QTL Analysis - rz transformed age of onset (rz.age) [ICI vs EOI] (corrected phenotype with outliers removed if any)
Belinda Cornes
2022-04-14
Last updated: 2022-04-14
Checks: 5 2
Knit directory: Serreze-T1D_Workflow/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20220210) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /Users/corneb/Documents/MyJax/CS/Projects/Serreze/qc/workflowr/Serreze-T1D_Workflow | . |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version a8c2d1a. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: analysis/.DS_Store
Ignored: data/.DS_Store
Untracked files:
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_0.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_52.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_mis_0.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_mis_52.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_mis_vo.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_5.batches_vo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.eoi_snpsqc_dis_no-x_updated_5.batches_11.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.eoi_snpsqc_dis_no-x_updated_5.batches_mis_11.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_1.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_5.batches_mis_1.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_1.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_11.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_5.batches_mis_11.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_changed.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_changed.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_scanone_5.batches.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_scanone_5.batches_mis.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_oops.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.4.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.4.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.4_miss.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.4_miss.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.5.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.5.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.5_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_test.with.5_0.Rmd.R
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_vo.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis_vo1.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_vo.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_vo1.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_52.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_0.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis_52.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.eoi_5.batches_0.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.eoi_5.batches_52.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.eoi_5.batches_mis_0.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.eoi_5.batches_mis_52.Rmd
Untracked: analysis/index_5.batches_additional_vo.Rmd
Untracked: data/GM_covar.csv
Untracked: data/GM_covar_BC312.csv
Untracked: data/bad_markers_all_4.batches.RData
Untracked: data/bad_markers_all_5.batches.RData
Untracked: data/blup_sub_chr10_lod.drop-1.5_5.batches.csv
Untracked: data/blup_sub_chr3_lod.drop-1.5.csv
Untracked: data/blup_sub_chr3_lod.drop-1.5_5.batches.csv
Untracked: data/blup_sub_chr4_lod.drop-1.5.csv
Untracked: data/blup_sub_chr4_lod.drop-1.5_5.batches.csv
Untracked: data/covar_cleaned_ici.vs.eoi.csv
Untracked: data/covar_cleaned_ici.vs.pbs.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_0.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_52.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_mis.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_mis_0.csv
Untracked: data/covar_corrected.cleaned_ici-early.vs.pbs_5.batches_mis_52.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi1.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_0.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_52.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_mis.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_mis_0.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi_5.batches_mis_52.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs1.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_0.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_52.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_mis.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_mis_0.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_5.batches_mis_52.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_0.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_52.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_mis.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_mis_0.csv
Untracked: data/covar_corrected_ici-early.vs.pbs_5.batches_mis_52.csv
Untracked: data/covar_corrected_ici.vs.eoi.csv
Untracked: data/covar_corrected_ici.vs.eoi1.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_0.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_52.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_mis.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_mis_0.csv
Untracked: data/covar_corrected_ici.vs.eoi_5.batches_mis_52.csv
Untracked: data/covar_corrected_ici.vs.pbs.csv
Untracked: data/covar_corrected_ici.vs.pbs1.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_0.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_52.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_mis.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_mis_0.csv
Untracked: data/covar_corrected_ici.vs.pbs_5.batches_mis_52.csv
Untracked: data/e.RData
Untracked: data/e_BC312.RData
Untracked: data/e_snpg_samqc_4.batches.RData
Untracked: data/e_snpg_samqc_4.batches_bc.RData
Untracked: data/e_snpg_samqc_5.batches.RData
Untracked: data/errors_ind_4.batches.RData
Untracked: data/errors_ind_4.batches_bc.RData
Untracked: data/errors_ind_5.batches.RData
Untracked: data/files.to.sync.txt
Untracked: data/fitqtl_chr3.peak_chr4.peak_additive.txt
Untracked: data/fitqtl_chr3.peak_chr4.peak_interacting.txt
Untracked: data/fitqtl_chr3.peak_chr4.peak_sex_additive.txt
Untracked: data/fitqtl_chr3.peak_chr4.peak_sex_interacting.txt
Untracked: data/g2blup_effects.csv
Untracked: data/g2blup_effects.xlsx
Untracked: data/genes_chr10_lod.drop-1.5_5.batches.csv
Untracked: data/genes_chr3_lod.drop-1.5.csv
Untracked: data/genes_chr3_lod.drop-1.5_5.batches.csv
Untracked: data/genes_chr4_lod.drop-1.5.csv
Untracked: data/genetic_map.csv
Untracked: data/genetic_map_BC312.csv
Untracked: data/genotype_errors_marker_4.batches.RData
Untracked: data/genotype_errors_marker_5.batches.RData
Untracked: data/genotype_freq_marker_4.batches.RData
Untracked: data/genotype_freq_marker_5.batches.RData
Untracked: data/gm_allqc_4.batches.RData
Untracked: data/gm_allqc_5.batches.RData
Untracked: data/gm_allqc_5.batches_mis.RData
Untracked: data/gm_samqc_3.batches.RData
Untracked: data/gm_samqc_4.batches.RData
Untracked: data/gm_samqc_4.batches_bc.RData
Untracked: data/gm_samqc_5.batches.RData
Untracked: data/gm_serreze.192.RData
Untracked: data/gm_serreze.BC312.RData
Untracked: data/ici-early.vs.pbs_age.of.onset-additive.covariates_blup_sub_chr-7_peak.marker-UNC13388811_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-additive.covariates_genes_chr-7_peak.marker-UNC13388811_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-15_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-15_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-15_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-17_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-17_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-17_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_5.batches.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici-early.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici-early.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici-early.vs.pbs_scanone_5.batches.Rdata
Untracked: data/ici-early.vs.pbs_scanone_5.batches_mis.Rdata
Untracked: data/ici-early.vs.pbs_scanone_snpsqc_5.batches.Rdata
Untracked: data/ici-early.vs.pbs_scanone_snpsqc_5.batches_mis.Rdata
Untracked: data/ici-early.vs.pbs_scanone_snpsqc_dis_no-x_updated_5.batches.Rdata
Untracked: data/ici-early.vs.pbs_scanone_snpsqc_dis_no-x_updated_5.batches_mis.Rdata
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00020646_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00292499_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-JAX00294019_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18216614_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18240977_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18343181_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18363544_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18363544_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18363544_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-11_peak.marker-UNC20090524_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-2_peak.marker-UNCHS008007_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-2_peak.marker-UNCHS008007_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR5263_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-ICR5263_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00105649_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00105649_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00106210_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00107193_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00516812_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00516812_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00522922_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00524205_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-JAX00535764_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4901083_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4901083_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4909519_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4909519_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4915446_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4915446_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4938565_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4938565_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4959266_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4959266_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4960612_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4960612_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4961542_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4961542_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5046545_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5055259_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5062524_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5068215_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5076935_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5082757_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5173293_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5193970_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5257268_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5264696_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5267813_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5270225_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5277061_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5278229_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5278229_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5282259_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5287977_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5301460_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5309136_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5321559_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5322455_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5329171_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5360055_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5778977_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNC5916997_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008229_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008229_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008234_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008234_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008242_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008260_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008260_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008281_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008281_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008338_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008338_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008363_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008363_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008409_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008432_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008609_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008614_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008627_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008659_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008725_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009066_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009067_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009068_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009112_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009693_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009821_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001276_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-sanger2496q_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-3_peak.marker-sanger2496q_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-4_peak.marker-UNC8250659_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-4_peak.marker-UNC8439633_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-4_peak.marker-UNC8439633_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-4_peak.marker-UNCHS012955_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-6_peak.marker-UNC11108920_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-6_peak.marker-UNC11108920_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00020646_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00292499_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-JAX00294019_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18216614_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18240977_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18343181_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18363544_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18363544_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18363544_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-11_peak.marker-UNC20090524_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-2_peak.marker-UNCHS008007_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-2_peak.marker-UNCHS008007_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR5263_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-ICR5263_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00105649_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00105649_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00106210_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00107193_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00516812_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00516812_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00522922_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00524205_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-JAX00535764_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4901083_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4901083_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4909519_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4909519_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4915446_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4915446_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4938565_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4938565_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4959266_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4959266_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4960612_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4960612_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4961542_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4961542_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5046545_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5055259_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5062524_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5068215_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5076935_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5082757_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5173293_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5193970_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5257268_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5264696_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5267813_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5270225_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5277061_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5278229_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5278229_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5282259_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5287977_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5301460_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5309136_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5321559_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5322455_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5329171_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5360055_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5778977_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNC5916997_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008229_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008229_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008234_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008234_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008242_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008260_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008260_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008281_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008281_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008338_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008338_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008363_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008363_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008409_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008432_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008609_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008614_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008627_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008659_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008725_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009066_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009067_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009068_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009112_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009693_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCHS009821_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-UNCJPD001276_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-sanger2496q_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-3_peak.marker-sanger2496q_lod.drop-1.5_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-4_peak.marker-UNC8250659_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-4_peak.marker-UNC8439633_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-4_peak.marker-UNC8439633_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-4_peak.marker-UNCHS012955_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-6_peak.marker-UNC11108920_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-6_peak.marker-UNC11108920_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_blup.full_chr-10_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-11_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-12_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-13_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-14_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-15_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-16_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-17_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-18_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-19_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-1_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-2_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-3_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-4_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-6_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-7_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-8_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-9_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.full_chr-X_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.full_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-15_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-17_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-ICR4295_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-JAX00292499_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18154330_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18195765_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18216614_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18240977_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNC18856953_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNCJPD004316_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-10_peak.marker-UNCJPD009532_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNC19155926_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNC19481366_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-11_peak.marker-UNCJPD004792_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-12_peak.marker-ICR4497_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-13_peak.marker-UNCHS036964_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-18_peak.marker-UNCHS045344_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-19_peak.marker-UNCJPD007087_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-1_peak.marker-ICR010_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-1_peak.marker-ICR010_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-2_peak.marker-UNC4609660_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-B6_rs31740628_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR1269_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR5325_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR5347_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-ICR5348_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00104971_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00105059_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00105203_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00107930_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00107980_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00108068_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00108114_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00109750_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00189245_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00517218_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00517906_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00518418_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00518684_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00520122_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00524717_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00524828_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00525380_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00525709_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00527291_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00527587_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00529707_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00531601_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-JAX00532122_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4673471_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4681411_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4793677_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4799475_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4909830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4914130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4964062_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4985064_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC4991565_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5001723_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5042361_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5055259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5076935_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5082757_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5173293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5182782_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5193970_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5240484_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5256922_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5266664_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5266711_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5267813_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5274554_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5277061_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5282259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5287977_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5301460_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5321559_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5322455_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5328596_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5350384_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5354724_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5375208_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5384072_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5400314_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5433945_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5435874_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5466669_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5517972_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5522257_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5523265_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5532424_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5553882_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5572051_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5582753_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5589260_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5595555_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5623239_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5639830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5680234_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5682314_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5725356_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5727351_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5734420_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5743257_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5751567_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5752623_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5776974_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5778977_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5788507_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5792468_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5794598_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5795561_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5802298_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5806628_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5817478_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5836999_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5841501_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5847927_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNC5966417_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008129_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008173_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008231_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008235_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008242_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008253_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008256_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008311_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008317_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008372_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008410_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008415_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008431_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008609_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008614_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008649_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008651_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008723_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008802_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008803_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008808_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008834_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008838_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008928_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008948_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008959_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008973_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS008987_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009001_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009025_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009034_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009087_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009109_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009142_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009207_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009216_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009306_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009316_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009364_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009377_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009382_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009387_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009395_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009413_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009428_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009463_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009503_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009532_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009541_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009560_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCHS009574_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001276_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001315_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001355_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001373_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-3_peak.marker-UNCJPD001382_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC6851205_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8250659_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8299912_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8300499_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8381981_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8436434_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8439633_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNC8530763_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS012948_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS012976_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013139_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-4_peak.marker-UNCHS013304_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-5_peak.marker-JAX00590770_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-ICR1830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-UNC13170794_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-UNCHS019480_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-7_peak.marker-cr31snv87_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-UNC15430711_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-9_peak.marker-UNC17203329_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-9_peak.marker-UNCHS024593_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-X_peak.marker-UNC30627130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-X_peak.marker-UNCHS048313_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_snpsqc_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_snpsqc_dis_no-x_updated_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_additive_snpsqc_dis_no-x_updated_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_snpsqc_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_snpsqc_dis_no-x_updated_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_interacting_snpsqc_dis_no-x_updated_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_snpsqc_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_snpsqc_dis_no-x_updated_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_additive_snpsqc_dis_no-x_updated_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_5.batches_mis.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_snpsqc_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_snpsqc_dis_no-x_updated_5.batches.txt
Untracked: data/ici.vs.eoi_fitqtl_peaks_sex_interacting_snpsqc_dis_no-x_updated_5.batches_mis.mis.txt
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-ICR4295_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-JAX00292499_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18154330_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18195765_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18216614_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18240977_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNC18856953_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNCJPD004316_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-10_peak.marker-UNCJPD009532_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNC19155926_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNC19481366_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-11_peak.marker-UNCJPD004792_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-12_peak.marker-ICR4497_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-13_peak.marker-UNCHS036964_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-18_peak.marker-UNCHS045344_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-19_peak.marker-UNCJPD007087_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-1_peak.marker-ICR010_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-1_peak.marker-ICR010_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-2_peak.marker-UNC4609660_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-B6_rs31740628_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR1269_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR5325_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR5347_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-ICR5348_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00104971_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00105059_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00105203_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00107930_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00107980_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00108068_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00108114_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00109750_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00189245_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00517218_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00517674_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00517906_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00518418_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00518684_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00518911r_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00520122_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00524717_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00524828_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00525380_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00525709_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00527291_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00527587_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00529707_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00529707r_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00531601_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-JAX00532122_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4673471_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4681411_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4793677_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4799475_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4809586_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4820379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4875653_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4909830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4912288_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4914130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4923398_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4945186_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4947492_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4957859_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4964062_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4979914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4985064_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4990111_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC4991565_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5001723_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5042361_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5055259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5076935_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5082757_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5173293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5182782_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5193970_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5240484_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5256922_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5266664_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5266711_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5267813_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5274554_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5277061_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5282259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5287977_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5301460_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5321559_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5322455_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5328596_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5350384_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5354724_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5375208_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5384072_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5400314_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5433945_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5435874_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5466669_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5517972_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5522257_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5523265_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5532424_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5553882_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5572051_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5582753_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5589260_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5595555_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5623239_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5639830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5680234_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5682314_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5725356_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5727351_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5734420_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5743257_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5751567_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5752623_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5776974_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5778977_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5788507_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5792468_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5794598_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5795561_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5802298_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5806628_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5817478_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5836999_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5841501_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5847927_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNC5966417_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008129_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008173_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008193_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008209_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008215_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008219_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008231_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008235_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008242_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008253_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008256_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008259_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008271_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008311_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008317_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008372_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008379_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008410_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008415_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008431_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008609_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008614_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008649_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008651_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008656_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008723_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008802_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008803_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008808_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008834_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008838_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008914_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008928_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008948_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008959_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008973_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS008987_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009001_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009025_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009034_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009087_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009109_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009142_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009207_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009216_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009224_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009306_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009316_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009364_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009377_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009382_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009387_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009395_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009413_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009428_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009463_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009503_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009532_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009541_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009560_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCHS009574_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001170_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001198_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001276_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001315_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001355_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001373_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-3_peak.marker-UNCJPD001382_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC6851205_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8250659_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8299912_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8300499_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8381981_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8436434_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8439633_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNC8530763_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS012948_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS012976_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013118_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013139_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-4_peak.marker-UNCHS013304_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-5_peak.marker-JAX00590770_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-ICR1830_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-UNC13170794_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-UNCHS019480_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-7_peak.marker-cr31snv87_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-UNC15430711_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-9_peak.marker-UNC17203329_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-9_peak.marker-UNCHS024593_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-X_peak.marker-UNC30627130_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-X_peak.marker-UNCHS048313_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_gm_qtl_5.batches.csv
Untracked: data/ici.vs.eoi_gm_qtl_5.batches_mis.csv
Untracked: data/ici.vs.eoi_gm_qtl_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_gm_qtl_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.eoi_gm_qtl_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.eoi_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_52.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_rz.age-additive.covariates_genes_chr-7_peak.marker-UNC13239362_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18343181_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18376338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNC18382395_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-10_peak.marker-UNCHS029024_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-1_peak.marker-UNC228500_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-1_peak.marker-UNC228500_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-ICR2047_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00112499_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00219554_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-JAX00529707_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4837069_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5006948_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5266664_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5300878_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5360055_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5679155_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC5945286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNC6227106_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008595_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009067_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCHS009693_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-UNCJPD001341_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-3_peak.marker-sanger2623_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-4_peak.marker-UNCHS013125_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-4_peak.marker-UNCHS013125_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-6_peak.marker-UNCHS018170_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-6_peak.marker-UNCHS018231_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-JAX00292927_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18311938_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18343181_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18376338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18376338_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNC18382395_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNCHS028236_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNCHS028536_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-10_peak.marker-UNCHS029024_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-11_peak.marker-UNC19970181_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-12_peak.marker-ICR499_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-13_peak.marker-UNCHS036773_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-15_peak.marker-UNC26070435_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-17_peak.marker-UNCHS044241_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-17_peak.marker-UNCrs47191360_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-1_peak.marker-UNC228500_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-1_peak.marker-UNC228500_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-2_peak.marker-UNC4609527_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-ICR1338_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-ICR2047_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00106957_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00112499_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00219554_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00220631_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00514662_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-JAX00529707_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4813186_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4836550_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4837069_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4912039_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4924377_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4956618_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC4957168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5006948_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5008656_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5083168_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5192582_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5213888_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5262472_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5266664_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5300878_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5360055_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5454839_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5645844_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5679155_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5808246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC5945286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNC6227106_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008182_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008286_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008487_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008511_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008595_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008613_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS008815_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS009067_lod.drop-1.5_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS009383_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCHS009693_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCJPD001246_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-UNCJPD001341_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-3_peak.marker-sanger2623_lod.drop-1.5_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-4_peak.marker-UNCHS013125_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-4_peak.marker-UNCHS013125_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-6_peak.marker-UNCHS018170_lod.drop-1.5_snpsqc_5.batches_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-6_peak.marker-UNCHS018231_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-7_peak.marker-UNCHS020066_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-9_peak.marker-UNC17203597_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_0.csv
Untracked: data/ici.vs.eoi_rz.age-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_52.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.geno.freq.removed.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.probs.freq.removed.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.eoi_scanone_5.batches.Rdata
Untracked: data/ici.vs.eoi_scanone_5.batches_mis.Rdata
Untracked: data/ici.vs.eoi_scanone_snpsqc_5.batches.Rdata
Untracked: data/ici.vs.eoi_scanone_snpsqc_5.batches_mis.Rdata
Untracked: data/ici.vs.eoi_scanone_snpsqc_dis_no-x_updated_5.batches.Rdata
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNC28655293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-5_peak.marker-UNC8675939_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_blup_sub_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-14_peak.marker-UNC24056202_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-17_peak.marker-UNCJPD006614_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNC28655293_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-5_peak.marker-UNC10044126_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-5_peak.marker-UNC8675939_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-8_peak.marker-JAX00667121_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-8_peak.marker-UNC15524531_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_age.of.onset-no.covariates_genes_chr-X_peak.marker-UNCHS048314_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_bad.markers_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-15_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-17_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.full_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-10_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-11_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-12_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-13_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-14_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-15_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-16_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-17_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-18_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-19_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-1_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-2_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-3_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-4_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-6_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-7_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-8_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_lod.drop-1.5_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-9_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup.qc_chr-X_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-10_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-11_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-12_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-13_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-14_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-15_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-15_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-15_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-16_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-17_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-17_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-17_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-18_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-19_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-1_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-2_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-3_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-4_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-5_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-6_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-7_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-8_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-9_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_blup_chr-X_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-13_peak.marker-UNC22384241_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-19_peak.marker-UNC29893228_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-19_peak.marker-UNC29990033_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-19_peak.marker-UNCHS047192_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-8_peak.marker-UNCHS023353_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-8_peak.marker-UNCHS023353_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_blup_sub_chr-8_peak.marker-UNCHS023364_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_fitqtl_peaks_additive_snpsqc_dis_no-x_updated_5.batches_mis_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_additive_snpsqc_dis_no-x_updated_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_additive_snpsqc_snpsqc_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_interacting_snpsqc_dis_no-x_updated_5.batches_mis_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_interacting_snpsqc_dis_no-x_updated_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_interacting_snpsqc_snpsqc_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_additive_snpsqc_dis_no-x_updated_5.batches_mis_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_additive_snpsqc_dis_no-x_updated_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_additive_snpsqc_snpsqc_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_interacting_snpsqc_dis_no-x_updated_5.batches_mis_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_interacting_snpsqc_dis_no-x_updated_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_fitqtl_peaks_sex_interacting_snpsqc_snpsqc_5.batches_peaks.txt
Untracked: data/ici.vs.pbs_genes_chr-13_peak.marker-UNC22384241_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-18_peak.marker-UNCHS045343_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_genes_chr-19_peak.marker-UNC29893228_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-19_peak.marker-UNC29990033_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-19_peak.marker-UNCHS047192_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-8_peak.marker-UNCHS023353_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-8_peak.marker-UNCHS023353_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_genes_chr-8_peak.marker-UNCHS023364_lod.drop-1.5_snpsqc_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches.csv
Untracked: data/ici.vs.pbs_gm_qtl_5.batches_mis.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_mis_peaks.csv
Untracked: data/ici.vs.pbs_gm_qtl_snpsqc_dis_no-x_updated_5.batches_peaks.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_5.batches_mis.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-JAX00153527_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-JAX00153527_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNCHS020443_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNCHS020443_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_blup_sub_chr-7_peak.marker-UNCHS020486_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-JAX00153527_lod.drop-1.5_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-JAX00153527_lod.drop-1.5_snpsqc_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNCHS020443_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNCHS020443_lod.drop-1.5_snpsqc_dis_no-x_updated_5.batches_mis.csv
Untracked: data/ici.vs.pbs_rz.age-additive.covariates_genes_chr-7_peak.marker-UNCHS020486_lod.drop-1.5_snpsqc_5.batches_mis.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.pbs_scanone_5.batches.Rdata
Untracked: data/ici.vs.pbs_scanone_5.batches_mis.Rdata
Untracked: data/ici.vs.pbs_scanone_snpsqc_5.batches.Rdata
Untracked: data/ici.vs.pbs_scanone_snpsqc_5.batches_mis.Rdata
Untracked: data/ici.vs.pbs_scanone_snpsqc_dis_no-x_updated_5.batches.Rdata
Untracked: data/ici.vs.pbs_scanone_snpsqc_dis_no-x_updated_5.batches_mis.Rdata
Untracked: data/percent_missing_id_3.batches.RData
Untracked: data/percent_missing_id_4.batches.RData
Untracked: data/percent_missing_id_4.batches_bc.RData
Untracked: data/percent_missing_id_5.batches.RData
Untracked: data/percent_missing_marker_4.batches.RData
Untracked: data/percent_missing_marker_5.batches.RData
Untracked: data/pheno.csv
Untracked: data/pheno_BC312.csv
Untracked: data/physical_map.csv
Untracked: data/physical_map_BC312.csv
Untracked: data/qc_info_bad_sample_3.batches.RData
Untracked: data/qc_info_bad_sample_4.batches.RData
Untracked: data/qc_info_bad_sample_4.batches_bc.RData
Untracked: data/qc_info_bad_sample_5.batches.RData
Untracked: data/remaining.markers_geno.freq.xlsx
Untracked: data/sample_geno.csv
Untracked: data/sample_geno_AHB_BC312.csv
Untracked: data/sample_geno_bc.csv
Untracked: data/sample_geno_bc_BC312.csv
Untracked: data/serreze_probs.rds
Untracked: data/serreze_probs_BC312.rds
Untracked: data/serreze_probs_allqc.rds
Untracked: data/serreze_probs_allqc_5.batches.rds
Untracked: data/serreze_probs_allqc_5.batches_mis.rds
Untracked: data/summary.cg_3.batches.RData
Untracked: data/summary.cg_4.batches.RData
Untracked: data/summary.cg_4.batches_bc.RData
Untracked: data/summary.cg_5.batches.RData
Untracked: output/Percent_missing_genotype_data_5.batches.pdf
Untracked: output/Percent_missing_genotype_data_per_marker_5.batches.pdf
Untracked: output/Proportion_matching_genotypes_before_removal_of_bad_samples_5.batches.pdf
Untracked: output/genotype_error_marker_5.batches.pdf
Untracked: output/genotype_frequency_marker_5.batches.pdf
Unstaged changes:
Modified: analysis/4.1.1_qtl.analysis_binary_ici.vs.eoi_snpsqc_dis_no-x_updated.Rmd
Modified: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici-early.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.eoi_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_5.batches_mis.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches.Rmd
Modified: analysis/4.1.2_qtl.analysis_cont_rz.age_ici.vs.pbs_snpsqc_pheno.corrected.cleaned_dis_no-xk_5.batches_mis.Rmd
Modified: analysis/index_5.batches.Rmd
Modified: analysis/index_5.batches_additional.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with wflow_publish() to start tracking its development.
Loading Data
load("data/gm_allqc_5.batches.RData")
#gm_allqc
gm=gm_allqc
gmObject of class cross2 (crosstype "bc")
Total individuals 308
No. genotyped individuals 308
No. phenotyped individuals 308
No. with both geno & pheno 308
No. phenotypes 1
No. covariates 6
No. phenotype covariates 0
No. chromosomes 20
Total markers 34537
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2643 2629 1857 1890 1774 1941 1672 1627 1878 1176 1871 1300 1549 1578 1257 935
17 18 19 X
501 913 1014 4532 #pr <- readRDS("data/serreze_probs_allqc.rds")
#pr <- readRDS("data/serreze_probs.rds")
##extracting animals with ici and eoi group status
miceinfo <- gm$covar[gm$covar$group == "EOI" | gm$covar$group == "ICI",]
table(miceinfo$group)
EOI ICI
164 104 mice.ids <- rownames(miceinfo)
gm <- gm[mice.ids]
gmObject of class cross2 (crosstype "bc")
Total individuals 268
No. genotyped individuals 268
No. phenotyped individuals 268
No. with both geno & pheno 268
No. phenotypes 1
No. covariates 6
No. phenotype covariates 0
No. chromosomes 20
Total markers 34537
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2643 2629 1857 1890 1774 1941 1672 1627 1878 1176 1871 1300 1549 1578 1257 935
17 18 19 X
501 913 1014 4532 table(gm$covar$group)
EOI ICI
164 104 gm.full <- gm
covars <- read_csv("data/covar_corrected.cleaned_ici.vs.eoi_5.batches_0.csv")
#removing any missing info
missing_ids <- covars[which(covars$"rz.age"==""),]$Mouse.ID
length(missing_ids)[1] 1missing_ids[1] "NG00522-ICI-LATE"if(length(missing_ids) != 0){
gm <- gm[paste0("-",as.character(missing_ids)),]
gm
table(gm$covar$group)
#keeping only mouseids in covars
covars <- covars[gm$covar$id,]
table(covars$group)
}
EOI ICI
164 103 #covars[covars$age.of.onset==0,]$age.of.onset <- ''
query_variants <- create_variant_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/cc_variants.sqlite")
query_genes <- create_gene_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/mouse_genes_mgi.sqlite")
##removing problmetic marker
gm <- drop_markers(gm, "UNCHS013106")
#pmap_interp = interp_map(map, gm$gmap, gm$pmap)
markers <- marker_names(gm)
gmapdf <- read.csv("/Users/corneb/Documents/MyJax/CS/Projects/Serreze/haplotype.reconstruction/output_5.batches/genetic_map.csv")
pmapdf <- read.csv("/Users/corneb/Documents/MyJax/CS/Projects/Serreze/haplotype.reconstruction/output_5.batches/physical_map.csv")
#mapdf <- merge(gmapdf,pmapdf, by=c("marker","chr"), all=T)
#rownames(mapdf) <- mapdf$marker
#mapdf <- mapdf[markers,]
#names(mapdf) <- c('marker','chr','gmapdf','pmapdf')
#mapdfnd <- mapdf[!duplicated(mapdf[c(2:3)]),]
pr.qc <- calc_genoprob(gm)
gmObject of class cross2 (crosstype "bc")
Total individuals 267
No. genotyped individuals 267
No. phenotyped individuals 267
No. with both geno & pheno 267
No. phenotypes 1
No. covariates 6
No. phenotype covariates 0
No. chromosomes 20
Total markers 34537
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2643 2629 1857 1890 1774 1941 1672 1627 1878 1176 1871 1300 1549 1578 1257 935
17 18 19 X
501 913 1014 4532 Preparing necessary info
covars$rz.age <- as.numeric(covars$rz.age)
Xcovar <- get_x_covar(gm)
addcovar = model.matrix(~ICI.vs.EOI, data = covars)[,-1]
#addcovar.i = model.matrix(~ICI.vs.EOI, data = covars)[,-1]
K <- calc_kinship(pr.qc, type = "loco")
#heatmap(K[[1]])
K.overall <- calc_kinship(pr.qc, type = "overall")
#heatmap(K.overall)
kinship <- calc_kinship(pr.qc)
heatmap(kinship)
No additive covariates
Significance thresholds
operm <- scan1perm(pr.qc, covars["rz.age"], model="normal", n_perm=10, perm_Xsp=TRUE, chr_lengths=chr_lengths(gm$gmap))
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
names(summary_table) <- c("autosomes","X")
summary_table$significance.level <- rownames(summary_table)
rownames(summary_table) <- NULL
summary_table[c(3,1:2)] %>%
kable(escape = F,align = c("ccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| significance.level | autosomes | X |
|---|---|---|
| 0.01 | 3.103325 | 5.005135 |
| 0.05 | 2.936908 | 4.711528 |
| 0.1 | 2.728368 | 4.327585 |
Genome-wide scan
plot_lod<-function(out,map){
for (i in 1:dim(out)[2]){
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - ICI vs. EOI (no covariates) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
print("with no kinship")[1] “with no kinship”
outf <- scan1(pr.qc, covars["rz.age"], Xcovar=Xcovar, model="normal")
plot_lod(outf,gm$gmap)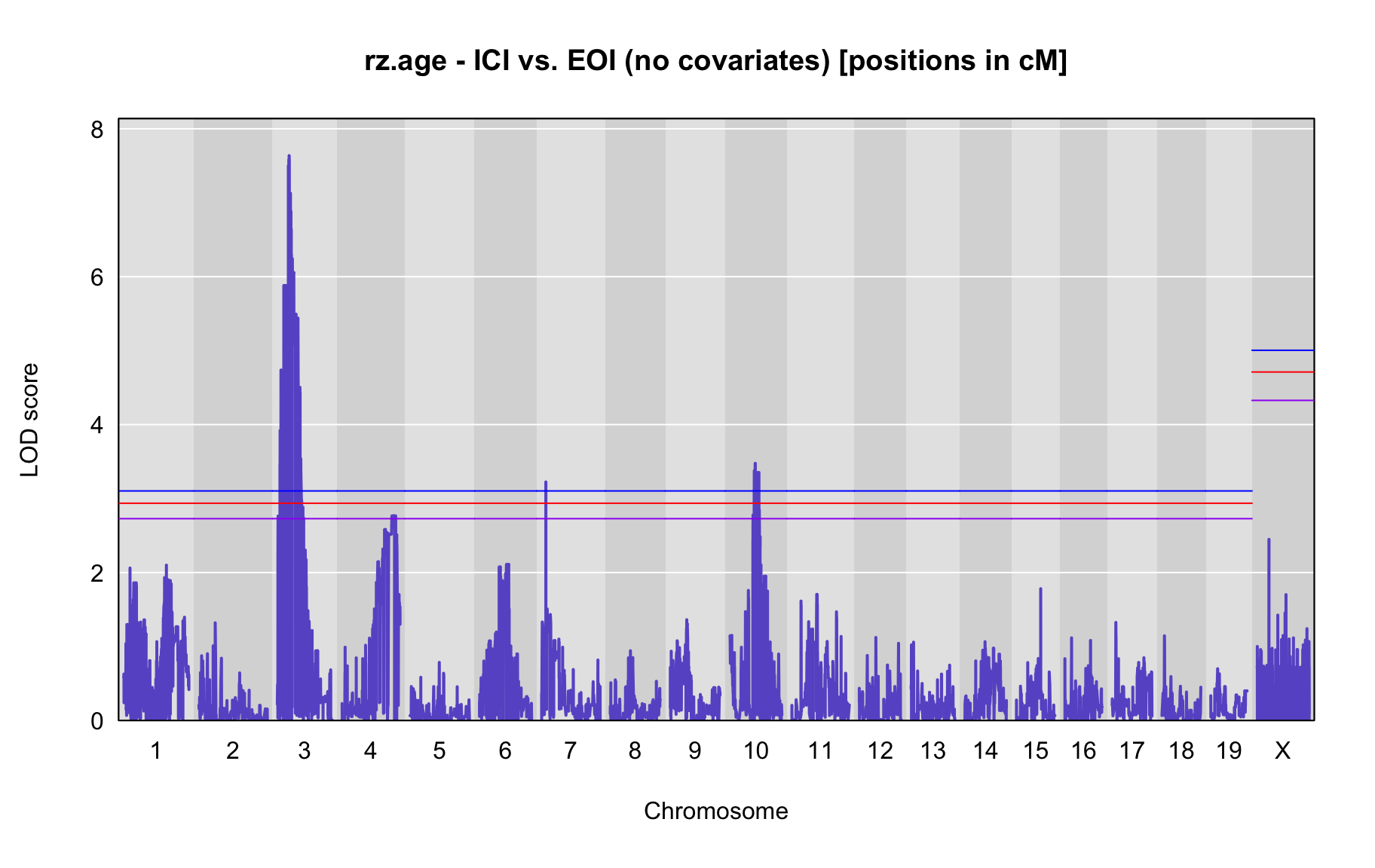
print("with normal kinship")[1] “with normal kinship”
outk <- scan1(pr.qc, covars["rz.age"], Xcovar=Xcovar, model="normal", kinship=kinship)
plot_lod(outk,gm$gmap)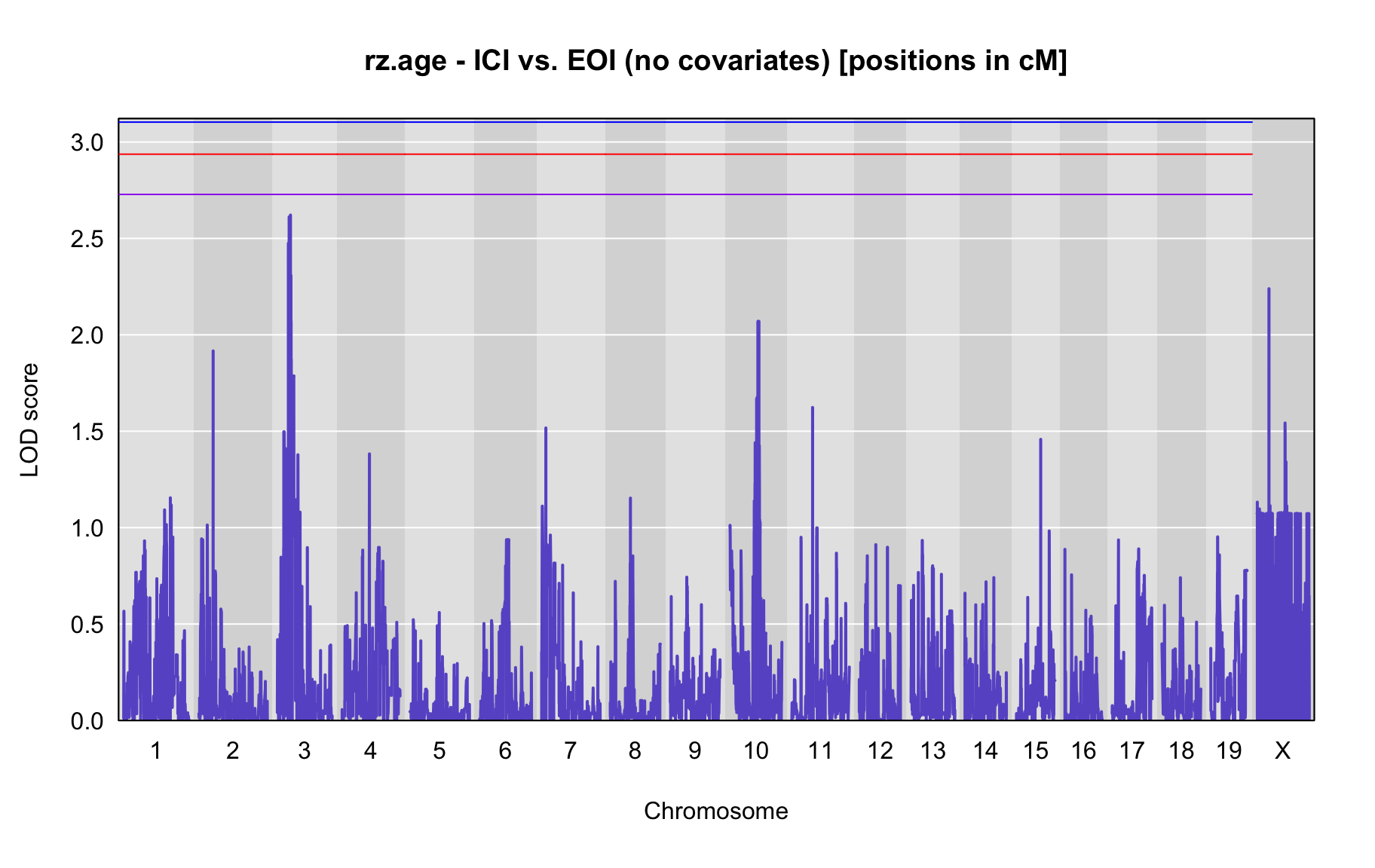
print("with loco kinship")[1] “with loco kinship”
out <- scan1(pr.qc, covars["rz.age"], Xcovar=Xcovar, model="normal", kinship=K)
plot_lod(out,gm$gmap)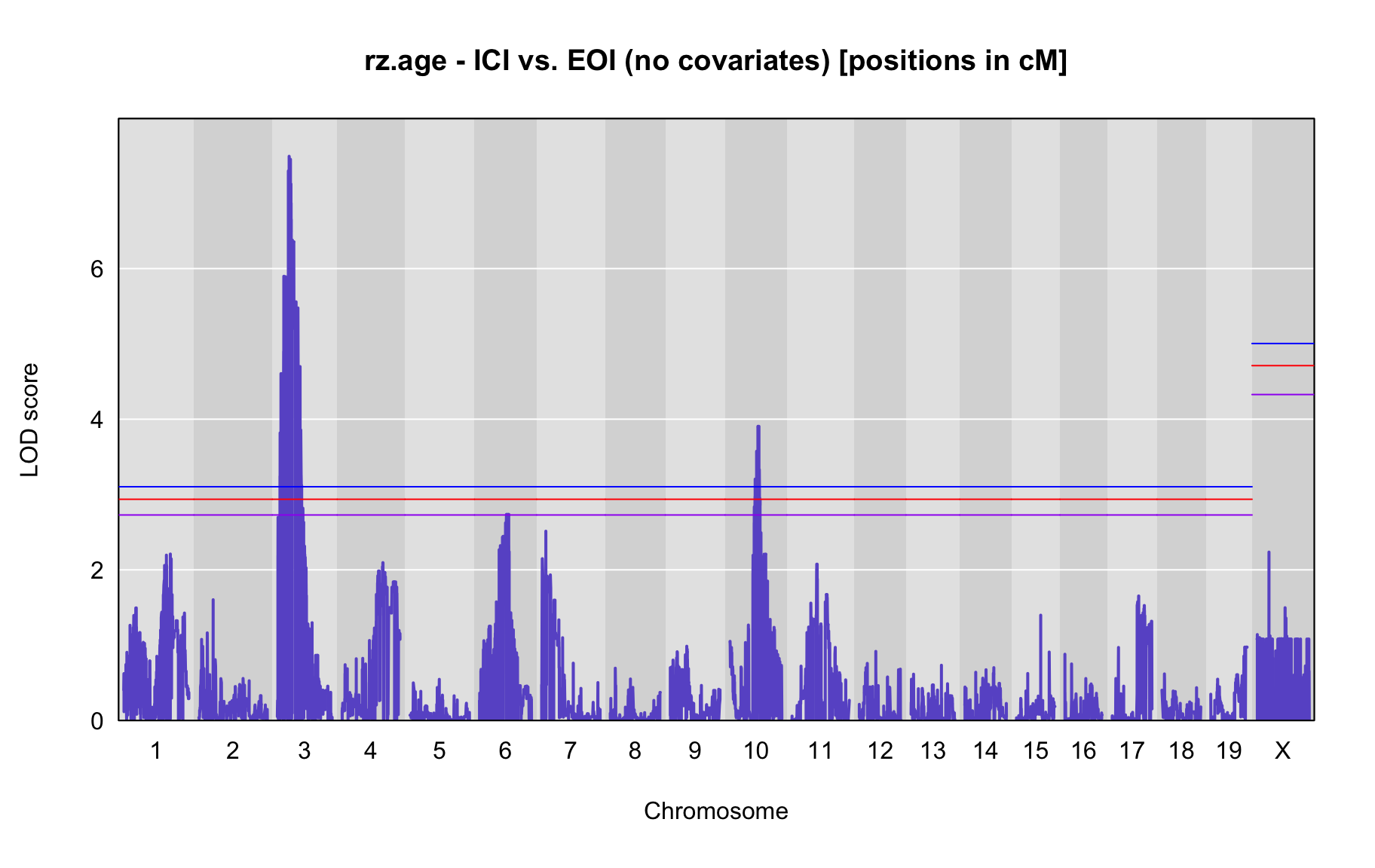
print(paste0("number of samples in analysis = ", as.data.frame(attributes(out)$sample_size)[1,]))[1] “number of samples in analysis = 267”
LOD peaks
Centimorgan (cM)
peaks<-find_peaks(out, gm$gmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) > 0){
peaks$marker <- find_marker(gm$gmap, chr=peaks$chr,pos=peaks$pos)
names(peaks)[2] <- c("phenotype")
peaks <- peaks[-1]
rownames(peaks) <- NULL
print(kable(peaks, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
#plot only peak chromosomes
plot_lod_chr<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " - ICI vs. EOI (no covariates) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}| phenotype | chr | pos | lod | ci_lo | ci_hi | marker |
|---|---|---|---|---|---|---|
| rz.age | 3 | 5.562000 | 3.818750 | 2.124 | 5.822 | UNC4813186 |
| rz.age | 3 | 6.943333 | 4.605390 | 6.356 | 7.572 | UNC4837069 |
| rz.age | 3 | 9.465000 | 4.521028 | 7.603 | 10.190 | JAX00219554 |
| rz.age | 3 | 10.815000 | 5.897553 | 10.282 | 10.970 | UNC4912039 |
| rz.age | 3 | 11.391000 | 5.897554 | 10.982 | 11.971 | UNC4924377 |
| rz.age | 3 | 12.290000 | 5.782550 | 12.254 | 12.989 | UNCHS008286 |
| rz.age | 3 | 13.715000 | 5.891007 | 13.052 | 15.109 | UNC4957168 |
| rz.age | 3 | 16.414000 | 5.813801 | 15.290 | 16.966 | UNC5008656 |
| rz.age | 3 | 18.746000 | 7.491257 | 17.482 | 19.077 | UNC5083168 |
| rz.age | 3 | 20.718000 | 7.451219 | 20.613 | 20.774 | JAX00106957 |
| rz.age | 3 | 20.879000 | 7.124942 | 20.784 | 21.549 | UNC5192582 |
| rz.age | 3 | 21.687333 | 6.867859 | 21.580 | 22.187 | UNCHS008613 |
| rz.age | 3 | 23.137000 | 6.299792 | 22.489 | 23.199 | UNC5262472 |
| rz.age | 3 | 23.437000 | 6.378744 | 23.380 | 27.894 | UNC5266664 |
| rz.age | 3 | 29.002000 | 5.558554 | 28.262 | 30.167 | UNCHS008815 |
| rz.age | 3 | 31.805000 | 5.477456 | 30.203 | 32.529 | UNC5454839 |
| rz.age | 3 | 32.823000 | 4.485128 | 32.607 | 33.376 | ICR1338 |
| rz.age | 3 | 33.724000 | 4.499413 | 33.391 | 33.741 | UNCHS009067 |
| rz.age | 3 | 34.841000 | 4.699274 | 33.825 | 35.168 | JAX00220631 |
| rz.age | 3 | 36.068000 | 3.857035 | 36.023 | 40.163 | UNC5645844 |
| rz.age | 10 | 38.425000 | 3.204739 | 34.740 | 38.502 | JAX00292927 |
| rz.age | 10 | 40.502000 | 3.575038 | 40.301 | 41.292 | UNC18311938 |
| rz.age | 10 | 41.494000 | 3.567013 | 41.319 | 41.720 | UNC18343181 |
| rz.age | 10 | 43.651000 | 3.905120 | 42.028 | 43.919 | UNC18376338 |
| rz.age | 10 | 44.382000 | 3.331703 | 44.060 | 56.696 | UNCHS029024 |
if(nrow(peaks) < 50 & nrow(peaks) > 0){
for(i in unique(peaks$chr)){
#for (i in 1:nrow(peaks)){
#plot_lod_chr(out,gm$gmap, peaks$chr[i])
plot_lod_chr(out,gm$gmap, i)
}
} else {
print(paste0("There are too many peaks (",nrow(peaks)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}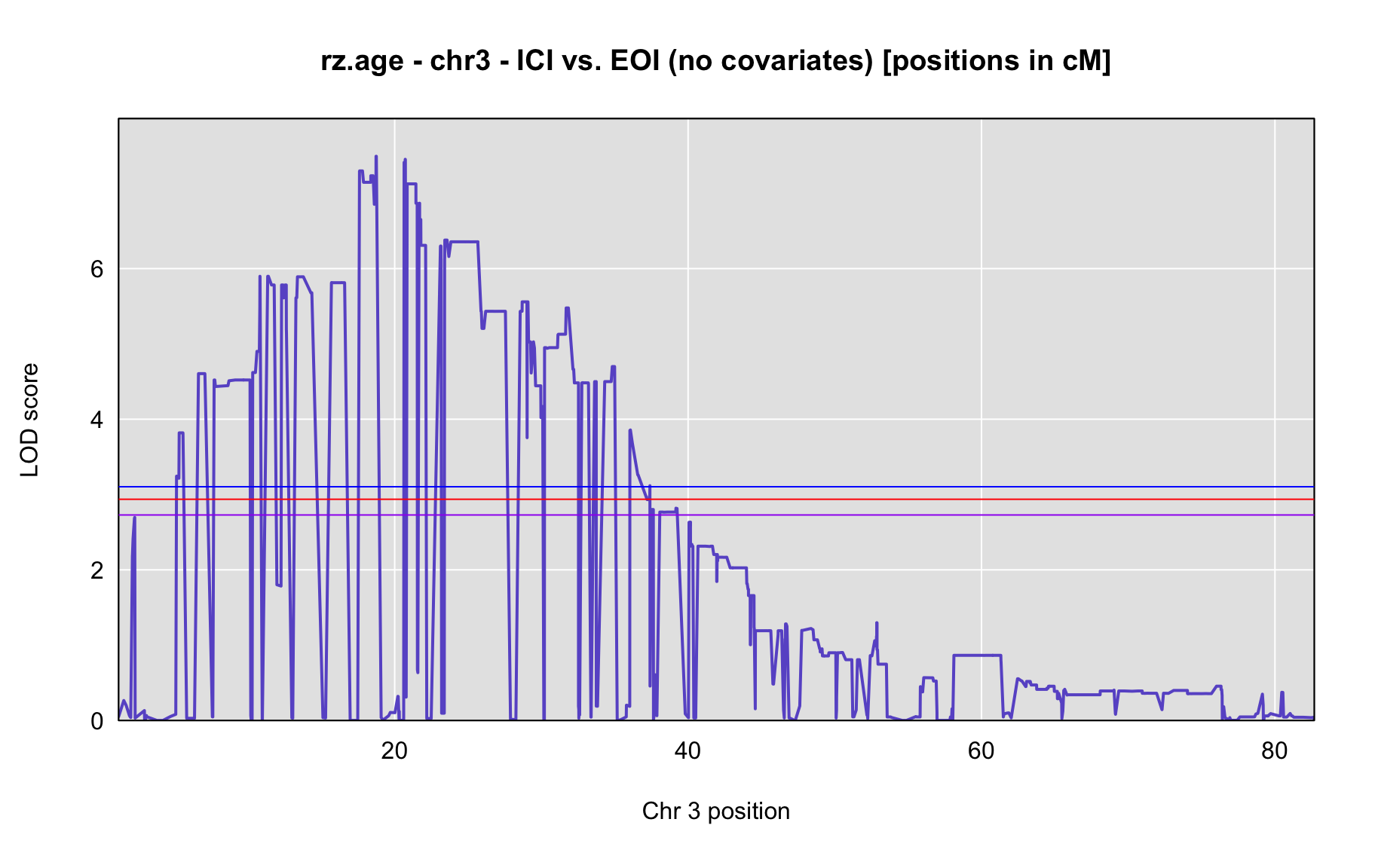

Megabase (MB)
peaks_mba<-find_peaks(out, gm$pmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) > 0){
peaks_mba$marker <- find_marker(gm$pmap, chr=peaks_mba$chr,pos=peaks_mba$pos)
names(peaks_mba)[2] <- c("phenotype")
peaks_mba <- peaks_mba[-1]
rownames(peaks_mba) <- NULL
print(kable(peaks_mba, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
plot_lod_chr_mb<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " - ICI vs. EOI (no covariates) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}| phenotype | chr | pos | lod | ci_lo | ci_hi | marker |
|---|---|---|---|---|---|---|
| rz.age | 3 | 19.48622 | 3.818750 | 7.917057 | 19.72964 | UNC4813186 |
| rz.age | 3 | 21.08393 | 4.605390 | 20.305499 | 22.03573 | UNC4837069 |
| rz.age | 3 | 24.17913 | 4.521028 | 22.177798 | 25.28624 | JAX00219554 |
| rz.age | 3 | 27.03622 | 5.897553 | 25.426933 | 27.55791 | UNCHS008242 |
| rz.age | 3 | 28.01969 | 5.897554 | 27.583922 | 28.41152 | UNC4924377 |
| rz.age | 3 | 28.70005 | 5.782550 | 28.647890 | 30.20381 | UNCHS008286 |
| rz.age | 3 | 30.41146 | 5.891007 | 30.221767 | 31.08157 | UNC4957168 |
| rz.age | 3 | 34.01961 | 5.813801 | 31.479710 | 34.95587 | UNC5008656 |
| rz.age | 3 | 38.94808 | 7.491257 | 36.524146 | 39.72051 | UNC5083168 |
| rz.age | 3 | 46.39933 | 7.451219 | 46.093688 | 46.77335 | JAX00106957 |
| rz.age | 3 | 47.51865 | 7.124942 | 47.267935 | 48.88766 | UNC5192582 |
| rz.age | 3 | 49.60906 | 6.867859 | 48.950852 | 51.24592 | UNCHS008613 |
| rz.age | 3 | 52.22783 | 6.299792 | 51.504201 | 52.28869 | UNC5262472 |
| rz.age | 3 | 52.52321 | 6.378744 | 52.466515 | 57.56748 | UNC5266664 |
| rz.age | 3 | 59.18647 | 5.558554 | 58.021842 | 64.96586 | UNCHS008815 |
| rz.age | 3 | 68.53602 | 5.477456 | 65.480179 | 70.64883 | UNC5454839 |
| rz.age | 3 | 73.26997 | 4.485128 | 72.338635 | 73.73010 | ICR1338 |
| rz.age | 3 | 75.30888 | 4.499413 | 73.742144 | 75.51898 | UNCHS009068 |
| rz.age | 3 | 78.26134 | 4.699274 | 75.789940 | 79.93182 | JAX00220631 |
| rz.age | 3 | 81.94948 | 3.857035 | 81.864954 | 92.67753 | UNC5645844 |
| rz.age | 10 | 74.69315 | 3.204739 | 66.318309 | 75.00161 | JAX00292927 |
| rz.age | 10 | 82.34722 | 3.575038 | 81.661568 | 83.57383 | UNC18311938 |
| rz.age | 10 | 84.67176 | 3.567013 | 84.093735 | 85.65568 | UNC18343181 |
| rz.age | 10 | 87.54037 | 3.905120 | 86.109709 | 88.25130 | UNC18376338 |
| rz.age | 10 | 88.92297 | 3.331703 | 88.459680 | 108.74474 | UNCHS029024 |
if(nrow(peaks_mba) < 50 & nrow(peaks_mba) > 0){
for(i in unique(peaks_mba$chr)){
#for (i in 1:nrow(peaks_mba)){
#plot_lod_chr_mb(out,gm$pmap, peaks_mba$chr[i])
plot_lod_chr_mb(out,gm$pmap,i)
}
} else {
print(paste0("There are too many peaks (",nrow(peaks_mba)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}

QTL effects
For each peak LOD location we give a list of gene
if(nrow(peaks) < 50 & nrow(peaks) > 0){
for (i in 1:nrow(peaks)){
#for (i in 1:1){
#Plot 1
g <- maxmarg(pr.qc, gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i], return_char=TRUE)
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/","qtl_effect_", i, ".png"))
#par(mar=c(4.1, 4.1, 1.5, 0.6))
plot_pxg(g, covars[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
##dev.off()
chr = peaks$chr[i]
# Plot 2
pr_sub <- pull_genoprobint(pr.qc, gm$gmap, chr, c(peaks$ci_lo[i], peaks$ci_hi[i]))
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar)
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], Xcovar=Xcovar)
#coeff <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
#coeff_sub <- scan1coef(pr_sub[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
blup <- scan1blup(pr.qc[,chr], covars[peaks$phenotype[i]])
blup_sub <- scan1blup(pr_sub[,chr], covars[peaks$phenotype[i]])
write.csv(as.data.frame(blup_sub), paste0("data/ici.vs.eoi_",peaks$phenotype[i],"-no.covariates_blup_sub_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_5.batches_0.csv"), quote=F)
#plot_coef(coeff,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i]," [scan1coeff; positions in cM] )")
# )
#plot_coef(coeff_sub,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i],"; 1.5 LOD drop interval [scan1coeff; positions in cM] ) ")
# )
plot_coef(blup,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," [scan1blup; positions in cM] )")
)
plot_coef(blup_sub,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i],"; 1.5 LOD drop interval [scan1blup; positions in cM] )")
)
# Plot 3
#c2effB <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary", contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
#c2effBb <- scan1blup(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]],Xcovar=Xcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
#plot(c2effB, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
#plot(c2effBb, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
##last_coef <- unclass(c2effB)[nrow(c2effB),2:3] # last two coefficients
##for(t in seq(along=last_coef))
## axis(side=4, at=last_coef[t], names(last_coef)[t], tick=FALSE)
#Table 1
chr = peaks_mba$chr[i]
start=as.numeric(peaks_mba$ci_lo[i])
end=as.numeric(peaks_mba$ci_hi[i])
genesgss = query_genes(chr, start, end)
write.csv(genesgss, file=paste0("data/ici.vs.eoi_",peaks$phenotype[i],"-no.covariates_genes_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_5.batches_0.csv"), quote=F)
rownames(genesgss) <- NULL
genesgss$strand_old = genesgss$strand
genesgss$strand[genesgss$strand=="+"] <- "positive"
genesgss$strand[genesgss$strand=="-"] <- "negative"
#genesgss <-
#table <-
#genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")] %>%
#kable(escape = F,align = c("ccccccccccc")) %>%
#kable_styling("striped", full_width = T) #%>%
#cat #%>%
#column_spec(1, bold=TRUE)
#
#print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], escape = F,align = c("ccccccccccc")))
print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], "html") %>% kable_styling("striped", full_width = T))
#table
}
} else {
print(paste0("There are too many peaks (",nrow(peaks_mba)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}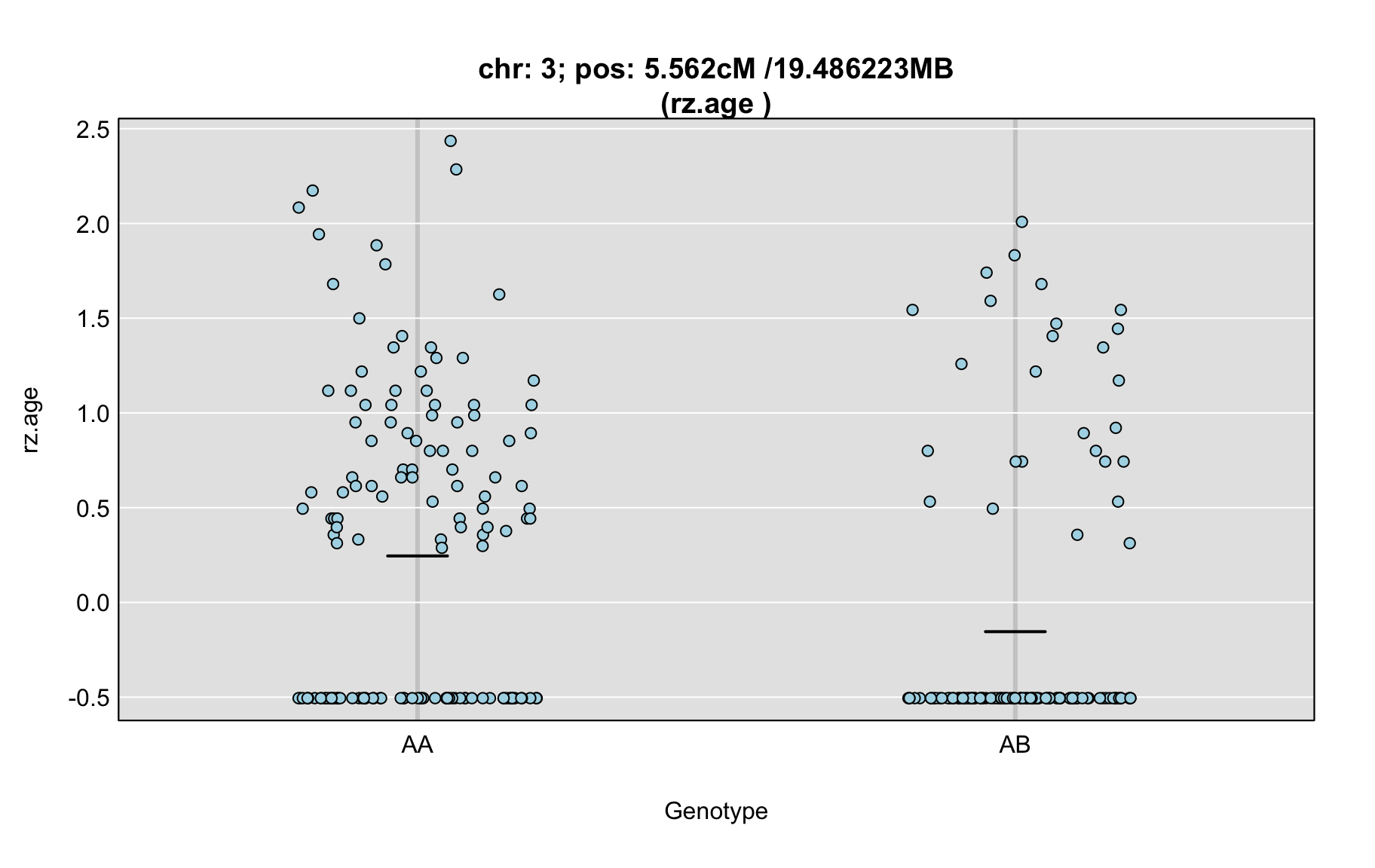


| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | gene | 7.925303 | 7.954889 | negative | MGI_C57BL6J_1922726 | 1700010I02Rik | NCBI_Gene:75476,ENSEMBL:ENSMUSG00000100010 | MGI:1922726 | lncRNA gene | RIKEN cDNA 1700010I02 gene |
| 3 | gene | 8.019669 | 8.022831 | negative | MGI_C57BL6J_3642332 | 5330432E05Rik | NCBI_Gene:383820 | MGI:3642332 | unclassified gene | RIKEN cDNA 5330432E05 gene |
| 3 | pseudogene | 8.214330 | 8.216183 | positive | MGI_C57BL6J_3646860 | Gm7103 | NCBI_Gene:633072,ENSEMBL:ENSMUSG00000103725 | MGI:3646860 | pseudogene | predicted gene 7103 |
| 3 | gene | 8.245438 | 8.245537 | negative | MGI_C57BL6J_3629711 | Gt(pU21)108Imeg | NA | NA | unclassified gene | gene trap 108%2c Institute of Molecular Embryology and Genetics |
| 3 | pseudogene | 8.327322 | 8.329907 | positive | MGI_C57BL6J_3645248 | Gm5841 | NCBI_Gene:545500,ENSEMBL:ENSMUSG00000103921 | MGI:3645248 | pseudogene | predicted gene 5841 |
| 3 | gene | 8.429158 | 8.429264 | positive | MGI_C57BL6J_5454391 | Gm24614 | ENSEMBL:ENSMUSG00000065203 | MGI:5454391 | snRNA gene | predicted gene, 24614 |
| 3 | pseudogene | 8.461387 | 8.463098 | negative | MGI_C57BL6J_3704094 | Gm6194 | NCBI_Gene:620966 | MGI:3704094 | pseudogene | predicted gene 6194 |
| 3 | pseudogene | 8.462228 | 8.463035 | negative | MGI_C57BL6J_1919464 | 1700003N22Rik | ENSEMBL:ENSMUSG00000102463 | MGI:1919464 | pseudogene | RIKEN cDNA 1700003N22 gene |
| 3 | gene | 8.494369 | 8.499902 | negative | MGI_C57BL6J_5624878 | Gm41993 | NCBI_Gene:105246751 | MGI:5624878 | lncRNA gene | predicted gene, 41993 |
| 3 | gene | 8.509360 | 8.561606 | positive | MGI_C57BL6J_98241 | Stmn2 | NCBI_Gene:20257,ENSEMBL:ENSMUSG00000027500 | MGI:98241 | protein coding gene | stathmin-like 2 |
| 3 | pseudogene | 8.615074 | 8.616522 | negative | MGI_C57BL6J_5011007 | Gm18822 | NCBI_Gene:100417782,ENSEMBL:ENSMUSG00000098047 | MGI:5011007 | pseudogene | predicted gene, 18822 |
| 3 | gene | 8.628051 | 8.632454 | positive | MGI_C57BL6J_5591655 | Gm32496 | NCBI_Gene:102635065,ENSEMBL:ENSMUSG00000104102 | MGI:5591655 | lncRNA gene | predicted gene, 32496 |
| 3 | gene | 8.663359 | 8.667256 | negative | MGI_C57BL6J_1341800 | Hey1 | NCBI_Gene:15213,ENSEMBL:ENSMUSG00000040289 | MGI:1341800 | protein coding gene | hairy/enhancer-of-split related with YRPW motif 1 |
| 3 | pseudogene | 8.673410 | 8.673722 | positive | MGI_C57BL6J_5611235 | Gm38007 | ENSEMBL:ENSMUSG00000103814 | MGI:5611235 | pseudogene | predicted gene, 38007 |
| 3 | gene | 8.679359 | 8.679477 | negative | MGI_C57BL6J_5453447 | Gm23670 | ENSEMBL:ENSMUSG00000088094 | MGI:5453447 | snoRNA gene | predicted gene, 23670 |
| 3 | gene | 8.679438 | 8.693098 | negative | MGI_C57BL6J_5591720 | Gm32561 | NCBI_Gene:102635149 | MGI:5591720 | lncRNA gene | predicted gene, 32561 |
| 3 | gene | 8.732188 | 8.736639 | negative | MGI_C57BL6J_2686488 | C230057A21Rik | NCBI_Gene:102635252,ENSEMBL:ENSMUSG00000103346 | MGI:2686488 | lncRNA gene | RIKEN cDNA C230057A21 gene |
| 3 | gene | 8.743677 | 8.746824 | positive | MGI_C57BL6J_1923855 | 1700042O13Rik | NA | NA | lncRNA gene | RIKEN cDNA 1700042O13 gene |
| 3 | gene | 8.743682 | 8.746824 | negative | MGI_C57BL6J_5610875 | Gm37647 | ENSEMBL:ENSMUSG00000103256 | MGI:5610875 | unclassified gene | predicted gene, 37647 |
| 3 | gene | 8.802146 | 8.923918 | negative | MGI_C57BL6J_1913480 | Mrps28 | NCBI_Gene:66230,ENSEMBL:ENSMUSG00000040269 | MGI:1913480 | protein coding gene | mitochondrial ribosomal protein S28 |
| 3 | gene | 8.866119 | 8.881665 | positive | MGI_C57BL6J_5624880 | Gm41995 | NCBI_Gene:105246753 | MGI:5624880 | lncRNA gene | predicted gene, 41995 |
| 3 | pseudogene | 8.881199 | 8.881643 | positive | MGI_C57BL6J_3705388 | Gm15467 | ENSEMBL:ENSMUSG00000081594 | MGI:3705388 | pseudogene | predicted gene 15467 |
| 3 | pseudogene | 8.881990 | 8.882945 | negative | MGI_C57BL6J_3705491 | Gm15466 | NCBI_Gene:100504173,ENSEMBL:ENSMUSG00000081657 | MGI:3705491 | pseudogene | predicted gene 15466 |
| 3 | gene | 8.925590 | 8.928038 | positive | MGI_C57BL6J_1918835 | 9130403I23Rik | NA | NA | unclassified gene | RIKEN cDNA 9130403I23 gene |
| 3 | gene | 8.925593 | 9.004771 | negative | MGI_C57BL6J_107749 | Tpd52 | NCBI_Gene:21985,ENSEMBL:ENSMUSG00000027506 | MGI:107749 | protein coding gene | tumor protein D52 |
| 3 | gene | 9.027570 | 9.029901 | positive | MGI_C57BL6J_5826469 | Gm46832 | NCBI_Gene:108168912 | MGI:5826469 | lncRNA gene | predicted gene, 46832 |
| 3 | pseudogene | 9.047171 | 9.047492 | negative | MGI_C57BL6J_5610844 | Gm37616 | ENSEMBL:ENSMUSG00000103461 | MGI:5610844 | pseudogene | predicted gene, 37616 |
| 3 | gene | 9.061964 | 9.071076 | positive | MGI_C57BL6J_1922399 | 4930539M17Rik | NCBI_Gene:75149,ENSEMBL:ENSMUSG00000103116 | MGI:1922399 | lncRNA gene | RIKEN cDNA 4930539M17 gene |
| 3 | pseudogene | 9.072767 | 9.073211 | positive | MGI_C57BL6J_3648010 | Gm8860 | ENSEMBL:ENSMUSG00000103011 | MGI:3648010 | pseudogene | predicted gene 8860 |
| 3 | gene | 9.157271 | 9.174711 | negative | MGI_C57BL6J_5624881 | Gm41996 | NCBI_Gene:105246754 | MGI:5624881 | lncRNA gene | predicted gene, 41996 |
| 3 | gene | 9.177692 | 9.182437 | negative | MGI_C57BL6J_5592177 | Gm33018 | NCBI_Gene:102635764 | MGI:5592177 | lncRNA gene | predicted gene, 33018 |
| 3 | gene | 9.250567 | 9.285333 | positive | MGI_C57BL6J_2139883 | Zbtb10 | NCBI_Gene:229055,ENSEMBL:ENSMUSG00000069114 | MGI:2139883 | protein coding gene | zinc finger and BTB domain containing 10 |
| 3 | gene | 9.302688 | 9.343835 | negative | MGI_C57BL6J_5624882 | Gm41997 | NCBI_Gene:105246755 | MGI:5624882 | lncRNA gene | predicted gene, 41997 |
| 3 | gene | 9.402175 | 9.402485 | positive | MGI_C57BL6J_5452381 | Gm22604 | ENSEMBL:ENSMUSG00000088229 | MGI:5452381 | unclassified non-coding RNA gene | predicted gene, 22604 |
| 3 | gene | 9.403064 | 9.437233 | positive | MGI_C57BL6J_2444519 | C030034L19Rik | NCBI_Gene:320088,ENSEMBL:ENSMUSG00000097365 | MGI:2444519 | lncRNA gene | RIKEN cDNA C030034L19 gene |
| 3 | gene | 9.427010 | 9.610085 | negative | MGI_C57BL6J_2180715 | Zfp704 | NCBI_Gene:170753,ENSEMBL:ENSMUSG00000040209 | MGI:2180715 | protein coding gene | zinc finger protein 704 |
| 3 | gene | 9.496168 | 9.499928 | positive | MGI_C57BL6J_5611229 | Gm38001 | ENSEMBL:ENSMUSG00000103485 | MGI:5611229 | unclassified gene | predicted gene, 38001 |
| 3 | gene | 9.646323 | 9.663118 | negative | MGI_C57BL6J_5592328 | Gm33169 | NCBI_Gene:102635968 | MGI:5592328 | lncRNA gene | predicted gene, 33169 |
| 3 | pseudogene | 9.656520 | 9.656915 | positive | MGI_C57BL6J_5009992 | Gm17806 | NCBI_Gene:100286840,ENSEMBL:ENSMUSG00000103876 | MGI:5009992 | pseudogene | predicted gene, 17806 |
| 3 | gene | 9.687479 | 9.833679 | negative | MGI_C57BL6J_2443160 | Pag1 | NCBI_Gene:94212,ENSEMBL:ENSMUSG00000027508 | MGI:2443160 | protein coding gene | phosphoprotein associated with glycosphingolipid microdomains 1 |
| 3 | gene | 9.757607 | 9.786142 | positive | MGI_C57BL6J_3840131 | Gm16337 | ENSEMBL:ENSMUSG00000089794 | MGI:3840131 | lncRNA gene | predicted gene 16337 |
| 3 | gene | 9.829620 | 9.829728 | negative | MGI_C57BL6J_5454563 | Gm24786 | ENSEMBL:ENSMUSG00000088004 | MGI:5454563 | rRNA gene | predicted gene, 24786 |
| 3 | gene | 9.880628 | 9.885288 | negative | MGI_C57BL6J_5592432 | Gm33273 | NCBI_Gene:102636109 | MGI:5592432 | lncRNA gene | predicted gene, 33273 |
| 3 | pseudogene | 9.899430 | 9.902377 | positive | MGI_C57BL6J_3779577 | Gm6236 | NCBI_Gene:100534370,ENSEMBL:ENSMUSG00000102795 | MGI:3779577 | pseudogene | predicted gene 6236 |
| 3 | pseudogene | 9.970503 | 9.972010 | negative | MGI_C57BL6J_5010062 | Gm17877 | NCBI_Gene:100416024,ENSEMBL:ENSMUSG00000103698 | MGI:5010062 | pseudogene | predicted gene, 17877 |
| 3 | gene | 10.012548 | 10.016610 | positive | MGI_C57BL6J_101790 | Fabp5 | NCBI_Gene:16592,ENSEMBL:ENSMUSG00000027533 | MGI:101790 | protein coding gene | fatty acid binding protein 5, epidermal |
| 3 | gene | 10.022311 | 10.025539 | negative | MGI_C57BL6J_5611563 | Gm38335 | ENSEMBL:ENSMUSG00000104000 | MGI:5611563 | lncRNA gene | predicted gene, 38335 |
| 3 | gene | 10.043293 | 10.064335 | negative | MGI_C57BL6J_5592524 | Gm33365 | NCBI_Gene:102636246 | MGI:5592524 | lncRNA gene | predicted gene, 33365 |
| 3 | gene | 10.088110 | 10.092565 | positive | MGI_C57BL6J_3641855 | Gm9833 | NCBI_Gene:100041480,ENSEMBL:ENSMUSG00000049230 | MGI:3641855 | protein coding gene | predicted gene 9833 |
| 3 | pseudogene | 10.089856 | 10.089955 | negative | MGI_C57BL6J_5610536 | Gm37308 | ENSEMBL:ENSMUSG00000103045 | MGI:5610536 | pseudogene | predicted gene, 37308 |
| 3 | gene | 10.179851 | 10.183929 | negative | MGI_C57BL6J_102667 | Pmp2 | NCBI_Gene:18857,ENSEMBL:ENSMUSG00000052468 | MGI:102667 | protein coding gene | peripheral myelin protein 2 |
| 3 | gene | 10.179852 | 10.197262 | negative | MGI_C57BL6J_5610617 | Gm37389 | ENSEMBL:ENSMUSG00000103124 | MGI:5610617 | protein coding gene | predicted gene, 37389 |
| 3 | gene | 10.193621 | 10.197283 | negative | MGI_C57BL6J_1194881 | Fabp9 | NCBI_Gene:21884,ENSEMBL:ENSMUSG00000027528 | MGI:1194881 | protein coding gene | fatty acid binding protein 9, testis |
| 3 | gene | 10.204088 | 10.208602 | negative | MGI_C57BL6J_88038 | Fabp4 | NCBI_Gene:11770,ENSEMBL:ENSMUSG00000062515 | MGI:88038 | protein coding gene | fatty acid binding protein 4, adipocyte |
| 3 | gene | 10.220995 | 10.223433 | negative | MGI_C57BL6J_1921296 | 4632432E15Rik | ENSEMBL:ENSMUSG00000102705 | MGI:1921296 | unclassified gene | RIKEN cDNA 4632432E15 gene |
| 3 | pseudogene | 10.232180 | 10.232311 | positive | MGI_C57BL6J_5610649 | Gm37421 | ENSEMBL:ENSMUSG00000104434 | MGI:5610649 | pseudogene | predicted gene, 37421 |
| 3 | gene | 10.241812 | 10.242308 | positive | MGI_C57BL6J_1919533 | 1700029B24Rik | ENSEMBL:ENSMUSG00000102834 | MGI:1919533 | unclassified gene | RIKEN cDNA 1700029B24 gene |
| 3 | gene | 10.244209 | 10.301528 | negative | MGI_C57BL6J_1922747 | Fabp12 | NCBI_Gene:75497,ENSEMBL:ENSMUSG00000027530 | MGI:1922747 | protein coding gene | fatty acid binding protein 12 |
| 3 | gene | 10.296945 | 10.305120 | positive | MGI_C57BL6J_1925848 | A930014E01Rik | NCBI_Gene:102636393,ENSEMBL:ENSMUSG00000104092 | MGI:1925848 | lncRNA gene | RIKEN cDNA A930014E01 gene |
| 3 | pseudogene | 10.309048 | 10.309226 | positive | MGI_C57BL6J_5611592 | Gm38364 | ENSEMBL:ENSMUSG00000103592 | MGI:5611592 | pseudogene | predicted gene, 38364 |
| 3 | gene | 10.311956 | 10.331439 | negative | MGI_C57BL6J_1933158 | Impa1 | NCBI_Gene:55980,ENSEMBL:ENSMUSG00000027531 | MGI:1933158 | protein coding gene | inositol (myo)-1(or 4)-monophosphatase 1 |
| 3 | gene | 10.321712 | 10.335681 | negative | MGI_C57BL6J_5611531 | Gm38303 | ENSEMBL:ENSMUSG00000103392 | MGI:5611531 | protein coding gene | predicted gene, 38303 |
| 3 | gene | 10.331734 | 10.335656 | negative | MGI_C57BL6J_2685251 | Slc10a5 | NCBI_Gene:241877,ENSEMBL:ENSMUSG00000058921 | MGI:2685251 | protein coding gene | solute carrier family 10 (sodium/bile acid cotransporter family), member 5 |
| 3 | gene | 10.339953 | 10.351338 | negative | MGI_C57BL6J_1913611 | Zfand1 | NCBI_Gene:66361,ENSEMBL:ENSMUSG00000039795 | MGI:1913611 | protein coding gene | zinc finger, AN1-type domain 1 |
| 3 | gene | 10.366907 | 10.428856 | positive | MGI_C57BL6J_1913621 | Chmp4c | NCBI_Gene:66371,ENSEMBL:ENSMUSG00000027536 | MGI:1913621 | protein coding gene | charged multivesicular body protein 4C |
| 3 | gene | 10.417817 | 10.440130 | negative | MGI_C57BL6J_1921968 | Snx16 | NCBI_Gene:74718,ENSEMBL:ENSMUSG00000027534 | MGI:1921968 | protein coding gene | sorting nexin 16 |
| 3 | gene | 10.426594 | 10.428866 | positive | MGI_C57BL6J_2679522 | 5430403G19Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 5430403G19 gene |
| 3 | gene | 10.430764 | 10.434207 | negative | MGI_C57BL6J_2445184 | A630040H13Rik | NA | NA | unclassified gene | RIKEN cDNA A630040H13 gene |
| 3 | gene | 10.646316 | 10.648102 | positive | MGI_C57BL6J_5611059 | Gm37831 | ENSEMBL:ENSMUSG00000104451 | MGI:5611059 | unclassified gene | predicted gene, 37831 |
| 3 | pseudogene | 10.753893 | 10.754428 | negative | MGI_C57BL6J_5010877 | Gm18692 | NCBI_Gene:100417569,ENSEMBL:ENSMUSG00000104185 | MGI:5010877 | pseudogene | predicted gene, 18692 |
| 3 | gene | 10.772919 | 10.774899 | negative | MGI_C57BL6J_5610472 | Gm37244 | ENSEMBL:ENSMUSG00000102470 | MGI:5610472 | unclassified gene | predicted gene, 37244 |
| 3 | gene | 10.848895 | 10.857537 | positive | MGI_C57BL6J_5592710 | Gm33551 | NCBI_Gene:102636504 | MGI:5592710 | lncRNA gene | predicted gene, 33551 |
| 3 | gene | 10.975469 | 10.975586 | negative | MGI_C57BL6J_5452572 | Gm22795 | ENSEMBL:ENSMUSG00000087858 | MGI:5452572 | snoRNA gene | predicted gene, 22795 |
| 3 | pseudogene | 11.008846 | 11.009603 | negative | MGI_C57BL6J_5434986 | Gm21631 | NCBI_Gene:100862302,ENSEMBL:ENSMUSG00000103323 | MGI:5434986 | pseudogene | predicted gene, 21631 |
| 3 | pseudogene | 11.013514 | 11.015199 | negative | MGI_C57BL6J_3643310 | Gm8910 | NCBI_Gene:667978 | MGI:3643310 | pseudogene | predicted gene 8910 |
| 3 | pseudogene | 11.044170 | 11.044358 | negative | MGI_C57BL6J_5611060 | Gm37832 | ENSEMBL:ENSMUSG00000081537 | MGI:5611060 | pseudogene | predicted gene, 37832 |
| 3 | gene | 11.154337 | 11.154437 | positive | MGI_C57BL6J_5452324 | Gm22547 | ENSEMBL:ENSMUSG00000095289 | MGI:5452324 | miRNA gene | predicted gene, 22547 |
| 3 | pseudogene | 11.658301 | 11.659519 | negative | MGI_C57BL6J_3643562 | Gm8919 | NCBI_Gene:667999,ENSEMBL:ENSMUSG00000098085 | MGI:3643562 | pseudogene | predicted gene 8919 |
| 3 | gene | 11.823229 | 11.844359 | negative | MGI_C57BL6J_3642274 | Gm10745 | NCBI_Gene:100038545,ENSEMBL:ENSMUSG00000104015 | MGI:3642274 | lncRNA gene | predicted gene 10745 |
| 3 | pseudogene | 12.384243 | 12.389663 | negative | MGI_C57BL6J_5611224 | Gm37996 | NCBI_Gene:108168848,ENSEMBL:ENSMUSG00000103339 | MGI:5611224 | pseudogene | predicted gene, 37996 |
| 3 | pseudogene | 12.521465 | 12.522489 | positive | MGI_C57BL6J_5010616 | Gm18431 | NCBI_Gene:100417159 | MGI:5010616 | pseudogene | predicted gene, 18431 |
| 3 | pseudogene | 12.631258 | 12.632025 | positive | MGI_C57BL6J_3780596 | Gm2429 | NCBI_Gene:100039798,ENSEMBL:ENSMUSG00000104177 | MGI:3780596 | pseudogene | predicted gene 2429 |
| 3 | pseudogene | 12.829051 | 12.831568 | positive | MGI_C57BL6J_5010417 | Gm18232 | NCBI_Gene:100416756 | MGI:5010417 | pseudogene | predicted gene, 18232 |
| 3 | pseudogene | 12.855810 | 12.858633 | positive | MGI_C57BL6J_3643401 | Gm8935 | NCBI_Gene:668033,ENSEMBL:ENSMUSG00000095661 | MGI:3643401 | pseudogene | predicted pseudogene 8935 |
| 3 | pseudogene | 12.892450 | 12.893414 | negative | MGI_C57BL6J_3643399 | Gm8939 | NCBI_Gene:668037,ENSEMBL:ENSMUSG00000103950 | MGI:3643399 | pseudogene | predicted gene 8939 |
| 3 | pseudogene | 12.975359 | 12.975701 | positive | MGI_C57BL6J_5611545 | Gm38317 | ENSEMBL:ENSMUSG00000103735 | MGI:5611545 | pseudogene | predicted gene, 38317 |
| 3 | gene | 13.288197 | 13.291891 | positive | MGI_C57BL6J_5611041 | Gm37813 | ENSEMBL:ENSMUSG00000102815 | MGI:5611041 | unclassified gene | predicted gene, 37813 |
| 3 | pseudogene | 13.429558 | 13.429958 | positive | MGI_C57BL6J_5610425 | Gm37197 | ENSEMBL:ENSMUSG00000104065 | MGI:5610425 | pseudogene | predicted gene, 37197 |
| 3 | gene | 13.470439 | 13.474596 | negative | MGI_C57BL6J_3780631 | Gm2464 | ENSEMBL:ENSMUSG00000097338,ENSEMBL:ENSMUSG00000102215 | MGI:3780631 | lncRNA gene | predicted gene 2464 |
| 3 | gene | 13.471655 | 14.182287 | positive | MGI_C57BL6J_1924147 | Ralyl | NCBI_Gene:76897,ENSEMBL:ENSMUSG00000039717 | MGI:1924147 | protein coding gene | RALY RNA binding protein-like |
| 3 | gene | 13.597139 | 13.652865 | negative | MGI_C57BL6J_1922453 | 4930539N22Rik | NCBI_Gene:75203 | MGI:1922453 | lncRNA gene | RIKEN cDNA 4930539N22 gene |
| 3 | pseudogene | 13.669509 | 13.670145 | negative | MGI_C57BL6J_5610392 | Gm37164 | ENSEMBL:ENSMUSG00000104514 | MGI:5610392 | pseudogene | predicted gene, 37164 |
| 3 | pseudogene | 13.749938 | 13.750512 | positive | MGI_C57BL6J_3780641 | Gm2474 | NCBI_Gene:100039882,ENSEMBL:ENSMUSG00000103856 | MGI:3780641 | pseudogene | predicted gene 2474 |
| 3 | pseudogene | 13.838875 | 13.840343 | negative | MGI_C57BL6J_5011299 | Gm19114 | NCBI_Gene:100418280,ENSEMBL:ENSMUSG00000102891 | MGI:5011299 | pseudogene | predicted gene, 19114 |
| 3 | gene | 13.845686 | 13.845980 | positive | MGI_C57BL6J_5455758 | Gm25981 | ENSEMBL:ENSMUSG00000088546 | MGI:5455758 | unclassified non-coding RNA gene | predicted gene, 25981 |
| 3 | gene | 13.847735 | 13.850864 | positive | MGI_C57BL6J_2444502 | C230073O14Rik | NA | NA | unclassified gene | RIKEN cDNA C230073O14 gene |
| 3 | gene | 13.914161 | 13.934435 | negative | MGI_C57BL6J_5624883 | Gm41998 | NCBI_Gene:105246756 | MGI:5624883 | lncRNA gene | predicted gene, 41998 |
| 3 | gene | 14.029482 | 14.029762 | negative | MGI_C57BL6J_5451803 | Gm22026 | ENSEMBL:ENSMUSG00000084684 | MGI:5451803 | unclassified non-coding RNA gene | predicted gene, 22026 |
| 3 | pseudogene | 14.098042 | 14.099016 | positive | MGI_C57BL6J_5010846 | Gm18661 | NCBI_Gene:100417513 | MGI:5010846 | pseudogene | predicted gene, 18661 |
| 3 | pseudogene | 14.150147 | 14.150785 | positive | MGI_C57BL6J_5011252 | Gm19067 | NCBI_Gene:100418194 | MGI:5011252 | pseudogene | predicted gene, 19067 |
| 3 | pseudogene | 14.163685 | 14.164632 | negative | MGI_C57BL6J_5010844 | Gm18659 | NCBI_Gene:100417511 | MGI:5010844 | pseudogene | predicted gene, 18659 |
| 3 | gene | 14.267315 | 14.267439 | negative | MGI_C57BL6J_5452889 | Gm23112 | ENSEMBL:ENSMUSG00000089231 | MGI:5452889 | snoRNA gene | predicted gene, 23112 |
| 3 | pseudogene | 14.330227 | 14.330391 | positive | MGI_C57BL6J_5611539 | Gm38311 | ENSEMBL:ENSMUSG00000102707 | MGI:5611539 | pseudogene | predicted gene, 38311 |
| 3 | pseudogene | 14.363117 | 14.377587 | positive | MGI_C57BL6J_3648364 | Gm6300 | NCBI_Gene:622229,ENSEMBL:ENSMUSG00000104375 | MGI:3648364 | pseudogene | predicted gene 6300 |
| 3 | pseudogene | 14.383901 | 14.393393 | negative | MGI_C57BL6J_5610586 | Gm37358 | NCBI_Gene:108168849,ENSEMBL:ENSMUSG00000103626 | MGI:5610586 | pseudogene | predicted gene, 37358 |
| 3 | pseudogene | 14.428283 | 14.435027 | negative | MGI_C57BL6J_3645918 | Gm8955 | NCBI_Gene:668062,ENSEMBL:ENSMUSG00000098036 | MGI:3645918 | pseudogene | predicted gene 8955 |
| 3 | gene | 14.480691 | 14.505830 | positive | MGI_C57BL6J_2156159 | Slc7a12 | NCBI_Gene:140918,ENSEMBL:ENSMUSG00000039710 | MGI:2156159 | protein coding gene | solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| 3 | pseudogene | 14.510888 | 14.512113 | negative | MGI_C57BL6J_3779827 | Gm8957 | NCBI_Gene:668068,ENSEMBL:ENSMUSG00000098083 | MGI:3779827 | pseudogene | predicted gene 8957 |
| 3 | gene | 14.533788 | 14.572658 | positive | MGI_C57BL6J_1918960 | Lrrcc1 | NCBI_Gene:71710,ENSEMBL:ENSMUSG00000027550 | MGI:1918960 | protein coding gene | leucine rich repeat and coiled-coil domain containing 1 |
| 3 | pseudogene | 14.564402 | 14.564681 | positive | MGI_C57BL6J_4937946 | Gm17119 | ENSEMBL:ENSMUSG00000090300 | MGI:4937946 | pseudogene | predicted gene 17119 |
| 3 | gene | 14.578602 | 14.606309 | positive | MGI_C57BL6J_105091 | E2f5 | NCBI_Gene:13559,ENSEMBL:ENSMUSG00000027552 | MGI:105091 | protein coding gene | E2F transcription factor 5 |
| 3 | gene | 14.606284 | 14.611285 | negative | MGI_C57BL6J_1916376 | Rbis | NCBI_Gene:69126,ENSEMBL:ENSMUSG00000078784 | MGI:1916376 | protein coding gene | ribosomal biogenesis factor |
| 3 | pseudogene | 14.628952 | 14.629240 | negative | MGI_C57BL6J_5611379 | Gm38151 | ENSEMBL:ENSMUSG00000104249 | MGI:5611379 | pseudogene | predicted gene, 38151 |
| 3 | gene | 14.641727 | 14.663002 | positive | MGI_C57BL6J_1931322 | Car13 | NCBI_Gene:71934,ENSEMBL:ENSMUSG00000027555 | MGI:1931322 | protein coding gene | carbonic anhydrase 13 |
| 3 | pseudogene | 14.722415 | 14.729626 | positive | MGI_C57BL6J_3645250 | Gm5843 | NCBI_Gene:545507,ENSEMBL:ENSMUSG00000084092 | MGI:3645250 | pseudogene | predicted gene 5843 |
| 3 | gene | 14.766214 | 14.808368 | negative | MGI_C57BL6J_88268 | Car1 | NCBI_Gene:12346,ENSEMBL:ENSMUSG00000027556 | MGI:88268 | protein coding gene | carbonic anhydrase 1 |
| 3 | pseudogene | 14.802215 | 14.802632 | negative | MGI_C57BL6J_3645963 | Gm16399 | NCBI_Gene:100041785,ENSEMBL:ENSMUSG00000097790 | MGI:3645963 | pseudogene | predicted pseudogene 16399 |
| 3 | gene | 14.825509 | 14.864243 | negative | MGI_C57BL6J_5624884 | Gm41999 | NCBI_Gene:105246757 | MGI:5624884 | lncRNA gene | predicted gene, 41999 |
| 3 | gene | 14.863512 | 14.872523 | positive | MGI_C57BL6J_88270 | Car3 | NCBI_Gene:12350,ENSEMBL:ENSMUSG00000027559 | MGI:88270 | protein coding gene | carbonic anhydrase 3 |
| 3 | gene | 14.886273 | 14.900770 | positive | MGI_C57BL6J_88269 | Car2 | NCBI_Gene:12349,ENSEMBL:ENSMUSG00000027562 | MGI:88269 | protein coding gene | carbonic anhydrase 2 |
| 3 | gene | 14.971201 | 15.002727 | positive | MGI_C57BL6J_1924377 | A930001A20Rik | NCBI_Gene:77127,ENSEMBL:ENSMUSG00000098008 | MGI:1924377 | lncRNA gene | RIKEN cDNA A930001A20 gene |
| 3 | pseudogene | 14.973368 | 14.974570 | negative | MGI_C57BL6J_3647002 | Gm7132 | NCBI_Gene:634323,ENSEMBL:ENSMUSG00000098186 | MGI:3647002 | pseudogene | predicted gene 7132 |
| 3 | gene | 15.055968 | 15.061831 | positive | MGI_C57BL6J_5624885 | Gm42000 | NCBI_Gene:105246758 | MGI:5624885 | lncRNA gene | predicted gene, 42000 |
| 3 | gene | 15.153012 | 15.171442 | negative | MGI_C57BL6J_5592978 | Gm33819 | NCBI_Gene:102636866,ENSEMBL:ENSMUSG00000103912 | MGI:5592978 | lncRNA gene | predicted gene, 33819 |
| 3 | gene | 15.295103 | 15.332310 | negative | MGI_C57BL6J_3780136 | Gm9733 | NCBI_Gene:751864,ENSEMBL:ENSMUSG00000078783 | MGI:3780136 | protein coding gene | predicted gene 9733 |
| 3 | gene | 15.371653 | 15.426522 | negative | MGI_C57BL6J_2444824 | Sirpb1a | NCBI_Gene:320832,ENSEMBL:ENSMUSG00000095788 | MGI:2444824 | protein coding gene | signal-regulatory protein beta 1A |
| 3 | gene | 15.495751 | 15.575067 | negative | MGI_C57BL6J_3779828 | Sirpb1b | NCBI_Gene:668101,ENSEMBL:ENSMUSG00000095028 | MGI:3779828 | protein coding gene | signal-regulatory protein beta 1B |
| 3 | pseudogene | 15.514358 | 15.515637 | positive | MGI_C57BL6J_5434369 | Gm21014 | NCBI_Gene:100534286 | MGI:5434369 | pseudogene | predicted gene, 21014 |
| 3 | gene | 15.547479 | 15.548599 | positive | MGI_C57BL6J_5610696 | Gm37468 | ENSEMBL:ENSMUSG00000102155 | MGI:5610696 | lncRNA gene | predicted gene, 37468 |
| 3 | gene | 15.795145 | 15.848530 | negative | MGI_C57BL6J_3807521 | Sirpb1c | NCBI_Gene:100038947,ENSEMBL:ENSMUSG00000074677 | MGI:3807521 | protein coding gene | signal-regulatory protein beta 1C |
| 3 | pseudogene | 15.891327 | 15.892583 | negative | MGI_C57BL6J_3648569 | Gm6321 | NCBI_Gene:622425,ENSEMBL:ENSMUSG00000103688 | MGI:3648569 | pseudogene | predicted gene 6321 |
| 3 | pseudogene | 15.941772 | 15.942833 | negative | MGI_C57BL6J_3648493 | Gm7343 | NCBI_Gene:664795,ENSEMBL:ENSMUSG00000103769 | MGI:3648493 | pseudogene | predicted gene 7343 |
| 3 | gene | 15.946870 | 16.006437 | negative | MGI_C57BL6J_3779469 | Gm5150 | NCBI_Gene:381484,ENSEMBL:ENSMUSG00000078780 | MGI:3779469 | protein coding gene | predicted gene 5150 |
| 3 | gene | 16.013818 | 16.013927 | positive | MGI_C57BL6J_5454471 | Gm24694 | ENSEMBL:ENSMUSG00000094598 | MGI:5454471 | miRNA gene | predicted gene, 24694 |
| 3 | pseudogene | 16.078150 | 16.078324 | negative | MGI_C57BL6J_5611064 | Gm37836 | ENSEMBL:ENSMUSG00000103535 | MGI:5611064 | pseudogene | predicted gene, 37836 |
| 3 | gene | 16.115615 | 16.115943 | negative | MGI_C57BL6J_5455640 | Gm25863 | ENSEMBL:ENSMUSG00000088874 | MGI:5455640 | ribozyme gene | predicted gene, 25863 |
| 3 | gene | 16.183141 | 16.217037 | positive | MGI_C57BL6J_1918850 | Ythdf3 | NCBI_Gene:229096,ENSEMBL:ENSMUSG00000047213 | MGI:1918850 | protein coding gene | YTH N6-methyladenosine RNA binding protein 3 |
| 3 | gene | 16.210483 | 16.211928 | positive | MGI_C57BL6J_1923346 | 5830468K08Rik | NA | NA | unclassified gene | RIKEN cDNA 5830468K08 gene |
| 3 | gene | 16.817078 | 16.847955 | positive | MGI_C57BL6J_5589382 | Gm30223 | NCBI_Gene:102632048 | MGI:5589382 | lncRNA gene | predicted gene, 30223 |
| 3 | gene | 16.823249 | 16.823312 | negative | MGI_C57BL6J_5456262 | Gm26485 | ENSEMBL:ENSMUSG00000088644 | MGI:5456262 | snRNA gene | predicted gene, 26485 |
| 3 | gene | 16.862931 | 16.902974 | positive | MGI_C57BL6J_5625069 | Gm42184 | NCBI_Gene:105246998 | MGI:5625069 | lncRNA gene | predicted gene, 42184 |
| 3 | gene | 17.002671 | 17.069484 | negative | MGI_C57BL6J_5625070 | Gm42185 | NCBI_Gene:105246999 | MGI:5625070 | lncRNA gene | predicted gene, 42185 |
| 3 | gene | 17.117133 | 17.139762 | negative | MGI_C57BL6J_5625071 | Gm42186 | NCBI_Gene:105247001 | MGI:5625071 | lncRNA gene | predicted gene, 42186 |
| 3 | pseudogene | 17.230436 | 17.231051 | negative | MGI_C57BL6J_3643834 | Gm5283 | NCBI_Gene:383936,ENSEMBL:ENSMUSG00000095827 | MGI:3643834 | pseudogene | predicted gene 5283 |
| 3 | gene | 17.251301 | 17.251444 | negative | MGI_C57BL6J_5454740 | Gm24963 | ENSEMBL:ENSMUSG00000088541 | MGI:5454740 | snoRNA gene | predicted gene, 24963 |
| 3 | pseudogene | 17.269422 | 17.271236 | positive | MGI_C57BL6J_3643969 | Gm6350 | NCBI_Gene:622737,ENSEMBL:ENSMUSG00000103351 | MGI:3643969 | pseudogene | predicted gene 6350 |
| 3 | pseudogene | 17.283151 | 17.284141 | positive | MGI_C57BL6J_5011122 | Gm18937 | NCBI_Gene:100417989 | MGI:5011122 | pseudogene | predicted gene, 18937 |
| 3 | gene | 17.321506 | 17.785980 | negative | MGI_C57BL6J_5589499 | Gm30340 | NCBI_Gene:102632200,ENSEMBL:ENSMUSG00000104411 | MGI:5589499 | lncRNA gene | predicted gene, 30340 |
| 3 | pseudogene | 17.395820 | 17.397734 | positive | MGI_C57BL6J_5011879 | Gm19694 | NCBI_Gene:108168858,ENSEMBL:ENSMUSG00000102646 | MGI:5011879 | pseudogene | predicted gene, 19694 |
| 3 | pseudogene | 17.670688 | 17.671027 | positive | MGI_C57BL6J_5611382 | Gm38154 | ENSEMBL:ENSMUSG00000104242 | MGI:5611382 | pseudogene | predicted gene, 38154 |
| 3 | gene | 17.783514 | 17.786898 | negative | MGI_C57BL6J_3642301 | Gm10742 | NA | NA | unclassified gene | predicted gene 10742 |
| 3 | gene | 17.789921 | 17.808970 | positive | MGI_C57BL6J_1917691 | Mir124-2hg | NCBI_Gene:70441,ENSEMBL:ENSMUSG00000100252 | MGI:1917691 | lncRNA gene | Mir124-2 host gene (non-protein coding) |
| 3 | gene | 17.795662 | 17.795770 | positive | MGI_C57BL6J_3618700 | Mir124a-2 | miRBase:MI0000717,NCBI_Gene:723950,ENSEMBL:ENSMUSG00000093073 | MGI:3618700 | miRNA gene | microRNA 124a-2 |
| 3 | gene | 17.803621 | 17.803747 | negative | MGI_C57BL6J_5453218 | Gm23441 | ENSEMBL:ENSMUSG00000087893 | MGI:5453218 | snRNA gene | predicted gene, 23441 |
| 3 | gene | 17.818728 | 17.853402 | positive | MGI_C57BL6J_5625072 | Gm42187 | NCBI_Gene:105247002 | MGI:5625072 | lncRNA gene | predicted gene, 42187 |
| 3 | gene | 17.829276 | 17.831396 | positive | MGI_C57BL6J_1921828 | 4833408G04Rik | NA | NA | unclassified gene | RIKEN cDNA 4833408G04 gene |
| 3 | pseudogene | 17.923706 | 17.924626 | positive | MGI_C57BL6J_5011360 | Gm19175 | NCBI_Gene:100418385,ENSEMBL:ENSMUSG00000105262 | MGI:5011360 | pseudogene | predicted gene, 19175 |
| 3 | gene | 17.948410 | 17.948959 | positive | MGI_C57BL6J_1922689 | Cypt12 | NCBI_Gene:75439,ENSEMBL:ENSMUSG00000027564 | MGI:1922689 | protein coding gene | cysteine-rich perinuclear theca 12 |
| 3 | pseudogene | 18.008748 | 18.011501 | positive | MGI_C57BL6J_3645252 | Gm5844 | NCBI_Gene:545508,ENSEMBL:ENSMUSG00000082896 | MGI:3645252 | pseudogene | predicted gene 5844 |
| 3 | gene | 18.054174 | 18.057517 | positive | MGI_C57BL6J_1930001 | Bhlhe22 | NCBI_Gene:59058,ENSEMBL:ENSMUSG00000025128 | MGI:1930001 | protein coding gene | basic helix-loop-helix family, member e22 |
| 3 | gene | 18.069326 | 18.069392 | positive | MGI_C57BL6J_5453503 | Gm23726 | ENSEMBL:ENSMUSG00000094983 | MGI:5453503 | miRNA gene | predicted gene, 23726 |
| 3 | gene | 18.071944 | 18.243890 | negative | MGI_C57BL6J_104978 | Cyp7b1 | NCBI_Gene:13123,ENSEMBL:ENSMUSG00000039519 | MGI:104978 | protein coding gene | cytochrome P450, family 7, subfamily b, polypeptide 1 |
| 3 | gene | 18.177462 | 18.177625 | negative | MGI_C57BL6J_5453463 | Gm23686 | ENSEMBL:ENSMUSG00000096391 | MGI:5453463 | snRNA gene | predicted gene, 23686 |
| 3 | gene | 18.299310 | 18.304139 | positive | MGI_C57BL6J_5663971 | Gm43834 | ENSEMBL:ENSMUSG00000104801 | MGI:5663971 | unclassified gene | predicted gene 43834 |
| 3 | gene | 18.341667 | 18.461341 | negative | MGI_C57BL6J_5589826 | Gm30667 | NCBI_Gene:102632645,ENSEMBL:ENSMUSG00000102886 | MGI:5589826 | lncRNA gene | predicted gene, 30667 |
| 3 | gene | 18.468285 | 18.480477 | negative | MGI_C57BL6J_1921209 | 4930433B08Rik | NCBI_Gene:73959,ENSEMBL:ENSMUSG00000102699 | MGI:1921209 | lncRNA gene | RIKEN cDNA 4930433B08 gene |
| 3 | pseudogene | 18.494617 | 18.534289 | positive | MGI_C57BL6J_3644758 | Gm6369 | NCBI_Gene:622921,ENSEMBL:ENSMUSG00000040217 | MGI:3644758 | pseudogene | predicted gene 6369 |
| 3 | pseudogene | 18.564493 | 18.566138 | positive | MGI_C57BL6J_97558 | Pgk1-ps3 | NCBI_Gene:100416868,ENSEMBL:ENSMUSG00000105280 | MGI:97558 | pseudogene | phosphoglycerate kinase 1, pseudogene 3 |
| 3 | gene | 18.609475 | 18.617380 | negative | MGI_C57BL6J_5589882 | Gm30723 | NCBI_Gene:102632727 | MGI:5589882 | lncRNA gene | predicted gene, 30723 |
| 3 | pseudogene | 18.609808 | 18.610831 | negative | MGI_C57BL6J_5610545 | Gm37317 | ENSEMBL:ENSMUSG00000104091 | MGI:5610545 | pseudogene | predicted gene, 37317 |
| 3 | gene | 18.619549 | 18.643095 | negative | MGI_C57BL6J_5589948 | Gm30789 | NCBI_Gene:102632812 | MGI:5589948 | lncRNA gene | predicted gene, 30789 |
| 3 | gene | 18.622162 | 18.626141 | positive | MGI_C57BL6J_5663081 | Gm42944 | ENSEMBL:ENSMUSG00000104918 | MGI:5663081 | unclassified gene | predicted gene 42944 |
| 3 | pseudogene | 18.753076 | 18.753404 | positive | MGI_C57BL6J_5589500 | Gm30341 | NCBI_Gene:102632201,ENSEMBL:ENSMUSG00000103264 | MGI:5589500 | pseudogene | predicted gene, 30341 |
| 3 | gene | 19.131402 | 19.163090 | negative | MGI_C57BL6J_1921502 | Armc1 | NCBI_Gene:74252,ENSEMBL:ENSMUSG00000027599 | MGI:1921502 | protein coding gene | armadillo repeat containing 1 |
| 3 | gene | 19.143188 | 19.143899 | negative | MGI_C57BL6J_5611207 | Gm37979 | ENSEMBL:ENSMUSG00000103135 | MGI:5611207 | unclassified gene | predicted gene, 37979 |
| 3 | gene | 19.187314 | 19.220817 | positive | MGI_C57BL6J_1914722 | Mtfr1 | NCBI_Gene:67472,ENSEMBL:ENSMUSG00000027601 | MGI:1914722 | protein coding gene | mitochondrial fission regulator 1 |
| 3 | gene | 19.223108 | 19.311322 | negative | MGI_C57BL6J_1202402 | Pde7a | NCBI_Gene:18583,ENSEMBL:ENSMUSG00000069094 | MGI:1202402 | protein coding gene | phosphodiesterase 7A |
| 3 | gene | 19.223115 | 19.223693 | negative | MGI_C57BL6J_1921035 | 4930431H11Rik | NA | NA | unclassified gene | RIKEN cDNA 4930431H11 gene |
| 3 | gene | 19.227780 | 19.240987 | positive | MGI_C57BL6J_3801946 | Gm16093 | ENSEMBL:ENSMUSG00000087108 | MGI:3801946 | lncRNA gene | predicted gene 16093 |
| 3 | gene | 19.311660 | 19.313735 | positive | MGI_C57BL6J_1920375 | 3110037C07Rik | NA | NA | unclassified gene | RIKEN cDNA 3110037C07 gene |
| 3 | gene | 19.330773 | 19.331084 | positive | MGI_C57BL6J_5452059 | Gm22282 | ENSEMBL:ENSMUSG00000089149 | MGI:5452059 | unclassified non-coding RNA gene | predicted gene, 22282 |
| 3 | gene | 19.374179 | 19.387634 | positive | MGI_C57BL6J_5625073 | Gm42188 | NCBI_Gene:105247004 | MGI:5625073 | lncRNA gene | predicted gene, 42188 |
| 3 | gene | 19.508591 | 19.610862 | positive | MGI_C57BL6J_1913576 | Dnajc5b | NCBI_Gene:66326,ENSEMBL:ENSMUSG00000027606 | MGI:1913576 | protein coding gene | DnaJ heat shock protein family (Hsp40) member C5 beta |
| 3 | gene | 19.560253 | 19.560416 | positive | MGI_C57BL6J_5453107 | Gm23330 | ENSEMBL:ENSMUSG00000096211 | MGI:5453107 | snRNA gene | predicted gene, 23330 |
| 3 | pseudogene | 19.584791 | 19.585542 | positive | MGI_C57BL6J_3646012 | Gm6141 | NCBI_Gene:620290,ENSEMBL:ENSMUSG00000084339 | MGI:3646012 | pseudogene | predicted pseudogene 6141 |
| 3 | gene | 19.610368 | 19.629477 | negative | MGI_C57BL6J_1920674 | 1700064H15Rik | NCBI_Gene:102633032,ENSEMBL:ENSMUSG00000082766 | MGI:1920674 | protein coding gene | RIKEN cDNA 1700064H15 gene |
| 3 | gene | 19.627959 | 19.628031 | negative | MGI_C57BL6J_4413716 | n-TRacg3 | NCBI_Gene:102467637 | MGI:4413716 | tRNA gene | nuclear encoded tRNA arginine 3 (anticodon ACG) |
| 3 | gene | 19.628355 | 19.628447 | positive | MGI_C57BL6J_4414074 | n-TYgta1 | NCBI_Gene:102467442 | MGI:4414074 | tRNA gene | nuclear encoded tRNA tyrosine 1 (anticodon GTA) |
| 3 | gene | 19.628782 | 19.628870 | positive | MGI_C57BL6J_4414075 | n-TYgta2 | NCBI_Gene:102467395 | MGI:4414075 | tRNA gene | nuclear encoded tRNA tyrosine 2 (anticodon GTA) |
| 3 | gene | 19.628995 | 19.629067 | positive | MGI_C57BL6J_4413687 | n-TAagc14 | NCBI_Gene:102467233 | MGI:4413687 | tRNA gene | nuclear encoded tRNA alanine 14 (anticodon AGC) |
| 3 | gene | 19.644343 | 19.692421 | positive | MGI_C57BL6J_3036269 | Trim55 | NCBI_Gene:381485,ENSEMBL:ENSMUSG00000060913 | MGI:3036269 | protein coding gene | tripartite motif-containing 55 |
| 3 | gene | 19.685971 | 19.686568 | positive | MGI_C57BL6J_3642300 | BC002189 | ENSEMBL:ENSMUSG00000104465 | MGI:3642300 | lncRNA gene | cDNA sequence BC002189 |
| 3 | gene | 19.693401 | 19.695396 | negative | MGI_C57BL6J_88496 | Crh | NCBI_Gene:12918,ENSEMBL:ENSMUSG00000049796 | MGI:88496 | protein coding gene | corticotropin releasing hormone |
| 3 | gene | 19.694486 | 19.707354 | positive | MGI_C57BL6J_5625074 | Gm42189 | NCBI_Gene:105247005 | MGI:5625074 | lncRNA gene | predicted gene, 42189 |
| 3 | pseudogene | 19.708253 | 19.709320 | negative | MGI_C57BL6J_5011049 | Gm18864 | NCBI_Gene:100417853,ENSEMBL:ENSMUSG00000102261 | MGI:5011049 | pseudogene | predicted gene, 18864 |
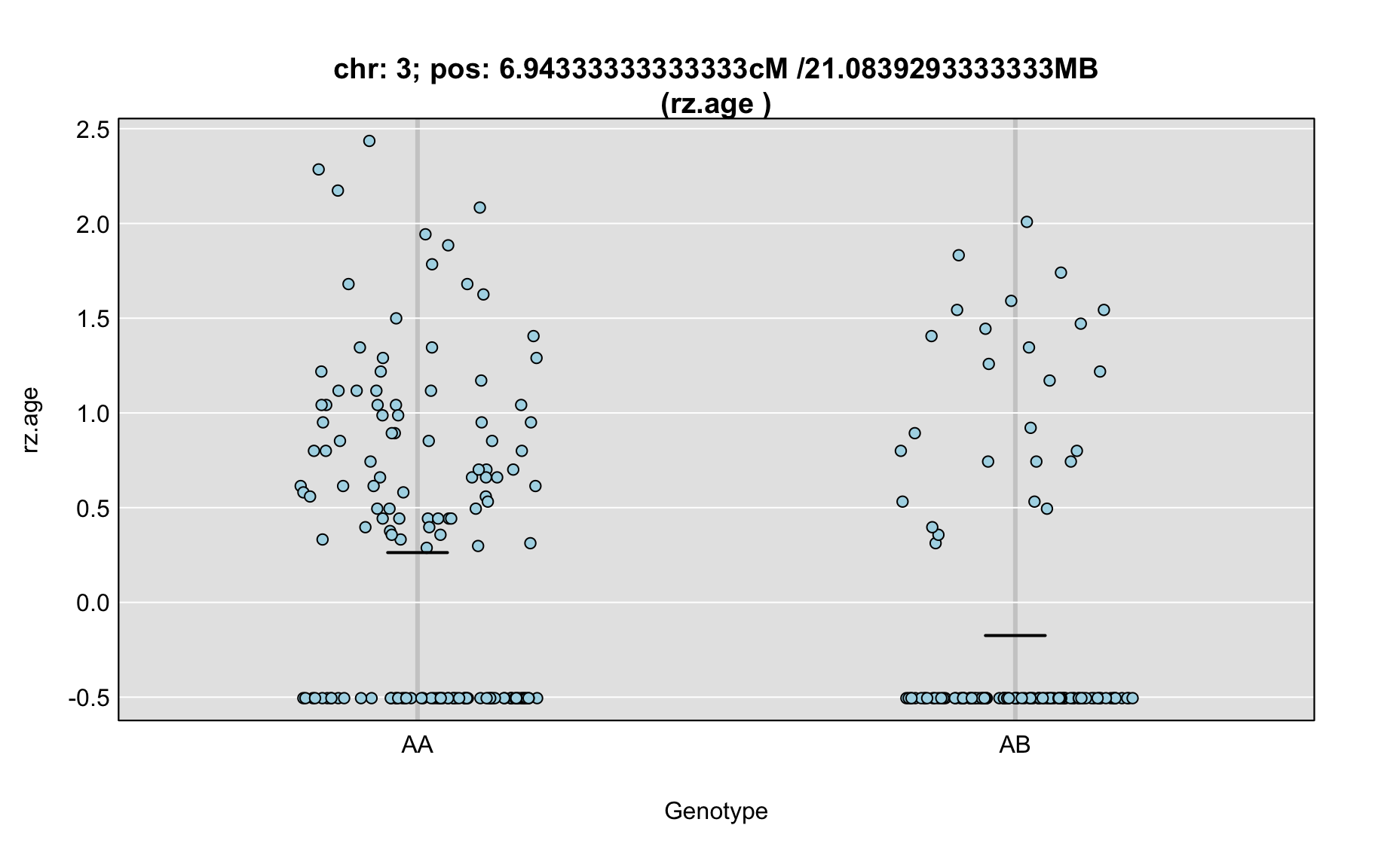

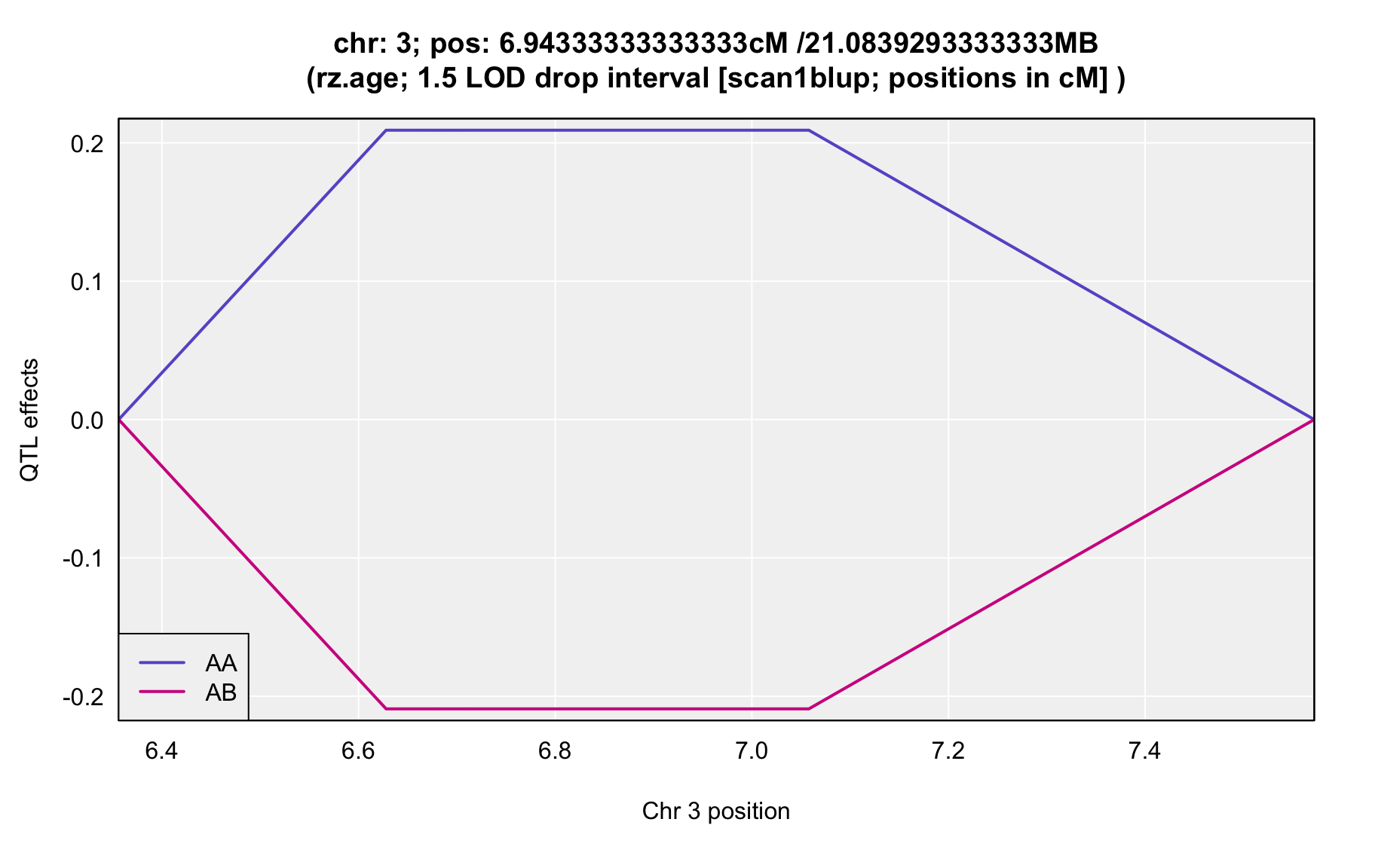
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | gene | 20.30841 | 20.30992 | positive | MGI_C57BL6J_5663662 | Gm43525 | ENSEMBL:ENSMUSG00000105208 | MGI:5663662 | lncRNA gene | predicted gene 43525 |
| 3 | gene | 20.31447 | 20.36718 | negative | MGI_C57BL6J_87965 | Agtr1b | NCBI_Gene:11608,ENSEMBL:ENSMUSG00000054988 | MGI:87965 | protein coding gene | angiotensin II receptor, type 1b |
| 3 | pseudogene | 20.39101 | 20.39198 | negative | MGI_C57BL6J_5010611 | Gm18426 | NCBI_Gene:100417153,ENSEMBL:ENSMUSG00000103573 | MGI:5010611 | pseudogene | predicted gene, 18426 |
| 3 | gene | 20.40250 | 20.50621 | negative | MGI_C57BL6J_5590625 | Gm31466 | NCBI_Gene:108168860,ENSEMBL:ENSMUSG00000102636 | MGI:5590625 | lncRNA gene | predicted gene, 31466 |
| 3 | gene | 20.40467 | 20.40491 | positive | MGI_C57BL6J_5662731 | Gm42594 | ENSEMBL:ENSMUSG00000106442 | MGI:5662731 | unclassified gene | predicted gene 42594 |
| 3 | pseudogene | 20.78247 | 20.78407 | negative | MGI_C57BL6J_5010676 | Gm18491 | NCBI_Gene:100417265,ENSEMBL:ENSMUSG00000103460 | MGI:5010676 | pseudogene | predicted gene, 18491 |
| 3 | pseudogene | 20.78794 | 20.78815 | positive | MGI_C57BL6J_5611551 | Gm38323 | ENSEMBL:ENSMUSG00000102986 | MGI:5611551 | pseudogene | predicted gene, 38323 |
| 3 | pseudogene | 20.80448 | 20.80609 | positive | MGI_C57BL6J_5010678 | Gm18493 | NCBI_Gene:100417268,ENSEMBL:ENSMUSG00000103533 | MGI:5010678 | pseudogene | predicted gene, 18493 |
| 3 | gene | 20.81639 | 20.82236 | negative | MGI_C57BL6J_5590711 | Gm31552 | NCBI_Gene:102633820 | MGI:5590711 | lncRNA gene | predicted gene, 31552 |
| 3 | gene | 20.81962 | 20.82369 | positive | MGI_C57BL6J_5610708 | Gm37480 | ENSEMBL:ENSMUSG00000102807 | MGI:5610708 | unclassified gene | predicted gene, 37480 |
| 3 | pseudogene | 21.04945 | 21.05053 | positive | MGI_C57BL6J_3647006 | Gm7488 | NCBI_Gene:665094,ENSEMBL:ENSMUSG00000039617 | MGI:3647006 | pseudogene | predicted gene 7488 |
| 3 | pseudogene | 21.11358 | 21.11400 | negative | MGI_C57BL6J_5663297 | Gm43160 | ENSEMBL:ENSMUSG00000106051 | MGI:5663297 | pseudogene | predicted gene 43160 |
| 3 | gene | 21.26837 | 21.26909 | negative | MGI_C57BL6J_5579843 | Gm29137 | ENSEMBL:ENSMUSG00000101897 | MGI:5579843 | lncRNA gene | predicted gene 29137 |
| 3 | gene | 21.42179 | 21.48966 | negative | MGI_C57BL6J_5590769 | Gm31610 | NCBI_Gene:102633894 | MGI:5590769 | lncRNA gene | predicted gene, 31610 |
| 3 | pseudogene | 21.60381 | 21.60551 | positive | MGI_C57BL6J_5663812 | Gm43675 | ENSEMBL:ENSMUSG00000106085 | MGI:5663812 | pseudogene | predicted gene 43675 |
| 3 | pseudogene | 21.64839 | 21.64918 | positive | MGI_C57BL6J_5010120 | Gm17935 | NCBI_Gene:100416137,ENSEMBL:ENSMUSG00000106460 | MGI:5010120 | pseudogene | predicted gene, 17935 |
| 3 | gene | 21.70282 | 21.71246 | positive | MGI_C57BL6J_5590852 | Gm31693 | NCBI_Gene:102634003,ENSEMBL:ENSMUSG00000105440 | MGI:5590852 | lncRNA gene | predicted gene, 31693 |
| 3 | gene | 21.75353 | 21.80080 | negative | MGI_C57BL6J_2442162 | 7530428D23Rik | NCBI_Gene:319506,ENSEMBL:ENSMUSG00000103441 | MGI:2442162 | lncRNA gene | RIKEN cDNA 7530428D23 gene |
| 3 | gene | 21.86417 | 21.96435 | negative | MGI_C57BL6J_5590972 | Gm31813 | NCBI_Gene:102634161 | MGI:5590972 | lncRNA gene | predicted gene, 31813 |
| 3 | gene | 21.99768 | 21.99847 | negative | MGI_C57BL6J_5663811 | Gm43674 | ENSEMBL:ENSMUSG00000106321 | MGI:5663811 | lncRNA gene | predicted gene 43674 |
| 3 | pseudogene | 22.02120 | 22.02163 | positive | MGI_C57BL6J_5663808 | Gm43671 | ENSEMBL:ENSMUSG00000106527 | MGI:5663808 | pseudogene | predicted gene 43671 |
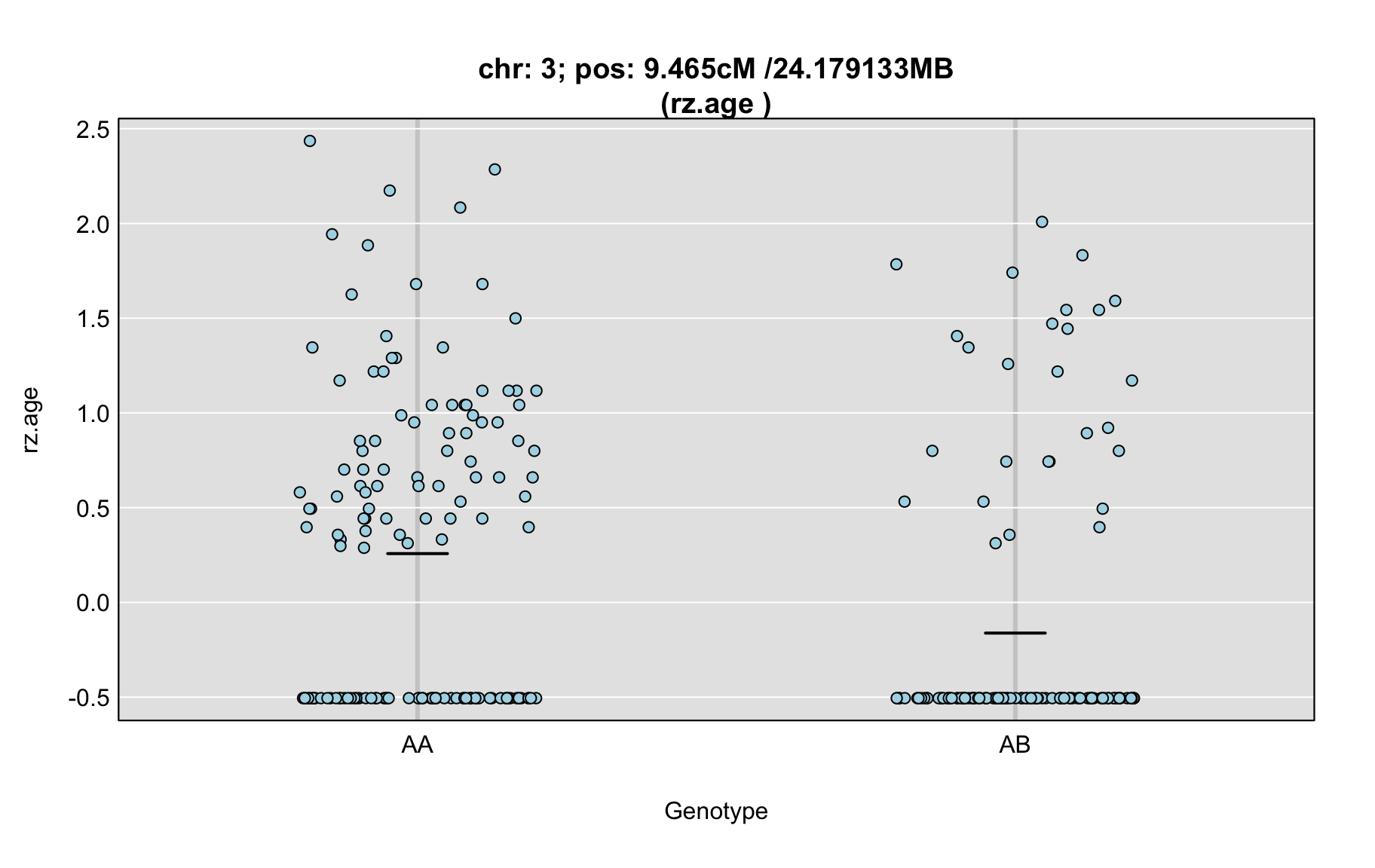
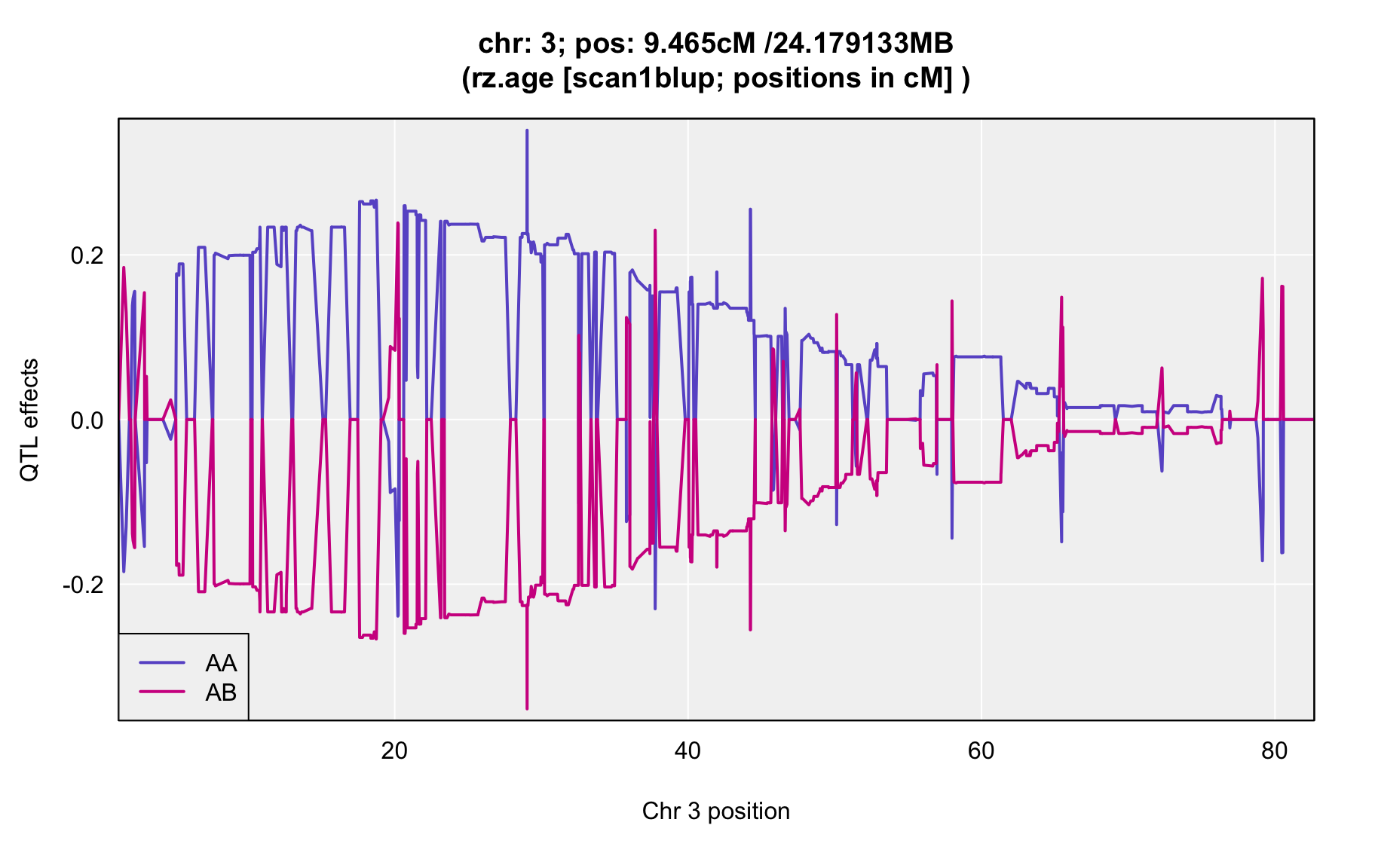

| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | gene | 22.07659 | 22.21659 | positive | MGI_C57BL6J_2441730 | Tbl1xr1 | NCBI_Gene:81004,ENSEMBL:ENSMUSG00000027630 | MGI:2441730 | protein coding gene | transducin (beta)-like 1X-linked receptor 1 |
| 3 | gene | 22.25137 | 22.25166 | positive | MGI_C57BL6J_105104 | Rprl2 | NCBI_Gene:19784,ENSEMBL:ENSMUSG00000087775 | MGI:105104 | RNase P RNA gene | ribonuclease P RNA-like 2 |
| 3 | gene | 22.28508 | 22.29783 | negative | MGI_C57BL6J_5611552 | Gm38324 | ENSEMBL:ENSMUSG00000102987 | MGI:5611552 | lncRNA gene | predicted gene, 38324 |
| 3 | pseudogene | 22.34885 | 22.34961 | negative | MGI_C57BL6J_5610864 | Gm37636 | NCBI_Gene:108168850,ENSEMBL:ENSMUSG00000102866 | MGI:5610864 | pseudogene | predicted gene, 37636 |
| 3 | gene | 22.40992 | 22.41005 | negative | MGI_C57BL6J_5456131 | Gm26354 | ENSEMBL:ENSMUSG00000088846 | MGI:5456131 | snoRNA gene | predicted gene, 26354 |
| 3 | pseudogene | 22.52501 | 22.52624 | positive | MGI_C57BL6J_3780056 | Spin3-ps | NCBI_Gene:675593,ENSEMBL:ENSMUSG00000103801 | MGI:3780056 | pseudogene | spindlin family, member 3, pseudogene |
| 3 | gene | 22.81221 | 22.81246 | positive | MGI_C57BL6J_5611117 | Gm37889 | ENSEMBL:ENSMUSG00000102233 | MGI:5611117 | unclassified gene | predicted gene, 37889 |
| 3 | pseudogene | 23.07242 | 23.07492 | positive | MGI_C57BL6J_5610211 | Gm36983 | ENSEMBL:ENSMUSG00000103275 | MGI:5610211 | pseudogene | predicted gene, 36983 |
| 3 | gene | 23.08598 | 23.08895 | positive | MGI_C57BL6J_5610249 | Gm37021 | ENSEMBL:ENSMUSG00000103607 | MGI:5610249 | lncRNA gene | predicted gene, 37021 |
| 3 | pseudogene | 23.37494 | 23.37532 | negative | MGI_C57BL6J_5610720 | Gm37492 | ENSEMBL:ENSMUSG00000102214 | MGI:5610720 | pseudogene | predicted gene, 37492 |
| 3 | gene | 23.46105 | 23.52140 | positive | MGI_C57BL6J_1924114 | 4930412E21Rik | NCBI_Gene:100039548,ENSEMBL:ENSMUSG00000102203 | MGI:1924114 | lncRNA gene | RIKEN cDNA 4930412E21 gene |
| 3 | gene | 23.79810 | 25.14426 | negative | MGI_C57BL6J_2685867 | Naaladl2 | NCBI_Gene:635702,ENSEMBL:ENSMUSG00000102758 | MGI:2685867 | protein coding gene | N-acetylated alpha-linked acidic dipeptidase-like 2 |
| 3 | pseudogene | 23.93096 | 23.93200 | positive | MGI_C57BL6J_5011009 | Gm18824 | NCBI_Gene:100417784 | MGI:5011009 | pseudogene | predicted gene, 18824 |
| 3 | gene | 23.94002 | 23.94593 | positive | MGI_C57BL6J_5625076 | Gm42191 | NCBI_Gene:105247007 | MGI:5625076 | lncRNA gene | predicted gene, 42191 |
| 3 | gene | 24.16303 | 24.16311 | negative | MGI_C57BL6J_4413931 | n-TLcag1 | NCBI_Gene:102467553 | MGI:4413931 | tRNA gene | nuclear encoded tRNA leucine 1 (anticodon CAG) |
| 3 | pseudogene | 24.33304 | 24.33355 | positive | MGI_C57BL6J_3645137 | Gm7536 | NCBI_Gene:665189,ENSEMBL:ENSMUSG00000057036 | MGI:3645137 | pseudogene | predicted gene 7536 |
| 3 | gene | 24.48671 | 24.48749 | positive | MGI_C57BL6J_1920306 | 2900080J11Rik | NA | NA | unclassified gene | RIKEN cDNA 2900080J11 gene |
| 3 | gene | 24.56207 | 24.56218 | positive | MGI_C57BL6J_5454481 | Gm24704 | ENSEMBL:ENSMUSG00000070227 | MGI:5454481 | snRNA gene | predicted gene, 24704 |
| 3 | gene | 24.71872 | 24.71898 | positive | MGI_C57BL6J_5453839 | Gm24062 | ENSEMBL:ENSMUSG00000088115 | MGI:5453839 | unclassified non-coding RNA gene | predicted gene, 24062 |
| 3 | pseudogene | 25.00742 | 25.00771 | negative | MGI_C57BL6J_5662911 | Gm42774 | ENSEMBL:ENSMUSG00000105009 | MGI:5662911 | pseudogene | predicted gene 42774 |
| 3 | gene | 25.14407 | 25.14426 | positive | MGI_C57BL6J_5610364 | Gm37136 | ENSEMBL:ENSMUSG00000102876 | MGI:5610364 | unclassified gene | predicted gene, 37136 |
| 3 | pseudogene | 25.18480 | 25.18498 | negative | MGI_C57BL6J_5611230 | Gm38002 | ENSEMBL:ENSMUSG00000102855 | MGI:5611230 | pseudogene | predicted gene, 38002 |
| 3 | pseudogene | 25.21076 | 25.21273 | negative | MGI_C57BL6J_3704211 | Gm10259 | NCBI_Gene:100039592,ENSEMBL:ENSMUSG00000069083 | MGI:3704211 | pseudogene | predicted pseudogene 10259 |

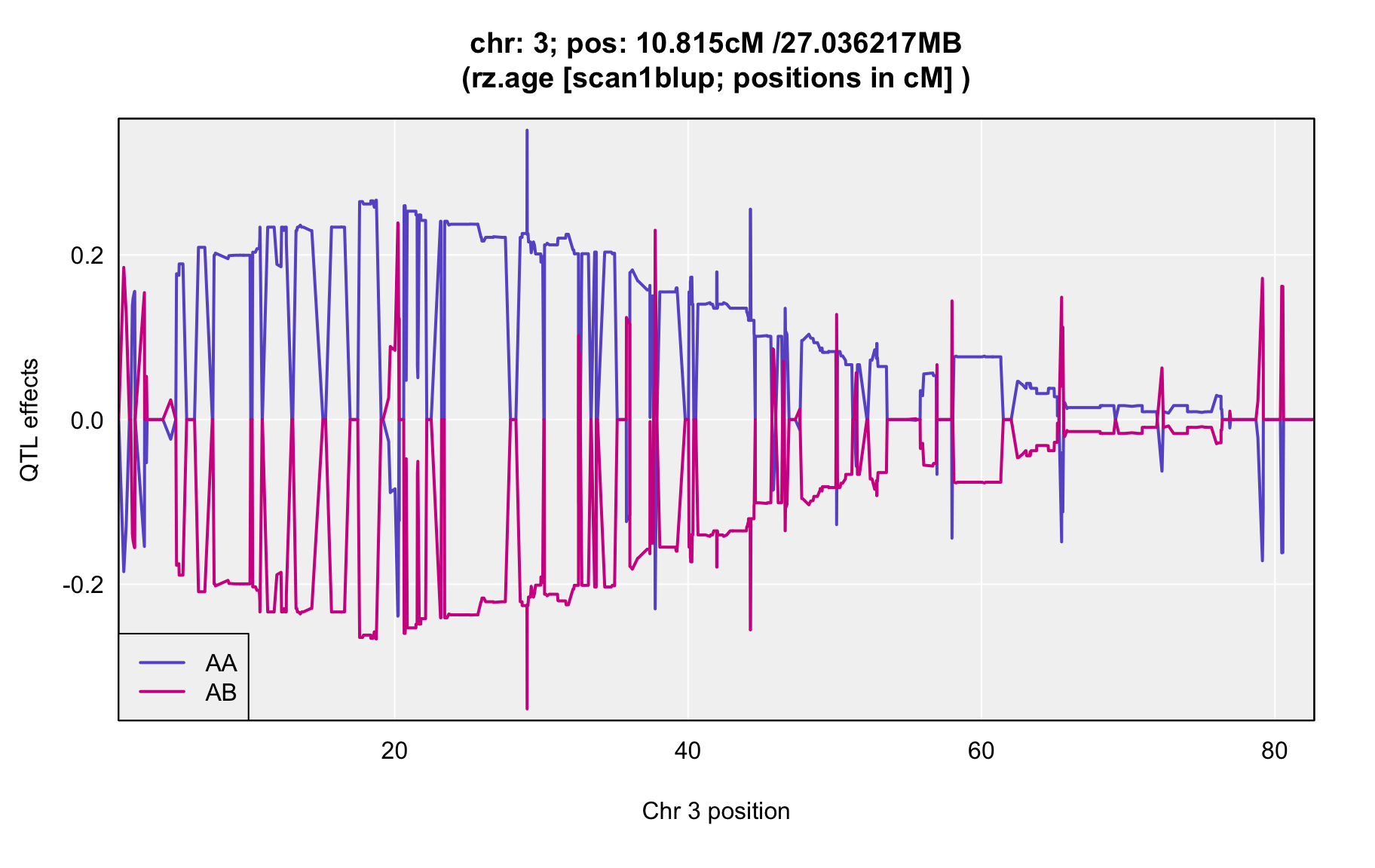
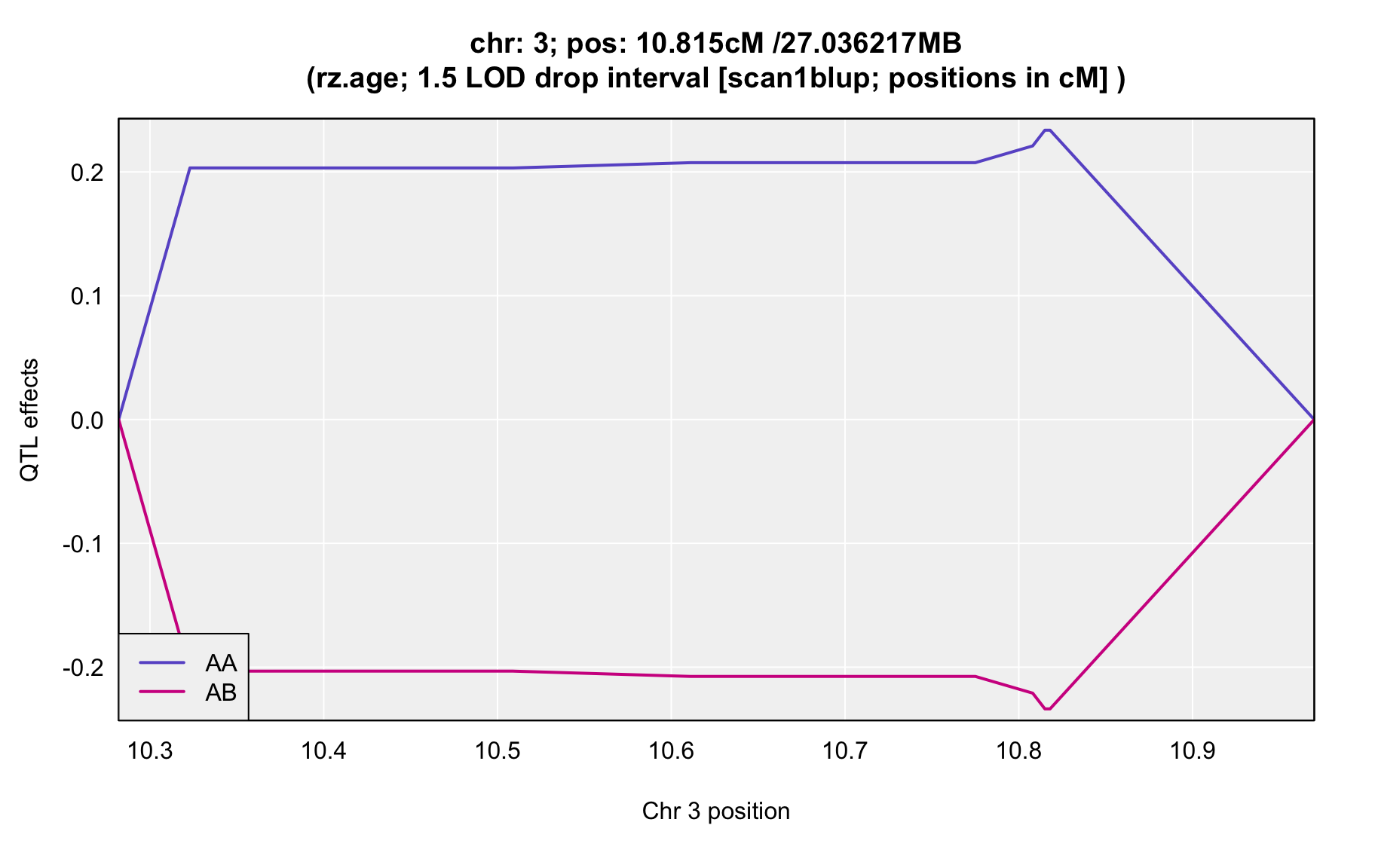
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | gene | 25.42621 | 26.33286 | negative | MGI_C57BL6J_2179435 | Nlgn1 | NCBI_Gene:192167,ENSEMBL:ENSMUSG00000063887 | MGI:2179435 | protein coding gene | neuroligin 1 |
| 3 | gene | 25.54602 | 25.54615 | positive | MGI_C57BL6J_5454027 | Gm24250 | ENSEMBL:ENSMUSG00000087945 | MGI:5454027 | rRNA gene | predicted gene, 24250 |
| 3 | gene | 25.64956 | 25.65077 | negative | MGI_C57BL6J_1920377 | 3110039C02Rik | NA | NA | unclassified gene | RIKEN cDNA 3110039C02 gene |
| 3 | gene | 25.69519 | 25.70030 | negative | MGI_C57BL6J_5610801 | Gm37573 | ENSEMBL:ENSMUSG00000102397 | MGI:5610801 | lncRNA gene | predicted gene, 37573 |
| 3 | pseudogene | 25.73123 | 25.73261 | positive | MGI_C57BL6J_3647151 | Gm6197 | NCBI_Gene:621017,ENSEMBL:ENSMUSG00000104295 | MGI:3647151 | pseudogene | predicted gene 6197 |
| 3 | gene | 25.86566 | 25.86752 | negative | MGI_C57BL6J_2443798 | A830010M09Rik | NA | NA | unclassified gene | RIKEN cDNA A830010M09 gene |
| 3 | gene | 26.33115 | 26.33688 | positive | MGI_C57BL6J_3698881 | A830092H15Rik | NCBI_Gene:100379095,ENSEMBL:ENSMUSG00000074664 | MGI:3698881 | lncRNA gene | RIKEN cDNA A830092H15 gene |
| 3 | pseudogene | 26.38044 | 26.38256 | positive | MGI_C57BL6J_3648363 | Gm4855 | NCBI_Gene:229152,ENSEMBL:ENSMUSG00000102274 | MGI:3648363 | pseudogene | predicted gene 4855 |
| 3 | gene | 26.62618 | 26.62631 | negative | MGI_C57BL6J_5455550 | Gm25773 | ENSEMBL:ENSMUSG00000080368 | MGI:5455550 | miRNA gene | predicted gene, 25773 |
| 3 | gene | 26.63762 | 26.98321 | positive | MGI_C57BL6J_1918112 | Spata16 | NCBI_Gene:70862,ENSEMBL:ENSMUSG00000039335 | MGI:1918112 | protein coding gene | spermatogenesis associated 16 |
| 3 | gene | 26.85020 | 26.87914 | negative | MGI_C57BL6J_5591617 | Gm32458 | NCBI_Gene:102635017 | MGI:5591617 | lncRNA gene | predicted gene, 32458 |
| 3 | pseudogene | 27.01276 | 27.01287 | positive | MGI_C57BL6J_5610887 | Gm37659 | ENSEMBL:ENSMUSG00000103712 | MGI:5610887 | pseudogene | predicted gene, 37659 |
| 3 | gene | 27.02642 | 27.02655 | negative | MGI_C57BL6J_5690856 | Gm44464 | ENSEMBL:ENSMUSG00000105670 | MGI:5690856 | miRNA gene | predicted gene, 44464 |
| 3 | pseudogene | 27.04674 | 27.04798 | negative | MGI_C57BL6J_3645515 | Gm7558 | NCBI_Gene:665258,ENSEMBL:ENSMUSG00000102908 | MGI:3645515 | pseudogene | predicted gene 7558 |
| 3 | gene | 27.09722 | 27.15391 | negative | MGI_C57BL6J_95281 | Ect2 | NCBI_Gene:13605,ENSEMBL:ENSMUSG00000027699 | MGI:95281 | protein coding gene | ect2 oncogene |
| 3 | gene | 27.10898 | 27.10909 | positive | MGI_C57BL6J_5454929 | Gm25152 | ENSEMBL:ENSMUSG00000087742 | MGI:5454929 | snoRNA gene | predicted gene, 25152 |
| 3 | gene | 27.15403 | 27.15702 | positive | MGI_C57BL6J_1923907 | 1700125G22Rik | NCBI_Gene:76657,ENSEMBL:ENSMUSG00000097584 | MGI:1923907 | lncRNA gene | RIKEN cDNA 1700125G22 gene |
| 3 | gene | 27.18296 | 27.28461 | positive | MGI_C57BL6J_2443191 | Nceh1 | NCBI_Gene:320024,ENSEMBL:ENSMUSG00000027698 | MGI:2443191 | protein coding gene | neutral cholesterol ester hydrolase 1 |
| 3 | gene | 27.26033 | 27.28446 | positive | MGI_C57BL6J_5625077 | Gm42192 | NCBI_Gene:105247008 | MGI:5625077 | lncRNA gene | predicted gene, 42192 |
| 3 | pseudogene | 27.27806 | 27.27845 | positive | MGI_C57BL6J_5610419 | Gm37191 | ENSEMBL:ENSMUSG00000102186 | MGI:5610419 | pseudogene | predicted gene, 37191 |
| 3 | pseudogene | 27.30841 | 27.30867 | negative | MGI_C57BL6J_5663482 | Gm43345 | ENSEMBL:ENSMUSG00000105806 | MGI:5663482 | pseudogene | predicted gene 43345 |
| 3 | gene | 27.31703 | 27.34243 | positive | MGI_C57BL6J_107414 | Tnfsf10 | NCBI_Gene:22035,ENSEMBL:ENSMUSG00000039304 | MGI:107414 | protein coding gene | tumor necrosis factor (ligand) superfamily, member 10 |
| 3 | gene | 27.37131 | 27.37923 | positive | MGI_C57BL6J_2441906 | Ghsr | NCBI_Gene:208188,ENSEMBL:ENSMUSG00000051136 | MGI:2441906 | protein coding gene | growth hormone secretagogue receptor |
| 3 | gene | 27.41511 | 27.41615 | negative | MGI_C57BL6J_1924904 | C430049A07Rik | NA | NA | unclassified gene | RIKEN cDNA C430049A07 gene |
| 3 | gene | 27.41616 | 27.71311 | negative | MGI_C57BL6J_1919257 | Fndc3b | NCBI_Gene:72007,ENSEMBL:ENSMUSG00000039286 | MGI:1919257 | protein coding gene | fibronectin type III domain containing 3B |
| 3 | gene | 27.44166 | 27.44411 | positive | MGI_C57BL6J_5625078 | Gm42193 | NCBI_Gene:105247009 | MGI:5625078 | lncRNA gene | predicted gene, 42193 |
| 3 | gene | 27.48228 | 27.48424 | positive | MGI_C57BL6J_5591897 | Gm32738 | NCBI_Gene:102635381 | MGI:5591897 | lncRNA gene | predicted gene, 32738 |
| 3 | gene | 27.54664 | 27.54787 | positive | MGI_C57BL6J_5663481 | Gm43344 | ENSEMBL:ENSMUSG00000106625 | MGI:5663481 | unclassified gene | predicted gene 43344 |
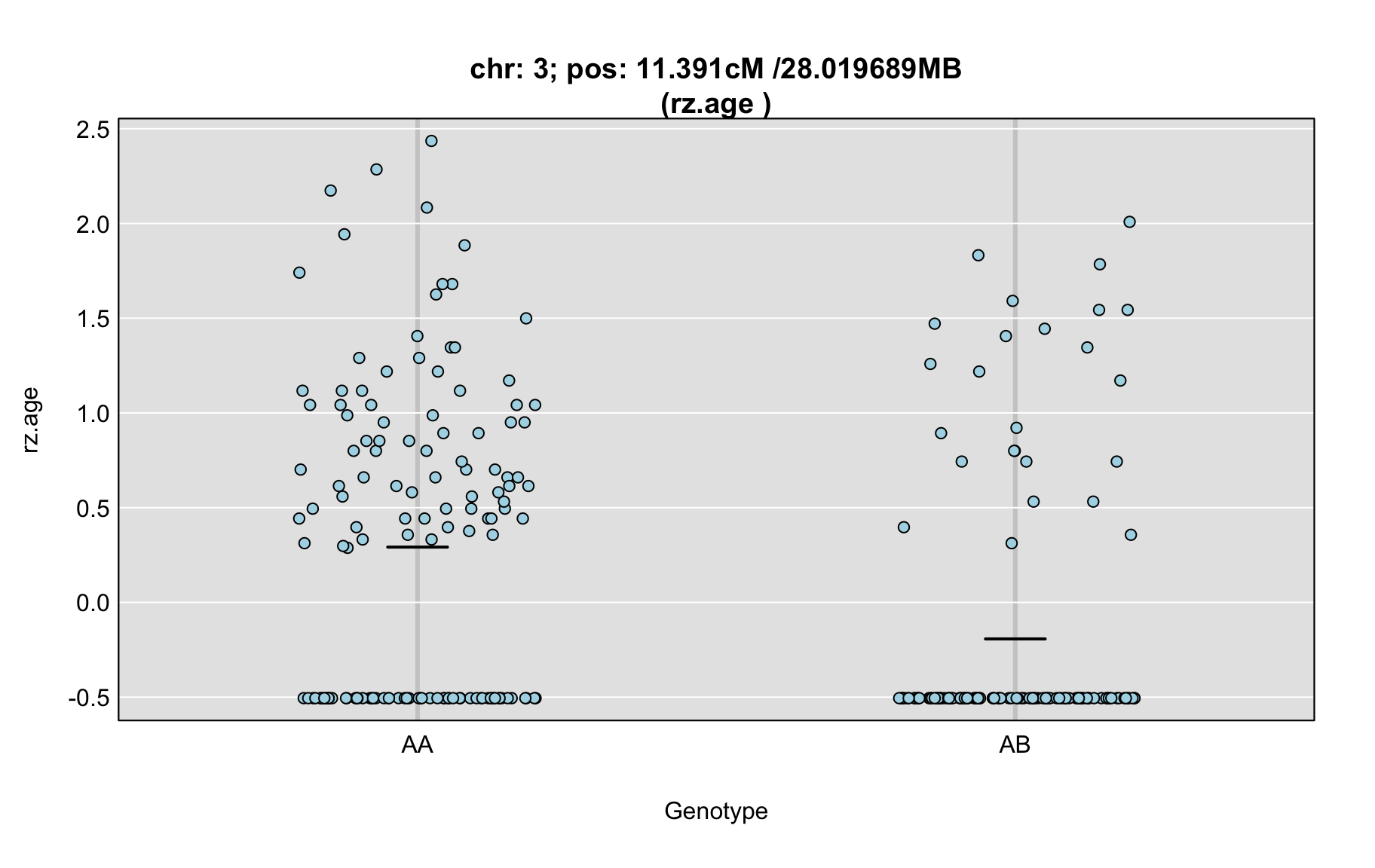
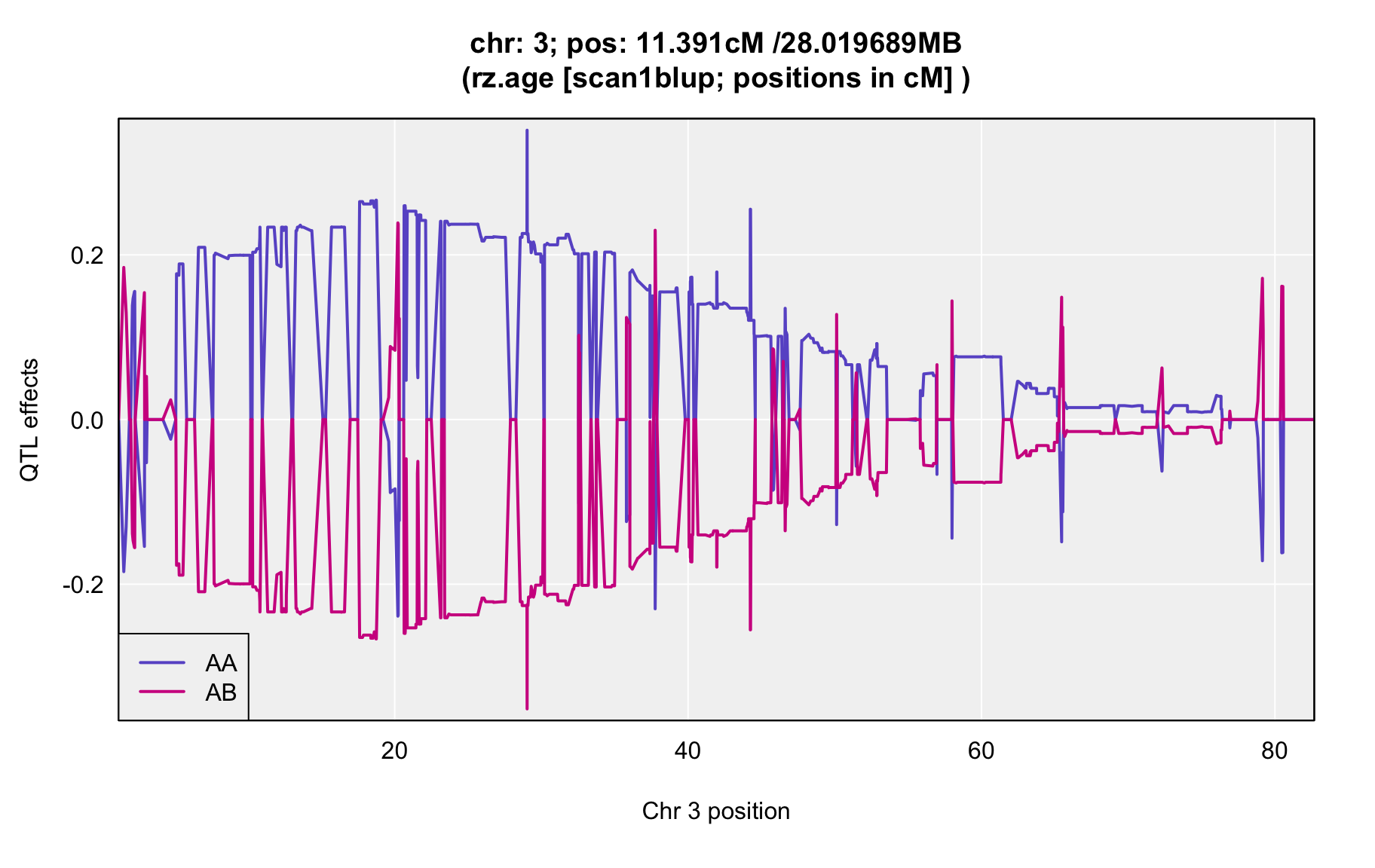

| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | gene | 27.41616 | 27.71311 | negative | MGI_C57BL6J_1919257 | Fndc3b | NCBI_Gene:72007,ENSEMBL:ENSMUSG00000039286 | MGI:1919257 | protein coding gene | fibronectin type III domain containing 3B |
| 3 | gene | 27.58490 | 27.58499 | negative | MGI_C57BL6J_4834306 | Mir3092 | miRBase:MI0014085,NCBI_Gene:100526525,ENSEMBL:ENSMUSG00000092695 | MGI:4834306 | miRNA gene | microRNA 3092 |
| 3 | gene | 27.77786 | 27.84092 | positive | MGI_C57BL6J_5625079 | Gm42194 | NCBI_Gene:105247010 | MGI:5625079 | lncRNA gene | predicted gene, 42194 |
| 3 | gene | 27.79788 | 27.81538 | negative | MGI_C57BL6J_5591954 | Gm32795 | NCBI_Gene:102635463 | MGI:5591954 | lncRNA gene | predicted gene, 32795 |
| 3 | gene | 27.86606 | 27.89639 | negative | MGI_C57BL6J_2685410 | Tmem212 | NCBI_Gene:208613,ENSEMBL:ENSMUSG00000043164 | MGI:2685410 | protein coding gene | transmembrane protein 212 |
| 3 | gene | 27.86811 | 27.86827 | positive | MGI_C57BL6J_5455817 | Gm26040 | ENSEMBL:ENSMUSG00000088772 | MGI:5455817 | snoRNA gene | predicted gene, 26040 |
| 3 | gene | 27.89680 | 27.90023 | negative | MGI_C57BL6J_5625080 | Gm42195 | NCBI_Gene:105247011 | MGI:5625080 | lncRNA gene | predicted gene, 42195 |
| 3 | gene | 27.93841 | 28.13336 | positive | MGI_C57BL6J_109585 | Pld1 | NCBI_Gene:18805,ENSEMBL:ENSMUSG00000027695 | MGI:109585 | protein coding gene | phospholipase D1 |
| 3 | gene | 28.17651 | 28.21221 | positive | MGI_C57BL6J_5625081 | Gm42196 | NCBI_Gene:105247012,ENSEMBL:ENSMUSG00000109716 | MGI:5625081 | lncRNA gene | predicted gene, 42196 |
| 3 | pseudogene | 28.24552 | 28.24577 | positive | MGI_C57BL6J_5610326 | Gm37098 | ENSEMBL:ENSMUSG00000103729 | MGI:5610326 | pseudogene | predicted gene, 37098 |
| 3 | gene | 28.26321 | 28.67586 | positive | MGI_C57BL6J_1916264 | Tnik | NCBI_Gene:665113,ENSEMBL:ENSMUSG00000027692 | MGI:1916264 | protein coding gene | TRAF2 and NCK interacting kinase |
| 3 | gene | 28.26928 | 28.26983 | positive | MGI_C57BL6J_2139599 | AA589472 | NA | NA | unclassified gene | expressed sequence AA589472 |
| 3 | gene | 28.32436 | 28.32609 | positive | MGI_C57BL6J_5610765 | Gm37537 | ENSEMBL:ENSMUSG00000103420 | MGI:5610765 | unclassified gene | predicted gene, 37537 |

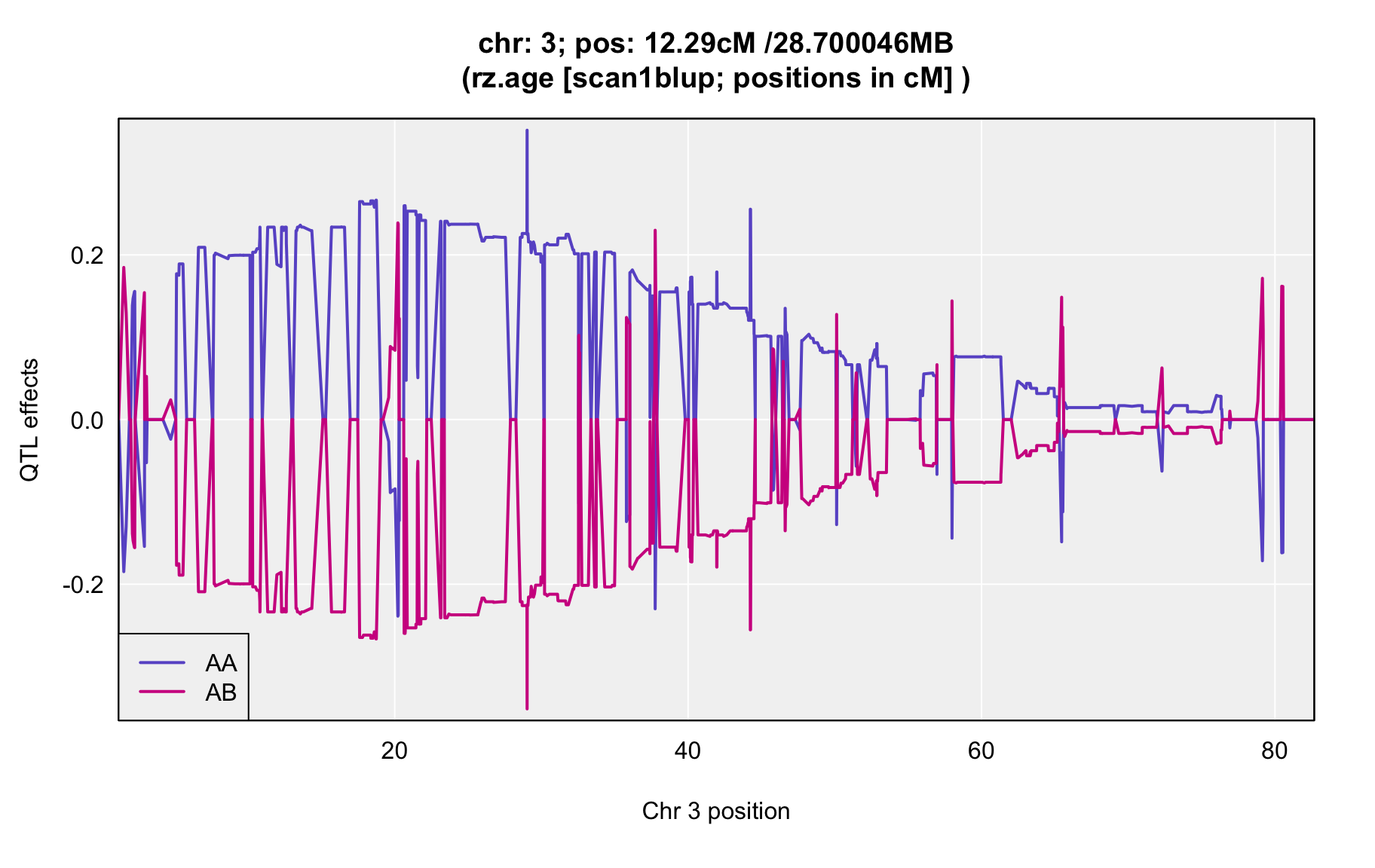

| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | gene | 28.26321 | 28.67586 | positive | MGI_C57BL6J_1916264 | Tnik | NCBI_Gene:665113,ENSEMBL:ENSMUSG00000027692 | MGI:1916264 | protein coding gene | TRAF2 and NCK interacting kinase |
| 3 | gene | 28.69790 | 28.73136 | positive | MGI_C57BL6J_1095438 | Slc2a2 | NCBI_Gene:20526,ENSEMBL:ENSMUSG00000027690 | MGI:1095438 | protein coding gene | solute carrier family 2 (facilitated glucose transporter), member 2 |
| 3 | gene | 28.70267 | 28.78111 | negative | MGI_C57BL6J_1920861 | 1700112D23Rik | NCBI_Gene:73611,ENSEMBL:ENSMUSG00000091329 | MGI:1920861 | lncRNA gene | RIKEN cDNA 1700112D23 gene |
| 3 | pseudogene | 28.76353 | 28.76539 | negative | MGI_C57BL6J_3648080 | Gm6505 | NCBI_Gene:624446,ENSEMBL:ENSMUSG00000070522 | MGI:3648080 | pseudogene | predicted pseudogene 6505 |
| 3 | gene | 28.78127 | 28.79885 | positive | MGI_C57BL6J_1933735 | Eif5a2 | NCBI_Gene:208691,ENSEMBL:ENSMUSG00000050192 | MGI:1933735 | protein coding gene | eukaryotic translation initiation factor 5A2 |
| 3 | gene | 28.79064 | 28.80542 | negative | MGI_C57BL6J_5592109 | Gm32950 | NCBI_Gene:102635671,ENSEMBL:ENSMUSG00000103827 | MGI:5592109 | lncRNA gene | predicted gene, 32950 |
| 3 | gene | 28.80544 | 28.80742 | positive | MGI_C57BL6J_1915278 | Rpl22l1 | NCBI_Gene:68028,ENSEMBL:ENSMUSG00000039221 | MGI:1915278 | protein coding gene | ribosomal protein L22 like 1 |
| 3 | gene | 28.80720 | 28.81618 | negative | MGI_C57BL6J_5592210 | Gm33051 | NCBI_Gene:102635804,ENSEMBL:ENSMUSG00000104444 | MGI:5592210 | lncRNA gene | predicted gene, 33051 |
| 3 | gene | 28.85203 | 28.85426 | negative | MGI_C57BL6J_5611095 | Gm37867 | ENSEMBL:ENSMUSG00000104275 | MGI:5611095 | lncRNA gene | predicted gene, 37867 |
| 3 | gene | 28.87981 | 28.88346 | positive | MGI_C57BL6J_5611152 | Gm37924 | ENSEMBL:ENSMUSG00000102482 | MGI:5611152 | unclassified gene | predicted gene, 37924 |
| 3 | gene | 28.88346 | 28.89728 | negative | MGI_C57BL6J_5592261 | Gm33102 | NCBI_Gene:102635876 | MGI:5592261 | lncRNA gene | predicted gene, 33102 |
| 3 | gene | 28.89262 | 28.92672 | positive | MGI_C57BL6J_2686373 | Gm1527 | NCBI_Gene:385263,ENSEMBL:ENSMUSG00000074655 | MGI:2686373 | protein coding gene | predicted gene 1527 |
| 3 | gene | 29.04377 | 29.04555 | negative | MGI_C57BL6J_5592313 | Gm33154 | NCBI_Gene:102635945 | MGI:5592313 | lncRNA gene | predicted gene, 33154 |
| 3 | gene | 29.08072 | 29.69121 | positive | MGI_C57BL6J_1922990 | Egfem1 | NCBI_Gene:75740,ENSEMBL:ENSMUSG00000063600 | MGI:1922990 | protein coding gene | EGF-like and EMI domain containing 1 |
| 3 | gene | 29.17484 | 29.17693 | positive | MGI_C57BL6J_5611257 | Gm38029 | ENSEMBL:ENSMUSG00000103992 | MGI:5611257 | unclassified gene | predicted gene, 38029 |
| 3 | gene | 29.20934 | 29.21198 | positive | MGI_C57BL6J_2442122 | D230021J17Rik | NA | NA | unclassified gene | RIKEN cDNA D230021J17 gene |
| 3 | gene | 29.21989 | 29.22125 | positive | MGI_C57BL6J_5611193 | Gm37965 | ENSEMBL:ENSMUSG00000102579 | MGI:5611193 | unclassified gene | predicted gene, 37965 |
| 3 | gene | 29.26628 | 29.26684 | negative | MGI_C57BL6J_2139582 | AA516838 | NA | NA | unclassified gene | expressed sequence AA516838 |
| 3 | gene | 29.41682 | 29.41692 | positive | MGI_C57BL6J_3691605 | Mir551b | miRBase:MI0004131,NCBI_Gene:791072,ENSEMBL:ENSMUSG00000076120 | MGI:3691605 | miRNA gene | microRNA 551b |
| 3 | gene | 29.63674 | 29.63685 | positive | MGI_C57BL6J_5531333 | Mir21b | miRBase:MI0021906,NCBI_Gene:102466637,ENSEMBL:ENSMUSG00000098315 | MGI:5531333 | miRNA gene | microRNA 21b |
| 3 | gene | 29.66251 | 29.69656 | negative | MGI_C57BL6J_5625082 | Gm42197 | NCBI_Gene:105247013 | MGI:5625082 | lncRNA gene | predicted gene, 42197 |
| 3 | gene | 29.76404 | 29.89323 | negative | MGI_C57BL6J_5592365 | Gm33206 | NCBI_Gene:102636014,ENSEMBL:ENSMUSG00000102574 | MGI:5592365 | lncRNA gene | predicted gene, 33206 |
| 3 | pseudogene | 29.77128 | 29.77142 | positive | MGI_C57BL6J_5610785 | Gm37557 | ENSEMBL:ENSMUSG00000103095 | MGI:5610785 | pseudogene | predicted gene, 37557 |
| 3 | gene | 29.89101 | 29.92419 | positive | MGI_C57BL6J_5564803 | Mannr | NCBI_Gene:102636083,ENSEMBL:ENSMUSG00000102590 | MGI:5564803 | lncRNA gene | Mecom adjacent non-protein coding RNA |
| 3 | pseudogene | 29.92654 | 29.92697 | positive | MGI_C57BL6J_5610509 | Gm37281 | ENSEMBL:ENSMUSG00000102884 | MGI:5610509 | pseudogene | predicted gene, 37281 |
| 3 | gene | 29.95130 | 30.54801 | negative | MGI_C57BL6J_95457 | Mecom | NCBI_Gene:14013,ENSEMBL:ENSMUSG00000027684 | MGI:95457 | protein coding gene | MDS1 and EVI1 complex locus |
| 3 | gene | 30.01238 | 30.05481 | positive | MGI_C57BL6J_4439646 | Mecomos | ENSEMBL:ENSMUSG00000085702 | MGI:4439646 | antisense lncRNA gene | MDS1 and EVI1 complex locus, opposite strand |
| 3 | gene | 30.16025 | 30.16189 | negative | MGI_C57BL6J_5611294 | Gm38066 | ENSEMBL:ENSMUSG00000104486 | MGI:5611294 | unclassified gene | predicted gene, 38066 |
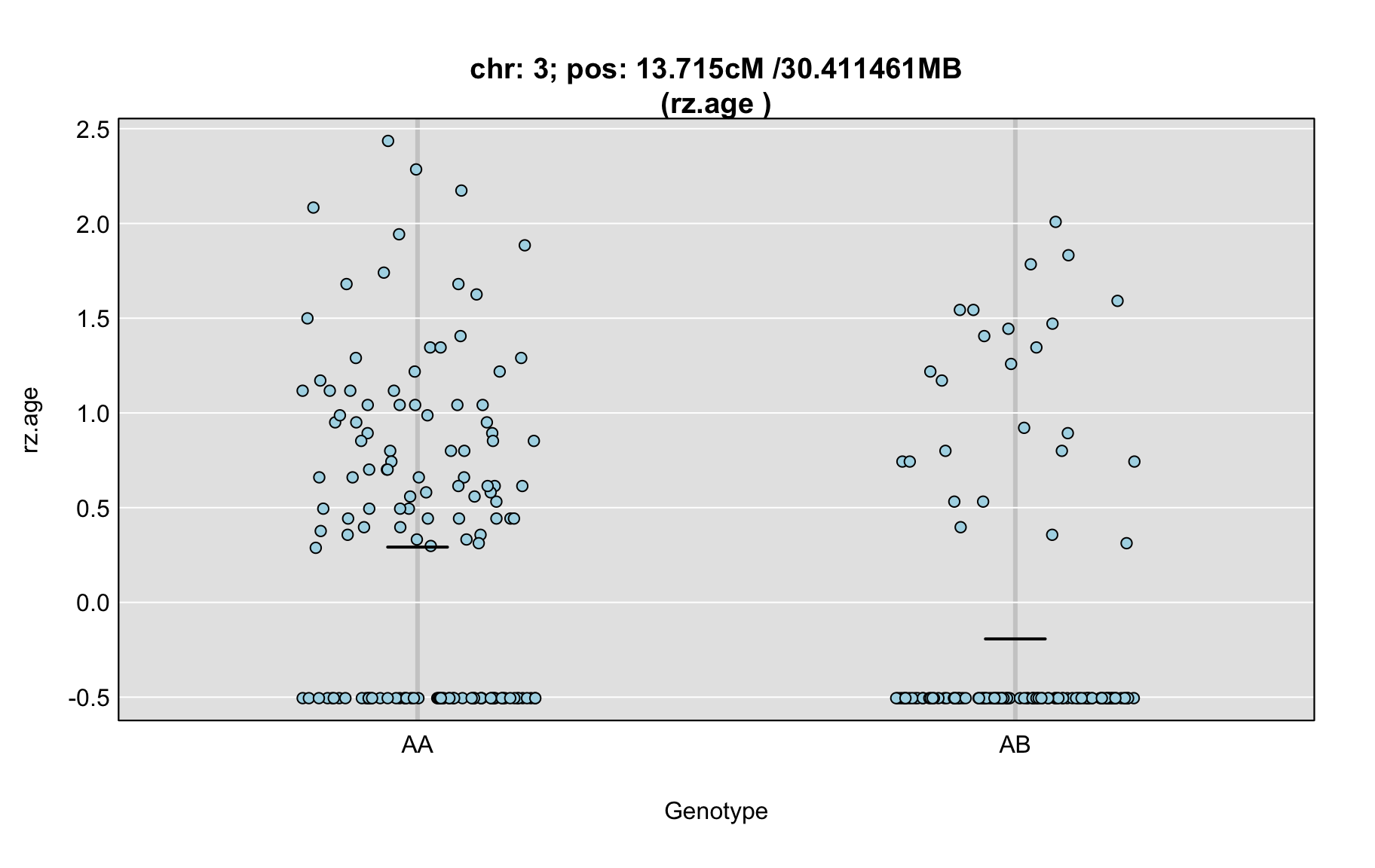
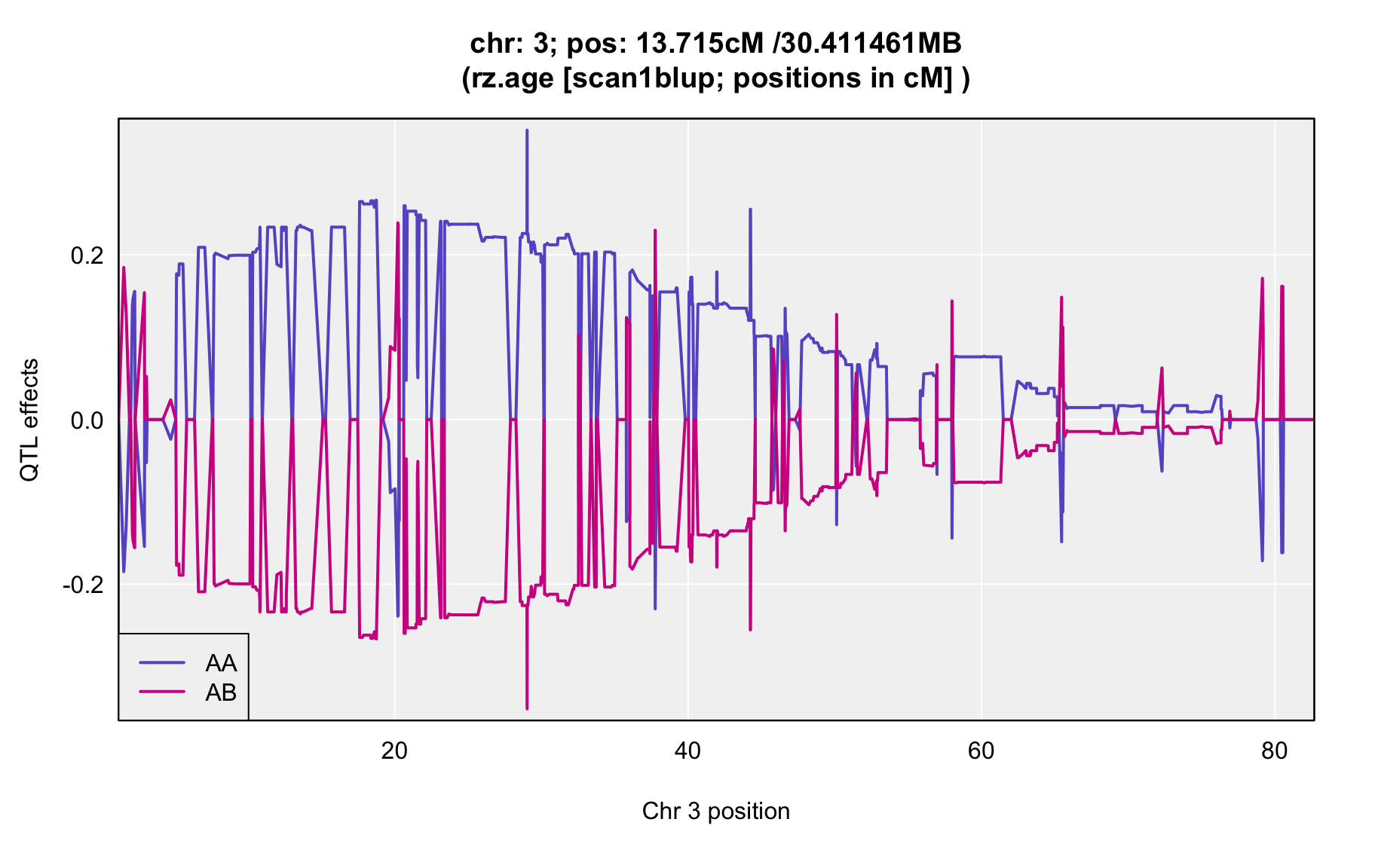
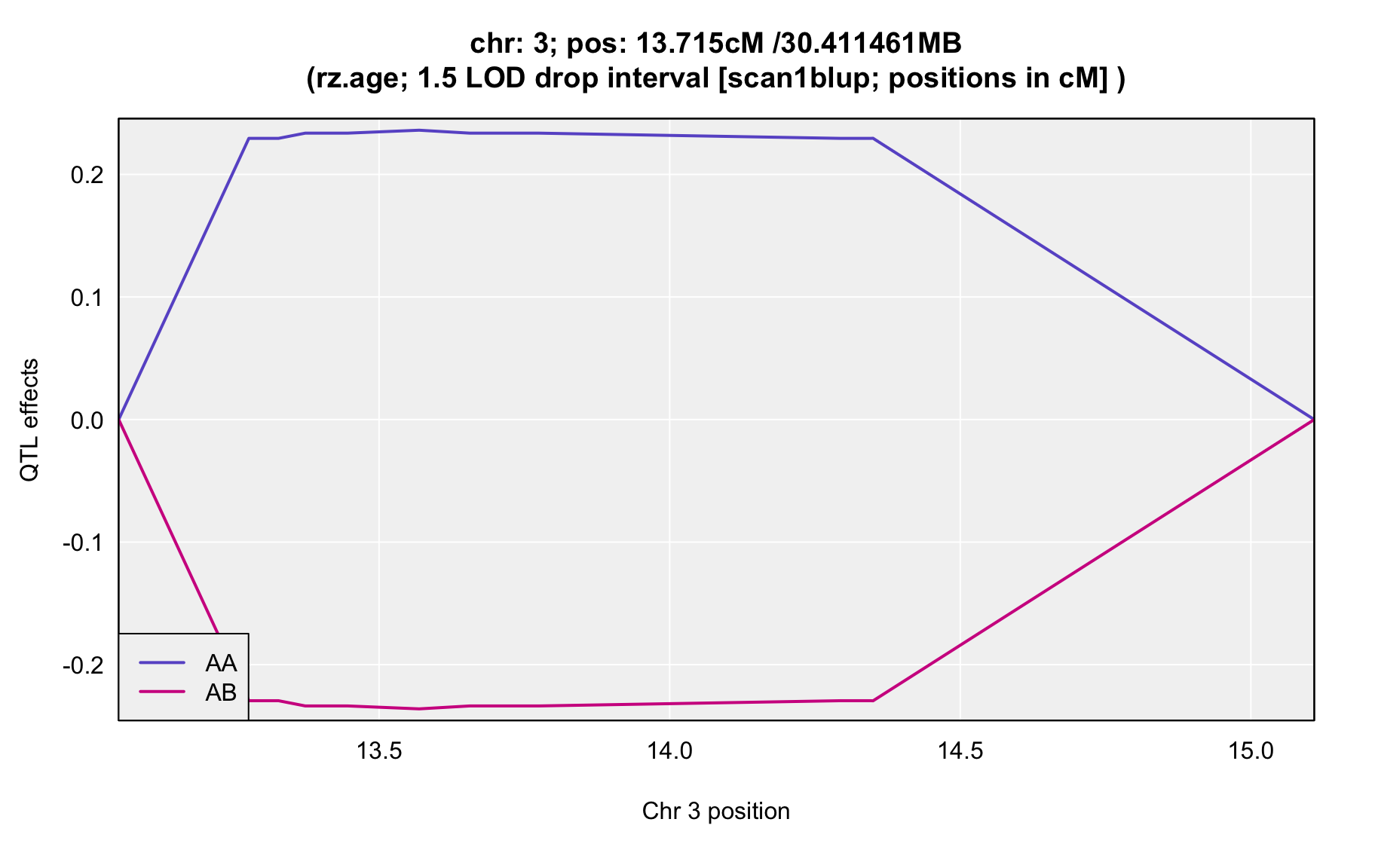
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | gene | 29.95130 | 30.54801 | negative | MGI_C57BL6J_95457 | Mecom | NCBI_Gene:14013,ENSEMBL:ENSMUSG00000027684 | MGI:95457 | protein coding gene | MDS1 and EVI1 complex locus |
| 3 | gene | 30.22557 | 30.22769 | negative | MGI_C57BL6J_5611425 | Gm38197 | ENSEMBL:ENSMUSG00000102460 | MGI:5611425 | unclassified gene | predicted gene, 38197 |
| 3 | gene | 30.26684 | 30.28019 | negative | MGI_C57BL6J_3648282 | Gm10258 | NCBI_Gene:545510,ENSEMBL:ENSMUSG00000069074 | MGI:3648282 | unclassified non-coding RNA gene | predicted gene 10258 |
| 3 | gene | 30.27232 | 30.27438 | negative | MGI_C57BL6J_5611590 | Gm38362 | ENSEMBL:ENSMUSG00000103870 | MGI:5611590 | unclassified gene | predicted gene, 38362 |
| 3 | gene | 30.34998 | 30.35132 | negative | MGI_C57BL6J_1924752 | 9530010C24Rik | NA | NA | unclassified gene | RIKEN cDNA 9530010C24 gene |
| 3 | gene | 30.48843 | 30.48929 | positive | MGI_C57BL6J_5610252 | Gm37024 | ENSEMBL:ENSMUSG00000104079 | MGI:5610252 | unclassified gene | predicted gene, 37024 |
| 3 | gene | 30.50172 | 30.50381 | negative | MGI_C57BL6J_5610880 | Gm37652 | ENSEMBL:ENSMUSG00000103738 | MGI:5610880 | unclassified gene | predicted gene, 37652 |
| 3 | gene | 30.55270 | 30.56424 | negative | MGI_C57BL6J_5611486 | Gm38258 | ENSEMBL:ENSMUSG00000102256 | MGI:5611486 | lncRNA gene | predicted gene, 38258 |
| 3 | pseudogene | 30.56166 | 30.56213 | negative | MGI_C57BL6J_5611381 | Gm38153 | ENSEMBL:ENSMUSG00000104243 | MGI:5611381 | pseudogene | predicted gene, 38153 |
| 3 | gene | 30.59484 | 30.59576 | negative | MGI_C57BL6J_1920499 | 1600017P15Rik | ENSEMBL:ENSMUSG00000102505,ENSEMBL:ENSMUSG00000065361 | MGI:1920499 | unclassified gene | RIKEN cDNA 1600017P15 gene |
| 3 | gene | 30.59707 | 30.59994 | negative | MGI_C57BL6J_1923902 | Actrt3 | NCBI_Gene:76652,ENSEMBL:ENSMUSG00000037737 | MGI:1923902 | protein coding gene | actin related protein T3 |
| 3 | gene | 30.60127 | 30.60134 | positive | MGI_C57BL6J_4414091 | n-TVaac8 | NCBI_Gene:102467402 | MGI:4414091 | tRNA gene | nuclear encoded tRNA valine 8 (anticodon AAC) |
| 3 | gene | 30.60206 | 30.61987 | positive | MGI_C57BL6J_1931415 | Mynn | NCBI_Gene:80732,ENSEMBL:ENSMUSG00000037730 | MGI:1931415 | protein coding gene | myoneurin |
| 3 | gene | 30.62427 | 30.64787 | negative | MGI_C57BL6J_1919077 | Lrrc34 | NCBI_Gene:71827,ENSEMBL:ENSMUSG00000027702 | MGI:1919077 | protein coding gene | leucine rich repeat containing 34 |
| 3 | gene | 30.64451 | 30.67243 | positive | MGI_C57BL6J_1915557 | Lrriq4 | NCBI_Gene:68307,ENSEMBL:ENSMUSG00000027703 | MGI:1915557 | protein coding gene | leucine-rich repeats and IQ motif containing 4 |
| 3 | gene | 30.67855 | 30.69984 | negative | MGI_C57BL6J_2443864 | Lrrc31 | NCBI_Gene:320352,ENSEMBL:ENSMUSG00000074653 | MGI:2443864 | protein coding gene | leucine rich repeat containing 31 |
| 3 | gene | 30.68594 | 30.69301 | positive | MGI_C57BL6J_3705271 | Gm15462 | NCBI_Gene:108168861,ENSEMBL:ENSMUSG00000086189 | MGI:3705271 | lncRNA gene | predicted gene 15462 |
| 3 | gene | 30.74629 | 30.76717 | positive | MGI_C57BL6J_1923203 | Samd7 | NCBI_Gene:75953,ENSEMBL:ENSMUSG00000051860 | MGI:1923203 | protein coding gene | sterile alpha motif domain containing 7 |
| 3 | gene | 30.75231 | 30.75665 | negative | MGI_C57BL6J_5592749 | Gm33590 | NCBI_Gene:102636556 | MGI:5592749 | lncRNA gene | predicted gene, 33590 |
| 3 | gene | 30.79257 | 30.79356 | negative | MGI_C57BL6J_1918530 | 4933429H19Rik | ENSEMBL:ENSMUSG00000097786 | MGI:1918530 | lncRNA gene | RIKEN cDNA 4933429H19 gene |
| 3 | gene | 30.79276 | 30.82126 | positive | MGI_C57BL6J_1916526 | Sec62 | NCBI_Gene:69276,ENSEMBL:ENSMUSG00000027706 | MGI:1916526 | protein coding gene | SEC62 homolog (S. cerevisiae) |
| 3 | pseudogene | 30.82857 | 30.82883 | negative | MGI_C57BL6J_5611308 | Gm38080 | ENSEMBL:ENSMUSG00000104361 | MGI:5611308 | pseudogene | predicted gene, 38080 |
| 3 | gene | 30.84154 | 30.85199 | negative | MGI_C57BL6J_5625083 | Gm42198 | NCBI_Gene:105247015 | MGI:5625083 | lncRNA gene | predicted gene, 42198 |
| 3 | gene | 30.85248 | 30.85586 | negative | MGI_C57BL6J_5625084 | Gm42199 | NCBI_Gene:105247016 | MGI:5625084 | lncRNA gene | predicted gene, 42199 |
| 3 | gene | 30.85587 | 30.89719 | positive | MGI_C57BL6J_1919112 | Gpr160 | NCBI_Gene:71862,ENSEMBL:ENSMUSG00000037661 | MGI:1919112 | protein coding gene | G protein-coupled receptor 160 |
| 3 | gene | 30.89929 | 30.96948 | negative | MGI_C57BL6J_2181434 | Phc3 | NCBI_Gene:241915,ENSEMBL:ENSMUSG00000037652 | MGI:2181434 | protein coding gene | polyhomeotic 3 |
| 3 | gene | 30.95733 | 30.95764 | negative | MGI_C57BL6J_1924590 | 9530022L04Rik | ENSEMBL:ENSMUSG00000102369 | MGI:1924590 | unclassified gene | RIKEN cDNA 9530022L04 gene |
| 3 | pseudogene | 30.97291 | 30.97511 | positive | MGI_C57BL6J_3781157 | Gm2979 | NCBI_Gene:100040809,ENSEMBL:ENSMUSG00000102741 | MGI:3781157 | pseudogene | predicted gene 2979 |
| 3 | gene | 30.99574 | 31.05296 | positive | MGI_C57BL6J_99260 | Prkci | NCBI_Gene:18759,ENSEMBL:ENSMUSG00000037643 | MGI:99260 | protein coding gene | protein kinase C, iota |
| 3 | gene | 31.03897 | 31.03903 | positive | MGI_C57BL6J_5531134 | Mir7008 | miRBase:MI0022857,NCBI_Gene:102465610,ENSEMBL:ENSMUSG00000098890 | MGI:5531134 | miRNA gene | microRNA 7008 |
| 3 | gene | 31.06130 | 31.06669 | positive | MGI_C57BL6J_5592800 | Gm33641 | NCBI_Gene:102636630 | MGI:5592800 | lncRNA gene | predicted gene, 33641 |
| 3 | gene | 31.06243 | 31.07994 | negative | MGI_C57BL6J_5592853 | Gm33694 | NCBI_Gene:102636694 | MGI:5592853 | lncRNA gene | predicted gene, 33694 |
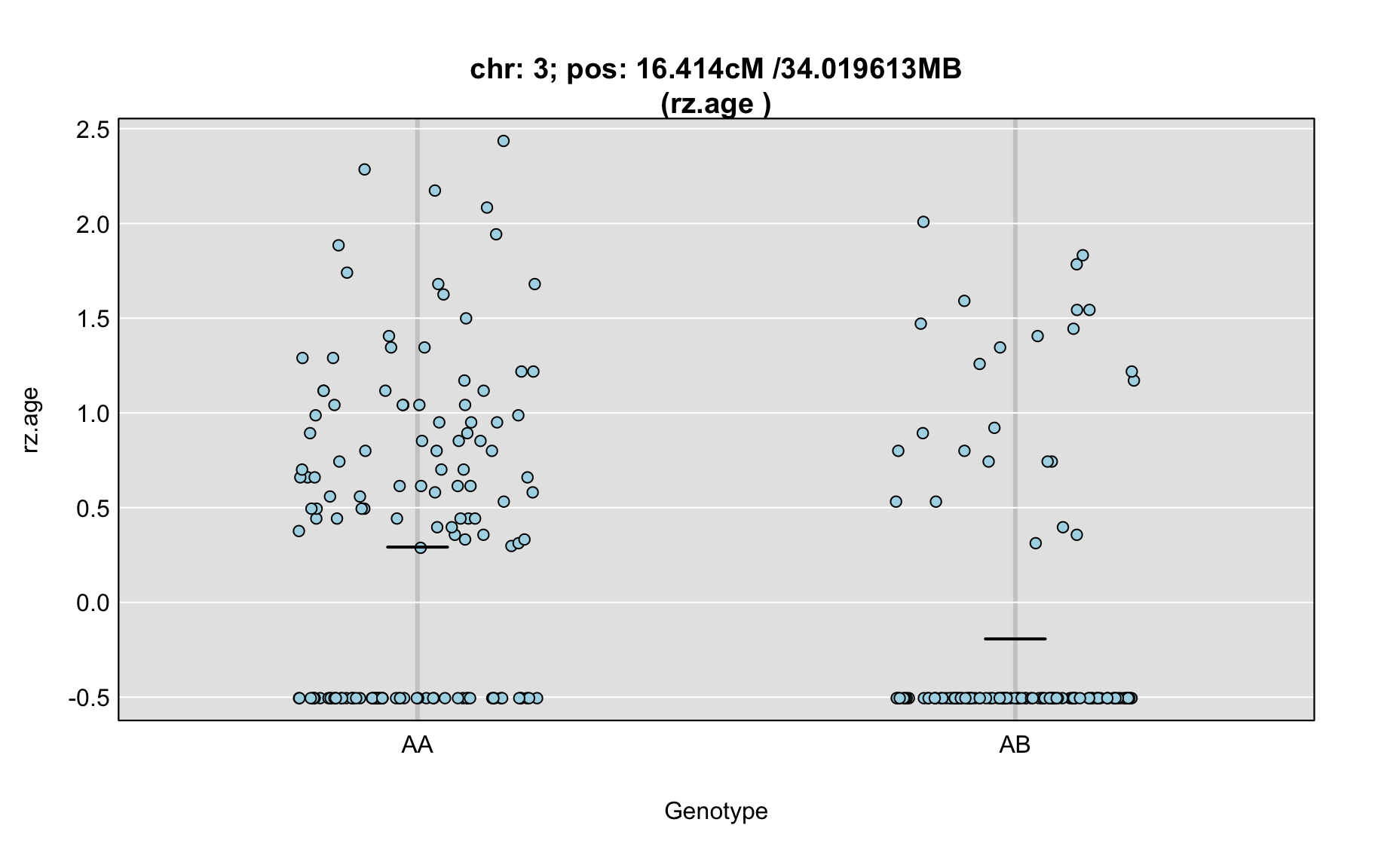
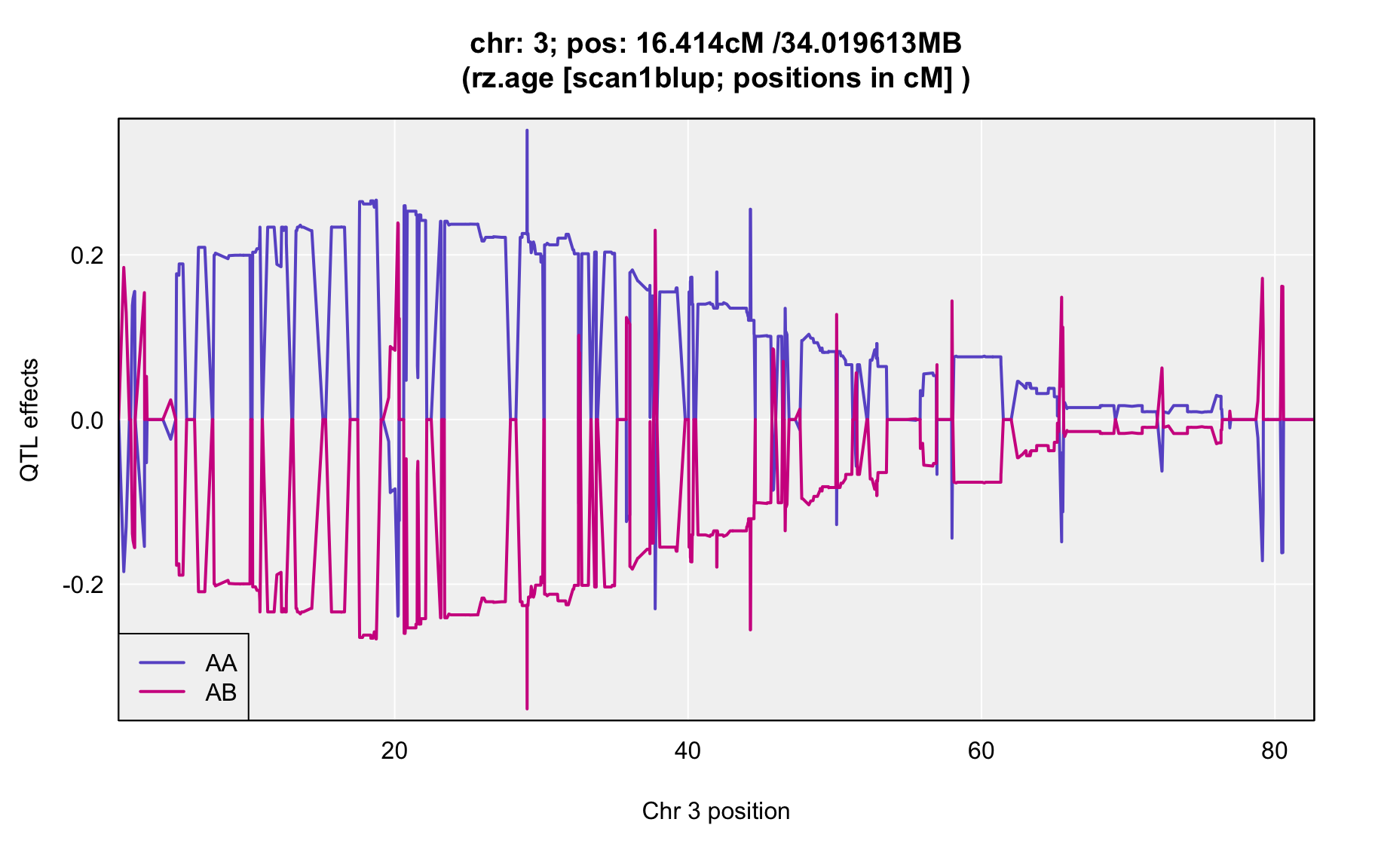
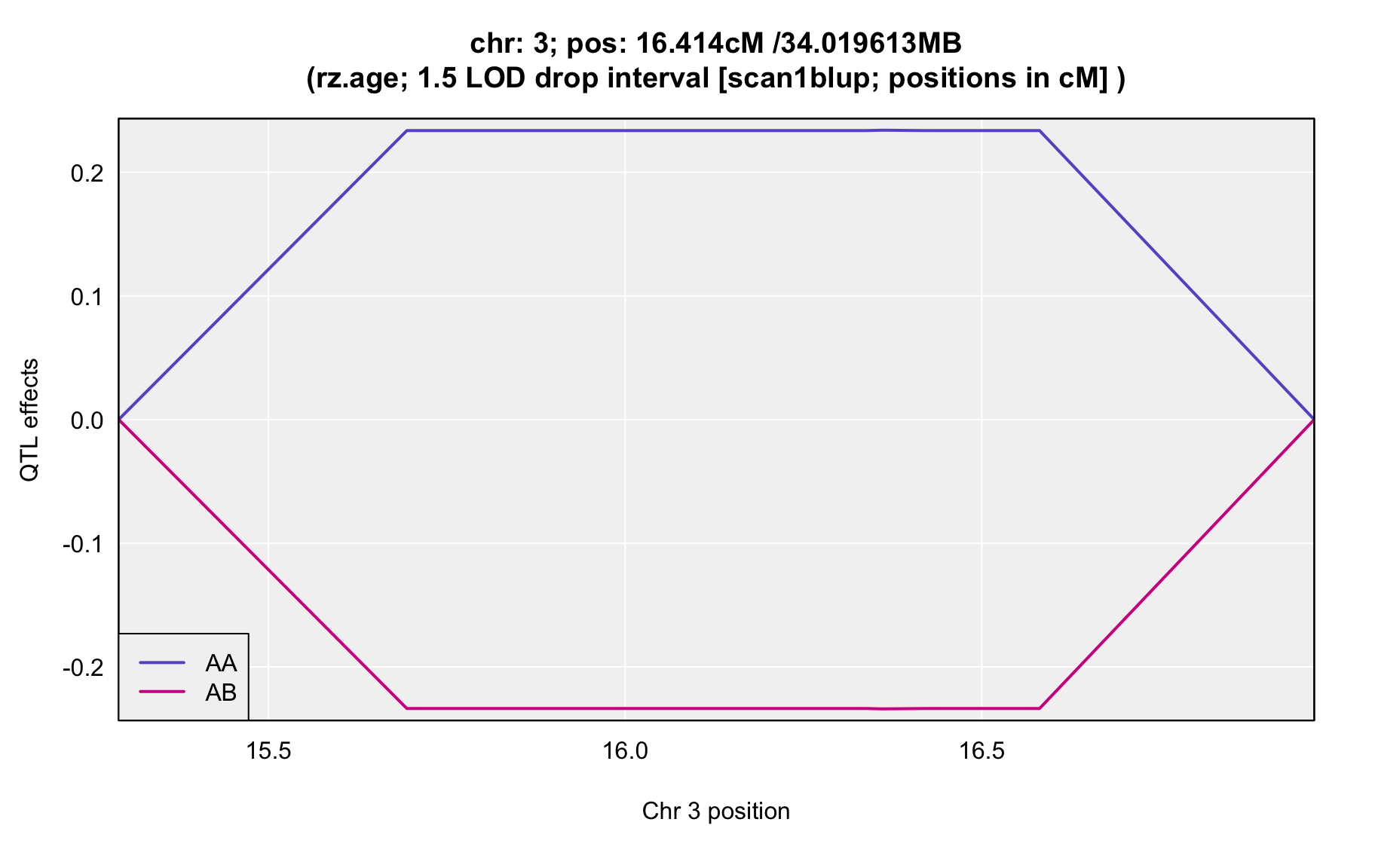
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | pseudogene | 31.64480 | 31.69058 | negative | MGI_C57BL6J_3779607 | Gm6558 | NCBI_Gene:625125,ENSEMBL:ENSMUSG00000103499 | MGI:3779607 | pseudogene | predicted gene 6558 |
| 3 | gene | 31.68642 | 31.69214 | positive | MGI_C57BL6J_5593045 | Gm33886 | NCBI_Gene:102636961 | MGI:5593045 | lncRNA gene | predicted gene, 33886 |
| 3 | gene | 31.89883 | 31.90212 | negative | MGI_C57BL6J_5593190 | Gm34031 | NCBI_Gene:102637147 | MGI:5593190 | lncRNA gene | predicted gene, 34031 |
| 3 | gene | 31.90213 | 32.20018 | positive | MGI_C57BL6J_1919663 | Kcnmb2 | NCBI_Gene:72413,ENSEMBL:ENSMUSG00000037610 | MGI:1919663 | protein coding gene | potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| 3 | gene | 31.95476 | 31.95780 | positive | MGI_C57BL6J_5611062 | Gm37834 | ENSEMBL:ENSMUSG00000103539 | MGI:5611062 | unclassified gene | predicted gene, 37834 |
| 3 | gene | 32.23753 | 32.32631 | negative | MGI_C57BL6J_5625086 | Gm42201 | NCBI_Gene:105247019 | MGI:5625086 | lncRNA gene | predicted gene, 42201 |
| 3 | pseudogene | 32.26033 | 32.26111 | positive | MGI_C57BL6J_5610998 | Gm37770 | NCBI_Gene:108168862,ENSEMBL:ENSMUSG00000103643 | MGI:5610998 | pseudogene | predicted gene, 37770 |
| 3 | gene | 32.33479 | 32.36601 | negative | MGI_C57BL6J_1195270 | Zmat3 | NCBI_Gene:22401,ENSEMBL:ENSMUSG00000027663 | MGI:1195270 | protein coding gene | zinc finger matrin type 3 |
| 3 | gene | 32.36549 | 32.36744 | positive | MGI_C57BL6J_1914826 | 4930429B21Rik | NCBI_Gene:67576 | MGI:1914826 | lncRNA gene | RIKEN cDNA 4930429B21 gene |
| 3 | gene | 32.37158 | 32.37299 | negative | MGI_C57BL6J_5610218 | Gm36990 | ENSEMBL:ENSMUSG00000102825 | MGI:5610218 | unclassified gene | predicted gene, 36990 |
| 3 | gene | 32.37898 | 32.38470 | negative | MGI_C57BL6J_5625087 | Gm42202 | NCBI_Gene:105247020 | MGI:5625087 | lncRNA gene | predicted gene, 42202 |
| 3 | gene | 32.39707 | 32.46849 | positive | MGI_C57BL6J_1206581 | Pik3ca | NCBI_Gene:18706,ENSEMBL:ENSMUSG00000027665 | MGI:1206581 | protein coding gene | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| 3 | gene | 32.45459 | 32.45576 | positive | MGI_C57BL6J_5610351 | Gm37123 | ENSEMBL:ENSMUSG00000103039 | MGI:5610351 | unclassified gene | predicted gene, 37123 |
| 3 | gene | 32.46778 | 32.50290 | negative | MGI_C57BL6J_3612244 | Kcnmb3 | NCBI_Gene:100502876,ENSEMBL:ENSMUSG00000091091 | MGI:3612244 | protein coding gene | potassium large conductance calcium-activated channel, subfamily M, beta member 3 |
| 3 | gene | 32.51026 | 32.52083 | positive | MGI_C57BL6J_1915028 | Zfp639 | NCBI_Gene:67778,ENSEMBL:ENSMUSG00000027667 | MGI:1915028 | protein coding gene | zinc finger protein 639 |
| 3 | gene | 32.51180 | 32.51234 | negative | MGI_C57BL6J_5610820 | Gm37592 | ENSEMBL:ENSMUSG00000104231 | MGI:5610820 | lncRNA gene | predicted gene, 37592 |
| 3 | gene | 32.52946 | 32.57924 | positive | MGI_C57BL6J_1914664 | Mfn1 | NCBI_Gene:67414,ENSEMBL:ENSMUSG00000027668 | MGI:1914664 | protein coding gene | mitofusin 1 |
| 3 | gene | 32.58033 | 32.61659 | negative | MGI_C57BL6J_104581 | Gnb4 | NCBI_Gene:14696,ENSEMBL:ENSMUSG00000027669 | MGI:104581 | protein coding gene | guanine nucleotide binding protein (G protein), beta 4 |
| 3 | gene | 32.61086 | 32.61351 | positive | MGI_C57BL6J_5610868 | Gm37640 | ENSEMBL:ENSMUSG00000102862 | MGI:5610868 | lncRNA gene | predicted gene, 37640 |
| 3 | gene | 32.70630 | 32.72697 | positive | MGI_C57BL6J_1861453 | Actl6a | NCBI_Gene:56456,ENSEMBL:ENSMUSG00000027671 | MGI:1861453 | protein coding gene | actin-like 6A |
| 3 | gene | 32.72178 | 32.72209 | negative | MGI_C57BL6J_5610581 | Gm37353 | ENSEMBL:ENSMUSG00000103590 | MGI:5610581 | unclassified gene | predicted gene, 37353 |
| 3 | gene | 32.72540 | 32.73761 | negative | MGI_C57BL6J_1921850 | Mrpl47 | NCBI_Gene:74600,ENSEMBL:ENSMUSG00000037531 | MGI:1921850 | protein coding gene | mitochondrial ribosomal protein L47 |
| 3 | gene | 32.73610 | 32.73761 | negative | MGI_C57BL6J_3704212 | Gm9859 | NA | NA | unclassified gene | predicted gene 9859 |
| 3 | gene | 32.73699 | 32.75157 | positive | MGI_C57BL6J_1913296 | Ndufb5 | NCBI_Gene:66046,ENSEMBL:ENSMUSG00000027673 | MGI:1913296 | protein coding gene | NADH:ubiquinone oxidoreductase subunit B5 |
| 3 | gene | 32.81702 | 32.93807 | positive | MGI_C57BL6J_1919857 | Usp13 | NCBI_Gene:72607,ENSEMBL:ENSMUSG00000056900 | MGI:1919857 | protein coding gene | ubiquitin specific peptidase 13 (isopeptidase T-3) |
| 3 | pseudogene | 32.88001 | 32.88061 | negative | MGI_C57BL6J_3783022 | Gm15574 | ENSEMBL:ENSMUSG00000081883 | MGI:3783022 | pseudogene | predicted gene 15574 |
| 3 | gene | 32.93931 | 32.98330 | positive | MGI_C57BL6J_5610687 | Gm37459 | ENSEMBL:ENSMUSG00000104283 | MGI:5610687 | lncRNA gene | predicted gene, 37459 |
| 3 | gene | 32.94793 | 33.14327 | negative | MGI_C57BL6J_1916672 | Pex5l | NCBI_Gene:58869,ENSEMBL:ENSMUSG00000027674 | MGI:1916672 | protein coding gene | peroxisomal biogenesis factor 5-like |
| 3 | gene | 33.01196 | 33.01211 | positive | MGI_C57BL6J_5452991 | Gm23214 | ENSEMBL:ENSMUSG00000084665 | MGI:5452991 | snRNA gene | predicted gene, 23214 |
| 3 | gene | 33.02478 | 33.08861 | positive | MGI_C57BL6J_1921124 | 4930419G24Rik | NCBI_Gene:73874,ENSEMBL:ENSMUSG00000087625 | MGI:1921124 | lncRNA gene | RIKEN cDNA 4930419G24 gene |
| 3 | gene | 33.03423 | 33.03705 | negative | MGI_C57BL6J_5610671 | Gm37443 | ENSEMBL:ENSMUSG00000102581 | MGI:5610671 | unclassified gene | predicted gene, 37443 |
| 3 | gene | 33.07392 | 33.07636 | negative | MGI_C57BL6J_5011630 | Gm19445 | ENSEMBL:ENSMUSG00000102498 | MGI:5011630 | unclassified gene | predicted gene, 19445 |
| 3 | pseudogene | 33.19080 | 33.19104 | positive | MGI_C57BL6J_5610356 | Gm37128 | ENSEMBL:ENSMUSG00000103896 | MGI:5610356 | pseudogene | predicted gene, 37128 |
| 3 | gene | 33.26072 | 33.26082 | negative | MGI_C57BL6J_5531356 | Gm27974 | ENSEMBL:ENSMUSG00000098622 | MGI:5531356 | miRNA gene | predicted gene, 27974 |
| 3 | gene | 33.28312 | 33.28565 | positive | MGI_C57BL6J_5610259 | Gm37031 | ENSEMBL:ENSMUSG00000104384 | MGI:5610259 | unclassified gene | predicted gene, 37031 |
| 3 | gene | 33.39306 | 33.39320 | negative | MGI_C57BL6J_5454557 | Gm24780 | ENSEMBL:ENSMUSG00000088260 | MGI:5454557 | snoRNA gene | predicted gene, 24780 |
| 3 | gene | 33.39338 | 33.39485 | negative | MGI_C57BL6J_5610626 | Gm37398 | ENSEMBL:ENSMUSG00000104203 | MGI:5610626 | unclassified gene | predicted gene, 37398 |
| 3 | gene | 33.54027 | 33.54342 | positive | MGI_C57BL6J_5593364 | Gm34205 | NCBI_Gene:102637377 | MGI:5593364 | lncRNA gene | predicted gene, 34205 |
| 3 | pseudogene | 33.62159 | 33.62245 | positive | MGI_C57BL6J_3649050 | Gm5845 | NCBI_Gene:545512,ENSEMBL:ENSMUSG00000103423 | MGI:3649050 | pseudogene | predicted gene 5845 |
| 3 | pseudogene | 33.68451 | 33.68487 | negative | MGI_C57BL6J_5662831 | Gm42694 | ENSEMBL:ENSMUSG00000104954 | MGI:5662831 | pseudogene | predicted gene 42694 |
| 3 | gene | 33.68696 | 33.69200 | negative | MGI_C57BL6J_5625088 | Gm42203 | NCBI_Gene:105247021 | MGI:5625088 | lncRNA gene | predicted gene, 42203 |
| 3 | gene | 33.70336 | 33.75962 | negative | MGI_C57BL6J_5663803 | Gm43666 | ENSEMBL:ENSMUSG00000104588 | MGI:5663803 | lncRNA gene | predicted gene 43666 |
| 3 | pseudogene | 33.73060 | 33.73353 | positive | MGI_C57BL6J_5010796 | Gm18611 | NCBI_Gene:100417436,ENSEMBL:ENSMUSG00000106231 | MGI:5010796 | pseudogene | predicted gene, 18611 |
| 3 | gene | 33.79983 | 33.81486 | positive | MGI_C57BL6J_1914370 | Ttc14 | NCBI_Gene:67120,ENSEMBL:ENSMUSG00000027677 | MGI:1914370 | protein coding gene | tetratricopeptide repeat domain 14 |
| 3 | gene | 33.81076 | 33.84432 | negative | MGI_C57BL6J_1289263 | Ccdc39 | NCBI_Gene:51938,ENSEMBL:ENSMUSG00000027676 | MGI:1289263 | protein coding gene | coiled-coil domain containing 39 |
| 3 | gene | 33.81726 | 33.81795 | positive | MGI_C57BL6J_5663277 | Gm43140 | ENSEMBL:ENSMUSG00000105286 | MGI:5663277 | lncRNA gene | predicted gene 43140 |
| 3 | gene | 33.86681 | 33.89758 | negative | MGI_C57BL6J_1921204 | 4930431L21Rik | ENSEMBL:ENSMUSG00000105475 | MGI:1921204 | lncRNA gene | RIKEN cDNA 4930431L21 gene |
| 3 | gene | 33.90684 | 33.90823 | positive | MGI_C57BL6J_5625089 | Gm42204 | NCBI_Gene:105247022 | MGI:5625089 | lncRNA gene | predicted gene, 42204 |
| 3 | gene | 33.95620 | 33.95715 | positive | MGI_C57BL6J_1922225 | 4930502C17Rik | ENSEMBL:ENSMUSG00000106584 | MGI:1922225 | unclassified gene | RIKEN cDNA 4930502C17 gene |
| 3 | gene | 33.95907 | 33.95920 | negative | MGI_C57BL6J_5452952 | Gm23175 | ENSEMBL:ENSMUSG00000077559 | MGI:5452952 | snoRNA gene | predicted gene, 23175 |
| 3 | gene | 33.97218 | 33.97545 | negative | MGI_C57BL6J_5826470 | Gm46833 | NCBI_Gene:108168913 | MGI:5826470 | lncRNA gene | predicted gene, 46833 |
| 3 | pseudogene | 34.00477 | 34.00551 | negative | MGI_C57BL6J_3642317 | Gm9791 | NCBI_Gene:100040232,ENSEMBL:ENSMUSG00000044434 | MGI:3642317 | pseudogene | predicted pseudogene 9791 |
| 3 | gene | 34.00880 | 34.01188 | negative | MGI_C57BL6J_5826435 | Gm46798 | NCBI_Gene:108168863 | MGI:5826435 | lncRNA gene | predicted gene, 46798 |
| 3 | gene | 34.01771 | 34.01988 | negative | MGI_C57BL6J_5663214 | Gm43077 | ENSEMBL:ENSMUSG00000105535 | MGI:5663214 | lncRNA gene | predicted gene 43077 |
| 3 | gene | 34.01994 | 34.07032 | positive | MGI_C57BL6J_104860 | Fxr1 | NCBI_Gene:14359,ENSEMBL:ENSMUSG00000027680 | MGI:104860 | protein coding gene | fragile X mental retardation gene 1, autosomal homolog |
| 3 | gene | 34.05434 | 34.05543 | negative | MGI_C57BL6J_5663804 | Gm43667 | ENSEMBL:ENSMUSG00000105135 | MGI:5663804 | unclassified gene | predicted gene 43667 |
| 3 | gene | 34.05602 | 34.08135 | negative | MGI_C57BL6J_1914963 | Dnajc19 | NCBI_Gene:67713,ENSEMBL:ENSMUSG00000027679 | MGI:1914963 | protein coding gene | DnaJ heat shock protein family (Hsp40) member C19 |
| 3 | gene | 34.07066 | 34.07600 | positive | MGI_C57BL6J_5663805 | Gm43668 | ENSEMBL:ENSMUSG00000105176 | MGI:5663805 | unclassified gene | predicted gene 43668 |
| 3 | gene | 34.10427 | 34.68262 | positive | MGI_C57BL6J_2444112 | Sox2ot | NCBI_Gene:320478,ENSEMBL:ENSMUSG00000105265 | MGI:2444112 | lncRNA gene | SOX2 overlapping transcript (non-protein coding) |
| 3 | pseudogene | 34.18331 | 34.18419 | positive | MGI_C57BL6J_3647478 | Gm5708 | NCBI_Gene:435727,ENSEMBL:ENSMUSG00000105739 | MGI:3647478 | pseudogene | predicted gene 5708 |
| 3 | gene | 34.28133 | 34.35199 | negative | MGI_C57BL6J_5621390 | Gm38505 | NCBI_Gene:102638008,ENSEMBL:ENSMUSG00000092196 | MGI:5621390 | lncRNA gene | predicted gene, 38505 |
| 3 | gene | 34.41544 | 34.41816 | positive | MGI_C57BL6J_5593758 | Gm34599 | NCBI_Gene:102637903,ENSEMBL:ENSMUSG00000105577 | MGI:5593758 | lncRNA gene | predicted gene, 34599 |
| 3 | gene | 34.42737 | 34.43510 | negative | MGI_C57BL6J_5141980 | Gm20515 | ENSEMBL:ENSMUSG00000092242 | MGI:5141980 | lncRNA gene | predicted gene 20515 |
| 3 | gene | 34.48143 | 34.48221 | negative | MGI_C57BL6J_5579841 | Gm29135 | ENSEMBL:ENSMUSG00000099437 | MGI:5579841 | lncRNA gene | predicted gene 29135 |
| 3 | gene | 34.56034 | 34.56055 | positive | MGI_C57BL6J_5530785 | Gm27403 | ENSEMBL:ENSMUSG00000098795 | MGI:5530785 | unclassified non-coding RNA gene | predicted gene, 27403 |
| 3 | gene | 34.56526 | 34.57220 | negative | MGI_C57BL6J_5662832 | Gm42695 | ENSEMBL:ENSMUSG00000106525 | MGI:5662832 | lncRNA gene | predicted gene 42695 |
| 3 | gene | 34.59782 | 34.60488 | negative | MGI_C57BL6J_5625090 | Gm42205 | NCBI_Gene:105247023,ENSEMBL:ENSMUSG00000105222 | MGI:5625090 | lncRNA gene | predicted gene, 42205 |
| 3 | gene | 34.63846 | 34.63854 | positive | MGI_C57BL6J_3811407 | Mir1897 | miRBase:MI0008314,NCBI_Gene:100316679,ENSEMBL:ENSMUSG00000104797 | MGI:3811407 | miRNA gene | microRNA 1897 |
| 3 | gene | 34.64326 | 34.64610 | positive | MGI_C57BL6J_5662829 | Gm42692 | ENSEMBL:ENSMUSG00000104901 | MGI:5662829 | unclassified gene | predicted gene 42692 |
| 3 | gene | 34.64999 | 34.65246 | positive | MGI_C57BL6J_98364 | Sox2 | NCBI_Gene:20674,ENSEMBL:ENSMUSG00000074637 | MGI:98364 | protein coding gene | SRY (sex determining region Y)-box 2 |
| 3 | gene | 34.65318 | 34.65328 | positive | MGI_C57BL6J_5530993 | Gm27611 | ENSEMBL:ENSMUSG00000098254 | MGI:5530993 | unclassified non-coding RNA gene | predicted gene, 27611 |
| 3 | gene | 34.66366 | 34.66429 | negative | MGI_C57BL6J_5662830 | Gm42693 | ENSEMBL:ENSMUSG00000105284 | MGI:5662830 | unclassified gene | predicted gene 42693 |
| 3 | gene | 34.68263 | 34.68275 | positive | MGI_C57BL6J_5663343 | Gm43206 | ENSEMBL:ENSMUSG00000105101 | MGI:5663343 | unclassified gene | predicted gene 43206 |
| 3 | gene | 34.68938 | 34.69292 | positive | MGI_C57BL6J_5594100 | Gm34941 | NCBI_Gene:102638356 | MGI:5594100 | lncRNA gene | predicted gene, 34941 |
| 3 | gene | 34.69964 | 34.71666 | negative | MGI_C57BL6J_3781322 | Gm3143 | NCBI_Gene:100041108,ENSEMBL:ENSMUSG00000105153 | MGI:3781322 | lncRNA gene | predicted gene 3143 |
| 3 | gene | 34.77208 | 34.78235 | positive | MGI_C57BL6J_5621394 | Gm38509 | NCBI_Gene:102638436,ENSEMBL:ENSMUSG00000106519 | MGI:5621394 | lncRNA gene | predicted gene, 38509 |
| 3 | gene | 34.82656 | 34.87928 | positive | MGI_C57BL6J_5434743 | Gm21388 | NCBI_Gene:100861998,ENSEMBL:ENSMUSG00000114832 | MGI:5434743 | lncRNA gene | predicted gene, 21388 |
| 3 | gene | 34.92254 | 34.92265 | negative | MGI_C57BL6J_5531088 | Mir6378 | miRBase:MI0021909,NCBI_Gene:102466155,ENSEMBL:ENSMUSG00000098862 | MGI:5531088 | miRNA gene | microRNA 6378 |
| 3 | gene | 34.95023 | 34.95953 | positive | MGI_C57BL6J_5594414 | Gm35255 | NCBI_Gene:102638771 | MGI:5594414 | lncRNA gene | predicted gene, 35255 |

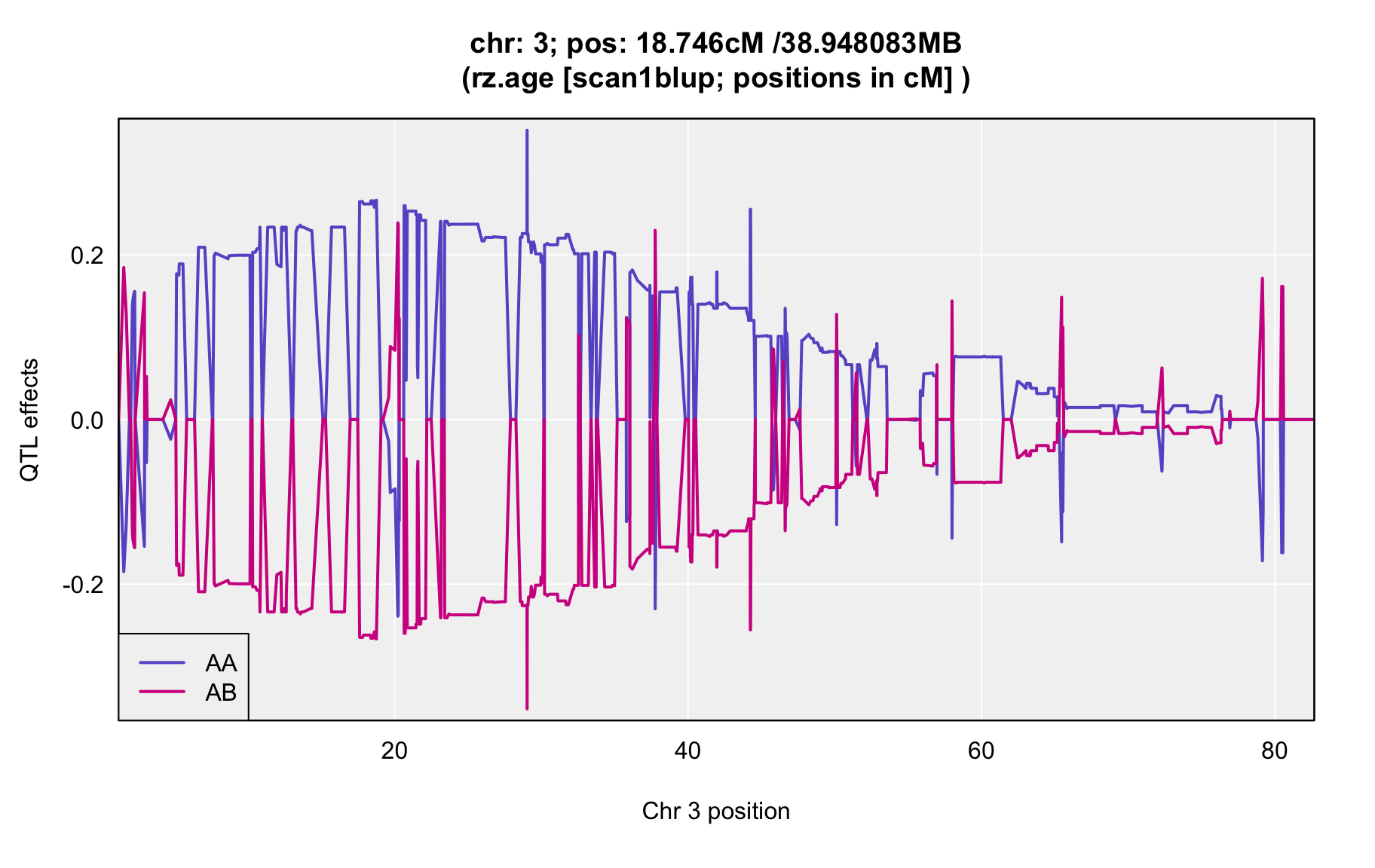
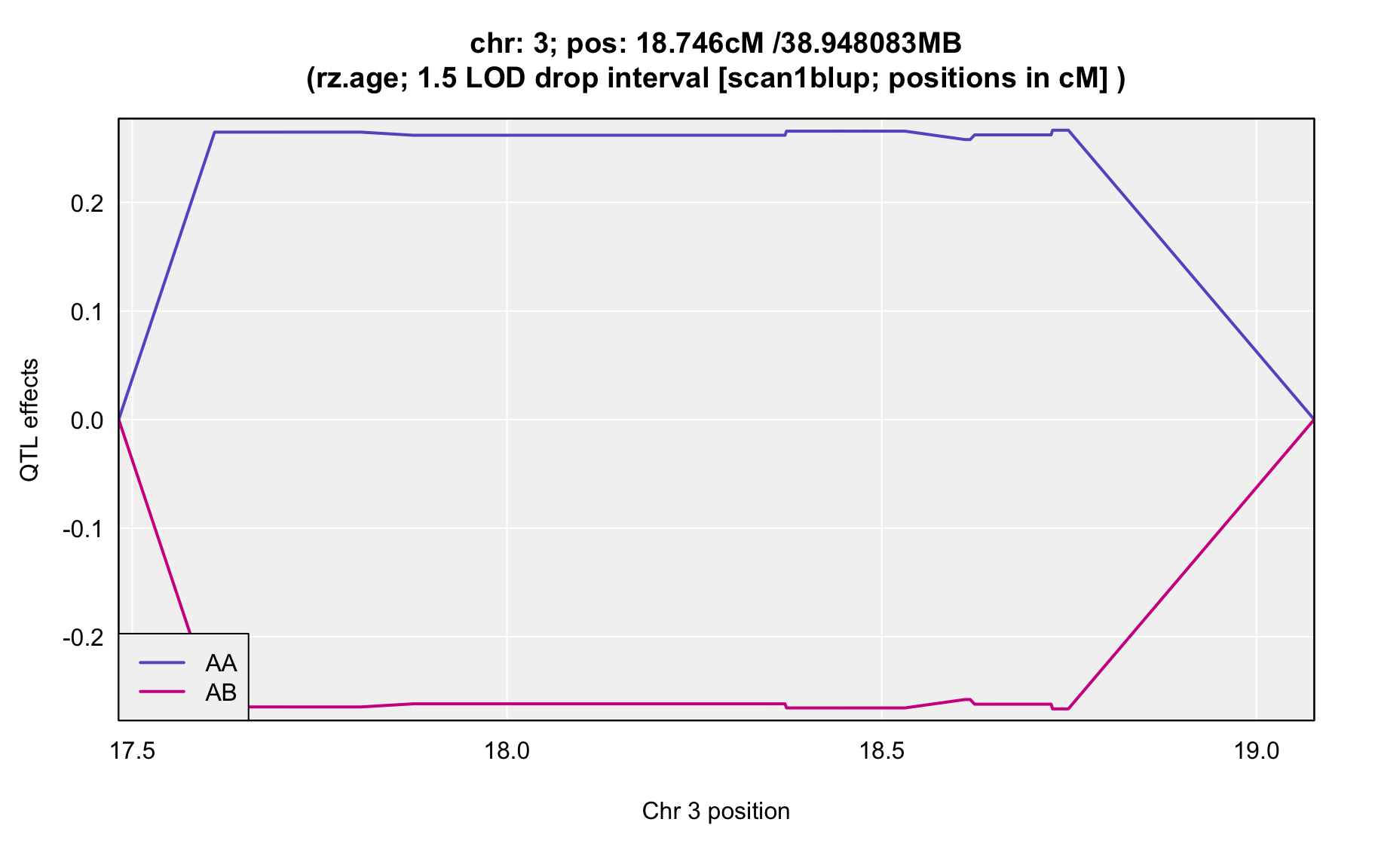
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | gene | 36.52922 | 36.53233 | negative | MGI_C57BL6J_5663674 | Gm43537 | ENSEMBL:ENSMUSG00000105022 | MGI:5663674 | unclassified gene | predicted gene 43537 |
| 3 | pseudogene | 36.53917 | 36.54010 | negative | MGI_C57BL6J_3650891 | Gm11550 | NCBI_Gene:665709,ENSEMBL:ENSMUSG00000083388 | MGI:3650891 | pseudogene | predicted gene 11550 |
| 3 | gene | 36.55261 | 36.56573 | positive | MGI_C57BL6J_1355319 | Exosc9 | NCBI_Gene:50911,ENSEMBL:ENSMUSG00000027714 | MGI:1355319 | protein coding gene | exosome component 9 |
| 3 | gene | 36.56486 | 36.57303 | negative | MGI_C57BL6J_108069 | Ccna2 | NCBI_Gene:12428,ENSEMBL:ENSMUSG00000027715 | MGI:108069 | protein coding gene | cyclin A2 |
| 3 | gene | 36.57314 | 36.61349 | negative | MGI_C57BL6J_1918742 | Bbs7 | NCBI_Gene:71492,ENSEMBL:ENSMUSG00000037325 | MGI:1918742 | protein coding gene | Bardet-Biedl syndrome 7 (human) |
| 3 | gene | 36.62048 | 36.69022 | negative | MGI_C57BL6J_109526 | Trpc3 | NCBI_Gene:22065,ENSEMBL:ENSMUSG00000027716 | MGI:109526 | protein coding gene | transient receptor potential cation channel, subfamily C, member 3 |
| 3 | gene | 36.65407 | 36.65501 | negative | MGI_C57BL6J_5595015 | Gm35856 | NCBI_Gene:102639576 | MGI:5595015 | lncRNA gene | predicted gene, 35856 |
| 3 | pseudogene | 36.69480 | 36.69505 | negative | MGI_C57BL6J_3651607 | Gm12377 | ENSEMBL:ENSMUSG00000082548 | MGI:3651607 | pseudogene | predicted gene 12377 |
| 3 | pseudogene | 36.82435 | 36.82506 | negative | MGI_C57BL6J_3651608 | Gm12376 | NCBI_Gene:100417988,ENSEMBL:ENSMUSG00000084027 | MGI:3651608 | pseudogene | predicted gene 12376 |
| 3 | gene | 36.86306 | 37.05303 | positive | MGI_C57BL6J_2444631 | 4932438A13Rik | NCBI_Gene:229227,ENSEMBL:ENSMUSG00000037270 | MGI:2444631 | protein coding gene | RIKEN cDNA 4932438A13 gene |
| 3 | gene | 37.05221 | 37.06003 | negative | MGI_C57BL6J_3650531 | Gm12531 | ENSEMBL:ENSMUSG00000086090 | MGI:3650531 | lncRNA gene | predicted gene 12531 |
| 3 | gene | 37.06309 | 37.12193 | positive | MGI_C57BL6J_103258 | Adad1 | NCBI_Gene:21744,ENSEMBL:ENSMUSG00000027719 | MGI:103258 | protein coding gene | adenosine deaminase domain containing 1 (testis specific) |
| 3 | pseudogene | 37.07756 | 37.07891 | positive | MGI_C57BL6J_3651489 | Gm12582 | NCBI_Gene:652987,ENSEMBL:ENSMUSG00000081705 | MGI:3651489 | pseudogene | predicted gene 12582 |
| 3 | pseudogene | 37.10808 | 37.10933 | positive | MGI_C57BL6J_3651485 | Gm12583 | NCBI_Gene:100416060,ENSEMBL:ENSMUSG00000082878 | MGI:3651485 | pseudogene | predicted gene 12583 |
| 3 | pseudogene | 37.11673 | 37.11784 | positive | MGI_C57BL6J_3651486 | Gm12584 | ENSEMBL:ENSMUSG00000083857 | MGI:3651486 | pseudogene | predicted gene 12584 |
| 3 | gene | 37.12052 | 37.12596 | negative | MGI_C57BL6J_96548 | Il2 | NCBI_Gene:16183,ENSEMBL:ENSMUSG00000027720 | MGI:96548 | protein coding gene | interleukin 2 |
| 3 | gene | 37.16972 | 37.17665 | negative | MGI_C57BL6J_5595159 | Gm36000 | NCBI_Gene:102639768 | MGI:5595159 | lncRNA gene | predicted gene, 36000 |
| 3 | gene | 37.18218 | 37.18860 | positive | MGI_C57BL6J_3650618 | Gm12532 | NCBI_Gene:102639848,ENSEMBL:ENSMUSG00000084754 | MGI:3650618 | lncRNA gene | predicted gene 12532 |
| 3 | gene | 37.22276 | 37.23264 | negative | MGI_C57BL6J_1890474 | Il21 | NCBI_Gene:60505,ENSEMBL:ENSMUSG00000027718 | MGI:1890474 | protein coding gene | interleukin 21 |
| 3 | gene | 37.22538 | 37.22711 | positive | MGI_C57BL6J_5595327 | Gm36168 | NCBI_Gene:102639985 | MGI:5595327 | lncRNA gene | predicted gene, 36168 |
| 3 | gene | 37.23133 | 37.23247 | positive | MGI_C57BL6J_3650303 | Gm12534 | ENSEMBL:ENSMUSG00000086517 | MGI:3650303 | lncRNA gene | predicted gene 12534 |
| 3 | gene | 37.23287 | 37.23334 | positive | MGI_C57BL6J_5663959 | Gm43822 | ENSEMBL:ENSMUSG00000104998 | MGI:5663959 | unclassified gene | predicted gene 43822 |
| 3 | gene | 37.25678 | 37.25689 | negative | MGI_C57BL6J_5453946 | Gm24169 | ENSEMBL:ENSMUSG00000094007 | MGI:5453946 | miRNA gene | predicted gene, 24169 |
| 3 | gene | 37.30775 | 37.31271 | negative | MGI_C57BL6J_2677454 | Cetn4 | NCBI_Gene:207175,ENSEMBL:ENSMUSG00000045031 | MGI:2677454 | protein coding gene | centrin 4 |
| 3 | gene | 37.31249 | 37.32145 | positive | MGI_C57BL6J_2686651 | Bbs12 | NCBI_Gene:241950,ENSEMBL:ENSMUSG00000051444 | MGI:2686651 | protein coding gene | Bardet-Biedl syndrome 12 (human) |
| 3 | gene | 37.31252 | 37.40487 | positive | MGI_C57BL6J_5663576 | Gm43439 | ENSEMBL:ENSMUSG00000106407 | MGI:5663576 | lncRNA gene | predicted gene 43439 |
| 3 | pseudogene | 37.31253 | 37.32919 | positive | MGI_C57BL6J_3651849 | Gm12540 | NCBI_Gene:102640067,ENSEMBL:ENSMUSG00000083353 | MGI:3651849 | pseudogene | predicted gene 12540 |
| 3 | gene | 37.33329 | 37.35103 | negative | MGI_C57BL6J_3649376 | Fgf2os | NCBI_Gene:329626,ENSEMBL:ENSMUSG00000054779 | MGI:3649376 | antisense lncRNA gene | fibroblast growth factor 2, opposite strand |
| 3 | gene | 37.33902 | 37.34152 | positive | MGI_C57BL6J_5663625 | Gm43488 | ENSEMBL:ENSMUSG00000105063 | MGI:5663625 | unclassified gene | predicted gene 43488 |
| 3 | gene | 37.34835 | 37.41011 | positive | MGI_C57BL6J_95516 | Fgf2 | NCBI_Gene:14173,ENSEMBL:ENSMUSG00000037225 | MGI:95516 | protein coding gene | fibroblast growth factor 2 |
| 3 | gene | 37.39221 | 37.39510 | negative | MGI_C57BL6J_5663060 | Gm42923 | ENSEMBL:ENSMUSG00000105134 | MGI:5663060 | unclassified gene | predicted gene 42923 |
| 3 | gene | 37.40491 | 37.42021 | negative | MGI_C57BL6J_2387618 | Nudt6 | NCBI_Gene:229228,ENSEMBL:ENSMUSG00000050174 | MGI:2387618 | protein coding gene | nudix (nucleoside diphosphate linked moiety X)-type motif 6 |
| 3 | gene | 37.41990 | 37.57910 | positive | MGI_C57BL6J_1927170 | Spata5 | NCBI_Gene:57815,ENSEMBL:ENSMUSG00000027722 | MGI:1927170 | protein coding gene | spermatogenesis associated 5 |
| 3 | gene | 37.48883 | 37.49479 | positive | MGI_C57BL6J_5663621 | Gm43484 | ENSEMBL:ENSMUSG00000106166 | MGI:5663621 | unclassified gene | predicted gene 43484 |
| 3 | gene | 37.54188 | 37.60239 | negative | MGI_C57BL6J_5595571 | Gm36412 | NCBI_Gene:102640314 | MGI:5595571 | lncRNA gene | predicted gene, 36412 |
| 3 | pseudogene | 37.55618 | 37.55650 | negative | MGI_C57BL6J_3652207 | Gm12564 | ENSEMBL:ENSMUSG00000080723 | MGI:3652207 | pseudogene | predicted gene 12564 |
| 3 | pseudogene | 37.59942 | 37.60007 | negative | MGI_C57BL6J_3652206 | Gm12565 | NCBI_Gene:665778,ENSEMBL:ENSMUSG00000081093 | MGI:3652206 | pseudogene | predicted gene 12565 |
| 3 | gene | 37.60989 | 37.61528 | positive | MGI_C57BL6J_1923197 | 4930594O21Rik | NCBI_Gene:75947,ENSEMBL:ENSMUSG00000085677 | MGI:1923197 | lncRNA gene | RIKEN cDNA 4930594O21 gene |
| 3 | gene | 37.63858 | 37.64066 | negative | MGI_C57BL6J_5663059 | Gm42922 | ENSEMBL:ENSMUSG00000104605 | MGI:5663059 | unclassified gene | predicted gene 42922 |
| 3 | gene | 37.63995 | 37.64460 | positive | MGI_C57BL6J_1345139 | Spry1 | NCBI_Gene:24063,ENSEMBL:ENSMUSG00000037211 | MGI:1345139 | protein coding gene | sprouty RTK signaling antagonist 1 |
| 3 | gene | 37.64461 | 37.64795 | positive | MGI_C57BL6J_5663957 | Gm43820 | ENSEMBL:ENSMUSG00000106062 | MGI:5663957 | unclassified gene | predicted gene 43820 |
| 3 | gene | 37.65268 | 37.65652 | negative | MGI_C57BL6J_5826436 | Gm46799 | NCBI_Gene:108168864 | MGI:5826436 | lncRNA gene | predicted gene, 46799 |
| 3 | gene | 37.69046 | 37.69486 | negative | MGI_C57BL6J_5625096 | Gm42211 | NCBI_Gene:105247029 | MGI:5625096 | lncRNA gene | predicted gene, 42211 |
| 3 | gene | 37.70802 | 37.71121 | positive | MGI_C57BL6J_5663551 | Gm43414 | ENSEMBL:ENSMUSG00000104935 | MGI:5663551 | unclassified gene | predicted gene 43414 |
| 3 | gene | 37.71419 | 37.72449 | negative | MGI_C57BL6J_3646006 | Gm5148 | NCBI_Gene:381438,ENSEMBL:ENSMUSG00000058174 | MGI:3646006 | protein coding gene | predicted gene 5148 |
| 3 | pseudogene | 37.71471 | 37.71522 | positive | MGI_C57BL6J_3705429 | Rps23-ps1 | NCBI_Gene:100038875,ENSEMBL:ENSMUSG00000100755 | MGI:3705429 | pseudogene | ribosomal protein S23, pseudogene 1 |
| 3 | gene | 37.72423 | 37.74132 | positive | MGI_C57BL6J_5595779 | Gm36620 | NCBI_Gene:102640592 | MGI:5595779 | lncRNA gene | predicted gene, 36620 |
| 3 | pseudogene | 37.73468 | 37.73514 | positive | MGI_C57BL6J_3643738 | Rpl31-ps15 | NCBI_Gene:626367,ENSEMBL:ENSMUSG00000097304 | MGI:3643738 | pseudogene | ribosomal protein L31, pseudogene 15 |
| 3 | gene | 37.75320 | 37.77440 | positive | MGI_C57BL6J_5625095 | Gm42210 | NCBI_Gene:105247028 | MGI:5625095 | lncRNA gene | predicted gene, 42210 |
| 3 | gene | 37.76104 | 37.76730 | positive | MGI_C57BL6J_5663058 | Gm42921 | ENSEMBL:ENSMUSG00000106441 | MGI:5663058 | lncRNA gene | predicted gene 42921 |
| 3 | gene | 37.80254 | 37.80265 | negative | MGI_C57BL6J_5456181 | Gm26404 | ENSEMBL:ENSMUSG00000089123 | MGI:5456181 | snoRNA gene | predicted gene, 26404 |
| 3 | gene | 37.82822 | 37.84829 | positive | MGI_C57BL6J_1925294 | 4930556A17Rik | NCBI_Gene:102640520 | MGI:1925294 | lncRNA gene | RIKEN cDNA 4930556A17 gene |
| 3 | gene | 37.83847 | 37.83957 | positive | MGI_C57BL6J_5662889 | Gm42752 | ENSEMBL:ENSMUSG00000106621 | MGI:5662889 | unclassified gene | predicted gene 42752 |
| 3 | gene | 37.84180 | 37.84307 | positive | MGI_C57BL6J_5662890 | Gm42753 | ENSEMBL:ENSMUSG00000105252 | MGI:5662890 | unclassified gene | predicted gene 42753 |
| 3 | gene | 37.89881 | 37.99273 | positive | MGI_C57BL6J_5434111 | Gm20755 | NCBI_Gene:626410,ENSEMBL:ENSMUSG00000106461 | MGI:5434111 | lncRNA gene | predicted gene, 20755 |
| 3 | pseudogene | 37.95961 | 37.96009 | negative | MGI_C57BL6J_3643378 | Gm7799 | ENSEMBL:ENSMUSG00000087457 | MGI:3643378 | pseudogene | predicted gene 7799 |
| 3 | gene | 37.98279 | 37.98335 | positive | MGI_C57BL6J_5662888 | Gm42751 | ENSEMBL:ENSMUSG00000105099 | MGI:5662888 | unclassified gene | predicted gene 42751 |
| 3 | gene | 37.99511 | 37.99524 | negative | MGI_C57BL6J_5452676 | Gm22899 | ENSEMBL:ENSMUSG00000077728 | MGI:5452676 | snoRNA gene | predicted gene, 22899 |
| 3 | gene | 38.09463 | 38.13019 | negative | MGI_C57BL6J_5663958 | Gm43821 | ENSEMBL:ENSMUSG00000105219 | MGI:5663958 | lncRNA gene | predicted gene 43821 |
| 3 | pseudogene | 38.10123 | 38.10798 | negative | MGI_C57BL6J_3643358 | Rpl21-ps10 | NCBI_Gene:665821,ENSEMBL:ENSMUSG00000105359 | MGI:3643358 | pseudogene | ribosomal protein L21, pseudogene 10 |
| 3 | gene | 38.15191 | 38.15915 | positive | MGI_C57BL6J_5595839 | Gm36680 | NCBI_Gene:102640666 | MGI:5595839 | lncRNA gene | predicted gene, 36680 |
| 3 | gene | 38.20315 | 38.20812 | negative | MGI_C57BL6J_1918592 | 5430434I15Rik | NCBI_Gene:71342,ENSEMBL:ENSMUSG00000106002 | MGI:1918592 | lncRNA gene | RIKEN cDNA 5430434I15 gene |
| 3 | pseudogene | 38.22345 | 38.22382 | positive | MGI_C57BL6J_3781143 | Gm2965 | NCBI_Gene:100040785,ENSEMBL:ENSMUSG00000105943 | MGI:3781143 | pseudogene | predicted gene 2965 |
| 3 | pseudogene | 38.25402 | 38.25480 | positive | MGI_C57BL6J_5010140 | Gm17955 | NCBI_Gene:100416169 | MGI:5010140 | pseudogene | predicted gene, 17955 |
| 3 | gene | 38.26784 | 38.26808 | negative | MGI_C57BL6J_5455351 | Gm25574 | ENSEMBL:ENSMUSG00000092986 | MGI:5455351 | unclassified non-coding RNA gene | predicted gene, 25574 |
| 3 | gene | 38.36615 | 38.36888 | negative | MGI_C57BL6J_5625098 | Gm42213 | NCBI_Gene:105247031 | MGI:5625098 | lncRNA gene | predicted gene, 42213 |
| 3 | gene | 38.36940 | 38.37364 | positive | MGI_C57BL6J_5663057 | Gm42920 | ENSEMBL:ENSMUSG00000106008 | MGI:5663057 | lncRNA gene | predicted gene 42920 |
| 3 | gene | 38.41897 | 38.42015 | negative | MGI_C57BL6J_1917958 | 3830422I06Rik | ENSEMBL:ENSMUSG00000106485 | MGI:1917958 | unclassified gene | RIKEN cDNA 3830422I06 gene |
| 3 | gene | 38.42655 | 38.44387 | negative | MGI_C57BL6J_5595982 | Gm36823 | NCBI_Gene:102640855,ENSEMBL:ENSMUSG00000105516 | MGI:5595982 | lncRNA gene | predicted gene, 36823 |
| 3 | pseudogene | 38.43895 | 38.43984 | negative | MGI_C57BL6J_3646285 | Gm7815 | NCBI_Gene:665838,ENSEMBL:ENSMUSG00000106201 | MGI:3646285 | pseudogene | predicted gene 7815 |
| 3 | gene | 38.44926 | 38.48486 | negative | MGI_C57BL6J_2139777 | Ankrd50 | NCBI_Gene:99696,ENSEMBL:ENSMUSG00000044864 | MGI:2139777 | protein coding gene | ankyrin repeat domain 50 |
| 3 | pseudogene | 38.55797 | 38.55886 | negative | MGI_C57BL6J_3779766 | Gm7824 | NCBI_Gene:665855,ENSEMBL:ENSMUSG00000105360 | MGI:3779766 | pseudogene | predicted gene 7824 |
| 3 | gene | 38.55811 | 38.55821 | negative | MGI_C57BL6J_5454239 | Gm24462 | ENSEMBL:ENSMUSG00000076385 | MGI:5454239 | miRNA gene | predicted gene, 24462 |
| 3 | gene | 38.61425 | 38.61556 | positive | MGI_C57BL6J_5663675 | Gm43538 | ENSEMBL:ENSMUSG00000106168 | MGI:5663675 | lncRNA gene | predicted gene 43538 |
| 3 | gene | 38.66361 | 38.70922 | positive | MGI_C57BL6J_5589038 | Gm29879 | NCBI_Gene:102631576 | MGI:5589038 | lncRNA gene | predicted gene, 29879 |
| 3 | gene | 38.76048 | 38.76195 | negative | MGI_C57BL6J_5589118 | Gm29959 | NCBI_Gene:102631683 | MGI:5589118 | lncRNA gene | predicted gene, 29959 |
| 3 | gene | 38.87457 | 38.88826 | negative | MGI_C57BL6J_2441892 | C230034O21Rik | NCBI_Gene:102632538,ENSEMBL:ENSMUSG00000086181 | MGI:2441892 | lncRNA gene | RIKEN cDNA C230034O21 gene |
| 3 | gene | 38.88563 | 39.01199 | positive | MGI_C57BL6J_3045256 | Fat4 | NCBI_Gene:329628,ENSEMBL:ENSMUSG00000046743 | MGI:3045256 | protein coding gene | FAT atypical cadherin 4 |
| 3 | gene | 39.00860 | 39.00968 | negative | MGI_C57BL6J_5663676 | Gm43539 | ENSEMBL:ENSMUSG00000105910 | MGI:5663676 | lncRNA gene | predicted gene 43539 |
| 3 | pseudogene | 39.12853 | 39.12891 | negative | MGI_C57BL6J_5663445 | Gm43308 | ENSEMBL:ENSMUSG00000104792 | MGI:5663445 | pseudogene | predicted gene 43308 |
| 3 | gene | 39.15083 | 39.15967 | negative | MGI_C57BL6J_5589170 | Gm30011 | NCBI_Gene:102631750 | MGI:5589170 | lncRNA gene | predicted gene, 30011 |
| 3 | pseudogene | 39.21756 | 39.21785 | negative | MGI_C57BL6J_5663145 | Gm43008 | ENSEMBL:ENSMUSG00000107333 | MGI:5663145 | pseudogene | predicted gene 43008 |
| 3 | gene | 39.27713 | 39.30593 | positive | MGI_C57BL6J_5625099 | Gm42214 | NCBI_Gene:105247032 | MGI:5625099 | lncRNA gene | predicted gene, 42214 |
| 3 | pseudogene | 39.35737 | 39.35913 | negative | MGI_C57BL6J_3704215 | Gm9845 | NCBI_Gene:100040853,ENSEMBL:ENSMUSG00000050533 | MGI:3704215 | pseudogene | predicted pseudogene 9845 |
| 3 | pseudogene | 39.44389 | 39.44435 | positive | MGI_C57BL6J_5295673 | Gm20566 | NCBI_Gene:100047050 | MGI:5295673 | pseudogene | predicted gene, 20566 |
| 3 | gene | 39.46078 | 39.59865 | positive | MGI_C57BL6J_5589222 | Gm30063 | NCBI_Gene:102631823 | MGI:5589222 | lncRNA gene | predicted gene, 30063 |
| 3 | gene | 39.52552 | 39.53415 | negative | MGI_C57BL6J_5625100 | Gm42215 | NCBI_Gene:105247033 | MGI:5625100 | lncRNA gene | predicted gene, 42215 |
| 3 | gene | 39.63596 | 39.63608 | negative | MGI_C57BL6J_4422060 | n-R5s195 | ENSEMBL:ENSMUSG00000093952 | MGI:4422060 | rRNA gene | nuclear encoded rRNA 5S 195 |
| 3 | gene | 39.63606 | 39.63878 | positive | MGI_C57BL6J_5662918 | Gm42781 | ENSEMBL:ENSMUSG00000106786 | MGI:5662918 | unclassified gene | predicted gene 42781 |
| 3 | gene | 39.66629 | 39.66645 | positive | MGI_C57BL6J_5453573 | Gm23796 | ENSEMBL:ENSMUSG00000065945 | MGI:5453573 | snRNA gene | predicted gene, 23796 |
| 3 | gene | 39.71315 | 39.72565 | positive | MGI_C57BL6J_5826471 | Gm46834 | NCBI_Gene:108168914 | MGI:5826471 | lncRNA gene | predicted gene, 46834 |

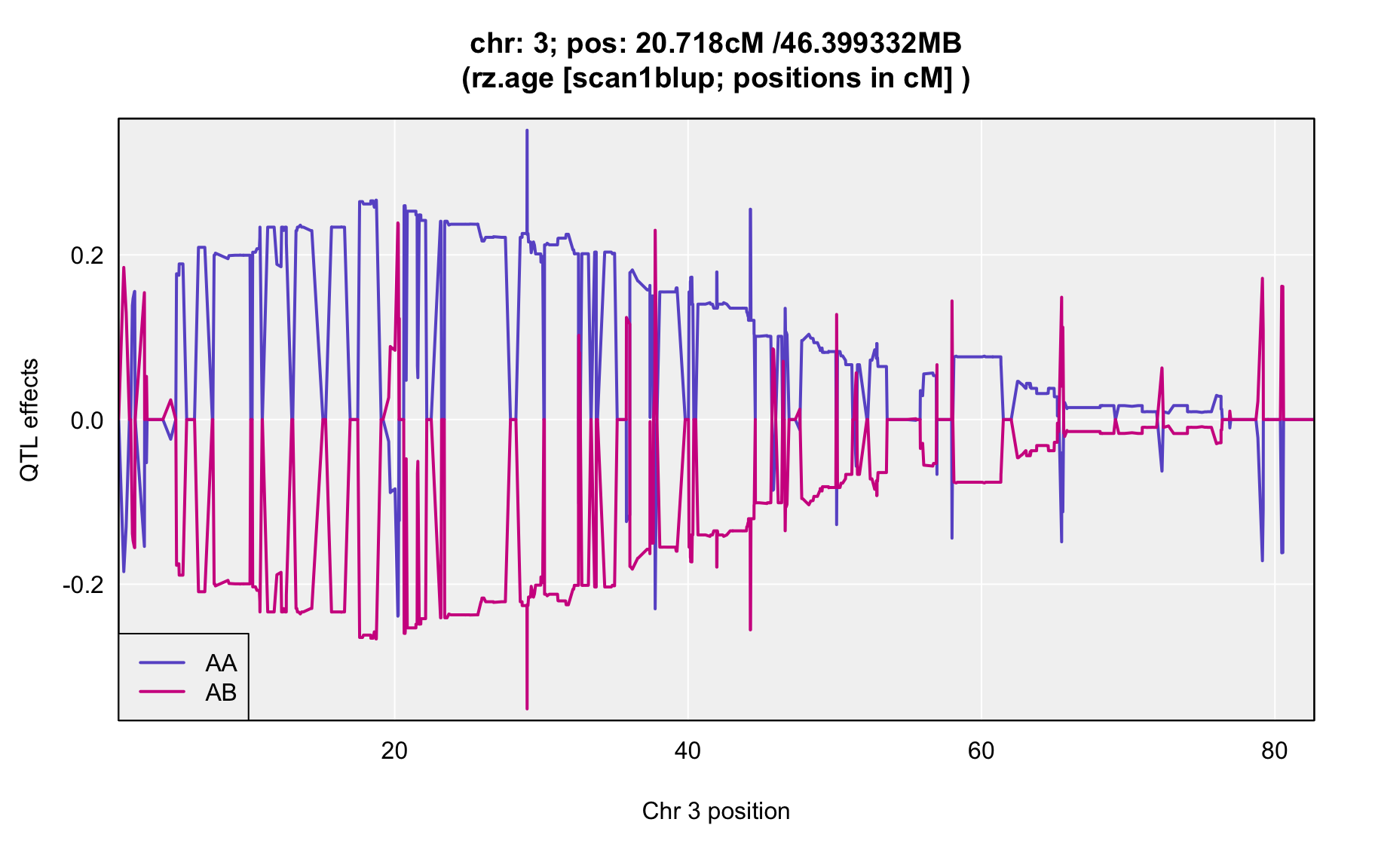

| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | gene | 46.28983 | 46.28996 | positive | MGI_C57BL6J_5455858 | Gm26081 | ENSEMBL:ENSMUSG00000077256 | MGI:5455858 | snoRNA gene | predicted gene, 26081 |
| 3 | pseudogene | 46.41340 | 46.41412 | positive | MGI_C57BL6J_5011028 | Gm18843 | NCBI_Gene:100417811,ENSEMBL:ENSMUSG00000104511 | MGI:5011028 | pseudogene | predicted gene, 18843 |
| 3 | gene | 46.44085 | 46.44822 | negative | MGI_C57BL6J_3643087 | Pabpc4l | NCBI_Gene:241989,ENSEMBL:ENSMUSG00000090919 | MGI:3643087 | protein coding gene | poly(A) binding protein, cytoplasmic 4-like |
| 3 | pseudogene | 46.62112 | 46.62136 | positive | MGI_C57BL6J_5610673 | Gm37445 | ENSEMBL:ENSMUSG00000102589 | MGI:5610673 | pseudogene | predicted gene, 37445 |
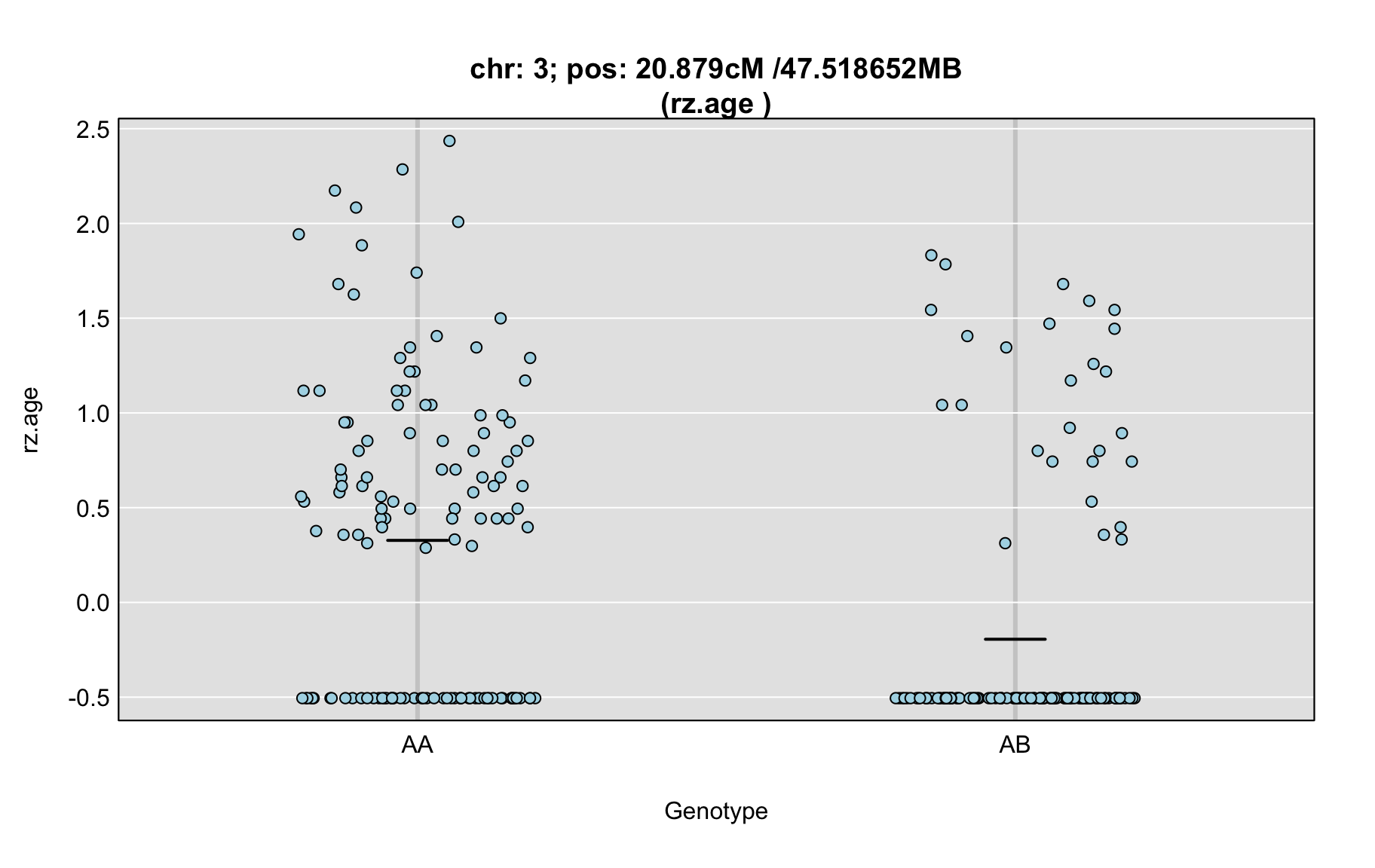
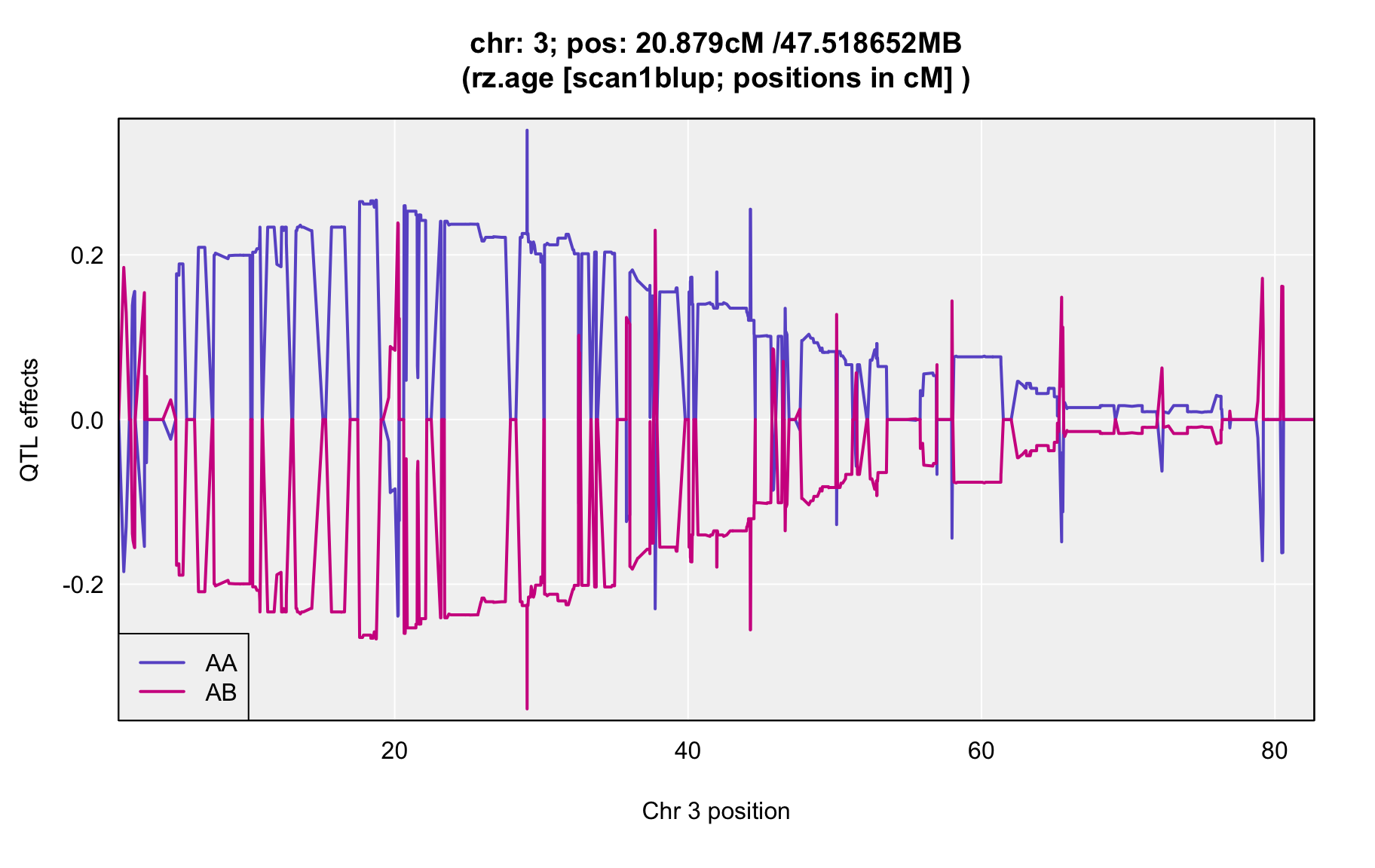

| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | pseudogene | 47.45961 | 47.46034 | negative | MGI_C57BL6J_3780399 | Gm2229 | NCBI_Gene:100039426,ENSEMBL:ENSMUSG00000103798 | MGI:3780399 | pseudogene | predicted gene 2229 |
| 3 | pseudogene | 48.02601 | 48.02676 | negative | MGI_C57BL6J_3648898 | Gm7977 | NCBI_Gene:666202,ENSEMBL:ENSMUSG00000103257 | MGI:3648898 | pseudogene | predicted gene 7977 |
| 3 | gene | 48.07834 | 48.07845 | positive | MGI_C57BL6J_5610523 | Gm37295 | ENSEMBL:ENSMUSG00000104110 | MGI:5610523 | unclassified gene | predicted gene, 37295 |
| 3 | gene | 48.54642 | 48.57947 | positive | MGI_C57BL6J_5826472 | Gm46835 | NCBI_Gene:108168915 | MGI:5826472 | lncRNA gene | predicted gene, 46835 |
| 3 | gene | 48.60573 | 48.60910 | negative | MGI_C57BL6J_1913582 | 1700018B24Rik | NCBI_Gene:66332,ENSEMBL:ENSMUSG00000037910 | MGI:1913582 | lncRNA gene | RIKEN cDNA 1700018B24 gene |
| 3 | gene | 48.74851 | 48.76095 | negative | MGI_C57BL6J_5610418 | Gm37190 | ENSEMBL:ENSMUSG00000102181 | MGI:5610418 | lncRNA gene | predicted gene, 37190 |

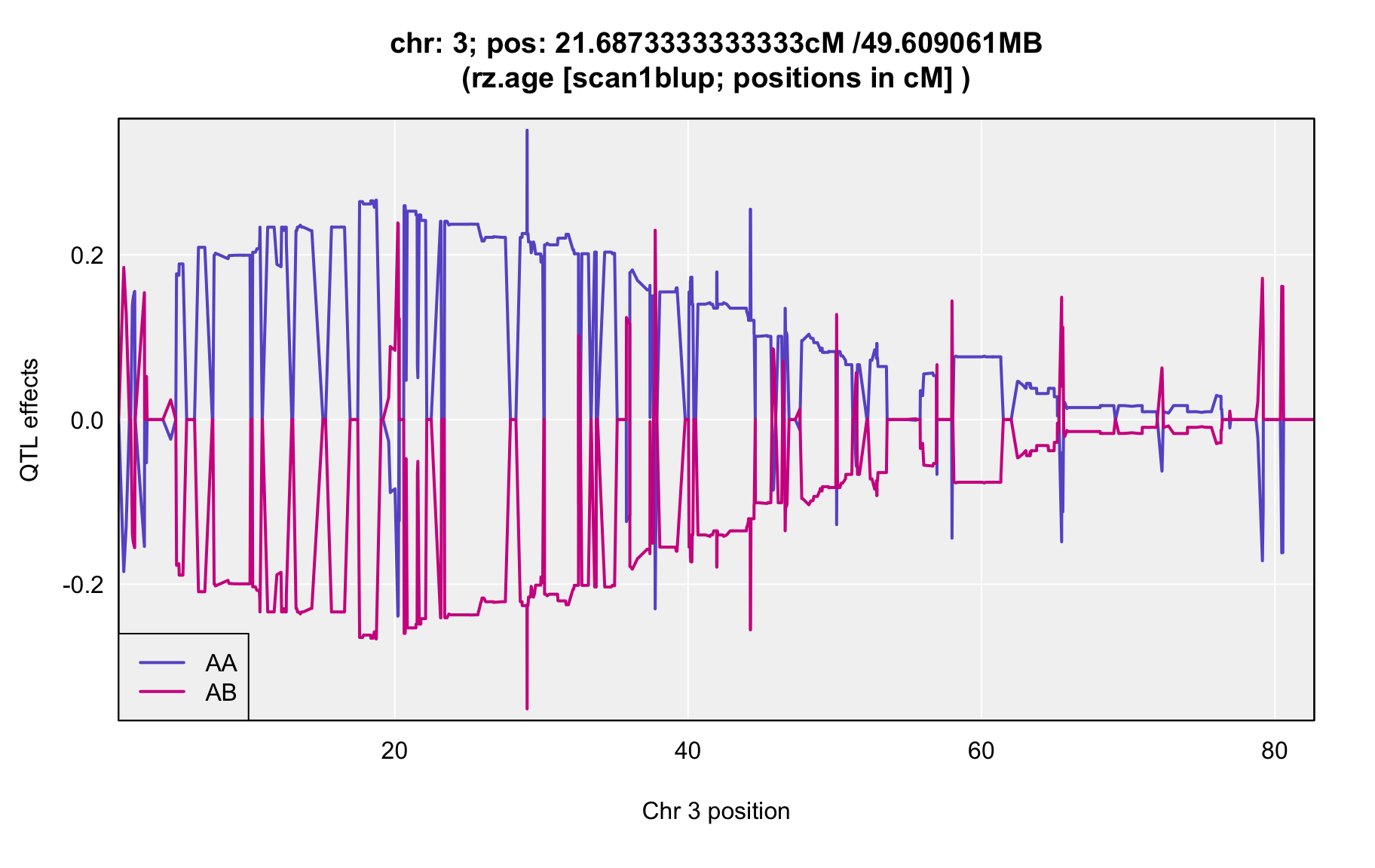
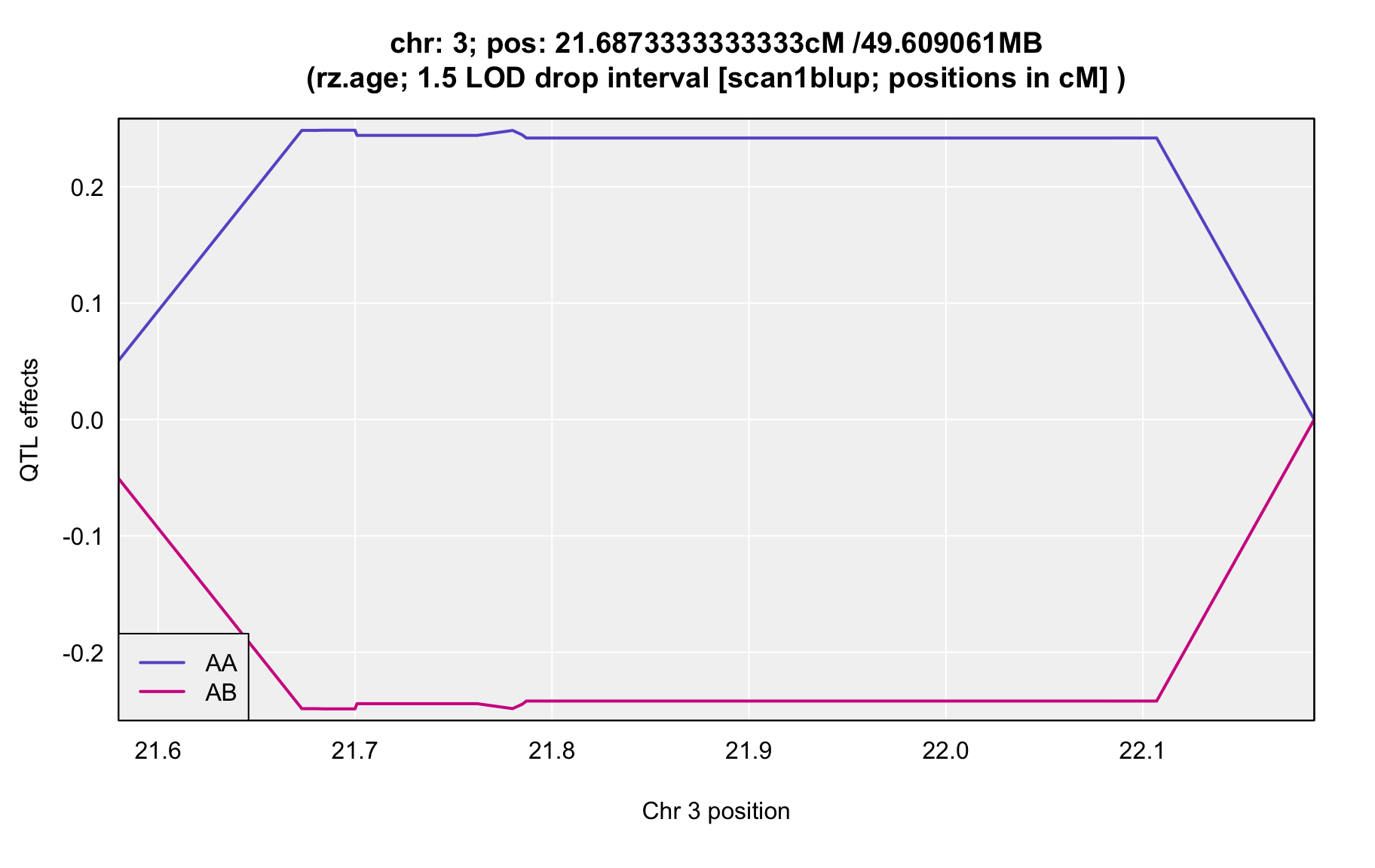
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | gene | 48.95472 | 48.95488 | positive | MGI_C57BL6J_5453288 | Gm23511 | ENSEMBL:ENSMUSG00000065018 | MGI:5453288 | snRNA gene | predicted gene, 23511 |
| 3 | gene | 49.06237 | 49.09636 | positive | MGI_C57BL6J_5590574 | Gm31415 | NCBI_Gene:102633632,ENSEMBL:ENSMUSG00000102263 | MGI:5590574 | lncRNA gene | predicted gene, 31415 |
| 3 | gene | 49.13746 | 49.13853 | positive | MGI_C57BL6J_5611047 | Gm37819 | ENSEMBL:ENSMUSG00000104186 | MGI:5611047 | unclassified gene | predicted gene, 37819 |
| 3 | pseudogene | 49.20771 | 49.20807 | negative | MGI_C57BL6J_5610912 | Gm37684 | ENSEMBL:ENSMUSG00000103027 | MGI:5610912 | pseudogene | predicted gene, 37684 |
| 3 | gene | 49.32568 | 49.36736 | positive | MGI_C57BL6J_5295664 | Gm20557 | NCBI_Gene:329633,ENSEMBL:ENSMUSG00000102679 | MGI:5295664 | lncRNA gene | predicted gene, 20557 |
| 3 | pseudogene | 49.38115 | 49.38285 | negative | MGI_C57BL6J_3643624 | Gm5846 | NCBI_Gene:545519,ENSEMBL:ENSMUSG00000104309 | MGI:3643624 | pseudogene | predicted gene 5846 |
| 3 | pseudogene | 49.40082 | 49.46744 | negative | MGI_C57BL6J_3780091 | Gm9683 | NCBI_Gene:676568 | MGI:3780091 | pseudogene | predicted gene 9683 |
| 3 | gene | 49.49135 | 49.49148 | positive | MGI_C57BL6J_5452257 | Gm22480 | ENSEMBL:ENSMUSG00000084517 | MGI:5452257 | snoRNA gene | predicted gene, 22480 |
| 3 | pseudogene | 49.53075 | 49.53131 | positive | MGI_C57BL6J_5610652 | Gm37424 | ENSEMBL:ENSMUSG00000104437 | MGI:5610652 | pseudogene | predicted gene, 37424 |
| 3 | pseudogene | 49.60356 | 49.60388 | positive | MGI_C57BL6J_5611489 | Gm38261 | ENSEMBL:ENSMUSG00000102731 | MGI:5611489 | pseudogene | predicted gene, 38261 |
| 3 | pseudogene | 49.60423 | 49.60447 | positive | MGI_C57BL6J_5611316 | Gm38088 | ENSEMBL:ENSMUSG00000103249 | MGI:5611316 | pseudogene | predicted gene, 38088 |
| 3 | pseudogene | 49.63244 | 49.63297 | negative | MGI_C57BL6J_3780498 | Gm2328 | NCBI_Gene:100039587,ENSEMBL:ENSMUSG00000104134 | MGI:3780498 | pseudogene | predicted gene 2328 |
| 3 | pseudogene | 49.64348 | 49.64413 | positive | MGI_C57BL6J_5588900 | Gm29741 | NCBI_Gene:101055903,ENSEMBL:ENSMUSG00000103568 | MGI:5588900 | pseudogene | predicted gene, 29741 |
| 3 | gene | 49.65876 | 49.65887 | positive | MGI_C57BL6J_5452873 | Gm23096 | ENSEMBL:ENSMUSG00000065381 | MGI:5452873 | snRNA gene | predicted gene, 23096 |
| 3 | gene | 49.74329 | 49.75738 | negative | MGI_C57BL6J_1920423 | Pcdh18 | NCBI_Gene:73173,ENSEMBL:ENSMUSG00000037892 | MGI:1920423 | protein coding gene | protocadherin 18 |
| 3 | pseudogene | 49.78043 | 49.78089 | negative | MGI_C57BL6J_5610778 | Gm37550 | ENSEMBL:ENSMUSG00000102943 | MGI:5610778 | pseudogene | predicted gene, 37550 |
| 3 | gene | 49.89253 | 50.49909 | negative | MGI_C57BL6J_1347355 | Slc7a11 | NCBI_Gene:26570,ENSEMBL:ENSMUSG00000027737 | MGI:1347355 | protein coding gene | solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| 3 | gene | 50.00197 | 50.01078 | positive | MGI_C57BL6J_5611082 | Gm37854 | ENSEMBL:ENSMUSG00000102892 | MGI:5611082 | lncRNA gene | predicted gene, 37854 |
| 3 | gene | 50.01177 | 50.02624 | negative | MGI_C57BL6J_5622927 | Gm40042 | NCBI_Gene:105244434 | MGI:5622927 | lncRNA gene | predicted gene, 40042 |
| 3 | pseudogene | 50.02967 | 50.02999 | positive | MGI_C57BL6J_5611054 | Gm37826 | ENSEMBL:ENSMUSG00000104457 | MGI:5611054 | pseudogene | predicted gene, 37826 |
| 3 | pseudogene | 50.16666 | 50.16689 | negative | MGI_C57BL6J_5610920 | Gm37692 | ENSEMBL:ENSMUSG00000104349 | MGI:5610920 | pseudogene | predicted gene, 37692 |
| 3 | pseudogene | 50.25485 | 50.25544 | positive | MGI_C57BL6J_3780515 | Gm2345 | NCBI_Gene:100039628,ENSEMBL:ENSMUSG00000102167 | MGI:3780515 | pseudogene | predicted gene 2345 |
| 3 | gene | 50.40028 | 50.40428 | negative | MGI_C57BL6J_5610726 | Gm37498 | ENSEMBL:ENSMUSG00000103907 | MGI:5610726 | unclassified gene | predicted gene, 37498 |
| 3 | gene | 50.51640 | 50.60731 | negative | MGI_C57BL6J_3643374 | Gm6209 | NCBI_Gene:621304,ENSEMBL:ENSMUSG00000102715 | MGI:3643374 | lncRNA gene | predicted gene 6209 |
| 3 | gene | 50.57519 | 50.57652 | negative | MGI_C57BL6J_1923259 | 5830415G21Rik | ENSEMBL:ENSMUSG00000102895 | MGI:1923259 | unclassified gene | RIKEN cDNA 5830415G21 gene |
| 3 | pseudogene | 50.59799 | 50.59850 | negative | MGI_C57BL6J_5611465 | Gm38237 | NCBI_Gene:108168851,ENSEMBL:ENSMUSG00000103745 | MGI:5611465 | pseudogene | predicted gene, 38237 |
| 3 | gene | 50.62435 | 50.62841 | negative | MGI_C57BL6J_5610427 | Gm37199 | ENSEMBL:ENSMUSG00000104068 | MGI:5610427 | unclassified gene | predicted gene, 37199 |
| 3 | gene | 50.67970 | 50.68340 | negative | MGI_C57BL6J_5610689 | Gm37461 | ENSEMBL:ENSMUSG00000104281 | MGI:5610689 | unclassified gene | predicted gene, 37461 |
| 3 | gene | 50.70690 | 50.71189 | negative | MGI_C57BL6J_5622928 | Gm40043 | NCBI_Gene:105244435 | MGI:5622928 | lncRNA gene | predicted gene, 40043 |
| 3 | gene | 50.83858 | 50.83872 | negative | MGI_C57BL6J_5454280 | Gm24503 | ENSEMBL:ENSMUSG00000089079 | MGI:5454280 | snoRNA gene | predicted gene, 24503 |
| 3 | gene | 50.88324 | 50.88889 | negative | MGI_C57BL6J_5610776 | Gm37548 | NCBI_Gene:105244436,ENSEMBL:ENSMUSG00000103012 | MGI:5610776 | lncRNA gene | predicted gene, 37548 |
| 3 | gene | 50.91889 | 50.92153 | negative | MGI_C57BL6J_5611071 | Gm37843 | ENSEMBL:ENSMUSG00000102614 | MGI:5611071 | unclassified gene | predicted gene, 37843 |
| 3 | gene | 50.96656 | 50.96677 | positive | MGI_C57BL6J_2139603 | AA672641 | NA | NA | unclassified gene | expressed sequence AA672641 |
| 3 | gene | 50.97754 | 50.97881 | negative | MGI_C57BL6J_5610437 | Gm37209 | ENSEMBL:ENSMUSG00000103652 | MGI:5610437 | unclassified gene | predicted gene, 37209 |
| 3 | gene | 51.03335 | 51.04284 | negative | MGI_C57BL6J_5594915 | Gm35756 | NCBI_Gene:102639442 | MGI:5594915 | lncRNA gene | predicted gene, 35756 |
| 3 | gene | 51.05891 | 51.08934 | negative | MGI_C57BL6J_5579936 | Gm29230 | NCBI_Gene:102639521,ENSEMBL:ENSMUSG00000102067 | MGI:5579936 | lncRNA gene | predicted gene 29230 |
| 3 | gene | 51.10366 | 51.10517 | positive | MGI_C57BL6J_5611474 | Gm38246 | ENSEMBL:ENSMUSG00000103464 | MGI:5611474 | lncRNA gene | predicted gene, 38246 |
| 3 | gene | 51.12669 | 51.12894 | negative | MGI_C57BL6J_5595050 | Gm35891 | NCBI_Gene:102639626 | MGI:5595050 | lncRNA gene | predicted gene, 35891 |
| 3 | gene | 51.21847 | 51.22033 | negative | MGI_C57BL6J_5622929 | Gm40044 | NCBI_Gene:105244437 | MGI:5622929 | lncRNA gene | predicted gene, 40044 |
| 3 | gene | 51.22445 | 51.25165 | positive | MGI_C57BL6J_109382 | Noct | NCBI_Gene:12457,ENSEMBL:ENSMUSG00000023087 | MGI:109382 | protein coding gene | nocturnin |
| 3 | gene | 51.23192 | 51.23438 | positive | MGI_C57BL6J_5611585 | Gm38357 | ENSEMBL:ENSMUSG00000103313 | MGI:5611585 | unclassified gene | predicted gene, 38357 |
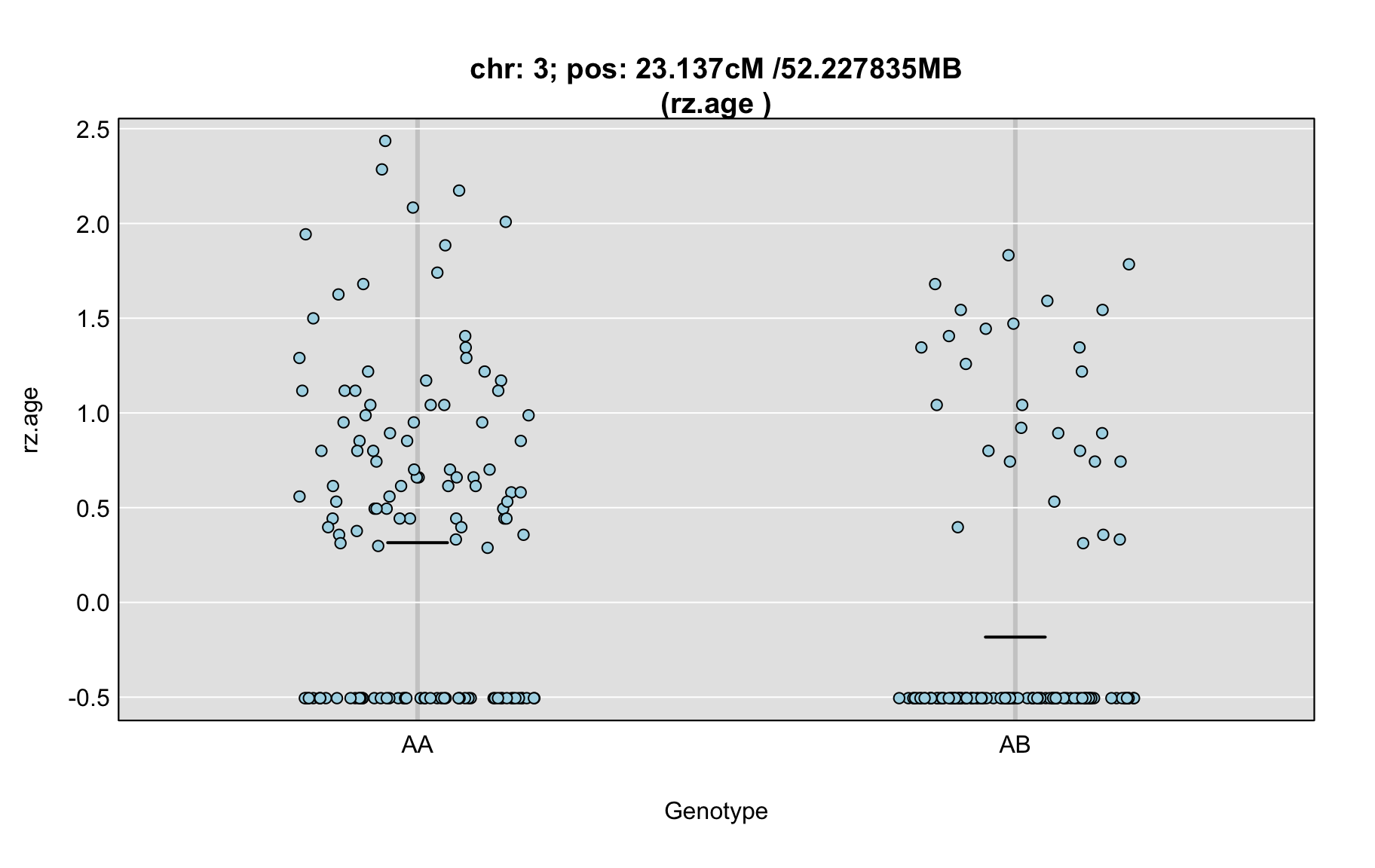

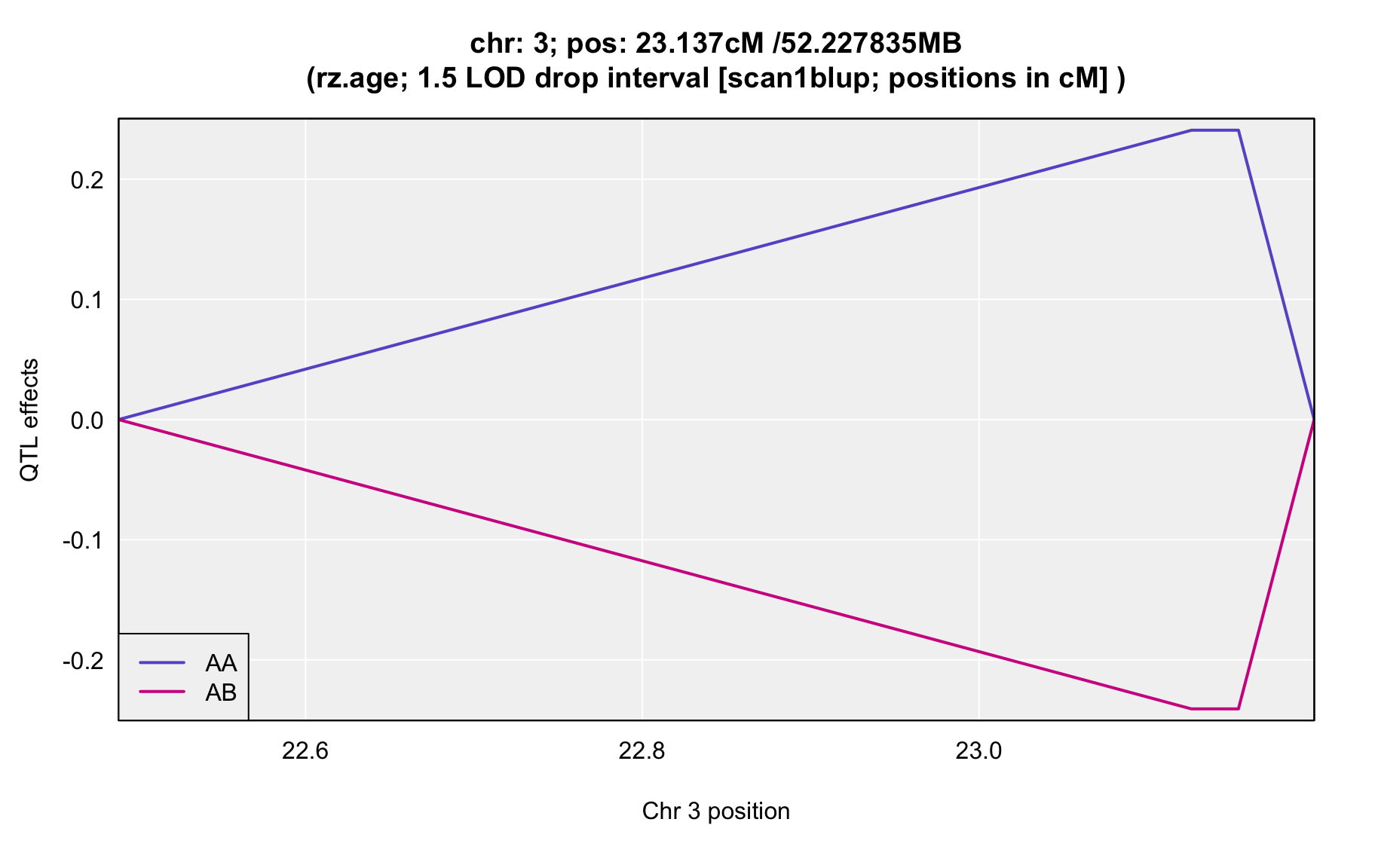
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | gene | 51.51099 | 51.51430 | positive | MGI_C57BL6J_5595167 | Gm36008 | NCBI_Gene:102639777 | MGI:5595167 | lncRNA gene | predicted gene, 36008 |
| 3 | gene | 51.51532 | 51.56088 | negative | MGI_C57BL6J_1920501 | Setd7 | NCBI_Gene:73251,ENSEMBL:ENSMUSG00000037111 | MGI:1920501 | protein coding gene | SET domain containing (lysine methyltransferase) 7 |
| 3 | gene | 51.55754 | 51.55965 | negative | MGI_C57BL6J_5611428 | Gm38200 | ENSEMBL:ENSMUSG00000102466 | MGI:5611428 | unclassified gene | predicted gene, 38200 |
| 3 | gene | 51.55976 | 51.56712 | positive | MGI_C57BL6J_1923230 | 5031434O11Rik | NCBI_Gene:100039684,ENSEMBL:ENSMUSG00000097885 | MGI:1923230 | lncRNA gene | RIKEN cDNA 5031434O11 gene |
| 3 | pseudogene | 51.57111 | 51.57142 | negative | MGI_C57BL6J_5611562 | Gm38334 | ENSEMBL:ENSMUSG00000104001 | MGI:5611562 | pseudogene | predicted gene, 38334 |
| 3 | gene | 51.58231 | 51.58285 | positive | MGI_C57BL6J_1918620 | 5430433H01Rik | ENSEMBL:ENSMUSG00000104075 | MGI:1918620 | unclassified gene | RIKEN cDNA 5430433H01 gene |
| 3 | pseudogene | 51.63649 | 51.63705 | negative | MGI_C57BL6J_5611475 | Gm38247 | NCBI_Gene:108168852,ENSEMBL:ENSMUSG00000104166 | MGI:5611475 | pseudogene | predicted gene, 38247 |
| 3 | gene | 51.66036 | 51.68268 | positive | MGI_C57BL6J_2448481 | Mgst2 | NCBI_Gene:211666,ENSEMBL:ENSMUSG00000074604 | MGI:2448481 | protein coding gene | microsomal glutathione S-transferase 2 |
| 3 | gene | 51.67280 | 51.67390 | positive | MGI_C57BL6J_5610489 | Gm37261 | ENSEMBL:ENSMUSG00000103527 | MGI:5610489 | unclassified gene | predicted gene, 37261 |
| 3 | gene | 51.68591 | 52.10509 | negative | MGI_C57BL6J_2389461 | Maml3 | NCBI_Gene:433586,ENSEMBL:ENSMUSG00000061143 | MGI:2389461 | protein coding gene | mastermind like transcriptional coactivator 3 |
| 3 | gene | 51.69372 | 51.69509 | positive | MGI_C57BL6J_3642905 | Gm10729 | ENSEMBL:ENSMUSG00000074603 | MGI:3642905 | protein coding gene | predicted gene 10729 |
| 3 | gene | 51.70772 | 51.71225 | positive | MGI_C57BL6J_5826440 | Gm46803 | NCBI_Gene:108168869 | MGI:5826440 | lncRNA gene | predicted gene, 46803 |
| 3 | gene | 51.75712 | 51.76209 | negative | MGI_C57BL6J_5610570 | Gm37342 | ENSEMBL:ENSMUSG00000102732 | MGI:5610570 | unclassified gene | predicted gene, 37342 |
| 3 | gene | 51.81295 | 51.81931 | positive | MGI_C57BL6J_5610457 | Gm37229 | ENSEMBL:ENSMUSG00000102539 | MGI:5610457 | lncRNA gene | predicted gene, 37229 |
| 3 | gene | 51.82891 | 51.82903 | positive | MGI_C57BL6J_4422061 | n-R5s196 | ENSEMBL:ENSMUSG00000065070 | MGI:4422061 | rRNA gene | nuclear encoded rRNA 5S 196 |
| 3 | gene | 51.82939 | 51.83040 | negative | MGI_C57BL6J_1920491 | 3110080O07Rik | ENSEMBL:ENSMUSG00000103284 | MGI:1920491 | unclassified gene | RIKEN cDNA 3110080O07 gene |
| 3 | gene | 51.86735 | 51.86856 | negative | MGI_C57BL6J_5611228 | Gm38000 | ENSEMBL:ENSMUSG00000103484 | MGI:5611228 | unclassified gene | predicted gene, 38000 |
| 3 | gene | 51.87299 | 51.87584 | negative | MGI_C57BL6J_5610903 | Gm37675 | ENSEMBL:ENSMUSG00000102620 | MGI:5610903 | unclassified gene | predicted gene, 37675 |
| 3 | gene | 51.88428 | 51.88689 | negative | MGI_C57BL6J_5611480 | Gm38252 | ENSEMBL:ENSMUSG00000102786 | MGI:5611480 | unclassified gene | predicted gene, 38252 |
| 3 | pseudogene | 51.90699 | 51.90882 | negative | MGI_C57BL6J_3708672 | Gm10728 | ENSEMBL:ENSMUSG00000103701 | MGI:3708672 | pseudogene | predicted gene 10728 |
| 3 | gene | 51.98094 | 51.99007 | positive | MGI_C57BL6J_5012274 | Gm20089 | ENSEMBL:ENSMUSG00000104161 | MGI:5012274 | lncRNA gene | predicted gene, 20089 |
| 3 | gene | 51.98763 | 51.98985 | negative | MGI_C57BL6J_5611283 | Gm38055 | ENSEMBL:ENSMUSG00000102147 | MGI:5611283 | unclassified gene | predicted gene, 38055 |
| 3 | gene | 51.99134 | 51.99196 | negative | MGI_C57BL6J_5610754 | Gm37526 | ENSEMBL:ENSMUSG00000103784 | MGI:5610754 | unclassified gene | predicted gene, 37526 |
| 3 | gene | 52.00074 | 52.00402 | negative | MGI_C57BL6J_5610693 | Gm37465 | ENSEMBL:ENSMUSG00000102423 | MGI:5610693 | unclassified gene | predicted gene, 37465 |
| 3 | gene | 52.05729 | 52.06424 | negative | MGI_C57BL6J_2444420 | C130089K02Rik | ENSEMBL:ENSMUSG00000104339 | MGI:2444420 | unclassified gene | RIKEN cDNA C130089K02 gene |
| 3 | gene | 52.07159 | 52.07339 | negative | MGI_C57BL6J_1921741 | 5430420F09Rik | ENSEMBL:ENSMUSG00000102808 | MGI:1921741 | unclassified gene | RIKEN cDNA 5430420F09 gene |
| 3 | gene | 52.13405 | 52.22200 | negative | MGI_C57BL6J_5610430 | Gm37202 | ENSEMBL:ENSMUSG00000103655 | MGI:5610430 | lncRNA gene | predicted gene, 37202 |
| 3 | gene | 52.17108 | 52.18013 | negative | MGI_C57BL6J_5589233 | Gm30074 | NCBI_Gene:102631843,ENSEMBL:ENSMUSG00000103726 | MGI:5589233 | lncRNA gene | predicted gene, 30074 |
| 3 | gene | 52.18657 | 52.18796 | positive | MGI_C57BL6J_5611360 | Gm38132 | ENSEMBL:ENSMUSG00000104055 | MGI:5611360 | lncRNA gene | predicted gene, 38132 |
| 3 | gene | 52.19825 | 52.20049 | positive | MGI_C57BL6J_5610398 | Gm37170 | ENSEMBL:ENSMUSG00000103170 | MGI:5610398 | lncRNA gene | predicted gene, 37170 |
| 3 | pseudogene | 52.23684 | 52.24694 | negative | MGI_C57BL6J_3031232 | Olfr1398-ps1 | NCBI_Gene:404507,ENSEMBL:ENSMUSG00000102510 | MGI:3031232 | pseudogene | olfactory receptor 1398, pseudogene 1 |
| 3 | gene | 52.23745 | 52.25650 | positive | MGI_C57BL6J_5589284 | Gm30125 | NCBI_Gene:102631910 | MGI:5589284 | lncRNA gene | predicted gene, 30125 |
| 3 | gene | 52.26834 | 52.35322 | positive | MGI_C57BL6J_1890077 | Foxo1 | NCBI_Gene:56458,ENSEMBL:ENSMUSG00000044167 | MGI:1890077 | protein coding gene | forkhead box O1 |
| 3 | gene | 52.26843 | 52.26944 | negative | MGI_C57BL6J_5141867 | Gm20402 | ENSEMBL:ENSMUSG00000092405 | MGI:5141867 | lncRNA gene | predicted gene 20402 |
| 3 | gene | 52.28224 | 52.28431 | positive | MGI_C57BL6J_5611262 | Gm38034 | ENSEMBL:ENSMUSG00000102844 | MGI:5611262 | unclassified gene | predicted gene, 38034 |
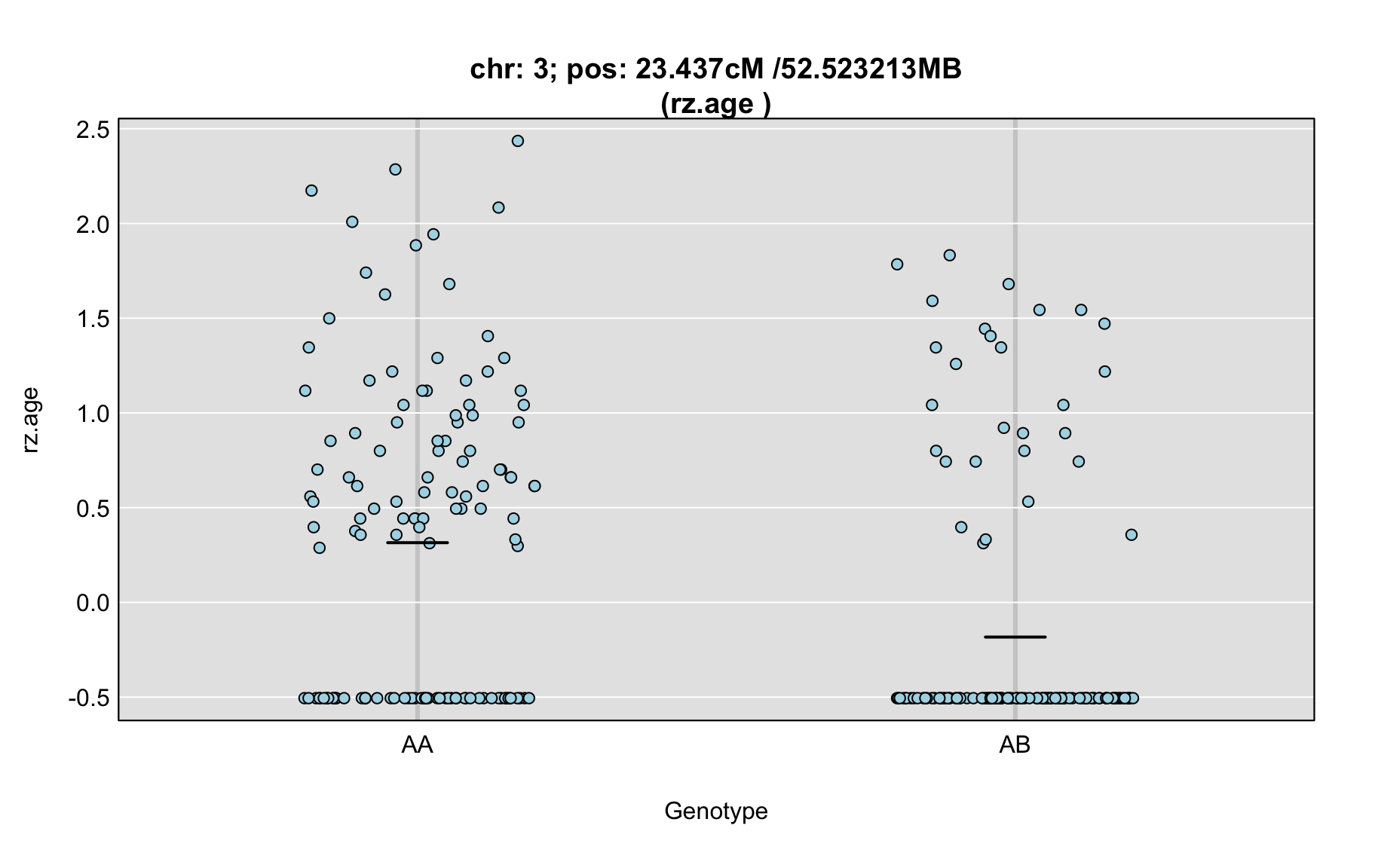

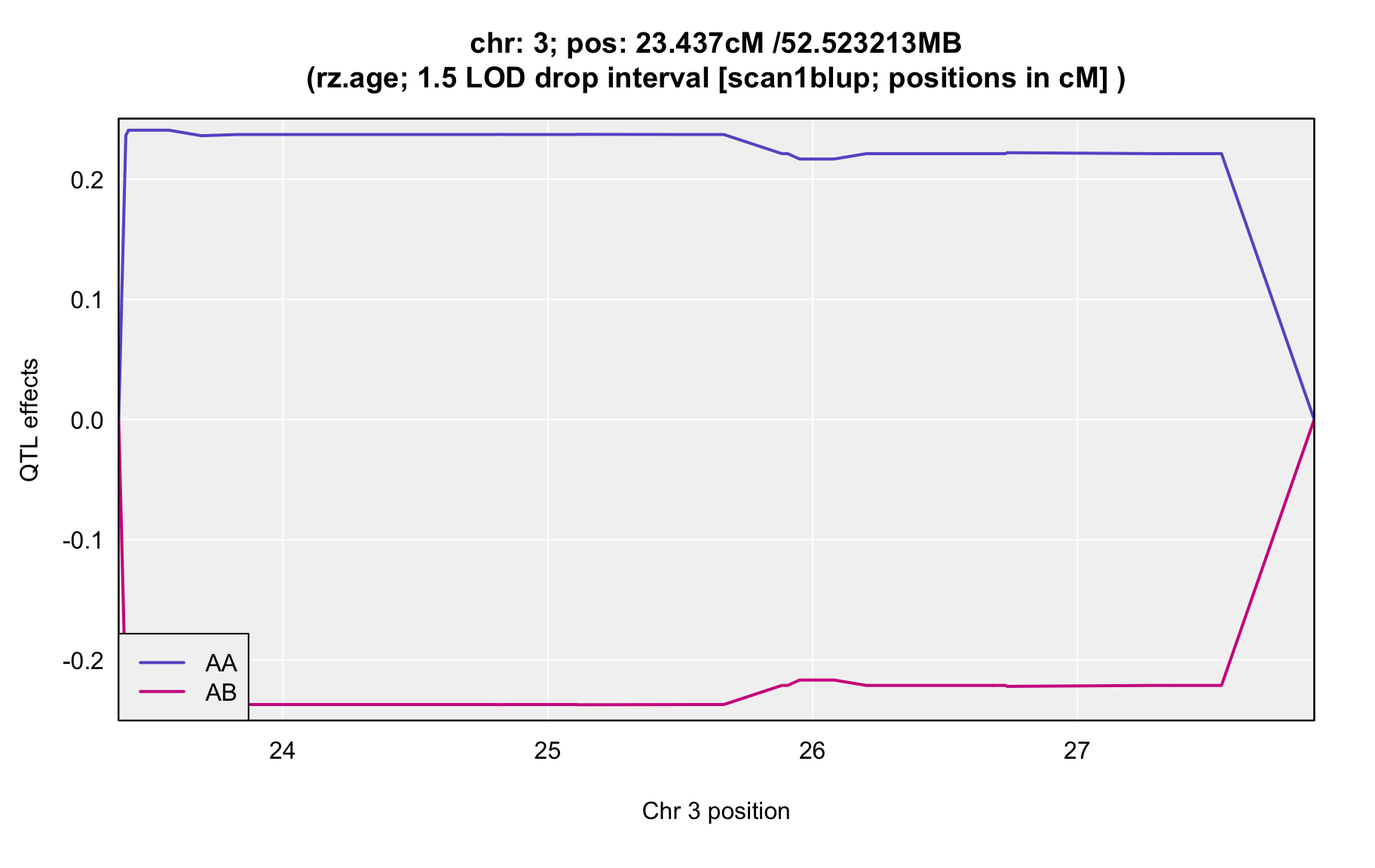
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | gene | 52.46717 | 52.49327 | positive | MGI_C57BL6J_5621422 | Gm38537 | NCBI_Gene:102641152 | MGI:5621422 | lncRNA gene | predicted gene, 38537 |
| 3 | gene | 52.50277 | 52.55022 | positive | MGI_C57BL6J_5589332 | Gm30173 | NCBI_Gene:102631981,ENSEMBL:ENSMUSG00000103965 | MGI:5589332 | lncRNA gene | predicted gene, 30173 |
| 3 | gene | 52.58013 | 52.58907 | positive | MGI_C57BL6J_5622932 | Gm40047 | NCBI_Gene:105244440 | MGI:5622932 | lncRNA gene | predicted gene, 40047 |
| 3 | pseudogene | 52.61282 | 52.61402 | positive | MGI_C57BL6J_3704216 | Gm10293 | NCBI_Gene:100039762,ENSEMBL:ENSMUSG00000070490 | MGI:3704216 | pseudogene | predicted pseudogene 10293 |
| 3 | gene | 52.64989 | 52.70971 | positive | MGI_C57BL6J_5589394 | Gm30235 | NCBI_Gene:102632062 | MGI:5589394 | lncRNA gene | predicted gene, 30235 |
| 3 | gene | 52.72265 | 52.72275 | negative | MGI_C57BL6J_5452575 | Gm22798 | ENSEMBL:ENSMUSG00000065926 | MGI:5452575 | snRNA gene | predicted gene, 22798 |
| 3 | gene | 52.73972 | 52.77666 | positive | MGI_C57BL6J_3780614 | Gm2447 | NCBI_Gene:329639,ENSEMBL:ENSMUSG00000102785 | MGI:3780614 | lncRNA gene | predicted gene 2447 |
| 3 | gene | 52.78900 | 52.79781 | negative | MGI_C57BL6J_5589451 | Gm30292 | NCBI_Gene:102632139,ENSEMBL:ENSMUSG00000104109 | MGI:5589451 | lncRNA gene | predicted gene, 30292 |
| 3 | gene | 52.90676 | 52.92767 | negative | MGI_C57BL6J_5434106 | Gm20750 | NCBI_Gene:622124,ENSEMBL:ENSMUSG00000104401 | MGI:5434106 | lncRNA gene | predicted gene, 20750 |
| 3 | gene | 52.93325 | 52.95628 | positive | MGI_C57BL6J_5589510 | Gm30351 | NCBI_Gene:102632211 | MGI:5589510 | lncRNA gene | predicted gene, 30351 |
| 3 | gene | 52.95618 | 52.95789 | negative | MGI_C57BL6J_5589561 | Gm30402 | NCBI_Gene:102632287 | MGI:5589561 | lncRNA gene | predicted gene, 30402 |
| 3 | pseudogene | 52.96005 | 52.96037 | negative | MGI_C57BL6J_5610235 | Gm37007 | ENSEMBL:ENSMUSG00000103886 | MGI:5610235 | pseudogene | predicted gene, 37007 |
| 3 | gene | 52.98188 | 53.01724 | negative | MGI_C57BL6J_1914792 | Cog6 | NCBI_Gene:67542,ENSEMBL:ENSMUSG00000027742 | MGI:1914792 | protein coding gene | component of oligomeric golgi complex 6 |
| 3 | gene | 53.03858 | 53.04176 | negative | MGI_C57BL6J_5663038 | Gm42901 | ENSEMBL:ENSMUSG00000104850 | MGI:5663038 | lncRNA gene | predicted gene 42901 |
| 3 | gene | 53.04153 | 53.26168 | positive | MGI_C57BL6J_1920048 | Lhfp | NCBI_Gene:108927,ENSEMBL:ENSMUSG00000048332 | MGI:1920048 | protein coding gene | lipoma HMGIC fusion partner |
| 3 | gene | 53.07216 | 53.07524 | negative | MGI_C57BL6J_5663035 | Gm42898 | ENSEMBL:ENSMUSG00000104828 | MGI:5663035 | unclassified gene | predicted gene 42898 |
| 3 | gene | 53.08322 | 53.08588 | positive | MGI_C57BL6J_5663036 | Gm42899 | ENSEMBL:ENSMUSG00000104687 | MGI:5663036 | unclassified gene | predicted gene 42899 |
| 3 | gene | 53.09900 | 53.10318 | positive | MGI_C57BL6J_2443940 | C130075A20Rik | ENSEMBL:ENSMUSG00000104735 | MGI:2443940 | unclassified gene | RIKEN cDNA C130075A20 gene |
| 3 | gene | 53.10967 | 53.11012 | positive | MGI_C57BL6J_5663941 | Gm43804 | ENSEMBL:ENSMUSG00000104615 | MGI:5663941 | unclassified gene | predicted gene 43804 |
| 3 | gene | 53.11040 | 53.11341 | negative | MGI_C57BL6J_5663940 | Gm43803 | ENSEMBL:ENSMUSG00000104968 | MGI:5663940 | lncRNA gene | predicted gene 43803 |
| 3 | gene | 53.11217 | 53.11299 | positive | MGI_C57BL6J_5663942 | Gm43805 | ENSEMBL:ENSMUSG00000105971 | MGI:5663942 | unclassified gene | predicted gene 43805 |
| 3 | pseudogene | 53.34649 | 53.34723 | negative | MGI_C57BL6J_5663609 | Gm43472 | ENSEMBL:ENSMUSG00000106660 | MGI:5663609 | pseudogene | predicted gene 43472 |
| 3 | gene | 53.39575 | 53.39850 | positive | MGI_C57BL6J_5622933 | Gm40048 | NCBI_Gene:105244441 | MGI:5622933 | lncRNA gene | predicted gene, 40048 |
| 3 | pseudogene | 53.40161 | 53.42457 | negative | MGI_C57BL6J_5663608 | Gm43471 | NCBI_Gene:108168853,ENSEMBL:ENSMUSG00000105397 | MGI:5663608 | pseudogene | predicted gene 43471 |
| 3 | gene | 53.44858 | 53.46333 | negative | MGI_C57BL6J_2444520 | Nhlrc3 | NCBI_Gene:212114,ENSEMBL:ENSMUSG00000042997 | MGI:2444520 | protein coding gene | NHL repeat containing 3 |
| 3 | gene | 53.45262 | 53.45805 | positive | MGI_C57BL6J_3801870 | Gm16206 | NCBI_Gene:102632669,ENSEMBL:ENSMUSG00000085174 | MGI:3801870 | lncRNA gene | predicted gene 16206 |
| 3 | gene | 53.46367 | 53.48175 | positive | MGI_C57BL6J_1919933 | Proser1 | NCBI_Gene:212127,ENSEMBL:ENSMUSG00000049504 | MGI:1919933 | protein coding gene | proline and serine rich 1 |
| 3 | gene | 53.48865 | 53.50850 | positive | MGI_C57BL6J_2388072 | Stoml3 | NCBI_Gene:229277,ENSEMBL:ENSMUSG00000027744 | MGI:2388072 | protein coding gene | stomatin (Epb7.2)-like 3 |
| 3 | gene | 53.50893 | 53.51010 | positive | MGI_C57BL6J_1924132 | 6430500D05Rik | ENSEMBL:ENSMUSG00000105481 | MGI:1924132 | unclassified gene | RIKEN cDNA 6430500D05 gene |
| 3 | gene | 53.51200 | 53.51213 | positive | MGI_C57BL6J_5453452 | Gm23675 | ENSEMBL:ENSMUSG00000087936 | MGI:5453452 | snoRNA gene | predicted gene, 23675 |
| 3 | gene | 53.51394 | 53.65736 | negative | MGI_C57BL6J_2444465 | Frem2 | NCBI_Gene:242022,ENSEMBL:ENSMUSG00000037016 | MGI:2444465 | protein coding gene | Fras1 related extracellular matrix protein 2 |
| 3 | pseudogene | 53.66666 | 53.66739 | negative | MGI_C57BL6J_3648425 | Gm6073 | NCBI_Gene:606547,ENSEMBL:ENSMUSG00000105840 | MGI:3648425 | pseudogene | predicted gene 6073 |
| 3 | pseudogene | 53.69237 | 53.69292 | negative | MGI_C57BL6J_3649038 | Gm6204 | NCBI_Gene:621155,ENSEMBL:ENSMUSG00000105879 | MGI:3649038 | pseudogene | predicted gene 6204 |
| 3 | gene | 53.70304 | 53.73576 | positive | MGI_C57BL6J_5589894 | Gm30735 | NCBI_Gene:102632743,ENSEMBL:ENSMUSG00000105168 | MGI:5589894 | lncRNA gene | predicted gene, 30735 |
| 3 | gene | 53.73622 | 53.73783 | positive | MGI_C57BL6J_3642187 | Gm10727 | ENSEMBL:ENSMUSG00000105444 | MGI:3642187 | unclassified gene | predicted gene 10727 |
| 3 | gene | 53.75673 | 53.77295 | positive | MGI_C57BL6J_2443105 | C820005J03Rik | ENSEMBL:ENSMUSG00000106270 | MGI:2443105 | lncRNA gene | RIKEN cDNA C820005J03 gene |
| 3 | gene | 53.84341 | 53.84351 | positive | MGI_C57BL6J_5454628 | Gm24851 | ENSEMBL:ENSMUSG00000088653 | MGI:5454628 | snRNA gene | predicted gene, 24851 |
| 3 | gene | 53.84509 | 53.84528 | positive | MGI_C57BL6J_3779200 | Gm10985 | ENSEMBL:ENSMUSG00000078742 | MGI:3779200 | protein coding gene | predicted gene 10985 |
| 3 | gene | 53.85338 | 53.86383 | negative | MGI_C57BL6J_1915140 | Ufm1 | NCBI_Gene:67890,ENSEMBL:ENSMUSG00000027746 | MGI:1915140 | protein coding gene | ubiquitin-fold modifier 1 |
| 3 | gene | 53.92484 | 53.94353 | positive | MGI_C57BL6J_5590185 | Gm31026 | NCBI_Gene:102633119,ENSEMBL:ENSMUSG00000106577 | MGI:5590185 | lncRNA gene | predicted gene, 31026 |
| 3 | pseudogene | 54.02115 | 54.02195 | positive | MGI_C57BL6J_3645533 | Gm8109 | NCBI_Gene:666444,ENSEMBL:ENSMUSG00000103155 | MGI:3645533 | pseudogene | predicted gene 8109 |
| 3 | pseudogene | 54.02988 | 54.03041 | positive | MGI_C57BL6J_5010203 | Gm18018 | NCBI_Gene:100416282,ENSEMBL:ENSMUSG00000105067 | MGI:5010203 | pseudogene | predicted gene, 18018 |
| 3 | gene | 54.15604 | 54.31847 | positive | MGI_C57BL6J_109525 | Trpc4 | NCBI_Gene:22066,ENSEMBL:ENSMUSG00000027748 | MGI:109525 | protein coding gene | transient receptor potential cation channel, subfamily C, member 4 |
| 3 | gene | 54.35927 | 54.39104 | positive | MGI_C57BL6J_1926321 | Postn | NCBI_Gene:50706,ENSEMBL:ENSMUSG00000027750 | MGI:1926321 | protein coding gene | periostin, osteoblast specific factor |
| 3 | gene | 54.35966 | 54.36101 | negative | MGI_C57BL6J_5590254 | Gm31095 | NCBI_Gene:102633208 | MGI:5590254 | lncRNA gene | predicted gene, 31095 |
| 3 | pseudogene | 54.48141 | 54.48293 | positive | MGI_C57BL6J_3645731 | Gm5641 | NCBI_Gene:434807,ENSEMBL:ENSMUSG00000069014 | MGI:3645731 | pseudogene | predicted gene 5641 |
| 3 | gene | 54.48856 | 54.53489 | negative | MGI_C57BL6J_5590313 | Gm31154 | NCBI_Gene:102633293 | MGI:5590313 | lncRNA gene | predicted gene, 31154 |
| 3 | gene | 54.59055 | 54.60139 | negative | MGI_C57BL6J_5622935 | Gm40050 | NCBI_Gene:105244443 | MGI:5622935 | lncRNA gene | predicted gene, 40050 |
| 3 | gene | 54.63144 | 54.63463 | positive | MGI_C57BL6J_5477172 | Gm26678 | ENSEMBL:ENSMUSG00000097599 | MGI:5477172 | lncRNA gene | predicted gene, 26678 |
| 3 | pseudogene | 54.63288 | 54.63457 | negative | MGI_C57BL6J_3588285 | B020017C02Rik | NCBI_Gene:100503708,ENSEMBL:ENSMUSG00000104831 | MGI:3588285 | pseudogene | RIKEN cDNA B020017C02 gene |
| 3 | gene | 54.69276 | 54.72877 | positive | MGI_C57BL6J_1929651 | Supt20 | NCBI_Gene:56790,ENSEMBL:ENSMUSG00000027751 | MGI:1929651 | protein coding gene | SPT20 SAGA complex component |
| 3 | gene | 54.71318 | 54.71435 | negative | MGI_C57BL6J_5439427 | Gm21958 | ENSEMBL:ENSMUSG00000094487 | MGI:5439427 | protein coding gene | predicted gene, 21958 |
| 3 | gene | 54.72868 | 54.73539 | negative | MGI_C57BL6J_1916889 | Exosc8 | NCBI_Gene:69639,ENSEMBL:ENSMUSG00000027752 | MGI:1916889 | protein coding gene | exosome component 8 |
| 3 | gene | 54.73554 | 54.75132 | positive | MGI_C57BL6J_1913498 | Alg5 | NCBI_Gene:66248,ENSEMBL:ENSMUSG00000036632 | MGI:1913498 | protein coding gene | asparagine-linked glycosylation 5 (dolichyl-phosphate beta-glucosyltransferase) |
| 3 | gene | 54.75545 | 54.80166 | positive | MGI_C57BL6J_1859993 | Smad9 | NCBI_Gene:55994,ENSEMBL:ENSMUSG00000027796 | MGI:1859993 | protein coding gene | SMAD family member 9 |
| 3 | gene | 54.80311 | 54.80779 | negative | MGI_C57BL6J_2180854 | Rfxap | NCBI_Gene:170767,ENSEMBL:ENSMUSG00000036615 | MGI:2180854 | protein coding gene | regulatory factor X-associated protein |
| 3 | pseudogene | 54.80997 | 54.81140 | negative | MGI_C57BL6J_3708673 | Gm10254 | NCBI_Gene:545524,ENSEMBL:ENSMUSG00000069011 | MGI:3708673 | pseudogene | predicted gene 10254 |
| 3 | gene | 54.89707 | 54.91589 | negative | MGI_C57BL6J_3607715 | Sertm1 | NCBI_Gene:329641,ENSEMBL:ENSMUSG00000056306 | MGI:3607715 | protein coding gene | serine rich and transmembrane domain containing 1 |
| 3 | gene | 54.91602 | 54.95326 | positive | MGI_C57BL6J_5590517 | Gm31358 | NCBI_Gene:102633562 | MGI:5590517 | lncRNA gene | predicted gene, 31358 |
| 3 | gene | 55.04547 | 55.05550 | negative | MGI_C57BL6J_108042 | Ccna1 | NCBI_Gene:12427,ENSEMBL:ENSMUSG00000027793 | MGI:108042 | protein coding gene | cyclin A1 |
| 3 | gene | 55.05524 | 55.08400 | positive | MGI_C57BL6J_1918220 | 4931419H13Rik | NCBI_Gene:70970,ENSEMBL:ENSMUSG00000106589 | MGI:1918220 | lncRNA gene | RIKEN cDNA 4931419H13 gene |
| 3 | pseudogene | 55.05735 | 55.05766 | negative | MGI_C57BL6J_5663692 | Gm43555 | ENSEMBL:ENSMUSG00000105977 | MGI:5663692 | pseudogene | predicted gene 43555 |
| 3 | gene | 55.11207 | 55.13733 | positive | MGI_C57BL6J_2139806 | Spg20 | NCBI_Gene:229285,ENSEMBL:ENSMUSG00000036580 | MGI:2139806 | protein coding gene | spastic paraplegia 20, spartin (Troyer syndrome) homolog (human) |
| 3 | gene | 55.13734 | 55.17525 | positive | MGI_C57BL6J_2444356 | Ccdc169 | NCBI_Gene:320604,ENSEMBL:ENSMUSG00000048655 | MGI:2444356 | protein coding gene | coiled-coil domain containing 169 |
| 3 | gene | 55.16231 | 55.16243 | negative | MGI_C57BL6J_5451806 | Gm22029 | ENSEMBL:ENSMUSG00000080590 | MGI:5451806 | miRNA gene | predicted gene, 22029 |
| 3 | gene | 55.18203 | 55.20996 | positive | MGI_C57BL6J_1921684 | Sohlh2 | NCBI_Gene:74434,ENSEMBL:ENSMUSG00000027794 | MGI:1921684 | protein coding gene | spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| 3 | gene | 55.24236 | 55.53907 | positive | MGI_C57BL6J_1330861 | Dclk1 | NCBI_Gene:13175,ENSEMBL:ENSMUSG00000027797 | MGI:1330861 | protein coding gene | doublecortin-like kinase 1 |
| 3 | gene | 55.29094 | 55.29194 | positive | MGI_C57BL6J_1924494 | 9430012M22Rik | NA | NA | unclassified gene | RIKEN cDNA 9430012M22 gene |
| 3 | gene | 55.30532 | 55.30542 | positive | MGI_C57BL6J_5454909 | Gm25132 | ENSEMBL:ENSMUSG00000064586 | MGI:5454909 | snRNA gene | predicted gene, 25132 |
| 3 | gene | 55.33084 | 55.33297 | positive | MGI_C57BL6J_5012002 | Gm19817 | ENSEMBL:ENSMUSG00000105302 | MGI:5012002 | unclassified gene | predicted gene, 19817 |
| 3 | gene | 55.35240 | 55.38079 | negative | MGI_C57BL6J_5622936 | Gm40051 | NCBI_Gene:105244444 | MGI:5622936 | lncRNA gene | predicted gene, 40051 |
| 3 | gene | 55.37746 | 55.40312 | negative | MGI_C57BL6J_5662746 | Gm42609 | ENSEMBL:ENSMUSG00000105975 | MGI:5662746 | lncRNA gene | predicted gene 42609 |
| 3 | gene | 55.39181 | 55.40420 | negative | MGI_C57BL6J_3642047 | Gm9831 | NCBI_Gene:101056032 | MGI:3642047 | protein coding gene | predicted gene 9831 |
| 3 | gene | 55.46198 | 55.47221 | negative | MGI_C57BL6J_5439423 | Gm21954 | ENSEMBL:ENSMUSG00000094962 | MGI:5439423 | protein coding gene | predicted gene, 21954 |
| 3 | gene | 55.47609 | 55.48162 | negative | MGI_C57BL6J_5590602 | Gm31443 | NCBI_Gene:102633676 | MGI:5590602 | lncRNA gene | predicted gene, 31443 |
| 3 | gene | 55.48379 | 55.48718 | positive | MGI_C57BL6J_5662744 | Gm42607 | ENSEMBL:ENSMUSG00000106354 | MGI:5662744 | unclassified gene | predicted gene 42607 |
| 3 | gene | 55.51757 | 55.51959 | positive | MGI_C57BL6J_5662745 | Gm42608 | ENSEMBL:ENSMUSG00000106651 | MGI:5662745 | unclassified gene | predicted gene 42608 |
| 3 | gene | 55.54045 | 55.54338 | negative | MGI_C57BL6J_5590955 | Gm31796 | NCBI_Gene:102634140 | MGI:5590955 | lncRNA gene | predicted gene, 31796 |
| 3 | gene | 55.54354 | 55.54855 | positive | MGI_C57BL6J_5455181 | Gm25404 | NCBI_Gene:102634218,ENSEMBL:ENSMUSG00000064904 | MGI:5455181 | snRNA gene | predicted gene, 25404 |
| 3 | gene | 55.57492 | 55.57727 | positive | MGI_C57BL6J_5663685 | Gm43548 | ENSEMBL:ENSMUSG00000105627 | MGI:5663685 | unclassified gene | predicted gene 43548 |
| 3 | gene | 55.57811 | 55.58273 | negative | MGI_C57BL6J_5663686 | Gm43549 | NCBI_Gene:108168870,ENSEMBL:ENSMUSG00000106334 | MGI:5663686 | lncRNA gene | predicted gene 43549 |
| 3 | gene | 55.62519 | 56.18427 | negative | MGI_C57BL6J_1347075 | Nbea | NCBI_Gene:26422,ENSEMBL:ENSMUSG00000027799 | MGI:1347075 | protein coding gene | neurobeachin |
| 3 | gene | 55.70589 | 55.71565 | positive | MGI_C57BL6J_5826441 | Gm46804 | NCBI_Gene:108168871 | MGI:5826441 | lncRNA gene | predicted gene, 46804 |
| 3 | gene | 55.72593 | 55.72844 | negative | MGI_C57BL6J_3609650 | D430004P15Rik | NA | NA | unclassified gene | Riken cDNA D430004P15 gene |
| 3 | gene | 55.77534 | 55.78795 | negative | MGI_C57BL6J_3698047 | 9630024C22Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 9630024C22 gene |
| 3 | gene | 55.78251 | 55.78529 | positive | MGI_C57BL6J_1333773 | Mab21l1 | NCBI_Gene:17116,ENSEMBL:ENSMUSG00000056947 | MGI:1333773 | protein coding gene | mab-21-like 1 |
| 3 | gene | 55.83367 | 55.83446 | negative | MGI_C57BL6J_1924979 | 6720451E02Rik | NA | NA | unclassified gene | RIKEN cDNA 6720451E02 gene |
| 3 | pseudogene | 55.84710 | 55.84840 | positive | MGI_C57BL6J_1918432 | 4933417G07Rik | NCBI_Gene:71182,ENSEMBL:ENSMUSG00000106572 | MGI:1918432 | pseudogene | RIKEN cDNA 4933417G07 gene |
| 3 | gene | 55.85525 | 55.85615 | negative | MGI_C57BL6J_1926125 | B230206L23Rik | NA | NA | unclassified gene | RIKEN cDNA B230206L23 gene |
| 3 | gene | 55.88327 | 56.00032 | positive | MGI_C57BL6J_5591073 | Gm31914 | NCBI_Gene:102634293 | MGI:5591073 | lncRNA gene | predicted gene, 31914 |
| 3 | gene | 55.89031 | 55.93670 | positive | MGI_C57BL6J_5663513 | Gm43376 | ENSEMBL:ENSMUSG00000104677 | MGI:5663513 | lncRNA gene | predicted gene 43376 |
| 3 | gene | 55.95678 | 55.95691 | negative | MGI_C57BL6J_5452561 | Gm22784 | ENSEMBL:ENSMUSG00000077276 | MGI:5452561 | snoRNA gene | predicted gene, 22784 |
| 3 | gene | 56.01492 | 56.01717 | negative | MGI_C57BL6J_5662977 | Gm42840 | ENSEMBL:ENSMUSG00000104938 | MGI:5662977 | unclassified gene | predicted gene 42840 |
| 3 | gene | 56.12316 | 56.12616 | negative | MGI_C57BL6J_5662979 | Gm42842 | ENSEMBL:ENSMUSG00000106211 | MGI:5662979 | unclassified gene | predicted gene 42842 |
| 3 | gene | 56.12860 | 56.13053 | negative | MGI_C57BL6J_5662978 | Gm42841 | ENSEMBL:ENSMUSG00000106528 | MGI:5662978 | unclassified gene | predicted gene 42841 |
| 3 | gene | 56.16395 | 56.16757 | negative | MGI_C57BL6J_5662980 | Gm42843 | ENSEMBL:ENSMUSG00000105293 | MGI:5662980 | unclassified gene | predicted gene 42843 |
| 3 | gene | 56.21791 | 56.22124 | negative | MGI_C57BL6J_5622937 | Gm40052 | NCBI_Gene:105244445 | MGI:5622937 | lncRNA gene | predicted gene, 40052 |
| 3 | gene | 56.25590 | 56.26994 | negative | MGI_C57BL6J_5622938 | Gm40053 | NCBI_Gene:105244446 | MGI:5622938 | lncRNA gene | predicted gene, 40053 |
| 3 | gene | 56.34703 | 56.35755 | negative | MGI_C57BL6J_5622939 | Gm40054 | NCBI_Gene:105244447 | MGI:5622939 | lncRNA gene | predicted gene, 40054 |
| 3 | gene | 56.35224 | 56.35230 | positive | MGI_C57BL6J_102792 | Mcptl | NA | NA | protein coding gene | mast cell protease-like |
| 3 | gene | 56.43600 | 56.43875 | negative | MGI_C57BL6J_5591336 | Gm32177 | NCBI_Gene:102634641 | MGI:5591336 | lncRNA gene | predicted gene, 32177 |
| 3 | gene | 56.44528 | 56.44541 | negative | MGI_C57BL6J_5455504 | Gm25727 | ENSEMBL:ENSMUSG00000077710 | MGI:5455504 | snoRNA gene | predicted gene, 25727 |
| 3 | gene | 56.69842 | 56.70506 | positive | MGI_C57BL6J_5591396 | Gm32237 | NCBI_Gene:102634719 | MGI:5591396 | lncRNA gene | predicted gene, 32237 |
| 3 | pseudogene | 56.82832 | 56.82884 | negative | MGI_C57BL6J_3780790 | Gm2622 | NCBI_Gene:100040139,ENSEMBL:ENSMUSG00000105113 | MGI:3780790 | pseudogene | predicted gene 2622 |
| 3 | gene | 56.88806 | 56.89133 | negative | MGI_C57BL6J_3588185 | F830104G03Rik | NA | NA | unclassified gene | RIKEN cDNA F830104G03 gene |
| 3 | gene | 57.05415 | 57.05428 | positive | MGI_C57BL6J_5452046 | Gm22269 | ENSEMBL:ENSMUSG00000077190 | MGI:5452046 | snoRNA gene | predicted gene, 22269 |
| 3 | pseudogene | 57.13549 | 57.13683 | negative | MGI_C57BL6J_3805962 | Gm8177 | NCBI_Gene:666585,ENSEMBL:ENSMUSG00000097366 | MGI:3805962 | pseudogene | predicted gene 8177 |
| 3 | gene | 57.22772 | 57.22918 | negative | MGI_C57BL6J_1918389 | 4933408K01Rik | NA | NA | unclassified gene | RIKEN cDNA 4933408K01 gene |
| 3 | pseudogene | 57.24925 | 57.24950 | positive | MGI_C57BL6J_5663561 | Gm43424 | ENSEMBL:ENSMUSG00000106286 | MGI:5663561 | pseudogene | predicted gene 43424 |
| 3 | pseudogene | 57.27174 | 57.27237 | negative | MGI_C57BL6J_5010845 | Gm18660 | NCBI_Gene:100417512,ENSEMBL:ENSMUSG00000104534 | MGI:5010845 | pseudogene | predicted gene, 18660 |
| 3 | gene | 57.28561 | 57.30199 | negative | MGI_C57BL6J_104678 | Tm4sf1 | NCBI_Gene:17112,ENSEMBL:ENSMUSG00000027800 | MGI:104678 | protein coding gene | transmembrane 4 superfamily member 1 |
| 3 | pseudogene | 57.36225 | 57.36361 | positive | MGI_C57BL6J_3646382 | Gm5276 | NCBI_Gene:383862,ENSEMBL:ENSMUSG00000105452 | MGI:3646382 | pseudogene | predicted gene 5276 |
| 3 | gene | 57.37929 | 57.39455 | negative | MGI_C57BL6J_5591526 | Gm32367 | NCBI_Gene:102634892 | MGI:5591526 | lncRNA gene | predicted gene, 32367 |
| 3 | gene | 57.42531 | 57.44168 | positive | MGI_C57BL6J_2385173 | Tm4sf4 | NCBI_Gene:229302,ENSEMBL:ENSMUSG00000027801 | MGI:2385173 | protein coding gene | transmembrane 4 superfamily member 4 |
| 3 | gene | 57.45564 | 57.57591 | negative | MGI_C57BL6J_1917649 | Wwtr1 | NCBI_Gene:97064,ENSEMBL:ENSMUSG00000027803 | MGI:1917649 | protein coding gene | WW domain containing transcription regulator 1 |
| 3 | pseudogene | 57.52655 | 57.53048 | positive | MGI_C57BL6J_3802068 | Gm16016 | NCBI_Gene:105244448,ENSEMBL:ENSMUSG00000083538 | MGI:3802068 | pseudogene | predicted gene 16016 |


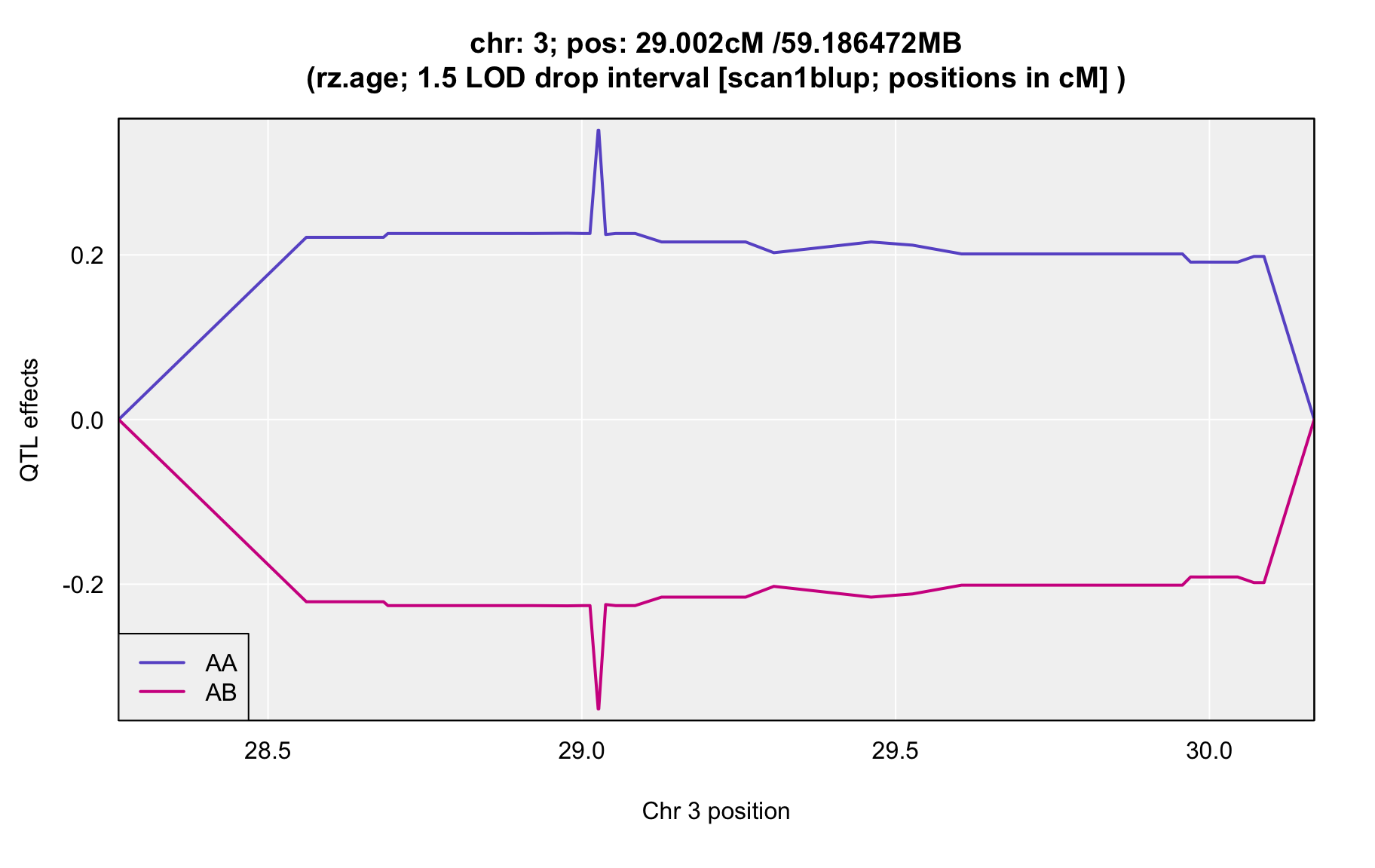
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | gene | 57.95198 | 58.03171 | negative | MGI_C57BL6J_5622940 | Gm40055 | NCBI_Gene:105244449,ENSEMBL:ENSMUSG00000105003 | MGI:5622940 | lncRNA gene | predicted gene, 40055 |
| 3 | gene | 58.07972 | 58.08183 | positive | MGI_C57BL6J_5662650 | Gm42513 | ENSEMBL:ENSMUSG00000105065 | MGI:5662650 | unclassified gene | predicted gene 42513 |
| 3 | gene | 58.08960 | 58.16235 | positive | MGI_C57BL6J_5622941 | Gm40056 | NCBI_Gene:105244450 | MGI:5622941 | lncRNA gene | predicted gene, 40056 |
| 3 | pseudogene | 58.10256 | 58.10430 | negative | MGI_C57BL6J_3643179 | Gm5537 | NCBI_Gene:433594,ENSEMBL:ENSMUSG00000069008 | MGI:3643179 | pseudogene | predicted gene 5537 |
| 3 | gene | 58.11705 | 58.16762 | positive | MGI_C57BL6J_1925506 | 4921539H07Rik | ENSEMBL:ENSMUSG00000104586 | MGI:1925506 | lncRNA gene | RIKEN cDNA 4921539H07 gene |
| 3 | gene | 58.14171 | 58.16381 | negative | MGI_C57BL6J_1915100 | 1700007F19Rik | NCBI_Gene:67850,ENSEMBL:ENSMUSG00000100666 | MGI:1915100 | lncRNA gene | RIKEN cDNA 1700007F19 gene |
| 3 | gene | 58.21192 | 58.21337 | positive | MGI_C57BL6J_5622942 | Gm40057 | NCBI_Gene:105244451 | MGI:5622942 | lncRNA gene | predicted gene, 40057 |
| 3 | gene | 58.29025 | 58.29036 | negative | MGI_C57BL6J_5455943 | Gm26166 | ENSEMBL:ENSMUSG00000065722 | MGI:5455943 | snRNA gene | predicted gene, 26166 |
| 3 | gene | 58.30186 | 58.31762 | positive | MGI_C57BL6J_5592006 | Gm32847 | NCBI_Gene:102635539 | MGI:5592006 | lncRNA gene | predicted gene, 32847 |
| 3 | pseudogene | 58.31494 | 58.31531 | negative | MGI_C57BL6J_5663867 | Gm43730 | ENSEMBL:ENSMUSG00000106176 | MGI:5663867 | pseudogene | predicted gene 43730 |
| 3 | pseudogene | 58.40465 | 58.40512 | positive | MGI_C57BL6J_5012313 | Gm20128 | NCBI_Gene:100504238,ENSEMBL:ENSMUSG00000105154 | MGI:5012313 | pseudogene | predicted gene, 20128 |
| 3 | gene | 58.41472 | 58.46679 | positive | MGI_C57BL6J_1919283 | Tsc22d2 | NCBI_Gene:72033,ENSEMBL:ENSMUSG00000027806 | MGI:1919283 | protein coding gene | TSC22 domain family, member 2 |
| 3 | gene | 58.43268 | 58.43301 | positive | MGI_C57BL6J_1338064 | Gdap9 | NA | NA | unclassified gene | ganglioside-induced differentiation-associated-protein 9 |
| 3 | gene | 58.51982 | 58.52589 | negative | MGI_C57BL6J_92638 | Serp1 | NCBI_Gene:28146,ENSEMBL:ENSMUSG00000027808 | MGI:92638 | protein coding gene | stress-associated endoplasmic reticulum protein 1 |
| 3 | gene | 58.52582 | 58.55750 | positive | MGI_C57BL6J_1098684 | Eif2a | NCBI_Gene:229317,ENSEMBL:ENSMUSG00000027810 | MGI:1098684 | protein coding gene | eukaryotic translation initiation factor 2A |
| 3 | gene | 58.52693 | 58.52812 | positive | MGI_C57BL6J_1925148 | 6720482G16Rik | ENSEMBL:ENSMUSG00000106553 | MGI:1925148 | unclassified gene | RIKEN cDNA 6720482G16 gene |
| 3 | gene | 58.57664 | 58.59354 | positive | MGI_C57BL6J_1916477 | Selenot | NCBI_Gene:69227,ENSEMBL:ENSMUSG00000075700 | MGI:1916477 | protein coding gene | selenoprotein T |
| 3 | gene | 58.61630 | 58.63721 | negative | MGI_C57BL6J_3588212 | Erich6 | NCBI_Gene:545527,ENSEMBL:ENSMUSG00000070471 | MGI:3588212 | protein coding gene | glutamate rich 6 |
| 3 | gene | 58.63671 | 58.63749 | positive | MGI_C57BL6J_5826443 | Gm46806 | NCBI_Gene:108168873 | MGI:5826443 | lncRNA gene | predicted gene, 46806 |
| 3 | pseudogene | 58.65204 | 58.65404 | positive | MGI_C57BL6J_3779790 | Gm8234 | NCBI_Gene:666680,ENSEMBL:ENSMUSG00000105021 | MGI:3779790 | pseudogene | predicted gene 8234 |
| 3 | pseudogene | 58.66547 | 58.66664 | negative | MGI_C57BL6J_2139888 | AU022133 | NCBI_Gene:100417266,ENSEMBL:ENSMUSG00000105336 | MGI:2139888 | pseudogene | expressed sequence AU022133 |
| 3 | gene | 58.67494 | 58.69244 | negative | MGI_C57BL6J_108062 | Siah2 | NCBI_Gene:20439,ENSEMBL:ENSMUSG00000036432 | MGI:108062 | protein coding gene | siah E3 ubiquitin protein ligase 2 |
| 3 | gene | 58.69259 | 58.79733 | positive | MGI_C57BL6J_1923132 | 4930593A02Rik | NCBI_Gene:71301,ENSEMBL:ENSMUSG00000097725 | MGI:1923132 | lncRNA gene | RIKEN cDNA 4930593A02 gene |
| 3 | gene | 58.69756 | 58.69867 | positive | MGI_C57BL6J_1925070 | A930028O11Rik | ENSEMBL:ENSMUSG00000105226 | MGI:1925070 | unclassified gene | RIKEN cDNA A930028O11 gene |
| 3 | pseudogene | 58.76149 | 58.76275 | negative | MGI_C57BL6J_5592251 | Gm33092 | NCBI_Gene:102635856 | MGI:5592251 | pseudogene | predicted gene, 33092 |
| 3 | pseudogene | 58.78841 | 58.82231 | negative | MGI_C57BL6J_2140018 | Mindy4b-ps | NCBI_Gene:100040293,ENSEMBL:ENSMUSG00000101860 | MGI:2140018 | pseudogene | MINDY lysine 48 deubiquitinase 4B, pseudogene |
| 3 | gene | 58.82499 | 58.83590 | positive | MGI_C57BL6J_5622944 | Gm40059 | NCBI_Gene:105244453 | MGI:5622944 | lncRNA gene | predicted gene, 40059 |
| 3 | gene | 58.84210 | 58.86675 | positive | MGI_C57BL6J_5622945 | Gm40060 | NCBI_Gene:105244454 | MGI:5622945 | lncRNA gene | predicted gene, 40060 |
| 3 | gene | 58.84403 | 58.88534 | negative | MGI_C57BL6J_2388124 | Clrn1 | NCBI_Gene:229320,ENSEMBL:ENSMUSG00000043850 | MGI:2388124 | protein coding gene | clarin 1 |
| 3 | gene | 58.97563 | 58.97573 | positive | MGI_C57BL6J_5452268 | Gm22491 | ENSEMBL:ENSMUSG00000087815 | MGI:5452268 | snRNA gene | predicted gene, 22491 |
| 3 | gene | 58.97886 | 58.97903 | negative | MGI_C57BL6J_5455349 | Gm25572 | ENSEMBL:ENSMUSG00000088152 | MGI:5455349 | snRNA gene | predicted gene, 25572 |
| 3 | pseudogene | 58.98713 | 58.98786 | positive | MGI_C57BL6J_3642685 | Rpl13-ps6 | NCBI_Gene:100040416,ENSEMBL:ENSMUSG00000059776 | MGI:3642685 | pseudogene | ribosomal protein L13, pseudogene 6 |
| 3 | gene | 59.00162 | 59.00169 | positive | MGI_C57BL6J_5531175 | Gm27793 | ENSEMBL:ENSMUSG00000099052 | MGI:5531175 | unclassified non-coding RNA gene | predicted gene, 27793 |
| 3 | gene | 59.00247 | 59.00616 | negative | MGI_C57BL6J_5663707 | Gm43570 | ENSEMBL:ENSMUSG00000105945 | MGI:5663707 | lncRNA gene | predicted gene 43570 |
| 3 | gene | 59.00559 | 59.31868 | positive | MGI_C57BL6J_2139916 | Med12l | NCBI_Gene:329650,ENSEMBL:ENSMUSG00000056476 | MGI:2139916 | protein coding gene | mediator complex subunit 12-like |
| 3 | gene | 59.09645 | 59.10182 | negative | MGI_C57BL6J_2442043 | Gpr171 | NCBI_Gene:229323,ENSEMBL:ENSMUSG00000050075 | MGI:2442043 | protein coding gene | G protein-coupled receptor 171 |
| 3 | gene | 59.11386 | 59.15362 | negative | MGI_C57BL6J_2155705 | P2ry14 | NCBI_Gene:140795,ENSEMBL:ENSMUSG00000036381 | MGI:2155705 | protein coding gene | purinergic receptor P2Y, G-protein coupled, 14 |
| 3 | gene | 59.14630 | 59.15363 | negative | MGI_C57BL6J_2444074 | F630111L10Rik | NCBI_Gene:320463 | MGI:2444074 | lncRNA gene | RIKEN cDNA F630111L10 gene |
| 3 | gene | 59.17890 | 59.19547 | negative | MGI_C57BL6J_1934133 | Gpr87 | NCBI_Gene:84111,ENSEMBL:ENSMUSG00000051431 | MGI:1934133 | protein coding gene | G protein-coupled receptor 87 |
| 3 | gene | 59.20789 | 59.21088 | negative | MGI_C57BL6J_1921441 | P2ry13 | NCBI_Gene:74191,ENSEMBL:ENSMUSG00000036362 | MGI:1921441 | protein coding gene | purinergic receptor P2Y, G-protein coupled 13 |
| 3 | gene | 59.21627 | 59.26301 | negative | MGI_C57BL6J_1918089 | P2ry12 | NCBI_Gene:70839,ENSEMBL:ENSMUSG00000036353 | MGI:1918089 | protein coding gene | purinergic receptor P2Y, G-protein coupled 12 |
| 3 | gene | 59.26979 | 59.27131 | positive | MGI_C57BL6J_5663726 | Gm43589 | ENSEMBL:ENSMUSG00000104550 | MGI:5663726 | unclassified gene | predicted gene 43589 |
| 3 | gene | 59.31674 | 59.34444 | negative | MGI_C57BL6J_1923481 | Igsf10 | NCBI_Gene:242050,ENSEMBL:ENSMUSG00000036334 | MGI:1923481 | protein coding gene | immunoglobulin superfamily, member 10 |
| 3 | gene | 59.34427 | 59.35117 | positive | MGI_C57BL6J_5611414 | Gm38186 | ENSEMBL:ENSMUSG00000103668 | MGI:5611414 | lncRNA gene | predicted gene, 38186 |
| 3 | pseudogene | 59.42453 | 59.42476 | positive | MGI_C57BL6J_5610943 | Gm37715 | ENSEMBL:ENSMUSG00000102247 | MGI:5610943 | pseudogene | predicted gene, 37715 |
| 3 | pseudogene | 59.43154 | 59.43335 | positive | MGI_C57BL6J_3648019 | Gm8276 | NCBI_Gene:666756,ENSEMBL:ENSMUSG00000102550 | MGI:3648019 | pseudogene | predicted gene 8276 |
| 3 | pseudogene | 59.46967 | 59.47409 | negative | MGI_C57BL6J_5611516 | Gm38288 | ENSEMBL:ENSMUSG00000103211 | MGI:5611516 | pseudogene | predicted gene, 38288 |
| 3 | gene | 59.47493 | 59.47500 | negative | MGI_C57BL6J_4414092 | n-TVcac1 | NCBI_Gene:102467620 | MGI:4414092 | tRNA gene | nuclear encoded tRNA valine 1 (anticodon CAC) |
| 3 | pseudogene | 59.48302 | 59.48329 | positive | MGI_C57BL6J_5610999 | Gm37771 | ENSEMBL:ENSMUSG00000104038 | MGI:5610999 | pseudogene | predicted gene, 37771 |
| 3 | pseudogene | 59.50558 | 59.53523 | negative | MGI_C57BL6J_3644725 | Gm4856 | NCBI_Gene:229330,ENSEMBL:ENSMUSG00000103598 | MGI:3644725 | pseudogene | predicted gene 4856 |
| 3 | pseudogene | 59.60245 | 59.63576 | negative | MGI_C57BL6J_3644280 | Gm5709 | NCBI_Gene:435732,ENSEMBL:ENSMUSG00000095128 | MGI:3644280 | pseudogene | predicted gene 5709 |
| 3 | gene | 59.66286 | 59.66528 | positive | MGI_C57BL6J_5611154 | Gm37926 | ENSEMBL:ENSMUSG00000102480 | MGI:5611154 | unclassified gene | predicted gene, 37926 |
| 3 | gene | 59.72979 | 59.75240 | positive | MGI_C57BL6J_3779495 | Aadacl2fm2 | NCBI_Gene:433597,ENSEMBL:ENSMUSG00000090527 | MGI:3779495 | protein coding gene | AADACL2 family member 2 |
| 3 | pseudogene | 59.73675 | 59.73686 | negative | MGI_C57BL6J_5611436 | Gm38208 | ENSEMBL:ENSMUSG00000104471 | MGI:5611436 | pseudogene | predicted gene, 38208 |
| 3 | pseudogene | 59.73718 | 59.73732 | negative | MGI_C57BL6J_5610807 | Gm37579 | ENSEMBL:ENSMUSG00000102390 | MGI:5610807 | pseudogene | predicted gene, 37579 |
| 3 | pseudogene | 59.76721 | 59.76891 | positive | MGI_C57BL6J_3644451 | Gm5539 | NCBI_Gene:433598,ENSEMBL:ENSMUSG00000103986 | MGI:3644451 | pseudogene | predicted gene 5539 |
| 3 | gene | 59.77781 | 59.84682 | positive | MGI_C57BL6J_1921922 | 4930449A18Rik | ENSEMBL:ENSMUSG00000074589 | MGI:1921922 | protein coding gene | RIKEN cDNA 4930449A18 gene |
| 3 | pseudogene | 59.78322 | 59.78863 | negative | MGI_C57BL6J_3780925 | Gm2756 | NCBI_Gene:100040407,ENSEMBL:ENSMUSG00000102757 | MGI:3780925 | pseudogene | predicted gene 2756 |
| 3 | gene | 59.83837 | 59.84682 | positive | MGI_C57BL6J_4361133 | Gm16527 | NCBI_Gene:669780 | MGI:4361133 | protein coding gene | predicted gene, 16527 |
| 3 | gene | 59.86105 | 59.87731 | positive | MGI_C57BL6J_3643798 | Aadacl2fm3 | NCBI_Gene:666803,ENSEMBL:ENSMUSG00000095522 | MGI:3643798 | protein coding gene | AADACL2 family member 3 |
| 3 | gene | 59.92521 | 59.93795 | positive | MGI_C57BL6J_3028051 | Aadacl2fm1 | NCBI_Gene:229333,ENSEMBL:ENSMUSG00000036951 | MGI:3028051 | protein coding gene | AADACL2 family member 1 |
| 3 | pseudogene | 59.95231 | 59.97359 | positive | MGI_C57BL6J_3819177 | Gm9696 | NCBI_Gene:676914,ENSEMBL:ENSMUSG00000070465 | MGI:3819177 | pseudogene | predicted gene 9696 |
| 3 | gene | 60.00674 | 60.02542 | positive | MGI_C57BL6J_3646333 | Aadacl2 | NCBI_Gene:639634,ENSEMBL:ENSMUSG00000091376 | MGI:3646333 | protein coding gene | arylacetamide deacetylase like 2 |
| 3 | pseudogene | 60.02036 | 60.02087 | negative | MGI_C57BL6J_5010612 | Gm18427 | NCBI_Gene:100417154,ENSEMBL:ENSMUSG00000103177 | MGI:5010612 | pseudogene | predicted gene, 18427 |
| 3 | gene | 60.02572 | 60.04016 | positive | MGI_C57BL6J_1915008 | Aadac | NCBI_Gene:67758,ENSEMBL:ENSMUSG00000027761 | MGI:1915008 | protein coding gene | arylacetamide deacetylase |
| 3 | gene | 60.08187 | 60.08757 | positive | MGI_C57BL6J_1934135 | Sucnr1 | NCBI_Gene:84112,ENSEMBL:ENSMUSG00000027762 | MGI:1934135 | protein coding gene | succinate receptor 1 |
| 3 | gene | 60.12764 | 60.12775 | negative | MGI_C57BL6J_5454159 | Gm24382 | ENSEMBL:ENSMUSG00000084647 | MGI:5454159 | snRNA gene | predicted gene, 24382 |
| 3 | gene | 60.29342 | 60.30941 | negative | MGI_C57BL6J_5592809 | Gm33650 | NCBI_Gene:102636642 | MGI:5592809 | lncRNA gene | predicted gene, 33650 |
| 3 | pseudogene | 60.34219 | 60.34309 | negative | MGI_C57BL6J_3779880 | Tgif1-ps | NCBI_Gene:669823,ENSEMBL:ENSMUSG00000107270 | MGI:3779880 | pseudogene | TGFB-induced factor homeobox 1, pseudogene |
| 3 | gene | 60.47279 | 60.62975 | positive | MGI_C57BL6J_1928482 | Mbnl1 | NCBI_Gene:56758,ENSEMBL:ENSMUSG00000027763 | MGI:1928482 | protein coding gene | muscleblind like splicing factor 1 |
| 3 | gene | 60.53575 | 60.53974 | positive | MGI_C57BL6J_5610817 | Gm37589 | ENSEMBL:ENSMUSG00000104235 | MGI:5610817 | unclassified gene | predicted gene, 37589 |
| 3 | gene | 60.57770 | 60.58136 | positive | MGI_C57BL6J_5012001 | Gm19816 | NA | NA | unclassified gene | predicted gene%2c 19816 |
| 3 | gene | 60.60438 | 60.60776 | positive | MGI_C57BL6J_5610716 | Gm37488 | ENSEMBL:ENSMUSG00000104125 | MGI:5610716 | unclassified gene | predicted gene, 37488 |
| 3 | pseudogene | 60.73763 | 60.73779 | negative | MGI_C57BL6J_5611508 | Gm38280 | ENSEMBL:ENSMUSG00000104337 | MGI:5611508 | pseudogene | predicted gene, 38280 |
| 3 | pseudogene | 60.76958 | 60.77164 | negative | MGI_C57BL6J_5592866 | Gm33707 | NCBI_Gene:102636711,ENSEMBL:ENSMUSG00000102298 | MGI:5592866 | pseudogene | predicted gene, 33707 |
| 3 | gene | 60.78148 | 61.00379 | negative | MGI_C57BL6J_5610263 | Gm37035 | NCBI_Gene:105244455,ENSEMBL:ENSMUSG00000102564 | MGI:5610263 | lncRNA gene | predicted gene, 37035 |
| 3 | gene | 60.78836 | 60.79473 | positive | MGI_C57BL6J_5611554 | Gm38326 | NCBI_Gene:102642162,ENSEMBL:ENSMUSG00000103587 | MGI:5611554 | lncRNA gene | predicted gene, 38326 |
| 3 | pseudogene | 60.86071 | 60.86109 | positive | MGI_C57BL6J_5610213 | Gm36985 | ENSEMBL:ENSMUSG00000102490 | MGI:5610213 | pseudogene | predicted gene, 36985 |
| 3 | pseudogene | 60.87700 | 60.87884 | positive | MGI_C57BL6J_3644852 | Gm8325 | NCBI_Gene:666853,ENSEMBL:ENSMUSG00000027694 | MGI:3644852 | pseudogene | predicted pseudogene 8325 |
| 3 | gene | 61.00279 | 61.00898 | positive | MGI_C57BL6J_105049 | P2ry1 | NCBI_Gene:18441,ENSEMBL:ENSMUSG00000027765 | MGI:105049 | protein coding gene | purinergic receptor P2Y, G-protein coupled 1 |
| 3 | pseudogene | 61.29116 | 61.29145 | negative | MGI_C57BL6J_5610947 | Gm37719 | ENSEMBL:ENSMUSG00000102242 | MGI:5610947 | pseudogene | predicted gene, 37719 |
| 3 | pseudogene | 61.32849 | 61.32923 | negative | MGI_C57BL6J_5012086 | Gm19901 | NCBI_Gene:100503798,ENSEMBL:ENSMUSG00000102667 | MGI:5012086 | pseudogene | predicted gene, 19901 |
| 3 | gene | 61.35855 | 61.36190 | negative | MGI_C57BL6J_5610924 | Gm37696 | ENSEMBL:ENSMUSG00000103473 | MGI:5610924 | lncRNA gene | predicted gene, 37696 |
| 3 | gene | 61.36164 | 61.36870 | positive | MGI_C57BL6J_1921262 | Rap2b | NCBI_Gene:74012,ENSEMBL:ENSMUSG00000036894 | MGI:1921262 | protein coding gene | RAP2B, member of RAS oncogene family |
| 3 | gene | 61.36225 | 61.36595 | negative | MGI_C57BL6J_3697707 | B430305J03Rik | ENSEMBL:ENSMUSG00000053706 | MGI:3697707 | protein coding gene | RIKEN cDNA B430305J03 gene |
| 3 | gene | 61.39215 | 61.41303 | positive | MGI_C57BL6J_5622946 | Gm40061 | NCBI_Gene:105244456 | MGI:5622946 | lncRNA gene | predicted gene, 40061 |
| 3 | pseudogene | 61.46166 | 61.46288 | negative | MGI_C57BL6J_3643135 | Gm8349 | NCBI_Gene:666891,ENSEMBL:ENSMUSG00000098208 | MGI:3643135 | pseudogene | predicted gene 8349 |
| 3 | pseudogene | 62.06754 | 62.06979 | negative | MGI_C57BL6J_3780107 | Gm9700 | NCBI_Gene:676965,ENSEMBL:ENSMUSG00000104489 | MGI:3780107 | pseudogene | predicted gene 9700 |
| 3 | gene | 62.12321 | 62.12356 | positive | MGI_C57BL6J_5610632 | Gm37404 | ENSEMBL:ENSMUSG00000103322 | MGI:5610632 | unclassified gene | predicted gene, 37404 |
| 3 | gene | 62.33752 | 62.34053 | negative | MGI_C57BL6J_3704218 | 9330121J05Rik | ENSEMBL:ENSMUSG00000103502 | MGI:3704218 | lncRNA gene | RIKEN cDNA 9330121J05 gene |
| 3 | gene | 62.33834 | 62.46222 | positive | MGI_C57BL6J_1918053 | Arhgef26 | NCBI_Gene:622434,ENSEMBL:ENSMUSG00000036885 | MGI:1918053 | protein coding gene | Rho guanine nucleotide exchange factor (GEF) 26 |
| 3 | gene | 62.34139 | 62.35052 | negative | MGI_C57BL6J_5593082 | Gm33923 | NCBI_Gene:102637008 | MGI:5593082 | lncRNA gene | predicted gene, 33923 |
| 3 | gene | 62.46801 | 62.50703 | negative | MGI_C57BL6J_1919412 | Dhx36 | NCBI_Gene:72162,ENSEMBL:ENSMUSG00000027770 | MGI:1919412 | protein coding gene | DEAH (Asp-Glu-Ala-His) box polypeptide 36 |
| 3 | gene | 62.52908 | 62.60544 | negative | MGI_C57BL6J_2443628 | Gpr149 | NCBI_Gene:229357,ENSEMBL:ENSMUSG00000043441 | MGI:2443628 | protein coding gene | G protein-coupled receptor 149 |
| 3 | gene | 62.60493 | 62.76393 | positive | MGI_C57BL6J_5593207 | Gm34048 | NCBI_Gene:102637165 | MGI:5593207 | lncRNA gene | predicted gene, 34048 |
| 3 | pseudogene | 62.76144 | 62.76202 | positive | MGI_C57BL6J_5010567 | Gm18382 | NCBI_Gene:100417044,ENSEMBL:ENSMUSG00000103764 | MGI:5010567 | pseudogene | predicted gene, 18382 |
| 3 | gene | 62.77198 | 62.86064 | positive | MGI_C57BL6J_5622947 | Gm40062 | NCBI_Gene:105244457 | MGI:5622947 | lncRNA gene | predicted gene, 40062 |
| 3 | gene | 62.86768 | 62.88900 | positive | MGI_C57BL6J_5622948 | Gm40063 | NCBI_Gene:105244458 | MGI:5622948 | lncRNA gene | predicted gene, 40063 |
| 3 | pseudogene | 62.92376 | 62.92426 | negative | MGI_C57BL6J_3780108 | Gm9701 | NCBI_Gene:676984,ENSEMBL:ENSMUSG00000063001 | MGI:3780108 | pseudogene | predicted gene 9701 |
| 3 | gene | 63.15222 | 63.15236 | positive | MGI_C57BL6J_5452210 | Gm22433 | ENSEMBL:ENSMUSG00000089175 | MGI:5452210 | snoRNA gene | predicted gene, 22433 |
| 3 | pseudogene | 63.17311 | 63.17386 | negative | MGI_C57BL6J_3645001 | Gm8388 | NCBI_Gene:666967,ENSEMBL:ENSMUSG00000102270 | MGI:3645001 | pseudogene | predicted gene 8388 |
| 3 | gene | 63.23282 | 63.24545 | negative | MGI_C57BL6J_5622949 | Gm40064 | NCBI_Gene:105244459 | MGI:5622949 | lncRNA gene | predicted gene, 40064 |
| 3 | gene | 63.24154 | 63.38603 | positive | MGI_C57BL6J_97004 | Mme | NCBI_Gene:17380,ENSEMBL:ENSMUSG00000027820 | MGI:97004 | protein coding gene | membrane metallo endopeptidase |
| 3 | gene | 63.43925 | 63.45790 | positive | MGI_C57BL6J_5593399 | Gm34240 | NCBI_Gene:102637430,ENSEMBL:ENSMUSG00000104336 | MGI:5593399 | lncRNA gene | predicted gene, 34240 |
| 3 | gene | 63.48110 | 63.48391 | negative | MGI_C57BL6J_5593461 | Strit1 | NCBI_Gene:102637511,ENSEMBL:ENSMUSG00000103476 | MGI:5593461 | protein coding gene | small transmembrane regulator of ion transport 1 |
| 3 | gene | 63.48388 | 63.48437 | positive | MGI_C57BL6J_5610507 | Gm37279 | ENSEMBL:ENSMUSG00000102883 | MGI:5610507 | unclassified gene | predicted gene, 37279 |
| 3 | gene | 63.48899 | 63.48910 | positive | MGI_C57BL6J_5454723 | Gm24946 | ENSEMBL:ENSMUSG00000088957 | MGI:5454723 | snRNA gene | predicted gene, 24946 |
| 3 | gene | 63.69620 | 63.89972 | negative | MGI_C57BL6J_2683547 | Plch1 | NCBI_Gene:269437,ENSEMBL:ENSMUSG00000036834 | MGI:2683547 | protein coding gene | phospholipase C, eta 1 |
| 3 | gene | 63.77021 | 63.77031 | positive | MGI_C57BL6J_5453277 | Gm23500 | ENSEMBL:ENSMUSG00000088360 | MGI:5453277 | snRNA gene | predicted gene, 23500 |
| 3 | gene | 63.79610 | 63.83685 | positive | MGI_C57BL6J_5826444 | Gm46807 | NCBI_Gene:108168874 | MGI:5826444 | lncRNA gene | predicted gene, 46807 |
| 3 | pseudogene | 63.82579 | 63.82644 | positive | MGI_C57BL6J_3802047 | Gm16162 | ENSEMBL:ENSMUSG00000090131 | MGI:3802047 | pseudogene | predicted gene 16162 |
| 3 | pseudogene | 63.84312 | 63.84407 | positive | MGI_C57BL6J_3801947 | Gm16129 | ENSEMBL:ENSMUSG00000085290 | MGI:3801947 | pseudogene | predicted gene 16129 |
| 3 | pseudogene | 63.90543 | 63.90573 | positive | MGI_C57BL6J_5593538 | Gm34379 | NCBI_Gene:102637617,ENSEMBL:ENSMUSG00000103905 | MGI:5593538 | pseudogene | predicted gene, 34379 |
| 3 | gene | 63.91468 | 63.92947 | negative | MGI_C57BL6J_3607716 | E130311K13Rik | NCBI_Gene:329659,ENSEMBL:ENSMUSG00000048581 | MGI:3607716 | protein coding gene | RIKEN cDNA E130311K13 gene |
| 3 | gene | 63.93351 | 63.96477 | negative | MGI_C57BL6J_1332247 | Slc33a1 | NCBI_Gene:11416,ENSEMBL:ENSMUSG00000027822 | MGI:1332247 | protein coding gene | solute carrier family 33 (acetyl-CoA transporter), member 1 |
| 3 | gene | 63.96363 | 63.96866 | positive | MGI_C57BL6J_5477344 | Gm26850 | ENSEMBL:ENSMUSG00000097724 | MGI:5477344 | lncRNA gene | predicted gene, 26850 |
| 3 | gene | 63.96482 | 64.02258 | positive | MGI_C57BL6J_2448526 | Gmps | NCBI_Gene:229363,ENSEMBL:ENSMUSG00000027823 | MGI:2448526 | protein coding gene | guanine monophosphate synthetase |
| 3 | pseudogene | 64.05303 | 64.05561 | negative | MGI_C57BL6J_3757653 | Vmn2r-ps3 | NCBI_Gene:100124546,ENSEMBL:ENSMUSG00000090303 | MGI:3757653 | pseudogene | vomeronasal 2, receptor, pseudogene 3 |
| 3 | gene | 64.07977 | 64.22085 | positive | MGI_C57BL6J_3645892 | Vmn2r1 | NCBI_Gene:56544,ENSEMBL:ENSMUSG00000027824 | MGI:3645892 | protein coding gene | vomeronasal 2, receptor 1 |
| 3 | gene | 64.10963 | 64.10974 | positive | MGI_C57BL6J_5451942 | Gm22165 | ENSEMBL:ENSMUSG00000089039 | MGI:5451942 | snRNA gene | predicted gene, 22165 |
| 3 | gene | 64.11528 | 64.17092 | negative | MGI_C57BL6J_3757666 | Vmn2r2 | NCBI_Gene:100125586,ENSEMBL:ENSMUSG00000043897 | MGI:3757666 | protein coding gene | vomeronasal 2, receptor 2 |
| 3 | pseudogene | 64.16339 | 64.16385 | positive | MGI_C57BL6J_3779465 | Gm5139 | NCBI_Gene:380739,ENSEMBL:ENSMUSG00000093447 | MGI:3779465 | pseudogene | predicted gene 5139 |
| 3 | pseudogene | 64.19557 | 64.21368 | negative | MGI_C57BL6J_5010479 | Gm18294 | NCBI_Gene:100416861,ENSEMBL:ENSMUSG00000093416 | MGI:5010479 | pseudogene | predicted gene, 18294 |
| 3 | gene | 64.23373 | 64.23546 | positive | MGI_C57BL6J_5593621 | Gm34462 | NCBI_Gene:102637724 | MGI:5593621 | lncRNA gene | predicted gene, 34462 |
| 3 | gene | 64.25722 | 64.28971 | negative | MGI_C57BL6J_3643995 | Vmn2r3 | NCBI_Gene:637004,ENSEMBL:ENSMUSG00000091572 | MGI:3643995 | protein coding gene | vomeronasal 2, receptor 3 |
| 3 | gene | 64.27582 | 64.27619 | negative | MGI_C57BL6J_1345615 | Ifld3 | NA | NA | unclassified gene | induced in fatty liver dystrophy 3 |
| 3 | gene | 64.31841 | 64.31856 | negative | MGI_C57BL6J_5453674 | Gm23897 | ENSEMBL:ENSMUSG00000088735 | MGI:5453674 | snoRNA gene | predicted gene, 23897 |
| 3 | pseudogene | 64.32866 | 64.33285 | positive | MGI_C57BL6J_3757669 | Vmn2r-ps4 | NCBI_Gene:100124547,ENSEMBL:ENSMUSG00000093503 | MGI:3757669 | pseudogene | vomeronasal 2, receptor, pseudogene 4 |
| 3 | pseudogene | 64.33956 | 64.35772 | negative | MGI_C57BL6J_3645404 | Vmn2r-ps5 | NCBI_Gene:667031,ENSEMBL:ENSMUSG00000091967 | MGI:3645404 | polymorphic pseudogene | vomeronasal 2, receptor, pseudogene 5 |
| 3 | gene | 64.38304 | 64.41537 | negative | MGI_C57BL6J_3648229 | Vmn2r4 | NCBI_Gene:637053,ENSEMBL:ENSMUSG00000092049 | MGI:3648229 | protein coding gene | vomeronasal 2, receptor 4 |
| 3 | pseudogene | 64.43688 | 64.43829 | negative | MGI_C57BL6J_3757678 | Vmn2r-ps6 | NCBI_Gene:100124548,ENSEMBL:ENSMUSG00000093395 | MGI:3757678 | pseudogene | vomeronasal 2, receptor, pseudogene 6 |
| 3 | pseudogene | 64.44260 | 64.44397 | negative | MGI_C57BL6J_3757679 | Vmn2r-ps7 | NCBI_Gene:672951,ENSEMBL:ENSMUSG00000093546 | MGI:3757679 | pseudogene | vomeronasal 2, receptor, pseudogene 7 |
| 3 | pseudogene | 64.44797 | 64.46009 | positive | MGI_C57BL6J_5313103 | Gm20656 | ENSEMBL:ENSMUSG00000093671 | MGI:5313103 | pseudogene | predicted gene 20656 |
| 3 | pseudogene | 64.47702 | 64.47932 | positive | MGI_C57BL6J_3757680 | Vmn2r-ps8 | NCBI_Gene:100124549,ENSEMBL:ENSMUSG00000094482 | MGI:3757680 | pseudogene | vomeronasal 2, receptor, pseudogene 8 |
| 3 | gene | 64.48861 | 64.50984 | negative | MGI_C57BL6J_3649074 | Vmn2r5 | NCBI_Gene:667060,ENSEMBL:ENSMUSG00000068999 | MGI:3649074 | protein coding gene | vomeronasal 2, receptor 5 |
| 3 | gene | 64.53131 | 64.56583 | negative | MGI_C57BL6J_3649068 | Vmn2r6 | NCBI_Gene:667069,ENSEMBL:ENSMUSG00000090581 | MGI:3649068 | protein coding gene | vomeronasal 2, receptor 6 |
| 3 | pseudogene | 64.59152 | 64.59290 | negative | MGI_C57BL6J_3757686 | Vmn2r-ps9 | NCBI_Gene:100124550,ENSEMBL:ENSMUSG00000093699 | MGI:3757686 | pseudogene | vomeronasal 2, receptor, pseudogene 9 |
| 3 | pseudogene | 64.59691 | 64.60577 | positive | MGI_C57BL6J_5313104 | Gm20657 | ENSEMBL:ENSMUSG00000093484 | MGI:5313104 | pseudogene | predicted gene 20657 |
| 3 | pseudogene | 64.60043 | 64.60114 | positive | MGI_C57BL6J_3781218 | Gm3040 | NCBI_Gene:100416170 | MGI:3781218 | pseudogene | predicted gene 3040 |
| 3 | pseudogene | 64.62169 | 64.62399 | positive | MGI_C57BL6J_3757943 | Vmn2r-ps10 | NCBI_Gene:100124554,ENSEMBL:ENSMUSG00000095107 | MGI:3757943 | pseudogene | vomeronasal 2, receptor, pseudogene 10 |
| 3 | pseudogene | 64.63257 | 64.84474 | negative | MGI_C57BL6J_2441682 | Vmn2r-ps11 | NCBI_Gene:319210,ENSEMBL:ENSMUSG00000093434 | MGI:2441682 | pseudogene | vomeronasal 2, receptor, pseudogene 11 |
| 3 | gene | 64.63286 | 64.84474 | negative | MGI_C57BL6J_5504054 | Gm26939 | ENSEMBL:ENSMUSG00000097538 | MGI:5504054 | lncRNA gene | predicted gene, 26939 |
| 3 | gene | 64.68504 | 64.84476 | negative | MGI_C57BL6J_2441693 | Vmn2r7 | NCBI_Gene:319217,ENSEMBL:ENSMUSG00000062200 | MGI:2441693 | protein coding gene | vomeronasal 2, receptor 7 |
| 3 | pseudogene | 64.74475 | 64.74564 | negative | MGI_C57BL6J_3757944 | Vmn2r-ps12 | NCBI_Gene:100125273,ENSEMBL:ENSMUSG00000093474 | MGI:3757944 | pseudogene | vomeronasal 2, receptor, pseudogene 12 |
| 3 | gene | 64.89447 | 64.89460 | negative | MGI_C57BL6J_5454999 | Gm25222 | ENSEMBL:ENSMUSG00000088824 | MGI:5454999 | snoRNA gene | predicted gene, 25222 |
| 3 | pseudogene | 64.89885 | 64.89976 | positive | MGI_C57BL6J_3757946 | Vmn2r-ps13 | NCBI_Gene:670110,ENSEMBL:ENSMUSG00000093767 | MGI:3757946 | pseudogene | vomeronasal 2, receptor, pseudogene 13 |
| 3 | gene | 64.94905 | 65.37823 | positive | MGI_C57BL6J_109155 | Kcnab1 | NCBI_Gene:16497,ENSEMBL:ENSMUSG00000027827 | MGI:109155 | protein coding gene | potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| 3 | gene | 64.94922 | 65.22370 | positive | MGI_C57BL6J_5611276 | Gm38048 | ENSEMBL:ENSMUSG00000102437 | MGI:5611276 | lncRNA gene | predicted gene, 38048 |
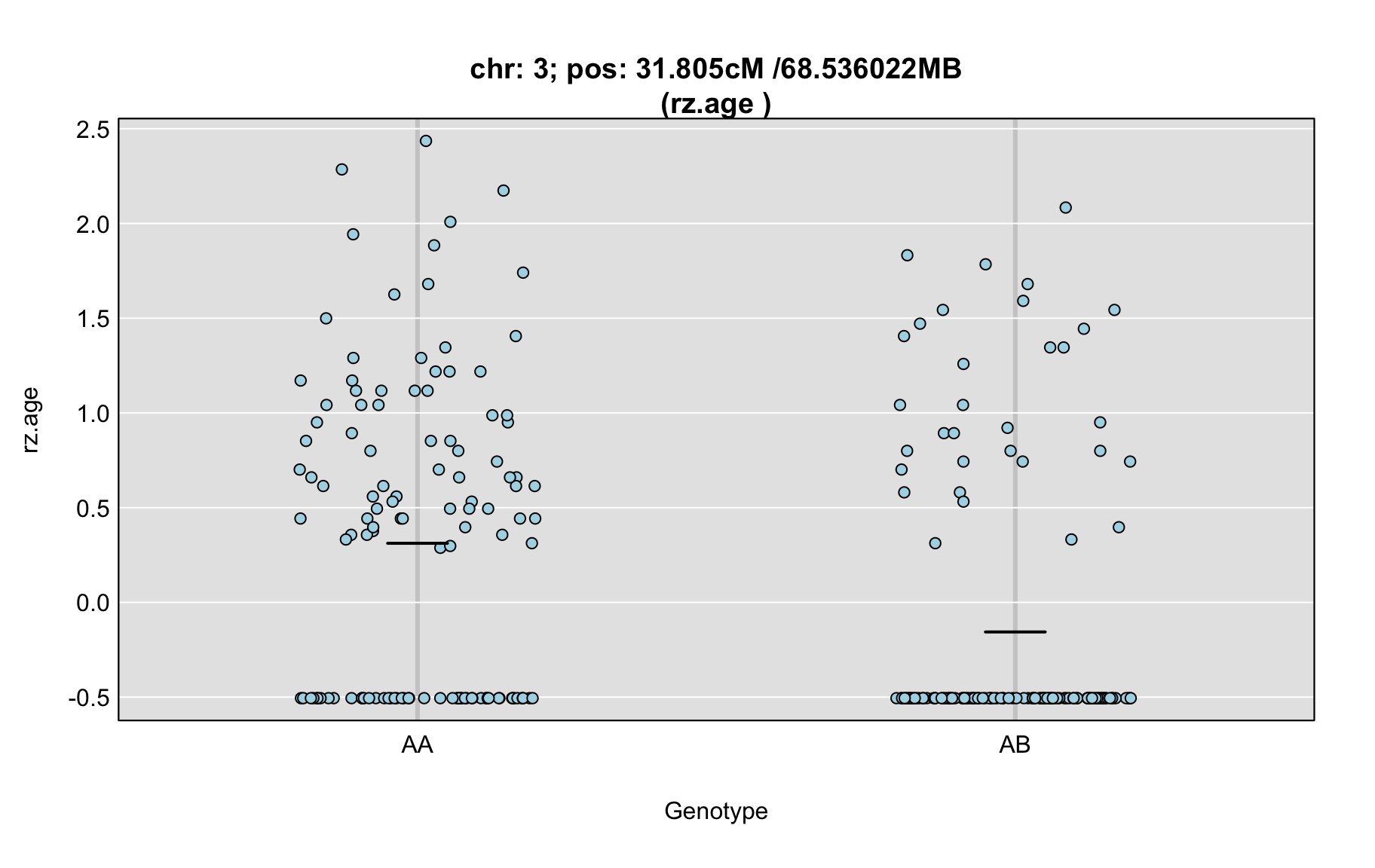
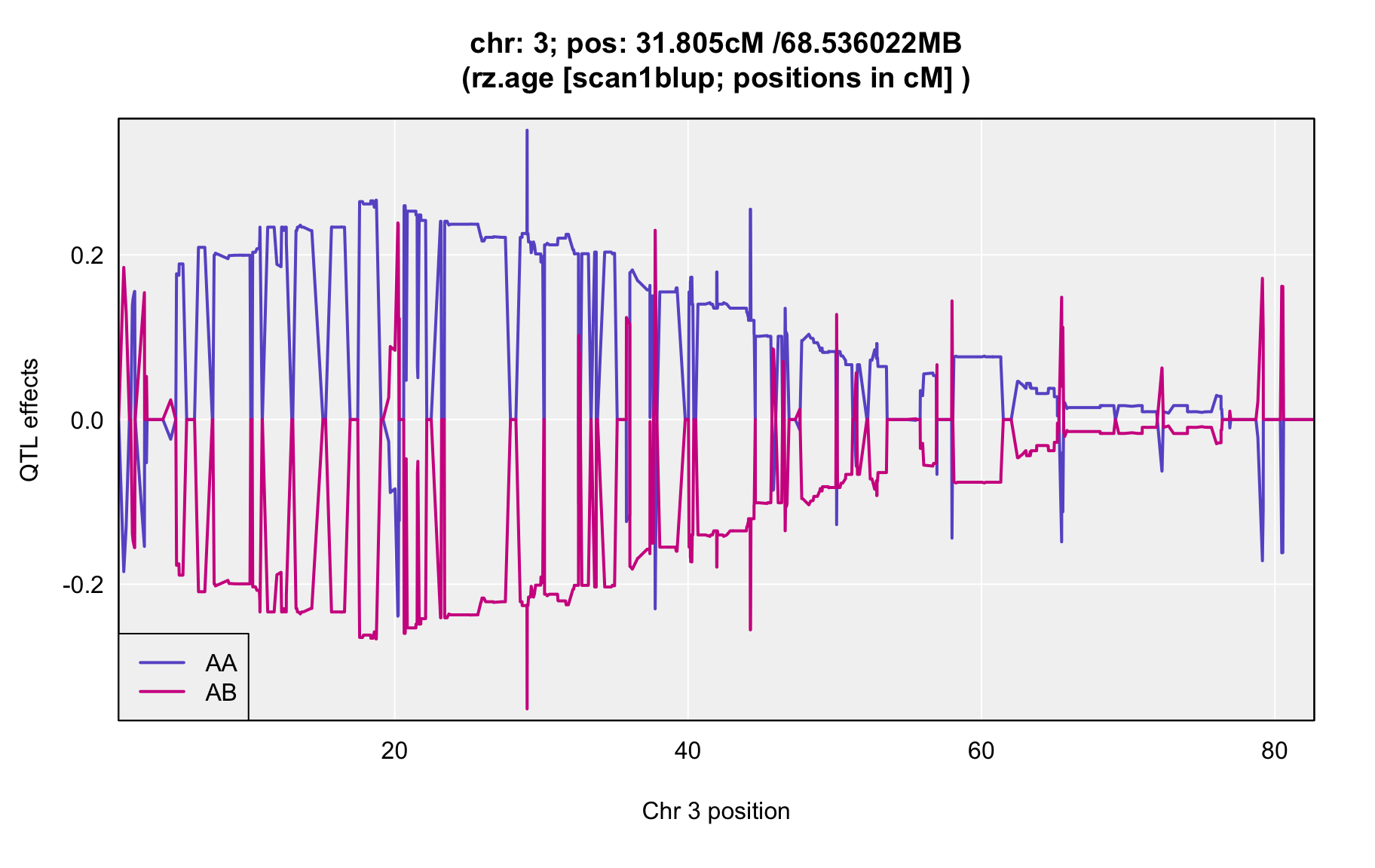
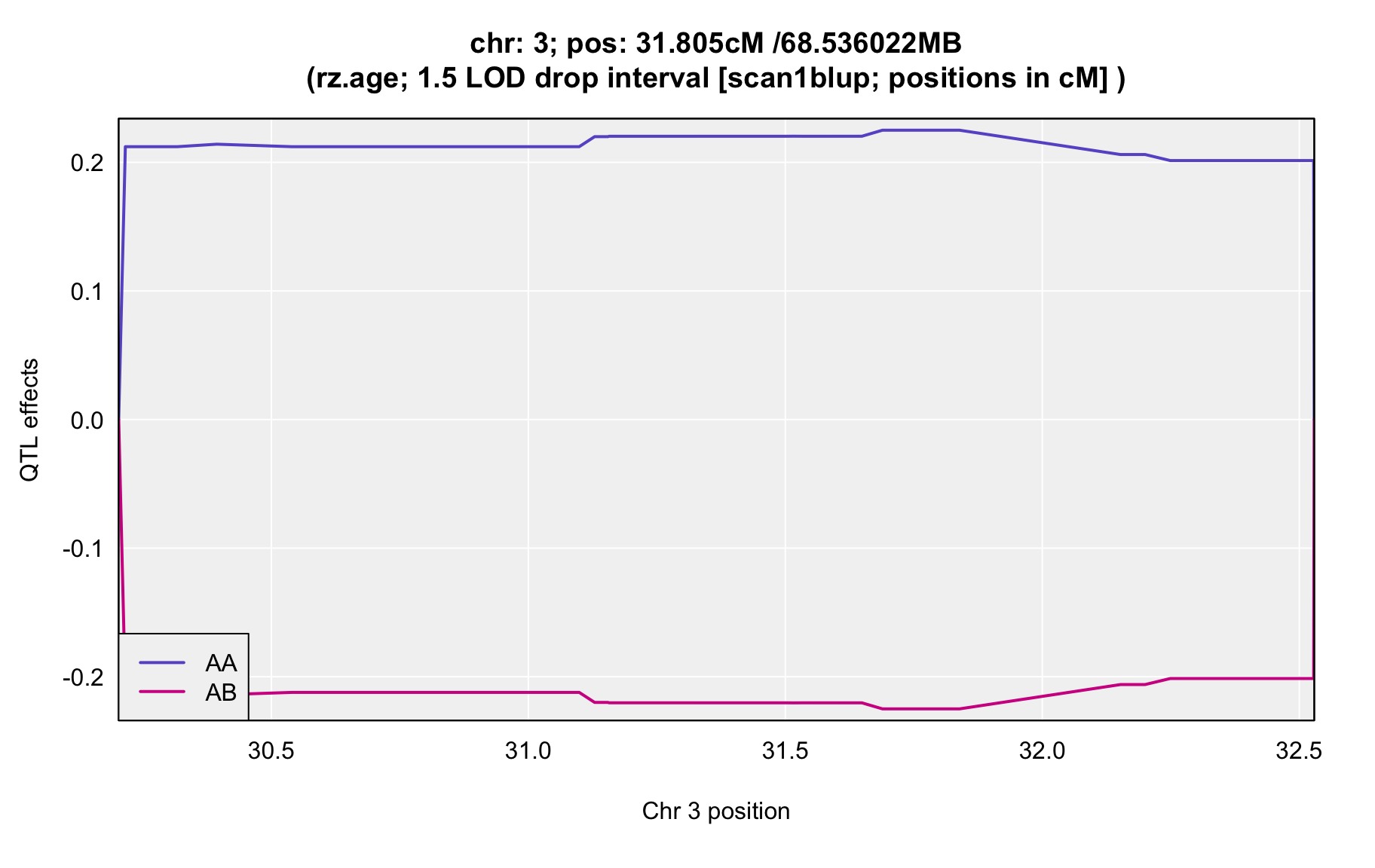
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | gene | 65.49409 | 65.49563 | negative | MGI_C57BL6J_5593939 | Gm34780 | NCBI_Gene:102638146,ENSEMBL:ENSMUSG00000103513 | MGI:5593939 | lncRNA gene | predicted gene, 34780 |
| 3 | gene | 65.49517 | 65.51729 | positive | MGI_C57BL6J_5622951 | Gm40066 | NCBI_Gene:105244461 | MGI:5622951 | lncRNA gene | predicted gene, 40066 |
| 3 | pseudogene | 65.50628 | 65.50664 | negative | MGI_C57BL6J_5610237 | Gm37009 | ENSEMBL:ENSMUSG00000103880 | MGI:5610237 | pseudogene | predicted gene, 37009 |
| 3 | gene | 65.50646 | 65.52004 | negative | MGI_C57BL6J_5621388 | Gm38503 | NCBI_Gene:102637953 | MGI:5621388 | lncRNA gene | predicted gene, 38503 |
| 3 | gene | 65.50726 | 65.52941 | negative | MGI_C57BL6J_1918254 | 4931440P22Rik | NCBI_Gene:71004,ENSEMBL:ENSMUSG00000074580 | MGI:1918254 | lncRNA gene | RIKEN cDNA 4931440P22 gene |
| 3 | gene | 65.52570 | 65.52579 | positive | MGI_C57BL6J_5452056 | Gm22279 | ENSEMBL:ENSMUSG00000087825 | MGI:5452056 | snRNA gene | predicted gene, 22279 |
| 3 | gene | 65.52841 | 65.55552 | positive | MGI_C57BL6J_2159210 | Tiparp | NCBI_Gene:99929,ENSEMBL:ENSMUSG00000034640 | MGI:2159210 | protein coding gene | TCDD-inducible poly(ADP-ribose) polymerase |
| 3 | gene | 65.57522 | 65.58085 | negative | MGI_C57BL6J_5594083 | Gm34924 | NCBI_Gene:102638334 | MGI:5594083 | lncRNA gene | predicted gene, 34924 |
| 3 | pseudogene | 65.61350 | 65.61532 | positive | MGI_C57BL6J_3648046 | Gm5847 | NCBI_Gene:545530,ENSEMBL:ENSMUSG00000082280 | MGI:3648046 | pseudogene | predicted gene 5847 |
| 3 | pseudogene | 65.61537 | 65.61617 | negative | MGI_C57BL6J_5611461 | Gm38233 | ENSEMBL:ENSMUSG00000103294 | MGI:5611461 | pseudogene | predicted gene, 38233 |
| 3 | gene | 65.65929 | 65.65943 | positive | MGI_C57BL6J_5531019 | Mir8120 | miRBase:MI0026052,NCBI_Gene:102465905,ENSEMBL:ENSMUSG00000098944 | MGI:5531019 | miRNA gene | microRNA 8120 |
| 3 | gene | 65.65954 | 65.83248 | positive | MGI_C57BL6J_3645902 | Lekr1 | NCBI_Gene:624866,ENSEMBL:ENSMUSG00000074579 | MGI:3645902 | protein coding gene | leucine, glutamate and lysine rich 1 |
| 3 | gene | 65.67839 | 65.68088 | positive | MGI_C57BL6J_5611004 | Gm37776 | ENSEMBL:ENSMUSG00000104037 | MGI:5611004 | unclassified gene | predicted gene, 37776 |
| 3 | gene | 65.77151 | 65.77162 | negative | MGI_C57BL6J_5455532 | Gm25755 | ENSEMBL:ENSMUSG00000065330 | MGI:5455532 | snRNA gene | predicted gene, 25755 |
| 3 | gene | 65.82912 | 65.87228 | negative | MGI_C57BL6J_5610587 | Gm37359 | ENSEMBL:ENSMUSG00000103620 | MGI:5610587 | lncRNA gene | predicted gene, 37359 |
| 3 | gene | 65.86327 | 65.86483 | negative | MGI_C57BL6J_5610359 | Gm37131 | ENSEMBL:ENSMUSG00000103440 | MGI:5610359 | unclassified gene | predicted gene, 37131 |
| 3 | gene | 65.92610 | 65.92828 | negative | MGI_C57BL6J_5594198 | Gm35039 | NCBI_Gene:102638487 | MGI:5594198 | lncRNA gene | predicted gene, 35039 |
| 3 | pseudogene | 65.93349 | 65.93391 | negative | MGI_C57BL6J_5610265 | Gm37037 | ENSEMBL:ENSMUSG00000102566 | MGI:5610265 | pseudogene | predicted gene, 37037 |
| 3 | gene | 65.94591 | 65.95839 | negative | MGI_C57BL6J_1922664 | Ccnl1 | NCBI_Gene:56706,ENSEMBL:ENSMUSG00000027829 | MGI:1922664 | protein coding gene | cyclin L1 |
| 3 | gene | 65.95776 | 65.96204 | positive | MGI_C57BL6J_5610533 | Gm37305 | ENSEMBL:ENSMUSG00000103041 | MGI:5610533 | lncRNA gene | predicted gene, 37305 |
| 3 | pseudogene | 65.96873 | 65.97028 | negative | MGI_C57BL6J_5594265 | Gm35106 | NCBI_Gene:105244463,ENSEMBL:ENSMUSG00000102914 | MGI:5594265 | pseudogene | predicted gene, 35106 |
| 3 | gene | 65.96991 | 65.97192 | positive | MGI_C57BL6J_5610306 | Gm37078 | ENSEMBL:ENSMUSG00000102652 | MGI:5610306 | unclassified gene | predicted gene, 37078 |
| 3 | pseudogene | 65.98716 | 65.98975 | negative | MGI_C57BL6J_5011209 | Gm19024 | NCBI_Gene:100418133,ENSEMBL:ENSMUSG00000103641 | MGI:5011209 | pseudogene | predicted gene, 19024 |
| 3 | pseudogene | 66.02950 | 66.03152 | negative | MGI_C57BL6J_3643872 | Gm6546 | NCBI_Gene:625048,ENSEMBL:ENSMUSG00000103369 | MGI:3643872 | pseudogene | predicted gene 6546 |
| 3 | gene | 66.05356 | 66.29707 | negative | MGI_C57BL6J_1920039 | Veph1 | NCBI_Gene:72789,ENSEMBL:ENSMUSG00000027831 | MGI:1920039 | protein coding gene | ventricular zone expressed PH domain-containing 1 |
| 3 | gene | 66.08668 | 66.08779 | positive | MGI_C57BL6J_5611050 | Gm37822 | ENSEMBL:ENSMUSG00000104189 | MGI:5611050 | unclassified gene | predicted gene, 37822 |
| 3 | gene | 66.08811 | 66.08826 | negative | MGI_C57BL6J_5456219 | Gm26442 | ENSEMBL:ENSMUSG00000064522 | MGI:5456219 | snRNA gene | predicted gene, 26442 |
| 3 | pseudogene | 66.09981 | 66.10006 | positive | MGI_C57BL6J_5610201 | Gm36973 | ENSEMBL:ENSMUSG00000104307 | MGI:5610201 | pseudogene | predicted gene, 36973 |
| 3 | pseudogene | 66.12840 | 66.12975 | negative | MGI_C57BL6J_98800 | Tpi-rs2 | NCBI_Gene:111205,ENSEMBL:ENSMUSG00000097968 | MGI:98800 | pseudogene | triosephosphate isomerase related sequence 2 |
| 3 | pseudogene | 66.19109 | 66.19124 | negative | MGI_C57BL6J_5610558 | Gm37330 | ENSEMBL:ENSMUSG00000103208 | MGI:5610558 | pseudogene | predicted gene, 37330 |
| 3 | gene | 66.21989 | 66.22581 | positive | MGI_C57BL6J_104641 | Ptx3 | NCBI_Gene:19288,ENSEMBL:ENSMUSG00000027832 | MGI:104641 | protein coding gene | pentraxin related gene |
| 3 | pseudogene | 66.36454 | 66.36463 | positive | MGI_C57BL6J_6275305 | Gm50014 | ENSEMBL:ENSMUSG00000117623 | MGI:6275305 | pseudogene | predicted gene, 50014 |
| 3 | gene | 66.37430 | 66.38810 | positive | MGI_C57BL6J_5594343 | Gm35184 | NCBI_Gene:102638679 | MGI:5594343 | lncRNA gene | predicted gene, 35184 |
| 3 | gene | 66.40041 | 66.40607 | positive | MGI_C57BL6J_5594395 | Gm35236 | NCBI_Gene:102638746 | MGI:5594395 | lncRNA gene | predicted gene, 35236 |
| 3 | pseudogene | 66.56278 | 66.56355 | negative | MGI_C57BL6J_5010137 | Gm17952 | NCBI_Gene:100416166,ENSEMBL:ENSMUSG00000103600 | MGI:5010137 | pseudogene | predicted gene, 17952 |
| 3 | pseudogene | 66.88235 | 66.88314 | positive | MGI_C57BL6J_3646021 | Gm6555 | NCBI_Gene:625115,ENSEMBL:ENSMUSG00000103700 | MGI:3646021 | pseudogene | predicted gene 6555 |
| 3 | pseudogene | 66.90736 | 66.90777 | negative | MGI_C57BL6J_5611584 | Gm38356 | ENSEMBL:ENSMUSG00000103318 | MGI:5611584 | pseudogene | predicted gene, 38356 |
| 3 | pseudogene | 66.95354 | 66.95386 | positive | MGI_C57BL6J_5663653 | Gm43516 | ENSEMBL:ENSMUSG00000105794 | MGI:5663653 | pseudogene | predicted gene 43516 |
| 3 | gene | 66.97172 | 66.98177 | negative | MGI_C57BL6J_1201673 | Shox2 | NCBI_Gene:20429,ENSEMBL:ENSMUSG00000027833 | MGI:1201673 | protein coding gene | short stature homeobox 2 |
| 3 | gene | 66.98139 | 67.35840 | positive | MGI_C57BL6J_1914130 | Rsrc1 | NCBI_Gene:66880,ENSEMBL:ENSMUSG00000034544 | MGI:1914130 | protein coding gene | arginine/serine-rich coiled-coil 1 |
| 3 | gene | 67.05596 | 67.05606 | positive | MGI_C57BL6J_5452072 | Gm22295 | ENSEMBL:ENSMUSG00000065245 | MGI:5452072 | snRNA gene | predicted gene, 22295 |
| 3 | gene | 67.36546 | 67.37516 | negative | MGI_C57BL6J_4937036 | Gm17402 | ENSEMBL:ENSMUSG00000090408 | MGI:4937036 | protein coding gene | predicted gene, 17402 |
| 3 | gene | 67.37410 | 67.40000 | positive | MGI_C57BL6J_1341819 | Mlf1 | NCBI_Gene:17349,ENSEMBL:ENSMUSG00000048416 | MGI:1341819 | protein coding gene | myeloid leukemia factor 1 |
| 3 | gene | 67.43010 | 67.47653 | positive | MGI_C57BL6J_107339 | Gfm1 | NCBI_Gene:28030,ENSEMBL:ENSMUSG00000027774 | MGI:107339 | protein coding gene | G elongation factor, mitochondrial 1 |
| 3 | gene | 67.45800 | 67.46393 | negative | MGI_C57BL6J_107633 | Lxn | NCBI_Gene:17035,ENSEMBL:ENSMUSG00000047557 | MGI:107633 | protein coding gene | latexin |
| 3 | gene | 67.47698 | 67.47882 | positive | MGI_C57BL6J_5662705 | Gm42568 | ENSEMBL:ENSMUSG00000103703 | MGI:5662705 | unclassified gene | predicted gene 42568 |
| 3 | gene | 67.47889 | 67.51552 | negative | MGI_C57BL6J_1924461 | Rarres1 | NCBI_Gene:109222,ENSEMBL:ENSMUSG00000049404 | MGI:1924461 | protein coding gene | retinoic acid receptor responder (tazarotene induced) 1 |
| 3 | gene | 67.51567 | 67.51966 | positive | MGI_C57BL6J_5622952 | Gm40067 | NCBI_Gene:105244464 | MGI:5622952 | lncRNA gene | predicted gene, 40067 |
| 3 | gene | 67.52266 | 67.55343 | negative | MGI_C57BL6J_5594458 | Gm35299 | NCBI_Gene:102638825,ENSEMBL:ENSMUSG00000103718 | MGI:5594458 | lncRNA gene | predicted gene, 35299 |
| 3 | gene | 67.57057 | 67.57282 | negative | MGI_C57BL6J_1921179 | 4930402C01Rik | ENSEMBL:ENSMUSG00000104382 | MGI:1921179 | lncRNA gene | RIKEN cDNA 4930402C01 gene |
| 3 | gene | 67.57316 | 67.57329 | negative | MGI_C57BL6J_5455696 | Gm25919 | ENSEMBL:ENSMUSG00000077459 | MGI:5455696 | snoRNA gene | predicted gene, 25919 |
| 3 | gene | 67.58274 | 67.60424 | positive | MGI_C57BL6J_1914118 | Mfsd1 | NCBI_Gene:66868,ENSEMBL:ENSMUSG00000027775 | MGI:1914118 | protein coding gene | major facilitator superfamily domain containing 1 |
| 3 | pseudogene | 67.60575 | 67.60785 | negative | MGI_C57BL6J_3648243 | Gm8515 | NCBI_Gene:667207,ENSEMBL:ENSMUSG00000103206 | MGI:3648243 | pseudogene | predicted gene 8515 |
| 3 | pseudogene | 67.61978 | 67.62073 | negative | MGI_C57BL6J_2685258 | Gm412 | NCBI_Gene:208424,ENSEMBL:ENSMUSG00000103606 | MGI:2685258 | pseudogene | predicted gene 412 |
| 3 | pseudogene | 67.68436 | 67.68475 | negative | MGI_C57BL6J_5610829 | Gm37601 | ENSEMBL:ENSMUSG00000102746 | MGI:5610829 | pseudogene | predicted gene, 37601 |
| 3 | pseudogene | 67.74197 | 67.74330 | negative | MGI_C57BL6J_5010902 | Gm18717 | NCBI_Gene:100417618,ENSEMBL:ENSMUSG00000104477 | MGI:5010902 | pseudogene | predicted gene, 18717 |
| 3 | gene | 67.75360 | 67.75565 | positive | MGI_C57BL6J_5594566 | Gm35407 | NCBI_Gene:102638975 | MGI:5594566 | lncRNA gene | predicted gene, 35407 |
| 3 | gene | 67.75566 | 67.76628 | positive | MGI_C57BL6J_5622953 | Gm40068 | NCBI_Gene:105244465 | MGI:5622953 | lncRNA gene | predicted gene, 40068 |
| 3 | gene | 67.89222 | 68.05659 | positive | MGI_C57BL6J_3644166 | Iqcj | NCBI_Gene:208426,ENSEMBL:ENSMUSG00000051777 | MGI:3644166 | protein coding gene | IQ motif containing J |
| 3 | gene | 67.89222 | 68.62648 | positive | MGI_C57BL6J_5439400 | Iqschfp | NCBI_Gene:100505386,ENSEMBL:ENSMUSG00000102422 | MGI:5439400 | protein coding gene | Iqcj and Schip1 fusion protein |
| 3 | gene | 68.06480 | 68.62648 | positive | MGI_C57BL6J_1353557 | Schip1 | NCBI_Gene:30953,ENSEMBL:ENSMUSG00000027777 | MGI:1353557 | protein coding gene | schwannomin interacting protein 1 |
| 3 | pseudogene | 68.30377 | 68.31036 | negative | MGI_C57BL6J_3648047 | Gm5848 | NCBI_Gene:545531 | MGI:3648047 | pseudogene | predicted pseudogene 5848 |
| 3 | pseudogene | 68.33319 | 68.33378 | positive | MGI_C57BL6J_3642851 | Gm10292 | NCBI_Gene:631379,ENSEMBL:ENSMUSG00000102846 | MGI:3642851 | pseudogene | predicted gene 10292 |
| 3 | gene | 68.40778 | 68.41470 | positive | MGI_C57BL6J_3641783 | Gm10723 | ENSEMBL:ENSMUSG00000103345 | MGI:3641783 | unclassified gene | predicted gene 10723 |
| 3 | gene | 68.54752 | 68.54771 | negative | MGI_C57BL6J_5451850 | Gm22073 | ENSEMBL:ENSMUSG00000084617 | MGI:5451850 | snRNA gene | predicted gene, 22073 |
| 3 | gene | 68.69064 | 68.69855 | positive | MGI_C57BL6J_96539 | Il12a | NCBI_Gene:16159,ENSEMBL:ENSMUSG00000027776 | MGI:96539 | protein coding gene | interleukin 12a |
| 3 | pseudogene | 68.73314 | 68.73808 | positive | MGI_C57BL6J_3708676 | Gm10040 | ENSEMBL:ENSMUSG00000058360 | MGI:3708676 | pseudogene | predicted gene 10040 |
| 3 | gene | 68.75991 | 68.76143 | negative | MGI_C57BL6J_1922468 | 4930535E02Rik | ENSEMBL:ENSMUSG00000102677 | MGI:1922468 | lncRNA gene | RIKEN cDNA 4930535E02 gene |
| 3 | gene | 68.76136 | 68.76267 | positive | MGI_C57BL6J_5594743 | Gm35584 | NCBI_Gene:102639225,ENSEMBL:ENSMUSG00000102936 | MGI:5594743 | lncRNA gene | predicted gene, 35584 |
| 3 | pseudogene | 68.84608 | 68.84741 | positive | MGI_C57BL6J_3644317 | Gm7270 | NCBI_Gene:639530,ENSEMBL:ENSMUSG00000103218,ENSEMBL:ENSMUSG00000102853 | MGI:3644317 | pseudogene | predicted gene 7270 |
| 3 | gene | 68.86792 | 68.87027 | negative | MGI_C57BL6J_4937275 | Gm17641 | ENSEMBL:ENSMUSG00000091272 | MGI:4937275 | protein coding gene | predicted gene, 17641 |
| 3 | gene | 68.86959 | 68.87216 | positive | MGI_C57BL6J_1915975 | 1110032F04Rik | NCBI_Gene:68725,ENSEMBL:ENSMUSG00000046999 | MGI:1915975 | protein coding gene | RIKEN cDNA 1110032F04 gene |
| 3 | gene | 68.89250 | 69.00464 | negative | MGI_C57BL6J_1915509 | Ift80 | NCBI_Gene:68259,ENSEMBL:ENSMUSG00000027778 | MGI:1915509 | protein coding gene | intraflagellar transport 80 |
| 3 | pseudogene | 68.90017 | 68.90045 | negative | MGI_C57BL6J_5313136 | Gm20689 | ENSEMBL:ENSMUSG00000093384 | MGI:5313136 | pseudogene | predicted gene 20689 |
| 3 | pseudogene | 68.90181 | 68.90216 | positive | MGI_C57BL6J_5313137 | Gm20690 | ENSEMBL:ENSMUSG00000093730,ENSEMBL:ENSMUSG00000103007 | MGI:5313137 | pseudogene | predicted gene 20690 |
| 3 | pseudogene | 68.90216 | 68.90278 | negative | MGI_C57BL6J_5010773 | Gm18588 | NCBI_Gene:100417394,ENSEMBL:ENSMUSG00000093402 | MGI:5010773 | pseudogene | predicted gene, 18588 |
| 3 | pseudogene | 68.90382 | 68.90415 | negative | MGI_C57BL6J_5313138 | Gm20691 | ENSEMBL:ENSMUSG00000093505 | MGI:5313138 | pseudogene | predicted gene 20691 |
| 3 | gene | 68.91650 | 68.91732 | negative | MGI_C57BL6J_6097561 | Gm48183 | ENSEMBL:ENSMUSG00000111756 | MGI:6097561 | unclassified gene | predicted gene, 48183 |
| 3 | gene | 69.00474 | 69.03462 | positive | MGI_C57BL6J_1917349 | Smc4 | NCBI_Gene:70099,ENSEMBL:ENSMUSG00000034349 | MGI:1917349 | protein coding gene | structural maintenance of chromosomes 4 |
| 3 | gene | 69.00977 | 69.00983 | positive | MGI_C57BL6J_2676842 | Mir15b | miRBase:MI0000140,NCBI_Gene:387175,ENSEMBL:ENSMUSG00000065580 | MGI:2676842 | miRNA gene | microRNA 15b |
| 3 | gene | 69.00990 | 69.01000 | positive | MGI_C57BL6J_3618690 | Mir16-2 | miRBase:MI0000566,NCBI_Gene:723949,ENSEMBL:ENSMUSG00000065606 | MGI:3618690 | miRNA gene | microRNA 16-2 |
| 3 | gene | 69.03529 | 69.04640 | negative | MGI_C57BL6J_1914199 | Trim59 | NCBI_Gene:66949,ENSEMBL:ENSMUSG00000034317 | MGI:1914199 | protein coding gene | tripartite motif-containing 59 |
| 3 | gene | 69.06065 | 69.06525 | positive | MGI_C57BL6J_2442898 | 9630025H16Rik | NCBI_Gene:105244467 | MGI:2442898 | lncRNA gene | RIKEN cDNA 9630025H16 gene |
| 3 | gene | 69.06715 | 69.12711 | negative | MGI_C57BL6J_1100848 | Kpna4 | NCBI_Gene:16649,ENSEMBL:ENSMUSG00000027782 | MGI:1100848 | protein coding gene | karyopherin (importin) alpha 4 |
| 3 | gene | 69.08510 | 69.08545 | negative | MGI_C57BL6J_5451786 | Gm22009 | ENSEMBL:ENSMUSG00000089417 | MGI:5451786 | snoRNA gene | predicted gene, 22009 |
| 3 | gene | 69.11974 | 69.12272 | negative | MGI_C57BL6J_5610786 | Gm37558 | ENSEMBL:ENSMUSG00000103094 | MGI:5610786 | unclassified gene | predicted gene, 37558 |
| 3 | gene | 69.13123 | 69.15718 | positive | MGI_C57BL6J_2686493 | Gm1647 | NCBI_Gene:381452,ENSEMBL:ENSMUSG00000094763 | MGI:2686493 | lncRNA gene | predicted gene 1647 |
| 3 | gene | 69.22242 | 69.22362 | positive | MGI_C57BL6J_1918869 | Arl14 | NCBI_Gene:71619,ENSEMBL:ENSMUSG00000098207 | MGI:1918869 | protein coding gene | ADP-ribosylation factor-like 14 |
| 3 | gene | 69.25461 | 69.26383 | positive | MGI_C57BL6J_5622955 | Gm40070 | NCBI_Gene:105244468 | MGI:5622955 | lncRNA gene | predicted gene, 40070 |
| 3 | gene | 69.31686 | 69.56080 | positive | MGI_C57BL6J_2139740 | Ppm1l | NCBI_Gene:242083,ENSEMBL:ENSMUSG00000027784 | MGI:2139740 | protein coding gene | protein phosphatase 1 (formerly 2C)-like |
| 3 | gene | 69.31816 | 69.31929 | negative | MGI_C57BL6J_1920514 | 1700037N05Rik | NA | NA | unclassified gene | RIKEN cDNA 1700037N05 gene |
| 3 | pseudogene | 69.32001 | 69.32062 | negative | MGI_C57BL6J_4938041 | Gm17214 | NCBI_Gene:100416279,ENSEMBL:ENSMUSG00000091315 | MGI:4938041 | pseudogene | predicted gene 17214 |
| 3 | pseudogene | 69.33519 | 69.33579 | negative | MGI_C57BL6J_4938039 | Gm17212 | NCBI_Gene:100416280,ENSEMBL:ENSMUSG00000090941 | MGI:4938039 | pseudogene | predicted gene 17212 |
| 3 | gene | 69.37640 | 69.38370 | positive | MGI_C57BL6J_2443158 | A330078L11Rik | NA | NA | unclassified gene | RIKEN cDNA A330078L11 gene |
| 3 | pseudogene | 69.43101 | 69.43238 | negative | MGI_C57BL6J_4938040 | Gm17213 | NCBI_Gene:100417908,ENSEMBL:ENSMUSG00000090743 | MGI:4938040 | pseudogene | predicted gene 17213 |
| 3 | pseudogene | 69.56624 | 69.56747 | negative | MGI_C57BL6J_3644126 | Gm8565 | NCBI_Gene:667309,ENSEMBL:ENSMUSG00000098006 | MGI:3644126 | pseudogene | predicted gene 8565 |
| 3 | gene | 69.57392 | 69.59896 | negative | MGI_C57BL6J_1349405 | B3galnt1 | NCBI_Gene:26879,ENSEMBL:ENSMUSG00000043300 | MGI:1349405 | protein coding gene | UDP-GalNAc:betaGlcNAc beta 1,3-galactosaminyltransferase, polypeptide 1 |
| 3 | pseudogene | 69.66177 | 69.66216 | positive | MGI_C57BL6J_5610608 | Gm37380 | ENSEMBL:ENSMUSG00000102342 | MGI:5610608 | pseudogene | predicted gene, 37380 |
| 3 | gene | 69.68547 | 69.68558 | positive | MGI_C57BL6J_5455398 | Gm25621 | ENSEMBL:ENSMUSG00000087848 | MGI:5455398 | rRNA gene | predicted gene, 25621 |
| 3 | pseudogene | 69.71694 | 69.71744 | negative | MGI_C57BL6J_98039 | Rpl32-ps | NCBI_Gene:19952,ENSEMBL:ENSMUSG00000068969 | MGI:98039 | pseudogene | ribosomal protein L32, pseudogene |
| 3 | gene | 69.72199 | 69.75637 | positive | MGI_C57BL6J_2140103 | Nmd3 | NCBI_Gene:97112,ENSEMBL:ENSMUSG00000027787 | MGI:2140103 | protein coding gene | NMD3 ribosome export adaptor |
| 3 | gene | 69.75311 | 69.75637 | positive | MGI_C57BL6J_2442565 | 9530051E23Rik | NA | NA | unclassified gene | RIKEN cDNA 9530051E23 gene |
| 3 | pseudogene | 69.80846 | 69.80902 | negative | MGI_C57BL6J_5011240 | Gm19055 | NCBI_Gene:100418175 | MGI:5011240 | pseudogene | predicted gene, 19055 |
| 3 | gene | 69.81954 | 69.85994 | negative | MGI_C57BL6J_1913433 | Sptssb | NCBI_Gene:66183,ENSEMBL:ENSMUSG00000043461 | MGI:1913433 | protein coding gene | serine palmitoyltransferase, small subunit B |
| 3 | gene | 69.88247 | 69.88411 | positive | MGI_C57BL6J_5594976 | Gm35817 | NCBI_Gene:102639522 | MGI:5594976 | lncRNA gene | predicted gene, 35817 |
| 3 | pseudogene | 69.95481 | 69.95568 | negative | MGI_C57BL6J_5010080 | Gm17895 | NCBI_Gene:100416058,ENSEMBL:ENSMUSG00000103549 | MGI:5010080 | pseudogene | predicted gene, 17895 |
| 3 | gene | 69.96232 | 69.96245 | negative | MGI_C57BL6J_5453261 | Gm23484 | ENSEMBL:ENSMUSG00000077366 | MGI:5453261 | snoRNA gene | predicted gene, 23484 |
| 3 | pseudogene | 69.96328 | 69.96347 | positive | MGI_C57BL6J_5611012 | Gm37784 | ENSEMBL:ENSMUSG00000102525 | MGI:5611012 | pseudogene | predicted gene, 37784 |
| 3 | gene | 70.00761 | 70.02871 | positive | MGI_C57BL6J_2685260 | Otol1 | NCBI_Gene:229389,ENSEMBL:ENSMUSG00000027788 | MGI:2685260 | protein coding gene | otolin 1 |
| 3 | pseudogene | 70.05975 | 70.06047 | positive | MGI_C57BL6J_5610909 | Gm37681 | ENSEMBL:ENSMUSG00000103020 | MGI:5610909 | pseudogene | predicted gene, 37681 |
| 3 | pseudogene | 70.11625 | 70.11653 | negative | MGI_C57BL6J_5610813 | Gm37585 | ENSEMBL:ENSMUSG00000103113 | MGI:5610813 | pseudogene | predicted gene, 37585 |
| 3 | gene | 70.22875 | 70.22887 | negative | MGI_C57BL6J_5453254 | Gm23477 | ENSEMBL:ENSMUSG00000089507 | MGI:5453254 | snoRNA gene | predicted gene, 23477 |
| 3 | pseudogene | 70.41328 | 70.41388 | positive | MGI_C57BL6J_3644892 | Gm6631 | NCBI_Gene:625868,ENSEMBL:ENSMUSG00000102663 | MGI:3644892 | pseudogene | predicted gene 6631 |
| 3 | pseudogene | 70.56626 | 70.56730 | negative | MGI_C57BL6J_5610441 | Gm37213 | ENSEMBL:ENSMUSG00000104023 | MGI:5610441 | pseudogene | predicted gene, 37213 |

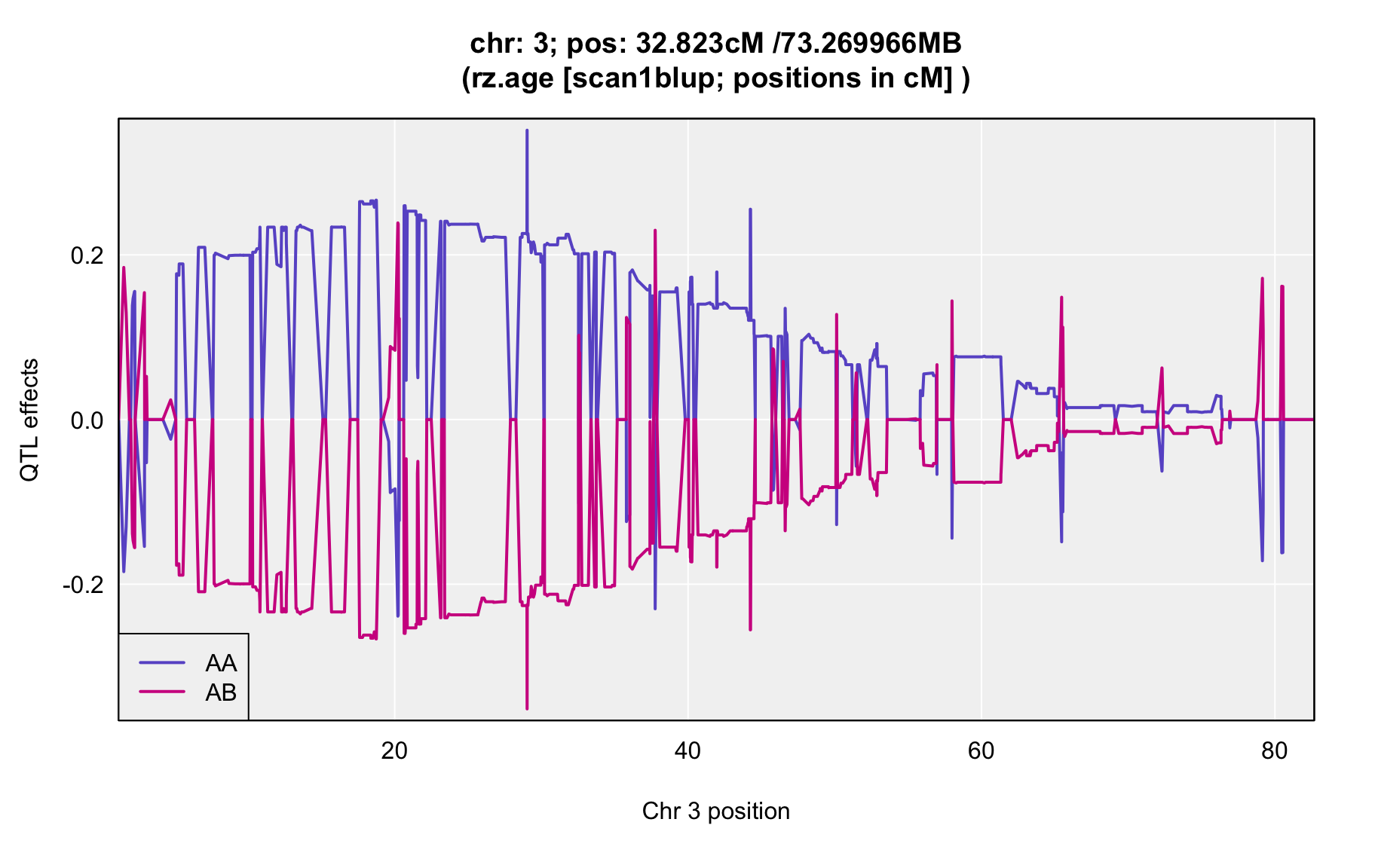

| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | pseudogene | 72.60384 | 72.60435 | negative | MGI_C57BL6J_5611129 | Gm37901 | ENSEMBL:ENSMUSG00000103078 | MGI:5611129 | pseudogene | predicted gene, 37901 |
| 3 | gene | 72.73783 | 72.73798 | negative | MGI_C57BL6J_5452228 | Gm22451 | ENSEMBL:ENSMUSG00000084752 | MGI:5452228 | snRNA gene | predicted gene, 22451 |
| 3 | gene | 72.88855 | 72.96786 | negative | MGI_C57BL6J_1917233 | Sis | NCBI_Gene:69983,ENSEMBL:ENSMUSG00000027790 | MGI:1917233 | protein coding gene | sucrase isomaltase (alpha-glucosidase) |
| 3 | gene | 73.04197 | 73.04384 | negative | MGI_C57BL6J_5610284 | Gm37056 | ENSEMBL:ENSMUSG00000102753 | MGI:5610284 | unclassified gene | predicted gene, 37056 |
| 3 | gene | 73.04725 | 73.05788 | negative | MGI_C57BL6J_2679447 | Slitrk3 | NCBI_Gene:386750,ENSEMBL:ENSMUSG00000048304 | MGI:2679447 | protein coding gene | SLIT and NTRK-like family, member 3 |
| 3 | gene | 73.06632 | 73.59483 | positive | MGI_C57BL6J_5434110 | Gm20754 | NCBI_Gene:626082,ENSEMBL:ENSMUSG00000103428 | MGI:5434110 | lncRNA gene | predicted gene, 20754 |
| 3 | gene | 73.14685 | 73.14696 | positive | MGI_C57BL6J_5454321 | Gm24544 | ENSEMBL:ENSMUSG00000077050 | MGI:5454321 | miRNA gene | predicted gene, 24544 |
| 3 | gene | 73.16409 | 73.16621 | positive | MGI_C57BL6J_5610953 | Gm37725 | ENSEMBL:ENSMUSG00000102916 | MGI:5610953 | unclassified gene | predicted gene, 37725 |
| 3 | gene | 73.20770 | 73.20800 | negative | MGI_C57BL6J_5452315 | Gm22538 | ENSEMBL:ENSMUSG00000088794 | MGI:5452315 | unclassified non-coding RNA gene | predicted gene, 22538 |
| 3 | gene | 73.32943 | 73.47001 | positive | MGI_C57BL6J_5611581 | Gm38353 | ENSEMBL:ENSMUSG00000102820 | MGI:5611581 | lncRNA gene | predicted gene, 38353 |
| 3 | gene | 73.35121 | 73.48165 | negative | MGI_C57BL6J_1921943 | 4930509J09Rik | NCBI_Gene:74693,ENSEMBL:ENSMUSG00000102362 | MGI:1921943 | lncRNA gene | RIKEN cDNA 4930509J09 gene |
| 3 | gene | 73.63581 | 73.70844 | negative | MGI_C57BL6J_894278 | Bche | NCBI_Gene:12038,ENSEMBL:ENSMUSG00000027792 | MGI:894278 | protein coding gene | butyrylcholinesterase |
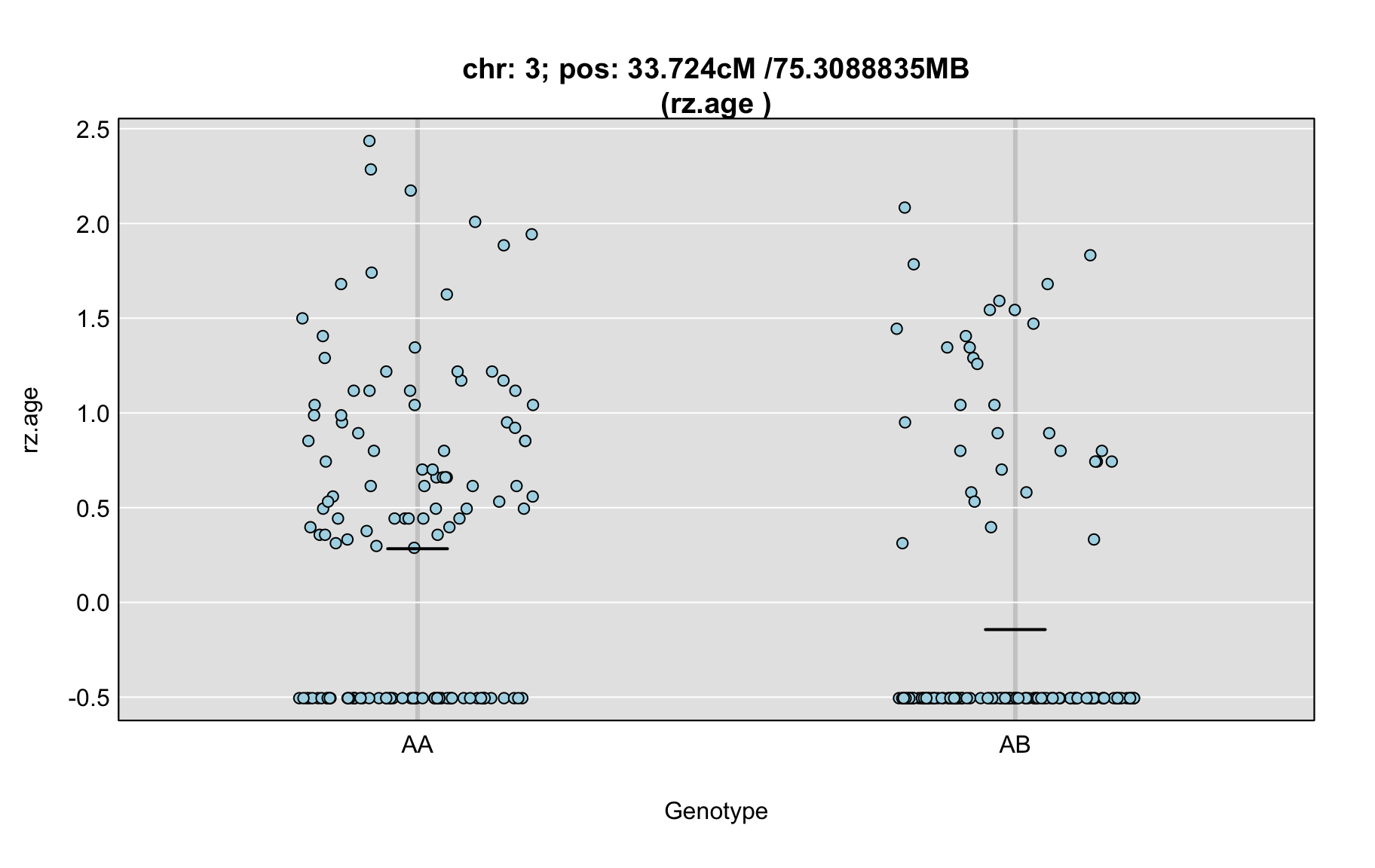
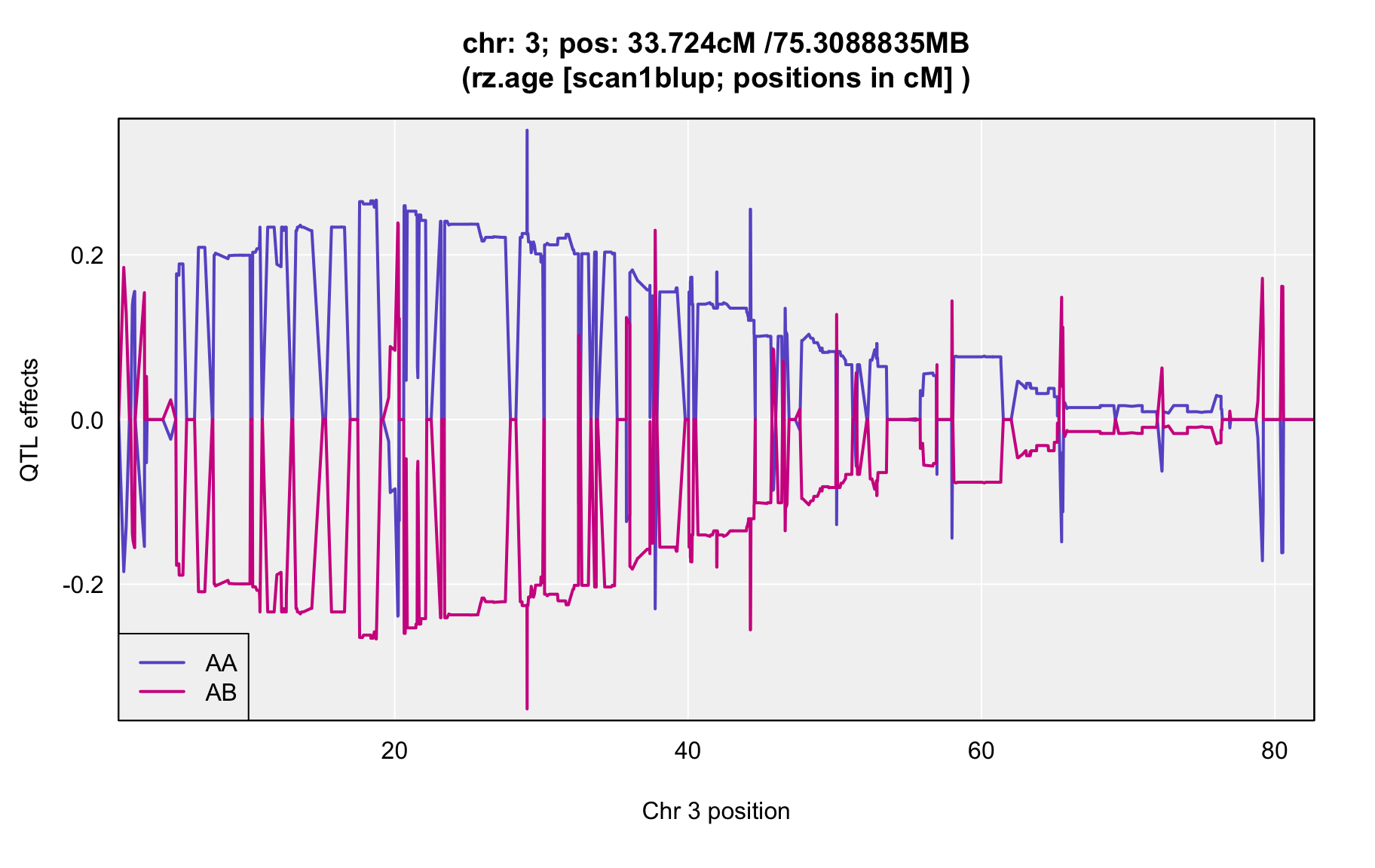
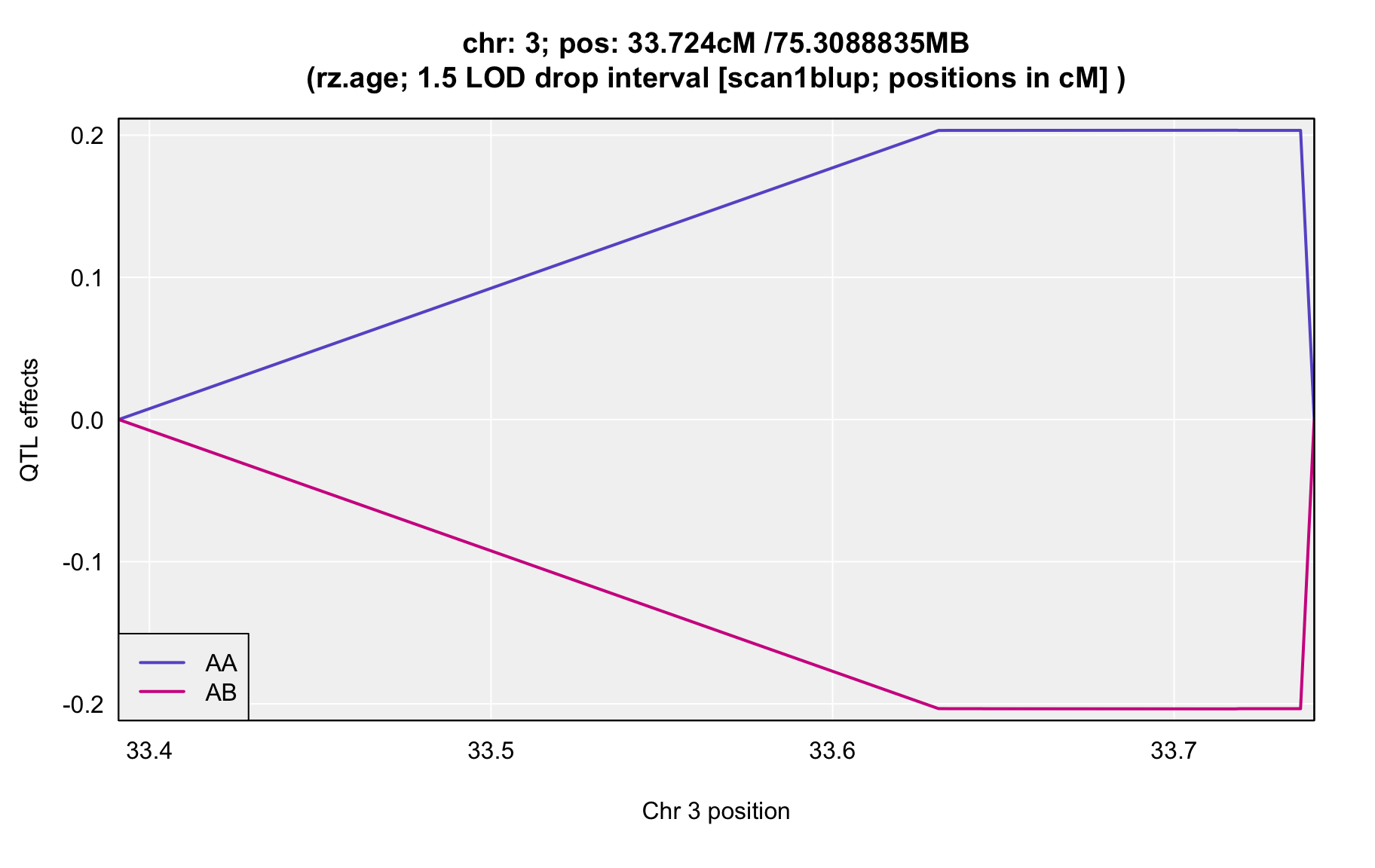
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | pseudogene | 73.93305 | 73.93412 | positive | MGI_C57BL6J_3644686 | Gm8625 | NCBI_Gene:667425,ENSEMBL:ENSMUSG00000102859 | MGI:3644686 | pseudogene | predicted gene 8625 |
| 3 | gene | 74.00922 | 74.01728 | negative | MGI_C57BL6J_5012541 | Gm20356 | NCBI_Gene:100504692,ENSEMBL:ENSMUSG00000104096 | MGI:5012541 | lncRNA gene | predicted gene, 20356 |
| 3 | gene | 74.10541 | 74.12645 | negative | MGI_C57BL6J_5595228 | Gm36069 | NCBI_Gene:102639855 | MGI:5595228 | lncRNA gene | predicted gene, 36069 |
| 3 | pseudogene | 74.24226 | 74.29687 | negative | MGI_C57BL6J_3779552 | Gm6098 | NCBI_Gene:100861784,ENSEMBL:ENSMUSG00000096400 | MGI:3779552 | pseudogene | predicted gene 6098 |
| 3 | pseudogene | 74.32353 | 74.32399 | negative | MGI_C57BL6J_5610278 | Gm37050 | ENSEMBL:ENSMUSG00000104227 | MGI:5610278 | pseudogene | predicted gene, 37050 |
| 3 | pseudogene | 74.81472 | 74.81534 | positive | MGI_C57BL6J_3647458 | Gm8643 | NCBI_Gene:108168917,ENSEMBL:ENSMUSG00000102809 | MGI:3647458 | pseudogene | predicted gene 8643 |
| 3 | gene | 75.02240 | 75.02461 | negative | MGI_C57BL6J_5610692 | Gm37464 | ENSEMBL:ENSMUSG00000102420 | MGI:5610692 | unclassified gene | predicted gene, 37464 |
| 3 | gene | 75.03790 | 75.16505 | negative | MGI_C57BL6J_2674085 | Zbbx | NCBI_Gene:213234,ENSEMBL:ENSMUSG00000034151 | MGI:2674085 | protein coding gene | zinc finger, B-box domain containing |
| 3 | gene | 75.16506 | 75.20808 | positive | MGI_C57BL6J_5622956 | Gm40071 | NCBI_Gene:105244469 | MGI:5622956 | lncRNA gene | predicted gene, 40071 |
| 3 | gene | 75.24236 | 75.27008 | negative | MGI_C57BL6J_1915181 | Serpini2 | NCBI_Gene:67931,ENSEMBL:ENSMUSG00000034139 | MGI:1915181 | protein coding gene | serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| 3 | gene | 75.27393 | 75.48216 | negative | MGI_C57BL6J_3645287 | Wdr49 | NCBI_Gene:213248,ENSEMBL:ENSMUSG00000104301 | MGI:3645287 | protein coding gene | WD repeat domain 49 |
| 3 | pseudogene | 75.39132 | 75.39264 | positive | MGI_C57BL6J_5010021 | Gm17836 | NCBI_Gene:100415953,ENSEMBL:ENSMUSG00000103444 | MGI:5010021 | pseudogene | predicted gene, 17836 |
| 3 | gene | 75.51649 | 75.55686 | negative | MGI_C57BL6J_1928396 | Pdcd10 | NCBI_Gene:56426,ENSEMBL:ENSMUSG00000027835 | MGI:1928396 | protein coding gene | programmed cell death 10 |
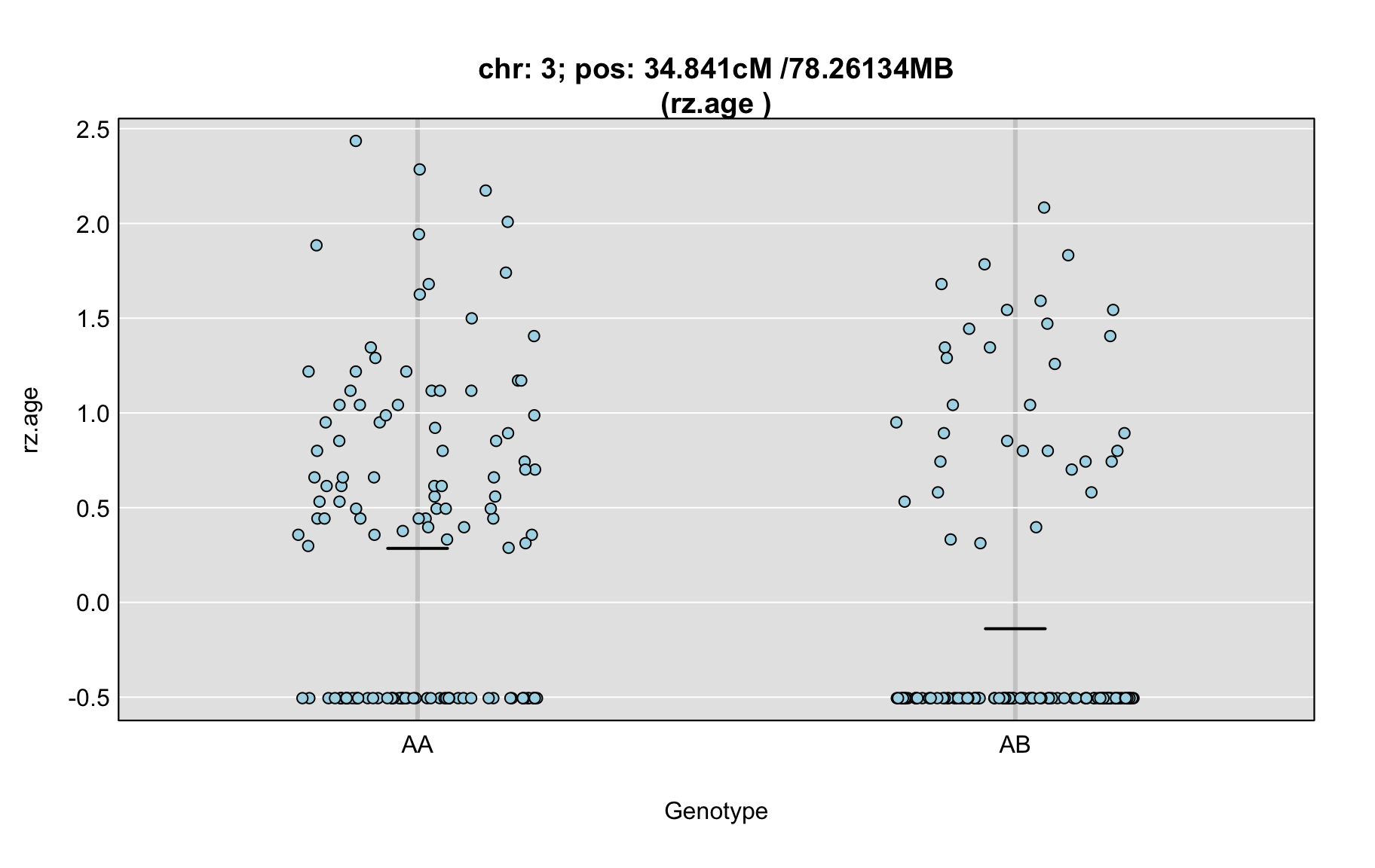
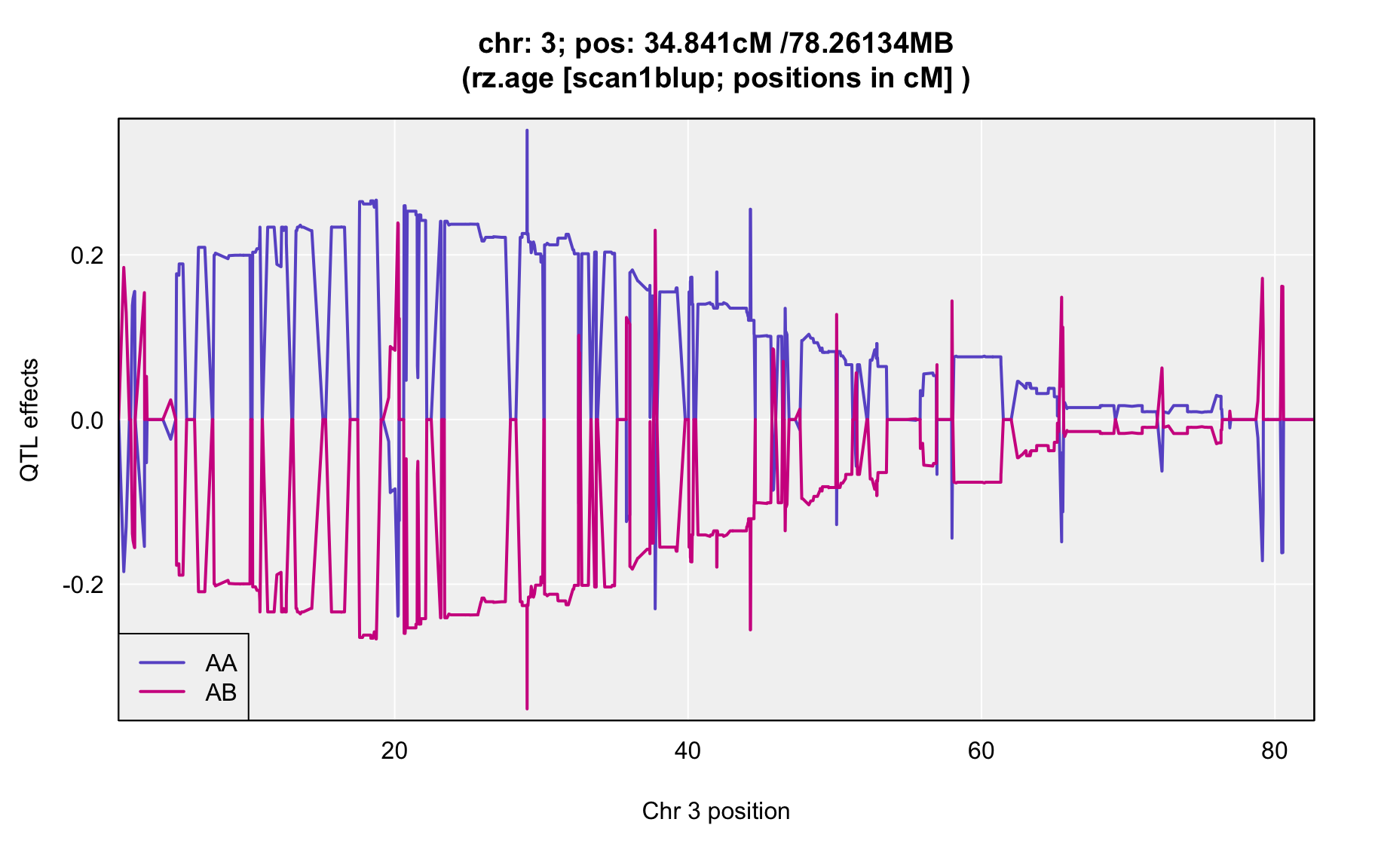

| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | gene | 75.87508 | 75.95695 | negative | MGI_C57BL6J_1920374 | Golim4 | NCBI_Gene:73124,ENSEMBL:ENSMUSG00000034109 | MGI:1920374 | protein coding gene | golgi integral membrane protein 4 |
| 3 | gene | 75.95212 | 75.95430 | positive | MGI_C57BL6J_5610913 | Gm37685 | ENSEMBL:ENSMUSG00000103024 | MGI:5610913 | unclassified gene | predicted gene, 37685 |
| 3 | gene | 75.95610 | 76.00114 | positive | MGI_C57BL6J_5610484 | Gm37256 | ENSEMBL:ENSMUSG00000104448 | MGI:5610484 | lncRNA gene | predicted gene, 37256 |
| 3 | pseudogene | 76.04092 | 76.04172 | negative | MGI_C57BL6J_5010613 | Gm18428 | NCBI_Gene:100417155,ENSEMBL:ENSMUSG00000104223 | MGI:5010613 | pseudogene | predicted gene, 18428 |
| 3 | gene | 76.07427 | 76.71002 | positive | MGI_C57BL6J_2442179 | Fstl5 | NCBI_Gene:213262,ENSEMBL:ENSMUSG00000034098 | MGI:2442179 | protein coding gene | follistatin-like 5 |
| 3 | gene | 76.54669 | 76.55025 | positive | MGI_C57BL6J_5611125 | Gm37897 | ENSEMBL:ENSMUSG00000103920 | MGI:5611125 | unclassified gene | predicted gene, 37897 |
| 3 | pseudogene | 76.82206 | 76.82240 | positive | MGI_C57BL6J_5610518 | Gm37290 | ENSEMBL:ENSMUSG00000103702 | MGI:5610518 | pseudogene | predicted gene, 37290 |
| 3 | pseudogene | 76.88450 | 76.88533 | positive | MGI_C57BL6J_5011416 | Gm19231 | NCBI_Gene:100418471,ENSEMBL:ENSMUSG00000102216 | MGI:5011416 | pseudogene | predicted gene, 19231 |
| 3 | gene | 77.10521 | 77.10528 | negative | MGI_C57BL6J_5453421 | Gm23644 | ENSEMBL:ENSMUSG00000088481 | MGI:5453421 | snoRNA gene | predicted gene, 23644 |
| 3 | pseudogene | 77.56081 | 77.56394 | negative | MGI_C57BL6J_2674154 | Dnmt3a-ps1 | NCBI_Gene:445261,ENSEMBL:ENSMUSG00000102489 | MGI:2674154 | pseudogene | DNA methyltransferase 3A, pseudogene 1 |
| 3 | gene | 77.56496 | 77.57134 | negative | MGI_C57BL6J_5611034 | Gm37806 | ENSEMBL:ENSMUSG00000103849 | MGI:5611034 | unclassified gene | predicted gene, 37806 |
| 3 | pseudogene | 77.69137 | 77.69223 | negative | MGI_C57BL6J_5010480 | Gm18295 | NCBI_Gene:100416866 | MGI:5010480 | pseudogene | predicted gene, 18295 |
| 3 | gene | 77.70289 | 77.70439 | negative | MGI_C57BL6J_5611423 | Gm38195 | ENSEMBL:ENSMUSG00000102872 | MGI:5611423 | unclassified gene | predicted gene, 38195 |
| 3 | pseudogene | 77.87278 | 77.87315 | negative | MGI_C57BL6J_5610804 | Gm37576 | ENSEMBL:ENSMUSG00000102393 | MGI:5610804 | pseudogene | predicted gene, 37576 |
| 3 | pseudogene | 77.96138 | 77.96280 | positive | MGI_C57BL6J_5010904 | Gm18719 | NCBI_Gene:100417620,ENSEMBL:ENSMUSG00000105026 | MGI:5010904 | pseudogene | predicted gene, 18719 |
| 3 | pseudogene | 78.00646 | 78.00698 | positive | MGI_C57BL6J_3648953 | Gm6677 | NCBI_Gene:626461,ENSEMBL:ENSMUSG00000103312 | MGI:3648953 | pseudogene | predicted gene 6677 |
| 3 | pseudogene | 78.08583 | 78.08740 | negative | MGI_C57BL6J_3643677 | Gm8684 | NCBI_Gene:667526,ENSEMBL:ENSMUSG00000102637 | MGI:3643677 | pseudogene | predicted gene 8684 |
| 3 | pseudogene | 78.13446 | 78.13558 | positive | MGI_C57BL6J_3645908 | Gm6680 | NCBI_Gene:626479,ENSEMBL:ENSMUSG00000103817 | MGI:3645908 | pseudogene | predicted gene 6680 |
| 3 | gene | 78.40430 | 78.40594 | negative | MGI_C57BL6J_5663463 | Gm43326 | ENSEMBL:ENSMUSG00000104782 | MGI:5663463 | unclassified gene | predicted gene 43326 |
| 3 | gene | 78.40616 | 78.41077 | negative | MGI_C57BL6J_5595447 | Gm36288 | NCBI_Gene:102640153 | MGI:5595447 | lncRNA gene | predicted gene, 36288 |
| 3 | pseudogene | 78.48658 | 78.48868 | negative | MGI_C57BL6J_3705708 | Gm15442 | NCBI_Gene:100041691,ENSEMBL:ENSMUSG00000084370 | MGI:3705708 | pseudogene | predicted gene 15442 |
| 3 | pseudogene | 78.75862 | 78.75946 | positive | MGI_C57BL6J_5011137 | Gm18952 | NCBI_Gene:100418022,ENSEMBL:ENSMUSG00000102341 | MGI:5011137 | pseudogene | predicted gene, 18952 |
| 3 | gene | 78.78973 | 78.79223 | negative | MGI_C57BL6J_5595500 | Gm36341 | NCBI_Gene:102640221 | MGI:5595500 | lncRNA gene | predicted gene, 36341 |
| 3 | gene | 78.84222 | 78.84257 | positive | MGI_C57BL6J_5454526 | Gm24749 | ENSEMBL:ENSMUSG00000088463 | MGI:5454526 | unclassified non-coding RNA gene | predicted gene, 24749 |
| 3 | pseudogene | 78.86489 | 78.86557 | negative | MGI_C57BL6J_5010138 | Gm17953 | NCBI_Gene:100416167,ENSEMBL:ENSMUSG00000102438 | MGI:5010138 | pseudogene | predicted gene, 17953 |
| 3 | pseudogene | 78.89183 | 78.89247 | negative | MGI_C57BL6J_3646590 | Gm5277 | NCBI_Gene:383883,ENSEMBL:ENSMUSG00000091361 | MGI:3646590 | pseudogene | predicted gene 5277 |
| 3 | pseudogene | 78.91724 | 78.91846 | positive | MGI_C57BL6J_3641638 | Gm10291 | NCBI_Gene:100041748,ENSEMBL:ENSMUSG00000070443 | MGI:3641638 | pseudogene | predicted pseudogene 10291 |
| 3 | gene | 78.91846 | 78.93631 | negative | MGI_C57BL6J_5621427 | Gm38542 | NCBI_Gene:102641233 | MGI:5621427 | lncRNA gene | predicted gene, 38542 |
| 3 | pseudogene | 78.96549 | 78.96715 | negative | MGI_C57BL6J_3704220 | Gm9762 | NCBI_Gene:100041766,ENSEMBL:ENSMUSG00000037096 | MGI:3704220 | pseudogene | predicted pseudogene 9762 |
| 3 | gene | 78.97992 | 78.98005 | positive | MGI_C57BL6J_5453217 | Gm23440 | ENSEMBL:ENSMUSG00000077487 | MGI:5453217 | snoRNA gene | predicted gene, 23440 |
| 3 | gene | 79.06251 | 79.28724 | negative | MGI_C57BL6J_2659071 | Rapgef2 | NCBI_Gene:76089,ENSEMBL:ENSMUSG00000062232 | MGI:2659071 | protein coding gene | Rap guanine nucleotide exchange factor (GEF) 2 |
| 3 | gene | 79.19637 | 79.19648 | positive | MGI_C57BL6J_5453927 | Gm24150 | ENSEMBL:ENSMUSG00000064693 | MGI:5453927 | snRNA gene | predicted gene, 24150 |
| 3 | gene | 79.21292 | 79.25391 | positive | MGI_C57BL6J_1918174 | 4921511C10Rik | NCBI_Gene:70924,ENSEMBL:ENSMUSG00000104585 | MGI:1918174 | lncRNA gene | RIKEN cDNA 4921511C10 gene |
| 3 | gene | 79.23228 | 79.23413 | negative | MGI_C57BL6J_2443462 | B930012P20Rik | NA | NA | unclassified gene | RIKEN cDNA B930012P20 gene |
| 3 | gene | 79.28717 | 79.28985 | positive | MGI_C57BL6J_2686494 | 6430573P05Rik | ENSEMBL:ENSMUSG00000102545 | MGI:2686494 | lncRNA gene | RIKEN cDNA 6430573P05 gene |
| 3 | gene | 79.33420 | 79.33658 | negative | MGI_C57BL6J_5622957 | Gm40072 | NCBI_Gene:105244470 | MGI:5622957 | lncRNA gene | predicted gene, 40072 |
| 3 | gene | 79.33666 | 79.46413 | positive | MGI_C57BL6J_4936993 | Gm17359 | NCBI_Gene:100233207,ENSEMBL:ENSMUSG00000091685 | MGI:4936993 | protein coding gene | predicted gene, 17359 |
| 3 | gene | 79.45597 | 79.56780 | negative | MGI_C57BL6J_2683054 | Fnip2 | NCBI_Gene:329679,ENSEMBL:ENSMUSG00000061175 | MGI:2683054 | protein coding gene | folliculin interacting protein 2 |
| 3 | gene | 79.50979 | 79.51210 | negative | MGI_C57BL6J_5595601 | Gm36442 | NCBI_Gene:102640362 | MGI:5595601 | lncRNA gene | predicted gene, 36442 |
| 3 | gene | 79.55131 | 79.55219 | negative | MGI_C57BL6J_3781690 | Gm3513 | ENSEMBL:ENSMUSG00000089746 | MGI:3781690 | lncRNA gene | predicted gene 3513 |
| 3 | gene | 79.56819 | 79.57332 | positive | MGI_C57BL6J_1923129 | 4930589L23Rik | NCBI_Gene:75879,ENSEMBL:ENSMUSG00000068957 | MGI:1923129 | lncRNA gene | RIKEN cDNA 4930589L23 gene |
| 3 | pseudogene | 79.58259 | 79.58323 | positive | MGI_C57BL6J_5594226 | Gm35067 | ENSEMBL:ENSMUSG00000103564 | MGI:5594226 | pseudogene | predicted gene, 35067 |
| 3 | gene | 79.59134 | 79.60365 | positive | MGI_C57BL6J_1914988 | Ppid | NCBI_Gene:67738,ENSEMBL:ENSMUSG00000027804 | MGI:1914988 | protein coding gene | peptidylprolyl isomerase D (cyclophilin D) |
| 3 | gene | 79.60379 | 79.62950 | negative | MGI_C57BL6J_106100 | Etfdh | NCBI_Gene:66841,ENSEMBL:ENSMUSG00000027809 | MGI:106100 | protein coding gene | electron transferring flavoprotein, dehydrogenase |
| 3 | gene | 79.61054 | 79.61183 | negative | MGI_C57BL6J_5611201 | Gm37973 | ENSEMBL:ENSMUSG00000102370 | MGI:5611201 | unclassified gene | predicted gene, 37973 |
| 3 | gene | 79.62908 | 79.63282 | positive | MGI_C57BL6J_1923189 | 4930579G24Rik | NCBI_Gene:75939,ENSEMBL:ENSMUSG00000027811 | MGI:1923189 | protein coding gene | RIKEN cDNA 4930579G24 gene |
| 3 | gene | 79.64160 | 79.73795 | negative | MGI_C57BL6J_2682211 | Rxfp1 | NCBI_Gene:381489,ENSEMBL:ENSMUSG00000034009 | MGI:2682211 | protein coding gene | relaxin/insulin-like family peptide receptor 1 |
| 3 | gene | 79.67337 | 79.67688 | negative | MGI_C57BL6J_5504099 | Gm26984 | ENSEMBL:ENSMUSG00000098204 | MGI:5504099 | lncRNA gene | predicted gene, 26984 |
| 3 | gene | 79.81256 | 79.85279 | negative | MGI_C57BL6J_1917902 | Tmem144 | NCBI_Gene:70652,ENSEMBL:ENSMUSG00000027956 | MGI:1917902 | protein coding gene | transmembrane protein 144 |
| 3 | gene | 79.83493 | 79.83504 | positive | MGI_C57BL6J_5456197 | Gm26420 | ENSEMBL:ENSMUSG00000077461 | MGI:5456197 | snRNA gene | predicted gene, 26420 |
| 3 | gene | 79.85287 | 79.85666 | positive | MGI_C57BL6J_5622958 | Gm40073 | NCBI_Gene:105244471 | MGI:5622958 | lncRNA gene | predicted gene, 40073 |
| 3 | gene | 79.85929 | 79.88681 | negative | MGI_C57BL6J_5595728 | Gm36569 | NCBI_Gene:102640530,ENSEMBL:ENSMUSG00000102191 | MGI:5595728 | lncRNA gene | predicted gene, 36569 |
| 3 | gene | 79.88447 | 79.94628 | positive | MGI_C57BL6J_1915909 | Gask1b | NCBI_Gene:68659,ENSEMBL:ENSMUSG00000027955 | MGI:1915909 | protein coding gene | golgi associated kinase 1B |
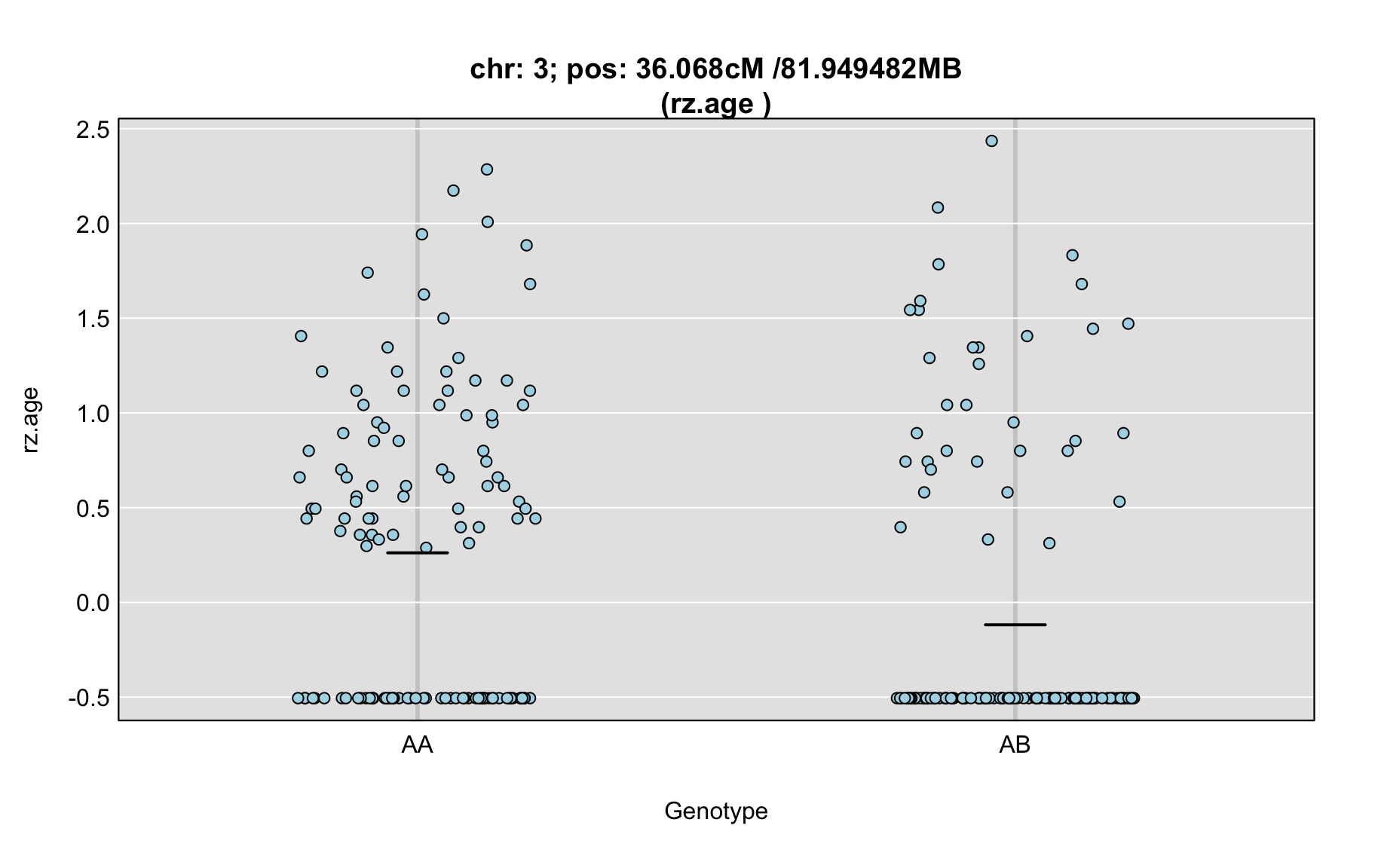

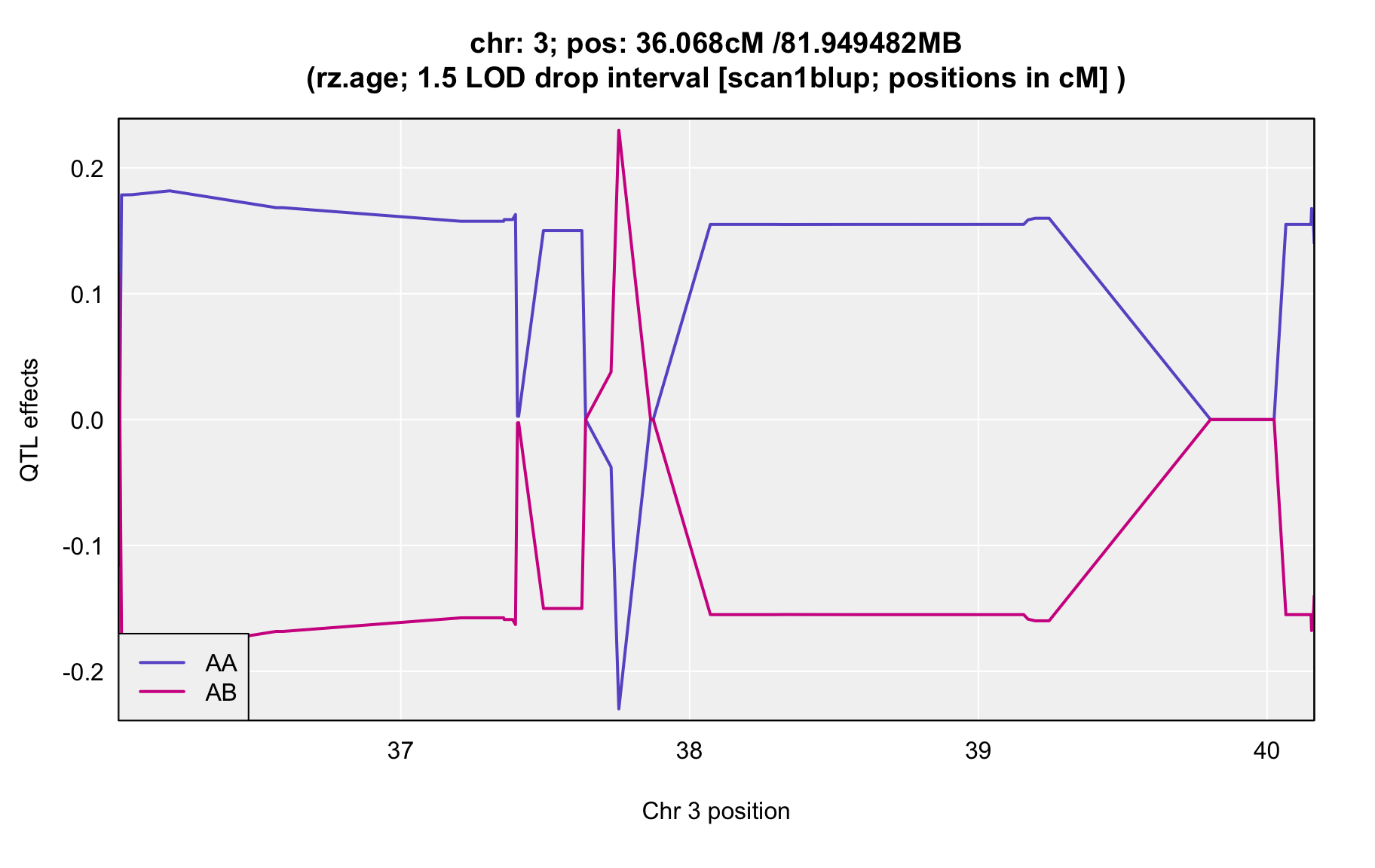
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | gene | 81.88073 | 81.88087 | positive | MGI_C57BL6J_5452308 | Gm22531 | ENSEMBL:ENSMUSG00000089444 | MGI:5452308 | snoRNA gene | predicted gene, 22531 |
| 3 | gene | 81.91659 | 81.92467 | negative | MGI_C57BL6J_3642361 | Gm9989 | NCBI_Gene:102640985,ENSEMBL:ENSMUSG00000056023 | MGI:3642361 | lncRNA gene | predicted gene 9989 |
| 3 | gene | 81.93260 | 81.95673 | positive | MGI_C57BL6J_2139628 | Ctso | NCBI_Gene:229445,ENSEMBL:ENSMUSG00000028015 | MGI:2139628 | protein coding gene | cathepsin O |
| 3 | gene | 81.95709 | 81.97620 | negative | MGI_C57BL6J_1928486 | Tdo2 | NCBI_Gene:56720,ENSEMBL:ENSMUSG00000028011 | MGI:1928486 | protein coding gene | tryptophan 2,3-dioxygenase |
| 3 | gene | 81.98229 | 82.02123 | positive | MGI_C57BL6J_1929259 | Asic5 | NCBI_Gene:58170,ENSEMBL:ENSMUSG00000028008 | MGI:1929259 | protein coding gene | acid-sensing (proton-gated) ion channel family member 5 |
| 3 | gene | 82.03200 | 82.07471 | negative | MGI_C57BL6J_1860604 | Gucy1b1 | NCBI_Gene:54195,ENSEMBL:ENSMUSG00000028005 | MGI:1860604 | protein coding gene | guanylate cyclase 1, soluble, beta 1 |
| 3 | gene | 82.09243 | 82.14588 | negative | MGI_C57BL6J_1926562 | Gucy1a1 | NCBI_Gene:60596,ENSEMBL:ENSMUSG00000033910 | MGI:1926562 | protein coding gene | guanylate cyclase 1, soluble, alpha 1 |
| 3 | gene | 82.10625 | 82.10631 | negative | MGI_C57BL6J_5531411 | Mir7010 | miRBase:MI0022859,NCBI_Gene:102466991,ENSEMBL:ENSMUSG00000098712 | MGI:5531411 | miRNA gene | microRNA 7010 |
| 3 | gene | 82.14640 | 82.23680 | negative | MGI_C57BL6J_5589080 | Gm29921 | NCBI_Gene:102631626 | MGI:5589080 | lncRNA gene | predicted gene, 29921 |
| 3 | gene | 82.35803 | 82.39527 | positive | MGI_C57BL6J_2442208 | Map9 | NCBI_Gene:213582,ENSEMBL:ENSMUSG00000033900 | MGI:2442208 | protein coding gene | microtubule-associated protein 9 |
| 3 | gene | 82.53838 | 82.54899 | negative | MGI_C57BL6J_108418 | Npy2r | NCBI_Gene:18167,ENSEMBL:ENSMUSG00000028004 | MGI:108418 | protein coding gene | neuropeptide Y receptor Y2 |
| 3 | pseudogene | 82.57740 | 82.57809 | positive | MGI_C57BL6J_5594482 | Gm35323 | NCBI_Gene:102638861,ENSEMBL:ENSMUSG00000102514 | MGI:5594482 | pseudogene | predicted gene, 35323 |
| 3 | gene | 82.66038 | 82.66490 | negative | MGI_C57BL6J_5589158 | Gm29999 | NCBI_Gene:102631733,ENSEMBL:ENSMUSG00000102629 | MGI:5589158 | lncRNA gene | predicted gene, 29999 |
| 3 | pseudogene | 82.67199 | 82.67257 | negative | MGI_C57BL6J_5663484 | Gm43347 | ENSEMBL:ENSMUSG00000104772 | MGI:5663484 | pseudogene | predicted gene 43347 |
| 3 | gene | 82.75452 | 82.75986 | positive | MGI_C57BL6J_5589202 | Gm30043 | ENSEMBL:ENSMUSG00000104898 | MGI:5589202 | lncRNA gene | predicted gene, 30043 |
| 3 | gene | 82.79366 | 82.79390 | negative | MGI_C57BL6J_5451815 | Gm22038 | ENSEMBL:ENSMUSG00000087902 | MGI:5451815 | unclassified non-coding RNA gene | predicted gene, 22038 |
| 3 | gene | 82.79497 | 82.81223 | negative | MGI_C57BL6J_5663485 | Gm43348 | ENSEMBL:ENSMUSG00000105034 | MGI:5663485 | lncRNA gene | predicted gene 43348 |
| 3 | gene | 82.83652 | 82.87648 | negative | MGI_C57BL6J_3645057 | Rbm46 | NCBI_Gene:633285,ENSEMBL:ENSMUSG00000033882 | MGI:3645057 | protein coding gene | RNA binding motif protein 46 |
| 3 | gene | 82.87671 | 82.94364 | positive | MGI_C57BL6J_1925462 | Rbm46os | NCBI_Gene:78212,ENSEMBL:ENSMUSG00000086273 | MGI:1925462 | antisense lncRNA gene | RNA binding motif protein 46, opposite strand |
| 3 | gene | 82.89258 | 82.90397 | negative | MGI_C57BL6J_1891259 | Lrat | NCBI_Gene:79235,ENSEMBL:ENSMUSG00000028003 | MGI:1891259 | protein coding gene | lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase) |
| 3 | gene | 82.92666 | 83.00849 | negative | MGI_C57BL6J_5589256 | Gm30097 | NCBI_Gene:102631873,ENSEMBL:ENSMUSG00000102595 | MGI:5589256 | lncRNA gene | predicted gene, 30097 |
| 3 | gene | 83.00772 | 83.01506 | positive | MGI_C57BL6J_95526 | Fgg | NCBI_Gene:99571,ENSEMBL:ENSMUSG00000033860 | MGI:95526 | protein coding gene | fibrinogen gamma chain |
| 3 | gene | 83.02608 | 83.03363 | positive | MGI_C57BL6J_1316726 | Fga | NCBI_Gene:14161,ENSEMBL:ENSMUSG00000028001 | MGI:1316726 | protein coding gene | fibrinogen alpha chain |
| 3 | gene | 83.03708 | 83.03897 | negative | MGI_C57BL6J_5622960 | Gm40075 | NCBI_Gene:105244473 | MGI:5622960 | lncRNA gene | predicted gene, 40075 |
| 3 | gene | 83.04014 | 83.04986 | negative | MGI_C57BL6J_99501 | Fgb | NCBI_Gene:110135,ENSEMBL:ENSMUSG00000033831 | MGI:99501 | protein coding gene | fibrinogen beta chain |
| 3 | gene | 83.05552 | 83.07236 | positive | MGI_C57BL6J_1858197 | Plrg1 | NCBI_Gene:53317,ENSEMBL:ENSMUSG00000027998 | MGI:1858197 | protein coding gene | pleiotropic regulator 1 |
| 3 | gene | 83.12528 | 83.12925 | negative | MGI_C57BL6J_3642896 | Gm10710 | NCBI_Gene:100038599,ENSEMBL:ENSMUSG00000074517 | MGI:3642896 | lncRNA gene | predicted gene 10710 |
| 3 | gene | 83.12795 | 83.35721 | positive | MGI_C57BL6J_2685263 | Dchs2 | NCBI_Gene:100534287,ENSEMBL:ENSMUSG00000102692 | MGI:2685263 | protein coding gene | dachsous cadherin related 2 |
| 3 | gene | 83.40340 | 83.40579 | positive | MGI_C57BL6J_5611324 | Gm38096 | ENSEMBL:ENSMUSG00000103459 | MGI:5611324 | lncRNA gene | predicted gene, 38096 |
| 3 | gene | 83.55536 | 83.57412 | negative | MGI_C57BL6J_1916710 | 1700028M03Rik | NCBI_Gene:69460,ENSEMBL:ENSMUSG00000101334 | MGI:1916710 | lncRNA gene | RIKEN cDNA 1700028M03 gene |
| 3 | gene | 83.59633 | 83.60109 | negative | MGI_C57BL6J_5622961 | Gm40076 | NCBI_Gene:105244474 | MGI:5622961 | lncRNA gene | predicted gene, 40076 |
| 3 | gene | 83.63458 | 83.63487 | negative | MGI_C57BL6J_5452119 | Gm22342 | ENSEMBL:ENSMUSG00000084596 | MGI:5452119 | unclassified non-coding RNA gene | predicted gene, 22342 |
| 3 | pseudogene | 83.68668 | 83.68727 | positive | MGI_C57BL6J_3781837 | Rpl28-ps2 | NCBI_Gene:100042091,ENSEMBL:ENSMUSG00000102137 | MGI:3781837 | pseudogene | ribosomal protein L28, pseudogene 2 |
| 3 | gene | 83.70225 | 83.70236 | positive | MGI_C57BL6J_5454361 | Gm24584 | ENSEMBL:ENSMUSG00000088597 | MGI:5454361 | snRNA gene | predicted gene, 24584 |
| 3 | gene | 83.71936 | 83.72737 | negative | MGI_C57BL6J_5589452 | Gm30293 | NCBI_Gene:102632140 | MGI:5589452 | lncRNA gene | predicted gene, 30293 |
| 3 | gene | 83.76616 | 83.77432 | positive | MGI_C57BL6J_108078 | Sfrp2 | NCBI_Gene:20319,ENSEMBL:ENSMUSG00000027996 | MGI:108078 | protein coding gene | secreted frizzled-related protein 2 |
| 3 | gene | 83.77385 | 83.78996 | negative | MGI_C57BL6J_5477265 | Gm26771 | ENSEMBL:ENSMUSG00000097250 | MGI:5477265 | lncRNA gene | predicted gene, 26771 |
| 3 | pseudogene | 83.78973 | 83.83222 | positive | MGI_C57BL6J_3646388 | Gm7115 | NCBI_Gene:633522,ENSEMBL:ENSMUSG00000104082 | MGI:3646388 | pseudogene | predicted gene 7115 |
| 3 | gene | 83.83627 | 83.84182 | negative | MGI_C57BL6J_1346060 | Tlr2 | NCBI_Gene:24088,ENSEMBL:ENSMUSG00000027995 | MGI:1346060 | protein coding gene | toll-like receptor 2 |
| 3 | gene | 83.89766 | 84.04018 | negative | MGI_C57BL6J_2443399 | Tmem131l | NCBI_Gene:229473,ENSEMBL:ENSMUSG00000033767 | MGI:2443399 | protein coding gene | transmembrane 131 like |
| 3 | gene | 84.02844 | 84.02928 | positive | MGI_C57BL6J_1922302 | 4930509B17Rik | NA | NA | unclassified gene | RIKEN cDNA 4930509B17 gene |
| 3 | gene | 84.04022 | 84.05210 | positive | MGI_C57BL6J_5589511 | Gm30352 | NCBI_Gene:102632212 | MGI:5589511 | lncRNA gene | predicted gene, 30352 |
| 3 | gene | 84.05897 | 84.06345 | negative | MGI_C57BL6J_5610209 | Gm36981 | ENSEMBL:ENSMUSG00000103261 | MGI:5610209 | unclassified non-coding RNA gene | predicted gene, 36981 |
| 3 | gene | 84.06174 | 84.06277 | positive | MGI_C57BL6J_5589617 | Gm30458 | NCBI_Gene:102632366 | MGI:5589617 | lncRNA gene | predicted gene, 30458 |
| 3 | gene | 84.08793 | 84.15579 | negative | MGI_C57BL6J_1924165 | Mnd1 | NCBI_Gene:76915,ENSEMBL:ENSMUSG00000033752 | MGI:1924165 | protein coding gene | meiotic nuclear divisions 1 |
| 3 | gene | 84.16044 | 84.30688 | negative | MGI_C57BL6J_1933163 | Trim2 | NCBI_Gene:80890,ENSEMBL:ENSMUSG00000027993 | MGI:1933163 | protein coding gene | tripartite motif-containing 2 |
| 3 | gene | 84.17410 | 84.17519 | positive | MGI_C57BL6J_3648757 | Gm6525 | NCBI_Gene:624713,ENSEMBL:ENSMUSG00000104043 | MGI:3648757 | protein coding gene | predicted pseudogene 6525 |
| 3 | gene | 84.22869 | 84.24704 | positive | MGI_C57BL6J_5622963 | Gm40078 | NCBI_Gene:105244477 | MGI:5622963 | lncRNA gene | predicted gene, 40078 |
| 3 | gene | 84.22901 | 84.22908 | negative | MGI_C57BL6J_4413878 | n-TGgcc9 | NCBI_Gene:102467313 | MGI:4413878 | tRNA gene | nuclear encoded tRNA glycine 9 (anticodon GCC) |
| 3 | gene | 84.30298 | 84.30542 | negative | MGI_C57BL6J_2443303 | C630001G18Rik | NA | NA | unclassified gene | RIKEN cDNA C630001G18 gene |
| 3 | gene | 84.34938 | 84.37992 | negative | MGI_C57BL6J_1922559 | 4930565D16Rik | NCBI_Gene:75309,ENSEMBL:ENSMUSG00000104004 | MGI:1922559 | lncRNA gene | RIKEN cDNA 4930565D16 gene |
| 3 | gene | 84.44219 | 84.48044 | negative | MGI_C57BL6J_2684972 | Fhdc1 | NCBI_Gene:229474,ENSEMBL:ENSMUSG00000041842 | MGI:2684972 | protein coding gene | FH2 domain containing 1 |
| 3 | gene | 84.49609 | 84.58283 | negative | MGI_C57BL6J_1277120 | Arfip1 | NCBI_Gene:99889,ENSEMBL:ENSMUSG00000074513 | MGI:1277120 | protein coding gene | ADP-ribosylation factor interacting protein 1 |
| 3 | gene | 84.49643 | 85.88752 | negative | MGI_C57BL6J_5610468 | Gm37240 | ENSEMBL:ENSMUSG00000102805 | MGI:5610468 | protein coding gene | predicted gene, 37240 |
| 3 | pseudogene | 84.55266 | 84.55348 | negative | MGI_C57BL6J_3645282 | Gm8813 | NCBI_Gene:667788,ENSEMBL:ENSMUSG00000085625 | MGI:3645282 | pseudogene | predicted gene 8813 |
| 3 | pseudogene | 84.55689 | 84.55779 | negative | MGI_C57BL6J_5611469 | Gm38241 | ENSEMBL:ENSMUSG00000103741 | MGI:5611469 | pseudogene | predicted gene, 38241 |
| 3 | gene | 84.58269 | 84.58510 | positive | MGI_C57BL6J_5594569 | Gm35410 | NA | NA | protein coding gene | predicted gene%2c 35410 |
| 3 | gene | 84.59357 | 84.59703 | positive | MGI_C57BL6J_2685264 | Tigd4 | NCBI_Gene:403175,ENSEMBL:ENSMUSG00000047819 | MGI:2685264 | protein coding gene | tigger transposable element derived 4 |
| 3 | gene | 84.60406 | 84.61617 | positive | MGI_C57BL6J_5826473 | Gm46836 | NCBI_Gene:108168918 | MGI:5826473 | lncRNA gene | predicted gene, 46836 |
| 3 | gene | 84.66619 | 84.70458 | positive | MGI_C57BL6J_2444725 | Tmem154 | NCBI_Gene:320782,ENSEMBL:ENSMUSG00000056498 | MGI:2444725 | protein coding gene | transmembrane protein 154 |
| 3 | gene | 84.74497 | 84.79221 | positive | MGI_C57BL6J_5622964 | Gm40079 | NCBI_Gene:105244478 | MGI:5622964 | lncRNA gene | predicted gene, 40079 |
| 3 | gene | 84.80979 | 84.81252 | negative | MGI_C57BL6J_5594645 | Gm35486 | NA | NA | protein coding gene | predicted gene%2c 35486 |
| 3 | gene | 84.81216 | 84.97920 | positive | MGI_C57BL6J_1354695 | Fbxw7 | NCBI_Gene:50754,ENSEMBL:ENSMUSG00000028086 | MGI:1354695 | protein coding gene | F-box and WD-40 domain protein 7 |
| 3 | gene | 84.84197 | 84.88484 | negative | MGI_C57BL6J_5622965 | Gm40080 | NCBI_Gene:105244479 | MGI:5622965 | lncRNA gene | predicted gene, 40080 |
| 3 | gene | 84.87389 | 84.87778 | positive | MGI_C57BL6J_5610954 | Gm37726 | ENSEMBL:ENSMUSG00000102919 | MGI:5610954 | unclassified gene | predicted gene, 37726 |
| 3 | pseudogene | 84.91290 | 84.91343 | negative | MGI_C57BL6J_5826445 | Gm46808 | NCBI_Gene:108168875 | MGI:5826445 | pseudogene | predicted gene, 46808 |
| 3 | gene | 84.96506 | 84.96558 | negative | MGI_C57BL6J_3614654 | Fbxw7as1 | NCBI_Gene:654362 | MGI:3614654 | antisense lncRNA gene | F-box and WD-40 domain protein 7 antisense transcript 1 |
| 3 | gene | 85.02169 | 85.02857 | positive | MGI_C57BL6J_5590103 | Gm30944 | NCBI_Gene:102633014 | MGI:5590103 | lncRNA gene | predicted gene, 30944 |
| 3 | gene | 85.04914 | 85.10790 | positive | MGI_C57BL6J_5590150 | Gm30991 | NCBI_Gene:102633077 | MGI:5590150 | lncRNA gene | predicted gene, 30991 |
| 3 | gene | 85.11498 | 85.12251 | negative | MGI_C57BL6J_5590255 | Gm31096 | NCBI_Gene:102633209 | MGI:5590255 | lncRNA gene | predicted gene, 31096 |
| 3 | pseudogene | 85.15204 | 85.15270 | negative | MGI_C57BL6J_5611269 | Gm38041 | ENSEMBL:ENSMUSG00000104290 | MGI:5611269 | pseudogene | predicted gene, 38041 |
| 3 | gene | 85.19518 | 85.19645 | negative | MGI_C57BL6J_5611541 | Gm38313 | ENSEMBL:ENSMUSG00000102703 | MGI:5611541 | unclassified gene | predicted gene, 38313 |
| 3 | gene | 85.31752 | 85.33203 | positive | MGI_C57BL6J_1920533 | 1700036G14Rik | NCBI_Gene:73283,ENSEMBL:ENSMUSG00000099995 | MGI:1920533 | lncRNA gene | RIKEN cDNA 1700036G14 gene |
| 3 | gene | 85.34212 | 85.34341 | positive | MGI_C57BL6J_5590369 | Gm31210 | NCBI_Gene:102633368 | MGI:5590369 | lncRNA gene | predicted gene, 31210 |
| 3 | gene | 85.39154 | 85.40210 | negative | MGI_C57BL6J_5590518 | Gm31359 | NCBI_Gene:102633563 | MGI:5590518 | lncRNA gene | predicted gene, 31359 |
| 3 | gene | 85.39752 | 85.42813 | negative | MGI_C57BL6J_5662950 | Gm42813 | ENSEMBL:ENSMUSG00000105012 | MGI:5662950 | lncRNA gene | predicted gene 42813 |
| 3 | gene | 85.40563 | 85.41035 | positive | MGI_C57BL6J_5590463 | Gm31304 | NCBI_Gene:102633491 | MGI:5590463 | lncRNA gene | predicted gene, 31304 |
| 3 | gene | 85.42727 | 85.49180 | negative | MGI_C57BL6J_5590578 | Gm31419 | NCBI_Gene:102633638 | MGI:5590578 | lncRNA gene | predicted gene, 31419 |
| 3 | gene | 85.43519 | 85.46613 | negative | MGI_C57BL6J_5804905 | Gm45790 | ENSEMBL:ENSMUSG00000110441 | MGI:5804905 | lncRNA gene | predicted gene 45790 |
| 3 | gene | 85.46738 | 85.46750 | positive | MGI_C57BL6J_3783377 | Mir466k | miRBase:MI0006292,NCBI_Gene:100316670,ENSEMBL:ENSMUSG00000080424 | MGI:3783377 | miRNA gene | microRNA 466k |
| 3 | gene | 85.52989 | 85.57399 | negative | MGI_C57BL6J_3781915 | Gm3740 | NCBI_Gene:100042237 | MGI:3781915 | unclassified gene | predicted gene 3740 |
| 3 | gene | 85.54432 | 85.55339 | negative | MGI_C57BL6J_5662949 | Gm42812 | ENSEMBL:ENSMUSG00000106422 | MGI:5662949 | lncRNA gene | predicted gene 42812 |
| 3 | gene | 85.55605 | 85.57394 | negative | MGI_C57BL6J_3782983 | Gm15535 | ENSEMBL:ENSMUSG00000085527 | MGI:3782983 | lncRNA gene | predicted gene 15535 |
| 3 | gene | 85.57410 | 85.65562 | positive | MGI_C57BL6J_2442496 | Gatb | NCBI_Gene:229487,ENSEMBL:ENSMUSG00000028085 | MGI:2442496 | protein coding gene | glutamyl-tRNA(Gln) amidotransferase, subunit B |
| 3 | gene | 85.57751 | 85.58983 | positive | MGI_C57BL6J_5826446 | Gm46809 | NCBI_Gene:108168876 | MGI:5826446 | lncRNA gene | predicted gene, 46809 |
| 3 | gene | 85.66006 | 85.81729 | negative | MGI_C57BL6J_2444746 | Fam160a1 | NCBI_Gene:229488,ENSEMBL:ENSMUSG00000051000 | MGI:2444746 | protein coding gene | family with sequence similarity 160, member A1 |
| 3 | gene | 85.71273 | 85.71776 | positive | MGI_C57BL6J_5622967 | Gm40082 | NCBI_Gene:105244481 | MGI:5622967 | lncRNA gene | predicted gene, 40082 |
| 3 | pseudogene | 85.78523 | 85.78600 | positive | MGI_C57BL6J_5011253 | Gm19068 | NCBI_Gene:100418195 | MGI:5011253 | pseudogene | predicted gene, 19068 |
| 3 | gene | 85.86985 | 85.88752 | negative | MGI_C57BL6J_2443773 | Glt28d2 | NCBI_Gene:320302,ENSEMBL:ENSMUSG00000031286 | MGI:2443773 | protein coding gene | glycosyltransferase 28 domain containing 2 |
| 3 | gene | 85.88769 | 85.88923 | positive | MGI_C57BL6J_1918440 | 4933425M03Rik | ENSEMBL:ENSMUSG00000106508 | MGI:1918440 | unclassified gene | RIKEN cDNA 4933425M03 gene |
| 3 | gene | 85.90835 | 85.90846 | negative | MGI_C57BL6J_5455981 | Gm26204 | ENSEMBL:ENSMUSG00000064840 | MGI:5455981 | snRNA gene | predicted gene, 26204 |
| 3 | pseudogene | 85.91464 | 85.91512 | positive | MGI_C57BL6J_5611258 | Gm38030 | ENSEMBL:ENSMUSG00000103993 | MGI:5611258 | pseudogene | predicted gene, 38030 |
| 3 | pseudogene | 85.91571 | 85.91617 | positive | MGI_C57BL6J_3704221 | Gm9790 | NCBI_Gene:100042265,ENSEMBL:ENSMUSG00000044330 | MGI:3704221 | pseudogene | predicted gene 9790 |
| 3 | gene | 85.95089 | 85.95101 | positive | MGI_C57BL6J_5455653 | Gm25876 | ENSEMBL:ENSMUSG00000077517 | MGI:5455653 | snoRNA gene | predicted gene, 25876 |
| 3 | gene | 85.96277 | 85.96283 | negative | MGI_C57BL6J_5454278 | Gm24501 | ENSEMBL:ENSMUSG00000089077 | MGI:5454278 | snRNA gene | predicted gene, 24501 |
| 3 | gene | 85.96285 | 85.96291 | negative | MGI_C57BL6J_5531193 | Gm27811 | ENSEMBL:ENSMUSG00000098444 | MGI:5531193 | snRNA gene | predicted gene, 27811 |
| 3 | gene | 85.97095 | 86.13053 | positive | MGI_C57BL6J_1350923 | Sh3d19 | NCBI_Gene:27059,ENSEMBL:ENSMUSG00000028082 | MGI:1350923 | protein coding gene | SH3 domain protein D19 |
| 3 | gene | 85.99371 | 86.00912 | negative | MGI_C57BL6J_2685865 | Prss48 | NCBI_Gene:368202,ENSEMBL:ENSMUSG00000049013 | MGI:2685865 | protein coding gene | protease, serine 48 |
| 3 | gene | 86.10053 | 86.10066 | positive | MGI_C57BL6J_5454965 | Gm25188 | ENSEMBL:ENSMUSG00000065847 | MGI:5454965 | snoRNA gene | predicted gene, 25188 |
| 3 | gene | 86.13794 | 86.14270 | negative | MGI_C57BL6J_1202063 | Rps3a1 | NCBI_Gene:20091,ENSEMBL:ENSMUSG00000028081 | MGI:1202063 | protein coding gene | ribosomal protein S3A1 |
| 3 | gene | 86.13879 | 86.13886 | negative | MGI_C57BL6J_1321388 | Snord73a | NCBI_Gene:19870,ENSEMBL:ENSMUSG00000064984 | MGI:1321388 | snoRNA gene | small nucleolar RNA, C/D box U73A |
| 3 | gene | 86.14062 | 86.14069 | negative | MGI_C57BL6J_1321387 | Rnu73b | NCBI_Gene:19871,ENSEMBL:ENSMUSG00000064390 | MGI:1321387 | snoRNA gene | U73B small nuclear RNA |
| 3 | gene | 86.14276 | 86.14554 | positive | MGI_C57BL6J_5611161 | Gm37933 | ENSEMBL:ENSMUSG00000103408 | MGI:5611161 | lncRNA gene | predicted gene, 37933 |
| 3 | pseudogene | 86.14494 | 86.14537 | negative | MGI_C57BL6J_5610721 | Gm37493 | ENSEMBL:ENSMUSG00000102218 | MGI:5610721 | pseudogene | predicted gene, 37493 |
| 3 | gene | 86.19142 | 86.19153 | positive | MGI_C57BL6J_5452802 | Gm23025 | ENSEMBL:ENSMUSG00000089394 | MGI:5452802 | snRNA gene | predicted gene, 23025 |
| 3 | gene | 86.22390 | 86.78269 | positive | MGI_C57BL6J_1933162 | Lrba | NCBI_Gene:80877,ENSEMBL:ENSMUSG00000028080 | MGI:1933162 | protein coding gene | LPS-responsive beige-like anchor |
| 3 | pseudogene | 86.26324 | 86.26358 | negative | MGI_C57BL6J_5610516 | Gm37288 | ENSEMBL:ENSMUSG00000103759 | MGI:5610516 | pseudogene | predicted gene, 37288 |
| 3 | gene | 86.26765 | 86.26867 | positive | MGI_C57BL6J_1924676 | 9430099H24Rik | NA | NA | unclassified gene | RIKEN cDNA 9430099H24 gene |
| 3 | gene | 86.27523 | 86.27536 | negative | MGI_C57BL6J_5456120 | Gm26343 | ENSEMBL:ENSMUSG00000077744 | MGI:5456120 | snoRNA gene | predicted gene, 26343 |
| 3 | gene | 86.35202 | 86.36244 | negative | MGI_C57BL6J_5622968 | Gm40083 | NCBI_Gene:105244482 | MGI:5622968 | lncRNA gene | predicted gene, 40083 |
| 3 | gene | 86.36249 | 86.36358 | negative | MGI_C57BL6J_1924912 | 9030218A15Rik | NA | NA | unclassified gene | RIKEN cDNA 9030218A15 gene |
| 3 | gene | 86.36446 | 86.36463 | positive | MGI_C57BL6J_5453627 | Gm23850 | ENSEMBL:ENSMUSG00000065764 | MGI:5453627 | snRNA gene | predicted gene, 23850 |
| 3 | gene | 86.38425 | 86.38435 | positive | MGI_C57BL6J_5454816 | Gm25039 | ENSEMBL:ENSMUSG00000064867 | MGI:5454816 | snRNA gene | predicted gene, 25039 |
| 3 | gene | 86.52442 | 86.55872 | negative | MGI_C57BL6J_5590836 | Gm31677 | NCBI_Gene:102633984 | MGI:5590836 | lncRNA gene | predicted gene, 31677 |
| 3 | gene | 86.54558 | 86.54863 | negative | MGI_C57BL6J_1346022 | Mab21l2 | NCBI_Gene:23937,ENSEMBL:ENSMUSG00000057777 | MGI:1346022 | protein coding gene | mab-21-like 2 |
| 3 | pseudogene | 86.67177 | 86.67224 | negative | MGI_C57BL6J_3781961 | Gm3788 | NCBI_Gene:100042319,ENSEMBL:ENSMUSG00000094392 | MGI:3781961 | pseudogene | predicted gene 3788 |
| 3 | gene | 86.76807 | 86.77865 | negative | MGI_C57BL6J_5611104 | Gm37876 | ENSEMBL:ENSMUSG00000104108 | MGI:5611104 | lncRNA gene | predicted gene, 37876 |
| 3 | gene | 86.78615 | 86.92088 | negative | MGI_C57BL6J_1918012 | Dclk2 | NCBI_Gene:70762,ENSEMBL:ENSMUSG00000028078 | MGI:1918012 | protein coding gene | doublecortin-like kinase 2 |
| 3 | gene | 86.82565 | 86.82710 | negative | MGI_C57BL6J_5826447 | Gm46810 | NCBI_Gene:108168877 | MGI:5826447 | lncRNA gene | predicted gene, 46810 |
| 3 | pseudogene | 86.86449 | 86.86532 | positive | MGI_C57BL6J_5610253 | Gm37025 | ENSEMBL:ENSMUSG00000104074 | MGI:5610253 | pseudogene | predicted gene, 37025 |
| 3 | gene | 86.93074 | 86.93090 | positive | MGI_C57BL6J_5453153 | Gm23376 | ENSEMBL:ENSMUSG00000064872 | MGI:5453153 | snRNA gene | predicted gene, 23376 |
| 3 | pseudogene | 86.93337 | 86.93366 | negative | MGI_C57BL6J_5610435 | Gm37207 | ENSEMBL:ENSMUSG00000103650 | MGI:5610435 | pseudogene | predicted gene, 37207 |
| 3 | pseudogene | 86.98655 | 86.98978 | positive | MGI_C57BL6J_107675 | Cd1d2 | NCBI_Gene:12480,ENSEMBL:ENSMUSG00000041750 | MGI:107675 | polymorphic pseudogene | CD1d2 antigen |
| 3 | gene | 86.99583 | 86.99944 | negative | MGI_C57BL6J_107674 | Cd1d1 | NCBI_Gene:12479,ENSEMBL:ENSMUSG00000028076 | MGI:107674 | protein coding gene | CD1d1 antigen |
| 3 | pseudogene | 87.02286 | 87.02396 | negative | MGI_C57BL6J_3644826 | Gm8869 | NCBI_Gene:667897,ENSEMBL:ENSMUSG00000104265 | MGI:3644826 | pseudogene | predicted gene 8869 |
| 3 | pseudogene | 87.05436 | 87.05500 | positive | MGI_C57BL6J_5010588 | Gm18403 | NCBI_Gene:100417112,ENSEMBL:ENSMUSG00000103913 | MGI:5010588 | pseudogene | predicted gene, 18403 |
| 3 | pseudogene | 87.06012 | 87.06043 | positive | MGI_C57BL6J_5610803 | Gm37575 | ENSEMBL:ENSMUSG00000102394 | MGI:5610803 | pseudogene | predicted gene, 37575 |
| 3 | gene | 87.06689 | 87.06702 | positive | MGI_C57BL6J_5454249 | Gm24472 | ENSEMBL:ENSMUSG00000077672 | MGI:5454249 | snoRNA gene | predicted gene, 24472 |
| 3 | gene | 87.07859 | 87.17475 | negative | MGI_C57BL6J_1891396 | Kirrel | NCBI_Gene:170643,ENSEMBL:ENSMUSG00000041734 | MGI:1891396 | protein coding gene | kirre like nephrin family adhesion molecule 1 |
| 3 | gene | 87.11060 | 87.11222 | negative | MGI_C57BL6J_5011567 | Gm19382 | NA | NA | unclassified gene | predicted gene%2c 19382 |
| 3 | gene | 87.15769 | 87.16245 | positive | MGI_C57BL6J_5611083 | Gm37855 | ENSEMBL:ENSMUSG00000102893 | MGI:5611083 | lncRNA gene | predicted gene, 37855 |
| 3 | pseudogene | 87.20734 | 87.20803 | negative | MGI_C57BL6J_3708678 | Gm10705 | NCBI_Gene:100042355,ENSEMBL:ENSMUSG00000074506 | MGI:3708678 | pseudogene | predicted gene 10705 |
| 3 | gene | 87.25076 | 87.26377 | negative | MGI_C57BL6J_1933397 | Fcrls | NCBI_Gene:80891,ENSEMBL:ENSMUSG00000015852 | MGI:1933397 | protein coding gene | Fc receptor-like S, scavenger receptor |
| 3 | gene | 87.29034 | 87.30091 | positive | MGI_C57BL6J_5591038 | Gm31879 | NCBI_Gene:102634249 | MGI:5591038 | lncRNA gene | predicted gene, 31879 |
| 3 | gene | 87.35400 | 87.37760 | negative | MGI_C57BL6J_5591099 | Gm31940 | NCBI_Gene:102634332 | MGI:5591099 | lncRNA gene | predicted gene, 31940 |
| 3 | gene | 87.35788 | 87.37107 | positive | MGI_C57BL6J_1334419 | Cd5l | NCBI_Gene:11801,ENSEMBL:ENSMUSG00000015854 | MGI:1334419 | protein coding gene | CD5 antigen-like |
| 3 | gene | 87.37634 | 87.40397 | positive | MGI_C57BL6J_2442862 | Fcrl1 | NCBI_Gene:229499,ENSEMBL:ENSMUSG00000059994 | MGI:2442862 | protein coding gene | Fc receptor-like 1 |
| 3 | gene | 87.41141 | 87.41479 | positive | MGI_C57BL6J_5622969 | Gm40084 | NCBI_Gene:105244483 | MGI:5622969 | lncRNA gene | predicted gene, 40084 |
| 3 | gene | 87.43577 | 87.50068 | positive | MGI_C57BL6J_3053558 | Fcrl5 | NCBI_Gene:329693,ENSEMBL:ENSMUSG00000048031 | MGI:3053558 | protein coding gene | Fc receptor-like 5 |
| 3 | gene | 87.48702 | 87.51219 | negative | MGI_C57BL6J_5622970 | Gm40085 | NCBI_Gene:105244484 | MGI:5622970 | lncRNA gene | predicted gene, 40085 |
| 3 | gene | 87.50853 | 87.51318 | positive | MGI_C57BL6J_5591157 | Gm31998 | NCBI_Gene:102634410 | MGI:5591157 | lncRNA gene | predicted gene, 31998 |
| 3 | gene | 87.52541 | 87.54016 | positive | MGI_C57BL6J_1350926 | Etv3 | NCBI_Gene:27049,ENSEMBL:ENSMUSG00000003382 | MGI:1350926 | protein coding gene | ets variant 3 |
| 3 | gene | 87.54988 | 87.56110 | positive | MGI_C57BL6J_3646099 | Etv3l | NCBI_Gene:546801,ENSEMBL:ENSMUSG00000104058 | MGI:3646099 | protein coding gene | ets variant 3-like |
| 3 | gene | 87.58267 | 87.58279 | negative | MGI_C57BL6J_5454944 | Gm25167 | ENSEMBL:ENSMUSG00000077376 | MGI:5454944 | snoRNA gene | predicted gene, 25167 |
| 3 | gene | 87.61740 | 87.61750 | positive | MGI_C57BL6J_5453886 | Gm24109 | ENSEMBL:ENSMUSG00000093230 | MGI:5453886 | miRNA gene | predicted gene, 24109 |
| 3 | gene | 87.61755 | 87.73803 | positive | MGI_C57BL6J_2441869 | Arhgef11 | NCBI_Gene:213498,ENSEMBL:ENSMUSG00000041977 | MGI:2441869 | protein coding gene | Rho guanine nucleotide exchange factor (GEF) 11 |
| 3 | pseudogene | 87.64843 | 87.64867 | negative | MGI_C57BL6J_5011427 | Gm19242 | NCBI_Gene:100462734 | MGI:5011427 | pseudogene | predicted gene, 19242 |
| 3 | gene | 87.67964 | 87.68003 | negative | MGI_C57BL6J_3644100 | Gm6570 | NCBI_Gene:625281 | MGI:3644100 | protein coding gene | predicted gene 6570 |
| 3 | gene | 87.73692 | 87.74863 | negative | MGI_C57BL6J_1921735 | Lrrc71 | NCBI_Gene:74485,ENSEMBL:ENSMUSG00000023084 | MGI:1921735 | protein coding gene | leucine rich repeat containing 71 |
| 3 | gene | 87.74789 | 87.82431 | negative | MGI_C57BL6J_1920432 | Pear1 | NCBI_Gene:73182,ENSEMBL:ENSMUSG00000028073 | MGI:1920432 | protein coding gene | platelet endothelial aggregation receptor 1 |
| 3 | gene | 87.77824 | 87.79524 | negative | MGI_C57BL6J_97383 | Ntrk1 | NCBI_Gene:18211,ENSEMBL:ENSMUSG00000028072 | MGI:97383 | protein coding gene | neurotrophic tyrosine kinase, receptor, type 1 |
| 3 | gene | 87.79691 | 87.81610 | positive | MGI_C57BL6J_1346037 | Insrr | NCBI_Gene:23920,ENSEMBL:ENSMUSG00000005640 | MGI:1346037 | protein coding gene | insulin receptor-related receptor |
| 3 | gene | 87.82054 | 87.82307 | positive | MGI_C57BL6J_5622971 | Gm40086 | NCBI_Gene:105244485 | MGI:5622971 | lncRNA gene | predicted gene, 40086 |
| 3 | gene | 87.82475 | 87.83127 | positive | MGI_C57BL6J_5591629 | Gm32470 | NCBI_Gene:105244486 | MGI:5591629 | lncRNA gene | predicted gene, 32470 |
| 3 | gene | 87.84676 | 87.85572 | positive | MGI_C57BL6J_1351596 | Sh2d2a | NCBI_Gene:27371,ENSEMBL:ENSMUSG00000028071 | MGI:1351596 | protein coding gene | SH2 domain containing 2A |
| 3 | gene | 87.85890 | 87.88561 | negative | MGI_C57BL6J_2137738 | Prcc | NCBI_Gene:94315,ENSEMBL:ENSMUSG00000004895 | MGI:2137738 | protein coding gene | papillary renal cell carcinoma (translocation-associated) |
| 3 | gene | 87.90632 | 87.91613 | positive | MGI_C57BL6J_1194494 | Hdgf | NCBI_Gene:15191,ENSEMBL:ENSMUSG00000004897 | MGI:1194494 | protein coding gene | heparin binding growth factor |
| 3 | gene | 87.91813 | 87.92367 | positive | MGI_C57BL6J_1914957 | Mrpl24 | NCBI_Gene:67707,ENSEMBL:ENSMUSG00000019710 | MGI:1914957 | protein coding gene | mitochondrial ribosomal protein L24 |
| 3 | gene | 87.92260 | 87.93072 | negative | MGI_C57BL6J_2387197 | Rrnad1 | NCBI_Gene:229503,ENSEMBL:ENSMUSG00000004896 | MGI:2387197 | protein coding gene | ribosomal RNA adenine dimethylase domain containing 1 |
| 3 | gene | 87.93031 | 87.94069 | positive | MGI_C57BL6J_2140076 | Isg20l2 | NCBI_Gene:229504,ENSEMBL:ENSMUSG00000048039 | MGI:2140076 | protein coding gene | interferon stimulated exonuclease gene 20-like 2 |
| 3 | gene | 87.94867 | 87.95338 | positive | MGI_C57BL6J_88491 | Crabp2 | NCBI_Gene:12904,ENSEMBL:ENSMUSG00000004885 | MGI:88491 | protein coding gene | cellular retinoic acid binding protein II |
| 3 | gene | 87.96008 | 87.96573 | positive | MGI_C57BL6J_5591677 | Gm32518 | NCBI_Gene:102635095 | MGI:5591677 | lncRNA gene | predicted gene, 32518 |
| 3 | pseudogene | 87.96676 | 87.96869 | positive | MGI_C57BL6J_3781920 | Gm3745 | NCBI_Gene:100042242,ENSEMBL:ENSMUSG00000103792 | MGI:3781920 | pseudogene | predicted gene 3745 |
| 3 | gene | 87.97108 | 87.98045 | positive | MGI_C57BL6J_101784 | Nes | NCBI_Gene:18008,ENSEMBL:ENSMUSG00000004891 | MGI:101784 | protein coding gene | nestin |
| 3 | gene | 87.98753 | 88.00089 | negative | MGI_C57BL6J_1096385 | Bcan | NCBI_Gene:12032,ENSEMBL:ENSMUSG00000004892 | MGI:1096385 | protein coding gene | brevican |
| 3 | gene | 88.02056 | 88.02758 | negative | MGI_C57BL6J_2137300 | Hapln2 | NCBI_Gene:73940,ENSEMBL:ENSMUSG00000004894 | MGI:2137300 | protein coding gene | hyaluronan and proteoglycan link protein 2 |
| 3 | gene | 88.03078 | 88.03621 | positive | MGI_C57BL6J_5011624 | Gm19439 | NCBI_Gene:105244487,ENSEMBL:ENSMUSG00000105843 | MGI:5011624 | lncRNA gene | predicted gene, 19439 |
| 3 | gene | 88.04311 | 88.05599 | positive | MGI_C57BL6J_1913864 | Gpatch4 | NCBI_Gene:66614,ENSEMBL:ENSMUSG00000028069 | MGI:1913864 | protein coding gene | G patch domain containing 4 |
| 3 | gene | 88.05652 | 88.05854 | negative | MGI_C57BL6J_2180167 | Naxe | NCBI_Gene:246703,ENSEMBL:ENSMUSG00000028070 | MGI:2180167 | protein coding gene | NAD(P)HX epimerase |
| 3 | gene | 88.06941 | 88.07830 | negative | MGI_C57BL6J_2443841 | Ttc24 | NCBI_Gene:214191,ENSEMBL:ENSMUSG00000051036 | MGI:2443841 | protein coding gene | tetratricopeptide repeat domain 24 |
| 3 | gene | 88.08197 | 88.12105 | positive | MGI_C57BL6J_3028642 | Iqgap3 | NCBI_Gene:404710,ENSEMBL:ENSMUSG00000028068 | MGI:3028642 | protein coding gene | IQ motif containing GTPase activating protein 3 |
| 3 | gene | 88.13352 | 88.13842 | positive | MGI_C57BL6J_5622972 | Gm40087 | NCBI_Gene:105244489 | MGI:5622972 | lncRNA gene | predicted gene, 40087 |
| 3 | gene | 88.14197 | 88.17209 | positive | MGI_C57BL6J_99533 | Mef2d | NCBI_Gene:17261,ENSEMBL:ENSMUSG00000001419 | MGI:99533 | protein coding gene | myocyte enhancer factor 2D |
| 3 | gene | 88.17156 | 88.17779 | negative | MGI_C57BL6J_1923892 | 1700113A16Rik | NCBI_Gene:76642,ENSEMBL:ENSMUSG00000096935 | MGI:1923892 | lncRNA gene | RIKEN cDNA 1700113A16 gene |
| 3 | gene | 88.19910 | 88.20459 | negative | MGI_C57BL6J_2139818 | AI849053 | ENSEMBL:ENSMUSG00000097280 | MGI:2139818 | lncRNA gene | expressed sequence AI849053 |
| 3 | gene | 88.20020 | 88.20348 | positive | MGI_C57BL6J_2139951 | AW047730 | NCBI_Gene:99870,ENSEMBL:ENSMUSG00000097428 | MGI:2139951 | lncRNA gene | expressed sequence AW047730 |
| 3 | gene | 88.20577 | 88.22996 | positive | MGI_C57BL6J_3781938 | Gm3764 | NCBI_Gene:100042277,ENSEMBL:ENSMUSG00000097156 | MGI:3781938 | lncRNA gene | predicted gene 3764 |
| 3 | gene | 88.21517 | 88.21526 | positive | MGI_C57BL6J_4834307 | Mir3093 | miRBase:MI0014086,NCBI_Gene:100526540,ENSEMBL:ENSMUSG00000092988 | MGI:4834307 | miRNA gene | microRNA 3093 |
| 3 | gene | 88.21559 | 88.21570 | negative | MGI_C57BL6J_5455418 | Gm25641 | ENSEMBL:ENSMUSG00000092876 | MGI:5455418 | miRNA gene | predicted gene, 25641 |
| 3 | gene | 88.21560 | 88.21569 | positive | MGI_C57BL6J_2676911 | Mir9-1 | miRBase:MI0000720,NCBI_Gene:387133,ENSEMBL:ENSMUSG00000106493 | MGI:2676911 | miRNA gene | microRNA 9-1 |
| 3 | gene | 88.22741 | 88.23234 | negative | MGI_C57BL6J_1918213 | 4931402H11Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 4931402H11 gene |
| 3 | gene | 88.24287 | 88.25474 | negative | MGI_C57BL6J_1927379 | Rhbg | NCBI_Gene:58176,ENSEMBL:ENSMUSG00000104445 | MGI:1927379 | protein coding gene | Rhesus blood group-associated B glycoprotein |
| 3 | gene | 88.24338 | 88.28721 | negative | MGI_C57BL6J_5613632 | Gm38392 | ENSEMBL:ENSMUSG00000103766 | MGI:5613632 | protein coding gene | predicted gene, 38392 |
| 3 | gene | 88.28275 | 88.29701 | negative | MGI_C57BL6J_1924177 | Tsacc | NCBI_Gene:76927,ENSEMBL:ENSMUSG00000010538 | MGI:1924177 | protein coding gene | TSSK6 activating co-chaperone |
| 3 | pseudogene | 88.28461 | 88.28475 | negative | MGI_C57BL6J_5610344 | Gm37116 | ENSEMBL:ENSMUSG00000103574 | MGI:5610344 | pseudogene | predicted gene, 37116 |
| 3 | gene | 88.29712 | 88.32177 | positive | MGI_C57BL6J_104708 | Cct3 | NCBI_Gene:12462,ENSEMBL:ENSMUSG00000001416 | MGI:104708 | protein coding gene | chaperonin containing Tcp1, subunit 3 (gamma) |
| 3 | gene | 88.32502 | 88.33131 | positive | MGI_C57BL6J_1913318 | Glmp | NCBI_Gene:56700,ENSEMBL:ENSMUSG00000001418 | MGI:1913318 | protein coding gene | glycosylated lysosomal membrane protein |
| 3 | gene | 88.32865 | 88.33615 | negative | MGI_C57BL6J_1919163 | Tmem79 | NCBI_Gene:71913,ENSEMBL:ENSMUSG00000001420 | MGI:1919163 | protein coding gene | transmembrane protein 79 |
| 3 | gene | 88.33474 | 88.33618 | negative | MGI_C57BL6J_3040675 | BC038331 | NA | NA | unclassified gene | cDNA sequence BC038331 |
| 3 | gene | 88.33626 | 88.36234 | positive | MGI_C57BL6J_2447364 | Smg5 | NCBI_Gene:229512,ENSEMBL:ENSMUSG00000001415 | MGI:2447364 | protein coding gene | Smg-5 homolog, nonsense mediated mRNA decay factor (C. elegans) |
| 3 | gene | 88.36407 | 88.36854 | positive | MGI_C57BL6J_1916207 | Paqr6 | NCBI_Gene:68957,ENSEMBL:ENSMUSG00000041423 | MGI:1916207 | protein coding gene | progestin and adipoQ receptor family member VI |
| 3 | gene | 88.36862 | 88.37274 | negative | MGI_C57BL6J_88155 | Bglap3 | NCBI_Gene:12095,ENSEMBL:ENSMUSG00000074489 | MGI:88155 | protein coding gene | bone gamma-carboxyglutamate protein 3 |
| 3 | pseudogene | 88.37581 | 88.38293 | positive | MGI_C57BL6J_5439425 | Gm21956 | NCBI_Gene:101055651,NCBI_Gene:108168878,ENSEMBL:ENSMUSG00000096792 | MGI:5439425 | pseudogene | predicted gene, 21956 |
| 3 | gene | 88.37774 | 88.37870 | negative | MGI_C57BL6J_88157 | Bglap2 | NCBI_Gene:12097,ENSEMBL:ENSMUSG00000074486 | MGI:88157 | protein coding gene | bone gamma-carboxyglutamate protein 2 |
| 3 | pseudogene | 88.38163 | 88.38293 | positive | MGI_C57BL6J_3779634 | Gm6821 | NCBI_Gene:101055651,ENSEMBL:ENSMUSG00000078691 | MGI:3779634 | pseudogene | predicted gene 6821 |
| 3 | gene | 88.38349 | 88.38447 | negative | MGI_C57BL6J_88156 | Bglap | NCBI_Gene:12096,ENSEMBL:ENSMUSG00000074483 | MGI:88156 | protein coding gene | bone gamma carboxyglutamate protein |
| 3 | gene | 88.39414 | 88.41040 | negative | MGI_C57BL6J_1914287 | Pmf1 | NCBI_Gene:67037,ENSEMBL:ENSMUSG00000028066 | MGI:1914287 | protein coding gene | polyamine-modulated factor 1 |
| 3 | gene | 88.41049 | 88.42514 | negative | MGI_C57BL6J_2444391 | Slc25a44 | NCBI_Gene:229517,ENSEMBL:ENSMUSG00000050144 | MGI:2444391 | protein coding gene | solute carrier family 25, member 44 |
| 3 | gene | 88.42508 | 88.42867 | positive | MGI_C57BL6J_5592672 | Gm33513 | NCBI_Gene:102636453 | MGI:5592672 | lncRNA gene | predicted gene, 33513 |
| 3 | pseudogene | 88.42894 | 88.42939 | positive | MGI_C57BL6J_5662951 | Gm42814 | ENSEMBL:ENSMUSG00000106568 | MGI:5662951 | pseudogene | predicted gene 42814 |
| 3 | gene | 88.43596 | 88.46118 | negative | MGI_C57BL6J_107560 | Sema4a | NCBI_Gene:20351,ENSEMBL:ENSMUSG00000028064 | MGI:107560 | protein coding gene | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| 3 | gene | 88.44088 | 88.44095 | negative | MGI_C57BL6J_5531059 | Mir7011 | miRBase:MI0022860,NCBI_Gene:102465611,ENSEMBL:ENSMUSG00000098655 | MGI:5531059 | miRNA gene | microRNA 7011 |
| 3 | gene | 88.46209 | 88.46613 | negative | MGI_C57BL6J_5622974 | Gm40089 | NCBI_Gene:105244492 | MGI:5622974 | lncRNA gene | predicted gene, 40089 |
| 3 | pseudogene | 88.47454 | 88.47535 | positive | MGI_C57BL6J_3643641 | Gm5979 | NCBI_Gene:546802,ENSEMBL:ENSMUSG00000103304 | MGI:3643641 | pseudogene | predicted gene 5979 |
| 3 | gene | 88.48015 | 88.50996 | negative | MGI_C57BL6J_96794 | Lmna | NCBI_Gene:16905,ENSEMBL:ENSMUSG00000028063 | MGI:96794 | protein coding gene | lamin A |
| 3 | gene | 88.52607 | 88.52736 | negative | MGI_C57BL6J_5610812 | Gm37584 | ENSEMBL:ENSMUSG00000103115 | MGI:5610812 | lncRNA gene | predicted gene, 37584 |
| 3 | gene | 88.52927 | 88.53179 | negative | MGI_C57BL6J_5621503 | Gm38618 | NCBI_Gene:102642612 | MGI:5621503 | lncRNA gene | predicted gene, 38618 |
| 3 | gene | 88.53239 | 88.54140 | positive | MGI_C57BL6J_1919890 | Mex3a | NCBI_Gene:72640,ENSEMBL:ENSMUSG00000074480 | MGI:1919890 | protein coding gene | mex3 RNA binding family member A |
| 3 | gene | 88.53630 | 88.53638 | negative | MGI_C57BL6J_3811408 | Mir1905 | miRBase:MI0008315,NCBI_Gene:100316808,ENSEMBL:ENSMUSG00000084728 | MGI:3811408 | miRNA gene | microRNA 1905 |
| 3 | gene | 88.54203 | 88.54830 | negative | MGI_C57BL6J_1858203 | Rab25 | NCBI_Gene:53868,ENSEMBL:ENSMUSG00000008601 | MGI:1858203 | protein coding gene | RAB25, member RAS oncogene family |
| 3 | gene | 88.54982 | 88.55307 | negative | MGI_C57BL6J_1932697 | Lamtor2 | NCBI_Gene:83409,ENSEMBL:ENSMUSG00000028062 | MGI:1932697 | protein coding gene | late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| 3 | gene | 88.55368 | 88.56973 | positive | MGI_C57BL6J_2150152 | Ubqln4 | NCBI_Gene:94232,ENSEMBL:ENSMUSG00000008604 | MGI:2150152 | protein coding gene | ubiquilin 4 |
| 3 | gene | 88.57588 | 88.58842 | positive | MGI_C57BL6J_1913506 | Ssr2 | NCBI_Gene:66256,ENSEMBL:ENSMUSG00000041355 | MGI:1913506 | protein coding gene | signal sequence receptor, beta |
| 3 | pseudogene | 88.57624 | 88.57720 | negative | MGI_C57BL6J_3642446 | Rtraf-ps | NCBI_Gene:545536,ENSEMBL:ENSMUSG00000074479 | MGI:3642446 | pseudogene | RNA transcription, translation and transport factor, pseudogene |
| 3 | gene | 88.59311 | 88.64953 | positive | MGI_C57BL6J_103264 | Arhgef2 | NCBI_Gene:16800,ENSEMBL:ENSMUSG00000028059 | MGI:103264 | protein coding gene | rho/rac guanine nucleotide exchange factor (GEF) 2 |
| 3 | gene | 88.60667 | 88.60722 | negative | MGI_C57BL6J_5622975 | Gm40090 | NCBI_Gene:105244493 | MGI:5622975 | lncRNA gene | predicted gene, 40090 |
| 3 | gene | 88.61646 | 88.62111 | negative | MGI_C57BL6J_5313099 | Gm20652 | NCBI_Gene:102636610,ENSEMBL:ENSMUSG00000093390 | MGI:5313099 | lncRNA gene | predicted gene 20652 |
| 3 | gene | 88.65190 | 88.65314 | negative | MGI_C57BL6J_2182926 | Rxfp4 | NCBI_Gene:242093,ENSEMBL:ENSMUSG00000049741 | MGI:2182926 | protein coding gene | relaxin family peptide receptor 4 |
| 3 | gene | 88.68502 | 88.68640 | negative | MGI_C57BL6J_5663851 | Gm43714 | ENSEMBL:ENSMUSG00000105512 | MGI:5663851 | lncRNA gene | predicted gene 43714 |
| 3 | gene | 88.68578 | 88.71293 | positive | MGI_C57BL6J_1921450 | Khdc4 | NCBI_Gene:74200,ENSEMBL:ENSMUSG00000028060 | MGI:1921450 | protein coding gene | KH domain containing 4, pre-mRNA splicing factor |
| 3 | gene | 88.69393 | 88.69406 | positive | MGI_C57BL6J_5455597 | Gm25820 | ENSEMBL:ENSMUSG00000105288,ENSEMBL:ENSMUSG00000105448 | MGI:5455597 | snoRNA gene | predicted gene, 25820 |
| 3 | gene | 88.70716 | 88.70729 | positive | MGI_C57BL6J_5455722 | Gm25945 | ENSEMBL:ENSMUSG00000084433 | MGI:5455722 | snoRNA gene | predicted gene, 25945 |
| 3 | gene | 88.71684 | 88.73105 | positive | MGI_C57BL6J_108053 | Rit1 | NCBI_Gene:19769,ENSEMBL:ENSMUSG00000028057 | MGI:108053 | protein coding gene | Ras-like without CAAX 1 |
| 3 | gene | 88.72209 | 88.72267 | positive | MGI_C57BL6J_4937973 | Gm17146 | ENSEMBL:ENSMUSG00000091881 | MGI:4937973 | lncRNA gene | predicted gene 17146 |
| 3 | gene | 88.73875 | 88.73969 | negative | MGI_C57BL6J_3641796 | Gm10253 | ENSEMBL:ENSMUSG00000104333 | MGI:3641796 | unclassified gene | predicted gene 10253 |
| 3 | gene | 88.74470 | 88.77516 | negative | MGI_C57BL6J_1859547 | Syt11 | NCBI_Gene:229521,ENSEMBL:ENSMUSG00000068923 | MGI:1859547 | protein coding gene | synaptotagmin XI |
| 3 | pseudogene | 88.77522 | 88.82937 | positive | MGI_C57BL6J_1923272 | 5830417I10Rik | NCBI_Gene:100302730,ENSEMBL:ENSMUSG00000078684 | MGI:1923272 | pseudogene | RIKEN cDNA 5830417I10 gene |
| 3 | gene | 88.78294 | 88.83255 | negative | MGI_C57BL6J_2442808 | 1500004A13Rik | NCBI_Gene:319830,ENSEMBL:ENSMUSG00000098912 | MGI:2442808 | lncRNA gene | RIKEN cDNA 1500004A13 gene |
| 3 | gene | 88.83097 | 88.91010 | positive | MGI_C57BL6J_1917579 | Gon4l | NCBI_Gene:76022,ENSEMBL:ENSMUSG00000054199 | MGI:1917579 | protein coding gene | gon-4-like (C.elegans) |
| 3 | gene | 88.84503 | 88.90000 | negative | MGI_C57BL6J_5663850 | Gm43713 | ENSEMBL:ENSMUSG00000106218 | MGI:5663850 | lncRNA gene | predicted gene 43713 |
| 3 | gene | 88.86292 | 88.86308 | positive | MGI_C57BL6J_5452831 | Gm23054 | ENSEMBL:ENSMUSG00000064738 | MGI:5452831 | snRNA gene | predicted gene, 23054 |
| 3 | gene | 88.88092 | 88.89994 | negative | MGI_C57BL6J_5622976 | Gm40091 | NCBI_Gene:105244494 | MGI:5622976 | lncRNA gene | predicted gene, 40091 |
| 3 | gene | 88.90511 | 88.91400 | negative | MGI_C57BL6J_2385175 | Msto1 | NCBI_Gene:229524,ENSEMBL:ENSMUSG00000068922 | MGI:2385175 | protein coding gene | misato 1, mitochondrial distribution and morphology regulator |
| 3 | gene | 88.90651 | 88.90663 | positive | MGI_C57BL6J_4422062 | n-R5s197 | ENSEMBL:ENSMUSG00000065236 | MGI:4422062 | rRNA gene | nuclear encoded rRNA 5S 197 |
| 3 | gene | 88.92080 | 88.95120 | negative | MGI_C57BL6J_1929538 | Dap3 | NCBI_Gene:65111,ENSEMBL:ENSMUSG00000068921 | MGI:1929538 | protein coding gene | death associated protein 3 |
| 3 | gene | 88.95049 | 89.07937 | positive | MGI_C57BL6J_2183158 | Ash1l | NCBI_Gene:192195,ENSEMBL:ENSMUSG00000028053 | MGI:2183158 | protein coding gene | ASH1 like histone lysine methyltransferase |
| 3 | gene | 88.97062 | 88.97073 | negative | MGI_C57BL6J_5456242 | Gm26465 | ENSEMBL:ENSMUSG00000095712 | MGI:5456242 | snRNA gene | predicted gene, 26465 |
| 3 | gene | 89.04900 | 89.04913 | negative | MGI_C57BL6J_5454180 | Gm24403 | ENSEMBL:ENSMUSG00000077804 | MGI:5454180 | snoRNA gene | predicted gene, 24403 |
| 3 | gene | 89.08194 | 89.10110 | negative | MGI_C57BL6J_5663875 | Gm43738 | ENSEMBL:ENSMUSG00000105204 | MGI:5663875 | protein coding gene | predicted gene 43738 |
| 3 | gene | 89.08397 | 89.09336 | negative | MGI_C57BL6J_1919546 | Rusc1 | NCBI_Gene:72296,ENSEMBL:ENSMUSG00000041263 | MGI:1919546 | protein coding gene | RUN and SH3 domain containing 1 |
| 3 | gene | 89.09359 | 89.10197 | negative | MGI_C57BL6J_104888 | Fdps | NCBI_Gene:110196,ENSEMBL:ENSMUSG00000059743 | MGI:104888 | protein coding gene | farnesyl diphosphate synthetase |
| 3 | gene | 89.09791 | 89.09801 | negative | MGI_C57BL6J_5452712 | Gm22935 | ENSEMBL:ENSMUSG00000077148 | MGI:5452712 | snRNA gene | predicted gene, 22935 |
| 3 | pseudogene | 89.11669 | 89.11702 | positive | MGI_C57BL6J_5588863 | Gm29704 | NCBI_Gene:101055641,ENSEMBL:ENSMUSG00000105654 | MGI:5588863 | pseudogene | predicted gene, 29704 |
| 3 | gene | 89.13613 | 89.14678 | positive | MGI_C57BL6J_97604 | Pklr | NCBI_Gene:18770,ENSEMBL:ENSMUSG00000041237 | MGI:97604 | protein coding gene | pyruvate kinase liver and red blood cell |
| 3 | gene | 89.14607 | 89.16023 | negative | MGI_C57BL6J_1298211 | Hcn3 | NCBI_Gene:15168,ENSEMBL:ENSMUSG00000028051 | MGI:1298211 | protein coding gene | hyperpolarization-activated, cyclic nucleotide-gated K+ 3 |
| 3 | gene | 89.16479 | 89.17709 | positive | MGI_C57BL6J_1098669 | Clk2 | NCBI_Gene:12748,ENSEMBL:ENSMUSG00000068917 | MGI:1098669 | protein coding gene | CDC-like kinase 2 |
| 3 | gene | 89.16479 | 89.18277 | positive | MGI_C57BL6J_5825564 | Gm45927 | NCBI_Gene:105943584 | MGI:5825564 | protein coding gene | predicted gene, 45927 |
| 3 | gene | 89.17739 | 89.18277 | positive | MGI_C57BL6J_1346346 | Scamp3 | NCBI_Gene:24045,ENSEMBL:ENSMUSG00000028049 | MGI:1346346 | protein coding gene | secretory carrier membrane protein 3 |
| 3 | gene | 89.18076 | 89.18410 | negative | MGI_C57BL6J_3801823 | Gm16069 | NCBI_Gene:102636781,ENSEMBL:ENSMUSG00000086718 | MGI:3801823 | lncRNA gene | predicted gene 16069 |
| 3 | gene | 89.18314 | 89.18930 | positive | MGI_C57BL6J_1915771 | Fam189b | NCBI_Gene:68521,ENSEMBL:ENSMUSG00000032657 | MGI:1915771 | protein coding gene | family with sequence similarity 189, member B |
| 3 | gene | 89.20117 | 89.20127 | negative | MGI_C57BL6J_5453046 | Gm23269 | ENSEMBL:ENSMUSG00000094489 | MGI:5453046 | snRNA gene | predicted gene, 23269 |
| 3 | gene | 89.20291 | 89.20897 | positive | MGI_C57BL6J_95665 | Gba | NCBI_Gene:14466,ENSEMBL:ENSMUSG00000028048 | MGI:95665 | protein coding gene | glucosidase, beta, acid |
| 3 | gene | 89.20908 | 89.22709 | negative | MGI_C57BL6J_103025 | Mtx1 | NCBI_Gene:17827,ENSEMBL:ENSMUSG00000064068 | MGI:103025 | protein coding gene | metaxin 1 |
| 3 | gene | 89.21043 | 89.21316 | positive | MGI_C57BL6J_5663874 | Gm43737 | ENSEMBL:ENSMUSG00000105692 | MGI:5663874 | unclassified gene | predicted gene 43737 |
| 3 | gene | 89.21481 | 89.22684 | positive | MGI_C57BL6J_98739 | Thbs3 | NCBI_Gene:21827,ENSEMBL:ENSMUSG00000028047 | MGI:98739 | protein coding gene | thrombospondin 3 |
| 3 | gene | 89.22712 | 89.22720 | negative | MGI_C57BL6J_3718577 | Mir92b | miRBase:MI0005521,NCBI_Gene:100124470,ENSEMBL:ENSMUSG00000076255 | MGI:3718577 | miRNA gene | microRNA 92b |
| 3 | gene | 89.22906 | 89.23338 | positive | MGI_C57BL6J_97231 | Muc1 | NCBI_Gene:17829,ENSEMBL:ENSMUSG00000042784 | MGI:97231 | protein coding gene | mucin 1, transmembrane |
| 3 | gene | 89.23417 | 89.24631 | negative | MGI_C57BL6J_2673000 | Trim46 | NCBI_Gene:360213,ENSEMBL:ENSMUSG00000042766 | MGI:2673000 | protein coding gene | tripartite motif-containing 46 |
| 3 | gene | 89.24597 | 89.24991 | positive | MGI_C57BL6J_1913309 | Krtcap2 | NCBI_Gene:66059,ENSEMBL:ENSMUSG00000042747 | MGI:1913309 | protein coding gene | keratinocyte associated protein 2 |
| 3 | gene | 89.25926 | 89.26708 | positive | MGI_C57BL6J_1915813 | Dpm3 | NCBI_Gene:68563,ENSEMBL:ENSMUSG00000042737 | MGI:1915813 | protein coding gene | dolichyl-phosphate mannosyltransferase polypeptide 3 |
| 3 | gene | 89.26825 | 89.27057 | negative | MGI_C57BL6J_107417 | Slc50a1 | NCBI_Gene:19729,ENSEMBL:ENSMUSG00000027953 | MGI:107417 | protein coding gene | solute carrier family 50 (sugar transporter), member 1 |
| 3 | gene | 89.27173 | 89.28114 | negative | MGI_C57BL6J_103236 | Efna1 | NCBI_Gene:13636,ENSEMBL:ENSMUSG00000027954 | MGI:103236 | protein coding gene | ephrin A1 |
| 3 | gene | 89.29054 | 89.30290 | negative | MGI_C57BL6J_5593004 | Gm33845 | NCBI_Gene:102636898 | MGI:5593004 | lncRNA gene | predicted gene, 33845 |
| 3 | gene | 89.30248 | 89.30273 | positive | MGI_C57BL6J_5452419 | Gm22642 | ENSEMBL:ENSMUSG00000093093 | MGI:5452419 | unclassified non-coding RNA gene | predicted gene, 22642 |
| 3 | gene | 89.30324 | 89.31422 | positive | MGI_C57BL6J_5593173 | Gm34014 | NCBI_Gene:102637122 | MGI:5593173 | lncRNA gene | predicted gene, 34014 |
| 3 | gene | 89.31390 | 89.32296 | negative | MGI_C57BL6J_106644 | Efna3 | NCBI_Gene:13638,ENSEMBL:ENSMUSG00000028039 | MGI:106644 | protein coding gene | ephrin A3 |
| 3 | gene | 89.31544 | 89.31891 | positive | MGI_C57BL6J_3802094 | Gm15998 | ENSEMBL:ENSMUSG00000086389 | MGI:3802094 | lncRNA gene | predicted gene 15998 |
| 3 | gene | 89.32590 | 89.33117 | negative | MGI_C57BL6J_5593277 | Gm34118 | NCBI_Gene:102637253 | MGI:5593277 | lncRNA gene | predicted gene, 34118 |
| 3 | gene | 89.33339 | 89.33803 | negative | MGI_C57BL6J_106643 | Efna4 | NCBI_Gene:13639,ENSEMBL:ENSMUSG00000028040 | MGI:106643 | protein coding gene | ephrin A4 |
| 3 | gene | 89.33785 | 89.34783 | positive | MGI_C57BL6J_3704222 | 4731419I09Rik | ENSEMBL:ENSMUSG00000097004 | MGI:3704222 | lncRNA gene | RIKEN cDNA 4731419I09 gene |
| 3 | gene | 89.33854 | 89.35003 | negative | MGI_C57BL6J_1333882 | Adam15 | NCBI_Gene:11490,ENSEMBL:ENSMUSG00000028041 | MGI:1333882 | protein coding gene | a disintegrin and metallopeptidase domain 15 (metargidin) |
| 3 | gene | 89.35022 | 89.36525 | negative | MGI_C57BL6J_1925022 | Dcst1 | NCBI_Gene:77772,ENSEMBL:ENSMUSG00000042672 | MGI:1925022 | protein coding gene | DC-STAMP domain containing 1 |
| 3 | gene | 89.35042 | 89.37764 | positive | MGI_C57BL6J_2685606 | Dcst2 | NCBI_Gene:329702,ENSEMBL:ENSMUSG00000109293 | MGI:2685606 | protein coding gene | DC-STAMP domain containing 2 |
| 3 | pseudogene | 89.35363 | 89.35490 | positive | MGI_C57BL6J_3708679 | Gm10702 | ENSEMBL:ENSMUSG00000074467 | MGI:3708679 | pseudogene | predicted gene 10702 |
| 3 | gene | 89.37764 | 89.39481 | negative | MGI_C57BL6J_102755 | Zbtb7b | NCBI_Gene:22724,ENSEMBL:ENSMUSG00000028042 | MGI:102755 | protein coding gene | zinc finger and BTB domain containing 7B |
| 3 | gene | 89.39185 | 89.39878 | positive | MGI_C57BL6J_3642531 | Gm15417 | NCBI_Gene:545539,ENSEMBL:ENSMUSG00000074466 | MGI:3642531 | lncRNA gene | predicted gene 15417 |
| 3 | gene | 89.40099 | 89.40279 | negative | MGI_C57BL6J_1930020 | Lenep | NCBI_Gene:57275,ENSEMBL:ENSMUSG00000078173 | MGI:1930020 | protein coding gene | lens epithelial protein |
| 3 | gene | 89.40100 | 89.41187 | negative | MGI_C57BL6J_2443030 | Flad1 | NCBI_Gene:319945,ENSEMBL:ENSMUSG00000042642 | MGI:2443030 | protein coding gene | flavin adenine dinucleotide synthetase 1 |
| 3 | gene | 89.41547 | 89.41838 | negative | MGI_C57BL6J_1889208 | Cks1b | NCBI_Gene:54124,ENSEMBL:ENSMUSG00000028044 | MGI:1889208 | protein coding gene | CDC28 protein kinase 1b |
| 3 | gene | 89.41840 | 89.43003 | positive | MGI_C57BL6J_98296 | Shc1 | NCBI_Gene:20416,ENSEMBL:ENSMUSG00000042626 | MGI:98296 | protein coding gene | src homology 2 domain-containing transforming protein C1 |
| 3 | gene | 89.43012 | 89.43513 | positive | MGI_C57BL6J_1916161 | Pygo2 | NCBI_Gene:68911,ENSEMBL:ENSMUSG00000047824 | MGI:1916161 | protein coding gene | pygopus 2 |
| 3 | gene | 89.43667 | 89.45095 | positive | MGI_C57BL6J_2441670 | Pbxip1 | NCBI_Gene:229534,ENSEMBL:ENSMUSG00000042613 | MGI:2441670 | protein coding gene | pre B cell leukemia transcription factor interacting protein 1 |
| 3 | gene | 89.45077 | 89.45094 | positive | MGI_C57BL6J_1916805 | 2310033F14Rik | NA | NA | unclassified gene | RIKEN cDNA 2310033F14 gene |
| 3 | gene | 89.45454 | 89.46901 | positive | MGI_C57BL6J_1915853 | Pmvk | NCBI_Gene:68603,ENSEMBL:ENSMUSG00000027952 | MGI:1915853 | protein coding gene | phosphomevalonate kinase |
| 3 | gene | 89.52016 | 89.67513 | positive | MGI_C57BL6J_2153183 | Kcnn3 | NCBI_Gene:140493,ENSEMBL:ENSMUSG00000000794 | MGI:2153183 | protein coding gene | potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| 3 | gene | 89.71502 | 89.75346 | positive | MGI_C57BL6J_1889575 | Adar | NCBI_Gene:56417,ENSEMBL:ENSMUSG00000027951 | MGI:1889575 | protein coding gene | adenosine deaminase, RNA-specific |
| 3 | gene | 89.74620 | 89.76463 | negative | MGI_C57BL6J_87891 | Chrnb2 | NCBI_Gene:11444,ENSEMBL:ENSMUSG00000027950 | MGI:87891 | protein coding gene | cholinergic receptor, nicotinic, beta polypeptide 2 (neuronal) |
| 3 | gene | 89.76751 | 89.77340 | negative | MGI_C57BL6J_1921284 | 4632404H12Rik | NCBI_Gene:74034,ENSEMBL:ENSMUSG00000042579 | MGI:1921284 | lncRNA gene | RIKEN cDNA 4632404H12 gene |
| 3 | gene | 89.77361 | 89.78400 | positive | MGI_C57BL6J_1917343 | Ube2q1 | NCBI_Gene:70093,ENSEMBL:ENSMUSG00000042572 | MGI:1917343 | protein coding gene | ubiquitin-conjugating enzyme E2Q family member 1 |
| 3 | pseudogene | 89.81948 | 89.82168 | positive | MGI_C57BL6J_3645290 | Gm8969 | NCBI_Gene:668087,ENSEMBL:ENSMUSG00000106564 | MGI:3645290 | pseudogene | predicted gene 8969 |
| 3 | gene | 89.83137 | 89.85885 | positive | MGI_C57BL6J_1099462 | She | NCBI_Gene:214547,ENSEMBL:ENSMUSG00000046280 | MGI:1099462 | protein coding gene | src homology 2 domain-containing transforming protein E |
| 3 | gene | 89.86406 | 89.91320 | negative | MGI_C57BL6J_105304 | Il6ra | NCBI_Gene:16194,ENSEMBL:ENSMUSG00000027947 | MGI:105304 | protein coding gene | interleukin 6 receptor, alpha |
| 3 | gene | 89.87208 | 89.89024 | positive | MGI_C57BL6J_5826448 | Gm46811 | NCBI_Gene:108168879 | MGI:5826448 | lncRNA gene | predicted gene, 46811 |
| 3 | gene | 89.91292 | 89.91405 | positive | MGI_C57BL6J_5662946 | Gm42809 | ENSEMBL:ENSMUSG00000105941 | MGI:5662946 | lncRNA gene | predicted gene 42809 |
| 3 | gene | 89.93948 | 89.96351 | negative | MGI_C57BL6J_1859660 | Atp8b2 | NCBI_Gene:54667,ENSEMBL:ENSMUSG00000060671 | MGI:1859660 | protein coding gene | ATPase, class I, type 8B, member 2 |
| 3 | pseudogene | 89.96344 | 89.96973 | negative | MGI_C57BL6J_4356425 | Aqp10-ps | NCBI_Gene:435743,ENSEMBL:ENSMUSG00000105082 | MGI:4356425 | pseudogene | aquaporin 10, pseudogene |
| 3 | gene | 89.96800 | 89.96816 | negative | MGI_C57BL6J_5453823 | Gm24046 | ENSEMBL:ENSMUSG00000065045 | MGI:5453823 | snRNA gene | predicted gene, 24046 |
| 3 | gene | 89.99545 | 89.99878 | negative | MGI_C57BL6J_1346319 | Hax1 | NCBI_Gene:23897,ENSEMBL:ENSMUSG00000027944 | MGI:1346319 | protein coding gene | HCLS1 associated X-1 |
| 3 | gene | 89.99876 | 90.02144 | positive | MGI_C57BL6J_5011895 | Gm19710 | NCBI_Gene:100503469,ENSEMBL:ENSMUSG00000105552 | MGI:5011895 | lncRNA gene | predicted gene, 19710 |
| 3 | gene | 89.99959 | 90.05263 | negative | MGI_C57BL6J_1921633 | Ubap2l | NCBI_Gene:74383,ENSEMBL:ENSMUSG00000042520 | MGI:1921633 | protein coding gene | ubiquitin-associated protein 2-like |
| 3 | gene | 90.01059 | 90.01073 | negative | MGI_C57BL6J_5454385 | Gm24608 | ENSEMBL:ENSMUSG00000077569 | MGI:5454385 | snoRNA gene | predicted gene, 24608 |
| 3 | gene | 90.01115 | 90.01121 | negative | MGI_C57BL6J_5531378 | Mir7669 | miRBase:MI0025009,NCBI_Gene:102465771,ENSEMBL:ENSMUSG00000098397 | MGI:5531378 | miRNA gene | microRNA 7669 |
| 3 | gene | 90.05164 | 90.06835 | positive | MGI_C57BL6J_1914027 | 4933434E20Rik | NCBI_Gene:99650,ENSEMBL:ENSMUSG00000027942 | MGI:1914027 | protein coding gene | RIKEN cDNA 4933434E20 gene |
| 3 | gene | 90.05726 | 90.05838 | positive | MGI_C57BL6J_4414960 | Gm16540 | ENSEMBL:ENSMUSG00000090005 | MGI:4414960 | lncRNA gene | predicted gene 16540 |
| 3 | gene | 90.06280 | 90.06835 | positive | MGI_C57BL6J_1920795 | 1700094D03Rik | NCBI_Gene:73545 | MGI:1920795 | protein coding gene | RIKEN cDNA 1700094D03 gene |
| 3 | gene | 90.07002 | 90.07010 | positive | MGI_C57BL6J_3718457 | Mir190b | miRBase:MI0005478,NCBI_Gene:100124481,ENSEMBL:ENSMUSG00000077996 | MGI:3718457 | miRNA gene | microRNA 190b |
| 3 | gene | 90.07265 | 90.10090 | positive | MGI_C57BL6J_1890149 | Tpm3 | NCBI_Gene:59069,ENSEMBL:ENSMUSG00000027940 | MGI:1890149 | protein coding gene | tropomyosin 3, gamma |
| 3 | gene | 90.10157 | 90.21215 | positive | MGI_C57BL6J_1924845 | Nup210l | NCBI_Gene:77595,ENSEMBL:ENSMUSG00000027939 | MGI:1924845 | protein coding gene | nucleoporin 210-like |
| 3 | gene | 90.12465 | 90.12712 | positive | MGI_C57BL6J_5594024 | Gm34865 | NCBI_Gene:102638261 | MGI:5594024 | lncRNA gene | predicted gene, 34865 |
| 3 | gene | 90.12654 | 90.12667 | positive | MGI_C57BL6J_5453500 | Gm23723 | ENSEMBL:ENSMUSG00000064930 | MGI:5453500 | snoRNA gene | predicted gene, 23723 |
| 3 | pseudogene | 90.12677 | 90.12706 | positive | MGI_C57BL6J_5663849 | Gm43712 | ENSEMBL:ENSMUSG00000104649 | MGI:5663849 | pseudogene | predicted gene 43712 |
| 3 | gene | 90.19868 | 90.19879 | positive | MGI_C57BL6J_5453962 | Gm24185 | ENSEMBL:ENSMUSG00000084527 | MGI:5453962 | snRNA gene | predicted gene, 24185 |
| 3 | gene | 90.21252 | 90.21365 | negative | MGI_C57BL6J_1888676 | Rps27 | NCBI_Gene:57294,ENSEMBL:ENSMUSG00000090733 | MGI:1888676 | protein coding gene | ribosomal protein S27 |
| 3 | gene | 90.21370 | 90.22639 | positive | MGI_C57BL6J_1927232 | Rab13 | NCBI_Gene:68328,ENSEMBL:ENSMUSG00000027935 | MGI:1927232 | protein coding gene | RAB13, member RAS oncogene family |
| 3 | gene | 90.23160 | 90.23584 | positive | MGI_C57BL6J_1346082 | Jtb | NCBI_Gene:23922,ENSEMBL:ENSMUSG00000027937 | MGI:1346082 | protein coding gene | jumping translocation breakpoint |
| 3 | gene | 90.23750 | 90.24445 | negative | MGI_C57BL6J_1916603 | Creb3l4 | NCBI_Gene:78284,ENSEMBL:ENSMUSG00000027938 | MGI:1916603 | protein coding gene | cAMP responsive element binding protein 3-like 4 |
| 3 | gene | 90.24817 | 90.25361 | positive | MGI_C57BL6J_1353474 | Slc39a1 | NCBI_Gene:30791,ENSEMBL:ENSMUSG00000052310 | MGI:1353474 | protein coding gene | solute carrier family 39 (zinc transporter), member 1 |
| 3 | gene | 90.25386 | 90.26413 | positive | MGI_C57BL6J_1921593 | Crtc2 | NCBI_Gene:74343,ENSEMBL:ENSMUSG00000027936 | MGI:1921593 | protein coding gene | CREB regulated transcription coactivator 2 |
| 3 | gene | 90.26492 | 90.28067 | positive | MGI_C57BL6J_2446201 | Dennd4b | NCBI_Gene:229541,ENSEMBL:ENSMUSG00000042404 | MGI:2446201 | protein coding gene | DENN/MADD domain containing 4B |
| 3 | gene | 90.27015 | 90.27021 | positive | MGI_C57BL6J_5530752 | Mir7012 | miRBase:MI0022861,NCBI_Gene:102465612,ENSEMBL:ENSMUSG00000099199 | MGI:5530752 | miRNA gene | microRNA 7012 |
| 3 | gene | 90.28999 | 90.29073 | positive | MGI_C57BL6J_1918721 | 8430422J01Rik | NA | NA | unclassified gene | RIKEN cDNA 8430422J01 gene |
| 3 | gene | 90.29230 | 90.29398 | negative | MGI_C57BL6J_1923075 | 4930537H20Rik | ENSEMBL:ENSMUSG00000105350 | MGI:1923075 | lncRNA gene | RIKEN cDNA 4930537H20 gene |
| 3 | gene | 90.29318 | 90.36341 | positive | MGI_C57BL6J_2443225 | Gatad2b | NCBI_Gene:229542,ENSEMBL:ENSMUSG00000042390 | MGI:2443225 | protein coding gene | GATA zinc finger domain containing 2B |
| 3 | gene | 90.31152 | 90.31701 | positive | MGI_C57BL6J_5791313 | Gm45477 | ENSEMBL:ENSMUSG00000108394 | MGI:5791313 | unclassified gene | predicted gene 45477 |
| 3 | gene | 90.33540 | 90.33795 | positive | MGI_C57BL6J_5791190 | Gm45354 | ENSEMBL:ENSMUSG00000108689 | MGI:5791190 | unclassified gene | predicted gene 45354 |
| 3 | gene | 90.38523 | 90.39005 | negative | MGI_C57BL6J_1347358 | Slc27a3 | NCBI_Gene:26568,ENSEMBL:ENSMUSG00000027932 | MGI:1347358 | protein coding gene | solute carrier family 27 (fatty acid transporter), member 3 |
| 3 | gene | 90.39138 | 90.43364 | negative | MGI_C57BL6J_2140050 | Ints3 | NCBI_Gene:229543,ENSEMBL:ENSMUSG00000027933 | MGI:2140050 | protein coding gene | integrator complex subunit 3 |
| 3 | gene | 90.45059 | 90.46593 | negative | MGI_C57BL6J_97371 | Npr1 | NCBI_Gene:18160,ENSEMBL:ENSMUSG00000027931 | MGI:97371 | protein coding gene | natriuretic peptide receptor 1 |
| 3 | gene | 90.45158 | 90.45709 | positive | MGI_C57BL6J_3801855 | Gm16048 | ENSEMBL:ENSMUSG00000087411 | MGI:3801855 | lncRNA gene | predicted gene 16048 |
| 3 | pseudogene | 90.46890 | 90.47096 | negative | MGI_C57BL6J_5663732 | Gm43595 | ENSEMBL:ENSMUSG00000105746 | MGI:5663732 | pseudogene | predicted gene 43595 |
| 3 | gene | 90.47586 | 90.47593 | negative | MGI_C57BL6J_4413988 | n-TMcat11 | NCBI_Gene:102467358 | MGI:4413988 | tRNA gene | nuclear encoded tRNA methionine 11 (anticodon CAT) |
| 3 | gene | 90.47613 | 90.48838 | positive | MGI_C57BL6J_1915031 | Ilf2 | NCBI_Gene:67781,ENSEMBL:ENSMUSG00000001016 | MGI:1915031 | protein coding gene | interleukin enhancer binding factor 2 |
| 3 | gene | 90.48803 | 90.49103 | negative | MGI_C57BL6J_1333745 | Snapin | NCBI_Gene:20615,ENSEMBL:ENSMUSG00000001018 | MGI:1333745 | protein coding gene | SNAP-associated protein |
| 3 | gene | 90.49854 | 90.50954 | negative | MGI_C57BL6J_1913761 | Chtop | NCBI_Gene:66511,ENSEMBL:ENSMUSG00000001017 | MGI:1913761 | protein coding gene | chromatin target of PRMT1 |
| 3 | gene | 90.51103 | 90.51439 | negative | MGI_C57BL6J_1338917 | S100a1 | NCBI_Gene:20193,ENSEMBL:ENSMUSG00000044080 | MGI:1338917 | protein coding gene | S100 calcium binding protein A1 |
| 3 | gene | 90.51444 | 90.52458 | positive | MGI_C57BL6J_109581 | S100a13 | NCBI_Gene:20196,ENSEMBL:ENSMUSG00000042312 | MGI:109581 | protein coding gene | S100 calcium binding protein A13 |
| 3 | gene | 90.52685 | 90.52884 | positive | MGI_C57BL6J_1913416 | S100a14 | NCBI_Gene:66166,ENSEMBL:ENSMUSG00000042306 | MGI:1913416 | protein coding gene | S100 calcium binding protein A14 |
| 3 | gene | 90.53725 | 90.54315 | positive | MGI_C57BL6J_1915110 | S100a16 | NCBI_Gene:67860,ENSEMBL:ENSMUSG00000074457 | MGI:1915110 | protein coding gene | S100 calcium binding protein A16 |
| 3 | gene | 90.54862 | 90.54872 | positive | MGI_C57BL6J_5454370 | Gm24593 | ENSEMBL:ENSMUSG00000084456 | MGI:5454370 | snRNA gene | predicted gene, 24593 |
| 3 | gene | 90.56036 | 90.60271 | positive | MGI_C57BL6J_1338849 | S100a3 | NCBI_Gene:20197,ENSEMBL:ENSMUSG00000001021 | MGI:1338849 | protein coding gene | S100 calcium binding protein A3 |
| 3 | gene | 90.56036 | 90.59153 | positive | MGI_C57BL6J_3510999 | S100a2 | NCBI_Gene:628324,ENSEMBL:ENSMUSG00000094018 | MGI:3510999 | protein coding gene | S100 calcium binding protein A2 |
| 3 | gene | 90.56056 | 90.56070 | negative | MGI_C57BL6J_5453241 | Gm23464 | ENSEMBL:ENSMUSG00000065079 | MGI:5453241 | snRNA gene | predicted gene, 23464 |
| 3 | gene | 90.60190 | 90.60589 | positive | MGI_C57BL6J_5662811 | Gm42674 | ENSEMBL:ENSMUSG00000105518 | MGI:5662811 | protein coding gene | predicted gene 42674 |
| 3 | gene | 90.60377 | 90.60604 | positive | MGI_C57BL6J_1330282 | S100a4 | NCBI_Gene:20198,ENSEMBL:ENSMUSG00000001020 | MGI:1330282 | protein coding gene | S100 calcium binding protein A4 |
| 3 | gene | 90.60648 | 90.61178 | positive | MGI_C57BL6J_1338915 | S100a5 | NCBI_Gene:20199,ENSEMBL:ENSMUSG00000001023 | MGI:1338915 | protein coding gene | S100 calcium binding protein A5 |
| 3 | gene | 90.61288 | 90.62418 | positive | MGI_C57BL6J_1339467 | S100a6 | NCBI_Gene:20200,ENSEMBL:ENSMUSG00000001025 | MGI:1339467 | protein coding gene | S100 calcium binding protein A6 (calcyclin) |
| 3 | pseudogene | 90.65165 | 90.65187 | negative | MGI_C57BL6J_5011924 | Gm19739 | NCBI_Gene:102638335,ENSEMBL:ENSMUSG00000105644 | MGI:5011924 | pseudogene | predicted gene, 19739 |
| 3 | gene | 90.65183 | 90.65867 | positive | MGI_C57BL6J_2687194 | S100a7a | NCBI_Gene:381493,ENSEMBL:ENSMUSG00000063767 | MGI:2687194 | protein coding gene | S100 calcium binding protein A7A |
| 3 | gene | 90.66898 | 90.67003 | positive | MGI_C57BL6J_88244 | S100a8 | NCBI_Gene:20201,ENSEMBL:ENSMUSG00000056054 | MGI:88244 | protein coding gene | S100 calcium binding protein A8 (calgranulin A) |
| 3 | gene | 90.69263 | 90.69572 | negative | MGI_C57BL6J_1338947 | S100a9 | NCBI_Gene:20202,ENSEMBL:ENSMUSG00000056071 | MGI:1338947 | protein coding gene | S100 calcium binding protein A9 (calgranulin B) |
| 3 | pseudogene | 90.70884 | 90.70961 | negative | MGI_C57BL6J_5662716 | Gm42579 | NCBI_Gene:108168854,ENSEMBL:ENSMUSG00000105819 | MGI:5662716 | pseudogene | predicted gene 42579 |
| 3 | pseudogene | 90.72257 | 90.72351 | negative | MGI_C57BL6J_3646209 | Gm5284 | NCBI_Gene:383949,ENSEMBL:ENSMUSG00000106117 | MGI:3646209 | pseudogene | predicted gene 5284 |
| 3 | gene | 90.72691 | 90.74152 | positive | MGI_C57BL6J_2686324 | Pglyrp4 | NCBI_Gene:384997,ENSEMBL:ENSMUSG00000042250 | MGI:2686324 | protein coding gene | peptidoglycan recognition protein 4 |
| 3 | pseudogene | 90.75873 | 90.75956 | positive | MGI_C57BL6J_5594172 | Gm35013 | ENSEMBL:ENSMUSG00000105892 | MGI:5594172 | pseudogene | predicted gene, 35013 |
| 3 | pseudogene | 90.75937 | 90.76131 | positive | MGI_C57BL6J_3645549 | Gm5104 | NCBI_Gene:329706 | MGI:3645549 | pseudogene | predicted gene 5104 |
| 3 | gene | 90.77582 | 90.77788 | negative | MGI_C57BL6J_3645655 | Gm5849 | ENSEMBL:ENSMUSG00000096621 | MGI:3645655 | protein coding gene | predicted gene 5849 |
| 3 | pseudogene | 90.84637 | 90.84825 | negative | MGI_C57BL6J_5663372 | Gm43235 | ENSEMBL:ENSMUSG00000106174 | MGI:5663372 | pseudogene | predicted gene 43235 |
| 3 | pseudogene | 91.00976 | 91.01718 | negative | MGI_C57BL6J_3643784 | Gm9023 | NCBI_Gene:668179,ENSEMBL:ENSMUSG00000106257 | MGI:3643784 | pseudogene | predicted gene 9023 |
| 3 | pseudogene | 91.03957 | 91.03987 | positive | MGI_C57BL6J_3779688 | Gm7166 | ENSEMBL:ENSMUSG00000105919 | MGI:3779688 | pseudogene | predicted gene 7166 |
| 3 | pseudogene | 91.07166 | 91.07519 | positive | MGI_C57BL6J_1922392 | 4930529C04Rik | NCBI_Gene:619677,ENSEMBL:ENSMUSG00000086377 | MGI:1922392 | pseudogene | RIKEN cDNA 4930529C04 gene |
| 3 | gene | 91.08814 | 91.10743 | negative | MGI_C57BL6J_2684973 | S100a7l2 | NCBI_Gene:229550,ENSEMBL:ENSMUSG00000091175 | MGI:2684973 | protein coding gene | S100 calcium binding protein A7 like 2 |
| 3 | gene | 91.62856 | 91.63070 | negative | MGI_C57BL6J_5595482 | Gm36323 | NCBI_Gene:102640196 | MGI:5595482 | protein coding gene | predicted gene, 36323 |
| 3 | gene | 91.73835 | 91.74049 | negative | MGI_C57BL6J_5594344 | Gm35185 | NCBI_Gene:102638680 | MGI:5594344 | protein coding gene | predicted gene, 35185 |
| 3 | pseudogene | 91.91438 | 91.91653 | positive | MGI_C57BL6J_5826474 | Gm46837 | NCBI_Gene:108168919 | MGI:5826474 | pseudogene | predicted gene, 46837 |
| 3 | pseudogene | 91.99611 | 91.99725 | positive | MGI_C57BL6J_5009818 | Gm17742 | NCBI_Gene:383887 | MGI:5009818 | pseudogene | predicted gene, 17742 |
| 3 | gene | 92.01076 | 92.01086 | negative | MGI_C57BL6J_5453474 | Gm23697 | ENSEMBL:ENSMUSG00000077085 | MGI:5453474 | miRNA gene | predicted gene, 23697 |
| 3 | gene | 92.01248 | 92.03267 | positive | MGI_C57BL6J_2685266 | Pglyrp3 | NCBI_Gene:242100,ENSEMBL:ENSMUSG00000042244 | MGI:2685266 | protein coding gene | peptidoglycan recognition protein 3 |
| 3 | pseudogene | 92.02449 | 92.02503 | positive | MGI_C57BL6J_103164 | Mif-ps3 | NCBI_Gene:619750,ENSEMBL:ENSMUSG00000082826 | MGI:103164 | pseudogene | macrophage migration inhibitory factor, pseudogene 3 |
| 3 | gene | 92.06140 | 92.06151 | negative | MGI_C57BL6J_5454922 | Gm25145 | ENSEMBL:ENSMUSG00000096133 | MGI:5454922 | miRNA gene | predicted gene, 25145 |
| 3 | gene | 92.08027 | 92.08314 | negative | MGI_C57BL6J_96816 | Lor | NCBI_Gene:16939,ENSEMBL:ENSMUSG00000043165 | MGI:96816 | protein coding gene | loricrin |
| 3 | gene | 92.09735 | 92.09963 | positive | MGI_C57BL6J_5663873 | Gm43736 | ENSEMBL:ENSMUSG00000104981 | MGI:5663873 | lncRNA gene | predicted gene 43736 |
| 3 | gene | 92.10258 | 92.14274 | negative | MGI_C57BL6J_1922313 | 4930511M18Rik | ENSEMBL:ENSMUSG00000104791 | MGI:1922313 | lncRNA gene | RIKEN cDNA 4930511M18 gene |
| 3 | gene | 92.11319 | 92.11932 | positive | MGI_C57BL6J_5663872 | Gm43735 | ENSEMBL:ENSMUSG00000106143 | MGI:5663872 | lncRNA gene | predicted gene 43735 |
| 3 | gene | 92.12220 | 92.12395 | negative | MGI_C57BL6J_1925680 | Prr9 | NCBI_Gene:109314,ENSEMBL:ENSMUSG00000056270 | MGI:1925680 | protein coding gene | proline rich 9 |
| 3 | gene | 92.13499 | 92.14275 | negative | MGI_C57BL6J_1916582 | Lelp1 | NCBI_Gene:69332,ENSEMBL:ENSMUSG00000027927 | MGI:1916582 | protein coding gene | late cornified envelope-like proline-rich 1 |
| 3 | gene | 92.17019 | 92.18323 | positive | MGI_C57BL6J_5594598 | Gm35439 | NCBI_Gene:102639022,ENSEMBL:ENSMUSG00000105476 | MGI:5594598 | lncRNA gene | predicted gene, 35439 |
| 3 | gene | 92.21580 | 92.22249 | positive | MGI_C57BL6J_1330350 | Sprr2a1 | NCBI_Gene:20755,ENSEMBL:ENSMUSG00000078664 | MGI:1330350 | protein coding gene | small proline-rich protein 2A1 |
| 3 | gene | 92.21592 | 92.25730 | positive | MGI_C57BL6J_3845026 | Sprr2a2 | NCBI_Gene:100303744,ENSEMBL:ENSMUSG00000068893 | MGI:3845026 | protein coding gene | small proline-rich protein 2A2 |
| 3 | pseudogene | 92.22741 | 92.22870 | negative | MGI_C57BL6J_3645657 | Gm5850 | NCBI_Gene:545543,ENSEMBL:ENSMUSG00000103286 | MGI:3645657 | pseudogene | predicted gene 5850 |
| 3 | gene | 92.24293 | 92.24624 | negative | MGI_C57BL6J_5610315 | Gm37087 | ENSEMBL:ENSMUSG00000103187 | MGI:5610315 | unclassified gene | predicted gene, 37087 |
| 3 | pseudogene | 92.26222 | 92.26350 | negative | MGI_C57BL6J_3645652 | Gm5851 | NCBI_Gene:545545,ENSEMBL:ENSMUSG00000104028 | MGI:3645652 | pseudogene | predicted gene 5851 |
| 3 | gene | 92.27773 | 92.28103 | negative | MGI_C57BL6J_5610980 | Gm37752 | ENSEMBL:ENSMUSG00000103385 | MGI:5610980 | unclassified gene | predicted gene, 37752 |
| 3 | gene | 92.28530 | 92.29140 | positive | MGI_C57BL6J_3845028 | Sprr2a3 | NCBI_Gene:100042514,ENSEMBL:ENSMUSG00000074445 | MGI:3845028 | protein coding gene | small proline-rich protein 2A3 |
| 3 | gene | 92.31662 | 92.31808 | positive | MGI_C57BL6J_1330352 | Sprr2b | NCBI_Gene:20756,ENSEMBL:ENSMUSG00000050092 | MGI:1330352 | protein coding gene | small proline-rich protein 2B |
| 3 | pseudogene | 92.32091 | 92.32105 | positive | MGI_C57BL6J_5610656 | Gm37428 | ENSEMBL:ENSMUSG00000103518 | MGI:5610656 | pseudogene | predicted gene, 37428 |
| 3 | pseudogene | 92.32660 | 92.32690 | positive | MGI_C57BL6J_1330351 | Sprr2c-ps | NCBI_Gene:20757,ENSEMBL:ENSMUSG00000103221 | MGI:1330351 | pseudogene | small proline-rich protein 2C, pseudogene |
| 3 | gene | 92.33914 | 92.34087 | positive | MGI_C57BL6J_1330347 | Sprr2d | NCBI_Gene:20758,ENSEMBL:ENSMUSG00000042212 | MGI:1330347 | protein coding gene | small proline-rich protein 2D |
| 3 | gene | 92.35214 | 92.35345 | positive | MGI_C57BL6J_1330346 | Sprr2e | NCBI_Gene:20759,ENSEMBL:ENSMUSG00000055030 | MGI:1330346 | protein coding gene | small proline-rich protein 2E |
| 3 | gene | 92.36519 | 92.36644 | positive | MGI_C57BL6J_1330349 | Sprr2f | NCBI_Gene:20760,ENSEMBL:ENSMUSG00000050635 | MGI:1330349 | protein coding gene | small proline-rich protein 2F |
| 3 | pseudogene | 92.37391 | 92.37523 | positive | MGI_C57BL6J_1330348 | Sprr2g | NCBI_Gene:20761,ENSEMBL:ENSMUSG00000046203 | MGI:1330348 | polymorphic pseudogene | small proline-rich protein 2G |
| 3 | gene | 92.38568 | 92.38732 | positive | MGI_C57BL6J_1330343 | Sprr2h | NCBI_Gene:20762,ENSEMBL:ENSMUSG00000046259 | MGI:1330343 | protein coding gene | small proline-rich protein 2H |
| 3 | gene | 92.40799 | 92.40927 | positive | MGI_C57BL6J_1330309 | Sprr2i | NCBI_Gene:20763,ENSEMBL:ENSMUSG00000042157 | MGI:1330309 | protein coding gene | small proline-rich protein 2I |
| 3 | pseudogene | 92.41808 | 92.41940 | positive | MGI_C57BL6J_1330345 | Sprr2j-ps | NCBI_Gene:20764,ENSEMBL:ENSMUSG00000027925 | MGI:1330345 | pseudogene | small proline-rich protein 2J, pseudogene |
| 3 | gene | 92.42803 | 92.42942 | negative | MGI_C57BL6J_3642386 | Gm9774 | NCBI_Gene:100043022,ENSEMBL:ENSMUSG00000042165 | MGI:3642386 | protein coding gene | predicted pseudogene 9774 |
| 3 | gene | 92.43258 | 92.43393 | positive | MGI_C57BL6J_1330344 | Sprr2k | NCBI_Gene:20765,ENSEMBL:ENSMUSG00000054215 | MGI:1330344 | protein coding gene | small proline-rich protein 2K |
| 3 | gene | 92.43681 | 92.43882 | negative | MGI_C57BL6J_106659 | Sprr1b | NCBI_Gene:20754,ENSEMBL:ENSMUSG00000048455 | MGI:106659 | protein coding gene | small proline-rich protein 1B |
| 3 | gene | 92.45650 | 92.45872 | negative | MGI_C57BL6J_1330237 | Sprr3 | NCBI_Gene:20766,ENSEMBL:ENSMUSG00000045539 | MGI:1330237 | protein coding gene | small proline-rich protein 3 |
| 3 | pseudogene | 92.47324 | 92.47354 | negative | MGI_C57BL6J_5610605 | Gm37377 | ENSEMBL:ENSMUSG00000102345 | MGI:5610605 | pseudogene | predicted gene, 37377 |
| 3 | gene | 92.48395 | 92.48589 | negative | MGI_C57BL6J_106660 | Sprr1a | NCBI_Gene:20753,ENSEMBL:ENSMUSG00000050359 | MGI:106660 | protein coding gene | small proline-rich protein 1A |
| 3 | gene | 92.50026 | 92.50049 | negative | MGI_C57BL6J_2654508 | Sprr4 | NCBI_Gene:229562,ENSEMBL:ENSMUSG00000045566 | MGI:2654508 | protein coding gene | small proline-rich protein 4 |
| 3 | gene | 92.53251 | 92.53451 | negative | MGI_C57BL6J_1924218 | 2310046K23Rik | ENSEMBL:ENSMUSG00000102308 | MGI:1924218 | protein coding gene | RIKEN cDNA 2310046K23 gene |
| 3 | gene | 92.57090 | 92.57379 | negative | MGI_C57BL6J_96626 | Ivl | NCBI_Gene:16447,ENSEMBL:ENSMUSG00000049128 | MGI:96626 | protein coding gene | involucrin |
| 3 | gene | 92.57387 | 92.62033 | positive | MGI_C57BL6J_5594798 | Gm35639 | NCBI_Gene:102639293 | MGI:5594798 | lncRNA gene | predicted gene, 35639 |
| 3 | gene | 92.58387 | 92.58902 | negative | MGI_C57BL6J_96945 | Smcp | NCBI_Gene:17235,ENSEMBL:ENSMUSG00000074435 | MGI:96945 | protein coding gene | sperm mitochondria-associated cysteine-rich protein |
| 3 | gene | 92.60472 | 92.60749 | negative | MGI_C57BL6J_3588251 | 2210017I01Rik | ENSEMBL:ENSMUSG00000103523 | MGI:3588251 | protein coding gene | RIKEN cDNA 2210017I01 gene |
| 3 | gene | 92.60476 | 92.62166 | negative | MGI_C57BL6J_1925632 | Lce6a | NCBI_Gene:78382,ENSEMBL:ENSMUSG00000086848 | MGI:1925632 | protein coding gene | late cornified envelope 6A |
| 3 | gene | 92.62034 | 92.64644 | positive | MGI_C57BL6J_5826449 | Gm46812 | NCBI_Gene:108168880 | MGI:5826449 | lncRNA gene | predicted gene, 46812 |
| 3 | gene | 92.63906 | 92.64007 | negative | MGI_C57BL6J_5594858 | Gm35699 | NCBI_Gene:102639369 | MGI:5594858 | lncRNA gene | predicted gene, 35699 |
| 3 | gene | 92.64653 | 92.64831 | negative | MGI_C57BL6J_1914377 | Lce1a1 | NCBI_Gene:67127,ENSEMBL:ENSMUSG00000057609 | MGI:1914377 | protein coding gene | late cornified envelope 1A1 |
| 3 | gene | 92.65565 | 92.65693 | negative | MGI_C57BL6J_1915970 | Lce1b | NCBI_Gene:68720,ENSEMBL:ENSMUSG00000027923 | MGI:1915970 | protein coding gene | late cornified envelope 1B |
| 3 | pseudogene | 92.66419 | 92.66490 | positive | MGI_C57BL6J_3648077 | Rpl13-ps4 | NCBI_Gene:620325,ENSEMBL:ENSMUSG00000104215 | MGI:3648077 | pseudogene | ribosomal protein L13, pseudogene 4 |
| 3 | gene | 92.66861 | 92.67032 | negative | MGI_C57BL6J_1920972 | Lce1a2 | NCBI_Gene:73722,ENSEMBL:ENSMUSG00000068890 | MGI:1920972 | protein coding gene | late cornified envelope 1A2 |


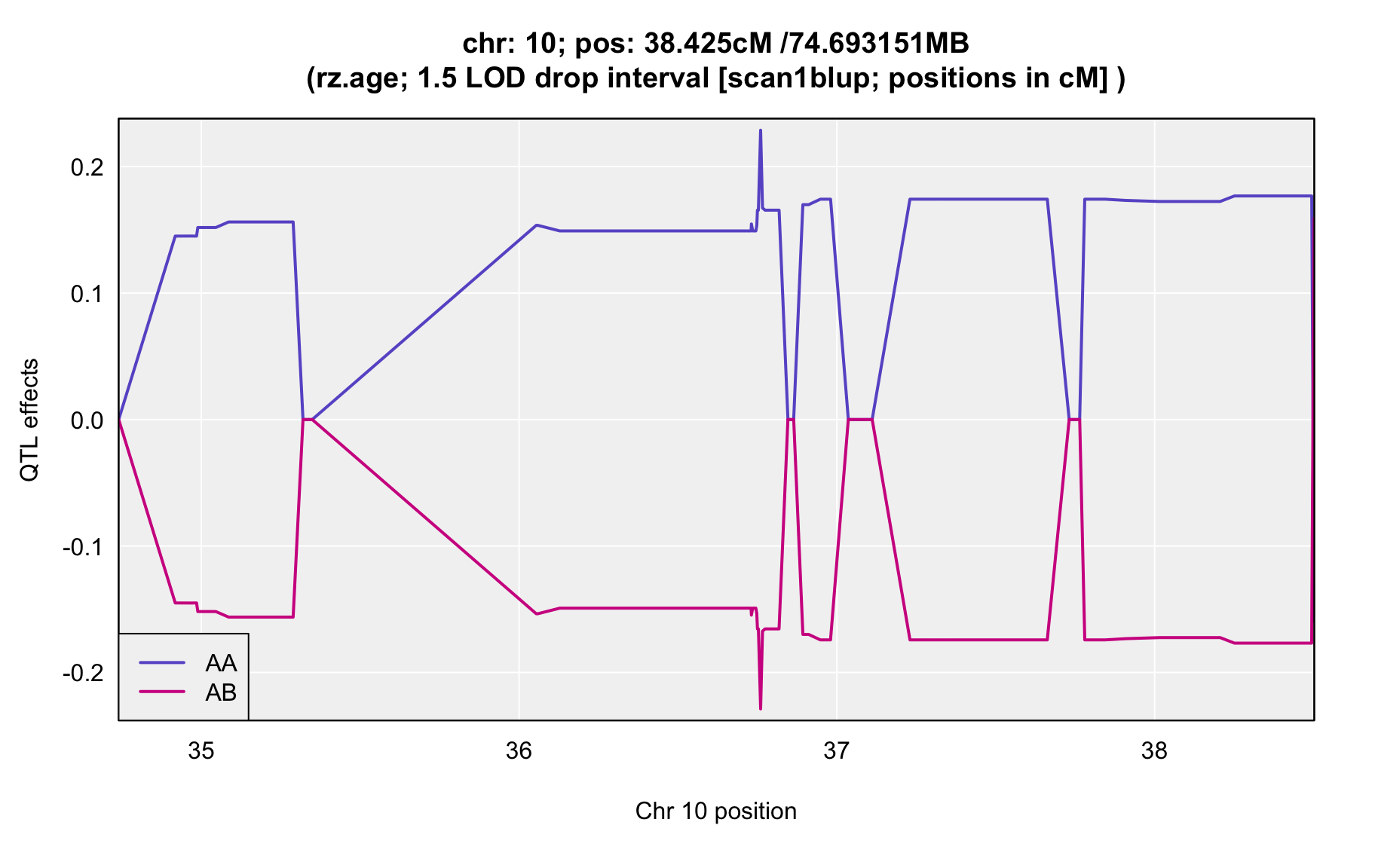
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 10 | gene | 66.29328 | 66.51251 | negative | MGI_C57BL6J_5623562 | Gm40677 | NCBI_Gene:105245190 | MGI:5623562 | lncRNA gene | predicted gene, 40677 |
| 10 | gene | 66.48355 | 66.48368 | positive | MGI_C57BL6J_5452371 | Gm22594 | ENSEMBL:ENSMUSG00000077439 | MGI:5452371 | snoRNA gene | predicted gene, 22594 |
| 10 | gene | 66.64092 | 66.64186 | positive | MGI_C57BL6J_5578845 | Gm28139 | ENSEMBL:ENSMUSG00000101338 | MGI:5578845 | lncRNA gene | predicted gene 28139 |
| 10 | pseudogene | 66.69134 | 66.69161 | negative | MGI_C57BL6J_6097127 | Gm47894 | ENSEMBL:ENSMUSG00000112247 | MGI:6097127 | pseudogene | predicted gene, 47894 |
| 10 | gene | 66.69824 | 66.70596 | positive | MGI_C57BL6J_5590340 | Gm31181 | ENSEMBL:ENSMUSG00000112202 | MGI:5590340 | lncRNA gene | predicted gene, 31181 |
| 10 | pseudogene | 66.74029 | 66.74116 | negative | MGI_C57BL6J_5010962 | Gm18777 | NCBI_Gene:100417714,ENSEMBL:ENSMUSG00000112159 | MGI:5010962 | pseudogene | predicted gene, 18777 |
| 10 | gene | 66.74093 | 66.74559 | negative | MGI_C57BL6J_5590399 | Gm31240 | NCBI_Gene:102633406 | MGI:5590399 | lncRNA gene | predicted gene, 31240 |
| 10 | gene | 66.75945 | 66.76210 | positive | MGI_C57BL6J_6097128 | Gm47895 | ENSEMBL:ENSMUSG00000112383 | MGI:6097128 | lncRNA gene | predicted gene, 47895 |
| 10 | gene | 66.80395 | 66.82967 | negative | MGI_C57BL6J_5623563 | Gm40678 | NCBI_Gene:105245191 | MGI:5623563 | lncRNA gene | predicted gene, 40678 |
| 10 | gene | 66.80984 | 66.82235 | positive | MGI_C57BL6J_6097130 | Gm47896 | ENSEMBL:ENSMUSG00000112425 | MGI:6097130 | lncRNA gene | predicted gene, 47896 |
| 10 | gene | 66.82417 | 66.82740 | positive | MGI_C57BL6J_6097132 | Gm47897 | ENSEMBL:ENSMUSG00000112514 | MGI:6097132 | lncRNA gene | predicted gene, 47897 |
| 10 | gene | 66.83214 | 66.91156 | negative | MGI_C57BL6J_5590575 | Gm31416 | NCBI_Gene:102633633 | MGI:5590575 | lncRNA gene | predicted gene, 31416 |
| 10 | gene | 66.83760 | 66.83950 | positive | MGI_C57BL6J_6097137 | Gm47900 | ENSEMBL:ENSMUSG00000112379 | MGI:6097137 | lncRNA gene | predicted gene, 47900 |
| 10 | gene | 66.88967 | 66.89161 | negative | MGI_C57BL6J_6097142 | Gm47903 | ENSEMBL:ENSMUSG00000112267 | MGI:6097142 | lncRNA gene | predicted gene, 47903 |
| 10 | gene | 66.91219 | 66.92062 | negative | MGI_C57BL6J_1915738 | 1110002J07Rik | NCBI_Gene:68488,ENSEMBL:ENSMUSG00000097134 | MGI:1915738 | lncRNA gene | RIKEN cDNA 1110002J07 gene |
| 10 | gene | 66.92030 | 66.92360 | positive | MGI_C57BL6J_5477070 | Gm26576 | ENSEMBL:ENSMUSG00000097921 | MGI:5477070 | lncRNA gene | predicted gene, 26576 |
| 10 | gene | 67.00502 | 67.04133 | positive | MGI_C57BL6J_5590922 | Gm31763 | NCBI_Gene:102634101,ENSEMBL:ENSMUSG00000112925 | MGI:5590922 | lncRNA gene | predicted gene, 31763 |
| 10 | gene | 67.00919 | 67.09699 | negative | MGI_C57BL6J_88930 | Reep3 | NCBI_Gene:28193,ENSEMBL:ENSMUSG00000019873 | MGI:88930 | protein coding gene | receptor accessory protein 3 |
| 10 | gene | 67.09613 | 67.25633 | positive | MGI_C57BL6J_1918614 | Jmjd1c | NCBI_Gene:108829,ENSEMBL:ENSMUSG00000037876 | MGI:1918614 | protein coding gene | jumonji domain containing 1C |
| 10 | gene | 67.12053 | 67.12478 | negative | MGI_C57BL6J_5825865 | Gm46228 | NCBI_Gene:108167815 | MGI:5825865 | lncRNA gene | predicted gene, 46228 |
| 10 | gene | 67.17778 | 67.18395 | positive | MGI_C57BL6J_2443021 | A630074N13Rik | NA | NA | unclassified gene | RIKEN cDNA A630074N13 gene |
| 10 | gene | 67.23586 | 67.23596 | negative | MGI_C57BL6J_5456136 | Gm26359 | ENSEMBL:ENSMUSG00000089498 | MGI:5456136 | snRNA gene | predicted gene, 26359 |
| 10 | gene | 67.26669 | 67.28535 | negative | MGI_C57BL6J_1354950 | Nrbf2 | NCBI_Gene:641340,ENSEMBL:ENSMUSG00000075000 | MGI:1354950 | protein coding gene | nuclear receptor binding factor 2 |
| 10 | gene | 67.28543 | 67.29316 | positive | MGI_C57BL6J_5825837 | Gm46200 | NCBI_Gene:108167782 | MGI:5825837 | lncRNA gene | predicted gene, 46200 |
| 10 | gene | 67.29741 | 67.41893 | positive | MGI_C57BL6J_6155108 | Gm49454 | ENSEMBL:ENSMUSG00000112314 | MGI:6155108 | lncRNA gene | predicted gene, 49454 |
| 10 | gene | 67.30440 | 67.31363 | positive | MGI_C57BL6J_5825866 | Gm46229 | NCBI_Gene:108167816 | MGI:5825866 | lncRNA gene | predicted gene, 46229 |
| 10 | pseudogene | 67.34880 | 67.34896 | negative | MGI_C57BL6J_6096511 | Gm47516 | ENSEMBL:ENSMUSG00000111908 | MGI:6096511 | pseudogene | predicted gene, 47516 |
| 10 | gene | 67.34998 | 67.39721 | positive | MGI_C57BL6J_5825867 | Gm46230 | NCBI_Gene:108167817 | MGI:5825867 | lncRNA gene | predicted gene, 46230 |
| 10 | gene | 67.39968 | 67.41929 | positive | MGI_C57BL6J_5591121 | Gm31962 | NCBI_Gene:102634362 | MGI:5591121 | lncRNA gene | predicted gene, 31962 |
| 10 | gene | 67.42005 | 67.42533 | positive | MGI_C57BL6J_5591182 | Gm32023 | NCBI_Gene:102634442,ENSEMBL:ENSMUSG00000112848 | MGI:5591182 | lncRNA gene | predicted gene, 32023 |
| 10 | gene | 67.43609 | 67.44343 | negative | MGI_C57BL6J_6096535 | Gm47531 | ENSEMBL:ENSMUSG00000112726 | MGI:6096535 | lncRNA gene | predicted gene, 47531 |
| 10 | gene | 67.48439 | 67.48605 | negative | MGI_C57BL6J_1923024 | 4930403L11Rik | NA | NA | unclassified gene | RIKEN cDNA 4930403L11 gene |
| 10 | gene | 67.49636 | 67.53534 | negative | MGI_C57BL6J_5591414 | Gm32255 | NCBI_Gene:102634748,ENSEMBL:ENSMUSG00000112120 | MGI:5591414 | lncRNA gene | predicted gene, 32255 |
| 10 | gene | 67.50114 | 67.50314 | negative | MGI_C57BL6J_5623564 | Gm40679 | NCBI_Gene:105245192 | MGI:5623564 | lncRNA gene | predicted gene, 40679 |
| 10 | gene | 67.50912 | 67.51154 | positive | MGI_C57BL6J_5591335 | Gm32176 | NCBI_Gene:102634639 | MGI:5591335 | lncRNA gene | predicted gene, 32176 |
| 10 | gene | 67.53445 | 67.54749 | positive | MGI_C57BL6J_95296 | Egr2 | NCBI_Gene:13654,ENSEMBL:ENSMUSG00000037868 | MGI:95296 | protein coding gene | early growth response 2 |
| 10 | gene | 67.54451 | 67.54896 | negative | MGI_C57BL6J_2685083 | Ado | NCBI_Gene:211488,ENSEMBL:ENSMUSG00000057134 | MGI:2685083 | protein coding gene | 2-aminoethanethiol (cysteamine) dioxygenase |
| 10 | gene | 67.56763 | 67.57199 | negative | MGI_C57BL6J_1922576 | 4930563J15Rik | NCBI_Gene:105245193,ENSEMBL:ENSMUSG00000112141 | MGI:1922576 | lncRNA gene | RIKEN cDNA 4930563J15 gene |
| 10 | gene | 67.57028 | 67.59659 | positive | MGI_C57BL6J_5591523 | Gm32364 | NCBI_Gene:102634888,ENSEMBL:ENSMUSG00000112558 | MGI:5591523 | lncRNA gene | predicted gene, 32364 |
| 10 | gene | 67.57204 | 67.57426 | positive | MGI_C57BL6J_3642860 | Gm10797 | ENSEMBL:ENSMUSG00000074999 | MGI:3642860 | protein coding gene | predicted gene 10797 |
| 10 | gene | 67.66463 | 67.66744 | positive | MGI_C57BL6J_5591674 | Gm32515 | NCBI_Gene:102635092,ENSEMBL:ENSMUSG00000112097 | MGI:5591674 | lncRNA gene | predicted gene, 32515 |
| 10 | gene | 67.69352 | 67.70392 | positive | MGI_C57BL6J_5591597 | Gm32438 | NCBI_Gene:102634989 | MGI:5591597 | lncRNA gene | predicted gene, 32438 |
| 10 | gene | 67.70523 | 67.71064 | positive | MGI_C57BL6J_5623565 | Gm40680 | NCBI_Gene:105245194 | MGI:5623565 | lncRNA gene | predicted gene, 40680 |
| 10 | gene | 67.70784 | 67.70947 | positive | MGI_C57BL6J_6096606 | Gm47574 | ENSEMBL:ENSMUSG00000112396 | MGI:6096606 | unclassified gene | predicted gene, 47574 |
| 10 | gene | 67.70859 | 67.71804 | negative | MGI_C57BL6J_1916765 | 1700030E10Rik | ENSEMBL:ENSMUSG00000112619 | MGI:1916765 | lncRNA gene | RIKEN cDNA 1700030E10 gene |
| 10 | gene | 67.74915 | 67.77838 | negative | MGI_C57BL6J_5623566 | Gm40681 | NCBI_Gene:105245195 | MGI:5623566 | lncRNA gene | predicted gene, 40681 |
| 10 | gene | 67.78555 | 67.78883 | positive | MGI_C57BL6J_1922475 | 4930543I11Rik | NCBI_Gene:102635169,ENSEMBL:ENSMUSG00000112537 | MGI:1922475 | lncRNA gene | RIKEN cDNA 4930543I11 gene |
| 10 | gene | 67.78575 | 67.83640 | positive | MGI_C57BL6J_6215275 | Gm49766 | ENSEMBL:ENSMUSG00000116971 | MGI:6215275 | lncRNA gene | predicted gene, 49766 |
| 10 | pseudogene | 67.79273 | 67.79326 | negative | MGI_C57BL6J_5588933 | Gm29774 | NCBI_Gene:101056081,ENSEMBL:ENSMUSG00000111956 | MGI:5588933 | pseudogene | predicted gene, 29774 |
| 10 | gene | 67.88610 | 67.91266 | negative | MGI_C57BL6J_2143676 | Zfp365 | NCBI_Gene:216049,ENSEMBL:ENSMUSG00000037855 | MGI:2143676 | protein coding gene | zinc finger protein 365 |
| 10 | gene | 67.96248 | 67.97371 | negative | MGI_C57BL6J_1922317 | 4930521O17Rik | NCBI_Gene:75067,ENSEMBL:ENSMUSG00000112899 | MGI:1922317 | lncRNA gene | RIKEN cDNA 4930521O17 gene |
| 10 | gene | 67.97957 | 68.05974 | positive | MGI_C57BL6J_2158417 | Rtkn2 | NCBI_Gene:170799,ENSEMBL:ENSMUSG00000037846 | MGI:2158417 | protein coding gene | rhotekin 2 |
| 10 | gene | 68.02545 | 68.02663 | negative | MGI_C57BL6J_3801951 | Gm16212 | ENSEMBL:ENSMUSG00000086838 | MGI:3801951 | lncRNA gene | predicted gene 16212 |
| 10 | gene | 68.09252 | 68.27874 | negative | MGI_C57BL6J_2175912 | Arid5b | NCBI_Gene:71371,ENSEMBL:ENSMUSG00000019947 | MGI:2175912 | protein coding gene | AT rich interactive domain 5B (MRF1-like) |
| 10 | gene | 68.31301 | 68.32698 | positive | MGI_C57BL6J_6095801 | Gm47078 | ENSEMBL:ENSMUSG00000112275 | MGI:6095801 | lncRNA gene | predicted gene, 47078 |
| 10 | gene | 68.31906 | 68.32114 | negative | MGI_C57BL6J_1922398 | 4930545H06Rik | NCBI_Gene:75148,ENSEMBL:ENSMUSG00000112157 | MGI:1922398 | lncRNA gene | RIKEN cDNA 4930545H06 gene |
| 10 | gene | 68.32564 | 68.34030 | negative | MGI_C57BL6J_5591907 | Gm32748 | NCBI_Gene:102635395 | MGI:5591907 | lncRNA gene | predicted gene, 32748 |
| 10 | gene | 68.38659 | 68.43979 | positive | MGI_C57BL6J_5623568 | Gm40683 | NCBI_Gene:105245197 | MGI:5623568 | lncRNA gene | predicted gene, 40683 |
| 10 | gene | 68.43095 | 68.54192 | negative | MGI_C57BL6J_1920537 | Cabcoco1 | NCBI_Gene:73287,ENSEMBL:ENSMUSG00000019945 | MGI:1920537 | protein coding gene | ciliary associated calcium binding coiled-coil 1 |
| 10 | gene | 68.53009 | 68.53394 | positive | MGI_C57BL6J_5592031 | Gm32872 | NCBI_Gene:102635578,ENSEMBL:ENSMUSG00000112793 | MGI:5592031 | lncRNA gene | predicted gene, 32872 |
| 10 | pseudogene | 68.69150 | 68.69237 | negative | MGI_C57BL6J_3647263 | Gm7594 | NCBI_Gene:665342,ENSEMBL:ENSMUSG00000112434 | MGI:3647263 | pseudogene | predicted gene 7594 |
| 10 | gene | 68.72365 | 68.78266 | positive | MGI_C57BL6J_2143537 | Tmem26 | NCBI_Gene:327766,ENSEMBL:ENSMUSG00000060044 | MGI:2143537 | protein coding gene | transmembrane protein 26 |
| 10 | gene | 68.79890 | 68.82282 | negative | MGI_C57BL6J_5592090 | Gm32931 | NCBI_Gene:102635644 | MGI:5592090 | lncRNA gene | predicted gene, 32931 |
| 10 | gene | 68.90280 | 68.90450 | negative | MGI_C57BL6J_1920605 | 1700048P04Rik | ENSEMBL:ENSMUSG00000112333 | MGI:1920605 | lncRNA gene | RIKEN cDNA 1700048P04 gene |
| 10 | gene | 69.02026 | 69.04451 | negative | MGI_C57BL6J_5592194 | Gm33035 | NCBI_Gene:102635787,ENSEMBL:ENSMUSG00000112331 | MGI:5592194 | lncRNA gene | predicted gene, 33035 |
| 10 | gene | 69.07235 | 69.08988 | negative | MGI_C57BL6J_5592289 | Gm33130 | NCBI_Gene:102635915 | MGI:5592289 | lncRNA gene | predicted gene, 33130 |
| 10 | gene | 69.09666 | 69.09787 | negative | MGI_C57BL6J_6095844 | Gm47107 | ENSEMBL:ENSMUSG00000112083 | MGI:6095844 | lncRNA gene | predicted gene, 47107 |
| 10 | gene | 69.11245 | 69.11744 | positive | MGI_C57BL6J_5623569 | Gm40684 | NCBI_Gene:105245198 | MGI:5623569 | lncRNA gene | predicted gene, 40684 |
| 10 | gene | 69.13288 | 69.13300 | positive | MGI_C57BL6J_5453949 | Gm24172 | ENSEMBL:ENSMUSG00000077315 | MGI:5453949 | snoRNA gene | predicted gene, 24172 |
| 10 | gene | 69.14775 | 69.15113 | negative | MGI_C57BL6J_5592422 | Gm33263 | NCBI_Gene:102636096,ENSEMBL:ENSMUSG00000111941 | MGI:5592422 | lncRNA gene | predicted gene, 33263 |
| 10 | gene | 69.15128 | 69.29179 | positive | MGI_C57BL6J_1916538 | Rhobtb1 | NCBI_Gene:69288,ENSEMBL:ENSMUSG00000019944 | MGI:1916538 | protein coding gene | Rho-related BTB domain containing 1 |
| 10 | gene | 69.20946 | 69.21316 | negative | MGI_C57BL6J_2444562 | A930033H14Rik | NCBI_Gene:320700,ENSEMBL:ENSMUSG00000090622 | MGI:2444562 | protein coding gene | RIKEN cDNA A930033H14 gene |
| 10 | gene | 69.31204 | 69.31542 | positive | MGI_C57BL6J_5623570 | Gm40685 | NCBI_Gene:105245199,ENSEMBL:ENSMUSG00000112798 | MGI:5623570 | lncRNA gene | predicted gene, 40685 |
| 10 | pseudogene | 69.32520 | 69.32567 | positive | MGI_C57BL6J_3780816 | Rpl30-ps7 | NCBI_Gene:100040182 | MGI:3780816 | pseudogene | ribosomal protein L30, pseudogene 7 |
| 10 | gene | 69.32636 | 69.32821 | negative | MGI_C57BL6J_1921141 | 4930423B08Rik | NCBI_Gene:102636160 | MGI:1921141 | lncRNA gene | RIKEN cDNA 4930423B08 gene |
| 10 | gene | 69.33515 | 69.35294 | negative | MGI_C57BL6J_88351 | Cdk1 | NCBI_Gene:12534,ENSEMBL:ENSMUSG00000019942 | MGI:88351 | protein coding gene | cyclin-dependent kinase 1 |
| 10 | pseudogene | 69.37269 | 69.37331 | negative | MGI_C57BL6J_6096016 | Gm47211 | ENSEMBL:ENSMUSG00000112509 | MGI:6096016 | pseudogene | predicted gene, 47211 |
| 10 | gene | 69.39877 | 69.60214 | positive | MGI_C57BL6J_3036232 | 4833431D13Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 4833431D13 gene |
| 10 | gene | 69.39877 | 70.02744 | positive | MGI_C57BL6J_88026 | Ank3 | NCBI_Gene:11735,ENSEMBL:ENSMUSG00000069601 | MGI:88026 | protein coding gene | ankyrin 3, epithelial |
| 10 | pseudogene | 69.42268 | 69.42375 | negative | MGI_C57BL6J_5010821 | Gm18636 | NCBI_Gene:100417479,ENSEMBL:ENSMUSG00000111899 | MGI:5010821 | pseudogene | predicted gene, 18636 |
| 10 | gene | 69.43900 | 69.43974 | negative | MGI_C57BL6J_1925199 | A930011B18Rik | NA | NA | unclassified gene | RIKEN cDNA A930011B18 gene |
| 10 | gene | 69.54109 | 69.55651 | negative | MGI_C57BL6J_5592623 | Gm33464 | NCBI_Gene:102636382 | MGI:5592623 | lncRNA gene | predicted gene, 33464 |
| 10 | pseudogene | 69.56567 | 69.56693 | negative | MGI_C57BL6J_5825868 | Gm46231 | NCBI_Gene:108167818,ENSEMBL:ENSMUSG00000112729 | MGI:5825868 | pseudogene | predicted gene, 46231 |
| 10 | gene | 69.68908 | 69.69018 | positive | MGI_C57BL6J_1924544 | 9430064K01Rik | NA | NA | unclassified gene | RIKEN cDNA 9430064K01 gene |
| 10 | gene | 69.86268 | 69.86639 | negative | MGI_C57BL6J_5592575 | Gm33416 | NCBI_Gene:102636314 | MGI:5592575 | lncRNA gene | predicted gene, 33416 |
| 10 | gene | 69.90705 | 69.90931 | negative | MGI_C57BL6J_5592520 | Gm33361 | NCBI_Gene:102636238 | MGI:5592520 | lncRNA gene | predicted gene, 33361 |
| 10 | gene | 69.90718 | 69.90798 | positive | MGI_C57BL6J_1920163 | 2900006A17Rik | NA | NA | unclassified gene | RIKEN cDNA 2900006A17 gene |
| 10 | gene | 70.09688 | 70.19320 | positive | MGI_C57BL6J_1923801 | Ccdc6 | NCBI_Gene:76551,ENSEMBL:ENSMUSG00000048701 | MGI:1923801 | protein coding gene | coiled-coil domain containing 6 |
| 10 | gene | 70.20466 | 70.21971 | positive | MGI_C57BL6J_1916813 | Mrln | NCBI_Gene:69563,ENSEMBL:ENSMUSG00000019933 | MGI:1916813 | protein coding gene | myoregulin |
| 10 | gene | 70.24510 | 70.28597 | positive | MGI_C57BL6J_1914109 | Slc16a9 | NCBI_Gene:66859,ENSEMBL:ENSMUSG00000037762 | MGI:1914109 | protein coding gene | solute carrier family 16 (monocarboxylic acid transporters), member 9 |
| 10 | gene | 70.28423 | 70.28495 | negative | MGI_C57BL6J_3779671 | Gm7097 | ENSEMBL:ENSMUSG00000089835 | MGI:3779671 | lncRNA gene | predicted gene 7097 |
| 10 | pseudogene | 70.28555 | 70.28818 | negative | MGI_C57BL6J_3708538 | Gm9877 | ENSEMBL:ENSMUSG00000052426 | MGI:3708538 | pseudogene | predicted gene 9877 |
| 10 | gene | 70.28977 | 70.30845 | positive | MGI_C57BL6J_5623572 | Gm40687 | NCBI_Gene:105245201 | MGI:5623572 | lncRNA gene | predicted gene, 40687 |
| 10 | gene | 70.29340 | 70.29406 | positive | MGI_C57BL6J_5592863 | Gm33704 | NCBI_Gene:102636706 | MGI:5592863 | lncRNA gene | predicted gene, 33704 |
| 10 | pseudogene | 70.33830 | 70.33847 | positive | MGI_C57BL6J_5012293 | Gm20108 | NCBI_Gene:102636776,ENSEMBL:ENSMUSG00000112676 | MGI:5012293 | pseudogene | predicted gene, 20108 |
| 10 | gene | 70.41246 | 70.41558 | negative | MGI_C57BL6J_6097213 | Gm47946 | ENSEMBL:ENSMUSG00000112884 | MGI:6097213 | lncRNA gene | predicted gene, 47946 |
| 10 | gene | 70.44008 | 70.55874 | positive | MGI_C57BL6J_1918971 | Fam13c | NCBI_Gene:71721,ENSEMBL:ENSMUSG00000043259 | MGI:1918971 | protein coding gene | family with sequence similarity 13, member C |
| 10 | gene | 70.50119 | 70.50150 | negative | MGI_C57BL6J_5452097 | Gm22320 | ENSEMBL:ENSMUSG00000087774 | MGI:5452097 | unclassified non-coding RNA gene | predicted gene, 22320 |
| 10 | pseudogene | 70.50477 | 70.50734 | negative | MGI_C57BL6J_5588942 | Gm29783 | NCBI_Gene:101056119,ENSEMBL:ENSMUSG00000112273 | MGI:5588942 | pseudogene | predicted gene, 29783 |
| 10 | gene | 70.55768 | 70.65596 | negative | MGI_C57BL6J_1918161 | Phyhipl | NCBI_Gene:70911,ENSEMBL:ENSMUSG00000037747 | MGI:1918161 | protein coding gene | phytanoyl-CoA hydroxylase interacting protein-like |
| 10 | gene | 70.59978 | 70.68021 | positive | MGI_C57BL6J_5825869 | Gm46232 | NCBI_Gene:108167819 | MGI:5825869 | lncRNA gene | predicted gene, 46232 |
| 10 | gene | 70.59983 | 70.60048 | positive | MGI_C57BL6J_2143903 | BB187676 | NA | NA | unclassified gene | expressed sequence BB187676 |
| 10 | gene | 70.60105 | 70.65596 | negative | MGI_C57BL6J_2442462 | D630013N20Rik | NCBI_Gene:319646 | MGI:2442462 | lncRNA gene | RIKEN cDNA D630013N20 gene |
| 10 | gene | 70.62673 | 70.63526 | positive | MGI_C57BL6J_5623573 | Gm40688 | NCBI_Gene:105245202 | MGI:5623573 | lncRNA gene | predicted gene, 40688 |
| 10 | gene | 70.71493 | 70.74045 | negative | MGI_C57BL6J_5593138 | Gm33979 | NCBI_Gene:102637077,ENSEMBL:ENSMUSG00000112607 | MGI:5593138 | lncRNA gene | predicted gene, 33979 |
| 10 | gene | 70.80724 | 70.81382 | positive | MGI_C57BL6J_6097330 | Gm48021 | ENSEMBL:ENSMUSG00000112752 | MGI:6097330 | lncRNA gene | predicted gene, 48021 |
| 10 | gene | 70.81385 | 70.81605 | positive | MGI_C57BL6J_6097332 | Gm48022 | ENSEMBL:ENSMUSG00000112320 | MGI:6097332 | lncRNA gene | predicted gene, 48022 |
| 10 | gene | 70.83075 | 70.84294 | positive | MGI_C57BL6J_5623574 | Gm40689 | NCBI_Gene:105245203,ENSEMBL:ENSMUSG00000112870 | MGI:5623574 | lncRNA gene | predicted gene, 40689 |
| 10 | gene | 70.84702 | 70.85373 | positive | MGI_C57BL6J_6097334 | Gm48023 | ENSEMBL:ENSMUSG00000112883 | MGI:6097334 | lncRNA gene | predicted gene, 48023 |
| 10 | gene | 70.86340 | 70.86845 | negative | MGI_C57BL6J_6097336 | Gm48024 | ENSEMBL:ENSMUSG00000112125 | MGI:6097336 | lncRNA gene | predicted gene, 48024 |
| 10 | gene | 70.86862 | 70.93432 | positive | MGI_C57BL6J_1922439 | 4930533K18Rik | NCBI_Gene:75189,ENSEMBL:ENSMUSG00000047692 | MGI:1922439 | lncRNA gene | RIKEN cDNA 4930533K18 gene |
| 10 | gene | 70.87000 | 70.87242 | negative | MGI_C57BL6J_6097351 | Gm48036 | ENSEMBL:ENSMUSG00000112032 | MGI:6097351 | lncRNA gene | predicted gene, 48036 |
| 10 | gene | 70.88210 | 70.88221 | positive | MGI_C57BL6J_5454263 | Gm24486 | ENSEMBL:ENSMUSG00000094703 | MGI:5454263 | snRNA gene | predicted gene, 24486 |
| 10 | gene | 70.92283 | 71.15970 | negative | MGI_C57BL6J_1933388 | Bicc1 | NCBI_Gene:83675,ENSEMBL:ENSMUSG00000014329 | MGI:1933388 | protein coding gene | BicC family RNA binding protein 1 |
| 10 | gene | 70.97551 | 70.99304 | negative | MGI_C57BL6J_6097338 | Gm48025 | ENSEMBL:ENSMUSG00000112745 | MGI:6097338 | lncRNA gene | predicted gene, 48025 |
| 10 | gene | 71.08288 | 71.08365 | positive | MGI_C57BL6J_3783091 | Gm15647 | ENSEMBL:ENSMUSG00000090212 | MGI:3783091 | lncRNA gene | predicted gene 15647 |
| 10 | gene | 71.12566 | 71.14157 | positive | MGI_C57BL6J_5593296 | Gm34137 | NCBI_Gene:102637280 | MGI:5593296 | lncRNA gene | predicted gene, 34137 |
| 10 | pseudogene | 71.14816 | 71.14981 | positive | MGI_C57BL6J_5591184 | Gm32025 | NCBI_Gene:102634444,ENSEMBL:ENSMUSG00000112566 | MGI:5591184 | pseudogene | predicted gene, 32025 |
| 10 | pseudogene | 71.16291 | 71.16338 | positive | MGI_C57BL6J_3643973 | Gm6331 | NCBI_Gene:622551,ENSEMBL:ENSMUSG00000112748 | MGI:3643973 | pseudogene | predicted gene 6331 |
| 10 | gene | 71.16874 | 71.18205 | negative | MGI_C57BL6J_2442303 | C730027H18Rik | NCBI_Gene:319572,ENSEMBL:ENSMUSG00000112189,ENSEMBL:ENSMUSG00000112366 | MGI:2442303 | lncRNA gene | RIKEN cDNA C730027H18 gene |
| 10 | gene | 71.18104 | 71.20449 | positive | MGI_C57BL6J_1925888 | 1700113B09Rik | NCBI_Gene:78638,ENSEMBL:ENSMUSG00000112028 | MGI:1925888 | lncRNA gene | RIKEN cDNA 1700113B09 gene |
| 10 | gene | 71.18117 | 71.20114 | positive | MGI_C57BL6J_5593487 | Gm34328 | NCBI_Gene:102637547 | MGI:5593487 | lncRNA gene | predicted gene, 34328 |
| 10 | gene | 71.22546 | 71.23833 | negative | MGI_C57BL6J_107810 | Tfam | NCBI_Gene:21780,ENSEMBL:ENSMUSG00000003923 | MGI:107810 | protein coding gene | transcription factor A, mitochondrial |
| 10 | gene | 71.25498 | 71.28526 | negative | MGI_C57BL6J_2384911 | Ube2d1 | NCBI_Gene:216080,ENSEMBL:ENSMUSG00000019927 | MGI:2384911 | protein coding gene | ubiquitin-conjugating enzyme E2D 1 |
| 10 | gene | 71.28773 | 71.29248 | positive | MGI_C57BL6J_5593535 | Gm34376 | NCBI_Gene:102637612,ENSEMBL:ENSMUSG00000112813 | MGI:5593535 | lncRNA gene | predicted gene, 34376 |
| 10 | gene | 71.31074 | 71.31398 | positive | MGI_C57BL6J_5313056 | Gm20609 | NCBI_Gene:102637684,ENSEMBL:ENSMUSG00000093732 | MGI:5313056 | lncRNA gene | predicted gene 20609 |
| 10 | pseudogene | 71.31873 | 71.31943 | positive | MGI_C57BL6J_3643734 | Rpl19-ps2 | NCBI_Gene:665472,ENSEMBL:ENSMUSG00000112582 | MGI:3643734 | pseudogene | ribosomal protein L19, pseudogene 2 |
| 10 | gene | 71.33048 | 71.34495 | negative | MGI_C57BL6J_1261855 | Cisd1 | NCBI_Gene:52637,ENSEMBL:ENSMUSG00000037710 | MGI:1261855 | protein coding gene | CDGSH iron sulfur domain 1 |
| 10 | gene | 71.33670 | 71.33962 | negative | MGI_C57BL6J_2143791 | 4932439E07Rik | NA | NA | unclassified gene | RIKEN cDNA 4932439E07 gene |
| 10 | gene | 71.34776 | 71.43333 | positive | MGI_C57BL6J_1916968 | Ipmk | NCBI_Gene:69718,ENSEMBL:ENSMUSG00000060733 | MGI:1916968 | protein coding gene | inositol polyphosphate multikinase |
| 10 | gene | 71.47640 | 71.48552 | negative | MGI_C57BL6J_1922563 | 4930551I15Rik | ENSEMBL:ENSMUSG00000112108 | MGI:1922563 | lncRNA gene | RIKEN cDNA 4930551I15 gene |
| 10 | pseudogene | 71.48250 | 71.48364 | positive | MGI_C57BL6J_5825870 | Gm46233 | NCBI_Gene:108167820,ENSEMBL:ENSMUSG00000112608 | MGI:5825870 | pseudogene | predicted gene, 46233 |
| 10 | gene | 71.52044 | 71.52230 | negative | MGI_C57BL6J_6095721 | Gm47028 | ENSEMBL:ENSMUSG00000112416 | MGI:6095721 | lncRNA gene | predicted gene, 47028 |
| 10 | gene | 71.52727 | 71.56537 | positive | MGI_C57BL6J_5593645 | Gm34486 | NCBI_Gene:102637756 | MGI:5593645 | lncRNA gene | predicted gene, 34486 |
| 10 | gene | 71.52728 | 71.67884 | positive | MGI_C57BL6J_5593703 | Gm34544 | NCBI_Gene:102637834,ENSEMBL:ENSMUSG00000112126 | MGI:5593703 | lncRNA gene | predicted gene, 34544 |
| 10 | pseudogene | 71.59585 | 71.59713 | negative | MGI_C57BL6J_3644095 | Gm4797 | NCBI_Gene:216081,ENSEMBL:ENSMUSG00000111965 | MGI:3644095 | pseudogene | predicted gene 4797 |
| 10 | pseudogene | 71.61165 | 71.61307 | negative | MGI_C57BL6J_5011259 | Gm19074 | NCBI_Gene:100418206,ENSEMBL:ENSMUSG00000112624 | MGI:5011259 | pseudogene | predicted gene, 19074 |
| 10 | pseudogene | 71.70372 | 71.70417 | positive | MGI_C57BL6J_6095749 | Gm47045 | ENSEMBL:ENSMUSG00000112152 | MGI:6095749 | pseudogene | predicted gene, 47045 |
| 10 | pseudogene | 71.70429 | 71.70786 | positive | MGI_C57BL6J_3644096 | Gm4798 | NCBI_Gene:216082,ENSEMBL:ENSMUSG00000112714 | MGI:3644096 | pseudogene | predicted pseudogene 4798 |
| 10 | pseudogene | 71.71678 | 71.71802 | negative | MGI_C57BL6J_3644102 | Gm6362 | NCBI_Gene:622812,ENSEMBL:ENSMUSG00000112717 | MGI:3644102 | pseudogene | predicted gene 6362 |
| 10 | pseudogene | 71.79895 | 71.79991 | negative | MGI_C57BL6J_3648383 | Gm5080 | NCBI_Gene:327767,ENSEMBL:ENSMUSG00000111999 | MGI:3648383 | pseudogene | predicted gene 5080 |
| 10 | gene | 71.89804 | 71.89817 | positive | MGI_C57BL6J_5452714 | Gm22937 | ENSEMBL:ENSMUSG00000077142 | MGI:5452714 | snoRNA gene | predicted gene, 22937 |
| 10 | gene | 71.96261 | 72.00535 | positive | MGI_C57BL6J_5623575 | Gm40690 | NCBI_Gene:105245205 | MGI:5623575 | lncRNA gene | predicted gene, 40690 |
| 10 | pseudogene | 71.97988 | 71.98069 | negative | MGI_C57BL6J_1920633 | 1700049L16Rik | NCBI_Gene:108950,ENSEMBL:ENSMUSG00000043859 | MGI:1920633 | pseudogene | RIKEN cDNA 1700049L16 gene |
| 10 | pseudogene | 72.01921 | 72.01952 | positive | MGI_C57BL6J_6095770 | Gm47060 | ENSEMBL:ENSMUSG00000112185 | MGI:6095770 | pseudogene | predicted gene, 47060 |
| 10 | gene | 72.02698 | 72.11244 | positive | MGI_C57BL6J_5593768 | Gm34609 | NCBI_Gene:102637916,ENSEMBL:ENSMUSG00000112414 | MGI:5593768 | lncRNA gene | predicted gene, 34609 |
| 10 | pseudogene | 72.30920 | 72.31184 | positive | MGI_C57BL6J_3704365 | Gm9923 | NCBI_Gene:100040505,ENSEMBL:ENSMUSG00000053830 | MGI:3704365 | pseudogene | predicted pseudogene 9923 |
| 10 | gene | 72.52755 | 72.52768 | positive | MGI_C57BL6J_5454761 | Gm24984 | ENSEMBL:ENSMUSG00000077642 | MGI:5454761 | snoRNA gene | predicted gene, 24984 |
| 10 | gene | 72.62307 | 72.64304 | negative | MGI_C57BL6J_5593818 | Gm34659 | NCBI_Gene:102637980 | MGI:5593818 | lncRNA gene | predicted gene, 34659 |
| 10 | gene | 72.65484 | 72.67496 | positive | MGI_C57BL6J_1289227 | Zwint | NCBI_Gene:52696,ENSEMBL:ENSMUSG00000019923 | MGI:1289227 | protein coding gene | ZW10 interactor |
| 10 | gene | 72.65790 | 72.66177 | positive | MGI_C57BL6J_1916486 | 2610034E01Rik | NA | NA | unclassified gene | RIKEN cDNA 2610034E01 gene |
| 10 | pseudogene | 72.71793 | 72.71849 | positive | MGI_C57BL6J_6095826 | Gm47094 | ENSEMBL:ENSMUSG00000112862 | MGI:6095826 | pseudogene | predicted gene, 47094 |
| 10 | gene | 72.93994 | 72.97316 | negative | MGI_C57BL6J_5593935 | Gm34776 | NCBI_Gene:102638141,ENSEMBL:ENSMUSG00000112168 | MGI:5593935 | lncRNA gene | predicted gene, 34776 |
| 10 | gene | 72.97330 | 73.08670 | negative | MGI_C57BL6J_3041175 | A330049N07Rik | NCBI_Gene:327768,ENSEMBL:ENSMUSG00000111994 | MGI:3041175 | lncRNA gene | RIKEN cDNA A330049N07 gene |
| 10 | gene | 73.09627 | 74.64983 | positive | MGI_C57BL6J_1891428 | Pcdh15 | NCBI_Gene:11994,ENSEMBL:ENSMUSG00000052613 | MGI:1891428 | protein coding gene | protocadherin 15 |
| 10 | gene | 73.18293 | 73.18332 | negative | MGI_C57BL6J_5610666 | Gm37438 | ENSEMBL:ENSMUSG00000102584 | MGI:5610666 | unclassified gene | predicted gene, 37438 |
| 10 | gene | 73.20140 | 73.32717 | negative | MGI_C57BL6J_3705254 | Gm15398 | ENSEMBL:ENSMUSG00000085456 | MGI:3705254 | lncRNA gene | predicted gene 15398 |
| 10 | pseudogene | 73.20153 | 73.20277 | positive | MGI_C57BL6J_3646275 | Gm5778 | NCBI_Gene:544705,ENSEMBL:ENSMUSG00000091825 | MGI:3646275 | pseudogene | predicted gene 5778 |
| 10 | pseudogene | 73.27574 | 73.27609 | positive | MGI_C57BL6J_6095833 | Gm47098 | ENSEMBL:ENSMUSG00000112838 | MGI:6095833 | pseudogene | predicted gene, 47098 |
| 10 | pseudogene | 73.37233 | 73.37603 | negative | MGI_C57BL6J_5011353 | Gm19168 | NCBI_Gene:100418377,ENSEMBL:ENSMUSG00000112427 | MGI:5011353 | pseudogene | predicted gene, 19168 |
| 10 | gene | 73.48997 | 73.51839 | positive | MGI_C57BL6J_3705298 | Gm15397 | ENSEMBL:ENSMUSG00000085176 | MGI:3705298 | lncRNA gene | predicted gene 15397 |
| 10 | pseudogene | 73.69010 | 73.69101 | negative | MGI_C57BL6J_3642965 | Gm6407 | NCBI_Gene:623191,ENSEMBL:ENSMUSG00000093548 | MGI:3642965 | pseudogene | predicted gene 6407 |
| 10 | gene | 73.88745 | 73.88812 | positive | MGI_C57BL6J_1925054 | A930019H14Rik | NA | NA | unclassified gene | RIKEN cDNA A930019H14 gene |
| 10 | gene | 74.12754 | 74.13688 | negative | MGI_C57BL6J_5623576 | Gm40691 | NCBI_Gene:105245206 | MGI:5623576 | lncRNA gene | predicted gene, 40691 |
| 10 | pseudogene | 74.82063 | 74.82149 | positive | MGI_C57BL6J_3644650 | Gm7034 | NCBI_Gene:630453,ENSEMBL:ENSMUSG00000112686 | MGI:3644650 | pseudogene | predicted gene 7034 |
| 10 | pseudogene | 74.86286 | 74.86351 | negative | MGI_C57BL6J_3647564 | Gm6419 | NCBI_Gene:623346,ENSEMBL:ENSMUSG00000072407 | MGI:3647564 | pseudogene | predicted gene 6419 |
| 10 | gene | 74.95748 | 75.03435 | negative | MGI_C57BL6J_1918486 | Rsph14 | NCBI_Gene:71236,ENSEMBL:ENSMUSG00000009070 | MGI:1918486 | protein coding gene | radial spoke head homolog 14 (Chlamydomonas) |
| 10 | gene | 74.96709 | 75.01691 | positive | MGI_C57BL6J_95780 | Gnaz | NCBI_Gene:14687,ENSEMBL:ENSMUSG00000040009 | MGI:95780 | protein coding gene | guanine nucleotide binding protein, alpha z subunit |
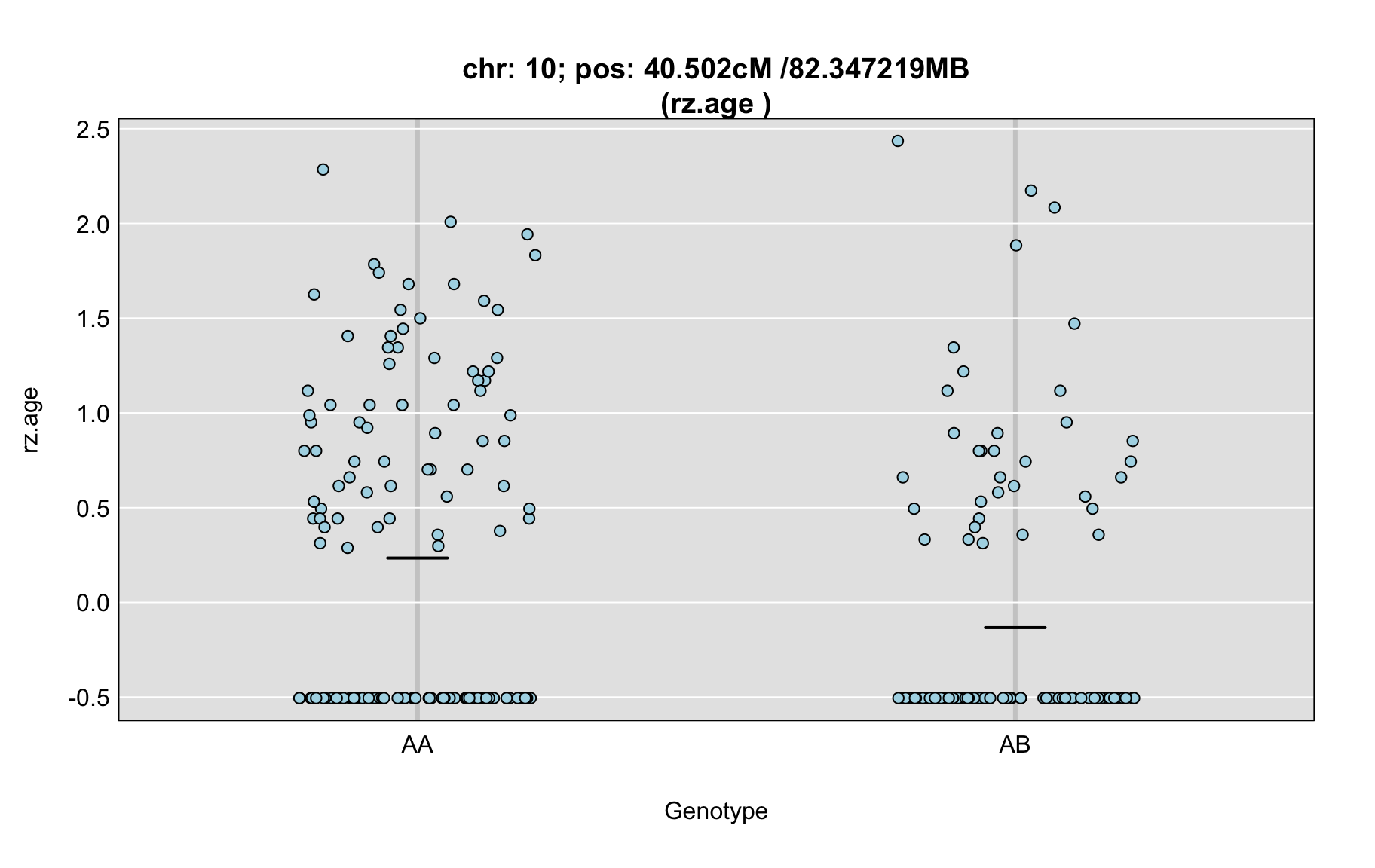

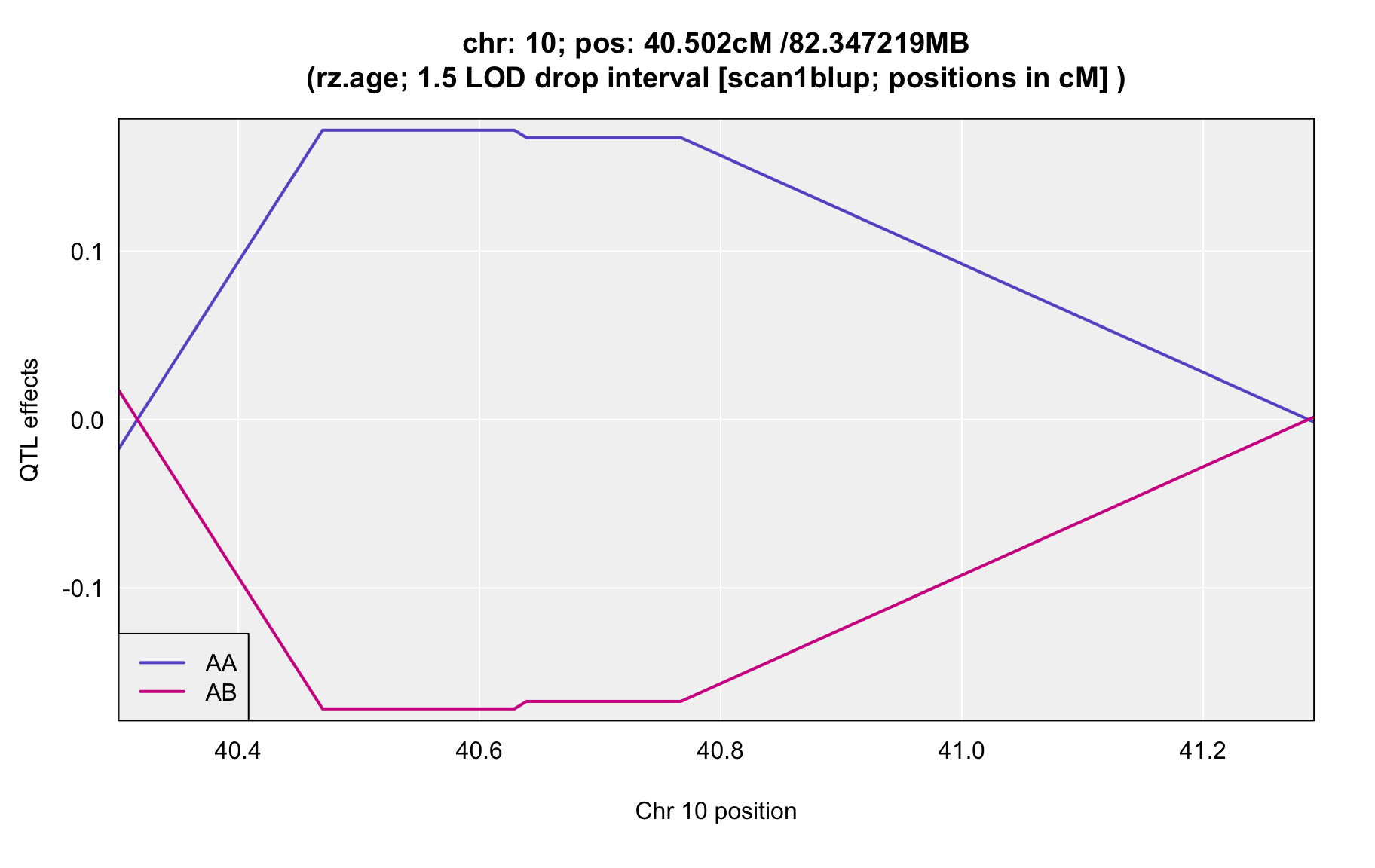
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 10 | gene | 81.64906 | 81.66736 | positive | MGI_C57BL6J_3809656 | Gm10778 | NCBI_Gene:100233208,ENSEMBL:ENSMUSG00000096856 | MGI:3809656 | protein coding gene | predicted gene 10778 |
| 10 | pseudogene | 81.67912 | 81.68051 | positive | MGI_C57BL6J_6096529 | Gm47527 | ENSEMBL:ENSMUSG00000112585 | MGI:6096529 | pseudogene | predicted gene, 47527 |
| 10 | pseudogene | 81.69368 | 81.69524 | negative | MGI_C57BL6J_3645531 | Gm8112 | NCBI_Gene:108167775,ENSEMBL:ENSMUSG00000112001 | MGI:3645531 | pseudogene | predicted gene 8112 |
| 10 | gene | 81.70419 | 81.72669 | positive | MGI_C57BL6J_1920860 | Zfp433 | NCBI_Gene:73610,ENSEMBL:ENSMUSG00000096795 | MGI:1920860 | protein coding gene | zinc finger protein 433 |
| 10 | pseudogene | 81.71460 | 81.71650 | positive | MGI_C57BL6J_3647617 | Gm8249 | NCBI_Gene:105245234,ENSEMBL:ENSMUSG00000112061 | MGI:3647617 | pseudogene | predicted gene 8249 |
| 10 | pseudogene | 81.71704 | 81.71756 | negative | MGI_C57BL6J_6096532 | Gm47529 | ENSEMBL:ENSMUSG00000112339 | MGI:6096532 | pseudogene | predicted gene, 47529 |
| 10 | pseudogene | 81.72790 | 81.72909 | positive | MGI_C57BL6J_3642939 | Gm5173 | NCBI_Gene:382388,ENSEMBL:ENSMUSG00000112621 | MGI:3642939 | pseudogene | predicted gene 5173 |
| 10 | gene | 81.74282 | 81.77116 | negative | MGI_C57BL6J_3809197 | Gm3055 | NCBI_Gene:100040944,ENSEMBL:ENSMUSG00000094622 | MGI:3809197 | protein coding gene | predicted gene 3055 |
| 10 | pseudogene | 81.80443 | 81.80570 | positive | MGI_C57BL6J_6096545 | Gm47537 | ENSEMBL:ENSMUSG00000112877 | MGI:6096545 | pseudogene | predicted gene, 47537 |
| 10 | gene | 81.81194 | 81.82649 | positive | MGI_C57BL6J_3644409 | Gm4767 | NCBI_Gene:210583,ENSEMBL:ENSMUSG00000094076 | MGI:3644409 | protein coding gene | predicted gene 4767 |
| 10 | pseudogene | 81.81843 | 81.81898 | negative | MGI_C57BL6J_3647363 | Gm8264 | NCBI_Gene:666738,ENSEMBL:ENSMUSG00000112740 | MGI:3647363 | pseudogene | predicted gene 8264 |
| 10 | pseudogene | 81.85277 | 81.85332 | negative | MGI_C57BL6J_3647467 | Gm8137 | NCBI_Gene:666501,ENSEMBL:ENSMUSG00000112697 | MGI:3647467 | pseudogene | predicted gene 8137 |
| 10 | gene | 81.86344 | 81.88291 | positive | MGI_C57BL6J_5591846 | Gm32687 | NCBI_Gene:102635315,ENSEMBL:ENSMUSG00000112640 | MGI:5591846 | protein coding gene | predicted gene, 32687 |
| 10 | pseudogene | 81.87371 | 81.87557 | positive | MGI_C57BL6J_3648016 | Gm8274 | NCBI_Gene:105245237,ENSEMBL:ENSMUSG00000112196 | MGI:3648016 | pseudogene | predicted gene 8274 |
| 10 | pseudogene | 81.87615 | 81.87667 | negative | MGI_C57BL6J_6096547 | Gm47539 | ENSEMBL:ENSMUSG00000112760 | MGI:6096547 | pseudogene | predicted gene, 47539 |
| 10 | pseudogene | 81.88699 | 81.88818 | positive | MGI_C57BL6J_3644502 | Gm8153 | NCBI_Gene:666537,ENSEMBL:ENSMUSG00000112311 | MGI:3644502 | pseudogene | predicted gene 8153 |
| 10 | gene | 81.90191 | 81.93127 | negative | MGI_C57BL6J_3696710 | Zfp781 | NCBI_Gene:331188,ENSEMBL:ENSMUSG00000096718 | MGI:3696710 | protein coding gene | zinc finger protein 781 |
| 10 | pseudogene | 81.96380 | 81.96509 | positive | MGI_C57BL6J_6096560 | Gm47546 | ENSEMBL:ENSMUSG00000112205 | MGI:6096560 | pseudogene | predicted gene, 47546 |
| 10 | gene | 81.97809 | 81.99536 | positive | MGI_C57BL6J_5011987 | Gm19802 | NCBI_Gene:105245236 | MGI:5011987 | protein coding gene | predicted gene, 19802 |
| 10 | pseudogene | 81.98590 | 81.98645 | negative | MGI_C57BL6J_3647883 | Gm8158 | NCBI_Gene:666548,ENSEMBL:ENSMUSG00000112051 | MGI:3647883 | pseudogene | predicted gene 8158 |
| 10 | gene | 81.99597 | 82.01164 | positive | MGI_C57BL6J_3642231 | Gm10777 | NCBI_Gene:102635133 | MGI:3642231 | lncRNA gene | predicted gene 10777 |
| 10 | pseudogene | 82.00829 | 82.00981 | positive | MGI_C57BL6J_3648461 | Gm8290 | NCBI_Gene:666790,ENSEMBL:ENSMUSG00000111985 | MGI:3648461 | pseudogene | predicted gene 8290 |
| 10 | pseudogene | 82.00948 | 82.00980 | negative | MGI_C57BL6J_5825831 | Gm46194 | NCBI_Gene:108167776,ENSEMBL:ENSMUSG00000112502 | MGI:5825831 | pseudogene | predicted gene, 46194 |
| 10 | gene | 82.01170 | 82.01723 | positive | MGI_C57BL6J_5592537 | Gm33378 | NCBI_Gene:102636264,ENSEMBL:ENSMUSG00000112790 | MGI:5592537 | protein coding gene | predicted gene, 33378 |
| 10 | pseudogene | 82.02053 | 82.02171 | positive | MGI_C57BL6J_3644928 | Gm8170 | NCBI_Gene:666571,ENSEMBL:ENSMUSG00000112381 | MGI:3644928 | pseudogene | predicted gene 8170 |
| 10 | gene | 82.04464 | 82.04804 | negative | MGI_C57BL6J_3034336 | AL117821 | NA | NA | unclassified gene | expressed sequence AL117821 |
| 10 | gene | 82.04645 | 82.04805 | negative | MGI_C57BL6J_5825832 | Gm46195 | NCBI_Gene:108167777 | MGI:5825832 | lncRNA gene | predicted gene, 46195 |
| 10 | gene | 82.04809 | 82.07773 | positive | MGI_C57BL6J_3040689 | Zfp873 | NCBI_Gene:408062,ENSEMBL:ENSMUSG00000061371 | MGI:3040689 | protein coding gene | zinc finger protein 873 |
| 10 | pseudogene | 82.05611 | 82.05724 | negative | MGI_C57BL6J_5010741 | Gm18556 | NCBI_Gene:100417356,ENSEMBL:ENSMUSG00000112373 | MGI:5010741 | pseudogene | predicted gene, 18556 |
| 10 | gene | 82.08301 | 82.08362 | negative | MGI_C57BL6J_3041223 | D430050E20Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA D430050E20 gene |
| 10 | gene | 82.08332 | 82.11122 | positive | MGI_C57BL6J_2670978 | BC024063 | NCBI_Gene:666584,ENSEMBL:ENSMUSG00000112160 | MGI:2670978 | protein coding gene | cDNA sequence BC024063 |
| 10 | gene | 82.08341 | 82.08567 | positive | MGI_C57BL6J_3041160 | 4933439G19Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 4933439G19 gene |
| 10 | gene | 82.12668 | 82.12829 | negative | MGI_C57BL6J_1918064 | 4921506L19Rik | NA | NA | unclassified gene | RIKEN cDNA 4921506L19 gene |
| 10 | gene | 82.12801 | 82.15306 | positive | MGI_C57BL6J_2143755 | AU041133 | NCBI_Gene:216177,ENSEMBL:ENSMUSG00000078435 | MGI:2143755 | protein coding gene | expressed sequence AU041133 |
| 10 | gene | 82.15710 | 82.16046 | positive | MGI_C57BL6J_6096602 | Gm47572 | ENSEMBL:ENSMUSG00000112922 | MGI:6096602 | unclassified gene | predicted gene, 47572 |
| 10 | gene | 82.16425 | 82.16678 | positive | MGI_C57BL6J_6096604 | Gm47573 | ENSEMBL:ENSMUSG00000112847 | MGI:6096604 | unclassified gene | predicted gene, 47573 |
| 10 | gene | 82.17763 | 82.19129 | positive | MGI_C57BL6J_5623605 | Gm40720 | NCBI_Gene:105245239 | MGI:5623605 | lncRNA gene | predicted gene, 40720 |
| 10 | gene | 82.17824 | 82.20149 | positive | MGI_C57BL6J_3801804 | Gm15608 | NCBI_Gene:105245241,ENSEMBL:ENSMUSG00000112110 | MGI:3801804 | lncRNA gene | predicted gene 15608 |
| 10 | gene | 82.17842 | 82.18109 | positive | MGI_C57BL6J_3642233 | Gm10776 | NA | NA | unclassified gene | predicted gene 10776 |
| 10 | pseudogene | 82.21905 | 82.22446 | positive | MGI_C57BL6J_3782155 | Gm3982 | NCBI_Gene:105245240 | MGI:3782155 | pseudogene | predicted gene 3982 |
| 10 | gene | 82.22457 | 82.24128 | negative | MGI_C57BL6J_3621440 | Zfp938 | NCBI_Gene:237411,ENSEMBL:ENSMUSG00000062931 | MGI:3621440 | protein coding gene | zinc finger protein 938 |
| 10 | pseudogene | 82.26645 | 82.26687 | negative | MGI_C57BL6J_6096302 | Gm47382 | ENSEMBL:ENSMUSG00000111930 | MGI:6096302 | pseudogene | predicted gene, 47382 |
| 10 | gene | 82.28211 | 82.31658 | negative | MGI_C57BL6J_3045298 | 4932415D10Rik | NCBI_Gene:102635990,ENSEMBL:ENSMUSG00000044581 | MGI:3045298 | protein coding gene | RIKEN cDNA 4932415D10 gene |
| 10 | gene | 82.31538 | 82.31656 | negative | MGI_C57BL6J_1918439 | 4933426I03Rik | NA | NA | unclassified gene | RIKEN cDNA 4933426I03 gene |
| 10 | gene | 82.35422 | 82.42288 | positive | MGI_C57BL6J_3643133 | Gm4924 | NCBI_Gene:237412,ENSEMBL:ENSMUSG00000073427 | MGI:3643133 | protein coding gene | predicted gene 4924 |
| 10 | pseudogene | 82.43070 | 82.43239 | positive | MGI_C57BL6J_3646272 | Gm8188 | NCBI_Gene:666611,ENSEMBL:ENSMUSG00000112163 | MGI:3646272 | pseudogene | predicted gene 8188 |
| 10 | pseudogene | 82.44262 | 82.44294 | positive | MGI_C57BL6J_6096993 | Gm47809 | ENSEMBL:ENSMUSG00000112070 | MGI:6096993 | pseudogene | predicted gene, 47809 |
| 10 | gene | 82.48653 | 82.49262 | negative | MGI_C57BL6J_2686399 | Gm1553 | NCBI_Gene:432480,ENSEMBL:ENSMUSG00000094186 | MGI:2686399 | protein coding gene | predicted gene 1553 |
| 10 | pseudogene | 82.56677 | 82.56751 | positive | MGI_C57BL6J_5011371 | Gm19186 | NCBI_Gene:100418403,ENSEMBL:ENSMUSG00000112560 | MGI:5011371 | pseudogene | predicted gene, 19186 |
| 10 | pseudogene | 82.58538 | 82.58754 | positive | MGI_C57BL6J_3645307 | Ap3m1-ps | NCBI_Gene:544716,ENSEMBL:ENSMUSG00000094683 | MGI:3645307 | pseudogene | adaptor-related protein complex 3, mu 1 subunit, pseudogene |
| 10 | gene | 82.61326 | 82.61895 | negative | MGI_C57BL6J_5623606 | Gm40721 | NCBI_Gene:105245242 | MGI:5623606 | lncRNA gene | predicted gene, 40721 |
| 10 | gene | 82.61985 | 82.62323 | negative | MGI_C57BL6J_3698433 | 1190007I07Rik | NCBI_Gene:544717,ENSEMBL:ENSMUSG00000063320 | MGI:3698433 | protein coding gene | RIKEN cDNA 1190007I07 gene |
| 10 | gene | 82.62982 | 82.65080 | positive | MGI_C57BL6J_108247 | Tdg | NCBI_Gene:21665,ENSEMBL:ENSMUSG00000034674 | MGI:108247 | protein coding gene | thymine DNA glycosylase |
| 10 | gene | 82.65043 | 82.69412 | negative | MGI_C57BL6J_1922032 | Glt8d2 | NCBI_Gene:74782,ENSEMBL:ENSMUSG00000020251 | MGI:1922032 | protein coding gene | glycosyltransferase 8 domain containing 2 |
| 10 | gene | 82.69616 | 82.79587 | positive | MGI_C57BL6J_1915183 | Hcfc2 | NCBI_Gene:67933,ENSEMBL:ENSMUSG00000020246 | MGI:1915183 | protein coding gene | host cell factor C2 |
| 10 | gene | 82.74870 | 82.76414 | negative | MGI_C57BL6J_97317 | Nfyb | NCBI_Gene:18045,ENSEMBL:ENSMUSG00000020248 | MGI:97317 | protein coding gene | nuclear transcription factor-Y beta |
| 10 | pseudogene | 82.76974 | 82.76990 | negative | MGI_C57BL6J_6096583 | Gm47560 | ENSEMBL:ENSMUSG00000112107 | MGI:6096583 | pseudogene | predicted gene, 47560 |
| 10 | gene | 82.77132 | 82.77778 | positive | MGI_C57BL6J_6096584 | Gm47561 | ENSEMBL:ENSMUSG00000112286 | MGI:6096584 | lncRNA gene | predicted gene, 47561 |
| 10 | gene | 82.81212 | 82.82424 | positive | MGI_C57BL6J_1917253 | 1700028I16Rik | NCBI_Gene:70003,ENSEMBL:ENSMUSG00000056821 | MGI:1917253 | lncRNA gene | RIKEN cDNA 1700028I16 gene |
| 10 | gene | 82.83395 | 82.89772 | positive | MGI_C57BL6J_1354175 | Txnrd1 | NCBI_Gene:50493,ENSEMBL:ENSMUSG00000020250 | MGI:1354175 | protein coding gene | thioredoxin reductase 1 |
| 10 | gene | 82.84534 | 82.85921 | negative | MGI_C57BL6J_5621445 | Gm38560 | NCBI_Gene:102641476,ENSEMBL:ENSMUSG00000112375 | MGI:5621445 | lncRNA gene | predicted gene, 38560 |
| 10 | gene | 82.85125 | 82.85265 | negative | MGI_C57BL6J_6095792 | Gm47073 | ENSEMBL:ENSMUSG00000112889 | MGI:6095792 | unclassified gene | predicted gene, 47073 |
| 10 | gene | 82.86663 | 82.86793 | positive | MGI_C57BL6J_1913591 | Eid3 | NCBI_Gene:66341,ENSEMBL:ENSMUSG00000109864 | MGI:1913591 | protein coding gene | EP300 interacting inhibitor of differentiation 3 |
| 10 | gene | 82.89138 | 82.89502 | positive | MGI_C57BL6J_2442548 | 9130221J17Rik | NA | NA | unclassified gene | RIKEN cDNA 9130221J17 gene |
| 10 | gene | 82.92700 | 82.93464 | negative | MGI_C57BL6J_5592940 | Gm33781 | NCBI_Gene:102636812 | MGI:5592940 | lncRNA gene | predicted gene, 33781 |
| 10 | pseudogene | 82.95433 | 82.95508 | positive | MGI_C57BL6J_3648850 | Gm4799 | NCBI_Gene:216185,ENSEMBL:ENSMUSG00000071151 | MGI:3648850 | pseudogene | predicted gene 4799 |
| 10 | gene | 82.98538 | 83.19590 | positive | MGI_C57BL6J_1927166 | Chst11 | NCBI_Gene:58250,ENSEMBL:ENSMUSG00000034612 | MGI:1927166 | protein coding gene | carbohydrate sulfotransferase 11 |
| 10 | gene | 83.02776 | 83.03169 | negative | MGI_C57BL6J_3641797 | Gm10773 | ENSEMBL:ENSMUSG00000112084 | MGI:3641797 | protein coding gene | predicted gene 10773 |
| 10 | gene | 83.13915 | 83.14204 | negative | MGI_C57BL6J_1916665 | 1700025N21Rik | ENSEMBL:ENSMUSG00000090200 | MGI:1916665 | lncRNA gene | RIKEN cDNA 1700025N21 gene |
| 10 | gene | 83.15802 | 83.15813 | negative | MGI_C57BL6J_5452899 | Gm23122 | ENSEMBL:ENSMUSG00000088270 | MGI:5452899 | snRNA gene | predicted gene, 23122 |
| 10 | gene | 83.23085 | 83.33833 | negative | MGI_C57BL6J_2442940 | Slc41a2 | NCBI_Gene:338365,ENSEMBL:ENSMUSG00000034591 | MGI:2442940 | protein coding gene | solute carrier family 41, member 2 |
| 10 | pseudogene | 83.33272 | 83.33336 | positive | MGI_C57BL6J_6095950 | Gm47170 | ENSEMBL:ENSMUSG00000112577 | MGI:6095950 | pseudogene | predicted gene, 47170 |
| 10 | gene | 83.33870 | 83.33963 | positive | MGI_C57BL6J_5623607 | Gm40722 | NCBI_Gene:105245243 | MGI:5623607 | lncRNA gene | predicted gene, 40722 |
| 10 | gene | 83.34395 | 83.35139 | positive | MGI_C57BL6J_5623608 | Gm40723 | NCBI_Gene:105245244,ENSEMBL:ENSMUSG00000112134 | MGI:5623608 | lncRNA gene | predicted gene, 40723 |
| 10 | gene | 83.36022 | 83.48851 | positive | MGI_C57BL6J_106381 | D10Wsu102e | NCBI_Gene:28109,ENSEMBL:ENSMUSG00000020255 | MGI:106381 | protein coding gene | DNA segment, Chr 10, Wayne State University 102, expressed |
| 10 | pseudogene | 83.47800 | 83.48125 | negative | MGI_C57BL6J_3648990 | Gm8270 | NCBI_Gene:102636894 | MGI:3648990 | pseudogene | predicted gene 8270 |
| 10 | gene | 83.48745 | 83.53415 | negative | MGI_C57BL6J_2444680 | Aldh1l2 | NCBI_Gene:216188,ENSEMBL:ENSMUSG00000020256 | MGI:2444680 | protein coding gene | aldehyde dehydrogenase 1 family, member L2 |
| 10 | gene | 83.54375 | 83.59647 | positive | MGI_C57BL6J_2441787 | Washc4 | NCBI_Gene:319277,ENSEMBL:ENSMUSG00000034560 | MGI:2441787 | protein coding gene | WASH complex subunit 4 |
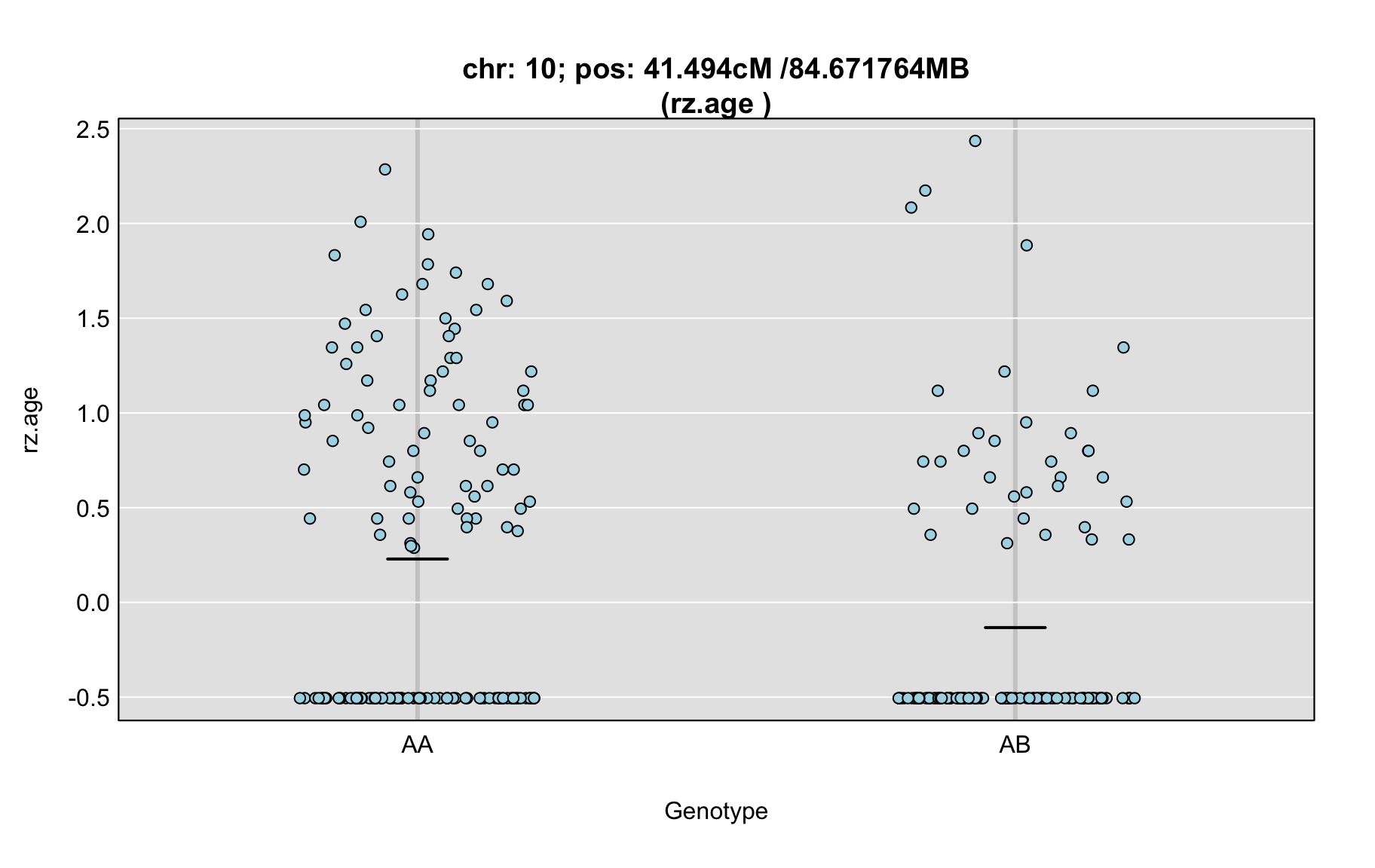


| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 10 | gene | 84.03656 | 84.10088 | negative | MGI_C57BL6J_5593343 | Gm34184 | NCBI_Gene:102637349,ENSEMBL:ENSMUSG00000111049 | MGI:5593343 | lncRNA gene | predicted gene, 34184 |
| 10 | gene | 84.08463 | 84.09625 | positive | MGI_C57BL6J_5593272 | Gm34113 | NCBI_Gene:102637247,ENSEMBL:ENSMUSG00000111524 | MGI:5593272 | lncRNA gene | predicted gene, 34113 |
| 10 | gene | 84.10438 | 84.11042 | negative | MGI_C57BL6J_5623612 | Gm40727 | NCBI_Gene:105245248 | MGI:5623612 | lncRNA gene | predicted gene, 40727 |
| 10 | gene | 84.19182 | 84.22206 | negative | MGI_C57BL6J_6096082 | Gm47253 | ENSEMBL:ENSMUSG00000111062 | MGI:6096082 | lncRNA gene | predicted gene, 47253 |
| 10 | pseudogene | 84.30887 | 84.30936 | positive | MGI_C57BL6J_3643901 | Gm5425 | NCBI_Gene:432482,ENSEMBL:ENSMUSG00000111875 | MGI:3643901 | pseudogene | predicted gene 5425 |
| 10 | gene | 84.31732 | 84.33575 | negative | MGI_C57BL6J_5825838 | Gm46201 | NCBI_Gene:108167783 | MGI:5825838 | lncRNA gene | predicted gene, 46201 |
| 10 | gene | 84.37090 | 84.44060 | negative | MGI_C57BL6J_1925226 | Nuak1 | NCBI_Gene:77976,ENSEMBL:ENSMUSG00000020032 | MGI:1925226 | protein coding gene | NUAK family, SNF1-like kinase, 1 |
| 10 | gene | 84.44083 | 84.46701 | positive | MGI_C57BL6J_6097239 | Gm47962 | ENSEMBL:ENSMUSG00000111583 | MGI:6097239 | lncRNA gene | predicted gene, 47962 |
| 10 | gene | 84.48829 | 84.49768 | positive | MGI_C57BL6J_1923054 | 4930463O16Rik | NCBI_Gene:75804,ENSEMBL:ENSMUSG00000020033 | MGI:1923054 | lncRNA gene | RIKEN cDNA 4930463O16 gene |
| 10 | gene | 84.49887 | 84.49963 | positive | MGI_C57BL6J_1921541 | 1700092E16Rik | ENSEMBL:ENSMUSG00000111487 | MGI:1921541 | unclassified non-coding RNA gene | RIKEN cDNA 1700092E16 gene |
| 10 | gene | 84.50004 | 84.51710 | negative | MGI_C57BL6J_5593564 | Gm34405 | NCBI_Gene:102637645 | MGI:5593564 | lncRNA gene | predicted gene, 34405 |
| 10 | pseudogene | 84.50203 | 84.50256 | negative | MGI_C57BL6J_5010436 | Gm18251 | NCBI_Gene:100416788,ENSEMBL:ENSMUSG00000111478 | MGI:5010436 | pseudogene | predicted gene, 18251 |
| 10 | gene | 84.51805 | 84.52327 | positive | MGI_C57BL6J_3045388 | E230014E18Rik | ENSEMBL:ENSMUSG00000111514 | MGI:3045388 | lncRNA gene | RIKEN cDNA E230014E18 gene |
| 10 | gene | 84.52630 | 84.53406 | negative | MGI_C57BL6J_2444926 | Ckap4 | NCBI_Gene:216197,ENSEMBL:ENSMUSG00000046841 | MGI:2444926 | protein coding gene | cytoskeleton-associated protein 4 |
| 10 | gene | 84.54742 | 84.55334 | negative | MGI_C57BL6J_3045387 | Platr7 | NCBI_Gene:442847,ENSEMBL:ENSMUSG00000097076 | MGI:3045387 | lncRNA gene | pluripotency associated transcript 7 |
| 10 | gene | 84.57232 | 84.57507 | positive | MGI_C57BL6J_5593674 | Gm34515 | NCBI_Gene:102637793 | MGI:5593674 | lncRNA gene | predicted gene, 34515 |
| 10 | gene | 84.57663 | 84.61436 | positive | MGI_C57BL6J_2444679 | Tcp11l2 | NCBI_Gene:216198,ENSEMBL:ENSMUSG00000020034 | MGI:2444679 | protein coding gene | t-complex 11 (mouse) like 2 |
| 10 | gene | 84.61472 | 84.61481 | positive | MGI_C57BL6J_5455434 | Gm25657 | ENSEMBL:ENSMUSG00000089096 | MGI:5455434 | snRNA gene | predicted gene, 25657 |
| 10 | gene | 84.61834 | 84.62753 | negative | MGI_C57BL6J_4936883 | Gm17249 | NCBI_Gene:102637873,ENSEMBL:ENSMUSG00000097309 | MGI:4936883 | lncRNA gene | predicted gene, 17249 |
| 10 | gene | 84.62229 | 84.72718 | positive | MGI_C57BL6J_1917678 | Polr3b | NCBI_Gene:70428,ENSEMBL:ENSMUSG00000034453 | MGI:1917678 | protein coding gene | polymerase (RNA) III (DNA directed) polypeptide B |
| 10 | gene | 84.75605 | 84.90654 | positive | MGI_C57BL6J_1918387 | Rfx4 | NCBI_Gene:71137,ENSEMBL:ENSMUSG00000020037 | MGI:1918387 | protein coding gene | regulatory factor X, 4 (influences HLA class II expression) |
| 10 | gene | 84.79795 | 84.86329 | negative | MGI_C57BL6J_5623613 | Gm40728 | NCBI_Gene:105245249 | MGI:5623613 | lncRNA gene | predicted gene, 40728 |
| 10 | gene | 84.81109 | 84.81118 | negative | MGI_C57BL6J_5454003 | Gm24226 | ENSEMBL:ENSMUSG00000089451 | MGI:5454003 | snRNA gene | predicted gene, 24226 |
| 10 | gene | 84.91725 | 85.01834 | positive | MGI_C57BL6J_2682307 | Ric8b | NCBI_Gene:237422,ENSEMBL:ENSMUSG00000035620 | MGI:2682307 | protein coding gene | RIC8 guanine nucleotide exchange factor B |
| 10 | gene | 84.98771 | 85.00365 | negative | MGI_C57BL6J_5593792 | Gm34633 | NCBI_Gene:102637948 | MGI:5593792 | lncRNA gene | predicted gene, 34633 |
| 10 | gene | 85.09629 | 85.10250 | negative | MGI_C57BL6J_1338765 | Fhl4 | NCBI_Gene:14202,ENSEMBL:ENSMUSG00000050035 | MGI:1338765 | protein coding gene | four and a half LIM domains 4 |
| 10 | gene | 85.10263 | 85.12062 | positive | MGI_C57BL6J_2143652 | Tmem263 | NCBI_Gene:103266,ENSEMBL:ENSMUSG00000060935 | MGI:2143652 | protein coding gene | transmembrane protein 263 |
| 10 | gene | 85.11943 | 85.12807 | negative | MGI_C57BL6J_1921488 | Mterf2 | NCBI_Gene:74238,ENSEMBL:ENSMUSG00000049038 | MGI:1921488 | protein coding gene | mitochondrial transcription termination factor 2 |
| 10 | gene | 85.13170 | 85.18506 | negative | MGI_C57BL6J_1270841 | Cry1 | NCBI_Gene:12952,ENSEMBL:ENSMUSG00000020038 | MGI:1270841 | protein coding gene | cryptochrome 1 (photolyase-like) |
| 10 | gene | 85.22094 | 85.26639 | negative | MGI_C57BL6J_5621460 | Gm38575 | ENSEMBL:ENSMUSG00000111752 | MGI:5621460 | lncRNA gene | predicted gene, 38575 |
| 10 | gene | 85.28423 | 85.28433 | negative | MGI_C57BL6J_5530774 | Gm27392 | ENSEMBL:ENSMUSG00000098852 | MGI:5530774 | miRNA gene | predicted gene, 27392 |
| 10 | pseudogene | 85.31347 | 85.31444 | positive | MGI_C57BL6J_3647964 | Gm8394 | NCBI_Gene:666974,ENSEMBL:ENSMUSG00000050490 | MGI:3647964 | pseudogene | predicted gene 8394 |
| 10 | gene | 85.33269 | 85.34757 | negative | MGI_C57BL6J_5593871 | Gm34712 | NCBI_Gene:102638057 | MGI:5593871 | lncRNA gene | predicted gene, 34712 |
| 10 | gene | 85.33390 | 85.33400 | positive | MGI_C57BL6J_5455058 | Gm25281 | ENSEMBL:ENSMUSG00000087895 | MGI:5455058 | miRNA gene | predicted gene, 25281 |
| 10 | gene | 85.33901 | 85.33995 | positive | MGI_C57BL6J_5623615 | Gm40730 | NCBI_Gene:105245251 | MGI:5623615 | lncRNA gene | predicted gene, 40730 |
| 10 | gene | 85.38677 | 85.66029 | positive | MGI_C57BL6J_1921257 | Btbd11 | NCBI_Gene:74007,ENSEMBL:ENSMUSG00000020042 | MGI:1921257 | protein coding gene | BTB (POZ) domain containing 11 |
| 10 | gene | 85.57888 | 85.58399 | negative | MGI_C57BL6J_5623616 | Gm40731 | NCBI_Gene:105245252 | MGI:5623616 | lncRNA gene | predicted gene, 40731 |
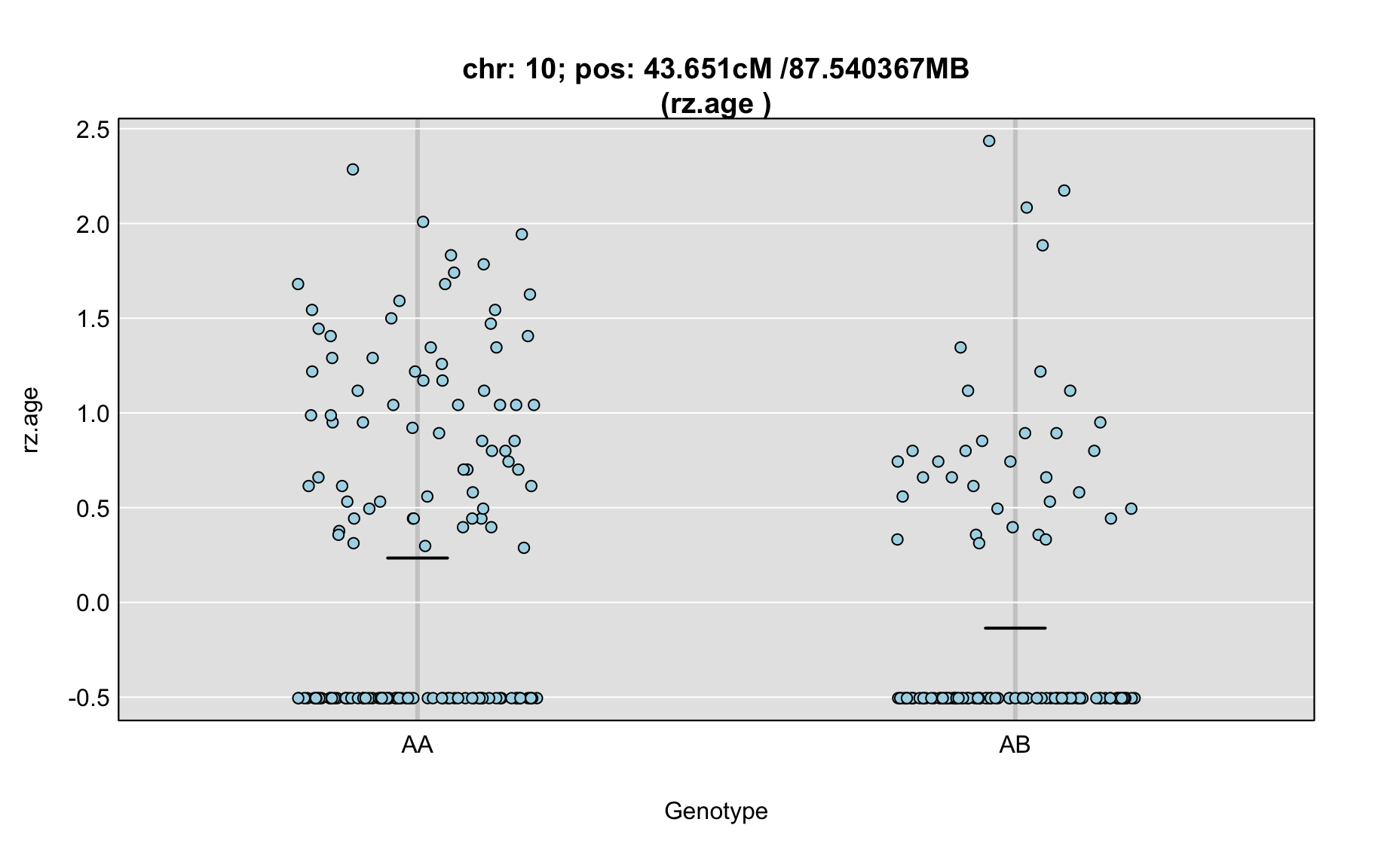

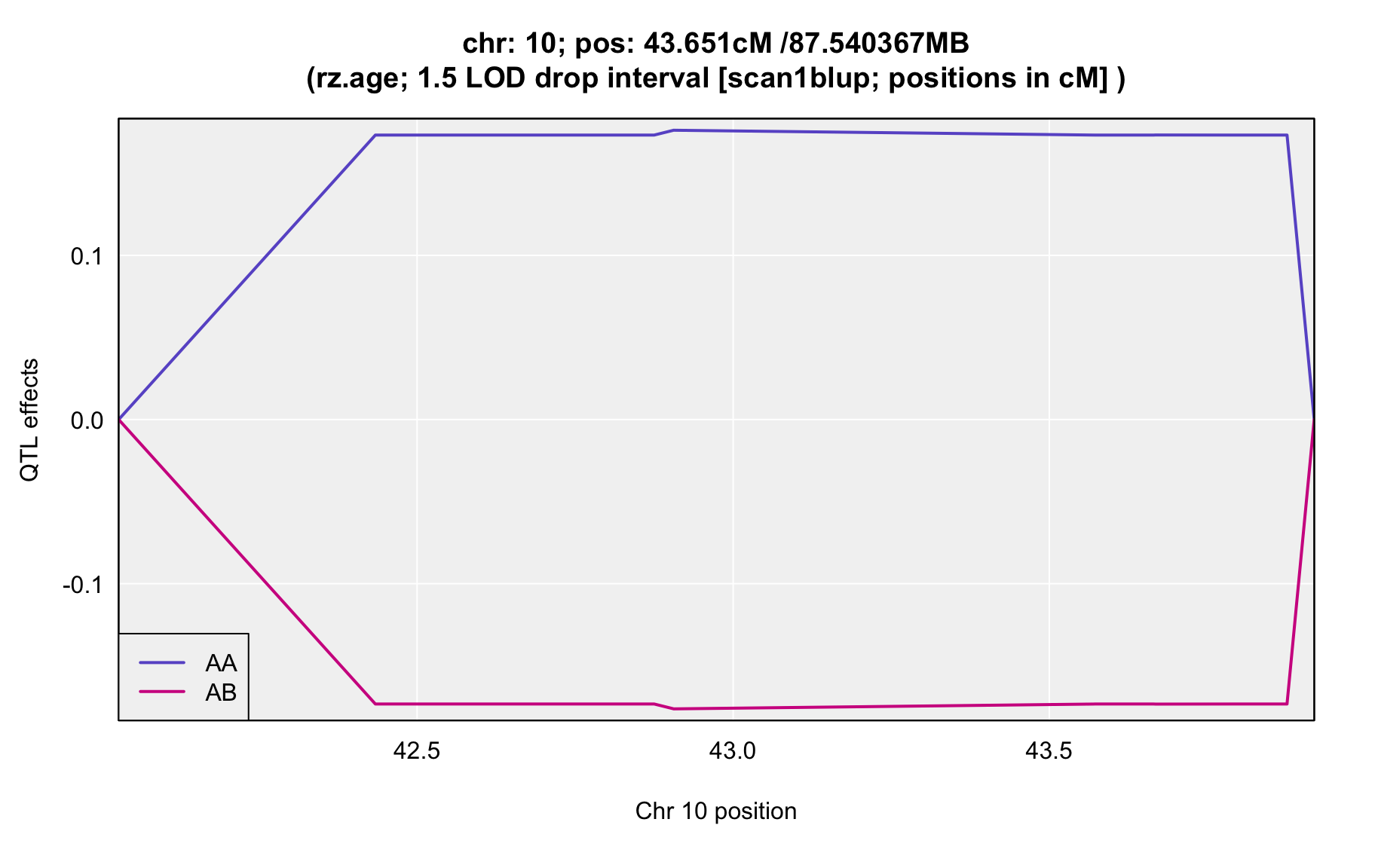
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 10 | gene | 86.04875 | 86.49903 | negative | MGI_C57BL6J_1351334 | Syn3 | NCBI_Gene:27204,ENSEMBL:ENSMUSG00000059602 | MGI:1351334 | protein coding gene | synapsin III |
| 10 | gene | 86.09585 | 86.11359 | positive | MGI_C57BL6J_1925420 | 4930486F22Rik | NCBI_Gene:78170,ENSEMBL:ENSMUSG00000112493 | MGI:1925420 | lncRNA gene | RIKEN cDNA 4930486F22 gene |
| 10 | gene | 86.12583 | 86.12985 | positive | MGI_C57BL6J_3801993 | Gm15990 | ENSEMBL:ENSMUSG00000087256 | MGI:3801993 | lncRNA gene | predicted gene 15990 |
| 10 | gene | 86.15613 | 86.18954 | positive | MGI_C57BL6J_5594795 | Gm35636 | NCBI_Gene:102639289 | MGI:5594795 | lncRNA gene | predicted gene, 35636 |
| 10 | gene | 86.30037 | 86.34951 | positive | MGI_C57BL6J_98754 | Timp3 | NCBI_Gene:21859,ENSEMBL:ENSMUSG00000020044 | MGI:98754 | protein coding gene | tissue inhibitor of metalloproteinase 3 |
| 10 | gene | 86.32703 | 86.32930 | positive | MGI_C57BL6J_2443425 | B930095M22Rik | NA | NA | unclassified gene | RIKEN cDNA B930095M22 gene |
| 10 | gene | 86.52630 | 86.54187 | negative | MGI_C57BL6J_3646339 | Gm6729 | NCBI_Gene:627035,ENSEMBL:ENSMUSG00000090816,ENSEMBL:ENSMUSG00000112586 | MGI:3646339 | protein coding gene | predicted gene 6729 |
| 10 | pseudogene | 86.55081 | 86.55266 | negative | MGI_C57BL6J_3779991 | Gm9583 | NCBI_Gene:672902,ENSEMBL:ENSMUSG00000112893 | MGI:3779991 | pseudogene | predicted gene 9583 |
| 10 | pseudogene | 86.55495 | 86.55618 | negative | MGI_C57BL6J_5434382 | Gm21027 | NCBI_Gene:100534314,ENSEMBL:ENSMUSG00000112115 | MGI:5434382 | pseudogene | predicted gene, 21027 |
| 10 | pseudogene | 86.56993 | 86.57168 | positive | MGI_C57BL6J_6097960 | Gm48448 | ENSEMBL:ENSMUSG00000112207 | MGI:6097960 | pseudogene | predicted gene, 48448 |
| 10 | pseudogene | 86.57176 | 86.57297 | positive | MGI_C57BL6J_6097963 | Gm48450 | ENSEMBL:ENSMUSG00000112579 | MGI:6097963 | pseudogene | predicted gene, 48450 |
| 10 | pseudogene | 86.57304 | 86.57425 | positive | MGI_C57BL6J_6097964 | Gm48451 | ENSEMBL:ENSMUSG00000112562 | MGI:6097964 | pseudogene | predicted gene, 48451 |
| 10 | pseudogene | 86.57432 | 86.57553 | positive | MGI_C57BL6J_6097968 | Gm48454 | ENSEMBL:ENSMUSG00000112325 | MGI:6097968 | pseudogene | predicted gene, 48454 |
| 10 | pseudogene | 86.57562 | 86.57585 | positive | MGI_C57BL6J_6097974 | Gm48460 | ENSEMBL:ENSMUSG00000112268 | MGI:6097974 | pseudogene | predicted gene, 48460 |
| 10 | pseudogene | 86.59235 | 86.59366 | positive | MGI_C57BL6J_5825828 | Gm46191 | NCBI_Gene:108167772,ENSEMBL:ENSMUSG00000112735 | MGI:5825828 | pseudogene | predicted gene, 46191 |
| 10 | pseudogene | 86.60092 | 86.60144 | positive | MGI_C57BL6J_6097975 | Gm48461 | ENSEMBL:ENSMUSG00000112010 | MGI:6097975 | pseudogene | predicted gene, 48461 |
| 10 | pseudogene | 86.60157 | 86.60174 | positive | MGI_C57BL6J_6097978 | Gm48463 | ENSEMBL:ENSMUSG00000112638 | MGI:6097978 | pseudogene | predicted gene, 48463 |
| 10 | pseudogene | 86.60178 | 86.60293 | positive | MGI_C57BL6J_6097979 | Gm48464 | ENSEMBL:ENSMUSG00000112834 | MGI:6097979 | pseudogene | predicted gene, 48464 |
| 10 | pseudogene | 86.60301 | 86.60420 | positive | MGI_C57BL6J_6097980 | Gm48465 | ENSEMBL:ENSMUSG00000111998 | MGI:6097980 | pseudogene | predicted gene, 48465 |
| 10 | pseudogene | 86.60429 | 86.60476 | positive | MGI_C57BL6J_6097981 | Gm48466 | ENSEMBL:ENSMUSG00000112078 | MGI:6097981 | pseudogene | predicted gene, 48466 |
| 10 | pseudogene | 86.62065 | 86.62093 | negative | MGI_C57BL6J_6097988 | Gm48473 | ENSEMBL:ENSMUSG00000112565 | MGI:6097988 | pseudogene | predicted gene, 48473 |
| 10 | pseudogene | 86.62098 | 86.62213 | negative | MGI_C57BL6J_6097989 | Gm48474 | ENSEMBL:ENSMUSG00000112699 | MGI:6097989 | pseudogene | predicted gene, 48474 |
| 10 | pseudogene | 86.62226 | 86.62400 | negative | MGI_C57BL6J_6097990 | Gm48475 | ENSEMBL:ENSMUSG00000112359 | MGI:6097990 | pseudogene | predicted gene, 48475 |
| 10 | pseudogene | 86.63714 | 86.63839 | positive | MGI_C57BL6J_5012174 | Gm19989 | NCBI_Gene:100534315,ENSEMBL:ENSMUSG00000112282 | MGI:5012174 | pseudogene | predicted gene, 19989 |
| 10 | pseudogene | 86.64068 | 86.64253 | positive | MGI_C57BL6J_5011061 | Gm18876 | NCBI_Gene:100417881,ENSEMBL:ENSMUSG00000112098 | MGI:5011061 | pseudogene | predicted gene, 18876 |
| 10 | gene | 86.65543 | 86.67102 | positive | MGI_C57BL6J_3646116 | Gm5174 | NCBI_Gene:382395,ENSEMBL:ENSMUSG00000090308 | MGI:3646116 | protein coding gene | predicted gene 5174 |
| 10 | gene | 86.65740 | 86.67102 | positive | MGI_C57BL6J_6098005 | Gm48485 | ENSEMBL:ENSMUSG00000112666 | MGI:6098005 | lncRNA gene | predicted gene, 48485 |
| 10 | pseudogene | 86.68552 | 86.69481 | positive | MGI_C57BL6J_1913513 | 1810014B01Rik | NCBI_Gene:66263,ENSEMBL:ENSMUSG00000097412 | MGI:1913513 | pseudogene | RIKEN cDNA 1810014B01 gene |
| 10 | gene | 86.69021 | 86.70551 | negative | MGI_C57BL6J_98817 | Hsp90b1 | NCBI_Gene:22027,ENSEMBL:ENSMUSG00000020048 | MGI:98817 | protein coding gene | heat shock protein 90, beta (Grp94), member 1 |
| 10 | gene | 86.69212 | 86.69309 | negative | MGI_C57BL6J_3708098 | Gm15344 | ENSEMBL:ENSMUSG00000084899 | MGI:3708098 | lncRNA gene | predicted gene 15344 |
| 10 | gene | 86.70532 | 86.70545 | negative | MGI_C57BL6J_5455522 | Gm25745 | ENSEMBL:ENSMUSG00000092686 | MGI:5455522 | miRNA gene | predicted gene, 25745 |
| 10 | gene | 86.70562 | 86.77684 | positive | MGI_C57BL6J_2387653 | Ttc41 | NCBI_Gene:103220,ENSEMBL:ENSMUSG00000044937 | MGI:2387653 | protein coding gene | tetratricopeptide repeat domain 41 |
| 10 | pseudogene | 86.73175 | 86.73239 | negative | MGI_C57BL6J_101929 | Fabp3-ps1 | NCBI_Gene:14074,ENSEMBL:ENSMUSG00000056366 | MGI:101929 | pseudogene | fatty acid binding protein 3, muscle and heart, pseudogene 1 |
| 10 | gene | 86.76397 | 86.83397 | positive | MGI_C57BL6J_6121566 | Gm49358 | ENSEMBL:ENSMUSG00000112743 | MGI:6121566 | protein coding gene | predicted gene, 49358 |
| 10 | gene | 86.77899 | 86.83839 | positive | MGI_C57BL6J_3513266 | Nt5dc3 | NCBI_Gene:103466,ENSEMBL:ENSMUSG00000054027 | MGI:3513266 | protein coding gene | 5’-nucleotidase domain containing 3 |
| 10 | gene | 86.83909 | 86.87495 | positive | MGI_C57BL6J_3826557 | Gm16268 | ENSEMBL:ENSMUSG00000089959 | MGI:3826557 | lncRNA gene | predicted gene 16268 |
| 10 | gene | 86.84120 | 87.00803 | negative | MGI_C57BL6J_2178743 | Stab2 | NCBI_Gene:192188,ENSEMBL:ENSMUSG00000035459 | MGI:2178743 | protein coding gene | stabilin 2 |
| 10 | gene | 86.90876 | 86.91001 | positive | MGI_C57BL6J_3809105 | Gm16280 | ENSEMBL:ENSMUSG00000089881 | MGI:3809105 | lncRNA gene | predicted gene 16280 |
| 10 | pseudogene | 86.90948 | 86.91272 | positive | MGI_C57BL6J_3841260 | Gm16270 | NCBI_Gene:100416105,ENSEMBL:ENSMUSG00000089775 | MGI:3841260 | pseudogene | predicted gene 16270 |
| 10 | gene | 86.91350 | 86.91468 | positive | MGI_C57BL6J_1890547 | Tex18 | NA | NA | unclassified non-coding RNA gene | testis expressed gene 18 |
| 10 | gene | 86.97222 | 87.02801 | positive | MGI_C57BL6J_3826610 | Gm16271 | ENSEMBL:ENSMUSG00000089656 | MGI:3826610 | lncRNA gene | predicted gene 16271 |
| 10 | pseudogene | 86.99996 | 87.00073 | positive | MGI_C57BL6J_3826558 | Gm16269 | NCBI_Gene:108167826,ENSEMBL:ENSMUSG00000089666 | MGI:3826558 | pseudogene | predicted gene 16269 |
| 10 | gene | 87.01893 | 87.05883 | negative | MGI_C57BL6J_5595966 | Gm36807 | NCBI_Gene:102640833 | MGI:5595966 | protein coding gene | predicted gene, 36807 |
| 10 | gene | 87.05805 | 87.23060 | positive | MGI_C57BL6J_1923890 | 1700113H08Rik | NCBI_Gene:76640,ENSEMBL:ENSMUSG00000047129 | MGI:1923890 | protein coding gene | RIKEN cDNA 1700113H08 gene |
| 10 | gene | 87.23076 | 87.29756 | positive | MGI_C57BL6J_1922532 | 4930555G07Rik | ENSEMBL:ENSMUSG00000112213 | MGI:1922532 | lncRNA gene | RIKEN cDNA 4930555G07 gene |
| 10 | gene | 87.28949 | 87.29183 | positive | MGI_C57BL6J_3641850 | Gm10764 | ENSEMBL:ENSMUSG00000103862 | MGI:3641850 | unclassified gene | predicted gene 10764 |
| 10 | gene | 87.36404 | 87.36415 | negative | MGI_C57BL6J_5452968 | Gm23191 | ENSEMBL:ENSMUSG00000096692 | MGI:5452968 | snRNA gene | predicted gene, 23191 |
| 10 | gene | 87.49082 | 87.49366 | negative | MGI_C57BL6J_96919 | Ascl1 | NCBI_Gene:17172,ENSEMBL:ENSMUSG00000020052 | MGI:96919 | protein coding gene | achaete-scute family bHLH transcription factor 1 |
| 10 | gene | 87.50283 | 87.50862 | negative | MGI_C57BL6J_5589074 | Gm29915 | NCBI_Gene:102631620 | MGI:5589074 | lncRNA gene | predicted gene, 29915 |
| 10 | pseudogene | 87.50786 | 87.50848 | positive | MGI_C57BL6J_6097476 | Gm48120 | ENSEMBL:ENSMUSG00000112460 | MGI:6097476 | pseudogene | predicted gene, 48120 |
| 10 | gene | 87.52179 | 87.58414 | positive | MGI_C57BL6J_97473 | Pah | NCBI_Gene:18478,ENSEMBL:ENSMUSG00000020051 | MGI:97473 | protein coding gene | phenylalanine hydroxylase |
| 10 | gene | 87.57144 | 87.57175 | positive | MGI_C57BL6J_1925634 | 2210420L05Rik | NA | NA | unclassified gene | RIKEN cDNA 2210420L05 gene |
| 10 | pseudogene | 87.68858 | 87.68903 | positive | MGI_C57BL6J_6097580 | Gm48195 | ENSEMBL:ENSMUSG00000112536 | MGI:6097580 | pseudogene | predicted gene, 48195 |
| 10 | pseudogene | 87.75914 | 87.75957 | positive | MGI_C57BL6J_5012191 | Gm20006 | NCBI_Gene:100504000 | MGI:5012191 | pseudogene | predicted gene, 20006 |
| 10 | gene | 87.77724 | 87.79837 | negative | MGI_C57BL6J_5589232 | Gm30073 | NCBI_Gene:102631839 | MGI:5589232 | lncRNA gene | predicted gene, 30073 |
| 10 | gene | 87.82638 | 87.86472 | negative | MGI_C57BL6J_3841250 | Igf1os | NCBI_Gene:627270,ENSEMBL:ENSMUSG00000058934 | MGI:3841250 | antisense lncRNA gene | insulin-like growth factor 1, opposite strand |
| 10 | gene | 87.82989 | 87.82996 | negative | MGI_C57BL6J_4413968 | n-TKctt21 | NCBI_Gene:102467349 | MGI:4413968 | tRNA gene | nuclear encoded tRNA lysine 21 (anticodon TTT) |
| 10 | gene | 87.85827 | 87.93705 | positive | MGI_C57BL6J_96432 | Igf1 | NCBI_Gene:16000,ENSEMBL:ENSMUSG00000020053 | MGI:96432 | protein coding gene | insulin-like growth factor 1 |
| 10 | pseudogene | 87.96667 | 87.96802 | positive | MGI_C57BL6J_98879 | Tyms-ps | NCBI_Gene:22172,ENSEMBL:ENSMUSG00000112222 | MGI:98879 | pseudogene | thymidylate synthase, pseudogene |
| 10 | gene | 87.98046 | 87.98422 | positive | MGI_C57BL6J_6097752 | Gm48306 | ENSEMBL:ENSMUSG00000112183 | MGI:6097752 | lncRNA gene | predicted gene, 48306 |
| 10 | gene | 88.03822 | 88.03968 | positive | MGI_C57BL6J_6097754 | Gm48307 | ENSEMBL:ENSMUSG00000112235 | MGI:6097754 | unclassified gene | predicted gene, 48307 |
| 10 | gene | 88.03822 | 88.03968 | negative | MGI_C57BL6J_3641951 | Gm10763 | NA | NA | unclassified gene | predicted gene 10763 |
| 10 | gene | 88.06807 | 88.09035 | negative | MGI_C57BL6J_5589392 | Gm30233 | NCBI_Gene:102632060 | MGI:5589392 | lncRNA gene | predicted gene, 30233 |
| 10 | gene | 88.08820 | 88.08982 | positive | MGI_C57BL6J_6097756 | Gm48308 | ENSEMBL:ENSMUSG00000112932 | MGI:6097756 | unclassified gene | predicted gene, 48308 |
| 10 | gene | 88.09107 | 88.09238 | positive | MGI_C57BL6J_97629 | Pmch | NCBI_Gene:110312,ENSEMBL:ENSMUSG00000035383 | MGI:97629 | protein coding gene | pro-melanin-concentrating hormone |
| 10 | gene | 88.09140 | 88.14695 | negative | MGI_C57BL6J_1922567 | Parpbp | NCBI_Gene:75317,ENSEMBL:ENSMUSG00000035365 | MGI:1922567 | protein coding gene | PARP1 binding protein |
| 10 | gene | 88.13438 | 88.13758 | negative | MGI_C57BL6J_3647484 | Gm6653 | NCBI_Gene:626115,ENSEMBL:ENSMUSG00000112026 | MGI:3647484 | unclassified gene | predicted gene 6653 |
| 10 | gene | 88.13592 | 88.13753 | positive | MGI_C57BL6J_1918118 | 4921515L22Rik | ENSEMBL:ENSMUSG00000112406 | MGI:1918118 | lncRNA gene | RIKEN cDNA 4921515L22 gene |
| 10 | gene | 88.14697 | 88.17839 | positive | MGI_C57BL6J_1919964 | Nup37 | NCBI_Gene:69736,ENSEMBL:ENSMUSG00000035351 | MGI:1919964 | protein coding gene | nucleoporin 37 |
| 10 | gene | 88.16496 | 88.16507 | negative | MGI_C57BL6J_5456134 | Gm26357 | ENSEMBL:ENSMUSG00000088843 | MGI:5456134 | snRNA gene | predicted gene, 26357 |
| 10 | gene | 88.20109 | 88.26082 | positive | MGI_C57BL6J_1914532 | Washc3 | NCBI_Gene:67282,ENSEMBL:ENSMUSG00000020056 | MGI:1914532 | protein coding gene | WASH complex subunit 3 |
| 10 | pseudogene | 88.24742 | 88.24863 | positive | MGI_C57BL6J_3648865 | Gm8415 | NCBI_Gene:667000 | MGI:3648865 | pseudogene | predicted gene 8415 |

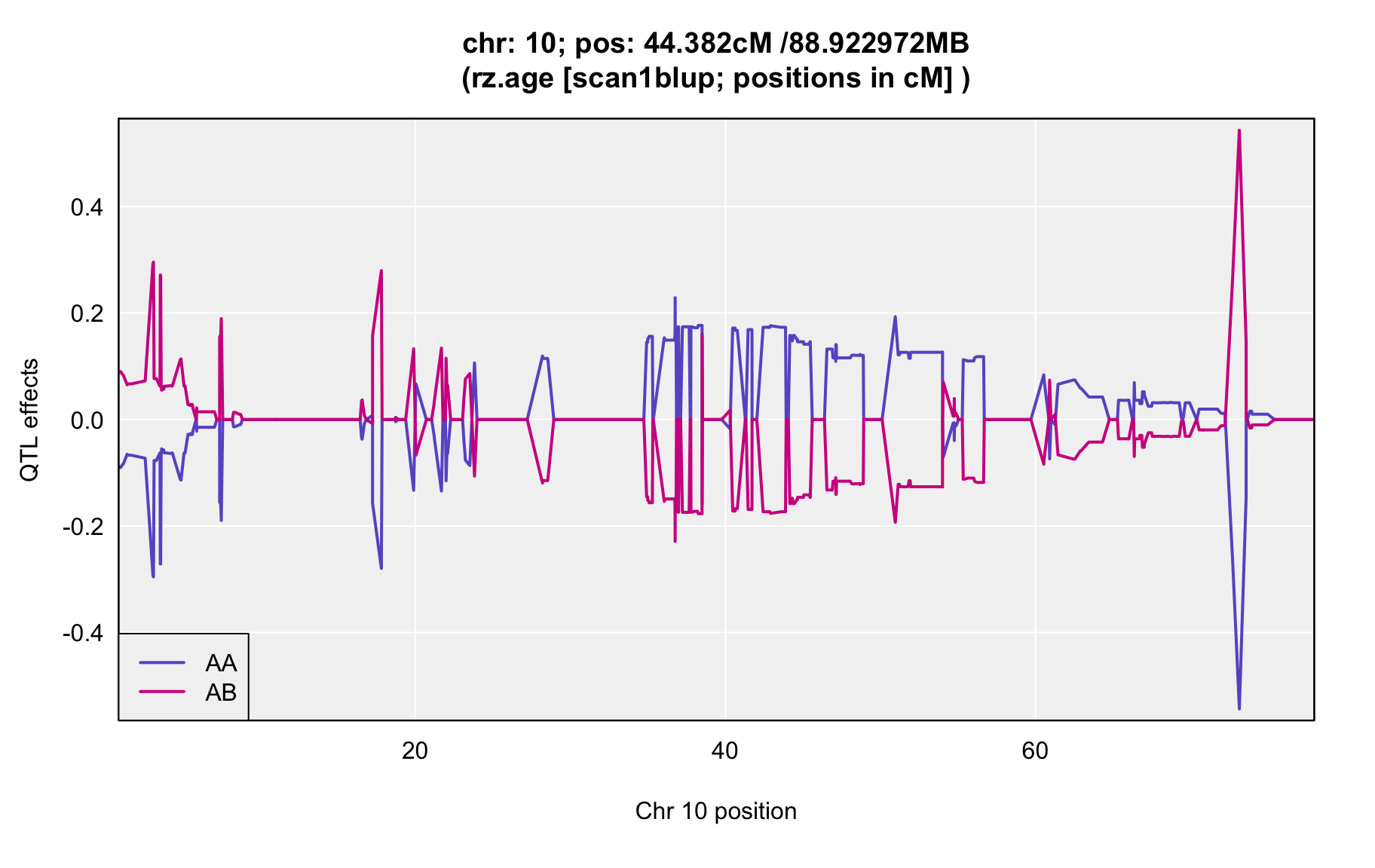
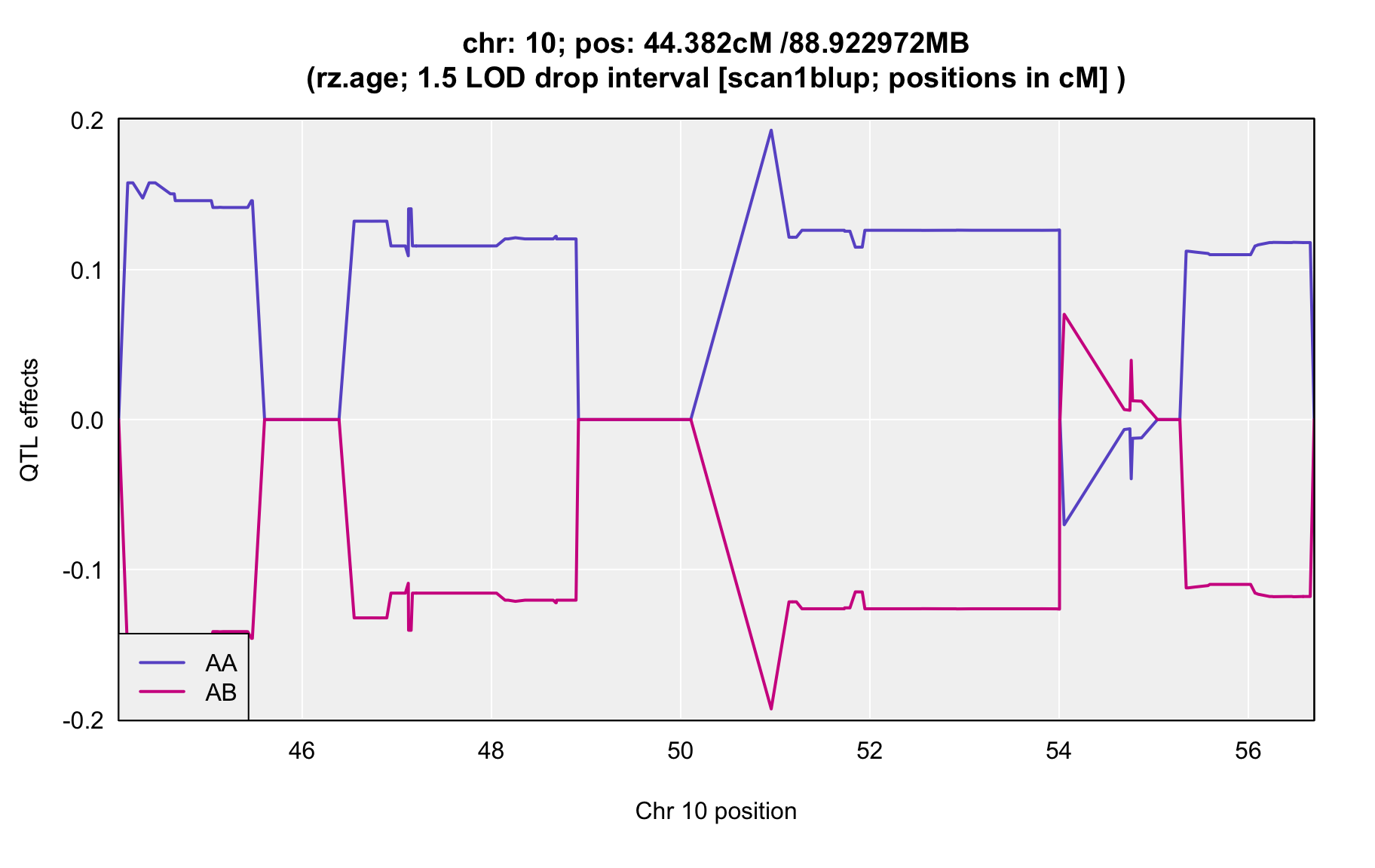
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 10 | gene | 88.45274 | 88.50407 | negative | MGI_C57BL6J_2384841 | Chpt1 | NCBI_Gene:212862,ENSEMBL:ENSMUSG00000060002 | MGI:2384841 | protein coding gene | choline phosphotransferase 1 |
| 10 | gene | 88.45957 | 88.47324 | positive | MGI_C57BL6J_109542 | Sycp3 | NCBI_Gene:20962,ENSEMBL:ENSMUSG00000020059 | MGI:109542 | protein coding gene | synaptonemal complex protein 3 |
| 10 | gene | 88.51828 | 88.60524 | negative | MGI_C57BL6J_1336213 | Mybpc1 | NCBI_Gene:109272,ENSEMBL:ENSMUSG00000020061 | MGI:1336213 | protein coding gene | myosin binding protein C, slow-type |
| 10 | gene | 88.60247 | 88.60534 | positive | MGI_C57BL6J_6098429 | Gm48752 | ENSEMBL:ENSMUSG00000112841 | MGI:6098429 | unclassified gene | predicted gene, 48752 |
| 10 | pseudogene | 88.64136 | 88.64221 | positive | MGI_C57BL6J_3643053 | Gm8435 | NCBI_Gene:667046,ENSEMBL:ENSMUSG00000112231 | MGI:3643053 | pseudogene | predicted gene 8435 |
| 10 | pseudogene | 88.65011 | 88.65823 | negative | MGI_C57BL6J_6097458 | Gm48106 | ENSEMBL:ENSMUSG00000112428 | MGI:6097458 | pseudogene | predicted gene, 48106 |
| 10 | pseudogene | 88.67050 | 88.67127 | negative | MGI_C57BL6J_6097461 | Gm48108 | ENSEMBL:ENSMUSG00000112869 | MGI:6097461 | pseudogene | predicted gene, 48108 |
| 10 | pseudogene | 88.67302 | 88.67339 | positive | MGI_C57BL6J_5588966 | Gm29807 | NCBI_Gene:101056238,ENSEMBL:ENSMUSG00000112885 | MGI:5588966 | pseudogene | predicted gene, 29807 |
| 10 | gene | 88.67477 | 88.68502 | negative | MGI_C57BL6J_1341168 | Spic | NCBI_Gene:20728,ENSEMBL:ENSMUSG00000004359 | MGI:1341168 | protein coding gene | Spi-C transcription factor (Spi-1/PU.1 related) |
| 10 | pseudogene | 88.69101 | 88.70511 | positive | MGI_C57BL6J_5825829 | Gm46192 | NCBI_Gene:108167773,ENSEMBL:ENSMUSG00000111919 | MGI:5825829 | pseudogene | predicted gene, 46192 |
| 10 | gene | 88.71001 | 88.71013 | negative | MGI_C57BL6J_4421927 | n-R5s79 | ENSEMBL:ENSMUSG00000065005 | MGI:4421927 | rRNA gene | nuclear encoded rRNA 5S 79 |
| 10 | pseudogene | 88.71565 | 88.71609 | negative | MGI_C57BL6J_6097511 | Gm48144 | ENSEMBL:ENSMUSG00000112467 | MGI:6097511 | pseudogene | predicted gene, 48144 |
| 10 | pseudogene | 88.72954 | 88.73110 | negative | MGI_C57BL6J_3643371 | Gm4925 | NCBI_Gene:237433,ENSEMBL:ENSMUSG00000069540 | MGI:3643371 | pseudogene | predicted gene 4925 |
| 10 | gene | 88.73086 | 88.74409 | positive | MGI_C57BL6J_99436 | Arl1 | NCBI_Gene:104303,ENSEMBL:ENSMUSG00000060904 | MGI:99436 | protein coding gene | ADP-ribosylation factor-like 1 |
| 10 | gene | 88.74661 | 88.82681 | negative | MGI_C57BL6J_1917933 | Utp20 | NCBI_Gene:70683,ENSEMBL:ENSMUSG00000004356 | MGI:1917933 | protein coding gene | UTP20 small subunit processome component |
| 10 | gene | 88.87460 | 88.87580 | negative | MGI_C57BL6J_1924528 | 9430024C03Rik | NA | NA | unclassified gene | RIKEN cDNA 9430024C03 gene |
| 10 | gene | 88.88599 | 88.92951 | positive | MGI_C57BL6J_2384916 | Slc5a8 | NCBI_Gene:216225,ENSEMBL:ENSMUSG00000020062 | MGI:2384916 | protein coding gene | solute carrier family 5 (iodide transporter), member 8 |
| 10 | gene | 88.94893 | 89.34508 | negative | MGI_C57BL6J_2443344 | Ano4 | NCBI_Gene:320091,ENSEMBL:ENSMUSG00000035189 | MGI:2443344 | protein coding gene | anoctamin 4 |
| 10 | gene | 89.25713 | 89.25914 | positive | MGI_C57BL6J_5589928 | Gm30769 | NCBI_Gene:102632785 | MGI:5589928 | lncRNA gene | predicted gene, 30769 |
| 10 | gene | 89.40882 | 89.44437 | negative | MGI_C57BL6J_1918780 | Gas2l3 | NCBI_Gene:237436,ENSEMBL:ENSMUSG00000074802 | MGI:1918780 | protein coding gene | growth arrest-specific 2 like 3 |
| 10 | gene | 89.45423 | 89.53365 | negative | MGI_C57BL6J_1352464 | Nr1h4 | NCBI_Gene:20186,ENSEMBL:ENSMUSG00000047638 | MGI:1352464 | protein coding gene | nuclear receptor subfamily 1, group H, member 4 |
| 10 | pseudogene | 89.55508 | 89.55572 | negative | MGI_C57BL6J_6097428 | Gm48087 | ENSEMBL:ENSMUSG00000112272 | MGI:6097428 | pseudogene | predicted gene, 48087 |
| 10 | gene | 89.57402 | 89.62125 | negative | MGI_C57BL6J_3039629 | Slc17a8 | NCBI_Gene:216227,ENSEMBL:ENSMUSG00000019935 | MGI:3039629 | protein coding gene | solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8 |
| 10 | gene | 89.63872 | 89.68629 | negative | MGI_C57BL6J_1289172 | Scyl2 | NCBI_Gene:213326,ENSEMBL:ENSMUSG00000069539 | MGI:1289172 | protein coding gene | SCY1-like 2 (S. cerevisiae) |
| 10 | gene | 89.68636 | 89.70087 | positive | MGI_C57BL6J_1916252 | 1500026H17Rik | NCBI_Gene:69002,ENSEMBL:ENSMUSG00000097383 | MGI:1916252 | lncRNA gene | RIKEN cDNA 1500026H17 gene |
| 10 | gene | 89.71197 | 89.73231 | negative | MGI_C57BL6J_1914269 | Actr6 | NCBI_Gene:67019,ENSEMBL:ENSMUSG00000019948 | MGI:1914269 | protein coding gene | ARP6 actin-related protein 6 |
| 10 | gene | 89.73678 | 89.74141 | negative | MGI_C57BL6J_5590122 | Gm30963 | NCBI_Gene:102633042 | MGI:5590122 | lncRNA gene | predicted gene, 30963 |
| 10 | gene | 89.74362 | 89.74582 | negative | MGI_C57BL6J_3642333 | E330037I15Rik | NA | NA | unclassified gene | RIKEN cDNA E330037I15 gene |
| 10 | gene | 89.74499 | 89.81989 | positive | MGI_C57BL6J_2442888 | Uhrf1bp1l | NCBI_Gene:75089,ENSEMBL:ENSMUSG00000019951 | MGI:2442888 | protein coding gene | UHRF1 (ICBP90) binding protein 1-like |
| 10 | gene | 89.81216 | 89.81389 | positive | MGI_C57BL6J_5590249 | Gm31090 | NCBI_Gene:102633203 | MGI:5590249 | lncRNA gene | predicted gene, 31090 |
| 10 | gene | 89.82134 | 89.82145 | negative | MGI_C57BL6J_5455957 | Gm26180 | ENSEMBL:ENSMUSG00000093967 | MGI:5455957 | snRNA gene | predicted gene, 26180 |
| 10 | gene | 89.82288 | 89.82654 | positive | MGI_C57BL6J_5825875 | Gm46238 | NCBI_Gene:108167827 | MGI:5825875 | protein coding gene | predicted gene, 46238 |
| 10 | gene | 89.87302 | 90.97330 | positive | MGI_C57BL6J_1924781 | Anks1b | NCBI_Gene:77531,ENSEMBL:ENSMUSG00000058589 | MGI:1924781 | protein coding gene | ankyrin repeat and sterile alpha motif domain containing 1B |
| 10 | gene | 89.87827 | 89.87860 | positive | MGI_C57BL6J_3782360 | Gm4184 | NCBI_Gene:100043039 | MGI:3782360 | protein coding gene | predicted gene 4184 |
| 10 | pseudogene | 90.13298 | 90.13482 | negative | MGI_C57BL6J_3645288 | Gm5780 | NCBI_Gene:544719,ENSEMBL:ENSMUSG00000112140 | MGI:3645288 | pseudogene | predicted gene 5780 |
| 10 | gene | 90.42659 | 90.42806 | positive | MGI_C57BL6J_2441893 | C230047L17Rik | NA | NA | unclassified gene | RIKEN cDNA C230047L17 gene |
| 10 | gene | 90.45091 | 90.45100 | positive | MGI_C57BL6J_5452379 | Gm22602 | ENSEMBL:ENSMUSG00000088221 | MGI:5452379 | snRNA gene | predicted gene, 22602 |
| 10 | gene | 90.59763 | 90.59916 | positive | MGI_C57BL6J_1920193 | 2900018E21Rik | NA | NA | unclassified gene | RIKEN cDNA 2900018E21 gene |
| 10 | gene | 90.72896 | 90.73633 | positive | MGI_C57BL6J_6096636 | Gm47591 | ENSEMBL:ENSMUSG00000112153 | MGI:6096636 | lncRNA gene | predicted gene, 47591 |
| 10 | pseudogene | 90.74999 | 90.75069 | negative | MGI_C57BL6J_5010855 | Gm18670 | NCBI_Gene:100417532,ENSEMBL:ENSMUSG00000111909 | MGI:5010855 | pseudogene | predicted gene, 18670 |
| 10 | pseudogene | 90.75087 | 90.75107 | negative | MGI_C57BL6J_6096633 | Gm47589 | ENSEMBL:ENSMUSG00000112507 | MGI:6096633 | pseudogene | predicted gene, 47589 |
| 10 | gene | 90.81308 | 90.81439 | positive | MGI_C57BL6J_6096638 | Gm47592 | ENSEMBL:ENSMUSG00000112015 | MGI:6096638 | unclassified gene | predicted gene, 47592 |
| 10 | gene | 90.85110 | 90.85407 | positive | MGI_C57BL6J_6096634 | Gm47590 | ENSEMBL:ENSMUSG00000112601 | MGI:6096634 | unclassified gene | predicted gene, 47590 |
| 10 | gene | 90.98931 | 91.08277 | negative | MGI_C57BL6J_1306796 | Apaf1 | NCBI_Gene:11783,ENSEMBL:ENSMUSG00000019979 | MGI:1306796 | protein coding gene | apoptotic peptidase activating factor 1 |
| 10 | gene | 91.00574 | 91.00584 | positive | MGI_C57BL6J_5455167 | Gm25390 | ENSEMBL:ENSMUSG00000087966 | MGI:5455167 | snRNA gene | predicted gene, 25390 |
| 10 | gene | 91.08269 | 91.10261 | positive | MGI_C57BL6J_1914704 | Ikbip | NCBI_Gene:67454,ENSEMBL:ENSMUSG00000019975 | MGI:1914704 | protein coding gene | IKBKB interacting protein |
| 10 | gene | 91.11378 | 91.11425 | negative | MGI_C57BL6J_1925088 | A930036M01Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA A930036M01 gene |
| 10 | gene | 91.11657 | 91.12406 | negative | MGI_C57BL6J_1353498 | Slc25a3 | NCBI_Gene:18674,ENSEMBL:ENSMUSG00000061904 | MGI:1353498 | protein coding gene | solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 3 |
| 10 | gene | 91.11829 | 91.11854 | negative | MGI_C57BL6J_5453896 | Gm24119 | ENSEMBL:ENSMUSG00000077167 | MGI:5453896 | snoRNA gene | predicted gene, 24119 |
| 10 | gene | 91.14757 | 91.18172 | negative | MGI_C57BL6J_106920 | Tmpo | NCBI_Gene:21917,ENSEMBL:ENSMUSG00000019961 | MGI:106920 | protein coding gene | thymopoietin |
| 10 | gene | 91.14951 | 91.15059 | positive | MGI_C57BL6J_5477259 | Gm26765 | ENSEMBL:ENSMUSG00000097664 | MGI:5477259 | lncRNA gene | predicted gene, 26765 |
| 10 | gene | 91.15008 | 91.15164 | negative | MGI_C57BL6J_1923371 | 5830485P09Rik | NA | NA | unclassified gene | RIKEN cDNA 5830485P09 gene |
| 10 | gene | 91.18141 | 91.18148 | negative | MGI_C57BL6J_4414067 | n-TWcca2 | NCBI_Gene:102467609 | MGI:4414067 | tRNA gene | nuclear encoded tRNA trytophan 2 (anticodon CCA) |
| 10 | gene | 91.18210 | 91.18217 | negative | MGI_C57BL6J_4413767 | n-TDgtc16 | NCBI_Gene:102467266 | MGI:4413767 | tRNA gene | nuclear encoded tRNA aspartic acid 16 (anticodon GTC) |
| 10 | gene | 91.20826 | 91.22064 | negative | MGI_C57BL6J_1922441 | 4930534H03Rik | NCBI_Gene:102633753,ENSEMBL:ENSMUSG00000111544 | MGI:1922441 | lncRNA gene | RIKEN cDNA 4930534H03 gene |
| 10 | pseudogene | 91.22819 | 91.22850 | positive | MGI_C57BL6J_6095807 | Gm47081 | ENSEMBL:ENSMUSG00000110870 | MGI:6095807 | pseudogene | predicted gene, 47081 |
| 10 | pseudogene | 91.25214 | 91.25950 | negative | MGI_C57BL6J_5010890 | Gm18705 | NCBI_Gene:100417593,ENSEMBL:ENSMUSG00000110847 | MGI:5010890 | pseudogene | predicted gene, 18705 |
| 10 | gene | 91.31034 | 91.31245 | positive | MGI_C57BL6J_6095808 | Gm47082 | ENSEMBL:ENSMUSG00000111525 | MGI:6095808 | lncRNA gene | predicted gene, 47082 |
| 10 | gene | 91.41060 | 91.43962 | positive | MGI_C57BL6J_5623621 | Gm40736 | NCBI_Gene:105245258 | MGI:5623621 | lncRNA gene | predicted gene, 40736 |
| 10 | pseudogene | 91.42335 | 91.42373 | positive | MGI_C57BL6J_6095810 | Gm47083 | ENSEMBL:ENSMUSG00000111591 | MGI:6095810 | pseudogene | predicted gene, 47083 |
| 10 | pseudogene | 91.44822 | 91.44838 | negative | MGI_C57BL6J_6095811 | Gm47084 | ENSEMBL:ENSMUSG00000111051 | MGI:6095811 | pseudogene | predicted gene, 47084 |
| 10 | pseudogene | 91.64070 | 91.64089 | negative | MGI_C57BL6J_6095821 | Gm47091 | ENSEMBL:ENSMUSG00000111782 | MGI:6095821 | pseudogene | predicted gene, 47091 |
| 10 | gene | 91.88859 | 91.95362 | positive | MGI_C57BL6J_5590751 | Gm31592 | NCBI_Gene:102633867,ENSEMBL:ENSMUSG00000111657 | MGI:5590751 | lncRNA gene | predicted gene, 31592 |
| 10 | gene | 91.98386 | 91.98743 | positive | MGI_C57BL6J_5590832 | Gm31673 | NCBI_Gene:102633980 | MGI:5590832 | lncRNA gene | predicted gene, 31673 |
| 10 | gene | 92.07104 | 92.16518 | negative | MGI_C57BL6J_1099806 | Rmst | NCBI_Gene:110333,ENSEMBL:ENSMUSG00000112117 | MGI:1099806 | lncRNA gene | rhabdomyosarcoma 2 associated transcript (non-coding RNA) |
| 10 | gene | 92.07120 | 92.07208 | negative | MGI_C57BL6J_3763716 | BI734849 | NA | NA | unclassified gene | expressed sequence BI734849 |
| 10 | gene | 92.07166 | 92.07182 | negative | MGI_C57BL6J_5530926 | Gm27544 | ENSEMBL:ENSMUSG00000098979 | MGI:5530926 | unclassified non-coding RNA gene | predicted gene, 27544 |
| 10 | gene | 92.07209 | 92.07219 | negative | MGI_C57BL6J_3618729 | Mir135a-2 | miRBase:MI0000715,NCBI_Gene:723955,ENSEMBL:ENSMUSG00000065524 | MGI:3618729 | miRNA gene | microRNA 135a-2 |
| 10 | gene | 92.07517 | 92.07536 | negative | MGI_C57BL6J_5531381 | Gm27999 | ENSEMBL:ENSMUSG00000098394 | MGI:5531381 | unclassified non-coding RNA gene | predicted gene, 27999 |
| 10 | gene | 92.07549 | 92.07565 | negative | MGI_C57BL6J_5530990 | Gm27608 | ENSEMBL:ENSMUSG00000099136 | MGI:5530990 | unclassified non-coding RNA gene | predicted gene, 27608 |
| 10 | gene | 92.10166 | 92.10193 | negative | MGI_C57BL6J_5530970 | Gm27588 | ENSEMBL:ENSMUSG00000098664 | MGI:5530970 | unclassified non-coding RNA gene | predicted gene, 27588 |
| 10 | gene | 92.10371 | 92.10391 | negative | MGI_C57BL6J_5531135 | Gm27753 | ENSEMBL:ENSMUSG00000098891 | MGI:5531135 | unclassified non-coding RNA gene | predicted gene, 27753 |
| 10 | gene | 92.13322 | 92.13335 | negative | MGI_C57BL6J_5530964 | Gm27582 | ENSEMBL:ENSMUSG00000098662 | MGI:5530964 | unclassified non-coding RNA gene | predicted gene, 27582 |
| 10 | gene | 92.13496 | 92.13507 | negative | MGI_C57BL6J_5530748 | Gm27366 | ENSEMBL:ENSMUSG00000098526 | MGI:5530748 | unclassified non-coding RNA gene | predicted gene, 27366 |
| 10 | gene | 92.13570 | 92.13581 | negative | MGI_C57BL6J_5530801 | Gm27419 | ENSEMBL:ENSMUSG00000099307 | MGI:5530801 | unclassified non-coding RNA gene | predicted gene, 27419 |
| 10 | gene | 92.13714 | 92.13722 | negative | MGI_C57BL6J_4834231 | Mir1251 | miRBase:MI0014021,NCBI_Gene:100526517,ENSEMBL:ENSMUSG00000093227 | MGI:4834231 | miRNA gene | microRNA 1251 |
| 10 | gene | 92.16210 | 92.16469 | positive | MGI_C57BL6J_5477194 | Gm26700 | ENSEMBL:ENSMUSG00000097201 | MGI:5477194 | lncRNA gene | predicted gene, 26700 |
| 10 | gene | 92.17130 | 92.37537 | negative | MGI_C57BL6J_5434113 | Gm20757 | NCBI_Gene:627800,ENSEMBL:ENSMUSG00000098040 | MGI:5434113 | lncRNA gene | predicted gene, 20757 |
| 10 | pseudogene | 92.22284 | 92.23768 | negative | MGI_C57BL6J_3648240 | Gm8512 | NCBI_Gene:667203,ENSEMBL:ENSMUSG00000111104 | MGI:3648240 | pseudogene | predicted gene 8512 |
| 10 | pseudogene | 92.25859 | 92.25928 | positive | MGI_C57BL6J_6095874 | Gm47124 | ENSEMBL:ENSMUSG00000111776 | MGI:6095874 | pseudogene | predicted gene, 47124 |
| 10 | gene | 92.38966 | 92.47119 | negative | MGI_C57BL6J_5623623 | Gm40738 | NCBI_Gene:105245260 | MGI:5623623 | lncRNA gene | predicted gene, 40738 |
| 10 | gene | 92.40101 | 92.41750 | positive | MGI_C57BL6J_1921071 | 4930401A07Rik | ENSEMBL:ENSMUSG00000110794 | MGI:1921071 | lncRNA gene | RIKEN cDNA 4930401A07 gene |
| 10 | pseudogene | 92.48421 | 92.48496 | positive | MGI_C57BL6J_5010228 | Gm18043 | NCBI_Gene:100416329,ENSEMBL:ENSMUSG00000111604 | MGI:5010228 | pseudogene | predicted gene, 18043 |
| 10 | gene | 92.48682 | 92.48688 | positive | MGI_C57BL6J_5455661 | Gm25884 | ENSEMBL:ENSMUSG00000088407 | MGI:5455661 | snRNA gene | predicted gene, 25884 |
| 10 | pseudogene | 92.54221 | 92.54374 | negative | MGI_C57BL6J_3646193 | Gm4800 | NCBI_Gene:216229,ENSEMBL:ENSMUSG00000111759 | MGI:3646193 | pseudogene | predicted gene 4800 |
| 10 | pseudogene | 92.58706 | 92.58737 | positive | MGI_C57BL6J_3781700 | Gm3523 | NCBI_Gene:102636707,ENSEMBL:ENSMUSG00000111121 | MGI:3781700 | pseudogene | predicted gene 3523 |
| 10 | gene | 92.58721 | 92.58731 | positive | MGI_C57BL6J_5452359 | Gm22582 | ENSEMBL:ENSMUSG00000076238 | MGI:5452359 | miRNA gene | predicted gene, 22582 |
| 10 | gene | 92.64409 | 92.65485 | positive | MGI_C57BL6J_5590981 | Gm31822 | NCBI_Gene:102634175,ENSEMBL:ENSMUSG00000111470 | MGI:5590981 | lncRNA gene | predicted gene, 31822 |
| 10 | gene | 92.68475 | 92.72368 | negative | MGI_C57BL6J_97293 | Nedd1 | NCBI_Gene:17997,ENSEMBL:ENSMUSG00000019988 | MGI:97293 | protein coding gene | neural precursor cell expressed, developmentally down-regulated gene 1 |
| 10 | gene | 92.68886 | 92.68901 | negative | MGI_C57BL6J_5454018 | Gm24241 | ENSEMBL:ENSMUSG00000088412 | MGI:5454018 | snoRNA gene | predicted gene, 24241 |
| 10 | pseudogene | 92.70608 | 92.70626 | negative | MGI_C57BL6J_6097289 | Gm47993 | ENSEMBL:ENSMUSG00000111432 | MGI:6097289 | pseudogene | predicted gene, 47993 |
| 10 | gene | 92.71751 | 92.72369 | negative | MGI_C57BL6J_6215303 | Gm49783 | ENSEMBL:ENSMUSG00000111740 | MGI:6215303 | unclassified gene | predicted gene, 49783 |
| 10 | gene | 92.77561 | 93.08359 | negative | MGI_C57BL6J_1922208 | Cfap54 | NCBI_Gene:380654,ENSEMBL:ENSMUSG00000020014 | MGI:1922208 | protein coding gene | cilia and flagella associated protein 54 |
| 10 | gene | 93.07481 | 93.07693 | positive | MGI_C57BL6J_5623624 | Gm40739 | NCBI_Gene:105245261 | MGI:5623624 | lncRNA gene | predicted gene, 40739 |
| 10 | gene | 93.08420 | 93.09373 | positive | MGI_C57BL6J_5591627 | Gm32468 | NCBI_Gene:102635028,ENSEMBL:ENSMUSG00000111013 | MGI:5591627 | lncRNA gene | predicted gene, 32468 |
| 10 | gene | 93.12642 | 93.12832 | positive | MGI_C57BL6J_5623625 | Gm40740 | NCBI_Gene:105245262 | MGI:5623625 | lncRNA gene | predicted gene, 40740 |
| 10 | gene | 93.14321 | 93.14547 | positive | MGI_C57BL6J_5623626 | Gm40741 | NCBI_Gene:105245263 | MGI:5623626 | lncRNA gene | predicted gene, 40741 |
| 10 | gene | 93.16088 | 93.26707 | positive | MGI_C57BL6J_97517 | Cdk17 | NCBI_Gene:237459,ENSEMBL:ENSMUSG00000020015 | MGI:97517 | protein coding gene | cyclin-dependent kinase 17 |
| 10 | gene | 93.16278 | 93.16290 | positive | MGI_C57BL6J_3836967 | Mir1931 | miRBase:MI0009920,NCBI_Gene:100316689,ENSEMBL:ENSMUSG00000089214 | MGI:3836967 | miRNA gene | microRNA 1931 |
| 10 | gene | 93.24741 | 93.31116 | negative | MGI_C57BL6J_101762 | Elk3 | NCBI_Gene:13713,ENSEMBL:ENSMUSG00000008398 | MGI:101762 | protein coding gene | ELK3, member of ETS oncogene family |
| 10 | gene | 93.27554 | 93.27881 | positive | MGI_C57BL6J_5591763 | Gm32604 | NA | NA | lncRNA gene | predicted gene%2c 32604 |
| 10 | gene | 93.28498 | 93.28947 | positive | MGI_C57BL6J_5591710 | Gm32551 | NCBI_Gene:102635134 | MGI:5591710 | lncRNA gene | predicted gene, 32551 |
| 10 | gene | 93.31733 | 93.32273 | positive | MGI_C57BL6J_5591847 | Gm32688 | NCBI_Gene:102635316,ENSEMBL:ENSMUSG00000111531 | MGI:5591847 | lncRNA gene | predicted gene, 32688 |
| 10 | gene | 93.33539 | 93.34888 | positive | MGI_C57BL6J_5009823 | Gm17745 | NCBI_Gene:432488,ENSEMBL:ENSMUSG00000110647 | MGI:5009823 | lncRNA gene | predicted gene, 17745 |
| 10 | pseudogene | 93.39709 | 93.39727 | negative | MGI_C57BL6J_6096240 | Gm47344 | ENSEMBL:ENSMUSG00000111316 | MGI:6096240 | pseudogene | predicted gene, 47344 |
| 10 | gene | 93.43028 | 93.43196 | negative | MGI_C57BL6J_5623627 | Gm40742 | NCBI_Gene:105245264 | MGI:5623627 | lncRNA gene | predicted gene, 40742 |
| 10 | gene | 93.43311 | 93.43316 | positive | MGI_C57BL6J_5531256 | Mir7688 | miRBase:MI0025041,NCBI_Gene:102465791,ENSEMBL:ENSMUSG00000098679 | MGI:5531256 | miRNA gene | microRNA 7688 |
| 10 | gene | 93.45298 | 93.45305 | negative | MGI_C57BL6J_4413759 | n-TDgtc8 | NCBI_Gene:102467629 | MGI:4413759 | tRNA gene | nuclear encoded tRNA aspartic acid 8 (anticodon GTC) |
| 10 | gene | 93.45340 | 93.48490 | positive | MGI_C57BL6J_96836 | Lta4h | NCBI_Gene:16993,ENSEMBL:ENSMUSG00000015889 | MGI:96836 | protein coding gene | leukotriene A4 hydrolase |
| 10 | gene | 93.48877 | 93.51930 | positive | MGI_C57BL6J_96010 | Hal | NCBI_Gene:15109,ENSEMBL:ENSMUSG00000020017 | MGI:96010 | protein coding gene | histidine ammonia lyase |
| 10 | gene | 93.52334 | 93.54004 | negative | MGI_C57BL6J_1919011 | Amdhd1 | NCBI_Gene:71761,ENSEMBL:ENSMUSG00000015890 | MGI:1919011 | protein coding gene | amidohydrolase domain containing 1 |
| 10 | gene | 93.54057 | 93.58433 | positive | MGI_C57BL6J_2444738 | Ccdc38 | NCBI_Gene:237465,ENSEMBL:ENSMUSG00000036168 | MGI:2444738 | protein coding gene | coiled-coil domain containing 38 |
| 10 | gene | 93.58303 | 93.58971 | negative | MGI_C57BL6J_1917128 | Snrpf | NCBI_Gene:69878,ENSEMBL:ENSMUSG00000020018 | MGI:1917128 | protein coding gene | small nuclear ribonucleoprotein polypeptide F |
| 10 | gene | 93.58941 | 93.60524 | positive | MGI_C57BL6J_1921704 | 4933408J17Rik | NCBI_Gene:74454,ENSEMBL:ENSMUSG00000097407 | MGI:1921704 | lncRNA gene | RIKEN cDNA 4933408J17 gene |
| 10 | gene | 93.61886 | 93.63432 | positive | MGI_C57BL6J_6095926 | Gm47156 | ENSEMBL:ENSMUSG00000111366 | MGI:6095926 | lncRNA gene | predicted gene, 47156 |
| 10 | gene | 93.64068 | 93.74721 | positive | MGI_C57BL6J_1888978 | Ntn4 | NCBI_Gene:57764,ENSEMBL:ENSMUSG00000020019 | MGI:1888978 | protein coding gene | netrin 4 |
| 10 | pseudogene | 93.66524 | 93.66585 | negative | MGI_C57BL6J_3647055 | Gm8580 | NCBI_Gene:667338,ENSEMBL:ENSMUSG00000090159 | MGI:3647055 | pseudogene | predicted gene 8580 |
| 10 | gene | 93.67422 | 93.68332 | negative | MGI_C57BL6J_3801763 | Gm15915 | NCBI_Gene:100504069,ENSEMBL:ENSMUSG00000085723 | MGI:3801763 | lncRNA gene | predicted gene 15915 |
| 10 | gene | 93.72673 | 93.72821 | negative | MGI_C57BL6J_3802140 | Gm15963 | ENSEMBL:ENSMUSG00000086891 | MGI:3802140 | lncRNA gene | predicted gene 15963 |
| 10 | pseudogene | 93.77861 | 93.77977 | positive | MGI_C57BL6J_3781748 | Gm3571 | NCBI_Gene:100041907,ENSEMBL:ENSMUSG00000090610 | MGI:3781748 | pseudogene | predicted gene 3571 |
| 10 | gene | 93.80730 | 93.80740 | positive | MGI_C57BL6J_5456016 | Gm26239 | ENSEMBL:ENSMUSG00000096598 | MGI:5456016 | snRNA gene | predicted gene, 26239 |
| 10 | gene | 93.81997 | 93.85943 | positive | MGI_C57BL6J_3045318 | Usp44 | NCBI_Gene:327799,ENSEMBL:ENSMUSG00000020020 | MGI:3045318 | protein coding gene | ubiquitin specific peptidase 44 |
| 10 | gene | 93.85849 | 93.89709 | negative | MGI_C57BL6J_1929701 | Metap2 | NCBI_Gene:56307,ENSEMBL:ENSMUSG00000036112 | MGI:1929701 | protein coding gene | methionine aminopeptidase 2 |
| 10 | gene | 93.89716 | 93.90196 | positive | MGI_C57BL6J_1922173 | 4930471D02Rik | ENSEMBL:ENSMUSG00000097689 | MGI:1922173 | lncRNA gene | RIKEN cDNA 4930471D02 gene |
| 10 | gene | 93.91617 | 93.91634 | negative | MGI_C57BL6J_5455739 | Gm25962 | ENSEMBL:ENSMUSG00000087798 | MGI:5455739 | snoRNA gene | predicted gene, 25962 |
| 10 | pseudogene | 93.91909 | 93.93627 | positive | MGI_C57BL6J_6097484 | Gm48126 | ENSEMBL:ENSMUSG00000114757 | MGI:6097484 | pseudogene | predicted gene, 48126 |
| 10 | gene | 93.93917 | 94.03582 | negative | MGI_C57BL6J_2143698 | Vezt | NCBI_Gene:215008,ENSEMBL:ENSMUSG00000036099 | MGI:2143698 | protein coding gene | vezatin, adherens junctions transmembrane protein |
| 10 | gene | 93.96152 | 93.96493 | negative | MGI_C57BL6J_2143686 | A630071D13Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA A630071D13 gene |
| 10 | gene | 93.96377 | 93.96386 | negative | MGI_C57BL6J_3619347 | Mir331 | miRBase:MI0000609,NCBI_Gene:723908,ENSEMBL:ENSMUSG00000065607 | MGI:3619347 | miRNA gene | microRNA 331 |
| 10 | pseudogene | 94.02808 | 94.02836 | positive | MGI_C57BL6J_6097573 | Gm48191 | ENSEMBL:ENSMUSG00000111349 | MGI:6097573 | pseudogene | predicted gene, 48191 |
| 10 | gene | 94.03401 | 94.03580 | negative | MGI_C57BL6J_6215053 | Gm49628 | ENSEMBL:ENSMUSG00000110869 | MGI:6215053 | unclassified gene | predicted gene, 49628 |
| 10 | gene | 94.03600 | 94.14534 | positive | MGI_C57BL6J_1261419 | Fgd6 | NCBI_Gene:13998,ENSEMBL:ENSMUSG00000020021 | MGI:1261419 | protein coding gene | FYVE, RhoGEF and PH domain containing 6 |
| 10 | pseudogene | 94.09748 | 94.09815 | negative | MGI_C57BL6J_5010576 | Gm18391 | NCBI_Gene:100417060,ENSEMBL:ENSMUSG00000110753 | MGI:5010576 | pseudogene | predicted gene, 18391 |
| 10 | gene | 94.10874 | 94.11115 | positive | MGI_C57BL6J_6097846 | Gm48372 | ENSEMBL:ENSMUSG00000111539 | MGI:6097846 | unclassified gene | predicted gene, 48372 |
| 10 | gene | 94.14751 | 94.19722 | positive | MGI_C57BL6J_1352465 | Nr2c1 | NCBI_Gene:22025,ENSEMBL:ENSMUSG00000005897 | MGI:1352465 | protein coding gene | nuclear receptor subfamily 2, group C, member 1 |
| 10 | gene | 94.18449 | 94.18457 | negative | MGI_C57BL6J_5455899 | Gm26122 | ENSEMBL:ENSMUSG00000065296 | MGI:5455899 | snRNA gene | predicted gene, 26122 |
| 10 | gene | 94.19872 | 94.22144 | positive | MGI_C57BL6J_1913664 | Ndufa12 | NCBI_Gene:66414,ENSEMBL:ENSMUSG00000020022 | MGI:1913664 | protein coding gene | NADH:ubiquinone oxidoreductase subunit A12 |
| 10 | gene | 94.23056 | 94.23861 | negative | MGI_C57BL6J_5591908 | Gm32749 | NCBI_Gene:102635396 | MGI:5591908 | lncRNA gene | predicted gene, 32749 |
| 10 | gene | 94.29366 | 94.29870 | negative | MGI_C57BL6J_3648918 | Gm4792 | NCBI_Gene:215472,ENSEMBL:ENSMUSG00000053420 | MGI:3648918 | protein coding gene | predicted gene 4792 |
| 10 | gene | 94.31191 | 94.59096 | positive | MGI_C57BL6J_2442900 | Tmcc3 | NCBI_Gene:319880,ENSEMBL:ENSMUSG00000020023 | MGI:2442900 | protein coding gene | transmembrane and coiled coil domains 3 |
| 10 | gene | 94.40889 | 94.41073 | positive | MGI_C57BL6J_3801971 | Gm16155 | ENSEMBL:ENSMUSG00000085409 | MGI:3801971 | lncRNA gene | predicted gene 16155 |
| 10 | gene | 94.41796 | 94.42863 | negative | MGI_C57BL6J_5591986 | Gm32827 | NCBI_Gene:102635509 | MGI:5591986 | lncRNA gene | predicted gene, 32827 |
| 10 | gene | 94.51285 | 94.51478 | negative | MGI_C57BL6J_5623628 | Gm40743 | NCBI_Gene:105245265 | MGI:5623628 | lncRNA gene | predicted gene, 40743 |
| 10 | gene | 94.56783 | 94.61208 | negative | MGI_C57BL6J_3801736 | Tmcc3os | ENSEMBL:ENSMUSG00000086269 | MGI:3801736 | antisense lncRNA gene | transmembrane and coiled coil domains 3, opposite strand |
| 10 | gene | 94.58614 | 94.58620 | negative | MGI_C57BL6J_5530918 | Mir7211 | miRBase:MI0023706,NCBI_Gene:102466817,ENSEMBL:ENSMUSG00000098544 | MGI:5530918 | miRNA gene | microRNA 7211 |
| 10 | gene | 94.60005 | 94.62370 | negative | MGI_C57BL6J_5592057 | Gm32898 | NCBI_Gene:102635607 | MGI:5592057 | lncRNA gene | predicted gene, 32898 |
| 10 | gene | 94.62054 | 94.64866 | positive | MGI_C57BL6J_6098271 | Gm48657 | ENSEMBL:ENSMUSG00000112477 | MGI:6098271 | lncRNA gene | predicted gene, 48657 |
| 10 | pseudogene | 94.65280 | 94.65305 | negative | MGI_C57BL6J_6098273 | Gm48658 | ENSEMBL:ENSMUSG00000112473 | MGI:6098273 | pseudogene | predicted gene, 48658 |
| 10 | gene | 94.67103 | 94.68861 | negative | MGI_C57BL6J_1914973 | Cep83os | NCBI_Gene:67723,ENSEMBL:ENSMUSG00000097164 | MGI:1914973 | bidirectional promoter lncRNA gene | centrosomal protein 83, opposite strand |
| 10 | gene | 94.68570 | 94.68862 | positive | MGI_C57BL6J_6118596 | Gm49167 | ENSEMBL:ENSMUSG00000112203 | MGI:6118596 | unclassified gene | predicted gene, 49167 |
| 10 | gene | 94.68850 | 94.79085 | positive | MGI_C57BL6J_1924298 | Cep83 | NCBI_Gene:77048,ENSEMBL:ENSMUSG00000020024 | MGI:1924298 | protein coding gene | centrosomal protein 83 |
| 10 | gene | 94.78197 | 94.78716 | negative | MGI_C57BL6J_6098317 | Gm48689 | ENSEMBL:ENSMUSG00000112669 | MGI:6098317 | lncRNA gene | predicted gene, 48689 |
| 10 | gene | 94.79087 | 94.94527 | negative | MGI_C57BL6J_1890127 | Plxnc1 | NCBI_Gene:54712,ENSEMBL:ENSMUSG00000074785 | MGI:1890127 | protein coding gene | plexin C1 |
| 10 | gene | 94.82153 | 94.82166 | negative | MGI_C57BL6J_5453963 | Gm24186 | ENSEMBL:ENSMUSG00000088608 | MGI:5453963 | rRNA gene | predicted gene, 24186 |
| 10 | gene | 94.88131 | 94.88379 | negative | MGI_C57BL6J_6098369 | Gm48718 | ENSEMBL:ENSMUSG00000112736 | MGI:6098369 | unclassified gene | predicted gene, 48718 |
| 10 | gene | 94.94859 | 94.94889 | positive | MGI_C57BL6J_5455043 | Gm25266 | ENSEMBL:ENSMUSG00000089146 | MGI:5455043 | unclassified non-coding RNA gene | predicted gene, 25266 |
| 10 | gene | 94.96164 | 94.96524 | positive | MGI_C57BL6J_5623629 | Gm40744 | NCBI_Gene:105245266 | MGI:5623629 | lncRNA gene | predicted gene, 40744 |
| 10 | gene | 94.99395 | 94.99944 | negative | MGI_C57BL6J_6098611 | Gm48867 | ENSEMBL:ENSMUSG00000112277 | MGI:6098611 | lncRNA gene | predicted gene, 48867 |
| 10 | gene | 95.11638 | 95.11773 | positive | MGI_C57BL6J_6098613 | Gm48868 | ENSEMBL:ENSMUSG00000112677 | MGI:6098613 | unclassified gene | predicted gene, 48868 |
| 10 | gene | 95.12058 | 95.14583 | negative | MGI_C57BL6J_5588843 | Gm29684 | NCBI_Gene:628062,ENSEMBL:ENSMUSG00000112481 | MGI:5588843 | lncRNA gene | predicted gene, 29684 |
| 10 | gene | 95.13261 | 95.14062 | negative | MGI_C57BL6J_5592250 | Gm33091 | NCBI_Gene:102635855,ENSEMBL:ENSMUSG00000112501 | MGI:5592250 | lncRNA gene | predicted gene, 33091 |
| 10 | gene | 95.16036 | 95.16566 | positive | MGI_C57BL6J_6098623 | Gm48874 | ENSEMBL:ENSMUSG00000112682 | MGI:6098623 | unclassified gene | predicted gene, 48874 |
| 10 | gene | 95.17474 | 95.32413 | negative | MGI_C57BL6J_1336168 | Cradd | NCBI_Gene:12905,ENSEMBL:ENSMUSG00000045867 | MGI:1336168 | protein coding gene | CASP2 and RIPK1 domain containing adaptor with death domain |
| 10 | gene | 95.26123 | 95.26498 | negative | MGI_C57BL6J_2443745 | A630089K24Rik | NA | NA | unclassified gene | RIKEN cDNA A630089K24 gene |
| 10 | gene | 95.31287 | 95.31485 | negative | MGI_C57BL6J_6098634 | Gm48880 | ENSEMBL:ENSMUSG00000112744 | MGI:6098634 | unclassified gene | predicted gene, 48880 |
| 10 | gene | 95.31825 | 95.32436 | positive | MGI_C57BL6J_6098637 | Gm48882 | ENSEMBL:ENSMUSG00000112593 | MGI:6098637 | lncRNA gene | predicted gene, 48882 |
| 10 | gene | 95.33628 | 95.36620 | positive | MGI_C57BL6J_1916879 | 2310039L15Rik | NCBI_Gene:69629,ENSEMBL:ENSMUSG00000100550 | MGI:1916879 | lncRNA gene | RIKEN cDNA 2310039L15 gene |
| 10 | gene | 95.38390 | 95.41725 | negative | MGI_C57BL6J_1201787 | Socs2 | NCBI_Gene:216233,ENSEMBL:ENSMUSG00000020027 | MGI:1201787 | protein coding gene | suppressor of cytokine signaling 2 |
| 10 | gene | 95.41312 | 95.41460 | positive | MGI_C57BL6J_5592389 | Gm33230 | NCBI_Gene:102636052 | MGI:5592389 | lncRNA gene | predicted gene, 33230 |
| 10 | gene | 95.41738 | 95.42864 | positive | MGI_C57BL6J_1917773 | 5730420D15Rik | NCBI_Gene:70523,ENSEMBL:ENSMUSG00000097766 | MGI:1917773 | lncRNA gene | RIKEN cDNA 5730420D15 gene |
| 10 | gene | 95.44532 | 95.46122 | negative | MGI_C57BL6J_6096651 | Gm47599 | ENSEMBL:ENSMUSG00000112261 | MGI:6096651 | lncRNA gene | predicted gene, 47599 |
| 10 | pseudogene | 95.45467 | 95.45497 | negative | MGI_C57BL6J_6096653 | Gm47600 | ENSEMBL:ENSMUSG00000112725 | MGI:6096653 | pseudogene | predicted gene, 47600 |
| 10 | pseudogene | 95.47699 | 95.47841 | negative | MGI_C57BL6J_3781795 | Gm3619 | NCBI_Gene:100042004,ENSEMBL:ENSMUSG00000112450 | MGI:3781795 | pseudogene | predicted gene 3619 |
| 10 | gene | 95.48081 | 95.50199 | negative | MGI_C57BL6J_1333774 | Mrpl42 | NCBI_Gene:67270,ENSEMBL:ENSMUSG00000062981 | MGI:1333774 | protein coding gene | mitochondrial ribosomal protein L42 |
| 10 | gene | 95.51223 | 95.51479 | negative | MGI_C57BL6J_5825876 | Gm46239 | NCBI_Gene:108167829 | MGI:5825876 | lncRNA gene | predicted gene, 46239 |
| 10 | gene | 95.51515 | 95.54566 | positive | MGI_C57BL6J_1934835 | Ube2n | NCBI_Gene:93765,ENSEMBL:ENSMUSG00000074781 | MGI:1934835 | protein coding gene | ubiquitin-conjugating enzyme E2N |
| 10 | gene | 95.54701 | 95.56418 | negative | MGI_C57BL6J_1918457 | Nudt4 | NCBI_Gene:71207,ENSEMBL:ENSMUSG00000020029 | MGI:1918457 | protein coding gene | nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| 10 | gene | 95.55923 | 95.55932 | negative | MGI_C57BL6J_4834226 | Mir3058 | miRBase:MI0004418,NCBI_Gene:100526463,ENSEMBL:ENSMUSG00000092867 | MGI:4834226 | miRNA gene | microRNA 3058 |
| 10 | gene | 95.60379 | 95.60412 | positive | MGI_C57BL6J_5453553 | Gm23776 | ENSEMBL:ENSMUSG00000089618 | MGI:5453553 | unclassified non-coding RNA gene | predicted gene, 23776 |
| 10 | gene | 95.63024 | 95.63224 | negative | MGI_C57BL6J_5592495 | Gm33336 | NCBI_Gene:102636201,ENSEMBL:ENSMUSG00000112439 | MGI:5592495 | lncRNA gene | predicted gene, 33336 |
| 10 | gene | 95.63448 | 95.63643 | positive | MGI_C57BL6J_6096760 | Gm47667 | ENSEMBL:ENSMUSG00000112538 | MGI:6096760 | lncRNA gene | predicted gene, 47667 |
| 10 | pseudogene | 95.67269 | 95.67351 | negative | MGI_C57BL6J_3646302 | Anapc15-ps | NCBI_Gene:628119,ENSEMBL:ENSMUSG00000074780 | MGI:3646302 | pseudogene | anaphase prompoting complex C subunit 15, pseudogene |
| 10 | gene | 95.68081 | 95.68112 | positive | MGI_C57BL6J_1916857 | 2310011C19Rik | NA | NA | unclassified gene | RIKEN cDNA 2310011C19 gene |
| 10 | pseudogene | 95.70305 | 95.70346 | negative | MGI_C57BL6J_6096763 | Gm47669 | ENSEMBL:ENSMUSG00000112025 | MGI:6096763 | pseudogene | predicted gene, 47669 |
| 10 | gene | 95.71400 | 95.71751 | negative | MGI_C57BL6J_6096765 | Gm47671 | ENSEMBL:ENSMUSG00000112519 | MGI:6096765 | lncRNA gene | predicted gene, 47671 |
| 10 | gene | 95.71913 | 95.72250 | negative | MGI_C57BL6J_1915779 | 1110019B22Rik | ENSEMBL:ENSMUSG00000112289 | MGI:1915779 | lncRNA gene | RIKEN cDNA 1110019B22 gene |
| 10 | gene | 95.73018 | 95.73936 | positive | MGI_C57BL6J_6096827 | Gm47708 | ENSEMBL:ENSMUSG00000112188 | MGI:6096827 | lncRNA gene | predicted gene, 47708 |
| 10 | gene | 95.74608 | 95.79724 | positive | MGI_C57BL6J_3041208 | 4732465J04Rik | NCBI_Gene:414105,ENSEMBL:ENSMUSG00000101517 | MGI:3041208 | lncRNA gene | RIKEN cDNA 4732465J04 gene |
| 10 | gene | 95.81134 | 95.81789 | negative | MGI_C57BL6J_6096845 | Gm47718 | ENSEMBL:ENSMUSG00000112771 | MGI:6096845 | lncRNA gene | predicted gene, 47718 |
| 10 | gene | 95.81732 | 95.89681 | positive | MGI_C57BL6J_5592536 | Gm33377 | NCBI_Gene:102636263 | MGI:5592536 | lncRNA gene | predicted gene, 33377 |
| 10 | gene | 95.81757 | 95.84912 | positive | MGI_C57BL6J_6096847 | Gm47719 | ENSEMBL:ENSMUSG00000112599 | MGI:6096847 | lncRNA gene | predicted gene, 47719 |
| 10 | gene | 95.82397 | 95.84316 | negative | MGI_C57BL6J_5592702 | Gm33543 | NCBI_Gene:102636491,ENSEMBL:ENSMUSG00000110353 | MGI:5592702 | lncRNA gene | predicted gene, 33543 |
| 10 | pseudogene | 95.87048 | 95.87080 | negative | MGI_C57BL6J_6096855 | Gm47724 | ENSEMBL:ENSMUSG00000111992 | MGI:6096855 | pseudogene | predicted gene, 47724 |
| 10 | gene | 95.89344 | 95.89726 | positive | MGI_C57BL6J_6096856 | Gm47725 | ENSEMBL:ENSMUSG00000112400 | MGI:6096856 | lncRNA gene | predicted gene, 47725 |
| 10 | gene | 95.89860 | 95.89874 | positive | MGI_C57BL6J_5454210 | Gm24433 | ENSEMBL:ENSMUSG00000077268 | MGI:5454210 | snoRNA gene | predicted gene, 24433 |
| 10 | gene | 95.94065 | 96.04552 | positive | MGI_C57BL6J_2442192 | Eea1 | NCBI_Gene:216238,ENSEMBL:ENSMUSG00000036499 | MGI:2442192 | protein coding gene | early endosome antigen 1 |
| 10 | pseudogene | 96.04763 | 96.09601 | negative | MGI_C57BL6J_6270484 | Gm49817 | ENSEMBL:ENSMUSG00000117008 | MGI:6270484 | pseudogene | predicted gene, 49817 |
| 10 | pseudogene | 96.12798 | 96.12974 | negative | MGI_C57BL6J_6097923 | Gm48425 | ENSEMBL:ENSMUSG00000112316 | MGI:6097923 | pseudogene | predicted gene, 48425 |
| 10 | gene | 96.13655 | 96.13704 | positive | MGI_C57BL6J_3646788 | Gm5426 | NCBI_Gene:432491,ENSEMBL:ENSMUSG00000096486 | MGI:3646788 | protein coding gene | predicted pseudogene 5426 |
| 10 | gene | 96.16433 | 96.16443 | positive | MGI_C57BL6J_5452690 | Gm22913 | ENSEMBL:ENSMUSG00000094523 | MGI:5452690 | miRNA gene | predicted gene, 22913 |
| 10 | gene | 96.18092 | 96.18269 | positive | MGI_C57BL6J_5623631 | Gm40746 | NCBI_Gene:105245268 | MGI:5623631 | lncRNA gene | predicted gene, 40746 |
| 10 | gene | 96.22602 | 96.24887 | positive | MGI_C57BL6J_1922131 | 4930459C07Rik | NCBI_Gene:74881,ENSEMBL:ENSMUSG00000112762 | MGI:1922131 | lncRNA gene | RIKEN cDNA 4930459C07 gene |
| 10 | gene | 96.22633 | 96.25891 | negative | MGI_C57BL6J_5623632 | Gm40747 | NCBI_Gene:105245269 | MGI:5623632 | lncRNA gene | predicted gene, 40747 |
| 10 | gene | 96.30550 | 96.30873 | negative | MGI_C57BL6J_5623633 | Gm40748 | NCBI_Gene:105245270 | MGI:5623633 | lncRNA gene | predicted gene, 40748 |
| 10 | gene | 96.30908 | 96.36253 | positive | MGI_C57BL6J_5592941 | Gm33782 | NCBI_Gene:102636813,ENSEMBL:ENSMUSG00000112290 | MGI:5592941 | lncRNA gene | predicted gene, 33782 |
| 10 | gene | 96.30934 | 96.32072 | positive | MGI_C57BL6J_6097926 | Gm48427 | ENSEMBL:ENSMUSG00000112234 | MGI:6097926 | lncRNA gene | predicted gene, 48427 |
| 10 | gene | 96.33609 | 96.34582 | negative | MGI_C57BL6J_6097928 | Gm48428 | ENSEMBL:ENSMUSG00000112749 | MGI:6097928 | lncRNA gene | predicted gene, 48428 |
| 10 | pseudogene | 96.40873 | 96.40913 | negative | MGI_C57BL6J_5012276 | Gm20091 | NCBI_Gene:100504157,ENSEMBL:ENSMUSG00000094249 | MGI:5012276 | pseudogene | predicted gene, 20091 |
| 10 | pseudogene | 96.47647 | 96.47753 | positive | MGI_C57BL6J_3643183 | Gm8601 | NCBI_Gene:667380,ENSEMBL:ENSMUSG00000097939 | MGI:3643183 | pseudogene | predicted gene 8601 |
| 10 | gene | 96.49572 | 96.52398 | negative | MGI_C57BL6J_6098035 | Gm48505 | ENSEMBL:ENSMUSG00000112264 | MGI:6098035 | lncRNA gene | predicted gene, 48505 |
| 10 | gene | 96.51916 | 96.59874 | negative | MGI_C57BL6J_5593002 | Gm33843 | NCBI_Gene:102636895,ENSEMBL:ENSMUSG00000112354 | MGI:5593002 | lncRNA gene | predicted gene, 33843 |
| 10 | gene | 96.61700 | 96.62281 | positive | MGI_C57BL6J_88215 | Btg1 | NCBI_Gene:12226,ENSEMBL:ENSMUSG00000036478 | MGI:88215 | protein coding gene | B cell translocation gene 1, anti-proliferative |
| 10 | pseudogene | 96.71181 | 96.71209 | positive | MGI_C57BL6J_6098038 | Gm48507 | ENSEMBL:ENSMUSG00000112362 | MGI:6098038 | pseudogene | predicted gene, 48507 |
| 10 | gene | 96.71576 | 96.72792 | positive | MGI_C57BL6J_5593056 | Gm33897 | NCBI_Gene:102636973 | MGI:5593056 | lncRNA gene | predicted gene, 33897 |
| 10 | pseudogene | 96.76848 | 96.76983 | negative | MGI_C57BL6J_3647629 | Gm6859 | NCBI_Gene:628256,ENSEMBL:ENSMUSG00000112588 | MGI:3647629 | pseudogene | predicted gene 6859 |
| 10 | pseudogene | 96.79839 | 96.79872 | positive | MGI_C57BL6J_6098041 | Gm48509 | ENSEMBL:ENSMUSG00000112293 | MGI:6098041 | pseudogene | predicted gene, 48509 |
| 10 | gene | 96.80104 | 96.88471 | positive | MGI_C57BL6J_6098057 | Gm48521 | ENSEMBL:ENSMUSG00000111912 | MGI:6098057 | lncRNA gene | predicted gene, 48521 |
| 10 | gene | 96.85558 | 96.85567 | negative | MGI_C57BL6J_5454364 | Gm24587 | ENSEMBL:ENSMUSG00000080347 | MGI:5454364 | miRNA gene | predicted gene, 24587 |
| 10 | gene | 96.90133 | 96.92019 | positive | MGI_C57BL6J_5825877 | Gm46240 | NCBI_Gene:108167830 | MGI:5825877 | protein coding gene | predicted gene, 46240 |
| 10 | gene | 96.90316 | 96.95887 | negative | MGI_C57BL6J_5593140 | Gm33981 | NCBI_Gene:102637079,ENSEMBL:ENSMUSG00000112488 | MGI:5593140 | lncRNA gene | predicted gene, 33981 |
| 10 | gene | 97.03577 | 97.06106 | positive | MGI_C57BL6J_1922551 | 4930556N09Rik | NCBI_Gene:75301,ENSEMBL:ENSMUSG00000111924 | MGI:1922551 | lncRNA gene | RIKEN cDNA 4930556N09 gene |
| 10 | gene | 97.05774 | 97.05825 | positive | MGI_C57BL6J_1920555 | 1700038C09Rik | NA | NA | unclassified gene | RIKEN cDNA 1700038C09 gene |
| 10 | pseudogene | 97.10549 | 97.10658 | negative | MGI_C57BL6J_6098109 | Gm48555 | ENSEMBL:ENSMUSG00000112631 | MGI:6098109 | pseudogene | predicted gene, 48555 |
| 10 | pseudogene | 97.19920 | 97.20045 | negative | MGI_C57BL6J_3648168 | Gm8613 | NCBI_Gene:667407,ENSEMBL:ENSMUSG00000112892 | MGI:3648168 | pseudogene | predicted gene 8613 |
| 10 | gene | 97.20739 | 97.23576 | negative | MGI_C57BL6J_5593203 | Gm34044 | NCBI_Gene:102637160 | MGI:5593203 | lncRNA gene | predicted gene, 34044 |
| 10 | pseudogene | 97.24191 | 97.24321 | positive | MGI_C57BL6J_5010700 | Gm18515 | NCBI_Gene:100417299,ENSEMBL:ENSMUSG00000111981 | MGI:5010700 | pseudogene | predicted gene, 18515 |
| 10 | gene | 97.38876 | 97.42858 | negative | MGI_C57BL6J_5593273 | Gm34114 | NCBI_Gene:102637248 | MGI:5593273 | lncRNA gene | predicted gene, 34114 |
| 10 | gene | 97.42348 | 97.42356 | positive | MGI_C57BL6J_4950405 | Mir3966 | miRBase:MI0016975,NCBI_Gene:100628616,ENSEMBL:ENSMUSG00000105557 | MGI:4950405 | miRNA gene | microRNA 3966 |
| 10 | gene | 97.47950 | 97.51816 | positive | MGI_C57BL6J_94872 | Dcn | NCBI_Gene:13179,ENSEMBL:ENSMUSG00000019929 | MGI:94872 | protein coding gene | decorin |
| 10 | gene | 97.56513 | 97.57270 | positive | MGI_C57BL6J_109347 | Lum | NCBI_Gene:17022,ENSEMBL:ENSMUSG00000036446 | MGI:109347 | protein coding gene | lumican |
| 10 | gene | 97.59098 | 97.59693 | negative | MGI_C57BL6J_1922275 | 4930473O22Rik | NCBI_Gene:75025,ENSEMBL:ENSMUSG00000112484 | MGI:1922275 | lncRNA gene | RIKEN cDNA 4930473O22 gene |
| 10 | gene | 97.60688 | 97.61369 | positive | MGI_C57BL6J_1202398 | Kera | NCBI_Gene:16545,ENSEMBL:ENSMUSG00000019932 | MGI:1202398 | protein coding gene | keratocan |
| 10 | gene | 97.62017 | 97.62326 | negative | MGI_C57BL6J_5623634 | Gm40749 | NCBI_Gene:105245271 | MGI:5623634 | lncRNA gene | predicted gene, 40749 |
| 10 | gene | 97.64407 | 97.68245 | positive | MGI_C57BL6J_107942 | Epyc | NCBI_Gene:13516,ENSEMBL:ENSMUSG00000019936 | MGI:107942 | protein coding gene | epiphycan |
| 10 | gene | 97.68141 | 97.96709 | negative | MGI_C57BL6J_3642925 | Gm10754 | NCBI_Gene:100038699,ENSEMBL:ENSMUSG00000074776 | MGI:3642925 | lncRNA gene | predicted gene 10754 |
| 10 | gene | 97.69306 | 97.69493 | positive | MGI_C57BL6J_1913966 | Ccer1 | NCBI_Gene:66716,ENSEMBL:ENSMUSG00000047025 | MGI:1913966 | protein coding gene | coiled-coil glutamate-rich protein 1 |
| 10 | gene | 97.71477 | 97.75370 | positive | MGI_C57BL6J_1925377 | 4930525C09Rik | NCBI_Gene:78127,ENSEMBL:ENSMUSG00000112497 | MGI:1925377 | lncRNA gene | RIKEN cDNA 4930525C09 gene |
| 10 | gene | 97.72487 | 97.72675 | negative | MGI_C57BL6J_3641867 | C030005K15Rik | ENSEMBL:ENSMUSG00000079183 | MGI:3641867 | protein coding gene | RIKEN cDNA C030005K15 gene |
| 10 | gene | 97.74476 | 97.74488 | negative | MGI_C57BL6J_5455435 | Gm25658 | ENSEMBL:ENSMUSG00000089097 | MGI:5455435 | snoRNA gene | predicted gene, 25658 |
| 10 | gene | 98.29993 | 98.35187 | negative | MGI_C57BL6J_3647678 | Gm8633 | NCBI_Gene:667438,ENSEMBL:ENSMUSG00000112896 | MGI:3647678 | lncRNA gene | predicted gene 8633 |
| 10 | pseudogene | 98.30102 | 98.30125 | positive | MGI_C57BL6J_6098315 | Gm48687 | ENSEMBL:ENSMUSG00000112457 | MGI:6098315 | pseudogene | predicted gene, 48687 |
| 10 | gene | 98.48651 | 98.48687 | negative | MGI_C57BL6J_5610859 | Gm37631 | ENSEMBL:ENSMUSG00000102310 | MGI:5610859 | unclassified gene | predicted gene, 37631 |
| 10 | gene | 98.53748 | 98.56984 | positive | MGI_C57BL6J_5593456 | Gm34297 | NCBI_Gene:102637506,ENSEMBL:ENSMUSG00000112874 | MGI:5593456 | lncRNA gene | predicted gene, 34297 |
| 10 | gene | 98.65380 | 98.69971 | negative | MGI_C57BL6J_3646787 | Gm5427 | ENSEMBL:ENSMUSG00000112647 | MGI:3646787 | lncRNA gene | predicted gene 5427 |
| 10 | gene | 98.80714 | 98.80947 | positive | MGI_C57BL6J_5623635 | Gm40750 | NCBI_Gene:105245273 | MGI:5623635 | lncRNA gene | predicted gene, 40750 |
| 10 | gene | 98.83401 | 98.88274 | negative | MGI_C57BL6J_5593505 | Gm34346 | NCBI_Gene:102637570 | MGI:5593505 | lncRNA gene | predicted gene, 34346 |
| 10 | gene | 98.91436 | 99.02614 | positive | MGI_C57BL6J_104653 | Atp2b1 | NCBI_Gene:67972,ENSEMBL:ENSMUSG00000019943 | MGI:104653 | protein coding gene | ATPase, Ca++ transporting, plasma membrane 1 |
| 10 | gene | 99.03947 | 99.04144 | positive | MGI_C57BL6J_6098443 | Gm48761 | ENSEMBL:ENSMUSG00000112660 | MGI:6098443 | unclassified gene | predicted gene, 48761 |
| 10 | gene | 99.10579 | 99.10808 | negative | MGI_C57BL6J_6098457 | Gm48768 | ENSEMBL:ENSMUSG00000112346 | MGI:6098457 | lncRNA gene | predicted gene, 48768 |
| 10 | gene | 99.10704 | 99.19807 | positive | MGI_C57BL6J_1918511 | Poc1b | NCBI_Gene:382406,ENSEMBL:ENSMUSG00000019952 | MGI:1918511 | protein coding gene | POC1 centriolar protein B |
| 10 | gene | 99.10814 | 99.11325 | positive | MGI_C57BL6J_894692 | Galnt4 | NCBI_Gene:14426,ENSEMBL:ENSMUSG00000090035 | MGI:894692 | protein coding gene | polypeptide N-acetylgalactosaminyltransferase 4 |
| 10 | gene | 99.12517 | 99.12632 | negative | MGI_C57BL6J_104524 | Phxr2 | ENSEMBL:ENSMUSG00000055108 | MGI:104524 | protein coding gene | per-hexamer repeat gene 2 |
| 10 | gene | 99.17918 | 99.18461 | negative | MGI_C57BL6J_3801746 | Gm16239 | ENSEMBL:ENSMUSG00000089994 | MGI:3801746 | lncRNA gene | predicted gene 16239 |
| 10 | gene | 99.21644 | 99.23209 | negative | MGI_C57BL6J_5593733 | Gm34574 | NCBI_Gene:102637874,ENSEMBL:ENSMUSG00000112805 | MGI:5593733 | lncRNA gene | predicted gene, 34574 |
| 10 | gene | 99.23279 | 99.23762 | negative | MGI_C57BL6J_6098641 | Gm48884 | ENSEMBL:ENSMUSG00000112902 | MGI:6098641 | lncRNA gene | predicted gene, 48884 |
| 10 | gene | 99.23307 | 99.24306 | positive | MGI_C57BL6J_5593936 | Gm34777 | NCBI_Gene:102638142,ENSEMBL:ENSMUSG00000111923 | MGI:5593936 | lncRNA gene | predicted gene, 34777 |
| 10 | gene | 99.26323 | 99.26749 | positive | MGI_C57BL6J_1914853 | Dusp6 | NCBI_Gene:67603,ENSEMBL:ENSMUSG00000019960 | MGI:1914853 | protein coding gene | dual specificity phosphatase 6 |
| 10 | gene | 99.26686 | 99.27091 | negative | MGI_C57BL6J_5594080 | Gm34921 | NCBI_Gene:102638331,ENSEMBL:ENSMUSG00000112384 | MGI:5594080 | lncRNA gene | predicted gene, 34921 |
| 10 | gene | 99.26790 | 99.27908 | positive | MGI_C57BL6J_6097430 | Gm48089 | ENSEMBL:ENSMUSG00000112544 | MGI:6097430 | lncRNA gene | predicted gene, 48089 |
| 10 | gene | 99.29889 | 99.42305 | positive | MGI_C57BL6J_2445145 | B530045E10Rik | ENSEMBL:ENSMUSG00000044633 | MGI:2445145 | lncRNA gene | RIKEN cDNA B530045E10 gene |
| 10 | pseudogene | 99.44384 | 99.44703 | positive | MGI_C57BL6J_95633 | Gad1-ps | NCBI_Gene:382407,ENSEMBL:ENSMUSG00000090665 | MGI:95633 | pseudogene | glutamate decarboxylase 1, pseudogene |
| 10 | gene | 99.45022 | 99.46204 | negative | MGI_C57BL6J_5594194 | Gm35035 | NCBI_Gene:102638482,ENSEMBL:ENSMUSG00000112082 | MGI:5594194 | lncRNA gene | predicted gene, 35035 |
| 10 | gene | 99.45689 | 99.49059 | positive | MGI_C57BL6J_5594142 | Gm34983 | NCBI_Gene:102638407,ENSEMBL:ENSMUSG00000112265 | MGI:5594142 | lncRNA gene | predicted gene, 34983 |
| 10 | pseudogene | 99.53762 | 99.53807 | negative | MGI_C57BL6J_6097425 | Gm48085 | ENSEMBL:ENSMUSG00000112180 | MGI:6097425 | pseudogene | predicted gene, 48085 |
| 10 | gene | 99.60917 | 99.65813 | positive | MGI_C57BL6J_5012295 | Gm20110 | NCBI_Gene:100504202,ENSEMBL:ENSMUSG00000111962 | MGI:5012295 | lncRNA gene | predicted gene, 20110 |
| 10 | pseudogene | 99.63872 | 99.64192 | negative | MGI_C57BL6J_5010594 | Gm18409 | NCBI_Gene:100417125,ENSEMBL:ENSMUSG00000112503 | MGI:5010594 | pseudogene | predicted gene, 18409 |
| 10 | gene | 99.66077 | 99.69201 | positive | MGI_C57BL6J_5594260 | Gm35101 | NCBI_Gene:102638567,ENSEMBL:ENSMUSG00000112810 | MGI:5594260 | lncRNA gene | predicted gene, 35101 |
| 10 | gene | 99.71914 | 99.89116 | positive | MGI_C57BL6J_6096613 | Gm47578 | ENSEMBL:ENSMUSG00000113032 | MGI:6096613 | lncRNA gene | predicted gene, 47578 |
| 10 | gene | 99.75771 | 99.75966 | negative | MGI_C57BL6J_1919082 | Csl | NCBI_Gene:71832,ENSEMBL:ENSMUSG00000046934 | MGI:1919082 | protein coding gene | citrate synthase like |
| 10 | gene | 99.75951 | 99.83056 | positive | MGI_C57BL6J_5594365 | Gm35206 | NCBI_Gene:102638712,ENSEMBL:ENSMUSG00000112304 | MGI:5594365 | lncRNA gene | predicted gene, 35206 |
| 10 | pseudogene | 99.86537 | 99.86602 | negative | MGI_C57BL6J_6096615 | Gm47579 | ENSEMBL:ENSMUSG00000112463 | MGI:6096615 | pseudogene | predicted gene, 47579 |
| 10 | gene | 99.90300 | 99.92554 | positive | MGI_C57BL6J_5594596 | Gm35437 | NCBI_Gene:102639018 | MGI:5594596 | lncRNA gene | predicted gene, 35437 |
| 10 | pseudogene | 99.98862 | 99.98979 | positive | MGI_C57BL6J_5010055 | Gm17870 | NCBI_Gene:100416012,ENSEMBL:ENSMUSG00000112299 | MGI:5010055 | pseudogene | predicted gene, 17870 |
| 10 | gene | 100.01563 | 100.10042 | positive | MGI_C57BL6J_96974 | Kitl | NCBI_Gene:17311,ENSEMBL:ENSMUSG00000019966 | MGI:96974 | protein coding gene | kit ligand |
| 10 | gene | 100.12987 | 100.14414 | negative | MGI_C57BL6J_5594692 | Gm35533 | NCBI_Gene:102639153,ENSEMBL:ENSMUSG00000112831 | MGI:5594692 | lncRNA gene | predicted gene, 35533 |
| 10 | gene | 100.13950 | 100.13961 | positive | MGI_C57BL6J_5452695 | Gm22918 | ENSEMBL:ENSMUSG00000096586 | MGI:5452695 | miRNA gene | predicted gene, 22918 |
| 10 | gene | 100.14425 | 100.14436 | positive | MGI_C57BL6J_5455064 | Gm25287 | ENSEMBL:ENSMUSG00000095539 | MGI:5455064 | miRNA gene | predicted gene, 25287 |
| 10 | gene | 100.20387 | 100.20434 | negative | MGI_C57BL6J_6096695 | Gm47627 | ENSEMBL:ENSMUSG00000112633 | MGI:6096695 | unclassified gene | predicted gene, 47627 |
| 10 | pseudogene | 100.29794 | 100.29865 | negative | MGI_C57BL6J_3646823 | Gm8689 | NCBI_Gene:667532,ENSEMBL:ENSMUSG00000112245 | MGI:3646823 | pseudogene | predicted gene 8689 |
| 10 | pseudogene | 100.30650 | 100.30719 | negative | MGI_C57BL6J_3779885 | Gm9476 | NCBI_Gene:670007,ENSEMBL:ENSMUSG00000112069 | MGI:3779885 | pseudogene | predicted gene 9476 |
| 10 | pseudogene | 100.31702 | 100.32027 | negative | MGI_C57BL6J_6096703 | Gm47632 | ENSEMBL:ENSMUSG00000112522 | MGI:6096703 | pseudogene | predicted gene, 47632 |
| 10 | pseudogene | 100.32031 | 100.32143 | negative | MGI_C57BL6J_6096697 | Gm47628 | ENSEMBL:ENSMUSG00000112122 | MGI:6096697 | pseudogene | predicted gene, 47628 |
| 10 | pseudogene | 100.32110 | 100.32137 | positive | MGI_C57BL6J_6096700 | Gm47630 | ENSEMBL:ENSMUSG00000112178 | MGI:6096700 | pseudogene | predicted gene, 47630 |
| 10 | pseudogene | 100.32546 | 100.32629 | positive | MGI_C57BL6J_3647354 | Gm4898 | NCBI_Gene:235966,ENSEMBL:ENSMUSG00000112737 | MGI:3647354 | pseudogene | predicted gene 4898 |
| 10 | pseudogene | 100.33054 | 100.33141 | positive | MGI_C57BL6J_3646142 | Gm8701 | NCBI_Gene:667556,ENSEMBL:ENSMUSG00000112408 | MGI:3646142 | pseudogene | predicted gene 8701 |
| 10 | gene | 100.33568 | 100.33659 | positive | MGI_C57BL6J_3782481 | Gm4301 | NCBI_Gene:100043224,ENSEMBL:ENSMUSG00000094314 | MGI:3782481 | protein coding gene | predicted gene 4301 |
| 10 | gene | 100.34084 | 100.34176 | positive | MGI_C57BL6J_3782482 | Gm4302 | NCBI_Gene:100043227,ENSEMBL:ENSMUSG00000091101 | MGI:3782482 | protein coding gene | predicted gene 4302 |
| 10 | gene | 100.34597 | 100.34688 | positive | MGI_C57BL6J_3782483 | Gm4303 | NCBI_Gene:100043231,ENSEMBL:ENSMUSG00000112931 | MGI:3782483 | protein coding gene | predicted gene 4303 |
| 10 | gene | 100.35109 | 100.35200 | positive | MGI_C57BL6J_3782485 | Gm4305 | NCBI_Gene:100043235,ENSEMBL:ENSMUSG00000112856 | MGI:3782485 | protein coding gene | predicted gene 4305 |
| 10 | pseudogene | 100.35623 | 100.35712 | positive | MGI_C57BL6J_3644351 | Gm8711 | NCBI_Gene:667580,ENSEMBL:ENSMUSG00000091552 | MGI:3644351 | pseudogene | predicted gene 8711 |
| 10 | gene | 100.36134 | 100.36225 | positive | MGI_C57BL6J_3782487 | Gm4307 | NCBI_Gene:100043239,ENSEMBL:ENSMUSG00000096744 | MGI:3782487 | protein coding gene | predicted gene 4307 |
| 10 | pseudogene | 100.36650 | 100.36741 | positive | MGI_C57BL6J_3782489 | Gm4308 | NCBI_Gene:100043242,ENSEMBL:ENSMUSG00000095673 | MGI:3782489 | pseudogene | predicted gene 4308 |
| 10 | pseudogene | 100.37125 | 100.37213 | negative | MGI_C57BL6J_3782491 | Gm4310 | NCBI_Gene:100043244,ENSEMBL:ENSMUSG00000112480 | MGI:3782491 | pseudogene | predicted gene 4310 |
| 10 | gene | 100.37286 | 100.38383 | negative | MGI_C57BL6J_6096701 | Gm47631 | ENSEMBL:ENSMUSG00000112485 | MGI:6096701 | lncRNA gene | predicted gene, 47631 |
| 10 | pseudogene | 100.37646 | 100.37733 | negative | MGI_C57BL6J_3647757 | Gm8723 | NCBI_Gene:667607,ENSEMBL:ENSMUSG00000112777 | MGI:3647757 | pseudogene | predicted gene 8723 |
| 10 | gene | 100.38165 | 100.38256 | negative | MGI_C57BL6J_3782493 | Gm4312 | NCBI_Gene:100043247,ENSEMBL:ENSMUSG00000096640 | MGI:3782493 | protein coding gene | predicted gene 4312 |
| 10 | pseudogene | 100.39629 | 100.39707 | negative | MGI_C57BL6J_3643164 | Gm4781 | NCBI_Gene:213051,ENSEMBL:ENSMUSG00000112453 | MGI:3643164 | pseudogene | predicted gene 4781 |
| 10 | gene | 100.43294 | 100.43357 | negative | MGI_C57BL6J_6096768 | Gm47673 | ENSEMBL:ENSMUSG00000112580 | MGI:6096768 | unclassified gene | predicted gene, 47673 |
| 10 | gene | 100.44390 | 100.48735 | negative | MGI_C57BL6J_3036255 | Tmtc3 | NCBI_Gene:237500,ENSEMBL:ENSMUSG00000036676 | MGI:3036255 | protein coding gene | transmembrane and tetratricopeptide repeat containing 3 |
| 10 | gene | 100.48755 | 100.57484 | positive | MGI_C57BL6J_2384917 | Cep290 | NCBI_Gene:216274,ENSEMBL:ENSMUSG00000019971 | MGI:2384917 | protein coding gene | centrosomal protein 290 |
| 10 | gene | 100.57227 | 100.59042 | negative | MGI_C57BL6J_1921197 | 4930430F08Rik | NCBI_Gene:68281,ENSEMBL:ENSMUSG00000046567 | MGI:1921197 | protein coding gene | RIKEN cDNA 4930430F08 gene |
| 10 | gene | 100.57743 | 100.58371 | positive | MGI_C57BL6J_6097228 | Gm47956 | ENSEMBL:ENSMUSG00000112000 | MGI:6097228 | lncRNA gene | predicted gene, 47956 |
| 10 | gene | 100.59048 | 100.61840 | positive | MGI_C57BL6J_1913855 | 1700017N19Rik | NCBI_Gene:66605,ENSEMBL:ENSMUSG00000056912 | MGI:1913855 | protein coding gene | RIKEN cDNA 1700017N19 gene |
| 10 | pseudogene | 100.65163 | 100.65194 | positive | MGI_C57BL6J_6096210 | Gm47325 | ENSEMBL:ENSMUSG00000112328 | MGI:6096210 | pseudogene | predicted gene, 47325 |
| 10 | gene | 100.70753 | 100.74164 | negative | MGI_C57BL6J_5594881 | Gm35722 | NCBI_Gene:102639396,ENSEMBL:ENSMUSG00000112104 | MGI:5594881 | lncRNA gene | predicted gene, 35722 |
| 10 | pseudogene | 101.27162 | 101.27262 | positive | MGI_C57BL6J_5011418 | Gm19233 | NCBI_Gene:100418475,ENSEMBL:ENSMUSG00000111906 | MGI:5011418 | pseudogene | predicted gene, 19233 |
| 10 | gene | 101.54418 | 101.54431 | positive | MGI_C57BL6J_5456055 | Gm26278 | ENSEMBL:ENSMUSG00000087723 | MGI:5456055 | snoRNA gene | predicted gene, 26278 |
| 10 | gene | 101.68149 | 102.39185 | positive | MGI_C57BL6J_1914819 | Mgat4c | NCBI_Gene:67569,ENSEMBL:ENSMUSG00000019888 | MGI:1914819 | protein coding gene | MGAT4 family, member C |
| 10 | gene | 101.73077 | 101.73156 | positive | MGI_C57BL6J_6095728 | Gm47032 | ENSEMBL:ENSMUSG00000111931 | MGI:6095728 | unclassified gene | predicted gene, 47032 |
| 10 | gene | 101.77269 | 101.77277 | positive | MGI_C57BL6J_4834225 | Mir3059 | miRBase:MI0004412,NCBI_Gene:100526516,ENSEMBL:ENSMUSG00000092952 | MGI:4834225 | miRNA gene | microRNA 3059 |
| 10 | gene | 102.11444 | 102.11646 | positive | MGI_C57BL6J_5608885 | B230210A14Rik | NA | NA | unclassified gene | Riken cDNA B230210A14 gene |
| 10 | gene | 102.15891 | 102.27354 | positive | MGI_C57BL6J_1921048 | 4930402I19Rik | NA | NA | unclassified gene | RIKEN cDNA 4930402I19 gene |
| 10 | gene | 102.27191 | 102.27441 | positive | MGI_C57BL6J_2444255 | D930019F10Rik | NA | NA | unclassified gene | RIKEN cDNA D930019F10 gene |
| 10 | gene | 102.31961 | 102.32189 | positive | MGI_C57BL6J_6095726 | Gm47031 | ENSEMBL:ENSMUSG00000112342 | MGI:6095726 | lncRNA gene | predicted gene, 47031 |
| 10 | gene | 102.41588 | 102.41801 | positive | MGI_C57BL6J_6095730 | Gm47033 | ENSEMBL:ENSMUSG00000111990 | MGI:6095730 | lncRNA gene | predicted gene, 47033 |
| 10 | gene | 102.43191 | 102.43234 | negative | MGI_C57BL6J_6095715 | Gm47025 | ENSEMBL:ENSMUSG00000112012 | MGI:6095715 | unclassified gene | predicted gene, 47025 |
| 10 | gene | 102.43527 | 102.43554 | positive | MGI_C57BL6J_6095836 | Gm47101 | ENSEMBL:ENSMUSG00000112073 | MGI:6095836 | lncRNA gene | predicted gene, 47101 |
| 10 | pseudogene | 102.46020 | 102.46114 | positive | MGI_C57BL6J_5504103 | Gm26988 | ENSEMBL:ENSMUSG00000098139 | MGI:5504103 | pseudogene | predicted gene, 26988 |
| 10 | gene | 102.48176 | 102.49049 | negative | MGI_C57BL6J_1328351 | Nts | NCBI_Gene:67405,ENSEMBL:ENSMUSG00000019890 | MGI:1328351 | protein coding gene | neurotensin |
| 10 | gene | 102.51222 | 102.54974 | positive | MGI_C57BL6J_2384307 | Rassf9 | NCBI_Gene:237504,ENSEMBL:ENSMUSG00000044921 | MGI:2384307 | protein coding gene | Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| 10 | pseudogene | 102.68307 | 102.68331 | negative | MGI_C57BL6J_6095904 | Gm47142 | ENSEMBL:ENSMUSG00000112221 | MGI:6095904 | pseudogene | predicted gene, 47142 |
| 10 | gene | 102.68327 | 102.69457 | positive | MGI_C57BL6J_6095905 | Gm47143 | ENSEMBL:ENSMUSG00000112845 | MGI:6095905 | lncRNA gene | predicted gene, 47143 |
| 10 | pseudogene | 102.76598 | 102.83866 | positive | MGI_C57BL6J_3647142 | Gm5175 | NCBI_Gene:382413,ENSEMBL:ENSMUSG00000112046 | MGI:3647142 | pseudogene | predicted gene 5175 |
| 10 | gene | 102.99871 | 103.03021 | negative | MGI_C57BL6J_104621 | Alx1 | NCBI_Gene:216285,ENSEMBL:ENSMUSG00000036602 | MGI:104621 | protein coding gene | ALX homeobox 1 |
| 10 | gene | 103.02899 | 103.03253 | positive | MGI_C57BL6J_5593457 | Gm34298 | NCBI_Gene:102637507 | MGI:5593457 | protein coding gene | predicted gene, 34298 |
| 10 | gene | 103.02948 | 103.03034 | negative | MGI_C57BL6J_4937855 | Gm17028 | ENSEMBL:ENSMUSG00000090638 | MGI:4937855 | lncRNA gene | predicted gene 17028 |
| 10 | gene | 103.04438 | 103.23633 | negative | MGI_C57BL6J_1922228 | Lrriq1 | NCBI_Gene:74978,ENSEMBL:ENSMUSG00000019892 | MGI:1922228 | protein coding gene | leucine-rich repeats and IQ motif containing 1 |
| 10 | gene | 103.12124 | 103.12137 | positive | MGI_C57BL6J_5455153 | Gm25376 | ENSEMBL:ENSMUSG00000077528 | MGI:5455153 | snoRNA gene | predicted gene, 25376 |
| 10 | gene | 103.20775 | 103.21344 | negative | MGI_C57BL6J_6095941 | Gm47164 | ENSEMBL:ENSMUSG00000112622 | MGI:6095941 | unclassified gene | predicted gene, 47164 |
| 10 | gene | 103.36719 | 103.36937 | negative | MGI_C57BL6J_2442852 | 6430590A10Rik | NCBI_Gene:100043277 | MGI:2442852 | lncRNA gene | RIKEN cDNA 6430590A10 gene |
| 10 | gene | 103.36746 | 103.41938 | positive | MGI_C57BL6J_2143484 | Slc6a15 | NCBI_Gene:103098,ENSEMBL:ENSMUSG00000019894 | MGI:2143484 | protein coding gene | solute carrier family 6 (neurotransmitter transporter), member 15 |
| 10 | pseudogene | 103.42989 | 103.43078 | positive | MGI_C57BL6J_3643663 | Gm8768 | NCBI_Gene:667697,ENSEMBL:ENSMUSG00000112923 | MGI:3643663 | pseudogene | predicted gene 8768 |
| 10 | gene | 103.50736 | 103.56656 | positive | MGI_C57BL6J_6096034 | Gm47223 | ENSEMBL:ENSMUSG00000112636 | MGI:6096034 | lncRNA gene | predicted gene, 47223 |
| 10 | gene | 103.51293 | 103.57169 | positive | MGI_C57BL6J_6096031 | Gm47221 | ENSEMBL:ENSMUSG00000112319 | MGI:6096031 | lncRNA gene | predicted gene, 47221 |
| 10 | pseudogene | 103.56765 | 103.56805 | positive | MGI_C57BL6J_6096030 | Gm47220 | ENSEMBL:ENSMUSG00000112644 | MGI:6096030 | pseudogene | predicted gene, 47220 |
| 10 | pseudogene | 103.56921 | 103.56940 | negative | MGI_C57BL6J_6096033 | Gm47222 | ENSEMBL:ENSMUSG00000112839 | MGI:6096033 | pseudogene | predicted gene, 47222 |
| 10 | pseudogene | 103.58140 | 103.58239 | negative | MGI_C57BL6J_5504038 | Gm26923 | ENSEMBL:ENSMUSG00000098173 | MGI:5504038 | pseudogene | predicted gene, 26923 |
| 10 | gene | 103.85627 | 103.85833 | negative | MGI_C57BL6J_6096036 | Gm47224 | ENSEMBL:ENSMUSG00000112554 | MGI:6096036 | lncRNA gene | predicted gene, 47224 |
| 10 | pseudogene | 103.89354 | 103.89381 | positive | MGI_C57BL6J_6096038 | Gm47225 | ENSEMBL:ENSMUSG00000112570 | MGI:6096038 | pseudogene | predicted gene, 47225 |
| 10 | gene | 103.95954 | 103.95965 | positive | MGI_C57BL6J_5454920 | Gm25143 | ENSEMBL:ENSMUSG00000064630 | MGI:5454920 | snRNA gene | predicted gene, 25143 |
| 10 | gene | 103.98699 | 103.98791 | positive | MGI_C57BL6J_6096039 | Gm47226 | ENSEMBL:ENSMUSG00000112281 | MGI:6096039 | lncRNA gene | predicted gene, 47226 |
| 10 | pseudogene | 104.08222 | 104.08234 | positive | MGI_C57BL6J_6096041 | Gm47227 | ENSEMBL:ENSMUSG00000112403 | MGI:6096041 | pseudogene | predicted gene, 47227 |
| 10 | pseudogene | 104.09632 | 104.09676 | negative | MGI_C57BL6J_6096042 | Gm47228 | ENSEMBL:ENSMUSG00000112382 | MGI:6096042 | pseudogene | predicted gene, 47228 |
| 10 | gene | 104.14299 | 104.15367 | positive | MGI_C57BL6J_5434648 | Gm21293 | NCBI_Gene:100861880,ENSEMBL:ENSMUSG00000091779 | MGI:5434648 | protein coding gene | predicted gene, 21293 |
| 10 | gene | 104.15150 | 104.15396 | positive | MGI_C57BL6J_3643573 | Gm6763 | NCBI_Gene:627488,ENSEMBL:ENSMUSG00000097427 | MGI:3643573 | protein coding gene | predicted gene 6763 |
| 10 | gene | 104.16001 | 104.16247 | positive | MGI_C57BL6J_3643666 | Gm8764 | NCBI_Gene:667692,ENSEMBL:ENSMUSG00000097878 | MGI:3643666 | protein coding gene | predicted gene 8764 |
| 10 | gene | 104.16851 | 104.17098 | positive | MGI_C57BL6J_5434659 | Gm21304 | NCBI_Gene:100861891,ENSEMBL:ENSMUSG00000097550 | MGI:5434659 | protein coding gene | predicted gene, 21304 |
| 10 | gene | 104.17702 | 104.17949 | positive | MGI_C57BL6J_5434667 | Gm21312 | NCBI_Gene:100861901,ENSEMBL:ENSMUSG00000094144 | MGI:5434667 | protein coding gene | predicted gene, 21312 |
| 10 | gene | 104.18553 | 104.18800 | positive | MGI_C57BL6J_5434121 | Gm20765 | NCBI_Gene:637578,ENSEMBL:ENSMUSG00000095022 | MGI:5434121 | protein coding gene | predicted gene, 20765 |
| 10 | gene | 104.19404 | 104.19652 | positive | MGI_C57BL6J_3782524 | Gm4340 | NCBI_Gene:100043292,ENSEMBL:ENSMUSG00000090854 | MGI:3782524 | protein coding gene | predicted gene 4340 |
| 10 | gene | 104.22484 | 104.22490 | negative | MGI_C57BL6J_5452116 | Gm22339 | ENSEMBL:ENSMUSG00000088343 | MGI:5452116 | snRNA gene | predicted gene, 22339 |
| 10 | gene | 104.28727 | 104.28738 | positive | MGI_C57BL6J_5531149 | Mir6411 | miRBase:MI0021948,NCBI_Gene:102465222,ENSEMBL:ENSMUSG00000099264 | MGI:5531149 | miRNA gene | microRNA 6411 |
| 10 | gene | 104.47105 | 104.47121 | negative | MGI_C57BL6J_5452722 | Gm22945 | ENSEMBL:ENSMUSG00000089182 | MGI:5452722 | snRNA gene | predicted gene, 22945 |
| 10 | gene | 104.86520 | 104.86531 | negative | MGI_C57BL6J_5455299 | Gm25522 | ENSEMBL:ENSMUSG00000089019 | MGI:5455299 | snRNA gene | predicted gene, 25522 |
| 10 | pseudogene | 104.87421 | 104.87448 | negative | MGI_C57BL6J_6097563 | Gm48184 | ENSEMBL:ENSMUSG00000112044 | MGI:6097563 | pseudogene | predicted gene, 48184 |
| 10 | gene | 104.92649 | 104.93659 | negative | MGI_C57BL6J_5623636 | Gm40751 | NCBI_Gene:105245274 | MGI:5623636 | lncRNA gene | predicted gene, 40751 |
| 10 | gene | 105.04027 | 105.04044 | positive | MGI_C57BL6J_5455364 | Gm25587 | ENSEMBL:ENSMUSG00000064432 | MGI:5455364 | snRNA gene | predicted gene, 25587 |
| 10 | gene | 105.04029 | 105.04381 | negative | MGI_C57BL6J_5623637 | Gm40752 | NCBI_Gene:105245275 | MGI:5623637 | lncRNA gene | predicted gene, 40752 |
| 10 | gene | 105.10282 | 105.10580 | negative | MGI_C57BL6J_5623638 | Gm40753 | NCBI_Gene:105245276 | MGI:5623638 | lncRNA gene | predicted gene, 40753 |
| 10 | gene | 105.14021 | 105.14033 | negative | MGI_C57BL6J_5455199 | Gm25422 | ENSEMBL:ENSMUSG00000077407 | MGI:5455199 | snoRNA gene | predicted gene, 25422 |
| 10 | gene | 105.14224 | 105.14294 | negative | MGI_C57BL6J_5610225 | Gm36997 | ENSEMBL:ENSMUSG00000103775 | MGI:5610225 | unclassified gene | predicted gene, 36997 |
| 10 | gene | 105.18766 | 105.57448 | negative | MGI_C57BL6J_1914057 | Tmtc2 | NCBI_Gene:278279,ENSEMBL:ENSMUSG00000036019 | MGI:1914057 | protein coding gene | transmembrane and tetratricopeptide repeat containing 2 |
| 10 | gene | 105.34025 | 105.34231 | positive | MGI_C57BL6J_6097597 | Gm48206 | ENSEMBL:ENSMUSG00000112193 | MGI:6097597 | unclassified gene | predicted gene, 48206 |
| 10 | gene | 105.37734 | 105.37748 | negative | MGI_C57BL6J_6097595 | Gm48205 | ENSEMBL:ENSMUSG00000112176 | MGI:6097595 | unclassified gene | predicted gene, 48205 |
| 10 | gene | 105.39250 | 105.39429 | negative | MGI_C57BL6J_6097591 | Gm48203 | ENSEMBL:ENSMUSG00000112512 | MGI:6097591 | unclassified gene | predicted gene, 48203 |
| 10 | gene | 105.44085 | 105.45253 | negative | MGI_C57BL6J_5825878 | Gm46241 | NCBI_Gene:108167831 | MGI:5825878 | lncRNA gene | predicted gene, 46241 |
| 10 | pseudogene | 105.50580 | 105.50860 | negative | MGI_C57BL6J_3783108 | Gm15666 | NCBI_Gene:100416011,ENSEMBL:ENSMUSG00000084399 | MGI:3783108 | pseudogene | predicted gene 15666 |
| 10 | gene | 105.51468 | 105.51665 | negative | MGI_C57BL6J_6097593 | Gm48204 | ENSEMBL:ENSMUSG00000112900 | MGI:6097593 | unclassified gene | predicted gene, 48204 |
| 10 | pseudogene | 105.52046 | 105.52064 | positive | MGI_C57BL6J_3783106 | Gm15664 | ENSEMBL:ENSMUSG00000089904 | MGI:3783106 | pseudogene | predicted gene 15664 |
| 10 | gene | 105.52404 | 105.52636 | negative | MGI_C57BL6J_1926067 | 4930588G05Rik | NA | NA | unclassified gene | RIKEN cDNA 4930588G05 gene |
| 10 | gene | 105.53298 | 105.53449 | negative | MGI_C57BL6J_6097599 | Gm48207 | ENSEMBL:ENSMUSG00000112571 | MGI:6097599 | unclassified gene | predicted gene, 48207 |
| 10 | gene | 105.57395 | 105.58387 | positive | MGI_C57BL6J_3783105 | Gm15663 | NCBI_Gene:327812,ENSEMBL:ENSMUSG00000085282 | MGI:3783105 | lncRNA gene | predicted gene 15663 |
| 10 | gene | 105.76318 | 105.84138 | negative | MGI_C57BL6J_3041259 | Mettl25 | NCBI_Gene:216292,ENSEMBL:ENSMUSG00000036009 | MGI:3041259 | protein coding gene | methyltransferase like 25 |
| 10 | gene | 105.80409 | 105.80420 | negative | MGI_C57BL6J_4421928 | n-R5s80 | ENSEMBL:ENSMUSG00000084545 | MGI:4421928 | rRNA gene | nuclear encoded rRNA 5S 80 |
| 10 | gene | 105.80654 | 105.80887 | negative | MGI_C57BL6J_6097740 | Gm48298 | ENSEMBL:ENSMUSG00000112820 | MGI:6097740 | unclassified gene | predicted gene, 48298 |
| 10 | gene | 105.83828 | 105.83964 | negative | MGI_C57BL6J_1917182 | 2810004I08Rik | NA | NA | unclassified gene | RIKEN cDNA 2810004I08 gene |
| 10 | gene | 105.84107 | 105.84783 | positive | MGI_C57BL6J_1289302 | Ccdc59 | NCBI_Gene:52713,ENSEMBL:ENSMUSG00000019897 | MGI:1289302 | protein coding gene | coiled-coil domain containing 59 |
| 10 | gene | 105.92961 | 105.92974 | negative | MGI_C57BL6J_5454014 | Gm24237 | ENSEMBL:ENSMUSG00000088415 | MGI:5454014 | snoRNA gene | predicted gene, 24237 |
| 10 | gene | 106.05596 | 106.05608 | negative | MGI_C57BL6J_5455119 | Gm25342 | ENSEMBL:ENSMUSG00000088084 | MGI:5455119 | snoRNA gene | predicted gene, 25342 |
| 10 | pseudogene | 106.06586 | 106.06679 | positive | MGI_C57BL6J_3643770 | Gm8796 | NCBI_Gene:667758,ENSEMBL:ENSMUSG00000111914 | MGI:3643770 | pseudogene | predicted gene 8796 |
| 10 | gene | 106.07423 | 106.07908 | positive | MGI_C57BL6J_6097833 | Gm48363 | ENSEMBL:ENSMUSG00000112034 | MGI:6097833 | unclassified gene | predicted gene, 48363 |
| 10 | pseudogene | 106.16678 | 106.16926 | negative | MGI_C57BL6J_6097853 | Gm48376 | ENSEMBL:ENSMUSG00000112668 | MGI:6097853 | pseudogene | predicted gene, 48376 |
| 10 | gene | 106.18302 | 106.18389 | positive | MGI_C57BL6J_6097854 | Gm48377 | ENSEMBL:ENSMUSG00000112523 | MGI:6097854 | unclassified gene | predicted gene, 48377 |
| 10 | pseudogene | 106.19234 | 106.19279 | negative | MGI_C57BL6J_3643034 | Rps15a-ps1 | NCBI_Gene:639162,ENSEMBL:ENSMUSG00000056877 | MGI:3643034 | pseudogene | ribosomal protein S15A, pseudogene 1 |
| 10 | pseudogene | 106.35832 | 106.35865 | positive | MGI_C57BL6J_6097857 | Gm48379 | ENSEMBL:ENSMUSG00000112520 | MGI:6097857 | pseudogene | predicted gene, 48379 |
| 10 | pseudogene | 106.44440 | 106.44510 | positive | MGI_C57BL6J_6097858 | Gm48380 | ENSEMBL:ENSMUSG00000112066 | MGI:6097858 | pseudogene | predicted gene, 48380 |
| 10 | gene | 106.46694 | 106.47077 | negative | MGI_C57BL6J_3697434 | C230021G24Rik | ENSEMBL:ENSMUSG00000097869 | MGI:3697434 | lncRNA gene | RIKEN cDNA C230021G24 gene |
| 10 | gene | 106.47027 | 106.93595 | positive | MGI_C57BL6J_2443834 | Ppfia2 | NCBI_Gene:327814,ENSEMBL:ENSMUSG00000053825 | MGI:2443834 | protein coding gene | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| 10 | pseudogene | 106.50271 | 106.50337 | positive | MGI_C57BL6J_3031390 | Olfr1556-ps1 | NCBI_Gene:259137,ENSEMBL:ENSMUSG00000108029 | MGI:3031390 | pseudogene | olfactory receptor 1556, pseudogene 1 |
| 10 | gene | 106.55519 | 106.60248 | negative | MGI_C57BL6J_1923083 | 4930532I03Rik | NCBI_Gene:75833,ENSEMBL:ENSMUSG00000112201 | MGI:1923083 | lncRNA gene | RIKEN cDNA 4930532I03 gene |
| 10 | pseudogene | 106.75381 | 106.75491 | negative | MGI_C57BL6J_5010033 | Gm17848 | NCBI_Gene:100415977 | MGI:5010033 | pseudogene | predicted gene, 17848 |
| 10 | gene | 106.93352 | 106.93595 | negative | MGI_C57BL6J_3028041 | B230114P17Rik | NA | NA | unclassified gene | RIKEN cDNA B230114P17 gene |
| 10 | gene | 106.93352 | 107.12367 | negative | MGI_C57BL6J_2685720 | Acss3 | NCBI_Gene:380660,ENSEMBL:ENSMUSG00000035948 | MGI:2685720 | protein coding gene | acyl-CoA synthetase short-chain family member 3 |
| 10 | pseudogene | 106.94930 | 106.95017 | positive | MGI_C57BL6J_5011192 | Gm19007 | NCBI_Gene:100418113,ENSEMBL:ENSMUSG00000112443 | MGI:5011192 | pseudogene | predicted gene, 19007 |
| 10 | gene | 107.03969 | 107.04319 | negative | MGI_C57BL6J_2442573 | E030002G19Rik | NA | NA | unclassified gene | RIKEN cDNA E030002G19 gene |
| 10 | pseudogene | 107.05852 | 107.05935 | negative | MGI_C57BL6J_5010225 | Gm18040 | NCBI_Gene:100416324,ENSEMBL:ENSMUSG00000112849 | MGI:5010225 | pseudogene | predicted gene, 18040 |
| 10 | pseudogene | 107.06104 | 107.06198 | negative | MGI_C57BL6J_5011366 | Gm19181 | NCBI_Gene:100418393,ENSEMBL:ENSMUSG00000112863 | MGI:5011366 | pseudogene | predicted gene, 19181 |
| 10 | gene | 107.13697 | 107.13706 | positive | MGI_C57BL6J_5454522 | Gm24745 | ENSEMBL:ENSMUSG00000088465 | MGI:5454522 | snoRNA gene | predicted gene, 24745 |
| 10 | gene | 107.19991 | 107.20578 | negative | MGI_C57BL6J_5588833 | Gm29674 | NCBI_Gene:432500,ENSEMBL:ENSMUSG00000112466 | MGI:5588833 | lncRNA gene | predicted gene, 29674 |
| 10 | gene | 107.27160 | 107.42515 | positive | MGI_C57BL6J_2135609 | Lin7a | NCBI_Gene:108030,ENSEMBL:ENSMUSG00000019906 | MGI:2135609 | protein coding gene | lin-7 homolog A (C. elegans) |
| 10 | gene | 107.33577 | 107.33595 | positive | MGI_C57BL6J_1925661 | 3110033O22Rik | NA | NA | unclassified gene | RIKEN cDNA 3110033O22 gene |
| 10 | gene | 107.43642 | 107.43736 | negative | MGI_C57BL6J_5611345 | Gm38117 | ENSEMBL:ENSMUSG00000102934 | MGI:5611345 | unclassified gene | predicted gene, 38117 |
| 10 | gene | 107.48041 | 107.48053 | positive | MGI_C57BL6J_5690795 | Gm44403 | ENSEMBL:ENSMUSG00000104558 | MGI:5690795 | miRNA gene | predicted gene, 44403 |
| 10 | gene | 107.48291 | 107.48693 | negative | MGI_C57BL6J_97252 | Myf5 | NCBI_Gene:17877,ENSEMBL:ENSMUSG00000000435 | MGI:97252 | protein coding gene | myogenic factor 5 |
| 10 | gene | 107.49285 | 107.49474 | negative | MGI_C57BL6J_97253 | Myf6 | NCBI_Gene:17878,ENSEMBL:ENSMUSG00000035923 | MGI:97253 | protein coding gene | myogenic factor 6 |
| 10 | gene | 107.49527 | 107.65940 | positive | MGI_C57BL6J_5595336 | Gm36177 | NCBI_Gene:102639996 | MGI:5595336 | lncRNA gene | predicted gene, 36177 |
| 10 | gene | 107.51436 | 107.72008 | negative | MGI_C57BL6J_1096349 | Ptprq | NCBI_Gene:237523,ENSEMBL:ENSMUSG00000035916 | MGI:1096349 | protein coding gene | protein tyrosine phosphatase, receptor type, Q |
| 10 | gene | 107.76053 | 107.91229 | negative | MGI_C57BL6J_3647600 | Otogl | NCBI_Gene:628870,ENSEMBL:ENSMUSG00000091455 | MGI:3647600 | protein coding gene | otogelin-like |
| 10 | gene | 107.94665 | 107.97783 | positive | MGI_C57BL6J_5588844 | Gm29685 | NCBI_Gene:628877,ENSEMBL:ENSMUSG00000112910 | MGI:5588844 | lncRNA gene | predicted gene, 29685 |
| 10 | pseudogene | 108.14169 | 108.14243 | negative | MGI_C57BL6J_6097298 | Gm47999 | ENSEMBL:ENSMUSG00000112401 | MGI:6097298 | pseudogene | predicted gene, 47999 |
| 10 | gene | 108.15050 | 108.16044 | negative | MGI_C57BL6J_5623639 | Gm40754 | NCBI_Gene:105245277 | MGI:5623639 | lncRNA gene | predicted gene, 40754 |
| 10 | gene | 108.16183 | 108.28448 | positive | MGI_C57BL6J_1309528 | Ppp1r12a | NCBI_Gene:17931,ENSEMBL:ENSMUSG00000019907 | MGI:1309528 | protein coding gene | protein phosphatase 1, regulatory subunit 12A |
| 10 | gene | 108.17981 | 108.18492 | positive | MGI_C57BL6J_2443885 | C430003N24Rik | NA | NA | unclassified gene | RIKEN cDNA C430003N24 gene |
| 10 | gene | 108.32237 | 108.33273 | negative | MGI_C57BL6J_5623640 | Gm40755 | NCBI_Gene:105245278 | MGI:5623640 | protein coding gene | predicted gene, 40755 |
| 10 | gene | 108.33212 | 108.41439 | positive | MGI_C57BL6J_2149961 | Pawr | NCBI_Gene:114774,ENSEMBL:ENSMUSG00000035873 | MGI:2149961 | protein coding gene | PRKC, apoptosis, WT1, regulator |
| 10 | gene | 108.36366 | 108.36380 | positive | MGI_C57BL6J_5452882 | Gm23105 | ENSEMBL:ENSMUSG00000065653 | MGI:5452882 | snoRNA gene | predicted gene, 23105 |
| 10 | gene | 108.41725 | 108.45330 | positive | MGI_C57BL6J_5595442 | Gm36283 | NCBI_Gene:102640148,ENSEMBL:ENSMUSG00000112532 | MGI:5595442 | lncRNA gene | predicted gene, 36283 |
| 10 | gene | 108.42807 | 108.42816 | positive | MGI_C57BL6J_5455368 | Gm25591 | ENSEMBL:ENSMUSG00000093762 | MGI:5455368 | miRNA gene | predicted gene, 25591 |
| 10 | gene | 108.44432 | 108.44481 | negative | MGI_C57BL6J_5595599 | Gm36440 | NCBI_Gene:102640357 | MGI:5595599 | lncRNA gene | predicted gene, 36440 |
| 10 | gene | 108.45576 | 108.51207 | positive | MGI_C57BL6J_5595760 | Gm36601 | NCBI_Gene:105245279 | MGI:5595760 | lncRNA gene | predicted gene, 36601 |
| 10 | pseudogene | 108.48265 | 108.48299 | positive | MGI_C57BL6J_6096432 | Gm47466 | ENSEMBL:ENSMUSG00000112065 | MGI:6096432 | pseudogene | predicted gene, 47466 |
| 10 | gene | 108.49765 | 109.01098 | negative | MGI_C57BL6J_99667 | Syt1 | NCBI_Gene:20979,ENSEMBL:ENSMUSG00000035864 | MGI:99667 | protein coding gene | synaptotagmin I |
| 10 | gene | 108.54576 | 108.54794 | negative | MGI_C57BL6J_2444139 | A830054O04Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA A830054O04 gene |
| 10 | gene | 108.63700 | 108.64008 | negative | MGI_C57BL6J_1925527 | 5330428N10Rik | NA | NA | unclassified gene | RIKEN cDNA 5330428N10 gene |
| 10 | gene | 108.68501 | 108.68579 | negative | MGI_C57BL6J_1924638 | C030018G05Rik | NA | NA | unclassified gene | RIKEN cDNA C030018G05 gene |
| 10 | gene | 108.69876 | 108.70016 | negative | MGI_C57BL6J_3647286 | Gm5136 | NCBI_Gene:368203,ENSEMBL:ENSMUSG00000112039 | MGI:3647286 | protein coding gene | predicted gene 5136 |
| 10 | gene | 108.71151 | 108.71228 | negative | MGI_C57BL6J_1924887 | C030044C18Rik | NA | NA | unclassified gene | RIKEN cDNA C030044C18 gene |
Additive covariates
Significance thresholds
operm <- scan1perm(pr.qc, covars["rz.age"], model="normal", addcovar=addcovar, n_perm=10, perm_Xsp=TRUE, chr_lengths=chr_lengths(gm$gmap))
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
names(summary_table) <- c("autosomes","X")
summary_table$significance.level <- rownames(summary_table)
rownames(summary_table) <- NULL
summary_table[c(3,1:2)] %>%
kable(escape = F,align = c("ccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| significance.level | autosomes | X |
|---|---|---|
| 0.01 | 3.402448 | 6.729276 |
| 0.05 | 3.229013 | 6.705621 |
| 0.1 | 3.011679 | 6.674687 |
Genome-wide scan
plot_lod<-function(out,map){
for (i in 1:dim(out)[2]){
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - ICI vs. EOI (with additive covariates) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
print("with no kinship")[1] “with no kinship”
outfa <- scan1(pr.qc, covars["rz.age"], Xcovar=Xcovar, model="normal", addcovar=addcovar)
plot_lod(outfa,gm$gmap)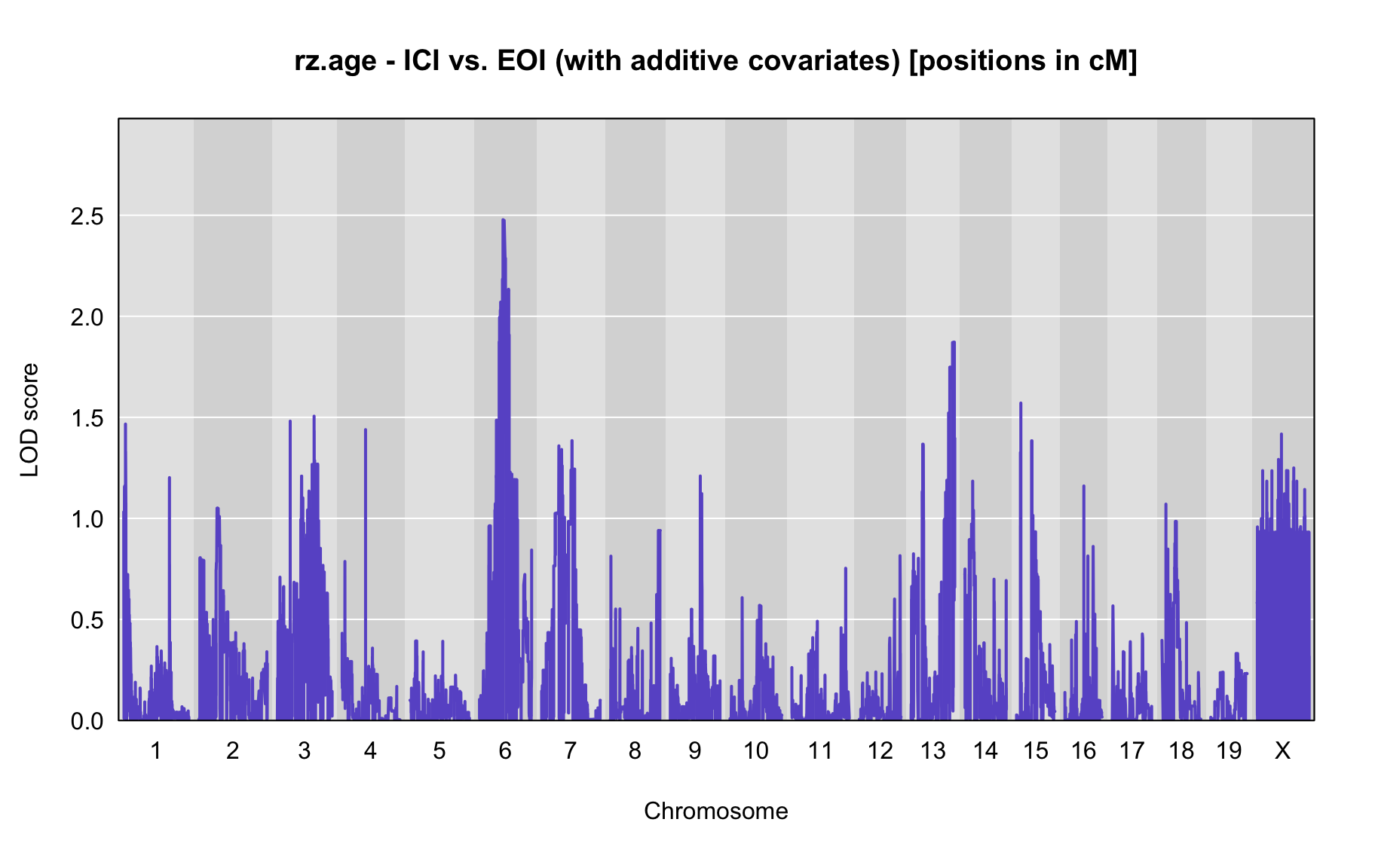
print("with normal kinship")[1] “with normal kinship”
outka <- scan1(pr.qc, covars["rz.age"], Xcovar=Xcovar, model="normal", addcovar=addcovar, kinship=kinship)
plot_lod(outka,gm$gmap)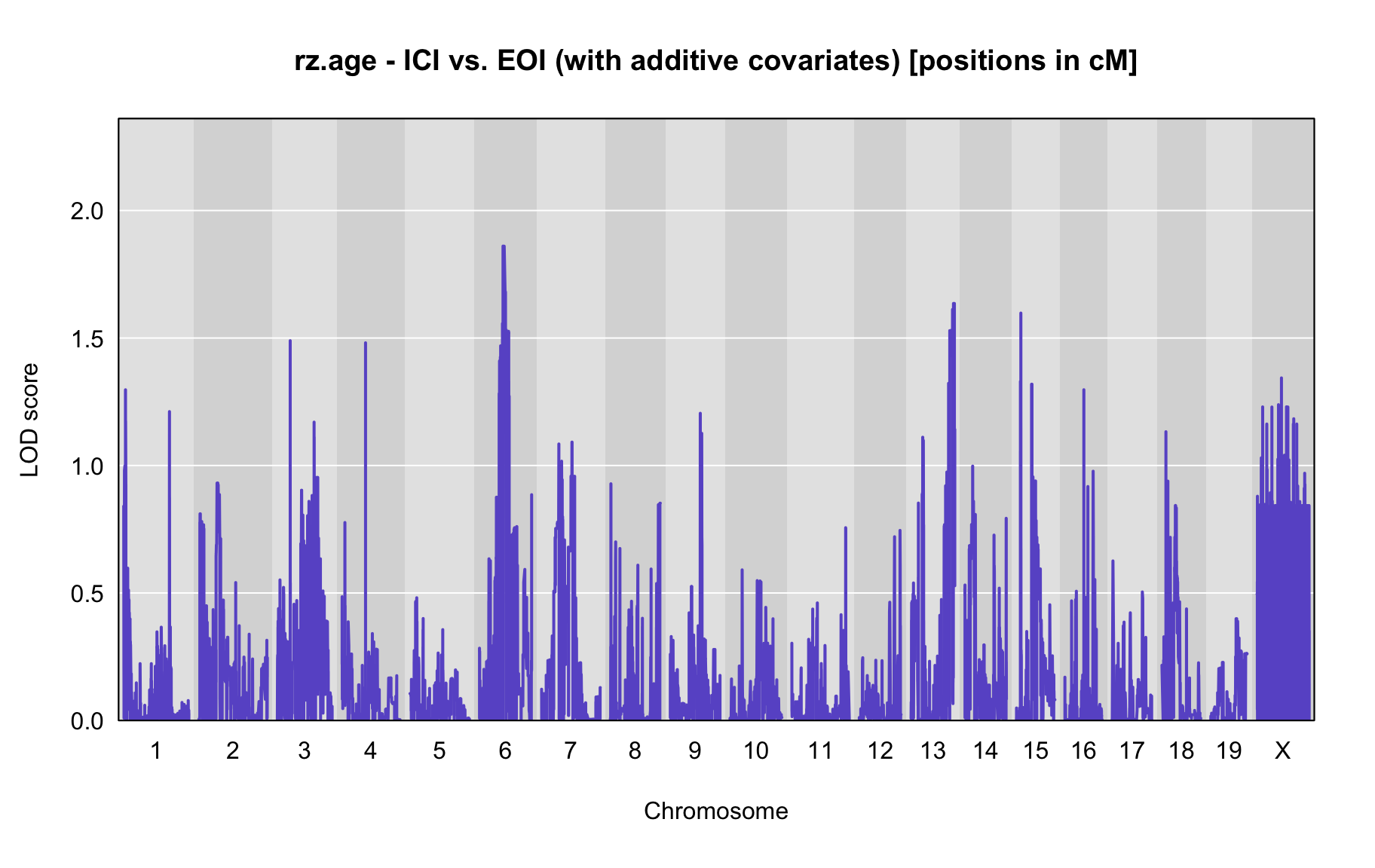
print("with loco kinship")[1] “with loco kinship”
outa <- scan1(pr.qc, covars["rz.age"], Xcovar=Xcovar, model="normal", addcovar=addcovar, kinship=K)
plot_lod(outa,gm$gmap)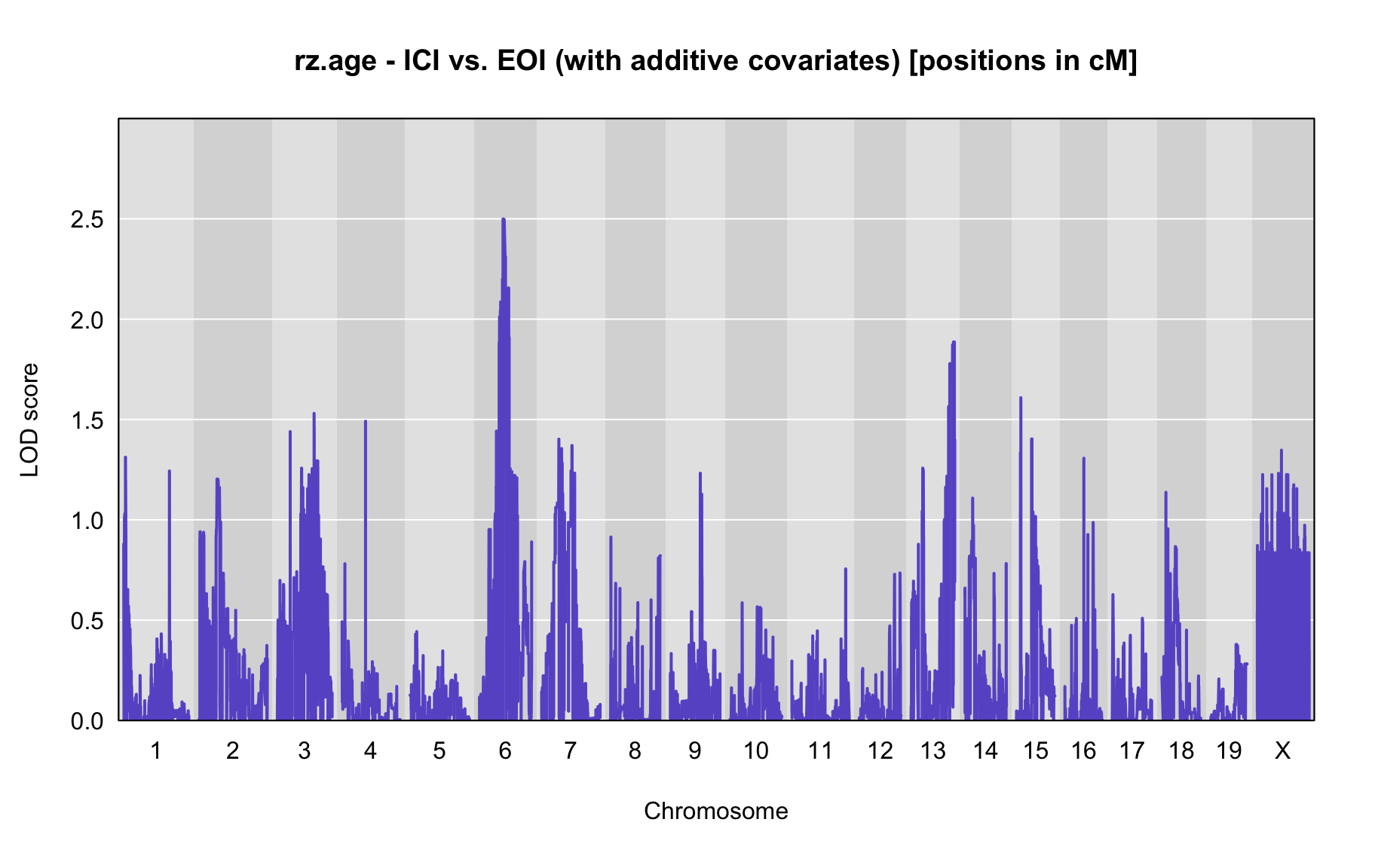
out = outa
print(paste0("number of samples in analysis = ", as.data.frame(attributes(out)$sample_size)[1,]))[1] “number of samples in analysis = 267”
LOD peaks
Centimorgan (cM)
peaks<-find_peaks(out, gm$gmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) > 0){
peaks$marker <- find_marker(gm$gmap, chr=peaks$chr,pos=peaks$pos)
names(peaks)[2] <- c("phenotype")
peaks <- peaks[-1]
rownames(peaks) <- NULL
print(kable(peaks, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
#plot only peak chromosomes
plot_lod_chr<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " - ICI vs. EOI (with additive covariates) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are no peaks that have a LOD that reaches suggestive (p<0.05) level of 3.22901302583048 [autosomes]/6.7056207215653 [x-chromosome]”
if(nrow(peaks) < 50 & nrow(peaks) > 0){
for(i in unique(peaks$chr)){
#for (i in 1:nrow(peaks)){
#plot_lod_chr(out,gm$gmap, peaks$chr[i])
plot_lod_chr(out,gm$gmap, i)
}
} else {
print(paste0("There are too many peaks (",nrow(peaks)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are too many peaks (0 peaks) that have a LOD that reaches suggestive (p<0.05) level of 3.22901302583048 [autosomes]/6.7056207215653 [x-chromosome]”
Megabase (MB)
peaks_mba<-find_peaks(out, gm$pmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) > 0){
peaks_mba$marker <- find_marker(gm$pmap, chr=peaks_mba$chr,pos=peaks_mba$pos)
names(peaks_mba)[2] <- c("phenotype")
peaks_mba <- peaks_mba[-1]
rownames(peaks_mba) <- NULL
print(kable(peaks_mba, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
plot_lod_chr_mb<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " - ICI vs. EOI (with additive covariates) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are no peaks that have a LOD that reaches suggestive (p<0.05) level of 3.22901302583048 [autosomes]/6.7056207215653 [x-chromosome]”
if(nrow(peaks_mba) < 50 & nrow(peaks_mba) > 0){
for(i in unique(peaks_mba$chr)){
#for (i in 1:nrow(peaks_mba)){
#plot_lod_chr_mb(out,gm$pmap, peaks_mba$chr[i])
plot_lod_chr_mb(out,gm$pmap,i)
}
} else {
print(paste0("There are too many peaks (",nrow(peaks_mba)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are too many peaks (0 peaks) that have a LOD that reaches suggestive (p<0.05) level of 3.22901302583048 [autosomes]/6.7056207215653 [x-chromosome]”
QTL effects
For each peak LOD location we give a list of gene
if(nrow(peaks) < 50 & nrow(peaks) > 0){
for (i in 1:nrow(peaks)){
#for (i in 1:1){
#Plot 1
g <- maxmarg(pr.qc, gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i], return_char=TRUE)
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/","qtl_effect_", i, ".png"))
#par(mar=c(4.1, 4.1, 1.5, 0.6))
plot_pxg(g, covars[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
##dev.off()
chr = peaks$chr[i]
# Plot 2
pr_sub <- pull_genoprobint(pr.qc, gm$gmap, chr, c(peaks$ci_lo[i], peaks$ci_hi[i]))
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar)
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], Xcovar=Xcovar)
#coeff <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
#coeff_sub <- scan1coef(pr_sub[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
blup <- scan1blup(pr.qc[,chr], covars[peaks$phenotype[i]])
blup_sub <- scan1blup(pr_sub[,chr], covars[peaks$phenotype[i]])
write.csv(as.data.frame(blup_sub), paste0("data/ici.vs.eoi_",peaks$phenotype[i],"-additive.covariates_blup_sub_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_5.batches_0.csv"), quote=F)
#plot_coef(coeff,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i]," [scan1coeff; positions in cM] )")
# )
#plot_coef(coeff_sub,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i],"; 1.5 LOD drop interval [scan1coeff; positions in cM] ) ")
# )
plot_coef(blup,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," [scan1blup; positions in cM] )")
)
plot_coef(blup_sub,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i],"; 1.5 LOD drop interval [scan1blup; positions in cM] )")
)
# Plot 3
#c2effB <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary", contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
#c2effBb <- scan1blup(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]],Xcovar=Xcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
#plot(c2effB, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
#plot(c2effBb, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
##last_coef <- unclass(c2effB)[nrow(c2effB),2:3] # last two coefficients
##for(t in seq(along=last_coef))
## axis(side=4, at=last_coef[t], names(last_coef)[t], tick=FALSE)
#Table 1
chr = peaks_mba$chr[i]
start=as.numeric(peaks_mba$ci_lo[i])
end=as.numeric(peaks_mba$ci_hi[i])
genesgss = query_genes(chr, start, end)
write.csv(genesgss, file=paste0("data/ici.vs.eoi_",peaks$phenotype[i],"-additive.covariates_genes_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_5.batches_0.csv"), quote=F)
rownames(genesgss) <- NULL
genesgss$strand_old = genesgss$strand
genesgss$strand[genesgss$strand=="+"] <- "positive"
genesgss$strand[genesgss$strand=="-"] <- "negative"
#genesgss <-
#table <-
#genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")] %>%
#kable(escape = F,align = c("ccccccccccc")) %>%
#kable_styling("striped", full_width = T) #%>%
#cat #%>%
#column_spec(1, bold=TRUE)
#
#print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], escape = F,align = c("ccccccccccc")))
print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], "html") %>% kable_styling("striped", full_width = T))
#table
}
} else {
print(paste0("There are too many peaks (",nrow(peaks_mba)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are too many peaks (0 peaks) that have a LOD that reaches suggestive (p<0.05) level of 3.22901302583048 [autosomes]/6.7056207215653 [x-chromosome]”
Interactive term
Significance thresholds
operm <- scan1perm(pr.qc, covars["rz.age"], model="normal", addcovar=addcovar, intcovar=addcovar, n_perm=10, perm_Xsp=TRUE, chr_lengths=chr_lengths(gm$gmap))
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
names(summary_table) <- c("autosomes","X")
summary_table$significance.level <- rownames(summary_table)
rownames(summary_table) <- NULL
summary_table[c(3,1:2)] %>%
kable(escape = F,align = c("ccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| significance.level | autosomes | X |
|---|---|---|
| 0.01 | 8.180585 | 7.720004 |
| 0.05 | 8.032968 | 7.490913 |
| 0.1 | 7.847986 | 7.191335 |
Genome-wide scan
plot_lod<-function(out,map){
for (i in 1:dim(out)[2]){
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - ICI vs. EOI (with interactive covariate) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
}
}
print("with no kinship")[1] “with no kinship”
outfi <- scan1(pr.qc, covars["rz.age"], Xcovar=Xcovar, model="normal", addcovar=addcovar, intcovar=addcovar)
plot_lod(outfi,gm$gmap)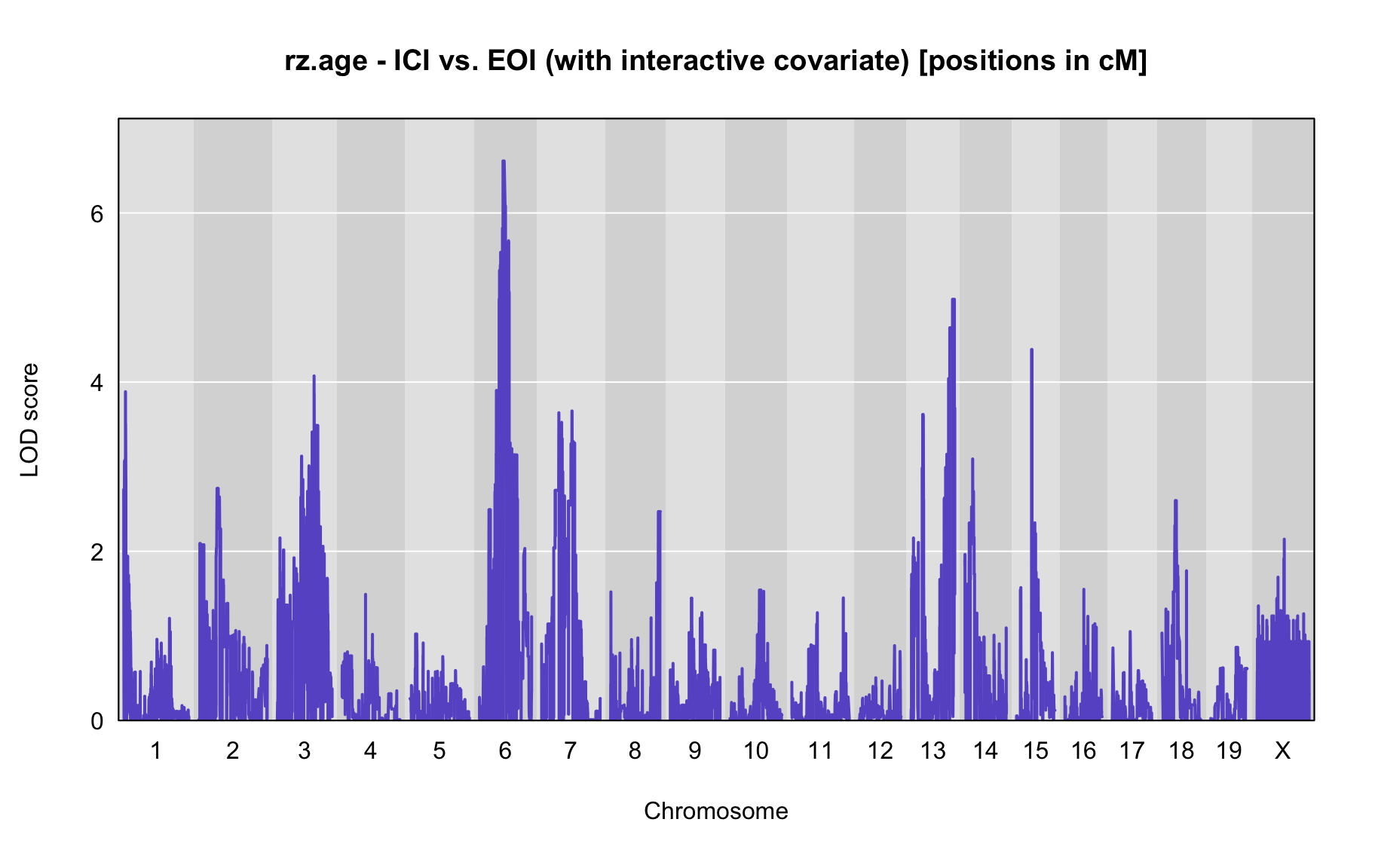
outf.new <- outfi-outfa
plot_lod(outf.new,gm$gmap)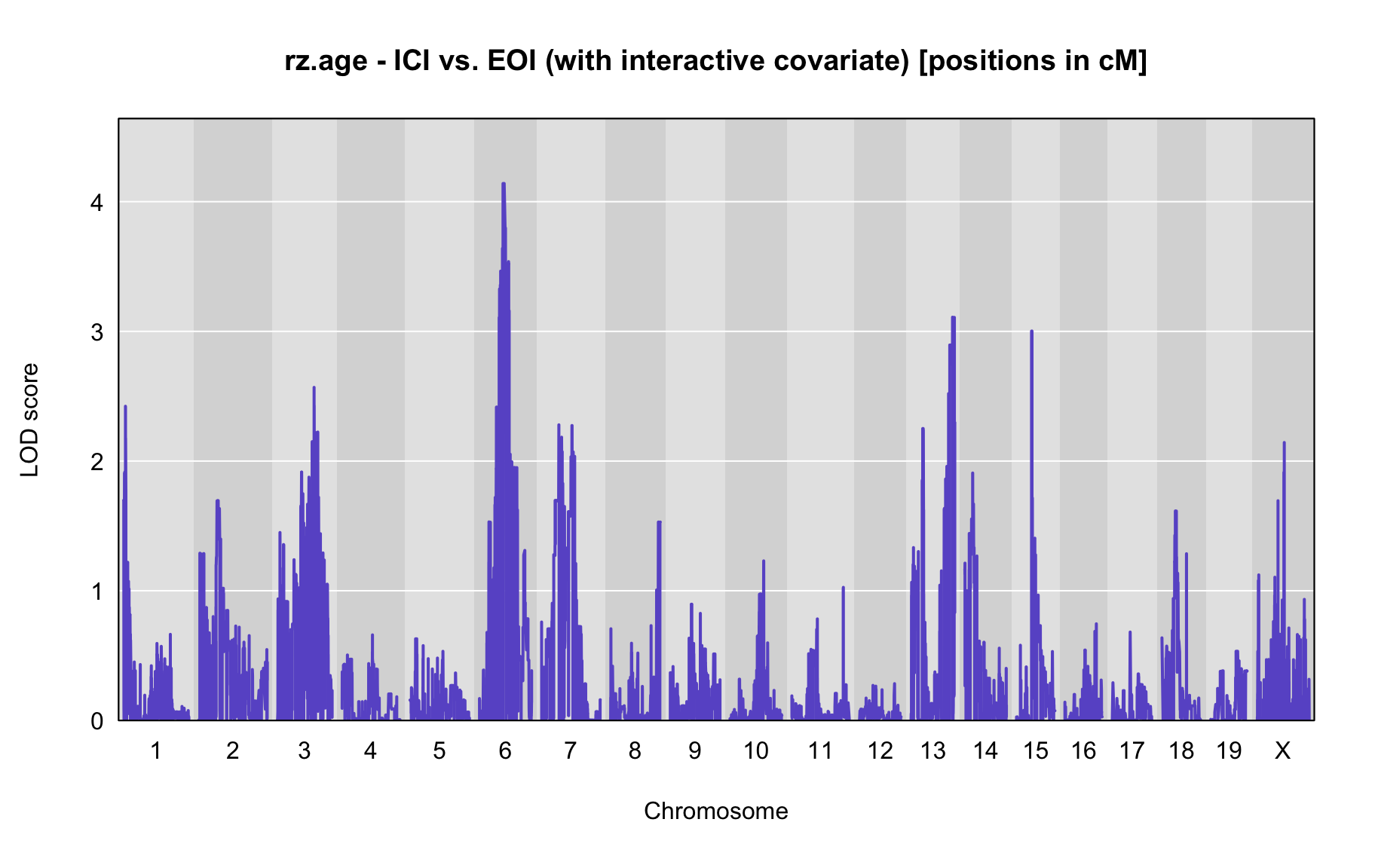
print("with normal kinship")[1] “with normal kinship”
outki <- scan1(pr.qc, covars["rz.age"], Xcovar=Xcovar, model="normal", addcovar=addcovar, intcovar=addcovar, kinship=kinship)
plot_lod(outki,gm$gmap)
outk.new <- outki-outka
plot_lod(outk.new,gm$gmap)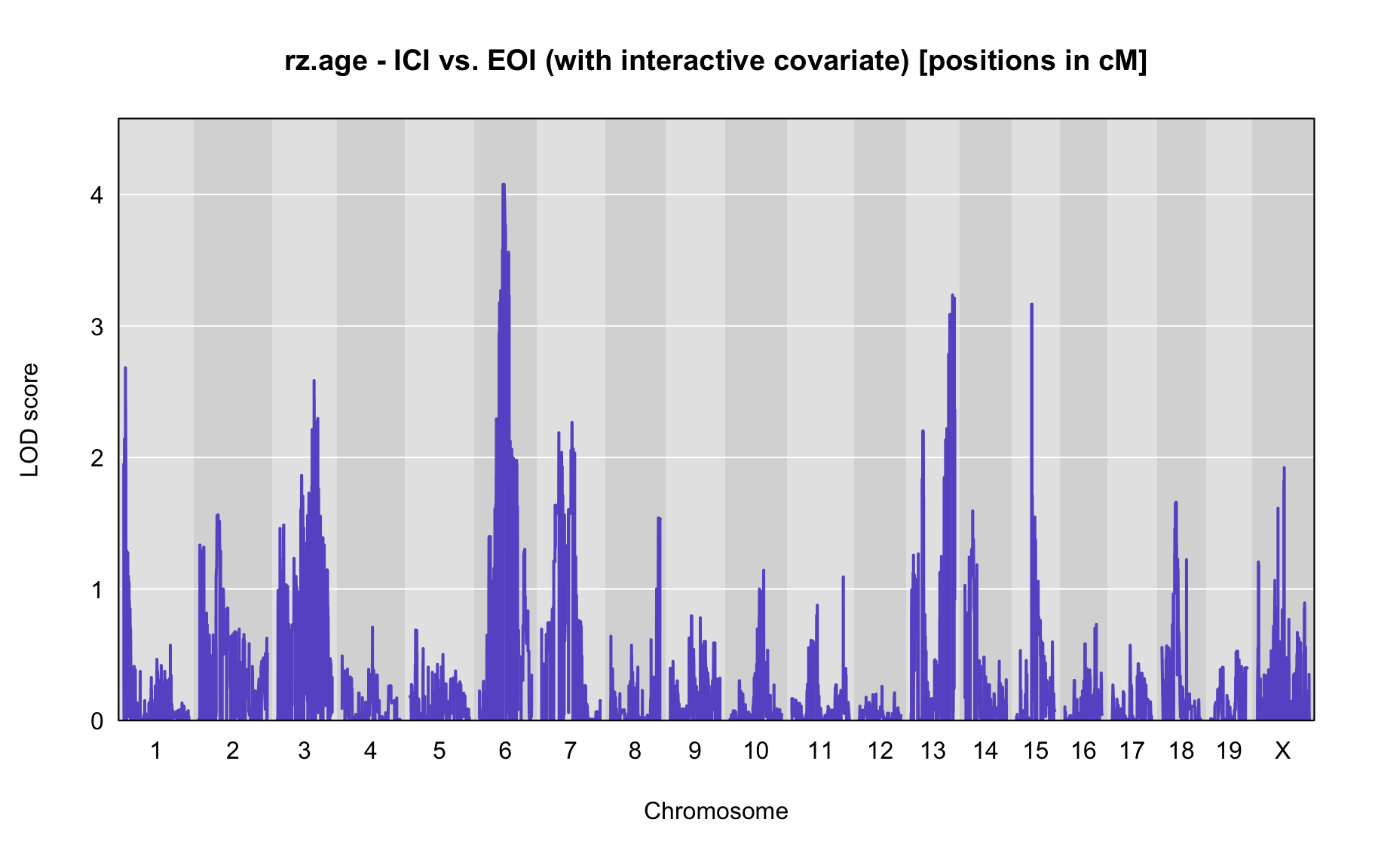
print("with loco kinship")[1] “with loco kinship”
outi <- scan1(pr.qc, covars["rz.age"], Xcovar=Xcovar, model="normal", addcovar=addcovar, intcovar=addcovar, kinship=K)
plot_lod(outi,gm$gmap)
outi.new <- outi-outa
plot_lod(outi.new,gm$gmap)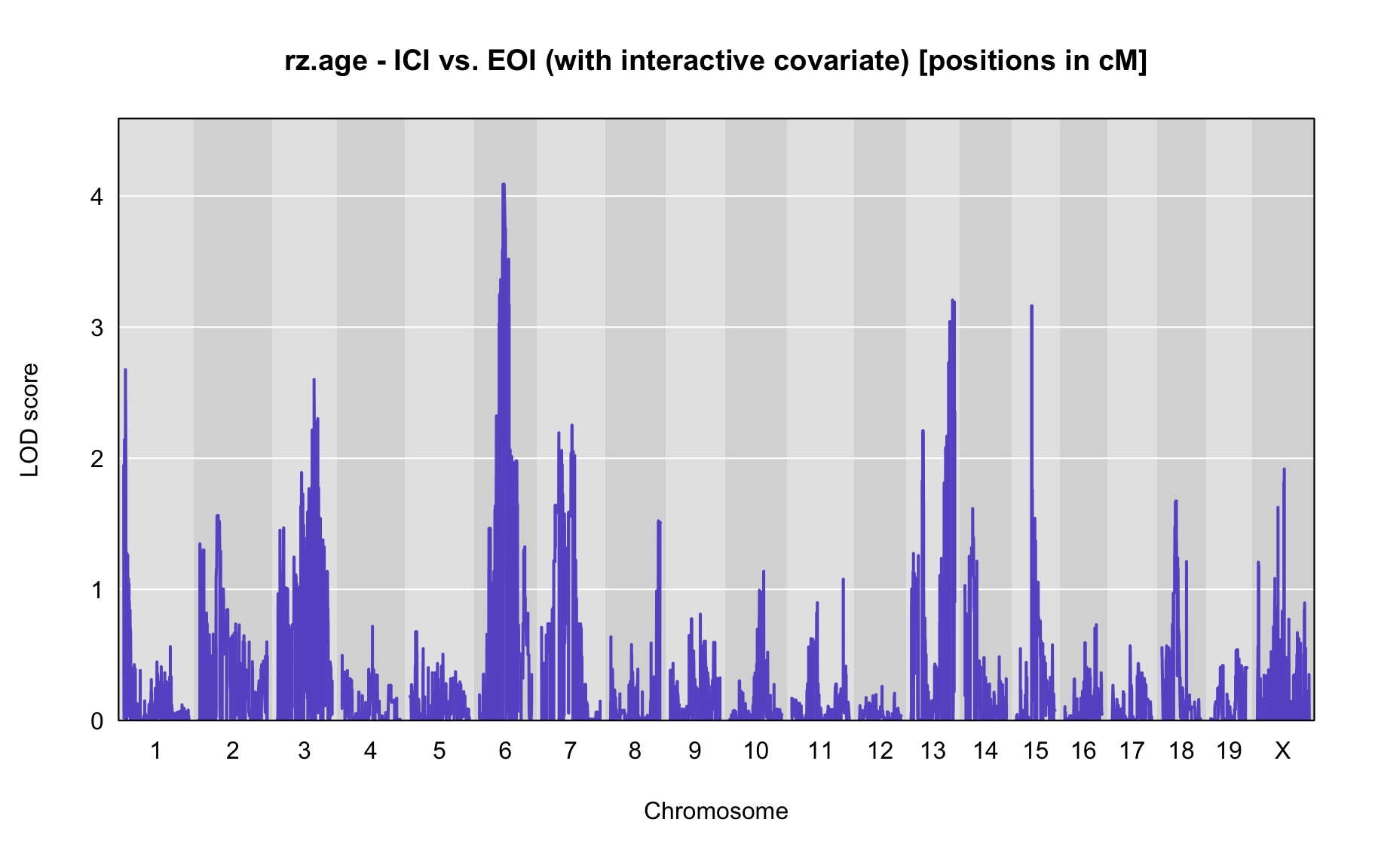
out = outi.new
print(paste0("number of samples in analysis = ", as.data.frame(attributes(out)$sample_size)[1,]))[1] “number of samples in analysis = 267”
LOD peaks
Centimorgan (cM)
peaks<-find_peaks(out, gm$gmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) > 0){
peaks$marker <- find_marker(gm$gmap, chr=peaks$chr,pos=peaks$pos)
names(peaks)[2] <- c("phenotype")
peaks <- peaks[-1]
rownames(peaks) <- NULL
print(kable(peaks, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
#plot only peak chromosomes
plot_lod_chr<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " - ICI vs. EOI (with interactive covariate) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are no peaks that have a LOD that reaches suggestive (p<0.05) level of 8.03296795029474 [autosomes]/7.49091276139169 [x-chromosome]”
if(nrow(peaks) < 50 & nrow(peaks) > 0){
for(i in unique(peaks$chr)){
#for (i in 1:nrow(peaks)){
#plot_lod_chr(out,gm$gmap, peaks$chr[i])
plot_lod_chr(out,gm$gmap, i)
}
} else {
print(paste0("There are too many peaks (",nrow(peaks)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are too many peaks (0 peaks) that have a LOD that reaches suggestive (p<0.05) level of 8.03296795029474 [autosomes]/7.49091276139169 [x-chromosome]”
Megabase (MB)
peaks_mba<-find_peaks(out, gm$pmap, threshold=summary(operm,alpha=0.05)$A, thresholdX = summary(operm,alpha=0.05)$X, peakdrop=3, drop=1.5)
if(nrow(peaks) > 0){
peaks_mba$marker <- find_marker(gm$pmap, chr=peaks_mba$chr,pos=peaks_mba$pos)
names(peaks_mba)[2] <- c("phenotype")
peaks_mba <- peaks_mba[-1]
rownames(peaks_mba) <- NULL
print(kable(peaks_mba, escape = F, align = c("cccccccc"), "html")
%>% kable_styling("striped", full_width = T)%>%
column_spec(1, bold=TRUE)
)
plot_lod_chr_mb<-function(out,map,chrom){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, map, chr = chrom, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
#legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = paste0(colnames(out)[i], " - chr", chrom, " - ICI vs. EOI (with interactive covariate) [positions in cM]"))
add_threshold(map, summary(operm,alpha=0.1), col = 'purple')
add_threshold(map, summary(operm, alpha=0.05), col = 'red')
add_threshold(map, summary(operm, alpha=0.01), col = 'blue')
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
} else {
print(paste0("There are no peaks that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are no peaks that have a LOD that reaches suggestive (p<0.05) level of 8.03296795029474 [autosomes]/7.49091276139169 [x-chromosome]”
if(nrow(peaks_mba) < 50 & nrow(peaks_mba) > 0){
for(i in unique(peaks_mba$chr)){
#for (i in 1:nrow(peaks_mba)){
#plot_lod_chr_mb(out,gm$pmap, peaks_mba$chr[i])
plot_lod_chr_mb(out,gm$pmap,i)
}
} else {
print(paste0("There are too many peaks (",nrow(peaks_mba)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are too many peaks (0 peaks) that have a LOD that reaches suggestive (p<0.05) level of 8.03296795029474 [autosomes]/7.49091276139169 [x-chromosome]”
QTL effects
For each peak LOD location we give a list of gene
if(nrow(peaks) < 50 & nrow(peaks) > 0){
for (i in 1:nrow(peaks)){
#for (i in 1:1){
#Plot 1
g <- maxmarg(pr.qc, gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i], return_char=TRUE)
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/","qtl_effect_", i, ".png"))
#par(mar=c(4.1, 4.1, 1.5, 0.6))
plot_pxg(g, covars[,peaks$phenotype[i]], ylab=peaks$phenotype[i], sort=FALSE)
title(main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," )"), line=0.2)
##dev.off()
chr = peaks$chr[i]
# Plot 2
pr_sub <- pull_genoprobint(pr.qc, gm$gmap, chr, c(peaks$ci_lo[i], peaks$ci_hi[i]))
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar)
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], Xcovar=Xcovar)
#coeff <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
#coeff_sub <- scan1coef(pr_sub[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
blup <- scan1blup(pr.qc[,chr], covars[peaks$phenotype[i]])
blup_sub <- scan1blup(pr_sub[,chr], covars[peaks$phenotype[i]])
write.csv(as.data.frame(blup_sub), paste0("data/ici.vs.eoi_",peaks$phenotype[i],"-interactive.covariate_blup_sub_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_5.batches_0.csv"), quote=F)
#plot_coef(coeff,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i]," [scan1coeff; positions in cM] )")
# )
#plot_coef(coeff_sub,
# gm$gmap, columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$lodcolumn[i],"; 1.5 LOD drop interval [scan1coeff; positions in cM] ) ")
# )
plot_coef(blup,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i]," [scan1blup; positions in cM] )")
)
plot_coef(blup_sub,
gm$gmap, columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste0("chr: ", chr=peaks$chr[i],"; pos: ", peaks$pos[i], "cM /",peaks_mba$pos[i],"MB\n(",peaks$phenotype[i],"; 1.5 LOD drop interval [scan1blup; positions in cM] )")
)
# Plot 3
#c2effB <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary", contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
#c2effBb <- scan1blup(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]],Xcovar=Xcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
#plot(c2effB, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
#plot(c2effBb, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
##last_coef <- unclass(c2effB)[nrow(c2effB),2:3] # last two coefficients
##for(t in seq(along=last_coef))
## axis(side=4, at=last_coef[t], names(last_coef)[t], tick=FALSE)
#Table 1
chr = peaks_mba$chr[i]
start=as.numeric(peaks_mba$ci_lo[i])
end=as.numeric(peaks_mba$ci_hi[i])
genesgss = query_genes(chr, start, end)
write.csv(genesgss, file=paste0("data/ici.vs.eoi_",peaks$phenotype[i],"-interactive.covariate_genes_chr-",chr,"_peak.marker-",peaks$marker[i],"_lod.drop-1.5_5.batches_0.csv"), quote=F)
rownames(genesgss) <- NULL
genesgss$strand_old = genesgss$strand
genesgss$strand[genesgss$strand=="+"] <- "positive"
genesgss$strand[genesgss$strand=="-"] <- "negative"
#genesgss <-
#table <-
#genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")] %>%
#kable(escape = F,align = c("ccccccccccc")) %>%
#kable_styling("striped", full_width = T) #%>%
#cat #%>%
#column_spec(1, bold=TRUE)
#
#print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], escape = F,align = c("ccccccccccc")))
print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], "html") %>% kable_styling("striped", full_width = T))
#table
}
} else {
print(paste0("There are too many peaks (",nrow(peaks_mba)," peaks) that have a LOD that reaches suggestive (p<0.05) level of ",summary(operm,alpha=0.05)$A, " [autosomes]/",summary(operm,alpha=0.05)$X, " [x-chromosome]"))
}[1] “There are too many peaks (0 peaks) that have a LOD that reaches suggestive (p<0.05) level of 8.03296795029474 [autosomes]/7.49091276139169 [x-chromosome]”
R version 3.6.2 (2019-12-12)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Catalina 10.15.7
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
locale:
[1] en_AU.UTF-8/en_AU.UTF-8/en_AU.UTF-8/C/en_AU.UTF-8/en_AU.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] abind_1.4-5 qtl2_0.22 reshape2_1.4.4 ggplot2_3.3.5
[5] tibble_3.1.2 psych_2.0.7 readxl_1.3.1 cluster_2.1.0
[9] dplyr_1.0.8 optparse_1.6.6 rhdf5_2.28.1 mclust_5.4.6
[13] tidyr_1.0.2 data.table_1.14.0 knitr_1.33 kableExtra_1.1.0
[17] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] httr_1.4.1 bit64_4.0.5 viridisLite_0.4.0 assertthat_0.2.1
[5] highr_0.9 blob_1.2.1 cellranger_1.1.0 yaml_2.2.1
[9] pillar_1.6.1 RSQLite_2.2.7 backports_1.2.1 lattice_0.20-38
[13] glue_1.4.2 digest_0.6.27 promises_1.1.0 rvest_0.3.5
[17] colorspace_2.0-2 htmltools_0.5.1.1 httpuv_1.5.2 plyr_1.8.6
[21] pkgconfig_2.0.3 purrr_0.3.4 scales_1.1.1 webshot_0.5.2
[25] getopt_1.20.3 later_1.0.0 git2r_0.26.1 generics_0.0.2
[29] ellipsis_0.3.2 cachem_1.0.5 withr_2.4.2 cli_3.0.0
[33] mnormt_1.5-7 magrittr_2.0.1 crayon_1.4.1 memoise_2.0.0
[37] evaluate_0.14 fs_1.4.1 fansi_0.5.0 nlme_3.1-142
[41] xml2_1.3.1 tools_3.6.2 hms_0.5.3 lifecycle_1.0.1
[45] stringr_1.4.0 Rhdf5lib_1.6.3 munsell_0.5.0 compiler_3.6.2
[49] rlang_1.0.2 grid_3.6.2 rstudioapi_0.13 rmarkdown_2.1
[53] gtable_0.3.0 DBI_1.1.1 R6_2.5.0 fastmap_1.1.0
[57] bit_4.0.4 utf8_1.2.1 rprojroot_1.3-2 readr_1.3.1
[61] stringi_1.7.2 parallel_3.6.2 Rcpp_1.0.7 vctrs_0.3.8
[65] tidyselect_1.1.2 xfun_0.24