Building the regulatory exome
GOPHER can generate a BED file with target definitions designed to generate a gene-panel with exons and regulatory features that can be used in conjunction with the capture Hi-C probe set. The regulatory regions are derived from the Ensembl regulatory build: https://useast.ensembl.org/info/genome/funcgen/regulatory_build.html.
After creating a capture Hi-C panel (set of viewpoints), users can export a BED file with definitions for a matching “regulatory exome”.
The regulatory exome feature is currently implemented for hg19 and hg38.
First, download the regulatory build file (Export > Download regulation data).
Following this, click on Export > Build regulatory exom and choose a location for GOPHER to export the regulatory exome BED file.
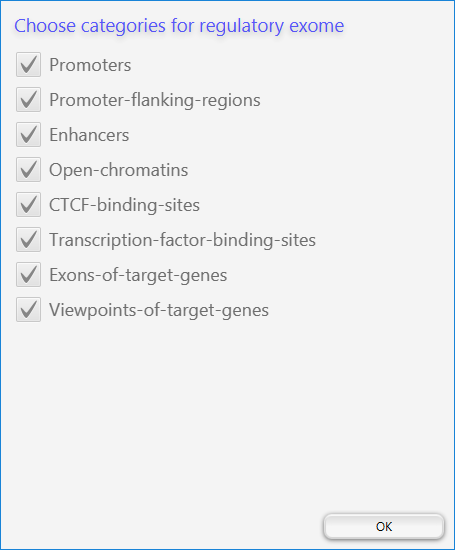
After you do this, a window will appear that allows you to choose the categories of regulatory element you would like to include.
By default, all categories are included. When you click OK, GOPHER will determine the locations for the regulatory exome and export the BED file.
GOPHER’s regulatory exome export function identifies the coding exons and regulatory regions for all target transcripts. It identifies exact duplicates (e.g., the same exon occuring in multiple transcripts) and removes them automatically so that just one copy of each duplicated sequence is present in the output file. In some cases, overlapping segments may be present in the output however. For instance, the following two segments overlap.
chr20 50170444 50170801 ENSR00000186386[OPEN_CHROMATIN]
chr20 50169801 50170800 ENSR00000388510[CTCF_BINDING_SITE]
Users may want to remove such overlaps from the BED file prior to ordering enrichment probes.
One easy way of doing this is with BEDtools.
Assuming that the output file was called regulatoryExomePanel.bed, then the following command will merge the overlapping regions.
$ bedtools merge -c 4 -o collapse -i regulatoryExomePanel.bed > mergedRegulatoryExomePanel.bed
The two lines shown above are now merged into a single line.
chr20 50169801 50170801 ENSR00000388510[CTCF_BINDING_SITE],ENSR00000186386[OPEN_CHROMATIN]