QTL Analysis - age of onset [ICI vs EOI] (corrected phenotype with outliers removed if any)
Belinda Cornes
2022-04-14
Last updated: 2022-04-14
Checks: 5 2
Knit directory: Serreze-T1D_Workflow/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20220210) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /Users/corneb/Documents/MyJax/CS/Projects/Serreze/qc/workflowr/Serreze-T1D_Workflow | . |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version a8c2d1a. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: analysis/.DS_Store
Ignored: data/.DS_Store
Untracked files:
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_0.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_52.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_mis_0.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_mis_52.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_mis_vo.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_vo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.eoi_snpsqc_dis_no-x_updated_5.batches_11.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.eoi_snpsqc_dis_no-x_updated_5.batches_mis_11.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_1.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_mis_1.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_1.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_11.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_11.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_changed.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_changed.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_scanone_5.batches.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_scanone_5.batches_mis.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_oops.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.4.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.4.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.4_miss.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.4_miss.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.5.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.5.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.5_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.5_0.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_vo.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_vo1.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_vo.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_vo1.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_52.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.eoi_5.batches_0.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.eoi_5.batches_52.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.eoi_5.batches_mis_0.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.eoi_5.batches_mis_52.Rmd
Untracked: analysis/index_5.batches_additional_vo.Rmd
Untracked: data/GM_covar.csv
Untracked: data/GM_covar_BC312.csv
Untracked: data/bad_markers_all_4.batches.RData
Untracked: data/bad_markers_all_5.batches.RData
Untracked: data/blup_sub_chr10_lod.drop-1.5_5.batches.csv
Untracked: data/blup_sub_chr3_lod.drop-1.5.csv
Untracked: data/blup_sub_chr3_lod.drop-1.5_5.batches.csv
Untracked: data/blup_sub_chr4_lod.drop-1.5.csv
Untracked: data/blup_sub_chr4_lod.drop-1.5_5.batches.csv
Untracked: data/covar_cleaned_ici.vs.eoi.csv
Untracked: data/covar_cleaned_ici.vs.pbs.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_0.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_52.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_mis.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_mis_0.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_mis_52.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi1.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_0.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_52.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_mis.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_mis_0.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_mis_52.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs1.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_0.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_52.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_mis.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_mis_0.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_mis_52.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_0.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_52.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_mis.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_mis_0.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_mis_52.csv
Untracked: data/covar_corrected_ici.vs.eoi.csv
Untracked: data/covar_corrected_ici.vs.eoi1.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_0.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_52.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_mis.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_mis_0.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_mis_52.csv
Untracked: data/covar_corrected_ici.vs.pbs.csv
Untracked: data/covar_corrected_ici.vs.pbs1.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_0.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_52.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_mis.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_mis_0.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_mis_52.csv
Untracked: data/e.RData
Untracked: data/e_BC312.RData
Untracked: data/e_snpg_samqc_4.batches.RData
Untracked: data/e_snpg_samqc_4.batches_bc.RData
Untracked: data/e_snpg_samqc_5.batches.RData
Untracked: data/errors_ind_4.batches.RData
Untracked: data/errors_ind_4.batches_bc.RData
Untracked: data/errors_ind_5.batches.RData
Untracked: data/files.to.sync.txt
Untracked: data/fitqtl_chr3.peak_chr4.peak_additive.txt
Untracked: data/fitqtl_chr3.peak_chr4.peak_interacting.txt
Untracked: data/fitqtl_chr3.peak_chr4.peak_sex_additive.txt
Untracked: data/fitqtl_chr3.peak_chr4.peak_sex_interacting.txt
Untracked: data/g2blup_effects.csv
Untracked: data/g2blup_effects.xlsx
Untracked: data/genes_chr10_lod.drop-1.5_5.batches.csv
Untracked: data/genes_chr3_lod.drop-1.5.csv
Untracked: data/genes_chr3_lod.drop-1.5_5.batches.csv
Untracked: data/genes_chr4_lod.drop-1.5.csv
Untracked: data/genetic_map.csv
Untracked: data/genetic_map_BC312.csv
Untracked: data/genotype_errors_marker_4.batches.RData
Untracked: data/genotype_errors_marker_5.batches.RData
Untracked: data/genotype_freq_marker_4.batches.RData
Untracked: data/genotype_freq_marker_5.batches.RData
Untracked: data/gm_allqc_4.batches.RData
Untracked: data/gm_allqc_5.batches.RData
Untracked: data/gm_allqc_5.batches_mis.RData
Untracked: data/gm_samqc_3.batches.RData
Untracked: data/gm_samqc_4.batches.RData
Untracked: data/gm_samqc_4.batches_bc.RData
Untracked: data/gm_samqc_5.batches.RData
Untracked: data/gm_serreze.192.RData
Untracked: data/gm_serreze.BC312.RData
Untracked: data/ici-early.vs.pbs_age.of.onset-additive.covariates_blup_sub_chr-7_peak.marker-UNC13388811_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-additive.covariates_genes_chr-7_peak.marker-UNC13388811_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-15_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-15_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-15_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-17_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-17_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-17_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_5.batches.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_scanone_5.batches.Rdata
Untracked: data/ici-early.vs.pbs_scanone_5.batches_mis.Rdata
Untracked: data/ici-early.vs.pbs_scanone_snpsqc_5.batches.Rdata
Untracked: data/ici-early.vs.pbs_scanone_snpsqc_5.batches_mis.Rdata
Untracked: data/ici-early.vs.pbs_scanone_snpsqc_dis_no-x_updated_5.batches.Rdata
Untracked: data/ici-early.vs.pbs_scanone_snpsqc_dis_no-x_updated_5.batches_mis.Rdata
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00020646_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00292499_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00294019_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18216614_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18240977_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18343181_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18363544_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18363544_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18363544_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-11_peak.marker-UNC20090524_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-2_peak.marker-UNCHS008007_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-2_peak.marker-UNCHS008007_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR5263_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR5263_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00105649_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00105649_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00106210_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00107193_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00516812_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00516812_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00522922_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00524205_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00535764_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4901083_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4901083_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4909519_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4909519_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4915446_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4915446_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4938565_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4938565_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4959266_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4959266_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4960612_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4960612_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4961542_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4961542_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5046545_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5055259_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5062524_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5068215_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5076935_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5082757_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5173293_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5193970_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5257268_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5264696_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5267813_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5270225_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5277061_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5278229_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5278229_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5282259_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5287977_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5301460_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5309136_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5321559_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5322455_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5329171_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5360055_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5778977_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5916997_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008229_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008229_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008234_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008234_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008242_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008260_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008260_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008281_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008281_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008338_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008338_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008363_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008363_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008409_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008432_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008609_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008614_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008627_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008659_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008725_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009066_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009067_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009068_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009112_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009693_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009821_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001276_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-sanger2496q_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-sanger2496q_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-4_peak.marker-UNC8250659_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-4_peak.marker-UNC8439633_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-4_peak.marker-UNC8439633_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-4_peak.marker-UNCHS012955_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-6_peak.marker-UNC11108920_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-6_peak.marker-UNC11108920_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00020646_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00292499_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00294019_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18216614_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18240977_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18343181_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18363544_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18363544_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18363544_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-11_peak.marker-UNC20090524_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-2_peak.marker-UNCHS008007_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-2_peak.marker-UNCHS008007_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR5263_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR5263_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00105649_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00105649_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00106210_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00107193_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00516812_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00516812_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00522922_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00524205_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00535764_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4901083_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4901083_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4909519_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4909519_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4915446_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4915446_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4938565_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4938565_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4959266_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4959266_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4960612_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4960612_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4961542_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4961542_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5046545_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5055259_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5062524_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5068215_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5076935_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5082757_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5173293_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5193970_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5257268_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5264696_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5267813_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5270225_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5277061_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5278229_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5278229_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5282259_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5287977_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5301460_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5309136_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5321559_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5322455_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5329171_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5360055_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5778977_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5916997_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008229_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008229_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008234_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008234_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008242_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008260_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008260_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008281_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008281_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008338_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008338_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008363_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008363_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008409_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008432_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008609_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008614_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008627_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008659_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008725_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009066_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009067_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009068_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009112_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009693_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009821_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001276_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-sanger2496q_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-sanger2496q_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-4_peak.marker-UNC8250659_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-4_peak.marker-UNC8439633_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-4_peak.marker-UNC8439633_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-4_peak.marker-UNCHS012955_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-6_peak.marker-UNC11108920_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-6_peak.marker-UNC11108920_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_blup.full_chr-10_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-11_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-12_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-13_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-14_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-15_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-16_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-17_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-18_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-19_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-1_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-2_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-3_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-4_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-6_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-7_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-8_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-9_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-X_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-ICR4295_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-JAX00292499_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18154330_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18195765_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18216614_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18240977_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18856953_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNCJPD004316_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNCJPD009532_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNC19155926_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNC19481366_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNCJPD004792_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-12_peak.marker-ICR4497_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-13_peak.marker-UNCHS036964_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-18_peak.marker-UNCHS045344_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-19_peak.marker-UNCJPD007087_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-1_peak.marker-ICR010_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-1_peak.marker-ICR010_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-2_peak.marker-UNC4609660_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-B6_rs31740628_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR1269_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR5325_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR5347_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR5348_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00104971_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00105059_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00105203_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00107930_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00107980_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00108068_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00108114_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00109750_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00189245_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00517218_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00517906_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00518418_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00518684_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00520122_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00524717_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00524828_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00525380_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00525709_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00527291_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00527587_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00529707_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00531601_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00532122_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4673471_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4681411_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4793677_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4799475_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4909830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4914130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4964062_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4985064_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4991565_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5001723_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5042361_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5055259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5076935_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5082757_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5173293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5182782_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5193970_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5240484_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5256922_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5266664_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5266711_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5267813_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5274554_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5277061_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5282259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5287977_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5301460_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5321559_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5322455_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5328596_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5350384_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5354724_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5375208_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5384072_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5400314_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5433945_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5435874_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5466669_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5517972_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5522257_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5523265_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5532424_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5553882_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5572051_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5582753_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5589260_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5595555_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5623239_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5639830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5680234_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5682314_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5725356_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5727351_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5734420_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5743257_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5751567_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5752623_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5776974_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5778977_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5788507_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5792468_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5794598_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5795561_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5802298_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5806628_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5817478_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5836999_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5841501_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5847927_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5966417_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008129_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008173_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008231_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008235_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008242_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008253_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008256_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008311_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008317_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008372_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008410_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008415_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008431_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008609_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008614_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008649_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008651_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008723_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008802_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008803_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008808_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008834_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008838_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008928_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008948_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008959_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008973_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008987_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009001_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009025_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009034_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009087_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009109_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009142_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009207_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009216_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009306_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009316_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009364_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009377_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009382_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009387_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009395_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009413_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009428_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009463_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009503_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009532_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009541_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009560_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009574_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001276_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001315_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001355_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001373_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001382_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC6851205_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8250659_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8299912_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8300499_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8381981_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8436434_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8439633_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8530763_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS012948_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS012976_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013139_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013304_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-5_peak.marker-JAX00590770_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-ICR1830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-UNC13170794_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-UNCHS019480_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-cr31snv87_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-UNC15430711_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-9_peak.marker-UNC17203329_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-9_peak.marker-UNCHS024593_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-X_peak.marker-UNC30627130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-X_peak.marker-UNCHS048313_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_snpsqc_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_snpsqc_dis_no-x_updated_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_snpsqc_dis_no-x_updated_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_snpsqc_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_snpsqc_dis_no-x_updated_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_snpsqc_dis_no-x_updated_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_snpsqc_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_snpsqc_dis_no-x_updated_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_snpsqc_dis_no-x_updated_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_snpsqc_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_snpsqc_dis_no-x_updated_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_snpsqc_dis_no-x_updated_5.batches_mis.mis.txt
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-ICR4295_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-JAX00292499_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18154330_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18195765_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18216614_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18240977_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18856953_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNCJPD004316_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNCJPD009532_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNC19155926_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNC19481366_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNCJPD004792_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-12_peak.marker-ICR4497_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-13_peak.marker-UNCHS036964_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-18_peak.marker-UNCHS045344_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-19_peak.marker-UNCJPD007087_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-1_peak.marker-ICR010_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-1_peak.marker-ICR010_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-2_peak.marker-UNC4609660_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-B6_rs31740628_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR1269_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR5325_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR5347_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR5348_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00104971_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00105059_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00105203_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00107930_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00107980_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00108068_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00108114_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00109750_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00189245_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00517218_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00517906_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00518418_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00518684_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00520122_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00524717_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00524828_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00525380_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00525709_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00527291_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00527587_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00529707_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00531601_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00532122_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4673471_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4681411_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4793677_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4799475_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4909830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4914130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4964062_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4985064_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4991565_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5001723_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5042361_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5055259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5076935_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5082757_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5173293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5182782_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5193970_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5240484_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5256922_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5266664_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5266711_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5267813_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5274554_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5277061_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5282259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5287977_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5301460_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5321559_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5322455_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5328596_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5350384_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5354724_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5375208_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5384072_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5400314_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5433945_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5435874_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5466669_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5517972_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5522257_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5523265_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5532424_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5553882_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5572051_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5582753_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5589260_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5595555_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5623239_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5639830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5680234_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5682314_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5725356_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5727351_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5734420_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5743257_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5751567_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5752623_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5776974_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5778977_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5788507_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5792468_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5794598_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5795561_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5802298_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5806628_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5817478_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5836999_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5841501_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5847927_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5966417_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008129_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008173_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008231_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008235_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008242_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008253_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008256_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008311_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008317_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008372_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008410_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008415_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008431_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008609_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008614_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008649_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008651_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008723_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008802_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008803_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008808_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008834_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008838_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008928_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008948_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008959_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008973_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008987_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009001_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009025_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009034_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009087_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009109_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009142_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009207_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009216_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009306_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009316_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009364_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009377_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009382_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009387_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009395_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009413_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009428_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009463_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009503_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009532_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009541_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009560_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009574_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001276_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001315_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001355_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001373_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001382_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC6851205_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8250659_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8299912_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8300499_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8381981_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8436434_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8439633_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8530763_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS012948_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS012976_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013139_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013304_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-5_peak.marker-JAX00590770_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-ICR1830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-UNC13170794_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-UNCHS019480_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-cr31snv87_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-UNC15430711_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-9_peak.marker-UNC17203329_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-9_peak.marker-UNCHS024593_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-X_peak.marker-UNC30627130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-X_peak.marker-UNCHS048313_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_gm_qtl_5.batches.csv
Untracked: data/ici.vs.eoi_gm_qtl_5.batches_mis.csv
Untracked: data/ici.vs.eoi_gm_qtl_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_gm_qtl_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_gm_qtl_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18343181_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18376338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18382395_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNCHS029024_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-1_peak.marker-UNC228500_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-1_peak.marker-UNC228500_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-ICR2047_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00112499_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00219554_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00529707_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4837069_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5006948_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5266664_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5300878_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5360055_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5679155_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5945286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC6227106_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008595_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009067_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009693_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001341_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-sanger2623_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-4_peak.marker-UNCHS013125_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-4_peak.marker-UNCHS013125_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-6_peak.marker-UNCHS018170_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-6_peak.marker-UNCHS018231_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18343181_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18376338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18382395_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNCHS029024_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-1_peak.marker-UNC228500_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-1_peak.marker-UNC228500_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-ICR2047_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00112499_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00219554_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00529707_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4837069_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5006948_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5266664_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5300878_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5360055_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5679155_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5945286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC6227106_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008595_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS009067_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS009693_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCJPD001246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCJPD001341_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-sanger2623_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-4_peak.marker-UNCHS013125_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-4_peak.marker-UNCHS013125_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-6_peak.marker-UNCHS018170_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-6_peak.marker-UNCHS018231_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.geno.freq.removed.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.probs.freq.removed.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.eoi_scanone_5.batches.Rdata
Untracked: data/ici.vs.eoi_scanone_5.batches_mis.Rdata
Untracked: data/ici.vs.eoi_scanone_snpsqc_5.batches.Rdata
Untracked: data/ici.vs.eoi_scanone_snpsqc_5.batches_mis.Rdata
Untracked: data/ici.vs.eoi_scanone_snpsqc_dis_no-x_updated_5.batches.Rdata
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNC28655293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-5_peak.marker-UNC8675939_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNC28655293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-5_peak.marker-UNC8675939_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_bad.markers_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-15_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-15_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-15_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-17_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-17_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-17_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-13_peak.marker-UNC22384241_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-19_peak.marker-UNC29893228_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-19_peak.marker-UNC29990033_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-19_peak.marker-UNCHS047192_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-8_peak.marker-UNCHS023353_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-8_peak.marker-UNCHS023353_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-8_peak.marker-UNCHS023364_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_fitqtl_peaks_additive_snpsqc_dis_no-x_updated_5.batches_mis_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_additive_snpsqc_dis_no-x_updated_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_additive_snpsqc_snpsqc_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_interacting_snpsqc_dis_no-x_updated_5.batches_mis_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_interacting_snpsqc_dis_no-x_updated_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_interacting_snpsqc_snpsqc_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_additive_snpsqc_dis_no-x_updated_5.batches_mis_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_additive_snpsqc_dis_no-x_updated_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_additive_snpsqc_snpsqc_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_interacting_snpsqc_dis_no-x_updated_5.batches_mis_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_interacting_snpsqc_dis_no-x_updated_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_interacting_snpsqc_snpsqc_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_genes_chr-13_peak.marker-UNC22384241_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_genes_chr-19_peak.marker-UNC29893228_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-19_peak.marker-UNC29990033_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-19_peak.marker-UNCHS047192_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-8_peak.marker-UNCHS023353_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-8_peak.marker-UNCHS023353_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-8_peak.marker-UNCHS023364_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_mis.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-JAX00153527_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-JAX00153527_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNCHS020443_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNCHS020443_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNCHS020486_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-JAX00153527_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-JAX00153527_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNCHS020443_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNCHS020443_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNCHS020486_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.pbs_scanone_5.batches.Rdata
Untracked: data/ici.vs.pbs_scanone_5.batches_mis.Rdata
Untracked: data/ici.vs.pbs_scanone_snpsqc_5.batches.Rdata
Untracked: data/ici.vs.pbs_scanone_snpsqc_5.batches_mis.Rdata
Untracked: data/ici.vs.pbs_scanone_snpsqc_dis_no-x_updated_5.batches.Rdata
Untracked: data/ici.vs.pbs_scanone_snpsqc_dis_no-x_updated_5.batches_mis.Rdata
Untracked: data/percent_missing_id_3.batches.RData
Untracked: data/percent_missing_id_4.batches.RData
Untracked: data/percent_missing_id_4.batches_bc.RData
Untracked: data/percent_missing_id_5.batches.RData
Untracked: data/percent_missing_marker_4.batches.RData
Untracked: data/percent_missing_marker_5.batches.RData
Untracked: data/pheno.csv
Untracked: data/pheno_BC312.csv
Untracked: data/physical_map.csv
Untracked: data/physical_map_BC312.csv
Untracked: data/qc_info_bad_sample_3.batches.RData
Untracked: data/qc_info_bad_sample_4.batches.RData
Untracked: data/qc_info_bad_sample_4.batches_bc.RData
Untracked: data/qc_info_bad_sample_5.batches.RData
Untracked: data/remaining.markers_geno.freq.xlsx
Untracked: data/sample_geno.csv
Untracked: data/sample_geno_AHB_BC312.csv
Untracked: data/sample_geno_bc.csv
Untracked: data/sample_geno_bc_BC312.csv
Untracked: data/serreze_probs.rds
Untracked: data/serreze_probs_BC312.rds
Untracked: data/serreze_probs_allqc.rds
Untracked: data/serreze_probs_allqc_5.batches.rds
Untracked: data/serreze_probs_allqc_5.batches_mis.rds
Untracked: data/summary.cg_3.batches.RData
Untracked: data/summary.cg_4.batches.RData
Untracked: data/summary.cg_4.batches_bc.RData
Untracked: data/summary.cg_5.batches.RData
Untracked: output/Percent_missing_genotype_data_5.batches.pdf
Untracked: output/Percent_missing_genotype_data_per_marker_5.batches.pdf
Untracked: output/Proportion_matching_genotypes_before_removal_of_bad_samples_5.batches.pdf
Untracked: output/genotype_error_marker_5.batches.pdf
Untracked: output/genotype_frequency_marker_5.batches.pdf
Unstaged changes:
Modified: analysis/4.1.1_qtl.analysis_binary_ici.vs.eoi_snpsqc_dis_no-x_updated.Rmd
Modified: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/index_5.batches.Rmd
Modified: analysis/index_5.batches_additional.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches.Rmd) and HTML (docs/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | a8c2d1a | Belinda Cornes | 2022-04-13 | additonal analysis - peaks and continuous variables |
| html | a8c2d1a | Belinda Cornes | 2022-04-13 | additonal analysis - peaks and continuous variables |
Loading Data
load("data/gm_allqc_5.batches.RData")
#gm_allqc
gm=gm_allqc
gmObject of class cross2 (crosstype "bc")
Total individuals 308
No. genotyped individuals 308
No. phenotyped individuals 308
No. with both geno & pheno 308
No. phenotypes 1
No. covariates 6
No. phenotype covariates 0
No. chromosomes 20
Total markers 34537
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2643 2629 1857 1890 1774 1941 1672 1627 1878 1176 1871 1300 1549 1578 1257 935
17 18 19 X
501 913 1014 4532 #pr <- readRDS("data/serreze_probs_allqc.rds")
#pr <- readRDS("data/serreze_probs.rds")
##extracting animals with ici and eoi group status
miceinfo <- gm$covar[gm$covar$group == "EOI" | gm$covar$group == "ICI",]
table(miceinfo$group)
EOI ICI
164 104 mice.ids <- rownames(miceinfo)
gm <- gm[mice.ids]
gmObject of class cross2 (crosstype "bc")
Total individuals 268
No. genotyped individuals 268
No. phenotyped individuals 268
No. with both geno & pheno 268
No. phenotypes 1
No. covariates 6
No. phenotype covariates 0
No. chromosomes 20
Total markers 34537
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2643 2629 1857 1890 1774 1941 1672 1627 1878 1176 1871 1300 1549 1578 1257 935
17 18 19 X
501 913 1014 4532 table(gm$covar$group)
EOI ICI
164 104 gm.full <- gm
covars <- read_csv("data/covar_corrected.cleaned_ici.vs.eoi_5.batches.csv")
#removing any missing info
missing_ids <- covars[which(covars$"age.of.onset"==""),]$Mouse.ID
length(missing_ids)[1] 3missing_ids[1] "NG00526-ICI-LATE" "NG00522-ICI-LATE" "NG01389-ICI-LATE"if(length(missing_ids) != 0){
gm <- gm[paste0("-",as.character(missing_ids)),]
gm
table(gm$covar$group)
#keeping only mouseids in covars
covars <- covars[gm$covar$id,]
table(covars$group)
}
EOI ICI
164 101 #covars[covars$age.of.onset==0,]$age.of.onset <- ''
##removing problmetic marker
gm <- drop_markers(gm, "UNCHS013106")
query_variants <- create_variant_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/cc_variants.sqlite")
query_genes <- create_gene_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/mouse_genes_mgi.sqlite")
##dropping monomorphic markers within the dataset
g <- do.call("cbind", gm$geno)
#g[1,1:ncol(g)]
gf_mar <- t(apply(g, 2, function(a) table(factor(a, 1:2))/sum(a != 0)))
#gn_mar <- t(apply(g, 2, function(a) table(factor(a, 1:2))))
gf_mar <- gf_mar[gf_mar[,2] != "NaN",]
count <- rowSums(gf_mar <=0.05)
low_freq_df <- merge(as.data.frame(gf_mar),as.data.frame(count), by="row.names",all=T)
low_freq_df[is.na(low_freq_df)] <- ''
low_freq_df <- low_freq_df[low_freq_df$count == 1,]
rownames(low_freq_df) <- low_freq_df$Row.names
low_freq <- find_markerpos(gm, rownames(low_freq_df))
low_freq$id <- rownames(low_freq)
nrow(low_freq)[1] 8397low_freq_bad <- merge(low_freq,low_freq_df, by="row.names",all=T)
names(low_freq_bad)[1] <- c("marker")
gf_mar <- gf_mar[gf_mar[,2] != "NaN",]
MAF <- apply(gf_mar, 1, function(x) min(x))
MAF <- as.data.frame(MAF)
MAF$index <- 1:nrow(gf_mar)
gf_mar_maf <- merge(gf_mar,as.data.frame(MAF), by="row.names")
gf_mar_maf <- gf_mar_maf[order(gf_mar_maf$index),]
gfmar <- NULL
gfmar$gfmar_mar_0 <- sum(gf_mar_maf$MAF==0)
gfmar$gfmar_mar_1 <- sum(gf_mar_maf$MAF< 0.01)
gfmar$gfmar_mar_5 <- sum(gf_mar_maf$MAF< 0.05)
gfmar$gfmar_mar_10 <- sum(gf_mar_maf$MAF< 0.10)
gfmar$gfmar_mar_15 <- sum(gf_mar_maf$MAF< 0.15)
gfmar$gfmar_mar_25 <- sum(gf_mar_maf$MAF< 0.25)
gfmar$gfmar_mar_50 <- sum(gf_mar_maf$MAF< 0.50)
gfmar$total_snps <- nrow(as.data.frame(gf_mar_maf))
gfmar <- t(as.data.frame(gfmar))
gfmar <- as.data.frame(gfmar)
gfmar$count <- gfmar$V1
gfmar[c(2)] %>%
kable(escape = F,align = c("ccccccccc"),linesep ="\\hline") %>%
kable_styling(full_width = F) %>%
kable_styling("striped", full_width = F) %>%
row_spec(8 ,bold=T,color= "white",background = "black")| count | |
|---|---|
| gfmar_mar_0 | 3816 |
| gfmar_mar_1 | 4125 |
| gfmar_mar_5 | 8396 |
| gfmar_mar_10 | 8556 |
| gfmar_mar_15 | 8620 |
| gfmar_mar_25 | 9026 |
| gfmar_mar_50 | 34342 |
| total_snps | 34537 |
gm_qc <- drop_markers(gm, low_freq_bad$marker)
gm_qc <- drop_nullmarkers(gm_qc)
gm = gm_qc
gmObject of class cross2 (crosstype "bc")
Total individuals 265
No. genotyped individuals 265
No. phenotyped individuals 265
No. with both geno & pheno 265
No. phenotypes 1
No. covariates 6
No. phenotype covariates 0
No. chromosomes 20
Total markers 26140
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2336 2342 1589 1661 1536 1681 1450 1438 1654 947 1651 1107 1359 1396 945 728
17 18 19 X
303 747 903 367 markers <- marker_names(gm)
gmapdf <- read.csv("/Users/corneb/Documents/MyJax/CS/Projects/Serreze/haplotype.reconstruction/output_5.batches/genetic_map.csv")
pmapdf <- read.csv("/Users/corneb/Documents/MyJax/CS/Projects/Serreze/haplotype.reconstruction/output_5.batches/physical_map.csv")
#mapdf <- merge(gmapdf,pmapdf, by=c("marker","chr"), all=T)
#rownames(mapdf) <- mapdf$marker
#mapdf <- mapdf[markers,]
#names(mapdf) <- c('marker','chr','gmapdf','pmapdf')
#mapdfnd <- mapdf[!duplicated(mapdf[c(2:3)]),]
pr.qc <- calc_genoprob(gm)Preparing necessary info
covars$age.of.onset <- as.numeric(covars$age.of.onset)
Xcovar <- get_x_covar(gm)
addcovar = model.matrix(~ICI.vs.EOI, data = covars)[,-1]
#addcovar.i = model.matrix(~ICI.vs.EOI, data = covars)[,-1]
K <- calc_kinship(pr.qc, type = "loco")
#heatmap(K[[1]])
K.overall <- calc_kinship(pr.qc, type = "overall")
#heatmap(K.overall)
kinship <- calc_kinship(pr.qc)
heatmap(kinship)
No additive covariates
Significance thresholds
operm <- scan1perm(pr.qc, covars["age.of.onset"], model="normal", n_perm=10, perm_Xsp=TRUE, chr_lengths=chr_lengths(gm$gmap))
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
names(summary_table) <- c("autosomes","X")
summary_table$significance.level <- rownames(summary_table)
rownames(summary_table) <- NULL
summary_table[c(3,1:2)] %>%
kable(escape = F,align = c("ccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| significance.level | autosomes | X |
|---|---|---|
| 0.01 | 2.126534 | 2.538558 |
| 0.05 | 2.027876 | 2.478866 |
| 0.1 | 1.904246 | 2.400809 |
Genome-wide scan
plot_lod<-function(out,map){
for (i in 1:dim(out)[2]){
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - ICI vs. EOI (no covariates) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
print("with no kinship")[1] “with no kinship”
outf <- scan1(pr.qc, covars["age.of.onset"], Xcovar=Xcovar, model="normal")
plot_lod(outf,gm$gmap)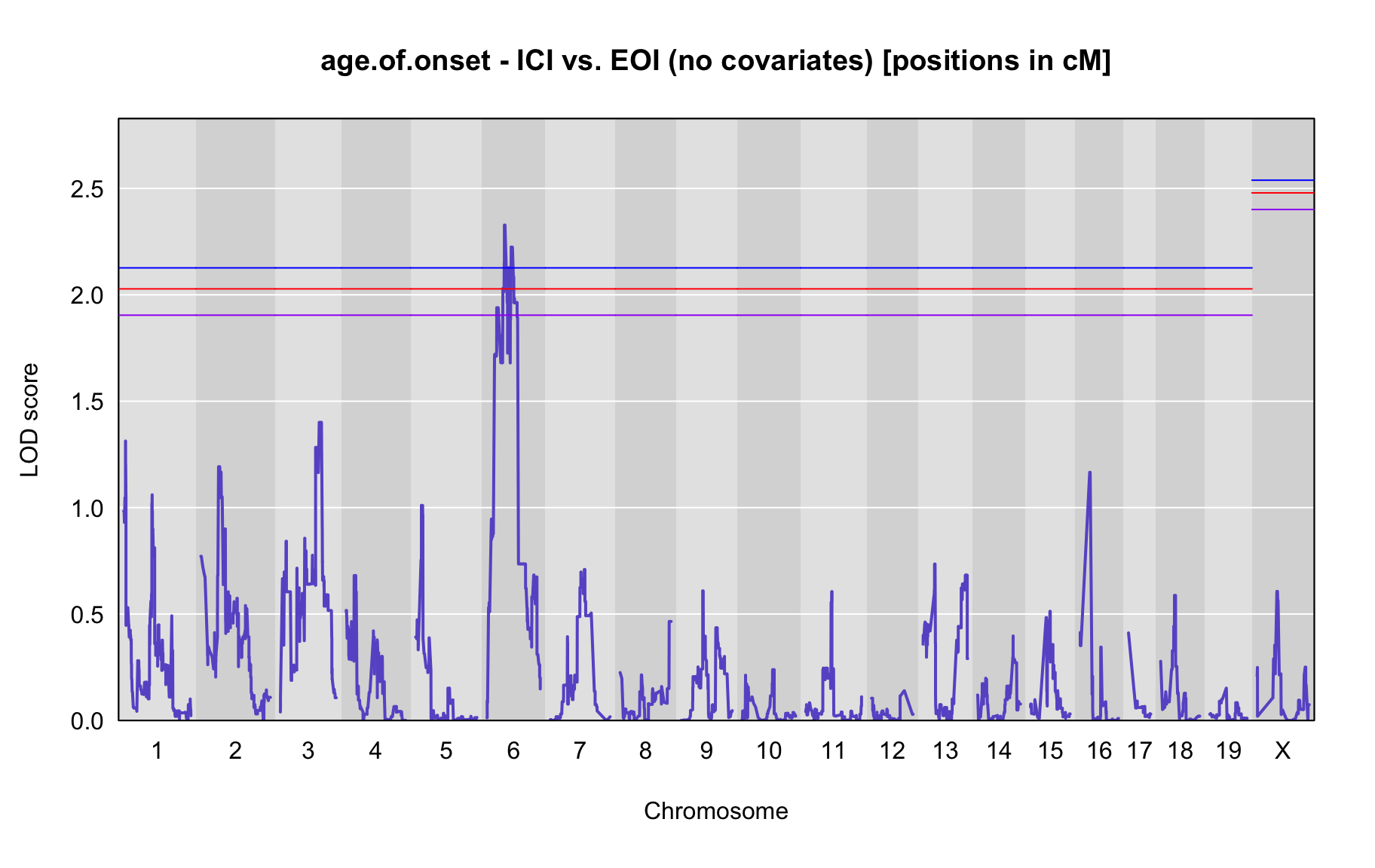
print("with normal kinship")[1] “with normal kinship”
outk <- scan1(pr.qc, covars["age.of.onset"], Xcovar=Xcovar, model="normal", kinship=kinship)
plot_lod(outk,gm$gmap)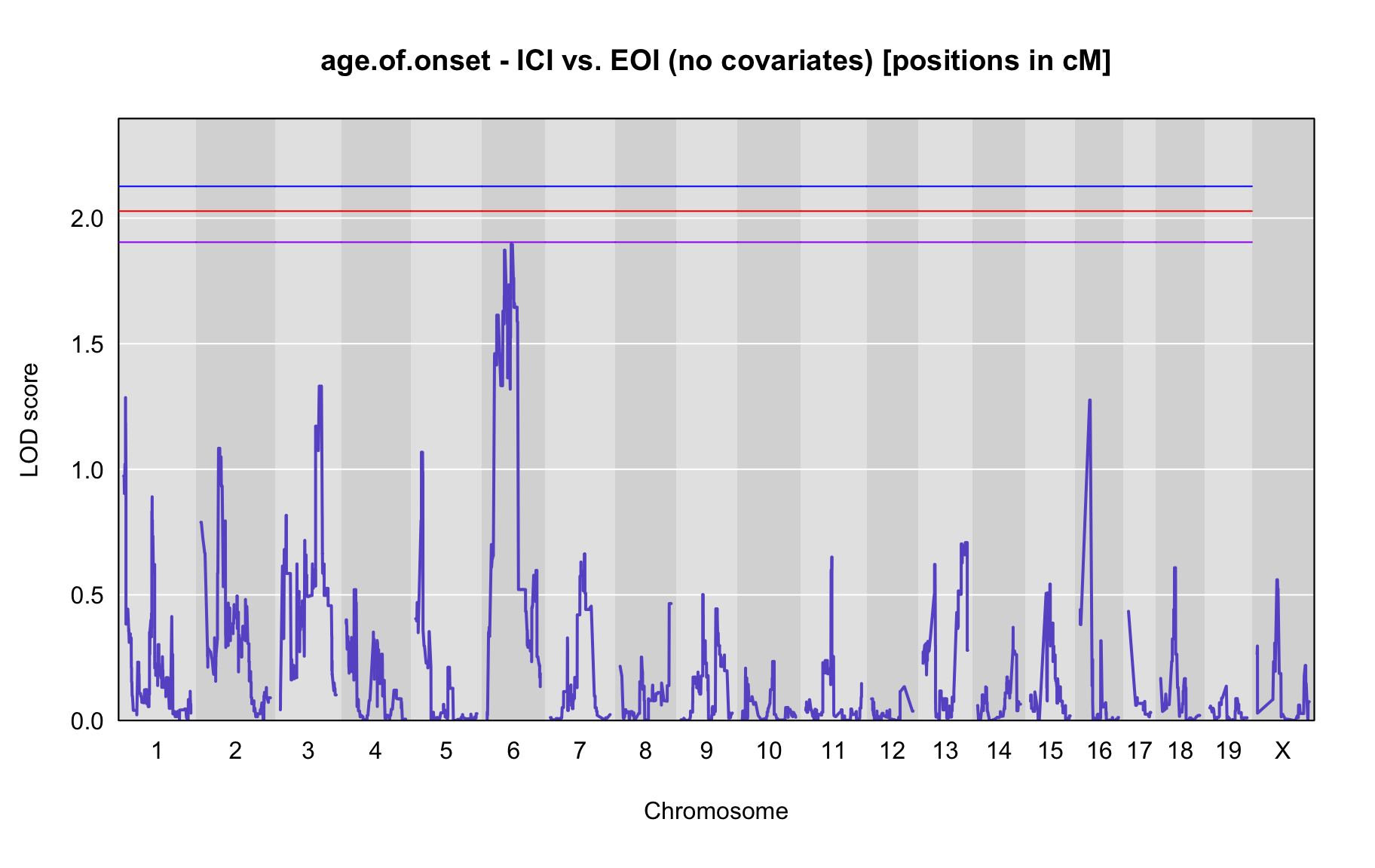
print("with loco kinship")[1] “with loco kinship”
out <- scan1(pr.qc, covars["age.of.onset"], Xcovar=Xcovar, model="normal", kinship=K)
plot_lod(out,gm$gmap)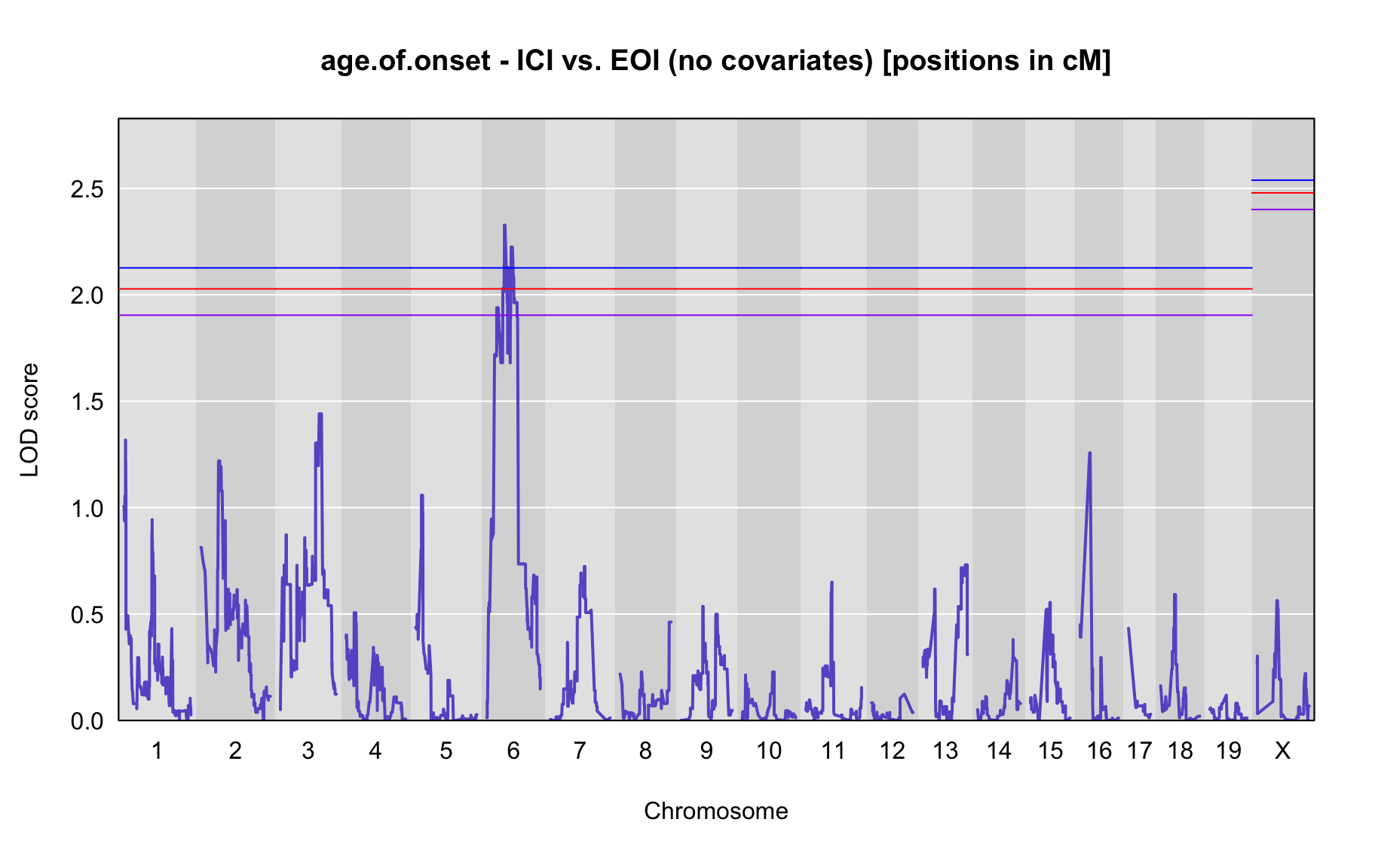
print(paste0("number of samples in analysis = ", as.data.frame(attributes(out)$sample_size)[1,]))[1] “number of samples in analysis = 101”
LOD peaks
Centimorgan (cM)
peaks<-find_peaks(out, gm$gmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) > 0){
peaks$marker <- find_marker(gm$gmap, chr=peaks$chr,pos=peaks$pos)
names(peaks)[2] <- c("phenotype")
peaks <- peaks[-1]
rownames(peaks) <- NULL
print(kable(peaks, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
#plot only peak chromosomes
plot_lod_chr<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " - ICI vs. EOI (no covariates) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}| phenotype | chr | pos | lod | ci_lo | ci_hi | marker |
|---|---|---|---|---|---|---|
| age.of.onset | 6 | 26.68 | 2.327825 | 5.377 | 46.914 | UNC11108920 |
if(nrow(peaks) < 50 & nrow(peaks) > 0){
for(i in unique(peaks$chr)){
#for (i in 1:nrow(peaks)){
#plot_lod_chr(out,gm$gmap, peaks$chr[i])
plot_lod_chr(out,gm$gmap, i)
}
} else {
print(paste0("There are too many peaks (",nrow(peaks)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}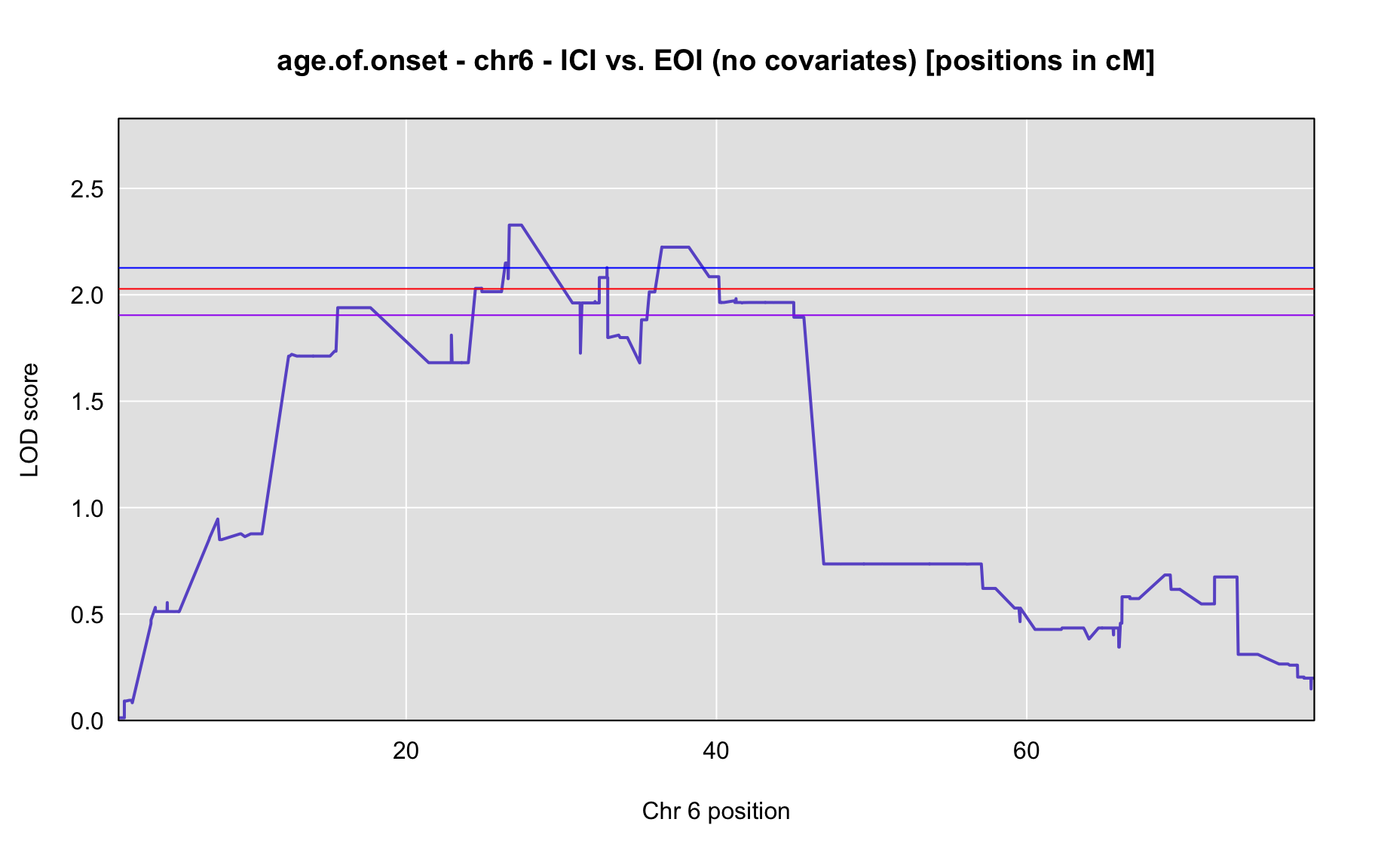
Megabase (MB)
peaks_mba<-find_peaks(out, gm$pmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) > 0){
peaks_mba$marker <- find_marker(gm$pmap, chr=peaks_mba$chr,pos=peaks_mba$pos)
names(peaks_mba)[2] <- c("phenotype")
peaks_mba <- peaks_mba[-1]
rownames(peaks_mba) <- NULL
print(kable(peaks_mba, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
plot_lod_chr_mb<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " - ICI vs. EOI (no covariates) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}| phenotype | chr | pos | lod | ci_lo | ci_hi | marker |
|---|---|---|---|---|---|---|
| age.of.onset | 6 | 54.12741 | 2.327825 | 12.98686 | 100.8239 | UNC11108920 |
if(nrow(peaks_mba) < 50 & nrow(peaks_mba) > 0){
for(i in unique(peaks_mba$chr)){
#for (i in 1:nrow(peaks_mba)){
#plot_lod_chr_mb(out,gm$pmap, peaks_mba$chr[i])
plot_lod_chr_mb(out,gm$pmap,i)
}
} else {
print(paste0("There are too many peaks (",nrow(peaks_mba)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}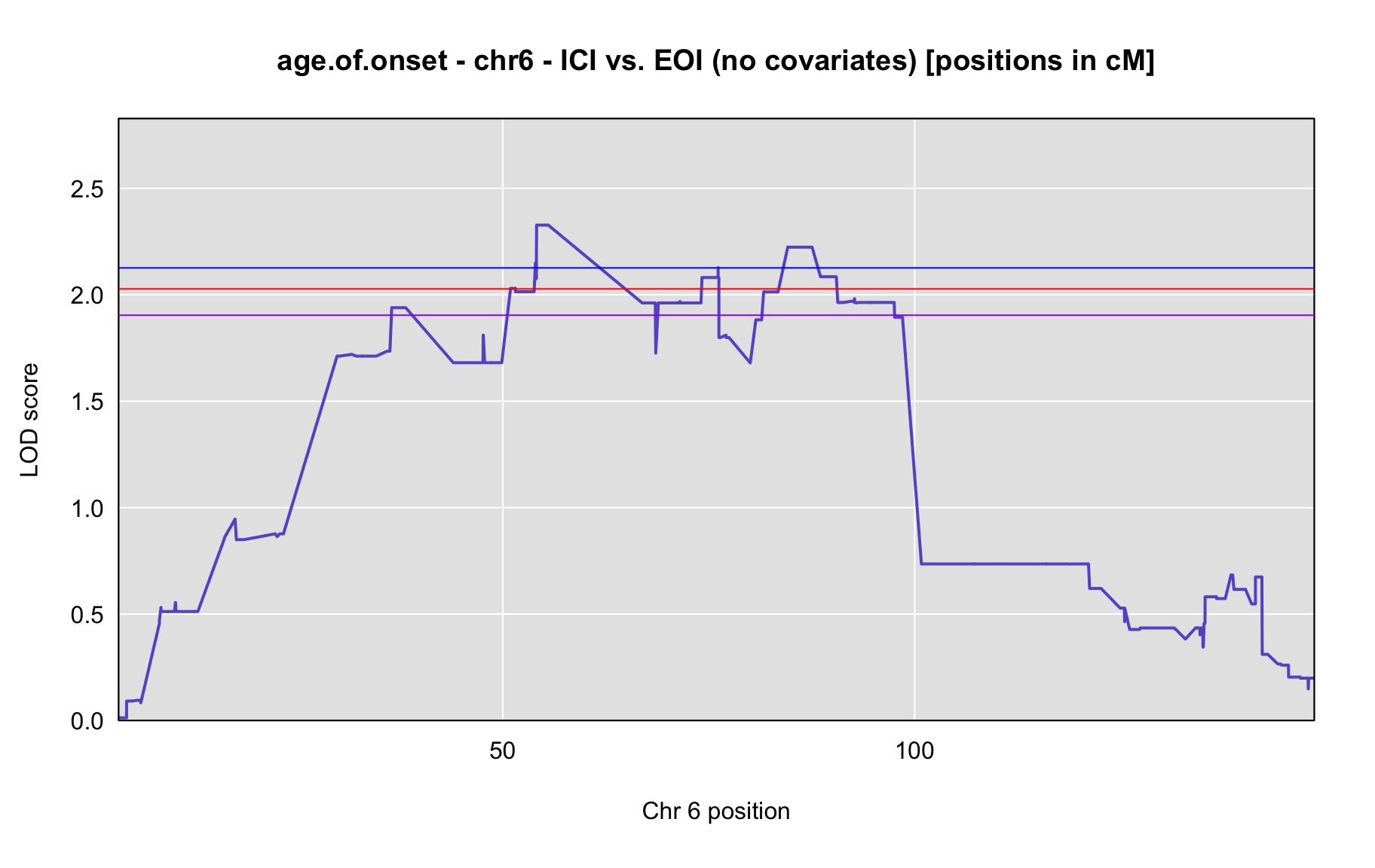
QTL effects
For each peak LOD location we give a list of gene
if(nrow(peaks) < 50 & nrow(peaks) > 0){
for (i in 1:nrow(peaks)){
#for (i in 1:1){
#Plot 1
g <- maxmarg(pr.qc, gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i], return_char=TRUE)
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/","qtl_effect_", i, ".png"))
#par(mar=c(4.1, 4.1, 1.5, 0.6))
plot_pxg(g, covars[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
##dev.off()
chr = peaks$chr[i]
# Plot 2
pr_sub <- pull_genoprobint(pr.qc, gm$gmap, chr, c(peaks$ci_lo[i], peaks$ci_hi[i]))
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar)
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], Xcovar=Xcovar)
#coeff <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
#coeff_sub <- scan1coef(pr_sub[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
blup <- scan1blup(pr.qc[,chr], covars[peaks$phenotype[i]])
blup_sub <- scan1blup(pr_sub[,chr], covars[peaks$phenotype[i]])
write.csv(as.data.frame(blup_sub), paste0("data/ici.vs.eoi_",peaks$phenotype[i],"-no.covariates_blup_sub_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_snpsqc_5.batches.csv"), quote=F)
#plot_coef(coeff,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i]," [scan1coeff; positions in cM] )")
# )
#plot_coef(coeff_sub,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i],"; 1.5 LOD drop interval [scan1coeff; positions in cM] ) ")
# )
plot_coef(blup,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," [scan1blup; positions in cM] )")
)
plot_coef(blup_sub,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i],"; 1.5 LOD drop interval [scan1blup; positions in cM] )")
)
# Plot 3
#c2effB <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary", contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
#c2effBb <- scan1blup(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]],Xcovar=Xcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
#plot(c2effB, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
#plot(c2effBb, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
##last_coef <- unclass(c2effB)[nrow(c2effB),2:3] # last two coefficients
##for(t in seq(along=last_coef))
## axis(side=4, at=last_coef[t], names(last_coef)[t], tick=FALSE)
#Table 1
chr = peaks_mba$chr[i]
start=as.numeric(peaks_mba$ci_lo[i])
end=as.numeric(peaks_mba$ci_hi[i])
genesgss = query_genes(chr, start, end)
write.csv(genesgss, file=paste0("data/ici.vs.eoi_",peaks$phenotype[i],"-no.covariates_genes_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_snpsqc_5.batches.csv"), quote=F)
rownames(genesgss) <- NULL
genesgss$strand_old = genesgss$strand
genesgss$strand[genesgss$strand=="+"] <- "positive"
genesgss$strand[genesgss$strand=="-"] <- "negative"
#genesgss <-
#table <-
#genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")] %>%
#kable(escape = F,align = c("ccccccccccc")) %>%
#kable_styling("striped", full_width = T) #%>%
#cat #%>%
#column_spec(1, bold=TRUE)
#
#print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], escape = F,align = c("ccccccccccc")))
print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], "html") %>% kable_styling("striped", full_width = T))
#table
}
} else {
print(paste0("There are too many peaks (",nrow(peaks_mba)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}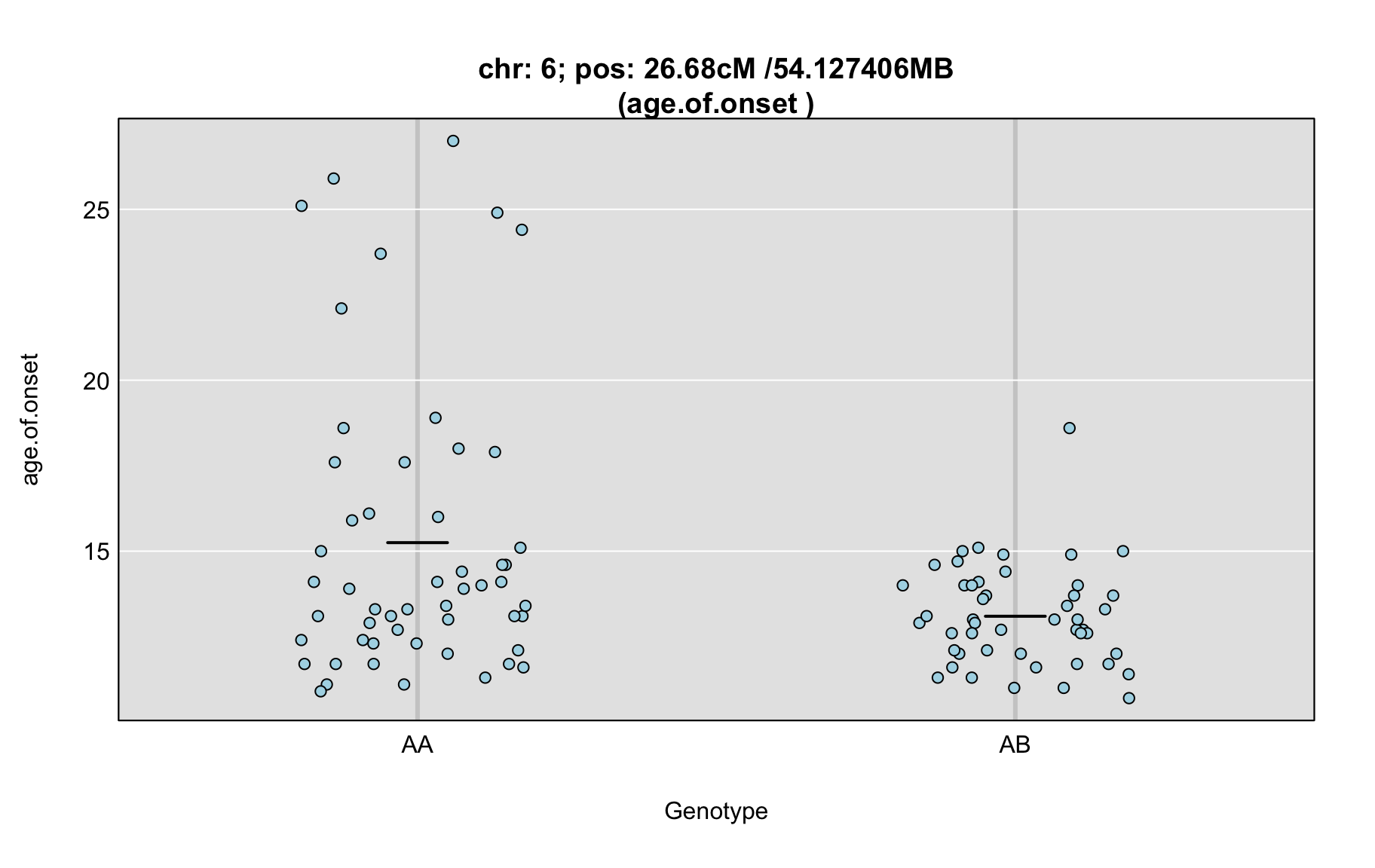
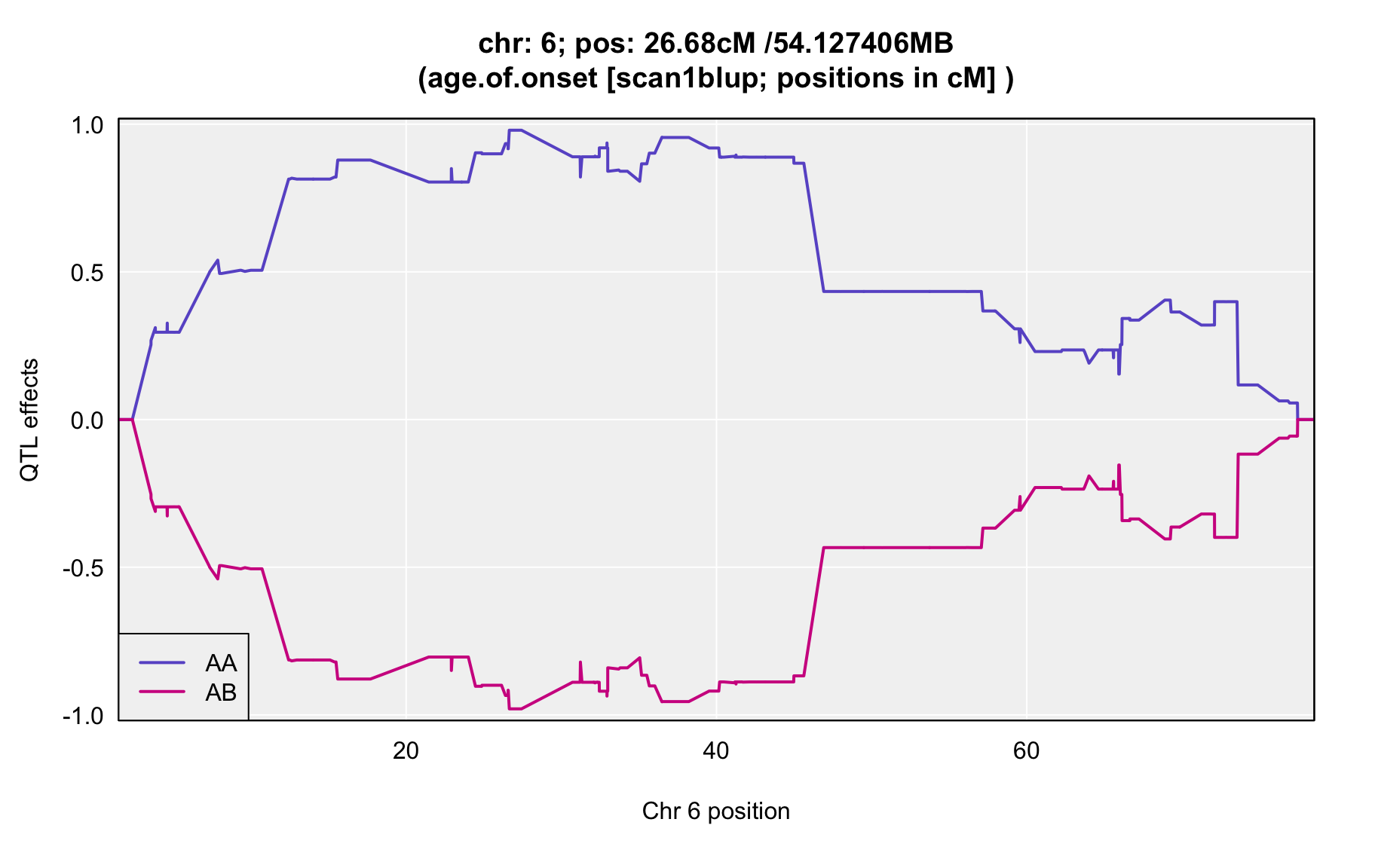
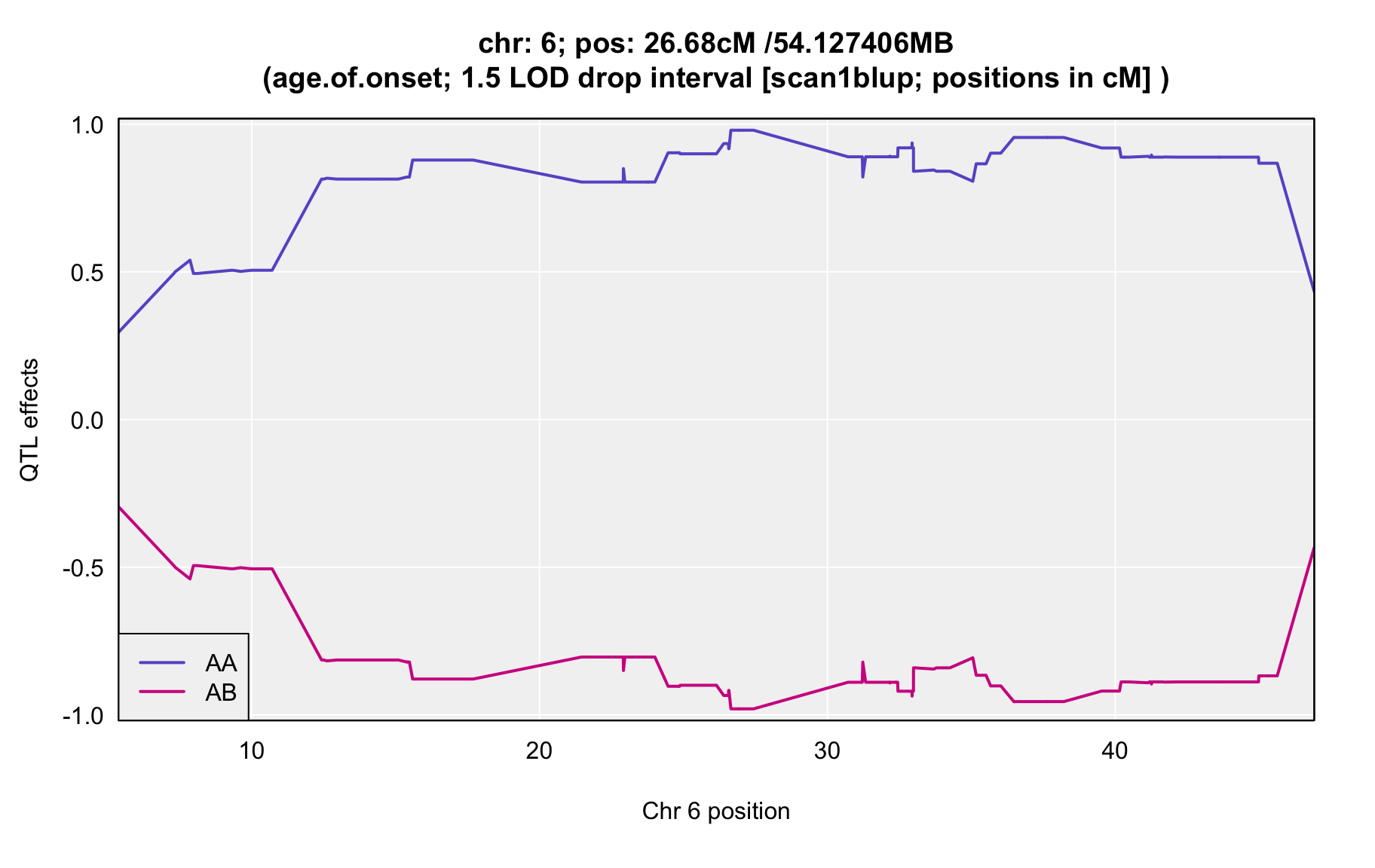
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 6 | pseudogene | 13.05540 | 13.05662 | negative | MGI_C57BL6J_3642954 | Gm5300 | NCBI_Gene:384350,ENSEMBL:ENSMUSG00000098052 | MGI:3642954 | pseudogene | predicted gene 5300 |
| 6 | gene | 13.06974 | 13.08927 | positive | MGI_C57BL6J_1919150 | Tmem106b | NCBI_Gene:71900,ENSEMBL:ENSMUSG00000029571 | MGI:1919150 | protein coding gene | transmembrane protein 106B |
| 6 | gene | 13.11130 | 13.11143 | positive | MGI_C57BL6J_5454762 | Gm24985 | ENSEMBL:ENSMUSG00000077643 | MGI:5454762 | snoRNA gene | predicted gene, 24985 |
| 6 | gene | 13.15630 | 13.22496 | negative | MGI_C57BL6J_2685313 | Vwde | NCBI_Gene:232585,ENSEMBL:ENSMUSG00000079679 | MGI:2685313 | protein coding gene | von Willebrand factor D and EGF domains |
| 6 | gene | 13.26370 | 13.26461 | positive | MGI_C57BL6J_2685571 | Gm725 | NCBI_Gene:277899 | MGI:2685571 | protein coding gene | predicted gene 725 |
| 6 | gene | 13.41334 | 13.41600 | positive | MGI_C57BL6J_1916663 | 1700016P04Rik | NCBI_Gene:69413,ENSEMBL:ENSMUSG00000101894 | MGI:1916663 | lncRNA gene | RIKEN cDNA 1700016P04 gene |
| 6 | pseudogene | 13.46140 | 13.46199 | negative | MGI_C57BL6J_3780965 | Gm2796 | NCBI_Gene:100040478,ENSEMBL:ENSMUSG00000107500 | MGI:3780965 | pseudogene | predicted gene 2796 |
| 6 | gene | 13.54951 | 13.54961 | positive | MGI_C57BL6J_5455514 | Gm25737 | ENSEMBL:ENSMUSG00000095489 | MGI:5455514 | snRNA gene | predicted gene, 25737 |
| 6 | gene | 13.58069 | 13.60822 | negative | MGI_C57BL6J_1921794 | Tmem168 | NCBI_Gene:101118,ENSEMBL:ENSMUSG00000029569 | MGI:1921794 | protein coding gene | transmembrane protein 168 |
| 6 | gene | 13.62567 | 13.67797 | negative | MGI_C57BL6J_2141466 | Bmt2 | NCBI_Gene:101148,ENSEMBL:ENSMUSG00000042742 | MGI:2141466 | protein coding gene | base methyltransferase of 25S rRNA 2 |
| 6 | gene | 13.74948 | 13.77671 | positive | MGI_C57BL6J_5623239 | Gm40354 | NCBI_Gene:105244809 | MGI:5623239 | lncRNA gene | predicted gene, 40354 |
| 6 | gene | 13.82416 | 13.83204 | negative | MGI_C57BL6J_5621651 | Gm38766 | NCBI_Gene:105242598 | MGI:5621651 | lncRNA gene | predicted gene, 38766 |
| 6 | gene | 13.83445 | 13.83994 | negative | MGI_C57BL6J_1927851 | Gpr85 | NCBI_Gene:64450,ENSEMBL:ENSMUSG00000048216 | MGI:1927851 | protein coding gene | G protein-coupled receptor 85 |
| 6 | gene | 13.86773 | 13.87152 | negative | MGI_C57BL6J_1913770 | 2610001J05Rik | NCBI_Gene:66520,ENSEMBL:ENSMUSG00000052419 | MGI:1913770 | protein coding gene | RIKEN cDNA 2610001J05 gene |
| 6 | gene | 13.87153 | 14.04439 | positive | MGI_C57BL6J_1923561 | 1110019D14Rik | NCBI_Gene:76311,ENSEMBL:ENSMUSG00000097616 | MGI:1923561 | lncRNA gene | RIKEN cDNA 1110019D14 gene |
| 6 | pseudogene | 13.91315 | 13.91345 | negative | MGI_C57BL6J_5690804 | Gm44412 | ENSEMBL:ENSMUSG00000107562 | MGI:5690804 | pseudogene | predicted gene, 44412 |
| 6 | pseudogene | 14.00601 | 14.00635 | positive | MGI_C57BL6J_5690803 | Gm44411 | ENSEMBL:ENSMUSG00000107942 | MGI:5690803 | pseudogene | predicted gene, 44411 |
| 6 | gene | 14.01853 | 14.04552 | positive | MGI_C57BL6J_5621652 | Gm38767 | NCBI_Gene:105242599 | MGI:5621652 | lncRNA gene | predicted gene, 38767 |
| 6 | pseudogene | 14.08103 | 14.08165 | negative | MGI_C57BL6J_5011437 | Gm19252 | NCBI_Gene:100462956,ENSEMBL:ENSMUSG00000108205 | MGI:5011437 | pseudogene | predicted gene, 19252 |
| 6 | gene | 14.11332 | 14.11343 | positive | MGI_C57BL6J_5453537 | Gm23760 | ENSEMBL:ENSMUSG00000088975 | MGI:5453537 | snoRNA gene | predicted gene, 23760 |
| 6 | pseudogene | 14.38948 | 14.39005 | negative | MGI_C57BL6J_3648638 | Gm8820 | NCBI_Gene:667797,ENSEMBL:ENSMUSG00000107403 | MGI:3648638 | pseudogene | predicted gene 8820 |
| 6 | gene | 14.45705 | 14.45715 | negative | MGI_C57BL6J_5452790 | Gm23013 | ENSEMBL:ENSMUSG00000077334 | MGI:5452790 | snRNA gene | predicted gene, 23013 |
| 6 | pseudogene | 14.57077 | 14.57103 | negative | MGI_C57BL6J_5690838 | Gm44446 | ENSEMBL:ENSMUSG00000107657 | MGI:5690838 | pseudogene | predicted gene, 44446 |
| 6 | gene | 14.60845 | 14.61164 | positive | MGI_C57BL6J_5621653 | Gm38768 | NCBI_Gene:105242600 | MGI:5621653 | lncRNA gene | predicted gene, 38768 |
| 6 | gene | 14.71382 | 14.75527 | negative | MGI_C57BL6J_2153588 | Ppp1r3a | NCBI_Gene:140491,ENSEMBL:ENSMUSG00000042717 | MGI:2153588 | protein coding gene | protein phosphatase 1, regulatory subunit 3A |
| 6 | gene | 14.78468 | 14.81921 | negative | MGI_C57BL6J_5595459 | Gm36300 | NCBI_Gene:102640167 | MGI:5595459 | lncRNA gene | predicted gene, 36300 |
| 6 | gene | 14.90135 | 15.44198 | positive | MGI_C57BL6J_2148705 | Foxp2 | NCBI_Gene:114142,ENSEMBL:ENSMUSG00000029563 | MGI:2148705 | protein coding gene | forkhead box P2 |
| 6 | pseudogene | 14.94852 | 14.94914 | positive | MGI_C57BL6J_3781017 | Gm2845 | NCBI_Gene:100040568 | MGI:3781017 | pseudogene | predicted gene 2845 |
| 6 | gene | 15.40498 | 15.40509 | positive | MGI_C57BL6J_5455247 | Gm25470 | ENSEMBL:ENSMUSG00000093317 | MGI:5455247 | miRNA gene | predicted gene, 25470 |
| 6 | gene | 15.63612 | 15.63837 | positive | MGI_C57BL6J_5690431 | Gm44039 | ENSEMBL:ENSMUSG00000107703 | MGI:5690431 | unclassified gene | predicted gene, 44039 |
| 6 | gene | 15.63813 | 15.63976 | negative | MGI_C57BL6J_5595511 | Gm36352 | NCBI_Gene:102640235 | MGI:5595511 | lncRNA gene | predicted gene, 36352 |
| 6 | gene | 15.72066 | 15.80217 | positive | MGI_C57BL6J_104611 | Mdfic | NCBI_Gene:16543,ENSEMBL:ENSMUSG00000041390 | MGI:104611 | protein coding gene | MyoD family inhibitor domain containing |
| 6 | gene | 15.81132 | 15.81306 | positive | MGI_C57BL6J_5690316 | Gm43924 | ENSEMBL:ENSMUSG00000108046 | MGI:5690316 | unclassified gene | predicted gene, 43924 |
| 6 | gene | 15.83609 | 15.83778 | positive | MGI_C57BL6J_5690382 | Gm43990 | ENSEMBL:ENSMUSG00000108048 | MGI:5690382 | unclassified gene | predicted gene, 43990 |
| 6 | pseudogene | 16.08969 | 16.08996 | negative | MGI_C57BL6J_5690383 | Gm43991 | ENSEMBL:ENSMUSG00000108201 | MGI:5690383 | pseudogene | predicted gene, 43991 |
| 6 | pseudogene | 16.23324 | 16.23349 | negative | MGI_C57BL6J_5690385 | Gm43993 | ENSEMBL:ENSMUSG00000107885 | MGI:5690385 | pseudogene | predicted gene, 43993 |
| 6 | pseudogene | 16.23763 | 16.23824 | positive | MGI_C57BL6J_3781327 | Gm3148 | NCBI_Gene:100041116,ENSEMBL:ENSMUSG00000108089 | MGI:3781327 | pseudogene | predicted gene 3148 |
| 6 | gene | 16.44401 | 16.46995 | negative | MGI_C57BL6J_5595565 | Gm36406 | NCBI_Gene:102640305,ENSEMBL:ENSMUSG00000108295 | MGI:5595565 | lncRNA gene | predicted gene, 36406 |
| 6 | gene | 16.52475 | 16.67438 | positive | MGI_C57BL6J_5595662 | Gm36503 | NCBI_Gene:102640443,ENSEMBL:ENSMUSG00000108025 | MGI:5595662 | lncRNA gene | predicted gene, 36503 |
| 6 | gene | 16.77752 | 16.78882 | positive | MGI_C57BL6J_5595828 | Gm36669 | NCBI_Gene:102640655,ENSEMBL:ENSMUSG00000107022 | MGI:5595828 | lncRNA gene | predicted gene, 36669 |
| 6 | gene | 16.83093 | 16.89847 | negative | MGI_C57BL6J_1333760 | Tfec | NCBI_Gene:21426,ENSEMBL:ENSMUSG00000029553 | MGI:1333760 | protein coding gene | transcription factor EC |
| 6 | pseudogene | 16.92523 | 16.92693 | positive | MGI_C57BL6J_3647813 | Gm5721 | NCBI_Gene:435881,ENSEMBL:ENSMUSG00000081744 | MGI:3647813 | pseudogene | predicted gene 5721 |
| 6 | gene | 17.04800 | 17.06515 | negative | MGI_C57BL6J_3705105 | Gm15473 | NCBI_Gene:100504308,ENSEMBL:ENSMUSG00000084772 | MGI:3705105 | lncRNA gene | predicted gene 15473 |
| 6 | gene | 17.06501 | 17.17183 | negative | MGI_C57BL6J_3647654 | Gm4876 | NCBI_Gene:232599,ENSEMBL:ENSMUSG00000054556 | MGI:3647654 | lncRNA gene | predicted gene 4876 |
| 6 | gene | 17.06515 | 17.10583 | positive | MGI_C57BL6J_105081 | Tes | NCBI_Gene:21753,ENSEMBL:ENSMUSG00000029552 | MGI:105081 | protein coding gene | testis derived transcript |
| 6 | gene | 17.19775 | 17.21015 | positive | MGI_C57BL6J_2442531 | D830026I12Rik | NCBI_Gene:319682,ENSEMBL:ENSMUSG00000085171 | MGI:2442531 | lncRNA gene | RIKEN cDNA D830026I12 gene |
| 6 | gene | 17.20682 | 17.38560 | negative | MGI_C57BL6J_3783029 | Gm15581 | ENSEMBL:ENSMUSG00000085264 | MGI:3783029 | lncRNA gene | predicted gene 15581 |
| 6 | gene | 17.28119 | 17.28913 | positive | MGI_C57BL6J_107571 | Cav2 | NCBI_Gene:12390,ENSEMBL:ENSMUSG00000000058 | MGI:107571 | protein coding gene | caveolin 2 |
| 6 | gene | 17.30634 | 17.34145 | positive | MGI_C57BL6J_102709 | Cav1 | NCBI_Gene:12389,ENSEMBL:ENSMUSG00000007655 | MGI:102709 | protein coding gene | caveolin 1, caveolae protein |
| 6 | gene | 17.34773 | 17.34834 | positive | MGI_C57BL6J_5580297 | Gm29591 | ENSEMBL:ENSMUSG00000100101 | MGI:5580297 | lncRNA gene | predicted gene 29591 |
| 6 | gene | 17.38191 | 17.38395 | positive | MGI_C57BL6J_5826590 | Gm46953 | NCBI_Gene:108169132 | MGI:5826590 | lncRNA gene | predicted gene, 46953 |
| 6 | gene | 17.43631 | 17.43641 | positive | MGI_C57BL6J_5454798 | Gm25021 | ENSEMBL:ENSMUSG00000093902 | MGI:5454798 | snRNA gene | predicted gene, 25021 |
| 6 | gene | 17.46335 | 17.57398 | positive | MGI_C57BL6J_96969 | Met | NCBI_Gene:17295,ENSEMBL:ENSMUSG00000009376 | MGI:96969 | protein coding gene | met proto-oncogene |
| 6 | gene | 17.57807 | 17.57883 | positive | MGI_C57BL6J_1925238 | D730047N16Rik | NA | NA | unclassified gene | RIKEN cDNA D730047N16 gene |
| 6 | gene | 17.63623 | 17.66697 | positive | MGI_C57BL6J_106222 | Capza2 | NCBI_Gene:12343,ENSEMBL:ENSMUSG00000015733 | MGI:106222 | protein coding gene | capping protein (actin filament) muscle Z-line, alpha 2 |
| 6 | gene | 17.69293 | 17.94302 | positive | MGI_C57BL6J_1927450 | St7 | NCBI_Gene:64213,ENSEMBL:ENSMUSG00000029534 | MGI:1927450 | protein coding gene | suppression of tumorigenicity 7 |
| 6 | gene | 17.69343 | 17.69356 | positive | MGI_C57BL6J_5531137 | Gm27755 | ENSEMBL:ENSMUSG00000098767 | MGI:5531137 | unclassified non-coding RNA gene | predicted gene, 27755 |
| 6 | gene | 17.69394 | 17.69404 | positive | MGI_C57BL6J_5531144 | Gm27762 | ENSEMBL:ENSMUSG00000098567 | MGI:5531144 | unclassified non-coding RNA gene | predicted gene, 27762 |
| 6 | gene | 17.69490 | 17.69508 | positive | MGI_C57BL6J_5531342 | Gm27960 | ENSEMBL:ENSMUSG00000098874 | MGI:5531342 | unclassified non-coding RNA gene | predicted gene, 27960 |
| 6 | gene | 17.69535 | 17.69553 | positive | MGI_C57BL6J_5530747 | Gm27365 | ENSEMBL:ENSMUSG00000098525 | MGI:5530747 | unclassified non-coding RNA gene | predicted gene, 27365 |
| 6 | gene | 17.69689 | 17.69715 | positive | MGI_C57BL6J_5530924 | Gm27542 | ENSEMBL:ENSMUSG00000099244 | MGI:5530924 | unclassified non-coding RNA gene | predicted gene, 27542 |
| 6 | gene | 17.70323 | 17.70342 | positive | MGI_C57BL6J_5531390 | Gm28008 | ENSEMBL:ENSMUSG00000098988 | MGI:5531390 | unclassified non-coding RNA gene | predicted gene, 28008 |
| 6 | pseudogene | 17.71060 | 17.71122 | negative | MGI_C57BL6J_3652332 | Gm13898 | NCBI_Gene:100417251,ENSEMBL:ENSMUSG00000086409 | MGI:3652332 | pseudogene | predicted gene 13898 |
| 6 | pseudogene | 17.71169 | 17.71294 | negative | MGI_C57BL6J_3652262 | Gm13899 | NCBI_Gene:100418218,ENSEMBL:ENSMUSG00000081077 | MGI:3652262 | pseudogene | predicted gene 13899 |
| 6 | gene | 17.71758 | 17.72051 | positive | MGI_C57BL6J_5663670 | Gm43533 | ENSEMBL:ENSMUSG00000106822 | MGI:5663670 | unclassified gene | predicted gene 43533 |
| 6 | gene | 17.74752 | 17.74890 | negative | MGI_C57BL6J_5662989 | Gm42852 | ENSEMBL:ENSMUSG00000106801 | MGI:5662989 | unclassified gene | predicted gene 42852 |
| 6 | gene | 17.83459 | 17.83518 | negative | MGI_C57BL6J_5477232 | Gm26738 | ENSEMBL:ENSMUSG00000097700 | MGI:5477232 | lncRNA gene | predicted gene, 26738 |
| 6 | gene | 17.90571 | 17.90584 | positive | MGI_C57BL6J_5530826 | Gm27444 | ENSEMBL:ENSMUSG00000098570 | MGI:5530826 | unclassified non-coding RNA gene | predicted gene, 27444 |
| 6 | gene | 17.90717 | 17.90729 | positive | MGI_C57BL6J_5530784 | Gm27402 | ENSEMBL:ENSMUSG00000098791 | MGI:5530784 | unclassified non-coding RNA gene | predicted gene, 27402 |
| 6 | gene | 17.91500 | 17.92060 | positive | MGI_C57BL6J_5663981 | Gm43844 | ENSEMBL:ENSMUSG00000106858 | MGI:5663981 | unclassified gene | predicted gene 43844 |
| 6 | gene | 17.98547 | 18.03275 | negative | MGI_C57BL6J_98954 | Wnt2 | NCBI_Gene:22413,ENSEMBL:ENSMUSG00000010797 | MGI:98954 | protein coding gene | wingless-type MMTV integration site family, member 2 |
| 6 | gene | 18.05096 | 18.10906 | negative | MGI_C57BL6J_1921318 | Asz1 | NCBI_Gene:74068,ENSEMBL:ENSMUSG00000010796 | MGI:1921318 | protein coding gene | ankyrin repeat, SAM and basic leucine zipper domain containing 1 |
| 6 | gene | 18.11014 | 18.11196 | negative | MGI_C57BL6J_5663982 | Gm43845 | ENSEMBL:ENSMUSG00000107177 | MGI:5663982 | unclassified gene | predicted gene 43845 |
| 6 | pseudogene | 18.14472 | 18.14594 | positive | MGI_C57BL6J_3783040 | Gm15593 | NCBI_Gene:386426,ENSEMBL:ENSMUSG00000084203 | MGI:3783040 | pseudogene | predicted gene 15593 |
| 6 | pseudogene | 18.15634 | 18.15712 | negative | MGI_C57BL6J_3783043 | Gm15596 | NCBI_Gene:636450,ENSEMBL:ENSMUSG00000082289 | MGI:3783043 | pseudogene | predicted gene 15596 |
| 6 | gene | 18.17026 | 18.32277 | positive | MGI_C57BL6J_88388 | Cftr | NCBI_Gene:12638,ENSEMBL:ENSMUSG00000041301 | MGI:88388 | protein coding gene | cystic fibrosis transmembrane conductance regulator |
| 6 | gene | 18.27014 | 18.28905 | negative | MGI_C57BL6J_3783039 | Gm15592 | ENSEMBL:ENSMUSG00000087389 | MGI:3783039 | lncRNA gene | predicted gene 15592 |
| 6 | pseudogene | 18.29426 | 18.29523 | positive | MGI_C57BL6J_3783042 | Gm15595 | NCBI_Gene:100417564,ENSEMBL:ENSMUSG00000080767 | MGI:3783042 | pseudogene | predicted gene 15595 |
| 6 | gene | 18.36647 | 18.51490 | negative | MGI_C57BL6J_1353467 | Cttnbp2 | NCBI_Gene:30785,ENSEMBL:ENSMUSG00000000416 | MGI:1353467 | protein coding gene | cortactin binding protein 2 |
| 6 | gene | 18.42468 | 18.42788 | positive | MGI_C57BL6J_3783041 | Gm15594 | ENSEMBL:ENSMUSG00000084947 | MGI:3783041 | lncRNA gene | predicted gene 15594 |
| 6 | gene | 18.42966 | 18.43125 | negative | MGI_C57BL6J_5663980 | Gm43843 | ENSEMBL:ENSMUSG00000106944 | MGI:5663980 | unclassified gene | predicted gene 43843 |
| 6 | gene | 18.43712 | 18.44824 | positive | MGI_C57BL6J_5623241 | Gm40356 | NCBI_Gene:105244811 | MGI:5623241 | lncRNA gene | predicted gene, 40356 |
| 6 | gene | 18.44927 | 18.44934 | negative | MGI_C57BL6J_5456010 | Gm26233 | ENSEMBL:ENSMUSG00000093271 | MGI:5456010 | miRNA gene | predicted gene, 26233 |
| 6 | gene | 18.72729 | 18.72740 | positive | MGI_C57BL6J_5453425 | Gm23648 | ENSEMBL:ENSMUSG00000095722 | MGI:5453425 | snRNA gene | predicted gene, 23648 |
| 6 | pseudogene | 18.79285 | 18.79311 | positive | MGI_C57BL6J_5663365 | Gm43228 | ENSEMBL:ENSMUSG00000107299 | MGI:5663365 | pseudogene | predicted gene 43228 |
| 6 | pseudogene | 18.82451 | 18.82558 | positive | MGI_C57BL6J_3648027 | Gm5874 | NCBI_Gene:545829,ENSEMBL:ENSMUSG00000071568 | MGI:3648027 | pseudogene | predicted gene 5874 |
| 6 | gene | 18.84422 | 18.84736 | positive | MGI_C57BL6J_5477303 | Gm26809 | ENSEMBL:ENSMUSG00000097815 | MGI:5477303 | lncRNA gene | predicted gene, 26809 |
| 6 | gene | 18.84591 | 18.84821 | negative | MGI_C57BL6J_5012371 | Gm20186 | ENSEMBL:ENSMUSG00000106874 | MGI:5012371 | lncRNA gene | predicted gene, 20186 |
| 6 | gene | 18.84858 | 18.85575 | positive | MGI_C57BL6J_1923772 | Lsm8 | NCBI_Gene:76522,ENSEMBL:ENSMUSG00000044155 | MGI:1923772 | protein coding gene | LSM8 homolog, U6 small nuclear RNA associated |
| 6 | gene | 18.85415 | 18.87959 | positive | MGI_C57BL6J_1922446 | Ankrd7 | NCBI_Gene:75196,ENSEMBL:ENSMUSG00000029517 | MGI:1922446 | protein coding gene | ankyrin repeat domain 7 |
| 6 | pseudogene | 18.88271 | 18.88420 | positive | MGI_C57BL6J_5663635 | Gm43498 | ENSEMBL:ENSMUSG00000106819 | MGI:5663635 | pseudogene | predicted gene 43498 |
| 6 | gene | 19.04684 | 19.04715 | positive | MGI_C57BL6J_5452495 | Gm22718 | ENSEMBL:ENSMUSG00000084650 | MGI:5452495 | unclassified non-coding RNA gene | predicted gene, 22718 |
| 6 | pseudogene | 19.11944 | 19.12117 | positive | MGI_C57BL6J_3645297 | Gm6655 | NCBI_Gene:626180,ENSEMBL:ENSMUSG00000107349 | MGI:3645297 | pseudogene | predicted gene 6655 |
| 6 | gene | 19.24790 | 19.27884 | negative | MGI_C57BL6J_1921975 | 4930524B17Rik | NCBI_Gene:74725,ENSEMBL:ENSMUSG00000107217 | MGI:1921975 | lncRNA gene | RIKEN cDNA 4930524B17 gene |
| 6 | pseudogene | 19.31722 | 19.31940 | negative | MGI_C57BL6J_103210 | Eif3s6-ps4 | NCBI_Gene:16345,ENSEMBL:ENSMUSG00000106885 | MGI:103210 | pseudogene | eukaryotic translation initiation factor 3, subunit 6, pseudogene 4 |
| 6 | gene | 19.34133 | 19.34159 | negative | MGI_C57BL6J_5453990 | Gm24213 | ENSEMBL:ENSMUSG00000088880 | MGI:5453990 | unclassified non-coding RNA gene | predicted gene, 24213 |
| 6 | gene | 19.55534 | 19.55546 | positive | MGI_C57BL6J_5530862 | Mir6370 | miRBase:MI0021899,NCBI_Gene:102465192,ENSEMBL:ENSMUSG00000099149 | MGI:5530862 | miRNA gene | microRNA 6370 |
| 6 | gene | 19.62461 | 19.62471 | positive | MGI_C57BL6J_5452605 | Gm22828 | ENSEMBL:ENSMUSG00000065702 | MGI:5452605 | snRNA gene | predicted gene, 22828 |
| 6 | pseudogene | 19.77396 | 19.77755 | negative | MGI_C57BL6J_5010667 | Gm18482 | NCBI_Gene:100417252,ENSEMBL:ENSMUSG00000107100 | MGI:5010667 | pseudogene | predicted gene, 18482 |
| 6 | pseudogene | 19.78469 | 19.78479 | positive | MGI_C57BL6J_5663638 | Gm43501 | ENSEMBL:ENSMUSG00000106814 | MGI:5663638 | pseudogene | predicted gene 43501 |
| 6 | pseudogene | 19.78805 | 19.78902 | positive | MGI_C57BL6J_5662718 | Gm42581 | ENSEMBL:ENSMUSG00000107067 | MGI:5662718 | pseudogene | predicted gene 42581 |
| 6 | pseudogene | 20.49054 | 20.49581 | positive | MGI_C57BL6J_5588957 | Gm29798 | NCBI_Gene:101056208 | MGI:5588957 | pseudogene | predicted gene, 29798 |
| 6 | gene | 20.95015 | 20.97038 | negative | MGI_C57BL6J_5621657 | Gm38772 | NCBI_Gene:105242604 | MGI:5621657 | lncRNA gene | predicted gene, 38772 |
| 6 | pseudogene | 20.97031 | 20.97122 | negative | MGI_C57BL6J_5011287 | Gm19102 | NCBI_Gene:100418256,ENSEMBL:ENSMUSG00000106763 | MGI:5011287 | pseudogene | predicted gene, 19102 |
| 6 | gene | 21.01311 | 21.01324 | negative | MGI_C57BL6J_5453737 | Gm23960 | ENSEMBL:ENSMUSG00000087831 | MGI:5453737 | snoRNA gene | predicted gene, 23960 |
| 6 | gene | 21.12816 | 21.12826 | positive | MGI_C57BL6J_5456150 | Gm26373 | ENSEMBL:ENSMUSG00000088458 | MGI:5456150 | snRNA gene | predicted gene, 26373 |
| 6 | gene | 21.21550 | 21.72980 | positive | MGI_C57BL6J_102663 | Kcnd2 | NCBI_Gene:16508,ENSEMBL:ENSMUSG00000060882 | MGI:102663 | protein coding gene | potassium voltage-gated channel, Shal-related family, member 2 |
| 6 | gene | 21.23559 | 21.23617 | negative | MGI_C57BL6J_2141497 | 6430519N07Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 6430519N07 gene |
| 6 | gene | 21.25892 | 21.26562 | positive | MGI_C57BL6J_5662720 | Gm42583 | ENSEMBL:ENSMUSG00000106826 | MGI:5662720 | unclassified gene | predicted gene 42583 |
| 6 | gene | 21.27497 | 21.27657 | positive | MGI_C57BL6J_1923983 | 6330436F06Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 6330436F06 gene |
| 6 | gene | 21.28210 | 21.28356 | positive | MGI_C57BL6J_1923408 | 6330510M09Rik | NA | NA | unclassified gene | RIKEN cDNA 6330510M09 gene |
| 6 | gene | 21.35212 | 21.35460 | positive | MGI_C57BL6J_2443190 | 6720467L15Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 6720467L15 gene |
| 6 | pseudogene | 21.49166 | 21.49188 | positive | MGI_C57BL6J_5662721 | Gm42584 | ENSEMBL:ENSMUSG00000107097 | MGI:5662721 | pseudogene | predicted gene 42584 |
| 6 | gene | 21.68130 | 21.68538 | positive | MGI_C57BL6J_2444650 | 5330437M03Rik | ENSEMBL:ENSMUSG00000106855 | MGI:2444650 | unclassified gene | RIKEN cDNA 5330437M03 gene |
| 6 | gene | 21.77139 | 21.87955 | negative | MGI_C57BL6J_1889818 | Tspan12 | NCBI_Gene:269831,ENSEMBL:ENSMUSG00000029669 | MGI:1889818 | protein coding gene | tetraspanin 12 |
| 6 | gene | 21.85242 | 21.85378 | positive | MGI_C57BL6J_5589429 | Gm30270 | NCBI_Gene:102632112,ENSEMBL:ENSMUSG00000106709 | MGI:5589429 | lncRNA gene | predicted gene, 30270 |
| 6 | gene | 21.85298 | 21.85818 | negative | MGI_C57BL6J_3781467 | Gm3289 | ENSEMBL:ENSMUSG00000106871 | MGI:3781467 | lncRNA gene | predicted gene 3289 |
| 6 | gene | 21.94957 | 21.97604 | positive | MGI_C57BL6J_1919027 | Ing3 | NCBI_Gene:71777,ENSEMBL:ENSMUSG00000029670 | MGI:1919027 | protein coding gene | inhibitor of growth family, member 3 |
| 6 | gene | 21.98570 | 22.25641 | positive | MGI_C57BL6J_2444814 | Cped1 | NCBI_Gene:214642,ENSEMBL:ENSMUSG00000062980 | MGI:2444814 | protein coding gene | cadherin-like and PC-esterase domain containing 1 |
| 6 | gene | 21.98591 | 22.06769 | negative | MGI_C57BL6J_5477213 | Gm26719 | ENSEMBL:ENSMUSG00000097364 | MGI:5477213 | lncRNA gene | predicted gene, 26719 |
| 6 | pseudogene | 22.18253 | 22.18517 | positive | MGI_C57BL6J_3644121 | Gm8564 | NCBI_Gene:105244812 | MGI:3644121 | pseudogene | predicted gene 8564 |
| 6 | pseudogene | 22.24544 | 22.24595 | negative | MGI_C57BL6J_5662710 | Gm42573 | NCBI_Gene:108169133,ENSEMBL:ENSMUSG00000106736 | MGI:5662710 | pseudogene | predicted gene 42573 |
| 6 | gene | 22.28817 | 22.29852 | positive | MGI_C57BL6J_2136018 | Wnt16 | NCBI_Gene:93735,ENSEMBL:ENSMUSG00000029671 | MGI:2136018 | protein coding gene | wingless-type MMTV integration site family, member 16 |
| 6 | gene | 22.30652 | 22.35624 | negative | MGI_C57BL6J_107892 | Fam3c | NCBI_Gene:27999,ENSEMBL:ENSMUSG00000029672 | MGI:107892 | protein coding gene | family with sequence similarity 3, member C |
| 6 | gene | 22.38961 | 22.39208 | negative | MGI_C57BL6J_5621658 | Gm38773 | NCBI_Gene:105242605 | MGI:5621658 | lncRNA gene | predicted gene, 38773 |
| 6 | gene | 22.54315 | 22.63414 | negative | MGI_C57BL6J_5621659 | Gm38774 | NCBI_Gene:105242606 | MGI:5621659 | lncRNA gene | predicted gene, 38774 |
| 6 | pseudogene | 22.62292 | 22.62369 | positive | MGI_C57BL6J_5663767 | Gm43630 | ENSEMBL:ENSMUSG00000107282 | MGI:5663767 | pseudogene | predicted gene 43630 |
| 6 | pseudogene | 22.70343 | 22.70463 | positive | MGI_C57BL6J_3643156 | Gm8927 | NCBI_Gene:668018,ENSEMBL:ENSMUSG00000098187 | MGI:3643156 | pseudogene | predicted gene 8927 |
| 6 | pseudogene | 22.73373 | 22.73459 | positive | MGI_C57BL6J_3646325 | Gm8930 | NCBI_Gene:668022,ENSEMBL:ENSMUSG00000106803 | MGI:3646325 | pseudogene | predicted gene 8930 |
| 6 | pseudogene | 22.75940 | 22.76140 | positive | MGI_C57BL6J_5011046 | Gm18861 | NCBI_Gene:100417850,ENSEMBL:ENSMUSG00000107303 | MGI:5011046 | pseudogene | predicted gene, 18861 |
| 6 | gene | 22.76811 | 22.76820 | negative | MGI_C57BL6J_5453738 | Gm23961 | ENSEMBL:ENSMUSG00000096079 | MGI:5453738 | miRNA gene | predicted gene, 23961 |
| 6 | gene | 22.78982 | 22.78993 | positive | MGI_C57BL6J_5455719 | Gm25942 | ENSEMBL:ENSMUSG00000096721 | MGI:5455719 | miRNA gene | predicted gene, 25942 |
| 6 | pseudogene | 22.80463 | 22.80533 | negative | MGI_C57BL6J_5663766 | Gm43629 | ENSEMBL:ENSMUSG00000107059 | MGI:5663766 | pseudogene | predicted gene 43629 |
| 6 | gene | 22.87546 | 23.05292 | positive | MGI_C57BL6J_97816 | Ptprz1 | NCBI_Gene:19283,ENSEMBL:ENSMUSG00000068748 | MGI:97816 | protein coding gene | protein tyrosine phosphatase, receptor type Z, polypeptide 1 |
| 6 | gene | 22.97909 | 22.98610 | negative | MGI_C57BL6J_5589652 | Gm30493 | NCBI_Gene:102632417 | MGI:5589652 | lncRNA gene | predicted gene, 30493 |
| 6 | gene | 22.98820 | 22.99163 | positive | MGI_C57BL6J_5826591 | Gm46954 | NCBI_Gene:108169134 | MGI:5826591 | lncRNA gene | predicted gene, 46954 |
| 6 | gene | 23.07217 | 23.13299 | negative | MGI_C57BL6J_1353573 | Aass | NCBI_Gene:30956,ENSEMBL:ENSMUSG00000029695 | MGI:1353573 | protein coding gene | aminoadipate-semialdehyde synthase |
| 6 | pseudogene | 23.15593 | 23.15662 | negative | MGI_C57BL6J_3645708 | Gm8945 | NCBI_Gene:668044,ENSEMBL:ENSMUSG00000117306 | MGI:3645708 | pseudogene | predicted gene 8945 |
| 6 | pseudogene | 23.15799 | 23.16006 | negative | MGI_C57BL6J_3643846 | Gm5989 | NCBI_Gene:546894,ENSEMBL:ENSMUSG00000106989 | MGI:3643846 | pseudogene | predicted pseudogene 5989 |
| 6 | pseudogene | 23.16337 | 23.16409 | negative | MGI_C57BL6J_3648955 | Gm8950 | NCBI_Gene:668053,ENSEMBL:ENSMUSG00000117101 | MGI:3648955 | pseudogene | predicted gene 8950 |
| 6 | pseudogene | 23.16534 | 23.16741 | negative | MGI_C57BL6J_3642953 | Gm5301 | NCBI_Gene:384356,ENSEMBL:ENSMUSG00000107032 | MGI:3642953 | pseudogene | predicted pseudogene 5301 |
| 6 | gene | 23.18048 | 23.18153 | negative | MGI_C57BL6J_1922499 | 4930554P06Rik | NCBI_Gene:102632493 | MGI:1922499 | lncRNA gene | RIKEN cDNA 4930554P06 gene |
| 6 | pseudogene | 23.23483 | 23.23648 | positive | MGI_C57BL6J_5010118 | Gm17933 | NCBI_Gene:100416135 | MGI:5010118 | pseudogene | predicted gene, 17933 |
| 6 | gene | 23.24504 | 23.25246 | negative | MGI_C57BL6J_1920441 | Fezf1 | NCBI_Gene:73191,ENSEMBL:ENSMUSG00000029697 | MGI:1920441 | protein coding gene | Fez family zinc finger 1 |
| 6 | gene | 23.25113 | 23.25336 | positive | MGI_C57BL6J_2443208 | C130093G08Rik | ENSEMBL:ENSMUSG00000106775 | MGI:2443208 | lncRNA gene | RIKEN cDNA C130093G08 gene |
| 6 | gene | 23.26277 | 23.83942 | negative | MGI_C57BL6J_2443963 | Cadps2 | NCBI_Gene:320405,ENSEMBL:ENSMUSG00000017978 | MGI:2443963 | protein coding gene | Ca2+-dependent activator protein for secretion 2 |
| 6 | gene | 23.51975 | 23.51986 | positive | MGI_C57BL6J_5531341 | Gm27959 | ENSEMBL:ENSMUSG00000098877 | MGI:5531341 | rRNA gene | predicted gene, 27959 |
| 6 | gene | 23.57343 | 23.58335 | positive | MGI_C57BL6J_5621660 | Gm38775 | NCBI_Gene:105242607 | MGI:5621660 | lncRNA gene | predicted gene, 38775 |
| 6 | gene | 23.63982 | 23.63996 | negative | MGI_C57BL6J_5453994 | Gm24217 | ENSEMBL:ENSMUSG00000088885 | MGI:5453994 | rRNA gene | predicted gene, 24217 |
| 6 | gene | 23.64887 | 23.65030 | negative | MGI_C57BL6J_2677436 | Rnf133 | NCBI_Gene:386611,ENSEMBL:ENSMUSG00000051956 | MGI:2677436 | protein coding gene | ring finger protein 133 |
| 6 | gene | 23.65389 | 23.65514 | negative | MGI_C57BL6J_1918550 | Rnf148 | NCBI_Gene:71300,ENSEMBL:ENSMUSG00000078179 | MGI:1918550 | protein coding gene | ring finger protein 148 |
| 6 | pseudogene | 23.73650 | 23.73813 | positive | MGI_C57BL6J_5011151 | Gm18966 | NCBI_Gene:100418047 | MGI:5011151 | pseudogene | predicted gene, 18966 |
| 6 | gene | 23.96916 | 23.97006 | negative | MGI_C57BL6J_2681247 | Tas2r118 | NCBI_Gene:387347,ENSEMBL:ENSMUSG00000043865 | MGI:2681247 | protein coding gene | taste receptor, type 2, member 118 |
| 6 | gene | 24.08828 | 24.16812 | negative | MGI_C57BL6J_1859937 | Slc13a1 | NCBI_Gene:55961,ENSEMBL:ENSMUSG00000029700 | MGI:1859937 | protein coding gene | solute carrier family 13 (sodium/sulfate symporters), member 1 |
| 6 | pseudogene | 24.13832 | 24.14172 | negative | MGI_C57BL6J_5313153 | Gm20706 | NCBI_Gene:102634112,ENSEMBL:ENSMUSG00000093580 | MGI:5313153 | pseudogene | predicted gene 20706 |
| 6 | gene | 24.14337 | 24.14347 | positive | MGI_C57BL6J_5456163 | Gm26386 | ENSEMBL:ENSMUSG00000096522 | MGI:5456163 | miRNA gene | predicted gene, 26386 |
| 6 | pseudogene | 24.34325 | 24.34459 | positive | MGI_C57BL6J_3781313 | Gm3136 | NCBI_Gene:100041094 | MGI:3781313 | pseudogene | predicted gene 3136 |
| 6 | gene | 24.44487 | 24.51856 | negative | MGI_C57BL6J_3041159 | Iqub | NCBI_Gene:214704,ENSEMBL:ENSMUSG00000046192 | MGI:3041159 | protein coding gene | IQ motif and ubiquitin domain containing |
| 6 | gene | 24.51866 | 24.52801 | negative | MGI_C57BL6J_1915452 | Ndufa5 | NCBI_Gene:68202,ENSEMBL:ENSMUSG00000023089 | MGI:1915452 | protein coding gene | NADH:ubiquinone oxidoreductase subunit A5 |
| 6 | gene | 24.52814 | 24.57316 | positive | MGI_C57BL6J_1926160 | Asb15 | NCBI_Gene:78910,ENSEMBL:ENSMUSG00000029685 | MGI:1926160 | protein coding gene | ankyrin repeat and SOCS box-containing 15 |
| 6 | pseudogene | 24.59453 | 24.59524 | positive | MGI_C57BL6J_5662826 | Gm42689 | ENSEMBL:ENSMUSG00000107189 | MGI:5662826 | pseudogene | predicted gene 42689 |
| 6 | gene | 24.59776 | 24.60541 | positive | MGI_C57BL6J_2135672 | Lmod2 | NCBI_Gene:93677,ENSEMBL:ENSMUSG00000029683 | MGI:2135672 | protein coding gene | leiomodin 2 (cardiac) |
| 6 | gene | 24.61380 | 24.66501 | negative | MGI_C57BL6J_1920428 | Wasl | NCBI_Gene:73178,ENSEMBL:ENSMUSG00000029684 | MGI:1920428 | protein coding gene | Wiskott-Aldrich syndrome-like (human) |
| 6 | gene | 24.65752 | 24.66037 | negative | MGI_C57BL6J_5662993 | Gm42856 | ENSEMBL:ENSMUSG00000106946 | MGI:5662993 | unclassified gene | predicted gene 42856 |
| 6 | gene | 24.73325 | 24.74545 | positive | MGI_C57BL6J_1921659 | Hyal6 | NCBI_Gene:74409,ENSEMBL:ENSMUSG00000029679 | MGI:1921659 | protein coding gene | hyaluronoglucosaminidase 6 |
| 6 | gene | 24.74833 | 24.76766 | positive | MGI_C57BL6J_1924292 | Hyal4 | NCBI_Gene:77042,ENSEMBL:ENSMUSG00000029680 | MGI:1924292 | protein coding gene | hyaluronoglucosaminidase 4 |
| 6 | gene | 24.76628 | 24.77525 | negative | MGI_C57BL6J_5621661 | Gm38776 | NCBI_Gene:105242608 | MGI:5621661 | lncRNA gene | predicted gene, 38776 |
| 6 | gene | 24.79119 | 24.80105 | positive | MGI_C57BL6J_109335 | Spam1 | NCBI_Gene:20690,ENSEMBL:ENSMUSG00000029682 | MGI:109335 | protein coding gene | sperm adhesion molecule 1 |
| 6 | pseudogene | 24.83729 | 24.83842 | positive | MGI_C57BL6J_5010512 | Gm18327 | NCBI_Gene:100416932,ENSEMBL:ENSMUSG00000107223 | MGI:5010512 | pseudogene | predicted gene, 18327 |
| 6 | gene | 24.85796 | 24.89196 | positive | MGI_C57BL6J_1921718 | Hyal5 | NCBI_Gene:74468,ENSEMBL:ENSMUSG00000029678 | MGI:1921718 | protein coding gene | hyaluronoglucosaminidase 5 |
| 6 | gene | 24.89467 | 24.89480 | negative | MGI_C57BL6J_5452044 | Gm22267 | ENSEMBL:ENSMUSG00000077196 | MGI:5452044 | snoRNA gene | predicted gene, 22267 |
| 6 | gene | 24.95114 | 24.95630 | negative | MGI_C57BL6J_2442812 | Tmem229a | NCBI_Gene:319832,ENSEMBL:ENSMUSG00000048022 | MGI:2442812 | protein coding gene | transmembrane protein 229A |
| 6 | gene | 25.03314 | 25.05747 | positive | MGI_C57BL6J_5621662 | Gm38777 | NCBI_Gene:105242609 | MGI:5621662 | lncRNA gene | predicted gene, 38777 |
| 6 | pseudogene | 25.04646 | 25.04732 | negative | MGI_C57BL6J_3642956 | Gm5302 | NCBI_Gene:384358,ENSEMBL:ENSMUSG00000107212 | MGI:3642956 | pseudogene | predicted gene 5302 |
| 6 | gene | 25.07877 | 25.07890 | negative | MGI_C57BL6J_5454220 | Gm24443 | ENSEMBL:ENSMUSG00000087867 | MGI:5454220 | snoRNA gene | predicted gene, 24443 |
| 6 | gene | 25.12699 | 25.18399 | negative | MGI_C57BL6J_5589817 | Gm30658 | NCBI_Gene:102632632 | MGI:5589817 | lncRNA gene | predicted gene, 30658 |
| 6 | gene | 25.19993 | 25.20005 | negative | MGI_C57BL6J_5452306 | Gm22529 | ENSEMBL:ENSMUSG00000089442 | MGI:5452306 | snoRNA gene | predicted gene, 22529 |
| 6 | gene | 25.66588 | 25.69073 | negative | MGI_C57BL6J_1313297 | Gpr37 | NCBI_Gene:14763,ENSEMBL:ENSMUSG00000039904 | MGI:1313297 | protein coding gene | G protein-coupled receptor 37 |
| 6 | gene | 25.73780 | 25.74055 | negative | MGI_C57BL6J_1925221 | A930037O16Rik | NA | NA | unclassified gene | RIKEN cDNA A930037O16 gene |
| 6 | gene | 25.74374 | 25.80928 | negative | MGI_C57BL6J_2141503 | Pot1a | NCBI_Gene:101185,ENSEMBL:ENSMUSG00000029676 | MGI:2141503 | protein coding gene | protection of telomeres 1A |
| 6 | gene | 25.93143 | 25.97723 | positive | MGI_C57BL6J_5434112 | Gm20756 | NCBI_Gene:626904,ENSEMBL:ENSMUSG00000106799 | MGI:5434112 | lncRNA gene | predicted gene, 20756 |
| 6 | pseudogene | 25.97661 | 25.97715 | negative | MGI_C57BL6J_3647632 | Gm6719 | NCBI_Gene:626912,ENSEMBL:ENSMUSG00000107127 | MGI:3647632 | pseudogene | predicted gene 6719 |
| 6 | gene | 26.46029 | 26.46043 | negative | MGI_C57BL6J_5454468 | Gm24691 | ENSEMBL:ENSMUSG00000088387 | MGI:5454468 | snRNA gene | predicted gene, 24691 |
| 6 | gene | 26.73978 | 26.76687 | negative | MGI_C57BL6J_5589938 | Gm30779 | NCBI_Gene:102632801 | MGI:5589938 | lncRNA gene | predicted gene, 30779 |
| 6 | pseudogene | 26.83042 | 26.83116 | positive | MGI_C57BL6J_5662611 | Gm42474 | ENSEMBL:ENSMUSG00000106742 | MGI:5662611 | pseudogene | predicted gene 42474 |
| 6 | gene | 27.20502 | 27.21848 | positive | MGI_C57BL6J_5621663 | Gm38778 | NCBI_Gene:105242610 | MGI:5621663 | lncRNA gene | predicted gene, 38778 |
| 6 | gene | 27.27512 | 28.13591 | negative | MGI_C57BL6J_1351345 | Grm8 | NCBI_Gene:14823,ENSEMBL:ENSMUSG00000024211 | MGI:1351345 | protein coding gene | glutamate receptor, metabotropic 8 |
| 6 | gene | 27.44109 | 27.44395 | negative | MGI_C57BL6J_5662938 | Gm42801 | ENSEMBL:ENSMUSG00000107377 | MGI:5662938 | unclassified gene | predicted gene 42801 |
| 6 | gene | 27.66809 | 27.66820 | positive | MGI_C57BL6J_5456087 | Gm26310 | ENSEMBL:ENSMUSG00000088391 | MGI:5456087 | snoRNA gene | predicted gene, 26310 |
| 6 | gene | 27.90555 | 27.90723 | negative | MGI_C57BL6J_1923475 | 6430711C07Rik | NA | NA | unclassified gene | RIKEN cDNA 6430711C07 gene |
| 6 | gene | 27.93665 | 27.93675 | negative | MGI_C57BL6J_3629661 | Mir592 | miRBase:MI0004127,NCBI_Gene:735266,ENSEMBL:ENSMUSG00000076067 | MGI:3629661 | miRNA gene | microRNA 592 |
| 6 | gene | 28.21055 | 28.39805 | negative | MGI_C57BL6J_1889334 | Zfp800 | NCBI_Gene:627049,ENSEMBL:ENSMUSG00000039841 | MGI:1889334 | protein coding gene | zinc finger protein 800 |
| 6 | gene | 28.21101 | 28.21480 | negative | MGI_C57BL6J_5662684 | Gm42547 | ENSEMBL:ENSMUSG00000106990 | MGI:5662684 | unclassified gene | predicted gene 42547 |
| 6 | gene | 28.21555 | 28.26086 | positive | MGI_C57BL6J_1923474 | 6530409C15Rik | ENSEMBL:ENSMUSG00000043340 | MGI:1923474 | lncRNA gene | RIKEN cDNA 6530409C15 gene |
| 6 | gene | 28.21917 | 28.22617 | negative | MGI_C57BL6J_5662686 | Gm42549 | ENSEMBL:ENSMUSG00000107320 | MGI:5662686 | unclassified gene | predicted gene 42549 |
| 6 | gene | 28.23308 | 28.23701 | negative | MGI_C57BL6J_5662685 | Gm42548 | ENSEMBL:ENSMUSG00000106959 | MGI:5662685 | unclassified gene | predicted gene 42548 |
| 6 | pseudogene | 28.36171 | 28.36317 | negative | MGI_C57BL6J_3647388 | Gm5303 | NCBI_Gene:384360,ENSEMBL:ENSMUSG00000083107 | MGI:3647388 | pseudogene | predicted gene 5303 |
| 6 | gene | 28.41609 | 28.42839 | negative | MGI_C57BL6J_1921625 | Gcc1 | NCBI_Gene:74375,ENSEMBL:ENSMUSG00000029708 | MGI:1921625 | protein coding gene | golgi coiled coil 1 |
| 6 | gene | 28.42356 | 28.42660 | positive | MGI_C57BL6J_99434 | Arf5 | NCBI_Gene:11844,ENSEMBL:ENSMUSG00000020440 | MGI:99434 | protein coding gene | ADP-ribosylation factor 5 |
| 6 | gene | 28.42779 | 28.43940 | positive | MGI_C57BL6J_1890386 | Fscn3 | NCBI_Gene:56223,ENSEMBL:ENSMUSG00000029707 | MGI:1890386 | protein coding gene | fascin actin-bundling protein 3 |
| 6 | gene | 28.44233 | 28.44935 | negative | MGI_C57BL6J_97488 | Pax4 | NCBI_Gene:18506,ENSEMBL:ENSMUSG00000029706 | MGI:97488 | protein coding gene | paired box 4 |
| 6 | pseudogene | 28.46557 | 28.46698 | negative | MGI_C57BL6J_5663401 | Gm43264 | ENSEMBL:ENSMUSG00000107130 | MGI:5663401 | pseudogene | predicted gene 43264 |
| 6 | gene | 28.47514 | 28.93516 | positive | MGI_C57BL6J_1929266 | Snd1 | NCBI_Gene:56463,ENSEMBL:ENSMUSG00000001424 | MGI:1929266 | protein coding gene | staphylococcal nuclease and tudor domain containing 1 |
| 6 | gene | 28.56349 | 28.56536 | positive | MGI_C57BL6J_5610549 | Gm37321 | ENSEMBL:ENSMUSG00000104325 | MGI:5610549 | unclassified gene | predicted gene, 37321 |
| 6 | gene | 28.56807 | 28.57099 | positive | MGI_C57BL6J_5611206 | Gm37978 | ENSEMBL:ENSMUSG00000103132 | MGI:5611206 | unclassified gene | predicted gene, 37978 |
| 6 | gene | 28.66183 | 28.83175 | negative | MGI_C57BL6J_2182081 | Lrrc4 | NCBI_Gene:192198,ENSEMBL:ENSMUSG00000049939 | MGI:2182081 | protein coding gene | leucine rich repeat containing 4 |
| 6 | gene | 28.79646 | 28.79971 | positive | MGI_C57BL6J_5611168 | Gm37940 | ENSEMBL:ENSMUSG00000103899 | MGI:5611168 | unclassified gene | predicted gene, 37940 |
| 6 | gene | 28.95739 | 28.96604 | negative | MGI_C57BL6J_5621664 | Gm38779 | NCBI_Gene:105242611 | MGI:5621664 | lncRNA gene | predicted gene, 38779 |
| 6 | gene | 28.96480 | 28.96641 | positive | MGI_C57BL6J_5623242 | Gm40357 | NCBI_Gene:105244813 | MGI:5623242 | lncRNA gene | predicted gene, 40357 |
| 6 | gene | 28.98163 | 29.02892 | positive | MGI_C57BL6J_3781472 | Gm3294 | ENSEMBL:ENSMUSG00000092090 | MGI:3781472 | lncRNA gene | predicted gene 3294 |
| 6 | gene | 29.02262 | 29.02269 | positive | MGI_C57BL6J_2676815 | Mir129-1 | miRBase:MI0000222,NCBI_Gene:387237,ENSEMBL:ENSMUSG00000065469 | MGI:2676815 | miRNA gene | microRNA 129-1 |
| 6 | gene | 29.03095 | 29.03953 | positive | MGI_C57BL6J_5589997 | Lnclep | NCBI_Gene:102632876 | MGI:5589997 | lncRNA gene | long noncoding RNA upstream of leptin |
| 6 | gene | 29.06020 | 29.07388 | positive | MGI_C57BL6J_104663 | Lep | NCBI_Gene:16846,ENSEMBL:ENSMUSG00000059201 | MGI:104663 | protein coding gene | leptin |
| 6 | gene | 29.11344 | 29.11361 | negative | MGI_C57BL6J_5455366 | Gm25589 | ENSEMBL:ENSMUSG00000064436 | MGI:5455366 | snRNA gene | predicted gene, 25589 |
| 6 | gene | 29.12357 | 29.16501 | negative | MGI_C57BL6J_2655711 | Rbm28 | NCBI_Gene:68272,ENSEMBL:ENSMUSG00000029701 | MGI:2655711 | protein coding gene | RNA binding motif protein 28 |
| 6 | gene | 29.15055 | 29.15068 | negative | MGI_C57BL6J_5454080 | Gm24303 | ENSEMBL:ENSMUSG00000095596 | MGI:5454080 | miRNA gene | predicted gene, 24303 |
| 6 | gene | 29.16923 | 29.17958 | negative | MGI_C57BL6J_2141677 | Prrt4 | NCBI_Gene:101359,ENSEMBL:ENSMUSG00000079654 | MGI:2141677 | protein coding gene | proline-rich transmembrane protein 4 |
| 6 | gene | 29.20043 | 29.21636 | negative | MGI_C57BL6J_96567 | Impdh1 | NCBI_Gene:23917,ENSEMBL:ENSMUSG00000003500 | MGI:96567 | protein coding gene | inosine monophosphate dehydrogenase 1 |
| 6 | gene | 29.23443 | 29.23456 | positive | MGI_C57BL6J_5455798 | Gm26021 | ENSEMBL:ENSMUSG00000089289 | MGI:5455798 | snoRNA gene | predicted gene, 26021 |
| 6 | gene | 29.25029 | 29.26496 | negative | MGI_C57BL6J_5623243 | Gm40358 | NCBI_Gene:105244814 | MGI:5623243 | lncRNA gene | predicted gene, 40358 |
| 6 | gene | 29.27249 | 29.27545 | positive | MGI_C57BL6J_1916823 | Hilpda | NCBI_Gene:69573,ENSEMBL:ENSMUSG00000043421 | MGI:1916823 | protein coding gene | hypoxia inducible lipid droplet associated |
| 6 | gene | 29.27580 | 29.29068 | positive | MGI_C57BL6J_2141439 | Fam71f2 | NCBI_Gene:245884,ENSEMBL:ENSMUSG00000079652 | MGI:2141439 | protein coding gene | family with sequence similarity 71, member F2 |
| 6 | gene | 29.31077 | 29.31222 | negative | MGI_C57BL6J_1921908 | 4930445G23Rik | NA | NA | unclassified gene | RIKEN cDNA 4930445G23 gene |
| 6 | gene | 29.31348 | 29.33602 | positive | MGI_C57BL6J_3032524 | Fam71f1 | NCBI_Gene:330277,ENSEMBL:ENSMUSG00000039742 | MGI:3032524 | protein coding gene | family with sequence similarity 71, member F1 |
| 6 | gene | 29.32280 | 29.32321 | negative | MGI_C57BL6J_4937999 | Gm17172 | ENSEMBL:ENSMUSG00000090692 | MGI:4937999 | lncRNA gene | predicted gene 17172 |
| 6 | gene | 29.34807 | 29.37711 | positive | MGI_C57BL6J_1097158 | Calu | NCBI_Gene:12321,ENSEMBL:ENSMUSG00000029767 | MGI:1097158 | protein coding gene | calumenin |
| 6 | gene | 29.37667 | 29.38847 | negative | MGI_C57BL6J_99438 | Opn1sw | NCBI_Gene:12057,ENSEMBL:ENSMUSG00000058831 | MGI:99438 | protein coding gene | opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) |
| 6 | gene | 29.38882 | 29.38890 | positive | MGI_C57BL6J_4414008 | n-TPagg6 | NCBI_Gene:102467586 | MGI:4414008 | tRNA gene | nuclear encoded tRNA proline 6 (anticodon AGG) |
| 6 | gene | 29.39631 | 29.42700 | positive | MGI_C57BL6J_1918128 | Ccdc136 | NCBI_Gene:232664,ENSEMBL:ENSMUSG00000029769 | MGI:1918128 | protein coding gene | coiled-coil domain containing 136 |
| 6 | gene | 29.39662 | 29.39671 | positive | MGI_C57BL6J_5455997 | Gm26220 | ENSEMBL:ENSMUSG00000092956 | MGI:5455997 | miRNA gene | predicted gene, 26220 |
| 6 | gene | 29.39743 | 29.39758 | negative | MGI_C57BL6J_5690746 | Gm44354 | ENSEMBL:ENSMUSG00000104796 | MGI:5690746 | miRNA gene | predicted gene, 44354 |
| 6 | gene | 29.43315 | 29.46189 | positive | MGI_C57BL6J_95557 | Flnc | NCBI_Gene:68794,ENSEMBL:ENSMUSG00000068699 | MGI:95557 | protein coding gene | filamin C, gamma |
| 6 | gene | 29.46772 | 29.47051 | positive | MGI_C57BL6J_1913394 | Atp6v1f | NCBI_Gene:66144,ENSEMBL:ENSMUSG00000004285 | MGI:1913394 | protein coding gene | ATPase, H+ transporting, lysosomal V1 subunit F |
| 6 | gene | 29.47144 | 29.47347 | positive | MGI_C57BL6J_3644212 | Atp6v1fnb | NCBI_Gene:668210,ENSEMBL:ENSMUSG00000090685 | MGI:3644212 | protein coding gene | Atp6v1f neighbor |
| 6 | gene | 29.47316 | 29.50801 | negative | MGI_C57BL6J_2141640 | Kcp | NCBI_Gene:333088,ENSEMBL:ENSMUSG00000059022 | MGI:2141640 | protein coding gene | kielin/chordin-like protein |
| 6 | gene | 29.48772 | 29.48782 | negative | MGI_C57BL6J_5456261 | Gm26484 | ENSEMBL:ENSMUSG00000088643 | MGI:5456261 | snRNA gene | predicted gene, 26484 |
| 6 | gene | 29.50724 | 29.52470 | positive | MGI_C57BL6J_5477121 | Gm26627 | NCBI_Gene:105242612,ENSEMBL:ENSMUSG00000097137 | MGI:5477121 | lncRNA gene | predicted gene, 26627 |
| 6 | gene | 29.52613 | 29.54187 | positive | MGI_C57BL6J_1350924 | Irf5 | NCBI_Gene:27056,ENSEMBL:ENSMUSG00000029771 | MGI:1350924 | protein coding gene | interferon regulatory factor 5 |
| 6 | gene | 29.54083 | 29.61011 | negative | MGI_C57BL6J_1196412 | Tnpo3 | NCBI_Gene:320938,ENSEMBL:ENSMUSG00000012535 | MGI:1196412 | protein coding gene | transportin 3 |
| 6 | gene | 29.54660 | 29.54814 | negative | MGI_C57BL6J_5663166 | Gm43029 | ENSEMBL:ENSMUSG00000106904 | MGI:5663166 | unclassified gene | predicted gene 43029 |
| 6 | gene | 29.67596 | 29.67753 | positive | MGI_C57BL6J_5590055 | Gm30896 | NCBI_Gene:102632951 | MGI:5590055 | lncRNA gene | predicted gene, 30896 |
| 6 | gene | 29.68690 | 29.68872 | negative | MGI_C57BL6J_5621665 | Gm38780 | NCBI_Gene:105242613 | MGI:5621665 | lncRNA gene | predicted gene, 38780 |
| 6 | gene | 29.69421 | 29.71856 | positive | MGI_C57BL6J_1919012 | Tspan33 | NCBI_Gene:232670,ENSEMBL:ENSMUSG00000001763 | MGI:1919012 | protein coding gene | tetraspanin 33 |
| 6 | gene | 29.73550 | 29.76137 | positive | MGI_C57BL6J_108075 | Smo | NCBI_Gene:319757,ENSEMBL:ENSMUSG00000001761 | MGI:108075 | protein coding gene | smoothened, frizzled class receptor |
| 6 | pseudogene | 29.75017 | 29.75151 | positive | MGI_C57BL6J_3650430 | Gm13717 | NCBI_Gene:102633026,ENSEMBL:ENSMUSG00000081564 | MGI:3650430 | pseudogene | predicted gene 13717 |
| 6 | gene | 29.76801 | 29.91231 | positive | MGI_C57BL6J_1921590 | Ahcyl2 | NCBI_Gene:74340,ENSEMBL:ENSMUSG00000029772 | MGI:1921590 | protein coding gene | S-adenosylhomocysteine hydrolase-like 2 |
| 6 | gene | 29.91202 | 29.91563 | negative | MGI_C57BL6J_1922474 | 4930528J11Rik | ENSEMBL:ENSMUSG00000107012 | MGI:1922474 | unclassified gene | RIKEN cDNA 4930528J11 gene |
| 6 | gene | 29.91698 | 29.95968 | positive | MGI_C57BL6J_2444363 | Strip2 | NCBI_Gene:320609,ENSEMBL:ENSMUSG00000039629 | MGI:2444363 | protein coding gene | striatin interacting protein 2 |
| 6 | pseudogene | 29.96095 | 29.96215 | positive | MGI_C57BL6J_3651278 | Gm13724 | NCBI_Gene:100416933,ENSEMBL:ENSMUSG00000082036 | MGI:3651278 | pseudogene | predicted gene 13724 |
| 6 | gene | 29.96355 | 29.96366 | positive | MGI_C57BL6J_5454313 | Gm24536 | ENSEMBL:ENSMUSG00000065078 | MGI:5454313 | snRNA gene | predicted gene, 24536 |
| 6 | pseudogene | 29.96364 | 29.96385 | positive | MGI_C57BL6J_3702114 | Gm13725 | ENSEMBL:ENSMUSG00000083961 | MGI:3702114 | pseudogene | predicted gene 13725 |
| 6 | gene | 29.96525 | 29.96856 | positive | MGI_C57BL6J_5590262 | Gm31103 | NCBI_Gene:102633221 | MGI:5590262 | lncRNA gene | predicted gene, 31103 |
| 6 | pseudogene | 29.98523 | 29.99378 | positive | MGI_C57BL6J_1923669 | Smkr-ps | NCBI_Gene:76419,ENSEMBL:ENSMUSG00000045709 | MGI:1923669 | pseudogene | smal lysine rich protein 1, pseudogene |
| 6 | gene | 29.98839 | 29.98849 | positive | MGI_C57BL6J_5452826 | Gm23049 | ENSEMBL:ENSMUSG00000095989 | MGI:5452826 | miRNA gene | predicted gene, 23049 |
| 6 | gene | 30.04740 | 30.04914 | negative | MGI_C57BL6J_1920783 | 1700080G18Rik | NCBI_Gene:73533 | MGI:1920783 | protein coding gene | RIKEN cDNA 1700080G18 gene |
| 6 | gene | 30.04799 | 30.15346 | positive | MGI_C57BL6J_1332235 | Nrf1 | NCBI_Gene:18181,ENSEMBL:ENSMUSG00000058440 | MGI:1332235 | protein coding gene | nuclear respiratory factor 1 |
| 6 | gene | 30.14380 | 30.14413 | negative | MGI_C57BL6J_5455357 | Gm25580 | ENSEMBL:ENSMUSG00000065343 | MGI:5455357 | unclassified non-coding RNA gene | predicted gene, 25580 |
| 6 | gene | 30.15864 | 30.17413 | negative | MGI_C57BL6J_5475487 | Mirc35hg | NCBI_Gene:101447999 | MGI:5475487 | lncRNA gene | microRNA cluster 35, host gene |
| 6 | gene | 30.16592 | 30.16599 | negative | MGI_C57BL6J_2676846 | Mir182 | miRBase:MI0000224,NCBI_Gene:387177,ENSEMBL:ENSMUSG00000076361 | MGI:2676846 | miRNA gene | microRNA 182 |
| 6 | gene | 30.16945 | 30.16955 | negative | MGI_C57BL6J_3619440 | Mir96 | miRBase:MI0000583,NCBI_Gene:723886,ENSEMBL:ENSMUSG00000065586 | MGI:3619440 | miRNA gene | microRNA 96 |
| 6 | gene | 30.16967 | 30.16974 | negative | MGI_C57BL6J_2676847 | Mir183 | miRBase:MI0000225,NCBI_Gene:387178,ENSEMBL:ENSMUSG00000065619 | MGI:2676847 | miRNA gene | microRNA 183 |
| 6 | gene | 30.17289 | 30.17513 | positive | MGI_C57BL6J_5610274 | Rncr4 | ENSEMBL:ENSMUSG00000103108 | MGI:5610274 | lncRNA gene | retina expressed non-coding RNA 4 |
| 6 | gene | 30.17662 | 30.17805 | positive | MGI_C57BL6J_3034268 | BB283400 | NCBI_Gene:100463519 | MGI:3034268 | lncRNA gene | expressed sequence BB283400 |
| 6 | gene | 30.17816 | 30.18183 | positive | MGI_C57BL6J_5590380 | Gm31221 | NCBI_Gene:102633386 | MGI:5590380 | lncRNA gene | predicted gene, 31221 |
| 6 | gene | 30.21129 | 30.30454 | negative | MGI_C57BL6J_104632 | Ube2h | NCBI_Gene:22214,ENSEMBL:ENSMUSG00000039159 | MGI:104632 | protein coding gene | ubiquitin-conjugating enzyme E2H |
| 6 | gene | 30.30460 | 30.33178 | positive | MGI_C57BL6J_1920824 | 1700095J07Rik | ENSEMBL:ENSMUSG00000084861 | MGI:1920824 | lncRNA gene | RIKEN cDNA 1700095J07 gene |
| 6 | gene | 30.34737 | 30.34747 | negative | MGI_C57BL6J_5455402 | Gm25625 | ENSEMBL:ENSMUSG00000087846 | MGI:5455402 | rRNA gene | predicted gene, 25625 |
| 6 | gene | 30.36638 | 30.39104 | negative | MGI_C57BL6J_1916023 | Zc3hc1 | NCBI_Gene:232679,ENSEMBL:ENSMUSG00000039130 | MGI:1916023 | protein coding gene | zinc finger, C3HC type 1 |
| 6 | gene | 30.37910 | 30.37921 | positive | MGI_C57BL6J_4422025 | n-R5s161 | ENSEMBL:ENSMUSG00000065662 | MGI:4422025 | rRNA gene | nuclear encoded rRNA 5S 161 |
| 6 | gene | 30.40186 | 30.45518 | positive | MGI_C57BL6J_1924038 | Klhdc10 | NCBI_Gene:76788,ENSEMBL:ENSMUSG00000029775 | MGI:1924038 | protein coding gene | kelch domain containing 10 |
| 6 | gene | 30.46013 | 30.46331 | positive | MGI_C57BL6J_1916587 | 1700012C14Rik | NCBI_Gene:69337,ENSEMBL:ENSMUSG00000085470 | MGI:1916587 | lncRNA gene | RIKEN cDNA 1700012C14 gene |
| 6 | gene | 30.47905 | 30.50978 | negative | MGI_C57BL6J_1919899 | Tmem209 | NCBI_Gene:72649,ENSEMBL:ENSMUSG00000029782 | MGI:1919899 | protein coding gene | transmembrane protein 209 |
| 6 | gene | 30.50985 | 30.52025 | positive | MGI_C57BL6J_1922897 | Ssmem1 | NCBI_Gene:75647,ENSEMBL:ENSMUSG00000029784 | MGI:1922897 | protein coding gene | serine-rich single-pass membrane protein 1 |
| 6 | gene | 30.54158 | 30.56448 | positive | MGI_C57BL6J_3617840 | Cpa2 | NCBI_Gene:232680,ENSEMBL:ENSMUSG00000071553 | MGI:3617840 | protein coding gene | carboxypeptidase A2, pancreatic |
| 6 | gene | 30.54764 | 30.55079 | negative | MGI_C57BL6J_5826592 | Gm46955 | NCBI_Gene:108169136 | MGI:5826592 | lncRNA gene | predicted gene, 46955 |
| 6 | gene | 30.56837 | 30.59242 | positive | MGI_C57BL6J_1919041 | Cpa4 | NCBI_Gene:71791,ENSEMBL:ENSMUSG00000039070 | MGI:1919041 | protein coding gene | carboxypeptidase A4 |
| 6 | gene | 30.57315 | 30.57604 | negative | MGI_C57BL6J_3649768 | Gm13781 | ENSEMBL:ENSMUSG00000086854 | MGI:3649768 | lncRNA gene | predicted gene 13781 |
| 6 | gene | 30.59414 | 30.61093 | negative | MGI_C57BL6J_5590612 | Gm31453 | NCBI_Gene:102633690 | MGI:5590612 | lncRNA gene | predicted gene, 31453 |
| 6 | gene | 30.61101 | 30.63174 | positive | MGI_C57BL6J_1921899 | Cpa5 | NCBI_Gene:74649,ENSEMBL:ENSMUSG00000029788 | MGI:1921899 | protein coding gene | carboxypeptidase A5 |
| 6 | gene | 30.63922 | 30.64536 | positive | MGI_C57BL6J_88478 | Cpa1 | NCBI_Gene:109697,ENSEMBL:ENSMUSG00000054446 | MGI:88478 | protein coding gene | carboxypeptidase A1, pancreatic |
| 6 | gene | 30.65346 | 30.71052 | negative | MGI_C57BL6J_1891414 | Cep41 | NCBI_Gene:83922,ENSEMBL:ENSMUSG00000029790 | MGI:1891414 | protein coding gene | centrosomal protein 41 |
| 6 | gene | 30.66745 | 30.68385 | positive | MGI_C57BL6J_3649363 | Gm13782 | NCBI_Gene:102633925,ENSEMBL:ENSMUSG00000086542 | MGI:3649363 | lncRNA gene | predicted gene 13782 |
| 6 | pseudogene | 30.71987 | 30.72010 | positive | MGI_C57BL6J_3705486 | Gm14528 | ENSEMBL:ENSMUSG00000083221 | MGI:3705486 | pseudogene | predicted gene 14528 |
| 6 | gene | 30.72355 | 30.74847 | positive | MGI_C57BL6J_96968 | Mest | NCBI_Gene:17294,ENSEMBL:ENSMUSG00000051855 | MGI:96968 | protein coding gene | mesoderm specific transcript |
| 6 | gene | 30.73442 | 30.73465 | positive | MGI_C57BL6J_5530904 | Gm27522 | ENSEMBL:ENSMUSG00000098260 | MGI:5530904 | unclassified non-coding RNA gene | predicted gene, 27522 |
| 6 | gene | 30.73503 | 30.73592 | negative | MGI_C57BL6J_5690688 | Gm44296 | ENSEMBL:ENSMUSG00000107825 | MGI:5690688 | lncRNA gene | predicted gene, 44296 |
| 6 | gene | 30.73596 | 30.73606 | positive | MGI_C57BL6J_5530950 | Gm27568 | ENSEMBL:ENSMUSG00000099073 | MGI:5530950 | unclassified non-coding RNA gene | predicted gene, 27568 |
| 6 | gene | 30.74130 | 30.74140 | positive | MGI_C57BL6J_3619348 | Mir335 | miRBase:MI0000817,NCBI_Gene:723930,ENSEMBL:ENSMUSG00000065501 | MGI:3619348 | miRNA gene | microRNA 335 |
| 6 | gene | 30.74755 | 30.89683 | negative | MGI_C57BL6J_1858683 | Copg2 | NCBI_Gene:54160,ENSEMBL:ENSMUSG00000025607 | MGI:1858683 | protein coding gene | coatomer protein complex, subunit gamma 2 |
| 6 | gene | 30.80478 | 30.80883 | positive | MGI_C57BL6J_1858731 | Copg2os2 | NA | NA | unclassified non-coding RNA gene | coatomer protein complex%2c subunit gamma 2%2c opposite strand 2 |
| 6 | gene | 30.88567 | 30.89450 | positive | MGI_C57BL6J_5621666 | Gm38781 | NCBI_Gene:105242616 | MGI:5621666 | lncRNA gene | predicted gene, 38781 |
| 6 | gene | 30.89690 | 30.94347 | positive | MGI_C57BL6J_1921177 | 4930412F09Rik | NCBI_Gene:102633576,ENSEMBL:ENSMUSG00000085321 | MGI:1921177 | lncRNA gene | RIKEN cDNA 4930412F09 gene |
| 6 | gene | 30.89698 | 30.92515 | negative | MGI_C57BL6J_1891413 | Tsga13 | NCBI_Gene:116732,ENSEMBL:ENSMUSG00000039032 | MGI:1891413 | protein coding gene | testis specific gene A13 |
| 6 | gene | 30.95602 | 30.95908 | negative | MGI_C57BL6J_3577024 | Klf14 | NCBI_Gene:619665,ENSEMBL:ENSMUSG00000073209 | MGI:3577024 | protein coding gene | Kruppel-like factor 14 |
| 6 | gene | 31.01007 | 31.01556 | negative | MGI_C57BL6J_5623244 | Gm40359 | NCBI_Gene:105244822 | MGI:5623244 | lncRNA gene | predicted gene, 40359 |
| 6 | gene | 31.06266 | 31.06275 | negative | MGI_C57BL6J_2676904 | Mir29a | miRBase:MI0000576,NCBI_Gene:387222,ENSEMBL:ENSMUSG00000065610 | MGI:2676904 | miRNA gene | microRNA 29a |
| 6 | gene | 31.06302 | 31.06309 | negative | MGI_C57BL6J_2676905 | Mir29b-1 | miRBase:MI0000143,NCBI_Gene:387223,ENSEMBL:ENSMUSG00000065604 | MGI:2676905 | miRNA gene | microRNA 29b-1 |
| 6 | gene | 31.06309 | 31.06349 | negative | MGI_C57BL6J_5504134 | Gm27019 | ENSEMBL:ENSMUSG00000097998 | MGI:5504134 | lncRNA gene | predicted gene, 27019 |
| 6 | gene | 31.08256 | 31.22049 | negative | MGI_C57BL6J_2673128 | Lncpint | NCBI_Gene:232685,ENSEMBL:ENSMUSG00000044471 | MGI:2673128 | lncRNA gene | long non-protein coding RNA, Trp53 induced transcript |
| 6 | gene | 31.10285 | 31.10465 | positive | MGI_C57BL6J_3650920 | Gm13833 | ENSEMBL:ENSMUSG00000087312 | MGI:3650920 | lncRNA gene | predicted gene 13833 |
| 6 | gene | 31.10883 | 31.11092 | negative | MGI_C57BL6J_1923921 | 5330406M23Rik | ENSEMBL:ENSMUSG00000104030 | MGI:1923921 | unclassified gene | RIKEN cDNA 5330406M23 gene |
| 6 | pseudogene | 31.14186 | 31.14273 | negative | MGI_C57BL6J_3650921 | Gm13835 | NCBI_Gene:100038973,ENSEMBL:ENSMUSG00000086922 | MGI:3650921 | pseudogene | predicted gene 13835 |
| 6 | gene | 31.14513 | 31.14742 | negative | MGI_C57BL6J_5610956 | Gm37728 | ENSEMBL:ENSMUSG00000103220 | MGI:5610956 | unclassified gene | predicted gene, 37728 |
| 6 | gene | 31.22035 | 31.33740 | positive | MGI_C57BL6J_1920902 | 2210408F21Rik | NCBI_Gene:73652,ENSEMBL:ENSMUSG00000087380 | MGI:1920902 | lncRNA gene | RIKEN cDNA 2210408F21 gene |
| 6 | gene | 31.24058 | 31.24396 | negative | MGI_C57BL6J_5621667 | Gm38782 | NCBI_Gene:105242618 | MGI:5621667 | lncRNA gene | predicted gene, 38782 |
| 6 | gene | 31.28896 | 31.29169 | positive | MGI_C57BL6J_5663937 | Gm43800 | ENSEMBL:ENSMUSG00000106867 | MGI:5663937 | unclassified gene | predicted gene 43800 |
| 6 | gene | 31.33063 | 31.33173 | positive | MGI_C57BL6J_1925667 | 9530034D02Rik | NA | NA | unclassified gene | RIKEN cDNA 9530034D02 gene |
| 6 | gene | 31.35633 | 31.37369 | positive | MGI_C57BL6J_3705266 | Gm14532 | NCBI_Gene:102633651,ENSEMBL:ENSMUSG00000086931 | MGI:3705266 | lncRNA gene | predicted gene 14532 |
| 6 | gene | 31.36688 | 31.39877 | negative | MGI_C57BL6J_3651306 | Mkln1os | NCBI_Gene:100503869,ENSEMBL:ENSMUSG00000086212 | MGI:3651306 | lncRNA gene | muskelin 1, intracellular mediator containing kelch motifs, opposite strand |
| 6 | gene | 31.37289 | 31.38388 | positive | MGI_C57BL6J_3651305 | Gm13844 | ENSEMBL:ENSMUSG00000086855 | MGI:3651305 | lncRNA gene | predicted gene 13844 |
| 6 | gene | 31.39873 | 31.51681 | positive | MGI_C57BL6J_1351638 | Mkln1 | NCBI_Gene:27418,ENSEMBL:ENSMUSG00000025609 | MGI:1351638 | protein coding gene | muskelin 1, intracellular mediator containing kelch motifs |
| 6 | gene | 31.51949 | 31.56398 | negative | MGI_C57BL6J_1351317 | Podxl | NCBI_Gene:27205,ENSEMBL:ENSMUSG00000025608 | MGI:1351317 | protein coding gene | podocalyxin-like |
| 6 | gene | 31.57473 | 31.57672 | positive | MGI_C57BL6J_3647032 | Gm6117 | NCBI_Gene:105242621,ENSEMBL:ENSMUSG00000086732 | MGI:3647032 | lncRNA gene | predicted gene 6117 |
| 6 | gene | 31.60514 | 31.63577 | positive | MGI_C57BL6J_5590645 | Gm31486 | NCBI_Gene:102633732 | MGI:5590645 | lncRNA gene | predicted gene, 31486 |
| 6 | gene | 31.60519 | 31.79805 | positive | MGI_C57BL6J_5663291 | Gm43154 | ENSEMBL:ENSMUSG00000086804 | MGI:5663291 | lncRNA gene | predicted gene 43154 |
| 6 | gene | 31.65098 | 31.74309 | positive | MGI_C57BL6J_3651995 | Gm13846 | NCBI_Gene:105242622 | MGI:3651995 | lncRNA gene | predicted gene 13846 |
| 6 | gene | 31.65817 | 31.66613 | negative | MGI_C57BL6J_3651996 | Gm13848 | NCBI_Gene:434003,ENSEMBL:ENSMUSG00000084973 | MGI:3651996 | lncRNA gene | predicted gene 13848 |
| 6 | gene | 31.66640 | 31.68070 | positive | MGI_C57BL6J_3651994 | Gm13847 | NCBI_Gene:102634036 | MGI:3651994 | lncRNA gene | predicted gene 13847 |
| 6 | gene | 31.70225 | 31.70699 | negative | MGI_C57BL6J_5663430 | Gm43293 | ENSEMBL:ENSMUSG00000107010 | MGI:5663430 | lncRNA gene | predicted gene 43293 |
| 6 | gene | 31.78667 | 31.92227 | positive | MGI_C57BL6J_3651992 | Gm13849 | ENSEMBL:ENSMUSG00000087379 | MGI:3651992 | lncRNA gene | predicted gene 13849 |
| 6 | gene | 32.01800 | 32.04093 | positive | MGI_C57BL6J_5591053 | Gm31894 | NCBI_Gene:102634267 | MGI:5591053 | lncRNA gene | predicted gene, 31894 |
| 6 | gene | 32.05025 | 32.05892 | positive | MGI_C57BL6J_1923632 | 1700012A03Rik | NCBI_Gene:76382,ENSEMBL:ENSMUSG00000029766 | MGI:1923632 | protein coding gene | RIKEN cDNA 1700012A03 gene |
| 6 | gene | 32.11210 | 32.11753 | positive | MGI_C57BL6J_3651993 | Gm13850 | NCBI_Gene:105242623,ENSEMBL:ENSMUSG00000085616 | MGI:3651993 | lncRNA gene | predicted gene 13850 |
| 6 | gene | 32.14427 | 32.58819 | negative | MGI_C57BL6J_2179061 | Plxna4 | NCBI_Gene:243743,ENSEMBL:ENSMUSG00000029765 | MGI:2179061 | protein coding gene | plexin A4 |
| 6 | gene | 32.30383 | 32.31661 | positive | MGI_C57BL6J_3651753 | Plxna4os3 | ENSEMBL:ENSMUSG00000086720 | MGI:3651753 | antisense lncRNA gene | plexin A4, opposite strand 3 |
| 6 | gene | 32.45117 | 32.45475 | positive | MGI_C57BL6J_3045237 | Plxna4os2 | ENSEMBL:ENSMUSG00000086675 | MGI:3045237 | antisense lncRNA gene | plexin A4, opposite strand 2 |
| 6 | gene | 32.51107 | 32.51558 | positive | MGI_C57BL6J_2442851 | Plxna4os1 | NCBI_Gene:319849,ENSEMBL:ENSMUSG00000086763 | MGI:2442851 | antisense lncRNA gene | plexin A4, opposite strand 1 |
| 6 | gene | 32.59825 | 32.63512 | negative | MGI_C57BL6J_5826593 | Gm46956 | NCBI_Gene:108169137 | MGI:5826593 | lncRNA gene | predicted gene, 46956 |
| 6 | pseudogene | 32.61012 | 32.61036 | positive | MGI_C57BL6J_3651754 | Gm13852 | ENSEMBL:ENSMUSG00000084386 | MGI:3651754 | pseudogene | predicted gene 13852 |
| 6 | gene | 32.63600 | 32.65722 | negative | MGI_C57BL6J_5591166 | Gm32007 | NCBI_Gene:102634422 | MGI:5591166 | lncRNA gene | predicted gene, 32007 |
| 6 | gene | 32.75163 | 32.75229 | negative | MGI_C57BL6J_3039568 | BC049739 | ENSEMBL:ENSMUSG00000106873 | MGI:3039568 | lncRNA gene | cDNA sequence BC049739 |
| 6 | gene | 32.79098 | 33.06028 | negative | MGI_C57BL6J_1913325 | Chchd3 | NCBI_Gene:66075,ENSEMBL:ENSMUSG00000053768 | MGI:1913325 | protein coding gene | coiled-coil-helix-coiled-coil-helix domain containing 3 |
| 6 | gene | 32.95395 | 32.95495 | negative | MGI_C57BL6J_1924889 | C030041B07Rik | NA | NA | unclassified gene | RIKEN cDNA C030041B07 gene |
| 6 | gene | 33.03847 | 33.04322 | positive | MGI_C57BL6J_3705140 | Gm14540 | ENSEMBL:ENSMUSG00000085847 | MGI:3705140 | lncRNA gene | predicted gene 14540 |
| 6 | gene | 33.03912 | 33.04212 | negative | MGI_C57BL6J_5663032 | Gm42895 | ENSEMBL:ENSMUSG00000106676 | MGI:5663032 | unclassified gene | predicted gene 42895 |
| 6 | gene | 33.08865 | 33.08875 | negative | MGI_C57BL6J_5452098 | Gm22321 | ENSEMBL:ENSMUSG00000087777 | MGI:5452098 | snRNA gene | predicted gene, 22321 |
| 6 | pseudogene | 33.24727 | 33.24811 | negative | MGI_C57BL6J_2686247 | Gm1401 | NCBI_Gene:102634733,ENSEMBL:ENSMUSG00000082217 | MGI:2686247 | pseudogene | predicted gene 1401 |
| 6 | gene | 33.24909 | 33.97398 | positive | MGI_C57BL6J_1096376 | Exoc4 | NCBI_Gene:20336,ENSEMBL:ENSMUSG00000029763 | MGI:1096376 | protein coding gene | exocyst complex component 4 |
| 6 | pseudogene | 33.37511 | 33.37745 | negative | MGI_C57BL6J_3651023 | Gm13854 | NCBI_Gene:100416262,ENSEMBL:ENSMUSG00000083728 | MGI:3651023 | pseudogene | predicted gene 13854 |
| 6 | gene | 33.43301 | 33.43462 | positive | MGI_C57BL6J_5663305 | Gm43168 | ENSEMBL:ENSMUSG00000107358 | MGI:5663305 | unclassified gene | predicted gene 43168 |
| 6 | gene | 33.46412 | 33.47484 | positive | MGI_C57BL6J_5621670 | Gm38785 | NCBI_Gene:105242624 | MGI:5621670 | lncRNA gene | predicted gene, 38785 |
| 6 | gene | 33.69003 | 33.69215 | negative | MGI_C57BL6J_5663301 | Gm43164 | ENSEMBL:ENSMUSG00000107103 | MGI:5663301 | unclassified gene | predicted gene 43164 |
| 6 | gene | 33.84311 | 33.84495 | positive | MGI_C57BL6J_1921750 | 5430439C14Rik | NA | NA | unclassified gene | RIKEN cDNA 5430439C14 gene |
| 6 | gene | 33.98181 | 33.99248 | negative | MGI_C57BL6J_3649279 | Gm13853 | NCBI_Gene:102634837,ENSEMBL:ENSMUSG00000087441 | MGI:3649279 | lncRNA gene | predicted gene 13853 |
| 6 | gene | 34.02241 | 34.02972 | negative | MGI_C57BL6J_5621671 | Gm38786 | NCBI_Gene:105242625 | MGI:5621671 | lncRNA gene | predicted gene, 38786 |
| 6 | gene | 34.02899 | 34.13403 | positive | MGI_C57BL6J_1921604 | Lrguk | NCBI_Gene:74354,ENSEMBL:ENSMUSG00000056215 | MGI:1921604 | protein coding gene | leucine-rich repeats and guanylate kinase domain containing |
| 6 | gene | 34.15193 | 34.17424 | positive | MGI_C57BL6J_3652199 | Gm13856 | ENSEMBL:ENSMUSG00000087356 | MGI:3652199 | lncRNA gene | predicted gene 13856 |
| 6 | gene | 34.15338 | 34.17711 | negative | MGI_C57BL6J_1931249 | Slc35b4 | NCBI_Gene:58246,ENSEMBL:ENSMUSG00000018999 | MGI:1931249 | protein coding gene | solute carrier family 35, member B4 |
| 6 | gene | 34.17717 | 34.18639 | positive | MGI_C57BL6J_3652198 | Gm13855 | NCBI_Gene:105242626,ENSEMBL:ENSMUSG00000087613 | MGI:3652198 | lncRNA gene | predicted gene 13855 |
| 6 | gene | 34.19717 | 34.22708 | positive | MGI_C57BL6J_5591692 | Gm32533 | NCBI_Gene:102635113 | MGI:5591692 | lncRNA gene | predicted gene, 32533 |
| 6 | gene | 34.29026 | 34.32425 | positive | MGI_C57BL6J_3705210 | Gm14546 | ENSEMBL:ENSMUSG00000086762 | MGI:3705210 | lncRNA gene | predicted gene 14546 |
| 6 | gene | 34.30243 | 34.31749 | negative | MGI_C57BL6J_1353494 | Akr1b3 | NCBI_Gene:11677,ENSEMBL:ENSMUSG00000001642 | MGI:1353494 | protein coding gene | aldo-keto reductase family 1, member B3 (aldose reductase) |
| 6 | gene | 34.35412 | 34.36846 | positive | MGI_C57BL6J_107673 | Akr1b8 | NCBI_Gene:14187,ENSEMBL:ENSMUSG00000029762 | MGI:107673 | protein coding gene | aldo-keto reductase family 1, member B8 |
| 6 | pseudogene | 34.36245 | 34.38045 | negative | MGI_C57BL6J_3649681 | Gm13858 | NCBI_Gene:664942,ENSEMBL:ENSMUSG00000081196 | MGI:3649681 | pseudogene | predicted gene 13858 |
| 6 | pseudogene | 34.37369 | 34.38079 | negative | MGI_C57BL6J_3649465 | Gm13859 | NCBI_Gene:100416263,ENSEMBL:ENSMUSG00000081991 | MGI:3649465 | pseudogene | predicted gene 13859 |
| 6 | gene | 34.38421 | 34.39696 | positive | MGI_C57BL6J_1915111 | Akr1b10 | NCBI_Gene:67861,ENSEMBL:ENSMUSG00000061758 | MGI:1915111 | protein coding gene | aldo-keto reductase family 1, member B10 (aldose reductase) |
| 6 | gene | 34.41233 | 34.42314 | positive | MGI_C57BL6J_101918 | Akr1b7 | NCBI_Gene:11997,ENSEMBL:ENSMUSG00000052131 | MGI:101918 | protein coding gene | aldo-keto reductase family 1, member B7 |
| 6 | pseudogene | 34.41940 | 34.44544 | negative | MGI_C57BL6J_3649688 | Gm13857 | NCBI_Gene:102635489,ENSEMBL:ENSMUSG00000084965 | MGI:3649688 | pseudogene | predicted gene 13857 |
| 6 | gene | 34.47621 | 34.50561 | positive | MGI_C57BL6J_1098242 | Bpgm | NCBI_Gene:12183,ENSEMBL:ENSMUSG00000038871 | MGI:1098242 | protein coding gene | 2,3-bisphosphoglycerate mutase |
| 6 | gene | 34.53355 | 34.54139 | positive | MGI_C57BL6J_3649464 | Gm13860 | ENSEMBL:ENSMUSG00000085187 | MGI:3649464 | lncRNA gene | predicted gene 13860 |
| 6 | gene | 34.53406 | 34.53691 | negative | MGI_C57BL6J_5592101 | Gm32942 | NCBI_Gene:102635662 | MGI:5592101 | lncRNA gene | predicted gene, 32942 |
| 6 | gene | 34.59850 | 34.77547 | positive | MGI_C57BL6J_88250 | Cald1 | NCBI_Gene:109624,ENSEMBL:ENSMUSG00000029761 | MGI:88250 | protein coding gene | caldesmon 1 |
| 6 | gene | 34.65135 | 34.65433 | negative | MGI_C57BL6J_3649466 | Gm13861 | NCBI_Gene:100039184,ENSEMBL:ENSMUSG00000085382 | MGI:3649466 | lncRNA gene | predicted gene 13861 |
| 6 | gene | 34.71229 | 34.71316 | negative | MGI_C57BL6J_5621672 | Gm38787 | NCBI_Gene:105242628 | MGI:5621672 | lncRNA gene | predicted gene, 38787 |
| 6 | gene | 34.71480 | 34.71666 | positive | MGI_C57BL6J_5012151 | Gm19966 | NA | NA | unclassified gene | predicted gene%2c 19966 |
| 6 | gene | 34.72667 | 34.72691 | negative | MGI_C57BL6J_104972 | Npn2 | ENSEMBL:ENSMUSG00000060411 | MGI:104972 | lncRNA gene | neoplastic progression 2 |
| 6 | gene | 34.72722 | 34.73539 | negative | MGI_C57BL6J_3705289 | Gm14547 | ENSEMBL:ENSMUSG00000086834 | MGI:3705289 | lncRNA gene | predicted gene 14547 |
| 6 | gene | 34.78043 | 34.85946 | positive | MGI_C57BL6J_1923473 | Agbl3 | NCBI_Gene:76223,ENSEMBL:ENSMUSG00000038836 | MGI:1923473 | protein coding gene | ATP/GTP binding protein-like 3 |
| 6 | pseudogene | 34.78666 | 34.78751 | positive | MGI_C57BL6J_3651491 | Gm13864 | NCBI_Gene:100384877,ENSEMBL:ENSMUSG00000082920 | MGI:3651491 | pseudogene | predicted gene 13864 |
| 6 | pseudogene | 34.81063 | 34.81083 | negative | MGI_C57BL6J_3651503 | Gm13862 | ENSEMBL:ENSMUSG00000082118 | MGI:3651503 | pseudogene | predicted gene 13862 |
| 6 | pseudogene | 34.82020 | 34.82090 | negative | MGI_C57BL6J_3652088 | Gm13863 | ENSEMBL:ENSMUSG00000082446 | MGI:3652088 | pseudogene | predicted gene 13863 |
| 6 | gene | 34.84134 | 34.84954 | negative | MGI_C57BL6J_5826594 | Gm46957 | NCBI_Gene:108169138 | MGI:5826594 | lncRNA gene | predicted gene, 46957 |
| 6 | gene | 34.85970 | 34.86094 | positive | MGI_C57BL6J_5663885 | Gm43748 | ENSEMBL:ENSMUSG00000106707 | MGI:5663885 | unclassified gene | predicted gene 43748 |
| 6 | gene | 34.86311 | 34.87495 | positive | MGI_C57BL6J_1915737 | Tmem140 | NCBI_Gene:68487,ENSEMBL:ENSMUSG00000057137 | MGI:1915737 | protein coding gene | transmembrane protein 140 |
| 6 | gene | 34.87177 | 34.87806 | negative | MGI_C57BL6J_1925662 | Cyren | NCBI_Gene:78412,ENSEMBL:ENSMUSG00000046806 | MGI:1925662 | protein coding gene | cell cycle regulator of NHEJ |
| 6 | gene | 34.87759 | 34.91089 | negative | MGI_C57BL6J_2141558 | Wdr91 | NCBI_Gene:101240,ENSEMBL:ENSMUSG00000058486 | MGI:2141558 | protein coding gene | WD repeat domain 91 |
| 6 | gene | 34.92016 | 34.93934 | positive | MGI_C57BL6J_107917 | Stra8 | NCBI_Gene:20899,ENSEMBL:ENSMUSG00000029848 | MGI:107917 | protein coding gene | stimulated by retinoic acid gene 8 |
| 6 | gene | 34.94528 | 34.98850 | negative | MGI_C57BL6J_1917272 | Slc23a4 | NCBI_Gene:243753,ENSEMBL:ENSMUSG00000029847 | MGI:1917272 | protein coding gene | solute carrier family 23 member 4 |
| 6 | gene | 35.02206 | 35.13374 | negative | MGI_C57BL6J_1859026 | Cnot4 | NCBI_Gene:53621,ENSEMBL:ENSMUSG00000038784 | MGI:1859026 | protein coding gene | CCR4-NOT transcription complex, subunit 4 |
| 6 | gene | 35.12152 | 35.12393 | negative | MGI_C57BL6J_2443917 | A230071A22Rik | NA | NA | unclassified gene | RIKEN cDNA A230071A22 gene |
| 6 | gene | 35.13361 | 35.15214 | positive | MGI_C57BL6J_5592205 | Gm33046 | NCBI_Gene:102635798 | MGI:5592205 | lncRNA gene | predicted gene, 33046 |
| 6 | gene | 35.17742 | 35.24760 | positive | MGI_C57BL6J_2141625 | Nup205 | NCBI_Gene:70699,ENSEMBL:ENSMUSG00000038759 | MGI:2141625 | protein coding gene | nucleoporin 205 |
| 6 | gene | 35.24359 | 35.24375 | positive | MGI_C57BL6J_5453212 | Gm23435 | ENSEMBL:ENSMUSG00000087808 | MGI:5453212 | snRNA gene | predicted gene, 23435 |
| 6 | gene | 35.25265 | 35.26350 | positive | MGI_C57BL6J_1914955 | 1810058I24Rik | NCBI_Gene:67705,ENSEMBL:ENSMUSG00000073155 | MGI:1914955 | lncRNA gene | RIKEN cDNA 1810058I24 gene |
| 6 | gene | 35.26795 | 35.30813 | negative | MGI_C57BL6J_2442367 | Slc13a4 | NCBI_Gene:243755,ENSEMBL:ENSMUSG00000029843 | MGI:2442367 | protein coding gene | solute carrier family 13 (sodium/sulfate symporters), member 4 |
| 6 | gene | 35.31267 | 35.32614 | negative | MGI_C57BL6J_3039626 | Fam180a | NCBI_Gene:208164,ENSEMBL:ENSMUSG00000047420 | MGI:3039626 | protein coding gene | family with sequence similarity 180, member A |
| 6 | gene | 35.31315 | 35.32232 | positive | MGI_C57BL6J_5621673 | Gm38788 | NCBI_Gene:105242629 | MGI:5621673 | lncRNA gene | predicted gene, 38788 |
| 6 | gene | 35.33085 | 35.33683 | positive | MGI_C57BL6J_1920659 | 1700065J11Rik | NCBI_Gene:73409,ENSEMBL:ENSMUSG00000107341 | MGI:1920659 | lncRNA gene | RIKEN cDNA 1700065J11 gene |
| 6 | gene | 35.41280 | 35.41901 | positive | MGI_C57BL6J_5621674 | Gm38789 | NCBI_Gene:105242630 | MGI:5621674 | lncRNA gene | predicted gene, 38789 |
| 6 | gene | 35.50882 | 35.53989 | negative | MGI_C57BL6J_99445 | Mtpn | NCBI_Gene:14489,ENSEMBL:ENSMUSG00000029840 | MGI:99445 | protein coding gene | myotrophin |
| 6 | gene | 35.52454 | 35.52464 | negative | MGI_C57BL6J_5452830 | Gm23053 | ENSEMBL:ENSMUSG00000095986 | MGI:5452830 | miRNA gene | predicted gene, 23053 |
| 6 | gene | 35.61109 | 35.61567 | negative | MGI_C57BL6J_5663068 | Gm42931 | ENSEMBL:ENSMUSG00000107110 | MGI:5663068 | lncRNA gene | predicted gene 42931 |
| 6 | gene | 35.88328 | 35.88544 | negative | MGI_C57BL6J_1921788 | 9030409K20Rik | NA | NA | unclassified gene | RIKEN cDNA 9030409K20 gene |
| 6 | pseudogene | 35.92671 | 35.92712 | positive | MGI_C57BL6J_5663579 | Gm43442 | ENSEMBL:ENSMUSG00000107145 | MGI:5663579 | pseudogene | predicted gene 43442 |
| 6 | gene | 35.96588 | 35.99970 | negative | MGI_C57BL6J_5621675 | Gm38790 | NCBI_Gene:105242631 | MGI:5621675 | lncRNA gene | predicted gene, 38790 |
| 6 | gene | 35.97533 | 35.97546 | positive | MGI_C57BL6J_5453050 | Gm23273 | ENSEMBL:ENSMUSG00000077246 | MGI:5453050 | snoRNA gene | predicted gene, 23273 |
| 6 | gene | 36.18671 | 36.18910 | negative | MGI_C57BL6J_5592279 | Gm33120 | NA | NA | unclassified non-coding RNA gene | predicted gene%2c 33120 |
| 6 | gene | 36.18671 | 36.18910 | negative | MGI_C57BL6J_5663580 | Gm43443 | ENSEMBL:ENSMUSG00000107089 | MGI:5663580 | unclassified gene | predicted gene 43443 |
| 6 | gene | 36.20962 | 36.39019 | negative | MGI_C57BL6J_2442094 | 9330158H04Rik | NCBI_Gene:319472,ENSEMBL:ENSMUSG00000073154 | MGI:2442094 | lncRNA gene | RIKEN cDNA 9330158H04 gene |
| 6 | gene | 36.38808 | 36.52864 | positive | MGI_C57BL6J_88397 | Chrm2 | NCBI_Gene:243764,ENSEMBL:ENSMUSG00000045613 | MGI:88397 | protein coding gene | cholinergic receptor, muscarinic 2, cardiac |
| 6 | gene | 36.42174 | 36.42182 | positive | MGI_C57BL6J_3629662 | Mir490 | miRBase:MI0005002,NCBI_Gene:735279,ENSEMBL:ENSMUSG00000070075 | MGI:3629662 | miRNA gene | microRNA 490 |
| 6 | gene | 36.52502 | 36.52864 | positive | MGI_C57BL6J_2444158 | 9330162L04Rik | NA | NA | unclassified gene | RIKEN cDNA 9330162L04 gene |
| 6 | pseudogene | 36.58778 | 36.58798 | positive | MGI_C57BL6J_5663578 | Gm43441 | ENSEMBL:ENSMUSG00000106817 | MGI:5663578 | pseudogene | predicted gene 43441 |
| 6 | pseudogene | 36.63981 | 36.64027 | negative | MGI_C57BL6J_3780359 | Gm2189 | NCBI_Gene:100039370,ENSEMBL:ENSMUSG00000106872 | MGI:3780359 | pseudogene | predicted gene 2189 |
| 6 | gene | 36.69940 | 36.69954 | positive | MGI_C57BL6J_5454888 | Gm25111 | ENSEMBL:ENSMUSG00000080336 | MGI:5454888 | miRNA gene | predicted gene, 25111 |
| 6 | gene | 36.71492 | 36.81136 | negative | MGI_C57BL6J_97804 | Ptn | NCBI_Gene:19242,ENSEMBL:ENSMUSG00000029838 | MGI:97804 | protein coding gene | pleiotrophin |
| 6 | gene | 36.83738 | 37.30018 | negative | MGI_C57BL6J_2443430 | Dgki | NCBI_Gene:320127,ENSEMBL:ENSMUSG00000038665 | MGI:2443430 | protein coding gene | diacylglycerol kinase, iota |
| 6 | gene | 36.84021 | 36.84399 | negative | MGI_C57BL6J_2443216 | 6430604M11Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 6430604M11 gene |
| 6 | gene | 36.93705 | 36.98578 | positive | MGI_C57BL6J_1920835 | 1700111E14Rik | ENSEMBL:ENSMUSG00000029837 | MGI:1920835 | lncRNA gene | RIKEN cDNA 1700111E14 gene |
| 6 | pseudogene | 36.95665 | 36.96043 | positive | MGI_C57BL6J_3645151 | Gm7452 | NCBI_Gene:665024,ENSEMBL:ENSMUSG00000107310 | MGI:3645151 | pseudogene | predicted pseudogene 7452 |
| 6 | gene | 37.32726 | 37.44219 | negative | MGI_C57BL6J_2442695 | Creb3l2 | NCBI_Gene:208647,ENSEMBL:ENSMUSG00000038648 | MGI:2442695 | protein coding gene | cAMP responsive element binding protein 3-like 2 |
| 6 | gene | 37.41819 | 37.41945 | positive | MGI_C57BL6J_5592434 | Gm33275 | NCBI_Gene:102636112 | MGI:5592434 | lncRNA gene | predicted gene, 33275 |
| 6 | gene | 37.44250 | 37.44741 | positive | MGI_C57BL6J_5592477 | Gm33318 | NCBI_Gene:102636178 | MGI:5592477 | lncRNA gene | predicted gene, 33318 |
| 6 | gene | 37.52643 | 37.53140 | negative | MGI_C57BL6J_5592588 | Gm33429 | NCBI_Gene:102636331 | MGI:5592588 | lncRNA gene | predicted gene, 33429 |
| 6 | gene | 37.52998 | 37.57493 | positive | MGI_C57BL6J_2384785 | Akr1d1 | NCBI_Gene:208665,ENSEMBL:ENSMUSG00000038641 | MGI:2384785 | protein coding gene | aldo-keto reductase family 1, member D1 |
| 6 | pseudogene | 37.54141 | 37.54199 | positive | MGI_C57BL6J_3645336 | Gm7463 | NCBI_Gene:665046,ENSEMBL:ENSMUSG00000106889 | MGI:3645336 | pseudogene | predicted gene 7463 |
| 6 | pseudogene | 37.68564 | 37.68731 | negative | MGI_C57BL6J_2137671 | Ybx1-ps2 | NCBI_Gene:94283,ENSEMBL:ENSMUSG00000107035 | MGI:2137671 | pseudogene | Y box protein 1, pseudogene 2 |
| 6 | gene | 37.71466 | 37.73438 | positive | MGI_C57BL6J_5621676 | Gm38791 | NCBI_Gene:105242632 | MGI:5621676 | lncRNA gene | predicted gene, 38791 |
| 6 | pseudogene | 37.80597 | 37.80711 | positive | MGI_C57BL6J_2151816 | Atp6v0c-ps2 | NCBI_Gene:100039636,ENSEMBL:ENSMUSG00000080242 | MGI:2151816 | pseudogene | ATPase, H+ transporting, lysosomal V0 subunit C, pseudogene 2 |
| 6 | pseudogene | 37.86584 | 37.86656 | negative | MGI_C57BL6J_3648381 | Gm7492 | NCBI_Gene:665102,ENSEMBL:ENSMUSG00000106853 | MGI:3648381 | pseudogene | predicted gene 7492 |
| 6 | gene | 37.86988 | 37.96845 | positive | MGI_C57BL6J_109275 | Trim24 | NCBI_Gene:21848,ENSEMBL:ENSMUSG00000029833 | MGI:109275 | protein coding gene | tripartite motif-containing 24 |
| 6 | gene | 37.94102 | 37.94241 | positive | MGI_C57BL6J_1918761 | 9030601B04Rik | NA | NA | unclassified gene | RIKEN cDNA 9030601B04 gene |
| 6 | gene | 37.98374 | 38.04701 | negative | MGI_C57BL6J_2444335 | Svopl | NCBI_Gene:320590,ENSEMBL:ENSMUSG00000029830 | MGI:2444335 | protein coding gene | SV2 related protein homolog (rat)-like |
| 6 | pseudogene | 37.99466 | 37.99532 | negative | MGI_C57BL6J_5010586 | Gm18401 | NCBI_Gene:100417109 | MGI:5010586 | pseudogene | predicted gene, 18401 |
| 6 | gene | 38.03266 | 38.03551 | positive | MGI_C57BL6J_5592653 | Gm33494 | NCBI_Gene:102636429 | MGI:5592653 | lncRNA gene | predicted gene, 33494 |
| 6 | gene | 38.04848 | 38.12459 | negative | MGI_C57BL6J_2153480 | Atp6v0a4 | NCBI_Gene:140494,ENSEMBL:ENSMUSG00000038600 | MGI:2153480 | protein coding gene | ATPase, H+ transporting, lysosomal V0 subunit A4 |
| 6 | gene | 38.10925 | 38.11581 | positive | MGI_C57BL6J_1924772 | Tmem213 | NCBI_Gene:77522,ENSEMBL:ENSMUSG00000029829 | MGI:1924772 | protein coding gene | transmembrane protein 213 |
| 6 | gene | 38.11838 | 38.11849 | positive | MGI_C57BL6J_5453008 | Gm23231 | ENSEMBL:ENSMUSG00000087924 | MGI:5453008 | snRNA gene | predicted gene, 23231 |
| 6 | gene | 38.12317 | 38.25401 | negative | MGI_C57BL6J_2669829 | D630045J12Rik | NCBI_Gene:330286,ENSEMBL:ENSMUSG00000063455 | MGI:2669829 | protein coding gene | RIKEN cDNA D630045J12 gene |
| 6 | pseudogene | 38.28444 | 38.28534 | positive | MGI_C57BL6J_3643241 | Gm7504 | NCBI_Gene:665125,ENSEMBL:ENSMUSG00000107026 | MGI:3643241 | pseudogene | predicted gene 7504 |
| 6 | gene | 38.28739 | 38.29926 | negative | MGI_C57BL6J_2443387 | Zc3hav1l | NCBI_Gene:209032,ENSEMBL:ENSMUSG00000047749 | MGI:2443387 | protein coding gene | zinc finger CCCH-type, antiviral 1-like |
| 6 | gene | 38.30529 | 38.35557 | negative | MGI_C57BL6J_1926031 | Zc3hav1 | NCBI_Gene:78781,ENSEMBL:ENSMUSG00000029826 | MGI:1926031 | protein coding gene | zinc finger CCCH type, antiviral 1 |
| 6 | pseudogene | 38.36523 | 38.36590 | negative | MGI_C57BL6J_5663715 | Gm43578 | ENSEMBL:ENSMUSG00000106836 | MGI:5663715 | pseudogene | predicted gene 43578 |
| 6 | pseudogene | 38.37674 | 38.37719 | positive | MGI_C57BL6J_1321399 | Rpl30-ps1 | NCBI_Gene:19947,ENSEMBL:ENSMUSG00000057696 | MGI:1321399 | pseudogene | ribosomal protein L30, pseudogene 1 |
| 6 | gene | 38.38147 | 38.42765 | positive | MGI_C57BL6J_2444853 | Ttc26 | NCBI_Gene:264134,ENSEMBL:ENSMUSG00000056832 | MGI:2444853 | protein coding gene | tetratricopeptide repeat domain 26 |
| 6 | pseudogene | 38.39627 | 38.39669 | positive | MGI_C57BL6J_4414983 | Gm16563 | ENSEMBL:ENSMUSG00000089905 | MGI:4414983 | pseudogene | predicted gene 16563 |
| 6 | gene | 38.43304 | 38.43378 | negative | MGI_C57BL6J_1919478 | 1700021L22Rik | NA | NA | unclassified gene | RIKEN cDNA 1700021L22 gene |
| 6 | gene | 38.43393 | 38.52482 | positive | MGI_C57BL6J_2444236 | Ubn2 | NCBI_Gene:320538,ENSEMBL:ENSMUSG00000038538 | MGI:2444236 | protein coding gene | ubinuclein 2 |
| 6 | gene | 38.44536 | 38.44705 | positive | MGI_C57BL6J_2443652 | E430026H10Rik | NA | NA | unclassified gene | RIKEN cDNA E430026H10 gene |
| 6 | gene | 38.50733 | 38.50739 | positive | MGI_C57BL6J_5531089 | Mir7670 | miRBase:MI0025010,NCBI_Gene:102466843,ENSEMBL:ENSMUSG00000098285 | MGI:5531089 | miRNA gene | microRNA 7670 |
| 6 | gene | 38.53258 | 38.53388 | positive | MGI_C57BL6J_1920489 | 3110054G05Rik | NA | NA | unclassified gene | RIKEN cDNA 3110054G05 gene |
| 6 | gene | 38.53350 | 38.53945 | positive | MGI_C57BL6J_1913367 | Fmc1 | NCBI_Gene:66117,ENSEMBL:ENSMUSG00000019689 | MGI:1913367 | protein coding gene | formation of mitochondrial complex V assembly factor 1 |
| 6 | gene | 38.53393 | 38.53400 | positive | MGI_C57BL6J_4413723 | n-TRcct1 | NCBI_Gene:102467248 | MGI:4413723 | tRNA gene | nuclear encoded tRNA arginine 1 (anticodon CCT) |
| 6 | pseudogene | 38.54230 | 38.54610 | negative | MGI_C57BL6J_3783138 | Gm15697 | NCBI_Gene:621228,ENSEMBL:ENSMUSG00000081670 | MGI:3783138 | pseudogene | predicted gene 15697 |
| 6 | gene | 38.55042 | 38.55125 | negative | MGI_C57BL6J_2443093 | D630014H12Rik | NA | NA | unclassified gene | RIKEN cDNA D630014H12 gene |
| 6 | gene | 38.55133 | 38.60995 | positive | MGI_C57BL6J_2183260 | Luc7l2 | NCBI_Gene:192196,ENSEMBL:ENSMUSG00000029823 | MGI:2183260 | protein coding gene | LUC7-like 2 (S. cerevisiae) |
| 6 | gene | 38.55153 | 38.55401 | negative | MGI_C57BL6J_5477193 | Gm26699 | ENSEMBL:ENSMUSG00000097206 | MGI:5477193 | lncRNA gene | predicted gene, 26699 |
| 6 | gene | 38.58236 | 38.58449 | negative | MGI_C57BL6J_5012207 | Gm20022 | NA | NA | unclassified gene | predicted gene%2c 20022 |
| 6 | gene | 38.61005 | 38.61096 | positive | MGI_C57BL6J_1924759 | 9330153N18Rik | NA | NA | unclassified gene | RIKEN cDNA 9330153N18 gene |
| 6 | gene | 38.62141 | 38.63771 | negative | MGI_C57BL6J_1921503 | Klrg2 | NCBI_Gene:74253,ENSEMBL:ENSMUSG00000071537 | MGI:1921503 | protein coding gene | killer cell lectin-like receptor subfamily G, member 2 |
| 6 | gene | 38.66307 | 38.68086 | positive | MGI_C57BL6J_2141402 | Clec2l | NCBI_Gene:665180,ENSEMBL:ENSMUSG00000079598 | MGI:2141402 | protein coding gene | C-type lectin domain family 2, member L |
| 6 | gene | 38.68751 | 38.87619 | negative | MGI_C57BL6J_1314872 | Hipk2 | NCBI_Gene:15258,ENSEMBL:ENSMUSG00000061436 | MGI:1314872 | protein coding gene | homeodomain interacting protein kinase 2 |
| 6 | gene | 38.69974 | 38.74190 | positive | MGI_C57BL6J_5621381 | Gm38496 | NCBI_Gene:102637181 | MGI:5621381 | lncRNA gene | predicted gene, 38496 |
| 6 | gene | 38.75224 | 38.78048 | positive | MGI_C57BL6J_2444621 | 4930502C15Rik | NCBI_Gene:320733 | MGI:2444621 | lncRNA gene | RIKEN cDNA 4930502C15 gene |
| 6 | gene | 38.87538 | 39.08459 | positive | MGI_C57BL6J_98497 | Tbxas1 | NCBI_Gene:21391,ENSEMBL:ENSMUSG00000029925 | MGI:98497 | protein coding gene | thromboxane A synthase 1, platelet |
| 6 | gene | 38.87611 | 38.88505 | positive | MGI_C57BL6J_5826625 | Gm46988 | NCBI_Gene:108169201 | MGI:5826625 | protein coding gene | predicted gene, 46988 |
| 6 | gene | 38.91710 | 38.98432 | negative | MGI_C57BL6J_5621678 | Gm38793 | NA | NA | lncRNA gene | predicted gene%2c 38793 |
| 6 | gene | 38.91710 | 38.98432 | negative | MGI_C57BL6J_5663099 | Gm42962 | ENSEMBL:ENSMUSG00000106737 | MGI:5663099 | lncRNA gene | predicted gene 42962 |
| 6 | gene | 38.94437 | 38.95028 | negative | MGI_C57BL6J_5592926 | Gm33767 | NCBI_Gene:102636794 | MGI:5592926 | lncRNA gene | predicted gene, 33767 |
| 6 | gene | 38.98884 | 39.00644 | negative | MGI_C57BL6J_5621573 | Gm38688 | NCBI_Gene:105242415 | MGI:5621573 | lncRNA gene | predicted gene, 38688 |
| 6 | gene | 39.01367 | 39.01379 | positive | MGI_C57BL6J_5452787 | Gm23010 | ENSEMBL:ENSMUSG00000077339 | MGI:5452787 | snoRNA gene | predicted gene, 23010 |
| 6 | gene | 39.01920 | 39.04383 | negative | MGI_C57BL6J_5621374 | Gm38489 | NCBI_Gene:102636584 | MGI:5621374 | lncRNA gene | predicted gene, 38489 |
| 6 | gene | 39.04417 | 39.05652 | negative | MGI_C57BL6J_5826626 | Gm46989 | NCBI_Gene:108169202 | MGI:5826626 | lncRNA gene | predicted gene, 46989 |
| 6 | gene | 39.06388 | 39.06760 | negative | MGI_C57BL6J_1917236 | 1700025N23Rik | NCBI_Gene:69986,ENSEMBL:ENSMUSG00000100121 | MGI:1917236 | lncRNA gene | RIKEN cDNA 1700025N23 gene |
| 6 | gene | 39.08641 | 39.11835 | negative | MGI_C57BL6J_2143990 | Parp12 | NCBI_Gene:243771,ENSEMBL:ENSMUSG00000038507 | MGI:2143990 | protein coding gene | poly (ADP-ribose) polymerase family, member 12 |
| 6 | gene | 39.11847 | 39.14831 | positive | MGI_C57BL6J_1922629 | 4930599N23Rik | NCBI_Gene:75379,ENSEMBL:ENSMUSG00000073144 | MGI:1922629 | lncRNA gene | RIKEN cDNA 4930599N23 gene |
| 6 | gene | 39.12517 | 39.12638 | negative | MGI_C57BL6J_5663100 | Gm42963 | ENSEMBL:ENSMUSG00000106976 | MGI:5663100 | unclassified gene | predicted gene 42963 |
| 6 | gene | 39.13662 | 39.20679 | negative | MGI_C57BL6J_2443388 | Kdm7a | NCBI_Gene:338523,ENSEMBL:ENSMUSG00000042599 | MGI:2443388 | protein coding gene | lysine (K)-specific demethylase 7A |
| 6 | gene | 39.20707 | 39.20761 | positive | MGI_C57BL6J_6118414 | Gm49041 | ENSEMBL:ENSMUSG00000115026 | MGI:6118414 | lncRNA gene | predicted gene, 49041 |
| 6 | gene | 39.22756 | 39.24535 | positive | MGI_C57BL6J_5621679 | Gm38794 | NCBI_Gene:105242635 | MGI:5621679 | lncRNA gene | predicted gene, 38794 |
| 6 | gene | 39.24011 | 39.24254 | positive | MGI_C57BL6J_5663616 | Gm43479 | ENSEMBL:ENSMUSG00000107365 | MGI:5663616 | lncRNA gene | predicted gene 43479 |
| 6 | gene | 39.24615 | 39.26561 | negative | MGI_C57BL6J_5593306 | Gm34147 | NCBI_Gene:102637300 | MGI:5593306 | lncRNA gene | predicted gene, 34147 |
| 6 | gene | 39.26565 | 39.29957 | negative | MGI_C57BL6J_5455785 | Gm26008 | NCBI_Gene:102637404,ENSEMBL:ENSMUSG00000089422 | MGI:5455785 | snRNA gene | predicted gene, 26008 |
| 6 | gene | 39.33477 | 39.37772 | negative | MGI_C57BL6J_1919394 | Slc37a3 | NCBI_Gene:72144,ENSEMBL:ENSMUSG00000029924 | MGI:1919394 | protein coding gene | solute carrier family 37 (glycerol-3-phosphate transporter), member 3 |
| 6 | gene | 39.38117 | 39.39067 | positive | MGI_C57BL6J_103292 | Rab19 | NCBI_Gene:19331,ENSEMBL:ENSMUSG00000029923 | MGI:103292 | protein coding gene | RAB19, member RAS oncogene family |
| 6 | gene | 39.39780 | 39.42330 | negative | MGI_C57BL6J_1859353 | Mkrn1 | NCBI_Gene:54484,ENSEMBL:ENSMUSG00000029922 | MGI:1859353 | protein coding gene | makorin, ring finger protein, 1 |
| 6 | gene | 39.41989 | 39.42129 | positive | MGI_C57BL6J_3642504 | Gm10244 | NCBI_Gene:621427,ENSEMBL:ENSMUSG00000068601 | MGI:3642504 | lncRNA gene | predicted gene 10244 |
| 6 | gene | 39.42229 | 39.42242 | negative | MGI_C57BL6J_5455179 | Gm25402 | ENSEMBL:ENSMUSG00000064900 | MGI:5455179 | snoRNA gene | predicted gene, 25402 |
| 6 | gene | 39.46237 | 39.55817 | negative | MGI_C57BL6J_2444961 | Dennd2a | NCBI_Gene:209773,ENSEMBL:ENSMUSG00000038456 | MGI:2444961 | protein coding gene | DENN/MADD domain containing 2A |
| 6 | gene | 39.52751 | 39.54449 | positive | MGI_C57BL6J_1924459 | 8030453O22Rik | NCBI_Gene:77209,ENSEMBL:ENSMUSG00000085058 | MGI:1924459 | lncRNA gene | RIKEN cDNA 8030453O22 gene |
| 6 | gene | 39.57384 | 39.58877 | positive | MGI_C57BL6J_1889336 | Adck2 | NCBI_Gene:57869,ENSEMBL:ENSMUSG00000046947 | MGI:1889336 | protein coding gene | aarF domain containing kinase 2 |
| 6 | gene | 39.58409 | 39.59278 | negative | MGI_C57BL6J_3826603 | Gm16272 | ENSEMBL:ENSMUSG00000086582 | MGI:3826603 | lncRNA gene | predicted gene 16272 |
| 6 | gene | 39.58575 | 39.59660 | positive | MGI_C57BL6J_5649067 | Gm42420 | ENSEMBL:ENSMUSG00000107071 | MGI:5649067 | protein coding gene | predicted gene, 42420 |
| 6 | gene | 39.59255 | 39.60338 | positive | MGI_C57BL6J_1915448 | Ndufb2 | NCBI_Gene:68198,ENSEMBL:ENSMUSG00000002416 | MGI:1915448 | protein coding gene | NADH:ubiquinone oxidoreductase subunit B2 |
| 6 | gene | 39.60323 | 39.72566 | negative | MGI_C57BL6J_88190 | Braf | NCBI_Gene:109880,ENSEMBL:ENSMUSG00000002413 | MGI:88190 | protein coding gene | Braf transforming gene |
| 6 | gene | 39.61650 | 39.62507 | positive | MGI_C57BL6J_5593548 | Gm34389 | NCBI_Gene:102637627 | MGI:5593548 | lncRNA gene | predicted gene, 34389 |
| 6 | pseudogene | 39.65330 | 39.65396 | negative | MGI_C57BL6J_104594 | Fth-ps2 | NCBI_Gene:14321,ENSEMBL:ENSMUSG00000091313 | MGI:104594 | pseudogene | ferritin heavy chain, pseudogene 2 |
| 6 | gene | 39.80180 | 39.81099 | negative | MGI_C57BL6J_1338046 | Mrps33 | NCBI_Gene:14548,ENSEMBL:ENSMUSG00000029918 | MGI:1338046 | protein coding gene | mitochondrial ribosomal protein S33 |
| 6 | gene | 39.81018 | 39.81065 | negative | MGI_C57BL6J_1925337 | 5730402E23Rik | NA | NA | unclassified gene | RIKEN cDNA 5730402E23 gene |
| 6 | pseudogene | 39.82815 | 39.82881 | negative | MGI_C57BL6J_3646122 | Gm7554 | NCBI_Gene:665247,ENSEMBL:ENSMUSG00000106936 | MGI:3646122 | pseudogene | predicted gene 7554 |
| 6 | pseudogene | 39.84302 | 39.84448 | negative | MGI_C57BL6J_3645467 | Gm7556 | NCBI_Gene:665254,ENSEMBL:ENSMUSG00000106827 | MGI:3645467 | pseudogene | predicted gene 7556 |
| 6 | gene | 39.87068 | 40.25515 | positive | MGI_C57BL6J_3647581 | Tmem178b | NCBI_Gene:434008,ENSEMBL:ENSMUSG00000057716 | MGI:3647581 | protein coding gene | transmembrane protein 178B |
| 6 | gene | 39.87153 | 39.87197 | negative | MGI_C57BL6J_5477327 | Gm26833 | ENSEMBL:ENSMUSG00000097406 | MGI:5477327 | lncRNA gene | predicted gene, 26833 |
| 6 | gene | 40.02689 | 40.02861 | positive | MGI_C57BL6J_5611223 | Gm37995 | ENSEMBL:ENSMUSG00000103331 | MGI:5611223 | unclassified gene | predicted gene, 37995 |
| 6 | gene | 40.09872 | 40.10117 | positive | MGI_C57BL6J_5610901 | Gm37673 | ENSEMBL:ENSMUSG00000102623 | MGI:5610901 | unclassified gene | predicted gene, 37673 |
| 6 | gene | 40.11878 | 40.12948 | positive | MGI_C57BL6J_5477132 | Gm26638 | ENSEMBL:ENSMUSG00000097563 | MGI:5477132 | lncRNA gene | predicted gene, 26638 |
| 6 | gene | 40.20330 | 40.20926 | negative | MGI_C57BL6J_5477301 | Gm26807 | ENSEMBL:ENSMUSG00000097484 | MGI:5477301 | lncRNA gene | predicted gene, 26807 |
| 6 | gene | 40.22622 | 40.22904 | positive | MGI_C57BL6J_5610231 | Gm37003 | ENSEMBL:ENSMUSG00000102287 | MGI:5610231 | unclassified gene | predicted gene, 37003 |
| 6 | gene | 40.32516 | 40.39756 | positive | MGI_C57BL6J_1917173 | Agk | NCBI_Gene:69923,ENSEMBL:ENSMUSG00000029916 | MGI:1917173 | protein coding gene | acylglycerol kinase |
| 6 | gene | 40.40138 | 40.43649 | negative | MGI_C57BL6J_2444256 | E330009J07Rik | NCBI_Gene:243780,ENSEMBL:ENSMUSG00000037172 | MGI:2444256 | protein coding gene | RIKEN cDNA E330009J07 gene |
| 6 | gene | 40.43540 | 40.46681 | positive | MGI_C57BL6J_3027899 | Wee2 | NCBI_Gene:381759,ENSEMBL:ENSMUSG00000037159 | MGI:3027899 | protein coding gene | WEE1 homolog 2 (S. pombe) |
| 6 | gene | 40.47054 | 40.47128 | negative | MGI_C57BL6J_1925080 | A930033M24Rik | NA | NA | unclassified gene | RIKEN cDNA A930033M24 gene |
| 6 | gene | 40.47135 | 40.48470 | positive | MGI_C57BL6J_1920040 | Ssbp1 | NCBI_Gene:381760,ENSEMBL:ENSMUSG00000029911 | MGI:1920040 | protein coding gene | single-stranded DNA binding protein 1 |
| 6 | gene | 40.49123 | 40.49231 | positive | MGI_C57BL6J_3606604 | Tas2r137 | NCBI_Gene:574417,ENSEMBL:ENSMUSG00000052850 | MGI:3606604 | protein coding gene | taste receptor, type 2, member 137 |
| 6 | gene | 40.49353 | 40.49454 | positive | MGI_C57BL6J_2681210 | Tas2r108 | NCBI_Gene:57253,ENSEMBL:ENSMUSG00000037140 | MGI:2681210 | protein coding gene | taste receptor, type 2, member 108 |
| 6 | gene | 40.51482 | 40.51955 | negative | MGI_C57BL6J_1914940 | Prss37 | NCBI_Gene:67690,ENSEMBL:ENSMUSG00000029909 | MGI:1914940 | protein coding gene | protease, serine 37 |
| 6 | pseudogene | 40.52280 | 40.52322 | negative | MGI_C57BL6J_5663432 | Gm43295 | ENSEMBL:ENSMUSG00000107359 | MGI:5663432 | pseudogene | predicted gene 43295 |
| 6 | gene | 40.54217 | 40.55505 | negative | MGI_C57BL6J_3030295 | Olfr461 | NCBI_Gene:258380,ENSEMBL:ENSMUSG00000068259 | MGI:3030295 | protein coding gene | olfactory receptor 461 |
| 6 | gene | 40.56915 | 40.57434 | positive | MGI_C57BL6J_3030294 | Olfr460 | NCBI_Gene:258381,ENSEMBL:ENSMUSG00000045514 | MGI:3030294 | protein coding gene | olfactory receptor 460 |
| 6 | gene | 40.57489 | 40.58582 | negative | MGI_C57BL6J_1345151 | Clec5a | NCBI_Gene:23845,ENSEMBL:ENSMUSG00000029915 | MGI:1345151 | protein coding gene | C-type lectin domain family 5, member a |
| 6 | gene | 40.61232 | 40.61331 | negative | MGI_C57BL6J_2681306 | Tas2r138 | NCBI_Gene:387513,ENSEMBL:ENSMUSG00000058250 | MGI:2681306 | protein coding gene | taste receptor, type 2, member 138 |
| 6 | gene | 40.62883 | 40.76912 | positive | MGI_C57BL6J_1203495 | Mgam | NCBI_Gene:232714,ENSEMBL:ENSMUSG00000068587 | MGI:1203495 | protein coding gene | maltase-glucoamylase |
| 6 | pseudogene | 40.77871 | 40.86816 | positive | MGI_C57BL6J_3645497 | Mgam2-ps | NCBI_Gene:638904,ENSEMBL:ENSMUSG00000102802 | MGI:3645497 | pseudogene | maltase-glucoamylase 2, pseudogene |
| 6 | gene | 40.87879 | 40.88749 | negative | MGI_C57BL6J_2388042 | Moxd2 | NCBI_Gene:194357,ENSEMBL:ENSMUSG00000029885 | MGI:2388042 | protein coding gene | monooxygenase, DBH-like 2 |
| 6 | gene | 40.88953 | 40.89087 | negative | MGI_C57BL6J_5611504 | Gm38276 | ENSEMBL:ENSMUSG00000104084 | MGI:5611504 | unclassified gene | predicted gene, 38276 |
| 6 | gene | 40.89123 | 40.89189 | positive | MGI_C57BL6J_98594 | Trbv1 | NCBI_Gene:621968,ENSEMBL:ENSMUSG00000076461 | MGI:98594 | gene segment | T cell receptor beta, variable 1 |
| 6 | gene | 40.89526 | 40.90039 | negative | MGI_C57BL6J_3608323 | Prss58 | NCBI_Gene:232717,ENSEMBL:ENSMUSG00000051936 | MGI:3608323 | protein coding gene | protease, serine 58 |
| 6 | gene | 40.92044 | 40.94097 | negative | MGI_C57BL6J_1920731 | 1700074P13Rik | NCBI_Gene:73481,ENSEMBL:ENSMUSG00000029883 | MGI:1920731 | protein coding gene | RIKEN cDNA 1700074P13 gene |
| 6 | gene | 40.94837 | 40.95435 | negative | MGI_C57BL6J_3643181 | Gm4744 | NCBI_Gene:194360,ENSEMBL:ENSMUSG00000063252 | MGI:3643181 | protein coding gene | predicted gene 4744 |
| 6 | gene | 40.95022 | 40.95052 | positive | MGI_C57BL6J_5580294 | Gm29588 | ENSEMBL:ENSMUSG00000101629 | MGI:5580294 | lncRNA gene | predicted gene 29588 |
| 6 | gene | 40.96475 | 40.96843 | positive | MGI_C57BL6J_1920876 | 1810009J06Rik | NCBI_Gene:73626,ENSEMBL:ENSMUSG00000094808 | MGI:1920876 | protein coding gene | RIKEN cDNA 1810009J06 gene |
| 6 | gene | 40.99582 | 40.99948 | negative | MGI_C57BL6J_3780832 | Gm2663 | NCBI_Gene:100040208,ENSEMBL:ENSMUSG00000096525 | MGI:3780832 | protein coding gene | predicted gene 2663 |
| 6 | pseudogene | 41.01118 | 41.01137 | negative | MGI_C57BL6J_5611243 | Gm38015 | ENSEMBL:ENSMUSG00000104408 | MGI:5611243 | pseudogene | predicted gene, 38015 |
| 6 | pseudogene | 41.02124 | 41.02188 | negative | MGI_C57BL6J_5611106 | Gm37878 | ENSEMBL:ENSMUSG00000104105 | MGI:5611106 | pseudogene | predicted gene, 37878 |
| 6 | pseudogene | 41.02627 | 41.02665 | positive | MGI_C57BL6J_5611522 | Gm38294 | ENSEMBL:ENSMUSG00000103157 | MGI:5611522 | pseudogene | predicted gene, 38294 |
| 6 | gene | 41.03027 | 41.03551 | negative | MGI_C57BL6J_1914623 | 2210010C04Rik | NCBI_Gene:67373,ENSEMBL:ENSMUSG00000029882 | MGI:1914623 | protein coding gene | RIKEN cDNA 2210010C04 gene |
| 6 | gene | 41.04734 | 41.04800 | positive | MGI_C57BL6J_98599 | Trbv2 | NCBI_Gene:100124659,ENSEMBL:ENSMUSG00000076462 | MGI:98599 | gene segment | T cell receptor beta, variable 2 |
| 6 | gene | 41.04829 | 41.04883 | positive | MGI_C57BL6J_98590 | Trbv3 | NCBI_Gene:100039790,ENSEMBL:ENSMUSG00000076463 | MGI:98590 | gene segment | T cell receptor beta, variable 3 |
| 6 | gene | 41.05588 | 41.06310 | positive | MGI_C57BL6J_1923265 | 5830405F06Rik | ENSEMBL:ENSMUSG00000103586 | MGI:1923265 | lncRNA gene | RIKEN cDNA 5830405F06 gene |
| 6 | gene | 41.05939 | 41.05989 | positive | MGI_C57BL6J_98584 | Trbv4 | NCBI_Gene:100124660,ENSEMBL:ENSMUSG00000107486 | MGI:98584 | gene segment | T cell receptor beta, variable 10 |
| 6 | gene | 41.06233 | 41.06280 | positive | MGI_C57BL6J_98583 | Trbv5 | NCBI_Gene:100124661,ENSEMBL:ENSMUSG00000076465 | MGI:98583 | gene segment | T cell receptor beta, variable 5 |
| 6 | pseudogene | 41.06687 | 41.06715 | positive | MGI_C57BL6J_5009962 | Trbv6 | NCBI_Gene:100124662,ENSEMBL:ENSMUSG00000103807 | MGI:5009962 | pseudogenic gene segment | T cell receptor beta variable 6 |
| 6 | pseudogene | 41.08025 | 41.08065 | positive | MGI_C57BL6J_5009963 | Trbv7 | NCBI_Gene:100124663,ENSEMBL:ENSMUSG00000102622 | MGI:5009963 | pseudogenic gene segment | T cell receptor beta variable 7 |
| 6 | gene | 41.08159 | 41.09198 | positive | MGI_C57BL6J_6270568 | Gm49885 | ENSEMBL:ENSMUSG00000103833 | MGI:6270568 | lncRNA gene | predicted gene, 49885 |
| 6 | pseudogene | 41.08467 | 41.08511 | positive | MGI_C57BL6J_5009964 | Trbv8 | NCBI_Gene:100124664,ENSEMBL:ENSMUSG00000102821 | MGI:5009964 | pseudogenic gene segment | T cell receptor beta variable 8 |
| 6 | pseudogene | 41.08799 | 41.08829 | positive | MGI_C57BL6J_5009965 | Trbv9 | NCBI_Gene:100124665,ENSEMBL:ENSMUSG00000104198 | MGI:5009965 | pseudogenic gene segment | T cell receptor beta variable 9 |
| 6 | pseudogene | 41.09193 | 41.09223 | positive | MGI_C57BL6J_4937160 | Trbv10 | NCBI_Gene:100124670,ENSEMBL:ENSMUSG00000091671 | MGI:4937160 | pseudogenic gene segment | T cell receptor beta variable 10 |
| 6 | pseudogene | 41.10681 | 41.10730 | positive | MGI_C57BL6J_5009966 | Trbv11 | NCBI_Gene:100124671,ENSEMBL:ENSMUSG00000102903 | MGI:5009966 | pseudogenic gene segment | T cell receptor beta variable 11 |
| 6 | gene | 41.11355 | 41.11407 | positive | MGI_C57BL6J_98602 | Trbv12-1 | NCBI_Gene:100124672,ENSEMBL:ENSMUSG00000095574 | MGI:98602 | gene segment | T cell receptor beta, variable 12-1 |
| 6 | gene | 41.11601 | 41.11647 | positive | MGI_C57BL6J_98609 | Trbv13-1 | NCBI_Gene:100124673,ENSEMBL:ENSMUSG00000076467 | MGI:98609 | gene segment | T cell receptor beta, variable 13-1 |
| 6 | gene | 41.11885 | 41.11936 | positive | MGI_C57BL6J_98601 | Trbv12-2 | NCBI_Gene:100124674,ENSEMBL:ENSMUSG00000094525 | MGI:98601 | gene segment | T cell receptor beta, variable 12-2 |
| 6 | gene | 41.12140 | 41.12183 | positive | MGI_C57BL6J_98608 | Trbv13-2 | NCBI_Gene:100124675,ENSEMBL:ENSMUSG00000076469 | MGI:98608 | gene segment | T cell receptor beta, variable 13-2 |
| 6 | pseudogene | 41.12759 | 41.12808 | positive | MGI_C57BL6J_98603 | Trbv12-3 | NCBI_Gene:100124676,ENSEMBL:ENSMUSG00000102327 | MGI:98603 | pseudogenic gene segment | T cell receptor beta, variable 12-3 |
| 6 | gene | 41.13015 | 41.13059 | positive | MGI_C57BL6J_98607 | Trbv13-3 | NCBI_Gene:100124677,ENSEMBL:ENSMUSG00000076470 | MGI:98607 | gene segment | T cell receptor beta, variable 13-3 |
| 6 | gene | 41.13515 | 41.13562 | positive | MGI_C57BL6J_98587 | Trbv14 | NCBI_Gene:100124678,ENSEMBL:ENSMUSG00000076471 | MGI:98587 | gene segment | T cell receptor beta, variable 14 |
| 6 | gene | 41.14119 | 41.14166 | positive | MGI_C57BL6J_98586 | Trbv15 | NCBI_Gene:100124679,ENSEMBL:ENSMUSG00000076472 | MGI:98586 | gene segment | T cell receptor beta, variable 15 |
| 6 | gene | 41.15174 | 41.15223 | positive | MGI_C57BL6J_98585 | Trbv16 | NCBI_Gene:100124680,ENSEMBL:ENSMUSG00000076473 | MGI:98585 | gene segment | T cell receptor beta, variable 16 |
| 6 | gene | 41.16306 | 41.16356 | positive | MGI_C57BL6J_98610 | Trbv17 | NCBI_Gene:100124681,ENSEMBL:ENSMUSG00000076474 | MGI:98610 | gene segment | T cell receptor beta, variable 17 |
| 6 | pseudogene | 41.17514 | 41.17561 | positive | MGI_C57BL6J_5009967 | Trbv18 | NCBI_Gene:100124682,ENSEMBL:ENSMUSG00000102409 | MGI:5009967 | pseudogenic gene segment | T cell receptor beta variable 18 |
| 6 | gene | 41.17838 | 41.17905 | positive | MGI_C57BL6J_98604 | Trbv19 | NCBI_Gene:100124684,ENSEMBL:ENSMUSG00000076475 | MGI:98604 | gene segment | T cell receptor beta, variable 19 |
| 6 | gene | 41.18827 | 41.18898 | positive | MGI_C57BL6J_98589 | Trbv20 | NCBI_Gene:100124685,ENSEMBL:ENSMUSG00000076476 | MGI:98589 | gene segment | T cell receptor beta, variable 20 |
| 6 | gene | 41.20255 | 41.20304 | positive | MGI_C57BL6J_98593 | Trbv21 | NCBI_Gene:100124686,ENSEMBL:ENSMUSG00000076477 | MGI:98593 | gene segment | T cell receptor beta, variable 21 |
| 6 | pseudogene | 41.21207 | 41.21253 | positive | MGI_C57BL6J_5009968 | Trbv22 | NCBI_Gene:100124687,ENSEMBL:ENSMUSG00000102988 | MGI:5009968 | pseudogenic gene segment | T cell receptor beta variable 22 |
| 6 | gene | 41.21607 | 41.21653 | positive | MGI_C57BL6J_98595 | Trbv23 | NCBI_Gene:100124688,ENSEMBL:ENSMUSG00000076478 | MGI:98595 | gene segment | T cell receptor beta, variable V23 |
| 6 | gene | 41.21809 | 41.21856 | positive | MGI_C57BL6J_98591 | Trbv24 | NCBI_Gene:100124689,ENSEMBL:ENSMUSG00000102955 | MGI:98591 | gene segment | T cell receptor beta, variable 24 |
| 6 | pseudogene | 41.22167 | 41.22221 | positive | MGI_C57BL6J_5009969 | Trbv25 | NCBI_Gene:100124690,ENSEMBL:ENSMUSG00000104473 | MGI:5009969 | pseudogenic gene segment | T cell receptor beta variable 25 |
| 6 | gene | 41.22751 | 41.22799 | positive | MGI_C57BL6J_98596 | Trbv26 | NCBI_Gene:110315,ENSEMBL:ENSMUSG00000076479 | MGI:98596 | gene segment | T cell receptor beta, variable 26 |
| 6 | pseudogene | 41.25323 | 41.25433 | positive | MGI_C57BL6J_3780888 | Gm2719 | NCBI_Gene:100040333,ENSEMBL:ENSMUSG00000103042 | MGI:3780888 | pseudogene | predicted pseudogene 2719 |
| 6 | pseudogene | 41.25812 | 41.25858 | positive | MGI_C57BL6J_5009970 | Trbv27 | NCBI_Gene:100124693,ENSEMBL:ENSMUSG00000104254 | MGI:5009970 | pseudogenic gene segment | T cell receptor beta variable 27 |
| 6 | pseudogene | 41.26670 | 41.26715 | positive | MGI_C57BL6J_5009971 | Trbv28 | NCBI_Gene:100124694,ENSEMBL:ENSMUSG00000104326 | MGI:5009971 | pseudogenic gene segment | T cell receptor beta variable 28 |
| 6 | pseudogene | 41.27139 | 41.27188 | positive | MGI_C57BL6J_98605 | Trbv29 | NCBI_Gene:100124698,ENSEMBL:ENSMUSG00000076480 | MGI:98605 | pseudogenic gene segment | T cell receptor beta, variable 29 |
| 6 | gene | 41.28138 | 41.28199 | positive | MGI_C57BL6J_98592 | Trbv30 | NCBI_Gene:100124699,ENSEMBL:ENSMUSG00000076481 | MGI:98592 | gene segment | T cell receptor beta, variable 30 |
| 6 | gene | 41.30227 | 41.30553 | positive | MGI_C57BL6J_102757 | Try4 | NCBI_Gene:22074,ENSEMBL:ENSMUSG00000054106 | MGI:102757 | protein coding gene | trypsin 4 |
| 6 | pseudogene | 41.30620 | 41.30676 | negative | MGI_C57BL6J_5011149 | Gm18964 | NCBI_Gene:100418045,ENSEMBL:ENSMUSG00000103797 | MGI:5011149 | pseudogene | predicted gene, 18964 |
| 6 | gene | 41.31123 | 41.31684 | negative | MGI_C57BL6J_102756 | Try5 | NCBI_Gene:103964,ENSEMBL:ENSMUSG00000036938 | MGI:102756 | protein coding gene | trypsin 5 |
| 6 | gene | 41.35410 | 41.35794 | positive | MGI_C57BL6J_3687012 | Try10 | NCBI_Gene:436522,ENSEMBL:ENSMUSG00000071521 | MGI:3687012 | protein coding gene | trypsin 10 |
| 6 | gene | 41.37376 | 41.37768 | negative | MGI_C57BL6J_102758 | Prss3 | NCBI_Gene:22073,ENSEMBL:ENSMUSG00000071519 | MGI:102758 | protein coding gene | protease, serine 3 |
| 6 | gene | 41.39236 | 41.39726 | positive | MGI_C57BL6J_3646222 | Gm5771 | NCBI_Gene:436523,ENSEMBL:ENSMUSG00000058119 | MGI:3646222 | protein coding gene | predicted gene 5771 |
| 6 | pseudogene | 41.40508 | 41.41963 | positive | MGI_C57BL6J_3643903 | Gm5409 | NCBI_Gene:386551,ENSEMBL:ENSMUSG00000059014 | MGI:3643903 | pseudogene | predicted pseudogene 5409 |
| 6 | pseudogene | 41.40691 | 41.40727 | positive | MGI_C57BL6J_5611534 | Gm38306 | ENSEMBL:ENSMUSG00000103397 | MGI:5611534 | pseudogene | predicted gene, 38306 |
| 6 | gene | 41.44221 | 41.44615 | negative | MGI_C57BL6J_3641889 | Gm10334 | NCBI_Gene:100040233,ENSEMBL:ENSMUSG00000071517 | MGI:3641889 | protein coding gene | predicted gene 10334 |
| 6 | gene | 41.45893 | 41.46379 | positive | MGI_C57BL6J_98839 | Prss1 | NCBI_Gene:114228,ENSEMBL:ENSMUSG00000062751 | MGI:98839 | protein coding gene | protease, serine 1 (trypsin 1) |
| 6 | pseudogene | 41.48051 | 41.48172 | positive | MGI_C57BL6J_5610529 | Gm37301 | ENSEMBL:ENSMUSG00000102221 | MGI:5610529 | pseudogene | predicted gene, 37301 |
| 6 | pseudogene | 41.49338 | 41.49515 | negative | MGI_C57BL6J_5588972 | Gm29813 | NCBI_Gene:101954205,ENSEMBL:ENSMUSG00000104513 | MGI:5588972 | pseudogene | predicted gene, 29813 |
| 6 | pseudogene | 41.49693 | 41.49730 | positive | MGI_C57BL6J_5611069 | Gm37841 | ENSEMBL:ENSMUSG00000102616 | MGI:5611069 | pseudogene | predicted gene, 37841 |
| 6 | pseudogene | 41.50412 | 41.50565 | positive | MGI_C57BL6J_5611246 | Gm38018 | ENSEMBL:ENSMUSG00000103699 | MGI:5611246 | pseudogene | predicted gene, 38018 |
| 6 | gene | 41.52178 | 41.52508 | positive | MGI_C57BL6J_102759 | Prss2 | NCBI_Gene:22072,ENSEMBL:ENSMUSG00000057163 | MGI:102759 | protein coding gene | protease, serine 2 |
| 6 | gene | 41.53320 | 41.53321 | positive | MGI_C57BL6J_4439571 | Trbd1 | NCBI_Gene:100125246,ENSEMBL:ENSMUSG00000095668 | MGI:4439571 | gene segment | T cell receptor beta, D region 1 |
| 6 | gene | 41.53386 | 41.53391 | positive | MGI_C57BL6J_4439570 | Trbj1-1 | NCBI_Gene:100125248,ENSEMBL:ENSMUSG00000076483 | MGI:4439570 | gene segment | T cell receptor beta joining 1-1 |
| 6 | gene | 41.53400 | 41.53405 | positive | MGI_C57BL6J_4439569 | Trbj1-2 | NCBI_Gene:100125249,ENSEMBL:ENSMUSG00000076484 | MGI:4439569 | gene segment | T cell receptor beta joining 1-2 |
| 6 | gene | 41.53432 | 41.53438 | positive | MGI_C57BL6J_4439568 | Trbj1-3 | NCBI_Gene:100125250,ENSEMBL:ENSMUSG00000076485 | MGI:4439568 | gene segment | T cell receptor beta joining 1-3 |
| 6 | gene | 41.53481 | 41.53486 | positive | MGI_C57BL6J_4439567 | Trbj1-4 | NCBI_Gene:100125251,ENSEMBL:ENSMUSG00000076486 | MGI:4439567 | gene segment | T cell receptor beta joining 1-4 |
| 6 | gene | 41.53508 | 41.53513 | positive | MGI_C57BL6J_4439566 | Trbj1-5 | NCBI_Gene:100125252,ENSEMBL:ENSMUSG00000076487 | MGI:4439566 | gene segment | T cell receptor beta joining 1-5 |
| 6 | gene | 41.53555 | 41.53561 | positive | MGI_C57BL6J_4439575 | Trbj1-6 | NCBI_Gene:100125253,ENSEMBL:ENSMUSG00000076488 | MGI:4439575 | gene segment | T cell receptor beta joining 1-6 |
| 6 | gene | 41.53564 | 41.53569 | positive | MGI_C57BL6J_4439574 | Trbj1-7 | NCBI_Gene:100125254,ENSEMBL:ENSMUSG00000076489 | MGI:4439574 | gene segment | T cell receptor beta joining 1-7 |
| 6 | gene | 41.53822 | 41.53988 | positive | MGI_C57BL6J_4439726 | Trbc1 | NCBI_Gene:100125262,ENSEMBL:ENSMUSG00000076490 | MGI:4439726 | gene segment | T cell receptor beta, constant region 1 |
| 6 | gene | 41.54216 | 41.54218 | positive | MGI_C57BL6J_4439727 | Trbd2 | NCBI_Gene:100125247,ENSEMBL:ENSMUSG00000094569 | MGI:4439727 | gene segment | T cell receptor beta, D region 2 |
| 6 | gene | 41.54275 | 41.54280 | positive | MGI_C57BL6J_4439728 | Trbj2-1 | NCBI_Gene:100125255,ENSEMBL:ENSMUSG00000076492 | MGI:4439728 | gene segment | T cell receptor beta joining 2-1 |
| 6 | gene | 41.54296 | 41.54301 | positive | MGI_C57BL6J_4439729 | Trbj2-2 | NCBI_Gene:100125256,ENSEMBL:ENSMUSG00000076493 | MGI:4439729 | gene segment | T cell receptor beta joining 2-2 |
| 6 | gene | 41.54322 | 41.54327 | positive | MGI_C57BL6J_4439898 | Trbj2-3 | NCBI_Gene:100125257,ENSEMBL:ENSMUSG00000076494 | MGI:4439898 | gene segment | T cell receptor beta joining 2-3 |
| 6 | gene | 41.54336 | 41.54341 | positive | MGI_C57BL6J_4439730 | Trbj2-4 | NCBI_Gene:100125258,ENSEMBL:ENSMUSG00000076495 | MGI:4439730 | gene segment | T cell receptor beta joining 2-4 |
| 6 | gene | 41.54345 | 41.54350 | positive | MGI_C57BL6J_4439899 | Trbj2-5 | NCBI_Gene:100125259,ENSEMBL:ENSMUSG00000076496 | MGI:4439899 | gene segment | T cell receptor beta joining 2-5 |
| 6 | pseudogene | 41.54360 | 41.54364 | positive | MGI_C57BL6J_4937236 | Trbj2-6 | NCBI_Gene:100125260,ENSEMBL:ENSMUSG00000090818 | MGI:4937236 | pseudogenic gene segment | T cell receptor beta joining 2-6 |
| 6 | gene | 41.54381 | 41.54386 | positive | MGI_C57BL6J_4439731 | Trbj2-7 | NCBI_Gene:100125261,ENSEMBL:ENSMUSG00000076497 | MGI:4439731 | gene segment | T cell receptor beta joining 2-7 |
| 6 | gene | 41.54673 | 41.54835 | positive | MGI_C57BL6J_4835227 | Trbc2 | NCBI_Gene:100125263,ENSEMBL:ENSMUSG00000076498 | MGI:4835227 | gene segment | T cell receptor beta, constant 2 |
| 6 | gene | 41.55769 | 41.55845 | negative | MGI_C57BL6J_98588 | Trbv31 | NCBI_Gene:269846,ENSEMBL:ENSMUSG00000076499 | MGI:98588 | gene segment | T cell receptor beta, variable 31 |
| 6 | gene | 41.55849 | 41.60345 | positive | MGI_C57BL6J_5621467 | Gm38582 | NCBI_Gene:102641876 | MGI:5621467 | lncRNA gene | predicted gene, 38582 |
| 6 | gene | 41.56729 | 41.57071 | positive | MGI_C57BL6J_2444452 | B630006K09Rik | NA | NA | unclassified gene | RIKEN cDNA B630006K09 gene |
| 6 | gene | 41.60224 | 41.60231 | negative | MGI_C57BL6J_5530931 | Gm27549 | ENSEMBL:ENSMUSG00000098970 | MGI:5530931 | snRNA gene | predicted gene, 27549 |
| 6 | gene | 41.60504 | 41.62051 | positive | MGI_C57BL6J_1096338 | Ephb6 | NCBI_Gene:13848,ENSEMBL:ENSMUSG00000029869 | MGI:1096338 | protein coding gene | Eph receptor B6 |
| 6 | gene | 41.62062 | 41.63641 | negative | MGI_C57BL6J_1927259 | Trpv6 | NCBI_Gene:64177,ENSEMBL:ENSMUSG00000029868 | MGI:1927259 | protein coding gene | transient receptor potential cation channel, subfamily V, member 6 |
| 6 | gene | 41.62178 | 41.62406 | positive | MGI_C57BL6J_5621680 | Gm38795 | NCBI_Gene:105242636 | MGI:5621680 | lncRNA gene | predicted gene, 38795 |
| 6 | gene | 41.64173 | 41.65880 | positive | MGI_C57BL6J_5593801 | Gm34642 | NCBI_Gene:102637960 | MGI:5593801 | lncRNA gene | predicted gene, 34642 |
| 6 | gene | 41.65166 | 41.68145 | negative | MGI_C57BL6J_2429764 | Trpv5 | NCBI_Gene:194352,ENSEMBL:ENSMUSG00000036899 | MGI:2429764 | protein coding gene | transient receptor potential cation channel, subfamily V, member 5 |
| 6 | gene | 41.68435 | 41.68572 | positive | MGI_C57BL6J_1923856 | Llcfc1 | NCBI_Gene:76606,ENSEMBL:ENSMUSG00000029867 | MGI:1923856 | protein coding gene | LLLL and CFNLAS motif containing 1 |
| 6 | gene | 41.68633 | 41.70434 | negative | MGI_C57BL6J_1346053 | Kel | NCBI_Gene:23925,ENSEMBL:ENSMUSG00000029866 | MGI:1346053 | protein coding gene | Kell blood group |
| 6 | gene | 41.69264 | 41.69619 | negative | MGI_C57BL6J_5610755 | Gm37527 | ENSEMBL:ENSMUSG00000103788 | MGI:5610755 | unclassified gene | predicted gene, 37527 |
| 6 | gene | 41.70619 | 41.71014 | positive | MGI_C57BL6J_5621681 | Gm38796 | NCBI_Gene:105242637 | MGI:5621681 | lncRNA gene | predicted gene, 38796 |
| 6 | gene | 41.76877 | 41.77518 | negative | MGI_C57BL6J_3030293 | Olfr459 | NCBI_Gene:258569,ENSEMBL:ENSMUSG00000045479 | MGI:3030293 | protein coding gene | olfactory receptor 459 |
| 6 | gene | 41.84754 | 41.85207 | positive | MGI_C57BL6J_102696 | Pip | NCBI_Gene:18716,ENSEMBL:ENSMUSG00000058499 | MGI:102696 | protein coding gene | prolactin induced protein |
| 6 | gene | 41.85299 | 41.86441 | positive | MGI_C57BL6J_1934254 | Sval2 | NCBI_Gene:84543,ENSEMBL:ENSMUSG00000014104 | MGI:1934254 | protein coding gene | seminal vesicle antigen-like 2 |
| 6 | gene | 41.95163 | 41.95610 | positive | MGI_C57BL6J_1918828 | Sval1 | NCBI_Gene:71578,ENSEMBL:ENSMUSG00000029865 | MGI:1918828 | protein coding gene | seminal vesicle antigen-like 1 |
| 6 | gene | 41.96814 | 41.97309 | positive | MGI_C57BL6J_3047714 | Sval3 | NCBI_Gene:387564,ENSEMBL:ENSMUSG00000062833 | MGI:3047714 | protein coding gene | seminal vesicle antigen-like 3 |
| 6 | gene | 42.00990 | 42.03309 | positive | MGI_C57BL6J_2141598 | AU046084 | NA | NA | unclassified gene | expressed sequence AU046084 |
| 6 | gene | 42.03839 | 42.04285 | positive | MGI_C57BL6J_102785 | Sva | NCBI_Gene:20939,ENSEMBL:ENSMUSG00000023289 | MGI:102785 | protein coding gene | seminal vesicle antigen |
| 6 | gene | 42.10570 | 42.11452 | negative | MGI_C57BL6J_5593860 | Gm34701 | NCBI_Gene:102638036,ENSEMBL:ENSMUSG00000107543 | MGI:5593860 | lncRNA gene | predicted gene, 34701 |
| 6 | gene | 42.14094 | 42.14189 | positive | MGI_C57BL6J_2681308 | Tas2r139 | NCBI_Gene:353148,ENSEMBL:ENSMUSG00000047102 | MGI:2681308 | protein coding gene | taste receptor, type 2, member 139 |
| 6 | gene | 42.21533 | 42.21629 | positive | MGI_C57BL6J_2681312 | Tas2r144 | NCBI_Gene:387515,ENSEMBL:ENSMUSG00000051917 | MGI:2681312 | protein coding gene | taste receptor, type 2, member 144 |
| 6 | gene | 42.24594 | 42.25045 | positive | MGI_C57BL6J_1923513 | Gstk1 | NCBI_Gene:76263,ENSEMBL:ENSMUSG00000029864 | MGI:1923513 | protein coding gene | glutathione S-transferase kappa 1 |
| 6 | gene | 42.26192 | 42.26456 | positive | MGI_C57BL6J_1924444 | Tmem139 | NCBI_Gene:109218,ENSEMBL:ENSMUSG00000071506 | MGI:1924444 | protein coding gene | transmembrane protein 139 |
| 6 | gene | 42.26490 | 42.28251 | positive | MGI_C57BL6J_97295 | Casp2 | NCBI_Gene:12366,ENSEMBL:ENSMUSG00000029863 | MGI:97295 | protein coding gene | caspase 2 |
| 6 | gene | 42.27565 | 42.27573 | positive | MGI_C57BL6J_5454885 | Gm25108 | ENSEMBL:ENSMUSG00000084422 | MGI:5454885 | snoRNA gene | predicted gene, 25108 |
| 6 | gene | 42.28260 | 42.31576 | positive | MGI_C57BL6J_88417 | Clcn1 | NCBI_Gene:12723,ENSEMBL:ENSMUSG00000029862 | MGI:88417 | protein coding gene | chloride channel, voltage-sensitive 1 |
| 6 | gene | 42.31015 | 42.31303 | negative | MGI_C57BL6J_5621682 | Gm38797 | NCBI_Gene:105242638 | MGI:5621682 | lncRNA gene | predicted gene, 38797 |
| 6 | gene | 42.31531 | 42.32464 | negative | MGI_C57BL6J_1923406 | Fam131b | NCBI_Gene:76156,ENSEMBL:ENSMUSG00000029861 | MGI:1923406 | protein coding gene | family with sequence similarity 131, member B |
| 6 | gene | 42.32871 | 42.35086 | negative | MGI_C57BL6J_5690676 | Gm44284 | ENSEMBL:ENSMUSG00000108033 | MGI:5690676 | lncRNA gene | predicted gene, 44284 |
| 6 | gene | 42.33768 | 42.33778 | negative | MGI_C57BL6J_5454388 | Gm24611 | ENSEMBL:ENSMUSG00000065206 | MGI:5454388 | snRNA gene | predicted gene, 24611 |
| 6 | gene | 42.34962 | 42.36021 | positive | MGI_C57BL6J_103072 | Zyx | NCBI_Gene:22793,ENSEMBL:ENSMUSG00000029860 | MGI:103072 | protein coding gene | zyxin |
| 6 | gene | 42.35849 | 42.37331 | negative | MGI_C57BL6J_107381 | Epha1 | NCBI_Gene:13835,ENSEMBL:ENSMUSG00000029859 | MGI:107381 | protein coding gene | Eph receptor A1 |
| 6 | gene | 42.37067 | 42.38121 | positive | MGI_C57BL6J_1919381 | 2010310C07Rik | NCBI_Gene:72131,ENSEMBL:ENSMUSG00000101268 | MGI:1919381 | lncRNA gene | RIKEN cDNA 2010310C07 gene |
| 6 | gene | 42.40024 | 42.40112 | positive | MGI_C57BL6J_2681310 | Tas2r143 | NCBI_Gene:387514,ENSEMBL:ENSMUSG00000046652 | MGI:2681310 | protein coding gene | taste receptor, type 2, member 143 |
| 6 | gene | 42.40543 | 42.40653 | positive | MGI_C57BL6J_2681302 | Tas2r135 | NCBI_Gene:387512,ENSEMBL:ENSMUSG00000056203 | MGI:2681302 | protein coding gene | taste receptor, type 2, member 135 |
| 6 | gene | 42.43453 | 42.43546 | positive | MGI_C57BL6J_2681273 | Tas2r126 | NCBI_Gene:387353,ENSEMBL:ENSMUSG00000048284 | MGI:2681273 | protein coding gene | taste receptor, type 2, member 126 |
| 6 | pseudogene | 42.44379 | 42.44393 | negative | MGI_C57BL6J_5690862 | Gm44470 | ENSEMBL:ENSMUSG00000108150 | MGI:5690862 | pseudogene | predicted gene, 44470 |
| 6 | pseudogene | 42.44524 | 42.44591 | negative | MGI_C57BL6J_5593977 | Gm34818 | NCBI_Gene:102638200 | MGI:5593977 | pseudogene | predicted gene, 34818 |
| 6 | gene | 42.45998 | 42.46520 | negative | MGI_C57BL6J_3030292 | Olfr458 | NCBI_Gene:258436,ENSEMBL:ENSMUSG00000068574 | MGI:3030292 | protein coding gene | olfactory receptor 458 |
| 6 | gene | 42.46802 | 42.47633 | negative | MGI_C57BL6J_3030291 | Olfr457 | NCBI_Gene:258989,ENSEMBL:ENSMUSG00000091983 | MGI:3030291 | protein coding gene | olfactory receptor 457 |
| 6 | gene | 42.48616 | 42.48721 | negative | MGI_C57BL6J_3030290 | Olfr456 | NCBI_Gene:259144,ENSEMBL:ENSMUSG00000090631 | MGI:3030290 | protein coding gene | olfactory receptor 456 |
| 6 | gene | 42.50963 | 42.52147 | positive | MGI_C57BL6J_5593911 | Gm34752 | NCBI_Gene:102638113 | MGI:5593911 | lncRNA gene | predicted gene, 34752 |
| 6 | pseudogene | 42.51947 | 42.52033 | negative | MGI_C57BL6J_3779444 | Gm4877 | NCBI_Gene:232745,ENSEMBL:ENSMUSG00000078177 | MGI:3779444 | pseudogene | predicted gene 4877 |
| 6 | gene | 42.53566 | 42.54759 | negative | MGI_C57BL6J_3030289 | Olfr455 | NCBI_Gene:546896,ENSEMBL:ENSMUSG00000071494 | MGI:3030289 | protein coding gene | olfactory receptor 455 |
| 6 | gene | 42.57823 | 42.58178 | negative | MGI_C57BL6J_5621683 | Gm38798 | NCBI_Gene:105242639 | MGI:5621683 | lncRNA gene | predicted gene, 38798 |
| 6 | gene | 42.58487 | 42.59769 | negative | MGI_C57BL6J_3042585 | Tcaf3 | NCBI_Gene:403088,ENSEMBL:ENSMUSG00000018656 | MGI:3042585 | protein coding gene | TRPM8 channel-associated factor 3 |
| 6 | gene | 42.62302 | 42.64525 | negative | MGI_C57BL6J_2385258 | Tcaf2 | NCBI_Gene:232748,ENSEMBL:ENSMUSG00000029851 | MGI:2385258 | protein coding gene | TRPM8 channel-associated factor 2 |
| 6 | gene | 42.66800 | 42.71011 | negative | MGI_C57BL6J_1914665 | Tcaf1 | NCBI_Gene:77574,ENSEMBL:ENSMUSG00000036667 | MGI:1914665 | protein coding gene | TRPM8 channel-associated factor 1 |
| 6 | gene | 42.69392 | 42.69576 | negative | MGI_C57BL6J_5608918 | B930083L10Rik | NA | NA | unclassified gene | Riken cDNA B930083L10 gene |
| 6 | pseudogene | 42.72134 | 42.72226 | positive | MGI_C57BL6J_3030288 | Olfr454-ps1 | NCBI_Gene:257777,ENSEMBL:ENSMUSG00000107986 | MGI:3030288 | pseudogene | olfactory receptor 454, pseudogene 1 |
| 6 | pseudogene | 42.72559 | 42.72602 | positive | MGI_C57BL6J_5690422 | Gm44030 | ENSEMBL:ENSMUSG00000107416 | MGI:5690422 | pseudogene | predicted gene, 44030 |
| 6 | pseudogene | 42.73393 | 42.73410 | negative | MGI_C57BL6J_5690424 | Gm44032 | ENSEMBL:ENSMUSG00000107650 | MGI:5690424 | pseudogene | predicted gene, 44032 |
| 6 | gene | 42.73937 | 42.74696 | positive | MGI_C57BL6J_3030287 | Olfr453 | NCBI_Gene:258016,ENSEMBL:ENSMUSG00000095831 | MGI:3030287 | protein coding gene | olfactory receptor 453 |
| 6 | pseudogene | 42.74519 | 42.74525 | positive | MGI_C57BL6J_5690426 | Gm44034 | ENSEMBL:ENSMUSG00000107818 | MGI:5690426 | pseudogene | predicted gene, 44034 |
| 6 | gene | 42.75640 | 42.76323 | positive | MGI_C57BL6J_1313140 | Olfr38 | NCBI_Gene:258988,ENSEMBL:ENSMUSG00000095236 | MGI:1313140 | protein coding gene | olfactory receptor 38 |
| 6 | gene | 42.77557 | 42.77568 | negative | MGI_C57BL6J_5452140 | Gm22363 | ENSEMBL:ENSMUSG00000065034 | MGI:5452140 | snRNA gene | predicted gene, 22363 |
| 6 | gene | 42.78509 | 42.79162 | positive | MGI_C57BL6J_3030286 | Olfr452 | NCBI_Gene:258207,ENSEMBL:ENSMUSG00000094192 | MGI:3030286 | protein coding gene | olfactory receptor 452 |
| 6 | pseudogene | 42.80074 | 42.80168 | positive | MGI_C57BL6J_3030285 | Olfr451-ps1 | NCBI_Gene:258017,ENSEMBL:ENSMUSG00000108058 | MGI:3030285 | polymorphic pseudogene | olfactory receptor 451, pseudogene 1 |
| 6 | gene | 42.81194 | 42.81908 | positive | MGI_C57BL6J_3030284 | Olfr450 | NCBI_Gene:258437,ENSEMBL:ENSMUSG00000054431 | MGI:3030284 | protein coding gene | olfactory receptor 450 |
| 6 | gene | 42.83278 | 42.83288 | positive | MGI_C57BL6J_5453759 | Gm23982 | ENSEMBL:ENSMUSG00000087766 | MGI:5453759 | snRNA gene | predicted gene, 23982 |
| 6 | gene | 42.83438 | 42.83952 | positive | MGI_C57BL6J_3030283 | Olfr449 | NCBI_Gene:259067,ENSEMBL:ENSMUSG00000049168 | MGI:3030283 | protein coding gene | olfactory receptor 449 |
| 6 | gene | 42.89044 | 42.89908 | positive | MGI_C57BL6J_3030282 | Olfr448 | NCBI_Gene:258270,ENSEMBL:ENSMUSG00000043119 | MGI:3030282 | protein coding gene | olfactory receptor 448 |
| 6 | gene | 42.90888 | 42.91427 | positive | MGI_C57BL6J_3030281 | Olfr447 | NCBI_Gene:258990,ENSEMBL:ENSMUSG00000045708 | MGI:3030281 | protein coding gene | olfactory receptor 447 |
| 6 | gene | 42.92138 | 42.93114 | positive | MGI_C57BL6J_3030280 | Olfr446 | NCBI_Gene:258292,ENSEMBL:ENSMUSG00000073111 | MGI:3030280 | protein coding gene | olfactory receptor 446 |
| 6 | pseudogene | 42.93407 | 42.93494 | positive | MGI_C57BL6J_3030279 | Olrf445-ps1 | NCBI_Gene:257781,ENSEMBL:ENSMUSG00000108157 | MGI:3030279 | pseudogene | olfactory receptor 445, pseudogene 1 |
| 6 | gene | 42.94907 | 42.95676 | positive | MGI_C57BL6J_3030278 | Olfr444 | NCBI_Gene:258650,ENSEMBL:ENSMUSG00000073110 | MGI:3030278 | protein coding gene | olfactory receptor 444 |
| 6 | gene | 42.98069 | 42.98115 | positive | MGI_C57BL6J_4937106 | Gm17472 | ENSEMBL:ENSMUSG00000091008 | MGI:4937106 | gene segment | predicted gene, 17472 |
| 6 | gene | 43.01450 | 43.06690 | negative | MGI_C57BL6J_5594064 | Gm34905 | NCBI_Gene:102638311 | MGI:5594064 | lncRNA gene | predicted gene, 34905 |
| 6 | pseudogene | 43.06731 | 43.09558 | positive | MGI_C57BL6J_3030277 | Olfr443-ps1 | NCBI_Gene:258106,ENSEMBL:ENSMUSG00000091386 | MGI:3030277 | pseudogene | olfactory receptor 443, pseudogene 1 |
| 6 | gene | 43.06746 | 43.09046 | positive | MGI_C57BL6J_5621684 | Gm38799 | NCBI_Gene:105242640 | MGI:5621684 | lncRNA gene | predicted gene, 38799 |
| 6 | gene | 43.08152 | 43.08210 | negative | MGI_C57BL6J_5313039 | Gm20730 | ENSEMBL:ENSMUSG00000076500 | MGI:5313039 | gene segment | predicted gene, 20730 |
| 6 | pseudogene | 43.09911 | 43.09972 | positive | MGI_C57BL6J_3030276 | Olfr442-ps1 | NCBI_Gene:257780,ENSEMBL:ENSMUSG00000108193 | MGI:3030276 | pseudogene | olfactory receptor 442, pseudogene 1 |
| 6 | gene | 43.10637 | 43.11780 | positive | MGI_C57BL6J_3030275 | Olfr441 | NCBI_Gene:258649,ENSEMBL:ENSMUSG00000094669 | MGI:3030275 | protein coding gene | olfactory receptor 441 |
| 6 | pseudogene | 43.12235 | 43.12327 | positive | MGI_C57BL6J_3030274 | Olfr440-ps1 | NCBI_Gene:258107,ENSEMBL:ENSMUSG00000107889 | MGI:3030274 | pseudogene | olfactory receptor 440, pseudogene 1 |
| 6 | pseudogene | 43.13783 | 43.13844 | positive | MGI_C57BL6J_3030273 | Olfr439-ps1 | NCBI_Gene:257779,ENSEMBL:ENSMUSG00000108149 | MGI:3030273 | pseudogene | olfactory receptor 439, pseudogene 1 |
| 6 | gene | 43.14657 | 43.15717 | positive | MGI_C57BL6J_3030071 | Olfr237-ps1 | NCBI_Gene:258648,ENSEMBL:ENSMUSG00000094200 | MGI:3030071 | protein coding gene | olfactory receptor 237, pseudogene 1 |
| 6 | gene | 43.16245 | 43.17486 | negative | MGI_C57BL6J_1922770 | 1700024N05Rik | NCBI_Gene:75520 | MGI:1922770 | lncRNA gene | RIKEN cDNA 1700024N05 gene |
| 6 | gene | 43.16328 | 43.16986 | positive | MGI_C57BL6J_3030271 | Olfr437 | NCBI_Gene:258293,ENSEMBL:ENSMUSG00000071481 | MGI:3030271 | protein coding gene | olfactory receptor 437 |
| 6 | gene | 43.17214 | 43.17746 | positive | MGI_C57BL6J_104812 | Olfr13 | NCBI_Gene:18310,ENSEMBL:ENSMUSG00000043605 | MGI:104812 | protein coding gene | olfactory receptor 13 |
| 6 | pseudogene | 43.18665 | 43.18770 | positive | MGI_C57BL6J_3030270 | Olfr436-ps1 | NCBI_Gene:257778,ENSEMBL:ENSMUSG00000107469 | MGI:3030270 | pseudogene | olfactory receptor 436, pseudogene 1 |
| 6 | gene | 43.19715 | 43.20363 | positive | MGI_C57BL6J_3030269 | Olfr435 | NCBI_Gene:258647,ENSEMBL:ENSMUSG00000048693 | MGI:3030269 | protein coding gene | olfactory receptor 435 |
| 6 | pseudogene | 43.21033 | 43.21075 | positive | MGI_C57BL6J_5433991 | Gm21827 | ENSEMBL:ENSMUSG00000096158 | MGI:5433991 | pseudogene | predicted gene, 21827 |
| 6 | gene | 43.21302 | 43.21811 | positive | MGI_C57BL6J_3030268 | Olfr434 | NCBI_Gene:258366,ENSEMBL:ENSMUSG00000059411 | MGI:3030268 | protein coding gene | olfactory receptor 434 |
| 6 | gene | 43.23153 | 43.23696 | positive | MGI_C57BL6J_1333821 | Olfr47 | NCBI_Gene:18346,ENSEMBL:ENSMUSG00000061210 | MGI:1333821 | protein coding gene | olfactory receptor 47 |
| 6 | gene | 43.26454 | 43.26586 | negative | MGI_C57BL6J_5753307 | Gm44731 | ENSEMBL:ENSMUSG00000108190 | MGI:5753307 | lncRNA gene | predicted gene 44731 |
| 6 | gene | 43.26558 | 43.28932 | positive | MGI_C57BL6J_1858952 | Arhgef5 | NCBI_Gene:54324,ENSEMBL:ENSMUSG00000033542 | MGI:1858952 | protein coding gene | Rho guanine nucleotide exchange factor (GEF) 5 |
| 6 | gene | 43.27775 | 43.27838 | positive | MGI_C57BL6J_5690645 | Gm44253 | ENSEMBL:ENSMUSG00000107861 | MGI:5690645 | unclassified gene | predicted gene, 44253 |
| 6 | gene | 43.30367 | 43.31007 | negative | MGI_C57BL6J_108011 | Nobox | NCBI_Gene:18291,ENSEMBL:ENSMUSG00000029736 | MGI:108011 | protein coding gene | NOBOX oogenesis homeobox |
| 6 | gene | 43.34500 | 43.66649 | negative | MGI_C57BL6J_1352500 | Tpk1 | NCBI_Gene:29807,ENSEMBL:ENSMUSG00000029735 | MGI:1352500 | protein coding gene | thiamine pyrophosphokinase |
| 6 | pseudogene | 43.37380 | 43.37555 | positive | MGI_C57BL6J_3782999 | Gm15550 | NCBI_Gene:665747,ENSEMBL:ENSMUSG00000084325 | MGI:3782999 | pseudogene | predicted gene 15550 |
| 6 | gene | 43.37609 | 43.39253 | positive | MGI_C57BL6J_5594272 | Gm35113 | NCBI_Gene:102638584 | MGI:5594272 | lncRNA gene | predicted gene, 35113 |
| 6 | gene | 43.41594 | 43.41867 | positive | MGI_C57BL6J_5621685 | Gm38800 | NCBI_Gene:105242642 | MGI:5621685 | lncRNA gene | predicted gene, 38800 |
| 6 | gene | 43.44243 | 43.47432 | positive | MGI_C57BL6J_2442776 | 9430018G01Rik | NCBI_Gene:319812,ENSEMBL:ENSMUSG00000107709 | MGI:2442776 | lncRNA gene | RIKEN cDNA 9430018G01 gene |
| 6 | gene | 43.67881 | 43.67972 | negative | MGI_C57BL6J_5826621 | Gm46984 | NCBI_Gene:108169189 | MGI:5826621 | lncRNA gene | predicted gene, 46984 |
| 6 | pseudogene | 43.76656 | 43.76732 | negative | MGI_C57BL6J_3648301 | Gm5990 | NCBI_Gene:546897,ENSEMBL:ENSMUSG00000107465 | MGI:3648301 | pseudogene | predicted gene 5990 |
| 6 | gene | 43.83926 | 43.85238 | positive | MGI_C57BL6J_5594322 | Gm35163 | NCBI_Gene:102638650 | MGI:5594322 | lncRNA gene | predicted gene, 35163 |
| 6 | pseudogene | 43.85608 | 43.85692 | negative | MGI_C57BL6J_3643099 | Gm7783 | NCBI_Gene:665779,ENSEMBL:ENSMUSG00000107920 | MGI:3643099 | pseudogene | predicted gene 7783 |
| 6 | gene | 43.88719 | 43.88732 | positive | MGI_C57BL6J_5452716 | Gm22939 | ENSEMBL:ENSMUSG00000077146 | MGI:5452716 | snoRNA gene | predicted gene, 22939 |
| 6 | gene | 44.08049 | 44.08338 | negative | MGI_C57BL6J_5594375 | Gm35216 | NCBI_Gene:102638722,ENSEMBL:ENSMUSG00000107598 | MGI:5594375 | lncRNA gene | predicted gene, 35216 |
| 6 | pseudogene | 44.66729 | 44.66743 | negative | MGI_C57BL6J_5690603 | Gm44211 | ENSEMBL:ENSMUSG00000107953 | MGI:5690603 | pseudogene | predicted gene, 44211 |
| 6 | pseudogene | 44.82157 | 44.82247 | positive | MGI_C57BL6J_5690604 | Gm44212 | ENSEMBL:ENSMUSG00000107845 | MGI:5690604 | pseudogene | predicted gene, 44212 |
| 6 | pseudogene | 44.96592 | 44.96663 | positive | MGI_C57BL6J_5690605 | Gm44213 | ENSEMBL:ENSMUSG00000107904 | MGI:5690605 | pseudogene | predicted gene, 44213 |
| 6 | gene | 45.05936 | 47.30421 | positive | MGI_C57BL6J_1914047 | Cntnap2 | NCBI_Gene:66797,ENSEMBL:ENSMUSG00000039419 | MGI:1914047 | protein coding gene | contactin associated protein-like 2 |
| 6 | gene | 45.07357 | 45.07479 | positive | MGI_C57BL6J_1925234 | B230112I24Rik | ENSEMBL:ENSMUSG00000107729 | MGI:1925234 | unclassified gene | RIKEN cDNA B230112I24 gene |
| 6 | gene | 45.08075 | 45.08451 | positive | MGI_C57BL6J_5690271 | Gm43879 | ENSEMBL:ENSMUSG00000107858 | MGI:5690271 | unclassified gene | predicted gene, 43879 |
| 6 | gene | 45.13527 | 45.13748 | positive | MGI_C57BL6J_5690264 | Gm43872 | ENSEMBL:ENSMUSG00000108070 | MGI:5690264 | unclassified gene | predicted gene, 43872 |
| 6 | gene | 45.19488 | 45.19742 | positive | MGI_C57BL6J_5690277 | Gm43885 | ENSEMBL:ENSMUSG00000107643 | MGI:5690277 | unclassified gene | predicted gene, 43885 |
| 6 | gene | 45.20381 | 45.20749 | positive | MGI_C57BL6J_5690281 | Gm43889 | ENSEMBL:ENSMUSG00000108146 | MGI:5690281 | unclassified gene | predicted gene, 43889 |
| 6 | gene | 45.22979 | 45.23377 | positive | MGI_C57BL6J_2441759 | 9630039A02Rik | NA | NA | unclassified gene | RIKEN cDNA 9630039A02 gene |
| 6 | gene | 45.24564 | 45.24843 | positive | MGI_C57BL6J_5690266 | Gm43874 | ENSEMBL:ENSMUSG00000107779 | MGI:5690266 | unclassified gene | predicted gene, 43874 |
| 6 | gene | 45.30110 | 45.30251 | positive | MGI_C57BL6J_5690261 | Gm43869 | ENSEMBL:ENSMUSG00000108008 | MGI:5690261 | unclassified gene | predicted gene, 43869 |
| 6 | gene | 45.30612 | 45.30739 | positive | MGI_C57BL6J_1918013 | 6330419E04Rik | ENSEMBL:ENSMUSG00000107414 | MGI:1918013 | unclassified gene | RIKEN cDNA 6330419E04 gene |
| 6 | gene | 45.32640 | 45.32867 | positive | MGI_C57BL6J_5690274 | Gm43882 | ENSEMBL:ENSMUSG00000107433 | MGI:5690274 | unclassified gene | predicted gene, 43882 |
| 6 | gene | 45.32924 | 45.32980 | positive | MGI_C57BL6J_5690276 | Gm43884 | ENSEMBL:ENSMUSG00000108123 | MGI:5690276 | unclassified gene | predicted gene, 43884 |
| 6 | gene | 45.41324 | 45.41540 | positive | MGI_C57BL6J_5690272 | Gm43880 | ENSEMBL:ENSMUSG00000107886 | MGI:5690272 | unclassified gene | predicted gene, 43880 |
| 6 | gene | 45.52908 | 45.53231 | positive | MGI_C57BL6J_5690268 | Gm43876 | ENSEMBL:ENSMUSG00000108055 | MGI:5690268 | unclassified gene | predicted gene, 43876 |
| 6 | pseudogene | 45.89447 | 45.89503 | negative | MGI_C57BL6J_5010668 | Gm18483 | NCBI_Gene:100417253 | MGI:5010668 | pseudogene | predicted gene, 18483 |
| 6 | gene | 45.90194 | 45.90432 | positive | MGI_C57BL6J_5690278 | Gm43886 | ENSEMBL:ENSMUSG00000107515 | MGI:5690278 | unclassified gene | predicted gene, 43886 |
| 6 | gene | 46.03778 | 46.03808 | positive | MGI_C57BL6J_5452616 | Gm22839 | ENSEMBL:ENSMUSG00000089061 | MGI:5452616 | unclassified non-coding RNA gene | predicted gene, 22839 |
| 6 | gene | 46.04195 | 46.04391 | positive | MGI_C57BL6J_5690263 | Gm43871 | ENSEMBL:ENSMUSG00000107913 | MGI:5690263 | unclassified gene | predicted gene, 43871 |
| 6 | gene | 46.04514 | 46.04908 | positive | MGI_C57BL6J_5690265 | Gm43873 | ENSEMBL:ENSMUSG00000107606 | MGI:5690265 | unclassified gene | predicted gene, 43873 |
| 6 | gene | 46.04916 | 46.05139 | positive | MGI_C57BL6J_5690269 | Gm43877 | ENSEMBL:ENSMUSG00000107571 | MGI:5690269 | unclassified gene | predicted gene, 43877 |
| 6 | gene | 46.07241 | 46.07500 | positive | MGI_C57BL6J_5690280 | Gm43888 | ENSEMBL:ENSMUSG00000107782 | MGI:5690280 | unclassified gene | predicted gene, 43888 |
| 6 | gene | 46.07618 | 46.07936 | positive | MGI_C57BL6J_5690279 | Gm43887 | ENSEMBL:ENSMUSG00000108189 | MGI:5690279 | unclassified gene | predicted gene, 43887 |
| 6 | gene | 46.13410 | 46.13847 | positive | MGI_C57BL6J_5690262 | Gm43870 | ENSEMBL:ENSMUSG00000107712 | MGI:5690262 | unclassified gene | predicted gene, 43870 |
| 6 | gene | 46.41376 | 46.41629 | positive | MGI_C57BL6J_5690273 | Gm43881 | ENSEMBL:ENSMUSG00000107442 | MGI:5690273 | unclassified gene | predicted gene, 43881 |
| 6 | gene | 46.48171 | 46.48184 | negative | MGI_C57BL6J_5454938 | Gm25161 | ENSEMBL:ENSMUSG00000089538 | MGI:5454938 | snoRNA gene | predicted gene, 25161 |
| 6 | gene | 46.48664 | 46.48912 | positive | MGI_C57BL6J_5690267 | Gm43875 | ENSEMBL:ENSMUSG00000107454 | MGI:5690267 | unclassified gene | predicted gene, 43875 |
| 6 | pseudogene | 46.52935 | 46.53079 | negative | MGI_C57BL6J_96525 | Rbpj-ps3 | NCBI_Gene:19667,ENSEMBL:ENSMUSG00000079575 | MGI:96525 | pseudogene | recombination signal binding protein for immunoglobulin kappa J region, pseudogene 3 |
| 6 | pseudogene | 46.63725 | 46.63830 | negative | MGI_C57BL6J_5010669 | Gm18484 | NCBI_Gene:100417254,ENSEMBL:ENSMUSG00000107725 | MGI:5010669 | pseudogene | predicted gene, 18484 |
| 6 | pseudogene | 46.77073 | 46.77965 | negative | MGI_C57BL6J_5595090 | Gm35931 | NCBI_Gene:105242644,ENSEMBL:ENSMUSG00000107532 | MGI:5595090 | pseudogene | predicted gene, 35931 |
| 6 | gene | 46.78595 | 46.78970 | positive | MGI_C57BL6J_5690275 | Gm43883 | ENSEMBL:ENSMUSG00000108217 | MGI:5690275 | unclassified gene | predicted gene, 43883 |
| 6 | gene | 46.95238 | 46.95823 | negative | MGI_C57BL6J_5621686 | Gm38801 | NCBI_Gene:105242643 | MGI:5621686 | lncRNA gene | predicted gene, 38801 |
| 6 | gene | 47.00655 | 47.04080 | negative | MGI_C57BL6J_5595034 | Gm35875 | NCBI_Gene:102639602 | MGI:5595034 | lncRNA gene | predicted gene, 35875 |
| 6 | pseudogene | 47.04516 | 47.04629 | positive | MGI_C57BL6J_3647816 | Gm6352 | NCBI_Gene:622745,ENSEMBL:ENSMUSG00000107952 | MGI:3647816 | pseudogene | predicted gene 6352 |
| 6 | gene | 47.12001 | 47.12496 | negative | MGI_C57BL6J_5594957 | Gm35798 | NCBI_Gene:102639497 | MGI:5594957 | lncRNA gene | predicted gene, 35798 |
| 6 | gene | 47.13921 | 47.16423 | negative | MGI_C57BL6J_1915519 | 4930432J01Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 4930432J01 gene |
| 6 | gene | 47.19138 | 47.25916 | negative | MGI_C57BL6J_5594439 | Gm35280 | NCBI_Gene:102638803 | MGI:5594439 | lncRNA gene | predicted gene, 35280 |
| 6 | gene | 47.22750 | 47.23058 | positive | MGI_C57BL6J_5594724 | Gm35565 | NCBI_Gene:102639203 | MGI:5594724 | lncRNA gene | predicted gene, 35565 |
| 6 | pseudogene | 47.43212 | 47.43309 | negative | MGI_C57BL6J_5010769 | Gm18584 | NCBI_Gene:100417389,ENSEMBL:ENSMUSG00000107853 | MGI:5010769 | pseudogene | predicted gene, 18584 |
| 6 | gene | 47.45340 | 47.52614 | positive | MGI_C57BL6J_1349658 | Cul1 | NCBI_Gene:26965,ENSEMBL:ENSMUSG00000029686 | MGI:1349658 | protein coding gene | cullin 1 |
| 6 | gene | 47.45378 | 47.45432 | negative | MGI_C57BL6J_2445106 | A930035D04Rik | ENSEMBL:ENSMUSG00000052241 | MGI:2445106 | lncRNA gene | RIKEN cDNA A930035D04 gene |
| 6 | gene | 47.53014 | 47.59534 | negative | MGI_C57BL6J_107940 | Ezh2 | NCBI_Gene:14056,ENSEMBL:ENSMUSG00000029687 | MGI:107940 | protein coding gene | enhancer of zeste 2 polycomb repressive complex 2 subunit |
| 6 | gene | 47.54667 | 47.55170 | positive | MGI_C57BL6J_5477119 | Gm26625 | ENSEMBL:ENSMUSG00000097138 | MGI:5477119 | lncRNA gene | predicted gene, 26625 |
| 6 | pseudogene | 47.58522 | 47.58541 | positive | MGI_C57BL6J_5690674 | Gm44282 | ENSEMBL:ENSMUSG00000107628 | MGI:5690674 | pseudogene | predicted gene, 44282 |
| 6 | gene | 47.59519 | 47.59753 | positive | MGI_C57BL6J_2442048 | F830010H11Rik | NA | NA | unclassified gene | RIKEN cDNA F830010H11 gene |
| 6 | gene | 47.63530 | 47.63688 | positive | MGI_C57BL6J_5690533 | Gm44141 | ENSEMBL:ENSMUSG00000108180 | MGI:5690533 | unclassified gene | predicted gene, 44141 |
| 6 | gene | 47.63714 | 47.63722 | positive | MGI_C57BL6J_5455060 | Gm25283 | ENSEMBL:ENSMUSG00000087897 | MGI:5455060 | unclassified non-coding RNA gene | predicted gene, 25283 |
| 6 | gene | 47.64461 | 47.65185 | negative | MGI_C57BL6J_5623256 | Gm40371 | NCBI_Gene:105244835 | MGI:5623256 | lncRNA gene | predicted gene, 40371 |
| 6 | gene | 47.65213 | 47.65612 | positive | MGI_C57BL6J_5623254 | Gm40369 | NCBI_Gene:105244833 | MGI:5623254 | protein coding gene | predicted gene, 40369 |
| 6 | gene | 47.65492 | 47.65509 | negative | MGI_C57BL6J_97946 | Rn4.5s | NCBI_Gene:19799 | MGI:97946 | rRNA gene | 4.5S RNA |
| 6 | gene | 47.65637 | 47.66043 | positive | MGI_C57BL6J_5623253 | Gm40368 | NCBI_Gene:105244832 | MGI:5623253 | protein coding gene | predicted gene, 40368 |
| 6 | gene | 47.66068 | 47.66479 | positive | MGI_C57BL6J_5623252 | Gm40367 | NCBI_Gene:105244831 | MGI:5623252 | protein coding gene | predicted gene, 40367 |
| 6 | gene | 47.66504 | 47.66912 | positive | MGI_C57BL6J_5595663 | Gm36504 | NCBI_Gene:102640444 | MGI:5595663 | protein coding gene | predicted gene, 36504 |
| 6 | gene | 47.66937 | 47.67345 | positive | MGI_C57BL6J_5623250 | Gm40365 | NCBI_Gene:105244829 | MGI:5623250 | protein coding gene | predicted gene, 40365 |
| 6 | gene | 47.67369 | 47.67777 | positive | MGI_C57BL6J_5595486 | Gm36327 | NCBI_Gene:102640203 | MGI:5595486 | protein coding gene | predicted gene, 36327 |
| 6 | gene | 47.67802 | 47.68209 | positive | MGI_C57BL6J_5623249 | Gm40364 | NCBI_Gene:105244828 | MGI:5623249 | protein coding gene | predicted gene, 40364 |
| 6 | gene | 47.68234 | 47.73833 | positive | MGI_C57BL6J_5623248 | Gm40363 | NCBI_Gene:105244826 | MGI:5623248 | protein coding gene | predicted gene, 40363 |
| 6 | gene | 47.76221 | 47.77309 | positive | MGI_C57BL6J_5595832 | Gm36673 | NCBI_Gene:102640659 | MGI:5595832 | lncRNA gene | predicted gene, 36673 |
| 6 | gene | 47.78162 | 47.78173 | positive | MGI_C57BL6J_107304 | Rny3 | NCBI_Gene:19874,ENSEMBL:ENSMUSG00000064945 | MGI:107304 | scRNA gene | RNA, Y3 small cytoplasmic (associated with Ro protein) |
| 6 | pseudogene | 47.78596 | 47.78637 | negative | MGI_C57BL6J_3648986 | Rpl31-ps7 | NCBI_Gene:384379,ENSEMBL:ENSMUSG00000097255 | MGI:3648986 | pseudogene | ribosomal protein L31, pseudogene 7 |
| 6 | gene | 47.78807 | 47.78818 | negative | MGI_C57BL6J_97995 | Rny1 | NCBI_Gene:19872,ENSEMBL:ENSMUSG00000065701 | MGI:97995 | scRNA gene | RNA, Y1 small cytoplasmic, Ro-associated |
| 6 | gene | 47.79609 | 47.81351 | negative | MGI_C57BL6J_104864 | Pdia4 | NCBI_Gene:12304,ENSEMBL:ENSMUSG00000025823 | MGI:104864 | protein coding gene | protein disulfide isomerase associated 4 |
| 6 | gene | 47.80358 | 47.80365 | negative | MGI_C57BL6J_3629672 | Mir704 | miRBase:MI0004688,NCBI_Gene:735289,ENSEMBL:ENSMUSG00000076060 | MGI:3629672 | miRNA gene | microRNA 704 |
| 6 | pseudogene | 47.81526 | 47.81569 | negative | MGI_C57BL6J_3781346 | Rpl35a-ps7 | NCBI_Gene:100041154,ENSEMBL:ENSMUSG00000094557 | MGI:3781346 | pseudogene | ribosomal protein L35A, pseudogene 7 |
| 6 | gene | 47.81926 | 47.83556 | negative | MGI_C57BL6J_3026883 | Zfp786 | NCBI_Gene:330301,ENSEMBL:ENSMUSG00000051499 | MGI:3026883 | protein coding gene | zinc finger protein 786 |
| 6 | gene | 47.83566 | 47.87354 | positive | MGI_C57BL6J_1917856 | Zfp398 | NCBI_Gene:272347,ENSEMBL:ENSMUSG00000062519 | MGI:1917856 | protein coding gene | zinc finger protein 398 |
| 6 | pseudogene | 47.84774 | 47.84814 | positive | MGI_C57BL6J_5690433 | Gm44041 | ENSEMBL:ENSMUSG00000107604 | MGI:5690433 | pseudogene | predicted gene, 44041 |
| 6 | gene | 47.87720 | 47.90848 | positive | MGI_C57BL6J_2141413 | Zfp282 | NCBI_Gene:101095,ENSEMBL:ENSMUSG00000025821 | MGI:2141413 | protein coding gene | zinc finger protein 282 |
| 6 | gene | 47.92048 | 47.93264 | positive | MGI_C57BL6J_2682609 | Zfp212 | NCBI_Gene:232784,ENSEMBL:ENSMUSG00000052763 | MGI:2682609 | protein coding gene | Zinc finger protein 212 |
| 6 | pseudogene | 47.94317 | 47.95591 | positive | MGI_C57BL6J_3040704 | Zfp783 | NCBI_Gene:232785,ENSEMBL:ENSMUSG00000072653 | MGI:3040704 | pseudogene | zinc finger protein 783 |
| 6 | gene | 47.95339 | 47.96530 | positive | MGI_C57BL6J_2141515 | Zfp956 | NCBI_Gene:101197,ENSEMBL:ENSMUSG00000045466 | MGI:2141515 | protein coding gene | zinc finger protein 956 |
| 6 | pseudogene | 47.97699 | 47.97790 | negative | MGI_C57BL6J_3646734 | Gm6443 | NCBI_Gene:623601,ENSEMBL:ENSMUSG00000107824 | MGI:3646734 | pseudogene | predicted gene 6443 |
| 6 | gene | 47.97900 | 47.97907 | positive | MGI_C57BL6J_4413824 | n-TCgca57 | NCBI_Gene:102467643 | MGI:4413824 | tRNA gene | nuclear encoded tRNA cysteine 57 (anticodon GCA) |
| 6 | gene | 48.01766 | 48.05000 | negative | MGI_C57BL6J_1919556 | Zfp777 | NCBI_Gene:72306,ENSEMBL:ENSMUSG00000071477 | MGI:1919556 | protein coding gene | zinc finger protein 777 |
| 6 | gene | 48.04555 | 48.04595 | negative | MGI_C57BL6J_5804853 | Gm45738 | ENSEMBL:ENSMUSG00000108101 | MGI:5804853 | lncRNA gene | predicted gene 45738 |
| 6 | gene | 48.04870 | 48.04879 | positive | MGI_C57BL6J_5454340 | Gm24563 | ENSEMBL:ENSMUSG00000092928 | MGI:5454340 | miRNA gene | predicted gene, 24563 |
| 6 | gene | 48.06239 | 48.08669 | negative | MGI_C57BL6J_1916478 | Zfp746 | NCBI_Gene:69228,ENSEMBL:ENSMUSG00000057691 | MGI:1916478 | protein coding gene | zinc finger protein 746 |
| 6 | gene | 48.06595 | 48.06605 | positive | MGI_C57BL6J_3629706 | Gt(pU21)138Imeg | NA | NA | unclassified gene | gene trap 138%2c Institute of Molecular Embryology and Genetics |
| 6 | gene | 48.12866 | 48.39541 | negative | MGI_C57BL6J_4439554 | Gm16630 | NCBI_Gene:100503622,ENSEMBL:ENSMUSG00000097434 | MGI:4439554 | lncRNA gene | predicted gene, 16630 |
| 6 | gene | 48.13047 | 48.14608 | positive | MGI_C57BL6J_5589594 | Gm30435 | NCBI_Gene:102632336 | MGI:5589594 | lncRNA gene | predicted gene, 30435 |
| 6 | gene | 48.13394 | 48.13401 | positive | MGI_C57BL6J_4413804 | n-TCgca37 | NCBI_Gene:102467630 | MGI:4413804 | tRNA gene | nuclear encoded tRNA cysteine 37 (anticodon GCA) |
| 6 | gene | 48.13951 | 48.13958 | negative | MGI_C57BL6J_4413777 | n-TCgca10 | NCBI_Gene:102467491 | MGI:4413777 | tRNA gene | nuclear encoded tRNA cysteine 10 (anticodon GCA) |
| 6 | gene | 48.14048 | 48.14055 | negative | MGI_C57BL6J_4413771 | n-TCgca4 | NCBI_Gene:102467488 | MGI:4413771 | tRNA gene | nuclear encoded tRNA cysteine 4 (anticodon GCA) |
| 6 | gene | 48.14336 | 48.14343 | positive | MGI_C57BL6J_4413780 | n-TCgca13 | NCBI_Gene:102467492 | MGI:4413780 | tRNA gene | nuclear encoded tRNA cysteine 13 (anticodon GCA) |
| 6 | gene | 48.14493 | 48.14500 | negative | MGI_C57BL6J_4413803 | n-TCgca36 | NCBI_Gene:102467282 | MGI:4413803 | tRNA gene | nuclear encoded tRNA cysteine 36 (anticodon GCA) |
| 6 | gene | 48.15471 | 48.15478 | positive | MGI_C57BL6J_4413805 | n-TCgca38 | NCBI_Gene:102467502 | MGI:4413805 | tRNA gene | nuclear encoded tRNA cysteine 38 (anticodon GCA) |
| 6 | gene | 48.15628 | 48.15635 | negative | MGI_C57BL6J_4413802 | n-TCgca35 | NCBI_Gene:102467501 | MGI:4413802 | tRNA gene | nuclear encoded tRNA cysteine 35 (anticodon GCA) |
| 6 | gene | 48.16018 | 48.16025 | positive | MGI_C57BL6J_4413806 | n-TCgca39 | NCBI_Gene:102467642 | MGI:4413806 | tRNA gene | nuclear encoded tRNA cysteine 39 (anticodon GCA) |
| 6 | gene | 48.16410 | 48.16417 | positive | MGI_C57BL6J_4413807 | n-TCgca40 | NCBI_Gene:109705 | MGI:4413807 | tRNA gene | nuclear encoded tRNA cysteine 40 (anticodon GCA) |
| 6 | gene | 48.18632 | 48.18639 | positive | MGI_C57BL6J_4413775 | n-TCgca8 | NCBI_Gene:102467490 | MGI:4413775 | tRNA gene | nuclear encoded tRNA cysteine 8 (anticodon GCA) |
| 6 | gene | 48.19703 | 48.19710 | positive | MGI_C57BL6J_4413808 | n-TCgca41 | NCBI_Gene:102467283 | MGI:4413808 | tRNA gene | nuclear encoded tRNA cysteine 41 (anticodon GCA) |
| 6 | gene | 48.20507 | 48.20514 | positive | MGI_C57BL6J_4413768 | n-TCgca1 | NCBI_Gene:102467487 | MGI:4413768 | tRNA gene | nuclear encoded tRNA cysteine 1 (anticodon GCA) |
| 6 | gene | 48.20745 | 48.20752 | negative | MGI_C57BL6J_4413801 | n-TCgca34 | NCBI_Gene:102467281 | MGI:4413801 | tRNA gene | nuclear encoded tRNA cysteine 34 (anticodon GCA) |
| 6 | gene | 48.20984 | 48.20991 | negative | MGI_C57BL6J_4413816 | n-TCgca49 | NCBI_Gene:102467286 | MGI:4413816 | tRNA gene | nuclear encoded tRNA cysteine 49 (anticodon GCA) |
| 6 | gene | 48.22218 | 48.24739 | positive | MGI_C57BL6J_5621687 | Gm38802 | NCBI_Gene:105242646 | MGI:5621687 | lncRNA gene | predicted gene, 38802 |
| 6 | gene | 48.22309 | 48.22317 | negative | MGI_C57BL6J_4413800 | n-TCgca33 | NCBI_Gene:102467500 | MGI:4413800 | tRNA gene | nuclear encoded tRNA cysteine 33 (anticodon GCA) |
| 6 | gene | 48.22433 | 48.22440 | negative | MGI_C57BL6J_4413815 | n-TCgca48 | NCBI_Gene:102467426 | MGI:4413815 | tRNA gene | nuclear encoded tRNA cysteine 48 (anticodon GCA) |
| 6 | gene | 48.24432 | 48.24439 | negative | MGI_C57BL6J_4413799 | n-TCgca32 | NCBI_Gene:102467280 | MGI:4413799 | tRNA gene | nuclear encoded tRNA cysteine 32 (anticodon GCA) |
| 6 | gene | 48.25210 | 48.25217 | negative | MGI_C57BL6J_4413798 | n-TCgca31 | NCBI_Gene:102467499 | MGI:4413798 | tRNA gene | nuclear encoded tRNA cysteine 31 (anticodon GCA) |
| 6 | gene | 48.25308 | 48.25315 | negative | MGI_C57BL6J_4413782 | n-TCgca15 | NCBI_Gene:102467412 | MGI:4413782 | tRNA gene | nuclear encoded tRNA cysteine 15 (anticodon GCA) |
| 6 | gene | 48.26172 | 48.26179 | negative | MGI_C57BL6J_4413797 | n-TCgca30 | NCBI_Gene:102467425 | MGI:4413797 | tRNA gene | nuclear encoded tRNA cysteine 30 (anticodon GCA) |
| 6 | gene | 48.26846 | 48.26853 | negative | MGI_C57BL6J_4413796 | n-TCgca29 | NCBI_Gene:102467279 | MGI:4413796 | tRNA gene | nuclear encoded tRNA cysteine 29 (anticodon GCA) |
| 6 | gene | 48.27193 | 48.27200 | positive | MGI_C57BL6J_4413809 | n-TCgca42 | NCBI_Gene:102467503 | MGI:4413809 | tRNA gene | nuclear encoded tRNA cysteine 42 (anticodon GCA) |
| 6 | gene | 48.27368 | 48.27375 | positive | MGI_C57BL6J_4413810 | n-TCgca43 | NCBI_Gene:102467504 | MGI:4413810 | tRNA gene | nuclear encoded tRNA cysteine 43 (anticodon GCA) |
| 6 | gene | 48.27524 | 48.33816 | positive | MGI_C57BL6J_5589521 | Gm30362 | NCBI_Gene:102632227 | MGI:5589521 | lncRNA gene | predicted gene, 30362 |
| 6 | gene | 48.27879 | 48.27886 | negative | MGI_C57BL6J_4413770 | n-TCgca3 | NCBI_Gene:102467640 | MGI:4413770 | tRNA gene | nuclear encoded tRNA cysteine 3 (anticodon GCA) |
| 6 | gene | 48.27977 | 48.27984 | negative | MGI_C57BL6J_4413795 | n-TCgca28 | NCBI_Gene:102467498 | MGI:4413795 | tRNA gene | nuclear encoded tRNA cysteine 28 (anticodon GCA) |
| 6 | gene | 48.28888 | 48.28895 | negative | MGI_C57BL6J_4413779 | n-TCgca12 | NCBI_Gene:102467424 | MGI:4413779 | tRNA gene | nuclear encoded tRNA cysteine 12 (anticodon GCA) |
| 6 | gene | 48.28923 | 48.28943 | positive | MGI_C57BL6J_5451899 | Gm22122 | ENSEMBL:ENSMUSG00000092888 | MGI:5451899 | snRNA gene | predicted gene, 22122 |
| 6 | gene | 48.29301 | 48.29308 | negative | MGI_C57BL6J_4413817 | n-TCgca50 | NCBI_Gene:102467287 | MGI:4413817 | tRNA gene | nuclear encoded tRNA cysteine 50 (anticodon GCA) |
| 6 | gene | 48.29743 | 48.29750 | positive | MGI_C57BL6J_4413811 | n-Tcgca44 | NCBI_Gene:102467284 | MGI:4413811 | tRNA gene | nuclear encoded tRNA cysteine 44 (anticodon GCA) |
| 6 | gene | 48.29895 | 48.29902 | negative | MGI_C57BL6J_4413794 | n-TCgca27 | NCBI_Gene:102467278 | MGI:4413794 | tRNA gene | nuclear encoded tRNA cysteine 27 (anticodon GCA) |
| 6 | gene | 48.30246 | 48.30253 | positive | MGI_C57BL6J_4413773 | n-TCgca6 | NCBI_Gene:102467489 | MGI:4413773 | tRNA gene | nuclear encoded tRNA cysteine 6 (anticodon GCA) |
| 6 | gene | 48.30347 | 48.30354 | positive | MGI_C57BL6J_4413784 | n-TCgca17 | NCBI_Gene:102467273 | MGI:4413784 | tRNA gene | nuclear encoded tRNA cysteine 17 (anticodon GCA) |
| 6 | gene | 48.30546 | 48.30553 | negative | MGI_C57BL6J_4413774 | n-TCgca7 | NCBI_Gene:102467269 | MGI:4413774 | tRNA gene | nuclear encoded tRNA cysteine 7 (anticodon GCA) |
| 6 | gene | 48.30873 | 48.30880 | negative | MGI_C57BL6J_4413783 | n-TCgca16 | NCBI_Gene:102467493 | MGI:4413783 | tRNA gene | nuclear encoded tRNA cysteine 16 (anticodon GCA) |
| 6 | gene | 48.30971 | 48.30978 | negative | MGI_C57BL6J_4413793 | n-TCgca26 | NCBI_Gene:102467497 | MGI:4413793 | tRNA gene | nuclear encoded tRNA cysteine 26 (anticodon GCA) |
| 6 | gene | 48.31150 | 48.31157 | positive | MGI_C57BL6J_4413776 | n-TCgca9 | NCBI_Gene:102467270 | MGI:4413776 | tRNA gene | nuclear encoded tRNA cysteine 9 (anticodon GCA) |
| 6 | gene | 48.31302 | 48.31309 | negative | MGI_C57BL6J_4413792 | n-TCgca25 | NCBI_Gene:102467277 | MGI:4413792 | tRNA gene | nuclear encoded tRNA cysteine 25 (anticodon GCA) |
| 6 | gene | 48.31654 | 48.31661 | positive | MGI_C57BL6J_4413769 | n-TCgca2 | NCBI_Gene:102467267 | MGI:4413769 | tRNA gene | nuclear encoded tRNA cysteine 2 (anticodon GCA) |
| 6 | gene | 48.31752 | 48.31759 | positive | MGI_C57BL6J_4413812 | n-TCgca45 | NCBI_Gene:102467505 | MGI:4413812 | tRNA gene | nuclear encoded tRNA cysteine 45 (anticodon GCA) |
| 6 | gene | 48.33284 | 48.33291 | negative | MGI_C57BL6J_4413791 | n-TCgca24 | NCBI_Gene:102467276 | MGI:4413791 | tRNA gene | nuclear encoded tRNA cysteine 24 (anticodon GCA) |
| 6 | gene | 48.33611 | 48.33618 | negative | MGI_C57BL6J_4413790 | n-TCgca23 | NCBI_Gene:102467496 | MGI:4413790 | tRNA gene | nuclear encoded tRNA cysteine 23 (anticodon GCA) |
| 6 | gene | 48.33709 | 48.33716 | negative | MGI_C57BL6J_4413789 | n-TCgca22 | NCBI_Gene:102467275 | MGI:4413789 | tRNA gene | nuclear encoded tRNA cysteine 22 (anticodon GCA) |
| 6 | gene | 48.34061 | 48.34068 | positive | MGI_C57BL6J_4413781 | n-TCgca14 | NCBI_Gene:102467272 | MGI:4413781 | tRNA gene | nuclear encoded tRNA cysteine 14 (anticodon GCA) |
| 6 | gene | 48.34159 | 48.34166 | positive | MGI_C57BL6J_4413772 | n-TCgca5 | NCBI_Gene:102467268 | MGI:4413772 | tRNA gene | nuclear encoded tRNA cysteine 5 (anticodon GCA) |
| 6 | gene | 48.34976 | 48.34983 | negative | MGI_C57BL6J_4413788 | n-TCgca21 | NCBI_Gene:102467641 | MGI:4413788 | tRNA gene | nuclear encoded tRNA cysteine 21 (anticodon GCA) |
| 6 | gene | 48.35402 | 48.35409 | negative | MGI_C57BL6J_4413787 | n-TCgca20 | NCBI_Gene:102467495 | MGI:4413787 | tRNA gene | nuclear encoded tRNA cysteine 20 (anticodon GCA) |
| 6 | gene | 48.35585 | 48.35593 | positive | MGI_C57BL6J_4413813 | n-TCgca46 | NCBI_Gene:102467285 | MGI:4413813 | tRNA gene | nuclear encoded tRNA cysteine 46 (anticodon GCA) |
| 6 | gene | 48.35685 | 48.35692 | positive | MGI_C57BL6J_4413778 | n-TCgca11 | NCBI_Gene:102467271 | MGI:4413778 | tRNA gene | nuclear encoded tRNA cysteine 11 (anticodon GCA) |
| 6 | gene | 48.36353 | 48.36360 | negative | MGI_C57BL6J_4413786 | n-TCgca19 | NCBI_Gene:102467274 | MGI:4413786 | tRNA gene | nuclear encoded tRNA cysteine 19 (anticodon GCA) |
| 6 | gene | 48.37414 | 48.39541 | negative | MGI_C57BL6J_5753272 | Gm44696 | ENSEMBL:ENSMUSG00000107875 | MGI:5753272 | lncRNA gene | predicted gene 44696 |
| 6 | pseudogene | 48.38116 | 48.38207 | negative | MGI_C57BL6J_3645672 | Gm7880 | NCBI_Gene:665990,ENSEMBL:ENSMUSG00000108275 | MGI:3645672 | pseudogene | predicted gene 7880 |
| 6 | gene | 48.39559 | 48.41985 | positive | MGI_C57BL6J_1925077 | Krba1 | NCBI_Gene:77827,ENSEMBL:ENSMUSG00000042810 | MGI:1925077 | protein coding gene | KRAB-A domain containing 1 |
| 6 | gene | 48.39618 | 48.39672 | negative | MGI_C57BL6J_1922119 | 4930447G04Rik | NA | NA | unclassified gene | RIKEN cDNA 4930447G04 gene |
| 6 | gene | 48.42077 | 48.42084 | positive | MGI_C57BL6J_5454102 | Gm24325 | ENSEMBL:ENSMUSG00000093085 | MGI:5454102 | miRNA gene | predicted gene, 24325 |
| 6 | gene | 48.42236 | 48.42756 | positive | MGI_C57BL6J_5621688 | Gm38803 | NCBI_Gene:105242647 | MGI:5621688 | lncRNA gene | predicted gene, 38803 |
| 6 | gene | 48.42769 | 48.44603 | negative | MGI_C57BL6J_1916160 | Zfp467 | NCBI_Gene:68910,ENSEMBL:ENSMUSG00000068551 | MGI:1916160 | protein coding gene | zinc finger protein 467 |
| 6 | gene | 48.44623 | 48.50126 | positive | MGI_C57BL6J_2674311 | Sspo | NCBI_Gene:243369,ENSEMBL:ENSMUSG00000029797 | MGI:2674311 | protein coding gene | SCO-spondin |
| 6 | pseudogene | 48.50434 | 48.53483 | positive | MGI_C57BL6J_1889827 | Zfp862-ps | NCBI_Gene:58894,ENSEMBL:ENSMUSG00000107476 | MGI:1889827 | pseudogene | zinc finger protein 862, pseudogene |
| 6 | pseudogene | 48.52346 | 48.52388 | positive | MGI_C57BL6J_3704266 | Gm10243 | NCBI_Gene:100302576,ENSEMBL:ENSMUSG00000095526 | MGI:3704266 | pseudogene | predicted gene 10243 |
| 6 | pseudogene | 48.52392 | 48.52513 | negative | MGI_C57BL6J_3648251 | Gm7887 | NCBI_Gene:666005,ENSEMBL:ENSMUSG00000044211 | MGI:3648251 | pseudogene | predicted gene 7887 |
| 6 | gene | 48.53658 | 48.53747 | negative | MGI_C57BL6J_1916682 | 1700026J14Rik | NCBI_Gene:69432,ENSEMBL:ENSMUSG00000097716 | MGI:1916682 | lncRNA gene | RIKEN cDNA 1700026J14 gene |
| 6 | gene | 48.53757 | 48.54180 | positive | MGI_C57BL6J_1923502 | Atp6v0e2 | NCBI_Gene:76252,ENSEMBL:ENSMUSG00000039347 | MGI:1923502 | protein coding gene | ATPase, H+ transporting, lysosomal V0 subunit E2 |
| 6 | gene | 48.55478 | 48.57072 | positive | MGI_C57BL6J_2652848 | Lrrc61 | NCBI_Gene:243371,ENSEMBL:ENSMUSG00000073096 | MGI:2652848 | protein coding gene | leucine rich repeat containing 61 |
| 6 | gene | 48.56970 | 48.57285 | negative | MGI_C57BL6J_1918910 | Rarres2 | NCBI_Gene:71660,ENSEMBL:ENSMUSG00000009281 | MGI:1918910 | protein coding gene | retinoic acid receptor responder (tazarotene induced) 2 |
| 6 | gene | 48.58944 | 48.59058 | positive | MGI_C57BL6J_3645688 | Gm5111 | NCBI_Gene:330305,ENSEMBL:ENSMUSG00000052730 | MGI:3645688 | protein coding gene | predicted gene 5111 |
| 6 | gene | 48.58961 | 48.61325 | positive | MGI_C57BL6J_5753597 | Gm45021 | ENSEMBL:ENSMUSG00000107588 | MGI:5753597 | protein coding gene | predicted gene 45021 |
| 6 | gene | 48.59388 | 48.59908 | positive | MGI_C57BL6J_1889817 | Repin1 | NCBI_Gene:58887,ENSEMBL:ENSMUSG00000052751 | MGI:1889817 | protein coding gene | replication initiator 1 |
| 6 | gene | 48.59394 | 48.61928 | positive | MGI_C57BL6J_5753541 | Gm44965 | ENSEMBL:ENSMUSG00000107789 | MGI:5753541 | protein coding gene | predicted gene 44965 |
| 6 | gene | 48.60200 | 48.62323 | positive | MGI_C57BL6J_2683557 | Zfp775 | NCBI_Gene:243372,ENSEMBL:ENSMUSG00000007216 | MGI:2683557 | protein coding gene | zinc finger protein 775 |
| 6 | gene | 48.61127 | 48.62236 | negative | MGI_C57BL6J_5621689 | Gm38804 | NCBI_Gene:105242648,ENSEMBL:ENSMUSG00000107640 | MGI:5621689 | lncRNA gene | predicted gene, 38804 |
| 6 | gene | 48.62676 | 48.63369 | positive | MGI_C57BL6J_2141510 | AI854703 | NCBI_Gene:243373,ENSEMBL:ENSMUSG00000053297 | MGI:2141510 | protein coding gene | expressed sequence AI854703 |
| 6 | gene | 48.64719 | 48.66087 | positive | MGI_C57BL6J_2685303 | Gimap8 | NCBI_Gene:243374,ENSEMBL:ENSMUSG00000064262 | MGI:2685303 | protein coding gene | GTPase, IMAP family member 8 |
| 6 | pseudogene | 48.66540 | 48.66586 | negative | MGI_C57BL6J_5690654 | Gm44262 | ENSEMBL:ENSMUSG00000108117 | MGI:5690654 | pseudogene | predicted gene, 44262 |
| 6 | gene | 48.67583 | 48.67694 | negative | MGI_C57BL6J_5826595 | Gm46958 | NCBI_Gene:108169139 | MGI:5826595 | lncRNA gene | predicted gene, 46958 |
| 6 | gene | 48.67612 | 48.67912 | positive | MGI_C57BL6J_3511744 | Gimap9 | NCBI_Gene:317758,ENSEMBL:ENSMUSG00000051124 | MGI:3511744 | protein coding gene | GTPase, IMAP family member 9 |
| 6 | gene | 48.68455 | 48.69206 | positive | MGI_C57BL6J_1349656 | Gimap4 | NCBI_Gene:107526,ENSEMBL:ENSMUSG00000054435 | MGI:1349656 | protein coding gene | GTPase, IMAP family member 4 |
| 6 | gene | 48.70158 | 48.70824 | negative | MGI_C57BL6J_1918876 | Gimap6 | NCBI_Gene:231931,ENSEMBL:ENSMUSG00000047867 | MGI:1918876 | protein coding gene | GTPase, IMAP family member 6 |
| 6 | gene | 48.71861 | 48.75264 | positive | MGI_C57BL6J_5547789 | Gm28053 | ENSEMBL:ENSMUSG00000098715 | MGI:5547789 | protein coding gene | predicted gene, 28053 |
| 6 | gene | 48.71862 | 48.72464 | positive | MGI_C57BL6J_1349657 | Gimap7 | NCBI_Gene:231932,ENSEMBL:ENSMUSG00000043931 | MGI:1349657 | protein coding gene | GTPase, IMAP family member 7 |
| 6 | gene | 48.72928 | 48.74144 | negative | MGI_C57BL6J_3781523 | Gimap1os | NCBI_Gene:100041450,ENSEMBL:ENSMUSG00000044867 | MGI:3781523 | lncRNA gene | GTPase, IMAP family member 1, opposite strand |
| 6 | gene | 48.72955 | 48.73367 | positive | MGI_C57BL6J_1889832 | A130009E19Rik | NA | NA | unclassified gene | RIKEN cDNA A130009E19 gene |
| 6 | gene | 48.73905 | 48.74379 | positive | MGI_C57BL6J_109368 | Gimap1 | NCBI_Gene:16205,ENSEMBL:ENSMUSG00000090019 | MGI:109368 | protein coding gene | GTPase, IMAP family member 1 |
| 6 | gene | 48.74614 | 48.75421 | positive | MGI_C57BL6J_2442232 | Gimap5 | NCBI_Gene:317757,ENSEMBL:ENSMUSG00000043505 | MGI:2442232 | protein coding gene | GTPase, IMAP family member 5 |
| 6 | gene | 48.75423 | 48.75599 | negative | MGI_C57BL6J_5690618 | Gm44226 | ENSEMBL:ENSMUSG00000108298 | MGI:5690618 | lncRNA gene | predicted gene, 44226 |
| 6 | gene | 48.75534 | 48.75637 | positive | MGI_C57BL6J_1921153 | 4833403J16Rik | ENSEMBL:ENSMUSG00000107481 | MGI:1921153 | lncRNA gene | RIKEN cDNA 4833403J16 gene |
| 6 | gene | 48.76446 | 48.77085 | negative | MGI_C57BL6J_1932723 | Gimap3 | NCBI_Gene:83408,ENSEMBL:ENSMUSG00000039264 | MGI:1932723 | protein coding gene | GTPase, IMAP family member 3 |
| 6 | gene | 48.79100 | 48.79378 | negative | MGI_C57BL6J_5589873 | Gm30714 | NCBI_Gene:102632718 | MGI:5589873 | lncRNA gene | predicted gene, 30714 |
| 6 | gene | 48.79809 | 48.79885 | negative | MGI_C57BL6J_5589940 | Gm30781 | NCBI_Gene:102632803,ENSEMBL:ENSMUSG00000108130 | MGI:5589940 | lncRNA gene | predicted gene, 30781 |
| 6 | pseudogene | 48.81902 | 48.82047 | positive | MGI_C57BL6J_5010019 | Gm17834 | NCBI_Gene:100415951,ENSEMBL:ENSMUSG00000107574 | MGI:5010019 | pseudogene | predicted gene, 17834 |
| 6 | gene | 48.83381 | 48.84595 | negative | MGI_C57BL6J_1916348 | Tmem176b | NCBI_Gene:65963,ENSEMBL:ENSMUSG00000029810 | MGI:1916348 | protein coding gene | transmembrane protein 176B |
| 6 | gene | 48.84092 | 48.84707 | positive | MGI_C57BL6J_1913308 | Tmem176a | NCBI_Gene:66058,ENSEMBL:ENSMUSG00000023367 | MGI:1913308 | protein coding gene | transmembrane protein 176A |
| 6 | gene | 48.84766 | 48.90306 | negative | MGI_C57BL6J_5753340 | Gm44764 | NCBI_Gene:108169140,ENSEMBL:ENSMUSG00000107597 | MGI:5753340 | lncRNA gene | predicted gene 44764 |
| 6 | gene | 48.85990 | 48.86608 | positive | MGI_C57BL6J_3643446 | Gm7932 | NCBI_Gene:666105,ENSEMBL:ENSMUSG00000108084 | MGI:3643446 | lncRNA gene | predicted gene 7932 |
| 6 | gene | 48.87290 | 48.90919 | positive | MGI_C57BL6J_1923757 | Aoc1 | NCBI_Gene:76507,ENSEMBL:ENSMUSG00000029811 | MGI:1923757 | protein coding gene | amine oxidase, copper-containing 1 |
| 6 | gene | 48.92990 | 48.93369 | positive | MGI_C57BL6J_1917011 | Doxl1 | NCBI_Gene:69761,ENSEMBL:ENSMUSG00000029813 | MGI:1917011 | protein coding gene | diamine oxidase-like protein 1 |
| 6 | gene | 48.93479 | 48.93492 | positive | MGI_C57BL6J_5454125 | Gm24348 | ENSEMBL:ENSMUSG00000089335 | MGI:5454125 | snoRNA gene | predicted gene, 24348 |
| 6 | pseudogene | 48.93975 | 48.94405 | negative | MGI_C57BL6J_3642310 | Gm10242 | NCBI_Gene:100040589,ENSEMBL:ENSMUSG00000068537 | MGI:3642310 | pseudogene | predicted gene 10242 |
| 6 | pseudogene | 48.95626 | 48.95690 | positive | MGI_C57BL6J_5010723 | Gm18538 | NCBI_Gene:100417335,ENSEMBL:ENSMUSG00000107943 | MGI:5010723 | pseudogene | predicted gene, 18538 |
| 6 | pseudogene | 48.96279 | 48.96417 | positive | MGI_C57BL6J_5011357 | Gm19172 | NCBI_Gene:100418382,ENSEMBL:ENSMUSG00000107692 | MGI:5011357 | pseudogene | predicted gene, 19172 |
| 6 | gene | 48.97481 | 48.97875 | positive | MGI_C57BL6J_3618290 | Doxl2 | NCBI_Gene:243376,ENSEMBL:ENSMUSG00000068536 | MGI:3618290 | protein coding gene | diamine oxidase-like protein 2 |
| 6 | pseudogene | 48.98110 | 48.98221 | positive | MGI_C57BL6J_5010724 | Gm18539 | NCBI_Gene:100417336,ENSEMBL:ENSMUSG00000108017 | MGI:5010724 | pseudogene | predicted gene, 18539 |
| 6 | gene | 48.98662 | 48.99962 | negative | MGI_C57BL6J_1922597 | 4930563H07Rik | NCBI_Gene:75347,ENSEMBL:ENSMUSG00000107656 | MGI:1922597 | lncRNA gene | RIKEN cDNA 4930563H07 gene |
| 6 | gene | 48.98686 | 48.99173 | positive | MGI_C57BL6J_2682321 | Svs1 | NCBI_Gene:243377,ENSEMBL:ENSMUSG00000039215 | MGI:2682321 | protein coding gene | seminal vesicle secretory protein 1 |
| 6 | pseudogene | 48.99521 | 48.99765 | positive | MGI_C57BL6J_5011358 | Gm19173 | NCBI_Gene:100418383,ENSEMBL:ENSMUSG00000107457 | MGI:5011358 | pseudogene | predicted gene, 19173 |
| 6 | gene | 49.03652 | 49.07093 | positive | MGI_C57BL6J_1934765 | Gpnmb | NCBI_Gene:93695,ENSEMBL:ENSMUSG00000029816 | MGI:1934765 | protein coding gene | glycoprotein (transmembrane) nmb |
| 6 | pseudogene | 49.07162 | 49.07216 | positive | MGI_C57BL6J_5011438 | Gm19253 | NCBI_Gene:100462958,ENSEMBL:ENSMUSG00000107758 | MGI:5011438 | pseudogene | predicted gene, 19253 |
| 6 | gene | 49.07379 | 49.13914 | positive | MGI_C57BL6J_1922843 | Malsu1 | NCBI_Gene:75593,ENSEMBL:ENSMUSG00000029815 | MGI:1922843 | protein coding gene | mitochondrial assembly of ribosomal large subunit 1 |
| 6 | pseudogene | 49.07995 | 49.08117 | negative | MGI_C57BL6J_3645227 | Gm7890 | NCBI_Gene:666011,ENSEMBL:ENSMUSG00000107990 | MGI:3645227 | pseudogene | predicted gene 7890 |
| 6 | gene | 49.08522 | 49.21501 | negative | MGI_C57BL6J_1890359 | Igf2bp3 | NCBI_Gene:140488,ENSEMBL:ENSMUSG00000029814 | MGI:1890359 | protein coding gene | insulin-like growth factor 2 mRNA binding protein 3 |
| 6 | pseudogene | 49.17267 | 49.17352 | positive | MGI_C57BL6J_5010195 | Gm18010 | NCBI_Gene:100416268,ENSEMBL:ENSMUSG00000107727 | MGI:5010195 | pseudogene | predicted gene, 18010 |
| 6 | gene | 49.20581 | 49.21248 | negative | MGI_C57BL6J_2442568 | D030074P21Rik | NCBI_Gene:102642435 | MGI:2442568 | lncRNA gene | RIKEN cDNA D030074P21 gene |
| 6 | gene | 49.24392 | 49.26425 | negative | MGI_C57BL6J_1933972 | Tra2a | NCBI_Gene:101214,ENSEMBL:ENSMUSG00000029817 | MGI:1933972 | protein coding gene | transformer 2 alpha |
| 6 | gene | 49.25485 | 49.25646 | negative | MGI_C57BL6J_2441851 | B230214O09Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA B230214O09 gene |
| 6 | gene | 49.25485 | 49.25646 | positive | MGI_C57BL6J_5690372 | Gm43980 | ENSEMBL:ENSMUSG00000107962 | MGI:5690372 | unclassified gene | predicted gene, 43980 |
| 6 | pseudogene | 49.28655 | 49.28729 | positive | MGI_C57BL6J_5010196 | Gm18011 | NCBI_Gene:100416269,ENSEMBL:ENSMUSG00000107916 | MGI:5010196 | pseudogene | predicted gene, 18011 |
| 6 | gene | 49.31927 | 49.34159 | positive | MGI_C57BL6J_1889376 | Ccdc126 | NCBI_Gene:57895,ENSEMBL:ENSMUSG00000050786 | MGI:1889376 | protein coding gene | coiled-coil domain containing 126 |
| 6 | gene | 49.36722 | 49.39054 | positive | MGI_C57BL6J_2442161 | Fam221a | NCBI_Gene:231946,ENSEMBL:ENSMUSG00000047115 | MGI:2442161 | protein coding gene | family with sequence similarity 221, member A |
| 6 | gene | 49.39548 | 49.46951 | positive | MGI_C57BL6J_1924735 | Stk31 | NCBI_Gene:77485,ENSEMBL:ENSMUSG00000023403 | MGI:1924735 | protein coding gene | serine threonine kinase 31 |
| 6 | pseudogene | 49.42375 | 49.42497 | negative | MGI_C57BL6J_4937887 | Gm17060 | NCBI_Gene:100416089,ENSEMBL:ENSMUSG00000090624 | MGI:4937887 | pseudogene | predicted gene 17060 |
| 6 | pseudogene | 49.54298 | 49.56317 | negative | MGI_C57BL6J_3644734 | Gm5305 | NCBI_Gene:102633308,ENSEMBL:ENSMUSG00000108266 | MGI:3644734 | pseudogene | predicted gene 5305 |
| 6 | pseudogene | 49.59624 | 49.59976 | positive | MGI_C57BL6J_3645840 | Rpl19-ps8 | NCBI_Gene:666210 | MGI:3645840 | pseudogene | ribosomal protein L19, pseudogene 8 |
| 6 | gene | 49.63363 | 49.67845 | positive | MGI_C57BL6J_5590385 | Gm31226 | NCBI_Gene:102633391 | MGI:5590385 | lncRNA gene | predicted gene, 31226 |
| 6 | pseudogene | 49.72725 | 49.72752 | negative | MGI_C57BL6J_5690611 | Gm44219 | ENSEMBL:ENSMUSG00000107982 | MGI:5690611 | pseudogene | predicted gene, 44219 |
| 6 | gene | 49.82271 | 49.82951 | positive | MGI_C57BL6J_97374 | Npy | NCBI_Gene:109648,ENSEMBL:ENSMUSG00000029819 | MGI:97374 | protein coding gene | neuropeptide Y |
| 6 | pseudogene | 49.85800 | 49.85885 | negative | MGI_C57BL6J_5010192 | Gm18007 | NCBI_Gene:100416264,ENSEMBL:ENSMUSG00000107664 | MGI:5010192 | pseudogene | predicted gene, 18007 |
| 6 | gene | 50.05119 | 50.10763 | positive | MGI_C57BL6J_3781631 | Gm3455 | ENSEMBL:ENSMUSG00000108071 | MGI:3781631 | lncRNA gene | predicted gene 3455 |
| 6 | gene | 50.10766 | 50.11016 | negative | MGI_C57BL6J_5621690 | Gm38805 | NCBI_Gene:105242649 | MGI:5621690 | lncRNA gene | predicted gene, 38805 |
| 6 | gene | 50.11024 | 50.19894 | positive | MGI_C57BL6J_1927340 | Mpp6 | NCBI_Gene:56524,ENSEMBL:ENSMUSG00000038388 | MGI:1927340 | protein coding gene | membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
| 6 | gene | 50.18889 | 50.26386 | negative | MGI_C57BL6J_1889850 | Gsdme | NCBI_Gene:54722,ENSEMBL:ENSMUSG00000029821 | MGI:1889850 | protein coding gene | gasdermin E |
| 6 | gene | 50.29333 | 50.45635 | negative | MGI_C57BL6J_1918970 | Osbpl3 | NCBI_Gene:71720,ENSEMBL:ENSMUSG00000029822 | MGI:1918970 | protein coding gene | oxysterol binding protein-like 3 |
| 6 | gene | 50.49955 | 50.50572 | negative | MGI_C57BL6J_5623257 | Gm40372 | NCBI_Gene:105244836 | MGI:5623257 | lncRNA gene | predicted gene, 40372 |
| 6 | gene | 50.50156 | 50.53802 | positive | MGI_C57BL6J_5590438 | Gm31279 | NCBI_Gene:102633461 | MGI:5590438 | lncRNA gene | predicted gene, 31279 |
| 6 | gene | 50.53990 | 50.54003 | negative | MGI_C57BL6J_5456066 | Gm26289 | ENSEMBL:ENSMUSG00000077357 | MGI:5456066 | snoRNA gene | predicted gene, 26289 |
| 6 | gene | 50.54076 | 50.54298 | positive | MGI_C57BL6J_5621691 | Gm38806 | NCBI_Gene:105242651 | MGI:5621691 | lncRNA gene | predicted gene, 38806 |
| 6 | gene | 50.56256 | 50.56654 | negative | MGI_C57BL6J_88578 | Cycs | NCBI_Gene:13063,ENSEMBL:ENSMUSG00000063694 | MGI:88578 | protein coding gene | cytochrome c, somatic |
| 6 | gene | 50.56664 | 50.59486 | positive | MGI_C57BL6J_1918631 | 5430402O13Rik | NCBI_Gene:73183,ENSEMBL:ENSMUSG00000085776 | MGI:1918631 | lncRNA gene | RIKEN cDNA 5430402O13 gene |
| 6 | gene | 50.57240 | 50.57251 | positive | MGI_C57BL6J_5530758 | Mir6371 | miRBase:MI0021900,NCBI_Gene:102465193,ENSEMBL:ENSMUSG00000099221 | MGI:5530758 | miRNA gene | microRNA 6371 |
| 6 | gene | 50.57330 | 50.59691 | negative | MGI_C57BL6J_1918071 | 4921507P07Rik | NCBI_Gene:70821,ENSEMBL:ENSMUSG00000029828 | MGI:1918071 | protein coding gene | RIKEN cDNA 4921507P07 gene |
| 6 | pseudogene | 50.59987 | 50.60024 | positive | MGI_C57BL6J_5690446 | Gm44054 | ENSEMBL:ENSMUSG00000107447 | MGI:5690446 | pseudogene | predicted gene, 44054 |
| 6 | gene | 50.60154 | 50.60528 | negative | MGI_C57BL6J_5621692 | Gm38807 | NCBI_Gene:105242652 | MGI:5621692 | lncRNA gene | predicted gene, 38807 |
| 6 | pseudogene | 50.60864 | 50.61069 | positive | MGI_C57BL6J_3646036 | Gm4782 | NCBI_Gene:213320,ENSEMBL:ENSMUSG00000107418 | MGI:3646036 | pseudogene | predicted gene 4782 |
| 6 | pseudogene | 50.61785 | 50.61854 | negative | MGI_C57BL6J_3644146 | Gm8035 | NCBI_Gene:666302,ENSEMBL:ENSMUSG00000108135 | MGI:3644146 | pseudogene | predicted gene 8035 |
| 6 | pseudogene | 50.61984 | 50.62188 | negative | MGI_C57BL6J_3644148 | Gm8038 | NCBI_Gene:666308,ENSEMBL:ENSMUSG00000107821 | MGI:3644148 | pseudogene | predicted gene 8038 |
| 6 | pseudogene | 50.62574 | 50.62646 | negative | MGI_C57BL6J_3647078 | Gm8039 | NCBI_Gene:666310,ENSEMBL:ENSMUSG00000108016 | MGI:3647078 | pseudogene | predicted gene 8039 |
| 6 | pseudogene | 50.62768 | 50.62834 | negative | MGI_C57BL6J_5010950 | Gm18765 | NCBI_Gene:100417688,ENSEMBL:ENSMUSG00000107740 | MGI:5010950 | pseudogene | predicted gene, 18765 |
| 6 | gene | 50.65067 | 50.65444 | negative | MGI_C57BL6J_1926488 | Npvf | NCBI_Gene:60531,ENSEMBL:ENSMUSG00000029831 | MGI:1926488 | protein coding gene | neuropeptide VF precursor |
| 6 | gene | 50.65985 | 50.67948 | negative | MGI_C57BL6J_5621693 | Gm38808 | NCBI_Gene:105242653 | MGI:5621693 | lncRNA gene | predicted gene, 38808 |
| 6 | pseudogene | 50.74668 | 50.74781 | negative | MGI_C57BL6J_3781195 | Gm3017 | NCBI_Gene:100040877,ENSEMBL:ENSMUSG00000107672 | MGI:3781195 | pseudogene | predicted gene 3017 |
| 6 | gene | 50.77611 | 50.81503 | positive | MGI_C57BL6J_2443109 | C530044C16Rik | NCBI_Gene:319981,ENSEMBL:ENSMUSG00000107667 | MGI:2443109 | lncRNA gene | RIKEN cDNA C530044C16 gene |
| 6 | gene | 50.79907 | 50.80303 | negative | MGI_C57BL6J_5690501 | Gm44109 | ENSEMBL:ENSMUSG00000107978 | MGI:5690501 | lncRNA gene | predicted gene, 44109 |
| 6 | gene | 50.80427 | 50.82229 | negative | MGI_C57BL6J_5590614 | Gm31455 | NCBI_Gene:102633693 | MGI:5590614 | lncRNA gene | predicted gene, 31455 |
| 6 | gene | 50.84671 | 50.87808 | negative | MGI_C57BL6J_3588242 | G930045G22Rik | ENSEMBL:ENSMUSG00000107993 | MGI:3588242 | lncRNA gene | RIKEN cDNA G930045G22 gene |
| 6 | gene | 50.88089 | 50.88262 | negative | MGI_C57BL6J_5825559 | Gm45922 | NCBI_Gene:105247310 | MGI:5825559 | lncRNA gene | predicted gene, 45922 |
| 6 | gene | 50.90046 | 50.90898 | negative | MGI_C57BL6J_5590790 | Gm31631 | NCBI_Gene:102633926 | MGI:5590790 | lncRNA gene | predicted gene, 31631 |
| 6 | gene | 50.90640 | 50.91074 | positive | MGI_C57BL6J_5590738 | Gm31579 | NCBI_Gene:102633851,ENSEMBL:ENSMUSG00000107488 | MGI:5590738 | lncRNA gene | predicted gene, 31579 |
| 6 | gene | 50.93500 | 50.94438 | negative | MGI_C57BL6J_5590845 | Gm31686 | NCBI_Gene:102633996 | MGI:5590845 | lncRNA gene | predicted gene, 31686 |
| 6 | pseudogene | 50.96727 | 50.96751 | positive | MGI_C57BL6J_5690794 | Gm44402 | ENSEMBL:ENSMUSG00000108153 | MGI:5690794 | pseudogene | predicted gene, 44402 |
| 6 | gene | 50.97338 | 50.99037 | negative | MGI_C57BL6J_5621694 | Gm38809 | NCBI_Gene:105242654 | MGI:5621694 | lncRNA gene | predicted gene, 38809 |
| 6 | gene | 50.98740 | 51.06801 | positive | MGI_C57BL6J_5826596 | Gm46959 | NCBI_Gene:108169141 | MGI:5826596 | lncRNA gene | predicted gene, 46959 |
| 6 | gene | 51.03294 | 51.06126 | negative | MGI_C57BL6J_5590905 | Gm31746 | NCBI_Gene:102634074 | MGI:5590905 | lncRNA gene | predicted gene, 31746 |
| 6 | gene | 51.09651 | 51.10067 | negative | MGI_C57BL6J_5591349 | Gm32190 | NCBI_Gene:102634656 | MGI:5591349 | lncRNA gene | predicted gene, 32190 |
| 6 | gene | 51.12475 | 51.16645 | negative | MGI_C57BL6J_5591487 | Gm32328 | NCBI_Gene:102634838 | MGI:5591487 | lncRNA gene | predicted gene, 32328 |
| 6 | gene | 51.14201 | 51.14608 | positive | MGI_C57BL6J_5591405 | Gm32246 | NCBI_Gene:102634734 | MGI:5591405 | lncRNA gene | predicted gene, 32246 |
| 6 | gene | 51.15428 | 51.15493 | positive | MGI_C57BL6J_5621695 | Gm38810 | NCBI_Gene:105242655 | MGI:5621695 | lncRNA gene | predicted gene, 38810 |
| 6 | gene | 51.20092 | 51.20735 | negative | MGI_C57BL6J_5591562 | Gm32403 | NCBI_Gene:102634943 | MGI:5591562 | lncRNA gene | predicted gene, 32403 |
| 6 | gene | 51.22296 | 51.28057 | negative | MGI_C57BL6J_5621706 | Gm38821 | NCBI_Gene:105242668 | MGI:5621706 | protein coding gene | predicted gene, 38821 |
| 6 | gene | 51.26981 | 51.26991 | negative | MGI_C57BL6J_2676833 | Mir148a | miRBase:MI0000550,NCBI_Gene:387166,ENSEMBL:ENSMUSG00000065505 | MGI:2676833 | miRNA gene | microRNA 148a |
| 6 | gene | 51.28353 | 51.30864 | negative | MGI_C57BL6J_5591638 | Gm32479 | NCBI_Gene:102635043,ENSEMBL:ENSMUSG00000107619 | MGI:5591638 | lncRNA gene | predicted gene, 32479 |
| 6 | gene | 51.33049 | 51.33371 | positive | MGI_C57BL6J_2679266 | BC022713 | NA | NA | unclassified gene | cDNA sequence BC022713 |
| 6 | gene | 51.37971 | 51.39278 | negative | MGI_C57BL6J_5477171 | Gm26677 | ENSEMBL:ENSMUSG00000097598 | MGI:5477171 | lncRNA gene | predicted gene, 26677 |
| 6 | gene | 51.37971 | 51.39279 | positive | MGI_C57BL6J_3643041 | Gm6559 | NCBI_Gene:625167,ENSEMBL:ENSMUSG00000107660 | MGI:3643041 | lncRNA gene | predicted gene 6559 |
| 6 | gene | 51.40097 | 51.40222 | negative | MGI_C57BL6J_1921343 | 0610033M10Rik | ENSEMBL:ENSMUSG00000108236 | MGI:1921343 | lncRNA gene | RIKEN cDNA 0610033M10 gene |
| 6 | gene | 51.43267 | 51.45877 | positive | MGI_C57BL6J_1339958 | Nfe2l3 | NCBI_Gene:18025,ENSEMBL:ENSMUSG00000029832 | MGI:1339958 | protein coding gene | nuclear factor, erythroid derived 2, like 3 |
| 6 | gene | 51.46043 | 51.47031 | negative | MGI_C57BL6J_104819 | Hnrnpa2b1 | NCBI_Gene:53379,ENSEMBL:ENSMUSG00000004980 | MGI:104819 | protein coding gene | heterogeneous nuclear ribonucleoprotein A2/B1 |
| 6 | gene | 51.47036 | 51.48370 | positive | MGI_C57BL6J_108515 | Cbx3 | NCBI_Gene:12417,ENSEMBL:ENSMUSG00000029836 | MGI:108515 | protein coding gene | chromobox 3 |
| 6 | gene | 51.52390 | 51.59068 | positive | MGI_C57BL6J_1919232 | Snx10 | NCBI_Gene:71982,ENSEMBL:ENSMUSG00000038301 | MGI:1919232 | protein coding gene | sorting nexin 10 |
| 6 | gene | 51.62163 | 51.62171 | positive | MGI_C57BL6J_5452691 | Gm22914 | ENSEMBL:ENSMUSG00000094528 | MGI:5452691 | miRNA gene | predicted gene, 22914 |
| 6 | gene | 51.69431 | 51.71108 | negative | MGI_C57BL6J_5621696 | Gm38811 | NCBI_Gene:105242656,ENSEMBL:ENSMUSG00000108145 | MGI:5621696 | lncRNA gene | predicted gene, 38811 |
| 6 | gene | 51.85742 | 52.01255 | negative | MGI_C57BL6J_1889206 | Skap2 | NCBI_Gene:54353,ENSEMBL:ENSMUSG00000059182 | MGI:1889206 | protein coding gene | src family associated phosphoprotein 2 |
| 6 | gene | 52.01260 | 52.02699 | positive | MGI_C57BL6J_5690469 | Gm44077 | ENSEMBL:ENSMUSG00000108244 | MGI:5690469 | lncRNA gene | predicted gene, 44077 |
| 6 | gene | 52.10295 | 52.11368 | positive | MGI_C57BL6J_3705267 | Halr1 | NCBI_Gene:102635415,ENSEMBL:ENSMUSG00000085412 | MGI:3705267 | lncRNA gene | Hoxa adjacent long noncoding RNA 1 |
| 6 | gene | 52.10480 | 52.10616 | positive | MGI_C57BL6J_5636487 | Halr1os1 | NA | NA | unclassified non-coding RNA gene | Hoxa adjacent long noncoding RNA 1%2c opposite strand 1 |
| 6 | pseudogene | 52.11822 | 52.11894 | positive | MGI_C57BL6J_3646130 | Rps8-ps3 | NCBI_Gene:666391,ENSEMBL:ENSMUSG00000081979 | MGI:3646130 | pseudogene | ribosomal protein S8, pseudogene 3 |
| 6 | pseudogene | 52.13178 | 52.13205 | negative | MGI_C57BL6J_3705565 | Rps19-ps14 | ENSEMBL:ENSMUSG00000082781 | MGI:3705565 | pseudogene | ribosomal protein S19, pseudogene 14 |
| 6 | gene | 52.15537 | 52.15929 | negative | MGI_C57BL6J_96170 | Hoxa1 | NCBI_Gene:15394,ENSEMBL:ENSMUSG00000029844 | MGI:96170 | protein coding gene | homeobox A1 |
| 6 | gene | 52.15690 | 52.15853 | positive | MGI_C57BL6J_5636492 | Hotairm2 | NA | NA | unclassified non-coding RNA gene | Hoxa transcript antisense RNA%2c myeloid-specific 2 |
| 6 | gene | 52.15690 | 52.16212 | positive | MGI_C57BL6J_3705155 | Hotairm1 | NCBI_Gene:105242658,ENSEMBL:ENSMUSG00000087658 | MGI:3705155 | antisense lncRNA gene | Hoxa transcript antisense RNA, myeloid-specific 1 |
| 6 | gene | 52.15843 | 52.15856 | positive | MGI_C57BL6J_5531249 | Gm27867 | ENSEMBL:ENSMUSG00000098537 | MGI:5531249 | unclassified non-coding RNA gene | predicted gene, 27867 |
| 6 | gene | 52.16104 | 52.16125 | positive | MGI_C57BL6J_5531243 | Gm27861 | ENSEMBL:ENSMUSG00000099068 | MGI:5531243 | unclassified non-coding RNA gene | predicted gene, 27861 |
| 6 | gene | 52.16201 | 52.16211 | positive | MGI_C57BL6J_5531250 | Gm27868 | ENSEMBL:ENSMUSG00000098536 | MGI:5531250 | unclassified non-coding RNA gene | predicted gene, 27868 |
| 6 | gene | 52.16213 | 52.16227 | positive | MGI_C57BL6J_5530859 | Gm27477 | ENSEMBL:ENSMUSG00000098749 | MGI:5530859 | unclassified non-coding RNA gene | predicted gene, 27477 |
| 6 | gene | 52.16242 | 52.16483 | negative | MGI_C57BL6J_96174 | Hoxa2 | NCBI_Gene:15399,ENSEMBL:ENSMUSG00000014704 | MGI:96174 | protein coding gene | homeobox A2 |
| 6 | gene | 52.16336 | 52.21334 | negative | MGI_C57BL6J_5579014 | Gm28308 | ENSEMBL:ENSMUSG00000101298 | MGI:5579014 | protein coding gene | predicted gene 28308 |
| 6 | gene | 52.16567 | 52.16956 | positive | MGI_C57BL6J_1913890 | Hoxaas2 | NCBI_Gene:105242657,ENSEMBL:ENSMUSG00000056445 | MGI:1913890 | lncRNA gene | Hoxa cluster antisense RNA 2 |
| 6 | gene | 52.16611 | 52.16679 | negative | MGI_C57BL6J_5826597 | Gm46960 | NCBI_Gene:108169142 | MGI:5826597 | lncRNA gene | predicted gene, 46960 |
| 6 | gene | 52.16906 | 52.21334 | negative | MGI_C57BL6J_96175 | Hoxa3 | NCBI_Gene:15400,ENSEMBL:ENSMUSG00000079560 | MGI:96175 | protein coding gene | homeobox A3 |
| 6 | gene | 52.17285 | 52.17406 | positive | MGI_C57BL6J_3705180 | Gm15050 | NCBI_Gene:100504154,ENSEMBL:ENSMUSG00000087626 | MGI:3705180 | lncRNA gene | predicted gene 15050 |
| 6 | gene | 52.17522 | 52.21508 | positive | MGI_C57BL6J_1919878 | Hoxaas3 | NCBI_Gene:72628,ENSEMBL:ENSMUSG00000085696 | MGI:1919878 | antisense lncRNA gene | Hoxa cluster antisense RNA 3 |
| 6 | gene | 52.17694 | 52.18085 | positive | MGI_C57BL6J_1924830 | 5730596B20Rik | NCBI_Gene:77580,ENSEMBL:ENSMUSG00000056468 | MGI:1924830 | lncRNA gene | RIKEN cDNA 5730596B20 gene |
| 6 | gene | 52.18311 | 52.18993 | positive | MGI_C57BL6J_5580136 | Gm29430 | ENSEMBL:ENSMUSG00000101451 | MGI:5580136 | lncRNA gene | predicted gene 29430 |
| 6 | gene | 52.18967 | 52.19175 | negative | MGI_C57BL6J_96176 | Hoxa4 | NCBI_Gene:15401,ENSEMBL:ENSMUSG00000000942 | MGI:96176 | protein coding gene | homeobox A4 |
| 6 | gene | 52.20175 | 52.20459 | negative | MGI_C57BL6J_96177 | Hoxa5 | NCBI_Gene:15402,ENSEMBL:ENSMUSG00000038253 | MGI:96177 | protein coding gene | homeobox A5 |
| 6 | gene | 52.20629 | 52.20872 | negative | MGI_C57BL6J_96178 | Hoxa6 | NCBI_Gene:15403,ENSEMBL:ENSMUSG00000043219 | MGI:96178 | protein coding gene | homeobox A6 |
| 6 | gene | 52.21449 | 52.21529 | negative | MGI_C57BL6J_5433747 | Mira | NCBI_Gene:100736249 | MGI:5433747 | lncRNA gene | mistral long non-coding RNA |
| 6 | gene | 52.21449 | 52.22185 | negative | MGI_C57BL6J_96179 | Hoxa7 | NCBI_Gene:15404,ENSEMBL:ENSMUSG00000038236 | MGI:96179 | protein coding gene | homeobox A7 |
| 6 | gene | 52.22310 | 52.23109 | negative | MGI_C57BL6J_96180 | Hoxa9 | NCBI_Gene:15405,ENSEMBL:ENSMUSG00000038227 | MGI:96180 | protein coding gene | homeobox A9 |
| 6 | gene | 52.23008 | 52.23016 | negative | MGI_C57BL6J_3618741 | Mir196b | miRBase:MI0001151,NCBI_Gene:723820,ENSEMBL:ENSMUSG00000065443 | MGI:3618741 | miRNA gene | microRNA 196b |
| 6 | gene | 52.23120 | 52.24085 | negative | MGI_C57BL6J_96171 | Hoxa10 | NCBI_Gene:15395,ENSEMBL:ENSMUSG00000000938 | MGI:96171 | protein coding gene | homeobox A10 |
| 6 | gene | 52.24211 | 52.24581 | negative | MGI_C57BL6J_96172 | Hoxa11 | NCBI_Gene:15396,ENSEMBL:ENSMUSG00000038210 | MGI:96172 | protein coding gene | homeobox A11 |
| 6 | gene | 52.24439 | 52.24980 | positive | MGI_C57BL6J_107208 | Hoxa11os | NCBI_Gene:15397,ENSEMBL:ENSMUSG00000086427 | MGI:107208 | lncRNA gene | homeobox A11, opposite strand |
| 6 | gene | 52.24601 | 52.24610 | positive | MGI_C57BL6J_5531414 | Gm28032 | ENSEMBL:ENSMUSG00000098717 | MGI:5531414 | unclassified non-coding RNA gene | predicted gene, 28032 |
| 6 | gene | 52.24640 | 52.24659 | positive | MGI_C57BL6J_5530874 | Gm27492 | ENSEMBL:ENSMUSG00000098435 | MGI:5530874 | unclassified non-coding RNA gene | predicted gene, 27492 |
| 6 | gene | 52.24672 | 52.24681 | positive | MGI_C57BL6J_5531077 | Gm27695 | ENSEMBL:ENSMUSG00000098969 | MGI:5531077 | unclassified non-coding RNA gene | predicted gene, 27695 |
| 6 | gene | 52.24742 | 52.24765 | positive | MGI_C57BL6J_5531199 | Gm27817 | ENSEMBL:ENSMUSG00000098683 | MGI:5531199 | unclassified non-coding RNA gene | predicted gene, 27817 |
| 6 | gene | 52.24833 | 52.24853 | positive | MGI_C57BL6J_5530836 | Gm27454 | ENSEMBL:ENSMUSG00000099270 | MGI:5530836 | unclassified non-coding RNA gene | predicted gene, 27454 |
| 6 | gene | 52.24930 | 52.24947 | positive | MGI_C57BL6J_5530925 | Gm27543 | ENSEMBL:ENSMUSG00000099248 | MGI:5530925 | unclassified non-coding RNA gene | predicted gene, 27543 |
| 6 | gene | 52.25622 | 52.25746 | negative | MGI_C57BL6J_1924645 | 9530018H14Rik | ENSEMBL:ENSMUSG00000102373 | MGI:1924645 | unclassified gene | RIKEN cDNA 9530018H14 gene |
| 6 | gene | 52.25769 | 52.26088 | negative | MGI_C57BL6J_96173 | Hoxa13 | NCBI_Gene:15398,ENSEMBL:ENSMUSG00000038203 | MGI:96173 | protein coding gene | homeobox A13 |
| 6 | gene | 52.26115 | 52.26121 | positive | MGI_C57BL6J_5530662 | Gm27280 | ENSEMBL:ENSMUSG00000098484 | MGI:5530662 | unclassified non-coding RNA gene | predicted gene, 27280 |
| 6 | gene | 52.26259 | 52.26297 | positive | MGI_C57BL6J_5531367 | Gm27985 | ENSEMBL:ENSMUSG00000099173 | MGI:5531367 | unclassified non-coding RNA gene | predicted gene, 27985 |
| 6 | gene | 52.26277 | 52.26760 | positive | MGI_C57BL6J_3642509 | Hottip | NCBI_Gene:791364,ENSEMBL:ENSMUSG00000055408 | MGI:3642509 | lncRNA gene | Hoxa distal transcript antisense RNA |
| 6 | gene | 52.26306 | 52.26342 | positive | MGI_C57BL6J_5530765 | Gm27383 | ENSEMBL:ENSMUSG00000099229 | MGI:5530765 | unclassified non-coding RNA gene | predicted gene, 27383 |
| 6 | gene | 52.26658 | 52.26675 | positive | MGI_C57BL6J_5531248 | Gm27866 | ENSEMBL:ENSMUSG00000098538 | MGI:5531248 | unclassified non-coding RNA gene | predicted gene, 27866 |
| 6 | gene | 52.29712 | 52.29785 | positive | MGI_C57BL6J_5690686 | Gm44294 | ENSEMBL:ENSMUSG00000107463 | MGI:5690686 | unclassified gene | predicted gene, 44294 |
| 6 | gene | 52.30839 | 52.31483 | negative | MGI_C57BL6J_1917843 | Evx1os | NCBI_Gene:70593,ENSEMBL:ENSMUSG00000086126 | MGI:1917843 | antisense lncRNA gene | even skipped homeotic gene 1, opposite strand |
| 6 | gene | 52.31350 | 52.31839 | positive | MGI_C57BL6J_95461 | Evx1 | NCBI_Gene:14028,ENSEMBL:ENSMUSG00000005503 | MGI:95461 | protein coding gene | even-skipped homeobox 1 |
| 6 | pseudogene | 52.34669 | 52.34700 | negative | MGI_C57BL6J_3646863 | Gm8129 | NCBI_Gene:666487,ENSEMBL:ENSMUSG00000059159 | MGI:3646863 | pseudogene | predicted pseudogene 8129 |
| 6 | pseudogene | 52.35754 | 52.35782 | negative | MGI_C57BL6J_3705761 | Gm15054 | ENSEMBL:ENSMUSG00000084282 | MGI:3705761 | pseudogene | predicted gene 15054 |
| 6 | gene | 52.49245 | 52.50009 | positive | MGI_C57BL6J_1921554 | 1700094M24Rik | NCBI_Gene:74304,ENSEMBL:ENSMUSG00000107895 | MGI:1921554 | lncRNA gene | RIKEN cDNA 1700094M24 gene |
| 6 | gene | 52.50989 | 52.51404 | positive | MGI_C57BL6J_5690420 | Gm44028 | ENSEMBL:ENSMUSG00000108062 | MGI:5690420 | lncRNA gene | predicted gene, 44028 |
| 6 | gene | 52.54623 | 52.64039 | negative | MGI_C57BL6J_1889802 | Hibadh | NCBI_Gene:58875,ENSEMBL:ENSMUSG00000029776 | MGI:1889802 | protein coding gene | 3-hydroxyisobutyrate dehydrogenase |
| 6 | gene | 52.56055 | 52.56323 | positive | MGI_C57BL6J_5690837 | Gm44445 | ENSEMBL:ENSMUSG00000107647 | MGI:5690837 | unclassified gene | predicted gene, 44445 |
| 6 | gene | 52.56596 | 52.57981 | negative | MGI_C57BL6J_5592206 | Gm33047 | NCBI_Gene:102635799 | MGI:5592206 | lncRNA gene | predicted gene, 33047 |
| 6 | gene | 52.59950 | 52.60325 | negative | MGI_C57BL6J_5690826 | Gm44434 | ENSEMBL:ENSMUSG00000107813 | MGI:5690826 | unclassified gene | predicted gene, 44434 |
| 6 | gene | 52.63923 | 52.63936 | negative | MGI_C57BL6J_5454132 | Gm24355 | ENSEMBL:ENSMUSG00000064792 | MGI:5454132 | snoRNA gene | predicted gene, 24355 |
| 6 | gene | 52.71334 | 52.71395 | negative | MGI_C57BL6J_5690358 | Gm43966 | ENSEMBL:ENSMUSG00000108170 | MGI:5690358 | unclassified gene | predicted gene, 43966 |
| 6 | gene | 52.71357 | 52.76678 | positive | MGI_C57BL6J_1289308 | Tax1bp1 | NCBI_Gene:52440,ENSEMBL:ENSMUSG00000004535 | MGI:1289308 | protein coding gene | Tax1 (human T cell leukemia virus type I) binding protein 1 |
| 6 | pseudogene | 52.72263 | 52.72293 | negative | MGI_C57BL6J_5690359 | Gm43967 | ENSEMBL:ENSMUSG00000108036 | MGI:5690359 | pseudogene | predicted gene, 43967 |
| 6 | gene | 52.76806 | 53.06863 | negative | MGI_C57BL6J_2141450 | Jazf1 | NCBI_Gene:231986,ENSEMBL:ENSMUSG00000063568 | MGI:2141450 | protein coding gene | JAZF zinc finger 1 |
| 6 | pseudogene | 52.84311 | 52.84370 | negative | MGI_C57BL6J_3783020 | Gm15572 | ENSEMBL:ENSMUSG00000081872 | MGI:3783020 | pseudogene | predicted gene 15572 |
| 6 | gene | 52.96516 | 52.97656 | negative | MGI_C57BL6J_5592378 | Gm33219 | NCBI_Gene:102636035 | MGI:5592378 | lncRNA gene | predicted gene, 33219 |
| 6 | gene | 53.01039 | 53.01053 | negative | MGI_C57BL6J_5455992 | Gm26215 | ENSEMBL:ENSMUSG00000089209 | MGI:5455992 | snoRNA gene | predicted gene, 26215 |
| 6 | pseudogene | 53.04562 | 53.04705 | positive | MGI_C57BL6J_3783021 | Gm15573 | NCBI_Gene:108169143,ENSEMBL:ENSMUSG00000080759 | MGI:3783021 | pseudogene | predicted gene 15573 |
| 6 | gene | 53.06009 | 53.06595 | negative | MGI_C57BL6J_5592256 | Gm33097 | NCBI_Gene:102635867 | MGI:5592256 | lncRNA gene | predicted gene, 33097 |
| 6 | pseudogene | 53.17055 | 53.17174 | negative | MGI_C57BL6J_3644270 | Gm8143 | NCBI_Gene:666516,ENSEMBL:ENSMUSG00000108028 | MGI:3644270 | pseudogene | predicted gene 8143 |
| 6 | gene | 53.22116 | 53.22660 | positive | MGI_C57BL6J_3779443 | Gm4872 | NCBI_Gene:231989,ENSEMBL:ENSMUSG00000107935 | MGI:3779443 | lncRNA gene | predicted gene 4872 |
| 6 | gene | 53.26248 | 53.26823 | negative | MGI_C57BL6J_5592507 | Gm33348 | NCBI_Gene:102636219 | MGI:5592507 | lncRNA gene | predicted gene, 33348 |
| 6 | gene | 53.28722 | 53.70038 | positive | MGI_C57BL6J_2443973 | Creb5 | NCBI_Gene:231991,ENSEMBL:ENSMUSG00000053007 | MGI:2443973 | protein coding gene | cAMP responsive element binding protein 5 |
| 6 | gene | 53.43397 | 53.43414 | positive | MGI_C57BL6J_5455595 | Gm25818 | ENSEMBL:ENSMUSG00000096147 | MGI:5455595 | snRNA gene | predicted gene, 25818 |
| 6 | gene | 53.46951 | 53.47175 | negative | MGI_C57BL6J_5621700 | Gm38815 | NCBI_Gene:105242662 | MGI:5621700 | lncRNA gene | predicted gene, 38815 |
| 6 | gene | 53.49354 | 53.49828 | positive | MGI_C57BL6J_5621699 | Gm38814 | NCBI_Gene:105242661 | MGI:5621699 | lncRNA gene | predicted gene, 38814 |
| 6 | gene | 53.58135 | 53.59288 | negative | MGI_C57BL6J_5592551 | Gm33392 | NCBI_Gene:102636280 | MGI:5592551 | lncRNA gene | predicted gene, 33392 |
| 6 | gene | 53.66777 | 53.66998 | positive | MGI_C57BL6J_5690472 | Gm44080 | ENSEMBL:ENSMUSG00000108004 | MGI:5690472 | unclassified gene | predicted gene, 44080 |
| 6 | pseudogene | 53.77736 | 53.77818 | positive | MGI_C57BL6J_3644732 | Gm5306 | NCBI_Gene:384386,ENSEMBL:ENSMUSG00000107984 | MGI:3644732 | pseudogene | predicted gene 5306 |
| 6 | gene | 53.81547 | 53.82083 | negative | MGI_C57BL6J_1914123 | Tril | NCBI_Gene:66873,ENSEMBL:ENSMUSG00000043496 | MGI:1914123 | protein coding gene | TLR4 interactor with leucine-rich repeats |
| 6 | gene | 53.81874 | 53.86818 | positive | MGI_C57BL6J_3704267 | Gm16499 | NCBI_Gene:100049166,ENSEMBL:ENSMUSG00000108255,ENSEMBL:ENSMUSG00000108175 | MGI:3704267 | lncRNA gene | predicted gene 16499 |
| 6 | gene | 53.84417 | 53.84506 | negative | MGI_C57BL6J_1923135 | 4930566K11Rik | NA | NA | unclassified gene | RIKEN cDNA 4930566K11 gene |
| 6 | gene | 53.87202 | 53.87215 | negative | MGI_C57BL6J_5452687 | Gm22910 | ENSEMBL:ENSMUSG00000094527 | MGI:5452687 | snoRNA gene | predicted gene, 22910 |
| 6 | gene | 53.87328 | 53.97867 | negative | MGI_C57BL6J_1918537 | Cpvl | NCBI_Gene:71287,ENSEMBL:ENSMUSG00000052955 | MGI:1918537 | protein coding gene | carboxypeptidase, vitellogenic-like |
| 6 | gene | 53.88076 | 53.88310 | positive | MGI_C57BL6J_5592875 | Gm33716 | NCBI_Gene:102636723 | MGI:5592875 | lncRNA gene | predicted gene, 33716 |
| 6 | gene | 53.97869 | 54.01803 | positive | MGI_C57BL6J_2443531 | 4921529L05Rik | NCBI_Gene:320178,ENSEMBL:ENSMUSG00000108060 | MGI:2443531 | lncRNA gene | RIKEN cDNA 4921529L05 gene |
| 6 | gene | 54.03955 | 54.30181 | positive | MGI_C57BL6J_1917243 | Chn2 | NCBI_Gene:69993,ENSEMBL:ENSMUSG00000004633 | MGI:1917243 | protein coding gene | chimerin 2 |
| 6 | gene | 54.13952 | 54.15144 | negative | MGI_C57BL6J_1925866 | 9530036M11Rik | NCBI_Gene:78616,ENSEMBL:ENSMUSG00000087670 | MGI:1925866 | lncRNA gene | RIKEN cDNA 9530036M11 gene |
| 6 | pseudogene | 54.17720 | 54.17773 | negative | MGI_C57BL6J_3782973 | Gm15526 | NCBI_Gene:108169144,ENSEMBL:ENSMUSG00000080205 | MGI:3782973 | pseudogene | predicted gene 15526 |
| 6 | gene | 54.24574 | 54.27908 | negative | MGI_C57BL6J_3782974 | Gm15527 | ENSEMBL:ENSMUSG00000086946 | MGI:3782974 | lncRNA gene | predicted gene 15527 |
| 6 | gene | 54.26968 | 54.43022 | negative | MGI_C57BL6J_1918824 | 9130019P16Rik | NCBI_Gene:100042056,ENSEMBL:ENSMUSG00000073067 | MGI:1918824 | lncRNA gene | RIKEN cDNA 9130019P16 gene |
| 6 | gene | 54.32583 | 54.32812 | negative | MGI_C57BL6J_1921781 | 9030405F24Rik | NA | NA | unclassified gene | RIKEN cDNA 9030405F24 gene |
| 6 | gene | 54.32583 | 54.33020 | positive | MGI_C57BL6J_1925254 | Prr15 | NCBI_Gene:78004,ENSEMBL:ENSMUSG00000045725 | MGI:1925254 | protein coding gene | proline rich 15 |
| 6 | pseudogene | 54.32964 | 54.34344 | positive | MGI_C57BL6J_5593328 | Gm34169 | NCBI_Gene:102637330 | MGI:5593328 | pseudogene | predicted gene, 34169 |
| 6 | gene | 54.42922 | 54.50377 | positive | MGI_C57BL6J_3044681 | Wipf3 | NCBI_Gene:330319,ENSEMBL:ENSMUSG00000086040 | MGI:3044681 | protein coding gene | WAS/WASL interacting protein family, member 3 |
| 6 | gene | 54.46027 | 54.47468 | negative | MGI_C57BL6J_5690418 | Gm44026 | NCBI_Gene:108169145,ENSEMBL:ENSMUSG00000108081 | MGI:5690418 | lncRNA gene | predicted gene, 44026 |
| 6 | gene | 54.47479 | 54.56652 | negative | MGI_C57BL6J_1917188 | Scrn1 | NCBI_Gene:69938,ENSEMBL:ENSMUSG00000019124 | MGI:1917188 | protein coding gene | secernin 1 |
| 6 | gene | 54.57760 | 54.59731 | negative | MGI_C57BL6J_2387639 | Fkbp14 | NCBI_Gene:231997,ENSEMBL:ENSMUSG00000038074 | MGI:2387639 | protein coding gene | FK506 binding protein 14 |
| 6 | gene | 54.59511 | 54.64584 | positive | MGI_C57BL6J_2681164 | Plekha8 | NCBI_Gene:231999,ENSEMBL:ENSMUSG00000005225 | MGI:2681164 | protein coding gene | pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| 6 | gene | 54.65944 | 54.66256 | positive | MGI_C57BL6J_5593408 | Gm34249 | NCBI_Gene:102637443 | MGI:5593408 | lncRNA gene | predicted gene, 34249 |
| 6 | gene | 54.68093 | 54.68192 | negative | MGI_C57BL6J_5579108 | Gm28402 | ENSEMBL:ENSMUSG00000101651 | MGI:5579108 | lncRNA gene | predicted gene 28402 |
| 6 | gene | 54.68162 | 54.70385 | positive | MGI_C57BL6J_1915485 | Mturn | NCBI_Gene:68235,ENSEMBL:ENSMUSG00000038065 | MGI:1915485 | protein coding gene | maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| 6 | gene | 54.69365 | 54.69406 | positive | MGI_C57BL6J_5690400 | Gm44008 | ENSEMBL:ENSMUSG00000108283 | MGI:5690400 | unclassified gene | predicted gene, 44008 |
| 6 | gene | 54.81375 | 54.81765 | negative | MGI_C57BL6J_1917699 | 2610209C05Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 2610209C05 gene |
| 6 | gene | 54.81692 | 54.89350 | positive | MGI_C57BL6J_1196246 | Znrf2 | NCBI_Gene:387524,ENSEMBL:ENSMUSG00000058446 | MGI:1196246 | protein coding gene | zinc and ring finger 2 |
| 6 | gene | 54.90242 | 54.90573 | positive | MGI_C57BL6J_5690577 | Gm44185 | ENSEMBL:ENSMUSG00000107841 | MGI:5690577 | unclassified gene | predicted gene, 44185 |
| 6 | gene | 54.92394 | 54.97267 | negative | MGI_C57BL6J_1341839 | Nod1 | NCBI_Gene:107607,ENSEMBL:ENSMUSG00000038058 | MGI:1341839 | protein coding gene | nucleotide-binding oligomerization domain containing 1 |
| 6 | gene | 54.97465 | 54.97478 | positive | MGI_C57BL6J_5454007 | Gm24230 | ENSEMBL:ENSMUSG00000077501 | MGI:5454007 | snoRNA gene | predicted gene, 24230 |
| 6 | gene | 54.98258 | 54.99296 | negative | MGI_C57BL6J_95700 | Ggct | NCBI_Gene:110175,ENSEMBL:ENSMUSG00000002797 | MGI:95700 | protein coding gene | gamma-glutamyl cyclotransferase |
| 6 | gene | 55.00212 | 55.00400 | positive | MGI_C57BL6J_5621424 | Gm38539 | NCBI_Gene:102641179 | MGI:5621424 | lncRNA gene | predicted gene, 38539 |
| 6 | gene | 55.03800 | 55.07950 | positive | MGI_C57BL6J_2449057 | Gars | NCBI_Gene:353172,ENSEMBL:ENSMUSG00000029777 | MGI:2449057 | protein coding gene | glycyl-tRNA synthetase |
| 6 | gene | 55.09005 | 55.13302 | negative | MGI_C57BL6J_894312 | Crhr2 | NCBI_Gene:12922,ENSEMBL:ENSMUSG00000003476 | MGI:894312 | protein coding gene | corticotropin releasing hormone receptor 2 |
| 6 | gene | 55.15443 | 55.16721 | positive | MGI_C57BL6J_5593517 | Gm34358 | NCBI_Gene:102637587,ENSEMBL:ENSMUSG00000108285 | MGI:5593517 | lncRNA gene | predicted gene, 34358 |
| 6 | gene | 55.17063 | 55.17504 | negative | MGI_C57BL6J_102963 | Inmt | NCBI_Gene:21743,ENSEMBL:ENSMUSG00000003477 | MGI:102963 | protein coding gene | indolethylamine N-methyltransferase |
| 6 | gene | 55.20334 | 55.32022 | positive | MGI_C57BL6J_3583959 | Mindy4 | NCBI_Gene:330323,ENSEMBL:ENSMUSG00000038022 | MGI:3583959 | protein coding gene | MINDY lysine 48 deubiquitinase 4 |
| 6 | gene | 55.29626 | 55.30789 | negative | MGI_C57BL6J_5690465 | Gm44073 | ENSEMBL:ENSMUSG00000107766 | MGI:5690465 | lncRNA gene | predicted gene, 44073 |
| 6 | gene | 55.30880 | 55.30891 | negative | MGI_C57BL6J_5455235 | Gm25458 | ENSEMBL:ENSMUSG00000093878 | MGI:5455235 | miRNA gene | predicted gene, 25458 |
| 6 | gene | 55.32564 | 55.33084 | negative | MGI_C57BL6J_5621704 | Gm38819 | NCBI_Gene:105242666 | MGI:5621704 | lncRNA gene | predicted gene, 38819 |
| 6 | gene | 55.33630 | 55.34855 | positive | MGI_C57BL6J_103201 | Aqp1 | NCBI_Gene:11826,ENSEMBL:ENSMUSG00000004655 | MGI:103201 | protein coding gene | aquaporin 1 |
| 6 | gene | 55.37629 | 55.38853 | positive | MGI_C57BL6J_95710 | Ghrhr | NCBI_Gene:14602,ENSEMBL:ENSMUSG00000004654 | MGI:95710 | protein coding gene | growth hormone releasing hormone receptor |
| 6 | gene | 55.39689 | 55.41228 | positive | MGI_C57BL6J_5439415 | 6430584L05Rik | NCBI_Gene:330324,ENSEMBL:ENSMUSG00000108228 | MGI:5439415 | lncRNA gene | RIKEN cDNA 6430584L05 gene |
| 6 | gene | 55.45150 | 55.50145 | positive | MGI_C57BL6J_108449 | Adcyap1r1 | NCBI_Gene:11517,ENSEMBL:ENSMUSG00000029778 | MGI:108449 | protein coding gene | adenylate cyclase activating polypeptide 1 receptor 1 |
| 6 | gene | 55.53351 | 55.53361 | positive | MGI_C57BL6J_5690744 | Gm44352 | ENSEMBL:ENSMUSG00000104746 | MGI:5690744 | miRNA gene | predicted gene, 44352 |
| 6 | gene | 55.53704 | 55.65178 | positive | MGI_C57BL6J_5593600 | Gm34441 | NCBI_Gene:102637698 | MGI:5593600 | lncRNA gene | predicted gene, 34441 |
| 6 | pseudogene | 55.62508 | 55.62561 | positive | MGI_C57BL6J_3781904 | Gm3729 | NCBI_Gene:100416934 | MGI:3781904 | pseudogene | predicted gene 3729 |
| 6 | gene | 55.67782 | 55.68126 | negative | MGI_C57BL6J_106593 | Neurod6 | NCBI_Gene:11922,ENSEMBL:ENSMUSG00000037984 | MGI:106593 | protein coding gene | neurogenic differentiation 6 |
| 6 | gene | 55.68457 | 55.69457 | positive | MGI_C57BL6J_3781457 | Gm3279 | NCBI_Gene:100041339,ENSEMBL:ENSMUSG00000108063 | MGI:3781457 | lncRNA gene | predicted gene 3279 |
| 6 | gene | 55.70057 | 55.70589 | positive | MGI_C57BL6J_5593835 | Gm34676 | NCBI_Gene:102638001 | MGI:5593835 | lncRNA gene | predicted gene, 34676 |
| 6 | gene | 55.78959 | 55.80470 | negative | MGI_C57BL6J_5621705 | Gm38820 | NCBI_Gene:105242667 | MGI:5621705 | lncRNA gene | predicted gene, 38820 |
| 6 | gene | 55.80463 | 55.97874 | positive | MGI_C57BL6J_2685304 | Itprid1 | NCBI_Gene:232016,ENSEMBL:ENSMUSG00000037973 | MGI:2685304 | protein coding gene | ITPR interacting domain containing 1 |
| 6 | gene | 56.01750 | 56.03269 | positive | MGI_C57BL6J_1333876 | Ppp1r17 | NCBI_Gene:19051,ENSEMBL:ENSMUSG00000002930 | MGI:1333876 | protein coding gene | protein phosphatase 1, regulatory subunit 17 |
| 6 | pseudogene | 56.06094 | 56.06148 | negative | MGI_C57BL6J_5010284 | Gm18099 | NCBI_Gene:100416405 | MGI:5010284 | pseudogene | predicted gene, 18099 |
| 6 | gene | 56.06980 | 56.65251 | negative | MGI_C57BL6J_108413 | Pde1c | NCBI_Gene:18575,ENSEMBL:ENSMUSG00000004347 | MGI:108413 | protein coding gene | phosphodiesterase 1C |
| 6 | gene | 56.17293 | 56.17303 | negative | MGI_C57BL6J_5690805 | Gm44413 | ENSEMBL:ENSMUSG00000106152 | MGI:5690805 | miRNA gene | predicted gene, 44413 |
| 6 | gene | 56.36971 | 56.37194 | positive | MGI_C57BL6J_5593912 | Gm34753 | NA | NA | unclassified non-coding RNA gene | predicted gene%2c 34753 |
| 6 | gene | 56.58120 | 56.65249 | negative | MGI_C57BL6J_3644042 | Gm8239 | ENSEMBL:ENSMUSG00000107494 | MGI:3644042 | lncRNA gene | predicted gene 8239 |
| 6 | gene | 56.58371 | 56.58385 | negative | MGI_C57BL6J_5454486 | Gm24709 | ENSEMBL:ENSMUSG00000096087 | MGI:5454486 | snoRNA gene | predicted gene, 24709 |
| 6 | gene | 56.60082 | 56.60095 | negative | MGI_C57BL6J_5452270 | Gm22493 | ENSEMBL:ENSMUSG00000077682 | MGI:5452270 | snoRNA gene | predicted gene, 22493 |
| 6 | gene | 56.70106 | 56.70471 | negative | MGI_C57BL6J_1913623 | Lsm5 | NCBI_Gene:66373,ENSEMBL:ENSMUSG00000091625 | MGI:1913623 | protein coding gene | LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| 6 | gene | 56.71450 | 56.71573 | negative | MGI_C57BL6J_1918466 | 4933427C19Rik | NA | NA | unclassified gene | RIKEN cDNA 4933427C19 gene |
| 6 | gene | 56.71490 | 56.76191 | positive | MGI_C57BL6J_1926187 | Avl9 | NCBI_Gene:78937,ENSEMBL:ENSMUSG00000029787 | MGI:1926187 | protein coding gene | AVL9 cell migration associated |
| 6 | gene | 56.77582 | 56.77735 | negative | MGI_C57BL6J_1920419 | 3110035G12Rik | NA | NA | unclassified gene | RIKEN cDNA 3110035G12 gene |
| 6 | gene | 56.77752 | 56.79827 | negative | MGI_C57BL6J_2384811 | Kbtbd2 | NCBI_Gene:210973,ENSEMBL:ENSMUSG00000059486 | MGI:2384811 | protein coding gene | kelch repeat and BTB (POZ) domain containing 2 |
| 6 | gene | 56.79759 | 56.79920 | positive | MGI_C57BL6J_3641874 | Gm10209 | ENSEMBL:ENSMUSG00000108080 | MGI:3641874 | protein coding gene | predicted gene 10209 |
| 6 | gene | 56.83206 | 56.87936 | positive | MGI_C57BL6J_1350921 | Fkbp9 | NCBI_Gene:27055,ENSEMBL:ENSMUSG00000029781 | MGI:1350921 | protein coding gene | FK506 binding protein 9 |
| 6 | gene | 56.88240 | 56.92393 | negative | MGI_C57BL6J_1927186 | Nt5c3 | NCBI_Gene:107569,ENSEMBL:ENSMUSG00000029780 | MGI:1927186 | protein coding gene | 5’-nucleotidase, cytosolic III |
| 6 | gene | 56.88443 | 56.88611 | positive | MGI_C57BL6J_5609018 | AK013988 | NA | NA | unclassified non-coding RNA gene | expressed sequence AK013988 |
| 6 | gene | 56.90591 | 56.90658 | negative | MGI_C57BL6J_3781966 | Gm3793 | ENSEMBL:ENSMUSG00000108260 | MGI:3781966 | unclassified gene | predicted gene 3793 |
| 6 | gene | 56.92401 | 56.95810 | positive | MGI_C57BL6J_2159457 | Vmn1r4 | NCBI_Gene:171194,ENSEMBL:ENSMUSG00000071893 | MGI:2159457 | protein coding gene | vomeronasal 1 receptor 4 |
| 6 | gene | 56.94184 | 56.94482 | negative | MGI_C57BL6J_5690441 | Gm44049 | ENSEMBL:ENSMUSG00000108021 | MGI:5690441 | unclassified gene | predicted gene, 44049 |
| 6 | gene | 56.97027 | 56.98839 | positive | MGI_C57BL6J_2159455 | Vmn1r5 | NCBI_Gene:171192,ENSEMBL:ENSMUSG00000090346 | MGI:2159455 | protein coding gene | vomeronasal 1 receptor 5 |
| 6 | gene | 56.99724 | 57.00982 | positive | MGI_C57BL6J_2159456 | Vmn1r6 | NCBI_Gene:171193,ENSEMBL:ENSMUSG00000115701 | MGI:2159456 | protein coding gene | vomeronasal 1 receptor 6 |
| 6 | gene | 57.02411 | 57.02532 | negative | MGI_C57BL6J_2159467 | Vmn1r7 | NCBI_Gene:434016,ENSEMBL:ENSMUSG00000093696 | MGI:2159467 | protein coding gene | vomeronasal 1 receptor 7 |
| 6 | gene | 57.03228 | 57.03739 | positive | MGI_C57BL6J_2159469 | Vmn1r8 | NCBI_Gene:171205,ENSEMBL:ENSMUSG00000061208 | MGI:2159469 | protein coding gene | vomeronasal 1 receptor 8 |
| 6 | gene | 57.06679 | 57.07491 | positive | MGI_C57BL6J_2159466 | Vmn1r9 | NCBI_Gene:171203,ENSEMBL:ENSMUSG00000091541 | MGI:2159466 | protein coding gene | vomeronasal 1 receptor 9 |
| 6 | gene | 57.10945 | 57.11503 | positive | MGI_C57BL6J_2148522 | Vmn1r10 | NCBI_Gene:113858,ENSEMBL:ENSMUSG00000115181 | MGI:2148522 | protein coding gene | vomeronasal 1 receptor 10 |
| 6 | gene | 57.13345 | 57.13962 | positive | MGI_C57BL6J_2148524 | Vmn1r11 | NCBI_Gene:113860,ENSEMBL:ENSMUSG00000115236 | MGI:2148524 | protein coding gene | vomeronasal 1 receptor 11 |
| 6 | gene | 57.15692 | 57.16533 | positive | MGI_C57BL6J_3645555 | Vmn1r12 | NCBI_Gene:626397,ENSEMBL:ENSMUSG00000057981 | MGI:3645555 | protein coding gene | vomeronasal 1 receptor 12 |
| 6 | pseudogene | 57.18849 | 57.18944 | positive | MGI_C57BL6J_2148525 | Vmn1r-ps8 | NCBI_Gene:113861,ENSEMBL:ENSMUSG00000093728 | MGI:2148525 | pseudogene | vomeronasal 1 receptor, pseudogene 8 |
| 6 | gene | 57.20090 | 57.21480 | positive | MGI_C57BL6J_2148526 | Vmn1r13 | NCBI_Gene:113862,ENSEMBL:ENSMUSG00000064259 | MGI:2148526 | protein coding gene | vomeronasal 1 receptor 13 |
| 6 | gene | 57.22615 | 57.24088 | positive | MGI_C57BL6J_2148528 | Vmn1r14 | NCBI_Gene:113864,ENSEMBL:ENSMUSG00000114982 | MGI:2148528 | protein coding gene | vomeronasal 1 receptor 14 |
| 6 | gene | 57.25795 | 57.25997 | positive | MGI_C57BL6J_2148527 | Vmn1r15 | NCBI_Gene:113863,ENSEMBL:ENSMUSG00000115199 | MGI:2148527 | protein coding gene | vomeronasal 1 receptor 15 |
| 6 | pseudogene | 57.27802 | 57.27867 | positive | MGI_C57BL6J_3781521 | Vmn1r-ps9 | NCBI_Gene:100041445,ENSEMBL:ENSMUSG00000093520 | MGI:3781521 | pseudogene | vomeronasal 1 receptor, pseudogene 9 |
| 6 | pseudogene | 57.31294 | 57.31383 | positive | MGI_C57BL6J_2148536 | Vmn1r-ps10 | NCBI_Gene:113874,ENSEMBL:ENSMUSG00000093621 | MGI:2148536 | pseudogene | vomeronasal 1 receptor, pseudogene 10 |
| 6 | gene | 57.31805 | 57.32949 | negative | MGI_C57BL6J_2159465 | Vmn1r16 | NCBI_Gene:171202,ENSEMBL:ENSMUSG00000115792 | MGI:2159465 | protein coding gene | vomeronasal 1 receptor 16 |
| 6 | gene | 57.35924 | 57.36373 | negative | MGI_C57BL6J_2159452 | Vmn1r17 | NCBI_Gene:171189,ENSEMBL:ENSMUSG00000115644 | MGI:2159452 | protein coding gene | vomeronasal 1 receptor 17 |
| 6 | gene | 57.37666 | 57.39578 | negative | MGI_C57BL6J_2159462 | Vmn1r18 | NCBI_Gene:171199,ENSEMBL:ENSMUSG00000091382 | MGI:2159462 | protein coding gene | vomeronasal 1 receptor 18 |
| 6 | gene | 57.40378 | 57.40603 | positive | MGI_C57BL6J_2159463 | Vmn1r19 | NCBI_Gene:171200,ENSEMBL:ENSMUSG00000115799 | MGI:2159463 | protein coding gene | vomeronasal 1 receptor 19 |
| 6 | gene | 57.42893 | 57.43526 | positive | MGI_C57BL6J_3644381 | Vmn1r20 | NCBI_Gene:434017,ENSEMBL:ENSMUSG00000115253 | MGI:3644381 | protein coding gene | vomeronasal 1 receptor 20 |
| 6 | pseudogene | 57.44736 | 57.44833 | negative | MGI_C57BL6J_3643544 | Vmn1r-ps11 | NCBI_Gene:666822,ENSEMBL:ENSMUSG00000093398 | MGI:3643544 | pseudogene | vomeronasal 1 receptor, pseudogene 11 |
| 6 | gene | 57.46407 | 57.49953 | positive | MGI_C57BL6J_3612486 | 4930533I22Rik | NCBI_Gene:434018,ENSEMBL:ENSMUSG00000093535 | MGI:3612486 | lncRNA gene | RIKEN cDNA 4930533I22 gene |
| 6 | gene | 57.48512 | 57.62603 | positive | MGI_C57BL6J_5753769 | Gm45193 | ENSEMBL:ENSMUSG00000108112 | MGI:5753769 | lncRNA gene | predicted gene 45193 |
| 6 | gene | 57.50650 | 57.53553 | negative | MGI_C57BL6J_2442111 | Ppm1k | NCBI_Gene:243382,ENSEMBL:ENSMUSG00000037826 | MGI:2442111 | protein coding gene | protein phosphatase 1K (PP2C domain containing) |
| 6 | gene | 57.56579 | 57.56611 | positive | MGI_C57BL6J_5531278 | Gm27896 | ENSEMBL:ENSMUSG00000092903 | MGI:5531278 | unclassified non-coding RNA gene | predicted gene, 27896 |
| 6 | gene | 57.58099 | 57.66514 | positive | MGI_C57BL6J_1914388 | Herc6 | NCBI_Gene:67138,ENSEMBL:ENSMUSG00000029798 | MGI:1914388 | protein coding gene | hect domain and RLD 6 |
| 6 | pseudogene | 57.61697 | 57.61731 | negative | MGI_C57BL6J_5011429 | Gm19244 | NCBI_Gene:100462737,ENSEMBL:ENSMUSG00000107627 | MGI:5011429 | pseudogene | predicted gene, 19244 |
| 6 | gene | 57.68474 | 57.68758 | negative | MGI_C57BL6J_5610625 | Gm37397 | ENSEMBL:ENSMUSG00000104204 | MGI:5610625 | unclassified gene | predicted gene, 37397 |
| 6 | gene | 57.68474 | 57.69208 | negative | MGI_C57BL6J_1913709 | Pyurf | NCBI_Gene:66459,ENSEMBL:ENSMUSG00000043162 | MGI:1913709 | protein coding gene | Pigy upstream reading frame |
| 6 | pseudogene | 57.68943 | 57.68965 | negative | MGI_C57BL6J_5477206 | Gm26712 | ENSEMBL:ENSMUSG00000097656 | MGI:5477206 | pseudogene | predicted gene, 26712 |
| 6 | gene | 57.70246 | 57.73945 | positive | MGI_C57BL6J_1919085 | Lancl2 | NCBI_Gene:71835,ENSEMBL:ENSMUSG00000062190 | MGI:1919085 | protein coding gene | LanC (bacterial lantibiotic synthetase component C)-like 2 |
| 6 | gene | 57.72781 | 57.82537 | negative | MGI_C57BL6J_2141658 | Vopp1 | NCBI_Gene:232023,ENSEMBL:ENSMUSG00000037788 | MGI:2141658 | protein coding gene | vesicular, overexpressed in cancer, prosurvival protein 1 |
| 6 | gene | 57.84283 | 57.85034 | negative | MGI_C57BL6J_2159464 | Vmn1r21 | NCBI_Gene:171201,ENSEMBL:ENSMUSG00000115343 | MGI:2159464 | protein coding gene | vomeronasal 1 receptor 21 |
| 6 | gene | 57.84559 | 57.84831 | positive | MGI_C57BL6J_5690506 | Gm44114 | ENSEMBL:ENSMUSG00000107431 | MGI:5690506 | unclassified gene | predicted gene, 44114 |
| 6 | gene | 57.85982 | 57.85995 | negative | MGI_C57BL6J_5455503 | Gm25726 | ENSEMBL:ENSMUSG00000077717 | MGI:5455503 | snoRNA gene | predicted gene, 25726 |
| 6 | pseudogene | 57.87241 | 57.87292 | negative | MGI_C57BL6J_3643065 | Vmn1r-ps12 | NCBI_Gene:626582,ENSEMBL:ENSMUSG00000093530 | MGI:3643065 | pseudogene | vomeronasal 1 receptor, pseudogene 12 |
| 6 | gene | 57.89813 | 57.90803 | negative | MGI_C57BL6J_2159459 | Vmn1r22 | NCBI_Gene:171196,ENSEMBL:ENSMUSG00000115091 | MGI:2159459 | protein coding gene | vomeronasal 1 receptor 22 |
| 6 | gene | 57.92584 | 57.92684 | negative | MGI_C57BL6J_2159460 | Vmn1r23 | NCBI_Gene:171197,ENSEMBL:ENSMUSG00000093376 | MGI:2159460 | protein coding gene | vomeronasal 1 receptor 23 |
| 6 | pseudogene | 57.94968 | 57.94997 | negative | MGI_C57BL6J_5313107 | Gm20660 | ENSEMBL:ENSMUSG00000093541 | MGI:5313107 | pseudogene | predicted gene 20660 |
| 6 | pseudogene | 57.95007 | 57.95047 | positive | MGI_C57BL6J_3781560 | Gm3382 | NCBI_Gene:100041514 | MGI:3781560 | pseudogene | predicted gene 3382 |
| 6 | gene | 57.95471 | 57.96150 | negative | MGI_C57BL6J_2159454 | Vmn1r24 | NCBI_Gene:171191,ENSEMBL:ENSMUSG00000115507 | MGI:2159454 | protein coding gene | vomeronasal 1 receptor 24 |
| 6 | gene | 57.97830 | 57.98081 | negative | MGI_C57BL6J_2148529 | Vmn1r25 | NCBI_Gene:113865,ENSEMBL:ENSMUSG00000115668 | MGI:2148529 | protein coding gene | vomeronasal 1 receptor 25 |
| 6 | pseudogene | 57.99299 | 57.99358 | negative | MGI_C57BL6J_3852363 | Vmn1r-ps13 | NCBI_Gene:100312486,ENSEMBL:ENSMUSG00000107678 | MGI:3852363 | pseudogene | vomeronasal 1 receptor, pseudogene 13 |
| 6 | gene | 58.00493 | 58.01516 | negative | MGI_C57BL6J_2159453 | Vmn1r26 | NCBI_Gene:171190,ENSEMBL:ENSMUSG00000048697 | MGI:2159453 | protein coding gene | vomeronasal 1 receptor 26 |
| 6 | pseudogene | 58.01748 | 58.01794 | positive | MGI_C57BL6J_3647234 | Rps15-ps2 | NCBI_Gene:633683,ENSEMBL:ENSMUSG00000071419 | MGI:3647234 | pseudogene | ribosomal protein S15, pseudogene 2 |
| 6 | pseudogene | 58.04645 | 58.04990 | negative | MGI_C57BL6J_3645438 | Vmn1r-ps14 | NCBI_Gene:626649,ENSEMBL:ENSMUSG00000093382 | MGI:3645438 | pseudogene | vomeronasal 1 receptor, pseudogene 14 |
| 6 | pseudogene | 58.07360 | 58.07913 | negative | MGI_C57BL6J_3648676 | Vmn1r-ps15 | NCBI_Gene:626659,ENSEMBL:ENSMUSG00000093666 | MGI:3648676 | pseudogene | vomeronasal 1 receptor, pseudogene 15 |
| 6 | pseudogene | 58.09108 | 58.09267 | positive | MGI_C57BL6J_3648885 | Gm6695 | NCBI_Gene:626672,ENSEMBL:ENSMUSG00000093560 | MGI:3648885 | pseudogene | predicted gene 6695 |
| 6 | pseudogene | 58.10802 | 58.10825 | negative | MGI_C57BL6J_3852364 | Vmn1r-ps16 | NCBI_Gene:100312487,ENSEMBL:ENSMUSG00000107530 | MGI:3852364 | pseudogene | vomeronasal 1 receptor, pseudogene 16 |
| 6 | pseudogene | 58.11573 | 58.11663 | negative | MGI_C57BL6J_3643137 | Vmn1r-ps17 | NCBI_Gene:666893,ENSEMBL:ENSMUSG00000093743 | MGI:3643137 | pseudogene | vomeronasal 1 receptor, pseudogene 17 |
| 6 | pseudogene | 58.13589 | 58.13614 | positive | MGI_C57BL6J_5313165 | Gm20718 | ENSEMBL:ENSMUSG00000093708 | MGI:5313165 | pseudogene | predicted gene 20718 |
| 6 | gene | 58.19232 | 58.19476 | positive | MGI_C57BL6J_5690386 | Gm43994 | ENSEMBL:ENSMUSG00000107521 | MGI:5690386 | lncRNA gene | predicted gene, 43994 |
| 6 | pseudogene | 58.20569 | 58.20700 | positive | MGI_C57BL6J_3781590 | Vmn1r-ps18 | NCBI_Gene:100041571,ENSEMBL:ENSMUSG00000093731 | MGI:3781590 | pseudogene | vomeronasal 1 receptor, pseudogene 18 |
| 6 | gene | 58.21309 | 58.21882 | negative | MGI_C57BL6J_2159470 | Vmn1r27 | NCBI_Gene:171206,ENSEMBL:ENSMUSG00000071428 | MGI:2159470 | protein coding gene | vomeronasal 1 receptor 27 |
| 6 | pseudogene | 58.25662 | 58.25755 | negative | MGI_C57BL6J_3647074 | Vmn1r-ps19 | NCBI_Gene:626724,ENSEMBL:ENSMUSG00000093522 | MGI:3647074 | pseudogene | vomeronasal 1 receptor, pseudogene 19 |
| 6 | gene | 58.26204 | 58.27564 | positive | MGI_C57BL6J_2159461 | Vmn1r28 | NCBI_Gene:171198,ENSEMBL:ENSMUSG00000115705 | MGI:2159461 | protein coding gene | vomeronasal 1 receptor 28 |
| 6 | gene | 58.30155 | 58.31168 | positive | MGI_C57BL6J_2148523 | Vmn1r29 | NCBI_Gene:113859,ENSEMBL:ENSMUSG00000091734 | MGI:2148523 | protein coding gene | vomeronasal 1 receptor 29 |
| 6 | pseudogene | 58.39405 | 58.39442 | positive | MGI_C57BL6J_5313121 | Gm20674 | ENSEMBL:ENSMUSG00000093547 | MGI:5313121 | pseudogene | predicted gene 20674 |
| 6 | pseudogene | 58.40641 | 58.40736 | negative | MGI_C57BL6J_3645210 | Vmn1r-ps20 | NCBI_Gene:666928,ENSEMBL:ENSMUSG00000093702 | MGI:3645210 | pseudogene | vomeronasal 1 receptor, pseudogene 20 |
| 6 | gene | 58.43416 | 58.44362 | negative | MGI_C57BL6J_2159458 | Vmn1r30 | NCBI_Gene:171195,ENSEMBL:ENSMUSG00000095670 | MGI:2159458 | protein coding gene | vomeronasal 1 receptor 30 |
| 6 | pseudogene | 58.43870 | 58.43886 | negative | MGI_C57BL6J_5690408 | Gm44016 | ENSEMBL:ENSMUSG00000107518 | MGI:5690408 | pseudogene | predicted gene, 44016 |
| 6 | pseudogene | 58.45507 | 58.45599 | negative | MGI_C57BL6J_3852365 | Vmn1r-ps21 | NCBI_Gene:100312488,ENSEMBL:ENSMUSG00000093461 | MGI:3852365 | pseudogene | vomeronasal 1 receptor, pseudogene 21 |
| 6 | gene | 58.45787 | 58.53369 | negative | MGI_C57BL6J_5594236 | Gm35077 | NCBI_Gene:102638540,ENSEMBL:ENSMUSG00000107960 | MGI:5594236 | lncRNA gene | predicted gene, 35077 |
| 6 | pseudogene | 58.46111 | 58.46154 | positive | MGI_C57BL6J_5313120 | Gm20673 | ENSEMBL:ENSMUSG00000093747 | MGI:5313120 | pseudogene | predicted gene 20673 |
| 6 | gene | 58.47094 | 58.47533 | negative | MGI_C57BL6J_3649162 | Vmn1r31 | NCBI_Gene:626828,ENSEMBL:ENSMUSG00000115404 | MGI:3649162 | protein coding gene | vomeronasal 1 receptor 31 |
| 6 | pseudogene | 58.49434 | 58.49525 | negative | MGI_C57BL6J_3647778 | Vmn1r-ps22 | NCBI_Gene:666954,ENSEMBL:ENSMUSG00000093518 | MGI:3647778 | pseudogene | vomeronasal 1 receptor, pseudogene 22 |
| 6 | pseudogene | 58.50444 | 58.50474 | negative | MGI_C57BL6J_3852366 | Vmn1r-ps23 | NCBI_Gene:100312489,ENSEMBL:ENSMUSG00000093588 | MGI:3852366 | pseudogene | vomeronasal 1 receptor, pseudogene 23 |
| 6 | pseudogene | 58.51227 | 58.51253 | positive | MGI_C57BL6J_5313119 | Gm20672 | ENSEMBL:ENSMUSG00000093741 | MGI:5313119 | pseudogene | predicted gene 20672 |
| 6 | gene | 58.53397 | 58.58425 | negative | MGI_C57BL6J_5825580 | Gm45943 | NCBI_Gene:108167332 | MGI:5825580 | protein coding gene | predicted gene, 45943 |
| 6 | gene | 58.58449 | 58.69568 | positive | MGI_C57BL6J_1347061 | Abcg2 | NCBI_Gene:26357,ENSEMBL:ENSMUSG00000029802 | MGI:1347061 | protein coding gene | ATP binding cassette subfamily G member 2 (Junior blood group) |
| 6 | gene | 58.58575 | 58.58656 | positive | MGI_C57BL6J_1921131 | 4930430M16Rik | NA | NA | unclassified gene | RIKEN cDNA 4930430M16 gene |
| 6 | gene | 58.67971 | 58.68064 | negative | MGI_C57BL6J_1917453 | 2310061B05Rik | NA | NA | unclassified gene | RIKEN cDNA 2310061B05 gene |
| 6 | gene | 58.75891 | 58.81375 | negative | MGI_C57BL6J_5621707 | Gm38822 | NCBI_Gene:105242670 | MGI:5621707 | lncRNA gene | predicted gene, 38822 |
| 6 | pseudogene | 58.78169 | 58.78194 | negative | MGI_C57BL6J_5690291 | Gm43899 | ENSEMBL:ENSMUSG00000108164 | MGI:5690291 | pseudogene | predicted gene, 43899 |
| 6 | gene | 58.83145 | 58.92040 | positive | MGI_C57BL6J_1921248 | Herc3 | NCBI_Gene:73998,ENSEMBL:ENSMUSG00000029804 | MGI:1921248 | protein coding gene | hect domain and RLD 3 |
| 6 | gene | 58.90523 | 58.90713 | negative | MGI_C57BL6J_1923555 | Nap1l5 | NCBI_Gene:58243,ENSEMBL:ENSMUSG00000055430 | MGI:1923555 | protein coding gene | nucleosome assembly protein 1-like 5 |
| 6 | gene | 58.93209 | 59.02455 | negative | MGI_C57BL6J_1889842 | Fam13a | NCBI_Gene:58909,ENSEMBL:ENSMUSG00000037709 | MGI:1889842 | protein coding gene | family with sequence similarity 13, member A |
| 6 | gene | 58.96487 | 58.97223 | positive | MGI_C57BL6J_5753636 | Gm45060 | ENSEMBL:ENSMUSG00000107439 | MGI:5753636 | lncRNA gene | predicted gene 45060 |
| 6 | gene | 58.96522 | 58.99231 | positive | MGI_C57BL6J_1918734 | 8430415O14Rik | NCBI_Gene:71484 | MGI:1918734 | lncRNA gene | RIKEN cDNA 8430415O14 gene |
| 6 | gene | 58.96565 | 59.00208 | positive | MGI_C57BL6J_3034340 | BB365896 | ENSEMBL:ENSMUSG00000092323 | MGI:3034340 | lncRNA gene | expressed sequence BB365896 |
| 6 | gene | 58.97320 | 58.97529 | positive | MGI_C57BL6J_5753637 | Gm45061 | ENSEMBL:ENSMUSG00000108075 | MGI:5753637 | unclassified gene | predicted gene 45061 |
| 6 | pseudogene | 59.03689 | 59.03938 | positive | MGI_C57BL6J_5011350 | Gm19165 | NCBI_Gene:100418369,ENSEMBL:ENSMUSG00000107449 | MGI:5011350 | pseudogene | predicted gene, 19165 |
| 6 | gene | 59.09925 | 59.09938 | positive | MGI_C57BL6J_5452144 | Gm22367 | ENSEMBL:ENSMUSG00000096371 | MGI:5452144 | snoRNA gene | predicted gene, 22367 |
| 6 | pseudogene | 59.19305 | 59.19325 | negative | MGI_C57BL6J_5690297 | Gm43905 | ENSEMBL:ENSMUSG00000107987 | MGI:5690297 | pseudogene | predicted gene, 43905 |
| 6 | gene | 59.20887 | 59.21203 | positive | MGI_C57BL6J_1915390 | Tigd2 | NCBI_Gene:68140,ENSEMBL:ENSMUSG00000049232 | MGI:1915390 | protein coding gene | tigger transposable element derived 2 |
| 6 | gene | 59.34450 | 59.42629 | negative | MGI_C57BL6J_1924785 | Gprin3 | NCBI_Gene:243385,ENSEMBL:ENSMUSG00000045441 | MGI:1924785 | protein coding gene | GPRIN family member 3 |
| 6 | gene | 59.69817 | 59.70590 | positive | MGI_C57BL6J_5594354 | Gm35195 | NCBI_Gene:102638695 | MGI:5594354 | lncRNA gene | predicted gene, 35195 |
| 6 | gene | 59.70575 | 59.70587 | negative | MGI_C57BL6J_5452399 | Gm22622 | ENSEMBL:ENSMUSG00000094183 | MGI:5452399 | snoRNA gene | predicted gene, 22622 |
| 6 | pseudogene | 59.94233 | 59.94553 | positive | MGI_C57BL6J_5010197 | Gm18012 | NCBI_Gene:100416270,ENSEMBL:ENSMUSG00000108110 | MGI:5010197 | pseudogene | predicted gene, 18012 |
| 6 | gene | 60.03197 | 60.15004 | positive | MGI_C57BL6J_3644382 | Gm5570 | NCBI_Gene:434019,ENSEMBL:ENSMUSG00000073045 | MGI:3644382 | lncRNA gene | predicted gene 5570 |
| 6 | gene | 60.07106 | 60.07119 | negative | MGI_C57BL6J_5452500 | Gm22723 | ENSEMBL:ENSMUSG00000095176 | MGI:5452500 | snoRNA gene | predicted gene, 22723 |
| 6 | gene | 60.17914 | 60.18178 | positive | MGI_C57BL6J_5826622 | Gm46985 | NCBI_Gene:108169190 | MGI:5826622 | protein coding gene | predicted gene, 46985 |
| 6 | gene | 60.25079 | 60.25091 | positive | MGI_C57BL6J_4422026 | n-R5s162 | ENSEMBL:ENSMUSG00000064515 | MGI:4422026 | rRNA gene | nuclear encoded rRNA 5S 162 |
| 6 | gene | 60.27904 | 60.40371 | negative | MGI_C57BL6J_2443347 | A530053G22Rik | NCBI_Gene:208079,ENSEMBL:ENSMUSG00000046764 | MGI:2443347 | lncRNA gene | RIKEN cDNA A530053G22 gene |
| 6 | gene | 60.33980 | 60.37427 | positive | MGI_C57BL6J_5594494 | Gm35335 | NCBI_Gene:102638876 | MGI:5594494 | lncRNA gene | predicted gene, 35335 |
| 6 | gene | 60.41120 | 60.44969 | positive | MGI_C57BL6J_5621708 | Gm38823 | NCBI_Gene:105242671 | MGI:5621708 | lncRNA gene | predicted gene, 38823 |
| 6 | pseudogene | 60.46193 | 60.46211 | negative | MGI_C57BL6J_5690781 | Gm44389 | ENSEMBL:ENSMUSG00000107863 | MGI:5690781 | pseudogene | predicted gene, 44389 |
| 6 | gene | 60.48996 | 60.49045 | positive | MGI_C57BL6J_5690802 | Gm44410 | ENSEMBL:ENSMUSG00000108192 | MGI:5690802 | lncRNA gene | predicted gene, 44410 |
| 6 | gene | 60.58677 | 60.59155 | positive | MGI_C57BL6J_5594545 | Gm35386 | NCBI_Gene:102638951,ENSEMBL:ENSMUSG00000108003 | MGI:5594545 | lncRNA gene | predicted gene, 35386 |
| 6 | gene | 60.73157 | 60.82986 | negative | MGI_C57BL6J_1277151 | Snca | NCBI_Gene:20617,ENSEMBL:ENSMUSG00000025889 | MGI:1277151 | protein coding gene | synuclein, alpha |
| 6 | pseudogene | 60.76952 | 60.77118 | negative | MGI_C57BL6J_3701602 | Mageb16-ps2 | NCBI_Gene:625863,ENSEMBL:ENSMUSG00000107755 | MGI:3701602 | pseudogene | melanoma antigen family B, 16, pseudogene 2 |
| 6 | gene | 60.79184 | 60.79587 | negative | MGI_C57BL6J_5690256 | Gm43864 | ENSEMBL:ENSMUSG00000108168 | MGI:5690256 | unclassified gene | predicted gene, 43864 |
| 6 | pseudogene | 60.88244 | 60.88296 | negative | MGI_C57BL6J_5690259 | Gm43867 | ENSEMBL:ENSMUSG00000108262 | MGI:5690259 | pseudogene | predicted gene, 43867 |
| 6 | gene | 60.92498 | 60.98938 | positive | MGI_C57BL6J_1918195 | Mmrn1 | NCBI_Gene:70945,ENSEMBL:ENSMUSG00000054641 | MGI:1918195 | protein coding gene | multimerin 1 |
| 6 | pseudogene | 61.01685 | 61.01774 | negative | MGI_C57BL6J_5011023 | Gm18838 | NCBI_Gene:100417805,ENSEMBL:ENSMUSG00000108098 | MGI:5011023 | pseudogene | predicted gene, 18838 |
| 6 | pseudogene | 61.04473 | 61.04499 | negative | MGI_C57BL6J_5690284 | Gm43892 | ENSEMBL:ENSMUSG00000108299 | MGI:5690284 | pseudogene | predicted gene, 43892 |
| 6 | gene | 61.06470 | 61.06925 | negative | MGI_C57BL6J_5690285 | Gm43893 | ENSEMBL:ENSMUSG00000108087 | MGI:5690285 | lncRNA gene | predicted gene, 43893 |
| 6 | gene | 61.14722 | 61.14791 | negative | MGI_C57BL6J_5690286 | Gm43894 | ENSEMBL:ENSMUSG00000107652 | MGI:5690286 | lncRNA gene | predicted gene, 43894 |
| 6 | gene | 61.17229 | 61.18081 | negative | MGI_C57BL6J_2444999 | A730020E08Rik | NCBI_Gene:100504262,ENSEMBL:ENSMUSG00000097924 | MGI:2444999 | lncRNA gene | RIKEN cDNA A730020E08 gene |
| 6 | gene | 61.18032 | 62.38287 | positive | MGI_C57BL6J_3045354 | Ccser1 | NCBI_Gene:232035,ENSEMBL:ENSMUSG00000039578 | MGI:3045354 | protein coding gene | coiled-coil serine rich 1 |
| 6 | gene | 61.23173 | 61.23446 | positive | MGI_C57BL6J_3026947 | B230204D01Rik | NA | NA | unclassified gene | RIKEN cDNA B230204D01 gene |
| 6 | pseudogene | 61.40061 | 61.40103 | positive | MGI_C57BL6J_5690290 | Gm43898 | ENSEMBL:ENSMUSG00000107757 | MGI:5690290 | pseudogene | predicted gene, 43898 |
| 6 | gene | 61.97510 | 61.97713 | positive | MGI_C57BL6J_5690288 | Gm43896 | ENSEMBL:ENSMUSG00000108246 | MGI:5690288 | unclassified gene | predicted gene, 43896 |
| 6 | gene | 61.99305 | 61.99639 | positive | MGI_C57BL6J_2443660 | A730075L09Rik | NA | NA | unclassified gene | RIKEN cDNA A730075L09 gene |
| 6 | pseudogene | 62.29370 | 62.29401 | negative | MGI_C57BL6J_5690289 | Gm43897 | ENSEMBL:ENSMUSG00000107590 | MGI:5690289 | pseudogene | predicted gene, 43897 |
| 6 | pseudogene | 62.30773 | 62.30926 | negative | MGI_C57BL6J_5690287 | Gm43895 | ENSEMBL:ENSMUSG00000107908 | MGI:5690287 | pseudogene | predicted gene, 43895 |
| 6 | pseudogene | 62.58207 | 62.58320 | positive | MGI_C57BL6J_5011422 | Gm19237 | NCBI_Gene:100418479,ENSEMBL:ENSMUSG00000108160 | MGI:5011422 | pseudogene | predicted gene, 19237 |
| 6 | pseudogene | 62.63058 | 62.63082 | positive | MGI_C57BL6J_5690480 | Gm44088 | ENSEMBL:ENSMUSG00000107957 | MGI:5690480 | pseudogene | predicted gene, 44088 |
| 6 | pseudogene | 62.75680 | 62.75705 | positive | MGI_C57BL6J_5690484 | Gm44092 | ENSEMBL:ENSMUSG00000108174 | MGI:5690484 | pseudogene | predicted gene, 44092 |
| 6 | pseudogene | 62.82010 | 62.82063 | positive | MGI_C57BL6J_5010725 | Gm18540 | NCBI_Gene:100417337,ENSEMBL:ENSMUSG00000108007 | MGI:5010725 | pseudogene | predicted gene, 18540 |
| 6 | pseudogene | 62.92910 | 62.92948 | positive | MGI_C57BL6J_3643923 | Gm5001 | NCBI_Gene:257772,ENSEMBL:ENSMUSG00000107854 | MGI:3643923 | pseudogene | predicted gene 5001 |
| 6 | pseudogene | 63.04496 | 63.04817 | negative | MGI_C57BL6J_5011316 | Gm19131 | NCBI_Gene:100418306,ENSEMBL:ENSMUSG00000107708 | MGI:5011316 | pseudogene | predicted gene, 19131 |
| 6 | pseudogene | 63.16027 | 63.16108 | positive | MGI_C57BL6J_2153023 | Nasp-ps1 | NCBI_Gene:667099,ENSEMBL:ENSMUSG00000107484 | MGI:2153023 | pseudogene | nuclear autoantigenic sperm protein (histone-binding), pseudogene 1 |
| 6 | gene | 63.20004 | 63.20015 | negative | MGI_C57BL6J_5451989 | Gm22212 | ENSEMBL:ENSMUSG00000088721 | MGI:5451989 | snRNA gene | predicted gene, 22212 |
| 6 | gene | 63.24582 | 63.24923 | positive | MGI_C57BL6J_5826627 | Gm46990 | NCBI_Gene:108169203 | MGI:5826627 | lncRNA gene | predicted gene, 46990 |
| 6 | gene | 63.24849 | 63.25762 | negative | MGI_C57BL6J_1924758 | 9330118I20Rik | ENSEMBL:ENSMUSG00000108242 | MGI:1924758 | lncRNA gene | RIKEN cDNA 9330118I20 gene |
| 6 | gene | 63.25588 | 64.70432 | positive | MGI_C57BL6J_95813 | Grid2 | NCBI_Gene:14804,ENSEMBL:ENSMUSG00000071424 | MGI:95813 | protein coding gene | glutamate receptor, ionotropic, delta 2 |
| 6 | gene | 63.32467 | 63.32844 | negative | MGI_C57BL6J_5594864 | Gm35705 | NA | NA | lncRNA gene | predicted gene%2c 35705 |
| 6 | gene | 63.32467 | 63.32844 | negative | MGI_C57BL6J_5690463 | Gm44071 | ENSEMBL:ENSMUSG00000107525 | MGI:5690463 | lncRNA gene | predicted gene, 44071 |
| 6 | gene | 63.63750 | 63.64046 | positive | MGI_C57BL6J_2442993 | 9630021D06Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 9630021D06 gene |
| 6 | pseudogene | 63.78385 | 63.78407 | negative | MGI_C57BL6J_5690467 | Gm44075 | ENSEMBL:ENSMUSG00000108068 | MGI:5690467 | pseudogene | predicted gene, 44075 |
| 6 | gene | 64.04043 | 64.04156 | negative | MGI_C57BL6J_5690464 | Gm44072 | ENSEMBL:ENSMUSG00000107864 | MGI:5690464 | lncRNA gene | predicted gene, 44072 |
| 6 | pseudogene | 64.09208 | 64.09231 | positive | MGI_C57BL6J_5012329 | Gm20144 | NCBI_Gene:102639204,ENSEMBL:ENSMUSG00000107700 | MGI:5012329 | pseudogene | predicted gene, 20144 |
| 6 | gene | 64.41365 | 64.41375 | positive | MGI_C57BL6J_5454982 | Gm25205 | ENSEMBL:ENSMUSG00000064775 | MGI:5454982 | snRNA gene | predicted gene, 25205 |
| 6 | gene | 64.70432 | 64.72886 | negative | MGI_C57BL6J_5595010 | Gm35851 | NCBI_Gene:102639571 | MGI:5595010 | lncRNA gene | predicted gene, 35851 |
| 6 | gene | 64.72912 | 64.73125 | positive | MGI_C57BL6J_104654 | Atoh1 | NCBI_Gene:11921,ENSEMBL:ENSMUSG00000073043 | MGI:104654 | protein coding gene | atonal bHLH transcription factor 1 |
| 6 | pseudogene | 64.87845 | 64.87913 | negative | MGI_C57BL6J_5011024 | Gm18839 | NCBI_Gene:100417806,ENSEMBL:ENSMUSG00000107424 | MGI:5011024 | pseudogene | predicted gene, 18839 |
| 6 | pseudogene | 64.89695 | 64.89944 | positive | MGI_C57BL6J_1100855 | Lt1 | NCBI_Gene:16991 | MGI:1100855 | pseudogene | lurcher transcript 1 |
| 6 | pseudogene | 64.97852 | 64.97940 | positive | MGI_C57BL6J_5690430 | Gm44038 | ENSEMBL:ENSMUSG00000107783 | MGI:5690430 | pseudogene | predicted gene, 44038 |
| 6 | gene | 64.98370 | 64.98716 | positive | MGI_C57BL6J_2441913 | C530040J15Rik | NA | NA | unclassified gene | RIKEN cDNA C530040J15 gene |
| 6 | pseudogene | 64.99095 | 64.99151 | positive | MGI_C57BL6J_3704268 | Gm15534 | NCBI_Gene:667115,ENSEMBL:ENSMUSG00000086176 | MGI:3704268 | pseudogene | predicted gene 15534 |
| 6 | gene | 65.04258 | 65.11606 | positive | MGI_C57BL6J_95453 | Smarcad1 | NCBI_Gene:13990,ENSEMBL:ENSMUSG00000029920 | MGI:95453 | protein coding gene | SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| 6 | gene | 65.11729 | 65.14502 | negative | MGI_C57BL6J_1859384 | Hpgds | NCBI_Gene:54486,ENSEMBL:ENSMUSG00000029919 | MGI:1859384 | protein coding gene | hematopoietic prostaglandin D synthase |
| 6 | gene | 65.12241 | 65.12732 | positive | MGI_C57BL6J_5595065 | Gm35906 | NCBI_Gene:102639643 | MGI:5595065 | lncRNA gene | predicted gene, 35906 |
| 6 | pseudogene | 65.12734 | 65.12910 | positive | MGI_C57BL6J_4937974 | Gm17147 | NCBI_Gene:102632836,ENSEMBL:ENSMUSG00000090939 | MGI:4937974 | pseudogene | predicted gene 17147 |
| 6 | pseudogene | 65.15100 | 65.15122 | positive | MGI_C57BL6J_5690811 | Gm44419 | ENSEMBL:ENSMUSG00000107968 | MGI:5690811 | pseudogene | predicted gene, 44419 |
| 6 | pseudogene | 65.16916 | 65.16996 | positive | MGI_C57BL6J_3781726 | Gm3549 | NCBI_Gene:100041858,ENSEMBL:ENSMUSG00000108230 | MGI:3781726 | pseudogene | predicted gene 3549 |
| 6 | pseudogene | 65.21357 | 65.21497 | negative | MGI_C57BL6J_5690450 | Gm44058 | ENSEMBL:ENSMUSG00000107490 | MGI:5690450 | pseudogene | predicted gene, 44058 |
| 6 | pseudogene | 65.23271 | 65.23306 | negative | MGI_C57BL6J_5690451 | Gm44059 | ENSEMBL:ENSMUSG00000108252 | MGI:5690451 | pseudogene | predicted gene, 44059 |
| 6 | pseudogene | 65.24727 | 65.24769 | positive | MGI_C57BL6J_5012362 | Gm20177 | NCBI_Gene:100504330,ENSEMBL:ENSMUSG00000107609 | MGI:5012362 | pseudogene | predicted gene, 20177 |
| 6 | pseudogene | 65.27150 | 65.27191 | negative | MGI_C57BL6J_5690452 | Gm44060 | ENSEMBL:ENSMUSG00000108097 | MGI:5690452 | pseudogene | predicted gene, 44060 |
| 6 | pseudogene | 65.27295 | 65.27414 | negative | MGI_C57BL6J_5690453 | Gm44061 | ENSEMBL:ENSMUSG00000107630 | MGI:5690453 | pseudogene | predicted gene, 44061 |
| 6 | pseudogene | 65.32810 | 65.32917 | positive | MGI_C57BL6J_3647865 | Gm8479 | NCBI_Gene:667141,ENSEMBL:ENSMUSG00000108052 | MGI:3647865 | pseudogene | predicted gene 8479 |
| 6 | gene | 65.38111 | 65.45815 | positive | MGI_C57BL6J_2441881 | Qrfprl | NCBI_Gene:243407,ENSEMBL:ENSMUSG00000029917 | MGI:2441881 | protein coding gene | pyroglutamylated RFamide peptide receptor like |
| 6 | pseudogene | 65.41602 | 65.41762 | positive | MGI_C57BL6J_5010901 | Gm18716 | NCBI_Gene:100417616,ENSEMBL:ENSMUSG00000107584 | MGI:5010901 | pseudogene | predicted gene, 18716 |
| 6 | pseudogene | 65.43205 | 65.43400 | positive | MGI_C57BL6J_5011098 | Gm18913 | NCBI_Gene:100417946,ENSEMBL:ENSMUSG00000107596 | MGI:5011098 | pseudogene | predicted gene, 18913 |
| 6 | gene | 65.52531 | 65.63404 | positive | MGI_C57BL6J_3041165 | Tnip3 | NCBI_Gene:414084,ENSEMBL:ENSMUSG00000044162 | MGI:3041165 | protein coding gene | TNFAIP3 interacting protein 3 |
| 6 | gene | 65.66205 | 65.67125 | negative | MGI_C57BL6J_5595303 | Gm36144 | NCBI_Gene:102639953 | MGI:5595303 | lncRNA gene | predicted gene, 36144 |
| 6 | gene | 65.67159 | 65.71233 | positive | MGI_C57BL6J_1915419 | Ndnf | NCBI_Gene:68169,ENSEMBL:ENSMUSG00000049001 | MGI:1915419 | protein coding gene | neuron-derived neurotrophic factor |
| 6 | pseudogene | 65.75794 | 65.75832 | positive | MGI_C57BL6J_3782936 | Gm15490 | ENSEMBL:ENSMUSG00000082115 | MGI:3782936 | pseudogene | predicted gene 15490 |
| 6 | gene | 65.76854 | 65.77925 | negative | MGI_C57BL6J_4439762 | Gm16838 | ENSEMBL:ENSMUSG00000085689 | MGI:4439762 | lncRNA gene | predicted gene, 16838 |
| 6 | gene | 65.77875 | 65.93762 | positive | MGI_C57BL6J_1918029 | Prdm5 | NCBI_Gene:70779,ENSEMBL:ENSMUSG00000029913 | MGI:1918029 | protein coding gene | PR domain containing 5 |
| 6 | gene | 65.86417 | 65.86428 | negative | MGI_C57BL6J_5453320 | Gm23543 | ENSEMBL:ENSMUSG00000088832 | MGI:5453320 | snRNA gene | predicted gene, 23543 |
| 6 | pseudogene | 65.88278 | 65.88334 | positive | MGI_C57BL6J_5141910 | Gm20445 | ENSEMBL:ENSMUSG00000092394 | MGI:5141910 | pseudogene | predicted gene 20445 |
| 6 | gene | 65.95257 | 65.95401 | positive | MGI_C57BL6J_1914903 | 4930544G11Rik | NCBI_Gene:67653,ENSEMBL:ENSMUSG00000036463 | MGI:1914903 | protein coding gene | RIKEN cDNA 4930544G11 gene |
| 6 | pseudogene | 65.98187 | 65.98344 | negative | MGI_C57BL6J_5690796 | Gm44404 | ENSEMBL:ENSMUSG00000107649 | MGI:5690796 | pseudogene | predicted gene, 44404 |
| 6 | gene | 65.98964 | 65.98974 | negative | MGI_C57BL6J_5453402 | Gm23625 | ENSEMBL:ENSMUSG00000064391 | MGI:5453402 | snRNA gene | predicted gene, 23625 |
| 6 | pseudogene | 65.99563 | 65.99653 | positive | MGI_C57BL6J_3647377 | Gm5876 | NCBI_Gene:545843,ENSEMBL:ENSMUSG00000108100 | MGI:3647377 | pseudogene | predicted gene 5876 |
| 6 | gene | 66.14844 | 66.27236 | negative | MGI_C57BL6J_5595567 | Gm36408 | NCBI_Gene:102640309 | MGI:5595567 | lncRNA gene | predicted gene, 36408 |
| 6 | pseudogene | 66.25474 | 66.25821 | positive | MGI_C57BL6J_5690681 | Gm44289 | ENSEMBL:ENSMUSG00000107695 | MGI:5690681 | pseudogene | predicted gene, 44289 |
| 6 | pseudogene | 66.26416 | 66.28485 | positive | MGI_C57BL6J_5590679 | Gm31520 | NCBI_Gene:102633773,ENSEMBL:ENSMUSG00000107653 | MGI:5590679 | pseudogene | predicted gene, 31520 |
| 6 | gene | 66.31101 | 66.31568 | negative | MGI_C57BL6J_5621709 | Gm38824 | NCBI_Gene:105242673 | MGI:5621709 | lncRNA gene | predicted gene, 38824 |
| 6 | pseudogene | 66.39855 | 66.39872 | negative | MGI_C57BL6J_5690673 | Gm44281 | ENSEMBL:ENSMUSG00000107837 | MGI:5690673 | pseudogene | predicted gene, 44281 |
| 6 | pseudogene | 66.39875 | 66.39897 | positive | MGI_C57BL6J_5690625 | Gm44233 | ENSEMBL:ENSMUSG00000107631 | MGI:5690625 | pseudogene | predicted gene, 44233 |
| 6 | gene | 66.45194 | 66.46648 | negative | MGI_C57BL6J_1919734 | 2610300M13Rik | NCBI_Gene:72484,ENSEMBL:ENSMUSG00000107479 | MGI:1919734 | lncRNA gene | RIKEN cDNA 2610300M13 gene |
| 6 | pseudogene | 66.49525 | 66.51654 | positive | MGI_C57BL6J_5011949 | Gm19764 | NCBI_Gene:100503550,ENSEMBL:ENSMUSG00000107775 | MGI:5011949 | pseudogene | predicted gene, 19764 |
| 6 | pseudogene | 66.51589 | 66.51773 | negative | MGI_C57BL6J_99709 | Tpm3-rs3 | NCBI_Gene:110376 | MGI:99709 | pseudogene | tropomyosin 3, related sequence 3 |
| 6 | pseudogene | 66.52701 | 66.52893 | positive | MGI_C57BL6J_3644036 | Gm6786 | NCBI_Gene:627751,ENSEMBL:ENSMUSG00000108086 | MGI:3644036 | pseudogene | predicted gene 6786 |
| 6 | gene | 66.53523 | 66.54722 | positive | MGI_C57BL6J_1860374 | Mad2l1 | NCBI_Gene:56150,ENSEMBL:ENSMUSG00000029910 | MGI:1860374 | protein coding gene | MAD2 mitotic arrest deficient-like 1 |
| 6 | gene | 66.55218 | 66.56010 | negative | MGI_C57BL6J_2159451 | Vmn1r32 | NCBI_Gene:171188,ENSEMBL:ENSMUSG00000062905 | MGI:2159451 | protein coding gene | vomeronasal 1 receptor 32 |
| 6 | pseudogene | 66.57704 | 66.57856 | positive | MGI_C57BL6J_3647907 | Gm5307 | NCBI_Gene:384398,ENSEMBL:ENSMUSG00000107710 | MGI:3647907 | pseudogene | predicted gene 5307 |
| 6 | gene | 66.61054 | 66.61828 | negative | MGI_C57BL6J_2159450 | Vmn1r33 | NCBI_Gene:171187,ENSEMBL:ENSMUSG00000059375 | MGI:2159450 | protein coding gene | vomeronasal 1 receptor 33 |
| 6 | gene | 66.63594 | 66.64388 | negative | MGI_C57BL6J_3644800 | Vmn1r34 | NCBI_Gene:546901,ENSEMBL:ENSMUSG00000091012 | MGI:3644800 | protein coding gene | vomeronasal 1 receptor 34 |
| 6 | gene | 66.67807 | 66.68591 | negative | MGI_C57BL6J_2159444 | Vmn1r35 | NCBI_Gene:171185,ENSEMBL:ENSMUSG00000060699 | MGI:2159444 | protein coding gene | vomeronasal 1 receptor 35 |
| 6 | pseudogene | 66.71168 | 66.71219 | positive | MGI_C57BL6J_3646483 | Gm6794 | NCBI_Gene:627833,ENSEMBL:ENSMUSG00000107591 | MGI:3646483 | pseudogene | predicted gene 6794 |
| 6 | gene | 66.71526 | 66.71917 | negative | MGI_C57BL6J_2159443 | Vmn1r36 | NCBI_Gene:171184,ENSEMBL:ENSMUSG00000115482 | MGI:2159443 | protein coding gene | vomeronasal 1 receptor 36 |
| 6 | gene | 66.72938 | 66.73304 | positive | MGI_C57BL6J_2159442 | Vmn1r37 | NCBI_Gene:171183,ENSEMBL:ENSMUSG00000115467 | MGI:2159442 | protein coding gene | vomeronasal 1 receptor 37 |
| 6 | pseudogene | 66.73397 | 66.73418 | negative | MGI_C57BL6J_5690653 | Gm44261 | ENSEMBL:ENSMUSG00000107721 | MGI:5690653 | pseudogene | predicted gene, 44261 |
| 6 | pseudogene | 66.74596 | 66.74641 | positive | MGI_C57BL6J_3782220 | Gm4045 | NCBI_Gene:100042816,ENSEMBL:ENSMUSG00000108231 | MGI:3782220 | pseudogene | predicted gene 4045 |
| 6 | pseudogene | 66.75228 | 66.76004 | negative | MGI_C57BL6J_3644801 | Vmn1r-ps24 | NCBI_Gene:546902,ENSEMBL:ENSMUSG00000093504 | MGI:3644801 | pseudogene | vomeronasal 1 receptor, pseudogene 24 |
| 6 | gene | 66.77400 | 66.78263 | negative | MGI_C57BL6J_2159445 | Vmn1r38 | NCBI_Gene:171186,ENSEMBL:ENSMUSG00000115170 | MGI:2159445 | protein coding gene | vomeronasal 1 receptor 38 |
| 6 | gene | 66.80330 | 66.81061 | negative | MGI_C57BL6J_3644802 | Vmn1r39 | NCBI_Gene:546903,ENSEMBL:ENSMUSG00000093755 | MGI:3644802 | protein coding gene | vomeronasal 1 receptor 39 |
| 6 | gene | 66.84001 | 66.84298 | negative | MGI_C57BL6J_5595769 | Gm36610 | NCBI_Gene:102640580 | MGI:5595769 | lncRNA gene | predicted gene, 36610 |
| 6 | pseudogene | 66.87526 | 66.87577 | negative | MGI_C57BL6J_3642653 | Gm9794 | NCBI_Gene:100042008,ENSEMBL:ENSMUSG00000107176 | MGI:3642653 | pseudogene | predicted pseudogene 9794 |
| 6 | gene | 66.89465 | 66.89652 | negative | MGI_C57BL6J_1922623 | 4930597O21Rik | ENSEMBL:ENSMUSG00000053442 | MGI:1922623 | lncRNA gene | RIKEN cDNA 4930597O21 gene |
| 6 | gene | 66.89640 | 67.02136 | positive | MGI_C57BL6J_1336171 | Gng12 | NCBI_Gene:14701,ENSEMBL:ENSMUSG00000036402 | MGI:1336171 | protein coding gene | guanine nucleotide binding protein (G protein), gamma 12 |
| 6 | gene | 66.98432 | 67.01039 | negative | MGI_C57BL6J_5595975 | Gm36816 | NCBI_Gene:102640846,ENSEMBL:ENSMUSG00000107634 | MGI:5595975 | lncRNA gene | predicted gene, 36816 |
| 6 | pseudogene | 67.01248 | 67.01287 | positive | MGI_C57BL6J_3783088 | Gm15644 | ENSEMBL:ENSMUSG00000084154 | MGI:3783088 | pseudogene | predicted gene 15644 |
| 6 | pseudogene | 67.02142 | 67.02448 | positive | MGI_C57BL6J_5589007 | Gm29848 | NCBI_Gene:102631535,ENSEMBL:ENSMUSG00000107869 | MGI:5589007 | pseudogene | predicted gene, 29848 |
| 6 | gene | 67.02704 | 67.02718 | positive | MGI_C57BL6J_5455037 | Gm25260 | ENSEMBL:ENSMUSG00000064987 | MGI:5455037 | snRNA gene | predicted gene, 25260 |
| 6 | gene | 67.03510 | 67.03746 | negative | MGI_C57BL6J_107799 | Gadd45a | NCBI_Gene:13197,ENSEMBL:ENSMUSG00000036390 | MGI:107799 | protein coding gene | growth arrest and DNA-damage-inducible 45 alpha |
| 6 | gene | 67.03660 | 67.08065 | positive | MGI_C57BL6J_2443524 | E230016M11Rik | NCBI_Gene:320172,ENSEMBL:ENSMUSG00000087231 | MGI:2443524 | lncRNA gene | RIKEN cDNA E230016M11 gene |
| 6 | gene | 67.09193 | 67.10326 | positive | MGI_C57BL6J_5589087 | Gm29928 | NCBI_Gene:102631637 | MGI:5589087 | lncRNA gene | predicted gene, 29928 |
| 6 | gene | 67.11567 | 67.16843 | positive | MGI_C57BL6J_2442501 | A430010J10Rik | NCBI_Gene:319665,ENSEMBL:ENSMUSG00000097603 | MGI:2442501 | lncRNA gene | RIKEN cDNA A430010J10 gene |
| 6 | gene | 67.17868 | 67.23971 | negative | MGI_C57BL6J_5621710 | Gm38825 | NCBI_Gene:105242674,ENSEMBL:ENSMUSG00000107799 | MGI:5621710 | lncRNA gene | predicted gene, 38825 |
| 6 | pseudogene | 67.20552 | 67.20617 | positive | MGI_C57BL6J_3647296 | Gm8566 | NCBI_Gene:667310,ENSEMBL:ENSMUSG00000047905 | MGI:3647296 | pseudogene | predicted pseudogene 8566 |
| 6 | pseudogene | 67.22190 | 67.22254 | negative | MGI_C57BL6J_5690444 | Gm44052 | ENSEMBL:ENSMUSG00000108013 | MGI:5690444 | pseudogene | predicted gene, 44052 |
| 6 | gene | 67.23818 | 67.29774 | positive | MGI_C57BL6J_1914120 | Serbp1 | NCBI_Gene:66870,ENSEMBL:ENSMUSG00000036371 | MGI:1914120 | protein coding gene | serpine1 mRNA binding protein 1 |
| 6 | pseudogene | 67.25203 | 67.25293 | negative | MGI_C57BL6J_5690442 | Gm44050 | ENSEMBL:ENSMUSG00000108292 | MGI:5690442 | pseudogene | predicted gene, 44050 |
| 6 | gene | 67.29132 | 67.37629 | negative | MGI_C57BL6J_1270861 | Il12rb2 | NCBI_Gene:16162,ENSEMBL:ENSMUSG00000018341 | MGI:1270861 | protein coding gene | interleukin 12 receptor, beta 2 |
| 6 | pseudogene | 67.38442 | 67.38534 | negative | MGI_C57BL6J_3643884 | Gm8574 | NCBI_Gene:667324,ENSEMBL:ENSMUSG00000107907 | MGI:3643884 | pseudogene | predicted gene 8574 |
| 6 | pseudogene | 67.40649 | 67.40691 | negative | MGI_C57BL6J_5690600 | Gm44208 | ENSEMBL:ENSMUSG00000108000 | MGI:5690600 | pseudogene | predicted gene, 44208 |
| 6 | pseudogene | 67.41443 | 67.41501 | negative | MGI_C57BL6J_3648855 | Gm4761 | NCBI_Gene:209589,ENSEMBL:ENSMUSG00000096086 | MGI:3648855 | pseudogene | predicted gene 4761 |
| 6 | pseudogene | 67.41767 | 67.41805 | negative | MGI_C57BL6J_5690475 | Gm44083 | NCBI_Gene:108169191,ENSEMBL:ENSMUSG00000107508 | MGI:5690475 | pseudogene | predicted gene, 44083 |
| 6 | gene | 67.42293 | 67.49186 | negative | MGI_C57BL6J_2181693 | Il23r | NCBI_Gene:209590,ENSEMBL:ENSMUSG00000049093 | MGI:2181693 | protein coding gene | interleukin 23 receptor |
| 6 | gene | 67.53249 | 67.55620 | positive | MGI_C57BL6J_5663026 | Gm42889 | ENSEMBL:ENSMUSG00000105650 | MGI:5663026 | lncRNA gene | predicted gene 42889 |
| 6 | gene | 67.53406 | 67.53582 | negative | MGI_C57BL6J_1861606 | Tacstd2 | NCBI_Gene:56753,ENSEMBL:ENSMUSG00000051397 | MGI:1861606 | protein coding gene | tumor-associated calcium signal transducer 2 |
| 6 | gene | 67.55550 | 67.55622 | positive | MGI_C57BL6J_4439879 | Igkv2-137 | NCBI_Gene:692187,ENSEMBL:ENSMUSG00000076501 | MGI:4439879 | gene segment | immunoglobulin kappa chain variable 2-137 |
| 6 | pseudogene | 67.56519 | 67.56645 | negative | MGI_C57BL6J_1922320 | 4930515G16Rik | NCBI_Gene:75070,ENSEMBL:ENSMUSG00000104819 | MGI:1922320 | pseudogene | RIKEN cDNA 4930515G16 gene |
| 6 | pseudogene | 67.59388 | 67.59431 | positive | MGI_C57BL6J_4947614 | Igkv1-136 | NCBI_Gene:692186,ENSEMBL:ENSMUSG00000104931 | MGI:4947614 | pseudogenic gene segment | immunoglobulin kappa chain variable 1-136 |
| 6 | gene | 67.60971 | 67.61051 | positive | MGI_C57BL6J_3819952 | Igkv1-135 | NCBI_Gene:243420,ENSEMBL:ENSMUSG00000096336 | MGI:3819952 | gene segment | immunoglobulin kappa variable 1-135 |
| 6 | pseudogene | 67.63290 | 67.63414 | negative | MGI_C57BL6J_3780132 | Gm9728 | NCBI_Gene:723992,ENSEMBL:ENSMUSG00000106248 | MGI:3780132 | pseudogene | predicted gene 9728 |
| 6 | pseudogene | 67.64150 | 67.64170 | negative | MGI_C57BL6J_3648164 | Rpl18a-ps2 | NCBI_Gene:628006,ENSEMBL:ENSMUSG00000105464 | MGI:3648164 | pseudogene | ribosomal protein L18A, pseudogene 2 |
| 6 | pseudogene | 67.71117 | 67.71163 | negative | MGI_C57BL6J_5009883 | Igkv14-134-1 | NCBI_Gene:692185,ENSEMBL:ENSMUSG00000104883 | MGI:5009883 | pseudogenic gene segment | immunoglobulin kappa chain variable 14-134-1 |
| 6 | gene | 67.71774 | 67.72144 | negative | MGI_C57BL6J_5621458 | Gm38573 | NCBI_Gene:102641749 | MGI:5621458 | lncRNA gene | predicted gene, 38573 |
| 6 | pseudogene | 67.72073 | 67.72123 | negative | MGI_C57BL6J_4936953 | Igkv17-134 | NCBI_Gene:692184,ENSEMBL:ENSMUSG00000091268 | MGI:4936953 | pseudogenic gene segment | immunoglobulin kappa chain variable 17-134 |
| 6 | gene | 67.72490 | 67.72566 | positive | MGI_C57BL6J_3648380 | Igkv1-133 | NCBI_Gene:628027,ENSEMBL:ENSMUSG00000094491 | MGI:3648380 | gene segment | immunoglobulin kappa variable 1-133 |
| 6 | pseudogene | 67.73407 | 67.73560 | negative | MGI_C57BL6J_3646418 | Gm5308 | NCBI_Gene:384401,ENSEMBL:ENSMUSG00000104843 | MGI:3646418 | pseudogene | predicted gene 5308 |
| 6 | pseudogene | 67.74852 | 67.74873 | negative | MGI_C57BL6J_3648591 | Rpl18a-ps3 | NCBI_Gene:628042,ENSEMBL:ENSMUSG00000104584 | MGI:3648591 | pseudogene | ribosomal protein L18A, pseudogene 3 |
| 6 | pseudogene | 67.75475 | 67.75500 | positive | MGI_C57BL6J_5663421 | Gm43284 | ENSEMBL:ENSMUSG00000106401 | MGI:5663421 | pseudogenic gene segment | predicted gene 43284 |
| 6 | gene | 67.75970 | 67.76041 | positive | MGI_C57BL6J_3648800 | Igkv1-132 | NCBI_Gene:243423,ENSEMBL:ENSMUSG00000096580 | MGI:3648800 | gene segment | immunoglobulin kappa variable 1-132 |
| 6 | gene | 67.76604 | 67.76677 | negative | MGI_C57BL6J_3645551 | Igkv1-131 | NCBI_Gene:628056,ENSEMBL:ENSMUSG00000076505 | MGI:3645551 | gene segment | immunoglobulin kappa variable 1-131 |
| 6 | pseudogene | 67.77825 | 67.77933 | negative | MGI_C57BL6J_5011165 | Gm18980 | NCBI_Gene:100418068,ENSEMBL:ENSMUSG00000104744 | MGI:5011165 | pseudogene | predicted gene, 18980 |
| 6 | gene | 67.79105 | 67.79151 | positive | MGI_C57BL6J_3645770 | Igkv14-130 | NCBI_Gene:628072,ENSEMBL:ENSMUSG00000096461 | MGI:3645770 | gene segment | immunoglobulin kappa variable 14-130 |
| 6 | gene | 67.79244 | 67.79258 | negative | MGI_C57BL6J_5452202 | Gm22425 | ENSEMBL:ENSMUSG00000089157 | MGI:5452202 | snoRNA gene | predicted gene, 22425 |
| 6 | gene | 67.80437 | 67.80451 | negative | MGI_C57BL6J_5454688 | Gm24911 | ENSEMBL:ENSMUSG00000087917 | MGI:5454688 | snoRNA gene | predicted gene, 24911 |
| 6 | gene | 67.83979 | 67.84027 | positive | MGI_C57BL6J_3525017 | Igkv9-129 | NCBI_Gene:692182,ENSEMBL:ENSMUSG00000093906 | MGI:3525017 | gene segment | immunoglobulin kappa variable 9-129 |
| 6 | pseudogene | 67.84741 | 67.84787 | positive | MGI_C57BL6J_3809196 | Igkv9-128 | NCBI_Gene:692181,ENSEMBL:ENSMUSG00000104742 | MGI:3809196 | pseudogenic gene segment | immunoglobulin kappa chain variable 9-128 |
| 6 | gene | 67.86115 | 67.86166 | positive | MGI_C57BL6J_3646891 | Igkv17-127 | NCBI_Gene:243433,ENSEMBL:ENSMUSG00000076508 | MGI:3646891 | gene segment | immunoglobulin kappa variable 17-127 |
| 6 | pseudogene | 67.87601 | 67.87648 | positive | MGI_C57BL6J_5009882 | Igkv14-126-1 | NCBI_Gene:692180,ENSEMBL:ENSMUSG00000106372 | MGI:5009882 | pseudogenic gene segment | immunoglobulin kappa chain variable 14-126-1 |
| 6 | pseudogene | 67.88070 | 67.88361 | negative | MGI_C57BL6J_3780133 | Gm9729 | NCBI_Gene:723993,ENSEMBL:ENSMUSG00000105005 | MGI:3780133 | pseudogene | predicted gene 9729 |
| 6 | pseudogene | 67.89617 | 67.89664 | positive | MGI_C57BL6J_3643131 | Igkv14-126 | NCBI_Gene:628127,ENSEMBL:ENSMUSG00000094345 | MGI:3643131 | pseudogenic gene segment | immunoglobulin kappa variable 14-126 |
| 6 | gene | 67.89706 | 67.89720 | negative | MGI_C57BL6J_5455687 | Gm25910 | ENSEMBL:ENSMUSG00000088245 | MGI:5455687 | snoRNA gene | predicted gene, 25910 |
| 6 | gene | 67.91357 | 67.91405 | positive | MGI_C57BL6J_3642338 | Igkv11-125 | NCBI_Gene:243428,ENSEMBL:ENSMUSG00000095737 | MGI:3642338 | gene segment | immunoglobulin kappa variable 11-125 |
| 6 | gene | 67.94207 | 67.94254 | negative | MGI_C57BL6J_3646892 | Igkv9-124 | NCBI_Gene:243431,ENSEMBL:ENSMUSG00000096632 | MGI:3646892 | gene segment | immunoglobulin kappa chain variable 9-124 |
| 6 | gene | 67.95423 | 67.95471 | negative | MGI_C57BL6J_3643848 | Igkv9-123 | NCBI_Gene:628144,ENSEMBL:ENSMUSG00000076512 | MGI:3643848 | gene segment | immunoglobulin kappa variable 9-123 |
| 6 | gene | 68.01672 | 68.01749 | positive | MGI_C57BL6J_4439722 | Igkv1-122 | NCBI_Gene:434024,ENSEMBL:ENSMUSG00000095497 | MGI:4439722 | gene segment | immunoglobulin kappa chain variable 1-122 |
| 6 | gene | 68.03681 | 68.03732 | positive | MGI_C57BL6J_3647671 | Igkv17-121 | NCBI_Gene:667435,ENSEMBL:ENSMUSG00000076514 | MGI:3647671 | gene segment | immunoglobulin kappa variable 17-121 |
| 6 | gene | 68.04998 | 68.05046 | positive | MGI_C57BL6J_3647784 | Igkv9-120 | NCBI_Gene:434025,ENSEMBL:ENSMUSG00000094872 | MGI:3647784 | gene segment | immunoglobulin kappa chain variable 9-120 |
| 6 | pseudogene | 68.05634 | 68.05681 | positive | MGI_C57BL6J_5009881 | Igkv9-119 | NCBI_Gene:692177,ENSEMBL:ENSMUSG00000104575 | MGI:5009881 | pseudogenic gene segment | immunoglobulin kappa chain variable 9-119 |
| 6 | pseudogene | 68.06637 | 68.06683 | positive | MGI_C57BL6J_5009880 | Igkv14-118-2 | NCBI_Gene:692176,ENSEMBL:ENSMUSG00000106098 | MGI:5009880 | pseudogenic gene segment | immunoglobulin kappa chain variable 14-118-2 |
| 6 | pseudogene | 68.08255 | 68.08315 | positive | MGI_C57BL6J_5009879 | Igkv14-118-1 | NCBI_Gene:692175,ENSEMBL:ENSMUSG00000106480 | MGI:5009879 | pseudogenic gene segment | immunoglobulin kappa chain variable 14-118-1 |
| 6 | pseudogene | 68.10057 | 68.10097 | positive | MGI_C57BL6J_5663876 | Gm43739 | ENSEMBL:ENSMUSG00000104532 | MGI:5663876 | pseudogenic gene segment | predicted gene 43739 |
| 6 | pseudogene | 68.10279 | 68.10347 | negative | MGI_C57BL6J_3646419 | Gm5309 | NCBI_Gene:384405,ENSEMBL:ENSMUSG00000106331 | MGI:3646419 | pseudogene | predicted gene 5309 |
| 6 | pseudogene | 68.10541 | 68.10588 | positive | MGI_C57BL6J_5009878 | Igkv11-118 | NCBI_Gene:692174,ENSEMBL:ENSMUSG00000104934 | MGI:5009878 | pseudogenic gene segment | immunoglobulin kappa chain variable 11-118 |
| 6 | gene | 68.12105 | 68.12183 | positive | MGI_C57BL6J_4439721 | Igkv1-117 | NCBI_Gene:16098,ENSEMBL:ENSMUSG00000094335 | MGI:4439721 | gene segment | immunoglobulin kappa variable 1-117 |
| 6 | pseudogene | 68.14640 | 68.14669 | negative | MGI_C57BL6J_5663308 | Gm43171 | ENSEMBL:ENSMUSG00000105306 | MGI:5663308 | pseudogene | predicted gene 43171 |
| 6 | pseudogene | 68.14712 | 68.14785 | positive | MGI_C57BL6J_5663307 | Gm43170 | ENSEMBL:ENSMUSG00000105007 | MGI:5663307 | pseudogene | predicted gene 43170 |
| 6 | pseudogene | 68.15188 | 68.15262 | positive | MGI_C57BL6J_3647785 | Igkv2-116 | NCBI_Gene:434026,ENSEMBL:ENSMUSG00000106630 | MGI:3647785 | pseudogenic gene segment | immunoglobulin kappa variable 2-116 |
| 6 | pseudogene | 68.16080 | 68.16172 | positive | MGI_C57BL6J_3644873 | Igkv1-115 | NCBI_Gene:628206,ENSEMBL:ENSMUSG00000091800 | MGI:3644873 | pseudogenic gene segment | immunoglobulin kappa variable 1-115 |
| 6 | pseudogene | 68.16446 | 68.16492 | positive | MGI_C57BL6J_5009877 | Igkv11-114 | NCBI_Gene:692173,ENSEMBL:ENSMUSG00000105363 | MGI:5009877 | pseudogenic gene segment | immunoglobulin kappa chain variable |
| 6 | pseudogene | 68.17880 | 68.17956 | positive | MGI_C57BL6J_5009876 | Igkv2-113 | NCBI_Gene:692172,ENSEMBL:ENSMUSG00000105463 | MGI:5009876 | pseudogenic gene segment | immunoglobulin kappa chain variable 2-113 |
| 6 | pseudogene | 68.21093 | 68.21537 | negative | MGI_C57BL6J_3648522 | Gm4873 | NCBI_Gene:232047,ENSEMBL:ENSMUSG00000105163 | MGI:3648522 | pseudogene | predicted gene 4873 |
| 6 | gene | 68.21998 | 68.22071 | positive | MGI_C57BL6J_3644894 | Igkv2-112 | NCBI_Gene:381776,ENSEMBL:ENSMUSG00000076518 | MGI:3644894 | gene segment | immunoglobulin kappa variable 2-112 |
| 6 | gene | 68.25639 | 68.25687 | positive | MGI_C57BL6J_4439863 | Igkv14-111 | NCBI_Gene:545847,ENSEMBL:ENSMUSG00000095771 | MGI:4439863 | gene segment | immunoglobulin kappa variable 14-111 |
| 6 | gene | 68.25739 | 68.25754 | negative | MGI_C57BL6J_5455213 | Gm25436 | ENSEMBL:ENSMUSG00000088900 | MGI:5455213 | snoRNA gene | predicted gene, 25436 |
| 6 | gene | 68.27048 | 68.27127 | positive | MGI_C57BL6J_4439558 | Igkv1-110 | NCBI_Gene:381777,ENSEMBL:ENSMUSG00000093861 | MGI:4439558 | gene segment | immunoglobulin kappa variable 1-110 |
| 6 | pseudogene | 68.29700 | 68.29729 | negative | MGI_C57BL6J_5663144 | Gm43007 | ENSEMBL:ENSMUSG00000104722 | MGI:5663144 | pseudogene | predicted gene 43007 |
| 6 | pseudogene | 68.29776 | 68.29849 | positive | MGI_C57BL6J_5662749 | Gm42612 | ENSEMBL:ENSMUSG00000105684 | MGI:5662749 | pseudogene | predicted gene 42612 |
| 6 | gene | 68.30243 | 68.30316 | positive | MGI_C57BL6J_3642626 | Igkv2-109 | NCBI_Gene:628268,ENSEMBL:ENSMUSG00000105606 | MGI:3642626 | gene segment | immunoglobulin kappa variable 2-109 |
| 6 | pseudogene | 68.31205 | 68.31236 | positive | MGI_C57BL6J_3644576 | Igkv1-108 | NCBI_Gene:628272,ENSEMBL:ENSMUSG00000105043 | MGI:3644576 | pseudogenic gene segment | immunoglobulin kappa variable 1-108 |
| 6 | pseudogene | 68.32610 | 68.32675 | positive | MGI_C57BL6J_3645630 | Igkv2-107 | NCBI_Gene:628283,ENSEMBL:ENSMUSG00000106494 | MGI:3645630 | pseudogenic gene segment | immunoglobulin kappa variable 2-107 |
| 6 | pseudogene | 68.33953 | 68.34000 | positive | MGI_C57BL6J_3648899 | Igkv11-106 | NCBI_Gene:628293,ENSEMBL:ENSMUSG00000076530 | MGI:3648899 | pseudogenic gene segment | immunoglobulin kappa variable 11-106 |
| 6 | pseudogene | 68.34865 | 68.34944 | positive | MGI_C57BL6J_5009874 | Igkv2-105 | NCBI_Gene:692170,ENSEMBL:ENSMUSG00000106066 | MGI:5009874 | pseudogenic gene segment | immunoglobulin kappa chain variable 2-105 |
| 6 | pseudogene | 68.38634 | 68.38919 | negative | MGI_C57BL6J_3646417 | Gm5310 | NCBI_Gene:384408,ENSEMBL:ENSMUSG00000105248 | MGI:3646417 | pseudogene | predicted gene 5310 |
| 6 | gene | 68.42557 | 68.42607 | positive | MGI_C57BL6J_2685913 | Igkv16-104 | NCBI_Gene:381778,ENSEMBL:ENSMUSG00000076522 | MGI:2685913 | gene segment | immunoglobulin kappa variable 16-104 |
| 6 | gene | 68.43744 | 68.43793 | positive | MGI_C57BL6J_96513 | Igkv15-103 | NCBI_Gene:692169,ENSEMBL:ENSMUSG00000076523 | MGI:96513 | gene segment | immunoglobulin kappa chain variable 15-103 |
| 6 | pseudogene | 68.46613 | 68.46659 | negative | MGI_C57BL6J_5009875 | Igkv15-102 | NCBI_Gene:692171,ENSEMBL:ENSMUSG00000104832 | MGI:5009875 | pseudogenic gene segment | immunoglobulin kappa chain variable 15-102 |
| 6 | gene | 68.47461 | 68.47510 | positive | MGI_C57BL6J_5009884 | Igkv20-101-2 | NCBI_Gene:724035,ENSEMBL:ENSMUSG00000106403 | MGI:5009884 | gene segment | immunoglobulin kappa chain variable 20-101-2 |
| 6 | pseudogene | 68.47908 | 68.47957 | positive | MGI_C57BL6J_5009872 | Igkv15-101-1 | NCBI_Gene:692167,ENSEMBL:ENSMUSG00000106187 | MGI:5009872 | pseudogenic gene segment | immunoglobulin kappa chain variable 15-101-1 |
| 6 | pseudogene | 68.48142 | 68.48172 | negative | MGI_C57BL6J_5009873 | Igkv15-101 | NCBI_Gene:692168,ENSEMBL:ENSMUSG00000105319 | MGI:5009873 | pseudogenic gene segment | immunoglobulin kappa chain variable 15-101 |
| 6 | gene | 68.51901 | 68.51948 | positive | MGI_C57BL6J_4439559 | Igkv14-100 | NCBI_Gene:243439,ENSEMBL:ENSMUSG00000096515 | MGI:4439559 | gene segment | immunoglobulin kappa chain variable 14-100 |
| 6 | gene | 68.53274 | 68.53288 | positive | MGI_C57BL6J_5455610 | Gm25833 | ENSEMBL:ENSMUSG00000077325 | MGI:5455610 | snoRNA gene | predicted gene, 25833 |
| 6 | gene | 68.54164 | 68.54242 | positive | MGI_C57BL6J_4439724 | Igkv1-99 | NCBI_Gene:434028,ENSEMBL:ENSMUSG00000076525 | MGI:4439724 | gene segment | immunoglobulin kappa variable 1-99 |
| 6 | gene | 68.57076 | 68.57124 | positive | MGI_C57BL6J_4439560 | Igkv12-98 | NCBI_Gene:435900,ENSEMBL:ENSMUSG00000076526 | MGI:4439560 | gene segment | immunoglobulin kappa variable 12-98 |
| 6 | pseudogene | 68.59137 | 68.59183 | positive | MGI_C57BL6J_5009871 | Igkv15-97 | NCBI_Gene:692166,ENSEMBL:ENSMUSG00000105817 | MGI:5009871 | pseudogenic gene segment | immunoglobulin kappa chain variable 15-97 |
| 6 | gene | 68.63196 | 68.63244 | negative | MGI_C57BL6J_4439561 | Igkv10-96 | NCBI_Gene:692165,ENSEMBL:ENSMUSG00000094420 | MGI:4439561 | gene segment | immunoglobulin kappa variable 10-96 |
| 6 | pseudogene | 68.64741 | 68.64853 | negative | MGI_C57BL6J_4937167 | Igkv2-95-2 | NCBI_Gene:692164,ENSEMBL:ENSMUSG00000090781 | MGI:4937167 | pseudogenic gene segment | immunoglobulin kappa variable 2-95-2 |
| 6 | pseudogene | 68.67075 | 68.67188 | positive | MGI_C57BL6J_5009870 | Igkv2-95-1 | NCBI_Gene:692163,ENSEMBL:ENSMUSG00000106381 | MGI:5009870 | pseudogenic gene segment | immunoglobulin kappa chain variable |
| 6 | gene | 68.68038 | 68.68085 | positive | MGI_C57BL6J_3644598 | Igkv10-95 | NCBI_Gene:434031,ENSEMBL:ENSMUSG00000094902 | MGI:3644598 | gene segment | immunoglobulin kappa variable 10-95 |
| 6 | gene | 68.70451 | 68.70498 | negative | MGI_C57BL6J_3646140 | Igkv10-94 | NCBI_Gene:667550,ENSEMBL:ENSMUSG00000096490 | MGI:3646140 | gene segment | immunoglobulin kappa variable 10-94 |
| 6 | pseudogene | 68.71237 | 68.71351 | negative | MGI_C57BL6J_5009869 | Igkv2-93-1 | NCBI_Gene:692162,ENSEMBL:ENSMUSG00000106318 | MGI:5009869 | pseudogenic gene segment | immunoglobulin kappa chain variable 2-93-1 |
| 6 | pseudogene | 68.73418 | 68.73466 | negative | MGI_C57BL6J_5623299 | Gm40414 | ENSEMBL:ENSMUSG00000106309 | MGI:5623299 | pseudogene | predicted gene, 40414 |
| 6 | gene | 68.73630 | 68.73678 | negative | MGI_C57BL6J_107617 | Igkv19-93 | NCBI_Gene:692161,ENSEMBL:ENSMUSG00000098814 | MGI:107617 | gene segment | immunoglobulin kappa chain variable 19-93 |
| 6 | gene | 68.75504 | 68.75559 | negative | MGI_C57BL6J_2686254 | Igkv4-92 | NCBI_Gene:384410,ENSEMBL:ENSMUSG00000076531 | MGI:2686254 | gene segment | immunoglobulin kappa variable 4-92 |
| 6 | pseudogene | 68.76328 | 68.76369 | negative | MGI_C57BL6J_5663062 | Gm42925 | ENSEMBL:ENSMUSG00000104995 | MGI:5663062 | pseudogenic gene segment | predicted gene 42925 |
| 6 | gene | 68.76856 | 68.76910 | negative | MGI_C57BL6J_3642277 | Igkv4-91 | NCBI_Gene:434033,ENSEMBL:ENSMUSG00000076532 | MGI:3642277 | gene segment | immunoglobulin kappa chain variable 4-91 |
| 6 | gene | 68.79908 | 68.80246 | positive | MGI_C57BL6J_1922585 | 4930549O18Rik | NA | NA | unclassified gene | RIKEN cDNA 4930549O18 gene |
| 6 | gene | 68.80718 | 68.80771 | negative | MGI_C57BL6J_4439830 | Igkv4-90 | NCBI_Gene:434034,ENSEMBL:ENSMUSG00000076533 | MGI:4439830 | gene segment | immunoglobulin kappa chain variable 4-90 |
| 6 | pseudogene | 68.81050 | 68.81054 | positive | MGI_C57BL6J_5009868 | Igkv13-89-1 | NCBI_Gene:692155 | MGI:5009868 | pseudogenic gene segment | immunoglobulin kappa chain variable 13-89-1 |
| 6 | pseudogene | 68.81067 | 68.81085 | positive | MGI_C57BL6J_5663061 | Gm42924 | ENSEMBL:ENSMUSG00000104849 | MGI:5663061 | pseudogenic gene segment | predicted gene 42924 |
| 6 | gene | 68.83485 | 68.83531 | negative | MGI_C57BL6J_4439829 | Igkv12-89 | NCBI_Gene:384411,ENSEMBL:ENSMUSG00000076534 | MGI:4439829 | gene segment | immunoglobulin kappa chain variable 12-89 |
| 6 | gene | 68.86226 | 68.86303 | negative | MGI_C57BL6J_4439828 | Igkv1-88 | NCBI_Gene:692154,ENSEMBL:ENSMUSG00000076535 | MGI:4439828 | gene segment | immunoglobulin kappa chain variable 1-88 |
| 6 | pseudogene | 68.87543 | 68.87582 | negative | MGI_C57BL6J_5662676 | Gm42539 | ENSEMBL:ENSMUSG00000106124 | MGI:5662676 | pseudogenic gene segment | predicted gene 42539 |
| 6 | gene | 68.88018 | 68.88067 | negative | MGI_C57BL6J_5662680 | Gm42543 | ENSEMBL:ENSMUSG00000105630 | MGI:5662680 | gene segment | predicted gene 42543 |
| 6 | pseudogene | 68.89358 | 68.89398 | negative | MGI_C57BL6J_5662679 | Gm42542 | ENSEMBL:ENSMUSG00000105267 | MGI:5662679 | pseudogenic gene segment | predicted gene 42542 |
| 6 | pseudogene | 68.90280 | 68.90327 | positive | MGI_C57BL6J_3779399 | Igkv13-87 | NCBI_Gene:692153,ENSEMBL:ENSMUSG00000079544 | MGI:3779399 | pseudogenic gene segment | immunoglobulin kappa variable 13-87 |
| 6 | pseudogene | 68.90941 | 68.90986 | positive | MGI_C57BL6J_5663847 | Gm43710 | ENSEMBL:ENSMUSG00000104838 | MGI:5663847 | pseudogene | predicted gene 43710 |
| 6 | gene | 68.91041 | 68.91097 | negative | MGI_C57BL6J_2685305 | Igkv4-86 | NCBI_Gene:243451,ENSEMBL:ENSMUSG00000076536 | MGI:2685305 | gene segment | immunoglobulin kappa variable 4-86 |
| 6 | gene | 68.93027 | 68.93073 | negative | MGI_C57BL6J_4439827 | Igkv13-85 | NCBI_Gene:434036,ENSEMBL:ENSMUSG00000079543 | MGI:4439827 | gene segment | immunoglobulin kappa chain variable 13-85 |
| 6 | gene | 68.93960 | 68.94007 | positive | MGI_C57BL6J_96514 | Igkv13-84 | NCBI_Gene:692152,ENSEMBL:ENSMUSG00000076538 | MGI:96514 | gene segment | immunoglobulin kappa chain variable 13-84 |
| 6 | pseudogene | 68.96138 | 68.96202 | negative | MGI_C57BL6J_5009867 | Igkv4-83 | NCBI_Gene:692145,ENSEMBL:ENSMUSG00000076557 | MGI:5009867 | pseudogenic gene segment | immunoglobulin kappa chain variable 4-83 |
| 6 | pseudogene | 68.97920 | 68.97966 | positive | MGI_C57BL6J_5009866 | Igkv13-82 | NCBI_Gene:692144,ENSEMBL:ENSMUSG00000105752 | MGI:5009866 | pseudogenic gene segment | immunoglobulin kappa chain variable 13-82 |
| 6 | gene | 68.99076 | 68.99129 | negative | MGI_C57BL6J_2685306 | Igkv4-81 | NCBI_Gene:243453,ENSEMBL:ENSMUSG00000076539 | MGI:2685306 | gene segment | immunoglobulin kappa variable 4-81 |
| 6 | pseudogene | 69.00914 | 69.00951 | positive | MGI_C57BL6J_5009865 | Igkv13-80-1 | NCBI_Gene:692143,ENSEMBL:ENSMUSG00000105539 | MGI:5009865 | pseudogenic gene segment | immunoglobulin kappa chain variable 13-80-1 |
| 6 | gene | 69.01656 | 69.01708 | negative | MGI_C57BL6J_4439653 | Igkv4-80 | NCBI_Gene:545848,ENSEMBL:ENSMUSG00000076540 | MGI:4439653 | gene segment | immunoglobulin kappa variable 4-80 |
| 6 | gene | 69.04297 | 69.04350 | negative | MGI_C57BL6J_2685040 | Igkv4-79 | NCBI_Gene:213684,ENSEMBL:ENSMUSG00000076541 | MGI:2685040 | gene segment | immunoglobulin kappa variable 4-79 |
| 6 | pseudogene | 69.05144 | 69.05182 | positive | MGI_C57BL6J_3647961 | Igkv13-78-1 | NCBI_Gene:628498,ENSEMBL:ENSMUSG00000105480 | MGI:3647961 | pseudogenic gene segment | immunoglobulin kappa variable 13-78-1 |
| 6 | gene | 69.05969 | 69.06022 | negative | MGI_C57BL6J_3819775 | Igkv4-78 | NCBI_Gene:545849,ENSEMBL:ENSMUSG00000094315 | MGI:3819775 | gene segment | immunoglobulin kappa variable 4-78 |
| 6 | pseudogene | 69.08619 | 69.08649 | positive | MGI_C57BL6J_5663720 | Gm43583 | ENSEMBL:ENSMUSG00000105313 | MGI:5663720 | pseudogenic gene segment | predicted gene 43583 |
| 6 | pseudogene | 69.11090 | 69.11143 | negative | MGI_C57BL6J_3647937 | Igkv4-77 | NCBI_Gene:667621,ENSEMBL:ENSMUSG00000105462 | MGI:3647937 | pseudogenic gene segment | immunoglobulin kappa variable 4-77 |
| 6 | pseudogene | 69.13787 | 69.13833 | positive | MGI_C57BL6J_5009864 | Igkv13-76 | NCBI_Gene:692142,ENSEMBL:ENSMUSG00000104609 | MGI:5009864 | pseudogenic gene segment | immunoglobulin kappa chain variable 13-76 |
| 6 | pseudogene | 69.15612 | 69.15666 | negative | MGI_C57BL6J_5009863 | Igkv4-75 | NCBI_Gene:692141,ENSEMBL:ENSMUSG00000104712 | MGI:5009863 | pseudogenic gene segment | immunoglobulin kappa chain variable 4-75 |
| 6 | pseudogene | 69.17742 | 69.17779 | positive | MGI_C57BL6J_5009862 | Igkv13-74-1 | NCBI_Gene:692140,ENSEMBL:ENSMUSG00000104922 | MGI:5009862 | pseudogenic gene segment | immunoglobulin kappa chain variable 13-74-1 |
| 6 | gene | 69.18483 | 69.18540 | negative | MGI_C57BL6J_3779447 | Igkv4-74 | NCBI_Gene:236047,ENSEMBL:ENSMUSG00000076543 | MGI:3779447 | gene segment | immunoglobulin kappa variable 4-74 |
| 6 | pseudogene | 69.19007 | 69.19028 | positive | MGI_C57BL6J_5009861 | Igkv13-73-1 | NCBI_Gene:692139,ENSEMBL:ENSMUSG00000105989 | MGI:5009861 | pseudogenic gene segment | immunoglobulin kappa chain variable 13-73-1 |
| 6 | gene | 69.19758 | 69.19812 | negative | MGI_C57BL6J_3645825 | Igkv4-73 | NCBI_Gene:628516,ENSEMBL:ENSMUSG00000106387 | MGI:3645825 | gene segment | immunoglobulin kappa variable 4-73 |
| 6 | gene | 69.22685 | 69.22743 | negative | MGI_C57BL6J_2686345 | Igkv4-72 | NCBI_Gene:385109,ENSEMBL:ENSMUSG00000076545 | MGI:2686345 | gene segment | immunoglobulin kappa chain variable 4-72 |
| 6 | pseudogene | 69.23576 | 69.23616 | positive | MGI_C57BL6J_5009860 | Igkv13-71-1 | NCBI_Gene:692138,ENSEMBL:ENSMUSG00000105968 | MGI:5009860 | pseudogenic gene segment | immunoglobulin kappa chain variable 13-71-1 |
| 6 | gene | 69.24316 | 69.24369 | negative | MGI_C57BL6J_4439654 | Igkv4-71 | NCBI_Gene:434586,ENSEMBL:ENSMUSG00000073028 | MGI:4439654 | gene segment | immunoglobulin kappa chain variable 4-71 |
| 6 | gene | 69.26789 | 69.26844 | negative | MGI_C57BL6J_2686348 | Igkv4-70 | NCBI_Gene:385120,ENSEMBL:ENSMUSG00000076547 | MGI:2686348 | gene segment | immunoglobulin kappa chain variable 4-70 |
| 6 | gene | 69.28379 | 69.28432 | negative | MGI_C57BL6J_3648668 | Igkv4-69 | NCBI_Gene:628541,ENSEMBL:ENSMUSG00000076548 | MGI:3648668 | gene segment | immunoglobulin kappa variable 4-69 |
| 6 | gene | 69.30483 | 69.30540 | negative | MGI_C57BL6J_2686265 | Igkv4-68 | NCBI_Gene:384515,ENSEMBL:ENSMUSG00000076549 | MGI:2686265 | gene segment | immunoglobulin kappa variable 4-68 |
| 6 | pseudogene | 69.32418 | 69.32465 | negative | MGI_C57BL6J_3645399 | Igkv12-67 | NCBI_Gene:628551,ENSEMBL:ENSMUSG00000104697 | MGI:3645399 | pseudogenic gene segment | immunoglobulin kappa chain variable 12-67 |
| 6 | gene | 69.32684 | 69.32717 | negative | MGI_C57BL6J_105105 | Rprl1 | NCBI_Gene:19783,ENSEMBL:ENSMUSG00000088675,ENSEMBL:ENSMUSG00000106222 | MGI:105105 | RNase P RNA gene | ribonuclease P RNA-like 1 |
| 6 | pseudogene | 69.33460 | 69.33508 | negative | MGI_C57BL6J_5009859 | Igkv12-66 | NCBI_Gene:692133,ENSEMBL:ENSMUSG00000105110 | MGI:5009859 | pseudogenic gene segment | immunoglobulin kappa chain variable 12-66 |
| 6 | pseudogene | 69.34247 | 69.34300 | negative | MGI_C57BL6J_3644339 | Igkv4-65 | NCBI_Gene:667670,ENSEMBL:ENSMUSG00000105883 | MGI:3644339 | pseudogenic gene segment | immunoglobulin kappa chain variable 4-65 |
| 6 | pseudogene | 69.36272 | 69.36319 | positive | MGI_C57BL6J_5009834 | Igkv13-64 | NCBI_Gene:628564,ENSEMBL:ENSMUSG00000105747 | MGI:5009834 | pseudogenic gene segment | immunoglobulin kappa chain variable 13-64 |
| 6 | gene | 69.37794 | 69.37847 | negative | MGI_C57BL6J_3645235 | Igkv4-63 | NCBI_Gene:628571,ENSEMBL:ENSMUSG00000076550 | MGI:3645235 | gene segment | immunoglobulin kappa variable 4-63 |
| 6 | pseudogene | 69.39242 | 69.39287 | positive | MGI_C57BL6J_5009858 | Igkv13-62-1 | NCBI_Gene:692119,ENSEMBL:ENSMUSG00000105323 | MGI:5009858 | pseudogenic gene segment | immunoglobulin kappa chain variable 13-62-1 |
| 6 | gene | 69.39981 | 69.40034 | negative | MGI_C57BL6J_3643587 | Igkv4-62 | NCBI_Gene:636752,ENSEMBL:ENSMUSG00000094262 | MGI:3643587 | gene segment | immunoglobulin kappa variable 4-62 |
| 6 | pseudogene | 69.40950 | 69.40993 | positive | MGI_C57BL6J_5009857 | Igkv13-61-1 | NCBI_Gene:692118,ENSEMBL:ENSMUSG00000106216 | MGI:5009857 | pseudogenic gene segment | immunoglobulin kappa chain variable 13-61-1 |
| 6 | gene | 69.41689 | 69.41746 | negative | MGI_C57BL6J_4439819 | Igkv4-61 | NCBI_Gene:546244,ENSEMBL:ENSMUSG00000076552 | MGI:4439819 | gene segment | immunoglobulin kappa chain variable 4-61 |
| 6 | gene | 69.43822 | 69.43876 | negative | MGI_C57BL6J_3646808 | Igkv4-59 | NCBI_Gene:667683,ENSEMBL:ENSMUSG00000094006 | MGI:3646808 | gene segment | immunoglobulin kappa variable 4-59 |
| 6 | gene | 69.46319 | 69.46372 | negative | MGI_C57BL6J_3708737 | Igkv4-60 | NCBI_Gene:385277,ENSEMBL:ENSMUSG00000104679 | MGI:3708737 | gene segment | immunoglobulin kappa variable 4-60 |
| 6 | gene | 69.50025 | 69.50079 | negative | MGI_C57BL6J_2685923 | Igkv4-58 | NCBI_Gene:381831,ENSEMBL:ENSMUSG00000095633 | MGI:2685923 | gene segment | immunoglobulin kappa variable 4-58 |
| 6 | gene | 69.51634 | 69.51663 | positive | MGI_C57BL6J_97956 | Rn7s6 | ENSEMBL:ENSMUSG00000092746 | MGI:97956 | SRP RNA gene | 7S RNA 6 |
| 6 | gene | 69.51648 | 69.51655 | positive | MGI_C57BL6J_6096173 | Gm47304 | ENSEMBL:ENSMUSG00000104690 | MGI:6096173 | miRNA gene | predicted gene, 47304 |
| 6 | pseudogene | 69.52355 | 69.52402 | positive | MGI_C57BL6J_5009885 | Igkv13-57-2 | NCBI_Gene:724106,ENSEMBL:ENSMUSG00000104551 | MGI:5009885 | pseudogenic gene segment | immunoglobulin kappa variable 13-57-2 |
| 6 | gene | 69.54436 | 69.54491 | negative | MGI_C57BL6J_2686264 | Igkv4-57-1 | NCBI_Gene:384514,ENSEMBL:ENSMUSG00000076555 | MGI:2686264 | gene segment | immunoglobulin kappa variable 4-57-1 |
| 6 | pseudogene | 69.56974 | 69.57016 | positive | MGI_C57BL6J_5009856 | Igkv13-57-1 | NCBI_Gene:692114,ENSEMBL:ENSMUSG00000105369 | MGI:5009856 | pseudogenic gene segment | immunoglobulin kappa chain variable 13-57-1 |
| 6 | gene | 69.57597 | 69.57650 | negative | MGI_C57BL6J_2685035 | Igkv4-57 | NCBI_Gene:235952,ENSEMBL:ENSMUSG00000076556 | MGI:2685035 | gene segment | immunoglobulin kappa variable 4-57 |
| 6 | pseudogene | 69.58124 | 69.58145 | positive | MGI_C57BL6J_5009855 | Igkv13-56-1 | NCBI_Gene:692113,ENSEMBL:ENSMUSG00000106546 | MGI:5009855 | pseudogenic gene segment | immunoglobulin kappa chain variable 13-56-1 |
| 6 | pseudogene | 69.58729 | 69.58781 | negative | MGI_C57BL6J_4439916 | Igkv4-56 | NCBI_Gene:619743,ENSEMBL:ENSMUSG00000106016 | MGI:4439916 | pseudogenic gene segment | immunoglobulin kappa chain variable 4-56 |
| 6 | pseudogene | 69.59989 | 69.60032 | positive | MGI_C57BL6J_5009854 | Igkv13-55-1 | NCBI_Gene:692112,ENSEMBL:ENSMUSG00000106213 | MGI:5009854 | pseudogenic gene segment | immunoglobulin kappa chain variable 13-55-1 |
| 6 | gene | 69.60728 | 69.60783 | negative | MGI_C57BL6J_2686370 | Igkv4-55 | NCBI_Gene:385253,ENSEMBL:ENSMUSG00000096833 | MGI:2686370 | gene segment | immunoglobulin kappa variable 4-55 |
| 6 | pseudogene | 69.61705 | 69.61748 | positive | MGI_C57BL6J_5009853 | Igkv13-54-1 | NCBI_Gene:692105,ENSEMBL:ENSMUSG00000105301 | MGI:5009853 | pseudogenic gene segment | immunoglobulin kappa chain variable 13-54-1 |
| 6 | gene | 69.63165 | 69.63218 | negative | MGI_C57BL6J_5009821 | Igkv4-54 | NCBI_Gene:385291,ENSEMBL:ENSMUSG00000094319 | MGI:5009821 | gene segment | immunoglobulin kappa chain variable 4-54 |
| 6 | gene | 69.64882 | 69.64936 | negative | MGI_C57BL6J_2686266 | Igkv4-53 | NCBI_Gene:546213,ENSEMBL:ENSMUSG00000095753 | MGI:2686266 | gene segment | immunoglobulin kappa variable 4-53 |
| 6 | pseudogene | 69.66109 | 69.66147 | positive | MGI_C57BL6J_5662743 | Gm42606 | ENSEMBL:ENSMUSG00000079542 | MGI:5662743 | pseudogenic gene segment | predicted gene 42606 |
| 6 | gene | 69.68141 | 69.68197 | negative | MGI_C57BL6J_5009829 | Igkv4-51 | NCBI_Gene:619806,ENSEMBL:ENSMUSG00000094993 | MGI:5009829 | gene segment | immunoglobulin kappa chain variable 4-51 |
| 6 | pseudogene | 69.69330 | 69.69361 | positive | MGI_C57BL6J_5663095 | Gm42958 | ENSEMBL:ENSMUSG00000106298 | MGI:5663095 | pseudogenic gene segment | predicted gene 42958 |
| 6 | gene | 69.70077 | 69.70129 | negative | MGI_C57BL6J_2685915 | Igkv4-50 | NCBI_Gene:381782,ENSEMBL:ENSMUSG00000076562 | MGI:2685915 | gene segment | immunoglobulin kappa variable 4-50 |
| 6 | pseudogene | 69.71650 | 69.71696 | negative | MGI_C57BL6J_5009830 | Igkv12-49 | NCBI_Gene:619824,ENSEMBL:ENSMUSG00000105725 | MGI:5009830 | pseudogenic gene segment | immunoglobulin kappa chain variable 12-49 |
| 6 | gene | 69.72657 | 69.72714 | negative | MGI_C57BL6J_3642817 | Igkv5-48 | NCBI_Gene:619846,ENSEMBL:ENSMUSG00000076563 | MGI:3642817 | gene segment | immunoglobulin kappa variable 5-48 |
| 6 | pseudogene | 69.75085 | 69.75133 | negative | MGI_C57BL6J_3644595 | Igkv12-47 | NCBI_Gene:434037,ENSEMBL:ENSMUSG00000106256 | MGI:3644595 | pseudogenic gene segment | immunoglobulin kappa variable 12-47 |
| 6 | gene | 69.76452 | 69.76502 | negative | MGI_C57BL6J_4439773 | Igkv12-46 | NCBI_Gene:692245,ENSEMBL:ENSMUSG00000076564 | MGI:4439773 | gene segment | immunoglobulin kappa variable 12-46 |
| 6 | gene | 69.77575 | 69.77634 | negative | MGI_C57BL6J_4439774 | Igkv5-45 | NCBI_Gene:545850,ENSEMBL:ENSMUSG00000094094 | MGI:4439774 | gene segment | immunoglobulin kappa chain variable 5-45 |
| 6 | gene | 69.81463 | 69.81515 | negative | MGI_C57BL6J_4439775 | Igkv12-44 | NCBI_Gene:545851,ENSEMBL:ENSMUSG00000096422 | MGI:4439775 | gene segment | immunoglobulin kappa variable 12-44 |
| 6 | gene | 69.82336 | 69.82394 | negative | MGI_C57BL6J_4943320 | Igkv5-43 | NCBI_Gene:381783,ENSEMBL:ENSMUSG00000094433 | MGI:4943320 | gene segment | immunoglobulin kappa chain variable 5-43 |
| 6 | pseudogene | 69.83479 | 69.83525 | negative | MGI_C57BL6J_5009852 | Igkv12-42 | NCBI_Gene:677861,ENSEMBL:ENSMUSG00000105915 | MGI:5009852 | pseudogenic gene segment | immunoglobulin kappa chain variable 12-42 |
| 6 | gene | 69.85842 | 69.85888 | negative | MGI_C57BL6J_4439772 | Igkv12-41 | NCBI_Gene:619960,ENSEMBL:ENSMUSG00000095007 | MGI:4439772 | gene segment | immunoglobulin kappa chain variable 12-41 |
| 6 | pseudogene | 69.86859 | 69.86919 | negative | MGI_C57BL6J_5009831 | Igkv5-40-1 | NCBI_Gene:619974,ENSEMBL:ENSMUSG00000104533 | MGI:5009831 | pseudogenic gene segment | immunoglobulin kappa chain variable 5-40-1 |
| 6 | pseudogene | 69.87935 | 69.87982 | negative | MGI_C57BL6J_3643951 | Igkv12-40 | NCBI_Gene:435904,ENSEMBL:ENSMUSG00000105955 | MGI:3643951 | pseudogenic gene segment | immunoglobulin kappa chain variable 12-40 |
| 6 | gene | 69.90042 | 69.90098 | negative | MGI_C57BL6J_2686255 | Igkv5-39 | NCBI_Gene:620017,ENSEMBL:ENSMUSG00000076569 | MGI:2686255 | gene segment | immunoglobulin kappa variable 5-39 |
| 6 | pseudogene | 69.92762 | 69.92790 | negative | MGI_C57BL6J_5663357 | Gm43220 | ENSEMBL:ENSMUSG00000105451 | MGI:5663357 | pseudogenic gene segment | predicted gene 43220 |
| 6 | gene | 69.94319 | 69.94365 | negative | MGI_C57BL6J_4439614 | Igkv12-38 | NCBI_Gene:620050,ENSEMBL:ENSMUSG00000076570 | MGI:4439614 | gene segment | immunoglobulin kappa chain variable 12-38 |
| 6 | gene | 69.96331 | 69.96387 | negative | MGI_C57BL6J_2686256 | Igkv5-37 | NCBI_Gene:384417,ENSEMBL:ENSMUSG00000076571 | MGI:2686256 | gene segment | immunoglobulin kappa variable 5-37 |
| 6 | gene | 69.99246 | 69.99298 | negative | MGI_C57BL6J_4439613 | Igkv18-36 | NCBI_Gene:620088,ENSEMBL:ENSMUSG00000076572 | MGI:4439613 | gene segment | immunoglobulin kappa chain variable 18-36 |
| 6 | gene | 70.01095 | 70.01167 | negative | MGI_C57BL6J_4439612 | Igkv1-35 | NCBI_Gene:620105,ENSEMBL:ENSMUSG00000076573 | MGI:4439612 | gene segment | immunoglobulin kappa variable 1-35 |
| 6 | gene | 70.02798 | 70.02809 | negative | MGI_C57BL6J_5453471 | Gm23694 | ENSEMBL:ENSMUSG00000094641 | MGI:5453471 | miRNA gene | predicted gene, 23694 |
| 6 | pseudogene | 70.04034 | 70.04065 | negative | MGI_C57BL6J_5663723 | Gm43586 | ENSEMBL:ENSMUSG00000106226 | MGI:5663723 | pseudogenic gene segment | predicted gene 43586 |
| 6 | gene | 70.04411 | 70.04468 | negative | MGI_C57BL6J_4949914 | Igkv8-34 | NCBI_Gene:620126,ENSEMBL:ENSMUSG00000104769 | MGI:4949914 | gene segment | immunoglobulin kappa variable 8-34 |
| 6 | gene | 70.05863 | 70.05920 | negative | MGI_C57BL6J_3577282 | Igkv7-33 | NCBI_Gene:243461,ENSEMBL:ENSMUSG00000076575 | MGI:3577282 | gene segment | immunoglobulin kappa chain variable 7-33 |
| 6 | gene | 70.07402 | 70.07460 | negative | MGI_C57BL6J_3641634 | Igkv6-32 | NCBI_Gene:434039,ENSEMBL:ENSMUSG00000076576 | MGI:3641634 | gene segment | immunoglobulin kappa variable 6-32 |
| 6 | pseudogene | 70.10586 | 70.10641 | negative | MGI_C57BL6J_3644798 | Igkv8-31 | NCBI_Gene:546905,ENSEMBL:ENSMUSG00000105781 | MGI:3644798 | pseudogenic gene segment | immunoglobulin kappa chain variable 8-31 |
| 6 | gene | 70.11706 | 70.11762 | negative | MGI_C57BL6J_3642250 | Igkv8-30 | NCBI_Gene:384419,ENSEMBL:ENSMUSG00000076577 | MGI:3642250 | gene segment | immunoglobulin kappa chain variable 8-30 |
| 6 | pseudogene | 70.12965 | 70.13064 | positive | MGI_C57BL6J_5662955 | Gm42818 | ENSEMBL:ENSMUSG00000106192 | MGI:5662955 | pseudogene | predicted gene 42818 |
| 6 | pseudogene | 70.13068 | 70.13250 | positive | MGI_C57BL6J_3644437 | Amd-ps2 | NCBI_Gene:545852 | MGI:3644437 | pseudogene | S-adenosylmethionine decarboxylase, pseudogene 2 |
| 6 | gene | 70.13846 | 70.13911 | negative | MGI_C57BL6J_4439616 | Igkv6-29 | NCBI_Gene:620191,ENSEMBL:ENSMUSG00000076578 | MGI:4439616 | gene segment | immunoglobulin kappa chain variable 6-29 |
| 6 | gene | 70.14359 | 70.14417 | negative | MGI_C57BL6J_3642251 | Igkv8-28 | NCBI_Gene:434040,ENSEMBL:ENSMUSG00000094356 | MGI:3642251 | gene segment | immunoglobulin kappa variable 8-28 |
| 6 | gene | 70.14794 | 70.14804 | negative | MGI_C57BL6J_5453873 | Gm24096 | ENSEMBL:ENSMUSG00000096560 | MGI:5453873 | miRNA gene | predicted gene, 24096 |
| 6 | gene | 70.17181 | 70.17227 | negative | MGI_C57BL6J_4439868 | Igkv8-27 | NCBI_Gene:434041,ENSEMBL:ENSMUSG00000076580 | MGI:4439868 | gene segment | immunoglobulin kappa chain variable 8-27 |
| 6 | gene | 70.19323 | 70.19380 | positive | MGI_C57BL6J_4439869 | Igkv8-26 | NCBI_Gene:620240,ENSEMBL:ENSMUSG00000076581 | MGI:4439869 | gene segment | immunoglobulin kappa variable 8-26 |
| 6 | pseudogene | 70.20953 | 70.21516 | negative | MGI_C57BL6J_3643936 | Gm8828 | NCBI_Gene:667812,ENSEMBL:ENSMUSG00000105727 | MGI:3643936 | pseudogene | predicted gene 8828 |
| 6 | gene | 70.21540 | 70.21596 | positive | MGI_C57BL6J_4439867 | Igkv6-25 | NCBI_Gene:381784,ENSEMBL:ENSMUSG00000094930 | MGI:4439867 | gene segment | immunoglobulin kappa chain variable 6-25 |
| 6 | gene | 70.21686 | 70.21742 | negative | MGI_C57BL6J_4947958 | Igkv8-24 | NCBI_Gene:677858,ENSEMBL:ENSMUSG00000076583 | MGI:4947958 | gene segment | immunoglobulin kappa chain variable 8-24 |
| 6 | gene | 70.21885 | 70.22070 | negative | MGI_C57BL6J_5826599 | Gm46962 | NCBI_Gene:108169149 | MGI:5826599 | lncRNA gene | predicted gene, 46962 |
| 6 | gene | 70.24043 | 70.24071 | positive | MGI_C57BL6J_4947960 | Igkv8-23-1 | NCBI_Gene:434685 | MGI:4947960 | gene segment | immunoglobulin kappa variable 8-23-1 |
| 6 | gene | 70.24043 | 70.24097 | negative | MGI_C57BL6J_5663355 | Gm43218 | ENSEMBL:ENSMUSG00000105432 | MGI:5663355 | gene segment | predicted gene 43218 |
| 6 | gene | 70.26041 | 70.26095 | negative | MGI_C57BL6J_3711980 | Igkv6-23 | NCBI_Gene:637227,ENSEMBL:ENSMUSG00000095630 | MGI:3711980 | gene segment | immunoglobulin kappa variable 6-23 |
| 6 | pseudogene | 70.26195 | 70.26393 | positive | MGI_C57BL6J_3646852 | Gm8836 | NCBI_Gene:667829,ENSEMBL:ENSMUSG00000106080 | MGI:3646852 | pseudogene | predicted gene 8836 |
| 6 | pseudogene | 70.29518 | 70.29541 | positive | MGI_C57BL6J_3779291 | Gm11065 | ENSEMBL:ENSMUSG00000079230 | MGI:3779291 | pseudogene | predicted gene 11065 |
| 6 | pseudogene | 70.29755 | 70.29812 | negative | MGI_C57BL6J_5009833 | Igkv8-22 | NCBI_Gene:620387,ENSEMBL:ENSMUSG00000105668 | MGI:5009833 | pseudogenic gene segment | immunoglobulin kappa chain variable 8-22 |
| 6 | gene | 70.31490 | 70.31547 | negative | MGI_C57BL6J_1330840 | Igkv8-21 | NCBI_Gene:620400,ENSEMBL:ENSMUSG00000076586 | MGI:1330840 | gene segment | immunoglobulin kappa variable 8-21 |
| 6 | pseudogene | 70.32654 | 70.32989 | positive | MGI_C57BL6J_3647577 | Amd-ps5 | NCBI_Gene:667840,ENSEMBL:ENSMUSG00000105114 | MGI:3647577 | pseudogene | S-adenosylmethionine decarboxylase, pseudogene 5 |
| 6 | gene | 70.33584 | 70.33651 | negative | MGI_C57BL6J_1330836 | Igkv6-20 | NCBI_Gene:108024,ENSEMBL:ENSMUSG00000076587 | MGI:1330836 | gene segment | immunoglobulin kappa variable 6-20 |
| 6 | gene | 70.34088 | 70.34145 | negative | MGI_C57BL6J_1330844 | Igkv8-19 | NCBI_Gene:232065,ENSEMBL:ENSMUSG00000096594 | MGI:1330844 | gene segment | immunoglobulin kappa variable 8-19 |
| 6 | gene | 70.35585 | 70.35644 | positive | MGI_C57BL6J_1330845 | Igkv8-18 | NCBI_Gene:667859,ENSEMBL:ENSMUSG00000076589 | MGI:1330845 | gene segment | immunoglobulin kappa variable 8-18 |
| 6 | pseudogene | 70.36844 | 70.36980 | negative | MGI_C57BL6J_3647327 | Gm8848 | NCBI_Gene:667863,ENSEMBL:ENSMUSG00000106031 | MGI:3647327 | pseudogene | predicted gene 8848 |
| 6 | gene | 70.37144 | 70.37199 | positive | MGI_C57BL6J_1330833 | Igkv6-17 | NCBI_Gene:667865,ENSEMBL:ENSMUSG00000095794 | MGI:1330833 | gene segment | immunoglobulin kappa variable 6-17 |
| 6 | gene | 70.38667 | 70.38725 | negative | MGI_C57BL6J_1330843 | Igkv8-16 | NCBI_Gene:640340,ENSEMBL:ENSMUSG00000076591 | MGI:1330843 | gene segment | immunoglobulin kappa variable 8-16 |
| 6 | pseudogene | 70.40570 | 70.40593 | negative | MGI_C57BL6J_5662804 | Gm42667 | ENSEMBL:ENSMUSG00000106428 | MGI:5662804 | pseudogenic gene segment | predicted gene 42667 |
| 6 | gene | 70.40647 | 70.40699 | negative | MGI_C57BL6J_1330831 | Igkv6-15 | NCBI_Gene:108022,ENSEMBL:ENSMUSG00000094797 | MGI:1330831 | gene segment | immunoglobulin kappa variable 6-15 |
| 6 | pseudogene | 70.40863 | 70.41001 | positive | MGI_C57BL6J_3644164 | Gm8853 | NCBI_Gene:667872,ENSEMBL:ENSMUSG00000106624 | MGI:3644164 | pseudogene | predicted gene 8853 |
| 6 | pseudogene | 70.42416 | 70.42443 | positive | MGI_C57BL6J_3708738 | Gm10360 | ENSEMBL:ENSMUSG00000072477 | MGI:3708738 | pseudogenic gene segment | predicted gene 10360 |
| 6 | pseudogene | 70.42892 | 70.43121 | negative | MGI_C57BL6J_3779561 | Gm6157 | NCBI_Gene:620497,ENSEMBL:ENSMUSG00000105687 | MGI:3779561 | pseudogene | predicted gene 6157 |
| 6 | gene | 70.43495 | 70.43548 | negative | MGI_C57BL6J_1330830 | Igkv6-14 | NCBI_Gene:667881,ENSEMBL:ENSMUSG00000096844 | MGI:1330830 | gene segment | immunoglobulin kappa variable 6-14 |
| 6 | pseudogene | 70.43711 | 70.43847 | positive | MGI_C57BL6J_3648004 | Gm8862 | NCBI_Gene:667889,ENSEMBL:ENSMUSG00000104643 | MGI:3648004 | pseudogene | predicted gene 8862 |
| 6 | gene | 70.45750 | 70.45804 | negative | MGI_C57BL6J_1330829 | Igkv6-13 | NCBI_Gene:667899,ENSEMBL:ENSMUSG00000076594 | MGI:1330829 | gene segment | immunoglobulin kappa variable 6-13 |
| 6 | pseudogene | 70.45966 | 70.46102 | positive | MGI_C57BL6J_3645636 | Gm8872 | NCBI_Gene:667902,ENSEMBL:ENSMUSG00000105305 | MGI:3645636 | pseudogene | predicted gene 8872 |
| 6 | pseudogene | 70.49336 | 70.49366 | negative | MGI_C57BL6J_5662857 | Gm42720 | ENSEMBL:ENSMUSG00000106315 | MGI:5662857 | pseudogenic gene segment | predicted gene 42720 |
| 6 | pseudogene | 70.49538 | 70.49553 | positive | MGI_C57BL6J_5662910 | Gm42773 | ENSEMBL:ENSMUSG00000104692 | MGI:5662910 | pseudogene | predicted gene 42773 |
| 6 | pseudogene | 70.49766 | 70.49834 | negative | MGI_C57BL6J_3645632 | Igkv3-12-1 | NCBI_Gene:667908,ENSEMBL:ENSMUSG00000105862 | MGI:3645632 | pseudogenic gene segment | immunoglobulin kappa chain variable 3-12-1 |
| 6 | gene | 70.51825 | 70.51885 | positive | MGI_C57BL6J_1330815 | Igkv3-12 | NCBI_Gene:667914,ENSEMBL:ENSMUSG00000094117 | MGI:1330815 | gene segment | immunoglobulin kappa variable 3-12 |
| 6 | pseudogene | 70.55324 | 70.55444 | positive | MGI_C57BL6J_1330820 | Igkv3-11 | NCBI_Gene:667917,ENSEMBL:ENSMUSG00000105499 | MGI:1330820 | pseudogenic gene segment | immunoglobulin kappa variable 3-11 |
| 6 | gene | 70.57263 | 70.57323 | positive | MGI_C57BL6J_1330821 | Igkv3-10 | NCBI_Gene:667924,ENSEMBL:ENSMUSG00000076596 | MGI:1330821 | gene segment | immunoglobulin kappa variable 3-10 |
| 6 | gene | 70.58819 | 70.58878 | positive | MGI_C57BL6J_1330856 | Igkv3-9 | NCBI_Gene:667928,ENSEMBL:ENSMUSG00000095338 | MGI:1330856 | gene segment | immunoglobulin kappa variable 3-9 |
| 6 | pseudogene | 70.58887 | 70.58945 | positive | MGI_C57BL6J_1330857 | Igkv3-8 | NCBI_Gene:108010,ENSEMBL:ENSMUSG00000104760 | MGI:1330857 | pseudogenic gene segment | immunoglobulin kappa variable 3-8 |
| 6 | gene | 70.60744 | 70.60804 | positive | MGI_C57BL6J_1330852 | Igkv3-7 | NCBI_Gene:108005,ENSEMBL:ENSMUSG00000076598 | MGI:1330852 | gene segment | immunoglobulin kappa variable 3-7 |
| 6 | pseudogene | 70.64240 | 70.64356 | positive | MGI_C57BL6J_1330853 | Igkv3-6 | NCBI_Gene:667936,ENSEMBL:ENSMUSG00000106667 | MGI:1330853 | pseudogenic gene segment | immunoglobulin kappa variable 3-6 |
| 6 | gene | 70.66327 | 70.66389 | positive | MGI_C57BL6J_1330854 | Igkv3-5 | NCBI_Gene:667940,ENSEMBL:ENSMUSG00000095335 | MGI:1330854 | gene segment | immunoglobulin kappa chain variable 3-5 |
| 6 | gene | 70.67179 | 70.67238 | positive | MGI_C57BL6J_1330855 | Igkv3-4 | NCBI_Gene:626347,ENSEMBL:ENSMUSG00000096715 | MGI:1330855 | gene segment | immunoglobulin kappa variable 3-4 |
| 6 | gene | 70.68695 | 70.68753 | positive | MGI_C57BL6J_1330849 | Igkv3-3 | NCBI_Gene:667946,ENSEMBL:ENSMUSG00000094478 | MGI:1330849 | gene segment | immunoglobulin kappa variable 3-3 |
| 6 | gene | 70.69845 | 70.69907 | positive | MGI_C57BL6J_1330850 | Igkv3-2 | NCBI_Gene:626583,ENSEMBL:ENSMUSG00000095351 | MGI:1330850 | gene segment | immunoglobulin kappa variable 3-2 |
| 6 | gene | 70.70358 | 70.70418 | positive | MGI_C57BL6J_1330851 | Igkv3-1 | NCBI_Gene:108004,ENSEMBL:ENSMUSG00000095682 | MGI:1330851 | gene segment | immunoglobulin kappa variable 3-1 |
| 6 | gene | 70.71308 | 70.71928 | negative | MGI_C57BL6J_5826598 | Gm46961 | NCBI_Gene:108169148 | MGI:5826598 | lncRNA gene | predicted gene, 46961 |
| 6 | gene | 70.71907 | 70.75053 | positive | MGI_C57BL6J_5589370 | Gm30211 | NCBI_Gene:102632036,ENSEMBL:ENSMUSG00000105646 | MGI:5589370 | lncRNA gene | predicted gene, 30211 |
| 6 | gene | 70.72256 | 70.72260 | positive | MGI_C57BL6J_1316689 | Igkj1 | NCBI_Gene:110759,ENSEMBL:ENSMUSG00000076604 | MGI:1316689 | gene segment | immunoglobulin kappa joining 1 |
| 6 | gene | 70.72292 | 70.72296 | positive | MGI_C57BL6J_1316690 | Igkj2 | NCBI_Gene:110760,ENSEMBL:ENSMUSG00000076605 | MGI:1316690 | gene segment | immunoglobulin kappa joining 2 |
| 6 | gene | 70.72322 | 70.72326 | positive | MGI_C57BL6J_1316691 | Igkj3 | NCBI_Gene:110761,ENSEMBL:ENSMUSG00000076606 | MGI:1316691 | gene segment | immunoglobulin kappa joining 3 |
| 6 | gene | 70.72355 | 70.72358 | positive | MGI_C57BL6J_1316692 | Igkj4 | NCBI_Gene:110762,ENSEMBL:ENSMUSG00000076607 | MGI:1316692 | gene segment | immunoglobulin kappa joining 4 |
| 6 | gene | 70.72389 | 70.72393 | positive | MGI_C57BL6J_1316738 | Igkj5 | NCBI_Gene:110763,ENSEMBL:ENSMUSG00000076608 | MGI:1316738 | gene segment | immunoglobulin kappa joining 5 |
| 6 | gene | 70.72643 | 70.72697 | positive | MGI_C57BL6J_96495 | Igkc | NCBI_Gene:16071,ENSEMBL:ENSMUSG00000076609 | MGI:96495 | gene segment | immunoglobulin kappa constant |
| 6 | gene | 70.76572 | 70.79223 | negative | MGI_C57BL6J_103254 | Rpia | NCBI_Gene:19895,ENSEMBL:ENSMUSG00000053604 | MGI:103254 | protein coding gene | ribose 5-phosphate isomerase A |
| 6 | gene | 70.80699 | 70.81479 | negative | MGI_C57BL6J_5589490 | Gm30331 | NCBI_Gene:102632191 | MGI:5589490 | lncRNA gene | predicted gene, 30331 |
| 6 | gene | 70.84448 | 70.90524 | positive | MGI_C57BL6J_1341830 | Eif2ak3 | NCBI_Gene:13666,ENSEMBL:ENSMUSG00000031668 | MGI:1341830 | protein coding gene | eukaryotic translation initiation factor 2 alpha kinase 3 |
| 6 | gene | 70.85544 | 70.85558 | positive | MGI_C57BL6J_5453123 | Gm23346 | ENSEMBL:ENSMUSG00000065750 | MGI:5453123 | snoRNA gene | predicted gene, 23346 |
| 6 | gene | 70.88335 | 70.88580 | negative | MGI_C57BL6J_5663429 | Gm43292 | ENSEMBL:ENSMUSG00000105776 | MGI:5663429 | unclassified gene | predicted gene 43292 |
| 6 | gene | 70.91309 | 70.95115 | negative | MGI_C57BL6J_1921471 | Tex37 | NCBI_Gene:74221,ENSEMBL:ENSMUSG00000051896 | MGI:1921471 | protein coding gene | testis expressed 37 |
| 6 | gene | 70.95653 | 70.96107 | positive | MGI_C57BL6J_3511278 | Foxi3 | NCBI_Gene:232077,ENSEMBL:ENSMUSG00000055874 | MGI:3511278 | protein coding gene | forkhead box I3 |
| 6 | gene | 71.03958 | 71.03973 | negative | MGI_C57BL6J_5453262 | Gm23485 | ENSEMBL:ENSMUSG00000077369 | MGI:5453262 | snRNA gene | predicted gene, 23485 |
| 6 | gene | 71.08778 | 71.10663 | negative | MGI_C57BL6J_5621711 | Gm38826 | NCBI_Gene:105242675 | MGI:5621711 | lncRNA gene | predicted gene, 38826 |
| 6 | gene | 71.10561 | 71.10651 | negative | MGI_C57BL6J_5690298 | Gm43906 | ENSEMBL:ENSMUSG00000107446 | MGI:5690298 | unclassified gene | predicted gene, 43906 |
| 6 | pseudogene | 71.12459 | 71.12500 | positive | MGI_C57BL6J_3704270 | Rpl34-ps1 | NCBI_Gene:619547,ENSEMBL:ENSMUSG00000068396 | MGI:3704270 | pseudogene | ribosomal protein L34, pseudogene 1 |
| 6 | gene | 71.12817 | 71.14444 | negative | MGI_C57BL6J_3041254 | Thnsl2 | NCBI_Gene:232078,ENSEMBL:ENSMUSG00000054474 | MGI:3041254 | protein coding gene | threonine synthase-like 2 (bacterial) |
| 6 | gene | 71.14859 | 71.15369 | positive | MGI_C57BL6J_5589550 | Gm30391 | NCBI_Gene:102632265 | MGI:5589550 | lncRNA gene | predicted gene, 30391 |
| 6 | gene | 71.15411 | 71.15797 | positive | MGI_C57BL6J_5621712 | Gm38827 | NCBI_Gene:105242676 | MGI:5621712 | lncRNA gene | predicted gene, 38827 |
| 6 | gene | 71.16480 | 71.16612 | positive | MGI_C57BL6J_5690819 | Gm44427 | ENSEMBL:ENSMUSG00000108206 | MGI:5690819 | lncRNA gene | predicted gene, 44427 |
| 6 | gene | 71.19983 | 71.20502 | positive | MGI_C57BL6J_95479 | Fabp1 | NCBI_Gene:14080,ENSEMBL:ENSMUSG00000054422 | MGI:95479 | protein coding gene | fatty acid binding protein 1, liver |
| 6 | gene | 71.21394 | 71.32223 | negative | MGI_C57BL6J_104790 | Smyd1 | NCBI_Gene:12180,ENSEMBL:ENSMUSG00000055027 | MGI:104790 | protein coding gene | SET and MYND domain containing 1 |
| 6 | gene | 71.27157 | 71.28532 | positive | MGI_C57BL6J_1889377 | Krcc1 | NCBI_Gene:57896,ENSEMBL:ENSMUSG00000053012 | MGI:1889377 | protein coding gene | lysine-rich coiled-coil 1 |
| 6 | gene | 71.27167 | 71.27180 | positive | MGI_C57BL6J_5531121 | Mir8112 | miRBase:MI0026044,NCBI_Gene:102465900,ENSEMBL:ENSMUSG00000098623 | MGI:5531121 | miRNA gene | microRNA 8112 |
| 6 | gene | 71.27649 | 71.27935 | positive | MGI_C57BL6J_5690713 | Gm44321 | ENSEMBL:ENSMUSG00000107749 | MGI:5690713 | unclassified gene | predicted gene, 44321 |
| 6 | gene | 71.29349 | 71.30288 | positive | MGI_C57BL6J_2685916 | Gm1070 | NCBI_Gene:381785,ENSEMBL:ENSMUSG00000073018 | MGI:2685916 | lncRNA gene | predicted gene 1070 |
| 6 | gene | 71.31221 | 71.32268 | negative | MGI_C57BL6J_5621713 | Gm38828 | NCBI_Gene:105242677 | MGI:5621713 | lncRNA gene | predicted gene, 38828 |
| 6 | gene | 71.32279 | 71.33749 | positive | MGI_C57BL6J_88347 | Cd8b1 | NCBI_Gene:12526,ENSEMBL:ENSMUSG00000053044 | MGI:88347 | protein coding gene | CD8 antigen, beta chain 1 |
| 6 | gene | 71.33889 | 71.33953 | negative | MGI_C57BL6J_5690502 | Gm44110 | ENSEMBL:ENSMUSG00000107855 | MGI:5690502 | lncRNA gene | predicted gene, 44110 |
| 6 | pseudogene | 71.34933 | 71.34973 | positive | MGI_C57BL6J_5690503 | Gm44111 | ENSEMBL:ENSMUSG00000107959 | MGI:5690503 | pseudogene | predicted gene, 44111 |
| 6 | gene | 71.35304 | 71.37328 | positive | MGI_C57BL6J_5690566 | Gm44174 | ENSEMBL:ENSMUSG00000108289 | MGI:5690566 | lncRNA gene | predicted gene, 44174 |
| 6 | gene | 71.35435 | 71.36083 | negative | MGI_C57BL6J_5690567 | Gm44175 | ENSEMBL:ENSMUSG00000108132 | MGI:5690567 | lncRNA gene | predicted gene, 44175 |
| 6 | gene | 71.35715 | 71.36083 | negative | MGI_C57BL6J_5621714 | Gm38829 | NA | NA | lncRNA gene | predicted gene%2c 38829 |
| 6 | gene | 71.37343 | 71.37917 | positive | MGI_C57BL6J_88346 | Cd8a | NCBI_Gene:12525,ENSEMBL:ENSMUSG00000053977 | MGI:88346 | protein coding gene | CD8 antigen, alpha chain |
| 6 | gene | 71.38863 | 71.44064 | negative | MGI_C57BL6J_1915727 | Rmnd5a | NCBI_Gene:68477,ENSEMBL:ENSMUSG00000002222 | MGI:1915727 | protein coding gene | required for meiotic nuclear division 5 homolog A |
| 6 | gene | 71.43210 | 71.43511 | negative | MGI_C57BL6J_5690522 | Gm44130 | ENSEMBL:ENSMUSG00000108211 | MGI:5690522 | unclassified gene | predicted gene, 44130 |
| 6 | gene | 71.45199 | 71.45900 | positive | MGI_C57BL6J_5690564 | Gm44172 | ENSEMBL:ENSMUSG00000108213 | MGI:5690564 | lncRNA gene | predicted gene, 44172 |
| 6 | gene | 71.45750 | 71.47915 | negative | MGI_C57BL6J_5589741 | Gm30582 | NCBI_Gene:102632531 | MGI:5589741 | lncRNA gene | predicted gene, 30582 |
| 6 | gene | 71.46331 | 71.46606 | negative | MGI_C57BL6J_5690578 | Gm44186 | ENSEMBL:ENSMUSG00000107460 | MGI:5690578 | unclassified gene | predicted gene, 44186 |
| 6 | gene | 71.49388 | 71.51088 | positive | MGI_C57BL6J_109483 | Rnf103 | NCBI_Gene:22644,ENSEMBL:ENSMUSG00000052656 | MGI:109483 | protein coding gene | ring finger protein 103 |
| 6 | gene | 71.53322 | 71.53787 | positive | MGI_C57BL6J_5621715 | Gm38830 | NCBI_Gene:105242679 | MGI:5621715 | lncRNA gene | predicted gene, 38830 |
| 6 | gene | 71.53851 | 71.54174 | negative | MGI_C57BL6J_5690586 | Gm44194 | ENSEMBL:ENSMUSG00000108278 | MGI:5690586 | unclassified gene | predicted gene, 44194 |
| 6 | gene | 71.54319 | 71.54411 | negative | MGI_C57BL6J_1920544 | 1700040L08Rik | ENSEMBL:ENSMUSG00000107406 | MGI:1920544 | lncRNA gene | RIKEN cDNA 1700040L08 gene |
| 6 | gene | 71.54380 | 71.58261 | positive | MGI_C57BL6J_1913950 | Chmp3 | NCBI_Gene:66700,ENSEMBL:ENSMUSG00000053119 | MGI:1913950 | protein coding gene | charged multivesicular body protein 3 |
| 6 | gene | 71.58897 | 71.63301 | negative | MGI_C57BL6J_98847 | Kdm3a | NCBI_Gene:104263,ENSEMBL:ENSMUSG00000053470 | MGI:98847 | protein coding gene | lysine (K)-specific demethylase 3A |
| 6 | pseudogene | 71.64141 | 71.64214 | negative | MGI_C57BL6J_3645395 | Gm6063 | NCBI_Gene:606532,ENSEMBL:ENSMUSG00000108772 | MGI:3645395 | pseudogene | predicted gene 6063 |
| 6 | pseudogene | 71.67209 | 71.67270 | negative | MGI_C57BL6J_5753344 | Gm44768 | NCBI_Gene:108169193,ENSEMBL:ENSMUSG00000108496 | MGI:5753344 | pseudogene | predicted gene 44768 |
| 6 | pseudogene | 71.68327 | 71.68398 | positive | MGI_C57BL6J_3643398 | Rpl19-ps9 | NCBI_Gene:668034,ENSEMBL:ENSMUSG00000108813 | MGI:3643398 | pseudogene | ribosomal protein L19, pseudogene 9 |
| 6 | gene | 71.70756 | 71.81071 | positive | MGI_C57BL6J_1098827 | Reep1 | NCBI_Gene:52250,ENSEMBL:ENSMUSG00000052852 | MGI:1098827 | protein coding gene | receptor accessory protein 1 |
| 6 | gene | 71.79067 | 71.79348 | positive | MGI_C57BL6J_5753345 | Gm44769 | ENSEMBL:ENSMUSG00000108802 | MGI:5753345 | unclassified gene | predicted gene 44769 |
| 6 | gene | 71.80289 | 71.80415 | positive | MGI_C57BL6J_5753346 | Gm44770 | ENSEMBL:ENSMUSG00000108594 | MGI:5753346 | unclassified gene | predicted gene 44770 |
| 6 | gene | 71.81300 | 71.82697 | negative | MGI_C57BL6J_1913473 | Mrpl35 | NCBI_Gene:66223,ENSEMBL:ENSMUSG00000052962 | MGI:1913473 | protein coding gene | mitochondrial ribosomal protein L35 |
| 6 | gene | 71.83109 | 71.83171 | negative | MGI_C57BL6J_5753347 | Gm44771 | ENSEMBL:ENSMUSG00000108466 | MGI:5753347 | unclassified gene | predicted gene 44771 |
| 6 | gene | 71.83109 | 71.87739 | positive | MGI_C57BL6J_1923864 | Immt | NCBI_Gene:76614,ENSEMBL:ENSMUSG00000052337 | MGI:1923864 | protein coding gene | inner membrane protein, mitochondrial |
| 6 | pseudogene | 71.86917 | 71.87002 | negative | MGI_C57BL6J_5010010 | Gm17825 | NCBI_Gene:100384878,ENSEMBL:ENSMUSG00000108681 | MGI:5010010 | pseudogene | predicted gene, 17825 |
| 6 | gene | 71.88064 | 71.90877 | negative | MGI_C57BL6J_1917206 | Ptcd3 | NCBI_Gene:69956,ENSEMBL:ENSMUSG00000063884 | MGI:1917206 | protein coding gene | pentatricopeptide repeat domain 3 |
| 6 | gene | 71.88256 | 71.88269 | negative | MGI_C57BL6J_5452263 | Gm22486 | ENSEMBL:ENSMUSG00000080465 | MGI:5452263 | snoRNA gene | predicted gene, 22486 |
| 6 | gene | 71.90905 | 71.98493 | positive | MGI_C57BL6J_1096397 | Polr1a | NCBI_Gene:20019,ENSEMBL:ENSMUSG00000049553 | MGI:1096397 | protein coding gene | polymerase (RNA) I polypeptide A |
| 6 | pseudogene | 71.91890 | 71.91984 | negative | MGI_C57BL6J_5011263 | Gm19078 | NCBI_Gene:100418220,ENSEMBL:ENSMUSG00000108305 | MGI:5011263 | pseudogene | predicted gene, 19078 |
| 6 | gene | 71.96115 | 71.96377 | negative | MGI_C57BL6J_5477122 | Gm26628 | ENSEMBL:ENSMUSG00000097133 | MGI:5477122 | lncRNA gene | predicted gene, 26628 |
| 6 | gene | 72.01390 | 72.04253 | negative | MGI_C57BL6J_4840488 | 9130221F21Rik | NCBI_Gene:330350,ENSEMBL:ENSMUSG00000108356 | MGI:4840488 | lncRNA gene | RIKEN cDNA 9130221F21 gene |
| 6 | pseudogene | 72.07305 | 72.07415 | negative | MGI_C57BL6J_3648959 | Gm8952 | NCBI_Gene:668057,ENSEMBL:ENSMUSG00000108344 | MGI:3648959 | pseudogene | predicted gene 8952 |
| 6 | pseudogene | 72.08736 | 72.08785 | negative | MGI_C57BL6J_5753180 | Gm44604 | ENSEMBL:ENSMUSG00000108355 | MGI:5753180 | pseudogene | predicted gene 44604 |
| 6 | gene | 72.09755 | 72.15457 | positive | MGI_C57BL6J_1339963 | St3gal5 | NCBI_Gene:20454,ENSEMBL:ENSMUSG00000056091 | MGI:1339963 | protein coding gene | ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| 6 | gene | 72.10420 | 72.10681 | positive | MGI_C57BL6J_5580144 | Gm29438 | ENSEMBL:ENSMUSG00000101823 | MGI:5580144 | sense intronic lncRNA gene | predicted gene 29438 |
| 6 | gene | 72.12021 | 72.12320 | negative | MGI_C57BL6J_1918515 | 4933431G14Rik | NCBI_Gene:71265,ENSEMBL:ENSMUSG00000108709 | MGI:1918515 | lncRNA gene | RIKEN cDNA 4933431G14 gene |
| 6 | gene | 72.16235 | 72.16783 | positive | MGI_C57BL6J_5621717 | Gm38832 | NCBI_Gene:105242682,ENSEMBL:ENSMUSG00000108371 | MGI:5621717 | lncRNA gene | predicted gene, 38832 |
| 6 | gene | 72.20617 | 72.23582 | negative | MGI_C57BL6J_1918343 | Atoh8 | NCBI_Gene:71093,ENSEMBL:ENSMUSG00000037621 | MGI:1918343 | protein coding gene | atonal bHLH transcription factor 8 |
| 6 | gene | 72.25010 | 72.25499 | negative | MGI_C57BL6J_5589941 | Gm30782 | NCBI_Gene:102632804 | MGI:5589941 | lncRNA gene | predicted gene, 30782 |
| 6 | gene | 72.30461 | 72.31437 | positive | MGI_C57BL6J_109516 | Sftpb | NCBI_Gene:20388,ENSEMBL:ENSMUSG00000056370 | MGI:109516 | protein coding gene | surfactant associated protein B |
| 6 | gene | 72.31868 | 72.34521 | negative | MGI_C57BL6J_107622 | Usp39 | NCBI_Gene:28035,ENSEMBL:ENSMUSG00000056305 | MGI:107622 | protein coding gene | ubiquitin specific peptidase 39 |
| 6 | gene | 72.34703 | 72.34846 | negative | MGI_C57BL6J_5753627 | Gm45051 | ENSEMBL:ENSMUSG00000108435 | MGI:5753627 | lncRNA gene | predicted gene 45051 |
| 6 | gene | 72.34720 | 72.35316 | positive | MGI_C57BL6J_1915614 | 0610030E20Rik | NCBI_Gene:68364,ENSEMBL:ENSMUSG00000058706 | MGI:1915614 | protein coding gene | RIKEN cDNA 0610030E20 gene |
| 6 | gene | 72.35545 | 72.35976 | positive | MGI_C57BL6J_2385244 | Tmem150a | NCBI_Gene:232086,ENSEMBL:ENSMUSG00000055912 | MGI:2385244 | protein coding gene | transmembrane protein 150A |
| 6 | gene | 72.35971 | 72.36697 | negative | MGI_C57BL6J_1913760 | Rnf181 | NCBI_Gene:66510,ENSEMBL:ENSMUSG00000055850 | MGI:1913760 | protein coding gene | ring finger protein 181 |
| 6 | gene | 72.36805 | 72.38047 | negative | MGI_C57BL6J_1858622 | Vamp5 | NCBI_Gene:53620,ENSEMBL:ENSMUSG00000073002 | MGI:1858622 | protein coding gene | vesicle-associated membrane protein 5 |
| 6 | gene | 72.37280 | 72.37471 | negative | MGI_C57BL6J_3040670 | BC019510 | NA | NA | unclassified gene | cDNA sequence BC019510 |
| 6 | gene | 72.37593 | 72.37699 | negative | MGI_C57BL6J_1925206 | A930026B05Rik | NA | NA | unclassified gene | RIKEN cDNA A930026B05 gene |
| 6 | gene | 72.38522 | 72.39070 | negative | MGI_C57BL6J_1336882 | Vamp8 | NCBI_Gene:22320,ENSEMBL:ENSMUSG00000050732 | MGI:1336882 | protein coding gene | vesicle-associated membrane protein 8 |
| 6 | gene | 72.41429 | 72.43111 | positive | MGI_C57BL6J_1927655 | Ggcx | NCBI_Gene:56316,ENSEMBL:ENSMUSG00000053460 | MGI:1927655 | protein coding gene | gamma-glutamyl carboxylase |
| 6 | gene | 72.43280 | 72.43956 | negative | MGI_C57BL6J_2443731 | Mat2a | NCBI_Gene:232087,ENSEMBL:ENSMUSG00000053907 | MGI:2443731 | protein coding gene | methionine adenosyltransferase II, alpha |
| 6 | gene | 72.43868 | 72.44061 | positive | MGI_C57BL6J_1925358 | Particl | NCBI_Gene:78108,ENSEMBL:ENSMUSG00000108591 | MGI:1925358 | lncRNA gene | promoter of Mat2a antisense radiation induced circulating long non-coding RNA |
| 6 | gene | 72.51364 | 72.53421 | negative | MGI_C57BL6J_1918380 | Sh2d6 | NCBI_Gene:71130,ENSEMBL:ENSMUSG00000052631 | MGI:1918380 | protein coding gene | SH2 domain containing 6 |
| 6 | gene | 72.53828 | 72.56298 | positive | MGI_C57BL6J_1098259 | Capg | NCBI_Gene:12332,ENSEMBL:ENSMUSG00000056737 | MGI:1098259 | protein coding gene | capping protein (actin filament), gelsolin-like |
| 6 | gene | 72.56592 | 72.59841 | negative | MGI_C57BL6J_2445168 | Elmod3 | NCBI_Gene:232089,ENSEMBL:ENSMUSG00000056698 | MGI:2445168 | protein coding gene | ELMO/CED-12 domain containing 3 |
| 6 | gene | 72.59847 | 72.60842 | positive | MGI_C57BL6J_1914692 | Retsat | NCBI_Gene:67442,ENSEMBL:ENSMUSG00000056666 | MGI:1914692 | protein coding gene | retinol saturase (all trans retinol 13,14 reductase) |
| 6 | gene | 72.60300 | 72.60550 | negative | MGI_C57BL6J_5753629 | Gm45053 | ENSEMBL:ENSMUSG00000108368 | MGI:5753629 | unclassified gene | predicted gene 45053 |
| 6 | gene | 72.60842 | 72.61700 | negative | MGI_C57BL6J_105080 | Tgoln1 | NCBI_Gene:22134,ENSEMBL:ENSMUSG00000056429 | MGI:105080 | protein coding gene | trans-golgi network protein |
| 6 | gene | 72.60984 | 72.61619 | positive | MGI_C57BL6J_5142001 | Gm20536 | ENSEMBL:ENSMUSG00000092386 | MGI:5142001 | lncRNA gene | predicted gene 20536 |
| 6 | gene | 72.61096 | 72.61675 | negative | MGI_C57BL6J_105079 | Tgoln2 | NA | NA | protein coding gene | trans-golgi network protein 2 |
| 6 | gene | 72.62637 | 72.78925 | negative | MGI_C57BL6J_1202876 | Tcf7l1 | NCBI_Gene:21415,ENSEMBL:ENSMUSG00000055799 | MGI:1202876 | protein coding gene | transcription factor 7 like 1 (T cell specific, HMG box) |
| 6 | gene | 72.62929 | 72.63772 | positive | MGI_C57BL6J_3642201 | Gm15401 | NCBI_Gene:100038714,ENSEMBL:ENSMUSG00000072999 | MGI:3642201 | lncRNA gene | predicted gene 15401 |
| 6 | gene | 72.63515 | 72.63855 | negative | MGI_C57BL6J_5611197 | Gm37969 | ENSEMBL:ENSMUSG00000102577 | MGI:5611197 | unclassified gene | predicted gene, 37969 |
| 6 | gene | 72.64285 | 72.64409 | negative | MGI_C57BL6J_5611342 | Gm38114 | ENSEMBL:ENSMUSG00000102931 | MGI:5611342 | unclassified gene | predicted gene, 38114 |
| 6 | gene | 72.64556 | 72.68653 | positive | MGI_C57BL6J_5590550 | Gm31391 | NCBI_Gene:102633607 | MGI:5590550 | lncRNA gene | predicted gene, 31391 |
| 6 | gene | 72.65232 | 72.65360 | positive | MGI_C57BL6J_5611548 | Gm38320 | ENSEMBL:ENSMUSG00000102983 | MGI:5611548 | unclassified gene | predicted gene, 38320 |
| 6 | pseudogene | 72.68688 | 72.69836 | negative | MGI_C57BL6J_5590474 | Gm31315 | NCBI_Gene:105244842 | MGI:5590474 | pseudogene | predicted gene, 31315 |
| 6 | gene | 72.69921 | 72.70509 | positive | MGI_C57BL6J_3707435 | Gm15402 | ENSEMBL:ENSMUSG00000079524 | MGI:3707435 | lncRNA gene | predicted gene 15402 |
| 6 | gene | 72.74179 | 72.75002 | positive | MGI_C57BL6J_5590408 | Gm31249 | NCBI_Gene:102633417 | MGI:5590408 | lncRNA gene | predicted gene, 31249 |
| 6 | gene | 72.74911 | 72.75191 | negative | MGI_C57BL6J_5610964 | Gm37736 | ENSEMBL:ENSMUSG00000104501 | MGI:5610964 | unclassified gene | predicted gene, 37736 |
| 6 | gene | 72.84111 | 72.89998 | negative | MGI_C57BL6J_1921537 | Kcmf1 | NCBI_Gene:74287,ENSEMBL:ENSMUSG00000055239 | MGI:1921537 | protein coding gene | potassium channel modulatory factor 1 |
| 6 | gene | 72.88361 | 72.88546 | negative | MGI_C57BL6J_2141724 | 6030456E01Rik | NA | NA | unclassified gene | RIKEN cDNA 6030456E01 gene |
| 6 | gene | 72.89496 | 72.89592 | negative | MGI_C57BL6J_1926117 | D530018E20Rik | ENSEMBL:ENSMUSG00000107610 | MGI:1926117 | unclassified gene | RIKEN cDNA D530018E20 gene |
| 6 | gene | 72.95324 | 72.95725 | positive | MGI_C57BL6J_5477134 | Gm26640 | ENSEMBL:ENSMUSG00000097569 | MGI:5477134 | lncRNA gene | predicted gene, 26640 |
| 6 | gene | 72.95735 | 72.95875 | negative | MGI_C57BL6J_109146 | Tmsb10 | NCBI_Gene:19240,ENSEMBL:ENSMUSG00000079523 | MGI:109146 | protein coding gene | thymosin, beta 10 |
| 6 | pseudogene | 72.98248 | 72.98306 | negative | MGI_C57BL6J_5010587 | Gm18402 | NCBI_Gene:100417110,ENSEMBL:ENSMUSG00000107661 | MGI:5010587 | pseudogene | predicted gene, 18402 |
| 6 | gene | 73.00710 | 73.01073 | positive | MGI_C57BL6J_5590706 | Gm31547 | NCBI_Gene:102633814 | MGI:5590706 | lncRNA gene | predicted gene, 31547 |
| 6 | gene | 73.01192 | 73.03352 | positive | MGI_C57BL6J_5295667 | Gm20560 | NCBI_Gene:330354,ENSEMBL:ENSMUSG00000107546 | MGI:5295667 | lncRNA gene | predicted gene, 20560 |
| 6 | gene | 73.01761 | 73.22240 | negative | MGI_C57BL6J_107744 | Dnah6 | NCBI_Gene:330355,ENSEMBL:ENSMUSG00000052861 | MGI:107744 | protein coding gene | dynein, axonemal, heavy chain 6 |
| 6 | gene | 73.05065 | 73.05117 | positive | MGI_C57BL6J_1918715 | 8430413B15Rik | NA | NA | unclassified gene | RIKEN cDNA 8430413B15 gene |
| 6 | gene | 73.14149 | 73.14168 | negative | MGI_C57BL6J_5454310 | Gm24533 | ENSEMBL:ENSMUSG00000065072 | MGI:5454310 | snoRNA gene | predicted gene, 24533 |
| 6 | gene | 73.22280 | 73.22589 | positive | MGI_C57BL6J_5623262 | Gm40377 | NCBI_Gene:105244843,ENSEMBL:ENSMUSG00000107440 | MGI:5623262 | lncRNA gene | predicted gene, 40377 |
| 6 | gene | 73.24838 | 73.27691 | positive | MGI_C57BL6J_1927234 | Suclg1 | NCBI_Gene:56451,ENSEMBL:ENSMUSG00000052738 | MGI:1927234 | protein coding gene | succinate-CoA ligase, GDP-forming, alpha subunit |
| 6 | gene | 73.24876 | 73.25449 | positive | MGI_C57BL6J_5621719 | Gm38834 | NCBI_Gene:105242685 | MGI:5621719 | lncRNA gene | predicted gene, 38834 |
| 6 | gene | 73.28760 | 73.28984 | negative | MGI_C57BL6J_5623263 | Gm40378 | NCBI_Gene:105244844 | MGI:5623263 | protein coding gene | predicted gene, 40378 |
| 6 | pseudogene | 73.31022 | 73.31064 | negative | MGI_C57BL6J_5690867 | Gm44475 | ENSEMBL:ENSMUSG00000107931 | MGI:5690867 | pseudogene | predicted gene, 44475 |
| 6 | gene | 73.39214 | 73.39224 | positive | MGI_C57BL6J_5454147 | Gm24370 | ENSEMBL:ENSMUSG00000093965 | MGI:5454147 | miRNA gene | predicted gene, 24370 |
| 6 | pseudogene | 73.42031 | 73.42122 | negative | MGI_C57BL6J_3030044 | Olfr210-ps1 | NCBI_Gene:257960,ENSEMBL:ENSMUSG00000108032 | MGI:3030044 | pseudogene | olfactory receptor 210, pseudogene 1 |
| 6 | gene | 73.43697 | 73.47102 | negative | MGI_C57BL6J_1920658 | 1700065L07Rik | NCBI_Gene:73408,ENSEMBL:ENSMUSG00000107408 | MGI:1920658 | lncRNA gene | RIKEN cDNA 1700065L07 gene |
| 6 | gene | 73.46857 | 73.46967 | negative | MGI_C57BL6J_1913990 | 4931417E11Rik | NCBI_Gene:66740,ENSEMBL:ENSMUSG00000056197 | MGI:1913990 | protein coding gene | RIKEN cDNA 4931417E11 gene |
| 6 | pseudogene | 73.52881 | 73.52924 | negative | MGI_C57BL6J_3782559 | Gm4374 | ENSEMBL:ENSMUSG00000107639 | MGI:3782559 | pseudogene | predicted gene 4374 |
| 6 | gene | 73.58611 | 73.60867 | negative | MGI_C57BL6J_5590906 | Gm31747 | NCBI_Gene:102634075,ENSEMBL:ENSMUSG00000107441 | MGI:5590906 | lncRNA gene | predicted gene, 31747 |
| 6 | pseudogene | 73.88566 | 73.88585 | negative | MGI_C57BL6J_5690617 | Gm44225 | ENSEMBL:ENSMUSG00000107646 | MGI:5690617 | pseudogene | predicted gene, 44225 |
| 6 | gene | 74.34095 | 74.34108 | positive | MGI_C57BL6J_5455956 | Gm26179 | ENSEMBL:ENSMUSG00000093964 | MGI:5455956 | snoRNA gene | predicted gene, 26179 |
| 6 | gene | 74.45180 | 74.45190 | negative | MGI_C57BL6J_5453701 | Gm23924 | ENSEMBL:ENSMUSG00000064408 | MGI:5453701 | snRNA gene | predicted gene, 23924 |
| 6 | gene | 74.50505 | 74.50516 | negative | MGI_C57BL6J_5452031 | Gm22254 | ENSEMBL:ENSMUSG00000096282 | MGI:5452031 | miRNA gene | predicted gene, 22254 |
| 6 | pseudogene | 75.18739 | 75.18849 | negative | MGI_C57BL6J_3644228 | Gm8992 | NCBI_Gene:668132,ENSEMBL:ENSMUSG00000107467 | MGI:3644228 | pseudogene | predicted gene 8992 |
| 6 | pseudogene | 75.20199 | 75.20297 | positive | MGI_C57BL6J_3644225 | Gm8993 | NCBI_Gene:668136,ENSEMBL:ENSMUSG00000097957 | MGI:3644225 | pseudogene | predicted gene 8993 |
| 6 | pseudogene | 75.29604 | 75.29814 | negative | MGI_C57BL6J_3643375 | Gm6210 | NCBI_Gene:621306,ENSEMBL:ENSMUSG00000107951 | MGI:3643375 | pseudogene | predicted gene 6210 |
| 6 | pseudogene | 75.38305 | 75.38402 | negative | MGI_C57BL6J_3647166 | Gm8999 | NCBI_Gene:668143,ENSEMBL:ENSMUSG00000107918 | MGI:3647166 | pseudogene | predicted gene 8999 |
| 6 | gene | 75.42745 | 75.47498 | negative | MGI_C57BL6J_5621720 | Gm38835 | NCBI_Gene:105242686 | MGI:5621720 | lncRNA gene | predicted gene, 38835 |
| 6 | gene | 75.57209 | 75.57346 | negative | MGI_C57BL6J_1918549 | 4933439N06Rik | ENSEMBL:ENSMUSG00000107830 | MGI:1918549 | unclassified gene | RIKEN cDNA 4933439N06 gene |
| 6 | pseudogene | 75.78216 | 75.78278 | negative | MGI_C57BL6J_3647168 | Gm9001 | NCBI_Gene:668145,ENSEMBL:ENSMUSG00000080824 | MGI:3647168 | pseudogene | predicted gene 9001 |
| 6 | gene | 75.90359 | 75.90581 | negative | MGI_C57BL6J_5590965 | Gm31806 | NCBI_Gene:102634151 | MGI:5590965 | lncRNA gene | predicted gene, 31806 |
| 6 | gene | 75.96805 | 75.97498 | negative | MGI_C57BL6J_5690505 | Gm44113 | ENSEMBL:ENSMUSG00000107880 | MGI:5690505 | lncRNA gene | predicted gene, 44113 |
| 6 | pseudogene | 76.12163 | 76.12290 | positive | MGI_C57BL6J_3644799 | Gm5995 | NCBI_Gene:546907,ENSEMBL:ENSMUSG00000107685 | MGI:3644799 | pseudogene | predicted gene 5995 |
| 6 | gene | 76.18171 | 76.18182 | positive | MGI_C57BL6J_5690726 | Gm44334 | ENSEMBL:ENSMUSG00000106544 | MGI:5690726 | miRNA gene | predicted gene, 44334 |
| 6 | gene | 76.20012 | 76.20028 | negative | MGI_C57BL6J_5452060 | Gm22283 | ENSEMBL:ENSMUSG00000064490 | MGI:5452060 | snRNA gene | predicted gene, 22283 |
| 6 | pseudogene | 76.28442 | 76.28493 | positive | MGI_C57BL6J_5690518 | Gm44126 | ENSEMBL:ENSMUSG00000107770 | MGI:5690518 | pseudogene | predicted gene, 44126 |
| 6 | pseudogene | 76.45531 | 76.45592 | negative | MGI_C57BL6J_5690555 | Gm44163 | NCBI_Gene:108169180,ENSEMBL:ENSMUSG00000107612 | MGI:5690555 | pseudogene | predicted gene, 44163 |
| 6 | gene | 76.49543 | 76.49778 | negative | MGI_C57BL6J_3644000 | Gm9008 | NCBI_Gene:668155,ENSEMBL:ENSMUSG00000072476 | MGI:3644000 | protein coding gene | predicted pseudogene 9008 |
| 6 | gene | 76.49608 | 76.49752 | positive | MGI_C57BL6J_5690558 | Gm44166 | ENSEMBL:ENSMUSG00000107651 | MGI:5690558 | lncRNA gene | predicted gene, 44166 |
| 6 | gene | 76.69545 | 76.71158 | negative | MGI_C57BL6J_5621721 | Gm38836 | NCBI_Gene:105242687 | MGI:5621721 | lncRNA gene | predicted gene, 38836 |
| 6 | gene | 76.81034 | 76.81555 | negative | MGI_C57BL6J_5591019 | Gm31860 | NCBI_Gene:102634227 | MGI:5591019 | lncRNA gene | predicted gene, 31860 |
| 6 | gene | 76.88164 | 77.97970 | negative | MGI_C57BL6J_88275 | Ctnna2 | NCBI_Gene:12386,ENSEMBL:ENSMUSG00000063063 | MGI:88275 | protein coding gene | catenin (cadherin associated protein), alpha 2 |
| 6 | gene | 76.98798 | 76.98965 | negative | MGI_C57BL6J_1923313 | 5830431M20Rik | NA | NA | unclassified gene | RIKEN cDNA 5830431M20 gene |
| 6 | gene | 77.24248 | 77.25992 | positive | MGI_C57BL6J_2389173 | Lrrtm1 | NCBI_Gene:74342,ENSEMBL:ENSMUSG00000060780 | MGI:2389173 | protein coding gene | leucine rich repeat transmembrane neuronal 1 |
| 6 | gene | 77.32391 | 77.32480 | negative | MGI_C57BL6J_5690626 | Gm44234 | ENSEMBL:ENSMUSG00000108057 | MGI:5690626 | unclassified gene | predicted gene, 44234 |
| 6 | gene | 77.38026 | 77.38247 | negative | MGI_C57BL6J_1918003 | 6330415B21Rik | NCBI_Gene:70753,ENSEMBL:ENSMUSG00000108077 | MGI:1918003 | lncRNA gene | RIKEN cDNA 6330415B21 gene |
| 6 | gene | 77.41230 | 77.41642 | negative | MGI_C57BL6J_5690827 | Gm44435 | ENSEMBL:ENSMUSG00000108005 | MGI:5690827 | unclassified gene | predicted gene, 44435 |
| 6 | gene | 77.42868 | 77.43351 | negative | MGI_C57BL6J_5690825 | Gm44433 | ENSEMBL:ENSMUSG00000107512 | MGI:5690825 | unclassified gene | predicted gene, 44433 |
| 6 | gene | 77.47114 | 77.47419 | negative | MGI_C57BL6J_5690627 | Gm44235 | ENSEMBL:ENSMUSG00000107917 | MGI:5690627 | unclassified gene | predicted gene, 44235 |
| 6 | gene | 77.53985 | 77.54308 | negative | MGI_C57BL6J_5690830 | Gm44438 | ENSEMBL:ENSMUSG00000107947 | MGI:5690830 | unclassified gene | predicted gene, 44438 |
| 6 | gene | 77.58700 | 77.58848 | negative | MGI_C57BL6J_5690678 | Gm44286 | ENSEMBL:ENSMUSG00000107707 | MGI:5690678 | unclassified gene | predicted gene, 44286 |
| 6 | gene | 77.59874 | 77.60517 | negative | MGI_C57BL6J_5621722 | Gm38837 | NCBI_Gene:105242688 | MGI:5621722 | lncRNA gene | predicted gene, 38837 |
| 6 | gene | 77.69298 | 77.69577 | negative | MGI_C57BL6J_5690680 | Gm44288 | ENSEMBL:ENSMUSG00000107483 | MGI:5690680 | unclassified gene | predicted gene, 44288 |
| 6 | gene | 77.74574 | 77.74978 | negative | MGI_C57BL6J_5690829 | Gm44437 | ENSEMBL:ENSMUSG00000108102 | MGI:5690829 | unclassified gene | predicted gene, 44437 |
| 6 | gene | 77.98452 | 77.98657 | positive | MGI_C57BL6J_5690647 | Gm44255 | ENSEMBL:ENSMUSG00000107473 | MGI:5690647 | unclassified gene | predicted gene, 44255 |
| 6 | pseudogene | 78.06857 | 78.06959 | negative | MGI_C57BL6J_5434400 | Gm21045 | NCBI_Gene:100534366,ENSEMBL:ENSMUSG00000097964 | MGI:5434400 | pseudogene | predicted gene, 21045 |
| 6 | pseudogene | 78.21588 | 78.21717 | positive | MGI_C57BL6J_3644832 | Gm5576 | NCBI_Gene:434050,ENSEMBL:ENSMUSG00000108045 | MGI:3644832 | pseudogene | predicted pseudogene 5576 |
| 6 | gene | 78.37066 | 78.37347 | positive | MGI_C57BL6J_97478 | Reg3b | NCBI_Gene:18489,ENSEMBL:ENSMUSG00000071356 | MGI:97478 | protein coding gene | regenerating islet-derived 3 beta |
| 6 | gene | 78.37587 | 78.37887 | negative | MGI_C57BL6J_1353426 | Reg3d | NCBI_Gene:30053,ENSEMBL:ENSMUSG00000068341 | MGI:1353426 | protein coding gene | regenerating islet-derived 3 delta |
| 6 | gene | 78.38071 | 78.38384 | positive | MGI_C57BL6J_109408 | Reg3a | NCBI_Gene:19694,ENSEMBL:ENSMUSG00000079516 | MGI:109408 | protein coding gene | regenerating islet-derived 3 alpha |
| 6 | gene | 78.39562 | 78.40811 | positive | MGI_C57BL6J_97896 | Reg2 | NCBI_Gene:19693,ENSEMBL:ENSMUSG00000023140 | MGI:97896 | protein coding gene | regenerating islet-derived 2 |
| 6 | gene | 78.42271 | 78.42867 | positive | MGI_C57BL6J_97895 | Reg1 | NCBI_Gene:19692,ENSEMBL:ENSMUSG00000059654 | MGI:97895 | protein coding gene | regenerating islet-derived 1 |
| 6 | gene | 78.46627 | 78.46887 | negative | MGI_C57BL6J_109406 | Reg3g | NCBI_Gene:19695,ENSEMBL:ENSMUSG00000030017 | MGI:109406 | protein coding gene | regenerating islet-derived 3 gamma |
| 6 | pseudogene | 78.48269 | 78.48366 | positive | MGI_C57BL6J_5434747 | Gm21392 | ENSEMBL:ENSMUSG00000097940 | MGI:5434747 | pseudogene | predicted gene, 21392 |
| 6 | pseudogene | 78.49676 | 78.49747 | negative | MGI_C57BL6J_3648423 | Gm6072 | NCBI_Gene:606545,ENSEMBL:ENSMUSG00000108073 | MGI:3648423 | pseudogene | predicted gene 6072 |
| 6 | gene | 78.91117 | 78.91133 | positive | MGI_C57BL6J_5453504 | Gm23727 | ENSEMBL:ENSMUSG00000094980 | MGI:5453504 | snRNA gene | predicted gene, 23727 |
| 6 | gene | 78.91260 | 78.91292 | negative | MGI_C57BL6J_5579711 | Gm29005 | ENSEMBL:ENSMUSG00000101720 | MGI:5579711 | lncRNA gene | predicted gene 29005 |
| 6 | gene | 79.03204 | 79.03587 | positive | MGI_C57BL6J_5012547 | Gm20362 | NCBI_Gene:100504701,ENSEMBL:ENSMUSG00000108072 | MGI:5012547 | lncRNA gene | predicted gene, 20362 |
| 6 | pseudogene | 79.04811 | 79.04899 | positive | MGI_C57BL6J_3646896 | Gm5311 | NCBI_Gene:384440,ENSEMBL:ENSMUSG00000107772 | MGI:3646896 | pseudogene | predicted gene 5311 |
| 6 | pseudogene | 79.54119 | 79.54206 | positive | MGI_C57BL6J_3648928 | Gm6261 | NCBI_Gene:621815,ENSEMBL:ENSMUSG00000107618 | MGI:3648928 | pseudogene | predicted gene 6261 |
| 6 | gene | 79.68885 | 79.71091 | positive | MGI_C57BL6J_5690663 | Gm44271 | ENSEMBL:ENSMUSG00000108027 | MGI:5690663 | lncRNA gene | predicted gene, 44271 |
| 6 | gene | 79.81792 | 79.81836 | positive | MGI_C57BL6J_5295700 | Gm20594 | NCBI_Gene:100463512,ENSEMBL:ENSMUSG00000096887 | MGI:5295700 | protein coding gene | predicted gene, 20594 |
| 6 | gene | 79.92641 | 80.02316 | negative | MGI_C57BL6J_3782594 | Gm4409 | ENSEMBL:ENSMUSG00000086739 | MGI:3782594 | lncRNA gene | predicted gene 4409 |
| 6 | gene | 80.01886 | 80.81016 | positive | MGI_C57BL6J_2389180 | Lrrtm4 | NCBI_Gene:243499,ENSEMBL:ENSMUSG00000052581 | MGI:2389180 | protein coding gene | leucine rich repeat transmembrane neuronal 4 |
| 6 | gene | 80.04868 | 80.05133 | positive | MGI_C57BL6J_5690632 | Gm44240 | ENSEMBL:ENSMUSG00000107498 | MGI:5690632 | unclassified gene | predicted gene, 44240 |
| 6 | gene | 80.14374 | 80.16666 | positive | MGI_C57BL6J_5591250 | Gm32091 | NCBI_Gene:102634534 | MGI:5591250 | lncRNA gene | predicted gene, 32091 |
| 6 | gene | 80.21745 | 80.21948 | positive | MGI_C57BL6J_5690620 | Gm44228 | ENSEMBL:ENSMUSG00000107996 | MGI:5690620 | unclassified gene | predicted gene, 44228 |
| 6 | gene | 80.26850 | 80.27205 | positive | MGI_C57BL6J_5690622 | Gm44230 | ENSEMBL:ENSMUSG00000107905 | MGI:5690622 | unclassified gene | predicted gene, 44230 |
| 6 | gene | 80.30261 | 80.30437 | positive | MGI_C57BL6J_5012556 | Gm20371 | ENSEMBL:ENSMUSG00000108264 | MGI:5012556 | unclassified gene | predicted gene, 20371 |
| 6 | gene | 80.31919 | 80.32085 | positive | MGI_C57BL6J_5690633 | Gm44241 | ENSEMBL:ENSMUSG00000107840 | MGI:5690633 | unclassified gene | predicted gene, 44241 |
| 6 | gene | 80.44535 | 80.44784 | positive | MGI_C57BL6J_5690621 | Gm44229 | ENSEMBL:ENSMUSG00000108088 | MGI:5690621 | unclassified gene | predicted gene, 44229 |
| 6 | gene | 80.50487 | 80.50535 | positive | MGI_C57BL6J_1924775 | C030013C02Rik | NA | NA | unclassified gene | RIKEN cDNA C030013C02 gene |
| 6 | gene | 80.51459 | 80.51731 | positive | MGI_C57BL6J_5690634 | Gm44242 | ENSEMBL:ENSMUSG00000107826 | MGI:5690634 | unclassified gene | predicted gene, 44242 |
| 6 | gene | 80.65503 | 80.65910 | positive | MGI_C57BL6J_5690623 | Gm44231 | ENSEMBL:ENSMUSG00000108173 | MGI:5690623 | unclassified gene | predicted gene, 44231 |
| 6 | gene | 80.82714 | 80.82981 | positive | MGI_C57BL6J_5690292 | Gm43900 | ENSEMBL:ENSMUSG00000108040 | MGI:5690292 | unclassified gene | predicted gene, 43900 |
| 6 | pseudogene | 81.42081 | 81.42126 | negative | MGI_C57BL6J_5690293 | Gm43901 | ENSEMBL:ENSMUSG00000107691 | MGI:5690293 | pseudogene | predicted gene, 43901 |
| 6 | gene | 81.42900 | 81.44626 | negative | MGI_C57BL6J_5621723 | Gm38838 | NCBI_Gene:105242690 | MGI:5621723 | lncRNA gene | predicted gene, 38838 |
| 6 | gene | 81.63831 | 81.63847 | positive | MGI_C57BL6J_5456041 | Gm26264 | ENSEMBL:ENSMUSG00000065212 | MGI:5456041 | snRNA gene | predicted gene, 26264 |
| 6 | gene | 81.67595 | 81.71620 | negative | MGI_C57BL6J_5621724 | Gm38839 | NCBI_Gene:105242691,ENSEMBL:ENSMUSG00000107419 | MGI:5621724 | lncRNA gene | predicted gene, 38839 |
| 6 | gene | 81.71846 | 81.72933 | negative | MGI_C57BL6J_5591382 | Gm32223 | NCBI_Gene:102634701 | MGI:5591382 | lncRNA gene | predicted gene, 32223 |
| 6 | gene | 81.80679 | 81.80691 | positive | MGI_C57BL6J_5452307 | Gm22530 | ENSEMBL:ENSMUSG00000089445 | MGI:5452307 | snoRNA gene | predicted gene, 22530 |
| 6 | pseudogene | 81.84067 | 81.84136 | negative | MGI_C57BL6J_3647948 | Gm4874 | NCBI_Gene:232143,ENSEMBL:ENSMUSG00000108202 | MGI:3647948 | pseudogene | predicted gene 4874 |
| 6 | pseudogene | 81.86424 | 81.87591 | negative | MGI_C57BL6J_5621725 | Gm38840 | NCBI_Gene:105242692,ENSEMBL:ENSMUSG00000107817 | MGI:5621725 | pseudogene | predicted gene, 38840 |
| 6 | gene | 81.89382 | 81.89534 | negative | MGI_C57BL6J_1920811 | 1700097M23Rik | ENSEMBL:ENSMUSG00000108194 | MGI:1920811 | unclassified gene | RIKEN cDNA 1700097M23 gene |
| 6 | gene | 81.89559 | 81.89566 | negative | MGI_C57BL6J_5562768 | Mir7232 | miRBase:MI0023727,NCBI_Gene:102465711,ENSEMBL:ENSMUSG00000106154 | MGI:5562768 | miRNA gene | microRNA 7232 |
| 6 | gene | 81.89660 | 81.89668 | negative | MGI_C57BL6J_3619414 | Mir468 | miRBase:MI0002403,NCBI_Gene:723871,ENSEMBL:ENSMUSG00000106253 | MGI:3619414 | miRNA gene | microRNA 468 |
| 6 | gene | 81.90046 | 81.91080 | negative | MGI_C57BL6J_1921470 | 1700009C05Rik | NCBI_Gene:74220,ENSEMBL:ENSMUSG00000101404 | MGI:1921470 | lncRNA gene | RIKEN cDNA 1700009C05 gene |
| 6 | gene | 81.91359 | 81.92043 | positive | MGI_C57BL6J_5621726 | Gm38841 | NCBI_Gene:105242693 | MGI:5621726 | lncRNA gene | predicted gene, 38841 |
| 6 | gene | 81.92364 | 81.95991 | positive | MGI_C57BL6J_2141656 | Gcfc2 | NCBI_Gene:330361,ENSEMBL:ENSMUSG00000035125 | MGI:2141656 | protein coding gene | GC-rich sequence DNA binding factor 2 |
| 6 | gene | 81.95783 | 81.96596 | negative | MGI_C57BL6J_1926274 | Mrpl19 | NCBI_Gene:56284,ENSEMBL:ENSMUSG00000030045 | MGI:1926274 | protein coding gene | mitochondrial ribosomal protein L19 |
| 6 | gene | 81.97160 | 81.97296 | negative | MGI_C57BL6J_5621727 | Gm38842 | NCBI_Gene:105242694 | MGI:5621727 | lncRNA gene | predicted gene, 38842 |
| 6 | gene | 81.98432 | 81.99243 | positive | MGI_C57BL6J_5012568 | Gm20383 | NCBI_Gene:100504740,ENSEMBL:ENSMUSG00000107412 | MGI:5012568 | lncRNA gene | predicted gene, 20383 |
| 6 | gene | 81.98575 | 81.98674 | negative | MGI_C57BL6J_5690594 | Gm44202 | ENSEMBL:ENSMUSG00000108288 | MGI:5690594 | lncRNA gene | predicted gene, 44202 |
| 6 | gene | 82.02588 | 82.03372 | negative | MGI_C57BL6J_5591537 | Gm32378 | NCBI_Gene:102634905 | MGI:5591537 | lncRNA gene | predicted gene, 32378 |
| 6 | gene | 82.04104 | 82.09310 | positive | MGI_C57BL6J_2385247 | Eva1a | NCBI_Gene:232146,ENSEMBL:ENSMUSG00000035104 | MGI:2385247 | protein coding gene | eva-1 homolog A (C. elegans) |
| 6 | gene | 82.05045 | 82.05258 | negative | MGI_C57BL6J_3802115 | Gm15864 | ENSEMBL:ENSMUSG00000087136 | MGI:3802115 | lncRNA gene | predicted gene 15864 |
| 6 | gene | 82.11977 | 82.12607 | positive | MGI_C57BL6J_5623266 | Gm40381 | NCBI_Gene:105244847 | MGI:5623266 | lncRNA gene | predicted gene, 40381 |
| 6 | pseudogene | 82.24217 | 82.24285 | positive | MGI_C57BL6J_5591020 | Gm31861 | ENSEMBL:ENSMUSG00000108224 | MGI:5591020 | pseudogene | predicted gene, 31861 |
| 6 | gene | 82.31883 | 82.32298 | negative | MGI_C57BL6J_5690601 | Gm44209 | ENSEMBL:ENSMUSG00000107809 | MGI:5690601 | unclassified gene | predicted gene, 44209 |
| 6 | gene | 82.40200 | 82.56010 | positive | MGI_C57BL6J_98475 | Tacr1 | NCBI_Gene:21336,ENSEMBL:ENSMUSG00000030043 | MGI:98475 | protein coding gene | tachykinin receptor 1 |
| 6 | gene | 82.61892 | 82.70537 | negative | MGI_C57BL6J_1914229 | Pole4 | NCBI_Gene:66979,ENSEMBL:ENSMUSG00000030042 | MGI:1914229 | protein coding gene | polymerase (DNA-directed), epsilon 4 (p12 subunit) |
| 6 | gene | 82.67967 | 82.70628 | positive | MGI_C57BL6J_5591584 | Gm32425 | NCBI_Gene:102634973 | MGI:5591584 | lncRNA gene | predicted gene, 32425 |
| 6 | gene | 82.68013 | 82.69482 | positive | MGI_C57BL6J_5690537 | Gm44145 | ENSEMBL:ENSMUSG00000107453 | MGI:5690537 | lncRNA gene | predicted gene, 44145 |
| 6 | gene | 82.71074 | 82.71481 | negative | MGI_C57BL6J_5591693 | Gm32534 | NCBI_Gene:102635114 | MGI:5591693 | lncRNA gene | predicted gene, 32534 |
| 6 | gene | 82.71438 | 82.71597 | positive | MGI_C57BL6J_5591639 | Gm32480 | NCBI_Gene:102635044 | MGI:5591639 | lncRNA gene | predicted gene, 32480 |
| 6 | gene | 82.72502 | 82.77445 | negative | MGI_C57BL6J_1315197 | Hk2 | NCBI_Gene:15277,ENSEMBL:ENSMUSG00000000628 | MGI:1315197 | protein coding gene | hexokinase 2 |
| 6 | gene | 82.76428 | 82.76729 | positive | MGI_C57BL6J_4937861 | Gm17034 | ENSEMBL:ENSMUSG00000090293 | MGI:4937861 | lncRNA gene | predicted gene 17034 |
| 6 | gene | 82.80377 | 82.80518 | negative | MGI_C57BL6J_5621728 | Gm38843 | NCBI_Gene:105242696,ENSEMBL:ENSMUSG00000109556 | MGI:5621728 | lncRNA gene | predicted gene, 38843 |
| 6 | gene | 82.82321 | 82.84139 | positive | MGI_C57BL6J_5591750 | Gm32591 | NCBI_Gene:102635189,ENSEMBL:ENSMUSG00000108015 | MGI:5591750 | lncRNA gene | predicted gene, 32591 |
| 6 | gene | 82.87786 | 82.88185 | negative | MGI_C57BL6J_1916902 | 2310069B03Rik | NCBI_Gene:69652,ENSEMBL:ENSMUSG00000100291 | MGI:1916902 | lncRNA gene | RIKEN cDNA 2310069B03 gene |
| 6 | pseudogene | 82.88726 | 82.88872 | negative | MGI_C57BL6J_3646278 | Gm6312 | NCBI_Gene:622318,ENSEMBL:ENSMUSG00000107401 | MGI:3646278 | pseudogene | predicted gene 6312 |
| 6 | gene | 82.89687 | 82.90521 | negative | MGI_C57BL6J_5753721 | Gm45145 | ENSEMBL:ENSMUSG00000109076 | MGI:5753721 | lncRNA gene | predicted gene 45145 |
| 6 | gene | 82.91188 | 82.93977 | negative | MGI_C57BL6J_1340055 | Sema4f | NCBI_Gene:20355,ENSEMBL:ENSMUSG00000000627 | MGI:1340055 | protein coding gene | sema domain, immunoglobulin domain (Ig), TM domain, and short cytoplasmic domain |
| 6 | gene | 82.94690 | 83.03031 | positive | MGI_C57BL6J_1315200 | M1ap | NCBI_Gene:110958,ENSEMBL:ENSMUSG00000030041 | MGI:1315200 | protein coding gene | meiosis 1 associated protein |
| 6 | pseudogene | 82.95278 | 82.95314 | positive | MGI_C57BL6J_5011472 | Gm19287 | NCBI_Gene:102635339,ENSEMBL:ENSMUSG00000107701 | MGI:5011472 | pseudogene | predicted gene, 19287 |
| 6 | pseudogene | 82.99082 | 82.99158 | negative | MGI_C57BL6J_5690377 | Gm43985 | ENSEMBL:ENSMUSG00000107829 | MGI:5690377 | pseudogene | predicted gene, 43985 |
| 6 | gene | 82.99165 | 82.99311 | positive | MGI_C57BL6J_5690373 | Gm43981 | ENSEMBL:ENSMUSG00000107599 | MGI:5690373 | unclassified gene | predicted gene, 43981 |
| 6 | gene | 83.00818 | 83.02290 | negative | MGI_C57BL6J_5591803 | Gm32644 | NCBI_Gene:102635258 | MGI:5591803 | lncRNA gene | predicted gene, 32644 |
| 6 | gene | 83.03093 | 83.03347 | negative | MGI_C57BL6J_893587 | Dok1 | NCBI_Gene:13448,ENSEMBL:ENSMUSG00000068335 | MGI:893587 | protein coding gene | docking protein 1 |
| 6 | gene | 83.03417 | 83.05256 | positive | MGI_C57BL6J_1337004 | Loxl3 | NCBI_Gene:16950,ENSEMBL:ENSMUSG00000000693 | MGI:1337004 | protein coding gene | lysyl oxidase-like 3 |
| 6 | gene | 83.04971 | 83.04977 | positive | MGI_C57BL6J_5530871 | Mir7040 | miRBase:MI0022889,NCBI_Gene:102465630,ENSEMBL:ENSMUSG00000099035 | MGI:5530871 | miRNA gene | microRNA 7040 |
| 6 | gene | 83.05127 | 83.05527 | negative | MGI_C57BL6J_1928676 | Htra2 | NCBI_Gene:64704,ENSEMBL:ENSMUSG00000068329 | MGI:1928676 | protein coding gene | HtrA serine peptidase 2 |
| 6 | gene | 83.05452 | 83.05781 | positive | MGI_C57BL6J_107789 | Aup1 | NCBI_Gene:11993,ENSEMBL:ENSMUSG00000068328 | MGI:107789 | protein coding gene | ancient ubiquitous protein 1 |
| 6 | gene | 83.05775 | 83.06732 | positive | MGI_C57BL6J_2136388 | Dqx1 | NCBI_Gene:93838,ENSEMBL:ENSMUSG00000009145 | MGI:2136388 | protein coding gene | DEAQ RNA-dependent ATPase |
| 6 | gene | 83.06832 | 83.07029 | negative | MGI_C57BL6J_1350935 | Tlx2 | NCBI_Gene:21909,ENSEMBL:ENSMUSG00000068327 | MGI:1350935 | protein coding gene | T cell leukemia, homeobox 2 |
| 6 | gene | 83.07460 | 83.07612 | negative | MGI_C57BL6J_5610320 | Gm37092 | ENSEMBL:ENSMUSG00000103189 | MGI:5610320 | unclassified gene | predicted gene, 37092 |
| 6 | gene | 83.07787 | 83.08085 | positive | MGI_C57BL6J_1917087 | Pcgf1 | NCBI_Gene:69837,ENSEMBL:ENSMUSG00000069678 | MGI:1917087 | protein coding gene | polycomb group ring finger 1 |
| 6 | gene | 83.08637 | 83.08824 | positive | MGI_C57BL6J_1342288 | Lbx2 | NCBI_Gene:16815,ENSEMBL:ENSMUSG00000034968 | MGI:1342288 | protein coding gene | ladybird homeobox 2 |
| 6 | gene | 83.09031 | 83.09039 | negative | MGI_C57BL6J_4441434 | Mir3470a | miRBase:MI0014696,NCBI_Gene:100499518,ENSEMBL:ENSMUSG00000095323 | MGI:4441434 | miRNA gene | microRNA 3470a |
| 6 | gene | 83.10152 | 83.10912 | positive | MGI_C57BL6J_3045292 | Ccdc142 | NCBI_Gene:243510,ENSEMBL:ENSMUSG00000107499 | MGI:3045292 | protein coding gene | coiled-coil domain containing 142 |
| 6 | gene | 83.10162 | 83.10994 | positive | MGI_C57BL6J_5662825 | Gm42688 | ENSEMBL:ENSMUSG00000079511 | MGI:5662825 | protein coding gene | predicted gene 42688 |
| 6 | gene | 83.10661 | 83.10905 | negative | MGI_C57BL6J_3783052 | Ccdc142os | ENSEMBL:ENSMUSG00000087578 | MGI:3783052 | antisense lncRNA gene | coiled-coil domain containing 142, opposite strand |
| 6 | gene | 83.10904 | 83.10994 | positive | MGI_C57BL6J_1915749 | Mrpl53 | NCBI_Gene:68499,ENSEMBL:ENSMUSG00000030037 | MGI:1915749 | protein coding gene | mitochondrial ribosomal protein L53 |
| 6 | gene | 83.11550 | 83.11890 | positive | MGI_C57BL6J_1929872 | Mogs | NCBI_Gene:57377,ENSEMBL:ENSMUSG00000030036 | MGI:1929872 | protein coding gene | mannosyl-oligosaccharide glucosidase |
| 6 | gene | 83.11904 | 83.12156 | negative | MGI_C57BL6J_104710 | Wbp1 | NCBI_Gene:22377,ENSEMBL:ENSMUSG00000030035 | MGI:104710 | protein coding gene | WW domain binding protein 1 |
| 6 | gene | 83.12176 | 83.12543 | negative | MGI_C57BL6J_1917270 | Ino80b | NCBI_Gene:70020,ENSEMBL:ENSMUSG00000030034 | MGI:1917270 | protein coding gene | INO80 complex subunit B |
| 6 | gene | 83.12252 | 83.12440 | positive | MGI_C57BL6J_5690679 | Gm44287 | ENSEMBL:ENSMUSG00000108109 | MGI:5690679 | lncRNA gene | predicted gene, 44287 |
| 6 | gene | 83.13443 | 83.15258 | positive | MGI_C57BL6J_107371 | Rtkn | NCBI_Gene:20166,ENSEMBL:ENSMUSG00000034930 | MGI:107371 | protein coding gene | rhotekin |
| 6 | gene | 83.14936 | 83.15644 | negative | MGI_C57BL6J_1922909 | Wdr54 | NCBI_Gene:75659,ENSEMBL:ENSMUSG00000030032 | MGI:1922909 | protein coding gene | WD repeat domain 54 |
| 6 | gene | 83.15640 | 83.16298 | positive | MGI_C57BL6J_1919087 | 1700003E16Rik | NCBI_Gene:71837,ENSEMBL:ENSMUSG00000030030 | MGI:1919087 | protein coding gene | RIKEN cDNA 1700003E16 gene |
| 6 | gene | 83.16591 | 83.20012 | positive | MGI_C57BL6J_107745 | Dctn1 | NCBI_Gene:13191,ENSEMBL:ENSMUSG00000031865 | MGI:107745 | protein coding gene | dynactin 1 |
| 6 | gene | 83.16615 | 83.17526 | negative | MGI_C57BL6J_3783068 | Gm15624 | NCBI_Gene:102635553,ENSEMBL:ENSMUSG00000085782 | MGI:3783068 | lncRNA gene | predicted gene 15624 |
| 6 | gene | 83.20758 | 83.30495 | positive | MGI_C57BL6J_2443220 | Slc4a5 | NCBI_Gene:232156,ENSEMBL:ENSMUSG00000068323 | MGI:2443220 | protein coding gene | solute carrier family 4, sodium bicarbonate cotransporter, member 5 |
| 6 | gene | 83.30569 | 83.32604 | negative | MGI_C57BL6J_1338850 | Mthfd2 | NCBI_Gene:17768,ENSEMBL:ENSMUSG00000005667 | MGI:1338850 | protein coding gene | methylenetetrahydrofolate dehydrogenase (NAD+ dependent), methenyltetrahydrofolate cyclohydrolase |
| 6 | gene | 83.31414 | 83.31694 | negative | MGI_C57BL6J_5690282 | Gm43890 | ENSEMBL:ENSMUSG00000108053 | MGI:5690282 | unclassified gene | predicted gene, 43890 |
| 6 | gene | 83.32602 | 83.34378 | positive | MGI_C57BL6J_2442631 | Mob1a | NCBI_Gene:232157,ENSEMBL:ENSMUSG00000043131 | MGI:2442631 | protein coding gene | MOB kinase activator 1A |
| 6 | gene | 83.34915 | 83.36014 | positive | MGI_C57BL6J_1925903 | Bola3 | NCBI_Gene:78653,ENSEMBL:ENSMUSG00000045160 | MGI:1925903 | protein coding gene | bolA-like 3 (E. coli) |
| 6 | gene | 83.36237 | 83.45908 | negative | MGI_C57BL6J_2446229 | Tet3 | NCBI_Gene:194388,ENSEMBL:ENSMUSG00000034832 | MGI:2446229 | protein coding gene | tet methylcytosine dioxygenase 3 |
| 6 | gene | 83.40100 | 83.40110 | negative | MGI_C57BL6J_5530674 | Mir6374 | miRBase:MI0021903,NCBI_Gene:102465194,ENSEMBL:ENSMUSG00000099045 | MGI:5530674 | miRNA gene | microRNA 6374 |
| 6 | gene | 83.41744 | 83.42099 | negative | MGI_C57BL6J_1925345 | 5430434F05Rik | ENSEMBL:ENSMUSG00000108780 | MGI:1925345 | unclassified gene | RIKEN cDNA 5430434F05 gene |
| 6 | gene | 83.44176 | 83.44832 | positive | MGI_C57BL6J_2444713 | B230319C09Rik | NCBI_Gene:320775,ENSEMBL:ENSMUSG00000051844 | MGI:2444713 | lncRNA gene | RIKEN cDNA B230319C09 gene |
| 6 | gene | 83.48021 | 83.50697 | negative | MGI_C57BL6J_1351602 | Dguok | NCBI_Gene:27369,ENSEMBL:ENSMUSG00000014554 | MGI:1351602 | protein coding gene | deoxyguanosine kinase |
| 6 | gene | 83.51291 | 83.53627 | negative | MGI_C57BL6J_104589 | Actg2 | NCBI_Gene:11468,ENSEMBL:ENSMUSG00000059430 | MGI:104589 | protein coding gene | actin, gamma 2, smooth muscle, enteric |
| 6 | gene | 83.53168 | 83.54724 | positive | MGI_C57BL6J_5621729 | Gm38844 | NCBI_Gene:105242698 | MGI:5621729 | lncRNA gene | predicted gene, 38844 |
| 6 | gene | 83.54321 | 83.57577 | negative | MGI_C57BL6J_1917777 | Stambp | NCBI_Gene:70527,ENSEMBL:ENSMUSG00000006906 | MGI:1917777 | protein coding gene | STAM binding protein |
| 6 | pseudogene | 83.54886 | 83.55038 | negative | MGI_C57BL6J_5010722 | Gm18537 | NCBI_Gene:100417334,ENSEMBL:ENSMUSG00000108484 | MGI:5010722 | pseudogene | predicted gene, 18537 |
| 6 | gene | 83.56804 | 83.58813 | positive | MGI_C57BL6J_5434639 | Gm21284 | NCBI_Gene:100861870,ENSEMBL:ENSMUSG00000108660 | MGI:5434639 | lncRNA gene | predicted gene, 21284 |
| 6 | gene | 83.59182 | 83.59214 | negative | MGI_C57BL6J_5454767 | Gm24990 | ENSEMBL:ENSMUSG00000088031 | MGI:5454767 | unclassified non-coding RNA gene | predicted gene, 24990 |
| 6 | pseudogene | 83.60076 | 83.60175 | negative | MGI_C57BL6J_3644728 | Gm7424 | NCBI_Gene:664956,ENSEMBL:ENSMUSG00000108609 | MGI:3644728 | pseudogene | predicted gene 7424 |
| 6 | gene | 83.64454 | 83.65619 | negative | MGI_C57BL6J_1859834 | Clec4f | NCBI_Gene:51811,ENSEMBL:ENSMUSG00000014542 | MGI:1859834 | protein coding gene | C-type lectin domain family 4, member f |
| 6 | gene | 83.67121 | 83.67786 | negative | MGI_C57BL6J_2180021 | Cd207 | NCBI_Gene:246278,ENSEMBL:ENSMUSG00000034783 | MGI:2180021 | protein coding gene | CD207 antigen |
| 6 | gene | 83.69281 | 83.71220 | negative | MGI_C57BL6J_3583301 | Vax2os | NCBI_Gene:574519,ENSEMBL:ENSMUSG00000085794 | MGI:3583301 | antisense lncRNA gene | ventral anterior homeobox 2, opposite strand |
| 6 | gene | 83.70249 | 83.70268 | positive | MGI_C57BL6J_5531154 | Gm27772 | ENSEMBL:ENSMUSG00000099263 | MGI:5531154 | unclassified non-coding RNA gene | predicted gene, 27772 |
| 6 | gene | 83.70352 | 83.70398 | positive | MGI_C57BL6J_5531013 | Gm27631 | ENSEMBL:ENSMUSG00000098591 | MGI:5531013 | unclassified non-coding RNA gene | predicted gene, 27631 |
| 6 | gene | 83.71074 | 83.71093 | positive | MGI_C57BL6J_5531275 | Gm27893 | ENSEMBL:ENSMUSG00000098848 | MGI:5531275 | unclassified non-coding RNA gene | predicted gene, 27893 |
| 6 | gene | 83.71126 | 83.73831 | positive | MGI_C57BL6J_1346018 | Vax2 | NCBI_Gene:24113,ENSEMBL:ENSMUSG00000034777 | MGI:1346018 | protein coding gene | ventral anterior homeobox 2 |
| 6 | gene | 83.71280 | 83.71638 | negative | MGI_C57BL6J_5592102 | Gm32943 | NCBI_Gene:102635663 | MGI:5592102 | lncRNA gene | predicted gene, 32943 |
| 6 | gene | 83.72130 | 83.72139 | positive | MGI_C57BL6J_5454831 | Gm25054 | ENSEMBL:ENSMUSG00000087995 | MGI:5454831 | snRNA gene | predicted gene, 25054 |
| 6 | gene | 83.74299 | 83.75885 | positive | MGI_C57BL6J_103285 | Atp6v1b1 | NCBI_Gene:110935,ENSEMBL:ENSMUSG00000006269 | MGI:103285 | protein coding gene | ATPase, H+ transporting, lysosomal V1 subunit B1 |
| 6 | gene | 83.76176 | 83.76287 | negative | MGI_C57BL6J_1915184 | 1700124L16Rik | NCBI_Gene:67934,ENSEMBL:ENSMUSG00000099354 | MGI:1915184 | lncRNA gene | RIKEN cDNA 1700124L16 gene |
| 6 | gene | 83.76264 | 83.76833 | positive | MGI_C57BL6J_1922555 | Ankrd53 | NCBI_Gene:75305,ENSEMBL:ENSMUSG00000014747 | MGI:1922555 | protein coding gene | ankyrin repeat domain 53 |
| 6 | gene | 83.76888 | 83.77582 | negative | MGI_C57BL6J_1096575 | Tex261 | NCBI_Gene:21766,ENSEMBL:ENSMUSG00000014748 | MGI:1096575 | protein coding gene | testis expressed gene 261 |
| 6 | pseudogene | 83.79260 | 83.79427 | negative | MGI_C57BL6J_3644945 | Gm7443 | NCBI_Gene:665004,ENSEMBL:ENSMUSG00000108525 | MGI:3644945 | pseudogene | predicted gene 7443 |
| 6 | gene | 83.79498 | 83.80402 | positive | MGI_C57BL6J_1860418 | Nagk | NCBI_Gene:56174,ENSEMBL:ENSMUSG00000034744 | MGI:1860418 | protein coding gene | N-acetylglucosamine kinase |
| 6 | gene | 83.80540 | 83.83174 | negative | MGI_C57BL6J_2386865 | Paip2b | NCBI_Gene:232164,ENSEMBL:ENSMUSG00000045896 | MGI:2386865 | protein coding gene | poly(A) binding protein interacting protein 2B |
| 6 | gene | 83.83694 | 83.83707 | negative | MGI_C57BL6J_5452791 | Gm23014 | ENSEMBL:ENSMUSG00000077332 | MGI:5452791 | snoRNA gene | predicted gene, 23014 |
| 6 | pseudogene | 83.84165 | 83.84227 | positive | MGI_C57BL6J_3643752 | Gm5312 | NCBI_Gene:384450,ENSEMBL:ENSMUSG00000108107 | MGI:3643752 | pseudogene | predicted gene 5312 |
| 6 | pseudogene | 83.84433 | 83.84512 | negative | MGI_C57BL6J_3645065 | Gm6342 | NCBI_Gene:622683,ENSEMBL:ENSMUSG00000107644 | MGI:3645065 | pseudogene | predicted gene 6342 |
| 6 | gene | 83.86710 | 83.98955 | positive | MGI_C57BL6J_1203484 | Zfp638 | NCBI_Gene:18139,ENSEMBL:ENSMUSG00000030016 | MGI:1203484 | protein coding gene | zinc finger protein 638 |
| 6 | gene | 83.87006 | 83.87229 | positive | MGI_C57BL6J_5753326 | Gm44750 | ENSEMBL:ENSMUSG00000108750 | MGI:5753326 | unclassified gene | predicted gene 44750 |
| 6 | pseudogene | 83.87420 | 83.87540 | negative | MGI_C57BL6J_3779464 | Gm5138 | NCBI_Gene:380687,ENSEMBL:ENSMUSG00000079501 | MGI:3779464 | pseudogene | predicted gene 5138 |
| 6 | pseudogene | 83.89696 | 83.89732 | positive | MGI_C57BL6J_5690311 | Gm43919 | ENSEMBL:ENSMUSG00000107991 | MGI:5690311 | pseudogene | predicted gene, 43919 |
| 6 | gene | 83.96228 | 83.96505 | positive | MGI_C57BL6J_5690312 | Gm43920 | ENSEMBL:ENSMUSG00000107534 | MGI:5690312 | lncRNA gene | predicted gene, 43920 |
| 6 | gene | 84.00831 | 84.21106 | positive | MGI_C57BL6J_1349385 | Dysf | NCBI_Gene:26903,ENSEMBL:ENSMUSG00000033788 | MGI:1349385 | protein coding gene | dysferlin |
| 6 | gene | 84.00884 | 84.01246 | negative | MGI_C57BL6J_5592183 | Gm33024 | NCBI_Gene:102635771,ENSEMBL:ENSMUSG00000107682 | MGI:5592183 | lncRNA gene | predicted gene, 33024 |
| 6 | pseudogene | 84.03159 | 84.03267 | positive | MGI_C57BL6J_3646544 | Gm7498 | NCBI_Gene:665114,ENSEMBL:ENSMUSG00000108142 | MGI:3646544 | pseudogene | predicted gene 7498 |
| 6 | gene | 84.09377 | 84.10701 | negative | MGI_C57BL6J_3705288 | Gm15475 | ENSEMBL:ENSMUSG00000087001 | MGI:3705288 | lncRNA gene | predicted gene 15475 |
| 6 | gene | 84.22525 | 84.28660 | positive | MGI_C57BL6J_5690519 | Gm44127 | ENSEMBL:ENSMUSG00000107538 | MGI:5690519 | lncRNA gene | predicted gene, 44127 |
| 6 | gene | 84.41294 | 84.42384 | negative | MGI_C57BL6J_3642570 | Gm10445 | NCBI_Gene:100038556,ENSEMBL:ENSMUSG00000107797 | MGI:3642570 | lncRNA gene | predicted gene 10445 |
| 6 | gene | 84.47984 | 84.52204 | negative | MGI_C57BL6J_5621730 | Gm38845 | NCBI_Gene:105242699 | MGI:5621730 | lncRNA gene | predicted gene, 38845 |
| 6 | gene | 84.53588 | 84.53632 | positive | MGI_C57BL6J_1925267 | 4930504D19Rik | ENSEMBL:ENSMUSG00000107926 | MGI:1925267 | lncRNA gene | RIKEN cDNA 4930504D19 gene |
| 6 | gene | 84.57141 | 84.59391 | negative | MGI_C57BL6J_2176159 | Cyp26b1 | NCBI_Gene:232174,ENSEMBL:ENSMUSG00000063415 | MGI:2176159 | protein coding gene | cytochrome P450, family 26, subfamily b, polypeptide 1 |
| 6 | gene | 84.61849 | 85.07026 | negative | MGI_C57BL6J_1923164 | Exoc6b | NCBI_Gene:75914,ENSEMBL:ENSMUSG00000033769 | MGI:1923164 | protein coding gene | exocyst complex component 6B |
| 6 | pseudogene | 84.81222 | 84.81345 | positive | MGI_C57BL6J_3643248 | Gm7507 | NCBI_Gene:665129,ENSEMBL:ENSMUSG00000098142 | MGI:3643248 | pseudogene | predicted gene 7507 |
| 6 | gene | 84.89816 | 84.89863 | positive | MGI_C57BL6J_3528153 | AW125320 | NA | NA | unclassified gene | cDNA sequence AW125320 |
| 6 | gene | 84.92112 | 84.92500 | positive | MGI_C57BL6J_5690404 | Gm44012 | ENSEMBL:ENSMUSG00000108012 | MGI:5690404 | lncRNA gene | predicted gene, 44012 |
| 6 | pseudogene | 85.06257 | 85.06297 | positive | MGI_C57BL6J_5690403 | Gm44011 | ENSEMBL:ENSMUSG00000107629 | MGI:5690403 | pseudogene | predicted gene, 44011 |
| 6 | gene | 85.06618 | 85.06644 | positive | MGI_C57BL6J_5456130 | Gm26353 | ENSEMBL:ENSMUSG00000088849 | MGI:5456130 | unclassified non-coding RNA gene | predicted gene, 26353 |
| 6 | pseudogene | 85.07614 | 85.07713 | positive | MGI_C57BL6J_894683 | Npm3-ps1 | NCBI_Gene:108176,ENSEMBL:ENSMUSG00000081669 | MGI:894683 | pseudogene | nucleoplasmin 3, pseudogene 1 |
| 6 | pseudogene | 85.09224 | 85.09278 | positive | MGI_C57BL6J_5690346 | Gm43954 | ENSEMBL:ENSMUSG00000107409 | MGI:5690346 | pseudogene | predicted gene, 43954 |
| 6 | pseudogene | 85.09693 | 85.09707 | positive | MGI_C57BL6J_5690347 | Gm43955 | ENSEMBL:ENSMUSG00000107536 | MGI:5690347 | pseudogene | predicted gene, 43955 |
| 6 | pseudogene | 85.10281 | 85.10357 | positive | MGI_C57BL6J_5690349 | Gm43957 | ENSEMBL:ENSMUSG00000107925 | MGI:5690349 | pseudogene | predicted gene, 43957 |
| 6 | gene | 85.11142 | 85.12613 | negative | MGI_C57BL6J_3647625 | Gm5878 | NCBI_Gene:545861,ENSEMBL:ENSMUSG00000072952 | MGI:3647625 | protein coding gene | predicted gene 5878 |
| 6 | pseudogene | 85.11169 | 85.11227 | positive | MGI_C57BL6J_5690351 | Gm43959 | ENSEMBL:ENSMUSG00000107769 | MGI:5690351 | pseudogene | predicted gene, 43959 |
| 6 | gene | 85.13018 | 85.13777 | negative | MGI_C57BL6J_103078 | Spr | NCBI_Gene:20751,ENSEMBL:ENSMUSG00000033735 | MGI:103078 | protein coding gene | sepiapterin reductase |
| 6 | pseudogene | 85.13801 | 85.14372 | positive | MGI_C57BL6J_3642569 | Gm10444 | NCBI_Gene:102635901,ENSEMBL:ENSMUSG00000072950 | MGI:3642569 | pseudogene | predicted pseudogene 10444 |
| 6 | pseudogene | 85.14545 | 85.14638 | negative | MGI_C57BL6J_5690677 | Gm44285 | ENSEMBL:ENSMUSG00000107922 | MGI:5690677 | pseudogene | predicted gene, 44285 |
| 6 | gene | 85.15034 | 85.15045 | positive | MGI_C57BL6J_5452670 | Gm22893 | ENSEMBL:ENSMUSG00000089262 | MGI:5452670 | snoRNA gene | predicted gene, 22893 |
| 6 | pseudogene | 85.15262 | 85.15622 | positive | MGI_C57BL6J_1345187 | Spr-ps1 | NCBI_Gene:665089,ENSEMBL:ENSMUSG00000068303 | MGI:1345187 | pseudogene | sepiapterin reductase pseudogene 1 |
| 6 | gene | 85.16584 | 85.16737 | positive | MGI_C57BL6J_5592359 | Gm33200 | NCBI_Gene:102636005 | MGI:5592359 | lncRNA gene | predicted gene, 33200 |
| 6 | gene | 85.18744 | 85.20446 | positive | MGI_C57BL6J_95387 | Emx1 | NCBI_Gene:13796,ENSEMBL:ENSMUSG00000033726 | MGI:95387 | protein coding gene | empty spiracles homeobox 1 |
| 6 | gene | 85.21305 | 85.33348 | negative | MGI_C57BL6J_2137681 | Sfxn5 | NCBI_Gene:94282,ENSEMBL:ENSMUSG00000033720 | MGI:2137681 | protein coding gene | sideroflexin 5 |
| 6 | gene | 85.24386 | 85.24666 | negative | MGI_C57BL6J_2444182 | D930014O13Rik | NA | NA | unclassified gene | RIKEN cDNA D930014O13 gene |
| 6 | gene | 85.33496 | 85.37465 | negative | MGI_C57BL6J_1098586 | Rab11fip5 | NCBI_Gene:52055,ENSEMBL:ENSMUSG00000051343 | MGI:1098586 | protein coding gene | RAB11 family interacting protein 5 (class I) |
| 6 | gene | 85.33629 | 85.33637 | negative | MGI_C57BL6J_3629673 | Mir705 | miRBase:MI0004689,NCBI_Gene:735267,ENSEMBL:ENSMUSG00000076012 | MGI:3629673 | miRNA gene | microRNA 705 |
| 6 | gene | 85.37485 | 85.38697 | positive | MGI_C57BL6J_1918407 | 4933423K11Rik | NA | NA | lncRNA gene | RIKEN cDNA 4933423K11 gene |
| 6 | gene | 85.42389 | 85.42888 | positive | MGI_C57BL6J_3053002 | Noto | NCBI_Gene:384452,ENSEMBL:ENSMUSG00000068302 | MGI:3053002 | protein coding gene | notochord homeobox |
| 6 | gene | 85.43198 | 85.44643 | positive | MGI_C57BL6J_108048 | Smyd5 | NCBI_Gene:232187,ENSEMBL:ENSMUSG00000033706 | MGI:108048 | protein coding gene | SET and MYND domain containing 5 |
| 6 | gene | 85.44679 | 85.45197 | negative | MGI_C57BL6J_1920577 | Pradc1 | NCBI_Gene:73327,ENSEMBL:ENSMUSG00000030008 | MGI:1920577 | protein coding gene | protease-associated domain containing 1 |
| 6 | gene | 85.45150 | 85.46848 | positive | MGI_C57BL6J_107184 | Cct7 | NCBI_Gene:12468,ENSEMBL:ENSMUSG00000030007 | MGI:107184 | protein coding gene | chaperonin containing Tcp1, subunit 7 (eta) |
| 6 | gene | 85.46957 | 85.50299 | negative | MGI_C57BL6J_1261912 | Fbxo41 | NCBI_Gene:330369,ENSEMBL:ENSMUSG00000047013 | MGI:1261912 | protein coding gene | F-box protein 41 |
| 6 | gene | 85.51112 | 85.51359 | negative | MGI_C57BL6J_99252 | Egr4 | NCBI_Gene:13656,ENSEMBL:ENSMUSG00000071341 | MGI:99252 | protein coding gene | early growth response 4 |
| 6 | pseudogene | 85.52945 | 85.52992 | positive | MGI_C57BL6J_5690415 | Gm44023 | ENSEMBL:ENSMUSG00000107410 | MGI:5690415 | pseudogene | predicted gene, 44023 |
| 6 | gene | 85.58750 | 85.72661 | positive | MGI_C57BL6J_1934606 | Alms1 | NCBI_Gene:236266,ENSEMBL:ENSMUSG00000063810 | MGI:1934606 | protein coding gene | ALMS1, centrosome and basal body associated |
| 6 | gene | 85.63286 | 85.63516 | positive | MGI_C57BL6J_5663725 | Gm43588 | ENSEMBL:ENSMUSG00000107370 | MGI:5663725 | unclassified gene | predicted gene 43588 |
| 6 | gene | 85.67928 | 85.68286 | positive | MGI_C57BL6J_5662737 | Gm42600 | ENSEMBL:ENSMUSG00000107278 | MGI:5662737 | unclassified gene | predicted gene 42600 |
| 6 | gene | 85.70671 | 85.71321 | negative | MGI_C57BL6J_3782661 | Nat8f7 | NCBI_Gene:100043497,ENSEMBL:ENSMUSG00000089694 | MGI:3782661 | protein coding gene | N-acetyltransferase 8 (GCN5-related) family member 7 |
| 6 | pseudogene | 85.72192 | 85.75664 | positive | MGI_C57BL6J_3646394 | Alms1-ps1 | NCBI_Gene:100043493,ENSEMBL:ENSMUSG00000090256 | MGI:3646394 | pseudogene | ALMS1, centrosome and basal body associated, pseudogene 1 |
| 6 | gene | 85.73251 | 85.76575 | negative | MGI_C57BL6J_2136449 | Nat8f3 | NCBI_Gene:93674,ENSEMBL:ENSMUSG00000051262 | MGI:2136449 | protein coding gene | N-acetyltransferase 8 (GCN5-related) family member 3 |
| 6 | gene | 85.74352 | 85.74361 | negative | MGI_C57BL6J_5530699 | Gm27317 | ENSEMBL:ENSMUSG00000098691 | MGI:5530699 | miRNA gene | predicted gene, 27317 |
| 6 | pseudogene | 85.77439 | 85.80406 | positive | MGI_C57BL6J_3779593 | Alms1-ps2 | NCBI_Gene:623273,ENSEMBL:ENSMUSG00000090098 | MGI:3779593 | pseudogene | ALMS1, centrosome and basal body associated, pseudogene 2 |
| 6 | gene | 85.79014 | 85.79023 | negative | MGI_C57BL6J_5562769 | Mir6375 | miRBase:MI0021904,NCBI_Gene:102465195,ENSEMBL:ENSMUSG00000105002 | MGI:5562769 | miRNA gene | microRNA 6375 |
| 6 | gene | 85.80802 | 85.82095 | negative | MGI_C57BL6J_3779382 | Nat8f6 | NCBI_Gene:100504710,ENSEMBL:ENSMUSG00000079495 | MGI:3779382 | protein coding gene | N-acetyltransferase 8 (GCN5-related) family member 6 |
| 6 | gene | 85.81722 | 85.82097 | negative | MGI_C57BL6J_1916299 | Nat8f5 | NCBI_Gene:69049,ENSEMBL:ENSMUSG00000079494 | MGI:1916299 | protein coding gene | N-acetyltransferase 8 (GCN5-related) family member 5 |
| 6 | gene | 85.83039 | 85.83749 | negative | MGI_C57BL6J_1915646 | Nat8 | NCBI_Gene:68396,ENSEMBL:ENSMUSG00000030004 | MGI:1915646 | protein coding gene | N-acetyltransferase 8 (GCN5-related) |
| 6 | pseudogene | 85.84382 | 85.84539 | positive | MGI_C57BL6J_3704271 | Ptges3-ps | NCBI_Gene:100043508,ENSEMBL:ENSMUSG00000040078 | MGI:3704271 | pseudogene | prostaglandin E synthase 3, pseudogene |
| 6 | gene | 85.85631 | 85.86113 | positive | MGI_C57BL6J_5621732 | Gm38847 | NCBI_Gene:105242703 | MGI:5621732 | lncRNA gene | predicted gene, 38847 |
| 6 | gene | 85.86542 | 85.86916 | negative | MGI_C57BL6J_2136446 | Nat8f2 | NCBI_Gene:93673,ENSEMBL:ENSMUSG00000033634 | MGI:2136446 | protein coding gene | N-acetyltransferase 8 (GCN5-related) family member 2 |
| 6 | pseudogene | 85.88932 | 85.88999 | positive | MGI_C57BL6J_5663446 | Gm43309 | NCBI_Gene:108169194,ENSEMBL:ENSMUSG00000106784 | MGI:5663446 | pseudogene | predicted gene 43309 |
| 6 | gene | 85.89905 | 85.90943 | negative | MGI_C57BL6J_1922791 | Nat8f4 | NCBI_Gene:75541,ENSEMBL:ENSMUSG00000068299 | MGI:1922791 | protein coding gene | N-acetyltransferase 8 (GCN5-related) family member 4 |
| 6 | gene | 85.91015 | 85.91569 | negative | MGI_C57BL6J_1913366 | Nat8f1 | NCBI_Gene:66116,ENSEMBL:ENSMUSG00000057103 | MGI:1913366 | protein coding gene | N-acetyltransferase 8 (GCN5-related) family member 1 |
| 6 | gene | 85.91176 | 85.93028 | positive | MGI_C57BL6J_1917036 | Tprkb | NCBI_Gene:69786,ENSEMBL:ENSMUSG00000054226 | MGI:1917036 | protein coding gene | Tp53rk binding protein |
| 6 | pseudogene | 85.92057 | 85.92142 | negative | MGI_C57BL6J_5010475 | Gm18290 | NCBI_Gene:100416843,ENSEMBL:ENSMUSG00000106965 | MGI:5010475 | pseudogene | predicted gene, 18290 |
| 6 | pseudogene | 85.93256 | 85.93338 | negative | MGI_C57BL6J_3644831 | Nat8b-ps | NCBI_Gene:434057,ENSEMBL:ENSMUSG00000089634 | MGI:3644831 | pseudogene | N-acetyltransferase 8B, pseudogene |
| 6 | pseudogene | 85.93844 | 85.93922 | positive | MGI_C57BL6J_3645071 | Gm7542 | NCBI_Gene:665213,ENSEMBL:ENSMUSG00000107203 | MGI:3645071 | pseudogene | predicted gene 7542 |
| 6 | gene | 85.94227 | 85.96205 | negative | MGI_C57BL6J_1919352 | Dusp11 | NCBI_Gene:72102,ENSEMBL:ENSMUSG00000030002 | MGI:1919352 | protein coding gene | dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) |
| 6 | gene | 85.96162 | 86.00193 | positive | MGI_C57BL6J_5011450 | Gm19265 | ENSEMBL:ENSMUSG00000107230 | MGI:5011450 | lncRNA gene | predicted gene, 19265 |
| 6 | gene | 85.98901 | 86.00350 | negative | MGI_C57BL6J_5592478 | Gm33319 | NCBI_Gene:102636179 | MGI:5592478 | lncRNA gene | predicted gene, 33319 |
| 6 | gene | 86.00438 | 86.00935 | positive | MGI_C57BL6J_1922605 | 4930553P18Rik | NCBI_Gene:75355,ENSEMBL:ENSMUSG00000106791 | MGI:1922605 | lncRNA gene | RIKEN cDNA 4930553P18 gene |
| 6 | gene | 86.01617 | 86.02100 | positive | MGI_C57BL6J_1349421 | Figla | NCBI_Gene:26910,ENSEMBL:ENSMUSG00000030001 | MGI:1349421 | protein coding gene | folliculogenesis specific basic helix-loop-helix |
| 6 | gene | 86.02868 | 86.12441 | positive | MGI_C57BL6J_87919 | Add2 | NCBI_Gene:11519,ENSEMBL:ENSMUSG00000030000 | MGI:87919 | protein coding gene | adducin 2 (beta) |
| 6 | gene | 86.03993 | 86.04177 | positive | MGI_C57BL6J_5690482 | Gm44090 | ENSEMBL:ENSMUSG00000107927 | MGI:5690482 | unclassified gene | predicted gene, 44090 |
| 6 | gene | 86.08437 | 86.08708 | negative | MGI_C57BL6J_5690481 | Gm44089 | ENSEMBL:ENSMUSG00000107842 | MGI:5690481 | lncRNA gene | predicted gene, 44089 |
| 6 | pseudogene | 86.12607 | 86.14825 | positive | MGI_C57BL6J_5011781 | Gm19596 | NCBI_Gene:100503219,ENSEMBL:ENSMUSG00000107857 | MGI:5011781 | pseudogene | predicted gene, 19596 |
| 6 | gene | 86.19522 | 86.27574 | positive | MGI_C57BL6J_98724 | Tgfa | NCBI_Gene:21802,ENSEMBL:ENSMUSG00000029999 | MGI:98724 | protein coding gene | transforming growth factor alpha |
| 6 | gene | 86.26156 | 86.26391 | negative | MGI_C57BL6J_5690309 | Gm43917 | ENSEMBL:ENSMUSG00000107745 | MGI:5690309 | unclassified gene | predicted gene, 43917 |
| 6 | pseudogene | 86.28487 | 86.28610 | positive | MGI_C57BL6J_3644882 | Gm7545 | NCBI_Gene:665233,ENSEMBL:ENSMUSG00000098175 | MGI:3644882 | pseudogene | predicted gene 7545 |
| 6 | pseudogene | 86.33401 | 86.33432 | positive | MGI_C57BL6J_3704272 | Gm10443 | NCBI_Gene:100043527,ENSEMBL:ENSMUSG00000072940 | MGI:3704272 | pseudogene | predicted pseudogene 10443 |
| 6 | pseudogene | 86.35184 | 86.35298 | positive | MGI_C57BL6J_5011047 | Gm18862 | NCBI_Gene:100417851,ENSEMBL:ENSMUSG00000107819 | MGI:5011047 | pseudogene | predicted gene, 18862 |
| 6 | gene | 86.36545 | 86.37006 | positive | MGI_C57BL6J_1913738 | Fam136a | NCBI_Gene:66488,ENSEMBL:ENSMUSG00000057497 | MGI:1913738 | protein coding gene | family with sequence similarity 136, member A |
| 6 | gene | 86.37130 | 86.37933 | positive | MGI_C57BL6J_1915261 | Snrpg | NCBI_Gene:68011,ENSEMBL:ENSMUSG00000057278 | MGI:1915261 | protein coding gene | small nuclear ribonucleoprotein polypeptide G |
| 6 | gene | 86.37254 | 86.37529 | negative | MGI_C57BL6J_5753517 | Gm44941 | ENSEMBL:ENSMUSG00000107509 | MGI:5753517 | lncRNA gene | predicted gene 44941 |
| 6 | gene | 86.38601 | 86.39716 | negative | MGI_C57BL6J_1914131 | Pcyox1 | NCBI_Gene:66881,ENSEMBL:ENSMUSG00000029998 | MGI:1914131 | protein coding gene | prenylcysteine oxidase 1 |
| 6 | gene | 86.40389 | 86.40396 | positive | MGI_C57BL6J_4413869 | n-TGccc4 | NCBI_Gene:102467309 | MGI:4413869 | tRNA gene | nuclear encoded tRNA glycine 4 (anticodon CCC) |
| 6 | gene | 86.40422 | 86.43340 | positive | MGI_C57BL6J_107914 | Tia1 | NCBI_Gene:21841,ENSEMBL:ENSMUSG00000071337 | MGI:107914 | protein coding gene | cytotoxic granule-associated RNA binding protein 1 |
| 6 | gene | 86.42666 | 86.44563 | positive | MGI_C57BL6J_5753366 | Gm44790 | ENSEMBL:ENSMUSG00000107539 | MGI:5753366 | protein coding gene | predicted gene 44790 |
| 6 | gene | 86.43733 | 86.47350 | positive | MGI_C57BL6J_2141787 | C87436 | NCBI_Gene:232196,ENSEMBL:ENSMUSG00000046679 | MGI:2141787 | protein coding gene | expressed sequence C87436 |
| 6 | gene | 86.47028 | 86.48083 | negative | MGI_C57BL6J_2444132 | A430078I02Rik | NCBI_Gene:320486,ENSEMBL:ENSMUSG00000085589 | MGI:2444132 | lncRNA gene | RIKEN cDNA A430078I02 gene |
| 6 | gene | 86.47290 | 86.47350 | positive | MGI_C57BL6J_5621734 | Gm38849 | NCBI_Gene:105242705 | MGI:5621734 | lncRNA gene | predicted gene, 38849 |
| 6 | gene | 86.48338 | 86.48823 | negative | MGI_C57BL6J_1916897 | 2310040G24Rik | NCBI_Gene:381792,ENSEMBL:ENSMUSG00000101655 | MGI:1916897 | lncRNA gene | RIKEN cDNA 2310040G24 gene |
| 6 | gene | 86.49322 | 86.49391 | negative | MGI_C57BL6J_5690328 | Gm43936 | ENSEMBL:ENSMUSG00000107876 | MGI:5690328 | lncRNA gene | predicted gene, 43936 |
| 6 | gene | 86.49465 | 86.49780 | positive | MGI_C57BL6J_5579426 | Gm28720 | ENSEMBL:ENSMUSG00000101210 | MGI:5579426 | lincRNA gene | predicted gene 28720 |
| 6 | gene | 86.49878 | 86.50054 | positive | MGI_C57BL6J_5579425 | Gm28719 | ENSEMBL:ENSMUSG00000100174 | MGI:5579425 | lincRNA gene | predicted gene 28719 |
| 6 | gene | 86.51389 | 86.51627 | negative | MGI_C57BL6J_5690778 | Gm44386 | ENSEMBL:ENSMUSG00000107689 | MGI:5690778 | lncRNA gene | predicted gene, 44386 |
| 6 | gene | 86.52449 | 86.52632 | negative | MGI_C57BL6J_1345635 | Pcbp1 | NCBI_Gene:23983,ENSEMBL:ENSMUSG00000051695 | MGI:1345635 | protein coding gene | poly(rC) binding protein 1 |
| 6 | gene | 86.52625 | 86.58247 | positive | MGI_C57BL6J_1919262 | 1600020E01Rik | NCBI_Gene:72012,ENSEMBL:ENSMUSG00000097048 | MGI:1919262 | lncRNA gene | RIKEN cDNA 1600020E01 gene |
| 6 | gene | 86.52876 | 86.53121 | positive | MGI_C57BL6J_5621561 | Gm38676 | NCBI_Gene:105242392 | MGI:5621561 | lncRNA gene | predicted gene, 38676 |
| 6 | gene | 86.57410 | 86.57416 | negative | MGI_C57BL6J_5451961 | Gm22184 | ENSEMBL:ENSMUSG00000093162 | MGI:5451961 | miRNA gene | predicted gene, 22184 |
| 6 | gene | 86.59143 | 86.59360 | positive | MGI_C57BL6J_5690761 | Gm44369 | ENSEMBL:ENSMUSG00000108059 | MGI:5690761 | unclassified gene | predicted gene, 44369 |
| 6 | gene | 86.60442 | 86.62786 | positive | MGI_C57BL6J_5826600 | Gm46963 | NCBI_Gene:108169150 | MGI:5826600 | protein coding gene | predicted gene, 46963 |
| 6 | gene | 86.62816 | 86.62971 | positive | MGI_C57BL6J_1915105 | Asprv1 | NCBI_Gene:67855,ENSEMBL:ENSMUSG00000033508 | MGI:1915105 | protein coding gene | aspartic peptidase, retroviral-like 1 |
| 6 | gene | 86.64704 | 86.66933 | negative | MGI_C57BL6J_96908 | Mxd1 | NCBI_Gene:17119,ENSEMBL:ENSMUSG00000001156 | MGI:96908 | protein coding gene | MAX dimerization protein 1 |
| 6 | gene | 86.65395 | 86.66306 | negative | MGI_C57BL6J_5621735 | Gm38850 | NCBI_Gene:105242706 | MGI:5621735 | lncRNA gene | predicted gene, 38850 |
| 6 | gene | 86.66319 | 86.66555 | negative | MGI_C57BL6J_5690684 | Gm44292 | ENSEMBL:ENSMUSG00000108291 | MGI:5690684 | unclassified gene | predicted gene, 44292 |
| 6 | gene | 86.67515 | 86.68452 | negative | MGI_C57BL6J_1913868 | Snrnp27 | NCBI_Gene:66618,ENSEMBL:ENSMUSG00000001158 | MGI:1913868 | protein coding gene | small nuclear ribonucleoprotein 27 (U4/U6.U5) |
| 6 | gene | 86.68782 | 86.68843 | negative | MGI_C57BL6J_5690606 | Gm44214 | ENSEMBL:ENSMUSG00000108197 | MGI:5690606 | lncRNA gene | predicted gene, 44214 |
| 6 | gene | 86.69177 | 86.73340 | negative | MGI_C57BL6J_1345156 | Gmcl1 | NCBI_Gene:23885,ENSEMBL:ENSMUSG00000001157 | MGI:1345156 | protein coding gene | germ cell-less, spermatogenesis associated 1 |
| 6 | gene | 86.73349 | 86.73707 | positive | MGI_C57BL6J_5690428 | Gm44036 | ENSEMBL:ENSMUSG00000107961 | MGI:5690428 | lncRNA gene | predicted gene, 44036 |
| 6 | gene | 86.73684 | 86.79359 | negative | MGI_C57BL6J_88030 | Anxa4 | NCBI_Gene:11746,ENSEMBL:ENSMUSG00000029994 | MGI:88030 | protein coding gene | annexin A4 |
| 6 | gene | 86.84840 | 86.84944 | negative | MGI_C57BL6J_1914420 | 2610306M01Rik | NCBI_Gene:67170,ENSEMBL:ENSMUSG00000100164 | MGI:1914420 | lncRNA gene | RIKEN cDNA 2610306M01 gene |
| 6 | gene | 86.84948 | 87.00323 | positive | MGI_C57BL6J_1098687 | Aak1 | NCBI_Gene:269774,ENSEMBL:ENSMUSG00000057230 | MGI:1098687 | protein coding gene | AP2 associated kinase 1 |
| 6 | gene | 86.86287 | 86.86445 | positive | MGI_C57BL6J_5690545 | Gm44153 | ENSEMBL:ENSMUSG00000108216 | MGI:5690545 | unclassified gene | predicted gene, 44153 |
| 6 | gene | 86.87379 | 86.87548 | positive | MGI_C57BL6J_5690544 | Gm44152 | ENSEMBL:ENSMUSG00000108152 | MGI:5690544 | unclassified gene | predicted gene, 44152 |
| 6 | gene | 86.96164 | 86.96344 | positive | MGI_C57BL6J_5690485 | Gm44093 | ENSEMBL:ENSMUSG00000107576 | MGI:5690485 | unclassified gene | predicted gene, 44093 |
| 6 | gene | 86.97653 | 86.97718 | positive | MGI_C57BL6J_5690483 | Gm44091 | ENSEMBL:ENSMUSG00000107688 | MGI:5690483 | lncRNA gene | predicted gene, 44091 |
| 6 | gene | 86.99498 | 87.00323 | positive | MGI_C57BL6J_5621380 | Gm38495 | NCBI_Gene:102637099 | MGI:5621380 | protein coding gene | predicted gene, 38495 |
| 6 | gene | 87.00924 | 87.02846 | positive | MGI_C57BL6J_1913290 | Nfu1 | NCBI_Gene:56748,ENSEMBL:ENSMUSG00000029993 | MGI:1913290 | protein coding gene | NFU1 iron-sulfur cluster scaffold |
| 6 | gene | 87.04285 | 87.09221 | positive | MGI_C57BL6J_95698 | Gfpt1 | NCBI_Gene:14583,ENSEMBL:ENSMUSG00000029992 | MGI:95698 | protein coding gene | glutamine fructose-6-phosphate transaminase 1 |
| 6 | gene | 87.10046 | 87.11300 | positive | MGI_C57BL6J_1261919 | D6Ertd527e | NCBI_Gene:52372,ENSEMBL:ENSMUSG00000090891 | MGI:1261919 | protein coding gene | DNA segment, Chr 6, ERATO Doi 527, expressed |
| 6 | gene | 87.12704 | 87.13274 | negative | MGI_C57BL6J_5623267 | Gm40382 | NCBI_Gene:105244848 | MGI:5623267 | lncRNA gene | predicted gene, 40382 |
| 6 | gene | 87.13385 | 87.33582 | negative | MGI_C57BL6J_1916788 | Antxr1 | NCBI_Gene:69538,ENSEMBL:ENSMUSG00000033420 | MGI:1916788 | protein coding gene | anthrax toxin receptor 1 |
| 6 | gene | 87.13874 | 87.14085 | negative | MGI_C57BL6J_5690807 | Gm44415 | ENSEMBL:ENSMUSG00000108195 | MGI:5690807 | unclassified gene | predicted gene, 44415 |
| 6 | gene | 87.17194 | 87.17401 | negative | MGI_C57BL6J_5690808 | Gm44416 | ENSEMBL:ENSMUSG00000107436 | MGI:5690808 | unclassified gene | predicted gene, 44416 |
| 6 | gene | 87.22125 | 87.26745 | positive | MGI_C57BL6J_5621736 | Gm38851 | NCBI_Gene:105242708 | MGI:5621736 | lncRNA gene | predicted gene, 38851 |
| 6 | gene | 87.25225 | 87.25561 | negative | MGI_C57BL6J_5690809 | Gm44417 | ENSEMBL:ENSMUSG00000107865 | MGI:5690809 | unclassified gene | predicted gene, 44417 |
| 6 | gene | 87.27774 | 87.27975 | positive | MGI_C57BL6J_5690810 | Gm44418 | ENSEMBL:ENSMUSG00000108006 | MGI:5690810 | lncRNA gene | predicted gene, 44418 |
| 6 | gene | 87.28115 | 87.28126 | positive | MGI_C57BL6J_4422028 | n-R5s164 | ENSEMBL:ENSMUSG00000084613 | MGI:4422028 | rRNA gene | nuclear encoded rRNA 5S 164 |
| 6 | gene | 87.28277 | 87.28545 | positive | MGI_C57BL6J_5593329 | Gm34170 | NCBI_Gene:102637331 | MGI:5593329 | lncRNA gene | predicted gene, 34170 |
| 6 | gene | 87.32156 | 87.32234 | negative | MGI_C57BL6J_1924658 | 9530013L04Rik | ENSEMBL:ENSMUSG00000107780 | MGI:1924658 | unclassified gene | RIKEN cDNA 9530013L04 gene |
| 6 | gene | 87.32307 | 87.33204 | negative | MGI_C57BL6J_5593249 | Gm34090 | NCBI_Gene:102637221 | MGI:5593249 | lncRNA gene | predicted gene, 34090 |
| 6 | gene | 87.33275 | 87.33486 | negative | MGI_C57BL6J_5690806 | Gm44414 | ENSEMBL:ENSMUSG00000108143 | MGI:5690806 | unclassified gene | predicted gene, 44414 |
| 6 | gene | 87.34565 | 87.35094 | negative | MGI_C57BL6J_1913533 | Gkn1 | NCBI_Gene:66283,ENSEMBL:ENSMUSG00000030050 | MGI:1913533 | protein coding gene | gastrokine 1 |
| 6 | gene | 87.37336 | 87.37950 | positive | MGI_C57BL6J_1913534 | Gkn2 | NCBI_Gene:66284,ENSEMBL:ENSMUSG00000030049 | MGI:1913534 | protein coding gene | gastrokine 2 |
| 6 | gene | 87.38326 | 87.38894 | negative | MGI_C57BL6J_1916138 | Gkn3 | NCBI_Gene:68888,ENSEMBL:ENSMUSG00000030048 | MGI:1916138 | protein coding gene | gastrokine 3 |
| 6 | gene | 87.42899 | 87.43768 | positive | MGI_C57BL6J_1338820 | Bmp10 | NCBI_Gene:12154,ENSEMBL:ENSMUSG00000030046 | MGI:1338820 | protein coding gene | bone morphogenetic protein 10 |
| 6 | gene | 87.45854 | 87.53326 | negative | MGI_C57BL6J_2443687 | Arhgap25 | NCBI_Gene:232201,ENSEMBL:ENSMUSG00000030047 | MGI:2443687 | protein coding gene | Rho GTPase activating protein 25 |
| 6 | gene | 87.53341 | 87.53604 | positive | MGI_C57BL6J_5690590 | Gm44198 | ENSEMBL:ENSMUSG00000108184 | MGI:5690590 | unclassified gene | predicted gene, 44198 |
| 6 | gene | 87.57859 | 87.59077 | negative | MGI_C57BL6J_1929676 | Prokr1 | NCBI_Gene:58182,ENSEMBL:ENSMUSG00000049409 | MGI:1929676 | protein coding gene | prokineticin receptor 1 |
| 6 | gene | 87.58185 | 87.58780 | positive | MGI_C57BL6J_5593381 | Gm34222 | NCBI_Gene:102637405 | MGI:5593381 | lncRNA gene | predicted gene, 34222 |
| 6 | gene | 87.59756 | 87.59919 | positive | MGI_C57BL6J_5593471 | Gm34312 | NCBI_Gene:102637526,ENSEMBL:ENSMUSG00000108118 | MGI:5593471 | lncRNA gene | predicted gene, 34312 |
| 6 | gene | 87.62842 | 87.67220 | negative | MGI_C57BL6J_1919353 | Aplf | NCBI_Gene:72103,ENSEMBL:ENSMUSG00000030051 | MGI:1919353 | protein coding gene | aprataxin and PNKP like factor |
| 6 | gene | 87.63699 | 87.63984 | positive | MGI_C57BL6J_5826601 | Gm46964 | NCBI_Gene:108169151 | MGI:5826601 | lncRNA gene | predicted gene, 46964 |
| 6 | gene | 87.65109 | 87.65370 | negative | MGI_C57BL6J_5621737 | Gm38852 | NCBI_Gene:105242709 | MGI:5621737 | lncRNA gene | predicted gene, 38852 |
| 6 | gene | 87.65775 | 87.66382 | positive | MGI_C57BL6J_5690489 | Gm44097 | ENSEMBL:ENSMUSG00000107593 | MGI:5690489 | lncRNA gene | predicted gene, 44097 |
| 6 | pseudogene | 87.67198 | 87.68109 | positive | MGI_C57BL6J_2443140 | E230015B07Rik | NCBI_Gene:320001,ENSEMBL:ENSMUSG00000071332 | MGI:2443140 | pseudogene | RIKEN cDNA E230015B07 gene |
| 6 | gene | 87.67559 | 87.72280 | negative | MGI_C57BL6J_3586838 | 1810020O05Rik | NCBI_Gene:613264,ENSEMBL:ENSMUSG00000089997 | MGI:3586838 | lncRNA gene | Riken cDNA 1810020O05 gene |
| 6 | gene | 87.72996 | 87.73115 | negative | MGI_C57BL6J_5690602 | Gm44210 | ENSEMBL:ENSMUSG00000108079 | MGI:5690602 | lncRNA gene | predicted gene, 44210 |
| 6 | gene | 87.73082 | 87.76242 | positive | MGI_C57BL6J_3611451 | Efcc1 | NCBI_Gene:58229,ENSEMBL:ENSMUSG00000068263 | MGI:3611451 | protein coding gene | EF hand and coiled-coil domain containing 1 |
| 6 | gene | 87.74104 | 87.74919 | negative | MGI_C57BL6J_5593573 | Gm34414 | NCBI_Gene:102637660 | MGI:5593573 | lncRNA gene | predicted gene, 34414 |
| 6 | pseudogene | 87.76488 | 87.76698 | negative | MGI_C57BL6J_3647624 | Gm5879 | NCBI_Gene:545864,ENSEMBL:ENSMUSG00000068262 | MGI:3647624 | pseudogene | predicted gene 5879 |
| 6 | gene | 87.77612 | 87.77979 | positive | MGI_C57BL6J_1860137 | Gp9 | NCBI_Gene:54368,ENSEMBL:ENSMUSG00000030054 | MGI:1860137 | protein coding gene | glycoprotein 9 (platelet) |
| 6 | gene | 87.77614 | 87.77748 | positive | MGI_C57BL6J_5690296 | Gm43904 | ENSEMBL:ENSMUSG00000107849 | MGI:5690296 | lncRNA gene | predicted gene, 43904 |
| 6 | gene | 87.78885 | 87.81216 | negative | MGI_C57BL6J_1917084 | Rab43 | NCBI_Gene:69834,ENSEMBL:ENSMUSG00000030055 | MGI:1917084 | protein coding gene | RAB43, member RAS oncogene family |
| 6 | gene | 87.78886 | 87.83580 | negative | MGI_C57BL6J_5753716 | Gm45140 | ENSEMBL:ENSMUSG00000107928 | MGI:5753716 | protein coding gene | predicted gene 45140 |
| 6 | gene | 87.79607 | 87.79923 | negative | MGI_C57BL6J_5690815 | Gm44423 | ENSEMBL:ENSMUSG00000108064 | MGI:5690815 | unclassified gene | predicted gene, 44423 |
| 6 | gene | 87.80547 | 87.80634 | negative | MGI_C57BL6J_1924996 | 9930120I10Rik | ENSEMBL:ENSMUSG00000108120 | MGI:1924996 | unclassified gene | RIKEN cDNA 9930120I10 gene |
| 6 | gene | 87.81427 | 87.83880 | negative | MGI_C57BL6J_1923310 | Isy1 | NCBI_Gene:57905,ENSEMBL:ENSMUSG00000030056 | MGI:1923310 | protein coding gene | ISY1 splicing factor homolog |
| 6 | gene | 87.83027 | 87.83287 | negative | MGI_C57BL6J_2442518 | B130021K23Rik | ENSEMBL:ENSMUSG00000108042 | MGI:2442518 | unclassified gene | RIKEN cDNA B130021K23 gene |
| 6 | gene | 87.84261 | 87.85111 | negative | MGI_C57BL6J_88431 | Cnbp | NCBI_Gene:12785,ENSEMBL:ENSMUSG00000030057 | MGI:88431 | protein coding gene | cellular nucleic acid binding protein |
| 6 | gene | 87.85121 | 87.85229 | positive | MGI_C57BL6J_5690456 | Gm44064 | ENSEMBL:ENSMUSG00000107798 | MGI:5690456 | unclassified gene | predicted gene, 44064 |
| 6 | gene | 87.88781 | 87.91360 | positive | MGI_C57BL6J_1858696 | Copg1 | NCBI_Gene:54161,ENSEMBL:ENSMUSG00000030058 | MGI:1858696 | protein coding gene | coatomer protein complex, subunit gamma 1 |
| 6 | gene | 87.90884 | 87.91446 | negative | MGI_C57BL6J_5477130 | Gm26636 | ENSEMBL:ENSMUSG00000097763 | MGI:5477130 | lncRNA gene | predicted gene, 26636 |
| 6 | gene | 87.91391 | 87.93663 | positive | MGI_C57BL6J_1914053 | Hmces | NCBI_Gene:232210,ENSEMBL:ENSMUSG00000030060 | MGI:1914053 | protein coding gene | 5-hydroxymethylcytosine (hmC) binding, ES cell specific |
| 6 | pseudogene | 87.92887 | 87.93028 | negative | MGI_C57BL6J_5011223 | Gm19038 | NCBI_Gene:100418152 | MGI:5011223 | pseudogene | predicted gene, 19038 |
| 6 | pseudogene | 87.94844 | 87.94944 | negative | MGI_C57BL6J_3641929 | Rps19-ps9 | NCBI_Gene:665416,ENSEMBL:ENSMUSG00000068689 | MGI:3641929 | pseudogene | ribosomal protein S19, pseudogene 9 |
| 6 | gene | 87.98042 | 87.98164 | negative | MGI_C57BL6J_2685307 | H1f10 | NCBI_Gene:243529,ENSEMBL:ENSMUSG00000044927 | MGI:2685307 | protein coding gene | H1.10 linker histone |
| 6 | gene | 87.98088 | 88.00547 | positive | MGI_C57BL6J_3648213 | Gm5577 | NCBI_Gene:434064,ENSEMBL:ENSMUSG00000084950 | MGI:3648213 | lncRNA gene | predicted gene 5577 |
| 6 | gene | 87.98161 | 87.98174 | negative | MGI_C57BL6J_6096137 | Gm47286 | ENSEMBL:ENSMUSG00000106591 | MGI:6096137 | miRNA gene | predicted gene, 47286 |
| 6 | gene | 87.98619 | 87.98973 | positive | MGI_C57BL6J_5690560 | Gm44168 | ENSEMBL:ENSMUSG00000108049 | MGI:5690560 | unclassified gene | predicted gene, 44168 |
| 6 | gene | 87.99314 | 87.99418 | negative | MGI_C57BL6J_1921328 | 4933412L11Rik | ENSEMBL:ENSMUSG00000108227 | MGI:1921328 | unclassified gene | RIKEN cDNA 4933412L11 gene |
| 6 | gene | 87.99911 | 88.04527 | negative | MGI_C57BL6J_105068 | Rab7 | NCBI_Gene:19349,ENSEMBL:ENSMUSG00000079477 | MGI:105068 | protein coding gene | RAB7, member RAS oncogene family |
| 6 | gene | 88.00520 | 88.01065 | positive | MGI_C57BL6J_5594031 | Gm34872 | NCBI_Gene:102638270 | MGI:5594031 | lncRNA gene | predicted gene, 34872 |
| 6 | gene | 88.04054 | 88.04247 | negative | MGI_C57BL6J_5690579 | Gm44187 | ENSEMBL:ENSMUSG00000108268 | MGI:5690579 | unclassified gene | predicted gene, 44187 |
| 6 | gene | 88.04814 | 88.05384 | negative | MGI_C57BL6J_5621740 | Gm38855 | NCBI_Gene:105242712 | MGI:5621740 | lncRNA gene | predicted gene, 38855 |
| 6 | gene | 88.08447 | 88.10530 | positive | MGI_C57BL6J_98084 | Rpn1 | NCBI_Gene:103963,ENSEMBL:ENSMUSG00000030062 | MGI:98084 | protein coding gene | ribophorin I |
| 6 | gene | 88.10413 | 88.10426 | negative | MGI_C57BL6J_5531160 | Mir6376 | miRBase:MI0021905,NCBI_Gene:102465196,ENSEMBL:ENSMUSG00000099095 | MGI:5531160 | miRNA gene | microRNA 6376 |
| 6 | gene | 88.14185 | 88.19828 | negative | MGI_C57BL6J_5621593 | Gm38708 | NCBI_Gene:105242443,ENSEMBL:ENSMUSG00000108010 | MGI:5621593 | lncRNA gene | predicted gene, 38708 |
| 6 | gene | 88.15156 | 88.15500 | positive | MGI_C57BL6J_5621741 | Gm38856 | NCBI_Gene:105242713 | MGI:5621741 | lncRNA gene | predicted gene, 38856 |
| 6 | gene | 88.19344 | 88.20703 | positive | MGI_C57BL6J_95662 | Gata2 | NCBI_Gene:14461,ENSEMBL:ENSMUSG00000015053 | MGI:95662 | protein coding gene | GATA binding protein 2 |
| 6 | gene | 88.21625 | 88.21650 | negative | MGI_C57BL6J_5690780 | Gm44388 | ENSEMBL:ENSMUSG00000107776 | MGI:5690780 | unclassified gene | predicted gene, 44388 |
| 6 | gene | 88.22227 | 88.25441 | positive | MGI_C57BL6J_1922801 | Dnajb8 | NCBI_Gene:56691,ENSEMBL:ENSMUSG00000048206 | MGI:1922801 | protein coding gene | DnaJ heat shock protein family (Hsp40) member B8 |
| 6 | gene | 88.22260 | 88.22568 | negative | MGI_C57BL6J_1920567 | 1700031F10Rik | ENSEMBL:ENSMUSG00000047046 | MGI:1920567 | lncRNA gene | RIKEN cDNA 1700031F10 gene |
| 6 | gene | 88.25733 | 88.44656 | negative | MGI_C57BL6J_2137092 | Eefsec | NCBI_Gene:65967,ENSEMBL:ENSMUSG00000033216 | MGI:2137092 | protein coding gene | eukaryotic elongation factor, selenocysteine-tRNA-specific |
| 6 | gene | 88.35314 | 88.35361 | negative | MGI_C57BL6J_5690657 | Gm44265 | ENSEMBL:ENSMUSG00000107472 | MGI:5690657 | lncRNA gene | predicted gene, 44265 |
| 6 | gene | 88.40598 | 88.40630 | positive | MGI_C57BL6J_5454955 | Gm25178 | ENSEMBL:ENSMUSG00000084582 | MGI:5454955 | unclassified non-coding RNA gene | predicted gene, 25178 |
| 6 | gene | 88.44569 | 88.44651 | positive | MGI_C57BL6J_5690656 | Gm44264 | ENSEMBL:ENSMUSG00000107561 | MGI:5690656 | lncRNA gene | predicted gene, 44264 |
| 6 | gene | 88.46541 | 88.49757 | positive | MGI_C57BL6J_1928760 | Ruvbl1 | NCBI_Gene:56505,ENSEMBL:ENSMUSG00000030079 | MGI:1928760 | protein coding gene | RuvB-like protein 1 |
| 6 | gene | 88.49162 | 88.49640 | positive | MGI_C57BL6J_5826602 | Gm46965 | NCBI_Gene:108169152 | MGI:5826602 | protein coding gene | predicted gene, 46965 |
| 6 | gene | 88.50358 | 88.51891 | negative | MGI_C57BL6J_1858417 | Sec61a1 | NCBI_Gene:53421,ENSEMBL:ENSMUSG00000030082 | MGI:1858417 | protein coding gene | Sec61 alpha 1 subunit (S. cerevisiae) |
| 6 | gene | 88.50360 | 88.50428 | positive | MGI_C57BL6J_5690562 | Gm44170 | ENSEMBL:ENSMUSG00000107659 | MGI:5690562 | lncRNA gene | predicted gene, 44170 |
| 6 | gene | 88.51901 | 88.52001 | positive | MGI_C57BL6J_5690822 | Gm44430 | ENSEMBL:ENSMUSG00000107698 | MGI:5690822 | unclassified gene | predicted gene, 44430 |
| 6 | gene | 88.54511 | 88.63795 | negative | MGI_C57BL6J_1918481 | Kbtbd12 | NCBI_Gene:74589,ENSEMBL:ENSMUSG00000033182 | MGI:1918481 | protein coding gene | kelch repeat and BTB (POZ) domain containing 12 |
| 6 | gene | 88.71642 | 88.72734 | negative | MGI_C57BL6J_5477082 | Gm26588 | NCBI_Gene:102639841,ENSEMBL:ENSMUSG00000097730 | MGI:5477082 | lncRNA gene | predicted gene, 26588 |
| 6 | gene | 88.72441 | 88.82836 | positive | MGI_C57BL6J_1346042 | Mgll | NCBI_Gene:23945,ENSEMBL:ENSMUSG00000033174 | MGI:1346042 | protein coding gene | monoglyceride lipase |
| 6 | gene | 88.73819 | 88.74136 | positive | MGI_C57BL6J_5690397 | Gm44005 | ENSEMBL:ENSMUSG00000107624 | MGI:5690397 | unclassified gene | predicted gene, 44005 |
| 6 | gene | 88.73861 | 88.74419 | negative | MGI_C57BL6J_5621744 | Gm38859 | NCBI_Gene:105242716 | MGI:5621744 | lncRNA gene | predicted gene, 38859 |
| 6 | gene | 88.74273 | 88.74562 | positive | MGI_C57BL6J_5690391 | Gm43999 | ENSEMBL:ENSMUSG00000107835 | MGI:5690391 | unclassified gene | predicted gene, 43999 |
| 6 | gene | 88.78433 | 88.78745 | negative | MGI_C57BL6J_5690329 | Gm43937 | ENSEMBL:ENSMUSG00000107719 | MGI:5690329 | unclassified gene | predicted gene, 43937 |
| 6 | gene | 88.80224 | 88.80602 | positive | MGI_C57BL6J_5690393 | Gm44001 | ENSEMBL:ENSMUSG00000107966 | MGI:5690393 | unclassified gene | predicted gene, 44001 |
| 6 | gene | 88.83591 | 88.84238 | negative | MGI_C57BL6J_1933148 | Abtb1 | NCBI_Gene:80283,ENSEMBL:ENSMUSG00000030083 | MGI:1933148 | protein coding gene | ankyrin repeat and BTB (POZ) domain containing 1 |
| 6 | gene | 88.84256 | 88.87505 | negative | MGI_C57BL6J_2442488 | Podxl2 | NCBI_Gene:319655,ENSEMBL:ENSMUSG00000033152 | MGI:2442488 | protein coding gene | podocalyxin-like 2 |
| 6 | gene | 88.84285 | 88.84727 | positive | MGI_C57BL6J_3641714 | Gm15612 | NCBI_Gene:100038356,ENSEMBL:ENSMUSG00000085828 | MGI:3641714 | lncRNA gene | predicted gene 15612 |
| 6 | pseudogene | 88.88265 | 88.88318 | positive | MGI_C57BL6J_5595348 | Gm36189 | ENSEMBL:ENSMUSG00000107432 | MGI:5595348 | pseudogene | predicted gene, 36189 |
| 6 | gene | 88.88347 | 88.89878 | negative | MGI_C57BL6J_105380 | Mcm2 | NCBI_Gene:17216,ENSEMBL:ENSMUSG00000002870 | MGI:105380 | protein coding gene | minichromosome maintenance complex component 2 |
| 6 | gene | 88.90225 | 88.91224 | positive | MGI_C57BL6J_1345190 | Tpra1 | NCBI_Gene:24100,ENSEMBL:ENSMUSG00000002871 | MGI:1345190 | protein coding gene | transmembrane protein, adipocyte asscociated 1 |
| 6 | gene | 88.91479 | 88.91710 | negative | MGI_C57BL6J_5595427 | Gm36268 | NCBI_Gene:102640127 | MGI:5595427 | lncRNA gene | predicted gene, 36268 |
| 6 | gene | 88.91971 | 88.92264 | positive | MGI_C57BL6J_5690570 | Gm44178 | ENSEMBL:ENSMUSG00000107676 | MGI:5690570 | unclassified gene | predicted gene, 44178 |
| 6 | gene | 88.93992 | 88.94648 | positive | MGI_C57BL6J_5595513 | Gm36354 | NCBI_Gene:102640238 | MGI:5595513 | lncRNA gene | predicted gene, 36354 |
| 6 | gene | 88.95068 | 89.11004 | positive | MGI_C57BL6J_3026922 | 4933427D06Rik | NCBI_Gene:232217,ENSEMBL:ENSMUSG00000055403 | MGI:3026922 | lncRNA gene | RIKEN cDNA 4933427D06 gene |
| 6 | gene | 88.96465 | 88.97389 | negative | MGI_C57BL6J_5595568 | Gm36409 | NCBI_Gene:102640310 | MGI:5595568 | lncRNA gene | predicted gene, 36409 |
| 6 | gene | 88.99798 | 89.00046 | positive | MGI_C57BL6J_5595664 | Gm36505 | NCBI_Gene:102640445 | MGI:5595664 | lncRNA gene | predicted gene, 36505 |
| 6 | gene | 89.02384 | 89.02591 | positive | MGI_C57BL6J_5595798 | Gm36639 | NCBI_Gene:102640620 | MGI:5595798 | lncRNA gene | predicted gene, 36639 |
| 6 | gene | 89.14099 | 89.14739 | positive | MGI_C57BL6J_3584270 | Gm1965 | NCBI_Gene:434065,ENSEMBL:ENSMUSG00000090254 | MGI:3584270 | lncRNA gene | predicted gene 1965 |
| 6 | gene | 89.18458 | 89.18603 | negative | MGI_C57BL6J_3645279 | Gm6507 | NCBI_Gene:101056056,ENSEMBL:ENSMUSG00000090013 | MGI:3645279 | lncRNA gene | predicted gene 6507 |
| 6 | pseudogene | 89.20125 | 89.20156 | positive | MGI_C57BL6J_5690595 | Gm44203 | ENSEMBL:ENSMUSG00000108221 | MGI:5690595 | pseudogene | predicted gene, 44203 |
| 6 | gene | 89.21116 | 89.21624 | negative | MGI_C57BL6J_2685685 | Gm839 | NCBI_Gene:330379,ENSEMBL:ENSMUSG00000060416 | MGI:2685685 | lncRNA gene | predicted gene 839 |
| 6 | gene | 89.24947 | 89.24960 | negative | MGI_C57BL6J_5455738 | Gm25961 | ENSEMBL:ENSMUSG00000087799 | MGI:5455738 | rRNA gene | predicted gene, 25961 |
| 6 | gene | 89.25782 | 89.25914 | negative | MGI_C57BL6J_5690596 | Gm44204 | ENSEMBL:ENSMUSG00000107732 | MGI:5690596 | lncRNA gene | predicted gene, 44204 |
| 6 | gene | 89.26951 | 89.28572 | negative | MGI_C57BL6J_1922381 | 4930512J16Rik | NCBI_Gene:75131,ENSEMBL:ENSMUSG00000107622 | MGI:1922381 | lncRNA gene | RIKEN cDNA 4930512J16 gene |
| 6 | gene | 89.27330 | 89.27633 | positive | MGI_C57BL6J_5595894 | Gm36735 | NCBI_Gene:102640739 | MGI:5595894 | lncRNA gene | predicted gene, 36735 |
| 6 | gene | 89.30463 | 89.32762 | positive | MGI_C57BL6J_5477305 | Gm26811 | ENSEMBL:ENSMUSG00000097816 | MGI:5477305 | lncRNA gene | predicted gene, 26811 |
| 6 | gene | 89.31631 | 89.36262 | negative | MGI_C57BL6J_107685 | Plxna1 | NCBI_Gene:18844,ENSEMBL:ENSMUSG00000030084 | MGI:107685 | protein coding gene | plexin A1 |
| 6 | gene | 89.34514 | 89.34656 | positive | MGI_C57BL6J_5690599 | Gm44207 | ENSEMBL:ENSMUSG00000107570 | MGI:5690599 | unclassified gene | predicted gene, 44207 |
| 6 | gene | 89.36136 | 89.36240 | positive | MGI_C57BL6J_5623300 | Gm40415 | NCBI_Gene:105244887 | MGI:5623300 | protein coding gene | predicted gene, 40415 |
| 6 | gene | 89.38315 | 89.59565 | negative | MGI_C57BL6J_1913348 | Chchd6 | NCBI_Gene:66098,ENSEMBL:ENSMUSG00000030086 | MGI:1913348 | protein coding gene | coiled-coil-helix-coiled-coil-helix domain containing 6 |
| 6 | gene | 89.45286 | 89.46738 | positive | MGI_C57BL6J_5826603 | Gm46966 | NCBI_Gene:108169153 | MGI:5826603 | lncRNA gene | predicted gene, 46966 |
| 6 | gene | 89.52684 | 89.52746 | negative | MGI_C57BL6J_5690295 | Gm43903 | ENSEMBL:ENSMUSG00000107680 | MGI:5690295 | unclassified gene | predicted gene, 43903 |
| 6 | pseudogene | 89.59930 | 89.59967 | negative | MGI_C57BL6J_5012081 | Gm19896 | NCBI_Gene:100503789 | MGI:5012081 | pseudogene | predicted gene, 19896 |
| 6 | gene | 89.64399 | 89.67553 | positive | MGI_C57BL6J_2386711 | Txnrd3 | NCBI_Gene:232223,ENSEMBL:ENSMUSG00000000811 | MGI:2386711 | protein coding gene | thioredoxin reductase 3 |
| 6 | pseudogene | 89.69866 | 89.69952 | negative | MGI_C57BL6J_2148531 | Vmn1r-ps25 | NCBI_Gene:113869,ENSEMBL:ENSMUSG00000100128 | MGI:2148531 | pseudogene | vomeronasal 1 receptor, pseudogene 25 |
| 6 | gene | 89.70754 | 89.71666 | positive | MGI_C57BL6J_2148518 | Vmn1r40 | NCBI_Gene:113855,ENSEMBL:ENSMUSG00000096051 | MGI:2148518 | protein coding gene | vomeronasal 1 receptor 40 |
| 6 | gene | 89.74077 | 89.74765 | positive | MGI_C57BL6J_2148520 | Vmn1r41 | NCBI_Gene:113857,ENSEMBL:ENSMUSG00000094586 | MGI:2148520 | protein coding gene | vomeronasal 1 receptor 41 |
| 6 | pseudogene | 89.78945 | 89.79041 | positive | MGI_C57BL6J_2148532 | Vmn1r-ps26 | NCBI_Gene:113870,ENSEMBL:ENSMUSG00000092311 | MGI:2148532 | pseudogene | vomeronasal 1 receptor, pseudogene 26 |
| 6 | pseudogene | 89.80283 | 89.80610 | negative | MGI_C57BL6J_2148533 | Vmn1r-ps27 | NCBI_Gene:113871,ENSEMBL:ENSMUSG00000092237 | MGI:2148533 | pseudogene | vomeronasal 1 receptor, pseudogene 27 |
| 6 | gene | 89.84257 | 89.87641 | negative | MGI_C57BL6J_2148511 | Vmn1r42 | NCBI_Gene:113848,ENSEMBL:ENSMUSG00000068232 | MGI:2148511 | protein coding gene | vomeronasal 1 receptor 42 |
| 6 | gene | 89.85893 | 89.87641 | negative | MGI_C57BL6J_2148510 | Vmn1r43 | NCBI_Gene:113847,ENSEMBL:ENSMUSG00000068231 | MGI:2148510 | protein coding gene | vomeronasal 1 receptor 43 |
| 6 | gene | 89.88366 | 89.89489 | positive | MGI_C57BL6J_2148517 | Vmn1r44 | NCBI_Gene:113854,ENSEMBL:ENSMUSG00000068234 | MGI:2148517 | protein coding gene | vomeronasal 1 receptor 44 |
| 6 | pseudogene | 89.90768 | 89.90862 | positive | MGI_C57BL6J_2148534 | Vmn1r-ps28 | NCBI_Gene:113872,ENSEMBL:ENSMUSG00000092228 | MGI:2148534 | pseudogene | vomeronasal 1 receptor, pseudogene 28 |
| 6 | gene | 89.93165 | 89.94060 | negative | MGI_C57BL6J_1333762 | Vmn1r45 | NCBI_Gene:22297,ENSEMBL:ENSMUSG00000044248 | MGI:1333762 | protein coding gene | vomeronasal 1 receptor 45 |
| 6 | gene | 89.96930 | 89.98672 | positive | MGI_C57BL6J_2148519 | Vmn1r46 | NCBI_Gene:113856,ENSEMBL:ENSMUSG00000061653 | MGI:2148519 | protein coding gene | vomeronasal 1 receptor 46 |
| 6 | pseudogene | 89.99761 | 90.00459 | positive | MGI_C57BL6J_3852367 | Vmn1r-ps29 | NCBI_Gene:100312490,ENSEMBL:ENSMUSG00000092303 | MGI:3852367 | pseudogene | vomeronasal 1 receptor, pseudogene 29 |
| 6 | gene | 90.01615 | 90.02748 | positive | MGI_C57BL6J_2148509 | Vmn1r47 | NCBI_Gene:113846,ENSEMBL:ENSMUSG00000060724 | MGI:2148509 | protein coding gene | vomeronasal 1 receptor 47 |
| 6 | gene | 90.03593 | 90.03684 | negative | MGI_C57BL6J_2148508 | Vmn1r48 | NCBI_Gene:113845,ENSEMBL:ENSMUSG00000057592 | MGI:2148508 | protein coding gene | vomeronasal 1 receptor 48 |
| 6 | pseudogene | 90.05641 | 90.05669 | negative | MGI_C57BL6J_3852370 | Vmn1r-ps31 | NCBI_Gene:100312492,ENSEMBL:ENSMUSG00000092450 | MGI:3852370 | pseudogene | vomeronasal 1 receptor, pseudogene 31 |
| 6 | pseudogene | 90.05663 | 90.05680 | positive | MGI_C57BL6J_5141999 | Gm20534 | ENSEMBL:ENSMUSG00000092588 | MGI:5141999 | pseudogene | predicted gene 20534 |
| 6 | gene | 90.07189 | 90.07851 | negative | MGI_C57BL6J_1344384 | Vmn1r49 | NCBI_Gene:24112,ENSEMBL:ENSMUSG00000095932 | MGI:1344384 | protein coding gene | vomeronasal 1, receptor 49 |
| 6 | gene | 90.10143 | 90.10911 | positive | MGI_C57BL6J_2148515 | Vmn1r50 | NCBI_Gene:113852,ENSEMBL:ENSMUSG00000094553 | MGI:2148515 | protein coding gene | vomeronasal 1 receptor 50 |
| 6 | gene | 90.12260 | 90.13226 | positive | MGI_C57BL6J_1333759 | Vmn1r51 | NCBI_Gene:22296,ENSEMBL:ENSMUSG00000062818 | MGI:1333759 | protein coding gene | vomeronasal 1 receptor 51 |
| 6 | pseudogene | 90.13037 | 90.23365 | negative | MGI_C57BL6J_5141891 | Gm20426 | NCBI_Gene:108169154,ENSEMBL:ENSMUSG00000092247 | MGI:5141891 | pseudogene | predicted gene 20426 |
| 6 | pseudogene | 90.14643 | 90.15564 | positive | MGI_C57BL6J_3646782 | Vmn1r-ps32 | NCBI_Gene:625050,ENSEMBL:ENSMUSG00000061843 | MGI:3646782 | pseudogene | vomeronasal 1 receptor, pseudogene 32 |
| 6 | gene | 90.17481 | 90.18125 | positive | MGI_C57BL6J_2148512 | Vmn1r52 | NCBI_Gene:113849,ENSEMBL:ENSMUSG00000060816 | MGI:2148512 | protein coding gene | vomeronasal 1 receptor 52 |
| 6 | gene | 90.20279 | 90.20368 | positive | MGI_C57BL6J_2148513 | V1ra8 | NCBI_Gene:113850,ENSEMBL:ENSMUSG00000062546 | MGI:2148513 | protein coding gene | vomeronasal 1 receptor, A8 |
| 6 | gene | 90.22332 | 90.22444 | negative | MGI_C57BL6J_2148516 | Vmn1r53 | NCBI_Gene:113853,ENSEMBL:ENSMUSG00000057697 | MGI:2148516 | protein coding gene | vomeronasal 1 receptor 53 |
| 6 | pseudogene | 90.23569 | 90.23627 | positive | MGI_C57BL6J_3782465 | Gm4287 | NCBI_Gene:100043191,ENSEMBL:ENSMUSG00000092552 | MGI:3782465 | pseudogene | predicted gene 4287 |
| 6 | pseudogene | 90.24227 | 90.24324 | positive | MGI_C57BL6J_2148535 | Vmn1r-ps34 | NCBI_Gene:113873,ENSEMBL:ENSMUSG00000099754 | MGI:2148535 | pseudogene | vomeronasal 1 receptor, pseudogene 34 |
| 6 | gene | 90.24630 | 90.27121 | positive | MGI_C57BL6J_2148514 | Vmn1r54 | NCBI_Gene:113851,ENSEMBL:ENSMUSG00000047203 | MGI:2148514 | protein coding gene | vomeronasal 1 receptor 54 |
| 6 | gene | 90.29622 | 90.29933 | positive | MGI_C57BL6J_5690283 | Gm43891 | ENSEMBL:ENSMUSG00000108119 | MGI:5690283 | unclassified gene | predicted gene, 43891 |
| 6 | gene | 90.30122 | 90.30545 | positive | MGI_C57BL6J_2679261 | BC048671 | NCBI_Gene:243535,ENSEMBL:ENSMUSG00000049694 | MGI:2679261 | protein coding gene | cDNA sequence BC048671 |
| 6 | gene | 90.30835 | 90.32519 | negative | MGI_C57BL6J_1919047 | Chst13 | NCBI_Gene:71797,ENSEMBL:ENSMUSG00000056643 | MGI:1919047 | protein coding gene | carbohydrate sulfotransferase 13 |
| 6 | gene | 90.33324 | 90.36455 | positive | MGI_C57BL6J_2385332 | Uroc1 | NCBI_Gene:243537,ENSEMBL:ENSMUSG00000034456 | MGI:2385332 | protein coding gene | urocanase domain containing 1 |
| 6 | gene | 90.36948 | 90.40349 | positive | MGI_C57BL6J_1933108 | Zxdc | NCBI_Gene:80292,ENSEMBL:ENSMUSG00000034430 | MGI:1933108 | protein coding gene | ZXD family zinc finger C |
| 6 | gene | 90.38681 | 90.38936 | positive | MGI_C57BL6J_1924668 | C030015A19Rik | ENSEMBL:ENSMUSG00000108181 | MGI:1924668 | unclassified gene | RIKEN cDNA C030015A19 gene |
| 6 | gene | 90.40348 | 90.42977 | negative | MGI_C57BL6J_2141635 | Cfap100 | NCBI_Gene:243538,ENSEMBL:ENSMUSG00000048794 | MGI:2141635 | protein coding gene | cilia and flagella associated protein 100 |
| 6 | gene | 90.45971 | 90.46147 | negative | MGI_C57BL6J_5690509 | Gm44117 | ENSEMBL:ENSMUSG00000107605 | MGI:5690509 | lncRNA gene | predicted gene, 44117 |
| 6 | gene | 90.46255 | 90.47917 | positive | MGI_C57BL6J_1929988 | Klf15 | NCBI_Gene:66277,ENSEMBL:ENSMUSG00000030087 | MGI:1929988 | protein coding gene | Kruppel-like factor 15 |
| 6 | gene | 90.47847 | 90.47969 | positive | MGI_C57BL6J_5690813 | Gm44421 | ENSEMBL:ENSMUSG00000107451 | MGI:5690813 | lncRNA gene | predicted gene, 44421 |
| 6 | gene | 90.48639 | 90.51948 | positive | MGI_C57BL6J_5805016 | Gm45901 | ENSEMBL:ENSMUSG00000108111 | MGI:5805016 | lncRNA gene | predicted gene 45901 |
| 6 | gene | 90.48643 | 90.60020 | positive | MGI_C57BL6J_1340024 | Aldh1l1 | NCBI_Gene:107747,ENSEMBL:ENSMUSG00000030088 | MGI:1340024 | protein coding gene | aldehyde dehydrogenase 1 family, member L1 |
| 6 | gene | 90.51330 | 90.51381 | positive | MGI_C57BL6J_5690628 | Gm44236 | ENSEMBL:ENSMUSG00000108271 | MGI:5690628 | unclassified gene | predicted gene, 44236 |
| 6 | gene | 90.59668 | 90.60020 | negative | MGI_C57BL6J_3783199 | Gm15756 | NCBI_Gene:102631993,ENSEMBL:ENSMUSG00000089781 | MGI:3783199 | lncRNA gene | predicted gene 15756 |
| 6 | gene | 90.60210 | 90.60693 | negative | MGI_C57BL6J_5825574 | Gm45937 | NCBI_Gene:108167323 | MGI:5825574 | lncRNA gene | predicted gene, 45937 |
| 6 | gene | 90.60473 | 90.64641 | positive | MGI_C57BL6J_1918949 | Slc41a3 | NCBI_Gene:71699,ENSEMBL:ENSMUSG00000030089 | MGI:1918949 | protein coding gene | solute carrier family 41, member 3 |
| 6 | gene | 90.65609 | 90.98869 | negative | MGI_C57BL6J_1196356 | Iqsec1 | NCBI_Gene:232227,ENSEMBL:ENSMUSG00000034312 | MGI:1196356 | protein coding gene | IQ motif and Sec7 domain 1 |
| 6 | gene | 90.68137 | 90.68152 | negative | MGI_C57BL6J_5454593 | Gm24816 | ENSEMBL:ENSMUSG00000093052 | MGI:5454593 | miRNA gene | predicted gene, 24816 |
| 6 | gene | 90.68973 | 90.69083 | positive | MGI_C57BL6J_5690515 | Gm44123 | ENSEMBL:ENSMUSG00000107999 | MGI:5690515 | lncRNA gene | predicted gene, 44123 |
| 6 | gene | 90.77774 | 90.78208 | negative | MGI_C57BL6J_2444127 | E130314K07Rik | NA | NA | unclassified gene | RIKEN cDNA E130314K07 gene |
| 6 | gene | 90.79390 | 90.79448 | negative | MGI_C57BL6J_5690497 | Gm44105 | ENSEMBL:ENSMUSG00000107795 | MGI:5690497 | lncRNA gene | predicted gene, 44105 |
| 6 | pseudogene | 90.84492 | 90.84549 | negative | MGI_C57BL6J_3648214 | Rpl21-ps12 | NCBI_Gene:100043646,ENSEMBL:ENSMUSG00000058700 | MGI:3648214 | pseudogene | ribosomal protein L21, pseudogene 12 |
| 6 | gene | 90.85754 | 90.85763 | positive | MGI_C57BL6J_5454962 | Gm25185 | ENSEMBL:ENSMUSG00000088379 | MGI:5454962 | snoRNA gene | predicted gene, 25185 |
| 6 | pseudogene | 90.90556 | 90.90712 | positive | MGI_C57BL6J_3647847 | Ppp1r2-ps2 | NCBI_Gene:665685,ENSEMBL:ENSMUSG00000099662 | MGI:3647847 | pseudogene | protein phosphatase 1, regulatory (inhibitor) subunit 2, pseudogene 2 |
| 6 | gene | 91.00997 | 91.01067 | positive | MGI_C57BL6J_5621745 | Gm38860 | NCBI_Gene:105242717 | MGI:5621745 | lncRNA gene | predicted gene, 38860 |
| 6 | gene | 91.01307 | 91.11701 | negative | MGI_C57BL6J_1859555 | Nup210 | NCBI_Gene:54563,ENSEMBL:ENSMUSG00000030091 | MGI:1859555 | protein coding gene | nucleoporin 210 |
| 6 | gene | 91.11710 | 91.12697 | positive | MGI_C57BL6J_5589523 | Gm30364 | NCBI_Gene:102632229 | MGI:5589523 | lncRNA gene | predicted gene, 30364 |
| 6 | gene | 91.11982 | 91.12094 | negative | MGI_C57BL6J_5690670 | Gm44278 | ENSEMBL:ENSMUSG00000107882 | MGI:5690670 | lncRNA gene | predicted gene, 44278 |
| 6 | gene | 91.15667 | 91.17469 | positive | MGI_C57BL6J_2385252 | Hdac11 | NCBI_Gene:232232,ENSEMBL:ENSMUSG00000034245 | MGI:2385252 | protein coding gene | histone deacetylase 11 |
| 6 | gene | 91.15871 | 91.15926 | negative | MGI_C57BL6J_3705150 | Gm14573 | ENSEMBL:ENSMUSG00000085624 | MGI:3705150 | lncRNA gene | predicted gene 14573 |
| 6 | gene | 91.17342 | 91.17402 | negative | MGI_C57BL6J_5690394 | Gm44002 | ENSEMBL:ENSMUSG00000108158 | MGI:5690394 | unclassified gene | predicted gene, 44002 |
| 6 | gene | 91.20722 | 91.21004 | negative | MGI_C57BL6J_1926044 | 4930402H05Rik | ENSEMBL:ENSMUSG00000107397 | MGI:1926044 | unclassified gene | RIKEN cDNA 4930402H05 gene |
| 6 | gene | 91.21246 | 91.27254 | positive | MGI_C57BL6J_95488 | Fbln2 | NCBI_Gene:14115,ENSEMBL:ENSMUSG00000064080 | MGI:95488 | protein coding gene | fibulin 2 |
| 6 | pseudogene | 91.32057 | 91.32103 | negative | MGI_C57BL6J_5826623 | Gm46986 | NCBI_Gene:108169195 | MGI:5826623 | pseudogene | predicted gene, 46986 |
| 6 | gene | 91.36398 | 91.41137 | negative | MGI_C57BL6J_98961 | Wnt7a | NCBI_Gene:22421,ENSEMBL:ENSMUSG00000030093 | MGI:98961 | protein coding gene | wingless-type MMTV integration site family, member 7A |
| 6 | gene | 91.41151 | 91.41272 | positive | MGI_C57BL6J_5589596 | Gm30437 | NCBI_Gene:102632338,ENSEMBL:ENSMUSG00000108804 | MGI:5589596 | lncRNA gene | predicted gene, 30437 |
| 6 | pseudogene | 91.42827 | 91.42881 | negative | MGI_C57BL6J_3852371 | Vmn1r-ps35 | NCBI_Gene:100312494 | MGI:3852371 | pseudogene | vomeronasal 1 receptor, pseudogene 35 |
| 6 | gene | 91.42831 | 91.44091 | negative | MGI_C57BL6J_1922150 | 4930471M09Rik | NCBI_Gene:75787,ENSEMBL:ENSMUSG00000097693 | MGI:1922150 | lncRNA gene | RIKEN cDNA 4930471M09 gene |
| 6 | pseudogene | 91.42887 | 91.43006 | positive | MGI_C57BL6J_3782758 | Gm4575 | NCBI_Gene:100043659,ENSEMBL:ENSMUSG00000098240 | MGI:3782758 | pseudogene | predicted gene 4575 |
| 6 | pseudogene | 91.43008 | 91.43048 | negative | MGI_C57BL6J_3852372 | Vmn1r-ps36 | NCBI_Gene:100312495 | MGI:3852372 | pseudogene | vomeronasal 1 receptor, pseudogene 36 |
| 6 | gene | 91.44099 | 91.44176 | positive | MGI_C57BL6J_1917048 | 1810044D09Rik | NCBI_Gene:69798,ENSEMBL:ENSMUSG00000100680 | MGI:1917048 | lncRNA gene | RIKEN cDNA 1810044D09 gene |
| 6 | pseudogene | 91.44503 | 91.44687 | negative | MGI_C57BL6J_5753205 | Gm44629 | ENSEMBL:ENSMUSG00000108810 | MGI:5753205 | pseudogene | predicted gene 44629 |
| 6 | gene | 91.46217 | 91.47355 | negative | MGI_C57BL6J_1919420 | Chchd4 | NCBI_Gene:72170,ENSEMBL:ENSMUSG00000034203 | MGI:1919420 | protein coding gene | coiled-coil-helix-coiled-coil-helix domain containing 4 |
| 6 | gene | 91.47370 | 91.48846 | positive | MGI_C57BL6J_1921372 | Tmem43 | NCBI_Gene:74122,ENSEMBL:ENSMUSG00000030095 | MGI:1921372 | protein coding gene | transmembrane protein 43 |
| 6 | gene | 91.48931 | 91.51589 | negative | MGI_C57BL6J_103557 | Xpc | NCBI_Gene:22591,ENSEMBL:ENSMUSG00000030094 | MGI:103557 | protein coding gene | xeroderma pigmentosum, complementation group C |
| 6 | gene | 91.51593 | 91.52263 | positive | MGI_C57BL6J_1914928 | Lsm3 | NCBI_Gene:67678,ENSEMBL:ENSMUSG00000034192 | MGI:1914928 | protein coding gene | LSM3 homolog, U6 small nuclear RNA and mRNA degradation associated |
| 6 | gene | 91.52241 | 91.53238 | positive | MGI_C57BL6J_5753792 | Gm45216 | ENSEMBL:ENSMUSG00000108584 | MGI:5753792 | lncRNA gene | predicted gene 45216 |
| 6 | pseudogene | 91.59476 | 91.59515 | positive | MGI_C57BL6J_5753791 | Gm45215 | ENSEMBL:ENSMUSG00000108335 | MGI:5753791 | pseudogene | predicted gene 45215 |
| 6 | gene | 91.59923 | 91.60551 | positive | MGI_C57BL6J_5753794 | Gm45218 | ENSEMBL:ENSMUSG00000108308 | MGI:5753794 | lncRNA gene | predicted gene 45218 |
| 6 | gene | 91.66074 | 91.67968 | positive | MGI_C57BL6J_5623272 | Gm40387 | NCBI_Gene:105244853 | MGI:5623272 | lncRNA gene | predicted gene, 40387 |
| 6 | gene | 91.66341 | 91.67962 | negative | MGI_C57BL6J_5589787 | Gm30628 | NCBI_Gene:102632595 | MGI:5589787 | lncRNA gene | predicted gene, 30628 |
| 6 | gene | 91.67820 | 91.67922 | negative | MGI_C57BL6J_5753793 | Gm45217 | ENSEMBL:ENSMUSG00000108751 | MGI:5753793 | unclassified gene | predicted gene 45217 |
| 6 | gene | 91.68405 | 91.75907 | positive | MGI_C57BL6J_98488 | Slc6a6 | NCBI_Gene:21366,ENSEMBL:ENSMUSG00000030096 | MGI:98488 | protein coding gene | solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| 6 | gene | 91.68993 | 91.69161 | negative | MGI_C57BL6J_1921977 | 4930517G19Rik | ENSEMBL:ENSMUSG00000108784 | MGI:1921977 | unclassified gene | RIKEN cDNA 4930517G19 gene |
| 6 | gene | 91.70941 | 91.71124 | positive | MGI_C57BL6J_5804943 | Gm45828 | ENSEMBL:ENSMUSG00000108526 | MGI:5804943 | unclassified gene | predicted gene 45828 |
| 6 | gene | 91.71134 | 91.71328 | negative | MGI_C57BL6J_5753557 | Gm44981 | ENSEMBL:ENSMUSG00000108574 | MGI:5753557 | unclassified gene | predicted gene 44981 |
| 6 | gene | 91.76151 | 91.84395 | negative | MGI_C57BL6J_2681173 | Grip2 | NCBI_Gene:243547,ENSEMBL:ENSMUSG00000030098 | MGI:2681173 | protein coding gene | glutamate receptor interacting protein 2 |
| 6 | gene | 91.87691 | 91.87798 | negative | MGI_C57BL6J_5753288 | Gm44712 | ENSEMBL:ENSMUSG00000108310 | MGI:5753288 | unclassified gene | predicted gene 44712 |
| 6 | gene | 91.87765 | 91.89985 | positive | MGI_C57BL6J_2444652 | Ccdc174 | NCBI_Gene:232236,ENSEMBL:ENSMUSG00000034083 | MGI:2444652 | protein coding gene | coiled-coil domain containing 174 |
| 6 | gene | 91.90269 | 91.95174 | positive | MGI_C57BL6J_2685917 | 4930590J08Rik | NCBI_Gene:381798,ENSEMBL:ENSMUSG00000034063 | MGI:2685917 | protein coding gene | RIKEN cDNA 4930590J08 gene |
| 6 | gene | 91.93825 | 91.93951 | negative | MGI_C57BL6J_5753287 | Gm44711 | ENSEMBL:ENSMUSG00000108625 | MGI:5753287 | unclassified gene | predicted gene 44711 |
| 6 | gene | 91.97882 | 92.07600 | positive | MGI_C57BL6J_2443369 | Fgd5 | NCBI_Gene:232237,ENSEMBL:ENSMUSG00000034037 | MGI:2443369 | protein coding gene | FYVE, RhoGEF and PH domain containing 5 |
| 6 | gene | 92.00755 | 92.00948 | positive | MGI_C57BL6J_5690487 | Gm44095 | ENSEMBL:ENSMUSG00000107794 | MGI:5690487 | unclassified gene | predicted gene, 44095 |
| 6 | gene | 92.01239 | 92.01612 | negative | MGI_C57BL6J_1923066 | 4930466I24Rik | ENSEMBL:ENSMUSG00000087317 | MGI:1923066 | lncRNA gene | RIKEN cDNA 4930466I24 gene |
| 6 | gene | 92.09139 | 92.17429 | positive | MGI_C57BL6J_1352466 | Nr2c2 | NCBI_Gene:22026,ENSEMBL:ENSMUSG00000005893 | MGI:1352466 | protein coding gene | nuclear receptor subfamily 2, group C, member 2 |
| 6 | gene | 92.14449 | 92.14629 | negative | MGI_C57BL6J_5690641 | Gm44249 | ENSEMBL:ENSMUSG00000108199 | MGI:5690641 | unclassified gene | predicted gene, 44249 |
| 6 | gene | 92.15101 | 92.15741 | negative | MGI_C57BL6J_5826604 | Gm46967 | NCBI_Gene:108169155 | MGI:5826604 | lncRNA gene | predicted gene, 46967 |
| 6 | gene | 92.16599 | 92.16702 | negative | MGI_C57BL6J_1924500 | 9430019H13Rik | NA | NA | unclassified gene | RIKEN cDNA 9430019H13 gene |
| 6 | gene | 92.16951 | 92.18403 | negative | MGI_C57BL6J_1928140 | Mrps25 | NCBI_Gene:64658,ENSEMBL:ENSMUSG00000014551 | MGI:1928140 | protein coding gene | mitochondrial ribosomal protein S25 |
| 6 | gene | 92.18671 | 92.21492 | negative | MGI_C57BL6J_1925537 | Rbsn | NCBI_Gene:78287,ENSEMBL:ENSMUSG00000014550 | MGI:1925537 | protein coding gene | rabenosyn, RAB effector |
| 6 | gene | 92.24206 | 92.24465 | negative | MGI_C57BL6J_98823 | Trh | NCBI_Gene:22044,ENSEMBL:ENSMUSG00000005892 | MGI:98823 | protein coding gene | thyrotropin releasing hormone |
| 6 | gene | 92.26922 | 92.31581 | positive | MGI_C57BL6J_5589847 | Gm30688 | NCBI_Gene:102632679 | MGI:5589847 | lncRNA gene | predicted gene, 30688 |
| 6 | pseudogene | 92.27134 | 92.27170 | negative | MGI_C57BL6J_5753379 | Gm44803 | NCBI_Gene:108169156,ENSEMBL:ENSMUSG00000108140 | MGI:5753379 | pseudogene | predicted gene 44803 |
| 6 | pseudogene | 92.28092 | 92.28139 | negative | MGI_C57BL6J_5690416 | Gm44024 | ENSEMBL:ENSMUSG00000108279 | MGI:5690416 | pseudogene | predicted gene, 44024 |
| 6 | gene | 92.37089 | 92.70634 | negative | MGI_C57BL6J_1925144 | Prickle2 | NCBI_Gene:243548,ENSEMBL:ENSMUSG00000030020 | MGI:1925144 | protein coding gene | prickle planar cell polarity protein 2 |
| 6 | gene | 92.40358 | 92.42232 | positive | MGI_C57BL6J_5621747 | Gm38862 | NCBI_Gene:105242720 | MGI:5621747 | lncRNA gene | predicted gene, 38862 |
| 6 | gene | 92.42245 | 92.42469 | positive | MGI_C57BL6J_5621746 | Gm38861 | NCBI_Gene:105242719 | MGI:5621746 | lncRNA gene | predicted gene, 38861 |
| 6 | pseudogene | 92.56176 | 92.56296 | positive | MGI_C57BL6J_5009983 | Gm17797 | NCBI_Gene:100151689,ENSEMBL:ENSMUSG00000098130 | MGI:5009983 | pseudogene | predicted gene, 17797 |
| 6 | pseudogene | 92.56443 | 92.56598 | negative | MGI_C57BL6J_3647623 | Gm5880 | NCBI_Gene:545866 | MGI:3647623 | pseudogene | predicted gene 5880 |
| 6 | gene | 92.77270 | 92.94405 | negative | MGI_C57BL6J_1916320 | Adamts9 | NCBI_Gene:101401,ENSEMBL:ENSMUSG00000030022 | MGI:1916320 | protein coding gene | a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 9 |
| 6 | gene | 92.81648 | 92.84718 | positive | MGI_C57BL6J_1921766 | A730049H05Rik | NCBI_Gene:74516,ENSEMBL:ENSMUSG00000048636 | MGI:1921766 | protein coding gene | RIKEN cDNA A730049H05 gene |
| 6 | gene | 92.86936 | 92.88441 | positive | MGI_C57BL6J_3783179 | Gm15737 | ENSEMBL:ENSMUSG00000079462 | MGI:3783179 | protein coding gene | predicted gene 15737 |
| 6 | gene | 92.94033 | 93.27298 | positive | MGI_C57BL6J_1924659 | 9530026P05Rik | NCBI_Gene:330385,ENSEMBL:ENSMUSG00000097462 | MGI:1924659 | lncRNA gene | RIKEN cDNA 9530026P05 gene |
| 6 | gene | 92.96385 | 92.96466 | positive | MGI_C57BL6J_2441979 | D630004L18Rik | NA | NA | unclassified gene | RIKEN cDNA D630004L18 gene |
| 6 | gene | 93.07911 | 93.08057 | positive | MGI_C57BL6J_5690616 | Gm44224 | ENSEMBL:ENSMUSG00000108286 | MGI:5690616 | unclassified gene | predicted gene, 44224 |
| 6 | pseudogene | 93.13680 | 93.13801 | negative | MGI_C57BL6J_3646641 | Gm5313 | NCBI_Gene:384461,ENSEMBL:ENSMUSG00000097996 | MGI:3646641 | pseudogene | predicted gene 5313 |
| 6 | gene | 93.19413 | 93.23900 | positive | MGI_C57BL6J_5621748 | Gm38863 | NCBI_Gene:105242721 | MGI:5621748 | lncRNA gene | predicted gene, 38863 |
| 6 | gene | 93.19494 | 93.19785 | positive | MGI_C57BL6J_5690315 | Gm43923 | ENSEMBL:ENSMUSG00000108256 | MGI:5690315 | unclassified gene | predicted gene, 43923 |
| 6 | gene | 93.24172 | 93.24421 | positive | MGI_C57BL6J_5690612 | Gm44220 | ENSEMBL:ENSMUSG00000107655 | MGI:5690612 | unclassified gene | predicted gene, 44220 |
| 6 | pseudogene | 93.27803 | 93.27971 | positive | MGI_C57BL6J_5434407 | Gm21052 | NCBI_Gene:100534392 | MGI:5434407 | pseudogene | predicted gene, 21052 |
| 6 | gene | 93.29915 | 93.31473 | negative | MGI_C57BL6J_5826605 | Gm46968 | NCBI_Gene:108169157 | MGI:5826605 | lncRNA gene | predicted gene, 46968 |
| 6 | gene | 93.33546 | 93.33559 | negative | MGI_C57BL6J_5454871 | Gm25094 | ENSEMBL:ENSMUSG00000077180 | MGI:5454871 | snoRNA gene | predicted gene, 25094 |
| 6 | gene | 93.45507 | 93.45556 | negative | MGI_C57BL6J_5690573 | Gm44181 | ENSEMBL:ENSMUSG00000107848 | MGI:5690573 | unclassified gene | predicted gene, 44181 |
| 6 | gene | 93.50957 | 93.50970 | positive | MGI_C57BL6J_5452089 | Gm22312 | ENSEMBL:ENSMUSG00000088630 | MGI:5452089 | snoRNA gene | predicted gene, 22312 |
| 6 | pseudogene | 93.64935 | 93.65023 | negative | MGI_C57BL6J_3646293 | Gm7812 | NCBI_Gene:665831,ENSEMBL:ENSMUSG00000107739 | MGI:3646293 | pseudogene | predicted gene 7812 |
| 6 | gene | 93.67545 | 94.28392 | negative | MGI_C57BL6J_1203522 | Magi1 | NCBI_Gene:14924,ENSEMBL:ENSMUSG00000045095 | MGI:1203522 | protein coding gene | membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| 6 | gene | 93.79377 | 93.79388 | positive | MGI_C57BL6J_5452812 | Gm23035 | ENSEMBL:ENSMUSG00000088627 | MGI:5452812 | snRNA gene | predicted gene, 23035 |
| 6 | gene | 93.89061 | 93.89075 | positive | MGI_C57BL6J_5452617 | Gm22840 | ENSEMBL:ENSMUSG00000089069 | MGI:5452617 | snoRNA gene | predicted gene, 22840 |
| 6 | gene | 93.91777 | 93.92078 | negative | MGI_C57BL6J_3028056 | B430316J06Rik | NA | NA | unclassified gene | RIKEN cDNA B430316J06 gene |
| 6 | gene | 93.98880 | 93.98974 | negative | MGI_C57BL6J_1924419 | 8030459D09Rik | NA | NA | unclassified gene | RIKEN cDNA 8030459D09 gene |
| 6 | gene | 94.12951 | 94.13266 | positive | MGI_C57BL6J_5590880 | Gm31721 | NCBI_Gene:102634038 | MGI:5590880 | lncRNA gene | predicted gene, 31721 |
| 6 | gene | 94.13292 | 94.23744 | negative | MGI_C57BL6J_1925428 | 4930511A08Rik | NCBI_Gene:78178 | MGI:1925428 | lncRNA gene | RIKEN cDNA 4930511A08 gene |
| 6 | gene | 94.28403 | 94.29209 | positive | MGI_C57BL6J_5590820 | Gm31661 | NCBI_Gene:102633964 | MGI:5590820 | lncRNA gene | predicted gene, 31661 |
| 6 | pseudogene | 94.32327 | 94.32403 | negative | MGI_C57BL6J_5690699 | Gm44307 | ENSEMBL:ENSMUSG00000107545 | MGI:5690699 | pseudogene | predicted gene, 44307 |
| 6 | pseudogene | 94.34158 | 94.35424 | positive | MGI_C57BL6J_5012093 | Gm19908 | NCBI_Gene:100503811,ENSEMBL:ENSMUSG00000107435 | MGI:5012093 | pseudogene | predicted gene, 19908 |
| 6 | pseudogene | 94.41780 | 94.41824 | positive | MGI_C57BL6J_3646507 | Gm7825 | NCBI_Gene:665856,ENSEMBL:ENSMUSG00000107616 | MGI:3646507 | pseudogene | predicted gene 7825 |
| 6 | pseudogene | 94.48418 | 94.48440 | negative | MGI_C57BL6J_5690707 | Gm44315 | ENSEMBL:ENSMUSG00000107781 | MGI:5690707 | pseudogene | predicted gene, 44315 |
| 6 | pseudogene | 94.49543 | 94.49597 | negative | MGI_C57BL6J_5011264 | Gm19079 | NCBI_Gene:100418221 | MGI:5011264 | pseudogene | predicted gene, 19079 |
| 6 | gene | 94.50001 | 94.60467 | positive | MGI_C57BL6J_1914832 | Slc25a26 | NCBI_Gene:67582,ENSEMBL:ENSMUSG00000045100 | MGI:1914832 | protein coding gene | solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 26 |
| 6 | gene | 94.60453 | 94.70025 | negative | MGI_C57BL6J_107935 | Lrig1 | NCBI_Gene:16206,ENSEMBL:ENSMUSG00000030029 | MGI:107935 | protein coding gene | leucine-rich repeats and immunoglobulin-like domains 1 |
| 6 | gene | 94.60636 | 94.60641 | negative | MGI_C57BL6J_5531254 | Mir7041 | miRBase:MI0022890,NCBI_Gene:102466992,ENSEMBL:ENSMUSG00000098531 | MGI:5531254 | miRNA gene | microRNA 7041 |
| 6 | pseudogene | 94.72217 | 94.72280 | positive | MGI_C57BL6J_3779619 | Gm6638 | NCBI_Gene:625941 | MGI:3779619 | pseudogene | predicted gene 6638 |
| 6 | gene | 94.79943 | 94.80044 | positive | MGI_C57BL6J_5690389 | Gm43997 | ENSEMBL:ENSMUSG00000107716 | MGI:5690389 | unclassified gene | predicted gene, 43997 |
| 6 | pseudogene | 94.82051 | 94.82172 | positive | MGI_C57BL6J_3643837 | Gm7833 | NCBI_Gene:665865,ENSEMBL:ENSMUSG00000097943 | MGI:3643837 | pseudogene | predicted gene 7833 |
| 6 | gene | 94.82373 | 94.82380 | positive | MGI_C57BL6J_5453141 | Gm23364 | ENSEMBL:ENSMUSG00000092767 | MGI:5453141 | miRNA gene | predicted gene, 23364 |
| 6 | gene | 94.82678 | 94.82692 | negative | MGI_C57BL6J_5454378 | Gm24601 | ENSEMBL:ENSMUSG00000088595 | MGI:5454378 | rRNA gene | predicted gene, 24601 |
| 6 | pseudogene | 94.84840 | 94.84883 | negative | MGI_C57BL6J_5690492 | Gm44100 | ENSEMBL:ENSMUSG00000107940 | MGI:5690492 | pseudogene | predicted gene, 44100 |
| 6 | gene | 94.94383 | 94.95154 | positive | MGI_C57BL6J_1926023 | 4930511E03Rik | NCBI_Gene:78773,ENSEMBL:ENSMUSG00000108187 | MGI:1926023 | lncRNA gene | RIKEN cDNA 4930511E03 gene |
| 6 | gene | 95.09201 | 95.09508 | negative | MGI_C57BL6J_1920900 | 2410024F20Rik | NA | NA | unclassified gene | RIKEN cDNA 2410024F20 gene |
| 6 | gene | 95.10020 | 95.10031 | negative | MGI_C57BL6J_5456278 | Gm26501 | ENSEMBL:ENSMUSG00000093999 | MGI:5456278 | snRNA gene | predicted gene, 26501 |
| 6 | gene | 95.11401 | 95.11795 | negative | MGI_C57BL6J_1919467 | Kbtbd8os | NCBI_Gene:102634268,ENSEMBL:ENSMUSG00000085413 | MGI:1919467 | antisense lncRNA gene | kelch repeat and BTB (POZ) domain containing 8, opposite strand |
| 6 | gene | 95.11724 | 95.12979 | positive | MGI_C57BL6J_2661430 | Kbtbd8 | NCBI_Gene:243574,ENSEMBL:ENSMUSG00000030031 | MGI:2661430 | protein coding gene | kelch repeat and BTB (POZ) domain containing 8 |
| 6 | pseudogene | 95.23992 | 95.24061 | negative | MGI_C57BL6J_5010007 | Gm17822 | NCBI_Gene:100328567,ENSEMBL:ENSMUSG00000107934 | MGI:5010007 | pseudogene | predicted gene, 17822 |
| 6 | pseudogene | 95.25030 | 95.25451 | positive | MGI_C57BL6J_3647796 | Gm5723 | NCBI_Gene:435912,ENSEMBL:ENSMUSG00000107900 | MGI:3647796 | pseudogene | predicted gene 5723 |
| 6 | gene | 95.25573 | 95.33399 | negative | MGI_C57BL6J_3041245 | AY512915 | NCBI_Gene:414067,ENSEMBL:ENSMUSG00000068180 | MGI:3041245 | lncRNA gene | cDNA sequence AY512915 |
| 6 | gene | 95.32211 | 95.32221 | positive | MGI_C57BL6J_3642207 | Gm10234 | ENSEMBL:ENSMUSG00000068181 | MGI:3642207 | protein coding gene | predicted gene 10234 |
| 6 | gene | 95.47301 | 95.71886 | negative | MGI_C57BL6J_1306824 | Suclg2 | NCBI_Gene:20917,ENSEMBL:ENSMUSG00000061838 | MGI:1306824 | protein coding gene | succinate-Coenzyme A ligase, GDP-forming, beta subunit |
| 6 | gene | 95.84160 | 95.89211 | positive | MGI_C57BL6J_5591168 | Gm32009 | NCBI_Gene:102634424 | MGI:5591168 | lncRNA gene | predicted gene, 32009 |
| 6 | gene | 95.90523 | 95.92297 | positive | MGI_C57BL6J_5621749 | Gm38864 | NCBI_Gene:105242722 | MGI:5621749 | lncRNA gene | predicted gene, 38864 |
| 6 | pseudogene | 95.92574 | 95.92693 | negative | MGI_C57BL6J_3644048 | Gm7838 | NCBI_Gene:665886,ENSEMBL:ENSMUSG00000107856 | MGI:3644048 | pseudogene | predicted gene 7838 |
| 6 | gene | 96.02263 | 96.11705 | negative | MGI_C57BL6J_1920261 | 2900060L22Rik | NCBI_Gene:102634502 | MGI:1920261 | unclassified non-coding RNA gene | RIKEN cDNA 2900060L22 gene |
| 6 | pseudogene | 96.03300 | 96.03337 | negative | MGI_C57BL6J_3779532 | Gm5881 | NCBI_Gene:545867,ENSEMBL:ENSMUSG00000107747 | MGI:3779532 | pseudogene | predicted gene 5881 |
| 6 | gene | 96.03543 | 96.03659 | negative | MGI_C57BL6J_1925169 | A330102K18Rik | NA | NA | unclassified gene | RIKEN cDNA A330102K18 gene |
| 6 | gene | 96.11315 | 96.65721 | positive | MGI_C57BL6J_2443695 | Tafa1 | NCBI_Gene:320265,ENSEMBL:ENSMUSG00000059187 | MGI:2443695 | protein coding gene | TAFA chemokine like family member 1 |
| 6 | gene | 96.16450 | 96.16624 | negative | MGI_C57BL6J_1925732 | 1700123L14Rik | NCBI_Gene:78482,ENSEMBL:ENSMUSG00000072878 | MGI:1925732 | protein coding gene | RIKEN cDNA 1700123L14 gene |
| 6 | gene | 96.46944 | 96.46953 | negative | MGI_C57BL6J_5452748 | Gm22971 | ENSEMBL:ENSMUSG00000065239 | MGI:5452748 | snRNA gene | predicted gene, 22971 |
| 6 | gene | 96.56076 | 96.56086 | negative | MGI_C57BL6J_5455788 | Gm26011 | ENSEMBL:ENSMUSG00000077598 | MGI:5455788 | snRNA gene | predicted gene, 26011 |
| 6 | gene | 96.83120 | 97.06041 | negative | MGI_C57BL6J_2444563 | Tafa4 | NCBI_Gene:320701,ENSEMBL:ENSMUSG00000046500 | MGI:2444563 | protein coding gene | TAFA chemokine like family member 4 |
| 6 | gene | 97.11002 | 97.14970 | negative | MGI_C57BL6J_2141669 | Eogt | NCBI_Gene:101351,ENSEMBL:ENSMUSG00000035245 | MGI:2141669 | protein coding gene | EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| 6 | gene | 97.14819 | 97.15172 | positive | MGI_C57BL6J_5690457 | Gm44065 | ENSEMBL:ENSMUSG00000107502 | MGI:5690457 | unclassified gene | predicted gene, 44065 |
| 6 | gene | 97.15195 | 97.17912 | negative | MGI_C57BL6J_2684999 | Tmf1 | NCBI_Gene:232286,ENSEMBL:ENSMUSG00000030059 | MGI:2684999 | protein coding gene | TATA element modulatory factor 1 |
| 6 | gene | 97.16282 | 97.16295 | negative | MGI_C57BL6J_5455952 | Gm26175 | ENSEMBL:ENSMUSG00000065728 | MGI:5455952 | snoRNA gene | predicted gene, 26175 |
| 6 | gene | 97.18363 | 97.20565 | negative | MGI_C57BL6J_1341217 | Uba3 | NCBI_Gene:22200,ENSEMBL:ENSMUSG00000030061 | MGI:1341217 | protein coding gene | ubiquitin-like modifier activating enzyme 3 |
| 6 | gene | 97.21069 | 97.23332 | positive | MGI_C57BL6J_1929501 | Arl6ip5 | NCBI_Gene:65106,ENSEMBL:ENSMUSG00000035199 | MGI:1929501 | protein coding gene | ADP-ribosylation factor-like 6 interacting protein 5 |
| 6 | gene | 97.23853 | 97.25310 | negative | MGI_C57BL6J_2444169 | Lmod3 | NCBI_Gene:320502,ENSEMBL:ENSMUSG00000044086 | MGI:2444169 | protein coding gene | leiomodin 3 (fetal) |
| 6 | gene | 97.25305 | 97.27045 | positive | MGI_C57BL6J_5591350 | Gm32191 | NCBI_Gene:102634657 | MGI:5591350 | lncRNA gene | predicted gene, 32191 |
| 6 | gene | 97.28687 | 97.61766 | negative | MGI_C57BL6J_2141794 | Frmd4b | NCBI_Gene:232288,ENSEMBL:ENSMUSG00000030064 | MGI:2141794 | protein coding gene | FERM domain containing 4B |
| 6 | gene | 97.39691 | 97.40069 | positive | MGI_C57BL6J_5591539 | Gm32380 | NCBI_Gene:102634907 | MGI:5591539 | lncRNA gene | predicted gene, 32380 |
| 6 | pseudogene | 97.44958 | 97.45149 | positive | MGI_C57BL6J_5011838 | Gm19653 | NCBI_Gene:100503356,ENSEMBL:ENSMUSG00000108185 | MGI:5011838 | pseudogene | predicted gene, 19653 |
| 6 | gene | 97.51457 | 97.53817 | positive | MGI_C57BL6J_5826607 | Gm46970 | NCBI_Gene:108169159 | MGI:5826607 | lncRNA gene | predicted gene, 46970 |
| 6 | gene | 97.54744 | 97.54757 | negative | MGI_C57BL6J_5455629 | Gm25852 | ENSEMBL:ENSMUSG00000064724 | MGI:5455629 | snoRNA gene | predicted gene, 25852 |
| 6 | gene | 97.56610 | 97.57746 | positive | MGI_C57BL6J_5826606 | Gm46969 | NCBI_Gene:108169158 | MGI:5826606 | lncRNA gene | predicted gene, 46969 |
| 6 | gene | 97.57208 | 97.57224 | negative | MGI_C57BL6J_5453287 | Gm23510 | ENSEMBL:ENSMUSG00000065017 | MGI:5453287 | snRNA gene | predicted gene, 23510 |
| 6 | gene | 97.57796 | 97.65640 | positive | MGI_C57BL6J_5591406 | Gm32247 | NCBI_Gene:102634735 | MGI:5591406 | lncRNA gene | predicted gene, 32247 |
| 6 | gene | 97.57943 | 97.58109 | negative | MGI_C57BL6J_5690494 | Gm44102 | ENSEMBL:ENSMUSG00000108093 | MGI:5690494 | unclassified gene | predicted gene, 44102 |
| 6 | gene | 97.61968 | 97.62042 | positive | MGI_C57BL6J_1925782 | C030006F08Rik | NA | NA | unclassified gene | RIKEN cDNA C030006F08 gene |
| 6 | gene | 97.69392 | 97.71040 | positive | MGI_C57BL6J_5591640 | Gm32481 | NCBI_Gene:102635045 | MGI:5591640 | lncRNA gene | predicted gene, 32481 |
| 6 | gene | 97.73044 | 97.73055 | positive | MGI_C57BL6J_5454342 | Gm24565 | ENSEMBL:ENSMUSG00000094510 | MGI:5454342 | snRNA gene | predicted gene, 24565 |
| 6 | gene | 97.79051 | 97.79065 | positive | MGI_C57BL6J_5453480 | Gm23703 | ENSEMBL:ENSMUSG00000089464 | MGI:5453480 | snoRNA gene | predicted gene, 23703 |
| 6 | gene | 97.80700 | 98.02136 | positive | MGI_C57BL6J_104554 | Mitf | NCBI_Gene:17342,ENSEMBL:ENSMUSG00000035158 | MGI:104554 | protein coding gene | melanogenesis associated transcription factor |
| 6 | pseudogene | 97.89023 | 97.89057 | negative | MGI_C57BL6J_3782979 | Gm15531 | NCBI_Gene:100043692,ENSEMBL:ENSMUSG00000083086 | MGI:3782979 | pseudogene | predicted gene 15531 |
| 6 | gene | 98.12180 | 98.25131 | positive | MGI_C57BL6J_5591751 | Gm32592 | NCBI_Gene:102635190,ENSEMBL:ENSMUSG00000107902 | MGI:5591751 | lncRNA gene | predicted gene, 32592 |
| 6 | gene | 98.23571 | 98.34275 | negative | MGI_C57BL6J_2685611 | Gm765 | NCBI_Gene:330390,ENSEMBL:ENSMUSG00000090667 | MGI:2685611 | protein coding gene | predicted gene 765 |
| 6 | gene | 98.25829 | 98.26637 | positive | MGI_C57BL6J_5591694 | Gm32535 | NCBI_Gene:102635115 | MGI:5591694 | lncRNA gene | predicted gene, 32535 |
| 6 | pseudogene | 98.27554 | 98.27611 | positive | MGI_C57BL6J_3645233 | Gm7892 | NCBI_Gene:666019 | MGI:3645233 | pseudogene | predicted gene 7892 |
| 6 | gene | 98.40274 | 98.42310 | positive | MGI_C57BL6J_5591805 | Gm32646 | NCBI_Gene:102635260 | MGI:5591805 | lncRNA gene | predicted gene, 32646 |
| 6 | gene | 98.50772 | 98.51039 | positive | MGI_C57BL6J_5591864 | Gm32705 | NCBI_Gene:102635340 | MGI:5591864 | lncRNA gene | predicted gene, 32705 |
| 6 | gene | 98.50773 | 98.69767 | negative | MGI_C57BL6J_5690588 | Gm44196 | ENSEMBL:ENSMUSG00000107614 | MGI:5690588 | lncRNA gene | predicted gene, 44196 |
| 6 | gene | 98.51764 | 98.52195 | positive | MGI_C57BL6J_5591942 | Gm32783 | NCBI_Gene:102635448 | MGI:5591942 | lncRNA gene | predicted gene, 32783 |
| 6 | gene | 98.53138 | 98.55218 | negative | MGI_C57BL6J_1925309 | 4930595L18Rik | NCBI_Gene:78059,ENSEMBL:ENSMUSG00000107929 | MGI:1925309 | lncRNA gene | RIKEN cDNA 4930595L18 gene |
| 6 | pseudogene | 98.75532 | 98.75803 | positive | MGI_C57BL6J_5591467 | Gm32308 | NCBI_Gene:102634810,ENSEMBL:ENSMUSG00000107938 | MGI:5591467 | pseudogene | predicted gene, 32308 |
| 6 | pseudogene | 98.86034 | 98.86082 | negative | MGI_C57BL6J_5591540 | Gm32381 | ENSEMBL:ENSMUSG00000107514 | MGI:5591540 | pseudogene | predicted gene, 32381 |
| 6 | gene | 98.92534 | 99.52282 | negative | MGI_C57BL6J_1914004 | Foxp1 | NCBI_Gene:108655,ENSEMBL:ENSMUSG00000030067 | MGI:1914004 | protein coding gene | forkhead box P1 |
| 6 | gene | 98.96770 | 98.97193 | negative | MGI_C57BL6J_1921805 | 9130401L11Rik | NA | NA | unclassified gene | RIKEN cDNA 9130401L11 gene |
| 6 | gene | 98.99764 | 98.99861 | negative | MGI_C57BL6J_1924976 | 6030492E11Rik | NA | NA | unclassified gene | RIKEN cDNA 6030492E11 gene |
| 6 | gene | 99.07591 | 99.07606 | negative | MGI_C57BL6J_6121547 | Gm49348 | ENSEMBL:ENSMUSG00000105902 | MGI:6121547 | miRNA gene | predicted gene, 49348 |
| 6 | gene | 99.09767 | 99.10832 | negative | MGI_C57BL6J_5011850 | Gm19665 | NCBI_Gene:100503384 | MGI:5011850 | unclassified gene | predicted gene, 19665 |
| 6 | gene | 99.11834 | 99.12753 | negative | MGI_C57BL6J_5621750 | Gm38865 | NCBI_Gene:105242725 | MGI:5621750 | lncRNA gene | predicted gene, 38865 |
| 6 | gene | 99.25748 | 99.66680 | negative | MGI_C57BL6J_5313143 | Gm20696 | ENSEMBL:ENSMUSG00000030068 | MGI:5313143 | protein coding gene | predicted gene 20696 |
| 6 | gene | 99.31849 | 99.32341 | positive | MGI_C57BL6J_5592041 | Gm32882 | NCBI_Gene:102635590 | MGI:5592041 | lncRNA gene | predicted gene, 32882 |
| 6 | gene | 99.38461 | 99.38850 | negative | MGI_C57BL6J_5313152 | Gm20705 | ENSEMBL:ENSMUSG00000093559 | MGI:5313152 | lncRNA gene | predicted gene 20705 |
| 6 | gene | 99.48459 | 99.48470 | negative | MGI_C57BL6J_5452105 | Gm22328 | ENSEMBL:ENSMUSG00000088699 | MGI:5452105 | miRNA gene | predicted gene, 22328 |
| 6 | gene | 99.52245 | 99.52417 | negative | MGI_C57BL6J_3045275 | B930049P21Rik | NA | NA | unclassified gene | RIKEN cDNA B930049P21 gene |
| 6 | gene | 99.52775 | 99.53367 | negative | MGI_C57BL6J_5826628 | Gm46991 | NCBI_Gene:108169204 | MGI:5826628 | lncRNA gene | predicted gene, 46991 |
| 6 | gene | 99.57589 | 99.57614 | positive | MGI_C57BL6J_5454713 | Gm24936 | ENSEMBL:ENSMUSG00000084547 | MGI:5454713 | unclassified non-coding RNA gene | predicted gene, 24936 |
| 6 | gene | 99.62514 | 99.66677 | negative | MGI_C57BL6J_1914142 | Eif4e3 | NCBI_Gene:66892,ENSEMBL:ENSMUSG00000093661 | MGI:1914142 | protein coding gene | eukaryotic translation initiation factor 4E member 3 |
| 6 | gene | 99.65844 | 99.66222 | negative | MGI_C57BL6J_5690496 | Gm44104 | ENSEMBL:ENSMUSG00000107583 | MGI:5690496 | unclassified gene | predicted gene, 44104 |
| 6 | gene | 99.69268 | 99.69507 | positive | MGI_C57BL6J_1202299 | Gpr27 | NCBI_Gene:14761,ENSEMBL:ENSMUSG00000072875 | MGI:1202299 | protein coding gene | G protein-coupled receptor 27 |
| 6 | gene | 99.71130 | 99.72639 | negative | MGI_C57BL6J_1354178 | Prok2 | NCBI_Gene:50501,ENSEMBL:ENSMUSG00000030069 | MGI:1354178 | protein coding gene | prokineticin 2 |
| 6 | gene | 99.72577 | 99.73261 | positive | MGI_C57BL6J_5477242 | Gm26748 | NCBI_Gene:102635772,ENSEMBL:ENSMUSG00000097548 | MGI:5477242 | lncRNA gene | predicted gene, 26748 |
| 6 | pseudogene | 99.76252 | 99.77127 | positive | MGI_C57BL6J_3646639 | Gm5314 | NCBI_Gene:384466,ENSEMBL:ENSMUSG00000108082 | MGI:3646639 | pseudogene | predicted gene 5314 |
| 6 | pseudogene | 99.83966 | 99.84035 | negative | MGI_C57BL6J_3643996 | Gm6681 | NCBI_Gene:626503,ENSEMBL:ENSMUSG00000107713 | MGI:3643996 | pseudogene | predicted gene 6681 |
| 6 | gene | 99.86507 | 99.86677 | positive | MGI_C57BL6J_5592302 | Gm33143 | NCBI_Gene:102635932 | MGI:5592302 | lncRNA gene | predicted gene, 33143 |
| 6 | pseudogene | 99.87761 | 99.87843 | negative | MGI_C57BL6J_2664997 | Tpt1-ps3 | NCBI_Gene:100043703,ENSEMBL:ENSMUSG00000084319 | MGI:2664997 | pseudogene | tumor protein, translationally-controlled, pseudogene 3 |
| 6 | gene | 99.88440 | 99.89684 | negative | MGI_C57BL6J_5663204 | Gm43067 | ENSEMBL:ENSMUSG00000107204 | MGI:5663204 | lncRNA gene | predicted gene 43067 |
| 6 | gene | 99.89323 | 99.89355 | negative | MGI_C57BL6J_5454025 | Gm24248 | ENSEMBL:ENSMUSG00000087946 | MGI:5454025 | unclassified non-coding RNA gene | predicted gene, 24248 |
| 6 | gene | 99.92825 | 99.95093 | negative | MGI_C57BL6J_5690834 | Gm44442 | ENSEMBL:ENSMUSG00000107528 | MGI:5690834 | lncRNA gene | predicted gene, 44442 |
| 6 | gene | 99.96652 | 99.99121 | negative | MGI_C57BL6J_5592360 | Gm33201 | NCBI_Gene:102636006,ENSEMBL:ENSMUSG00000108151 | MGI:5592360 | lncRNA gene | predicted gene, 33201 |
| 6 | gene | 100.06190 | 100.08724 | positive | MGI_C57BL6J_5592402 | Gm33243 | NCBI_Gene:102636070 | MGI:5592402 | lncRNA gene | predicted gene, 33243 |
| 6 | gene | 100.09503 | 100.09961 | positive | MGI_C57BL6J_5621751 | Gm38866 | NCBI_Gene:105242726 | MGI:5621751 | lncRNA gene | predicted gene, 38866 |
| 6 | gene | 100.09962 | 100.10141 | positive | MGI_C57BL6J_5592456 | Gm33297 | NCBI_Gene:102636145 | MGI:5592456 | lncRNA gene | predicted gene, 33297 |
| 6 | gene | 100.14333 | 100.14708 | negative | MGI_C57BL6J_1917515 | 2010109P13Rik | NCBI_Gene:108169160,ENSEMBL:ENSMUSG00000107558 | MGI:1917515 | lncRNA gene | RIKEN cDNA 2010109P13 gene |
| 6 | gene | 100.15513 | 100.18173 | negative | MGI_C57BL6J_5592607 | Gm33448 | NCBI_Gene:102636361 | MGI:5592607 | lncRNA gene | predicted gene, 33448 |
| 6 | gene | 100.19600 | 100.20123 | positive | MGI_C57BL6J_5592713 | Gm33554 | NCBI_Gene:102636508 | MGI:5592713 | lncRNA gene | predicted gene, 33554 |
| 6 | gene | 100.20533 | 100.20914 | positive | MGI_C57BL6J_5621752 | Gm38867 | NCBI_Gene:105242727 | MGI:5621752 | lncRNA gene | predicted gene, 38867 |
| 6 | gene | 100.22857 | 100.28749 | negative | MGI_C57BL6J_1929059 | Rybp | NCBI_Gene:56353,ENSEMBL:ENSMUSG00000072872 | MGI:1929059 | protein coding gene | RING1 and YY1 binding protein |
| 6 | pseudogene | 100.24723 | 100.25034 | positive | MGI_C57BL6J_5010607 | Gm18422 | NCBI_Gene:100417147,ENSEMBL:ENSMUSG00000108269 | MGI:5010607 | pseudogene | predicted gene, 18422 |
| 6 | gene | 100.25671 | 100.25923 | negative | MGI_C57BL6J_5690437 | Gm44045 | ENSEMBL:ENSMUSG00000107461 | MGI:5690437 | unclassified gene | predicted gene, 44045 |
| 6 | gene | 100.27261 | 100.27370 | negative | MGI_C57BL6J_5690436 | Gm44044 | ENSEMBL:ENSMUSG00000107690 | MGI:5690436 | unclassified gene | predicted gene, 44044 |
| 6 | gene | 100.46924 | 100.47124 | positive | MGI_C57BL6J_5621753 | Gm38868 | NCBI_Gene:105242728 | MGI:5621753 | lncRNA gene | predicted gene, 38868 |
| 6 | gene | 100.49468 | 100.49481 | negative | MGI_C57BL6J_5453011 | Gm23234 | ENSEMBL:ENSMUSG00000087929 | MGI:5453011 | snoRNA gene | predicted gene, 23234 |
| 6 | gene | 100.49851 | 100.49991 | positive | MGI_C57BL6J_5690499 | Gm44107 | ENSEMBL:ENSMUSG00000107663 | MGI:5690499 | lncRNA gene | predicted gene, 44107 |
| 6 | gene | 100.50419 | 100.51003 | negative | MGI_C57BL6J_5592900 | Gm33741 | NCBI_Gene:102636759 | MGI:5592900 | lncRNA gene | predicted gene, 33741 |
| 6 | gene | 100.52740 | 100.53343 | positive | MGI_C57BL6J_1920645 | 1700049E22Rik | NCBI_Gene:73395,ENSEMBL:ENSMUSG00000101553 | MGI:1920645 | lncRNA gene | RIKEN cDNA 1700049E22 gene |
| 6 | gene | 100.56826 | 100.67118 | negative | MGI_C57BL6J_1919421 | Shq1 | NCBI_Gene:72171,ENSEMBL:ENSMUSG00000035378 | MGI:1919421 | protein coding gene | SHQ1 homolog (S. cerevisiae) |
| 6 | gene | 100.56826 | 100.56973 | negative | MGI_C57BL6J_1917782 | 5730433K22Rik | NA | NA | unclassified gene | RIKEN cDNA 5730433K22 gene |
| 6 | pseudogene | 100.60456 | 100.60641 | positive | MGI_C57BL6J_3644755 | Gm6565 | NCBI_Gene:100503412,ENSEMBL:ENSMUSG00000079224 | MGI:3644755 | pseudogene | predicted gene 6565 |
| 6 | pseudogene | 100.62652 | 100.62694 | negative | MGI_C57BL6J_5690500 | Gm44108 | ENSEMBL:ENSMUSG00000108019 | MGI:5690500 | pseudogene | predicted gene, 44108 |
| 6 | gene | 100.66094 | 100.66246 | negative | MGI_C57BL6J_5690508 | Gm44116 | ENSEMBL:ENSMUSG00000108122 | MGI:5690508 | unclassified gene | predicted gene, 44116 |
| 6 | gene | 100.70429 | 100.81091 | positive | MGI_C57BL6J_2682940 | Gxylt2 | NCBI_Gene:232313,ENSEMBL:ENSMUSG00000030074 | MGI:2682940 | protein coding gene | glucoside xylosyltransferase 2 |
| 6 | pseudogene | 100.72617 | 100.72701 | positive | MGI_C57BL6J_3783024 | Gm15576 | NCBI_Gene:100416942,ENSEMBL:ENSMUSG00000084157 | MGI:3783024 | pseudogene | predicted gene 15576 |
| 6 | gene | 100.79183 | 100.79391 | positive | MGI_C57BL6J_5011877 | Gm19692 | ENSEMBL:ENSMUSG00000107989 | MGI:5011877 | unclassified gene | predicted gene, 19692 |
Additive covariates
Significance thresholds
operm <- scan1perm(pr.qc, covars["age.of.onset"], model="normal", addcovar=addcovar, n_perm=10, perm_Xsp=TRUE, chr_lengths=chr_lengths(gm$gmap))
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
names(summary_table) <- c("autosomes","X")
summary_table$significance.level <- rownames(summary_table)
rownames(summary_table) <- NULL
summary_table[c(3,1:2)] %>%
kable(escape = F,align = c("ccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| significance.level | autosomes | X |
|---|---|---|
| 0.01 | 2.885451 | 3.379466 |
| 0.05 | 2.881192 | 3.124444 |
| 0.1 | 2.875854 | 2.790959 |
Genome-wide scan
plot_lod<-function(out,map){
for (i in 1:dim(out)[2]){
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - ICI vs. EOI (ith additive covariates) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
print("with no kinship")[1] “with no kinship”
outfa <- scan1(pr.qc, covars["age.of.onset"], Xcovar=Xcovar, model="normal", addcovar=addcovar)
plot_lod(outfa,gm$gmap)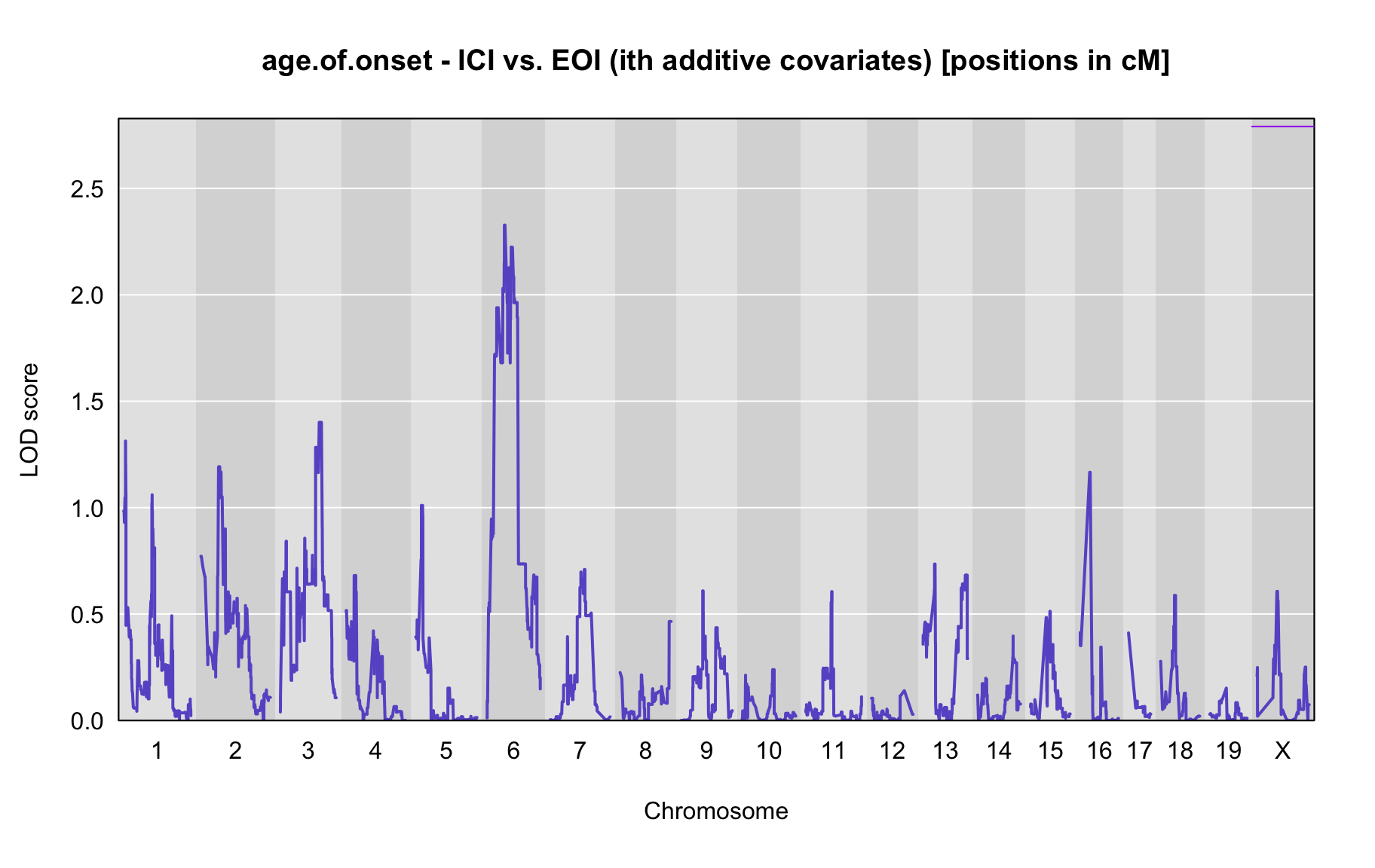
print("with normal kinship")[1] “with normal kinship”
outka <- scan1(pr.qc, covars["age.of.onset"], Xcovar=Xcovar, model="normal", addcovar=addcovar, kinship=kinship)
plot_lod(outka,gm$gmap)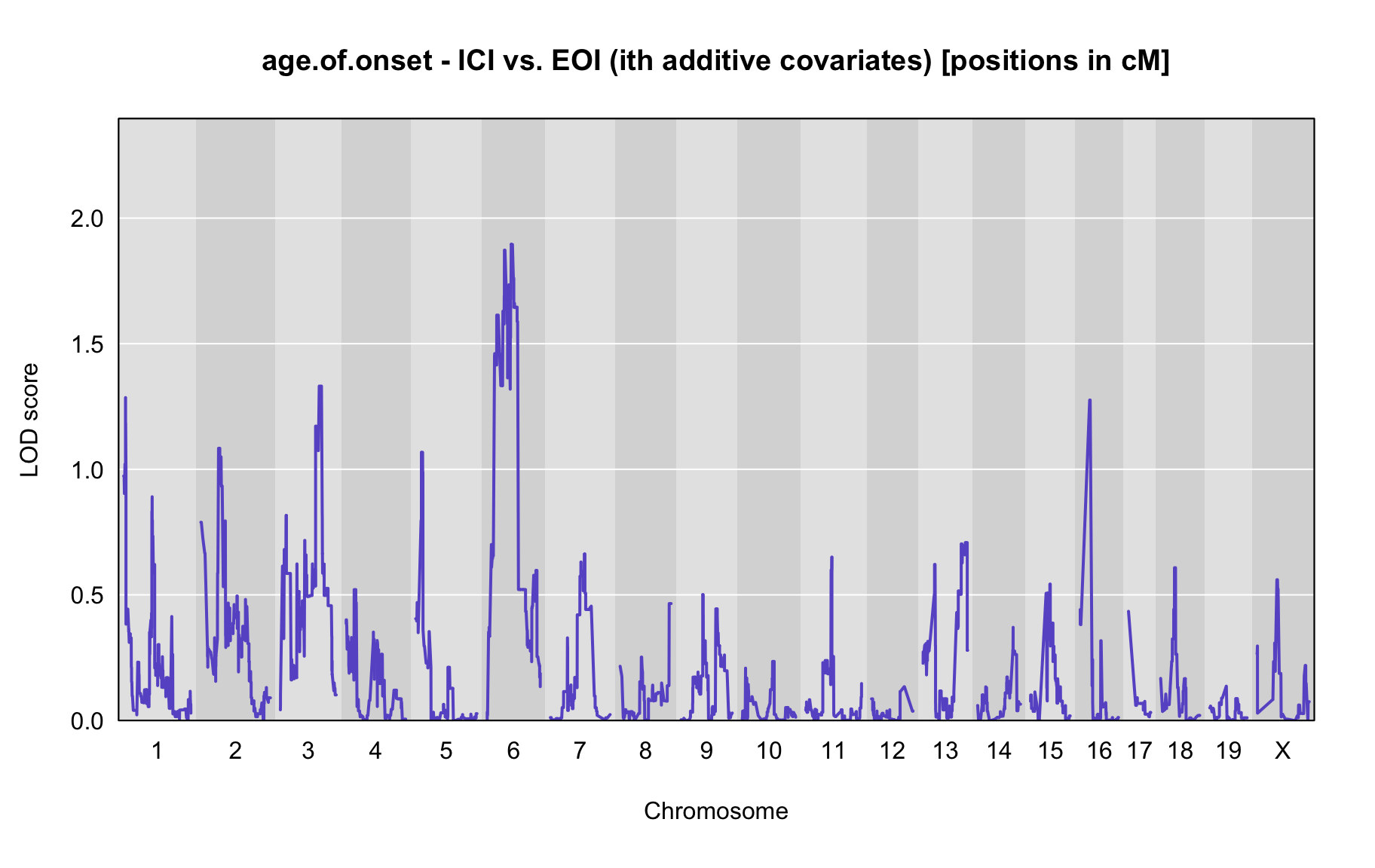
print("with loco kinship")[1] “with loco kinship”
outa <- scan1(pr.qc, covars["age.of.onset"], Xcovar=Xcovar, model="normal", addcovar=addcovar, kinship=K)
plot_lod(outa,gm$gmap)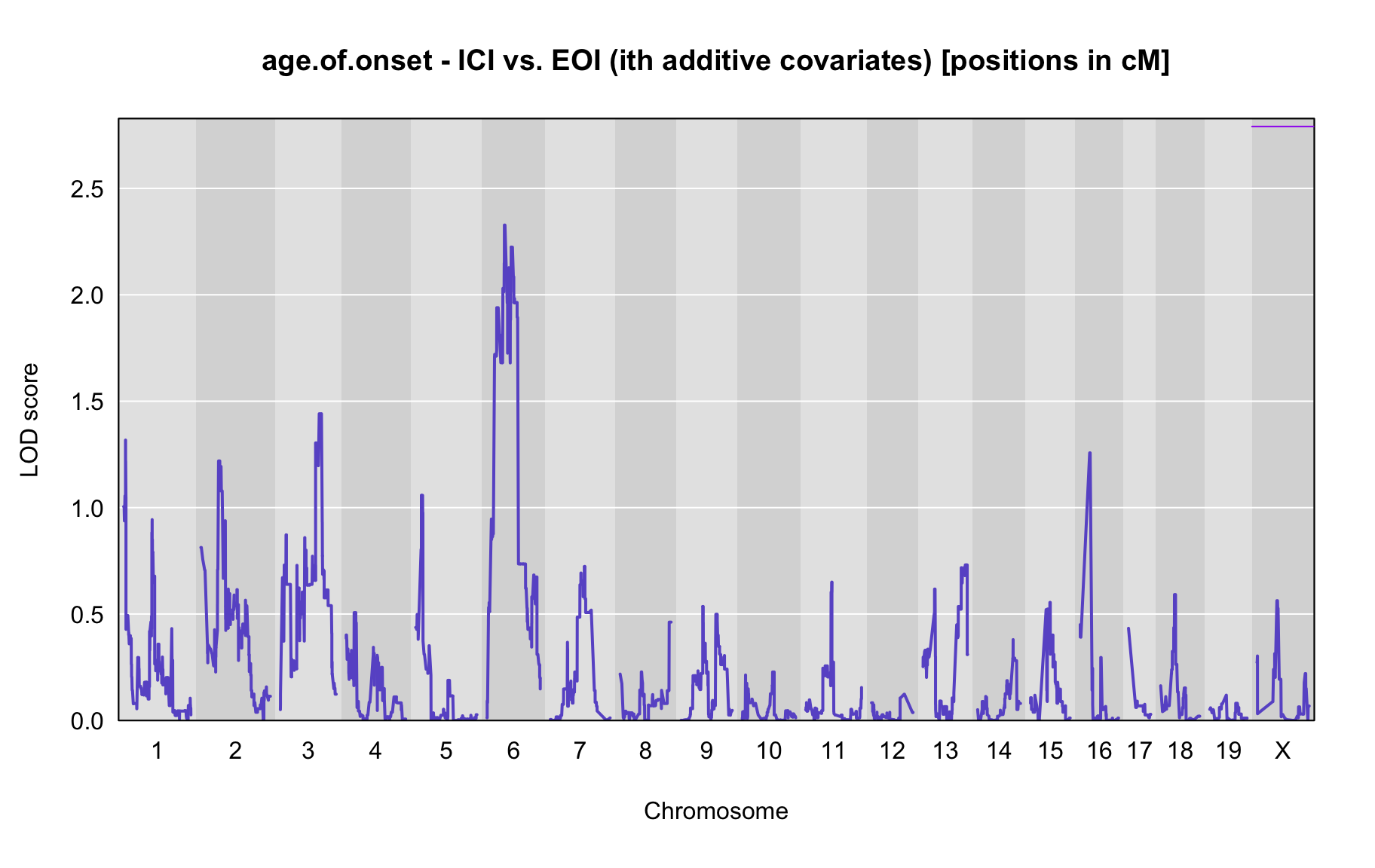
out = outa
print(paste0("number of samples in analysis = ", as.data.frame(attributes(out)$sample_size)[1,]))[1] “number of samples in analysis = 101”
LOD peaks
Centimorgan (cM)
peaks<-find_peaks(out, gm$gmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) > 0){
peaks$marker <- find_marker(gm$gmap, chr=peaks$chr,pos=peaks$pos)
names(peaks)[2] <- c("phenotype")
peaks <- peaks[-1]
rownames(peaks) <- NULL
print(kable(peaks, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
#plot only peak chromosomes
plot_lod_chr<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " - ICI vs. EOI (with additive covariates) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are no peaks that have a LOD that reaches suggestive (p<0.05) level of 2.88119166563917 [autosomes]/3.12444410138338 [x-chromosome]”
if(nrow(peaks) < 50 & nrow(peaks) > 0){
for(i in unique(peaks$chr)){
#for (i in 1:nrow(peaks)){
#plot_lod_chr(out,gm$gmap, peaks$chr[i])
plot_lod_chr(out,gm$gmap, i)
}
} else {
print(paste0("There are too many peaks (",nrow(peaks)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are too many peaks (0 peaks) that have a LOD that reaches suggestive (p<0.05) level of 2.88119166563917 [autosomes]/3.12444410138338 [x-chromosome]”
Megabase (MB)
peaks_mba<-find_peaks(out, gm$pmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) > 0){
peaks_mba$marker <- find_marker(gm$pmap, chr=peaks_mba$chr,pos=peaks_mba$pos)
names(peaks_mba)[2] <- c("phenotype")
peaks_mba <- peaks_mba[-1]
rownames(peaks_mba) <- NULL
print(kable(peaks_mba, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
plot_lod_chr_mb<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " - ICI vs. EOI (with additive covariates) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are no peaks that have a LOD that reaches suggestive (p<0.05) level of 2.88119166563917 [autosomes]/3.12444410138338 [x-chromosome]”
if(nrow(peaks_mba) < 50 & nrow(peaks_mba) > 0){
for(i in unique(peaks_mba$chr)){
#for (i in 1:nrow(peaks_mba)){
#plot_lod_chr_mb(out,gm$pmap, peaks_mba$chr[i])
plot_lod_chr_mb(out,gm$pmap,i)
}
} else {
print(paste0("There are too many peaks (",nrow(peaks_mba)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are too many peaks (0 peaks) that have a LOD that reaches suggestive (p<0.05) level of 2.88119166563917 [autosomes]/3.12444410138338 [x-chromosome]”
QTL effects
For each peak LOD location we give a list of gene
if(nrow(peaks) < 50 & nrow(peaks) > 0){
for (i in 1:nrow(peaks)){
#for (i in 1:1){
#Plot 1
g <- maxmarg(pr.qc, gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i], return_char=TRUE)
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/","qtl_effect_", i, ".png"))
#par(mar=c(4.1, 4.1, 1.5, 0.6))
plot_pxg(g, covars[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
##dev.off()
chr = peaks$chr[i]
# Plot 2
pr_sub <- pull_genoprobint(pr.qc, gm$gmap, chr, c(peaks$ci_lo[i], peaks$ci_hi[i]))
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar)
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], Xcovar=Xcovar)
#coeff <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
#coeff_sub <- scan1coef(pr_sub[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
blup <- scan1blup(pr.qc[,chr], covars[peaks$phenotype[i]])
blup_sub <- scan1blup(pr_sub[,chr], covars[peaks$phenotype[i]])
write.csv(as.data.frame(blup_sub), paste0("data/ici.vs.eoi_",peaks$phenotype[i],"-additive.covariates_blup_sub_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_snpsqc_5.batches.csv"), quote=F)
#plot_coef(coeff,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i]," [scan1coeff; positions in cM] )")
# )
#plot_coef(coeff_sub,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i],"; 1.5 LOD drop interval [scan1coeff; positions in cM] ) ")
# )
plot_coef(blup,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," [scan1blup; positions in cM] )")
)
plot_coef(blup_sub,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i],"; 1.5 LOD drop interval [scan1blup; positions in cM] )")
)
# Plot 3
#c2effB <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary", contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
#c2effBb <- scan1blup(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]],Xcovar=Xcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
#plot(c2effB, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
#plot(c2effBb, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
##last_coef <- unclass(c2effB)[nrow(c2effB),2:3] # last two coefficients
##for(t in seq(along=last_coef))
## axis(side=4, at=last_coef[t], names(last_coef)[t], tick=FALSE)
#Table 1
chr = peaks_mba$chr[i]
start=as.numeric(peaks_mba$ci_lo[i])
end=as.numeric(peaks_mba$ci_hi[i])
genesgss = query_genes(chr, start, end)
write.csv(genesgss, file=paste0("data/ici.vs.eoi_",peaks$phenotype[i],"-additive.covariates_genes_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_snpsqc_5.batches.csv"), quote=F)
rownames(genesgss) <- NULL
genesgss$strand_old = genesgss$strand
genesgss$strand[genesgss$strand=="+"] <- "positive"
genesgss$strand[genesgss$strand=="-"] <- "negative"
#genesgss <-
#table <-
#genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")] %>%
#kable(escape = F,align = c("ccccccccccc")) %>%
#kable_styling("striped", full_width = T) #%>%
#cat #%>%
#column_spec(1, bold=TRUE)
#
#print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], escape = F,align = c("ccccccccccc")))
print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], "html") %>% kable_styling("striped", full_width = T))
#table
}
} else {
print(paste0("There are too many peaks (",nrow(peaks_mba)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are too many peaks (0 peaks) that have a LOD that reaches suggestive (p<0.05) level of 2.88119166563917 [autosomes]/3.12444410138338 [x-chromosome]”
Interactive term
Significance thresholds
operm <- scan1perm(pr.qc, covars["age.of.onset"], model="normal", addcovar=addcovar, intcovar=addcovar, n_perm=10, perm_Xsp=TRUE, chr_lengths=chr_lengths(gm$gmap))
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
names(summary_table) <- c("autosomes","X")
summary_table$significance.level <- rownames(summary_table)
rownames(summary_table) <- NULL
summary_table[c(3,1:2)] %>%
kable(escape = F,align = c("ccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| significance.level | autosomes | X |
|---|---|---|
| 0.01 | 2.123784 | 3.089406 |
| 0.05 | 2.077429 | 2.942496 |
| 0.1 | 2.019340 | 2.750385 |
Genome-wide scan
plot_lod<-function(out,map){
for (i in 1:dim(out)[2]){
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - ICI vs. EOI (with interactive covariate) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
print("with no kinship")[1] “with no kinship”
outfi <- scan1(pr.qc, covars["age.of.onset"], Xcovar=Xcovar, model="normal", addcovar=addcovar, intcovar=addcovar)
plot_lod(outfi,gm$gmap)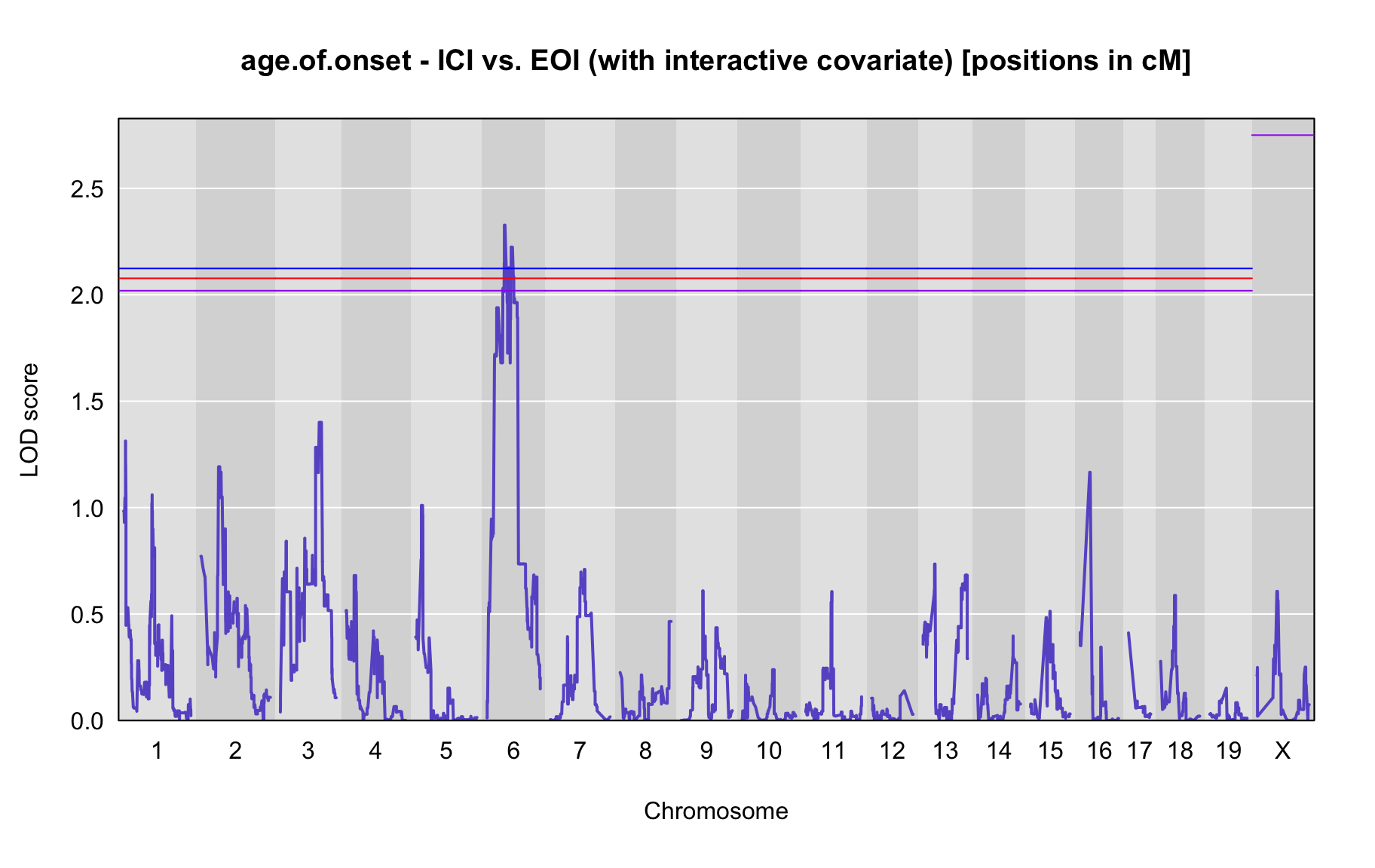
outf.new <- outfi-outfa
plot_lod(outf.new,gm$gmap)
print("with normal kinship")[1] “with normal kinship”
outki <- scan1(pr.qc, covars["age.of.onset"], Xcovar=Xcovar, model="normal", addcovar=addcovar, intcovar=addcovar, kinship=kinship)
plot_lod(outki,gm$gmap)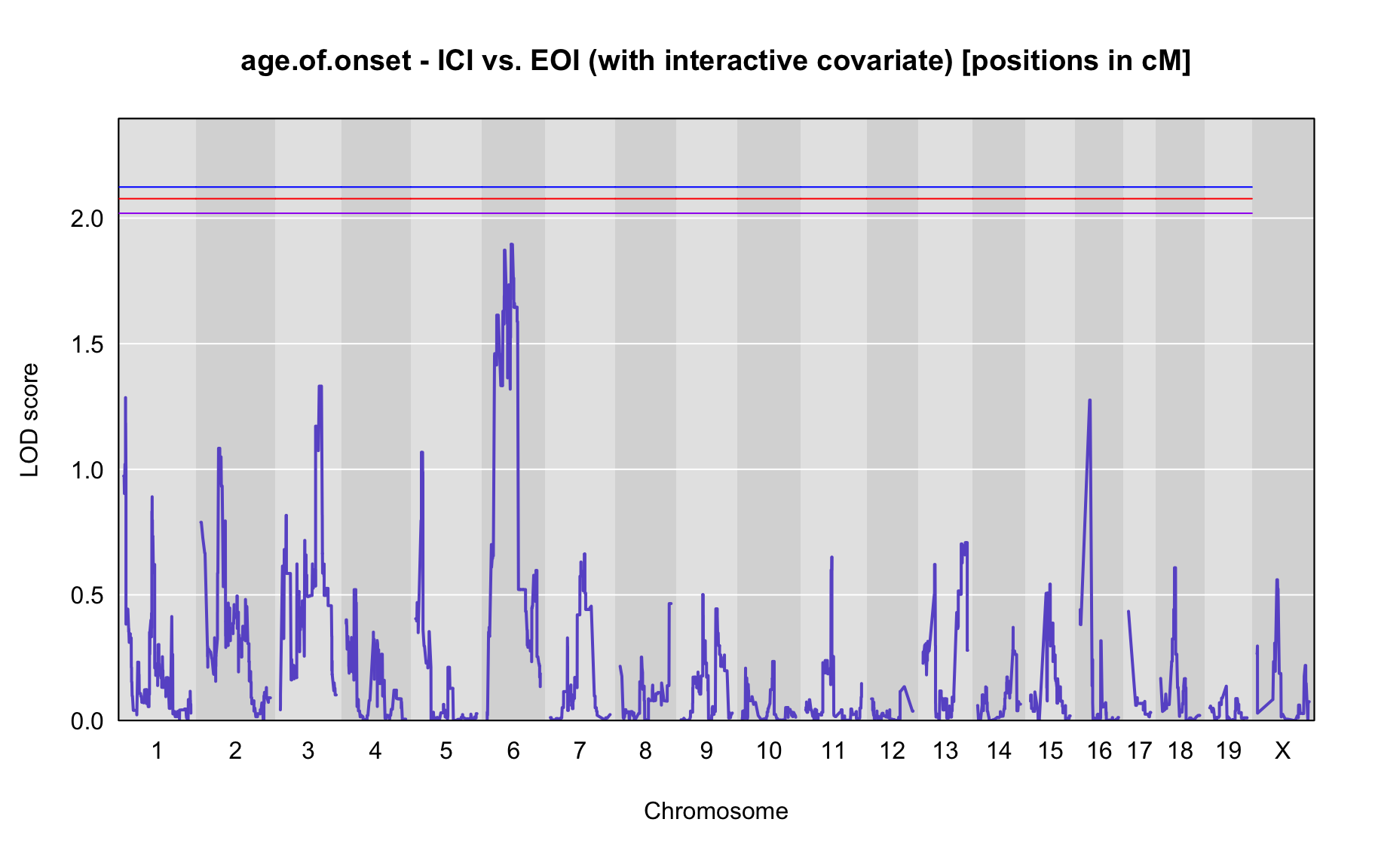
outk.new <- outki-outka
plot_lod(outk.new,gm$gmap)
print("with loco kinship")[1] “with loco kinship”
outi <- scan1(pr.qc, covars["age.of.onset"], Xcovar=Xcovar, model="normal", addcovar=addcovar, intcovar=addcovar, kinship=K)
plot_lod(outi,gm$gmap)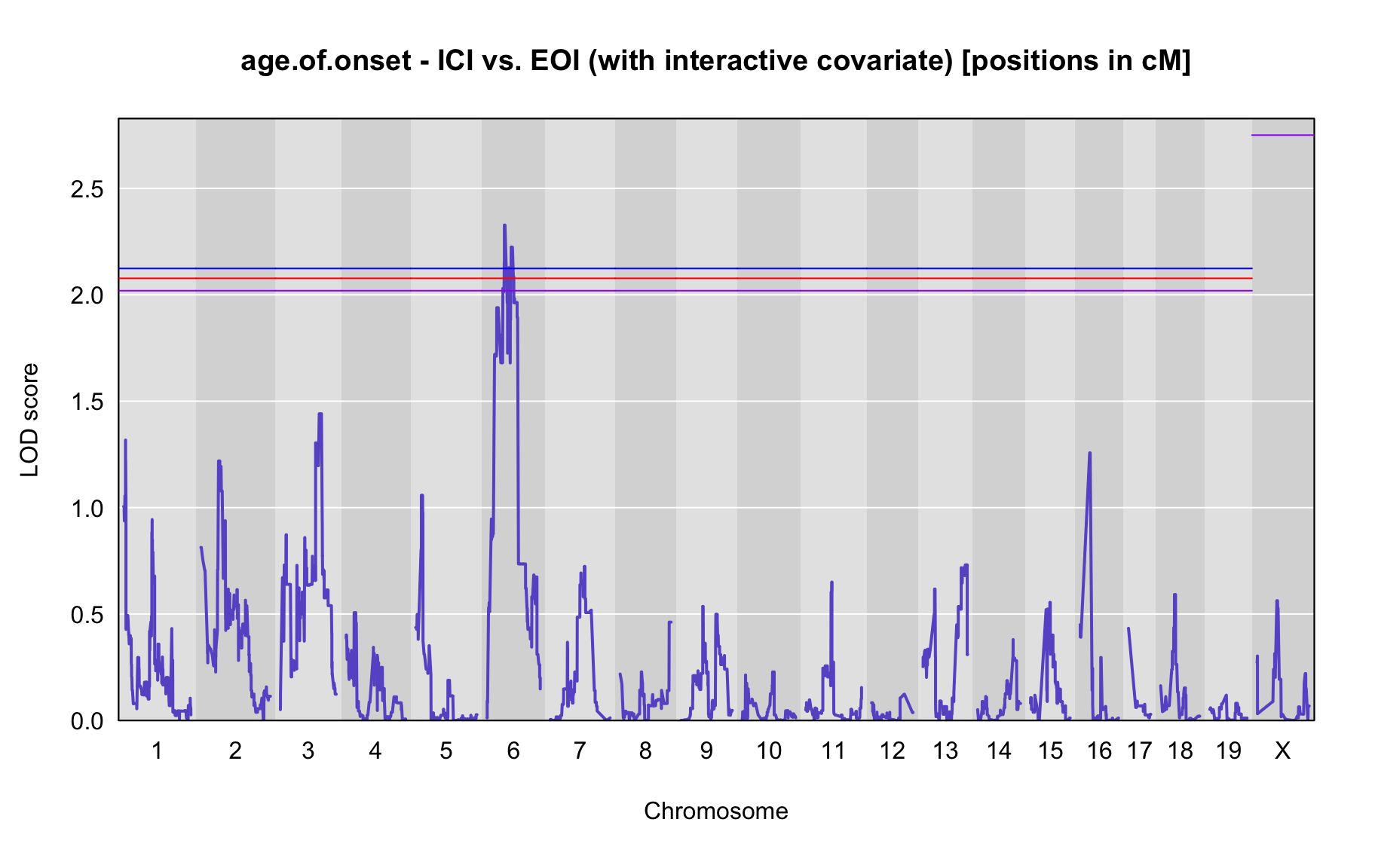
outi.new <- outi-outa
plot_lod(outi.new,gm$gmap)
out = outi.new
print(paste0("number of samples in analysis = ", as.data.frame(attributes(out)$sample_size)[1,]))[1] “number of samples in analysis = 101”
LOD peaks
Centimorgan (cM)
peaks<-find_peaks(out, gm$gmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) > 0){
peaks$marker <- find_marker(gm$gmap, chr=peaks$chr,pos=peaks$pos)
names(peaks)[2] <- c("phenotype")
peaks <- peaks[-1]
rownames(peaks) <- NULL
print(kable(peaks, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
#plot only peak chromosomes
plot_lod_chr<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " - ICI vs. EOI (with interactive covariate) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are no peaks that have a LOD that reaches suggestive (p<0.05) level of 2.07742869819588 [autosomes]/2.94249586915186 [x-chromosome]”
if(nrow(peaks) < 50 & nrow(peaks) > 0){
for(i in unique(peaks$chr)){
#for (i in 1:nrow(peaks)){
#plot_lod_chr(out,gm$gmap, peaks$chr[i])
plot_lod_chr(out,gm$gmap, i)
}
} else {
print(paste0("There are too many peaks (",nrow(peaks)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are too many peaks (0 peaks) that have a LOD that reaches suggestive (p<0.05) level of 2.07742869819588 [autosomes]/2.94249586915186 [x-chromosome]”
Megabase (MB)
peaks_mba<-find_peaks(out, gm$pmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) > 0){
peaks_mba$marker <- find_marker(gm$pmap, chr=peaks_mba$chr,pos=peaks_mba$pos)
names(peaks_mba)[2] <- c("phenotype")
peaks_mba <- peaks_mba[-1]
rownames(peaks_mba) <- NULL
print(kable(peaks_mba, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
plot_lod_chr_mb<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " - ICI vs. EOI (with interactive covariate) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are no peaks that have a LOD that reaches suggestive (p<0.05) level of 2.07742869819588 [autosomes]/2.94249586915186 [x-chromosome]”
if(nrow(peaks_mba) < 50 & nrow(peaks_mba) > 0){
for(i in unique(peaks_mba$chr)){
#for (i in 1:nrow(peaks_mba)){
#plot_lod_chr_mb(out,gm$pmap, peaks_mba$chr[i])
plot_lod_chr_mb(out,gm$pmap,i)
}
} else {
print(paste0("There are too many peaks (",nrow(peaks_mba)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are too many peaks (0 peaks) that have a LOD that reaches suggestive (p<0.05) level of 2.07742869819588 [autosomes]/2.94249586915186 [x-chromosome]”
QTL effects
For each peak LOD location we give a list of gene
if(nrow(peaks) < 50 & nrow(peaks) > 0){
for (i in 1:nrow(peaks)){
#for (i in 1:1){
#Plot 1
g <- maxmarg(pr.qc, gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i], return_char=TRUE)
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/","qtl_effect_", i, ".png"))
#par(mar=c(4.1, 4.1, 1.5, 0.6))
plot_pxg(g, covars[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
##dev.off()
chr = peaks$chr[i]
# Plot 2
pr_sub <- pull_genoprobint(pr.qc, gm$gmap, chr, c(peaks$ci_lo[i], peaks$ci_hi[i]))
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar)
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], Xcovar=Xcovar)
#coeff <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
#coeff_sub <- scan1coef(pr_sub[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
blup <- scan1blup(pr.qc[,chr], covars[peaks$phenotype[i]])
blup_sub <- scan1blup(pr_sub[,chr], covars[peaks$phenotype[i]])
write.csv(as.data.frame(blup_sub), paste0("data/ici.vs.eoi_",peaks$phenotype[i],"-interactive.covariate_blup_sub_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_snpsqc_5.batches.csv"), quote=F)
#plot_coef(coeff,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i]," [scan1coeff; positions in cM] )")
# )
#plot_coef(coeff_sub,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i],"; 1.5 LOD drop interval [scan1coeff; positions in cM] ) ")
# )
plot_coef(blup,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," [scan1blup; positions in cM] )")
)
plot_coef(blup_sub,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i],"; 1.5 LOD drop interval [scan1blup; positions in cM] )")
)
# Plot 3
#c2effB <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary", contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
#c2effBb <- scan1blup(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]],Xcovar=Xcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
#plot(c2effB, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
#plot(c2effBb, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
##last_coef <- unclass(c2effB)[nrow(c2effB),2:3] # last two coefficients
##for(t in seq(along=last_coef))
## axis(side=4, at=last_coef[t], names(last_coef)[t], tick=FALSE)
#Table 1
chr = peaks_mba$chr[i]
start=as.numeric(peaks_mba$ci_lo[i])
end=as.numeric(peaks_mba$ci_hi[i])
genesgss = query_genes(chr, start, end)
write.csv(genesgss, file=paste0("data/ici.vs.eoi_",peaks$phenotype[i],"-interactive.covariate_genes_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_snpsqc_5.batches.csv"), quote=F)
rownames(genesgss) <- NULL
genesgss$strand_old = genesgss$strand
genesgss$strand[genesgss$strand=="+"] <- "positive"
genesgss$strand[genesgss$strand=="-"] <- "negative"
#genesgss <-
#table <-
#genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")] %>%
#kable(escape = F,align = c("ccccccccccc")) %>%
#kable_styling("striped", full_width = T) #%>%
#cat #%>%
#column_spec(1, bold=TRUE)
#
#print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], escape = F,align = c("ccccccccccc")))
print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], "html") %>% kable_styling("striped", full_width = T))
#table
}
} else {
print(paste0("There are too many peaks (",nrow(peaks_mba)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are too many peaks (0 peaks) that have a LOD that reaches suggestive (p<0.05) level of 2.07742869819588 [autosomes]/2.94249586915186 [x-chromosome]”
R version 3.6.2 (2019-12-12)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Catalina 10.15.7
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
locale:
[1] en_AU.UTF-8/en_AU.UTF-8/en_AU.UTF-8/C/en_AU.UTF-8/en_AU.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] abind_1.4-5 qtl2_0.22 reshape2_1.4.4 ggplot2_3.3.5
[5] tibble_3.1.2 psych_2.0.7 readxl_1.3.1 cluster_2.1.0
[9] dplyr_1.0.8 optparse_1.6.6 rhdf5_2.28.1 mclust_5.4.6
[13] tidyr_1.0.2 data.table_1.14.0 knitr_1.33 kableExtra_1.1.0
[17] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] httr_1.4.1 bit64_4.0.5 viridisLite_0.4.0 assertthat_0.2.1
[5] highr_0.9 blob_1.2.1 cellranger_1.1.0 yaml_2.2.1
[9] pillar_1.6.1 RSQLite_2.2.7 backports_1.2.1 lattice_0.20-38
[13] glue_1.4.2 digest_0.6.27 promises_1.1.0 rvest_0.3.5
[17] colorspace_2.0-2 htmltools_0.5.1.1 httpuv_1.5.2 plyr_1.8.6
[21] pkgconfig_2.0.3 purrr_0.3.4 scales_1.1.1 webshot_0.5.2
[25] whisker_0.4 getopt_1.20.3 later_1.0.0 git2r_0.26.1
[29] generics_0.0.2 ellipsis_0.3.2 cachem_1.0.5 withr_2.4.2
[33] cli_3.0.0 mnormt_1.5-7 magrittr_2.0.1 crayon_1.4.1
[37] memoise_2.0.0 evaluate_0.14 fs_1.4.1 fansi_0.5.0
[41] nlme_3.1-142 xml2_1.3.1 tools_3.6.2 hms_0.5.3
[45] lifecycle_1.0.1 stringr_1.4.0 Rhdf5lib_1.6.3 munsell_0.5.0
[49] compiler_3.6.2 rlang_1.0.2 grid_3.6.2 rstudioapi_0.13
[53] rmarkdown_2.1 gtable_0.3.0 DBI_1.1.1 R6_2.5.0
[57] fastmap_1.1.0 bit_4.0.4 utf8_1.2.1 rprojroot_1.3-2
[61] readr_1.3.1 stringi_1.7.2 parallel_3.6.2 Rcpp_1.0.7
[65] vctrs_0.3.8 tidyselect_1.1.2 xfun_0.24