QTL Analysis - age of onset [ICI vs EOI] (corrected phenotype with outliers removed if any)
Belinda Cornes
2022-04-14
Last updated: 2022-04-14
Checks: 5 2
Knit directory: Serreze-T1D_Workflow/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20220210) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /Users/corneb/Documents/MyJax/CS/Projects/Serreze/qc/workflowr/Serreze-T1D_Workflow | . |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version a8c2d1a. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: analysis/.DS_Store
Ignored: data/.DS_Store
Untracked files:
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_0.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_52.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_mis_0.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_mis_52.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_mis_vo.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_vo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.eoi_snpsqc_dis_no-x_updated_5.batches_11.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.eoi_snpsqc_dis_no-x_updated_5.batches_mis_11.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_1.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_mis_1.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_1.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_11.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_11.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_changed.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_changed.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_scanone_5.batches.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_scanone_5.batches_mis.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_oops.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.4.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.4.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.4_miss.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.4_miss.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.5.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.5.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.5_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.5_0.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_vo.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_vo1.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_vo.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_vo1.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_52.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.eoi_5.batches_0.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.eoi_5.batches_52.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.eoi_5.batches_mis_0.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.eoi_5.batches_mis_52.Rmd
Untracked: analysis/index_5.batches_additional_vo.Rmd
Untracked: data/GM_covar.csv
Untracked: data/GM_covar_BC312.csv
Untracked: data/bad_markers_all_4.batches.RData
Untracked: data/bad_markers_all_5.batches.RData
Untracked: data/blup_sub_chr10_lod.drop-1.5_5.batches.csv
Untracked: data/blup_sub_chr3_lod.drop-1.5.csv
Untracked: data/blup_sub_chr3_lod.drop-1.5_5.batches.csv
Untracked: data/blup_sub_chr4_lod.drop-1.5.csv
Untracked: data/blup_sub_chr4_lod.drop-1.5_5.batches.csv
Untracked: data/covar_cleaned_ici.vs.eoi.csv
Untracked: data/covar_cleaned_ici.vs.pbs.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_0.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_52.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_mis.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_mis_0.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_mis_52.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi1.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_0.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_52.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_mis.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_mis_0.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_mis_52.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs1.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_0.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_52.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_mis.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_mis_0.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_mis_52.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_0.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_52.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_mis.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_mis_0.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_mis_52.csv
Untracked: data/covar_corrected_ici.vs.eoi.csv
Untracked: data/covar_corrected_ici.vs.eoi1.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_0.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_52.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_mis.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_mis_0.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_mis_52.csv
Untracked: data/covar_corrected_ici.vs.pbs.csv
Untracked: data/covar_corrected_ici.vs.pbs1.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_0.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_52.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_mis.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_mis_0.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_mis_52.csv
Untracked: data/e.RData
Untracked: data/e_BC312.RData
Untracked: data/e_snpg_samqc_4.batches.RData
Untracked: data/e_snpg_samqc_4.batches_bc.RData
Untracked: data/e_snpg_samqc_5.batches.RData
Untracked: data/errors_ind_4.batches.RData
Untracked: data/errors_ind_4.batches_bc.RData
Untracked: data/errors_ind_5.batches.RData
Untracked: data/files.to.sync.txt
Untracked: data/fitqtl_chr3.peak_chr4.peak_additive.txt
Untracked: data/fitqtl_chr3.peak_chr4.peak_interacting.txt
Untracked: data/fitqtl_chr3.peak_chr4.peak_sex_additive.txt
Untracked: data/fitqtl_chr3.peak_chr4.peak_sex_interacting.txt
Untracked: data/g2blup_effects.csv
Untracked: data/g2blup_effects.xlsx
Untracked: data/genes_chr10_lod.drop-1.5_5.batches.csv
Untracked: data/genes_chr3_lod.drop-1.5.csv
Untracked: data/genes_chr3_lod.drop-1.5_5.batches.csv
Untracked: data/genes_chr4_lod.drop-1.5.csv
Untracked: data/genetic_map.csv
Untracked: data/genetic_map_BC312.csv
Untracked: data/genotype_errors_marker_4.batches.RData
Untracked: data/genotype_errors_marker_5.batches.RData
Untracked: data/genotype_freq_marker_4.batches.RData
Untracked: data/genotype_freq_marker_5.batches.RData
Untracked: data/gm_allqc_4.batches.RData
Untracked: data/gm_allqc_5.batches.RData
Untracked: data/gm_allqc_5.batches_mis.RData
Untracked: data/gm_samqc_3.batches.RData
Untracked: data/gm_samqc_4.batches.RData
Untracked: data/gm_samqc_4.batches_bc.RData
Untracked: data/gm_samqc_5.batches.RData
Untracked: data/gm_serreze.192.RData
Untracked: data/gm_serreze.BC312.RData
Untracked: data/ici-early.vs.pbs_age.of.onset-additive.covariates_blup_sub_chr-7_peak.marker-UNC13388811_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-additive.covariates_genes_chr-7_peak.marker-UNC13388811_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-15_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-15_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-15_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-17_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-17_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-17_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_5.batches.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_scanone_5.batches.Rdata
Untracked: data/ici-early.vs.pbs_scanone_5.batches_mis.Rdata
Untracked: data/ici-early.vs.pbs_scanone_snpsqc_5.batches.Rdata
Untracked: data/ici-early.vs.pbs_scanone_snpsqc_5.batches_mis.Rdata
Untracked: data/ici-early.vs.pbs_scanone_snpsqc_dis_no-x_updated_5.batches.Rdata
Untracked: data/ici-early.vs.pbs_scanone_snpsqc_dis_no-x_updated_5.batches_mis.Rdata
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00020646_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00292499_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00294019_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18216614_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18240977_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18343181_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18363544_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18363544_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18363544_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-11_peak.marker-UNC20090524_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-2_peak.marker-UNCHS008007_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-2_peak.marker-UNCHS008007_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR5263_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR5263_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00105649_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00105649_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00106210_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00107193_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00516812_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00516812_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00522922_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00524205_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00535764_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4901083_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4901083_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4909519_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4909519_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4915446_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4915446_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4938565_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4938565_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4959266_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4959266_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4960612_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4960612_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4961542_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4961542_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5046545_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5055259_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5062524_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5068215_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5076935_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5082757_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5173293_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5193970_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5257268_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5264696_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5267813_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5270225_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5277061_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5278229_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5278229_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5282259_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5287977_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5301460_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5309136_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5321559_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5322455_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5329171_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5360055_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5778977_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5916997_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008229_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008229_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008234_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008234_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008242_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008260_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008260_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008281_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008281_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008338_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008338_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008363_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008363_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008409_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008432_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008609_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008614_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008627_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008659_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008725_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009066_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009067_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009068_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009112_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009693_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009821_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001276_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-sanger2496q_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-sanger2496q_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-4_peak.marker-UNC8250659_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-4_peak.marker-UNC8439633_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-4_peak.marker-UNC8439633_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-4_peak.marker-UNCHS012955_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-6_peak.marker-UNC11108920_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-6_peak.marker-UNC11108920_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00020646_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00292499_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00294019_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18216614_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18240977_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18343181_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18363544_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18363544_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18363544_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-11_peak.marker-UNC20090524_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-2_peak.marker-UNCHS008007_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-2_peak.marker-UNCHS008007_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR5263_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR5263_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00105649_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00105649_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00106210_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00107193_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00516812_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00516812_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00522922_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00524205_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00535764_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4901083_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4901083_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4909519_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4909519_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4915446_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4915446_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4938565_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4938565_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4959266_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4959266_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4960612_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4960612_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4961542_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4961542_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5046545_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5055259_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5062524_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5068215_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5076935_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5082757_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5173293_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5193970_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5257268_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5264696_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5267813_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5270225_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5277061_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5278229_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5278229_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5282259_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5287977_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5301460_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5309136_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5321559_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5322455_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5329171_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5360055_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5778977_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5916997_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008229_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008229_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008234_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008234_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008242_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008260_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008260_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008281_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008281_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008338_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008338_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008363_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008363_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008409_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008432_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008609_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008614_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008627_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008659_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008725_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009066_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009067_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009068_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009112_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009693_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009821_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001276_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-sanger2496q_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-sanger2496q_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-4_peak.marker-UNC8250659_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-4_peak.marker-UNC8439633_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-4_peak.marker-UNC8439633_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-4_peak.marker-UNCHS012955_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-6_peak.marker-UNC11108920_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-6_peak.marker-UNC11108920_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_blup.full_chr-10_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-11_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-12_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-13_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-14_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-15_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-16_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-17_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-18_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-19_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-1_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-2_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-3_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-4_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-6_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-7_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-8_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-9_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-X_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-ICR4295_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-JAX00292499_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18154330_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18195765_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18216614_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18240977_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18856953_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNCJPD004316_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNCJPD009532_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNC19155926_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNC19481366_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNCJPD004792_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-12_peak.marker-ICR4497_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-13_peak.marker-UNCHS036964_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-18_peak.marker-UNCHS045344_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-19_peak.marker-UNCJPD007087_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-1_peak.marker-ICR010_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-1_peak.marker-ICR010_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-2_peak.marker-UNC4609660_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-B6_rs31740628_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR1269_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR5325_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR5347_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR5348_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00104971_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00105059_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00105203_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00107930_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00107980_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00108068_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00108114_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00109750_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00189245_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00517218_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00517906_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00518418_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00518684_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00520122_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00524717_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00524828_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00525380_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00525709_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00527291_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00527587_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00529707_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00531601_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00532122_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4673471_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4681411_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4793677_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4799475_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4909830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4914130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4964062_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4985064_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4991565_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5001723_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5042361_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5055259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5076935_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5082757_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5173293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5182782_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5193970_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5240484_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5256922_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5266664_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5266711_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5267813_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5274554_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5277061_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5282259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5287977_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5301460_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5321559_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5322455_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5328596_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5350384_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5354724_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5375208_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5384072_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5400314_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5433945_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5435874_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5466669_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5517972_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5522257_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5523265_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5532424_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5553882_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5572051_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5582753_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5589260_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5595555_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5623239_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5639830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5680234_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5682314_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5725356_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5727351_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5734420_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5743257_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5751567_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5752623_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5776974_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5778977_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5788507_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5792468_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5794598_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5795561_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5802298_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5806628_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5817478_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5836999_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5841501_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5847927_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5966417_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008129_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008173_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008231_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008235_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008242_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008253_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008256_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008311_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008317_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008372_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008410_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008415_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008431_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008609_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008614_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008649_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008651_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008723_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008802_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008803_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008808_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008834_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008838_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008928_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008948_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008959_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008973_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008987_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009001_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009025_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009034_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009087_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009109_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009142_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009207_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009216_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009306_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009316_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009364_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009377_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009382_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009387_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009395_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009413_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009428_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009463_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009503_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009532_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009541_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009560_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009574_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001276_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001315_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001355_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001373_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001382_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC6851205_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8250659_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8299912_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8300499_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8381981_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8436434_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8439633_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8530763_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS012948_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS012976_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013139_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013304_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-5_peak.marker-JAX00590770_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-ICR1830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-UNC13170794_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-UNCHS019480_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-cr31snv87_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-UNC15430711_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-9_peak.marker-UNC17203329_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-9_peak.marker-UNCHS024593_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-X_peak.marker-UNC30627130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-X_peak.marker-UNCHS048313_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_snpsqc_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_snpsqc_dis_no-x_updated_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_snpsqc_dis_no-x_updated_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_snpsqc_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_snpsqc_dis_no-x_updated_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_snpsqc_dis_no-x_updated_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_snpsqc_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_snpsqc_dis_no-x_updated_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_snpsqc_dis_no-x_updated_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_snpsqc_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_snpsqc_dis_no-x_updated_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_snpsqc_dis_no-x_updated_5.batches_mis.mis.txt
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-ICR4295_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-JAX00292499_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18154330_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18195765_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18216614_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18240977_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18856953_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNCJPD004316_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNCJPD009532_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNC19155926_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNC19481366_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNCJPD004792_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-12_peak.marker-ICR4497_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-13_peak.marker-UNCHS036964_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-18_peak.marker-UNCHS045344_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-19_peak.marker-UNCJPD007087_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-1_peak.marker-ICR010_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-1_peak.marker-ICR010_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-2_peak.marker-UNC4609660_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-B6_rs31740628_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR1269_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR5325_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR5347_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR5348_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00104971_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00105059_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00105203_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00107930_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00107980_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00108068_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00108114_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00109750_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00189245_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00517218_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00517906_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00518418_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00518684_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00520122_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00524717_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00524828_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00525380_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00525709_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00527291_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00527587_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00529707_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00531601_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00532122_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4673471_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4681411_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4793677_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4799475_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4909830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4914130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4964062_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4985064_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4991565_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5001723_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5042361_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5055259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5076935_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5082757_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5173293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5182782_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5193970_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5240484_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5256922_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5266664_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5266711_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5267813_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5274554_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5277061_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5282259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5287977_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5301460_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5321559_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5322455_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5328596_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5350384_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5354724_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5375208_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5384072_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5400314_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5433945_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5435874_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5466669_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5517972_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5522257_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5523265_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5532424_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5553882_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5572051_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5582753_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5589260_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5595555_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5623239_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5639830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5680234_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5682314_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5725356_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5727351_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5734420_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5743257_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5751567_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5752623_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5776974_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5778977_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5788507_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5792468_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5794598_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5795561_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5802298_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5806628_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5817478_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5836999_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5841501_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5847927_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5966417_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008129_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008173_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008231_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008235_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008242_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008253_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008256_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008311_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008317_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008372_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008410_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008415_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008431_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008609_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008614_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008649_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008651_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008723_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008802_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008803_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008808_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008834_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008838_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008928_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008948_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008959_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008973_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008987_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009001_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009025_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009034_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009087_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009109_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009142_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009207_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009216_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009306_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009316_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009364_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009377_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009382_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009387_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009395_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009413_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009428_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009463_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009503_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009532_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009541_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009560_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009574_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001276_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001315_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001355_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001373_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001382_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC6851205_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8250659_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8299912_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8300499_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8381981_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8436434_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8439633_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8530763_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS012948_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS012976_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013139_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013304_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-5_peak.marker-JAX00590770_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-ICR1830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-UNC13170794_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-UNCHS019480_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-cr31snv87_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-UNC15430711_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-9_peak.marker-UNC17203329_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-9_peak.marker-UNCHS024593_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-X_peak.marker-UNC30627130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-X_peak.marker-UNCHS048313_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_gm_qtl_5.batches.csv
Untracked: data/ici.vs.eoi_gm_qtl_5.batches_mis.csv
Untracked: data/ici.vs.eoi_gm_qtl_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_gm_qtl_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_gm_qtl_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18343181_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18376338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18382395_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNCHS029024_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-1_peak.marker-UNC228500_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-1_peak.marker-UNC228500_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-ICR2047_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00112499_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00219554_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00529707_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4837069_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5006948_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5266664_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5300878_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5360055_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5679155_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5945286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC6227106_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008595_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009067_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009693_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001341_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-sanger2623_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-4_peak.marker-UNCHS013125_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-4_peak.marker-UNCHS013125_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-6_peak.marker-UNCHS018170_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-6_peak.marker-UNCHS018231_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18343181_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18376338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18382395_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNCHS029024_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-1_peak.marker-UNC228500_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-1_peak.marker-UNC228500_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-ICR2047_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00112499_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00219554_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00529707_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4837069_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5006948_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5266664_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5300878_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5360055_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5679155_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5945286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC6227106_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008595_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS009067_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS009693_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCJPD001246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCJPD001341_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-sanger2623_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-4_peak.marker-UNCHS013125_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-4_peak.marker-UNCHS013125_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-6_peak.marker-UNCHS018170_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-6_peak.marker-UNCHS018231_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.geno.freq.removed.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.probs.freq.removed.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.eoi_scanone_5.batches.Rdata
Untracked: data/ici.vs.eoi_scanone_5.batches_mis.Rdata
Untracked: data/ici.vs.eoi_scanone_snpsqc_5.batches.Rdata
Untracked: data/ici.vs.eoi_scanone_snpsqc_5.batches_mis.Rdata
Untracked: data/ici.vs.eoi_scanone_snpsqc_dis_no-x_updated_5.batches.Rdata
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNC28655293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-5_peak.marker-UNC8675939_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNC28655293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-5_peak.marker-UNC8675939_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_bad.markers_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-15_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-15_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-15_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-17_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-17_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-17_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-13_peak.marker-UNC22384241_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-19_peak.marker-UNC29893228_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-19_peak.marker-UNC29990033_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-19_peak.marker-UNCHS047192_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-8_peak.marker-UNCHS023353_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-8_peak.marker-UNCHS023353_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-8_peak.marker-UNCHS023364_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_fitqtl_peaks_additive_snpsqc_dis_no-x_updated_5.batches_mis_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_additive_snpsqc_dis_no-x_updated_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_additive_snpsqc_snpsqc_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_interacting_snpsqc_dis_no-x_updated_5.batches_mis_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_interacting_snpsqc_dis_no-x_updated_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_interacting_snpsqc_snpsqc_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_additive_snpsqc_dis_no-x_updated_5.batches_mis_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_additive_snpsqc_dis_no-x_updated_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_additive_snpsqc_snpsqc_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_interacting_snpsqc_dis_no-x_updated_5.batches_mis_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_interacting_snpsqc_dis_no-x_updated_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_interacting_snpsqc_snpsqc_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_genes_chr-13_peak.marker-UNC22384241_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_genes_chr-19_peak.marker-UNC29893228_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-19_peak.marker-UNC29990033_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-19_peak.marker-UNCHS047192_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-8_peak.marker-UNCHS023353_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-8_peak.marker-UNCHS023353_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-8_peak.marker-UNCHS023364_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_mis.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-JAX00153527_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-JAX00153527_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNCHS020443_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNCHS020443_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNCHS020486_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-JAX00153527_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-JAX00153527_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNCHS020443_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNCHS020443_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNCHS020486_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.pbs_scanone_5.batches.Rdata
Untracked: data/ici.vs.pbs_scanone_5.batches_mis.Rdata
Untracked: data/ici.vs.pbs_scanone_snpsqc_5.batches.Rdata
Untracked: data/ici.vs.pbs_scanone_snpsqc_5.batches_mis.Rdata
Untracked: data/ici.vs.pbs_scanone_snpsqc_dis_no-x_updated_5.batches.Rdata
Untracked: data/ici.vs.pbs_scanone_snpsqc_dis_no-x_updated_5.batches_mis.Rdata
Untracked: data/percent_missing_id_3.batches.RData
Untracked: data/percent_missing_id_4.batches.RData
Untracked: data/percent_missing_id_4.batches_bc.RData
Untracked: data/percent_missing_id_5.batches.RData
Untracked: data/percent_missing_marker_4.batches.RData
Untracked: data/percent_missing_marker_5.batches.RData
Untracked: data/pheno.csv
Untracked: data/pheno_BC312.csv
Untracked: data/physical_map.csv
Untracked: data/physical_map_BC312.csv
Untracked: data/qc_info_bad_sample_3.batches.RData
Untracked: data/qc_info_bad_sample_4.batches.RData
Untracked: data/qc_info_bad_sample_4.batches_bc.RData
Untracked: data/qc_info_bad_sample_5.batches.RData
Untracked: data/remaining.markers_geno.freq.xlsx
Untracked: data/sample_geno.csv
Untracked: data/sample_geno_AHB_BC312.csv
Untracked: data/sample_geno_bc.csv
Untracked: data/sample_geno_bc_BC312.csv
Untracked: data/serreze_probs.rds
Untracked: data/serreze_probs_BC312.rds
Untracked: data/serreze_probs_allqc.rds
Untracked: data/serreze_probs_allqc_5.batches.rds
Untracked: data/serreze_probs_allqc_5.batches_mis.rds
Untracked: data/summary.cg_3.batches.RData
Untracked: data/summary.cg_4.batches.RData
Untracked: data/summary.cg_4.batches_bc.RData
Untracked: data/summary.cg_5.batches.RData
Untracked: output/Percent_missing_genotype_data_5.batches.pdf
Untracked: output/Percent_missing_genotype_data_per_marker_5.batches.pdf
Untracked: output/Proportion_matching_genotypes_before_removal_of_bad_samples_5.batches.pdf
Untracked: output/genotype_error_marker_5.batches.pdf
Untracked: output/genotype_frequency_marker_5.batches.pdf
Unstaged changes:
Modified: analysis/4.1.1_qtl.analysis_binary_ici.vs.eoi_snpsqc_dis_no-x_updated.Rmd
Modified: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/index_5.batches.Rmd
Modified: analysis/index_5.batches_additional.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches.Rmd) and HTML (docs/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | a8c2d1a | Belinda Cornes | 2022-04-13 | additonal analysis - peaks and continuous variables |
| html | a8c2d1a | Belinda Cornes | 2022-04-13 | additonal analysis - peaks and continuous variables |
Loading Data
load("data/gm_allqc_5.batches.RData")
#gm_allqc
gm=gm_allqc
gmObject of class cross2 (crosstype "bc")
Total individuals 308
No. genotyped individuals 308
No. phenotyped individuals 308
No. with both geno & pheno 308
No. phenotypes 1
No. covariates 6
No. phenotype covariates 0
No. chromosomes 20
Total markers 34537
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2643 2629 1857 1890 1774 1941 1672 1627 1878 1176 1871 1300 1549 1578 1257 935
17 18 19 X
501 913 1014 4532 #pr <- readRDS("data/serreze_probs_allqc.rds")
#pr <- readRDS("data/serreze_probs.rds")
##extracting animals with ici and eoi group status
miceinfo <- gm$covar[gm$covar$group == "EOI" | gm$covar$group == "ICI",]
table(miceinfo$group)
EOI ICI
164 104 mice.ids <- rownames(miceinfo)
gm <- gm[mice.ids]
gmObject of class cross2 (crosstype "bc")
Total individuals 268
No. genotyped individuals 268
No. phenotyped individuals 268
No. with both geno & pheno 268
No. phenotypes 1
No. covariates 6
No. phenotype covariates 0
No. chromosomes 20
Total markers 34537
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2643 2629 1857 1890 1774 1941 1672 1627 1878 1176 1871 1300 1549 1578 1257 935
17 18 19 X
501 913 1014 4532 table(gm$covar$group)
EOI ICI
164 104 gm.full <- gm
covars <- read_csv("data/covar_corrected.cleaned_ici.vs.eoi_5.batches.csv")
#removing any missing info
missing_ids <- covars[which(covars$"age.of.onset"==""),]$Mouse.ID
length(missing_ids)[1] 3missing_ids[1] "NG00526-ICI-LATE" "NG00522-ICI-LATE" "NG01389-ICI-LATE"if(length(missing_ids) != 0){
gm <- gm[paste0("-",as.character(missing_ids)),]
gm
table(gm$covar$group)
#keeping only mouseids in covars
covars <- covars[gm$covar$id,]
table(covars$group)
}
EOI ICI
164 101 #covars[covars$age.of.onset==0,]$age.of.onset <- ''
query_variants <- create_variant_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/cc_variants.sqlite")
query_genes <- create_gene_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/mouse_genes_mgi.sqlite")
##removing problmetic marker
gm <- drop_markers(gm, "UNCHS013106")
#pmap_interp = interp_map(map, gm$gmap, gm$pmap)
markers <- marker_names(gm)
gmapdf <- read.csv("/Users/corneb/Documents/MyJax/CS/Projects/Serreze/haplotype.reconstruction/output_5.batches/genetic_map.csv")
pmapdf <- read.csv("/Users/corneb/Documents/MyJax/CS/Projects/Serreze/haplotype.reconstruction/output_5.batches/physical_map.csv")
#mapdf <- merge(gmapdf,pmapdf, by=c("marker","chr"), all=T)
#rownames(mapdf) <- mapdf$marker
#mapdf <- mapdf[markers,]
#names(mapdf) <- c('marker','chr','gmapdf','pmapdf')
#mapdfnd <- mapdf[!duplicated(mapdf[c(2:3)]),]
pr.qc <- calc_genoprob(gm)
gmObject of class cross2 (crosstype "bc")
Total individuals 265
No. genotyped individuals 265
No. phenotyped individuals 265
No. with both geno & pheno 265
No. phenotypes 1
No. covariates 6
No. phenotype covariates 0
No. chromosomes 20
Total markers 34537
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2643 2629 1857 1890 1774 1941 1672 1627 1878 1176 1871 1300 1549 1578 1257 935
17 18 19 X
501 913 1014 4532 Preparing necessary info
covars$age.of.onset <- as.numeric(covars$age.of.onset)
Xcovar <- get_x_covar(gm)
addcovar = model.matrix(~ICI.vs.EOI, data = covars)[,-1]
#addcovar.i = model.matrix(~ICI.vs.EOI, data = covars)[,-1]
K <- calc_kinship(pr.qc, type = "loco")
#heatmap(K[[1]])
K.overall <- calc_kinship(pr.qc, type = "overall")
#heatmap(K.overall)
kinship <- calc_kinship(pr.qc)
heatmap(kinship)
No additive covariates
Significance thresholds
operm <- scan1perm(pr.qc, covars["age.of.onset"], model="normal", n_perm=10, perm_Xsp=TRUE, chr_lengths=chr_lengths(gm$gmap))
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
names(summary_table) <- c("autosomes","X")
summary_table$significance.level <- rownames(summary_table)
rownames(summary_table) <- NULL
summary_table[c(3,1:2)] %>%
kable(escape = F,align = c("ccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| significance.level | autosomes | X |
|---|---|---|
| 0.01 | 5.960676 | 8.929044 |
| 0.05 | 5.017067 | 8.688022 |
| 0.1 | 3.834614 | 8.372843 |
Genome-wide scan
plot_lod<-function(out,map){
for (i in 1:dim(out)[2]){
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - ICI vs. EOI (no covariates) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
print("with no kinship")[1] “with no kinship”
outf <- scan1(pr.qc, covars["age.of.onset"], Xcovar=Xcovar, model="normal")
plot_lod(outf,gm$gmap)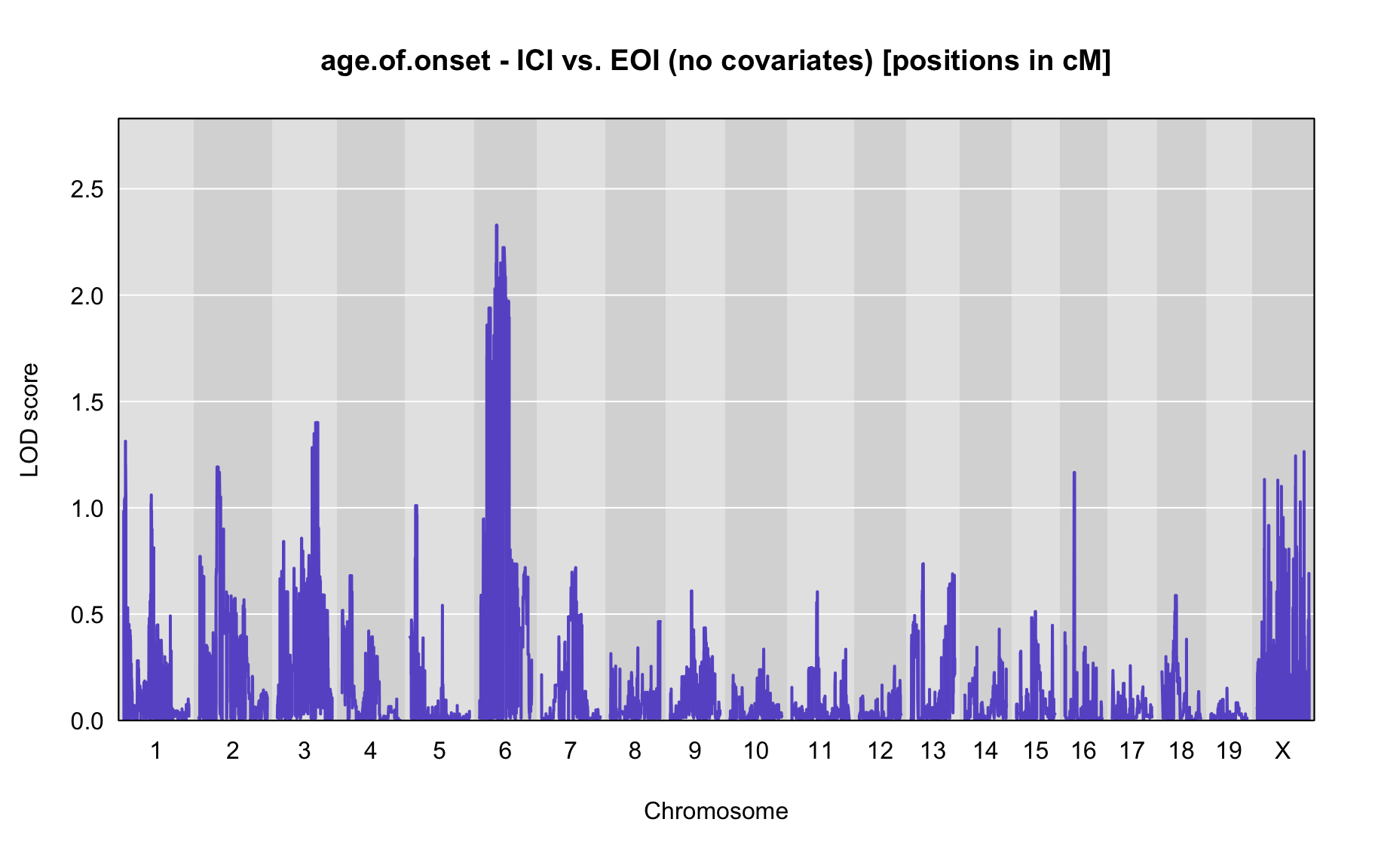
print("with normal kinship")[1] “with normal kinship”
outk <- scan1(pr.qc, covars["age.of.onset"], Xcovar=Xcovar, model="normal", kinship=kinship)
plot_lod(outk,gm$gmap)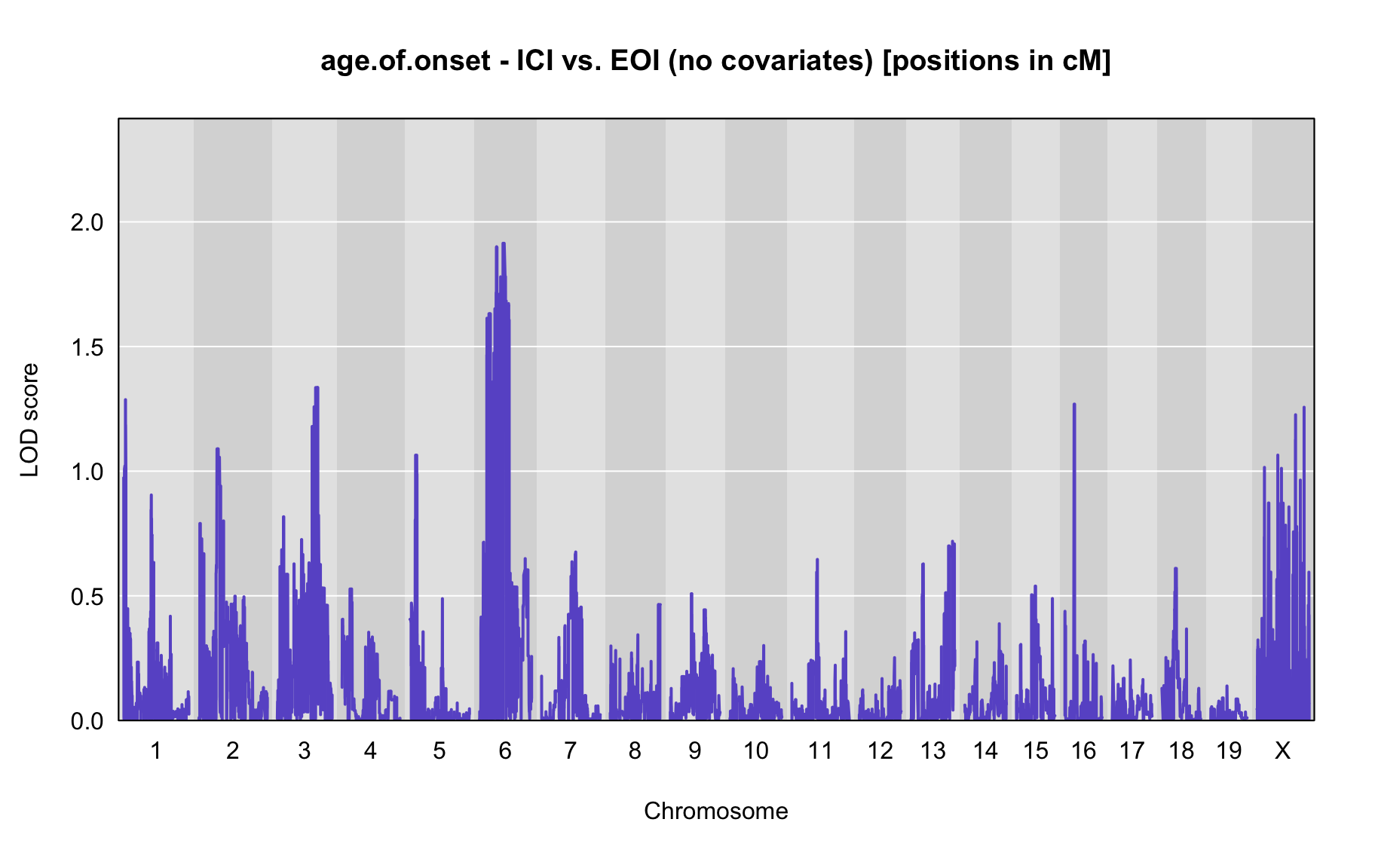
print("with loco kinship")[1] “with loco kinship”
out <- scan1(pr.qc, covars["age.of.onset"], Xcovar=Xcovar, model="normal", kinship=K)
plot_lod(out,gm$gmap)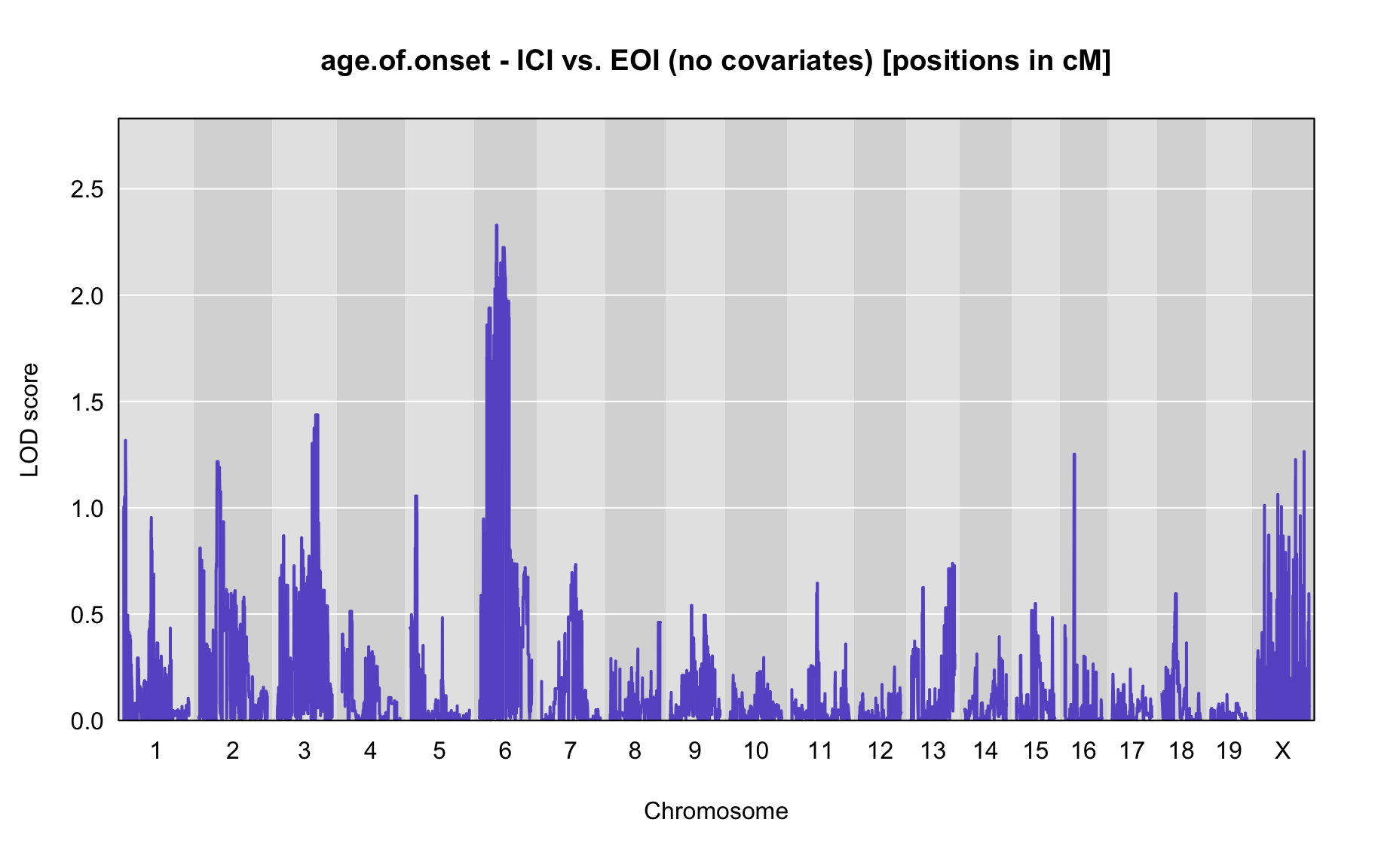
print(paste0("number of samples in analysis = ", as.data.frame(attributes(out)$sample_size)[1,]))[1] “number of samples in analysis = 101”
LOD peaks
Centimorgan (cM)
peaks<-find_peaks(out, gm$gmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) > 0){
peaks$marker <- find_marker(gm$gmap, chr=peaks$chr,pos=peaks$pos)
names(peaks)[2] <- c("phenotype")
peaks <- peaks[-1]
rownames(peaks) <- NULL
print(kable(peaks, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
#plot only peak chromosomes
plot_lod_chr<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " - ICI vs. EOI (no covariates) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are no peaks that have a LOD that reaches suggestive (p<0.05) level of 5.01706726976089 [autosomes]/8.68802183193601 [x-chromosome]”
if(nrow(peaks) < 50 & nrow(peaks) > 0){
for(i in unique(peaks$chr)){
#for (i in 1:nrow(peaks)){
#plot_lod_chr(out,gm$gmap, peaks$chr[i])
plot_lod_chr(out,gm$gmap, i)
}
} else {
print(paste0("There are too many peaks (",nrow(peaks)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are too many peaks (0 peaks) that have a LOD that reaches suggestive (p<0.05) level of 5.01706726976089 [autosomes]/8.68802183193601 [x-chromosome]”
Megabase (MB)
peaks_mba<-find_peaks(out, gm$pmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) > 0){
peaks_mba$marker <- find_marker(gm$pmap, chr=peaks_mba$chr,pos=peaks_mba$pos)
names(peaks_mba)[2] <- c("phenotype")
peaks_mba <- peaks_mba[-1]
rownames(peaks_mba) <- NULL
print(kable(peaks_mba, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
plot_lod_chr_mb<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " - ICI vs. EOI (no covariates) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are no peaks that have a LOD that reaches suggestive (p<0.05) level of 5.01706726976089 [autosomes]/8.68802183193601 [x-chromosome]”
if(nrow(peaks_mba) < 50 & nrow(peaks_mba) > 0){
for(i in unique(peaks_mba$chr)){
#for (i in 1:nrow(peaks_mba)){
#plot_lod_chr_mb(out,gm$pmap, peaks_mba$chr[i])
plot_lod_chr_mb(out,gm$pmap,i)
}
} else {
print(paste0("There are too many peaks (",nrow(peaks_mba)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are too many peaks (0 peaks) that have a LOD that reaches suggestive (p<0.05) level of 5.01706726976089 [autosomes]/8.68802183193601 [x-chromosome]”
QTL effects
For each peak LOD location we give a list of gene
if(nrow(peaks) < 50 & nrow(peaks) > 0){
for (i in 1:nrow(peaks)){
#for (i in 1:1){
#Plot 1
g <- maxmarg(pr.qc, gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i], return_char=TRUE)
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/","qtl_effect_", i, ".png"))
#par(mar=c(4.1, 4.1, 1.5, 0.6))
plot_pxg(g, covars[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
##dev.off()
chr = peaks$chr[i]
# Plot 2
pr_sub <- pull_genoprobint(pr.qc, gm$gmap, chr, c(peaks$ci_lo[i], peaks$ci_hi[i]))
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar)
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], Xcovar=Xcovar)
#coeff <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
#coeff_sub <- scan1coef(pr_sub[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
blup <- scan1blup(pr.qc[,chr], covars[peaks$phenotype[i]])
blup_sub <- scan1blup(pr_sub[,chr], covars[peaks$phenotype[i]])
write.csv(as.data.frame(blup_sub), paste0("data/ici.vs.eoi_",peaks$phenotype[i],"-no.covariates_blup_sub_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_5.batches.csv"), quote=F)
#plot_coef(coeff,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i]," [scan1coeff; positions in cM] )")
# )
#plot_coef(coeff_sub,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i],"; 1.5 LOD drop interval [scan1coeff; positions in cM] ) ")
# )
plot_coef(blup,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," [scan1blup; positions in cM] )")
)
plot_coef(blup_sub,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i],"; 1.5 LOD drop interval [scan1blup; positions in cM] )")
)
# Plot 3
#c2effB <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary", contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
#c2effBb <- scan1blup(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]],Xcovar=Xcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
#plot(c2effB, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
#plot(c2effBb, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
##last_coef <- unclass(c2effB)[nrow(c2effB),2:3] # last two coefficients
##for(t in seq(along=last_coef))
## axis(side=4, at=last_coef[t], names(last_coef)[t], tick=FALSE)
#Table 1
chr = peaks_mba$chr[i]
start=as.numeric(peaks_mba$ci_lo[i])
end=as.numeric(peaks_mba$ci_hi[i])
genesgss = query_genes(chr, start, end)
write.csv(genesgss, file=paste0("data/ici.vs.eoi_",peaks$phenotype[i],"-no.covariates_genes_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_5.batches.csv"), quote=F)
rownames(genesgss) <- NULL
genesgss$strand_old = genesgss$strand
genesgss$strand[genesgss$strand=="+"] <- "positive"
genesgss$strand[genesgss$strand=="-"] <- "negative"
#genesgss <-
#table <-
#genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")] %>%
#kable(escape = F,align = c("ccccccccccc")) %>%
#kable_styling("striped", full_width = T) #%>%
#cat #%>%
#column_spec(1, bold=TRUE)
#
#print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], escape = F,align = c("ccccccccccc")))
print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], "html") %>% kable_styling("striped", full_width = T))
#table
}
} else {
print(paste0("There are too many peaks (",nrow(peaks_mba)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are too many peaks (0 peaks) that have a LOD that reaches suggestive (p<0.05) level of 5.01706726976089 [autosomes]/8.68802183193601 [x-chromosome]”
Additive covariates
Significance thresholds
operm <- scan1perm(pr.qc, covars["age.of.onset"], model="normal", addcovar=addcovar, n_perm=10, perm_Xsp=TRUE, chr_lengths=chr_lengths(gm$gmap))
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
names(summary_table) <- c("autosomes","X")
summary_table$significance.level <- rownames(summary_table)
rownames(summary_table) <- NULL
summary_table[c(3,1:2)] %>%
kable(escape = F,align = c("ccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| significance.level | autosomes | X |
|---|---|---|
| 0.01 | 6.037390 | 8.258475 |
| 0.05 | 5.500254 | 8.245282 |
| 0.1 | 4.827158 | 8.228030 |
Genome-wide scan
plot_lod<-function(out,map){
for (i in 1:dim(out)[2]){
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - ICI vs. EOI (ith additive covariates) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
print("with no kinship")[1] “with no kinship”
outfa <- scan1(pr.qc, covars["age.of.onset"], Xcovar=Xcovar, model="normal", addcovar=addcovar)
plot_lod(outfa,gm$gmap)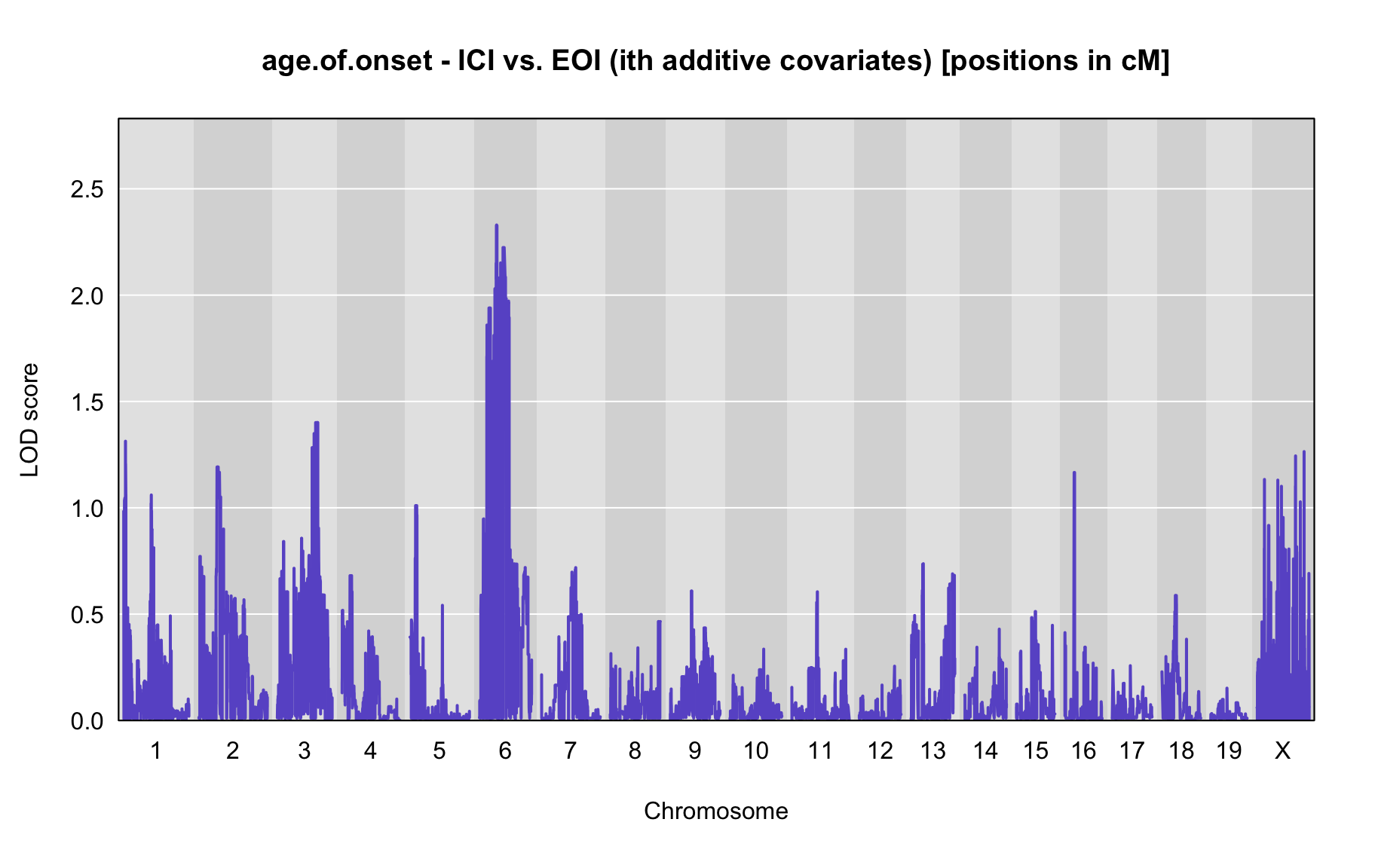
print("with normal kinship")[1] “with normal kinship”
outka <- scan1(pr.qc, covars["age.of.onset"], Xcovar=Xcovar, model="normal", addcovar=addcovar, kinship=kinship)
plot_lod(outka,gm$gmap)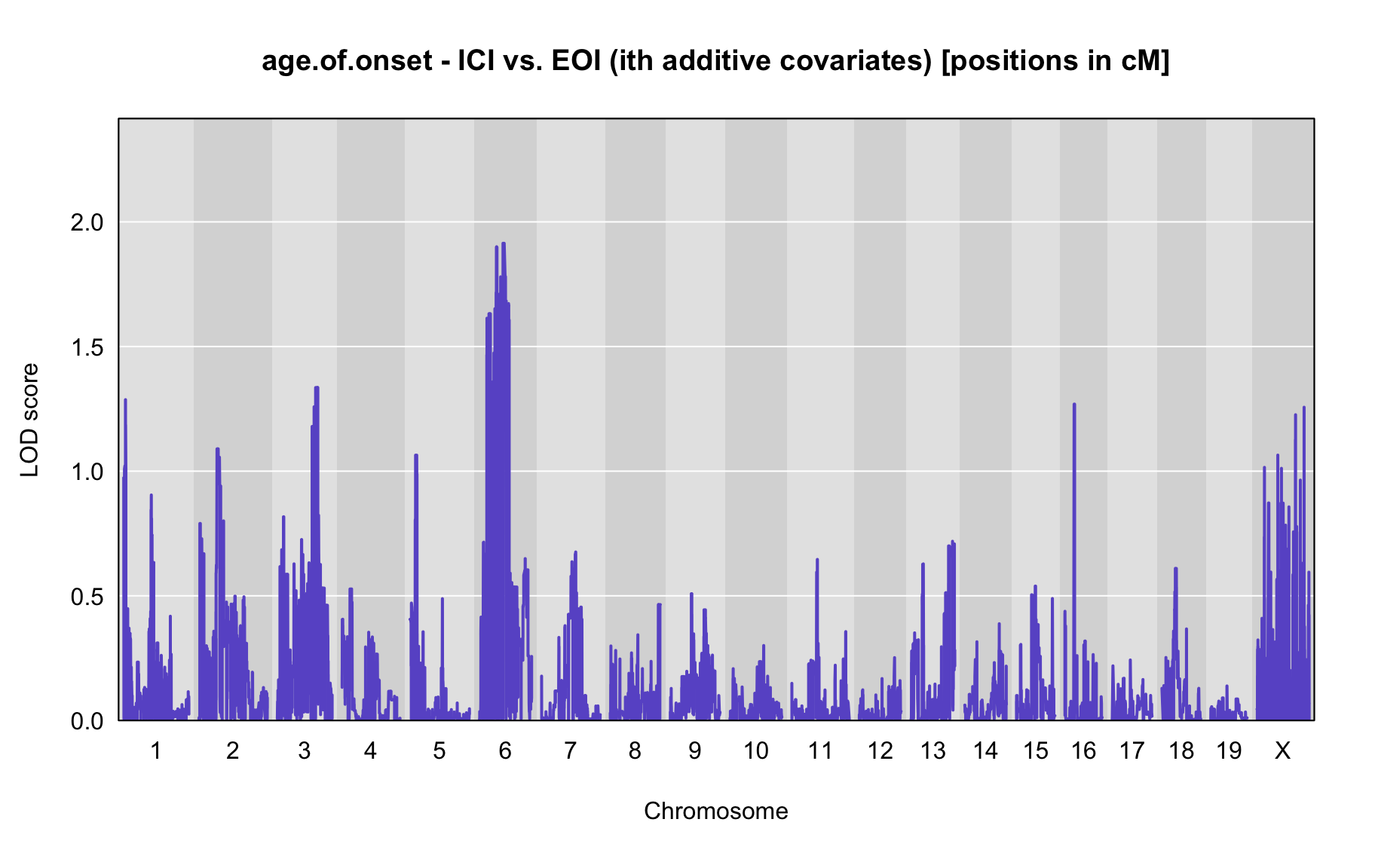
print("with loco kinship")[1] “with loco kinship”
outa <- scan1(pr.qc, covars["age.of.onset"], Xcovar=Xcovar, model="normal", addcovar=addcovar, kinship=K)
plot_lod(outa,gm$gmap)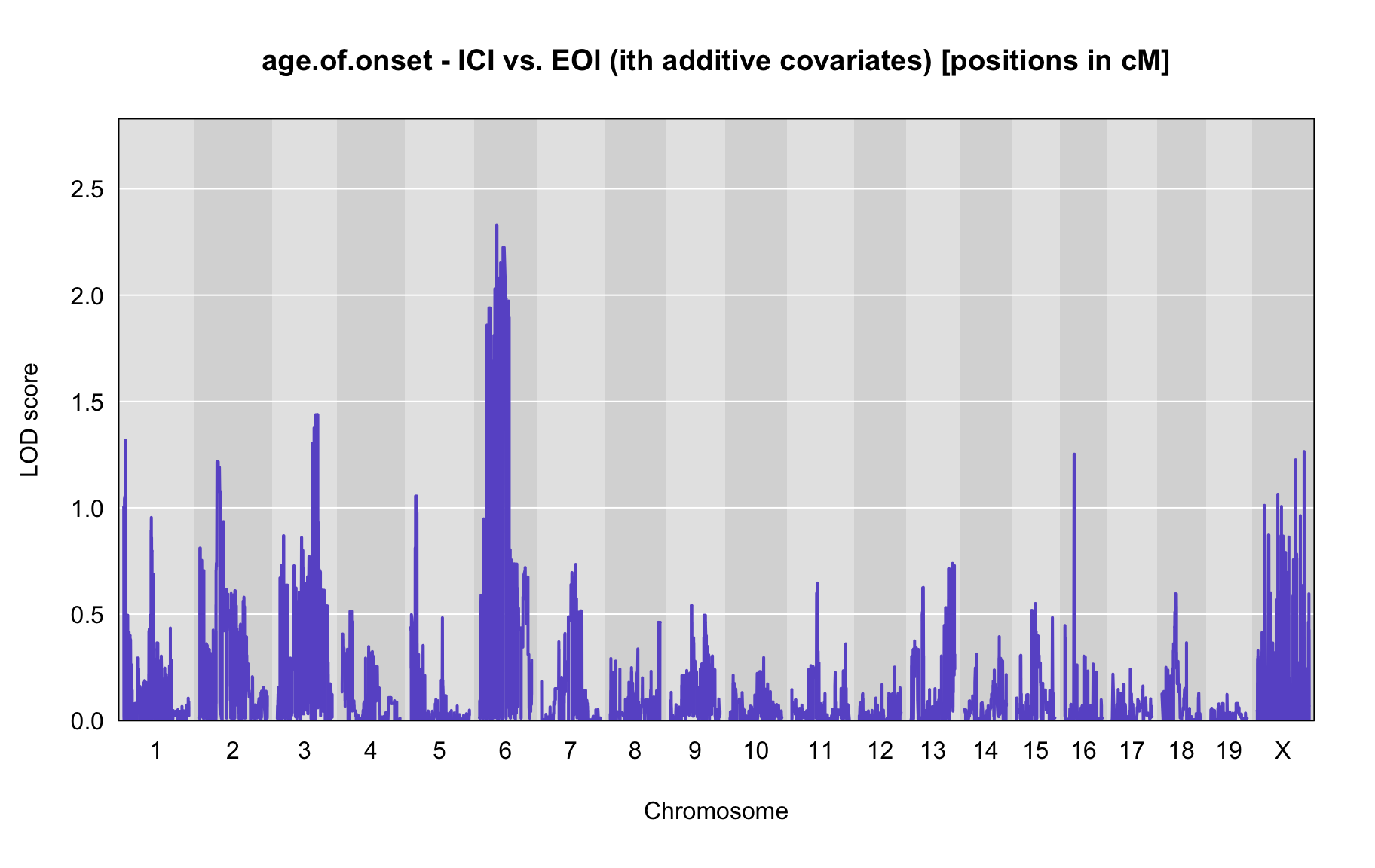
out = outa
print(paste0("number of samples in analysis = ", as.data.frame(attributes(out)$sample_size)[1,]))[1] “number of samples in analysis = 101”
LOD peaks
Centimorgan (cM)
peaks<-find_peaks(out, gm$gmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) > 0){
peaks$marker <- find_marker(gm$gmap, chr=peaks$chr,pos=peaks$pos)
names(peaks)[2] <- c("phenotype")
peaks <- peaks[-1]
rownames(peaks) <- NULL
print(kable(peaks, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
#plot only peak chromosomes
plot_lod_chr<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " - ICI vs. EOI (with additive covariates) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are no peaks that have a LOD that reaches suggestive (p<0.05) level of 5.50025389368799 [autosomes]/8.24528197447556 [x-chromosome]”
if(nrow(peaks) < 50 & nrow(peaks) > 0){
for(i in unique(peaks$chr)){
#for (i in 1:nrow(peaks)){
#plot_lod_chr(out,gm$gmap, peaks$chr[i])
plot_lod_chr(out,gm$gmap, i)
}
} else {
print(paste0("There are too many peaks (",nrow(peaks)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are too many peaks (0 peaks) that have a LOD that reaches suggestive (p<0.05) level of 5.50025389368799 [autosomes]/8.24528197447556 [x-chromosome]”
Megabase (MB)
peaks_mba<-find_peaks(out, gm$pmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) > 0){
peaks_mba$marker <- find_marker(gm$pmap, chr=peaks_mba$chr,pos=peaks_mba$pos)
names(peaks_mba)[2] <- c("phenotype")
peaks_mba <- peaks_mba[-1]
rownames(peaks_mba) <- NULL
print(kable(peaks_mba, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
plot_lod_chr_mb<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " - ICI vs. EOI (with additive covariates) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are no peaks that have a LOD that reaches suggestive (p<0.05) level of 5.50025389368799 [autosomes]/8.24528197447556 [x-chromosome]”
if(nrow(peaks_mba) < 50 & nrow(peaks_mba) > 0){
for(i in unique(peaks_mba$chr)){
#for (i in 1:nrow(peaks_mba)){
#plot_lod_chr_mb(out,gm$pmap, peaks_mba$chr[i])
plot_lod_chr_mb(out,gm$pmap,i)
}
} else {
print(paste0("There are too many peaks (",nrow(peaks_mba)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are too many peaks (0 peaks) that have a LOD that reaches suggestive (p<0.05) level of 5.50025389368799 [autosomes]/8.24528197447556 [x-chromosome]”
QTL effects
For each peak LOD location we give a list of gene
if(nrow(peaks) < 50 & nrow(peaks) > 0){
for (i in 1:nrow(peaks)){
#for (i in 1:1){
#Plot 1
g <- maxmarg(pr.qc, gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i], return_char=TRUE)
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/","qtl_effect_", i, ".png"))
#par(mar=c(4.1, 4.1, 1.5, 0.6))
plot_pxg(g, covars[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
##dev.off()
chr = peaks$chr[i]
# Plot 2
pr_sub <- pull_genoprobint(pr.qc, gm$gmap, chr, c(peaks$ci_lo[i], peaks$ci_hi[i]))
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar)
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], Xcovar=Xcovar)
#coeff <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
#coeff_sub <- scan1coef(pr_sub[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
blup <- scan1blup(pr.qc[,chr], covars[peaks$phenotype[i]])
blup_sub <- scan1blup(pr_sub[,chr], covars[peaks$phenotype[i]])
write.csv(as.data.frame(blup_sub), paste0("data/ici.vs.eoi_",peaks$phenotype[i],"-additive.covariates_blup_sub_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_5.batches.csv"), quote=F)
#plot_coef(coeff,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i]," [scan1coeff; positions in cM] )")
# )
#plot_coef(coeff_sub,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i],"; 1.5 LOD drop interval [scan1coeff; positions in cM] ) ")
# )
plot_coef(blup,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," [scan1blup; positions in cM] )")
)
plot_coef(blup_sub,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i],"; 1.5 LOD drop interval [scan1blup; positions in cM] )")
)
# Plot 3
#c2effB <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary", contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
#c2effBb <- scan1blup(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]],Xcovar=Xcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
#plot(c2effB, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
#plot(c2effBb, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
##last_coef <- unclass(c2effB)[nrow(c2effB),2:3] # last two coefficients
##for(t in seq(along=last_coef))
## axis(side=4, at=last_coef[t], names(last_coef)[t], tick=FALSE)
#Table 1
chr = peaks_mba$chr[i]
start=as.numeric(peaks_mba$ci_lo[i])
end=as.numeric(peaks_mba$ci_hi[i])
genesgss = query_genes(chr, start, end)
write.csv(genesgss, file=paste0("data/ici.vs.eoi_",peaks$phenotype[i],"-additive.covariates_genes_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_5.batches.csv"), quote=F)
rownames(genesgss) <- NULL
genesgss$strand_old = genesgss$strand
genesgss$strand[genesgss$strand=="+"] <- "positive"
genesgss$strand[genesgss$strand=="-"] <- "negative"
#genesgss <-
#table <-
#genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")] %>%
#kable(escape = F,align = c("ccccccccccc")) %>%
#kable_styling("striped", full_width = T) #%>%
#cat #%>%
#column_spec(1, bold=TRUE)
#
#print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], escape = F,align = c("ccccccccccc")))
print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], "html") %>% kable_styling("striped", full_width = T))
#table
}
} else {
print(paste0("There are too many peaks (",nrow(peaks_mba)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are too many peaks (0 peaks) that have a LOD that reaches suggestive (p<0.05) level of 5.50025389368799 [autosomes]/8.24528197447556 [x-chromosome]”
Interactive term
Significance thresholds
operm <- scan1perm(pr.qc, covars["age.of.onset"], model="normal", addcovar=addcovar, intcovar=addcovar, n_perm=10, perm_Xsp=TRUE, chr_lengths=chr_lengths(gm$gmap))
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
names(summary_table) <- c("autosomes","X")
summary_table$significance.level <- rownames(summary_table)
rownames(summary_table) <- NULL
summary_table[c(3,1:2)] %>%
kable(escape = F,align = c("ccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| significance.level | autosomes | X |
|---|---|---|
| 0.01 | 4.312812 | 6.152544 |
| 0.05 | 3.988720 | 6.004072 |
| 0.1 | 3.582594 | 5.809919 |
Genome-wide scan
plot_lod<-function(out,map){
for (i in 1:dim(out)[2]){
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - ICI vs. EOI (with interactive covariate) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
print("with no kinship")[1] “with no kinship”
outfi <- scan1(pr.qc, covars["age.of.onset"], Xcovar=Xcovar, model="normal", addcovar=addcovar, intcovar=addcovar)
plot_lod(outfi,gm$gmap)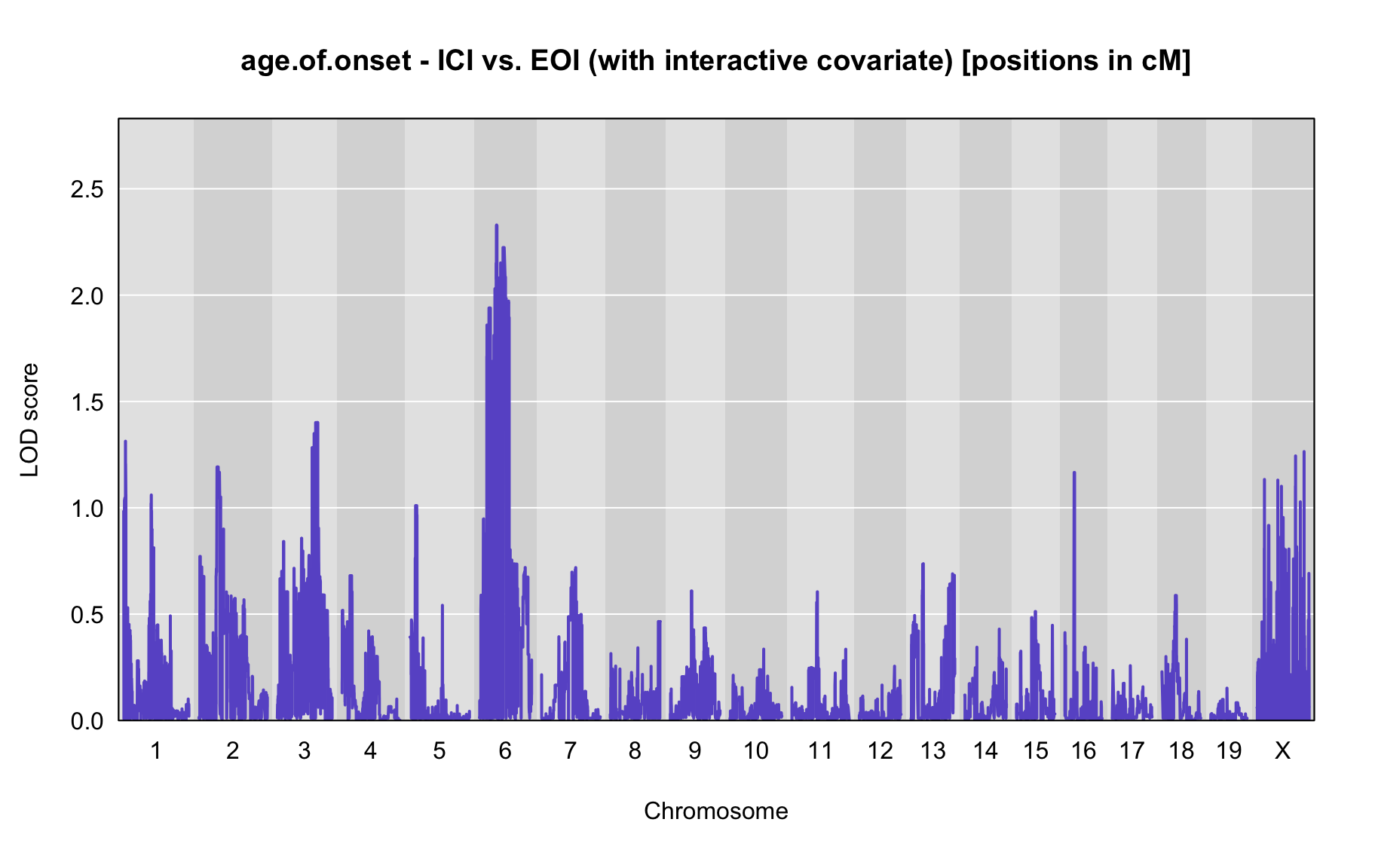
outf.new <- outfi-outfa
plot_lod(outf.new,gm$gmap)
print("with normal kinship")[1] “with normal kinship”
outki <- scan1(pr.qc, covars["age.of.onset"], Xcovar=Xcovar, model="normal", addcovar=addcovar, intcovar=addcovar, kinship=kinship)
plot_lod(outki,gm$gmap)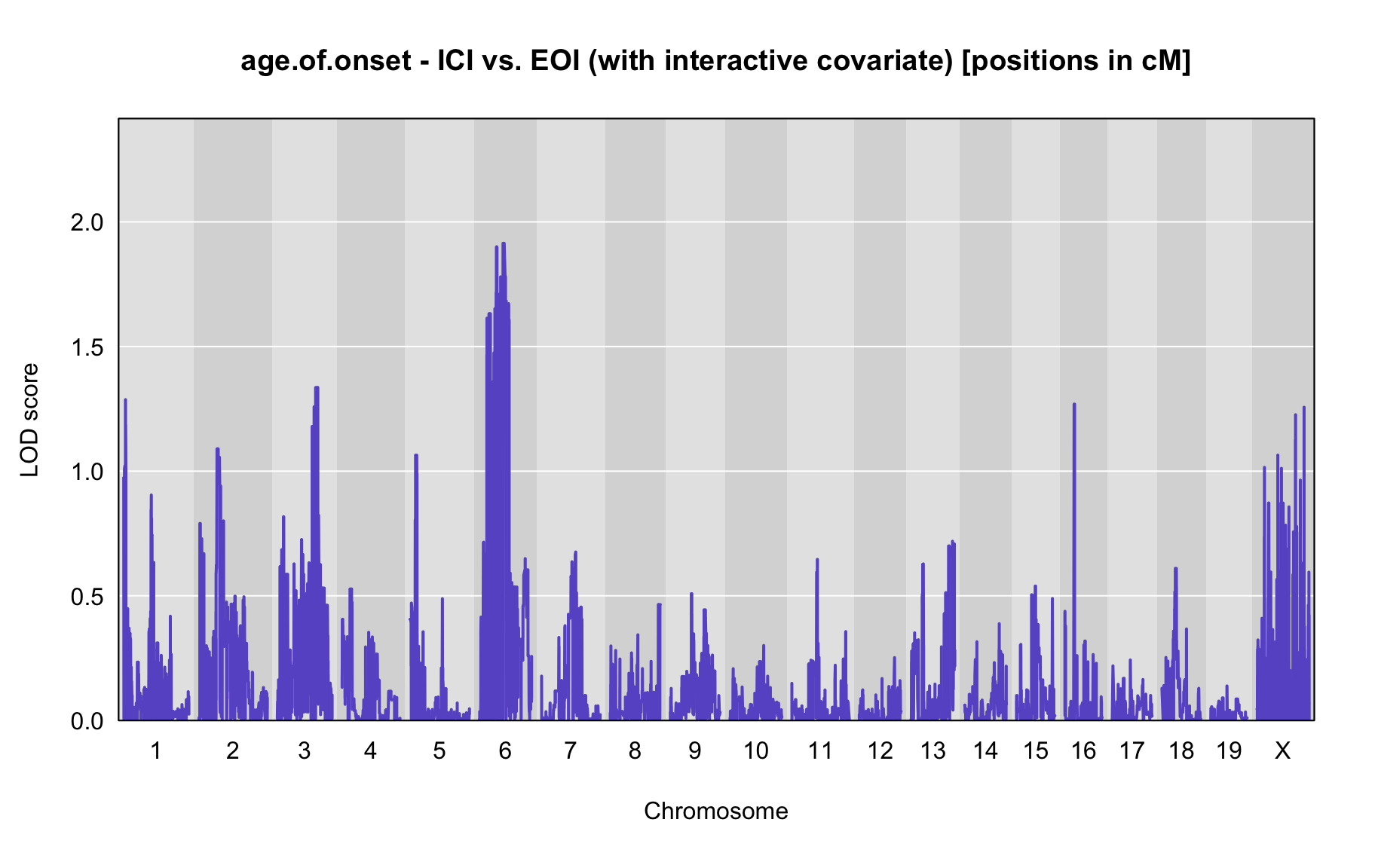
outk.new <- outki-outka
plot_lod(outk.new,gm$gmap)
print("with loco kinship")[1] “with loco kinship”
outi <- scan1(pr.qc, covars["age.of.onset"], Xcovar=Xcovar, model="normal", addcovar=addcovar, intcovar=addcovar, kinship=K)
plot_lod(outi,gm$gmap)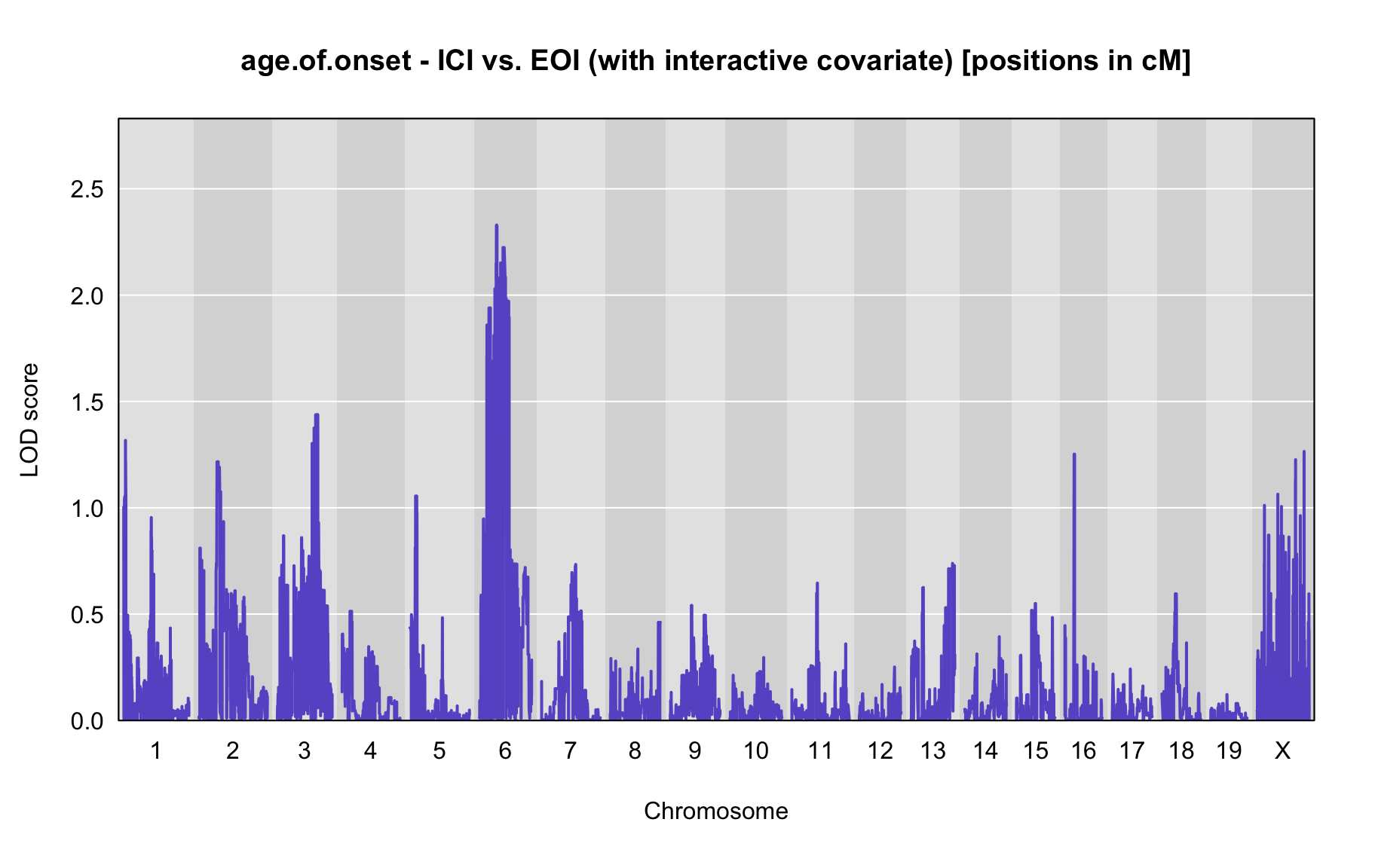
outi.new <- outi-outa
plot_lod(outi.new,gm$gmap)
out = outi.new
print(paste0("number of samples in analysis = ", as.data.frame(attributes(out)$sample_size)[1,]))[1] “number of samples in analysis = 101”
LOD peaks
Centimorgan (cM)
peaks<-find_peaks(out, gm$gmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) > 0){
peaks$marker <- find_marker(gm$gmap, chr=peaks$chr,pos=peaks$pos)
names(peaks)[2] <- c("phenotype")
peaks <- peaks[-1]
rownames(peaks) <- NULL
print(kable(peaks, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
#plot only peak chromosomes
plot_lod_chr<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " - ICI vs. EOI (with interactive covariate) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are no peaks that have a LOD that reaches suggestive (p<0.05) level of 3.98872008636779 [autosomes]/6.00407230801965 [x-chromosome]”
if(nrow(peaks) < 50 & nrow(peaks) > 0){
for(i in unique(peaks$chr)){
#for (i in 1:nrow(peaks)){
#plot_lod_chr(out,gm$gmap, peaks$chr[i])
plot_lod_chr(out,gm$gmap, i)
}
} else {
print(paste0("There are too many peaks (",nrow(peaks)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are too many peaks (0 peaks) that have a LOD that reaches suggestive (p<0.05) level of 3.98872008636779 [autosomes]/6.00407230801965 [x-chromosome]”
Megabase (MB)
peaks_mba<-find_peaks(out, gm$pmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) > 0){
peaks_mba$marker <- find_marker(gm$pmap, chr=peaks_mba$chr,pos=peaks_mba$pos)
names(peaks_mba)[2] <- c("phenotype")
peaks_mba <- peaks_mba[-1]
rownames(peaks_mba) <- NULL
print(kable(peaks_mba, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
plot_lod_chr_mb<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " - ICI vs. EOI (with interactive covariate) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are no peaks that have a LOD that reaches suggestive (p<0.05) level of 3.98872008636779 [autosomes]/6.00407230801965 [x-chromosome]”
if(nrow(peaks_mba) < 50 & nrow(peaks_mba) > 0){
for(i in unique(peaks_mba$chr)){
#for (i in 1:nrow(peaks_mba)){
#plot_lod_chr_mb(out,gm$pmap, peaks_mba$chr[i])
plot_lod_chr_mb(out,gm$pmap,i)
}
} else {
print(paste0("There are too many peaks (",nrow(peaks_mba)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are too many peaks (0 peaks) that have a LOD that reaches suggestive (p<0.05) level of 3.98872008636779 [autosomes]/6.00407230801965 [x-chromosome]”
QTL effects
For each peak LOD location we give a list of gene
if(nrow(peaks) < 50 & nrow(peaks) > 0){
for (i in 1:nrow(peaks)){
#for (i in 1:1){
#Plot 1
g <- maxmarg(pr.qc, gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i], return_char=TRUE)
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/","qtl_effect_", i, ".png"))
#par(mar=c(4.1, 4.1, 1.5, 0.6))
plot_pxg(g, covars[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
##dev.off()
chr = peaks$chr[i]
# Plot 2
pr_sub <- pull_genoprobint(pr.qc, gm$gmap, chr, c(peaks$ci_lo[i], peaks$ci_hi[i]))
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar)
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], Xcovar=Xcovar)
#coeff <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
#coeff_sub <- scan1coef(pr_sub[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
blup <- scan1blup(pr.qc[,chr], covars[peaks$phenotype[i]])
blup_sub <- scan1blup(pr_sub[,chr], covars[peaks$phenotype[i]])
write.csv(as.data.frame(blup_sub), paste0("data/ici.vs.eoi_",peaks$phenotype[i],"-interactive.covariate_blup_sub_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_5.batches.csv"), quote=F)
#plot_coef(coeff,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i]," [scan1coeff; positions in cM] )")
# )
#plot_coef(coeff_sub,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i],"; 1.5 LOD drop interval [scan1coeff; positions in cM] ) ")
# )
plot_coef(blup,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," [scan1blup; positions in cM] )")
)
plot_coef(blup_sub,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i],"; 1.5 LOD drop interval [scan1blup; positions in cM] )")
)
# Plot 3
#c2effB <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary", contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
#c2effBb <- scan1blup(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]],Xcovar=Xcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
#plot(c2effB, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
#plot(c2effBb, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
##last_coef <- unclass(c2effB)[nrow(c2effB),2:3] # last two coefficients
##for(t in seq(along=last_coef))
## axis(side=4, at=last_coef[t], names(last_coef)[t], tick=FALSE)
#Table 1
chr = peaks_mba$chr[i]
start=as.numeric(peaks_mba$ci_lo[i])
end=as.numeric(peaks_mba$ci_hi[i])
genesgss = query_genes(chr, start, end)
write.csv(genesgss, file=paste0("data/ici.vs.eoi_",peaks$phenotype[i],"-interactive.covariate_genes_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_5.batches.csv"), quote=F)
rownames(genesgss) <- NULL
genesgss$strand_old = genesgss$strand
genesgss$strand[genesgss$strand=="+"] <- "positive"
genesgss$strand[genesgss$strand=="-"] <- "negative"
#genesgss <-
#table <-
#genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")] %>%
#kable(escape = F,align = c("ccccccccccc")) %>%
#kable_styling("striped", full_width = T) #%>%
#cat #%>%
#column_spec(1, bold=TRUE)
#
#print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], escape = F,align = c("ccccccccccc")))
print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], "html") %>% kable_styling("striped", full_width = T))
#table
}
} else {
print(paste0("There are too many peaks (",nrow(peaks_mba)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are too many peaks (0 peaks) that have a LOD that reaches suggestive (p<0.05) level of 3.98872008636779 [autosomes]/6.00407230801965 [x-chromosome]”
R version 3.6.2 (2019-12-12)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Catalina 10.15.7
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
locale:
[1] en_AU.UTF-8/en_AU.UTF-8/en_AU.UTF-8/C/en_AU.UTF-8/en_AU.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] abind_1.4-5 qtl2_0.22 reshape2_1.4.4 ggplot2_3.3.5
[5] tibble_3.1.2 psych_2.0.7 readxl_1.3.1 cluster_2.1.0
[9] dplyr_1.0.8 optparse_1.6.6 rhdf5_2.28.1 mclust_5.4.6
[13] tidyr_1.0.2 data.table_1.14.0 knitr_1.33 kableExtra_1.1.0
[17] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] httr_1.4.1 bit64_4.0.5 viridisLite_0.4.0 assertthat_0.2.1
[5] highr_0.9 blob_1.2.1 cellranger_1.1.0 yaml_2.2.1
[9] pillar_1.6.1 RSQLite_2.2.7 backports_1.2.1 lattice_0.20-38
[13] glue_1.4.2 digest_0.6.27 promises_1.1.0 rvest_0.3.5
[17] colorspace_2.0-2 htmltools_0.5.1.1 httpuv_1.5.2 plyr_1.8.6
[21] pkgconfig_2.0.3 purrr_0.3.4 scales_1.1.1 webshot_0.5.2
[25] whisker_0.4 getopt_1.20.3 later_1.0.0 git2r_0.26.1
[29] generics_0.0.2 ellipsis_0.3.2 cachem_1.0.5 withr_2.4.2
[33] cli_3.0.0 mnormt_1.5-7 magrittr_2.0.1 crayon_1.4.1
[37] memoise_2.0.0 evaluate_0.14 fs_1.4.1 fansi_0.5.0
[41] nlme_3.1-142 xml2_1.3.1 tools_3.6.2 hms_0.5.3
[45] lifecycle_1.0.1 stringr_1.4.0 Rhdf5lib_1.6.3 munsell_0.5.0
[49] compiler_3.6.2 rlang_1.0.2 grid_3.6.2 rstudioapi_0.13
[53] rmarkdown_2.1 gtable_0.3.0 DBI_1.1.1 R6_2.5.0
[57] fastmap_1.1.0 bit_4.0.4 utf8_1.2.1 rprojroot_1.3-2
[61] readr_1.3.1 stringi_1.7.2 parallel_3.6.2 Rcpp_1.0.7
[65] vctrs_0.3.8 tidyselect_1.1.2 xfun_0.24