |
| 13 | pseudogene | 49.944117 | 49.945430 | negative | MGI_C57BL6J_3642059 | Gm10784 | NCBI_Gene:100040099,ENSEMBL:ENSMUSG00000094886 | MGI:3642059 | pseudogene | predicted pseudogene 10784 |
| 13 | gene | 49.970001 | 49.974260 | positive | MGI_C57BL6J_6097370 | Gm48051 | ENSEMBL:ENSMUSG00000114175 | MGI:6097370 | lncRNA gene | predicted gene, 48051 |
| 13 | gene | 49.976258 | 49.977074 | negative | MGI_C57BL6J_4937245 | Gm17611 | ENSEMBL:ENSMUSG00000090497 | MGI:4937245 | protein coding gene | predicted gene, 17611 |
| 13 | pseudogene | 50.000347 | 50.002424 | negative | MGI_C57BL6J_3646019 | Gm6554 | NCBI_Gene:625110,ENSEMBL:ENSMUSG00000113963 | MGI:3646019 | pseudogene | predicted gene 6554 |
| 13 | gene | 50.008262 | 50.052848 | positive | MGI_C57BL6J_5589679 | Gm30520 | NCBI_Gene:102632452 | MGI:5589679 | lncRNA gene | predicted gene, 30520 |
| 13 | pseudogene | 50.049004 | 50.051005 | negative | MGI_C57BL6J_6097372 | Gm48052 | ENSEMBL:ENSMUSG00000113312 | MGI:6097372 | pseudogene | predicted gene, 48052 |
| 13 | pseudogene | 50.054762 | 50.069184 | negative | MGI_C57BL6J_3642967 | LOC667541 | NCBI_Gene:667541,ENSEMBL:ENSMUSG00000113888 | MGI:3642967 | pseudogene | hypothetical LOC667541 |
| 13 | gene | 50.098968 | 50.104382 | negative | MGI_C57BL6J_3779809 | Gm8700 | NCBI_Gene:667552,ENSEMBL:ENSMUSG00000113007 | MGI:3779809 | protein coding gene | predicted gene 8700 |
| 13 | gene | 50.118006 | 50.118114 | positive | MGI_C57BL6J_5454077 | Gm24300 | ENSEMBL:ENSMUSG00000095849 | MGI:5454077 | miRNA gene | predicted gene, 24300 |
| 13 | pseudogene | 50.133670 | 50.134510 | negative | MGI_C57BL6J_6097375 | Gm48054 | ENSEMBL:ENSMUSG00000113921 | MGI:6097375 | pseudogene | predicted gene, 48054 |
| 13 | gene | 50.136993 | 50.155392 | positive | MGI_C57BL6J_1918502 | 4933433N18Rik | NCBI_Gene:71252,ENSEMBL:ENSMUSG00000113227 | MGI:1918502 | lncRNA gene | RIKEN cDNA 4933433N18 gene |
| 13 | gene | 50.161821 | 50.163398 | negative | MGI_C57BL6J_5011655 | Gm19470 | NCBI_Gene:100502955 | MGI:5011655 | protein coding gene | predicted gene, 19470 |
| 13 | pseudogene | 50.165573 | 50.167849 | negative | MGI_C57BL6J_6096348 | Gm47412 | ENSEMBL:ENSMUSG00000113437 | MGI:6096348 | pseudogene | predicted gene, 47412 |
| 13 | pseudogene | 50.195183 | 50.198131 | positive | MGI_C57BL6J_5011669 | Gm19484 | NCBI_Gene:100502978 | MGI:5011669 | pseudogene | predicted gene, 19484 |
| 13 | pseudogene | 50.197941 | 50.201627 | positive | MGI_C57BL6J_5621338 | Gm38453 | NCBI_Gene:102633452 | MGI:5621338 | pseudogene | predicted gene, 38453 |
| 13 | pseudogene | 50.206785 | 50.208099 | negative | MGI_C57BL6J_3780813 | Gm2645 | NCBI_Gene:100040176,ENSEMBL:ENSMUSG00000113718 | MGI:3780813 | pseudogene | predicted gene 2645 |
| 13 | gene | 50.218358 | 50.228101 | positive | MGI_C57BL6J_5590376 | Gm31217 | NCBI_Gene:102633381 | MGI:5590376 | lncRNA gene | predicted gene, 31217 |
| 13 | pseudogene | 50.235111 | 50.238756 | negative | MGI_C57BL6J_5588928 | Gm29769 | NCBI_Gene:101056055 | MGI:5588928 | pseudogene | predicted gene, 29769 |
| 13 | gene | 50.238757 | 50.253444 | negative | MGI_C57BL6J_2685752 | Gm906 | NCBI_Gene:380882,ENSEMBL:ENSMUSG00000095300 | MGI:2685752 | protein coding gene | predicted gene 906 |
| 13 | pseudogene | 50.241709 | 50.241957 | positive | MGI_C57BL6J_6096349 | Gm47413 | ENSEMBL:ENSMUSG00000113480 | MGI:6096349 | pseudogene | predicted gene, 47413 |
| 13 | gene | 50.261815 | 50.262869 | positive | MGI_C57BL6J_1916461 | 2310081J21Rik | NCBI_Gene:69211 | MGI:1916461 | lncRNA gene | RIKEN cDNA 2310081J21 gene |
| 13 | gene | 50.262394 | 50.262502 | positive | MGI_C57BL6J_5453794 | Gm24017 | ENSEMBL:ENSMUSG00000095027 | MGI:5453794 | miRNA gene | predicted gene, 24017 |
| 13 | gene | 50.267847 | 50.289832 | positive | MGI_C57BL6J_5590285 | Gm31126 | NCBI_Gene:102633258,ENSEMBL:ENSMUSG00000114035 | MGI:5590285 | lncRNA gene | predicted gene, 31126 |
| 13 | pseudogene | 50.291081 | 50.293188 | negative | MGI_C57BL6J_3780106 | Gm9699 | NCBI_Gene:676954 | MGI:3780106 | pseudogene | predicted gene 9699 |
| 13 | pseudogene | 50.300800 | 50.302602 | negative | MGI_C57BL6J_5826021 | Gm46384 | NCBI_Gene:108168041 | MGI:5826021 | pseudogene | predicted gene, 46384 |
| 13 | pseudogene | 50.300899 | 50.342216 | negative | MGI_C57BL6J_5826022 | Gm46385 | NCBI_Gene:108168042 | MGI:5826022 | pseudogene | predicted gene, 46385 |
| 13 | gene | 50.343583 | 50.491741 | negative | MGI_C57BL6J_3644775 | Gm8739 | NCBI_Gene:667632,ENSEMBL:ENSMUSG00000101136 | MGI:3644775 | lncRNA gene | predicted gene 8739 |
| 13 | gene | 50.399335 | 50.400423 | negative | MGI_C57BL6J_5011721 | Gm19536 | NCBI_Gene:100503071 | MGI:5011721 | protein coding gene | predicted gene, 19536 |
| 13 | pseudogene | 50.412062 | 50.414576 | negative | MGI_C57BL6J_3644780 | Gm8742 | NCBI_Gene:667637 | MGI:3644780 | pseudogene | predicted gene 8742 |
| 13 | gene | 50.417834 | 50.433781 | positive | MGI_C57BL6J_1923584 | Fbxw17 | NCBI_Gene:109082,ENSEMBL:ENSMUSG00000037816 | MGI:1923584 | protein coding gene | F-box and WD-40 domain protein 17 |
| 13 | gene | 50.443408 | 50.443912 | negative | MGI_C57BL6J_6096375 | Gm47429 | ENSEMBL:ENSMUSG00000114559 | MGI:6096375 | protein coding gene | predicted gene, 47429 |
| 13 | gene | 50.448942 | 50.465268 | negative | MGI_C57BL6J_5623831 | Gm40946 | NCBI_Gene:105245498 | MGI:5623831 | lncRNA gene | predicted gene, 40946 |
| 13 | gene | 50.467307 | 50.475355 | positive | MGI_C57BL6J_2685652 | Nutm2 | NCBI_Gene:328250,ENSEMBL:ENSMUSG00000071909 | MGI:2685652 | protein coding gene | NUT family member 2 |
| 13 | pseudogene | 50.490585 | 50.491435 | positive | MGI_C57BL6J_3648502 | Gm7259 | NCBI_Gene:639044 | MGI:3648502 | pseudogene | predicted gene 7259 |
| 13 | gene | 50.516647 | 50.539528 | negative | MGI_C57BL6J_5590994 | Gm31835 | NCBI_Gene:102634193 | MGI:5590994 | lncRNA gene | predicted gene, 31835 |
| 13 | gene | 50.544626 | 50.544734 | negative | MGI_C57BL6J_3629908 | Mir683-1 | miRBase:MI0004646,NCBI_Gene:751559,ENSEMBL:ENSMUSG00000095697 | MGI:3629908 | miRNA gene | microRNA 683-1 |
| 13 | pseudogene | 50.561473 | 50.570906 | positive | MGI_C57BL6J_5434145 | Gm20789 | NCBI_Gene:676982,ENSEMBL:ENSMUSG00000113354 | MGI:5434145 | pseudogene | predicted gene, 20789 |
| 13 | gene | 50.572802 | 50.595600 | negative | MGI_C57BL6J_5590871 | Gm31712 | NCBI_Gene:102634028 | MGI:5590871 | lncRNA gene | predicted gene, 31712 |
| 13 | gene | 50.600972 | 50.601080 | negative | MGI_C57BL6J_3719634 | Mir683-2 | miRBase:MI0010690,NCBI_Gene:100124649,ENSEMBL:ENSMUSG00000094933 | MGI:3719634 | miRNA gene | microRNA 683-2 |
| 13 | pseudogene | 50.610205 | 50.613242 | negative | MGI_C57BL6J_5591049 | Gm31890 | NCBI_Gene:102634263 | MGI:5591049 | pseudogene | predicted gene, 31890 |
| 13 | pseudogene | 50.610733 | 50.618160 | negative | MGI_C57BL6J_5588936 | Gm29777 | NCBI_Gene:101056092 | MGI:5588936 | pseudogene | predicted gene, 29777 |
| 13 | gene | 50.642548 | 50.646368 | positive | MGI_C57BL6J_2685750 | Gm904 | NCBI_Gene:380845,ENSEMBL:ENSMUSG00000096641 | MGI:2685750 | protein coding gene | predicted gene 904 |
| 13 | pseudogene | 50.647276 | 50.649746 | positive | MGI_C57BL6J_5011848 | Gm19663 | NCBI_Gene:100503376 | MGI:5011848 | pseudogene | predicted gene, 19663 |
| 13 | gene | 50.651705 | 50.673401 | negative | MGI_C57BL6J_5591197 | Gm32038 | NCBI_Gene:102634460 | MGI:5591197 | lncRNA gene | predicted gene, 32038 |
| 13 | gene | 50.676613 | 50.680284 | negative | MGI_C57BL6J_5453729 | Gm23952 | NCBI_Gene:102634376,ENSEMBL:ENSMUSG00000095380 | MGI:5453729 | miRNA gene | predicted gene, 23952 |
| 13 | gene | 50.697269 | 50.703435 | positive | MGI_C57BL6J_3704126 | Gm8765 | NCBI_Gene:667693,ENSEMBL:ENSMUSG00000094918 | MGI:3704126 | protein coding gene | predicted gene 8765 |
| 13 | gene | 50.753391 | 50.765248 | negative | MGI_C57BL6J_1918904 | 4930518P08Rik | NCBI_Gene:71654,ENSEMBL:ENSMUSG00000113652 | MGI:1918904 | lncRNA gene | RIKEN cDNA 4930518P08 gene |
| 13 | pseudogene | 50.758754 | 50.759668 | positive | MGI_C57BL6J_5009851 | Gm17761 | NCBI_Gene:676992 | MGI:5009851 | pseudogene | predicted gene, 17761 |
| 13 | pseudogene | 50.785649 | 50.787735 | positive | MGI_C57BL6J_3780822 | Gm2654 | NCBI_Gene:100040190,ENSEMBL:ENSMUSG00000112976 | MGI:3780822 | pseudogene | predicted gene 2654 |
| 13 | gene | 50.854923 | 50.878273 | negative | MGI_C57BL6J_5623832 | Gm40947 | NCBI_Gene:105245499 | MGI:5623832 | lncRNA gene | predicted gene, 40947 |
| 13 | pseudogene | 50.859334 | 50.860156 | negative | MGI_C57BL6J_5011194 | Gm19009 | NCBI_Gene:100418116,ENSEMBL:ENSMUSG00000113509 | MGI:5011194 | pseudogene | predicted gene, 19009 |
| 13 | gene | 50.885270 | 50.891424 | positive | MGI_C57BL6J_3643286 | Gm8777 | NCBI_Gene:667712 | MGI:3643286 | protein coding gene | predicted gene 8777 |
| 13 | gene | 50.913817 | 50.934152 | negative | MGI_C57BL6J_1920715 | 1700048B10Rik | NCBI_Gene:73465 | MGI:1920715 | lncRNA gene | RIKEN cDNA 1700048B10 gene |
| 13 | pseudogene | 50.936043 | 50.937246 | negative | MGI_C57BL6J_3780109 | Gm9702 | NCBI_Gene:676997 | MGI:3780109 | pseudogene | predicted gene 9702 |
| 13 | pseudogene | 50.950222 | 50.951017 | positive | MGI_C57BL6J_3643288 | Gm8779 | NCBI_Gene:667717 | MGI:3643288 | pseudogene | predicted gene 8779 |
| 13 | gene | 50.981488 | 50.991416 | negative | MGI_C57BL6J_5591772 | Gm32613 | NCBI_Gene:102635220 | MGI:5591772 | lncRNA gene | predicted gene, 32613 |
| 13 | gene | 51.014086 | 51.014211 | positive | MGI_C57BL6J_5451976 | Gm22199 | ENSEMBL:ENSMUSG00000084609 | MGI:5451976 | unclassified non-coding RNA gene | predicted gene, 22199 |
| 13 | gene | 51.097738 | 51.100727 | negative | MGI_C57BL6J_1924678 | 9430083A17Rik | NCBI_Gene:77428 | MGI:1924678 | lncRNA gene | RIKEN cDNA 9430083A17 gene |
| 13 | gene | 51.100508 | 51.152562 | positive | MGI_C57BL6J_109242 | Spin1 | NCBI_Gene:20729,ENSEMBL:ENSMUSG00000021395 | MGI:109242 | protein coding gene | spindlin 1 |
| 13 | gene | 51.170111 | 51.175191 | positive | MGI_C57BL6J_1922374 | Nxnl2 | NCBI_Gene:75124,ENSEMBL:ENSMUSG00000021396 | MGI:1922374 | protein coding gene | nucleoredoxin-like 2 |
| 13 | pseudogene | 51.200084 | 51.203066 | negative | MGI_C57BL6J_3646032 | Hist1h2al | NCBI_Gene:667728,ENSEMBL:ENSMUSG00000091383 | MGI:3646032 | pseudogene | histone cluster 1, H2al |
| 13 | gene | 51.202698 | 51.202835 | positive | MGI_C57BL6J_5690733 | Gm44341 | ENSEMBL:ENSMUSG00000106200 | MGI:5690733 | miRNA gene | predicted gene, 44341 |
| 13 | pseudogene | 51.204696 | 51.206162 | negative | MGI_C57BL6J_5588946 | Gm29787 | NCBI_Gene:101056142,ENSEMBL:ENSMUSG00000113521 | MGI:5588946 | pseudogene | predicted gene, 29787 |
| 13 | gene | 51.208641 | 51.372636 | positive | MGI_C57BL6J_5826051 | Gm46414 | NCBI_Gene:108168083 | MGI:5826051 | lncRNA gene | predicted gene, 46414 |
| 13 | pseudogene | 51.338921 | 51.339930 | positive | MGI_C57BL6J_3648619 | Gm6056 | NCBI_Gene:606521,ENSEMBL:ENSMUSG00000113426 | MGI:3648619 | pseudogene | predicted gene 6056 |
| 13 | pseudogene | 51.344996 | 51.345598 | positive | MGI_C57BL6J_6096574 | Gm47554 | ENSEMBL:ENSMUSG00000113162 | MGI:6096574 | pseudogene | predicted gene, 47554 |
| 13 | gene | 51.378136 | 51.418686 | negative | MGI_C57BL6J_5591993 | Gm32834 | NCBI_Gene:102635522,ENSEMBL:ENSMUSG00000112959 | MGI:5591993 | lncRNA gene | predicted gene, 32834 |
| 13 | gene | 51.408618 | 51.422797 | positive | MGI_C57BL6J_1339365 | S1pr3 | NCBI_Gene:13610,ENSEMBL:ENSMUSG00000067586 | MGI:1339365 | protein coding gene | sphingosine-1-phosphate receptor 3 |
| 13 | gene | 51.431041 | 51.569419 | negative | MGI_C57BL6J_106179 | Shc3 | NCBI_Gene:20418,ENSEMBL:ENSMUSG00000021448 | MGI:106179 | protein coding gene | src homology 2 domain-containing transforming protein C3 |
| 13 | gene | 51.645232 | 51.650662 | positive | MGI_C57BL6J_1913447 | Cks2 | NCBI_Gene:66197,ENSEMBL:ENSMUSG00000062248 | MGI:1913447 | protein coding gene | CDC28 protein kinase regulatory subunit 2 |
| 13 | gene | 51.648199 | 51.648334 | positive | MGI_C57BL6J_5452583 | Gm22806 | ENSEMBL:ENSMUSG00000064672 | MGI:5452583 | snoRNA gene | predicted gene, 22806 |
| 13 | gene | 51.650828 | 51.684044 | positive | MGI_C57BL6J_1922670 | Secisbp2 | NCBI_Gene:75420,ENSEMBL:ENSMUSG00000035139 | MGI:1922670 | protein coding gene | SECIS binding protein 2 |
| 13 | gene | 51.685529 | 51.793747 | negative | MGI_C57BL6J_109244 | Sema4d | NCBI_Gene:20354,ENSEMBL:ENSMUSG00000021451 | MGI:109244 | protein coding gene | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
| 13 | gene | 51.685534 | 51.686045 | negative | MGI_C57BL6J_3642353 | Gm15440 | NA | NA | protein coding gene | predicted gene 15440 |
| 13 | gene | 51.765294 | 51.765401 | positive | MGI_C57BL6J_5453343 | Gm23566 | ENSEMBL:ENSMUSG00000092648 | MGI:5453343 | miRNA gene | predicted gene, 23566 |
| 13 | gene | 51.766720 | 51.769365 | negative | MGI_C57BL6J_5011466 | Gm19281 | NA | NA | unclassified gene | predicted gene%2c 19281 |
| 13 | gene | 51.846675 | 51.848475 | positive | MGI_C57BL6J_1346325 | Gadd45g | NCBI_Gene:23882,ENSEMBL:ENSMUSG00000021453 | MGI:1346325 | protein coding gene | growth arrest and DNA-damage-inducible 45 gamma |
| 13 | gene | 51.973214 | 51.976583 | positive | MGI_C57BL6J_5477145 | Gm26651 | ENSEMBL:ENSMUSG00000097495 | MGI:5477145 | lncRNA gene | predicted gene, 26651 |
| 13 | gene | 52.019774 | 52.021927 | negative | MGI_C57BL6J_5611100 | Gm37872 | ENSEMBL:ENSMUSG00000102173 | MGI:5611100 | unclassified gene | predicted gene, 37872 |
| 13 | gene | 52.082639 | 52.083607 | negative | MGI_C57BL6J_5623833 | Gm40948 | NCBI_Gene:105245500 | MGI:5623833 | lncRNA gene | predicted gene, 40948 |
| 13 | gene | 52.087607 | 52.096668 | negative | MGI_C57BL6J_6097571 | Gm48190 | ENSEMBL:ENSMUSG00000113090 | MGI:6097571 | lncRNA gene | predicted gene, 48190 |
| 13 | gene | 52.096741 | 52.124160 | positive | MGI_C57BL6J_1921300 | 4921525O09Rik | NCBI_Gene:74050,ENSEMBL:ENSMUSG00000113211 | MGI:1921300 | lncRNA gene | RIKEN cDNA 4921525O09 gene |
| 13 | gene | 52.180411 | 52.181637 | positive | MGI_C57BL6J_6097585 | Gm48199 | ENSEMBL:ENSMUSG00000113399 | MGI:6097585 | lncRNA gene | predicted gene, 48199 |
| 13 | gene | 52.222868 | 52.227977 | positive | MGI_C57BL6J_5592200 | Gm33041 | NCBI_Gene:102635793 | MGI:5592200 | lncRNA gene | predicted gene, 33041 |
| 13 | gene | 52.233830 | 52.245034 | positive | MGI_C57BL6J_5592275 | Gm33116 | NCBI_Gene:102635896 | MGI:5592275 | lncRNA gene | predicted gene, 33116 |
| 13 | gene | 52.504375 | 52.531279 | negative | MGI_C57BL6J_1915453 | Diras2 | NCBI_Gene:68203,ENSEMBL:ENSMUSG00000047842 | MGI:1915453 | protein coding gene | DIRAS family, GTP-binding RAS-like 2 |
| 13 | gene | 52.550956 | 52.557396 | positive | MGI_C57BL6J_5592327 | Gm33168 | NCBI_Gene:102635967 | MGI:5592327 | lncRNA gene | predicted gene, 33168 |
| 13 | gene | 52.569880 | 52.573225 | negative | MGI_C57BL6J_3781020 | Gm2848 | NCBI_Gene:100040572 | MGI:3781020 | lncRNA gene | predicted gene 2848 |
| 13 | gene | 52.582329 | 52.648792 | positive | MGI_C57BL6J_99515 | Syk | NCBI_Gene:20963,ENSEMBL:ENSMUSG00000021457 | MGI:99515 | protein coding gene | spleen tyrosine kinase |
| 13 | gene | 52.729370 | 52.757205 | negative | MGI_C57BL6J_2145475 | BB123696 | NCBI_Gene:105404,ENSEMBL:ENSMUSG00000113737 | MGI:2145475 | lncRNA gene | expressed sequence BB123696 |
| 13 | gene | 52.828125 | 52.845595 | positive | MGI_C57BL6J_5592474 | Gm33315 | NCBI_Gene:102636175 | MGI:5592474 | lncRNA gene | predicted gene, 33315 |
| 13 | gene | 52.835107 | 52.929705 | negative | MGI_C57BL6J_1338011 | Auh | NCBI_Gene:11992,ENSEMBL:ENSMUSG00000021460 | MGI:1338011 | protein coding gene | AU RNA binding protein/enoyl-coenzyme A hydratase |
| 13 | gene | 52.954388 | 52.967120 | positive | MGI_C57BL6J_5623835 | Gm40950 | NCBI_Gene:105245502 | MGI:5623835 | lncRNA gene | predicted gene, 40950 |
| 13 | gene | 52.964621 | 52.964732 | negative | MGI_C57BL6J_4421899 | n-R5s54 | ENSEMBL:ENSMUSG00000065771 | MGI:4421899 | rRNA gene | nuclear encoded rRNA 5S 54 |
| 13 | gene | 52.967209 | 52.982789 | negative | MGI_C57BL6J_109495 | Nfil3 | NCBI_Gene:18030,ENSEMBL:ENSMUSG00000056749 | MGI:109495 | protein coding gene | nuclear factor, interleukin 3, regulated |
| 13 | gene | 52.985965 | 53.018481 | positive | MGI_C57BL6J_5592583 | Gm33424 | NCBI_Gene:102636324,ENSEMBL:ENSMUSG00000113101 | MGI:5592583 | lncRNA gene | predicted gene, 33424 |
| 13 | gene | 53.008090 | 53.018019 | negative | MGI_C57BL6J_5592678 | Gm33519 | NCBI_Gene:102636460 | MGI:5592678 | lncRNA gene | predicted gene, 33519 |
| 13 | gene | 53.018058 | 53.018427 | negative | MGI_C57BL6J_1201795 | Oit4 | NA | NA | unclassified gene | oncoprotein induced transcript 4 |
| 13 | gene | 53.028918 | 53.035090 | positive | MGI_C57BL6J_6097796 | Gm48336 | ENSEMBL:ENSMUSG00000112952 | MGI:6097796 | lncRNA gene | predicted gene, 48336 |
| 13 | gene | 53.109312 | 53.286124 | negative | MGI_C57BL6J_1347521 | Ror2 | NCBI_Gene:26564,ENSEMBL:ENSMUSG00000021464 | MGI:1347521 | protein coding gene | receptor tyrosine kinase-like orphan receptor 2 |
| 13 | gene | 53.185866 | 53.186940 | positive | MGI_C57BL6J_1925781 | C330026N13Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA C330026N13 gene |
| 13 | gene | 53.286244 | 53.294035 | positive | MGI_C57BL6J_5592733 | Gm33574 | NCBI_Gene:102636536 | MGI:5592733 | lncRNA gene | predicted gene, 33574 |
| 13 | gene | 53.308672 | 53.309751 | negative | MGI_C57BL6J_5592790 | Gm33631 | NCBI_Gene:102636619 | MGI:5592790 | lncRNA gene | predicted gene, 33631 |
| 13 | gene | 53.332748 | 53.377417 | negative | MGI_C57BL6J_1099431 | Sptlc1 | NCBI_Gene:268656,ENSEMBL:ENSMUSG00000021468 | MGI:1099431 | protein coding gene | serine palmitoyltransferase, long chain base subunit 1 |
| 13 | gene | 53.393657 | 53.455630 | positive | MGI_C57BL6J_3780931 | Gm2762 | NCBI_Gene:100040421,ENSEMBL:ENSMUSG00000107008 | MGI:3780931 | lncRNA gene | predicted gene 2762 |
| 13 | gene | 53.403950 | 53.414830 | negative | MGI_C57BL6J_1920673 | 1700058M13Rik | NA | NA | lncRNA gene | RIKEN cDNA 1700058M13 gene |
| 13 | gene | 53.403950 | 53.414830 | negative | MGI_C57BL6J_5663399 | Gm43262 | ENSEMBL:ENSMUSG00000107305 | MGI:5663399 | lncRNA gene | predicted gene 43262 |
| 13 | gene | 53.415943 | 53.419345 | negative | MGI_C57BL6J_5621449 | Gm38564 | NCBI_Gene:102641557 | MGI:5621449 | protein coding gene | predicted gene, 38564 |
| 13 | gene | 53.461317 | 53.478244 | positive | MGI_C57BL6J_5592922 | Gm33763 | NCBI_Gene:102636789 | MGI:5592922 | lncRNA gene | predicted gene, 33763 |
| 13 | gene | 53.466881 | 53.473074 | negative | MGI_C57BL6J_97169 | Msx2 | NCBI_Gene:17702,ENSEMBL:ENSMUSG00000021469 | MGI:97169 | protein coding gene | msh homeobox 2 |
| 13 | gene | 53.478351 | 53.481973 | positive | MGI_C57BL6J_5623836 | Gm40951 | NCBI_Gene:105245503 | MGI:5623836 | lncRNA gene | predicted gene, 40951 |
| 13 | gene | 53.482624 | 53.505790 | positive | MGI_C57BL6J_5593032 | Gm33873 | NCBI_Gene:102636939 | MGI:5593032 | lncRNA gene | predicted gene, 33873 |
| 13 | gene | 53.501857 | 53.504386 | negative | MGI_C57BL6J_5623837 | Gm40952 | NCBI_Gene:105245504 | MGI:5623837 | lncRNA gene | predicted gene, 40952 |
| 13 | gene | 53.503606 | 53.534643 | negative | MGI_C57BL6J_5593181 | Gm34022 | NCBI_Gene:102637135,ENSEMBL:ENSMUSG00000113834 | MGI:5593181 | lncRNA gene | predicted gene, 34022 |
| 13 | pseudogene | 53.525671 | 53.526132 | positive | MGI_C57BL6J_3643792 | Gm5449 | NCBI_Gene:432760,ENSEMBL:ENSMUSG00000063166 | MGI:3643792 | pseudogene | predicted pseudogene 5449 |
| 13 | gene | 53.635892 | 53.639201 | negative | MGI_C57BL6J_1924648 | 9530014B07Rik | NCBI_Gene:77398,ENSEMBL:ENSMUSG00000113599 | MGI:1924648 | lncRNA gene | RIKEN cDNA 9530014B07 gene |
| 13 | gene | 53.648585 | 53.650081 | positive | MGI_C57BL6J_1922502 | 4930555G21Rik | NCBI_Gene:75252,ENSEMBL:ENSMUSG00000113329 | MGI:1922502 | lncRNA gene | RIKEN cDNA 4930555G21 gene |
| 13 | gene | 53.681609 | 53.700474 | negative | MGI_C57BL6J_5593437 | Gm34278 | NCBI_Gene:102637481,ENSEMBL:ENSMUSG00000112965 | MGI:5593437 | lncRNA gene | predicted gene, 34278 |
| 13 | gene | 53.688767 | 53.690351 | positive | MGI_C57BL6J_5593491 | Gm34332 | NCBI_Gene:102637554 | MGI:5593491 | lncRNA gene | predicted gene, 34332 |
| 13 | gene | 53.748391 | 53.787724 | negative | MGI_C57BL6J_5593598 | Gm34439 | NCBI_Gene:102637696 | MGI:5593598 | lncRNA gene | predicted gene, 34439 |
| 13 | gene | 53.776627 | 53.778973 | positive | MGI_C57BL6J_6098098 | Gm48548 | ENSEMBL:ENSMUSG00000114143 | MGI:6098098 | lncRNA gene | predicted gene, 48548 |
| 13 | gene | 53.839281 | 53.850088 | negative | MGI_C57BL6J_5623839 | Gm40954 | NCBI_Gene:105245506 | MGI:5623839 | lncRNA gene | predicted gene, 40954 |
| 13 | pseudogene | 53.888534 | 53.890855 | positive | MGI_C57BL6J_5011159 | Gm18974 | NCBI_Gene:100418060,ENSEMBL:ENSMUSG00000114012 | MGI:5011159 | pseudogene | predicted gene, 18974 |
| 13 | gene | 53.912263 | 53.916257 | negative | MGI_C57BL6J_1925800 | E130119H09Rik | NCBI_Gene:78550,ENSEMBL:ENSMUSG00000114069 | MGI:1925800 | lncRNA gene | RIKEN cDNA E130119H09 gene |
| 13 | gene | 53.921313 | 53.935283 | negative | MGI_C57BL6J_5593716 | Gm34557 | NCBI_Gene:102637851,ENSEMBL:ENSMUSG00000113982 | MGI:5593716 | lncRNA gene | predicted gene, 34557 |
| 13 | gene | 53.968944 | 53.976666 | positive | MGI_C57BL6J_6098101 | Gm48550 | ENSEMBL:ENSMUSG00000113882 | MGI:6098101 | lncRNA gene | predicted gene, 48550 |
| 13 | gene | 54.051183 | 54.055705 | negative | MGI_C57BL6J_99578 | Drd1 | NCBI_Gene:13488,ENSEMBL:ENSMUSG00000021478 | MGI:99578 | protein coding gene | dopamine receptor D1 |
| 13 | gene | 54.071825 | 54.108346 | positive | MGI_C57BL6J_2137677 | Sfxn1 | NCBI_Gene:14057,ENSEMBL:ENSMUSG00000021474 | MGI:2137677 | protein coding gene | sideroflexin 1 |
| 13 | pseudogene | 54.140933 | 54.141802 | positive | MGI_C57BL6J_5010658 | Gm18473 | NCBI_Gene:100417234,ENSEMBL:ENSMUSG00000114042 | MGI:5010658 | pseudogene | predicted gene, 18473 |
| 13 | gene | 54.177791 | 54.181769 | negative | MGI_C57BL6J_5593799 | Gm34640 | NCBI_Gene:102637957 | MGI:5593799 | lncRNA gene | predicted gene, 34640 |
| 13 | gene | 54.191970 | 54.236182 | positive | MGI_C57BL6J_108482 | Hrh2 | NCBI_Gene:15466,ENSEMBL:ENSMUSG00000034987 | MGI:108482 | protein coding gene | histamine receptor H2 |
| 13 | gene | 54.215385 | 54.223023 | negative | MGI_C57BL6J_4414998 | Gm16578 | NCBI_Gene:105245508,ENSEMBL:ENSMUSG00000090016 | MGI:4414998 | lncRNA gene | predicted gene 16578 |
| 13 | gene | 54.245296 | 54.245584 | positive | MGI_C57BL6J_6098206 | Gm48615 | ENSEMBL:ENSMUSG00000113327 | MGI:6098206 | lncRNA gene | predicted gene, 48615 |
| 13 | gene | 54.283242 | 54.289078 | positive | MGI_C57BL6J_5623840 | Gm40955 | NCBI_Gene:105245507 | MGI:5623840 | lncRNA gene | predicted gene, 40955 |
| 13 | gene | 54.301959 | 54.312797 | positive | MGI_C57BL6J_6098220 | Gm48623 | ENSEMBL:ENSMUSG00000112941 | MGI:6098220 | lncRNA gene | predicted gene, 48623 |
| 13 | gene | 54.301964 | 54.383917 | positive | MGI_C57BL6J_104726 | Cplx2 | NCBI_Gene:12890,ENSEMBL:ENSMUSG00000025867 | MGI:104726 | protein coding gene | complexin 2 |
| 13 | gene | 54.309905 | 54.327880 | negative | MGI_C57BL6J_6098218 | Gm48622 | ENSEMBL:ENSMUSG00000113404 | MGI:6098218 | lncRNA gene | predicted gene, 48622 |
| 13 | gene | 54.399092 | 54.399334 | negative | MGI_C57BL6J_3826528 | Gm16248 | ENSEMBL:ENSMUSG00000086137 | MGI:3826528 | lncRNA gene | predicted gene 16248 |
| 13 | gene | 54.429441 | 54.429700 | negative | MGI_C57BL6J_5452445 | Gm22668 | ENSEMBL:ENSMUSG00000089341 | MGI:5452445 | unclassified non-coding RNA gene | predicted gene, 22668 |
| 13 | gene | 54.458837 | 54.468849 | negative | MGI_C57BL6J_1920916 | Thoc3 | NCBI_Gene:73666,ENSEMBL:ENSMUSG00000025872 | MGI:1920916 | protein coding gene | THO complex 3 |
| 13 | pseudogene | 54.499578 | 54.503877 | negative | MGI_C57BL6J_3781002 | Gm2830 | NCBI_Gene:100040543,ENSEMBL:ENSMUSG00000086567 | MGI:3781002 | pseudogene | predicted gene 2830 |
| 13 | gene | 54.503739 | 54.551290 | positive | MGI_C57BL6J_2442599 | Simc1 | NCBI_Gene:319719,ENSEMBL:ENSMUSG00000043183 | MGI:2442599 | protein coding gene | SUMO-interacting motifs containing 1 |
| 13 | gene | 54.551218 | 54.565435 | negative | MGI_C57BL6J_1921162 | 4833439L19Rik | NCBI_Gene:97820,ENSEMBL:ENSMUSG00000025871 | MGI:1921162 | protein coding gene | RIKEN cDNA 4833439L19 gene |
| 13 | gene | 54.575000 | 54.582765 | positive | MGI_C57BL6J_1930788 | Arl10 | NCBI_Gene:56795,ENSEMBL:ENSMUSG00000025870 | MGI:1930788 | protein coding gene | ADP-ribosylation factor-like 10 |
| 13 | gene | 54.578113 | 54.578249 | positive | MGI_C57BL6J_5453972 | Gm24195 | ENSEMBL:ENSMUSG00000080454 | MGI:5453972 | miRNA gene | predicted gene, 24195 |
| 13 | gene | 54.584185 | 54.590090 | negative | MGI_C57BL6J_107862 | Nop16 | NCBI_Gene:28126,ENSEMBL:ENSMUSG00000025869 | MGI:107862 | protein coding gene | NOP16 nucleolar protein |
| 13 | gene | 54.590207 | 54.591158 | positive | MGI_C57BL6J_1914294 | Higd2a | NCBI_Gene:67044,ENSEMBL:ENSMUSG00000025868 | MGI:1914294 | protein coding gene | HIG1 domain family, member 2A |
| 13 | gene | 54.592401 | 54.611405 | negative | MGI_C57BL6J_1921575 | Cltb | NCBI_Gene:74325,ENSEMBL:ENSMUSG00000047547 | MGI:1921575 | protein coding gene | clathrin, light polypeptide (Lcb) |
| 13 | gene | 54.621784 | 54.664063 | positive | MGI_C57BL6J_1923827 | Faf2 | NCBI_Gene:76577,ENSEMBL:ENSMUSG00000025873 | MGI:1923827 | protein coding gene | Fas associated factor family member 2 |
| 13 | pseudogene | 54.656909 | 54.657002 | negative | MGI_C57BL6J_97944 | Rn4.5s-ps3 | NCBI_Gene:19802 | MGI:97944 | pseudogene | 4.5s RNA, pseudogene 3 |
| 13 | gene | 54.679399 | 54.693994 | negative | MGI_C57BL6J_2145310 | Rnf44 | NCBI_Gene:105239,ENSEMBL:ENSMUSG00000034928 | MGI:2145310 | protein coding gene | ring finger protein 44 |
| 13 | gene | 54.701461 | 54.736662 | positive | MGI_C57BL6J_2687323 | Cdhr2 | NCBI_Gene:268663,ENSEMBL:ENSMUSG00000034918 | MGI:2687323 | protein coding gene | cadherin-related family member 2 |
| 13 | gene | 54.736671 | 54.749874 | negative | MGI_C57BL6J_1349455 | Gprin1 | NCBI_Gene:26913,ENSEMBL:ENSMUSG00000069227 | MGI:1349455 | protein coding gene | G protein-regulated inducer of neurite outgrowth 1 |
| 13 | gene | 54.741952 | 54.742332 | positive | MGI_C57BL6J_1924681 | 9530001D17Rik | NA | NA | unclassified gene | RIKEN cDNA 9530001D17 gene |
| 13 | gene | 54.758860 | 54.766613 | negative | MGI_C57BL6J_1889011 | Sncb | NCBI_Gene:104069,ENSEMBL:ENSMUSG00000034891 | MGI:1889011 | protein coding gene | synuclein, beta |
| 13 | gene | 54.777514 | 54.783091 | negative | MGI_C57BL6J_3826524 | Gm16249 | NCBI_Gene:102633259,ENSEMBL:ENSMUSG00000084949 | MGI:3826524 | lncRNA gene | predicted gene 16249 |
| 13 | gene | 54.783928 | 54.788459 | positive | MGI_C57BL6J_2685119 | Eif4e1b | NCBI_Gene:218268,ENSEMBL:ENSMUSG00000074895 | MGI:2685119 | protein coding gene | eukaryotic translation initiation factor 4E family member 1B |
| 13 | gene | 54.789351 | 54.796776 | positive | MGI_C57BL6J_1921507 | Tspan17 | NCBI_Gene:74257,ENSEMBL:ENSMUSG00000025875 | MGI:1921507 | protein coding gene | tetraspanin 17 |
| 13 | pseudogene | 54.840649 | 54.840904 | negative | MGI_C57BL6J_3826525 | Gm16250 | ENSEMBL:ENSMUSG00000086097 | MGI:3826525 | pseudogene | predicted gene 16250 |
| 13 | gene | 54.858135 | 54.874869 | negative | MGI_C57BL6J_5623844 | Gm40959 | NCBI_Gene:105245512 | MGI:5623844 | lncRNA gene | predicted gene, 40959 |
| 13 | gene | 54.876135 | 54.878598 | positive | MGI_C57BL6J_5590347 | Gm31188 | NCBI_Gene:102633340 | MGI:5590347 | lncRNA gene | predicted gene, 31188 |
| 13 | gene | 54.887470 | 54.888013 | positive | MGI_C57BL6J_5580137 | Gm29431 | ENSEMBL:ENSMUSG00000100728 | MGI:5580137 | lncRNA gene | predicted gene 29431 |
| 13 | gene | 54.913235 | 54.914479 | positive | MGI_C57BL6J_1925321 | 5330429C05Rik | ENSEMBL:ENSMUSG00000085896 | MGI:1925321 | lncRNA gene | RIKEN cDNA 5330429C05 gene |
| 13 | gene | 54.926763 | 54.930753 | negative | MGI_C57BL6J_1922443 | 4930526F13Rik | NCBI_Gene:75193,ENSEMBL:ENSMUSG00000087335 | MGI:1922443 | lncRNA gene | RIKEN cDNA 4930526F13 gene |
| 13 | gene | 54.933420 | 54.949336 | negative | MGI_C57BL6J_5590472 | Gm31313 | NCBI_Gene:102633503 | MGI:5590472 | lncRNA gene | predicted gene, 31313 |
| 13 | gene | 54.949411 | 55.006018 | positive | MGI_C57BL6J_894682 | Unc5a | NCBI_Gene:107448,ENSEMBL:ENSMUSG00000025876 | MGI:894682 | protein coding gene | unc-5 netrin receptor A |
| 13 | gene | 55.005985 | 55.021476 | negative | MGI_C57BL6J_2670962 | Hk3 | NCBI_Gene:212032,ENSEMBL:ENSMUSG00000025877 | MGI:2670962 | protein coding gene | hexokinase 3 |
| 13 | gene | 55.027880 | 55.100309 | negative | MGI_C57BL6J_103185 | Uimc1 | NCBI_Gene:20184,ENSEMBL:ENSMUSG00000025878 | MGI:103185 | protein coding gene | ubiquitin interaction motif containing 1 |
| 13 | pseudogene | 55.085752 | 55.086373 | positive | MGI_C57BL6J_3779586 | Gm6344 | NCBI_Gene:622707,ENSEMBL:ENSMUSG00000094446 | MGI:3779586 | pseudogene | predicted gene 6344 |
| 13 | gene | 55.105270 | 55.135071 | positive | MGI_C57BL6J_1349417 | Zfp346 | NCBI_Gene:26919,ENSEMBL:ENSMUSG00000021481 | MGI:1349417 | protein coding gene | zinc finger protein 346 |
| 13 | gene | 55.109795 | 55.112340 | positive | MGI_C57BL6J_1924277 | 4432414F05Rik | NA | NA | unclassified gene | RIKEN cDNA 4432414F05 gene |
| 13 | gene | 55.124748 | 55.126631 | positive | MGI_C57BL6J_2443167 | A130082J08Rik | NA | NA | unclassified gene | RIKEN cDNA A130082J08 gene |
| 13 | gene | 55.127687 | 55.128277 | negative | MGI_C57BL6J_5826052 | Gm46415 | NCBI_Gene:108168084,ENSEMBL:ENSMUSG00000114481 | MGI:5826052 | protein coding gene | predicted gene, 46415 |
| 13 | gene | 55.132962 | 55.133548 | negative | MGI_C57BL6J_4937251 | Gm17617 | ENSEMBL:ENSMUSG00000091768 | MGI:4937251 | protein coding gene | predicted gene, 17617 |
| 13 | gene | 55.152640 | 55.168759 | positive | MGI_C57BL6J_95525 | Fgfr4 | NCBI_Gene:14186,ENSEMBL:ENSMUSG00000005320 | MGI:95525 | protein coding gene | fibroblast growth factor receptor 4 |
| 13 | gene | 55.184771 | 55.186647 | positive | MGI_C57BL6J_1924820 | 3930401B19Rik | NA | NA | unclassified gene | RIKEN cDNA 3930401B19 gene |
| 13 | gene | 55.209782 | 55.318325 | positive | MGI_C57BL6J_1276545 | Nsd1 | NCBI_Gene:18193,ENSEMBL:ENSMUSG00000021488 | MGI:1276545 | protein coding gene | nuclear receptor-binding SET-domain protein 1 |
| 13 | pseudogene | 55.253225 | 55.254401 | negative | MGI_C57BL6J_6097003 | Gm47816 | ENSEMBL:ENSMUSG00000114359 | MGI:6097003 | pseudogene | predicted gene, 47816 |
| 13 | gene | 55.319223 | 55.321980 | negative | MGI_C57BL6J_105065 | Rab24 | NCBI_Gene:19336,ENSEMBL:ENSMUSG00000034789 | MGI:105065 | protein coding gene | RAB24, member RAS oncogene family |
| 13 | gene | 55.320500 | 55.325272 | positive | MGI_C57BL6J_1913744 | Prelid1 | NCBI_Gene:66494,ENSEMBL:ENSMUSG00000021486 | MGI:1913744 | protein coding gene | PRELI domain containing 1 |
| 13 | gene | 55.325168 | 55.330046 | negative | MGI_C57BL6J_104987 | Mxd3 | NCBI_Gene:17121,ENSEMBL:ENSMUSG00000021485 | MGI:104987 | protein coding gene | Max dimerization protein 3 |
| 13 | gene | 55.343833 | 55.362783 | negative | MGI_C57BL6J_1914140 | Lman2 | NCBI_Gene:66890,ENSEMBL:ENSMUSG00000021484 | MGI:1914140 | protein coding gene | lectin, mannose-binding 2 |
| 13 | gene | 55.369712 | 55.384691 | positive | MGI_C57BL6J_1859709 | Rgs14 | NCBI_Gene:51791,ENSEMBL:ENSMUSG00000052087 | MGI:1859709 | protein coding gene | regulator of G-protein signaling 14 |
| 13 | gene | 55.398187 | 55.415592 | positive | MGI_C57BL6J_1345284 | Slc34a1 | NCBI_Gene:20505,ENSEMBL:ENSMUSG00000021490 | MGI:1345284 | protein coding gene | solute carrier family 34 (sodium phosphate), member 1 |
| 13 | gene | 55.414688 | 55.415232 | negative | MGI_C57BL6J_2178800 | Pfn3 | NCBI_Gene:75477,ENSEMBL:ENSMUSG00000044444 | MGI:2178800 | protein coding gene | profilin 3 |
| 13 | gene | 55.414901 | 55.415592 | positive | MGI_C57BL6J_3704403 | BC038268 | NA | NA | protein coding gene | cDNA sequence BC038268 |
| 13 | gene | 55.417958 | 55.426804 | negative | MGI_C57BL6J_1891012 | F12 | NCBI_Gene:58992,ENSEMBL:ENSMUSG00000021492 | MGI:1891012 | protein coding gene | coagulation factor XII (Hageman factor) |
| 13 | gene | 55.445072 | 55.460927 | positive | MGI_C57BL6J_1347078 | Grk6 | NCBI_Gene:26385,ENSEMBL:ENSMUSG00000074886 | MGI:1347078 | protein coding gene | G protein-coupled receptor kinase 6 |
| 13 | gene | 55.453106 | 55.453171 | positive | MGI_C57BL6J_5531173 | Mir6943 | miRBase:MI0022790,NCBI_Gene:102466770,ENSEMBL:ENSMUSG00000099058 | MGI:5531173 | miRNA gene | microRNA 6943 |
| 13 | gene | 55.464267 | 55.473162 | positive | MGI_C57BL6J_3487246 | Prr7 | NCBI_Gene:432763,ENSEMBL:ENSMUSG00000034686 | MGI:3487246 | protein coding gene | proline rich 7 (synaptic) |
| 13 | gene | 55.469208 | 55.470398 | positive | MGI_C57BL6J_1925119 | E130112B07Rik | NA | NA | unclassified gene | RIKEN cDNA E130112B07 gene |
| 13 | gene | 55.473428 | 55.488111 | negative | MGI_C57BL6J_1931838 | Dbn1 | NCBI_Gene:56320,ENSEMBL:ENSMUSG00000034675 | MGI:1931838 | protein coding gene | drebrin 1 |
| 13 | gene | 55.477681 | 55.477776 | negative | MGI_C57BL6J_5531092 | Mir6944 | miRBase:MI0022791,NCBI_Gene:102465569,ENSEMBL:ENSMUSG00000098341 | MGI:5531092 | miRNA gene | microRNA 6944 |
| 13 | gene | 55.495795 | 55.513676 | negative | MGI_C57BL6J_1914649 | Pdlim7 | NCBI_Gene:67399,ENSEMBL:ENSMUSG00000021493 | MGI:1914649 | protein coding gene | PDZ and LIM domain 7 |
| 13 | gene | 55.507625 | 55.507692 | negative | MGI_C57BL6J_5531161 | Mir6945 | miRBase:MI0022792,NCBI_Gene:102465570,ENSEMBL:ENSMUSG00000099096 | MGI:5531161 | miRNA gene | microRNA 6945 |
| 13 | gene | 55.523231 | 55.530345 | negative | MGI_C57BL6J_1351490 | Dok3 | NCBI_Gene:27261,ENSEMBL:ENSMUSG00000035711 | MGI:1351490 | protein coding gene | docking protein 3 |
| 13 | gene | 55.530410 | 55.536658 | negative | MGI_C57BL6J_1920185 | Ddx41 | NCBI_Gene:72935,ENSEMBL:ENSMUSG00000021494 | MGI:1920185 | protein coding gene | DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 |
| 13 | gene | 55.535995 | 55.546848 | positive | MGI_C57BL6J_5826053 | Gm46416 | NCBI_Gene:108168085,ENSEMBL:ENSMUSG00000114633 | MGI:5826053 | lncRNA gene | predicted gene, 46416 |
| 13 | gene | 55.539316 | 55.571120 | negative | MGI_C57BL6J_2385851 | Fam193b | NCBI_Gene:212483,ENSEMBL:ENSMUSG00000021495 | MGI:2385851 | protein coding gene | family with sequence similarity 193, member B |
| 13 | gene | 55.547571 | 55.548859 | positive | MGI_C57BL6J_5826054 | Gm46417 | NCBI_Gene:108168086 | MGI:5826054 | lncRNA gene | predicted gene, 46417 |
| 13 | gene | 55.571216 | 55.574475 | positive | MGI_C57BL6J_5590523 | Gm31364 | NCBI_Gene:102633572 | MGI:5590523 | lncRNA gene | predicted gene, 31364 |
| 13 | gene | 55.573732 | 55.597697 | positive | MGI_C57BL6J_1914761 | Tmed9 | NCBI_Gene:67511,ENSEMBL:ENSMUSG00000058569 | MGI:1914761 | protein coding gene | transmembrane p24 trafficking protein 9 |
| 13 | gene | 55.599896 | 55.610443 | positive | MGI_C57BL6J_2384987 | B4galt7 | NCBI_Gene:218271,ENSEMBL:ENSMUSG00000021504 | MGI:2384987 | protein coding gene | xylosylprotein beta1,4-galactosyltransferase, polypeptide 7 (galactosyltransferase I) |
| 13 | gene | 55.602182 | 55.604783 | negative | MGI_C57BL6J_3802139 | Gm15911 | ENSEMBL:ENSMUSG00000086506 | MGI:3802139 | lncRNA gene | predicted gene 15911 |
| 13 | gene | 55.623005 | 55.632416 | positive | MGI_C57BL6J_104728 | Caml | NCBI_Gene:12328,ENSEMBL:ENSMUSG00000021501 | MGI:104728 | protein coding gene | calcium modulating ligand |
| 13 | gene | 55.634974 | 55.681267 | positive | MGI_C57BL6J_1920895 | Ddx46 | NCBI_Gene:212880,ENSEMBL:ENSMUSG00000021500 | MGI:1920895 | protein coding gene | DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 |
| 13 | gene | 55.693124 | 55.703500 | positive | MGI_C57BL6J_1925771 | B230219D22Rik | NCBI_Gene:78521,ENSEMBL:ENSMUSG00000045767 | MGI:1925771 | protein coding gene | RIKEN cDNA B230219D22 gene |
| 13 | gene | 55.714650 | 55.726227 | positive | MGI_C57BL6J_1916922 | Txndc15 | NCBI_Gene:69672,ENSEMBL:ENSMUSG00000021497 | MGI:1916922 | protein coding gene | thioredoxin domain containing 15 |
| 13 | gene | 55.727116 | 55.776830 | positive | MGI_C57BL6J_1919812 | Pcbd2 | NCBI_Gene:72562,ENSEMBL:ENSMUSG00000021496 | MGI:1919812 | protein coding gene | pterin 4 alpha carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2 |
| 13 | gene | 55.775136 | 55.784505 | negative | MGI_C57BL6J_1925361 | 4930451E10Rik | NCBI_Gene:78111,ENSEMBL:ENSMUSG00000069223 | MGI:1925361 | lncRNA gene | RIKEN cDNA 4930451E10 gene |
| 13 | gene | 55.784568 | 55.824942 | positive | MGI_C57BL6J_1924106 | Catsper3 | NCBI_Gene:76856,ENSEMBL:ENSMUSG00000021499 | MGI:1924106 | protein coding gene | cation channel, sperm associated 3 |
| 13 | gene | 55.825044 | 55.836193 | negative | MGI_C57BL6J_107374 | Pitx1 | NCBI_Gene:18740,ENSEMBL:ENSMUSG00000021506 | MGI:107374 | protein coding gene | paired-like homeodomain transcription factor 1 |
| 13 | gene | 55.826250 | 56.084849 | positive | MGI_C57BL6J_6095789 | Gm47071 | ENSEMBL:ENSMUSG00000114493 | MGI:6095789 | lncRNA gene | predicted gene, 47071 |
| 13 | gene | 55.835360 | 55.836095 | negative | MGI_C57BL6J_5579466 | Gm28760 | ENSEMBL:ENSMUSG00000100096 | MGI:5579466 | lncRNA gene | predicted gene 28760 |
| 13 | gene | 55.904608 | 55.910101 | negative | MGI_C57BL6J_5590701 | Gm31542 | NCBI_Gene:102633807 | MGI:5590701 | lncRNA gene | predicted gene, 31542 |
| 13 | pseudogene | 56.027384 | 56.030234 | negative | MGI_C57BL6J_6095791 | Gm47072 | ENSEMBL:ENSMUSG00000114229 | MGI:6095791 | pseudogene | predicted gene, 47072 |
| 13 | gene | 56.073619 | 56.136361 | negative | MGI_C57BL6J_1349392 | Macroh2a1 | NCBI_Gene:26914,ENSEMBL:ENSMUSG00000015937 | MGI:1349392 | protein coding gene | macroH2A.1 histone |
| 13 | gene | 56.110546 | 56.116812 | negative | MGI_C57BL6J_5590644 | Gm31485 | NCBI_Gene:102633730 | MGI:5590644 | lncRNA gene | predicted gene, 31485 |
| 13 | gene | 56.166600 | 56.193544 | positive | MGI_C57BL6J_5590816 | Gm31657 | NCBI_Gene:102633960 | MGI:5590816 | lncRNA gene | predicted gene, 31657 |
| 13 | gene | 56.167600 | 56.167722 | positive | MGI_C57BL6J_5454925 | Gm25148 | ENSEMBL:ENSMUSG00000077799 | MGI:5454925 | snoRNA gene | predicted gene, 25148 |
| 13 | gene | 56.173703 | 56.179013 | negative | MGI_C57BL6J_2385852 | Tifab | NCBI_Gene:212937,ENSEMBL:ENSMUSG00000049625 | MGI:2385852 | protein coding gene | TRAF-interacting protein with forkhead-associated domain, family member B |
| 13 | pseudogene | 56.202231 | 56.202937 | negative | MGI_C57BL6J_6095840 | Gm47104 | ENSEMBL:ENSMUSG00000114341 | MGI:6095840 | pseudogene | predicted gene, 47104 |
| 13 | gene | 56.222224 | 56.243940 | negative | MGI_C57BL6J_1922495 | 4930550C17Rik | NCBI_Gene:75245,ENSEMBL:ENSMUSG00000097361 | MGI:1922495 | lncRNA gene | RIKEN cDNA 4930550C17 gene |
| 13 | gene | 56.250478 | 56.252163 | negative | MGI_C57BL6J_107754 | Neurog1 | NCBI_Gene:18014,ENSEMBL:ENSMUSG00000048904 | MGI:107754 | protein coding gene | neurogenin 1 |
| 13 | gene | 56.288643 | 56.296794 | negative | MGI_C57BL6J_1888514 | Cxcl14 | NCBI_Gene:57266,ENSEMBL:ENSMUSG00000021508 | MGI:1888514 | protein coding gene | chemokine (C-X-C motif) ligand 14 |
| 13 | pseudogene | 56.318325 | 56.320334 | negative | MGI_C57BL6J_5010063 | Gm17878 | NCBI_Gene:100416027,ENSEMBL:ENSMUSG00000114911 | MGI:5010063 | pseudogene | predicted gene, 17878 |
| 13 | gene | 56.337706 | 56.339976 | negative | MGI_C57BL6J_1921654 | 4933404M09Rik | NA | NA | unclassified gene | RIKEN cDNA 4933404M09 gene |
| 13 | gene | 56.342425 | 56.342528 | negative | MGI_C57BL6J_5454493 | Gm24716 | ENSEMBL:ENSMUSG00000088149 | MGI:5454493 | snRNA gene | predicted gene, 24716 |
| 13 | gene | 56.345760 | 56.348847 | positive | MGI_C57BL6J_5591162 | Gm32003 | NCBI_Gene:102634418 | MGI:5591162 | lncRNA gene | predicted gene, 32003 |
| 13 | gene | 56.346559 | 56.357181 | negative | MGI_C57BL6J_5623845 | Gm40960 | NCBI_Gene:105245513 | MGI:5623845 | lncRNA gene | predicted gene, 40960 |
| 13 | gene | 56.362900 | 56.368857 | positive | MGI_C57BL6J_3642506 | Gm10782 | NCBI_Gene:100038494,ENSEMBL:ENSMUSG00000074885 | MGI:3642506 | lncRNA gene | predicted gene 10782 |
| 13 | gene | 56.382708 | 56.423812 | positive | MGI_C57BL6J_5826055 | Gm46418 | NCBI_Gene:108168087 | MGI:5826055 | lncRNA gene | predicted gene, 46418 |
| 13 | gene | 56.425680 | 56.434099 | positive | MGI_C57BL6J_3781223 | Gm3045 | NCBI_Gene:100040926,ENSEMBL:ENSMUSG00000110477 | MGI:3781223 | protein coding gene | predicted gene 3045 |
| 13 | gene | 56.437964 | 56.474239 | positive | MGI_C57BL6J_2145373 | Slc25a48 | NCBI_Gene:328258,ENSEMBL:ENSMUSG00000021509 | MGI:2145373 | protein coding gene | solute carrier family 25, member 48 |
| 13 | gene | 56.479277 | 56.482246 | negative | MGI_C57BL6J_96563 | Il9 | NCBI_Gene:16198,ENSEMBL:ENSMUSG00000021538 | MGI:96563 | protein coding gene | interleukin 9 |
| 13 | gene | 56.522388 | 56.537898 | positive | MGI_C57BL6J_2442921 | Fbxl21 | NCBI_Gene:213311,ENSEMBL:ENSMUSG00000035509 | MGI:2442921 | protein coding gene | F-box and leucine-rich repeat protein 21 |
| 13 | gene | 56.526112 | 56.527002 | negative | MGI_C57BL6J_5621487 | Gm38602 | NCBI_Gene:102642281 | MGI:5621487 | lncRNA gene | predicted gene, 38602 |
| 13 | gene | 56.528959 | 56.548563 | negative | MGI_C57BL6J_1278342 | Lect2 | NCBI_Gene:16841,ENSEMBL:ENSMUSG00000021539 | MGI:1278342 | protein coding gene | leukocyte cell-derived chemotaxin 2 |
| 13 | gene | 56.585690 | 56.591754 | positive | MGI_C57BL6J_5591918 | Gm32759 | NCBI_Gene:102635412 | MGI:5591918 | lncRNA gene | predicted gene, 32759 |
| 13 | gene | 56.595481 | 56.601674 | positive | MGI_C57BL6J_1913797 | 2010203P06Rik | NCBI_Gene:66547,ENSEMBL:ENSMUSG00000114604 | MGI:1913797 | lncRNA gene | RIKEN cDNA 2010203P06 gene |
| 13 | gene | 56.601419 | 56.602563 | positive | MGI_C57BL6J_3642505 | Gm10781 | NA | NA | protein coding gene | predicted gene 10781 |
| 13 | gene | 56.609523 | 56.639562 | positive | MGI_C57BL6J_99959 | Tgfbi | NCBI_Gene:21810,ENSEMBL:ENSMUSG00000035493 | MGI:99959 | protein coding gene | transforming growth factor, beta induced |
| 13 | gene | 56.663677 | 56.665463 | negative | MGI_C57BL6J_5623846 | Gm40961 | NCBI_Gene:105245514 | MGI:5623846 | lncRNA gene | predicted gene, 40961 |
| 13 | gene | 56.671045 | 56.678624 | negative | MGI_C57BL6J_5592124 | Gm32965 | NCBI_Gene:102635693 | MGI:5592124 | lncRNA gene | predicted gene, 32965 |
| 13 | gene | 56.678032 | 56.679772 | positive | MGI_C57BL6J_5592069 | Gm32910 | NCBI_Gene:102635621 | MGI:5592069 | lncRNA gene | predicted gene, 32910 |
| 13 | gene | 56.682604 | 56.690296 | positive | MGI_C57BL6J_5592178 | Gm33019 | NCBI_Gene:102635765 | MGI:5592178 | lncRNA gene | predicted gene, 33019 |
| 13 | gene | 56.699930 | 56.700178 | positive | MGI_C57BL6J_5531080 | Gm27698 | ENSEMBL:ENSMUSG00000098962 | MGI:5531080 | unclassified non-coding RNA gene | predicted gene, 27698 |
| 13 | gene | 56.702990 | 56.742379 | positive | MGI_C57BL6J_1328787 | Smad5 | NCBI_Gene:17129,ENSEMBL:ENSMUSG00000021540 | MGI:1328787 | protein coding gene | SMAD family member 5 |
| 13 | gene | 56.703641 | 56.703767 | positive | MGI_C57BL6J_5530683 | Gm27301 | ENSEMBL:ENSMUSG00000098614 | MGI:5530683 | unclassified non-coding RNA gene | predicted gene, 27301 |
| 13 | gene | 56.703881 | 56.704005 | positive | MGI_C57BL6J_5531216 | Gm27834 | ENSEMBL:ENSMUSG00000098732 | MGI:5531216 | unclassified non-coding RNA gene | predicted gene, 27834 |
| 13 | gene | 56.704030 | 56.704199 | positive | MGI_C57BL6J_5530653 | Gm27271 | ENSEMBL:ENSMUSG00000098328 | MGI:5530653 | unclassified non-coding RNA gene | predicted gene, 27271 |
| 13 | gene | 56.752371 | 56.753827 | negative | MGI_C57BL6J_5791459 | Gm45623 | ENSEMBL:ENSMUSG00000110086 | MGI:5791459 | protein coding gene | predicted gene 45623 |
| 13 | gene | 56.773098 | 56.895993 | negative | MGI_C57BL6J_1349470 | Trpc7 | NCBI_Gene:26946,ENSEMBL:ENSMUSG00000021541 | MGI:1349470 | protein coding gene | transient receptor potential cation channel, subfamily C, member 7 |
| 13 | gene | 56.930820 | 56.930948 | negative | MGI_C57BL6J_5452554 | Gm22777 | ENSEMBL:ENSMUSG00000077279 | MGI:5452554 | snoRNA gene | predicted gene, 22777 |
| 13 | gene | 56.963730 | 56.964660 | positive | MGI_C57BL6J_5623847 | Gm40962 | NCBI_Gene:105245515 | MGI:5623847 | lncRNA gene | predicted gene, 40962 |
| 13 | gene | 56.993712 | 57.003774 | positive | MGI_C57BL6J_5592299 | Gm33140 | NCBI_Gene:102635927 | MGI:5592299 | lncRNA gene | predicted gene, 33140 |
| 13 | pseudogene | 57.239847 | 57.240255 | positive | MGI_C57BL6J_6097550 | Gm48174 | ENSEMBL:ENSMUSG00000114363 | MGI:6097550 | pseudogene | predicted gene, 48174 |
| 13 | gene | 57.242452 | 57.246455 | negative | MGI_C57BL6J_5592356 | Gm33197 | NCBI_Gene:102636002 | MGI:5592356 | lncRNA gene | predicted gene, 33197 |
| 13 | pseudogene | 57.276678 | 57.277128 | positive | MGI_C57BL6J_6097552 | Gm48176 | ENSEMBL:ENSMUSG00000114411 | MGI:6097552 | pseudogene | predicted gene, 48176 |
| 13 | gene | 57.282363 | 57.283266 | positive | MGI_C57BL6J_1921528 | 1700072F12Rik | NCBI_Gene:105245516 | MGI:1921528 | lncRNA gene | RIKEN cDNA 1700072F12 gene |
| 13 | gene | 57.294128 | 57.440352 | positive | MGI_C57BL6J_5592453 | Gm33294 | NCBI_Gene:102636141 | MGI:5592453 | lncRNA gene | predicted gene, 33294 |
| 13 | gene | 57.421195 | 57.908332 | negative | MGI_C57BL6J_105371 | Spock1 | NCBI_Gene:20745,ENSEMBL:ENSMUSG00000056222 | MGI:105371 | protein coding gene | sparc/osteonectin, cwcv and kazal-like domains proteoglycan 1 |
| 13 | gene | 57.445000 | 57.450938 | positive | MGI_C57BL6J_5592399 | Gm33240 | NCBI_Gene:102636067 | MGI:5592399 | lncRNA gene | predicted gene, 33240 |
| 13 | gene | 57.644150 | 57.645380 | negative | MGI_C57BL6J_1920284 | 2900064B16Rik | NA | NA | unclassified gene | RIKEN cDNA 2900064B16 gene |
| 13 | gene | 58.000228 | 58.123479 | negative | MGI_C57BL6J_2445185 | Klhl3 | NCBI_Gene:100503085,ENSEMBL:ENSMUSG00000014164 | MGI:2445185 | protein coding gene | kelch-like 3 |
| 13 | gene | 58.023125 | 58.023200 | negative | MGI_C57BL6J_3718564 | Mir874 | miRBase:MI0005479,NCBI_Gene:100124491,ENSEMBL:ENSMUSG00000077962 | MGI:3718564 | miRNA gene | microRNA 874 |
| 13 | gene | 58.071690 | 58.113135 | positive | MGI_C57BL6J_5592548 | Gm33389 | NCBI_Gene:102636277 | MGI:5592548 | lncRNA gene | predicted gene, 33389 |
| 13 | gene | 58.098288 | 58.099340 | negative | MGI_C57BL6J_1925965 | D530015H24Rik | NA | NA | unclassified gene | RIKEN cDNA D530015H24 gene |
| 13 | gene | 58.125879 | 58.128556 | negative | MGI_C57BL6J_1924384 | Hnrnpa0 | NCBI_Gene:77134,ENSEMBL:ENSMUSG00000007836 | MGI:1924384 | protein coding gene | heterogeneous nuclear ribonucleoprotein A0 |
| 13 | gene | 58.130752 | 58.140093 | positive | MGI_C57BL6J_5623849 | Gm40964 | NCBI_Gene:105245518 | MGI:5623849 | lncRNA gene | predicted gene, 40964 |
| 13 | gene | 58.146949 | 58.149692 | positive | MGI_C57BL6J_1914589 | 1700066J03Rik | NCBI_Gene:67339,ENSEMBL:ENSMUSG00000097717 | MGI:1914589 | lncRNA gene | RIKEN cDNA 1700066J03 gene |
| 13 | gene | 58.155141 | 58.157575 | negative | MGI_C57BL6J_6097735 | Gm48295 | ENSEMBL:ENSMUSG00000114309 | MGI:6097735 | lncRNA gene | predicted gene, 48295 |
| 13 | gene | 58.157649 | 58.164693 | positive | MGI_C57BL6J_1922981 | Idnk | NCBI_Gene:75731,ENSEMBL:ENSMUSG00000050002 | MGI:1922981 | protein coding gene | idnK gluconokinase homolog (E. coli) |
| 13 | gene | 58.176156 | 58.215653 | negative | MGI_C57BL6J_1860276 | Ubqln1 | NCBI_Gene:56085,ENSEMBL:ENSMUSG00000005312 | MGI:1860276 | protein coding gene | ubiquilin 1 |
| 13 | gene | 58.184896 | 58.191318 | negative | MGI_C57BL6J_6097824 | Gm48357 | ENSEMBL:ENSMUSG00000114524 | MGI:6097824 | lncRNA gene | predicted gene, 48357 |
| 13 | pseudogene | 58.218719 | 58.219276 | positive | MGI_C57BL6J_6097934 | Gm48432 | ENSEMBL:ENSMUSG00000114886 | MGI:6097934 | pseudogene | predicted gene, 48432 |
| 13 | gene | 58.232143 | 58.236750 | positive | MGI_C57BL6J_6097949 | Gm48441 | ENSEMBL:ENSMUSG00000114929 | MGI:6097949 | lncRNA gene | predicted gene, 48441 |
| 13 | gene | 58.233346 | 58.275699 | negative | MGI_C57BL6J_1891694 | Gkap1 | NCBI_Gene:56278,ENSEMBL:ENSMUSG00000021552 | MGI:1891694 | protein coding gene | G kinase anchoring protein 1 |
| 13 | gene | 58.274899 | 58.311362 | positive | MGI_C57BL6J_5477049 | Gm26555 | NCBI_Gene:102636582,ENSEMBL:ENSMUSG00000097399 | MGI:5477049 | lncRNA gene | predicted gene, 26555 |
| 13 | gene | 58.284727 | 58.364746 | negative | MGI_C57BL6J_1922300 | Kif27 | NCBI_Gene:75050,ENSEMBL:ENSMUSG00000060176 | MGI:1922300 | protein coding gene | kinesin family member 27 |
| 13 | gene | 58.313044 | 58.313149 | negative | MGI_C57BL6J_5530997 | Mir6369 | miRBase:MI0021898,NCBI_Gene:102465191,ENSEMBL:ENSMUSG00000098258 | MGI:5530997 | miRNA gene | microRNA 6369 |
| 13 | gene | 58.325267 | 58.325408 | positive | MGI_C57BL6J_5454061 | Gm24284 | ENSEMBL:ENSMUSG00000077428 | MGI:5454061 | snoRNA gene | predicted gene, 24284 |
| 13 | gene | 58.379817 | 58.385225 | negative | MGI_C57BL6J_1917403 | 2210016F16Rik | NCBI_Gene:70153,ENSEMBL:ENSMUSG00000021550 | MGI:1917403 | protein coding gene | RIKEN cDNA 2210016F16 gene |
| 13 | gene | 58.391132 | 58.403343 | negative | MGI_C57BL6J_99894 | Hnrnpk | NCBI_Gene:15387,ENSEMBL:ENSMUSG00000021546 | MGI:99894 | protein coding gene | heterogeneous nuclear ribonucleoprotein K |
| 13 | gene | 58.392779 | 58.392886 | negative | MGI_C57BL6J_3619436 | Mir7-1 | miRBase:MI0000728,NCBI_Gene:723902,ENSEMBL:ENSMUSG00000065434 | MGI:3619436 | miRNA gene | microRNA 7-1 |
| 13 | gene | 58.402248 | 58.411149 | positive | MGI_C57BL6J_1921636 | Rmi1 | NCBI_Gene:74386,ENSEMBL:ENSMUSG00000035367 | MGI:1921636 | protein coding gene | RecQ mediated genome instability 1 |
| 13 | gene | 58.443021 | 58.444580 | positive | MGI_C57BL6J_6097170 | Gm47918 | ENSEMBL:ENSMUSG00000114654 | MGI:6097170 | lncRNA gene | predicted gene, 47918 |
| 13 | gene | 58.443406 | 58.451773 | negative | MGI_C57BL6J_5593034 | Gm33875 | NCBI_Gene:102636942 | MGI:5593034 | lncRNA gene | predicted gene, 33875 |
| 13 | gene | 58.470620 | 58.477449 | negative | MGI_C57BL6J_5623851 | Gm40966 | NCBI_Gene:105245520 | MGI:5623851 | lncRNA gene | predicted gene, 40966 |
| 13 | gene | 58.542808 | 58.546790 | negative | MGI_C57BL6J_5621379 | Gm38494 | NCBI_Gene:102637097 | MGI:5621379 | lncRNA gene | predicted gene, 38494 |
| 13 | gene | 58.545399 | 58.610877 | negative | MGI_C57BL6J_2137361 | Slc28a3 | NCBI_Gene:114304,ENSEMBL:ENSMUSG00000021553 | MGI:2137361 | protein coding gene | solute carrier family 28 (sodium-coupled nucleoside transporter), member 3 |
| 13 | pseudogene | 58.546028 | 58.546502 | positive | MGI_C57BL6J_3781308 | Gm3131 | NCBI_Gene:100041086,ENSEMBL:ENSMUSG00000114234 | MGI:3781308 | pseudogene | predicted gene 3131 |
| 13 | pseudogene | 58.668624 | 58.668839 | negative | MGI_C57BL6J_6097208 | Gm47942 | ENSEMBL:ENSMUSG00000114380 | MGI:6097208 | pseudogene | predicted gene, 47942 |
| 13 | gene | 58.683672 | 58.688405 | negative | MGI_C57BL6J_5593217 | Gm34058 | NCBI_Gene:102637179 | MGI:5593217 | lncRNA gene | predicted gene, 34058 |
| 13 | gene | 58.736408 | 58.762360 | negative | MGI_C57BL6J_1914835 | 4930455J16Rik | NCBI_Gene:67585,ENSEMBL:ENSMUSG00000114384 | MGI:1914835 | lncRNA gene | RIKEN cDNA 4930455J16 gene |
| 13 | pseudogene | 58.756511 | 58.757204 | positive | MGI_C57BL6J_3645614 | Rpl13-ps2 | NCBI_Gene:435361,ENSEMBL:ENSMUSG00000114325 | MGI:3645614 | pseudogene | ribosomal protein L13, pseudogene 2 |
| 13 | gene | 58.762385 | 58.776543 | positive | MGI_C57BL6J_6097215 | Gm47947 | ENSEMBL:ENSMUSG00000114808 | MGI:6097215 | lncRNA gene | predicted gene, 47947 |
| 13 | gene | 58.775458 | 58.797450 | negative | MGI_C57BL6J_5623853 | Gm40968 | NCBI_Gene:105245522,ENSEMBL:ENSMUSG00000114231 | MGI:5623853 | lncRNA gene | predicted gene, 40968 |
| 13 | gene | 58.806569 | 59.133970 | positive | MGI_C57BL6J_97384 | Ntrk2 | NCBI_Gene:18212,ENSEMBL:ENSMUSG00000055254 | MGI:97384 | protein coding gene | neurotrophic tyrosine kinase, receptor, type 2 |
| 13 | gene | 58.812153 | 58.814363 | positive | MGI_C57BL6J_1925962 | C630044B11Rik | ENSEMBL:ENSMUSG00000114448 | MGI:1925962 | unclassified gene | RIKEN cDNA C630044B11 gene |
| 13 | gene | 58.824218 | 58.826994 | negative | MGI_C57BL6J_6096364 | Gm47423 | ENSEMBL:ENSMUSG00000114218 | MGI:6096364 | lncRNA gene | predicted gene, 47423 |
| 13 | gene | 59.054896 | 59.084093 | negative | MGI_C57BL6J_1921130 | 4930415C11Rik | ENSEMBL:ENSMUSG00000114437 | MGI:1921130 | lncRNA gene | RIKEN cDNA 4930415C11 gene |
| 13 | gene | 59.065802 | 59.083249 | negative | MGI_C57BL6J_5593404 | Gm34245 | NCBI_Gene:102637438,ENSEMBL:ENSMUSG00000114896 | MGI:5593404 | lncRNA gene | predicted gene, 34245 |
| 13 | gene | 59.144257 | 59.145855 | negative | MGI_C57BL6J_5593466 | Gm34307 | NCBI_Gene:102637520,ENSEMBL:ENSMUSG00000114208 | MGI:5593466 | lncRNA gene | predicted gene, 34307 |
| 13 | gene | 59.145873 | 59.204327 | positive | MGI_C57BL6J_5593513 | Gm34354 | NCBI_Gene:102637583,ENSEMBL:ENSMUSG00000114854 | MGI:5593513 | lncRNA gene | predicted gene, 34354 |
| 13 | gene | 59.170633 | 59.182816 | negative | MGI_C57BL6J_6096266 | Gm47359 | ENSEMBL:ENSMUSG00000114393 | MGI:6096266 | lncRNA gene | predicted gene, 47359 |
| 13 | gene | 59.188016 | 59.217172 | negative | MGI_C57BL6J_5593717 | Gm34558 | NCBI_Gene:102637852,ENSEMBL:ENSMUSG00000114478 | MGI:5593717 | lncRNA gene | predicted gene, 34558 |
| 13 | gene | 59.235750 | 59.255722 | negative | MGI_C57BL6J_5593831 | Gm34672 | NCBI_Gene:102637997,ENSEMBL:ENSMUSG00000114743 | MGI:5593831 | lncRNA gene | predicted gene, 34672 |
| 13 | gene | 59.294819 | 59.330243 | negative | MGI_C57BL6J_6096268 | Gm47360 | ENSEMBL:ENSMUSG00000114240 | MGI:6096268 | lncRNA gene | predicted gene, 47360 |
| 13 | gene | 59.405486 | 59.409301 | negative | MGI_C57BL6J_5593880 | Gm34721 | NCBI_Gene:102638071,ENSEMBL:ENSMUSG00000114200 | MGI:5593880 | lncRNA gene | predicted gene, 34721 |
| 13 | gene | 59.411618 | 59.419254 | negative | MGI_C57BL6J_5593947 | Gm34788 | NCBI_Gene:102638158,ENSEMBL:ENSMUSG00000114623 | MGI:5593947 | lncRNA gene | predicted gene, 34788 |
| 13 | gene | 59.419108 | 59.423161 | positive | MGI_C57BL6J_1926168 | 4930455M05Rik | ENSEMBL:ENSMUSG00000114787 | MGI:1926168 | lncRNA gene | RIKEN cDNA 4930455M05 gene |
| 13 | gene | 59.445742 | 59.585227 | negative | MGI_C57BL6J_2159437 | Agtpbp1 | NCBI_Gene:67269,ENSEMBL:ENSMUSG00000021557 | MGI:2159437 | protein coding gene | ATP/GTP binding protein 1 |
| 13 | gene | 59.557566 | 59.558337 | positive | MGI_C57BL6J_1925051 | A930015P04Rik | ENSEMBL:ENSMUSG00000114526 | MGI:1925051 | unclassified gene | RIKEN cDNA A930015P04 gene |
| 13 | gene | 59.580176 | 59.585223 | negative | MGI_C57BL6J_2442089 | A230056J06Rik | NCBI_Gene:319469 | MGI:2442089 | lncRNA gene | RIKEN cDNA A230056J06 gene |
| 13 | gene | 59.585259 | 59.635922 | positive | MGI_C57BL6J_1925939 | Naa35 | NCBI_Gene:78689,ENSEMBL:ENSMUSG00000021555 | MGI:1925939 | protein coding gene | N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| 13 | gene | 59.603309 | 59.605662 | positive | MGI_C57BL6J_6096070 | Gm47246 | ENSEMBL:ENSMUSG00000114535 | MGI:6096070 | unclassified gene | predicted gene, 47246 |
| 13 | gene | 59.634626 | 59.675811 | negative | MGI_C57BL6J_1917329 | Golm1 | NCBI_Gene:105348,ENSEMBL:ENSMUSG00000021556 | MGI:1917329 | protein coding gene | golgi membrane protein 1 |
| 13 | gene | 59.687402 | 59.694108 | negative | MGI_C57BL6J_1918150 | 4921517D22Rik | NCBI_Gene:70900,ENSEMBL:ENSMUSG00000049902 | MGI:1918150 | protein coding gene | RIKEN cDNA 4921517D22 gene |
| 13 | pseudogene | 59.688371 | 59.688791 | positive | MGI_C57BL6J_6096106 | Gm47269 | ENSEMBL:ENSMUSG00000114727 | MGI:6096106 | pseudogene | predicted gene, 47269 |
| 13 | gene | 59.699806 | 59.710330 | negative | MGI_C57BL6J_1919469 | Spata31d1a | NCBI_Gene:72219,ENSEMBL:ENSMUSG00000050876 | MGI:1919469 | protein coding gene | spermatogenesis associated 31 subfamily D, member 1A |
| 13 | gene | 59.706687 | 59.711214 | negative | MGI_C57BL6J_5623857 | Gm40972 | NCBI_Gene:105245526 | MGI:5623857 | lncRNA gene | predicted gene, 40972 |
| 13 | gene | 59.712284 | 59.719295 | positive | MGI_C57BL6J_3646080 | Spata31d1b | NCBI_Gene:238662,ENSEMBL:ENSMUSG00000091311 | MGI:3646080 | protein coding gene | spermatogenesis associated 31 subfamily D, member 1B |
| 13 | gene | 59.725925 | 59.731752 | negative | MGI_C57BL6J_3045260 | Spata31d1d | NCBI_Gene:238663,ENSEMBL:ENSMUSG00000043986 | MGI:3045260 | protein coding gene | spermatogenesis associated 31 subfamily D, member 1D |
| 13 | gene | 59.726724 | 59.751235 | positive | MGI_C57BL6J_5826056 | Gm46419 | NCBI_Gene:108168088,ENSEMBL:ENSMUSG00000114903 | MGI:5826056 | lncRNA gene | predicted gene, 46419 |
| 13 | gene | 59.733630 | 59.739185 | positive | MGI_C57BL6J_1922473 | 4930528D03Rik | NCBI_Gene:75223,ENSEMBL:ENSMUSG00000097502 | MGI:1922473 | lncRNA gene | RIKEN cDNA 4930528D03 gene |
| 13 | gene | 59.740269 | 59.747800 | negative | MGI_C57BL6J_1921474 | 1700014D04Rik | NCBI_Gene:102638268,ENSEMBL:ENSMUSG00000051054 | MGI:1921474 | protein coding gene | RIKEN cDNA 1700014D04 gene |
| 13 | gene | 59.755409 | 59.769810 | negative | MGI_C57BL6J_1916296 | Isca1 | NCBI_Gene:69046,ENSEMBL:ENSMUSG00000044792 | MGI:1916296 | protein coding gene | iron-sulfur cluster assembly 1 |
| 13 | gene | 59.758935 | 59.782245 | negative | MGI_C57BL6J_6121558 | Gm49354 | ENSEMBL:ENSMUSG00000114763 | MGI:6121558 | protein coding gene | predicted gene, 49354 |
| 13 | gene | 59.769497 | 59.775407 | positive | MGI_C57BL6J_1261417 | Etohd2 | NCBI_Gene:13996,ENSEMBL:ENSMUSG00000089875 | MGI:1261417 | lncRNA gene | ethanol decreased 2 |
| 13 | gene | 59.771561 | 59.823169 | negative | MGI_C57BL6J_2387179 | Tut7 | NCBI_Gene:214290,ENSEMBL:ENSMUSG00000035248 | MGI:2387179 | protein coding gene | terminal uridylyl transferase 7 |
| 13 | gene | 59.781062 | 59.781578 | negative | MGI_C57BL6J_6097578 | Gm48194 | ENSEMBL:ENSMUSG00000114835 | MGI:6097578 | lncRNA gene | predicted gene, 48194 |
| 13 | gene | 59.823266 | 59.833239 | positive | MGI_C57BL6J_5594120 | Gm34961 | NCBI_Gene:102638381,ENSEMBL:ENSMUSG00000114354 | MGI:5594120 | lncRNA gene | predicted gene, 34961 |
| 13 | gene | 59.873642 | 59.874964 | positive | MGI_C57BL6J_6097865 | Gm48384 | ENSEMBL:ENSMUSG00000114669 | MGI:6097865 | lncRNA gene | predicted gene, 48384 |
| 13 | gene | 59.885642 | 59.915925 | negative | MGI_C57BL6J_5012051 | Gm19866 | NCBI_Gene:100503743,ENSEMBL:ENSMUSG00000114490 | MGI:5012051 | lncRNA gene | predicted gene, 19866 |
| 13 | pseudogene | 59.919438 | 59.920340 | positive | MGI_C57BL6J_6097873 | Gm48390 | ENSEMBL:ENSMUSG00000114308 | MGI:6097873 | pseudogene | predicted gene, 48390 |
| 13 | gene | 59.990668 | 59.998541 | positive | MGI_C57BL6J_5594302 | Gm35143 | NCBI_Gene:102638620 | MGI:5594302 | lncRNA gene | predicted gene, 35143 |
| 13 | gene | 60.015570 | 60.016153 | positive | MGI_C57BL6J_2145492 | C77681 | NA | NA | unclassified gene | expressed sequence C77681 |
| 13 | gene | 60.029711 | 60.116669 | positive | MGI_C57BL6J_5439407 | A530065N20Rik | NCBI_Gene:328263,ENSEMBL:ENSMUSG00000114531 | MGI:5439407 | lncRNA gene | RIKEN cDNA A530046M15 gene |
| 13 | gene | 60.041125 | 60.044998 | negative | MGI_C57BL6J_5594352 | Gm35193 | NCBI_Gene:102638692 | MGI:5594352 | lncRNA gene | predicted gene, 35193 |
| 13 | gene | 60.053684 | 60.057625 | negative | MGI_C57BL6J_6097881 | Gm48396 | ENSEMBL:ENSMUSG00000114795 | MGI:6097881 | lncRNA gene | predicted gene, 48396 |
| 13 | gene | 60.068641 | 60.074017 | positive | MGI_C57BL6J_6097883 | Gm48397 | ENSEMBL:ENSMUSG00000114734 | MGI:6097883 | lncRNA gene | predicted gene, 48397 |
| 13 | gene | 60.114526 | 60.126416 | negative | MGI_C57BL6J_5826057 | Gm46420 | NCBI_Gene:108168089 | MGI:5826057 | lncRNA gene | predicted gene, 46420 |
| 13 | gene | 60.174405 | 60.177535 | negative | MGI_C57BL6J_95655 | Gas1 | NCBI_Gene:14451,ENSEMBL:ENSMUSG00000052957 | MGI:95655 | protein coding gene | growth arrest specific 1 |
| 13 | gene | 60.176682 | 60.182862 | positive | MGI_C57BL6J_6098010 | Gm48488 | ENSEMBL:ENSMUSG00000114433 | MGI:6098010 | lncRNA gene | predicted gene, 48488 |
| 13 | gene | 60.205471 | 60.216023 | positive | MGI_C57BL6J_3647835 | Gm5084 | NCBI_Gene:100503759,ENSEMBL:ENSMUSG00000050765 | MGI:3647835 | lncRNA gene | predicted gene 5084 |
| 13 | pseudogene | 60.248566 | 60.249006 | positive | MGI_C57BL6J_5011434 | Gm19249 | NCBI_Gene:100462933 | MGI:5011434 | pseudogene | predicted gene, 19249 |
| 13 | gene | 60.293672 | 60.293772 | positive | MGI_C57BL6J_5454776 | Gm24999 | ENSEMBL:ENSMUSG00000080668 | MGI:5454776 | miRNA gene | predicted gene, 24999 |
| 13 | gene | 60.331449 | 60.333751 | positive | MGI_C57BL6J_3642421 | Gm10779 | ENSEMBL:ENSMUSG00000114460 | MGI:3642421 | protein coding gene | predicted gene 10779 |
| 13 | gene | 60.422754 | 60.434983 | negative | MGI_C57BL6J_5594492 | Gm35333 | NCBI_Gene:102638874,ENSEMBL:ENSMUSG00000114274 | MGI:5594492 | lncRNA gene | predicted gene, 35333 |
| 13 | gene | 60.463586 | 60.481298 | negative | MGI_C57BL6J_6098026 | Gm48500 | ENSEMBL:ENSMUSG00000114566 | MGI:6098026 | lncRNA gene | predicted gene, 48500 |
| 13 | gene | 60.483504 | 60.508840 | negative | MGI_C57BL6J_3045361 | A530001N23Rik | NCBI_Gene:328265,ENSEMBL:ENSMUSG00000114452 | MGI:3045361 | lncRNA gene | RIKEN cDNA A530001N23 gene |
| 13 | gene | 60.529296 | 60.529409 | negative | MGI_C57BL6J_5454777 | Gm25000 | ENSEMBL:ENSMUSG00000080666 | MGI:5454777 | miRNA gene | predicted gene, 25000 |
| 13 | gene | 60.601110 | 60.602485 | negative | MGI_C57BL6J_5623859 | Gm40974 | NCBI_Gene:105245529 | MGI:5623859 | lncRNA gene | predicted gene, 40974 |
| 13 | gene | 60.601947 | 60.763191 | positive | MGI_C57BL6J_1916885 | Dapk1 | NCBI_Gene:69635,ENSEMBL:ENSMUSG00000021559 | MGI:1916885 | protein coding gene | death associated protein kinase 1 |
| 13 | gene | 60.622967 | 60.629395 | positive | MGI_C57BL6J_6098150 | Gm48582 | ENSEMBL:ENSMUSG00000114501 | MGI:6098150 | unclassified gene | predicted gene, 48582 |
| 13 | pseudogene | 60.641790 | 60.642009 | positive | MGI_C57BL6J_6098152 | Gm48583 | ENSEMBL:ENSMUSG00000114277 | MGI:6098152 | pseudogene | predicted gene, 48583 |
| 13 | gene | 60.685966 | 60.688276 | positive | MGI_C57BL6J_5594699 | Gm35540 | NCBI_Gene:102639165 | MGI:5594699 | lncRNA gene | predicted gene, 35540 |
| 13 | pseudogene | 60.771675 | 60.772163 | positive | MGI_C57BL6J_3642243 | Gm10312 | ENSEMBL:ENSMUSG00000096874 | MGI:3642243 | pseudogene | predicted gene 10312 |
| 13 | gene | 60.781887 | 60.786364 | negative | MGI_C57BL6J_2682300 | BC051665 | NCBI_Gene:218275,ENSEMBL:ENSMUSG00000042243 | MGI:2682300 | protein coding gene | cDNA sequence BC051665 |
| 13 | gene | 60.798250 | 60.802849 | negative | MGI_C57BL6J_1917452 | Ctsll3 | NCBI_Gene:70202,ENSEMBL:ENSMUSG00000056728 | MGI:1917452 | protein coding gene | cathepsin L-like 3 |
| 13 | gene | 60.842612 | 60.864475 | negative | MGI_C57BL6J_1922258 | 4930486L24Rik | NCBI_Gene:214639,ENSEMBL:ENSMUSG00000050345 | MGI:1922258 | protein coding gene | RIKEN cDNA 4930486L24 gene |
| 13 | gene | 60.842621 | 60.896894 | negative | MGI_C57BL6J_6121621 | Gm49392 | ENSEMBL:ENSMUSG00000114635 | MGI:6121621 | protein coding gene | predicted gene, 49392 |
| 13 | gene | 60.853230 | 60.935590 | negative | MGI_C57BL6J_6121623 | Gm49393 | ENSEMBL:ENSMUSG00000114487 | MGI:6121623 | protein coding gene | predicted gene, 49393 |
| 13 | gene | 60.895350 | 60.897934 | negative | MGI_C57BL6J_88555 | Ctla2b | NCBI_Gene:13025,ENSEMBL:ENSMUSG00000074874 | MGI:88555 | protein coding gene | cytotoxic T lymphocyte-associated protein 2 beta |
| 13 | gene | 60.895350 | 60.936571 | negative | MGI_C57BL6J_6121619 | Gm49391 | ENSEMBL:ENSMUSG00000114432 | MGI:6121619 | protein coding gene | predicted gene, 49391 |
| 13 | gene | 60.901188 | 60.904880 | negative | MGI_C57BL6J_2151721 | Tpbpb | NCBI_Gene:116913,ENSEMBL:ENSMUSG00000062705 | MGI:2151721 | protein coding gene | trophoblast specific protein beta |
| 13 | gene | 60.928086 | 60.928184 | positive | MGI_C57BL6J_5454371 | Gm24594 | ENSEMBL:ENSMUSG00000084451 | MGI:5454371 | snRNA gene | predicted gene, 24594 |
| 13 | gene | 60.934155 | 60.936625 | negative | MGI_C57BL6J_88554 | Ctla2a | NCBI_Gene:13024,ENSEMBL:ENSMUSG00000044258 | MGI:88554 | protein coding gene | cytotoxic T lymphocyte-associated protein 2 alpha |
| 13 | gene | 60.938492 | 60.941984 | negative | MGI_C57BL6J_98795 | Tpbpa | NCBI_Gene:21984,ENSEMBL:ENSMUSG00000033834 | MGI:98795 | protein coding gene | trophoblast specific protein alpha |
| 13 | gene | 60.943621 | 60.962931 | negative | MGI_C57BL6J_5623861 | Gm40976 | NCBI_Gene:105245531,ENSEMBL:ENSMUSG00000114663 | MGI:5623861 | lncRNA gene | predicted gene, 40976 |
| 13 | gene | 60.954099 | 60.959948 | positive | MGI_C57BL6J_5623860 | Gm40975 | NCBI_Gene:105245530,ENSEMBL:ENSMUSG00000114502 | MGI:5623860 | lncRNA gene | predicted gene, 40975 |
| 13 | gene | 61.000179 | 61.005959 | negative | MGI_C57BL6J_1349426 | Ctsj | NCBI_Gene:26898,ENSEMBL:ENSMUSG00000055298 | MGI:1349426 | protein coding gene | cathepsin J |
| 13 | gene | 61.035024 | 61.040631 | negative | MGI_C57BL6J_2137385 | Ctsq | NCBI_Gene:104002,ENSEMBL:ENSMUSG00000021439 | MGI:2137385 | protein coding gene | cathepsin Q |
| 13 | pseudogene | 61.146195 | 61.149807 | negative | MGI_C57BL6J_5010249 | Gm18064 | NCBI_Gene:100416353,ENSEMBL:ENSMUSG00000114643 | MGI:5010249 | pseudogene | predicted gene, 18064 |
| 13 | gene | 61.159204 | 61.164256 | negative | MGI_C57BL6J_1861723 | Ctsr | NCBI_Gene:56835,ENSEMBL:ENSMUSG00000055679 | MGI:1861723 | protein coding gene | cathepsin R |
| 13 | gene | 61.195132 | 61.203410 | negative | MGI_C57BL6J_1889619 | Cts6 | NCBI_Gene:58518,ENSEMBL:ENSMUSG00000021441 | MGI:1889619 | protein coding gene | cathepsin 6 |
| 13 | gene | 61.246745 | 61.255358 | negative | MGI_C57BL6J_1860275 | Cts8 | NCBI_Gene:56094,ENSEMBL:ENSMUSG00000057446 | MGI:1860275 | protein coding gene | cathepsin 8 |
| 13 | gene | 61.247878 | 61.288839 | negative | MGI_C57BL6J_6121633 | Gm49398 | ENSEMBL:ENSMUSG00000114473 | MGI:6121633 | protein coding gene | predicted gene, 49398 |
| 13 | pseudogene | 61.281380 | 61.288850 | negative | MGI_C57BL6J_2151708 | Cts8-ps | NCBI_Gene:116911,ENSEMBL:ENSMUSG00000114867 | MGI:2151708 | pseudogene | cathepsin 8, pseudogene |
| 13 | pseudogene | 61.298206 | 61.305727 | negative | MGI_C57BL6J_3645615 | Gm5664 | NCBI_Gene:435362,ENSEMBL:ENSMUSG00000114495 | MGI:3645615 | pseudogene | predicted gene 5664 |
| 13 | gene | 61.336455 | 61.336561 | positive | MGI_C57BL6J_5531086 | Gm27704 | ENSEMBL:ENSMUSG00000098860 | MGI:5531086 | miRNA gene | predicted gene, 27704 |
| 13 | gene | 61.352461 | 61.358290 | negative | MGI_C57BL6J_1860262 | Cts7 | NCBI_Gene:56092,ENSEMBL:ENSMUSG00000021440 | MGI:1860262 | protein coding gene | cathepsin 7 |
| 13 | pseudogene | 61.435838 | 61.439870 | negative | MGI_C57BL6J_2151716 | Cts7-ps | NCBI_Gene:116909,ENSEMBL:ENSMUSG00000114802 | MGI:2151716 | pseudogene | cathepsin 7, pseudogene |
| 13 | pseudogene | 61.443965 | 61.444195 | positive | MGI_C57BL6J_6097019 | Gm47825 | ENSEMBL:ENSMUSG00000114405 | MGI:6097019 | pseudogene | predicted gene, 47825 |
| 13 | pseudogene | 61.482408 | 61.490329 | negative | MGI_C57BL6J_3643776 | Ctsm-ps2 | NCBI_Gene:624056,ENSEMBL:ENSMUSG00000114667 | MGI:3643776 | pseudogene | cathepsin M, pseudogene 2 |
| 13 | pseudogene | 61.510288 | 61.511518 | positive | MGI_C57BL6J_6097054 | Gm47848 | ENSEMBL:ENSMUSG00000114928 | MGI:6097054 | pseudogene | predicted gene, 47848 |
| 13 | pseudogene | 61.531840 | 61.532673 | negative | MGI_C57BL6J_3779760 | Gm7756 | NCBI_Gene:665717 | MGI:3779760 | pseudogene | predicted gene 7756 |
| 13 | gene | 61.535739 | 61.541881 | negative | MGI_C57BL6J_1927229 | Ctsm | NCBI_Gene:64139,ENSEMBL:ENSMUSG00000074871 | MGI:1927229 | protein coding gene | cathepsin M |
| 13 | gene | 61.538390 | 61.568174 | negative | MGI_C57BL6J_6121554 | Gm49352 | ENSEMBL:ENSMUSG00000114904 | MGI:6121554 | protein coding gene | predicted gene, 49352 |
| 13 | gene | 61.564630 | 61.570163 | negative | MGI_C57BL6J_2151929 | Cts3 | NCBI_Gene:117066,ENSEMBL:ENSMUSG00000074870 | MGI:2151929 | protein coding gene | cathepsin 3 |
| 13 | gene | 61.630023 | 61.636282 | positive | MGI_C57BL6J_5439384 | Gm21933 | NCBI_Gene:108168055 | MGI:5439384 | protein coding gene | predicted gene, 21933 |
| 13 | gene | 61.655303 | 61.743770 | positive | MGI_C57BL6J_6121564 | Gm49357 | ENSEMBL:ENSMUSG00000113295 | MGI:6121564 | lncRNA gene | predicted gene, 49357 |
| 13 | pseudogene | 61.655310 | 61.700833 | positive | MGI_C57BL6J_5295705 | Gm20599 | NCBI_Gene:100503368 | MGI:5295705 | pseudogene | predicted gene, 20599 |
| 13 | pseudogene | 61.665886 | 61.667200 | positive | MGI_C57BL6J_3647115 | Gm7622 | NCBI_Gene:665401,ENSEMBL:ENSMUSG00000113728 | MGI:3647115 | pseudogene | predicted pseudogene 7622 |
| 13 | pseudogene | 61.687135 | 61.688009 | negative | MGI_C57BL6J_3648182 | Gm6474 | NCBI_Gene:624140,ENSEMBL:ENSMUSG00000114023 | MGI:3648182 | pseudogene | predicted gene 6474 |
| 13 | gene | 61.709556 | 61.711238 | positive | MGI_C57BL6J_6097471 | Gm48116 | ENSEMBL:ENSMUSG00000113471 | MGI:6097471 | lncRNA gene | predicted gene, 48116 |
| 13 | pseudogene | 61.710582 | 61.712979 | negative | MGI_C57BL6J_3644667 | Gm6436 | NCBI_Gene:623505,ENSEMBL:ENSMUSG00000061330 | MGI:3644667 | pseudogene | predicted gene 6436 |
| 13 | pseudogene | 61.712995 | 61.746500 | positive | MGI_C57BL6J_3780004 | Gm9595 | NCBI_Gene:100861651,ENSEMBL:ENSMUSG00000113899 | MGI:3780004 | pseudogene | predicted gene 9595 |
| 13 | pseudogene | 61.771580 | 61.787282 | positive | MGI_C57BL6J_3583898 | BC052688 | NCBI_Gene:432812 | MGI:3583898 | pseudogene | cDNA sequence BC052688 |
| 13 | pseudogene | 61.799816 | 61.800543 | positive | MGI_C57BL6J_5012146 | Selenok-ps4 | NCBI_Gene:100503914,ENSEMBL:ENSMUSG00000095905 | MGI:5012146 | pseudogene | selenoprotein K, pseudogene 4 |
| 13 | gene | 61.804141 | 61.804248 | negative | MGI_C57BL6J_5453516 | Gm23739 | ENSEMBL:ENSMUSG00000096212 | MGI:5453516 | miRNA gene | predicted gene, 23739 |
| 13 | pseudogene | 61.804257 | 61.916728 | positive | MGI_C57BL6J_3643235 | Gm7240 | NCBI_Gene:638502,ENSEMBL:ENSMUSG00000113626 | MGI:3643235 | pseudogene | predicted gene 7240 |
| 13 | gene | 61.804312 | 61.826524 | positive | MGI_C57BL6J_1917252 | 1700028E11Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 1700028E11 gene |
| 13 | gene | 61.823544 | 61.823664 | negative | MGI_C57BL6J_5453970 | Gm24193 | ENSEMBL:ENSMUSG00000080451 | MGI:5453970 | miRNA gene | predicted gene, 24193 |
| 13 | pseudogene | 61.915959 | 61.918672 | positive | MGI_C57BL6J_5594778 | Gm35619 | NCBI_Gene:102639269,ENSEMBL:ENSMUSG00000113887 | MGI:5594778 | pseudogene | predicted gene, 35619 |
| 13 | pseudogene | 61.984004 | 61.993289 | positive | MGI_C57BL6J_5439386 | Gm21935 | NCBI_Gene:100861666 | MGI:5439386 | pseudogene | predicted gene, 21935 |
| 13 | pseudogene | 62.006581 | 62.007311 | positive | MGI_C57BL6J_5295670 | Selenok-ps8 | NCBI_Gene:100046282,ENSEMBL:ENSMUSG00000113014 | MGI:5295670 | pseudogene | selenoprotein K, pseudogene 8 |
| 13 | gene | 62.006640 | 62.126102 | negative | MGI_C57BL6J_6097631 | Gm48228 | ENSEMBL:ENSMUSG00000114036 | MGI:6097631 | lncRNA gene | predicted gene, 48228 |
| 13 | gene | 62.013035 | 62.013144 | negative | MGI_C57BL6J_5455426 | Gm25649 | ENSEMBL:ENSMUSG00000094096 | MGI:5455426 | miRNA gene | predicted gene, 25649 |
| 13 | gene | 62.015605 | 62.053940 | positive | MGI_C57BL6J_3781503 | Gm3325 | NCBI_Gene:100041420,ENSEMBL:ENSMUSG00000113836 | MGI:3781503 | protein coding gene | predicted gene 3325 |
| 13 | pseudogene | 62.035149 | 62.036163 | positive | MGI_C57BL6J_3647766 | Gm6478 | NCBI_Gene:624180,ENSEMBL:ENSMUSG00000074868 | MGI:3647766 | pseudogene | predicted pseudogene 6478 |
| 13 | pseudogene | 62.097748 | 62.112047 | positive | MGI_C57BL6J_5623862 | Gm40977 | NCBI_Gene:105245532,ENSEMBL:ENSMUSG00000113566 | MGI:5623862 | pseudogene | predicted gene, 40977 |
| 13 | pseudogene | 62.125642 | 62.126370 | positive | MGI_C57BL6J_5012170 | Selenok-ps5 | NCBI_Gene:100503951,ENSEMBL:ENSMUSG00000113504 | MGI:5012170 | pseudogene | selenoprotein K, pseudogene 5 |
| 13 | gene | 62.129886 | 62.236144 | positive | MGI_C57BL6J_3704127 | Zfp808 | NCBI_Gene:630579,ENSEMBL:ENSMUSG00000074867 | MGI:3704127 | protein coding gene | zinc finger protein 808 |
| 13 | gene | 62.136762 | 62.136831 | positive | MGI_C57BL6J_5453490 | Gm23713 | ENSEMBL:ENSMUSG00000095111 | MGI:5453490 | snoRNA gene | predicted gene, 23713 |
| 13 | gene | 62.136930 | 62.137062 | positive | MGI_C57BL6J_5454316 | Gm24539 | ENSEMBL:ENSMUSG00000093874 | MGI:5454316 | snoRNA gene | predicted gene, 24539 |
| 13 | pseudogene | 62.150849 | 62.155349 | positive | MGI_C57BL6J_5595660 | Gm36501 | NCBI_Gene:102640441,ENSEMBL:ENSMUSG00000113690 | MGI:5595660 | pseudogene | predicted gene, 36501 |
| 13 | pseudogene | 62.199609 | 62.200482 | negative | MGI_C57BL6J_3644246 | Gm7640 | NCBI_Gene:665455 | MGI:3644246 | pseudogene | predicted gene 7640 |
| 13 | pseudogene | 62.217501 | 62.217986 | negative | MGI_C57BL6J_4936986 | Gm17352 | NCBI_Gene:630612 | MGI:4936986 | pseudogene | predicted gene, 17352 |
| 13 | gene | 62.232674 | 62.237254 | positive | MGI_C57BL6J_1923621 | 2810408B13Rik | NCBI_Gene:76371 | MGI:1923621 | unclassified non-coding RNA gene | RIKEN cDNA 2810408B13 gene |
| 13 | pseudogene | 62.263566 | 62.275253 | positive | MGI_C57BL6J_3781511 | Gm3333 | NCBI_Gene:100041430,ENSEMBL:ENSMUSG00000113081 | MGI:3781511 | pseudogene | predicted gene 3333 |
| 13 | pseudogene | 62.322394 | 62.322667 | positive | MGI_C57BL6J_3781516 | Gm3338 | NCBI_Gene:102640921,ENSEMBL:ENSMUSG00000113107 | MGI:3781516 | pseudogene | predicted gene 3338 |
| 13 | pseudogene | 62.336463 | 62.355152 | negative | MGI_C57BL6J_5011977 | Gm19792 | NCBI_Gene:100503600,ENSEMBL:ENSMUSG00000113495 | MGI:5011977 | pseudogene | predicted gene, 19792 |
| 13 | gene | 62.367716 | 62.413623 | negative | MGI_C57BL6J_3781781 | Gm3604 | NCBI_Gene:100041979,ENSEMBL:ENSMUSG00000094942 | MGI:3781781 | protein coding gene | predicted gene 3604 |
| 13 | gene | 62.383237 | 62.383344 | positive | MGI_C57BL6J_5455710 | Gm25933 | ENSEMBL:ENSMUSG00000095876 | MGI:5455710 | miRNA gene | predicted gene, 25933 |
| 13 | gene | 62.384918 | 62.385052 | positive | MGI_C57BL6J_5453632 | Gm23855 | ENSEMBL:ENSMUSG00000089002 | MGI:5453632 | snoRNA gene | predicted gene, 23855 |
| 13 | pseudogene | 62.385104 | 62.385416 | positive | MGI_C57BL6J_6098319 | Gm48690 | ENSEMBL:ENSMUSG00000113151 | MGI:6098319 | pseudogene | predicted gene, 48690 |
| 13 | pseudogene | 62.387292 | 62.388017 | negative | MGI_C57BL6J_5012188 | Selenok-ps6 | NCBI_Gene:100503994,ENSEMBL:ENSMUSG00000113865 | MGI:5012188 | pseudogene | selenoprotein K, pseudogene 6 |
| 13 | pseudogene | 62.402493 | 62.417043 | negative | MGI_C57BL6J_6098320 | Gm48691 | ENSEMBL:ENSMUSG00000113623 | MGI:6098320 | pseudogene | predicted gene, 48691 |
| 13 | gene | 62.432866 | 62.434390 | negative | MGI_C57BL6J_6098334 | Gm48699 | ENSEMBL:ENSMUSG00000113812 | MGI:6098334 | unclassified gene | predicted gene, 48699 |
| 13 | gene | 62.449317 | 62.451609 | negative | MGI_C57BL6J_6098338 | Gm48701 | ENSEMBL:ENSMUSG00000113841 | MGI:6098338 | unclassified gene | predicted gene, 48701 |
| 13 | gene | 62.453016 | 62.466832 | negative | MGI_C57BL6J_6121568 | Gm49359 | ENSEMBL:ENSMUSG00000113450 | MGI:6121568 | protein coding gene | predicted gene, 49359 |
| 13 | gene | 62.453016 | 62.558511 | negative | MGI_C57BL6J_1918758 | Zfp935 | NCBI_Gene:71508,ENSEMBL:ENSMUSG00000055228 | MGI:1918758 | protein coding gene | zinc finger protein 935 |
| 13 | gene | 62.466865 | 62.466971 | positive | MGI_C57BL6J_5452033 | Gm22256 | ENSEMBL:ENSMUSG00000076246 | MGI:5452033 | miRNA gene | predicted gene, 22256 |
| 13 | gene | 62.479827 | 62.579320 | negative | MGI_C57BL6J_1924367 | Zfp934 | NCBI_Gene:77117,ENSEMBL:ENSMUSG00000074865 | MGI:1924367 | protein coding gene | zinc finger protein 934 |
| 13 | gene | 62.486263 | 62.498749 | negative | MGI_C57BL6J_5434500 | Gm21145 | NCBI_Gene:100861698 | MGI:5434500 | lncRNA gene | predicted gene, 21145 |
| 13 | pseudogene | 62.524788 | 62.532670 | negative | MGI_C57BL6J_3645656 | Gm7664 | NCBI_Gene:665503,ENSEMBL:ENSMUSG00000113428 | MGI:3645656 | pseudogene | predicted gene 7664 |
| 13 | gene | 62.543203 | 62.543335 | negative | MGI_C57BL6J_5453872 | Gm24095 | ENSEMBL:ENSMUSG00000096563 | MGI:5453872 | snoRNA gene | predicted gene, 24095 |
| 13 | gene | 62.543434 | 62.543503 | negative | MGI_C57BL6J_5452392 | Gm22615 | ENSEMBL:ENSMUSG00000094815 | MGI:5452392 | snoRNA gene | predicted gene, 22615 |
| 13 | gene | 62.552641 | 62.555638 | negative | MGI_C57BL6J_5621524 | Gm38639 | NCBI_Gene:102642842 | MGI:5621524 | lncRNA gene | predicted gene, 38639 |
| 13 | gene | 62.603013 | 62.636081 | negative | MGI_C57BL6J_2443901 | 6720489N17Rik | NCBI_Gene:211378,ENSEMBL:ENSMUSG00000072066 | MGI:2443901 | protein coding gene | RIKEN cDNA 6720489N17 gene |
| 13 | gene | 62.624194 | 62.624301 | positive | MGI_C57BL6J_5453797 | Gm24020 | ENSEMBL:ENSMUSG00000095020 | MGI:5453797 | miRNA gene | predicted gene, 24020 |
| 13 | gene | 62.630175 | 62.633775 | negative | MGI_C57BL6J_5826058 | Gm46421 | NCBI_Gene:108168091 | MGI:5826058 | lncRNA gene | predicted gene, 46421 |
| 13 | pseudogene | 62.639454 | 62.653262 | negative | MGI_C57BL6J_5595457 | Gm36298 | NCBI_Gene:102640165,ENSEMBL:ENSMUSG00000114172 | MGI:5595457 | pseudogene | predicted gene, 36298 |
| 13 | pseudogene | 62.669752 | 62.671981 | negative | MGI_C57BL6J_3779655 | Gm7045 | NCBI_Gene:630857,ENSEMBL:ENSMUSG00000113473 | MGI:3779655 | pseudogene | predicted gene 7045 |
| 13 | gene | 62.671784 | 62.706256 | negative | MGI_C57BL6J_5825588 | Gm45951 | NCBI_Gene:108167344 | MGI:5825588 | protein coding gene | predicted gene, 45951 |
| 13 | gene | 62.671784 | 62.760709 | negative | MGI_C57BL6J_3645613 | Platr25 | NCBI_Gene:435366,ENSEMBL:ENSMUSG00000074863 | MGI:3645613 | lncRNA gene | pluripotency associated transcript 25 |
| 13 | gene | 62.681192 | 62.682501 | negative | MGI_C57BL6J_6098399 | Gm48735 | ENSEMBL:ENSMUSG00000113601 | MGI:6098399 | unclassified gene | predicted gene, 48735 |
| 13 | gene | 62.734907 | 62.737195 | negative | MGI_C57BL6J_6098397 | Gm48734 | ENSEMBL:ENSMUSG00000113078 | MGI:6098397 | unclassified gene | predicted gene, 48734 |
| 13 | gene | 62.739042 | 62.761999 | negative | MGI_C57BL6J_5623864 | Gm40979 | NCBI_Gene:105245534 | MGI:5623864 | lncRNA gene | predicted gene, 40979 |
| 13 | gene | 62.760739 | 62.760846 | positive | MGI_C57BL6J_3629909 | Mir713 | miRBase:MI0004698,NCBI_Gene:751548,ENSEMBL:ENSMUSG00000096098 | MGI:3629909 | miRNA gene | microRNA 713 |
| 13 | gene | 62.760803 | 62.761714 | positive | MGI_C57BL6J_1924368 | 6720490N10Rik | NA | NA | unclassified gene | RIKEN cDNA 6720490N10 gene |
| 13 | gene | 62.772200 | 62.785808 | negative | MGI_C57BL6J_3779466 | Gm5141 | NCBI_Gene:380850,ENSEMBL:ENSMUSG00000091183 | MGI:3779466 | protein coding gene | predicted gene 5141 |
| 13 | pseudogene | 62.775871 | 62.779544 | positive | MGI_C57BL6J_5439432 | Gm21963 | NCBI_Gene:108168090,ENSEMBL:ENSMUSG00000095895 | MGI:5439432 | pseudogene | predicted gene, 21963 |
| 13 | gene | 62.779594 | 62.779676 | positive | MGI_C57BL6J_5454365 | Gm24588 | ENSEMBL:ENSMUSG00000080346 | MGI:5454365 | miRNA gene | predicted gene, 24588 |
| 13 | gene | 62.791507 | 62.791613 | positive | MGI_C57BL6J_5455225 | Gm25448 | ENSEMBL:ENSMUSG00000095825 | MGI:5455225 | snRNA gene | predicted gene, 25448 |
| 13 | pseudogene | 62.812916 | 62.815515 | positive | MGI_C57BL6J_3646400 | Gm7049 | NCBI_Gene:630951,ENSEMBL:ENSMUSG00000113637 | MGI:3646400 | pseudogene | predicted gene 7049 |
| 13 | gene | 62.813969 | 62.815523 | negative | MGI_C57BL6J_6098498 | Gm48795 | ENSEMBL:ENSMUSG00000114117 | MGI:6098498 | lncRNA gene | predicted gene, 48795 |
| 13 | gene | 62.836877 | 62.858422 | negative | MGI_C57BL6J_95491 | Fbp2 | NCBI_Gene:14120,ENSEMBL:ENSMUSG00000021456 | MGI:95491 | protein coding gene | fructose bisphosphatase 2 |
| 13 | gene | 62.864753 | 62.888298 | negative | MGI_C57BL6J_95492 | Fbp1 | NCBI_Gene:14121,ENSEMBL:ENSMUSG00000069805 | MGI:95492 | protein coding gene | fructose bisphosphatase 1 |
| 13 | gene | 62.878667 | 62.885078 | negative | MGI_C57BL6J_5621564 | Gm38679 | NCBI_Gene:105242395 | MGI:5621564 | lncRNA gene | predicted gene, 38679 |
| 13 | gene | 62.912315 | 62.917835 | negative | MGI_C57BL6J_5623866 | Gm40981 | NCBI_Gene:105245536 | MGI:5623866 | lncRNA gene | predicted gene, 40981 |
| 13 | gene | 62.944561 | 62.949442 | negative | MGI_C57BL6J_6098525 | Gm48812 | ENSEMBL:ENSMUSG00000113057 | MGI:6098525 | lncRNA gene | predicted gene, 48812 |
| 13 | gene | 62.964893 | 63.326096 | positive | MGI_C57BL6J_1919311 | Aopep | NCBI_Gene:72061,ENSEMBL:ENSMUSG00000021458 | MGI:1919311 | protein coding gene | aminopeptidase O |
| 13 | pseudogene | 62.967346 | 62.968657 | negative | MGI_C57BL6J_3647236 | Tubb2a-ps1 | NCBI_Gene:665524 | MGI:3647236 | pseudogene | tubulin, beta 2a, pseudogene 1 |
| 13 | gene | 63.009959 | 63.011240 | positive | MGI_C57BL6J_6096623 | Gm47584 | ENSEMBL:ENSMUSG00000113708 | MGI:6096623 | unclassified gene | predicted gene, 47584 |
| 13 | gene | 63.090776 | 63.094751 | negative | MGI_C57BL6J_5826059 | Gm46422 | NCBI_Gene:108168092 | MGI:5826059 | lncRNA gene | predicted gene, 46422 |
| 13 | gene | 63.116585 | 63.134513 | positive | MGI_C57BL6J_5589568 | Gm30409 | NCBI_Gene:102632297,ENSEMBL:ENSMUSG00000113257 | MGI:5589568 | lncRNA gene | predicted gene, 30409 |
| 13 | gene | 63.142494 | 63.143934 | positive | MGI_C57BL6J_6096629 | Gm47587 | ENSEMBL:ENSMUSG00000113203 | MGI:6096629 | lncRNA gene | predicted gene, 47587 |
| 13 | gene | 63.210950 | 63.214374 | positive | MGI_C57BL6J_6096621 | Gm47583 | ENSEMBL:ENSMUSG00000113853 | MGI:6096621 | unclassified gene | predicted gene, 47583 |
| 13 | gene | 63.244221 | 63.246248 | positive | MGI_C57BL6J_6096627 | Gm47586 | ENSEMBL:ENSMUSG00000113326 | MGI:6096627 | unclassified gene | predicted gene, 47586 |
| 13 | gene | 63.253024 | 63.255564 | negative | MGI_C57BL6J_6096658 | Gm47603 | ENSEMBL:ENSMUSG00000113665 | MGI:6096658 | unclassified gene | predicted gene, 47603 |
| 13 | gene | 63.273600 | 63.275911 | positive | MGI_C57BL6J_6096625 | Gm47585 | ENSEMBL:ENSMUSG00000114104 | MGI:6096625 | unclassified gene | predicted gene, 47585 |
| 13 | gene | 63.276905 | 63.279224 | negative | MGI_C57BL6J_6096656 | Gm47602 | ENSEMBL:ENSMUSG00000113472 | MGI:6096656 | unclassified gene | predicted gene, 47602 |
| 13 | gene | 63.285043 | 63.497294 | negative | MGI_C57BL6J_95480 | Fancc | NCBI_Gene:14088,ENSEMBL:ENSMUSG00000021461 | MGI:95480 | protein coding gene | Fanconi anemia, complementation group C |
| 13 | gene | 63.289154 | 63.297147 | negative | MGI_C57BL6J_4439831 | Gm16907 | NCBI_Gene:100504114,ENSEMBL:ENSMUSG00000097242 | MGI:4439831 | lncRNA gene | predicted gene, 16907 |
| 13 | gene | 63.300484 | 63.300557 | positive | MGI_C57BL6J_2676898 | Mir23b | miRBase:MI0000141,NCBI_Gene:387217,ENSEMBL:ENSMUSG00000065599 | MGI:2676898 | miRNA gene | microRNA 23b |
| 13 | gene | 63.300712 | 63.300784 | positive | MGI_C57BL6J_2676903 | Mir27b | miRBase:MI0000142,NCBI_Gene:387221,ENSEMBL:ENSMUSG00000065475 | MGI:2676903 | miRNA gene | microRNA 27b |
| 13 | gene | 63.301199 | 63.301283 | negative | MGI_C57BL6J_4834247 | Mir3074-1 | miRBase:MI0014037,NCBI_Gene:100526474,ENSEMBL:ENSMUSG00000092741 | MGI:4834247 | miRNA gene | microRNA 3074-1 |
| 13 | gene | 63.301208 | 63.301275 | positive | MGI_C57BL6J_2676899 | Mir24-1 | miRBase:MI0000231,NCBI_Gene:387142,ENSEMBL:ENSMUSG00000105904 | MGI:2676899 | miRNA gene | microRNA 24-1 |
| 13 | gene | 63.329556 | 63.339961 | positive | MGI_C57BL6J_3802155 | Gm16133 | ENSEMBL:ENSMUSG00000089854 | MGI:3802155 | lncRNA gene | predicted gene 16133 |
| 13 | gene | 63.376761 | 63.381045 | negative | MGI_C57BL6J_3802156 | Gm16132 | ENSEMBL:ENSMUSG00000090232 | MGI:3802156 | lncRNA gene | predicted gene 16132 |
| 13 | gene | 63.410918 | 63.414767 | negative | MGI_C57BL6J_6096631 | Gm47588 | ENSEMBL:ENSMUSG00000113126 | MGI:6096631 | unclassified gene | predicted gene, 47588 |
| 13 | pseudogene | 63.446930 | 63.447265 | negative | MGI_C57BL6J_3802157 | Gm16134 | ENSEMBL:ENSMUSG00000089868 | MGI:3802157 | pseudogene | predicted gene 16134 |
| 13 | gene | 63.508328 | 63.573616 | negative | MGI_C57BL6J_105373 | Ptch1 | NCBI_Gene:19206,ENSEMBL:ENSMUSG00000021466 | MGI:105373 | protein coding gene | patched 1 |
| 13 | gene | 63.567098 | 63.568054 | negative | MGI_C57BL6J_1925083 | A930032L01Rik | ENSEMBL:ENSMUSG00000102282 | MGI:1925083 | unclassified gene | RIKEN cDNA A930032L01 gene |
| 13 | gene | 63.575051 | 63.615298 | positive | MGI_C57BL6J_5589814 | Gm30655 | NCBI_Gene:102632415,ENSEMBL:ENSMUSG00000113459 | MGI:5589814 | lncRNA gene | predicted gene, 30655 |
| 13 | gene | 63.621500 | 63.627379 | negative | MGI_C57BL6J_5589868 | Gm30709 | NCBI_Gene:102632712,ENSEMBL:ENSMUSG00000113453 | MGI:5589868 | lncRNA gene | predicted gene, 30709 |
| 13 | pseudogene | 63.661797 | 63.662161 | positive | MGI_C57BL6J_6096307 | Gm47387 | ENSEMBL:ENSMUSG00000113700 | MGI:6096307 | pseudogene | predicted gene, 47387 |
| 13 | gene | 63.675756 | 63.734298 | negative | MGI_C57BL6J_1922772 | 1700024I08Rik | NCBI_Gene:75522,ENSEMBL:ENSMUSG00000113825 | MGI:1922772 | lncRNA gene | RIKEN cDNA 1700024I08 gene |
| 13 | gene | 63.730149 | 63.734599 | positive | MGI_C57BL6J_6096312 | Gm47390 | ENSEMBL:ENSMUSG00000114071 | MGI:6096312 | lncRNA gene | predicted gene, 47390 |
| 13 | pseudogene | 63.748751 | 63.749268 | positive | MGI_C57BL6J_6096356 | Gm47417 | ENSEMBL:ENSMUSG00000114015 | MGI:6096356 | pseudogene | predicted gene, 47417 |
| 13 | gene | 63.794106 | 63.795428 | positive | MGI_C57BL6J_6096357 | Gm47418 | ENSEMBL:ENSMUSG00000114092 | MGI:6096357 | lncRNA gene | predicted gene, 47418 |
| 13 | gene | 63.813311 | 63.815560 | negative | MGI_C57BL6J_1924579 | C030010C08Rik | NA | NA | unclassified gene | RIKEN cDNA C030010C08 gene |
| 13 | gene | 63.815058 | 63.900302 | positive | MGI_C57BL6J_1923501 | Ercc6l2 | NCBI_Gene:76251,ENSEMBL:ENSMUSG00000021470 | MGI:1923501 | protein coding gene | excision repair cross-complementing rodent repair deficiency, complementation group 6 like 2 |
| 13 | gene | 63.861082 | 63.862291 | positive | MGI_C57BL6J_6096366 | Gm47424 | ENSEMBL:ENSMUSG00000114095 | MGI:6096366 | lncRNA gene | predicted gene, 47424 |
| 13 | gene | 63.900364 | 63.908682 | negative | MGI_C57BL6J_5826060 | Gm46423 | NCBI_Gene:108168093 | MGI:5826060 | lncRNA gene | predicted gene, 46423 |
| 13 | gene | 63.927755 | 63.938637 | positive | MGI_C57BL6J_5590109 | Gm30950 | NCBI_Gene:102633025 | MGI:5590109 | lncRNA gene | predicted gene, 30950 |
| 13 | pseudogene | 63.945845 | 63.946564 | negative | MGI_C57BL6J_3649134 | Gm7695 | NCBI_Gene:665575,ENSEMBL:ENSMUSG00000113340 | MGI:3649134 | pseudogene | predicted gene 7695 |
| 13 | gene | 64.056253 | 64.057242 | positive | MGI_C57BL6J_5590158 | Gm30999 | NCBI_Gene:102633088 | MGI:5590158 | lncRNA gene | predicted gene, 30999 |
| 13 | gene | 64.058266 | 64.089262 | negative | MGI_C57BL6J_107177 | Hsd17b3 | NCBI_Gene:15487,ENSEMBL:ENSMUSG00000033122 | MGI:107177 | protein coding gene | hydroxysteroid (17-beta) dehydrogenase 3 |
| 13 | gene | 64.096308 | 64.129385 | negative | MGI_C57BL6J_1917734 | Slc35d2 | NCBI_Gene:70484,ENSEMBL:ENSMUSG00000033114 | MGI:1917734 | protein coding gene | solute carrier family 35, member D2 |
| 13 | gene | 64.133018 | 64.153202 | negative | MGI_C57BL6J_2442266 | Zfp367 | NCBI_Gene:238673,ENSEMBL:ENSMUSG00000044934 | MGI:2442266 | protein coding gene | zinc finger protein 367 |
| 13 | gene | 64.152581 | 64.154596 | positive | MGI_C57BL6J_6096505 | Gm47513 | ENSEMBL:ENSMUSG00000114165 | MGI:6096505 | unclassified gene | predicted gene, 47513 |
| 13 | gene | 64.161824 | 64.186537 | positive | MGI_C57BL6J_1891713 | Habp4 | NCBI_Gene:56541,ENSEMBL:ENSMUSG00000021476 | MGI:1891713 | protein coding gene | hyaluronic acid binding protein 4 |
| 13 | gene | 64.189268 | 64.275290 | negative | MGI_C57BL6J_2441808 | Cdc14b | NCBI_Gene:218294,ENSEMBL:ENSMUSG00000033102 | MGI:2441808 | protein coding gene | CDC14 cell division cycle 14B |
| 13 | pseudogene | 64.210340 | 64.210798 | positive | MGI_C57BL6J_5826061 | Gm46424 | NCBI_Gene:108168094,ENSEMBL:ENSMUSG00000113804 | MGI:5826061 | pseudogene | predicted gene, 46424 |
| 13 | gene | 64.248700 | 64.271852 | positive | MGI_C57BL6J_1913753 | 1810034E14Rik | NCBI_Gene:66503,ENSEMBL:ENSMUSG00000097101 | MGI:1913753 | lncRNA gene | RIKEN cDNA 1810034E14 gene |
| 13 | gene | 64.262114 | 64.264374 | positive | MGI_C57BL6J_6095681 | Gm47004 | ENSEMBL:ENSMUSG00000113603 | MGI:6095681 | unclassified gene | predicted gene, 47004 |
| 13 | gene | 64.275280 | 64.312710 | negative | MGI_C57BL6J_1913379 | Prxl2c | NCBI_Gene:66129,ENSEMBL:ENSMUSG00000021482 | MGI:1913379 | protein coding gene | peroxiredoxin like 2C |
| 13 | gene | 64.291077 | 64.298039 | positive | MGI_C57BL6J_6095679 | Gm47003 | ENSEMBL:ENSMUSG00000113816 | MGI:6095679 | lncRNA gene | predicted gene, 47003 |
| 13 | gene | 64.296437 | 64.296524 | negative | MGI_C57BL6J_5455431 | Gm25654 | ENSEMBL:ENSMUSG00000089092 | MGI:5455431 | snRNA gene | predicted gene, 25654 |
| 13 | gene | 64.316550 | 64.343787 | negative | MGI_C57BL6J_5590287 | Gm31128 | NCBI_Gene:102633261 | MGI:5590287 | lncRNA gene | predicted gene, 31128 |
| 13 | gene | 64.359337 | 64.370890 | negative | MGI_C57BL6J_88564 | Ctsl | NCBI_Gene:13039,ENSEMBL:ENSMUSG00000021477 | MGI:88564 | protein coding gene | cathepsin L |
| 13 | gene | 64.361052 | 64.363022 | positive | MGI_C57BL6J_6095872 | Gm47123 | ENSEMBL:ENSMUSG00000113683 | MGI:6095872 | lncRNA gene | predicted gene, 47123 |
| 13 | pseudogene | 64.361343 | 64.361737 | negative | MGI_C57BL6J_6118691 | Gm49230 | ENSEMBL:ENSMUSG00000114072 | MGI:6118691 | pseudogene | predicted gene, 49230 |
| 13 | gene | 64.427780 | 64.432436 | negative | MGI_C57BL6J_1922782 | 1700015C15Rik | NCBI_Gene:75532,ENSEMBL:ENSMUSG00000113696 | MGI:1922782 | lncRNA gene | RIKEN cDNA 1700015C15 gene |
| 13 | gene | 64.432314 | 64.441773 | positive | MGI_C57BL6J_2145349 | Cdk20 | NCBI_Gene:105278,ENSEMBL:ENSMUSG00000021483 | MGI:2145349 | protein coding gene | cyclin-dependent kinase 20 |
| 13 | gene | 64.449437 | 64.508611 | positive | MGI_C57BL6J_5590377 | Gm31218 | NCBI_Gene:102633383,ENSEMBL:ENSMUSG00000113244 | MGI:5590377 | lncRNA gene | predicted gene, 31218 |
| 13 | pseudogene | 64.454413 | 64.458923 | negative | MGI_C57BL6J_3646255 | Gm7712 | NCBI_Gene:665612,ENSEMBL:ENSMUSG00000113109 | MGI:3646255 | pseudogene | predicted gene 7712 |
| 13 | gene | 64.481246 | 64.497792 | negative | MGI_C57BL6J_1916130 | Fam240b | NCBI_Gene:68880,ENSEMBL:ENSMUSG00000096537 | MGI:1916130 | protein coding gene | family with sequence similarity 240 member B |
| 13 | pseudogene | 64.509488 | 64.511096 | positive | MGI_C57BL6J_3648106 | Gm4810 | NCBI_Gene:218298,ENSEMBL:ENSMUSG00000094597 | MGI:3648106 | pseudogene | predicted gene 4810 |
| 13 | pseudogene | 64.515443 | 64.515854 | negative | MGI_C57BL6J_6095982 | Gm47190 | ENSEMBL:ENSMUSG00000113609 | MGI:6095982 | pseudogene | predicted gene, 47190 |
| 13 | pseudogene | 64.611860 | 64.619498 | positive | MGI_C57BL6J_6095983 | Gm47191 | ENSEMBL:ENSMUSG00000113850 | MGI:6095983 | pseudogene | predicted gene, 47191 |
| 13 | gene | 64.619550 | 64.625002 | positive | MGI_C57BL6J_6095985 | Gm47193 | ENSEMBL:ENSMUSG00000113124 | MGI:6095985 | lncRNA gene | predicted gene, 47193 |
| 13 | pseudogene | 64.624067 | 64.624802 | negative | MGI_C57BL6J_3648515 | Gm5791 | NCBI_Gene:544940,ENSEMBL:ENSMUSG00000113753 | MGI:3648515 | pseudogene | predicted gene 5791 |
| 13 | pseudogene | 64.673796 | 64.675084 | positive | MGI_C57BL6J_3779450 | Gm4935 | NCBI_Gene:238678,ENSEMBL:ENSMUSG00000113405 | MGI:3779450 | pseudogene | predicted gene 4935 |
| 13 | gene | 64.674539 | 64.685275 | negative | MGI_C57BL6J_6095987 | Gm47194 | ENSEMBL:ENSMUSG00000114168 | MGI:6095987 | lncRNA gene | predicted gene, 47194 |
| 13 | pseudogene | 64.703141 | 64.703685 | positive | MGI_C57BL6J_3647309 | Gm7065 | NCBI_Gene:631470,ENSEMBL:ENSMUSG00000113469 | MGI:3647309 | pseudogene | predicted gene 7065 |
| 13 | gene | 64.736182 | 64.904029 | negative | MGI_C57BL6J_3588199 | Cntnap3 | NCBI_Gene:238680,ENSEMBL:ENSMUSG00000033063 | MGI:3588199 | protein coding gene | contactin associated protein-like 3 |
| 13 | pseudogene | 64.913895 | 64.915116 | positive | MGI_C57BL6J_3643793 | Eif1-ps2 | NCBI_Gene:432767,ENSEMBL:ENSMUSG00000114179 | MGI:3643793 | pseudogene | eukaryotic translation initiation factor 1, pseudogene 2 |
| 13 | gene | 64.917406 | 64.923195 | positive | MGI_C57BL6J_1925374 | Spata31 | NCBI_Gene:78124,ENSEMBL:ENSMUSG00000056223 | MGI:1925374 | protein coding gene | spermatogenesis associated 31 |
| 13 | pseudogene | 64.927940 | 64.929162 | negative | MGI_C57BL6J_3648985 | Gm4811 | NCBI_Gene:218301,ENSEMBL:ENSMUSG00000113263 | MGI:3648985 | pseudogene | predicted gene 4811 |
| 13 | pseudogene | 64.998833 | 65.000358 | positive | MGI_C57BL6J_3779638 | Gm6888 | NCBI_Gene:628515,ENSEMBL:ENSMUSG00000113807 | MGI:3779638 | pseudogene | predicted gene 6888 |
| 13 | gene | 65.033058 | 65.038004 | positive | MGI_C57BL6J_3588241 | Spata31d1c | NCBI_Gene:238683,ENSEMBL:ENSMUSG00000074849 | MGI:3588241 | protein coding gene | spermatogenesis associated 31 subfamily D, member 1C |
| 13 | gene | 65.044606 | 65.052986 | negative | MGI_C57BL6J_2685120 | Prss47 | NCBI_Gene:218304,ENSEMBL:ENSMUSG00000090658 | MGI:2685120 | protein coding gene | protease, serine 47 |
| 13 | gene | 65.064663 | 65.113002 | negative | MGI_C57BL6J_1913881 | Mfsd14b | NCBI_Gene:66631,ENSEMBL:ENSMUSG00000038212 | MGI:1913881 | protein coding gene | major facilitator superfamily domain containing 14B |
| 13 | pseudogene | 65.125876 | 65.126180 | positive | MGI_C57BL6J_3781958 | Gm3785 | NCBI_Gene:102633809,ENSEMBL:ENSMUSG00000114162 | MGI:3781958 | pseudogene | predicted gene 3785 |
| 13 | pseudogene | 65.138089 | 65.146047 | positive | MGI_C57BL6J_3030299 | Olfr465-ps1 | NCBI_Gene:258419,ENSEMBL:ENSMUSG00000108426 | MGI:3030299 | pseudogene | olfactory receptor 465, pseudogene 1 |
| 13 | gene | 65.150240 | 65.156152 | positive | MGI_C57BL6J_3030300 | Olfr466 | NCBI_Gene:258816,ENSEMBL:ENSMUSG00000049806 | MGI:3030300 | protein coding gene | olfactory receptor 466 |
| 13 | gene | 65.158258 | 65.158408 | positive | MGI_C57BL6J_5453907 | Gm24130 | ENSEMBL:ENSMUSG00000089161 | MGI:5453907 | snoRNA gene | predicted gene, 24130 |
| 13 | gene | 65.177111 | 65.205977 | negative | MGI_C57BL6J_2145528 | Nlrp4f | NCBI_Gene:97895,ENSEMBL:ENSMUSG00000032999 | MGI:2145528 | protein coding gene | NLR family, pyrin domain containing 4F |
| 13 | gene | 65.212111 | 65.214903 | positive | MGI_C57BL6J_6096075 | Gm47249 | ENSEMBL:ENSMUSG00000113500 | MGI:6096075 | lncRNA gene | predicted gene, 47249 |
| 13 | gene | 65.241753 | 65.250154 | positive | MGI_C57BL6J_5595604 | Gm36445 | ENSEMBL:ENSMUSG00000110393 | MGI:5595604 | lncRNA gene | predicted gene, 36445 |
| 13 | pseudogene | 65.243717 | 65.243867 | negative | MGI_C57BL6J_6096077 | Gm47250 | ENSEMBL:ENSMUSG00000113946 | MGI:6096077 | pseudogene | predicted gene, 47250 |
| 13 | gene | 65.252704 | 65.253703 | negative | MGI_C57BL6J_6096078 | Gm47251 | ENSEMBL:ENSMUSG00000113238 | MGI:6096078 | lncRNA gene | predicted gene, 47251 |
| 13 | gene | 65.259539 | 65.260826 | positive | MGI_C57BL6J_3642324 | Gm10775 | ENSEMBL:ENSMUSG00000074847 | MGI:3642324 | protein coding gene | predicted gene 10775 |
| 13 | pseudogene | 65.264677 | 65.264878 | negative | MGI_C57BL6J_6096084 | Gm47254 | ENSEMBL:ENSMUSG00000112960 | MGI:6096084 | pseudogene | predicted gene, 47254 |
| 13 | gene | 65.278519 | 65.304221 | positive | MGI_C57BL6J_2176229 | Zfp369 | NCBI_Gene:170936,ENSEMBL:ENSMUSG00000021514 | MGI:2176229 | protein coding gene | zinc finger protein 369 |
| 13 | gene | 65.289453 | 65.290794 | positive | MGI_C57BL6J_6096089 | Gm47258 | ENSEMBL:ENSMUSG00000114016 | MGI:6096089 | unclassified gene | predicted gene, 47258 |
| 13 | pseudogene | 65.313716 | 65.314351 | positive | MGI_C57BL6J_3781973 | Gm3800 | NCBI_Gene:100042347,ENSEMBL:ENSMUSG00000114194 | MGI:3781973 | pseudogene | predicted gene 3800 |
| 13 | pseudogene | 65.332907 | 65.333113 | negative | MGI_C57BL6J_6096098 | Gm47263 | ENSEMBL:ENSMUSG00000113990 | MGI:6096098 | pseudogene | predicted gene, 47263 |
| 13 | gene | 65.336222 | 65.348597 | negative | MGI_C57BL6J_3779761 | Gm7762 | NCBI_Gene:665738,ENSEMBL:ENSMUSG00000113406 | MGI:3779761 | lncRNA gene | predicted gene 7762 |
| 13 | pseudogene | 65.345127 | 65.345282 | positive | MGI_C57BL6J_6096099 | Gm47264 | ENSEMBL:ENSMUSG00000114142 | MGI:6096099 | pseudogene | predicted gene, 47264 |
| 13 | pseudogene | 65.353134 | 65.367140 | negative | MGI_C57BL6J_3761347 | Vmn2r-ps95 | NCBI_Gene:665742,ENSEMBL:ENSMUSG00000113145 | MGI:3761347 | pseudogene | vomeronasal 2, receptor, pseudogene 95 |
| 13 | pseudogene | 65.368150 | 65.368746 | negative | MGI_C57BL6J_3649111 | Gm7768 | NCBI_Gene:665750,ENSEMBL:ENSMUSG00000113551 | MGI:3649111 | pseudogene | predicted gene 7768 |
| 13 | gene | 65.379007 | 65.382097 | positive | MGI_C57BL6J_5623868 | Gm40983 | NCBI_Gene:105245538,ENSEMBL:ENSMUSG00000112935 | MGI:5623868 | lncRNA gene | predicted gene, 40983 |
| 13 | gene | 65.381228 | 65.381866 | negative | MGI_C57BL6J_5623867 | Gm40982 | NCBI_Gene:105245537 | MGI:5623867 | lncRNA gene | predicted gene, 40982 |
| 13 | pseudogene | 65.396007 | 65.396138 | negative | MGI_C57BL6J_6097453 | Gm48103 | ENSEMBL:ENSMUSG00000113301 | MGI:6097453 | pseudogene | predicted gene, 48103 |
| 13 | gene | 65.396483 | 65.415122 | negative | MGI_C57BL6J_5590873 | Gm31714 | NCBI_Gene:102634030 | MGI:5590873 | protein coding gene | predicted gene, 31714 |
| 13 | pseudogene | 65.396555 | 65.397678 | negative | MGI_C57BL6J_5439382 | Gm21930 | NCBI_Gene:100534321,ENSEMBL:ENSMUSG00000113911 | MGI:5439382 | pseudogene | predicted gene, 21930 |
| 13 | pseudogene | 65.404296 | 65.408136 | positive | MGI_C57BL6J_6097464 | Gm48111 | ENSEMBL:ENSMUSG00000113388 | MGI:6097464 | pseudogene | predicted gene, 48111 |
| 13 | pseudogene | 65.416514 | 65.417147 | positive | MGI_C57BL6J_3647433 | Cbx3-ps5 | NCBI_Gene:544944,ENSEMBL:ENSMUSG00000113207 | MGI:3647433 | pseudogene | chromobox 3, pseudogene 5 |
| 13 | pseudogene | 65.421635 | 65.435539 | negative | MGI_C57BL6J_3761348 | Vmn2r-ps96 | NCBI_Gene:665767,ENSEMBL:ENSMUSG00000114065 | MGI:3761348 | pseudogene | vomeronasal 2, receptor, pseudogene 96 |
| 13 | pseudogene | 65.436448 | 65.437243 | negative | MGI_C57BL6J_3643106 | Hmgb1-ps11 | NCBI_Gene:665777,ENSEMBL:ENSMUSG00000113709 | MGI:3643106 | pseudogene | high mobility group box 1, pseudogene 11 |
| 13 | gene | 65.461756 | 65.469447 | positive | MGI_C57BL6J_5623870 | Gm40985 | NCBI_Gene:108168095 | MGI:5623870 | lncRNA gene | predicted gene, 40985 |
| 13 | pseudogene | 65.478385 | 65.479292 | negative | MGI_C57BL6J_6097499 | Gm48136 | ENSEMBL:ENSMUSG00000114031 | MGI:6097499 | pseudogene | predicted gene, 48136 |
| 13 | pseudogene | 65.487439 | 65.624203 | positive | MGI_C57BL6J_6097538 | Gm48165 | ENSEMBL:ENSMUSG00000113468 | MGI:6097538 | pseudogene | predicted gene, 48165 |
| 13 | gene | 65.491789 | 65.496793 | negative | MGI_C57BL6J_6097542 | Gm48168 | ENSEMBL:ENSMUSG00000113955 | MGI:6097542 | lncRNA gene | predicted gene, 48168 |
| 13 | gene | 65.493490 | 65.630774 | positive | MGI_C57BL6J_6097539 | Gm48166 | ENSEMBL:ENSMUSG00000113618 | MGI:6097539 | lncRNA gene | predicted gene, 48166 |
| 13 | gene | 65.511862 | 65.512932 | positive | MGI_C57BL6J_3642815 | Gm10139 | NCBI_Gene:102634499,ENSEMBL:ENSMUSG00000063905 | MGI:3642815 | protein coding gene | predicted gene 10139 |
| 13 | pseudogene | 65.514187 | 65.514978 | positive | MGI_C57BL6J_6097547 | Gm48171 | ENSEMBL:ENSMUSG00000113008 | MGI:6097547 | pseudogene | predicted gene, 48171 |
| 13 | pseudogene | 65.518174 | 65.518412 | positive | MGI_C57BL6J_6097544 | Gm48169 | ENSEMBL:ENSMUSG00000113661 | MGI:6097544 | pseudogene | predicted gene, 48169 |
| 13 | pseudogene | 65.541990 | 65.542205 | negative | MGI_C57BL6J_6097541 | Gm48167 | ENSEMBL:ENSMUSG00000113482 | MGI:6097541 | pseudogene | predicted gene, 48167 |
| 13 | gene | 65.545966 | 65.713454 | negative | MGI_C57BL6J_5011613 | Gm19428 | NCBI_Gene:100502880,ENSEMBL:ENSMUSG00000113260 | MGI:5011613 | protein coding gene | predicted gene, 19428 |
| 13 | pseudogene | 65.559168 | 65.559711 | positive | MGI_C57BL6J_1890539 | Cbx3-ps2 | ENSEMBL:ENSMUSG00000113565 | MGI:1890539 | pseudogene | chromobox 3, pseudogene 2 |
| 13 | pseudogene | 65.564110 | 65.578050 | negative | MGI_C57BL6J_3761349 | Vmn2r-ps97 | NCBI_Gene:665809,ENSEMBL:ENSMUSG00000113381 | MGI:3761349 | pseudogene | vomeronasal 2, receptor, pseudogene 97 |
| 13 | pseudogene | 65.579040 | 65.579659 | negative | MGI_C57BL6J_3782001 | Gm3829 | NCBI_Gene:100042404,ENSEMBL:ENSMUSG00000113118 | MGI:3782001 | pseudogene | predicted gene 3829 |
| 13 | gene | 65.590293 | 65.591561 | negative | MGI_C57BL6J_5477133 | Gm26639 | ENSEMBL:ENSMUSG00000097562 | MGI:5477133 | lncRNA gene | predicted gene, 26639 |
| 13 | gene | 65.615824 | 65.620243 | negative | MGI_C57BL6J_6097545 | Gm48170 | ENSEMBL:ENSMUSG00000113135 | MGI:6097545 | lncRNA gene | predicted gene, 48170 |
| 13 | pseudogene | 65.623404 | 65.624052 | positive | MGI_C57BL6J_6097537 | Gm48164 | ENSEMBL:ENSMUSG00000113641 | MGI:6097537 | pseudogene | predicted gene, 48164 |
| 13 | gene | 65.639768 | 65.660118 | positive | MGI_C57BL6J_5011633 | Gm19448 | NCBI_Gene:102634730 | MGI:5011633 | lncRNA gene | predicted gene, 19448 |
| 13 | pseudogene | 65.649558 | 65.650349 | positive | MGI_C57BL6J_6097564 | Gm48185 | ENSEMBL:ENSMUSG00000113870 | MGI:6097564 | pseudogene | predicted gene, 48185 |
| 13 | pseudogene | 65.653504 | 65.653742 | positive | MGI_C57BL6J_6097565 | Gm48186 | ENSEMBL:ENSMUSG00000113461 | MGI:6097565 | pseudogene | predicted gene, 48186 |
| 13 | pseudogene | 65.697004 | 65.762538 | positive | MGI_C57BL6J_5011303 | Gm19118 | NCBI_Gene:100418286,ENSEMBL:ENSMUSG00000113513 | MGI:5011303 | pseudogene | predicted gene, 19118 |
| 13 | pseudogene | 65.727793 | 65.805531 | negative | MGI_C57BL6J_3761350 | Vmn2r-ps98 | NCBI_Gene:628722,ENSEMBL:ENSMUSG00000117836 | MGI:3761350 | pseudogene | vomeronasal 2, receptor, pseudogene 98 |
| 13 | pseudogene | 65.761225 | 65.762695 | negative | MGI_C57BL6J_5621313 | Gm38428 | NCBI_Gene:100534322,ENSEMBL:ENSMUSG00000113712 | MGI:5621313 | pseudogene | predicted gene, 38428 |
| 13 | gene | 65.780905 | 65.867305 | positive | MGI_C57BL6J_5477252 | Platr2 | ENSEMBL:ENSMUSG00000097066 | MGI:5477252 | lncRNA gene | pluripotency associated transcript 2 |
| 13 | gene | 65.849063 | 65.863393 | negative | MGI_C57BL6J_5623871 | Gm40986 | NCBI_Gene:105245541 | MGI:5623871 | lncRNA gene | predicted gene, 40986 |
| 13 | pseudogene | 65.874239 | 65.874834 | positive | MGI_C57BL6J_3646510 | Gm7822 | NCBI_Gene:665851,ENSEMBL:ENSMUSG00000113954 | MGI:3646510 | pseudogene | predicted gene 7822 |
| 13 | pseudogene | 65.875841 | 65.889733 | positive | MGI_C57BL6J_3761351 | Vmn2r-ps99 | NCBI_Gene:628808,ENSEMBL:ENSMUSG00000113945 | MGI:3761351 | pseudogene | vomeronasal 2, receptor, pseudogene 99 |
| 13 | pseudogene | 65.894215 | 65.894759 | negative | MGI_C57BL6J_1890538 | Cbx3-ps1 | ENSEMBL:ENSMUSG00000074841 | MGI:1890538 | pseudogene | chromobox 3, pseudogene 1 |
| 13 | gene | 65.897183 | 65.907988 | positive | MGI_C57BL6J_4936987 | Gm17353 | NCBI_Gene:100502941 | MGI:4936987 | protein coding gene | predicted gene, 17353 |
| 13 | pseudogene | 65.908223 | 65.908904 | positive | MGI_C57BL6J_6097615 | Gm48218 | ENSEMBL:ENSMUSG00000114625 | MGI:6097615 | pseudogene | predicted gene, 48218 |
| 13 | pseudogene | 65.912526 | 65.912759 | positive | MGI_C57BL6J_6097616 | Gm48219 | ENSEMBL:ENSMUSG00000114013 | MGI:6097616 | pseudogene | predicted gene, 48219 |
| 13 | gene | 65.935670 | 65.950327 | negative | MGI_C57BL6J_5591635 | Gm32476 | NCBI_Gene:102635039 | MGI:5591635 | lncRNA gene | predicted gene, 32476 |
| 13 | pseudogene | 65.944036 | 65.948582 | positive | MGI_C57BL6J_3761519 | Vmn2r-ps100 | NCBI_Gene:100041855,ENSEMBL:ENSMUSG00000113723 | MGI:3761519 | pseudogene | vomeronasal 2, receptor, pseudogene 100 |
| 13 | gene | 65.950589 | 65.958927 | positive | MGI_C57BL6J_5826062 | Gm46425 | NCBI_Gene:108168096 | MGI:5826062 | protein coding gene | predicted gene, 46425 |
| 13 | gene | 65.976343 | 65.990996 | negative | MGI_C57BL6J_5623872 | Gm40987 | NCBI_Gene:105245542,ENSEMBL:ENSMUSG00000113559 | MGI:5623872 | lncRNA gene | predicted gene, 40987 |
| 13 | pseudogene | 65.994038 | 65.995128 | positive | MGI_C57BL6J_6097617 | Gm48220 | ENSEMBL:ENSMUSG00000113653 | MGI:6097617 | pseudogene | predicted gene, 48220 |
| 13 | gene | 65.994049 | 65.995400 | negative | MGI_C57BL6J_6097618 | Gm48221 | ENSEMBL:ENSMUSG00000113173 | MGI:6097618 | lncRNA gene | predicted gene, 48221 |
| 13 | pseudogene | 66.007598 | 66.009319 | positive | MGI_C57BL6J_3648043 | Rybp-ps | NCBI_Gene:628746,ENSEMBL:ENSMUSG00000055763 | MGI:3648043 | pseudogene | RING1 and YY1 binding protein, pseudogene |
| 13 | gene | 66.032209 | 66.040247 | positive | MGI_C57BL6J_6097620 | Gm48222 | ENSEMBL:ENSMUSG00000113760 | MGI:6097620 | lncRNA gene | predicted gene, 48222 |
| 13 | pseudogene | 66.040567 | 66.042029 | positive | MGI_C57BL6J_5439383 | Gm21931 | NCBI_Gene:100534326 | MGI:5439383 | pseudogene | predicted gene, 21931 |
| 13 | gene | 66.113436 | 66.123487 | positive | MGI_C57BL6J_3642406 | Gm10324 | NCBI_Gene:628709,ENSEMBL:ENSMUSG00000095909 | MGI:3642406 | protein coding gene | predicted gene 10324 |
| 13 | gene | 66.119040 | 66.129693 | negative | MGI_C57BL6J_6324765 | Gm50489 | ENSEMBL:ENSMUSG00000118427 | MGI:6324765 | lncRNA gene | predicted gene, 50489 |
| 13 | gene | 66.141540 | 66.207074 | negative | MGI_C57BL6J_4937148 | Gm17514 | NCBI_Gene:100418283,ENSEMBL:ENSMUSG00000097118 | MGI:4937148 | lncRNA gene | predicted gene, 17514 |
| 13 | gene | 66.156616 | 66.165062 | positive | MGI_C57BL6J_5825602 | Gm45965 | NCBI_Gene:108167370,ENSEMBL:ENSMUSG00000113477 | MGI:5825602 | protein coding gene | predicted gene, 45965 |
| 13 | gene | 66.177347 | 66.178100 | positive | MGI_C57BL6J_6097622 | Gm48223 | ENSEMBL:ENSMUSG00000113156 | MGI:6097622 | lncRNA gene | predicted gene, 48223 |
| 13 | pseudogene | 66.192322 | 66.198556 | positive | MGI_C57BL6J_3761521 | Vmn2r-ps102 | NCBI_Gene:100041892,ENSEMBL:ENSMUSG00000113350 | MGI:3761521 | pseudogene | vomeronasal 2, receptor, pseudogene 102 |
| 13 | pseudogene | 66.221378 | 66.222470 | negative | MGI_C57BL6J_6097651 | Gm48240 | ENSEMBL:ENSMUSG00000113713 | MGI:6097651 | pseudogene | predicted gene, 48240 |
| 13 | gene | 66.227124 | 66.236670 | negative | MGI_C57BL6J_3704404 | Gm10772 | NCBI_Gene:105245577,ENSEMBL:ENSMUSG00000091347 | MGI:3704404 | protein coding gene | predicted gene 10772 |
| 13 | gene | 66.227497 | 66.227575 | negative | MGI_C57BL6J_4937083 | Gm17449 | NA | NA | protein coding gene | predicted gene%2c 17449 |
| 13 | pseudogene | 66.240777 | 66.240929 | positive | MGI_C57BL6J_6097652 | Gm48241 | ENSEMBL:ENSMUSG00000113253 | MGI:6097652 | pseudogene | predicted gene, 48241 |
| 13 | gene | 66.244270 | 66.451277 | positive | MGI_C57BL6J_5477248 | Gm26754 | ENSEMBL:ENSMUSG00000114616 | MGI:5477248 | lncRNA gene | predicted gene, 26754 |
| 13 | pseudogene | 66.249281 | 66.255539 | negative | MGI_C57BL6J_3761522 | Vmn2r-ps103 | NCBI_Gene:665974,ENSEMBL:ENSMUSG00000114027 | MGI:3761522 | pseudogene | vomeronasal 2, receptor, pseudogene 103 |
| 13 | pseudogene | 66.279919 | 66.280057 | negative | MGI_C57BL6J_6097657 | Gm48245 | ENSEMBL:ENSMUSG00000114239 | MGI:6097657 | pseudogene | predicted gene, 48245 |
| 13 | pseudogene | 66.280468 | 66.281580 | negative | MGI_C57BL6J_5434384 | Gm21029 | NCBI_Gene:100534324 | MGI:5434384 | pseudogene | predicted gene, 21029 |
| 13 | gene | 66.281136 | 66.284067 | negative | MGI_C57BL6J_5826063 | Gm46426 | NCBI_Gene:108168097,ENSEMBL:ENSMUSG00000114550 | MGI:5826063 | protein coding gene | predicted gene, 46426 |
| 13 | pseudogene | 66.308304 | 66.309296 | negative | MGI_C57BL6J_5010445 | Gm18260 | NCBI_Gene:100416799,ENSEMBL:ENSMUSG00000114776 | MGI:5010445 | pseudogene | predicted gene, 18260 |
| 13 | pseudogene | 66.334682 | 66.334879 | negative | MGI_C57BL6J_6097658 | Gm48246 | ENSEMBL:ENSMUSG00000114328 | MGI:6097658 | pseudogene | predicted gene, 48246 |
| 13 | pseudogene | 66.339451 | 66.340569 | negative | MGI_C57BL6J_5826032 | Gm46395 | NCBI_Gene:108168056 | MGI:5826032 | pseudogene | predicted gene, 46395 |
| 13 | pseudogene | 66.340586 | 66.342935 | negative | MGI_C57BL6J_1925949 | C330022B21Rik | ENSEMBL:ENSMUSG00000114729 | MGI:1925949 | pseudogene | RIKEN cDNA C330022B21 gene |
| 13 | gene | 66.349396 | 66.412059 | positive | MGI_C57BL6J_5477209 | Gm26715 | NCBI_Gene:102635928,ENSEMBL:ENSMUSG00000097659 | MGI:5477209 | lncRNA gene | predicted gene, 26715 |
| 13 | pseudogene | 66.350919 | 66.401873 | negative | MGI_C57BL6J_3761523 | Vmn2r-ps104 | NCBI_Gene:100041915,ENSEMBL:ENSMUSG00000094499 | MGI:3761523 | pseudogene | vomeronasal 2, receptor, pseudogene 104 |
| 13 | gene | 66.368462 | 66.752983 | negative | MGI_C57BL6J_3040710 | C330013J06Rik | NA | NA | unclassified gene | RIKEN cDNA C330013J06 gene |
| 13 | gene | 66.368764 | 66.370696 | negative | MGI_C57BL6J_5592202 | Gm33043 | NCBI_Gene:102635795 | MGI:5592202 | lncRNA gene | predicted gene, 33043 |
| 13 | pseudogene | 66.393128 | 66.393323 | negative | MGI_C57BL6J_6097659 | Gm48247 | ENSEMBL:ENSMUSG00000114377 | MGI:6097659 | pseudogene | predicted gene, 48247 |
| 13 | pseudogene | 66.396552 | 66.397532 | negative | MGI_C57BL6J_6097656 | Gm48244 | ENSEMBL:ENSMUSG00000114629 | MGI:6097656 | pseudogene | predicted gene, 48244 |
| 13 | pseudogene | 66.398531 | 66.399255 | positive | MGI_C57BL6J_5011304 | Gm19119 | NCBI_Gene:100418287,ENSEMBL:ENSMUSG00000114825 | MGI:5011304 | pseudogene | predicted gene, 19119 |
| 13 | pseudogene | 66.412482 | 66.412857 | positive | MGI_C57BL6J_6097655 | Gm48243 | ENSEMBL:ENSMUSG00000114261 | MGI:6097655 | pseudogene | predicted gene, 48243 |
| 13 | gene | 66.418114 | 66.441118 | negative | MGI_C57BL6J_1924053 | 2410141K09Rik | NCBI_Gene:76803,ENSEMBL:ENSMUSG00000074832 | MGI:1924053 | protein coding gene | RIKEN cDNA 2410141K09 gene |
| 13 | pseudogene | 66.454648 | 66.455354 | negative | MGI_C57BL6J_6097776 | Gm48322 | ENSEMBL:ENSMUSG00000114343 | MGI:6097776 | pseudogene | predicted gene, 48322 |
| 13 | pseudogene | 66.490947 | 66.491147 | positive | MGI_C57BL6J_6097777 | Gm48323 | ENSEMBL:ENSMUSG00000114406 | MGI:6097777 | pseudogene | predicted gene, 48323 |
| 13 | gene | 66.503160 | 66.519887 | negative | MGI_C57BL6J_5477300 | Gm26806 | ENSEMBL:ENSMUSG00000097483 | MGI:5477300 | lncRNA gene | predicted gene, 26806 |
| 13 | pseudogene | 66.512195 | 66.517704 | positive | MGI_C57BL6J_3040709 | Vmn2r-ps105 | NCBI_Gene:382770,ENSEMBL:ENSMUSG00000114684 | MGI:3040709 | pseudogene | vomeronasal 2, receptor, pseudogene 105 |
| 13 | gene | 66.519349 | 66.526631 | positive | MGI_C57BL6J_5826065 | Gm46428 | NCBI_Gene:108168099 | MGI:5826065 | lncRNA gene | predicted gene, 46428 |
| 13 | gene | 66.525979 | 66.529383 | positive | MGI_C57BL6J_5826064 | Gm46427 | NCBI_Gene:108168098,ENSEMBL:ENSMUSG00000114777 | MGI:5826064 | protein coding gene | predicted gene, 46427 |
| 13 | pseudogene | 66.534025 | 66.534222 | positive | MGI_C57BL6J_6097816 | Gm48351 | ENSEMBL:ENSMUSG00000114685 | MGI:6097816 | pseudogene | predicted gene, 48351 |
| 13 | pseudogene | 66.549713 | 66.551220 | negative | MGI_C57BL6J_6097829 | Gm48360 | ENSEMBL:ENSMUSG00000114434 | MGI:6097829 | pseudogene | predicted gene, 48360 |
| 13 | pseudogene | 66.565936 | 66.592059 | negative | MGI_C57BL6J_5011302 | Gm19117 | NCBI_Gene:100418284 | MGI:5011302 | pseudogene | predicted gene, 19117 |
| 13 | pseudogene | 66.565945 | 66.566038 | negative | MGI_C57BL6J_6155232 | Gm49531 | ENSEMBL:ENSMUSG00000114645 | MGI:6155232 | pseudogene | predicted gene, 49531 |
| 13 | pseudogene | 66.572921 | 66.575911 | positive | MGI_C57BL6J_3646733 | Gm10769 | NCBI_Gene:665874,ENSEMBL:ENSMUSG00000114426 | MGI:3646733 | pseudogene | predicted pseudogene 10769 |
| 13 | gene | 66.586810 | 66.586916 | negative | MGI_C57BL6J_5530883 | Gm27501 | ENSEMBL:ENSMUSG00000098452 | MGI:5530883 | miRNA gene | predicted gene, 27501 |
| 13 | gene | 66.589169 | 66.598733 | positive | MGI_C57BL6J_5825603 | Gm45966 | NCBI_Gene:108167371 | MGI:5825603 | protein coding gene | predicted gene, 45966 |
| 13 | gene | 66.589186 | 66.589285 | positive | MGI_C57BL6J_1925563 | 2410006F04Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 2410006F04 gene |
| 13 | pseudogene | 66.591913 | 66.592013 | negative | MGI_C57BL6J_6155233 | Gm49532 | ENSEMBL:ENSMUSG00000114262 | MGI:6155233 | pseudogene | predicted gene, 49532 |
| 13 | pseudogene | 66.621686 | 66.622905 | negative | MGI_C57BL6J_6097830 | Gm48361 | ENSEMBL:ENSMUSG00000114814 | MGI:6097830 | pseudogene | predicted gene, 48361 |
| 13 | gene | 66.633005 | 66.634986 | positive | MGI_C57BL6J_5826066 | Gm46429 | NCBI_Gene:108168100 | MGI:5826066 | lncRNA gene | predicted gene, 46429 |
| 13 | pseudogene | 66.668920 | 66.669864 | negative | MGI_C57BL6J_5010597 | Gm18412 | NCBI_Gene:100417129 | MGI:5010597 | pseudogene | predicted gene, 18412 |
| 13 | pseudogene | 66.668946 | 66.698243 | negative | MGI_C57BL6J_2678025 | Zfp640 | NCBI_Gene:102636110,ENSEMBL:ENSMUSG00000074830 | MGI:2678025 | pseudogene | zinc finger protein 640 |
| 13 | gene | 66.680954 | 66.711165 | positive | MGI_C57BL6J_5623873 | Gm40988 | NCBI_Gene:105245543,ENSEMBL:ENSMUSG00000114327 | MGI:5623873 | lncRNA gene | predicted gene, 40988 |
| 13 | pseudogene | 66.682839 | 66.698457 | negative | MGI_C57BL6J_3761525 | Vmn2r-ps106 | NCBI_Gene:100041938,ENSEMBL:ENSMUSG00000114549 | MGI:3761525 | pseudogene | vomeronasal 2, receptor, pseudogene 106 |
| 13 | gene | 66.738371 | 66.753012 | negative | MGI_C57BL6J_5623874 | Gm40989 | NCBI_Gene:105245544,ENSEMBL:ENSMUSG00000114839 | MGI:5623874 | lncRNA gene | predicted gene, 40989 |
| 13 | pseudogene | 66.756334 | 66.756486 | negative | MGI_C57BL6J_6097893 | Gm48404 | ENSEMBL:ENSMUSG00000114423 | MGI:6097893 | pseudogene | predicted gene, 48404 |
| 13 | gene | 66.777441 | 66.779142 | positive | MGI_C57BL6J_6097904 | Gm48412 | ENSEMBL:ENSMUSG00000114226 | MGI:6097904 | lncRNA gene | predicted gene, 48412 |
| 13 | gene | 66.792070 | 66.802563 | negative | MGI_C57BL6J_3779770 | Gm7896 | NCBI_Gene:105245578,ENSEMBL:ENSMUSG00000114353 | MGI:3779770 | protein coding gene | predicted gene 7869 |
| 13 | pseudogene | 66.799297 | 66.799390 | positive | MGI_C57BL6J_6097906 | Gm48413 | ENSEMBL:ENSMUSG00000114314 | MGI:6097906 | pseudogene | predicted gene, 48413 |
| 13 | pseudogene | 66.808949 | 66.815219 | negative | MGI_C57BL6J_3761527 | Vmn2r-ps108 | NCBI_Gene:100041947,ENSEMBL:ENSMUSG00000117784 | MGI:3761527 | pseudogene | vomeronasal 2, receptor, pseudogene 108 |
| 13 | pseudogene | 66.845824 | 66.846036 | negative | MGI_C57BL6J_6097907 | Gm48414 | ENSEMBL:ENSMUSG00000114793 | MGI:6097907 | pseudogene | predicted gene, 48414 |
| 13 | pseudogene | 66.849651 | 66.850772 | negative | MGI_C57BL6J_5621314 | Gm38429 | NCBI_Gene:100534325 | MGI:5621314 | pseudogene | predicted gene, 38429 |
| 13 | gene | 66.849651 | 66.862122 | negative | MGI_C57BL6J_3809062 | Gm10323 | NCBI_Gene:105245545,ENSEMBL:ENSMUSG00000114816 | MGI:3809062 | protein coding gene | predicted gene 10323 |
| 13 | pseudogene | 66.864444 | 66.866167 | positive | MGI_C57BL6J_3645820 | Cbx3-ps4 | NCBI_Gene:382769,ENSEMBL:ENSMUSG00000071294 | MGI:3645820 | pseudogene | chromobox 3, pseudogene 4 |
| 13 | pseudogene | 66.869457 | 66.873358 | negative | MGI_C57BL6J_3761528 | Vmn2r-ps109 | NCBI_Gene:666052,ENSEMBL:ENSMUSG00000114489 | MGI:3761528 | pseudogene | vomeronasal 2, receptor, pseudogene 109 |
| 13 | pseudogene | 66.874063 | 66.874689 | positive | MGI_C57BL6J_3643790 | Gm5451 | NCBI_Gene:108168114,ENSEMBL:ENSMUSG00000095847 | MGI:3643790 | pseudogene | predicted gene 5451 |
| 13 | pseudogene | 66.875088 | 66.875710 | negative | MGI_C57BL6J_3644791 | Gm7911 | NCBI_Gene:666061,ENSEMBL:ENSMUSG00000114374 | MGI:3644791 | pseudogene | predicted gene 7911 |
| 13 | gene | 66.900617 | 66.905378 | negative | MGI_C57BL6J_1914780 | Uqcrb | NCBI_Gene:67530,ENSEMBL:ENSMUSG00000021520 | MGI:1914780 | protein coding gene | ubiquinol-cytochrome c reductase binding protein |
| 13 | gene | 66.904914 | 66.909236 | positive | MGI_C57BL6J_3642136 | Gm10767 | NCBI_Gene:100038538,ENSEMBL:ENSMUSG00000074826 | MGI:3642136 | protein coding gene | predicted gene 10767 |
| 13 | gene | 66.905456 | 66.933088 | negative | MGI_C57BL6J_1913660 | Mterf3 | NCBI_Gene:66410,ENSEMBL:ENSMUSG00000021519 | MGI:1913660 | protein coding gene | mitochondrial transcription termination factor 3 |
| 13 | gene | 66.932830 | 66.998401 | positive | MGI_C57BL6J_1276575 | Ptdss1 | NCBI_Gene:19210,ENSEMBL:ENSMUSG00000021518 | MGI:1276575 | protein coding gene | phosphatidylserine synthase 1 |
| 13 | gene | 66.996818 | 67.032408 | negative | MGI_C57BL6J_1918476 | 4933433G19Rik | NCBI_Gene:71226,ENSEMBL:ENSMUSG00000114362 | MGI:1918476 | lncRNA gene | RIKEN cDNA 4933433G19 gene |
| 13 | gene | 67.038594 | 67.061170 | negative | MGI_C57BL6J_1925501 | Zfp712 | NCBI_Gene:78251,ENSEMBL:ENSMUSG00000090641 | MGI:1925501 | protein coding gene | zinc finger protein 712 |
| 13 | pseudogene | 67.050735 | 67.051025 | positive | MGI_C57BL6J_6098346 | Gm48705 | ENSEMBL:ENSMUSG00000114834 | MGI:6098346 | pseudogene | predicted gene, 48705 |
| 13 | gene | 67.060505 | 67.092042 | positive | MGI_C57BL6J_5826077 | Gm46440 | NCBI_Gene:108168115,ENSEMBL:ENSMUSG00000114516 | MGI:5826077 | lncRNA gene | predicted gene, 46440 |
| 13 | gene | 67.069398 | 67.097976 | negative | MGI_C57BL6J_3040674 | Zfp708 | NCBI_Gene:432769,ENSEMBL:ENSMUSG00000058883 | MGI:3040674 | protein coding gene | zinc finger protein 708 |
| 13 | gene | 67.069399 | 67.108604 | negative | MGI_C57BL6J_5579263 | Gm28557 | ENSEMBL:ENSMUSG00000100235 | MGI:5579263 | protein coding gene | predicted gene 28557 |
| 13 | pseudogene | 67.089475 | 67.090225 | positive | MGI_C57BL6J_5010123 | Gm17938 | NCBI_Gene:100416142,ENSEMBL:ENSMUSG00000114603 | MGI:5010123 | pseudogene | predicted gene, 17938 |
| 13 | gene | 67.096613 | 67.116263 | negative | MGI_C57BL6J_5433745 | Rslcan18 | NCBI_Gene:432770,ENSEMBL:ENSMUSG00000074824 | MGI:5433745 | protein coding gene | regulator of sex-limitation candidate 18 |
| 13 | gene | 67.121660 | 67.142404 | positive | MGI_C57BL6J_2446280 | Zfp759 | NCBI_Gene:268670,ENSEMBL:ENSMUSG00000057396 | MGI:2446280 | protein coding gene | zinc finger protein 759 |
| 13 | gene | 67.143672 | 67.145366 | negative | MGI_C57BL6J_6098394 | Gm48732 | ENSEMBL:ENSMUSG00000114568 | MGI:6098394 | lncRNA gene | predicted gene, 48732 |
| 13 | pseudogene | 67.167781 | 67.168778 | positive | MGI_C57BL6J_6098396 | Gm48733 | ENSEMBL:ENSMUSG00000114677 | MGI:6098396 | pseudogene | predicted gene, 48733 |
| 13 | gene | 67.173182 | 67.184044 | positive | MGI_C57BL6J_3044162 | Rsl1 | NCBI_Gene:380855,ENSEMBL:ENSMUSG00000058900 | MGI:3044162 | protein coding gene | regulator of sex limited protein 1 |
| 13 | pseudogene | 67.185379 | 67.189778 | positive | MGI_C57BL6J_3780034 | Gm9626 | NCBI_Gene:102636462,ENSEMBL:ENSMUSG00000114805 | MGI:3780034 | pseudogene | predicted gene 9626 |
| 13 | pseudogene | 67.187098 | 67.187319 | positive | MGI_C57BL6J_6215078 | Gm49646 | ENSEMBL:ENSMUSG00000114870 | MGI:6215078 | pseudogene | predicted gene, 49646 |
| 13 | gene | 67.194506 | 67.209298 | positive | MGI_C57BL6J_3040708 | Zfp455 | NCBI_Gene:218311,ENSEMBL:ENSMUSG00000051037 | MGI:3040708 | protein coding gene | zinc finger protein 455 |
| 13 | gene | 67.225137 | 67.278466 | negative | MGI_C57BL6J_3040691 | Zfp458 | NCBI_Gene:238690,ENSEMBL:ENSMUSG00000055480 | MGI:3040691 | protein coding gene | zinc finger protein 458 |
| 13 | gene | 67.226997 | 67.229969 | negative | MGI_C57BL6J_6118443 | Gm49064 | ENSEMBL:ENSMUSG00000114771 | MGI:6118443 | lncRNA gene | predicted gene, 49064 |
| 13 | gene | 67.278577 | 67.283361 | positive | MGI_C57BL6J_3642133 | F630042J09Rik | NCBI_Gene:100038627,ENSEMBL:ENSMUSG00000114442 | MGI:3642133 | lncRNA gene | RIKEN cDNA F630042J09 gene |
| 13 | gene | 67.288137 | 67.306485 | negative | MGI_C57BL6J_2664334 | Zfp457 | NCBI_Gene:431706,ENSEMBL:ENSMUSG00000055341 | MGI:2664334 | protein coding gene | zinc finger protein 457 |
| 13 | gene | 67.307334 | 67.308907 | negative | MGI_C57BL6J_6098544 | Gm48824 | ENSEMBL:ENSMUSG00000114863 | MGI:6098544 | lncRNA gene | predicted gene, 48824 |
| 13 | gene | 67.312998 | 67.342947 | negative | MGI_C57BL6J_3040707 | Zfp595 | NCBI_Gene:218314,ENSEMBL:ENSMUSG00000057842 | MGI:3040707 | protein coding gene | zinc finger protein 595 |
| 13 | gene | 67.317942 | 67.360536 | negative | MGI_C57BL6J_5547780 | Gm28044 | ENSEMBL:ENSMUSG00000098781 | MGI:5547780 | protein coding gene | predicted gene, 28044 |
| 13 | gene | 67.339306 | 67.360605 | negative | MGI_C57BL6J_3612873 | Zfp953 | NCBI_Gene:629016,ENSEMBL:ENSMUSG00000098905 | MGI:3612873 | protein coding gene | zinc finger protein 953 |
| 13 | gene | 67.345246 | 67.375733 | negative | MGI_C57BL6J_5547777 | Gm28041 | ENSEMBL:ENSMUSG00000098692 | MGI:5547777 | protein coding gene | predicted gene, 28041 |
| 13 | gene | 67.360671 | 67.365682 | positive | MGI_C57BL6J_4937866 | Gm17039 | NCBI_Gene:102636903,ENSEMBL:ENSMUSG00000092103 | MGI:4937866 | lncRNA gene | predicted gene 17039 |
| 13 | gene | 67.362113 | 67.375810 | negative | MGI_C57BL6J_3040694 | Zfp456 | NCBI_Gene:408065,ENSEMBL:ENSMUSG00000078995 | MGI:3040694 | protein coding gene | zinc finger protein 456 |
| 13 | gene | 67.387905 | 67.399819 | negative | MGI_C57BL6J_1920057 | Zfp429 | NCBI_Gene:72807,ENSEMBL:ENSMUSG00000078994 | MGI:1920057 | protein coding gene | zinc finger protein 429 |
| 13 | gene | 67.390475 | 67.408858 | positive | MGI_C57BL6J_6098667 | Gm48900 | ENSEMBL:ENSMUSG00000114726 | MGI:6098667 | lncRNA gene | predicted gene, 48900 |
| 13 | gene | 67.405033 | 67.440341 | negative | MGI_C57BL6J_3040701 | Zfp459 | NCBI_Gene:328274,ENSEMBL:ENSMUSG00000055560 | MGI:3040701 | protein coding gene | zinc finger protein 459 |
| 13 | gene | 67.421442 | 67.424481 | positive | MGI_C57BL6J_5592946 | Gm33787 | NCBI_Gene:102636824 | MGI:5592946 | lncRNA gene | predicted gene, 33787 |
| 13 | gene | 67.424549 | 67.451624 | negative | MGI_C57BL6J_3040703 | Zfp874a | NCBI_Gene:238692,ENSEMBL:ENSMUSG00000069206 | MGI:3040703 | protein coding gene | zinc finger protein 874a |
| 13 | gene | 67.451658 | 67.453497 | positive | MGI_C57BL6J_5611535 | Gm38307 | ENSEMBL:ENSMUSG00000103398 | MGI:5611535 | unclassified gene | predicted gene, 38307 |
| 13 | pseudogene | 67.459668 | 67.460765 | positive | MGI_C57BL6J_3646067 | Gm7928 | NCBI_Gene:666098,ENSEMBL:ENSMUSG00000097244 | MGI:3646067 | pseudogene | predicted gene 7928 |
| 13 | gene | 67.471513 | 67.486241 | negative | MGI_C57BL6J_3040702 | Zfp874b | NCBI_Gene:408067,ENSEMBL:ENSMUSG00000059839 | MGI:3040702 | protein coding gene | zinc finger protein 874b |
| 13 | gene | 67.472478 | 67.473500 | positive | MGI_C57BL6J_6098665 | Gm48899 | ENSEMBL:ENSMUSG00000114788 | MGI:6098665 | lncRNA gene | predicted gene, 48899 |
| 13 | gene | 67.490167 | 67.500555 | negative | MGI_C57BL6J_99205 | Zfp58 | NCBI_Gene:238693,ENSEMBL:ENSMUSG00000071291 | MGI:99205 | protein coding gene | zinc finger protein 58 |
| 13 | gene | 67.491152 | 67.526151 | negative | MGI_C57BL6J_5504080 | Gm26965 | ENSEMBL:ENSMUSG00000097565 | MGI:5504080 | protein coding gene | predicted gene, 26965 |
| 13 | pseudogene | 67.505524 | 67.506446 | negative | MGI_C57BL6J_5010446 | Gm18261 | NCBI_Gene:100416800 | MGI:5010446 | pseudogene | predicted gene, 18261 |
| 13 | gene | 67.515781 | 67.526231 | negative | MGI_C57BL6J_107768 | Zfp87 | NCBI_Gene:170763,ENSEMBL:ENSMUSG00000097333 | MGI:107768 | protein coding gene | zinc finger protein 87 |
| 13 | gene | 67.538641 | 67.553830 | negative | MGI_C57BL6J_1916455 | Zfp748 | NCBI_Gene:212276,ENSEMBL:ENSMUSG00000095432 | MGI:1916455 | protein coding gene | zinc finger protein 748 |
| 13 | gene | 67.553035 | 67.570892 | positive | MGI_C57BL6J_1924606 | 9430065F17Rik | NCBI_Gene:77857,ENSEMBL:ENSMUSG00000097360 | MGI:1924606 | lncRNA gene | RIKEN cDNA 9430065F17 gene |
| 13 | gene | 67.589439 | 67.609707 | negative | MGI_C57BL6J_2145180 | Zfp729b | NCBI_Gene:100416706,ENSEMBL:ENSMUSG00000058093 | MGI:2145180 | protein coding gene | zinc finger protein 729b |
| 13 | gene | 67.592159 | 67.637769 | negative | MGI_C57BL6J_6121541 | Gm49345 | ENSEMBL:ENSMUSG00000114923 | MGI:6121541 | protein coding gene | predicted gene, 49345 |
| 13 | gene | 67.612763 | 67.638063 | negative | MGI_C57BL6J_3036250 | Zfp729a | NCBI_Gene:212281,ENSEMBL:ENSMUSG00000021510 | MGI:3036250 | protein coding gene | zinc finger protein 729a |
| 13 | pseudogene | 67.642545 | 67.646495 | positive | MGI_C57BL6J_5826076 | Gm46439 | NCBI_Gene:108168113 | MGI:5826076 | pseudogene | predicted gene, 46439 |
| 13 | gene | 67.658669 | 67.667943 | positive | MGI_C57BL6J_6097442 | Gm48095 | ENSEMBL:ENSMUSG00000114264 | MGI:6097442 | lncRNA gene | predicted gene, 48095 |
| 13 | gene | 67.658685 | 67.687071 | negative | MGI_C57BL6J_3040706 | Zfp738 | NCBI_Gene:408068,ENSEMBL:ENSMUSG00000048280 | MGI:3040706 | protein coding gene | zinc finger protein 738 |
| 13 | pseudogene | 67.660457 | 67.661445 | positive | MGI_C57BL6J_5477081 | Gm26587 | ENSEMBL:ENSMUSG00000097470 | MGI:5477081 | pseudogene | predicted gene, 26587 |
| 13 | pseudogene | 67.699082 | 67.700031 | positive | MGI_C57BL6J_5477369 | Gm26875 | ENSEMBL:ENSMUSG00000097371 | MGI:5477369 | pseudogene | predicted gene, 26875 |
| 13 | gene | 67.705306 | 67.729173 | negative | MGI_C57BL6J_107769 | Zfp65 | NCBI_Gene:235907,ENSEMBL:ENSMUSG00000071281 | MGI:107769 | protein coding gene | zinc finger protein 65 |
| 13 | pseudogene | 67.725060 | 67.725438 | negative | MGI_C57BL6J_6098658 | Gm48894 | ENSEMBL:ENSMUSG00000114887 | MGI:6098658 | pseudogene | predicted gene, 48894 |
| 13 | gene | 67.729262 | 67.756838 | positive | MGI_C57BL6J_1915521 | Zfp85os | NCBI_Gene:68271,ENSEMBL:ENSMUSG00000044081 | MGI:1915521 | antisense lncRNA gene | zinc finger protein 85, opposite strand |
| 13 | gene | 67.747800 | 67.755228 | negative | MGI_C57BL6J_107767 | Zfp85 | NCBI_Gene:22746,ENSEMBL:ENSMUSG00000058331 | MGI:107767 | protein coding gene | zinc finger protein 85 |
| 13 | pseudogene | 67.762714 | 67.773451 | negative | MGI_C57BL6J_3642213 | Gm9894 | NCBI_Gene:100417934,ENSEMBL:ENSMUSG00000052909 | MGI:3642213 | pseudogene | predicted gene 9894 |
| 13 | gene | 67.779693 | 67.792512 | positive | MGI_C57BL6J_1920208 | Zfp493 | NCBI_Gene:72958,ENSEMBL:ENSMUSG00000090659 | MGI:1920208 | protein coding gene | zinc finger protein 493 |
| 13 | gene | 67.796594 | 67.830985 | negative | MGI_C57BL6J_2443505 | 4930525G20Rik | NCBI_Gene:320163,ENSEMBL:ENSMUSG00000097276 | MGI:2443505 | lncRNA gene | RIKEN cDNA 4930525G20 gene |
| 13 | pseudogene | 67.810172 | 67.811271 | positive | MGI_C57BL6J_3780033 | Gm9625 | NCBI_Gene:674597,ENSEMBL:ENSMUSG00000097906 | MGI:3780033 | pseudogene | predicted gene 9625 |
| 13 | gene | 67.813740 | 67.827002 | positive | MGI_C57BL6J_3036278 | Zfp273 | NCBI_Gene:212569,ENSEMBL:ENSMUSG00000030446 | MGI:3036278 | protein coding gene | zinc finger protein 273 |
| 13 | gene | 67.831036 | 67.844879 | positive | MGI_C57BL6J_3645096 | Gm10037 | NCBI_Gene:102637366,ENSEMBL:ENSMUSG00000058246 | MGI:3645096 | protein coding gene | predicted gene 10037 |
| 13 | gene | 67.863326 | 67.863925 | positive | MGI_C57BL6J_3040680 | BC048507 | NCBI_Gene:408058,ENSEMBL:ENSMUSG00000064063 | MGI:3040680 | protein coding gene | cDNA sequence BC048507 |
| 13 | gene | 67.885454 | 67.889982 | negative | MGI_C57BL6J_5623876 | Gm40991 | NCBI_Gene:105245547 | MGI:5623876 | protein coding gene | predicted gene, 40991 |
| 13 | gene | 67.949751 | 67.954507 | positive | MGI_C57BL6J_3779771 | Gm7969 | NCBI_Gene:666185,ENSEMBL:ENSMUSG00000113095 | MGI:3779771 | lncRNA gene | predicted gene 7969 |
| 13 | gene | 67.965483 | 67.970239 | positive | MGI_C57BL6J_6097912 | Gm48419 | ENSEMBL:ENSMUSG00000112984 | MGI:6097912 | lncRNA gene | predicted gene, 48419 |
| 13 | gene | 67.981215 | 67.985971 | positive | MGI_C57BL6J_6097920 | Gm48423 | ENSEMBL:ENSMUSG00000113273 | MGI:6097920 | lncRNA gene | predicted gene, 48423 |
| 13 | pseudogene | 68.060456 | 68.060608 | negative | MGI_C57BL6J_6097922 | Gm48424 | ENSEMBL:ENSMUSG00000113365 | MGI:6097922 | pseudogene | predicted gene, 48424 |
| 13 | gene | 68.181294 | 68.183102 | positive | MGI_C57BL6J_2142939 | AA414992 | NCBI_Gene:102402,ENSEMBL:ENSMUSG00000113342 | MGI:2142939 | unclassified gene | expressed sequence AA414992 |
| 13 | gene | 68.228432 | 68.234731 | negative | MGI_C57BL6J_1932124 | Urml | NCBI_Gene:101056219 | MGI:1932124 | lncRNA gene | up-regulated in Myc liver |
| 13 | gene | 68.353364 | 68.356904 | negative | MGI_C57BL6J_3645625 | Gm8016 | NCBI_Gene:666272,ENSEMBL:ENSMUSG00000113050 | MGI:3645625 | lncRNA gene | predicted gene 8016 |
| 13 | gene | 68.444523 | 68.447939 | negative | MGI_C57BL6J_5826023 | Gm46386 | NCBI_Gene:108168045 | MGI:5826023 | protein coding gene | predicted gene, 46386 |
| 13 | pseudogene | 68.469680 | 68.469832 | negative | MGI_C57BL6J_6097930 | Gm48429 | ENSEMBL:ENSMUSG00000113382 | MGI:6097930 | pseudogene | predicted gene, 48429 |
| 13 | gene | 68.488998 | 68.491389 | negative | MGI_C57BL6J_6097940 | Gm48436 | ENSEMBL:ENSMUSG00000113394 | MGI:6097940 | lncRNA gene | predicted gene, 48436 |
| 13 | gene | 68.560780 | 68.582169 | negative | MGI_C57BL6J_1891037 | Mtrr | NCBI_Gene:210009,ENSEMBL:ENSMUSG00000034617 | MGI:1891037 | protein coding gene | 5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| 13 | gene | 68.582100 | 68.592338 | positive | MGI_C57BL6J_1916827 | Fastkd3 | NCBI_Gene:69577,ENSEMBL:ENSMUSG00000021532 | MGI:1916827 | protein coding gene | FAST kinase domains 3 |
| 13 | gene | 68.590481 | 68.594722 | negative | MGI_C57BL6J_6098110 | Gm48556 | ENSEMBL:ENSMUSG00000112985 | MGI:6098110 | lncRNA gene | predicted gene, 48556 |
| 13 | gene | 68.597421 | 68.614231 | positive | MGI_C57BL6J_1916565 | 1700001L19Rik | NCBI_Gene:69315,ENSEMBL:ENSMUSG00000021534 | MGI:1916565 | protein coding gene | RIKEN cDNA 1700001L19 gene |
| 13 | gene | 68.601553 | 68.603793 | negative | MGI_C57BL6J_5594062 | Gm34903 | NCBI_Gene:102638309 | MGI:5594062 | lncRNA gene | predicted gene, 34903 |
| 13 | gene | 68.620043 | 68.999541 | negative | MGI_C57BL6J_99676 | Adcy2 | NCBI_Gene:210044,ENSEMBL:ENSMUSG00000021536 | MGI:99676 | protein coding gene | adenylate cyclase 2 |
| 13 | gene | 68.657460 | 68.665243 | positive | MGI_C57BL6J_5594152 | Gm34993 | NCBI_Gene:102638422 | MGI:5594152 | lncRNA gene | predicted gene, 34993 |
| 13 | pseudogene | 68.810080 | 68.810832 | positive | MGI_C57BL6J_5504044 | Gm26929 | ENSEMBL:ENSMUSG00000098242 | MGI:5504044 | pseudogene | predicted gene, 26929 |
| 13 | pseudogene | 68.856483 | 68.857615 | positive | MGI_C57BL6J_3648893 | Gm7979 | NCBI_Gene:666205,ENSEMBL:ENSMUSG00000113859 | MGI:3648893 | pseudogene | predicted gene 7979 |
| 13 | gene | 68.995537 | 68.998019 | negative | MGI_C57BL6J_6098166 | Gm48593 | ENSEMBL:ENSMUSG00000113505 | MGI:6098166 | unclassified gene | predicted gene, 48593 |
| 13 | gene | 68.998793 | 69.031555 | positive | MGI_C57BL6J_5594320 | Gm35161 | NCBI_Gene:102638647,ENSEMBL:ENSMUSG00000113121 | MGI:5594320 | lncRNA gene | predicted gene, 35161 |
| 13 | gene | 69.005920 | 69.070001 | negative | MGI_C57BL6J_5594208 | Gm35049 | NCBI_Gene:102638499 | MGI:5594208 | lncRNA gene | predicted gene, 35049 |
| 13 | gene | 69.016622 | 69.018188 | positive | MGI_C57BL6J_5610350 | Gm37122 | ENSEMBL:ENSMUSG00000103035 | MGI:5610350 | unclassified gene | predicted gene, 37122 |
| 13 | gene | 69.141218 | 69.143965 | positive | MGI_C57BL6J_5477338 | Gm26844 | NCBI_Gene:102638947,ENSEMBL:ENSMUSG00000097522 | MGI:5477338 | lncRNA gene | predicted gene, 26844 |
| 13 | gene | 69.275394 | 69.291978 | negative | MGI_C57BL6J_5594603 | Gm35444 | NCBI_Gene:102639031 | MGI:5594603 | lncRNA gene | predicted gene, 35444 |
| 13 | pseudogene | 69.345655 | 69.346647 | positive | MGI_C57BL6J_3779437 | Gm4812 | NCBI_Gene:102640696,ENSEMBL:ENSMUSG00000098227 | MGI:3779437 | pseudogene | predicted gene 4812 |
| 13 | gene | 69.395547 | 69.397565 | positive | MGI_C57BL6J_5594673 | Gm35514 | NCBI_Gene:102639128,ENSEMBL:ENSMUSG00000113595 | MGI:5594673 | lncRNA gene | predicted gene, 35514 |
| 13 | gene | 69.401248 | 69.401397 | negative | MGI_C57BL6J_5690767 | Gm44375 | ENSEMBL:ENSMUSG00000106556 | MGI:5690767 | miRNA gene | predicted gene, 44375 |
| 13 | gene | 69.497952 | 69.534617 | negative | MGI_C57BL6J_2682295 | Tent4a | NCBI_Gene:210106,ENSEMBL:ENSMUSG00000034575 | MGI:2682295 | protein coding gene | terminal nucleotidyltransferase 4A |
| 13 | pseudogene | 69.514000 | 69.514176 | positive | MGI_C57BL6J_6098296 | Gm48676 | ENSEMBL:ENSMUSG00000113486 | MGI:6098296 | pseudogene | predicted gene, 48676 |
| 13 | gene | 69.533746 | 69.635780 | positive | MGI_C57BL6J_107252 | Nsun2 | NCBI_Gene:28114,ENSEMBL:ENSMUSG00000021595 | MGI:107252 | protein coding gene | NOL1/NOP2/Sun domain family member 2 |
| 13 | gene | 69.534549 | 69.535451 | positive | MGI_C57BL6J_1925067 | A930018O16Rik | NCBI_Gene:105245580 | MGI:1925067 | protein coding gene | RIKEN cDNA A930018O16 gene |
| 13 | gene | 69.543046 | 69.544074 | negative | MGI_C57BL6J_2685376 | A530095I07Rik | NCBI_Gene:210373,ENSEMBL:ENSMUSG00000051805 | MGI:2685376 | lncRNA gene | RIKEN cDNA A530095I07 gene |
| 13 | gene | 69.548557 | 69.553730 | negative | MGI_C57BL6J_5623877 | Gm40992 | NCBI_Gene:105245548 | MGI:5623877 | lncRNA gene | predicted gene, 40992 |
| 13 | pseudogene | 69.548956 | 69.549762 | positive | MGI_C57BL6J_5011067 | Gm18882 | NCBI_Gene:100417887 | MGI:5011067 | pseudogene | predicted gene, 18882 |
| 13 | gene | 69.573449 | 69.611907 | negative | MGI_C57BL6J_98400 | Srd5a1 | NCBI_Gene:78925,ENSEMBL:ENSMUSG00000021594 | MGI:98400 | protein coding gene | steroid 5 alpha-reductase 1 |
| 13 | gene | 69.610498 | 69.614901 | negative | MGI_C57BL6J_4937935 | Gm17108 | ENSEMBL:ENSMUSG00000091133 | MGI:4937935 | lncRNA gene | predicted gene 17108 |
| 13 | pseudogene | 69.654773 | 69.655521 | negative | MGI_C57BL6J_5010057 | Gm17872 | NCBI_Gene:100416014,ENSEMBL:ENSMUSG00000113790 | MGI:5010057 | pseudogene | predicted gene, 17872 |
| 13 | pseudogene | 69.662055 | 69.662420 | positive | MGI_C57BL6J_6098531 | Gm48816 | ENSEMBL:ENSMUSG00000113635 | MGI:6098531 | pseudogene | predicted gene, 48816 |
| 13 | pseudogene | 69.664483 | 69.665833 | negative | MGI_C57BL6J_6098532 | Gm48817 | ENSEMBL:ENSMUSG00000114037 | MGI:6098532 | pseudogene | predicted gene, 48817 |
| 13 | pseudogene | 69.666718 | 69.668944 | negative | MGI_C57BL6J_5010498 | Gm18313 | NCBI_Gene:100416910,ENSEMBL:ENSMUSG00000114171 | MGI:5010498 | pseudogene | predicted gene, 18313 |
| 13 | gene | 69.680046 | 69.697777 | negative | MGI_C57BL6J_5594777 | Gm35618 | NCBI_Gene:102639267,ENSEMBL:ENSMUSG00000113299 | MGI:5594777 | lncRNA gene | predicted gene, 35618 |
| 13 | gene | 69.681550 | 69.687492 | positive | MGI_C57BL6J_5623879 | Gm40994 | NCBI_Gene:105245550 | MGI:5623879 | lncRNA gene | predicted gene, 40994 |
| 13 | gene | 69.688078 | 69.695153 | positive | MGI_C57BL6J_5623878 | Gm40993 | NCBI_Gene:105245549 | MGI:5623878 | lncRNA gene | predicted gene, 40993 |
| 13 | gene | 69.697811 | 69.700895 | positive | MGI_C57BL6J_6098534 | Gm48819 | ENSEMBL:ENSMUSG00000113324 | MGI:6098534 | lncRNA gene | predicted gene, 48819 |
| 13 | gene | 69.702832 | 69.739897 | negative | MGI_C57BL6J_1924230 | Ube2ql1 | NCBI_Gene:76980,ENSEMBL:ENSMUSG00000052981 | MGI:1924230 | protein coding gene | ubiquitin-conjugating enzyme E2Q family-like 1 |
| 13 | gene | 69.771518 | 69.772714 | negative | MGI_C57BL6J_1922528 | 4930547H16Rik | NCBI_Gene:75278,ENSEMBL:ENSMUSG00000112957 | MGI:1922528 | lncRNA gene | RIKEN cDNA 4930547H16 gene |
| 13 | pseudogene | 69.775436 | 69.775619 | positive | MGI_C57BL6J_5012473 | Gm20288 | NCBI_Gene:102639453,ENSEMBL:ENSMUSG00000113639 | MGI:5012473 | pseudogene | predicted gene, 20288 |
| 13 | gene | 69.802347 | 69.808825 | positive | MGI_C57BL6J_1916660 | 1700025O18Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 1700025O18 gene |
| 13 | gene | 69.802348 | 69.816104 | positive | MGI_C57BL6J_106331 | Med10 | NCBI_Gene:28077,ENSEMBL:ENSMUSG00000021598 | MGI:106331 | protein coding gene | mediator complex subunit 10 |
| 13 | gene | 69.809914 | 69.816094 | negative | MGI_C57BL6J_5477313 | Gm26819 | ENSEMBL:ENSMUSG00000097033 | MGI:5477313 | lncRNA gene | predicted gene, 26819 |
| 13 | gene | 69.957962 | 69.963437 | positive | MGI_C57BL6J_1914800 | 4933416O17Rik | NCBI_Gene:67550,ENSEMBL:ENSMUSG00000113802 | MGI:1914800 | lncRNA gene | RIKEN cDNA 4933416O17 gene |
| 13 | gene | 69.983596 | 69.996461 | positive | MGI_C57BL6J_5595178 | Gm36019 | NCBI_Gene:102639789 | MGI:5595178 | lncRNA gene | predicted gene, 36019 |
| 13 | gene | 70.004050 | 70.028348 | negative | MGI_C57BL6J_1921530 | 1700084F23Rik | NCBI_Gene:74280,ENSEMBL:ENSMUSG00000099639 | MGI:1921530 | lncRNA gene | RIKEN cDNA 1700084F23 gene |
| 13 | pseudogene | 70.007231 | 70.007614 | negative | MGI_C57BL6J_6096739 | Gm47655 | ENSEMBL:ENSMUSG00000113980 | MGI:6096739 | pseudogene | predicted gene, 47655 |
| 13 | gene | 70.021959 | 70.119949 | positive | MGI_C57BL6J_3781946 | Gm3772 | NCBI_Gene:100042291,ENSEMBL:ENSMUSG00000113074 | MGI:3781946 | lncRNA gene | predicted gene 3772 |
| 13 | gene | 70.061558 | 70.063922 | positive | MGI_C57BL6J_6096740 | Gm47656 | ENSEMBL:ENSMUSG00000112998 | MGI:6096740 | unclassified gene | predicted gene, 47656 |
| 13 | gene | 70.149800 | 70.205972 | positive | MGI_C57BL6J_2145342 | AU017674 | ENSEMBL:ENSMUSG00000113573 | MGI:2145342 | lncRNA gene | expressed sequence AU017674 |
| 13 | gene | 70.168832 | 70.168959 | positive | MGI_C57BL6J_5455795 | Gm26018 | ENSEMBL:ENSMUSG00000077594 | MGI:5455795 | snoRNA gene | predicted gene, 26018 |
| 13 | gene | 70.232822 | 70.248122 | positive | MGI_C57BL6J_1922336 | 4930520P13Rik | NCBI_Gene:75086,ENSEMBL:ENSMUSG00000113463 | MGI:1922336 | lncRNA gene | RIKEN cDNA 4930520P13 gene |
| 13 | gene | 70.278705 | 70.290082 | positive | MGI_C57BL6J_5595375 | Gm36216 | NCBI_Gene:102640057 | MGI:5595375 | lncRNA gene | predicted gene, 36216 |
| 13 | pseudogene | 70.422491 | 70.422781 | negative | MGI_C57BL6J_6096824 | Gm47706 | ENSEMBL:ENSMUSG00000114054 | MGI:6096824 | pseudogene | predicted gene, 47706 |
| 13 | gene | 70.550859 | 70.574289 | negative | MGI_C57BL6J_5595426 | Gm36267 | NCBI_Gene:102640125 | MGI:5595426 | lncRNA gene | predicted gene, 36267 |
| 13 | gene | 70.551707 | 70.637840 | negative | MGI_C57BL6J_2385865 | Ice1 | NCBI_Gene:218333,ENSEMBL:ENSMUSG00000034525 | MGI:2385865 | protein coding gene | interactor of little elongation complex ELL subunit 1 |
| 13 | gene | 70.573241 | 70.576110 | positive | MGI_C57BL6J_1920834 | 1700100L14Rik | NCBI_Gene:73584,ENSEMBL:ENSMUSG00000101051 | MGI:1920834 | lncRNA gene | RIKEN cDNA 1700100L14 gene |
| 13 | gene | 70.610786 | 70.614925 | negative | MGI_C57BL6J_5623881 | Gm40996 | NCBI_Gene:105245552 | MGI:5623881 | lncRNA gene | predicted gene, 40996 |
| 13 | gene | 70.650144 | 70.655979 | positive | MGI_C57BL6J_3026977 | D030007L05Rik | ENSEMBL:ENSMUSG00000113334 | MGI:3026977 | lncRNA gene | RIKEN cDNA D030007L05 gene |
| 13 | gene | 70.650586 | 70.656706 | negative | MGI_C57BL6J_5595536 | Gm36377 | NCBI_Gene:102640266,ENSEMBL:ENSMUSG00000113909 | MGI:5595536 | lncRNA gene | predicted gene, 36377 |
| 13 | gene | 70.673122 | 70.679943 | positive | MGI_C57BL6J_5595633 | Gm36474 | NCBI_Gene:102640409 | MGI:5595633 | lncRNA gene | predicted gene, 36474 |
| 13 | gene | 70.676716 | 70.681966 | negative | MGI_C57BL6J_5595585 | Gm36426 | ENSEMBL:ENSMUSG00000113796 | MGI:5595585 | lncRNA gene | predicted gene, 36426 |
| 13 | gene | 70.706152 | 70.735643 | positive | MGI_C57BL6J_5595766 | Gm36607 | NCBI_Gene:102640576,ENSEMBL:ENSMUSG00000113794 | MGI:5595766 | lncRNA gene | predicted gene, 36607 |
| 13 | gene | 70.727798 | 70.841851 | negative | MGI_C57BL6J_2429637 | Adamts16 | NCBI_Gene:271127,ENSEMBL:ENSMUSG00000049538 | MGI:2429637 | protein coding gene | a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 16 |
| 13 | gene | 70.840423 | 70.859615 | positive | MGI_C57BL6J_5595688 | Gm36529 | NCBI_Gene:102640479,ENSEMBL:ENSMUSG00000113771 | MGI:5595688 | lncRNA gene | predicted gene, 36529 |
| 13 | gene | 70.881916 | 70.884912 | positive | MGI_C57BL6J_1924416 | 8030423J24Rik | NCBI_Gene:77166,ENSEMBL:ENSMUSG00000021600 | MGI:1924416 | protein coding gene | RIKEN cDNA 8030423J24 gene |
| 13 | gene | 71.018326 | 71.020421 | negative | MGI_C57BL6J_5595893 | Gm36734 | NCBI_Gene:102640737 | MGI:5595893 | lncRNA gene | predicted gene, 36734 |
| 13 | gene | 71.068480 | 71.071726 | negative | MGI_C57BL6J_5611419 | Gm38191 | ENSEMBL:ENSMUSG00000102303 | MGI:5611419 | unclassified gene | predicted gene, 38191 |
| 13 | gene | 71.069601 | 71.076179 | positive | MGI_C57BL6J_5595944 | Gm36785 | NCBI_Gene:102640803 | MGI:5595944 | lncRNA gene | predicted gene, 36785 |
| 13 | gene | 71.079422 | 71.106427 | negative | MGI_C57BL6J_5623882 | Gm40997 | NCBI_Gene:105245553 | MGI:5623882 | lncRNA gene | predicted gene, 40997 |
| 13 | gene | 71.107089 | 71.107209 | positive | MGI_C57BL6J_3783374 | Mir466f-4 | miRBase:MI0006291,NCBI_Gene:100316804,ENSEMBL:ENSMUSG00000080580 | MGI:3783374 | miRNA gene | microRNA 466f-4 |
| 13 | gene | 71.372533 | 71.394296 | negative | MGI_C57BL6J_5623883 | Gm40998 | NCBI_Gene:105245554 | MGI:5623883 | lncRNA gene | predicted gene, 40998 |
| 13 | gene | 71.487607 | 71.612710 | positive | MGI_C57BL6J_1920833 | 1700112M02Rik | NCBI_Gene:102640882,ENSEMBL:ENSMUSG00000113358 | MGI:1920833 | lncRNA gene | RIKEN cDNA 1700112M02 gene |
| 13 | pseudogene | 71.624610 | 71.624974 | negative | MGI_C57BL6J_6096995 | Gm47811 | ENSEMBL:ENSMUSG00000113305 | MGI:6096995 | pseudogene | predicted gene, 47811 |
| 13 | pseudogene | 71.625028 | 71.625143 | positive | MGI_C57BL6J_6096996 | Gm47812 | ENSEMBL:ENSMUSG00000113034 | MGI:6096996 | pseudogene | predicted gene, 47812 |
| 13 | gene | 71.709055 | 71.956552 | negative | MGI_C57BL6J_5623884 | Gm40999 | NCBI_Gene:105245555,ENSEMBL:ENSMUSG00000114038 | MGI:5623884 | lncRNA gene | predicted gene, 40999 |
| 13 | gene | 71.957921 | 71.963723 | negative | MGI_C57BL6J_1197515 | Irx1 | NCBI_Gene:16371,ENSEMBL:ENSMUSG00000060969 | MGI:1197515 | protein coding gene | Iroquois homeobox 1 |
| 13 | pseudogene | 72.350221 | 72.350674 | positive | MGI_C57BL6J_3782227 | Gm4052 | NCBI_Gene:100042828,ENSEMBL:ENSMUSG00000113594 | MGI:3782227 | pseudogene | predicted gene 4052 |
| 13 | gene | 72.367326 | 72.371647 | negative | MGI_C57BL6J_6097035 | Gm47836 | ENSEMBL:ENSMUSG00000113172 | MGI:6097035 | unclassified gene | predicted gene, 47836 |
| 13 | pseudogene | 72.454661 | 72.455378 | negative | MGI_C57BL6J_3704410 | Rpl9-ps4 | NCBI_Gene:100042832,ENSEMBL:ENSMUSG00000094989 | MGI:3704410 | pseudogene | ribosomal protein L9, pseudogene 4 |
| 13 | gene | 72.467977 | 72.471803 | positive | MGI_C57BL6J_6097038 | Gm47838 | ENSEMBL:ENSMUSG00000113375 | MGI:6097038 | unclassified gene | predicted gene, 47838 |
| 13 | pseudogene | 72.472190 | 72.472569 | negative | MGI_C57BL6J_3782232 | Gm4057 | NCBI_Gene:100042836,ENSEMBL:ENSMUSG00000113858 | MGI:3782232 | pseudogene | predicted gene 4057 |
| 13 | gene | 72.532484 | 72.537564 | negative | MGI_C57BL6J_5623885 | Gm41000 | NCBI_Gene:105245556 | MGI:5623885 | lncRNA gene | predicted gene, 41000 |
| 13 | gene | 72.542765 | 72.601064 | negative | MGI_C57BL6J_5621465 | Gm38580 | NCBI_Gene:102641841 | MGI:5621465 | lncRNA gene | predicted gene, 38580 |
| 13 | gene | 72.623466 | 72.628564 | negative | MGI_C57BL6J_5295661 | Gm20554 | NCBI_Gene:328287,ENSEMBL:ENSMUSG00000093587 | MGI:5295661 | lncRNA gene | predicted gene, 20554 |
| 13 | gene | 72.628797 | 72.634198 | positive | MGI_C57BL6J_1197526 | Irx2 | NCBI_Gene:16372,ENSEMBL:ENSMUSG00000001504 | MGI:1197526 | protein coding gene | Iroquois homeobox 2 |
| 13 | gene | 72.669467 | 72.676554 | positive | MGI_C57BL6J_5623886 | Gm41001 | NCBI_Gene:105245557 | MGI:5623886 | lncRNA gene | predicted gene, 41001 |
| 13 | gene | 72.702849 | 72.705392 | positive | MGI_C57BL6J_6097122 | Gm47890 | ENSEMBL:ENSMUSG00000113697 | MGI:6097122 | unclassified gene | predicted gene, 47890 |
| 13 | gene | 72.769431 | 72.786342 | positive | MGI_C57BL6J_6097140 | Gm47902 | ENSEMBL:ENSMUSG00000113837 | MGI:6097140 | lncRNA gene | predicted gene, 47902 |
| 13 | gene | 72.809601 | 72.816763 | negative | MGI_C57BL6J_1925696 | D730050B12Rik | NCBI_Gene:78446,ENSEMBL:ENSMUSG00000113082 | MGI:1925696 | lncRNA gene | RIKEN cDNA D730050B12 gene |
| 13 | gene | 72.932327 | 72.939787 | negative | MGI_C57BL6J_5589208 | Gm30049 | NCBI_Gene:102631807 | MGI:5589208 | lncRNA gene | predicted gene, 30049 |
| 13 | gene | 73.132998 | 73.155973 | positive | MGI_C57BL6J_5589313 | Gm30154 | NCBI_Gene:102631955 | MGI:5589313 | lncRNA gene | predicted gene, 30154 |
| 13 | gene | 73.206091 | 73.229100 | negative | MGI_C57BL6J_1922611 | 4930555K05Rik | NCBI_Gene:75361 | MGI:1922611 | lncRNA gene | RIKEN cDNA 4930555K05 gene |
| 13 | pseudogene | 73.233335 | 73.233874 | positive | MGI_C57BL6J_3779573 | Rpl31-ps2 | NCBI_Gene:621486,ENSEMBL:ENSMUSG00000113952 | MGI:3779573 | pseudogene | ribosomal protein L31, pseudogene 2 |
| 13 | gene | 73.259590 | 73.269622 | positive | MGI_C57BL6J_1355275 | Irx4 | NCBI_Gene:50916,ENSEMBL:ENSMUSG00000021604 | MGI:1355275 | protein coding gene | Iroquois homeobox 4 |
| 13 | gene | 73.273230 | 73.273732 | positive | MGI_C57BL6J_6097299 | Gm48000 | ENSEMBL:ENSMUSG00000113485 | MGI:6097299 | unclassified gene | predicted gene, 48000 |
| 13 | pseudogene | 73.317828 | 73.318145 | positive | MGI_C57BL6J_3642825 | Gm10263 | NCBI_Gene:100042864,ENSEMBL:ENSMUSG00000066407 | MGI:3642825 | pseudogene | predicted gene 10263 |
| 13 | gene | 73.319838 | 73.328542 | negative | MGI_C57BL6J_107932 | Ndufs6 | NCBI_Gene:407785,ENSEMBL:ENSMUSG00000021606 | MGI:107932 | protein coding gene | NADH:ubiquinone oxidoreductase core subunit S6 |
| 13 | gene | 73.328513 | 73.332178 | positive | MGI_C57BL6J_2137228 | Mrpl36 | NCBI_Gene:94066,ENSEMBL:ENSMUSG00000021607 | MGI:2137228 | protein coding gene | mitochondrial ribosomal protein L36 |
| 13 | gene | 73.344665 | 73.347384 | negative | MGI_C57BL6J_3583897 | Eprn | NCBI_Gene:432777,ENSEMBL:ENSMUSG00000113346 | MGI:3583897 | lncRNA gene | ephemeron, early developmental lncRNA |
| 13 | gene | 73.467197 | 73.516422 | positive | MGI_C57BL6J_2384812 | Lpcat1 | NCBI_Gene:210992,ENSEMBL:ENSMUSG00000021608 | MGI:2384812 | protein coding gene | lysophosphatidylcholine acyltransferase 1 |
| 13 | gene | 73.535529 | 73.578672 | positive | MGI_C57BL6J_94862 | Slc6a3 | NCBI_Gene:13162,ENSEMBL:ENSMUSG00000021609 | MGI:94862 | protein coding gene | solute carrier family 6 (neurotransmitter transporter, dopamine), member 3 |
| 13 | gene | 73.604002 | 73.620639 | positive | MGI_C57BL6J_2442892 | Clptm1l | NCBI_Gene:218335,ENSEMBL:ENSMUSG00000021610 | MGI:2442892 | protein coding gene | CLPTM1-like |
| 13 | gene | 73.626179 | 73.652366 | positive | MGI_C57BL6J_1202709 | Tert | NCBI_Gene:21752,ENSEMBL:ENSMUSG00000021611 | MGI:1202709 | protein coding gene | telomerase reverse transcriptase |
| 13 | gene | 73.661750 | 73.678023 | negative | MGI_C57BL6J_1336892 | Slc6a18 | NCBI_Gene:22598,ENSEMBL:ENSMUSG00000021612 | MGI:1336892 | protein coding gene | solute carrier family 6 (neurotransmitter transporter), member 18 |
| 13 | gene | 73.679745 | 73.704865 | negative | MGI_C57BL6J_1921588 | Slc6a19 | NCBI_Gene:74338,ENSEMBL:ENSMUSG00000021565 | MGI:1921588 | protein coding gene | solute carrier family 6 (neurotransmitter transporter), member 19 |
| 13 | gene | 73.699826 | 73.709856 | positive | MGI_C57BL6J_1918640 | Slc6a19os | NCBI_Gene:71390,ENSEMBL:ENSMUSG00000021566 | MGI:1918640 | antisense lncRNA gene | solute carrier family 6 (neurotransmitter transporter), member 19, opposite strand |
| 13 | gene | 73.733052 | 73.816754 | positive | MGI_C57BL6J_1342283 | Slc12a7 | NCBI_Gene:20499,ENSEMBL:ENSMUSG00000017756 | MGI:1342283 | protein coding gene | solute carrier family 12, member 7 |
| 13 | gene | 73.818529 | 73.847631 | negative | MGI_C57BL6J_1919543 | Nkd2 | NCBI_Gene:72293,ENSEMBL:ENSMUSG00000021567 | MGI:1919543 | protein coding gene | naked cuticle 2 |
| 13 | pseudogene | 73.829747 | 73.830507 | positive | MGI_C57BL6J_3643433 | Gm8062 | NCBI_Gene:102632224,ENSEMBL:ENSMUSG00000066714 | MGI:3643433 | pseudogene | predicted pseudogene 8062 |
| 13 | gene | 73.847727 | 73.853107 | positive | MGI_C57BL6J_5589462 | Gm30303 | NCBI_Gene:102632153 | MGI:5589462 | lncRNA gene | predicted gene, 30303 |
| 13 | gene | 73.858455 | 73.860338 | positive | MGI_C57BL6J_5589591 | Gm30432 | NCBI_Gene:102632333 | MGI:5589591 | lncRNA gene | predicted gene, 30432 |
| 13 | gene | 73.911347 | 73.937810 | negative | MGI_C57BL6J_1916966 | Trip13 | NCBI_Gene:69716,ENSEMBL:ENSMUSG00000021569 | MGI:1916966 | protein coding gene | thyroid hormone receptor interactor 13 |
| 13 | gene | 73.924585 | 73.924710 | negative | MGI_C57BL6J_5454974 | Gm25197 | ENSEMBL:ENSMUSG00000095943 | MGI:5454974 | miRNA gene | predicted gene, 25197 |
| 13 | gene | 73.937811 | 73.960895 | positive | MGI_C57BL6J_2145317 | Brd9 | NCBI_Gene:105246,ENSEMBL:ENSMUSG00000057649 | MGI:2145317 | protein coding gene | bromodomain containing 9 |
| 13 | gene | 73.958913 | 73.963843 | negative | MGI_C57BL6J_5623887 | Gm41002 | NCBI_Gene:105245558,ENSEMBL:ENSMUSG00000113274 | MGI:5623887 | lncRNA gene | predicted gene, 41002 |
| 13 | gene | 73.963847 | 73.992990 | positive | MGI_C57BL6J_1918414 | Zdhhc11 | NCBI_Gene:71164,ENSEMBL:ENSMUSG00000069189 | MGI:1918414 | protein coding gene | zinc finger, DHHC domain containing 11 |
| 13 | pseudogene | 73.988866 | 73.989165 | negative | MGI_C57BL6J_3809072 | Gm10126 | NCBI_Gene:100045766,ENSEMBL:ENSMUSG00000113923 | MGI:3809072 | pseudogene | predicted gene 10126 |
| 13 | gene | 74.008009 | 74.009354 | negative | MGI_C57BL6J_3801895 | Gm15912 | ENSEMBL:ENSMUSG00000085881 | MGI:3801895 | lncRNA gene | predicted gene 15912 |
| 13 | gene | 74.009407 | 74.035753 | positive | MGI_C57BL6J_1920198 | Tppp | NCBI_Gene:72948,ENSEMBL:ENSMUSG00000021573 | MGI:1920198 | protein coding gene | tubulin polymerization promoting protein |
| 13 | pseudogene | 74.018595 | 74.020312 | negative | MGI_C57BL6J_3646097 | Gm6263 | NCBI_Gene:621824,ENSEMBL:ENSMUSG00000080966 | MGI:3646097 | pseudogene | predicted gene 6263 |
| 13 | gene | 74.036495 | 74.062315 | negative | MGI_C57BL6J_1921720 | Cep72 | NCBI_Gene:74470,ENSEMBL:ENSMUSG00000021572 | MGI:1921720 | protein coding gene | centrosomal protein 72 |
| 13 | gene | 74.086253 | 74.169442 | positive | MGI_C57BL6J_105064 | Slc9a3 | NCBI_Gene:105243,ENSEMBL:ENSMUSG00000036123 | MGI:105064 | protein coding gene | solute carrier family 9 (sodium/hydrogen exchanger), member 3 |
| 13 | gene | 74.120827 | 74.121997 | negative | MGI_C57BL6J_1915626 | 0610040A22Rik | NA | NA | unclassified gene | RIKEN cDNA 0610040A22 gene |
| 13 | gene | 74.122101 | 74.131263 | positive | MGI_C57BL6J_5623888 | Gm41003 | NCBI_Gene:105245559 | MGI:5623888 | lncRNA gene | predicted gene, 41003 |
| 13 | gene | 74.169488 | 74.208732 | negative | MGI_C57BL6J_2443972 | Exoc3 | NCBI_Gene:211446,ENSEMBL:ENSMUSG00000034152 | MGI:2443972 | protein coding gene | exocyst complex component 3 |
| 13 | gene | 74.211117 | 74.292331 | negative | MGI_C57BL6J_1333776 | Ahrr | NCBI_Gene:11624,ENSEMBL:ENSMUSG00000021575 | MGI:1333776 | protein coding gene | aryl-hydrocarbon receptor repressor |
| 13 | gene | 74.303121 | 74.317580 | negative | MGI_C57BL6J_109283 | Pdcd6 | NCBI_Gene:18570,ENSEMBL:ENSMUSG00000021576 | MGI:109283 | protein coding gene | programmed cell death 6 |
| 13 | gene | 74.322254 | 74.350280 | negative | MGI_C57BL6J_1914195 | Sdha | NCBI_Gene:66945,ENSEMBL:ENSMUSG00000021577 | MGI:1914195 | protein coding gene | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| 13 | pseudogene | 74.340539 | 74.340877 | positive | MGI_C57BL6J_3802000 | Gm16125 | ENSEMBL:ENSMUSG00000090004 | MGI:3802000 | pseudogene | predicted gene 16125 |
| 13 | gene | 74.350309 | 74.365783 | positive | MGI_C57BL6J_1914683 | Ccdc127 | NCBI_Gene:67433,ENSEMBL:ENSMUSG00000021578 | MGI:1914683 | protein coding gene | coiled-coil domain containing 127 |
| 13 | gene | 74.359578 | 74.364005 | negative | MGI_C57BL6J_2145269 | Lrrc14b | NCBI_Gene:432779,ENSEMBL:ENSMUSG00000021579 | MGI:2145269 | protein coding gene | leucine rich repeat containing 14B |
| 13 | gene | 74.369321 | 74.390861 | negative | MGI_C57BL6J_3033356 | Zfp72 | NCBI_Gene:238722,ENSEMBL:ENSMUSG00000069184 | MGI:3033356 | protein coding gene | zinc finger protein 72 |
| 13 | pseudogene | 74.381629 | 74.382209 | negative | MGI_C57BL6J_3648589 | Gm8227 | NCBI_Gene:666670,ENSEMBL:ENSMUSG00000113315 | MGI:3648589 | pseudogene | predicted gene 8227 |
| 13 | pseudogene | 74.399694 | 74.407346 | positive | MGI_C57BL6J_3779109 | Ftl1-ps1 | NCBI_Gene:634386,ENSEMBL:ENSMUSG00000062382 | MGI:3779109 | pseudogene | ferritin light polypeptide 1, pseudogene 1 |
| 13 | gene | 74.404183 | 74.406737 | negative | MGI_C57BL6J_6096373 | Gm47428 | ENSEMBL:ENSMUSG00000113311 | MGI:6096373 | lncRNA gene | predicted gene, 47428 |
| 13 | gene | 74.407065 | 74.407173 | positive | MGI_C57BL6J_3719623 | Mir692-3 | NCBI_Gene:100124641,ENSEMBL:ENSMUSG00000076033 | MGI:3719623 | miRNA gene | microRNA 692-3 |
| 13 | gene | 74.408984 | 74.416668 | negative | MGI_C57BL6J_5623889 | Gm41004 | NCBI_Gene:105245561 | MGI:5623889 | lncRNA gene | predicted gene, 41004 |
| 13 | gene | 74.433049 | 74.463820 | negative | MGI_C57BL6J_3649010 | E430024I08Rik | NCBI_Gene:622175,ENSEMBL:ENSMUSG00000112964 | MGI:3649010 | lncRNA gene | RIKEN cDNA E430024I08 gene |
| 13 | pseudogene | 74.440166 | 74.440532 | positive | MGI_C57BL6J_6096386 | Gm47435 | ENSEMBL:ENSMUSG00000113212 | MGI:6096386 | pseudogene | predicted gene, 47435 |
| 13 | gene | 74.474643 | 74.476068 | positive | MGI_C57BL6J_6215268 | Gm49762 | ENSEMBL:ENSMUSG00000116765 | MGI:6215268 | lncRNA gene | predicted gene, 49762 |
| 13 | gene | 74.478483 | 74.495167 | negative | MGI_C57BL6J_2385315 | Zfp825 | NCBI_Gene:235956,ENSEMBL:ENSMUSG00000069208 | MGI:2385315 | protein coding gene | zinc finger protein 825 |
| 13 | gene | 74.494143 | 74.498470 | positive | MGI_C57BL6J_6215270 | Gm49763 | ENSEMBL:ENSMUSG00000116646 | MGI:6215270 | lncRNA gene | predicted gene, 49763 |
| 13 | pseudogene | 74.505022 | 74.570556 | negative | MGI_C57BL6J_5295696 | Gm20590 | NCBI_Gene:100418285 | MGI:5295696 | pseudogene | predicted gene, 20590 |
| 13 | gene | 74.506810 | 74.508367 | negative | MGI_C57BL6J_1924428 | 5930412I24Rik | NA | NA | unclassified gene | RIKEN cDNA 5930412I24 gene |
| 13 | gene | 74.509598 | 74.522461 | negative | MGI_C57BL6J_6096433 | Gm47467 | ENSEMBL:ENSMUSG00000113019 | MGI:6096433 | lncRNA gene | predicted gene, 47467 |
| 13 | pseudogene | 74.533523 | 74.534722 | positive | MGI_C57BL6J_3779505 | Gm5626 | NCBI_Gene:434490,ENSEMBL:ENSMUSG00000113660 | MGI:3779505 | pseudogene | predicted gene 5626 |
| 13 | gene | 74.553910 | 74.564256 | negative | MGI_C57BL6J_6096437 | Gm47469 | ENSEMBL:ENSMUSG00000113047 | MGI:6096437 | lncRNA gene | predicted gene, 47469 |
| 13 | pseudogene | 74.570447 | 74.570555 | negative | MGI_C57BL6J_6096439 | Gm47470 | ENSEMBL:ENSMUSG00000113279 | MGI:6096439 | pseudogene | predicted gene, 47470 |
| 13 | gene | 74.579358 | 74.618221 | negative | MGI_C57BL6J_5826067 | Gm46430 | NCBI_Gene:108168101,ENSEMBL:ENSMUSG00000113204 | MGI:5826067 | lncRNA gene | predicted gene, 46430 |
| 13 | gene | 74.639568 | 74.693201 | positive | MGI_C57BL6J_1933403 | Erap1 | NCBI_Gene:80898,ENSEMBL:ENSMUSG00000021583 | MGI:1933403 | protein coding gene | endoplasmic reticulum aminopeptidase 1 |
| 13 | gene | 74.692366 | 74.808874 | negative | MGI_C57BL6J_1098236 | Cast | NCBI_Gene:12380,ENSEMBL:ENSMUSG00000021585 | MGI:1098236 | protein coding gene | calpastatin |
| 13 | pseudogene | 74.767580 | 74.768923 | negative | MGI_C57BL6J_3648150 | Gm6296 | NCBI_Gene:622204,ENSEMBL:ENSMUSG00000113015 | MGI:3648150 | pseudogene | predicted gene 6296 |
| 13 | pseudogene | 74.864232 | 74.865733 | positive | MGI_C57BL6J_3644479 | Hnrnpa1l2-ps | NCBI_Gene:544954,ENSEMBL:ENSMUSG00000086743 | MGI:3644479 | pseudogene | heterogeneous nuclear ribonucleoprotein A1-like 2, pseudogene |
| 13 | gene | 74.995715 | 75.012756 | negative | MGI_C57BL6J_5590026 | Gm30867 | NCBI_Gene:102632915 | MGI:5590026 | lncRNA gene | predicted gene, 30867 |
| 13 | gene | 75.089420 | 75.134861 | positive | MGI_C57BL6J_97511 | Pcsk1 | NCBI_Gene:18548,ENSEMBL:ENSMUSG00000021587 | MGI:97511 | protein coding gene | proprotein convertase subtilisin/kexin type 1 |
| 13 | pseudogene | 75.199214 | 75.199944 | positive | MGI_C57BL6J_5011280 | Gm19095 | NCBI_Gene:100418242,ENSEMBL:ENSMUSG00000113264 | MGI:5011280 | pseudogene | predicted gene, 19095 |
| 13 | pseudogene | 75.479195 | 75.479387 | negative | MGI_C57BL6J_6097642 | Gm48234 | ENSEMBL:ENSMUSG00000113821 | MGI:6097642 | pseudogene | predicted gene, 48234 |
| 13 | pseudogene | 75.509279 | 75.510047 | positive | MGI_C57BL6J_6097643 | Gm48235 | ENSEMBL:ENSMUSG00000113270 | MGI:6097643 | pseudogene | predicted gene, 48235 |
| 13 | pseudogene | 75.515320 | 75.518414 | negative | MGI_C57BL6J_3644857 | Gm8278 | NCBI_Gene:666764,ENSEMBL:ENSMUSG00000113773 | MGI:3644857 | pseudogene | predicted gene 8278 |
| 13 | pseudogene | 75.523226 | 75.524348 | negative | MGI_C57BL6J_3782100 | Gm3926 | NCBI_Gene:100042606,ENSEMBL:ENSMUSG00000113929 | MGI:3782100 | pseudogene | predicted gene 3926 |
| 13 | pseudogene | 75.644865 | 75.645228 | positive | MGI_C57BL6J_3782325 | Gm4149 | NCBI_Gene:100653392,ENSEMBL:ENSMUSG00000074800 | MGI:3782325 | pseudogene | predicted pseudogene 4149 |
| 13 | gene | 75.645046 | 75.645141 | positive | MGI_C57BL6J_3629911 | Mir682 | miRBase:MI0004644,NCBI_Gene:751556,ENSEMBL:ENSMUSG00000076236 | MGI:3629911 | miRNA gene | microRNA 682 |
| 13 | gene | 75.653653 | 75.707250 | negative | MGI_C57BL6J_1914566 | 1700037F03Rik | NCBI_Gene:105245562,ENSEMBL:ENSMUSG00000114125 | MGI:1914566 | lncRNA gene | RIKEN cDNA 1700037F03 gene |
| 13 | gene | 75.691972 | 75.692054 | positive | MGI_C57BL6J_5452789 | Gm23012 | ENSEMBL:ENSMUSG00000077337 | MGI:5452789 | snoRNA gene | predicted gene, 23012 |
| 13 | gene | 75.706757 | 75.772364 | positive | MGI_C57BL6J_2183438 | Ell2 | NCBI_Gene:192657,ENSEMBL:ENSMUSG00000001542 | MGI:2183438 | protein coding gene | elongation factor RNA polymerase II 2 |
| 13 | gene | 75.727279 | 75.729162 | positive | MGI_C57BL6J_6097746 | Gm48302 | ENSEMBL:ENSMUSG00000113041 | MGI:6097746 | lncRNA gene | predicted gene, 48302 |
| 13 | gene | 75.768068 | 75.768197 | positive | MGI_C57BL6J_5452904 | Gm23127 | ENSEMBL:ENSMUSG00000077449 | MGI:5452904 | snoRNA gene | predicted gene, 23127 |
| 13 | pseudogene | 75.802705 | 75.803298 | positive | MGI_C57BL6J_3645459 | Gm8288 | NCBI_Gene:666786,ENSEMBL:ENSMUSG00000113409 | MGI:3645459 | pseudogene | predicted gene 8288 |
| 13 | gene | 75.818226 | 75.847161 | negative | MGI_C57BL6J_5623890 | Gm41005 | NCBI_Gene:105245563 | MGI:5623890 | lncRNA gene | predicted gene, 41005 |
| 13 | gene | 75.839868 | 75.850154 | positive | MGI_C57BL6J_2135625 | Glrx | NCBI_Gene:93692,ENSEMBL:ENSMUSG00000021591 | MGI:2135625 | protein coding gene | glutaredoxin |
| 13 | gene | 75.869537 | 75.943925 | negative | MGI_C57BL6J_1920546 | Rhobtb3 | NCBI_Gene:73296,ENSEMBL:ENSMUSG00000021589 | MGI:1920546 | protein coding gene | Rho-related BTB domain containing 3 |
| 13 | gene | 75.905147 | 75.905278 | positive | MGI_C57BL6J_5453940 | Gm24163 | ENSEMBL:ENSMUSG00000070170 | MGI:5453940 | snoRNA gene | predicted gene, 24163 |
| 13 | pseudogene | 75.929311 | 75.930328 | negative | MGI_C57BL6J_3782975 | Gm15528 | NCBI_Gene:666553,ENSEMBL:ENSMUSG00000081816 | MGI:3782975 | pseudogene | predicted gene 15528 |
| 13 | pseudogene | 75.954110 | 75.955057 | positive | MGI_C57BL6J_3644735 | Gm6311 | NCBI_Gene:622313,ENSEMBL:ENSMUSG00000061833 | MGI:3644735 | pseudogene | predicted gene 6311 |
| 13 | gene | 75.959970 | 75.998987 | positive | MGI_C57BL6J_1922821 | Spata9 | NCBI_Gene:75571,ENSEMBL:ENSMUSG00000021590 | MGI:1922821 | protein coding gene | spermatogenesis associated 9 |
| 13 | gene | 75.963657 | 75.963787 | negative | MGI_C57BL6J_5453096 | Gm23319 | ENSEMBL:ENSMUSG00000077655 | MGI:5453096 | snoRNA gene | predicted gene, 23319 |
| 13 | gene | 76.001535 | 76.018719 | negative | MGI_C57BL6J_2145198 | Rfesd | NCBI_Gene:218341,ENSEMBL:ENSMUSG00000043190 | MGI:2145198 | protein coding gene | Rieske (Fe-S) domain containing |
| 13 | pseudogene | 76.019719 | 76.020639 | negative | MGI_C57BL6J_3782341 | Gm4165 | NCBI_Gene:102633176,ENSEMBL:ENSMUSG00000114366 | MGI:3782341 | pseudogene | predicted gene 4165 |
| 13 | gene | 76.054851 | 76.056996 | negative | MGI_C57BL6J_2441872 | Gpr150 | NCBI_Gene:238725,ENSEMBL:ENSMUSG00000045509 | MGI:2441872 | protein coding gene | G protein-coupled receptor 150 |
| 13 | gene | 76.060422 | 76.098667 | negative | MGI_C57BL6J_1924291 | Arsk | NCBI_Gene:77041,ENSEMBL:ENSMUSG00000021592 | MGI:1924291 | protein coding gene | arylsulfatase K |
| 13 | gene | 76.065583 | 76.066279 | positive | MGI_C57BL6J_5623891 | Gm41006 | NCBI_Gene:105245564 | MGI:5623891 | lncRNA gene | predicted gene, 41006 |
| 13 | gene | 76.068778 | 76.071938 | positive | MGI_C57BL6J_5590320 | Gm31161 | NCBI_Gene:102633302 | MGI:5590320 | lncRNA gene | predicted gene, 31161 |
| 13 | gene | 76.098730 | 76.190318 | positive | MGI_C57BL6J_2679923 | Ttc37 | NCBI_Gene:218343,ENSEMBL:ENSMUSG00000033991 | MGI:2679923 | protein coding gene | tetratricopeptide repeat domain 37 |
| 13 | gene | 76.198991 | 76.200866 | positive | MGI_C57BL6J_6098245 | Gm48639 | ENSEMBL:ENSMUSG00000114664 | MGI:6098245 | unclassified gene | predicted gene, 48639 |
| 13 | gene | 76.201708 | 76.305072 | negative | MGI_C57BL6J_2685122 | Fam81b | NCBI_Gene:238726,ENSEMBL:ENSMUSG00000109228 | MGI:2685122 | protein coding gene | family with sequence similarity 81, member B |
| 13 | gene | 76.246249 | 76.278968 | positive | MGI_C57BL6J_5590378 | Gm31219 | NCBI_Gene:102633384,ENSEMBL:ENSMUSG00000114242 | MGI:5590378 | lncRNA gene | predicted gene, 31219 |
| 13 | gene | 76.300825 | 76.308553 | positive | MGI_C57BL6J_5590675 | Gm31516 | NCBI_Gene:102633769 | MGI:5590675 | lncRNA gene | predicted gene, 31516 |
| 13 | gene | 76.321204 | 76.321279 | negative | MGI_C57BL6J_5453802 | Gm24025 | ENSEMBL:ENSMUSG00000092732 | MGI:5453802 | miRNA gene | predicted gene, 24025 |
| 13 | gene | 76.382499 | 76.385028 | negative | MGI_C57BL6J_3642546 | Gm10760 | NA | NA | protein coding gene | predicted gene 10760 |
| 13 | gene | 76.384108 | 77.033277 | positive | MGI_C57BL6J_1926021 | Mctp1 | NCBI_Gene:78771,ENSEMBL:ENSMUSG00000021596 | MGI:1926021 | protein coding gene | multiple C2 domains, transmembrane 1 |
| 13 | pseudogene | 76.414386 | 76.415344 | negative | MGI_C57BL6J_5010499 | Gm18314 | NCBI_Gene:100416911 | MGI:5010499 | pseudogene | predicted gene, 18314 |
| 13 | pseudogene | 76.439331 | 76.439670 | positive | MGI_C57BL6J_6097415 | Gm48078 | ENSEMBL:ENSMUSG00000114293 | MGI:6097415 | pseudogene | predicted gene, 48078 |
| 13 | gene | 76.507644 | 76.516272 | positive | MGI_C57BL6J_5826068 | Gm46431 | NCBI_Gene:108168102 | MGI:5826068 | lncRNA gene | predicted gene, 46431 |
| 13 | gene | 76.604319 | 76.604450 | negative | MGI_C57BL6J_5454296 | Gm24519 | ENSEMBL:ENSMUSG00000096675 | MGI:5454296 | snoRNA gene | predicted gene, 24519 |
| 13 | pseudogene | 76.683982 | 76.684433 | negative | MGI_C57BL6J_5439428 | Gm21959 | ENSEMBL:ENSMUSG00000096216 | MGI:5439428 | pseudogene | predicted gene, 21959 |
| 13 | pseudogene | 76.696619 | 76.697910 | positive | MGI_C57BL6J_3783182 | Gm15740 | NCBI_Gene:666603,ENSEMBL:ENSMUSG00000090027 | MGI:3783182 | pseudogene | predicted gene 15740 |
| 13 | pseudogene | 76.700328 | 76.702010 | negative | MGI_C57BL6J_5010748 | Gm18563 | NCBI_Gene:100417364 | MGI:5010748 | pseudogene | predicted gene, 18563 |
| 13 | pseudogene | 76.935088 | 76.935890 | positive | MGI_C57BL6J_3643116 | Gm8185 | NCBI_Gene:666607,ENSEMBL:ENSMUSG00000090119 | MGI:3643116 | pseudogene | predicted gene 8185 |
| 13 | gene | 77.043088 | 77.135509 | negative | MGI_C57BL6J_2145448 | Slf1 | NCBI_Gene:105377,ENSEMBL:ENSMUSG00000021597 | MGI:2145448 | protein coding gene | SMC5-SMC6 complex localization factor 1 |
| 13 | pseudogene | 77.135324 | 77.170127 | positive | MGI_C57BL6J_5589003 | Gm29844 | NCBI_Gene:102631529,ENSEMBL:ENSMUSG00000114256 | MGI:5589003 | pseudogene | predicted gene, 29844 |
| 13 | gene | 77.135536 | 77.613785 | positive | MGI_C57BL6J_1919621 | 2210408I21Rik | NCBI_Gene:72371,ENSEMBL:ENSMUSG00000071252 | MGI:1919621 | protein coding gene | RIKEN cDNA 2210408I21 gene |
| 13 | pseudogene | 77.553610 | 77.554373 | negative | MGI_C57BL6J_3780041 | Gm9634 | NCBI_Gene:675063,ENSEMBL:ENSMUSG00000114778 | MGI:3780041 | pseudogene | predicted gene 9634 |
| 13 | pseudogene | 77.669074 | 77.670475 | negative | MGI_C57BL6J_2145515 | Atp5c1-ps | NCBI_Gene:382823,ENSEMBL:ENSMUSG00000114809 | MGI:2145515 | pseudogene | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1, pseudogene |
| 13 | gene | 77.676821 | 77.676951 | positive | MGI_C57BL6J_5455865 | Gm26088 | ENSEMBL:ENSMUSG00000088521 | MGI:5455865 | snoRNA gene | predicted gene, 26088 |
| 13 | gene | 77.702033 | 78.166240 | positive | MGI_C57BL6J_1915925 | Fam172a | NCBI_Gene:68675,ENSEMBL:ENSMUSG00000064138 | MGI:1915925 | protein coding gene | family with sequence similarity 172, member A |
| 13 | gene | 77.837860 | 77.840517 | positive | MGI_C57BL6J_2441882 | C130063H03Rik | NA | NA | unclassified gene | RIKEN cDNA C130063H03 gene |
| 13 | gene | 78.024902 | 78.026296 | positive | MGI_C57BL6J_1922757 | Pou5f2 | NCBI_Gene:75507,ENSEMBL:ENSMUSG00000093668 | MGI:1922757 | protein coding gene | POU domain class 5, transcription factor 2 |
| 13 | gene | 78.033534 | 78.034751 | positive | MGI_C57BL6J_1925018 | A430105P17Rik | NA | NA | unclassified gene | RIKEN cDNA A430105P17 gene |
| 13 | pseudogene | 78.079388 | 78.079856 | positive | MGI_C57BL6J_6097669 | Gm48254 | ENSEMBL:ENSMUSG00000114812 | MGI:6097669 | pseudogene | predicted gene, 48254 |
| 13 | gene | 78.112482 | 78.116319 | positive | MGI_C57BL6J_2443856 | C130051F05Rik | ENSEMBL:ENSMUSG00000114605 | MGI:2443856 | unclassified gene | RIKEN cDNA C130051F05 gene |
| 13 | pseudogene | 78.121070 | 78.122718 | positive | MGI_C57BL6J_5011175 | Gm18990 | NCBI_Gene:100418084,ENSEMBL:ENSMUSG00000114622 | MGI:5011175 | pseudogene | predicted gene, 18990 |
| 13 | gene | 78.123379 | 78.185910 | negative | MGI_C57BL6J_5621489 | Gm38604 | NCBI_Gene:102642321,ENSEMBL:ENSMUSG00000114401 | MGI:5621489 | lncRNA gene | predicted gene, 38604 |
| 13 | gene | 78.161107 | 78.163920 | negative | MGI_C57BL6J_2441826 | A730087J23Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA A730087J23 gene |
| 13 | gene | 78.171630 | 78.182973 | positive | MGI_C57BL6J_1920343 | 3110006O06Rik | ENSEMBL:ENSMUSG00000114649 | MGI:1920343 | lncRNA gene | RIKEN cDNA 3110006O06 gene |
| 13 | gene | 78.188971 | 78.199757 | negative | MGI_C57BL6J_1352451 | Nr2f1 | NCBI_Gene:13865,ENSEMBL:ENSMUSG00000069171 | MGI:1352451 | protein coding gene | nuclear receptor subfamily 2, group F, member 1 |
| 13 | gene | 78.198017 | 78.279391 | positive | MGI_C57BL6J_2443527 | A830082K12Rik | NCBI_Gene:320174,ENSEMBL:ENSMUSG00000087143 | MGI:2443527 | lncRNA gene | RIKEN cDNA A830082K12 gene |
| 13 | gene | 78.199288 | 78.199956 | positive | MGI_C57BL6J_3644662 | Gm8327 | NA | NA | unclassified gene | predicted gene 8327 |
| 13 | pseudogene | 78.273394 | 78.273990 | negative | MGI_C57BL6J_3647790 | Gm5042 | NCBI_Gene:268676,ENSEMBL:ENSMUSG00000114592 | MGI:3647790 | pseudogene | predicted gene 5042 |
| 13 | pseudogene | 78.275043 | 78.283824 | negative | MGI_C57BL6J_5826024 | Gm46387 | NCBI_Gene:108168046,ENSEMBL:ENSMUSG00000114600 | MGI:5826024 | pseudogene | predicted gene, 46387 |
| 13 | pseudogene | 78.286093 | 78.287165 | negative | MGI_C57BL6J_3782137 | Gm3963 | NCBI_Gene:100042683,ENSEMBL:ENSMUSG00000114383 | MGI:3782137 | pseudogene | predicted gene 3963 |
| 13 | gene | 78.316330 | 78.319403 | positive | MGI_C57BL6J_5623892 | Gm41007 | NCBI_Gene:105245565 | MGI:5623892 | lncRNA gene | predicted gene, 41007 |
| 13 | pseudogene | 78.372786 | 78.379146 | negative | MGI_C57BL6J_3648927 | Gm6317 | NCBI_Gene:622365,ENSEMBL:ENSMUSG00000098003 | MGI:3648927 | pseudogene | predicted gene 6317 |
| 13 | gene | 78.374179 | 78.384198 | positive | MGI_C57BL6J_6097886 | Gm48399 | ENSEMBL:ENSMUSG00000114632 | MGI:6097886 | lncRNA gene | predicted gene, 48399 |
| 13 | gene | 78.426926 | 78.442524 | positive | MGI_C57BL6J_5591105 | Gm31946 | NCBI_Gene:102634341,ENSEMBL:ENSMUSG00000114639 | MGI:5591105 | lncRNA gene | predicted gene, 31946 |
| 13 | gene | 78.483354 | 78.636734 | positive | MGI_C57BL6J_5591226 | Gm32067 | NCBI_Gene:102634500,ENSEMBL:ENSMUSG00000114522 | MGI:5591226 | lncRNA gene | predicted gene, 32067 |
| 13 | gene | 78.638017 | 78.643064 | positive | MGI_C57BL6J_6097890 | Gm48402 | ENSEMBL:ENSMUSG00000114594 | MGI:6097890 | lncRNA gene | predicted gene, 48402 |
| 13 | pseudogene | 78.953082 | 78.954792 | negative | MGI_C57BL6J_3649106 | Gm8345 | NCBI_Gene:666883,ENSEMBL:ENSMUSG00000114557 | MGI:3649106 | pseudogene | predicted gene 8345 |
| 13 | gene | 79.161719 | 79.161862 | positive | MGI_C57BL6J_5455160 | Gm25383 | ENSEMBL:ENSMUSG00000088435 | MGI:5455160 | snoRNA gene | predicted gene, 25383 |
| 13 | gene | 79.285712 | 79.298242 | positive | MGI_C57BL6J_5580024 | Gm29318 | ENSEMBL:ENSMUSG00000101416 | MGI:5580024 | lncRNA gene | predicted gene 29318 |
| 13 | gene | 79.294878 | 79.295339 | negative | MGI_C57BL6J_5611577 | Gm38349 | ENSEMBL:ENSMUSG00000102355 | MGI:5611577 | unclassified gene | predicted gene, 38349 |
| 13 | pseudogene | 79.441143 | 79.441407 | negative | MGI_C57BL6J_6097985 | Gm48470 | ENSEMBL:ENSMUSG00000114874 | MGI:6097985 | pseudogene | predicted gene, 48470 |
| 13 | gene | 79.454970 | 79.455076 | positive | MGI_C57BL6J_5454640 | Gm24863 | ENSEMBL:ENSMUSG00000096005 | MGI:5454640 | snRNA gene | predicted gene, 24863 |
| 13 | pseudogene | 79.838077 | 79.838843 | positive | MGI_C57BL6J_6097986 | Gm48471 | ENSEMBL:ENSMUSG00000114505 | MGI:6097986 | pseudogene | predicted gene, 48471 |
| 13 | gene | 79.873085 | 80.130468 | negative | MGI_C57BL6J_5579232 | Gm28526 | ENSEMBL:ENSMUSG00000100168 | MGI:5579232 | lncRNA gene | predicted gene 28526 |
| 13 | pseudogene | 79.878462 | 79.878937 | positive | MGI_C57BL6J_5011505 | Gm19320 | NCBI_Gene:100502682,ENSEMBL:ENSMUSG00000114222 | MGI:5011505 | pseudogene | predicted gene, 19320 |
| 13 | pseudogene | 80.098222 | 80.099426 | negative | MGI_C57BL6J_3779472 | Gm5197 | NCBI_Gene:382780,ENSEMBL:ENSMUSG00000114287 | MGI:3779472 | pseudogene | predicted gene 5197 |
| 13 | gene | 80.158663 | 80.158790 | positive | MGI_C57BL6J_5454412 | Gm24635 | ENSEMBL:ENSMUSG00000084661 | MGI:5454412 | snRNA gene | predicted gene, 24635 |
| 13 | pseudogene | 80.161302 | 80.161918 | positive | MGI_C57BL6J_3644592 | Gm8371 | NCBI_Gene:666933,ENSEMBL:ENSMUSG00000114462 | MGI:3644592 | pseudogene | predicted gene 8371 |
| 13 | pseudogene | 80.485901 | 80.493487 | positive | MGI_C57BL6J_5826025 | Gm46388 | NCBI_Gene:108168047,ENSEMBL:ENSMUSG00000114528 | MGI:5826025 | pseudogene | predicted gene, 46388 |
| 13 | gene | 80.883338 | 80.896042 | positive | MGI_C57BL6J_2145242 | Arrdc3 | NCBI_Gene:105171,ENSEMBL:ENSMUSG00000074794 | MGI:2145242 | protein coding gene | arrestin domain containing 3 |
| 13 | gene | 80.884224 | 80.885524 | negative | MGI_C57BL6J_1916692 | 1700023H06Rik | ENSEMBL:ENSMUSG00000089827 | MGI:1916692 | lncRNA gene | RIKEN cDNA 1700023H06 gene |
| 13 | gene | 80.940403 | 80.948597 | positive | MGI_C57BL6J_1918666 | 5430425K12Rik | NCBI_Gene:71416,ENSEMBL:ENSMUSG00000114554 | MGI:1918666 | lncRNA gene | RIKEN cDNA 5430425K12 gene |
| 13 | gene | 80.957664 | 80.963730 | negative | MGI_C57BL6J_3782387 | Gm4211 | NCBI_Gene:100043074,ENSEMBL:ENSMUSG00000097146 | MGI:3782387 | lncRNA gene | predicted gene 4211 |
| 13 | gene | 80.962888 | 81.128864 | positive | MGI_C57BL6J_2443112 | 9330111N05Rik | NCBI_Gene:319983,ENSEMBL:ENSMUSG00000097098 | MGI:2443112 | lncRNA gene | RIKEN cDNA 9330111N05 gene |
| 13 | gene | 80.979470 | 80.983873 | positive | MGI_C57BL6J_6121594 | Gm49375 | ENSEMBL:ENSMUSG00000114479 | MGI:6121594 | lncRNA gene | predicted gene, 49375 |
| 13 | pseudogene | 80.992551 | 80.992800 | positive | MGI_C57BL6J_6098129 | Gm48568 | ENSEMBL:ENSMUSG00000114773 | MGI:6098129 | pseudogene | predicted gene, 48568 |
| 13 | pseudogene | 81.016196 | 81.016560 | negative | MGI_C57BL6J_6098100 | Gm48549 | ENSEMBL:ENSMUSG00000114801 | MGI:6098100 | pseudogene | predicted gene, 48549 |
| 13 | pseudogene | 81.029101 | 81.029906 | negative | MGI_C57BL6J_3648783 | Gm4813 | NCBI_Gene:218368,ENSEMBL:ENSMUSG00000114822 | MGI:3648783 | pseudogene | predicted gene 4813 |
| 13 | pseudogene | 81.064604 | 81.066200 | positive | MGI_C57BL6J_3647971 | Gm8399 | NCBI_Gene:666979,ENSEMBL:ENSMUSG00000043889 | MGI:3647971 | pseudogene | predicted gene 8399 |
| 13 | gene | 81.065477 | 81.065560 | positive | MGI_C57BL6J_5455018 | Gm25241 | ENSEMBL:ENSMUSG00000092650 | MGI:5455018 | miRNA gene | predicted gene, 25241 |
| 13 | pseudogene | 81.085391 | 81.087675 | negative | MGI_C57BL6J_5010702 | Gm18517 | NCBI_Gene:100417303,ENSEMBL:ENSMUSG00000114836 | MGI:5010702 | pseudogene | predicted gene, 18517 |
| 13 | gene | 81.095068 | 81.633388 | negative | MGI_C57BL6J_1274784 | Adgrv1 | NCBI_Gene:110789,ENSEMBL:ENSMUSG00000069170 | MGI:1274784 | protein coding gene | adhesion G protein-coupled receptor V1 |
| 13 | gene | 81.103406 | 81.128868 | positive | MGI_C57BL6J_5591798 | Gm32639 | NCBI_Gene:102635253 | MGI:5591798 | lncRNA gene | predicted gene, 32639 |
| 13 | gene | 81.239177 | 81.240784 | negative | MGI_C57BL6J_6098126 | Gm48566 | ENSEMBL:ENSMUSG00000114939 | MGI:6098126 | unclassified gene | predicted gene, 48566 |
| 13 | pseudogene | 81.500802 | 81.501456 | positive | MGI_C57BL6J_6098128 | Gm48567 | ENSEMBL:ENSMUSG00000114454 | MGI:6098128 | pseudogene | predicted gene, 48567 |
| 13 | gene | 81.512739 | 81.513867 | negative | MGI_C57BL6J_1917714 | 2610312I10Rik | NA | NA | unclassified gene | RIKEN cDNA 2610312I10 gene |
| 13 | gene | 81.657657 | 81.672871 | positive | MGI_C57BL6J_1915906 | Lysmd3 | NCBI_Gene:80289,ENSEMBL:ENSMUSG00000035840 | MGI:1915906 | protein coding gene | LysM, putative peptidoglycan-binding, domain containing 3 |
| 13 | gene | 81.673837 | 81.711013 | negative | MGI_C57BL6J_1914736 | Polr3g | NCBI_Gene:67486,ENSEMBL:ENSMUSG00000035834 | MGI:1914736 | protein coding gene | polymerase (RNA) III (DNA directed) polypeptide G |
| 13 | gene | 81.711341 | 81.753275 | positive | MGI_C57BL6J_1920102 | Mblac2 | NCBI_Gene:72852,ENSEMBL:ENSMUSG00000051098 | MGI:1920102 | protein coding gene | metallo-beta-lactamase domain containing 2 |
| 13 | pseudogene | 81.723990 | 81.725028 | negative | MGI_C57BL6J_6097466 | Gm48113 | ENSEMBL:ENSMUSG00000114905 | MGI:6097466 | pseudogene | predicted gene, 48113 |
| 13 | gene | 81.726900 | 81.727571 | positive | MGI_C57BL6J_1925252 | E130101A01Rik | NA | NA | unclassified gene | RIKEN cDNA E130101A01 gene |
| 13 | gene | 81.778773 | 81.783749 | negative | MGI_C57BL6J_4936893 | Gm17259 | ENSEMBL:ENSMUSG00000097793 | MGI:4936893 | lncRNA gene | predicted gene, 17259 |
| 13 | gene | 81.783077 | 81.797157 | positive | MGI_C57BL6J_1097706 | Cetn3 | NCBI_Gene:12626,ENSEMBL:ENSMUSG00000021537 | MGI:1097706 | protein coding gene | centrin 3 |
| 13 | gene | 81.825367 | 81.827218 | negative | MGI_C57BL6J_5591995 | Gm32836 | NCBI_Gene:102635524 | MGI:5591995 | lncRNA gene | predicted gene, 32836 |
| 13 | gene | 81.871469 | 81.873049 | positive | MGI_C57BL6J_5623893 | Gm41008 | NCBI_Gene:105245567 | MGI:5623893 | lncRNA gene | predicted gene, 41008 |
| 13 | gene | 81.927785 | 81.929687 | negative | MGI_C57BL6J_3588228 | F830210D05Rik | ENSEMBL:ENSMUSG00000114634 | MGI:3588228 | unclassified gene | RIKEN cDNA F830210D05 gene |
| 13 | gene | 82.001868 | 82.004830 | positive | MGI_C57BL6J_5623894 | Gm41009 | NCBI_Gene:105245568 | MGI:5623894 | lncRNA gene | predicted gene, 41009 |
| 13 | gene | 82.050751 | 82.050881 | positive | MGI_C57BL6J_5454072 | Gm24295 | ENSEMBL:ENSMUSG00000088923 | MGI:5454072 | snoRNA gene | predicted gene, 24295 |
| 13 | gene | 82.089757 | 82.258960 | positive | MGI_C57BL6J_6097523 | Gm48155 | ENSEMBL:ENSMUSG00000114461 | MGI:6097523 | lncRNA gene | predicted gene, 48155 |
| 13 | pseudogene | 82.378743 | 82.379674 | negative | MGI_C57BL6J_3648727 | Gm4936 | NCBI_Gene:238756,ENSEMBL:ENSMUSG00000114932 | MGI:3648727 | pseudogene | predicted gene 4936 |
| 13 | gene | 82.698275 | 82.698333 | negative | MGI_C57BL6J_4950395 | Mir3961 | miRBase:MI0016965,NCBI_Gene:100628621,ENSEMBL:ENSMUSG00000106655 | MGI:4950395 | miRNA gene | microRNA 3961 |
| 13 | gene | 83.065457 | 83.194347 | positive | MGI_C57BL6J_1917536 | 2310067P03Rik | ENSEMBL:ENSMUSG00000114372 | MGI:1917536 | lncRNA gene | RIKEN cDNA 2310067P03 gene |
| 13 | gene | 83.071680 | 83.120537 | negative | MGI_C57BL6J_5592232 | Gm33073 | NCBI_Gene:102635831 | MGI:5592232 | lncRNA gene | predicted gene, 33073 |
| 13 | pseudogene | 83.189565 | 83.189709 | negative | MGI_C57BL6J_6097525 | Gm48156 | ENSEMBL:ENSMUSG00000114735 | MGI:6097525 | pseudogene | predicted gene, 48156 |
| 13 | gene | 83.314533 | 83.356400 | negative | MGI_C57BL6J_5592376 | Gm33217 | NCBI_Gene:102636033 | MGI:5592376 | lncRNA gene | predicted gene, 33217 |
| 13 | gene | 83.429231 | 83.442790 | negative | MGI_C57BL6J_5623895 | Gm41010 | NCBI_Gene:105245569 | MGI:5623895 | lncRNA gene | predicted gene, 41010 |
| 13 | gene | 83.442914 | 83.446408 | negative | MGI_C57BL6J_5592433 | Gm33274 | NCBI_Gene:102636111 | MGI:5592433 | lncRNA gene | predicted gene, 33274 |
| 13 | gene | 83.498349 | 83.503901 | negative | MGI_C57BL6J_5592476 | Gm33317 | NCBI_Gene:102636177 | MGI:5592476 | lncRNA gene | predicted gene, 33317 |
| 13 | gene | 83.504034 | 83.667080 | positive | MGI_C57BL6J_99458 | Mef2c | NCBI_Gene:17260,ENSEMBL:ENSMUSG00000005583 | MGI:99458 | protein coding gene | myocyte enhancer factor 2C |
| 13 | gene | 83.601246 | 83.602668 | positive | MGI_C57BL6J_3708581 | Gm10759 | NA | NA | unclassified gene | predicted gene 10759 |
| 13 | pseudogene | 83.687222 | 83.687441 | positive | MGI_C57BL6J_6097396 | Gm48067 | ENSEMBL:ENSMUSG00000114254 | MGI:6097396 | pseudogene | predicted gene, 48067 |
| 13 | gene | 83.721381 | 83.884194 | positive | MGI_C57BL6J_2443574 | C130071C03Rik | NCBI_Gene:320203,ENSEMBL:ENSMUSG00000050334 | MGI:2443574 | lncRNA gene | RIKEN cDNA C130071C03 gene |
| 13 | gene | 83.723442 | 83.729582 | negative | MGI_C57BL6J_5477297 | Gm26803 | ENSEMBL:ENSMUSG00000097265 | MGI:5477297 | lncRNA gene | predicted gene, 26803 |
| 13 | gene | 83.734709 | 83.738535 | negative | MGI_C57BL6J_5592525 | Gm33366 | ENSEMBL:ENSMUSG00000102969 | MGI:5592525 | unclassified non-coding RNA gene | predicted gene, 33366 |
| 13 | gene | 83.738814 | 83.738885 | positive | MGI_C57BL6J_3619442 | Mir9-2 | miRBase:MI0000157,NCBI_Gene:723967,ENSEMBL:ENSMUSG00000065424 | MGI:3619442 | miRNA gene | microRNA 9-2 |
| 13 | pseudogene | 83.761132 | 83.762335 | negative | MGI_C57BL6J_3643659 | Gm6411 | NCBI_Gene:623241,ENSEMBL:ENSMUSG00000114588 | MGI:3643659 | pseudogene | predicted gene 6411 |
| 13 | pseudogene | 83.832241 | 83.833845 | positive | MGI_C57BL6J_3645281 | Gm5452 | NCBI_Gene:432789,ENSEMBL:ENSMUSG00000114687 | MGI:3645281 | pseudogene | predicted gene 5452 |
| 13 | gene | 83.891208 | 83.928710 | positive | MGI_C57BL6J_1919927 | 2810049E08Rik | NCBI_Gene:72677,ENSEMBL:ENSMUSG00000100891 | MGI:1919927 | lncRNA gene | RIKEN cDNA 2810049E08 gene |
| 13 | gene | 83.971355 | 83.987991 | negative | MGI_C57BL6J_3782418 | Gm4241 | ENSEMBL:ENSMUSG00000114379 | MGI:3782418 | lncRNA gene | predicted gene 4241 |
| 13 | pseudogene | 83.997456 | 83.998510 | negative | MGI_C57BL6J_3647954 | Gm8447 | NCBI_Gene:667081,ENSEMBL:ENSMUSG00000114930 | MGI:3647954 | pseudogene | predicted gene 8447 |
| 13 | gene | 84.025297 | 84.064774 | negative | MGI_C57BL6J_5009828 | Gm17750 | NCBI_Gene:553095,ENSEMBL:ENSMUSG00000098087 | MGI:5009828 | lncRNA gene | predicted gene, 17750 |
| 13 | pseudogene | 84.166453 | 84.166911 | positive | MGI_C57BL6J_3647453 | Gm8456 | NCBI_Gene:667101,ENSEMBL:ENSMUSG00000114335 | MGI:3647453 | pseudogene | predicted gene 8456 |
| 13 | gene | 84.182278 | 84.182344 | positive | MGI_C57BL6J_5454108 | Gm24331 | ENSEMBL:ENSMUSG00000092840 | MGI:5454108 | miRNA gene | predicted gene, 24331 |
| 13 | gene | 84.219580 | 84.222230 | negative | MGI_C57BL6J_2445042 | A230107N01Rik | NCBI_Gene:320919,ENSEMBL:ENSMUSG00000097292 | MGI:2445042 | lncRNA gene | RIKEN cDNA A230107N01 gene |
| 13 | gene | 84.222187 | 84.296141 | positive | MGI_C57BL6J_1919995 | Tmem161b | NCBI_Gene:72745,ENSEMBL:ENSMUSG00000035762 | MGI:1919995 | protein coding gene | transmembrane protein 161B |
| 13 | gene | 84.274844 | 84.280817 | positive | MGI_C57BL6J_6118679 | Gm49223 | ENSEMBL:ENSMUSG00000114602 | MGI:6118679 | lncRNA gene | predicted gene, 49223 |
| 13 | pseudogene | 84.281176 | 84.282125 | negative | MGI_C57BL6J_5826069 | Gm46432 | NCBI_Gene:108168103,ENSEMBL:ENSMUSG00000114827 | MGI:5826069 | pseudogene | predicted gene, 46432 |
| 13 | pseudogene | 84.339149 | 84.340113 | negative | MGI_C57BL6J_5504042 | Gm26927 | ENSEMBL:ENSMUSG00000098245 | MGI:5504042 | pseudogene | predicted gene, 26927 |
| 13 | gene | 84.347671 | 84.357771 | positive | MGI_C57BL6J_5623896 | Gm41011 | NCBI_Gene:105245571 | MGI:5623896 | lncRNA gene | predicted gene, 41011 |
| 13 | gene | 84.404839 | 84.405472 | positive | MGI_C57BL6J_4937252 | Gm17618 | NA | NA | protein coding gene | predicted gene%2c 17618 |
| 13 | gene | 84.573766 | 84.906167 | negative | MGI_C57BL6J_5593355 | Gm34196 | NCBI_Gene:102637367 | MGI:5593355 | lncRNA gene | predicted gene, 34196 |
| 13 | gene | 84.677177 | 84.699677 | positive | MGI_C57BL6J_5593405 | Gm34246 | NCBI_Gene:102637439 | MGI:5593405 | lncRNA gene | predicted gene, 34246 |
| 13 | pseudogene | 84.690210 | 84.690941 | negative | MGI_C57BL6J_5504028 | Gm26913 | ENSEMBL:ENSMUSG00000098081 | MGI:5504028 | pseudogene | predicted gene, 26913 |
| 13 | pseudogene | 84.973709 | 84.974317 | negative | MGI_C57BL6J_3782234 | Gm4059 | NCBI_Gene:100042841,ENSEMBL:ENSMUSG00000114338 | MGI:3782234 | pseudogene | predicted gene 4059 |
| 13 | pseudogene | 85.024162 | 85.025436 | negative | MGI_C57BL6J_3782241 | Gm4066 | NCBI_Gene:100042851,ENSEMBL:ENSMUSG00000114313 | MGI:3782241 | pseudogene | predicted pseudogene 4066 |
| 13 | pseudogene | 85.094335 | 85.095373 | positive | MGI_C57BL6J_6096889 | Gm47745 | ENSEMBL:ENSMUSG00000114614 | MGI:6096889 | pseudogene | predicted gene, 47745 |
| 13 | gene | 85.126550 | 85.127514 | negative | MGI_C57BL6J_3782251 | Gm4076 | ENSEMBL:ENSMUSG00000096449 | MGI:3782251 | protein coding gene | predicted gene 4076 |
| 13 | pseudogene | 85.140720 | 85.141102 | positive | MGI_C57BL6J_6096890 | Gm47746 | ENSEMBL:ENSMUSG00000114954 | MGI:6096890 | pseudogene | predicted gene, 47746 |
| 13 | gene | 85.189377 | 85.223469 | positive | MGI_C57BL6J_1913921 | Ccnh | NCBI_Gene:66671,ENSEMBL:ENSMUSG00000021548 | MGI:1913921 | protein coding gene | cyclin H |
| 13 | gene | 85.214699 | 85.289493 | negative | MGI_C57BL6J_97860 | Rasa1 | NCBI_Gene:218397,ENSEMBL:ENSMUSG00000021549 | MGI:97860 | protein coding gene | RAS p21 protein activator 1 |
| 13 | gene | 85.411681 | 85.411919 | negative | MGI_C57BL6J_5610869 | Gm37641 | ENSEMBL:ENSMUSG00000103196 | MGI:5610869 | unclassified gene | predicted gene, 37641 |
| 13 | pseudogene | 85.516402 | 85.516619 | positive | MGI_C57BL6J_6096708 | Gm47635 | ENSEMBL:ENSMUSG00000114211 | MGI:6096708 | pseudogene | predicted gene, 47635 |
| 13 | pseudogene | 85.572776 | 85.573677 | negative | MGI_C57BL6J_3648664 | Gm5666 | NCBI_Gene:435373,ENSEMBL:ENSMUSG00000114506 | MGI:3648664 | pseudogene | predicted gene 5666 |
| 13 | pseudogene | 85.865112 | 85.865333 | negative | MGI_C57BL6J_6096709 | Gm47636 | ENSEMBL:ENSMUSG00000114447 | MGI:6096709 | pseudogene | predicted gene, 47636 |
| 13 | pseudogene | 85.891742 | 85.892128 | positive | MGI_C57BL6J_5826033 | Gm46396 | NCBI_Gene:108168057,ENSEMBL:ENSMUSG00000114900 | MGI:5826033 | pseudogene | predicted gene, 46396 |
| 13 | gene | 86.017038 | 86.038907 | negative | MGI_C57BL6J_5593685 | Gm34526 | NCBI_Gene:102637807 | MGI:5593685 | lncRNA gene | predicted gene, 34526 |
| 13 | gene | 86.044798 | 86.046904 | negative | MGI_C57BL6J_103226 | Cox7c | NCBI_Gene:12867,ENSEMBL:ENSMUSG00000017778 | MGI:103226 | protein coding gene | cytochrome c oxidase subunit 7C |
| 13 | gene | 86.044989 | 86.045048 | negative | MGI_C57BL6J_5452351 | Gm22574 | ENSEMBL:ENSMUSG00000064917 | MGI:5452351 | snoRNA gene | predicted gene, 22574 |
| 13 | pseudogene | 86.235827 | 86.237727 | positive | MGI_C57BL6J_5504140 | Gm27025 | ENSEMBL:ENSMUSG00000098152 | MGI:5504140 | pseudogene | predicted gene, 27025 |
| 13 | gene | 86.725823 | 86.725932 | negative | MGI_C57BL6J_5455477 | Gm25700 | ENSEMBL:ENSMUSG00000095933 | MGI:5455477 | miRNA gene | predicted gene, 25700 |
| 13 | pseudogene | 87.234439 | 87.234594 | negative | MGI_C57BL6J_6096719 | Gm47642 | ENSEMBL:ENSMUSG00000114856 | MGI:6096719 | pseudogene | predicted gene, 47642 |
| 13 | pseudogene | 87.307726 | 87.308524 | positive | MGI_C57BL6J_5010450 | Gm18265 | NCBI_Gene:100416804,ENSEMBL:ENSMUSG00000114750 | MGI:5010450 | pseudogene | predicted gene, 18265 |
| 13 | gene | 87.456778 | 87.456887 | positive | MGI_C57BL6J_5454712 | Gm24935 | ENSEMBL:ENSMUSG00000076958 | MGI:5454712 | miRNA gene | predicted gene, 24935 |
| 13 | pseudogene | 87.782971 | 87.783976 | positive | MGI_C57BL6J_5434587 | Gm21232 | NCBI_Gene:108168058,ENSEMBL:ENSMUSG00000098011 | MGI:5434587 | pseudogene | predicted gene, 21232 |
| 13 | gene | 87.793851 | 87.799957 | negative | MGI_C57BL6J_5623897 | Gm41012 | NCBI_Gene:105245572 | MGI:5623897 | lncRNA gene | predicted gene, 41012 |
| 13 | pseudogene | 87.826538 | 87.827059 | positive | MGI_C57BL6J_5011424 | Gm19239 | NCBI_Gene:100418481,ENSEMBL:ENSMUSG00000114674 | MGI:5011424 | pseudogene | predicted gene, 19239 |
| 13 | gene | 87.977497 | 87.980299 | negative | MGI_C57BL6J_6096731 | Gm47649 | ENSEMBL:ENSMUSG00000114960 | MGI:6096731 | unclassified gene | predicted gene, 47649 |
| 13 | pseudogene | 88.184661 | 88.185456 | negative | MGI_C57BL6J_5504159 | Gm27044 | ENSEMBL:ENSMUSG00000098213 | MGI:5504159 | pseudogene | predicted gene, 27044 |
| 13 | pseudogene | 88.382346 | 88.382622 | positive | MGI_C57BL6J_6096734 | Gm47651 | ENSEMBL:ENSMUSG00000114565 | MGI:6096734 | pseudogene | predicted gene, 47651 |
| 13 | pseudogene | 88.467543 | 88.479056 | positive | MGI_C57BL6J_3645225 | Gm8526 | NCBI_Gene:667230,ENSEMBL:ENSMUSG00000057685 | MGI:3645225 | pseudogene | predicted gene 8526 |
| 13 | pseudogene | 88.480632 | 88.480791 | positive | MGI_C57BL6J_6096737 | Gm47653 | ENSEMBL:ENSMUSG00000114740 | MGI:6096737 | pseudogene | predicted gene, 47653 |
| 13 | pseudogene | 88.509280 | 88.511336 | negative | MGI_C57BL6J_5011324 | Gm19139 | NCBI_Gene:100418321,ENSEMBL:ENSMUSG00000114465 | MGI:5011324 | pseudogene | predicted gene, 19139 |
| 13 | gene | 88.521190 | 88.578168 | positive | MGI_C57BL6J_5593744 | Gm34585 | NCBI_Gene:102637889,ENSEMBL:ENSMUSG00000114273 | MGI:5593744 | lncRNA gene | predicted gene, 34585 |
| 13 | pseudogene | 88.547216 | 88.548270 | positive | MGI_C57BL6J_3648667 | Gm5667 | NCBI_Gene:435375,ENSEMBL:ENSMUSG00000114915 | MGI:3648667 | pseudogene | predicted gene 5667 |
| 13 | gene | 88.821472 | 89.323225 | positive | MGI_C57BL6J_1329025 | Edil3 | NCBI_Gene:13612,ENSEMBL:ENSMUSG00000034488 | MGI:1329025 | protein coding gene | EGF-like repeats and discoidin I-like domains 3 |
| 13 | gene | 88.824224 | 88.836120 | positive | MGI_C57BL6J_5621389 | Gm38504 | NCBI_Gene:102637959 | MGI:5621389 | lncRNA gene | predicted gene, 38504 |
| 13 | pseudogene | 88.843574 | 88.844576 | positive | MGI_C57BL6J_5010502 | Gm18317 | NCBI_Gene:100416914 | MGI:5010502 | pseudogene | predicted gene, 18317 |
| 13 | gene | 88.870746 | 88.880679 | positive | MGI_C57BL6J_5826070 | Gm46433 | NCBI_Gene:108168104 | MGI:5826070 | lncRNA gene | predicted gene, 46433 |
| 13 | gene | 89.499220 | 89.539669 | negative | MGI_C57BL6J_5593858 | Gm34699 | NCBI_Gene:102638034 | MGI:5593858 | lncRNA gene | predicted gene, 34699 |
| 13 | gene | 89.539796 | 89.611832 | positive | MGI_C57BL6J_1337006 | Hapln1 | NCBI_Gene:12950,ENSEMBL:ENSMUSG00000021613 | MGI:1337006 | protein coding gene | hyaluronan and proteoglycan link protein 1 |
| 13 | pseudogene | 89.577156 | 89.578419 | negative | MGI_C57BL6J_3647346 | Gm8546 | NCBI_Gene:100534278,ENSEMBL:ENSMUSG00000094463 | MGI:3647346 | pseudogene | predicted gene 8546 |
| 13 | gene | 89.655310 | 89.742694 | negative | MGI_C57BL6J_102889 | Vcan | NCBI_Gene:13003,ENSEMBL:ENSMUSG00000021614 | MGI:102889 | protein coding gene | versican |
| 13 | pseudogene | 89.662669 | 89.663573 | positive | MGI_C57BL6J_3826563 | Gm16318 | NCBI_Gene:100417016,ENSEMBL:ENSMUSG00000089972 | MGI:3826563 | pseudogene | predicted gene 16318 |
| 13 | gene | 89.734040 | 89.887166 | positive | MGI_C57BL6J_3782293 | Gm4117 | NCBI_Gene:100042940,ENSEMBL:ENSMUSG00000089940 | MGI:3782293 | lncRNA gene | predicted gene 4117 |
| 13 | gene | 89.774027 | 90.089608 | negative | MGI_C57BL6J_1333799 | Xrcc4 | NCBI_Gene:108138,ENSEMBL:ENSMUSG00000021615 | MGI:1333799 | protein coding gene | X-ray repair complementing defective repair in Chinese hamster cells 4 |
| 13 | gene | 89.902607 | 89.905712 | negative | MGI_C57BL6J_2443406 | A430063P04Rik | NA | NA | unclassified gene | RIKEN cDNA A430063P04 gene |
| 13 | gene | 89.927442 | 89.927543 | negative | MGI_C57BL6J_5454275 | Gm24498 | ENSEMBL:ENSMUSG00000089070 | MGI:5454275 | snRNA gene | predicted gene, 24498 |
| 13 | gene | 90.089123 | 90.143907 | positive | MGI_C57BL6J_1913324 | Tmem167 | NCBI_Gene:66074,ENSEMBL:ENSMUSG00000012422 | MGI:1913324 | protein coding gene | transmembrane protein 167 |
| 13 | gene | 90.098981 | 90.099114 | positive | MGI_C57BL6J_5454284 | Gm24507 | ENSEMBL:ENSMUSG00000088308 | MGI:5454284 | snoRNA gene | predicted gene, 24507 |
| 13 | gene | 90.157154 | 90.157972 | positive | MGI_C57BL6J_5610955 | Gm37727 | ENSEMBL:ENSMUSG00000103228 | MGI:5610955 | unclassified gene | predicted gene, 37727 |
| 13 | gene | 90.160746 | 90.163019 | negative | MGI_C57BL6J_5623898 | Gm41013 | NCBI_Gene:105245573 | MGI:5623898 | lncRNA gene | predicted gene, 41013 |
| 13 | gene | 90.179217 | 90.217142 | negative | MGI_C57BL6J_5588839 | Gm29680 | ENSEMBL:ENSMUSG00000114541 | MGI:5588839 | lncRNA gene | predicted gene, 29680 |
| 13 | gene | 90.238172 | 90.238714 | positive | MGI_C57BL6J_5610282 | Gm37054 | ENSEMBL:ENSMUSG00000102756 | MGI:5610282 | unclassified gene | predicted gene, 37054 |
| 13 | gene | 90.360130 | 90.418100 | negative | MGI_C57BL6J_6096517 | Gm47520 | ENSEMBL:ENSMUSG00000114204 | MGI:6096517 | lncRNA gene | predicted gene, 47520 |
| 13 | gene | 90.385663 | 90.386557 | negative | MGI_C57BL6J_5610936 | Gm37708 | ENSEMBL:ENSMUSG00000102793 | MGI:5610936 | unclassified gene | predicted gene, 37708 |
| 13 | gene | 90.466750 | 90.549165 | negative | MGI_C57BL6J_5594270 | Gm35111 | NCBI_Gene:102638581 | MGI:5594270 | lncRNA gene | predicted gene, 35111 |
| 13 | pseudogene | 90.583329 | 90.583720 | positive | MGI_C57BL6J_5433890 | Gm21726 | ENSEMBL:ENSMUSG00000094277 | MGI:5433890 | pseudogene | predicted gene, 21726 |
| 13 | gene | 90.634085 | 90.634497 | positive | MGI_C57BL6J_5610194 | Gm36966 | ENSEMBL:ENSMUSG00000103069 | MGI:5610194 | unclassified gene | predicted gene, 36966 |
| 13 | gene | 90.760112 | 90.760221 | positive | MGI_C57BL6J_5690730 | Gm44338 | ENSEMBL:ENSMUSG00000106360 | MGI:5690730 | miRNA gene | predicted gene, 44338 |
| 13 | pseudogene | 90.777825 | 90.778083 | negative | MGI_C57BL6J_6096519 | Gm47521 | ENSEMBL:ENSMUSG00000114265 | MGI:6096519 | pseudogene | predicted gene, 47521 |
| 13 | pseudogene | 90.867606 | 90.868607 | positive | MGI_C57BL6J_5010703 | Gm18518 | NCBI_Gene:100417305,ENSEMBL:ENSMUSG00000114589 | MGI:5010703 | pseudogene | predicted gene, 18518 |
| 13 | gene | 90.883433 | 90.905355 | negative | MGI_C57BL6J_3648665 | Atp6ap1l | NCBI_Gene:435376,ENSEMBL:ENSMUSG00000078958 | MGI:3648665 | protein coding gene | ATPase, H+ transporting, lysosomal accessory protein 1-like |
| 13 | gene | 90.922958 | 90.924950 | positive | MGI_C57BL6J_1913725 | Rps23 | NCBI_Gene:66475,ENSEMBL:ENSMUSG00000049517 | MGI:1913725 | protein coding gene | ribosomal protein S23 |
| 13 | gene | 90.929910 | 90.931681 | negative | MGI_C57BL6J_5826071 | Gm46434 | NCBI_Gene:108168106 | MGI:5826071 | lncRNA gene | predicted gene, 46434 |
| 13 | gene | 90.935348 | 91.224789 | negative | MGI_C57BL6J_1914045 | Atg10 | NCBI_Gene:66795,ENSEMBL:ENSMUSG00000021619 | MGI:1914045 | protein coding gene | autophagy related 10 |
| 13 | pseudogene | 91.220008 | 91.222703 | positive | MGI_C57BL6J_4937084 | Gm17450 | NCBI_Gene:102638648,ENSEMBL:ENSMUSG00000091419 | MGI:4937084 | pseudogene | predicted gene, 17450 |
| 13 | gene | 91.250388 | 91.278012 | negative | MGI_C57BL6J_5623899 | Gm41014 | NCBI_Gene:105245574 | MGI:5623899 | lncRNA gene | predicted gene, 41014 |
| 13 | gene | 91.259376 | 91.274954 | positive | MGI_C57BL6J_6096300 | Gm47381 | ENSEMBL:ENSMUSG00000114688 | MGI:6096300 | lncRNA gene | predicted gene, 47381 |
| 13 | pseudogene | 91.260932 | 91.262420 | negative | MGI_C57BL6J_3782306 | Gm4130 | NCBI_Gene:100042959,ENSEMBL:ENSMUSG00000114579 | MGI:3782306 | pseudogene | predicted gene 4130 |
| 13 | gene | 91.368990 | 91.388085 | negative | MGI_C57BL6J_2443762 | A830009L08Rik | NCBI_Gene:320297,ENSEMBL:ENSMUSG00000097182 | MGI:2443762 | lncRNA gene | RIKEN cDNA A830009L08 gene |
| 13 | gene | 91.373842 | 91.374294 | negative | MGI_C57BL6J_5580246 | Gm29540 | ENSEMBL:ENSMUSG00000099340 | MGI:5580246 | lncRNA gene | predicted gene 29540 |
| 13 | gene | 91.460283 | 91.706175 | positive | MGI_C57BL6J_1914220 | Ssbp2 | NCBI_Gene:66970,ENSEMBL:ENSMUSG00000003992 | MGI:1914220 | protein coding gene | single-stranded DNA binding protein 2 |
| 13 | gene | 91.534058 | 91.539333 | positive | MGI_C57BL6J_5623901 | Gm41016 | NCBI_Gene:105245576 | MGI:5623901 | lncRNA gene | predicted gene, 41016 |
| 13 | gene | 91.701665 | 91.741872 | negative | MGI_C57BL6J_3603826 | 4833422C13Rik | NCBI_Gene:373852,ENSEMBL:ENSMUSG00000074782 | MGI:3603826 | lncRNA gene | RIKEN cDNA 4833422C13 gene |
| 13 | gene | 91.723256 | 91.723383 | positive | MGI_C57BL6J_5452555 | Gm22778 | ENSEMBL:ENSMUSG00000077278 | MGI:5452555 | snoRNA gene | predicted gene, 22778 |
| 13 | gene | 91.741505 | 91.786148 | positive | MGI_C57BL6J_1921406 | Acot12 | NCBI_Gene:74156,ENSEMBL:ENSMUSG00000021620 | MGI:1921406 | protein coding gene | acyl-CoA thioesterase 12 |
| 13 | gene | 91.744705 | 91.745004 | negative | MGI_C57BL6J_5454374 | Gm24597 | ENSEMBL:ENSMUSG00000084452 | MGI:5454374 | unclassified non-coding RNA gene | predicted gene, 24597 |
| 13 | gene | 91.752106 | 91.752185 | positive | MGI_C57BL6J_5531038 | Gm27656 | ENSEMBL:ENSMUSG00000099004 | MGI:5531038 | miRNA gene | predicted gene, 27656 |
| 13 | gene | 91.780897 | 91.782355 | negative | MGI_C57BL6J_1918538 | 4933440K10Rik | NA | NA | unclassified gene | RIKEN cDNA 4933440K10 gene |
| 13 | gene | 91.796519 | 91.807997 | negative | MGI_C57BL6J_1916335 | Zcchc9 | NCBI_Gene:69085,ENSEMBL:ENSMUSG00000021621 | MGI:1916335 | protein coding gene | zinc finger, CCHC domain containing 9 |
| 13 | gene | 91.853387 | 91.876923 | negative | MGI_C57BL6J_1923972 | Ckmt2 | NCBI_Gene:76722,ENSEMBL:ENSMUSG00000021622 | MGI:1923972 | protein coding gene | creatine kinase, mitochondrial 2 |
| 13 | gene | 91.861432 | 91.862044 | positive | MGI_C57BL6J_1925729 | 1700119I11Rik | ENSEMBL:ENSMUSG00000082488 | MGI:1925729 | lncRNA gene | RIKEN cDNA 1700119I11 gene |
| 13 | gene | 91.880400 | 92.131656 | negative | MGI_C57BL6J_109137 | Rasgrf2 | NCBI_Gene:19418,ENSEMBL:ENSMUSG00000021708 | MGI:109137 | protein coding gene | RAS protein-specific guanine nucleotide-releasing factor 2 |
| 13 | gene | 91.923619 | 91.934105 | positive | MGI_C57BL6J_5826072 | Gm46435 | NCBI_Gene:108168107 | MGI:5826072 | lncRNA gene | predicted gene, 46435 |
| 13 | gene | 92.211872 | 92.355003 | negative | MGI_C57BL6J_109519 | Msh3 | NCBI_Gene:17686,ENSEMBL:ENSMUSG00000014850 | MGI:109519 | protein coding gene | mutS homolog 3 |
| 13 | gene | 92.304432 | 92.307504 | positive | MGI_C57BL6J_5012564 | Gm20379 | NCBI_Gene:105245585,ENSEMBL:ENSMUSG00000100622 | MGI:5012564 | protein coding gene | predicted gene, 20379 |
| 13 | gene | 92.354726 | 92.389053 | positive | MGI_C57BL6J_94890 | Dhfr | NCBI_Gene:13361,ENSEMBL:ENSMUSG00000021707 | MGI:94890 | protein coding gene | dihydrofolate reductase |
| 13 | pseudogene | 92.355706 | 92.357918 | negative | MGI_C57BL6J_5011493 | Gm19308 | NCBI_Gene:102633605 | MGI:5011493 | pseudogene | predicted gene, 19308 |
| 13 | gene | 92.415017 | 92.427224 | negative | MGI_C57BL6J_1924634 | C030017D09Rik | ENSEMBL:ENSMUSG00000090907 | MGI:1924634 | lncRNA gene | RIKEN cDNA C030017D09 gene |
| 13 | gene | 92.425642 | 92.441658 | positive | MGI_C57BL6J_2443245 | Ankrd34b | NCBI_Gene:218440,ENSEMBL:ENSMUSG00000045034 | MGI:2443245 | protein coding gene | ankyrin repeat domain 34B |
| 13 | gene | 92.449620 | 92.484031 | negative | MGI_C57BL6J_1921192 | Fam151b | NCBI_Gene:73942,ENSEMBL:ENSMUSG00000034334 | MGI:1921192 | protein coding gene | family with sequence similarity 151, member B |
| 13 | gene | 92.487101 | 92.530922 | negative | MGI_C57BL6J_2145181 | Zfyve16 | NCBI_Gene:218441,ENSEMBL:ENSMUSG00000021706 | MGI:2145181 | protein coding gene | zinc finger, FYVE domain containing 16 |
| 13 | gene | 92.574631 | 92.576232 | negative | MGI_C57BL6J_1930801 | Spz1 | NCBI_Gene:79401,ENSEMBL:ENSMUSG00000046957 | MGI:1930801 | protein coding gene | spermatogenic leucine zipper 1 |
| 13 | gene | 92.611091 | 92.711947 | positive | MGI_C57BL6J_2444223 | Serinc5 | NCBI_Gene:218442,ENSEMBL:ENSMUSG00000021703 | MGI:2444223 | protein coding gene | serine incorporator 5 |
| 13 | gene | 92.704730 | 92.707718 | negative | MGI_C57BL6J_5623903 | Gm41018 | NCBI_Gene:105245586 | MGI:5623903 | lncRNA gene | predicted gene, 41018 |
| 13 | pseudogene | 92.719349 | 92.720028 | positive | MGI_C57BL6J_3643997 | Gm5199 | NCBI_Gene:382792,ENSEMBL:ENSMUSG00000114631 | MGI:3643997 | pseudogene | predicted gene 5199 |
| 13 | gene | 92.751586 | 92.794818 | negative | MGI_C57BL6J_1101779 | Thbs4 | NCBI_Gene:21828,ENSEMBL:ENSMUSG00000021702 | MGI:1101779 | protein coding gene | thrombospondin 4 |
| 13 | gene | 92.844743 | 92.858230 | positive | MGI_C57BL6J_2686040 | Mtx3 | NCBI_Gene:382793,ENSEMBL:ENSMUSG00000021704 | MGI:2686040 | protein coding gene | metaxin 3 |
| 13 | gene | 92.883010 | 92.908723 | positive | MGI_C57BL6J_5590789 | Gm31630 | NCBI_Gene:102633924,ENSEMBL:ENSMUSG00000114823 | MGI:5590789 | lncRNA gene | predicted gene, 31630 |
| 13 | gene | 92.925353 | 92.927712 | negative | MGI_C57BL6J_3642923 | Gm10753 | NA | NA | protein coding gene | predicted gene 10753 |
| 13 | gene | 93.005828 | 93.023589 | positive | MGI_C57BL6J_5623904 | Gm41019 | NCBI_Gene:105245587 | MGI:5623904 | lncRNA gene | predicted gene, 41019 |
| 13 | gene | 93.040713 | 93.144724 | negative | MGI_C57BL6J_1923719 | Cmya5 | NCBI_Gene:76469,ENSEMBL:ENSMUSG00000047419 | MGI:1923719 | protein coding gene | cardiomyopathy associated 5 |
| 13 | gene | 93.055189 | 93.069524 | positive | MGI_C57BL6J_5590904 | Gm31745 | NCBI_Gene:102634073 | MGI:5590904 | lncRNA gene | predicted gene, 31745 |
| 13 | gene | 93.073304 | 93.074757 | positive | MGI_C57BL6J_3647983 | Gm4814 | NCBI_Gene:100502942,ENSEMBL:ENSMUSG00000114382 | MGI:3647983 | lncRNA gene | predicted gene 4814 |
| 13 | gene | 93.094833 | 93.098968 | positive | MGI_C57BL6J_5439422 | A630019I02Rik | NCBI_Gene:408254,ENSEMBL:ENSMUSG00000114968 | MGI:5439422 | lncRNA gene | RIKEN cDNA A630019I02 gene |
| 13 | gene | 93.146282 | 93.192385 | negative | MGI_C57BL6J_2140950 | Tent2 | NCBI_Gene:100715,ENSEMBL:ENSMUSG00000042167 | MGI:2140950 | protein coding gene | terminal nucleotidyltransferase 2 |
| 13 | gene | 93.157267 | 93.158065 | positive | MGI_C57BL6J_2145256 | D130076G13Rik | NA | NA | unclassified gene | RIKEN cDNA D130076G13 gene |
| 13 | gene | 93.197339 | 93.198690 | negative | MGI_C57BL6J_5623905 | Gm41020 | NCBI_Gene:105245588 | MGI:5623905 | lncRNA gene | predicted gene, 41020 |
| 13 | pseudogene | 93.293763 | 93.294947 | positive | MGI_C57BL6J_3780243 | Gm2076 | NCBI_Gene:100039163,ENSEMBL:ENSMUSG00000098092 | MGI:3780243 | pseudogene | predicted gene 2076 |
| 13 | gene | 93.299635 | 93.405129 | positive | MGI_C57BL6J_1347345 | Homer1 | NCBI_Gene:26556,ENSEMBL:ENSMUSG00000007617 | MGI:1347345 | protein coding gene | homer scaffolding protein 1 |
| 13 | gene | 93.301514 | 93.303368 | negative | MGI_C57BL6J_3026940 | A230091C14Rik | NA | NA | unclassified gene | RIKEN cDNA A230091C14 gene |
| 13 | gene | 93.312041 | 93.314055 | positive | MGI_C57BL6J_6095795 | Gm47075 | ENSEMBL:ENSMUSG00000114169 | MGI:6095795 | unclassified gene | predicted gene, 47075 |
| 13 | gene | 93.317427 | 93.319065 | positive | MGI_C57BL6J_6095816 | Gm47088 | ENSEMBL:ENSMUSG00000113076 | MGI:6095816 | unclassified gene | predicted gene, 47088 |
| 13 | gene | 93.358401 | 93.360525 | positive | MGI_C57BL6J_2444330 | C330006P03Rik | NA | NA | unclassified gene | RIKEN cDNA C330006P03 gene |
| 13 | gene | 93.410783 | 93.412957 | positive | MGI_C57BL6J_6095924 | Gm47155 | ENSEMBL:ENSMUSG00000114019 | MGI:6095924 | unclassified gene | predicted gene, 47155 |
| 13 | pseudogene | 93.420604 | 93.421325 | negative | MGI_C57BL6J_5011145 | Gm18960 | NCBI_Gene:100418038,ENSEMBL:ENSMUSG00000113514 | MGI:5011145 | pseudogene | predicted gene, 18960 |
| 13 | gene | 93.430097 | 93.499808 | negative | MGI_C57BL6J_1913096 | Jmy | NCBI_Gene:57748,ENSEMBL:ENSMUSG00000021690 | MGI:1913096 | protein coding gene | junction-mediating and regulatory protein |
| 13 | gene | 93.448413 | 93.448890 | positive | MGI_C57BL6J_3643032 | Gm6109 | NCBI_Gene:619883,ENSEMBL:ENSMUSG00000113237 | MGI:3643032 | protein coding gene | predicted gene 6109 |
| 13 | pseudogene | 93.461479 | 93.462184 | positive | MGI_C57BL6J_5589314 | Gm30155 | NCBI_Gene:102631956,ENSEMBL:ENSMUSG00000113247 | MGI:5589314 | pseudogene | predicted gene, 30155 |
| 13 | pseudogene | 93.585154 | 93.585697 | positive | MGI_C57BL6J_5010318 | Gm18133 | NCBI_Gene:100416505 | MGI:5010318 | pseudogene | predicted gene, 18133 |
| 13 | gene | 93.595723 | 93.598384 | negative | MGI_C57BL6J_5591136 | Gm31977 | NCBI_Gene:102634381 | MGI:5591136 | lncRNA gene | predicted gene, 31977 |
| 13 | gene | 93.616675 | 93.637961 | negative | MGI_C57BL6J_1339972 | Bhmt | NCBI_Gene:12116,ENSEMBL:ENSMUSG00000074768 | MGI:1339972 | protein coding gene | betaine-homocysteine methyltransferase |
| 13 | gene | 93.625382 | 93.628633 | positive | MGI_C57BL6J_3783067 | Gm15622 | ENSEMBL:ENSMUSG00000085834 | MGI:3783067 | lncRNA gene | predicted gene 15622 |
| 13 | gene | 93.655720 | 93.674302 | negative | MGI_C57BL6J_1891379 | Bhmt2 | NCBI_Gene:64918,ENSEMBL:ENSMUSG00000042118 | MGI:1891379 | protein coding gene | betaine-homocysteine methyltransferase 2 |
| 13 | gene | 93.674433 | 93.752833 | positive | MGI_C57BL6J_1921379 | Dmgdh | NCBI_Gene:74129,ENSEMBL:ENSMUSG00000042102 | MGI:1921379 | protein coding gene | dimethylglycine dehydrogenase precursor |
| 13 | gene | 93.706100 | 93.719175 | negative | MGI_C57BL6J_2145435 | AW495222 | NCBI_Gene:105364,ENSEMBL:ENSMUSG00000087334 | MGI:2145435 | lncRNA gene | expressed sequence AW495222 |
| 13 | pseudogene | 93.727363 | 93.727702 | negative | MGI_C57BL6J_3783065 | Gm15620 | ENSEMBL:ENSMUSG00000085202 | MGI:3783065 | pseudogene | predicted gene 15620 |
| 13 | gene | 93.754998 | 93.758525 | positive | MGI_C57BL6J_5623906 | Gm41021 | NCBI_Gene:105245589 | MGI:5623906 | lncRNA gene | predicted gene, 41021 |
| 13 | gene | 93.770950 | 93.772194 | negative | MGI_C57BL6J_6096014 | Gm47210 | ENSEMBL:ENSMUSG00000113647 | MGI:6096014 | lncRNA gene | predicted gene, 47210 |
| 13 | gene | 93.771630 | 93.943016 | positive | MGI_C57BL6J_88075 | Arsb | NCBI_Gene:11881,ENSEMBL:ENSMUSG00000042082 | MGI:88075 | protein coding gene | arylsulfatase B |
| 13 | gene | 93.790777 | 93.790837 | positive | MGI_C57BL6J_5454852 | Mir5624 | miRBase:MI0019193,NCBI_Gene:100885837,ENSEMBL:ENSMUSG00000093480 | MGI:5454852 | miRNA gene | microRNA 5624 |
| 13 | gene | 93.923187 | 93.923311 | negative | MGI_C57BL6J_5455311 | Gm25534 | ENSEMBL:ENSMUSG00000093106 | MGI:5455311 | miRNA gene | predicted gene, 25534 |
| 13 | gene | 93.940990 | 93.943015 | negative | MGI_C57BL6J_5477021 | Gm26527 | ENSEMBL:ENSMUSG00000097582 | MGI:5477021 | lncRNA gene | predicted gene, 26527 |
| 13 | gene | 93.956310 | 93.956409 | positive | MGI_C57BL6J_5454514 | Gm24737 | ENSEMBL:ENSMUSG00000089486 | MGI:5454514 | snRNA gene | predicted gene, 24737 |
| 13 | gene | 93.983195 | 93.991768 | negative | MGI_C57BL6J_6096024 | Gm47216 | ENSEMBL:ENSMUSG00000113791 | MGI:6096024 | lncRNA gene | predicted gene, 47216 |
| 13 | pseudogene | 94.053835 | 94.054679 | positive | MGI_C57BL6J_109498 | Cycs-ps3 | NCBI_Gene:13066,ENSEMBL:ENSMUSG00000114174 | MGI:109498 | pseudogene | cytochrome c, pseudogene 3 |
| 13 | gene | 94.057654 | 94.195410 | positive | MGI_C57BL6J_2145236 | Lhfpl2 | NCBI_Gene:218454,ENSEMBL:ENSMUSG00000045312 | MGI:2145236 | protein coding gene | lipoma HMGIC fusion partner-like 2 |
| 13 | gene | 94.137447 | 94.140117 | negative | MGI_C57BL6J_3801930 | Gm15907 | ENSEMBL:ENSMUSG00000087319 | MGI:3801930 | lncRNA gene | predicted gene 15907 |
| 13 | gene | 94.201310 | 94.285857 | negative | MGI_C57BL6J_1349480 | Scamp1 | NCBI_Gene:107767,ENSEMBL:ENSMUSG00000021687 | MGI:1349480 | protein coding gene | secretory carrier membrane protein 1 |
| 13 | gene | 94.224890 | 94.226025 | positive | MGI_C57BL6J_1914574 | 1700042D18Rik | NA | NA | unclassified gene | RIKEN cDNA 1700042D18 gene |
| 13 | gene | 94.289448 | 94.296741 | negative | MGI_C57BL6J_5591248 | Gm32089 | NCBI_Gene:102634530,ENSEMBL:ENSMUSG00000114055 | MGI:5591248 | lncRNA gene | predicted gene, 32089 |
| 13 | gene | 94.294017 | 94.301250 | positive | MGI_C57BL6J_6097451 | Gm48102 | ENSEMBL:ENSMUSG00000113494 | MGI:6097451 | lncRNA gene | predicted gene, 48102 |
| 13 | pseudogene | 94.298064 | 94.301287 | negative | MGI_C57BL6J_3648669 | Gm5668 | NCBI_Gene:105245651,ENSEMBL:ENSMUSG00000114056 | MGI:3648669 | pseudogene | predicted gene 5668 |
| 13 | gene | 94.303169 | 94.306838 | negative | MGI_C57BL6J_5621538 | Gm38653 | NCBI_Gene:102643051 | MGI:5621538 | lncRNA gene | predicted gene, 38653 |
| 13 | gene | 94.306850 | 94.310297 | positive | MGI_C57BL6J_5623907 | Gm41022 | NCBI_Gene:105245590 | MGI:5623907 | lncRNA gene | predicted gene, 41022 |
| 13 | gene | 94.355839 | 94.358923 | negative | MGI_C57BL6J_3641792 | Gm9776 | NCBI_Gene:328309,ENSEMBL:ENSMUSG00000042857 | MGI:3641792 | lncRNA gene | predicted gene 9776 |
| 13 | gene | 94.358580 | 94.566317 | positive | MGI_C57BL6J_1333879 | Ap3b1 | NCBI_Gene:11774,ENSEMBL:ENSMUSG00000021686 | MGI:1333879 | protein coding gene | adaptor-related protein complex 3, beta 1 subunit |
| 13 | gene | 94.517162 | 94.517275 | positive | MGI_C57BL6J_5454886 | Gm25109 | ENSEMBL:ENSMUSG00000084423 | MGI:5454886 | miRNA gene | predicted gene, 25109 |
| 13 | gene | 94.619280 | 94.619927 | positive | MGI_C57BL6J_5610219 | Gm36991 | ENSEMBL:ENSMUSG00000103438 | MGI:5610219 | unclassified gene | predicted gene, 36991 |
| 13 | gene | 94.645899 | 94.647344 | negative | MGI_C57BL6J_6097724 | Gm48287 | ENSEMBL:ENSMUSG00000113598 | MGI:6097724 | lncRNA gene | predicted gene, 48287 |
| 13 | gene | 94.685566 | 94.700356 | negative | MGI_C57BL6J_5591464 | Gm32305 | NCBI_Gene:102634807,ENSEMBL:ENSMUSG00000112934 | MGI:5591464 | lncRNA gene | predicted gene, 32305 |
| 13 | gene | 94.700733 | 94.703152 | negative | MGI_C57BL6J_5591510 | Gm32351 | NCBI_Gene:102634869,ENSEMBL:ENSMUSG00000113904 | MGI:5591510 | lncRNA gene | predicted gene, 32351 |
| 13 | pseudogene | 94.752270 | 94.752593 | positive | MGI_C57BL6J_6097769 | Gm48316 | ENSEMBL:ENSMUSG00000113006 | MGI:6097769 | pseudogene | predicted gene, 48316 |
| 13 | pseudogene | 94.754202 | 94.754593 | negative | MGI_C57BL6J_5826026 | Gm46389 | NCBI_Gene:108168048,ENSEMBL:ENSMUSG00000113547 | MGI:5826026 | pseudogene | predicted gene, 46389 |
| 13 | gene | 94.788910 | 94.842922 | positive | MGI_C57BL6J_107549 | Tbca | NCBI_Gene:21371,ENSEMBL:ENSMUSG00000042043 | MGI:107549 | protein coding gene | tubulin cofactor A |
| 13 | gene | 94.875602 | 94.885130 | positive | MGI_C57BL6J_99835 | Otp | NCBI_Gene:18420,ENSEMBL:ENSMUSG00000021685 | MGI:99835 | protein coding gene | orthopedia homeobox |
| 13 | gene | 94.976215 | 95.023316 | positive | MGI_C57BL6J_2445123 | Wdr41 | NCBI_Gene:218460,ENSEMBL:ENSMUSG00000042015 | MGI:2445123 | protein coding gene | WD repeat domain 41 |
| 13 | gene | 94.978597 | 94.982413 | positive | MGI_C57BL6J_5623909 | Gm41024 | NCBI_Gene:105245592 | MGI:5623909 | lncRNA gene | predicted gene, 41024 |
| 13 | pseudogene | 95.002358 | 95.002824 | positive | MGI_C57BL6J_6097813 | Gm48349 | ENSEMBL:ENSMUSG00000113871 | MGI:6097813 | pseudogene | predicted gene, 48349 |
| 13 | gene | 95.012681 | 95.023790 | negative | MGI_C57BL6J_6097814 | Gm48350 | ENSEMBL:ENSMUSG00000114014 | MGI:6097814 | lncRNA gene | predicted gene, 48350 |
| 13 | gene | 95.024105 | 95.250400 | negative | MGI_C57BL6J_2443999 | Pde8b | NCBI_Gene:218461,ENSEMBL:ENSMUSG00000021684 | MGI:2443999 | protein coding gene | phosphodiesterase 8B |
| 13 | gene | 95.024301 | 95.024900 | positive | MGI_C57BL6J_6097914 | Gm48420 | ENSEMBL:ENSMUSG00000113070 | MGI:6097914 | lncRNA gene | predicted gene, 48420 |
| 13 | gene | 95.058837 | 95.071701 | positive | MGI_C57BL6J_5623910 | Gm41025 | NCBI_Gene:105245593 | MGI:5623910 | lncRNA gene | predicted gene, 41025 |
| 13 | gene | 95.208112 | 95.208936 | positive | MGI_C57BL6J_3801905 | Gm16243 | ENSEMBL:ENSMUSG00000090033 | MGI:3801905 | lncRNA gene | predicted gene 16243 |
| 13 | gene | 95.218193 | 95.219690 | negative | MGI_C57BL6J_5012249 | Gm20064 | NA | NA | unclassified gene | predicted gene%2c 20064 |
| 13 | gene | 95.220327 | 95.222349 | negative | MGI_C57BL6J_3028046 | C030010F05Rik | NA | NA | unclassified gene | RIKEN cDNA C030010F05 gene |
| 13 | gene | 95.323605 | 95.337842 | positive | MGI_C57BL6J_1919364 | Zbed3 | NCBI_Gene:72114,ENSEMBL:ENSMUSG00000041995 | MGI:1919364 | protein coding gene | zinc finger, BED type containing 3 |
| 13 | gene | 95.330580 | 95.330683 | positive | MGI_C57BL6J_3836978 | Mir1940 | miRBase:MI0009929 | MGI:3836978 | miRNA gene | microRNA 1940 |
| 13 | gene | 95.330600 | 95.330737 | positive | MGI_C57BL6J_3819507 | Snora47 | NCBI_Gene:100217450,ENSEMBL:ENSMUSG00000088108 | MGI:3819507 | snoRNA gene | small nucleolar RNA, H/ACA box 47 |
| 13 | gene | 95.332618 | 95.332749 | positive | MGI_C57BL6J_5452438 | Gm22661 | ENSEMBL:ENSMUSG00000077380 | MGI:5452438 | snoRNA gene | predicted gene, 22661 |
| 13 | gene | 95.350683 | 95.375357 | negative | MGI_C57BL6J_1913799 | Aggf1 | NCBI_Gene:66549,ENSEMBL:ENSMUSG00000021681 | MGI:1913799 | protein coding gene | angiogenic factor with G patch and FHA domains 1 |
| 13 | pseudogene | 95.421217 | 95.421676 | negative | MGI_C57BL6J_6098358 | Gm48712 | ENSEMBL:ENSMUSG00000114122 | MGI:6098358 | pseudogene | predicted gene, 48712 |
| 13 | gene | 95.431371 | 95.444924 | negative | MGI_C57BL6J_88497 | Crhbp | NCBI_Gene:12919,ENSEMBL:ENSMUSG00000021680 | MGI:88497 | protein coding gene | corticotropin releasing hormone binding protein |
| 13 | gene | 95.477268 | 95.478721 | negative | MGI_C57BL6J_2685471 | S100z | NCBI_Gene:268686,ENSEMBL:ENSMUSG00000021679 | MGI:2685471 | protein coding gene | S100 calcium binding protein, zeta |
| 13 | gene | 95.511718 | 95.525240 | negative | MGI_C57BL6J_101910 | F2rl1 | NCBI_Gene:14063,ENSEMBL:ENSMUSG00000021678 | MGI:101910 | protein coding gene | coagulation factor II (thrombin) receptor-like 1 |
| 13 | gene | 95.535590 | 95.545200 | positive | MGI_C57BL6J_6098391 | Gm48730 | ENSEMBL:ENSMUSG00000113359 | MGI:6098391 | lncRNA gene | predicted gene, 48730 |
| 13 | gene | 95.544861 | 95.548820 | negative | MGI_C57BL6J_5591799 | Gm32640 | NCBI_Gene:102635254 | MGI:5591799 | lncRNA gene | predicted gene, 32640 |
| 13 | pseudogene | 95.578292 | 95.578976 | negative | MGI_C57BL6J_6098393 | Gm48731 | ENSEMBL:ENSMUSG00000113969 | MGI:6098393 | pseudogene | predicted gene, 48731 |
| 13 | gene | 95.601789 | 95.618487 | negative | MGI_C57BL6J_101802 | F2r | NCBI_Gene:14062,ENSEMBL:ENSMUSG00000048376 | MGI:101802 | protein coding gene | coagulation factor II (thrombin) receptor |
| 13 | gene | 95.627174 | 95.892483 | negative | MGI_C57BL6J_2449975 | Iqgap2 | NCBI_Gene:544963,ENSEMBL:ENSMUSG00000021676 | MGI:2449975 | protein coding gene | IQ motif containing GTPase activating protein 2 |
| 13 | gene | 95.672136 | 95.680237 | positive | MGI_C57BL6J_5592233 | Gm33074 | NCBI_Gene:102635832 | MGI:5592233 | lncRNA gene | predicted gene, 33074 |
| 13 | gene | 95.685145 | 95.686809 | positive | MGI_C57BL6J_6098416 | Gm48745 | ENSEMBL:ENSMUSG00000113631 | MGI:6098416 | unclassified gene | predicted gene, 48745 |
| 13 | gene | 95.696853 | 95.702768 | positive | MGI_C57BL6J_1298208 | F2rl2 | NCBI_Gene:14064,ENSEMBL:ENSMUSG00000021675 | MGI:1298208 | protein coding gene | coagulation factor II (thrombin) receptor-like 2 |
| 13 | gene | 95.707494 | 95.725547 | positive | MGI_C57BL6J_6324738 | Gm50469 | ENSEMBL:ENSMUSG00000118445 | MGI:6324738 | lncRNA gene | predicted gene, 50469 |
| 13 | gene | 95.736583 | 95.753638 | positive | MGI_C57BL6J_5592039 | Gm32880 | ENSEMBL:ENSMUSG00000118436 | MGI:5592039 | lncRNA gene | predicted gene, 32880 |
| 13 | gene | 95.763409 | 95.777960 | positive | MGI_C57BL6J_5591861 | Gm32702 | NCBI_Gene:102635336 | MGI:5591861 | lncRNA gene | predicted gene, 32702 |
| 13 | gene | 95.896962 | 95.897120 | negative | MGI_C57BL6J_5453809 | Gm24032 | ENSEMBL:ENSMUSG00000065866 | MGI:5453809 | snRNA gene | predicted gene, 24032 |
| 13 | gene | 95.954594 | 96.132577 | negative | MGI_C57BL6J_1922459 | Sv2c | NCBI_Gene:75209,ENSEMBL:ENSMUSG00000051111 | MGI:1922459 | protein coding gene | synaptic vesicle glycoprotein 2c |
| 13 | gene | 96.079216 | 96.081376 | negative | MGI_C57BL6J_5012260 | Gm20075 | NCBI_Gene:100504125,ENSEMBL:ENSMUSG00000114133 | MGI:5012260 | protein coding gene | predicted gene, 20075 |
| 13 | gene | 96.082054 | 96.083231 | positive | MGI_C57BL6J_4938017 | Gm17190 | NCBI_Gene:100417936,ENSEMBL:ENSMUSG00000099115 | MGI:4938017 | protein coding gene | predicted gene 17190 |
| 13 | gene | 96.104463 | 96.104562 | negative | MGI_C57BL6J_5453732 | Gm23955 | ENSEMBL:ENSMUSG00000087839 | MGI:5453732 | snRNA gene | predicted gene, 23955 |
| 13 | gene | 96.132700 | 96.134335 | positive | MGI_C57BL6J_5580249 | Gm29543 | NCBI_Gene:105245595,ENSEMBL:ENSMUSG00000100475 | MGI:5580249 | lncRNA gene | predicted gene 29543 |
| 13 | gene | 96.357111 | 96.357223 | positive | MGI_C57BL6J_5454990 | Gm25213 | ENSEMBL:ENSMUSG00000076343 | MGI:5454990 | miRNA gene | predicted gene, 25213 |
| 13 | pseudogene | 96.376867 | 96.377703 | positive | MGI_C57BL6J_3644353 | Gm8712 | NCBI_Gene:667582,ENSEMBL:ENSMUSG00000113743 | MGI:3644353 | pseudogene | predicted gene 8712 |
| 13 | gene | 96.388294 | 96.417737 | positive | MGI_C57BL6J_1914713 | Poc5 | NCBI_Gene:67463,ENSEMBL:ENSMUSG00000021671 | MGI:1914713 | protein coding gene | POC5 centriolar protein |
| 13 | gene | 96.416134 | 96.471354 | negative | MGI_C57BL6J_2444730 | Ankdd1b | NCBI_Gene:271144,ENSEMBL:ENSMUSG00000047117 | MGI:2444730 | protein coding gene | ankyrin repeat and death domain containing 1B |
| 13 | gene | 96.480689 | 96.542646 | negative | MGI_C57BL6J_1349767 | Polk | NCBI_Gene:27015,ENSEMBL:ENSMUSG00000021668 | MGI:1349767 | protein coding gene | polymerase (DNA directed), kappa |
| 13 | gene | 96.490733 | 96.491120 | positive | MGI_C57BL6J_4937256 | Gm17622 | ENSEMBL:ENSMUSG00000091109 | MGI:4937256 | protein coding gene | predicted gene, 17622 |
| 13 | gene | 96.542618 | 96.640167 | positive | MGI_C57BL6J_1915268 | Col4a3bp | NCBI_Gene:68018,ENSEMBL:ENSMUSG00000021669 | MGI:1915268 | protein coding gene | collagen, type IV, alpha 3 (Goodpasture antigen) binding protein |
| 13 | gene | 96.542993 | 96.543652 | negative | MGI_C57BL6J_6097494 | Gm48133 | ENSEMBL:ENSMUSG00000113068 | MGI:6097494 | lncRNA gene | predicted gene, 48133 |
| 13 | gene | 96.556232 | 96.559111 | positive | MGI_C57BL6J_6097502 | Gm48138 | ENSEMBL:ENSMUSG00000113842 | MGI:6097502 | unclassified gene | predicted gene, 48138 |
| 13 | gene | 96.573558 | 96.581487 | positive | MGI_C57BL6J_5826073 | Gm46436 | NCBI_Gene:108168109,ENSEMBL:ENSMUSG00000113379 | MGI:5826073 | lncRNA gene | predicted gene, 46436 |
| 13 | gene | 96.605677 | 96.609591 | positive | MGI_C57BL6J_6097504 | Gm48139 | ENSEMBL:ENSMUSG00000114193 | MGI:6097504 | unclassified gene | predicted gene, 48139 |
| 13 | gene | 96.648962 | 96.670936 | negative | MGI_C57BL6J_96159 | Hmgcr | NCBI_Gene:15357,ENSEMBL:ENSMUSG00000021670 | MGI:96159 | protein coding gene | 3-hydroxy-3-methylglutaryl-Coenzyme A reductase |
| 13 | pseudogene | 96.677469 | 96.678123 | negative | MGI_C57BL6J_6098141 | Gm48575 | ENSEMBL:ENSMUSG00000113351 | MGI:6098141 | pseudogene | predicted gene, 48575 |
| 13 | gene | 96.698023 | 96.703747 | positive | MGI_C57BL6J_5592330 | Gm33171 | NCBI_Gene:102635970 | MGI:5592330 | lncRNA gene | predicted gene, 33171 |
| 13 | gene | 96.748272 | 96.910039 | positive | MGI_C57BL6J_5006716 | Ankrd31 | NCBI_Gene:625662,ENSEMBL:ENSMUSG00000109561 | MGI:5006716 | protein coding gene | ankyrin repeat domain 31 |
| 13 | pseudogene | 96.776466 | 96.777930 | negative | MGI_C57BL6J_6098147 | Gm48580 | ENSEMBL:ENSMUSG00000113879 | MGI:6098147 | pseudogene | predicted gene, 48580 |
| 13 | pseudogene | 96.870300 | 96.870606 | negative | MGI_C57BL6J_6098146 | Gm48579 | ENSEMBL:ENSMUSG00000113046 | MGI:6098146 | pseudogene | predicted gene, 48579 |
| 13 | gene | 96.897276 | 96.897390 | positive | MGI_C57BL6J_5531174 | Gm27792 | ENSEMBL:ENSMUSG00000099053 | MGI:5531174 | rRNA gene | predicted gene, 27792 |
| 13 | gene | 96.924352 | 96.950914 | positive | MGI_C57BL6J_2684919 | Gcnt4 | NCBI_Gene:218476,ENSEMBL:ENSMUSG00000091387 | MGI:2684919 | protein coding gene | glucosaminyl (N-acetyl) transferase 4, core 2 (beta-1,6-N-acetylglucosaminyltransferase) |
| 13 | gene | 96.950372 | 96.950914 | negative | MGI_C57BL6J_6098174 | Gm48597 | ENSEMBL:ENSMUSG00000114154 | MGI:6098174 | lncRNA gene | predicted gene, 48597 |
| 13 | gene | 96.965977 | 96.984375 | negative | MGI_C57BL6J_5623912 | Gm41027 | NCBI_Gene:105245596 | MGI:5623912 | lncRNA gene | predicted gene, 41027 |
| 13 | pseudogene | 97.014885 | 97.015299 | negative | MGI_C57BL6J_6098195 | Gm48609 | ENSEMBL:ENSMUSG00000113934 | MGI:6098195 | pseudogene | predicted gene, 48609 |
| 13 | gene | 97.016271 | 97.059235 | positive | MGI_C57BL6J_5623913 | Gm41028 | NCBI_Gene:105245597 | MGI:5623913 | lncRNA gene | predicted gene, 41028 |
| 13 | gene | 97.021864 | 97.035254 | negative | MGI_C57BL6J_1913729 | 1700029F12Rik | NCBI_Gene:66479,ENSEMBL:ENSMUSG00000052075 | MGI:1913729 | protein coding gene | RIKEN cDNA 1700029F12 gene |
| 13 | gene | 97.067286 | 97.131013 | positive | MGI_C57BL6J_2444268 | Fam169a | NCBI_Gene:320557,ENSEMBL:ENSMUSG00000041817 | MGI:2444268 | protein coding gene | family with sequence similarity 169, member A |
| 13 | gene | 97.098208 | 97.099340 | negative | MGI_C57BL6J_3646298 | Gm6169 | NCBI_Gene:620648,ENSEMBL:ENSMUSG00000057762 | MGI:3646298 | protein coding gene | predicted gene 6169 |
| 13 | gene | 97.129424 | 97.137937 | negative | MGI_C57BL6J_1913883 | Nsa2 | NCBI_Gene:59050,ENSEMBL:ENSMUSG00000060739 | MGI:1913883 | protein coding gene | NSA2 ribosome biogenesis homolog |
| 13 | gene | 97.137937 | 97.181196 | positive | MGI_C57BL6J_2444783 | Gfm2 | NCBI_Gene:320806,ENSEMBL:ENSMUSG00000021666 | MGI:2444783 | protein coding gene | G elongation factor, mitochondrial 2 |
| 13 | gene | 97.176331 | 97.198357 | negative | MGI_C57BL6J_96074 | Hexb | NCBI_Gene:15212,ENSEMBL:ENSMUSG00000021665 | MGI:96074 | protein coding gene | hexosaminidase B |
| 13 | pseudogene | 97.202247 | 97.202975 | negative | MGI_C57BL6J_6098636 | Gm48881 | ENSEMBL:ENSMUSG00000114583 | MGI:6098636 | pseudogene | predicted gene, 48881 |
| 13 | gene | 97.241105 | 97.253040 | positive | MGI_C57BL6J_109610 | Enc1 | NCBI_Gene:13803,ENSEMBL:ENSMUSG00000041773 | MGI:109610 | protein coding gene | ectodermal-neural cortex 1 |
| 13 | gene | 97.283183 | 97.285560 | positive | MGI_C57BL6J_1924873 | C430039J16Rik | ENSEMBL:ENSMUSG00000097932 | MGI:1924873 | lncRNA gene | RIKEN cDNA C430039J16 gene |
| 13 | gene | 97.293714 | 97.299131 | negative | MGI_C57BL6J_5592549 | Gm33390 | NCBI_Gene:102636278 | MGI:5592549 | lncRNA gene | predicted gene, 33390 |
| 13 | gene | 97.376180 | 97.382156 | positive | MGI_C57BL6J_5592606 | Gm33447 | NCBI_Gene:102636359,ENSEMBL:ENSMUSG00000114425 | MGI:5592606 | lncRNA gene | predicted gene, 33447 |
| 13 | gene | 97.434970 | 97.497664 | positive | MGI_C57BL6J_3780541 | Lncenc1 | NCBI_Gene:100039691,ENSEMBL:ENSMUSG00000078952 | MGI:3780541 | lncRNA gene | long non-coding RNA, embryonic stem cells expressed 1 |
| 13 | pseudogene | 97.457947 | 97.459077 | negative | MGI_C57BL6J_3644333 | Gm8756 | NCBI_Gene:667676,ENSEMBL:ENSMUSG00000098238 | MGI:3644333 | pseudogene | predicted gene 8756 |
| 13 | gene | 97.511204 | 97.515730 | positive | MGI_C57BL6J_3780547 | Gm2379 | ENSEMBL:ENSMUSG00000114926 | MGI:3780547 | lncRNA gene | predicted gene 2379 |
| 13 | gene | 97.511500 | 97.513240 | positive | MGI_C57BL6J_2145469 | 9330199F22Rik | NA | NA | unclassified gene | RIKEN cDNA 9330199F22 gene |
| 13 | gene | 97.520827 | 97.527736 | negative | MGI_C57BL6J_5623915 | Gm41030 | NCBI_Gene:105245599,ENSEMBL:ENSMUSG00000114415 | MGI:5623915 | lncRNA gene | predicted gene, 41030 |
| 13 | gene | 97.532569 | 97.538675 | negative | MGI_C57BL6J_5623914 | Gm41029 | NCBI_Gene:105245598 | MGI:5623914 | lncRNA gene | predicted gene, 41029 |
| 13 | gene | 97.541712 | 97.543335 | negative | MGI_C57BL6J_6097432 | Gm48090 | ENSEMBL:ENSMUSG00000114837 | MGI:6097432 | unclassified gene | predicted gene, 48090 |
| 13 | gene | 97.559971 | 97.583996 | positive | MGI_C57BL6J_3644417 | Gm5086 | NCBI_Gene:328314,ENSEMBL:ENSMUSG00000110235 | MGI:3644417 | lncRNA gene | predicted gene 5086 |
| 13 | gene | 97.602610 | 97.622876 | negative | MGI_C57BL6J_5593246 | Gm34087 | NCBI_Gene:102637217 | MGI:5593246 | lncRNA gene | predicted gene, 34087 |
| 13 | pseudogene | 97.609536 | 97.609817 | positive | MGI_C57BL6J_6097400 | Gm48070 | ENSEMBL:ENSMUSG00000114599 | MGI:6097400 | pseudogene | predicted gene, 48070 |
| 13 | gene | 97.712315 | 97.778952 | negative | MGI_C57BL6J_2685378 | 5330416C01Rik | NCBI_Gene:403201,ENSEMBL:ENSMUSG00000054944 | MGI:2685378 | lncRNA gene | RIKEN cDNA 5330416C01 gene |
| 13 | pseudogene | 97.751112 | 97.760965 | positive | MGI_C57BL6J_5477113 | Rps18-ps4 | ENSEMBL:ENSMUSG00000097346 | MGI:5477113 | pseudogene | ribosomal protein S18, pseudogene 4 |
| 13 | gene | 97.760072 | 97.760632 | negative | MGI_C57BL6J_3642298 | Gm10260 | NCBI_Gene:100039740,ENSEMBL:ENSMUSG00000069117 | MGI:3642298 | protein coding gene | predicted gene 10260 |
| 13 | gene | 97.790564 | 97.793258 | positive | MGI_C57BL6J_6096611 | Gm47577 | ENSEMBL:ENSMUSG00000114294 | MGI:6096611 | unclassified gene | predicted gene, 47577 |
| 13 | gene | 97.796162 | 97.826055 | negative | MGI_C57BL6J_5623916 | Gm41031 | NCBI_Gene:105245600,ENSEMBL:ENSMUSG00000114348 | MGI:5623916 | lncRNA gene | predicted gene, 41031 |
| 13 | gene | 97.865811 | 97.894001 | negative | MGI_C57BL6J_5593547 | Gm34388 | NCBI_Gene:102637626,ENSEMBL:ENSMUSG00000114474 | MGI:5593547 | lncRNA gene | predicted gene, 34388 |
| 13 | gene | 97.898594 | 98.206624 | negative | MGI_C57BL6J_1346016 | Arhgef28 | NCBI_Gene:110596,ENSEMBL:ENSMUSG00000021662 | MGI:1346016 | protein coding gene | Rho guanine nucleotide exchange factor (GEF) 28 |
| 13 | gene | 97.980802 | 97.981442 | positive | MGI_C57BL6J_1922107 | 4930448O17Rik | NA | NA | unclassified gene | RIKEN cDNA 4930448O17 gene |
| 13 | gene | 98.170451 | 98.178479 | positive | MGI_C57BL6J_5593628 | Gm34469 | NCBI_Gene:102637733 | MGI:5593628 | lncRNA gene | predicted gene, 34469 |
| 13 | gene | 98.246845 | 98.263041 | negative | MGI_C57BL6J_2145443 | Utp15 | NCBI_Gene:105372,ENSEMBL:ENSMUSG00000041747 | MGI:2145443 | protein coding gene | UTP15 small subunit processome component |
| 13 | gene | 98.263074 | 98.274754 | positive | MGI_C57BL6J_1915808 | Ankra2 | NCBI_Gene:68558,ENSEMBL:ENSMUSG00000021661 | MGI:1915808 | protein coding gene | ankyrin repeat, family A (RFXANK-like), 2 |
| 13 | gene | 98.279816 | 98.307677 | positive | MGI_C57BL6J_5439445 | Gm21976 | NCBI_Gene:102637808,ENSEMBL:ENSMUSG00000096330 | MGI:5439445 | protein coding gene | predicted gene 21976 |
| 13 | gene | 98.309896 | 98.317006 | negative | MGI_C57BL6J_1202875 | Btf3 | NCBI_Gene:218490,ENSEMBL:ENSMUSG00000021660 | MGI:1202875 | protein coding gene | basic transcription factor 3 |
| 13 | gene | 98.316422 | 98.324415 | positive | MGI_C57BL6J_3642913 | Gm9828 | NCBI_Gene:105246056,ENSEMBL:ENSMUSG00000048603 | MGI:3642913 | lncRNA gene | predicted gene 9828 |
| 13 | gene | 98.354242 | 98.359305 | positive | MGI_C57BL6J_1347463 | Foxd1 | NCBI_Gene:15229,ENSEMBL:ENSMUSG00000078302 | MGI:1347463 | protein coding gene | forkhead box D1 |
| 13 | pseudogene | 98.401895 | 98.403101 | negative | MGI_C57BL6J_3780612 | Gm2445 | NCBI_Gene:100039827,ENSEMBL:ENSMUSG00000098113 | MGI:3780612 | pseudogene | predicted gene 2445 |
| 13 | gene | 98.424109 | 98.435065 | negative | MGI_C57BL6J_1917340 | 2310005E17Rik | NCBI_Gene:70090,ENSEMBL:ENSMUSG00000099446 | MGI:1917340 | lncRNA gene | RIKEN cDNA 2310005E17 gene |
| 13 | gene | 98.429333 | 98.430788 | positive | MGI_C57BL6J_5826074 | Gm46437 | NCBI_Gene:108168110 | MGI:5826074 | lncRNA gene | predicted gene, 46437 |
| 13 | gene | 98.476598 | 98.477459 | negative | MGI_C57BL6J_5580207 | Gm29501 | ENSEMBL:ENSMUSG00000100416 | MGI:5580207 | lncRNA gene | predicted gene 29501 |
| 13 | gene | 98.489348 | 98.492001 | negative | MGI_C57BL6J_3642323 | Gm10320 | NCBI_Gene:101055909,ENSEMBL:ENSMUSG00000092116 | MGI:3642323 | protein coding gene | predicted pseudogene 10320 |
| 13 | gene | 98.517364 | 98.517938 | negative | MGI_C57BL6J_5610653 | Gm37425 | ENSEMBL:ENSMUSG00000104430 | MGI:5610653 | unclassified gene | predicted gene, 37425 |
| 13 | gene | 98.529939 | 98.536167 | negative | MGI_C57BL6J_5623917 | Gm41032 | NCBI_Gene:105245601 | MGI:5623917 | lncRNA gene | predicted gene, 41032 |
| 13 | gene | 98.549116 | 98.575040 | negative | MGI_C57BL6J_5623918 | Gm41033 | NCBI_Gene:105245602 | MGI:5623918 | lncRNA gene | predicted gene, 41033 |
| 13 | pseudogene | 98.557761 | 98.558031 | positive | MGI_C57BL6J_6097367 | Gm48049 | ENSEMBL:ENSMUSG00000114912 | MGI:6097367 | pseudogene | predicted gene, 48049 |
| 13 | gene | 98.560354 | 98.560461 | positive | MGI_C57BL6J_5452475 | Gm22698 | ENSEMBL:ENSMUSG00000093116 | MGI:5452475 | miRNA gene | predicted gene, 22698 |
| 13 | gene | 98.608411 | 98.617662 | negative | MGI_C57BL6J_6097368 | Gm48050 | ENSEMBL:ENSMUSG00000114521 | MGI:6097368 | lncRNA gene | predicted gene, 48050 |
| 13 | pseudogene | 98.613501 | 98.614494 | positive | MGI_C57BL6J_3643439 | Gm4815 | NCBI_Gene:218501,ENSEMBL:ENSMUSG00000114733 | MGI:3643439 | pseudogene | predicted gene 4815 |
| 13 | gene | 98.634978 | 98.637410 | negative | MGI_C57BL6J_1915594 | Tmem174 | NCBI_Gene:68344,ENSEMBL:ENSMUSG00000046082 | MGI:1915594 | protein coding gene | transmembrane protein 174 |
| 13 | gene | 98.686235 | 98.695639 | negative | MGI_C57BL6J_2685751 | Tmem171 | NCBI_Gene:380863,ENSEMBL:ENSMUSG00000052485 | MGI:2685751 | protein coding gene | transmembrane protein 171 |
| 13 | gene | 98.713074 | 98.718270 | negative | MGI_C57BL6J_3779875 | Gm9465 | NCBI_Gene:669675,ENSEMBL:ENSMUSG00000114689 | MGI:3779875 | lncRNA gene | predicted gene 9465 |
| 13 | gene | 98.723403 | 98.815768 | negative | MGI_C57BL6J_3505790 | Fcho2 | NCBI_Gene:218503,ENSEMBL:ENSMUSG00000041685 | MGI:3505790 | protein coding gene | FCH domain only 2 |
| 13 | gene | 98.732986 | 98.734690 | positive | MGI_C57BL6J_6096567 | Gm47550 | ENSEMBL:ENSMUSG00000114612 | MGI:6096567 | unclassified gene | predicted gene, 47550 |
| 13 | gene | 98.798611 | 98.801410 | negative | MGI_C57BL6J_2441949 | D130062J10Rik | ENSEMBL:ENSMUSG00000114598 | MGI:2441949 | lncRNA gene | RIKEN cDNA D130062J10 gene |
| 13 | gene | 98.801172 | 98.803736 | positive | MGI_C57BL6J_6096569 | Gm47551 | ENSEMBL:ENSMUSG00000114458 | MGI:6096569 | lncRNA gene | predicted gene, 47551 |
| 13 | gene | 98.815741 | 98.832137 | positive | MGI_C57BL6J_5594154 | Gm34995 | NCBI_Gene:102638424 | MGI:5594154 | lncRNA gene | predicted gene, 34995 |
| 13 | pseudogene | 98.816752 | 98.817265 | positive | MGI_C57BL6J_5010319 | Gm18134 | NCBI_Gene:100416506,ENSEMBL:ENSMUSG00000114719 | MGI:5010319 | pseudogene | predicted gene, 18134 |
| 13 | gene | 98.839019 | 98.926384 | negative | MGI_C57BL6J_2681523 | Tnpo1 | NCBI_Gene:238799,ENSEMBL:ENSMUSG00000009470 | MGI:2681523 | protein coding gene | transportin 1 |
| 13 | gene | 98.871164 | 98.874492 | negative | MGI_C57BL6J_2444816 | 8030448K23Rik | NA | NA | unclassified gene | RIKEN cDNA 8030448K23 gene |
| 13 | pseudogene | 98.899989 | 98.904707 | positive | MGI_C57BL6J_3648523 | Gm5453 | NCBI_Gene:432798,ENSEMBL:ENSMUSG00000062461 | MGI:3648523 | pseudogene | predicted gene 5453 |
| 13 | gene | 98.936763 | 98.945925 | negative | MGI_C57BL6J_5594374 | Gm35215 | NCBI_Gene:102638721,ENSEMBL:ENSMUSG00000114569 | MGI:5594374 | lncRNA gene | predicted gene, 35215 |
| 13 | pseudogene | 98.973378 | 98.973742 | positive | MGI_C57BL6J_6095760 | Gm47052 | ENSEMBL:ENSMUSG00000114873 | MGI:6095760 | pseudogene | predicted gene, 47052 |
| 13 | gene | 98.984085 | 98.984565 | negative | MGI_C57BL6J_1916632 | 1700024P04Rik | NCBI_Gene:69382,ENSEMBL:ENSMUSG00000045022 | MGI:1916632 | protein coding gene | RIKEN cDNA 1700024P04 gene |
| 13 | gene | 98.987600 | 98.990984 | negative | MGI_C57BL6J_5623920 | Gm41035 | NCBI_Gene:105245604 | MGI:5623920 | protein coding gene | predicted gene, 41035 |
| 13 | gene | 98.994151 | 99.001471 | positive | MGI_C57BL6J_5594438 | Gm35279 | NCBI_Gene:102638802,ENSEMBL:ENSMUSG00000114203 | MGI:5594438 | lncRNA gene | predicted gene, 35279 |
| 13 | gene | 99.007099 | 99.021695 | positive | MGI_C57BL6J_1925197 | A930014D07Rik | ENSEMBL:ENSMUSG00000114414 | MGI:1925197 | lncRNA gene | RIKEN cDNA A930014D07 gene |
| 13 | pseudogene | 99.034309 | 99.034848 | negative | MGI_C57BL6J_6095766 | Gm47057 | ENSEMBL:ENSMUSG00000114283 | MGI:6095766 | pseudogene | predicted gene, 47057 |
| 13 | gene | 99.040398 | 99.040650 | negative | MGI_C57BL6J_5454510 | Gm24733 | ENSEMBL:ENSMUSG00000089488 | MGI:5454510 | unclassified non-coding RNA gene | predicted gene, 24733 |
| 13 | gene | 99.073062 | 99.074034 | positive | MGI_C57BL6J_5594652 | Gm35493 | NCBI_Gene:102639099 | MGI:5594652 | lncRNA gene | predicted gene, 35493 |
| 13 | gene | 99.076526 | 99.087921 | negative | MGI_C57BL6J_1916853 | 2310020H05Rik | NCBI_Gene:69603,ENSEMBL:ENSMUSG00000100410 | MGI:1916853 | lncRNA gene | RIKEN cDNA 2310020H05 gene |
| 13 | gene | 99.100694 | 99.107581 | positive | MGI_C57BL6J_2685653 | Gm807 | NCBI_Gene:328320,ENSEMBL:ENSMUSG00000097848 | MGI:2685653 | lncRNA gene | predicted gene 807 |
| 13 | gene | 99.110537 | 99.122458 | positive | MGI_C57BL6J_5825575 | Gm45938 | NCBI_Gene:108167324 | MGI:5825575 | lncRNA gene | predicted gene, 45938 |
| 13 | gene | 99.173120 | 99.176459 | negative | MGI_C57BL6J_5623921 | Gm41036 | NCBI_Gene:105245605 | MGI:5623921 | lncRNA gene | predicted gene, 41036 |
| 13 | gene | 99.184823 | 99.250656 | positive | MGI_C57BL6J_2178429 | Zfp366 | NCBI_Gene:238803,ENSEMBL:ENSMUSG00000050919 | MGI:2178429 | protein coding gene | zinc finger protein 366 |
| 13 | gene | 99.319647 | 99.344705 | negative | MGI_C57BL6J_1916177 | Ptcd2 | NCBI_Gene:68927,ENSEMBL:ENSMUSG00000021650 | MGI:1916177 | protein coding gene | pentatricopeptide repeat domain 2 |
| 13 | gene | 99.344767 | 99.415562 | positive | MGI_C57BL6J_1919064 | Mrps27 | NCBI_Gene:218506,ENSEMBL:ENSMUSG00000041632 | MGI:1919064 | protein coding gene | mitochondrial ribosomal protein S27 |
| 13 | gene | 99.354717 | 99.362958 | positive | MGI_C57BL6J_5623923 | Gm41038 | NCBI_Gene:105245607 | MGI:5623923 | lncRNA gene | predicted gene, 41038 |
| 13 | gene | 99.396390 | 99.409815 | negative | MGI_C57BL6J_5623922 | Gm41037 | NCBI_Gene:105245606 | MGI:5623922 | lncRNA gene | predicted gene, 41037 |
| 13 | gene | 99.396924 | 99.412876 | negative | MGI_C57BL6J_2444982 | 6430562O15Rik | NCBI_Gene:320893,ENSEMBL:ENSMUSG00000097828 | MGI:2444982 | lncRNA gene | RIKEN cDNA 6430562O15 gene |
| 13 | gene | 99.421446 | 99.516602 | negative | MGI_C57BL6J_1306778 | Map1b | NCBI_Gene:17755,ENSEMBL:ENSMUSG00000052727 | MGI:1306778 | protein coding gene | microtubule-associated protein 1B |
| 13 | gene | 99.502477 | 99.504439 | negative | MGI_C57BL6J_1915439 | 5330431K02Rik | ENSEMBL:ENSMUSG00000114282 | MGI:1915439 | unclassified gene | RIKEN cDNA 5330431K02 gene |
| 13 | gene | 99.516629 | 99.526040 | positive | MGI_C57BL6J_5477053 | Gm26559 | NCBI_Gene:102639339,ENSEMBL:ENSMUSG00000097027 | MGI:5477053 | lncRNA gene | predicted gene, 26559 |
| 13 | pseudogene | 99.561442 | 99.561919 | positive | MGI_C57BL6J_6095930 | Gm47158 | ENSEMBL:ENSMUSG00000114578 | MGI:6095930 | pseudogene | predicted gene, 47158 |
| 13 | gene | 99.641974 | 99.642803 | positive | MGI_C57BL6J_1922746 | 1700012J22Rik | ENSEMBL:ENSMUSG00000103545 | MGI:1922746 | unclassified gene | RIKEN cDNA 1700012J22 gene |
| 13 | gene | 99.692399 | 99.692529 | negative | MGI_C57BL6J_5454248 | Gm24471 | ENSEMBL:ENSMUSG00000077673 | MGI:5454248 | snoRNA gene | predicted gene, 24471 |
| 13 | gene | 99.898483 | 99.901091 | negative | MGI_C57BL6J_1351330 | Cartpt | NCBI_Gene:27220,ENSEMBL:ENSMUSG00000021647 | MGI:1351330 | protein coding gene | CART prepropeptide |
| 13 | gene | 99.933395 | 99.948322 | negative | MGI_C57BL6J_5623924 | Gm41039 | NCBI_Gene:105245608 | MGI:5623924 | lncRNA gene | predicted gene, 41039 |
| 13 | gene | 99.948530 | 100.015639 | negative | MGI_C57BL6J_1925288 | Mccc2 | NCBI_Gene:78038,ENSEMBL:ENSMUSG00000021646 | MGI:1925288 | protein coding gene | methylcrotonoyl-Coenzyme A carboxylase 2 (beta) |
| 13 | gene | 100.017994 | 100.104073 | negative | MGI_C57BL6J_1347077 | Bdp1 | NCBI_Gene:544971,ENSEMBL:ENSMUSG00000049658 | MGI:1347077 | protein coding gene | B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB |
| 13 | gene | 100.073217 | 100.073742 | negative | MGI_C57BL6J_3029305 | D430018B14Rik | NA | NA | unclassified gene | RIKEN cDNA D430018B14 gene |
| 13 | gene | 100.106795 | 100.108339 | negative | MGI_C57BL6J_3704411 | BC001981 | ENSEMBL:ENSMUSG00000097022 | MGI:3704411 | lncRNA gene | cDNA sequence BC001981 |
| 13 | gene | 100.108012 | 100.114571 | positive | MGI_C57BL6J_1337114 | Serf1 | NCBI_Gene:20365,ENSEMBL:ENSMUSG00000021643 | MGI:1337114 | protein coding gene | small EDRK-rich factor 1 |
| 13 | gene | 100.123205 | 100.137699 | positive | MGI_C57BL6J_109257 | Smn1 | NCBI_Gene:20595,ENSEMBL:ENSMUSG00000021645 | MGI:109257 | protein coding gene | survival motor neuron 1 |
| 13 | gene | 100.144063 | 100.202122 | negative | MGI_C57BL6J_1298226 | Naip2 | NCBI_Gene:17948,ENSEMBL:ENSMUSG00000078945 | MGI:1298226 | protein coding gene | NLR family, apoptosis inhibitory protein 2 |
| 13 | pseudogene | 100.153851 | 100.154505 | negative | MGI_C57BL6J_5010598 | Gm18413 | NCBI_Gene:100417130,ENSEMBL:ENSMUSG00000113292 | MGI:5010598 | pseudogene | predicted gene, 18413 |
| 13 | pseudogene | 100.188893 | 100.189155 | negative | MGI_C57BL6J_109439 | Naip1-rs1 | NCBI_Gene:17941 | MGI:109439 | pseudogene | NLR family, apoptosis inhibitory protein 1, related sequence 1 |
| 13 | gene | 100.211739 | 100.250927 | negative | MGI_C57BL6J_1298220 | Naip5 | NCBI_Gene:17951,ENSEMBL:ENSMUSG00000071203 | MGI:1298220 | protein coding gene | NLR family, apoptosis inhibitory protein 5 |
| 13 | gene | 100.246477 | 100.247051 | positive | MGI_C57BL6J_1298221 | Naip4 | NA | NA | protein coding gene | NLR family%2c apoptosis inhibitory protein 4 |
| 13 | pseudogene | 100.264609 | 100.283091 | negative | MGI_C57BL6J_1858253 | Naip3-ps1 | NCBI_Gene:53898,ENSEMBL:ENSMUSG00000114576 | MGI:1858253 | pseudogene | NLR family, apoptosis inhibitory protein 3, pseudogene 1 |
| 13 | gene | 100.281121 | 100.317674 | negative | MGI_C57BL6J_1298222 | Naip6 | NCBI_Gene:17952,ENSEMBL:ENSMUSG00000078942 | MGI:1298222 | protein coding gene | NLR family, apoptosis inhibitory protein 6 |
| 13 | gene | 100.283115 | 100.317688 | negative | MGI_C57BL6J_1858256 | Naip7 | NA | NA | protein coding gene | NLR family%2c apoptosis inhibitory protein 7 |
| 13 | pseudogene | 100.340005 | 100.389242 | negative | MGI_C57BL6J_1298225 | Naip3 | NCBI_Gene:667838,ENSEMBL:ENSMUSG00000082956 | MGI:1298225 | pseudogene | NLR family, apoptosis inhibitory protein 3 |
| 13 | pseudogene | 100.392726 | 100.394499 | positive | MGI_C57BL6J_5010786 | Gm18601 | NCBI_Gene:100417419,ENSEMBL:ENSMUSG00000112978 | MGI:5010786 | pseudogene | predicted gene, 18601 |
| 13 | gene | 100.407764 | 100.452869 | negative | MGI_C57BL6J_1298223 | Naip1 | NCBI_Gene:17940,ENSEMBL:ENSMUSG00000021640 | MGI:1298223 | protein coding gene | NLR family, apoptosis inhibitory protein 1 |
| 13 | pseudogene | 100.415565 | 100.415979 | positive | MGI_C57BL6J_3809036 | Rpl31-ps13 | NCBI_Gene:100504277,ENSEMBL:ENSMUSG00000051723 | MGI:3809036 | pseudogene | ribosomal protein L31, pseudogene 13 |
| 13 | gene | 100.460218 | 100.492611 | negative | MGI_C57BL6J_1345669 | Gtf2h2 | NCBI_Gene:23894,ENSEMBL:ENSMUSG00000021639 | MGI:1345669 | protein coding gene | general transcription factor II H, polypeptide 2 |
| 13 | pseudogene | 100.467932 | 100.468187 | positive | MGI_C57BL6J_6097965 | Gm48452 | ENSEMBL:ENSMUSG00000113596 | MGI:6097965 | pseudogene | predicted gene, 48452 |
| 13 | gene | 100.496504 | 100.553691 | negative | MGI_C57BL6J_106183 | Ocln | NCBI_Gene:18260,ENSEMBL:ENSMUSG00000021638 | MGI:106183 | protein coding gene | occludin |
| 13 | gene | 100.557426 | 100.560639 | positive | MGI_C57BL6J_6098172 | Gm48596 | ENSEMBL:ENSMUSG00000114360 | MGI:6098172 | lncRNA gene | predicted gene, 48596 |
| 13 | pseudogene | 100.559221 | 100.560032 | negative | MGI_C57BL6J_3647328 | Gm8847 | NCBI_Gene:667861,ENSEMBL:ENSMUSG00000114332 | MGI:3647328 | pseudogene | predicted gene 8847 |
| 13 | gene | 100.595957 | 100.616971 | negative | MGI_C57BL6J_2446166 | Marveld2 | NCBI_Gene:218518,ENSEMBL:ENSMUSG00000021636 | MGI:2446166 | protein coding gene | MARVEL (membrane-associating) domain containing 2 |
| 13 | gene | 100.604359 | 100.604462 | positive | MGI_C57BL6J_5454038 | Gm24261 | ENSEMBL:ENSMUSG00000065678 | MGI:5454038 | snRNA gene | predicted gene, 24261 |
| 13 | gene | 100.617164 | 100.651061 | negative | MGI_C57BL6J_1333807 | Rad17 | NCBI_Gene:19356,ENSEMBL:ENSMUSG00000021635 | MGI:1333807 | protein coding gene | RAD17 checkpoint clamp loader component |
| 13 | gene | 100.650979 | 100.666415 | positive | MGI_C57BL6J_5510732 | Ak6 | NCBI_Gene:102216272,ENSEMBL:ENSMUSG00000078941 | MGI:5510732 | protein coding gene | adenylate kinase 6 |
| 13 | gene | 100.651343 | 100.656060 | positive | MGI_C57BL6J_1888697 | Taf9 | NCBI_Gene:108143,ENSEMBL:ENSMUSG00000052293 | MGI:1888697 | protein coding gene | TATA-box binding protein associated factor 9 |
| 13 | gene | 100.668934 | 100.697240 | positive | MGI_C57BL6J_1923291 | Ccdc125 | NCBI_Gene:76041,ENSEMBL:ENSMUSG00000048924 | MGI:1923291 | protein coding gene | coiled-coil domain containing 125 |
| 13 | gene | 100.697024 | 100.730942 | negative | MGI_C57BL6J_102956 | Cdk7 | NCBI_Gene:12572,ENSEMBL:ENSMUSG00000069089 | MGI:102956 | protein coding gene | cyclin-dependent kinase 7 |
| 13 | gene | 100.735940 | 100.744659 | negative | MGI_C57BL6J_1913378 | Mrps36 | NCBI_Gene:66128,ENSEMBL:ENSMUSG00000061474 | MGI:1913378 | protein coding gene | mitochondrial ribosomal protein S36 |
| 13 | gene | 100.759674 | 100.775899 | negative | MGI_C57BL6J_1349448 | Cenph | NCBI_Gene:26886,ENSEMBL:ENSMUSG00000045273 | MGI:1349448 | protein coding gene | centromere protein H |
| 13 | pseudogene | 100.770464 | 100.770772 | positive | MGI_C57BL6J_3840152 | Gm16335 | ENSEMBL:ENSMUSG00000085340 | MGI:3840152 | pseudogene | predicted gene 16335 |
| 13 | gene | 100.778650 | 100.786570 | negative | MGI_C57BL6J_88302 | Ccnb1 | NCBI_Gene:268697,ENSEMBL:ENSMUSG00000041431 | MGI:88302 | protein coding gene | cyclin B1 |
| 13 | gene | 100.802645 | 100.833570 | negative | MGI_C57BL6J_1916298 | Slc30a5 | NCBI_Gene:69048,ENSEMBL:ENSMUSG00000021629 | MGI:1916298 | protein coding gene | solute carrier family 30 (zinc transporter), member 5 |
| 13 | gene | 100.847433 | 100.977915 | positive | MGI_C57BL6J_5580208 | Gm29502 | ENSEMBL:ENSMUSG00000100596 | MGI:5580208 | lncRNA gene | predicted gene 29502 |
| 13 | gene | 100.863402 | 100.863851 | negative | MGI_C57BL6J_5623925 | Gm41040 | NCBI_Gene:105245610 | MGI:5623925 | lncRNA gene | predicted gene, 41040 |
| 13 | gene | 100.872780 | 100.876195 | positive | MGI_C57BL6J_5611058 | Gm37830 | ENSEMBL:ENSMUSG00000104450 | MGI:5611058 | unclassified gene | predicted gene, 37830 |
| 13 | pseudogene | 100.909059 | 100.909897 | negative | MGI_C57BL6J_5623926 | Gm41041 | NCBI_Gene:105245611,ENSEMBL:ENSMUSG00000114738 | MGI:5623926 | pseudogene | predicted gene, 41041 |
| 13 | pseudogene | 100.945895 | 100.946929 | negative | MGI_C57BL6J_3644245 | Gm10257 | NCBI_Gene:544973,ENSEMBL:ENSMUSG00000114759 | MGI:3644245 | pseudogene | predicted gene 10257 |
| 13 | gene | 100.997184 | 101.080021 | positive | MGI_C57BL6J_5623927 | Gm41042 | NCBI_Gene:105245612 | MGI:5623927 | lncRNA gene | predicted gene, 41042 |
| 13 | pseudogene | 101.012837 | 101.013991 | negative | MGI_C57BL6J_3646517 | Gm6114 | NCBI_Gene:619917,ENSEMBL:ENSMUSG00000098005 | MGI:3646517 | pseudogene | predicted gene 6114 |
| 13 | pseudogene | 101.038599 | 101.040182 | negative | MGI_C57BL6J_96115 | Hmgb1-ps9 | NCBI_Gene:108168111,ENSEMBL:ENSMUSG00000096722 | MGI:96115 | pseudogene | high mobility group box 1, pseudogene 9 |
| 13 | gene | 101.226219 | 101.306017 | negative | MGI_C57BL6J_5595586 | Gm36427 | NCBI_Gene:102640336 | MGI:5595586 | lncRNA gene | predicted gene, 36427 |
| 13 | gene | 101.232127 | 101.247551 | positive | MGI_C57BL6J_5439408 | 5930438M14Rik | NCBI_Gene:328324,ENSEMBL:ENSMUSG00000114590 | MGI:5439408 | lncRNA gene | RIKEN cDNA 5930438M14 gene |
| 13 | gene | 101.254294 | 101.464160 | positive | MGI_C57BL6J_3512682 | 4932411K12Rik | NCBI_Gene:102639950,ENSEMBL:ENSMUSG00000100733 | MGI:3512682 | lncRNA gene | RIKEN cDNA 4932411K12 gene |
| 13 | gene | 101.254319 | 101.293396 | positive | MGI_C57BL6J_1918424 | 4933416M06Rik | NCBI_Gene:71174 | MGI:1918424 | lncRNA gene | RIKEN cDNA 4933416M06 gene |
| 13 | pseudogene | 101.348751 | 101.349181 | positive | MGI_C57BL6J_3780702 | Gm2534 | NCBI_Gene:100039983,ENSEMBL:ENSMUSG00000114295 | MGI:3780702 | pseudogene | predicted gene 2534 |
| 13 | gene | 101.391871 | 101.397580 | negative | MGI_C57BL6J_1925386 | 4922502H24Rik | NCBI_Gene:78136 | MGI:1925386 | lncRNA gene | RIKEN cDNA 4922502H24 gene |
| 13 | gene | 101.442760 | 101.443251 | negative | MGI_C57BL6J_5610222 | Gm36994 | ENSEMBL:ENSMUSG00000103437 | MGI:5610222 | unclassified gene | predicted gene, 36994 |
| 13 | gene | 101.514604 | 101.515800 | positive | MGI_C57BL6J_2444647 | A130049L09Rik | NA | NA | unclassified gene | RIKEN cDNA A130049L09 gene |
| 13 | gene | 101.537317 | 101.548152 | negative | MGI_C57BL6J_5623928 | Gm41043 | NCBI_Gene:105245613,ENSEMBL:ENSMUSG00000114652 | MGI:5623928 | lncRNA gene | predicted gene, 41043 |
| 13 | gene | 101.540559 | 101.566823 | positive | MGI_C57BL6J_5595634 | Gm36475 | NCBI_Gene:102640410 | MGI:5595634 | lncRNA gene | predicted gene, 36475 |
| 13 | gene | 101.545206 | 101.548279 | positive | MGI_C57BL6J_6096539 | Gm47533 | ENSEMBL:ENSMUSG00000114400 | MGI:6096539 | lncRNA gene | predicted gene, 47533 |
| 13 | pseudogene | 101.557219 | 101.557859 | negative | MGI_C57BL6J_5011195 | Gm19010 | NCBI_Gene:100418117,ENSEMBL:ENSMUSG00000114700 | MGI:5011195 | pseudogene | predicted gene, 19010 |
| 13 | gene | 101.589833 | 101.613282 | positive | MGI_C57BL6J_5595712 | Gm36553 | NCBI_Gene:102640512 | MGI:5595712 | lncRNA gene | predicted gene, 36553 |
| 13 | gene | 101.605618 | 101.606233 | negative | MGI_C57BL6J_5580047 | Gm29341 | ENSEMBL:ENSMUSG00000099774 | MGI:5580047 | lncRNA gene | predicted gene 29341 |
| 13 | gene | 101.680563 | 101.768217 | negative | MGI_C57BL6J_97583 | Pik3r1 | NCBI_Gene:18708,ENSEMBL:ENSMUSG00000041417 | MGI:97583 | protein coding gene | phosphoinositide-3-kinase regulatory subunit 1 |
| 13 | gene | 101.749720 | 101.753205 | negative | MGI_C57BL6J_5595797 | Gm36638 | ENSEMBL:ENSMUSG00000102635,ENSEMBL:ENSMUSG00000104214 | MGI:5595797 | unclassified non-coding RNA gene | predicted gene, 36638 |
| 13 | pseudogene | 101.841540 | 101.842306 | negative | MGI_C57BL6J_5011293 | Gm19108 | NCBI_Gene:100418265,ENSEMBL:ENSMUSG00000114223 | MGI:5011293 | pseudogene | predicted gene, 19108 |
| 13 | gene | 101.855491 | 101.856351 | positive | MGI_C57BL6J_6095685 | Gm47007 | ENSEMBL:ENSMUSG00000114762 | MGI:6095685 | lncRNA gene | predicted gene, 47007 |
| 13 | pseudogene | 101.920411 | 101.922391 | positive | MGI_C57BL6J_5010017 | Gm17832 | NCBI_Gene:100415946,ENSEMBL:ENSMUSG00000114717 | MGI:5010017 | pseudogene | predicted gene, 17832 |
| 13 | gene | 101.998661 | 102.014347 | negative | MGI_C57BL6J_5623929 | Gm41044 | NCBI_Gene:105245614 | MGI:5623929 | lncRNA gene | predicted gene, 41044 |
| 13 | gene | 102.114117 | 102.116035 | negative | MGI_C57BL6J_5611361 | Gm38133 | ENSEMBL:ENSMUSG00000102542 | MGI:5611361 | unclassified gene | predicted gene, 38133 |
| 13 | gene | 102.228669 | 102.228952 | positive | MGI_C57BL6J_5455585 | Gm25808 | ENSEMBL:ENSMUSG00000084733 | MGI:5455585 | unclassified non-coding RNA gene | predicted gene, 25808 |
| 13 | gene | 102.342557 | 102.351981 | negative | MGI_C57BL6J_5826034 | Gm46397 | NCBI_Gene:108168059 | MGI:5826034 | lncRNA gene | predicted gene, 46397 |
| 13 | gene | 102.349730 | 102.356684 | positive | MGI_C57BL6J_5595862 | Gm36703 | NCBI_Gene:102640697 | MGI:5595862 | lncRNA gene | predicted gene, 36703 |
| 13 | pseudogene | 102.366891 | 102.367152 | negative | MGI_C57BL6J_6095687 | Gm47008 | ENSEMBL:ENSMUSG00000114444 | MGI:6095687 | pseudogene | predicted gene, 47008 |
| 13 | gene | 102.452532 | 102.454304 | positive | MGI_C57BL6J_5596080 | Gm36921 | NCBI_Gene:102640995 | MGI:5596080 | lncRNA gene | predicted gene, 36921 |
| 13 | gene | 102.469333 | 102.479674 | negative | MGI_C57BL6J_5623930 | Gm41045 | NCBI_Gene:105245615 | MGI:5623930 | lncRNA gene | predicted gene, 41045 |
| 13 | gene | 102.482194 | 102.483584 | negative | MGI_C57BL6J_5589029 | Gm29870 | NCBI_Gene:102631566 | MGI:5589029 | lncRNA gene | predicted gene, 29870 |
| 13 | gene | 102.545734 | 102.592348 | negative | MGI_C57BL6J_5589086 | Gm29927 | NCBI_Gene:102631635,ENSEMBL:ENSMUSG00000114846 | MGI:5589086 | lncRNA gene | predicted gene, 29927 |
| 13 | pseudogene | 102.604533 | 102.606903 | positive | MGI_C57BL6J_6095696 | Gm47014 | ENSEMBL:ENSMUSG00000114730 | MGI:6095696 | pseudogene | predicted gene, 47014 |
| 13 | gene | 102.661408 | 102.666459 | negative | MGI_C57BL6J_5623931 | Gm41046 | NCBI_Gene:105245616 | MGI:5623931 | lncRNA gene | predicted gene, 41046 |
| 13 | gene | 102.693558 | 102.739629 | positive | MGI_C57BL6J_1194924 | Cd180 | NCBI_Gene:17079,ENSEMBL:ENSMUSG00000021624 | MGI:1194924 | protein coding gene | CD180 antigen |
| 13 | gene | 102.732486 | 103.335099 | negative | MGI_C57BL6J_1918885 | Mast4 | NCBI_Gene:328329,ENSEMBL:ENSMUSG00000034751 | MGI:1918885 | protein coding gene | microtubule associated serine/threonine kinase family member 4 |
| 13 | gene | 102.760255 | 102.761896 | positive | MGI_C57BL6J_1923874 | 1700099I09Rik | NCBI_Gene:76624,ENSEMBL:ENSMUSG00000091906 | MGI:1923874 | lncRNA gene | RIKEN cDNA 1700099I09 gene |
| 13 | gene | 102.762435 | 102.764785 | positive | MGI_C57BL6J_4937987 | Gm17160 | ENSEMBL:ENSMUSG00000090329 | MGI:4937987 | lncRNA gene | predicted gene 17160 |
| 13 | gene | 102.912096 | 102.957913 | positive | MGI_C57BL6J_5589265 | Gm30106 | NCBI_Gene:102631885 | MGI:5589265 | lncRNA gene | predicted gene, 30106 |
| 13 | gene | 103.097060 | 103.100912 | positive | MGI_C57BL6J_5589209 | Gm30050 | NCBI_Gene:102631808 | MGI:5589209 | lncRNA gene | predicted gene, 30050 |
| 13 | pseudogene | 103.159795 | 103.164069 | negative | MGI_C57BL6J_3646534 | Gm6211 | NCBI_Gene:621312,ENSEMBL:ENSMUSG00000091577 | MGI:3646534 | pseudogene | predicted gene 6211 |
| 13 | pseudogene | 103.356181 | 103.358567 | positive | MGI_C57BL6J_3648286 | Gm5454 | NCBI_Gene:432800,ENSEMBL:ENSMUSG00000047643 | MGI:3648286 | pseudogene | predicted gene 5454 |
| 13 | gene | 103.396202 | 103.396306 | positive | MGI_C57BL6J_4421901 | n-R5s56 | ENSEMBL:ENSMUSG00000064455 | MGI:4421901 | rRNA gene | nuclear encoded rRNA 5S 56 |
| 13 | pseudogene | 103.397070 | 103.397624 | positive | MGI_C57BL6J_3646635 | Gm16416 | NCBI_Gene:432801,ENSEMBL:ENSMUSG00000095940 | MGI:3646635 | pseudogene | predicted gene 16416 |
| 13 | gene | 103.420802 | 103.421082 | negative | MGI_C57BL6J_5454218 | Gm24441 | ENSEMBL:ENSMUSG00000087860 | MGI:5454218 | unclassified non-coding RNA gene | predicted gene, 24441 |
| 13 | gene | 103.430877 | 103.467279 | negative | MGI_C57BL6J_5623964 | Gm41079 | NCBI_Gene:105245653 | MGI:5623964 | lncRNA gene | predicted gene, 41079 |
| 13 | gene | 103.507258 | 103.520175 | positive | MGI_C57BL6J_5623932 | Gm41047 | NCBI_Gene:105245617 | MGI:5623932 | lncRNA gene | predicted gene, 41047 |
| 13 | gene | 103.562494 | 103.591359 | negative | MGI_C57BL6J_5826035 | Gm46398 | NCBI_Gene:108168060 | MGI:5826035 | lncRNA gene | predicted gene, 46398 |
| 13 | gene | 103.591395 | 103.707633 | negative | MGI_C57BL6J_5589710 | Gm30551 | NCBI_Gene:102632492,ENSEMBL:ENSMUSG00000114206 | MGI:5589710 | lncRNA gene | predicted gene, 30551 |
| 13 | gene | 103.641926 | 103.642032 | positive | MGI_C57BL6J_5454647 | Gm24870 | ENSEMBL:ENSMUSG00000096642 | MGI:5454647 | snRNA gene | predicted gene, 24870 |
| 13 | pseudogene | 103.657929 | 103.658571 | positive | MGI_C57BL6J_3645684 | Gm6080 | NCBI_Gene:606728 | MGI:3645684 | pseudogene | predicted gene 6080 |
| 13 | gene | 103.738565 | 103.774608 | negative | MGI_C57BL6J_2145245 | Srek1 | NCBI_Gene:218543,ENSEMBL:ENSMUSG00000032621 | MGI:2145245 | protein coding gene | splicing regulatory glutamine/lysine-rich protein 1 |
| 13 | gene | 103.818786 | 103.920586 | negative | MGI_C57BL6J_1890169 | Erbin | NCBI_Gene:59079,ENSEMBL:ENSMUSG00000021709 | MGI:1890169 | protein coding gene | Erbb2 interacting protein |
| 13 | gene | 103.897985 | 103.903205 | negative | MGI_C57BL6J_3780758 | Gm2590 | NA | NA | unclassified gene | predicted gene 2590 |
| 13 | gene | 103.920266 | 103.923396 | positive | MGI_C57BL6J_3027005 | D130037M23Rik | NCBI_Gene:328330 | MGI:3027005 | protein coding gene | RIKEN cDNA D130037M23 gene |
| 13 | gene | 103.945928 | 103.970124 | negative | MGI_C57BL6J_6097055 | Gm47849 | ENSEMBL:ENSMUSG00000114276 | MGI:6097055 | lncRNA gene | predicted gene, 47849 |
| 13 | gene | 103.976312 | 103.978330 | negative | MGI_C57BL6J_6097059 | Gm47851 | ENSEMBL:ENSMUSG00000114637 | MGI:6097059 | lncRNA gene | predicted gene, 47851 |
| 13 | gene | 104.023050 | 104.109660 | negative | MGI_C57BL6J_1923055 | Nln | NCBI_Gene:75805,ENSEMBL:ENSMUSG00000021710 | MGI:1923055 | protein coding gene | neurolysin (metallopeptidase M3 family) |
| 13 | gene | 104.037491 | 104.046225 | positive | MGI_C57BL6J_5590126 | Gm30967 | NCBI_Gene:102633048 | MGI:5590126 | lncRNA gene | predicted gene, 30967 |
| 13 | gene | 104.109724 | 104.142492 | positive | MGI_C57BL6J_2444615 | Sgtb | NCBI_Gene:218544,ENSEMBL:ENSMUSG00000042743 | MGI:2444615 | protein coding gene | small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| 13 | gene | 104.142149 | 104.178703 | negative | MGI_C57BL6J_1914225 | Trappc13 | NCBI_Gene:66975,ENSEMBL:ENSMUSG00000021711 | MGI:1914225 | protein coding gene | trafficking protein particle complex 13 |
| 13 | gene | 104.173721 | 104.178459 | negative | MGI_C57BL6J_6194609 | Shld3 | NA | NA | protein coding gene | shieldin complex subunit 3 |
| 13 | gene | 104.178774 | 104.203421 | positive | MGI_C57BL6J_1933161 | Trim23 | NCBI_Gene:81003,ENSEMBL:ENSMUSG00000021712 | MGI:1933161 | protein coding gene | tripartite motif-containing 23 |
| 13 | gene | 104.205117 | 104.228844 | negative | MGI_C57BL6J_2443069 | Ppwd1 | NCBI_Gene:238831,ENSEMBL:ENSMUSG00000021713 | MGI:2443069 | protein coding gene | peptidylprolyl isomerase domain and WD repeat containing 1 |
| 13 | gene | 104.228611 | 104.252392 | positive | MGI_C57BL6J_1926210 | Cenpk | NCBI_Gene:60411,ENSEMBL:ENSMUSG00000021714 | MGI:1926210 | protein coding gene | centromere protein K |
| 13 | gene | 104.287348 | 104.496695 | positive | MGI_C57BL6J_1347348 | Adamts6 | NCBI_Gene:108154,ENSEMBL:ENSMUSG00000046169 | MGI:1347348 | protein coding gene | a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 6 |
| 13 | pseudogene | 104.430931 | 104.432507 | negative | MGI_C57BL6J_3646580 | Gm8680 | NCBI_Gene:667518 | MGI:3646580 | pseudogene | predicted gene 8680 |
| 13 | gene | 104.631140 | 104.817142 | negative | MGI_C57BL6J_1914535 | Cwc27 | NCBI_Gene:67285,ENSEMBL:ENSMUSG00000021715 | MGI:1914535 | protein coding gene | CWC27 spliceosome-associated protein |
| 13 | gene | 104.684931 | 104.694994 | positive | MGI_C57BL6J_1919718 | 2610204G07Rik | ENSEMBL:ENSMUSG00000084860 | MGI:1919718 | lncRNA gene | RIKEN cDNA 2610204G07 gene |
| 13 | gene | 104.792484 | 104.839274 | positive | MGI_C57BL6J_1914538 | Srek1ip1 | NCBI_Gene:67288,ENSEMBL:ENSMUSG00000021716 | MGI:1914538 | protein coding gene | splicing regulatory glutamine/lysine-rich protein 1interacting protein 1 |
| 13 | gene | 104.845283 | 104.863893 | negative | MGI_C57BL6J_1925053 | Shisal2b | NCBI_Gene:77803,ENSEMBL:ENSMUSG00000042655 | MGI:1925053 | protein coding gene | shisa like 2B |
| 13 | gene | 104.914755 | 104.921688 | positive | MGI_C57BL6J_5623933 | Gm41048 | NCBI_Gene:105245618 | MGI:5623933 | lncRNA gene | predicted gene, 41048 |
| 13 | pseudogene | 104.918673 | 104.919547 | positive | MGI_C57BL6J_3645099 | Gm4938 | NCBI_Gene:238836,ENSEMBL:ENSMUSG00000114326 | MGI:3645099 | pseudogene | predicted pseudogene 4938 |
| 13 | gene | 104.945904 | 105.054930 | negative | MGI_C57BL6J_106334 | Rgs7bp | NCBI_Gene:52882,ENSEMBL:ENSMUSG00000021719 | MGI:106334 | protein coding gene | regulator of G-protein signalling 7 binding protein |
| 13 | gene | 105.082122 | 105.121782 | positive | MGI_C57BL6J_1914013 | Nt5el | NCBI_Gene:66763,ENSEMBL:ENSMUSG00000021718 | MGI:1914013 | protein coding gene | 5’ nucleotidase, ecto-like |
| 13 | gene | 105.130001 | 105.294818 | negative | MGI_C57BL6J_1919066 | Rnf180 | NCBI_Gene:71816,ENSEMBL:ENSMUSG00000021720 | MGI:1919066 | protein coding gene | ring finger protein 180 |
| 13 | gene | 105.223269 | 105.223398 | negative | MGI_C57BL6J_5455408 | Gm25631 | ENSEMBL:ENSMUSG00000077578 | MGI:5455408 | snoRNA gene | predicted gene, 25631 |
| 13 | gene | 105.311429 | 105.324918 | positive | MGI_C57BL6J_5623934 | Gm41049 | NCBI_Gene:105245619 | MGI:5623934 | lncRNA gene | predicted gene, 41049 |
| 13 | gene | 105.409655 | 105.432605 | positive | MGI_C57BL6J_5590289 | Gm31130 | NCBI_Gene:102633263 | MGI:5590289 | lncRNA gene | predicted gene, 31130 |
| 13 | gene | 105.443639 | 105.448133 | positive | MGI_C57BL6J_96273 | Htr1a | NCBI_Gene:15550,ENSEMBL:ENSMUSG00000021721 | MGI:96273 | protein coding gene | 5-hydroxytryptamine (serotonin) receptor 1A |
| 13 | gene | 105.486672 | 105.502918 | positive | MGI_C57BL6J_5623935 | Gm41050 | NCBI_Gene:105245620 | MGI:5623935 | lncRNA gene | predicted gene, 41050 |
| 13 | pseudogene | 105.568754 | 105.569963 | negative | MGI_C57BL6J_6097601 | Gm48208 | ENSEMBL:ENSMUSG00000114518 | MGI:6097601 | pseudogene | predicted gene, 48208 |
| 13 | pseudogene | 105.833453 | 105.833678 | negative | MGI_C57BL6J_6097606 | Gm48211 | ENSEMBL:ENSMUSG00000114671 | MGI:6097606 | pseudogene | predicted gene, 48211 |
| 13 | gene | 105.838547 | 106.019783 | positive | MGI_C57BL6J_3642385 | Gm10739 | ENSEMBL:ENSMUSG00000114564 | MGI:3642385 | lncRNA gene | predicted gene 10739 |
| 13 | pseudogene | 105.901764 | 105.902140 | positive | MGI_C57BL6J_6097607 | Gm48212 | ENSEMBL:ENSMUSG00000114840 | MGI:6097607 | pseudogene | predicted gene, 48212 |
| 13 | pseudogene | 106.042124 | 106.042481 | positive | MGI_C57BL6J_6097610 | Gm48214 | ENSEMBL:ENSMUSG00000114899 | MGI:6097610 | pseudogene | predicted gene, 48214 |
| 13 | pseudogene | 106.197216 | 106.197823 | negative | MGI_C57BL6J_3030551 | Olfr717-ps1 | NCBI_Gene:257877,ENSEMBL:ENSMUSG00000108515 | MGI:3030551 | pseudogene | olfactory receptor 717, pseudogene 1 |
| 13 | pseudogene | 106.475922 | 106.476755 | positive | MGI_C57BL6J_3779659 | Gm7054 | NCBI_Gene:631097,ENSEMBL:ENSMUSG00000114217 | MGI:3779659 | pseudogene | predicted gene 7054 |
| 13 | pseudogene | 106.530998 | 106.531817 | negative | MGI_C57BL6J_6097613 | Gm48216 | ENSEMBL:ENSMUSG00000114367 | MGI:6097613 | pseudogene | predicted gene, 48216 |
| 13 | pseudogene | 106.546497 | 106.547136 | negative | MGI_C57BL6J_3647154 | Dph3b-ps | NCBI_Gene:620949,ENSEMBL:ENSMUSG00000059877 | MGI:3647154 | pseudogene | diphthamide biosynthesis 3, pseudogene |
| 13 | gene | 106.712003 | 106.722044 | negative | MGI_C57BL6J_5825577 | Gm45940 | NCBI_Gene:108167328 | MGI:5825577 | lncRNA gene | predicted gene, 45940 |
| 13 | gene | 106.737780 | 106.761579 | negative | MGI_C57BL6J_5589570 | Gm30411 | NCBI_Gene:102632299,ENSEMBL:ENSMUSG00000114422 | MGI:5589570 | lncRNA gene | predicted gene, 30411 |
| 13 | gene | 106.780114 | 106.780220 | negative | MGI_C57BL6J_5454304 | Gm24527 | ENSEMBL:ENSMUSG00000064854 | MGI:5454304 | snRNA gene | predicted gene, 24527 |
| 13 | gene | 106.788070 | 106.796796 | positive | MGI_C57BL6J_5579695 | Gm28989 | ENSEMBL:ENSMUSG00000101308 | MGI:5579695 | lncRNA gene | predicted gene 28989 |
| 13 | gene | 106.794439 | 106.936960 | negative | MGI_C57BL6J_2442377 | Ipo11 | NCBI_Gene:76582,ENSEMBL:ENSMUSG00000042590 | MGI:2442377 | protein coding gene | importin 11 |
| 13 | gene | 106.833714 | 106.847267 | negative | MGI_C57BL6J_5317337 | Lrrc70 | ENSEMBL:ENSMUSG00000118423 | MGI:5317337 | protein coding gene | leucine rich repeat containing 70 |
| 13 | gene | 106.851900 | 106.853846 | positive | MGI_C57BL6J_5579694 | Gm28988 | ENSEMBL:ENSMUSG00000100833 | MGI:5579694 | lncRNA gene | predicted gene 28988 |
| 13 | pseudogene | 106.945223 | 106.945529 | negative | MGI_C57BL6J_6097971 | Gm48457 | ENSEMBL:ENSMUSG00000114572 | MGI:6097971 | pseudogene | predicted gene, 48457 |
| 13 | gene | 106.947040 | 106.960224 | positive | MGI_C57BL6J_1913504 | Dimt1 | NCBI_Gene:66254,ENSEMBL:ENSMUSG00000021692 | MGI:1913504 | protein coding gene | DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| 13 | gene | 106.958996 | 107.022228 | negative | MGI_C57BL6J_108390 | Kif2a | NCBI_Gene:16563,ENSEMBL:ENSMUSG00000021693 | MGI:108390 | protein coding gene | kinesin family member 2A |
| 13 | gene | 106.990402 | 106.990467 | negative | MGI_C57BL6J_5454208 | Gm24431 | ENSEMBL:ENSMUSG00000077261 | MGI:5454208 | snoRNA gene | predicted gene, 24431 |
| 13 | gene | 107.011618 | 107.012207 | positive | MGI_C57BL6J_2145331 | D930043N17Rik | NA | NA | unclassified gene | RIKEN cDNA D930043N17 gene |
| 13 | gene | 107.022553 | 107.055497 | positive | MGI_C57BL6J_1917940 | 3830408C21Rik | NCBI_Gene:100040322,ENSEMBL:ENSMUSG00000071181 | MGI:1917940 | lncRNA gene | RIKEN cDNA 3830408C21 gene |
| 13 | gene | 107.034284 | 107.035217 | positive | MGI_C57BL6J_6098310 | Gm48684 | ENSEMBL:ENSMUSG00000114893 | MGI:6098310 | lncRNA gene | predicted gene, 48684 |
| 13 | gene | 107.063641 | 107.148551 | positive | MGI_C57BL6J_5590611 | Gm31452 | NCBI_Gene:102633689,ENSEMBL:ENSMUSG00000114311 | MGI:5590611 | lncRNA gene | predicted gene, 31452 |
| 13 | gene | 107.121641 | 107.130108 | positive | MGI_C57BL6J_5623936 | Gm41051 | NCBI_Gene:105245621 | MGI:5623936 | lncRNA gene | predicted gene, 41051 |
| 13 | gene | 107.210533 | 107.212526 | positive | MGI_C57BL6J_5590874 | Gm31715 | NCBI_Gene:102634031 | MGI:5590874 | lncRNA gene | predicted gene, 31715 |
| 13 | pseudogene | 107.279615 | 107.280193 | negative | MGI_C57BL6J_3780895 | Gm2726 | NCBI_Gene:100040350,ENSEMBL:ENSMUSG00000114475 | MGI:3780895 | pseudogene | predicted gene 2726 |
| 13 | pseudogene | 107.332558 | 107.348561 | positive | MGI_C57BL6J_3780905 | Gm2736 | NCBI_Gene:100040366,ENSEMBL:ENSMUSG00000114459 | MGI:3780905 | pseudogene | predicted gene 2736 |
| 13 | pseudogene | 107.414144 | 107.414767 | negative | MGI_C57BL6J_3649039 | Apoo-ps | NCBI_Gene:621156,ENSEMBL:ENSMUSG00000049233 | MGI:3649039 | pseudogene | apolipoprotein O, pseudogene |
| 13 | gene | 107.469822 | 107.482718 | positive | MGI_C57BL6J_2145191 | AI197445 | NCBI_Gene:105120,ENSEMBL:ENSMUSG00000114780 | MGI:2145191 | lncRNA gene | expressed sequence AI197445 |
| 13 | gene | 107.493275 | 107.504526 | negative | MGI_C57BL6J_5590962 | Gm31803 | NCBI_Gene:102634148 | MGI:5590962 | lncRNA gene | predicted gene, 31803 |
| 13 | gene | 107.523632 | 107.566194 | negative | MGI_C57BL6J_5591163 | Gm32004 | NCBI_Gene:102634419,ENSEMBL:ENSMUSG00000114534 | MGI:5591163 | lncRNA gene | predicted gene, 32004 |
| 13 | gene | 107.540690 | 107.587943 | positive | MGI_C57BL6J_5591079 | Gm31920 | NCBI_Gene:102634303 | MGI:5591079 | lncRNA gene | predicted gene, 31920 |
| 13 | gene | 107.545031 | 107.547710 | positive | MGI_C57BL6J_3781162 | Gm2984 | NA | NA | unclassified gene | predicted gene 2984 |
| 13 | gene | 107.605284 | 107.664374 | negative | MGI_C57BL6J_5591249 | Gm32090 | NCBI_Gene:102634531,ENSEMBL:ENSMUSG00000114807 | MGI:5591249 | lncRNA gene | predicted gene, 32090 |
| 13 | gene | 107.635752 | 107.643271 | negative | MGI_C57BL6J_5591345 | Gm32186 | NCBI_Gene:102634652 | MGI:5591345 | lncRNA gene | predicted gene, 32186 |
| 13 | gene | 107.687397 | 107.692173 | positive | MGI_C57BL6J_1919507 | 1700006H21Rik | NCBI_Gene:72257,ENSEMBL:ENSMUSG00000102000 | MGI:1919507 | lncRNA gene | RIKEN cDNA 1700006H21 gene |
| 13 | gene | 107.724617 | 107.890064 | negative | MGI_C57BL6J_1914513 | Zswim6 | NCBI_Gene:67263,ENSEMBL:ENSMUSG00000032846 | MGI:1914513 | protein coding gene | zinc finger SWIM-type containing 6 |
| 13 | gene | 107.815467 | 107.816702 | negative | MGI_C57BL6J_1924332 | 2900011L18Rik | NA | NA | unclassified gene | RIKEN cDNA 2900011L18 gene |
| 13 | gene | 107.829110 | 107.829280 | negative | MGI_C57BL6J_5451895 | Gm22118 | ENSEMBL:ENSMUSG00000093334 | MGI:5451895 | miRNA gene | predicted gene, 22118 |
| 13 | gene | 107.851977 | 107.852741 | negative | MGI_C57BL6J_1924660 | 9530027D23Rik | NA | NA | unclassified gene | RIKEN cDNA 9530027D23 gene |
| 13 | gene | 107.863530 | 107.863768 | positive | MGI_C57BL6J_2442337 | 9930031P18Rik | NA | NA | unclassified gene | RIKEN cDNA 9930031P18 gene |
| 13 | gene | 107.877341 | 107.878372 | positive | MGI_C57BL6J_5623937 | Gm41052 | NCBI_Gene:105245622 | MGI:5623937 | lncRNA gene | predicted gene, 41052 |
| 13 | gene | 107.923411 | 107.930047 | positive | MGI_C57BL6J_5623938 | Gm41053 | NCBI_Gene:105245624 | MGI:5623938 | lncRNA gene | predicted gene, 41053 |
| 13 | gene | 107.944838 | 107.956079 | negative | MGI_C57BL6J_5621442 | Gm38557 | NCBI_Gene:102641408 | MGI:5621442 | lncRNA gene | predicted gene, 38557 |
| 13 | gene | 107.978143 | 107.981517 | negative | MGI_C57BL6J_5623939 | Gm41054 | NCBI_Gene:105245625 | MGI:5623939 | lncRNA gene | predicted gene, 41054 |
| 13 | gene | 107.996285 | 108.004669 | positive | MGI_C57BL6J_2444601 | B230220B15Rik | NCBI_Gene:102634835,ENSEMBL:ENSMUSG00000114627 | MGI:2444601 | lncRNA gene | RIKEN cDNA B230220B15 gene |
| 13 | gene | 108.002715 | 108.158655 | negative | MGI_C57BL6J_1922847 | Ndufaf2 | NCBI_Gene:75597,ENSEMBL:ENSMUSG00000068184 | MGI:1922847 | protein coding gene | NADH:ubiquinone oxidoreductase complex assembly factor 2 |
| 13 | gene | 108.018764 | 108.034753 | negative | MGI_C57BL6J_5623940 | Gm41055 | NCBI_Gene:105245626 | MGI:5623940 | lncRNA gene | predicted gene, 41055 |
| 13 | gene | 108.042121 | 108.044117 | negative | MGI_C57BL6J_1917930 | 3021401N23Rik | NA | NA | unclassified gene | RIKEN cDNA 3021401N23 gene |
| 13 | gene | 108.044446 | 108.049175 | positive | MGI_C57BL6J_1922866 | Smim15 | NCBI_Gene:75616,ENSEMBL:ENSMUSG00000071180 | MGI:1922866 | protein coding gene | small integral membrane protein 15 |
| 13 | gene | 108.158712 | 108.201204 | positive | MGI_C57BL6J_1919241 | Ercc8 | NCBI_Gene:71991,ENSEMBL:ENSMUSG00000021694 | MGI:1919241 | protein coding gene | excision repaiross-complementing rodent repair deficiency, complementation group 8 |
| 13 | gene | 108.159582 | 108.166383 | negative | MGI_C57BL6J_5591427 | Gm32268 | NCBI_Gene:102634764 | MGI:5591427 | lncRNA gene | predicted gene, 32268 |
| 13 | gene | 108.214326 | 108.287109 | positive | MGI_C57BL6J_1921809 | Elovl7 | NCBI_Gene:74559,ENSEMBL:ENSMUSG00000021696 | MGI:1921809 | protein coding gene | ELOVL family member 7, elongation of long chain fatty acids (yeast) |
| 13 | pseudogene | 108.218864 | 108.221309 | negative | MGI_C57BL6J_5591535 | Gm32376 | NCBI_Gene:102634902,ENSEMBL:ENSMUSG00000114519 | MGI:5591535 | pseudogene | predicted gene, 32376 |
| 13 | pseudogene | 108.231449 | 108.235694 | negative | MGI_C57BL6J_5589906 | Gm30747 | NCBI_Gene:102632758,ENSEMBL:ENSMUSG00000114507 | MGI:5589906 | pseudogene | predicted gene, 30747 |
| 13 | pseudogene | 108.307624 | 108.310986 | negative | MGI_C57BL6J_3647409 | Gm8990 | NCBI_Gene:668129,ENSEMBL:ENSMUSG00000114607 | MGI:3647409 | pseudogene | predicted gene 8990 |
| 13 | gene | 108.313099 | 108.316273 | negative | MGI_C57BL6J_3780992 | Gm2822 | ENSEMBL:ENSMUSG00000090358 | MGI:3780992 | lncRNA gene | predicted gene 2822 |
| 13 | gene | 108.316013 | 108.407782 | positive | MGI_C57BL6J_2145425 | Depdc1b | NCBI_Gene:218581,ENSEMBL:ENSMUSG00000021697 | MGI:2145425 | protein coding gene | DEP domain containing 1B |
| 13 | gene | 108.328429 | 108.329880 | negative | MGI_C57BL6J_1923297 | 5830433M15Rik | NA | NA | unclassified gene | RIKEN cDNA 5830433M15 gene |
| 13 | gene | 108.402067 | 108.409406 | positive | MGI_C57BL6J_5623941 | Gm41056 | NCBI_Gene:105245627 | MGI:5623941 | lncRNA gene | predicted gene, 41056 |
| 13 | gene | 108.437258 | 108.448897 | positive | MGI_C57BL6J_5591609 | Gm32450 | NCBI_Gene:102635006 | MGI:5591609 | lncRNA gene | predicted gene, 32450 |
| 13 | gene | 108.449948 | 109.955969 | positive | MGI_C57BL6J_99555 | Pde4d | NCBI_Gene:238871,ENSEMBL:ENSMUSG00000021699 | MGI:99555 | protein coding gene | phosphodiesterase 4D, cAMP specific |
| 13 | gene | 108.495384 | 108.498243 | negative | MGI_C57BL6J_3695756 | Part1 | NA | NA | unclassified gene | prostate androgen-regulated transcript 1 |
| 13 | gene | 108.495818 | 108.498170 | positive | MGI_C57BL6J_2442070 | D230040N21Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA D230040N21 gene |
| 13 | gene | 108.496869 | 108.498689 | negative | MGI_C57BL6J_5530954 | Gm27572 | NCBI_Gene:102635184,ENSEMBL:ENSMUSG00000098836 | MGI:5530954 | lncRNA gene | predicted gene, 27572 |
| 13 | gene | 108.498775 | 108.498970 | negative | MGI_C57BL6J_5531398 | Gm28016 | ENSEMBL:ENSMUSG00000099323 | MGI:5531398 | unclassified non-coding RNA gene | predicted gene, 28016 |
| 13 | pseudogene | 108.670580 | 108.671443 | positive | MGI_C57BL6J_3643406 | Rps3a3 | NCBI_Gene:668144,ENSEMBL:ENSMUSG00000059751 | MGI:3643406 | pseudogene | ribosomal protein S3A3 |
| 13 | pseudogene | 108.743694 | 108.744520 | negative | MGI_C57BL6J_3646308 | Gm6247 | NCBI_Gene:621663 | MGI:3646308 | pseudogene | predicted gene 6247 |
| 13 | pseudogene | 109.163840 | 109.164693 | positive | MGI_C57BL6J_5011108 | Gm18923 | NCBI_Gene:100417966 | MGI:5011108 | pseudogene | predicted gene, 18923 |
| 13 | gene | 109.324744 | 109.324824 | positive | MGI_C57BL6J_3718550 | Mir582 | miRBase:MI0006127,NCBI_Gene:100124489,ENSEMBL:ENSMUSG00000076946 | MGI:3718550 | miRNA gene | microRNA 582 |
| 13 | gene | 109.344633 | 109.351655 | positive | MGI_C57BL6J_2445216 | B930063P07Rik | NA | NA | unclassified gene | RIKEN cDNA B930063P07 gene |
| 13 | gene | 109.381471 | 109.400142 | negative | MGI_C57BL6J_5591972 | Gm32813 | NCBI_Gene:102635487 | MGI:5591972 | lncRNA gene | predicted gene, 32813 |
| 13 | gene | 109.425166 | 109.441193 | negative | MGI_C57BL6J_5591862 | Gm32703 | NCBI_Gene:102635337 | MGI:5591862 | lncRNA gene | predicted gene, 32703 |
| 13 | gene | 109.458602 | 109.463840 | positive | MGI_C57BL6J_2442381 | 9530083O12Rik | NA | NA | unclassified gene | RIKEN cDNA 9530083O12 gene |
| 13 | gene | 109.583992 | 109.584445 | positive | MGI_C57BL6J_1925329 | 9130219B18Rik | NA | NA | unclassified gene | RIKEN cDNA 9130219B18 gene |
| 13 | gene | 109.640920 | 109.643186 | positive | MGI_C57BL6J_2442371 | A730009E18Rik | NA | NA | unclassified gene | RIKEN cDNA A730009E18 gene |
| 13 | gene | 109.645656 | 109.647893 | positive | MGI_C57BL6J_1919624 | 2210416J07Rik | NA | NA | unclassified gene | RIKEN cDNA 2210416J07 gene |
| 13 | gene | 109.649924 | 109.651730 | positive | MGI_C57BL6J_1921825 | 4833406M21Rik | NA | NA | unclassified gene | RIKEN cDNA 4833406M21 gene |
| 13 | gene | 109.820819 | 109.822499 | negative | MGI_C57BL6J_2444062 | A630036H22Rik | NA | NA | unclassified gene | RIKEN cDNA A630036H22 gene |
| 13 | gene | 109.884222 | 109.884870 | positive | MGI_C57BL6J_1922510 | 4930562M23Rik | NA | NA | unclassified gene | RIKEN cDNA 4930562M23 gene |
| 13 | gene | 109.903809 | 109.903888 | positive | MGI_C57BL6J_3811419 | Mir1904 | miRBase:MI0008323,NCBI_Gene:100316683,ENSEMBL:ENSMUSG00000084651 | MGI:3811419 | miRNA gene | microRNA 1904 |
| 13 | gene | 110.006473 | 110.022710 | positive | MGI_C57BL6J_5592040 | Gm32881 | NCBI_Gene:102635588 | MGI:5592040 | lncRNA gene | predicted gene, 32881 |
| 13 | gene | 110.054187 | 110.281047 | negative | MGI_C57BL6J_1914545 | Rab3c | NCBI_Gene:67295,ENSEMBL:ENSMUSG00000021700 | MGI:1914545 | protein coding gene | RAB3C, member RAS oncogene family |
| 13 | gene | 110.181118 | 110.204666 | positive | MGI_C57BL6J_5623942 | Gm41057 | NCBI_Gene:105245628 | MGI:5623942 | lncRNA gene | predicted gene, 41057 |
| 13 | pseudogene | 110.303770 | 110.305599 | negative | MGI_C57BL6J_3646920 | Gm5455 | NCBI_Gene:432803,ENSEMBL:ENSMUSG00000045609 | MGI:3646920 | pseudogene | predicted pseudogene 5455 |
| 13 | gene | 110.352614 | 110.357279 | negative | MGI_C57BL6J_3608341 | Gapt | NCBI_Gene:238875,ENSEMBL:ENSMUSG00000046006 | MGI:3608341 | protein coding gene | Grb2-binding adaptor, transmembrane |
| 13 | gene | 110.358444 | 110.383294 | positive | MGI_C57BL6J_5623943 | Gm41058 | NCBI_Gene:105245629 | MGI:5623943 | lncRNA gene | predicted gene, 41058 |
| 13 | gene | 110.395044 | 110.400844 | positive | MGI_C57BL6J_1099790 | Plk2 | NCBI_Gene:20620,ENSEMBL:ENSMUSG00000021701 | MGI:1099790 | protein coding gene | polo like kinase 2 |
| 13 | gene | 110.568712 | 110.578225 | positive | MGI_C57BL6J_5592149 | Gm32990 | NCBI_Gene:102635727 | MGI:5592149 | lncRNA gene | predicted gene, 32990 |
| 13 | gene | 110.627242 | 110.642741 | positive | MGI_C57BL6J_5592204 | Gm33045 | NCBI_Gene:102635797,ENSEMBL:ENSMUSG00000109853 | MGI:5592204 | lncRNA gene | predicted gene, 33045 |
| 13 | gene | 110.642862 | 110.656095 | positive | MGI_C57BL6J_5623944 | Gm41059 | NCBI_Gene:105245630 | MGI:5623944 | lncRNA gene | predicted gene, 41059 |
| 13 | gene | 110.683826 | 110.686925 | positive | MGI_C57BL6J_5592331 | Gm33172 | NCBI_Gene:102635971,ENSEMBL:ENSMUSG00000114775 | MGI:5592331 | lncRNA gene | predicted gene, 33172 |
| 13 | gene | 110.743646 | 110.744673 | negative | MGI_C57BL6J_893593 | Tel-rs4 | NA | NA | unclassified gene | telomere%2c related sequence 4 |
| 13 | pseudogene | 110.842905 | 110.843121 | negative | MGI_C57BL6J_6098075 | Gm48533 | ENSEMBL:ENSMUSG00000114587 | MGI:6098075 | pseudogene | predicted gene, 48533 |
| 13 | pseudogene | 110.844574 | 110.844936 | negative | MGI_C57BL6J_6098076 | Gm48534 | ENSEMBL:ENSMUSG00000114275 | MGI:6098076 | pseudogene | predicted gene, 48534 |
| 13 | pseudogene | 110.860341 | 110.860671 | negative | MGI_C57BL6J_5589998 | Gm30839 | NCBI_Gene:102632877,ENSEMBL:ENSMUSG00000114320 | MGI:5589998 | pseudogene | predicted gene, 30839 |
| 13 | gene | 110.882411 | 110.903846 | negative | MGI_C57BL6J_1922389 | 4930526H09Rik | NCBI_Gene:102636142,ENSEMBL:ENSMUSG00000114307 | MGI:1922389 | lncRNA gene | RIKEN cDNA 4930526H09 gene |
| 13 | gene | 110.903499 | 110.908280 | positive | MGI_C57BL6J_5618690 | Gm38397 | NCBI_Gene:75139,ENSEMBL:ENSMUSG00000114472 | MGI:5618690 | lncRNA gene | predicted gene, 38397 |
| 13 | pseudogene | 111.017523 | 111.018821 | negative | MGI_C57BL6J_6098097 | Gm48547 | ENSEMBL:ENSMUSG00000114747 | MGI:6098097 | pseudogene | predicted gene, 48547 |
| 13 | pseudogene | 111.175142 | 111.176762 | negative | MGI_C57BL6J_5010500 | Gm18315 | NCBI_Gene:100416912 | MGI:5010500 | pseudogene | predicted gene, 18315 |
| 13 | gene | 111.211467 | 111.229431 | negative | MGI_C57BL6J_1920379 | 3110015C05Rik | NCBI_Gene:73129,ENSEMBL:ENSMUSG00000114824 | MGI:1920379 | lncRNA gene | RIKEN cDNA 3110015C05 gene |
| 13 | gene | 111.255013 | 111.257749 | positive | MGI_C57BL6J_2444552 | Actbl2 | NCBI_Gene:238880,ENSEMBL:ENSMUSG00000055194 | MGI:2444552 | protein coding gene | actin, beta-like 2 |
| 13 | pseudogene | 111.361134 | 111.361866 | negative | MGI_C57BL6J_3705624 | Gm15285 | NCBI_Gene:101055980,ENSEMBL:ENSMUSG00000081277 | MGI:3705624 | pseudogene | predicted gene 15285 |
| 13 | pseudogene | 111.361676 | 111.361919 | positive | MGI_C57BL6J_3705710 | Gm15289 | ENSEMBL:ENSMUSG00000082258 | MGI:3705710 | pseudogene | predicted gene 15289 |
| 13 | gene | 111.361840 | 111.368680 | negative | MGI_C57BL6J_3647282 | Gm6270 | ENSEMBL:ENSMUSG00000086201 | MGI:3647282 | lncRNA gene | predicted gene 6270 |
| 13 | gene | 111.425680 | 111.490125 | negative | MGI_C57BL6J_1920524 | Gpbp1 | NCBI_Gene:73274,ENSEMBL:ENSMUSG00000032745 | MGI:1920524 | protein coding gene | GC-rich promoter binding protein 1 |
| 13 | gene | 111.439229 | 111.549947 | negative | MGI_C57BL6J_6155177 | Gm49496 | ENSEMBL:ENSMUSG00000116016 | MGI:6155177 | protein coding gene | predicted gene, 49496 |
| 13 | gene | 111.522350 | 111.549977 | negative | MGI_C57BL6J_3705144 | Gm15290 | NCBI_Gene:102636218,ENSEMBL:ENSMUSG00000085634 | MGI:3705144 | lncRNA gene | predicted gene 15290 |
| 13 | gene | 111.561372 | 111.568553 | negative | MGI_C57BL6J_3705195 | Gm15288 | ENSEMBL:ENSMUSG00000086870 | MGI:3705195 | lncRNA gene | predicted gene 15288 |
| 13 | gene | 111.580202 | 111.644841 | positive | MGI_C57BL6J_3705204 | Gm15286 | NCBI_Gene:105245631,ENSEMBL:ENSMUSG00000086649 | MGI:3705204 | lncRNA gene | predicted gene 15286 |
| 13 | gene | 111.592785 | 111.592856 | positive | MGI_C57BL6J_3642607 | Gm10198 | ENSEMBL:ENSMUSG00000067122 | MGI:3642607 | protein coding gene | predicted gene 10198 |
| 13 | pseudogene | 111.593534 | 111.593611 | negative | MGI_C57BL6J_3704412 | Gm10736 | ENSEMBL:ENSMUSG00000096842 | MGI:3704412 | pseudogene | predicted gene 10736 |
| 13 | gene | 111.680979 | 111.718596 | positive | MGI_C57BL6J_2442317 | Mier3 | NCBI_Gene:218613,ENSEMBL:ENSMUSG00000032727 | MGI:2442317 | protein coding gene | MIER family member 3 |
| 13 | pseudogene | 111.721479 | 111.724334 | negative | MGI_C57BL6J_3705835 | Gm15488 | NCBI_Gene:102633089,ENSEMBL:ENSMUSG00000084217 | MGI:3705835 | pseudogene | predicted gene 15488 |
| 13 | gene | 111.725517 | 111.735124 | positive | MGI_C57BL6J_3705156 | Gm15287 | ENSEMBL:ENSMUSG00000086480 | MGI:3705156 | lncRNA gene | predicted gene 15287 |
| 13 | gene | 111.746428 | 111.818269 | negative | MGI_C57BL6J_1346872 | Map3k1 | NCBI_Gene:26401,ENSEMBL:ENSMUSG00000021754 | MGI:1346872 | protein coding gene | mitogen-activated protein kinase kinase kinase 1 |
| 13 | gene | 111.809137 | 111.811043 | positive | MGI_C57BL6J_3705295 | Gm15327 | NCBI_Gene:100504675,ENSEMBL:ENSMUSG00000085204 | MGI:3705295 | lncRNA gene | predicted gene 15327 |
| 13 | gene | 111.819812 | 111.838046 | positive | MGI_C57BL6J_5623945 | Gm41060 | NCBI_Gene:105245632 | MGI:5623945 | lncRNA gene | predicted gene, 41060 |
| 13 | gene | 111.850316 | 111.852622 | negative | MGI_C57BL6J_3705151 | Gm15325 | NCBI_Gene:102636540,ENSEMBL:ENSMUSG00000086861 | MGI:3705151 | lncRNA gene | predicted gene 15325 |
| 13 | gene | 111.856996 | 111.858505 | positive | MGI_C57BL6J_3705211 | Gm15324 | ENSEMBL:ENSMUSG00000086057 | MGI:3705211 | lncRNA gene | predicted gene 15324 |
| 13 | gene | 111.867936 | 111.874730 | positive | MGI_C57BL6J_3705205 | Gm15326 | NCBI_Gene:102636620,ENSEMBL:ENSMUSG00000086111 | MGI:3705205 | lncRNA gene | predicted gene 15326 |
| 13 | pseudogene | 111.882252 | 111.884419 | negative | MGI_C57BL6J_3646679 | Gm9025 | NCBI_Gene:668181,ENSEMBL:ENSMUSG00000114429,ENSEMBL:ENSMUSG00000082433 | MGI:3646679 | pseudogene | predicted gene 9025 |
| 13 | gene | 111.991466 | 112.048263 | positive | MGI_C57BL6J_3705208 | Gm15322 | NCBI_Gene:102636826,ENSEMBL:ENSMUSG00000086402 | MGI:3705208 | lncRNA gene | predicted gene 15322 |
| 13 | gene | 112.001009 | 112.017332 | positive | MGI_C57BL6J_3705285 | Gm15323 | NCBI_Gene:102636755,ENSEMBL:ENSMUSG00000086092 | MGI:3705285 | lncRNA gene | predicted gene 15323 |
| 13 | gene | 112.038313 | 112.041558 | negative | MGI_C57BL6J_5623946 | Gm41061 | NCBI_Gene:105245633 | MGI:5623946 | lncRNA gene | predicted gene, 41061 |
| 13 | gene | 112.100504 | 112.101699 | positive | MGI_C57BL6J_5623947 | Gm41062 | NCBI_Gene:105245634 | MGI:5623947 | lncRNA gene | predicted gene, 41062 |
| 13 | pseudogene | 112.138098 | 112.139335 | positive | MGI_C57BL6J_5590263 | Gm31104 | NCBI_Gene:102633222,ENSEMBL:ENSMUSG00000114361 | MGI:5590263 | pseudogene | predicted gene, 31104 |
| 13 | gene | 112.165458 | 112.172068 | negative | MGI_C57BL6J_6098510 | Gm48802 | ENSEMBL:ENSMUSG00000114745 | MGI:6098510 | lncRNA gene | predicted gene, 48802 |
| 13 | gene | 112.182285 | 112.186031 | positive | MGI_C57BL6J_5623948 | Gm41063 | NCBI_Gene:105245635 | MGI:5623948 | lncRNA gene | predicted gene, 41063 |
| 13 | gene | 112.186176 | 112.191342 | positive | MGI_C57BL6J_5593008 | Gm33849 | NCBI_Gene:102636904 | MGI:5593008 | lncRNA gene | predicted gene, 33849 |
| 13 | gene | 112.219193 | 112.220481 | positive | MGI_C57BL6J_5610655 | Gm37427 | ENSEMBL:ENSMUSG00000104438 | MGI:5610655 | unclassified gene | predicted gene, 37427 |
| 13 | gene | 112.288444 | 112.384002 | positive | MGI_C57BL6J_1924568 | Ankrd55 | NCBI_Gene:77318,ENSEMBL:ENSMUSG00000049985 | MGI:1924568 | protein coding gene | ankyrin repeat domain 55 |
| 13 | gene | 112.291121 | 112.291273 | positive | MGI_C57BL6J_5452545 | Gm22768 | ENSEMBL:ENSMUSG00000088991 | MGI:5452545 | snRNA gene | predicted gene, 22768 |
| 13 | gene | 112.351554 | 112.355805 | negative | MGI_C57BL6J_5623950 | Gm41065 | NCBI_Gene:105245637 | MGI:5623950 | lncRNA gene | predicted gene, 41065 |
| 13 | gene | 112.356925 | 112.367845 | negative | MGI_C57BL6J_6098566 | Gm48837 | ENSEMBL:ENSMUSG00000114536 | MGI:6098566 | lncRNA gene | predicted gene, 48837 |
| 13 | gene | 112.411142 | 112.423037 | positive | MGI_C57BL6J_5593064 | Gm33905 | NCBI_Gene:102636983 | MGI:5593064 | lncRNA gene | predicted gene, 33905 |
| 13 | gene | 112.414938 | 112.417365 | negative | MGI_C57BL6J_1917210 | 2810403G07Rik | ENSEMBL:ENSMUSG00000114656 | MGI:1917210 | lncRNA gene | RIKEN cDNA 2810403G07 gene |
| 13 | gene | 112.424222 | 112.426340 | negative | MGI_C57BL6J_6098626 | Gm48876 | ENSEMBL:ENSMUSG00000114869 | MGI:6098626 | lncRNA gene | predicted gene, 48876 |
| 13 | gene | 112.429434 | 112.436596 | negative | MGI_C57BL6J_6098632 | Gm48879 | ENSEMBL:ENSMUSG00000114468 | MGI:6098632 | lncRNA gene | predicted gene, 48879 |
| 13 | gene | 112.463096 | 112.463977 | negative | MGI_C57BL6J_1925149 | 6720470G18Rik | NA | NA | unclassified gene | RIKEN cDNA 6720470G18 gene |
| 13 | gene | 112.464070 | 112.510086 | positive | MGI_C57BL6J_96560 | Il6st | NCBI_Gene:16195,ENSEMBL:ENSMUSG00000021756 | MGI:96560 | protein coding gene | interleukin 6 signal transducer |
| 13 | gene | 112.519898 | 112.594360 | negative | MGI_C57BL6J_2180511 | Il31ra | NCBI_Gene:218624,ENSEMBL:ENSMUSG00000050377 | MGI:2180511 | protein coding gene | interleukin 31 receptor A |
| 13 | pseudogene | 112.593585 | 112.595140 | negative | MGI_C57BL6J_3804968 | Gm3226 | NCBI_Gene:100041240,ENSEMBL:ENSMUSG00000114547 | MGI:3804968 | pseudogene | predicted gene 3226 |
| 13 | gene | 112.598333 | 112.652633 | negative | MGI_C57BL6J_102670 | Ddx4 | NCBI_Gene:13206,ENSEMBL:ENSMUSG00000021758 | MGI:102670 | protein coding gene | DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| 13 | gene | 112.660691 | 112.738752 | positive | MGI_C57BL6J_1918839 | Slc38a9 | NCBI_Gene:268706,ENSEMBL:ENSMUSG00000047789 | MGI:1918839 | protein coding gene | solute carrier family 38, member 9 |
| 13 | gene | 112.669139 | 112.685696 | negative | MGI_C57BL6J_6097022 | Gm47827 | ENSEMBL:ENSMUSG00000114438 | MGI:6097022 | lncRNA gene | predicted gene, 47827 |
| 13 | pseudogene | 112.699430 | 112.700036 | positive | MGI_C57BL6J_5011068 | Gm18883 | NCBI_Gene:100417888,ENSEMBL:ENSMUSG00000114418 | MGI:5011068 | pseudogene | predicted gene, 18883 |
| 13 | gene | 112.764848 | 112.765616 | positive | MGI_C57BL6J_6097057 | Gm47850 | ENSEMBL:ENSMUSG00000114286 | MGI:6097057 | lncRNA gene | predicted gene, 47850 |
| 13 | gene | 112.768458 | 112.799371 | positive | MGI_C57BL6J_5623952 | Gm41067 | NCBI_Gene:105245639 | MGI:5623952 | lncRNA gene | predicted gene, 41067 |
| 13 | gene | 112.800379 | 112.800729 | negative | MGI_C57BL6J_1920886 | 1700125C05Rik | NA | NA | unclassified gene | RIKEN cDNA 1700125C05 gene |
| 13 | gene | 112.800777 | 112.867894 | positive | MGI_C57BL6J_108412 | Plpp1 | NCBI_Gene:19012,ENSEMBL:ENSMUSG00000021759 | MGI:108412 | protein coding gene | phospholipid phosphatase 1 |
| 13 | gene | 112.867418 | 112.927398 | negative | MGI_C57BL6J_1919448 | Mtrex | NCBI_Gene:72198,ENSEMBL:ENSMUSG00000016018 | MGI:1919448 | protein coding gene | Mtr4 exosome RNA helicase |
| 13 | pseudogene | 112.884226 | 112.885182 | positive | MGI_C57BL6J_5504058 | Gm26943 | ENSEMBL:ENSMUSG00000098067 | MGI:5504058 | pseudogene | predicted gene, 26943 |
| 13 | gene | 112.927454 | 112.969432 | positive | MGI_C57BL6J_2145374 | Dhx29 | NCBI_Gene:218629,ENSEMBL:ENSMUSG00000042426 | MGI:2145374 | protein coding gene | DEAH (Asp-Glu-Ala-His) box polypeptide 29 |
| 13 | pseudogene | 112.935604 | 112.939172 | positive | MGI_C57BL6J_3643764 | Gm8795 | NCBI_Gene:102637219,ENSEMBL:ENSMUSG00000114477 | MGI:3643764 | pseudogene | predicted gene 8795 |
| 13 | gene | 112.987802 | 112.990778 | positive | MGI_C57BL6J_2145534 | Ccno | NCBI_Gene:218630,ENSEMBL:ENSMUSG00000042417 | MGI:2145534 | protein coding gene | cyclin O |
| 13 | gene | 112.993472 | 113.000394 | positive | MGI_C57BL6J_3648807 | Mcidas | NCBI_Gene:622408,ENSEMBL:ENSMUSG00000074651 | MGI:3648807 | protein coding gene | multiciliate differentiation and DNA synthesis associated cell cycle protein |
| 13 | gene | 112.997786 | 112.999249 | negative | MGI_C57BL6J_1923984 | 6330437I11Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 6330437I11 gene |
| 13 | gene | 113.032954 | 113.121019 | positive | MGI_C57BL6J_3644472 | Cdc20b | NCBI_Gene:238896,ENSEMBL:ENSMUSG00000078926 | MGI:3644472 | protein coding gene | cell division cycle 20B |
| 13 | gene | 113.035983 | 113.036091 | positive | MGI_C57BL6J_3639505 | Mir449c | miRBase:MI0004645,NCBI_Gene:735309,ENSEMBL:ENSMUSG00000076146 | MGI:3639505 | miRNA gene | microRNA 449c |
| 13 | gene | 113.037419 | 113.037498 | positive | MGI_C57BL6J_3718803 | Mir449b | miRBase:MI0005547,NCBI_Gene:100190765,ENSEMBL:ENSMUSG00000078027 | MGI:3718803 | miRNA gene | microRNA 449b |
| 13 | gene | 113.037534 | 113.037624 | positive | MGI_C57BL6J_3619407 | Mir449a | miRBase:MI0001649,NCBI_Gene:723868,ENSEMBL:ENSMUSG00000065575 | MGI:3619407 | miRNA gene | microRNA 449a |
| 13 | gene | 113.039301 | 113.042243 | negative | MGI_C57BL6J_3642476 | Gm10735 | ENSEMBL:ENSMUSG00000074650 | MGI:3642476 | protein coding gene | predicted gene 10735 |
| 13 | gene | 113.042753 | 113.046428 | negative | MGI_C57BL6J_1916840 | Gpx8 | NCBI_Gene:69590,ENSEMBL:ENSMUSG00000021760 | MGI:1916840 | protein coding gene | glutathione peroxidase 8 (putative) |
| 13 | gene | 113.044674 | 113.045312 | positive | MGI_C57BL6J_1916511 | 2810411K19Rik | NA | NA | unclassified gene | RIKEN cDNA 2810411K19 gene |
| 13 | gene | 113.078106 | 113.079147 | negative | MGI_C57BL6J_1914918 | 4930563N14Rik | ENSEMBL:ENSMUSG00000116554 | MGI:1914918 | lncRNA gene | RIKEN cDNA 4930563N14 gene |
| 13 | gene | 113.093825 | 113.100981 | negative | MGI_C57BL6J_109266 | Gzma | NCBI_Gene:14938,ENSEMBL:ENSMUSG00000023132 | MGI:109266 | protein coding gene | granzyme A |
| 13 | pseudogene | 113.108073 | 113.108331 | positive | MGI_C57BL6J_3642450 | Gm9848 | ENSEMBL:ENSMUSG00000051002 | MGI:3642450 | pseudogene | predicted gene 9848 |
| 13 | gene | 113.136076 | 113.144012 | positive | MGI_C57BL6J_1923970 | 1700084D21Rik | NCBI_Gene:76720,ENSEMBL:ENSMUSG00000114333 | MGI:1923970 | lncRNA gene | RIKEN cDNA 1700084D21 gene |
| 13 | pseudogene | 113.150677 | 113.151450 | negative | MGI_C57BL6J_6214953 | Gm49564 | ENSEMBL:ENSMUSG00000116741 | MGI:6214953 | pseudogene | predicted gene, 49564 |
| 13 | gene | 113.171608 | 113.225908 | negative | MGI_C57BL6J_1298232 | Gzmk | NCBI_Gene:14945,ENSEMBL:ENSMUSG00000042385 | MGI:1298232 | protein coding gene | granzyme K |
| 13 | gene | 113.188439 | 113.225967 | negative | MGI_C57BL6J_5623954 | Gm41069 | NCBI_Gene:105245641 | MGI:5623954 | lncRNA gene | predicted gene, 41069 |
| 13 | gene | 113.209659 | 113.218104 | positive | MGI_C57BL6J_1918940 | Esm1 | NCBI_Gene:71690,ENSEMBL:ENSMUSG00000042379 | MGI:1918940 | protein coding gene | endothelial cell-specific molecule 1 |
| 13 | gene | 113.292423 | 113.379711 | positive | MGI_C57BL6J_3040697 | BC067074 | NCBI_Gene:408066,ENSEMBL:ENSMUSG00000021763 | MGI:3040697 | protein coding gene | cDNA sequence BC067074 |
| 13 | gene | 113.355672 | 113.365091 | negative | MGI_C57BL6J_5623955 | Gm41070 | NCBI_Gene:105245642 | MGI:5623955 | lncRNA gene | predicted gene, 41070 |
| 13 | gene | 113.403088 | 113.403188 | positive | MGI_C57BL6J_5452162 | Gm22385 | ENSEMBL:ENSMUSG00000088814 | MGI:5452162 | rRNA gene | predicted gene, 22385 |
| 13 | gene | 113.419550 | 113.423990 | negative | MGI_C57BL6J_5623956 | Gm41071 | NCBI_Gene:105245643,ENSEMBL:ENSMUSG00000114864 | MGI:5623956 | lncRNA gene | predicted gene, 41071 |
| 13 | gene | 113.446288 | 113.502844 | negative | MGI_C57BL6J_1918167 | 4921509O07Rik | NCBI_Gene:70917,ENSEMBL:ENSMUSG00000114790 | MGI:1918167 | lncRNA gene | RIKEN cDNA 4921509O07 gene |
| 13 | pseudogene | 113.479942 | 113.480305 | negative | MGI_C57BL6J_5590350 | Gm31191 | NCBI_Gene:102633343,ENSEMBL:ENSMUSG00000114659 | MGI:5590350 | pseudogene | predicted gene, 31191 |
| 13 | pseudogene | 113.488956 | 113.489135 | positive | MGI_C57BL6J_6096370 | Gm47426 | ENSEMBL:ENSMUSG00000114364 | MGI:6096370 | pseudogene | predicted gene, 47426 |
| 13 | gene | 113.507828 | 113.567767 | negative | MGI_C57BL6J_5593630 | Gm34471 | NCBI_Gene:102637735,ENSEMBL:ENSMUSG00000114715 | MGI:5593630 | lncRNA gene | predicted gene, 34471 |
| 13 | gene | 113.523554 | 113.527237 | negative | MGI_C57BL6J_5593687 | Gm34528 | NCBI_Gene:102637810 | MGI:5593687 | lncRNA gene | predicted gene, 34528 |
| 13 | gene | 113.592179 | 113.618564 | negative | MGI_C57BL6J_2137642 | Snx18 | NCBI_Gene:170625,ENSEMBL:ENSMUSG00000042364 | MGI:2137642 | protein coding gene | sorting nexin 18 |
| 13 | gene | 113.653201 | 113.686540 | positive | MGI_C57BL6J_5593745 | Gm34586 | NCBI_Gene:102637890,ENSEMBL:ENSMUSG00000114417 | MGI:5593745 | lncRNA gene | predicted gene, 34586 |
| 13 | gene | 113.662896 | 113.663676 | negative | MGI_C57BL6J_1928479 | Hspb3 | NCBI_Gene:56534,ENSEMBL:ENSMUSG00000051456 | MGI:1928479 | protein coding gene | heat shock protein 3 |
| 13 | gene | 113.736220 | 113.750531 | positive | MGI_C57BL6J_5590435 | Gm31276 | NCBI_Gene:102633457 | MGI:5590435 | lncRNA gene | predicted gene, 31276 |
| 13 | pseudogene | 113.761386 | 113.762136 | negative | MGI_C57BL6J_3647371 | Gm6343 | NCBI_Gene:622701,ENSEMBL:ENSMUSG00000114962 | MGI:3647371 | pseudogene | predicted gene 6343 |
| 13 | pseudogene | 113.764112 | 113.764277 | positive | MGI_C57BL6J_6096427 | Gm47462 | ENSEMBL:ENSMUSG00000114901 | MGI:6096427 | pseudogene | predicted gene, 47462 |
| 13 | pseudogene | 113.764615 | 113.764921 | positive | MGI_C57BL6J_6096428 | Gm47463 | ENSEMBL:ENSMUSG00000114781 | MGI:6096428 | pseudogene | predicted gene, 47463 |
| 13 | pseudogene | 113.791552 | 113.791777 | negative | MGI_C57BL6J_6096429 | Gm47464 | ENSEMBL:ENSMUSG00000114395 | MGI:6096429 | pseudogene | predicted gene, 47464 |
| 13 | gene | 113.794505 | 114.157462 | positive | MGI_C57BL6J_2442308 | Arl15 | NCBI_Gene:218639,ENSEMBL:ENSMUSG00000042348 | MGI:2442308 | protein coding gene | ADP-ribosylation factor-like 15 |
| 13 | gene | 113.823598 | 113.824178 | negative | MGI_C57BL6J_2145484 | C130040J23Rik | NA | NA | unclassified gene | RIKEN cDNA C130040J23 gene |
| 13 | gene | 113.972886 | 113.976592 | positive | MGI_C57BL6J_1925134 | 6720427H10Rik | NA | NA | unclassified gene | RIKEN cDNA 6720427H10 gene |
| 13 | gene | 114.096297 | 114.099563 | positive | MGI_C57BL6J_2145507 | B430203I24Rik | NA | NA | unclassified gene | RIKEN cDNA B430203I24 gene |
| 13 | gene | 114.118441 | 114.120416 | negative | MGI_C57BL6J_6096452 | Gm47479 | ENSEMBL:ENSMUSG00000114696 | MGI:6096452 | unclassified gene | predicted gene, 47479 |
| 13 | gene | 114.124348 | 114.151592 | negative | MGI_C57BL6J_2442754 | A430090L17Rik | NCBI_Gene:319803,ENSEMBL:ENSMUSG00000114213 | MGI:2442754 | lncRNA gene | RIKEN cDNA A430090L17 gene |
| 13 | gene | 114.208989 | 114.218138 | positive | MGI_C57BL6J_5623957 | Gm41072 | NCBI_Gene:105245644 | MGI:5623957 | lncRNA gene | predicted gene, 41072 |
| 13 | gene | 114.227111 | 114.237432 | positive | MGI_C57BL6J_5611135 | Gm37907 | ENSEMBL:ENSMUSG00000104314 | MGI:5611135 | unclassified gene | predicted gene, 37907 |
| 13 | gene | 114.240878 | 114.242640 | negative | MGI_C57BL6J_1918048 | 4631422I05Rik | ENSEMBL:ENSMUSG00000114655 | MGI:1918048 | unclassified gene | RIKEN cDNA 4631422I05 gene |
| 13 | gene | 114.281668 | 114.284381 | negative | MGI_C57BL6J_6096579 | Gm47558 | ENSEMBL:ENSMUSG00000114707 | MGI:6096579 | unclassified gene | predicted gene, 47558 |
| 13 | gene | 114.287795 | 114.388258 | negative | MGI_C57BL6J_1343135 | Ndufs4 | NCBI_Gene:17993,ENSEMBL:ENSMUSG00000021764 | MGI:1343135 | protein coding gene | NADH:ubiquinone oxidoreductase core subunit S4 |
| 13 | gene | 114.299766 | 114.303863 | negative | MGI_C57BL6J_2444238 | B330018G23Rik | NA | NA | unclassified gene | RIKEN cDNA B330018G23 gene |
| 13 | pseudogene | 114.435409 | 114.436699 | negative | MGI_C57BL6J_6096598 | Gm47569 | ENSEMBL:ENSMUSG00000114712 | MGI:6096598 | pseudogene | predicted gene, 47569 |
| 13 | pseudogene | 114.437790 | 114.438214 | negative | MGI_C57BL6J_5623958 | Gm41073 | NCBI_Gene:105245645,ENSEMBL:ENSMUSG00000114520 | MGI:5623958 | pseudogene | predicted gene, 41073 |
| 13 | gene | 114.452262 | 114.458951 | negative | MGI_C57BL6J_95586 | Fst | NCBI_Gene:14313,ENSEMBL:ENSMUSG00000021765 | MGI:95586 | protein coding gene | follistatin |
| 13 | gene | 114.456760 | 114.474219 | positive | MGI_C57BL6J_5623959 | Gm41074 | NCBI_Gene:105245646 | MGI:5623959 | lncRNA gene | predicted gene, 41074 |
| 13 | gene | 114.458663 | 114.462047 | positive | MGI_C57BL6J_6095741 | Gm47040 | ENSEMBL:ENSMUSG00000114935 | MGI:6095741 | lncRNA gene | predicted gene, 47040 |
| 13 | gene | 114.580495 | 114.580594 | positive | MGI_C57BL6J_5455727 | Gm25950 | ENSEMBL:ENSMUSG00000093072 | MGI:5455727 | miRNA gene | predicted gene, 25950 |
| 13 | gene | 114.586510 | 114.607124 | negative | MGI_C57BL6J_1922159 | 4930467J12Rik | NCBI_Gene:74909,ENSEMBL:ENSMUSG00000114879 | MGI:1922159 | lncRNA gene | RIKEN cDNA 4930467J12 gene |
| 13 | gene | 114.607228 | 114.697822 | positive | MGI_C57BL6J_1922411 | 4930544M13Rik | NCBI_Gene:75161,ENSEMBL:ENSMUSG00000051758 | MGI:1922411 | lncRNA gene | RIKEN cDNA 4930544M13 gene |
| 13 | gene | 114.635086 | 114.636852 | positive | MGI_C57BL6J_1921957 | 4930521G14Rik | NA | NA | unclassified gene | RIKEN cDNA 4930521G14 gene |
| 13 | gene | 114.784234 | 114.818061 | negative | MGI_C57BL6J_5593949 | Gm34790 | NCBI_Gene:102638161 | MGI:5593949 | lncRNA gene | predicted gene, 34790 |
| 13 | gene | 114.818236 | 114.832275 | positive | MGI_C57BL6J_1336894 | Mocs2 | NCBI_Gene:17434,ENSEMBL:ENSMUSG00000015536 | MGI:1336894 | protein coding gene | molybdenum cofactor synthesis 2 |
| 13 | gene | 114.833081 | 114.932100 | negative | MGI_C57BL6J_96600 | Itga2 | NCBI_Gene:16398,ENSEMBL:ENSMUSG00000015533 | MGI:96600 | protein coding gene | integrin alpha 2 |
| 13 | gene | 114.922899 | 114.927758 | positive | MGI_C57BL6J_5594004 | Gm34845 | NCBI_Gene:102638236 | MGI:5594004 | lncRNA gene | predicted gene, 34845 |
| 13 | pseudogene | 114.934642 | 114.935068 | negative | MGI_C57BL6J_6096937 | Gm47776 | ENSEMBL:ENSMUSG00000114703 | MGI:6096937 | pseudogene | predicted gene, 47776 |
| 13 | gene | 114.953096 | 115.101964 | negative | MGI_C57BL6J_96599 | Itga1 | NCBI_Gene:109700,ENSEMBL:ENSMUSG00000042284 | MGI:96599 | protein coding gene | integrin alpha 1 |
| 13 | gene | 114.953100 | 114.955732 | negative | MGI_C57BL6J_1915036 | 6230424C14Rik | NA | NA | unclassified gene | RIKEN cDNA 6230424C14 gene |
| 13 | gene | 115.035273 | 115.101921 | negative | MGI_C57BL6J_6121627 | Gm49395 | ENSEMBL:ENSMUSG00000114470 | MGI:6121627 | protein coding gene | predicted gene, 49395 |
| 13 | gene | 115.088355 | 115.090186 | negative | MGI_C57BL6J_2145154 | Pelo | NCBI_Gene:105083,ENSEMBL:ENSMUSG00000042275 | MGI:2145154 | protein coding gene | pelota mRNA surveillance and ribosome rescue factor |
| 13 | gene | 115.089501 | 115.090987 | positive | MGI_C57BL6J_3642559 | Gm10734 | ENSEMBL:ENSMUSG00000114255 | MGI:3642559 | lncRNA gene | predicted gene 10734 |
| 13 | gene | 115.109830 | 115.135838 | positive | MGI_C57BL6J_5623960 | Gm41075 | NCBI_Gene:105245647 | MGI:5623960 | lncRNA gene | predicted gene, 41075 |
| 13 | gene | 115.116127 | 115.116233 | positive | MGI_C57BL6J_5455762 | Gm25985 | ENSEMBL:ENSMUSG00000096021 | MGI:5455762 | snRNA gene | predicted gene, 25985 |
| 13 | pseudogene | 115.163607 | 115.164825 | negative | MGI_C57BL6J_5010944 | Gm18759 | NCBI_Gene:100417678,ENSEMBL:ENSMUSG00000114872 | MGI:5010944 | pseudogene | predicted gene, 18759 |
| 13 | pseudogene | 115.282201 | 115.282675 | positive | MGI_C57BL6J_6097124 | Gm47891 | ENSEMBL:ENSMUSG00000114471 | MGI:6097124 | pseudogene | predicted gene, 47891 |
| 13 | pseudogene | 115.528501 | 115.529797 | negative | MGI_C57BL6J_3643289 | Gm6035 | NCBI_Gene:547402,ENSEMBL:ENSMUSG00000114551 | MGI:3643289 | pseudogene | predicted gene 6035 |
| 13 | pseudogene | 115.719304 | 115.719499 | negative | MGI_C57BL6J_6097125 | Gm47892 | ENSEMBL:ENSMUSG00000114847 | MGI:6097125 | pseudogene | predicted gene, 47892 |
| 13 | pseudogene | 115.770936 | 115.771468 | positive | MGI_C57BL6J_5010320 | Gm18135 | NCBI_Gene:100416507,ENSEMBL:ENSMUSG00000114324 | MGI:5010320 | pseudogene | predicted gene, 18135 |
| 13 | pseudogene | 116.096035 | 116.096300 | positive | MGI_C57BL6J_6097126 | Gm47893 | ENSEMBL:ENSMUSG00000114956 | MGI:6097126 | pseudogene | predicted gene, 47893 |
| 13 | gene | 116.106913 | 116.107870 | negative | MGI_C57BL6J_3642560 | Gm10733 | NA | NA | unclassified gene | predicted gene 10733 |
| 13 | pseudogene | 116.153766 | 116.154172 | positive | MGI_C57BL6J_3648994 | Rpl34-ps2 | NCBI_Gene:623174,ENSEMBL:ENSMUSG00000061669 | MGI:3648994 | pseudogene | ribosomal protein L34, pseudogene 2 |
| 13 | gene | 116.298257 | 116.309721 | negative | MGI_C57BL6J_101791 | Isl1 | NCBI_Gene:16392,ENSEMBL:ENSMUSG00000042258 | MGI:101791 | protein coding gene | ISL1 transcription factor, LIM/homeodomain |
| 13 | pseudogene | 116.330726 | 116.331538 | negative | MGI_C57BL6J_5588923 | Gm29764 | NCBI_Gene:101056040,ENSEMBL:ENSMUSG00000114889 | MGI:5588923 | pseudogene | predicted gene, 29764 |
| 13 | pseudogene | 116.361198 | 116.361694 | negative | MGI_C57BL6J_6097159 | Gm47912 | ENSEMBL:ENSMUSG00000114451 | MGI:6097159 | pseudogene | predicted gene, 47912 |
| 13 | gene | 116.558311 | 116.677705 | negative | MGI_C57BL6J_6097162 | Gm47914 | ENSEMBL:ENSMUSG00000114731 | MGI:6097162 | lncRNA gene | predicted gene, 47914 |
| 13 | gene | 116.651448 | 116.663591 | negative | MGI_C57BL6J_6097160 | Gm47913 | ENSEMBL:ENSMUSG00000114347 | MGI:6097160 | lncRNA gene | predicted gene, 47913 |
| 13 | gene | 116.675878 | 116.675968 | positive | MGI_C57BL6J_5452975 | Gm23198 | ENSEMBL:ENSMUSG00000089436 | MGI:5452975 | rRNA gene | predicted gene, 23198 |
| 13 | gene | 116.802095 | 116.838178 | negative | MGI_C57BL6J_1921930 | 4930435F18Rik | NCBI_Gene:74680,ENSEMBL:ENSMUSG00000114290 | MGI:1921930 | lncRNA gene | RIKEN cDNA 4930435F18 gene |
| 13 | gene | 116.850842 | 117.025925 | negative | MGI_C57BL6J_1098713 | Parp8 | NCBI_Gene:52552,ENSEMBL:ENSMUSG00000021725 | MGI:1098713 | protein coding gene | poly (ADP-ribose) polymerase family, member 8 |
| 13 | gene | 116.850844 | 116.852228 | negative | MGI_C57BL6J_1925084 | A930041H05Rik | NA | NA | unclassified gene | RIKEN cDNA A930041H05 gene |
| 13 | gene | 117.017996 | 117.018568 | positive | MGI_C57BL6J_3026967 | 5930433N17Rik | NA | NA | unclassified gene | RIKEN cDNA 5930433N17 gene |
| 13 | pseudogene | 117.050734 | 117.063087 | positive | MGI_C57BL6J_5588926 | Gm29767 | NCBI_Gene:101056052,ENSEMBL:ENSMUSG00000114609 | MGI:5588926 | pseudogene | predicted gene, 29767 |
| 13 | pseudogene | 117.085631 | 117.085979 | negative | MGI_C57BL6J_6097340 | Gm48026 | ENSEMBL:ENSMUSG00000114876 | MGI:6097340 | pseudogene | predicted gene, 48026 |
| 13 | gene | 117.130025 | 117.135884 | positive | MGI_C57BL6J_3779595 | Gm6416 | NCBI_Gene:623312,ENSEMBL:ENSMUSG00000114792 | MGI:3779595 | lncRNA gene | predicted gene 6416 |
| 13 | pseudogene | 117.143106 | 117.144197 | positive | MGI_C57BL6J_5011130 | Gm18945 | NCBI_Gene:100418010,ENSEMBL:ENSMUSG00000114964 | MGI:5011130 | pseudogene | predicted gene, 18945 |
| 13 | gene | 117.208536 | 117.274415 | positive | MGI_C57BL6J_95321 | Emb | NCBI_Gene:13723,ENSEMBL:ENSMUSG00000021728 | MGI:95321 | protein coding gene | embigin |
| 13 | gene | 117.218701 | 117.221075 | negative | MGI_C57BL6J_4937143 | Gm17509 | ENSEMBL:ENSMUSG00000091423 | MGI:4937143 | protein coding gene | predicted gene, 17509 |
| 13 | pseudogene | 117.229355 | 117.230117 | positive | MGI_C57BL6J_5594303 | Gm35144 | NCBI_Gene:102638621 | MGI:5594303 | pseudogene | predicted gene, 35144 |
| 13 | gene | 117.265024 | 117.265615 | positive | MGI_C57BL6J_1925184 | A430106F12Rik | NA | NA | unclassified gene | RIKEN cDNA A430106F12 gene |
| 13 | pseudogene | 117.347745 | 117.358718 | negative | MGI_C57BL6J_3647311 | Gm6421 | NCBI_Gene:623365,ENSEMBL:ENSMUSG00000114540 | MGI:3647311 | pseudogene | predicted pseudogene 6421 |
| 13 | pseudogene | 117.457772 | 117.459126 | negative | MGI_C57BL6J_3646335 | Gm5200 | NCBI_Gene:382807,ENSEMBL:ENSMUSG00000114358 | MGI:3646335 | pseudogene | predicted gene 5200 |
| 13 | gene | 117.600271 | 117.602333 | negative | MGI_C57BL6J_5826075 | Gm46438 | NCBI_Gene:108168112 | MGI:5826075 | lncRNA gene | predicted gene, 46438 |
| 13 | gene | 117.602320 | 117.987418 | positive | MGI_C57BL6J_1096392 | Hcn1 | NCBI_Gene:15165,ENSEMBL:ENSMUSG00000021730 | MGI:1096392 | protein coding gene | hyperpolarization activated cyclic nucleotide gated potassium channel 1 |
| 13 | gene | 117.706764 | 117.720011 | negative | MGI_C57BL6J_1918350 | 4933413L06Rik | NCBI_Gene:71100,ENSEMBL:ENSMUSG00000079048 | MGI:1918350 | lncRNA gene | RIKEN cDNA 4933413L06 gene |
| 13 | pseudogene | 117.741095 | 117.741819 | negative | MGI_C57BL6J_5011174 | Gm18989 | NCBI_Gene:100418083 | MGI:5011174 | pseudogene | predicted gene, 18989 |
| 13 | gene | 117.822313 | 117.823322 | positive | MGI_C57BL6J_1924778 | C030014A21Rik | NA | NA | unclassified gene | RIKEN cDNA C030014A21 gene |
| 13 | gene | 117.943664 | 117.959078 | negative | MGI_C57BL6J_3781650 | Gm3474 | NCBI_Gene:105245648 | MGI:3781650 | lncRNA gene | predicted gene 3474 |
| 13 | gene | 117.985193 | 117.987410 | positive | MGI_C57BL6J_1923436 | 6330563C09Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 6330563C09 gene |
| 13 | gene | 118.036614 | 118.036743 | positive | MGI_C57BL6J_5456126 | Gm26349 | ENSEMBL:ENSMUSG00000088155 | MGI:5456126 | snoRNA gene | predicted gene, 26349 |
| 13 | pseudogene | 118.047292 | 118.047989 | positive | MGI_C57BL6J_3780040 | Gm9633 | NCBI_Gene:674926,ENSEMBL:ENSMUSG00000114713 | MGI:3780040 | pseudogene | predicted gene 9633 |
| 13 | gene | 118.378381 | 118.387252 | negative | MGI_C57BL6J_1926237 | Mrps30 | NCBI_Gene:59054,ENSEMBL:ENSMUSG00000021731 | MGI:1926237 | protein coding gene | mitochondrial ribosomal protein S30 |
| 13 | gene | 118.386630 | 118.388124 | positive | MGI_C57BL6J_3704413 | B430218F22Rik | ENSEMBL:ENSMUSG00000097411 | MGI:3704413 | protein coding gene | RIKEN cDNA B430218F22 gene |
| 13 | pseudogene | 118.510358 | 118.510913 | negative | MGI_C57BL6J_6096587 | Gm47563 | ENSEMBL:ENSMUSG00000114668 | MGI:6096587 | pseudogene | predicted gene, 47563 |
| 13 | gene | 118.555360 | 118.588704 | positive | MGI_C57BL6J_1916594 | 1700003P14Rik | NCBI_Gene:75890,ENSEMBL:ENSMUSG00000100727 | MGI:1916594 | lncRNA gene | RIKEN cDNA 1700003P14 gene |
| 13 | pseudogene | 118.568772 | 118.569783 | negative | MGI_C57BL6J_3648600 | Gm9098 | NCBI_Gene:668309,ENSEMBL:ENSMUSG00000114365 | MGI:3648600 | pseudogene | predicted gene 9098 |
| 13 | pseudogene | 118.589732 | 118.590257 | positive | MGI_C57BL6J_6096218 | Gm47330 | ENSEMBL:ENSMUSG00000114485 | MGI:6096218 | pseudogene | predicted gene, 47330 |
| 13 | pseudogene | 118.598039 | 118.598918 | negative | MGI_C57BL6J_5010546 | Gm18361 | NCBI_Gene:100417013,ENSEMBL:ENSMUSG00000114831 | MGI:5010546 | pseudogene | predicted gene, 18361 |
| 13 | gene | 118.603410 | 118.658743 | negative | MGI_C57BL6J_3642638 | Gm10732 | NCBI_Gene:100038690,ENSEMBL:ENSMUSG00000114613 | MGI:3642638 | lncRNA gene | predicted gene 10732 |
| 13 | gene | 118.669791 | 118.792573 | positive | MGI_C57BL6J_1099809 | Fgf10 | NCBI_Gene:14165,ENSEMBL:ENSMUSG00000021732 | MGI:1099809 | protein coding gene | fibroblast growth factor 10 |
| 13 | gene | 118.686120 | 118.716619 | negative | MGI_C57BL6J_3826536 | Gm16263 | NCBI_Gene:102638835,ENSEMBL:ENSMUSG00000086709 | MGI:3826536 | lncRNA gene | predicted gene 16263 |
| 13 | gene | 118.712701 | 118.714495 | negative | MGI_C57BL6J_3781425 | Gm3247 | NA | NA | unclassified gene | predicted gene 3247 |
| 13 | pseudogene | 118.858145 | 118.858414 | negative | MGI_C57BL6J_6096224 | Gm47335 | ENSEMBL:ENSMUSG00000114868 | MGI:6096224 | pseudogene | predicted gene, 47335 |
Chromsome 19
query_variants <- create_variant_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/cc_variants.sqlite")
query_genes <- create_gene_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/mouse_genes_mgi.sqlite")
if(nrow(peaks) >0){
for (i in 3){
#for (i in 1:1){
#Plot 1
#marker = find_marker(gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i])
#gp <- g[,marker]
#gp[gp==1] <- "AA"
#gp[gp==2] <- "AB"
#gp[gp==0] <- NA
#plot_pxg(gp, gm$covar[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
#title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
###dev.off()
g <- maxmarg(pr.qc, gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i], return_char=TRUE)
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/","qtl_effect_", i, ".png"))
#par(mar=c(4.1, 4.1, 1.5, 0.6))
plot_pxg(g, gm$covar[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
##dev.off()
chr = peaks$chr[i]
# Plot 2
pr_sub <- pull_genoprobint(pr.qc, gm$gmap, chr, c(peaks$ci_lo[i], peaks$ci_hi[i]))
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar)
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], Xcovar=Xcovar)
#coeff <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
#coeff_sub <- scan1coef(pr_sub[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
blup <- scan1blup(pr.qc[,chr], gm$covar[peaks$phenotype[i]])
blup_sub <- scan1blup(pr_sub[,chr], gm$covar[peaks$phenotype[i]])
write.csv(as.data.frame(blup_sub), paste0("data/ici.vs.pbs_blup_sub_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv"), quote=F)
#plot_coef(coeff,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i]," [scan1coeff; positions in cM])")
# )
#plot_coef(coeff_sub,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i],"; 1.5 LOD drop interval [scan1coeff; positions in cM] ) ")
# )
plot_coef(blup,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," [scan1blup; positions in cM])")
)
plot_coef(blup_sub,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i],"; 1.5 LOD drop interval [scan1blup; positions in cM])")
)
# Plot 3
#c2effB <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary", contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
#c2effBb <- scan1blup(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]],Xcovar=Xcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
#plot(c2effB, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
#plot(c2effBb, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
##last_coef <- unclass(c2effB)[nrow(c2effB),2:3] # last two coefficients
##for(t in seq(along=last_coef))
## axis(side=4, at=last_coef[t], names(last_coef)[t], tick=FALSE)
#Table 1
chr = peaks_mba$chr[i]
start=as.numeric(peaks_mba$ci_lo[i])
end=as.numeric(peaks_mba$ci_hi[i])
genesgss = query_genes(chr, start, end)
write.csv(genesgss, file=paste0("data/ici.vs.pbs_genes_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv"), quote=F)
rownames(genesgss) <- NULL
genesgss$strand_old = genesgss$strand
genesgss$strand[genesgss$strand=="+"] <- "positive"
genesgss$strand[genesgss$strand=="-"] <- "negative"
#genesgss <-
#table <-
#genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")] %>%
#kable(escape = F,align = c("ccccccccccc")) %>%
#kable_styling("striped", full_width = T) #%>%
#cat #%>%
#column_spec(1, bold=TRUE)
#
#print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], escape = F,align = c("ccccccccccc")))
print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], "html") %>% kable_styling("striped", full_width = T))
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}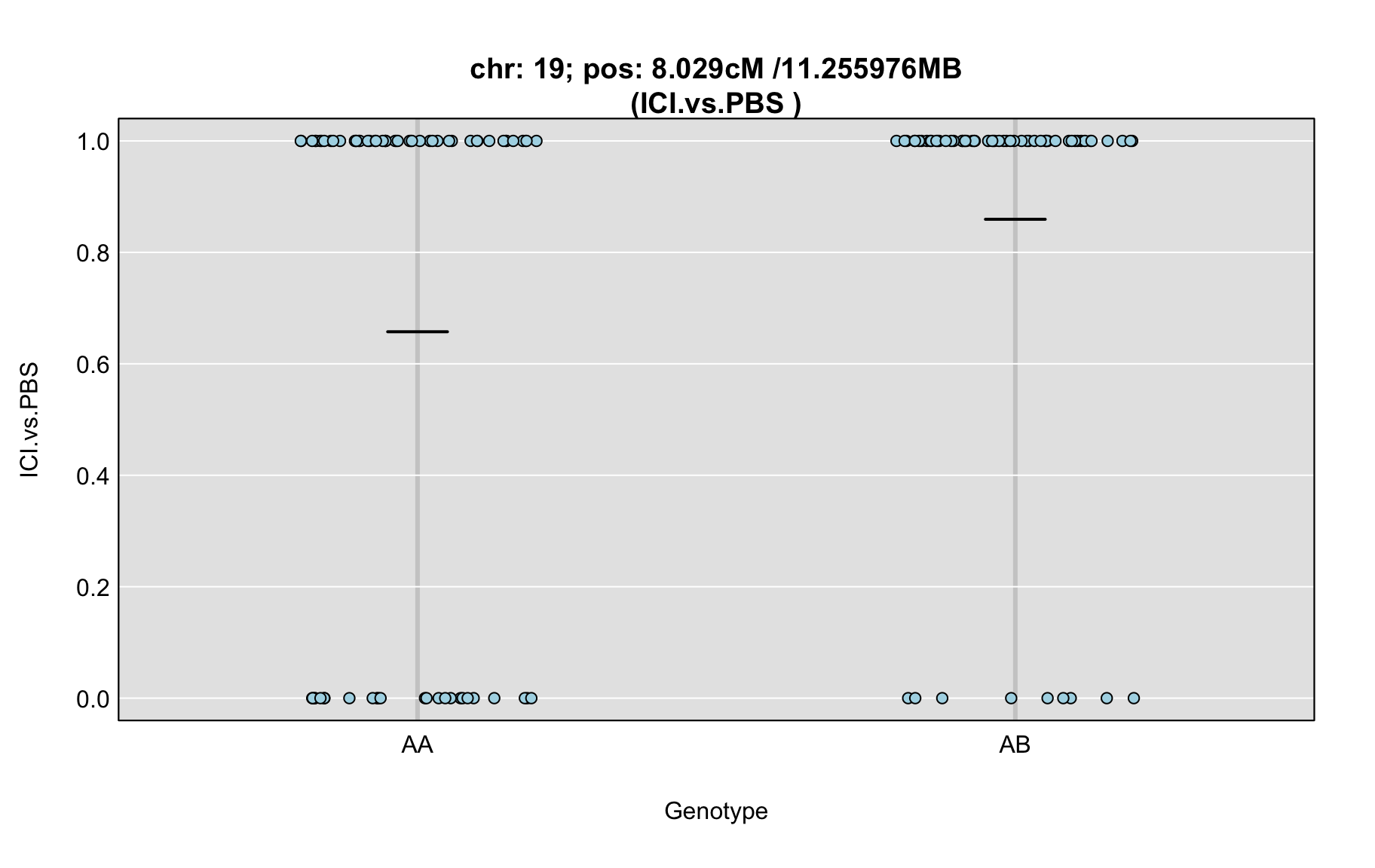

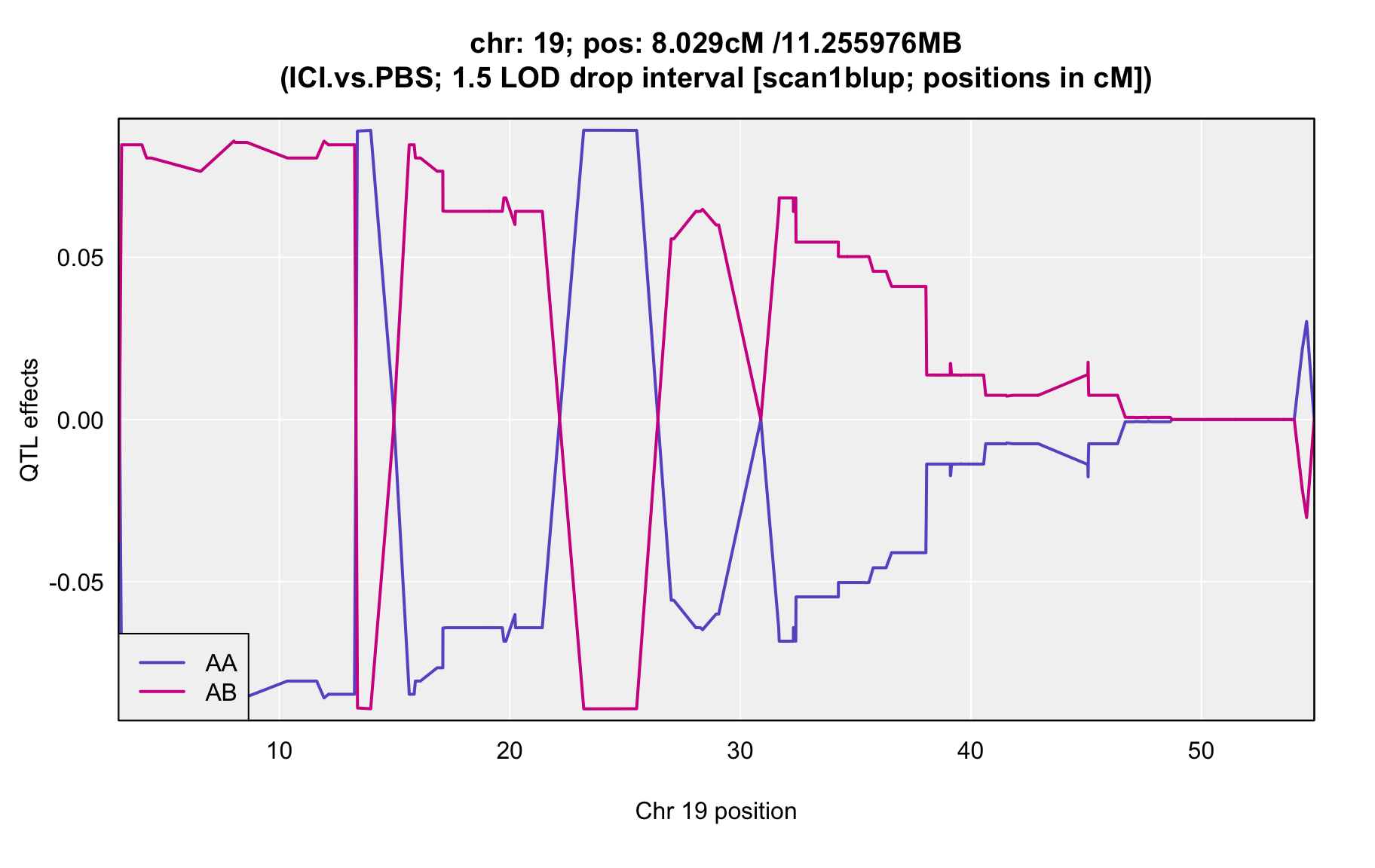
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 19 | gene | 3.259076 | 3.283017 | negative | MGI_C57BL6J_99954 | Ighmbp2 | NCBI_Gene:20589,ENSEMBL:ENSMUSG00000024831 | MGI:99954 | protein coding gene | immunoglobulin mu binding protein 2 |
| 19 | gene | 3.282901 | 3.292837 | positive | MGI_C57BL6J_2660674 | Mrpl21 | NCBI_Gene:353242,ENSEMBL:ENSMUSG00000024829 | MGI:2660674 | protein coding gene | mitochondrial ribosomal protein L21 |
| 19 | gene | 3.288920 | 3.288983 | positive | MGI_C57BL6J_5531153 | Mir6984 | miRBase:MI0022832,NCBI_Gene:102465594,ENSEMBL:ENSMUSG00000099261 | MGI:5531153 | miRNA gene | microRNA 6984 |
| 19 | pseudogene | 3.316782 | 3.317581 | negative | MGI_C57BL6J_6303159 | Gm50307 | ENSEMBL:ENSMUSG00000118213 | MGI:6303159 | pseudogene | predicted gene, 50307 |
| 19 | gene | 3.322325 | 3.385735 | positive | MGI_C57BL6J_1098296 | Cpt1a | NCBI_Gene:12894,ENSEMBL:ENSMUSG00000024900 | MGI:1098296 | protein coding gene | carnitine palmitoyltransferase 1a, liver |
| 19 | gene | 3.373448 | 3.378102 | negative | MGI_C57BL6J_5595691 | Gm36532 | NCBI_Gene:102640483 | MGI:5595691 | lncRNA gene | predicted gene, 36532 |
| 19 | gene | 3.386211 | 3.387996 | positive | MGI_C57BL6J_1921854 | 4833417J20Rik | NA | NA | unclassified gene | RIKEN cDNA 4833417J20 gene |
| 19 | gene | 3.388857 | 3.407823 | positive | MGI_C57BL6J_1340029 | Tesmin | NCBI_Gene:17771,ENSEMBL:ENSMUSG00000024905 | MGI:1340029 | protein coding gene | testis expressed metallothionein like |
| 19 | gene | 3.409915 | 3.414544 | negative | MGI_C57BL6J_95637 | Gal | NCBI_Gene:14419,ENSEMBL:ENSMUSG00000024907 | MGI:95637 | protein coding gene | galanin and GMAP prepropeptide |
| 19 | gene | 3.454928 | 3.575749 | negative | MGI_C57BL6J_1921807 | Ppp6r3 | NCBI_Gene:52036,ENSEMBL:ENSMUSG00000024908 | MGI:1921807 | protein coding gene | protein phosphatase 6, regulatory subunit 3 |
| 19 | gene | 3.486090 | 3.488014 | positive | MGI_C57BL6J_6098329 | Gm48696 | ENSEMBL:ENSMUSG00000114315 | MGI:6098329 | lncRNA gene | predicted gene, 48696 |
| 19 | gene | 3.490309 | 3.490413 | positive | MGI_C57BL6J_5453717 | Gm23940 | ENSEMBL:ENSMUSG00000075908 | MGI:5453717 | snRNA gene | predicted gene, 23940 |
| 19 | gene | 3.527104 | 3.528613 | positive | MGI_C57BL6J_6098308 | Gm48683 | ENSEMBL:ENSMUSG00000114302 | MGI:6098308 | lncRNA gene | predicted gene, 48683 |
| 19 | pseudogene | 3.567551 | 3.568448 | positive | MGI_C57BL6J_5011390 | Gm19205 | NCBI_Gene:100418426 | MGI:5011390 | pseudogene | predicted gene, 19205 |
| 19 | gene | 3.576263 | 3.576335 | positive | MGI_C57BL6J_4413682 | n-TAagc9 | NCBI_Gene:102467450 | MGI:4413682 | tRNA gene | nuclear encoded tRNA alanine 9 (anticodon AGC) |
| 19 | gene | 3.584825 | 3.686564 | negative | MGI_C57BL6J_1278315 | Lrp5 | NCBI_Gene:16973,ENSEMBL:ENSMUSG00000024913 | MGI:1278315 | protein coding gene | low density lipoprotein receptor-related protein 5 |
| 19 | gene | 3.686410 | 3.692435 | positive | MGI_C57BL6J_5595831 | Gm36672 | NCBI_Gene:102640658 | MGI:5595831 | protein coding gene | predicted gene, 36672 |
| 19 | gene | 3.689236 | 3.708226 | negative | MGI_C57BL6J_5595767 | Gm36608 | NCBI_Gene:102640578,ENSEMBL:ENSMUSG00000118249 | MGI:5595767 | lncRNA gene | predicted gene, 36608 |
| 19 | gene | 3.708293 | 3.717882 | positive | MGI_C57BL6J_1919306 | 1810055G02Rik | NCBI_Gene:72056,ENSEMBL:ENSMUSG00000035372 | MGI:1919306 | protein coding gene | RIKEN cDNA 1810055G02 gene |
| 19 | gene | 3.712777 | 3.744426 | negative | MGI_C57BL6J_5595946 | Gm36787 | NCBI_Gene:102640806,ENSEMBL:ENSMUSG00000117739 | MGI:5595946 | lncRNA gene | predicted gene, 36787 |
| 19 | gene | 3.719885 | 3.721369 | positive | MGI_C57BL6J_5596030 | Gm36871 | NCBI_Gene:102640924 | MGI:5596030 | lncRNA gene | predicted gene, 36871 |
| 19 | pseudogene | 3.730432 | 3.731047 | negative | MGI_C57BL6J_3780310 | Gm2141 | NCBI_Gene:100039289,ENSEMBL:ENSMUSG00000118231 | MGI:3780310 | pseudogene | predicted gene 2141 |
| 19 | pseudogene | 3.732094 | 3.732340 | negative | MGI_C57BL6J_6303269 | Gm50374 | ENSEMBL:ENSMUSG00000118096 | MGI:6303269 | pseudogene | predicted gene, 50374 |
| 19 | gene | 3.740225 | 3.742537 | negative | MGI_C57BL6J_6303270 | Gm50375 | ENSEMBL:ENSMUSG00000117773 | MGI:6303270 | lncRNA gene | predicted gene, 50375 |
| 19 | gene | 3.765976 | 3.818303 | positive | MGI_C57BL6J_2444557 | Kmt5b | NCBI_Gene:225888,ENSEMBL:ENSMUSG00000045098 | MGI:2444557 | protein coding gene | lysine methyltransferase 5B |
| 19 | gene | 3.798615 | 3.852419 | negative | MGI_C57BL6J_3801953 | Gm16066 | ENSEMBL:ENSMUSG00000082848 | MGI:3801953 | lncRNA gene | predicted gene 16066 |
| 19 | pseudogene | 3.821562 | 3.822389 | negative | MGI_C57BL6J_5011394 | Gm19209 | NCBI_Gene:100418430,ENSEMBL:ENSMUSG00000117779 | MGI:5011394 | pseudogene | predicted gene, 19209 |
| 19 | gene | 3.851585 | 3.894369 | positive | MGI_C57BL6J_107760 | Chka | NCBI_Gene:12660,ENSEMBL:ENSMUSG00000024843 | MGI:107760 | protein coding gene | choline kinase alpha |
| 19 | gene | 3.863568 | 3.867673 | negative | MGI_C57BL6J_6303282 | Gm50383 | ENSEMBL:ENSMUSG00000117912 | MGI:6303282 | lncRNA gene | predicted gene, 50383 |
| 19 | gene | 3.896050 | 3.907133 | negative | MGI_C57BL6J_1350931 | Tcirg1 | NCBI_Gene:27060,ENSEMBL:ENSMUSG00000001750 | MGI:1350931 | protein coding gene | T cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 protein A3 |
| 19 | gene | 3.908863 | 3.912774 | negative | MGI_C57BL6J_2385079 | Ndufs8 | NCBI_Gene:225887,ENSEMBL:ENSMUSG00000059734 | MGI:2385079 | protein coding gene | NADH:ubiquinone oxidoreductase core subunit S8 |
| 19 | gene | 3.913491 | 3.930860 | negative | MGI_C57BL6J_1914939 | Aldh3b1 | NCBI_Gene:67689,ENSEMBL:ENSMUSG00000024885 | MGI:1914939 | protein coding gene | aldehyde dehydrogenase 3 family, member B1 |
| 19 | gene | 3.919391 | 3.920826 | positive | MGI_C57BL6J_5624944 | Gm42059 | NCBI_Gene:105246837 | MGI:5624944 | lncRNA gene | predicted gene, 42059 |
| 19 | gene | 3.923864 | 3.924975 | positive | MGI_C57BL6J_5589005 | Gm29846 | NCBI_Gene:102631533 | MGI:5589005 | lncRNA gene | predicted gene, 29846 |
| 19 | pseudogene | 3.931795 | 3.932090 | negative | MGI_C57BL6J_6303191 | Gm50327 | ENSEMBL:ENSMUSG00000118129 | MGI:6303191 | pseudogene | predicted gene, 50327 |
| 19 | gene | 3.935186 | 3.949340 | positive | MGI_C57BL6J_1859307 | Unc93b1 | NCBI_Gene:54445,ENSEMBL:ENSMUSG00000036908 | MGI:1859307 | protein coding gene | unc-93 homolog B1, TLR signaling regulator |
| 19 | gene | 3.958645 | 3.972419 | positive | MGI_C57BL6J_1920708 | Aldh3b3 | NCBI_Gene:73458,ENSEMBL:ENSMUSG00000037263 | MGI:1920708 | protein coding gene | aldehyde dehydrogenase 3 family, member B3 |
| 19 | gene | 3.972328 | 3.981665 | positive | MGI_C57BL6J_2147613 | Aldh3b2 | NCBI_Gene:621603,ENSEMBL:ENSMUSG00000075296 | MGI:2147613 | protein coding gene | aldehyde dehydrogenase 3 family, member B2 |
| 19 | gene | 3.986570 | 3.990007 | positive | MGI_C57BL6J_1918920 | Acy3 | NCBI_Gene:71670,ENSEMBL:ENSMUSG00000024866 | MGI:1918920 | protein coding gene | aspartoacylase (aminoacylase) 3 |
| 19 | gene | 3.992752 | 3.999828 | positive | MGI_C57BL6J_1261436 | Tbx10 | NCBI_Gene:109575,ENSEMBL:ENSMUSG00000037477 | MGI:1261436 | protein coding gene | T-box 10 |
| 19 | gene | 3.999548 | 4.000542 | negative | MGI_C57BL6J_1921038 | 4833408A19Rik | ENSEMBL:ENSMUSG00000118132 | MGI:1921038 | lncRNA gene | RIKEN cDNA 4833408A19 gene |
| 19 | gene | 4.000580 | 4.002103 | positive | MGI_C57BL6J_1913637 | Nudt8 | NCBI_Gene:66387,ENSEMBL:ENSMUSG00000110949 | MGI:1913637 | protein coding gene | nudix (nucleoside diphosphate linked moiety X)-type motif 8 |
| 19 | gene | 4.000626 | 4.005816 | positive | MGI_C57BL6J_6155029 | Gm49405 | ENSEMBL:ENSMUSG00000024869 | MGI:6155029 | protein coding gene | predicted gene, 49405 |
| 19 | gene | 4.000758 | 4.001232 | negative | MGI_C57BL6J_3826559 | Gm16312 | ENSEMBL:ENSMUSG00000086107 | MGI:3826559 | lncRNA gene | predicted gene 16312 |
| 19 | gene | 4.003313 | 4.007005 | positive | MGI_C57BL6J_1926250 | Doc2g | NCBI_Gene:60425,ENSEMBL:ENSMUSG00000024871 | MGI:1926250 | protein coding gene | double C2, gamma |
| 19 | gene | 4.007384 | 4.012806 | negative | MGI_C57BL6J_107851 | Ndufv1 | NCBI_Gene:17995,ENSEMBL:ENSMUSG00000037916 | MGI:107851 | protein coding gene | NADH:ubiquinone oxidoreductase core subunit V1 |
| 19 | gene | 4.035407 | 4.037985 | negative | MGI_C57BL6J_95865 | Gstp1 | NCBI_Gene:14870,ENSEMBL:ENSMUSG00000060803 | MGI:95865 | protein coding gene | glutathione S-transferase, pi 1 |
| 19 | gene | 4.040285 | 4.046023 | negative | MGI_C57BL6J_95864 | Gstp2 | NCBI_Gene:14869,ENSEMBL:ENSMUSG00000038155 | MGI:95864 | protein coding gene | glutathione S-transferase, pi 2 |
| 19 | gene | 4.057470 | 4.059588 | negative | MGI_C57BL6J_2385078 | Gstp3 | NCBI_Gene:225884,ENSEMBL:ENSMUSG00000058216 | MGI:2385078 | protein coding gene | glutathione S-transferase pi 3 |
| 19 | gene | 4.067251 | 4.071445 | negative | MGI_C57BL6J_5624945 | Gm42060 | NCBI_Gene:105246838 | MGI:5624945 | lncRNA gene | predicted gene, 42060 |
| 19 | gene | 4.078297 | 4.078512 | negative | MGI_C57BL6J_5453704 | Gm23927 | ENSEMBL:ENSMUSG00000064400,ENSEMBL:ENSMUSG00000104921 | MGI:5453704 | snoRNA gene | predicted gene, 23927 |
| 19 | gene | 4.081469 | 4.087340 | positive | MGI_C57BL6J_1352749 | Cabp2 | NCBI_Gene:29866,ENSEMBL:ENSMUSG00000024857 | MGI:1352749 | protein coding gene | calcium binding protein 2 |
| 19 | gene | 4.096894 | 4.099019 | positive | MGI_C57BL6J_1098779 | Cdk2ap2 | NCBI_Gene:52004,ENSEMBL:ENSMUSG00000024856 | MGI:1098779 | protein coding gene | CDK2-associated protein 2 |
| 19 | gene | 4.099998 | 4.113965 | positive | MGI_C57BL6J_1197524 | Pitpnm1 | NCBI_Gene:18739,ENSEMBL:ENSMUSG00000024851 | MGI:1197524 | protein coding gene | phosphatidylinositol transfer protein, membrane-associated 1 |
| 19 | gene | 4.113756 | 4.127234 | negative | MGI_C57BL6J_109622 | Aip | NCBI_Gene:11632,ENSEMBL:ENSMUSG00000024847 | MGI:109622 | protein coding gene | aryl-hydrocarbon receptor-interacting protein |
| 19 | gene | 4.125934 | 4.132307 | positive | MGI_C57BL6J_1914240 | Tmem134 | NCBI_Gene:66990,ENSEMBL:ENSMUSG00000024845 | MGI:1914240 | protein coding gene | transmembrane protein 134 |
| 19 | gene | 4.135423 | 4.144033 | negative | MGI_C57BL6J_1920910 | Cabp4 | NCBI_Gene:73660,ENSEMBL:ENSMUSG00000024842 | MGI:1920910 | protein coding gene | calcium binding protein 4 |
| 19 | gene | 4.139787 | 4.145741 | positive | MGI_C57BL6J_2685519 | Gpr152 | NCBI_Gene:269053,ENSEMBL:ENSMUSG00000044724 | MGI:2685519 | protein coding gene | G protein-coupled receptor 152 |
| 19 | gene | 4.148612 | 4.154035 | positive | MGI_C57BL6J_1345963 | Coro1b | NCBI_Gene:23789,ENSEMBL:ENSMUSG00000024835 | MGI:1345963 | protein coding gene | coronin, actin binding protein 1B |
| 19 | gene | 4.153604 | 4.156751 | positive | MGI_C57BL6J_97811 | Ptprcap | NCBI_Gene:19265,ENSEMBL:ENSMUSG00000045826 | MGI:97811 | protein coding gene | protein tyrosine phosphatase, receptor type, C polypeptide-associated protein |
| 19 | gene | 4.153991 | 4.163387 | negative | MGI_C57BL6J_1927343 | Rps6kb2 | NCBI_Gene:58988,ENSEMBL:ENSMUSG00000024830 | MGI:1927343 | protein coding gene | ribosomal protein S6 kinase, polypeptide 2 |
| 19 | gene | 4.164324 | 4.175677 | negative | MGI_C57BL6J_2147595 | Carns1 | NCBI_Gene:107239,ENSEMBL:ENSMUSG00000075289 | MGI:2147595 | protein coding gene | carnosine synthase 1 |
| 19 | gene | 4.168742 | 4.191047 | negative | MGI_C57BL6J_6303033 | Gm50230 | ENSEMBL:ENSMUSG00000117905 | MGI:6303033 | lncRNA gene | predicted gene, 50230 |
| 19 | gene | 4.169566 | 4.174287 | positive | MGI_C57BL6J_4937186 | Gm17552 | NCBI_Gene:102631780,ENSEMBL:ENSMUSG00000097233 | MGI:4937186 | lncRNA gene | predicted gene, 17552 |
| 19 | gene | 4.183411 | 4.191284 | negative | MGI_C57BL6J_1922072 | Tbc1d10c | NCBI_Gene:108995,ENSEMBL:ENSMUSG00000040247 | MGI:1922072 | protein coding gene | TBC1 domain family, member 10c |
| 19 | gene | 4.192065 | 4.195420 | positive | MGI_C57BL6J_103016 | Ppp1ca | NCBI_Gene:19045,ENSEMBL:ENSMUSG00000040385 | MGI:103016 | protein coding gene | protein phosphatase 1 catalytic subunit alpha |
| 19 | gene | 4.192109 | 4.192782 | negative | MGI_C57BL6J_6303067 | Gm50251 | ENSEMBL:ENSMUSG00000117839 | MGI:6303067 | lncRNA gene | predicted gene, 50251 |
| 19 | gene | 4.195196 | 4.201662 | negative | MGI_C57BL6J_1328356 | Rad9a | NCBI_Gene:19367,ENSEMBL:ENSMUSG00000024824 | MGI:1328356 | protein coding gene | RAD9 checkpoint clamp component A |
| 19 | gene | 4.201706 | 4.203240 | positive | MGI_C57BL6J_5589241 | Gm30082 | NCBI_Gene:102631855,ENSEMBL:ENSMUSG00000118245 | MGI:5589241 | lncRNA gene | predicted gene, 30082 |
| 19 | gene | 4.214238 | 4.223505 | positive | MGI_C57BL6J_1930088 | Clcf1 | NCBI_Gene:56708,ENSEMBL:ENSMUSG00000040663 | MGI:1930088 | protein coding gene | cardiotrophin-like cytokine factor 1 |
| 19 | gene | 4.214238 | 4.233634 | positive | MGI_C57BL6J_5825565 | Gm45928 | NCBI_Gene:105948585,ENSEMBL:ENSMUSG00000117873 | MGI:5825565 | lncRNA gene | predicted gene, 45928 |
| 19 | gene | 4.215402 | 4.215488 | negative | MGI_C57BL6J_5455892 | Gm26115 | ENSEMBL:ENSMUSG00000093346 | MGI:5455892 | miRNA gene | predicted gene, 26115 |
| 19 | gene | 4.231893 | 4.233634 | positive | MGI_C57BL6J_1916995 | Pold4 | NCBI_Gene:69745,ENSEMBL:ENSMUSG00000024854 | MGI:1916995 | protein coding gene | polymerase (DNA-directed), delta 4 |
| 19 | gene | 4.254031 | 4.257651 | negative | MGI_C57BL6J_1925555 | 1500032F14Rik | ENSEMBL:ENSMUSG00000117718 | MGI:1925555 | lncRNA gene | RIKEN cDNA 1500032F14 gene |
| 19 | gene | 4.261668 | 4.269180 | negative | MGI_C57BL6J_2683546 | Ssh3 | NCBI_Gene:245857,ENSEMBL:ENSMUSG00000034616 | MGI:2683546 | protein coding gene | slingshot protein phosphatase 3 |
| 19 | gene | 4.263818 | 4.263878 | negative | MGI_C57BL6J_5531101 | Mir6985 | miRBase:MI0022833,NCBI_Gene:102465595,ENSEMBL:ENSMUSG00000099290 | MGI:5531101 | miRNA gene | microRNA 6985 |
| 19 | gene | 4.270180 | 4.283137 | negative | MGI_C57BL6J_1915673 | Ankrd13d | NCBI_Gene:68423,ENSEMBL:ENSMUSG00000005986 | MGI:1915673 | protein coding gene | ankyrin repeat domain 13 family, member D |
| 19 | gene | 4.273987 | 4.275643 | positive | MGI_C57BL6J_6302910 | Gm50152 | ENSEMBL:ENSMUSG00000118344 | MGI:6302910 | lncRNA gene | predicted gene, 50152 |
| 19 | gene | 4.285999 | 4.306222 | negative | MGI_C57BL6J_87940 | Grk2 | NCBI_Gene:110355,ENSEMBL:ENSMUSG00000024858 | MGI:87940 | protein coding gene | G protein-coupled receptor kinase 2 |
| 19 | gene | 4.314419 | 4.398596 | negative | MGI_C57BL6J_1354736 | Kdm2a | NCBI_Gene:225876,ENSEMBL:ENSMUSG00000054611 | MGI:1354736 | protein coding gene | lysine (K)-specific demethylase 2A |
| 19 | gene | 4.397714 | 4.399821 | positive | MGI_C57BL6J_3588259 | 9430078G10Rik | NA | NA | unclassified gene | RIKEN cDNA 9430078G10 gene |
| 19 | gene | 4.425457 | 4.439432 | negative | MGI_C57BL6J_108446 | Rhod | NCBI_Gene:11854,ENSEMBL:ENSMUSG00000041845 | MGI:108446 | protein coding gene | ras homolog family member D |
| 19 | gene | 4.439003 | 4.448973 | positive | MGI_C57BL6J_2441838 | A930001C03Rik | NCBI_Gene:319314,ENSEMBL:ENSMUSG00000087132 | MGI:2441838 | lncRNA gene | RIKEN cDNA A930001C03 gene |
| 19 | gene | 4.445908 | 4.477447 | negative | MGI_C57BL6J_2159601 | Syt12 | NCBI_Gene:171180,ENSEMBL:ENSMUSG00000049303 | MGI:2159601 | protein coding gene | synaptotagmin XII |
| 19 | gene | 4.496788 | 4.498583 | negative | MGI_C57BL6J_1917111 | 2010003K11Rik | NCBI_Gene:69861,ENSEMBL:ENSMUSG00000042041 | MGI:1917111 | protein coding gene | RIKEN cDNA 2010003K11 gene |
| 19 | gene | 4.510472 | 4.621752 | positive | MGI_C57BL6J_97520 | Pcx | NCBI_Gene:18563,ENSEMBL:ENSMUSG00000024892 | MGI:97520 | protein coding gene | pyruvate carboxylase |
| 19 | gene | 4.611784 | 4.615667 | negative | MGI_C57BL6J_2385612 | Lrfn4 | NCBI_Gene:225875,ENSEMBL:ENSMUSG00000045045 | MGI:2385612 | protein coding gene | leucine rich repeat and fibronectin type III domain containing 4 |
| 19 | gene | 4.622551 | 4.625641 | negative | MGI_C57BL6J_1336895 | Rce1 | NCBI_Gene:19671,ENSEMBL:ENSMUSG00000024889 | MGI:1336895 | protein coding gene | Ras converting CAAX endopeptidase 1 |
| 19 | gene | 4.623898 | 4.623955 | negative | MGI_C57BL6J_5531091 | Mir6986 | miRBase:MI0022834,NCBI_Gene:102466780,ENSEMBL:ENSMUSG00000098346 | MGI:5531091 | miRNA gene | microRNA 6986 |
| 19 | gene | 4.625395 | 4.629857 | positive | MGI_C57BL6J_1921792 | 9030625N01Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 9030625N01 gene |
| 19 | gene | 4.625734 | 4.698776 | negative | MGI_C57BL6J_2685806 | Gm960 | NCBI_Gene:381196,ENSEMBL:ENSMUSG00000071691 | MGI:2685806 | protein coding gene | predicted gene 960 |
| 19 | pseudogene | 4.680115 | 4.680616 | positive | MGI_C57BL6J_6303266 | Gm50372 | ENSEMBL:ENSMUSG00000117746 | MGI:6303266 | pseudogene | predicted gene, 50372 |
| 19 | gene | 4.711167 | 4.752360 | positive | MGI_C57BL6J_1313261 | Sptbn2 | NCBI_Gene:20743,ENSEMBL:ENSMUSG00000067889 | MGI:1313261 | protein coding gene | spectrin beta, non-erythrocytic 2 |
| 19 | gene | 4.711651 | 4.720886 | negative | MGI_C57BL6J_5624946 | Gm42061 | NCBI_Gene:105246839 | MGI:5624946 | protein coding gene | predicted gene, 42061 |
| 19 | gene | 4.742913 | 4.811634 | negative | MGI_C57BL6J_5439461 | Gm21992 | NCBI_Gene:102902673,ENSEMBL:ENSMUSG00000096370 | MGI:5439461 | protein coding gene | predicted gene 21992 |
| 19 | gene | 4.752883 | 4.767065 | positive | MGI_C57BL6J_1913954 | Rbm4b | NCBI_Gene:66704,ENSEMBL:ENSMUSG00000033760 | MGI:1913954 | protein coding gene | RNA binding motif protein 4B |
| 19 | gene | 4.762103 | 4.765171 | negative | MGI_C57BL6J_5610434 | Gm37206 | ENSEMBL:ENSMUSG00000103651 | MGI:5610434 | unclassified gene | predicted gene, 37206 |
| 19 | pseudogene | 4.778072 | 4.778697 | positive | MGI_C57BL6J_5011288 | Gm19103 | NCBI_Gene:100418257 | MGI:5011288 | pseudogene | predicted gene, 19103 |
| 19 | gene | 4.784293 | 4.794009 | negative | MGI_C57BL6J_1100865 | Rbm4 | NCBI_Gene:19653,ENSEMBL:ENSMUSG00000094936 | MGI:1100865 | protein coding gene | RNA binding motif protein 4 |
| 19 | gene | 4.794720 | 4.795956 | negative | MGI_C57BL6J_1918634 | 5430440L12Rik | NA | NA | unclassified gene | RIKEN cDNA 5430440L12 gene |
| 19 | gene | 4.800566 | 4.811634 | negative | MGI_C57BL6J_1929092 | Rbm14 | NCBI_Gene:56275,ENSEMBL:ENSMUSG00000006456 | MGI:1929092 | protein coding gene | RNA binding motif protein 14 |
| 19 | gene | 4.801512 | 4.805784 | positive | MGI_C57BL6J_5434008 | Gm21844 | ENSEMBL:ENSMUSG00000095913 | MGI:5434008 | lncRNA gene | predicted gene, 21844 |
| 19 | gene | 4.825366 | 4.839332 | negative | MGI_C57BL6J_1333783 | Ccs | NCBI_Gene:12460,ENSEMBL:ENSMUSG00000034108 | MGI:1333783 | protein coding gene | copper chaperone for superoxide dismutase |
| 19 | gene | 4.839366 | 4.842528 | positive | MGI_C57BL6J_3026882 | Ccdc87 | NCBI_Gene:399599,ENSEMBL:ENSMUSG00000067872 | MGI:3026882 | protein coding gene | coiled-coil domain containing 87 |
| 19 | gene | 4.855129 | 4.860918 | positive | MGI_C57BL6J_1861434 | Ctsf | NCBI_Gene:56464,ENSEMBL:ENSMUSG00000083282 | MGI:1861434 | protein coding gene | cathepsin F |
| 19 | gene | 4.861216 | 4.877909 | negative | MGI_C57BL6J_99678 | Actn3 | NCBI_Gene:11474,ENSEMBL:ENSMUSG00000006457 | MGI:99678 | protein coding gene | actinin alpha 3 |
| 19 | gene | 4.878668 | 4.885397 | positive | MGI_C57BL6J_1917855 | Zdhhc24 | NCBI_Gene:70605,ENSEMBL:ENSMUSG00000006463 | MGI:1917855 | protein coding gene | zinc finger, DHHC domain containing 24 |
| 19 | gene | 4.886878 | 4.906628 | negative | MGI_C57BL6J_1277215 | Bbs1 | NCBI_Gene:52028,ENSEMBL:ENSMUSG00000006464 | MGI:1277215 | protein coding gene | Bardet-Biedl syndrome 1 (human) |
| 19 | gene | 4.904673 | 4.904782 | positive | MGI_C57BL6J_5455111 | Gm25334 | ENSEMBL:ENSMUSG00000088661 | MGI:5455111 | snRNA gene | predicted gene, 25334 |
| 19 | gene | 4.907229 | 4.928287 | negative | MGI_C57BL6J_1922471 | Dpp3 | NCBI_Gene:75221,ENSEMBL:ENSMUSG00000063904 | MGI:1922471 | protein coding gene | dipeptidylpeptidase 3 |
| 19 | gene | 4.929717 | 4.943127 | negative | MGI_C57BL6J_1924963 | Peli3 | NCBI_Gene:240518,ENSEMBL:ENSMUSG00000024901 | MGI:1924963 | protein coding gene | pellino 3 |
| 19 | gene | 4.962147 | 4.966999 | positive | MGI_C57BL6J_2137215 | Mrpl11 | NCBI_Gene:66419,ENSEMBL:ENSMUSG00000024902 | MGI:2137215 | protein coding gene | mitochondrial ribosomal protein L11 |
| 19 | gene | 4.984353 | 5.001929 | negative | MGI_C57BL6J_2664186 | Npas4 | NCBI_Gene:225872,ENSEMBL:ENSMUSG00000045903 | MGI:2664186 | protein coding gene | neuronal PAS domain protein 4 |
| 19 | pseudogene | 5.009385 | 5.009799 | positive | MGI_C57BL6J_6303281 | Gm50382 | ENSEMBL:ENSMUSG00000117723 | MGI:6303281 | pseudogene | predicted gene, 50382 |
| 19 | gene | 5.017356 | 5.021765 | negative | MGI_C57BL6J_5624948 | Gm42063 | NCBI_Gene:105246841 | MGI:5624948 | lncRNA gene | predicted gene, 42063 |
| 19 | gene | 5.023860 | 5.031972 | positive | MGI_C57BL6J_1345278 | Slc29a2 | NCBI_Gene:13340,ENSEMBL:ENSMUSG00000024891 | MGI:1345278 | protein coding gene | solute carrier family 29 (nucleoside transporters), member 2 |
| 19 | gene | 5.037817 | 5.038805 | negative | MGI_C57BL6J_5589740 | Gm30581 | NCBI_Gene:102632530 | MGI:5589740 | lncRNA gene | predicted gene, 30581 |
| 19 | gene | 5.038304 | 5.038385 | negative | MGI_C57BL6J_4414040 | n-TSgct6 | NCBI_Gene:102467380 | MGI:4414040 | tRNA gene | nuclear encoded tRNA serine 6 (anticodon GCT) |
| 19 | gene | 5.038826 | 5.041134 | positive | MGI_C57BL6J_1919680 | B4gat1 | NCBI_Gene:108902,ENSEMBL:ENSMUSG00000047379 | MGI:1919680 | protein coding gene | beta-1,4-glucuronyltransferase 1 |
| 19 | gene | 5.038891 | 5.049912 | positive | MGI_C57BL6J_6303291 | Gm50388 | ENSEMBL:ENSMUSG00000117789 | MGI:6303291 | protein coding gene | predicted gene, 50388 |
| 19 | gene | 5.041232 | 5.049917 | positive | MGI_C57BL6J_2388804 | Brms1 | NCBI_Gene:107392,ENSEMBL:ENSMUSG00000080268 | MGI:2388804 | protein coding gene | breast cancer metastasis-suppressor 1 |
| 19 | gene | 5.050019 | 5.057072 | positive | MGI_C57BL6J_2385695 | Rin1 | NCBI_Gene:225870,ENSEMBL:ENSMUSG00000024883 | MGI:2385695 | protein coding gene | Ras and Rab interactor 1 |
| 19 | gene | 5.068078 | 5.070682 | positive | MGI_C57BL6J_1917695 | Cd248 | NCBI_Gene:70445,ENSEMBL:ENSMUSG00000056481 | MGI:1917695 | protein coding gene | CD248 antigen, endosialin |
| 19 | gene | 5.070830 | 5.072120 | negative | MGI_C57BL6J_5589786 | Gm30627 | NCBI_Gene:102632593 | MGI:5589786 | lncRNA gene | predicted gene, 30627 |
| 19 | gene | 5.070859 | 5.085531 | negative | MGI_C57BL6J_2147713 | Tmem151a | NCBI_Gene:381199,ENSEMBL:ENSMUSG00000061451 | MGI:2147713 | protein coding gene | transmembrane protein 151A |
| 19 | gene | 5.072130 | 5.075259 | positive | MGI_C57BL6J_5826295 | Gm46658 | NCBI_Gene:108168419,ENSEMBL:ENSMUSG00000117893 | MGI:5826295 | lncRNA gene | predicted gene, 46658 |
| 19 | gene | 5.088538 | 5.092881 | positive | MGI_C57BL6J_1915340 | Yif1a | NCBI_Gene:68090,ENSEMBL:ENSMUSG00000024875 | MGI:1915340 | protein coding gene | Yip1 interacting factor homolog A (S. cerevisiae) |
| 19 | gene | 5.092868 | 5.098521 | negative | MGI_C57BL6J_1277225 | Cnih2 | NCBI_Gene:12794,ENSEMBL:ENSMUSG00000024873 | MGI:1277225 | protein coding gene | cornichon family AMPA receptor auxiliary protein 2 |
| 19 | gene | 5.099205 | 5.107072 | negative | MGI_C57BL6J_1923558 | Rab1b | NCBI_Gene:76308,ENSEMBL:ENSMUSG00000024870 | MGI:1923558 | protein coding gene | RAB1B, member RAS oncogene family |
| 19 | gene | 5.107746 | 5.118573 | negative | MGI_C57BL6J_107953 | Klc2 | NCBI_Gene:16594,ENSEMBL:ENSMUSG00000024862 | MGI:107953 | protein coding gene | kinesin light chain 2 |
| 19 | gene | 5.118038 | 5.119329 | positive | MGI_C57BL6J_3704489 | Gm10817 | ENSEMBL:ENSMUSG00000117710 | MGI:3704489 | lncRNA gene | predicted gene 10817 |
| 19 | gene | 5.118844 | 5.126127 | positive | MGI_C57BL6J_5589999 | Gm30840 | NCBI_Gene:102632878 | MGI:5589999 | lncRNA gene | predicted gene, 30840 |
| 19 | pseudogene | 5.131740 | 5.132972 | negative | MGI_C57BL6J_3779478 | Gm5242 | NCBI_Gene:383424,ENSEMBL:ENSMUSG00000117729 | MGI:3779478 | pseudogene | predicted gene 5242 |
| 19 | gene | 5.133158 | 5.273119 | negative | MGI_C57BL6J_1277113 | Pacs1 | NCBI_Gene:107975,ENSEMBL:ENSMUSG00000024855 | MGI:1277113 | protein coding gene | phosphofurin acidic cluster sorting protein 1 |
| 19 | gene | 5.201198 | 5.201303 | negative | MGI_C57BL6J_5530850 | Gm27468 | ENSEMBL:ENSMUSG00000098803 | MGI:5530850 | miRNA gene | predicted gene, 27468 |
| 19 | pseudogene | 5.258750 | 5.258962 | positive | MGI_C57BL6J_6302964 | Gm50189 | ENSEMBL:ENSMUSG00000118104 | MGI:6302964 | pseudogene | predicted gene, 50189 |
| 19 | gene | 5.273921 | 5.295457 | negative | MGI_C57BL6J_2441856 | Sf3b2 | NCBI_Gene:319322,ENSEMBL:ENSMUSG00000024853 | MGI:2441856 | protein coding gene | splicing factor 3b, subunit 2 |
| 19 | gene | 5.295196 | 5.296028 | positive | MGI_C57BL6J_6302985 | Gm50203 | ENSEMBL:ENSMUSG00000118243 | MGI:6302985 | lncRNA gene | predicted gene, 50203 |
| 19 | gene | 5.298331 | 5.308739 | positive | MGI_C57BL6J_3617843 | Gal3st3 | NCBI_Gene:545276,ENSEMBL:ENSMUSG00000047658 | MGI:3617843 | protein coding gene | galactose-3-O-sulfotransferase 3 |
| 19 | gene | 5.305873 | 5.307416 | negative | MGI_C57BL6J_5624949 | Gm42064 | NCBI_Gene:105246843,ENSEMBL:ENSMUSG00000118263 | MGI:5624949 | lncRNA gene | predicted gene, 42064 |
| 19 | pseudogene | 5.321606 | 5.321798 | positive | MGI_C57BL6J_6302994 | Gm50208 | ENSEMBL:ENSMUSG00000118009 | MGI:6302994 | pseudogene | predicted gene, 50208 |
| 19 | pseudogene | 5.324014 | 5.325823 | positive | MGI_C57BL6J_3645420 | Gm7074 | NCBI_Gene:631868,ENSEMBL:ENSMUSG00000117807 | MGI:3645420 | pseudogene | predicted gene 7074 |
| 19 | gene | 5.334032 | 5.335433 | negative | MGI_C57BL6J_5792253 | Catsper1au | NA | NA | antisense lncRNA gene | Catsper1 antisense upstream transcript |
| 19 | gene | 5.335602 | 5.344281 | positive | MGI_C57BL6J_2179947 | Catsper1 | NCBI_Gene:225865,ENSEMBL:ENSMUSG00000038498 | MGI:2179947 | protein coding gene | cation channel, sperm associated 1 |
| 19 | gene | 5.344705 | 5.349574 | negative | MGI_C57BL6J_1920970 | Cst6 | NCBI_Gene:73720,ENSEMBL:ENSMUSG00000024846 | MGI:1920970 | protein coding gene | cystatin E/M |
| 19 | gene | 5.364633 | 5.367168 | negative | MGI_C57BL6J_1346330 | Banf1 | NCBI_Gene:23825,ENSEMBL:ENSMUSG00000024844 | MGI:1346330 | protein coding gene | barrier to autointegration factor 1 |
| 19 | gene | 5.366741 | 5.371526 | positive | MGI_C57BL6J_1917110 | Eif1ad | NCBI_Gene:69860,ENSEMBL:ENSMUSG00000024841 | MGI:1917110 | protein coding gene | eukaryotic translation initiation factor 1A domain containing |
| 19 | gene | 5.377523 | 5.389304 | negative | MGI_C57BL6J_1309453 | Sart1 | NCBI_Gene:20227,ENSEMBL:ENSMUSG00000039148 | MGI:1309453 | protein coding gene | squamous cell carcinoma antigen recognized by T cells 1 |
| 19 | gene | 5.388336 | 5.390069 | positive | MGI_C57BL6J_3041222 | D330050I16Rik | NCBI_Gene:414115,ENSEMBL:ENSMUSG00000117959 | MGI:3041222 | lncRNA gene | RIKEN cDNA D330050I16 gene |
| 19 | gene | 5.390047 | 5.402534 | negative | MGI_C57BL6J_1925556 | Tsga10ip | NCBI_Gene:78306,ENSEMBL:ENSMUSG00000039330 | MGI:1925556 | protein coding gene | testis specific 10 interacting protein |
| 19 | gene | 5.406740 | 5.422847 | positive | MGI_C57BL6J_1922181 | 4930481A15Rik | NCBI_Gene:74931,ENSEMBL:ENSMUSG00000086938 | MGI:1922181 | lncRNA gene | RIKEN cDNA 4930481A15 gene |
| 19 | pseudogene | 5.415382 | 5.415811 | negative | MGI_C57BL6J_3649011 | Gm6293 | NCBI_Gene:102633050,ENSEMBL:ENSMUSG00000051133 | MGI:3649011 | pseudogene | predicted pseudogene 6293 |
| 19 | gene | 5.422804 | 5.424979 | negative | MGI_C57BL6J_1913806 | Drap1 | NCBI_Gene:66556,ENSEMBL:ENSMUSG00000024914 | MGI:1913806 | protein coding gene | Dr1 associated protein 1 (negative cofactor 2 alpha) |
| 19 | gene | 5.425144 | 5.427317 | positive | MGI_C57BL6J_2147598 | AI837181 | NCBI_Gene:107242,ENSEMBL:ENSMUSG00000047423 | MGI:2147598 | protein coding gene | expressed sequence AI837181 |
| 19 | gene | 5.435900 | 5.435965 | positive | MGI_C57BL6J_6302848 | Gm50111 | ENSEMBL:ENSMUSG00000088909 | MGI:6302848 | snoRNA gene | predicted gene, 50111 |
| 19 | pseudogene | 5.435929 | 5.436800 | positive | MGI_C57BL6J_5455209 | Gm25432 | NCBI_Gene:102635008,ENSEMBL:ENSMUSG00000117771 | MGI:5455209 | pseudogene | predicted gene, 25432 |
| 19 | pseudogene | 5.439246 | 5.439521 | negative | MGI_C57BL6J_5590229 | Gm31070 | NCBI_Gene:102633178,ENSEMBL:ENSMUSG00000117845 | MGI:5590229 | pseudogene | predicted gene, 31070 |
| 19 | gene | 5.447547 | 5.455946 | positive | MGI_C57BL6J_107179 | Fosl1 | NCBI_Gene:14283,ENSEMBL:ENSMUSG00000024912 | MGI:107179 | protein coding gene | fos-like antigen 1 |
| 19 | gene | 5.453163 | 5.457894 | negative | MGI_C57BL6J_2147607 | Ccdc85b | NCBI_Gene:240514,ENSEMBL:ENSMUSG00000095098 | MGI:2147607 | protein coding gene | coiled-coil domain containing 85B |
| 19 | gene | 5.460600 | 5.465052 | positive | MGI_C57BL6J_1926233 | Fibp | NCBI_Gene:58249,ENSEMBL:ENSMUSG00000024911 | MGI:1926233 | protein coding gene | fibroblast growth factor (acidic) intracellular binding protein |
| 19 | gene | 5.465043 | 5.470389 | negative | MGI_C57BL6J_1338045 | Ctsw | NCBI_Gene:13041,ENSEMBL:ENSMUSG00000024910 | MGI:1338045 | protein coding gene | cathepsin W |
| 19 | gene | 5.473954 | 5.482517 | positive | MGI_C57BL6J_1891209 | Efemp2 | NCBI_Gene:58859,ENSEMBL:ENSMUSG00000024909 | MGI:1891209 | protein coding gene | epidermal growth factor-containing fibulin-like extracellular matrix protein 2 |
| 19 | gene | 5.482345 | 5.488410 | negative | MGI_C57BL6J_1918961 | Mus81 | NCBI_Gene:71711,ENSEMBL:ENSMUSG00000024906 | MGI:1918961 | protein coding gene | MUS81 structure-specific endonuclease subunit |
| 19 | gene | 5.488744 | 5.489198 | negative | MGI_C57BL6J_1920664 | 1700061A03Rik | ENSEMBL:ENSMUSG00000117966 | MGI:1920664 | unclassified gene | RIKEN cDNA 1700061A03 gene |
| 19 | gene | 5.490455 | 5.495201 | positive | MGI_C57BL6J_101757 | Cfl1 | NCBI_Gene:12631,ENSEMBL:ENSMUSG00000056201 | MGI:101757 | protein coding gene | cofilin 1, non-muscle |
| 19 | gene | 5.492235 | 5.510696 | negative | MGI_C57BL6J_2444704 | Snx32 | NCBI_Gene:225861,ENSEMBL:ENSMUSG00000056185 | MGI:2444704 | protein coding gene | sorting nexin 32 |
| 19 | gene | 5.502767 | 5.503787 | negative | MGI_C57BL6J_1922805 | 1700020D05Rik | NCBI_Gene:75555,ENSEMBL:ENSMUSG00000100937 | MGI:1922805 | protein coding gene | RIKEN cDNA 1700020D05 gene |
| 19 | pseudogene | 5.510783 | 5.550374 | positive | MGI_C57BL6J_5590325 | Gm31166 | NCBI_Gene:102633307,ENSEMBL:ENSMUSG00000109695 | MGI:5590325 | pseudogene | predicted gene, 31166 |
| 19 | gene | 5.549136 | 5.560646 | negative | MGI_C57BL6J_1330290 | Ovol1 | NCBI_Gene:18426,ENSEMBL:ENSMUSG00000024922 | MGI:1330290 | protein coding gene | ovo like zinc finger 1 |
| 19 | gene | 5.560250 | 5.571262 | positive | MGI_C57BL6J_2685808 | Ap5b1 | NCBI_Gene:381201,ENSEMBL:ENSMUSG00000049562 | MGI:2685808 | protein coding gene | adaptor-related protein complex 5, beta 1 subunit |
| 19 | gene | 5.560260 | 5.561677 | positive | MGI_C57BL6J_1923763 | 1810058N15Rik | ENSEMBL:ENSMUSG00000117861 | MGI:1923763 | lncRNA gene | RIKEN cDNA 1810058N15 gene |
| 19 | gene | 5.601873 | 5.607019 | positive | MGI_C57BL6J_1915459 | Rnaseh2c | NCBI_Gene:68209,ENSEMBL:ENSMUSG00000024925 | MGI:1915459 | protein coding gene | ribonuclease H2, subunit C |
| 19 | gene | 5.603014 | 5.610237 | negative | MGI_C57BL6J_1932051 | Kat5 | NCBI_Gene:81601,ENSEMBL:ENSMUSG00000024926 | MGI:1932051 | protein coding gene | K(lysine) acetyltransferase 5 |
| 19 | gene | 5.637440 | 5.648131 | positive | MGI_C57BL6J_103290 | Rela | NCBI_Gene:19697,ENSEMBL:ENSMUSG00000024927 | MGI:103290 | protein coding gene | v-rel reticuloendotheliosis viral oncogene homolog A (avian) |
| 19 | gene | 5.651185 | 5.663839 | negative | MGI_C57BL6J_107576 | Sipa1 | NCBI_Gene:20469,ENSEMBL:ENSMUSG00000056917 | MGI:107576 | protein coding gene | signal-induced proliferation associated gene 1 |
| 19 | gene | 5.664635 | 5.688908 | negative | MGI_C57BL6J_1861733 | Pcnx3 | NCBI_Gene:104401,ENSEMBL:ENSMUSG00000054874 | MGI:1861733 | protein coding gene | pecanex homolog 3 |
| 19 | gene | 5.679004 | 5.679076 | negative | MGI_C57BL6J_5531273 | Mir6987 | miRBase:MI0022835,NCBI_Gene:102465596,ENSEMBL:ENSMUSG00000098568 | MGI:5531273 | miRNA gene | microRNA 6987 |
| 19 | gene | 5.688740 | 5.702865 | positive | MGI_C57BL6J_1346880 | Map3k11 | NCBI_Gene:26403,ENSEMBL:ENSMUSG00000004054 | MGI:1346880 | protein coding gene | mitogen-activated protein kinase kinase kinase 11 |
| 19 | gene | 5.689236 | 5.690010 | negative | MGI_C57BL6J_6303171 | Gm50315 | ENSEMBL:ENSMUSG00000118077 | MGI:6303171 | lncRNA gene | predicted gene, 50315 |
| 19 | gene | 5.704367 | 5.707406 | positive | MGI_C57BL6J_1341841 | Kcnk7 | NCBI_Gene:16530,ENSEMBL:ENSMUSG00000024936 | MGI:1341841 | protein coding gene | potassium channel, subfamily K, member 7 |
| 19 | gene | 5.707369 | 5.726461 | negative | MGI_C57BL6J_3612340 | Ehbp1l1 | NCBI_Gene:114601,ENSEMBL:ENSMUSG00000024937 | MGI:3612340 | protein coding gene | EH domain binding protein 1-like 1 |
| 19 | gene | 5.727206 | 5.731627 | positive | MGI_C57BL6J_4414958 | Gm16538 | ENSEMBL:ENSMUSG00000089766 | MGI:4414958 | lncRNA gene | predicted gene 16538 |
| 19 | gene | 5.728087 | 5.729666 | negative | MGI_C57BL6J_106595 | Fam89b | NCBI_Gene:17826,ENSEMBL:ENSMUSG00000024939 | MGI:106595 | protein coding gene | family with sequence similarity 89, member B |
| 19 | gene | 5.728088 | 5.731713 | negative | MGI_C57BL6J_6303289 | Gm50387 | ENSEMBL:ENSMUSG00000118125 | MGI:6303289 | protein coding gene | predicted gene, 50387 |
| 19 | gene | 5.730303 | 5.731732 | negative | MGI_C57BL6J_1913482 | Znrd2 | NCBI_Gene:56390,ENSEMBL:ENSMUSG00000079478 | MGI:1913482 | protein coding gene | zinc ribbon domain containing 2 |
| 19 | gene | 5.738405 | 5.740762 | negative | MGI_C57BL6J_3642330 | Gm10815 | NA | NA | unclassified gene | predicted gene 10815 |
| 19 | gene | 5.740904 | 5.758532 | positive | MGI_C57BL6J_1101355 | Ltbp3 | NCBI_Gene:16998,ENSEMBL:ENSMUSG00000024940 | MGI:1101355 | protein coding gene | latent transforming growth factor beta binding protein 3 |
| 19 | gene | 5.758351 | 5.771419 | negative | MGI_C57BL6J_1931787 | Scyl1 | NCBI_Gene:78891,ENSEMBL:ENSMUSG00000024941 | MGI:1931787 | protein coding gene | SCY1-like 1 (S. cerevisiae) |
| 19 | gene | 5.795690 | 5.802672 | negative | MGI_C57BL6J_1919539 | Malat1 | NCBI_Gene:72289,ENSEMBL:ENSMUSG00000092341 | MGI:1919539 | lncRNA gene | metastasis associated lung adenocarcinoma transcript 1 (non-coding RNA) |
| 19 | gene | 5.799000 | 5.799085 | negative | MGI_C57BL6J_5531076 | Gm27694 | ENSEMBL:ENSMUSG00000098462 | MGI:5531076 | unclassified non-coding RNA gene | predicted gene, 27694 |
| 19 | gene | 5.801941 | 5.802393 | positive | MGI_C57BL6J_5610604 | Gm37376 | ENSEMBL:ENSMUSG00000102349 | MGI:5610604 | unclassified gene | predicted gene, 37376 |
| 19 | gene | 5.824652 | 5.824707 | negative | MGI_C57BL6J_5531084 | Gm27702 | ENSEMBL:ENSMUSG00000098865 | MGI:5531084 | unclassified non-coding RNA gene | predicted gene, 27702 |
| 19 | gene | 5.824708 | 5.845480 | negative | MGI_C57BL6J_1914211 | Neat1 | NCBI_Gene:66961,ENSEMBL:ENSMUSG00000092274 | MGI:1914211 | lncRNA gene | nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| 19 | gene | 5.844286 | 5.844437 | negative | MGI_C57BL6J_5530915 | Gm27533 | ENSEMBL:ENSMUSG00000098543 | MGI:5530915 | unclassified non-coding RNA gene | predicted gene, 27533 |
| 19 | gene | 5.844638 | 5.844747 | negative | MGI_C57BL6J_5530887 | Gm27505 | ENSEMBL:ENSMUSG00000098974 | MGI:5530887 | unclassified non-coding RNA gene | predicted gene, 27505 |
| 19 | gene | 5.844916 | 5.845037 | negative | MGI_C57BL6J_5531388 | Gm28006 | ENSEMBL:ENSMUSG00000098986 | MGI:5531388 | unclassified non-coding RNA gene | predicted gene, 28006 |
| 19 | gene | 5.845623 | 5.851388 | positive | MGI_C57BL6J_3704490 | Frmd8os | NCBI_Gene:69595,ENSEMBL:ENSMUSG00000043488 | MGI:3704490 | antisense lncRNA gene | FERM domain containing 8, opposite strand |
| 19 | gene | 5.849702 | 5.875274 | negative | MGI_C57BL6J_1914707 | Frmd8 | NCBI_Gene:67457,ENSEMBL:ENSMUSG00000024816 | MGI:1914707 | protein coding gene | FERM domain containing 8 |
| 19 | gene | 5.877799 | 5.885878 | positive | MGI_C57BL6J_2147731 | Slc25a45 | NCBI_Gene:107375,ENSEMBL:ENSMUSG00000024818 | MGI:2147731 | protein coding gene | solute carrier family 25, member 45 |
| 19 | gene | 5.891138 | 5.895169 | negative | MGI_C57BL6J_2681860 | Tigd3 | NCBI_Gene:332359,ENSEMBL:ENSMUSG00000044390 | MGI:2681860 | protein coding gene | tigger transposable element derived 3 |
| 19 | gene | 5.896516 | 5.913010 | negative | MGI_C57BL6J_109529 | Dpf2 | NCBI_Gene:19708,ENSEMBL:ENSMUSG00000024826 | MGI:109529 | protein coding gene | D4, zinc and double PHD fingers family 2 |
| 19 | gene | 5.912822 | 5.914831 | positive | MGI_C57BL6J_5624952 | Gm42067 | NCBI_Gene:105246846,ENSEMBL:ENSMUSG00000110156 | MGI:5624952 | lncRNA gene | predicted gene, 42067 |
| 19 | gene | 5.915636 | 5.924816 | negative | MGI_C57BL6J_1929744 | Cdc42ep2 | NCBI_Gene:104252,ENSEMBL:ENSMUSG00000045664 | MGI:1929744 | protein coding gene | CDC42 effector protein (Rho GTPase binding) 2 |
| 19 | gene | 5.940542 | 5.964216 | negative | MGI_C57BL6J_99690 | Pola2 | NCBI_Gene:18969,ENSEMBL:ENSMUSG00000024833 | MGI:99690 | protein coding gene | polymerase (DNA directed), alpha 2 |
| 19 | gene | 5.970234 | 5.986143 | negative | MGI_C57BL6J_2685809 | Slc22a20 | NCBI_Gene:381203,ENSEMBL:ENSMUSG00000037451 | MGI:2685809 | protein coding gene | solute carrier family 22 (organic anion transporter), member 20 |
| 19 | gene | 5.970486 | 5.984846 | positive | MGI_C57BL6J_5590878 | Gm31719 | NCBI_Gene:102634035 | MGI:5590878 | lncRNA gene | predicted gene, 31719 |
| 19 | gene | 5.988545 | 6.015897 | negative | MGI_C57BL6J_88263 | Capn1 | NCBI_Gene:12333,ENSEMBL:ENSMUSG00000024942 | MGI:88263 | protein coding gene | calpain 1 |
| 19 | gene | 6.012620 | 6.018462 | positive | MGI_C57BL6J_3642580 | Gm10814 | NCBI_Gene:100502985,ENSEMBL:ENSMUSG00000097404 | MGI:3642580 | lncRNA gene | predicted gene 10814 |
| 19 | pseudogene | 6.027533 | 6.028401 | negative | MGI_C57BL6J_5011690 | Gm19505 | NCBI_Gene:100503014,ENSEMBL:ENSMUSG00000118194 | MGI:5011690 | pseudogene | predicted gene, 19505 |
| 19 | pseudogene | 6.044442 | 6.044897 | negative | MGI_C57BL6J_3644144 | Gm8034 | NCBI_Gene:666300,ENSEMBL:ENSMUSG00000094281 | MGI:3644144 | pseudogene | predicted gene 8034 |
| 19 | gene | 6.045060 | 6.047019 | negative | MGI_C57BL6J_1925963 | D530017H19Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA D530017H19 gene |
| 19 | gene | 6.046576 | 6.053718 | positive | MGI_C57BL6J_1921376 | Syvn1 | NCBI_Gene:74126,ENSEMBL:ENSMUSG00000024807 | MGI:1921376 | protein coding gene | synovial apoptosis inhibitor 1, synoviolin |
| 19 | gene | 6.051334 | 6.051392 | positive | MGI_C57BL6J_5530975 | Mir6988 | miRBase:MI0022836,NCBI_Gene:102465597,ENSEMBL:ENSMUSG00000098928 | MGI:5530975 | miRNA gene | microRNA 6988 |
| 19 | gene | 6.053622 | 6.057785 | negative | MGI_C57BL6J_108180 | Mrpl49 | NCBI_Gene:18120,ENSEMBL:ENSMUSG00000007338 | MGI:108180 | protein coding gene | mitochondrial ribosomal protein L49 |
| 19 | gene | 6.057844 | 6.059524 | positive | MGI_C57BL6J_102547 | Fau | NCBI_Gene:14109,ENSEMBL:ENSMUSG00000038274 | MGI:102547 | protein coding gene | Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed (fox derived) |
| 19 | gene | 6.061192 | 6.062472 | positive | MGI_C57BL6J_1352481 | Znhit2 | NCBI_Gene:29805,ENSEMBL:ENSMUSG00000075227 | MGI:1352481 | protein coding gene | zinc finger, HIT domain containing 2 |
| 19 | gene | 6.062821 | 6.068373 | negative | MGI_C57BL6J_1920416 | Tm7sf2 | NCBI_Gene:73166,ENSEMBL:ENSMUSG00000024799 | MGI:1920416 | protein coding gene | transmembrane 7 superfamily member 2 |
| 19 | gene | 6.067842 | 6.077231 | negative | MGI_C57BL6J_1915755 | Vps51 | NCBI_Gene:68505,ENSEMBL:ENSMUSG00000024797 | MGI:1915755 | protein coding gene | VPS51 GARP complex subunit |
| 19 | gene | 6.076945 | 6.082256 | negative | MGI_C57BL6J_5579080 | Gm28374 | ENSEMBL:ENSMUSG00000100621 | MGI:5579080 | protein coding gene | predicted gene 28374 |
| 19 | gene | 6.077884 | 6.080788 | positive | MGI_C57BL6J_3690536 | Tmem262 | NCBI_Gene:433215,ENSEMBL:ENSMUSG00000047733 | MGI:3690536 | protein coding gene | transmembrane protein 262 |
| 19 | gene | 6.080757 | 6.084956 | negative | MGI_C57BL6J_1891017 | Zfpl1 | NCBI_Gene:81909,ENSEMBL:ENSMUSG00000024792 | MGI:1891017 | protein coding gene | zinc finger like protein 1 |
| 19 | gene | 6.084983 | 6.091777 | positive | MGI_C57BL6J_1915099 | Cdca5 | NCBI_Gene:67849,ENSEMBL:ENSMUSG00000024791 | MGI:1915099 | protein coding gene | cell division cycle associated 5 |
| 19 | gene | 6.093724 | 6.098241 | positive | MGI_C57BL6J_2685396 | Gm550 | NCBI_Gene:225852,ENSEMBL:ENSMUSG00000117666 | MGI:2685396 | protein coding gene | predicted gene 550 |
| 19 | gene | 6.105782 | 6.115792 | positive | MGI_C57BL6J_2685810 | Naaladl1 | NCBI_Gene:381204,ENSEMBL:ENSMUSG00000054999 | MGI:2685810 | protein coding gene | N-acetylated alpha-linked acidic dipeptidase-like 1 |
| 19 | gene | 6.116000 | 6.118650 | negative | MGI_C57BL6J_1913656 | Sac3d1 | NCBI_Gene:66406,ENSEMBL:ENSMUSG00000024790 | MGI:1913656 | protein coding gene | SAC3 domain containing 1 |
| 19 | gene | 6.119399 | 6.132563 | negative | MGI_C57BL6J_1916274 | Snx15 | NCBI_Gene:69024,ENSEMBL:ENSMUSG00000024787 | MGI:1916274 | protein coding gene | sorting nexin 15 |
| 19 | gene | 6.134374 | 6.141548 | negative | MGI_C57BL6J_1928393 | Arl2 | NCBI_Gene:56327,ENSEMBL:ENSMUSG00000024944 | MGI:1928393 | protein coding gene | ADP-ribosylation factor-like 2 |
| 19 | gene | 6.140983 | 6.172476 | positive | MGI_C57BL6J_1921731 | Batf2 | NCBI_Gene:74481,ENSEMBL:ENSMUSG00000039699 | MGI:1921731 | protein coding gene | basic leucine zipper transcription factor, ATF-like 2 |
| 19 | gene | 6.144732 | 6.144851 | positive | MGI_C57BL6J_4421753 | n-R5s19 | ENSEMBL:ENSMUSG00000084742 | MGI:4421753 | rRNA gene | nuclear encoded rRNA 5S 19 |
| 19 | pseudogene | 6.152666 | 6.153712 | positive | MGI_C57BL6J_3648472 | Gm5510 | NCBI_Gene:433216,ENSEMBL:ENSMUSG00000083458 | MGI:3648472 | pseudogene | predicted pseudogene 5510 |
| 19 | gene | 6.184328 | 6.225885 | positive | MGI_C57BL6J_1923913 | Majin | NCBI_Gene:622554,ENSEMBL:ENSMUSG00000024786 | MGI:1923913 | protein coding gene | membrane anchored junction protein |
| 19 | gene | 6.226380 | 6.227768 | positive | MGI_C57BL6J_2156541 | Gpha2 | NCBI_Gene:170458,ENSEMBL:ENSMUSG00000024784 | MGI:2156541 | protein coding gene | glycoprotein hormone alpha 2 |
| 19 | gene | 6.227765 | 6.235872 | negative | MGI_C57BL6J_2388480 | Ppp2r5b | NCBI_Gene:225849,ENSEMBL:ENSMUSG00000024777 | MGI:2388480 | protein coding gene | protein phosphatase 2, regulatory subunit B’, beta |
| 19 | gene | 6.241664 | 6.262335 | positive | MGI_C57BL6J_1916291 | Atg2a | NCBI_Gene:329015,ENSEMBL:ENSMUSG00000024773 | MGI:1916291 | protein coding gene | autophagy related 2A |
| 19 | gene | 6.264643 | 6.264728 | positive | MGI_C57BL6J_3618738 | Mir194-2 | miRBase:MI0000733,NCBI_Gene:723957,ENSEMBL:ENSMUSG00000065582 | MGI:3618738 | miRNA gene | microRNA 194-2 |
| 19 | gene | 6.264844 | 6.264932 | positive | MGI_C57BL6J_2676856 | Mir192 | miRBase:MI0000551,NCBI_Gene:387187,ENSEMBL:ENSMUSG00000065523 | MGI:2676856 | miRNA gene | microRNA 192 |
| 19 | gene | 6.268794 | 6.276607 | negative | MGI_C57BL6J_3705162 | Gm14963 | NCBI_Gene:105246847,ENSEMBL:ENSMUSG00000085196 | MGI:3705162 | lncRNA gene | predicted gene 14963 |
| 19 | gene | 6.276725 | 6.300096 | positive | MGI_C57BL6J_1341878 | Ehd1 | NCBI_Gene:13660,ENSEMBL:ENSMUSG00000024772 | MGI:1341878 | protein coding gene | EH-domain containing 1 |
| 19 | gene | 6.306456 | 6.325653 | positive | MGI_C57BL6J_2652845 | Cdc42bpg | NCBI_Gene:240505,ENSEMBL:ENSMUSG00000024769 | MGI:2652845 | protein coding gene | CDC42 binding protein kinase gamma (DMPK-like) |
| 19 | pseudogene | 6.331054 | 6.331453 | negative | MGI_C57BL6J_6302977 | Gm50198 | ENSEMBL:ENSMUSG00000117681 | MGI:6302977 | pseudogene | predicted gene, 50198 |
| 19 | gene | 6.334979 | 6.340894 | positive | MGI_C57BL6J_1316736 | Men1 | NCBI_Gene:17283,ENSEMBL:ENSMUSG00000024947 | MGI:1316736 | protein coding gene | multiple endocrine neoplasia 1 |
| 19 | gene | 6.341117 | 6.355619 | positive | MGI_C57BL6J_1346883 | Map4k2 | NCBI_Gene:26412,ENSEMBL:ENSMUSG00000024948 | MGI:1346883 | protein coding gene | mitogen-activated protein kinase kinase kinase kinase 2 |
| 19 | gene | 6.346480 | 6.346544 | positive | MGI_C57BL6J_5531267 | Mir6989 | miRBase:MI0022837,NCBI_Gene:102465598,ENSEMBL:ENSMUSG00000099230 | MGI:5531267 | miRNA gene | microRNA 6989 |
| 19 | gene | 6.355473 | 6.363759 | negative | MGI_C57BL6J_3705223 | Gm14966 | NCBI_Gene:102634533,ENSEMBL:ENSMUSG00000079467 | MGI:3705223 | lncRNA gene | predicted gene 14966 |
| 19 | gene | 6.363279 | 6.378038 | positive | MGI_C57BL6J_1095403 | Sf1 | NCBI_Gene:22668,ENSEMBL:ENSMUSG00000024949 | MGI:1095403 | protein coding gene | splicing factor 1 |
| 19 | gene | 6.384399 | 6.398464 | positive | MGI_C57BL6J_97830 | Pygm | NCBI_Gene:19309,ENSEMBL:ENSMUSG00000032648 | MGI:97830 | protein coding gene | muscle glycogen phosphorylase |
| 19 | gene | 6.399008 | 6.415216 | positive | MGI_C57BL6J_1333849 | Rasgrp2 | NCBI_Gene:19395,ENSEMBL:ENSMUSG00000032946 | MGI:1333849 | protein coding gene | RAS, guanyl releasing protein 2 |
| 19 | gene | 6.409400 | 6.418659 | negative | MGI_C57BL6J_3705214 | Gm14965 | NCBI_Gene:105246848,ENSEMBL:ENSMUSG00000084876 | MGI:3705214 | lncRNA gene | predicted gene 14965 |
| 19 | gene | 6.418731 | 6.544169 | positive | MGI_C57BL6J_1096362 | Nrxn2 | NCBI_Gene:18190,ENSEMBL:ENSMUSG00000033768 | MGI:1096362 | protein coding gene | neurexin II |
| 19 | gene | 6.457419 | 6.460107 | negative | MGI_C57BL6J_6302908 | Gm50151 | ENSEMBL:ENSMUSG00000117897 | MGI:6302908 | unclassified gene | predicted gene, 50151 |
| 19 | gene | 6.530711 | 6.532011 | negative | MGI_C57BL6J_3641621 | Gm14964 | ENSEMBL:ENSMUSG00000052188 | MGI:3641621 | lncRNA gene | predicted gene 14964 |
| 19 | gene | 6.535845 | 6.543070 | negative | MGI_C57BL6J_1195269 | Slc22a12 | NCBI_Gene:20521,ENSEMBL:ENSMUSG00000061742 | MGI:1195269 | protein coding gene | solute carrier family 22 (organic anion/cation transporter), member 12 |
| 19 | gene | 6.563780 | 6.563905 | positive | MGI_C57BL6J_5456237 | Gm26460 | ENSEMBL:ENSMUSG00000092802 | MGI:5456237 | miRNA gene | predicted gene, 26460 |
| 19 | gene | 6.567126 | 6.578552 | positive | MGI_C57BL6J_5591404 | Gm32245 | NCBI_Gene:102634732 | MGI:5591404 | lncRNA gene | predicted gene, 32245 |
| 19 | pseudogene | 6.569080 | 6.575362 | negative | MGI_C57BL6J_6303367 | Gm50435 | ENSEMBL:ENSMUSG00000118062 | MGI:6303367 | pseudogene | predicted gene, 50435 |
| 19 | pseudogene | 6.592099 | 6.593339 | negative | MGI_C57BL6J_3768540 | Hmgb1-ps4 | NCBI_Gene:629372,ENSEMBL:ENSMUSG00000084221 | MGI:3768540 | pseudogene | high mobility group box 1, pseudogene 4 |
| 19 | gene | 6.617671 | 6.674548 | negative | MGI_C57BL6J_1924061 | 2410152P15Rik | ENSEMBL:ENSMUSG00000084825 | MGI:1924061 | lncRNA gene | RIKEN cDNA 2410152P15 gene |
| 19 | gene | 6.645687 | 6.647172 | positive | MGI_C57BL6J_3705315 | Gm14968 | ENSEMBL:ENSMUSG00000085285 | MGI:3705315 | lncRNA gene | predicted gene 14968 |
| 19 | gene | 6.829084 | 6.840989 | negative | MGI_C57BL6J_1930076 | Rps6ka4 | NCBI_Gene:56613,ENSEMBL:ENSMUSG00000024952 | MGI:1930076 | protein coding gene | ribosomal protein S6 kinase, polypeptide 4 |
| 19 | gene | 6.829886 | 6.829944 | negative | MGI_C57BL6J_4950413 | Mir5046 | miRBase:MI0017931,NCBI_Gene:100628576,ENSEMBL:ENSMUSG00000104884 | MGI:4950413 | miRNA gene | microRNA 5046 |
| 19 | gene | 6.844623 | 6.858333 | negative | MGI_C57BL6J_1925567 | Ccdc88b | NCBI_Gene:78317,ENSEMBL:ENSMUSG00000047810 | MGI:1925567 | protein coding gene | coiled-coil domain containing 88B |
| 19 | gene | 6.906697 | 6.910106 | negative | MGI_C57BL6J_1859821 | Prdx5 | NCBI_Gene:54683,ENSEMBL:ENSMUSG00000024953 | MGI:1859821 | protein coding gene | peroxiredoxin 5 |
| 19 | gene | 6.909698 | 6.911049 | positive | MGI_C57BL6J_1914924 | Trmt112 | NCBI_Gene:67674,ENSEMBL:ENSMUSG00000038812 | MGI:1914924 | protein coding gene | tRNA methyltransferase 11-2 |
| 19 | gene | 6.910977 | 6.922048 | negative | MGI_C57BL6J_1346831 | Esrra | NCBI_Gene:26379,ENSEMBL:ENSMUSG00000024955 | MGI:1346831 | protein coding gene | estrogen related receptor, alpha |
| 19 | gene | 6.914196 | 6.914286 | negative | MGI_C57BL6J_5531108 | Mir6990 | miRBase:MI0022838,NCBI_Gene:102466213,ENSEMBL:ENSMUSG00000098413 | MGI:5531108 | miRNA gene | microRNA 6990 |
| 19 | gene | 6.922371 | 6.925380 | negative | MGI_C57BL6J_1914327 | Catsperz | NCBI_Gene:67077,ENSEMBL:ENSMUSG00000050623 | MGI:1914327 | protein coding gene | cation channel sperm associated auxiliary subunit zeta |
| 19 | gene | 6.923966 | 6.934772 | negative | MGI_C57BL6J_1298234 | Kcnk4 | NCBI_Gene:16528,ENSEMBL:ENSMUSG00000024957 | MGI:1298234 | protein coding gene | potassium channel, subfamily K, member 4 |
| 19 | gene | 6.927166 | 6.929518 | positive | MGI_C57BL6J_5826283 | Gm46646 | NCBI_Gene:108168407 | MGI:5826283 | lncRNA gene | predicted gene, 46646 |
| 19 | gene | 6.938057 | 6.942480 | negative | MGI_C57BL6J_2147529 | Gpr137 | NCBI_Gene:107173,ENSEMBL:ENSMUSG00000024958 | MGI:2147529 | protein coding gene | G protein-coupled receptor 137 |
| 19 | gene | 6.941861 | 6.951905 | positive | MGI_C57BL6J_1096330 | Bad | NCBI_Gene:12015,ENSEMBL:ENSMUSG00000024959 | MGI:1096330 | protein coding gene | BCL2-associated agonist of cell death |
| 19 | pseudogene | 6.945071 | 6.945620 | negative | MGI_C57BL6J_5011238 | Gm19053 | NCBI_Gene:100418171 | MGI:5011238 | pseudogene | predicted gene, 19053 |
| 19 | gene | 6.952325 | 6.976470 | negative | MGI_C57BL6J_104778 | Plcb3 | NCBI_Gene:18797,ENSEMBL:ENSMUSG00000024960 | MGI:104778 | protein coding gene | phospholipase C, beta 3 |
| 19 | gene | 6.974968 | 6.977324 | positive | MGI_C57BL6J_107682 | Ppp1r14b | NCBI_Gene:18938,ENSEMBL:ENSMUSG00000056612 | MGI:107682 | protein coding gene | protein phosphatase 1, regulatory inhibitor subunit 14B |
| 19 | gene | 6.977739 | 6.980501 | negative | MGI_C57BL6J_95542 | Fkbp2 | NCBI_Gene:14227,ENSEMBL:ENSMUSG00000056629 | MGI:95542 | protein coding gene | FK506 binding protein 2 |
| 19 | gene | 6.980528 | 6.984168 | positive | MGI_C57BL6J_5624688 | Gm41803 | NCBI_Gene:105246528 | MGI:5624688 | lncRNA gene | predicted gene, 41803 |
| 19 | gene | 6.982472 | 6.987651 | negative | MGI_C57BL6J_106199 | Vegfb | NCBI_Gene:22340,ENSEMBL:ENSMUSG00000024962 | MGI:106199 | protein coding gene | vascular endothelial growth factor B |
| 19 | gene | 6.987911 | 6.992697 | negative | MGI_C57BL6J_1927346 | Dnajc4 | NCBI_Gene:57431,ENSEMBL:ENSMUSG00000024963 | MGI:1927346 | protein coding gene | DnaJ heat shock protein family (Hsp40) member C4 |
| 19 | gene | 6.992674 | 6.993046 | positive | MGI_C57BL6J_5594997 | Gm35838 | NCBI_Gene:102639551 | MGI:5594997 | lncRNA gene | predicted gene, 35838 |
| 19 | gene | 6.993018 | 6.996117 | negative | MGI_C57BL6J_1915573 | Nudt22 | NCBI_Gene:68323,ENSEMBL:ENSMUSG00000037349 | MGI:1915573 | protein coding gene | nudix (nucleoside diphosphate linked moiety X)-type motif 22 |
| 19 | gene | 6.996131 | 6.999048 | positive | MGI_C57BL6J_1333115 | Trpt1 | NCBI_Gene:107328,ENSEMBL:ENSMUSG00000047656 | MGI:1333115 | protein coding gene | tRNA phosphotransferase 1 |
| 19 | gene | 6.998958 | 7.019469 | negative | MGI_C57BL6J_2147790 | Fermt3 | NCBI_Gene:108101,ENSEMBL:ENSMUSG00000024965 | MGI:2147790 | protein coding gene | fermitin family member 3 |
| 19 | gene | 7.020696 | 7.040026 | negative | MGI_C57BL6J_109130 | Stip1 | NCBI_Gene:20867,ENSEMBL:ENSMUSG00000024966 | MGI:109130 | protein coding gene | stress-induced phosphoprotein 1 |
| 19 | gene | 7.056713 | 7.198062 | positive | MGI_C57BL6J_2147583 | Macrod1 | NCBI_Gene:107227,ENSEMBL:ENSMUSG00000036278 | MGI:2147583 | protein coding gene | MACRO domain containing 1 |
| 19 | gene | 7.084710 | 7.090741 | positive | MGI_C57BL6J_1917744 | 5730409K12Rik | NCBI_Gene:70494 | MGI:1917744 | unclassified gene | RIKEN cDNA 5730409K12 gene |
| 19 | gene | 7.087464 | 7.166030 | negative | MGI_C57BL6J_3026647 | Flrt1 | NCBI_Gene:396184,ENSEMBL:ENSMUSG00000047787 | MGI:3026647 | protein coding gene | fibronectin leucine rich transmembrane protein 1 |
| 19 | gene | 7.131506 | 7.136039 | positive | MGI_C57BL6J_5621506 | Gm38621 | NCBI_Gene:102642633 | MGI:5621506 | lncRNA gene | predicted gene, 38621 |
| 19 | gene | 7.198202 | 7.206316 | negative | MGI_C57BL6J_2147616 | Otub1 | NCBI_Gene:107260,ENSEMBL:ENSMUSG00000024767 | MGI:2147616 | protein coding gene | OTU domain, ubiquitin aldehyde binding 1 |
| 19 | gene | 7.215153 | 7.217616 | negative | MGI_C57BL6J_105959 | Cox8a | NCBI_Gene:12868,ENSEMBL:ENSMUSG00000035885 | MGI:105959 | protein coding gene | cytochrome c oxidase subunit 8A |
| 19 | gene | 7.225668 | 7.241222 | negative | MGI_C57BL6J_1918249 | Naa40 | NCBI_Gene:70999,ENSEMBL:ENSMUSG00000024764 | MGI:1918249 | protein coding gene | N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| 19 | gene | 7.246709 | 7.246812 | positive | MGI_C57BL6J_5451890 | Gm22113 | ENSEMBL:ENSMUSG00000096774 | MGI:5451890 | snRNA gene | predicted gene, 22113 |
| 19 | gene | 7.259941 | 7.260061 | positive | MGI_C57BL6J_5453774 | Gm23997 | ENSEMBL:ENSMUSG00000088680 | MGI:5453774 | rRNA gene | predicted gene, 23997 |
| 19 | gene | 7.267325 | 7.275225 | positive | MGI_C57BL6J_1859854 | Rcor2 | NCBI_Gene:104383,ENSEMBL:ENSMUSG00000024968 | MGI:1859854 | protein coding gene | REST corepressor 2 |
| 19 | gene | 7.275396 | 7.341867 | negative | MGI_C57BL6J_99638 | Mark2 | NCBI_Gene:13728,ENSEMBL:ENSMUSG00000024969 | MGI:99638 | protein coding gene | MAP/microtubule affinity regulating kinase 2 |
| 19 | gene | 7.297654 | 7.304528 | positive | MGI_C57BL6J_4938054 | Gm17227 | NCBI_Gene:102639888,ENSEMBL:ENSMUSG00000091558 | MGI:4938054 | lncRNA gene | predicted gene 17227 |
| 19 | gene | 7.324971 | 7.327376 | negative | MGI_C57BL6J_2441993 | D830016O14Rik | NA | NA | unclassified gene | RIKEN cDNA D830016O14 gene |
| 19 | gene | 7.354078 | 7.383051 | negative | MGI_C57BL6J_2147611 | Spindoc | NCBI_Gene:68229,ENSEMBL:ENSMUSG00000024970 | MGI:2147611 | protein coding gene | spindlin interactor and repressor of chromatin binding |
| 19 | gene | 7.364515 | 7.370240 | positive | MGI_C57BL6J_6303036 | Gm50232 | ENSEMBL:ENSMUSG00000117743 | MGI:6303036 | lncRNA gene | predicted gene, 50232 |
| 19 | gene | 7.370339 | 7.372580 | negative | MGI_C57BL6J_5826284 | Gm46647 | NCBI_Gene:108168408 | MGI:5826284 | lncRNA gene | predicted gene, 46647 |
| 19 | gene | 7.382537 | 7.383561 | positive | MGI_C57BL6J_1915479 | 1700105P06Rik | NCBI_Gene:654802,ENSEMBL:ENSMUSG00000099923 | MGI:1915479 | lncRNA gene | RIKEN cDNA 1700105P06 gene |
| 19 | pseudogene | 7.397442 | 7.399173 | negative | MGI_C57BL6J_3643959 | Rab11b-ps2 | NCBI_Gene:665419,ENSEMBL:ENSMUSG00000095690 | MGI:3643959 | pseudogene | RAB11B, member RAS oncogene family, pseudogene 2 |
| 19 | pseudogene | 7.402834 | 7.403431 | negative | MGI_C57BL6J_5011027 | Gm18842 | NCBI_Gene:100417810,ENSEMBL:ENSMUSG00000118297 | MGI:5011027 | pseudogene | predicted gene, 18842 |
| 19 | gene | 7.417625 | 7.425904 | positive | MGI_C57BL6J_1919667 | 2700081O15Rik | NCBI_Gene:108899,ENSEMBL:ENSMUSG00000053080 | MGI:1919667 | protein coding gene | RIKEN cDNA 2700081O15 gene |
| 19 | gene | 7.422573 | 7.422642 | positive | MGI_C57BL6J_5531348 | Mir6991 | miRBase:MI0022839,NCBI_Gene:102466781,ENSEMBL:ENSMUSG00000098293 | MGI:5531348 | miRNA gene | microRNA 6991 |
| 19 | gene | 7.425895 | 7.483291 | negative | MGI_C57BL6J_1339970 | Rtn3 | NCBI_Gene:20168,ENSEMBL:ENSMUSG00000024758 | MGI:1339970 | protein coding gene | reticulon 3 |
| 19 | gene | 7.465335 | 7.469619 | negative | MGI_C57BL6J_2444344 | A830039H05Rik | NA | NA | unclassified gene | RIKEN cDNA A830039H05 gene |
| 19 | gene | 7.472709 | 7.476413 | negative | MGI_C57BL6J_2443568 | A830080L01Rik | NA | NA | unclassified gene | RIKEN cDNA A830080L01 gene |
| 19 | gene | 7.494026 | 7.539674 | positive | MGI_C57BL6J_1924270 | Atl3 | NCBI_Gene:109168,ENSEMBL:ENSMUSG00000024759 | MGI:1924270 | protein coding gene | atlastin GTPase 3 |
| 19 | gene | 7.557433 | 7.588545 | positive | MGI_C57BL6J_2179715 | Plaat3 | NCBI_Gene:225845,ENSEMBL:ENSMUSG00000060675 | MGI:2179715 | protein coding gene | phospholipase A and acyltransferase 3 |
| 19 | gene | 7.596656 | 7.607237 | negative | MGI_C57BL6J_1929094 | Lgals12 | NCBI_Gene:56072,ENSEMBL:ENSMUSG00000024972 | MGI:1929094 | protein coding gene | lectin, galactose binding, soluble 12 |
| 19 | gene | 7.612541 | 7.639642 | positive | MGI_C57BL6J_1913977 | Plaat5 | NCBI_Gene:66727,ENSEMBL:ENSMUSG00000024973 | MGI:1913977 | protein coding gene | phospholipase A and acyltransferase 5 |
| 19 | pseudogene | 7.668271 | 7.669148 | positive | MGI_C57BL6J_3644508 | Gm16437 | NCBI_Gene:433217,ENSEMBL:ENSMUSG00000024761 | MGI:3644508 | pseudogene | predicted pseudogene 16437 |
| 19 | gene | 7.673061 | 7.711310 | negative | MGI_C57BL6J_2442751 | Slc22a19 | NCBI_Gene:207151,ENSEMBL:ENSMUSG00000024757 | MGI:2442751 | protein coding gene | solute carrier family 22 (organic anion transporter), member 19 |
| 19 | pseudogene | 7.695899 | 7.697016 | negative | MGI_C57BL6J_3801808 | Gm15967 | NCBI_Gene:100418172,ENSEMBL:ENSMUSG00000083160 | MGI:3801808 | pseudogene | predicted gene 15967 |
| 19 | pseudogene | 7.703903 | 7.705127 | negative | MGI_C57BL6J_3801809 | Gm15968 | NCBI_Gene:100416946,ENSEMBL:ENSMUSG00000081596 | MGI:3801809 | pseudogene | predicted gene 15968 |
| 19 | pseudogene | 7.727182 | 7.727673 | negative | MGI_C57BL6J_3647695 | Gm8157 | NCBI_Gene:666543,ENSEMBL:ENSMUSG00000117937 | MGI:3647695 | pseudogene | predicted gene 8157 |
| 19 | pseudogene | 7.729370 | 7.729953 | positive | MGI_C57BL6J_5010729 | Gm18544 | NCBI_Gene:100417341 | MGI:5010729 | pseudogene | predicted gene, 18544 |
| 19 | gene | 7.780468 | 7.802669 | negative | MGI_C57BL6J_2385316 | Slc22a26 | NCBI_Gene:236149,ENSEMBL:ENSMUSG00000053303 | MGI:2385316 | protein coding gene | solute carrier family 22 (organic cation transporter), member 26 |
| 19 | pseudogene | 7.806297 | 7.848669 | negative | MGI_C57BL6J_3646592 | Gm7096 | NCBI_Gene:100534290,ENSEMBL:ENSMUSG00000118312 | MGI:3646592 | pseudogene | predicted gene 7096 |
| 19 | gene | 7.864388 | 7.966595 | negative | MGI_C57BL6J_3042283 | Slc22a27 | NCBI_Gene:171405,ENSEMBL:ENSMUSG00000067656 | MGI:3042283 | protein coding gene | solute carrier family 22, member 27 |
| 19 | gene | 7.959530 | 7.959638 | negative | MGI_C57BL6J_5454728 | Gm24951 | ENSEMBL:ENSMUSG00000076136 | MGI:5454728 | miRNA gene | predicted gene, 24951 |
| 19 | pseudogene | 7.959743 | 7.961107 | negative | MGI_C57BL6J_3779569 | Gm6192 | NCBI_Gene:620946,ENSEMBL:ENSMUSG00000118220 | MGI:3779569 | pseudogene | predicted gene 6192 |
| 19 | pseudogene | 7.975735 | 7.978339 | positive | MGI_C57BL6J_3643067 | Gm6386 | NCBI_Gene:623030,ENSEMBL:ENSMUSG00000118036 | MGI:3643067 | pseudogene | predicted gene 6386 |
| 19 | gene | 8.019555 | 8.034088 | positive | MGI_C57BL6J_1925675 | 9530053H05Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 9530053H05 gene |
| 19 | gene | 8.062209 | 8.131982 | negative | MGI_C57BL6J_3645714 | Slc22a28 | NCBI_Gene:434674,ENSEMBL:ENSMUSG00000063590 | MGI:3645714 | protein coding gene | solute carrier family 22, member 28 |
| 19 | gene | 8.160165 | 8.253522 | negative | MGI_C57BL6J_3605624 | Slc22a29 | NCBI_Gene:236293,ENSEMBL:ENSMUSG00000075044 | MGI:3605624 | protein coding gene | solute carrier family 22. member 29 |
| 19 | gene | 8.251312 | 8.317179 | positive | MGI_C57BL6J_5624689 | Gm41804 | NCBI_Gene:105246529,ENSEMBL:ENSMUSG00000118280 | MGI:5624689 | lncRNA gene | predicted gene, 41804 |
| 19 | pseudogene | 8.255938 | 8.257662 | negative | MGI_C57BL6J_3644446 | Gm7105 | NCBI_Gene:633120,ENSEMBL:ENSMUSG00000118345 | MGI:3644446 | pseudogene | predicted gene 7105 |
| 19 | pseudogene | 8.272397 | 8.275004 | positive | MGI_C57BL6J_3643112 | Gm8184 | NCBI_Gene:666601,ENSEMBL:ENSMUSG00000117877 | MGI:3643112 | pseudogene | predicted gene 8184 |
| 19 | gene | 8.335371 | 8.405111 | negative | MGI_C57BL6J_2442750 | Slc22a30 | NCBI_Gene:319800,ENSEMBL:ENSMUSG00000052562 | MGI:2442750 | protein coding gene | solute carrier family 22, member 30 |
| 19 | pseudogene | 8.473313 | 8.541800 | negative | MGI_C57BL6J_3779786 | Gm8189 | NCBI_Gene:100534389,ENSEMBL:ENSMUSG00000110298 | MGI:3779786 | pseudogene | predicted gene 8189 |
| 19 | pseudogene | 8.588211 | 8.589292 | negative | MGI_C57BL6J_3646016 | Gm6425 | NCBI_Gene:623402,ENSEMBL:ENSMUSG00000086174 | MGI:3646016 | pseudogene | predicted pseudogene 6425 |
| 19 | gene | 8.591254 | 8.611835 | positive | MGI_C57BL6J_1336187 | Slc22a8 | NCBI_Gene:19879,ENSEMBL:ENSMUSG00000063796 | MGI:1336187 | protein coding gene | solute carrier family 22 (organic anion transporter), member 8 |
| 19 | gene | 8.611932 | 8.621184 | negative | MGI_C57BL6J_5595553 | Gm36394 | NCBI_Gene:102640287 | MGI:5595553 | lncRNA gene | predicted gene, 36394 |
| 19 | gene | 8.617996 | 8.628299 | positive | MGI_C57BL6J_892001 | Slc22a6 | NCBI_Gene:18399,ENSEMBL:ENSMUSG00000024650 | MGI:892001 | protein coding gene | solute carrier family 22 (organic anion transporter), member 6 |
| 19 | gene | 8.651796 | 8.663335 | negative | MGI_C57BL6J_3028064 | 9830166K06Rik | NCBI_Gene:402757,ENSEMBL:ENSMUSG00000117763 | MGI:3028064 | lncRNA gene | RIKEN cDNA 9830166K06 gene |
| 19 | gene | 8.663789 | 8.683606 | positive | MGI_C57BL6J_88396 | Chrm1 | NCBI_Gene:12669,ENSEMBL:ENSMUSG00000032773 | MGI:88396 | protein coding gene | cholinergic receptor, muscarinic 1, CNS |
| 19 | gene | 8.681835 | 8.682163 | negative | MGI_C57BL6J_5452597 | Gm22820 | ENSEMBL:ENSMUSG00000087830 | MGI:5452597 | unclassified non-coding RNA gene | predicted gene, 22820 |
| 19 | gene | 8.706882 | 8.723369 | negative | MGI_C57BL6J_96955 | Slc3a2 | NCBI_Gene:17254,ENSEMBL:ENSMUSG00000010095 | MGI:96955 | protein coding gene | solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| 19 | gene | 8.723475 | 8.726443 | positive | MGI_C57BL6J_3763743 | Snhg1 | NCBI_Gene:83673,ENSEMBL:ENSMUSG00000108414 | MGI:3763743 | lncRNA gene | small nucleolar RNA host gene 1 |
| 19 | gene | 8.723993 | 8.724064 | positive | MGI_C57BL6J_5455632 | Gm25855 | ENSEMBL:ENSMUSG00000064721 | MGI:5455632 | snoRNA gene | predicted gene, 25855 |
| 19 | gene | 8.724251 | 8.724325 | positive | MGI_C57BL6J_5455671 | Gm25894 | ENSEMBL:ENSMUSG00000065392 | MGI:5455671 | snoRNA gene | predicted gene, 25894 |
| 19 | gene | 8.724531 | 8.724602 | positive | MGI_C57BL6J_5454229 | Gm24452 | ENSEMBL:ENSMUSG00000065281 | MGI:5454229 | snoRNA gene | predicted gene, 24452 |
| 19 | gene | 8.724868 | 8.724943 | positive | MGI_C57BL6J_5454230 | Gm24453 | ENSEMBL:ENSMUSG00000065280 | MGI:5454230 | snoRNA gene | predicted gene, 24453 |
| 19 | gene | 8.725092 | 8.725156 | positive | MGI_C57BL6J_5452521 | Gm22744 | ENSEMBL:ENSMUSG00000065378 | MGI:5452521 | snoRNA gene | predicted gene, 22744 |
| 19 | gene | 8.725341 | 8.725409 | positive | MGI_C57BL6J_5452457 | Gm22680 | ENSEMBL:ENSMUSG00000065782 | MGI:5452457 | snoRNA gene | predicted gene, 22680 |
| 19 | gene | 8.725594 | 8.725660 | positive | MGI_C57BL6J_5453023 | Gm23246 | ENSEMBL:ENSMUSG00000065305 | MGI:5453023 | snoRNA gene | predicted gene, 23246 |
| 19 | gene | 8.725866 | 8.725991 | positive | MGI_C57BL6J_1933385 | Snord22 | NCBI_Gene:100127111,ENSEMBL:ENSMUSG00000065087 | MGI:1933385 | snoRNA gene | small nucleolar RNA, C/D box 22 |
| 19 | gene | 8.735804 | 8.740624 | positive | MGI_C57BL6J_2147427 | Wdr74 | NCBI_Gene:107071,ENSEMBL:ENSMUSG00000042729 | MGI:2147427 | protein coding gene | WD repeat domain 74 |
| 19 | gene | 8.740718 | 8.741225 | positive | MGI_C57BL6J_1921557 | 1700092M07Rik | NCBI_Gene:74307,ENSEMBL:ENSMUSG00000090840 | MGI:1921557 | protein coding gene | RIKEN cDNA 1700092M07 gene |
| 19 | gene | 8.741413 | 8.756069 | positive | MGI_C57BL6J_1928483 | Stx5a | NCBI_Gene:56389,ENSEMBL:ENSMUSG00000010110 | MGI:1928483 | protein coding gene | syntaxin 5A |
| 19 | gene | 8.742971 | 8.743079 | positive | MGI_C57BL6J_5531288 | Mir6992 | miRBase:MI0022840,NCBI_Gene:102465599,ENSEMBL:ENSMUSG00000098844 | MGI:5531288 | miRNA gene | microRNA 6992 |
| 19 | gene | 8.754789 | 8.756969 | negative | MGI_C57BL6J_1916695 | 1700023D09Rik | NCBI_Gene:102640359 | MGI:1916695 | lncRNA gene | RIKEN cDNA 1700023D09 gene |
| 19 | gene | 8.757073 | 8.770922 | positive | MGI_C57BL6J_1858330 | Nxf1 | NCBI_Gene:53319,ENSEMBL:ENSMUSG00000010097 | MGI:1858330 | protein coding gene | nuclear RNA export factor 1 |
| 19 | gene | 8.770996 | 8.772475 | positive | MGI_C57BL6J_1914086 | Tmem223 | NCBI_Gene:66836,ENSEMBL:ENSMUSG00000117924 | MGI:1914086 | protein coding gene | transmembrane protein 223 |
| 19 | gene | 8.772522 | 8.774467 | negative | MGI_C57BL6J_1914956 | Tmem179b | NCBI_Gene:67706,ENSEMBL:ENSMUSG00000118346 | MGI:1914956 | protein coding gene | transmembrane protein 179B |
| 19 | gene | 8.774354 | 8.786417 | negative | MGI_C57BL6J_2444957 | Taf6l | NCBI_Gene:225895,ENSEMBL:ENSMUSG00000003680 | MGI:2444957 | protein coding gene | TATA-box binding protein associated factor 6 like |
| 19 | pseudogene | 8.774485 | 8.776962 | positive | MGI_C57BL6J_3780685 | Gm2518 | NCBI_Gene:100039957,ENSEMBL:ENSMUSG00000118261 | MGI:3780685 | pseudogene | predicted gene 2518 |
| 19 | pseudogene | 8.790085 | 8.790464 | negative | MGI_C57BL6J_6302874 | Gm50131 | ENSEMBL:ENSMUSG00000118305 | MGI:6302874 | pseudogene | predicted gene, 50131 |
| 19 | gene | 8.793127 | 8.798577 | negative | MGI_C57BL6J_1914960 | Polr2g | NCBI_Gene:67710,ENSEMBL:ENSMUSG00000071662 | MGI:1914960 | protein coding gene | polymerase (RNA) II (DNA directed) polypeptide G |
| 19 | gene | 8.801979 | 8.804854 | positive | MGI_C57BL6J_1922541 | Zbtb3 | NCBI_Gene:75291,ENSEMBL:ENSMUSG00000071661 | MGI:1922541 | protein coding gene | zinc finger and BTB domain containing 3 |
| 19 | gene | 8.809066 | 8.819455 | negative | MGI_C57BL6J_1917637 | Ttc9c | NCBI_Gene:70387,ENSEMBL:ENSMUSG00000071660 | MGI:1917637 | protein coding gene | tetratricopeptide repeat domain 9C |
| 19 | gene | 8.819401 | 8.834143 | positive | MGI_C57BL6J_1915943 | Hnrnpul2 | NCBI_Gene:68693,ENSEMBL:ENSMUSG00000071659 | MGI:1915943 | protein coding gene | heterogeneous nuclear ribonucleoprotein U-like 2 |
| 19 | gene | 8.829773 | 8.841137 | positive | MGI_C57BL6J_6302888 | Gm50139 | ENSEMBL:ENSMUSG00000118124 | MGI:6302888 | protein coding gene | predicted gene, 50139 |
| 19 | gene | 8.836929 | 8.839246 | negative | MGI_C57BL6J_102704 | Gng3 | NCBI_Gene:14704,ENSEMBL:ENSMUSG00000071658 | MGI:102704 | protein coding gene | guanine nucleotide binding protein (G protein), gamma 3 |
| 19 | gene | 8.837467 | 8.848683 | positive | MGI_C57BL6J_1298392 | Bscl2 | NCBI_Gene:14705,ENSEMBL:ENSMUSG00000071657 | MGI:1298392 | protein coding gene | Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| 19 | gene | 8.850194 | 8.853914 | positive | MGI_C57BL6J_1916102 | Lrrn4cl | NCBI_Gene:68852,ENSEMBL:ENSMUSG00000071656 | MGI:1916102 | protein coding gene | LRRN4 C-terminal like |
| 19 | gene | 8.871496 | 8.875863 | positive | MGI_C57BL6J_1289301 | Ubxn1 | NCBI_Gene:225896,ENSEMBL:ENSMUSG00000071655 | MGI:1289301 | protein coding gene | UBX domain protein 1 |
| 19 | gene | 8.880014 | 8.880933 | negative | MGI_C57BL6J_2147553 | Uqcc3 | NCBI_Gene:107197,ENSEMBL:ENSMUSG00000071654 | MGI:2147553 | protein coding gene | ubiquinol-cytochrome c reductase complex assembly factor 3 |
| 19 | gene | 8.883732 | 8.892627 | positive | MGI_C57BL6J_5516029 | Lbhd1 | NCBI_Gene:102308570,ENSEMBL:ENSMUSG00000096740 | MGI:5516029 | protein coding gene | LBH domain containing 1 |
| 19 | gene | 8.883930 | 8.887258 | positive | MGI_C57BL6J_6155026 | Gm49403 | ENSEMBL:ENSMUSG00000116166 | MGI:6155026 | protein coding gene | predicted gene, 49403 |
| 19 | gene | 8.888387 | 8.888770 | negative | MGI_C57BL6J_1914781 | 5730408K05Rik | NCBI_Gene:67531 | MGI:1914781 | lncRNA gene | RIKEN cDNA 5730408K05 gene |
| 19 | gene | 8.888538 | 8.888685 | negative | MGI_C57BL6J_5455599 | Snora57 | ENSEMBL:ENSMUSG00000070167 | MGI:5455599 | snoRNA gene | small nucleolar RNA, H/ACA box 57 |
| 19 | gene | 8.888699 | 8.888774 | negative | MGI_C57BL6J_4950461 | Mir5136 | miRBase:MI0018048,NCBI_Gene:100628605,ENSEMBL:ENSMUSG00000093314 | MGI:4950461 | miRNA gene | microRNA 5136 |
| 19 | gene | 8.888853 | 8.890881 | positive | MGI_C57BL6J_1913526 | 1810009A15Rik | NCBI_Gene:66276,ENSEMBL:ENSMUSG00000071653 | MGI:1913526 | protein coding gene | RIKEN cDNA 1810009A15 gene |
| 19 | gene | 8.888926 | 8.895227 | positive | MGI_C57BL6J_6155046 | Gm49416 | ENSEMBL:ENSMUSG00000116347 | MGI:6155046 | protein coding gene | predicted gene, 49416 |
| 19 | gene | 8.892987 | 8.897890 | positive | MGI_C57BL6J_1923578 | Ints5 | NCBI_Gene:109077,ENSEMBL:ENSMUSG00000071652 | MGI:1923578 | protein coding gene | integrator complex subunit 5 |
| 19 | gene | 8.898057 | 8.916742 | positive | MGI_C57BL6J_1097667 | Ganab | NCBI_Gene:14376,ENSEMBL:ENSMUSG00000071650 | MGI:1097667 | protein coding gene | alpha glucosidase 2 alpha neutral subunit |
| 19 | gene | 8.918937 | 8.921842 | negative | MGI_C57BL6J_5595788 | Gm36629 | NCBI_Gene:102640604 | MGI:5595788 | lncRNA gene | predicted gene, 36629 |
| 19 | gene | 8.920374 | 8.927236 | positive | MGI_C57BL6J_1919977 | B3gat3 | NCBI_Gene:72727,ENSEMBL:ENSMUSG00000071649 | MGI:1919977 | protein coding gene | beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| 19 | gene | 8.927382 | 8.929356 | negative | MGI_C57BL6J_97998 | Rom1 | NCBI_Gene:19881,ENSEMBL:ENSMUSG00000071648 | MGI:97998 | protein coding gene | rod outer segment membrane protein 1 |
| 19 | gene | 8.929552 | 8.941582 | positive | MGI_C57BL6J_2387612 | Eml3 | NCBI_Gene:225898,ENSEMBL:ENSMUSG00000071647 | MGI:2387612 | protein coding gene | echinoderm microtubule associated protein like 3 |
| 19 | gene | 8.936755 | 8.940896 | negative | MGI_C57BL6J_3704491 | Gm10353 | NCBI_Gene:102640526,ENSEMBL:ENSMUSG00000114220 | MGI:3704491 | lncRNA gene | predicted gene 10353 |
| 19 | gene | 8.941875 | 8.952303 | positive | MGI_C57BL6J_1346340 | Mta2 | NCBI_Gene:23942,ENSEMBL:ENSMUSG00000071646 | MGI:1346340 | protein coding gene | metastasis-associated gene family, member 2 |
| 19 | gene | 8.953847 | 8.966210 | positive | MGI_C57BL6J_1917294 | Tut1 | NCBI_Gene:70044,ENSEMBL:ENSMUSG00000071645 | MGI:1917294 | protein coding gene | terminal uridylyl transferase 1, U6 snRNA-specific |
| 19 | gene | 8.967041 | 8.978479 | positive | MGI_C57BL6J_1914410 | Eef1g | NCBI_Gene:67160,ENSEMBL:ENSMUSG00000071644 | MGI:1914410 | protein coding gene | eukaryotic translation elongation factor 1 gamma |
| 19 | gene | 8.989121 | 9.076935 | positive | MGI_C57BL6J_1316648 | Ahnak | NCBI_Gene:66395,ENSEMBL:ENSMUSG00000069833 | MGI:1316648 | protein coding gene | AHNAK nucleoprotein (desmoyokin) |
| 19 | gene | 9.083636 | 9.087958 | negative | MGI_C57BL6J_98919 | Scgb1a1 | NCBI_Gene:22287,ENSEMBL:ENSMUSG00000024653 | MGI:98919 | protein coding gene | secretoglobin, family 1A, member 1 (uteroglobin) |
| 19 | gene | 9.099766 | 9.102711 | positive | MGI_C57BL6J_5624690 | Gm41805 | NCBI_Gene:105246530 | MGI:5624690 | lncRNA gene | predicted gene, 41805 |
| 19 | gene | 9.109868 | 9.135636 | negative | MGI_C57BL6J_1913764 | Asrgl1 | NCBI_Gene:66514,ENSEMBL:ENSMUSG00000024654 | MGI:1913764 | protein coding gene | asparaginase like 1 |
| 19 | gene | 9.135157 | 9.279175 | positive | MGI_C57BL6J_3642669 | Gm9750 | NCBI_Gene:105246531,ENSEMBL:ENSMUSG00000024658 | MGI:3642669 | lncRNA gene | predicted gene 9750 |
| 19 | gene | 9.185225 | 9.187019 | negative | MGI_C57BL6J_5621541 | Gm38656 | NCBI_Gene:102643088 | MGI:5621541 | protein coding gene | predicted gene, 38656 |
| 19 | pseudogene | 9.216311 | 9.216995 | positive | MGI_C57BL6J_3644748 | Gm6252 | NCBI_Gene:621699,ENSEMBL:ENSMUSG00000117804 | MGI:3644748 | pseudogene | predicted gene 6252 |
| 19 | gene | 9.234897 | 9.234978 | negative | MGI_C57BL6J_5454320 | Gm24543 | ENSEMBL:ENSMUSG00000077055 | MGI:5454320 | miRNA gene | predicted gene, 24543 |
| 19 | pseudogene | 9.283238 | 9.284495 | positive | MGI_C57BL6J_97505 | Pcna-ps2 | NCBI_Gene:18540,ENSEMBL:ENSMUSG00000067608 | MGI:97505 | pseudogene | proliferating cell nuclear antigen pseudogene 2 |
| 19 | pseudogene | 9.323219 | 9.339986 | negative | MGI_C57BL6J_3780785 | Gm2617 | ENSEMBL:ENSMUSG00000101398 | MGI:3780785 | pseudogene | predicted gene 2617 |
| 19 | pseudogene | 9.557606 | 9.559248 | negative | MGI_C57BL6J_3586869 | Stxbp3-ps | NCBI_Gene:619371,ENSEMBL:ENSMUSG00000071640 | MGI:3586869 | pseudogene | syntaxin-binding protein 3, pseudogene |
| 19 | pseudogene | 9.605121 | 9.614034 | positive | MGI_C57BL6J_5579937 | Gm29231 | ENSEMBL:ENSMUSG00000096818 | MGI:5579937 | pseudogene | predicted gene 29231 |
| 19 | pseudogene | 9.606968 | 9.608500 | positive | MGI_C57BL6J_3779598 | Gm6445 | NCBI_Gene:623688,ENSEMBL:ENSMUSG00000100141 | MGI:3779598 | pseudogene | predicted gene 6445 |
| 19 | pseudogene | 9.827117 | 9.827236 | positive | MGI_C57BL6J_6303180 | Gm50320 | ENSEMBL:ENSMUSG00000117756 | MGI:6303180 | pseudogene | predicted gene, 50320 |
| 19 | gene | 9.828899 | 9.836897 | negative | MGI_C57BL6J_6303181 | Gm50321 | ENSEMBL:ENSMUSG00000118383 | MGI:6303181 | lncRNA gene | predicted gene, 50321 |
| 19 | gene | 9.847435 | 9.852808 | positive | MGI_C57BL6J_3780828 | Scgb2a2 | NCBI_Gene:102639117,ENSEMBL:ENSMUSG00000096872 | MGI:3780828 | protein coding gene | secretoglobin, family 2A, member 2 |
| 19 | gene | 9.855319 | 9.855422 | positive | MGI_C57BL6J_5452146 | Gm22369 | ENSEMBL:ENSMUSG00000076370 | MGI:5452146 | miRNA gene | predicted gene, 22369 |
| 19 | gene | 9.864924 | 9.868995 | negative | MGI_C57BL6J_6303183 | Gm50322 | ENSEMBL:ENSMUSG00000118138 | MGI:6303183 | lncRNA gene | predicted gene, 50322 |
| 19 | gene | 9.872296 | 9.899551 | negative | MGI_C57BL6J_1313288 | Incenp | NCBI_Gene:16319,ENSEMBL:ENSMUSG00000024660 | MGI:1313288 | protein coding gene | inner centromere protein |
| 19 | pseudogene | 9.888477 | 9.889169 | positive | MGI_C57BL6J_5011239 | Gm19054 | NCBI_Gene:100418173 | MGI:5011239 | pseudogene | predicted gene, 19054 |
| 19 | gene | 9.897500 | 9.898707 | negative | MGI_C57BL6J_6303187 | Gm50324 | ENSEMBL:ENSMUSG00000118325 | MGI:6303187 | unclassified gene | predicted gene, 50324 |
| 19 | gene | 9.899690 | 9.902661 | positive | MGI_C57BL6J_6303216 | Gm50340 | ENSEMBL:ENSMUSG00000118100 | MGI:6303216 | lncRNA gene | predicted gene, 50340 |
| 19 | gene | 9.908158 | 9.908278 | positive | MGI_C57BL6J_5454261 | Gm24484 | ENSEMBL:ENSMUSG00000080659 | MGI:5454261 | miRNA gene | predicted gene, 24484 |
| 19 | gene | 9.924510 | 9.947628 | negative | MGI_C57BL6J_5596072 | Gm36913 | NCBI_Gene:102640980,ENSEMBL:ENSMUSG00000118008 | MGI:5596072 | lncRNA gene | predicted gene, 36913 |
| 19 | pseudogene | 9.937669 | 9.937833 | positive | MGI_C57BL6J_6303221 | Gm50343 | ENSEMBL:ENSMUSG00000118065 | MGI:6303221 | pseudogene | predicted gene, 50343 |
| 19 | gene | 9.939266 | 9.940652 | positive | MGI_C57BL6J_6303223 | Gm50345 | ENSEMBL:ENSMUSG00000118183 | MGI:6303223 | unclassified gene | predicted gene, 50345 |
| 19 | gene | 9.941349 | 9.942832 | negative | MGI_C57BL6J_6303225 | Gm50346 | ENSEMBL:ENSMUSG00000118123 | MGI:6303225 | unclassified gene | predicted gene, 50346 |
| 19 | gene | 9.970121 | 9.971605 | positive | MGI_C57BL6J_5624691 | Gm41806 | NCBI_Gene:105246532 | MGI:5624691 | lncRNA gene | predicted gene, 41806 |
| 19 | gene | 9.980598 | 9.985111 | positive | MGI_C57BL6J_95588 | Fth1 | NCBI_Gene:14319,ENSEMBL:ENSMUSG00000024661 | MGI:95588 | protein coding gene | ferritin heavy polypeptide 1 |
| 19 | gene | 9.985172 | 10.001633 | negative | MGI_C57BL6J_1346332 | Best1 | NCBI_Gene:24115,ENSEMBL:ENSMUSG00000037418 | MGI:1346332 | protein coding gene | bestrophin 1 |
| 19 | gene | 10.001669 | 10.038380 | positive | MGI_C57BL6J_1922010 | Rab3il1 | NCBI_Gene:74760,ENSEMBL:ENSMUSG00000024663 | MGI:1922010 | protein coding gene | RAB3A interacting protein (rabin3)-like 1 |
| 19 | gene | 10.040855 | 10.060110 | positive | MGI_C57BL6J_1928740 | Fads3 | NCBI_Gene:60527,ENSEMBL:ENSMUSG00000024664 | MGI:1928740 | protein coding gene | fatty acid desaturase 3 |
| 19 | gene | 10.046264 | 10.047940 | negative | MGI_C57BL6J_5589076 | Gm29917 | NCBI_Gene:102631622 | MGI:5589076 | lncRNA gene | predicted gene, 29917 |
| 19 | gene | 10.062765 | 10.101746 | negative | MGI_C57BL6J_1930079 | Fads2 | NCBI_Gene:56473,ENSEMBL:ENSMUSG00000024665 | MGI:1930079 | protein coding gene | fatty acid desaturase 2 |
| 19 | pseudogene | 10.175854 | 10.176161 | negative | MGI_C57BL6J_6303246 | Gm50359 | ENSEMBL:ENSMUSG00000117700 | MGI:6303246 | pseudogene | predicted gene, 50359 |
| 19 | gene | 10.182888 | 10.196877 | positive | MGI_C57BL6J_1923517 | Fads1 | NCBI_Gene:76267,ENSEMBL:ENSMUSG00000010663 | MGI:1923517 | protein coding gene | fatty acid desaturase 1 |
| 19 | gene | 10.191375 | 10.191439 | positive | MGI_C57BL6J_5531024 | Mir6993 | miRBase:MI0022841,NCBI_Gene:102465600,ENSEMBL:ENSMUSG00000098363 | MGI:5531024 | miRNA gene | microRNA 6993 |
| 19 | gene | 10.198368 | 10.202384 | positive | MGI_C57BL6J_3704492 | Gm10143 | ENSEMBL:ENSMUSG00000064032 | MGI:3704492 | lncRNA gene | predicted gene 10143 |
| 19 | gene | 10.199132 | 10.204187 | negative | MGI_C57BL6J_102779 | Fen1 | NCBI_Gene:14156,ENSEMBL:ENSMUSG00000024742 | MGI:102779 | protein coding gene | flap structure specific endonuclease 1 |
| 19 | gene | 10.204014 | 10.207824 | positive | MGI_C57BL6J_1916288 | Tmem258 | NCBI_Gene:69038,ENSEMBL:ENSMUSG00000036372 | MGI:1916288 | protein coding gene | transmembrane protein 258 |
| 19 | gene | 10.208271 | 10.240781 | negative | MGI_C57BL6J_2684944 | Myrf | NCBI_Gene:225908,ENSEMBL:ENSMUSG00000036098 | MGI:2684944 | protein coding gene | myelin regulatory factor |
| 19 | gene | 10.245265 | 10.304877 | negative | MGI_C57BL6J_2677061 | Dagla | NCBI_Gene:269060,ENSEMBL:ENSMUSG00000035735 | MGI:2677061 | protein coding gene | diacylglycerol lipase, alpha |
| 19 | gene | 10.290223 | 10.296440 | positive | MGI_C57BL6J_5589201 | Gm30042 | NCBI_Gene:102631796,ENSEMBL:ENSMUSG00000118330 | MGI:5589201 | lncRNA gene | predicted gene, 30042 |
| 19 | gene | 10.362065 | 10.366277 | negative | MGI_C57BL6J_5589422 | Gm30263 | NCBI_Gene:102632100 | MGI:5589422 | lncRNA gene | predicted gene, 30263 |
| 19 | gene | 10.388781 | 10.453181 | positive | MGI_C57BL6J_1859545 | Syt7 | NCBI_Gene:54525,ENSEMBL:ENSMUSG00000024743 | MGI:1859545 | protein coding gene | synaptotagmin VII |
| 19 | gene | 10.389130 | 10.391735 | negative | MGI_C57BL6J_5477353 | Gm26859 | ENSEMBL:ENSMUSG00000097862 | MGI:5477353 | lncRNA gene | predicted gene, 26859 |
| 19 | gene | 10.439897 | 10.442652 | positive | MGI_C57BL6J_6303093 | Gm50268 | ENSEMBL:ENSMUSG00000118310 | MGI:6303093 | unclassified gene | predicted gene, 50268 |
| 19 | gene | 10.455371 | 10.457473 | negative | MGI_C57BL6J_2685551 | Lrrc10b | NCBI_Gene:278795,ENSEMBL:ENSMUSG00000090291 | MGI:2685551 | protein coding gene | leucine rich repeat containing 10B |
| 19 | gene | 10.474235 | 10.491452 | negative | MGI_C57BL6J_1915002 | Ppp1r32 | NCBI_Gene:67752,ENSEMBL:ENSMUSG00000035179 | MGI:1915002 | protein coding gene | protein phosphatase 1, regulatory subunit 32 |
| 19 | gene | 10.500512 | 10.526142 | negative | MGI_C57BL6J_1913322 | Sdhaf2 | NCBI_Gene:66072,ENSEMBL:ENSMUSG00000024668 | MGI:1913322 | protein coding gene | succinate dehydrogenase complex assembly factor 2 |
| 19 | gene | 10.525244 | 10.547735 | positive | MGI_C57BL6J_1917826 | Cpsf7 | NCBI_Gene:269061,ENSEMBL:ENSMUSG00000034820 | MGI:1917826 | protein coding gene | cleavage and polyadenylation specific factor 7 |
| 19 | gene | 10.533865 | 10.556303 | negative | MGI_C57BL6J_1920020 | Tmem216 | NCBI_Gene:68642,ENSEMBL:ENSMUSG00000024667 | MGI:1920020 | protein coding gene | transmembrane protein 216 |
| 19 | gene | 10.570478 | 10.577386 | negative | MGI_C57BL6J_1920232 | Tmem138 | NCBI_Gene:72982,ENSEMBL:ENSMUSG00000024666 | MGI:1920232 | protein coding gene | transmembrane protein 138 |
| 19 | gene | 10.573512 | 10.576545 | positive | MGI_C57BL6J_6302988 | Gm50205 | ENSEMBL:ENSMUSG00000117955 | MGI:6302988 | unclassified gene | predicted gene, 50205 |
| 19 | gene | 10.577157 | 10.595961 | positive | MGI_C57BL6J_2686925 | Cyb561a3 | NCBI_Gene:225912,ENSEMBL:ENSMUSG00000034445 | MGI:2686925 | protein coding gene | cytochrome b561 family, member A3 |
| 19 | gene | 10.587791 | 10.605654 | negative | MGI_C57BL6J_2385084 | Tkfc | NCBI_Gene:225913,ENSEMBL:ENSMUSG00000034371 | MGI:2385084 | protein coding gene | triokinase, FMN cyclase |
| 19 | gene | 10.605327 | 10.629822 | positive | MGI_C57BL6J_1202384 | Ddb1 | NCBI_Gene:13194,ENSEMBL:ENSMUSG00000024740 | MGI:1202384 | protein coding gene | damage specific DNA binding protein 1 |
| 19 | gene | 10.634213 | 10.666213 | positive | MGI_C57BL6J_1919018 | Vwce | NCBI_Gene:71768,ENSEMBL:ENSMUSG00000043789 | MGI:1919018 | protein coding gene | von Willebrand factor C and EGF domains |
| 19 | gene | 10.645690 | 10.648053 | negative | MGI_C57BL6J_1922346 | 4930524O05Rik | NCBI_Gene:75096,ENSEMBL:ENSMUSG00000118392 | MGI:1922346 | lncRNA gene | RIKEN cDNA 4930524O05 gene |
| 19 | gene | 10.668956 | 10.678071 | negative | MGI_C57BL6J_1915935 | Pga5 | NCBI_Gene:58803,ENSEMBL:ENSMUSG00000024738 | MGI:1915935 | protein coding gene | pepsinogen 5, group I |
| 19 | gene | 10.684793 | 10.685558 | positive | MGI_C57BL6J_1919555 | 2210404E10Rik | NCBI_Gene:72305,ENSEMBL:ENSMUSG00000114772 | MGI:1919555 | lncRNA gene | RIKEN cDNA 2210404E10 gene |
| 19 | gene | 10.688728 | 10.714628 | positive | MGI_C57BL6J_2147661 | Vps37c | NCBI_Gene:107305,ENSEMBL:ENSMUSG00000048832 | MGI:2147661 | protein coding gene | vacuolar protein sorting 37C |
| 19 | gene | 10.717116 | 10.739026 | negative | MGI_C57BL6J_88340 | Cd5 | NCBI_Gene:12507,ENSEMBL:ENSMUSG00000024669 | MGI:88340 | protein coding gene | CD5 antigen |
| 19 | gene | 10.740947 | 10.786043 | positive | MGI_C57BL6J_2685520 | A430093F15Rik | NCBI_Gene:403202,ENSEMBL:ENSMUSG00000067577 | MGI:2685520 | lncRNA gene | RIKEN cDNA A430093F15 gene |
| 19 | gene | 10.789339 | 10.830063 | negative | MGI_C57BL6J_103566 | Cd6 | NCBI_Gene:12511,ENSEMBL:ENSMUSG00000024670 | MGI:103566 | protein coding gene | CD6 antigen |
| 19 | gene | 10.839727 | 10.859362 | positive | MGI_C57BL6J_1929691 | Slc15a3 | NCBI_Gene:65221,ENSEMBL:ENSMUSG00000024737 | MGI:1929691 | protein coding gene | solute carrier family 15, member 3 |
| 19 | gene | 10.840163 | 10.841762 | negative | MGI_C57BL6J_3641658 | Gm10802 | ENSEMBL:ENSMUSG00000118027 | MGI:3641658 | lncRNA gene | predicted gene 10802 |
| 19 | gene | 10.857822 | 10.869940 | negative | MGI_C57BL6J_2147810 | Tmem132a | NCBI_Gene:98170,ENSEMBL:ENSMUSG00000024736 | MGI:2147810 | protein coding gene | transmembrane protein 132A |
| 19 | gene | 10.870657 | 10.882001 | negative | MGI_C57BL6J_1915789 | Tmem109 | NCBI_Gene:68539,ENSEMBL:ENSMUSG00000034659 | MGI:1915789 | protein coding gene | transmembrane protein 109 |
| 19 | gene | 10.895231 | 10.909559 | positive | MGI_C57BL6J_106247 | Prpf19 | NCBI_Gene:28000,ENSEMBL:ENSMUSG00000024735 | MGI:106247 | protein coding gene | pre-mRNA processing factor 19 |
| 19 | gene | 10.914287 | 10.920632 | negative | MGI_C57BL6J_103073 | Zp1 | NCBI_Gene:22786,ENSEMBL:ENSMUSG00000024734 | MGI:103073 | protein coding gene | zona pellucida glycoprotein 1 |
| 19 | gene | 10.927861 | 10.942511 | positive | MGI_C57BL6J_1330275 | Ptgdr2 | NCBI_Gene:14764,ENSEMBL:ENSMUSG00000034117 | MGI:1330275 | protein coding gene | prostaglandin D2 receptor 2 |
| 19 | gene | 10.941481 | 10.949266 | negative | MGI_C57BL6J_1277220 | Ccdc86 | NCBI_Gene:108673,ENSEMBL:ENSMUSG00000024732 | MGI:1277220 | protein coding gene | coiled-coil domain containing 86 |
| 19 | gene | 10.962289 | 10.974725 | negative | MGI_C57BL6J_1917076 | Ms4a10 | NCBI_Gene:69826,ENSEMBL:ENSMUSG00000024731 | MGI:1917076 | protein coding gene | membrane-spanning 4-domains, subfamily A, member 10 |
| 19 | gene | 10.978303 | 10.993518 | negative | MGI_C57BL6J_3617853 | Ms4a15 | NCBI_Gene:545279,ENSEMBL:ENSMUSG00000067571 | MGI:3617853 | protein coding gene | membrane-spanning 4-domains, subfamily A, member 15 |
| 19 | gene | 10.997024 | 11.018031 | negative | MGI_C57BL6J_1923252 | Ms4a18 | NCBI_Gene:76002,ENSEMBL:ENSMUSG00000094584 | MGI:1923252 | protein coding gene | membrane-spanning 4-domains, subfamily A, member 18 |
| 19 | gene | 11.035415 | 11.037635 | negative | MGI_C57BL6J_5579053 | Gm28347 | NCBI_Gene:102632179,ENSEMBL:ENSMUSG00000101264 | MGI:5579053 | lncRNA gene | predicted gene 28347 |
| 19 | gene | 11.047612 | 11.055808 | negative | MGI_C57BL6J_2147706 | AW112010 | NCBI_Gene:107350,ENSEMBL:ENSMUSG00000075010 | MGI:2147706 | lncRNA gene | expressed sequence AW112010 |
| 19 | gene | 11.067471 | 11.081103 | negative | MGI_C57BL6J_1927657 | Ms4a8a | NCBI_Gene:64381,ENSEMBL:ENSMUSG00000024730 | MGI:1927657 | protein coding gene | membrane-spanning 4-domains, subfamily A, member 8A |
| 19 | pseudogene | 11.090596 | 11.092077 | negative | MGI_C57BL6J_2685182 | Gm336 | NCBI_Gene:240539,ENSEMBL:ENSMUSG00000101439 | MGI:2685182 | pseudogene | predicted pseudogene 336 |
| 19 | gene | 11.096814 | 11.130889 | negative | MGI_C57BL6J_1916619 | 1700017D01Rik | NCBI_Gene:69369,ENSEMBL:ENSMUSG00000024729 | MGI:1916619 | protein coding gene | RIKEN cDNA 1700017D01 gene |
| 19 | pseudogene | 11.121344 | 11.122425 | positive | MGI_C57BL6J_3645570 | Gm6501 | NCBI_Gene:624395,ENSEMBL:ENSMUSG00000099998 | MGI:3645570 | pseudogene | predicted gene 6501 |
| 19 | gene | 11.139662 | 11.165628 | negative | MGI_C57BL6J_1916666 | 1700025F22Rik | NCBI_Gene:69416,ENSEMBL:ENSMUSG00000024728 | MGI:1916666 | protein coding gene | RIKEN cDNA 1700025F22 gene |
| 19 | gene | 11.169418 | 11.196934 | negative | MGI_C57BL6J_1920716 | Ms4a13 | NCBI_Gene:73466,ENSEMBL:ENSMUSG00000057240 | MGI:1920716 | protein coding gene | membrane-spanning 4-domains, subfamily A, member 13 |
| 19 | gene | 11.196817 | 11.299886 | positive | MGI_C57BL6J_1922388 | 4930526L06Rik | NCBI_Gene:75138,ENSEMBL:ENSMUSG00000097091 | MGI:1922388 | lncRNA gene | RIKEN cDNA 4930526L06 gene |
| 19 | gene | 11.214082 | 11.232482 | negative | MGI_C57BL6J_2685812 | Ms4a12 | NCBI_Gene:381213,ENSEMBL:ENSMUSG00000101031 | MGI:2685812 | protein coding gene | membrane-spanning 4-domains, subfamily A, member 12 |
| 19 | gene | 11.249675 | 11.266241 | negative | MGI_C57BL6J_88321 | Ms4a1 | NCBI_Gene:12482,ENSEMBL:ENSMUSG00000024673 | MGI:88321 | protein coding gene | membrane-spanning 4-domains, subfamily A, member 1 |
| 19 | gene | 11.273238 | 11.283895 | negative | MGI_C57BL6J_2670985 | Ms4a5 | NCBI_Gene:269063,ENSEMBL:ENSMUSG00000054523 | MGI:2670985 | protein coding gene | membrane-spanning 4-domains, subfamily A, member 5 |
| 19 | gene | 11.301249 | 11.315875 | negative | MGI_C57BL6J_2686122 | Ms4a14 | NCBI_Gene:383435,ENSEMBL:ENSMUSG00000099398 | MGI:2686122 | protein coding gene | membrane-spanning 4-domains, subfamily A, member 14 |
| 19 | gene | 11.321039 | 11.336146 | negative | MGI_C57BL6J_1918846 | Ms4a7 | NCBI_Gene:109225,ENSEMBL:ENSMUSG00000024672 | MGI:1918846 | protein coding gene | membrane-spanning 4-domains, subfamily A, member 7 |
| 19 | gene | 11.362278 | 11.363890 | positive | MGI_C57BL6J_5579641 | Gm28935 | ENSEMBL:ENSMUSG00000100392 | MGI:5579641 | lncRNA gene | predicted gene 28935 |
| 19 | gene | 11.375488 | 11.392915 | positive | MGI_C57BL6J_3643932 | Ms4a4a | NCBI_Gene:666907,ENSEMBL:ENSMUSG00000101389 | MGI:3643932 | protein coding gene | membrane-spanning 4-domains, subfamily A, member 4A |
| 19 | pseudogene | 11.403948 | 11.405004 | negative | MGI_C57BL6J_3646968 | Gm5692 | NCBI_Gene:435588,ENSEMBL:ENSMUSG00000101024 | MGI:3646968 | pseudogene | predicted gene 5692 |
| 19 | gene | 11.404717 | 11.427246 | positive | MGI_C57BL6J_1927656 | Ms4a4c | NCBI_Gene:64380,ENSEMBL:ENSMUSG00000024675 | MGI:1927656 | protein coding gene | membrane-spanning 4-domains, subfamily A, member 4C |
| 19 | pseudogene | 11.438029 | 11.438832 | positive | MGI_C57BL6J_3779492 | Gm5511 | NCBI_Gene:433221,ENSEMBL:ENSMUSG00000100615 | MGI:3779492 | pseudogene | predicted gene 5511 |
| 19 | gene | 11.443553 | 11.467055 | positive | MGI_C57BL6J_1913083 | Ms4a4b | NCBI_Gene:60361,ENSEMBL:ENSMUSG00000056290 | MGI:1913083 | protein coding gene | membrane-spanning 4-domains, subfamily A, member 4B |
| 19 | gene | 11.469041 | 11.487069 | positive | MGI_C57BL6J_2385644 | Ms4a6c | NCBI_Gene:73656,ENSEMBL:ENSMUSG00000079419 | MGI:2385644 | protein coding gene | membrane-spanning 4-domains, subfamily A, member 6C |
| 19 | gene | 11.470166 | 11.484578 | negative | MGI_C57BL6J_3035348 | BE692007 | NCBI_Gene:100504727,ENSEMBL:ENSMUSG00000099757 | MGI:3035348 | lncRNA gene | expressed sequence BE692007 |
| 19 | gene | 11.485938 | 11.512577 | positive | MGI_C57BL6J_3645380 | Gm8369 | NCBI_Gene:666926,ENSEMBL:ENSMUSG00000058470 | MGI:3645380 | protein coding gene | predicted gene 8369 |
| 19 | gene | 11.516211 | 11.516953 | negative | MGI_C57BL6J_5011446 | Gm19261 | ENSEMBL:ENSMUSG00000100529 | MGI:5011446 | lncRNA gene | predicted gene, 19261 |
| 19 | gene | 11.516512 | 11.531256 | positive | MGI_C57BL6J_1917024 | Ms4a6b | NCBI_Gene:69774,ENSEMBL:ENSMUSG00000024677 | MGI:1917024 | protein coding gene | membrane-spanning 4-domains, subfamily A, member 6B |
| 19 | gene | 11.536801 | 11.558467 | positive | MGI_C57BL6J_1913857 | Ms4a4d | NCBI_Gene:66607,ENSEMBL:ENSMUSG00000024678 | MGI:1913857 | protein coding gene | membrane-spanning 4-domains, subfamily A, member 4D |
| 19 | pseudogene | 11.569439 | 11.573112 | positive | MGI_C57BL6J_3641904 | Gm10212 | NCBI_Gene:101055836,ENSEMBL:ENSMUSG00000067555 | MGI:3641904 | pseudogene | predicted pseudogene 10212 |
| 19 | gene | 11.582826 | 11.584357 | negative | MGI_C57BL6J_5611503 | Gm38275 | ENSEMBL:ENSMUSG00000104088 | MGI:5611503 | unclassified gene | predicted gene, 38275 |
| 19 | gene | 11.586604 | 11.604849 | negative | MGI_C57BL6J_1916024 | Ms4a6d | NCBI_Gene:68774,ENSEMBL:ENSMUSG00000024679 | MGI:1916024 | protein coding gene | membrane-spanning 4-domains, subfamily A, member 6D |
| 19 | gene | 11.607389 | 11.607438 | negative | MGI_C57BL6J_5454376 | Gm24599 | ENSEMBL:ENSMUSG00000088598 | MGI:5454376 | snoRNA gene | predicted gene, 24599 |
| 19 | gene | 11.615517 | 11.623790 | negative | MGI_C57BL6J_95495 | Ms4a2 | NCBI_Gene:14126,ENSEMBL:ENSMUSG00000024680 | MGI:95495 | protein coding gene | membrane-spanning 4-domains, subfamily A, member 2 |
| 19 | gene | 11.629496 | 11.640851 | negative | MGI_C57BL6J_2158468 | Ms4a3 | NCBI_Gene:170813,ENSEMBL:ENSMUSG00000024681 | MGI:2158468 | protein coding gene | membrane-spanning 4-domains, subfamily A, member 3 |
| 19 | gene | 11.647279 | 11.660559 | negative | MGI_C57BL6J_2684945 | Oosp2 | NCBI_Gene:225922,ENSEMBL:ENSMUSG00000055895 | MGI:2684945 | protein coding gene | oocyte secreted protein 2 |
| 19 | gene | 11.667460 | 11.691150 | negative | MGI_C57BL6J_2149290 | Oosp1 | NCBI_Gene:170834,ENSEMBL:ENSMUSG00000041857 | MGI:2149290 | protein coding gene | oocyte secreted protein 1 |
| 19 | gene | 11.695041 | 11.695179 | positive | MGI_C57BL6J_5455960 | Gm26183 | ENSEMBL:ENSMUSG00000088310 | MGI:5455960 | snoRNA gene | predicted gene, 26183 |
| 19 | gene | 11.695267 | 11.695342 | positive | MGI_C57BL6J_5455220 | Gm25443 | ENSEMBL:ENSMUSG00000089625 | MGI:5455220 | snoRNA gene | predicted gene, 25443 |
| 19 | gene | 11.697055 | 11.711874 | positive | MGI_C57BL6J_2684943 | Oosp3 | NCBI_Gene:225923,ENSEMBL:ENSMUSG00000055933 | MGI:2684943 | protein coding gene | oocyte secreted protein 3 |
| 19 | pseudogene | 11.703309 | 11.704101 | negative | MGI_C57BL6J_5011409 | Gm19224 | NCBI_Gene:100418458,ENSEMBL:ENSMUSG00000100918 | MGI:5011409 | pseudogene | predicted gene, 19224 |
| 19 | gene | 11.747543 | 11.763447 | positive | MGI_C57BL6J_1202394 | Cblif | NCBI_Gene:14603,ENSEMBL:ENSMUSG00000024682 | MGI:1202394 | protein coding gene | cobalamin binding intrinsic factor |
| 19 | gene | 11.760943 | 11.763189 | negative | MGI_C57BL6J_5826292 | Gm46655 | NCBI_Gene:108168416 | MGI:5826292 | lncRNA gene | predicted gene, 46655 |
| 19 | gene | 11.770391 | 11.774960 | positive | MGI_C57BL6J_2137219 | Mrpl16 | NCBI_Gene:94063,ENSEMBL:ENSMUSG00000024683 | MGI:2137219 | protein coding gene | mitochondrial ribosomal protein L16 |
| 19 | gene | 11.775118 | 11.819886 | negative | MGI_C57BL6J_103077 | Stx3 | NCBI_Gene:20908,ENSEMBL:ENSMUSG00000041488 | MGI:103077 | protein coding gene | syntaxin 3 |
| 19 | gene | 11.818245 | 11.831258 | positive | MGI_C57BL6J_5624695 | Gm41810 | NA | NA | lncRNA gene | predicted gene%2c 41810 |
| 19 | gene | 11.826803 | 11.839219 | negative | MGI_C57BL6J_3031251 | Olfr1417 | NCBI_Gene:258938,ENSEMBL:ENSMUSG00000048292 | MGI:3031251 | protein coding gene | olfactory receptor 1417 |
| 19 | gene | 11.852587 | 11.861066 | negative | MGI_C57BL6J_3031252 | Olfr1418 | NCBI_Gene:258227,ENSEMBL:ENSMUSG00000060556 | MGI:3031252 | protein coding gene | olfactory receptor 1418 |
| 19 | pseudogene | 11.864436 | 11.864634 | negative | MGI_C57BL6J_5753695 | Gm45119 | ENSEMBL:ENSMUSG00000108966 | MGI:5753695 | pseudogene | predicted gene 45119 |
| 19 | gene | 11.868613 | 11.875106 | negative | MGI_C57BL6J_3031253 | Olfr1419 | NCBI_Gene:257938,ENSEMBL:ENSMUSG00000067545 | MGI:3031253 | protein coding gene | olfactory receptor 1419 |
| 19 | pseudogene | 11.876645 | 11.878668 | negative | MGI_C57BL6J_3643502 | Gm6365 | NCBI_Gene:622845,ENSEMBL:ENSMUSG00000051548 | MGI:3643502 | pseudogene | predicted pseudogene 6365 |
| 19 | gene | 11.886565 | 11.898079 | positive | MGI_C57BL6J_3031254 | Olfr1420 | NCBI_Gene:258405,ENSEMBL:ENSMUSG00000060878 | MGI:3031254 | protein coding gene | olfactory receptor 1420 |
| 19 | gene | 11.895522 | 11.912231 | negative | MGI_C57BL6J_5590251 | Gm31092 | NCBI_Gene:102633205 | MGI:5590251 | lncRNA gene | predicted gene, 31092 |
| 19 | gene | 11.912399 | 11.945096 | positive | MGI_C57BL6J_2147679 | Patl1 | NCBI_Gene:225929,ENSEMBL:ENSMUSG00000046139 | MGI:2147679 | protein coding gene | protein associated with topoisomerase II homolog 1 (yeast) |
| 19 | gene | 11.923728 | 11.923800 | positive | MGI_C57BL6J_5562752 | Mir6994 | miRBase:MI0022842,NCBI_Gene:102465601,ENSEMBL:ENSMUSG00000104979 | MGI:5562752 | miRNA gene | microRNA 6994 |
| 19 | gene | 11.933495 | 11.933628 | negative | MGI_C57BL6J_5452049 | Gm22272 | ENSEMBL:ENSMUSG00000077227 | MGI:5452049 | snoRNA gene | predicted gene, 22272 |
| 19 | pseudogene | 11.958619 | 11.958994 | positive | MGI_C57BL6J_5826280 | Gm46643 | NCBI_Gene:108168403 | MGI:5826280 | pseudogene | predicted gene, 46643 |
| 19 | gene | 11.965844 | 11.994114 | positive | MGI_C57BL6J_97447 | Osbp | NCBI_Gene:76303,ENSEMBL:ENSMUSG00000024687 | MGI:97447 | protein coding gene | oxysterol binding protein |
| 19 | gene | 12.002236 | 12.002308 | positive | MGI_C57BL6J_4414001 | n-TFgaa6 | NCBI_Gene:102467583 | MGI:4414001 | tRNA gene | nuclear encoded tRNA pheylalanine 6 (anticodon GAA) |
| 19 | gene | 12.006039 | 12.006111 | positive | MGI_C57BL6J_4413976 | n-TKctt29 | NCBI_Gene:102467572 | MGI:4413976 | tRNA gene | nuclear encoded tRNA lysine 29 (anticodon TTT) |
| 19 | gene | 12.008545 | 12.008617 | positive | MGI_C57BL6J_4413997 | n-TFgaa2 | NCBI_Gene:102467361 | MGI:4413997 | tRNA gene | nuclear encoded tRNA pheylalanine 2 (anticodon GAA) |
| 19 | gene | 12.009696 | 12.009768 | negative | MGI_C57BL6J_4413977 | n-TKctt30 | NCBI_Gene:102467353 | MGI:4413977 | tRNA gene | nuclear encoded tRNA lysine 30 (anticodon TTT) |
| 19 | gene | 12.010115 | 12.018404 | positive | MGI_C57BL6J_5590310 | Gm31151 | NCBI_Gene:102633290 | MGI:5590310 | lncRNA gene | predicted gene, 31151 |
| 19 | gene | 12.011086 | 12.011168 | negative | MGI_C57BL6J_4413944 | n-TLtaa4 | NCBI_Gene:102467632 | MGI:4413944 | tRNA gene | nuclear encoded tRNA leucine 4 (anticodon TAA) |
| 19 | gene | 12.011588 | 12.011673 | negative | MGI_C57BL6J_4413734 | n-TRtct1 | NCBI_Gene:102467472 | MGI:4413734 | tRNA gene | nuclear encoded tRNA arginine 1 (anticodon TCT) |
| 19 | gene | 12.011931 | 12.012003 | positive | MGI_C57BL6J_4414104 | n-TVtac1 | NCBI_Gene:102467626 | MGI:4414104 | tRNA gene | nuclear encoded tRNA valine 1 (anticodon TAC) |
| 19 | gene | 12.012266 | 12.012338 | positive | MGI_C57BL6J_4414105 | n-TVtac2 | NCBI_Gene:102467409 | MGI:4414105 | tRNA gene | nuclear encoded tRNA valine 2 (anticodon TAC) |
| 19 | pseudogene | 12.020175 | 12.020403 | positive | MGI_C57BL6J_3031255 | Olfr1421-ps1 | NCBI_Gene:404514,ENSEMBL:ENSMUSG00000109030 | MGI:3031255 | pseudogene | olfactory receptor 1421, pseudogene 1 |
| 19 | pseudogene | 12.020505 | 12.020562 | negative | MGI_C57BL6J_5753691 | Gm45115 | ENSEMBL:ENSMUSG00000109047 | MGI:5753691 | pseudogene | predicted gene 45115 |
| 19 | pseudogene | 12.020614 | 12.021763 | negative | MGI_C57BL6J_3031256 | Olfr1422-ps1 | NCBI_Gene:258005,ENSEMBL:ENSMUSG00000108947 | MGI:3031256 | pseudogene | olfactory receptor 1422, pseudogene 1 |
| 19 | pseudogene | 12.026646 | 12.026920 | negative | MGI_C57BL6J_5753692 | Gm45116 | ENSEMBL:ENSMUSG00000109223 | MGI:5753692 | pseudogene | predicted gene 45116 |
| 19 | gene | 12.031787 | 12.041317 | negative | MGI_C57BL6J_3031257 | Olfr1423 | NCBI_Gene:258675,ENSEMBL:ENSMUSG00000067529 | MGI:3031257 | protein coding gene | olfactory receptor 1423 |
| 19 | pseudogene | 12.037731 | 12.037824 | positive | MGI_C57BL6J_5753435 | Gm44859 | ENSEMBL:ENSMUSG00000109417 | MGI:5753435 | pseudogene | predicted gene 44859 |
| 19 | gene | 12.058624 | 12.062682 | negative | MGI_C57BL6J_3031258 | Olfr1424 | NCBI_Gene:258676,ENSEMBL:ENSMUSG00000067528 | MGI:3031258 | protein coding gene | olfactory receptor 1424 |
| 19 | gene | 12.073234 | 12.077731 | negative | MGI_C57BL6J_3031259 | Olfr1425 | NCBI_Gene:258155,ENSEMBL:ENSMUSG00000067526 | MGI:3031259 | protein coding gene | olfactory receptor 1425 |
| 19 | gene | 12.086746 | 12.092013 | negative | MGI_C57BL6J_3031260 | Olfr1426 | NCBI_Gene:258805,ENSEMBL:ENSMUSG00000044994 | MGI:3031260 | protein coding gene | olfactory receptor 1426 |
| 19 | gene | 12.098006 | 12.103334 | negative | MGI_C57BL6J_3031261 | Olfr1427 | NCBI_Gene:258674,ENSEMBL:ENSMUSG00000067525 | MGI:3031261 | protein coding gene | olfactory receptor 1427 |
| 19 | gene | 12.107716 | 12.115897 | negative | MGI_C57BL6J_3031262 | Olfr1428 | NCBI_Gene:258673,ENSEMBL:ENSMUSG00000067524 | MGI:3031262 | protein coding gene | olfactory receptor 1428 |
| 19 | gene | 12.117996 | 12.123203 | negative | MGI_C57BL6J_2153205 | Olfr76 | NCBI_Gene:258677,ENSEMBL:ENSMUSG00000067522 | MGI:2153205 | protein coding gene | olfactory receptor 76 |
| 19 | pseudogene | 12.164424 | 12.170112 | negative | MGI_C57BL6J_3031389 | Olfr1555-ps1 | NCBI_Gene:257954,ENSEMBL:ENSMUSG00000108889 | MGI:3031389 | pseudogene | olfactory receptor 1555, pseudogene 1 |
| 19 | pseudogene | 12.177607 | 12.178523 | negative | MGI_C57BL6J_3031264 | Olfr1430-ps1 | NCBI_Gene:257992,ENSEMBL:ENSMUSG00000109410 | MGI:3031264 | pseudogene | olfactory receptor 1430, pseudogene 1 |
| 19 | gene | 12.200634 | 12.212191 | positive | MGI_C57BL6J_3031265 | Olfr1431 | NCBI_Gene:258409,ENSEMBL:ENSMUSG00000094133 | MGI:3031265 | protein coding gene | olfactory receptor 1431 |
| 19 | gene | 12.227755 | 12.232596 | negative | MGI_C57BL6J_3031266 | Olfr1432 | NCBI_Gene:257961,ENSEMBL:ENSMUSG00000109022 | MGI:3031266 | protein coding gene | olfactory receptor 1432 |
| 19 | gene | 12.240721 | 12.241659 | negative | MGI_C57BL6J_3030096 | Olfr262 | NCBI_Gene:258683,ENSEMBL:ENSMUSG00000067519 | MGI:3030096 | protein coding gene | olfactory receptor 262 |
| 19 | pseudogene | 12.247491 | 12.247646 | negative | MGI_C57BL6J_5477358 | Gm26864 | ENSEMBL:ENSMUSG00000097646 | MGI:5477358 | pseudogene | predicted gene, 26864 |
| 19 | gene | 12.261032 | 12.270387 | positive | MGI_C57BL6J_3030069 | Olfr235 | NCBI_Gene:258681,ENSEMBL:ENSMUSG00000060049 | MGI:3030069 | protein coding gene | olfactory receptor 235 |
| 19 | pseudogene | 12.273011 | 12.275157 | positive | MGI_C57BL6J_5009988 | Gm17802 | NCBI_Gene:100286833,ENSEMBL:ENSMUSG00000118093 | MGI:5009988 | pseudogene | predicted gene, 17802 |
| 19 | gene | 12.279854 | 12.285626 | positive | MGI_C57BL6J_3031268 | Olfr1434 | NCBI_Gene:258680,ENSEMBL:ENSMUSG00000095640 | MGI:3031268 | protein coding gene | olfactory receptor 1434 |
| 19 | pseudogene | 12.289854 | 12.290306 | negative | MGI_C57BL6J_3031269 | Olfr1435-ps1 | NCBI_Gene:258004,ENSEMBL:ENSMUSG00000109433 | MGI:3031269 | pseudogene | olfactory receptor 1435, pseudogene 1 |
| 19 | gene | 12.298183 | 12.299130 | negative | MGI_C57BL6J_3031270 | Olfr1436 | NCBI_Gene:258682,ENSEMBL:ENSMUSG00000067513 | MGI:3031270 | protein coding gene | olfactory receptor 1436 |
| 19 | pseudogene | 12.314874 | 12.315027 | negative | MGI_C57BL6J_5753434 | Gm44858 | ENSEMBL:ENSMUSG00000109229 | MGI:5753434 | pseudogene | predicted gene 44858 |
| 19 | gene | 12.319207 | 12.325101 | negative | MGI_C57BL6J_3031271 | Olfr1437 | NCBI_Gene:258117,ENSEMBL:ENSMUSG00000096436 | MGI:3031271 | protein coding gene | olfactory receptor 1437 |
| 19 | pseudogene | 12.329422 | 12.335910 | negative | MGI_C57BL6J_3031272 | Olfr1438-ps1 | NCBI_Gene:258116,ENSEMBL:ENSMUSG00000109328 | MGI:3031272 | pseudogene | olfactory receptor 1438, pseudogene 1 |
| 19 | pseudogene | 12.339738 | 12.341142 | negative | MGI_C57BL6J_3031273 | Olfr1439-ps1 | NCBI_Gene:258015,ENSEMBL:ENSMUSG00000109007 | MGI:3031273 | pseudogene | olfactory receptor 1439, pseudogene 1 |
| 19 | gene | 12.387279 | 12.401584 | positive | MGI_C57BL6J_3031274 | Olfr1440 | NCBI_Gene:258679,ENSEMBL:ENSMUSG00000046650 | MGI:3031274 | protein coding gene | olfactory receptor 1440 |
| 19 | gene | 12.417433 | 12.423569 | positive | MGI_C57BL6J_3031275 | Olfr1441 | NCBI_Gene:258678,ENSEMBL:ENSMUSG00000050815 | MGI:3031275 | protein coding gene | olfactory receptor 1441 |
| 19 | gene | 12.427905 | 12.432110 | positive | MGI_C57BL6J_1860266 | Pfpl | NCBI_Gene:56093,ENSEMBL:ENSMUSG00000040065 | MGI:1860266 | protein coding gene | pore forming protein-like |
| 19 | gene | 12.460709 | 12.465285 | positive | MGI_C57BL6J_1333743 | Mpeg1 | NCBI_Gene:17476,ENSEMBL:ENSMUSG00000046805 | MGI:1333743 | protein coding gene | macrophage expressed gene 1 |
| 19 | gene | 12.466329 | 12.502201 | negative | MGI_C57BL6J_2672905 | Dtx4 | NCBI_Gene:207521,ENSEMBL:ENSMUSG00000039982 | MGI:2672905 | protein coding gene | deltex 4, E3 ubiquitin ligase |
| 19 | gene | 12.517915 | 12.596566 | negative | MGI_C57BL6J_3698434 | A330040F15Rik | NCBI_Gene:74333,ENSEMBL:ENSMUSG00000086213 | MGI:3698434 | lncRNA gene | RIKEN cDNA A330040F15 gene |
| 19 | gene | 12.517921 | 12.524338 | negative | MGI_C57BL6J_5590367 | Gm31208 | NCBI_Gene:102633365 | MGI:5590367 | lncRNA gene | predicted gene, 31208 |
| 19 | gene | 12.519800 | 12.523576 | positive | MGI_C57BL6J_6096066 | Gm47243 | ENSEMBL:ENSMUSG00000114644 | MGI:6096066 | lncRNA gene | predicted gene, 47243 |
| 19 | pseudogene | 12.528755 | 12.530540 | negative | MGI_C57BL6J_3643874 | Gm6545 | NCBI_Gene:625046,ENSEMBL:ENSMUSG00000111118 | MGI:3643874 | pseudogene | predicted gene 6545 |
| 19 | gene | 12.539588 | 12.540823 | negative | MGI_C57BL6J_6096064 | Gm47242 | ENSEMBL:ENSMUSG00000114761 | MGI:6096064 | lncRNA gene | predicted gene, 47242 |
| 19 | gene | 12.545740 | 12.589769 | positive | MGI_C57BL6J_1915508 | Fam111a | NCBI_Gene:107373,ENSEMBL:ENSMUSG00000024691 | MGI:1915508 | protein coding gene | family with sequence similarity 111, member A |
| 19 | pseudogene | 12.568188 | 12.568836 | negative | MGI_C57BL6J_3826552 | Gm16274 | NCBI_Gene:100418049,ENSEMBL:ENSMUSG00000084836 | MGI:3826552 | pseudogene | predicted gene 16274 |
| 19 | gene | 12.595749 | 12.598309 | positive | MGI_C57BL6J_1921583 | 4122401K19Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 4122401K19 gene |
| 19 | gene | 12.599974 | 12.628251 | positive | MGI_C57BL6J_3643569 | Gm4952 | NCBI_Gene:240549,ENSEMBL:ENSMUSG00000071633 | MGI:3643569 | protein coding gene | predicted gene 4952 |
| 19 | gene | 12.633308 | 12.653911 | positive | MGI_C57BL6J_2147502 | Glyat | NCBI_Gene:107146,ENSEMBL:ENSMUSG00000063683 | MGI:2147502 | protein coding gene | glycine-N-acyltransferase |
| 19 | gene | 12.665255 | 12.665361 | positive | MGI_C57BL6J_5454298 | Gm24521 | ENSEMBL:ENSMUSG00000065892 | MGI:5454298 | snRNA gene | predicted gene, 24521 |
| 19 | gene | 12.670439 | 12.677277 | positive | MGI_C57BL6J_3031276 | Olfr1442 | NCBI_Gene:258692,ENSEMBL:ENSMUSG00000044441 | MGI:3031276 | protein coding gene | olfactory receptor 1442 |
| 19 | gene | 12.678112 | 12.683851 | positive | MGI_C57BL6J_3031277 | Olfr1443 | NCBI_Gene:258693,ENSEMBL:ENSMUSG00000045030 | MGI:3031277 | protein coding gene | olfactory receptor 1443 |
| 19 | pseudogene | 12.681771 | 12.681915 | positive | MGI_C57BL6J_6215285 | Gm49772 | ENSEMBL:ENSMUSG00000109309 | MGI:6215285 | pseudogene | predicted gene, 49772 |
| 19 | gene | 12.695786 | 12.719902 | positive | MGI_C57BL6J_1928492 | Keg1 | NCBI_Gene:64697,ENSEMBL:ENSMUSG00000024694 | MGI:1928492 | protein coding gene | kidney expressed gene 1 |
| 19 | gene | 12.706055 | 12.721247 | negative | MGI_C57BL6J_3802141 | Gm15962 | ENSEMBL:ENSMUSG00000084981 | MGI:3802141 | lncRNA gene | predicted gene 15962 |
| 19 | pseudogene | 12.713227 | 12.713948 | negative | MGI_C57BL6J_5010728 | Gm18543 | NCBI_Gene:100417340 | MGI:5010728 | pseudogene | predicted gene, 18543 |
| 19 | gene | 12.763528 | 12.765632 | negative | MGI_C57BL6J_88439 | Cntf | NCBI_Gene:12803,ENSEMBL:ENSMUSG00000079415 | MGI:88439 | protein coding gene | ciliary neurotrophic factor |
| 19 | gene | 12.763528 | 12.796123 | negative | MGI_C57BL6J_3845910 | Gm44505 | NCBI_Gene:664779 | MGI:3845910 | lncRNA gene | predicted readthrough transcript (NMD candidate), 44505 |
| 19 | gene | 12.763660 | 12.796126 | negative | MGI_C57BL6J_104854 | Zfp91 | NCBI_Gene:109910,ENSEMBL:ENSMUSG00000024695 | MGI:104854 | protein coding gene | zinc finger protein 91 |
| 19 | gene | 12.796193 | 12.833808 | positive | MGI_C57BL6J_2147677 | Lpxn | NCBI_Gene:107321,ENSEMBL:ENSMUSG00000024696 | MGI:2147677 | protein coding gene | leupaxin |
| 19 | gene | 12.846682 | 12.847424 | positive | MGI_C57BL6J_3646958 | Gm5244 | NCBI_Gene:383436,ENSEMBL:ENSMUSG00000091014 | MGI:3646958 | protein coding gene | predicted pseudogene 5244 |
| 19 | gene | 12.857283 | 12.863440 | positive | MGI_C57BL6J_3031278 | Olfr1444 | NCBI_Gene:258697,ENSEMBL:ENSMUSG00000046272 | MGI:3031278 | protein coding gene | olfactory receptor 1444 |
| 19 | gene | 12.883855 | 12.884855 | positive | MGI_C57BL6J_3031279 | Olfr1445 | NCBI_Gene:258694,ENSEMBL:ENSMUSG00000045126 | MGI:3031279 | protein coding gene | olfactory receptor 1445 |
| 19 | gene | 12.888266 | 12.893524 | negative | MGI_C57BL6J_3031280 | Olfr1446 | NCBI_Gene:258699,ENSEMBL:ENSMUSG00000057817 | MGI:3031280 | protein coding gene | olfactory receptor 1446 |
| 19 | gene | 12.899434 | 12.903743 | negative | MGI_C57BL6J_3031281 | Olfr1447 | NCBI_Gene:258698,ENSEMBL:ENSMUSG00000060303 | MGI:3031281 | protein coding gene | olfactory receptor 1447 |
| 19 | pseudogene | 12.905231 | 12.906993 | positive | MGI_C57BL6J_3645436 | Gm5512 | NCBI_Gene:433224,ENSEMBL:ENSMUSG00000112693 | MGI:3645436 | pseudogene | predicted gene 5512 |
| 19 | gene | 12.916661 | 12.924419 | negative | MGI_C57BL6J_3031282 | Olfr1448 | NCBI_Gene:258696,ENSEMBL:ENSMUSG00000048456 | MGI:3031282 | protein coding gene | olfactory receptor 1448 |
| 19 | gene | 12.930840 | 12.935752 | positive | MGI_C57BL6J_3031283 | Olfr1449 | NCBI_Gene:258300,ENSEMBL:ENSMUSG00000049498 | MGI:3031283 | protein coding gene | olfactory receptor 1449 |
| 19 | gene | 12.950258 | 12.956406 | positive | MGI_C57BL6J_3031284 | Olfr1450 | NCBI_Gene:258368,ENSEMBL:ENSMUSG00000062892 | MGI:3031284 | protein coding gene | olfactory receptor 1450 |
| 19 | gene | 12.995014 | 13.000417 | positive | MGI_C57BL6J_3031285 | Olfr1451 | NCBI_Gene:258700,ENSEMBL:ENSMUSG00000046913 | MGI:3031285 | protein coding gene | olfactory receptor 1451 |
| 19 | pseudogene | 13.011704 | 13.012668 | negative | MGI_C57BL6J_5011317 | Gm19132 | NCBI_Gene:100418307 | MGI:5011317 | pseudogene | predicted gene, 19132 |
| 19 | pseudogene | 13.016225 | 13.017149 | positive | MGI_C57BL6J_3031286 | Olfr1452-ps1 | NCBI_Gene:258126,ENSEMBL:ENSMUSG00000109148 | MGI:3031286 | polymorphic pseudogene | olfactory receptor 1452, pseudogene 1 |
| 19 | gene | 13.027404 | 13.028327 | negative | MGI_C57BL6J_3031287 | Olfr1453 | NCBI_Gene:258695,ENSEMBL:ENSMUSG00000094755 | MGI:3031287 | protein coding gene | olfactory receptor 1453 |
| 19 | gene | 13.060610 | 13.067157 | positive | MGI_C57BL6J_3031288 | Olfr1454 | NCBI_Gene:258687,ENSEMBL:ENSMUSG00000094986 | MGI:3031288 | protein coding gene | olfactory receptor 1454 |
| 19 | pseudogene | 13.068904 | 13.069677 | positive | MGI_C57BL6J_3031289 | Olfr1455-ps1 | NCBI_Gene:404515,ENSEMBL:ENSMUSG00000109126 | MGI:3031289 | pseudogene | olfactory receptor 1455, pseudogene 1 |
| 19 | pseudogene | 13.078637 | 13.079335 | negative | MGI_C57BL6J_3031290 | Olfr1456-ps1 | NCBI_Gene:404516,ENSEMBL:ENSMUSG00000067482 | MGI:3031290 | pseudogene | olfactory receptor 1456, pseudogene 1 |
| 19 | gene | 13.093379 | 13.097891 | negative | MGI_C57BL6J_3031291 | Olfr1457 | NCBI_Gene:258568,ENSEMBL:ENSMUSG00000061637 | MGI:3031291 | protein coding gene | olfactory receptor 1457 |
| 19 | gene | 13.101593 | 13.106775 | negative | MGI_C57BL6J_3031292 | Olfr1458 | NCBI_Gene:667271,ENSEMBL:ENSMUSG00000062844 | MGI:3031292 | protein coding gene | olfactory receptor 1458 |
| 19 | gene | 13.138021 | 13.149549 | negative | MGI_C57BL6J_3031293 | Olfr1459 | NCBI_Gene:258684,ENSEMBL:ENSMUSG00000057503 | MGI:3031293 | protein coding gene | olfactory receptor 1459 |
| 19 | pseudogene | 13.158191 | 13.159316 | positive | MGI_C57BL6J_3031294 | Olfr1460-ps1 | NCBI_Gene:258118,ENSEMBL:ENSMUSG00000109415 | MGI:3031294 | pseudogene | olfactory receptor 1460, pseudogene 1 |
| 19 | gene | 13.163000 | 13.168130 | positive | MGI_C57BL6J_3031295 | Olfr1461 | NCBI_Gene:258299,ENSEMBL:ENSMUSG00000045883 | MGI:3031295 | protein coding gene | olfactory receptor 1461 |
| 19 | gene | 13.185734 | 13.193775 | positive | MGI_C57BL6J_3031296 | Olfr1462 | NCBI_Gene:258688,ENSEMBL:ENSMUSG00000094721 | MGI:3031296 | protein coding gene | olfactory receptor 1462 |
| 19 | gene | 13.231328 | 13.236400 | positive | MGI_C57BL6J_3031297 | Olfr1463 | NCBI_Gene:258120,ENSEMBL:ENSMUSG00000096365 | MGI:3031297 | protein coding gene | olfactory receptor 1463 |
| 19 | pseudogene | 13.282134 | 13.283056 | negative | MGI_C57BL6J_3031298 | Olfr1464-ps1 | NCBI_Gene:667300,ENSEMBL:ENSMUSG00000109100 | MGI:3031298 | polymorphic pseudogene | olfactory receptor 1464, pseudogene 1 |
| 19 | gene | 13.301001 | 13.301107 | negative | MGI_C57BL6J_5452220 | Gm22443 | ENSEMBL:ENSMUSG00000064824 | MGI:5452220 | snRNA gene | predicted gene, 22443 |
| 19 | gene | 13.312768 | 13.315541 | negative | MGI_C57BL6J_3031299 | Olfr1465 | NCBI_Gene:258121,ENSEMBL:ENSMUSG00000062199 | MGI:3031299 | protein coding gene | olfactory receptor 1465 |
| 19 | gene | 13.338862 | 13.343572 | positive | MGI_C57BL6J_3031300 | Olfr1466 | NCBI_Gene:258689,ENSEMBL:ENSMUSG00000096485 | MGI:3031300 | protein coding gene | olfactory receptor 1466 |
| 19 | gene | 13.362236 | 13.368054 | positive | MGI_C57BL6J_3031301 | Olfr1467 | NCBI_Gene:258686,ENSEMBL:ENSMUSG00000049015 | MGI:3031301 | protein coding gene | olfactory receptor 1467 |
| 19 | pseudogene | 13.371854 | 13.376066 | positive | MGI_C57BL6J_3031302 | Olfr1468-ps1 | NCBI_Gene:258192,ENSEMBL:ENSMUSG00000109544 | MGI:3031302 | polymorphic pseudogene | olfactory receptor 1468, pseudogene 1 |
| 19 | pseudogene | 13.403157 | 13.403640 | positive | MGI_C57BL6J_5753425 | Gm44849 | ENSEMBL:ENSMUSG00000108881 | MGI:5753425 | pseudogene | predicted gene 44849 |
| 19 | gene | 13.407849 | 13.414540 | positive | MGI_C57BL6J_3031303 | Olfr1469 | NCBI_Gene:258690,ENSEMBL:ENSMUSG00000063777 | MGI:3031303 | protein coding gene | olfactory receptor 1469 |
| 19 | pseudogene | 13.434147 | 13.435095 | negative | MGI_C57BL6J_3031304 | Olfr1470-ps1 | NCBI_Gene:258119,ENSEMBL:ENSMUSG00000109230 | MGI:3031304 | pseudogene | olfactory receptor 1470, pseudogene 1 |
| 19 | gene | 13.443409 | 13.451527 | positive | MGI_C57BL6J_3031305 | Olfr1471 | NCBI_Gene:258231,ENSEMBL:ENSMUSG00000096320 | MGI:3031305 | protein coding gene | olfactory receptor 1471 |
| 19 | gene | 13.453280 | 13.456086 | negative | MGI_C57BL6J_3031306 | Olfr1472 | NCBI_Gene:258685,ENSEMBL:ENSMUSG00000095189 | MGI:3031306 | protein coding gene | olfactory receptor 1472 |
| 19 | pseudogene | 13.462678 | 13.463635 | negative | MGI_C57BL6J_3031307 | Olfr1473-ps1 | NCBI_Gene:258124,ENSEMBL:ENSMUSG00000109573 | MGI:3031307 | pseudogene | olfactory receptor 1473, pseudogene 1 |
| 19 | gene | 13.469565 | 13.472157 | positive | MGI_C57BL6J_3031308 | Olfr1474 | NCBI_Gene:258123,ENSEMBL:ENSMUSG00000096273 | MGI:3031308 | protein coding gene | olfactory receptor 1474 |
| 19 | gene | 13.479252 | 13.480196 | negative | MGI_C57BL6J_3031309 | Olfr1475 | NCBI_Gene:258298,ENSEMBL:ENSMUSG00000096708 | MGI:3031309 | protein coding gene | olfactory receptor 1475 |
| 19 | pseudogene | 13.488309 | 13.488741 | positive | MGI_C57BL6J_3031310 | Olfr1476-ps1 | NCBI_Gene:404517,ENSEMBL:ENSMUSG00000109189 | MGI:3031310 | pseudogene | olfactory receptor 1476, pseudogene 1 |
| 19 | gene | 13.500546 | 13.506192 | positive | MGI_C57BL6J_3031311 | Olfr1477 | NCBI_Gene:258691,ENSEMBL:ENSMUSG00000071629 | MGI:3031311 | protein coding gene | olfactory receptor 1477 |
| 19 | pseudogene | 13.505325 | 13.505715 | positive | MGI_C57BL6J_3031312 | Olfr1478-ps1 | NCBI_Gene:404518,ENSEMBL:ENSMUSG00000109184 | MGI:3031312 | pseudogene | olfactory receptor 1478, pseudogene 1 |
| 19 | pseudogene | 13.511433 | 13.511931 | positive | MGI_C57BL6J_3031313 | Olfr1479-ps1 | NCBI_Gene:404519,ENSEMBL:ENSMUSG00000108887 | MGI:3031313 | pseudogene | olfactory receptor 1479, pseudogene 1 |
| 19 | pseudogene | 13.512553 | 13.515138 | negative | MGI_C57BL6J_3781367 | Gm3188 | NCBI_Gene:100041185,ENSEMBL:ENSMUSG00000118174 | MGI:3781367 | pseudogene | predicted gene 3188 |
| 19 | gene | 13.527913 | 13.531275 | positive | MGI_C57BL6J_3031314 | Olfr1480 | NCBI_Gene:404339,ENSEMBL:ENSMUSG00000095484 | MGI:3031314 | protein coding gene | olfactory receptor 1480 |
| 19 | pseudogene | 13.535390 | 13.535725 | positive | MGI_C57BL6J_3031315 | Olfr1481-ps1 | NCBI_Gene:404520,ENSEMBL:ENSMUSG00000108945 | MGI:3031315 | pseudogene | olfactory receptor 1481, pseudogene 1 |
| 19 | pseudogene | 13.563036 | 13.563213 | negative | MGI_C57BL6J_3031316 | Olfr1482-ps1 | NCBI_Gene:404521,ENSEMBL:ENSMUSG00000109319 | MGI:3031316 | pseudogene | olfactory receptor 1482, pseudogene 1 |
| 19 | pseudogene | 13.566758 | 13.567178 | positive | MGI_C57BL6J_3031317 | Olfr1483-ps1 | NCBI_Gene:404522,ENSEMBL:ENSMUSG00000108974 | MGI:3031317 | pseudogene | olfactory receptor 1483, pseudogene 1 |
| 19 | gene | 13.583511 | 13.588832 | positive | MGI_C57BL6J_3031318 | Olfr1484 | NCBI_Gene:258288,ENSEMBL:ENSMUSG00000096289 | MGI:3031318 | protein coding gene | olfactory receptor 1484 |
| 19 | pseudogene | 13.588290 | 13.588659 | positive | MGI_C57BL6J_3031319 | Olfr1485-ps1 | NCBI_Gene:404523,ENSEMBL:ENSMUSG00000109496 | MGI:3031319 | pseudogene | olfactory receptor 1485, pseudogene 1 |
| 19 | pseudogene | 13.593860 | 13.594231 | positive | MGI_C57BL6J_3031320 | Olfr1486-ps1 | NCBI_Gene:404524,ENSEMBL:ENSMUSG00000109343 | MGI:3031320 | pseudogene | olfactory receptor 1486, pseudogene 1 |
| 19 | pseudogene | 13.595018 | 13.596063 | negative | MGI_C57BL6J_3779948 | Gm9538 | NCBI_Gene:671653 | MGI:3779948 | pseudogene | predicted gene 9538 |
| 19 | gene | 13.599660 | 13.599768 | negative | MGI_C57BL6J_5452100 | Gm22323 | ENSEMBL:ENSMUSG00000094077 | MGI:5452100 | miRNA gene | predicted gene, 22323 |
| 19 | gene | 13.604999 | 13.605107 | negative | MGI_C57BL6J_5453708 | Gm23931 | ENSEMBL:ENSMUSG00000096298 | MGI:5453708 | miRNA gene | predicted gene, 23931 |
| 19 | gene | 13.617375 | 13.623072 | positive | MGI_C57BL6J_3031321 | Olfr1487 | NCBI_Gene:258629,ENSEMBL:ENSMUSG00000094846 | MGI:3031321 | protein coding gene | olfactory receptor 1487 |
| 19 | pseudogene | 13.625625 | 13.625944 | positive | MGI_C57BL6J_3031322 | Olfr1488-ps1 | NCBI_Gene:257768,ENSEMBL:ENSMUSG00000109285 | MGI:3031322 | pseudogene | olfactory receptor 1488, pseudogene 1 |
| 19 | gene | 13.631621 | 13.643987 | positive | MGI_C57BL6J_3031323 | Olfr1489 | NCBI_Gene:258628,ENSEMBL:ENSMUSG00000045678 | MGI:3031323 | protein coding gene | olfactory receptor 1489 |
| 19 | gene | 13.635583 | 13.635714 | negative | MGI_C57BL6J_5453901 | Gm24124 | ENSEMBL:ENSMUSG00000077291 | MGI:5453901 | snoRNA gene | predicted gene, 24124 |
| 19 | gene | 13.651091 | 13.659545 | positive | MGI_C57BL6J_3031324 | Olfr1490 | NCBI_Gene:258098,ENSEMBL:ENSMUSG00000061387 | MGI:3031324 | protein coding gene | olfactory receptor 1490 |
| 19 | gene | 13.697268 | 13.706127 | positive | MGI_C57BL6J_3031325 | Olfr1491 | NCBI_Gene:258342,ENSEMBL:ENSMUSG00000051156 | MGI:3031325 | protein coding gene | olfactory receptor 1491 |
| 19 | pseudogene | 13.718013 | 13.718877 | positive | MGI_C57BL6J_3031326 | Olfr1492-ps1 | NCBI_Gene:404526,ENSEMBL:ENSMUSG00000109527 | MGI:3031326 | pseudogene | olfactory receptor 1492, pseudogene 1 |
| 19 | pseudogene | 13.723278 | 13.727983 | positive | MGI_C57BL6J_3031327 | Olfr1493-ps1 | NCBI_Gene:258735,ENSEMBL:ENSMUSG00000109520 | MGI:3031327 | pseudogene | olfactory receptor 1493, pseudogene 1 |
| 19 | gene | 13.739728 | 13.750479 | positive | MGI_C57BL6J_3031328 | Olfr1494 | NCBI_Gene:258992,ENSEMBL:ENSMUSG00000050865 | MGI:3031328 | protein coding gene | olfactory receptor 1494 |
| 19 | gene | 13.764606 | 13.770248 | positive | MGI_C57BL6J_3031329 | Olfr1495 | NCBI_Gene:258341,ENSEMBL:ENSMUSG00000047207 | MGI:3031329 | protein coding gene | olfactory receptor 1495 |
| 19 | gene | 13.777878 | 13.782920 | positive | MGI_C57BL6J_3031330 | Olfr1496 | NCBI_Gene:258991,ENSEMBL:ENSMUSG00000048356 | MGI:3031330 | protein coding gene | olfactory receptor 1496 |
| 19 | pseudogene | 13.791965 | 13.792514 | positive | MGI_C57BL6J_5011403 | Gm19218 | NCBI_Gene:100418446 | MGI:5011403 | pseudogene | predicted gene, 19218 |
| 19 | gene | 13.794324 | 13.798202 | negative | MGI_C57BL6J_3031331 | Olfr1497 | NCBI_Gene:258736,ENSEMBL:ENSMUSG00000044040 | MGI:3031331 | protein coding gene | olfactory receptor 1497 |
| 19 | pseudogene | 13.804428 | 13.806257 | negative | MGI_C57BL6J_3031332 | Olfr1498-ps1 | NCBI_Gene:258217,ENSEMBL:ENSMUSG00000109530 | MGI:3031332 | pseudogene | olfactory receptor 1498, pseudogene 1 |
| 19 | gene | 13.814275 | 13.819447 | negative | MGI_C57BL6J_3031333 | Olfr1499 | NCBI_Gene:258792,ENSEMBL:ENSMUSG00000045395 | MGI:3031333 | protein coding gene | olfactory receptor 1499 |
| 19 | gene | 13.826882 | 13.830488 | negative | MGI_C57BL6J_3031334 | Olfr1500 | NCBI_Gene:258097,ENSEMBL:ENSMUSG00000054526 | MGI:3031334 | protein coding gene | olfactory receptor 1500 |
| 19 | gene | 13.835033 | 13.850692 | negative | MGI_C57BL6J_3031335 | Olfr1501 | NCBI_Gene:258626,ENSEMBL:ENSMUSG00000057270 | MGI:3031335 | protein coding gene | olfactory receptor 1501 |
| 19 | gene | 13.861795 | 13.862745 | positive | MGI_C57BL6J_3031336 | Olfr1502 | NCBI_Gene:258793,ENSEMBL:ENSMUSG00000056858 | MGI:3031336 | protein coding gene | olfactory receptor 1502 |
| 19 | pseudogene | 13.877451 | 13.878253 | positive | MGI_C57BL6J_3031337 | Olfr1503-ps1 | NCBI_Gene:404527,ENSEMBL:ENSMUSG00000058078 | MGI:3031337 | pseudogene | olfactory receptor 1503, pseudogene 1 |
| 19 | gene | 13.886920 | 13.897928 | negative | MGI_C57BL6J_3031338 | Olfr1504 | NCBI_Gene:258627,ENSEMBL:ENSMUSG00000059105 | MGI:3031338 | protein coding gene | olfactory receptor 1504 |
| 19 | gene | 13.913530 | 13.921866 | positive | MGI_C57BL6J_3031339 | Olfr1505 | NCBI_Gene:258151,ENSEMBL:ENSMUSG00000062314 | MGI:3031339 | protein coding gene | olfactory receptor 1505 |
| 19 | pseudogene | 13.938563 | 13.939288 | positive | MGI_C57BL6J_3761697 | Vmn2r-ps135 | NCBI_Gene:100124582 | MGI:3761697 | pseudogene | vomeronasal 2, receptor, pseudogene 135 |
| 19 | pseudogene | 14.017583 | 14.018371 | negative | MGI_C57BL6J_5504108 | Gm26993 | ENSEMBL:ENSMUSG00000098074 | MGI:5504108 | pseudogene | predicted gene, 26993 |
| 19 | gene | 14.083631 | 14.084378 | positive | MGI_C57BL6J_1922297 | 4930504B16Rik | NA | NA | unclassified gene | RIKEN cDNA 4930504B16 gene |
| 19 | gene | 14.090247 | 14.096731 | negative | MGI_C57BL6J_5590490 | Gm31331 | NCBI_Gene:102633524 | MGI:5590490 | lncRNA gene | predicted gene, 31331 |
| 19 | gene | 14.397487 | 14.435950 | negative | MGI_C57BL6J_3647677 | Gm8630 | NCBI_Gene:667433 | MGI:3647677 | unclassified gene | predicted gene 8630 |
| 19 | gene | 14.448072 | 14.598183 | negative | MGI_C57BL6J_104633 | Tle4 | NCBI_Gene:21888,ENSEMBL:ENSMUSG00000024642 | MGI:104633 | protein coding gene | transducin-like enhancer of split 4 |
| 19 | gene | 14.564458 | 14.566993 | positive | MGI_C57BL6J_5590600 | Gm31441 | NCBI_Gene:102633674 | MGI:5590600 | lncRNA gene | predicted gene, 31441 |
| 19 | gene | 14.658667 | 14.658933 | positive | MGI_C57BL6J_5611225 | Gm37997 | ENSEMBL:ENSMUSG00000103488 | MGI:5611225 | unclassified gene | predicted gene, 37997 |
| 19 | gene | 14.718358 | 14.718480 | negative | MGI_C57BL6J_5455803 | Gm26026 | ENSEMBL:ENSMUSG00000084675 | MGI:5455803 | snRNA gene | predicted gene, 26026 |
| 19 | pseudogene | 14.959002 | 14.960782 | negative | MGI_C57BL6J_3645437 | Gm5513 | NCBI_Gene:433225,ENSEMBL:ENSMUSG00000063586 | MGI:3645437 | pseudogene | predicted pseudogene 5513 |
| 19 | pseudogene | 15.193521 | 15.194233 | negative | MGI_C57BL6J_3647037 | Gm7159 | NCBI_Gene:635497 | MGI:3647037 | pseudogene | predicted gene 7159 |
| 19 | gene | 15.290167 | 15.340387 | negative | MGI_C57BL6J_5826282 | Gm46645 | NCBI_Gene:108168406 | MGI:5826282 | lncRNA gene | predicted gene, 46645 |
| 19 | pseudogene | 15.362335 | 15.362870 | positive | MGI_C57BL6J_104962 | Pomc-ps1 | NCBI_Gene:18977 | MGI:104962 | pseudogene | pro-opiomelanocortin, pseudogene 1 |
| 19 | pseudogene | 15.458964 | 15.459672 | negative | MGI_C57BL6J_5011482 | Selenok-ps3 | NCBI_Gene:100502633 | MGI:5011482 | pseudogene | selenoprotein K, pseudogene 3 |
| 19 | gene | 15.599131 | 15.632101 | positive | MGI_C57BL6J_5590753 | Gm31594 | NCBI_Gene:102633870 | MGI:5590753 | lncRNA gene | predicted gene, 31594 |
| 19 | gene | 15.636548 | 15.662923 | positive | MGI_C57BL6J_5826302 | Gm46665 | NCBI_Gene:108168427 | MGI:5826302 | lncRNA gene | predicted gene, 46665 |
| 19 | gene | 15.659946 | 15.664346 | negative | MGI_C57BL6J_5590892 | Gm31733 | NCBI_Gene:102634054 | MGI:5590892 | lncRNA gene | predicted gene, 31733 |
| 19 | gene | 15.680004 | 15.680110 | positive | MGI_C57BL6J_5454096 | Gm24319 | ENSEMBL:ENSMUSG00000094850 | MGI:5454096 | snRNA gene | predicted gene, 24319 |
| 19 | gene | 15.803058 | 15.807653 | positive | MGI_C57BL6J_6303228 | Gm50348 | ENSEMBL:ENSMUSG00000117995 | MGI:6303228 | lncRNA gene | predicted gene, 50348 |
| 19 | gene | 15.904678 | 15.947337 | negative | MGI_C57BL6J_2183441 | Psat1 | NCBI_Gene:107272,ENSEMBL:ENSMUSG00000024640 | MGI:2183441 | protein coding gene | phosphoserine aminotransferase 1 |
| 19 | pseudogene | 15.943610 | 15.944165 | positive | MGI_C57BL6J_3781507 | Gm3329 | ENSEMBL:ENSMUSG00000090147 | MGI:3781507 | pseudogene | predicted gene 3329 |
| 19 | gene | 15.955770 | 15.985016 | negative | MGI_C57BL6J_1924386 | Cep78 | NCBI_Gene:208518,ENSEMBL:ENSMUSG00000041491 | MGI:1924386 | protein coding gene | centrosomal protein 78 |
| 19 | gene | 15.985075 | 16.010912 | positive | MGI_C57BL6J_3041192 | C130060C02Rik | NCBI_Gene:100502653,ENSEMBL:ENSMUSG00000097557 | MGI:3041192 | lncRNA gene | RIKEN cDNA C130060C02 gene |
| 19 | pseudogene | 16.025451 | 16.025762 | positive | MGI_C57BL6J_3646957 | Rpl37-ps1 | NCBI_Gene:383438,ENSEMBL:ENSMUSG00000117711 | MGI:3646957 | pseudogene | ribosomal protein 37, pseudogene 1 |
| 19 | gene | 16.064113 | 16.090591 | positive | MGI_C57BL6J_5591185 | Gm32026 | NCBI_Gene:102634445 | MGI:5591185 | lncRNA gene | predicted gene, 32026 |
| 19 | gene | 16.132831 | 16.387463 | positive | MGI_C57BL6J_95776 | Gnaq | NCBI_Gene:14682,ENSEMBL:ENSMUSG00000024639 | MGI:95776 | protein coding gene | guanine nucleotide binding protein, alpha q polypeptide |
| 19 | pseudogene | 16.164098 | 16.170458 | negative | MGI_C57BL6J_2442009 | E030024N20Rik | NCBI_Gene:595139,ENSEMBL:ENSMUSG00000057990 | MGI:2442009 | pseudogene | RIKEN cDNA E030024N20 gene |
| 19 | pseudogene | 16.266432 | 16.267635 | negative | MGI_C57BL6J_3642135 | Gm10819 | NCBI_Gene:624438,ENSEMBL:ENSMUSG00000075268 | MGI:3642135 | pseudogene | predicted gene 10819 |
| 19 | pseudogene | 16.277779 | 16.278974 | positive | MGI_C57BL6J_4937895 | Gm17068 | NCBI_Gene:100158259,ENSEMBL:ENSMUSG00000078528 | MGI:4937895 | pseudogene | predicted gene 17068 |
| 19 | gene | 16.314892 | 16.315004 | negative | MGI_C57BL6J_5531410 | Mir496b | miRBase:MI0021942,NCBI_Gene:102465218,ENSEMBL:ENSMUSG00000098718 | MGI:5531410 | miRNA gene | microRNA 496b |
| 19 | pseudogene | 16.413431 | 16.414290 | positive | MGI_C57BL6J_5011497 | Gm19312 | NCBI_Gene:100502667,ENSEMBL:ENSMUSG00000118142 | MGI:5011497 | pseudogene | predicted gene, 19312 |
| 19 | gene | 16.415826 | 16.422829 | negative | MGI_C57BL6J_6303250 | Gm50362 | ENSEMBL:ENSMUSG00000118033 | MGI:6303250 | lncRNA gene | predicted gene, 50362 |
| 19 | gene | 16.435667 | 16.610820 | positive | MGI_C57BL6J_95769 | Gna14 | NCBI_Gene:14675,ENSEMBL:ENSMUSG00000024697 | MGI:95769 | protein coding gene | guanine nucleotide binding protein, alpha 14 |
| 19 | pseudogene | 16.477107 | 16.491122 | negative | MGI_C57BL6J_3643833 | Gm8222 | NCBI_Gene:666665,ENSEMBL:ENSMUSG00000117699 | MGI:3643833 | pseudogene | predicted gene 8222 |
| 19 | pseudogene | 16.572160 | 16.572468 | negative | MGI_C57BL6J_6303252 | Gm50363 | ENSEMBL:ENSMUSG00000118023 | MGI:6303252 | pseudogene | predicted gene, 50363 |
| 19 | gene | 16.580085 | 16.583462 | negative | MGI_C57BL6J_5624696 | Gm41811 | NCBI_Gene:105246537 | MGI:5624696 | lncRNA gene | predicted gene, 41811 |
| 19 | gene | 16.612196 | 16.614869 | negative | MGI_C57BL6J_2442532 | E230008J23Rik | NA | NA | unclassified gene | RIKEN cDNA E230008J23 gene |
| 19 | gene | 16.615366 | 16.781878 | negative | MGI_C57BL6J_2444304 | Vps13a | NCBI_Gene:271564,ENSEMBL:ENSMUSG00000046230 | MGI:2444304 | protein coding gene | vacuolar protein sorting 13A |
| 19 | pseudogene | 16.715686 | 16.716289 | positive | MGI_C57BL6J_5591415 | Gm32256 | NCBI_Gene:102634749,ENSEMBL:ENSMUSG00000114931 | MGI:5591415 | pseudogene | predicted gene, 32256 |
| 19 | pseudogene | 16.780547 | 16.781103 | positive | MGI_C57BL6J_3708646 | Gm9806 | ENSEMBL:ENSMUSG00000046388 | MGI:3708646 | pseudogene | predicted gene 9806 |
| 19 | gene | 16.871092 | 16.925014 | positive | MGI_C57BL6J_5591500 | Gm32341 | NCBI_Gene:102634857,ENSEMBL:ENSMUSG00000118126 | MGI:5591500 | lncRNA gene | predicted gene, 32341 |
| 19 | gene | 16.872316 | 16.873830 | negative | MGI_C57BL6J_1347468 | Foxb2 | NCBI_Gene:14240,ENSEMBL:ENSMUSG00000056829 | MGI:1347468 | protein coding gene | forkhead box B2 |
| 19 | gene | 16.955670 | 17.223932 | positive | MGI_C57BL6J_1925004 | Prune2 | NCBI_Gene:353211,ENSEMBL:ENSMUSG00000039126 | MGI:1925004 | protein coding gene | prune homolog 2 |
| 19 | gene | 17.056781 | 17.057187 | negative | MGI_C57BL6J_5531352 | Gm27970 | ENSEMBL:ENSMUSG00000098298 | MGI:5531352 | unclassified non-coding RNA gene | predicted gene, 27970 |
| 19 | gene | 17.060137 | 17.060397 | negative | MGI_C57BL6J_5531178 | Gm27796 | ENSEMBL:ENSMUSG00000099316 | MGI:5531178 | unclassified non-coding RNA gene | predicted gene, 27796 |
| 19 | gene | 17.238047 | 17.240669 | negative | MGI_C57BL6J_6303107 | Gm50276 | ENSEMBL:ENSMUSG00000117941 | MGI:6303107 | lncRNA gene | predicted gene, 50276 |
| 19 | gene | 17.244010 | 17.256488 | negative | MGI_C57BL6J_6303109 | Gm50277 | ENSEMBL:ENSMUSG00000118367 | MGI:6303109 | lncRNA gene | predicted gene, 50277 |
| 19 | pseudogene | 17.265984 | 17.266338 | negative | MGI_C57BL6J_6303114 | Gm50280 | ENSEMBL:ENSMUSG00000117946 | MGI:6303114 | pseudogene | predicted gene, 50280 |
| 19 | gene | 17.326141 | 17.372844 | negative | MGI_C57BL6J_95676 | Gcnt1 | NCBI_Gene:14537,ENSEMBL:ENSMUSG00000038843 | MGI:95676 | protein coding gene | glucosaminyl (N-acetyl) transferase 1, core 2 |
| 19 | gene | 17.394043 | 17.401349 | positive | MGI_C57BL6J_1914688 | Rfk | NCBI_Gene:54391,ENSEMBL:ENSMUSG00000024712 | MGI:1914688 | protein coding gene | riboflavin kinase |
| 19 | gene | 17.432314 | 17.837632 | negative | MGI_C57BL6J_97515 | Pcsk5 | NCBI_Gene:18552,ENSEMBL:ENSMUSG00000024713 | MGI:97515 | protein coding gene | proprotein convertase subtilisin/kexin type 5 |
| 19 | gene | 17.536046 | 17.555945 | positive | MGI_C57BL6J_5624698 | Gm41813 | NCBI_Gene:105246539 | MGI:5624698 | lncRNA gene | predicted gene, 41813 |
| 19 | gene | 17.573784 | 17.580645 | positive | MGI_C57BL6J_5624697 | Gm41812 | NCBI_Gene:105246538 | MGI:5624697 | lncRNA gene | predicted gene, 41812 |
| 19 | gene | 17.632559 | 17.633491 | negative | MGI_C57BL6J_1925784 | C430005N20Rik | NA | NA | unclassified gene | RIKEN cDNA C430005N20 gene |
| 19 | pseudogene | 17.692763 | 17.693534 | positive | MGI_C57BL6J_5010004 | Gm17819 | NCBI_Gene:100310868,ENSEMBL:ENSMUSG00000118270 | MGI:5010004 | pseudogene | predicted gene, 17819 |
| 19 | gene | 17.865317 | 17.865419 | negative | MGI_C57BL6J_5452657 | Gm22880 | ENSEMBL:ENSMUSG00000088370 | MGI:5452657 | snRNA gene | predicted gene, 22880 |
| 19 | gene | 17.873829 | 17.893500 | positive | MGI_C57BL6J_6302952 | Gm50181 | ENSEMBL:ENSMUSG00000118079 | MGI:6302952 | lncRNA gene | predicted gene, 50181 |
| 19 | gene | 17.891959 | 17.892051 | negative | MGI_C57BL6J_5452840 | Gm23063 | ENSEMBL:ENSMUSG00000076305 | MGI:5452840 | miRNA gene | predicted gene, 23063 |
| 19 | pseudogene | 17.926557 | 17.928613 | positive | MGI_C57BL6J_2388571 | Eef1a1-ps1 | NCBI_Gene:235503,ENSEMBL:ENSMUSG00000117822 | MGI:2388571 | pseudogene | eukaryotic translation elongation factor 1 alpha 1, pseudogene 1 |
| 19 | gene | 17.928111 | 17.928226 | positive | MGI_C57BL6J_5452838 | Gm23061 | ENSEMBL:ENSMUSG00000076303 | MGI:5452838 | miRNA gene | predicted gene, 23061 |
| 19 | gene | 18.028111 | 18.040803 | positive | MGI_C57BL6J_6302954 | Gm50182 | ENSEMBL:ENSMUSG00000117687 | MGI:6302954 | lncRNA gene | predicted gene, 50182 |
| 19 | pseudogene | 18.205148 | 18.205795 | negative | MGI_C57BL6J_5010795 | Gm18610 | NCBI_Gene:100417435,ENSEMBL:ENSMUSG00000117774 | MGI:5010795 | pseudogene | predicted gene, 18610 |
| 19 | pseudogene | 18.480076 | 18.480748 | positive | MGI_C57BL6J_6302956 | Gm50183 | ENSEMBL:ENSMUSG00000118192 | MGI:6302956 | pseudogene | predicted gene, 50183 |
| 19 | gene | 18.516137 | 18.631823 | negative | MGI_C57BL6J_700012 | Ostf1 | NCBI_Gene:20409,ENSEMBL:ENSMUSG00000024725 | MGI:700012 | protein coding gene | osteoclast stimulating factor 1 |
| 19 | pseudogene | 18.565138 | 18.565755 | positive | MGI_C57BL6J_3779480 | Rpl29-ps6 | NCBI_Gene:383440 | MGI:3779480 | pseudogene | ribosomal protein L29, pseudogene 6 |
| 19 | pseudogene | 18.581455 | 18.582228 | positive | MGI_C57BL6J_3647615 | Gm8250 | ENSEMBL:ENSMUSG00000080783 | MGI:3647615 | pseudogene | predicted gene 8250 |
| 19 | gene | 18.627447 | 18.627921 | negative | MGI_C57BL6J_2147638 | 1110034N17Rik | NA | NA | unclassified gene | RIKEN cDNA 1110034N17 gene |
| 19 | gene | 18.631950 | 18.652428 | positive | MGI_C57BL6J_2147434 | Nmrk1 | NCBI_Gene:225994,ENSEMBL:ENSMUSG00000037847 | MGI:2147434 | protein coding gene | nicotinamide riboside kinase 1 |
| 19 | gene | 18.670734 | 18.707201 | positive | MGI_C57BL6J_1914633 | Carnmt1 | NCBI_Gene:67383,ENSEMBL:ENSMUSG00000024726 | MGI:1914633 | protein coding gene | carnosine N-methyltransferase 1 |
| 19 | gene | 18.705798 | 18.707200 | positive | MGI_C57BL6J_1918901 | 4930447K04Rik | NA | NA | unclassified gene | RIKEN cDNA 4930447K04 gene |
| 19 | gene | 18.712849 | 18.718429 | positive | MGI_C57BL6J_3583960 | D030056L22Rik | NCBI_Gene:225995,ENSEMBL:ENSMUSG00000047044 | MGI:3583960 | protein coding gene | RIKEN cDNA D030056L22 gene |
| 19 | gene | 18.730177 | 18.892521 | positive | MGI_C57BL6J_2675603 | Trpm6 | NCBI_Gene:225997,ENSEMBL:ENSMUSG00000024727 | MGI:2675603 | protein coding gene | transient receptor potential cation channel, subfamily M, member 6 |
| 19 | gene | 18.731873 | 18.732776 | negative | MGI_C57BL6J_6302983 | Gm50202 | ENSEMBL:ENSMUSG00000117878 | MGI:6302983 | lncRNA gene | predicted gene, 50202 |
| 19 | gene | 18.732741 | 18.732904 | positive | MGI_C57BL6J_5453015 | Gm23238 | ENSEMBL:ENSMUSG00000064941 | MGI:5453015 | snRNA gene | predicted gene, 23238 |
| 19 | gene | 18.736739 | 18.749516 | negative | MGI_C57BL6J_5624699 | Gm41814 | NCBI_Gene:105246540 | MGI:5624699 | lncRNA gene | predicted gene, 41814 |
| 19 | gene | 18.776746 | 18.776961 | negative | MGI_C57BL6J_5453967 | Gm24190 | ENSEMBL:ENSMUSG00000088601 | MGI:5453967 | unclassified non-coding RNA gene | predicted gene, 24190 |
| 19 | gene | 18.930605 | 19.111196 | negative | MGI_C57BL6J_1343464 | Rorb | NCBI_Gene:225998,ENSEMBL:ENSMUSG00000036192 | MGI:1343464 | protein coding gene | RAR-related orphan receptor beta |
| 19 | gene | 19.244035 | 19.276348 | negative | MGI_C57BL6J_6303001 | Gm50212 | ENSEMBL:ENSMUSG00000117909 | MGI:6303001 | lncRNA gene | predicted gene, 50212 |
| 19 | gene | 19.275899 | 19.276030 | positive | MGI_C57BL6J_5455214 | Gm25437 | ENSEMBL:ENSMUSG00000088907 | MGI:5455214 | snoRNA gene | predicted gene, 25437 |
| 19 | gene | 19.276471 | 19.319651 | positive | MGI_C57BL6J_5624700 | Gm41815 | NCBI_Gene:105246541,ENSEMBL:ENSMUSG00000118109 | MGI:5624700 | lncRNA gene | predicted gene, 41815 |
| 19 | gene | 19.821035 | 19.979149 | positive | MGI_C57BL6J_5591628 | Gm32469 | NCBI_Gene:102635029 | MGI:5591628 | lncRNA gene | predicted gene, 32469 |
| 19 | gene | 19.871432 | 19.876563 | negative | MGI_C57BL6J_6303008 | Gm50216 | ENSEMBL:ENSMUSG00000118167 | MGI:6303008 | lncRNA gene | predicted gene, 50216 |
| 19 | gene | 20.005940 | 20.060758 | negative | MGI_C57BL6J_3781903 | Gm3728 | NCBI_Gene:100042218,ENSEMBL:ENSMUSG00000118150 | MGI:3781903 | lncRNA gene | predicted gene 3728 |
| 19 | gene | 20.033504 | 20.033635 | negative | MGI_C57BL6J_5452461 | Gm22684 | ENSEMBL:ENSMUSG00000064783 | MGI:5452461 | snoRNA gene | predicted gene, 22684 |
| 19 | gene | 20.117385 | 20.283256 | negative | MGI_C57BL6J_5591788 | Gm32629 | NCBI_Gene:102635239,ENSEMBL:ENSMUSG00000118067 | MGI:5591788 | lncRNA gene | predicted gene, 32629 |
| 19 | gene | 20.324856 | 20.444883 | positive | MGI_C57BL6J_1916244 | 1500015L24Rik | NCBI_Gene:68994,ENSEMBL:ENSMUSG00000094732 | MGI:1916244 | lncRNA gene | RIKEN cDNA 1500015L24 gene |
| 19 | gene | 20.373428 | 20.390944 | negative | MGI_C57BL6J_96819 | Anxa1 | NCBI_Gene:16952,ENSEMBL:ENSMUSG00000024659 | MGI:96819 | protein coding gene | annexin A1 |
| 19 | gene | 20.410544 | 20.552330 | negative | MGI_C57BL6J_2443493 | C730002L08Rik | NCBI_Gene:320155,ENSEMBL:ENSMUSG00000114469 | MGI:2443493 | lncRNA gene | RIKEN cDNA C730002L08 gene |
| 19 | gene | 20.424651 | 20.435261 | negative | MGI_C57BL6J_5591848 | Gm32689 | NCBI_Gene:102635317 | MGI:5591848 | lncRNA gene | predicted gene, 32689 |
| 19 | gene | 20.437754 | 20.441386 | positive | MGI_C57BL6J_5624701 | Gm41816 | NCBI_Gene:105246542 | MGI:5624701 | lncRNA gene | predicted gene, 41816 |
| 19 | gene | 20.492715 | 20.556410 | positive | MGI_C57BL6J_2443348 | E030003E18Rik | NCBI_Gene:320092 | MGI:2443348 | lncRNA gene | RIKEN cDNA E030003E18 gene |
| 19 | gene | 20.492715 | 20.643465 | positive | MGI_C57BL6J_1353450 | Aldh1a1 | NCBI_Gene:11668,ENSEMBL:ENSMUSG00000053279 | MGI:1353450 | protein coding gene | aldehyde dehydrogenase family 1, subfamily A1 |
| 19 | gene | 20.583524 | 20.585067 | positive | MGI_C57BL6J_1917662 | 2900002J02Rik | NA | NA | unclassified gene | RIKEN cDNA 2900002J02 gene |
| 19 | pseudogene | 20.585913 | 20.586826 | negative | MGI_C57BL6J_3643522 | Gm6684 | NCBI_Gene:626549,ENSEMBL:ENSMUSG00000047168 | MGI:3643522 | pseudogene | predicted pseudogene 6684 |
| 19 | gene | 20.689467 | 20.690527 | negative | MGI_C57BL6J_3641948 | C730037M02Rik | NA | NA | unclassified gene | RIKEN cDNA C730037M02 gene |
| 19 | gene | 20.692953 | 20.727562 | negative | MGI_C57BL6J_1347050 | Aldh1a7 | NCBI_Gene:26358,ENSEMBL:ENSMUSG00000024747 | MGI:1347050 | protein coding gene | aldehyde dehydrogenase family 1, subfamily A7 |
| 19 | gene | 20.783456 | 21.037147 | negative | MGI_C57BL6J_2151016 | Tmc1 | NCBI_Gene:13409,ENSEMBL:ENSMUSG00000024749 | MGI:2151016 | protein coding gene | transmembrane channel-like gene family 1 |
| 19 | gene | 20.922392 | 20.954762 | positive | MGI_C57BL6J_5591909 | Gm32750 | NCBI_Gene:102635397,ENSEMBL:ENSMUSG00000118293 | MGI:5591909 | lncRNA gene | predicted gene, 32750 |
| 19 | gene | 20.956541 | 20.961920 | positive | MGI_C57BL6J_5624702 | Gm41817 | NCBI_Gene:105246543 | MGI:5624702 | lncRNA gene | predicted gene, 41817 |
| 19 | gene | 21.104502 | 21.107157 | positive | MGI_C57BL6J_1922604 | 4930554I06Rik | NCBI_Gene:75354,ENSEMBL:ENSMUSG00000118047 | MGI:1922604 | lncRNA gene | RIKEN cDNA 4930554I06 gene |
| 19 | gene | 21.121191 | 21.125705 | negative | MGI_C57BL6J_3781949 | Gm3775 | NCBI_Gene:100042294 | MGI:3781949 | lncRNA gene | predicted gene 3775 |
| 19 | gene | 21.138695 | 21.146597 | positive | MGI_C57BL6J_5592138 | Gm32979 | NCBI_Gene:102635711 | MGI:5592138 | lncRNA gene | predicted gene, 32979 |
| 19 | gene | 21.165087 | 21.210686 | negative | MGI_C57BL6J_5825555 | Gm45918 | NCBI_Gene:105242408 | MGI:5825555 | lncRNA gene | predicted gene, 45918 |
| 19 | gene | 21.208477 | 21.211973 | positive | MGI_C57BL6J_6303041 | Gm50236 | ENSEMBL:ENSMUSG00000118090 | MGI:6303041 | lncRNA gene | predicted gene, 50236 |
| 19 | pseudogene | 21.261976 | 21.263212 | negative | MGI_C57BL6J_6303054 | Gm50243 | ENSEMBL:ENSMUSG00000118044 | MGI:6303054 | pseudogene | predicted gene, 50243 |
| 19 | gene | 21.271891 | 21.286840 | positive | MGI_C57BL6J_1278334 | Zfand5 | NCBI_Gene:22682,ENSEMBL:ENSMUSG00000024750 | MGI:1278334 | protein coding gene | zinc finger, AN1-type domain 5 |
| 19 | gene | 21.293946 | 21.295010 | positive | MGI_C57BL6J_1916228 | 1500002I01Rik | NA | NA | unclassified gene | RIKEN cDNA 1500002I01 gene |
| 19 | gene | 21.310376 | 21.313283 | negative | MGI_C57BL6J_6303075 | Gm50255 | ENSEMBL:ENSMUSG00000118342 | MGI:6303075 | lncRNA gene | predicted gene, 50255 |
| 19 | gene | 21.391307 | 21.473445 | negative | MGI_C57BL6J_95678 | Gda | NCBI_Gene:14544,ENSEMBL:ENSMUSG00000058624 | MGI:95678 | protein coding gene | guanine deaminase |
| 19 | gene | 21.487932 | 21.488152 | positive | MGI_C57BL6J_1917501 | 2010105D22Rik | NA | NA | unclassified gene | RIKEN cDNA 2010105D22 gene |
| 19 | gene | 21.552204 | 21.557867 | positive | MGI_C57BL6J_3781620 | Gm3443 | ENSEMBL:ENSMUSG00000079382 | MGI:3781620 | protein coding gene | predicted gene 3443 |
| 19 | gene | 21.577760 | 21.596179 | negative | MGI_C57BL6J_5621371 | Gm38486 | NCBI_Gene:102636055 | MGI:5621371 | lncRNA gene | predicted gene, 38486 |
| 19 | gene | 21.581202 | 21.652976 | negative | MGI_C57BL6J_1913456 | 1110059E24Rik | NCBI_Gene:66206,ENSEMBL:ENSMUSG00000035171 | MGI:1913456 | protein coding gene | RIKEN cDNA 1110059E24 gene |
| 19 | gene | 21.653185 | 21.685638 | positive | MGI_C57BL6J_1917816 | Abhd17b | NCBI_Gene:226016,ENSEMBL:ENSMUSG00000047368 | MGI:1917816 | protein coding gene | abhydrolase domain containing 17B |
| 19 | gene | 21.657796 | 21.661573 | positive | MGI_C57BL6J_1923222 | 5033423O07Rik | NA | NA | unclassified gene | RIKEN cDNA 5033423O07 gene |
| 19 | gene | 21.663158 | 21.666018 | positive | MGI_C57BL6J_2445142 | B230378D16Rik | NA | NA | unclassified gene | RIKEN cDNA B230378D16 gene |
| 19 | gene | 21.689292 | 21.691115 | positive | MGI_C57BL6J_3028050 | C130076G22Rik | NA | NA | unclassified gene | RIKEN cDNA C130076G22 gene |
| 19 | gene | 21.713818 | 21.732331 | positive | MGI_C57BL6J_5624703 | Gm41818 | NCBI_Gene:105246544 | MGI:5624703 | lncRNA gene | predicted gene, 41818 |
| 19 | pseudogene | 21.729270 | 21.729745 | positive | MGI_C57BL6J_6302871 | Gm50129 | ENSEMBL:ENSMUSG00000118055 | MGI:6302871 | pseudogene | predicted gene, 50129 |
| 19 | gene | 21.777511 | 21.858360 | positive | MGI_C57BL6J_1890373 | Cemip2 | NCBI_Gene:83921,ENSEMBL:ENSMUSG00000024754 | MGI:1890373 | protein coding gene | cell migration inducing hyaluronidase 2 |
| 19 | gene | 21.831451 | 21.833756 | negative | MGI_C57BL6J_6302872 | Gm50130 | ENSEMBL:ENSMUSG00000118021 | MGI:6302872 | lncRNA gene | predicted gene, 50130 |
| 19 | pseudogene | 21.837571 | 21.838711 | positive | MGI_C57BL6J_5011136 | Gm18951 | NCBI_Gene:100418019 | MGI:5011136 | pseudogene | predicted gene, 18951 |
| 19 | pseudogene | 21.937283 | 21.938567 | positive | MGI_C57BL6J_3645435 | Gm5514 | NCBI_Gene:433229,ENSEMBL:ENSMUSG00000045104 | MGI:3645435 | pseudogene | predicted gene 5514 |
| 19 | gene | 21.941978 | 21.950889 | negative | MGI_C57BL6J_6302877 | Gm50133 | ENSEMBL:ENSMUSG00000118237 | MGI:6302877 | lncRNA gene | predicted gene, 50133 |
| 19 | gene | 21.992573 | 22.021016 | positive | MGI_C57BL6J_5624704 | Gm41819 | ENSEMBL:ENSMUSG00000118273 | MGI:5624704 | lncRNA gene | predicted gene, 41819 |
| 19 | gene | 21.997331 | 22.000152 | positive | MGI_C57BL6J_6302879 | Gm50134 | ENSEMBL:ENSMUSG00000118226 | MGI:6302879 | lncRNA gene | predicted gene, 50134 |
| 19 | gene | 22.137798 | 22.995410 | positive | MGI_C57BL6J_2443101 | Trpm3 | NCBI_Gene:226025,ENSEMBL:ENSMUSG00000052387 | MGI:2443101 | protein coding gene | transient receptor potential cation channel, subfamily M, member 3 |
| 19 | gene | 22.235521 | 22.235618 | negative | MGI_C57BL6J_5452283 | Gm22506 | ENSEMBL:ENSMUSG00000088558 | MGI:5452283 | snoRNA gene | predicted gene, 22506 |
| 19 | pseudogene | 22.254745 | 22.262751 | positive | MGI_C57BL6J_5592914 | Gm33755 | NCBI_Gene:102636778 | MGI:5592914 | pseudogene | predicted gene, 33755 |
| 19 | gene | 22.436276 | 22.455241 | negative | MGI_C57BL6J_5520994 | Gm27151 | NCBI_Gene:102641617,ENSEMBL:ENSMUSG00000098739 | MGI:5520994 | lncRNA gene | predicted gene 27151 |
| 19 | pseudogene | 22.441007 | 22.443653 | positive | MGI_C57BL6J_3648670 | Nsa2-ps1 | NCBI_Gene:433230,ENSEMBL:ENSMUSG00000118353 | MGI:3648670 | pseudogene | NSA2 ribosome biogenesis homolog, pseudogene 1 |
| 19 | gene | 22.509995 | 22.517465 | positive | MGI_C57BL6J_5592808 | Gm33649 | NCBI_Gene:102636640 | MGI:5592808 | lncRNA gene | predicted gene, 33649 |
| 19 | gene | 22.552662 | 22.555198 | positive | MGI_C57BL6J_1925993 | 9630005C17Rik | NA | NA | unclassified gene | RIKEN cDNA 9630005C17 gene |
| 19 | gene | 22.750605 | 22.750672 | positive | MGI_C57BL6J_2676879 | Mir204 | miRBase:MI0000247,NCBI_Gene:387200,ENSEMBL:ENSMUSG00000065507 | MGI:2676879 | miRNA gene | microRNA 204 |
| 19 | gene | 22.776821 | 22.778324 | positive | MGI_C57BL6J_1920165 | 2900017F05Rik | NA | NA | unclassified gene | RIKEN cDNA 2900017F05 gene |
| 19 | gene | 22.978230 | 23.075924 | negative | MGI_C57BL6J_1924860 | C330002G04Rik | NCBI_Gene:77610,ENSEMBL:ENSMUSG00000097930 | MGI:1924860 | lncRNA gene | RIKEN cDNA C330002G04 gene |
| 19 | gene | 23.061454 | 23.064032 | positive | MGI_C57BL6J_6302882 | Gm50136 | ENSEMBL:ENSMUSG00000118155 | MGI:6302882 | lncRNA gene | predicted gene, 50136 |
| 19 | gene | 23.134498 | 23.137111 | positive | MGI_C57BL6J_1915498 | 2410080I02Rik | NCBI_Gene:102636896,ENSEMBL:ENSMUSG00000044387 | MGI:1915498 | lncRNA gene | RIKEN cDNA 2410080I02 gene |
| 19 | gene | 23.141226 | 23.168134 | positive | MGI_C57BL6J_1333856 | Klf9 | NCBI_Gene:16601,ENSEMBL:ENSMUSG00000033863 | MGI:1333856 | protein coding gene | Kruppel-like factor 9 |
| 19 | gene | 23.149431 | 23.149551 | positive | MGI_C57BL6J_3783365 | Mir1192 | miRBase:MI0006297,NCBI_Gene:100316672,ENSEMBL:ENSMUSG00000080626 | MGI:3783365 | miRNA gene | microRNA 1192 |
| 19 | gene | 23.206441 | 23.273914 | negative | MGI_C57BL6J_2385088 | Smc5 | NCBI_Gene:226026,ENSEMBL:ENSMUSG00000024943 | MGI:2385088 | protein coding gene | structural maintenance of chromosomes 5 |
| 19 | gene | 23.273925 | 23.274359 | positive | MGI_C57BL6J_5593109 | Gm33950 | NCBI_Gene:102637041 | MGI:5593109 | lncRNA gene | predicted gene, 33950 |
| 19 | gene | 23.282096 | 23.309919 | positive | MGI_C57BL6J_5624706 | Gm41821 | NCBI_Gene:105246547,ENSEMBL:ENSMUSG00000117837 | MGI:5624706 | lncRNA gene | predicted gene, 41821 |
| 19 | gene | 23.296329 | 23.296435 | positive | MGI_C57BL6J_5454040 | Gm24263 | ENSEMBL:ENSMUSG00000096249 | MGI:5454040 | snRNA gene | predicted gene, 24263 |
| 19 | gene | 23.302609 | 23.448498 | negative | MGI_C57BL6J_1918988 | Mamdc2 | NCBI_Gene:71738,ENSEMBL:ENSMUSG00000033207 | MGI:1918988 | protein coding gene | MAM domain containing 2 |
| 19 | gene | 23.418343 | 23.430928 | positive | MGI_C57BL6J_6303381 | Gm50445 | ENSEMBL:ENSMUSG00000118092 | MGI:6303381 | lncRNA gene | predicted gene, 50445 |
| 19 | pseudogene | 23.437256 | 23.437727 | negative | MGI_C57BL6J_6303379 | Gm50443 | ENSEMBL:ENSMUSG00000117850 | MGI:6303379 | pseudogene | predicted gene, 50443 |
| 19 | gene | 23.496764 | 23.523052 | positive | MGI_C57BL6J_5593171 | Gm34012 | NCBI_Gene:102637119 | MGI:5593171 | lncRNA gene | predicted gene, 34012 |
| 19 | gene | 23.558753 | 23.652812 | negative | MGI_C57BL6J_1914733 | 1700028P14Rik | NCBI_Gene:67483,ENSEMBL:ENSMUSG00000033053 | MGI:1914733 | protein coding gene | RIKEN cDNA 1700028P14 gene |
| 19 | pseudogene | 23.605898 | 23.606279 | positive | MGI_C57BL6J_6303347 | Gm50422 | ENSEMBL:ENSMUSG00000117944 | MGI:6303347 | pseudogene | predicted gene, 50422 |
| 19 | pseudogene | 23.616904 | 23.620368 | positive | MGI_C57BL6J_3779903 | Gm9493 | NCBI_Gene:102638448,ENSEMBL:ENSMUSG00000044424 | MGI:3779903 | pseudogene | predicted gene 9493 |
| 19 | gene | 23.643409 | 23.643510 | positive | MGI_C57BL6J_5451921 | Gm22144 | ENSEMBL:ENSMUSG00000088027 | MGI:5451921 | snoRNA gene | predicted gene, 22144 |
| 19 | pseudogene | 23.666591 | 23.667583 | positive | MGI_C57BL6J_5434897 | Gm21542 | ENSEMBL:ENSMUSG00000098010 | MGI:5434897 | pseudogene | predicted gene, 21542 |
| 19 | pseudogene | 23.675811 | 23.676735 | positive | MGI_C57BL6J_3646907 | Gm6563 | NCBI_Gene:625193,ENSEMBL:ENSMUSG00000051255 | MGI:3646907 | pseudogene | predicted pseudogene 6563 |
| 19 | gene | 23.686898 | 23.732885 | positive | MGI_C57BL6J_1921875 | Ptar1 | NCBI_Gene:72351,ENSEMBL:ENSMUSG00000074925 | MGI:1921875 | protein coding gene | protein prenyltransferase alpha subunit repeat containing 1 |
| 19 | gene | 23.723279 | 23.725034 | positive | MGI_C57BL6J_3641836 | Gm9938 | NA | NA | protein coding gene | predicted gene 9938 |
| 19 | gene | 23.728707 | 23.731661 | positive | MGI_C57BL6J_1920060 | 2810455D13Rik | NA | NA | unclassified gene | RIKEN cDNA 2810455D13 gene |
| 19 | gene | 23.745502 | 23.748001 | negative | MGI_C57BL6J_6303146 | Gm50299 | ENSEMBL:ENSMUSG00000117685 | MGI:6303146 | lncRNA gene | predicted gene, 50299 |
| 19 | gene | 23.758806 | 23.949598 | positive | MGI_C57BL6J_1860297 | Apba1 | NCBI_Gene:319924,ENSEMBL:ENSMUSG00000024897 | MGI:1860297 | protein coding gene | amyloid beta (A4) precursor protein binding, family A, member 1 |
| 19 | pseudogene | 23.800538 | 23.800817 | positive | MGI_C57BL6J_6303148 | Gm50300 | ENSEMBL:ENSMUSG00000118287 | MGI:6303148 | pseudogene | predicted gene, 50300 |
| 19 | gene | 23.870740 | 23.871826 | positive | MGI_C57BL6J_1925097 | B230104F01Rik | NA | NA | unclassified gene | RIKEN cDNA B230104F01 gene |
| 19 | gene | 23.905122 | 23.917546 | negative | MGI_C57BL6J_5593235 | Gm34076 | NCBI_Gene:102637203 | MGI:5593235 | lncRNA gene | predicted gene, 34076 |
| 19 | gene | 23.929825 | 23.941594 | negative | MGI_C57BL6J_6303149 | Gm50301 | ENSEMBL:ENSMUSG00000118340 | MGI:6303149 | lncRNA gene | predicted gene, 50301 |
| 19 | gene | 23.972750 | 24.031272 | negative | MGI_C57BL6J_2685813 | Fam189a2 | NCBI_Gene:381217,ENSEMBL:ENSMUSG00000071604 | MGI:2685813 | protein coding gene | family with sequence similarity 189, member A2 |
| 19 | pseudogene | 24.004780 | 24.005008 | positive | MGI_C57BL6J_6303156 | Gm50305 | ENSEMBL:ENSMUSG00000117820 | MGI:6303156 | pseudogene | predicted gene, 50305 |
| 19 | gene | 24.044383 | 24.047157 | negative | MGI_C57BL6J_5624708 | Gm41823 | NCBI_Gene:105246549 | MGI:5624708 | lncRNA gene | predicted gene, 41823 |
| 19 | gene | 24.064793 | 24.066452 | positive | MGI_C57BL6J_1922771 | 1700021P04Rik | ENSEMBL:ENSMUSG00000096796 | MGI:1922771 | lncRNA gene | RIKEN cDNA 1700021P04 gene |
| 19 | gene | 24.080931 | 24.090765 | positive | MGI_C57BL6J_5624709 | Gm41824 | NCBI_Gene:105246550 | MGI:5624709 | lncRNA gene | predicted gene, 41824 |
| 19 | gene | 24.094502 | 24.225130 | negative | MGI_C57BL6J_1341872 | Tjp2 | NCBI_Gene:21873,ENSEMBL:ENSMUSG00000024812 | MGI:1341872 | protein coding gene | tight junction protein 2 |
| 19 | pseudogene | 24.146897 | 24.147224 | negative | MGI_C57BL6J_6303163 | Gm50310 | ENSEMBL:ENSMUSG00000118277 | MGI:6303163 | pseudogene | predicted gene, 50310 |
| 19 | pseudogene | 24.154070 | 24.154601 | positive | MGI_C57BL6J_6303160 | Gm50308 | ENSEMBL:ENSMUSG00000118371 | MGI:6303160 | pseudogene | predicted gene, 50308 |
| 19 | gene | 24.261453 | 24.280605 | negative | MGI_C57BL6J_1096879 | Fxn | NCBI_Gene:14297,ENSEMBL:ENSMUSG00000059363 | MGI:1096879 | protein coding gene | frataxin |
| 19 | pseudogene | 24.289000 | 24.290908 | negative | MGI_C57BL6J_3646651 | Gm8789 | NCBI_Gene:667743 | MGI:3646651 | pseudogene | predicted gene 8789 |
| 19 | pseudogene | 24.289716 | 24.290826 | positive | MGI_C57BL6J_3801728 | Gm16216 | ENSEMBL:ENSMUSG00000082107 | MGI:3801728 | pseudogene | predicted gene 16216 |
| 19 | gene | 24.294794 | 24.555872 | negative | MGI_C57BL6J_107930 | Pip5k1b | NCBI_Gene:18719,ENSEMBL:ENSMUSG00000024867 | MGI:107930 | protein coding gene | phosphatidylinositol-4-phosphate 5-kinase, type 1 beta |
| 19 | gene | 24.298838 | 24.300938 | negative | MGI_C57BL6J_3782025 | Gm3853 | NA | NA | unclassified gene | predicted gene 3853 |
| 19 | gene | 24.413789 | 24.430694 | positive | MGI_C57BL6J_1922106 | Pip5k1bos | NCBI_Gene:102637283,ENSEMBL:ENSMUSG00000085003 | MGI:1922106 | antisense lncRNA gene | phosphatidylinositol-4-phosphate 5-kinase, type 1 beta, opposite strand |
| 19 | gene | 24.475779 | 24.477474 | negative | MGI_C57BL6J_1915284 | Fam122a | NCBI_Gene:68034,ENSEMBL:ENSMUSG00000074922 | MGI:1915284 | protein coding gene | family with sequence similarity 122, member A |
| 19 | gene | 24.541198 | 24.541455 | negative | MGI_C57BL6J_3641847 | E860004J03Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA E860004J03 gene |
| 19 | gene | 24.556352 | 24.557228 | positive | MGI_C57BL6J_6303218 | Gm50341 | ENSEMBL:ENSMUSG00000117979 | MGI:6303218 | lncRNA gene | predicted gene, 50341 |
| 19 | gene | 24.674008 | 24.682233 | positive | MGI_C57BL6J_3583948 | Tmem252 | NCBI_Gene:226040,ENSEMBL:ENSMUSG00000048572 | MGI:3583948 | protein coding gene | transmembrane protein 252 |
| 19 | gene | 24.678261 | 24.861855 | negative | MGI_C57BL6J_1925668 | Pgm5 | NCBI_Gene:226041,ENSEMBL:ENSMUSG00000041731 | MGI:1925668 | protein coding gene | phosphoglucomutase 5 |
| 19 | pseudogene | 24.692233 | 24.693391 | negative | MGI_C57BL6J_5009986 | Gm17800 | NCBI_Gene:100158260,ENSEMBL:ENSMUSG00000097950 | MGI:5009986 | pseudogene | predicted gene, 17800 |
| 19 | gene | 24.706836 | 24.713712 | positive | MGI_C57BL6J_5621466 | Gm38581 | NCBI_Gene:102641855 | MGI:5621466 | lncRNA gene | predicted gene, 38581 |
| 19 | pseudogene | 24.856196 | 24.856457 | positive | MGI_C57BL6J_6303220 | Gm50342 | ENSEMBL:ENSMUSG00000117934 | MGI:6303220 | pseudogene | predicted gene, 50342 |
| 19 | pseudogene | 24.875686 | 24.876875 | positive | MGI_C57BL6J_3704493 | Gm10053 | NCBI_Gene:672195,ENSEMBL:ENSMUSG00000058927 | MGI:3704493 | pseudogene | predicted gene 10053 |
| 19 | gene | 24.898965 | 24.901309 | negative | MGI_C57BL6J_1347467 | Foxd4 | NCBI_Gene:14237,ENSEMBL:ENSMUSG00000051490 | MGI:1347467 | protein coding gene | forkhead box D4 |
| 19 | gene | 24.919916 | 24.961616 | negative | MGI_C57BL6J_2385089 | Cbwd1 | NCBI_Gene:226043,ENSEMBL:ENSMUSG00000024878 | MGI:2385089 | protein coding gene | COBW domain containing 1 |
| 19 | gene | 24.942235 | 24.942303 | negative | MGI_C57BL6J_5562753 | Mir3084-1 | miRBase:MI0014047,NCBI_Gene:100526521,ENSEMBL:ENSMUSG00000104890 | MGI:5562753 | miRNA gene | microRNA 3084-1 |
| 19 | gene | 24.942236 | 24.942367 | negative | MGI_C57BL6J_5453435 | Gm23658 | ENSEMBL:ENSMUSG00000095097 | MGI:5453435 | snoRNA gene | predicted gene, 23658 |
| 19 | gene | 24.999511 | 25.202432 | positive | MGI_C57BL6J_1921396 | Dock8 | NCBI_Gene:76088,ENSEMBL:ENSMUSG00000052085 | MGI:1921396 | protein coding gene | dedicator of cytokinesis 8 |
| 19 | gene | 25.033108 | 25.033578 | positive | MGI_C57BL6J_3782046 | Ndufb4b | NCBI_Gene:100042503 | MGI:3782046 | protein coding gene | NADH:ubiquinone oxidoreductase subunit B4B |
| 19 | gene | 25.092866 | 25.097404 | negative | MGI_C57BL6J_5593394 | Gm34235 | NCBI_Gene:102637425 | MGI:5593394 | lncRNA gene | predicted gene, 34235 |
| 19 | gene | 25.153231 | 25.155877 | negative | MGI_C57BL6J_5593345 | Gm34186 | NCBI_Gene:102637351 | MGI:5593345 | lncRNA gene | predicted gene, 34186 |
| 19 | gene | 25.166085 | 25.166216 | negative | MGI_C57BL6J_5454029 | Gm24252 | ENSEMBL:ENSMUSG00000065634 | MGI:5454029 | snoRNA gene | predicted gene, 24252 |
| 19 | gene | 25.236732 | 25.434497 | positive | MGI_C57BL6J_2147707 | Kank1 | NCBI_Gene:107351,ENSEMBL:ENSMUSG00000032702 | MGI:2147707 | protein coding gene | KN motif and ankyrin repeat domains 1 |
| 19 | pseudogene | 25.346409 | 25.347243 | negative | MGI_C57BL6J_5593591 | Gm34432 | NCBI_Gene:102637686,ENSEMBL:ENSMUSG00000118238 | MGI:5593591 | pseudogene | predicted gene, 34432 |
| 19 | gene | 25.453960 | 25.454063 | negative | MGI_C57BL6J_5455984 | Gm26207 | ENSEMBL:ENSMUSG00000064843 | MGI:5455984 | snRNA gene | predicted gene, 26207 |
| 19 | pseudogene | 25.493833 | 25.496716 | positive | MGI_C57BL6J_3646471 | Gm5249 | NCBI_Gene:383474,ENSEMBL:ENSMUSG00000117720 | MGI:3646471 | pseudogene | predicted gene 5249 |
| 19 | gene | 25.505618 | 25.604329 | positive | MGI_C57BL6J_1354733 | Dmrt1 | NCBI_Gene:50796,ENSEMBL:ENSMUSG00000024837 | MGI:1354733 | protein coding gene | doublesex and mab-3 related transcription factor 1 |
| 19 | gene | 25.610246 | 25.623921 | positive | MGI_C57BL6J_2449470 | Dmrt3 | NCBI_Gene:240590,ENSEMBL:ENSMUSG00000042372 | MGI:2449470 | protein coding gene | doublesex and mab-3 related transcription factor 3 |
| 19 | gene | 25.671134 | 25.672253 | negative | MGI_C57BL6J_1919653 | 2610016A17Rik | NCBI_Gene:100503074,ENSEMBL:ENSMUSG00000101089 | MGI:1919653 | lncRNA gene | RIKEN cDNA 2610016A17 gene |
| 19 | gene | 25.672389 | 25.679013 | positive | MGI_C57BL6J_1330307 | Dmrt2 | NCBI_Gene:226049,ENSEMBL:ENSMUSG00000048138 | MGI:1330307 | protein coding gene | doublesex and mab-3 related transcription factor 2 |
| 19 | gene | 25.775487 | 25.946975 | positive | MGI_C57BL6J_5593675 | Gm34516 | NCBI_Gene:102637794 | MGI:5593675 | lncRNA gene | predicted gene, 34516 |
| 19 | pseudogene | 25.795612 | 25.796825 | negative | MGI_C57BL6J_3647098 | Gm8825 | NCBI_Gene:667806,ENSEMBL:ENSMUSG00000098105 | MGI:3647098 | pseudogene | predicted gene 8825 |
| 19 | gene | 26.030730 | 26.046626 | negative | MGI_C57BL6J_1920683 | 1700048O14Rik | NCBI_Gene:73433,ENSEMBL:ENSMUSG00000118016 | MGI:1920683 | lncRNA gene | RIKEN cDNA 1700048O14 gene |
| 19 | gene | 26.246024 | 26.249930 | negative | MGI_C57BL6J_6303272 | Gm50376 | ENSEMBL:ENSMUSG00000117950 | MGI:6303272 | lncRNA gene | predicted gene, 50376 |
| 19 | gene | 26.260369 | 26.260493 | negative | MGI_C57BL6J_5454953 | Gm25176 | ENSEMBL:ENSMUSG00000084589 | MGI:5454953 | snRNA gene | predicted gene, 25176 |
| 19 | gene | 26.277846 | 26.308707 | negative | MGI_C57BL6J_6303274 | Gm50377 | ENSEMBL:ENSMUSG00000118005 | MGI:6303274 | lncRNA gene | predicted gene, 50377 |
| 19 | gene | 26.462344 | 26.467779 | negative | MGI_C57BL6J_6303276 | Gm50378 | ENSEMBL:ENSMUSG00000117707 | MGI:6303276 | lncRNA gene | predicted gene, 50378 |
| 19 | gene | 26.482315 | 26.509084 | negative | MGI_C57BL6J_5593967 | Gm34808 | NCBI_Gene:102638185,ENSEMBL:ENSMUSG00000118001 | MGI:5593967 | lncRNA gene | predicted gene, 34808 |
| 19 | gene | 26.510339 | 26.527796 | positive | MGI_C57BL6J_5624713 | Gm41828 | NCBI_Gene:105246554 | MGI:5624713 | lncRNA gene | predicted gene, 41828 |
| 19 | pseudogene | 26.530752 | 26.530929 | positive | MGI_C57BL6J_6303278 | Gm50379 | ENSEMBL:ENSMUSG00000117803 | MGI:6303278 | pseudogene | predicted gene, 50379 |
| 19 | gene | 26.572996 | 26.584672 | positive | MGI_C57BL6J_6098469 | Gm48775 | ENSEMBL:ENSMUSG00000114682 | MGI:6098469 | lncRNA gene | predicted gene, 48775 |
| 19 | gene | 26.605050 | 26.778322 | positive | MGI_C57BL6J_99603 | Smarca2 | NCBI_Gene:67155,ENSEMBL:ENSMUSG00000024921 | MGI:99603 | protein coding gene | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| 19 | gene | 26.620749 | 26.623419 | negative | MGI_C57BL6J_5313063 | Gm20616 | NCBI_Gene:102638259,ENSEMBL:ENSMUSG00000093628 | MGI:5313063 | lncRNA gene | predicted gene 20616 |
| 19 | gene | 26.768801 | 26.823907 | negative | MGI_C57BL6J_1918192 | 4931403E22Rik | NCBI_Gene:70942,ENSEMBL:ENSMUSG00000093772 | MGI:1918192 | lncRNA gene | RIKEN cDNA 4931403E22 gene |
| 19 | pseudogene | 26.802040 | 26.803219 | positive | MGI_C57BL6J_5010565 | Gm18380 | NCBI_Gene:100417041,ENSEMBL:ENSMUSG00000118331 | MGI:5010565 | pseudogene | predicted gene, 18380 |
| 19 | gene | 26.823866 | 26.872509 | positive | MGI_C57BL6J_5826285 | Gm46648 | NCBI_Gene:108168409 | MGI:5826285 | lncRNA gene | predicted gene, 46648 |
| 19 | gene | 26.885881 | 26.888647 | positive | MGI_C57BL6J_2685661 | Gm815 | NCBI_Gene:329047,ENSEMBL:ENSMUSG00000074913 | MGI:2685661 | protein coding gene | predicted gene 815 |
| 19 | pseudogene | 27.025981 | 27.026314 | positive | MGI_C57BL6J_6302862 | Gm50121 | ENSEMBL:ENSMUSG00000118313 | MGI:6302862 | pseudogene | predicted gene, 50121 |
| 19 | gene | 27.101951 | 27.316698 | negative | MGI_C57BL6J_5594597 | Gm35438 | NCBI_Gene:102639019,ENSEMBL:ENSMUSG00000118020,ENSEMBL:ENSMUSG00000118209 | MGI:5594597 | lncRNA gene | predicted gene, 35438 |
| 19 | gene | 27.216484 | 27.254231 | positive | MGI_C57BL6J_98935 | Vldlr | NCBI_Gene:22359,ENSEMBL:ENSMUSG00000024924 | MGI:98935 | protein coding gene | very low density lipoprotein receptor |
| 19 | gene | 27.260735 | 27.262323 | negative | MGI_C57BL6J_3782090 | Gm3916 | NA | NA | unclassified gene | predicted gene 3916 |
| 19 | gene | 27.260735 | 27.262323 | negative | MGI_C57BL6J_6302852 | Gm50114 | ENSEMBL:ENSMUSG00000117692 | MGI:6302852 | lncRNA gene | predicted gene, 50114 |
| 19 | gene | 27.312632 | 27.321729 | positive | MGI_C57BL6J_5594427 | Gm35268 | NCBI_Gene:102638787 | MGI:5594427 | lncRNA gene | predicted gene, 35268 |
| 19 | gene | 27.322061 | 27.337179 | positive | MGI_C57BL6J_2670981 | Kcnv2 | NCBI_Gene:240595,ENSEMBL:ENSMUSG00000047298 | MGI:2670981 | protein coding gene | potassium channel, subfamily V, member 2 |
| 19 | gene | 27.344725 | 27.373932 | positive | MGI_C57BL6J_5594713 | Gm35554 | NCBI_Gene:102639185,ENSEMBL:ENSMUSG00000117752 | MGI:5594713 | lncRNA gene | predicted gene, 35554 |
| 19 | pseudogene | 27.366072 | 27.366567 | positive | MGI_C57BL6J_6302851 | Gm50113 | ENSEMBL:ENSMUSG00000117705 | MGI:6302851 | pseudogene | predicted gene, 50113 |
| 19 | gene | 27.388698 | 27.429908 | negative | MGI_C57BL6J_106253 | Pum3 | NCBI_Gene:52874,ENSEMBL:ENSMUSG00000041360 | MGI:106253 | protein coding gene | pumilio RNA-binding family member 3 |
| 19 | gene | 27.430037 | 27.432631 | positive | MGI_C57BL6J_2147728 | C030016D13Rik | NCBI_Gene:107372 | MGI:2147728 | lncRNA gene | RIKEN cDNA C030016D13 gene |
| 19 | pseudogene | 27.512508 | 27.513279 | positive | MGI_C57BL6J_6302831 | Gm50101 | ENSEMBL:ENSMUSG00000118072 | MGI:6302831 | pseudogene | predicted gene, 50101 |
| 19 | gene | 27.666428 | 27.668494 | negative | MGI_C57BL6J_5594856 | Gm35697 | NCBI_Gene:102639367 | MGI:5594856 | lncRNA gene | predicted gene, 35697 |
| 19 | gene | 27.703876 | 27.704007 | negative | MGI_C57BL6J_5454559 | Gm24782 | ENSEMBL:ENSMUSG00000088266 | MGI:5454559 | snoRNA gene | predicted gene, 24782 |
| 19 | gene | 27.761721 | 28.011693 | negative | MGI_C57BL6J_106582 | Rfx3 | NCBI_Gene:19726,ENSEMBL:ENSMUSG00000040929 | MGI:106582 | protein coding gene | regulatory factor X, 3 (influences HLA class II expression) |
| 19 | pseudogene | 27.858058 | 27.858497 | positive | MGI_C57BL6J_3647318 | Gm8412 | ENSEMBL:ENSMUSG00000092617 | MGI:3647318 | pseudogene | predicted gene 8412 |
| 19 | pseudogene | 27.961274 | 27.962110 | positive | MGI_C57BL6J_5010079 | Gm17894 | NCBI_Gene:100416057 | MGI:5010079 | pseudogene | predicted gene, 17894 |
| 19 | gene | 27.994197 | 27.995693 | positive | MGI_C57BL6J_5141872 | Gm20407 | ENSEMBL:ENSMUSG00000092182 | MGI:5141872 | lncRNA gene | predicted gene 20407 |
| 19 | pseudogene | 28.033455 | 28.034153 | positive | MGI_C57BL6J_3648671 | Gm5517 | NCBI_Gene:433235,ENSEMBL:ENSMUSG00000117788 | MGI:3648671 | pseudogene | predicted gene 5517 |
| 19 | gene | 28.088856 | 28.089764 | positive | MGI_C57BL6J_5578934 | Gm28228 | ENSEMBL:ENSMUSG00000101084 | MGI:5578934 | lncRNA gene | predicted gene 28228 |
| 19 | pseudogene | 28.145864 | 28.146319 | negative | MGI_C57BL6J_5594940 | Gm35781 | NCBI_Gene:102639477,ENSEMBL:ENSMUSG00000117862 | MGI:5594940 | pseudogene | predicted gene, 35781 |
| 19 | gene | 28.258851 | 28.680265 | negative | MGI_C57BL6J_2444289 | Glis3 | NCBI_Gene:226075,ENSEMBL:ENSMUSG00000052942 | MGI:2444289 | protein coding gene | GLIS family zinc finger 3 |
| 19 | gene | 28.362542 | 28.365815 | negative | MGI_C57BL6J_5595025 | Gm35866 | NCBI_Gene:102639589 | MGI:5595025 | lncRNA gene | predicted gene, 35866 |
| 19 | gene | 28.580533 | 28.583294 | negative | MGI_C57BL6J_1921702 | 4933413C19Rik | ENSEMBL:ENSMUSG00000089952 | MGI:1921702 | lncRNA gene | RIKEN cDNA 4933413C19 gene |
| 19 | gene | 28.678224 | 28.720027 | positive | MGI_C57BL6J_2443529 | D930032P07Rik | NCBI_Gene:320176,ENSEMBL:ENSMUSG00000097735 | MGI:2443529 | lncRNA gene | RIKEN cDNA D930032P07 gene |
| 19 | pseudogene | 28.742123 | 28.742568 | negative | MGI_C57BL6J_3643316 | Rps15a-ps2 | NCBI_Gene:434460,ENSEMBL:ENSMUSG00000067351 | MGI:3643316 | pseudogene | ribosomal protein S15A, pseudogene 2 |
| 19 | gene | 28.760033 | 28.788185 | positive | MGI_C57BL6J_5595081 | Gm35922 | NCBI_Gene:102639664 | MGI:5595081 | lncRNA gene | predicted gene, 35922 |
| 19 | pseudogene | 28.762865 | 28.763539 | negative | MGI_C57BL6J_3646480 | Gm6788 | NCBI_Gene:627788,ENSEMBL:ENSMUSG00000090737 | MGI:3646480 | pseudogene | predicted gene 6788 |
| 19 | pseudogene | 28.812406 | 28.813202 | negative | MGI_C57BL6J_5504057 | Gm26942 | ENSEMBL:ENSMUSG00000098069 | MGI:5504057 | pseudogene | predicted gene, 26942 |
| 19 | gene | 28.835049 | 28.913960 | positive | MGI_C57BL6J_105083 | Slc1a1 | NCBI_Gene:20510,ENSEMBL:ENSMUSG00000024935 | MGI:105083 | protein coding gene | solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
| 19 | pseudogene | 28.862012 | 28.863766 | negative | MGI_C57BL6J_3648836 | Gm5822 | NCBI_Gene:545262,ENSEMBL:ENSMUSG00000089993 | MGI:3648836 | pseudogene | predicted gene 5822 |
| 19 | gene | 28.893042 | 28.968356 | negative | MGI_C57BL6J_1918036 | 4430402I18Rik | NCBI_Gene:381218,ENSEMBL:ENSMUSG00000064202 | MGI:1918036 | protein coding gene | RIKEN cDNA 4430402I18 gene |
| 19 | gene | 28.963920 | 28.966811 | positive | MGI_C57BL6J_1921661 | Plpp6 | NCBI_Gene:74411,ENSEMBL:ENSMUSG00000040105 | MGI:1921661 | protein coding gene | phospholipid phosphatase 6 |
| 19 | pseudogene | 28.973039 | 28.974077 | positive | MGI_C57BL6J_3781797 | Gm3621 | NCBI_Gene:100042006,ENSEMBL:ENSMUSG00000118165 | MGI:3781797 | pseudogene | predicted gene 3621 |
| 19 | pseudogene | 28.974550 | 28.975633 | negative | MGI_C57BL6J_3643046 | Gm4844 | NCBI_Gene:226086 | MGI:3643046 | pseudogene | predicted gene 4844 |
| 19 | gene | 28.990318 | 29.026681 | positive | MGI_C57BL6J_1914322 | Cdc37l1 | NCBI_Gene:67072,ENSEMBL:ENSMUSG00000024780 | MGI:1914322 | protein coding gene | cell division cycle 37-like 1 |
| 19 | gene | 28.995379 | 29.048091 | negative | MGI_C57BL6J_1860835 | Ak3 | NCBI_Gene:56248,ENSEMBL:ENSMUSG00000024782 | MGI:1860835 | protein coding gene | adenylate kinase 3 |
| 19 | pseudogene | 29.003343 | 29.003875 | negative | MGI_C57BL6J_3704242 | Gm10136 | NCBI_Gene:672214,ENSEMBL:ENSMUSG00000063754 | MGI:3704242 | pseudogene | predicted pseudogene 10136 |
| 19 | gene | 29.046245 | 29.048729 | positive | MGI_C57BL6J_1914579 | 1700018L02Rik | NCBI_Gene:67329,ENSEMBL:ENSMUSG00000100075 | MGI:1914579 | lncRNA gene | RIKEN cDNA 1700018L02 gene |
| 19 | gene | 29.067301 | 29.069503 | positive | MGI_C57BL6J_3642802 | Gm9895 | NCBI_Gene:100503337,ENSEMBL:ENSMUSG00000118038 | MGI:3642802 | lncRNA gene | predicted gene 9895 |
| 19 | pseudogene | 29.087603 | 29.090255 | negative | MGI_C57BL6J_6303352 | Gm50425 | ENSEMBL:ENSMUSG00000118320 | MGI:6303352 | pseudogene | predicted gene, 50425 |
| 19 | gene | 29.101375 | 29.143929 | positive | MGI_C57BL6J_1913275 | Rcl1 | NCBI_Gene:59028,ENSEMBL:ENSMUSG00000024785 | MGI:1913275 | protein coding gene | RNA terminal phosphate cyclase-like 1 |
| 19 | gene | 29.135279 | 29.135375 | positive | MGI_C57BL6J_3618696 | Mir101b | miRBase:MI0000649,NCBI_Gene:724062,ENSEMBL:ENSMUSG00000065556 | MGI:3618696 | miRNA gene | microRNA 101b |
| 19 | gene | 29.147630 | 29.149431 | positive | MGI_C57BL6J_3642922 | Gm9883 | NA | NA | unclassified gene | predicted gene 9883 |
| 19 | pseudogene | 29.160000 | 29.161262 | positive | MGI_C57BL6J_3648673 | Gm5518 | NCBI_Gene:433238,ENSEMBL:ENSMUSG00000068466 | MGI:3648673 | pseudogene | predicted gene 5518 |
| 19 | gene | 29.238057 | 29.242703 | negative | MGI_C57BL6J_5624714 | Gm41829 | NCBI_Gene:105246555 | MGI:5624714 | lncRNA gene | predicted gene, 41829 |
| 19 | gene | 29.251708 | 29.313080 | positive | MGI_C57BL6J_96629 | Jak2 | NCBI_Gene:16452,ENSEMBL:ENSMUSG00000024789 | MGI:96629 | protein coding gene | Janus kinase 2 |
| 19 | gene | 29.321344 | 29.325356 | negative | MGI_C57BL6J_1351595 | Insl6 | NCBI_Gene:27356,ENSEMBL:ENSMUSG00000050957 | MGI:1351595 | protein coding gene | insulin-like 6 |
| 19 | gene | 29.331170 | 29.334670 | negative | MGI_C57BL6J_97931 | Rln1 | NCBI_Gene:19773,ENSEMBL:ENSMUSG00000039097 | MGI:97931 | protein coding gene | relaxin 1 |
| 19 | gene | 29.348599 | 29.367390 | negative | MGI_C57BL6J_1915009 | Plgrkt | NCBI_Gene:67759,ENSEMBL:ENSMUSG00000016495 | MGI:1915009 | protein coding gene | plasminogen receptor, C-terminal lysine transmembrane protein |
| 19 | gene | 29.367438 | 29.388095 | positive | MGI_C57BL6J_1926446 | Cd274 | NCBI_Gene:60533,ENSEMBL:ENSMUSG00000016496 | MGI:1926446 | protein coding gene | CD274 antigen |
| 19 | gene | 29.373417 | 29.385383 | negative | MGI_C57BL6J_5595202 | Gm36043 | NCBI_Gene:102639821,ENSEMBL:ENSMUSG00000117964 | MGI:5595202 | lncRNA gene | predicted gene, 36043 |
| 19 | gene | 29.410919 | 29.472927 | positive | MGI_C57BL6J_1930125 | Pdcd1lg2 | NCBI_Gene:58205,ENSEMBL:ENSMUSG00000016498 | MGI:1930125 | protein coding gene | programmed cell death 1 ligand 2 |
| 19 | gene | 29.464750 | 29.523159 | negative | MGI_C57BL6J_1925029 | A930007I19Rik | NCBI_Gene:77779,ENSEMBL:ENSMUSG00000097855 | MGI:1925029 | lncRNA gene | RIKEN cDNA A930007I19 gene |
| 19 | gene | 29.522282 | 29.606829 | positive | MGI_C57BL6J_1924893 | Ric1 | NCBI_Gene:226089,ENSEMBL:ENSMUSG00000038658 | MGI:1924893 | protein coding gene | RAB6A GEF complex partner 1 |
| 19 | gene | 29.608214 | 29.648510 | negative | MGI_C57BL6J_106250 | Ermp1 | NCBI_Gene:226090,ENSEMBL:ENSMUSG00000046324 | MGI:106250 | protein coding gene | endoplasmic reticulum metallopeptidase 1 |
| 19 | gene | 29.682177 | 29.684238 | negative | MGI_C57BL6J_5595285 | Gm36126 | NCBI_Gene:102639928 | MGI:5595285 | lncRNA gene | predicted gene, 36126 |
| 19 | gene | 29.697541 | 29.708634 | positive | MGI_C57BL6J_108454 | Mlana | NCBI_Gene:77836,ENSEMBL:ENSMUSG00000024806 | MGI:108454 | protein coding gene | melan-A |
| 19 | gene | 29.714402 | 29.808053 | negative | MGI_C57BL6J_2444398 | 9930021J03Rik | NCBI_Gene:240613,ENSEMBL:ENSMUSG00000046138 | MGI:2444398 | protein coding gene | RIKEN cDNA 9930021J03 gene |
| 19 | gene | 29.806663 | 29.810096 | positive | MGI_C57BL6J_6303267 | Gm50373 | ENSEMBL:ENSMUSG00000118251 | MGI:6303267 | lncRNA gene | predicted gene, 50373 |
| 19 | gene | 29.808108 | 29.812974 | negative | MGI_C57BL6J_2683212 | Ranbp6 | NCBI_Gene:240614,ENSEMBL:ENSMUSG00000074909 | MGI:2683212 | protein coding gene | RAN binding protein 6 |
| 19 | gene | 29.838453 | 29.927333 | negative | MGI_C57BL6J_5624717 | Gm41832 | NCBI_Gene:105246558 | MGI:5624717 | lncRNA gene | predicted gene, 41832 |
| 19 | pseudogene | 29.874191 | 29.875217 | negative | MGI_C57BL6J_6303279 | Gm50380 | ENSEMBL:ENSMUSG00000117883 | MGI:6303279 | pseudogene | predicted gene, 50380 |
| 19 | gene | 29.877449 | 29.884689 | positive | MGI_C57BL6J_5595368 | Gm36209 | NCBI_Gene:102640041 | MGI:5595368 | lncRNA gene | predicted gene, 36209 |
| 19 | gene | 29.893703 | 29.896235 | positive | MGI_C57BL6J_5624716 | Gm41831 | NCBI_Gene:105246557 | MGI:5624716 | lncRNA gene | predicted gene, 41831 |
| 19 | gene | 29.925114 | 29.960718 | positive | MGI_C57BL6J_1924375 | Il33 | NCBI_Gene:77125,ENSEMBL:ENSMUSG00000024810 | MGI:1924375 | protein coding gene | interleukin 33 |
| 19 | gene | 29.986099 | 29.986264 | positive | MGI_C57BL6J_5455823 | Gm26046 | ENSEMBL:ENSMUSG00000087937 | MGI:5455823 | snRNA gene | predicted gene, 26046 |
| 19 | gene | 30.003790 | 30.006020 | positive | MGI_C57BL6J_1913995 | Trpd52l3 | NCBI_Gene:66745,ENSEMBL:ENSMUSG00000024815 | MGI:1913995 | protein coding gene | tumor protein D52-like 3 |
| 19 | gene | 30.030435 | 30.093726 | positive | MGI_C57BL6J_1923718 | Uhrf2 | NCBI_Gene:109113,ENSEMBL:ENSMUSG00000024817 | MGI:1923718 | protein coding gene | ubiquitin-like, containing PHD and RING finger domains 2 |
| 19 | gene | 30.098441 | 30.175441 | negative | MGI_C57BL6J_1341155 | Gldc | NCBI_Gene:104174,ENSEMBL:ENSMUSG00000024827 | MGI:1341155 | protein coding gene | glycine decarboxylase |
| 19 | pseudogene | 30.160620 | 30.161030 | positive | MGI_C57BL6J_3644512 | Rpl31-ps20 | NCBI_Gene:625917,ENSEMBL:ENSMUSG00000058607 | MGI:3644512 | pseudogene | ribosomal protein L31, pseudogene 20 |
| 19 | gene | 30.164751 | 30.165769 | negative | MGI_C57BL6J_1924689 | 9530025L08Rik | NA | NA | unclassified gene | RIKEN cDNA 9530025L08 gene |
| 19 | gene | 30.232906 | 30.239687 | positive | MGI_C57BL6J_96924 | Mbl2 | NCBI_Gene:17195,ENSEMBL:ENSMUSG00000024863 | MGI:96924 | protein coding gene | mannose-binding lectin (protein C) 2 |
| 19 | gene | 30.261441 | 30.261517 | negative | MGI_C57BL6J_5453885 | Gm24108 | ENSEMBL:ENSMUSG00000093236 | MGI:5453885 | miRNA gene | predicted gene, 24108 |
| 19 | gene | 30.264880 | 30.265160 | positive | MGI_C57BL6J_5454660 | Gm24883 | ENSEMBL:ENSMUSG00000089016 | MGI:5454660 | unclassified non-coding RNA gene | predicted gene, 24883 |
| 19 | pseudogene | 30.352005 | 30.355618 | negative | MGI_C57BL6J_5595165 | Gm36006 | NCBI_Gene:102639775 | MGI:5595165 | pseudogene | predicted gene, 36006 |
| 19 | gene | 30.358217 | 30.360680 | negative | MGI_C57BL6J_1920607 | 1700048J15Rik | NA | NA | unclassified gene | RIKEN cDNA 1700048J15 gene |
| 19 | gene | 30.371465 | 30.395327 | negative | MGI_C57BL6J_5595478 | Gm36319 | NCBI_Gene:102640191 | MGI:5595478 | lncRNA gene | predicted gene, 36319 |
| 19 | pseudogene | 30.490703 | 30.491363 | negative | MGI_C57BL6J_5010070 | Gm17885 | NCBI_Gene:100416039,ENSEMBL:ENSMUSG00000118374 | MGI:5010070 | pseudogene | predicted gene, 17885 |
| 19 | pseudogene | 30.531476 | 30.531929 | negative | MGI_C57BL6J_6303088 | Gm50264 | ENSEMBL:ENSMUSG00000117953 | MGI:6303088 | pseudogene | predicted gene, 50264 |
| 19 | pseudogene | 30.538876 | 30.539679 | negative | MGI_C57BL6J_3645534 | Ppp1r2-ps3 | NCBI_Gene:546723,ENSEMBL:ENSMUSG00000058816 | MGI:3645534 | pseudogene | protein phosphatase 1, regulatory (inhibitor) subunit 2, pseudogene 3 |
| 19 | gene | 30.545863 | 30.549665 | negative | MGI_C57BL6J_1329040 | Dkk1 | NCBI_Gene:13380,ENSEMBL:ENSMUSG00000024868 | MGI:1329040 | protein coding gene | dickkopf WNT signaling pathway inhibitor 1 |
| 19 | gene | 30.564487 | 31.765309 | negative | MGI_C57BL6J_108174 | Prkg1 | NCBI_Gene:19091,ENSEMBL:ENSMUSG00000052920 | MGI:108174 | protein coding gene | protein kinase, cGMP-dependent, type I |
| 19 | gene | 30.704081 | 30.704398 | positive | MGI_C57BL6J_5453203 | Gm23426 | ENSEMBL:ENSMUSG00000084432 | MGI:5453203 | unclassified non-coding RNA gene | predicted gene, 23426 |
| 19 | pseudogene | 30.878834 | 30.879352 | positive | MGI_C57BL6J_5011208 | Gm19023 | NCBI_Gene:100418132 | MGI:5011208 | pseudogene | predicted gene, 19023 |
| 19 | pseudogene | 30.930380 | 30.930962 | negative | MGI_C57BL6J_3648551 | Gm6642 | NCBI_Gene:625995 | MGI:3648551 | pseudogene | predicted gene 6642 |
| 19 | gene | 31.082837 | 31.087069 | positive | MGI_C57BL6J_1932622 | Cstf2t | NCBI_Gene:83410,ENSEMBL:ENSMUSG00000053536 | MGI:1932622 | protein coding gene | cleavage stimulation factor, 3’ pre-RNA subunit 2, tau |
| 19 | pseudogene | 31.112546 | 31.113095 | positive | MGI_C57BL6J_6303091 | Gm50266 | ENSEMBL:ENSMUSG00000118011 | MGI:6303091 | pseudogene | predicted gene, 50266 |
| 19 | gene | 31.127685 | 31.137532 | positive | MGI_C57BL6J_5624719 | Gm41834 | NCBI_Gene:105246560 | MGI:5624719 | lncRNA gene | predicted gene, 41834 |
| 19 | gene | 31.196484 | 31.197357 | negative | MGI_C57BL6J_1920156 | 2900042A17Rik | NA | NA | unclassified gene | RIKEN cDNA 2900042A17 gene |
| 19 | gene | 31.213543 | 31.216396 | negative | MGI_C57BL6J_1925353 | 8430431K14Rik | NCBI_Gene:78103 | MGI:1925353 | lncRNA gene | RIKEN cDNA 8430431K14 gene |
| 19 | gene | 31.505322 | 31.507369 | negative | MGI_C57BL6J_1921789 | 9030425L15Rik | NA | NA | unclassified gene | RIKEN cDNA 9030425L15 gene |
| 19 | gene | 31.542570 | 31.550540 | negative | MGI_C57BL6J_2442795 | 9330185G11Rik | NA | NA | unclassified gene | RIKEN cDNA 9330185G11 gene |
| 19 | gene | 31.868755 | 31.952365 | positive | MGI_C57BL6J_1917115 | A1cf | NCBI_Gene:69865,ENSEMBL:ENSMUSG00000052595 | MGI:1917115 | protein coding gene | APOBEC1 complementation factor |
| 19 | pseudogene | 31.913644 | 31.914916 | positive | MGI_C57BL6J_5011426 | Gm19241 | NCBI_Gene:100418483,ENSEMBL:ENSMUSG00000114443 | MGI:5011426 | pseudogene | predicted gene, 19241 |
| 19 | gene | 31.950084 | 31.952361 | positive | MGI_C57BL6J_1918870 | 9130016M20Rik | ENSEMBL:ENSMUSG00000118366 | MGI:1918870 | lncRNA gene | RIKEN cDNA 9130016M20 gene |
| 19 | gene | 31.982597 | 32.108242 | negative | MGI_C57BL6J_1859310 | Asah2 | NCBI_Gene:54447,ENSEMBL:ENSMUSG00000024887 | MGI:1859310 | protein coding gene | N-acylsphingosine amidohydrolase 2 |
| 19 | pseudogene | 32.073335 | 32.074926 | positive | MGI_C57BL6J_3645706 | Gm8943 | NCBI_Gene:668042 | MGI:3645706 | pseudogene | predicted gene 8943 |
| 19 | gene | 32.122727 | 32.389714 | negative | MGI_C57BL6J_2444110 | Sgms1 | NCBI_Gene:208449,ENSEMBL:ENSMUSG00000040451 | MGI:2444110 | protein coding gene | sphingomyelin synthase 1 |
| 19 | gene | 32.204182 | 32.209273 | negative | MGI_C57BL6J_6302997 | Gm50210 | ENSEMBL:ENSMUSG00000118064 | MGI:6302997 | lncRNA gene | predicted gene, 50210 |
| 19 | gene | 32.389216 | 32.391190 | positive | MGI_C57BL6J_1914438 | 2700046G09Rik | NCBI_Gene:67188,ENSEMBL:ENSMUSG00000097787 | MGI:1914438 | lncRNA gene | RIKEN cDNA 2700046G09 gene |
| 19 | pseudogene | 32.423192 | 32.423582 | positive | MGI_C57BL6J_5621501 | Gm38616 | NCBI_Gene:102642581,ENSEMBL:ENSMUSG00000118355 | MGI:5621501 | pseudogene | predicted gene, 38616 |
| 19 | pseudogene | 32.439941 | 32.441024 | negative | MGI_C57BL6J_3779973 | Gm9563 | NCBI_Gene:672490,ENSEMBL:ENSMUSG00000118329 | MGI:3779973 | pseudogene | predicted gene 9563 |
| 19 | pseudogene | 32.465865 | 32.466578 | negative | MGI_C57BL6J_3642682 | Rpl9-ps6 | NCBI_Gene:100042823,ENSEMBL:ENSMUSG00000062456 | MGI:3642682 | pseudogene | ribosomal protein L9, pseudogene 6 |
| 19 | gene | 32.470601 | 32.483595 | negative | MGI_C57BL6J_5826296 | Gm46659 | NCBI_Gene:108168420 | MGI:5826296 | lncRNA gene | predicted gene, 46659 |
| 19 | gene | 32.485769 | 32.515370 | positive | MGI_C57BL6J_1336159 | Minpp1 | NCBI_Gene:17330,ENSEMBL:ENSMUSG00000024896 | MGI:1336159 | protein coding gene | multiple inositol polyphosphate histidine phosphatase 1 |
| 19 | gene | 32.543347 | 32.563611 | positive | MGI_C57BL6J_5595578 | Gm36419 | NCBI_Gene:102640323,ENSEMBL:ENSMUSG00000118168 | MGI:5595578 | lncRNA gene | predicted gene, 36419 |
| 19 | gene | 32.595645 | 32.667187 | positive | MGI_C57BL6J_1330223 | Papss2 | NCBI_Gene:23972,ENSEMBL:ENSMUSG00000024899 | MGI:1330223 | protein coding gene | 3’-phosphoadenosine 5’-phosphosulfate synthase 2 |
| 19 | pseudogene | 32.658479 | 32.660200 | negative | MGI_C57BL6J_5595651 | Gm36492 | NCBI_Gene:102640429 | MGI:5595651 | pseudogene | predicted gene, 36492 |
| 19 | gene | 32.671638 | 32.739786 | negative | MGI_C57BL6J_1915229 | Atad1 | NCBI_Gene:67979,ENSEMBL:ENSMUSG00000013662 | MGI:1915229 | protein coding gene | ATPase family, AAA domain containing 1 |
| 19 | gene | 32.754378 | 32.756612 | negative | MGI_C57BL6J_5595725 | Gm36566 | NCBI_Gene:102640527 | MGI:5595725 | protein coding gene | predicted gene, 36566 |
| 19 | gene | 32.757497 | 32.826160 | positive | MGI_C57BL6J_109583 | Pten | NCBI_Gene:19211,ENSEMBL:ENSMUSG00000013663 | MGI:109583 | protein coding gene | phosphatase and tensin homolog |
| 19 | gene | 32.933345 | 32.939008 | positive | MGI_C57BL6J_5595933 | Gm36774 | NCBI_Gene:102640791 | MGI:5595933 | lncRNA gene | predicted gene, 36774 |
| 19 | gene | 32.959987 | 32.976159 | positive | MGI_C57BL6J_5596073 | Gm36914 | NCBI_Gene:102640981 | MGI:5596073 | lncRNA gene | predicted gene, 36914 |
| 19 | gene | 32.966066 | 32.980390 | negative | MGI_C57BL6J_5596019 | Gm36860 | NCBI_Gene:102640908,ENSEMBL:ENSMUSG00000117664 | MGI:5596019 | lncRNA gene | predicted gene, 36860 |
| 19 | gene | 32.969950 | 32.970308 | negative | MGI_C57BL6J_5452259 | Gm22482 | ENSEMBL:ENSMUSG00000084512 | MGI:5452259 | unclassified non-coding RNA gene | predicted gene, 22482 |
| 19 | gene | 33.039471 | 33.074928 | negative | MGI_C57BL6J_5624720 | Gm41835 | NCBI_Gene:105246561 | MGI:5624720 | lncRNA gene | predicted gene, 41835 |
| 19 | gene | 33.070072 | 33.102167 | positive | MGI_C57BL6J_5589022 | Gm29863 | NCBI_Gene:102631554,ENSEMBL:ENSMUSG00000117914 | MGI:5589022 | lncRNA gene | predicted gene, 29863 |
| 19 | gene | 33.122647 | 33.134819 | positive | MGI_C57BL6J_5589105 | Gm29946 | NCBI_Gene:102631663,ENSEMBL:ENSMUSG00000118196 | MGI:5589105 | lncRNA gene | predicted gene, 29946 |
| 19 | pseudogene | 33.137744 | 33.392295 | negative | MGI_C57BL6J_1915045 | Rnls | NCBI_Gene:67795,ENSEMBL:ENSMUSG00000071573 | MGI:1915045 | polymorphic pseudogene | renalase, FAD-dependent amine oxidase |
| 19 | gene | 33.157112 | 33.157201 | positive | MGI_C57BL6J_4950409 | Mir3970 | miRBase:MI0016980,NCBI_Gene:100628575,ENSEMBL:ENSMUSG00000106278 | MGI:4950409 | miRNA gene | microRNA 3970 |
| 19 | pseudogene | 33.353066 | 33.354120 | negative | MGI_C57BL6J_5010027 | Gm17842 | NCBI_Gene:100415967 | MGI:5010027 | pseudogene | predicted gene, 17842 |
| 19 | gene | 33.392253 | 33.392745 | negative | MGI_C57BL6J_1914484 | 2810449D17Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 2810449D17 gene |
| 19 | gene | 33.392433 | 33.420370 | positive | MGI_C57BL6J_5589157 | Gm29998 | NCBI_Gene:102631731 | MGI:5589157 | lncRNA gene | predicted gene, 29998 |
| 19 | pseudogene | 33.405917 | 33.420642 | negative | MGI_C57BL6J_3779701 | Gm7237 | NCBI_Gene:638381,ENSEMBL:ENSMUSG00000087112 | MGI:3779701 | pseudogene | predicted gene 7237 |
| 19 | pseudogene | 33.447613 | 33.447798 | negative | MGI_C57BL6J_5011909 | Gm19724 | NCBI_Gene:102631943,ENSEMBL:ENSMUSG00000118289 | MGI:5011909 | pseudogene | predicted gene, 19724 |
| 19 | pseudogene | 33.459609 | 33.473191 | negative | MGI_C57BL6J_3648531 | Lipo5 | NCBI_Gene:668095,ENSEMBL:ENSMUSG00000086875 | MGI:3648531 | pseudogene | lipase, member O5 |
| 19 | gene | 33.498037 | 33.517780 | negative | MGI_C57BL6J_3779637 | Lipo4 | NCBI_Gene:628236,ENSEMBL:ENSMUSG00000079344 | MGI:3779637 | protein coding gene | lipase, member O4 |
| 19 | pseudogene | 33.550198 | 33.550397 | negative | MGI_C57BL6J_6303038 | Gm50233 | ENSEMBL:ENSMUSG00000118121 | MGI:6303038 | pseudogene | predicted gene, 50233 |
| 19 | gene | 33.555160 | 33.761951 | negative | MGI_C57BL6J_2147592 | Lipo3 | NCBI_Gene:381236,ENSEMBL:ENSMUSG00000024766 | MGI:2147592 | protein coding gene | lipase, member O3 |
| 19 | gene | 33.589971 | 33.590999 | positive | MGI_C57BL6J_1916554 | 1700001A13Rik | NA | NA | unclassified gene | RIKEN cDNA 1700001A13 gene |
| 19 | gene | 33.592607 | 33.592713 | negative | MGI_C57BL6J_5530822 | Gm27440 | ENSEMBL:ENSMUSG00000098770 | MGI:5530822 | miRNA gene | predicted gene, 27440 |
| 19 | gene | 33.612707 | 33.625252 | negative | MGI_C57BL6J_3647649 | Gm8978 | NCBI_Gene:668106,ENSEMBL:ENSMUSG00000086812 | MGI:3647649 | protein coding gene | predicted gene 8978 |
| 19 | pseudogene | 33.668266 | 33.668463 | negative | MGI_C57BL6J_5011940 | Gm19755 | NCBI_Gene:102632180,ENSEMBL:ENSMUSG00000118360 | MGI:5011940 | pseudogene | predicted gene, 19755 |
| 19 | pseudogene | 33.673329 | 33.673928 | negative | MGI_C57BL6J_5010478 | Gm18293 | NCBI_Gene:100416852,ENSEMBL:ENSMUSG00000117667 | MGI:5010478 | pseudogene | predicted gene, 18293 |
| 19 | pseudogene | 33.674073 | 33.688481 | negative | MGI_C57BL6J_3644463 | Gm8980 | NCBI_Gene:668112,ENSEMBL:ENSMUSG00000085924 | MGI:3644463 | pseudogene | predicted gene 8980 |
| 19 | gene | 33.719670 | 33.762109 | negative | MGI_C57BL6J_3644466 | Lipo2 | NCBI_Gene:101055671,ENSEMBL:ENSMUSG00000087303 | MGI:3644466 | protein coding gene | lipase, member O2 |
| 19 | gene | 33.775498 | 33.787459 | negative | MGI_C57BL6J_3647308 | Lipo1 | NCBI_Gene:329055,ENSEMBL:ENSMUSG00000079342 | MGI:3647308 | protein coding gene | lipase, member O1 |
| 19 | pseudogene | 33.822897 | 33.826711 | positive | MGI_C57BL6J_3645618 | Gm5519 | NCBI_Gene:433241,ENSEMBL:ENSMUSG00000037603 | MGI:3645618 | pseudogene | predicted pseudogene 5519 |
| 19 | pseudogene | 33.838827 | 33.842598 | negative | MGI_C57BL6J_3647410 | Gm8988 | ENSEMBL:ENSMUSG00000085934 | MGI:3647410 | pseudogene | predicted gene 8988 |
| 19 | pseudogene | 33.853164 | 33.872207 | negative | MGI_C57BL6J_6302861 | Gm50120 | ENSEMBL:ENSMUSG00000117821 | MGI:6302861 | pseudogene | predicted gene, 50120 |
| 19 | gene | 33.954235 | 33.954341 | positive | MGI_C57BL6J_5452872 | Gm23095 | ENSEMBL:ENSMUSG00000065380 | MGI:5452872 | snRNA gene | predicted gene, 23095 |
| 19 | gene | 33.961187 | 33.976813 | positive | MGI_C57BL6J_1914967 | Lipf | NCBI_Gene:67717,ENSEMBL:ENSMUSG00000024768 | MGI:1914967 | protein coding gene | lipase, gastric |
| 19 | gene | 34.008254 | 34.047903 | positive | MGI_C57BL6J_2679259 | Lipk | NCBI_Gene:240633,ENSEMBL:ENSMUSG00000024771 | MGI:2679259 | protein coding gene | lipase, family member K |
| 19 | gene | 34.056140 | 34.058360 | negative | MGI_C57BL6J_5590459 | Gm31300 | NCBI_Gene:102633486,ENSEMBL:ENSMUSG00000118302 | MGI:5590459 | lncRNA gene | predicted gene, 31300 |
| 19 | gene | 34.067058 | 34.085946 | positive | MGI_C57BL6J_1917416 | Lipn | NCBI_Gene:70166,ENSEMBL:ENSMUSG00000024770 | MGI:1917416 | protein coding gene | lipase, family member N |
| 19 | gene | 34.068931 | 34.069054 | positive | MGI_C57BL6J_5453111 | Gm23334 | ENSEMBL:ENSMUSG00000080678 | MGI:5453111 | miRNA gene | predicted gene, 23334 |
| 19 | gene | 34.100933 | 34.122687 | positive | MGI_C57BL6J_1926003 | Lipm | NCBI_Gene:78753,ENSEMBL:ENSMUSG00000056078 | MGI:1926003 | protein coding gene | lipase, family member M |
| 19 | gene | 34.121469 | 34.166053 | negative | MGI_C57BL6J_1277101 | Ankrd22 | NCBI_Gene:52024,ENSEMBL:ENSMUSG00000024774 | MGI:1277101 | protein coding gene | ankyrin repeat domain 22 |
| 19 | gene | 34.192186 | 34.240333 | positive | MGI_C57BL6J_1923880 | Stambpl1 | NCBI_Gene:76630,ENSEMBL:ENSMUSG00000024776 | MGI:1923880 | protein coding gene | STAM binding protein like 1 |
| 19 | gene | 34.240336 | 34.255590 | negative | MGI_C57BL6J_87909 | Acta2 | NCBI_Gene:11475,ENSEMBL:ENSMUSG00000035783 | MGI:87909 | protein coding gene | actin, alpha 2, smooth muscle, aorta |
| 19 | gene | 34.256636 | 34.278225 | negative | MGI_C57BL6J_5590514 | Gm31355 | NCBI_Gene:102633559 | MGI:5590514 | lncRNA gene | predicted gene, 31355 |
| 19 | pseudogene | 34.284280 | 34.284537 | negative | MGI_C57BL6J_6303047 | Gm50239 | ENSEMBL:ENSMUSG00000118066 | MGI:6303047 | pseudogene | predicted gene, 50239 |
| 19 | gene | 34.290659 | 34.327775 | positive | MGI_C57BL6J_95484 | Fas | NCBI_Gene:14102,ENSEMBL:ENSMUSG00000024778 | MGI:95484 | protein coding gene | Fas (TNF receptor superfamily member 6) |
| 19 | gene | 34.294137 | 34.294295 | positive | MGI_C57BL6J_5531200 | Gm27818 | ENSEMBL:ENSMUSG00000098670 | MGI:5531200 | unclassified non-coding RNA gene | predicted gene, 27818 |
| 19 | gene | 34.395647 | 34.395755 | negative | MGI_C57BL6J_5452837 | Gm23060 | ENSEMBL:ENSMUSG00000076300 | MGI:5452837 | miRNA gene | predicted gene, 23060 |
| 19 | gene | 34.473778 | 34.475164 | negative | MGI_C57BL6J_1333869 | Ch25h | NCBI_Gene:12642,ENSEMBL:ENSMUSG00000050370 | MGI:1333869 | protein coding gene | cholesterol 25-hydroxylase |
| 19 | gene | 34.474808 | 34.481546 | positive | MGI_C57BL6J_5477396 | Gm26902 | ENSEMBL:ENSMUSG00000097558 | MGI:5477396 | lncRNA gene | predicted gene, 26902 |
| 19 | gene | 34.492316 | 34.527474 | negative | MGI_C57BL6J_96789 | Lipa | NCBI_Gene:16889,ENSEMBL:ENSMUSG00000024781 | MGI:96789 | protein coding gene | lysosomal acid lipase A |
| 19 | gene | 34.543288 | 34.544729 | negative | MGI_C57BL6J_5590576 | Gm31417 | NCBI_Gene:102633635 | MGI:5590576 | lncRNA gene | predicted gene, 31417 |
| 19 | pseudogene | 34.549264 | 34.549392 | positive | MGI_C57BL6J_6303079 | Gm50257 | ENSEMBL:ENSMUSG00000118048 | MGI:6303079 | pseudogene | predicted gene, 50257 |
| 19 | gene | 34.550549 | 34.576534 | positive | MGI_C57BL6J_99449 | Ifit2 | NCBI_Gene:15958,ENSEMBL:ENSMUSG00000045932 | MGI:99449 | protein coding gene | interferon-induced protein with tetratricopeptide repeats 2 |
| 19 | gene | 34.583142 | 34.588982 | positive | MGI_C57BL6J_1101055 | Ifit3 | NCBI_Gene:15959,ENSEMBL:ENSMUSG00000074896 | MGI:1101055 | protein coding gene | interferon-induced protein with tetratricopeptide repeats 3 |
| 19 | gene | 34.592888 | 34.662997 | negative | MGI_C57BL6J_3650685 | Ifit1bl1 | NCBI_Gene:667373,ENSEMBL:ENSMUSG00000079339 | MGI:3650685 | protein coding gene | interferon induced protein with tetratricpeptide repeats 1B like 1 |
| 19 | gene | 34.607540 | 34.613409 | positive | MGI_C57BL6J_3698419 | Ifit3b | NCBI_Gene:667370,ENSEMBL:ENSMUSG00000062488 | MGI:3698419 | protein coding gene | interferon-induced protein with tetratricopeptide repeats 3B |
| 19 | gene | 34.617049 | 34.640743 | negative | MGI_C57BL6J_2148249 | Ifit1bl2 | NCBI_Gene:112419,ENSEMBL:ENSMUSG00000067297 | MGI:2148249 | protein coding gene | interferon induced protein with tetratricopeptide repeats 1B like 2 |
| 19 | pseudogene | 34.623630 | 34.624804 | positive | MGI_C57BL6J_5010061 | Cnn2-ps | NCBI_Gene:100416023,ENSEMBL:ENSMUSG00000118260 | MGI:5010061 | pseudogene | calponin2, pseudogene |
| 19 | gene | 34.640871 | 34.650009 | positive | MGI_C57BL6J_99450 | Ifit1 | NCBI_Gene:15957,ENSEMBL:ENSMUSG00000034459 | MGI:99450 | protein coding gene | interferon-induced protein with tetratricopeptide repeats 1 |
| 19 | gene | 34.668401 | 34.747823 | negative | MGI_C57BL6J_2147716 | Slc16a12 | NCBI_Gene:240638,ENSEMBL:ENSMUSG00000009378 | MGI:2147716 | protein coding gene | solute carrier family 16 (monocarboxylic acid transporters), member 12 |
| 19 | pseudogene | 34.682183 | 34.684822 | negative | MGI_C57BL6J_5826291 | Gm46654 | NCBI_Gene:108168415 | MGI:5826291 | pseudogene | predicted gene, 46654 |
| 19 | pseudogene | 34.710008 | 34.711016 | positive | MGI_C57BL6J_5010610 | Gm18425 | NCBI_Gene:100417152,ENSEMBL:ENSMUSG00000118098 | MGI:5010610 | pseudogene | predicted gene, 18425 |
| 19 | gene | 34.750510 | 34.750613 | positive | MGI_C57BL6J_5530858 | Gm27476 | ENSEMBL:ENSMUSG00000098742 | MGI:5530858 | miRNA gene | predicted gene, 27476 |
| 19 | pseudogene | 34.773881 | 34.774451 | positive | MGI_C57BL6J_6302944 | Gm50173 | ENSEMBL:ENSMUSG00000118083 | MGI:6302944 | pseudogene | predicted gene, 50173 |
| 19 | gene | 34.796178 | 34.796338 | positive | MGI_C57BL6J_5451859 | Gm22082 | ENSEMBL:ENSMUSG00000088212 | MGI:5451859 | snRNA gene | predicted gene, 22082 |
| 19 | pseudogene | 34.804262 | 34.804759 | positive | MGI_C57BL6J_6302856 | Gm50116 | ENSEMBL:ENSMUSG00000117945 | MGI:6302856 | pseudogene | predicted gene, 50116 |
| 19 | gene | 34.806935 | 34.879593 | negative | MGI_C57BL6J_1922985 | Pank1 | NCBI_Gene:75735,ENSEMBL:ENSMUSG00000033610 | MGI:1922985 | protein coding gene | pantothenate kinase 1 |
| 19 | gene | 34.820687 | 34.820773 | negative | MGI_C57BL6J_3619063 | Mir107 | miRBase:MI0000684,NCBI_Gene:723826,ENSEMBL:ENSMUSG00000065594 | MGI:3619063 | miRNA gene | microRNA 107 |
| 19 | gene | 34.922341 | 34.975750 | positive | MGI_C57BL6J_2444576 | Kif20b | NCBI_Gene:240641,ENSEMBL:ENSMUSG00000024795 | MGI:2444576 | protein coding gene | kinesin family member 20B |
| 19 | gene | 34.965396 | 34.965469 | positive | MGI_C57BL6J_3837023 | Mir1950 | miRBase:MI0009941,NCBI_Gene:100316775,ENSEMBL:ENSMUSG00000089151 | MGI:3837023 | miRNA gene | microRNA 1950 |
| 19 | pseudogene | 35.051830 | 35.052744 | negative | MGI_C57BL6J_3643322 | Gm5248 | NCBI_Gene:383464,ENSEMBL:ENSMUSG00000101966 | MGI:3643322 | pseudogene | predicted gene 5248 |
| 19 | gene | 35.054465 | 35.083440 | positive | MGI_C57BL6J_6302884 | Gm50137 | ENSEMBL:ENSMUSG00000117865 | MGI:6302884 | lncRNA gene | predicted gene, 50137 |
| 19 | gene | 35.117283 | 35.133451 | positive | MGI_C57BL6J_6302890 | Gm50140 | ENSEMBL:ENSMUSG00000117973 | MGI:6302890 | lncRNA gene | predicted gene, 50140 |
| 19 | gene | 35.151913 | 35.156629 | positive | MGI_C57BL6J_5590727 | Gm31568 | NCBI_Gene:102633838 | MGI:5590727 | lncRNA gene | predicted gene, 31568 |
| 19 | gene | 35.203235 | 35.203366 | positive | MGI_C57BL6J_5453783 | Gm24006 | ENSEMBL:ENSMUSG00000089087 | MGI:5453783 | snoRNA gene | predicted gene, 24006 |
| 19 | gene | 35.276453 | 35.276522 | negative | MGI_C57BL6J_5452852 | Gm23075 | ENSEMBL:ENSMUSG00000093373 | MGI:5452852 | miRNA gene | predicted gene, 23075 |
| 19 | gene | 35.605674 | 35.605804 | negative | MGI_C57BL6J_5452630 | Gm22853 | ENSEMBL:ENSMUSG00000092715 | MGI:5452630 | rRNA gene | predicted gene, 22853 |
| 19 | gene | 35.700349 | 35.766127 | negative | MGI_C57BL6J_5624721 | Gm41836 | NCBI_Gene:105246562,ENSEMBL:ENSMUSG00000118212 | MGI:5624721 | lncRNA gene | predicted gene, 41836 |
| 19 | gene | 35.759933 | 35.760065 | negative | MGI_C57BL6J_5455388 | Gm25611 | ENSEMBL:ENSMUSG00000077205 | MGI:5455388 | snoRNA gene | predicted gene, 25611 |
| 19 | gene | 35.835217 | 35.924735 | negative | MGI_C57BL6J_3028035 | A830019P07Rik | NCBI_Gene:329056,ENSEMBL:ENSMUSG00000094707 | MGI:3028035 | lncRNA gene | RIKEN cDNA A830019P07 gene |
| 19 | pseudogene | 35.870079 | 35.870772 | positive | MGI_C57BL6J_5010111 | Gm17926 | NCBI_Gene:100416119 | MGI:5010111 | pseudogene | predicted gene, 17926 |
| 19 | gene | 35.958415 | 36.058287 | negative | MGI_C57BL6J_99841 | Htr7 | NCBI_Gene:15566,ENSEMBL:ENSMUSG00000024798 | MGI:99841 | protein coding gene | 5-hydroxytryptamine (serotonin) receptor 7 |
| 19 | gene | 36.083689 | 36.104777 | positive | MGI_C57BL6J_1859683 | Rpp30 | NCBI_Gene:54364,ENSEMBL:ENSMUSG00000024800 | MGI:1859683 | protein coding gene | ribonuclease P/MRP 30 subunit |
| 19 | gene | 36.111961 | 36.120099 | negative | MGI_C57BL6J_1097717 | Ankrd1 | NCBI_Gene:107765,ENSEMBL:ENSMUSG00000024803 | MGI:1097717 | protein coding gene | ankyrin repeat domain 1 (cardiac muscle) |
| 19 | gene | 36.182580 | 36.221676 | positive | MGI_C57BL6J_5591240 | Gm32081 | NCBI_Gene:102634520,ENSEMBL:ENSMUSG00000117998 | MGI:5591240 | lncRNA gene | predicted gene, 32081 |
| 19 | gene | 36.215016 | 36.264437 | negative | MGI_C57BL6J_5591186 | Gm32027 | NCBI_Gene:102634446,ENSEMBL:ENSMUSG00000117990 | MGI:5591186 | lncRNA gene | predicted gene, 32027 |
| 19 | gene | 36.235979 | 36.243323 | positive | MGI_C57BL6J_5826301 | Gm46664 | NCBI_Gene:108168426 | MGI:5826301 | lncRNA gene | predicted gene, 46664 |
| 19 | gene | 36.251727 | 36.256336 | positive | MGI_C57BL6J_6302902 | Gm50147 | ENSEMBL:ENSMUSG00000118032 | MGI:6302902 | lncRNA gene | predicted gene, 50147 |
| 19 | pseudogene | 36.339044 | 36.339931 | negative | MGI_C57BL6J_5011345 | Gm19160 | NCBI_Gene:100418358 | MGI:5011345 | pseudogene | predicted gene, 19160 |
| 19 | gene | 36.347440 | 36.460970 | positive | MGI_C57BL6J_1923505 | Pcgf5 | NCBI_Gene:76073,ENSEMBL:ENSMUSG00000024805 | MGI:1923505 | protein coding gene | polycomb group ring finger 5 |
| 19 | gene | 36.347614 | 36.348298 | negative | MGI_C57BL6J_6096874 | Gm47735 | ENSEMBL:ENSMUSG00000114859 | MGI:6096874 | lncRNA gene | predicted gene, 47735 |
| 19 | pseudogene | 36.361343 | 36.361811 | positive | MGI_C57BL6J_6096932 | Gm47773 | ENSEMBL:ENSMUSG00000114285 | MGI:6096932 | pseudogene | predicted gene, 47773 |
| 19 | gene | 36.384792 | 36.384897 | negative | MGI_C57BL6J_5452947 | Gm23170 | ENSEMBL:ENSMUSG00000087783 | MGI:5452947 | snRNA gene | predicted gene, 23170 |
| 19 | pseudogene | 36.404404 | 36.406211 | positive | MGI_C57BL6J_3647150 | Gm9042 | NCBI_Gene:668201,ENSEMBL:ENSMUSG00000114409 | MGI:3647150 | pseudogene | predicted gene 9042 |
| 19 | gene | 36.448992 | 36.451157 | negative | MGI_C57BL6J_3642424 | F530104D19Rik | ENSEMBL:ENSMUSG00000114235 | MGI:3642424 | lncRNA gene | RIKEN cDNA F530104D19 gene |
| 19 | gene | 36.533923 | 36.538802 | negative | MGI_C57BL6J_5591302 | Gm32143 | NCBI_Gene:102634602 | MGI:5591302 | lncRNA gene | predicted gene, 32143 |
| 19 | gene | 36.552925 | 36.553977 | negative | MGI_C57BL6J_5624723 | Gm41838 | NCBI_Gene:105246565 | MGI:5624723 | lncRNA gene | predicted gene, 41838 |
| 19 | gene | 36.554053 | 36.621135 | positive | MGI_C57BL6J_2442663 | Hectd2 | NCBI_Gene:226098,ENSEMBL:ENSMUSG00000041180 | MGI:2442663 | protein coding gene | HECT domain E3 ubiquitin protein ligase 2 |
| 19 | gene | 36.614052 | 36.705641 | negative | MGI_C57BL6J_1919243 | Hectd2os | NCBI_Gene:668215,ENSEMBL:ENSMUSG00000087579 | MGI:1919243 | antisense lncRNA gene | Hectd2, opposite strand |
| 19 | gene | 36.731731 | 36.736653 | negative | MGI_C57BL6J_1858229 | Ppp1r3c | NCBI_Gene:53412,ENSEMBL:ENSMUSG00000067279 | MGI:1858229 | protein coding gene | protein phosphatase 1, regulatory subunit 3C |
| 19 | pseudogene | 36.818648 | 36.818930 | positive | MGI_C57BL6J_6302850 | Gm50112 | ENSEMBL:ENSMUSG00000117938 | MGI:6302850 | pseudogene | predicted gene, 50112 |
| 19 | gene | 36.834230 | 36.893477 | positive | MGI_C57BL6J_1921743 | Tnks2 | NCBI_Gene:74493,ENSEMBL:ENSMUSG00000024811 | MGI:1921743 | protein coding gene | tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 |
| 19 | gene | 36.917550 | 36.919615 | negative | MGI_C57BL6J_1919764 | Fgfbp3 | NCBI_Gene:72514,ENSEMBL:ENSMUSG00000047632 | MGI:1919764 | protein coding gene | fibroblast growth factor binding protein 3 |
| 19 | gene | 36.926079 | 37.014057 | positive | MGI_C57BL6J_2147538 | Btaf1 | NCBI_Gene:107182,ENSEMBL:ENSMUSG00000040565 | MGI:2147538 | protein coding gene | B-TFIID TATA-box binding protein associated factor 1 |
| 19 | gene | 37.019341 | 37.208601 | negative | MGI_C57BL6J_2443075 | Cpeb3 | NCBI_Gene:208922,ENSEMBL:ENSMUSG00000039652 | MGI:2443075 | protein coding gene | cytoplasmic polyadenylation element binding protein 3 |
| 19 | gene | 37.149085 | 37.149162 | negative | MGI_C57BL6J_5452491 | Gm22714 | ENSEMBL:ENSMUSG00000080544 | MGI:5452491 | ribozyme gene | predicted gene, 22714 |
| 19 | gene | 37.173843 | 37.196541 | positive | MGI_C57BL6J_2443002 | A330032B11Rik | NCBI_Gene:319931,ENSEMBL:ENSMUSG00000097591 | MGI:2443002 | lncRNA gene | RIKEN cDNA A330032B11 gene |
| 19 | gene | 37.179186 | 37.179291 | positive | MGI_C57BL6J_5453695 | Gm23918 | ENSEMBL:ENSMUSG00000092994 | MGI:5453695 | rRNA gene | predicted gene, 23918 |
| 19 | gene | 37.207527 | 37.224457 | positive | MGI_C57BL6J_1915207 | March5 | NCBI_Gene:69104,ENSEMBL:ENSMUSG00000023307 | MGI:1915207 | protein coding gene | membrane-associated ring finger (C3HC4) 5 |
| 19 | gene | 37.233382 | 37.233484 | negative | MGI_C57BL6J_5455045 | Gm25268 | ENSEMBL:ENSMUSG00000089143 | MGI:5455045 | snRNA gene | predicted gene, 25268 |
| 19 | pseudogene | 37.233603 | 37.234560 | positive | MGI_C57BL6J_3643992 | Gm9056 | NCBI_Gene:668230 | MGI:3643992 | pseudogene | predicted gene 9056 |
| 19 | gene | 37.258423 | 37.264095 | negative | MGI_C57BL6J_1924309 | 4931408D14Rik | NCBI_Gene:77059 | MGI:1924309 | lncRNA gene | RIKEN cDNA 4931408D14 gene |
| 19 | gene | 37.268741 | 37.337852 | negative | MGI_C57BL6J_96412 | Ide | NCBI_Gene:15925,ENSEMBL:ENSMUSG00000056999 | MGI:96412 | protein coding gene | insulin degrading enzyme |
| 19 | pseudogene | 37.295093 | 37.295837 | negative | MGI_C57BL6J_3782343 | Rpl10-ps6 | NCBI_Gene:100043010,ENSEMBL:ENSMUSG00000080885 | MGI:3782343 | pseudogene | ribosomal protein L10, pseudogene 6 |
| 19 | gene | 37.376403 | 37.421859 | positive | MGI_C57BL6J_1098231 | Kif11 | NCBI_Gene:16551,ENSEMBL:ENSMUSG00000012443 | MGI:1098231 | protein coding gene | kinesin family member 11 |
| 19 | pseudogene | 37.388339 | 37.389495 | positive | MGI_C57BL6J_3648745 | Gm8663 | NCBI_Gene:667490,ENSEMBL:ENSMUSG00000117988 | MGI:3648745 | pseudogene | predicted gene 8663 |
| 19 | pseudogene | 37.413424 | 37.413878 | negative | MGI_C57BL6J_5826286 | Gm46649 | NCBI_Gene:108168410,ENSEMBL:ENSMUSG00000118211 | MGI:5826286 | pseudogene | predicted gene, 46649 |
| 19 | gene | 37.432909 | 37.434656 | negative | MGI_C57BL6J_5611573 | Gm38345 | ENSEMBL:ENSMUSG00000102353 | MGI:5611573 | unclassified gene | predicted gene, 38345 |
| 19 | gene | 37.434810 | 37.440731 | positive | MGI_C57BL6J_96086 | Hhex | NCBI_Gene:15242,ENSEMBL:ENSMUSG00000024986 | MGI:96086 | protein coding gene | hematopoietically expressed homeobox |
| 19 | gene | 37.463746 | 37.463851 | positive | MGI_C57BL6J_5452803 | Gm23026 | ENSEMBL:ENSMUSG00000089397 | MGI:5452803 | snRNA gene | predicted gene, 23026 |
| 19 | gene | 37.465253 | 37.471834 | positive | MGI_C57BL6J_1925890 | 1700122C19Rik | NCBI_Gene:78640,ENSEMBL:ENSMUSG00000117800 | MGI:1925890 | lncRNA gene | RIKEN cDNA 1700122C19 gene |
| 19 | gene | 37.493599 | 37.495964 | positive | MGI_C57BL6J_5591453 | Gm32294 | NCBI_Gene:102634793 | MGI:5591453 | lncRNA gene | predicted gene, 32294 |
| 19 | gene | 37.536763 | 37.684059 | positive | MGI_C57BL6J_1351611 | Exoc6 | NCBI_Gene:107371,ENSEMBL:ENSMUSG00000053799 | MGI:1351611 | protein coding gene | exocyst complex component 6 |
| 19 | gene | 37.545104 | 37.546314 | negative | MGI_C57BL6J_5624727 | Gm41842 | NCBI_Gene:105246569 | MGI:5624727 | lncRNA gene | predicted gene, 41842 |
| 19 | gene | 37.677387 | 37.697564 | negative | MGI_C57BL6J_5591501 | Gm32342 | NCBI_Gene:102634858 | MGI:5591501 | lncRNA gene | predicted gene, 32342 |
| 19 | gene | 37.685195 | 37.693398 | positive | MGI_C57BL6J_2679699 | Cyp26c1 | NCBI_Gene:546726,ENSEMBL:ENSMUSG00000062432 | MGI:2679699 | protein coding gene | cytochrome P450, family 26, subfamily c, polypeptide 1 |
| 19 | gene | 37.697732 | 37.701536 | positive | MGI_C57BL6J_1096359 | Cyp26a1 | NCBI_Gene:13082,ENSEMBL:ENSMUSG00000024987 | MGI:1096359 | protein coding gene | cytochrome P450, family 26, subfamily a, polypeptide 1 |
| 19 | pseudogene | 37.722761 | 37.723516 | negative | MGI_C57BL6J_3647635 | Gm9066 | NCBI_Gene:668248,ENSEMBL:ENSMUSG00000117989 | MGI:3647635 | pseudogene | predicted gene 9066 |
| 19 | pseudogene | 37.784259 | 37.785765 | positive | MGI_C57BL6J_3644658 | Gm9067 | NCBI_Gene:668251,ENSEMBL:ENSMUSG00000117939 | MGI:3644658 | pseudogene | predicted gene 9067 |
| 19 | pseudogene | 37.870267 | 37.872256 | positive | MGI_C57BL6J_3647440 | Gm4757 | NCBI_Gene:209281,ENSEMBL:ENSMUSG00000118115 | MGI:3647440 | pseudogene | predicted gene 4757 |
| 19 | gene | 37.899036 | 38.043577 | negative | MGI_C57BL6J_1919192 | Myof | NCBI_Gene:226101,ENSEMBL:ENSMUSG00000048612 | MGI:1919192 | protein coding gene | myoferlin |
| 19 | gene | 38.051774 | 38.054528 | negative | MGI_C57BL6J_3588250 | I830134H01Rik | ENSEMBL:ENSMUSG00000069554 | MGI:3588250 | lncRNA gene | RIKEN cDNA I830134H01 gene |
| 19 | gene | 38.054981 | 38.074425 | positive | MGI_C57BL6J_1921357 | Cep55 | NCBI_Gene:74107,ENSEMBL:ENSMUSG00000024989 | MGI:1921357 | protein coding gene | centrosomal protein 55 |
| 19 | gene | 38.071929 | 38.072053 | positive | MGI_C57BL6J_5453077 | Gm23300 | ENSEMBL:ENSMUSG00000064646 | MGI:5453077 | snoRNA gene | predicted gene, 23300 |
| 19 | gene | 38.097071 | 38.114263 | positive | MGI_C57BL6J_2147577 | Ffar4 | NCBI_Gene:107221,ENSEMBL:ENSMUSG00000054200 | MGI:2147577 | protein coding gene | free fatty acid receptor 4 |
| 19 | gene | 38.116620 | 38.125321 | negative | MGI_C57BL6J_97879 | Rbp4 | NCBI_Gene:19662,ENSEMBL:ENSMUSG00000024990 | MGI:97879 | protein coding gene | retinol binding protein 4, plasma |
| 19 | gene | 38.125754 | 38.134496 | negative | MGI_C57BL6J_5591599 | Gm32440 | NCBI_Gene:102634991 | MGI:5591599 | lncRNA gene | predicted gene, 32440 |
| 19 | gene | 38.132781 | 38.183958 | positive | MGI_C57BL6J_105956 | Pde6c | NCBI_Gene:110855,ENSEMBL:ENSMUSG00000024992 | MGI:105956 | protein coding gene | phosphodiesterase 6C, cGMP specific, cone, alpha prime |
| 19 | gene | 38.188477 | 38.224517 | negative | MGI_C57BL6J_1917817 | Fra10ac1 | NCBI_Gene:70567,ENSEMBL:ENSMUSG00000054237 | MGI:1917817 | protein coding gene | FRA10AC1 homolog (human) |
| 19 | gene | 38.257836 | 38.260793 | negative | MGI_C57BL6J_5591733 | Gm32574 | NCBI_Gene:102635170 | MGI:5591733 | lncRNA gene | predicted gene, 32574 |
| 19 | gene | 38.257928 | 38.263285 | positive | MGI_C57BL6J_5591650 | Gm32491 | NCBI_Gene:102635059 | MGI:5591650 | lncRNA gene | predicted gene, 32491 |
| 19 | gene | 38.264535 | 38.272565 | negative | MGI_C57BL6J_5591930 | Gm32771 | NCBI_Gene:102635432 | MGI:5591930 | lncRNA gene | predicted gene, 32771 |
| 19 | gene | 38.264536 | 38.312214 | positive | MGI_C57BL6J_1861691 | Lgi1 | NCBI_Gene:56839,ENSEMBL:ENSMUSG00000067242 | MGI:1861691 | protein coding gene | leucine-rich repeat LGI family, member 1 |
| 19 | pseudogene | 38.287951 | 38.288224 | positive | MGI_C57BL6J_6302892 | Gm50141 | ENSEMBL:ENSMUSG00000118242 | MGI:6302892 | pseudogene | predicted gene, 50141 |
| 19 | gene | 38.300083 | 38.315270 | negative | MGI_C57BL6J_5662860 | Gm42723 | ENSEMBL:ENSMUSG00000105709 | MGI:5662860 | lncRNA gene | predicted gene 42723 |
| 19 | gene | 38.311058 | 38.327048 | negative | MGI_C57BL6J_3641826 | Gm9886 | NCBI_Gene:105246571 | MGI:3641826 | lncRNA gene | predicted gene 9886 |
| 19 | pseudogene | 38.342179 | 38.342614 | negative | MGI_C57BL6J_6302907 | Gm50150 | ENSEMBL:ENSMUSG00000118381 | MGI:6302907 | pseudogene | predicted gene, 50150 |
| 19 | gene | 38.348636 | 38.350079 | positive | MGI_C57BL6J_5592119 | Gm32960 | NCBI_Gene:102635685 | MGI:5592119 | lncRNA gene | predicted gene, 32960 |
| 19 | gene | 38.352239 | 38.358506 | negative | MGI_C57BL6J_5592195 | Gm33036 | NCBI_Gene:102635788 | MGI:5592195 | lncRNA gene | predicted gene, 33036 |
| 19 | gene | 38.379292 | 38.381215 | positive | MGI_C57BL6J_5624729 | Gm41844 | NCBI_Gene:105246572 | MGI:5624729 | protein coding gene | predicted gene, 41844 |
| 19 | pseudogene | 38.384566 | 38.385514 | negative | MGI_C57BL6J_6302915 | Gm50155 | ENSEMBL:ENSMUSG00000117920 | MGI:6302915 | pseudogene | predicted gene, 50155 |
| 19 | pseudogene | 38.386963 | 38.388161 | negative | MGI_C57BL6J_5011121 | Gm18936 | NCBI_Gene:100417987,ENSEMBL:ENSMUSG00000118335 | MGI:5011121 | pseudogene | predicted gene, 18936 |
| 19 | gene | 38.395980 | 38.405610 | positive | MGI_C57BL6J_2444789 | Slc35g1 | NCBI_Gene:240660,ENSEMBL:ENSMUSG00000044026 | MGI:2444789 | protein coding gene | solute carrier family 35, member G1 |
| 19 | gene | 38.458585 | 38.463554 | positive | MGI_C57BL6J_5826303 | Gm46666 | NCBI_Gene:108168429 | MGI:5826303 | lncRNA gene | predicted gene, 46666 |
| 19 | gene | 38.473120 | 38.473202 | negative | MGI_C57BL6J_5454707 | Gm24930 | ENSEMBL:ENSMUSG00000088940 | MGI:5454707 | unclassified non-coding RNA gene | predicted gene, 24930 |
| 19 | gene | 38.481109 | 38.785105 | positive | MGI_C57BL6J_1921305 | Plce1 | NCBI_Gene:74055,ENSEMBL:ENSMUSG00000024998 | MGI:1921305 | protein coding gene | phospholipase C, epsilon 1 |
| 19 | pseudogene | 38.703444 | 38.704438 | negative | MGI_C57BL6J_3644350 | Gm8717 | NCBI_Gene:667587,ENSEMBL:ENSMUSG00000117935 | MGI:3644350 | pseudogene | predicted gene 8717 |
| 19 | gene | 38.748760 | 38.752645 | negative | MGI_C57BL6J_5579697 | Gm28991 | ENSEMBL:ENSMUSG00000101079 | MGI:5579697 | lncRNA gene | predicted gene 28991 |
| 19 | gene | 38.788121 | 38.819950 | negative | MGI_C57BL6J_1932610 | Noc3l | NCBI_Gene:57753,ENSEMBL:ENSMUSG00000024999 | MGI:1932610 | protein coding gene | NOC3 like DNA replication regulator |
| 19 | gene | 38.836579 | 38.919923 | positive | MGI_C57BL6J_2384803 | Tbc1d12 | NCBI_Gene:209478,ENSEMBL:ENSMUSG00000048720 | MGI:2384803 | protein coding gene | TBC1D12: TBC1 domain family, member 12 |
| 19 | gene | 38.885242 | 38.885361 | positive | MGI_C57BL6J_5455615 | Gm25838 | ENSEMBL:ENSMUSG00000089545 | MGI:5455615 | unclassified non-coding RNA gene | predicted gene, 25838 |
| 19 | gene | 38.930915 | 38.971051 | positive | MGI_C57BL6J_106209 | Hells | NCBI_Gene:15201,ENSEMBL:ENSMUSG00000025001 | MGI:106209 | protein coding gene | helicase, lymphoid specific |
| 19 | pseudogene | 38.976647 | 39.003627 | positive | MGI_C57BL6J_3716994 | Cyp2c52-ps | NCBI_Gene:209753,ENSEMBL:ENSMUSG00000114610 | MGI:3716994 | pseudogene | cytochrome P450, family 2, subfamily c, polypeptide 52, pseudogene |
| 19 | gene | 39.006938 | 39.042693 | positive | MGI_C57BL6J_1919332 | Cyp2c55 | NCBI_Gene:72082,ENSEMBL:ENSMUSG00000025002 | MGI:1919332 | protein coding gene | cytochrome P450, family 2, subfamily c, polypeptide 55 |
| 19 | gene | 39.007026 | 39.007089 | negative | MGI_C57BL6J_5453577 | Gm23800 | ENSEMBL:ENSMUSG00000093303 | MGI:5453577 | miRNA gene | predicted gene, 23800 |
| 19 | pseudogene | 39.052630 | 39.054824 | negative | MGI_C57BL6J_3782048 | Gm3875 | NCBI_Gene:100042507 | MGI:3782048 | pseudogene | predicted gene 3875 |
| 19 | gene | 39.061006 | 39.093948 | positive | MGI_C57BL6J_1919553 | Cyp2c65 | NCBI_Gene:72303,ENSEMBL:ENSMUSG00000067231 | MGI:1919553 | protein coding gene | cytochrome P450, family 2, subfamily c, polypeptide 65 |
| 19 | gene | 39.113898 | 39.187072 | positive | MGI_C57BL6J_1917138 | Cyp2c66 | NCBI_Gene:69888,ENSEMBL:ENSMUSG00000067229 | MGI:1917138 | protein coding gene | cytochrome P450, family 2, subfamily c, polypeptide 66 |
| 19 | pseudogene | 39.229254 | 39.274985 | positive | MGI_C57BL6J_3646306 | Cyp2c53-ps | NCBI_Gene:638988,ENSEMBL:ENSMUSG00000093610 | MGI:3646306 | pseudogene | cytochrome P450, family 2, subfamily c, polypeptide 53-ps |
| 19 | gene | 39.269405 | 39.330713 | positive | MGI_C57BL6J_103238 | Cyp2c29 | NCBI_Gene:13095,ENSEMBL:ENSMUSG00000003053 | MGI:103238 | protein coding gene | cytochrome P450, family 2, subfamily c, polypeptide 29 |
| 19 | pseudogene | 39.347367 | 39.360403 | positive | MGI_C57BL6J_3721925 | Cyp2c72-ps | NCBI_Gene:100125395,ENSEMBL:ENSMUSG00000118146 | MGI:3721925 | pseudogene | cytochrome P450, family 2, subfamily c, polypeptide 72, pseudogene |
| 19 | gene | 39.389556 | 39.463089 | negative | MGI_C57BL6J_1306819 | Cyp2c38 | NCBI_Gene:13097,ENSEMBL:ENSMUSG00000032808 | MGI:1306819 | protein coding gene | cytochrome P450, family 2, subfamily c, polypeptide 38 |
| 19 | gene | 39.510822 | 39.568529 | positive | MGI_C57BL6J_1306818 | Cyp2c39 | NCBI_Gene:13098,ENSEMBL:ENSMUSG00000025003 | MGI:1306818 | protein coding gene | cytochrome P450, family 2, subfamily c, polypeptide 39 |
| 19 | gene | 39.608842 | 39.649079 | negative | MGI_C57BL6J_3612288 | Cyp2c67 | NCBI_Gene:545288,ENSEMBL:ENSMUSG00000062624 | MGI:3612288 | protein coding gene | cytochrome P450, family 2, subfamily c, polypeptide 67 |
| 19 | gene | 39.688834 | 39.741101 | negative | MGI_C57BL6J_3612287 | Cyp2c68 | NCBI_Gene:433247,ENSEMBL:ENSMUSG00000074882 | MGI:3612287 | protein coding gene | cytochrome P450, family 2, subfamily c, polypeptide 68 |
| 19 | gene | 39.767071 | 39.812814 | negative | MGI_C57BL6J_1306815 | Cyp2c40 | NCBI_Gene:13099,ENSEMBL:ENSMUSG00000025004 | MGI:1306815 | protein coding gene | cytochrome P450, family 2, subfamily c, polypeptide 40 |
| 19 | gene | 39.842660 | 39.886799 | negative | MGI_C57BL6J_3721049 | Cyp2c69 | NCBI_Gene:100043108,ENSEMBL:ENSMUSG00000092008 | MGI:3721049 | protein coding gene | cytochrome P450, family 2, subfamily c, polypeptide 69 |
| 19 | pseudogene | 39.928044 | 39.952861 | positive | MGI_C57BL6J_3721924 | Cyp2c71-ps | NCBI_Gene:100125394,ENSEMBL:ENSMUSG00000117738 | MGI:3721924 | pseudogene | cytochrome P450, family 2, subfamily c, polypeptide 71, pseudogene |
| 19 | gene | 39.992424 | 40.012243 | positive | MGI_C57BL6J_1306806 | Cyp2c37 | NCBI_Gene:13096,ENSEMBL:ENSMUSG00000042248 | MGI:1306806 | protein coding gene | cytochrome P450, family 2. subfamily c, polypeptide 37 |
| 19 | gene | 40.037940 | 40.073845 | negative | MGI_C57BL6J_3642960 | Cyp2c54 | NCBI_Gene:404195,ENSEMBL:ENSMUSG00000067225 | MGI:3642960 | protein coding gene | cytochrome P450, family 2, subfamily c, polypeptide 54 |
| 19 | gene | 40.089679 | 40.113955 | positive | MGI_C57BL6J_2147497 | Cyp2c50 | NCBI_Gene:107141,ENSEMBL:ENSMUSG00000054827 | MGI:2147497 | protein coding gene | cytochrome P450, family 2, subfamily c, polypeptide 50 |
| 19 | gene | 40.153353 | 40.187333 | negative | MGI_C57BL6J_2385878 | Cyp2c70 | NCBI_Gene:226105,ENSEMBL:ENSMUSG00000060613 | MGI:2385878 | protein coding gene | cytochrome P450, family 2, subfamily c, polypeptide 70 |
| 19 | pseudogene | 40.189990 | 40.194415 | positive | MGI_C57BL6J_3647389 | Gm5827 | NCBI_Gene:545289,ENSEMBL:ENSMUSG00000117785 | MGI:3647389 | pseudogene | predicted gene 5827 |
| 19 | pseudogene | 40.212364 | 40.213575 | positive | MGI_C57BL6J_3645079 | Gm16470 | NCBI_Gene:676923,ENSEMBL:ENSMUSG00000098091 | MGI:3645079 | pseudogene | predicted pseudogene 16470 |
| 19 | gene | 40.221173 | 40.271842 | negative | MGI_C57BL6J_1860611 | Pdlim1 | NCBI_Gene:54132,ENSEMBL:ENSMUSG00000055044 | MGI:1860611 | protein coding gene | PDZ and LIM domain 1 (elfin) |
| 19 | gene | 40.271701 | 40.274454 | positive | MGI_C57BL6J_5624730 | Gm41845 | NCBI_Gene:105246573 | MGI:5624730 | lncRNA gene | predicted gene, 41845 |
| 19 | pseudogene | 40.280951 | 40.282794 | positive | MGI_C57BL6J_3643821 | Gm5693 | NCBI_Gene:435599,ENSEMBL:ENSMUSG00000117793 | MGI:3643821 | pseudogene | predicted gene 5693 |
| 19 | gene | 40.292040 | 40.513874 | negative | MGI_C57BL6J_700014 | Sorbs1 | NCBI_Gene:20411,ENSEMBL:ENSMUSG00000025006 | MGI:700014 | protein coding gene | sorbin and SH3 domain containing 1 |
| 19 | gene | 40.297820 | 40.299555 | positive | MGI_C57BL6J_1925688 | A930028N01Rik | NCBI_Gene:78438,ENSEMBL:ENSMUSG00000114796 | MGI:1925688 | lncRNA gene | RIKEN cDNA A930028N01 gene |
| 19 | gene | 40.416249 | 40.416870 | negative | MGI_C57BL6J_1925672 | 9530039L23Rik | NA | NA | unclassified gene | RIKEN cDNA 9530039L23 gene |
| 19 | gene | 40.458213 | 40.458965 | negative | MGI_C57BL6J_3642414 | 3010085J16Rik | NA | NA | protein coding gene | RIKEN cDNA 3010085J16 gene |
| 19 | gene | 40.472755 | 40.474430 | negative | MGI_C57BL6J_1917735 | 5730409N24Rik | NA | NA | unclassified gene | RIKEN cDNA 5730409N24 gene |
| 19 | gene | 40.474599 | 40.476539 | negative | MGI_C57BL6J_1918838 | 9130009M17Rik | NA | NA | unclassified gene | RIKEN cDNA 9130009M17 gene |
| 19 | gene | 40.550257 | 40.588463 | negative | MGI_C57BL6J_1888908 | Aldh18a1 | NCBI_Gene:56454,ENSEMBL:ENSMUSG00000025007 | MGI:1888908 | protein coding gene | aldehyde dehydrogenase 18 family, member A1 |
| 19 | pseudogene | 40.558919 | 40.561755 | negative | MGI_C57BL6J_3779625 | Gm15801 | NCBI_Gene:627166,ENSEMBL:ENSMUSG00000096592 | MGI:3779625 | pseudogene | predicted gene 15801 |
| 19 | gene | 40.588535 | 40.594862 | positive | MGI_C57BL6J_5504157 | Gm27042 | NCBI_Gene:102636239,ENSEMBL:ENSMUSG00000097792 | MGI:5504157 | lncRNA gene | predicted gene, 27042 |
| 19 | gene | 40.596446 | 40.612281 | negative | MGI_C57BL6J_1914840 | Tctn3 | NCBI_Gene:67590,ENSEMBL:ENSMUSG00000025008 | MGI:1914840 | protein coding gene | tectonic family member 3 |
| 19 | gene | 40.611601 | 40.612996 | positive | MGI_C57BL6J_3712488 | E130009M23Rik | NA | NA | unclassified gene | RIKEN cDNA E130009M23 gene |
| 19 | gene | 40.612366 | 40.741602 | positive | MGI_C57BL6J_102805 | Entpd1 | NCBI_Gene:12495,ENSEMBL:ENSMUSG00000048120 | MGI:102805 | protein coding gene | ectonucleoside triphosphate diphosphohydrolase 1 |
| 19 | pseudogene | 40.615886 | 40.616758 | positive | MGI_C57BL6J_3801732 | Gm16027 | NCBI_Gene:100417906,ENSEMBL:ENSMUSG00000090041 | MGI:3801732 | pseudogene | predicted gene 16027 |
| 19 | pseudogene | 40.621776 | 40.624599 | positive | MGI_C57BL6J_3646027 | Gm8783 | NCBI_Gene:667723,ENSEMBL:ENSMUSG00000094388 | MGI:3646027 | pseudogene | predicted pseudogene 8783 |
| 19 | gene | 40.644146 | 40.654035 | positive | MGI_C57BL6J_6302875 | Gm50132 | ENSEMBL:ENSMUSG00000117952 | MGI:6302875 | lncRNA gene | predicted gene, 50132 |
| 19 | gene | 40.658272 | 40.661201 | negative | MGI_C57BL6J_5592464 | Gm33305 | NCBI_Gene:102636161 | MGI:5592464 | lncRNA gene | predicted gene, 33305 |
| 19 | gene | 40.729437 | 40.731663 | negative | MGI_C57BL6J_5624731 | Gm41846 | NCBI_Gene:105246574 | MGI:5624731 | lncRNA gene | predicted gene, 41846 |
| 19 | gene | 40.748753 | 40.829057 | positive | MGI_C57BL6J_3645359 | Cc2d2b | NCBI_Gene:668310,ENSEMBL:ENSMUSG00000108929 | MGI:3645359 | protein coding gene | coiled-coil and C2 domain containing 2B |
| 19 | gene | 40.816765 | 40.818268 | negative | MGI_C57BL6J_5592641 | Gm33482 | NCBI_Gene:102636408 | MGI:5592641 | lncRNA gene | predicted gene, 33482 |
| 19 | gene | 40.830522 | 40.831855 | negative | MGI_C57BL6J_3645360 | Gm9099 | NA | NA | unclassified gene | predicted gene 9099 |
| 19 | gene | 40.831279 | 40.848572 | positive | MGI_C57BL6J_2443297 | Ccnj | NCBI_Gene:240665,ENSEMBL:ENSMUSG00000025010 | MGI:2443297 | protein coding gene | cyclin J |
| 19 | gene | 40.868304 | 40.882639 | positive | MGI_C57BL6J_2686464 | E030044B06Rik | NCBI_Gene:381223,ENSEMBL:ENSMUSG00000097740 | MGI:2686464 | lncRNA gene | RIKEN cDNA E030044B06 gene |
| 19 | gene | 40.893933 | 40.917947 | positive | MGI_C57BL6J_1919922 | Zfp518a | NCBI_Gene:72672,ENSEMBL:ENSMUSG00000049164 | MGI:1919922 | protein coding gene | zinc finger protein 518A |
| 19 | gene | 40.894687 | 40.898391 | positive | MGI_C57BL6J_3053211 | E130314M14Rik | NA | NA | unclassified gene | RIKEN cDNA E130314M14 gene |
| 19 | gene | 40.894703 | 40.894789 | positive | MGI_C57BL6J_5530789 | Mir8092 | miRBase:MI0026019,NCBI_Gene:102466881,ENSEMBL:ENSMUSG00000099118 | MGI:5530789 | miRNA gene | microRNA 8092 |
| 19 | gene | 40.928927 | 40.994535 | negative | MGI_C57BL6J_96878 | Blnk | NCBI_Gene:17060,ENSEMBL:ENSMUSG00000061132 | MGI:96878 | protein coding gene | B cell linker |
| 19 | gene | 41.004815 | 41.014643 | negative | MGI_C57BL6J_1926115 | C330026H20Rik | NCBI_Gene:100861913 | MGI:1926115 | lncRNA gene | RIKEN cDNA C330026H20 gene |
| 19 | gene | 41.009103 | 41.011703 | negative | MGI_C57BL6J_2443821 | A130015J22Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA A130015J22 gene |
| 19 | pseudogene | 41.017683 | 41.020295 | positive | MGI_C57BL6J_3782446 | Gm4269 | NCBI_Gene:105246575 | MGI:3782446 | pseudogene | predicted gene 4269 |
| 19 | gene | 41.029275 | 41.059525 | positive | MGI_C57BL6J_98659 | Dntt | NCBI_Gene:21673,ENSEMBL:ENSMUSG00000025014 | MGI:98659 | protein coding gene | deoxynucleotidyltransferase, terminal |
| 19 | gene | 41.061710 | 41.077113 | negative | MGI_C57BL6J_2657025 | Opalin | NCBI_Gene:226115,ENSEMBL:ENSMUSG00000050121 | MGI:2657025 | protein coding gene | oligodendrocytic myelin paranodal and inner loop protein |
| 19 | gene | 41.080940 | 41.210848 | negative | MGI_C57BL6J_1346044 | Tll2 | NCBI_Gene:24087,ENSEMBL:ENSMUSG00000025013 | MGI:1346044 | protein coding gene | tolloid-like 2 |
| 19 | gene | 41.210842 | 41.264004 | negative | MGI_C57BL6J_1914262 | Tm9sf3 | NCBI_Gene:107358,ENSEMBL:ENSMUSG00000025016 | MGI:1914262 | protein coding gene | transmembrane 9 superfamily member 3 |
| 19 | gene | 41.259566 | 41.263709 | negative | MGI_C57BL6J_5826288 | Gm46651 | NCBI_Gene:108168412 | MGI:5826288 | lncRNA gene | predicted gene, 46651 |
| 19 | gene | 41.264112 | 41.265030 | positive | MGI_C57BL6J_1922881 | 1700008F19Rik | NA | NA | unclassified gene | RIKEN cDNA 1700008F19 gene |
| 19 | gene | 41.272377 | 41.385102 | negative | MGI_C57BL6J_1933177 | Pik3ap1 | NCBI_Gene:83490,ENSEMBL:ENSMUSG00000025017 | MGI:1933177 | protein coding gene | phosphoinositide-3-kinase adaptor protein 1 |
| 19 | gene | 41.443382 | 41.445082 | negative | MGI_C57BL6J_5593110 | Gm33951 | NCBI_Gene:102637042 | MGI:5593110 | lncRNA gene | predicted gene, 33951 |
| 19 | pseudogene | 41.459783 | 41.461451 | positive | MGI_C57BL6J_3704496 | Gm9788 | NCBI_Gene:100043167,ENSEMBL:ENSMUSG00000044065 | MGI:3704496 | pseudogene | predicted pseudogene 9788 |
| 19 | gene | 41.481254 | 41.482185 | negative | MGI_C57BL6J_1917137 | 2010100M03Rik | NA | NA | unclassified gene | RIKEN cDNA 2010100M03 gene |
| 19 | gene | 41.482583 | 41.596161 | positive | MGI_C57BL6J_2443930 | Lcor | NCBI_Gene:212391,ENSEMBL:ENSMUSG00000025019 | MGI:2443930 | protein coding gene | ligand dependent nuclear receptor corepressor |
| 19 | gene | 41.525252 | 41.526961 | positive | MGI_C57BL6J_3648734 | Morf4l1b | NCBI_Gene:627352 | MGI:3648734 | protein coding gene | mortality factor 4 like 1B |
| 19 | gene | 41.575867 | 41.589383 | positive | MGI_C57BL6J_2685186 | Gm340 | NA | NA | protein coding gene | predicted gene 340 |
| 19 | gene | 41.588651 | 41.588789 | positive | MGI_C57BL6J_5530957 | Mir8091 | miRBase:MI0026018,NCBI_Gene:102465884,ENSEMBL:ENSMUSG00000098286 | MGI:5530957 | miRNA gene | microRNA 8091 |
| 19 | gene | 41.593363 | 41.596158 | positive | MGI_C57BL6J_2147586 | AI606181 | NA | NA | protein coding gene | expressed sequence AI606181 |
| 19 | gene | 41.597824 | 41.747540 | negative | MGI_C57BL6J_1315203 | Slit1 | NCBI_Gene:20562,ENSEMBL:ENSMUSG00000025020 | MGI:1315203 | protein coding gene | slit guidance ligand 1 |
| 19 | gene | 41.691154 | 41.702436 | positive | MGI_C57BL6J_5593275 | Gm34116 | NCBI_Gene:102637251 | MGI:5593275 | lncRNA gene | predicted gene, 34116 |
| 19 | gene | 41.744804 | 41.745053 | positive | MGI_C57BL6J_5452378 | Gm22601 | ENSEMBL:ENSMUSG00000088220 | MGI:5452378 | unclassified non-coding RNA gene | predicted gene, 22601 |
| 19 | gene | 41.746961 | 41.750097 | positive | MGI_C57BL6J_5011609 | Gm19424 | NCBI_Gene:100502871 | MGI:5011609 | lncRNA gene | predicted gene, 19424 |
| 19 | gene | 41.766588 | 41.802084 | negative | MGI_C57BL6J_1918335 | Arhgap19 | NCBI_Gene:71085,ENSEMBL:ENSMUSG00000025154 | MGI:1918335 | protein coding gene | Rho GTPase activating protein 19 |
| 19 | gene | 41.807949 | 41.819642 | negative | MGI_C57BL6J_5624732 | Gm41847 | NCBI_Gene:105246576 | MGI:5624732 | lncRNA gene | predicted gene, 41847 |
| 19 | gene | 41.829970 | 41.832583 | positive | MGI_C57BL6J_109450 | Frat1 | NCBI_Gene:14296,ENSEMBL:ENSMUSG00000067199 | MGI:109450 | protein coding gene | frequently rearranged in advanced T cell lymphomas |
| 19 | gene | 41.845972 | 41.848132 | negative | MGI_C57BL6J_2673967 | Frat2 | NCBI_Gene:212398,ENSEMBL:ENSMUSG00000047604 | MGI:2673967 | protein coding gene | frequently rearranged in advanced T cell lymphomas 2 |
| 19 | gene | 41.862851 | 41.896173 | negative | MGI_C57BL6J_2147437 | Rrp12 | NCBI_Gene:107094,ENSEMBL:ENSMUSG00000035049 | MGI:2147437 | protein coding gene | ribosomal RNA processing 12 homolog (S. cerevisiae) |
| 19 | pseudogene | 41.900999 | 41.903231 | positive | MGI_C57BL6J_5826281 | Gm46644 | NCBI_Gene:108168404,ENSEMBL:ENSMUSG00000117675 | MGI:5826281 | pseudogene | predicted gene, 46644 |
| 19 | gene | 41.911871 | 41.918665 | positive | MGI_C57BL6J_97552 | Pgam1 | NCBI_Gene:18648,ENSEMBL:ENSMUSG00000011752 | MGI:97552 | protein coding gene | phosphoglycerate mutase 1 |
| 19 | gene | 41.919650 | 41.922399 | positive | MGI_C57BL6J_2443824 | B130024M06Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA B130024M06 gene |
| 19 | gene | 41.922292 | 41.933423 | negative | MGI_C57BL6J_1913833 | Exosc1 | NCBI_Gene:66583,ENSEMBL:ENSMUSG00000034321 | MGI:1913833 | protein coding gene | exosome component 1 |
| 19 | gene | 41.933472 | 41.944104 | positive | MGI_C57BL6J_1921418 | Zdhhc16 | NCBI_Gene:74168,ENSEMBL:ENSMUSG00000025157 | MGI:1921418 | protein coding gene | zinc finger, DHHC domain containing 16 |
| 19 | gene | 41.941086 | 41.981165 | negative | MGI_C57BL6J_1919449 | Mms19 | NCBI_Gene:72199,ENSEMBL:ENSMUSG00000025159 | MGI:1919449 | protein coding gene | MMS19 cytosolic iron-sulfur assembly component |
| 19 | gene | 41.972105 | 41.974522 | negative | MGI_C57BL6J_2443381 | B130065D12Rik | NA | NA | unclassified gene | RIKEN cDNA B130065D12 gene |
| 19 | gene | 41.975188 | 41.975286 | positive | MGI_C57BL6J_5452890 | Gm23113 | ENSEMBL:ENSMUSG00000089102 | MGI:5452890 | snRNA gene | predicted gene, 23113 |
| 19 | gene | 41.981763 | 42.034650 | positive | MGI_C57BL6J_2385092 | Ubtd1 | NCBI_Gene:226122,ENSEMBL:ENSMUSG00000025171 | MGI:2385092 | protein coding gene | ubiquitin domain containing 1 |
| 19 | gene | 41.986563 | 41.990400 | positive | MGI_C57BL6J_2443153 | E130107B13Rik | NA | NA | unclassified gene | RIKEN cDNA E130107B13 gene |
| 19 | gene | 42.035918 | 42.045111 | positive | MGI_C57BL6J_1861447 | Ankrd2 | NCBI_Gene:56642,ENSEMBL:ENSMUSG00000025172 | MGI:1861447 | protein coding gene | ankyrin repeat domain 2 (stretch responsive muscle) |
| 19 | gene | 42.045610 | 42.070953 | positive | MGI_C57BL6J_1914682 | Hoga1 | NCBI_Gene:67432,ENSEMBL:ENSMUSG00000025176 | MGI:1914682 | protein coding gene | 4-hydroxy-2-oxoglutarate aldolase 1 |
| 19 | gene | 42.052228 | 42.053628 | positive | MGI_C57BL6J_1914015 | 4933411K16Rik | NCBI_Gene:66765,ENSEMBL:ENSMUSG00000090369 | MGI:1914015 | protein coding gene | RIKEN cDNA 4933411K16 gene |
| 19 | gene | 42.074939 | 42.086370 | negative | MGI_C57BL6J_2449568 | Morn4 | NCBI_Gene:226123,ENSEMBL:ENSMUSG00000049670 | MGI:2449568 | protein coding gene | MORN repeat containing 4 |
| 19 | gene | 42.090145 | 42.122218 | positive | MGI_C57BL6J_1934031 | Pi4k2a | NCBI_Gene:84095,ENSEMBL:ENSMUSG00000025178 | MGI:1934031 | protein coding gene | phosphatidylinositol 4-kinase type 2 alpha |
| 19 | gene | 42.123273 | 42.129059 | negative | MGI_C57BL6J_1916784 | Avpi1 | NCBI_Gene:69534,ENSEMBL:ENSMUSG00000018821 | MGI:1916784 | protein coding gene | arginine vasopressin-induced 1 |
| 19 | gene | 42.147389 | 42.151703 | positive | MGI_C57BL6J_2147570 | Marveld1 | NCBI_Gene:277010,ENSEMBL:ENSMUSG00000044345 | MGI:2147570 | protein coding gene | MARVEL (membrane-associating) domain containing 1 |
| 19 | pseudogene | 42.159631 | 42.160768 | positive | MGI_C57BL6J_3647230 | Gm6937 | NCBI_Gene:628994,ENSEMBL:ENSMUSG00000117856 | MGI:3647230 | pseudogene | predicted pseudogene 6937 |
| 19 | gene | 42.163951 | 42.194593 | positive | MGI_C57BL6J_1919602 | Zfyve27 | NCBI_Gene:319740,ENSEMBL:ENSMUSG00000018820 | MGI:1919602 | protein coding gene | zinc finger, FYVE domain containing 27 |
| 19 | gene | 42.197971 | 42.202252 | negative | MGI_C57BL6J_1860298 | Sfrp5 | NCBI_Gene:54612,ENSEMBL:ENSMUSG00000018822 | MGI:1860298 | protein coding gene | secreted frizzled-related sequence protein 5 |
| 19 | gene | 42.247574 | 42.270348 | positive | MGI_C57BL6J_1918396 | Golga7b | NCBI_Gene:71146,ENSEMBL:ENSMUSG00000042532 | MGI:1918396 | protein coding gene | golgi autoantigen, golgin subfamily a, 7B |
| 19 | gene | 42.268287 | 42.431822 | negative | MGI_C57BL6J_1920082 | Crtac1 | NCBI_Gene:72832,ENSEMBL:ENSMUSG00000042401 | MGI:1920082 | protein coding gene | cartilage acidic protein 1 |
| 19 | gene | 42.274964 | 42.275062 | positive | MGI_C57BL6J_5456116 | Gm26339 | ENSEMBL:ENSMUSG00000076323 | MGI:5456116 | miRNA gene | predicted gene, 26339 |
| 19 | gene | 42.276887 | 42.314626 | positive | MGI_C57BL6J_5593506 | Gm34347 | NCBI_Gene:102637571 | MGI:5593506 | lncRNA gene | predicted gene, 34347 |
| 19 | gene | 42.280082 | 42.280170 | negative | MGI_C57BL6J_4834259 | Mir3085 | miRBase:MI0014048,NCBI_Gene:100526549,ENSEMBL:ENSMUSG00000093234 | MGI:4834259 | miRNA gene | microRNA 3085 |
| 19 | pseudogene | 42.378991 | 42.379101 | negative | MGI_C57BL6J_6302957 | Gm50184 | ENSEMBL:ENSMUSG00000117706 | MGI:6302957 | pseudogene | predicted gene, 50184 |
| 19 | gene | 42.387741 | 42.397551 | negative | MGI_C57BL6J_5624733 | Gm41848 | NCBI_Gene:105246577 | MGI:5624733 | lncRNA gene | predicted gene, 41848 |
| 19 | gene | 42.402212 | 42.414246 | positive | MGI_C57BL6J_5593458 | Gm34299 | NCBI_Gene:102637508,ENSEMBL:ENSMUSG00000117728 | MGI:5593458 | lncRNA gene | predicted gene, 34299 |
| 19 | gene | 42.444057 | 42.448376 | positive | MGI_C57BL6J_5624734 | Gm41849 | NCBI_Gene:105246578 | MGI:5624734 | lncRNA gene | predicted gene, 41849 |
| 19 | gene | 42.483124 | 42.486499 | negative | MGI_C57BL6J_5624735 | Gm41850 | NCBI_Gene:105246579 | MGI:5624735 | lncRNA gene | predicted gene, 41850 |
| 19 | gene | 42.509119 | 42.512197 | negative | MGI_C57BL6J_5624736 | Gm41851 | NCBI_Gene:105246580 | MGI:5624736 | lncRNA gene | predicted gene, 41851 |
| 19 | gene | 42.518315 | 42.592343 | positive | MGI_C57BL6J_1196316 | R3hcc1l | NCBI_Gene:52013,ENSEMBL:ENSMUSG00000025184 | MGI:1196316 | protein coding gene | R3H domain and coiled-coil containing 1 like |
| 19 | gene | 42.536850 | 42.563108 | negative | MGI_C57BL6J_4414961 | Gm16541 | ENSEMBL:ENSMUSG00000090117 | MGI:4414961 | lncRNA gene | predicted gene 16541 |
| 19 | gene | 42.551570 | 42.552164 | positive | MGI_C57BL6J_1926126 | B230214N19Rik | NA | NA | unclassified gene | RIKEN cDNA B230214N19 gene |
| 19 | gene | 42.566036 | 42.567962 | positive | MGI_C57BL6J_1924199 | 2810404I24Rik | NA | NA | unclassified gene | RIKEN cDNA 2810404I24 gene |
| 19 | gene | 42.592279 | 42.612813 | negative | MGI_C57BL6J_1914823 | Loxl4 | NCBI_Gene:67573,ENSEMBL:ENSMUSG00000025185 | MGI:1914823 | protein coding gene | lysyl oxidase-like 4 |
| 19 | gene | 42.699777 | 42.699902 | negative | MGI_C57BL6J_5454993 | Gm25216 | ENSEMBL:ENSMUSG00000077768 | MGI:5454993 | snoRNA gene | predicted gene, 25216 |
| 19 | gene | 42.725857 | 42.752824 | negative | MGI_C57BL6J_1921830 | Pyroxd2 | NCBI_Gene:74580,ENSEMBL:ENSMUSG00000060224 | MGI:1921830 | protein coding gene | pyridine nucleotide-disulphide oxidoreductase domain 2 |
| 19 | gene | 42.745691 | 42.752329 | positive | MGI_C57BL6J_5011622 | Gm19437 | NCBI_Gene:102637646,ENSEMBL:ENSMUSG00000118354 | MGI:5011622 | lncRNA gene | predicted gene, 19437 |
| 19 | gene | 42.755105 | 42.779996 | negative | MGI_C57BL6J_2177763 | Hps1 | NCBI_Gene:192236,ENSEMBL:ENSMUSG00000025188 | MGI:2177763 | protein coding gene | HPS1, biogenesis of lysosomal organelles complex 3 subunit 1 |
| 19 | gene | 42.778653 | 42.781574 | positive | MGI_C57BL6J_3802128 | Gm16244 | NCBI_Gene:102637720,ENSEMBL:ENSMUSG00000090235 | MGI:3802128 | lncRNA gene | predicted gene 16244 |
| 19 | gene | 42.786539 | 43.388455 | negative | MGI_C57BL6J_2685814 | Hpse2 | NCBI_Gene:545291,ENSEMBL:ENSMUSG00000074852 | MGI:2685814 | protein coding gene | heparanase 2 |
| 19 | gene | 43.160795 | 43.160902 | negative | MGI_C57BL6J_5451912 | Gm22135 | ENSEMBL:ENSMUSG00000089377 | MGI:5451912 | rRNA gene | predicted gene, 22135 |
| 19 | gene | 43.266936 | 43.277654 | positive | MGI_C57BL6J_5593676 | Gm34517 | ENSEMBL:ENSMUSG00000117880 | MGI:5593676 | lncRNA gene | predicted gene, 34517 |
| 19 | pseudogene | 43.278429 | 43.279303 | negative | MGI_C57BL6J_5010564 | Gm18379 | NCBI_Gene:100417040,ENSEMBL:ENSMUSG00000117682 | MGI:5010564 | pseudogene | predicted gene, 18379 |
| 19 | pseudogene | 43.280776 | 43.284225 | positive | MGI_C57BL6J_3647201 | Gm6776 | NCBI_Gene:627624,ENSEMBL:ENSMUSG00000047509 | MGI:3647201 | pseudogene | predicted pseudogene 6776 |
| 19 | gene | 43.386283 | 43.387399 | positive | MGI_C57BL6J_1918806 | 8430434A19Rik | NA | NA | unclassified gene | RIKEN cDNA 8430434A19 gene |
| 19 | gene | 43.424826 | 43.427960 | positive | MGI_C57BL6J_5621453 | Gm38568 | NCBI_Gene:102641642 | MGI:5621453 | lncRNA gene | predicted gene, 38568 |
| 19 | gene | 43.434610 | 43.440460 | negative | MGI_C57BL6J_6097198 | Gm47936 | ENSEMBL:ENSMUSG00000114742 | MGI:6097198 | lncRNA gene | predicted gene, 47936 |
| 19 | gene | 43.440350 | 43.497213 | positive | MGI_C57BL6J_1891366 | Cnnm1 | NCBI_Gene:83674,ENSEMBL:ENSMUSG00000025189 | MGI:1891366 | protein coding gene | cyclin M1 |
| 19 | gene | 43.457444 | 43.469758 | negative | MGI_C57BL6J_6097201 | Gm47938 | ENSEMBL:ENSMUSG00000114721 | MGI:6097201 | lncRNA gene | predicted gene, 47938 |
| 19 | gene | 43.499752 | 43.524605 | negative | MGI_C57BL6J_95791 | Got1 | NCBI_Gene:14718,ENSEMBL:ENSMUSG00000025190 | MGI:95791 | protein coding gene | glutamic-oxaloacetic transaminase 1, soluble |
| 19 | gene | 43.528312 | 43.566029 | positive | MGI_C57BL6J_6302970 | Gm50194 | ENSEMBL:ENSMUSG00000118221 | MGI:6302970 | lncRNA gene | predicted gene, 50194 |
| 19 | pseudogene | 43.544183 | 43.544962 | positive | MGI_C57BL6J_5011364 | Gm19179 | NCBI_Gene:100418390,ENSEMBL:ENSMUSG00000118308 | MGI:5011364 | pseudogene | predicted gene, 19179 |
| 19 | gene | 43.545927 | 43.573357 | negative | MGI_C57BL6J_5593795 | Gm34636 | NCBI_Gene:102637951,ENSEMBL:ENSMUSG00000117991 | MGI:5593795 | lncRNA gene | predicted gene, 34636 |
| 19 | gene | 43.609077 | 43.610717 | negative | MGI_C57BL6J_5141932 | Gm20467 | NCBI_Gene:102638028,ENSEMBL:ENSMUSG00000092250 | MGI:5141932 | lncRNA gene | predicted gene 20467 |
| 19 | gene | 43.610832 | 43.611890 | negative | MGI_C57BL6J_5826293 | Gm46656 | NCBI_Gene:108168417 | MGI:5826293 | lncRNA gene | predicted gene, 46656 |
| 19 | gene | 43.612325 | 43.615892 | positive | MGI_C57BL6J_97348 | Nkx2-3 | NCBI_Gene:18089,ENSEMBL:ENSMUSG00000044220 | MGI:97348 | protein coding gene | NK2 homeobox 3 |
| 19 | gene | 43.622446 | 43.622560 | positive | MGI_C57BL6J_5452423 | Gm22646 | ENSEMBL:ENSMUSG00000092871 | MGI:5452423 | miRNA gene | predicted gene, 22646 |
| 19 | gene | 43.637372 | 43.642404 | negative | MGI_C57BL6J_3782492 | Gm4311 | NCBI_Gene:100043245 | MGI:3782492 | unclassified gene | predicted gene 4311 |
| 19 | gene | 43.645554 | 43.655630 | positive | MGI_C57BL6J_5594081 | Gm34922 | NCBI_Gene:102638332,ENSEMBL:ENSMUSG00000118236 | MGI:5594081 | lncRNA gene | predicted gene, 34922 |
| 19 | gene | 43.663801 | 43.675510 | negative | MGI_C57BL6J_2180509 | Slc25a28 | NCBI_Gene:246696,ENSEMBL:ENSMUSG00000040414 | MGI:2180509 | protein coding gene | solute carrier family 25, member 28 |
| 19 | gene | 43.675178 | 43.677176 | positive | MGI_C57BL6J_2652856 | BC037704 | NCBI_Gene:100502982,ENSEMBL:ENSMUSG00000117698 | MGI:2652856 | lncRNA gene | cDNA sequence BC037704 |
| 19 | gene | 43.689672 | 43.733853 | positive | MGI_C57BL6J_2135885 | Entpd7 | NCBI_Gene:93685,ENSEMBL:ENSMUSG00000025192 | MGI:2135885 | protein coding gene | ectonucleoside triphosphate diphosphohydrolase 7 |
| 19 | gene | 43.733254 | 43.753000 | negative | MGI_C57BL6J_1920112 | Cox15 | NCBI_Gene:226139,ENSEMBL:ENSMUSG00000040018 | MGI:1920112 | protein coding gene | cytochrome c oxidase assembly protein 15 |
| 19 | gene | 43.752996 | 43.768638 | positive | MGI_C57BL6J_1913638 | Cutc | NCBI_Gene:66388,ENSEMBL:ENSMUSG00000025193 | MGI:1913638 | protein coding gene | cutC copper transporter |
| 19 | gene | 43.782192 | 43.840740 | positive | MGI_C57BL6J_1352447 | Abcc2 | NCBI_Gene:12780,ENSEMBL:ENSMUSG00000025194 | MGI:1352447 | protein coding gene | ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| 19 | gene | 43.838803 | 43.840845 | positive | MGI_C57BL6J_3642139 | Gm10768 | NCBI_Gene:100038628 | MGI:3642139 | lncRNA gene | predicted gene 10768 |
| 19 | gene | 43.846815 | 43.940323 | negative | MGI_C57BL6J_1917352 | Dnmbp | NCBI_Gene:71972,ENSEMBL:ENSMUSG00000025195 | MGI:1917352 | protein coding gene | dynamin binding protein |
| 19 | gene | 43.857245 | 43.857351 | negative | MGI_C57BL6J_5454177 | Gm24400 | ENSEMBL:ENSMUSG00000088490 | MGI:5454177 | snRNA gene | predicted gene, 24400 |
| 19 | gene | 43.920473 | 43.927928 | positive | MGI_C57BL6J_6303010 | Gm50217 | ENSEMBL:ENSMUSG00000118000 | MGI:6303010 | lncRNA gene | predicted gene, 50217 |
| 19 | gene | 43.932316 | 43.932432 | positive | MGI_C57BL6J_4421755 | n-R5s21 | ENSEMBL:ENSMUSG00000084641 | MGI:4421755 | rRNA gene | nuclear encoded rRNA 5S 21 |
| 19 | gene | 43.940037 | 43.940920 | positive | MGI_C57BL6J_1920770 | 1700084K02Rik | NA | NA | unclassified gene | RIKEN cDNA 1700084K02 gene |
| 19 | gene | 43.956307 | 43.986556 | negative | MGI_C57BL6J_2135874 | Cpn1 | NCBI_Gene:93721,ENSEMBL:ENSMUSG00000025196 | MGI:2135874 | protein coding gene | carboxypeptidase N, polypeptide 1 |
| 19 | gene | 44.005022 | 44.029256 | negative | MGI_C57BL6J_1888897 | Cyp2c23 | NCBI_Gene:226143,ENSEMBL:ENSMUSG00000025197 | MGI:1888897 | protein coding gene | cytochrome P450, family 2, subfamily c, polypeptide 23 |
| 19 | gene | 44.034943 | 44.070494 | negative | MGI_C57BL6J_2387613 | Erlin1 | NCBI_Gene:226144,ENSEMBL:ENSMUSG00000025198 | MGI:2387613 | protein coding gene | ER lipid raft associated 1 |
| 19 | pseudogene | 44.052234 | 44.052736 | negative | MGI_C57BL6J_5010415 | Gm18230 | NCBI_Gene:100416753 | MGI:5010415 | pseudogene | predicted gene, 18230 |
| 19 | gene | 44.073334 | 44.107509 | negative | MGI_C57BL6J_99484 | Chuk | NCBI_Gene:12675,ENSEMBL:ENSMUSG00000025199 | MGI:99484 | protein coding gene | conserved helix-loop-helix ubiquitous kinase |
| 19 | gene | 44.108637 | 44.135876 | negative | MGI_C57BL6J_1919752 | Cwf19l1 | NCBI_Gene:72502,ENSEMBL:ENSMUSG00000025200 | MGI:1919752 | protein coding gene | CWF19-like 1, cell cycle control (S. pombe) |
| 19 | gene | 44.113979 | 44.114124 | negative | MGI_C57BL6J_5454113 | Gm24336 | ENSEMBL:ENSMUSG00000077391 | MGI:5454113 | snoRNA gene | predicted gene, 24336 |
| 19 | gene | 44.139246 | 44.146470 | negative | MGI_C57BL6J_1920939 | Bloc1s2 | NCBI_Gene:73689,ENSEMBL:ENSMUSG00000057506 | MGI:1920939 | protein coding gene | biogenesis of lysosomal organelles complex-1, subunit 2 |
| 19 | gene | 44.147637 | 44.249352 | negative | MGI_C57BL6J_1352448 | Pkd2l1 | NCBI_Gene:329064,ENSEMBL:ENSMUSG00000037578 | MGI:1352448 | protein coding gene | polycystic kidney disease 2-like 1 |
| 19 | gene | 44.203269 | 44.244016 | positive | MGI_C57BL6J_1353437 | Scd3 | NCBI_Gene:30049,ENSEMBL:ENSMUSG00000025202 | MGI:1353437 | protein coding gene | stearoyl-coenzyme A desaturase 3 |
| 19 | gene | 44.225796 | 44.246570 | negative | MGI_C57BL6J_6303185 | Gm50323 | ENSEMBL:ENSMUSG00000117900 | MGI:6303185 | lncRNA gene | predicted gene, 50323 |
| 19 | pseudogene | 44.252517 | 44.261446 | positive | MGI_C57BL6J_3643963 | Gm6951 | NCBI_Gene:629152,ENSEMBL:ENSMUSG00000118217 | MGI:3643963 | pseudogene | predicted gene 6951 |
| 19 | gene | 44.272137 | 44.278228 | positive | MGI_C57BL6J_5594342 | Gm35183 | NCBI_Gene:102638677,ENSEMBL:ENSMUSG00000117958 | MGI:5594342 | lncRNA gene | predicted gene, 35183 |
| 19 | gene | 44.282343 | 44.283697 | positive | MGI_C57BL6J_5624738 | Gm41853 | NCBI_Gene:105246582 | MGI:5624738 | lncRNA gene | predicted gene, 41853 |
| 19 | gene | 44.293674 | 44.306864 | positive | MGI_C57BL6J_98240 | Scd2 | NCBI_Gene:20250,ENSEMBL:ENSMUSG00000025203 | MGI:98240 | protein coding gene | stearoyl-Coenzyme A desaturase 2 |
| 19 | gene | 44.303171 | 44.303231 | positive | MGI_C57BL6J_4950434 | Mir5114 | miRBase:MI0018023,NCBI_Gene:100628619,ENSEMBL:ENSMUSG00000093315 | MGI:4950434 | miRNA gene | microRNA 5114 |
| 19 | gene | 44.313772 | 44.317329 | positive | MGI_C57BL6J_6303204 | Gm50334 | ENSEMBL:ENSMUSG00000117957 | MGI:6303204 | lncRNA gene | predicted gene, 50334 |
| 19 | gene | 44.333092 | 44.346743 | positive | MGI_C57BL6J_2670997 | Scd4 | NCBI_Gene:329065,ENSEMBL:ENSMUSG00000050195 | MGI:2670997 | protein coding gene | stearoyl-coenzyme A desaturase 4 |
| 19 | gene | 44.394450 | 44.407879 | negative | MGI_C57BL6J_98239 | Scd1 | NCBI_Gene:20249,ENSEMBL:ENSMUSG00000037071 | MGI:98239 | protein coding gene | stearoyl-Coenzyme A desaturase 1 |
| 19 | gene | 44.419610 | 44.437886 | positive | MGI_C57BL6J_6303210 | Gm50337 | ENSEMBL:ENSMUSG00000118076 | MGI:6303210 | lncRNA gene | predicted gene, 50337 |
| 19 | gene | 44.453017 | 44.459853 | positive | MGI_C57BL6J_5594565 | Gm35406 | NCBI_Gene:102638973,ENSEMBL:ENSMUSG00000117796 | MGI:5594565 | lncRNA gene | predicted gene, 35406 |
| 19 | gene | 44.493472 | 44.514273 | positive | MGI_C57BL6J_109485 | Wnt8b | NCBI_Gene:22423,ENSEMBL:ENSMUSG00000036961 | MGI:109485 | protein coding gene | wingless-type MMTV integration site family, member 8B |
| 19 | gene | 44.516957 | 44.545864 | negative | MGI_C57BL6J_2685187 | Sec31b | NCBI_Gene:240667,ENSEMBL:ENSMUSG00000051984 | MGI:2685187 | protein coding gene | Sec31 homolog B (S. cerevisiae) |
| 19 | gene | 44.543722 | 44.552831 | negative | MGI_C57BL6J_5142003 | Gm20538 | ENSEMBL:ENSMUSG00000091471 | MGI:5142003 | protein coding gene | predicted gene 20538 |
| 19 | gene | 44.548572 | 44.555442 | negative | MGI_C57BL6J_1914514 | Ndufb8 | NCBI_Gene:67264,ENSEMBL:ENSMUSG00000025204 | MGI:1914514 | protein coding gene | NADH:ubiquinone oxidoreductase subunit B8 |
| 19 | gene | 44.562850 | 44.576274 | positive | MGI_C57BL6J_2442345 | Hif1an | NCBI_Gene:319594,ENSEMBL:ENSMUSG00000036450 | MGI:2442345 | protein coding gene | hypoxia-inducible factor 1, alpha subunit inhibitor |
| 19 | gene | 44.614740 | 44.646879 | positive | MGI_C57BL6J_5594619 | Gm35460 | NCBI_Gene:102639056,ENSEMBL:ENSMUSG00000117911 | MGI:5594619 | lncRNA gene | predicted gene, 35460 |
| 19 | gene | 44.672417 | 44.674865 | positive | MGI_C57BL6J_5624739 | Gm41854 | NCBI_Gene:105246583 | MGI:5624739 | lncRNA gene | predicted gene, 41854 |
| 19 | gene | 44.685753 | 44.695460 | negative | MGI_C57BL6J_5477138 | Gm26644 | NCBI_Gene:102639186,ENSEMBL:ENSMUSG00000097683 | MGI:5477138 | lncRNA gene | predicted gene, 26644 |
| 19 | gene | 44.739879 | 44.743444 | positive | MGI_C57BL6J_5594769 | Gm35610 | NCBI_Gene:102639257,ENSEMBL:ENSMUSG00000117823 | MGI:5594769 | lncRNA gene | predicted gene, 35610 |
| 19 | gene | 44.745693 | 44.837871 | positive | MGI_C57BL6J_97486 | Pax2 | NCBI_Gene:18504,ENSEMBL:ENSMUSG00000004231 | MGI:97486 | protein coding gene | paired box 2 |
| 19 | gene | 44.773762 | 44.784512 | negative | MGI_C57BL6J_5141860 | Gm20395 | ENSEMBL:ENSMUSG00000092469 | MGI:5141860 | lncRNA gene | predicted gene 20395 |
| 19 | gene | 44.808406 | 44.808529 | positive | MGI_C57BL6J_5530731 | Mir6405 | miRBase:MI0021941,NCBI_Gene:102466645,ENSEMBL:ENSMUSG00000098952 | MGI:5530731 | miRNA gene | microRNA 6405 |
| 19 | gene | 44.828493 | 44.835938 | negative | MGI_C57BL6J_1920572 | 1700039E22Rik | NCBI_Gene:73322,ENSEMBL:ENSMUSG00000092369 | MGI:1920572 | lncRNA gene | RIKEN cDNA 1700039E22 gene |
| 19 | pseudogene | 44.851009 | 44.851979 | positive | MGI_C57BL6J_3646252 | Gm5246 | NCBI_Gene:383450,ENSEMBL:ENSMUSG00000117967 | MGI:3646252 | pseudogene | predicted gene 5246 |
| 19 | gene | 44.918505 | 44.983787 | positive | MGI_C57BL6J_1924968 | Slf2 | NCBI_Gene:226151,ENSEMBL:ENSMUSG00000036097 | MGI:1924968 | protein coding gene | SMC5-SMC6 complex localization factor 2 |
| 19 | gene | 44.945022 | 44.945128 | positive | MGI_C57BL6J_5455259 | Gm25482 | ENSEMBL:ENSMUSG00000065159 | MGI:5455259 | snRNA gene | predicted gene, 25482 |
| 19 | gene | 44.960373 | 44.984550 | negative | MGI_C57BL6J_1921144 | 4930414N06Rik | NCBI_Gene:73894,ENSEMBL:ENSMUSG00000118204 | MGI:1921144 | lncRNA gene | RIKEN cDNA 4930414N06 gene |
| 19 | gene | 44.988966 | 45.003397 | positive | MGI_C57BL6J_1347047 | Sema4g | NCBI_Gene:26456,ENSEMBL:ENSMUSG00000025207 | MGI:1347047 | protein coding gene | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| 19 | gene | 45.004829 | 45.006442 | negative | MGI_C57BL6J_2137229 | Mrpl43 | NCBI_Gene:94067,ENSEMBL:ENSMUSG00000025208 | MGI:2137229 | protein coding gene | mitochondrial ribosomal protein L43 |
| 19 | gene | 45.005662 | 45.012763 | positive | MGI_C57BL6J_2137410 | Twnk | NCBI_Gene:226153,ENSEMBL:ENSMUSG00000025209 | MGI:2137410 | protein coding gene | twinkle mtDNA helicase |
| 19 | gene | 45.014414 | 45.027115 | positive | MGI_C57BL6J_2385095 | Lzts2 | NCBI_Gene:226154,ENSEMBL:ENSMUSG00000035342 | MGI:2385095 | protein coding gene | leucine zipper, putative tumor suppressor 2 |
| 19 | gene | 45.026906 | 45.058307 | negative | MGI_C57BL6J_3608325 | Pdzd7 | NCBI_Gene:100503041,ENSEMBL:ENSMUSG00000074818 | MGI:3608325 | protein coding gene | PDZ domain containing 7 |
| 19 | gene | 45.030404 | 45.031132 | positive | MGI_C57BL6J_1924586 | 9430030N17Rik | NA | NA | unclassified gene | RIKEN cDNA 9430030N17 gene |
| 19 | gene | 45.047241 | 45.056383 | positive | MGI_C57BL6J_2137679 | Sfxn3 | NCBI_Gene:94280,ENSEMBL:ENSMUSG00000025212 | MGI:2137679 | protein coding gene | sideroflexin 3 |
| 19 | gene | 45.058841 | 45.110342 | positive | MGI_C57BL6J_5595026 | Gm35867 | NCBI_Gene:102639590,ENSEMBL:ENSMUSG00000117848 | MGI:5595026 | lncRNA gene | predicted gene, 35867 |
| 19 | gene | 45.060632 | 45.075109 | negative | MGI_C57BL6J_5594941 | Gm35782 | NCBI_Gene:102639478 | MGI:5594941 | lncRNA gene | predicted gene, 35782 |
| 19 | gene | 45.075091 | 45.079289 | positive | MGI_C57BL6J_2147606 | Kazald1 | NCBI_Gene:107250,ENSEMBL:ENSMUSG00000025213 | MGI:2147606 | protein coding gene | Kazal-type serine peptidase inhibitor domain 1 |
| 19 | gene | 45.080008 | 45.080583 | positive | MGI_C57BL6J_1922558 | 4930557B21Rik | NA | NA | unclassified gene | RIKEN cDNA 4930557B21 gene |
| 19 | gene | 45.144743 | 45.146954 | negative | MGI_C57BL6J_5595082 | Gm35923 | NCBI_Gene:102639665 | MGI:5595082 | lncRNA gene | predicted gene, 35923 |
| 19 | gene | 45.150082 | 45.156943 | positive | MGI_C57BL6J_98769 | Tlx1 | NCBI_Gene:21908,ENSEMBL:ENSMUSG00000025215 | MGI:98769 | protein coding gene | T cell leukemia, homeobox 1 |
| 19 | gene | 45.154057 | 45.159304 | negative | MGI_C57BL6J_5141889 | Tlx1os | ENSEMBL:ENSMUSG00000092346 | MGI:5141889 | antisense lncRNA gene | T cell leukemia, homeobox 1, opposite strand |
| 19 | gene | 45.232684 | 45.237430 | negative | MGI_C57BL6J_104867 | Lbx1 | NCBI_Gene:16814,ENSEMBL:ENSMUSG00000025216 | MGI:104867 | protein coding gene | ladybird homeobox 1 |
| 19 | gene | 45.300455 | 45.338477 | negative | MGI_C57BL6J_5579284 | Gm28578 | NCBI_Gene:102639970,NCBI_Gene:102640042,ENSEMBL:ENSMUSG00000100417 | MGI:5579284 | lncRNA gene | predicted gene 28578 |
| 19 | gene | 45.300458 | 45.303246 | negative | MGI_C57BL6J_5580301 | Gm29595 | NCBI_Gene:102639746 | MGI:5580301 | lncRNA gene | predicted gene 29595 |
| 19 | gene | 45.311291 | 45.312882 | negative | MGI_C57BL6J_5580299 | Gm29593 | NCBI_Gene:108168425 | MGI:5580299 | lncRNA gene | predicted gene 29593 |
| 19 | gene | 45.317893 | 45.318429 | positive | MGI_C57BL6J_5580300 | Gm29594 | ENSEMBL:ENSMUSG00000102057 | MGI:5580300 | lncRNA gene | predicted gene 29594 |
| 19 | gene | 45.363213 | 45.363649 | negative | MGI_C57BL6J_1922903 | 1700016H03Rik | ENSEMBL:ENSMUSG00000114673 | MGI:1922903 | lncRNA gene | RIKEN cDNA 1700016H03 gene |
| 19 | gene | 45.363734 | 45.533343 | positive | MGI_C57BL6J_1338871 | Btrc | NCBI_Gene:12234,ENSEMBL:ENSMUSG00000025217 | MGI:1338871 | protein coding gene | beta-transducin repeat containing protein |
| 19 | pseudogene | 45.396765 | 45.397796 | negative | MGI_C57BL6J_5010289 | Gm18104 | NCBI_Gene:100416415 | MGI:5010289 | pseudogene | predicted gene, 18104 |
| 19 | pseudogene | 45.450276 | 45.451247 | negative | MGI_C57BL6J_3649145 | Gm6807 | NCBI_Gene:627889,ENSEMBL:ENSMUSG00000057605 | MGI:3649145 | pseudogene | predicted gene 6807 |
| 19 | gene | 45.542766 | 45.544997 | negative | MGI_C57BL6J_5624741 | Gm41856 | NCBI_Gene:105246585 | MGI:5624741 | lncRNA gene | predicted gene, 41856 |
| 19 | gene | 45.544596 | 45.546346 | positive | MGI_C57BL6J_5011824 | Gm19639 | NCBI_Gene:100503332 | MGI:5011824 | lncRNA gene | predicted gene, 19639 |
| 19 | gene | 45.552275 | 45.560543 | negative | MGI_C57BL6J_1889000 | Poll | NCBI_Gene:56626,ENSEMBL:ENSMUSG00000025218 | MGI:1889000 | protein coding gene | polymerase (DNA directed), lambda |
| 19 | gene | 45.560590 | 45.578287 | positive | MGI_C57BL6J_1924407 | Dpcd | NCBI_Gene:226162 | MGI:1924407 | protein coding gene | deleted in primary ciliary dyskinesia |
| 19 | gene | 45.560615 | 45.579763 | positive | MGI_C57BL6J_4820566 | Gm17018 | ENSEMBL:ENSMUSG00000041035 | MGI:4820566 | protein coding gene | predicted gene 17018 |
| 19 | gene | 45.578254 | 45.660539 | negative | MGI_C57BL6J_1354698 | Fbxw4 | NCBI_Gene:30838,ENSEMBL:ENSMUSG00000040913 | MGI:1354698 | protein coding gene | F-box and WD-40 domain protein 4 |
| 19 | gene | 45.726555 | 45.730558 | negative | MGI_C57BL6J_1918482 | 4933429K18Rik | NCBI_Gene:71232 | MGI:1918482 | lncRNA gene | RIKEN cDNA 4933429K18 gene |
| 19 | gene | 45.736798 | 45.742938 | negative | MGI_C57BL6J_99604 | Fgf8 | NCBI_Gene:14179,ENSEMBL:ENSMUSG00000025219 | MGI:99604 | protein coding gene | fibroblast growth factor 8 |
| 19 | gene | 45.747734 | 45.749591 | negative | MGI_C57BL6J_894653 | Npm3 | NCBI_Gene:18150,ENSEMBL:ENSMUSG00000056209 | MGI:894653 | protein coding gene | nucleoplasmin 3 |
| 19 | gene | 45.749508 | 45.752310 | positive | MGI_C57BL6J_3782937 | Gm15491 | ENSEMBL:ENSMUSG00000087367 | MGI:3782937 | lncRNA gene | predicted gene 15491 |
| 19 | gene | 45.750259 | 45.783837 | negative | MGI_C57BL6J_1932139 | Oga | NCBI_Gene:76055,ENSEMBL:ENSMUSG00000025220 | MGI:1932139 | protein coding gene | O-GlcNAcase |
| 19 | gene | 45.791839 | 45.816432 | negative | MGI_C57BL6J_2135916 | Kcnip2 | NCBI_Gene:80906,ENSEMBL:ENSMUSG00000025221 | MGI:2135916 | protein coding gene | Kv channel-interacting protein 2 |
| 19 | gene | 45.817364 | 45.998580 | negative | MGI_C57BL6J_1918867 | Armh3 | NCBI_Gene:71617,ENSEMBL:ENSMUSG00000039901 | MGI:1918867 | protein coding gene | armadillo-like helical domain containing 3 |
| 19 | gene | 45.843617 | 45.848296 | positive | MGI_C57BL6J_6302974 | Gm50196 | ENSEMBL:ENSMUSG00000117956 | MGI:6302974 | lncRNA gene | predicted gene, 50196 |
| 19 | pseudogene | 45.877611 | 45.878164 | negative | MGI_C57BL6J_3645759 | Gm6813 | NCBI_Gene:627939,ENSEMBL:ENSMUSG00000117972 | MGI:3645759 | pseudogene | predicted gene 6813 |
| 19 | gene | 45.997572 | 46.013387 | positive | MGI_C57BL6J_1922306 | 4930505N22Rik | NCBI_Gene:100174921,ENSEMBL:ENSMUSG00000097690 | MGI:1922306 | lncRNA gene | RIKEN cDNA 4930505N22 gene |
| 19 | gene | 46.003478 | 46.006173 | positive | MGI_C57BL6J_2181763 | Hps6 | NCBI_Gene:20170,ENSEMBL:ENSMUSG00000074811 | MGI:2181763 | protein coding gene | HPS6, biogenesis of lysosomal organelles complex 2 subunit 3 |
| 19 | gene | 46.031570 | 46.045515 | negative | MGI_C57BL6J_894762 | Ldb1 | NCBI_Gene:16825,ENSEMBL:ENSMUSG00000025223 | MGI:894762 | protein coding gene | LIM domain binding 1 |
| 19 | gene | 46.044886 | 46.072915 | positive | MGI_C57BL6J_2385096 | Pprc1 | NCBI_Gene:226169,ENSEMBL:ENSMUSG00000055491 | MGI:2385096 | protein coding gene | peroxisome proliferative activated receptor, gamma, coactivator-related 1 |
| 19 | gene | 46.075847 | 46.085543 | positive | MGI_C57BL6J_1918019 | Nolc1 | NCBI_Gene:70769,ENSEMBL:ENSMUSG00000015176 | MGI:1918019 | protein coding gene | nucleolar and coiled-body phosphoprotein 1 |
| 19 | gene | 46.124930 | 46.131471 | positive | MGI_C57BL6J_6303157 | Gm50306 | ENSEMBL:ENSMUSG00000117703 | MGI:6303157 | lncRNA gene | predicted gene, 50306 |
| 19 | gene | 46.131897 | 46.135694 | positive | MGI_C57BL6J_1195976 | Elovl3 | NCBI_Gene:12686,ENSEMBL:ENSMUSG00000038754 | MGI:1195976 | protein coding gene | elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 |
| 19 | gene | 46.135294 | 46.148326 | negative | MGI_C57BL6J_1100498 | Pitx3 | NCBI_Gene:18742,ENSEMBL:ENSMUSG00000025229 | MGI:1100498 | protein coding gene | paired-like homeodomain transcription factor 3 |
| 19 | gene | 46.152509 | 46.286510 | positive | MGI_C57BL6J_1861607 | Gbf1 | NCBI_Gene:107338,ENSEMBL:ENSMUSG00000025224 | MGI:1861607 | protein coding gene | golgi-specific brefeldin A-resistance factor 1 |
| 19 | gene | 46.180483 | 46.185809 | positive | MGI_C57BL6J_2444071 | D430033H22Rik | NA | NA | unclassified gene | RIKEN cDNA D430033H22 gene |
| 19 | pseudogene | 46.204141 | 46.205964 | negative | MGI_C57BL6J_3648532 | Gm8974 | NCBI_Gene:105246586,ENSEMBL:ENSMUSG00000093525 | MGI:3648532 | pseudogene | predicted gene 8974 |
| 19 | gene | 46.213099 | 46.215985 | positive | MGI_C57BL6J_2443201 | C130013I19Rik | NA | NA | unclassified gene | RIKEN cDNA C130013I19 gene |
| 19 | gene | 46.264539 | 46.267669 | negative | MGI_C57BL6J_4439650 | Gm16726 | ENSEMBL:ENSMUSG00000086816 | MGI:4439650 | lncRNA gene | predicted gene, 16726 |
| 19 | gene | 46.303590 | 46.305623 | negative | MGI_C57BL6J_2147599 | 4833438C02Rik | NCBI_Gene:100503392,ENSEMBL:ENSMUSG00000118087 | MGI:2147599 | lncRNA gene | RIKEN cDNA 4833438C02 gene |
| 19 | gene | 46.304320 | 46.312385 | positive | MGI_C57BL6J_1099800 | Nfkb2 | NCBI_Gene:18034,ENSEMBL:ENSMUSG00000025225 | MGI:1099800 | protein coding gene | nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| 19 | gene | 46.312085 | 46.327188 | negative | MGI_C57BL6J_1920978 | Psd | NCBI_Gene:73728,ENSEMBL:ENSMUSG00000037126 | MGI:1920978 | protein coding gene | pleckstrin and Sec7 domain containing |
| 19 | gene | 46.328119 | 46.330446 | positive | MGI_C57BL6J_1915681 | Fbxl15 | NCBI_Gene:68431,ENSEMBL:ENSMUSG00000025226 | MGI:1915681 | protein coding gene | F-box and leucine-rich repeat protein 15 |
| 19 | gene | 46.329812 | 46.338702 | negative | MGI_C57BL6J_1914366 | Cuedc2 | NCBI_Gene:67116,ENSEMBL:ENSMUSG00000036748 | MGI:1914366 | protein coding gene | CUE domain containing 2 |
| 19 | gene | 46.335287 | 46.338538 | positive | MGI_C57BL6J_5477286 | Gm26792 | ENSEMBL:ENSMUSG00000096996 | MGI:5477286 | lncRNA gene | predicted gene, 26792 |
| 19 | gene | 46.338776 | 46.344131 | positive | MGI_C57BL6J_5595652 | Gm36493 | NCBI_Gene:102640430 | MGI:5595652 | lncRNA gene | predicted gene, 36493 |
| 19 | gene | 46.341121 | 46.375254 | positive | MGI_C57BL6J_1922396 | Mfsd13a | NCBI_Gene:75146,ENSEMBL:ENSMUSG00000025227 | MGI:1922396 | protein coding gene | major facilitator superfamily domain containing 13a |
| 19 | gene | 46.342762 | 46.342870 | positive | MGI_C57BL6J_3629945 | Mir146b | miRBase:MI0004665,NCBI_Gene:751550,ENSEMBL:ENSMUSG00000070127 | MGI:3629945 | miRNA gene | microRNA 146b |
| 19 | gene | 46.348201 | 46.350302 | negative | MGI_C57BL6J_1922829 | 2310034G01Rik | NCBI_Gene:75579,ENSEMBL:ENSMUSG00000099655 | MGI:1922829 | lncRNA gene | RIKEN cDNA 2310034G01 gene |
| 19 | gene | 46.359036 | 46.359144 | positive | MGI_C57BL6J_5454387 | Gm24610 | ENSEMBL:ENSMUSG00000065204 | MGI:5454387 | snoRNA gene | predicted gene, 24610 |
| 19 | gene | 46.376811 | 46.395748 | negative | MGI_C57BL6J_1858964 | Actr1a | NCBI_Gene:54130,ENSEMBL:ENSMUSG00000025228 | MGI:1858964 | protein coding gene | ARP1 actin-related protein 1A, centractin alpha |
| 19 | gene | 46.396896 | 46.488804 | positive | MGI_C57BL6J_1345643 | Sufu | NCBI_Gene:24069,ENSEMBL:ENSMUSG00000025231 | MGI:1345643 | protein coding gene | SUFU negative regulator of hedgehog signaling |
| 19 | gene | 46.426855 | 46.427656 | positive | MGI_C57BL6J_1924501 | 9430020M11Rik | NA | NA | unclassified gene | RIKEN cDNA 9430020M11 gene |
| 19 | gene | 46.501648 | 46.502782 | negative | MGI_C57BL6J_3704497 | Gm9798 | NA | NA | unclassified gene | predicted gene 9798 |
| 19 | gene | 46.501648 | 46.516848 | positive | MGI_C57BL6J_1933302 | Trim8 | NCBI_Gene:93679,ENSEMBL:ENSMUSG00000025034 | MGI:1933302 | protein coding gene | tripartite motif-containing 8 |
| 19 | gene | 46.531096 | 46.573198 | negative | MGI_C57BL6J_1929699 | Arl3 | NCBI_Gene:56350,ENSEMBL:ENSMUSG00000025035 | MGI:1929699 | protein coding gene | ADP-ribosylation factor-like 3 |
| 19 | gene | 46.573323 | 46.598066 | positive | MGI_C57BL6J_2137678 | Sfxn2 | NCBI_Gene:94279,ENSEMBL:ENSMUSG00000025036 | MGI:2137678 | protein coding gene | sideroflexin 2 |
| 19 | gene | 46.599084 | 46.657389 | positive | MGI_C57BL6J_107577 | Wbp1l | NCBI_Gene:226178,ENSEMBL:ENSMUSG00000047731 | MGI:107577 | protein coding gene | WW domain binding protein 1 like |
| 19 | gene | 46.657734 | 46.666450 | negative | MGI_C57BL6J_5595761 | Gm36602 | NCBI_Gene:102640568,ENSEMBL:ENSMUSG00000118040 | MGI:5595761 | lncRNA gene | predicted gene, 36602 |
| 19 | gene | 46.667165 | 46.673172 | negative | MGI_C57BL6J_88586 | Cyp17a1 | NCBI_Gene:13074,ENSEMBL:ENSMUSG00000003555 | MGI:88586 | protein coding gene | cytochrome P450, family 17, subfamily a, polypeptide 1 |
| 19 | gene | 46.668680 | 46.676083 | positive | MGI_C57BL6J_5624742 | Gm41857 | NCBI_Gene:105246587 | MGI:5624742 | lncRNA gene | predicted gene, 41857 |
| 19 | pseudogene | 46.674086 | 46.675297 | negative | MGI_C57BL6J_3648147 | Gm6967 | NCBI_Gene:629359,ENSEMBL:ENSMUSG00000097937 | MGI:3648147 | pseudogene | predicted gene 6967 |
| 19 | gene | 46.689906 | 46.703382 | positive | MGI_C57BL6J_1913689 | Borcs7 | NCBI_Gene:66439,ENSEMBL:ENSMUSG00000062376 | MGI:1913689 | protein coding gene | BLOC-1 related complex subunit 7 |
| 19 | gene | 46.707359 | 46.741099 | positive | MGI_C57BL6J_1929882 | As3mt | NCBI_Gene:57344,ENSEMBL:ENSMUSG00000003559 | MGI:1929882 | protein coding gene | arsenic (+3 oxidation state) methyltransferase |
| 19 | gene | 46.759671 | 46.763467 | negative | MGI_C57BL6J_3704498 | Gm10199 | NA | NA | unclassified gene | predicted gene 10199 |
| 19 | gene | 46.761564 | 46.880192 | positive | MGI_C57BL6J_2151054 | Cnnm2 | NCBI_Gene:94219,ENSEMBL:ENSMUSG00000064105 | MGI:2151054 | protein coding gene | cyclin M2 |
| 19 | gene | 46.881495 | 46.883439 | positive | MGI_C57BL6J_6303362 | Gm50431 | ENSEMBL:ENSMUSG00000117814 | MGI:6303362 | lncRNA gene | predicted gene, 50431 |
| 19 | gene | 46.883090 | 46.883430 | negative | MGI_C57BL6J_1925220 | A930026I22Rik | NA | NA | unclassified gene | RIKEN cDNA A930026I22 gene |
| 19 | gene | 46.883317 | 47.015189 | negative | MGI_C57BL6J_2178563 | Nt5c2 | NCBI_Gene:76952,ENSEMBL:ENSMUSG00000025041 | MGI:2178563 | protein coding gene | 5’-nucleotidase, cytosolic II |
| 19 | gene | 46.889008 | 46.909575 | positive | MGI_C57BL6J_3704499 | Gm9751 | NA | NA | unclassified gene | predicted gene 9751 |
| 19 | pseudogene | 46.920507 | 46.921270 | negative | MGI_C57BL6J_5010675 | Gm18490 | NCBI_Gene:100417264 | MGI:5010675 | pseudogene | predicted gene, 18490 |
| 19 | pseudogene | 46.996290 | 46.997196 | positive | MGI_C57BL6J_5010842 | Gm18657 | NCBI_Gene:100417509 | MGI:5010842 | pseudogene | predicted gene, 18657 |
| 19 | gene | 47.014698 | 47.025327 | positive | MGI_C57BL6J_96568 | Ina | NCBI_Gene:226180,ENSEMBL:ENSMUSG00000034336 | MGI:96568 | protein coding gene | internexin neuronal intermediate filament protein, alpha |
| 19 | gene | 47.033617 | 47.051096 | negative | MGI_C57BL6J_1918291 | Pcgf6 | NCBI_Gene:71041,ENSEMBL:ENSMUSG00000025050 | MGI:1918291 | protein coding gene | polycomb group ring finger 6 |
| 19 | pseudogene | 47.054505 | 47.055248 | negative | MGI_C57BL6J_6303179 | Gm50319 | ENSEMBL:ENSMUSG00000117691 | MGI:6303179 | pseudogene | predicted gene, 50319 |
| 19 | gene | 47.058162 | 47.058268 | positive | MGI_C57BL6J_5452917 | Gm23140 | ENSEMBL:ENSMUSG00000094400 | MGI:5452917 | snRNA gene | predicted gene, 23140 |
| 19 | gene | 47.067746 | 47.083479 | positive | MGI_C57BL6J_2442144 | Taf5 | NCBI_Gene:226182,ENSEMBL:ENSMUSG00000025049 | MGI:2442144 | protein coding gene | TATA-box binding protein associated factor 5 |
| 19 | gene | 47.083464 | 47.090647 | negative | MGI_C57BL6J_1891435 | Atp5md | NCBI_Gene:66477,ENSEMBL:ENSMUSG00000071528 | MGI:1891435 | protein coding gene | ATP synthase membrane subunit DAPIT |
| 19 | gene | 47.090749 | 47.131865 | positive | MGI_C57BL6J_1341788 | Pdcd11 | NCBI_Gene:18572,ENSEMBL:ENSMUSG00000025047 | MGI:1341788 | protein coding gene | programmed cell death 11 |
| 19 | gene | 47.105353 | 47.138294 | negative | MGI_C57BL6J_1919941 | Calhm2 | NCBI_Gene:72691,ENSEMBL:ENSMUSG00000033033 | MGI:1919941 | protein coding gene | calcium homeostasis modulator family member 2 |
| 19 | gene | 47.126150 | 47.131251 | negative | MGI_C57BL6J_3704500 | D930027P08Rik | NA | NA | unclassified gene | RIKEN cDNA D930027P08 gene |
| 19 | gene | 47.140909 | 47.144254 | negative | MGI_C57BL6J_3643383 | Calhm1 | NCBI_Gene:546729,ENSEMBL:ENSMUSG00000079258 | MGI:3643383 | protein coding gene | calcium homeostasis modulator 1 |
| 19 | gene | 47.151609 | 47.157764 | negative | MGI_C57BL6J_3645665 | Calhm3 | NCBI_Gene:240669,ENSEMBL:ENSMUSG00000094219 | MGI:3645665 | protein coding gene | calcium homeostasis modulator 3 |
| 19 | pseudogene | 47.170426 | 47.171200 | negative | MGI_C57BL6J_3645322 | H1f11-ps | NCBI_Gene:629389,ENSEMBL:ENSMUSG00000091230 | MGI:3645322 | pseudogene | H1.11 linker histone, pseudogene |
| 19 | gene | 47.178820 | 47.259441 | positive | MGI_C57BL6J_1334263 | Neurl1a | NCBI_Gene:18011,ENSEMBL:ENSMUSG00000006435 | MGI:1334263 | protein coding gene | neuralized E3 ubiquitin protein ligase 1A |
| 19 | gene | 47.197881 | 47.206768 | negative | MGI_C57BL6J_5595852 | Gm36693 | NCBI_Gene:102640681,ENSEMBL:ENSMUSG00000118127 | MGI:5595852 | lncRNA gene | predicted gene, 36693 |
| 19 | gene | 47.220713 | 47.221909 | negative | MGI_C57BL6J_6303214 | Gm50339 | ENSEMBL:ENSMUSG00000117659 | MGI:6303214 | lncRNA gene | predicted gene, 50339 |
| 19 | gene | 47.260174 | 47.464613 | negative | MGI_C57BL6J_1298393 | Sh3pxd2a | NCBI_Gene:14218,ENSEMBL:ENSMUSG00000053617 | MGI:1298393 | protein coding gene | SH3 and PX domains 2A |
| 19 | gene | 47.273519 | 47.273589 | negative | MGI_C57BL6J_5530958 | Mir6995 | miRBase:MI0022843,NCBI_Gene:102466782,ENSEMBL:ENSMUSG00000098786 | MGI:5530958 | miRNA gene | microRNA 6995 |
| 19 | gene | 47.286725 | 47.293330 | positive | MGI_C57BL6J_5588993 | Gm29834 | NCBI_Gene:102631515 | MGI:5588993 | lncRNA gene | predicted gene, 29834 |
| 19 | gene | 47.426070 | 47.435135 | positive | MGI_C57BL6J_5596044 | Gm36885 | NCBI_Gene:102640939 | MGI:5596044 | lncRNA gene | predicted gene, 36885 |
| 19 | gene | 47.483840 | 47.485344 | negative | MGI_C57BL6J_5624743 | Gm41858 | NCBI_Gene:105246588 | MGI:5624743 | lncRNA gene | predicted gene, 41858 |
| 19 | gene | 47.501033 | 47.537507 | negative | MGI_C57BL6J_1915581 | Stn1 | NCBI_Gene:108689,ENSEMBL:ENSMUSG00000042694 | MGI:1915581 | protein coding gene | STN1, CST complex subunit |
| 19 | gene | 47.512982 | 47.516578 | positive | MGI_C57BL6J_5011742 | Gm19557 | NCBI_Gene:100503104,ENSEMBL:ENSMUSG00000097990 | MGI:5011742 | lncRNA gene | predicted gene, 19557 |
| 19 | gene | 47.579678 | 47.645246 | positive | MGI_C57BL6J_103241 | Slk | NCBI_Gene:20874,ENSEMBL:ENSMUSG00000025060 | MGI:103241 | protein coding gene | STE20-like kinase |
| 19 | gene | 47.597237 | 47.602329 | negative | MGI_C57BL6J_6303244 | Gm50358 | ENSEMBL:ENSMUSG00000118206 | MGI:6303244 | lncRNA gene | predicted gene, 50358 |
| 19 | gene | 47.605899 | 47.617106 | negative | MGI_C57BL6J_3802005 | Gm16068 | NCBI_Gene:102631588,ENSEMBL:ENSMUSG00000086121 | MGI:3802005 | lncRNA gene | predicted gene 16068 |
| 19 | gene | 47.646341 | 47.692094 | negative | MGI_C57BL6J_88450 | Col17a1 | NCBI_Gene:12821,ENSEMBL:ENSMUSG00000025064 | MGI:88450 | protein coding gene | collagen, type XVII, alpha 1 |
| 19 | gene | 47.694671 | 47.696533 | negative | MGI_C57BL6J_6303115 | Gm50281 | ENSEMBL:ENSMUSG00000117702 | MGI:6303115 | lncRNA gene | predicted gene, 50281 |
| 19 | gene | 47.710866 | 47.710981 | positive | MGI_C57BL6J_5455862 | Gm26085 | ENSEMBL:ENSMUSG00000088525 | MGI:5455862 | snoRNA gene | predicted gene, 26085 |
| 19 | gene | 47.714784 | 47.714887 | negative | MGI_C57BL6J_5454582 | Gm24805 | ENSEMBL:ENSMUSG00000096704 | MGI:5454582 | snRNA gene | predicted gene, 24805 |
| 19 | gene | 47.721035 | 47.723066 | positive | MGI_C57BL6J_6303117 | Gm50282 | ENSEMBL:ENSMUSG00000118149 | MGI:6303117 | lncRNA gene | predicted gene, 50282 |
| 19 | gene | 47.731682 | 47.735588 | positive | MGI_C57BL6J_1915038 | Sfr1 | NCBI_Gene:67788,ENSEMBL:ENSMUSG00000025066 | MGI:1915038 | protein coding gene | SWI5 dependent recombination repair 1 |
| 19 | gene | 47.735267 | 47.919287 | negative | MGI_C57BL6J_1289258 | Cfap43 | NCBI_Gene:100048534,ENSEMBL:ENSMUSG00000044948 | MGI:1289258 | protein coding gene | cilia and flagella associated protein 43 |
| 19 | gene | 47.755022 | 47.755157 | negative | MGI_C57BL6J_5531244 | Mir8090 | miRBase:MI0026017,NCBI_Gene:102465883,ENSEMBL:ENSMUSG00000099069 | MGI:5531244 | miRNA gene | microRNA 8090 |
| 19 | gene | 47.853111 | 47.854772 | negative | MGI_C57BL6J_5589180 | Gm30021 | NCBI_Gene:102631766 | MGI:5589180 | lncRNA gene | predicted gene, 30021 |
| 19 | gene | 47.854970 | 47.864790 | positive | MGI_C57BL6J_1342273 | Gsto1 | NCBI_Gene:14873,ENSEMBL:ENSMUSG00000025068 | MGI:1342273 | protein coding gene | glutathione S-transferase omega 1 |
| 19 | gene | 47.865534 | 47.886324 | positive | MGI_C57BL6J_1915464 | Gsto2 | NCBI_Gene:68214,ENSEMBL:ENSMUSG00000025069 | MGI:1915464 | protein coding gene | glutathione S-transferase omega 2 |
| 19 | gene | 47.894593 | 47.919519 | negative | MGI_C57BL6J_3042776 | Itprip | NCBI_Gene:414801,ENSEMBL:ENSMUSG00000117975 | MGI:3042776 | protein coding gene | inositol 1,4,5-triphosphate receptor interacting protein |
| 19 | gene | 47.919192 | 47.919273 | negative | MGI_C57BL6J_5452017 | Gm22240 | ENSEMBL:ENSMUSG00000092966 | MGI:5452017 | miRNA gene | predicted gene, 22240 |
| 19 | gene | 47.937675 | 48.035387 | positive | MGI_C57BL6J_2685815 | Cfap58 | NCBI_Gene:381229,ENSEMBL:ENSMUSG00000046585 | MGI:2685815 | protein coding gene | cilia and flagella associated protein 58 |
| 19 | gene | 48.117348 | 48.136082 | positive | MGI_C57BL6J_6303360 | Gm50430 | ENSEMBL:ENSMUSG00000117690 | MGI:6303360 | lncRNA gene | predicted gene, 50430 |
| 19 | gene | 48.206025 | 48.805505 | positive | MGI_C57BL6J_1913923 | Sorcs3 | NCBI_Gene:66673,ENSEMBL:ENSMUSG00000063434 | MGI:1913923 | protein coding gene | sortilin-related VPS10 domain containing receptor 3 |
| 19 | gene | 48.240535 | 48.240645 | negative | MGI_C57BL6J_5453634 | Gm23857 | ENSEMBL:ENSMUSG00000089004 | MGI:5453634 | rRNA gene | predicted gene, 23857 |
| 19 | gene | 48.240774 | 48.241114 | negative | MGI_C57BL6J_1924179 | 1700021N20Rik | NA | NA | unclassified gene | RIKEN cDNA 1700021N20 gene |
| 19 | gene | 48.811136 | 48.813495 | positive | MGI_C57BL6J_6303368 | Gm50436 | ENSEMBL:ENSMUSG00000117930 | MGI:6303368 | unclassified gene | predicted gene, 50436 |
| 19 | gene | 48.990469 | 48.991981 | negative | MGI_C57BL6J_1922433 | 4930535F04Rik | ENSEMBL:ENSMUSG00000118014 | MGI:1922433 | unclassified gene | RIKEN cDNA 4930535F04 gene |
| 19 | gene | 48.992084 | 48.996546 | positive | MGI_C57BL6J_6303375 | Gm50441 | ENSEMBL:ENSMUSG00000117996 | MGI:6303375 | lncRNA gene | predicted gene, 50441 |
| 19 | gene | 49.090673 | 49.107771 | positive | MGI_C57BL6J_6303377 | Gm50442 | ENSEMBL:ENSMUSG00000118111 | MGI:6303377 | lncRNA gene | predicted gene, 50442 |
| 19 | pseudogene | 49.554018 | 49.557406 | positive | MGI_C57BL6J_6303380 | Gm50444 | ENSEMBL:ENSMUSG00000117931 | MGI:6303380 | pseudogene | predicted gene, 50444 |
| 19 | pseudogene | 49.709393 | 49.710153 | negative | MGI_C57BL6J_3644403 | Gm6975 | NCBI_Gene:629448,ENSEMBL:ENSMUSG00000118059 | MGI:3644403 | pseudogene | predicted gene 6975 |
| 19 | gene | 49.709794 | 49.709872 | negative | MGI_C57BL6J_5531317 | Gm27935 | ENSEMBL:ENSMUSG00000098507 | MGI:5531317 | miRNA gene | predicted gene, 27935 |
| 19 | pseudogene | 49.868272 | 49.870143 | positive | MGI_C57BL6J_108066 | Siah1-ps1 | NCBI_Gene:20435,ENSEMBL:ENSMUSG00000117697 | MGI:108066 | pseudogene | siah E3 ubiquitin protein ligase 1, pseudogene 1 |
| 19 | pseudogene | 50.030100 | 50.030756 | negative | MGI_C57BL6J_3648883 | Rpl13a-ps1 | NCBI_Gene:433251,ENSEMBL:ENSMUSG00000062083 | MGI:3648883 | pseudogene | ribosomal protein 13A, pseudogene 1 |
| 19 | gene | 50.143295 | 50.679136 | negative | MGI_C57BL6J_1929666 | Sorcs1 | NCBI_Gene:58178,ENSEMBL:ENSMUSG00000043531 | MGI:1929666 | protein coding gene | sortilin-related VPS10 domain containing receptor 1 |
| 19 | gene | 50.144491 | 50.144615 | positive | MGI_C57BL6J_5531131 | Gm27749 | ENSEMBL:ENSMUSG00000098916 | MGI:5531131 | miRNA gene | predicted gene, 27749 |
| 19 | gene | 50.146938 | 50.180652 | positive | MGI_C57BL6J_5791188 | Gm45352 | ENSEMBL:ENSMUSG00000110007 | MGI:5791188 | lncRNA gene | predicted gene 45352 |
| 19 | gene | 50.327639 | 50.331581 | positive | MGI_C57BL6J_5589282 | Gm30123 | NA | NA | lncRNA gene | predicted gene%2c 30123 |
| 19 | gene | 50.665964 | 50.778662 | positive | MGI_C57BL6J_5477123 | Gm26629 | NCBI_Gene:102631841,ENSEMBL:ENSMUSG00000097446 | MGI:5477123 | lncRNA gene | predicted gene, 26629 |
| 19 | gene | 50.705722 | 50.713685 | negative | MGI_C57BL6J_6303151 | Gm50302 | ENSEMBL:ENSMUSG00000117758 | MGI:6303151 | lncRNA gene | predicted gene, 50302 |
| 19 | gene | 50.812324 | 50.812360 | positive | MGI_C57BL6J_5455436 | Gm25659 | ENSEMBL:ENSMUSG00000089094 | MGI:5455436 | rRNA gene | predicted gene, 25659 |
| 19 | gene | 51.144749 | 51.148034 | positive | MGI_C57BL6J_6303175 | Gm50317 | ENSEMBL:ENSMUSG00000118338 | MGI:6303175 | unclassified gene | predicted gene, 50317 |
| 19 | gene | 51.239690 | 51.239796 | negative | MGI_C57BL6J_5455421 | Gm25644 | ENSEMBL:ENSMUSG00000065143 | MGI:5455421 | snRNA gene | predicted gene, 25644 |
| 19 | pseudogene | 51.631549 | 51.632627 | positive | MGI_C57BL6J_6303189 | Gm50325 | ENSEMBL:ENSMUSG00000117806 | MGI:6303189 | pseudogene | predicted gene, 50325 |
| 19 | pseudogene | 51.674388 | 51.674650 | negative | MGI_C57BL6J_6303190 | Gm50326 | ENSEMBL:ENSMUSG00000118158 | MGI:6303190 | pseudogene | predicted gene, 50326 |
| 19 | pseudogene | 51.792226 | 51.792633 | positive | MGI_C57BL6J_3644718 | Rpl31-ps18 | NCBI_Gene:668536,ENSEMBL:ENSMUSG00000097719 | MGI:3644718 | pseudogene | ribosomal protein L31, pseudogene 18 |
| 19 | gene | 51.891724 | 51.920333 | negative | MGI_C57BL6J_6303196 | Gm50330 | ENSEMBL:ENSMUSG00000117867 | MGI:6303196 | lncRNA gene | predicted gene, 50330 |
| 19 | gene | 51.913963 | 51.984533 | positive | MGI_C57BL6J_6303194 | Gm50329 | ENSEMBL:ENSMUSG00000118173 | MGI:6303194 | lncRNA gene | predicted gene, 50329 |
| 19 | pseudogene | 51.972994 | 51.974073 | negative | MGI_C57BL6J_3782291 | Gm4115 | NCBI_Gene:100042936 | MGI:3782291 | pseudogene | predicted gene 4115 |
| 19 | gene | 52.264297 | 52.265476 | positive | MGI_C57BL6J_96572 | Ins1 | NCBI_Gene:16333,ENSEMBL:ENSMUSG00000035804 | MGI:96572 | protein coding gene | insulin I |
| 19 | gene | 52.303521 | 52.308652 | positive | MGI_C57BL6J_6303198 | Gm50331 | ENSEMBL:ENSMUSG00000118128 | MGI:6303198 | lncRNA gene | predicted gene, 50331 |
| 19 | gene | 52.337165 | 52.345933 | positive | MGI_C57BL6J_6303200 | Gm50332 | ENSEMBL:ENSMUSG00000118114 | MGI:6303200 | lncRNA gene | predicted gene, 50332 |
| 19 | gene | 52.372948 | 52.389660 | negative | MGI_C57BL6J_5624745 | Gm41860 | NCBI_Gene:105246590 | MGI:5624745 | lncRNA gene | predicted gene, 41860 |
| 19 | gene | 52.923181 | 52.941209 | positive | MGI_C57BL6J_3035466 | AA387883 | NCBI_Gene:100043450,ENSEMBL:ENSMUSG00000117896 | MGI:3035466 | lncRNA gene | expressed sequence AA387883 |
| 19 | gene | 52.931926 | 53.040214 | negative | MGI_C57BL6J_2180003 | Xpnpep1 | NCBI_Gene:170750,ENSEMBL:ENSMUSG00000025027 | MGI:2180003 | protein coding gene | X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| 19 | gene | 52.943109 | 52.945972 | negative | MGI_C57BL6J_5624746 | Gm41861 | NCBI_Gene:105246591 | MGI:5624746 | lncRNA gene | predicted gene, 41861 |
| 19 | gene | 53.010656 | 53.010756 | negative | MGI_C57BL6J_5531214 | Mir6407 | miRBase:MI0021944,NCBI_Gene:102465220,ENSEMBL:ENSMUSG00000098730 | MGI:5531214 | miRNA gene | microRNA 6407 |
| 19 | gene | 53.033648 | 53.084516 | positive | MGI_C57BL6J_1925707 | 1700054A03Rik | NCBI_Gene:78457,ENSEMBL:ENSMUSG00000099988 | MGI:1925707 | lncRNA gene | RIKEN cDNA 1700054A03 gene |
| 19 | gene | 53.050716 | 53.105907 | negative | MGI_C57BL6J_5589393 | Gm30234 | NCBI_Gene:102632061 | MGI:5589393 | lncRNA gene | predicted gene, 30234 |
| 19 | gene | 53.085037 | 53.088696 | positive | MGI_C57BL6J_5624747 | Gm41862 | NCBI_Gene:105246592 | MGI:5624747 | lncRNA gene | predicted gene, 41862 |
| 19 | gene | 53.127579 | 53.131793 | positive | MGI_C57BL6J_5589540 | Gm30381 | NCBI_Gene:102632253,ENSEMBL:ENSMUSG00000118233 | MGI:5589540 | lncRNA gene | predicted gene, 30381 |
| 19 | gene | 53.133413 | 53.139839 | negative | MGI_C57BL6J_3780026 | Gm9618 | NCBI_Gene:674277,ENSEMBL:ENSMUSG00000118003 | MGI:3780026 | lncRNA gene | predicted gene 9618 |
| 19 | gene | 53.140443 | 53.247399 | positive | MGI_C57BL6J_1351615 | Add3 | NCBI_Gene:27360,ENSEMBL:ENSMUSG00000025026 | MGI:1351615 | protein coding gene | adducin 3 (gamma) |
| 19 | gene | 53.159757 | 53.159863 | negative | MGI_C57BL6J_5454501 | Gm24724 | ENSEMBL:ENSMUSG00000088855 | MGI:5454501 | snRNA gene | predicted gene, 24724 |
| 19 | gene | 53.248736 | 53.265405 | negative | MGI_C57BL6J_1916569 | 1700001K23Rik | NCBI_Gene:69319,ENSEMBL:ENSMUSG00000118314 | MGI:1916569 | lncRNA gene | RIKEN cDNA 1700001K23 gene |
| 19 | pseudogene | 53.265747 | 53.266864 | positive | MGI_C57BL6J_5595498 | Gm36339 | NCBI_Gene:102640219,ENSEMBL:ENSMUSG00000118350 | MGI:5595498 | pseudogene | predicted gene, 36339 |
| 19 | gene | 53.310414 | 53.375810 | positive | MGI_C57BL6J_97245 | Mxi1 | NCBI_Gene:17859,ENSEMBL:ENSMUSG00000025025 | MGI:97245 | protein coding gene | MAX interactor 1, dimerization protein |
| 19 | gene | 53.317356 | 53.326998 | negative | MGI_C57BL6J_5589700 | Gm30541 | NCBI_Gene:102632479,ENSEMBL:ENSMUSG00000118369 | MGI:5589700 | lncRNA gene | predicted gene, 30541 |
| 19 | gene | 53.371566 | 53.371766 | negative | MGI_C57BL6J_3704501 | Gm10197 | ENSEMBL:ENSMUSG00000067085 | MGI:3704501 | protein coding gene | predicted gene 10197 |
| 19 | gene | 53.379214 | 53.390917 | negative | MGI_C57BL6J_1923729 | Smndc1 | NCBI_Gene:76479,ENSEMBL:ENSMUSG00000025024 | MGI:1923729 | protein coding gene | survival motor neuron domain containing 1 |
| 19 | gene | 53.390789 | 53.391795 | positive | MGI_C57BL6J_1924742 | 8030456M14Rik | ENSEMBL:ENSMUSG00000117768 | MGI:1924742 | lncRNA gene | RIKEN cDNA 8030456M14 gene |
| 19 | gene | 53.441212 | 53.464796 | negative | MGI_C57BL6J_1922001 | Mirt1 | NCBI_Gene:381232,ENSEMBL:ENSMUSG00000097636 | MGI:1922001 | lncRNA gene | myocardial infarction associated transcript 1 |
| 19 | gene | 53.460066 | 53.462731 | positive | MGI_C57BL6J_1921149 | 4833407H14Rik | NCBI_Gene:73899,ENSEMBL:ENSMUSG00000097779 | MGI:1921149 | lncRNA gene | RIKEN cDNA 4833407H14 gene |
| 19 | gene | 53.483494 | 53.486485 | positive | MGI_C57BL6J_6303297 | Gm50393 | ENSEMBL:ENSMUSG00000118378 | MGI:6303297 | unclassified gene | predicted gene, 50393 |
| 19 | gene | 53.488977 | 53.492649 | negative | MGI_C57BL6J_5589985 | Gm30826 | NCBI_Gene:102632863 | MGI:5589985 | lncRNA gene | predicted gene, 30826 |
| 19 | gene | 53.528609 | 53.529655 | negative | MGI_C57BL6J_6303299 | Gm50394 | ENSEMBL:ENSMUSG00000118210 | MGI:6303299 | lncRNA gene | predicted gene, 50394 |
| 19 | gene | 53.529109 | 53.542431 | positive | MGI_C57BL6J_2685183 | Dusp5 | NCBI_Gene:240672,ENSEMBL:ENSMUSG00000034765 | MGI:2685183 | protein coding gene | dual specificity phosphatase 5 |
| 19 | pseudogene | 53.588151 | 53.589068 | negative | MGI_C57BL6J_108008 | Nutf2-ps1 | NCBI_Gene:100043462,ENSEMBL:ENSMUSG00000071497 | MGI:108008 | pseudogene | nuclear transport factor 2, pseudogene 1 |
| 19 | gene | 53.598542 | 53.599411 | negative | MGI_C57BL6J_6303314 | Gm50402 | ENSEMBL:ENSMUSG00000118159 | MGI:6303314 | unclassified gene | predicted gene, 50402 |
| 19 | gene | 53.600396 | 53.645833 | positive | MGI_C57BL6J_1339795 | Smc3 | NCBI_Gene:13006,ENSEMBL:ENSMUSG00000024974 | MGI:1339795 | protein coding gene | structural maintenance of chromosomes 3 |
| 19 | gene | 53.660989 | 53.867083 | positive | MGI_C57BL6J_1920963 | Rbm20 | NCBI_Gene:73713,ENSEMBL:ENSMUSG00000043639 | MGI:1920963 | protein coding gene | RNA binding motif protein 20 |
| 19 | gene | 53.673178 | 53.675863 | negative | MGI_C57BL6J_5826290 | Gm46653 | NCBI_Gene:108168414 | MGI:5826290 | lncRNA gene | predicted gene, 46653 |
| 19 | gene | 53.754244 | 53.758239 | positive | MGI_C57BL6J_5590102 | Gm30943 | NCBI_Gene:102633013 | MGI:5590102 | lncRNA gene | predicted gene, 30943 |
| 19 | pseudogene | 53.767891 | 53.768310 | negative | MGI_C57BL6J_3826538 | Gm16298 | ENSEMBL:ENSMUSG00000089807 | MGI:3826538 | pseudogene | predicted gene 16298 |
| 19 | gene | 53.770365 | 53.788290 | negative | MGI_C57BL6J_5590049 | Gm30890 | NCBI_Gene:102632944 | MGI:5590049 | lncRNA gene | predicted gene, 30890 |
| 19 | gene | 53.819252 | 53.825401 | negative | MGI_C57BL6J_3826539 | Gm16299 | NCBI_Gene:102641509,ENSEMBL:ENSMUSG00000089679 | MGI:3826539 | lncRNA gene | predicted gene 16299 |
| 19 | gene | 53.855298 | 53.860220 | negative | MGI_C57BL6J_5624748 | Gm41863 | NCBI_Gene:105246593 | MGI:5624748 | lncRNA gene | predicted gene, 41863 |
| 19 | gene | 53.871785 | 53.903185 | negative | MGI_C57BL6J_5590149 | Gm30990 | NCBI_Gene:102633075 | MGI:5590149 | lncRNA gene | predicted gene, 30990 |
| 19 | pseudogene | 53.883367 | 53.883768 | positive | MGI_C57BL6J_6303101 | Gm50272 | ENSEMBL:ENSMUSG00000117693 | MGI:6303101 | pseudogene | predicted gene, 50272 |
| 19 | gene | 53.892231 | 53.929861 | positive | MGI_C57BL6J_107490 | Pdcd4 | NCBI_Gene:18569,ENSEMBL:ENSMUSG00000024975 | MGI:107490 | protein coding gene | programmed cell death 4 |
| 19 | gene | 53.903996 | 53.904958 | positive | MGI_C57BL6J_1922933 | 2310035P21Rik | NA | NA | unclassified gene | RIKEN cDNA 2310035P21 gene |
| 19 | gene | 53.929656 | 53.944643 | negative | MGI_C57BL6J_1913610 | Bbip1 | NCBI_Gene:100503572,ENSEMBL:ENSMUSG00000084957 | MGI:1913610 | protein coding gene | BBSome interacting protein 1 |
| 19 | gene | 53.944232 | 54.033280 | positive | MGI_C57BL6J_1927197 | Shoc2 | NCBI_Gene:56392,ENSEMBL:ENSMUSG00000024976 | MGI:1927197 | protein coding gene | Shoc2, leucine rich repeat scaffold protein |
| 19 | gene | 53.960355 | 53.962769 | negative | MGI_C57BL6J_6303102 | Gm50273 | ENSEMBL:ENSMUSG00000117704 | MGI:6303102 | unclassified gene | predicted gene, 50273 |
| 19 | gene | 53.976462 | 54.001442 | negative | MGI_C57BL6J_1922206 | 4930484I04Rik | ENSEMBL:ENSMUSG00000117981 | MGI:1922206 | lncRNA gene | RIKEN cDNA 4930484I04 gene |
| 19 | gene | 54.044151 | 54.048982 | positive | MGI_C57BL6J_87934 | Adra2a | NCBI_Gene:11551,ENSEMBL:ENSMUSG00000033717 | MGI:87934 | protein coding gene | adrenergic receptor, alpha 2a |
| 19 | gene | 54.084559 | 54.086658 | positive | MGI_C57BL6J_6302959 | Gm50186 | ENSEMBL:ENSMUSG00000117921 | MGI:6302959 | lncRNA gene | predicted gene, 50186 |
| 19 | gene | 54.154877 | 54.175117 | positive | MGI_C57BL6J_5624750 | Gm41865 | NCBI_Gene:105246595 | MGI:5624750 | lncRNA gene | predicted gene, 41865 |
| 19 | gene | 54.198385 | 54.201323 | negative | MGI_C57BL6J_5624751 | Gm41866 | NCBI_Gene:105246596 | MGI:5624751 | lncRNA gene | predicted gene, 41866 |
| 19 | gene | 54.198391 | 54.201454 | positive | MGI_C57BL6J_6302961 | Gm50187 | ENSEMBL:ENSMUSG00000118324 | MGI:6302961 | unclassified gene | predicted gene, 50187 |
| 19 | pseudogene | 54.444980 | 54.445204 | positive | MGI_C57BL6J_6302963 | Gm50188 | ENSEMBL:ENSMUSG00000117755 | MGI:6302963 | pseudogene | predicted gene, 50188 |
| 19 | gene | 54.465865 | 54.586424 | positive | MGI_C57BL6J_5590460 | Gm31301 | NCBI_Gene:102633487 | MGI:5590460 | lncRNA gene | predicted gene, 31301 |
| 19 | gene | 54.758648 | 54.760955 | negative | MGI_C57BL6J_5624752 | Gm41867 | NCBI_Gene:105246597 | MGI:5624752 | lncRNA gene | predicted gene, 41867 |
| 19 | gene | 54.923805 | 54.936724 | negative | MGI_C57BL6J_6302966 | Gm50191 | ENSEMBL:ENSMUSG00000118207 | MGI:6302966 | lncRNA gene | predicted gene, 50191 |
| 19 | gene | 55.067268 | 55.127238 | negative | MGI_C57BL6J_109162 | Gpam | NCBI_Gene:14732,ENSEMBL:ENSMUSG00000024978 | MGI:109162 | protein coding gene | glycerol-3-phosphate acyltransferase, mitochondrial |
| 19 | gene | 55.099502 | 55.118931 | positive | MGI_C57BL6J_5590515 | Gm31356 | NCBI_Gene:102633560,ENSEMBL:ENSMUSG00000118099 | MGI:5590515 | lncRNA gene | predicted gene, 31356 |
| 19 | gene | 55.127314 | 55.140840 | positive | MGI_C57BL6J_5590577 | Gm31418 | NCBI_Gene:102633636 | MGI:5590577 | lncRNA gene | predicted gene, 31418 |
| 19 | gene | 55.132642 | 55.134832 | negative | MGI_C57BL6J_3642924 | Gm10755 | NA | NA | unclassified gene | predicted gene 10755 |
| 19 | gene | 55.180733 | 55.196313 | positive | MGI_C57BL6J_109574 | Tectb | NCBI_Gene:21684,ENSEMBL:ENSMUSG00000024979 | MGI:109574 | protein coding gene | tectorin beta |
| 19 | gene | 55.192678 | 55.192734 | positive | MGI_C57BL6J_5530856 | Mir6715 | miRBase:MI0025026,NCBI_Gene:102465990,ENSEMBL:ENSMUSG00000098289 | MGI:5530856 | miRNA gene | microRNA 6715 |
| 19 | gene | 55.198098 | 55.241236 | negative | MGI_C57BL6J_106025 | Gucy2g | NCBI_Gene:73707,ENSEMBL:ENSMUSG00000055523 | MGI:106025 | protein coding gene | guanylate cyclase 2g |
| 19 | gene | 55.251884 | 55.297720 | positive | MGI_C57BL6J_1919129 | Acsl5 | NCBI_Gene:433256,ENSEMBL:ENSMUSG00000024981 | MGI:1919129 | protein coding gene | acyl-CoA synthetase long-chain family member 5 |
| 19 | gene | 55.262382 | 55.274479 | negative | MGI_C57BL6J_5590754 | Gm31595 | NCBI_Gene:102633871 | MGI:5590754 | lncRNA gene | predicted gene, 31595 |
| 19 | gene | 55.271291 | 55.316032 | negative | MGI_C57BL6J_1914230 | Zdhhc6 | NCBI_Gene:66980,ENSEMBL:ENSMUSG00000024982 | MGI:1914230 | protein coding gene | zinc finger, DHHC domain containing 6 |
| 19 | gene | 55.316057 | 55.627461 | positive | MGI_C57BL6J_1855699 | Vti1a | NCBI_Gene:53611,ENSEMBL:ENSMUSG00000024983 | MGI:1855699 | protein coding gene | vesicle transport through interaction with t-SNAREs 1A |
| 19 | gene | 55.480917 | 55.484622 | negative | MGI_C57BL6J_3782650 | Gm4466 | NA | NA | unclassified gene | predicted gene 4466 |
| 19 | gene | 55.555873 | 55.556183 | negative | MGI_C57BL6J_1917659 | 2310066F23Rik | NA | NA | unclassified gene | RIKEN cDNA 2310066F23 gene |
| 19 | gene | 55.640589 | 55.702204 | negative | MGI_C57BL6J_1924844 | 4930552P12Rik | NCBI_Gene:77594,ENSEMBL:ENSMUSG00000097632 | MGI:1924844 | lncRNA gene | RIKEN cDNA 4930552P12 gene |
| 19 | gene | 55.701143 | 55.702134 | positive | MGI_C57BL6J_5624756 | Gm41871 | NCBI_Gene:105246602 | MGI:5624756 | lncRNA gene | predicted gene, 41871 |
| 19 | gene | 55.734407 | 55.743225 | negative | MGI_C57BL6J_5590893 | Gm31734 | NCBI_Gene:102634055 | MGI:5590893 | lncRNA gene | predicted gene, 31734 |
| 19 | gene | 55.741785 | 55.933661 | positive | MGI_C57BL6J_1202879 | Tcf7l2 | NCBI_Gene:21416,ENSEMBL:ENSMUSG00000024985 | MGI:1202879 | protein coding gene | transcription factor 7 like 2, T cell specific, HMG box |
| 19 | gene | 55.841672 | 55.845897 | positive | MGI_C57BL6J_1349458 | Ppnr | ENSEMBL:ENSMUSG00000099746 | MGI:1349458 | lncRNA gene | per-pentamer repeat gene |
| 19 | gene | 55.940237 | 55.950980 | positive | MGI_C57BL6J_5624757 | Gm41872 | NCBI_Gene:105246603 | MGI:5624757 | lncRNA gene | predicted gene, 41872 |
| 19 | pseudogene | 56.012529 | 56.012925 | positive | MGI_C57BL6J_6302881 | Gm50135 | ENSEMBL:ENSMUSG00000117833 | MGI:6302881 | pseudogene | predicted gene, 50135 |
| 19 | gene | 56.035096 | 56.040941 | positive | MGI_C57BL6J_3041200 | D730002M21Rik | ENSEMBL:ENSMUSG00000117827 | MGI:3041200 | lncRNA gene | RIKEN cDNA D730002M21 gene |
| 19 | gene | 56.066128 | 56.066934 | positive | MGI_C57BL6J_1923871 | 1700106J12Rik | NA | NA | unclassified gene | RIKEN cDNA 1700106J12 gene |
| 19 | gene | 56.077954 | 56.081392 | positive | MGI_C57BL6J_3782654 | Gm4470 | NCBI_Gene:102634217 | MGI:3782654 | lncRNA gene | predicted gene 4470 |
| 19 | gene | 56.105689 | 56.140607 | positive | MGI_C57BL6J_5591071 | Gm31912 | NCBI_Gene:102634291,ENSEMBL:ENSMUSG00000117695 | MGI:5591071 | lncRNA gene | predicted gene, 31912 |
| 19 | gene | 56.155105 | 56.176419 | positive | MGI_C57BL6J_5591124 | Gm31965 | NCBI_Gene:102634365 | MGI:5591124 | lncRNA gene | predicted gene, 31965 |
| 19 | gene | 56.177735 | 56.199050 | positive | MGI_C57BL6J_5591187 | Gm32028 | NCBI_Gene:102634447 | MGI:5591187 | lncRNA gene | predicted gene, 32028 |
| 19 | gene | 56.263413 | 56.266917 | negative | MGI_C57BL6J_5591303 | Gm32144 | NCBI_Gene:102634603 | MGI:5591303 | lncRNA gene | predicted gene, 32144 |
| 19 | gene | 56.273194 | 56.292830 | negative | MGI_C57BL6J_5591416 | Gm32257 | NCBI_Gene:102634750 | MGI:5591416 | lncRNA gene | predicted gene, 32257 |
| 19 | gene | 56.277518 | 56.277645 | negative | MGI_C57BL6J_5452048 | Gm22271 | ENSEMBL:ENSMUSG00000077223 | MGI:5452048 | snoRNA gene | predicted gene, 22271 |
| 19 | gene | 56.287109 | 56.320822 | positive | MGI_C57BL6J_1196378 | Habp2 | NCBI_Gene:226243,ENSEMBL:ENSMUSG00000025075 | MGI:1196378 | protein coding gene | hyaluronic acid binding protein 2 |
| 19 | gene | 56.320035 | 56.390063 | negative | MGI_C57BL6J_1098765 | Nrap | NCBI_Gene:18175,ENSEMBL:ENSMUSG00000049134 | MGI:1098765 | protein coding gene | nebulin-related anchoring protein |
| 19 | gene | 56.356412 | 56.366241 | positive | MGI_C57BL6J_4938024 | Gm17197 | NCBI_Gene:102634685,ENSEMBL:ENSMUSG00000090990 | MGI:4938024 | lncRNA gene | predicted gene 17197 |
| 19 | gene | 56.396634 | 56.442348 | positive | MGI_C57BL6J_109383 | Casp7 | NCBI_Gene:12369,ENSEMBL:ENSMUSG00000025076 | MGI:109383 | protein coding gene | caspase 7 |
| 19 | gene | 56.442446 | 56.455000 | negative | MGI_C57BL6J_5826294 | Gm46657 | NCBI_Gene:108168418 | MGI:5826294 | lncRNA gene | predicted gene, 46657 |
| 19 | gene | 56.457275 | 56.486752 | positive | MGI_C57BL6J_2443041 | Plekhs1 | NCBI_Gene:226245,ENSEMBL:ENSMUSG00000035818 | MGI:2443041 | protein coding gene | pleckstrin homology domain containing, family S member 1 |
| 19 | gene | 56.529161 | 56.548284 | negative | MGI_C57BL6J_1930042 | Dclre1a | NCBI_Gene:55947,ENSEMBL:ENSMUSG00000025077 | MGI:1930042 | protein coding gene | DNA cross-link repair 1A |
| 19 | gene | 56.548261 | 56.603503 | positive | MGI_C57BL6J_1914116 | Nhlrc2 | NCBI_Gene:66866,ENSEMBL:ENSMUSG00000025078 | MGI:1914116 | protein coding gene | NHL repeat containing 2 |
| 19 | gene | 56.655970 | 56.664626 | negative | MGI_C57BL6J_5591600 | Gm32441 | NCBI_Gene:102634992,ENSEMBL:ENSMUSG00000117890 | MGI:5591600 | lncRNA gene | predicted gene, 32441 |
| 19 | gene | 56.669067 | 56.674549 | positive | MGI_C57BL6J_5591651 | Gm32492 | NCBI_Gene:102635060,ENSEMBL:ENSMUSG00000117859 | MGI:5591651 | lncRNA gene | predicted gene, 32492 |
| 19 | gene | 56.703751 | 56.714015 | positive | MGI_C57BL6J_5591734 | Gm32575 | NCBI_Gene:102635171 | MGI:5591734 | lncRNA gene | predicted gene, 32575 |
| 19 | gene | 56.722199 | 56.733113 | positive | MGI_C57BL6J_87937 | Adrb1 | NCBI_Gene:11554,ENSEMBL:ENSMUSG00000035283 | MGI:87937 | protein coding gene | adrenergic receptor, beta 1 |
| 19 | gene | 56.741029 | 56.746913 | negative | MGI_C57BL6J_6302829 | Gm50100 | ENSEMBL:ENSMUSG00000117696 | MGI:6302829 | lncRNA gene | predicted gene, 50100 |
| 19 | gene | 56.743045 | 56.765957 | positive | MGI_C57BL6J_5591877 | Gm32718 | NCBI_Gene:102635359 | MGI:5591877 | lncRNA gene | predicted gene, 32718 |
| 19 | pseudogene | 56.754878 | 56.755428 | negative | MGI_C57BL6J_3648860 | Gm6990 | NCBI_Gene:629643,ENSEMBL:ENSMUSG00000067063 | MGI:3648860 | pseudogene | predicted pseudogene 6990 |
| 19 | gene | 56.787481 | 56.822488 | negative | MGI_C57BL6J_2445022 | Ccdc186 | NCBI_Gene:213993,ENSEMBL:ENSMUSG00000035173 | MGI:2445022 | protein coding gene | coiled-coil domain containing 186 |
| 19 | gene | 56.821962 | 56.822539 | positive | MGI_C57BL6J_1922878 | 1700010L13Rik | NA | NA | protein coding gene | RIKEN cDNA 1700010L13 gene |
| 19 | gene | 56.826171 | 56.870012 | positive | MGI_C57BL6J_1933218 | Tdrd1 | NCBI_Gene:83561,ENSEMBL:ENSMUSG00000025081 | MGI:1933218 | protein coding gene | tudor domain containing 1 |
| 19 | gene | 56.874249 | 56.912078 | positive | MGI_C57BL6J_2684334 | Vwa2 | NCBI_Gene:240675,ENSEMBL:ENSMUSG00000025082 | MGI:2684334 | protein coding gene | von Willebrand factor A domain containing 2 |
| 19 | gene | 56.912354 | 57.008591 | negative | MGI_C57BL6J_2147658 | Afap1l2 | NCBI_Gene:226250,ENSEMBL:ENSMUSG00000025083 | MGI:2147658 | protein coding gene | actin filament associated protein 1-like 2 |
| 19 | gene | 56.977200 | 56.984849 | positive | MGI_C57BL6J_5621482 | Gm38597 | NCBI_Gene:102642146 | MGI:5621482 | lncRNA gene | predicted gene, 38597 |
| 19 | gene | 57.032733 | 57.315204 | negative | MGI_C57BL6J_1194500 | Ablim1 | NCBI_Gene:226251,ENSEMBL:ENSMUSG00000025085 | MGI:1194500 | protein coding gene | actin-binding LIM protein 1 |
| 19 | gene | 57.273960 | 57.277052 | positive | MGI_C57BL6J_1925370 | 4930449E18Rik | NCBI_Gene:78120,ENSEMBL:ENSMUSG00000117684 | MGI:1925370 | lncRNA gene | RIKEN cDNA 4930449E18 gene |
| 19 | gene | 57.323193 | 57.360899 | negative | MGI_C57BL6J_2444948 | B230217O12Rik | NCBI_Gene:320879,ENSEMBL:ENSMUSG00000097785 | MGI:2444948 | lncRNA gene | RIKEN cDNA B230217O12 gene |
| 19 | gene | 57.323602 | 57.331224 | positive | MGI_C57BL6J_6303097 | Gm50270 | ENSEMBL:ENSMUSG00000117670 | MGI:6303097 | lncRNA gene | predicted gene, 50270 |
| 19 | gene | 57.360680 | 57.389594 | positive | MGI_C57BL6J_2147545 | Fam160b1 | NCBI_Gene:226252,ENSEMBL:ENSMUSG00000033478 | MGI:2147545 | protein coding gene | family with sequence similarity 160, member B1 |
| 19 | gene | 57.404096 | 57.404275 | negative | MGI_C57BL6J_1924071 | 2600017C09Rik | NA | NA | unclassified gene | RIKEN cDNA 2600017C09 gene |
| 19 | gene | 57.404678 | 57.411272 | positive | MGI_C57BL6J_5624759 | Gm41874 | NCBI_Gene:105246606 | MGI:5624759 | lncRNA gene | predicted gene, 41874 |
| 19 | gene | 57.418291 | 57.423494 | negative | MGI_C57BL6J_6303099 | Gm50271 | ENSEMBL:ENSMUSG00000117927 | MGI:6303099 | lncRNA gene | predicted gene, 50271 |
| 19 | gene | 57.452892 | 57.491005 | positive | MGI_C57BL6J_1919383 | Trub1 | NCBI_Gene:72133,ENSEMBL:ENSMUSG00000025086 | MGI:1919383 | protein coding gene | TruB pseudouridine (psi) synthase family member 1 |
| 19 | gene | 57.459572 | 57.461553 | negative | MGI_C57BL6J_5477368 | Gm26874 | ENSEMBL:ENSMUSG00000097374 | MGI:5477368 | lncRNA gene | predicted gene, 26874 |
| 19 | gene | 57.497390 | 57.512788 | positive | MGI_C57BL6J_1925323 | 6720468P15Rik | NCBI_Gene:78073,ENSEMBL:ENSMUSG00000117655 | MGI:1925323 | lncRNA gene | RIKEN cDNA 6720468P15 gene |
| 19 | gene | 57.522180 | 57.533305 | positive | MGI_C57BL6J_5592091 | Gm32932 | NCBI_Gene:102635646,ENSEMBL:ENSMUSG00000118178 | MGI:5592091 | lncRNA gene | predicted gene, 32932 |
| 19 | pseudogene | 57.529582 | 57.529897 | negative | MGI_C57BL6J_6303113 | Gm50279 | ENSEMBL:ENSMUSG00000118248 | MGI:6303113 | pseudogene | predicted gene, 50279 |
| 19 | gene | 57.540466 | 57.544223 | negative | MGI_C57BL6J_6303122 | Gm50285 | ENSEMBL:ENSMUSG00000117770 | MGI:6303122 | lncRNA gene | predicted gene, 50285 |
| 19 | pseudogene | 57.584761 | 57.584956 | positive | MGI_C57BL6J_6303124 | Gm50286 | ENSEMBL:ENSMUSG00000117978 | MGI:6303124 | pseudogene | predicted gene, 50286 |
| 19 | gene | 57.610963 | 58.133343 | positive | MGI_C57BL6J_2147749 | Atrnl1 | NCBI_Gene:226255,ENSEMBL:ENSMUSG00000054843 | MGI:2147749 | protein coding gene | attractin like 1 |
| 19 | gene | 58.022571 | 58.031893 | negative | MGI_C57BL6J_5592139 | Gm32980 | NCBI_Gene:102635712 | MGI:5592139 | lncRNA gene | predicted gene, 32980 |
| 19 | gene | 58.051167 | 58.051236 | positive | MGI_C57BL6J_5455845 | Mir5623 | miRBase:MI0019191,NCBI_Gene:100885835,ENSEMBL:ENSMUSG00000093757 | MGI:5455845 | miRNA gene | microRNA 5623 |
| 19 | gene | 58.091814 | 58.096031 | negative | MGI_C57BL6J_6303125 | Gm50287 | ENSEMBL:ENSMUSG00000117871 | MGI:6303125 | lncRNA gene | predicted gene, 50287 |
| 19 | gene | 58.091818 | 58.094002 | positive | MGI_C57BL6J_2442561 | E430016L07Rik | NA | NA | unclassified gene | RIKEN cDNA E430016L07 gene |
| 19 | gene | 58.204139 | 58.236047 | positive | MGI_C57BL6J_3826543 | Gm16277 | ENSEMBL:ENSMUSG00000087002 | MGI:3826543 | lncRNA gene | predicted gene 16277 |
| 19 | gene | 58.231333 | 58.232284 | negative | MGI_C57BL6J_5624760 | Gm41875 | NCBI_Gene:105246608 | MGI:5624760 | lncRNA gene | predicted gene, 41875 |
| 19 | gene | 58.235581 | 58.455946 | negative | MGI_C57BL6J_1100842 | Gfra1 | NCBI_Gene:14585,ENSEMBL:ENSMUSG00000025089 | MGI:1100842 | protein coding gene | glial cell line derived neurotrophic factor family receptor alpha 1 |
| 19 | gene | 58.280564 | 58.294578 | negative | MGI_C57BL6J_5826287 | Gm46650 | NCBI_Gene:108168411 | MGI:5826287 | protein coding gene | predicted gene, 46650 |
| 19 | gene | 58.296204 | 58.300348 | positive | MGI_C57BL6J_3642351 | Gm10007 | NCBI_Gene:791347 | MGI:3642351 | lncRNA gene | predicted gene 10007 |
| 19 | gene | 58.456149 | 58.459592 | positive | MGI_C57BL6J_5624761 | Gm41876 | NCBI_Gene:105246609 | MGI:5624761 | lncRNA gene | predicted gene, 41876 |
| 19 | pseudogene | 58.489282 | 58.489603 | positive | MGI_C57BL6J_6302958 | Gm50185 | ENSEMBL:ENSMUSG00000118253 | MGI:6302958 | pseudogene | predicted gene, 50185 |
| 19 | gene | 58.500223 | 58.508119 | positive | MGI_C57BL6J_5592465 | Gm33306 | NCBI_Gene:102636162 | MGI:5592465 | lncRNA gene | predicted gene, 33306 |
| 19 | gene | 58.511993 | 58.553104 | positive | MGI_C57BL6J_1922895 | Ccdc172 | NCBI_Gene:75645,ENSEMBL:ENSMUSG00000025090 | MGI:1922895 | protein coding gene | coiled-coil domain containing 172 |
| 19 | gene | 58.606329 | 58.629753 | positive | MGI_C57BL6J_1916305 | 1810007D17Rik | NCBI_Gene:69055,ENSEMBL:ENSMUSG00000100001 | MGI:1916305 | lncRNA gene | RIKEN cDNA 1810007D17 gene |
| 19 | gene | 58.652498 | 58.681788 | positive | MGI_C57BL6J_97722 | Pnlip | NCBI_Gene:69060,ENSEMBL:ENSMUSG00000046008 | MGI:97722 | protein coding gene | pancreatic lipase |
| 19 | gene | 58.695506 | 58.696178 | positive | MGI_C57BL6J_1925578 | 1810035K13Rik | NCBI_Gene:78328,ENSEMBL:ENSMUSG00000118223 | MGI:1925578 | lncRNA gene | RIKEN cDNA 1810035K13 gene |
| 19 | gene | 58.698917 | 58.715770 | positive | MGI_C57BL6J_1921421 | 1810018F18Rik | NCBI_Gene:69166,ENSEMBL:ENSMUSG00000101088 | MGI:1921421 | lncRNA gene | RIKEN cDNA 1810018F18 gene |
| 19 | gene | 58.728887 | 58.744169 | positive | MGI_C57BL6J_97723 | Pnliprp1 | NCBI_Gene:18946,ENSEMBL:ENSMUSG00000042179 | MGI:97723 | protein coding gene | pancreatic lipase related protein 1 |
| 19 | gene | 58.742043 | 58.743761 | positive | MGI_C57BL6J_1917628 | 1810073G21Rik | NA | NA | unclassified gene | RIKEN cDNA 1810073G21 gene |
| 19 | gene | 58.759719 | 58.777534 | positive | MGI_C57BL6J_1336202 | Pnliprp2 | NCBI_Gene:18947,ENSEMBL:ENSMUSG00000025091 | MGI:1336202 | protein coding gene | pancreatic lipase-related protein 2 |
| 19 | gene | 58.785803 | 58.794461 | negative | MGI_C57BL6J_1914757 | 1700019N19Rik | NCBI_Gene:67507,ENSEMBL:ENSMUSG00000026931 | MGI:1914757 | protein coding gene | RIKEN cDNA 1700019N19 gene |
| 19 | gene | 58.786608 | 58.786691 | negative | MGI_C57BL6J_5452338 | Gm22561 | ENSEMBL:ENSMUSG00000087956 | MGI:5452338 | unclassified non-coding RNA gene | predicted gene, 22561 |
| 19 | pseudogene | 58.790195 | 58.790575 | positive | MGI_C57BL6J_4937977 | Gm17150 | ENSEMBL:ENSMUSG00000091410 | MGI:4937977 | pseudogene | predicted gene 17150 |
| 19 | gene | 58.794515 | 58.802121 | positive | MGI_C57BL6J_5624762 | Gm41877 | NCBI_Gene:105246610 | MGI:5624762 | lncRNA gene | predicted gene, 41877 |
| 19 | gene | 58.795750 | 58.943672 | negative | MGI_C57BL6J_1920692 | Hspa12a | NCBI_Gene:73442,ENSEMBL:ENSMUSG00000025092 | MGI:1920692 | protein coding gene | heat shock protein 12A |
| 19 | gene | 58.873930 | 58.875138 | negative | MGI_C57BL6J_1921900 | 4930433M22Rik | NA | NA | unclassified gene | RIKEN cDNA 4930433M22 gene |
| 19 | gene | 58.911673 | 58.911759 | negative | MGI_C57BL6J_4834260 | Mir3086 | miRBase:MI0014049,NCBI_Gene:100526480,ENSEMBL:ENSMUSG00000092813 | MGI:4834260 | miRNA gene | microRNA 3086 |
| 19 | gene | 58.935184 | 58.971421 | positive | MGI_C57BL6J_2441717 | Eno4 | NCBI_Gene:226265,ENSEMBL:ENSMUSG00000048029 | MGI:2441717 | protein coding gene | enolase 4 |
| 19 | gene | 58.973356 | 59.076124 | negative | MGI_C57BL6J_1918903 | Shtn1 | NCBI_Gene:71653,ENSEMBL:ENSMUSG00000041362 | MGI:1918903 | protein coding gene | shootin 1 |
| 19 | pseudogene | 59.087487 | 59.089034 | positive | MGI_C57BL6J_3643073 | Gm9276 | NCBI_Gene:668626,ENSEMBL:ENSMUSG00000118377 | MGI:3643073 | pseudogene | predicted gene 9276 |
| 19 | gene | 59.164480 | 59.170056 | negative | MGI_C57BL6J_1277163 | Vax1 | NCBI_Gene:22326,ENSEMBL:ENSMUSG00000006270 | MGI:1277163 | protein coding gene | ventral anterior homeobox 1 |
| 19 | gene | 59.171624 | 59.184456 | positive | MGI_C57BL6J_1923050 | 4930442E04Rik | NCBI_Gene:75800 | MGI:1923050 | lncRNA gene | RIKEN cDNA 4930442E04 gene |
| 19 | pseudogene | 59.180655 | 59.181432 | positive | MGI_C57BL6J_6303359 | Gm50429 | ENSEMBL:ENSMUSG00000117834 | MGI:6303359 | pseudogene | predicted gene, 50429 |
| 19 | gene | 59.219214 | 59.237374 | positive | MGI_C57BL6J_2685627 | Kcnk18 | NCBI_Gene:332396,ENSEMBL:ENSMUSG00000040901 | MGI:2685627 | protein coding gene | potassium channel, subfamily K, member 18 |
| 19 | gene | 59.259512 | 59.261419 | negative | MGI_C57BL6J_5579967 | Gm29261 | ENSEMBL:ENSMUSG00000101630 | MGI:5579967 | lncRNA gene | predicted gene 29261 |
| 19 | gene | 59.260878 | 59.296012 | positive | MGI_C57BL6J_106677 | Slc18a2 | NCBI_Gene:214084,ENSEMBL:ENSMUSG00000025094 | MGI:106677 | protein coding gene | solute carrier family 18 (vesicular monoamine), member 2 |
| 19 | gene | 59.296084 | 59.345780 | negative | MGI_C57BL6J_2677270 | Pdzd8 | NCBI_Gene:107368,ENSEMBL:ENSMUSG00000074746 | MGI:2677270 | protein coding gene | PDZ domain containing 8 |
| 19 | pseudogene | 59.322290 | 59.322784 | positive | MGI_C57BL6J_3704503 | Rps12-ps3 | NCBI_Gene:100034727,ENSEMBL:ENSMUSG00000067038 | MGI:3704503 | pseudogene | ribosomal protein S12, pseudogene 3 |
| 19 | pseudogene | 59.337576 | 59.337969 | positive | MGI_C57BL6J_6303374 | Gm50440 | ENSEMBL:ENSMUSG00000118276 | MGI:6303374 | pseudogene | predicted gene, 50440 |
| 19 | gene | 59.345908 | 59.346675 | positive | MGI_C57BL6J_1919460 | 1700022C07Rik | NA | NA | unclassified gene | RIKEN cDNA 1700022C07 gene |
R/qtl
scanone
gmObject of class cross2 (crosstype "bc")
Total individuals 138
No. genotyped individuals 138
No. phenotyped individuals 138
No. with both geno & pheno 138
No. phenotypes 1
No. covariates 7
No. phenotype covariates 0
No. chromosomes 20
Total markers 26548
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2374 2374 1623 1691 1568 1703 1464 1468 1685 974 1671 1134 1377 1411 876 751
17 18 19 X
330 770 920 384 #detach("package:qtl2", unload=TRUE)
#library(qtl)
cross <- qtl::read.cross("csv", file = "data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_peaks.csv",alleles=c("A","B")) --Read the following data:
138 individuals
26548 markers
3 phenotypes
--Cross type: bc cross <- qtl::jittermap(cross)
summary(cross) Backcross
No. individuals: 138
No. phenotypes: 3
Percent phenotyped: 100 100 100
No. chromosomes: 20
Autosomes: 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19
X chr: X
Total markers: 26548
No. markers: 2374 2374 1623 1691 1568 1703 1464 1468 1685 974 1671
1134 1377 1411 876 751 330 770 920 384
Percent genotyped: 99.5
Genotypes (%):
Autosomes: AA:54.4 AB:45.6
X chromosome: AA:53.3 AB:46.7 cross.probs <- qtl::calc.genoprob(cross)
print("method == hk")[1] "method == hk"scanone.hk <-qtl::scanone(cross.probs, pheno.col="ICI.vs.PBS" , model="binary", method="hk")
operm.hk <- qtl::scanone(cross.probs, method = "hk", pheno.col="ICI.vs.PBS", n.perm = 10, perm.Xsp = TRUE, model="binary", verbose=FALSE)
plot(operm.hk)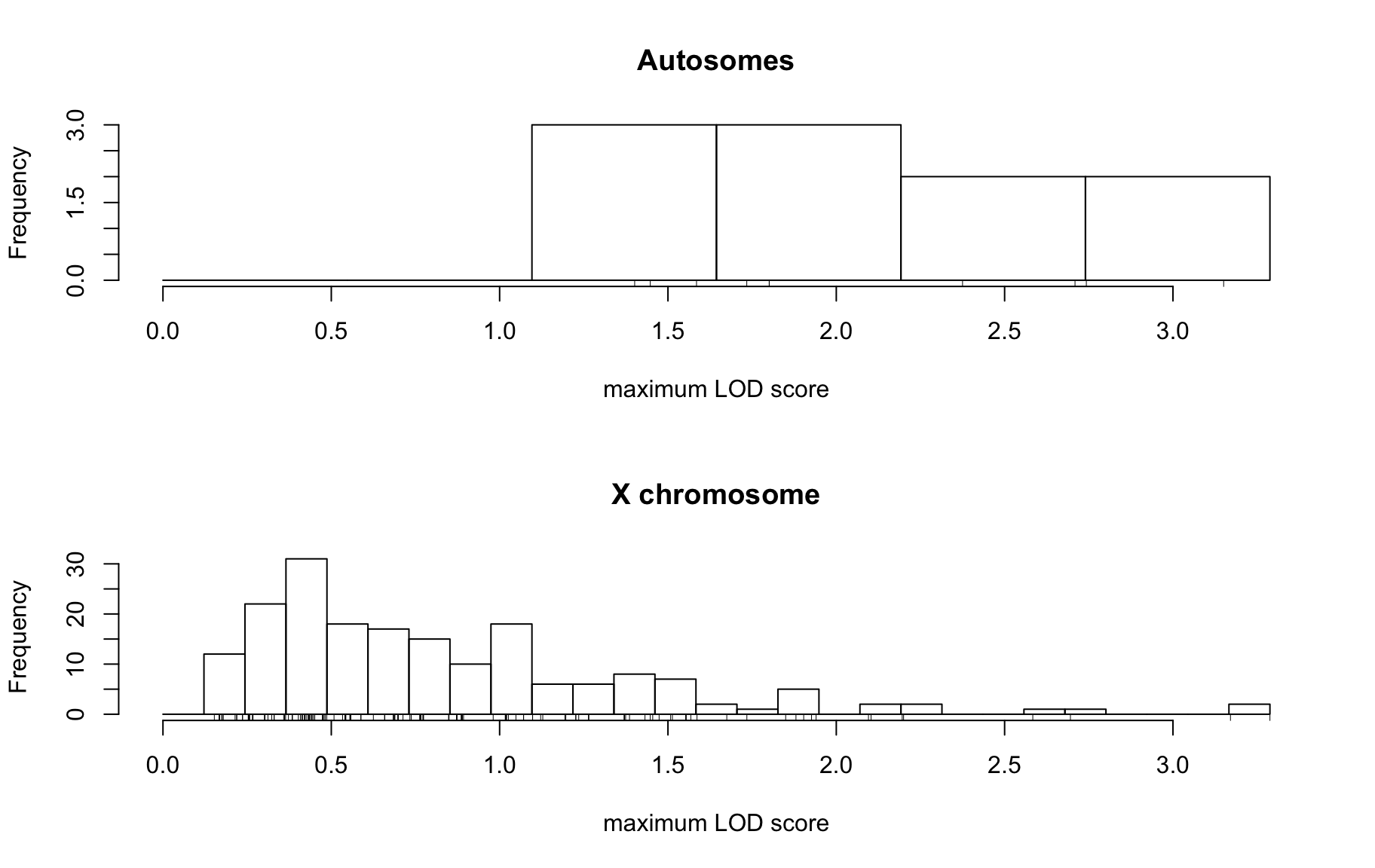
print(summary(operm.hk, alpha=c(0.01, 0.05, 0.1)))Autosome LOD thresholds (10 permutations)
lod
1% 3.12
5% 2.98
10% 2.80
X chromosome LOD thresholds (186 permutations)
lod
1% 3.28
5% 3.23
10% 3.17#plot(scanone.hk, bandcol = "grey90",lty=1, cex=1, col = "steelblue")
#qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.01, col = 'blue')
#qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.05, col = 'red')
#qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.1, col = 'purple')
ymx <- maxlod(out) # overall maximum LOD score
plot(scanone.hk, bandcol = "grey90",lty=1, cex=1, col = "slateblue", ylim=c(0, ymx+0.5))
title(main = paste0(colnames(out), " [positions in cM]"))
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.01, col = 'blue')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.05, col = 'red')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.1, col = 'purple')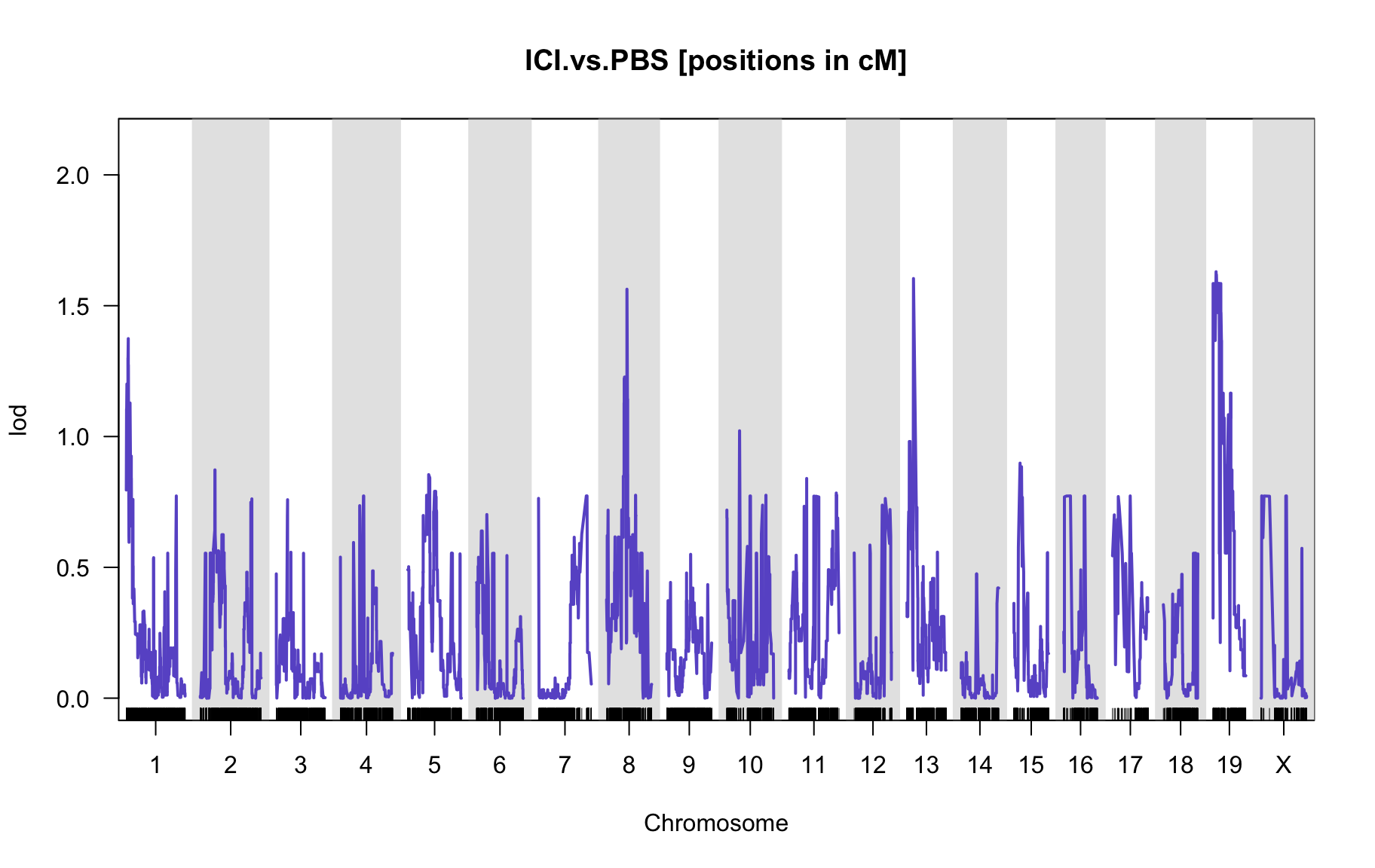
ymx <- 11
plot(scanone.hk, bandcol = "grey90",lty=1, cex=1, col = "slateblue", ylim=c(0, ymx+0.5))
title(main = paste0(colnames(out), " [positions in cM]\n(using same scale as eoi vs ici for easier comparison)"))
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.01, col = 'blue')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.05, col = 'red')
qtl::add.threshold(scanone.hk, perms= operm.hk, alpha=0.1, col = 'purple')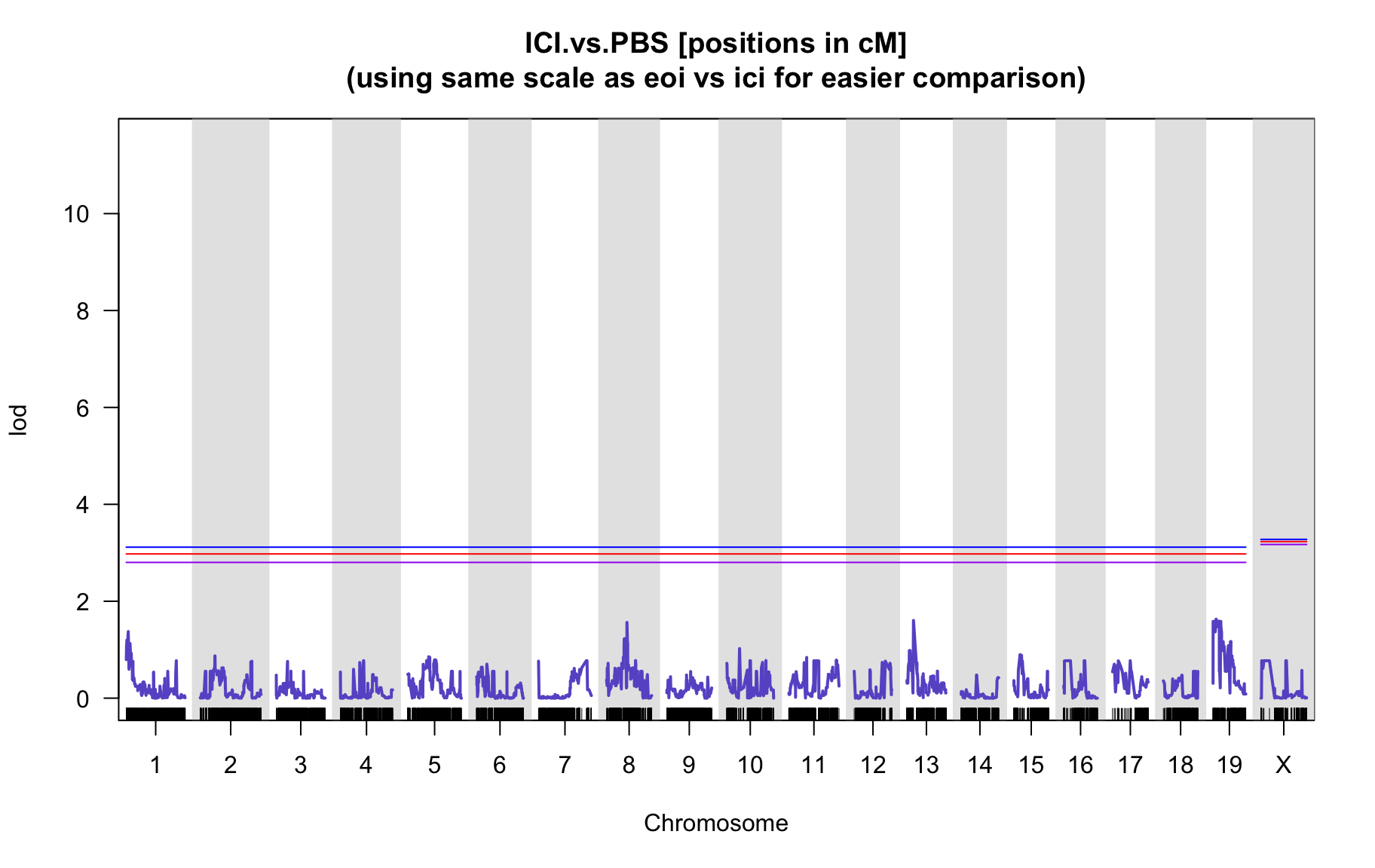
print(as.data.frame(summary(scanone.hk, perms=operm.hk, pvalues=TRUE, format="allpeaks"))) chr pos lod pval
UNC163443 1 4.774075 1.3742470 1.0000000
UNC2987704 2 27.689343 0.8724495 1.0000000
JAX00521745 3 20.613263 0.7585338 1.0000000
JAX00556948 4 41.038578 0.7729973 1.0000000
JAX00585101 5 36.216736 0.8544324 1.0000000
UNC10923091 6 18.321248 0.7018820 1.0000000
JAX00657315 7 81.940339 0.7731087 1.0000000
UNCHS023364 8 35.540812 1.5630473 0.8166471
UNCHS026281 9 40.415965 0.5497421 1.0000000
UNCHS028001 10 21.783253 1.0219015 1.0000000
UNCHS030712 11 32.135777 0.8396617 1.0000000
UNC21787084 12 53.288083 0.7631460 1.0000000
UNC22384241 13 13.831342 1.6038654 0.7188828
UNCHS038350 14 30.005599 0.4749227 1.0000000
UNC25285746 15 12.031111 0.8983676 1.0000000
UNC26465114 16 13.431084 0.7730271 1.0000000
JAX00077123 17 31.150016 0.7729946 1.0000000
UNC29709766 18 55.191705 0.5543284 1.0000000
UNC29893228 19 8.029054 1.6298590 0.7188828
JAX00176908 X 4.172023 0.7731463 0.9999705print("all peaks with a p-value less or equal to 0.05 (suggestive)")[1] "all peaks with a p-value less or equal to 0.05 (suggestive)"print(as.data.frame(summary(scanone.hk, perms=operm.hk, alpha=0.05, pvalues=TRUE, format="allpeaks")))[1] chr pos lod
<0 rows> (or 0-length row.names)print("all peaks with a threshold LOD greater than 1.5")[1] "all peaks with a threshold LOD greater than 1.5"print(as.data.frame(summary(scanone.hk, perms=operm.hk, threshold=1.5, pvalues=TRUE, format="allpeaks"))) chr pos lod pval
UNCHS023364 8 35.540812 1.563047 0.8166471
UNC22384241 13 13.831342 1.603865 0.7188828
UNC29893228 19 8.029054 1.629859 0.7188828#print("method == ehk")
#scanone.ehk <-qtl::scanone(cross.probs, pheno.col="ICI.vs.PBS" , model="binary", method="ehk")
#operm.ehk <- qtl::scanone(cross.probs, method = "ehk", pheno.col="ICI.vs.PBS", n.perm = 1000, perm.Xsp = TRUE, model="binary", verbose=FALSE)
#plot(operm.ehk)
#print(summary(operm.ehk, alpha=c(0.01, 0.05, 0.1)))
#plot(scanone.ehk, bandcol = "grey90",lty=1, cex=1, col = "steelblue")
#qtl::add.threshold(scanone.ehk, perms= operm.ehk, alpha=0.01, col = 'blue')
#qtl::add.threshold(scanone.ehk, perms= operm.ehk, alpha=0.05, col = 'red')
#qtl::add.threshold(scanone.ehk, perms= operm.ehk, alpha=0.1, col = 'purple')
#print(as.data.frame(summary(scanone.ehk)))
#print(as.data.frame(summary(scanone.ehk, perms=operm.ehk, alpha=0.05, pvalues=TRUE, format="allpeaks")))Multi-qtl (genoprobs, haley knott regression method)
gmObject of class cross2 (crosstype "bc")
Total individuals 138
No. genotyped individuals 138
No. phenotyped individuals 138
No. with both geno & pheno 138
No. phenotypes 1
No. covariates 7
No. phenotype covariates 0
No. chromosomes 20
Total markers 26548
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2374 2374 1623 1691 1568 1703 1464 1468 1685 974 1671 1134 1377 1411 876 751
17 18 19 X
330 770 920 384 detach("package:qtl2", unload=TRUE)
library(qtl)
cross <- qtl::read.cross("csv", file = "data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_peaks.csv",alleles=c("A","B")) --Read the following data:
138 individuals
26548 markers
3 phenotypes
--Cross type: bc cross <- qtl::jittermap(cross)
fitqtl.d1 = cross
#qc <- c(3, 4)
#qp <- c(29.03815, 69.1085558)
peaks[] <- lapply(peaks, as.character)
qc <- c(as.numeric(peaks$chr[1]), as.numeric(peaks$chr[2]),as.numeric(peaks$chr[3]))
qp <- c(as.numeric(peaks$pos[1]), as.numeric(peaks$pos[2]),as.numeric(peaks$chr[3]))
#itqtl.d1 <- subset(fitqtl.d1, chr=qc)
fitqtl.d1 <- qtl::calc.genoprob(fitqtl.d1)
qtl <- qtl::makeqtl(fitqtl.d1, qc, qp, what="prob")
# fit an additive QTL model
lod.add.1 <- qtl::fitqtl(fitqtl.d1, pheno.col="ICI.vs.PBS", qtl, formula=y~Q1+Q2+Q3,method="hk", model="binary", get.ests=FALSE)
print(summary(lod.add.1))
fitqtl summary
Method: Haley-Knott regression
Model: binary phenotype
Number of observations : 138
Full model result
----------------------------------
Model formula: y ~ Q1 + Q2 + Q3
df LOD %var Pvalue(Chi2)
Model 3 4.19339 13.05868 0.0002357288
Drop one QTL at a time ANOVA table:
----------------------------------
df LOD %var Pvalue(Chi2)
8@35.5 1 1.4419 4.286 0.00997 **
13@13.8 1 1.7960 5.370 0.00403 **
19@19.0 1 0.8833 2.601 0.04371 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1capture.output(summary(lod.add.1),file="data/ici.vs.pbs_fitqtl_peaks_additive_snpsqc_snpsqc_5.batches_peaks.txt", quote=F)
# fit model with 2 interacting QTLs interacting
lod.1 <- qtl::fitqtl(fitqtl.d1, pheno.col="ICI.vs.PBS", qtl, formula=y~Q1*Q2*Q3, method="hk", model="binary", get.ests=FALSE)
print(summary(lod.1))
fitqtl summary
Method: Haley-Knott regression
Model: binary phenotype
Number of observations : 138
Full model result
----------------------------------
Model formula: y ~ Q1 + Q2 + Q3 + Q1:Q2 + Q1:Q3 + Q2:Q3 + Q1:Q2:Q3
df LOD %var Pvalue(Chi2)
Model 7 5.544111 16.89052 0.0006105068
Drop one QTL at a time ANOVA table:
----------------------------------
df LOD %var Pvalue(Chi2)
8@35.5 4 2.0510 5.888 0.05088 .
13@13.8 4 3.1379 9.175 0.00599 **
19@19.0 4 1.7866 5.106 0.08359 .
8@35.5:13@13.8 2 0.6921 1.942 0.20318
8@35.5:19@19.0 2 0.4819 1.347 0.32969
13@13.8:19@19.0 2 0.9807 2.765 0.10454
8@35.5:13@13.8:19@19.0 1 0.4688 1.310 0.14174
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1capture.output(summary(lod.1),file="data/ici.vs.pbs_fitqtl_peaks_interacting_snpsqc_snpsqc_5.batches_peaks.txt", quote=F)
# fit the model including sex as an interacting covariate
Sex <- data.frame(Sex=qtl::pull.pheno(fitqtl.d1, "sex"))
# additive model
lod.sex.add.1 <- qtl::fitqtl(fitqtl.d1, pheno.col="ICI.vs.PBS", qtl, formula=y~Q1+Q2+Q3+Sex, cov=Sex, method="hk", model="binary", get.ests=FALSE)
print(summary(lod.sex.add.1))
fitqtl summary
Method: Haley-Knott regression
Model: binary phenotype
Number of observations : 138
Full model result
----------------------------------
Model formula: y ~ Q1 + Q2 + Q3 + Sex
df LOD %var Pvalue(Chi2)
Model 4 4.19339 13.05868 0.0006826371
Drop one QTL at a time ANOVA table:
----------------------------------
df LOD %var Pvalue(Chi2)
8@35.5 1 1.4419 4.286 0.00997 **
13@13.8 1 1.7960 5.370 0.00403 **
19@19.0 1 0.8833 2.601 0.04371 *
Sex 1 0.0000 0.000 1.00000
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1capture.output(summary(lod.sex.add.1),file="data/ici.vs.pbs_fitqtl_peaks_sex_additive_snpsqc_snpsqc_5.batches_peaks.txt", quote=F)
#interactive model
lod.sex.1 <- qtl::fitqtl(fitqtl.d1, pheno.col="ICI.vs.PBS", qtl, formula=y~Q1*Q2*Q3*Sex, cov=Sex, method="hk", model="binary", get.ests=FALSE)
print(summary(lod.sex.1))
fitqtl summary
Method: Haley-Knott regression
Model: binary phenotype
Number of observations : 138
Full model result
----------------------------------
Model formula: y ~ Q1 + Q2 + Q3 + Sex + Q1:Q2 + Q1:Q3 + Q1:Sex + Q2:Q3 + Q2:Sex
+ Q3:Sex + Q1:Q2:Q3 + Q1:Q2:Sex + Q1:Q3:Sex + Q2:Q3:Sex +
Q1:Q2:Q3:Sex
df LOD %var Pvalue(Chi2)
Model 15 5.544111 16.89052 0.04324422
Drop one QTL at a time ANOVA table:
----------------------------------
df LOD %var Pvalue(Chi2)
8@35.5 8 2.0510 5.888 0.3061
13@13.8 8 3.1379 9.175 0.0708 .
19@19.0 8 1.7866 5.106 0.4116
Sex 8 0.0000 0.000 1.0000
8@35.5:13@13.8 4 0.6921 1.942 0.5270
8@35.5:19@19.0 4 0.4819 1.347 0.6955
8@35.5:Sex 4 0.0000 0.000 1.0000
13@13.8:19@19.0 4 0.9807 2.765 0.3406
13@13.8:Sex 4 0.0000 0.000 1.0000
19@19.0:Sex 4 0.0000 0.000 1.0000
8@35.5:13@13.8:19@19.0 2 0.4688 1.310 0.3398
8@35.5:13@13.8:Sex 2 0.0000 0.000 1.0000
8@35.5:19@19.0:Sex 2 0.0000 0.000 1.0000
13@13.8:19@19.0:Sex 2 0.0000 0.000 1.0000
8@35.5:13@13.8:19@19.0:Sex 1 0.0000 0.000 1.0000
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1capture.output(summary(lod.sex.1),file="data/ici.vs.pbs_fitqtl_peaks_sex_interacting_snpsqc_snpsqc_5.batches_peaks.txt", quote=F)
#detach("package:qtl", unload=TRUE)
#library(qtl2)
R version 3.6.2 (2019-12-12)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Catalina 10.15.7
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
locale:
[1] en_AU.UTF-8/en_AU.UTF-8/en_AU.UTF-8/C/en_AU.UTF-8/en_AU.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] qtl_1.46-2 abind_1.4-5 reshape2_1.4.4 ggplot2_3.3.5
[5] tibble_3.1.2 psych_2.0.7 readxl_1.3.1 cluster_2.1.0
[9] dplyr_1.0.8 optparse_1.6.6 rhdf5_2.28.1 mclust_5.4.6
[13] tidyr_1.0.2 data.table_1.14.0 knitr_1.33 kableExtra_1.1.0
[17] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] httr_1.4.1 bit64_4.0.5 viridisLite_0.4.0 assertthat_0.2.1
[5] highr_0.9 blob_1.2.1 cellranger_1.1.0 yaml_2.2.1
[9] pillar_1.6.1 RSQLite_2.2.7 backports_1.2.1 lattice_0.20-38
[13] glue_1.4.2 digest_0.6.27 promises_1.1.0 rvest_0.3.5
[17] colorspace_2.0-2 htmltools_0.5.1.1 httpuv_1.5.2 plyr_1.8.6
[21] pkgconfig_2.0.3 purrr_0.3.4 scales_1.1.1 webshot_0.5.2
[25] getopt_1.20.3 later_1.0.0 git2r_0.26.1 generics_0.0.2
[29] ellipsis_0.3.2 cachem_1.0.5 withr_2.4.2 cli_3.0.0
[33] mnormt_1.5-7 magrittr_2.0.1 crayon_1.4.1 memoise_2.0.0
[37] evaluate_0.14 fs_1.4.1 fansi_0.5.0 nlme_3.1-142
[41] xml2_1.3.1 tools_3.6.2 hms_0.5.3 lifecycle_1.0.1
[45] stringr_1.4.0 Rhdf5lib_1.6.3 munsell_0.5.0 compiler_3.6.2
[49] rlang_1.0.2 grid_3.6.2 rstudioapi_0.13 rmarkdown_2.1
[53] gtable_0.3.0 DBI_1.1.1 R6_2.5.0 fastmap_1.1.0
[57] bit_4.0.4 utf8_1.2.1 rprojroot_1.3-2 readr_1.3.1
[61] stringi_1.7.2 parallel_3.6.2 Rcpp_1.0.7 vctrs_0.3.8
[65] tidyselect_1.1.2 xfun_0.24