Custom masks for sampling specific regions from images with ZarrDataset
import zarrdataset as zds
import zarr
# These are images from the Image Data Resource (IDR)
# https://idr.openmicroscopy.org/ that are publicly available and were
# converted to the OME-NGFF (Zarr) format by the OME group. More examples
# can be found at Public OME-Zarr data (Nov. 2020)
# https://www.openmicroscopy.org/2020/11/04/zarr-data.html
filenames = ["https://uk1s3.embassy.ebi.ac.uk/idr/zarr/v0.4/idr0073A/9798462.zarr"]
import random
import numpy as np
# For reproducibility
np.random.seed(478963)
random.seed(478965)
z_img = zarr.open(filenames[0], mode="r")
z_img["0"].info
| Name | /0 |
|---|---|
| Type | zarr.core.Array |
| Data type | uint8 |
| Shape | (1, 3, 1, 16433, 21115) |
| Chunk shape | (1, 1, 1, 1024, 1024) |
| Order | C |
| Read-only | True |
| Compressor | Blosc(cname='lz4', clevel=5, shuffle=SHUFFLE, blocksize=0) |
| Store type | zarr.storage.FSStore |
| No. bytes | 1040948385 (992.7M) |
| Chunks initialized | 0/1071 |
import numpy as np
import matplotlib.pyplot as plt
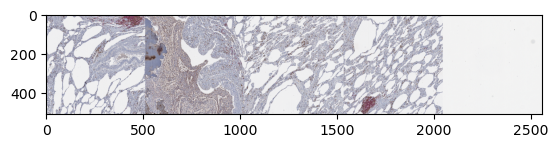
plt.imshow(np.moveaxis(z_img["4"][0, :, 0], 0, -1))
plt.show()
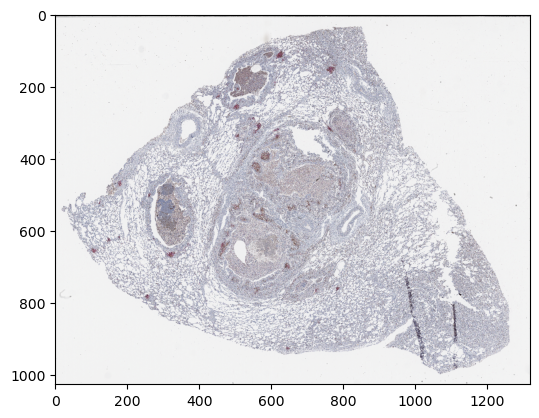
Define a mask from where patches can be extracted
from skimage import color, filters, morphology
im_gray = color.rgb2gray(z_img["4"][0, :, 0], channel_axis=0)
thresh = filters.threshold_otsu(im_gray)
mask = im_gray > thresh
mask = morphology.remove_small_objects(mask == 0, min_size=16 ** 2,
connectivity=2)
mask = morphology.remove_small_holes(mask, area_threshold=128)
mask = morphology.binary_erosion(mask, morphology.disk(8))
mask = morphology.binary_dilation(mask, morphology.disk(8))
plt.imshow(mask)
plt.show()
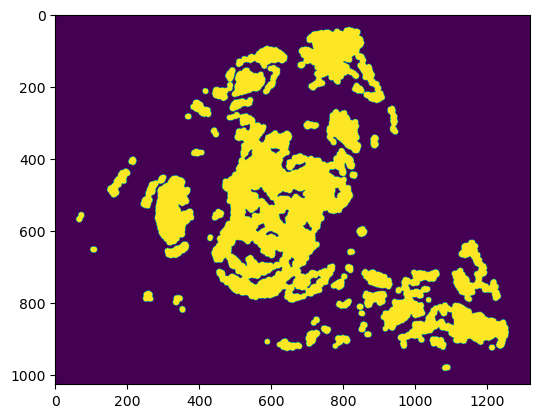
plt.imshow(np.moveaxis(z_img["4"][0, :, 0], 0, -1))
plt.imshow(mask, cmap="gray", alpha=1.0*(mask < 1))
plt.show()
Extract patches of size 512x512 pixels from a Whole Slide Image (WSI)
Sample the image uniformly in a squared grid pattern
patch_size = dict(Y=512, X=512)
patch_sampler = zds.PatchSampler(patch_size=patch_size)
Use the ZarrDataset class to enable extraction of samples from masked regions.
An extra dimension is added to the mask, so it matches the number of spatial axes in the image
image_specs = zds.ImagesDatasetSpecs(
filenames=filenames,
data_group="1",
source_axes="TCZYX",
)
# Use the MasksDatasetSpecs to add the specifications of the masks.
#masks_specs = zds.MasksDatasetSpecs(
masks_specs = zds.LabelsDatasetSpecs(
filenames=[mask],
source_axes="YX",
axes="ZYX",
modality="masks",
)
my_dataset = zds.ZarrDataset([image_specs, masks_specs],
patch_sampler=patch_sampler)
ds_iterator = iter(my_dataset)
sample = next(ds_iterator)
type(sample[0]), sample[0].shape, sample[0].dtype
type(sample[1]), sample[1].shape, sample[1].dtype
(numpy.ndarray, (1, 64, 65), dtype('bool'))
plt.imshow(np.moveaxis(sample[0][0, :, 0], 0, -1))
plt.show()
plt.imshow(sample[1][0])
plt.show()
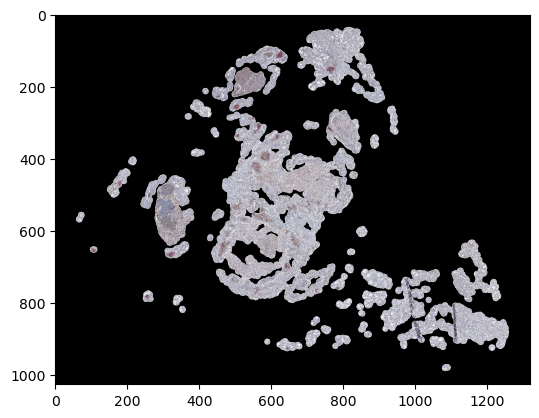
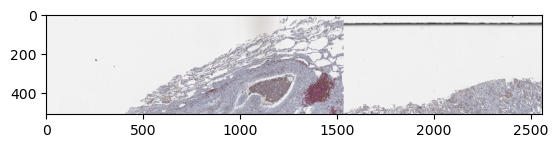
samples = []
labels = []
for i, sample in enumerate(my_dataset):
samples.append(np.moveaxis(sample[0][0, :, 0], 0, -1))
labels.append(sample[1][0])
if i >= 4:
# Take only five samples for illustration purposes
break
samples = np.hstack(samples)
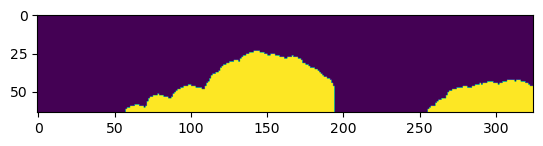
labels = np.hstack(labels)
plt.imshow(samples)
plt.show()
plt.imshow(labels)
plt.show()
Use a function to generate the masks for each image in the dataset
Get only patches that are covered by at least 1/16th of their area by the mask
patch_size = dict(Y=512, X=512)
patch_sampler = zds.PatchSampler(patch_size=patch_size, min_area=1/16)
Apply WSITissueMaskGenerator transform to each image in the dataset to define each sampling mask
mask_func = zds.WSITissueMaskGenerator(mask_scale=1,
min_size=16,
area_threshold=128,
axes="ZYX")
Because the input image (zarr group “1”) is large, computing the mask directly on that could require high computational resources.
For that reason, use a donwsampled version of that image instead by pointing mask_data_group="4" to use a 1:16 downsampled version of the input image.
The mask_axes should match the ones that WSITissueMaskGenerator requies as input (“YXC”). To do that, a ROI can be specified to take just the spatial and channel axes from the input image with mask_roi="(0,0,0,0,0):(1,-1,1,-1,-1)", and rearrange the output axes with mask_axes="YXC".
image_specs = zds.ImagesDatasetSpecs(
filenames=filenames,
data_group="1",
source_axes="TCZYX",
)
# Use the MasksDatasetSpecs to add the specifications of the masks.
# The mask generation function is added as `image_loader_func` parameter of the dataset specification for masks.
masks_specs = zds.MasksDatasetSpecs(
filenames=filenames,
data_group="4",
source_axes="TCZYX",
axes="YXC",
roi="(0,0,0,0,0):(1,-1,1,-1,-1)",
image_loader_func=mask_func,
)
my_dataset = zds.ZarrDataset([image_specs, masks_specs],
patch_sampler=patch_sampler,
shuffle=True)
samples = []
for i, sample in enumerate(my_dataset):
samples.append(np.moveaxis(sample[0, :, 0], 0, -1))
if i >= 4:
# Take only five samples for illustration purposes
break
samples = np.hstack(samples)
plt.imshow(samples)
plt.show()