Genotype Frequencies - PBS Myocarditis Yes vs PBS Myocarditis No
Belinda Cornes
2023-04-16
Last updated: 2023-04-16
Checks: 5 2
Knit directory: Serreze-T1D_Workflow/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R
Markdown file created these results, you’ll want to first commit it to
the Git repo. If you’re still working on the analysis, you can ignore
this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20220210) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /Users/corneb/Documents/MyJax/CS/Projects/Serreze/qc/workflowr/Serreze-T1D_Workflow | . |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 51e3845. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: analysis/.DS_Store
Untracked files:
Untracked: analysis/0.1.1_preparing.data_bqc_7.batches_myo.R
Untracked: analysis/0.1.1_preparing.data_bqc_7.batches_myo.Rmd
Untracked: analysis/0.1_samples_batch_20230124.Rmd
Untracked: analysis/0.1_samples_batch_20230225.Rmd
Untracked: analysis/0.1_samples_batch_20230313.Rmd
Untracked: analysis/0.2_haplotype_comparison_bqc_7.batches_myo_minprob.R
Untracked: analysis/0.2_haplotype_comparison_bqc_7.batches_myo_minprob.Rmd
Untracked: analysis/2.1_sample_bqc_7.batches_myo.R
Untracked: analysis/2.1_sample_bqc_7.batches_myo.Rmd
Untracked: analysis/2.1_sample_bqc_7.batches_myo.Rmd.R
Untracked: analysis/2.2.1_snp_qc_7.batches_myo.R
Untracked: analysis/2.2.1_snp_qc_7.batches_myo.Rmd
Untracked: analysis/2.2.1_snp_qc_7.batches_myo_mis.R
Untracked: analysis/2.2.1_snp_qc_7.batches_myo_mis.Rmd
Untracked: analysis/2.4_preparing.data_aqc_7.batches_myo.Rmd
Untracked: analysis/2.4_preparing.data_aqc_7.batches_myo_mis.Rmd
Untracked: analysis/2.4_preparing.data_aqc_7.batches_myo_misd.R
Untracked: analysis/3.1_phenotype.qc_corrected_7.batches_myo.Rmd
Untracked: analysis/3.1_phenotype.qc_corrected_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici-myo-yes.vs.het-ici-myo-no_snpsqc_7.batches_myo.R
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici-myo-yes.vs.het-ici-myo-no_snpsqc_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici-myo-yes.vs.het-ici-myo-no_snpsqc_7.batches_myo_mis.R
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici-myo-yes.vs.het-ici-myo-no_snpsqc_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici-myo-yes.vs.het-ici-myo-no_snpsqc_dis_no-x_updated_7.batches_myo.R
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici-myo-yes.vs.het-ici-myo-no_snpsqc_dis_no-x_updated_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici-myo-yes.vs.het-ici-myo-no_snpsqc_dis_no-x_updated_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici-myo-yes.vs.het-ici-myo-no_snpsqc_dis_no-x_updated_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici-myo-yes.vs.het-ici-myo-no_snpsqc_dis_no-x_updated_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici-myo-yes.vs.het-ici-myo-no_snpsqc_no-sex_7.batches_myo.R
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici-myo-yes.vs.het-ici-myo-no_snpsqc_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici-myo-yes.vs.het-ici-myo-no_snpsqc_no-sex_7.batches_myo_mis.R
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici-myo-yes.vs.het-ici-myo-no_snpsqc_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici.vs.het-pbs_snpsqc_7.batches_myo.R
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici.vs.het-pbs_snpsqc_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici.vs.het-pbs_snpsqc_7.batches_myo_mis.R
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici.vs.het-pbs_snpsqc_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici.vs.het-pbs_snpsqc_dis_no-x_updated_7.batches_myo.R
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici.vs.het-pbs_snpsqc_dis_no-x_updated_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici.vs.het-pbs_snpsqc_dis_no-x_updated_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici.vs.het-pbs_snpsqc_dis_no-x_updated_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici.vs.het-pbs_snpsqc_dis_no-x_updated_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici.vs.het-pbs_snpsqc_no-sex_7.batches_myo.R
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici.vs.het-pbs_snpsqc_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici.vs.het-pbs_snpsqc_no-sex_7.batches_myo_mis.R
Untracked: analysis/4.1.1_qtl.analysis_binary_het-ici.vs.het-pbs_snpsqc_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-myo-yes.vs.ici-myo-no_snpsqc_7.batches_myo.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-myo-yes.vs.ici-myo-no_snpsqc_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-myo-yes.vs.ici-myo-no_snpsqc_7.batches_myo_mis.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-myo-yes.vs.ici-myo-no_snpsqc_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-myo-yes.vs.ici-myo-no_snpsqc_dis_no-x_updated_7.batches_myo.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-myo-yes.vs.ici-myo-no_snpsqc_dis_no-x_updated_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-myo-yes.vs.ici-myo-no_snpsqc_dis_no-x_updated_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-myo-yes.vs.ici-myo-no_snpsqc_dis_no-x_updated_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-myo-yes.vs.ici-myo-no_snpsqc_dis_no-x_updated_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-myo-yes.vs.ici-myo-no_snpsqc_no-sex_7.batches_myo.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-myo-yes.vs.ici-myo-no_snpsqc_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-myo-yes.vs.ici-myo-no_snpsqc_no-sex_7.batches_myo_mis.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-myo-yes.vs.ici-myo-no_snpsqc_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-sick.vs.ici-eoi_snpsqc_7.batches_myo.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-sick.vs.ici-eoi_snpsqc_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-sick.vs.ici-eoi_snpsqc_7.batches_myo_mis.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-sick.vs.ici-eoi_snpsqc_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-sick.vs.ici-eoi_snpsqc_dis_no-x_updated_7.batches_myo.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-sick.vs.ici-eoi_snpsqc_dis_no-x_updated_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-sick.vs.ici-eoi_snpsqc_dis_no-x_updated_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-sick.vs.ici-eoi_snpsqc_dis_no-x_updated_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-sick.vs.ici-eoi_snpsqc_dis_no-x_updated_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-sick.vs.ici-eoi_snpsqc_no-sex_7.batches_myo.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-sick.vs.ici-eoi_snpsqc_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-sick.vs.ici-eoi_snpsqc_no-sex_7.batches_myo_mis.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici-sick.vs.ici-eoi_snpsqc_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_7.batches_myo.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_7.batches_myo_mis.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_7.batches_myo.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_dis_no-x_updated_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_no-sex_7.batches_myo.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_no-sex_7.batches_myo_mis.R
Untracked: analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_myo-yes.vs.myo-no_snpsqc_7.batches_myo.R
Untracked: analysis/4.1.1_qtl.analysis_binary_myo-yes.vs.myo-no_snpsqc_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_myo-yes.vs.myo-no_snpsqc_7.batches_myo_mis.R
Untracked: analysis/4.1.1_qtl.analysis_binary_myo-yes.vs.myo-no_snpsqc_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_myo-yes.vs.myo-no_snpsqc_dis_no-x_updated_7.batches_myo.R
Untracked: analysis/4.1.1_qtl.analysis_binary_myo-yes.vs.myo-no_snpsqc_dis_no-x_updated_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_myo-yes.vs.myo-no_snpsqc_dis_no-x_updated_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_myo-yes.vs.myo-no_snpsqc_dis_no-x_updated_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_myo-yes.vs.myo-no_snpsqc_dis_no-x_updated_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_myo-yes.vs.myo-no_snpsqc_no-sex_7.batches_myo.R
Untracked: analysis/4.1.1_qtl.analysis_binary_myo-yes.vs.myo-no_snpsqc_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_myo-yes.vs.myo-no_snpsqc_no-sex_7.batches_myo_mis.R
Untracked: analysis/4.1.1_qtl.analysis_binary_myo-yes.vs.myo-no_snpsqc_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_pbs-myo-yes.vs.pbs-myo-no_snpsqc_7.batches_myo.R
Untracked: analysis/4.1.1_qtl.analysis_binary_pbs-myo-yes.vs.pbs-myo-no_snpsqc_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_pbs-myo-yes.vs.pbs-myo-no_snpsqc_7.batches_myo_mis.R
Untracked: analysis/4.1.1_qtl.analysis_binary_pbs-myo-yes.vs.pbs-myo-no_snpsqc_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_pbs-myo-yes.vs.pbs-myo-no_snpsqc_dis_no-x_updated_7.batches_myo.R
Untracked: analysis/4.1.1_qtl.analysis_binary_pbs-myo-yes.vs.pbs-myo-no_snpsqc_dis_no-x_updated_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_pbs-myo-yes.vs.pbs-myo-no_snpsqc_dis_no-x_updated_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_pbs-myo-yes.vs.pbs-myo-no_snpsqc_dis_no-x_updated_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_pbs-myo-yes.vs.pbs-myo-no_snpsqc_dis_no-x_updated_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_pbs-myo-yes.vs.pbs-myo-no_snpsqc_no-sex_7.batches_myo.R
Untracked: analysis/4.1.1_qtl.analysis_binary_pbs-myo-yes.vs.pbs-myo-no_snpsqc_no-sex_7.batches_myo.Rmd
Untracked: analysis/4.1.1_qtl.analysis_binary_pbs-myo-yes.vs.pbs-myo-no_snpsqc_no-sex_7.batches_myo_mis.R
Untracked: analysis/4.1.1_qtl.analysis_binary_pbs-myo-yes.vs.pbs-myo-no_snpsqc_no-sex_7.batches_myo_mis.Rmd
Untracked: analysis/debugging_comparing_old_Rmd/
Untracked: analysis/frequency.plot.R
Untracked: analysis/genotype.frequencies_het-ici-myo-yes.vs.het-ici-myo-no_7.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_het-ici-myo-yes.vs.het-ici-myo-no_7.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_het-ici-myo-yes.vs.het-ici-myo-no_freq_7.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_het-ici-myo-yes.vs.het-ici-myo-no_freq_7.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_het-ici.vs.het-pbs_7.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_het-ici.vs.het-pbs_7.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_het-ici.vs.het-pbs_freq_7.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_het-ici.vs.het-pbs_freq_7.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_ici-myo-yes.vs.ici-myo-no_7.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_ici-myo-yes.vs.ici-myo-no_7.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_ici-myo-yes.vs.ici-myo-no_freq_7.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_ici-myo-yes.vs.ici-myo-no_freq_7.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_ici-sick.vs.ici-eoi_7.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_ici-sick.vs.ici-eoi_7.batches_myo.Rmd.r
Untracked: analysis/genotype.frequencies_ici-sick.vs.ici-eoi_7.batches_myo_edit.Rmd
Untracked: analysis/genotype.frequencies_ici-sick.vs.ici-eoi_7.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_ici-sick.vs.ici-eoi_freq_7.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_ici-sick.vs.ici-eoi_freq_7.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.pbs_7.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.pbs_7.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.pbs_freq_7.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_ici.vs.pbs_freq_7.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_myo-yes.vs.myo-no_7.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_myo-yes.vs.myo-no_7.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_myo-yes.vs.myo-no_freq_7.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_myo-yes.vs.myo-no_freq_7.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_pbs-myo-yes.vs.pbs-myo-no_7.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_pbs-myo-yes.vs.pbs-myo-no_7.batches_myo_mis.Rmd
Untracked: analysis/genotype.frequencies_pbs-myo-yes.vs.pbs-myo-no_freq_7.batches_myo.Rmd
Untracked: analysis/genotype.frequencies_pbs-myo-yes.vs.pbs-myo-no_freq_7.batches_myo_mis.Rmd
Untracked: analysis/index_7.batches_myo.Rmd
Untracked: analysis/power.analysis_het-ici-myo-yes.vs.het-ici-myo-no_7.batches_myo.Rmd
Untracked: analysis/power.analysis_het-ici-myo-yes.vs.het-ici-myo-no_7.batches_myo_mis.Rmd
Untracked: analysis/power.analysis_het-ici.vs.het-pbs_7.batches_myo.Rmd
Untracked: analysis/power.analysis_het-ici.vs.het-pbs_7.batches_myo_mis.Rmd
Untracked: analysis/power.analysis_ici-myo-yes.vs.ici-myo-no_7.batches_myo.Rmd
Untracked: analysis/power.analysis_ici-myo-yes.vs.ici-myo-no_7.batches_myo_mis.Rmd
Untracked: analysis/power.analysis_ici-sick.vs.ici-eoi_7.batches_myo.Rmd
Untracked: analysis/power.analysis_ici-sick.vs.ici-eoi_7.batches_myo_mis.Rmd
Untracked: analysis/power.analysis_ici.vs.pbs_7.batches_myo.Rmd
Untracked: analysis/power.analysis_ici.vs.pbs_7batches_myo_mis.Rmd
Untracked: analysis/power.analysis_myo-yes.vs.myo-no_7.batches_myo.Rmd
Untracked: analysis/power.analysis_myo-yes.vs.myo-no_7.batches_myo_mis.Rmd
Untracked: analysis/power.analysis_pbs-myo-yes.vs.pbs-myo-no_7.batches_myo.Rmd
Untracked: analysis/power.analysis_pbs-myo-yes.vs.pbs-myo-no_7.batches_myo_mis.Rmd
Untracked: data/GM_covar_4.batches_myo.csv
Untracked: data/GM_covar_7.batches_myo.csv
Untracked: data/bad_markers_all_4.batches_myo.RData
Untracked: data/bad_markers_all_7.batches_myo.RData
Untracked: data/covar_corrected.cleaned_het-ici-myo-yes.vs.het-ici-myo-no_7.batches_myo.csv
Untracked: data/covar_corrected.cleaned_het-ici-myo-yes.vs.het-ici-myo-no_7.batches_myo_mis.csv
Untracked: data/covar_corrected.cleaned_het-ici.vs.het-pbs_7.batches_myo.csv
Untracked: data/covar_corrected.cleaned_het-ici.vs.het-pbs_7.batches_myo_mis.csv
Untracked: data/covar_corrected.cleaned_ici-myo-yes.vs.ici-myo-no_7.batches_myo.csv
Untracked: data/covar_corrected.cleaned_ici-myo-yes.vs.ici-myo-no_7.batches_myo_mis.csv
Untracked: data/covar_corrected.cleaned_ici-sick.vs.ici-eoi_7.batches_myo.csv
Untracked: data/covar_corrected.cleaned_ici-sick.vs.ici-eoi_7.batches_myo_mis.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_7.batches_myo.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs_7.batches_myo_mis.csv
Untracked: data/covar_corrected.cleaned_myo-yes.vs.myo-no_7.batches_myo.csv
Untracked: data/covar_corrected.cleaned_myo-yes.vs.myo-no_7.batches_myo_mis.csv
Untracked: data/covar_corrected.cleaned_pbs-myo-yes.vs.pbs-myo-no_7.batches_myo.csv
Untracked: data/covar_corrected.cleaned_pbs-myo-yes.vs.pbs-myo-no_7.batches_myo_mis.csv
Untracked: data/covar_corrected_het-ici-myo-yes.vs.het-ici-myo-no_7.batches_myo.csv
Untracked: data/covar_corrected_het-ici-myo-yes.vs.het-ici-myo-no_7.batches_myo_mis.csv
Untracked: data/covar_corrected_het-ici.vs.het-pbs_7.batches_myo.csv
Untracked: data/covar_corrected_het-ici.vs.het-pbs_7.batches_myo_mis.csv
Untracked: data/covar_corrected_ici-myo-yes.vs.ici-myo-no_7.batches_myo.csv
Untracked: data/covar_corrected_ici-myo-yes.vs.ici-myo-no_7.batches_myo_mis.csv
Untracked: data/covar_corrected_ici-sick.vs.ici-eoi_7.batches_myo.csv
Untracked: data/covar_corrected_ici-sick.vs.ici-eoi_7.batches_myo_mis.csv
Untracked: data/covar_corrected_ici.vs.pbs_7.batches_myo.csv
Untracked: data/covar_corrected_ici.vs.pbs_7.batches_myo_mis.csv
Untracked: data/covar_corrected_myo-yes.vs.myo-no_7.batches_myo.csv
Untracked: data/covar_corrected_myo-yes.vs.myo-no_7.batches_myo_mis.csv
Untracked: data/covar_corrected_pbs-myo-yes.vs.pbs-myo-no_7.batches_myo.csv
Untracked: data/covar_corrected_pbs-myo-yes.vs.pbs-myo-no_7.batches_myo_mis.csv
Untracked: data/e_4.batches_myo.RData
Untracked: data/e_7.batches_myo.RData
Untracked: data/e_snpg_samqc_4.batches_myo.RData
Untracked: data/e_snpg_samqc_7.batches_myo.RData
Untracked: data/errors_ind_4.batches_myo.RData
Untracked: data/errors_ind_7.batches_myo.RData
Untracked: data/genetic_map_4.batches_myo.csv
Untracked: data/genetic_map_7.batches_myo.csv
Untracked: data/genotype_errors_marker_4.batches_myo.RData
Untracked: data/genotype_errors_marker_7.batches_myo.RData
Untracked: data/genotype_freq_marker_4.batches_myo.RData
Untracked: data/genotype_freq_marker_7.batches_myo.RData
Untracked: data/gm_allqc_4.batches_myo.RData
Untracked: data/gm_allqc_4.batches_myo_mis.RData
Untracked: data/gm_allqc_7.batches_myo.RData
Untracked: data/gm_allqc_7.batches_myo_mis.RData
Untracked: data/gm_samqc_4.batches_myo.RData
Untracked: data/gm_samqc_7.batches_myo.RData
Untracked: data/gm_serreze.BC312.RData
Untracked: data/gm_serreze.BC366.RData
Untracked: data/gm_serreze_7.batches_myo.RData
Untracked: data/gm_serreze_bc_7.batches_myo.RData
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_gm_qtl_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo_mis.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/het-ici.vs.het-pbs_gm_qtl_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/het-ici.vs.het-pbs_gm_qtl_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/het-ici.vs.het-pbs_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/het-ici.vs.het-pbs_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/het-ici.vs.het-pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/het-ici.vs.het-pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/het-ici.vs.het-pbs_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/het-ici.vs.het-pbs_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/het-ici.vs.het-pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/het-ici.vs.het-pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/het-ici.vs.het-pbs_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/het-ici.vs.het-pbs_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/het-ici.vs.het-pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/het-ici.vs.het-pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/het-ici.vs.het-pbs_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo.csv
Untracked: data/het-ici.vs.het-pbs_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo_mis.csv
Untracked: data/het-ici.vs.het-pbs_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/het-ici.vs.het-pbs_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_gm_qtl_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_gm_qtl_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo_mis.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-10_peak.marker-UNC18343990_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-10_peak.marker-UNC18343990_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-11_peak.marker-UNC20070077_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-11_peak.marker-UNC20070077_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-12_peak.marker-UNC21652584_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-12_peak.marker-UNC21652584_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-13_peak.marker-UNCHS036579_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-13_peak.marker-UNCHS036579_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-14_peak.marker-UNCHS037782_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-14_peak.marker-UNCHS037782_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-15_peak.marker-UNC26069905_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-15_peak.marker-UNCHS041223_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-16_peak.marker-JAX00070117_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-16_peak.marker-JAX00070117_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-17_peak.marker-UNC28542319_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-17_peak.marker-UNCJPD006670_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-18_peak.marker-UNC28776739_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-18_peak.marker-UNCHS045594_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-19_peak.marker-UNC30414168_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-19_peak.marker-UNC30426276_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-1_peak.marker-UNC2031646_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-1_peak.marker-UNCHS003700_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-2_peak.marker-ICR5131_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-2_peak.marker-UNCHS006420_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-3_peak.marker-UNC5667757_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-3_peak.marker-UNC5667757_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-4_peak.marker-UNC6759992_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-4_peak.marker-UNC6765178_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-5_peak.marker-UNC9889957_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-5_peak.marker-UNC9900273_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-6_peak.marker-UNC10800126_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-6_peak.marker-UNC10832076_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-7_peak.marker-UNCHS020903_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-7_peak.marker-UNCHS021163_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-8_peak.marker-UNC15471847_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-8_peak.marker-UNC15548888_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-9_peak.marker-UNC16231874_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-9_peak.marker-UNC16231874_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-X_peak.marker-UNCHS049472_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_blup_sub_chr-X_peak.marker-UNCHS049472_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-10_peak.marker-UNC18343990_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-10_peak.marker-UNC18343990_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-11_peak.marker-UNC20070077_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-11_peak.marker-UNC20070077_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-12_peak.marker-UNC21652584_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-12_peak.marker-UNC21652584_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-13_peak.marker-UNCHS036579_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-13_peak.marker-UNCHS036579_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-14_peak.marker-UNCHS037782_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-14_peak.marker-UNCHS037782_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-15_peak.marker-UNC26069905_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-15_peak.marker-UNCHS041223_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-16_peak.marker-JAX00070117_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-16_peak.marker-JAX00070117_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-17_peak.marker-UNC28542319_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-17_peak.marker-UNCJPD006670_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-18_peak.marker-UNC28776739_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-18_peak.marker-UNCHS045594_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-19_peak.marker-UNC30414168_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-19_peak.marker-UNC30426276_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-1_peak.marker-UNC2031646_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-1_peak.marker-UNCHS003700_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-2_peak.marker-ICR5131_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-2_peak.marker-UNCHS006420_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-3_peak.marker-UNC5667757_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-3_peak.marker-UNC5667757_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-4_peak.marker-UNC6759992_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-4_peak.marker-UNC6765178_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-5_peak.marker-UNC9889957_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-5_peak.marker-UNC9900273_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-6_peak.marker-UNC10800126_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-6_peak.marker-UNC10832076_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-7_peak.marker-UNCHS020903_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-7_peak.marker-UNCHS021163_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-8_peak.marker-UNC15471847_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-8_peak.marker-UNC15548888_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-9_peak.marker-UNC16231874_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-9_peak.marker-UNC16231874_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-X_peak.marker-UNCHS049472_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_genes_chr-X_peak.marker-UNCHS049472_lod.drop-1.5_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_gm_qtl_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_gm_qtl_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo_mis.csv
Untracked: data/ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo_mis.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_age.of.onset_het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_age.of.onset_het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_age.of.onset_het-ici.vs.het-pbs_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_het-ici.vs.het-pbs_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_age.of.onset_het-ici.vs.het-pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_het-ici.vs.het-pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_age.of.onset_ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_age.of.onset_ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_age.of.onset_ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_age.of.onset_ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_age.of.onset_ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_myo-yes.vs.myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_myo-yes.vs.myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_age.of.onset_myo-yes.vs.myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_myo-yes.vs.myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_age.of.onset_pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_age.of.onset_pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_age.of.onset_pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_group_het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_group_het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_het-ici-myo-yes.vs.het-ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_group_het-ici.vs.het-pbs_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_het-ici.vs.het-pbs_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_group_het-ici.vs.het-pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_het-ici.vs.het-pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_group_ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_group_ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_ici-myo-yes.vs.ici-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_group_ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_group_ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_ici-sick.vs.ici-eoi_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_group_ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_myo-yes.vs.myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_myo-yes.vs.myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_group_myo-yes.vs.myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_myo-yes.vs.myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_group_pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/mean.differences_group_pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/mean.differences_group_pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/myo-yes.vs.myo-no_gm_qtl_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/myo-yes.vs.myo-no_gm_qtl_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/myo-yes.vs.myo-no_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/myo-yes.vs.myo-no_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/myo-yes.vs.myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/myo-yes.vs.myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/myo-yes.vs.myo-no_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/myo-yes.vs.myo-no_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/myo-yes.vs.myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/myo-yes.vs.myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/myo-yes.vs.myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/myo-yes.vs.myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/myo-yes.vs.myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/myo-yes.vs.myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/myo-yes.vs.myo-no_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo.csv
Untracked: data/myo-yes.vs.myo-no_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo_mis.csv
Untracked: data/myo-yes.vs.myo-no_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/myo-yes.vs.myo-no_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_gm_qtl_snpsqc_dis_no-x_updated_7.batches_myo.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_gm_qtl_snpsqc_dis_no-x_updated_7.batches_myo_mis.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo_mis.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo_mis.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo_mis.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo.csv
Untracked: data/pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo_mis.csv
Untracked: data/percent_missing_id_4.batches_myo.RData
Untracked: data/percent_missing_id_7.batches_myo.RData
Untracked: data/percent_missing_marker_4.batches_myo.RData
Untracked: data/percent_missing_marker_7.batches_myo.RData
Untracked: data/pheno_4.batches_myo.csv
Untracked: data/pheno_7.batches_myo.csv
Untracked: data/physical_map_4.batches_myo.csv
Untracked: data/physical_map_7.batches_myo.csv
Untracked: data/probs_36state_bc_7.batches_myo.rds
Untracked: data/probs_8state_bc_7.batches_myo.rds
Untracked: data/qc_info_bad_sample_4.batches_myo.RData
Untracked: data/qc_info_bad_sample_7.batches_myo.RData
Untracked: data/sample_geno_AHB_4.batches_myo.csv
Untracked: data/sample_geno_AHB_7.batches_myo.csv
Untracked: data/sample_geno_AHB_7.batches_myo_orig.id.csv
Untracked: data/sample_geno_bc_4.batches_myo.csv
Untracked: data/sample_geno_bc_7.batches_myo.csv
Untracked: data/sample_geno_bc_7.batches_myo_orig.id.csv
Untracked: data/serreze_probs_4.batches_myo.rds
Untracked: data/serreze_probs_7.batches_myo.rds
Untracked: data/serreze_probs_allqc_4.batches_myo.rds
Untracked: data/serreze_probs_allqc_4.batches_myo_mis.rds
Untracked: data/serreze_probs_allqc_7.batches_myo.rds
Untracked: data/serreze_probs_allqc_7.batches_myo_mis.rds
Untracked: data/summary.cg_4.batches_myo.RData
Untracked: data/summary.cg_7.batches_myo.RData
Untracked: output/Percent_missing_genotype_data_4.batches_myo.pdf
Untracked: output/Percent_missing_genotype_data_7.batches_myo.pdf
Untracked: output/Percent_missing_genotype_data_per_marker_4.batches_myo.pdf
Untracked: output/Percent_missing_genotype_data_per_marker_7.batches_myo.pdf
Untracked: output/Proportion_matching_genotypes_before_removal_of_bad_samples_4.batches_myo.pdf
Untracked: output/Proportion_matching_genotypes_before_removal_of_bad_samples_7.batches_myo.pdf
Untracked: output/genotype_error_marker_4.batches_myo.pdf
Untracked: output/genotype_error_marker_7.batches_myo.pdf
Untracked: output/genotype_frequency_marker_4.batches_myo.pdf
Untracked: output/genotype_frequency_marker_7.batches_myo.pdf
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with
wflow_publish() to start tracking its development.
Allele Frequencies
All variants (before snp qc)
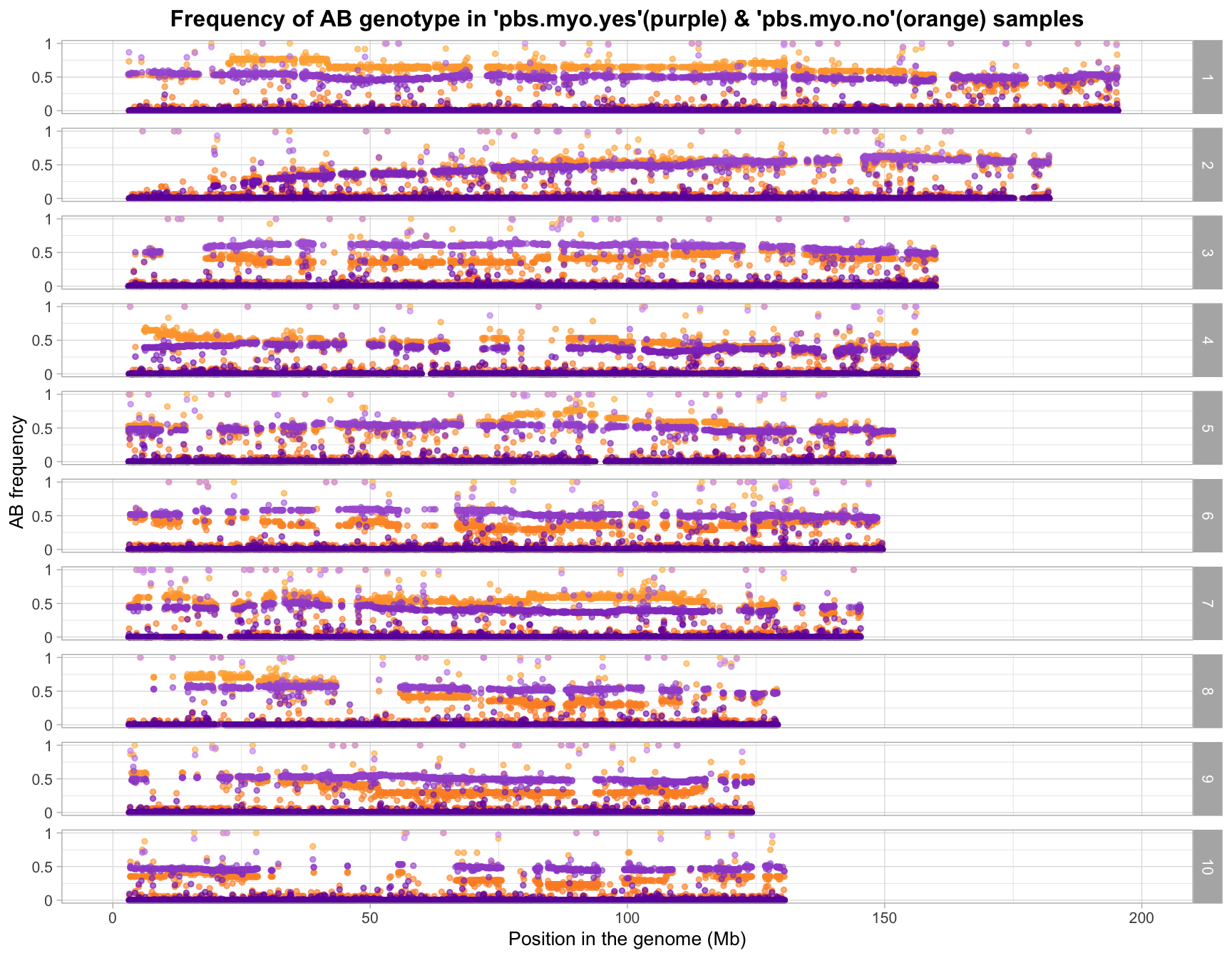
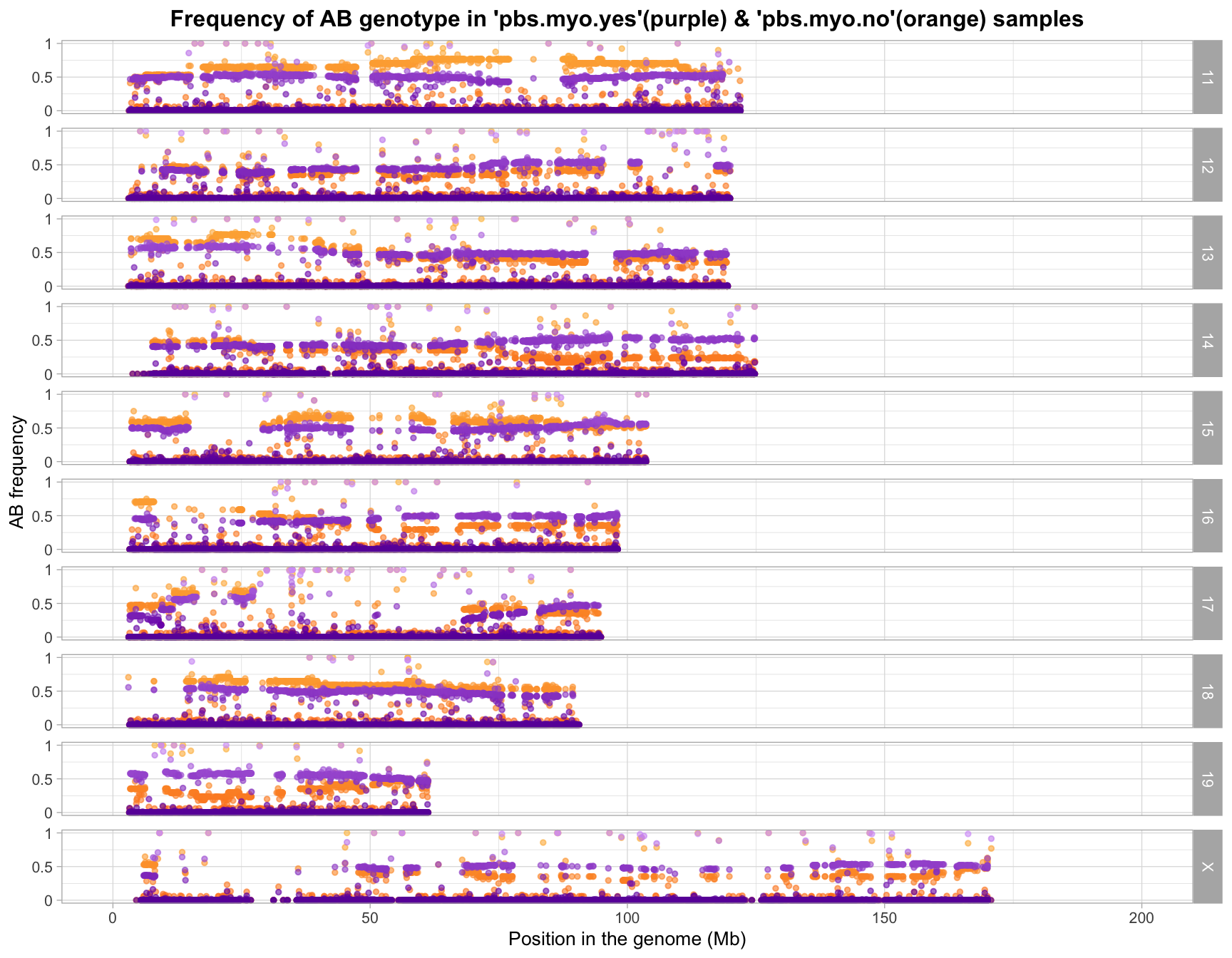
After SNP QC
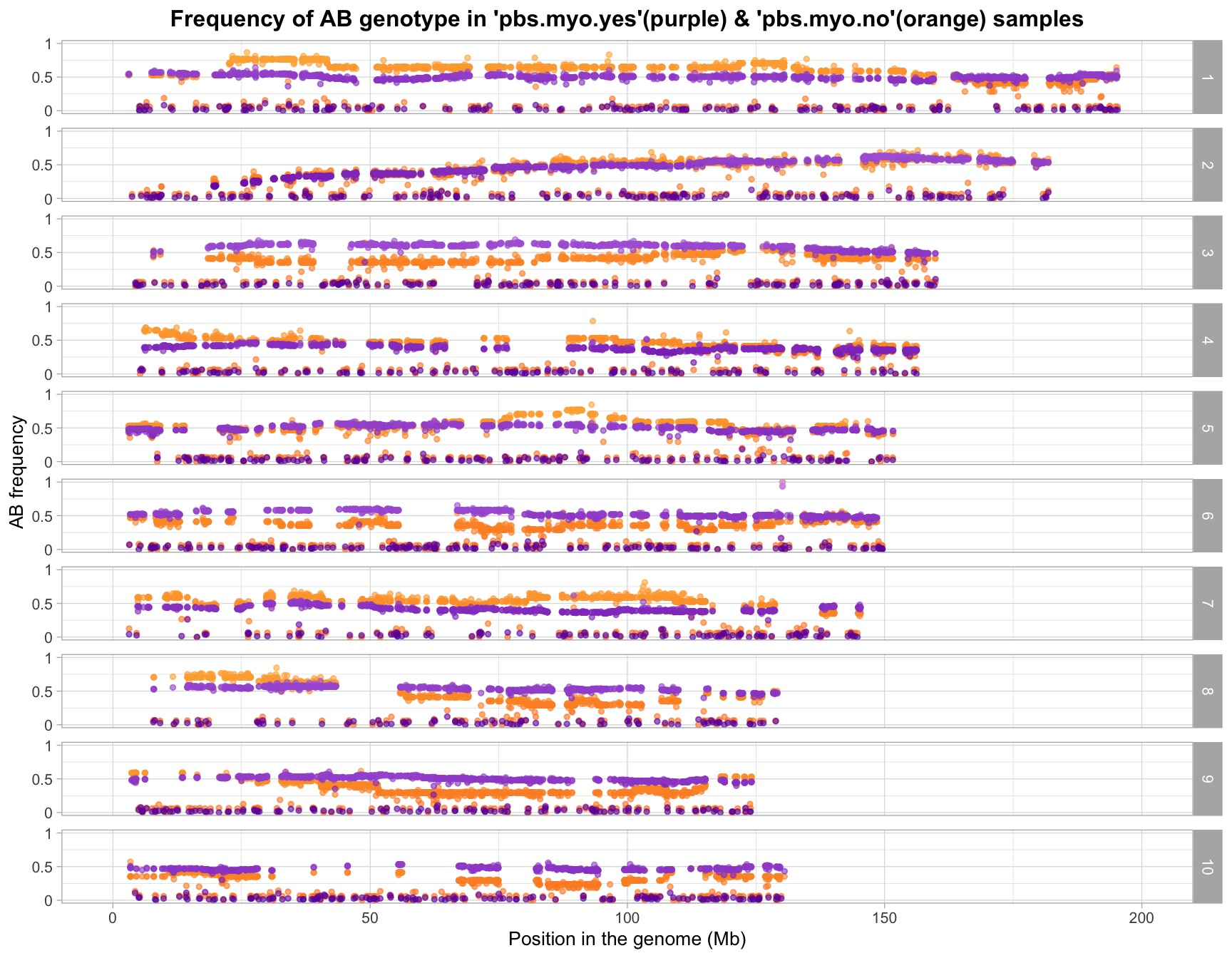

with sample outliers
load("data/gm_allqc_7.batches_myo.RData")
#gm_allqc
gm=gm_allqc
gmObject of class cross2 (crosstype "bc")
Total individuals 357
No. genotyped individuals 357
No. phenotyped individuals 357
No. with both geno & pheno 357
No. phenotypes 1
No. covariates 11
No. phenotype covariates 0
No. chromosomes 20
Total markers 32660
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2482 2412 1742 1772 1648 1842 1541 1517 1769 1092 1754 1230 1441 1505 1128 842
17 18 19 X
655 820 945 4523 pr <- readRDS("data/serreze_probs_allqc_7.batches_myo.rds")
#pr <- readRDS("data/serreze_probs.rds")
geno <- read.csv("data/sample_geno_bc_7.batches_myo.csv", as.is=T, check.names=FALSE)
#names(geno) <- gsub("^X","",names(geno))
#names(geno) <- gsub("\.","-",names(geno))
#names(geno)[2:ncol(geno)] <- samples$Unique.Sample.ID
rownames(geno) <- geno$marker## extracting animals with ici and pbs group status
#miceinfo <- gm$covar[gm$covar$group == "PBS" | gm$covar$group == "ICI",]
#table(miceinfo$group)
#mice.ids <- rownames(miceinfo)
#gm <- gm[mice.ids]
#gm
#table(gm$covar$group)
covars <- read_csv("data/covar_corrected_pbs-myo-yes.vs.pbs-myo-no_7.batches_myo.csv")
# removing any missing info
#covars <- subset(covars, covars$age.of.onset!='')
nrow(covars)[1] 96table(covars$"Myocarditis Status")
NO YES
17 79 table(covars$"Murine MHC KO Status")
HOM
96 table(covars$"Drug Treatment")
PBS
96 table(covars$"clinical pheno")
EOI SICK
64 32 # keeping only informative mice
gm <- gm[covars$Mouse.ID]
gmObject of class cross2 (crosstype "bc")
Total individuals 96
No. genotyped individuals 96
No. phenotyped individuals 96
No. with both geno & pheno 96
No. phenotypes 1
No. covariates 11
No. phenotype covariates 0
No. chromosomes 20
Total markers 32660
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2482 2412 1742 1772 1648 1842 1541 1517 1769 1092 1754 1230 1441 1505 1128 842
17 18 19 X
655 820 945 4523 table(gm$covar$"Myocarditis Status")
NO YES
17 79 table(gm$covar$"Murine MHC KO Status")
HOM
96 table(gm$covar$"Drug Treatment")
PBS
96 table(gm$covar$"clinical pheno")
EOI SICK
64 32 pr.qc.ids <- pr
for (i in 1:20){pr.qc.ids[[i]] = pr.qc.ids[[i]][covars$Mouse.ID,,]}
geno <- geno[,covars$Mouse.ID]
geno <- geno[marker_names(gm),]
dim(geno)[1] 32660 96## calculating genotype frequencies
### from geno genotypes
g <- do.call("cbind", gm$geno)
gf_mar_geno <- t(apply(g, 2, function(a) table(factor(a, 1:2))/sum(a != 0)))
gn_mar_geno <- t(apply(g, 2, function(a) table(factor(a, 0:2))))
#gf_mar_raw<- gf_mar_raw[gf_mar_raw[,2] != "NaN",]
colnames(gf_mar_geno) <- c("freq_AA_geno_table","freq_AB_geno_table")
colnames(gn_mar_geno) <- c("count_missing_geno_table","count_AA_geno_table","count_AB_geno_table")
gfn_mar_geno <- merge(as.data.frame(gn_mar_geno), as.data.frame(gf_mar_geno), by="row.names")
rownames(gfn_mar_geno) <- gfn_mar_geno[,1]
gfn_mar_geno <- gfn_mar_geno[-1]
### from raw using table function in R
#genosl <- list()
#for(i in 1:nrow(geno)){
##for(i in 1:3){
# genoi <- geno[i,]
# freqf <- table(factor(geno[i,], c("-","AA","AB")))
# genoi$count_AA_raw_rowSums <- rowSums(genoi == "AA")
# genoi$count_AB_raw_rowSums <- rowSums(genoi == "AB")
# genoi$count_missing_raw_rowSums <- rowSums(genoi == "-")
# freqf <- t(table(factor(geno[i,], c("-","AA","AB"))))
# freqf <- as.data.frame(t(freqf[1,]))
# rownames(freqf) <- rownames(genoi)
# colnames(freqf) <- c("count_missing_raw_table","count_AA_raw_table","count_AB_raw_table")
# genoif <- cbind(freqf,genoi[c("count_AA_raw_rowSums","count_AB_raw_rowSums","count_missing_raw_rowSums")])
# genosl[[i]] = genoif
#}
#gf_mar_raw <- do.call("rbind",genosl)
#gf_mar_raw <- gf_mar_raw[,c(1:3,6,4:5)]
#gf_mar_raw$index <- 1:nrow(gf_mar_raw)
### from probabilities
gf_mar_probs.1 <- calc_geno_freq(pr.qc.ids, by = "marker", omit_x = FALSE)
#gn_mar_probs <- calc_geno_freq(probs, by = "individual", omit_x = FALSE)
gf_mar_probs <- rbind(gf_mar_probs.1$A[,1:2], gf_mar_probs.1$X[,1:2])
colnames(gf_mar_probs) <- paste0("freq_",colnames(gf_mar_probs),"_probs")
gf_mar_probs <- as.data.frame(gf_mar_probs)
gf_mar_probs$index <- 1:nrow(gf_mar_probs)
### merging all genotype frequecies for all markers
#gf_mar.1 <- merge(as.data.frame(gf_mar_raw), as.data.frame(gfn_mar_geno), by="row.names")
#rownames(gf_mar.1) <- gf_mar.1[,1]
#gf_mar.1 <- gf_mar.1[-1]
#gf_mar <- merge(gf_mar.1,as.data.frame(gf_mar_probs), by="row.names")
gf_mar <- merge(as.data.frame(gfn_mar_geno),as.data.frame(gf_mar_probs), by="row.names")
rownames(gf_mar) <- gf_mar[,1]
gf_mar <- gf_mar[-1]
gf_mar <- gf_mar[order(gf_mar$index),]
dim(gf_mar)[1] 32660 8# Calculating ratio and flagging informative marker
gf_mar$ratio = as.numeric(gf_mar$freq_AA_geno_table)/as.numeric(gf_mar$freq_AB_geno_table)
gf_mar$Include = ifelse(gf_mar$ratio >= 0.90 & gf_mar$ratio <= 1.10, TRUE,FALSE)
table(gf_mar$Include)
FALSE TRUE
24849 7811 ## filtering out <= 0.05
gf_mar$count.geno <- rowSums(gf_mar[c("freq_AA_geno_table","freq_AB_geno_table")] <=0.05)
filtered_gf_mar_geno <- gf_mar[gf_mar$count.geno != 1,]
filtered_gf_mar_geno <- filtered_gf_mar_geno[,-which(names(filtered_gf_mar_geno) %in% c("count.geno","index"))]
dim(filtered_gf_mar_geno)[1] 26651 9table(filtered_gf_mar_geno$Include)
FALSE TRUE
18840 7811 gf_mar$count.probs <- rowSums(gf_mar[c("freq_AA_probs","freq_AB_probs")] <=0.05)
filtered_gf_mar_probs <- gf_mar[gf_mar$count.probs != 1,]
filtered_gf_mar_probs <- filtered_gf_mar_probs[,-which(names(filtered_gf_mar_probs) %in% c("count.geno","count.probs","index"))]
dim(filtered_gf_mar_probs)[1] 27553 9table(filtered_gf_mar_probs$Include)
FALSE TRUE
19754 7799 ## merging with sample_genos
filtered_gf_mar_geno_sample <- merge(geno,filtered_gf_mar_geno, by="row.names", all.y=T, sort=F)
#filtered_gf_mar_geno_sample <- filtered_gf_mar_geno_sample[order(filtered_gf_mar_geno_sample$index),]
#filtered_gf_mar_geno_sample <- filtered_gf_mar_geno_sample[,-which(names(filtered_gf_mar_geno_sample) %in% c("count.geno","index"))]
names(filtered_gf_mar_geno_sample)[1] <- c("marker")
dim(filtered_gf_mar_geno_sample)[1] 26651 106filtered_gf_mar_probs_sample <- merge(geno,filtered_gf_mar_probs, by="row.names", all.y=T, sort=F)
#filtered_gf_mar_probs_sample <- filtered_gf_mar_probs_sample[order(filtered_gf_mar_probs_sample$index),]
#filtered_gf_mar_probs_sample <- filtered_gf_mar_probs_sample[,-which(names(filtered_gf_mar_probs_sample) %in% c("count.geno","count.probs","index"))]
names(filtered_gf_mar_probs_sample)[1] <- c("marker")
dim(filtered_gf_mar_probs_sample)[1] 27553 106## saving files
write.csv(filtered_gf_mar_geno_sample, "data/pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_7.batches_myo.csv", quote=F)
write.csv(filtered_gf_mar_probs_sample, "data/pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.probs.freq.removed_7.batches_myo.csv", quote=F)
write.csv(filtered_gf_mar_geno, "data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.geno.freq.removed_geno.ratio_7.batches_myo.csv", quote=F)
write.csv(filtered_gf_mar_probs, "data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.probs.freq.removed_geno.ratio_7.batches_myo.csv", quote=F)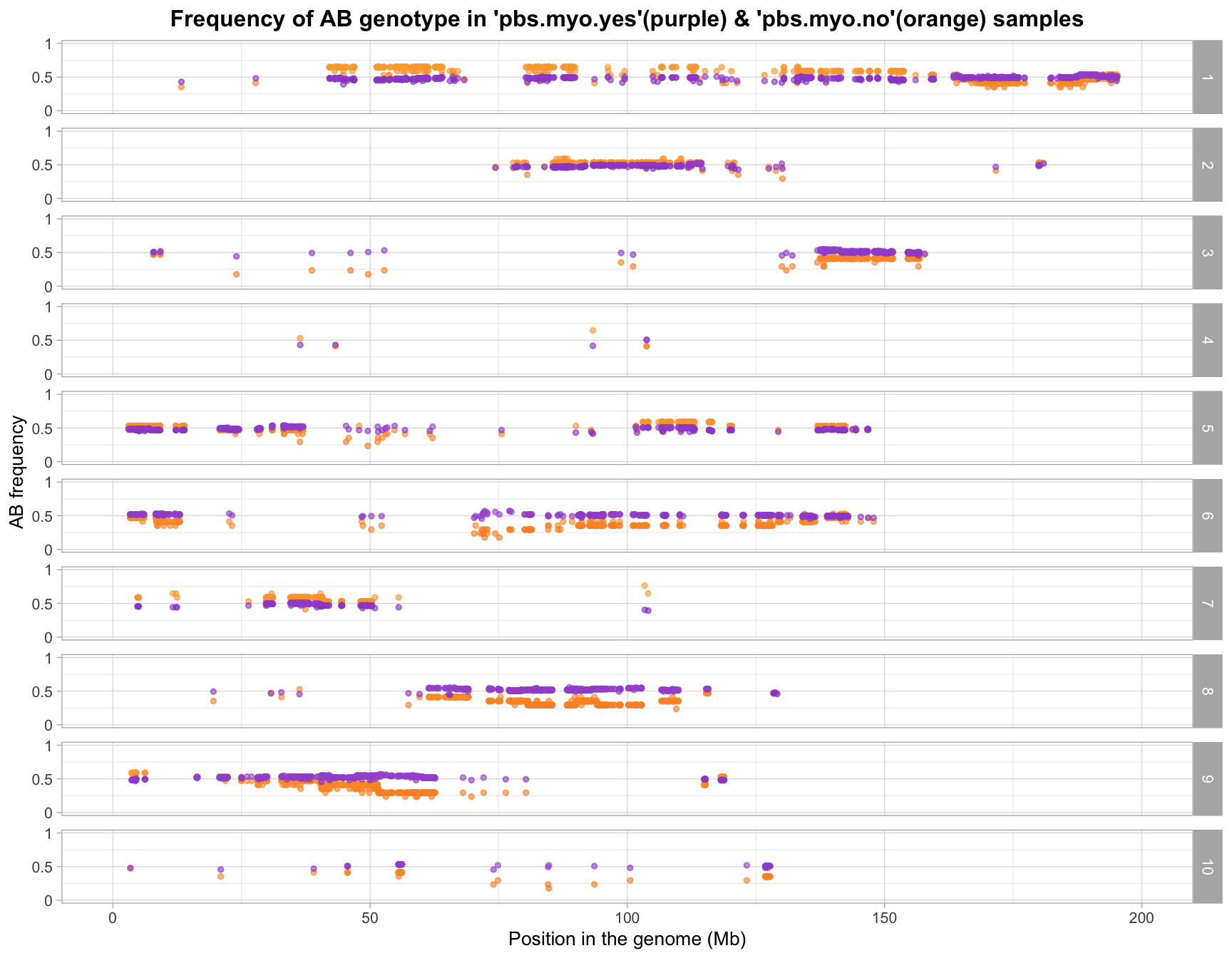
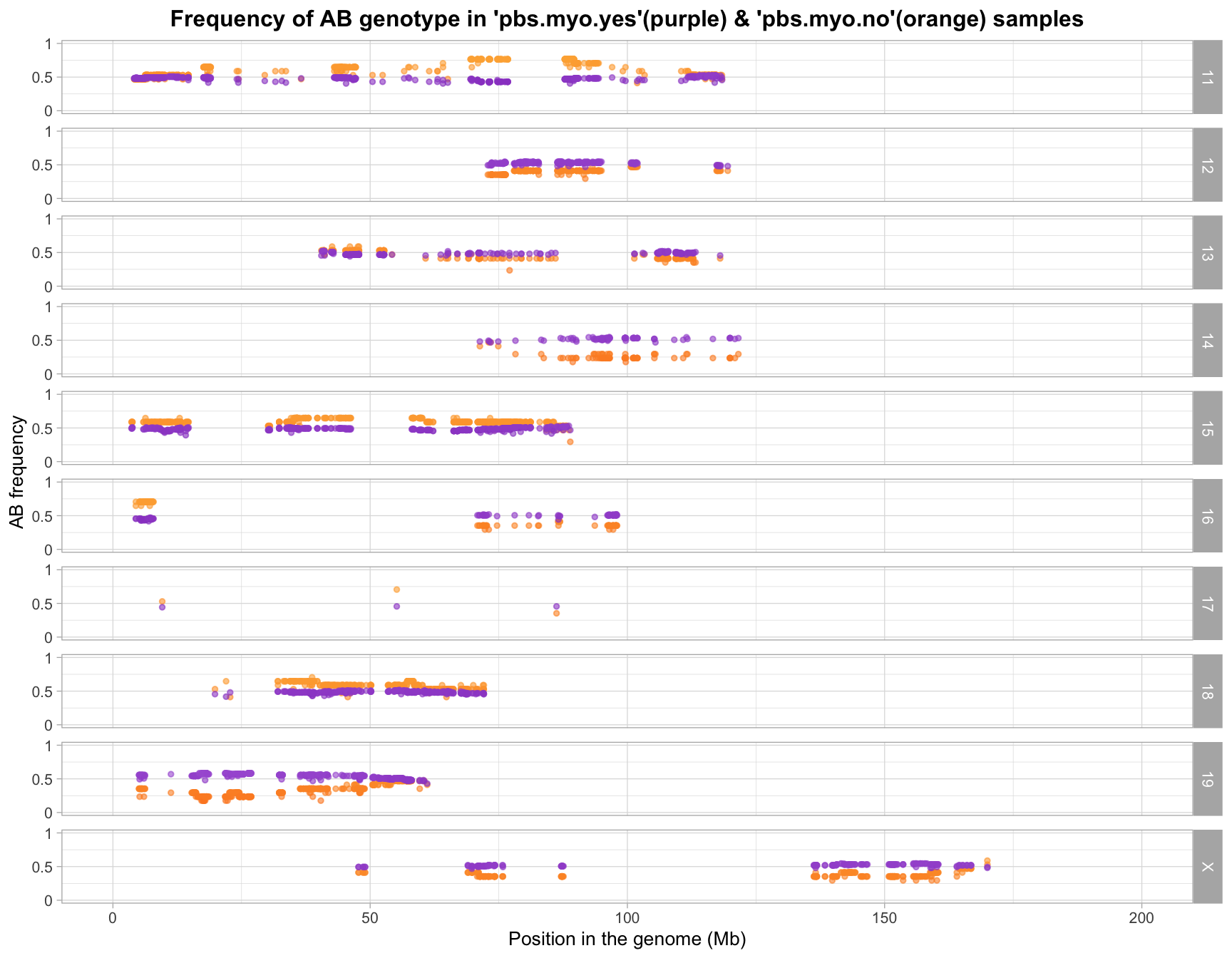
sample outliers removed
load("data/gm_allqc_7.batches_myo.RData")
#gm_allqc
gm=gm_allqc
gmObject of class cross2 (crosstype "bc")
Total individuals 357
No. genotyped individuals 357
No. phenotyped individuals 357
No. with both geno & pheno 357
No. phenotypes 1
No. covariates 11
No. phenotype covariates 0
No. chromosomes 20
Total markers 32660
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2482 2412 1742 1772 1648 1842 1541 1517 1769 1092 1754 1230 1441 1505 1128 842
17 18 19 X
655 820 945 4523 pr <- readRDS("data/serreze_probs_allqc_7.batches_myo.rds")
#pr <- readRDS("data/serreze_probs.rds")
geno <- read.csv("data/sample_geno_bc_7.batches_myo.csv", as.is=T, check.names=FALSE)
#names(geno) <- gsub("^X","",names(geno))
#names(geno) <- gsub("\.","-",names(geno))
#names(geno)[2:ncol(geno)] <- samples$Unique.Sample.ID
rownames(geno) <- geno$marker## extracting animals with ici and pbs group status
#miceinfo <- gm$covar[gm$covar$group == "PBS" | gm$covar$group == "ICI",]
#table(miceinfo$group)
#mice.ids <- rownames(miceinfo)
#gm <- gm[mice.ids]
#gm
#table(gm$covar$group)
covars <- read_csv("data/covar_corrected.cleaned_pbs-myo-yes.vs.pbs-myo-no_7.batches_myo.csv")
# removing any missing info
covars <- subset(covars, covars$out.age.of.onset=='Keep' & covars$out.rz.age=='Keep')
nrow(covars)[1] 92table(covars$"Myocarditis Status")
NO YES
16 76 table(covars$"Murine MHC KO Status")
HOM
92 table(covars$"Drug Treatment")
PBS
92 table(covars$"clinical pheno")
EOI SICK
64 28 # keeping only informative mice
gm <- gm[covars$Mouse.ID]
gmObject of class cross2 (crosstype "bc")
Total individuals 92
No. genotyped individuals 92
No. phenotyped individuals 92
No. with both geno & pheno 92
No. phenotypes 1
No. covariates 11
No. phenotype covariates 0
No. chromosomes 20
Total markers 32660
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2482 2412 1742 1772 1648 1842 1541 1517 1769 1092 1754 1230 1441 1505 1128 842
17 18 19 X
655 820 945 4523 table(gm$covar$"Myocarditis Status")
NO YES
16 76 table(gm$covar$"Murine MHC KO Status")
HOM
92 table(gm$covar$"Drug Treatment")
PBS
92 table(gm$covar$"clinical pheno")
EOI SICK
64 28 pr.qc.ids <- pr
for (i in 1:20){pr.qc.ids[[i]] = pr.qc.ids[[i]][covars$Mouse.ID,,]}
geno <- geno[,covars$Mouse.ID]
geno <- geno[marker_names(gm),]
dim(geno)[1] 32660 92## calculating genotype frequencies
### from geno genotypes
g <- do.call("cbind", gm$geno)
gf_mar_geno <- t(apply(g, 2, function(a) table(factor(a, 1:2))/sum(a != 0)))
gn_mar_geno <- t(apply(g, 2, function(a) table(factor(a, 0:2))))
#gf_mar_raw<- gf_mar_raw[gf_mar_raw[,2] != "NaN",]
colnames(gf_mar_geno) <- c("freq_AA_geno_table","freq_AB_geno_table")
colnames(gn_mar_geno) <- c("count_missing_geno_table","count_AA_geno_table","count_AB_geno_table")
gfn_mar_geno <- merge(as.data.frame(gn_mar_geno), as.data.frame(gf_mar_geno), by="row.names")
rownames(gfn_mar_geno) <- gfn_mar_geno[,1]
gfn_mar_geno <- gfn_mar_geno[-1]
### from raw using table function in R
#genosl <- list()
#for(i in 1:nrow(geno)){
##for(i in 1:3){
# genoi <- geno[i,]
# freqf <- table(factor(geno[i,], c("-","AA","AB")))
# genoi$count_AA_raw_rowSums <- rowSums(genoi == "AA")
# genoi$count_AB_raw_rowSums <- rowSums(genoi == "AB")
# genoi$count_missing_raw_rowSums <- rowSums(genoi == "-")
# freqf <- t(table(factor(geno[i,], c("-","AA","AB"))))
# freqf <- as.data.frame(t(freqf[1,]))
# rownames(freqf) <- rownames(genoi)
# colnames(freqf) <- c("count_missing_raw_table","count_AA_raw_table","count_AB_raw_table")
# genoif <- cbind(freqf,genoi[c("count_AA_raw_rowSums","count_AB_raw_rowSums","count_missing_raw_rowSums")])
# genosl[[i]] = genoif
#}
#gf_mar_raw <- do.call("rbind",genosl)
#gf_mar_raw <- gf_mar_raw[,c(1:3,6,4:5)]
#gf_mar_raw$index <- 1:nrow(gf_mar_raw)
### from probabilities
gf_mar_probs.1 <- calc_geno_freq(pr.qc.ids, by = "marker", omit_x = FALSE)
#gn_mar_probs <- calc_geno_freq(probs, by = "individual", omit_x = FALSE)
gf_mar_probs <- rbind(gf_mar_probs.1$A[,1:2], gf_mar_probs.1$X[,1:2])
colnames(gf_mar_probs) <- paste0("freq_",colnames(gf_mar_probs),"_probs")
gf_mar_probs <- as.data.frame(gf_mar_probs)
gf_mar_probs$index <- 1:nrow(gf_mar_probs)
### merging all genotype frequecies for all markers
#gf_mar.1 <- merge(as.data.frame(gf_mar_raw), as.data.frame(gfn_mar_geno), by="row.names")
#rownames(gf_mar.1) <- gf_mar.1[,1]
#gf_mar.1 <- gf_mar.1[-1]
#gf_mar <- merge(gf_mar.1,as.data.frame(gf_mar_probs), by="row.names")
gf_mar <- merge(as.data.frame(gfn_mar_geno),as.data.frame(gf_mar_probs), by="row.names")
rownames(gf_mar) <- gf_mar[,1]
gf_mar <- gf_mar[-1]
gf_mar <- gf_mar[order(gf_mar$index),]
dim(gf_mar)[1] 32660 8# Calculating ratio and flagging informative marker
gf_mar$ratio = as.numeric(gf_mar$freq_AA_geno_table)/as.numeric(gf_mar$freq_AB_geno_table)
gf_mar$Include = ifelse(gf_mar$ratio >= 0.90 & gf_mar$ratio <= 1.10, TRUE,FALSE)
table(gf_mar$Include)
FALSE TRUE
25079 7581 ## filtering out <= 0.05
gf_mar$count.geno <- rowSums(gf_mar[c("freq_AA_geno_table","freq_AB_geno_table")] <=0.05)
filtered_gf_mar_geno <- gf_mar[gf_mar$count.geno != 1,]
filtered_gf_mar_geno <- filtered_gf_mar_geno[,-which(names(filtered_gf_mar_geno) %in% c("count.geno","index"))]
dim(filtered_gf_mar_geno)[1] 26649 9table(filtered_gf_mar_geno$Include)
FALSE TRUE
19068 7581 gf_mar$count.probs <- rowSums(gf_mar[c("freq_AA_probs","freq_AB_probs")] <=0.05)
filtered_gf_mar_probs <- gf_mar[gf_mar$count.probs != 1,]
filtered_gf_mar_probs <- filtered_gf_mar_probs[,-which(names(filtered_gf_mar_probs) %in% c("count.geno","count.probs","index"))]
dim(filtered_gf_mar_probs)[1] 27576 9table(filtered_gf_mar_probs$Include)
FALSE TRUE
20006 7570 ## merging with sample_genos
filtered_gf_mar_geno_sample <- merge(geno,filtered_gf_mar_geno, by="row.names", all.y=T, sort=F)
#filtered_gf_mar_geno_sample <- filtered_gf_mar_geno_sample[order(filtered_gf_mar_geno_sample$index),]
#filtered_gf_mar_geno_sample <- filtered_gf_mar_geno_sample[,-which(names(filtered_gf_mar_geno_sample) %in% c("count.geno","index"))]
names(filtered_gf_mar_geno_sample)[1] <- c("marker")
dim(filtered_gf_mar_geno_sample)[1] 26649 102filtered_gf_mar_probs_sample <- merge(geno,filtered_gf_mar_probs, by="row.names", all.y=T, sort=F)
#filtered_gf_mar_probs_sample <- filtered_gf_mar_probs_sample[order(filtered_gf_mar_probs_sample$index),]
#filtered_gf_mar_probs_sample <- filtered_gf_mar_probs_sample[,-which(names(filtered_gf_mar_probs_sample) %in% c("count.geno","count.probs","index"))]
names(filtered_gf_mar_probs_sample)[1] <- c("marker")
dim(filtered_gf_mar_probs_sample)[1] 27576 102## saving files
write.csv(filtered_gf_mar_geno_sample, "data/pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed_7.batches_myo.csv", quote=F)
write.csv(filtered_gf_mar_probs_sample, "data/pbs-myo-yes.vs.pbs-myo-no_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed_7.batches_myo.csv", quote=F)
write.csv(filtered_gf_mar_geno, "data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv", quote=F)
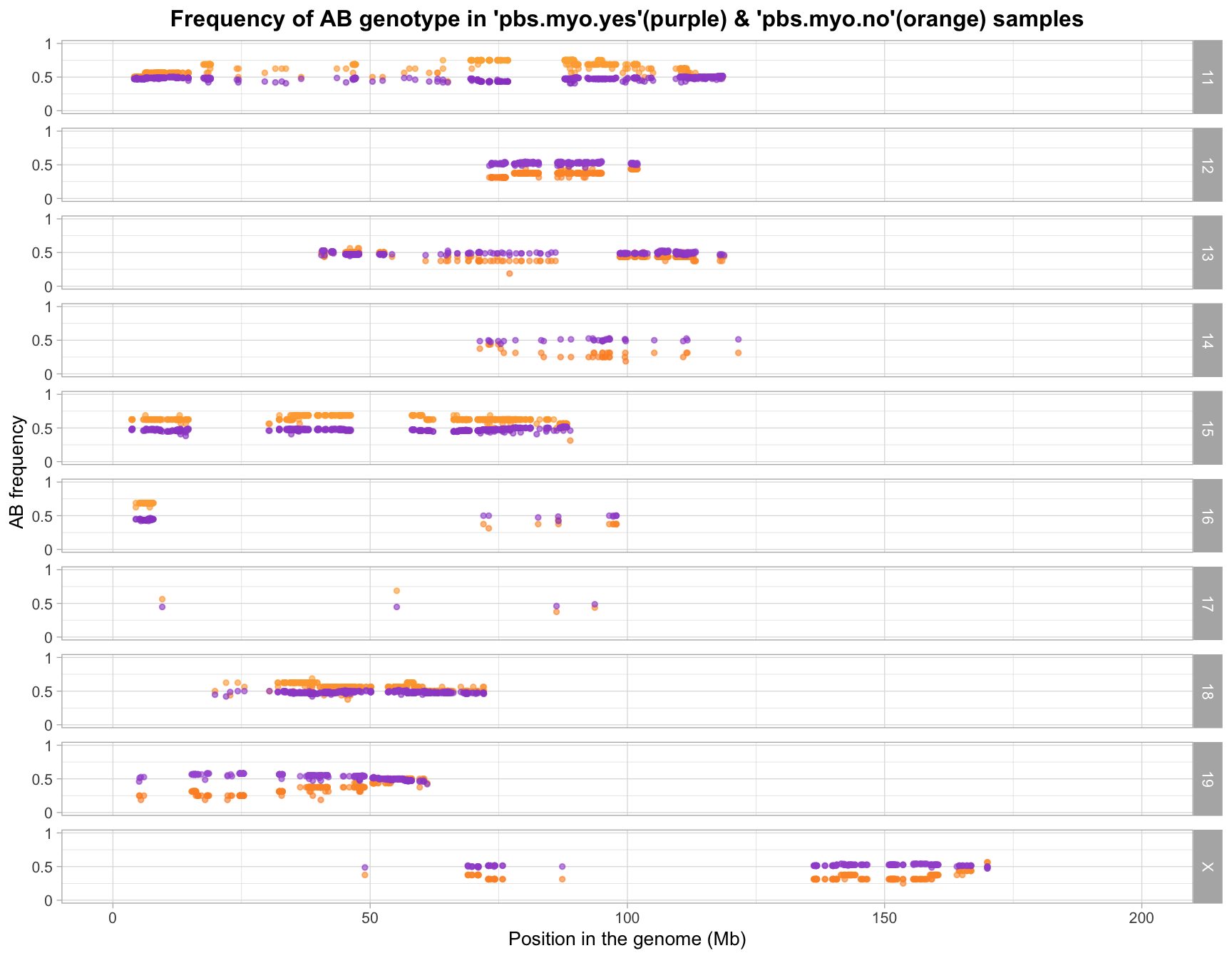
write.csv(filtered_gf_mar_probs, "data/pbs-myo-yes.vs.pbs-myo-no_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio_7.batches_myo.csv", quote=F)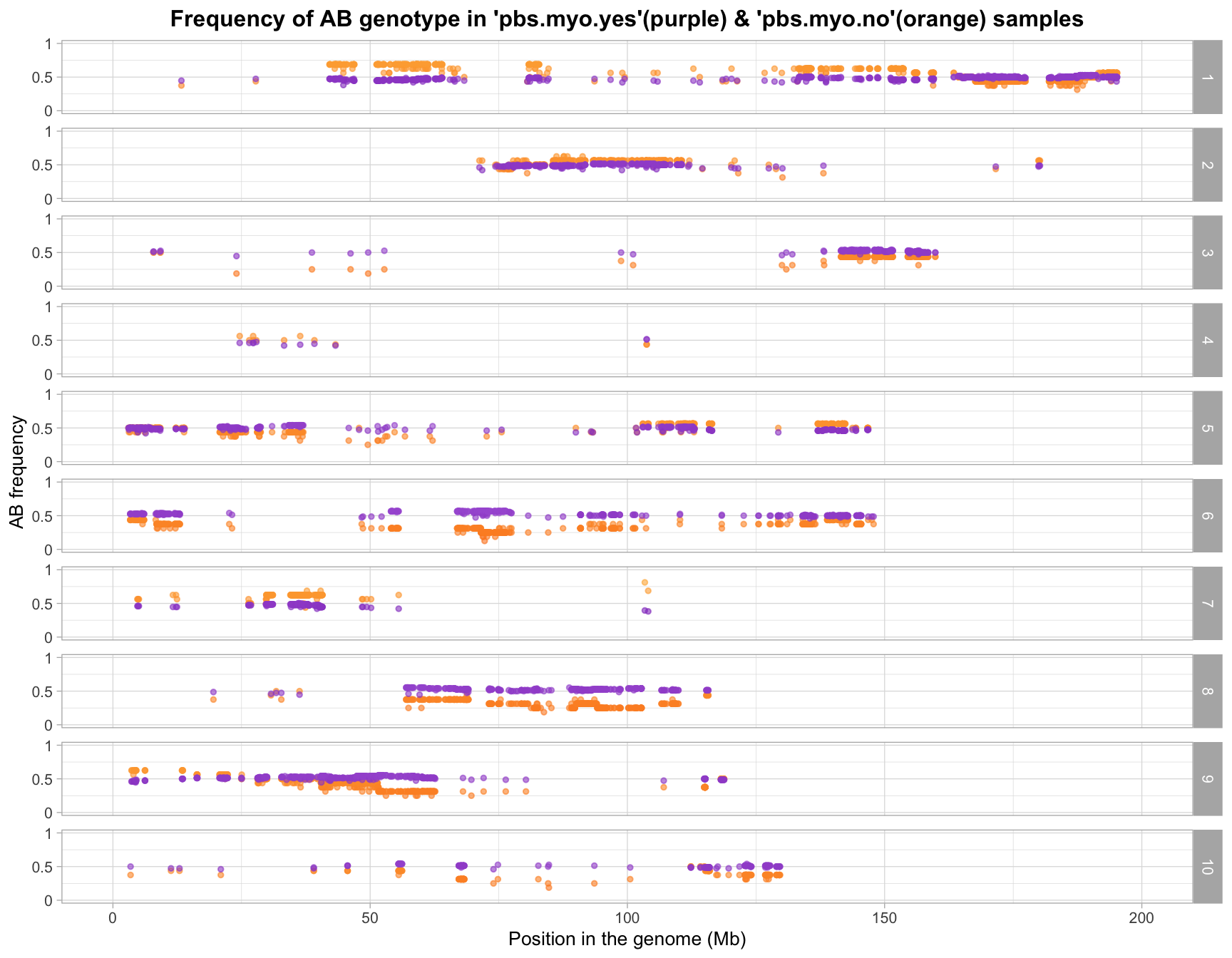
R version 4.2.2 (2022-10-31)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Big Sur ... 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ggdark_0.2.1 ggthemes_4.2.4 cowplot_1.1.1 ggnewscale_0.4.8
[5] abind_1.4-5 qtl2_0.30 reshape2_1.4.4 ggplot2_3.4.0
[9] tibble_3.1.8 psych_2.2.9 readxl_1.4.1 cluster_2.1.4
[13] dplyr_1.0.10 optparse_1.7.3 rhdf5_2.40.0 mclust_6.0.0
[17] tidyr_1.2.1 data.table_1.14.6 knitr_1.42 kableExtra_1.3.4
[21] workflowr_1.7.0
loaded via a namespace (and not attached):
[1] httr_1.4.4 sass_0.4.5 bit64_4.0.5 jsonlite_1.8.4
[5] viridisLite_0.4.1 bslib_0.4.2 assertthat_0.2.1 getPass_0.2-2
[9] highr_0.10 blob_1.2.3 cellranger_1.1.0 yaml_2.3.7
[13] pillar_1.8.1 RSQLite_2.2.19 lattice_0.20-45 glue_1.6.2
[17] digest_0.6.31 promises_1.2.0.1 rvest_1.0.3 colorspace_2.0-3
[21] htmltools_0.5.5 httpuv_1.6.6 plyr_1.8.8 pkgconfig_2.0.3
[25] purrr_0.3.5 scales_1.2.1 webshot_0.5.4 processx_3.8.0
[29] svglite_2.1.0 whisker_0.4.1 getopt_1.20.3 later_1.3.0
[33] git2r_0.30.1 farver_2.1.1 generics_0.1.3 cachem_1.0.7
[37] withr_2.5.0 cli_3.6.1 mnormt_2.1.1 magrittr_2.0.3
[41] memoise_2.0.1 evaluate_0.20 ps_1.7.2 fs_1.6.1
[45] fansi_1.0.3 nlme_3.1-161 xml2_1.3.3 tools_4.2.2
[49] lifecycle_1.0.3 stringr_1.5.0 Rhdf5lib_1.18.2 munsell_0.5.0
[53] callr_3.7.3 compiler_4.2.2 jquerylib_0.1.4 systemfonts_1.0.4
[57] rlang_1.1.0 grid_4.2.2 rhdf5filters_1.8.0 rstudioapi_0.14
[61] rmarkdown_2.21.2 gtable_0.3.1 DBI_1.1.3 R6_2.5.1
[65] bit_4.0.5 fastmap_1.1.1 utf8_1.2.2 rprojroot_2.0.3
[69] stringi_1.7.12 parallel_4.2.2 Rcpp_1.0.9 vctrs_0.6.1
[73] tidyselect_1.2.0 xfun_0.38