QTL Analysis - binary [ICI vs PBS]
Belinda Cornes
2023-04-16
Last updated: 2023-04-16
Checks: 5 2
Knit directory: Serreze-T1D_Workflow/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230413) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /projects/serreze-lab/USERS/corneb/qtl2/workflowr/2023.04.13/Serreze-T1D_Workflow | . |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 03e83ae. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Untracked files:
Untracked: analysis/0.1.1_preparing.data_bqc_7.batches_myo.Rmd
Untracked: analysis/0.1_samples_batch_20230124.Rmd
Untracked: analysis/0.1_samples_batch_20230225.Rmd
Untracked: analysis/0.1_samples_batch_20230313.Rmd
Untracked: analysis/0.2_haplotype_comparison_bqc_7.batches_myo_minprob.Rmd
Untracked: analysis/2.1_sample_bqc_7.batches_myo.Rmd
Untracked: analysis/2.2.1_snp_qc_7.batches_myo.Rmd
Untracked: analysis/2.2.1_snp_qc_7.batches_myo_mis.Rmd
Untracked: analysis/2.4_preparing.data_aqc_7.batches_myo.Rmd
Untracked: analysis/2.4_preparing.data_aqc_7.batches_myo_mis.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_7.batches_myo.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici-myo-yes.vs.het-ici-myo-no_snpsqc_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici-myo-yes.vs.het-ici-myo-no_snpsqc_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici-myo-yes.vs.het-ici-myo-no_snpsqc_dis_no-x_updated_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici-myo-yes.vs.het-ici-myo-no_snpsqc_dis_no-x_updated_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici-myo-yes.vs.het-ici-myo-no_snpsqc_dis_no-x_updated_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici-myo-yes.vs.het-ici-myo-no_snpsqc_dis_no-x_updated_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici-myo-yes.vs.het-ici-myo-no_snpsqc_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici-myo-yes.vs.het-ici-myo-no_snpsqc_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici.vs.het-pbs_snpsqc_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici.vs.het-pbs_snpsqc_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici.vs.het-pbs_snpsqc_dis_no-x_updated_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici.vs.het-pbs_snpsqc_dis_no-x_updated_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici.vs.het-pbs_snpsqc_dis_no-x_updated_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici.vs.het-pbs_snpsqc_dis_no-x_updated_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici.vs.het-pbs_snpsqc_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici.vs.het-pbs_snpsqc_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-myo-yes.vs.ici-myo-no_snpsqc_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-myo-yes.vs.ici-myo-no_snpsqc_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-myo-yes.vs.ici-myo-no_snpsqc_dis_no-x_updated_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-myo-yes.vs.ici-myo-no_snpsqc_dis_no-x_updated_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-myo-yes.vs.ici-myo-no_snpsqc_dis_no-x_updated_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-myo-yes.vs.ici-myo-no_snpsqc_dis_no-x_updated_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-myo-yes.vs.ici-myo-no_snpsqc_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-myo-yes.vs.ici-myo-no_snpsqc_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-sick.vs.ici-eoi_snpsqc_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-sick.vs.ici-eoi_snpsqc_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-sick.vs.ici-eoi_snpsqc_dis_no-x_updated_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-sick.vs.ici-eoi_snpsqc_dis_no-x_updated_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-sick.vs.ici-eoi_snpsqc_dis_no-x_updated_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-sick.vs.ici-eoi_snpsqc_dis_no-x_updated_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-sick.vs.ici-eoi_snpsqc_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-sick.vs.ici-eoi_snpsqc_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_myo-yes.vs.myo-no_snpsqc_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_myo-yes.vs.myo-no_snpsqc_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_myo-yes.vs.myo-no_snpsqc_dis_no-x_updated_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_myo-yes.vs.myo-no_snpsqc_dis_no-x_updated_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_myo-yes.vs.myo-no_snpsqc_dis_no-x_updated_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_myo-yes.vs.myo-no_snpsqc_dis_no-x_updated_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_myo-yes.vs.myo-no_snpsqc_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_myo-yes.vs.myo-no_snpsqc_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_pbs-myo-yes.vs.pbs-myo-no_snpsqc_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_pbs-myo-yes.vs.pbs-myo-no_snpsqc_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_pbs-myo-yes.vs.pbs-myo-no_snpsqc_dis_no-x_updated_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_pbs-myo-yes.vs.pbs-myo-no_snpsqc_dis_no-x_updated_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_pbs-myo-yes.vs.pbs-myo-no_snpsqc_dis_no-x_updated_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_pbs-myo-yes.vs.pbs-myo-no_snpsqc_dis_no-x_updated_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_pbs-myo-yes.vs.pbs-myo-no_snpsqc_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_pbs-myo-yes.vs.pbs-myo-no_snpsqc_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_het-ici-myo-yes.vs.het-ici-myo-no_7.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_het-ici-myo-yes.vs.het-ici-myo-no_7.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_het-ici.vs.het-pbs_7.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_het-ici.vs.het-pbs_7.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_ici-myo-yes.vs.ici-myo-no_7.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_ici-myo-yes.vs.ici-myo-no_7.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_ici-sick.vs.ici-eoi_7.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_ici-sick.vs.ici-eoi_7.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.pbs_7.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.pbs_7.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_myo-yes.vs.myo-no_7.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_myo-yes.vs.myo-no_7.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_pbs-myo-yes.vs.pbs-myo-no_7.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_pbs-myo-yes.vs.pbs-myo-no_7.batches_myo_mis.Rmd
Untracked: analysis/index_7.batches_myo.Rmd
Untracked: analysis/power.analysis_het-ici-myo-yes.vs.het-ici-myo-no_7.batches_myo.Rmd
Untracked: analysis/power.analysis_het-ici-myo-yes.vs.het-ici-myo-no_7.batches_myo_mis.Rmd
Untracked: analysis/power.analysis_het-ici.vs.het-pbs_7.batches_myo.Rmd
Untracked: analysis/power.analysis_het-ici.vs.het-pbs_7.batches_myo_mis.Rmd
Untracked: analysis/power.analysis_ici-myo-yes.vs.ici-myo-no_7.batches_myo.Rmd
Untracked: analysis/power.analysis_ici-myo-yes.vs.ici-myo-no_7.batches_myo_mis.Rmd
Untracked: analysis/power.analysis_ici-sick.vs.ici-eoi_7.batches_myo.Rmd
Untracked: analysis/power.analysis_ici-sick.vs.ici-eoi_7.batches_myo_mis.Rmd
Untracked: analysis/power.analysis_ici.vs.pbs_7.batches_myo.Rmd
Untracked: analysis/power.analysis_ici.vs.pbs_7batches_myo_mis.Rmd
Untracked: analysis/power.analysis_myo-yes.vs.myo-no_7.batches_myo.Rmd
Untracked: analysis/power.analysis_myo-yes.vs.myo-no_7.batches_myo_mis.Rmd
Untracked: analysis/power.analysis_pbs-myo-yes.vs.pbs-myo-no_7.batches_myo.Rmd
Untracked: analysis/power.analysis_pbs-myo-yes.vs.pbs-myo-no_7.batches_myo_mis.Rmd
Untracked: code/cc_variants.sqlite
Untracked: code/mouse_genes.sqlite
Untracked: code/mouse_genes_mgi.sqlite
Untracked: data/GM_covar_7.batches_myo.csv
Untracked: data/bad_markers_all_7.batches_myo.RData
Untracked: data/covar_corrected.cleaned_het-ici-myo-yes.vs.het-ici-myo-no_7.batches_myo.csv
Untracked: data/covar_corrected.cleaned_het-ici-myo-yes.vs.het-ici-myo-no_7.batches_myo_mis.csv
Untracked: data/covar_corrected.cleaned_het-ici.vs.het-pbs_7.batches_myo.csv
Untracked: data/covar_corrected.cleaned_het-ici.vs.het-pbs_7.batches_myo_mis.csv
Untracked: data/covar_corrected.cleaned_ici-myo-yes.vs.ici-myo-no_7.batches_myo.csv
Untracked: data/covar_corrected.cleaned_ici-myo-yes.vs.ici-myo-no_7.batches_myo_mis.csv
Untracked: data/covar_corrected.cleaned_ici-sick.vs.ici-eoi_7.batches_myo.csv
Untracked: data/covar_corrected.cleaned_ici-sick.vs.ici-eoi_7.batches_myo_mis.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_7.batches_myo.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_7.batches_myo_mis.csv
Untracked: data/covar_corrected.cleaned_myo-yes.vs.myo-no_7.batches_myo.csv
Untracked: data/covar_corrected.cleaned_myo-yes.vs.myo-no_7.batches_myo_mis.csv
Untracked: data/covar_corrected.cleaned_pbs-myo-yes.vs.pbs-myo-no_7.batches_myo.csv
Untracked: data/covar_corrected.cleaned_pbs-myo-yes.vs.pbs-myo-no_7.batches_myo_mis.csv
Untracked: data/covar_corrected_het-ici-myo-yes.vs.het-ici-myo-no_7.batches_myo.csv
Untracked: data/covar_corrected_het-ici-myo-yes.vs.het-ici-myo-no_7.batches_myo_mis.csv
Untracked: data/covar_corrected_het-ici.vs.het-pbs_7.batches_myo.csv
Untracked: data/covar_corrected_het-ici.vs.het-pbs_7.batches_myo_mis.csv
Untracked: data/covar_corrected_ici-myo-yes.vs.ici-myo-no_7.batches_myo.csv
Untracked: data/covar_corrected_ici-myo-yes.vs.ici-myo-no_7.batches_myo_mis.csv
Untracked: data/covar_corrected_ici-sick.vs.ici-eoi_7.batches_myo.csv
Untracked: data/covar_corrected_ici-sick.vs.ici-eoi_7.batches_myo_mis.csv
Untracked: data/covar_corrected_ici.vs.pbs_7.batches_myo.csv
Untracked: data/covar_corrected_ici.vs.pbs_7.batches_myo_mis.csv
Untracked: data/covar_corrected_myo-yes.vs.myo-no_7.batches_myo.csv
Untracked: data/covar_corrected_myo-yes.vs.myo-no_7.batches_myo_mis.csv
Untracked: data/covar_corrected_pbs-myo-yes.vs.pbs-myo-no_7.batches_myo.csv
Untracked: data/covar_corrected_pbs-myo-yes.vs.pbs-myo-no_7.batches_myo_mis.csv
Untracked: data/e_7.batches_myo.RData
Untracked: data/e_snpg_samqc_7.batches_myo.RData
Untracked: data/errors_ind_7.batches_myo.RData
Untracked: data/genetic_map_7.batches_myo.csv
Untracked: data/genotype_errors_marker_7.batches_myo.RData
Untracked: data/genotype_freq_marker_7.batches_myo.RData
Untracked: data/gm_allqc_7.batches_myo.RData
Untracked: data/gm_allqc_7.batches_myo_mis.RData
Untracked: data/gm_samqc_7.batches_myo.RData
Untracked: data/gm_serreze_7.batches_myo.RData
Untracked: data/gm_serreze_bc_7.batches_myo.RData
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_gm_qtl_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo_mis.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/het-ici.vs.het-pbs_gm_qtl_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/het-ici.vs.het-pbs_gm_qtl_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/het-ici.vs.het-pbs_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/het-ici.vs.het-pbs_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/het-ici.vs.het-pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/het-ici.vs.het-pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/het-ici.vs.het-pbs_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/het-ici.vs.het-pbs_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/het-ici.vs.het-pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/het-ici.vs.het-pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/het-ici.vs.het-pbs_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/het-ici.vs.het-pbs_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/het-ici.vs.het-pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/het-ici.vs.het-pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/het-ici.vs.het-pbs_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo.csv
Untracked: data/het-ici.vs.het-pbs_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo_mis.csv
Untracked: data/het-ici.vs.het-pbs_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/het-ici.vs.het-pbs_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_gm_qtl_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_gm_qtl_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo_mis.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-10_peak.marker-UNC18343990_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-10_peak.marker-UNC18343990_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-11_peak.marker-UNC20070077_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-11_peak.marker-UNC20070077_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-12_peak.marker-UNC21652584_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-12_peak.marker-UNC21652584_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-13_peak.marker-UNCHS036579_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-13_peak.marker-UNCHS036579_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-14_peak.marker-UNCHS037782_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-14_peak.marker-UNCHS037782_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-15_peak.marker-UNC26069905_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-15_peak.marker-UNCHS041223_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-16_peak.marker-JAX00070117_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-16_peak.marker-JAX00070117_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-17_peak.marker-UNC28542319_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-17_peak.marker-UNCJPD006670_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-18_peak.marker-UNC28776739_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-18_peak.marker-UNCHS045594_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-19_peak.marker-UNC30414168_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-19_peak.marker-UNC30426276_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-1_peak.marker-UNC2031646_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-1_peak.marker-UNCHS003700_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-2_peak.marker-ICR5131_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-2_peak.marker-UNCHS006420_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-3_peak.marker-UNC5667757_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-3_peak.marker-UNC5667757_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-4_peak.marker-UNC6759992_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-4_peak.marker-UNC6765178_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-5_peak.marker-UNC9889957_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-5_peak.marker-UNC9900273_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-6_peak.marker-UNC10800126_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-6_peak.marker-UNC10832076_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-7_peak.marker-UNCHS020903_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-7_peak.marker-UNCHS021163_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-8_peak.marker-UNC15471847_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-8_peak.marker-UNC15548888_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-9_peak.marker-UNC16231874_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-9_peak.marker-UNC16231874_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-X_peak.marker-UNCHS049472_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-X_peak.marker-UNCHS049472_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-10_peak.marker-UNC18343990_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-10_peak.marker-UNC18343990_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-11_peak.marker-UNC20070077_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-11_peak.marker-UNC20070077_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-12_peak.marker-UNC21652584_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-12_peak.marker-UNC21652584_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-13_peak.marker-UNCHS036579_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-13_peak.marker-UNCHS036579_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-14_peak.marker-UNCHS037782_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-14_peak.marker-UNCHS037782_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-15_peak.marker-UNC26069905_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-15_peak.marker-UNCHS041223_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-16_peak.marker-JAX00070117_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-16_peak.marker-JAX00070117_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-17_peak.marker-UNC28542319_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-17_peak.marker-UNCJPD006670_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-18_peak.marker-UNC28776739_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-18_peak.marker-UNCHS045594_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-19_peak.marker-UNC30414168_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-19_peak.marker-UNC30426276_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-1_peak.marker-UNC2031646_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-1_peak.marker-UNCHS003700_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-2_peak.marker-ICR5131_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-2_peak.marker-UNCHS006420_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-3_peak.marker-UNC5667757_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-3_peak.marker-UNC5667757_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-4_peak.marker-UNC6759992_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-4_peak.marker-UNC6765178_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-5_peak.marker-UNC9889957_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-5_peak.marker-UNC9900273_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-6_peak.marker-UNC10800126_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-6_peak.marker-UNC10832076_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-7_peak.marker-UNCHS020903_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-7_peak.marker-UNCHS021163_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-8_peak.marker-UNC15471847_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-8_peak.marker-UNC15548888_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-9_peak.marker-UNC16231874_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-9_peak.marker-UNC16231874_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-X_peak.marker-UNCHS049472_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-X_peak.marker-UNCHS049472_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_gm_qtl_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_gm_qtl_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-7_peak.marker-UNCHS020711_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-7_peak.marker-UNCHS020711_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici.vs.pbs_genes_chr-7_peak.marker-UNCHS020711_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici.vs.pbs_genes_chr-7_peak.marker-UNCHS020711_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo_mis.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_age.of.onset_het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_age.of.onset_het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_age.of.onset_het-ici.vs.het-pbs_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_het-ici.vs.het-pbs_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_age.of.onset_het-ici.vs.het-pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_het-ici.vs.het-pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_age.of.onset_ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_age.of.onset_ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_age.of.onset_ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_age.of.onset_ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_age.of.onset_ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_myo-yes.vs.myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_myo-yes.vs.myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_age.of.onset_myo-yes.vs.myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_myo-yes.vs.myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_age.of.onset_pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_age.of.onset_pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_group_het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_group_het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_group_het-ici.vs.het-pbs_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_het-ici.vs.het-pbs_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_group_het-ici.vs.het-pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_het-ici.vs.het-pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_group_ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_group_ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_group_ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_group_ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_group_ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_myo-yes.vs.myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_myo-yes.vs.myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_group_myo-yes.vs.myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_myo-yes.vs.myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_group_pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_group_pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/myo-yes.vs.myo-no_gm_qtl_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/myo-yes.vs.myo-no_gm_qtl_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/myo-yes.vs.myo-no_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/myo-yes.vs.myo-no_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/myo-yes.vs.myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/myo-yes.vs.myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/myo-yes.vs.myo-no_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/myo-yes.vs.myo-no_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/myo-yes.vs.myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/myo-yes.vs.myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/myo-yes.vs.myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/myo-yes.vs.myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/myo-yes.vs.myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/myo-yes.vs.myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/myo-yes.vs.myo-no_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo.csv
Untracked: data/myo-yes.vs.myo-no_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo_mis.csv
Untracked: data/myo-yes.vs.myo-no_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/myo-yes.vs.myo-no_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_gm_qtl_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_gm_qtl_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo_mis.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/percent_missing_id_7.batches_myo.RData
Untracked: data/percent_missing_marker_7.batches_myo.RData
Untracked: data/pheno_7.batches_myo.csv
Untracked: data/physical_map_7.batches_myo.csv
Untracked: data/probs_36state_bc_7.batches_myo.rds
Untracked: data/probs_8state_bc_7.batches_myo.rds
Untracked: data/qc_info_bad_sample_7.batches_myo.RData
Untracked: data/sample_geno_AHB_7.batches_myo.csv
Untracked: data/sample_geno_AHB_7.batches_myo_orig.id.csv
Untracked: data/sample_geno_bc_7.batches_myo.csv
Untracked: data/sample_geno_bc_7.batches_myo_orig.id.csv
Untracked: data/serreze_probs_7.batches_myo.rds
Untracked: data/serreze_probs_allqc_7.batches_myo.rds
Untracked: data/serreze_probs_allqc_7.batches_myo_mis.rds
Untracked: data/summary.cg_7.batches_myo.RData
Untracked: data/summary.counts_EOI_ici.sick_vs_ici.eoi
Untracked: data/summary.counts_SICK_ici.sick_vs_ici.eoi
Untracked: data/summary.frequency_EOI_ici.sick_vs_ici.eoi
Untracked: data/summary.frequency_SICK_ici.sick_vs_ici.eoi
Unstaged changes:
Modified: analysis/_site.yml
Modified: analysis/index.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with wflow_publish() to start tracking its development.
Data Information
- Myocarditis Status: Yes
- Murine MHC KO Status: Homo
- Drug Treatment(s): ICI, PBS
- Clinical Phenotype(s): Sick, EOI
- Covariates (Myocarditis Score, Age of Clinical Phenotype – if Null, use 40 age at which surviving mice were end of incidence without clinical symptoms, sex)
Loading Data
We will load the data and subset indivials out that are in the groups of interest.
load("data/gm_allqc_7.batches_myo.RData")
#gm_allqc
gm=gm_allqc
gmObject of class cross2 (crosstype "bc")
Total individuals 357
No. genotyped individuals 357
No. phenotyped individuals 357
No. with both geno & pheno 357
No. phenotypes 1
No. covariates 11
No. phenotype covariates 0
No. chromosomes 20
Total markers 32660
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2482 2412 1742 1772 1648 1842 1541 1517 1769 1092 1754 1230 1441 1505 1128 842
17 18 19 X
655 820 945 4523 #pr <- readRDS("data/serreze_probs_allqc.rds")
#pr <- readRDS("data/serreze_probs.rds")
##extracting animals with ici and pbs group status
#miceinfo <- covars[gm$covar$group == "PBS" | gm$covar$group == "ICI",]
#table(miceinfo$group)
#mice.ids <- rownames(miceinfo)
#gm <- gm[mice.ids]
#gm
#table(gm$covar$group)
#gm$covar$ici_vs_pbs <- ifelse(gm$covar$group == "PBS", 0, 1)
#gm.full <- gm
covars <- read_csv("data/covar_corrected_ici.vs.pbs_7.batches_myo.csv")
#removing any missing info
#covars <- subset(covars, covars$ici_vs_pbs!='')
nrow(covars)[1] 212table(covars$"Myocarditis Status")
YES
212 table(covars$"Murine MHC KO Status")
HOM
212 table(covars$"Drug Treatment")
ICI PBS
133 79 table(covars$"clinical pheno")
EOI SICK
74 138 #keeping only informative mice
gm <- gm[covars$Mouse.ID]
gmObject of class cross2 (crosstype "bc")
Total individuals 212
No. genotyped individuals 212
No. phenotyped individuals 212
No. with both geno & pheno 212
No. phenotypes 1
No. covariates 11
No. phenotype covariates 0
No. chromosomes 20
Total markers 32660
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2482 2412 1742 1772 1648 1842 1541 1517 1769 1092 1754 1230 1441 1505 1128 842
17 18 19 X
655 820 945 4523 table(gm$covar$"Myocarditis Status")
YES
212 table(gm$covar$"Murine MHC KO Status")
HOM
212 table(gm$covar$"Drug Treatment")
ICI PBS
133 79 table(gm$covar$"clinical pheno")
EOI SICK
74 138 #pr.qc.ids <- pr
#for (i in 1:20){pr.qc.ids[[i]] = pr.qc.ids[[i]][covars$Mouse.ID,,]}
##removing problmetic marker
#gm <- drop_markers(gm, "UNCHS013106")
##dropping monomorphic markers within the dataset
g <- do.call("cbind", gm$geno)
gf_mar <- t(apply(g, 2, function(a) table(factor(a, 1:2))/sum(a != 0)))
#gn_mar <- t(apply(g, 2, function(a) table(factor(a, 1:2))))
gf_mar <- gf_mar[gf_mar[,2] != "NaN",]
count <- rowSums(gf_mar <=0.05)
low_freq_df <- merge(as.data.frame(gf_mar),as.data.frame(count), by="row.names",all=T)
low_freq_df[is.na(low_freq_df)] <- ''
low_freq_df <- low_freq_df[low_freq_df$count == 1,]
rownames(low_freq_df) <- low_freq_df$Row.names
low_freq <- find_markerpos(gm, rownames(low_freq_df))
low_freq$id <- rownames(low_freq)
nrow(low_freq)[1] 6198low_freq_bad <- merge(low_freq,low_freq_df, by="row.names",all=T)
names(low_freq_bad)[1] <- c("marker")
gf_mar <- gf_mar[gf_mar[,2] != "NaN",]
MAF <- apply(gf_mar, 1, function(x) min(x))
MAF <- as.data.frame(MAF)
MAF$index <- 1:nrow(gf_mar)
gf_mar_maf <- merge(gf_mar,as.data.frame(MAF), by="row.names")
gf_mar_maf <- gf_mar_maf[order(gf_mar_maf$index),]
gfmar <- NULL
gfmar$gfmar_mar_0 <- sum(gf_mar_maf$MAF==0)
gfmar$gfmar_mar_1 <- sum(gf_mar_maf$MAF< 0.01)
gfmar$gfmar_mar_5 <- sum(gf_mar_maf$MAF< 0.05)
gfmar$gfmar_mar_10 <- sum(gf_mar_maf$MAF< 0.10)
gfmar$gfmar_mar_15 <- sum(gf_mar_maf$MAF< 0.15)
gfmar$gfmar_mar_25 <- sum(gf_mar_maf$MAF< 0.25)
gfmar$gfmar_mar_50 <- sum(gf_mar_maf$MAF< 0.50)
gfmar$total_snps <- nrow(as.data.frame(gf_mar_maf))
gfmar <- t(as.data.frame(gfmar))
gfmar <- as.data.frame(gfmar)
gfmar$count <- gfmar$V1
gfmar[c(2)] %>%
kable(escape = F,align = c("ccccccccc"),linesep ="\\hline") %>%
kable_styling(full_width = F) %>%
kable_styling("striped", full_width = F) %>%
row_spec(8 ,bold=T,color= "white",background = "black")| count | |
|---|---|
| gfmar_mar_0 | 3976 |
| gfmar_mar_1 | 4421 |
| gfmar_mar_5 | 6196 |
| gfmar_mar_10 | 6519 |
| gfmar_mar_15 | 6544 |
| gfmar_mar_25 | 6628 |
| gfmar_mar_50 | 31794 |
| total_snps | 32660 |
gm_qc <- drop_markers(gm, low_freq_bad$marker)
gm_qc <- drop_nullmarkers(gm_qc)
gm_qcObject of class cross2 (crosstype "bc")
Total individuals 212
No. genotyped individuals 212
No. phenotyped individuals 212
No. with both geno & pheno 212
No. phenotypes 1
No. covariates 11
No. phenotype covariates 0
No. chromosomes 20
Total markers 26462
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2324 2273 1597 1657 1517 1684 1447 1443 1664 958 1657 1120 1366 1407 1035 744
17 18 19 X
560 741 896 372 ## dropping disproportionate markers
dismark <- read.csv("data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo.csv")
nrow(dismark)[1] 26462names(dismark)[1] <- c("marker")
dismark <- dismark[!dismark$Include,]
nrow(dismark)[1] 14447gm_qc_dis <- drop_markers(gm_qc, dismark$marker)
gm_qc_dis <- drop_nullmarkers(gm_qc_dis)
gm = gm_qc_dis
gmObject of class cross2 (crosstype "bc")
Total individuals 212
No. genotyped individuals 212
No. phenotyped individuals 212
No. with both geno & pheno 212
No. phenotypes 1
No. covariates 11
No. phenotype covariates 0
No. chromosomes 20
Total markers 12015
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
1422 548 642 18 991 582 862 1344 1228 467 966 262 715 363 600 410
17 18 19 X
109 162 314 10 markers <- marker_names(gm)
gmapdf <- read.csv("data/genetic_map_7.batches_myo.csv")
pmapdf <- read.csv("data/physical_map_7.batches_myo.csv")
#mapdf <- merge(gmapdf,pmapdf, by=c("marker","chr"), all=T)
#rownames(mapdf) <- mapdf$marker
#mapdf <- mapdf[markers,]
#names(mapdf) <- c('marker','chr','gmapdf','pmapdf')
#mapdfnd <- mapdf[!duplicated(mapdf[c(2:3)]),]
pr.qc <- calc_genoprob(gm)
colnames(covars) <- gsub(" ", ".", colnames(covars))Genome-wide scan
For each of the phenotype analyzed, permutations were used for each model to obtain genome-wide LOD significance threshold for p < 0.01, p < 0.05, p < 0.10, respectively, separately for X and automsomes (A).
The table shows the estimated significance thresholds from permutation test.
We also looked at the kinship to see how correlated each sample is. Kinship values between pairs of samples range between 0 (no relationship) and 1.0 (completely identical). The darker the colour the more indentical the pairs are.
#Xcovar <- get_x_covar(gm)
addcovar = model.matrix(~age.of.onset+Histology.Score, data = covars)[,-1]
covars$ici_vs_pbs= as.numeric(covars$ici_vs_pbs)
kinship <- calc_kinship(pr.qc)
heatmap(kinship)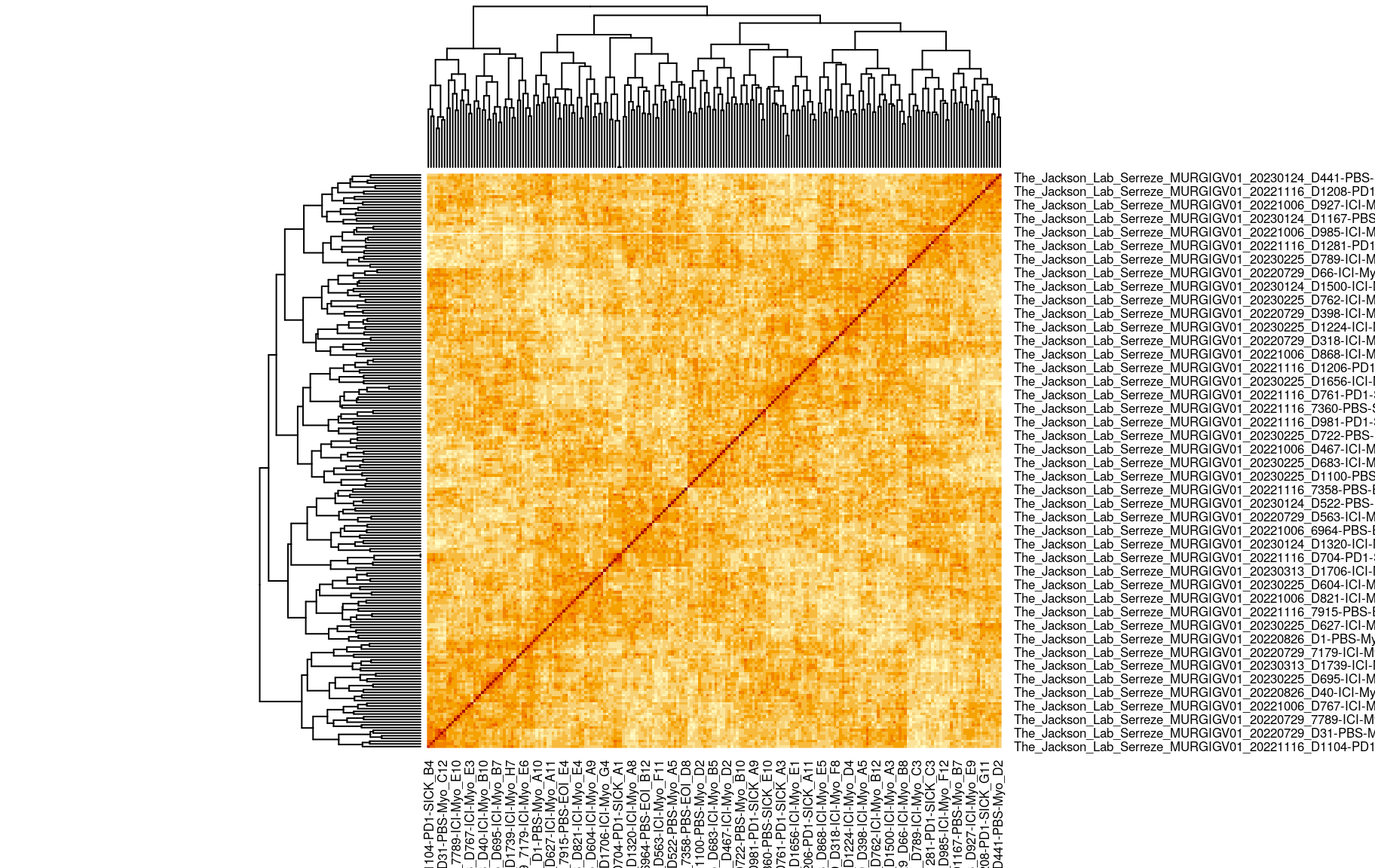
operm <- scan1perm(pr.qc, covars["ici_vs_pbs"], model="binary", addcovar=addcovar, n_perm=1000, perm_Xsp=TRUE, chr_lengths=chr_lengths(gm$gmap))
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
names(summary_table) <- c("autosomes","X")
summary_table$significance.level <- rownames(summary_table)
rownames(summary_table) <- NULL
summary_table[c(3,1:2)] %>%
kable(escape = F,align = c("ccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| significance.level | autosomes | X |
|---|---|---|
| 0.01 | 4.077915 | 4.307024 |
| 0.05 | 3.351758 | 3.754614 |
| 0.1 | 2.978898 | 3.403754 |
The figures below show QTL maps for each phenotype
#out <- scan1(pr.qc, covars["ici_vs_pbs"], Xcovar=Xcovar, model="binary")
out <- scan1(pr.qc, covars["ici_vs_pbs"], model="binary",addcovar=addcovar)
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
plot_lod<-function(out,map){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
##par(mar=c(5.1, 6.1, 1.1, 1.1))
#ymx <- 11 # overall maximum LOD score
#plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
##legend("topright", lwd=2, colnames(out)[i], bg="gray90")
#title(main = paste0(colnames(out)[i], " [positions in cM] \n(using same scale as eoi vs ici for easier comparison)"))
#add_threshold(map, summary(operm, alpha=0.1), col = 'purple')
#add_threshold(map, summary(operm, alpha=0.05), col = 'red')
#add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
##for (j in 1: dim(summary_table)[1]){
## abline(h=summary_table[j, i],col="red")
## text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
##}
##dev.off()
}
}
plot_lod(out,gm$gmap)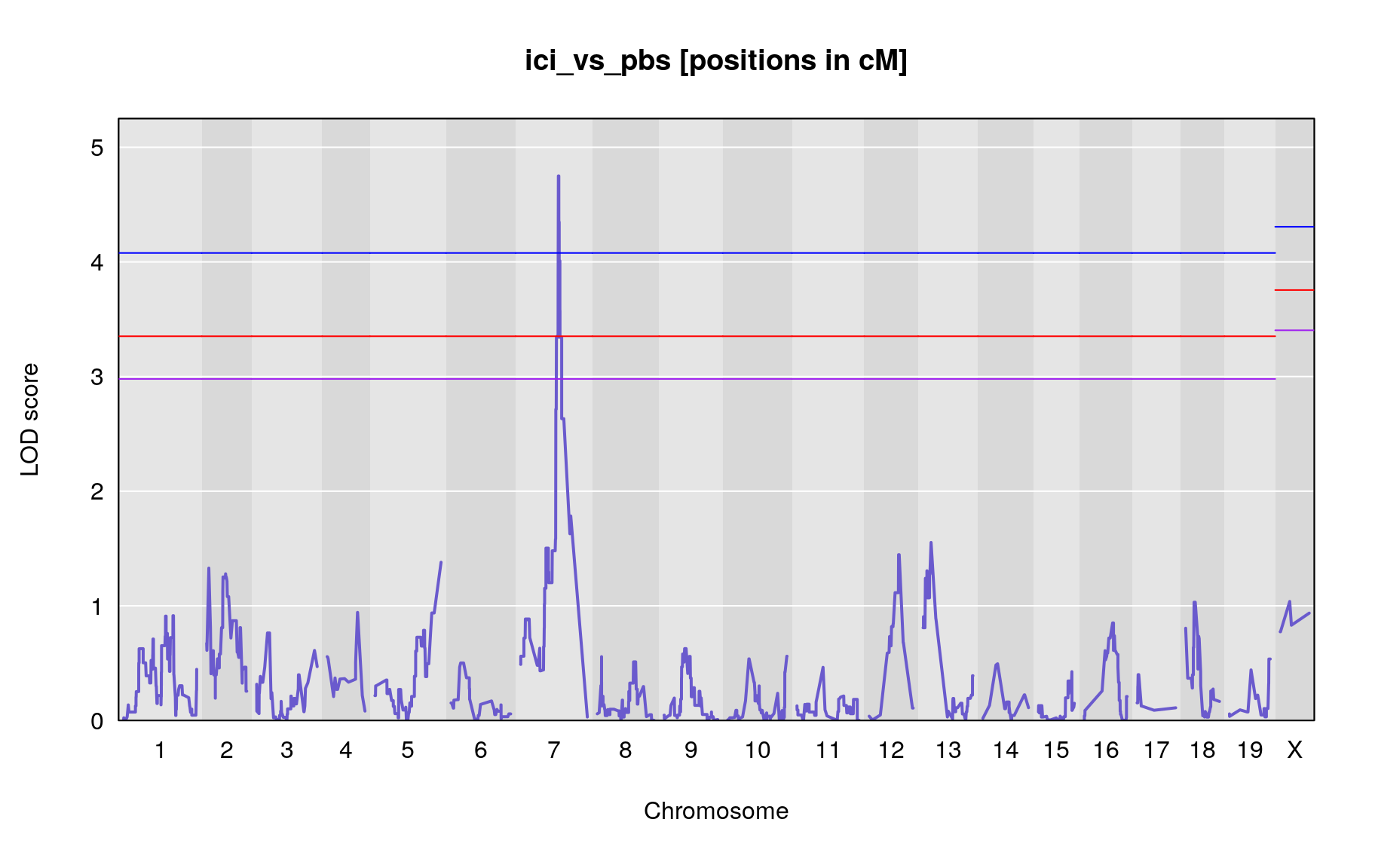
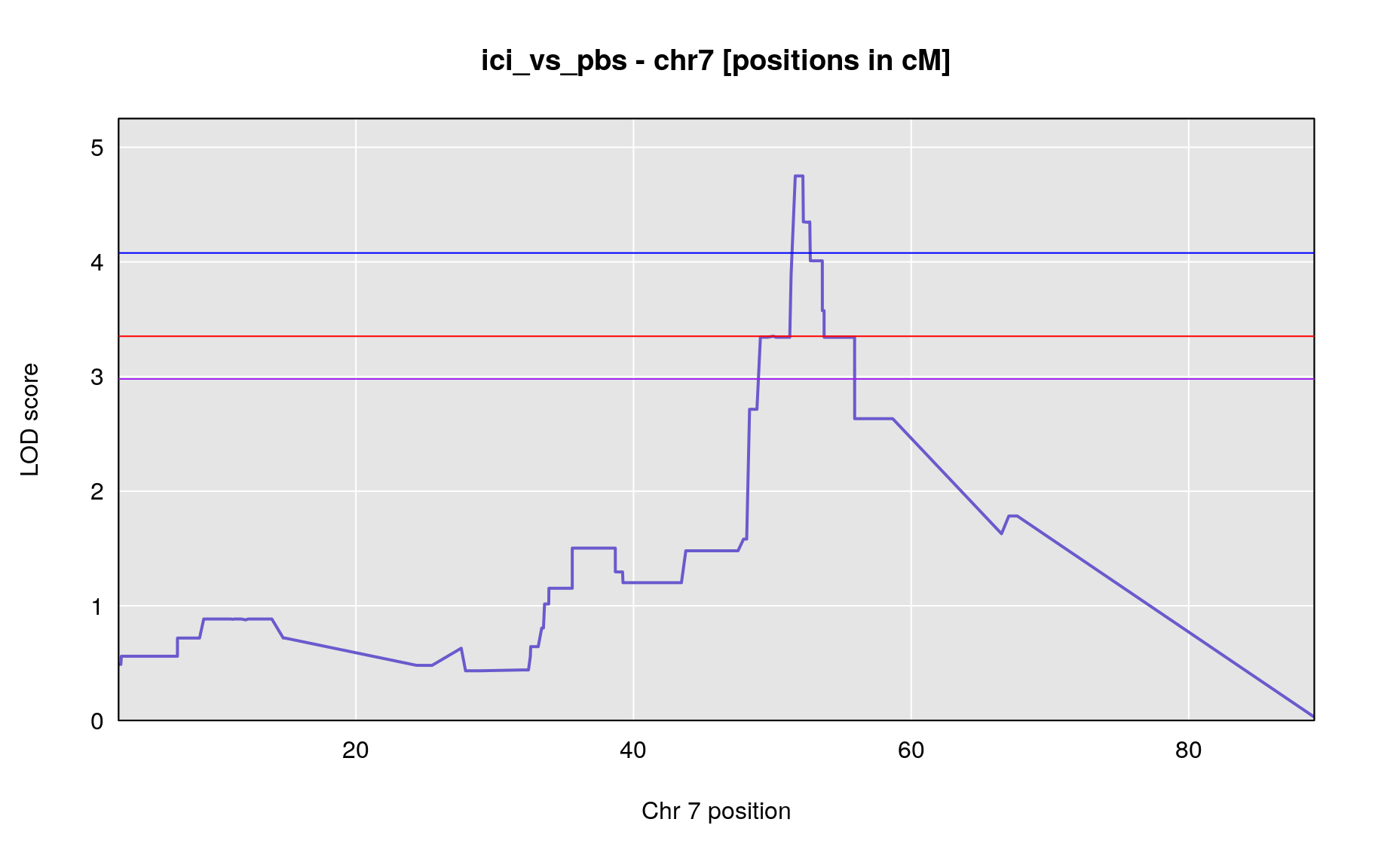
LOD peaks
The table below shows QTL peaks associated with the phenotype. We use the 95% threshold from the permutations to find peaks.
Centimorgan (cM)
peaks <- find_peaks(out, gm$gmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) >0){
peaks$marker <- find_marker(gm$gmap, chr=peaks$chr,pos=peaks$pos)
names(peaks)[2] <- c("phenotype")
peaks <- peaks[-1]
rownames(peaks) <- NULL
print(kable(peaks, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
#plot only peak chromosomes
plot_lod_chr<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
#ymx <- 11
#plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
##legend("topright", lwd=2, colnames(out)[i], bg="gray90")
#title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in cM]\n(using same scale as eoi vs. ici for easier comparison)"))
#add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
#add_threshold(map, summary(operm, alpha=0.05), col = 'red')
#add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
for(i in unique(peaks$chr)){
#for (i in 1:nrow(peaks)){
#plot_lod_chr(out,gm$gmap, peaks$chr[i])
plot_lod_chr(out,gm$gmap, i)
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}| phenotype | chr | pos | lod | ci_lo | ci_hi | marker |
|---|---|---|---|---|---|---|
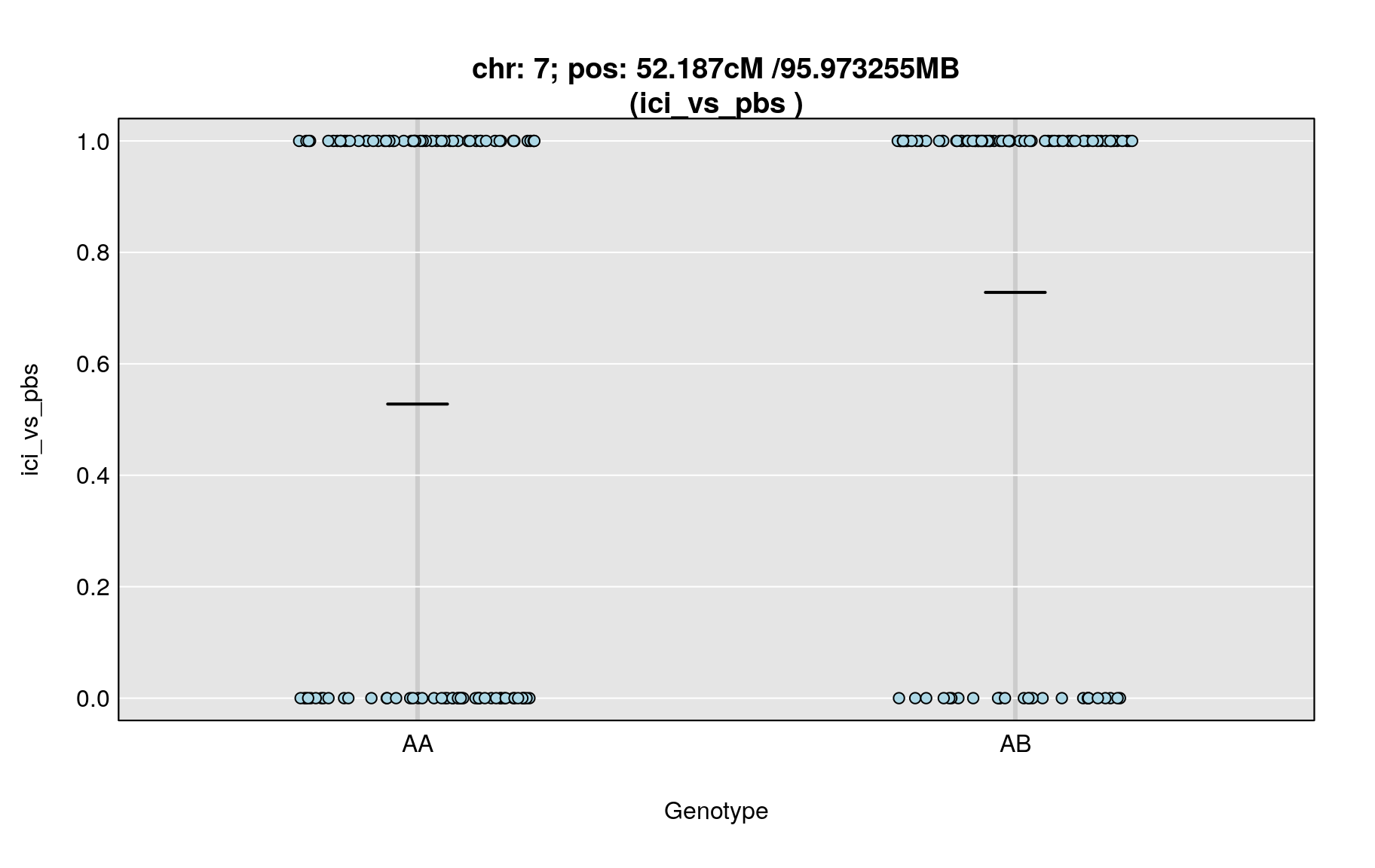
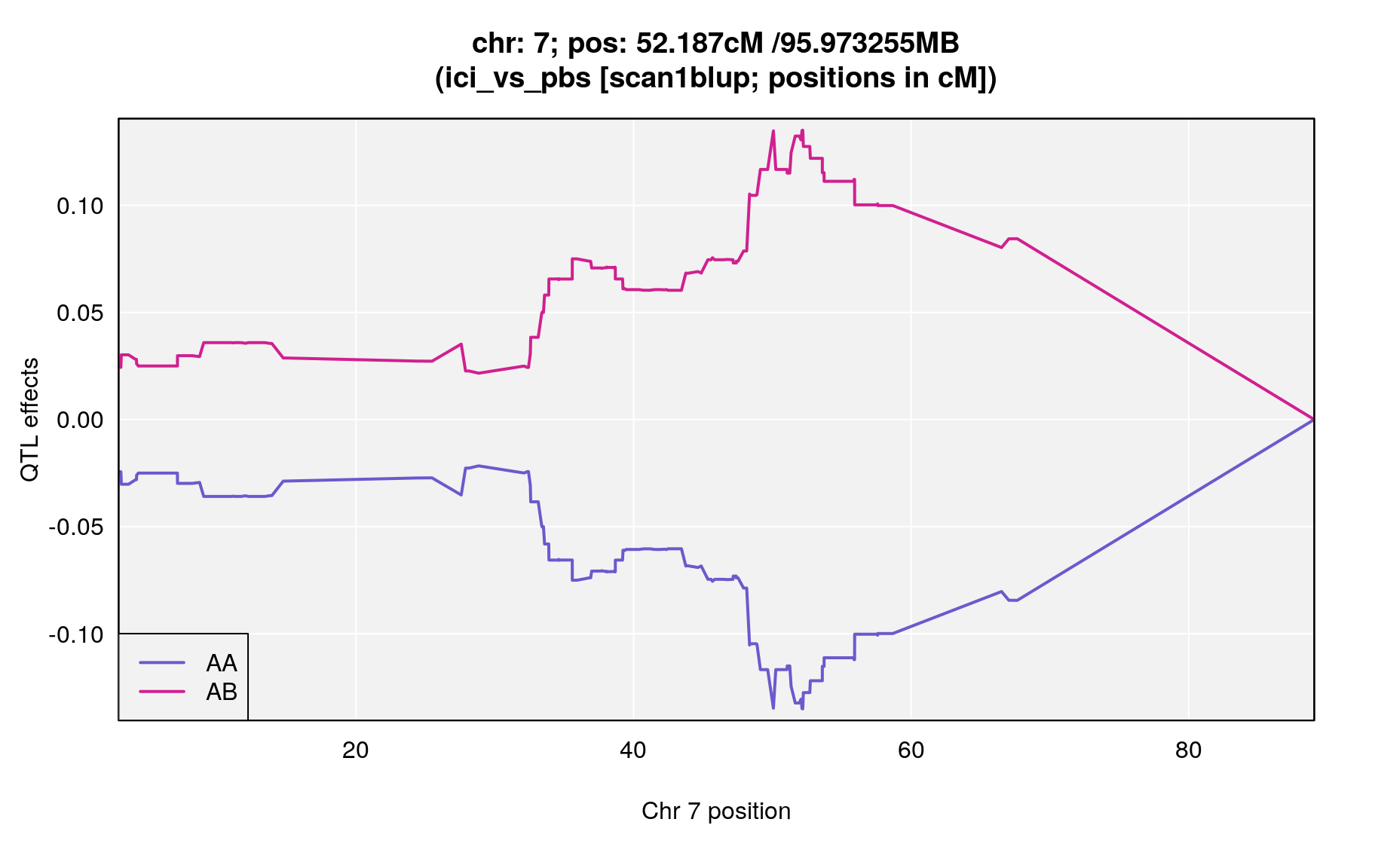
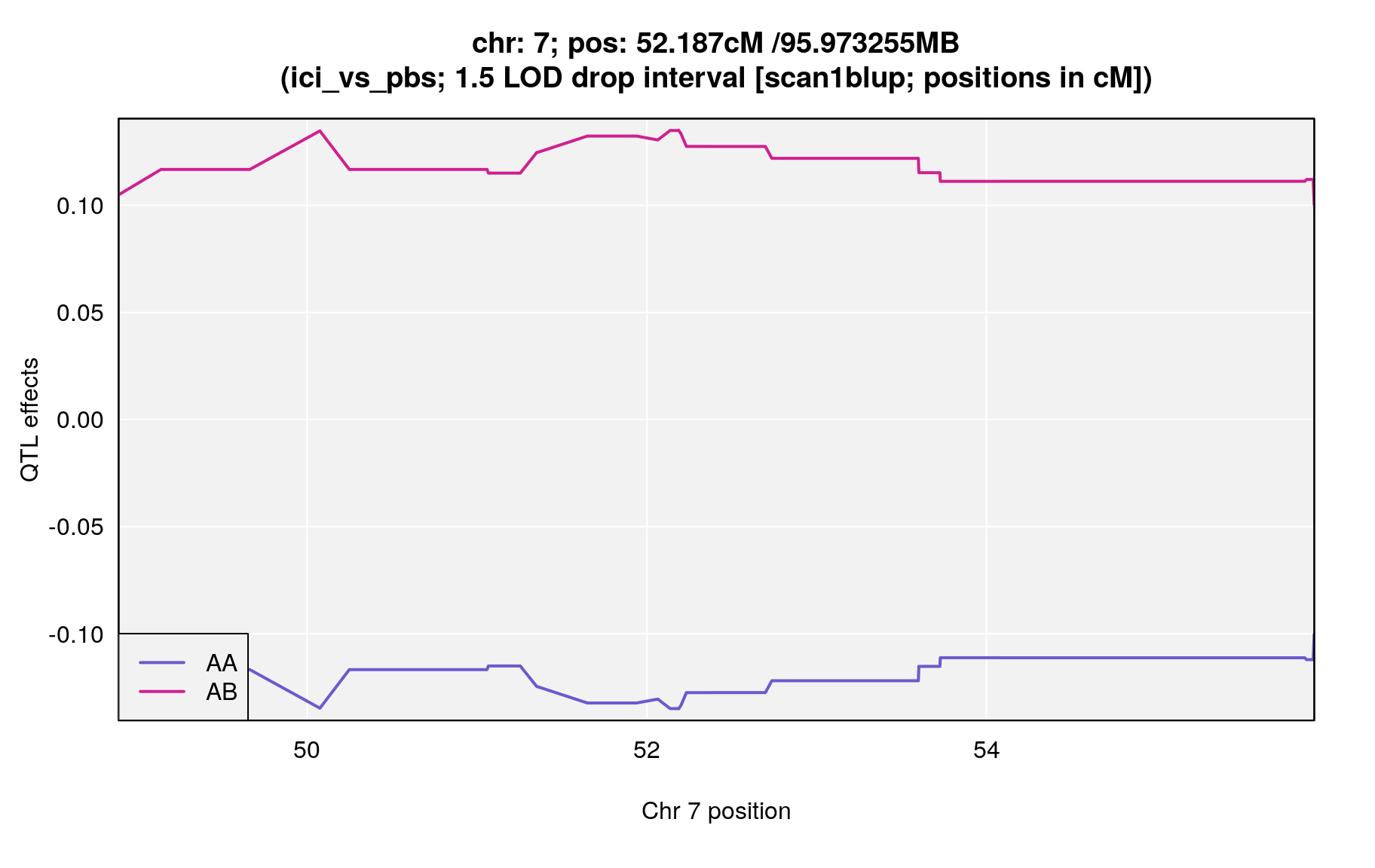
| ici_vs_pbs | 7 | 52.187 | 4.750019 | 48.89 | 55.93 | UNCHS020711 |
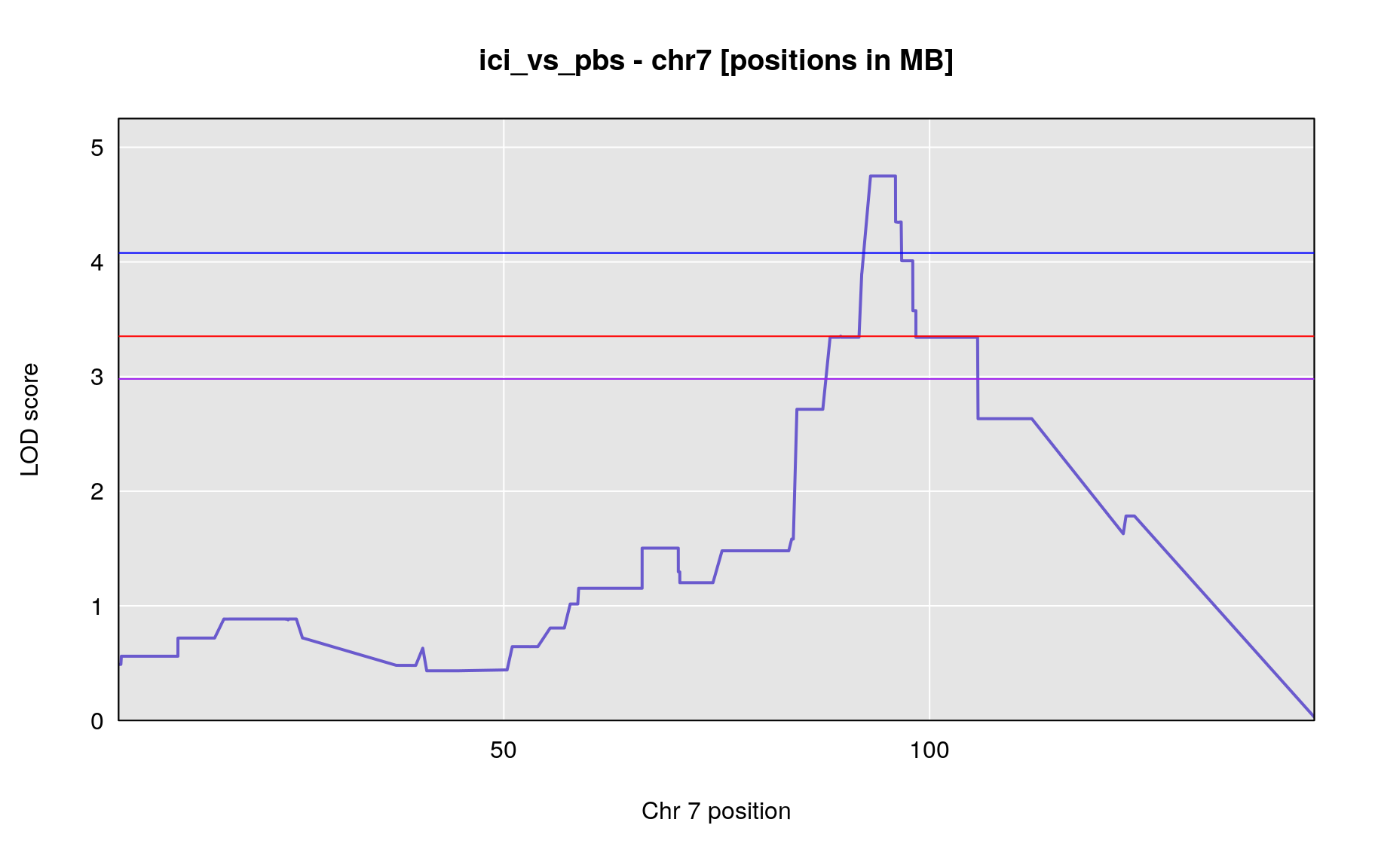
Megabase (MB)
print("peaks in MB positions")[1] “peaks in MB positions”
peaks_mba <- find_peaks(out, gm$pmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) >0){
peaks_mba$marker <- find_marker(gm$pmap, chr=peaks_mba$chr,pos=peaks_mba$pos)
names(peaks_mba)[2] <- c("phenotype")
peaks_mba <- peaks_mba[-1]
rownames(peaks_mba) <- NULL
print(kable(peaks_mba, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
plot_lod_chr_mb<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in MB]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
#ymx <- 11
#plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
##legend("topright", lwd=2, colnames(out)[i], bg="gray90")
#title(main = paste0(colnames(out)[i], " - chr", chrom, " [positions in MB]\n(using same scale as eoi vs. ici for easier comparison)"))
#add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
#add_threshold(map, summary(operm, alpha=0.05), col = 'red')
#add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
for(i in unique(peaks_mba$chr)){
#for (i in 1:nrow(peaks_mba)){
#plot_lod_chr_mb(out,gm$pmap, peaks_mba$chr[i])
plot_lod_chr_mb(out,gm$pmap,i)
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}| phenotype | chr | pos | lod | ci_lo | ci_hi | marker |
|---|---|---|---|---|---|---|
| ici_vs_pbs | 7 | 95.97325 | 4.750019 | 87.4604 | 105.688 | UNCHS020712 |
QTL effects
For each peak LOD location we give a list of gene
query_variants <- create_variant_query_func("code/cc_variants.sqlite")
query_genes <- create_gene_query_func("code/mouse_genes_mgi.sqlite")
if(nrow(peaks) >0){
for (i in 1:nrow(peaks)){
g <- maxmarg(pr.qc, gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i], return_char=TRUE)
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/","qtl_effect_", i, ".png"))
#par(mar=c(4.1, 4.1, 1.5, 0.6))
plot_pxg(g, covars[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
##dev.off()
chr = peaks$chr[i]
# Plot 2
pr_sub <- pull_genoprobint(pr.qc, gm$gmap, chr, c(peaks$ci_lo[i], peaks$ci_hi[i]))
blup <- scan1blup(pr.qc[,chr], covars[peaks$phenotype[i]],addcovar = addcovar)
blup_sub <- scan1blup(pr_sub[,chr], covars[peaks$phenotype[i]], addcovar = addcovar)
write.csv(as.data.frame(blup_sub), paste0("data/ici.vs.pbs_blup_sub_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_snpsqc_dis_no-x_updated_no-sex_7.batches_myo.csv"), quote=F)
plot_coef(blup,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," [scan1blup; positions in cM])")
)
plot_coef(blup_sub,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i],"; 1.5 LOD drop interval [scan1blup; positions in cM])")
)
#Table 1
chr = peaks_mba$chr[i]
start=as.numeric(peaks_mba$ci_lo[i])
end=as.numeric(peaks_mba$ci_hi[i])
genesgss = query_genes(chr, start, end)
write.csv(genesgss, file=paste0("data/ici.vs.pbs_genes_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_snpsqc_dis_no-x_updated_no-sex_7.batches_myo.csv"), quote=F)
rownames(genesgss) <- NULL
genesgss$strand_old = genesgss$strand
genesgss$strand[genesgss$strand=="+"] <- "positive"
genesgss$strand[genesgss$strand=="-"] <- "negative"
print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], "html") %>% kable_styling("striped", full_width = T))
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 7 | gene | 87.42477 | 87.49351 | negative | MGI_C57BL6J_98880 | Tyr | NCBI_Gene:22173,ENSEMBL:ENSMUSG00000004651 | MGI:98880 | protein coding gene | tyrosinase |
| 7 | gene | 87.58398 | 88.13506 | positive | MGI_C57BL6J_1351342 | Grm5 | NCBI_Gene:108071,ENSEMBL:ENSMUSG00000049583 | MGI:1351342 | protein coding gene | glutamate receptor, metabotropic 5 |
| 7 | gene | 87.58651 | 87.58672 | positive | MGI_C57BL6J_1924639 | C030032F19Rik | NA | NA | unclassified gene | RIKEN cDNA C030032F19 gene |
| 7 | pseudogene | 87.69782 | 87.69911 | negative | MGI_C57BL6J_5010639 | Gm18454 | NCBI_Gene:100417212 | MGI:5010639 | pseudogene | predicted gene, 18454 |
| 7 | gene | 87.77257 | 87.77439 | positive | MGI_C57BL6J_5753502 | Gm44926 | ENSEMBL:ENSMUSG00000108983 | MGI:5753502 | unclassified gene | predicted gene 44926 |
| 7 | gene | 87.89075 | 87.89146 | positive | MGI_C57BL6J_5753503 | Gm44927 | ENSEMBL:ENSMUSG00000108893 | MGI:5753503 | unclassified gene | predicted gene 44927 |
| 7 | gene | 88.06306 | 88.06629 | positive | MGI_C57BL6J_5753424 | Gm44848 | ENSEMBL:ENSMUSG00000109461 | MGI:5753424 | unclassified gene | predicted gene 44848 |
| 7 | gene | 88.08281 | 88.08480 | positive | MGI_C57BL6J_5753423 | Gm44847 | ENSEMBL:ENSMUSG00000108890 | MGI:5753423 | unclassified gene | predicted gene 44847 |
| 7 | gene | 88.08719 | 88.08725 | negative | MGI_C57BL6J_5454026 | Gm24249 | ENSEMBL:ENSMUSG00000087947 | MGI:5454026 | snRNA gene | predicted gene, 24249 |
| 7 | gene | 88.11916 | 88.12240 | positive | MGI_C57BL6J_5753422 | Gm44846 | ENSEMBL:ENSMUSG00000108993 | MGI:5753422 | unclassified gene | predicted gene 44846 |
| 7 | gene | 88.27802 | 88.31586 | positive | MGI_C57BL6J_109553 | Ctsc | NCBI_Gene:13032,ENSEMBL:ENSMUSG00000030560 | MGI:109553 | protein coding gene | cathepsin C |
| 7 | gene | 88.27879 | 88.28353 | negative | MGI_C57BL6J_5477016 | Gm26522 | ENSEMBL:ENSMUSG00000097423 | MGI:5477016 | lncRNA gene | predicted gene, 26522 |
| 7 | gene | 88.31148 | 88.31586 | positive | MGI_C57BL6J_5753327 | Gm44751 | ENSEMBL:ENSMUSG00000109244 | MGI:5753327 | lncRNA gene | predicted gene 44751 |
| 7 | gene | 88.34397 | 88.34410 | positive | MGI_C57BL6J_5455122 | Gm25345 | ENSEMBL:ENSMUSG00000077693 | MGI:5455122 | snoRNA gene | predicted gene, 25345 |
| 7 | gene | 88.34815 | 88.34906 | negative | MGI_C57BL6J_1924892 | C330001P17Rik | NA | NA | unclassified gene | RIKEN cDNA C330001P17 gene |
| 7 | pseudogene | 88.34931 | 88.35659 | negative | MGI_C57BL6J_3780575 | Gm2407 | NCBI_Gene:100039756 | MGI:3780575 | pseudogene | predicted gene 2407 |
| 7 | gene | 88.43027 | 88.49157 | positive | MGI_C57BL6J_1919683 | Rab38 | NCBI_Gene:72433,ENSEMBL:ENSMUSG00000030559 | MGI:1919683 | protein coding gene | RAB38, member RAS oncogene family |
| 7 | pseudogene | 88.44867 | 88.44923 | negative | MGI_C57BL6J_3783103 | Gm15661 | ENSEMBL:ENSMUSG00000080748 | MGI:3783103 | pseudogene | predicted gene 15661 |
| 7 | pseudogene | 88.53040 | 88.53092 | negative | MGI_C57BL6J_3704295 | Rps13-ps2 | NCBI_Gene:100039924,ENSEMBL:ENSMUSG00000069972 | MGI:3704295 | pseudogene | ribosomal protein S13, pseudogene 2 |
| 7 | pseudogene | 88.57167 | 88.57286 | negative | MGI_C57BL6J_3643325 | Gm8183 | NCBI_Gene:666598,ENSEMBL:ENSMUSG00000098174 | MGI:3643325 | pseudogene | predicted gene 8183 |
| 7 | pseudogene | 88.78192 | 88.78240 | negative | MGI_C57BL6J_3648708 | Gm6240 | NCBI_Gene:621562,ENSEMBL:ENSMUSG00000109135 | MGI:3648708 | pseudogene | predicted gene 6240 |
| 7 | gene | 88.85304 | 88.86489 | negative | MGI_C57BL6J_5753720 | Gm45144 | ENSEMBL:ENSMUSG00000109278 | MGI:5753720 | lncRNA gene | predicted gene 45144 |
| 7 | gene | 89.13971 | 89.40427 | negative | MGI_C57BL6J_1920009 | Tmem135 | NCBI_Gene:72759,ENSEMBL:ENSMUSG00000039428 | MGI:1920009 | protein coding gene | transmembrane protein 135 |
| 7 | gene | 89.21511 | 89.21710 | negative | MGI_C57BL6J_5753280 | Gm44704 | ENSEMBL:ENSMUSG00000109576 | MGI:5753280 | unclassified gene | predicted gene 44704 |
| 7 | gene | 89.29119 | 89.29130 | negative | MGI_C57BL6J_5454436 | Gm24659 | ENSEMBL:ENSMUSG00000094780 | MGI:5454436 | miRNA gene | predicted gene, 24659 |
| 7 | gene | 89.29575 | 89.29586 | negative | MGI_C57BL6J_5451948 | Gm22171 | ENSEMBL:ENSMUSG00000096631 | MGI:5451948 | miRNA gene | predicted gene, 22171 |
| 7 | gene | 89.33837 | 89.34629 | positive | MGI_C57BL6J_5753571 | Gm44995 | ENSEMBL:ENSMUSG00000109327 | MGI:5753571 | lncRNA gene | predicted gene 44995 |
| 7 | gene | 89.38834 | 89.39132 | negative | MGI_C57BL6J_5753572 | Gm44996 | ENSEMBL:ENSMUSG00000109199 | MGI:5753572 | unclassified gene | predicted gene 44996 |
| 7 | gene | 89.40435 | 89.41313 | positive | MGI_C57BL6J_108520 | Fzd4 | NCBI_Gene:14366,ENSEMBL:ENSMUSG00000049791 | MGI:108520 | protein coding gene | frizzled class receptor 4 |
| 7 | gene | 89.42631 | 89.44193 | positive | MGI_C57BL6J_2141898 | AI314278 | NCBI_Gene:101521,ENSEMBL:ENSMUSG00000109311 | MGI:2141898 | lncRNA gene | expressed sequence AI314278 |
| 7 | gene | 89.50778 | 89.52724 | negative | MGI_C57BL6J_1923703 | Prss23 | NCBI_Gene:76453,ENSEMBL:ENSMUSG00000039405 | MGI:1923703 | protein coding gene | protease, serine 23 |
| 7 | gene | 89.51001 | 89.51187 | positive | MGI_C57BL6J_1916654 | Prss23os | NCBI_Gene:108167445,ENSEMBL:ENSMUSG00000030623 | MGI:1916654 | antisense lncRNA gene | protease, serine 23, opposite strand |
| 7 | gene | 89.55237 | 89.56808 | positive | MGI_C57BL6J_5753640 | Gm45064 | ENSEMBL:ENSMUSG00000108875 | MGI:5753640 | lncRNA gene | predicted gene 45064 |
| 7 | pseudogene | 89.60769 | 89.60889 | positive | MGI_C57BL6J_3780714 | Gm2546 | NCBI_Gene:100040001,ENSEMBL:ENSMUSG00000098019 | MGI:3780714 | pseudogene | predicted gene 2546 |
| 7 | gene | 89.63210 | 89.63312 | negative | MGI_C57BL6J_3642744 | A230065N10Rik | ENSEMBL:ENSMUSG00000092071 | MGI:3642744 | lncRNA gene | RIKEN cDNA A230065N10 gene |
| 7 | gene | 89.63220 | 89.85436 | positive | MGI_C57BL6J_1916679 | Me3 | NCBI_Gene:109264,ENSEMBL:ENSMUSG00000030621 | MGI:1916679 | protein coding gene | malic enzyme 3, NADP(+)-dependent, mitochondrial |
| 7 | pseudogene | 89.76141 | 89.76217 | negative | MGI_C57BL6J_3783186 | Gm15744 | NCBI_Gene:100502804,ENSEMBL:ENSMUSG00000089938 | MGI:3783186 | pseudogene | predicted gene 15744 |
| 7 | gene | 89.82722 | 89.82856 | positive | MGI_C57BL6J_5804908 | Gm45793 | ENSEMBL:ENSMUSG00000109150 | MGI:5804908 | unclassified gene | predicted gene 45793 |
| 7 | gene | 89.86615 | 89.90363 | negative | MGI_C57BL6J_1918134 | Ccdc81 | NCBI_Gene:70884,ENSEMBL:ENSMUSG00000039391 | MGI:1918134 | protein coding gene | coiled-coil domain containing 81 |
| 7 | gene | 89.91753 | 89.94122 | negative | MGI_C57BL6J_96738 | Hikeshi | NCBI_Gene:67669,ENSEMBL:ENSMUSG00000062797 | MGI:96738 | protein coding gene | heat shock protein nuclear import factor |
| 7 | gene | 89.95465 | 89.98142 | negative | MGI_C57BL6J_95286 | Eed | NCBI_Gene:13626,ENSEMBL:ENSMUSG00000030619 | MGI:95286 | protein coding gene | embryonic ectoderm development |
| 7 | gene | 89.96499 | 89.96539 | negative | MGI_C57BL6J_5753696 | Gm45120 | ENSEMBL:ENSMUSG00000109038 | MGI:5753696 | unclassified gene | predicted gene 45120 |
| 7 | gene | 89.98072 | 90.04907 | positive | MGI_C57BL6J_2442580 | E230029C05Rik | NCBI_Gene:319711,ENSEMBL:ENSMUSG00000097585 | MGI:2442580 | lncRNA gene | RIKEN cDNA E230029C05 gene |
| 7 | gene | 90.03057 | 90.03292 | positive | MGI_C57BL6J_5753436 | Gm44860 | ENSEMBL:ENSMUSG00000109245 | MGI:5753436 | unclassified gene | predicted gene 44860 |
| 7 | gene | 90.04270 | 90.04392 | positive | MGI_C57BL6J_5753437 | Gm44861 | ENSEMBL:ENSMUSG00000108897 | MGI:5753437 | unclassified gene | predicted gene 44861 |
| 7 | gene | 90.04343 | 90.04752 | negative | MGI_C57BL6J_5825630 | Gm45993 | NCBI_Gene:108167446 | MGI:5825630 | lncRNA gene | predicted gene, 45993 |
| 7 | gene | 90.05537 | 90.06351 | negative | MGI_C57BL6J_5591323 | Gm32164 | NCBI_Gene:102634625 | MGI:5591323 | lncRNA gene | predicted gene, 32164 |
| 7 | pseudogene | 90.10722 | 90.10809 | negative | MGI_C57BL6J_3644965 | Gm5341 | NCBI_Gene:384719,ENSEMBL:ENSMUSG00000062235 | MGI:3644965 | pseudogene | predicted pseudogene 5341 |
| 7 | gene | 90.12482 | 90.12998 | negative | MGI_C57BL6J_1925579 | 2310010J17Rik | NCBI_Gene:78329,ENSEMBL:ENSMUSG00000097162 | MGI:1925579 | lncRNA gene | RIKEN cDNA 2310010J17 gene |
| 7 | gene | 90.13021 | 90.21346 | positive | MGI_C57BL6J_2385902 | Picalm | NCBI_Gene:233489,ENSEMBL:ENSMUSG00000039361 | MGI:2385902 | protein coding gene | phosphatidylinositol binding clathrin assembly protein |
| 7 | gene | 90.13249 | 90.13763 | positive | MGI_C57BL6J_5753799 | Gm45223 | ENSEMBL:ENSMUSG00000109429 | MGI:5753799 | unclassified gene | predicted gene 45223 |
| 7 | gene | 90.14595 | 90.15025 | positive | MGI_C57BL6J_5753798 | Gm45222 | ENSEMBL:ENSMUSG00000109498 | MGI:5753798 | unclassified gene | predicted gene 45222 |
| 7 | gene | 90.18593 | 90.18820 | positive | MGI_C57BL6J_5753797 | Gm45221 | ENSEMBL:ENSMUSG00000109005 | MGI:5753797 | unclassified gene | predicted gene 45221 |
| 7 | gene | 90.20128 | 90.20379 | positive | MGI_C57BL6J_5753796 | Gm45220 | ENSEMBL:ENSMUSG00000109279 | MGI:5753796 | unclassified gene | predicted gene 45220 |
| 7 | gene | 90.22350 | 90.26578 | negative | MGI_C57BL6J_1918255 | Ccdc83 | NCBI_Gene:75338,ENSEMBL:ENSMUSG00000030617 | MGI:1918255 | protein coding gene | coiled-coil domain containing 83 |
| 7 | gene | 90.29373 | 90.29385 | positive | MGI_C57BL6J_4422018 | n-R5s155 | ENSEMBL:ENSMUSG00000084480 | MGI:4422018 | rRNA gene | nuclear encoded rRNA 5S 155 |
| 7 | gene | 90.30221 | 90.41072 | positive | MGI_C57BL6J_1933366 | Sytl2 | NCBI_Gene:83671,ENSEMBL:ENSMUSG00000030616 | MGI:1933366 | protein coding gene | synaptotagmin-like 2 |
| 7 | gene | 90.42631 | 90.42867 | positive | MGI_C57BL6J_1917304 | Ccdc89 | NCBI_Gene:70054,ENSEMBL:ENSMUSG00000044362 | MGI:1917304 | protein coding gene | coiled-coil domain containing 89 |
| 7 | gene | 90.44273 | 90.44838 | positive | MGI_C57BL6J_2675296 | Crebzf | NCBI_Gene:233490,ENSEMBL:ENSMUSG00000051451 | MGI:2675296 | protein coding gene | CREB/ATF bZIP transcription factor |
| 7 | gene | 90.45070 | 90.45723 | negative | MGI_C57BL6J_1913521 | Tmem126a | NCBI_Gene:66271,ENSEMBL:ENSMUSG00000030615 | MGI:1913521 | protein coding gene | transmembrane protein 126A |
| 7 | gene | 90.45725 | 90.45888 | positive | MGI_C57BL6J_5753483 | Gm44907 | ENSEMBL:ENSMUSG00000109216 | MGI:5753483 | unclassified gene | predicted gene 44907 |
| 7 | gene | 90.46744 | 90.47600 | negative | MGI_C57BL6J_1915722 | Tmem126b | NCBI_Gene:68472,ENSEMBL:ENSMUSG00000030614 | MGI:1915722 | protein coding gene | transmembrane protein 126B |
| 7 | gene | 90.47618 | 92.44925 | positive | MGI_C57BL6J_1344351 | Dlg2 | NCBI_Gene:23859,ENSEMBL:ENSMUSG00000052572 | MGI:1344351 | protein coding gene | discs large MAGUK scaffold protein 2 |
| 7 | pseudogene | 90.81154 | 90.81216 | negative | MGI_C57BL6J_5753738 | Gm45162 | ENSEMBL:ENSMUSG00000109074 | MGI:5753738 | pseudogene | predicted gene 45162 |
| 7 | gene | 90.87160 | 90.87278 | positive | MGI_C57BL6J_1924466 | A930002H02Rik | ENSEMBL:ENSMUSG00000109242 | MGI:1924466 | unclassified gene | RIKEN cDNA A930002H02 gene |
| 7 | gene | 90.88544 | 90.88771 | negative | MGI_C57BL6J_5753737 | Gm45161 | ENSEMBL:ENSMUSG00000109431 | MGI:5753737 | unclassified gene | predicted gene 45161 |
| 7 | gene | 90.88707 | 90.94005 | positive | MGI_C57BL6J_5753735 | Gm45159 | ENSEMBL:ENSMUSG00000109125 | MGI:5753735 | lncRNA gene | predicted gene 45159 |
| 7 | gene | 90.99531 | 90.99942 | positive | MGI_C57BL6J_5753754 | Gm45178 | ENSEMBL:ENSMUSG00000109562 | MGI:5753754 | unclassified gene | predicted gene 45178 |
| 7 | gene | 91.01574 | 91.01630 | positive | MGI_C57BL6J_5621933 | Gm39048 | NCBI_Gene:105242994 | MGI:5621933 | lncRNA gene | predicted gene, 39048 |
| 7 | gene | 91.04900 | 91.05334 | positive | MGI_C57BL6J_5753755 | Gm45179 | ENSEMBL:ENSMUSG00000108956 | MGI:5753755 | unclassified gene | predicted gene 45179 |
| 7 | gene | 91.12300 | 91.12321 | negative | MGI_C57BL6J_5530852 | Gm27470 | ENSEMBL:ENSMUSG00000098805 | MGI:5530852 | unclassified non-coding RNA gene | predicted gene, 27470 |
| 7 | gene | 91.12635 | 91.12655 | negative | MGI_C57BL6J_5531042 | Gm27660 | ENSEMBL:ENSMUSG00000099001 | MGI:5531042 | unclassified non-coding RNA gene | predicted gene, 27660 |
| 7 | gene | 91.13184 | 91.13594 | positive | MGI_C57BL6J_5753752 | Gm45176 | ENSEMBL:ENSMUSG00000109239 | MGI:5753752 | unclassified gene | predicted gene 45176 |
| 7 | gene | 91.13996 | 91.14227 | positive | MGI_C57BL6J_5753753 | Gm45177 | ENSEMBL:ENSMUSG00000108819 | MGI:5753753 | unclassified gene | predicted gene 45177 |
| 7 | gene | 91.15860 | 91.16100 | positive | MGI_C57BL6J_5753758 | Gm45182 | ENSEMBL:ENSMUSG00000109387 | MGI:5753758 | unclassified gene | predicted gene 45182 |
| 7 | gene | 91.24299 | 91.24310 | positive | MGI_C57BL6J_5454329 | Gm24552 | ENSEMBL:ENSMUSG00000089271 | MGI:5454329 | snRNA gene | predicted gene, 24552 |
| 7 | gene | 91.30469 | 91.30619 | positive | MGI_C57BL6J_5753759 | Gm45183 | ENSEMBL:ENSMUSG00000109353 | MGI:5753759 | unclassified gene | predicted gene 45183 |
| 7 | gene | 91.34002 | 91.34279 | positive | MGI_C57BL6J_5753254 | Gm44678 | ENSEMBL:ENSMUSG00000108920 | MGI:5753254 | unclassified gene | predicted gene 44678 |
| 7 | gene | 91.39258 | 91.39269 | negative | MGI_C57BL6J_5451857 | Gm22080 | ENSEMBL:ENSMUSG00000088216 | MGI:5451857 | snRNA gene | predicted gene, 22080 |
| 7 | gene | 91.51911 | 91.52202 | positive | MGI_C57BL6J_5753255 | Gm44679 | ENSEMBL:ENSMUSG00000109320 | MGI:5753255 | unclassified gene | predicted gene 44679 |
| 7 | gene | 91.56437 | 91.56729 | positive | MGI_C57BL6J_5753251 | Gm44675 | ENSEMBL:ENSMUSG00000109370 | MGI:5753251 | unclassified gene | predicted gene 44675 |
| 7 | gene | 91.60896 | 91.61201 | positive | MGI_C57BL6J_5753252 | Gm44676 | ENSEMBL:ENSMUSG00000109217 | MGI:5753252 | unclassified gene | predicted gene 44676 |
| 7 | gene | 91.62727 | 91.63101 | positive | MGI_C57BL6J_5753253 | Gm44677 | ENSEMBL:ENSMUSG00000109505 | MGI:5753253 | unclassified gene | predicted gene 44677 |
| 7 | gene | 91.63524 | 91.63815 | positive | MGI_C57BL6J_5753256 | Gm44680 | ENSEMBL:ENSMUSG00000109044 | MGI:5753256 | unclassified gene | predicted gene 44680 |
| 7 | gene | 91.64771 | 91.64855 | positive | MGI_C57BL6J_5753257 | Gm44681 | ENSEMBL:ENSMUSG00000109256 | MGI:5753257 | unclassified gene | predicted gene 44681 |
| 7 | gene | 91.66715 | 91.66729 | negative | MGI_C57BL6J_5453705 | Gm23928 | ENSEMBL:ENSMUSG00000064403 | MGI:5453705 | snRNA gene | predicted gene, 23928 |
| 7 | gene | 91.68947 | 91.69021 | positive | MGI_C57BL6J_1924862 | C030038I04Rik | ENSEMBL:ENSMUSG00000109037 | MGI:1924862 | unclassified gene | RIKEN cDNA C030038I04 gene |
| 7 | gene | 91.81617 | 91.81955 | positive | MGI_C57BL6J_5753706 | Gm45130 | ENSEMBL:ENSMUSG00000109481 | MGI:5753706 | unclassified gene | predicted gene 45130 |
| 7 | gene | 91.90806 | 91.91029 | positive | MGI_C57BL6J_5753705 | Gm45129 | ENSEMBL:ENSMUSG00000108953 | MGI:5753705 | unclassified gene | predicted gene 45129 |
| 7 | gene | 91.93277 | 91.96076 | negative | MGI_C57BL6J_5591512 | Gm32353 | NCBI_Gene:102634874 | MGI:5591512 | lncRNA gene | predicted gene, 32353 |
| 7 | gene | 92.06459 | 92.06717 | positive | MGI_C57BL6J_5753707 | Gm45131 | ENSEMBL:ENSMUSG00000109191 | MGI:5753707 | unclassified gene | predicted gene 45131 |
| 7 | gene | 92.08109 | 92.08254 | negative | MGI_C57BL6J_1918245 | 4931412I15Rik | ENSEMBL:ENSMUSG00000108826 | MGI:1918245 | unclassified gene | RIKEN cDNA 4931412I15 gene |
| 7 | gene | 92.09221 | 92.11951 | negative | MGI_C57BL6J_1923095 | 4930567K12Rik | NCBI_Gene:75845,ENSEMBL:ENSMUSG00000109382 | MGI:1923095 | lncRNA gene | RIKEN cDNA 4930567K12 gene |
| 7 | gene | 92.40818 | 92.41137 | positive | MGI_C57BL6J_1926094 | B230206I08Rik | ENSEMBL:ENSMUSG00000109574 | MGI:1926094 | unclassified gene | RIKEN cDNA B230206I08 gene |
| 7 | gene | 92.43117 | 92.43385 | positive | MGI_C57BL6J_5753777 | Gm45201 | ENSEMBL:ENSMUSG00000109031 | MGI:5753777 | unclassified gene | predicted gene 45201 |
| 7 | pseudogene | 92.46923 | 92.47823 | positive | MGI_C57BL6J_5590650 | Gm31491 | NCBI_Gene:102633738 | MGI:5590650 | pseudogene | predicted gene, 31491 |
| 7 | gene | 92.56115 | 92.58229 | positive | MGI_C57BL6J_1913615 | Ccdc90b | NCBI_Gene:66365,ENSEMBL:ENSMUSG00000030613 | MGI:1913615 | protein coding gene | coiled-coil domain containing 90B |
| 7 | gene | 92.58172 | 92.63714 | negative | MGI_C57BL6J_1921095 | Ankrd42 | NCBI_Gene:73845,ENSEMBL:ENSMUSG00000041343 | MGI:1921095 | protein coding gene | ankyrin repeat domain 42 |
| 7 | gene | 92.63719 | 92.64830 | positive | MGI_C57BL6J_5504059 | Gm26944 | NCBI_Gene:102635071,ENSEMBL:ENSMUSG00000098066 | MGI:5504059 | lncRNA gene | predicted gene, 26944 |
| 7 | gene | 92.63721 | 92.63753 | positive | MGI_C57BL6J_5610236 | Gm37008 | ENSEMBL:ENSMUSG00000103887 | MGI:5610236 | unclassified gene | predicted gene, 37008 |
| 7 | gene | 92.63995 | 92.64378 | positive | MGI_C57BL6J_2443259 | 6430511E19Rik | ENSEMBL:ENSMUSG00000102555 | MGI:2443259 | unclassified gene | RIKEN cDNA 6430511E19 gene |
| 7 | gene | 92.64354 | 92.67005 | negative | MGI_C57BL6J_1919579 | Pcf11 | NCBI_Gene:74737,ENSEMBL:ENSMUSG00000041328 | MGI:1919579 | protein coding gene | PCF11 cleavage and polyadenylation factor subunit |
| 7 | pseudogene | 92.69368 | 92.69791 | negative | MGI_C57BL6J_3780719 | Gm2551 | NCBI_Gene:100040007 | MGI:3780719 | pseudogene | predicted gene 2551 |
| 7 | gene | 92.70677 | 92.70828 | positive | MGI_C57BL6J_5753775 | Gm45199 | ENSEMBL:ENSMUSG00000109395 | MGI:5753775 | unclassified gene | predicted gene 45199 |
| 7 | gene | 92.72984 | 92.73375 | positive | MGI_C57BL6J_5504096 | Gm26981 | ENSEMBL:ENSMUSG00000098041 | MGI:5504096 | lncRNA gene | predicted gene, 26981 |
| 7 | gene | 92.73417 | 92.74147 | negative | MGI_C57BL6J_1915436 | 4632427E13Rik | NCBI_Gene:666737,ENSEMBL:ENSMUSG00000074024 | MGI:1915436 | lncRNA gene | RIKEN cDNA 4632427E13 gene |
| 7 | gene | 92.74160 | 92.84453 | positive | MGI_C57BL6J_1923235 | Rab30 | NCBI_Gene:75985,ENSEMBL:ENSMUSG00000030643 | MGI:1923235 | protein coding gene | RAB30, member RAS oncogene family |
| 7 | gene | 92.77850 | 92.77942 | positive | MGI_C57BL6J_5753776 | Gm45200 | ENSEMBL:ENSMUSG00000109257 | MGI:5753776 | unclassified gene | predicted gene 45200 |
| 7 | gene | 92.82056 | 92.82077 | positive | MGI_C57BL6J_1917506 | 2010107C10Rik | NA | NA | unclassified gene | RIKEN cDNA 2010107C10 gene |
| 7 | gene | 92.85752 | 92.87429 | negative | MGI_C57BL6J_1921291 | Ddias | NCBI_Gene:74041,ENSEMBL:ENSMUSG00000030641 | MGI:1921291 | protein coding gene | DNA damage-induced apoptosis suppressor |
| 7 | gene | 92.87447 | 92.93458 | positive | MGI_C57BL6J_1919711 | Prcp | NCBI_Gene:72461,ENSEMBL:ENSMUSG00000061119 | MGI:1919711 | protein coding gene | prolylcarboxypeptidase (angiotensinase C) |
| 7 | gene | 92.87743 | 92.88999 | positive | MGI_C57BL6J_5623333 | Gm40448 | NCBI_Gene:105244926 | MGI:5623333 | lncRNA gene | predicted gene, 40448 |
| 7 | gene | 92.90218 | 92.90287 | positive | MGI_C57BL6J_1925814 | 9530078K11Rik | ENSEMBL:ENSMUSG00000109268 | MGI:1925814 | unclassified gene | RIKEN cDNA 9530078K11 gene |
| 7 | pseudogene | 92.97925 | 93.00654 | positive | MGI_C57BL6J_5590822 | Gm31663 | NCBI_Gene:102633966,ENSEMBL:ENSMUSG00000108994 | MGI:5590822 | pseudogene | predicted gene, 31663 |
| 7 | gene | 93.00698 | 93.10479 | positive | MGI_C57BL6J_5477356 | Gm26862 | ENSEMBL:ENSMUSG00000097644 | MGI:5477356 | lncRNA gene | predicted gene, 26862 |
| 7 | gene | 93.05208 | 93.08103 | negative | MGI_C57BL6J_3641747 | Gm9934 | ENSEMBL:ENSMUSG00000054061 | MGI:3641747 | lncRNA gene | predicted gene 9934 |
| 7 | gene | 93.07986 | 93.08187 | positive | MGI_C57BL6J_1930951 | Fam181b | NCBI_Gene:58238,ENSEMBL:ENSMUSG00000051515 | MGI:1930951 | protein coding gene | family with sequence similarity 181, member B |
| 7 | gene | 93.14785 | 93.14797 | negative | MGI_C57BL6J_5455637 | Gm25860 | ENSEMBL:ENSMUSG00000088877 | MGI:5455637 | snoRNA gene | predicted gene, 25860 |
| 7 | pseudogene | 93.16883 | 93.17267 | positive | MGI_C57BL6J_5753291 | Gm44715 | ENSEMBL:ENSMUSG00000109196 | MGI:5753291 | pseudogene | predicted gene 44715 |
| 7 | pseudogene | 93.17898 | 93.18418 | negative | MGI_C57BL6J_3704296 | Gm15501 | NCBI_Gene:100040298,ENSEMBL:ENSMUSG00000087412 | MGI:3704296 | pseudogene | predicted pseudogene 15501 |
| 7 | gene | 93.29070 | 93.32395 | negative | MGI_C57BL6J_5753523 | Gm44947 | ENSEMBL:ENSMUSG00000109175 | MGI:5753523 | lncRNA gene | predicted gene 44947 |
| 7 | gene | 93.36168 | 93.38086 | positive | MGI_C57BL6J_5753613 | Gm45037 | ENSEMBL:ENSMUSG00000109315 | MGI:5753613 | lncRNA gene | predicted gene 45037 |
| 7 | pseudogene | 93.52306 | 93.52690 | positive | MGI_C57BL6J_5753614 | Gm45038 | ENSEMBL:ENSMUSG00000109195 | MGI:5753614 | pseudogene | predicted gene 45038 |
| 7 | pseudogene | 93.56494 | 93.56570 | negative | MGI_C57BL6J_3645456 | Gm8285 | NCBI_Gene:666783,ENSEMBL:ENSMUSG00000109077 | MGI:3645456 | pseudogene | predicted gene 8285 |
| 7 | gene | 93.60484 | 93.60832 | negative | MGI_C57BL6J_5753615 | Gm45039 | ENSEMBL:ENSMUSG00000109226 | MGI:5753615 | unclassified gene | predicted gene 45039 |
| 7 | pseudogene | 93.66024 | 93.66056 | negative | MGI_C57BL6J_5052079 | Bc1-ps1 | NCBI_Gene:12031,ENSEMBL:ENSMUSG00000109440 | MGI:5052079 | pseudogene | brain cytoplasmic RNA 1, pseudogene 1 |
| 7 | pseudogene | 93.84690 | 93.84783 | negative | MGI_C57BL6J_5010382 | Gm18197 | NCBI_Gene:100416690 | MGI:5010382 | pseudogene | predicted gene, 18197 |
| 7 | gene | 94.04250 | 95.32643 | positive | MGI_C57BL6J_5591806 | Gm32647 | NCBI_Gene:102635262,ENSEMBL:ENSMUSG00000108532 | MGI:5591806 | lncRNA gene | predicted gene, 32647 |
| 7 | pseudogene | 94.19642 | 94.19756 | positive | MGI_C57BL6J_3643908 | Gm5899 | NCBI_Gene:545982,ENSEMBL:ENSMUSG00000096033 | MGI:3643908 | pseudogene | predicted pseudogene 5899 |
| 7 | gene | 94.36898 | 94.37257 | negative | MGI_C57BL6J_5753177 | Gm44601 | ENSEMBL:ENSMUSG00000108315 | MGI:5753177 | lncRNA gene | predicted gene 44601 |
| 7 | pseudogene | 94.47558 | 94.47571 | positive | MGI_C57BL6J_5753178 | Gm44602 | ENSEMBL:ENSMUSG00000108516 | MGI:5753178 | pseudogene | predicted gene 44602 |
| 7 | pseudogene | 94.60182 | 94.60263 | negative | MGI_C57BL6J_5011946 | Gm19761 | NCBI_Gene:100503542,ENSEMBL:ENSMUSG00000108433 | MGI:5011946 | pseudogene | predicted gene, 19761 |
| 7 | gene | 95.17186 | 95.17216 | positive | MGI_C57BL6J_5453661 | Gm23884 | ENSEMBL:ENSMUSG00000084636 | MGI:5453661 | unclassified non-coding RNA gene | predicted gene, 23884 |
| 7 | gene | 95.44235 | 95.44246 | negative | MGI_C57BL6J_5455461 | Gm25684 | ENSEMBL:ENSMUSG00000065875 | MGI:5455461 | snRNA gene | predicted gene, 25684 |
| 7 | pseudogene | 95.46191 | 95.46356 | positive | MGI_C57BL6J_3643545 | Gm8309 | NCBI_Gene:666823,ENSEMBL:ENSMUSG00000108786 | MGI:3643545 | pseudogene | predicted gene 8309 |
| 7 | pseudogene | 95.47495 | 95.47531 | positive | MGI_C57BL6J_5753179 | Gm44603 | ENSEMBL:ENSMUSG00000108429 | MGI:5753179 | pseudogene | predicted gene 44603 |
| 7 | gene | 95.77545 | 95.78117 | positive | MGI_C57BL6J_5621934 | Gm39049 | NCBI_Gene:105242997 | MGI:5621934 | lncRNA gene | predicted gene, 39049 |
| 7 | pseudogene | 95.91275 | 95.91339 | negative | MGI_C57BL6J_3648478 | Gm5037 | NCBI_Gene:260347,ENSEMBL:ENSMUSG00000108628 | MGI:3648478 | pseudogene | predicted gene 5037 |
| 7 | gene | 95.95821 | 95.96161 | positive | MGI_C57BL6J_3641871 | Gm9966 | NA | NA | protein coding gene | predicted gene 9966 |
| 7 | gene | 96.17124 | 96.91109 | positive | MGI_C57BL6J_2447063 | Tenm4 | NCBI_Gene:23966,ENSEMBL:ENSMUSG00000048078 | MGI:2447063 | protein coding gene | teneurin transmembrane protein 4 |
| 7 | pseudogene | 96.20239 | 96.20364 | negative | MGI_C57BL6J_3646693 | Gm8319 | NCBI_Gene:666836,ENSEMBL:ENSMUSG00000108528 | MGI:3646693 | pseudogene | predicted gene 8319 |
| 7 | gene | 96.24942 | 96.24953 | positive | MGI_C57BL6J_3629686 | Mir708 | miRBase:MI0004692,NCBI_Gene:735284,ENSEMBL:ENSMUSG00000076143 | MGI:3629686 | miRNA gene | microRNA 708 |
| 7 | gene | 96.30567 | 96.30576 | positive | MGI_C57BL6J_5531311 | Mir6394 | miRBase:MI0021928,NCBI_Gene:102465210,ENSEMBL:ENSMUSG00000099030 | MGI:5531311 | miRNA gene | microRNA 6394 |
| 7 | gene | 96.33948 | 96.34196 | negative | MGI_C57BL6J_3705100 | Gm15412 | NCBI_Gene:670727,ENSEMBL:ENSMUSG00000085751 | MGI:3705100 | lncRNA gene | predicted gene 15412 |
| 7 | pseudogene | 96.43482 | 96.43675 | negative | MGI_C57BL6J_3647437 | Rps11-ps5 | NCBI_Gene:102635492,ENSEMBL:ENSMUSG00000082780 | MGI:3647437 | pseudogene | ribosomal protein S11, pseudogene 5 |
| 7 | gene | 96.58581 | 96.58691 | positive | MGI_C57BL6J_1924457 | 8030425K09Rik | NA | NA | unclassified gene | RIKEN cDNA 8030425K09 gene |
| 7 | gene | 96.59665 | 96.62907 | negative | MGI_C57BL6J_3705307 | Gm15414 | ENSEMBL:ENSMUSG00000085792 | MGI:3705307 | lncRNA gene | predicted gene 15414 |
| 7 | gene | 96.79143 | 96.80155 | negative | MGI_C57BL6J_3642665 | Gm15413 | NCBI_Gene:791381,ENSEMBL:ENSMUSG00000053049 | MGI:3642665 | lncRNA gene | predicted gene 15413 |
| 7 | gene | 96.81243 | 96.81749 | negative | MGI_C57BL6J_3705308 | Gm15416 | ENSEMBL:ENSMUSG00000085443 | MGI:3705308 | lncRNA gene | predicted gene 15416 |
| 7 | gene | 96.81387 | 96.81831 | negative | MGI_C57BL6J_5753209 | Gm44633 | ENSEMBL:ENSMUSG00000108635 | MGI:5753209 | lncRNA gene | predicted gene 44633 |
| 7 | gene | 96.86329 | 96.86338 | negative | MGI_C57BL6J_5455489 | Gm25712 | ENSEMBL:ENSMUSG00000089259 | MGI:5455489 | rRNA gene | predicted gene, 25712 |
| 7 | gene | 96.90683 | 96.94703 | negative | MGI_C57BL6J_3713275 | Gm15415 | NA | NA | antisense lncRNA gene | predicted gene 15415 |
| 7 | gene | 96.90683 | 96.95189 | negative | MGI_C57BL6J_3697433 | C230038L03Rik | NCBI_Gene:108167512,ENSEMBL:ENSMUSG00000085560 | MGI:3697433 | lncRNA gene | RIKEN cDNA C230038L03 gene |
| 7 | gene | 96.91999 | 96.92010 | negative | MGI_C57BL6J_5452003 | Gm22226 | ENSEMBL:ENSMUSG00000087980 | MGI:5452003 | snRNA gene | predicted gene, 22226 |
| 7 | gene | 96.95150 | 97.06476 | positive | MGI_C57BL6J_2142075 | Nars2 | NCBI_Gene:244141,ENSEMBL:ENSMUSG00000018995 | MGI:2142075 | protein coding gene | asparaginyl-tRNA synthetase 2 (mitochondrial)(putative) |
| 7 | gene | 97.08159 | 97.30895 | positive | MGI_C57BL6J_1333854 | Gab2 | NCBI_Gene:14389,ENSEMBL:ENSMUSG00000004508 | MGI:1333854 | protein coding gene | growth factor receptor bound protein 2-associated protein 2 |
| 7 | gene | 97.30938 | 97.33229 | negative | MGI_C57BL6J_2685339 | Usp35 | NCBI_Gene:244144,ENSEMBL:ENSMUSG00000035713 | MGI:2685339 | protein coding gene | ubiquitin specific peptidase 35 |
| 7 | gene | 97.33232 | 97.35022 | positive | MGI_C57BL6J_3643121 | Kctd21 | NCBI_Gene:622320,ENSEMBL:ENSMUSG00000044952 | MGI:3643121 | protein coding gene | potassium channel tetramerisation domain containing 21 |
| 7 | gene | 97.37159 | 97.39218 | positive | MGI_C57BL6J_2141959 | Alg8 | NCBI_Gene:381903,ENSEMBL:ENSMUSG00000035704 | MGI:2141959 | protein coding gene | asparagine-linked glycosylation 8 (alpha-1,3-glucosyltransferase) |
| 7 | gene | 97.40000 | 97.40780 | positive | MGI_C57BL6J_1344370 | Ndufc2 | NCBI_Gene:68197,ENSEMBL:ENSMUSG00000030647 | MGI:1344370 | protein coding gene | NADH:ubiquinone oxidoreductase subunit C2 |
| 7 | pseudogene | 97.40614 | 97.40633 | positive | MGI_C57BL6J_3802033 | Gm16053 | ENSEMBL:ENSMUSG00000083974 | MGI:3802033 | pseudogene | predicted gene 16053 |
| 7 | gene | 97.41293 | 97.41773 | negative | MGI_C57BL6J_109126 | Thrsp | NCBI_Gene:21835,ENSEMBL:ENSMUSG00000035686 | MGI:109126 | protein coding gene | thyroid hormone responsive |
| 7 | gene | 97.41865 | 97.42014 | positive | MGI_C57BL6J_5592104 | Gm32945 | NCBI_Gene:102635665 | MGI:5592104 | lncRNA gene | predicted gene, 32945 |
| 7 | gene | 97.43638 | 97.45956 | positive | MGI_C57BL6J_1289222 | Kctd14 | NCBI_Gene:233529,ENSEMBL:ENSMUSG00000051727 | MGI:1289222 | protein coding gene | potassium channel tetramerisation domain containing 14 |
| 7 | gene | 97.45527 | 97.45671 | positive | MGI_C57BL6J_3028073 | 7030407A21Rik | NA | NA | unclassified gene | RIKEN cDNA 7030407A21 gene |
| 7 | gene | 97.48095 | 97.54140 | positive | MGI_C57BL6J_1917164 | Ints4 | NCBI_Gene:101861,ENSEMBL:ENSMUSG00000025133 | MGI:1917164 | protein coding gene | integrator complex subunit 4 |
| 7 | gene | 97.52181 | 97.52192 | positive | MGI_C57BL6J_5454189 | Gm24412 | ENSEMBL:ENSMUSG00000065360 | MGI:5454189 | snoRNA gene | predicted gene, 24412 |
| 7 | gene | 97.55033 | 97.57951 | negative | MGI_C57BL6J_1913523 | Aamdc | NCBI_Gene:66273,ENSEMBL:ENSMUSG00000035642 | MGI:1913523 | protein coding gene | adipogenesis associated Mth938 domain containing |
| 7 | gene | 97.57977 | 97.69278 | positive | MGI_C57BL6J_2682305 | Rsf1 | NCBI_Gene:233532,ENSEMBL:ENSMUSG00000035623 | MGI:2682305 | protein coding gene | remodeling and spacing factor 1 |
| 7 | pseudogene | 97.59149 | 97.59245 | negative | MGI_C57BL6J_5010024 | Ywhaq-ps1 | NCBI_Gene:100415958,ENSEMBL:ENSMUSG00000108405 | MGI:5010024 | pseudogene | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein theta, pseudogene 1 |
| 7 | pseudogene | 97.59484 | 97.59544 | negative | MGI_C57BL6J_5010232 | Rpl15-ps5 | NCBI_Gene:100416334,ENSEMBL:ENSMUSG00000108442 | MGI:5010232 | pseudogene | ribosomal protein L15, pseudogene 5 |
| 7 | gene | 97.59835 | 97.64511 | negative | MGI_C57BL6J_1922698 | Rsf1os1 | NCBI_Gene:75448,ENSEMBL:ENSMUSG00000086433 | MGI:1922698 | antisense lncRNA gene | remodeling and spacing factor 1, opposite strand 1 |
| 7 | gene | 97.64900 | 97.65232 | positive | MGI_C57BL6J_2441926 | C920008N22Rik | NA | NA | unclassified gene | RIKEN cDNA C920008N22 gene |
| 7 | gene | 97.65001 | 97.69683 | negative | MGI_C57BL6J_3642179 | Rsf1os2 | NCBI_Gene:102641859,ENSEMBL:ENSMUSG00000086993 | MGI:3642179 | antisense lncRNA gene | remodeling and spacing factor 1, opposite strand 2 |
| 7 | pseudogene | 97.65428 | 97.65475 | negative | MGI_C57BL6J_5753762 | Gm45186 | ENSEMBL:ENSMUSG00000108547 | MGI:5753762 | pseudogene | predicted gene 45186 |
| 7 | gene | 97.69663 | 97.72080 | positive | MGI_C57BL6J_109638 | Clns1a | NCBI_Gene:12729,ENSEMBL:ENSMUSG00000025439 | MGI:109638 | protein coding gene | chloride channel, nucleotide-sensitive, 1A |
| 7 | gene | 97.72084 | 97.72292 | positive | MGI_C57BL6J_3642272 | Gm9990 | NA | NA | protein coding gene | predicted gene 9990 |
| 7 | gene | 97.72401 | 97.73829 | negative | MGI_C57BL6J_1913583 | Aqp11 | NCBI_Gene:66333,ENSEMBL:ENSMUSG00000042797 | MGI:1913583 | protein coding gene | aquaporin 11 |
| 7 | gene | 97.75932 | 97.80116 | negative | MGI_C57BL6J_5621935 | Gm39050 | NCBI_Gene:105243000 | MGI:5621935 | lncRNA gene | predicted gene, 39050 |
| 7 | gene | 97.78854 | 97.91238 | positive | MGI_C57BL6J_1339975 | Pak1 | NCBI_Gene:18479,ENSEMBL:ENSMUSG00000030774 | MGI:1339975 | protein coding gene | p21 (RAC1) activated kinase 1 |
| 7 | pseudogene | 97.91824 | 97.91863 | positive | MGI_C57BL6J_5753166 | Gm44590 | ENSEMBL:ENSMUSG00000108424 | MGI:5753166 | pseudogene | predicted gene 44590 |
| 7 | gene | 97.91994 | 98.04966 | positive | MGI_C57BL6J_3606573 | Gdpd4 | NCBI_Gene:233537,ENSEMBL:ENSMUSG00000035582 | MGI:3606573 | protein coding gene | glycerophosphodiester phosphodiesterase domain containing 4 |
| 7 | pseudogene | 97.98456 | 97.98539 | negative | MGI_C57BL6J_5011389 | Gm19204 | NCBI_Gene:100418424 | MGI:5011389 | pseudogene | predicted gene, 19204 |
| 7 | gene | 98.05105 | 98.11952 | negative | MGI_C57BL6J_104510 | Myo7a | NCBI_Gene:17921,ENSEMBL:ENSMUSG00000030761 | MGI:104510 | protein coding gene | myosin VIIA |
| 7 | gene | 98.12156 | 98.17827 | negative | MGI_C57BL6J_1100859 | Capn5 | NCBI_Gene:12337,ENSEMBL:ENSMUSG00000035547 | MGI:1100859 | protein coding gene | calpain 5 |
| 7 | gene | 98.14336 | 98.14550 | negative | MGI_C57BL6J_97436 | Omp | NCBI_Gene:18378,ENSEMBL:ENSMUSG00000074006 | MGI:97436 | protein coding gene | olfactory marker protein |
| 7 | gene | 98.17719 | 98.18480 | positive | MGI_C57BL6J_4439862 | Gm16938 | NCBI_Gene:330599,ENSEMBL:ENSMUSG00000097749 | MGI:4439862 | lncRNA gene | predicted gene, 16938 |
| 7 | gene | 98.19241 | 98.19948 | negative | MGI_C57BL6J_3039603 | B3gnt6 | NCBI_Gene:272411,ENSEMBL:ENSMUSG00000074004 | MGI:3039603 | protein coding gene | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase) |
| 7 | gene | 98.20639 | 98.21038 | negative | MGI_C57BL6J_2142310 | E230006M18Rik | NA | NA | unclassified gene | RIKEN cDNA E230006M18 gene |
| 7 | gene | 98.20639 | 98.32121 | negative | MGI_C57BL6J_1913440 | Acer3 | NCBI_Gene:66190,ENSEMBL:ENSMUSG00000030760 | MGI:1913440 | protein coding gene | alkaline ceramidase 3 |
| 7 | gene | 98.35067 | 98.36139 | negative | MGI_C57BL6J_2443855 | Tsku | NCBI_Gene:244152,ENSEMBL:ENSMUSG00000049580 | MGI:2443855 | protein coding gene | tsukushi, small leucine rich proteoglycan |
| 7 | gene | 98.36699 | 98.41980 | negative | MGI_C57BL6J_5753083 | Gm44507 | NCBI_Gene:108167447,ENSEMBL:ENSMUSG00000108449 | MGI:5753083 | lncRNA gene | predicted gene 44507 |
| 7 | gene | 98.43348 | 98.43795 | positive | MGI_C57BL6J_5621938 | Gm39053 | NCBI_Gene:105243003 | MGI:5621938 | lncRNA gene | predicted gene, 39053 |
| 7 | pseudogene | 98.43721 | 98.43752 | negative | MGI_C57BL6J_5753556 | Gm44980 | ENSEMBL:ENSMUSG00000108346 | MGI:5753556 | pseudogene | predicted gene 44980 |
| 7 | gene | 98.44042 | 98.47748 | positive | MGI_C57BL6J_106030 | Gucy2d | NCBI_Gene:14918,ENSEMBL:ENSMUSG00000074003 | MGI:106030 | protein coding gene | guanylate cyclase 2d |
| 7 | pseudogene | 98.45509 | 98.45568 | positive | MGI_C57BL6J_3781104 | Gm2926 | NCBI_Gene:100040721,ENSEMBL:ENSMUSG00000108764 | MGI:3781104 | pseudogene | predicted gene 2926 |
| 7 | pseudogene | 98.46513 | 98.46648 | negative | MGI_C57BL6J_5753554 | Gm44978 | NCBI_Gene:108167471,ENSEMBL:ENSMUSG00000108508 | MGI:5753554 | pseudogene | predicted gene 44978 |
| 7 | gene | 98.48126 | 98.48622 | positive | MGI_C57BL6J_5621473 | Gm38588 | NCBI_Gene:102641966 | MGI:5621473 | lncRNA gene | predicted gene, 38588 |
| 7 | gene | 98.48928 | 98.50218 | positive | MGI_C57BL6J_93882 | Lrrc32 | NCBI_Gene:434215,ENSEMBL:ENSMUSG00000090958 | MGI:93882 | protein coding gene | leucine rich repeat containing 32 |
| 7 | gene | 98.50259 | 98.50665 | negative | MGI_C57BL6J_5592553 | Gm33394 | NCBI_Gene:102636286 | MGI:5592553 | lncRNA gene | predicted gene, 33394 |
| 7 | gene | 98.50695 | 98.50712 | positive | MGI_C57BL6J_5452817 | Gm23040 | ENSEMBL:ENSMUSG00000065803 | MGI:5452817 | snRNA gene | predicted gene, 23040 |
| 7 | gene | 98.54332 | 98.56299 | negative | MGI_C57BL6J_2442837 | A630091E08Rik | ENSEMBL:ENSMUSG00000055446 | MGI:2442837 | lncRNA gene | RIKEN cDNA A630091E08 gene |
| 7 | gene | 98.58713 | 98.65956 | negative | MGI_C57BL6J_1924203 | Emsy | NCBI_Gene:233545,ENSEMBL:ENSMUSG00000035401 | MGI:1924203 | protein coding gene | EMSY, BRCA2-interacting transcriptional repressor |
| 7 | gene | 98.65620 | 98.65783 | positive | MGI_C57BL6J_1923090 | 4930558N01Rik | NA | NA | unclassified gene | RIKEN cDNA 4930558N01 gene |
| 7 | gene | 98.67811 | 98.68065 | negative | MGI_C57BL6J_5753708 | Gm45132 | ENSEMBL:ENSMUSG00000109101 | MGI:5753708 | unclassified gene | predicted gene 45132 |
| 7 | gene | 98.68233 | 98.70293 | negative | MGI_C57BL6J_3782954 | Gm15506 | NCBI_Gene:100040769,ENSEMBL:ENSMUSG00000086477 | MGI:3782954 | lncRNA gene | predicted gene 15506 |
| 7 | gene | 98.70310 | 98.71806 | positive | MGI_C57BL6J_1920231 | Thap12 | NCBI_Gene:72981,ENSEMBL:ENSMUSG00000030753 | MGI:1920231 | protein coding gene | THAP domain containing 12 |
| 7 | gene | 98.71950 | 98.72876 | negative | MGI_C57BL6J_5011841 | Gm19656 | NCBI_Gene:105243004,ENSEMBL:ENSMUSG00000109149 | MGI:5011841 | lncRNA gene | predicted gene, 19656 |
| 7 | gene | 98.73475 | 98.73508 | negative | MGI_C57BL6J_5753510 | Gm44934 | ENSEMBL:ENSMUSG00000108988 | MGI:5753510 | lncRNA gene | predicted gene 44934 |
| 7 | pseudogene | 98.76148 | 98.76194 | positive | MGI_C57BL6J_5011435 | Gm19250 | NCBI_Gene:100462934 | MGI:5011435 | pseudogene | predicted gene, 19250 |
| 7 | gene | 98.76302 | 98.76984 | positive | MGI_C57BL6J_5592879 | Gm33720 | NCBI_Gene:102636728 | MGI:5592879 | lncRNA gene | predicted gene, 33720 |
| 7 | gene | 98.81542 | 98.81549 | positive | MGI_C57BL6J_4414020 | n-TPtgg8 | NCBI_Gene:102467371 | MGI:4414020 | tRNA gene | nuclear encoded tRNA proline 8 (anticodon TGG) |
| 7 | gene | 98.81568 | 98.81575 | negative | MGI_C57BL6J_4414009 | n-TPagg7 | NCBI_Gene:102467367 | MGI:4414009 | tRNA gene | nuclear encoded tRNA proline 7 (anticodon AGG) |
| 7 | gene | 98.81613 | 98.82193 | positive | MGI_C57BL6J_5621939 | Gm39054 | NCBI_Gene:105243005 | MGI:5621939 | lncRNA gene | predicted gene, 39054 |
| 7 | gene | 98.83066 | 98.83606 | negative | MGI_C57BL6J_5592929 | Gm33770 | NCBI_Gene:102636798 | MGI:5592929 | lncRNA gene | predicted gene, 33770 |
| 7 | gene | 98.83492 | 98.85519 | positive | MGI_C57BL6J_101948 | Wnt11 | NCBI_Gene:22411,ENSEMBL:ENSMUSG00000015957 | MGI:101948 | protein coding gene | wingless-type MMTV integration site family, member 11 |
| 7 | pseudogene | 98.85473 | 98.85510 | negative | MGI_C57BL6J_3648175 | Gm8398 | ENSEMBL:ENSMUSG00000091496 | MGI:3648175 | pseudogene | predicted gene 8398 |
| 7 | gene | 98.86926 | 98.87663 | negative | MGI_C57BL6J_5592986 | Gm33827 | NCBI_Gene:102636877 | MGI:5592986 | lncRNA gene | predicted gene, 33827 |
| 7 | gene | 98.88502 | 99.14117 | negative | MGI_C57BL6J_1925860 | Uvrag | NCBI_Gene:78610,ENSEMBL:ENSMUSG00000035354 | MGI:1925860 | protein coding gene | UV radiation resistance associated gene |
| 7 | gene | 98.89522 | 98.90649 | positive | MGI_C57BL6J_3647698 | Gm8149 | NCBI_Gene:666529 | MGI:3647698 | unclassified gene | predicted gene 8149 |
| 7 | gene | 98.89593 | 98.89955 | positive | MGI_C57BL6J_5753764 | Gm45188 | ENSEMBL:ENSMUSG00000109282 | MGI:5753764 | lncRNA gene | predicted gene 45188 |
| 7 | gene | 98.90427 | 98.90806 | positive | MGI_C57BL6J_5753763 | Gm45187 | ENSEMBL:ENSMUSG00000109130 | MGI:5753763 | lncRNA gene | predicted gene 45187 |
| 7 | gene | 98.90659 | 98.91100 | positive | MGI_C57BL6J_5621940 | Gm39055 | NCBI_Gene:105243006 | MGI:5621940 | lncRNA gene | predicted gene, 39055 |
| 7 | gene | 98.93589 | 98.93601 | positive | MGI_C57BL6J_5453256 | Gm23479 | ENSEMBL:ENSMUSG00000089501 | MGI:5453256 | snoRNA gene | predicted gene, 23479 |
| 7 | pseudogene | 99.08946 | 99.09088 | positive | MGI_C57BL6J_5593041 | Gm33882 | NCBI_Gene:102636953,ENSEMBL:ENSMUSG00000109492 | MGI:5593041 | pseudogene | predicted gene, 33882 |
| 7 | gene | 99.12207 | 99.12346 | negative | MGI_C57BL6J_5753551 | Gm44975 | ENSEMBL:ENSMUSG00000109073 | MGI:5753551 | unclassified gene | predicted gene 44975 |
| 7 | gene | 99.14159 | 99.14200 | positive | MGI_C57BL6J_5753761 | Gm45185 | ENSEMBL:ENSMUSG00000109468 | MGI:5753761 | unclassified gene | predicted gene 45185 |
| 7 | gene | 99.15366 | 99.18272 | negative | MGI_C57BL6J_1915050 | Dgat2 | NCBI_Gene:67800,ENSEMBL:ENSMUSG00000030747 | MGI:1915050 | protein coding gene | diacylglycerol O-acyltransferase 2 |
| 7 | gene | 99.20237 | 99.22708 | positive | MGI_C57BL6J_5753588 | Gm45012 | ENSEMBL:ENSMUSG00000109052 | MGI:5753588 | lncRNA gene | predicted gene 45012 |
| 7 | gene | 99.21908 | 99.23862 | negative | MGI_C57BL6J_2663253 | Mogat2 | NCBI_Gene:233549,ENSEMBL:ENSMUSG00000052396 | MGI:2663253 | protein coding gene | monoacylglycerol O-acyltransferase 2 |
| 7 | pseudogene | 99.24757 | 99.24897 | negative | MGI_C57BL6J_5011128 | Gm18943 | NCBI_Gene:100418007,ENSEMBL:ENSMUSG00000109472 | MGI:5011128 | pseudogene | predicted gene, 18943 |
| 7 | gene | 99.25151 | 99.25519 | positive | MGI_C57BL6J_5593221 | Gm34062 | NCBI_Gene:102637185 | MGI:5593221 | lncRNA gene | predicted gene, 34062 |
| 7 | gene | 99.26048 | 99.26722 | negative | MGI_C57BL6J_5477199 | Gm26705 | NCBI_Gene:381967,ENSEMBL:ENSMUSG00000097015 | MGI:5477199 | lncRNA gene | predicted gene, 26705 |
| 7 | gene | 99.26718 | 99.33714 | positive | MGI_C57BL6J_1201690 | Map6 | NCBI_Gene:17760,ENSEMBL:ENSMUSG00000055407 | MGI:1201690 | protein coding gene | microtubule-associated protein 6 |
| 7 | gene | 99.30027 | 99.30449 | negative | MGI_C57BL6J_5623334 | Gm40449 | NCBI_Gene:105244927 | MGI:5623334 | lncRNA gene | predicted gene, 40449 |
| 7 | gene | 99.32120 | 99.32246 | positive | MGI_C57BL6J_5791427 | Gm45591 | ENSEMBL:ENSMUSG00000109460 | MGI:5791427 | lncRNA gene | predicted gene 45591 |
| 7 | gene | 99.34538 | 99.35324 | negative | MGI_C57BL6J_88283 | Serpinh1 | NCBI_Gene:12406,ENSEMBL:ENSMUSG00000070436 | MGI:88283 | protein coding gene | serine (or cysteine) peptidase inhibitor, clade H, member 1 |
| 7 | gene | 99.38141 | 99.46188 | positive | MGI_C57BL6J_2686926 | Gdpd5 | NCBI_Gene:233552,ENSEMBL:ENSMUSG00000035314 | MGI:2686926 | protein coding gene | glycerophosphodiester phosphodiesterase domain containing 5 |
| 7 | gene | 99.46600 | 99.47402 | positive | MGI_C57BL6J_1919434 | Klhl35 | NCBI_Gene:72184,ENSEMBL:ENSMUSG00000035298 | MGI:1919434 | protein coding gene | kelch-like 35 |
| 7 | gene | 99.47790 | 99.48374 | negative | MGI_C57BL6J_1350917 | Rps3 | NCBI_Gene:27050,ENSEMBL:ENSMUSG00000030744 | MGI:1350917 | protein coding gene | ribosomal protein S3 |
| 7 | gene | 99.47956 | 99.47971 | negative | MGI_C57BL6J_3779515 | Snord15b | NCBI_Gene:449631,ENSEMBL:ENSMUSG00000064966 | MGI:3779515 | snoRNA gene | small nucleolar RNA, C/D box 14B |
| 7 | gene | 99.48279 | 99.48293 | negative | MGI_C57BL6J_3645887 | Snord15a | NCBI_Gene:449630,ENSEMBL:ENSMUSG00000065822 | MGI:3645887 | snoRNA gene | small nucleolar RNA, C/D box 15A |
| 7 | gene | 99.49772 | 99.49782 | positive | MGI_C57BL6J_4422019 | n-R5s156 | ENSEMBL:ENSMUSG00000065311 | MGI:4422019 | rRNA gene | nuclear encoded rRNA 5S 156 |
| 7 | gene | 99.50046 | 99.50594 | positive | MGI_C57BL6J_5621941 | Gm39056 | NCBI_Gene:105243007 | MGI:5621941 | lncRNA gene | predicted gene, 39056 |
| 7 | gene | 99.51165 | 99.51715 | positive | MGI_C57BL6J_5623335 | Gm40450 | NCBI_Gene:105244928 | MGI:5623335 | lncRNA gene | predicted gene, 40450 |
| 7 | gene | 99.51664 | 99.52964 | negative | MGI_C57BL6J_5593331 | Gm34172 | NCBI_Gene:102637333 | MGI:5593331 | lncRNA gene | predicted gene, 34172 |
| 7 | gene | 99.53547 | 99.60677 | positive | MGI_C57BL6J_99473 | Arrb1 | NCBI_Gene:109689,ENSEMBL:ENSMUSG00000018909 | MGI:99473 | protein coding gene | arrestin, beta 1 |
| 7 | gene | 99.54390 | 99.54397 | negative | MGI_C57BL6J_4413843 | n-TEctc3 | NCBI_Gene:102467644 | MGI:4413843 | tRNA gene | nuclear encoded tRNA glutamic acid 3 (anticodon CTC) |
| 7 | gene | 99.55227 | 99.55236 | positive | MGI_C57BL6J_3619338 | Mir326 | miRBase:MI0000598,NCBI_Gene:723840,ENSEMBL:ENSMUSG00000065571 | MGI:3619338 | miRNA gene | microRNA 326 |
| 7 | gene | 99.56812 | 99.57084 | negative | MGI_C57BL6J_5753490 | Gm44914 | ENSEMBL:ENSMUSG00000108850 | MGI:5753490 | unclassified gene | predicted gene 44914 |
| 7 | gene | 99.62407 | 99.62710 | negative | MGI_C57BL6J_3646425 | Tpbgl | NCBI_Gene:100503386,ENSEMBL:ENSMUSG00000096606 | MGI:3646425 | protein coding gene | trophoblast glycoprotein-like |
| 7 | gene | 99.62594 | 99.63171 | positive | MGI_C57BL6J_3642596 | Gm10605 | ENSEMBL:ENSMUSG00000097974 | MGI:3642596 | lncRNA gene | predicted gene 10605 |
| 7 | gene | 99.65369 | 99.65382 | negative | MGI_C57BL6J_5455186 | Gm25409 | ENSEMBL:ENSMUSG00000065698 | MGI:5455186 | snoRNA gene | predicted gene, 25409 |
| 7 | gene | 99.65780 | 99.71134 | negative | MGI_C57BL6J_1351872 | Slco2b1 | NCBI_Gene:101488,ENSEMBL:ENSMUSG00000030737 | MGI:1351872 | protein coding gene | solute carrier organic anion transporter family, member 2b1 |
| 7 | gene | 99.66013 | 99.66750 | positive | MGI_C57BL6J_3783079 | Gm15635 | ENSEMBL:ENSMUSG00000085095 | MGI:3783079 | lncRNA gene | predicted gene 15635 |
| 7 | gene | 99.69303 | 99.71423 | positive | MGI_C57BL6J_5593439 | Gm34280 | NCBI_Gene:102637485,ENSEMBL:ENSMUSG00000109559 | MGI:5593439 | lncRNA gene | predicted gene, 34280 |
| 7 | pseudogene | 99.72521 | 99.72538 | negative | MGI_C57BL6J_5753127 | Gm44551 | ENSEMBL:ENSMUSG00000108965 | MGI:5753127 | pseudogene | predicted gene 44551 |
| 7 | gene | 99.73027 | 99.73640 | positive | MGI_C57BL6J_3030354 | Olfr520 | NCBI_Gene:259066,ENSEMBL:ENSMUSG00000073998 | MGI:3030354 | protein coding gene | olfactory receptor 520 |
| 7 | pseudogene | 99.74021 | 99.74079 | negative | MGI_C57BL6J_5011367 | Gm19182 | NCBI_Gene:100418394,ENSEMBL:ENSMUSG00000109275 | MGI:5011367 | pseudogene | predicted gene, 19182 |
| 7 | gene | 99.76351 | 99.76932 | positive | MGI_C57BL6J_3030355 | Olfr521 | NCBI_Gene:258353,ENSEMBL:ENSMUSG00000073997 | MGI:3030355 | protein coding gene | olfactory receptor 521 |
| 7 | gene | 99.78000 | 99.78154 | negative | MGI_C57BL6J_3045385 | F730035P03Rik | ENSEMBL:ENSMUSG00000053360 | MGI:3045385 | lncRNA gene | RIKEN cDNA F730035P03 gene |
| 7 | gene | 99.81144 | 99.82842 | negative | MGI_C57BL6J_1355305 | Neu3 | NCBI_Gene:50877,ENSEMBL:ENSMUSG00000035239 | MGI:1355305 | protein coding gene | neuraminidase 3 |
| 7 | gene | 99.83757 | 99.87034 | negative | MGI_C57BL6J_1913874 | Spcs2 | NCBI_Gene:66624,ENSEMBL:ENSMUSG00000035227 | MGI:1913874 | protein coding gene | signal peptidase complex subunit 2 homolog (S. cerevisiae) |
| 7 | gene | 99.85912 | 99.91782 | positive | MGI_C57BL6J_2181647 | Xrra1 | NCBI_Gene:446101,ENSEMBL:ENSMUSG00000035211 | MGI:2181647 | protein coding gene | X-ray radiation resistance associated 1 |
| 7 | gene | 99.87067 | 99.91738 | negative | MGI_C57BL6J_5621290 | Gm38405 | NCBI_Gene:233561,ENSEMBL:ENSMUSG00000109002 | MGI:5621290 | lncRNA gene | predicted gene, 38405 |
| 7 | gene | 99.90019 | 99.90416 | negative | MGI_C57BL6J_1924443 | A930030B08Rik | ENSEMBL:ENSMUSG00000108985 | MGI:1924443 | unclassified gene | RIKEN cDNA A930030B08 gene |
| 7 | gene | 99.92025 | 99.98069 | negative | MGI_C57BL6J_1920257 | Rnf169 | NCBI_Gene:108937,ENSEMBL:ENSMUSG00000058761 | MGI:1920257 | protein coding gene | ring finger protein 169 |
| 7 | gene | 99.92965 | 99.93229 | negative | MGI_C57BL6J_3643844 | Gm5115 | NCBI_Gene:330602 | MGI:3643844 | unclassified gene | predicted gene 5115 |
| 7 | pseudogene | 99.94506 | 99.94572 | positive | MGI_C57BL6J_5011144 | Gm18959 | NCBI_Gene:100418037,ENSEMBL:ENSMUSG00000108823 | MGI:5011144 | pseudogene | predicted gene, 18959 |
| 7 | pseudogene | 99.99677 | 99.99741 | positive | MGI_C57BL6J_5010116 | Gm17931 | NCBI_Gene:100416129,ENSEMBL:ENSMUSG00000109384 | MGI:5010116 | pseudogene | predicted gene, 17931 |
| 7 | gene | 100.00617 | 100.03473 | positive | MGI_C57BL6J_1916371 | Chrdl2 | NCBI_Gene:69121,ENSEMBL:ENSMUSG00000030732 | MGI:1916371 | protein coding gene | chordin-like 2 |
| 7 | gene | 100.08211 | 100.12157 | negative | MGI_C57BL6J_1915217 | Pold3 | NCBI_Gene:67967,ENSEMBL:ENSMUSG00000030726 | MGI:1915217 | protein coding gene | polymerase (DNA-directed), delta 3, accessory subunit |
| 7 | gene | 100.13188 | 100.13749 | positive | MGI_C57BL6J_5593919 | Gm34760 | NCBI_Gene:102638122 | MGI:5593919 | lncRNA gene | predicted gene, 34760 |
| 7 | gene | 100.15928 | 100.16137 | positive | MGI_C57BL6J_1914414 | Lipt2 | NCBI_Gene:67164,ENSEMBL:ENSMUSG00000030725 | MGI:1914414 | protein coding gene | lipoyl(octanoyl) transferase 2 (putative) |
| 7 | gene | 100.16332 | 100.17116 | negative | MGI_C57BL6J_5621942 | Gm39057 | NCBI_Gene:105243008 | MGI:5621942 | lncRNA gene | predicted gene, 39057 |
| 7 | gene | 100.17142 | 100.22721 | negative | MGI_C57BL6J_5593980 | Gm34821 | NCBI_Gene:102638204,ENSEMBL:ENSMUSG00000109186 | MGI:5593980 | lncRNA gene | predicted gene, 34821 |
| 7 | gene | 100.17650 | 100.18487 | positive | MGI_C57BL6J_1891124 | Kcne3 | NCBI_Gene:57442,ENSEMBL:ENSMUSG00000035165 | MGI:1891124 | protein coding gene | potassium voltage-gated channel, Isk-related subfamily, gene 3 |
| 7 | gene | 100.18926 | 100.19267 | negative | MGI_C57BL6J_5825631 | Gm45994 | NCBI_Gene:108167448 | MGI:5825631 | lncRNA gene | predicted gene, 45994 |
| 7 | pseudogene | 100.19710 | 100.19835 | negative | MGI_C57BL6J_3779664 | Gm7067 | NCBI_Gene:631577,ENSEMBL:ENSMUSG00000109435 | MGI:3779664 | pseudogene | predicted gene 7067 |
| 7 | gene | 100.20078 | 100.22721 | negative | MGI_C57BL6J_5594036 | Gm34877 | NCBI_Gene:102638275 | MGI:5594036 | lncRNA gene | predicted gene, 34877 |
| 7 | gene | 100.22728 | 100.27887 | positive | MGI_C57BL6J_1918224 | Pgm2l1 | NCBI_Gene:70974,ENSEMBL:ENSMUSG00000030729 | MGI:1918224 | protein coding gene | phosphoglucomutase 2-like 1 |
| 7 | pseudogene | 100.26454 | 100.26556 | positive | MGI_C57BL6J_106627 | Gpx2-ps1 | NCBI_Gene:14777,ENSEMBL:ENSMUSG00000089987 | MGI:106627 | pseudogene | glutathione peroxidase 2, pseudogene 1 |
| 7 | gene | 100.28549 | 100.31970 | positive | MGI_C57BL6J_2444049 | P4ha3 | NCBI_Gene:320452,ENSEMBL:ENSMUSG00000051048 | MGI:2444049 | protein coding gene | procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide III |
| 7 | gene | 100.32674 | 100.37231 | negative | MGI_C57BL6J_1919840 | Ppme1 | NCBI_Gene:72590,ENSEMBL:ENSMUSG00000030718 | MGI:1919840 | protein coding gene | protein phosphatase methylesterase 1 |
| 7 | gene | 100.37223 | 100.47016 | positive | MGI_C57BL6J_2142166 | C2cd3 | NCBI_Gene:277939,ENSEMBL:ENSMUSG00000047248 | MGI:2142166 | protein coding gene | C2 calcium-dependent domain containing 3 |
| 7 | pseudogene | 100.38159 | 100.38231 | positive | MGI_C57BL6J_5753147 | Gm44571 | ENSEMBL:ENSMUSG00000109046 | MGI:5753147 | pseudogene | predicted gene 44571 |
| 7 | gene | 100.44674 | 100.47485 | negative | MGI_C57BL6J_3642592 | Gm10603 | NCBI_Gene:100038497,ENSEMBL:ENSMUSG00000099137 | MGI:3642592 | lncRNA gene | predicted gene 10603 |
| 7 | gene | 100.45846 | 100.45941 | positive | MGI_C57BL6J_5610944 | Gm37716 | ENSEMBL:ENSMUSG00000102241 | MGI:5610944 | unclassified gene | predicted gene, 37716 |
| 7 | gene | 100.47299 | 100.48643 | positive | MGI_C57BL6J_1099787 | Ucp3 | NCBI_Gene:22229,ENSEMBL:ENSMUSG00000032942 | MGI:1099787 | protein coding gene | uncoupling protein 3 (mitochondrial, proton carrier) |
| 7 | gene | 100.49334 | 100.50202 | positive | MGI_C57BL6J_109354 | Ucp2 | NCBI_Gene:22228,ENSEMBL:ENSMUSG00000033685 | MGI:109354 | protein coding gene | uncoupling protein 2 (mitochondrial, proton carrier) |
| 7 | gene | 100.50172 | 100.51496 | negative | MGI_C57BL6J_1916637 | Dnajb13 | NCBI_Gene:69387,ENSEMBL:ENSMUSG00000030708 | MGI:1916637 | protein coding gene | DnaJ heat shock protein family (Hsp40) member B13 |
| 7 | pseudogene | 100.52398 | 100.52467 | positive | MGI_C57BL6J_3644490 | Gm8463 | NCBI_Gene:667112,ENSEMBL:ENSMUSG00000109560 | MGI:3644490 | pseudogene | predicted gene 8463 |
| 7 | gene | 100.53559 | 100.53698 | negative | MGI_C57BL6J_5621943 | Gm39058 | NCBI_Gene:105243010 | MGI:5621943 | lncRNA gene | predicted gene, 39058 |
| 7 | gene | 100.53707 | 100.54037 | positive | MGI_C57BL6J_1915435 | Coa4 | NCBI_Gene:68185,ENSEMBL:ENSMUSG00000044881 | MGI:1915435 | protein coding gene | cytochrome c oxidase assembly factor 4 |
| 7 | gene | 100.54135 | 100.54412 | negative | MGI_C57BL6J_5621944 | Gm39059 | NCBI_Gene:105243011,ENSEMBL:ENSMUSG00000109123 | MGI:5621944 | lncRNA gene | predicted gene, 39059 |
| 7 | gene | 100.54574 | 100.54779 | negative | MGI_C57BL6J_3028054 | D630004N19Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA D630004N19 gene |
| 7 | gene | 100.54575 | 100.60830 | negative | MGI_C57BL6J_1289321 | Mrpl48 | NCBI_Gene:52443,ENSEMBL:ENSMUSG00000030706 | MGI:1289321 | protein coding gene | mitochondrial ribosomal protein L48 |
| 7 | pseudogene | 100.57192 | 100.57225 | positive | MGI_C57BL6J_5753224 | Gm44648 | ENSEMBL:ENSMUSG00000108990 | MGI:5753224 | pseudogene | predicted gene 44648 |
| 7 | pseudogene | 100.57841 | 100.58064 | positive | MGI_C57BL6J_3783125 | Cox20b | NCBI_Gene:108167449,ENSEMBL:ENSMUSG00000081359 | MGI:3783125 | pseudogene | cytochrome c oxidase assembly protein 20B |
| 7 | gene | 100.60741 | 100.64127 | positive | MGI_C57BL6J_894313 | Rab6a | NCBI_Gene:19346,ENSEMBL:ENSMUSG00000030704 | MGI:894313 | protein coding gene | RAB6A, member RAS oncogene family |
| 7 | gene | 100.61877 | 100.62126 | positive | MGI_C57BL6J_2442194 | D930046H04Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA D930046H04 gene |
| 7 | pseudogene | 100.62452 | 100.62495 | positive | MGI_C57BL6J_3650372 | Gm14382 | ENSEMBL:ENSMUSG00000082210 | MGI:3650372 | pseudogene | predicted gene 14382 |
| 7 | gene | 100.64289 | 100.66241 | negative | MGI_C57BL6J_1351469 | Plekhb1 | NCBI_Gene:27276,ENSEMBL:ENSMUSG00000030701 | MGI:1351469 | protein coding gene | pleckstrin homology domain containing, family B (evectins) member 1 |
| 7 | gene | 100.67499 | 100.67814 | positive | MGI_C57BL6J_5594241 | Gm35082 | NCBI_Gene:102638545,ENSEMBL:ENSMUSG00000108866 | MGI:5594241 | lncRNA gene | predicted gene, 35082 |
| 7 | gene | 100.70664 | 100.84166 | positive | MGI_C57BL6J_2442372 | Fam168a | NCBI_Gene:319604,ENSEMBL:ENSMUSG00000029461 | MGI:2442372 | protein coding gene | family with sequence similarity 168, member A |
| 7 | pseudogene | 100.73805 | 100.73863 | negative | MGI_C57BL6J_5753785 | Gm45209 | ENSEMBL:ENSMUSG00000109035 | MGI:5753785 | pseudogene | predicted gene 45209 |
| 7 | pseudogene | 100.76062 | 100.76182 | negative | MGI_C57BL6J_3781379 | Gm3200 | NCBI_Gene:100041204,ENSEMBL:ENSMUSG00000097388 | MGI:3781379 | pseudogene | predicted pseudogene 3200 |
| 7 | gene | 100.81135 | 100.81264 | positive | MGI_C57BL6J_5753529 | Gm44953 | ENSEMBL:ENSMUSG00000109109 | MGI:5753529 | unclassified gene | predicted gene 44953 |
| 7 | gene | 100.84585 | 100.86348 | negative | MGI_C57BL6J_2443373 | Relt | NCBI_Gene:320100,ENSEMBL:ENSMUSG00000008318 | MGI:2443373 | protein coding gene | RELT tumor necrosis factor receptor |
| 7 | gene | 100.86975 | 100.93216 | negative | MGI_C57BL6J_2673002 | Arhgef17 | NCBI_Gene:207212,ENSEMBL:ENSMUSG00000032875 | MGI:2673002 | protein coding gene | Rho guanine nucleotide exchange factor (GEF) 17 |
| 7 | gene | 100.88231 | 100.88241 | negative | MGI_C57BL6J_4834321 | Mir3102 | miRBase:MI0014099,NCBI_Gene:100526508,ENSEMBL:ENSMUSG00000093296 | MGI:4834321 | miRNA gene | microRNA 3102 |
| 7 | gene | 100.93763 | 100.97550 | negative | MGI_C57BL6J_2673874 | P2ry6 | NCBI_Gene:233571,ENSEMBL:ENSMUSG00000048779 | MGI:2673874 | protein coding gene | pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| 7 | gene | 100.95082 | 100.95156 | positive | MGI_C57BL6J_5753121 | Gm44545 | ENSEMBL:ENSMUSG00000109132 | MGI:5753121 | unclassified gene | predicted gene 44545 |
| 7 | gene | 100.99657 | 101.01287 | negative | MGI_C57BL6J_105107 | P2ry2 | NCBI_Gene:18442,ENSEMBL:ENSMUSG00000032860 | MGI:105107 | protein coding gene | purinergic receptor P2Y, G-protein coupled 2 |
| 7 | pseudogene | 101.01862 | 101.01887 | negative | MGI_C57BL6J_5753646 | Gm45070 | ENSEMBL:ENSMUSG00000108895 | MGI:5753646 | pseudogene | predicted gene 45070 |
| 7 | pseudogene | 101.06122 | 101.06148 | negative | MGI_C57BL6J_3645951 | Gm5735 | NCBI_Gene:435996,ENSEMBL:ENSMUSG00000109171 | MGI:3645951 | pseudogene | predicted gene 5735 |
| 7 | gene | 101.09286 | 101.28441 | positive | MGI_C57BL6J_2448475 | Fchsd2 | NCBI_Gene:207278,ENSEMBL:ENSMUSG00000030691 | MGI:2448475 | protein coding gene | FCH and double SH3 domains 2 |
| 7 | gene | 101.11355 | 101.11780 | positive | MGI_C57BL6J_2442785 | 6030496E16Rik | NA | NA | unclassified gene | RIKEN cDNA 6030496E16 gene |
| 7 | pseudogene | 101.15460 | 101.15514 | negative | MGI_C57BL6J_3643600 | Gm6341 | NCBI_Gene:622673,ENSEMBL:ENSMUSG00000083121 | MGI:3643600 | pseudogene | predicted pseudogene 6341 |
| 7 | gene | 101.20767 | 101.21103 | positive | MGI_C57BL6J_3026972 | 6430502G17Rik | NA | NA | unclassified gene | RIKEN cDNA 6430502G17 gene |
| 7 | pseudogene | 101.23822 | 101.23928 | positive | MGI_C57BL6J_6096209 | Gm47324 | ENSEMBL:ENSMUSG00000111425 | MGI:6096209 | pseudogene | predicted gene, 47324 |
| 7 | gene | 101.26303 | 101.30227 | negative | MGI_C57BL6J_1920933 | Atg16l2 | NCBI_Gene:73683,ENSEMBL:ENSMUSG00000047767 | MGI:1920933 | protein coding gene | autophagy related 16-like 2 (S. cerevisiae) |
| 7 | gene | 101.31709 | 101.34663 | positive | MGI_C57BL6J_1860093 | Stard10 | NCBI_Gene:56018,ENSEMBL:ENSMUSG00000030688 | MGI:1860093 | protein coding gene | START domain containing 10 |
| 7 | gene | 101.31772 | 101.32623 | negative | MGI_C57BL6J_5141941 | Gm20476 | ENSEMBL:ENSMUSG00000092364 | MGI:5141941 | lncRNA gene | predicted gene 20476 |
| 7 | pseudogene | 101.33813 | 101.33847 | positive | MGI_C57BL6J_5791180 | Gm45344 | ENSEMBL:ENSMUSG00000110099 | MGI:5791180 | pseudogene | predicted gene 45344 |
| 7 | gene | 101.34691 | 101.34798 | negative | MGI_C57BL6J_5791456 | Gm45620 | NCBI_Gene:108167329,ENSEMBL:ENSMUSG00000110239 | MGI:5791456 | lncRNA gene | predicted gene 45620 |
| 7 | gene | 101.34765 | 101.41259 | positive | MGI_C57BL6J_1916960 | Arap1 | NCBI_Gene:69710,ENSEMBL:ENSMUSG00000032812 | MGI:1916960 | protein coding gene | ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| 7 | gene | 101.34821 | 101.35542 | negative | MGI_C57BL6J_5594378 | Gm35219 | NCBI_Gene:102638725 | MGI:5594378 | lncRNA gene | predicted gene, 35219 |
| 7 | gene | 101.35127 | 101.35444 | negative | MGI_C57BL6J_5791384 | Gm45548 | ENSEMBL:ENSMUSG00000109656 | MGI:5791384 | unclassified gene | predicted gene 45548 |
| 7 | gene | 101.41089 | 101.51282 | positive | MGI_C57BL6J_5804952 | Gm45837 | ENSEMBL:ENSMUSG00000030653 | MGI:5804952 | protein coding gene | predicted gene 45837 |
| 7 | gene | 101.42169 | 101.51283 | positive | MGI_C57BL6J_2446107 | Pde2a | NCBI_Gene:207728,ENSEMBL:ENSMUSG00000110195 | MGI:2446107 | protein coding gene | phosphodiesterase 2A, cGMP-stimulated |
| 7 | gene | 101.47538 | 101.47544 | positive | MGI_C57BL6J_2676824 | Mir139 | miRBase:MI0000693,NCBI_Gene:387157,ENSEMBL:ENSMUSG00000065446 | MGI:2676824 | miRNA gene | microRNA 139 |
| 7 | gene | 101.51819 | 101.53866 | negative | MGI_C57BL6J_5594522 | Gm35363 | NCBI_Gene:102638917,ENSEMBL:ENSMUSG00000110301 | MGI:5594522 | lncRNA gene | predicted gene, 35363 |
| 7 | gene | 101.54787 | 101.55746 | positive | MGI_C57BL6J_5011950 | Gm19765 | NCBI_Gene:102638993 | MGI:5011950 | lncRNA gene | predicted gene, 19765 |
| 7 | pseudogene | 101.55245 | 101.56087 | negative | MGI_C57BL6J_107546 | Art2a | NCBI_Gene:11871,ENSEMBL:ENSMUSG00000092517 | MGI:107546 | polymorphic pseudogene | ADP-ribosyltransferase 2a |
| 7 | pseudogene | 101.57342 | 101.57436 | negative | MGI_C57BL6J_3644555 | Gm7027 | NCBI_Gene:630138,ENSEMBL:ENSMUSG00000067121 | MGI:3644555 | pseudogene | predicted gene 7027 |
| 7 | gene | 101.57573 | 101.58560 | negative | MGI_C57BL6J_107545 | Art2b | NCBI_Gene:11872,ENSEMBL:ENSMUSG00000030651 | MGI:107545 | protein coding gene | ADP-ribosyltransferase 2b |
| 7 | gene | 101.58159 | 101.58283 | positive | MGI_C57BL6J_5825632 | Gm45995 | NCBI_Gene:108167451 | MGI:5825632 | lncRNA gene | predicted gene, 45995 |
| 7 | gene | 101.66359 | 101.79551 | positive | MGI_C57BL6J_1100517 | Clpb | NCBI_Gene:20480,ENSEMBL:ENSMUSG00000001829 | MGI:1100517 | protein coding gene | ClpB caseinolytic peptidase B |
| 7 | pseudogene | 101.79054 | 101.79085 | positive | MGI_C57BL6J_5791216 | Gm45380 | ENSEMBL:ENSMUSG00000110360 | MGI:5791216 | pseudogene | predicted gene 45380 |
| 7 | gene | 101.81831 | 101.82273 | positive | MGI_C57BL6J_106633 | Phox2a | NCBI_Gene:11859,ENSEMBL:ENSMUSG00000007946 | MGI:106633 | protein coding gene | paired-like homeobox 2a |
| 7 | gene | 101.82263 | 101.83823 | negative | MGI_C57BL6J_1333787 | Inppl1 | NCBI_Gene:16332,ENSEMBL:ENSMUSG00000032737 | MGI:1333787 | protein coding gene | inositol polyphosphate phosphatase-like 1 |
| 7 | gene | 101.83650 | 101.84453 | positive | MGI_C57BL6J_3708759 | Gm10602 | NCBI_Gene:102639173,ENSEMBL:ENSMUSG00000073985 | MGI:3708759 | lncRNA gene | predicted gene 10602 |
| 7 | gene | 101.83999 | 101.85719 | negative | MGI_C57BL6J_95569 | Folr2 | NCBI_Gene:14276,ENSEMBL:ENSMUSG00000032725 | MGI:95569 | protein coding gene | folate receptor 2 (fetal) |
| 7 | pseudogene | 101.85296 | 101.85328 | positive | MGI_C57BL6J_5791155 | Gm45319 | ENSEMBL:ENSMUSG00000110295 | MGI:5791155 | pseudogene | predicted gene 45319 |
| 7 | gene | 101.85833 | 101.87079 | negative | MGI_C57BL6J_95568 | Folr1 | NCBI_Gene:14275,ENSEMBL:ENSMUSG00000001827 | MGI:95568 | protein coding gene | folate receptor 1 (adult) |
| 7 | gene | 101.86371 | 101.90185 | positive | MGI_C57BL6J_1922680 | Anapc15 | NCBI_Gene:75430,ENSEMBL:ENSMUSG00000030649 | MGI:1922680 | protein coding gene | anaphase prompoting complex C subunit 15 |
| 7 | gene | 101.87086 | 101.87958 | positive | MGI_C57BL6J_5594756 | Gm35597 | NCBI_Gene:102639241 | MGI:5594756 | lncRNA gene | predicted gene, 35597 |
| 7 | gene | 101.88118 | 101.88128 | positive | MGI_C57BL6J_5452432 | Gm22655 | ENSEMBL:ENSMUSG00000093388 | MGI:5452432 | miRNA gene | predicted gene, 22655 |
| 7 | gene | 101.89837 | 101.90641 | negative | MGI_C57BL6J_3769724 | Tomt | NCBI_Gene:791260,ENSEMBL:ENSMUSG00000078630 | MGI:3769724 | protein coding gene | transmembrane O-methyltransferase |
| 7 | gene | 101.90584 | 101.92668 | positive | MGI_C57BL6J_1913758 | Lamtor1 | NCBI_Gene:66508,ENSEMBL:ENSMUSG00000030842 | MGI:1913758 | protein coding gene | late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| 7 | gene | 101.91298 | 101.93392 | negative | MGI_C57BL6J_1916608 | Lrrc51 | NCBI_Gene:69358,ENSEMBL:ENSMUSG00000064307 | MGI:1916608 | protein coding gene | leucine rich repeat containing 51 |
| 7 | gene | 101.93400 | 102.01496 | positive | MGI_C57BL6J_2443665 | Numa1 | NCBI_Gene:101706,ENSEMBL:ENSMUSG00000066306 | MGI:2443665 | protein coding gene | nuclear mitotic apparatus protein 1 |
| 7 | pseudogene | 101.94596 | 101.94651 | negative | MGI_C57BL6J_3779801 | Gm8523 | NCBI_Gene:100417414,ENSEMBL:ENSMUSG00000110150 | MGI:3779801 | pseudogene | predicted gene 8523 |
| 7 | pseudogene | 101.95697 | 101.95728 | negative | MGI_C57BL6J_5791148 | Gm45312 | ENSEMBL:ENSMUSG00000109872 | MGI:5791148 | pseudogene | predicted gene 45312 |
| 7 | gene | 101.97966 | 101.98229 | positive | MGI_C57BL6J_2444705 | C030040A22Rik | NA | NA | unclassified gene | RIKEN cDNA C030040A22 gene |
| 7 | pseudogene | 101.98008 | 101.98026 | negative | MGI_C57BL6J_5791149 | Gm45313 | ENSEMBL:ENSMUSG00000110287 | MGI:5791149 | pseudogene | predicted gene 45313 |
| 7 | gene | 101.98867 | 101.99095 | negative | MGI_C57BL6J_5621946 | Gm39061 | NCBI_Gene:105243016 | MGI:5621946 | lncRNA gene | predicted gene, 39061 |
| 7 | gene | 102.01489 | 102.01869 | negative | MGI_C57BL6J_1333800 | Il18bp | NCBI_Gene:16068,ENSEMBL:ENSMUSG00000070427 | MGI:1333800 | protein coding gene | interleukin 18 binding protein |
| 7 | gene | 102.01914 | 102.07244 | negative | MGI_C57BL6J_1922462 | Rnf121 | NCBI_Gene:75212,ENSEMBL:ENSMUSG00000070426 | MGI:1922462 | protein coding gene | ring finger protein 121 |
| 7 | gene | 102.06549 | 102.08376 | positive | MGI_C57BL6J_5546359 | Xndc1 | NCBI_Gene:102443350,ENSEMBL:ENSMUSG00000099481 | MGI:5546359 | protein coding gene | Xrcc1 N-terminal domain containing 1 |
| 7 | gene | 102.06549 | 102.09686 | positive | MGI_C57BL6J_5546370 | Xntrpc | NCBI_Gene:102443351,ENSEMBL:ENSMUSG00000070425 | MGI:5546370 | protein coding gene | Xndc1-transient receptor potential cation channel, subfamily C, member 2 readthrough |
| 7 | gene | 102.08312 | 102.09686 | positive | MGI_C57BL6J_109527 | Trpc2 | NCBI_Gene:22064,ENSEMBL:ENSMUSG00000100254 | MGI:109527 | protein coding gene | transient receptor potential cation channel, subfamily C, member 2 |
| 7 | gene | 102.09688 | 102.10949 | negative | MGI_C57BL6J_107948 | Art5 | NCBI_Gene:11875,ENSEMBL:ENSMUSG00000070424 | MGI:107948 | protein coding gene | ADP-ribosyltransferase 5 |
| 7 | gene | 102.10171 | 102.11393 | positive | MGI_C57BL6J_107511 | Art1 | NCBI_Gene:11870,ENSEMBL:ENSMUSG00000030996 | MGI:107511 | protein coding gene | ADP-ribosyltransferase 1 |
| 7 | gene | 102.11126 | 102.11698 | negative | MGI_C57BL6J_3609260 | Chrna10 | NCBI_Gene:504186,ENSEMBL:ENSMUSG00000066279 | MGI:3609260 | protein coding gene | cholinergic receptor, nicotinic, alpha polypeptide 10 |
| 7 | gene | 102.11339 | 102.12206 | positive | MGI_C57BL6J_5621947 | Gm39062 | NCBI_Gene:105243018 | MGI:5621947 | lncRNA gene | predicted gene, 39062 |
| 7 | gene | 102.11940 | 102.21019 | negative | MGI_C57BL6J_109404 | Nup98 | NCBI_Gene:269966,ENSEMBL:ENSMUSG00000063550 | MGI:109404 | protein coding gene | nucleoporin 98 |
| 7 | gene | 102.21021 | 102.23857 | positive | MGI_C57BL6J_2385286 | Pgap2 | NCBI_Gene:233575,ENSEMBL:ENSMUSG00000030990 | MGI:2385286 | protein coding gene | post-GPI attachment to proteins 2 |
| 7 | gene | 102.23912 | 102.25605 | negative | MGI_C57BL6J_1928370 | Rhog | NCBI_Gene:56212,ENSEMBL:ENSMUSG00000073982 | MGI:1928370 | protein coding gene | ras homolog family member G |
| 7 | gene | 102.26151 | 102.43732 | positive | MGI_C57BL6J_107476 | Stim1 | NCBI_Gene:20866,ENSEMBL:ENSMUSG00000030987 | MGI:107476 | protein coding gene | stromal interaction molecule 1 |
| 7 | gene | 102.36097 | 102.36348 | positive | MGI_C57BL6J_2443525 | A630057J21Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA A630057J21 gene |
| 7 | pseudogene | 102.37750 | 102.37834 | negative | MGI_C57BL6J_5010440 | Gm18255 | NCBI_Gene:100416793,ENSEMBL:ENSMUSG00000110175 | MGI:5010440 | pseudogene | predicted gene, 18255 |
| 7 | gene | 102.44169 | 102.46977 | positive | MGI_C57BL6J_98180 | Rrm1 | NCBI_Gene:20133,ENSEMBL:ENSMUSG00000030978 | MGI:98180 | protein coding gene | ribonucleotide reductase M1 |
| 7 | gene | 102.45825 | 102.45926 | positive | MGI_C57BL6J_5791235 | Gm45399 | ENSEMBL:ENSMUSG00000109940 | MGI:5791235 | unclassified gene | predicted gene 45399 |
| 7 | gene | 102.47046 | 102.47668 | positive | MGI_C57BL6J_5825633 | Gm45996 | NCBI_Gene:108167452 | MGI:5825633 | lncRNA gene | predicted gene, 45996 |
| 7 | gene | 102.47658 | 102.48168 | negative | MGI_C57BL6J_3030377 | Olfr543 | NCBI_Gene:257947,ENSEMBL:ENSMUSG00000044814 | MGI:3030377 | protein coding gene | olfactory receptor 543 |
| 7 | gene | 102.48203 | 102.48831 | negative | MGI_C57BL6J_3030378 | Olfr544 | NCBI_Gene:257926,ENSEMBL:ENSMUSG00000043925 | MGI:3030378 | protein coding gene | olfactory receptor 544 |
| 7 | gene | 102.49147 | 102.49957 | negative | MGI_C57BL6J_3030379 | Olfr545 | NCBI_Gene:258837,ENSEMBL:ENSMUSG00000044824 | MGI:3030379 | protein coding gene | olfactory receptor 545 |
| 7 | gene | 102.49227 | 102.49372 | positive | MGI_C57BL6J_3039577 | BC049285 | NA | NA | unclassified gene | cDNA sequence BC049285 |
| 7 | gene | 102.50548 | 102.50642 | positive | MGI_C57BL6J_5621512 | Gm38627 | NCBI_Gene:102642720 | MGI:5621512 | lncRNA gene | predicted gene, 38627 |
| 7 | pseudogene | 102.52035 | 102.52106 | negative | MGI_C57BL6J_3649082 | Gm8556 | NCBI_Gene:667284,ENSEMBL:ENSMUSG00000109822 | MGI:3649082 | pseudogene | predicted gene 8556 |
| 7 | pseudogene | 102.52069 | 102.52166 | positive | MGI_C57BL6J_3030380 | Olfr546-ps1 | NCBI_Gene:404405,ENSEMBL:ENSMUSG00000109906 | MGI:3030380 | pseudogene | olfactory receptor 546, pseudogene 1 |
| 7 | gene | 102.53475 | 102.53569 | positive | MGI_C57BL6J_3030381 | Olfr547 | NCBI_Gene:259083,ENSEMBL:ENSMUSG00000073979 | MGI:3030381 | protein coding gene | olfactory receptor 547 |
| 7 | pseudogene | 102.54090 | 102.54589 | positive | MGI_C57BL6J_3030382 | Olfr548-ps1 | NCBI_Gene:258229,ENSEMBL:ENSMUSG00000073978 | MGI:3030382 | pseudogene | olfactory receptor 548, pseudogene 1 |
| 7 | gene | 102.54987 | 102.55584 | positive | MGI_C57BL6J_3030383 | Olfr549 | NCBI_Gene:259105,ENSEMBL:ENSMUSG00000073977 | MGI:3030383 | protein coding gene | olfactory receptor 549 |
| 7 | gene | 102.55792 | 102.56549 | negative | MGI_C57BL6J_106657 | Trim21 | NCBI_Gene:20821,ENSEMBL:ENSMUSG00000030966 | MGI:106657 | protein coding gene | tripartite motif-containing 21 |
| 7 | pseudogene | 102.56848 | 102.56973 | negative | MGI_C57BL6J_3645143 | Gm5339 | NCBI_Gene:384699,ENSEMBL:ENSMUSG00000109756 | MGI:3645143 | pseudogene | predicted gene 5339 |
| 7 | gene | 102.57143 | 102.58301 | positive | MGI_C57BL6J_3030384 | Olfr550 | NCBI_Gene:259108,ENSEMBL:ENSMUSG00000073975 | MGI:3030384 | protein coding gene | olfactory receptor 550 |
| 7 | gene | 102.58351 | 102.59189 | negative | MGI_C57BL6J_3030385 | Olfr551 | NCBI_Gene:258750,ENSEMBL:ENSMUSG00000073974 | MGI:3030385 | protein coding gene | olfactory receptor 551 |
| 7 | gene | 102.59777 | 102.60813 | positive | MGI_C57BL6J_3030386 | Olfr552 | NCBI_Gene:259106,ENSEMBL:ENSMUSG00000073973 | MGI:3030386 | protein coding gene | olfactory receptor 552 |
| 7 | gene | 102.61128 | 102.61822 | negative | MGI_C57BL6J_3030387 | Olfr553 | NCBI_Gene:233578,ENSEMBL:ENSMUSG00000073972 | MGI:3030387 | protein coding gene | olfactory receptor 553 |
| 7 | gene | 102.63843 | 102.64256 | positive | MGI_C57BL6J_3030388 | Olfr554 | NCBI_Gene:258322,ENSEMBL:ENSMUSG00000073971 | MGI:3030388 | protein coding gene | olfactory receptor 554 |
| 7 | pseudogene | 102.64763 | 102.64822 | negative | MGI_C57BL6J_3781174 | Gm2996 | NCBI_Gene:100040840,ENSEMBL:ENSMUSG00000110251 | MGI:3781174 | pseudogene | predicted gene 2996 |
| 7 | gene | 102.65882 | 102.65977 | positive | MGI_C57BL6J_3030389 | Olfr555 | NCBI_Gene:259107,ENSEMBL:ENSMUSG00000073970 | MGI:3030389 | protein coding gene | olfactory receptor 555 |
| 7 | gene | 102.66490 | 102.67327 | positive | MGI_C57BL6J_3030390 | Olfr556 | NCBI_Gene:258749,ENSEMBL:ENSMUSG00000073969 | MGI:3030390 | protein coding gene | olfactory receptor 556 |
| 7 | pseudogene | 102.67654 | 102.67669 | positive | MGI_C57BL6J_5791523 | Gm45687 | ENSEMBL:ENSMUSG00000110352 | MGI:5791523 | pseudogene | predicted gene 45687 |
| 7 | gene | 102.67758 | 102.68733 | negative | MGI_C57BL6J_2142077 | Trim68 | NCBI_Gene:101700,ENSEMBL:ENSMUSG00000073968 | MGI:2142077 | protein coding gene | tripartite motif-containing 68 |
| 7 | gene | 102.68545 | 102.68622 | negative | MGI_C57BL6J_5791363 | Gm45527 | ENSEMBL:ENSMUSG00000109618 | MGI:5791363 | unclassified gene | predicted gene 45527 |
| 7 | gene | 102.69591 | 102.70071 | positive | MGI_C57BL6J_3030391 | Olfr557 | NCBI_Gene:258358,ENSEMBL:ENSMUSG00000073967 | MGI:3030391 | protein coding gene | olfactory receptor 557 |
| 7 | gene | 102.70221 | 102.71206 | positive | MGI_C57BL6J_3030392 | Olfr558 | NCBI_Gene:259097,ENSEMBL:ENSMUSG00000070423 | MGI:3030392 | protein coding gene | olfactory receptor 558 |
| 7 | pseudogene | 102.71138 | 102.71148 | negative | MGI_C57BL6J_5791524 | Gm45688 | ENSEMBL:ENSMUSG00000110376 | MGI:5791524 | pseudogene | predicted gene 45688 |
| 7 | gene | 102.71215 | 102.71907 | negative | MGI_C57BL6J_109302 | Olfr33 | NCBI_Gene:18332,ENSEMBL:ENSMUSG00000066273 | MGI:109302 | protein coding gene | olfactory receptor 33 |
| 7 | gene | 102.72321 | 102.72749 | negative | MGI_C57BL6J_3030393 | Olfr559 | NCBI_Gene:259116,ENSEMBL:ENSMUSG00000066272 | MGI:3030393 | protein coding gene | olfactory receptor 559 |
| 7 | gene | 102.73458 | 102.73471 | negative | MGI_C57BL6J_5453987 | Gm24210 | ENSEMBL:ENSMUSG00000088888 | MGI:5453987 | snoRNA gene | predicted gene, 24210 |
| 7 | gene | 102.73851 | 102.75947 | negative | MGI_C57BL6J_2157548 | Olfr78 | NCBI_Gene:170639,ENSEMBL:ENSMUSG00000043366 | MGI:2157548 | protein coding gene | olfactory receptor 78 |
| 7 | gene | 102.75182 | 102.75937 | negative | MGI_C57BL6J_3030394 | Olfr560 | NCBI_Gene:259117,ENSEMBL:ENSMUSG00000110008 | MGI:3030394 | protein coding gene | olfactory receptor 560 |
| 7 | gene | 102.77122 | 102.77686 | positive | MGI_C57BL6J_3030395 | Olfr561 | NCBI_Gene:259096,ENSEMBL:ENSMUSG00000073966 | MGI:3030395 | protein coding gene | olfactory receptor 561 |
| 7 | pseudogene | 102.78148 | 102.78243 | positive | MGI_C57BL6J_3030396 | Olfr562-ps1 | NCBI_Gene:259090,ENSEMBL:ENSMUSG00000110122 | MGI:3030396 | polymorphic pseudogene | olfactory receptor 562, pseudogene 1 |
| 7 | pseudogene | 102.79125 | 102.79234 | positive | MGI_C57BL6J_3030397 | Olfr563-ps1 | NCBI_Gene:259157,ENSEMBL:ENSMUSG00000110074 | MGI:3030397 | pseudogene | olfactory receptor 563, pseudogene 1 |
| 7 | gene | 102.80348 | 102.80443 | positive | MGI_C57BL6J_3030398 | Olfr564 | NCBI_Gene:258356,ENSEMBL:ENSMUSG00000048469 | MGI:3030398 | protein coding gene | olfactory receptor 564 |
| 7 | gene | 102.81331 | 102.90171 | positive | MGI_C57BL6J_3030404 | Olfr570 | NCBI_Gene:259114,ENSEMBL:ENSMUSG00000073964 | MGI:3030404 | protein coding gene | olfactory receptor 570 |
| 7 | pseudogene | 102.81755 | 102.81849 | negative | MGI_C57BL6J_3030399 | Olfr565-ps1 | NCBI_Gene:258142,ENSEMBL:ENSMUSG00000109596 | MGI:3030399 | pseudogene | olfactory receptor 565, pseudogene 1 |
| 7 | gene | 102.85534 | 102.85877 | negative | MGI_C57BL6J_3030400 | Olfr566 | NCBI_Gene:258168,ENSEMBL:ENSMUSG00000060888 | MGI:3030400 | protein coding gene | olfactory receptor 566 |
| 7 | pseudogene | 102.86116 | 102.86129 | negative | MGI_C57BL6J_5791525 | Gm45689 | ENSEMBL:ENSMUSG00000110152 | MGI:5791525 | pseudogene | predicted gene 45689 |
| 7 | pseudogene | 102.86392 | 102.86478 | negative | MGI_C57BL6J_3030401 | Olfr567-ps1 | NCBI_Gene:258140,ENSEMBL:ENSMUSG00000109878 | MGI:3030401 | pseudogene | olfactory receptor 567, pseudogene 1 |
| 7 | gene | 102.87712 | 102.87806 | positive | MGI_C57BL6J_3030402 | Olfr568 | NCBI_Gene:259095,ENSEMBL:ENSMUSG00000073965 | MGI:3030402 | protein coding gene | olfactory receptor 568 |
| 7 | gene | 102.88371 | 102.89567 | negative | MGI_C57BL6J_3030403 | Olfr569 | NCBI_Gene:259092,ENSEMBL:ENSMUSG00000062142 | MGI:3030403 | protein coding gene | olfactory receptor 569 |
| 7 | gene | 102.90509 | 102.91754 | negative | MGI_C57BL6J_3030405 | Olfr571 | NCBI_Gene:259089,ENSEMBL:ENSMUSG00000043310 | MGI:3030405 | protein coding gene | olfactory receptor 571 |
| 7 | gene | 102.92441 | 102.92861 | positive | MGI_C57BL6J_3030406 | Olfr572 | NCBI_Gene:259093,ENSEMBL:ENSMUSG00000073963 | MGI:3030406 | protein coding gene | olfactory receptor 572 |
| 7 | pseudogene | 102.94164 | 102.94258 | negative | MGI_C57BL6J_3030407 | Olfr573-ps1 | NCBI_Gene:258230,ENSEMBL:ENSMUSG00000052785 | MGI:3030407 | pseudogene | olfactory receptor 573, pseudogene 1 |
| 7 | gene | 102.94847 | 102.94951 | positive | MGI_C57BL6J_3030408 | Olfr574 | NCBI_Gene:258357,ENSEMBL:ENSMUSG00000045824 | MGI:3030408 | protein coding gene | olfactory receptor 574 |
| 7 | gene | 102.95227 | 102.95878 | negative | MGI_C57BL6J_3030409 | Olfr575 | NCBI_Gene:259118,ENSEMBL:ENSMUSG00000066269 | MGI:3030409 | protein coding gene | olfactory receptor 575 |
| 7 | gene | 102.96266 | 102.97516 | positive | MGI_C57BL6J_3030410 | Olfr576 | NCBI_Gene:258248,ENSEMBL:ENSMUSG00000073962 | MGI:3030410 | protein coding gene | olfactory receptor 576 |
| 7 | gene | 102.97118 | 102.97594 | negative | MGI_C57BL6J_3030411 | Olfr577 | NCBI_Gene:259113,ENSEMBL:ENSMUSG00000043354 | MGI:3030411 | protein coding gene | olfactory receptor 577 |
| 7 | gene | 102.98161 | 102.98932 | negative | MGI_C57BL6J_3030412 | Olfr578 | NCBI_Gene:259119,ENSEMBL:ENSMUSG00000045792 | MGI:3030412 | protein coding gene | olfactory receptor 578 |
| 7 | pseudogene | 102.99893 | 102.99975 | negative | MGI_C57BL6J_3031361 | Olfr1527-ps1 | NCBI_Gene:259158 | MGI:3031361 | pseudogene | olfactory receptor 1527, pseudogene 1 |
| 7 | pseudogene | 103.00461 | 103.00520 | negative | MGI_C57BL6J_3030413 | Olfr579-ps1 | NCBI_Gene:258141 | MGI:3030413 | pseudogene | olfactory receptor 579, pseudogene 1 |
| 7 | pseudogene | 103.01314 | 103.01449 | negative | MGI_C57BL6J_3030414 | Olfr580-ps1 | NCBI_Gene:258213,ENSEMBL:ENSMUSG00000110143 | MGI:3030414 | pseudogene | olfactory receptor 580, pseudogene 1 |
| 7 | pseudogene | 103.02909 | 103.02965 | positive | MGI_C57BL6J_3030415 | Olfr581-ps1 | NCBI_Gene:404406,ENSEMBL:ENSMUSG00000109845 | MGI:3030415 | pseudogene | olfactory receptor 581, pseudogene 1 |
| 7 | gene | 103.03802 | 103.04404 | positive | MGI_C57BL6J_3030416 | Olfr582 | NCBI_Gene:259055,ENSEMBL:ENSMUSG00000073961 | MGI:3030416 | protein coding gene | olfactory receptor 582 |
| 7 | gene | 103.05130 | 103.05226 | positive | MGI_C57BL6J_3030417 | Olfr583 | NCBI_Gene:258752,ENSEMBL:ENSMUSG00000073960 | MGI:3030417 | protein coding gene | olfactory receptor 583 |
| 7 | gene | 103.08188 | 103.08831 | positive | MGI_C57BL6J_3030418 | Olfr584 | NCBI_Gene:259056,ENSEMBL:ENSMUSG00000073959 | MGI:3030418 | protein coding gene | olfactory receptor 584 |
| 7 | gene | 103.09772 | 103.09875 | positive | MGI_C57BL6J_3030419 | Olfr585 | NCBI_Gene:259091,ENSEMBL:ENSMUSG00000078080 | MGI:3030419 | protein coding gene | olfactory receptor 585 |
| 7 | gene | 103.11840 | 103.12564 | negative | MGI_C57BL6J_3030420 | Olfr586 | NCBI_Gene:259115,ENSEMBL:ENSMUSG00000066268 | MGI:3030420 | protein coding gene | olfactory receptor 586 |
| 7 | pseudogene | 103.13438 | 103.13532 | positive | MGI_C57BL6J_3030421 | Olfr587-ps1 | NCBI_Gene:404407,ENSEMBL:ENSMUSG00000080891 | MGI:3030421 | polymorphic pseudogene | olfactory receptor 587, pseudogene 1 |
| 7 | pseudogene | 103.13574 | 103.13604 | positive | MGI_C57BL6J_3705492 | Gm15120 | ENSEMBL:ENSMUSG00000082714 | MGI:3705492 | pseudogene | predicted gene 15120 |
| 7 | pseudogene | 103.14088 | 103.14786 | positive | MGI_C57BL6J_3030422 | Olfr588-ps1 | NCBI_Gene:259094,ENSEMBL:ENSMUSG00000084262 | MGI:3030422 | pseudogene | olfactory receptor 588, pseudogene 1 |
| 7 | pseudogene | 103.14799 | 103.14867 | negative | MGI_C57BL6J_3705687 | Gm15116 | NCBI_Gene:100417214,ENSEMBL:ENSMUSG00000083254 | MGI:3705687 | pseudogene | predicted gene 15116 |
| 7 | gene | 103.15479 | 103.15636 | negative | MGI_C57BL6J_3030423 | Olfr589 | NCBI_Gene:259054,ENSEMBL:ENSMUSG00000051362 | MGI:3030423 | protein coding gene | olfactory receptor 589 |
| 7 | pseudogene | 103.16585 | 103.16675 | positive | MGI_C57BL6J_3030424 | Olfr590-ps1 | NCBI_Gene:258170,ENSEMBL:ENSMUSG00000084356 | MGI:3030424 | pseudogene | olfactory receptor 590, pseudogene 1 |
| 7 | gene | 103.16707 | 103.17834 | negative | MGI_C57BL6J_3030425 | Olfr591 | NCBI_Gene:258139,ENSEMBL:ENSMUSG00000057461 | MGI:3030425 | protein coding gene | olfactory receptor 591 |
| 7 | gene | 103.18480 | 103.19297 | positive | MGI_C57BL6J_3030426 | Olfr592 | NCBI_Gene:404317,ENSEMBL:ENSMUSG00000073956 | MGI:3030426 | protein coding gene | olfactory receptor 592 |
| 7 | gene | 103.21186 | 103.21285 | positive | MGI_C57BL6J_3030427 | Olfr593 | NCBI_Gene:258378,ENSEMBL:ENSMUSG00000073955 | MGI:3030427 | protein coding gene | olfactory receptor 593 |
| 7 | gene | 103.21700 | 103.22231 | positive | MGI_C57BL6J_3030428 | Olfr594 | NCBI_Gene:258246,ENSEMBL:ENSMUSG00000073954 | MGI:3030428 | protein coding gene | olfactory receptor 594 |
| 7 | pseudogene | 103.23563 | 103.23659 | positive | MGI_C57BL6J_3030429 | Olfr595-ps1 | NCBI_Gene:258175,ENSEMBL:ENSMUSG00000083383 | MGI:3030429 | pseudogene | olfactory receptor 595, pseudogene 1 |
| 7 | gene | 103.24974 | 103.25250 | negative | MGI_C57BL6J_3051372 | Usp17ld | NCBI_Gene:384701,ENSEMBL:ENSMUSG00000057321 | MGI:3051372 | protein coding gene | ubiquitin specific peptidase 17-like D |
| 7 | gene | 103.26088 | 103.26100 | negative | MGI_C57BL6J_5454948 | Gm25171 | ENSEMBL:ENSMUSG00000095000 | MGI:5454948 | miRNA gene | predicted gene, 25171 |
| 7 | gene | 103.26712 | 103.26723 | negative | MGI_C57BL6J_5453327 | Gm23550 | ENSEMBL:ENSMUSG00000094572 | MGI:5453327 | miRNA gene | predicted gene, 23550 |
| 7 | gene | 103.27155 | 103.27187 | positive | MGI_C57BL6J_5453750 | Gm23973 | ENSEMBL:ENSMUSG00000065254 | MGI:5453750 | unclassified non-coding RNA gene | predicted gene, 23973 |
| 7 | pseudogene | 103.29700 | 103.30042 | positive | MGI_C57BL6J_5825651 | Gm46014 | NCBI_Gene:108167486 | MGI:5825651 | pseudogene | predicted gene, 46014 |
| 7 | gene | 103.30565 | 103.31278 | positive | MGI_C57BL6J_3030430 | Olfr596 | NCBI_Gene:100041187,ENSEMBL:ENSMUSG00000073953 | MGI:3030430 | protein coding gene | olfactory receptor 596 |
| 7 | gene | 103.31632 | 103.32280 | positive | MGI_C57BL6J_3030431 | Olfr597 | NCBI_Gene:258135,ENSEMBL:ENSMUSG00000073952 | MGI:3030431 | protein coding gene | olfactory receptor 597 |
| 7 | gene | 103.32357 | 103.32989 | positive | MGI_C57BL6J_3030432 | Olfr598 | NCBI_Gene:257975,ENSEMBL:ENSMUSG00000073951 | MGI:3030432 | protein coding gene | olfactory receptor 598 |
| 7 | gene | 103.33592 | 103.33956 | positive | MGI_C57BL6J_3030433 | Olfr599 | NCBI_Gene:258726,ENSEMBL:ENSMUSG00000073950 | MGI:3030433 | protein coding gene | olfactory receptor 599 |
| 7 | gene | 103.34348 | 103.34967 | negative | MGI_C57BL6J_3030434 | Olfr600 | NCBI_Gene:259048,ENSEMBL:ENSMUSG00000045540 | MGI:3030434 | protein coding gene | olfactory receptor 600 |
| 7 | gene | 103.35709 | 103.36266 | negative | MGI_C57BL6J_3030435 | Olfr601 | NCBI_Gene:258311,ENSEMBL:ENSMUSG00000109951 | MGI:3030435 | protein coding gene | olfactory receptor 601 |
| 7 | pseudogene | 103.36752 | 103.37244 | negative | MGI_C57BL6J_3030436 | Olfr602-ps1 | NCBI_Gene:257976,ENSEMBL:ENSMUSG00000082739 | MGI:3030436 | pseudogene | olfactory receptor 602, pseudogene 1 |
| 7 | gene | 103.38306 | 103.38618 | negative | MGI_C57BL6J_3030437 | Olfr603 | NCBI_Gene:259073,ENSEMBL:ENSMUSG00000059874 | MGI:3030437 | protein coding gene | olfactory receptor 603 |
| 7 | gene | 103.41507 | 103.41988 | positive | MGI_C57BL6J_107698 | Usp17lc | NCBI_Gene:13532,ENSEMBL:ENSMUSG00000058976 | MGI:107698 | protein coding gene | ubiquitin specific peptidase 17-like C |
| 7 | gene | 103.41581 | 103.41593 | positive | MGI_C57BL6J_5455551 | Gm25774 | ENSEMBL:ENSMUSG00000080367 | MGI:5455551 | miRNA gene | predicted gene, 25774 |
| 7 | gene | 103.44041 | 103.44544 | negative | MGI_C57BL6J_3030439 | Olfr605 | NCBI_Gene:258156,ENSEMBL:ENSMUSG00000109659 | MGI:3030439 | protein coding gene | olfactory receptor 605 |
| 7 | gene | 103.44905 | 103.45353 | positive | MGI_C57BL6J_3030440 | Olfr606 | NCBI_Gene:259098,ENSEMBL:ENSMUSG00000073949 | MGI:3030440 | protein coding gene | olfactory receptor 606 |
| 7 | gene | 103.45553 | 103.46415 | negative | MGI_C57BL6J_3030441 | Olfr607 | NCBI_Gene:546989,ENSEMBL:ENSMUSG00000081945 | MGI:3030441 | protein coding gene | olfactory receptor 607 |
| 7 | gene | 103.46988 | 103.47136 | positive | MGI_C57BL6J_3030442 | Olfr608 | NCBI_Gene:258751,ENSEMBL:ENSMUSG00000073948 | MGI:3030442 | protein coding gene | olfactory receptor 608 |
| 7 | gene | 103.49003 | 103.49806 | negative | MGI_C57BL6J_3030443 | Olfr609 | NCBI_Gene:259086,ENSEMBL:ENSMUSG00000046396 | MGI:3030443 | protein coding gene | olfactory receptor 609 |
| 7 | gene | 103.50538 | 103.51212 | negative | MGI_C57BL6J_3030444 | Olfr610 | NCBI_Gene:259085,ENSEMBL:ENSMUSG00000045584 | MGI:3030444 | protein coding gene | olfactory receptor 610 |
| 7 | gene | 103.51646 | 103.52085 | negative | MGI_C57BL6J_3030445 | Olfr611 | NCBI_Gene:258722,ENSEMBL:ENSMUSG00000096584 | MGI:3030445 | protein coding gene | olfactory receptor 611 |
| 7 | gene | 103.53722 | 103.54272 | negative | MGI_C57BL6J_3030446 | Olfr612 | NCBI_Gene:545985,ENSEMBL:ENSMUSG00000094119 | MGI:3030446 | protein coding gene | olfactory receptor 612 |
| 7 | gene | 103.54807 | 103.55551 | positive | MGI_C57BL6J_3030447 | Olfr613 | NCBI_Gene:259104,ENSEMBL:ENSMUSG00000078624 | MGI:3030447 | protein coding gene | olfactory receptor 613 |
| 7 | gene | 103.55608 | 103.56380 | positive | MGI_C57BL6J_3030449 | Olfr615 | NCBI_Gene:259084,ENSEMBL:ENSMUSG00000073947 | MGI:3030449 | protein coding gene | olfactory receptor 615 |
| 7 | gene | 103.56375 | 103.56914 | negative | MGI_C57BL6J_3030450 | Olfr616 | NCBI_Gene:259103,ENSEMBL:ENSMUSG00000047544 | MGI:3030450 | protein coding gene | olfactory receptor 616 |
| 7 | gene | 103.57306 | 103.58679 | positive | MGI_C57BL6J_3030451 | Olfr617 | NCBI_Gene:258838,ENSEMBL:ENSMUSG00000073946 | MGI:3030451 | protein coding gene | olfactory receptor 617 |
| 7 | gene | 103.58127 | 103.58138 | positive | MGI_C57BL6J_5456283 | Gm26506 | ENSEMBL:ENSMUSG00000096906 | MGI:5456283 | miRNA gene | predicted gene, 26506 |
| 7 | gene | 103.59010 | 103.60045 | positive | MGI_C57BL6J_3030452 | Olfr618 | NCBI_Gene:259049,ENSEMBL:ENSMUSG00000073945 | MGI:3030452 | protein coding gene | olfactory receptor 618 |
| 7 | gene | 103.60058 | 103.60486 | positive | MGI_C57BL6J_3030453 | Olfr619 | NCBI_Gene:259080,ENSEMBL:ENSMUSG00000073944 | MGI:3030453 | protein coding gene | olfactory receptor 619 |
| 7 | gene | 103.60489 | 103.61253 | negative | MGI_C57BL6J_3030454 | Olfr620 | NCBI_Gene:258808,ENSEMBL:ENSMUSG00000045132 | MGI:3030454 | protein coding gene | olfactory receptor 620 |
| 7 | pseudogene | 103.62902 | 103.62996 | negative | MGI_C57BL6J_3030455 | Olfr621-ps1 | NCBI_Gene:667639,ENSEMBL:ENSMUSG00000081315 | MGI:3030455 | pseudogene | olfactory receptor 621, pseudogene 1 |
| 7 | gene | 103.63845 | 103.64270 | negative | MGI_C57BL6J_3030456 | Olfr622 | NCBI_Gene:259087,ENSEMBL:ENSMUSG00000050085 | MGI:3030456 | protein coding gene | olfactory receptor 622 |
| 7 | gene | 103.65770 | 103.66370 | negative | MGI_C57BL6J_3030457 | Olfr623 | NCBI_Gene:259126,ENSEMBL:ENSMUSG00000099687 | MGI:3030457 | protein coding gene | olfactory receptor 623 |
| 7 | gene | 103.66546 | 103.67119 | negative | MGI_C57BL6J_3030458 | Olfr624 | NCBI_Gene:258189,ENSEMBL:ENSMUSG00000045780 | MGI:3030458 | protein coding gene | olfactory receptor 624 |
| 7 | pseudogene | 103.68272 | 103.68368 | positive | MGI_C57BL6J_3030459 | Olfr625-ps1 | NCBI_Gene:258169,ENSEMBL:ENSMUSG00000073943 | MGI:3030459 | pseudogene | olfactory receptor 625, pseudogene 1 |
| 7 | pseudogene | 103.69157 | 103.69212 | negative | MGI_C57BL6J_3030460 | Olfr626-ps1 | NCBI_Gene:404366,ENSEMBL:ENSMUSG00000081038 | MGI:3030460 | pseudogene | olfactory receptor 626, pseudogene 1 |
| 7 | pseudogene | 103.69958 | 103.70075 | negative | MGI_C57BL6J_3030094 | Olfr260-ps1 | NCBI_Gene:404365,ENSEMBL:ENSMUSG00000082077 | MGI:3030094 | pseudogene | olfactory receptor 260, pseudogene 1 |
| 7 | gene | 103.71467 | 103.72117 | positive | MGI_C57BL6J_3030077 | Olfr243 | NCBI_Gene:436002,ENSEMBL:ENSMUSG00000094822 | MGI:3030077 | protein coding gene | olfactory receptor 243 |
| 7 | gene | 103.73037 | 103.73332 | positive | MGI_C57BL6J_3030462 | Olfr628 | NCBI_Gene:259159,ENSEMBL:ENSMUSG00000096516 | MGI:3030462 | protein coding gene | olfactory receptor 628 |
| 7 | gene | 103.73977 | 103.74494 | negative | MGI_C57BL6J_3030463 | Olfr629 | NCBI_Gene:258818,ENSEMBL:ENSMUSG00000047545 | MGI:3030463 | protein coding gene | olfactory receptor 629 |
| 7 | gene | 103.75348 | 103.75735 | negative | MGI_C57BL6J_3030464 | Olfr630 | NCBI_Gene:259102,ENSEMBL:ENSMUSG00000050281 | MGI:3030464 | protein coding gene | olfactory receptor 630 |
| 7 | gene | 103.76581 | 103.77159 | negative | MGI_C57BL6J_1341789 | Olfr69 | NCBI_Gene:18370,ENSEMBL:ENSMUSG00000058662 | MGI:1341789 | protein coding gene | olfactory receptor 69 |
| 7 | gene | 103.77436 | 103.77970 | negative | MGI_C57BL6J_1341790 | Olfr68 | NCBI_Gene:18369,ENSEMBL:ENSMUSG00000061626 | MGI:1341790 | protein coding gene | olfactory receptor 68 |
| 7 | gene | 103.78459 | 103.79183 | negative | MGI_C57BL6J_1341911 | Olfr67 | NCBI_Gene:18368,ENSEMBL:ENSMUSG00000047535 | MGI:1341911 | protein coding gene | olfactory receptor 67 |
| 7 | gene | 103.81252 | 103.81400 | negative | MGI_C57BL6J_5474850 | Hbb-bt | NCBI_Gene:101488143,ENSEMBL:ENSMUSG00000073940 | MGI:5474850 | protein coding gene | hemoglobin, beta adult t chain |
| 7 | gene | 103.82652 | 103.82810 | negative | MGI_C57BL6J_5474852 | Hbb-bs | NCBI_Gene:100503605,ENSEMBL:ENSMUSG00000052305 | MGI:5474852 | protein coding gene | hemoglobin, beta adult s chain |
| 7 | pseudogene | 103.82959 | 103.82996 | positive | MGI_C57BL6J_3642219 | Gm15115 | NCBI_Gene:108167487,ENSEMBL:ENSMUSG00000083967 | MGI:3642219 | pseudogene | predicted gene 15115 |
| 7 | pseudogene | 103.83263 | 103.83373 | negative | MGI_C57BL6J_96026 | Hbb-bh3 | NCBI_Gene:15134,ENSEMBL:ENSMUSG00000083216 | MGI:96026 | pseudogene | hemoglobin beta, pseudogene bh3 |
| 7 | gene | 103.83880 | 103.84073 | negative | MGI_C57BL6J_96025 | Hbb-bh2 | NCBI_Gene:436003,ENSEMBL:ENSMUSG00000078621 | MGI:96025 | protein coding gene | hemoglobin beta, bh2 |
| 7 | gene | 103.84164 | 103.84316 | negative | MGI_C57BL6J_96024 | Hbb-bh1 | NCBI_Gene:15132,ENSEMBL:ENSMUSG00000052217 | MGI:96024 | protein coding gene | hemoglobin Z, beta-like embryonic chain |
| 7 | pseudogene | 103.85002 | 103.85015 | negative | MGI_C57BL6J_96023 | Hbb-bh0 | ENSEMBL:ENSMUSG00000085700 | MGI:96023 | pseudogene | hemoglobin, beta, pseudogene bh0 |
| 7 | gene | 103.85175 | 103.85322 | negative | MGI_C57BL6J_96027 | Hbb-y | NCBI_Gene:15135,ENSEMBL:ENSMUSG00000052187 | MGI:96027 | protein coding gene | hemoglobin Y, beta-like embryonic chain |
| 7 | gene | 103.87944 | 103.88625 | negative | MGI_C57BL6J_1341906 | Olfr66 | NCBI_Gene:18367,ENSEMBL:ENSMUSG00000058200 | MGI:1341906 | protein coding gene | olfactory receptor 66 |
| 7 | pseudogene | 103.88257 | 103.88310 | negative | MGI_C57BL6J_3709029 | Dnajc19-ps | NCBI_Gene:100503724,ENSEMBL:ENSMUSG00000083854 | MGI:3709029 | pseudogene | DnaJ heat shock protein family (Hsp40) member C19, pseudogene |
| 7 | gene | 103.88948 | 103.89449 | negative | MGI_C57BL6J_1341900 | Olfr64 | NCBI_Gene:18366,ENSEMBL:ENSMUSG00000063615 | MGI:1341900 | protein coding gene | olfactory receptor 64 |
| 7 | gene | 103.90349 | 103.90865 | positive | MGI_C57BL6J_1341910 | Olfr65 | NCBI_Gene:18365,ENSEMBL:ENSMUSG00000110259 | MGI:1341910 | protein coding gene | olfactory receptor 65 |
| 7 | gene | 103.91506 | 103.93067 | positive | MGI_C57BL6J_3030465 | Olfr631 | NCBI_Gene:258961,ENSEMBL:ENSMUSG00000042219 | MGI:3030465 | protein coding gene | olfactory receptor 631 |
| 7 | gene | 103.93435 | 103.94058 | positive | MGI_C57BL6J_3030466 | Olfr632 | NCBI_Gene:259123,ENSEMBL:ENSMUSG00000073938 | MGI:3030466 | protein coding gene | olfactory receptor 632 |
| 7 | pseudogene | 103.93975 | 103.93991 | positive | MGI_C57BL6J_5791491 | Gm45655 | ENSEMBL:ENSMUSG00000109837 | MGI:5791491 | pseudogene | predicted gene 45655 |
| 7 | gene | 103.94444 | 103.94896 | positive | MGI_C57BL6J_3030467 | Olfr633 | NCBI_Gene:258351,ENSEMBL:ENSMUSG00000073937 | MGI:3030467 | protein coding gene | olfactory receptor 633 |
| 7 | pseudogene | 103.95641 | 103.95737 | positive | MGI_C57BL6J_3030468 | Olfr634-ps1 | NCBI_Gene:257949,ENSEMBL:ENSMUSG00000080751 | MGI:3030468 | polymorphic pseudogene | olfactory receptor 634, pseudogene 1 |
| 7 | gene | 103.97115 | 103.98754 | positive | MGI_C57BL6J_3030469 | Olfr635 | NCBI_Gene:259122,ENSEMBL:ENSMUSG00000094520 | MGI:3030469 | protein coding gene | olfactory receptor 635 |
| 7 | pseudogene | 103.97336 | 103.97711 | negative | MGI_C57BL6J_3645929 | Gm4886 | NCBI_Gene:233614,ENSEMBL:ENSMUSG00000082281 | MGI:3645929 | pseudogene | predicted gene 4886 |
| 7 | pseudogene | 103.98910 | 103.99004 | negative | MGI_C57BL6J_3030470 | Olfr636-ps1 | NCBI_Gene:258144,ENSEMBL:ENSMUSG00000055621 | MGI:3030470 | pseudogene | olfactory receptor 636, pseudogene 1 |
| 7 | pseudogene | 103.99598 | 103.99664 | positive | MGI_C57BL6J_3030471 | Olfr637-ps1 | NCBI_Gene:258143,ENSEMBL:ENSMUSG00000084236 | MGI:3030471 | pseudogene | olfactory receptor 637, pseudogene 1 |
| 7 | gene | 103.99875 | 104.00436 | positive | MGI_C57BL6J_3030472 | Olfr638 | NCBI_Gene:259124,ENSEMBL:ENSMUSG00000094063 | MGI:3030472 | protein coding gene | olfactory receptor 638 |
| 7 | gene | 103.99880 | 104.13741 | negative | MGI_C57BL6J_3030477 | Olfr643 | NCBI_Gene:259081,ENSEMBL:ENSMUSG00000109824 | MGI:3030477 | protein coding gene | olfactory receptor 643 |
| 7 | gene | 104.00623 | 104.01730 | negative | MGI_C57BL6J_3030473 | Olfr639 | NCBI_Gene:259088,ENSEMBL:ENSMUSG00000066263 | MGI:3030473 | protein coding gene | olfactory receptor 639 |
| 7 | gene | 104.01869 | 104.02557 | negative | MGI_C57BL6J_3030474 | Olfr640 | NCBI_Gene:258819,ENSEMBL:ENSMUSG00000066262 | MGI:3030474 | protein coding gene | olfactory receptor 640 |
| 7 | gene | 104.03544 | 104.04207 | positive | MGI_C57BL6J_3030475 | Olfr641 | NCBI_Gene:259075,ENSEMBL:ENSMUSG00000073932 | MGI:3030475 | protein coding gene | olfactory receptor 641 |
| 7 | gene | 104.04653 | 104.05267 | negative | MGI_C57BL6J_3030476 | Olfr642 | NCBI_Gene:258326,ENSEMBL:ENSMUSG00000049797 | MGI:3030476 | protein coding gene | olfactory receptor 642 |
| 7 | gene | 104.05360 | 104.06145 | negative | MGI_C57BL6J_1924125 | 4930516K23Rik | ENSEMBL:ENSMUSG00000090219 | MGI:1924125 | protein coding gene | RIKEN cDNA 4930516K23 gene |
| 7 | gene | 104.06629 | 104.07187 | negative | MGI_C57BL6J_3030478 | Olfr644 | NCBI_Gene:259125,ENSEMBL:ENSMUSG00000110012 | MGI:3030478 | protein coding gene | olfactory receptor 644 |
| 7 | gene | 104.08179 | 104.08546 | negative | MGI_C57BL6J_3030479 | Olfr645 | NCBI_Gene:258247,ENSEMBL:ENSMUSG00000051340 | MGI:3030479 | protein coding gene | olfactory receptor 645 |
| 7 | gene | 104.09732 | 104.10977 | positive | MGI_C57BL6J_3030480 | Olfr646 | NCBI_Gene:259058,ENSEMBL:ENSMUSG00000073931 | MGI:3030480 | protein coding gene | olfactory receptor 646 |
| 7 | gene | 104.12791 | 104.12983 | negative | MGI_C57BL6J_1918230 | Ubqln5 | NCBI_Gene:70980,ENSEMBL:ENSMUSG00000055643 | MGI:1918230 | protein coding gene | ubiquilin 5 |
| 7 | gene | 104.14062 | 104.14332 | negative | MGI_C57BL6J_3045291 | Ubqln3 | NCBI_Gene:244178,ENSEMBL:ENSMUSG00000051618 | MGI:3045291 | protein coding gene | ubiquilin 3 |
| 7 | gene | 104.14826 | 104.15056 | negative | MGI_C57BL6J_2685336 | Ubqlnl | NCBI_Gene:244179,ENSEMBL:ENSMUSG00000051437 | MGI:2685336 | protein coding gene | ubiquilin-like |
| 7 | gene | 104.15301 | 104.16483 | negative | MGI_C57BL6J_2443346 | Olfm5 | NCBI_Gene:244180,ENSEMBL:ENSMUSG00000044265 | MGI:2443346 | protein coding gene | olfactomedin 5 |
| 7 | pseudogene | 104.16738 | 104.16834 | negative | MGI_C57BL6J_3030481 | Olfr647-ps1 | NCBI_Gene:258137,ENSEMBL:ENSMUSG00000083513 | MGI:3030481 | pseudogene | olfactory receptor 647, pseudogene 1 |
| 7 | gene | 104.17760 | 104.18339 | negative | MGI_C57BL6J_3030482 | Olfr648 | NCBI_Gene:258746,ENSEMBL:ENSMUSG00000042909 | MGI:3030482 | protein coding gene | olfactory receptor 648 |
| 7 | gene | 104.18899 | 104.19598 | negative | MGI_C57BL6J_3030483 | Olfr649 | NCBI_Gene:259057,ENSEMBL:ENSMUSG00000044899 | MGI:3030483 | protein coding gene | olfactory receptor 649 |
| 7 | pseudogene | 104.20344 | 104.20786 | positive | MGI_C57BL6J_3030484 | Olfr650-ps1 | NCBI_Gene:258212,ENSEMBL:ENSMUSG00000081859 | MGI:3030484 | polymorphic pseudogene | olfactory receptor 650, pseudogene 1 |
| 7 | gene | 104.21878 | 104.23515 | positive | MGI_C57BL6J_2137352 | Trim6 | NCBI_Gene:94088,ENSEMBL:ENSMUSG00000072244 | MGI:2137352 | protein coding gene | tripartite motif-containing 6 |
| 7 | pseudogene | 104.22159 | 104.22298 | negative | MGI_C57BL6J_3648748 | Gm8667 | NCBI_Gene:667497,ENSEMBL:ENSMUSG00000109712 | MGI:3648748 | pseudogene | predicted gene 8667 |
| 7 | gene | 104.24444 | 104.26224 | positive | MGI_C57BL6J_2137359 | Trim34a | NCBI_Gene:94094,ENSEMBL:ENSMUSG00000056144 | MGI:2137359 | protein coding gene | tripartite motif-containing 34A |
| 7 | gene | 104.26147 | 104.26217 | negative | MGI_C57BL6J_6096073 | Gm47248 | ENSEMBL:ENSMUSG00000111737 | MGI:6096073 | lncRNA gene | predicted gene, 47248 |
| 7 | gene | 104.26339 | 104.28842 | negative | MGI_C57BL6J_3646853 | Trim5 | NCBI_Gene:667823,ENSEMBL:ENSMUSG00000060441 | MGI:3646853 | protein coding gene | tripartite motif-containing 5 |
| 7 | gene | 104.26468 | 104.26479 | negative | MGI_C57BL6J_5454623 | Gm24846 | ENSEMBL:ENSMUSG00000088658 | MGI:5454623 | snRNA gene | predicted gene, 24846 |
| 7 | gene | 104.26522 | 104.26590 | negative | MGI_C57BL6J_4821182 | Trim12b | NA | NA | protein coding gene | tripartite motif-containing 12B |
| 7 | gene | 104.29989 | 104.31585 | negative | MGI_C57BL6J_1923931 | Trim12a | NCBI_Gene:76681,ENSEMBL:ENSMUSG00000066258 | MGI:1923931 | protein coding gene | tripartite motif-containing 12A |
| 7 | gene | 104.31051 | 104.31106 | negative | MGI_C57BL6J_1925028 | A430102O06Rik | NA | NA | unclassified gene | RIKEN cDNA A430102O06 gene |
| 7 | gene | 104.31502 | 104.32484 | positive | MGI_C57BL6J_3705170 | Gm15133 | NCBI_Gene:622070,ENSEMBL:ENSMUSG00000087175 | MGI:3705170 | lncRNA gene | predicted gene 15133 |
| 7 | pseudogene | 104.31907 | 104.32018 | negative | MGI_C57BL6J_5009952 | Gm17788 | NCBI_Gene:100048312 | MGI:5009952 | pseudogene | predicted gene, 17788 |
| 7 | gene | 104.32947 | 104.33691 | positive | MGI_C57BL6J_4821264 | Trim34b | NCBI_Gene:434218,ENSEMBL:ENSMUSG00000090215 | MGI:4821264 | protein coding gene | tripartite motif-containing 34B |
| 7 | gene | 104.33875 | 104.35336 | negative | MGI_C57BL6J_4821183 | Trim12c | NCBI_Gene:319236,ENSEMBL:ENSMUSG00000057143 | MGI:4821183 | protein coding gene | tripartite motif-containing 12C |
| 7 | gene | 104.34009 | 104.34019 | negative | MGI_C57BL6J_5531377 | Gm27995 | ENSEMBL:ENSMUSG00000098399 | MGI:5531377 | snRNA gene | predicted gene, 27995 |
| 7 | gene | 104.35182 | 104.35575 | positive | MGI_C57BL6J_5595589 | Gm36430 | NCBI_Gene:102640341 | MGI:5595589 | lncRNA gene | predicted gene, 36430 |
| 7 | gene | 104.35538 | 104.36988 | negative | MGI_C57BL6J_4821256 | Trim30b | NCBI_Gene:244183,ENSEMBL:ENSMUSG00000052749 | MGI:4821256 | protein coding gene | tripartite motif-containing 30B |
| 7 | gene | 104.35540 | 104.36988 | negative | MGI_C57BL6J_5621410 | Gm38525 | NCBI_Gene:102640171 | MGI:5621410 | protein coding gene | predicted gene, 38525 |
| 7 | gene | 104.35616 | 104.36269 | positive | MGI_C57BL6J_5825634 | Gm45997 | NCBI_Gene:108167453 | MGI:5825634 | lncRNA gene | predicted gene, 45997 |
| 7 | gene | 104.37912 | 104.40085 | negative | MGI_C57BL6J_4821257 | Trim30c | NCBI_Gene:434219,ENSEMBL:ENSMUSG00000078616 | MGI:4821257 | protein coding gene | tripartite motif-containing 30C |
| 7 | pseudogene | 104.40540 | 104.40653 | negative | MGI_C57BL6J_5791307 | Gm45471 | ENSEMBL:ENSMUSG00000110114 | MGI:5791307 | pseudogene | predicted gene 45471 |
| 7 | gene | 104.40902 | 104.46519 | negative | MGI_C57BL6J_98178 | Trim30a | NCBI_Gene:20128,ENSEMBL:ENSMUSG00000030921 | MGI:98178 | protein coding gene | tripartite motif-containing 30A |
| 7 | gene | 104.42327 | 104.42338 | positive | MGI_C57BL6J_5455182 | Gm25405 | ENSEMBL:ENSMUSG00000064907 | MGI:5455182 | snRNA gene | predicted gene, 25405 |
| 7 | gene | 104.47001 | 104.50785 | negative | MGI_C57BL6J_3035181 | Trim30d | NCBI_Gene:209387,ENSEMBL:ENSMUSG00000057596 | MGI:3035181 | protein coding gene | tripartite motif-containing 30D |
| 7 | pseudogene | 104.49906 | 104.50215 | negative | MGI_C57BL6J_3643716 | Gm16464 | NCBI_Gene:672495,ENSEMBL:ENSMUSG00000082154 | MGI:3643716 | pseudogene | predicted gene 16464 |
| 7 | pseudogene | 104.52861 | 104.52909 | negative | MGI_C57BL6J_3643499 | Gm6574 | NCBI_Gene:625305,ENSEMBL:ENSMUSG00000083248 | MGI:3643499 | pseudogene | predicted gene 6574 |
| 7 | pseudogene | 104.53296 | 104.53536 | negative | MGI_C57BL6J_4821258 | Trim30e-ps1 | NCBI_Gene:625321,ENSEMBL:ENSMUSG00000073929 | MGI:4821258 | pseudogene | tripartite motif-containing 30E, pseudogene 1 |
| 7 | gene | 104.55013 | 104.55442 | positive | MGI_C57BL6J_3030485 | Olfr651 | NCBI_Gene:258809,ENSEMBL:ENSMUSG00000073928 | MGI:3030485 | protein coding gene | olfactory receptor 651 |
| 7 | gene | 104.56090 | 104.56863 | positive | MGI_C57BL6J_3030486 | Olfr652 | NCBI_Gene:259050,ENSEMBL:ENSMUSG00000073927 | MGI:3030486 | protein coding gene | olfactory receptor 652 |
| 7 | gene | 104.57731 | 104.58412 | positive | MGI_C57BL6J_3030487 | Olfr653 | NCBI_Gene:57250,ENSEMBL:ENSMUSG00000073926 | MGI:3030487 | protein coding gene | olfactory receptor 653 |
| 7 | gene | 104.58702 | 104.59072 | positive | MGI_C57BL6J_3030488 | Olfr654 | NCBI_Gene:258377,ENSEMBL:ENSMUSG00000073925 | MGI:3030488 | protein coding gene | olfactory receptor 654 |
| 7 | gene | 104.59530 | 104.60178 | negative | MGI_C57BL6J_3030489 | Olfr655 | NCBI_Gene:258817,ENSEMBL:ENSMUSG00000051182 | MGI:3030489 | protein coding gene | olfactory receptor 655 |
| 7 | gene | 104.61396 | 104.62160 | positive | MGI_C57BL6J_3030490 | Olfr656 | NCBI_Gene:259078,ENSEMBL:ENSMUSG00000073924 | MGI:3030490 | protein coding gene | olfactory receptor 656 |
| 7 | gene | 104.62479 | 104.63748 | positive | MGI_C57BL6J_3030491 | Olfr657 | NCBI_Gene:258309,ENSEMBL:ENSMUSG00000073923 | MGI:3030491 | protein coding gene | olfactory receptor 657 |
| 7 | pseudogene | 104.63411 | 104.63462 | negative | MGI_C57BL6J_5588865 | Gm29706 | NCBI_Gene:101055662 | MGI:5588865 | pseudogene | predicted gene, 29706 |
| 7 | pseudogene | 104.63464 | 104.63522 | positive | MGI_C57BL6J_5010596 | Gm18411 | NCBI_Gene:100417128 | MGI:5010596 | pseudogene | predicted gene, 18411 |
| 7 | gene | 104.64288 | 104.64731 | negative | MGI_C57BL6J_3030492 | Olfr658 | NCBI_Gene:259051,ENSEMBL:ENSMUSG00000070421 | MGI:3030492 | protein coding gene | olfactory receptor 658 |
| 7 | pseudogene | 104.64725 | 104.65321 | positive | MGI_C57BL6J_3582591 | Usp17le-ps | NCBI_Gene:667882,ENSEMBL:ENSMUSG00000082084 | MGI:3582591 | pseudogene | ubiquitin specific peptidase 17-like E, pseudogene |
| 7 | gene | 104.66675 | 104.67495 | positive | MGI_C57BL6J_3030493 | Olfr659 | NCBI_Gene:259052,ENSEMBL:ENSMUSG00000073922 | MGI:3030493 | protein coding gene | olfactory receptor 659 |
| 7 | pseudogene | 104.67872 | 104.67909 | positive | MGI_C57BL6J_3030494 | Olfr660-ps1 | NCBI_Gene:257836,ENSEMBL:ENSMUSG00000081694 | MGI:3030494 | pseudogene | olfactory receptor 660, pseudogene 1 |
| 7 | gene | 104.68650 | 104.69135 | positive | MGI_C57BL6J_3030495 | Olfr661 | NCBI_Gene:258743,ENSEMBL:ENSMUSG00000073920 | MGI:3030495 | protein coding gene | olfactory receptor 661 |
| 7 | pseudogene | 104.69230 | 104.69343 | negative | MGI_C57BL6J_3644209 | Gm6593 | NCBI_Gene:625495,ENSEMBL:ENSMUSG00000109821 | MGI:3644209 | pseudogene | predicted gene 6593 |
| 7 | pseudogene | 104.70021 | 104.70065 | positive | MGI_C57BL6J_3030496 | Olfr662-ps1 | NCBI_Gene:404413,ENSEMBL:ENSMUSG00000109832 | MGI:3030496 | pseudogene | olfactory receptor 662, pseudogene 1 |
| 7 | pseudogene | 104.70357 | 104.70460 | positive | MGI_C57BL6J_3030497 | Olfr663 | NCBI_Gene:257914,ENSEMBL:ENSMUSG00000109806 | MGI:3030497 | polymorphic pseudogene | olfactory receptor 663 |
| 7 | pseudogene | 104.73338 | 104.73436 | negative | MGI_C57BL6J_3030498 | Olfr664 | NCBI_Gene:667918,ENSEMBL:ENSMUSG00000051885 | MGI:3030498 | polymorphic pseudogene | olfactory receptor 664 |
| 7 | gene | 104.76796 | 104.77747 | negative | MGI_C57BL6J_107697 | Usp17le | NCBI_Gene:625530,ENSEMBL:ENSMUSG00000043073 | MGI:107697 | protein coding gene | ubiquitin specific peptidase 17-like E |
| 7 | pseudogene | 104.82115 | 104.82228 | positive | MGI_C57BL6J_3645730 | Gm4887 | NCBI_Gene:233637,ENSEMBL:ENSMUSG00000086229 | MGI:3645730 | pseudogene | predicted gene 4887 |
| 7 | gene | 104.84026 | 104.84260 | negative | MGI_C57BL6J_3051498 | Usp17lb | NCBI_Gene:381944,ENSEMBL:ENSMUSG00000062369 | MGI:3051498 | protein coding gene | ubiquitin specific peptidase 17-like B |
| 7 | gene | 104.85701 | 104.86267 | positive | MGI_C57BL6J_107699 | Usp17la | NCBI_Gene:13531,ENSEMBL:ENSMUSG00000054568 | MGI:107699 | protein coding gene | ubiquitin specific peptidase 17-like A |
| 7 | gene | 104.86799 | 104.87177 | positive | MGI_C57BL6J_5791392 | Gm45556 | ENSEMBL:ENSMUSG00000109641 | MGI:5791392 | lncRNA gene | predicted gene 45556 |
| 7 | gene | 104.87803 | 104.88291 | positive | MGI_C57BL6J_3030499 | Olfr665 | NCBI_Gene:258810,ENSEMBL:ENSMUSG00000073917 | MGI:3030499 | protein coding gene | olfactory receptor 665 |
| 7 | gene | 104.89182 | 104.89595 | negative | MGI_C57BL6J_3030500 | Olfr666 | NCBI_Gene:259100,ENSEMBL:ENSMUSG00000063582 | MGI:3030500 | protein coding gene | olfactory receptor 666 |
| 7 | gene | 104.90875 | 104.91334 | positive | MGI_C57BL6J_5791108 | Gm45272 | ENSEMBL:ENSMUSG00000109734 | MGI:5791108 | lncRNA gene | predicted gene 45272 |
| 7 | pseudogene | 104.91101 | 104.91138 | positive | MGI_C57BL6J_5791111 | Gm45275 | ENSEMBL:ENSMUSG00000110330 | MGI:5791111 | pseudogene | predicted gene 45275 |
| 7 | gene | 104.91474 | 104.92148 | negative | MGI_C57BL6J_3030501 | Olfr667 | NCBI_Gene:259062,ENSEMBL:ENSMUSG00000056782 | MGI:3030501 | protein coding gene | olfactory receptor 667 |
| 7 | gene | 104.92229 | 104.92806 | negative | MGI_C57BL6J_3030502 | Olfr668 | NCBI_Gene:259061,ENSEMBL:ENSMUSG00000057770 | MGI:3030502 | protein coding gene | olfactory receptor 668 |
| 7 | gene | 104.93424 | 104.94048 | positive | MGI_C57BL6J_3030503 | Olfr669 | NCBI_Gene:259045,ENSEMBL:ENSMUSG00000073916 | MGI:3030503 | protein coding gene | olfactory receptor 669 |
| 7 | pseudogene | 104.94297 | 104.94325 | positive | MGI_C57BL6J_5791109 | Gm45273 | ENSEMBL:ENSMUSG00000110284 | MGI:5791109 | pseudogene | predicted gene 45273 |
| 7 | pseudogene | 104.94918 | 104.95043 | negative | MGI_C57BL6J_3643907 | Gm5900 | NCBI_Gene:545987,ENSEMBL:ENSMUSG00000094685 | MGI:3643907 | pseudogene | predicted pseudogene 5900 |
| 7 | gene | 104.95711 | 104.96262 | negative | MGI_C57BL6J_3030504 | Olfr670 | NCBI_Gene:384703,ENSEMBL:ENSMUSG00000044705 | MGI:3030504 | protein coding gene | olfactory receptor 670 |
| 7 | gene | 104.97248 | 104.97913 | negative | MGI_C57BL6J_3030505 | Olfr671 | NCBI_Gene:257910,ENSEMBL:ENSMUSG00000094531 | MGI:3030505 | protein coding gene | olfactory receptor 671 |
| 7 | gene | 104.99576 | 105.00203 | negative | MGI_C57BL6J_3030506 | Olfr672 | NCBI_Gene:258755,ENSEMBL:ENSMUSG00000051172 | MGI:3030506 | protein coding gene | olfactory receptor 672 |
| 7 | pseudogene | 105.00422 | 105.00517 | negative | MGI_C57BL6J_3030507 | Olfr673-ps1 | NCBI_Gene:257774,ENSEMBL:ENSMUSG00000110041 | MGI:3030507 | pseudogene | olfactory receptor 673, pseudogene 1 |
| 7 | pseudogene | 105.00631 | 105.00726 | negative | MGI_C57BL6J_3030508 | Olfr674-ps1 | NCBI_Gene:258172,ENSEMBL:ENSMUSG00000109671 | MGI:3030508 | pseudogene | olfactory receptor 674, pseudogene 1 |
| 7 | gene | 105.02137 | 105.02846 | negative | MGI_C57BL6J_3030509 | Olfr675 | NCBI_Gene:258147,ENSEMBL:ENSMUSG00000096773 | MGI:3030509 | protein coding gene | olfactory receptor 675 |
| 7 | gene | 105.03157 | 105.03775 | positive | MGI_C57BL6J_3030510 | Olfr676 | NCBI_Gene:259099,ENSEMBL:ENSMUSG00000073915 | MGI:3030510 | protein coding gene | olfactory receptor 676 |
| 7 | gene | 105.05137 | 105.05901 | positive | MGI_C57BL6J_3030511 | Olfr677 | NCBI_Gene:258355,ENSEMBL:ENSMUSG00000073914 | MGI:3030511 | protein coding gene | olfactory receptor 677 |
| 7 | pseudogene | 105.06023 | 105.06087 | positive | MGI_C57BL6J_3646463 | Gm8913 | NCBI_Gene:667987,ENSEMBL:ENSMUSG00000109676 | MGI:3646463 | pseudogene | predicted gene 8913 |
| 7 | gene | 105.06448 | 105.07164 | positive | MGI_C57BL6J_3030512 | Olfr678 | NCBI_Gene:258753,ENSEMBL:ENSMUSG00000073913 | MGI:3030512 | protein coding gene | olfactory receptor 678 |
| 7 | pseudogene | 105.07692 | 105.07818 | positive | MGI_C57BL6J_5010174 | Gm17989 | NCBI_Gene:100416234,ENSEMBL:ENSMUSG00000109972 | MGI:5010174 | pseudogene | predicted gene, 17989 |
| 7 | gene | 105.07843 | 105.08008 | negative | MGI_C57BL6J_5791393 | Gm45557 | ENSEMBL:ENSMUSG00000110293 | MGI:5791393 | lncRNA gene | predicted gene 45557 |
| 7 | gene | 105.08120 | 105.09064 | positive | MGI_C57BL6J_3030513 | Olfr679 | NCBI_Gene:259046,ENSEMBL:ENSMUSG00000096029 | MGI:3030513 | protein coding gene | olfactory receptor 679 |
| 7 | pseudogene | 105.09070 | 105.09282 | negative | MGI_C57BL6J_3030514 | Olfr680-ps1 | NCBI_Gene:257981,ENSEMBL:ENSMUSG00000041885 | MGI:3030514 | pseudogene | olfactory receptor 680, pseudogene 1 |
| 7 | pseudogene | 105.10603 | 105.10782 | positive | MGI_C57BL6J_5010175 | Gm17990 | NCBI_Gene:100416235,ENSEMBL:ENSMUSG00000110117 | MGI:5010175 | pseudogene | predicted gene, 17990 |
| 7 | gene | 105.10831 | 105.11020 | negative | MGI_C57BL6J_5791394 | Gm45558 | ENSEMBL:ENSMUSG00000109742 | MGI:5791394 | lncRNA gene | predicted gene 45558 |
| 7 | gene | 105.11684 | 105.12552 | positive | MGI_C57BL6J_3030515 | Olfr681 | NCBI_Gene:404318,ENSEMBL:ENSMUSG00000095248 | MGI:3030515 | protein coding gene | olfactory receptor 681 |
| 7 | pseudogene | 105.12637 | 105.12843 | negative | MGI_C57BL6J_3030516 | Olfr682-ps1 | NCBI_Gene:258187,ENSEMBL:ENSMUSG00000059768 | MGI:3030516 | polymorphic pseudogene | olfactory receptor 682, pseudogene 1 |
| 7 | gene | 105.14084 | 105.14701 | negative | MGI_C57BL6J_3030517 | Olfr683 | NCBI_Gene:259047,ENSEMBL:ENSMUSG00000044120 | MGI:3030517 | protein coding gene | olfactory receptor 683 |
| 7 | pseudogene | 105.14541 | 105.14555 | negative | MGI_C57BL6J_5791210 | Gm45374 | ENSEMBL:ENSMUSG00000109918 | MGI:5791210 | pseudogene | predicted gene 45374 |
| 7 | gene | 105.15469 | 105.16365 | negative | MGI_C57BL6J_3030518 | Olfr684 | NCBI_Gene:244187,ENSEMBL:ENSMUSG00000047225 | MGI:3030518 | protein coding gene | olfactory receptor 684 |
| 7 | gene | 105.17842 | 105.18601 | negative | MGI_C57BL6J_3030519 | Olfr685 | NCBI_Gene:258160,ENSEMBL:ENSMUSG00000047794 | MGI:3030519 | protein coding gene | olfactory receptor 685 |
| 7 | gene | 105.20186 | 105.20749 | negative | MGI_C57BL6J_3030520 | Olfr686 | NCBI_Gene:259072,ENSEMBL:ENSMUSG00000048425 | MGI:3030520 | protein coding gene | olfactory receptor 686 |
| 7 | gene | 105.22772 | 105.27654 | positive | MGI_C57BL6J_3030521 | Olfr687 | NCBI_Gene:668035,ENSEMBL:ENSMUSG00000073908 | MGI:3030521 | protein coding gene | olfactory receptor 687 |
| 7 | gene | 105.28809 | 105.28906 | positive | MGI_C57BL6J_3030522 | Olfr688 | NCBI_Gene:259161,ENSEMBL:ENSMUSG00000073909 | MGI:3030522 | protein coding gene | olfactory receptor 688 |
| 7 | gene | 105.31117 | 105.31966 | positive | MGI_C57BL6J_3030523 | Olfr689 | NCBI_Gene:258745,ENSEMBL:ENSMUSG00000073907 | MGI:3030523 | protein coding gene | olfactory receptor 689 |
| 7 | pseudogene | 105.32666 | 105.32721 | positive | MGI_C57BL6J_5791167 | Gm45331 | ENSEMBL:ENSMUSG00000110265 | MGI:5791167 | pseudogene | predicted gene 45331 |
| 7 | gene | 105.32823 | 105.33410 | negative | MGI_C57BL6J_3030524 | Olfr690 | NCBI_Gene:56860,ENSEMBL:ENSMUSG00000050266 | MGI:3030524 | protein coding gene | olfactory receptor 690 |
| 7 | gene | 105.33551 | 105.34227 | negative | MGI_C57BL6J_3030525 | Olfr691 | NCBI_Gene:259063,ENSEMBL:ENSMUSG00000043948 | MGI:3030525 | protein coding gene | olfactory receptor 691 |
| 7 | gene | 105.36823 | 105.36935 | positive | MGI_C57BL6J_3030526 | Olfr692 | NCBI_Gene:258352,ENSEMBL:ENSMUSG00000073906 | MGI:3030526 | protein coding gene | olfactory receptor 692 |
| 7 | gene | 105.37121 | 105.40005 | negative | MGI_C57BL6J_1921599 | Fam160a2 | NCBI_Gene:74349,ENSEMBL:ENSMUSG00000044465 | MGI:1921599 | protein coding gene | family with sequence similarity 160, member A2 |
| 7 | gene | 105.37504 | 105.37838 | positive | MGI_C57BL6J_3643909 | Gm5901 | NCBI_Gene:100503879,ENSEMBL:ENSMUSG00000078611 | MGI:3643909 | protein coding gene | predicted gene 5901 |
| 7 | gene | 105.39745 | 105.40910 | positive | MGI_C57BL6J_2664099 | Cnga4 | NCBI_Gene:233649,ENSEMBL:ENSMUSG00000030897 | MGI:2664099 | protein coding gene | cyclic nucleotide gated channel alpha 4 |
| 7 | gene | 105.40031 | 105.40189 | positive | MGI_C57BL6J_5805000 | Gm45885 | ENSEMBL:ENSMUSG00000110366 | MGI:5805000 | lncRNA gene | predicted gene 45885 |
| 7 | pseudogene | 105.41270 | 105.41544 | negative | MGI_C57BL6J_3643689 | Gm4972 | NCBI_Gene:244189,ENSEMBL:ENSMUSG00000109609 | MGI:3643689 | pseudogene | predicted gene 4972 |
| 7 | gene | 105.42556 | 105.47090 | positive | MGI_C57BL6J_99479 | Cckbr | NCBI_Gene:12426,ENSEMBL:ENSMUSG00000030898 | MGI:99479 | protein coding gene | cholecystokinin B receptor |
| 7 | gene | 105.48008 | 105.48230 | negative | MGI_C57BL6J_1923422 | Cavin3 | NCBI_Gene:109042,ENSEMBL:ENSMUSG00000037060 | MGI:1923422 | protein coding gene | caveolae associated 3 |
| 7 | gene | 105.48193 | 105.48324 | positive | MGI_C57BL6J_5791503 | Gm45667 | ENSEMBL:ENSMUSG00000109735 | MGI:5791503 | unclassified gene | predicted gene 45667 |
| 7 | gene | 105.48264 | 105.52267 | positive | MGI_C57BL6J_5596006 | Gm36847 | NCBI_Gene:102640889 | MGI:5596006 | lncRNA gene | predicted gene, 36847 |
| 7 | gene | 105.54490 | 105.55428 | negative | MGI_C57BL6J_5621948 | Gm39063 | NCBI_Gene:105243022 | MGI:5621948 | lncRNA gene | predicted gene, 39063 |
| 7 | gene | 105.55436 | 105.55839 | positive | MGI_C57BL6J_98325 | Smpd1 | NCBI_Gene:20597,ENSEMBL:ENSMUSG00000037049 | MGI:98325 | protein coding gene | sphingomyelin phosphodiesterase 1, acid lysosomal |
| 7 | gene | 105.55846 | 105.58192 | negative | MGI_C57BL6J_107765 | Apbb1 | NCBI_Gene:11785,ENSEMBL:ENSMUSG00000037032 | MGI:107765 | protein coding gene | amyloid beta (A4) precursor protein-binding, family B, member 1 |
| 7 | gene | 105.59161 | 105.60014 | negative | MGI_C57BL6J_105112 | Hpx | NCBI_Gene:15458,ENSEMBL:ENSMUSG00000030895 | MGI:105112 | protein coding gene | hemopexin |
| 7 | gene | 105.60446 | 105.63357 | negative | MGI_C57BL6J_1860040 | Trim3 | NCBI_Gene:55992,ENSEMBL:ENSMUSG00000036989 | MGI:1860040 | protein coding gene | tripartite motif-containing 3 |
| 7 | gene | 105.62282 | 105.62470 | negative | MGI_C57BL6J_5791468 | Gm45632 | ENSEMBL:ENSMUSG00000110077 | MGI:5791468 | unclassified gene | predicted gene 45632 |
| 7 | gene | 105.63420 | 105.64042 | negative | MGI_C57BL6J_1924182 | Arfip2 | NCBI_Gene:76932,ENSEMBL:ENSMUSG00000030881 | MGI:1924182 | protein coding gene | ADP-ribosylation factor interacting protein 2 |
| 7 | gene | 105.64006 | 105.65699 | positive | MGI_C57BL6J_1315196 | Timm10b | NCBI_Gene:14356,ENSEMBL:ENSMUSG00000089847 | MGI:1315196 | protein coding gene | translocase of inner mitochondrial membrane 10B |
| 7 | gene | 105.64055 | 105.68444 | positive | MGI_C57BL6J_5804914 | Gm45799 | ENSEMBL:ENSMUSG00000110234 | MGI:5804914 | protein coding gene | predicted gene 45799 |
| 7 | gene | 105.64234 | 105.64248 | negative | MGI_C57BL6J_5452281 | Gm22504 | ENSEMBL:ENSMUSG00000064897 | MGI:5452281 | snoRNA gene | predicted gene, 22504 |
| 7 | gene | 105.65081 | 105.72180 | positive | MGI_C57BL6J_1924755 | Dnhd1 | NCBI_Gene:77505,ENSEMBL:ENSMUSG00000030882 | MGI:1924755 | protein coding gene | dynein heavy chain domain 1 |
R/qtl
scanone
R version 4.0.3 (2020-10-10)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 20.04.5 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/liblapack.so.3
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] abind_1.4-5 qtl2_0.24 reshape2_1.4.4 ggplot2_3.3.3
[5] tibble_3.0.6 psych_2.0.12 readxl_1.3.1 cluster_2.1.0
[9] dplyr_1.0.4 optparse_1.6.6 rhdf5_2.34.0 mclust_5.4.7
[13] tidyr_1.1.2 data.table_1.13.6 knitr_1.31 kableExtra_1.3.1
[17] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rcpp_1.0.10 lattice_0.20-41 assertthat_0.2.1 rprojroot_2.0.2
[5] digest_0.6.27 R6_2.5.0 cellranger_1.1.0 plyr_1.8.6
[9] RSQLite_2.2.3 evaluate_0.14 httr_1.4.2 highr_0.8
[13] pillar_1.4.7 rlang_0.4.10 rstudioapi_0.13 blob_1.2.1
[17] rmarkdown_2.6 webshot_0.5.2 stringr_1.4.0 bit_4.0.4
[21] munsell_0.5.0 compiler_4.0.3 httpuv_1.5.5 xfun_0.21
[25] pkgconfig_2.0.3 mnormt_2.0.2 tmvnsim_1.0-2 htmltools_0.5.1.1
[29] tidyselect_1.1.0 viridisLite_0.3.0 crayon_1.4.1 withr_2.4.1
[33] later_1.1.0.1 rhdf5filters_1.2.1 grid_4.0.3 nlme_3.1-152
[37] gtable_0.3.0 lifecycle_0.2.0 DBI_1.1.1 git2r_0.28.0
[41] magrittr_2.0.1 scales_1.1.1 cachem_1.0.3 stringi_1.5.3
[45] fs_1.5.0 promises_1.1.1 getopt_1.20.3 xml2_1.3.2
[49] ellipsis_0.3.1 generics_0.1.0 vctrs_0.3.6 Rhdf5lib_1.12.1
[53] tools_4.0.3 bit64_4.0.5 glue_1.4.2 purrr_0.3.4
[57] fastmap_1.1.0 parallel_4.0.3 yaml_2.2.1 colorspace_2.0-0
[61] rvest_0.3.6 memoise_2.0.0