Overview
Teaching: 60 min
Exercises: 0 minQuestions
How to run a workflow?
Objectives
Use a series of command line tools to perform a alignment workflow
Group a series of sequential commands into a script to automate a workflow
Adapted from the materials developed by members of the teaching team at the Harvard Chan Bioinformatics Core (HBC). These are open access materials distributed under the terms of the Creative Commons Attribution license (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
- Some materials used in these lessons were derived from work that is Copyright © Data Carpentry (http://datacarpentry.org/). All Data Carpentry instructional material is made available under the Creative Commons Attribution license (CC BY 4.0). </sup>
A simple bioinformatics workflow
When working with NGS data, the raw reads you get off of the sequencer will need to pass through a number of different tools in order to generate your final desired output. The execution of this set of tools in a specified order is commonly referred to as a workflow or a pipeline.
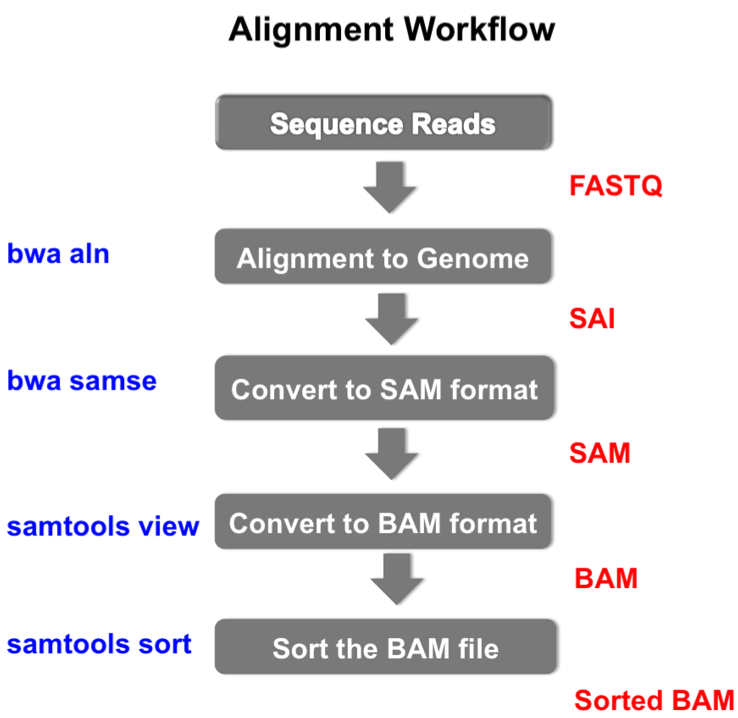
An example of the workflow we will be using for our analysis is provided below with a brief description of each step.
To get started with this lesson, make sure you are in the workflow directory first
cd ~/Desktop/data-shell/workflow
This directory contains some input data (reference genome and fastq files) and a shell script that details the series of commands used to run an alignment workflow.
workflow
├── fastqs
│ ├── SRR000011.fastq
│ ├── SRR000012.fastq
│ ├── SRR000013.fastq
│ ├── SRR000014.fastq
│ ├── SRR000015.fastq
│ └── SRR000016.fastq
└── ref_genome
└── ecoli_rel606.fasta
Without getting into the details yet, the workflow will do the following steps
- Align the reads to the reference genome
- Convert the output to the SAM format
- Convert the SAM file to BAM format
- Sort the BAM file
Let’s walk through the commands in the workflow
The first command is to change to our working directory so the script can find all the files it expects
$ cd ~/Desktop/data-shell/workflow
Assign the name/location of our reference genome to a variable ($genome)
$ genome=~/Desktop/data-shell/workflow/ref_genome/ecoli_rel606.fasta
bwa and samtools are programs that are pre-installed on our server. We need to load the module with module load
module load bwa
module load samtools
Create output paths for various intermediate and result files The -p option means mkdir will create the whole path if it does not exist (no error or message will give given if it does exist)
$ mkdir -p results/sai
$ mkdir -p results/sam
$ mkdir -p results/bam
In the script, it is a good idea to use echo for debugging/reporting to the screen
$ echo "working with file $fq"
But, wait! What about the ouput files? They are all named using the SRA ID from the original FASTQ file. We can use a unix command to extract the base name of the file (without the path and .fastq extension) and ue this for naming all of the output files. We’ll assign it to the $base variable:
This command will extract the base name of the file (without the path and .fastq extension) and assign it to the $base variable
$ base=$(basename $fq .fastq)
$ echo "base name is $base"
We will assign various file names to variables both for convenience but also to make it easier to see what is going on in the commands below.
$ sai=results/sai/$base\_aligned.sai
$ sam=results/sam/$base\_aligned.sam
$ bam=results/bam/$base\_aligned.bam
$ sorted_bam=results/bam/$base\_aligned_sorted.bam
Our data are now staged. The series of command below will run the steps of the analytical workflow
Align the reads to the reference genome
$ bwa aln $genome $fq > $sai
Convert the output to the SAM format
$ bwa samse $genome $sai $fq > $sam
Convert the SAM file to BAM format
$ samtools view -S -b $sam > $bam
Sort the BAM file
$ samtools sort -f $bam $sorted_bam
That was a lot of work, yes? But you have five more FASTQ files to go…
Remembering what commands and what parameters to type can be pretty daunting. What can you do to help yourself out in this regard? If you were to automate this process, what additional bits of information might you need? Automating this Workflow with a Bash Script
The easiest way for you to be able to repeat this process is to capture the steps that you’ve performed in a bash script. And you’ve already learned how to do this in previous lessons. So…
The shebang line
The line at the top (‘#!/bin/bash’) is commonly called the shebang line, which is a special kind of comment that tells the shell which program is to be used as the ‘intepreter’ that executes the code.
Usage and commenting
Finally, when you are writing a multi-step workflow that accepts command-line options (positional parameters), it is very important to have the usage right in the beginning of the script. In addition to the usage, it is a good practice to comment about the inputs, steps/tools and output briefly. It is easier to fill both of these in after your script is ready and you have done a test run. So even though this is at the top of the script, it may be the last few lines you add to the script.
Final Script (run_alignment.sh)
#!/bin/bash
# USAGE: sh run_workflow_call_on_file.sh <fastq file>
# This script accepts the location of a fastq file and performs the following steps on it:
## starting with alignment using bwa
## convert the .sai to SAM -> BAM -> sorted BAM
# load the modules
module load bwa samtools
# The first command is to change to our working directory
cd ~/Desktop/data-shell/workflow
# make directories to store output
mkdir -p results/sai
mkdir -p results/sam
mkdir -p results/bam
# Get input file and locations
fq=$1
# Grab base of filename for future naming
base=$(basename $fq .fastq)
echo "Starting analysis of" $base
# set up output filenames and locations
sai=results/sai/$base.aligned.sai
sam=results/sam/$base.aligned.sam
bam=results/bam/${base}.aligned.bam
sorted_bam=results/bam/$base.aligned.sorted
# location to genome reference FASTA file
genome=ref_genome/ecoli_rel606.fasta
### Alignmemt
# Align the reads to the reference genome
bwa aln $genome $fq > $sai
# Convert the output to the SAM format
bwa samse $genome $sai $fq > $sam
# Convert the SAM file to BAM format
samtools view -S -b $sam > $bam
# Sort the BAM file
samtools sort $bam -o $sorted_bam
Key Points
Automate a workflow by grouping a series of sequential commands into a script
Modify the script to grant it more flexibility