Basic ZarrDataset usage example
Import the “zarrdataset” package
import zarrdataset as zds
import zarr
Load data stored on S3 storage
# These are images from the Image Data Resource (IDR)
# https://idr.openmicroscopy.org/ that are publicly available and were
# converted to the OME-NGFF (Zarr) format by the OME group. More examples
# can be found at Public OME-Zarr data (Nov. 2020)
# https://www.openmicroscopy.org/2020/11/04/zarr-data.html
filenames = ["https://uk1s3.embassy.ebi.ac.uk/idr/zarr/v0.4/idr0073A/9798462.zarr"]
import random
import numpy as np
# For reproducibility
np.random.seed(478963)
random.seed(478965)
Inspect the image to sample
z_img = zarr.open(filenames[0], mode="r")
z_img["0"].info
| Name | /0 |
|---|---|
| Type | zarr.core.Array |
| Data type | uint8 |
| Shape | (1, 3, 1, 16433, 21115) |
| Chunk shape | (1, 1, 1, 1024, 1024) |
| Order | C |
| Read-only | True |
| Compressor | Blosc(cname='lz4', clevel=5, shuffle=SHUFFLE, blocksize=0) |
| Store type | zarr.storage.FSStore |
| No. bytes | 1040948385 (992.7M) |
| Chunks initialized | 0/1071 |
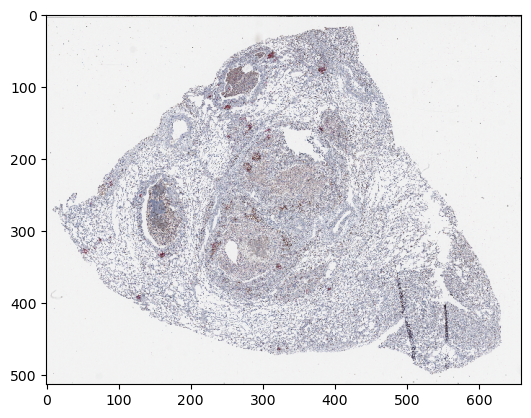
Display a downsampled version of the image
import matplotlib.pyplot as plt
plt.imshow(np.moveaxis(z_img["5"][0, :, 0], 0, -1))
plt.show()
Retrieving whole images
Create a ZarrDataset to handle the image dataset instead of opening all the dataset images by separate and hold them in memory until they are not used anymore.
my_dataset = zds.ZarrDataset()
Start by retrieving whole images, from a subsampled (pyramid) group (e.g. group 6) within the zarr image file, instead the full resolution image at group “0”. The source array axes should be specified in order to handle images properly, in this case Time-Channel-Depth-Height-Width (TCZYX).
my_dataset.add_modality(
modality="image",
filenames=filenames,
source_axes="TCZYX",
data_group="6"
)
The ZarrDataset class can be used as a Python’s generator, and can be accessed by iter and subsequently next operations.
ds_iterator = iter(my_dataset)
ds_iterator
<generator object ZarrDataset.__iter__ at 0x155501416c80>
sample = next(ds_iterator)
print(type(sample), sample.shape)
<class 'numpy.ndarray'> (1, 3, 1, 256, 329)
Compare the shape of the retreived sample with the shape of the original image in group “6”
z_img["6"].info
| Name | /6 |
|---|---|
| Type | zarr.core.Array |
| Data type | uint8 |
| Shape | (1, 3, 1, 256, 329) |
| Chunk shape | (1, 1, 1, 256, 329) |
| Order | C |
| Read-only | True |
| Compressor | Blosc(cname='lz4', clevel=5, shuffle=SHUFFLE, blocksize=0) |
| Store type | zarr.storage.FSStore |
| No. bytes | 252672 (246.8K) |
| Chunks initialized | 0/3 |
Extracting patches of size 512x512 pixels from a Whole Slide Image (WSI)
The PatchSampler class can be used along with ZarrDataset to retrieve patches from WSIs without having to tiling them in a pre-process step.
patch_size = dict(Y=512, X=512)
patch_sampler = zds.PatchSampler(patch_size=patch_size)
patch_sampler
<class 'zarrdataset._samplers.PatchSampler'> for sampling patches of size {'Z': 1, 'Y': 512, 'X': 512}.
Create a new dataset using the ZarrDataset class, and pass the PatchSampler as patch_sampler argument.
Because patches are being exracted instead of whole images, the full resolution image at group “0” can be used as input.
my_dataset = zds.ZarrDataset(patch_sampler=patch_sampler)
my_dataset.add_modality(
modality="image",
filenames=filenames,
source_axes="TCZYX",
data_group="0"
)
my_dataset
ZarrDataset (PyTorch support:True, tqdm support :False)
Modalities: image
Transforms order: []
Using image modality as reference.
Using <class 'zarrdataset._samplers.PatchSampler'> for sampling patches of size {'Z': 1, 'Y': 512, 'X': 512}.
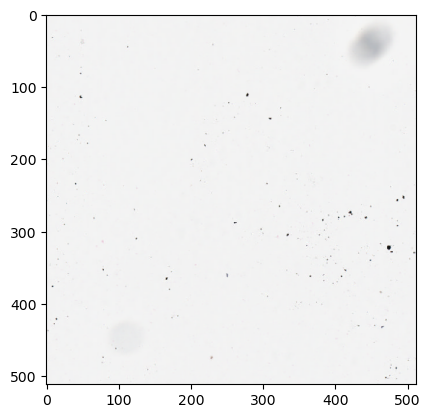
Create a generator from the dataset object and extract some patches
ds_iterator = iter(my_dataset)
sample = next(ds_iterator)
type(sample), sample.shape, sample.dtype
sample = next(ds_iterator)
type(sample), sample.shape, sample.dtype
sample = next(ds_iterator)
type(sample), sample.shape, sample.dtype
(numpy.ndarray, (1, 3, 1, 512, 512), dtype('uint8'))
plt.imshow(np.moveaxis(sample[0, :, 0], 0, -1))
plt.show()
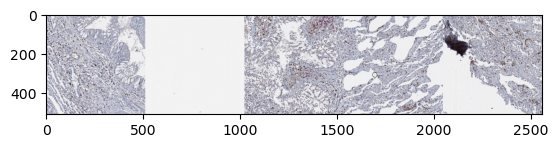
Using ZarrDataset in a for loop
ZarrDatasets can be used as generators, for example in for loops
samples = []
for i, sample in enumerate(my_dataset):
samples.append(np.moveaxis(sample[0, :, 0], 0, -1))
if i >= 4:
# Take only five samples for illustration purposes
break
samples_stack = np.hstack(samples)
plt.imshow(samples_stack)
plt.show()
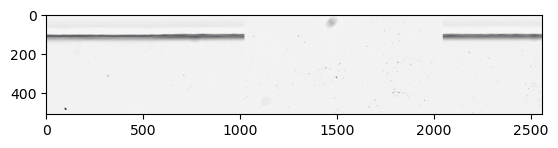
Create a ZarrDataset with all the dataset specifications.
Use a dictionary (or a list of them for multiple modalities) to define the dataset specifications. Alternatively, use a list of DatasetSpecs (or derived classes) to define the dataset specifications that ZarrDataset requires.
For example, ImagesDatasetSpecs can be used to define an image data modality. Other pre-defined modalities are LabelsDatasetSpecs for labels, and MaskDatasetSpecs for masks.
image_specs = zds.ImagesDatasetSpecs(
filenames=filenames,
data_group="0",
source_axes="TCZYX",
)
Also, try sampling patches from random locations by setting shuffle=True.
my_dataset = zds.ZarrDataset(dataset_specs=[image_specs],
patch_sampler=patch_sampler,
shuffle=True)
samples = []
for i, sample in enumerate(my_dataset):
samples.append(np.moveaxis(sample[0, :, 0], 0, -1))
if i >= 4:
# Take only five samples for illustration purposes
break
samples_stack = np.hstack(samples)
plt.imshow(samples_stack)
plt.show()