DO_morphine_gemma
Last updated: 2020-11-19
Checks: 6 1
Knit directory: DO_Opioid/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200504) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /projects/heh/DO_Opioid/data/combined/combined_batch | data/combined/combined_batch |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 622975f. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .RData
Ignored: analysis/Picture1.png
Ignored: data/output/
Untracked files:
Untracked: analysis/DDO_morphine1_second_set_69k.stdout
Untracked: analysis/DO_morphine1.R
Untracked: analysis/DO_morphine1.Rout
Untracked: analysis/DO_morphine1.sh
Untracked: analysis/DO_morphine1.stderr
Untracked: analysis/DO_morphine1.stdout
Untracked: analysis/DO_morphine1_SNP.R
Untracked: analysis/DO_morphine1_SNP.Rout
Untracked: analysis/DO_morphine1_SNP.sh
Untracked: analysis/DO_morphine1_SNP.stderr
Untracked: analysis/DO_morphine1_SNP.stdout
Untracked: analysis/DO_morphine1_combined.R
Untracked: analysis/DO_morphine1_combined.Rout
Untracked: analysis/DO_morphine1_combined.sh
Untracked: analysis/DO_morphine1_combined.stderr
Untracked: analysis/DO_morphine1_combined.stdout
Untracked: analysis/DO_morphine1_combined_69k.R
Untracked: analysis/DO_morphine1_combined_69k.Rout
Untracked: analysis/DO_morphine1_combined_69k.sh
Untracked: analysis/DO_morphine1_combined_69k.stderr
Untracked: analysis/DO_morphine1_combined_69k.stdout
Untracked: analysis/DO_morphine1_combined_69k_m2.R
Untracked: analysis/DO_morphine1_combined_69k_m2.Rout
Untracked: analysis/DO_morphine1_combined_69k_m2.sh
Untracked: analysis/DO_morphine1_combined_69k_m2.stderr
Untracked: analysis/DO_morphine1_combined_69k_m2.stdout
Untracked: analysis/DO_morphine1_combined_weight_DOB.R
Untracked: analysis/DO_morphine1_combined_weight_DOB.Rout
Untracked: analysis/DO_morphine1_combined_weight_DOB.err
Untracked: analysis/DO_morphine1_combined_weight_DOB.out
Untracked: analysis/DO_morphine1_combined_weight_DOB.sh
Untracked: analysis/DO_morphine1_combined_weight_DOB.stderr
Untracked: analysis/DO_morphine1_combined_weight_DOB.stdout
Untracked: analysis/DO_morphine1_combined_weight_age.R
Untracked: analysis/DO_morphine1_combined_weight_age.err
Untracked: analysis/DO_morphine1_combined_weight_age.out
Untracked: analysis/DO_morphine1_combined_weight_age.sh
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.R
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.err
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.out
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.sh
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.R
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.err
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.out
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.sh
Untracked: analysis/DO_morphine1_cph.R
Untracked: analysis/DO_morphine1_cph.Rout
Untracked: analysis/DO_morphine1_cph.sh
Untracked: analysis/DO_morphine1_second_set.R
Untracked: analysis/DO_morphine1_second_set.Rout
Untracked: analysis/DO_morphine1_second_set.sh
Untracked: analysis/DO_morphine1_second_set.stderr
Untracked: analysis/DO_morphine1_second_set.stdout
Untracked: analysis/DO_morphine1_second_set_69k.R
Untracked: analysis/DO_morphine1_second_set_69k.Rout
Untracked: analysis/DO_morphine1_second_set_69k.sh
Untracked: analysis/DO_morphine1_second_set_69k.stderr
Untracked: analysis/DO_morphine1_second_set_SNP.R
Untracked: analysis/DO_morphine1_second_set_SNP.Rout
Untracked: analysis/DO_morphine1_second_set_SNP.sh
Untracked: analysis/DO_morphine1_second_set_SNP.stderr
Untracked: analysis/DO_morphine1_second_set_SNP.stdout
Untracked: analysis/DO_morphine1_second_set_weight_DOB.R
Untracked: analysis/DO_morphine1_second_set_weight_DOB.Rout
Untracked: analysis/DO_morphine1_second_set_weight_DOB.err
Untracked: analysis/DO_morphine1_second_set_weight_DOB.out
Untracked: analysis/DO_morphine1_second_set_weight_DOB.sh
Untracked: analysis/DO_morphine1_second_set_weight_DOB.stderr
Untracked: analysis/DO_morphine1_second_set_weight_DOB.stdout
Untracked: analysis/DO_morphine1_second_set_weight_age.R
Untracked: analysis/DO_morphine1_second_set_weight_age.Rout
Untracked: analysis/DO_morphine1_second_set_weight_age.err
Untracked: analysis/DO_morphine1_second_set_weight_age.out
Untracked: analysis/DO_morphine1_second_set_weight_age.sh
Untracked: analysis/DO_morphine1_second_set_weight_age.stderr
Untracked: analysis/DO_morphine1_second_set_weight_age.stdout
Untracked: analysis/DO_morphine1_weight_DOB.R
Untracked: analysis/DO_morphine1_weight_DOB.sh
Untracked: analysis/DO_morphine1_weight_age.R
Untracked: analysis/DO_morphine1_weight_age.sh
Untracked: analysis/DO_morphine_gemma.R
Untracked: analysis/DO_morphine_gemma.err
Untracked: analysis/DO_morphine_gemma.out
Untracked: analysis/DO_morphine_gemma.sh
Untracked: analysis/DO_morphine_gemma_firstmin.R
Untracked: analysis/DO_morphine_gemma_firstmin.err
Untracked: analysis/DO_morphine_gemma_firstmin.out
Untracked: analysis/DO_morphine_gemma_firstmin.sh
Untracked: analysis/DO_morphine_gemma_withpermu.R
Untracked: analysis/DO_morphine_gemma_withpermu.err
Untracked: analysis/DO_morphine_gemma_withpermu.out
Untracked: analysis/DO_morphine_gemma_withpermu.sh
Untracked: analysis/Plot_DO_morphine1_SNP.R
Untracked: analysis/Plot_DO_morphine1_SNP.Rout
Untracked: analysis/Plot_DO_morphine1_SNP.sh
Untracked: analysis/Plot_DO_morphine1_SNP.stderr
Untracked: analysis/Plot_DO_morphine1_SNP.stdout
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.R
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.Rout
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.sh
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.stderr
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.stdout
Untracked: analysis/workflow_proc.R
Untracked: analysis/workflow_proc.sh
Untracked: analysis/workflow_proc.stderr
Untracked: analysis/workflow_proc.stdout
Untracked: code/cfw/
Untracked: code/gemma_plot.R
Untracked: data/69k_grid_pgmap.RData
Untracked: data/GM_covar.csv
Untracked: data/GM_covar_07092018_morphine.csv
Untracked: data/MasterMorphine Second Set DO w DOB2.xlsx
Untracked: data/MasterMorphine Second Set DO.xlsx
Untracked: data/cc_variants.sqlite
Untracked: data/combined/
Untracked: data/first/
Untracked: data/founder_geno.csv
Untracked: data/genetic_map.csv
Untracked: data/gm.json
Untracked: data/gwas.sh
Untracked: data/marker_grid_0.02cM_plus.txt
Untracked: data/mouse_genes_mgi.sqlite
Untracked: data/pheno.csv
Untracked: data/pheno_qtl2.csv
Untracked: data/pheno_qtl2_07092018_morphine.csv
Untracked: data/pheno_qtl2_w_dob.csv
Untracked: data/physical_map.csv
Untracked: data/sample_geno.csv
Untracked: data/second/
Untracked: output/DO_morphine_Min.depression.png
Untracked: output/DO_morphine_Min.depression22222_violin_chr5.pdf
Untracked: output/DO_morphine_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_Min.depression_coefplot_blup_chr5.png
Untracked: output/DO_morphine_Min.depression_coefplot_blup_chrX.png
Untracked: output/DO_morphine_Min.depression_coefplot_chr5.png
Untracked: output/DO_morphine_Min.depression_coefplot_chrX.png
Untracked: output/DO_morphine_Min.depression_peak_genes_chr5.png
Untracked: output/DO_morphine_Min.depression_violin_chr5.png
Untracked: output/DO_morphine_Recovery.Time.png
Untracked: output/DO_morphine_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr11.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr4.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr7.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr9.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr11.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr4.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr7.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr9.png
Untracked: output/DO_morphine_Status_bin.png
Untracked: output/DO_morphine_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_Survival.Time.png
Untracked: output/DO_morphine_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_Survival.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_Survival.Time_coefplot_blup_chr17.png
Untracked: output/DO_morphine_Survival.Time_coefplot_blup_chr8.png
Untracked: output/DO_morphine_Survival.Time_coefplot_chr17.png
Untracked: output/DO_morphine_Survival.Time_coefplot_chr8.png
Untracked: output/DO_morphine_combine_batch_peak_violin.pdf
Untracked: output/DO_morphine_combined_69k_m2_Min.depression.png
Untracked: output/DO_morphine_combined_69k_m2_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_combined_69k_m2_Recovery.Time.png
Untracked: output/DO_morphine_combined_69k_m2_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_combined_69k_m2_Status_bin.png
Untracked: output/DO_morphine_combined_69k_m2_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_combined_69k_m2_Survival.Time.png
Untracked: output/DO_morphine_combined_69k_m2_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Survival.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_coxph_24hrs_kinship_QTL.png
Untracked: output/DO_morphine_cphout.RData
Untracked: output/DO_morphine_first_batch_peak_in_second_batch_violin.pdf
Untracked: output/DO_morphine_first_batch_peak_in_second_batch_violin_sidebyside.pdf
Untracked: output/DO_morphine_first_batch_peak_violin.pdf
Untracked: output/DO_morphine_operm.cph.RData
Untracked: output/DO_morphine_second_batch_on_first_batch_peak_violin.pdf
Untracked: output/DO_morphine_second_batch_peak_ch6surv_on_first_batchviolin.pdf
Untracked: output/DO_morphine_second_batch_peak_ch6surv_on_first_batchviolin2.pdf
Untracked: output/DO_morphine_second_batch_peak_in_first_batch_violin.pdf
Untracked: output/DO_morphine_second_batch_peak_in_first_batch_violin_sidebyside.pdf
Untracked: output/DO_morphine_second_batch_peak_violin.pdf
Untracked: output/DO_morphine_secondbatch_69k_Min.depression.png
Untracked: output/DO_morphine_secondbatch_69k_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_69k_Recovery.Time.png
Untracked: output/DO_morphine_secondbatch_69k_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_69k_Status_bin.png
Untracked: output/DO_morphine_secondbatch_69k_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_69k_Survival.Time.png
Untracked: output/DO_morphine_secondbatch_69k_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Survival.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Min.depression.png
Untracked: output/DO_morphine_secondbatch_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Recovery.Time.png
Untracked: output/DO_morphine_secondbatch_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Status_bin.png
Untracked: output/DO_morphine_secondbatch_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Survival.Time.png
Untracked: output/DO_morphine_secondbatch_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Survival.Time_coefplot_blup.pdf
Untracked: output/apr_69kchr_combined.RData
Untracked: output/apr_69kchr_k_loco_combined.rds
Untracked: output/apr_69kchr_second_set.RData
Untracked: output/combine_batch_variation.RData
Untracked: output/combined_gm.RData
Untracked: output/combined_gm.k_loco.rds
Untracked: output/combined_gm.k_overall.rds
Untracked: output/combined_gm.probs_36state.rds
Untracked: output/combined_gm.probs_8state.rds
Untracked: output/coxph/
Untracked: output/do.morphine.RData
Untracked: output/do.morphine.k_loco.rds
Untracked: output/do.morphine.probs_36state.rds
Untracked: output/do.morphine.probs_8state.rds
Untracked: output/first_batch_variation.RData
Untracked: output/old_temp/
Untracked: output/pr_69k_combined.RData
Untracked: output/pr_69kchr_combined.RData
Untracked: output/pr_69kchr_second_set.RData
Untracked: output/qtl.morphine.69k.out.combined.RData
Untracked: output/qtl.morphine.69k.out.combined_m2.RData
Untracked: output/qtl.morphine.69k.out.second_set.RData
Untracked: output/qtl.morphine.operm.RData
Untracked: output/qtl.morphine.out.RData
Untracked: output/qtl.morphine.out.combined_gm.RData
Untracked: output/qtl.morphine.out.combined_weight_DOB.RData
Untracked: output/qtl.morphine.out.combined_weight_age.RData
Untracked: output/qtl.morphine.out.second_set.RData
Untracked: output/qtl.morphine.out.second_set.weight_DOB.RData
Untracked: output/qtl.morphine.out.second_set.weight_age.RData
Untracked: output/qtl.morphine.out.weight_DOB.RData
Untracked: output/qtl.morphine.out.weight_age.RData
Untracked: output/qtl.morphine1.snpout.RData
Untracked: output/qtl.morphine2.snpout.RData
Untracked: output/second_batch_variation.RData
Untracked: output/second_set_apr_69kchr_k_loco.rds
Untracked: output/second_set_gm.RData
Untracked: output/second_set_gm.k_loco.rds
Untracked: output/second_set_gm.probs_36state.rds
Untracked: output/second_set_gm.probs_8state.rds
Untracked: output/topSNP_chr5_mindepression.csv
Unstaged changes:
Modified: _workflowr.yml
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/DO_morphine_gemma.Rmd) and HTML (docs/DO_morphine_gemma.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 622975f | xhyuo | 2020-11-19 | gemma |
###library
library("ggplot2")
library("gridExtra")
library("parallel")
library("qtl2")
library(qtl)
#
# Attaching package: 'qtl'
# The following object is masked from 'package:qtl2':
#
# clean
library("tidyverse")
# ── Attaching packages ─────────────────────────────────────── tidyverse 1.3.0 ──
# ✔ tibble 3.0.1 ✔ dplyr 1.0.0
# ✔ tidyr 1.1.0 ✔ stringr 1.4.0
# ✔ readr 1.4.0 ✔ forcats 0.5.0
# ✔ purrr 0.3.4
# ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
# ✖ dplyr::combine() masks gridExtra::combine()
# ✖ dplyr::filter() masks stats::filter()
# ✖ dplyr::lag() masks stats::lag()
# ✖ readr::read_csv() masks qtl2::read_csv()
library("parallel")
library("survival")
#library("regress")
library("abind")
library("xlsx")
library(furrr)
# Loading required package: future
#
# Attaching package: 'future'
# The following object is masked from 'package:survival':
#
# cluster
library(cowplot)
theme_set(theme_cowplot())
source("code/gemma_plot.R")
source("code/cfw/R/gemma.R")
options(stringsAsFactors = FALSE)
#5 workers
# Set a "plan" for how the code should run.
#plan(multisession, workers = 5)
rz.transform <- function(y) {
rankY=rank(y, ties.method="average", na.last="keep")
rzT=qnorm(rankY/(length(na.exclude(rankY))+1))
return(rzT)
}
Pvaltolod <- function(p){
if(p == 0) p = 1e-10
y = ifelse(p >= 0.5, 0, qchisq(1-2*p, df=1)/(2*log(10)))
y
}
pheno.name <- c("Survival.Time", "Recovery.Time", "Min.depression", "Status_bin")
#GM_SNP
load(url("ftp://ftp.jax.org/MUGA/GM_snps.Rdata"))#GM_snps
###combined
#collecting the permutation results
# Load the results of the GEMMA association analysis, and get the minimum p-value.
readperm <- function(p=p){
print(p)
perms.gemma <- matrix(NA,1000,1)
colnames(perms.gemma) <- p
for(i in 1:1000){
gwscan <- read.gemma.assoc(paste0("data/combined/combined_batch_gwas_", p, "_", i, ".assoc.txt"))
gwscan <- gwscan[gwscan$chr != 0 & gwscan$chr != 23,]
perms.gemma[i] <- max(gwscan$log10p)
}
class(perms.gemma) <- c("scanoneperm","matrix")
return(perms.gemma)
}
p = list("Survival.Time", "Recovery.Time", "Min.depression", "Status_bin")
args1 <- list(p)
#perms <- args1 %>% future_pmap(readperm)
#names(perms) <- pheno.name
#save(perms, file = "data/combined/perms.RData")
load("data/combined/perms.RData")
#cross2 object
gm <- get(load("output/combined_gm.RData"))
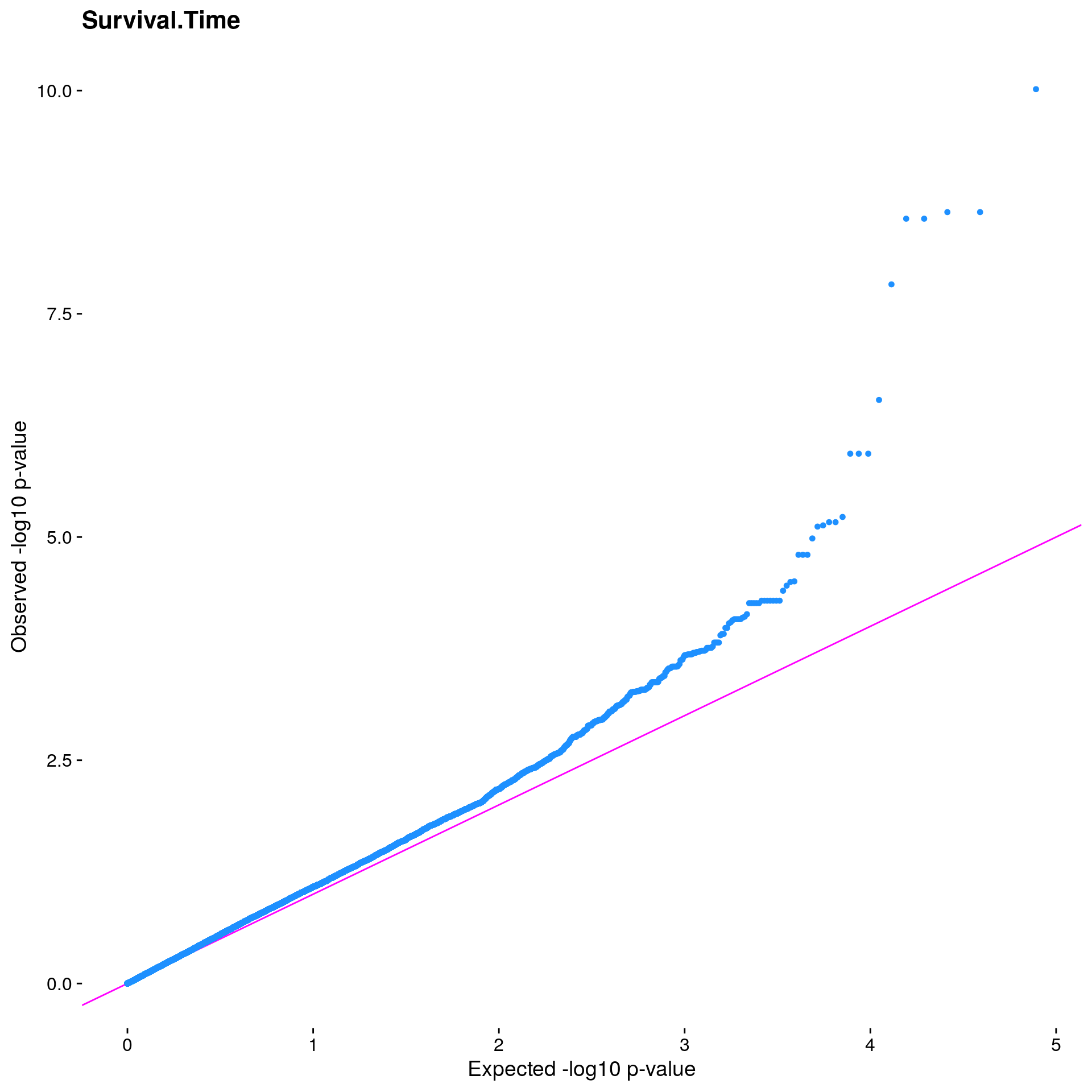
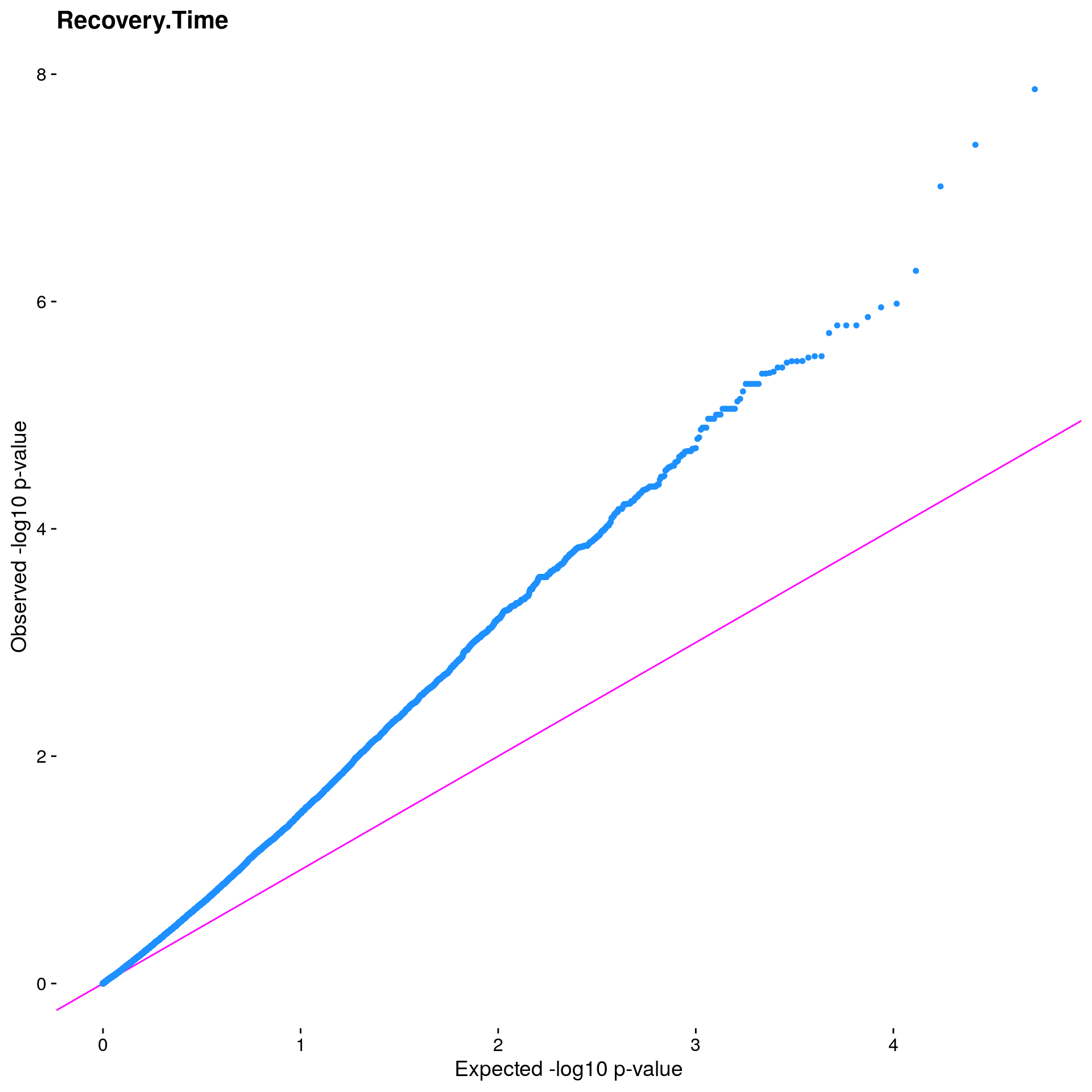
#The LMM is expected to reduce inflation of small *p*-values; a high
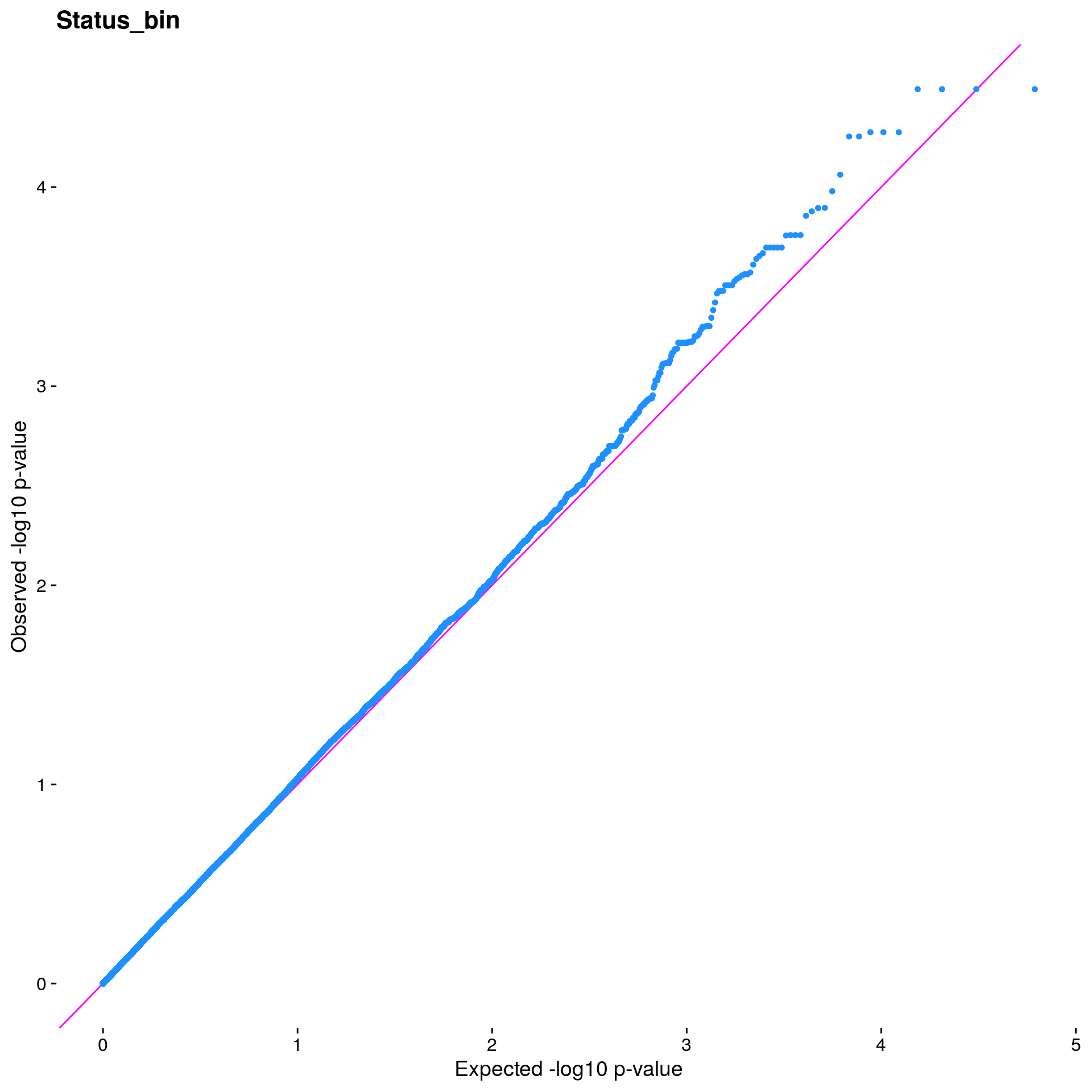
#level of inflation could indicate many false positive associations.
#The q-q plot is commonly used to assess inflation. This test is
#useful as a simple, heuristic diagnostic.
#import the results of the first GEMMA association analysis:
for(i in pheno.name){
print(i)
#load-gemma-pvalues}
gwscan <- read.table(paste0("data/combined/combined_batch_gwas_",i,".assoc.txt"),
header = TRUE, stringsAsFactors = F)
gwscan <- gwscan[gwscan$chr != 0 & gwscan$chr != 23,]
#add lod
gwscan$lod <- gwscan$p_lrt %>% map_dbl(Pvaltolod)
#left join with GM_snps to update pos
gwscan <- left_join(gwscan, GM_snps[,c(1:6,12)], by = c("rs" = "marker"))
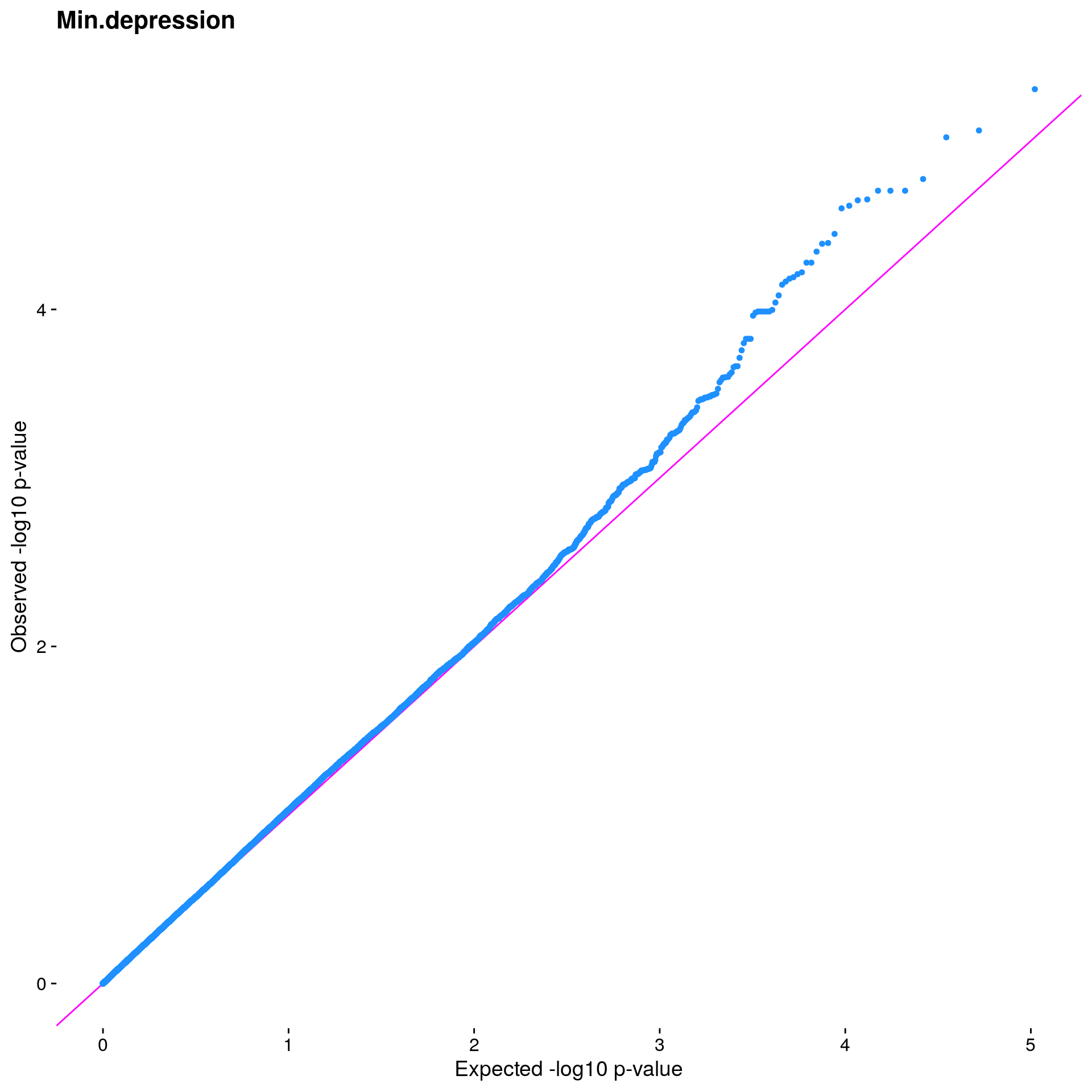
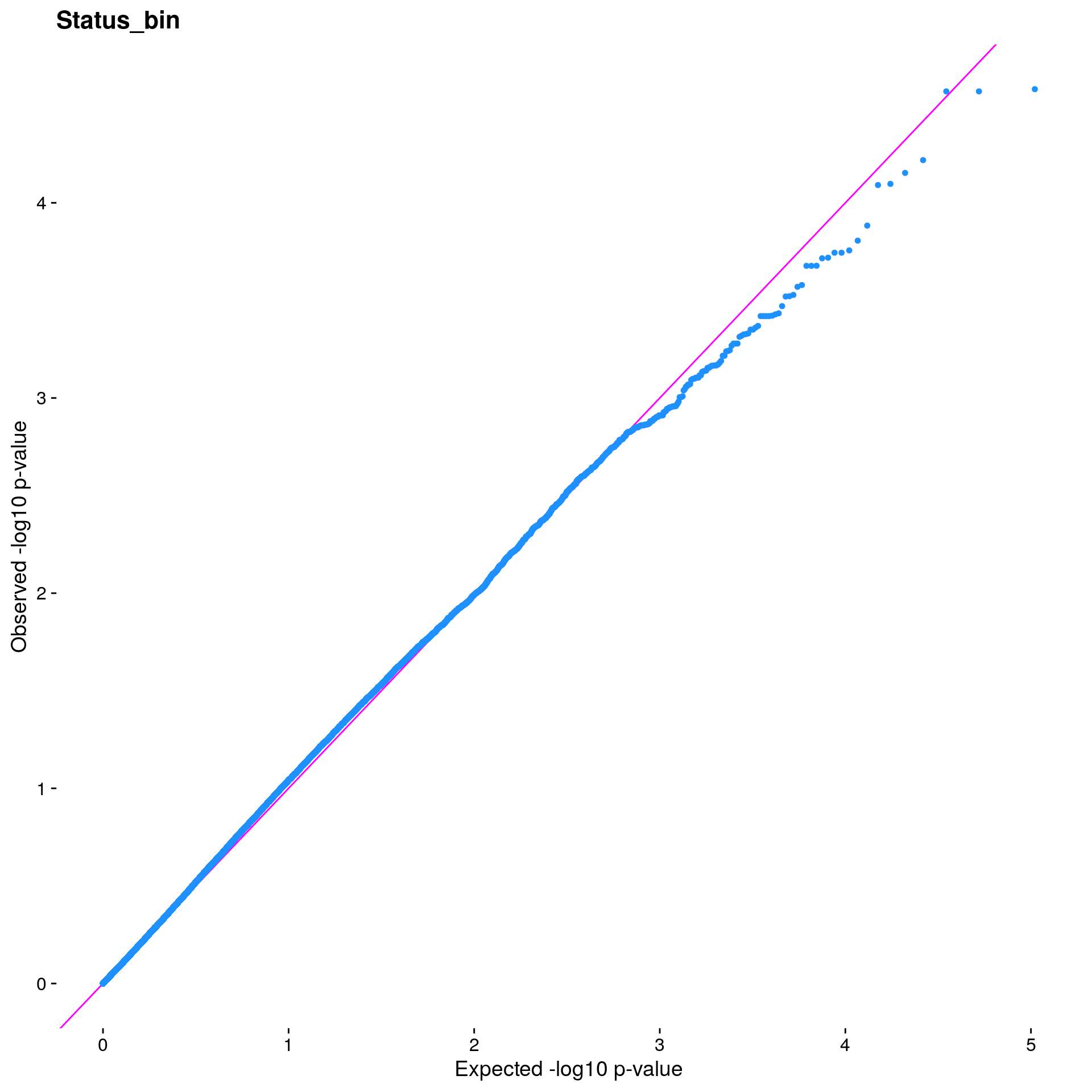
#Plot the observed *p*-values against the expected *p*-values under the
#null distribution
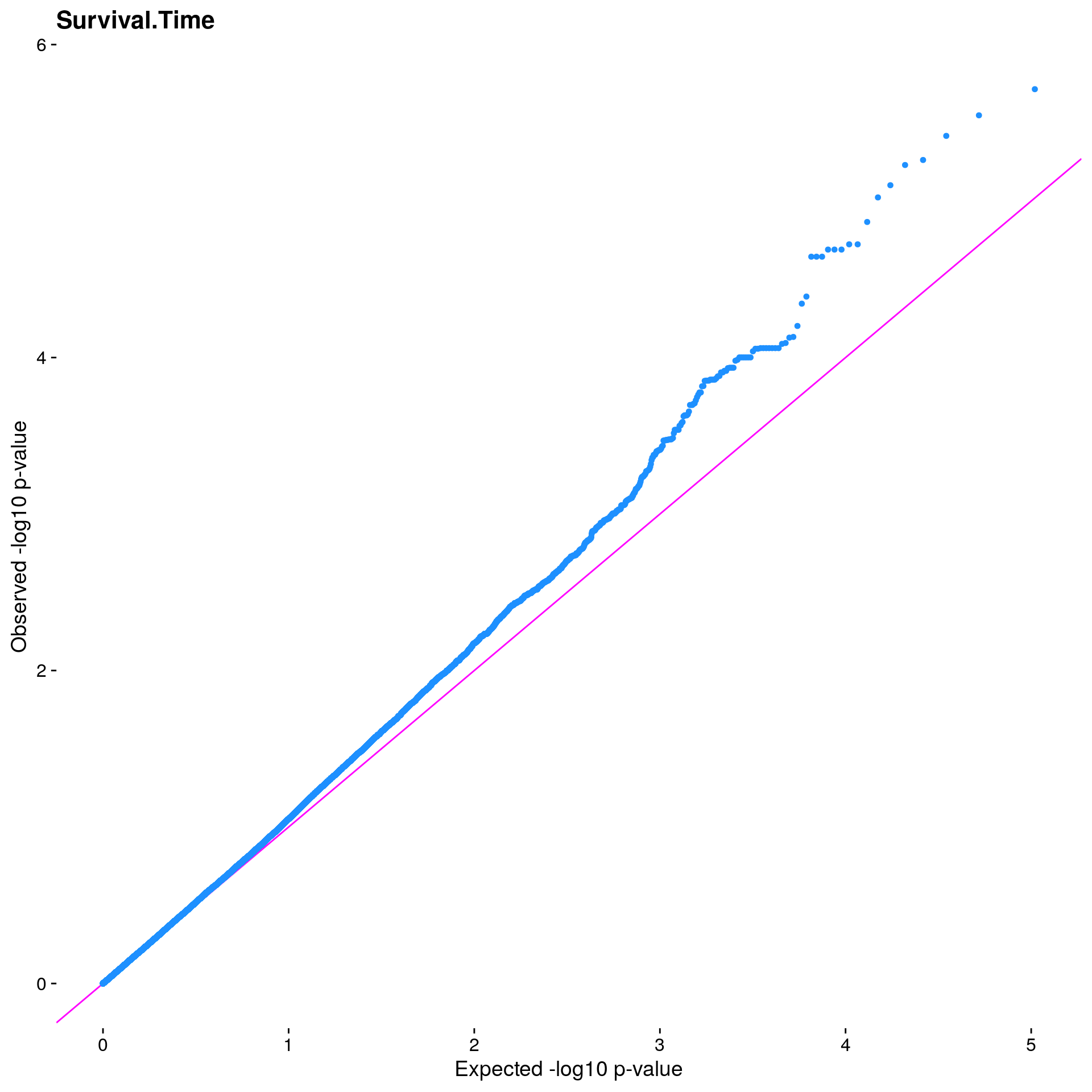
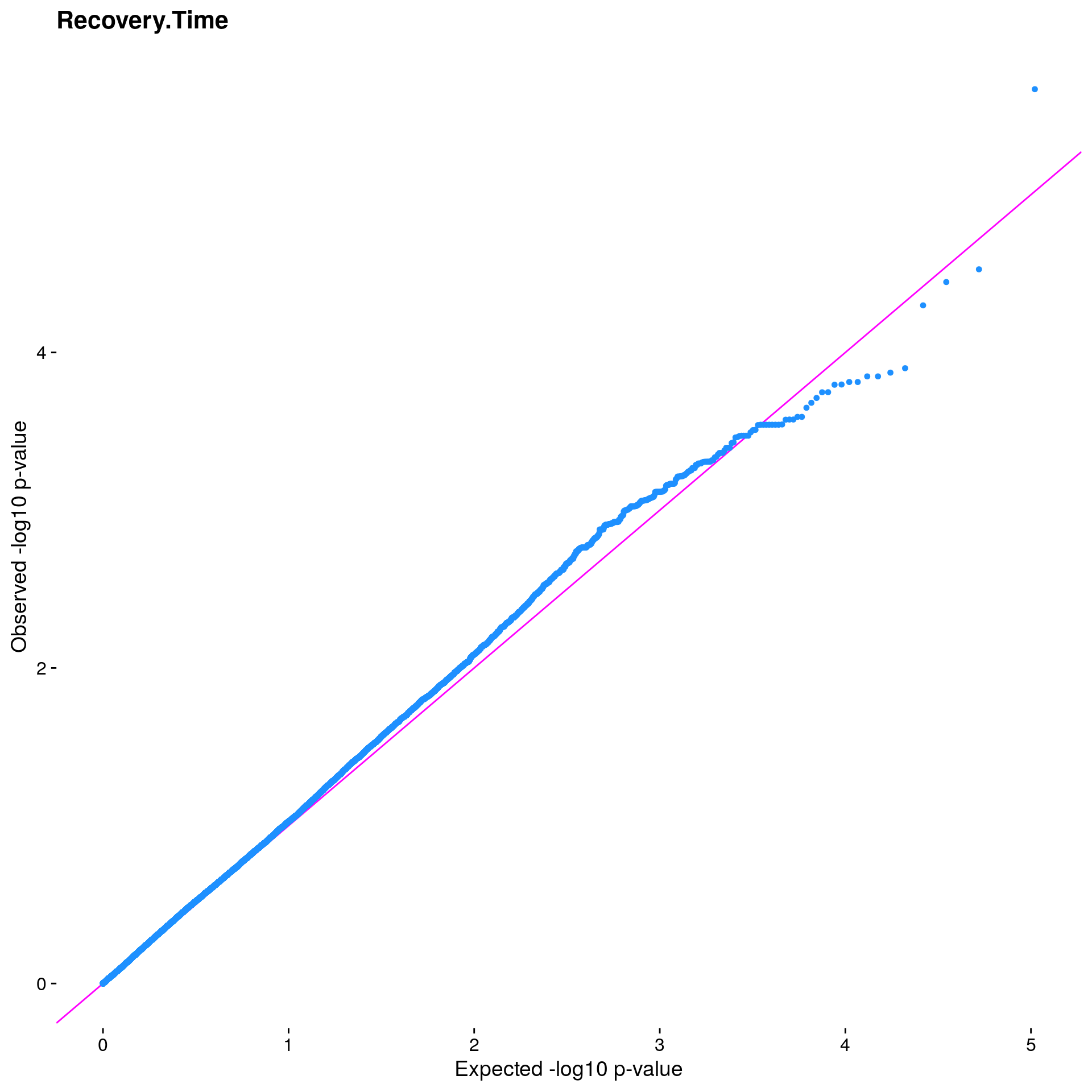
p1 <- plot.inflation(gwscan$p_lrt, title = i)
print(p1)
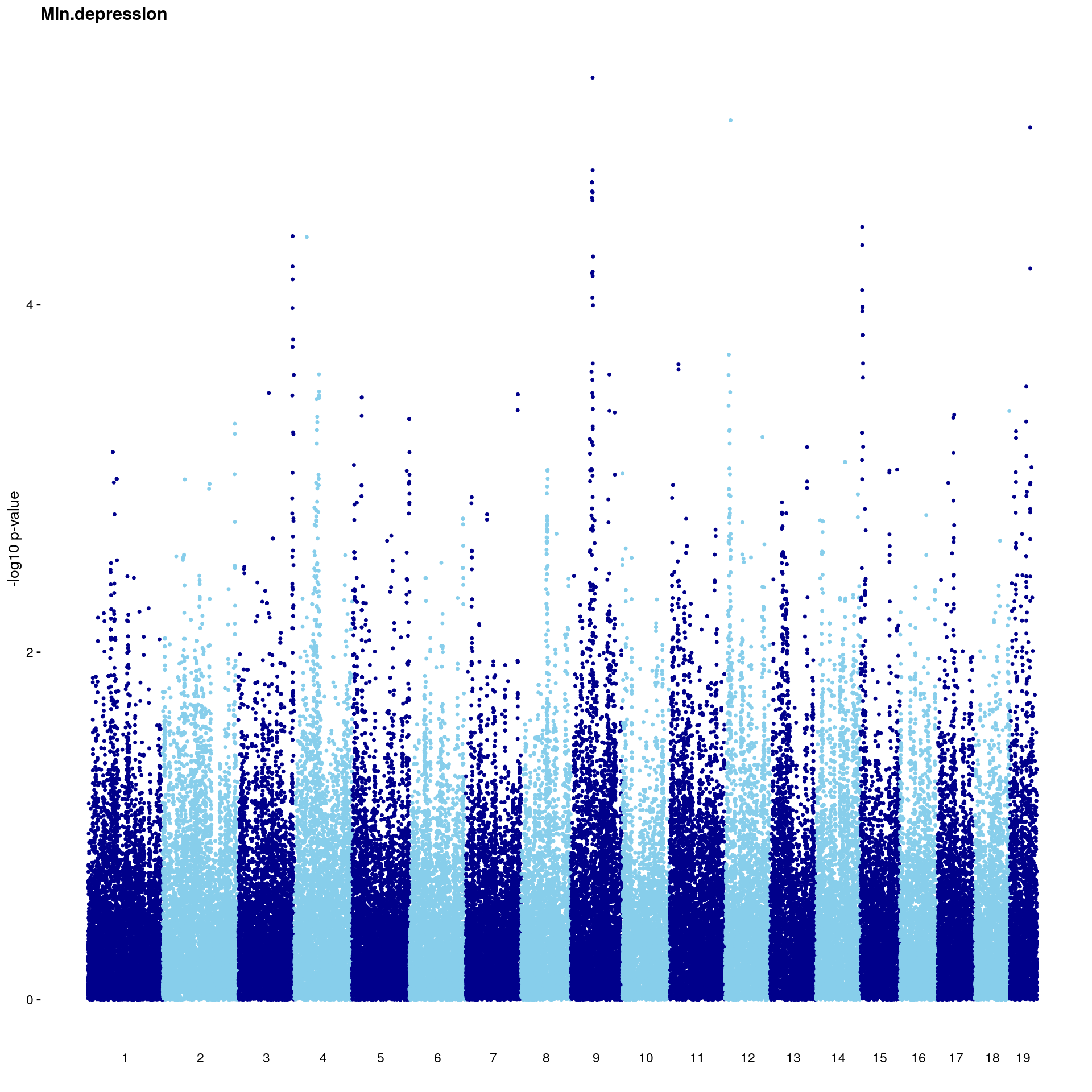
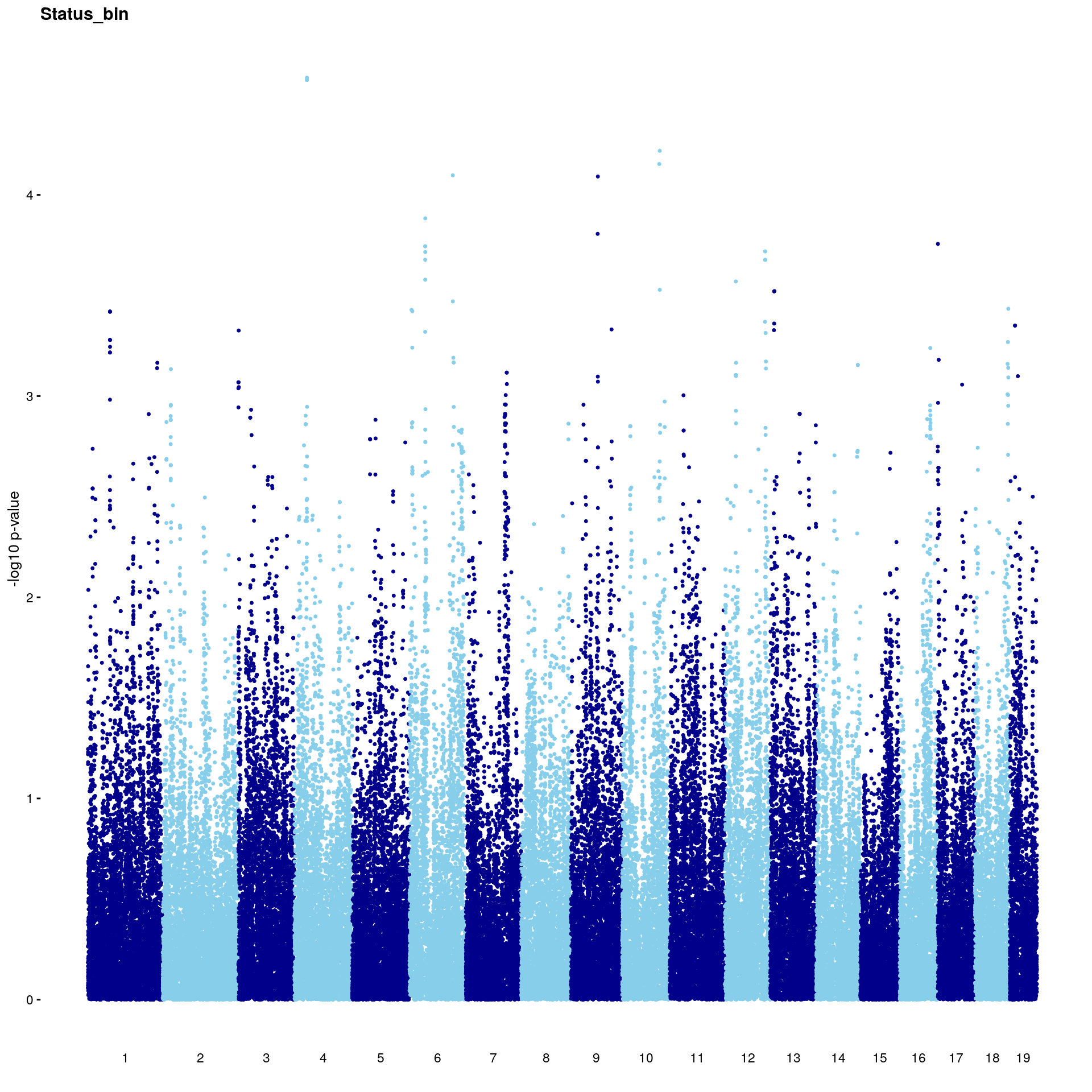
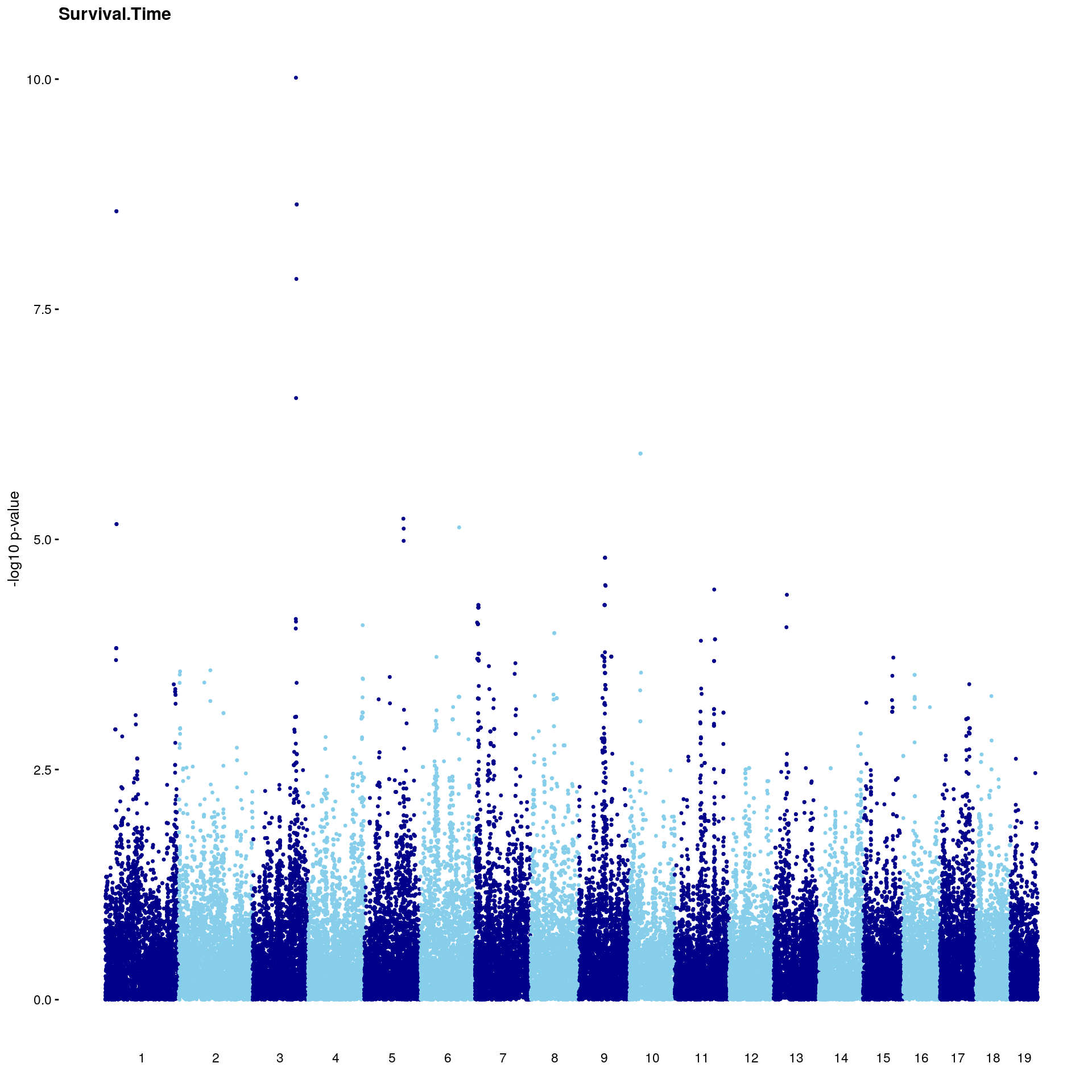
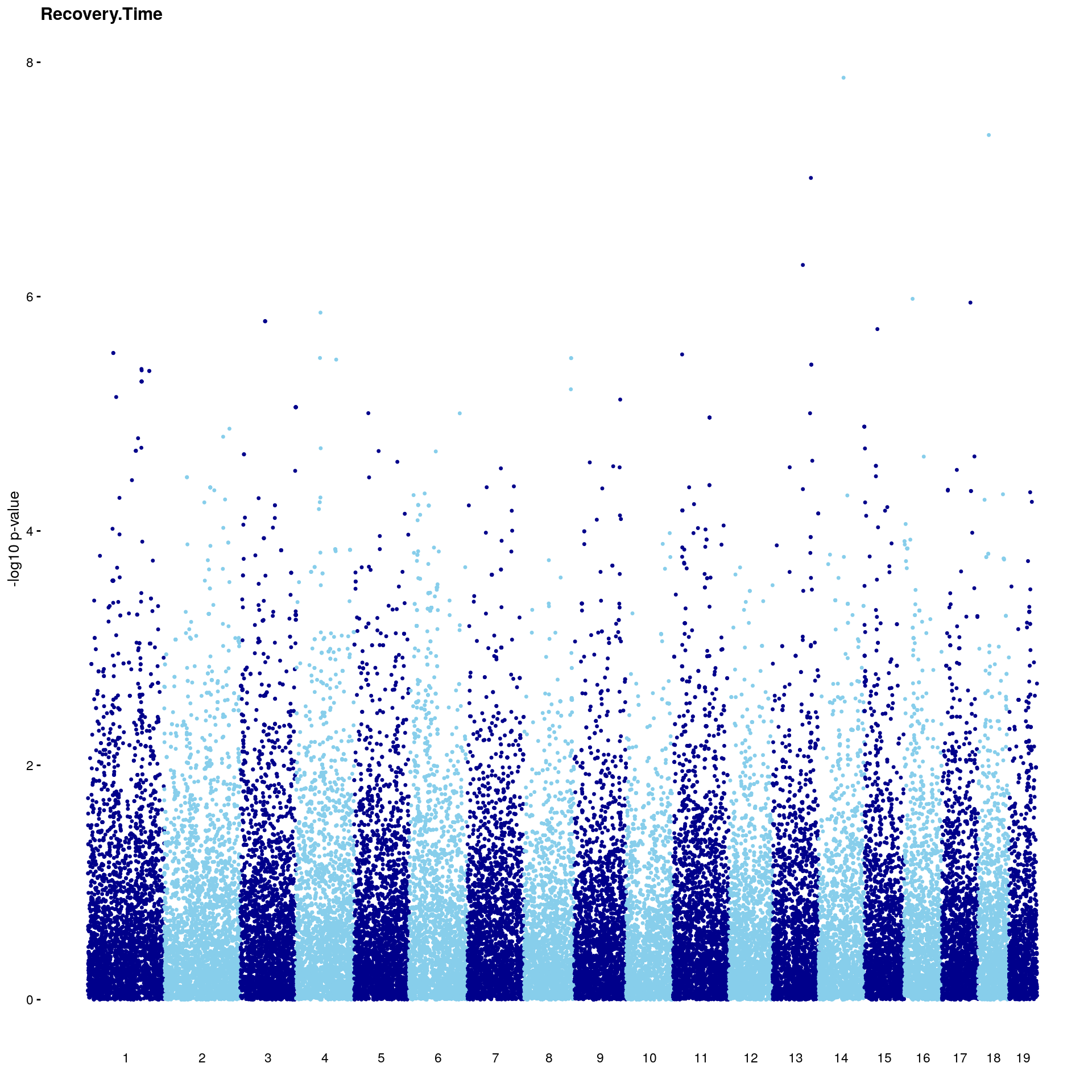
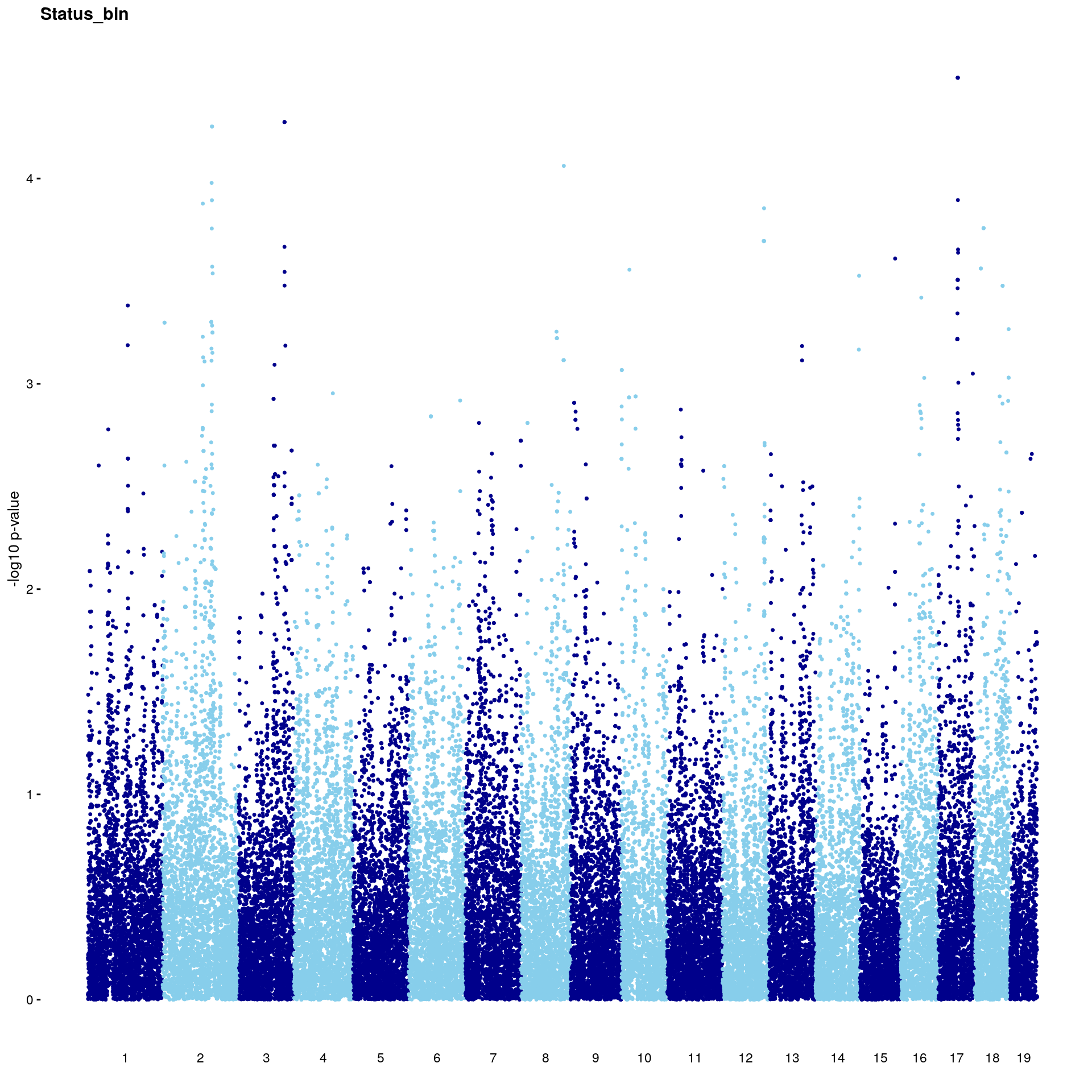
#Plot genome-wide scan
# Add a column with the marker index.
n <- nrow(gwscan)
gwscan <- cbind(gwscan,marker = 1:n)
# Convert the p-values to the -log10 scale.
gwscan <- transform(gwscan,logpp_lrt = -log10(p_lrt))
# Add column "odd.chr" to the table, and find the positions of the
# chromosomes along the x-axis.
gwscan <- transform(gwscan,odd.chr = (chr.x %% 2) == 1)
x.chr <- tapply(gwscan$marker,gwscan$chr.x,mean)
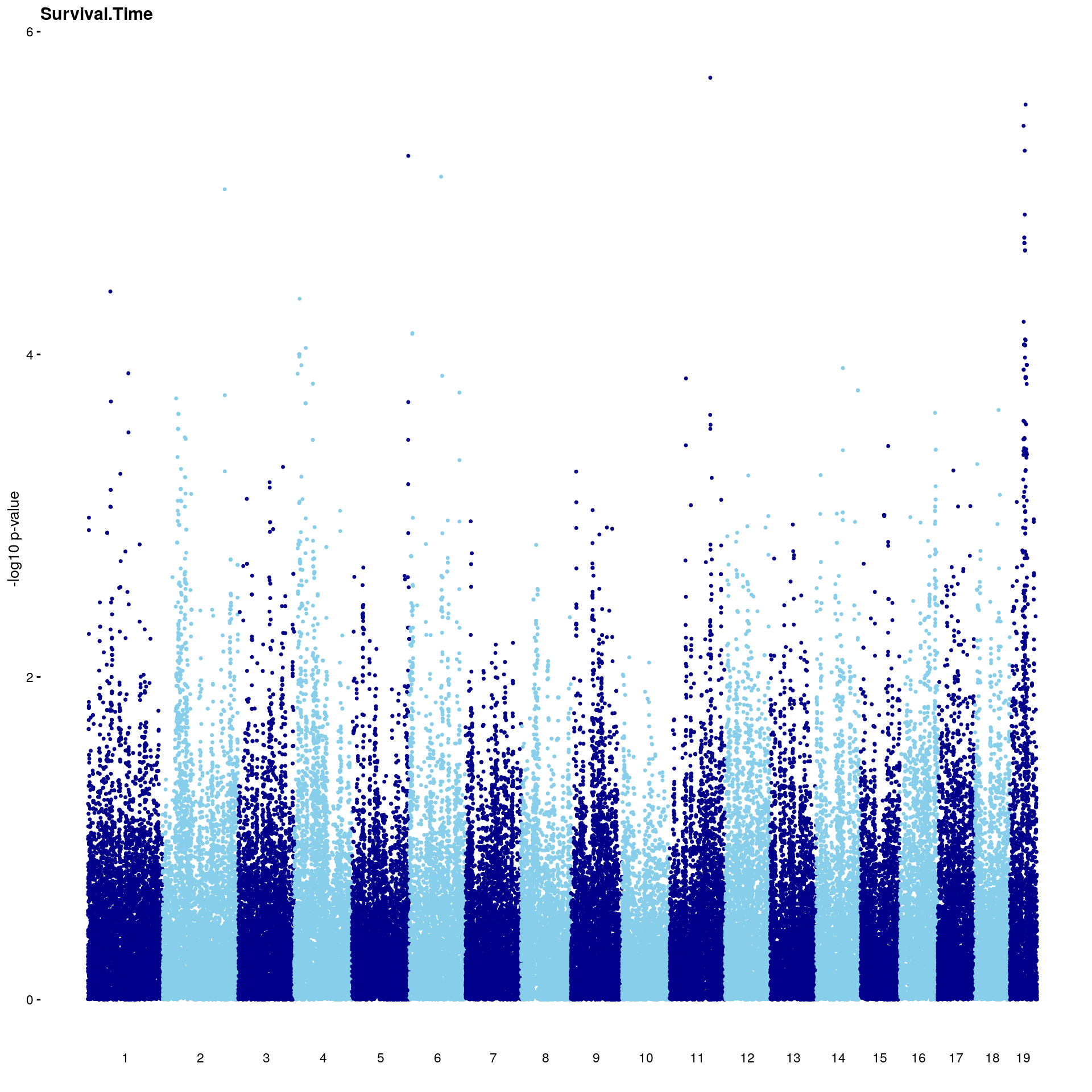
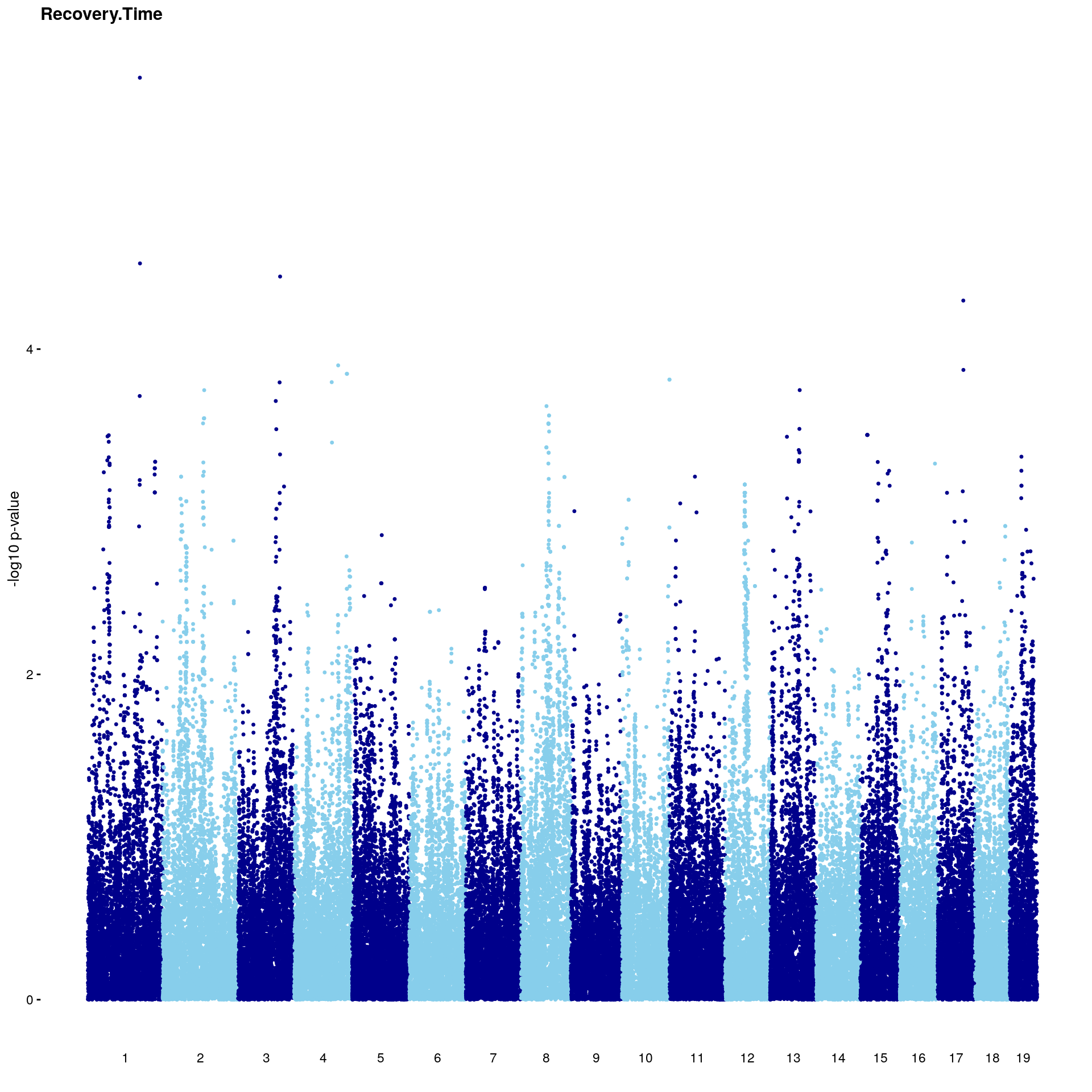
# Create the genome-wide scan ("Manhattan plot").
p2 <- ggplot(gwscan,aes(x = marker,y = logpp_lrt,color = odd.chr)) +
geom_point(size = 1,shape = 20) +
geom_hline(yintercept=summary(perms[[i]], c(0.1))[,1], color = "red") +
geom_hline(yintercept=summary(perms[[i]], c(0.05))[,1], linetype="dashed",color = "red") +
scale_x_continuous(breaks = x.chr,labels = 1:19) +
scale_color_manual(values = c("skyblue","darkblue"),guide = "none") +
labs(title= i, x = "",y = "-log10 p-value") +
theme_cowplot(font_size = 10)
print(summary(perms[[i]], c(0.05))[,1])
print(p2)
#qtl2 format
qtl.out <- matrix(gwscan$lod, ncol = 1)
rownames(qtl.out) <- gwscan$rs
names(qtl.out) <- "lod"
sub.gm <- pull_markers(gm, gwscan$rs)
#for get peak for each chr
peak <- gwscan %>%
group_by(chr.x) %>%
slice_min(p_lrt, n = 1) %>%
ungroup() %>%
arrange(p_lrt) %>%
slice(1) # top 1 peak
for(chr in peak$chr.x){
print(chr)
#Identify genomic region with lowest P-value
gwscan.lowest <- peak %>% filter(chr.x == chr)
print.data.frame(gwscan.lowest)
#1.5 drop region
drop.int <- lod_int(qtl.out, sub.gm$pmap, threshold = c(gwscan.lowest$lod-0.0001), drop = 1.5, chr = chr)
print(drop.int)
dat.region <- gwscan %>% filter(chr.x == chr, between(pos, drop.int[,1], drop.int[,3]))
#ld matrix
system(paste0("/projects/csna/csna_workflow/data/GCTA/plink -bfile ",
"/projects/heh/DO_Opioid/data/combined/combined_batch ",
" --r2 --ld-window-kb 10000000 --ld-window 99999 --ld-window-r2 0 --ld-snp ",
gwscan.lowest$rs,
" --out /projects/heh/DO_Opioid/data/combined/topsnp"))
Sys.sleep(5)
#ld
ld <- read.table("data/combined/topsnp.ld", header = T, sep = "")
ld <- ld[ld$SNP_B %in% dat.region$rs,]
#left join with dat.region
dat.region <- left_join(dat.region, ld, by = c("rs" = "SNP_B")) %>%
mutate(color = case_when(
R2 >= 0 & R2 <= 0.2 ~ "darkblue",
R2 > 0.2 & R2 <= 0.4 ~ "deepskyblue",
R2 > 0.4 & R2 <= 0.6 ~ "green",
R2 > 0.6 & R2 <= 0.8 ~ "orange",
R2 > 0.8 & R2 <= 1 ~ "red"
)) %>%
mutate(shape = case_when(
lod == max(lod) ~ 17,
TRUE ~ 20
))
#annotation
query_genes <- create_gene_query_func("data/mouse_genes_mgi.sqlite")
genes <- query_genes(chr,
min(dat.region$pos),
max(dat.region$pos))
#plot parameters
top_panel_prop = 0.5
old_mfrow <- par("mfrow")
old_mar <- par("mar")
on.exit(par(mfrow=old_mfrow, mar=old_mar))
layout(rbind(1,2), heights=c(top_panel_prop, 1-top_panel_prop))
top_mar <- bottom_mar <- old_mar
top_mar[1] <- 0.1
bottom_mar[3] <- 0.1
#top
par(mfrow = c(2,1))
par(mar=top_mar)
plot(dat.region$pos, dat.region$lod, frame.plot=TRUE, xaxt='n', col = dat.region[,"color"], pch = dat.region[,"shape"], xlab = "", ylab = "LOD score", ylim = c(0,7))
legend("topright", as.character(c(0.2, 0.4, 0.6, 0.8, 1)), col = c("darkblue","deepskyblue","green","orange","red"), pch = 20, title = "R2", ncol = 5, cex = 0.75)
#bottom
par(mar=bottom_mar)
plot_genes(genes, cex=1.5)
pdf(file = paste0("output/zoompeak_", i, "_", chr,".pdf"), width = 8, height = 12)
#top
par(mfrow = c(2,1))
par(mar=top_mar)
plot(dat.region$pos, dat.region$lod, frame.plot=TRUE, xaxt='n', col = dat.region[,"color"], pch = dat.region[,"shape"], xlab = "", ylab = "LOD score", ylim = c(0,7))
legend("topright", as.character(c(0.2, 0.4, 0.6, 0.8, 1)), col = c("darkblue","deepskyblue","green","orange","red"), pch = 20, title = "R2", ncol = 5, cex = 0.75)
#bottom
par(mar=bottom_mar)
plot_genes(genes, cex=1.5)
dev.off()
}
}
# [1] "Survival.Time"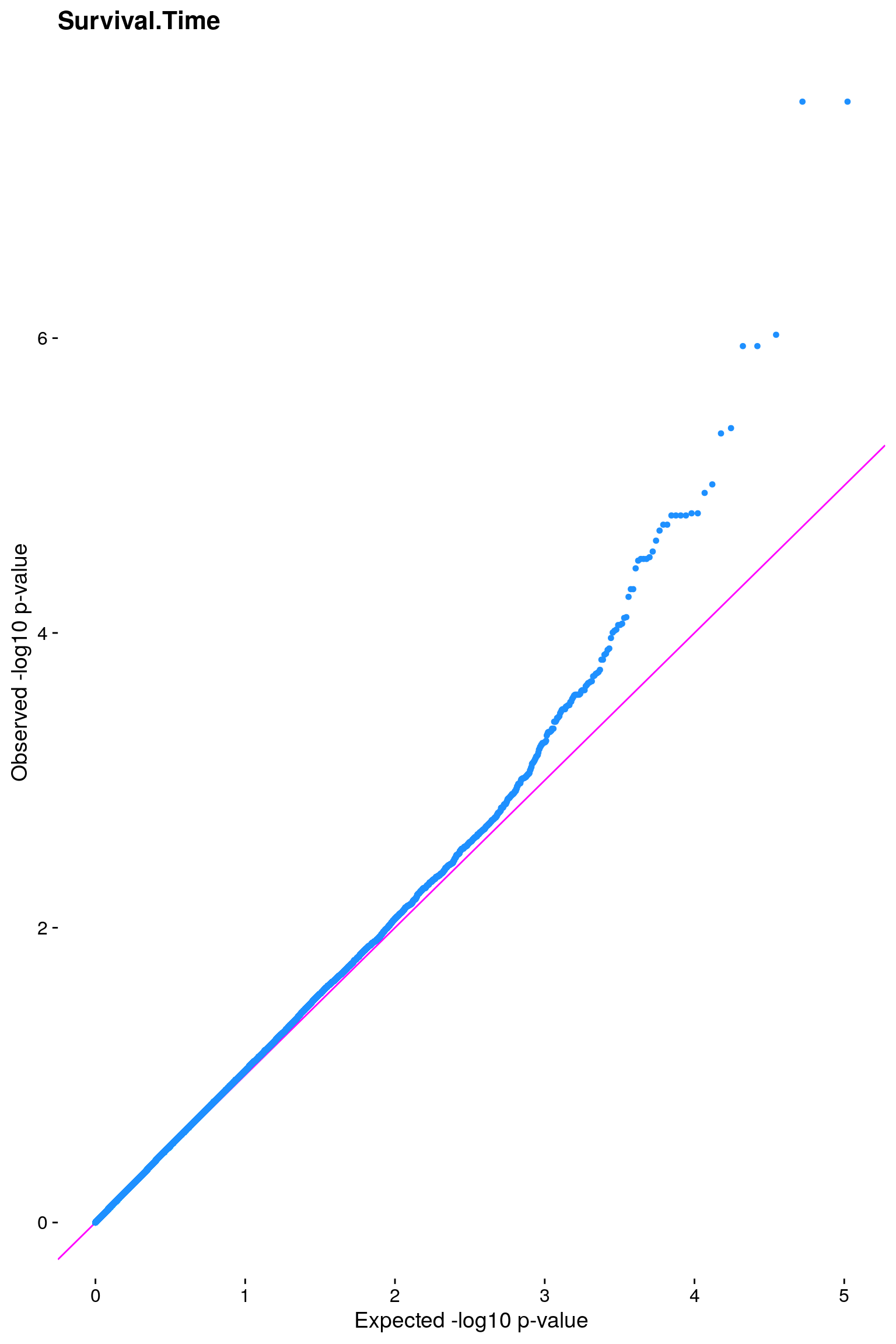
# [1] 7.276283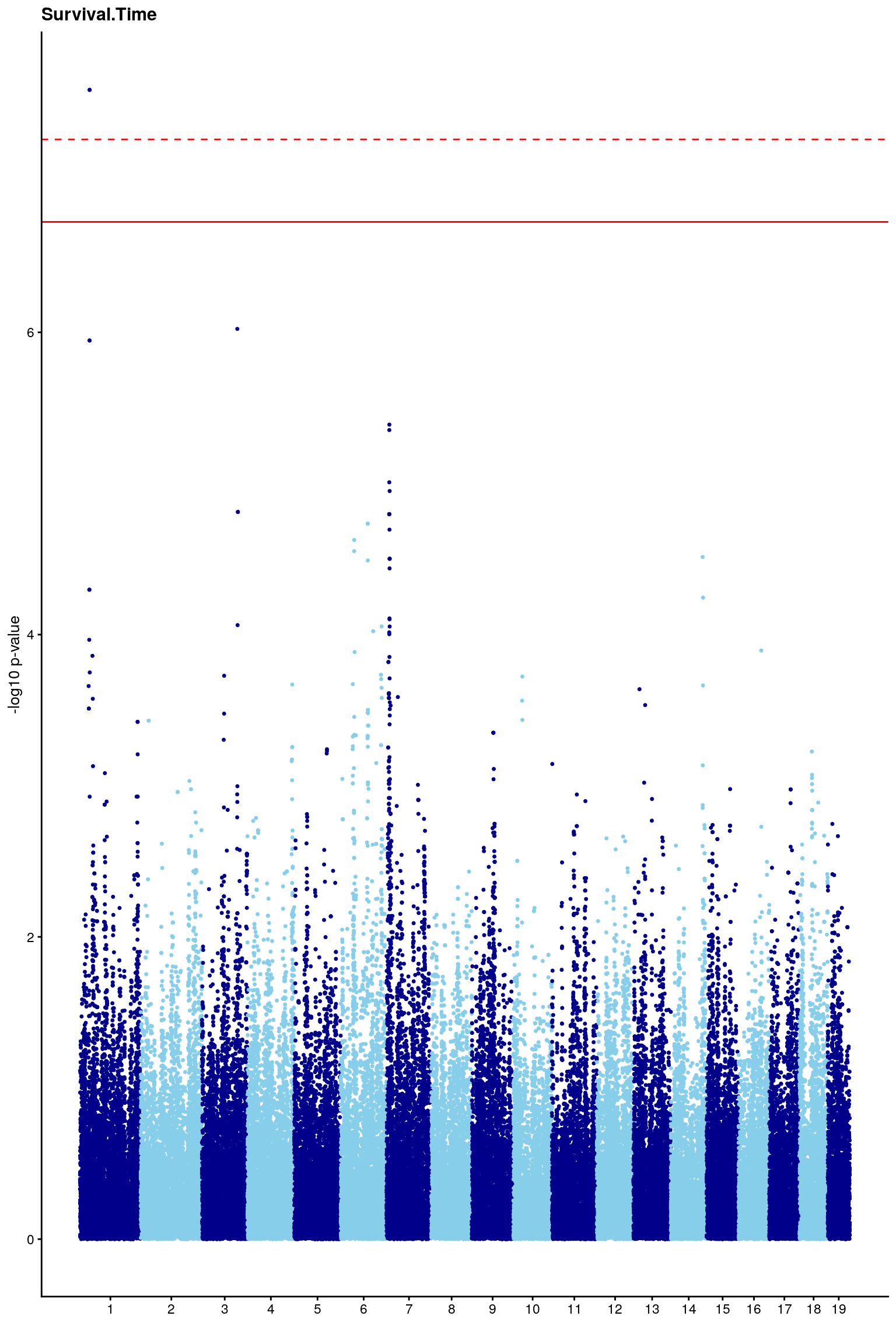
# [1] 1
# chr.x rs ps n_miss allele1 allele0 af logl_H1 l_mle
# 1 1 UNC503061 40069810 0 A G 0.041 -346.7855 0.3785897
# p_lrt lod chr.y pos cM A1 A2 rsID marker
# 1 2.487026e-08 6.455108 1 40.01297 15.55671 G A rs47122477 1308
# logpp_lrt odd.chr
# 1 7.60432 TRUE
# ci_lo pos ci_hi
# 1 40.00467 40.04955 40.09829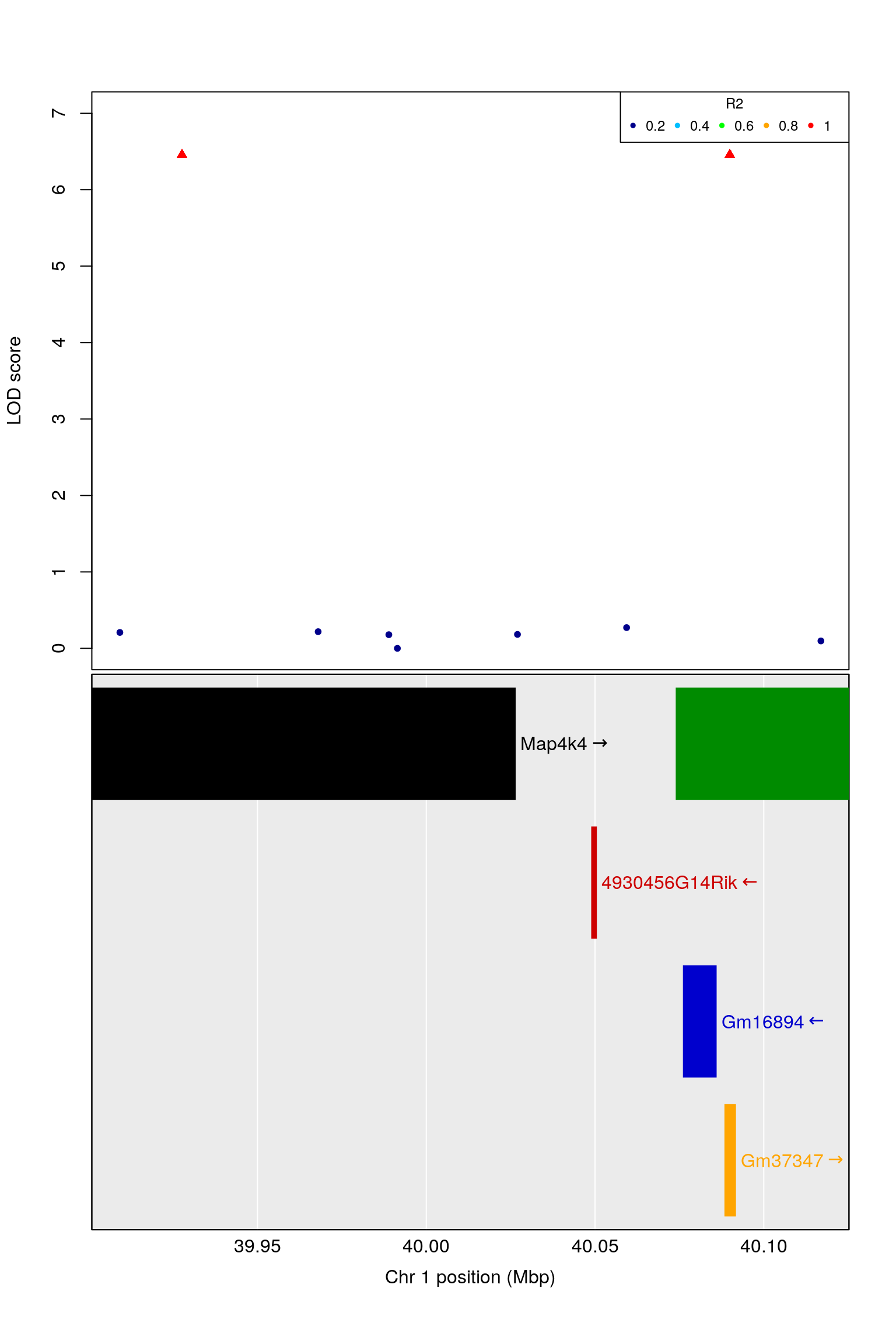
# [1] "Recovery.Time"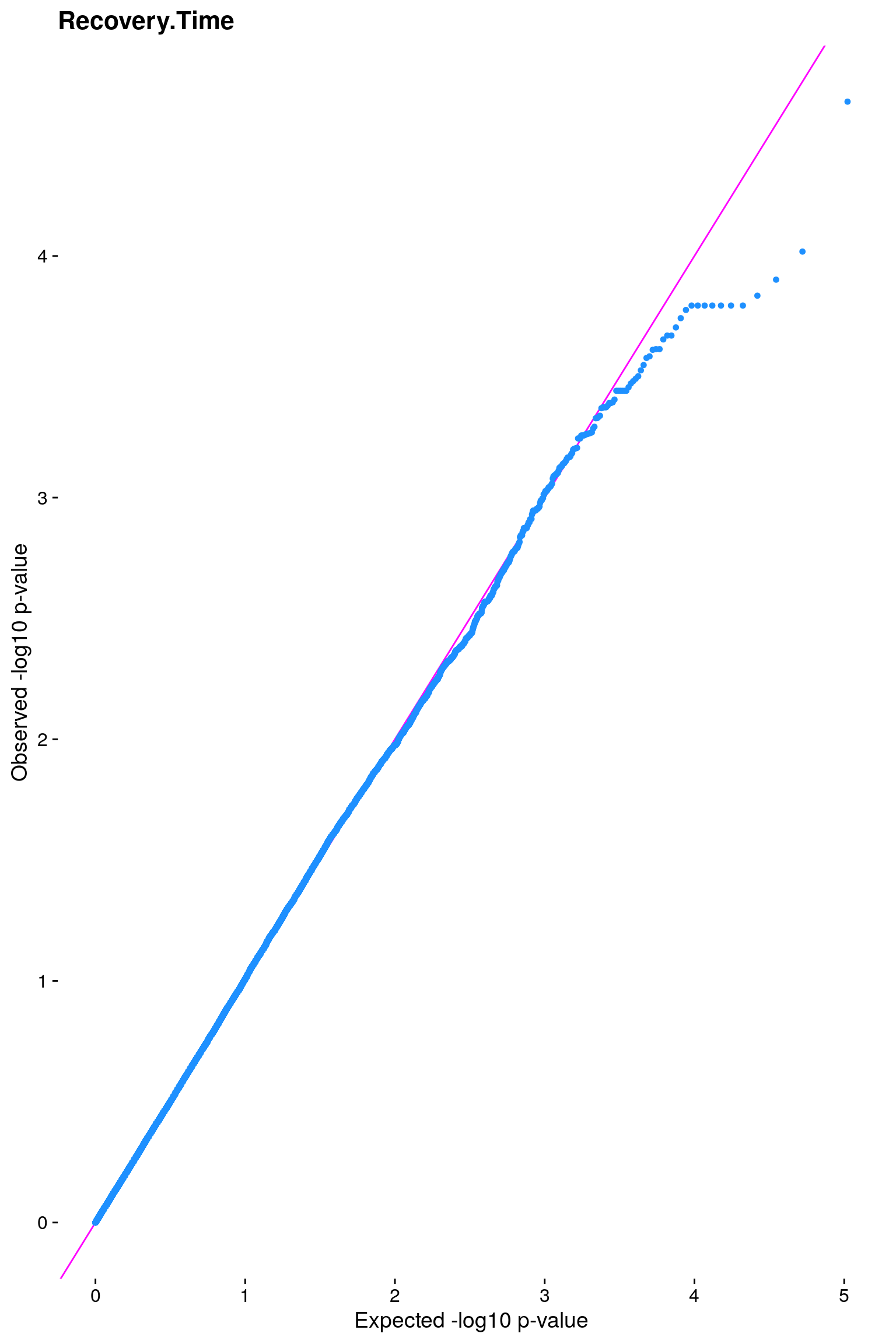
# [1] 6.279772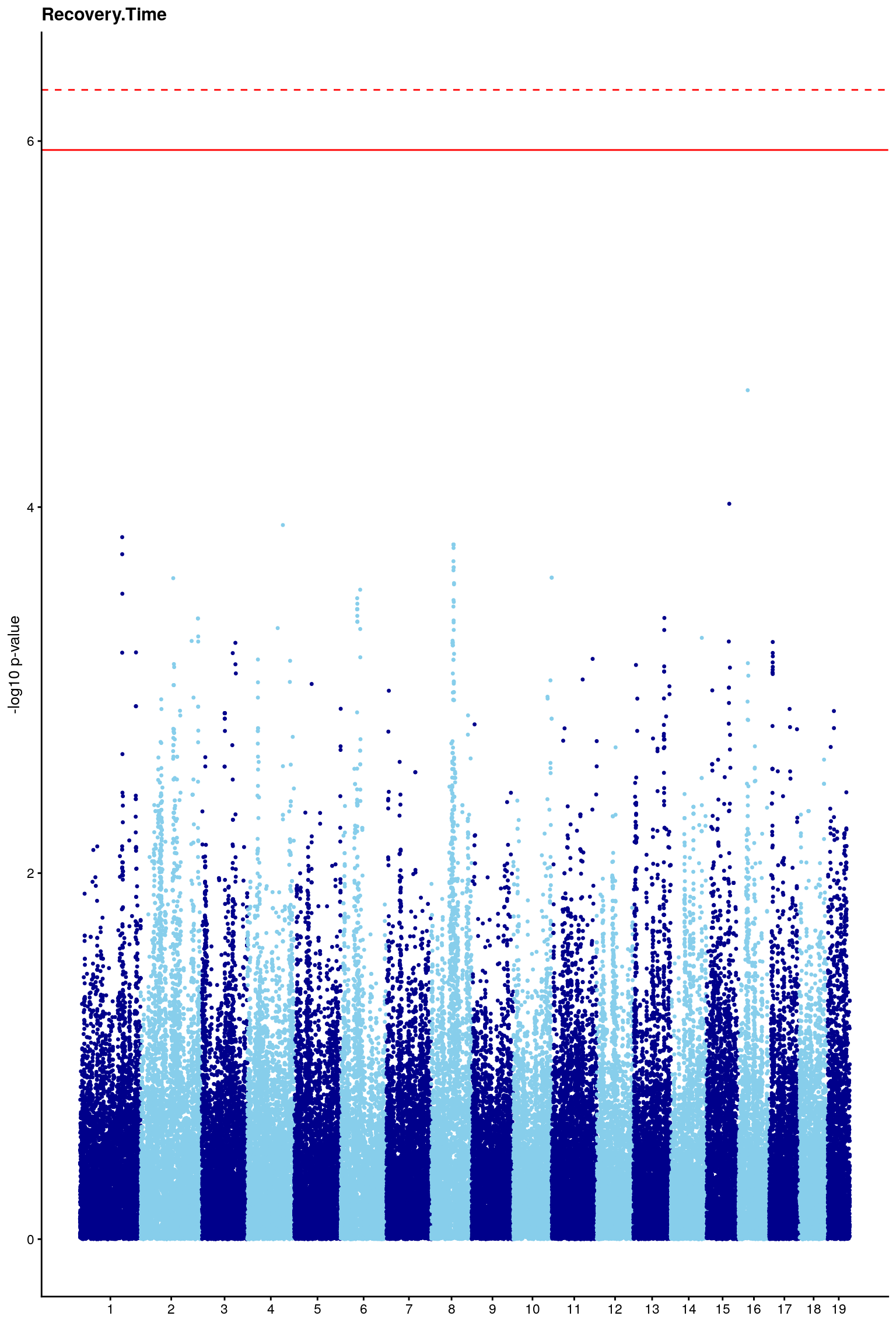
# [1] 16
# chr.x rs ps n_miss allele1 allele0 af logl_H1 l_mle
# 1 16 JAX00416833 29421922 0 A G 0.364 -981.0558 0.7356818
# p_lrt lod chr.y pos cM A1 A2 rsID marker
# 1 2.298313e-05 3.606296 16 29.42184 14.56337 A G rs31420717 91333
# logpp_lrt odd.chr
# 1 4.638591 FALSE
# ci_lo pos ci_hi
# 1 29.37 29.42184 30.25786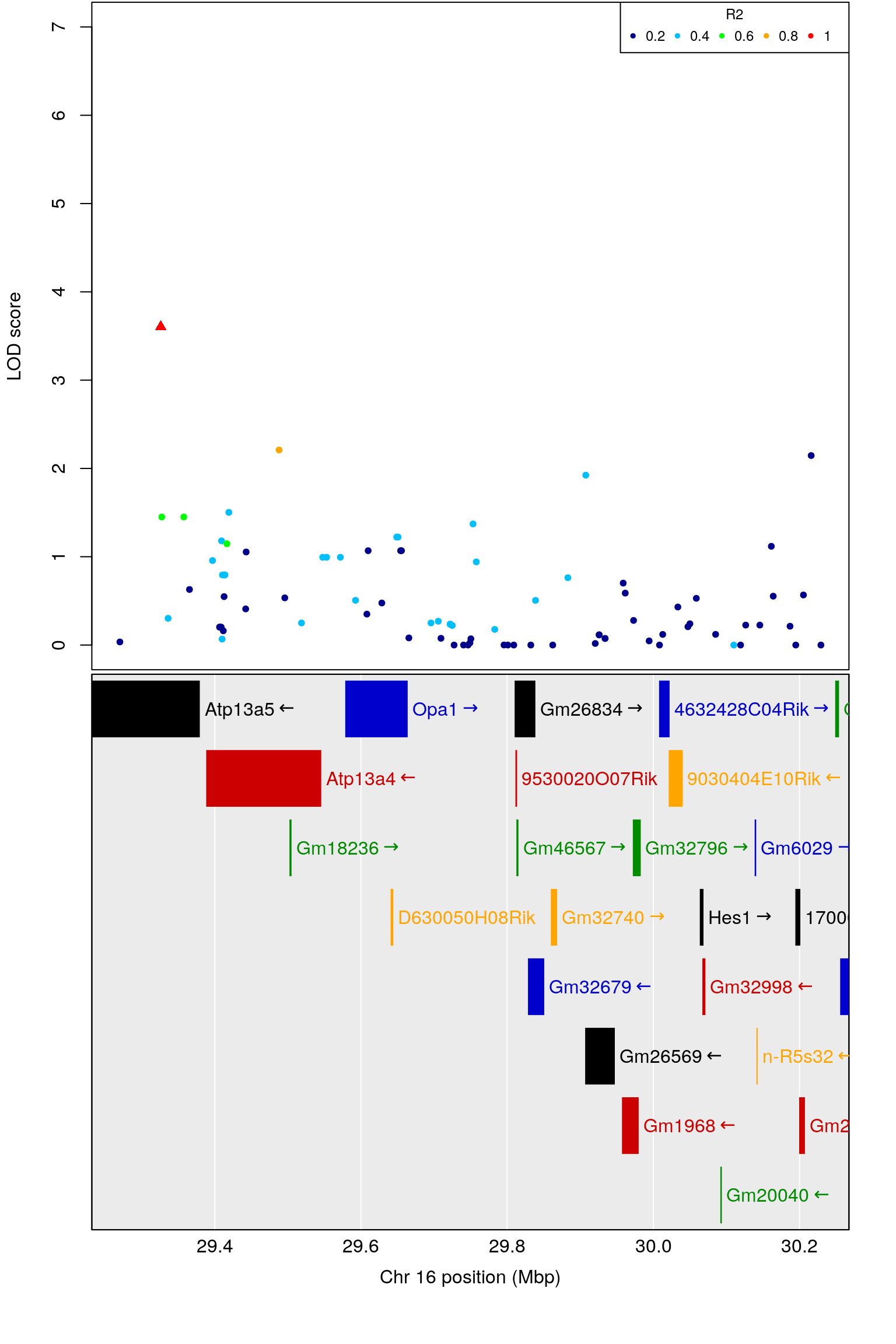
# [1] "Min.depression"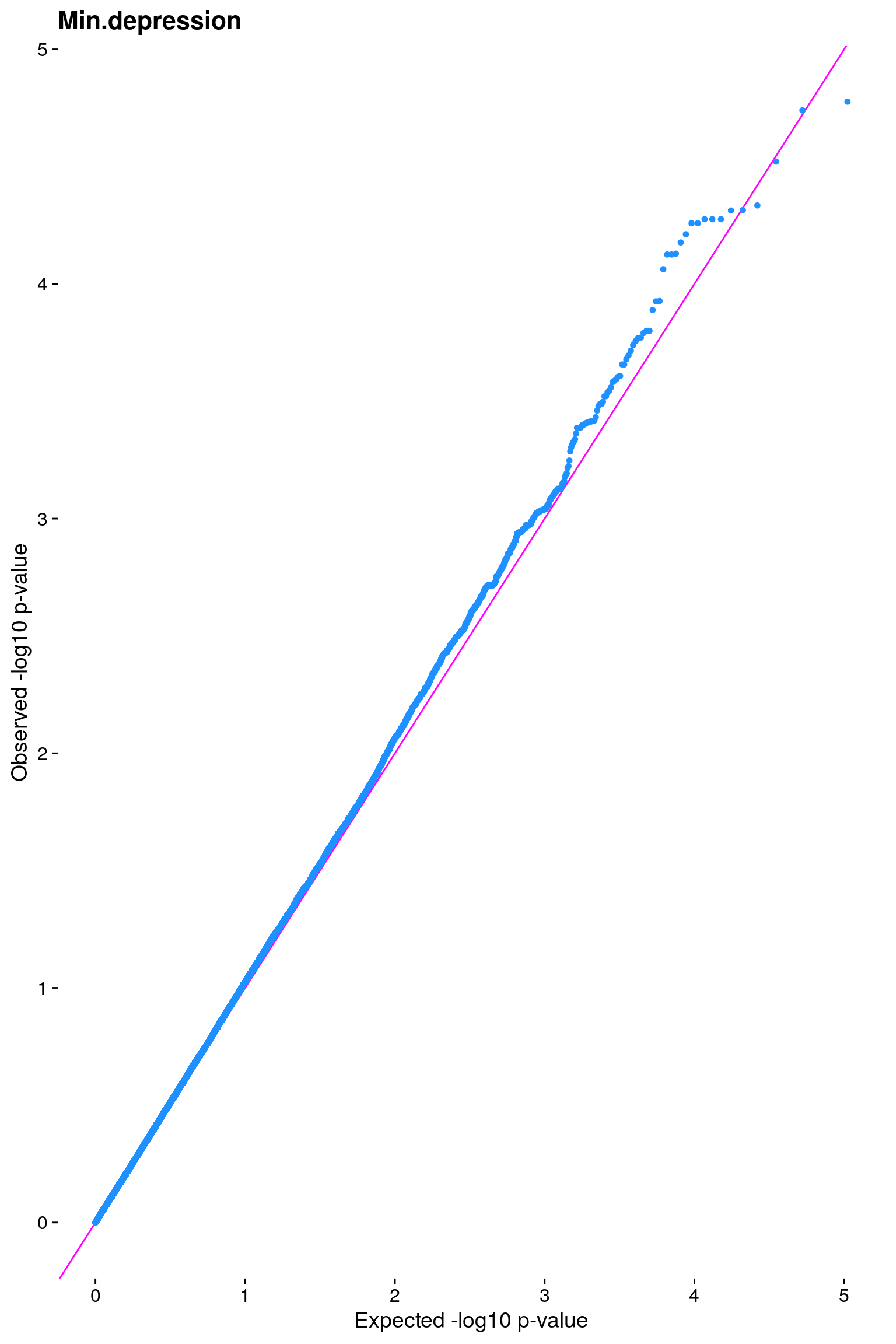
# [1] 5.851045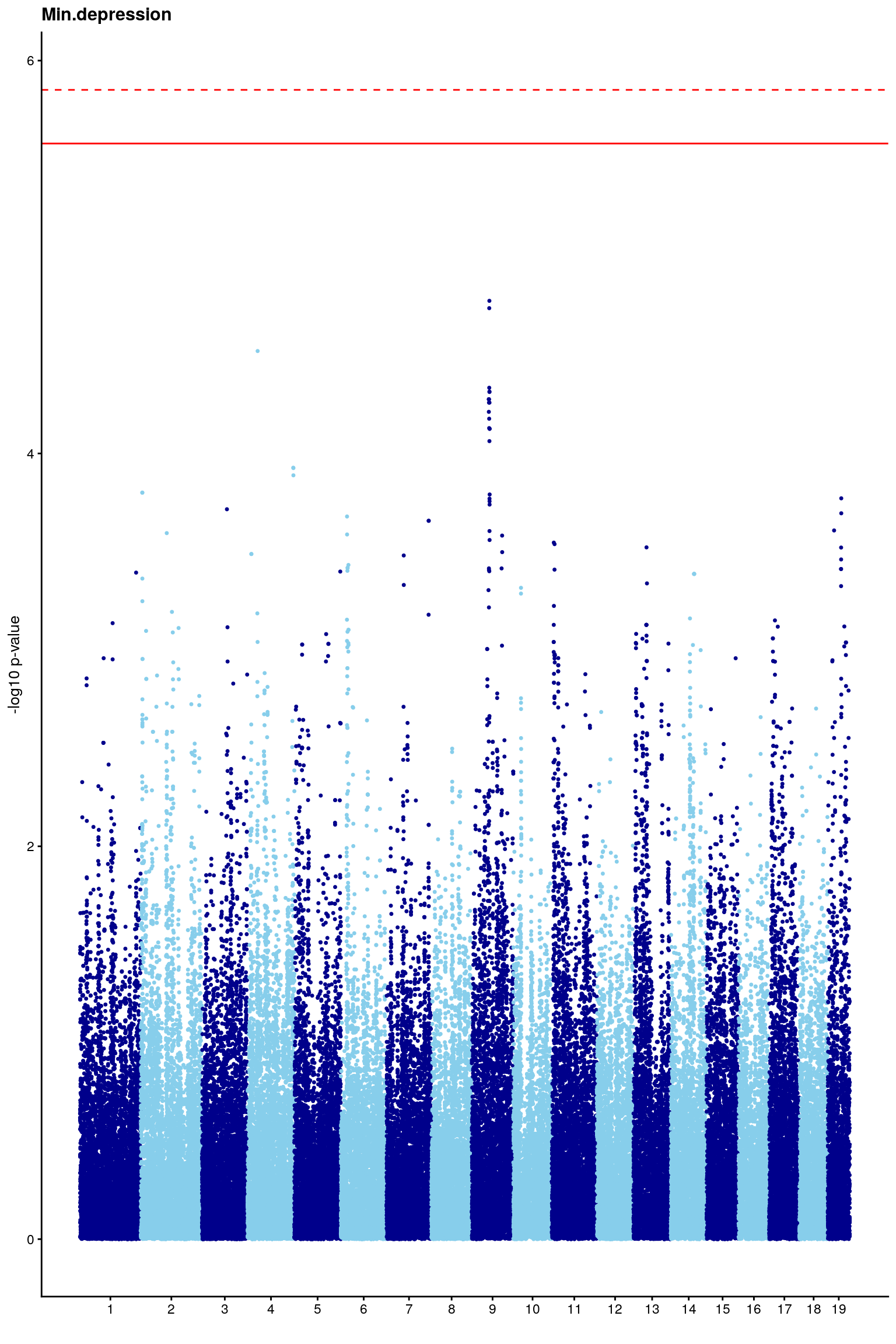
# [1] 9
# chr.x rs ps n_miss allele1 allele0 af logl_H1 l_mle
# 1 9 UNCHS025742 58286671 1 A C 0.226 539.765 0.5719939
# p_lrt lod chr.y pos cM A1 A2 rsID marker
# 1 1.670997e-05 3.737685 9 58.43886 28.1673 A C rs30310680 56007
# logpp_lrt odd.chr
# 1 4.777024 TRUE
# ci_lo pos ci_hi
# 1 55.97323 58.43886 98.00211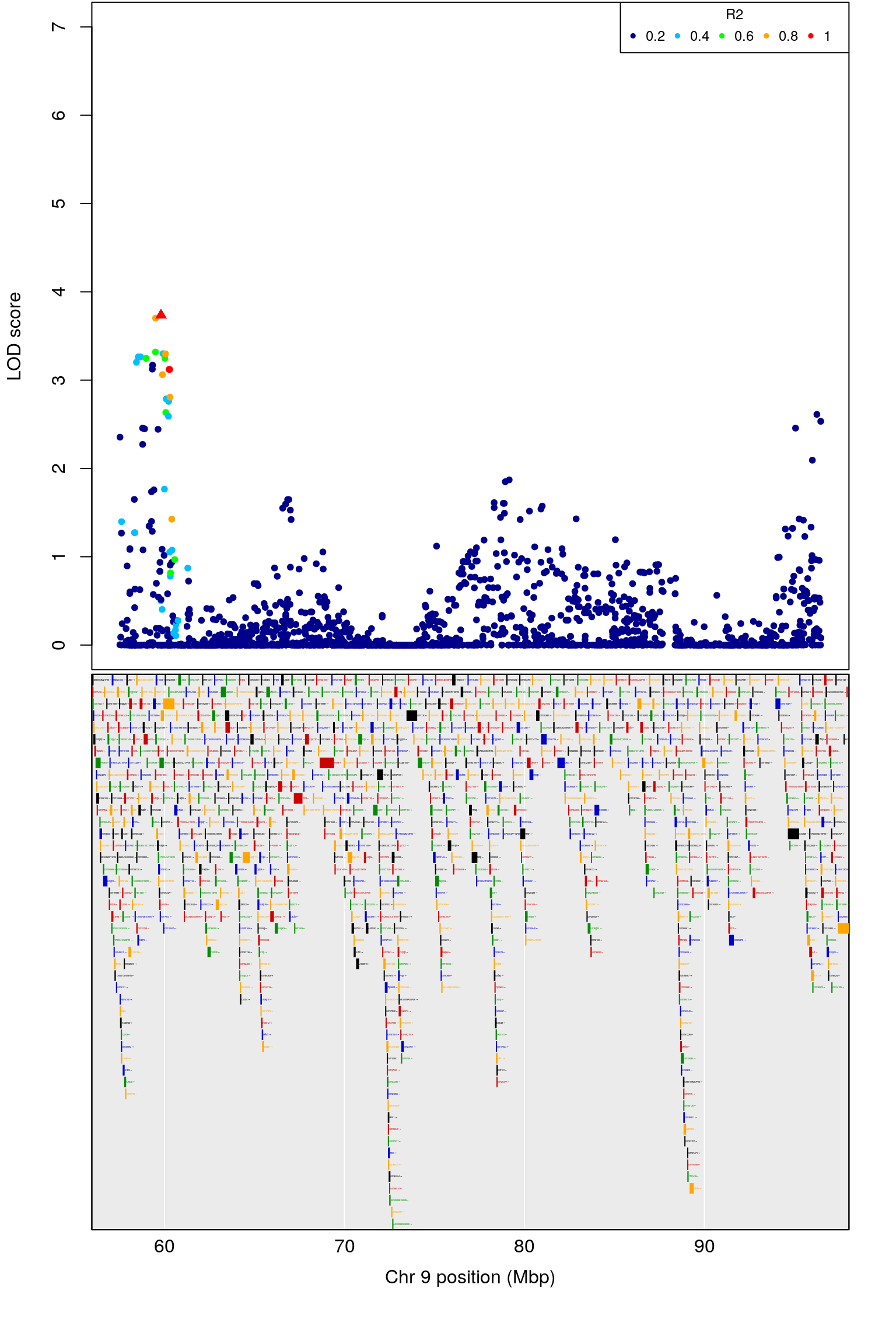
# [1] "Status_bin"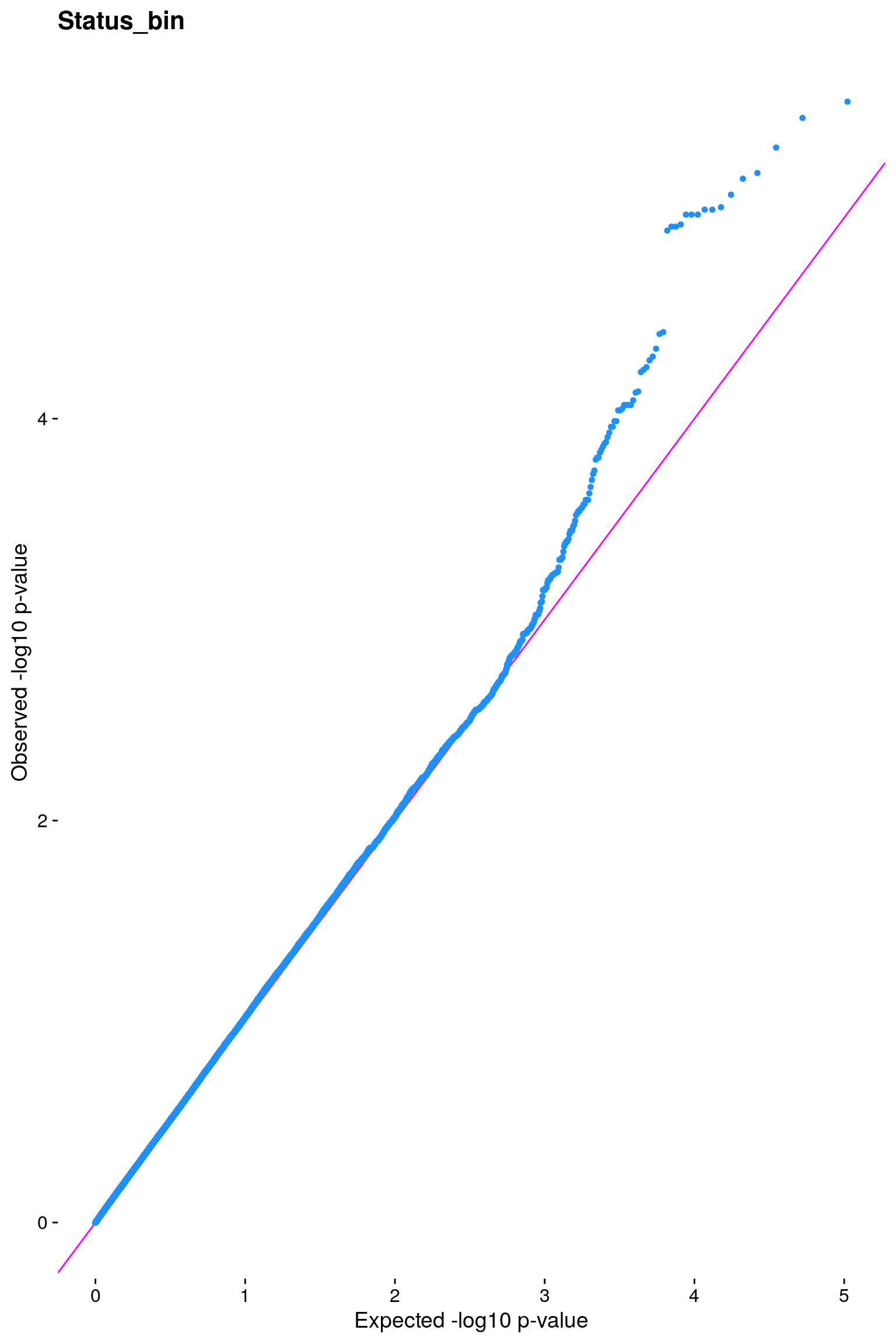
# [1] 5.921437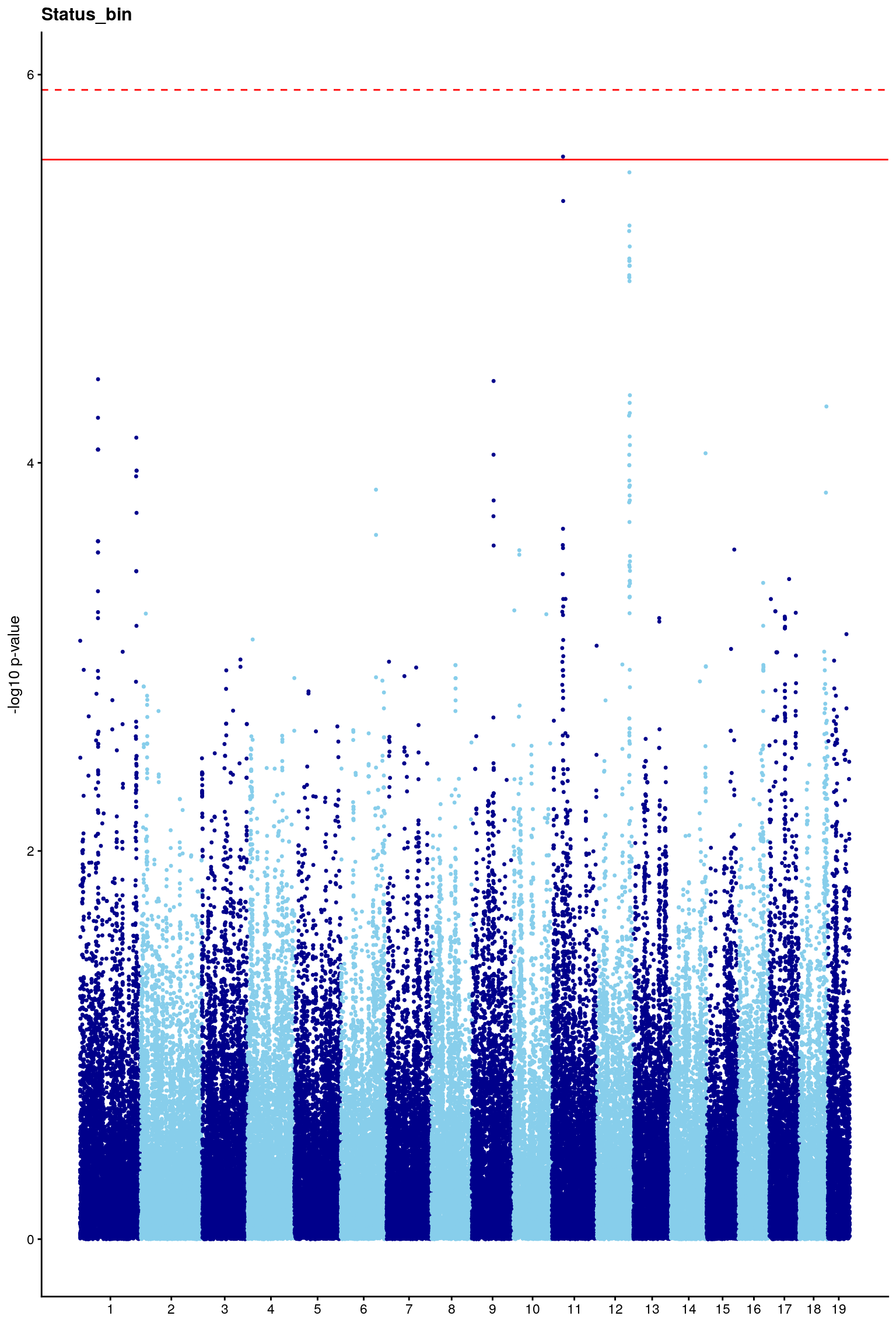
# [1] 11
# chr.x rs ps n_miss allele1 allele0 af logl_H1 l_mle
# 1 11 UNC19431523 37321071 2 A G 0.35 -403.3376 0.4443877
# p_lrt lod chr.y pos cM A1 A2 rsID marker
# 1 2.645205e-06 4.501281 11 37.50757 18.81608 A G rs26883853 66076
# logpp_lrt odd.chr
# 1 5.577541 TRUE
# ci_lo pos ci_hi
# 1 37.50252 37.50757 37.8321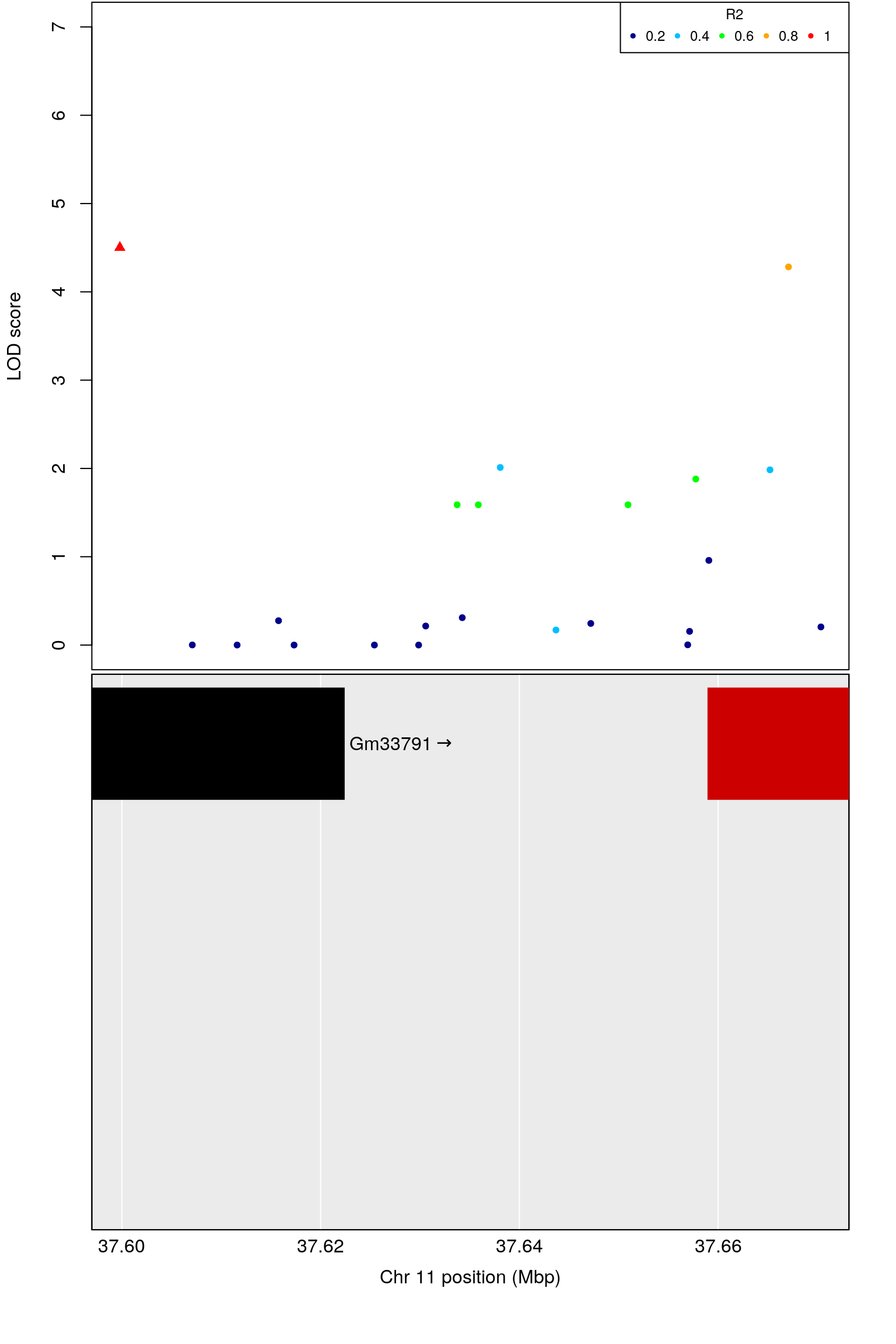
###second
for(i in pheno.name){
#load-gemma-pvalues}
gwscan1 <- read.table(paste0("data/second/second_batch_gwas_",i,".assoc.txt"),
header = TRUE, stringsAsFactors = F)
gwscan1 <- gwscan1[gwscan1$chr != 0 & gwscan1$chr != 23,]
#Plot the observed *p*-values against the expected *p*-values under the
#null distribution
p1 <- plot.inflation(gwscan1$p_lrt, title = i)
print(p1)
#Plot genome-wide scan
p2 <- plot.gwscan(gwscan1, title = i)
print(p2)
}
###first
for(i in pheno.name[-3]){
#load-gemma-pvalues}
gwscan1 <- read.table(paste0("data/first/first_batch_gwas_",i,".assoc.txt"),
header = TRUE, stringsAsFactors = F)
gwscan1 <- gwscan1[gwscan1$chr != 0 & gwscan1$chr != 23,]
#Plot the observed *p*-values against the expected *p*-values under the
#null distribution
p1 <- plot.inflation(gwscan1$p_lrt, title = i)
print(p1)
#Plot genome-wide scan
p2 <- plot.gwscan(gwscan1, title = i)
print(p2)
}
sessionInfo()
# R version 4.0.0 (2020-04-24)
# Platform: x86_64-pc-linux-gnu (64-bit)
# Running under: Ubuntu 18.04.4 LTS
#
# Matrix products: default
# BLAS/LAPACK: /usr/lib/x86_64-linux-gnu/libopenblasp-r0.2.20.so
#
# locale:
# [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
# [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
# [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=C
# [7] LC_PAPER=en_US.UTF-8 LC_NAME=en_US.UTF-8
# [9] LC_ADDRESS=en_US.UTF-8 LC_TELEPHONE=en_US.UTF-8
# [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=en_US.UTF-8
#
# attached base packages:
# [1] parallel stats graphics grDevices utils datasets methods
# [8] base
#
# other attached packages:
# [1] cowplot_1.1.0 furrr_0.2.1 future_1.20.1 xlsx_0.6.4.2
# [5] abind_1.4-5 survival_3.1-12 forcats_0.5.0 stringr_1.4.0
# [9] dplyr_1.0.0 purrr_0.3.4 readr_1.4.0 tidyr_1.1.0
# [13] tibble_3.0.1 tidyverse_1.3.0 qtl_1.46-2 qtl2_0.22-8
# [17] gridExtra_2.3 ggplot2_3.3.2 workflowr_1.6.2
#
# loaded via a namespace (and not attached):
# [1] fs_1.4.1 lubridate_1.7.9 bit64_0.9-7 httr_1.4.1
# [5] rprojroot_1.3-2 tools_4.0.0 backports_1.1.6 R6_2.4.1
# [9] DBI_1.1.0 colorspace_1.4-1 withr_2.2.0 tidyselect_1.1.0
# [13] bit_1.1-15.2 compiler_4.0.0 git2r_0.27.1 cli_2.0.2
# [17] rvest_0.3.6 xml2_1.3.2 labeling_0.4.2 scales_1.1.1
# [21] digest_0.6.25 rmarkdown_2.5 pkgconfig_2.0.3 htmltools_0.4.0
# [25] parallelly_1.21.0 dbplyr_2.0.0 highr_0.8 rlang_0.4.6
# [29] readxl_1.3.1 rstudioapi_0.11 RSQLite_2.2.0 generics_0.0.2
# [33] farver_2.0.3 jsonlite_1.6.1 magrittr_1.5 Matrix_1.2-18
# [37] Rcpp_1.0.4.6 munsell_0.5.0 fansi_0.4.1 lifecycle_0.2.0
# [41] stringi_1.4.6 whisker_0.4 yaml_2.2.1 grid_4.0.0
# [45] blob_1.2.1 listenv_0.8.0 promises_1.1.0 crayon_1.3.4
# [49] lattice_0.20-41 haven_2.3.1 splines_4.0.0 xlsxjars_0.6.1
# [53] hms_0.5.3 knitr_1.28 pillar_1.4.4 codetools_0.2-16
# [57] reprex_0.3.0 glue_1.4.0 evaluate_0.14 data.table_1.12.8
# [61] modelr_0.1.8 vctrs_0.3.1 httpuv_1.5.4 cellranger_1.1.0
# [65] gtable_0.3.0 assertthat_0.2.1 xfun_0.13 broom_0.7.2
# [69] later_1.0.0 rJava_0.9-13 memoise_1.1.0 globals_0.13.1
# [73] ellipsis_0.3.0This R Markdown site was created with workflowr