LIRICAL HTML Output
Sample information and list of differentials
The HTML output page begins with a summary of the sample name and a list of
the HPO terms
used to run the program. By default, LIRICAL shows a detailed output only for the top 10 differential
diagnoses (or more if more diagnoses have a posterior probability above the default threshold of 1%).
The minimum number of differential diagnoses to show can be changed with the -m option, and
the probability threshold can be changed with the -t option.
Disease evaluations
LIRICAL evaluates each of the diseases in the HPO database and estimates the probability that a disease explains the observed phenotypic abnormalities (and if applicable, the observed variants).
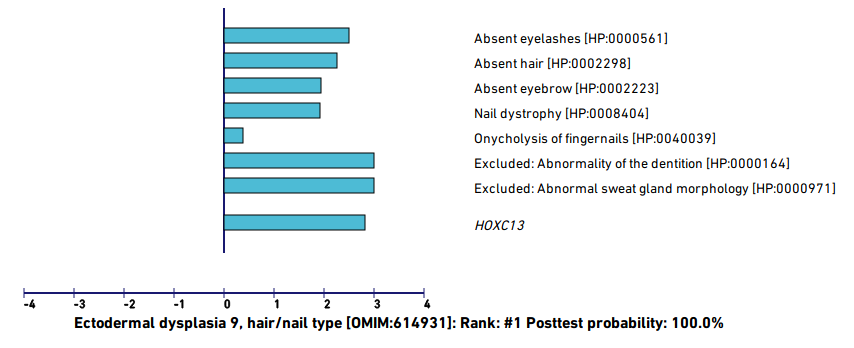
For example, the following figure shows the evaluation of a simulated case based on a published case report of an individual with pure hair and nail ectodermal dysplasia (ECTD9) related to a pathogenic variant in the HOXC13 gene (Khan et al., 2017).
LIRICAL has estimate the composite likelihood ratio score at 8.951 (note that this is expressed on a log10 scale, so that the likelihood ratio is actually 108.951). The posttest probability is close to 100%.
The contribution of each of the HPO terms entered for the proband is shown. The contribution of each term is indicated by the length of the blue bar (which shows the decadic logarithm of the likelihood ratio for the term. For instance, if the bar is 2 units long, then the likelihood ratio is 10<sup>2</sup>=100).
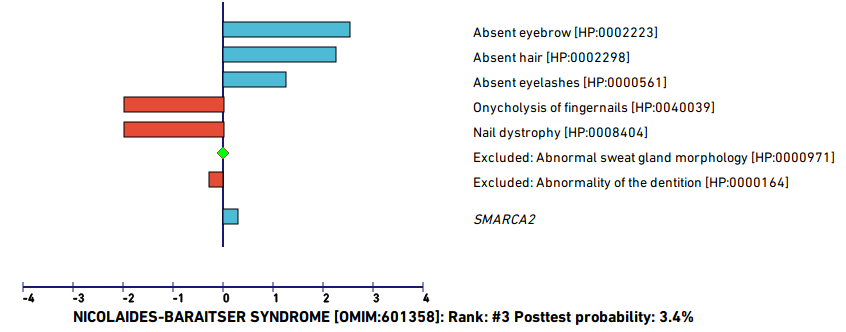
The user should inspect the top differentials. In this case, the posterior probability of the remaining differentials drops off quickly. For instance, the third best hit, Nicolaides Baraitser syndrome, has a posttest probability of only 3.6% and several of the observed phenotypes are not characteristic of this syndrome and thus reduce the match score (indicated as red bars).
Other information
LIRICAL shows a list of candidate diseases and genes with a low post-test probability in the section
Genes/Diseases with low posttest probability (Click on the Table to show details).
In some cases, we have observed that some variants are linked to gene entities that do not have an NCBI Gene ID. This
effectively means that LIRICAL will not further analyze these variants. If any such variants are found, they will
be shown in a section called Gene symbols that could not be annotated. If the section is not present, then
all variants were annotated. In practice, the gene symbols that cannot be linked to an NCBI ID represent accession numbers
that are not confirmed genes, e.g., CR627135, AX746851, AK096159, …. We have found that using refseq as the transcript option
means that all genes also have a Gene ID, but UCSC includes transcripts that do not have a Gene ID, and so users can
experiment with both options. This information is provided for completeness’ sake, but it is not diagnostically important.