Loading patches/windows from masked regions of images with ZarrDataset
Import the “zarrdataset” package
import zarrdataset as zds
import zarr
Load data stored on S3 storage
# These are images from the Image Data Resource (IDR)
# https://idr.openmicroscopy.org/ that are publicly available and were
# converted to the OME-NGFF (Zarr) format by the OME group. More examples
# can be found at Public OME-Zarr data (Nov. 2020)
# https://www.openmicroscopy.org/2020/11/04/zarr-data.html
filenames = ["https://uk1s3.embassy.ebi.ac.uk/idr/zarr/v0.4/idr0073A/9798462.zarr"]
import random
import numpy as np
# For reproducibility
np.random.seed(478963)
random.seed(478965)
z_img = zarr.open(filenames[0], mode="r")
z_img["0"].info
| Name | /0 |
|---|---|
| Type | zarr.core.Array |
| Data type | uint8 |
| Shape | (1, 3, 1, 16433, 21115) |
| Chunk shape | (1, 1, 1, 1024, 1024) |
| Order | C |
| Read-only | True |
| Compressor | Blosc(cname='lz4', clevel=5, shuffle=SHUFFLE, blocksize=0) |
| Store type | zarr.storage.FSStore |
| No. bytes | 1040948385 (992.7M) |
| Chunks initialized | 0/1071 |
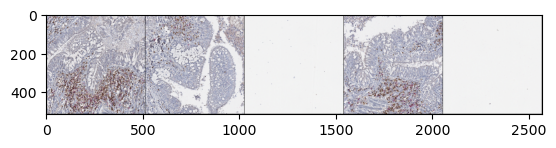
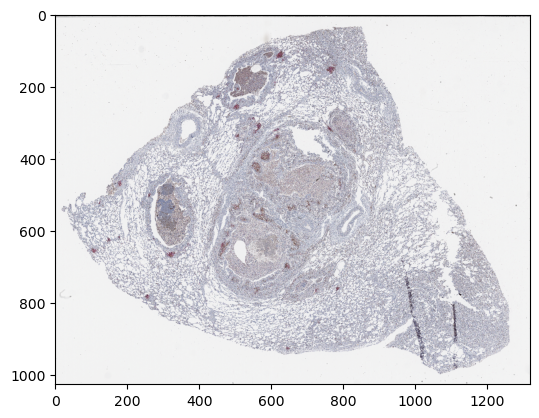
import matplotlib.pyplot as plt
plt.imshow(np.moveaxis(z_img["4"][0, :, 0], 0, -1))
plt.show()
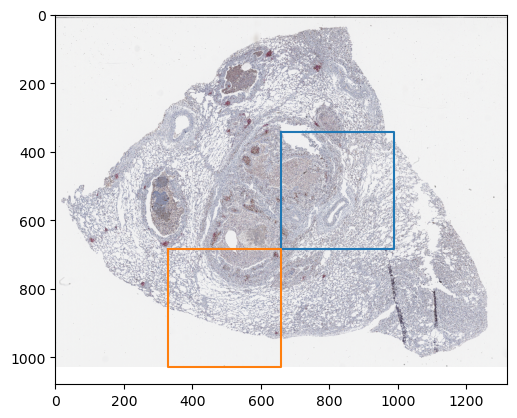
Define a mask from where patches will be extracted
mask = np.array([
[0, 0, 0, 0],
[0, 0, 1, 0],
[0, 1, 0, 0],
], dtype=bool)
ZarrDataset will match the size of the mask t the size of the image that is being sampled.
For that reason, it is not necessary for the mask to be of the same size of the image.
_, d, _, h, w = z_img["4"].shape
m_h, m_w = mask.shape
factor_h = h / m_h
factor_w = w / m_w
plt.imshow(np.moveaxis(z_img["4"][0, :, 0], 0, -1))
sampling_region = np.array([
[0, 0],
[0, factor_w],
[factor_h, factor_w],
[factor_h, 0],
[0, 0]
])
for m_y, m_x in zip(*np.nonzero(mask)):
offset_y = m_y * factor_h
offset_x = m_x * factor_w
plt.plot(sampling_region[:, 1] + offset_x,
sampling_region[:, 0] + offset_y)
plt.show()
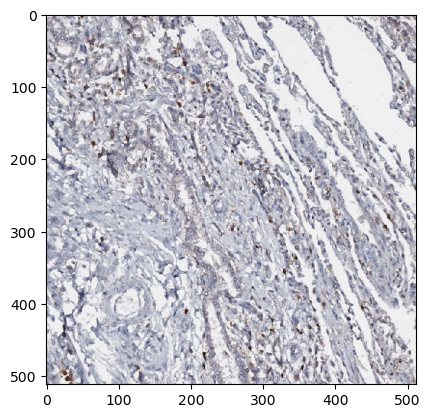
Extract patches of size 512x512 pixels from masked regiosn of a Whole Slide Image (WSI)
Sample the image uniformly in a squared grid pattern using a PatchSampler
patch_size = dict(Y=512, X=512)
patch_sampler = zds.PatchSampler(patch_size=patch_size)
Use the ZarrDataset class to enable extraction of samples from masked regions by specifying two modalities: images, and masks.
Enable sampling patched from random locations with shuffle=True
image_specs = zds.ImagesDatasetSpecs(
filenames=filenames,
data_group="0",
source_axes="TCZYX",
)
# Use the MasksDatasetSpecs to add the specifications of the masks.
# Filenames can receive different types of variables, in this case a list with a single mask for the only image in image_specs.
masks_specs = zds.MasksDatasetSpecs(
filenames=[mask],
source_axes="YX",
)
my_dataset = zds.ZarrDataset([image_specs, masks_specs],
patch_sampler=patch_sampler,
draw_same_chunk=False,
shuffle=True)
ds_iterator = iter(my_dataset)
sample = next(ds_iterator)
type(sample), sample.shape, sample.dtype
(numpy.ndarray, (1, 3, 1, 512, 512), dtype('uint8'))
plt.imshow(np.moveaxis(sample[0, :, 0], 0, -1))
plt.show()
samples = []
for i, sample in enumerate(my_dataset):
samples.append(np.pad(np.moveaxis(sample[0, :, 0], 0, -1),((1, 1), (1, 1), (0, 0))))
# Obtain only 5 samples
if i >= 4:
break
grid_samples = np.hstack(samples)
plt.imshow(grid_samples)
plt.show()