Plot_DO_morphine
Hao He
2023-02-21
Last updated: 2023-02-21
Checks: 7 0
Knit directory: DO_Opioid/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200504) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version cc3ae27. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/Picture1.png
Untracked files:
Untracked: Rplot_rz.png
Untracked: TIMBR.test.prop.bm.MN.RET.RData
Untracked: TIMBR.test.random.RData
Untracked: TIMBR.test.rz.transformed_TVb_ml.RData
Untracked: analysis/DDO_morphine1_second_set_69k.stdout
Untracked: analysis/DO_Fentanyl.R
Untracked: analysis/DO_Fentanyl.err
Untracked: analysis/DO_Fentanyl.out
Untracked: analysis/DO_Fentanyl.sh
Untracked: analysis/DO_Fentanyl_69k.R
Untracked: analysis/DO_Fentanyl_69k.err
Untracked: analysis/DO_Fentanyl_69k.out
Untracked: analysis/DO_Fentanyl_69k.sh
Untracked: analysis/DO_Fentanyl_Cohort2_GCTA_herit.R
Untracked: analysis/DO_Fentanyl_Cohort2_gemma.R
Untracked: analysis/DO_Fentanyl_Cohort2_mapping.R
Untracked: analysis/DO_Fentanyl_Cohort2_mapping.err
Untracked: analysis/DO_Fentanyl_Cohort2_mapping.out
Untracked: analysis/DO_Fentanyl_Cohort2_mapping.sh
Untracked: analysis/DO_Fentanyl_GCTA_herit.R
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.R
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.err
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.out
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.sh
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.R
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.err
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.out
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.sh
Untracked: analysis/DO_Fentanyl_array.R
Untracked: analysis/DO_Fentanyl_array.err
Untracked: analysis/DO_Fentanyl_array.out
Untracked: analysis/DO_Fentanyl_array.sh
Untracked: analysis/DO_Fentanyl_combining2Cohort_GCTA_herit.R
Untracked: analysis/DO_Fentanyl_combining2Cohort_gemma.R
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping.R
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping.err
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping.out
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping.sh
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping_CoxPH.R
Untracked: analysis/DO_Fentanyl_finalreport_to_plink.sh
Untracked: analysis/DO_Fentanyl_gemma.R
Untracked: analysis/DO_Fentanyl_gemma.err
Untracked: analysis/DO_Fentanyl_gemma.out
Untracked: analysis/DO_Fentanyl_gemma.sh
Untracked: analysis/DO_morphine1.R
Untracked: analysis/DO_morphine1.Rout
Untracked: analysis/DO_morphine1.sh
Untracked: analysis/DO_morphine1.stderr
Untracked: analysis/DO_morphine1.stdout
Untracked: analysis/DO_morphine1_SNP.R
Untracked: analysis/DO_morphine1_SNP.Rout
Untracked: analysis/DO_morphine1_SNP.sh
Untracked: analysis/DO_morphine1_SNP.stderr
Untracked: analysis/DO_morphine1_SNP.stdout
Untracked: analysis/DO_morphine1_combined.R
Untracked: analysis/DO_morphine1_combined.Rout
Untracked: analysis/DO_morphine1_combined.sh
Untracked: analysis/DO_morphine1_combined.stderr
Untracked: analysis/DO_morphine1_combined.stdout
Untracked: analysis/DO_morphine1_combined_69k.R
Untracked: analysis/DO_morphine1_combined_69k.Rout
Untracked: analysis/DO_morphine1_combined_69k.sh
Untracked: analysis/DO_morphine1_combined_69k.stderr
Untracked: analysis/DO_morphine1_combined_69k.stdout
Untracked: analysis/DO_morphine1_combined_69k_m2.R
Untracked: analysis/DO_morphine1_combined_69k_m2.Rout
Untracked: analysis/DO_morphine1_combined_69k_m2.sh
Untracked: analysis/DO_morphine1_combined_69k_m2.stderr
Untracked: analysis/DO_morphine1_combined_69k_m2.stdout
Untracked: analysis/DO_morphine1_combined_weight_DOB.R
Untracked: analysis/DO_morphine1_combined_weight_DOB.Rout
Untracked: analysis/DO_morphine1_combined_weight_DOB.err
Untracked: analysis/DO_morphine1_combined_weight_DOB.out
Untracked: analysis/DO_morphine1_combined_weight_DOB.sh
Untracked: analysis/DO_morphine1_combined_weight_DOB.stderr
Untracked: analysis/DO_morphine1_combined_weight_DOB.stdout
Untracked: analysis/DO_morphine1_combined_weight_age.R
Untracked: analysis/DO_morphine1_combined_weight_age.err
Untracked: analysis/DO_morphine1_combined_weight_age.out
Untracked: analysis/DO_morphine1_combined_weight_age.sh
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.R
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.err
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.out
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.sh
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.R
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.err
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.out
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.sh
Untracked: analysis/DO_morphine1_cph.R
Untracked: analysis/DO_morphine1_cph.Rout
Untracked: analysis/DO_morphine1_cph.sh
Untracked: analysis/DO_morphine1_second_set.R
Untracked: analysis/DO_morphine1_second_set.Rout
Untracked: analysis/DO_morphine1_second_set.sh
Untracked: analysis/DO_morphine1_second_set.stderr
Untracked: analysis/DO_morphine1_second_set.stdout
Untracked: analysis/DO_morphine1_second_set_69k.R
Untracked: analysis/DO_morphine1_second_set_69k.Rout
Untracked: analysis/DO_morphine1_second_set_69k.sh
Untracked: analysis/DO_morphine1_second_set_69k.stderr
Untracked: analysis/DO_morphine1_second_set_SNP.R
Untracked: analysis/DO_morphine1_second_set_SNP.Rout
Untracked: analysis/DO_morphine1_second_set_SNP.sh
Untracked: analysis/DO_morphine1_second_set_SNP.stderr
Untracked: analysis/DO_morphine1_second_set_SNP.stdout
Untracked: analysis/DO_morphine1_second_set_weight_DOB.R
Untracked: analysis/DO_morphine1_second_set_weight_DOB.Rout
Untracked: analysis/DO_morphine1_second_set_weight_DOB.err
Untracked: analysis/DO_morphine1_second_set_weight_DOB.out
Untracked: analysis/DO_morphine1_second_set_weight_DOB.sh
Untracked: analysis/DO_morphine1_second_set_weight_DOB.stderr
Untracked: analysis/DO_morphine1_second_set_weight_DOB.stdout
Untracked: analysis/DO_morphine1_second_set_weight_age.R
Untracked: analysis/DO_morphine1_second_set_weight_age.Rout
Untracked: analysis/DO_morphine1_second_set_weight_age.err
Untracked: analysis/DO_morphine1_second_set_weight_age.out
Untracked: analysis/DO_morphine1_second_set_weight_age.sh
Untracked: analysis/DO_morphine1_second_set_weight_age.stderr
Untracked: analysis/DO_morphine1_second_set_weight_age.stdout
Untracked: analysis/DO_morphine1_weight_DOB.R
Untracked: analysis/DO_morphine1_weight_DOB.sh
Untracked: analysis/DO_morphine1_weight_age.R
Untracked: analysis/DO_morphine1_weight_age.sh
Untracked: analysis/DO_morphine_gemma.R
Untracked: analysis/DO_morphine_gemma.err
Untracked: analysis/DO_morphine_gemma.out
Untracked: analysis/DO_morphine_gemma.sh
Untracked: analysis/DO_morphine_gemma_firstmin.R
Untracked: analysis/DO_morphine_gemma_firstmin.err
Untracked: analysis/DO_morphine_gemma_firstmin.out
Untracked: analysis/DO_morphine_gemma_firstmin.sh
Untracked: analysis/DO_morphine_gemma_withpermu.R
Untracked: analysis/DO_morphine_gemma_withpermu.err
Untracked: analysis/DO_morphine_gemma_withpermu.out
Untracked: analysis/DO_morphine_gemma_withpermu.sh
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.R
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.err
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.out
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.sh
Untracked: analysis/Plot_DO_morphine1_SNP.R
Untracked: analysis/Plot_DO_morphine1_SNP.Rout
Untracked: analysis/Plot_DO_morphine1_SNP.sh
Untracked: analysis/Plot_DO_morphine1_SNP.stderr
Untracked: analysis/Plot_DO_morphine1_SNP.stdout
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.R
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.Rout
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.sh
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.stderr
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.stdout
Untracked: analysis/download_GSE100356_sra.sh
Untracked: analysis/fentanyl_2cohorts_coxph.R
Untracked: analysis/fentanyl_2cohorts_coxph.err
Untracked: analysis/fentanyl_2cohorts_coxph.out
Untracked: analysis/fentanyl_2cohorts_coxph.sh
Untracked: analysis/fentanyl_scanone.cph.R
Untracked: analysis/fentanyl_scanone.cph.err
Untracked: analysis/fentanyl_scanone.cph.out
Untracked: analysis/fentanyl_scanone.cph.sh
Untracked: analysis/geo_rnaseq.R
Untracked: analysis/heritability_first_second_batch.R
Untracked: analysis/nf-rnaseq-b6.R
Untracked: analysis/plot_fentanyl_2cohorts_coxph.R
Untracked: analysis/scripts/
Untracked: analysis/tibmr.R
Untracked: analysis/timbr_demo.R
Untracked: analysis/workflow_proc.R
Untracked: analysis/workflow_proc.sh
Untracked: analysis/workflow_proc.stderr
Untracked: analysis/workflow_proc.stdout
Untracked: analysis/x.R
Untracked: code/cfw/
Untracked: code/gemma_plot.R
Untracked: code/process.sanger.snp.R
Untracked: code/reconst_utils.R
Untracked: data/69k_grid_pgmap.RData
Untracked: data/Composite Post Kevins Program Group 2 Fentanyl Prepped for Hao.xlsx
Untracked: data/DO_WBP_Data_JAB_to_map.xlsx
Untracked: data/Fentanyl_alternate_metrics.xlsx
Untracked: data/FinalReport/
Untracked: data/GM/
Untracked: data/GM_covar.csv
Untracked: data/GM_covar_07092018_morphine.csv
Untracked: data/Jackson_Lab_Bubier_MURGIGV01/
Untracked: data/MPD_Upload_October.csv
Untracked: data/MPD_Upload_October_updated_sex.csv
Untracked: data/Master Fentanyl DO Study Sheet.xlsx
Untracked: data/MasterMorphine Second Set DO w DOB2.xlsx
Untracked: data/MasterMorphine Second Set DO.xlsx
Untracked: data/Morphine CC DO mice Updated with Published inbred strains.csv
Untracked: data/Morphine_CC_DO_mice_Updated_with_Published_inbred_strains.csv
Untracked: data/cc_variants.sqlite
Untracked: data/combined/
Untracked: data/fentanyl/
Untracked: data/fentanyl2/
Untracked: data/fentanyl_1_2/
Untracked: data/fentanyl_2cohorts_coxph_data.Rdata
Untracked: data/first/
Untracked: data/founder_geno.csv
Untracked: data/genetic_map.csv
Untracked: data/gm.json
Untracked: data/gwas.sh
Untracked: data/marker_grid_0.02cM_plus.txt
Untracked: data/mouse_genes_mgi.sqlite
Untracked: data/pheno.csv
Untracked: data/pheno_qtl2.csv
Untracked: data/pheno_qtl2_07092018_morphine.csv
Untracked: data/pheno_qtl2_w_dob.csv
Untracked: data/physical_map.csv
Untracked: data/rnaseq/
Untracked: data/sample_geno.csv
Untracked: data/second/
Untracked: figure/
Untracked: glimma-plots/
Untracked: output/DO_Fentanyl_Cohort2_MinDepressionRR_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_MinDepressionRR_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_RRDepressionRateHrSLOPE_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_RRRecoveryRateHrSLOPE_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_RRRecoveryRateHrSLOPE_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_StartofRecoveryHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_StartofRecoveryHr_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_Statusbin_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_Statusbin_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_SteadyStateDepressionDurationHrINTERVAL_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoDead(Hr)_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoDeadHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoDeadHr_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoProjectedRecoveryHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoProjectedRecoveryHr_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoSteadyRRDepression(Hr)_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoSteadyRRDepressionHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoSteadyRRDepressionHr_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoThresholdRecoveryHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoThresholdRecoveryHr_coefplot_blup.pdf
Untracked: output/DO_morphine_Min.depression.png
Untracked: output/DO_morphine_Min.depression22222_violin_chr5.pdf
Untracked: output/DO_morphine_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_Min.depression_coefplot_blup_chr5.png
Untracked: output/DO_morphine_Min.depression_coefplot_blup_chrX.png
Untracked: output/DO_morphine_Min.depression_coefplot_chr5.png
Untracked: output/DO_morphine_Min.depression_coefplot_chrX.png
Untracked: output/DO_morphine_Min.depression_peak_genes_chr5.png
Untracked: output/DO_morphine_Min.depression_violin_chr5.png
Untracked: output/DO_morphine_Recovery.Time.png
Untracked: output/DO_morphine_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr11.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr4.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr7.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr9.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr11.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr4.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr7.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr9.png
Untracked: output/DO_morphine_Status_bin.png
Untracked: output/DO_morphine_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_Survival.Time.png
Untracked: output/DO_morphine_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_Survival.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_Survival.Time_coefplot_blup_chr17.png
Untracked: output/DO_morphine_Survival.Time_coefplot_blup_chr8.png
Untracked: output/DO_morphine_Survival.Time_coefplot_chr17.png
Untracked: output/DO_morphine_Survival.Time_coefplot_chr8.png
Untracked: output/DO_morphine_combine_batch_peak_violin.pdf
Untracked: output/DO_morphine_combined_69k_m2_Min.depression.png
Untracked: output/DO_morphine_combined_69k_m2_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_combined_69k_m2_Recovery.Time.png
Untracked: output/DO_morphine_combined_69k_m2_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_combined_69k_m2_Status_bin.png
Untracked: output/DO_morphine_combined_69k_m2_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_combined_69k_m2_Survival.Time.png
Untracked: output/DO_morphine_combined_69k_m2_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Survival.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_coxph_24hrs_kinship_QTL.png
Untracked: output/DO_morphine_cphout.RData
Untracked: output/DO_morphine_first_batch_peak_in_second_batch_violin.pdf
Untracked: output/DO_morphine_first_batch_peak_in_second_batch_violin_sidebyside.pdf
Untracked: output/DO_morphine_first_batch_peak_violin.pdf
Untracked: output/DO_morphine_operm.cph.RData
Untracked: output/DO_morphine_second_batch_on_first_batch_peak_violin.pdf
Untracked: output/DO_morphine_second_batch_peak_ch6surv_on_first_batchviolin.pdf
Untracked: output/DO_morphine_second_batch_peak_ch6surv_on_first_batchviolin2.pdf
Untracked: output/DO_morphine_second_batch_peak_in_first_batch_violin.pdf
Untracked: output/DO_morphine_second_batch_peak_in_first_batch_violin_sidebyside.pdf
Untracked: output/DO_morphine_second_batch_peak_violin.pdf
Untracked: output/DO_morphine_secondbatch_69k_Min.depression.png
Untracked: output/DO_morphine_secondbatch_69k_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_69k_Recovery.Time.png
Untracked: output/DO_morphine_secondbatch_69k_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_69k_Status_bin.png
Untracked: output/DO_morphine_secondbatch_69k_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_69k_Survival.Time.png
Untracked: output/DO_morphine_secondbatch_69k_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Survival.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Min.depression.png
Untracked: output/DO_morphine_secondbatch_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Recovery.Time.png
Untracked: output/DO_morphine_secondbatch_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Status_bin.png
Untracked: output/DO_morphine_secondbatch_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Survival.Time.png
Untracked: output/DO_morphine_secondbatch_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Survival.Time_coefplot_blup.pdf
Untracked: output/Fentanyl/
Untracked: output/KPNA3.pdf
Untracked: output/SSC4D.pdf
Untracked: output/TIMBR.test.RData
Untracked: output/apr_69kchr_combined.RData
Untracked: output/apr_69kchr_k_loco_combined.rds
Untracked: output/apr_69kchr_second_set.RData
Untracked: output/combine_batch_variation.RData
Untracked: output/combined_gm.RData
Untracked: output/combined_gm.k_loco.rds
Untracked: output/combined_gm.k_overall.rds
Untracked: output/combined_gm.probs_8state.rds
Untracked: output/coxph/
Untracked: output/do.morphine.RData
Untracked: output/do.morphine.k_loco.rds
Untracked: output/do.morphine.probs_36state.rds
Untracked: output/do.morphine.probs_8state.rds
Untracked: output/do_Fentanyl_combine2cohort_MeanDepressionBR_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_MeanDepressionBR_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_MinDepressionBR_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_MinDepressionBR_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_MinDepressionRR_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_MinDepressionRR_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_RRRecoveryRateHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_RRRecoveryRateHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_StartofRecoveryHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_StartofRecoveryHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_Statusbin_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_Statusbin_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_SteadyStateDepressionDurationHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_SteadyStateDepressionDurationHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_SurvivalTime_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_SurvivalTime_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoDeadHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoDeadHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoMostlyDeadHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoMostlyDeadHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoProjectedRecoveryHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoProjectedRecoveryHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoRecoveryHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoRecoveryHr_coefplot_blup.pdf
Untracked: output/first_batch_variation.RData
Untracked: output/first_second_survival_peak_chr.xlsx
Untracked: output/hsq_1_first_batch_herit_qtl2.RData
Untracked: output/hsq_2_second_batch_herit_qtl2.RData
Untracked: output/old_temp/
Untracked: output/pr_69kchr_combined.RData
Untracked: output/pr_69kchr_second_set.RData
Untracked: output/qtl.morphine.69k.out.combined.RData
Untracked: output/qtl.morphine.69k.out.combined_m2.RData
Untracked: output/qtl.morphine.69k.out.second_set.RData
Untracked: output/qtl.morphine.operm.RData
Untracked: output/qtl.morphine.out.RData
Untracked: output/qtl.morphine.out.combined_gm.RData
Untracked: output/qtl.morphine.out.combined_gm.female.RData
Untracked: output/qtl.morphine.out.combined_gm.male.RData
Untracked: output/qtl.morphine.out.combined_weight_DOB.RData
Untracked: output/qtl.morphine.out.combined_weight_age.RData
Untracked: output/qtl.morphine.out.female.RData
Untracked: output/qtl.morphine.out.male.RData
Untracked: output/qtl.morphine.out.second_set.RData
Untracked: output/qtl.morphine.out.second_set.female.RData
Untracked: output/qtl.morphine.out.second_set.male.RData
Untracked: output/qtl.morphine.out.second_set.weight_DOB.RData
Untracked: output/qtl.morphine.out.second_set.weight_age.RData
Untracked: output/qtl.morphine.out.weight_DOB.RData
Untracked: output/qtl.morphine.out.weight_age.RData
Untracked: output/qtl.morphine1.snpout.RData
Untracked: output/qtl.morphine2.snpout.RData
Untracked: output/second_batch_pheno.csv
Untracked: output/second_batch_variation.RData
Untracked: output/second_set_apr_69kchr_k_loco.rds
Untracked: output/second_set_gm.RData
Untracked: output/second_set_gm.k_loco.rds
Untracked: output/second_set_gm.probs_36state.rds
Untracked: output/second_set_gm.probs_8state.rds
Untracked: output/topSNP_chr5_mindepression.csv
Untracked: output/zoompeak_Min.depression_9.pdf
Untracked: output/zoompeak_Recovery.Time_16.pdf
Untracked: output/zoompeak_Status_bin_11.pdf
Untracked: output/zoompeak_Survival.Time_1.pdf
Untracked: output/zoompeak_fentanyl_Survival.Time_2.pdf
Untracked: sra-tools_v2.10.7.sif
Unstaged changes:
Modified: _workflowr.yml
Modified: analysis/Plot_DO_Fentanyl_Cohort2_mapping.Rmd
Modified: analysis/Plot_DO_Fentanyl_combining2Cohort_mapping.Rmd
Modified: analysis/Plot_DO_fentanyl.Rmd
Modified: analysis/marker_violin.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/Plot_DO_morphine.Rmd) and HTML (docs/Plot_DO_morphine.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | cc3ae27 | xhyuo | 2023-02-21 | update with sex specific morphine plot |
| html | 8fe8543 | xhyuo | 2023-02-20 | Build site. |
| Rmd | b46775c | xhyuo | 2023-02-20 | update with sex specific morphine |
| html | 422da68 | xhyuo | 2020-07-12 | Build site. |
| html | c9ed9ff | xhyuo | 2020-05-29 | Build site. |
| Rmd | 3277c6d | xhyuo | 2020-05-29 | Plot_DO_morphine at helix |
Last update: 2023-02-21
loading libraries
library(ggplot2)
library(gridExtra)
library(GGally)
library(parallel)
library(qtl2)
library(parallel)
library(survival)
library(regress)
library(abind)
rz.transform <- function(y) {
rankY=rank(y, ties.method="average", na.last="keep")
rzT=qnorm(rankY/(length(na.exclude(rankY))+1))
return(rzT)
}
load("code/do.colors.RData")###Read phenotype data
do.pheno <- read.csv("data/pheno_qtl2.csv")
do.pheno$Survival.Time <- as.numeric(do.pheno$Survival.Time)
do.pheno$Recovery.Time <- as.numeric(do.pheno$Recovery.Time)
do.pheno$Min.depression <- as.numeric(do.pheno$Min.depression)
#histogram
a <- ggplot(data=do.pheno, aes(do.pheno$Survival.Time)) +
geom_histogram() +
ylab("Number of DO mice") + xlab("Survival Time (h)")
b <- ggplot(data=do.pheno, aes(do.pheno$Recovery.Time)) +
geom_histogram() +
ylab("Number of DO mice") + xlab("Recovery Time (h)")
c <- ggplot(data=do.pheno, aes(do.pheno$Min.depression)) +
geom_histogram() +
ylab("Number of DO mice") + xlab("Respiratory Depression (% of baseline)")
grid.arrange(a,b,c)
# Warning: Use of `do.pheno$Survival.Time` is discouraged. Use `Survival.Time`
# instead.
# Warning: Removed 198 rows containing non-finite values (stat_bin).
# Warning: Use of `do.pheno$Recovery.Time` is discouraged. Use `Recovery.Time`
# instead.
# Warning: Removed 121 rows containing non-finite values (stat_bin).
# Warning: Use of `do.pheno$Min.depression` is discouraged. Use `Min.depression`
# instead.
# Warning: Removed 27 rows containing non-finite values (stat_bin).
d <- ggpairs(do.pheno[,2:4])
d
# Warning: Removed 198 rows containing non-finite values (stat_density).
# Warning in ggally_statistic(data = data, mapping = mapping, na.rm = na.rm, :
# Removed 215 rows containing missing values
# Warning: Removed 121 rows containing non-finite values (stat_density).
# Warning in ggally_statistic(data = data, mapping = mapping, na.rm = na.rm, :
# Removed 124 rows containing missing values
# Warning: Removed 215 rows containing missing values (geom_point).
# Warning: Removed 124 rows containing missing values (geom_point).
# Warning: Removed 27 rows containing non-finite values (stat_density).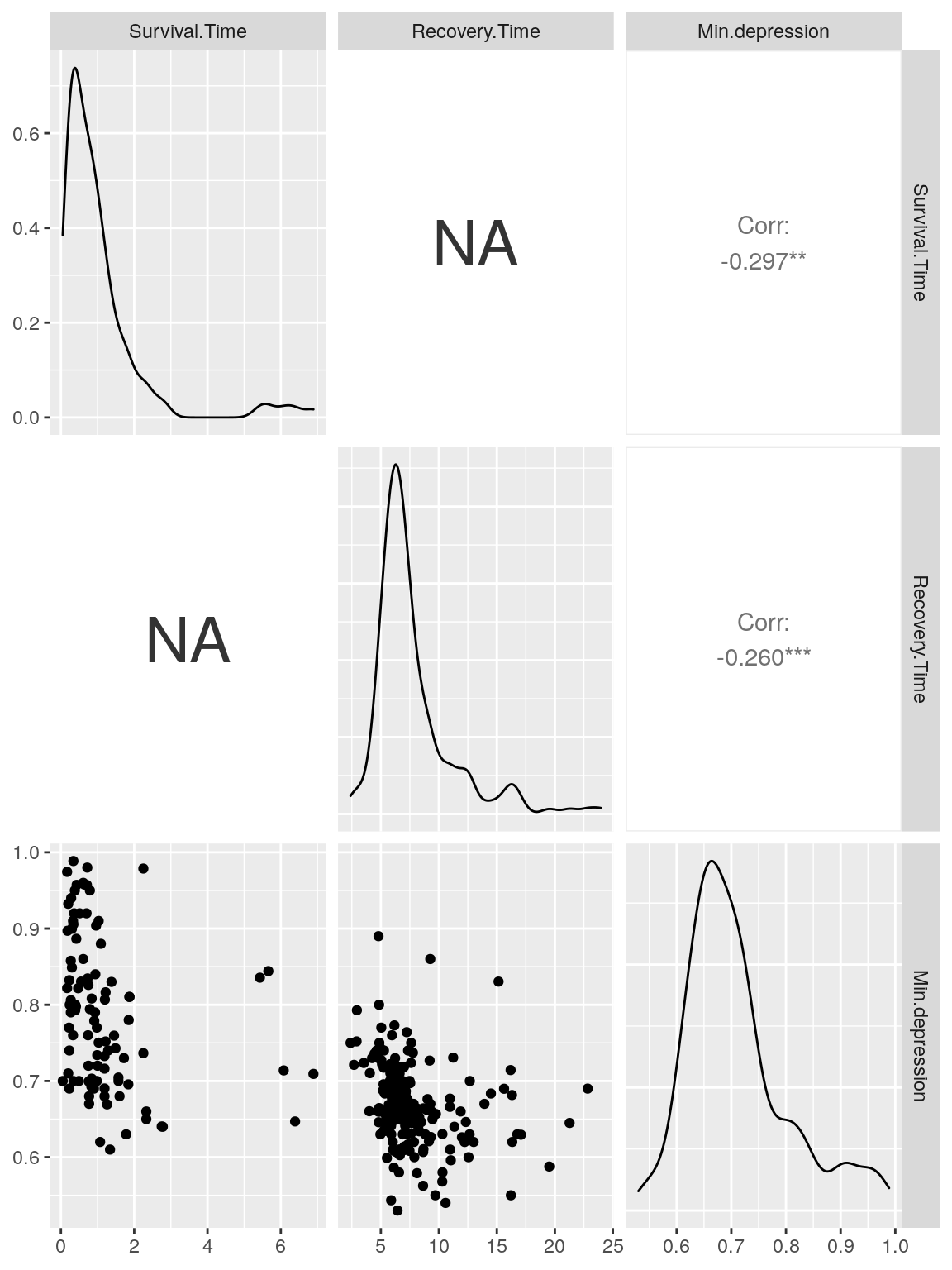
Plot qtl mapping
DO_morphine1.sh/DO_morphine1.R performs qtl mapping for DO mice in DO Opioid project. Plot by using its output qtl.morphine.out.RData
load("output/do.morphine.RData")
load("output/qtl.morphine.out.RData")
load("output/qtl.morphine.operm.RData") #the first permutation
#pmap and gmap
chr.names <- c("1","2","3","4","5","6","7","8","9","10","11","12","13","14","15","16","17","18","19", "X")
gmap <- list()
pmap <- list()
for (chr in chr.names){
gmap[[chr]] <- do.morphine$gmap[[chr]]
pmap[[chr]] <- do.morphine$pmap[[chr]]
}
attr(gmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
attr(pmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
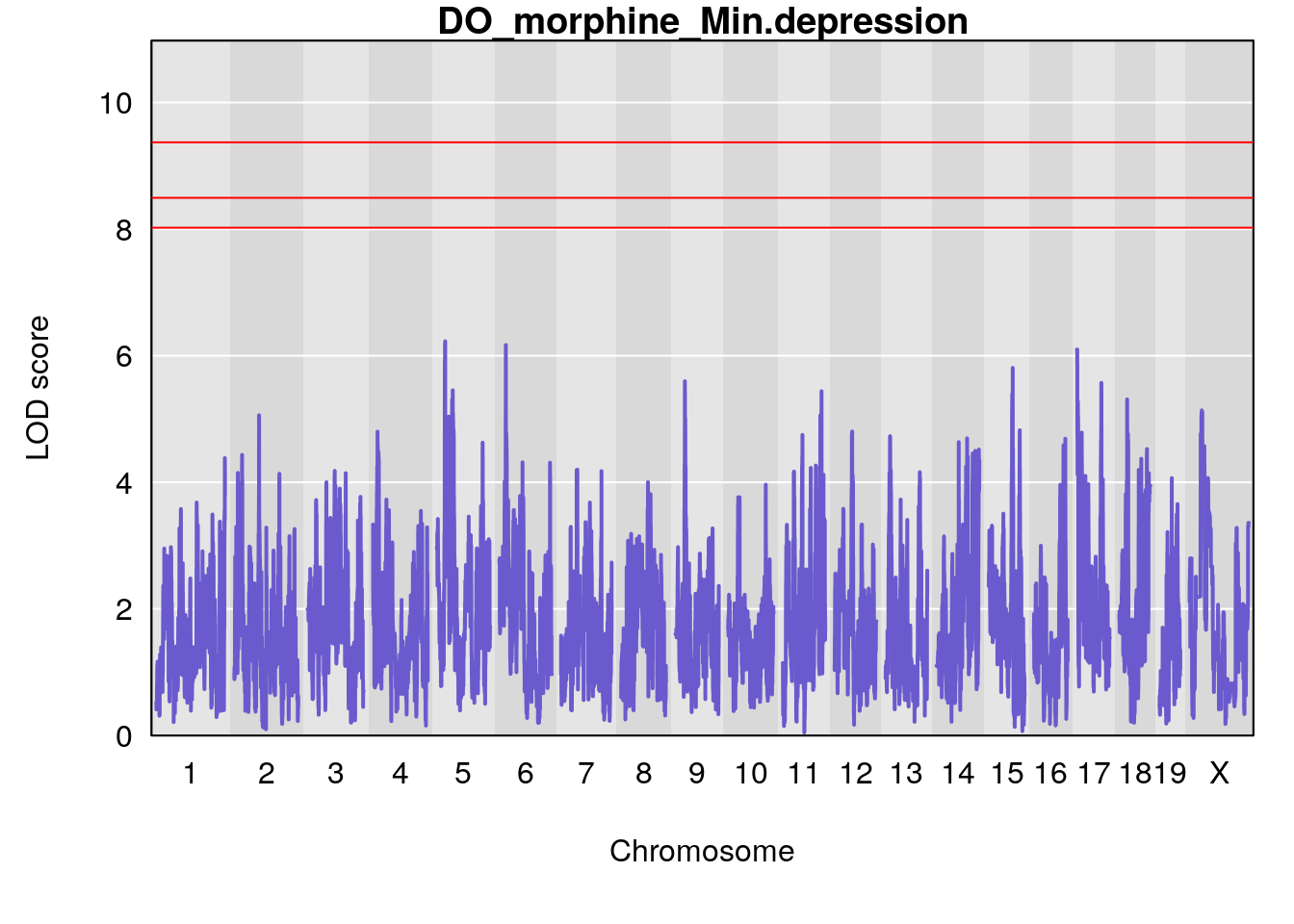
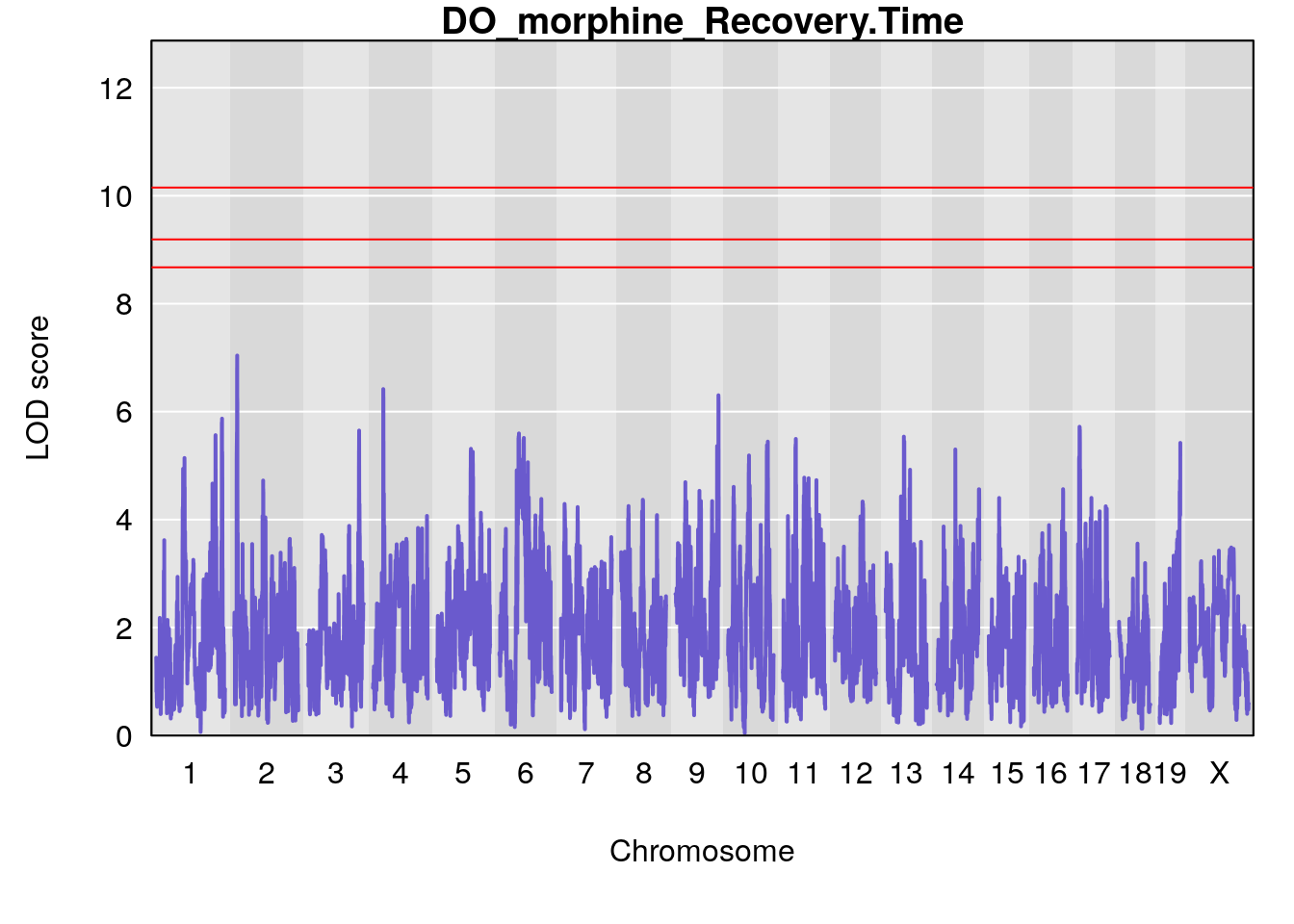
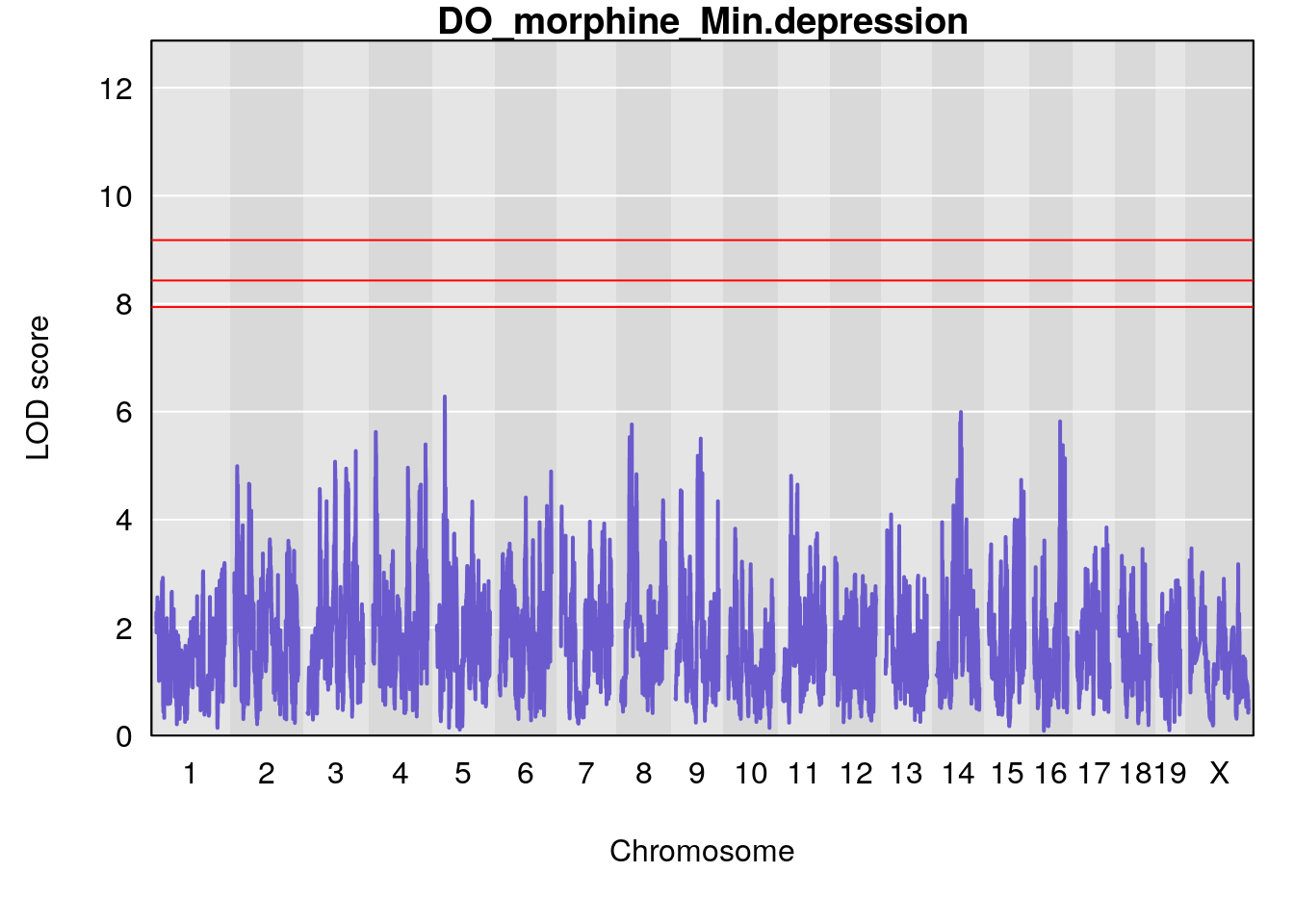
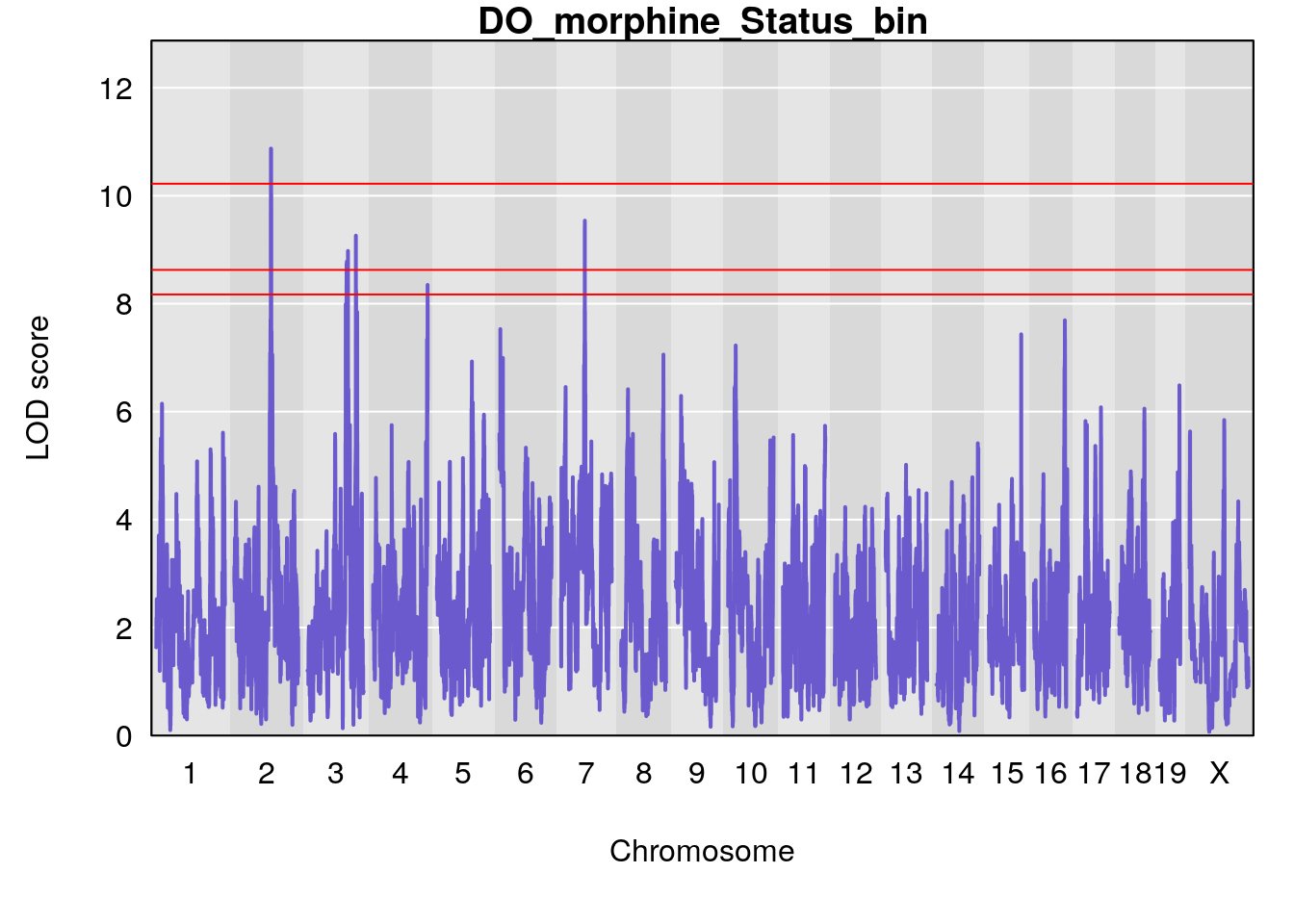
#genome-wide plot
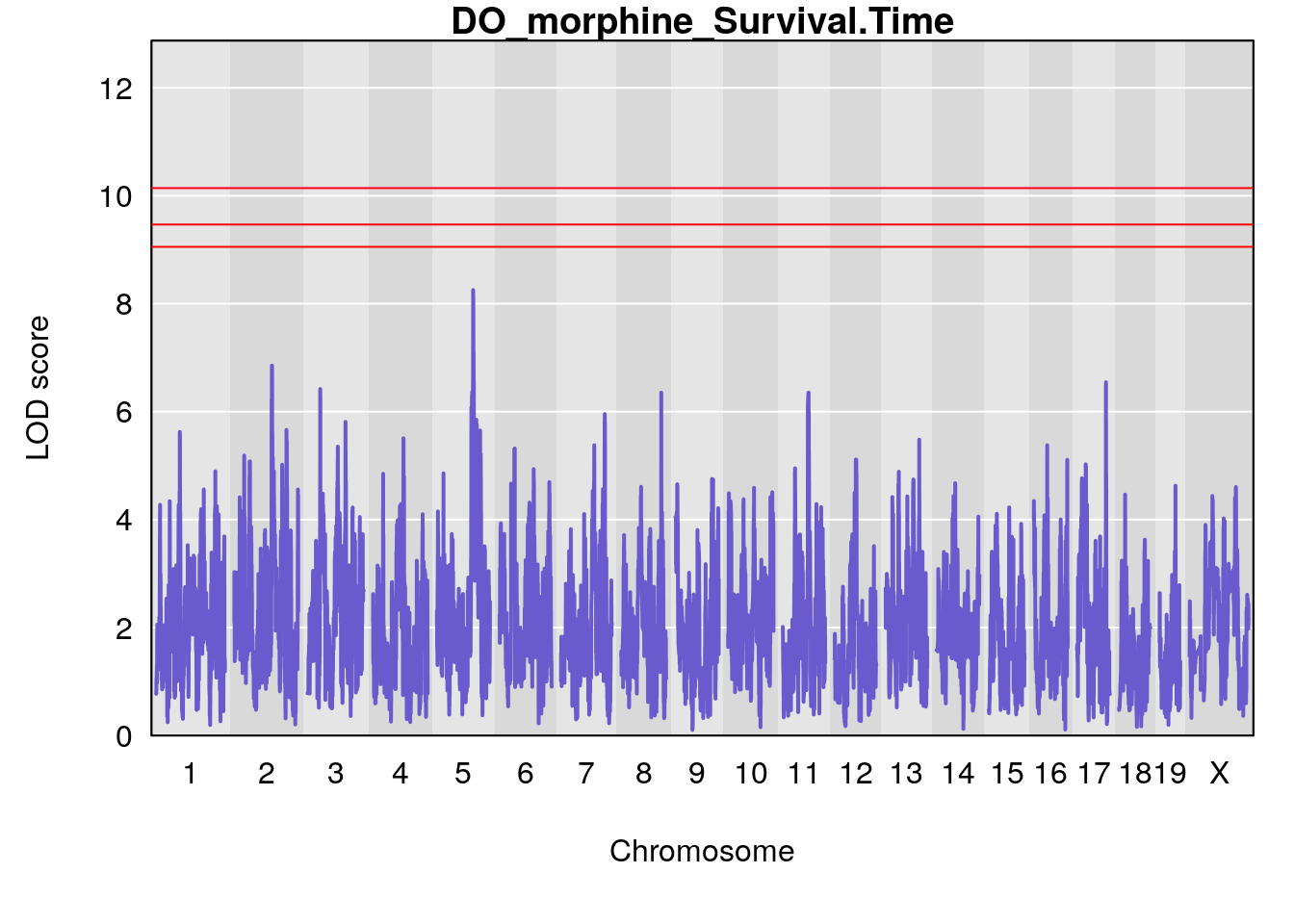
for(i in names(qtl.morphine.out)){
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.morphine.out[[i]]) # overall maximum LOD score
plot(qtl.morphine.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, 10))
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("DO_morphine_",i))
}
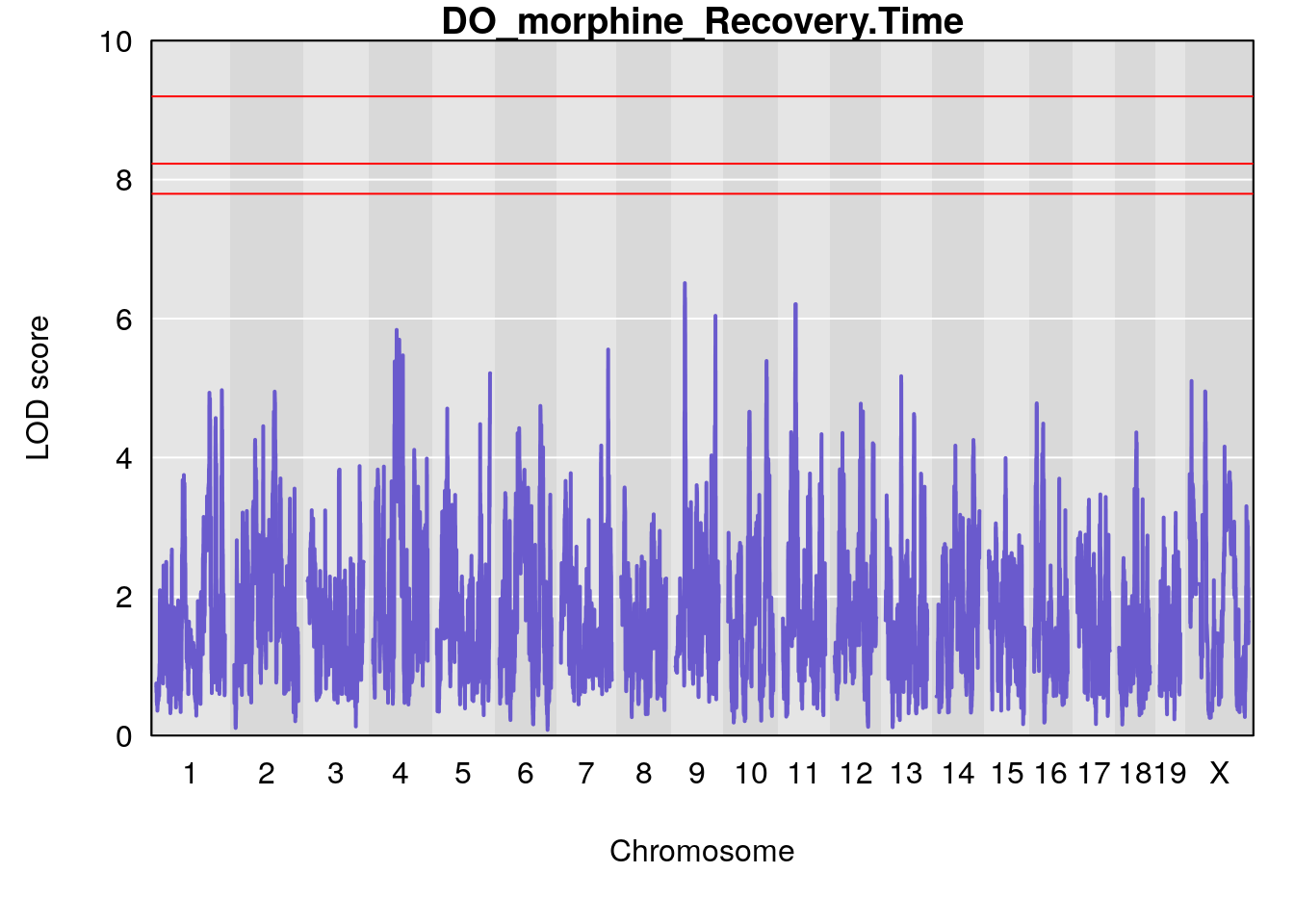

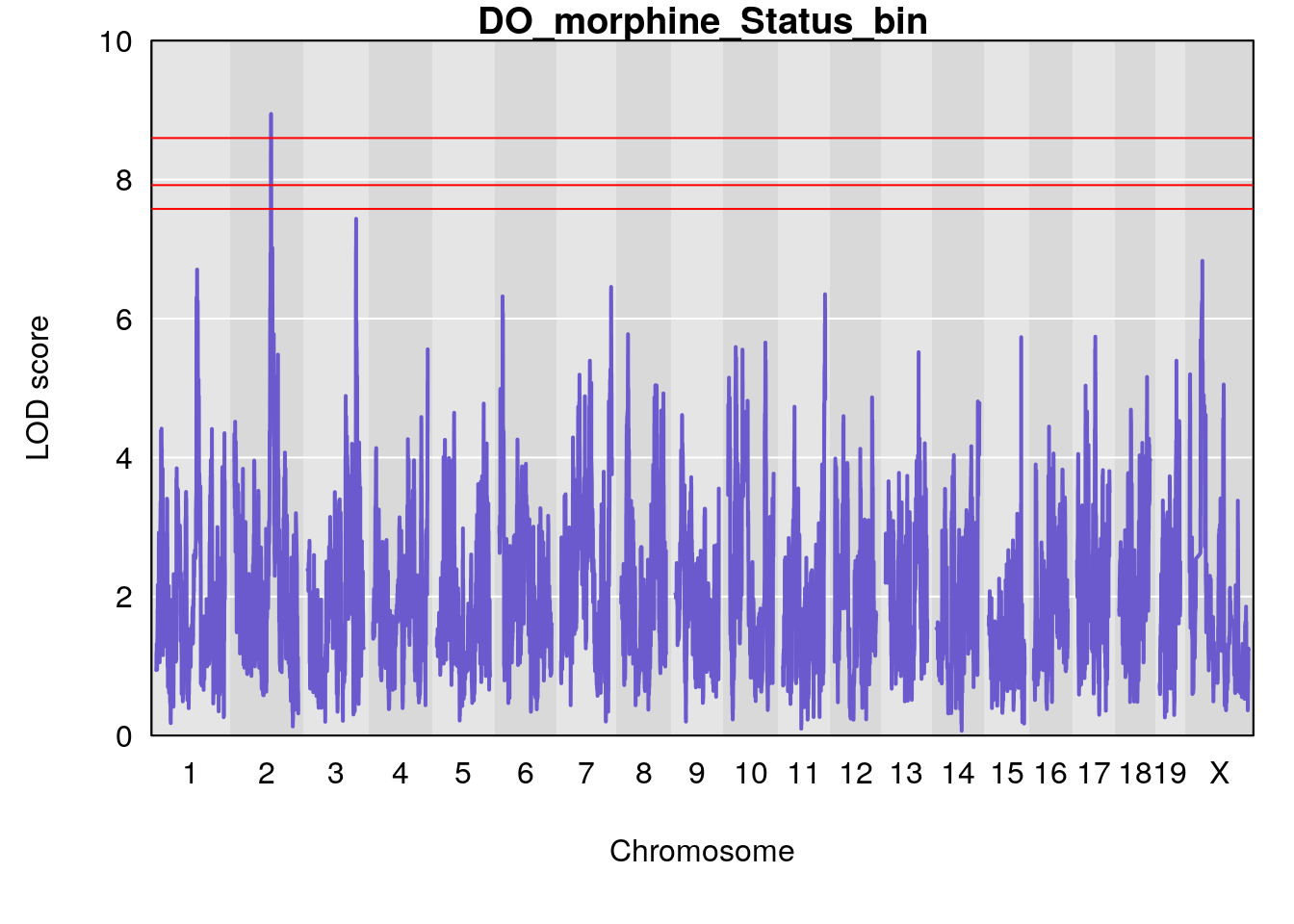
#save genome-wide plot
for(i in names(qtl.morphine.out)){
png(file = paste0("output/DO_morphine_", i, ".png"), width = 16, height =8, res=300, units = "in")
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.morphine.out[[i]]) # overall maximum LOD score
plot(qtl.morphine.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, 10))
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("DO_morphine_",i))
dev.off()
}
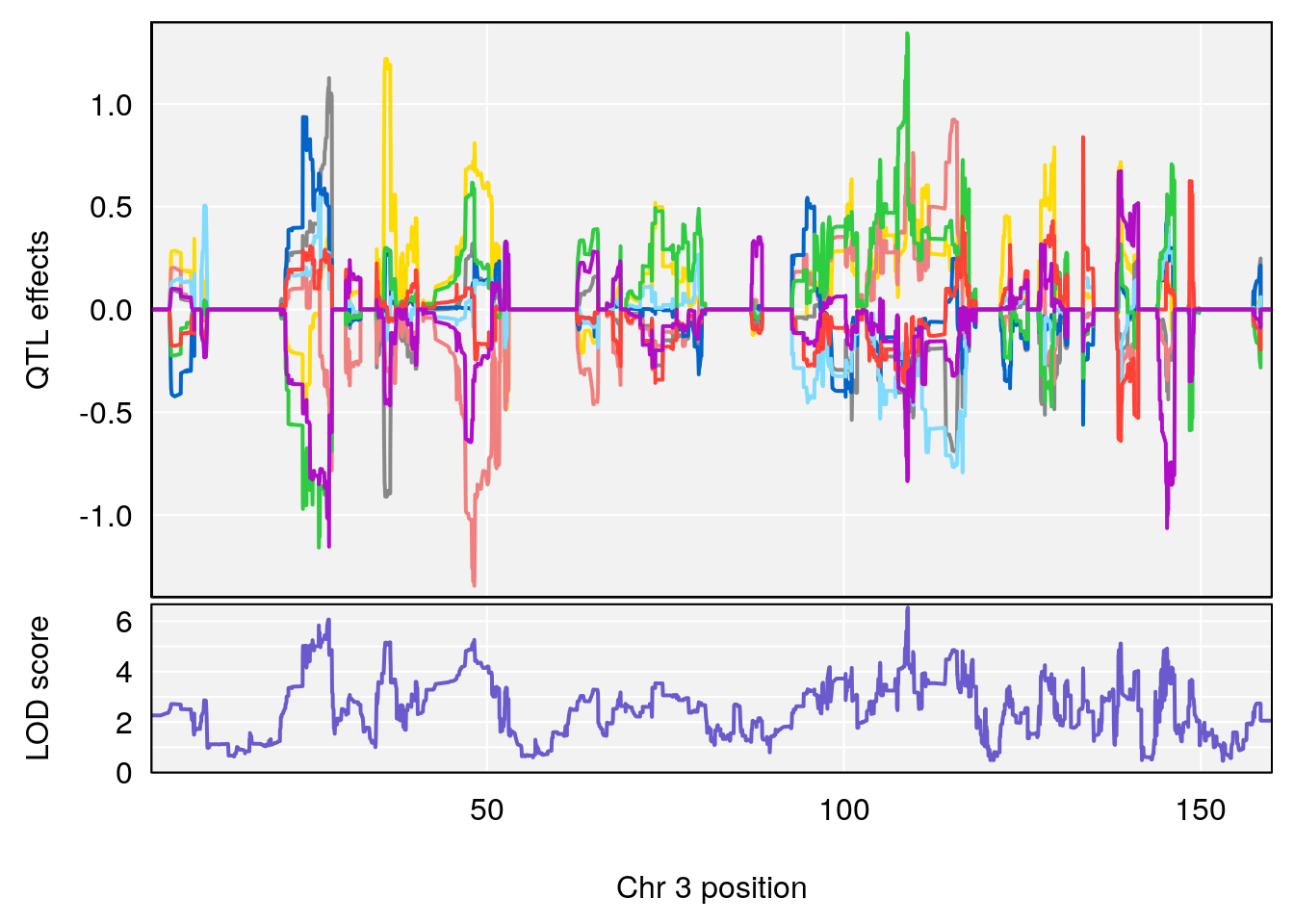
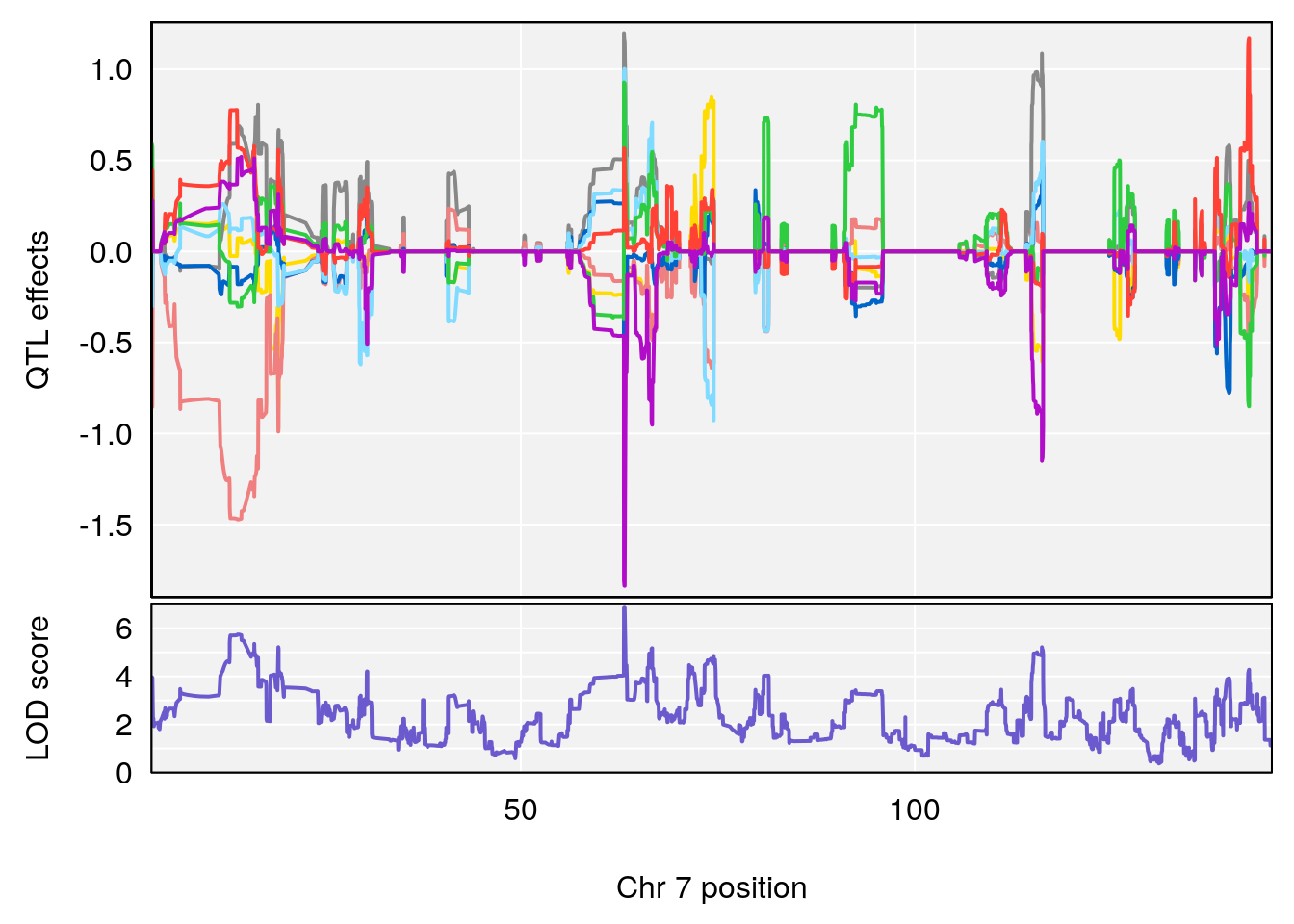
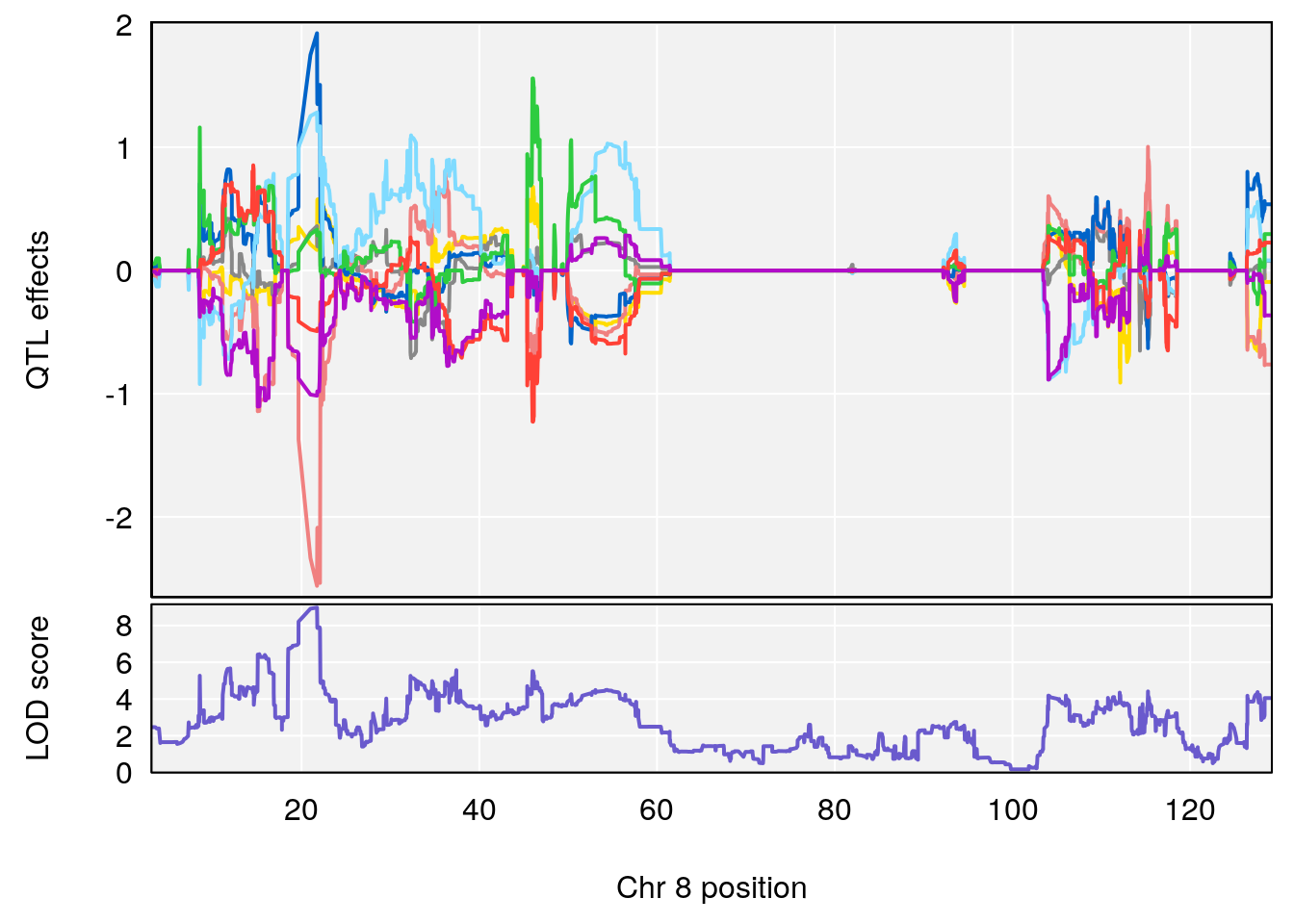
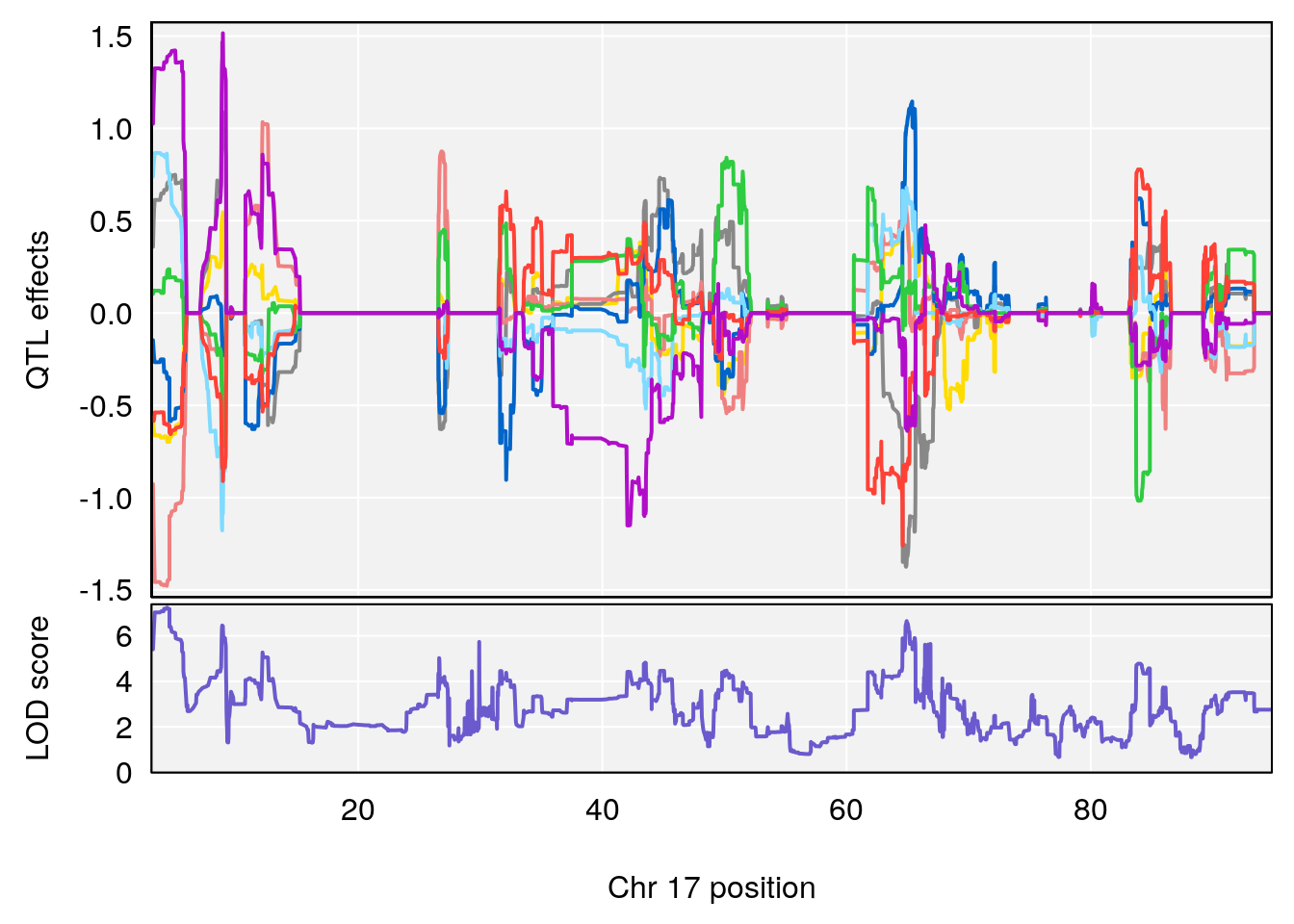
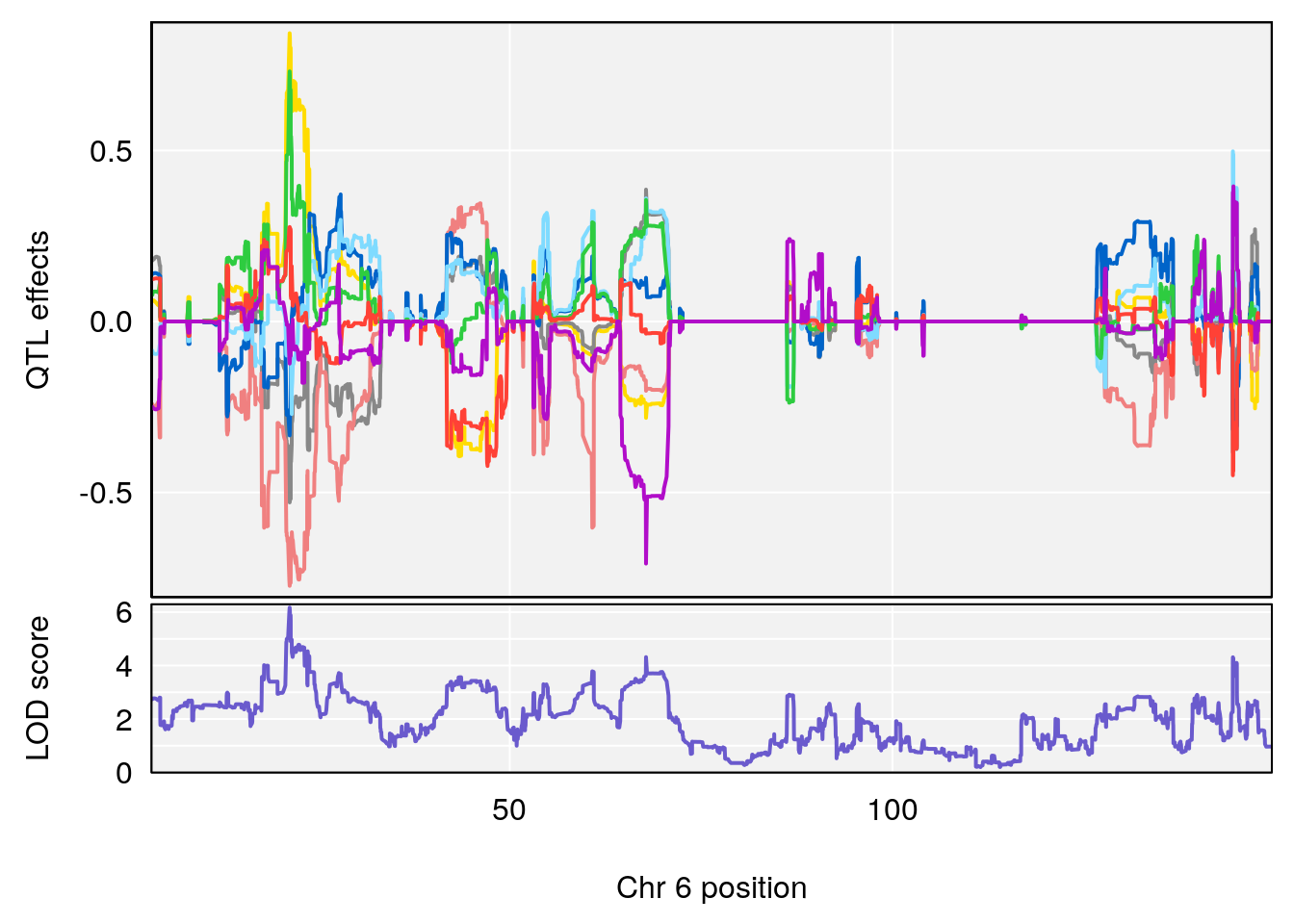
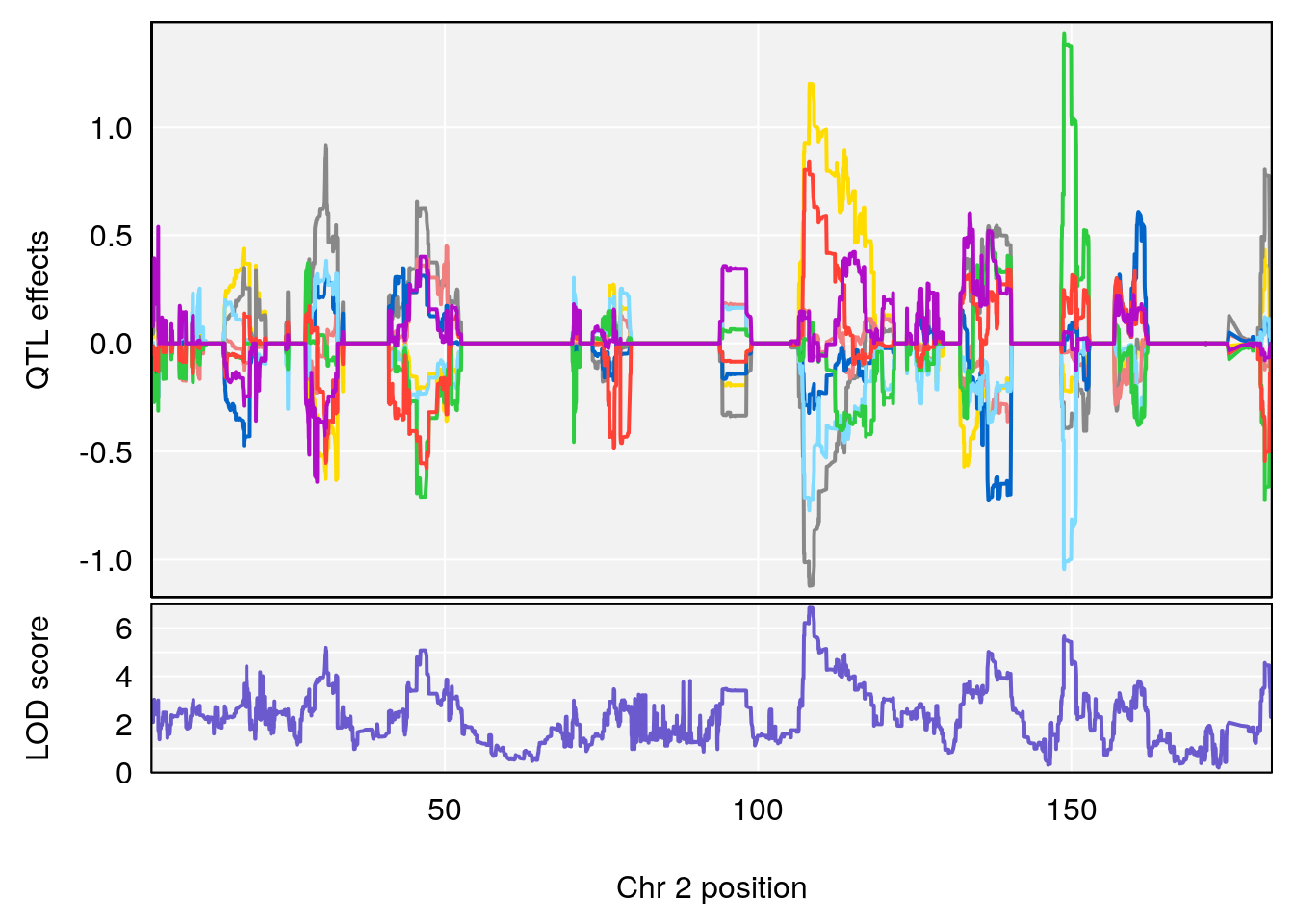
#peaks coeff plot
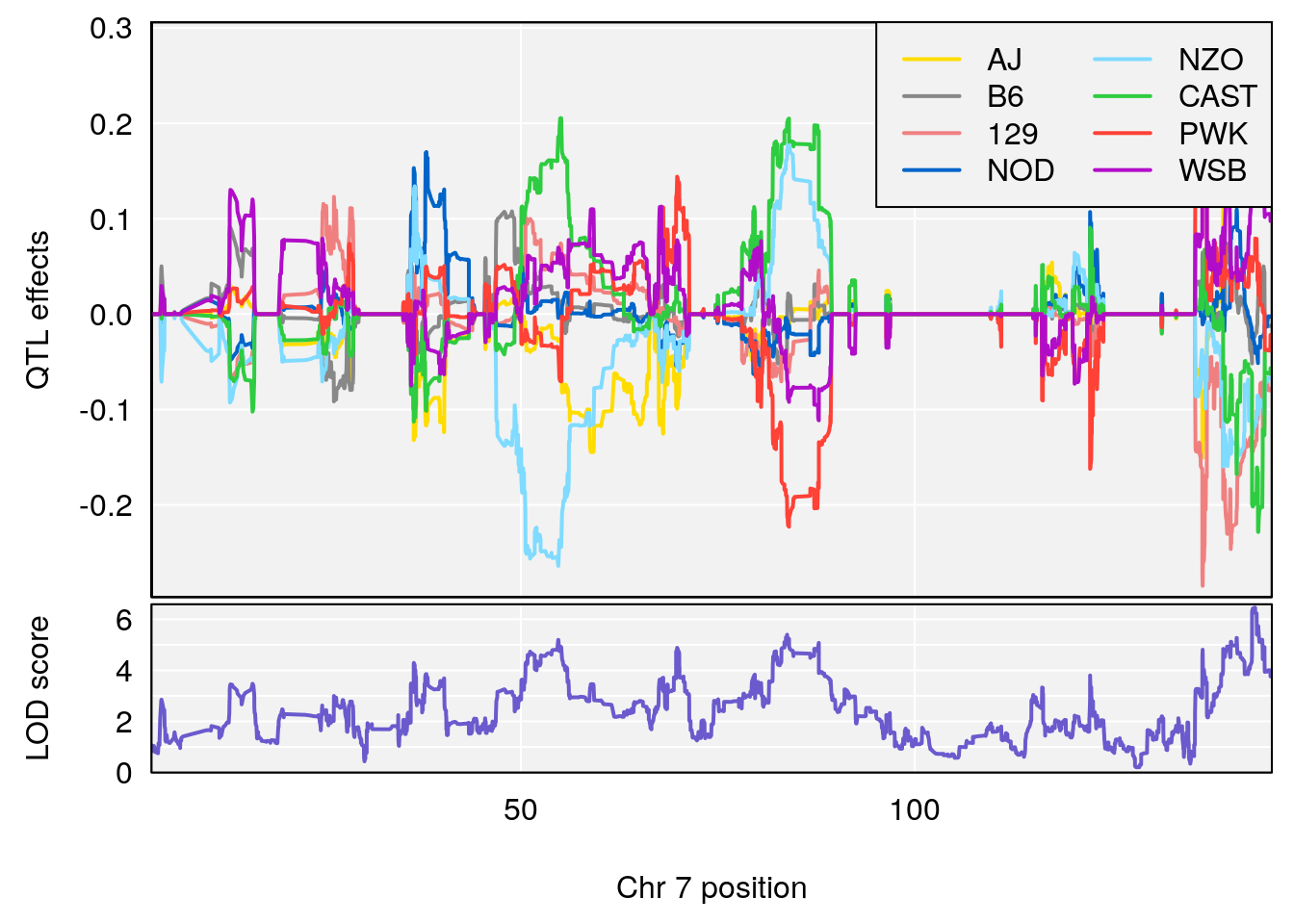
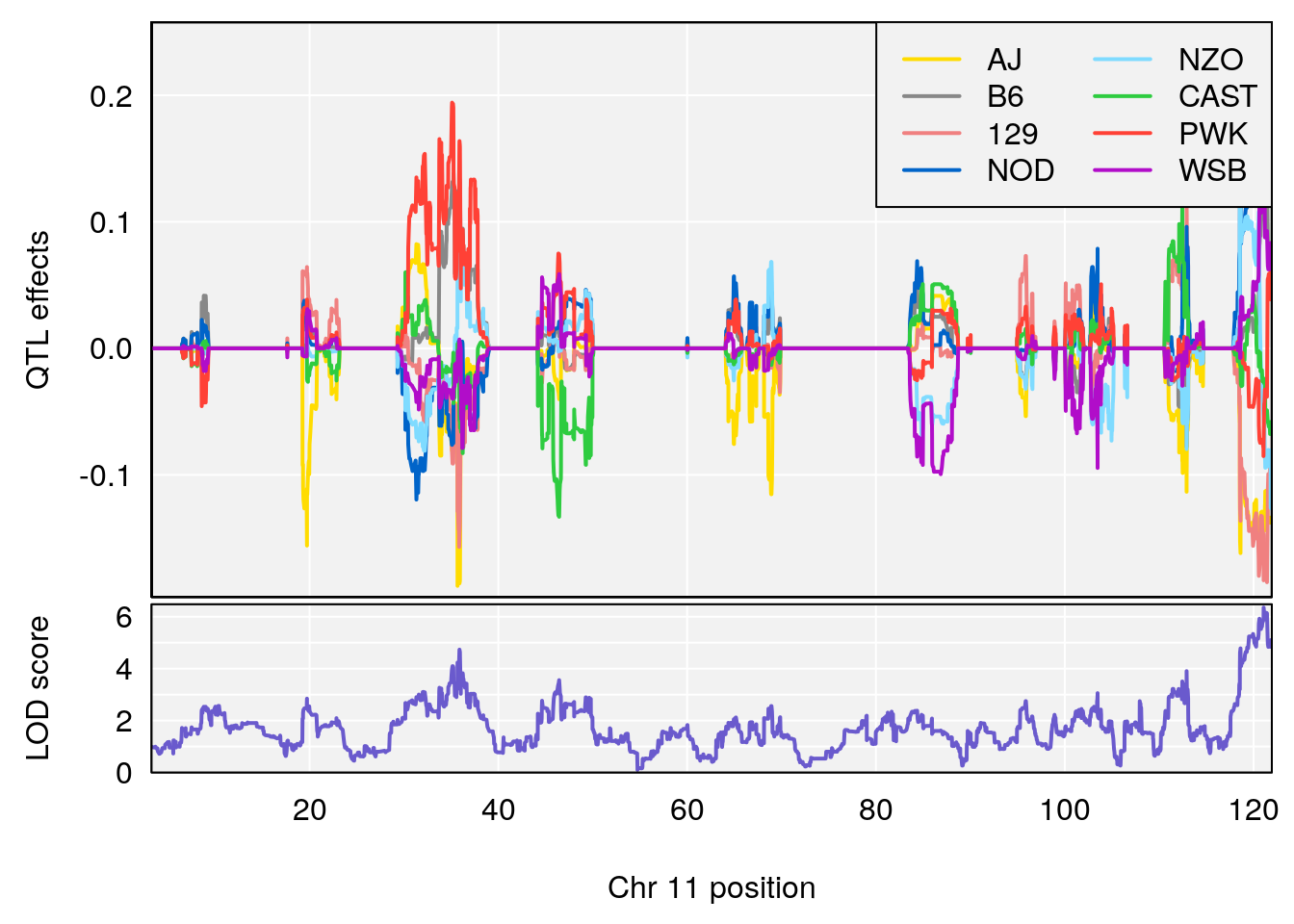
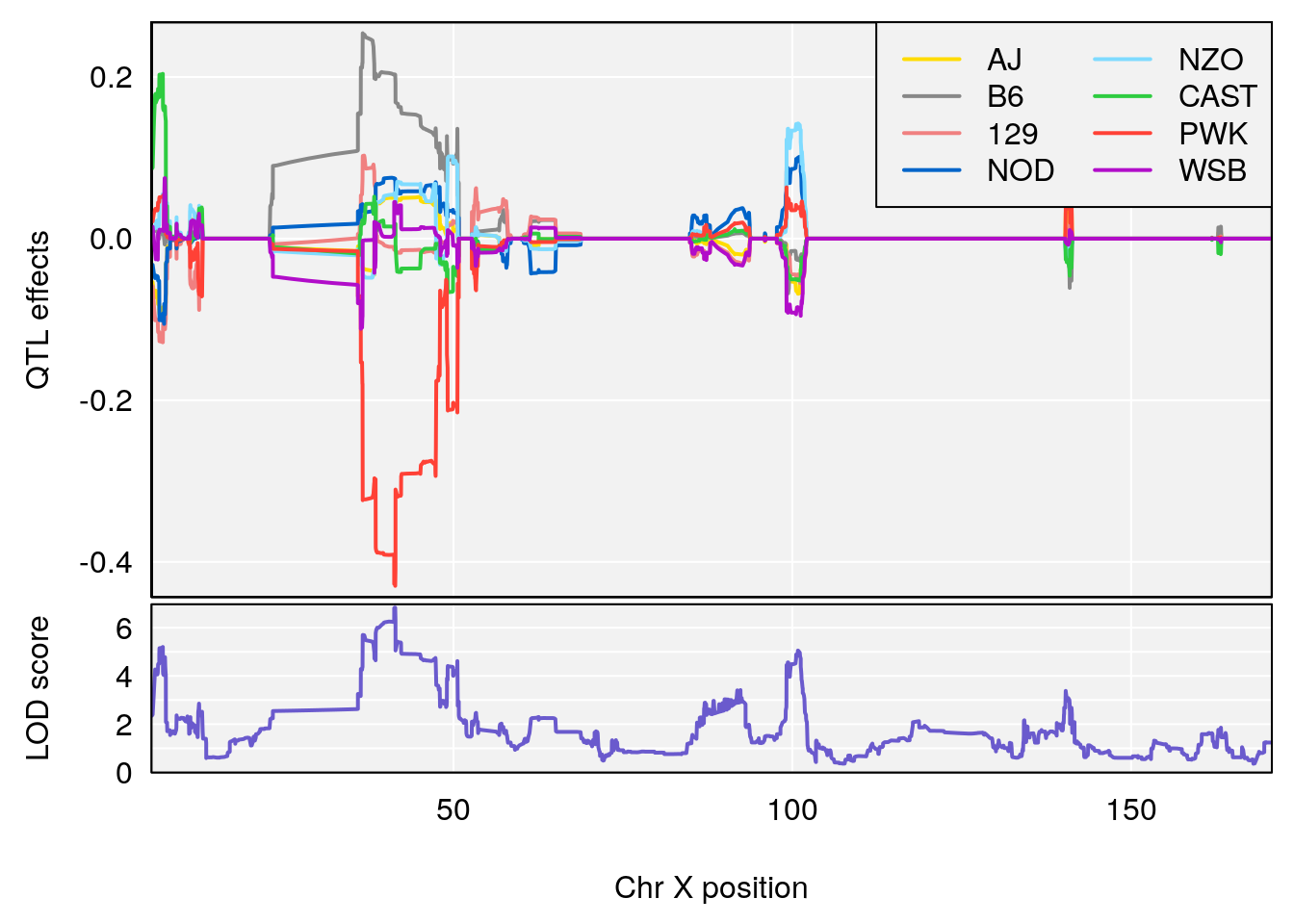
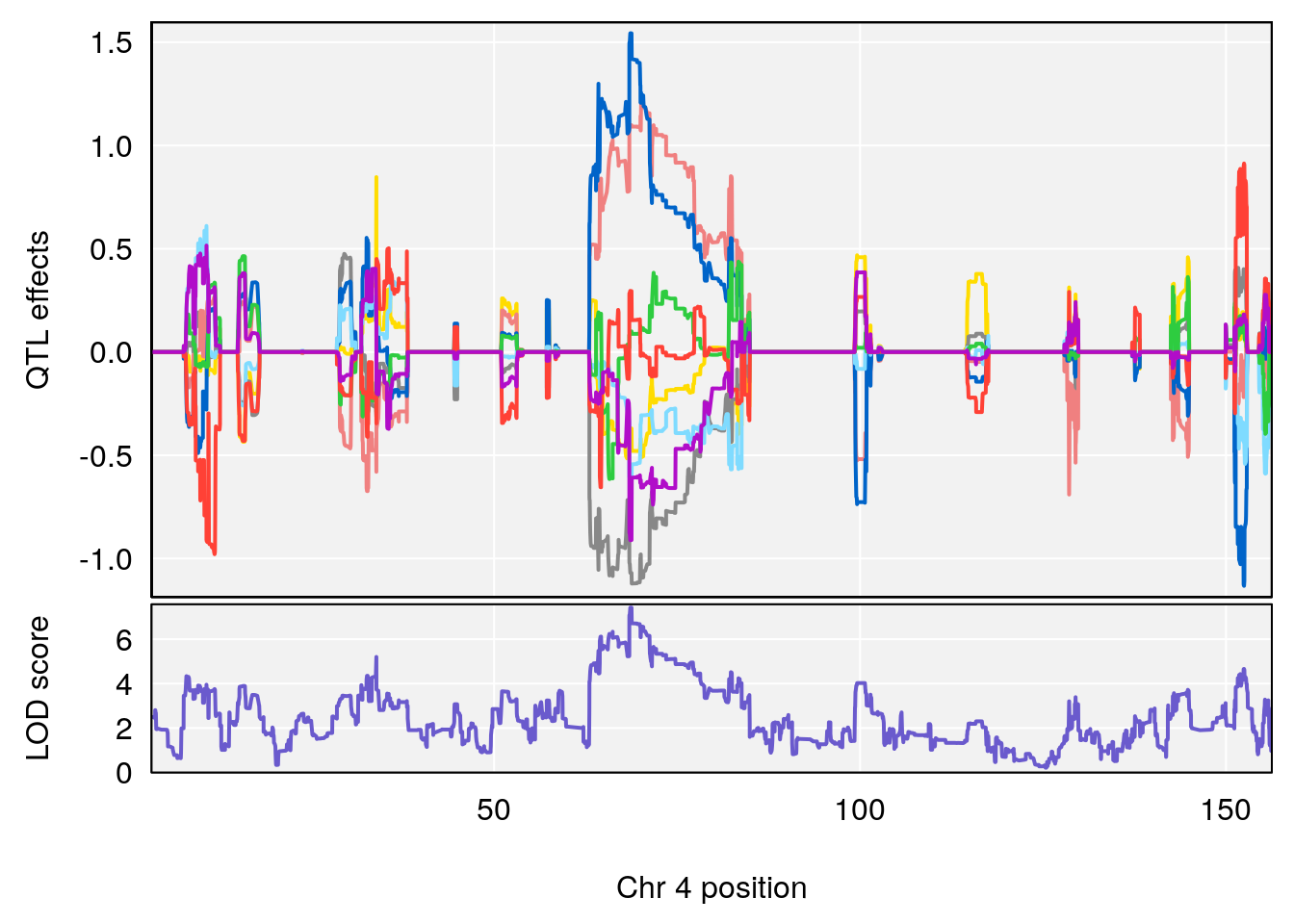
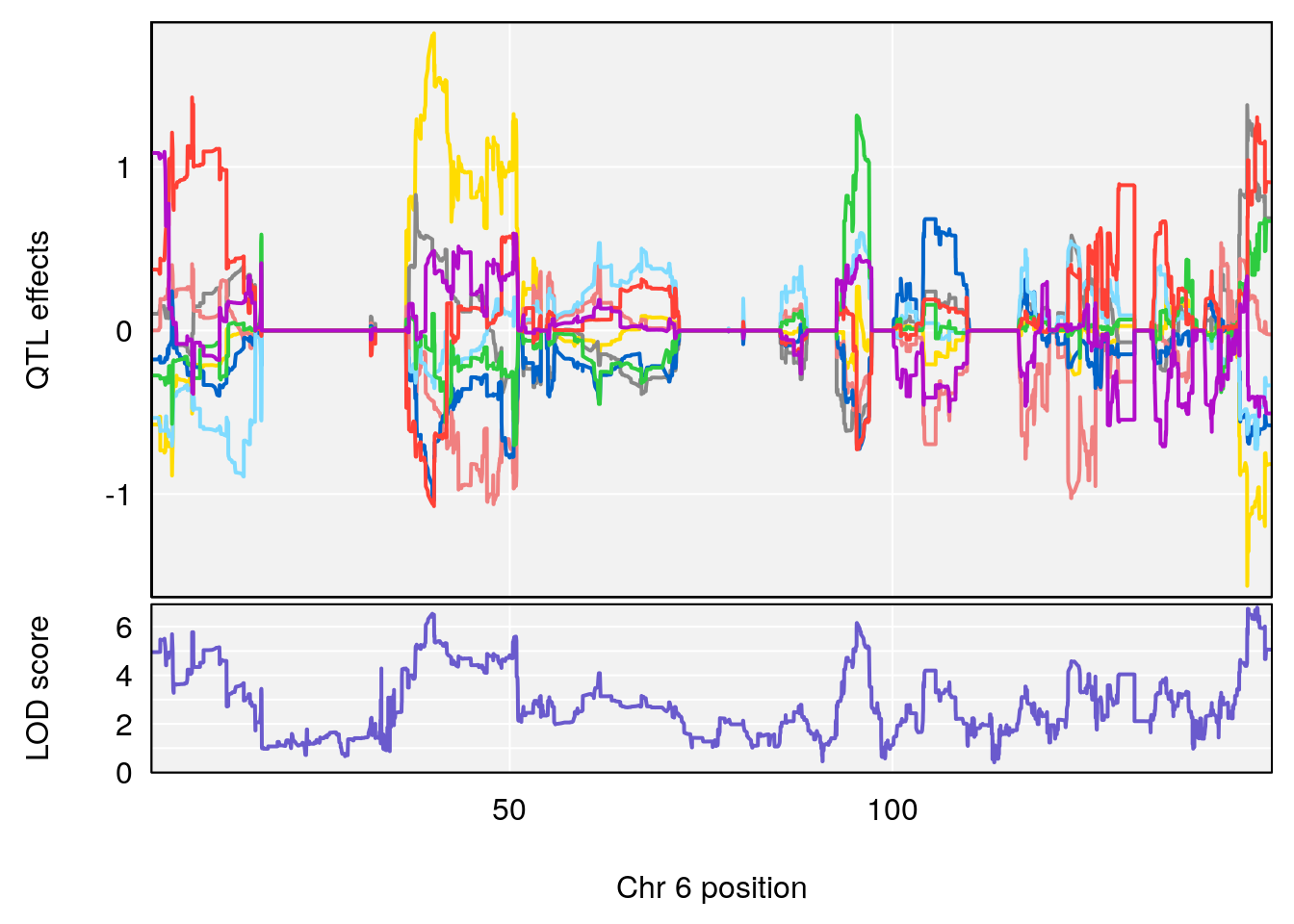
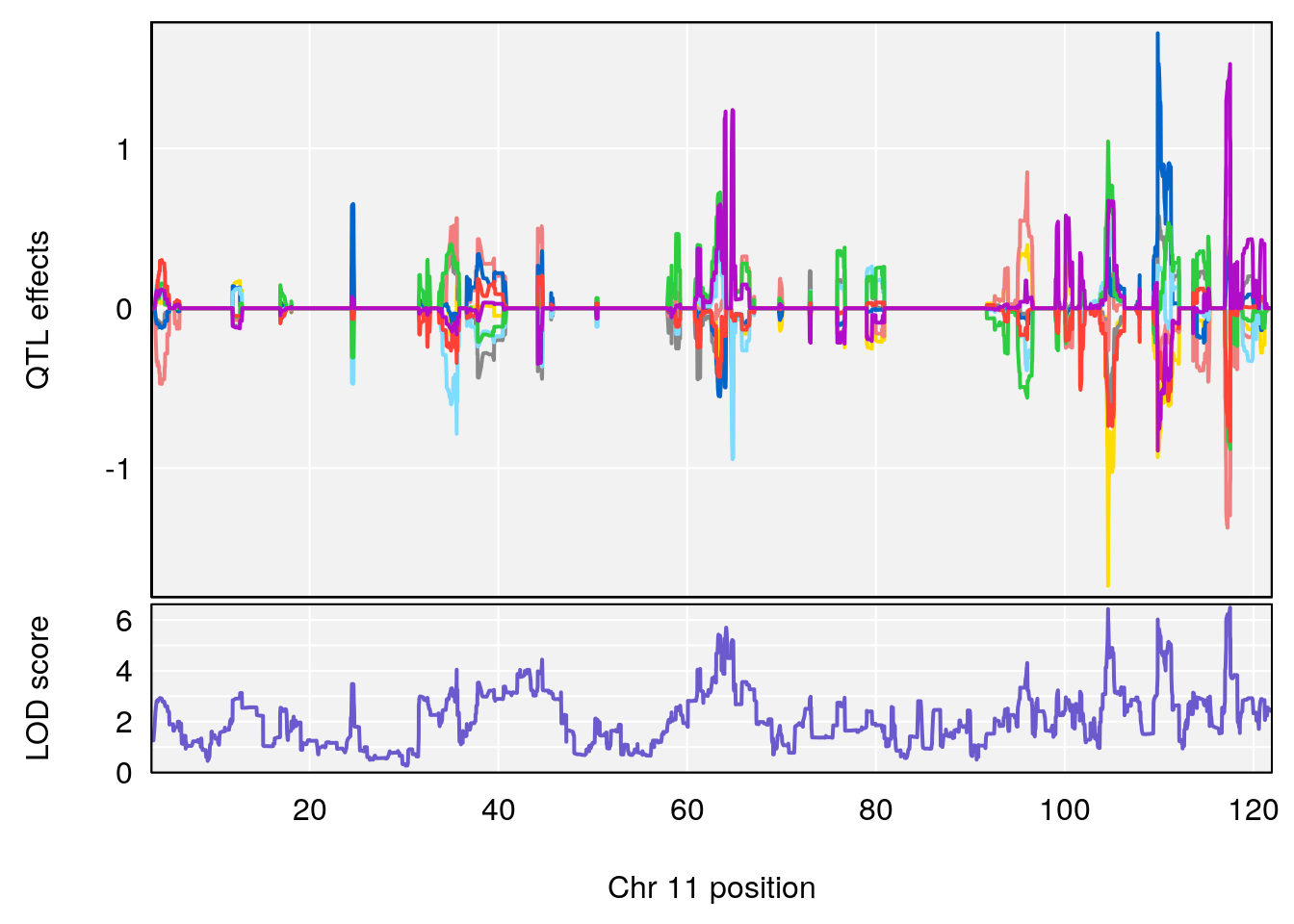
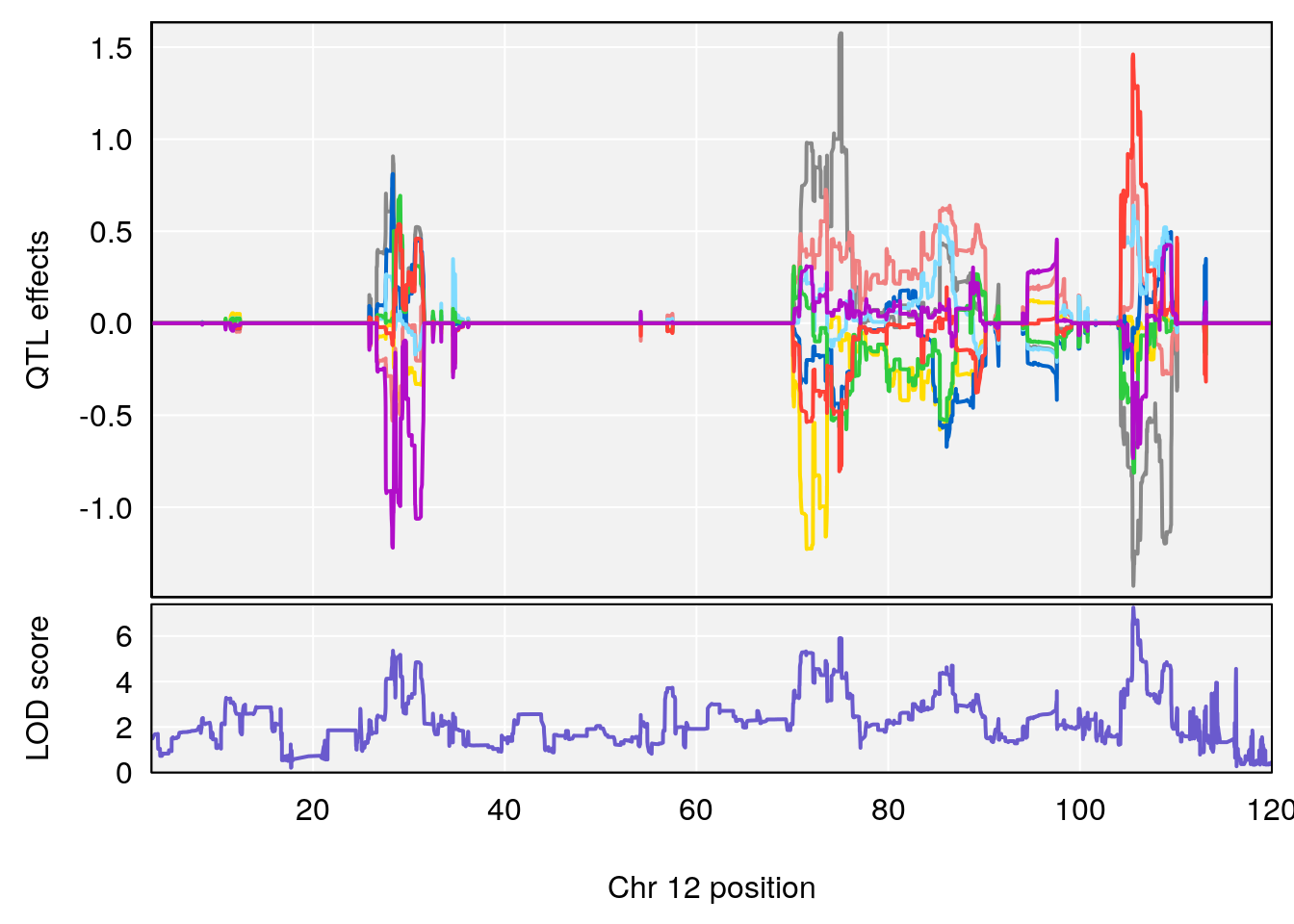
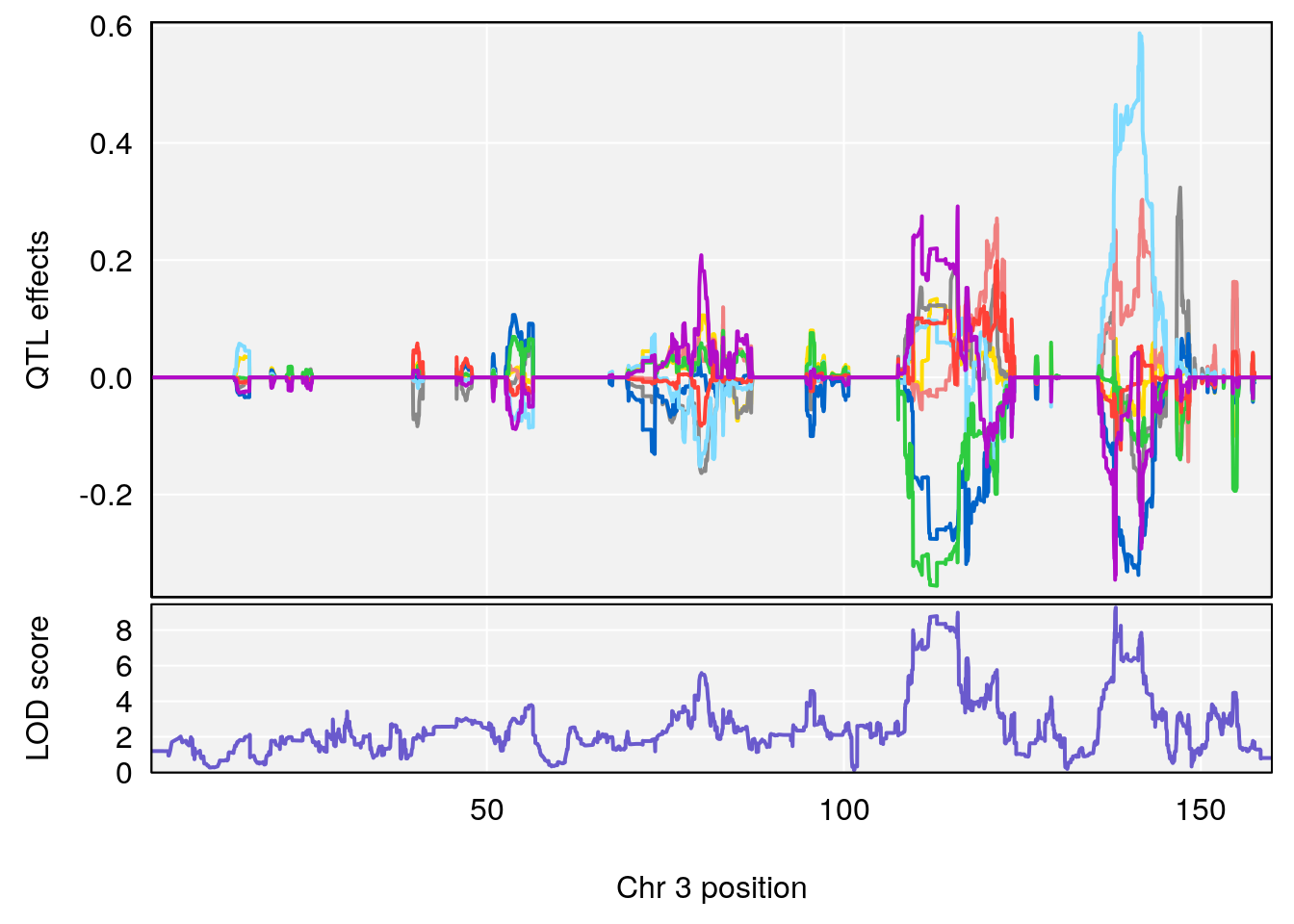
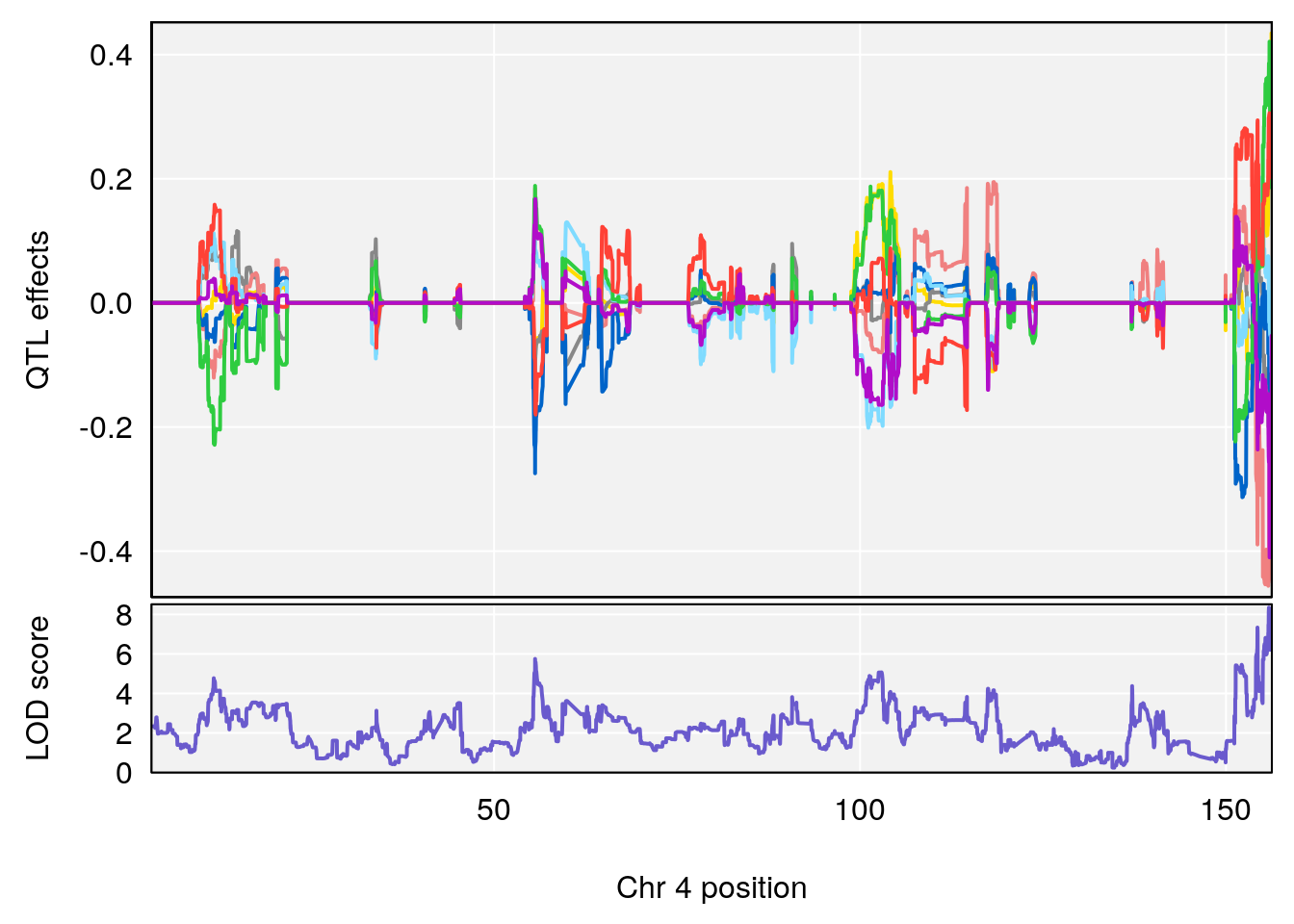
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
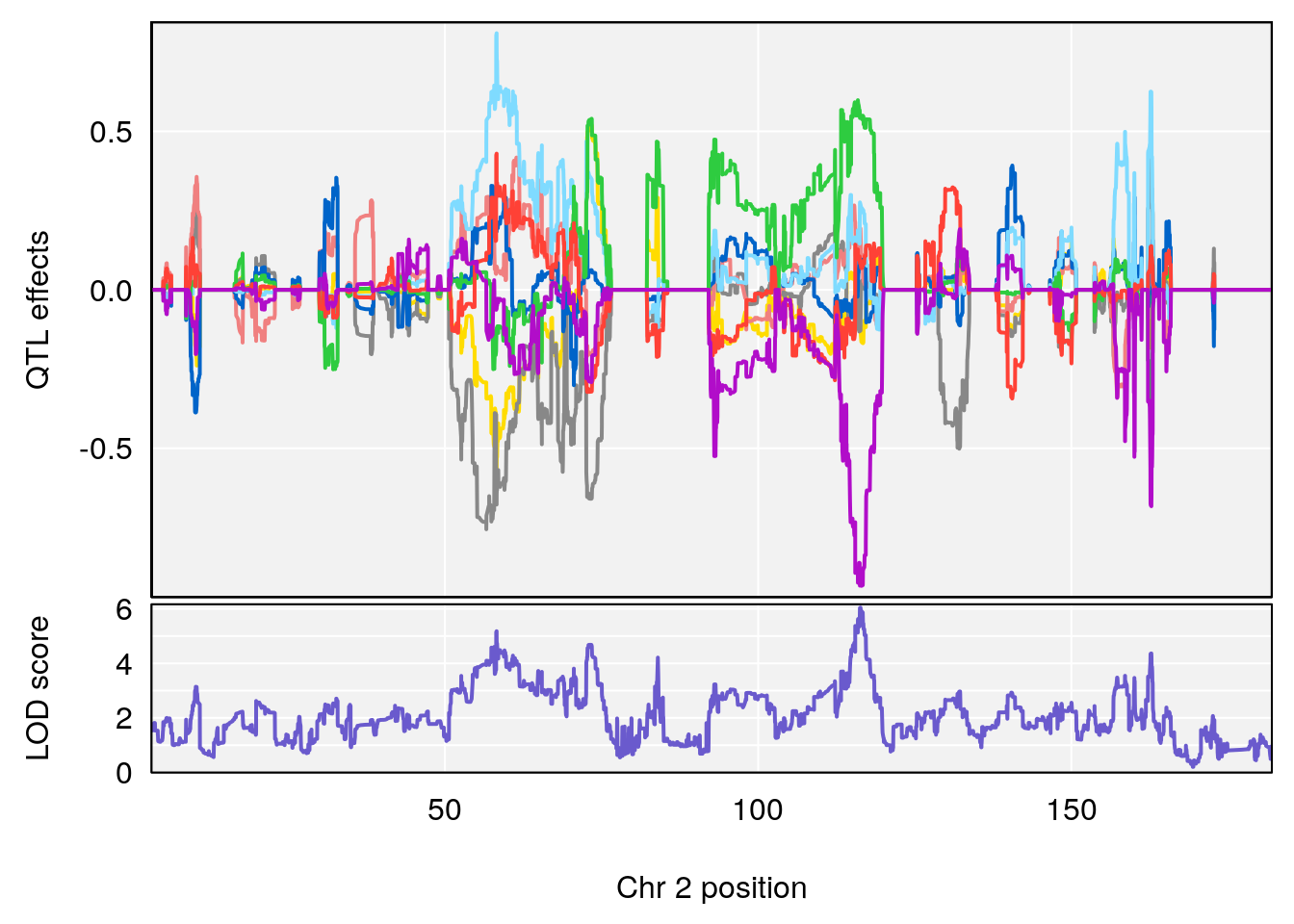
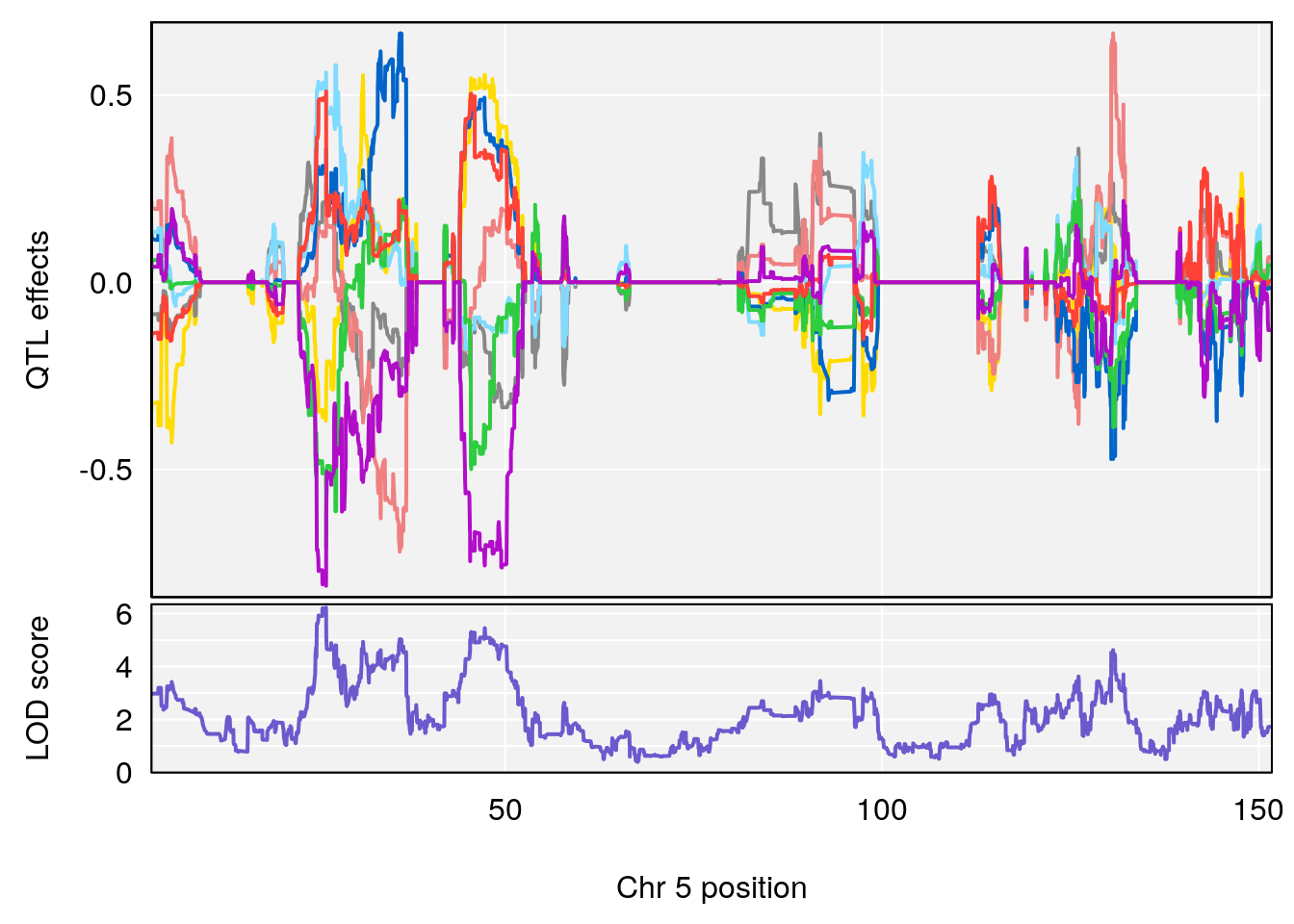
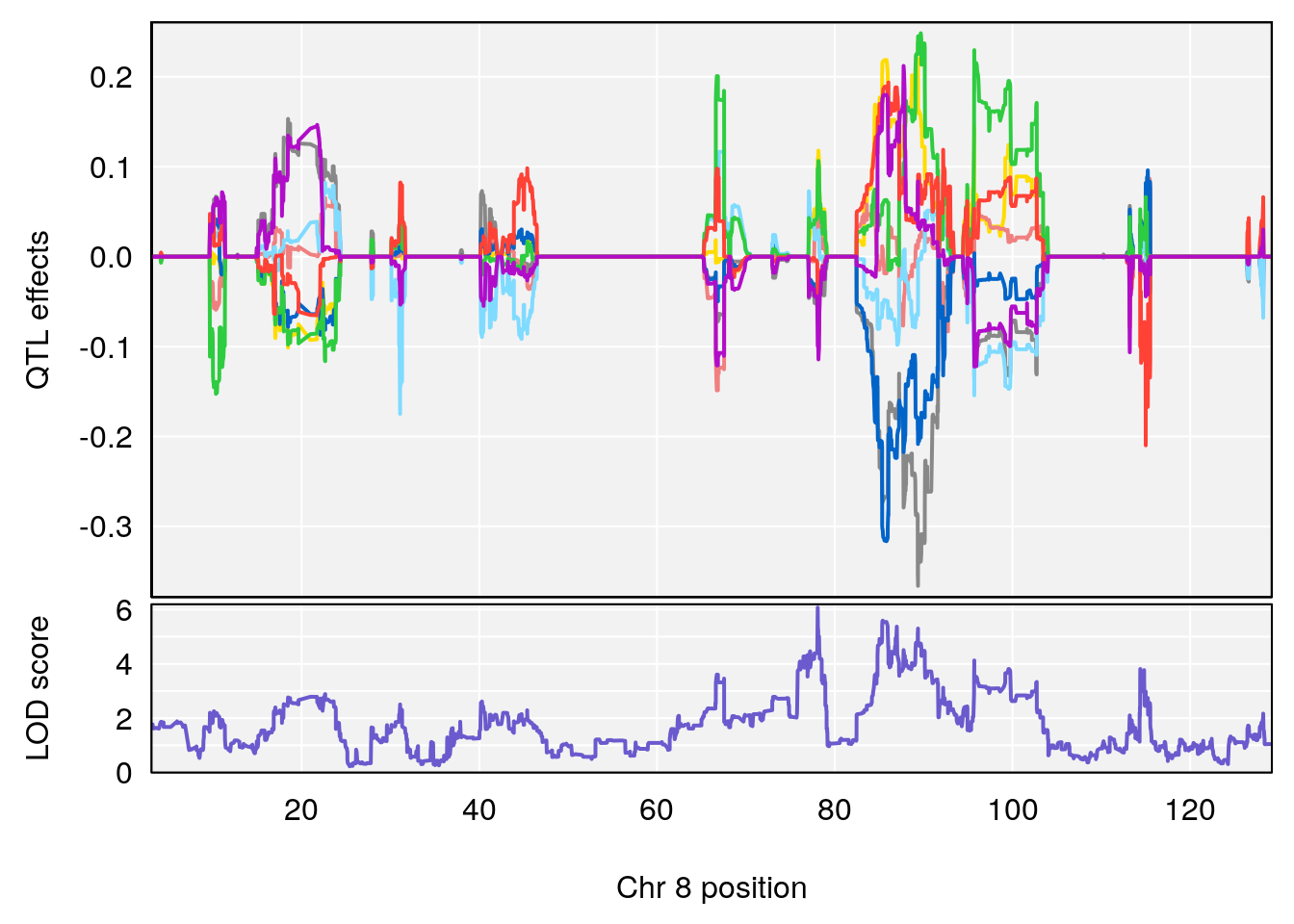
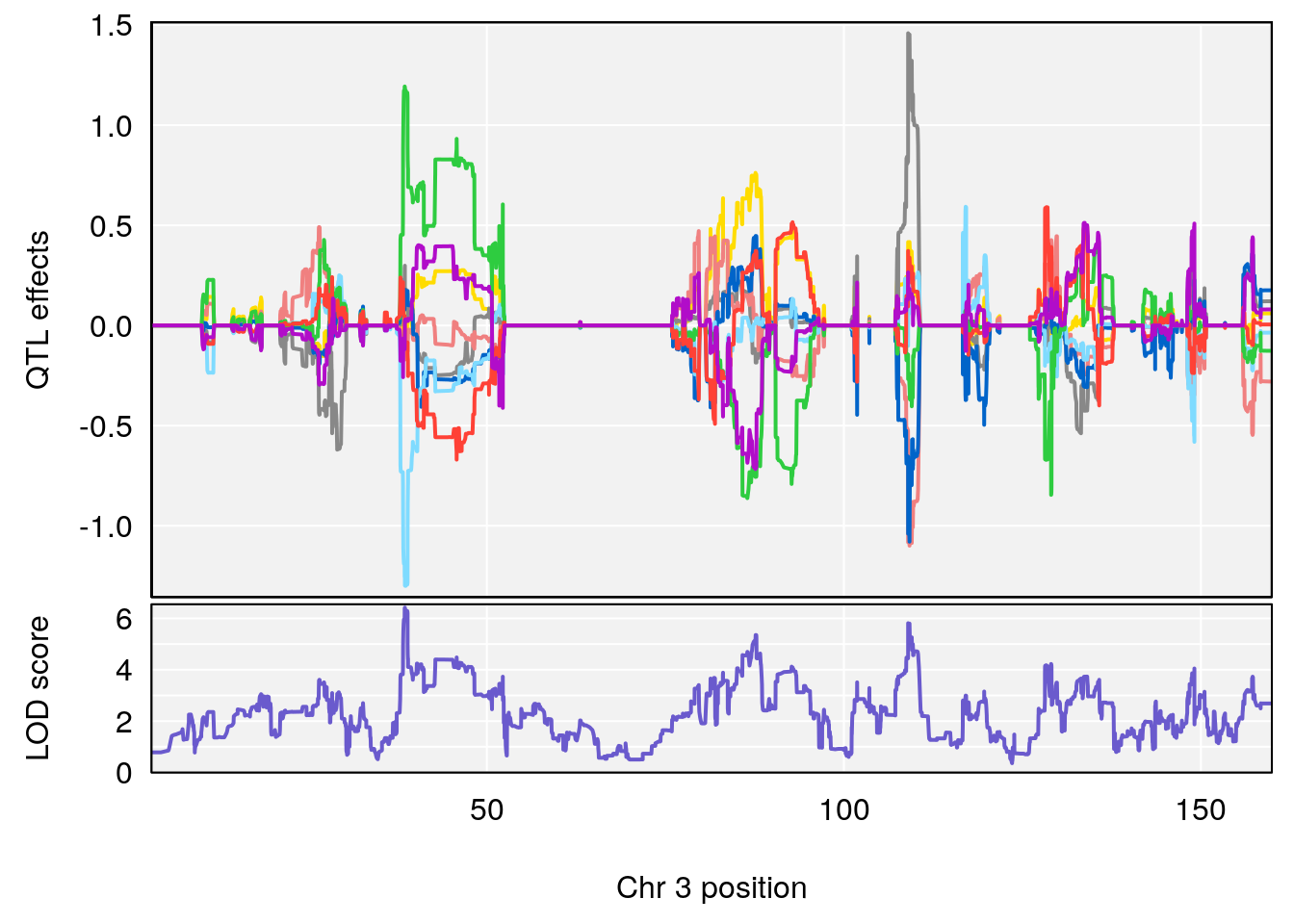
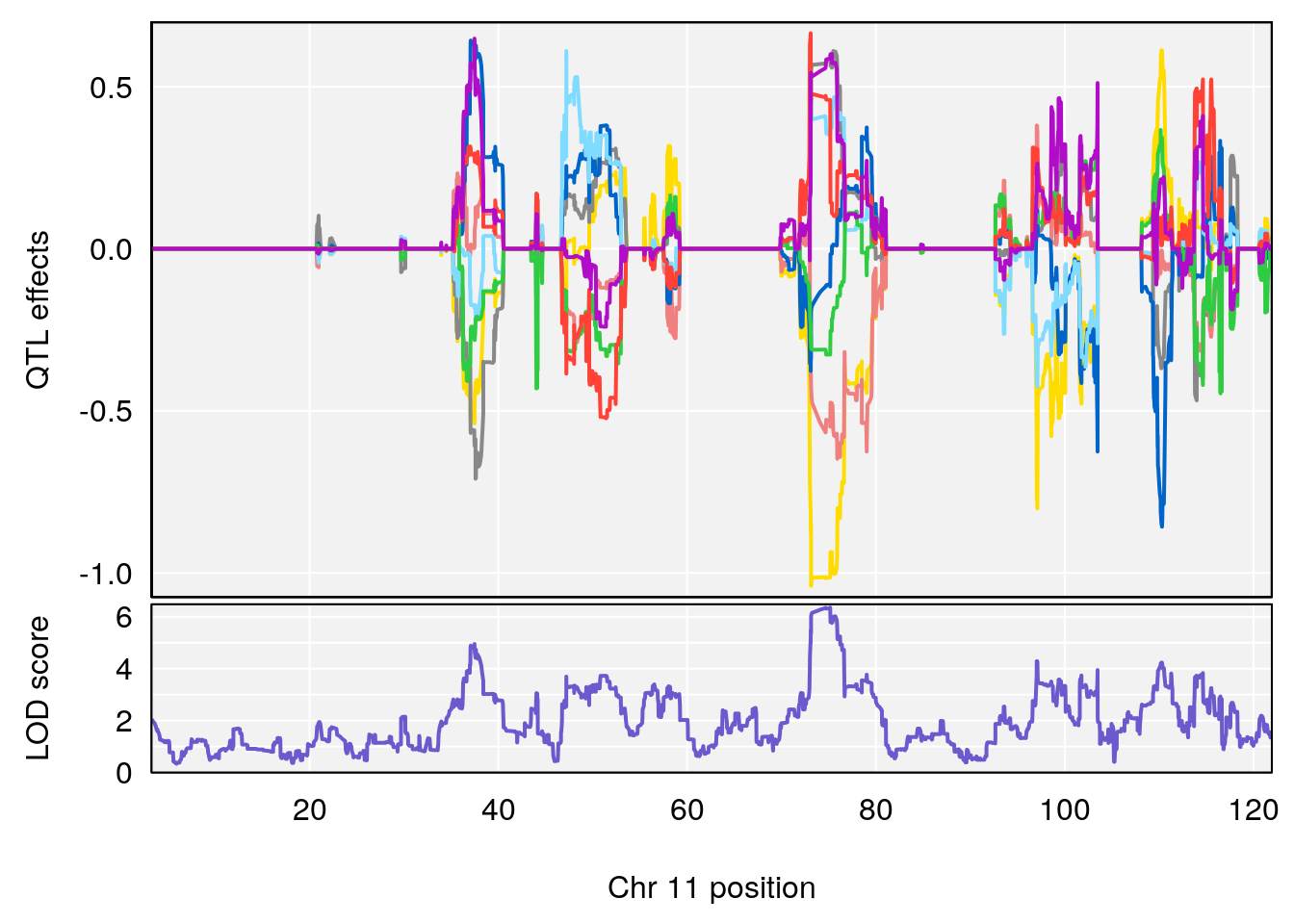
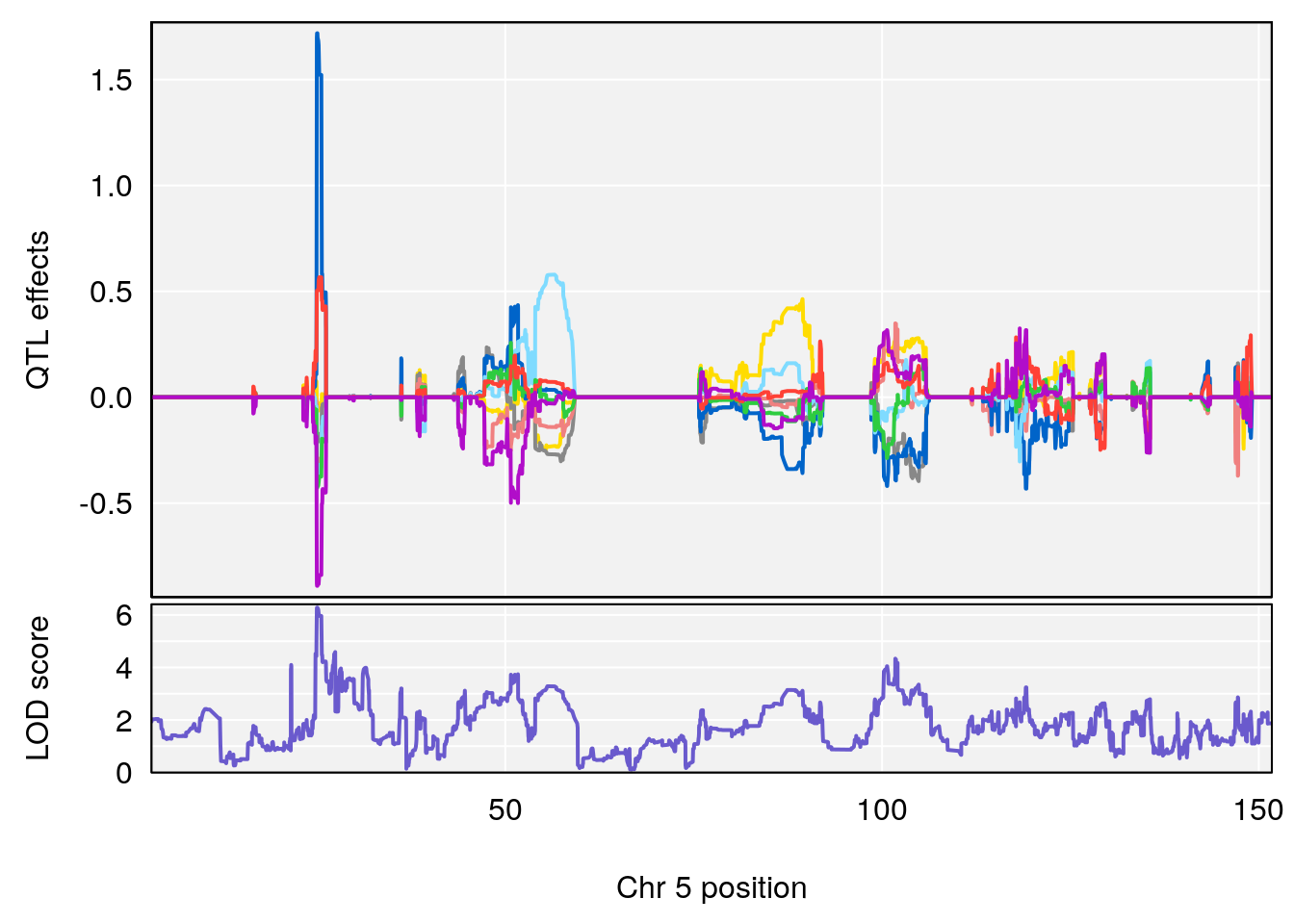
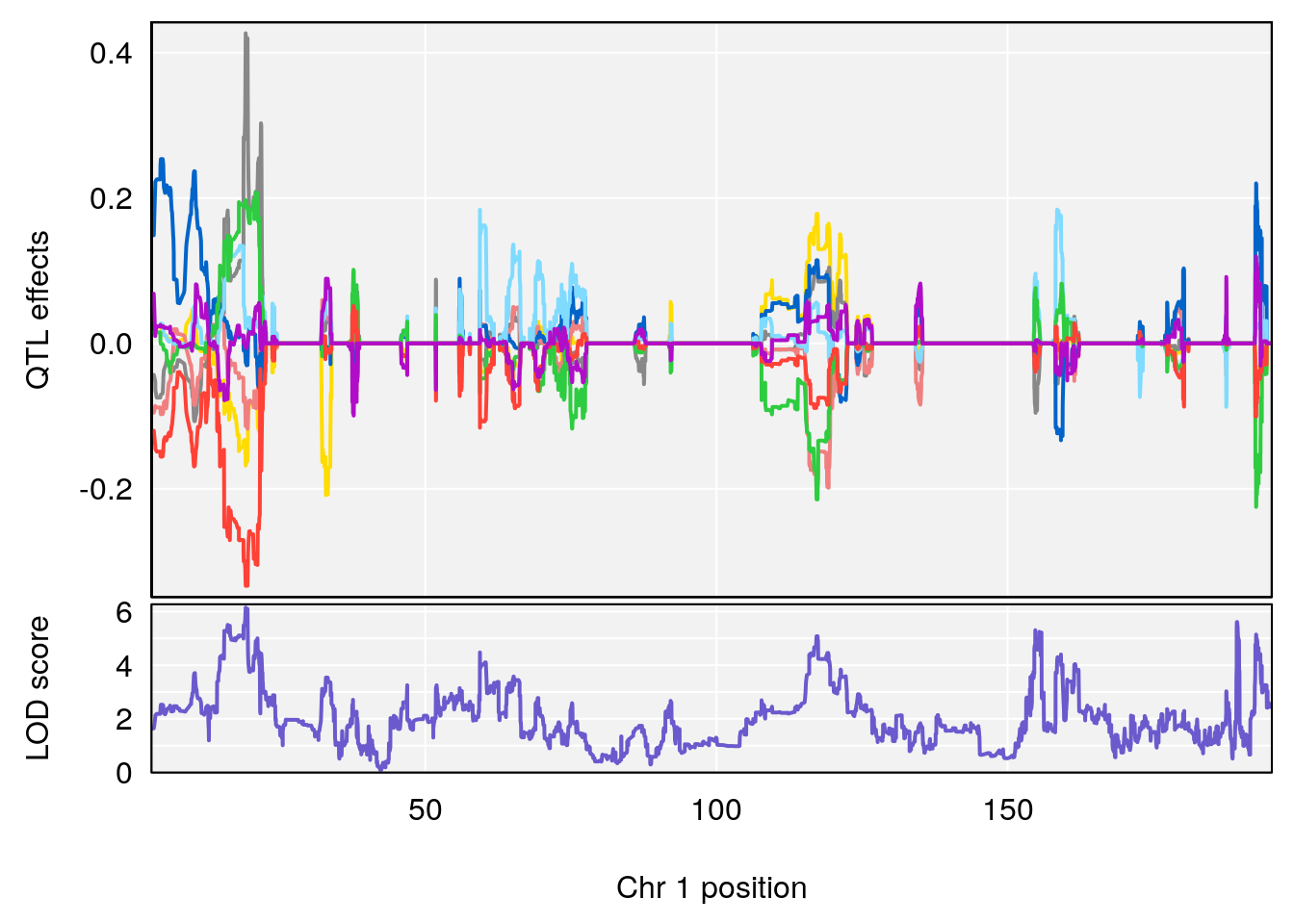
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend="topright")
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend="topright")
}
}
# [1] "Survival.Time"
# [1] 1

# [1] 2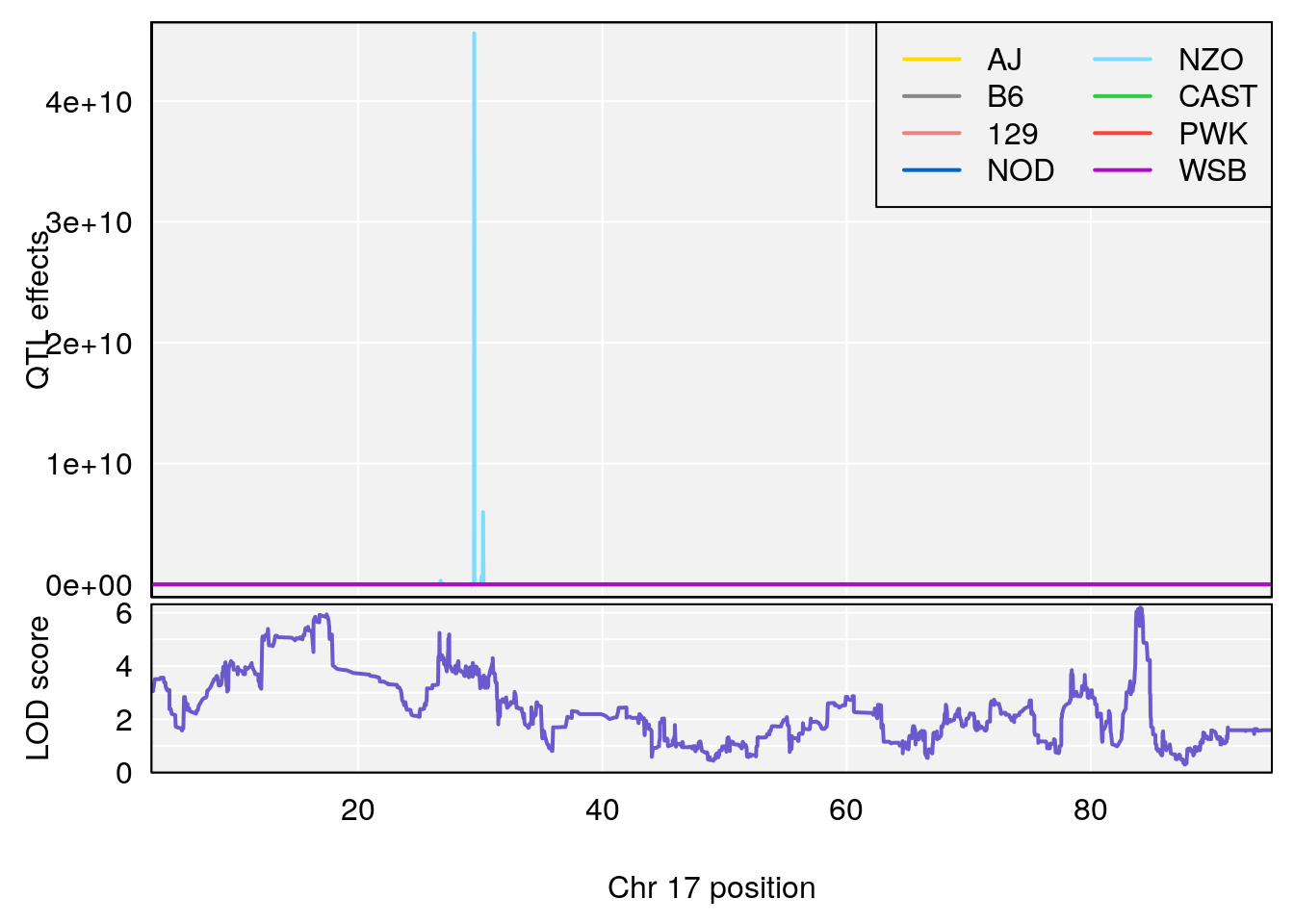
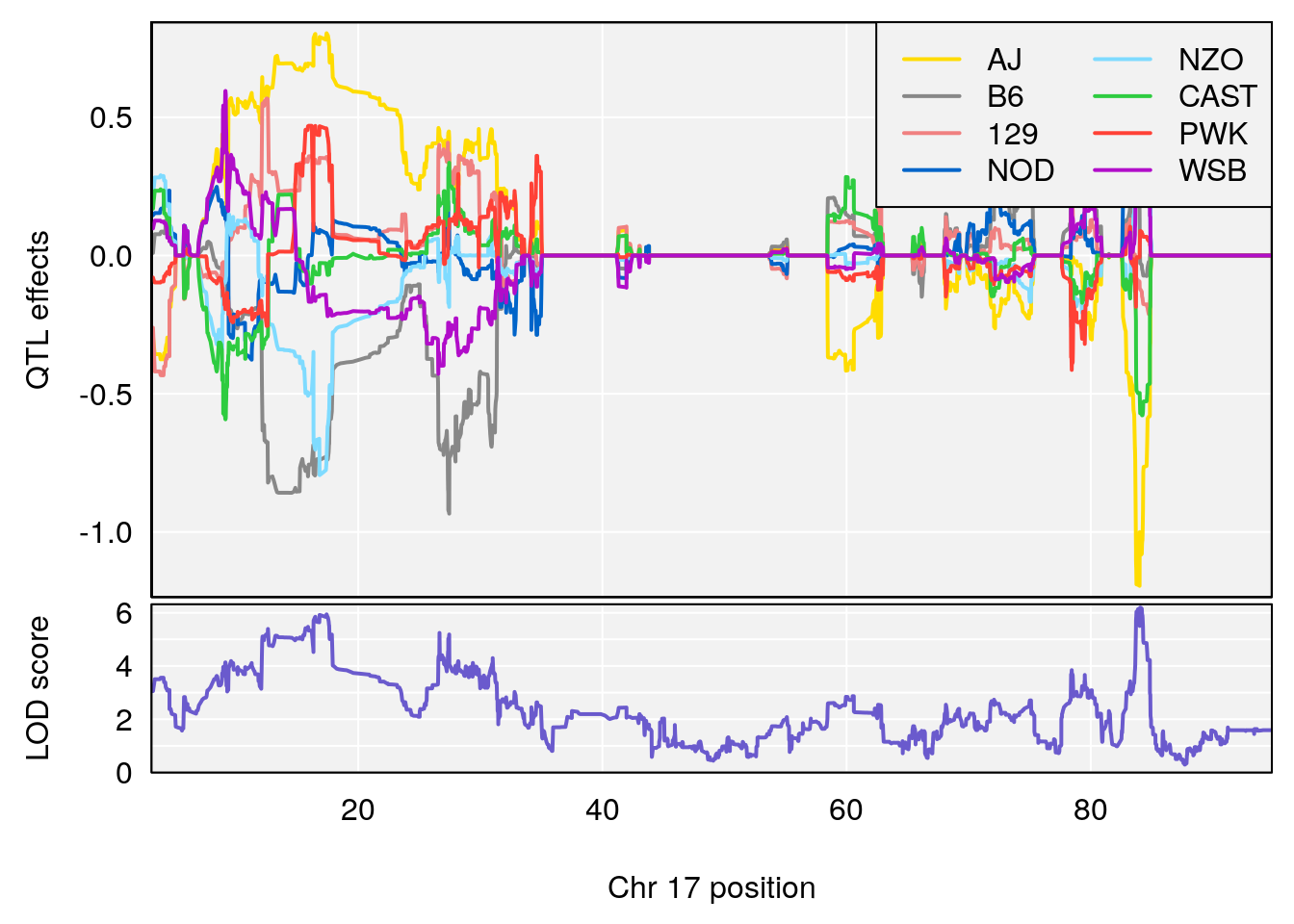
# [1] "Recovery.Time"
# [1] 1

# [1] 2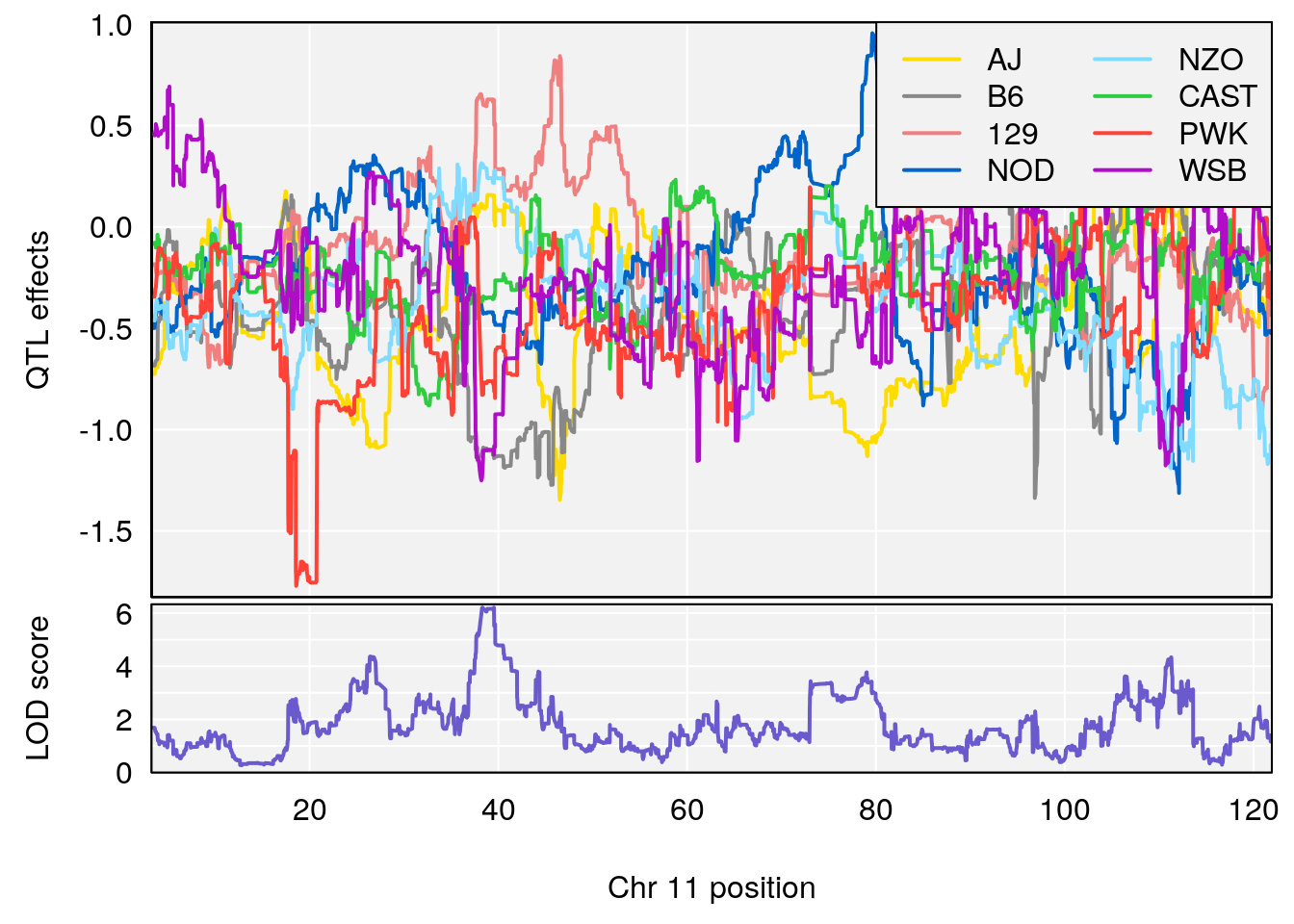

# [1] "Min.depression"
# [1] 1
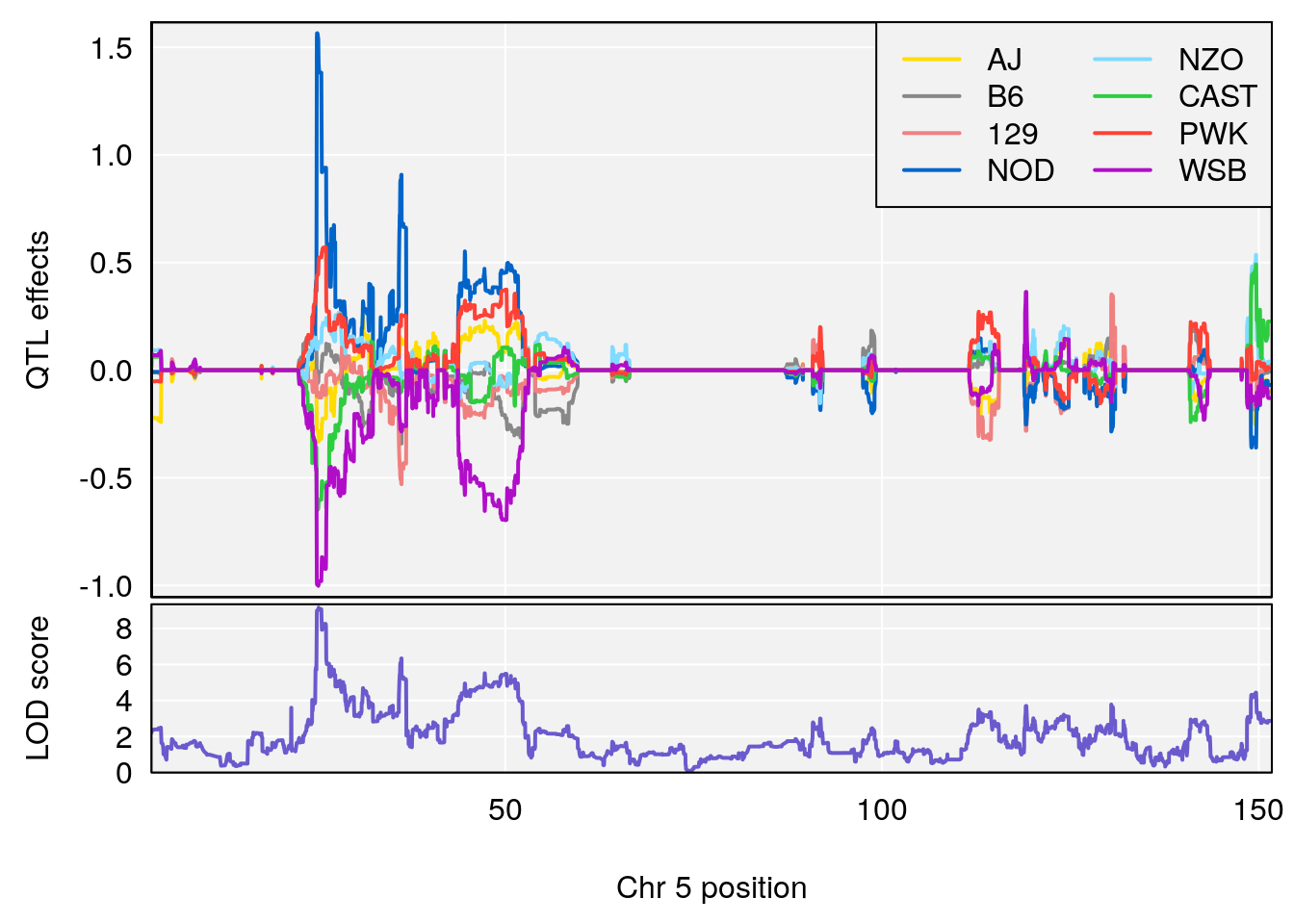
# [1] 2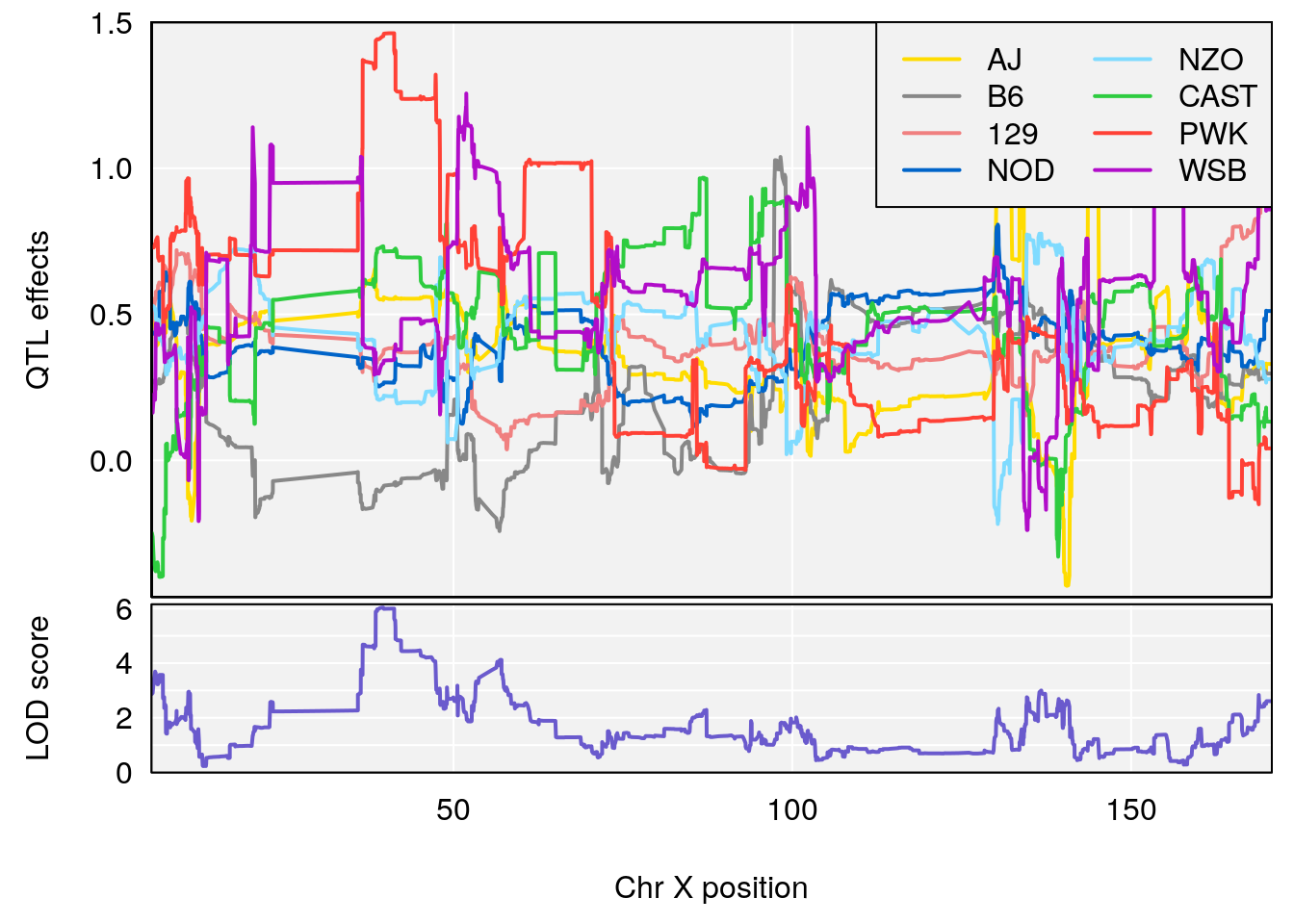

# [1] "Status_bin"
# [1] 1

# [1] 2

# [1] 3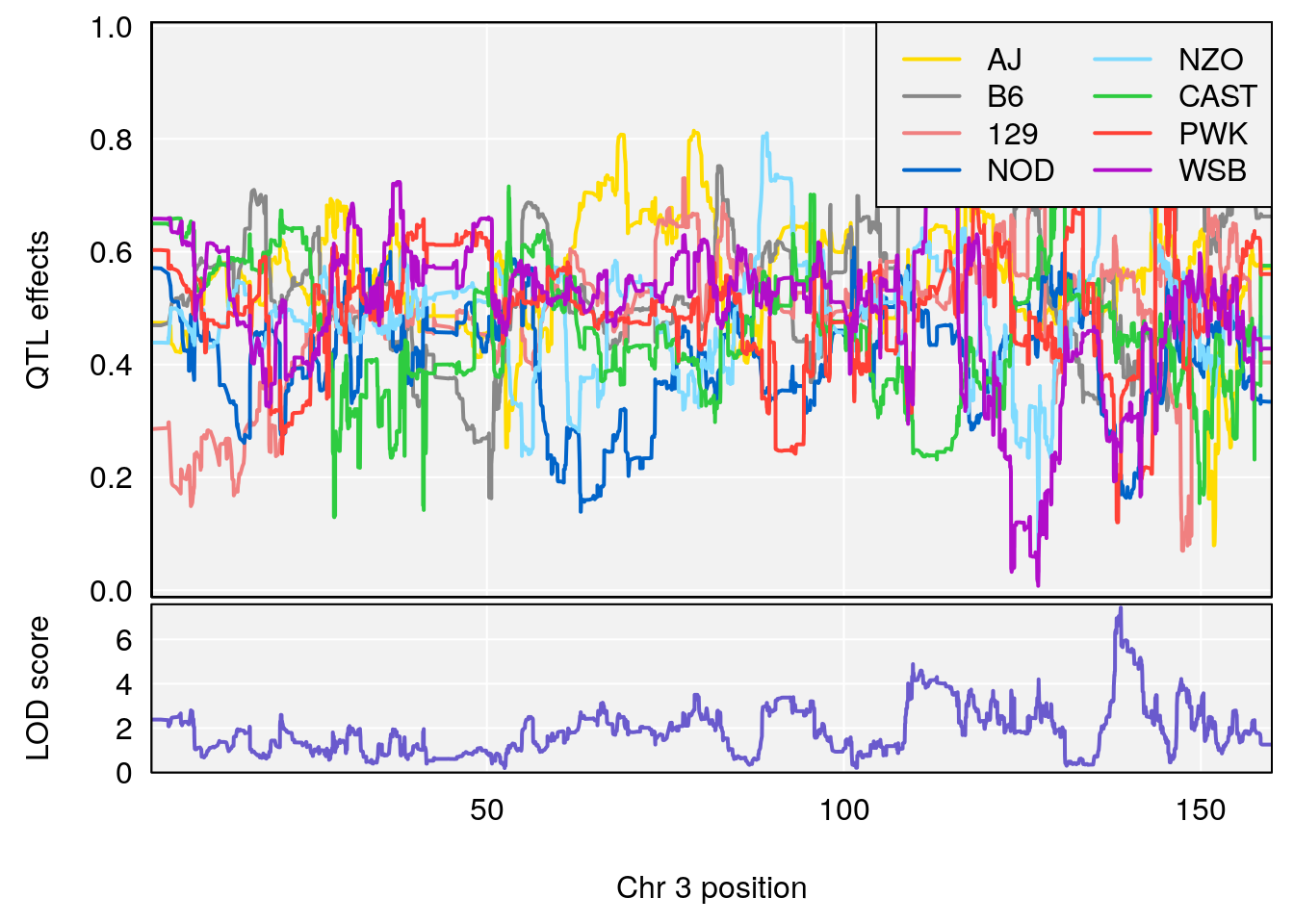
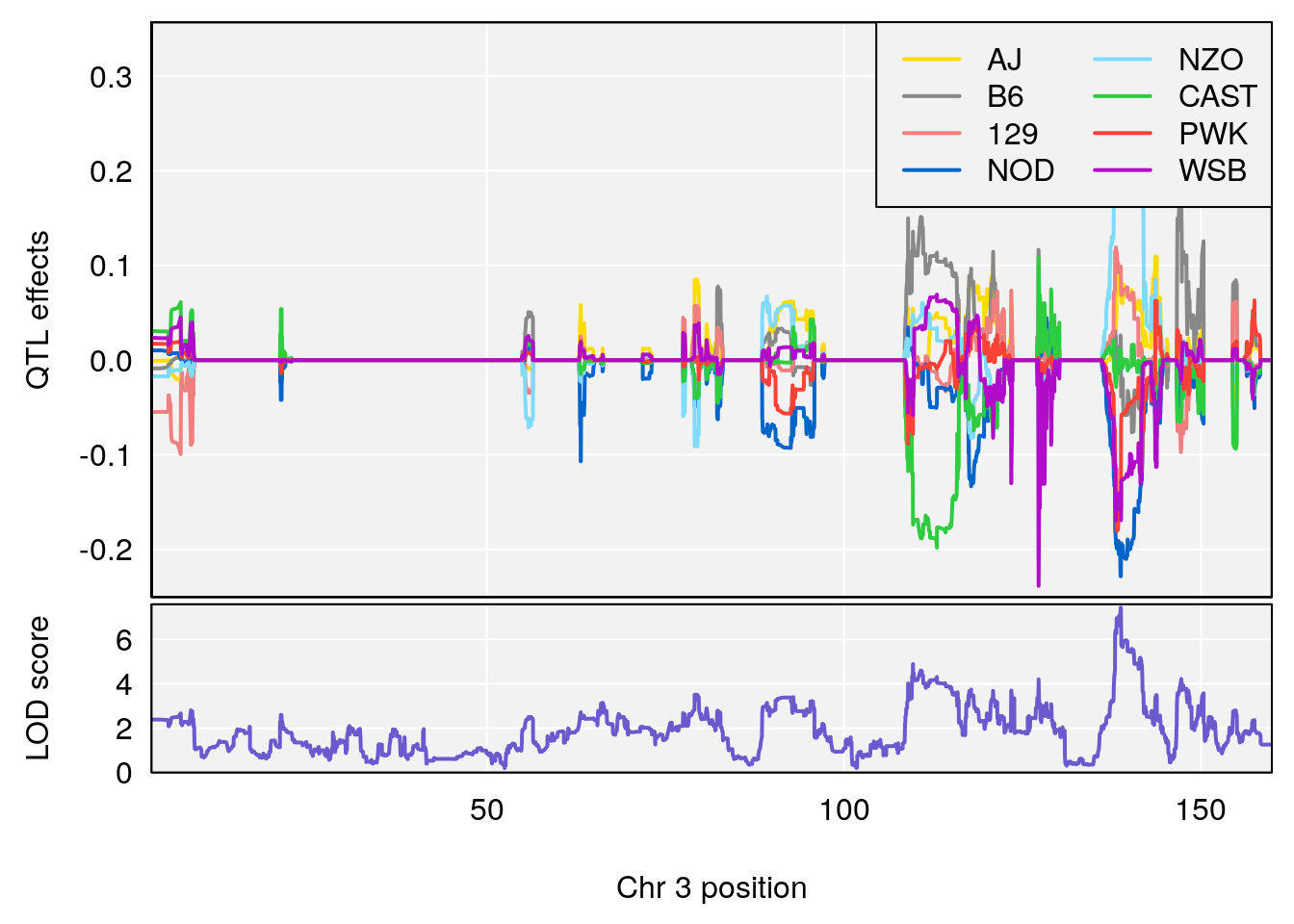
# [1] 4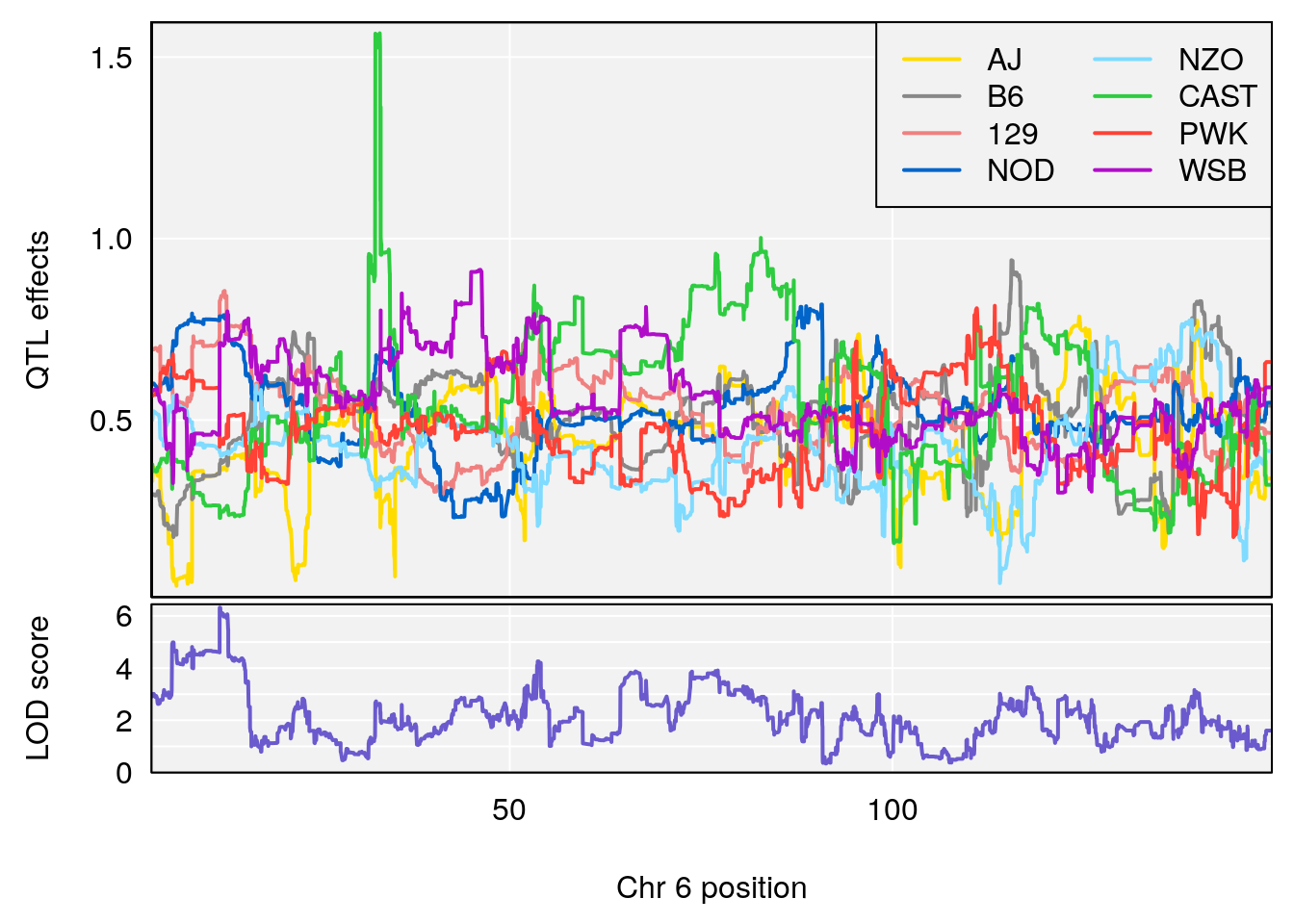

# [1] 5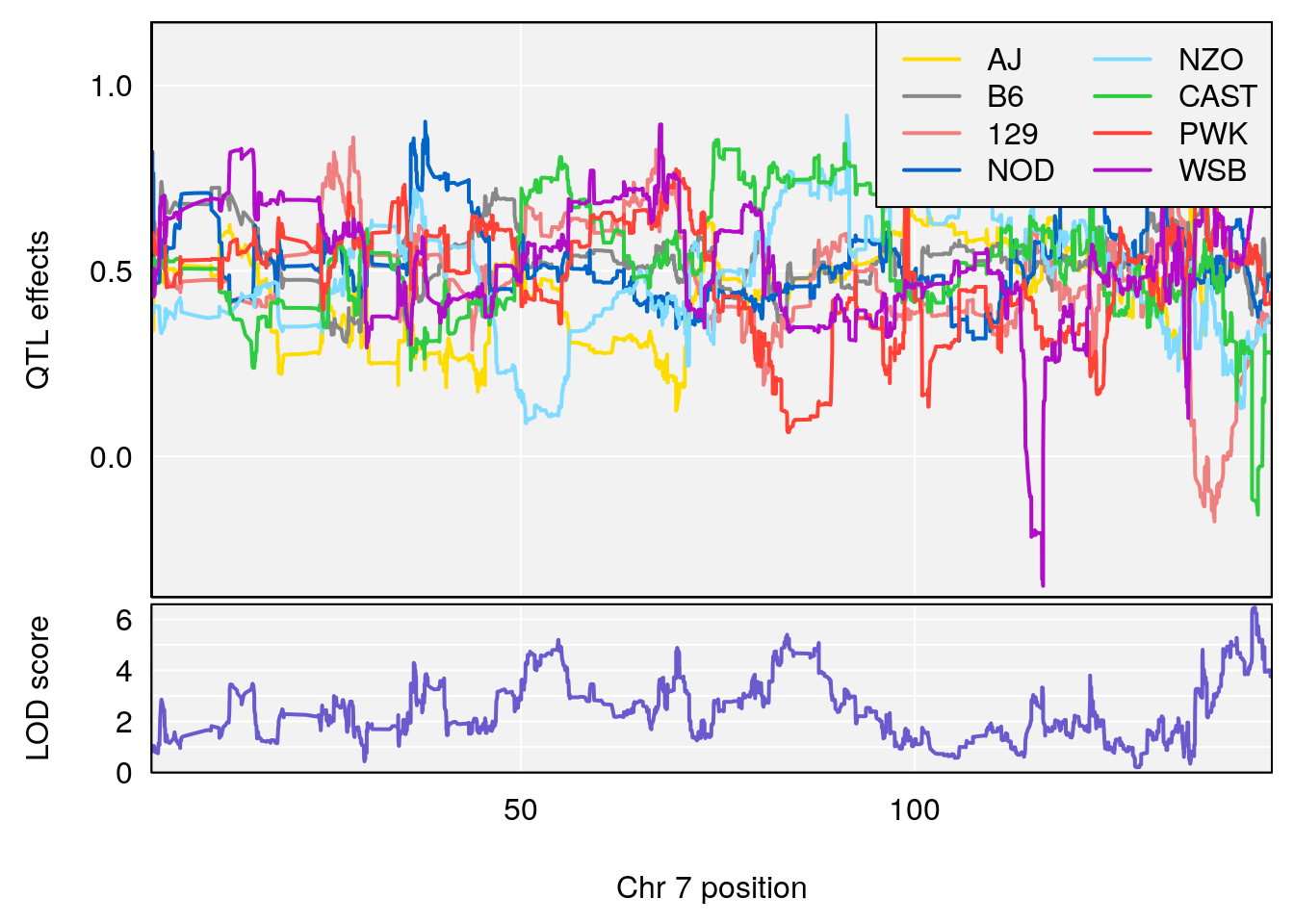
# [1] 6
# [1] 7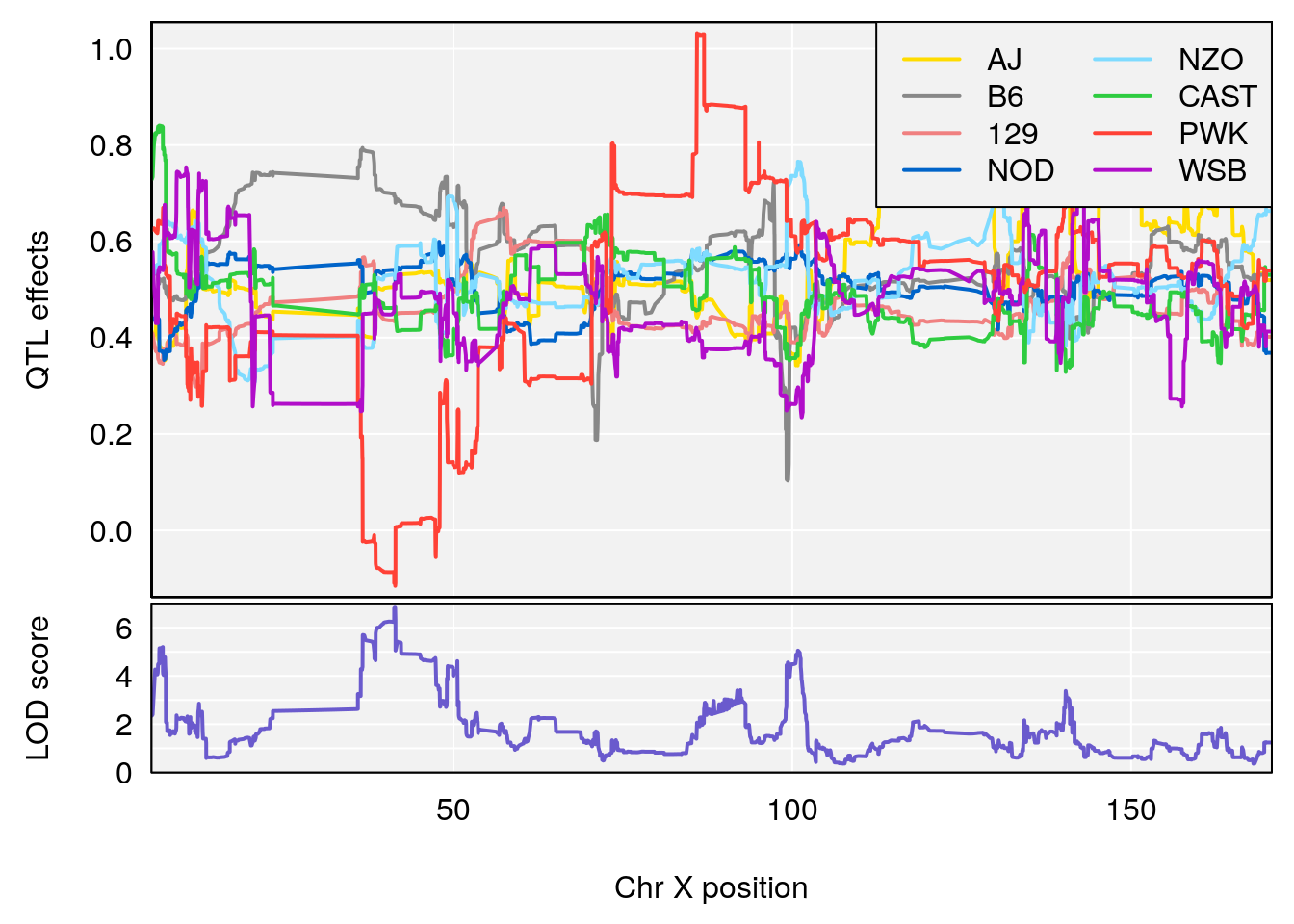
#save peaks coeff plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
fname <- paste("output/DO_morphine_",i,"_coefplot.pdf",sep="")
pdf(file = fname, width = 16, height =8)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend="topright")
}
dev.off()
}
# [1] "Survival.Time"
# [1] 1
# [1] 2
# [1] "Recovery.Time"
# [1] 1
# [1] 2
# [1] "Min.depression"
# [1] 1
# [1] 2
# [1] "Status_bin"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] 5
# [1] 6
# [1] 7
#save peaks coeff blup plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
fname <- paste("output/DO_morphine_",i,"_coefplot_blup.pdf",sep="")
pdf(file = fname, width = 16, height =8)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend="topright")
}
dev.off()
}
# [1] "Survival.Time"
# [1] 1
# [1] 2
# [1] "Recovery.Time"
# [1] 1
# [1] 2
# [1] "Min.depression"
# [1] 1
# [1] 2
# [1] "Status_bin"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] 5
# [1] 6
# [1] 7
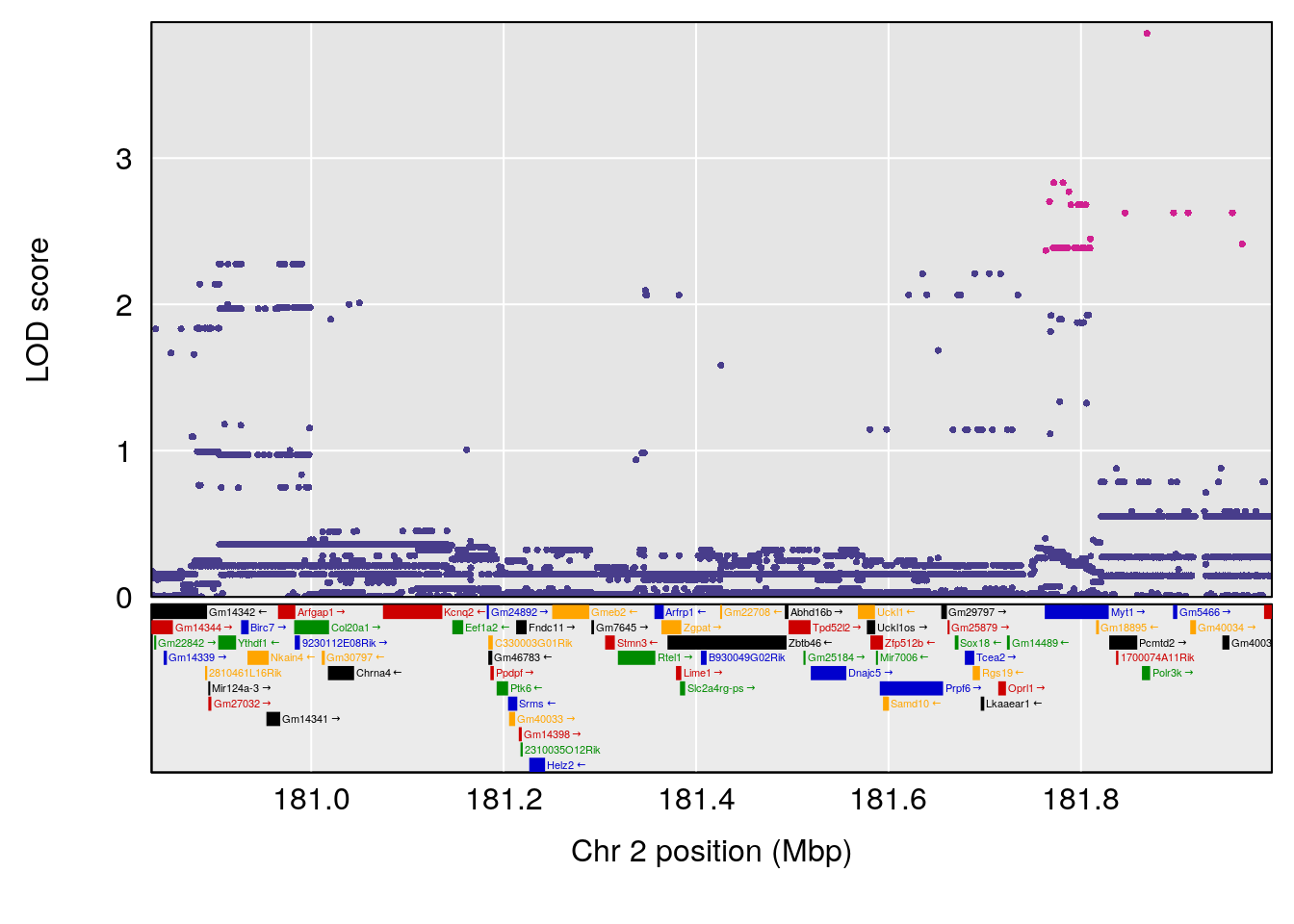
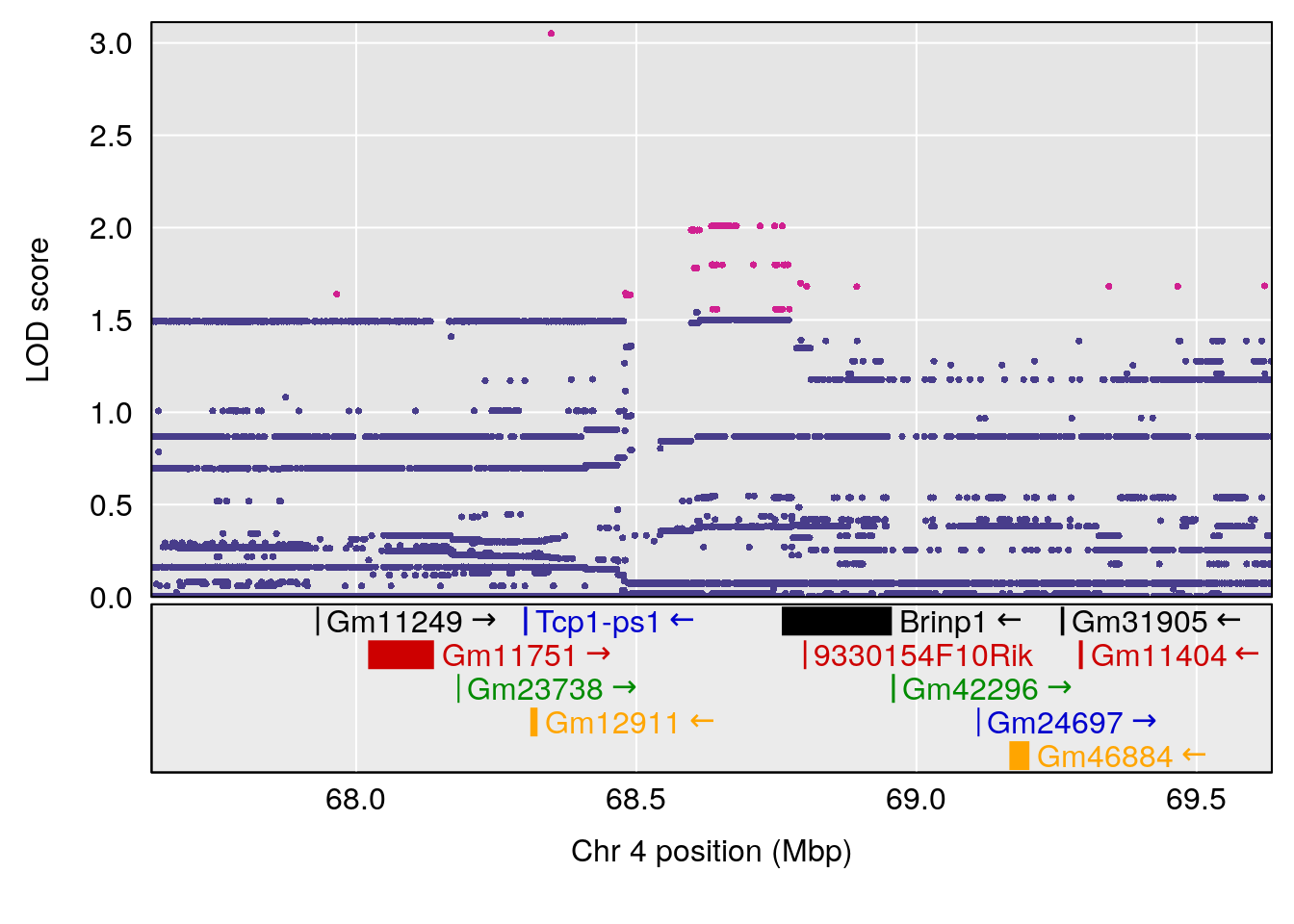
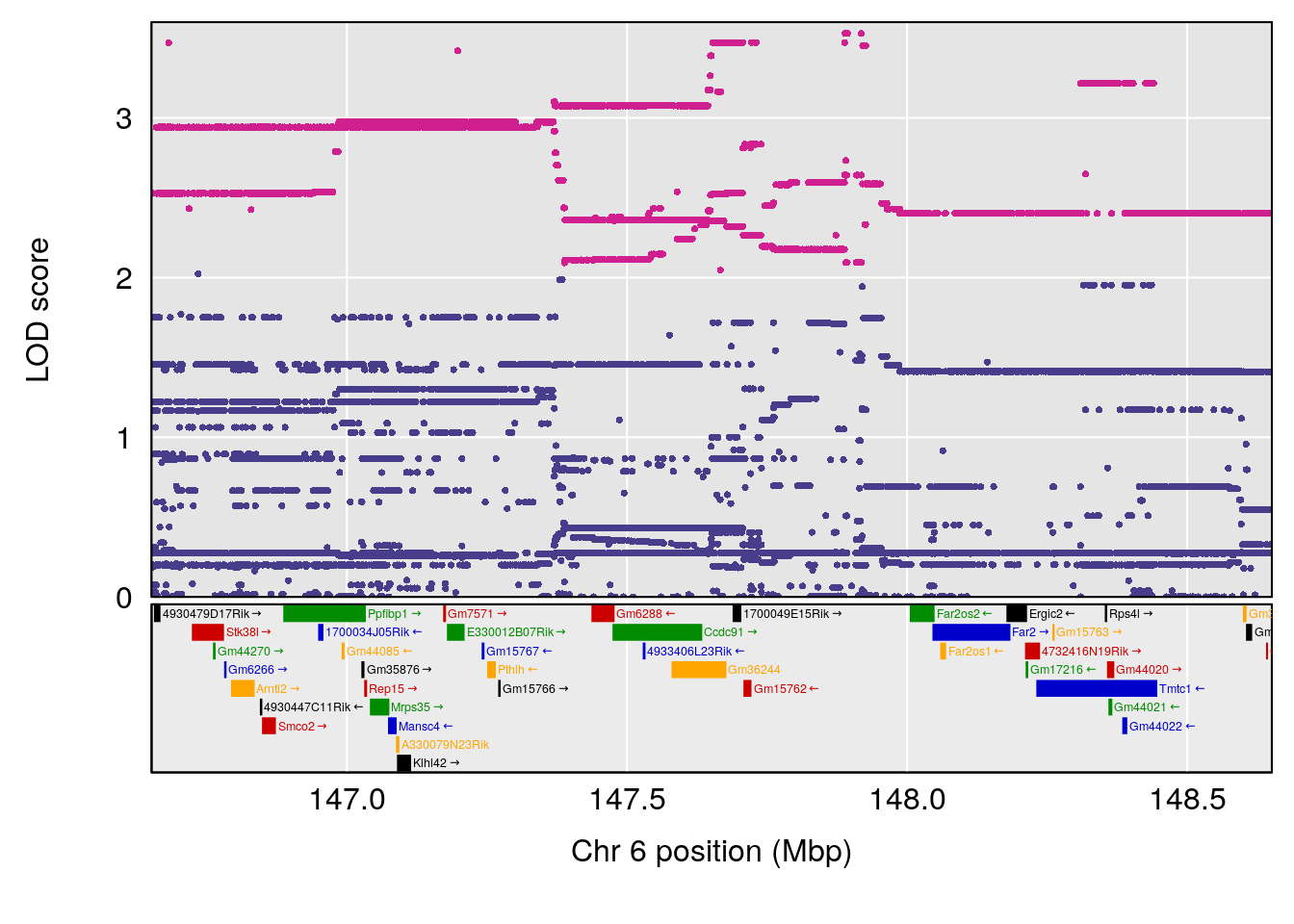
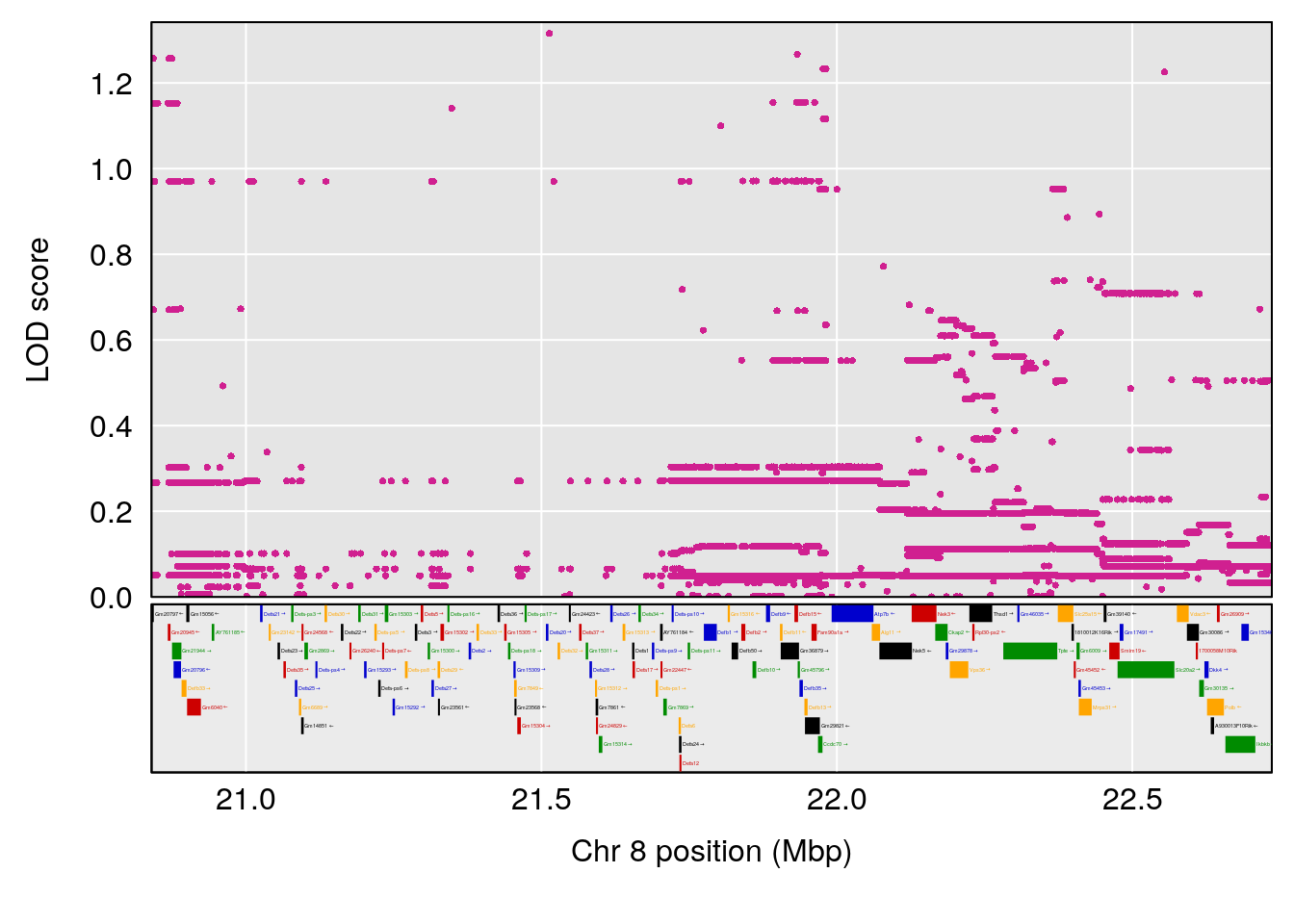
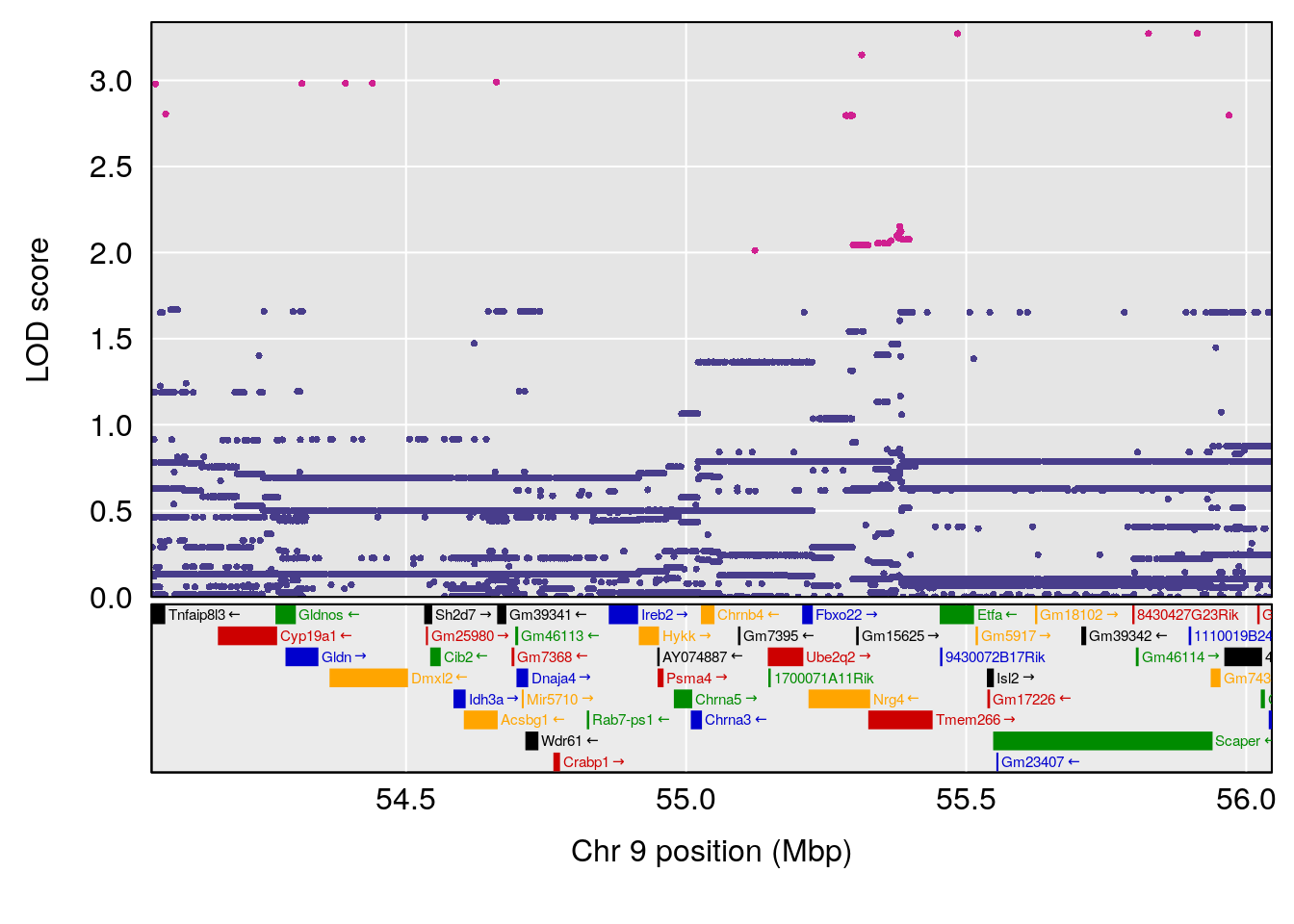
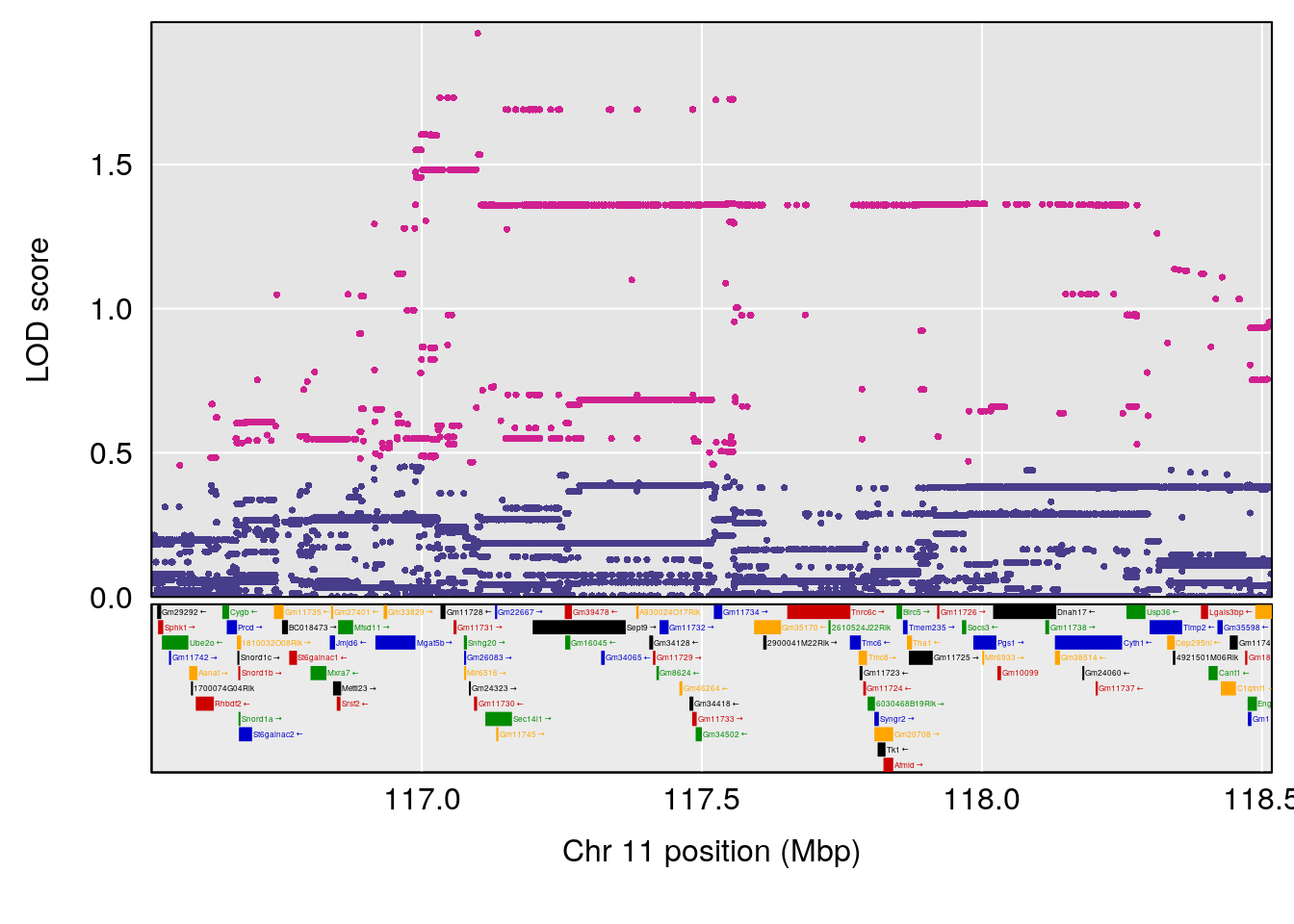
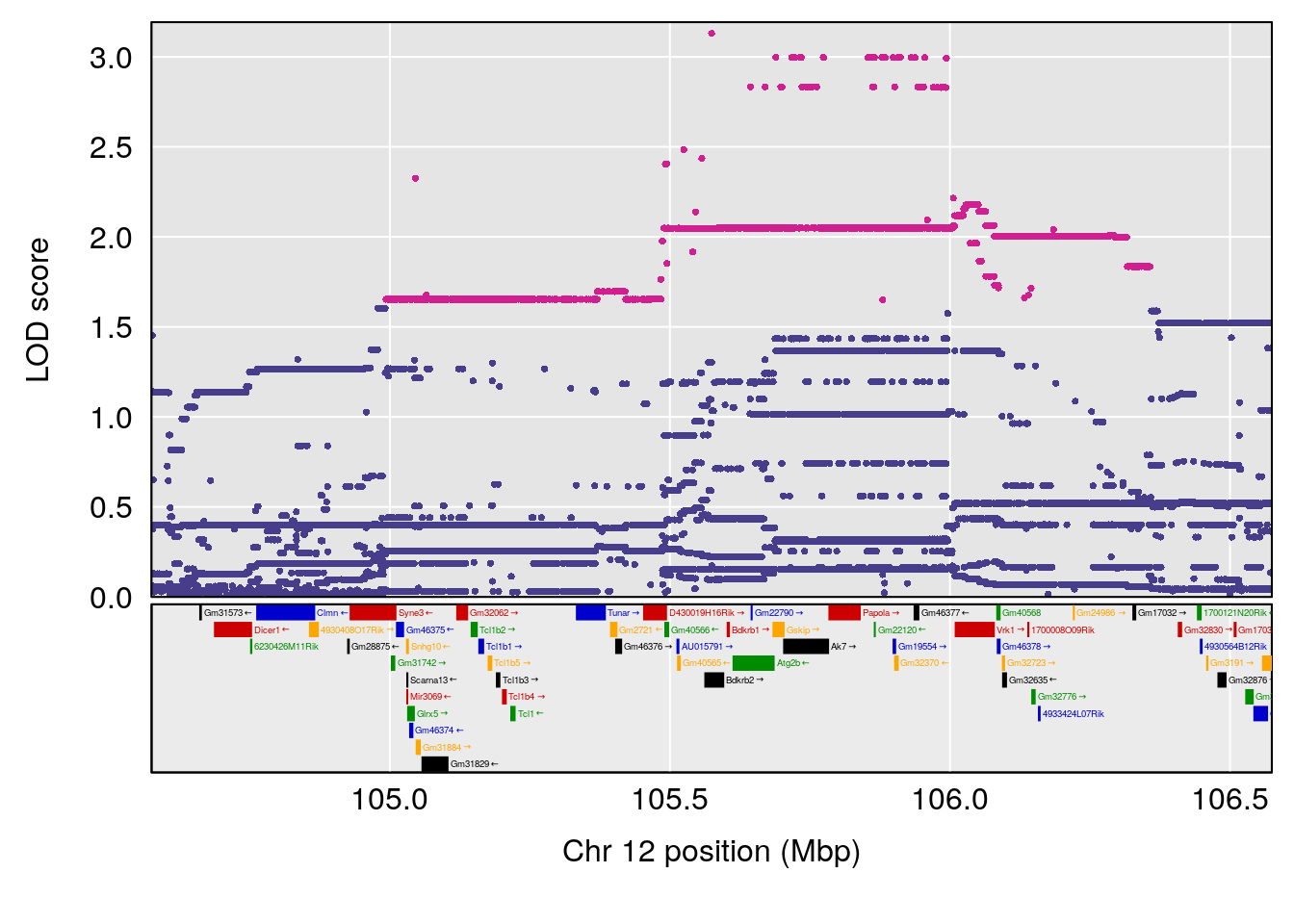
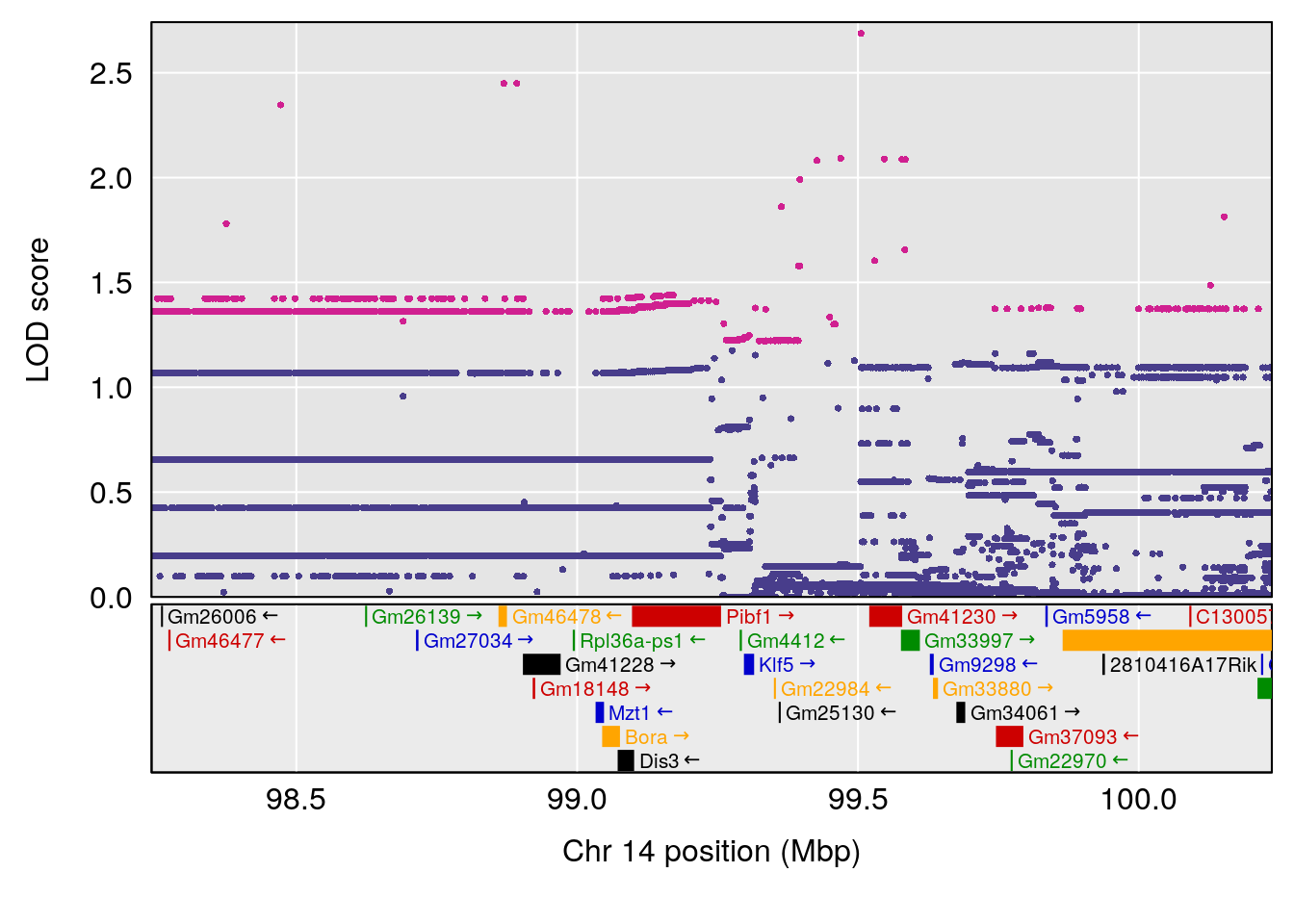
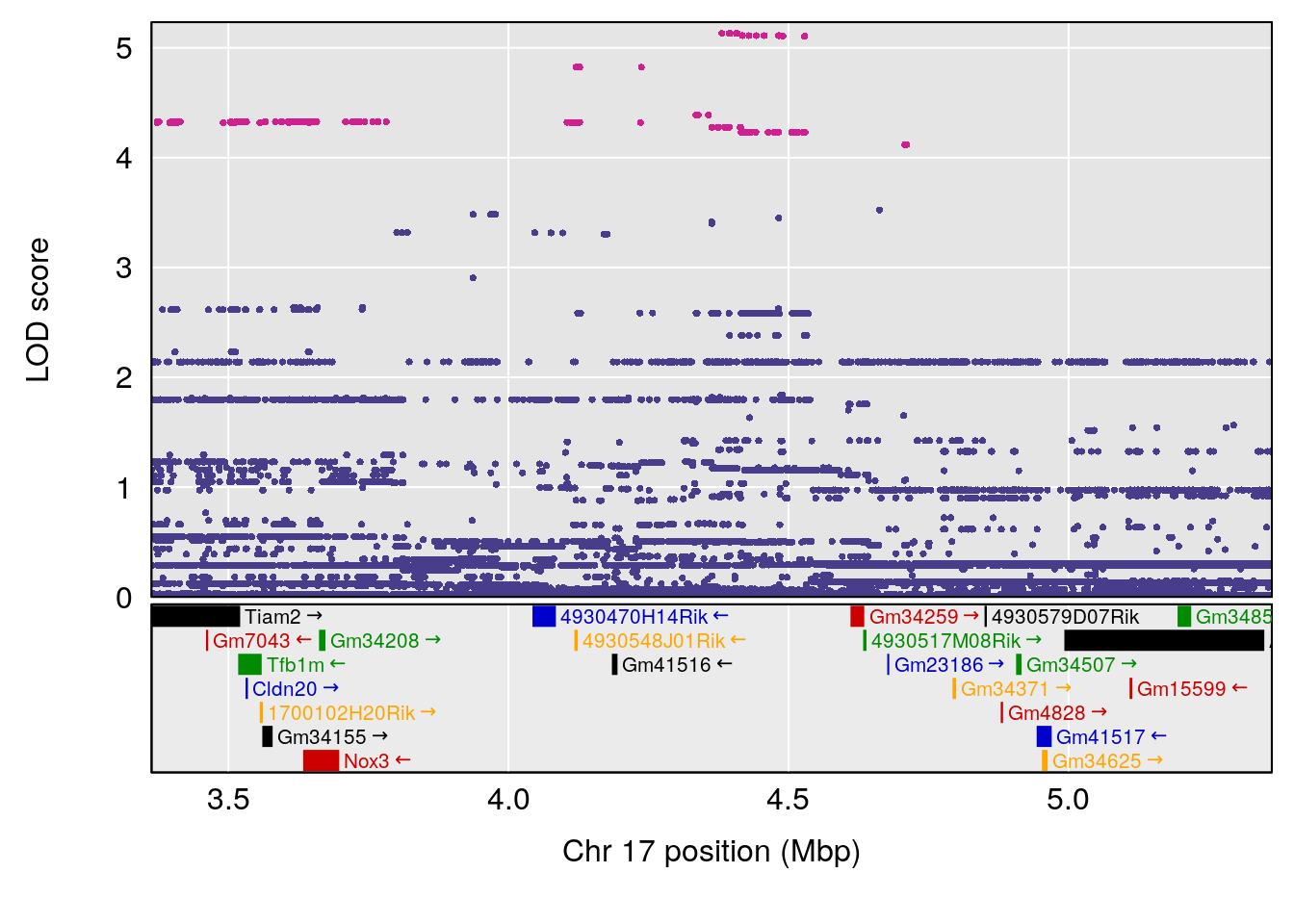
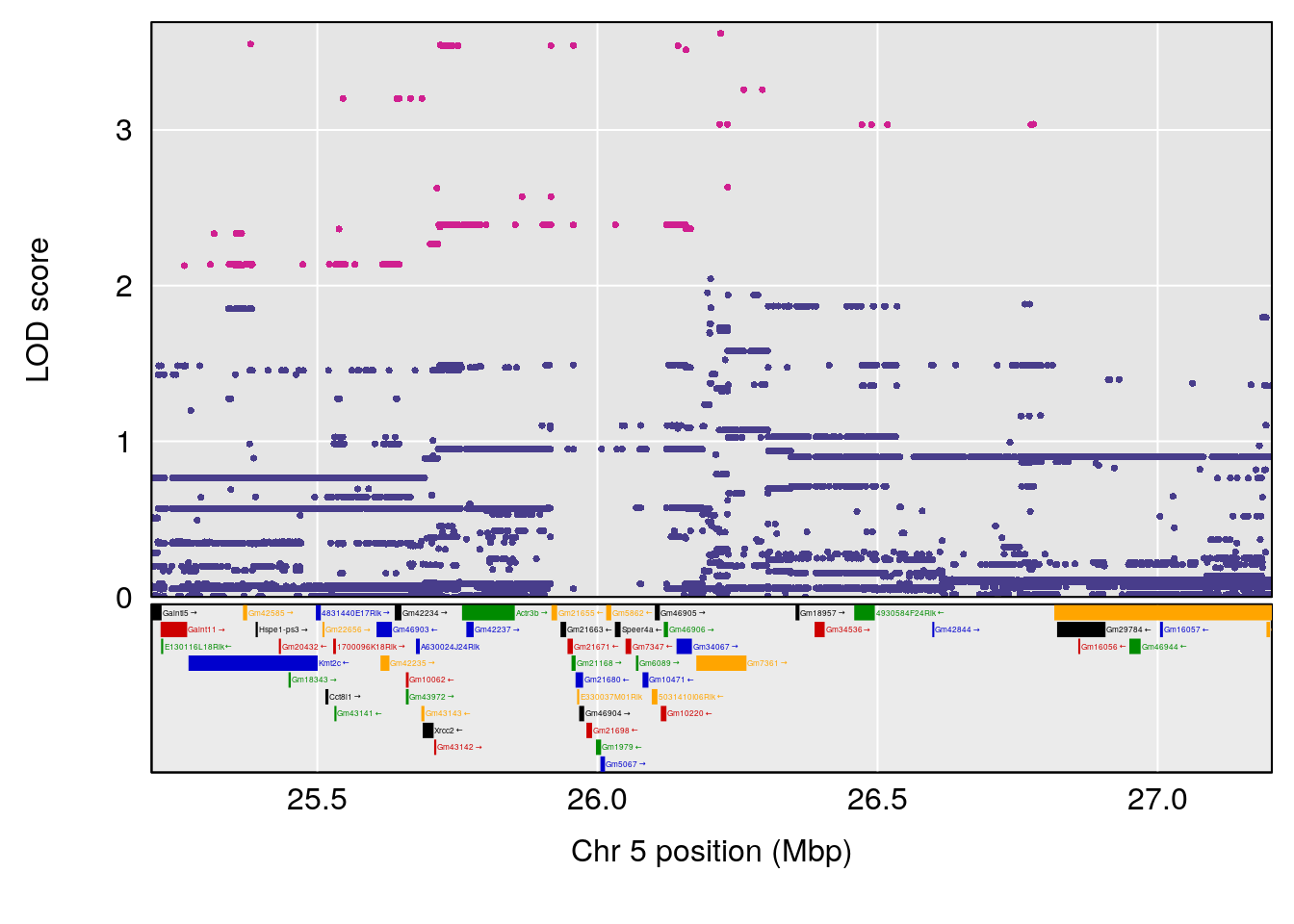
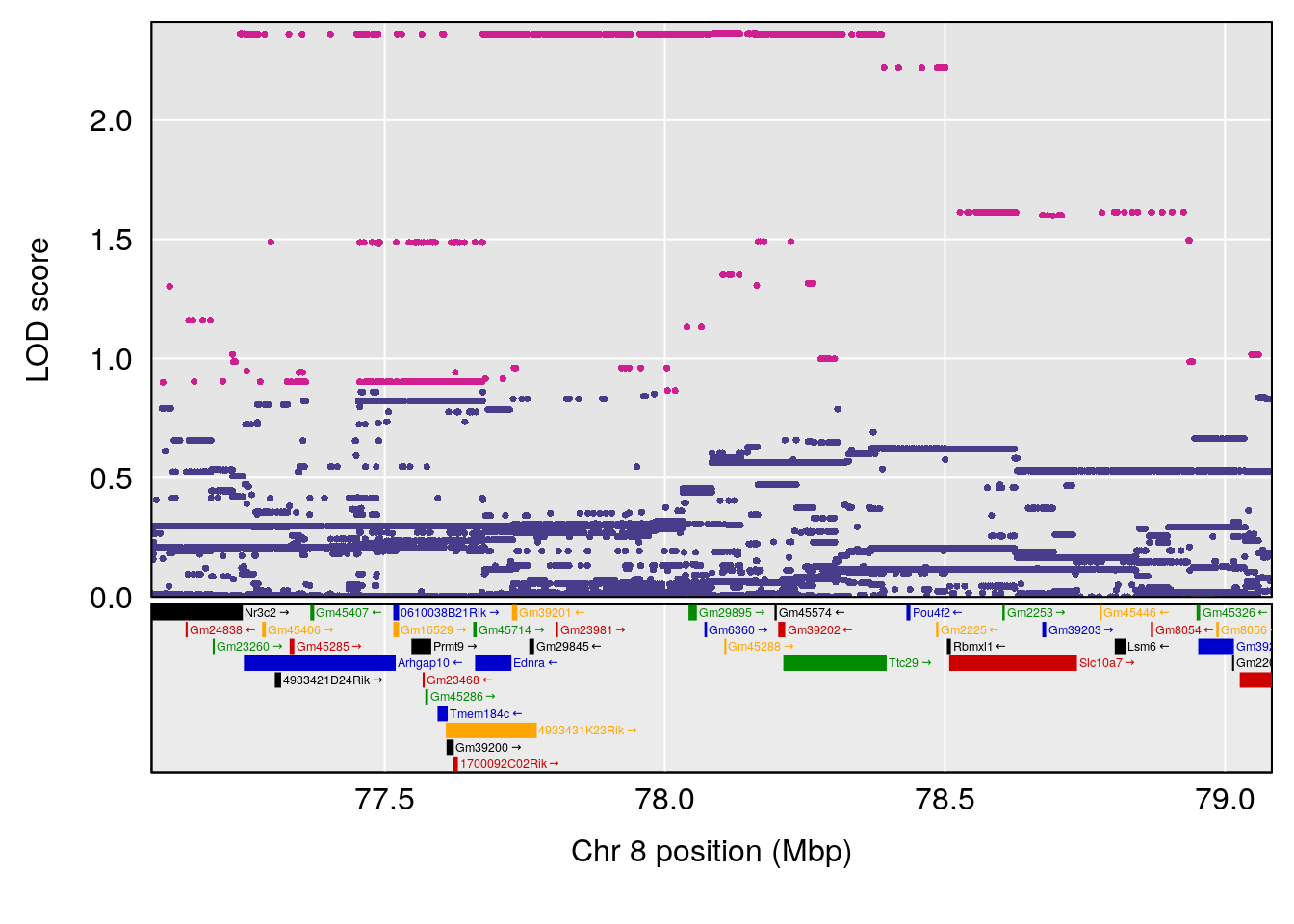
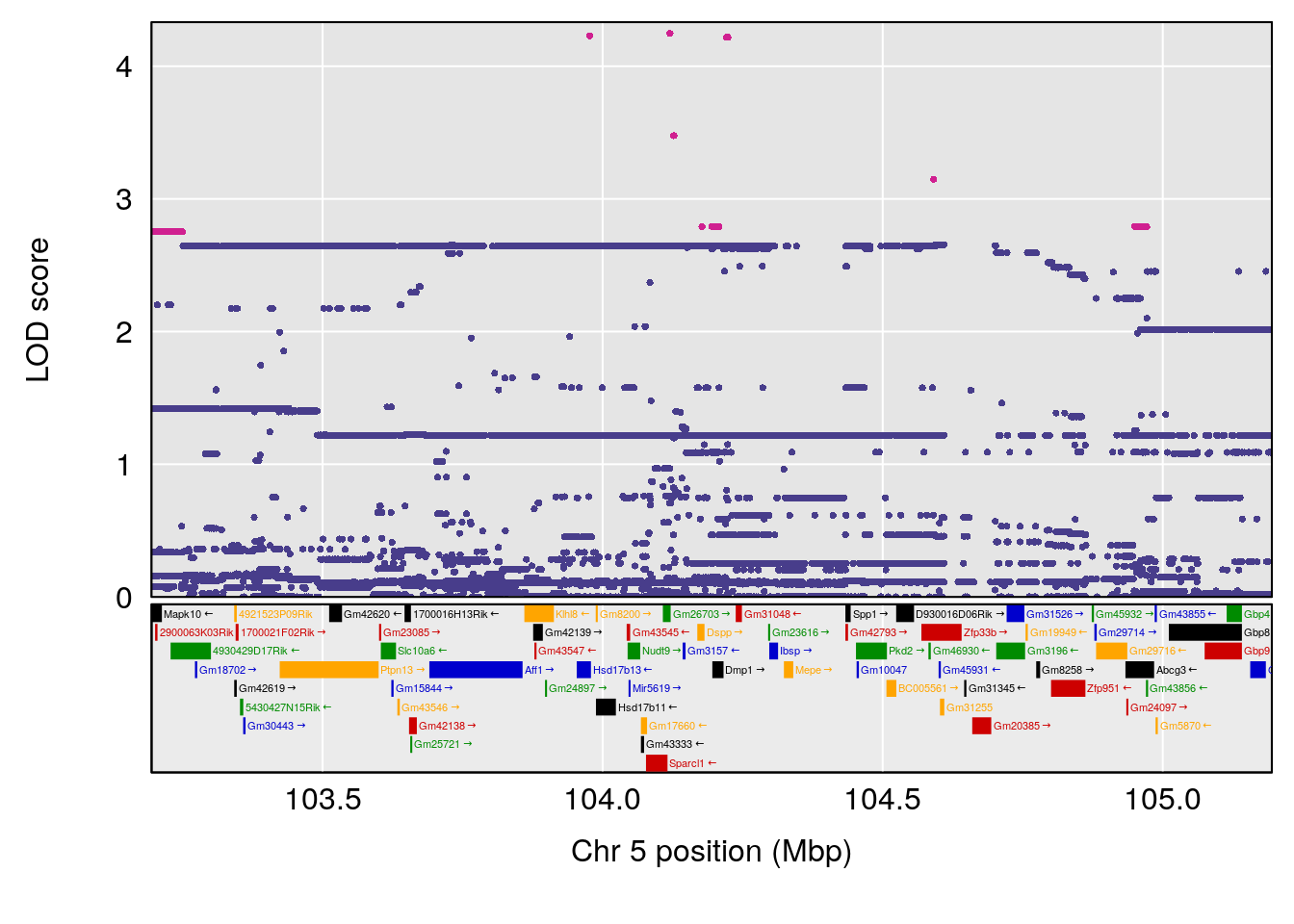
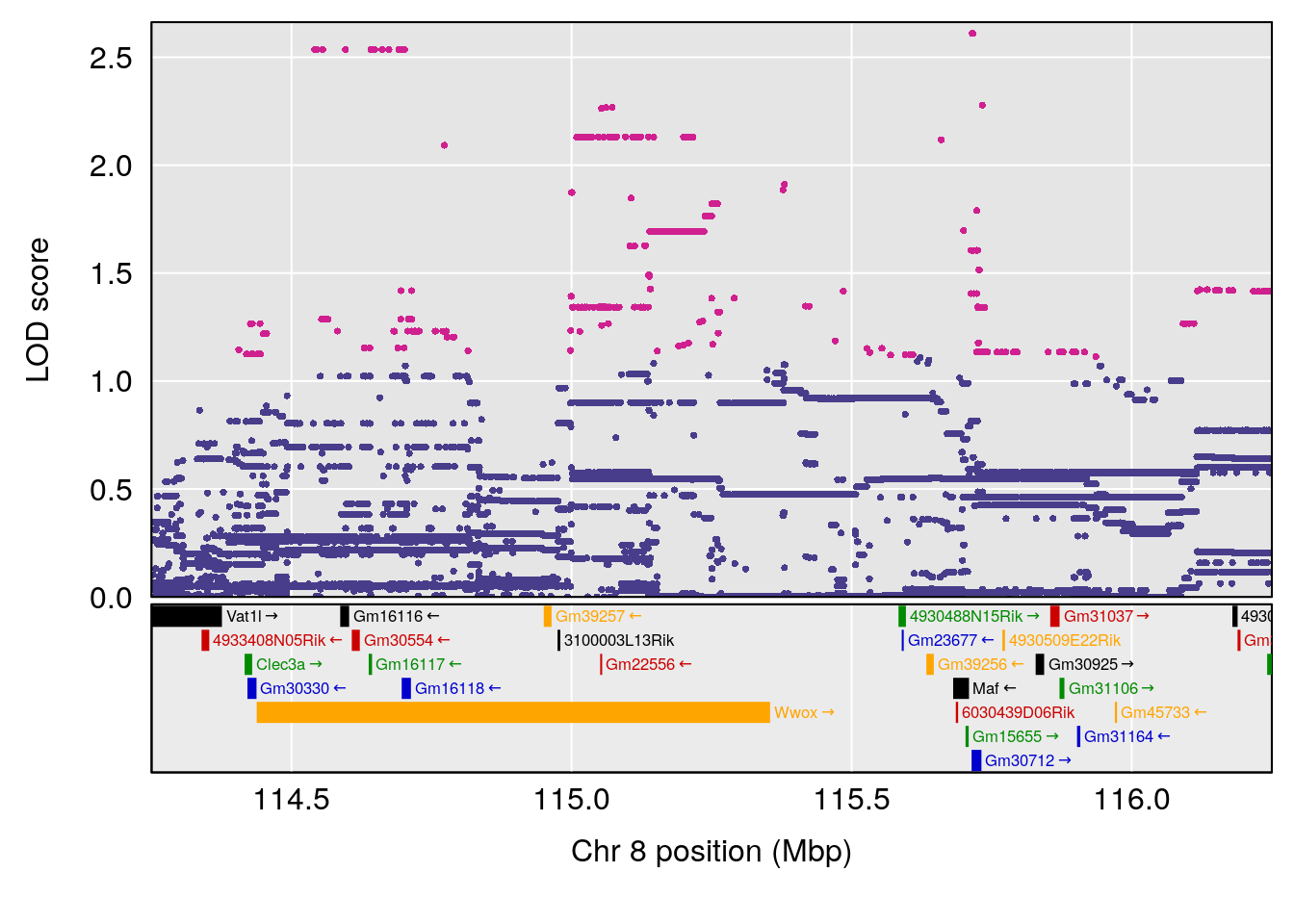
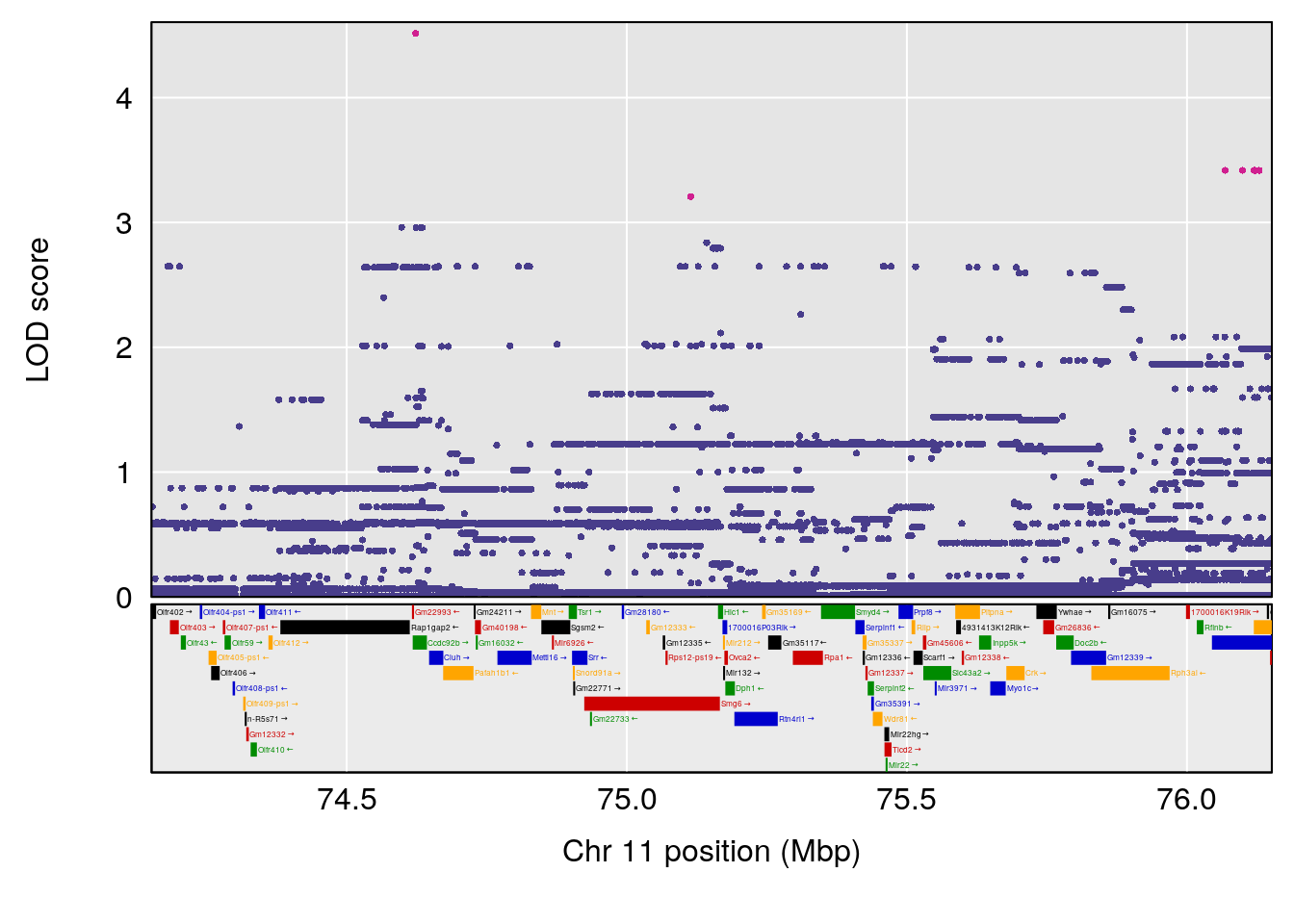
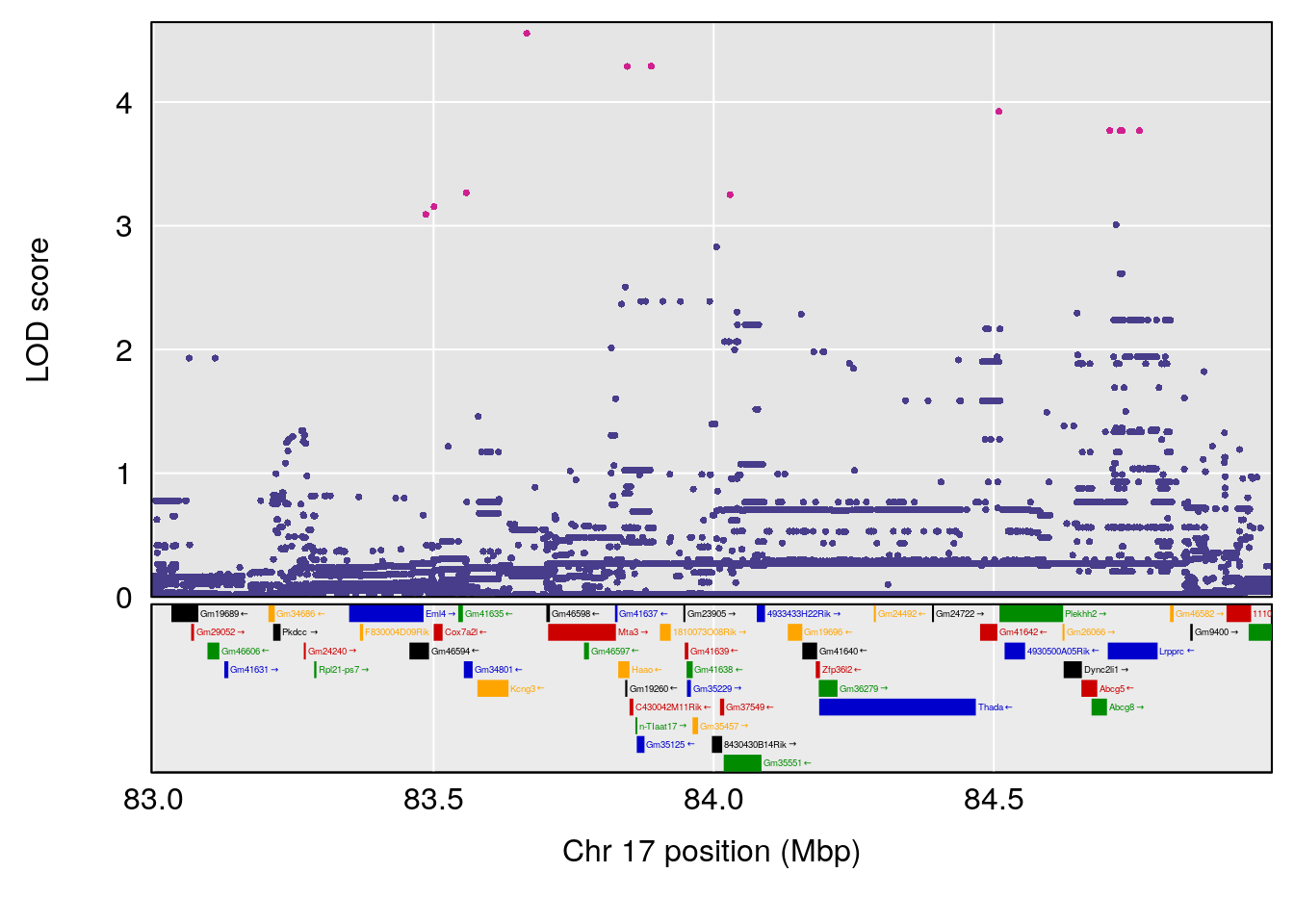
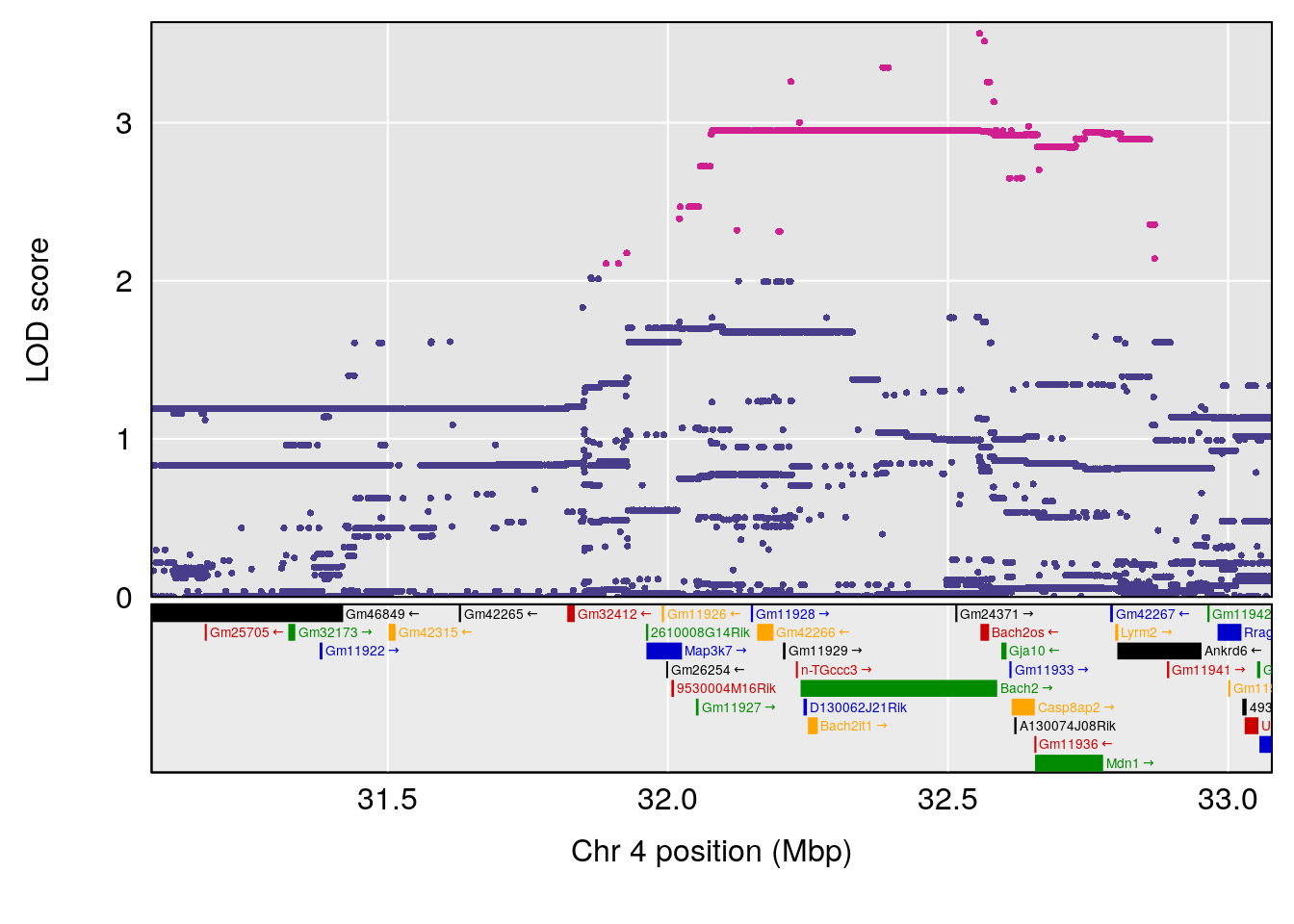
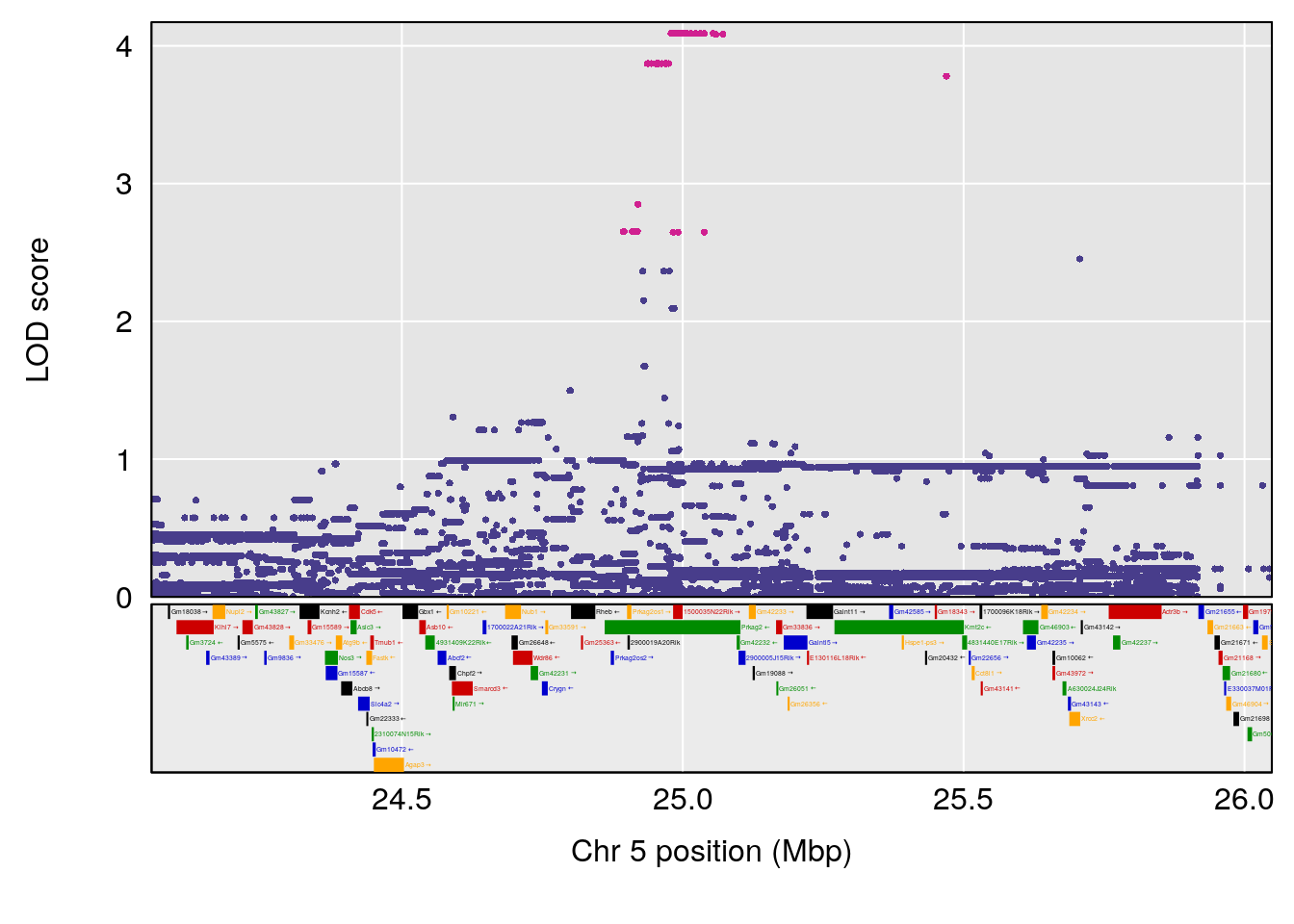
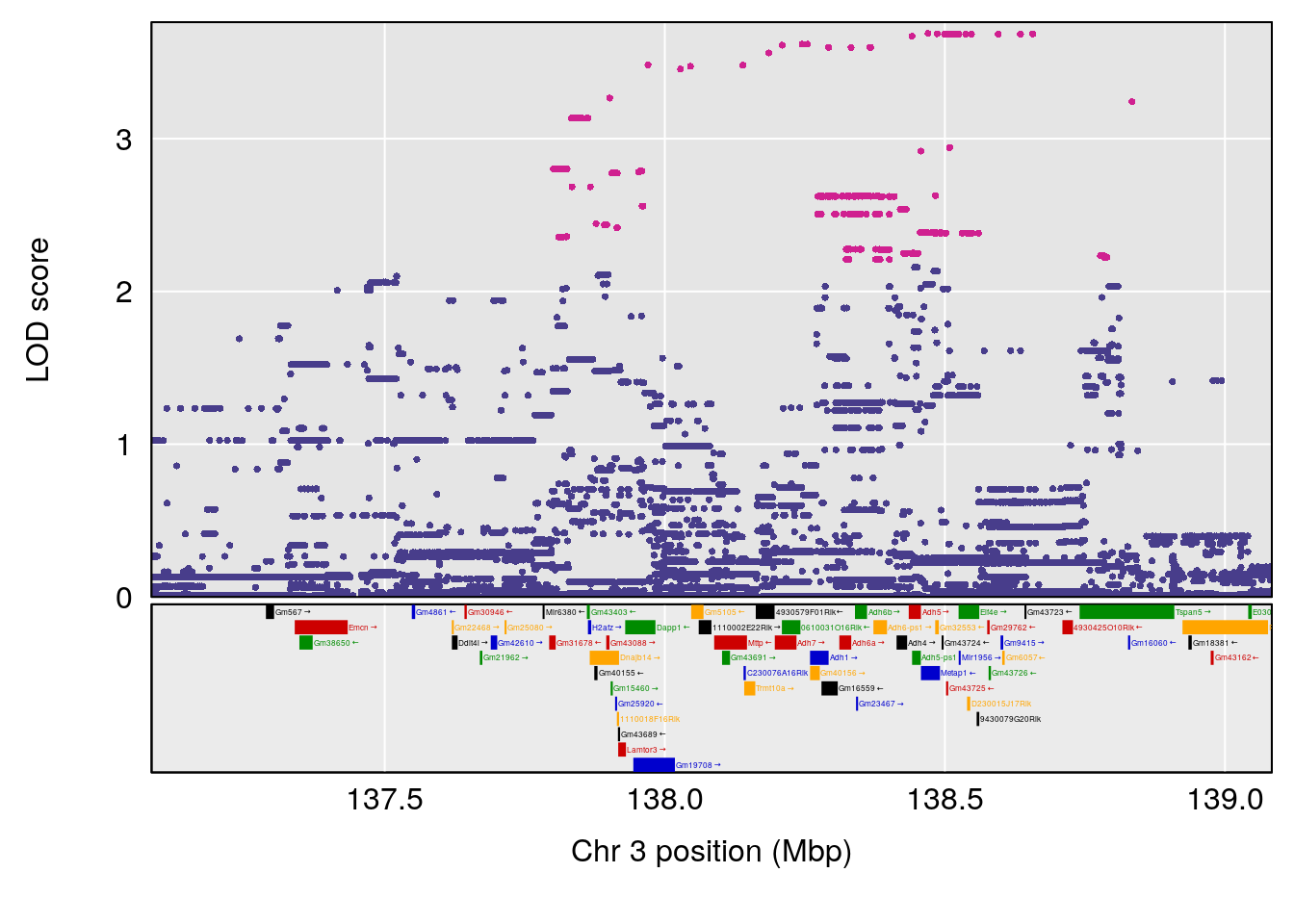
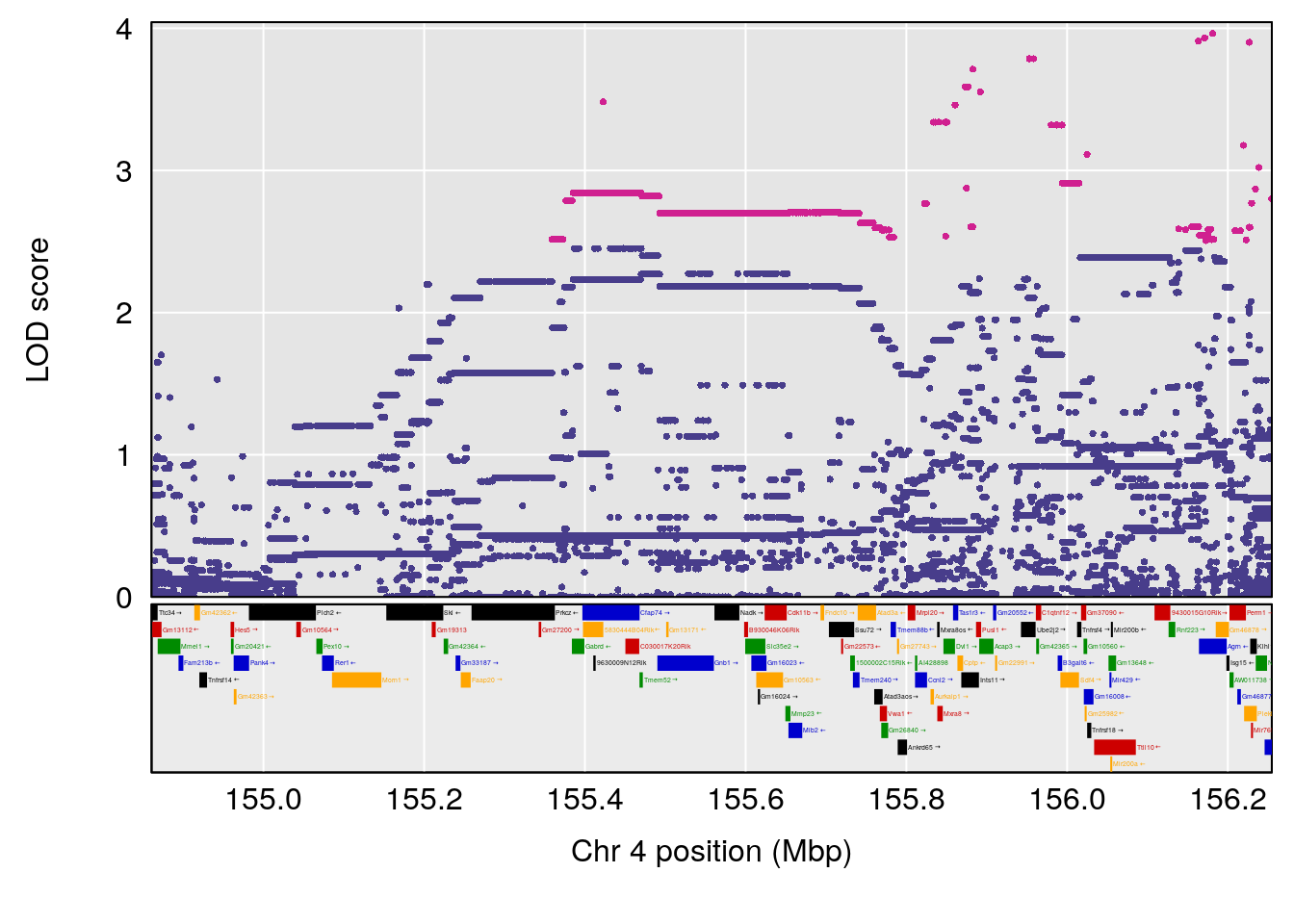
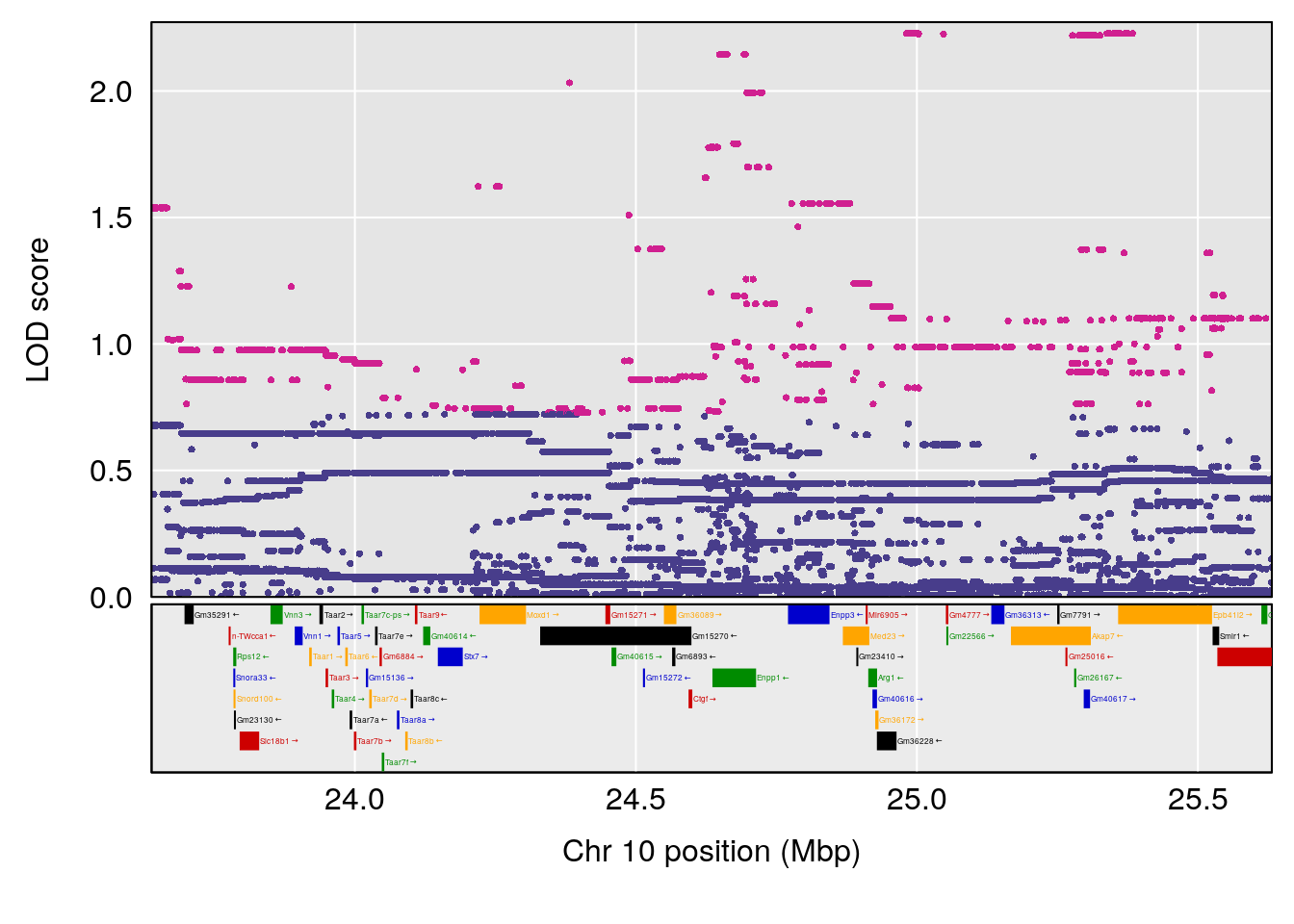
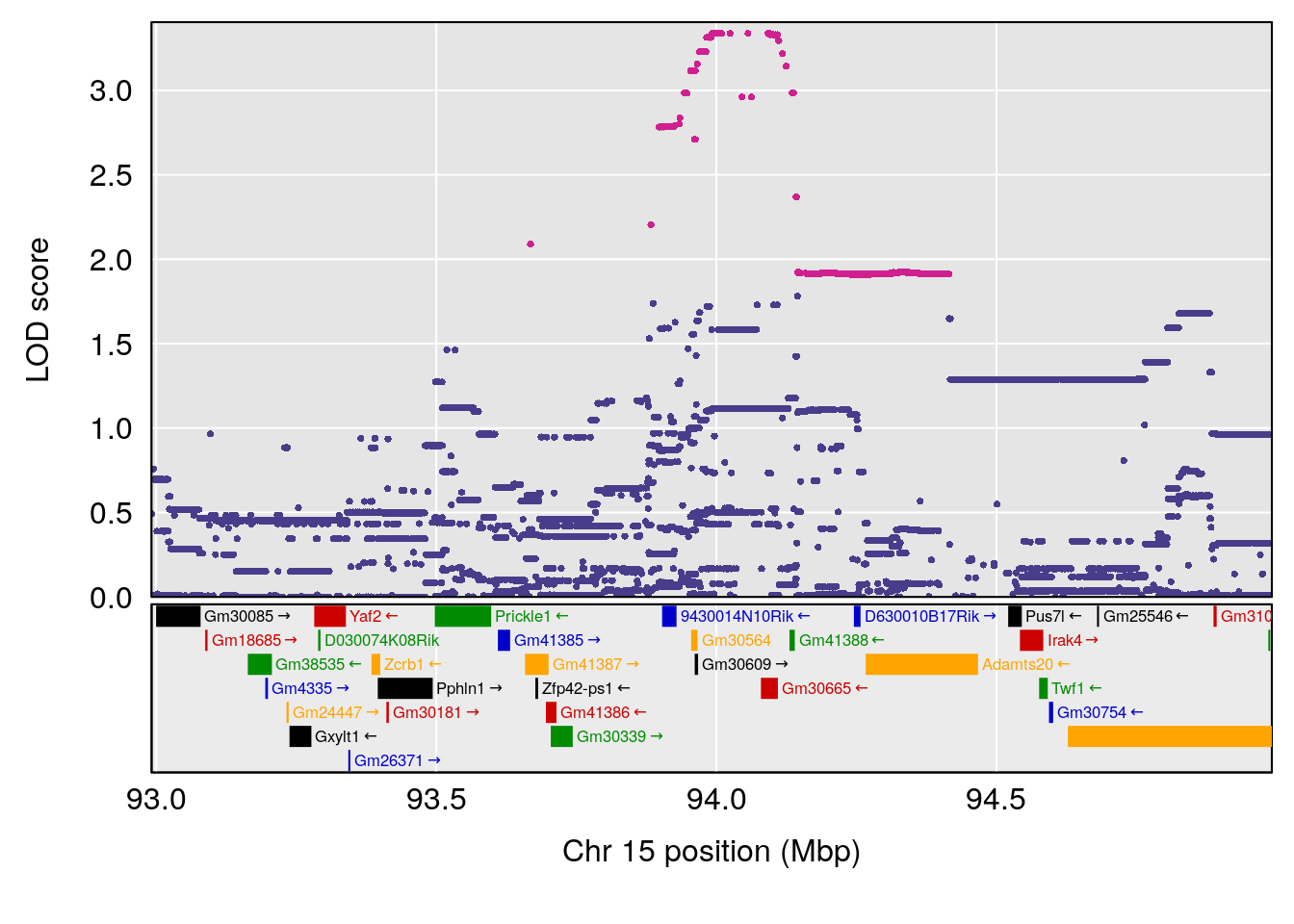
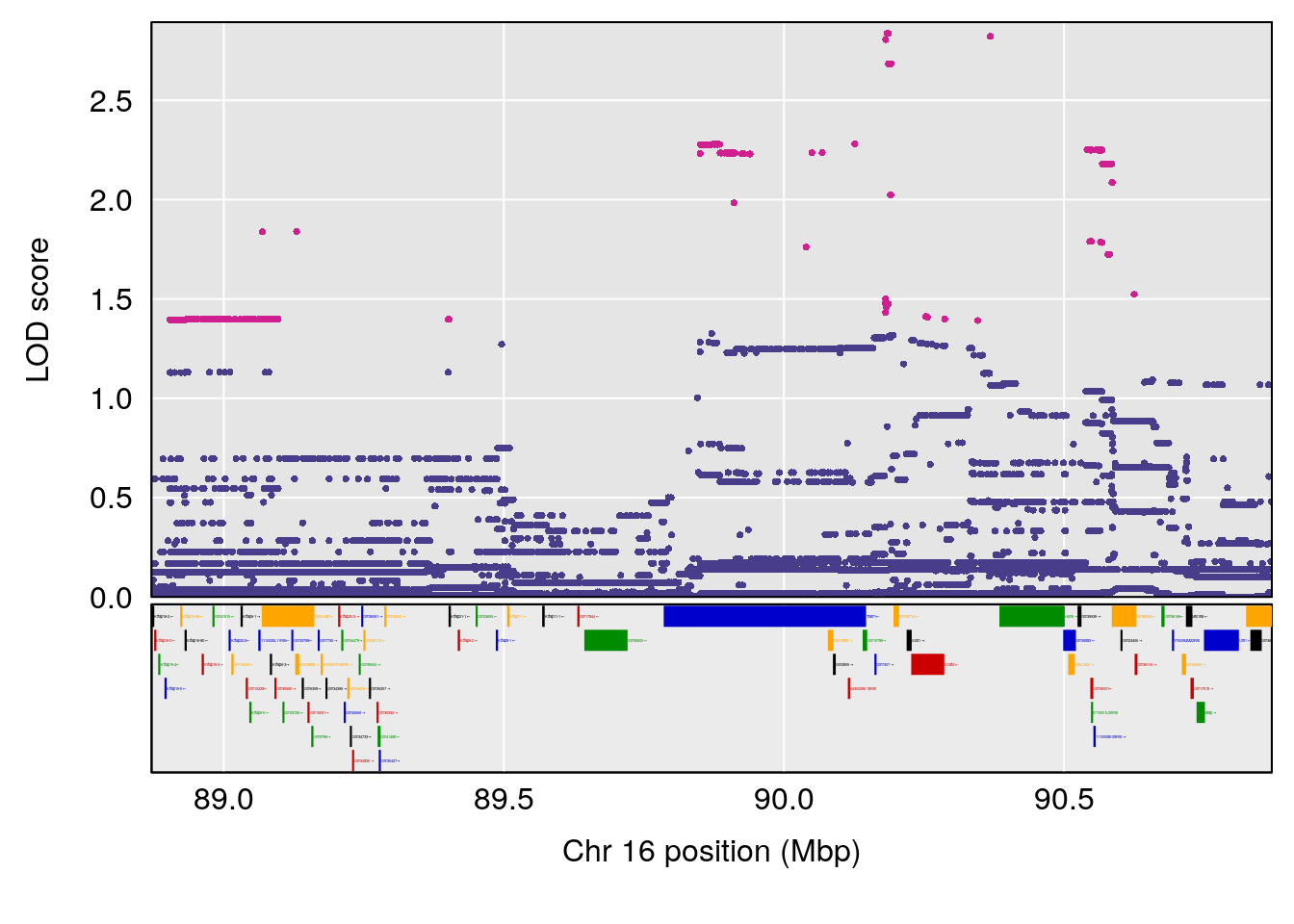
#SNP association of chr5 for Min.depression
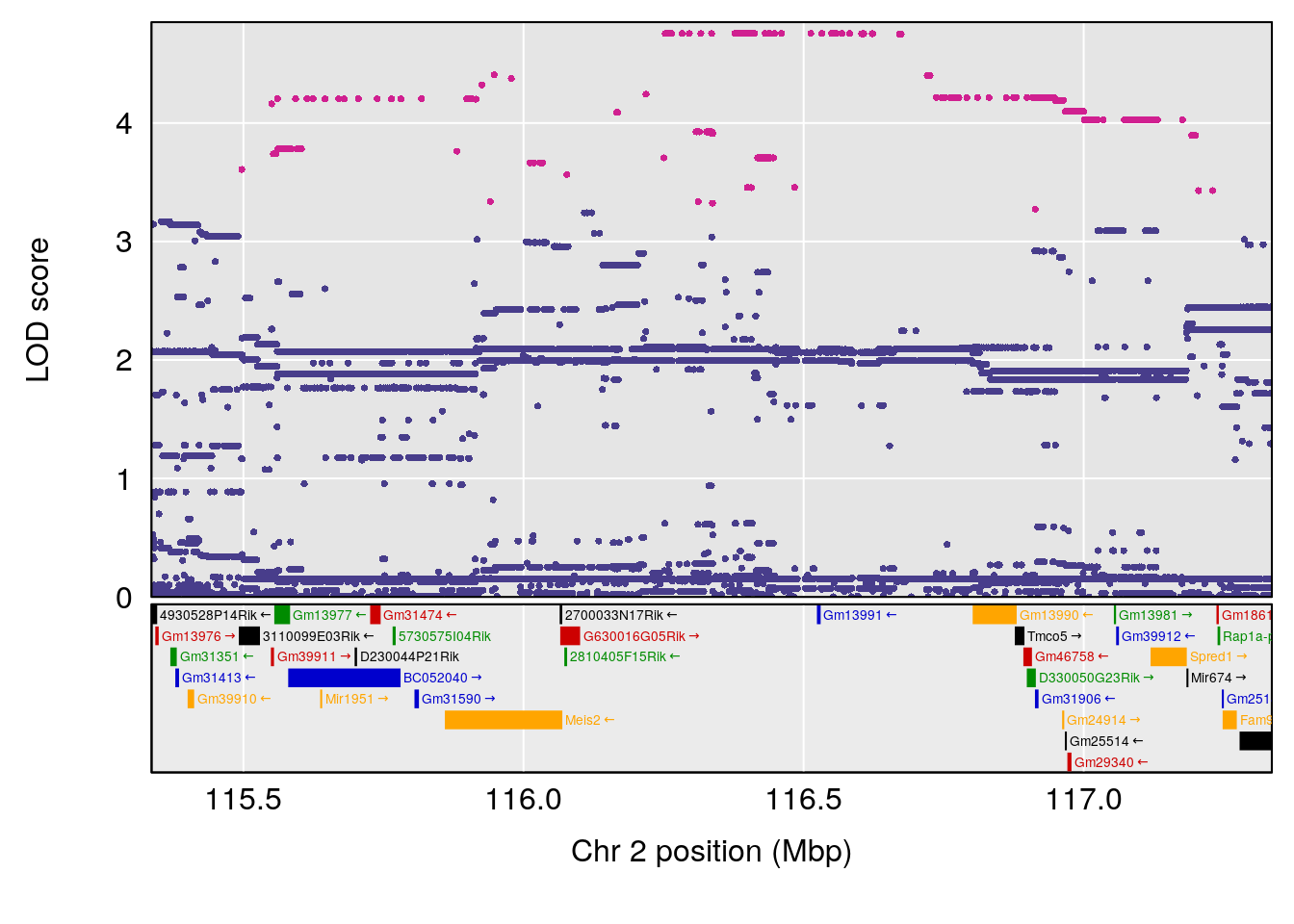
i = "Min.depression"
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot(out_snps[[i]][[1]]$lod, out_snps[[i]][[1]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[1]])
png(file = paste0("output/DO_morphine_", i, "_peak_genes_chr5.png"), width = 16, height =8, res=300, units = "in")
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot(out_snps[[i]][[1]]$lod, out_snps[[i]][[1]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[1]])
dev.off()
# png
# 2
#SNP table
top <- top_snps(out_snps[[i]][[1]]$lod, out_snps[[i]][[1]]$snpinfo,show_all_snps = T, drop = 1.5)
dim(top)
# [1] 406 20
write.csv(top, file = "output/topSNP_chr5_mindepression.csv", row.names = F)
#violin at chr5 for Min.depression
#need the apr
aprobs <- readRDS(file = "output/do.morphine.probs_8state.rds")
#morphine pheno
morphine.pheno <- read.csv("data/pheno_qtl2_07092018_morphine.csv",header = T)
rownames(morphine.pheno) <- gsub("morphine_DO_jbubier_","", morphine.pheno$X)
chr = 5
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
marker <- find_marker(pmap, chr, pos=peaks[peaks$chr==chr, "pos"])
peak_Mbp <- pmap[[chr]][marker]
g <- maxmarg(aprobs, do.morphine$gmap, chr=chr, minprob=0.4, pos=peaks[peaks$chr==chr, "pos"], return_char = TRUE)
stopifnot(all.equal(names(g), rownames(morphine.pheno)))
px <- cbind(g, morphine.pheno)
colnames(px)[1] <- "Strain"
px <- px[!is.na(px$Strain),]
png(paste("output/DO_morphine_Min.depression_violin_chr",chr,".png",sep=""),
width = 24, height =16, res=300, units = "in")
p <- ggplot(px, aes_string(x="Strain", y="Min.depression", group="Strain", fill="Strain")) +
geom_violin() +
geom_point() +
scale_fill_manual(labels = c("A/J", "C57BL/6J", "129S1/SvImJ", "NOD/ShiLtJ", "NZO/HlLtJ", "CAST/EiJ", "PWK/PhJ", "WSB/EiJ"),
values=as.vector(qtl2::CCcolors)) +
scale_x_discrete(labels=c("A" = "A/J",
"B" = "C57BL/6J",
"C" = "129S1/SvImJ",
"D" = "NOD/ShiLtJ",
"E" = "NZO/HlLtJ",
"F" = "CAST/EiJ",
"G" = "PWK/PhJ",
"H" = "WSB/EiJ")) +
xlab(paste0("marker:", marker, " chr", chr, ":", peak_Mbp, "Mbp")) +
theme(axis.text.x = element_text(angle = -45, hjust = 0.1, vjust = 0.1),
text = element_text(size=14))
print(p)
dev.off()
# png
# 2
p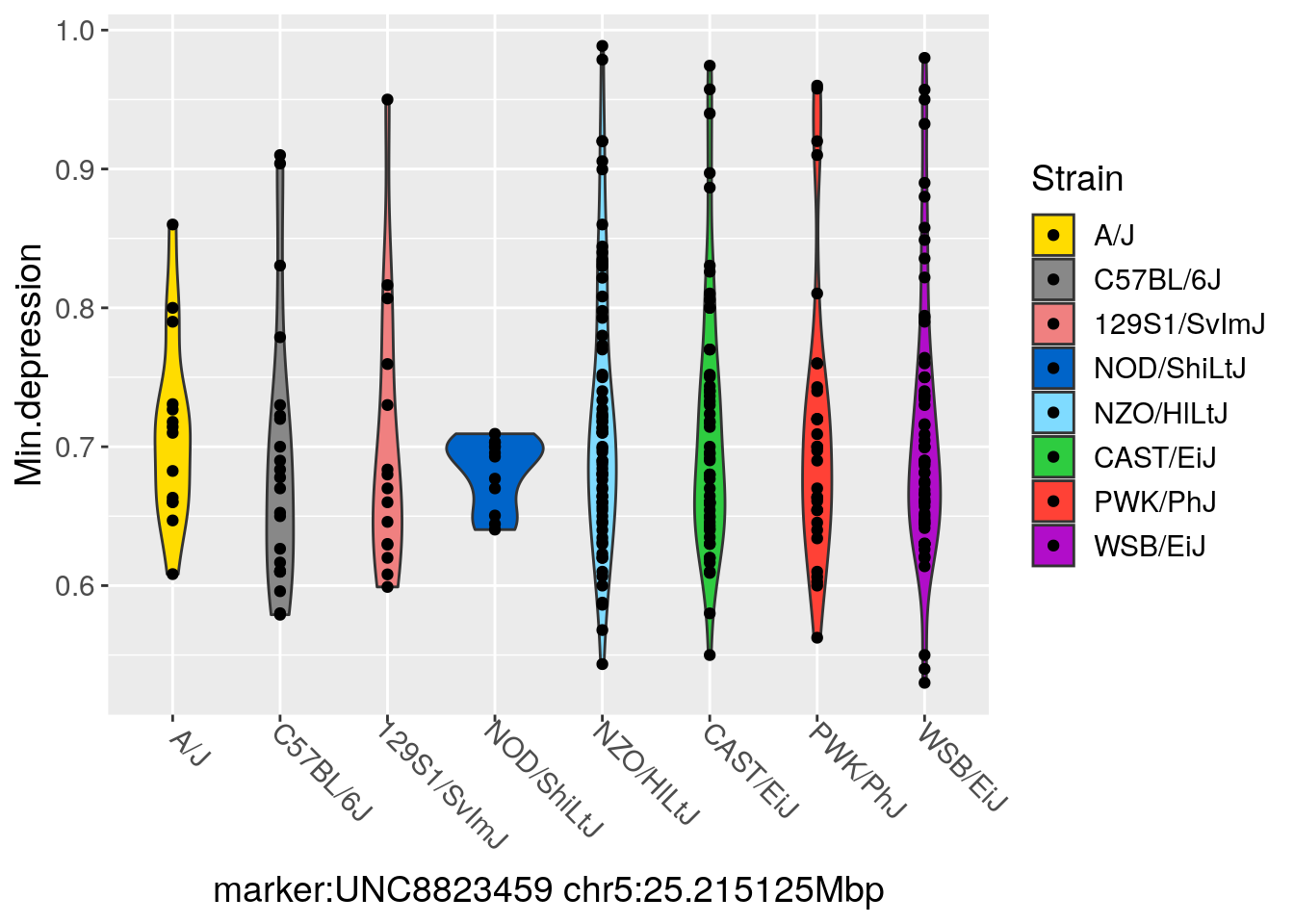
Plot CoxPH results
#markers
pmap_all <- read.csv("data/physical_map.csv", header = T,stringsAsFactors = F)
gmap_all <- read.csv("data/genetic_map.csv", header = T,stringsAsFactors = F)
markers <- data.frame(stringsAsFactors = F,
marker = as.character(pmap_all$marker),
chr = as.character(pmap_all$chr),
pos = as.numeric(pmap_all$pos)*10^6,
cM = as.numeric(gmap_all$pos))
rownames(markers) <- markers$marker
markers <- markers[marker_names(do.morphine),]
load("output/DO_morphine_cphout.RData")
out.cph2 <- as.matrix(out.cph[,1,drop=F])
attr(out.cph2, "class") <- c("scan1", "matrix")
#plot
plot(out.cph2, map = pmap, type = "l", lwd = 1, main = "Do_morphine: COX PH model")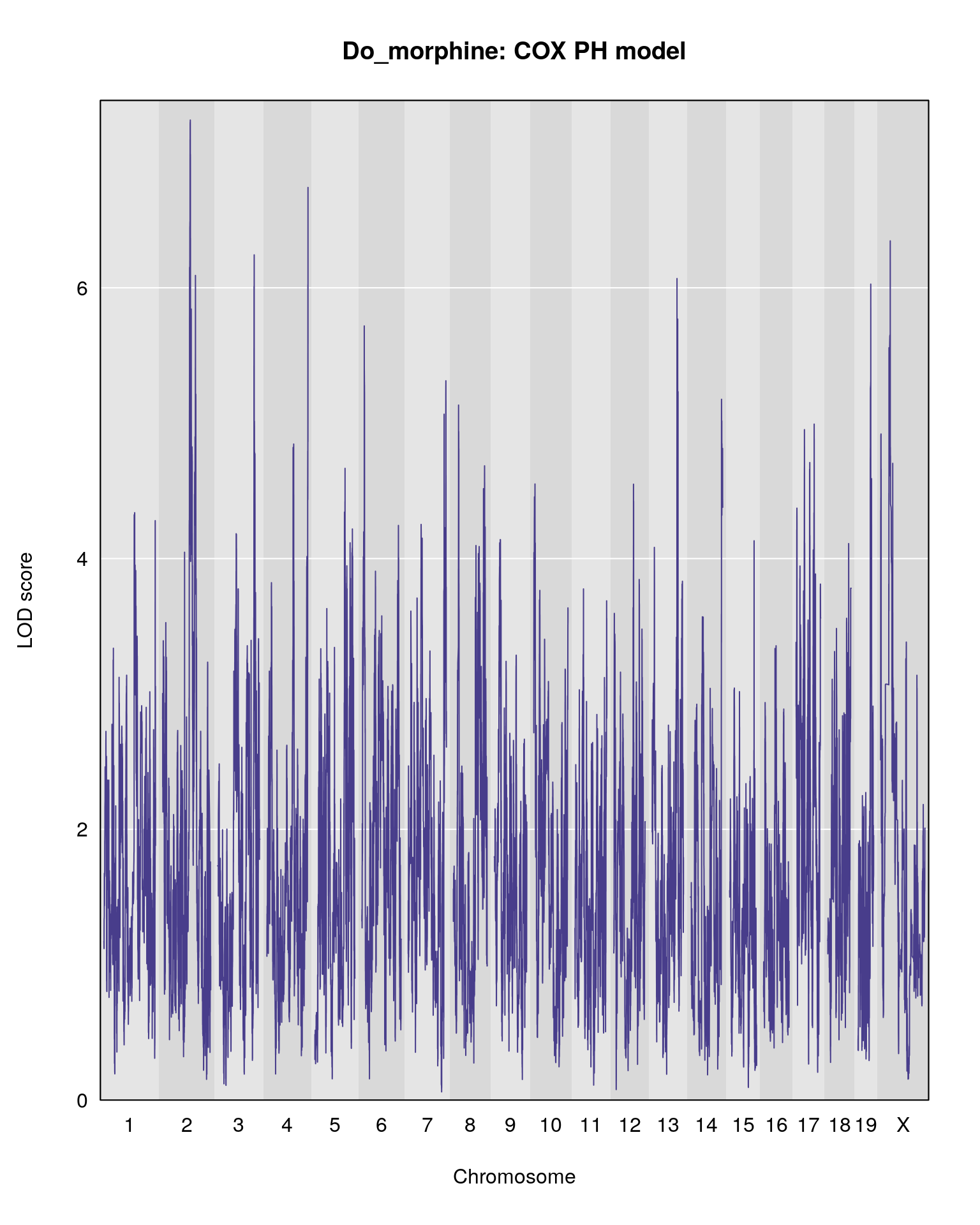
#save CoxPH plot
png("output/DO_morphine_coxph_24hrs_kinship_QTL.png", width = 1000, height = 800, res = 128)
plot(out.cph2, map = pmap, type = "l", lwd = 1, main = "Do_morphine: COX PH model")
dev.off()
# png
# 2
#coeff plot
coef <- out.cph[,c(-1:-9)]
lod <- out.cph[,1]
col = do.colors
col[1,3] = "#FFC000"
for(i in c(1:19, "X")) {
png(paste0("output/coxph/DO_morphine_coxph_24hrs_kinship_QTL_chr", i, ".png"), width = 1200,
height = 1000, res = 128)
rng = which(markers[,2] == i)
tmp = coef[rng,c(2:ncol(coef))]
tmp[,1] = 0
tmp = tmp - rowMeans(tmp)
colnames(tmp)[1] = "A"
layout(matrix(1:2, 2, 1), heights = c(0.8, 0.2))
par(plt = c(0.15, 0.99, 0, 0.9))
plot(markers[rng,3] * 1e-6, tmp[,1], lwd = 2, type = "l", col = col[1,3],
main = paste("Do_morphine_coxph_24hrs: COX PH model: Chr", i),
ylim = range(tmp), las = 1, xaxt = "n", ylab = "")
mtext(side = 2, line = 4, text = "Founder Effects")
for(j in 2:8) {
points(markers[rng,3] * 1e-6, tmp[,j], lwd = 2, type = "l", col = col[j,3])
} # for(j)
par(plt = c(0.15, 0.99, 0.25, 1))
plot(markers[rng,3] * 1e-6, lod[rng], type = "l", las = 1, ylab = "LOD")
dev.off()
} # for(i)
#remove some noisy markers in chr 2
i = 2
png(paste0("output/coxph/DO_morphine_coxph_24hrs_kinship_QTL_chr", i, ".removesome_markers.png"), width = 1200,height = 1000, res = 128)
rng = which(markers[,2] == i)
tmp = coef[rng,c(2:ncol(coef))]
tmp[,1] = 0
#remove some markers
tmp = tmp[!rownames(tmp) %in% names(which(abs(rowMeans(tmp)) >5000)),]
markers2 = markers[markers$marker %in% rownames(tmp),]
coef2 <- coef[markers2$marker,]
#redo
rng = which(markers2[,2] == i)
tmp = coef[rng,c(2:ncol(coef2))]
tmp[,1] = 0
tmp = tmp - rowMeans(tmp)
colnames(tmp)[1] = "A"
lod2 <- lod[markers2$marker]
print(all.equal(names(lod2),markers2$marker))
# [1] TRUE
layout(matrix(1:2, 2, 1), heights = c(0.8, 0.2))
par(plt = c(0.15, 0.99, 0, 0.9))
plot(markers2[rng,3] * 1e-6, tmp[,1], lwd = 2, type = "l", col = col[1,3],
main = paste("Do_morphine_coxph_24hrs: COX PH model: Chr", i),
ylim = range(tmp), las = 1, xaxt = "n", ylab = "")
mtext(side = 2, line = 4, text = "Founder Effects")
for(j in 2:8) {
points(markers2[rng,3] * 1e-6, tmp[,j], lwd = 2, type = "l", col = col[j,3])
} # for(j)
par(plt = c(0.15, 0.99, 0.25, 1))
plot(markers2[rng,3] * 1e-6, lod2[rng], type = "l", las = 1, ylab = "LOD")
dev.off()
# png
# 2
#plot
layout(matrix(1:2, 2, 1), heights = c(0.8, 0.2))
par(plt = c(0.15, 0.99, 0, 0.9))
plot(markers2[rng,3] * 1e-6, tmp[,1], lwd = 2, type = "l", col = col[1,3],
main = paste("Do_morphine_coxph_24hrs: COX PH model: Chr", i),
ylim = range(tmp), las = 1, xaxt = "n", ylab = "")
mtext(side = 2, line = 4, text = "Founder Effects")
for(j in 2:8) {
points(markers2[rng,3] * 1e-6, tmp[,j], lwd = 2, type = "l", col = col[j,3])
} # for(j)
par(plt = c(0.15, 0.99, 0.25, 1))
plot(markers2[rng,3] * 1e-6, lod2[rng], type = "l", las = 1, ylab = "LOD")
Plot qtl mapping on array male
load("output/qtl.morphine.out.male.RData")
#pmap and gmap
chr.names <- c("1","2","3","4","5","6","7","8","9","10","11","12","13","14","15","16","17","18","19", "X")
gmap <- list()
pmap <- list()
for (chr in chr.names){
gmap[[chr]] <- do.morphine$gmap[[chr]]
pmap[[chr]] <- do.morphine$pmap[[chr]]
}
attr(gmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
attr(pmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
#genome-wide plot
for(i in names(qtl.morphine.out)){
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.morphine.out[[i]]) # overall maximum LOD score
plot(qtl.morphine.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, max(sapply(qtl.morphine.out, maxlod)) +2))
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("DO_morphine_",i))
}
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |

| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
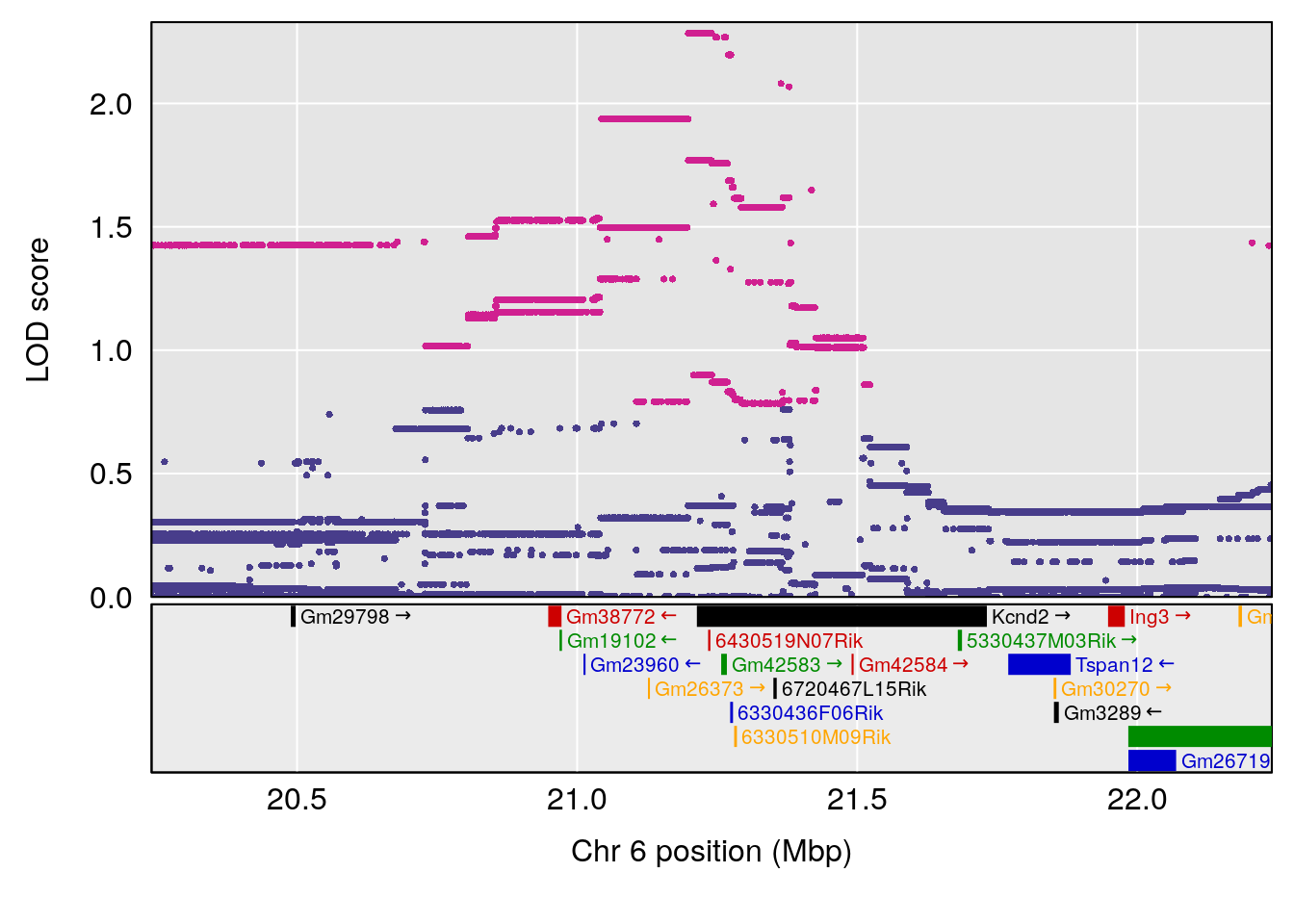
#peaks coeff plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
print(peaks)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
}
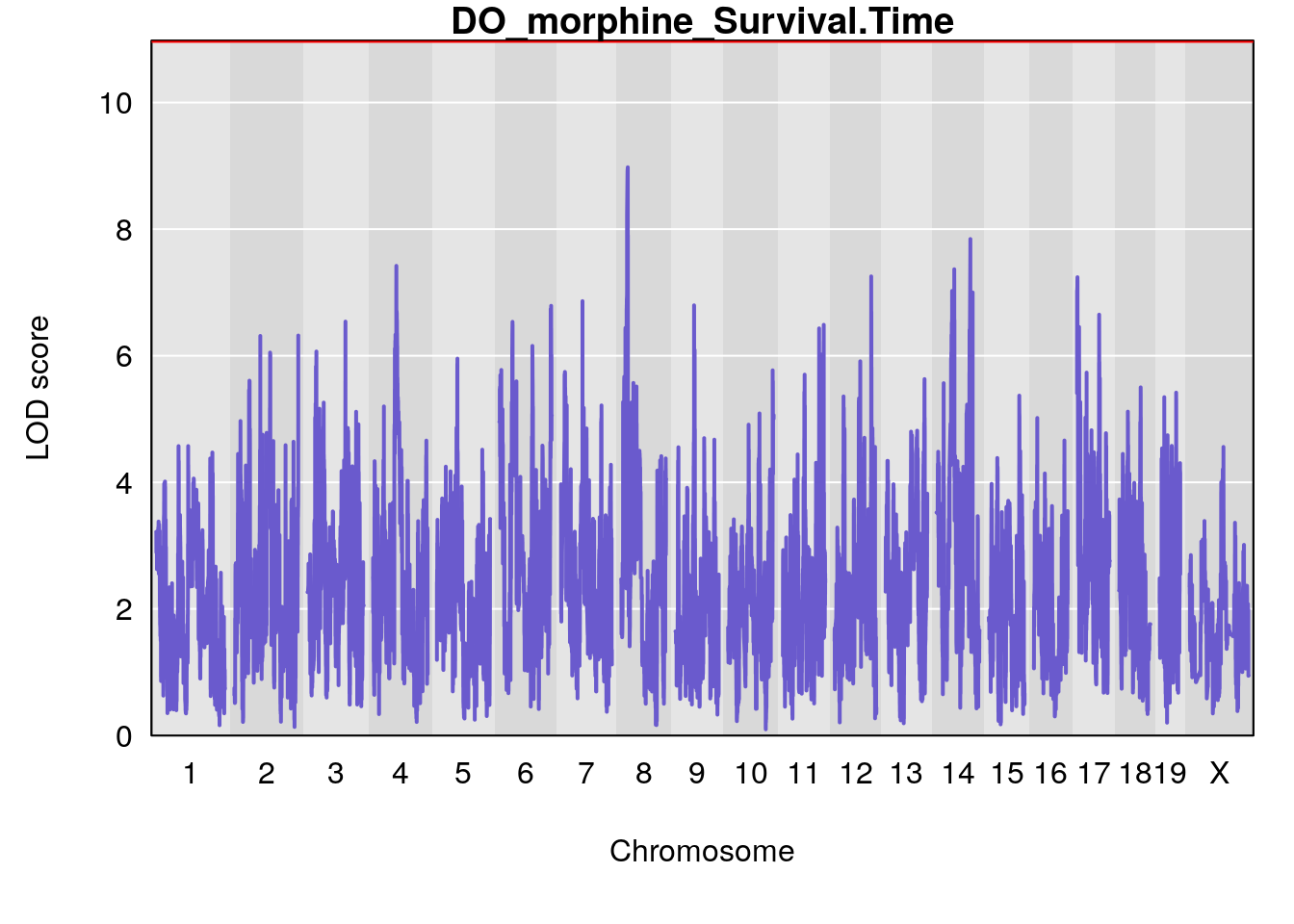
# [1] "Survival.Time"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 2 181.834074 6.319026 20.490494 181.99828
# 2 1 pheno1 3 108.967671 6.539246 25.209613 138.81008
# 3 1 pheno1 4 68.634564 7.419725 64.445383 71.27997
# 4 1 pheno1 6 147.651116 6.788338 4.328042 148.66752
# 5 1 pheno1 7 63.138787 6.862268 13.038070 63.27048
# 6 1 pheno1 8 21.736266 8.977322 19.654408 22.11891
# 7 1 pheno1 9 55.045695 6.795889 54.991021 56.84955
# 8 1 pheno1 11 117.517563 6.488320 63.237669 117.55926
# 9 1 pheno1 12 105.574622 7.253200 74.893565 106.31609
# 10 1 pheno1 14 99.236933 7.843284 47.332527 105.66663
# 11 1 pheno1 17 4.362966 7.240142 3.192903 65.62523
# [1] 1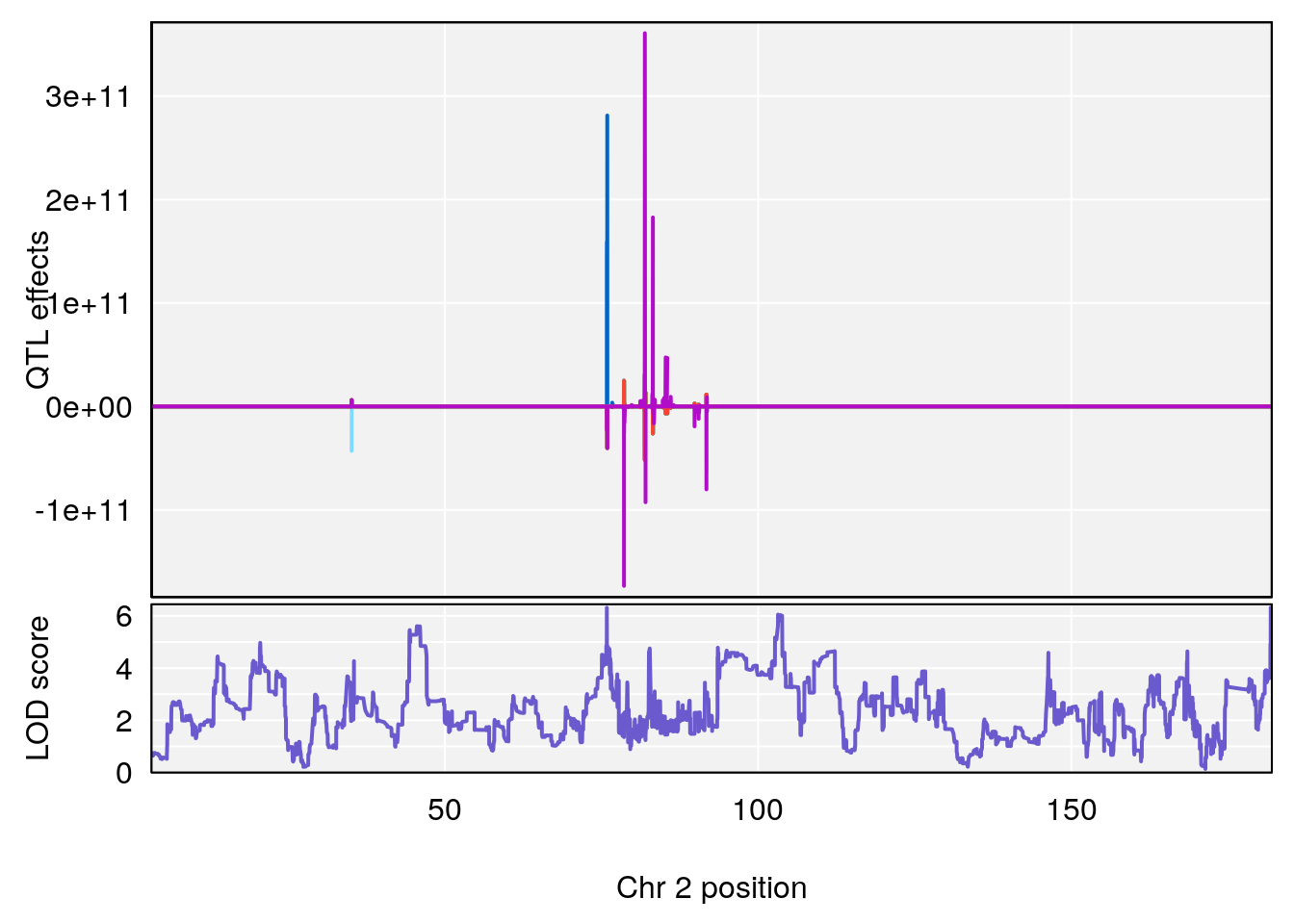
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |

| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 2
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |

| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 3
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 4
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 5
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |

| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 6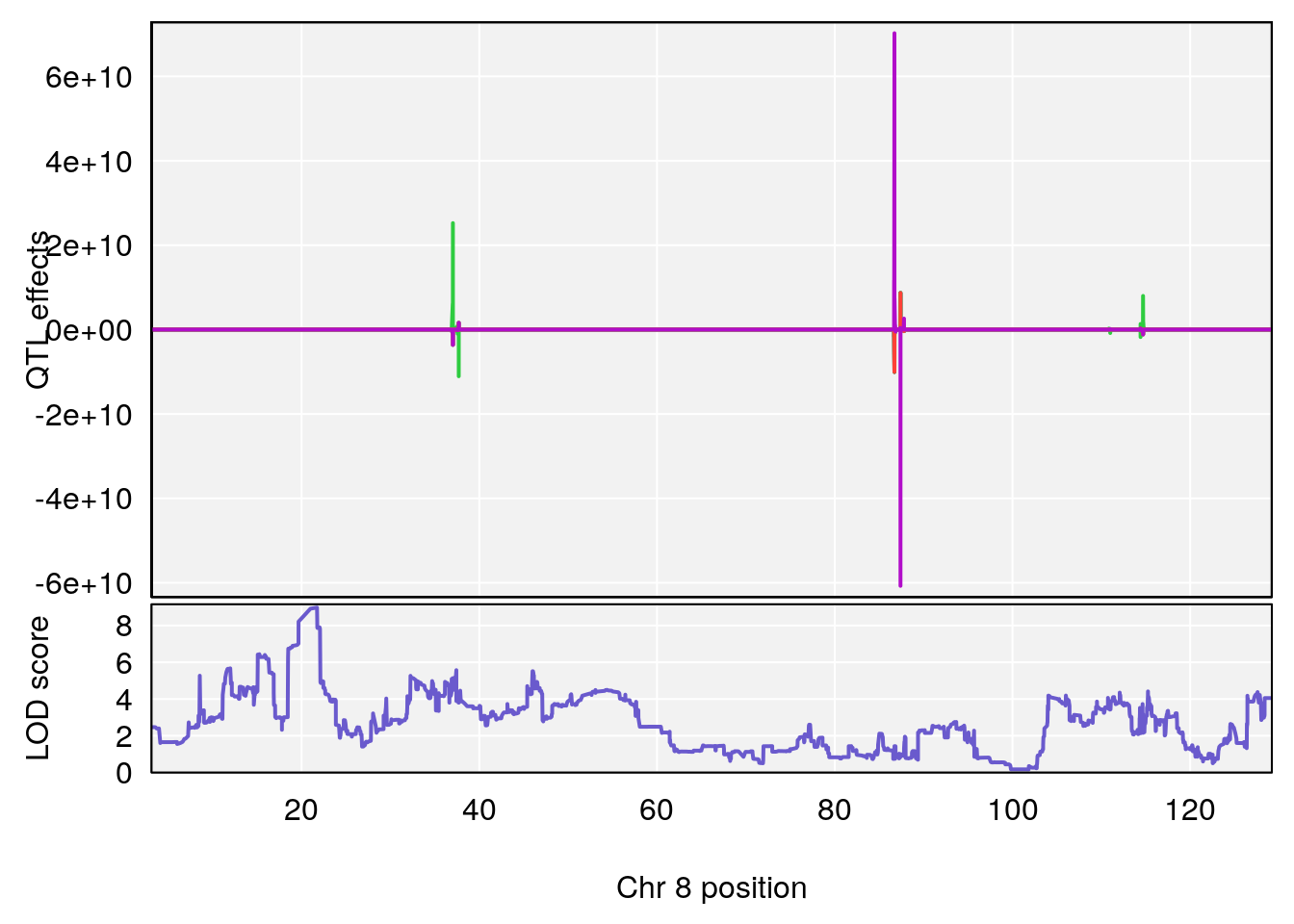
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 7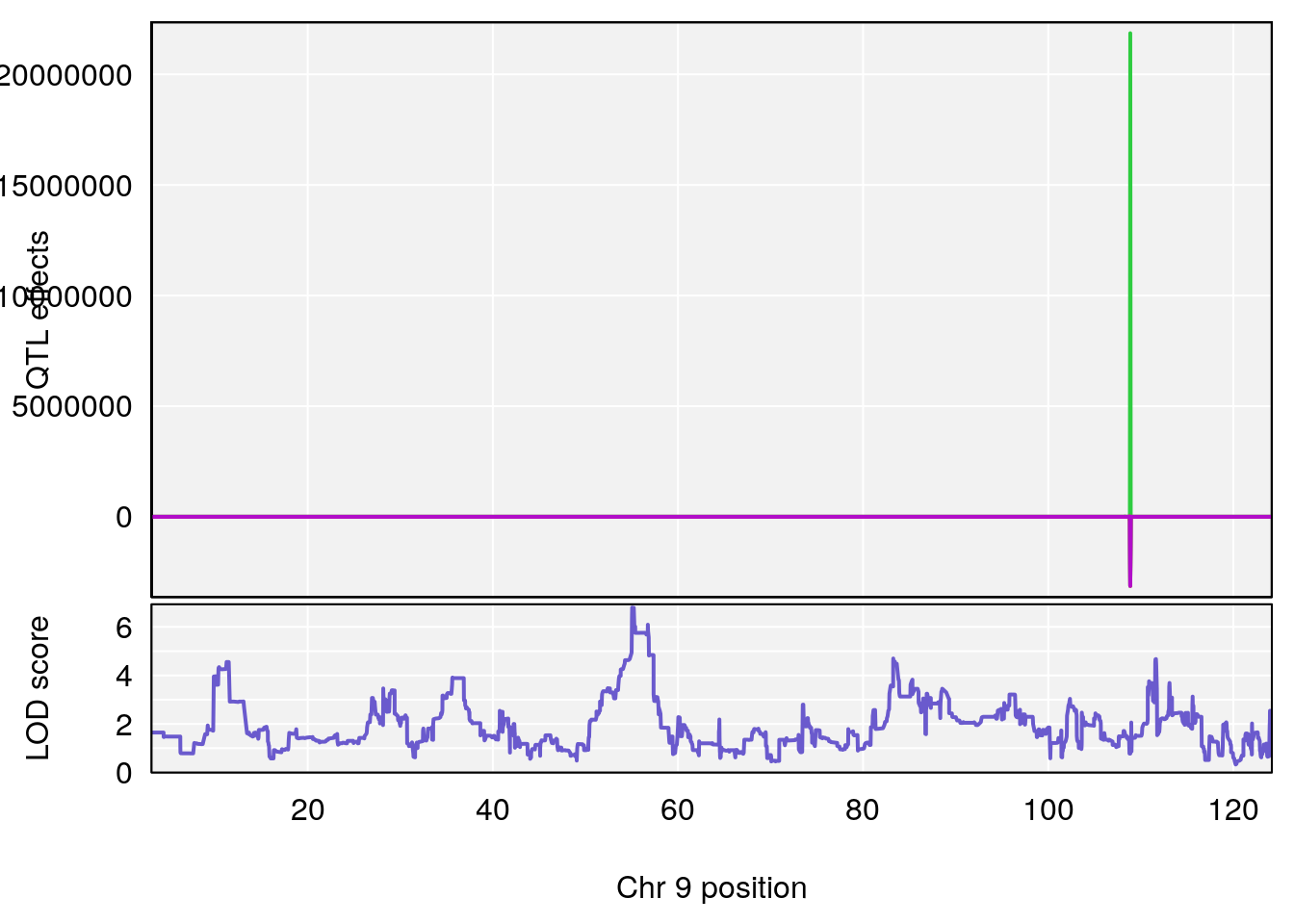
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |

| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 8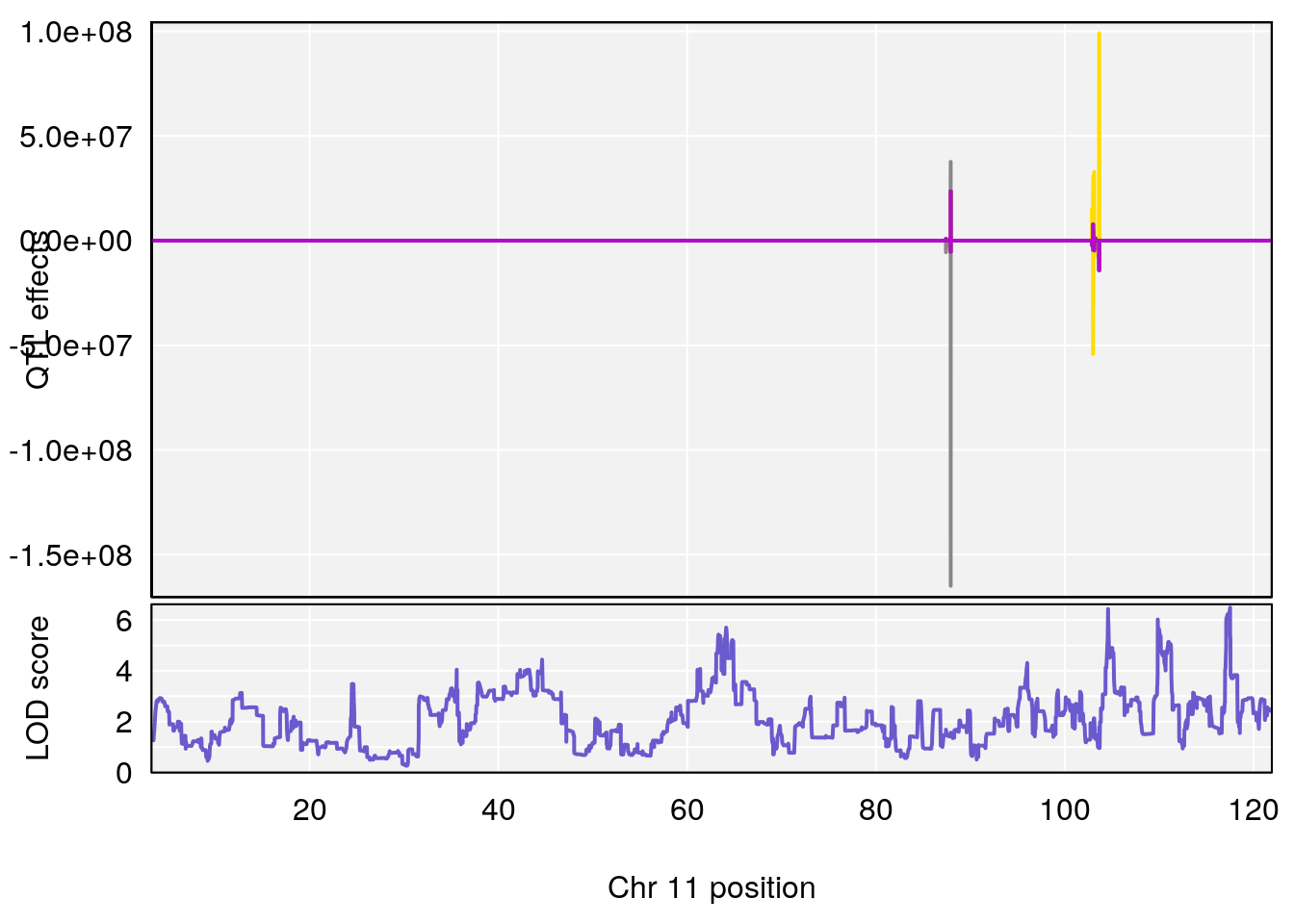
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 9
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 10
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |

| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 11
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
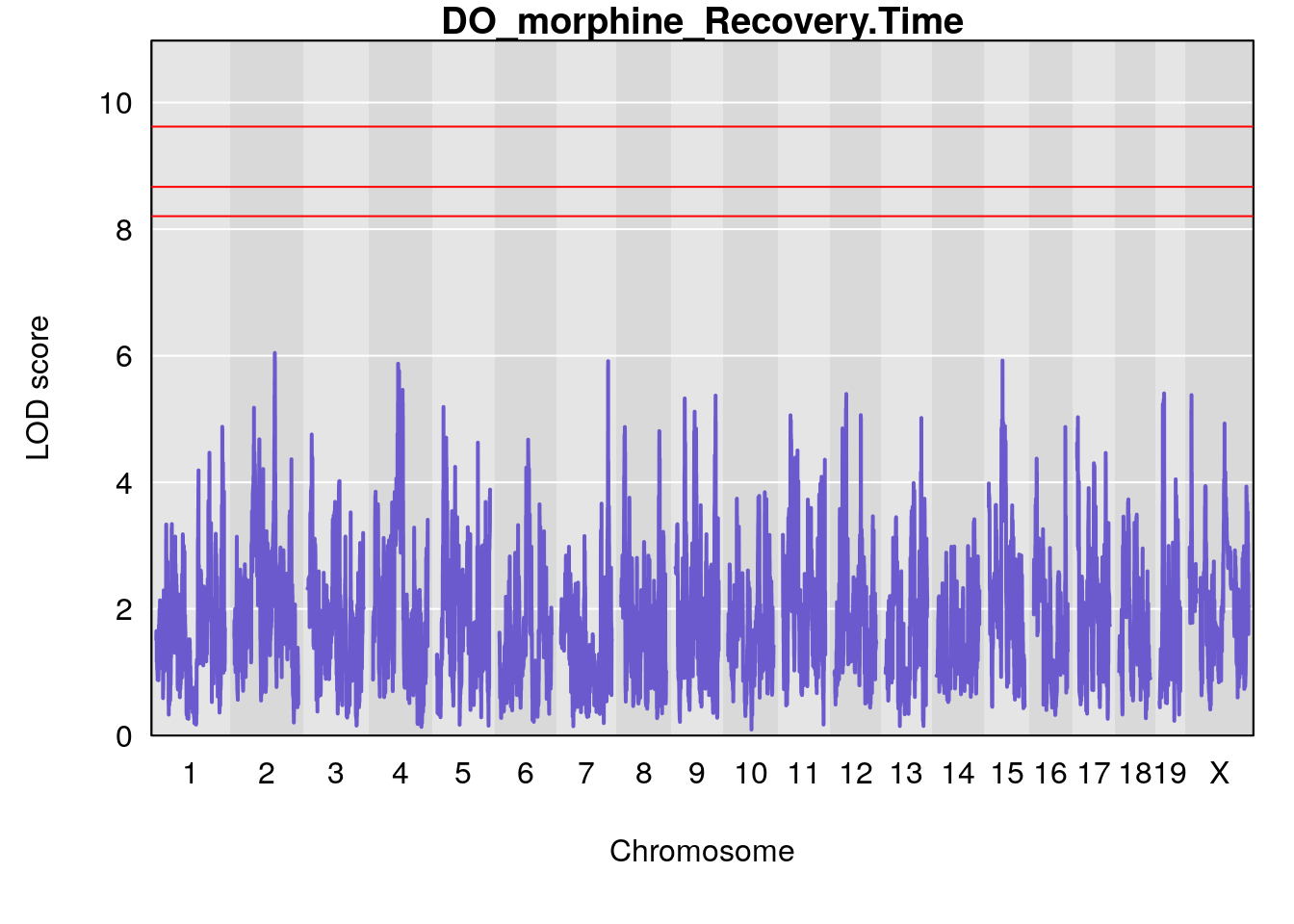
# [1] "Recovery.Time"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 2 116.336 6.044203 57.38935 117.2478
# [1] 1
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
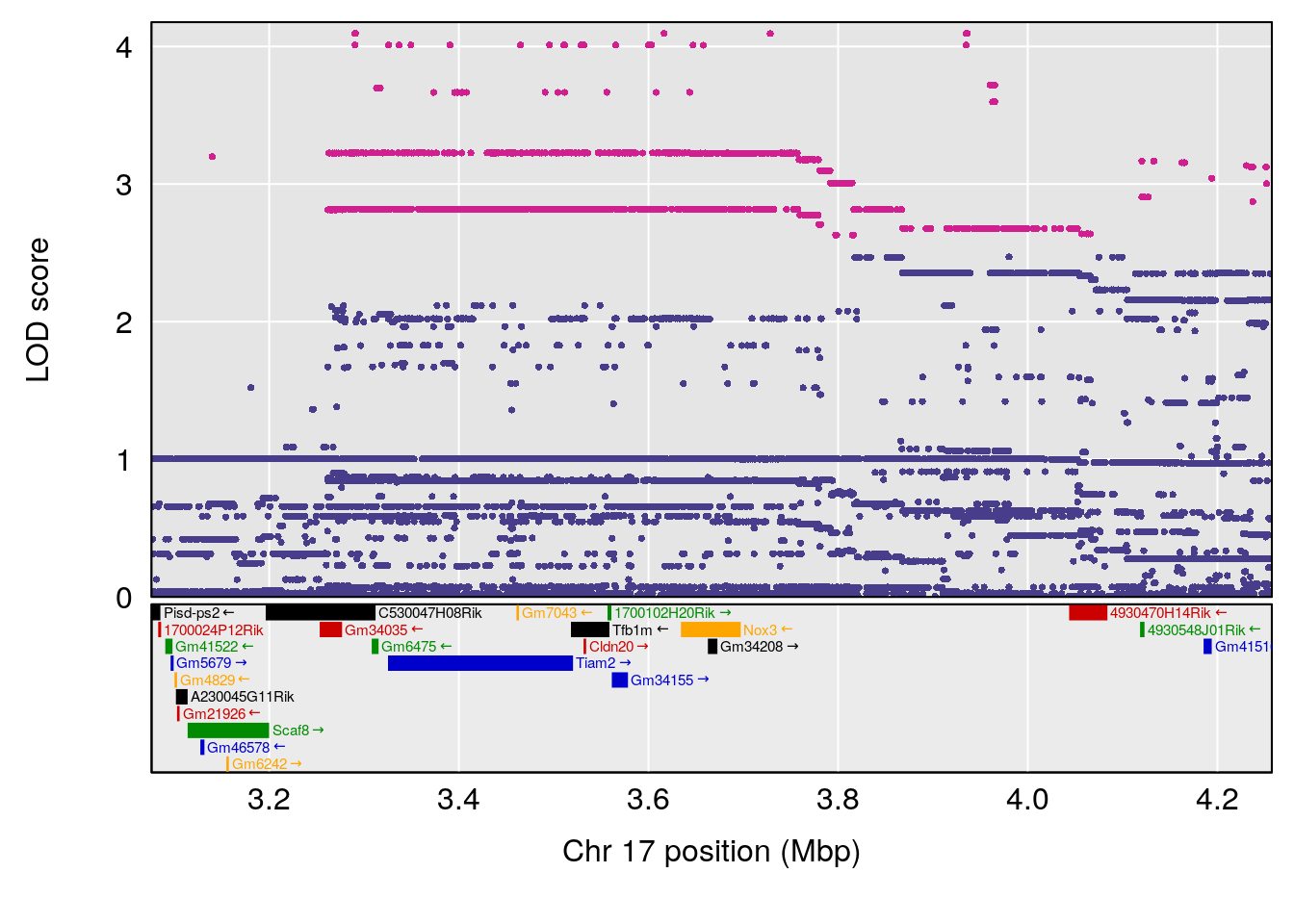
# [1] "Min.depression"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 5 26.203835 6.228551 24.841651 50.24141
# 2 1 pheno1 6 21.240236 6.168790 20.676183 23.20589
# 3 1 pheno1 17 3.257454 6.099197 3.075928 71.51265
# [1] 1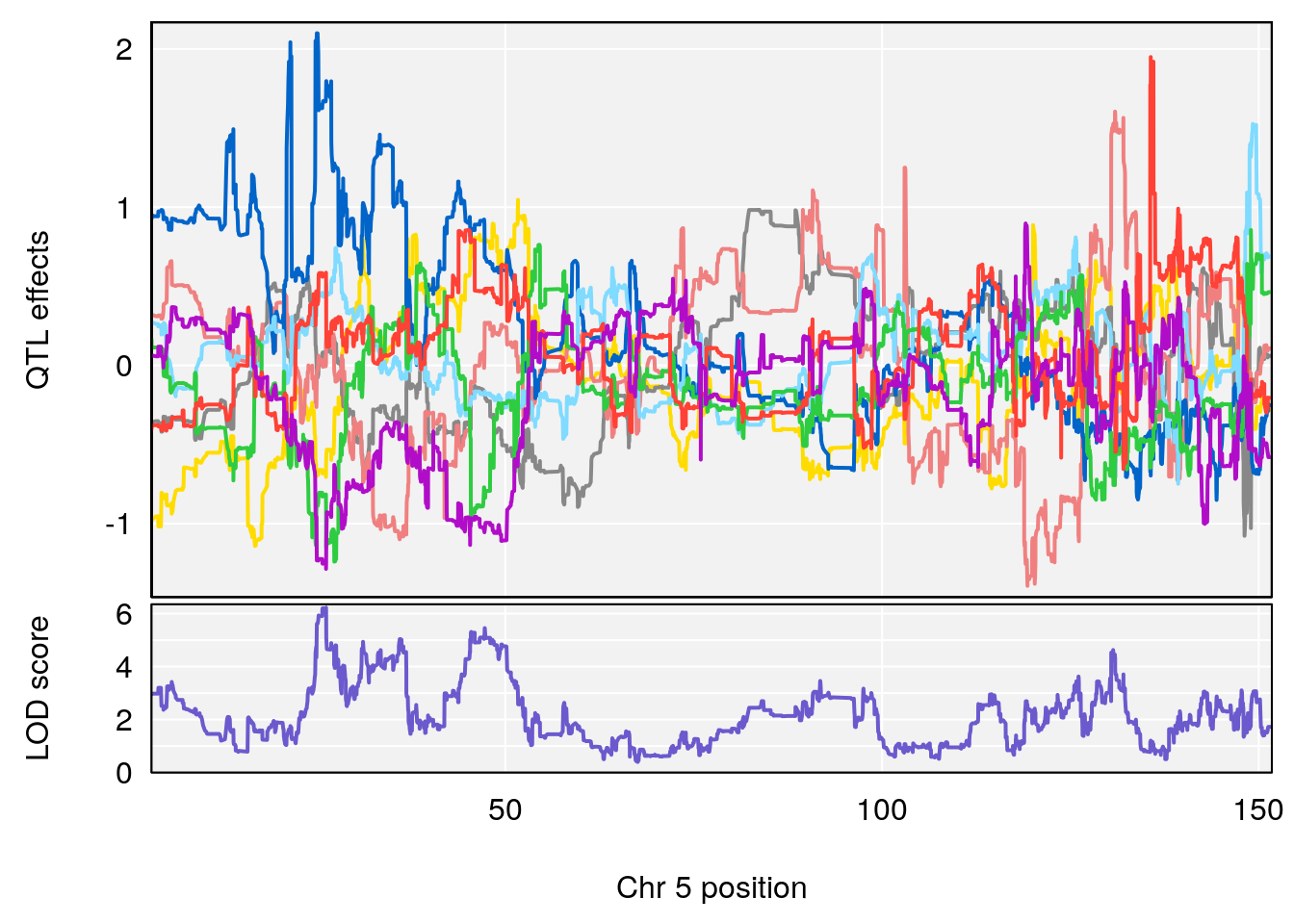
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 2
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 3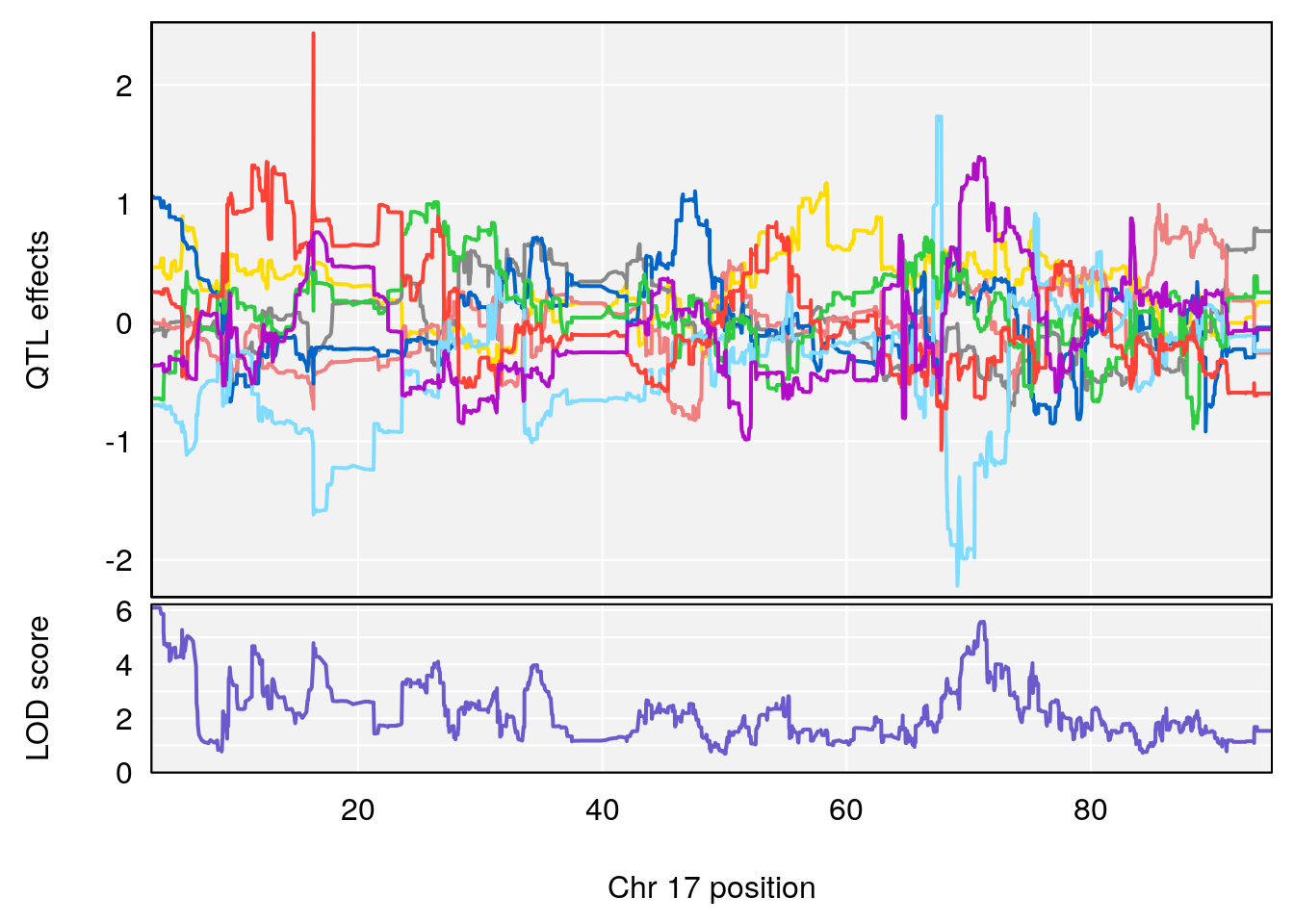
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
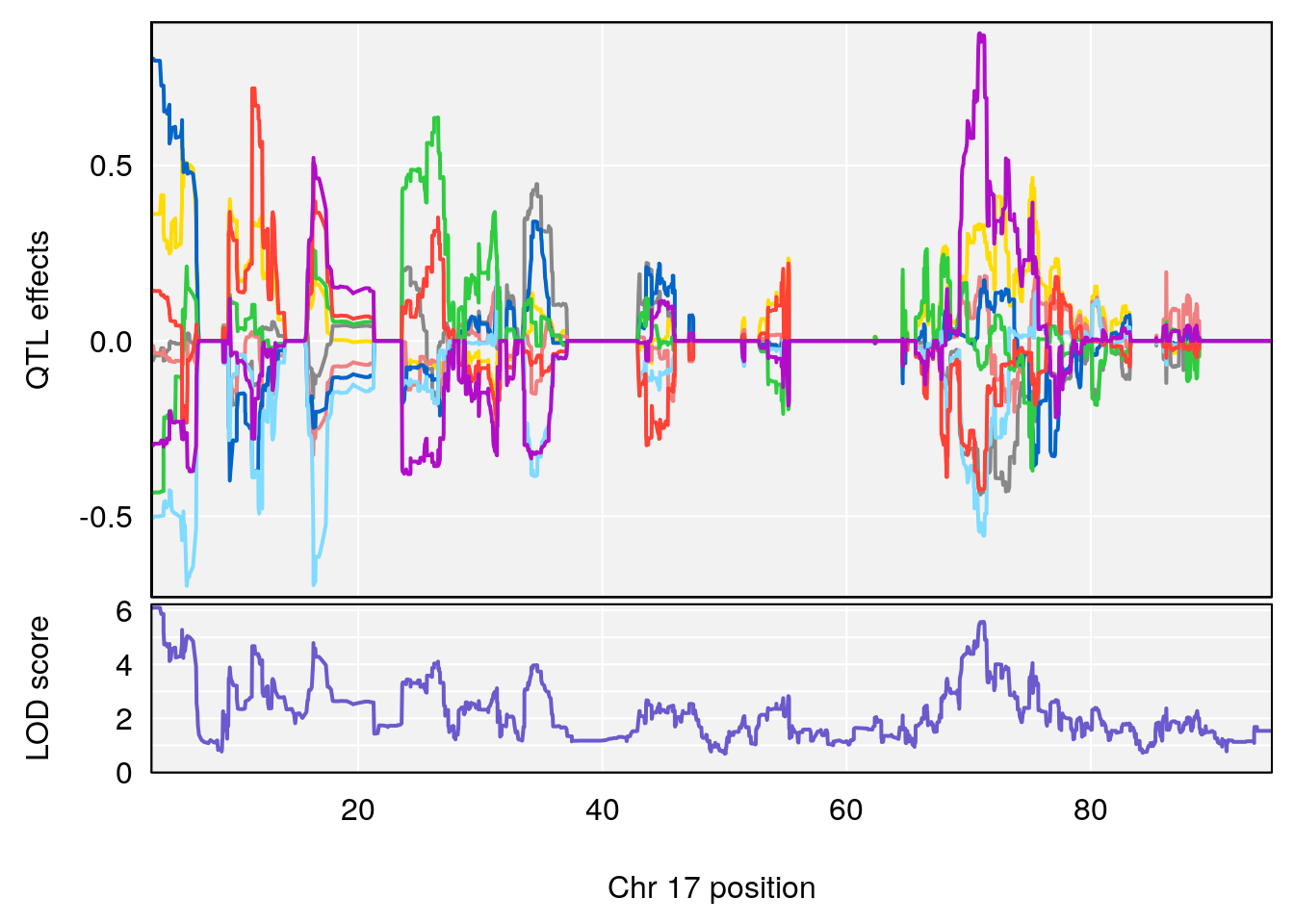
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
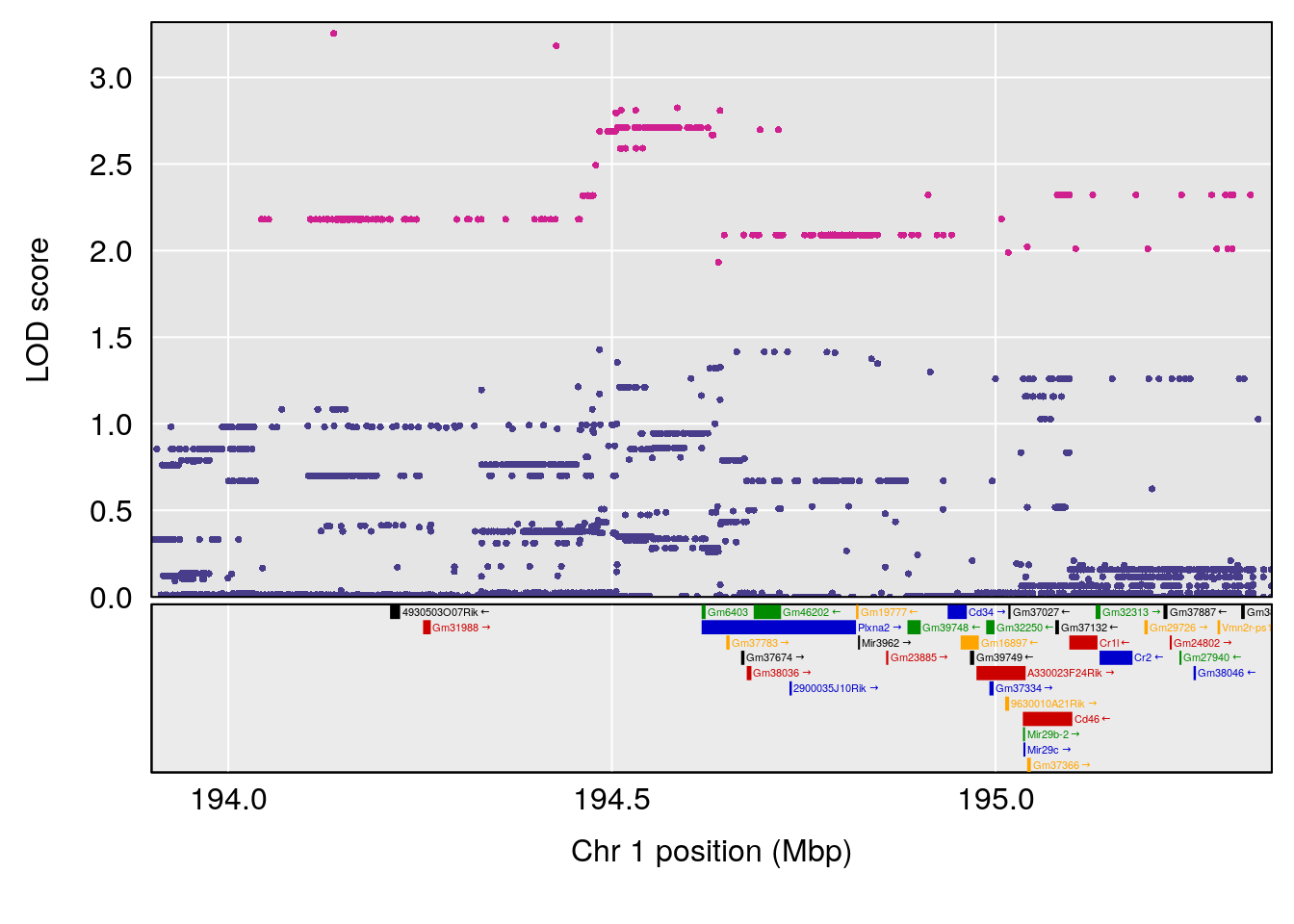
# [1] "Status_bin"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 194.90004 7.231262 194.63822 195.36002
# 2 1 pheno1 8 78.08382 6.071287 78.03203 89.74609
# [1] 1
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
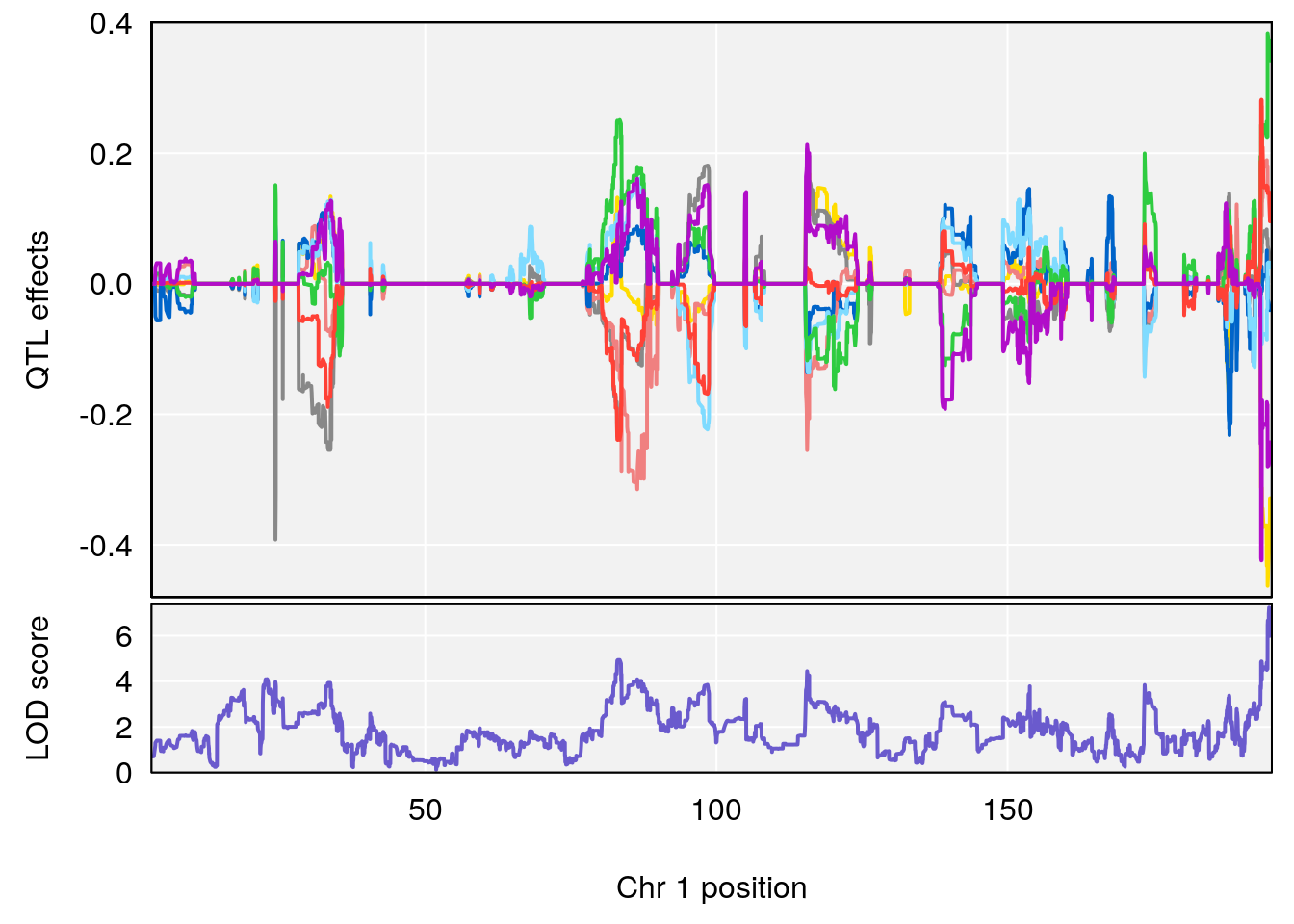
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 2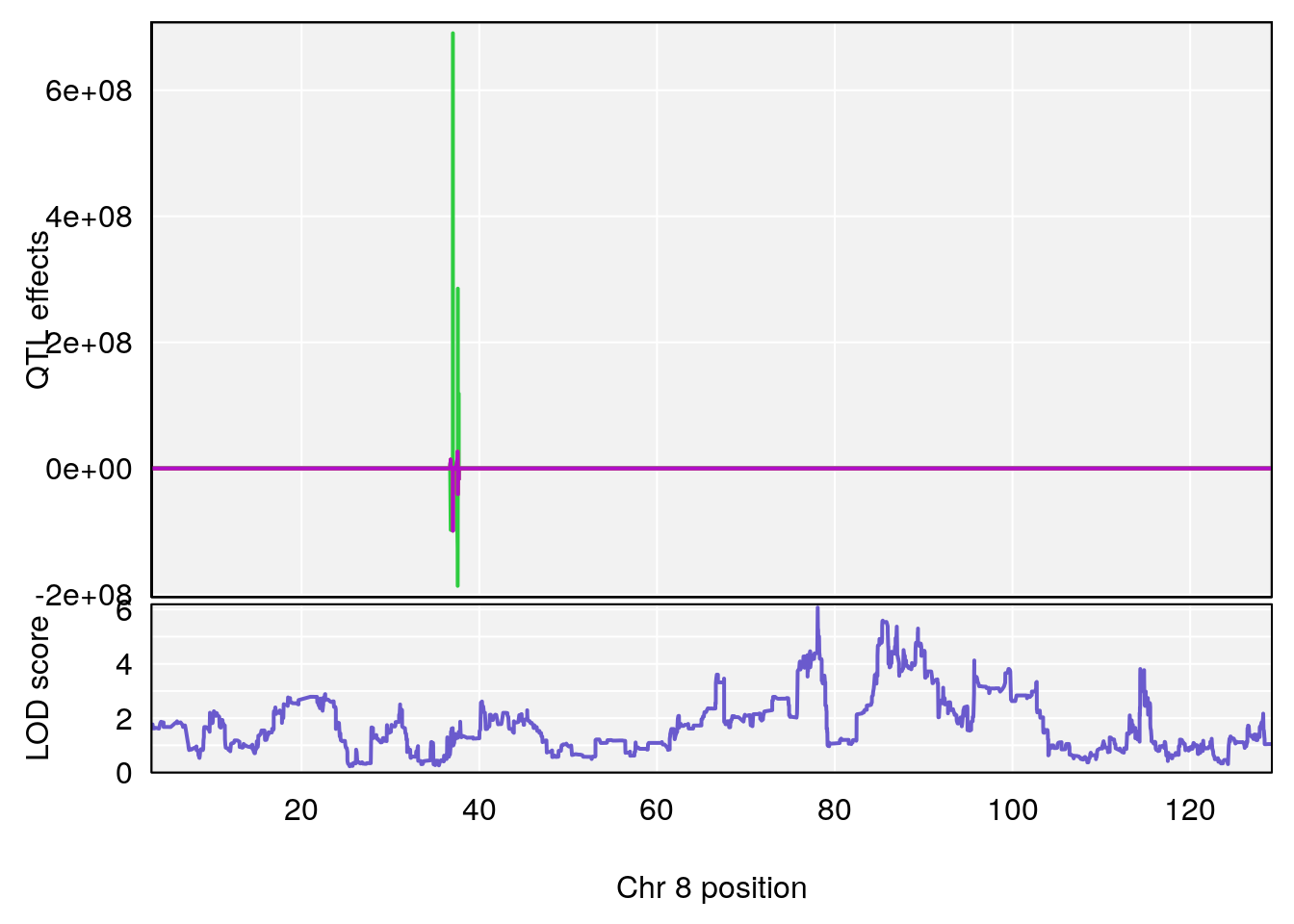
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
Plot qtl mapping on array female
load("output/qtl.morphine.out.female.RData")
#pmap and gmap
chr.names <- c("1","2","3","4","5","6","7","8","9","10","11","12","13","14","15","16","17","18","19", "X")
gmap <- list()
pmap <- list()
for (chr in chr.names){
gmap[[chr]] <- do.morphine$gmap[[chr]]
pmap[[chr]] <- do.morphine$pmap[[chr]]
}
attr(gmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
attr(pmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
#genome-wide plot
for(i in names(qtl.morphine.out)){
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.morphine.out[[i]]) # overall maximum LOD score
plot(qtl.morphine.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, max(sapply(qtl.morphine.out, maxlod)) +2))
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("DO_morphine_",i))
}
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
#peaks coeff plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
print(peaks)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
}
# [1] "Survival.Time"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 2 108.20689 6.854241 107.24263 150.00078
# 2 1 pheno1 3 38.52197 6.419152 38.25791 109.66640
# 3 1 pheno1 5 104.19473 8.252319 103.82013 105.66710
# 4 1 pheno1 8 115.25017 6.352567 114.99883 115.26932
# 5 1 pheno1 11 75.15149 6.353510 37.05275 76.42170
# 6 1 pheno1 17 83.99650 6.544571 83.63927 84.29922
# [1] 1
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |

| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 2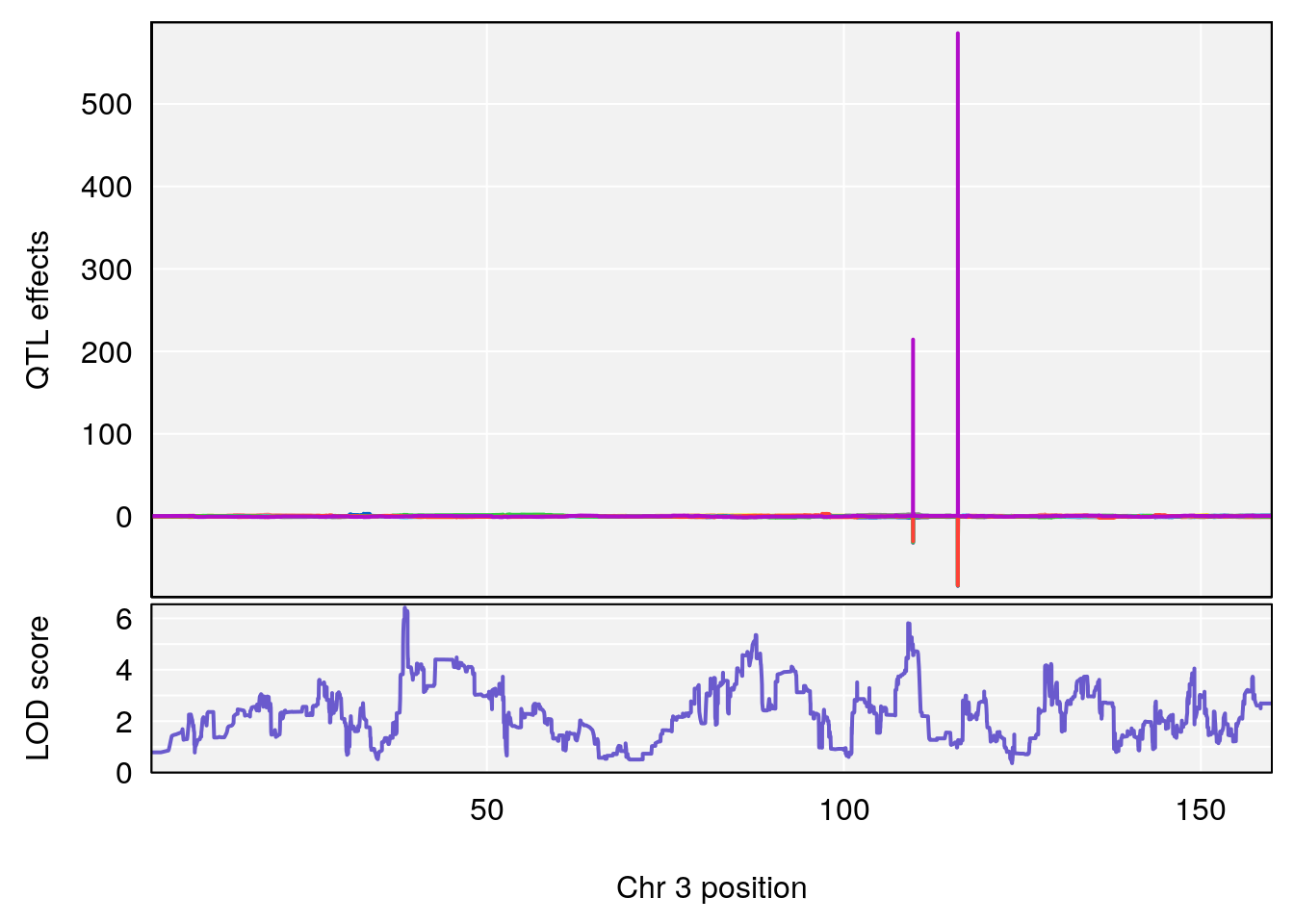
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |

| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 3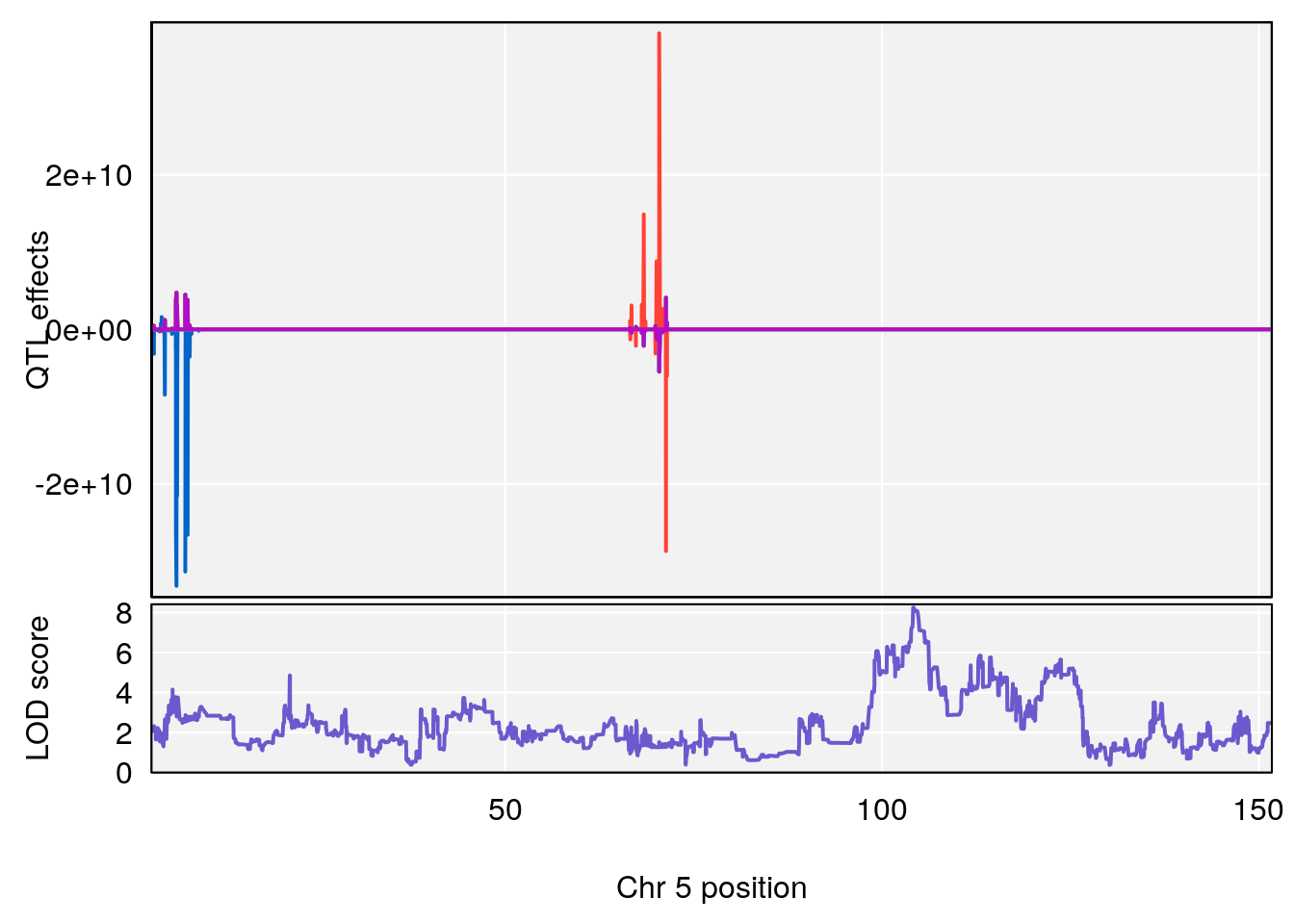
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
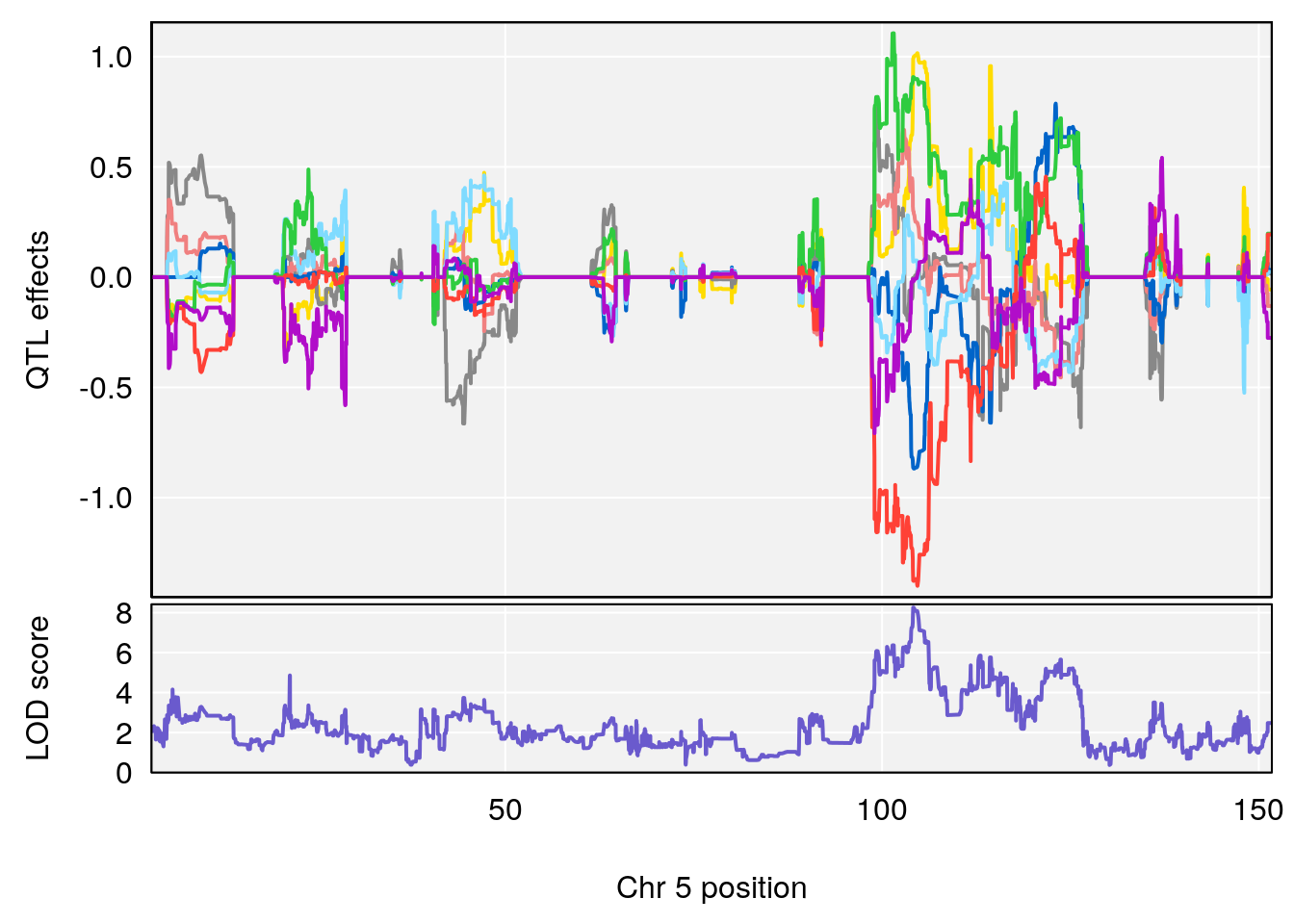
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 4
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
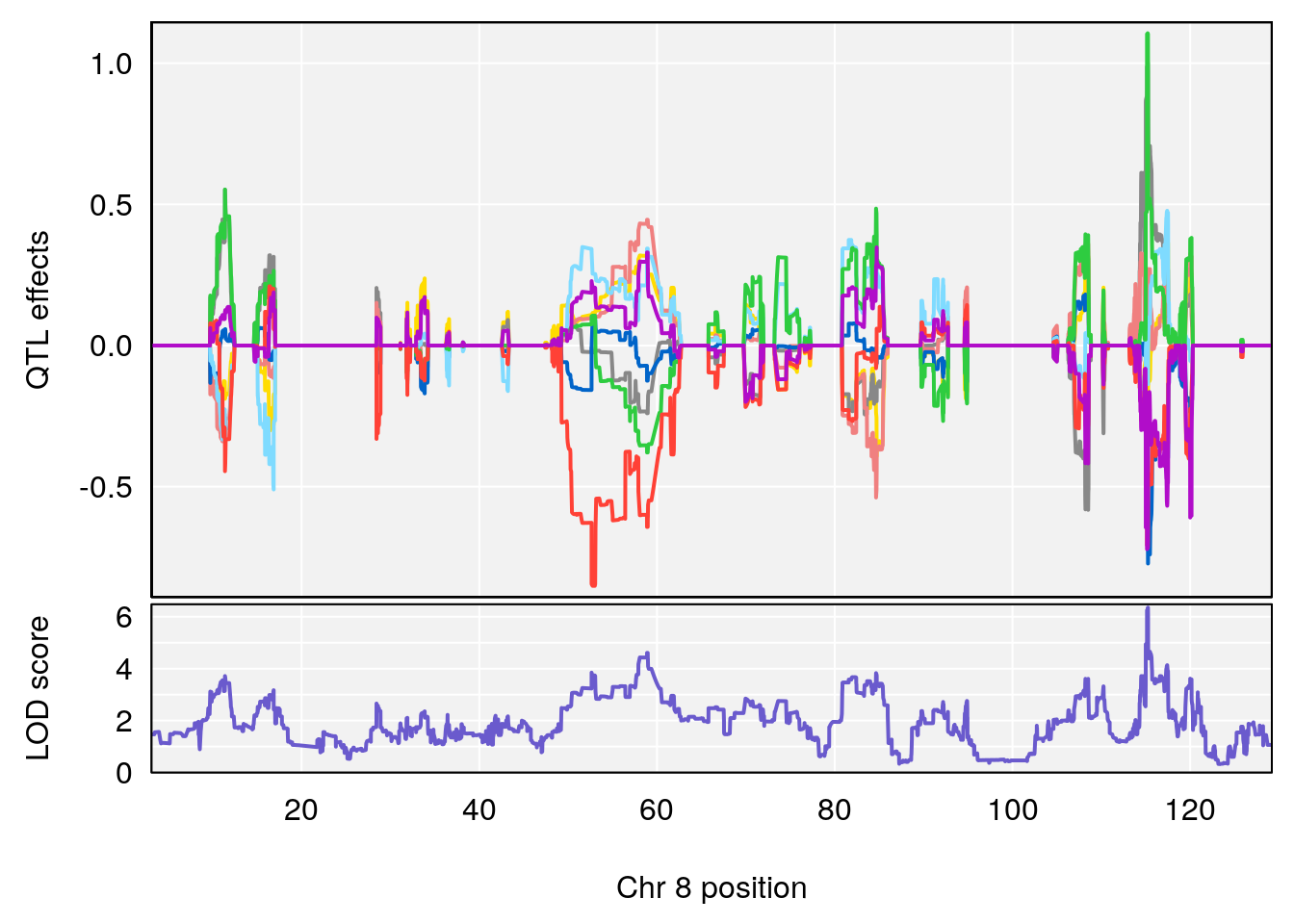
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 5
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 6
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |

| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
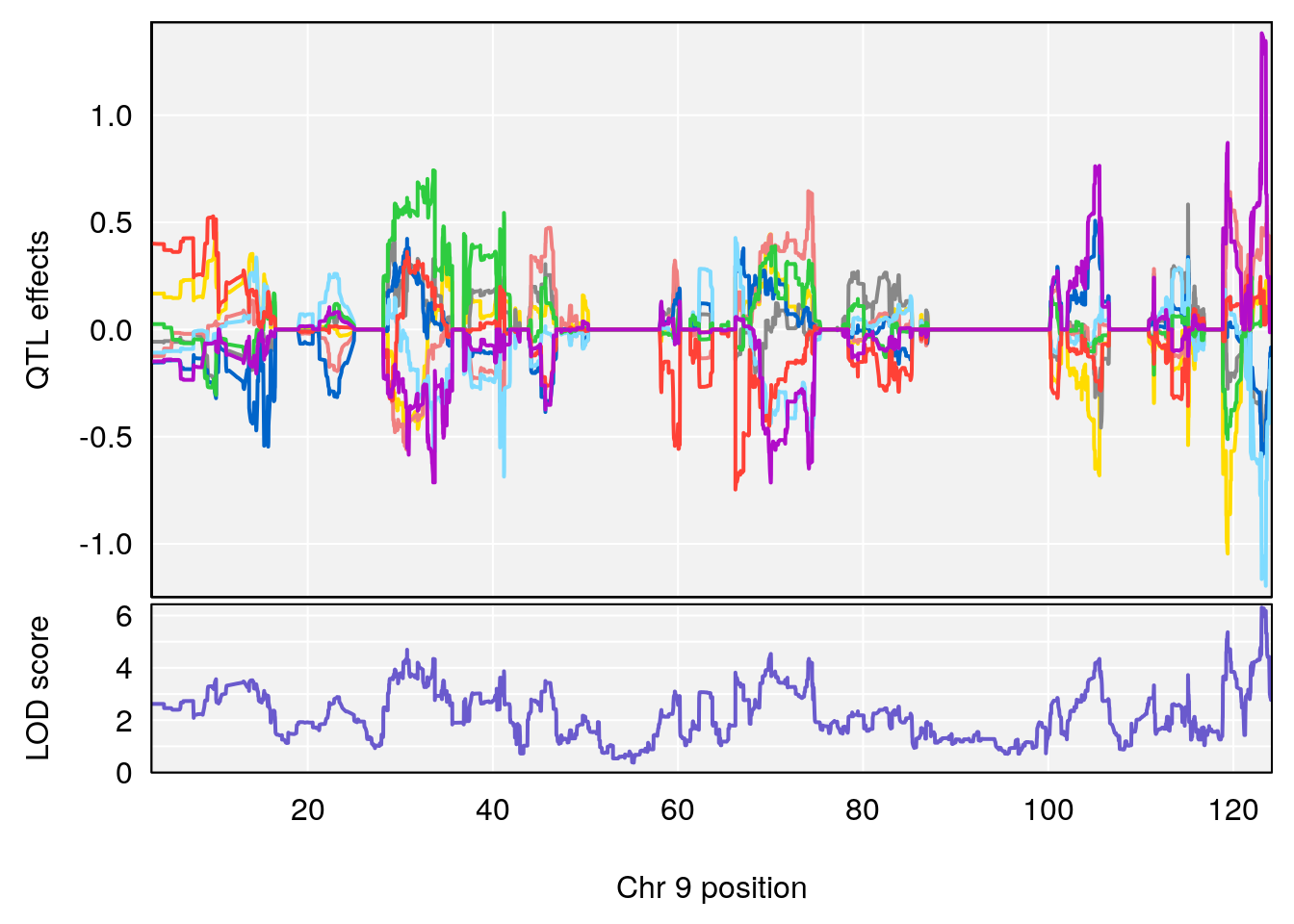
# [1] "Recovery.Time"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 2 11.42175 7.039471 10.46705 11.90852
# 2 1 pheno1 4 32.07822 6.418205 31.92851 32.86047
# 3 1 pheno1 9 123.06013 6.303715 119.26361 123.62391
# [1] 1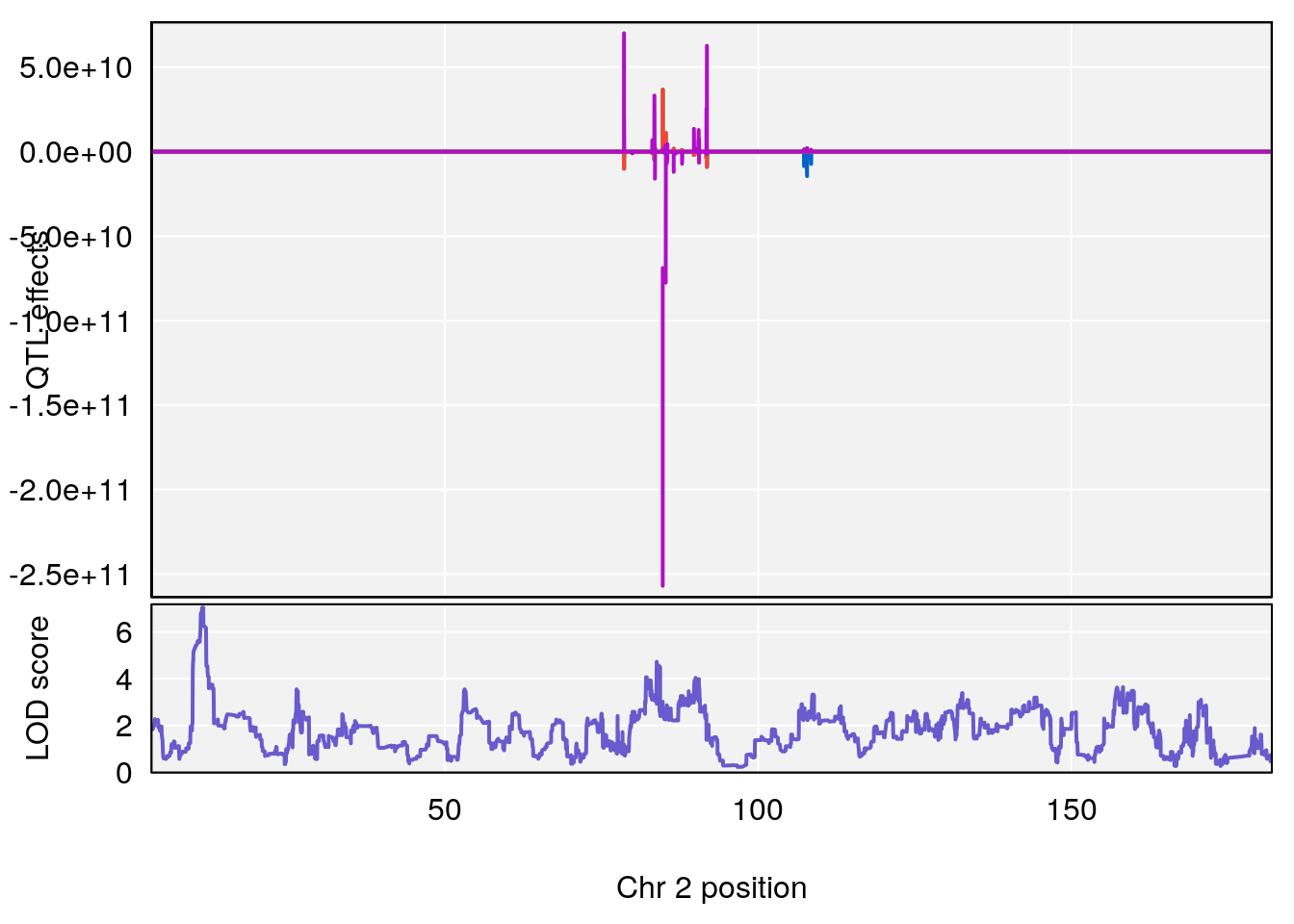
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
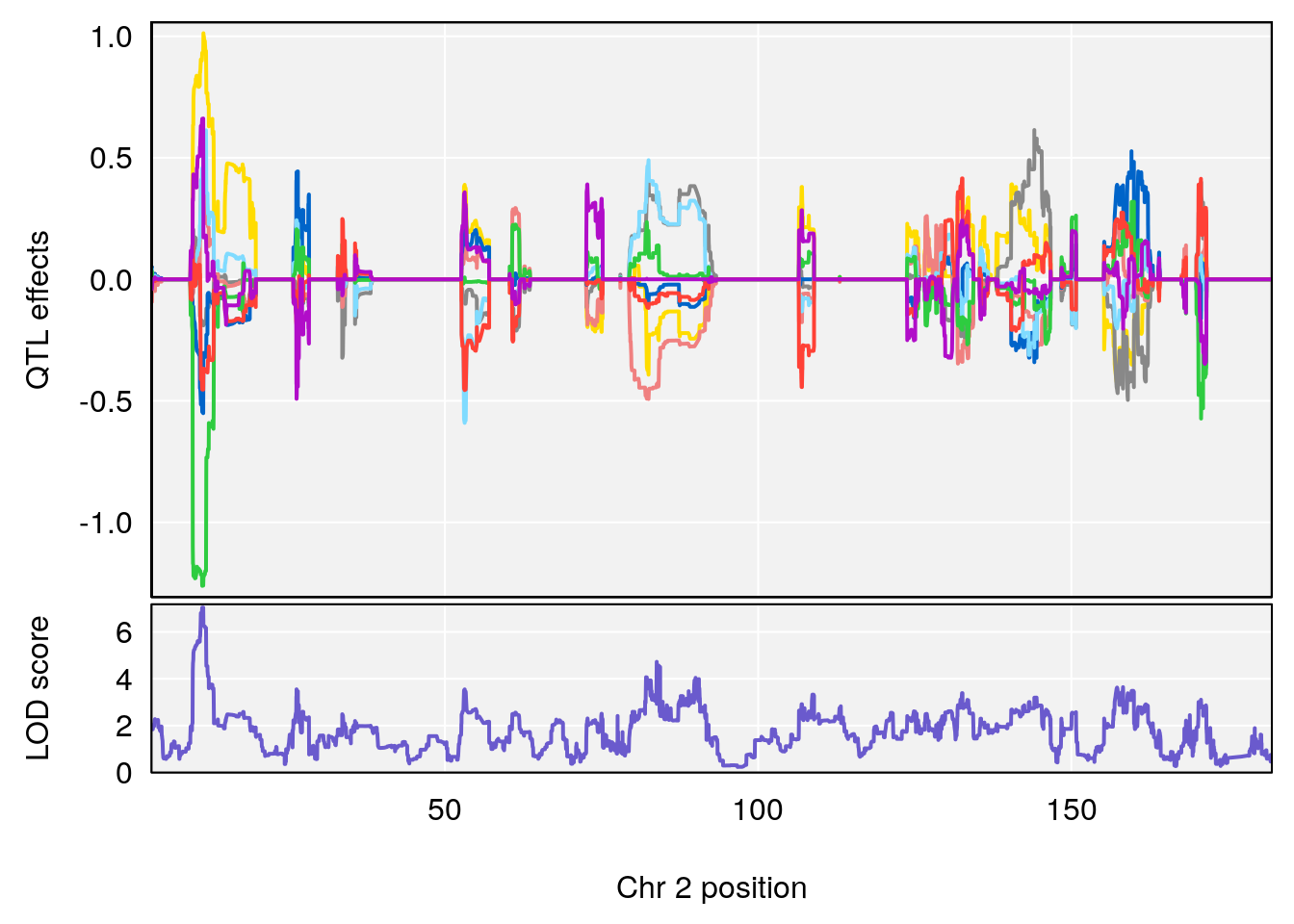
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |

| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 2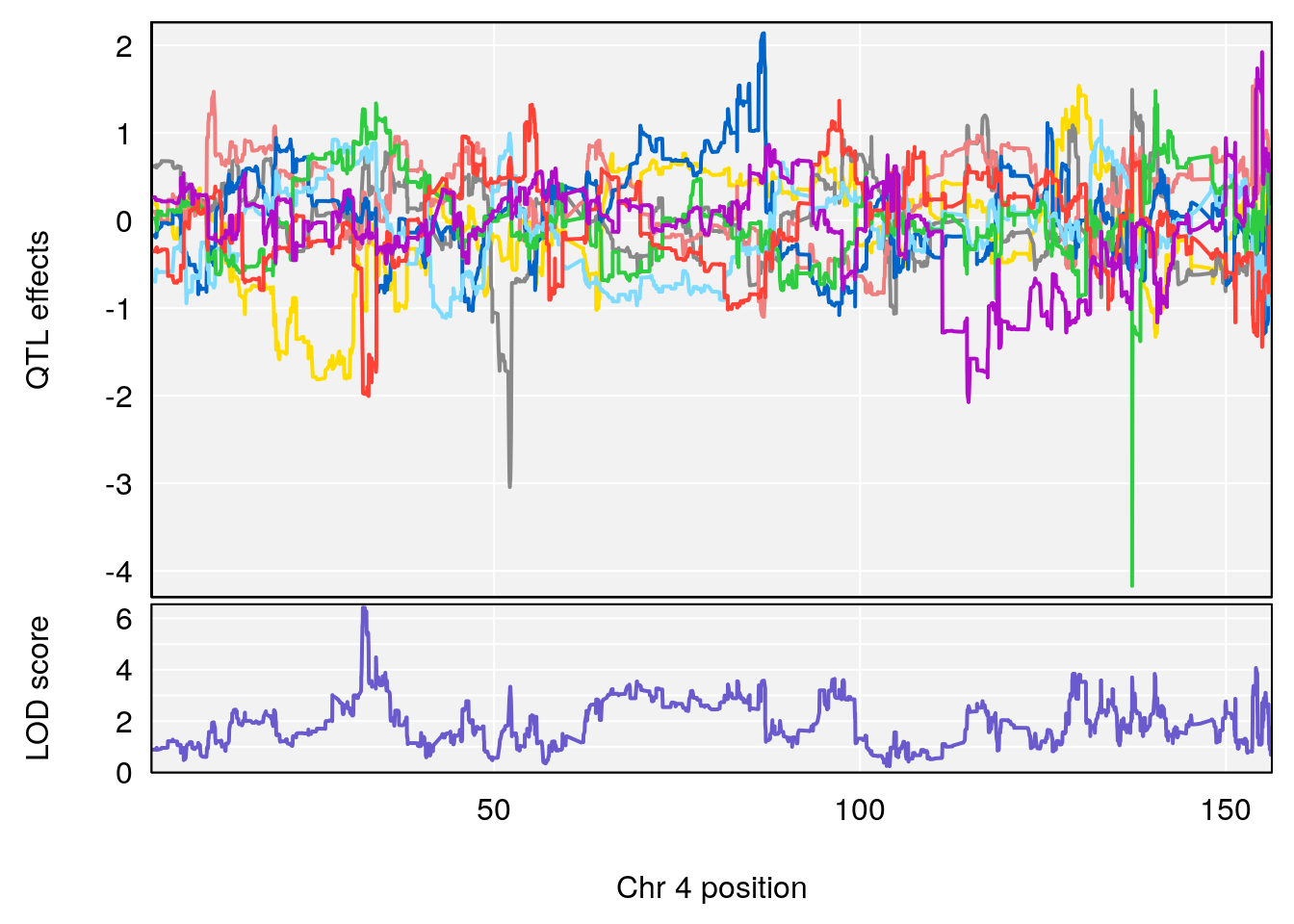
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
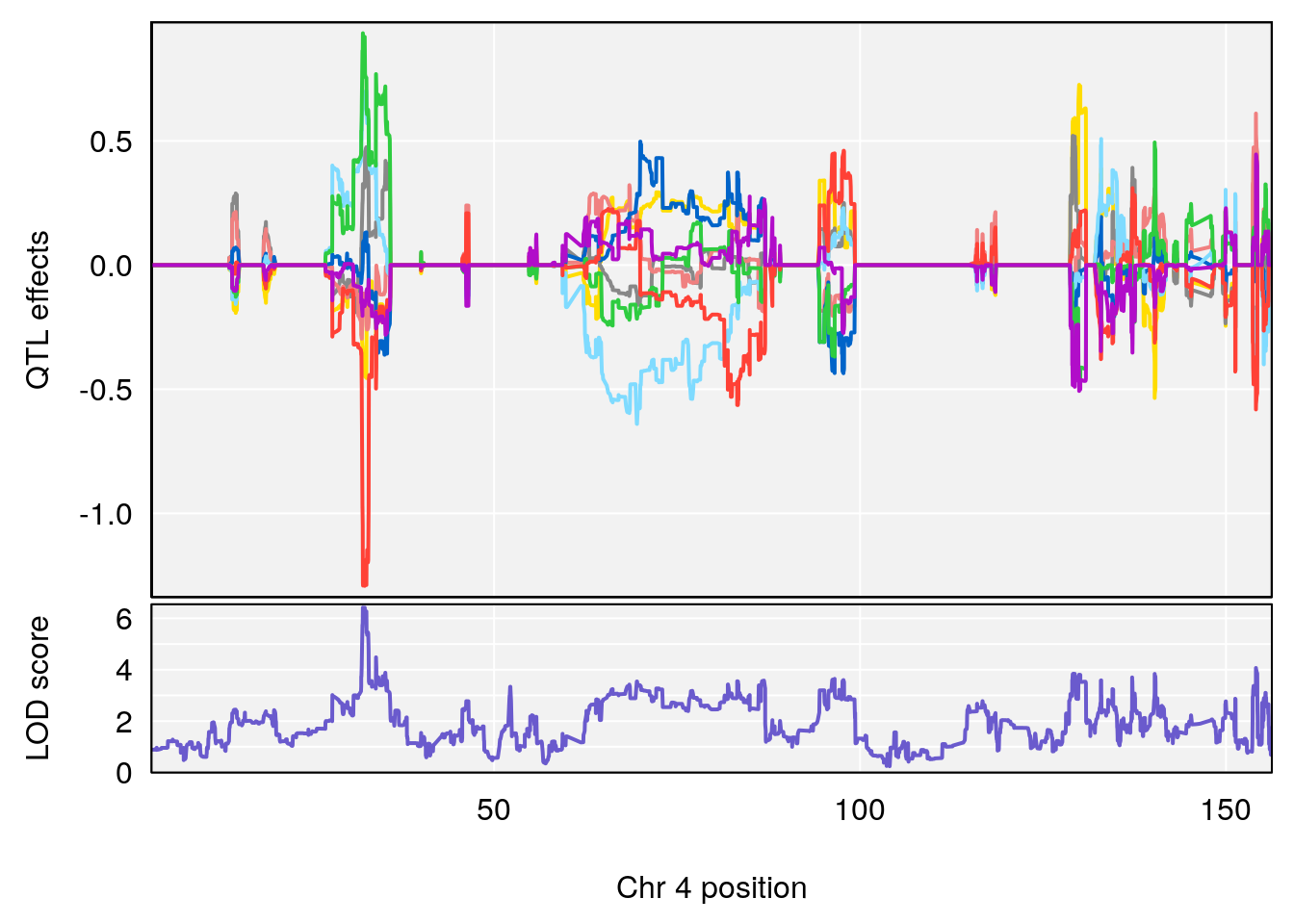
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 3
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
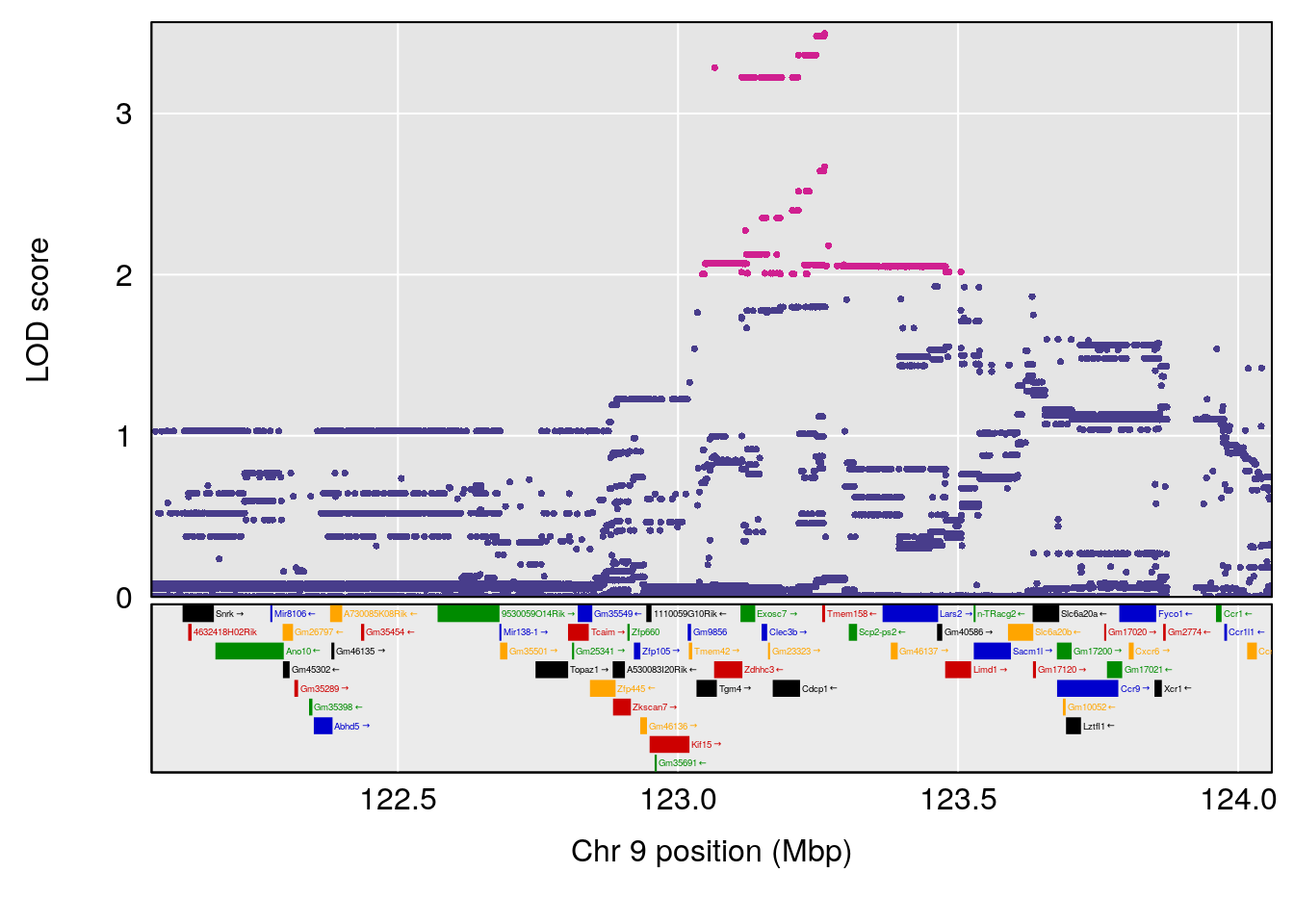
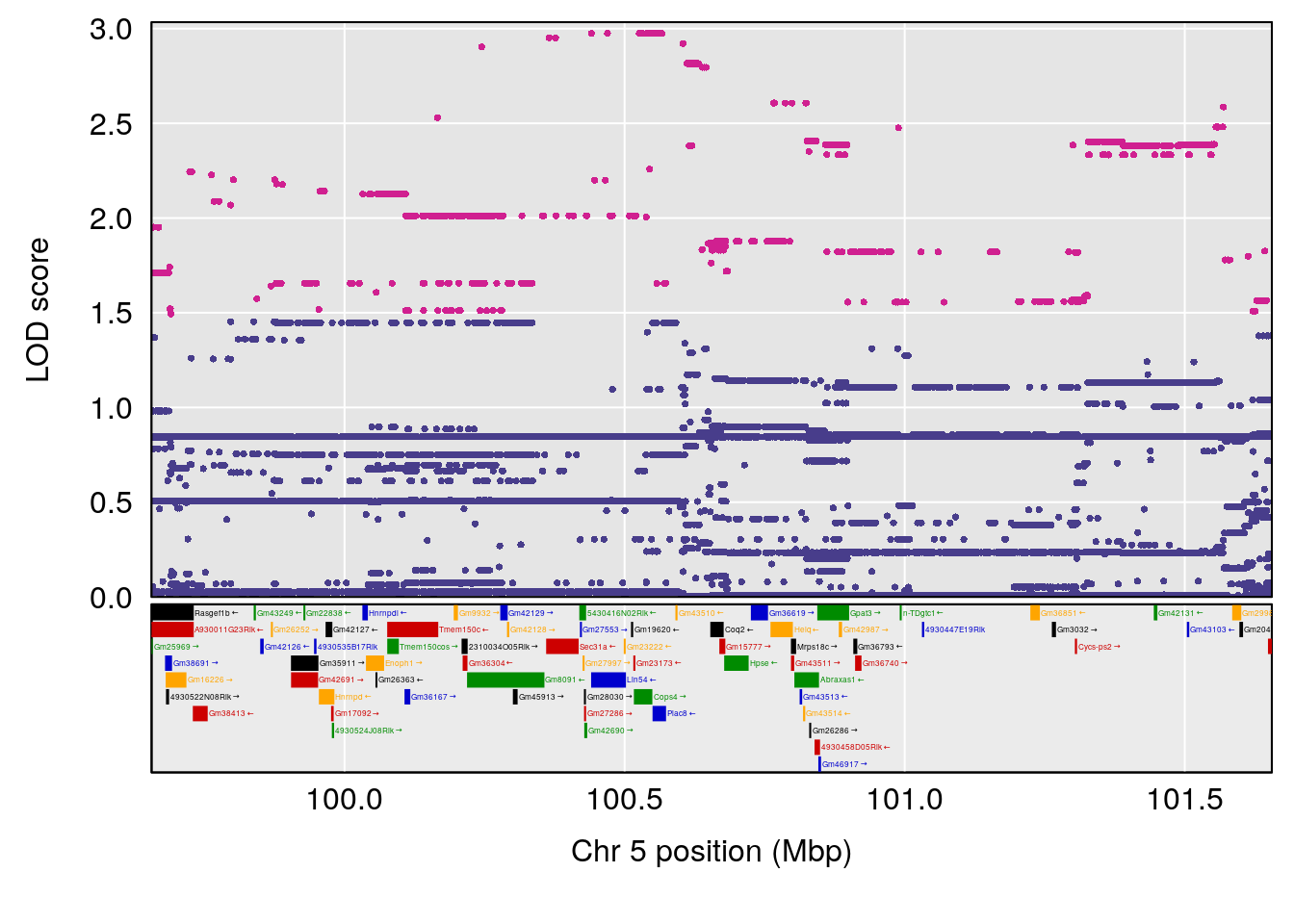
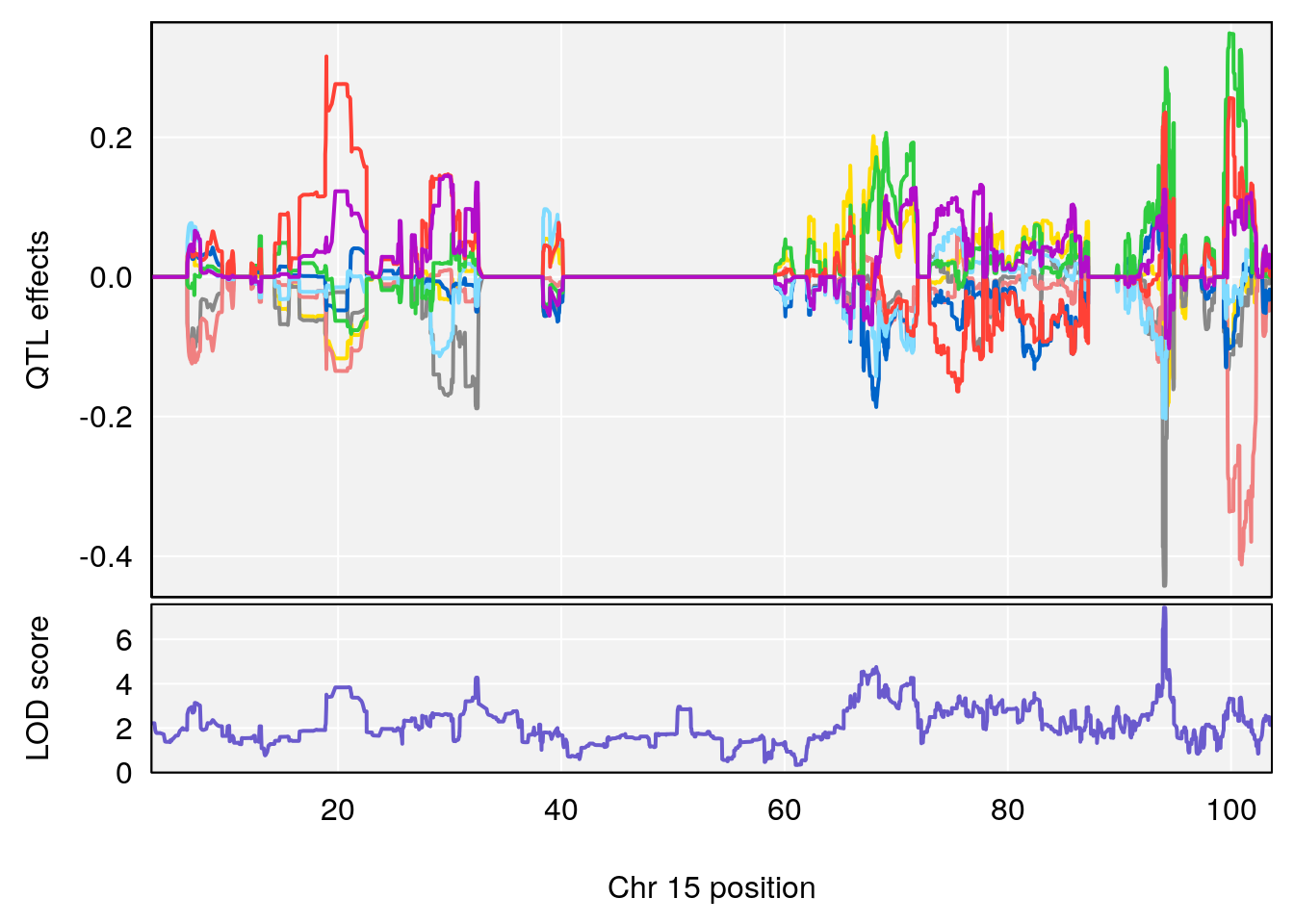
# [1] "Min.depression"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 5 25.05437 6.281007 24.9198 25.6421
# [1] 1
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
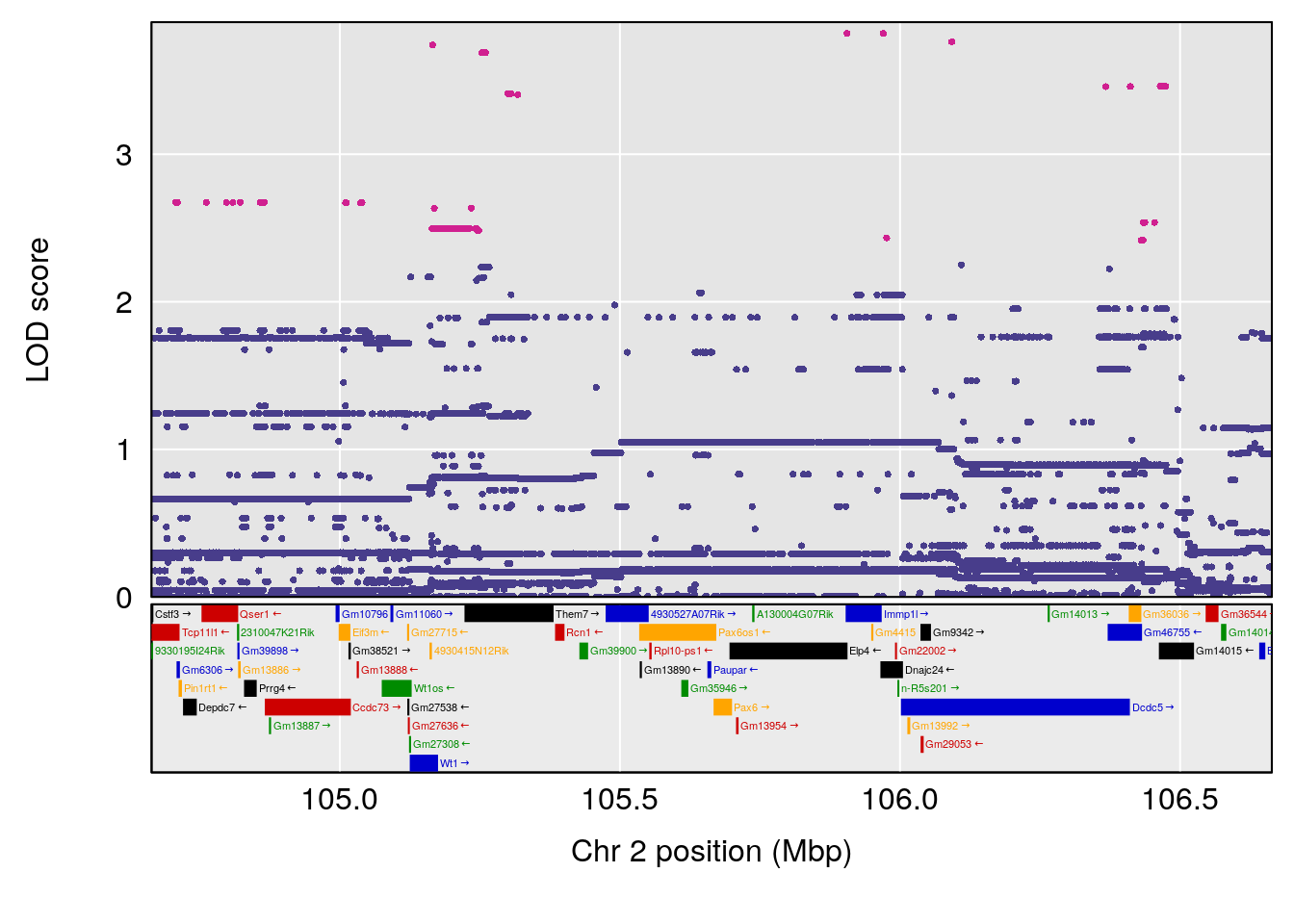
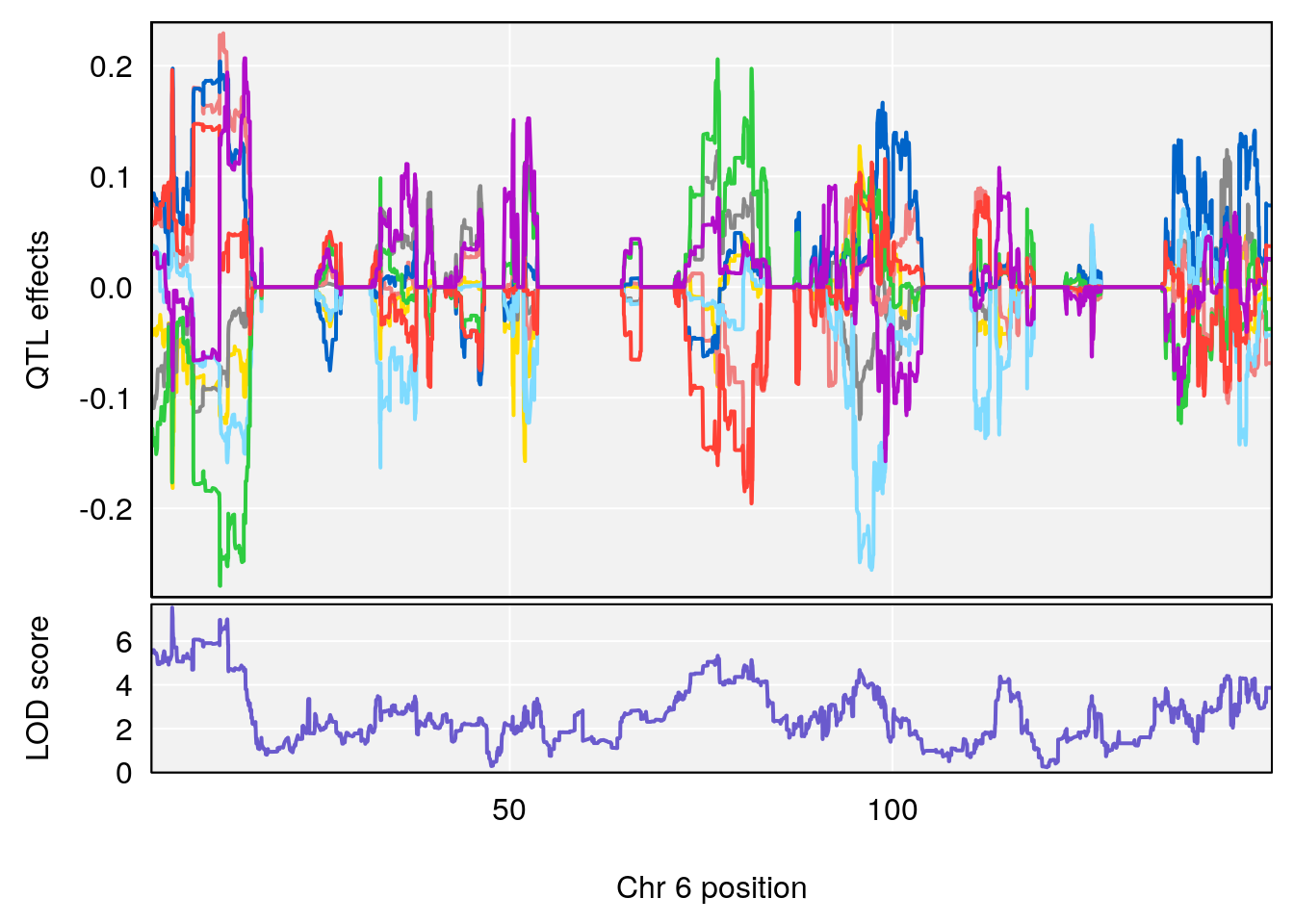
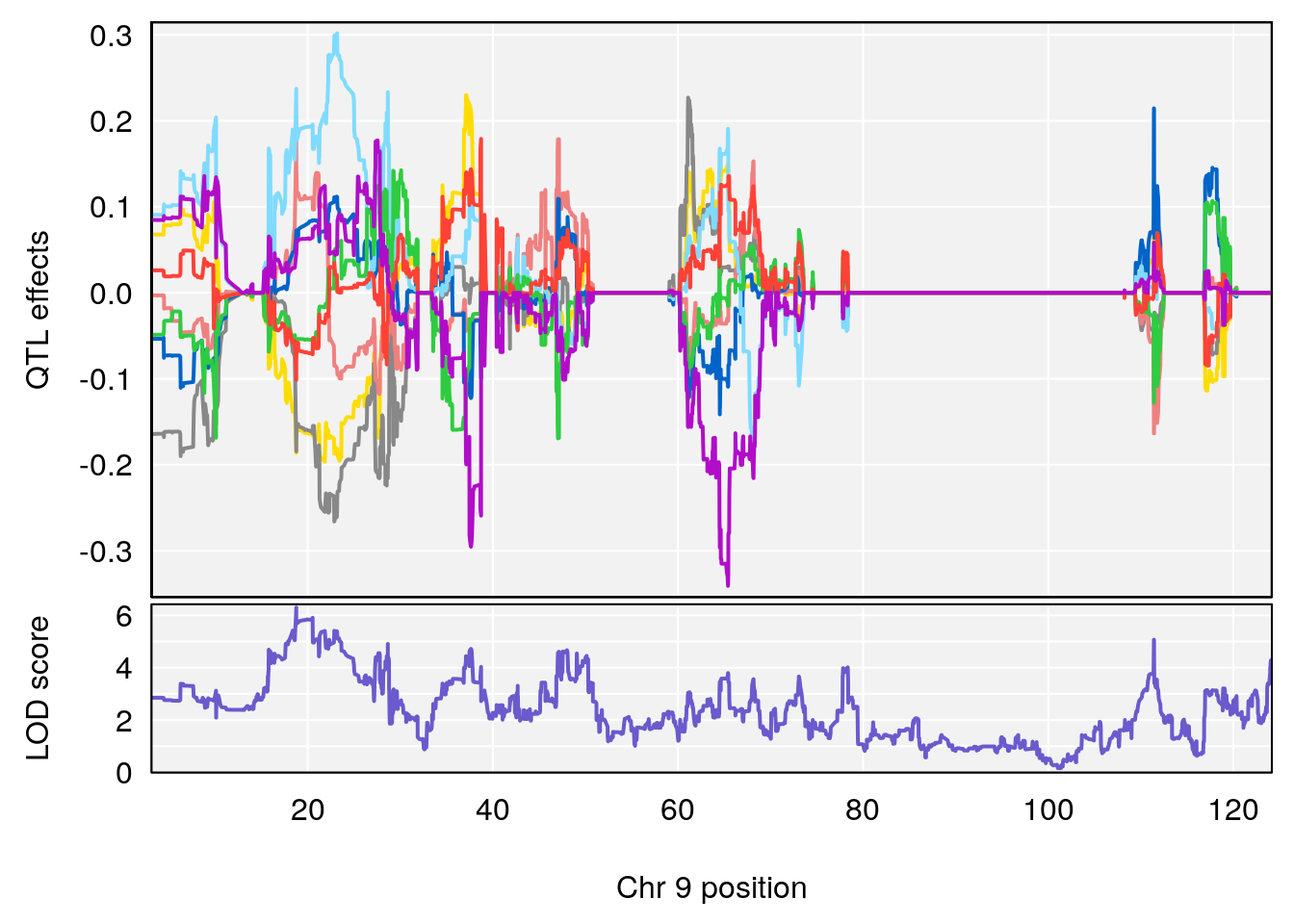
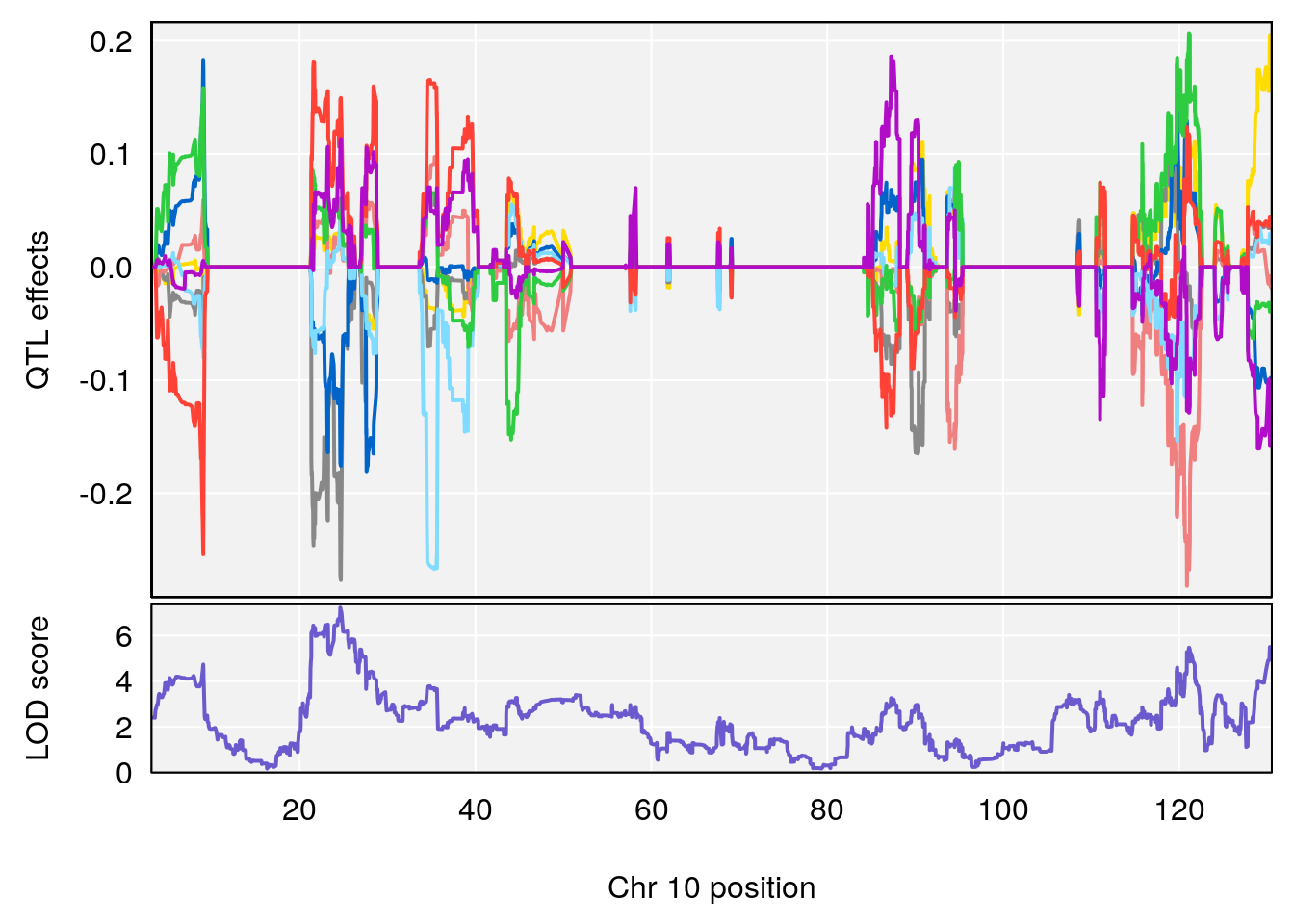
# [1] "Status_bin"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 19.164865 6.147268 15.439741 192.89712
# 2 1 pheno1 2 105.663925 10.873472 105.124103 106.49603
# 3 1 pheno1 3 138.083675 9.260545 109.677215 141.68062
# 4 1 pheno1 4 155.860646 8.348013 154.284934 156.25473
# 5 1 pheno1 5 100.655515 6.928981 99.534811 134.77861
# 6 1 pheno1 6 5.891843 7.528947 5.832646 13.19729
# 7 1 pheno1 7 69.702691 9.538679 69.411383 69.70587
# 8 1 pheno1 8 121.211859 7.057530 21.823677 121.48475
# 9 1 pheno1 9 18.763008 6.291868 17.554432 111.43643
# 10 1 pheno1 10 24.631651 7.226505 21.331286 26.28567
# 11 1 pheno1 15 93.991656 7.433259 93.880481 94.14353
# 12 1 pheno1 16 89.871019 7.692891 88.692381 90.42208
# 13 1 pheno1 17 69.904861 6.080928 26.641041 70.50038
# 14 1 pheno1 18 73.967036 6.055066 36.069954 74.54257
# 15 1 pheno1 19 58.739120 6.488041 58.674418 59.16199
# [1] 1
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |

| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 2
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |

| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 3
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 4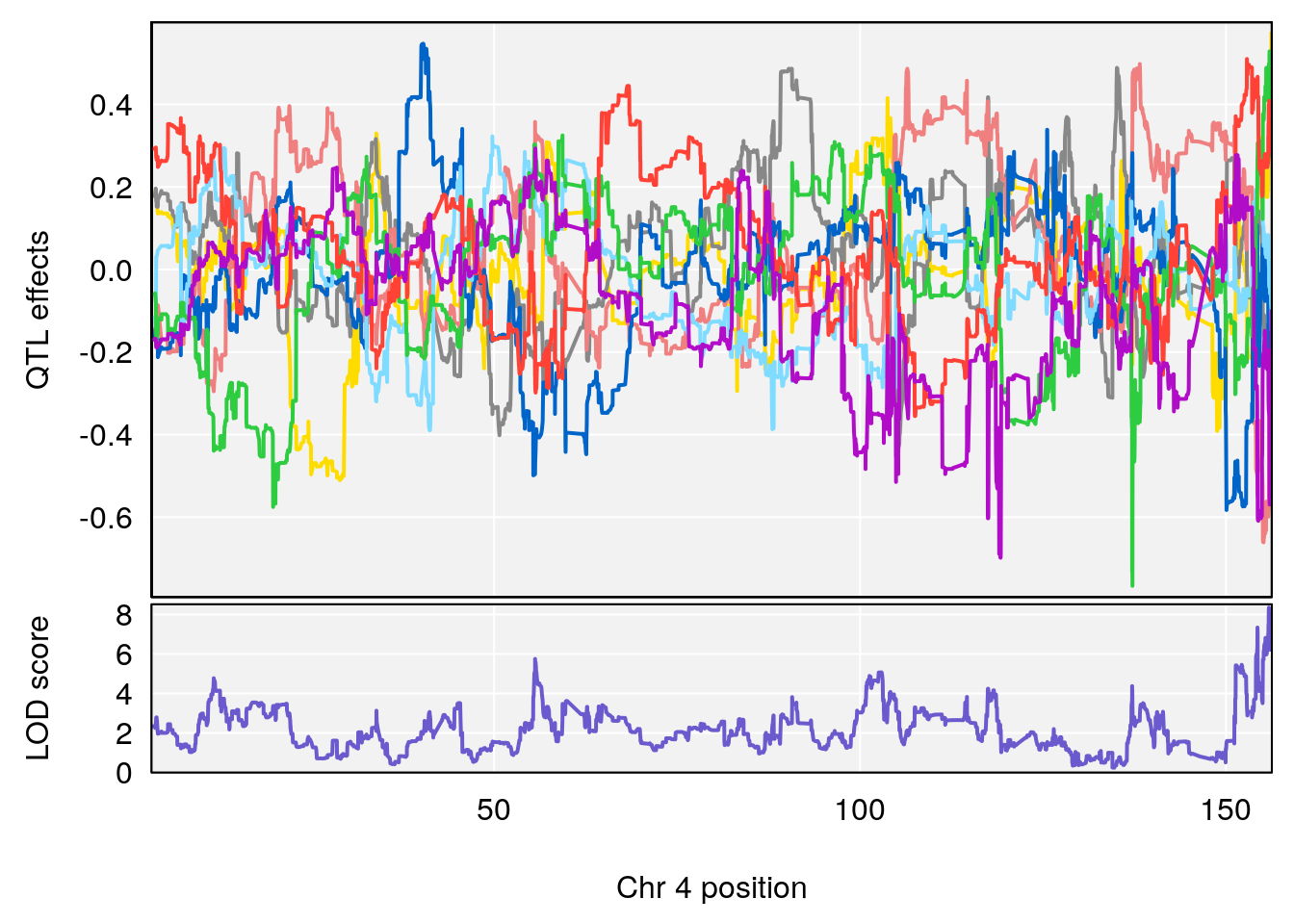
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 5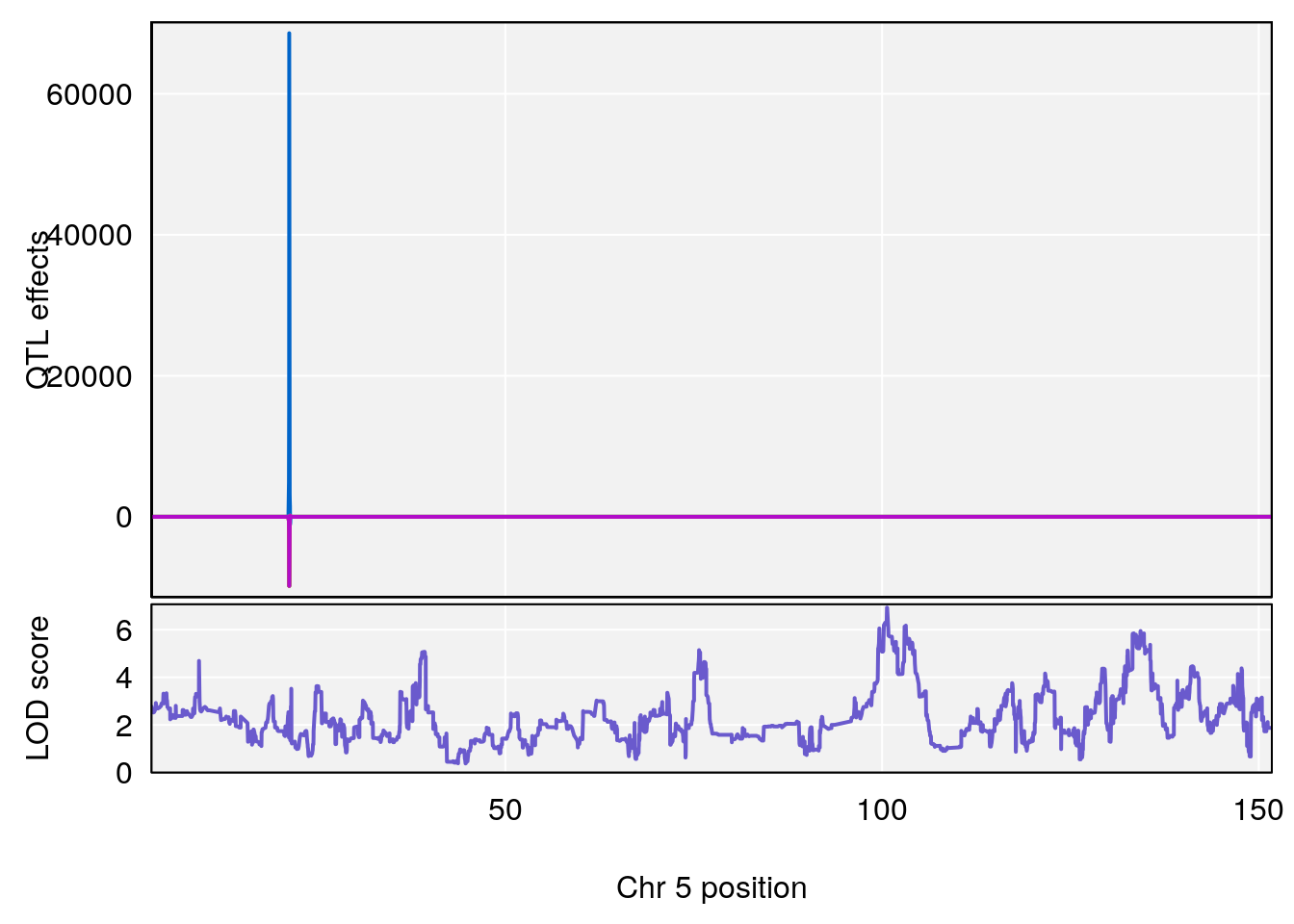
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |

| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 6
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |

| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 7
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
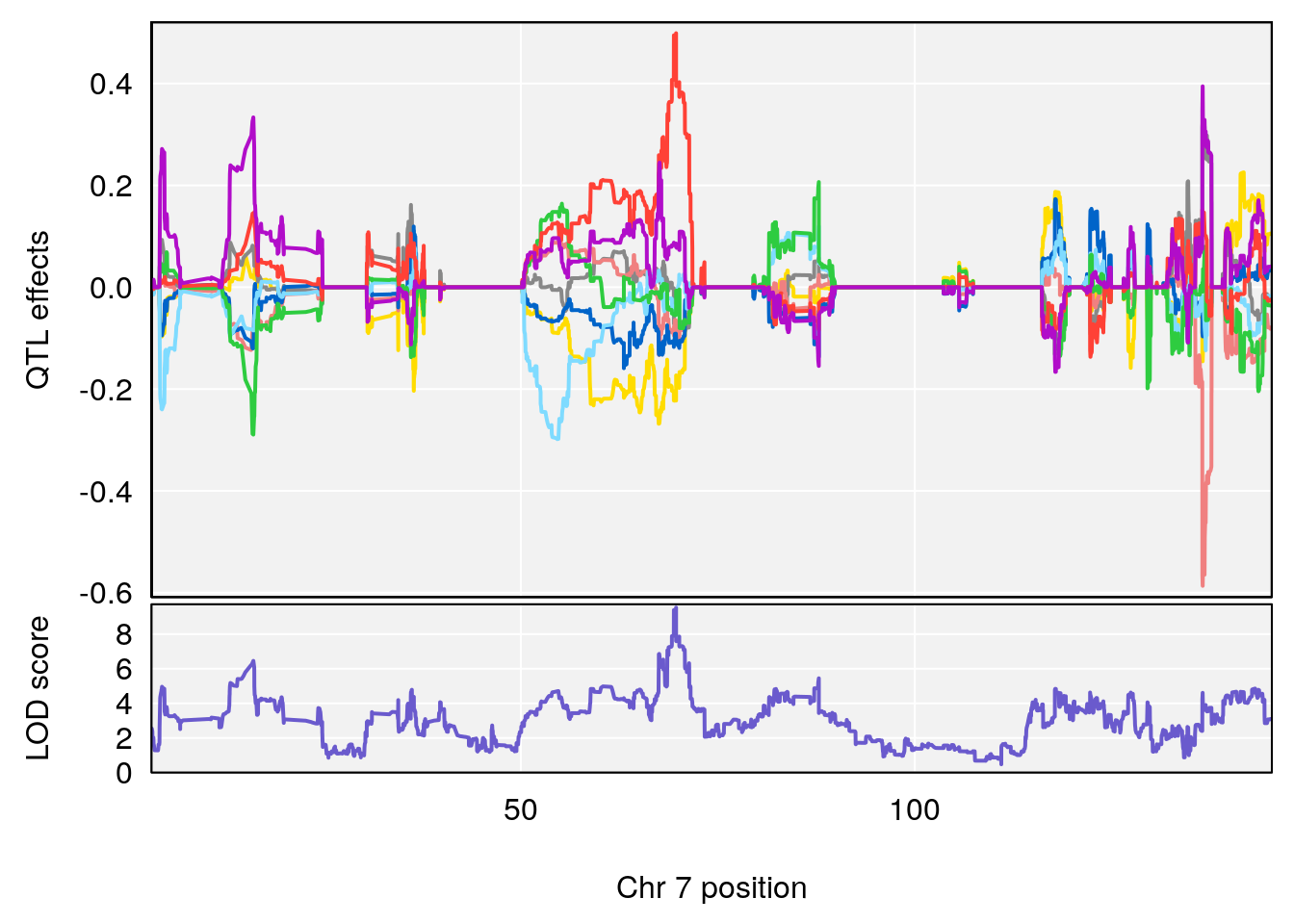
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |

| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 8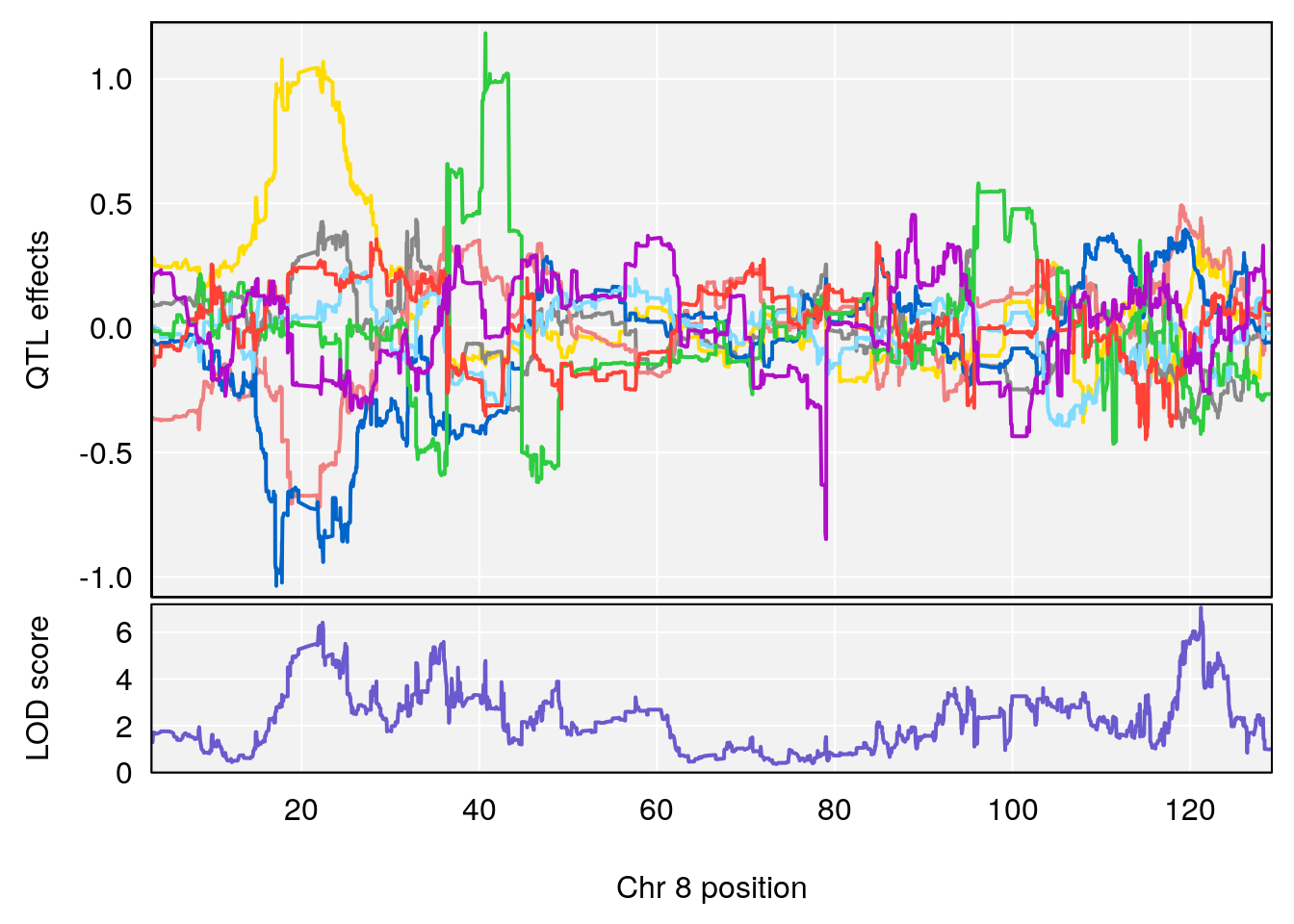
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |

| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
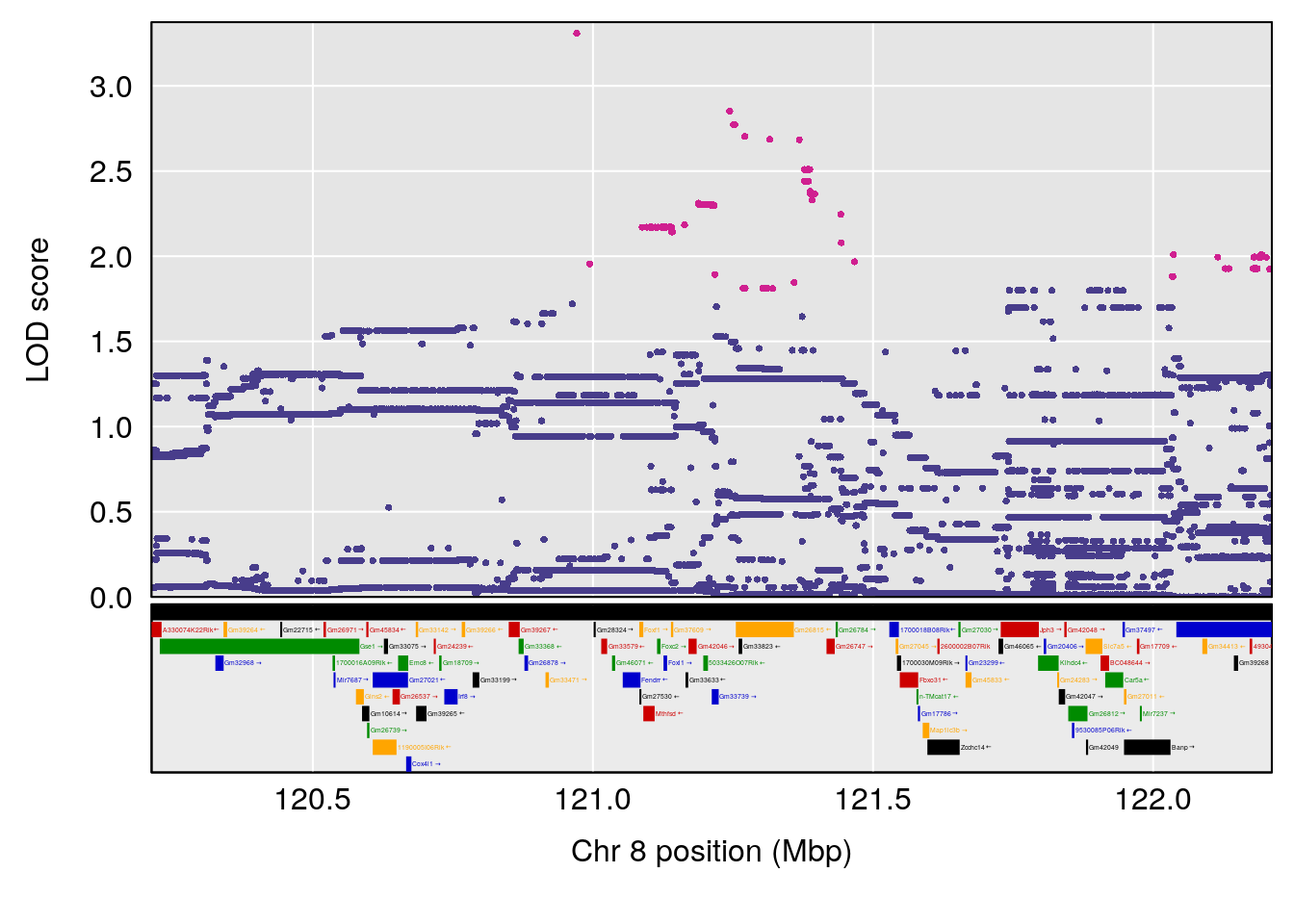
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 9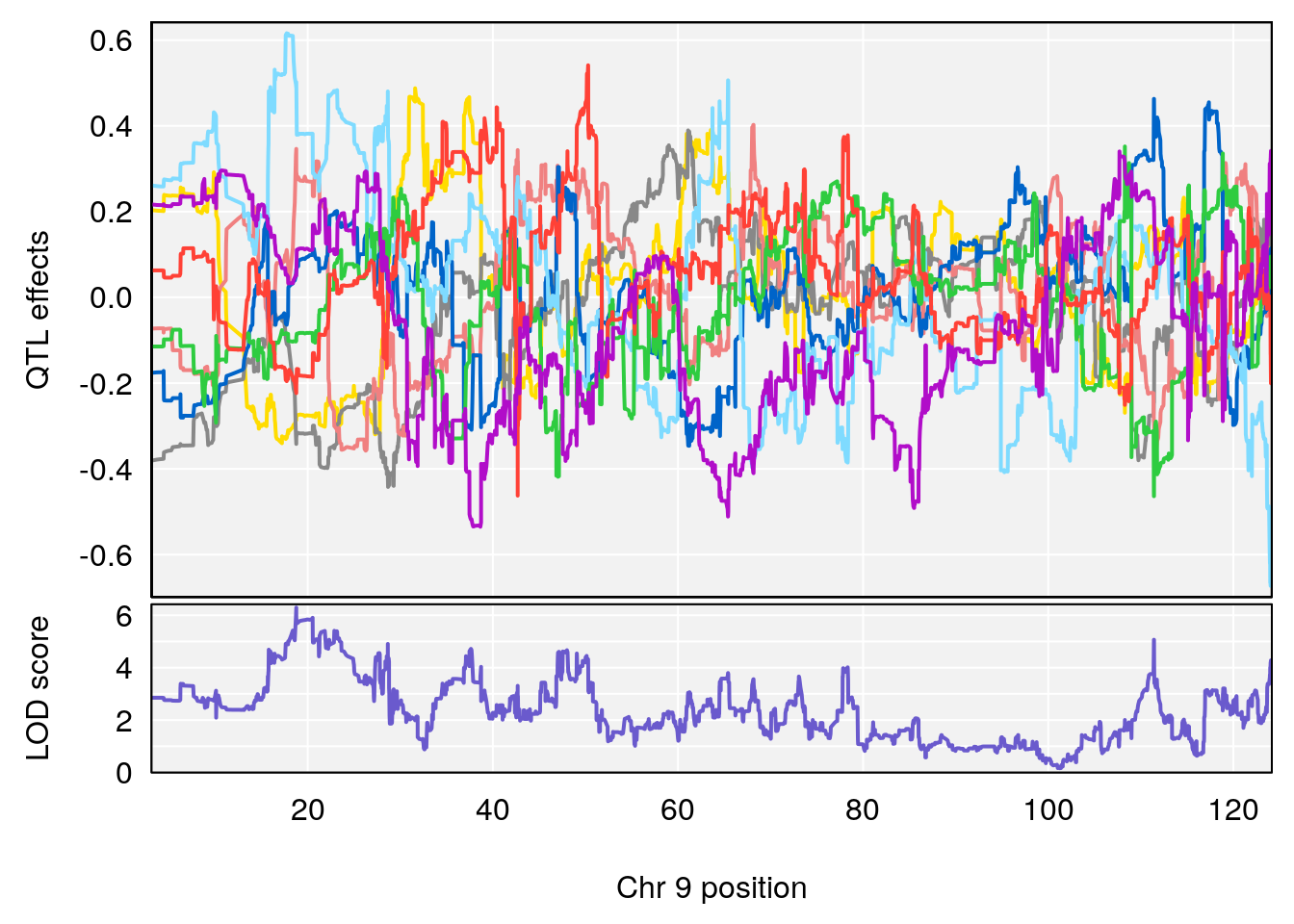
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |

| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 10
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 11
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 12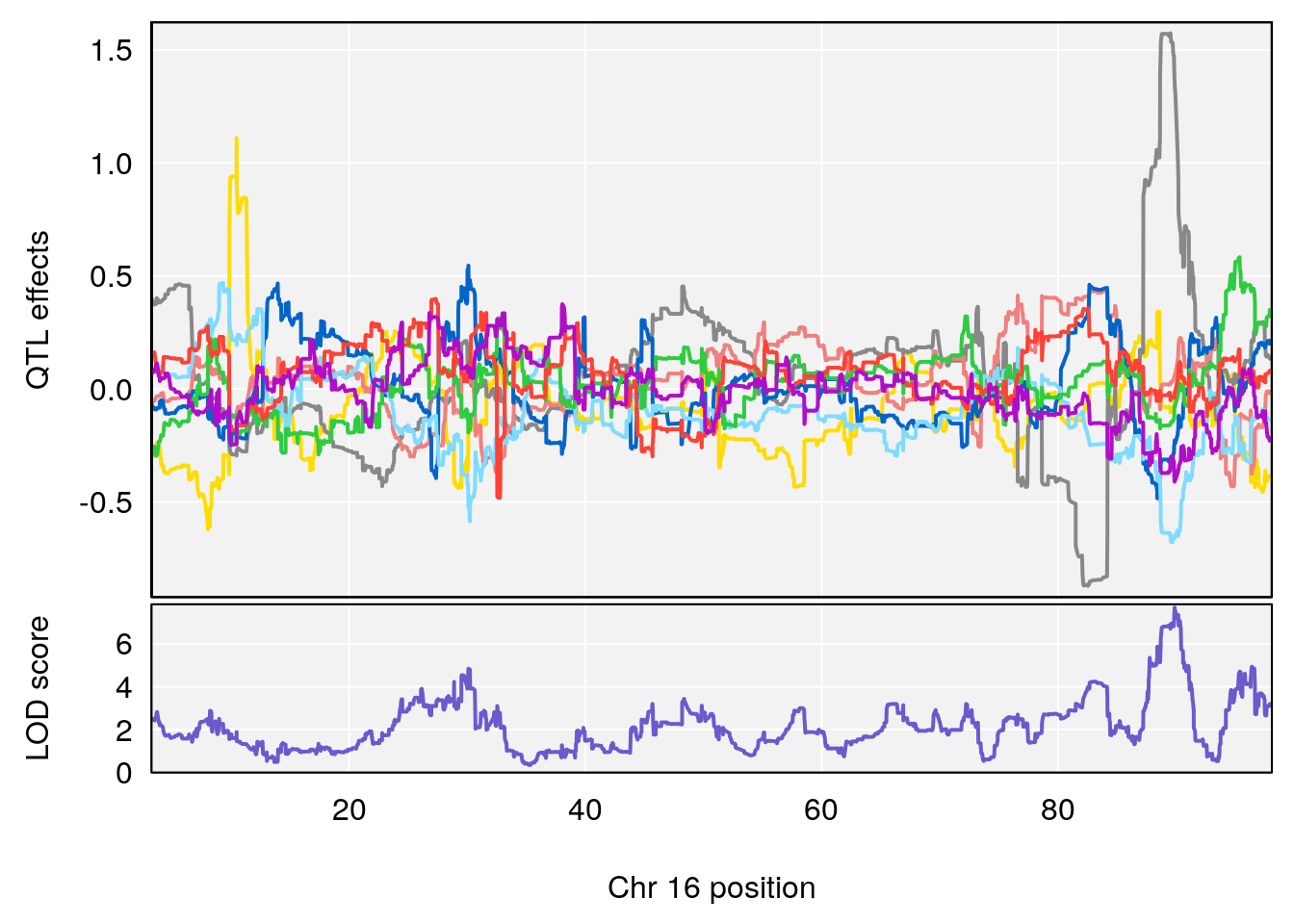
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |

| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 13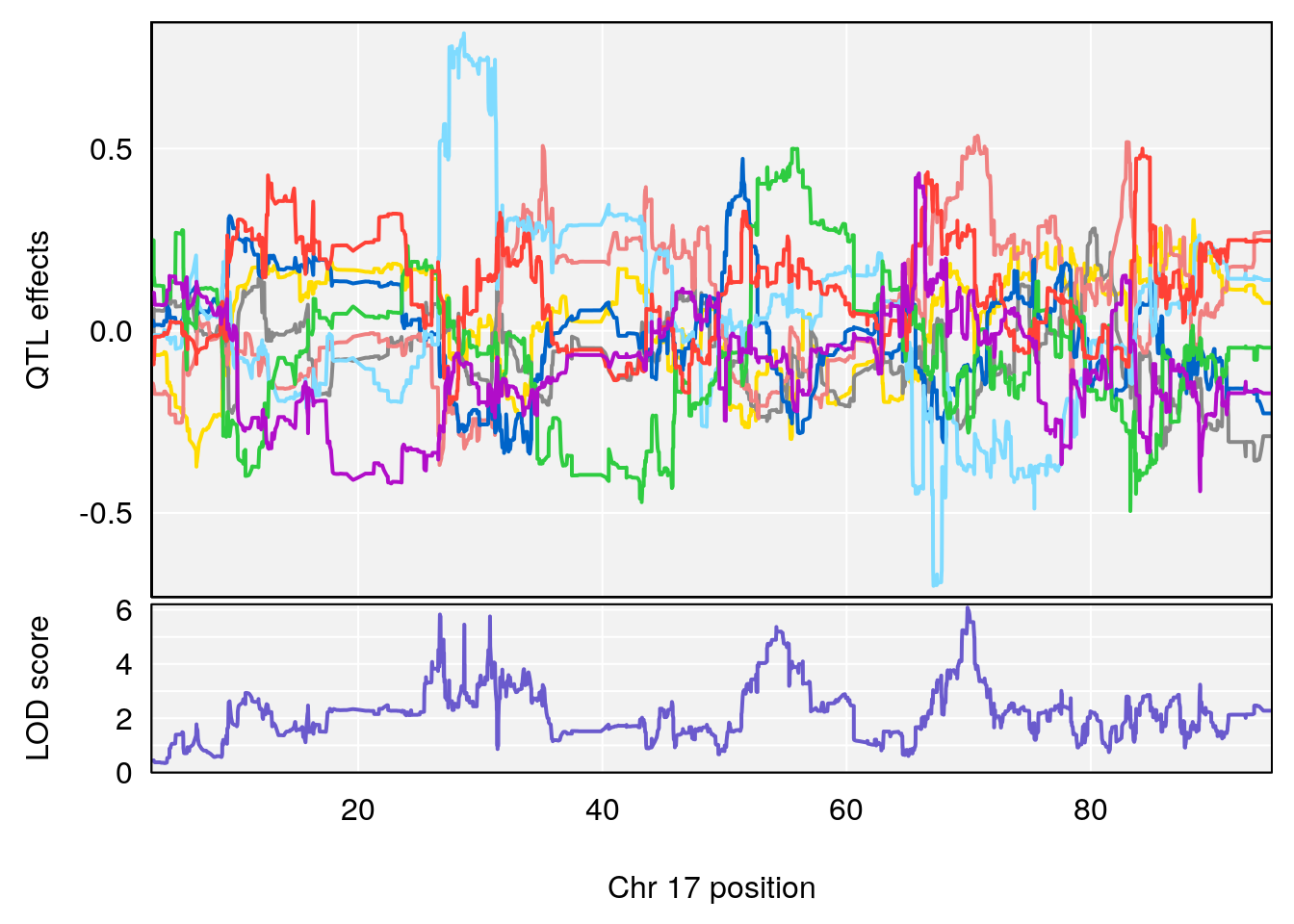
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |

| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
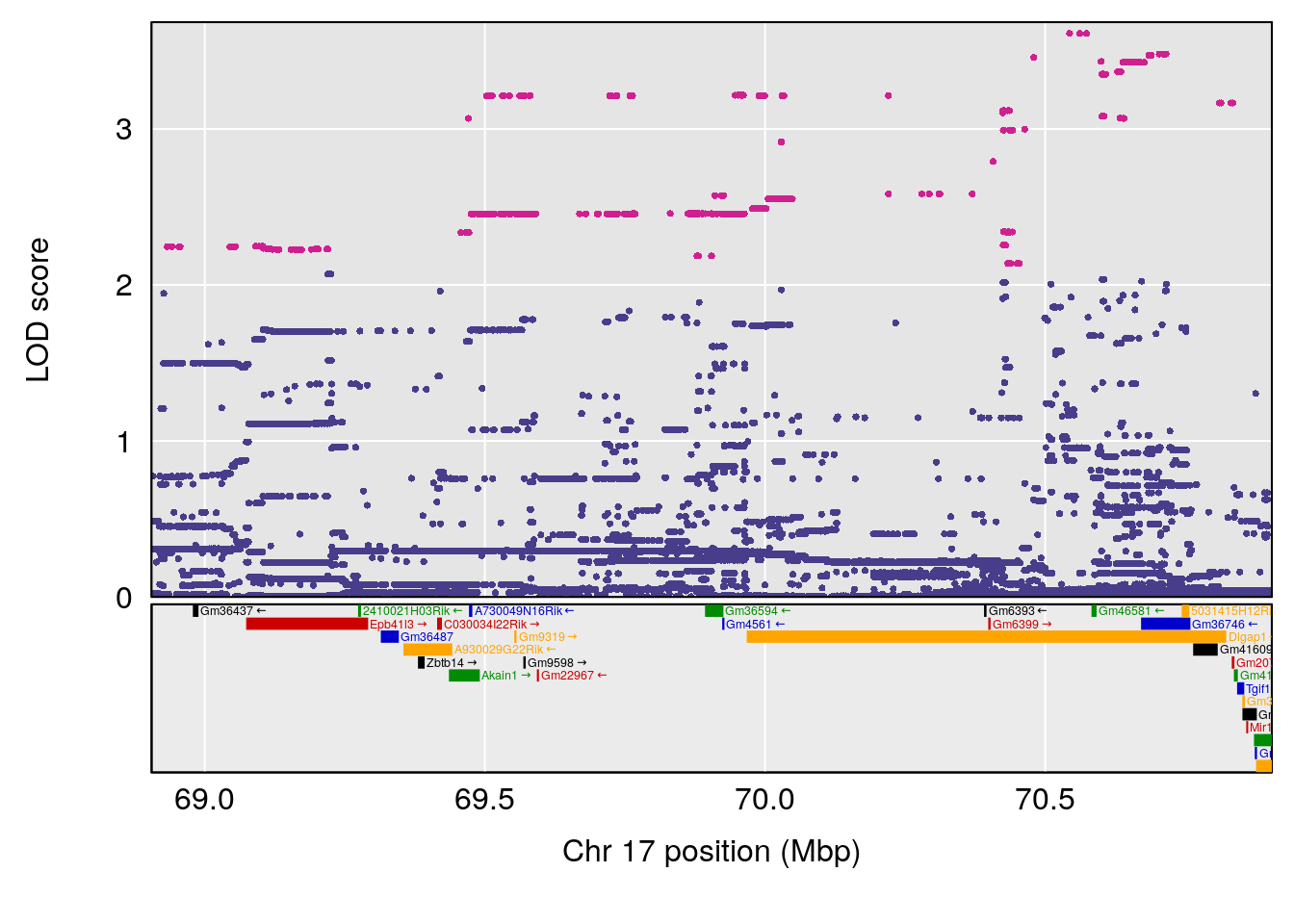
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 14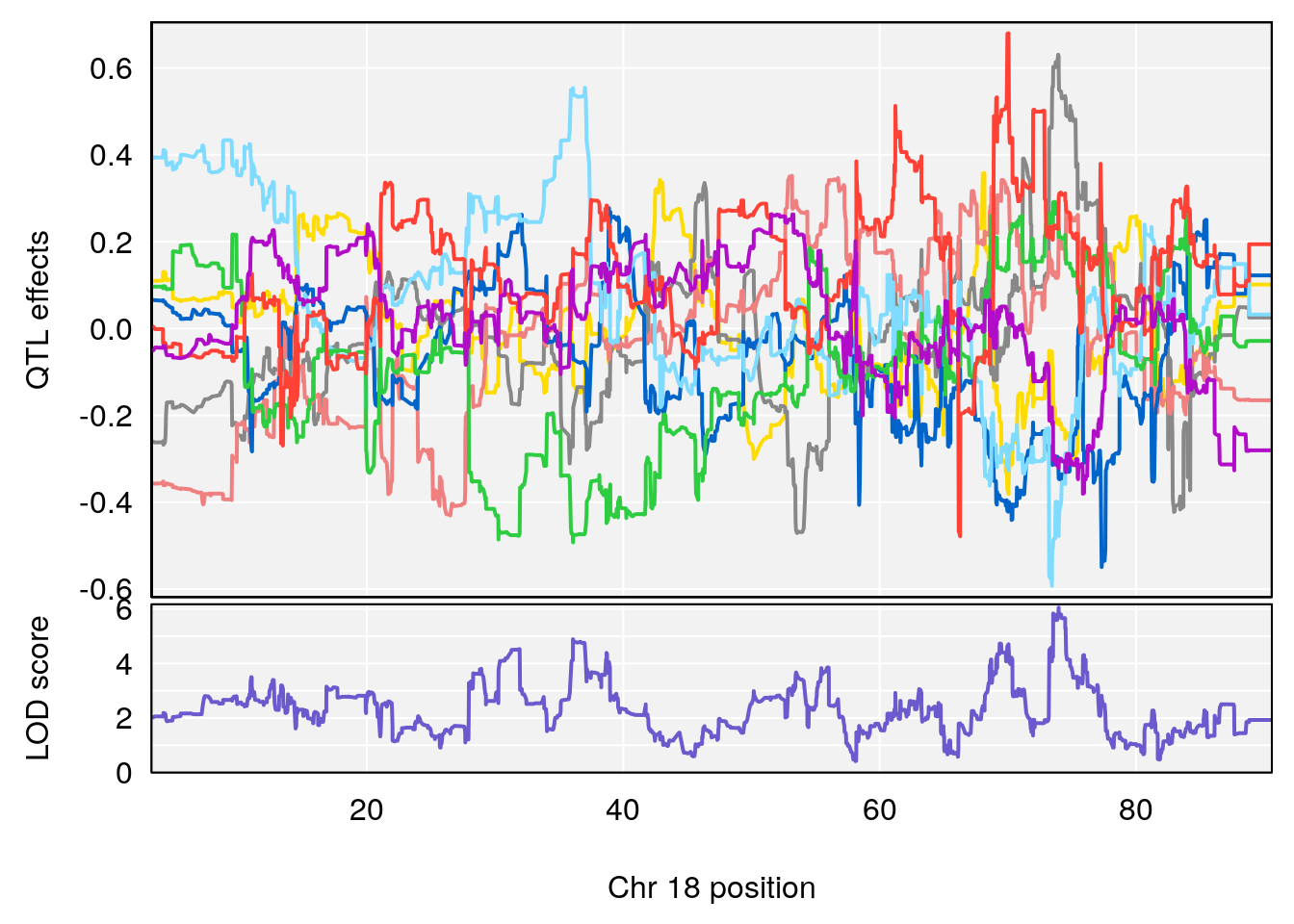
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
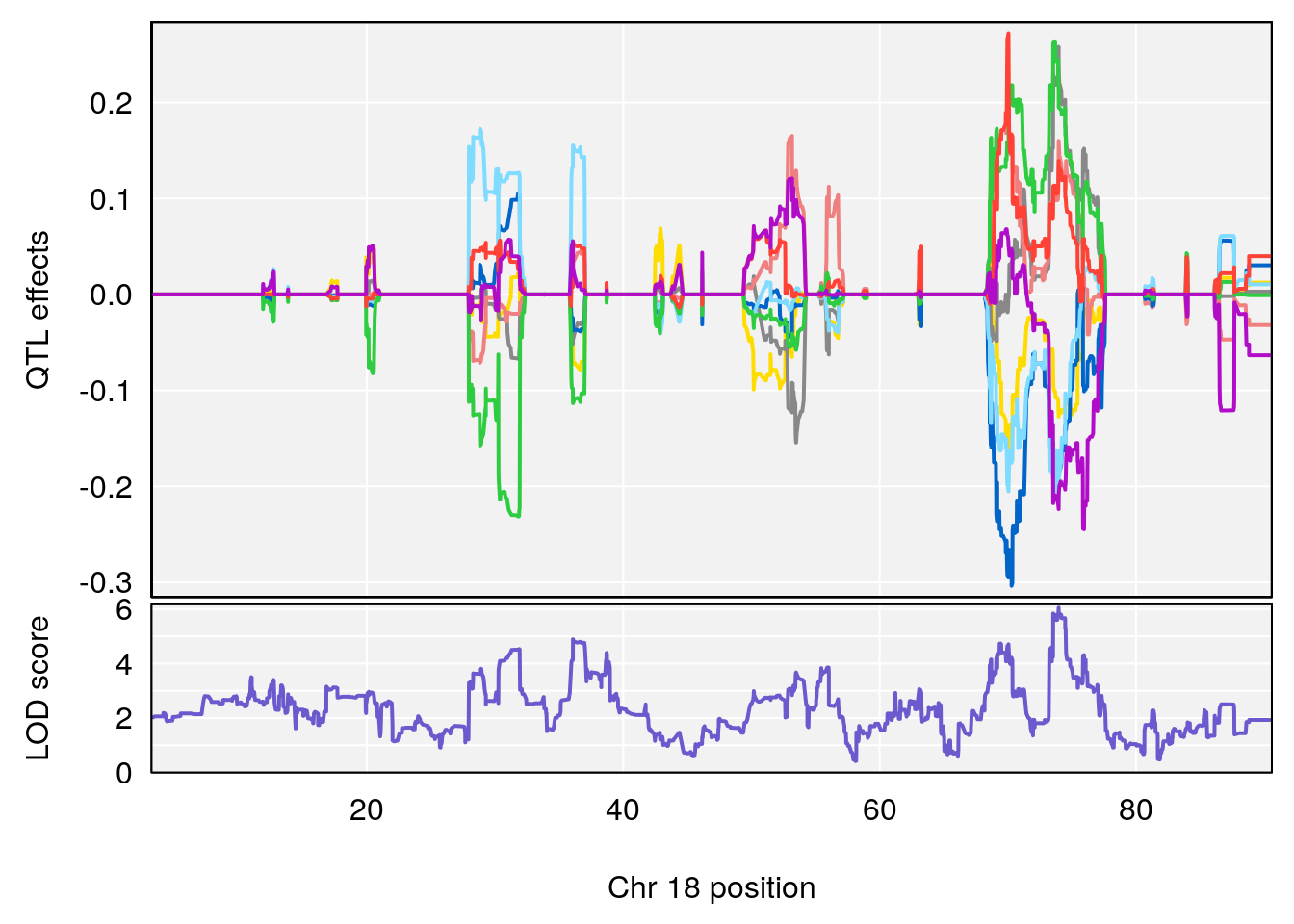
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
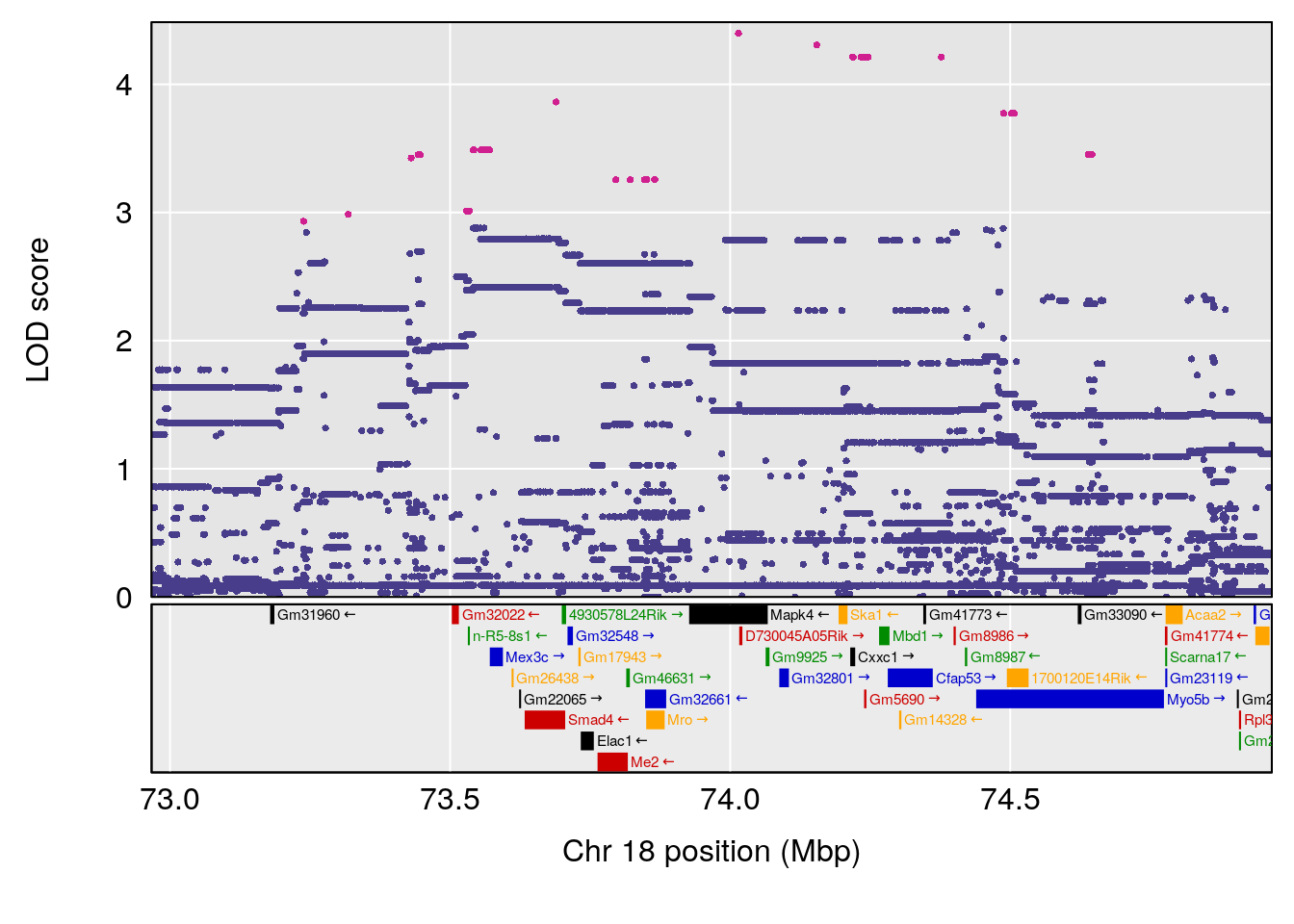
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 15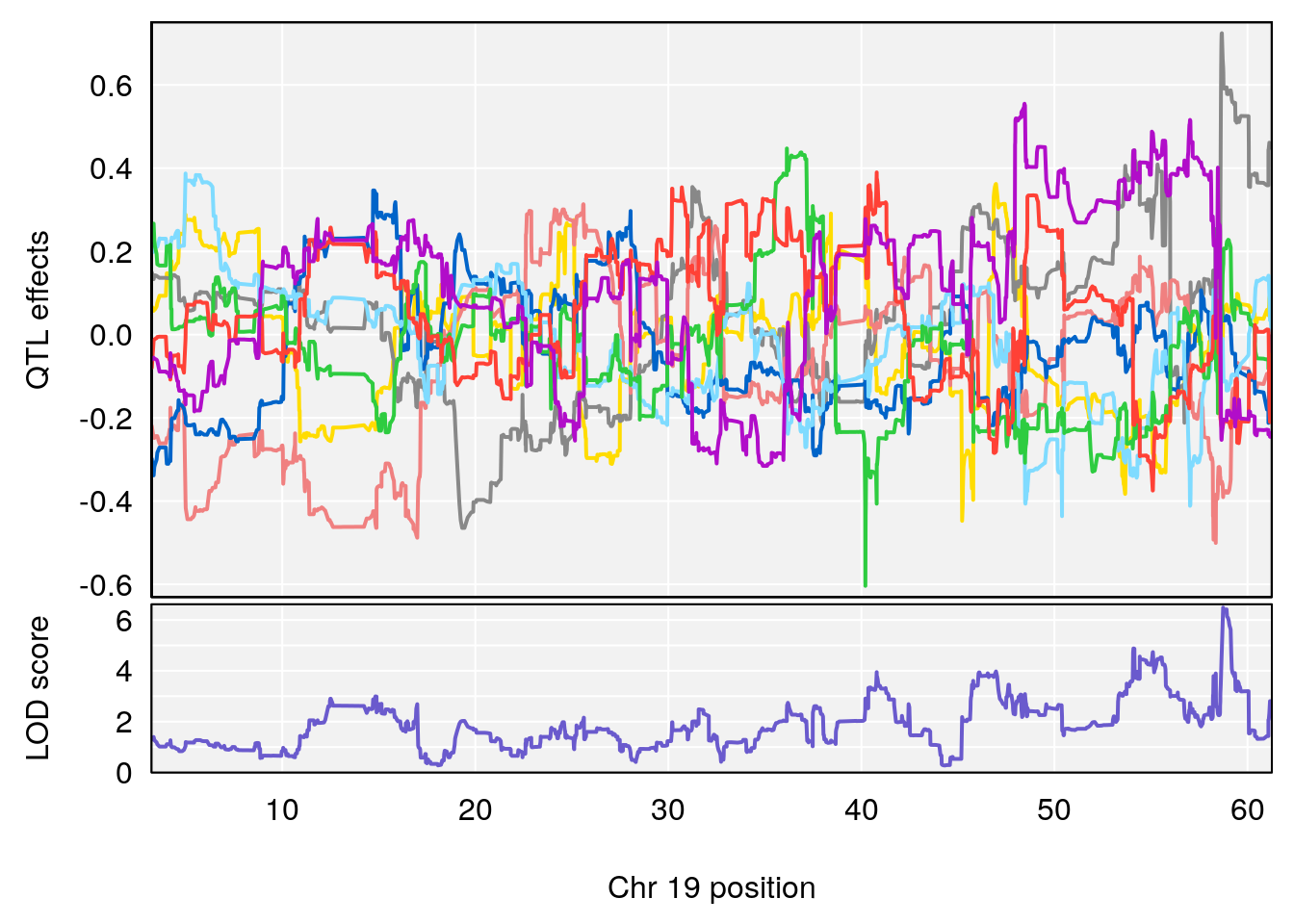
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |

| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
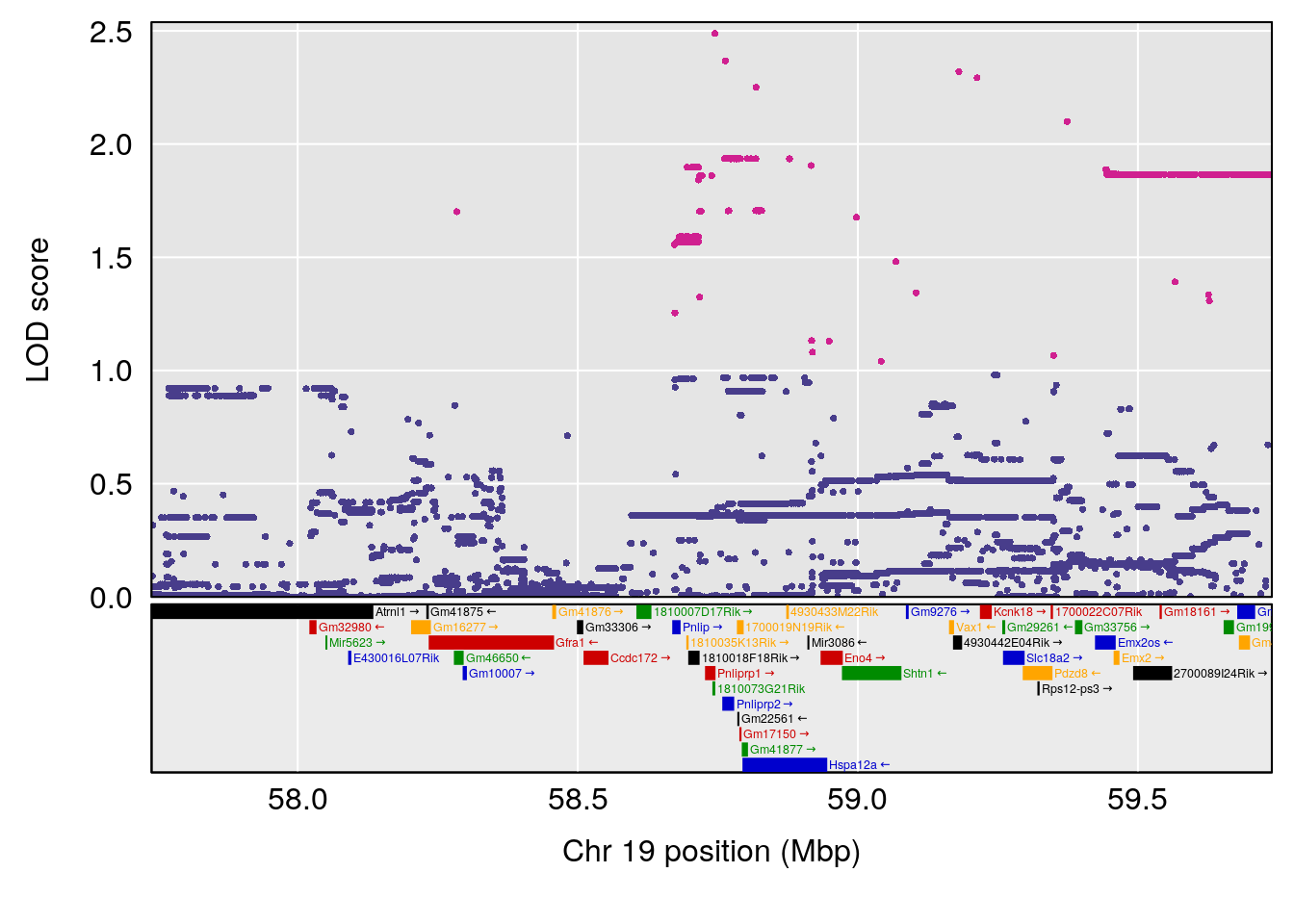
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
sessionInfo()
# R version 4.0.3 (2020-10-10)
# Platform: x86_64-pc-linux-gnu (64-bit)
# Running under: Ubuntu 20.04.2 LTS
#
# Matrix products: default
# BLAS/LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.8.so
#
# locale:
# [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
# [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
# [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=C
# [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
# [9] LC_ADDRESS=C LC_TELEPHONE=C
# [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
#
# attached base packages:
# [1] parallel stats graphics grDevices utils datasets methods
# [8] base
#
# other attached packages:
# [1] abind_1.4-5 regress_1.3-21 survival_3.2-7 qtl2_0.24
# [5] GGally_2.1.0 gridExtra_2.3 ggplot2_3.3.3 workflowr_1.6.2
#
# loaded via a namespace (and not attached):
# [1] tidyselect_1.1.0 xfun_0.21 purrr_0.3.4 lattice_0.20-41
# [5] splines_4.0.3 colorspace_2.0-0 vctrs_0.3.6 generics_0.1.0
# [9] htmltools_0.5.1.1 yaml_2.2.1 blob_1.2.1 rlang_1.0.2
# [13] later_1.1.0.1 pillar_1.4.7 glue_1.4.2 withr_2.4.1
# [17] DBI_1.1.1 bit64_4.0.5 RColorBrewer_1.1-2 lifecycle_1.0.0
# [21] plyr_1.8.6 stringr_1.4.0 munsell_0.5.0 gtable_0.3.0
# [25] evaluate_0.14 memoise_2.0.0 labeling_0.4.2 knitr_1.31
# [29] fastmap_1.1.0 httpuv_1.5.5 highr_0.8 Rcpp_1.0.6
# [33] promises_1.2.0.1 scales_1.1.1 cachem_1.0.4 farver_2.0.3
# [37] fs_1.5.0 bit_4.0.4 digest_0.6.27 stringi_1.5.3
# [41] dplyr_1.0.4 grid_4.0.3 rprojroot_2.0.2 cli_2.3.0
# [45] tools_4.0.3 magrittr_2.0.1 tibble_3.0.6 RSQLite_2.2.3
# [49] crayon_1.4.1 whisker_0.4 pkgconfig_2.0.3 Matrix_1.3-2
# [53] ellipsis_0.3.1 data.table_1.13.6 assertthat_0.2.1 rmarkdown_2.6
# [57] reshape_0.8.8 R6_2.5.0 git2r_0.28.0 compiler_4.0.3This R Markdown site was created with workflowr