Plot_DO_morphine_secondbatch
Hao He
2023-02-21
Last updated: 2023-02-21
Checks: 7 0
Knit directory: DO_Opioid/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200504) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version cc3ae27. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/Picture1.png
Untracked files:
Untracked: Rplot_rz.png
Untracked: TIMBR.test.prop.bm.MN.RET.RData
Untracked: TIMBR.test.random.RData
Untracked: TIMBR.test.rz.transformed_TVb_ml.RData
Untracked: analysis/DDO_morphine1_second_set_69k.stdout
Untracked: analysis/DO_Fentanyl.R
Untracked: analysis/DO_Fentanyl.err
Untracked: analysis/DO_Fentanyl.out
Untracked: analysis/DO_Fentanyl.sh
Untracked: analysis/DO_Fentanyl_69k.R
Untracked: analysis/DO_Fentanyl_69k.err
Untracked: analysis/DO_Fentanyl_69k.out
Untracked: analysis/DO_Fentanyl_69k.sh
Untracked: analysis/DO_Fentanyl_Cohort2_GCTA_herit.R
Untracked: analysis/DO_Fentanyl_Cohort2_gemma.R
Untracked: analysis/DO_Fentanyl_Cohort2_mapping.R
Untracked: analysis/DO_Fentanyl_Cohort2_mapping.err
Untracked: analysis/DO_Fentanyl_Cohort2_mapping.out
Untracked: analysis/DO_Fentanyl_Cohort2_mapping.sh
Untracked: analysis/DO_Fentanyl_GCTA_herit.R
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.R
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.err
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.out
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.sh
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.R
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.err
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.out
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.sh
Untracked: analysis/DO_Fentanyl_array.R
Untracked: analysis/DO_Fentanyl_array.err
Untracked: analysis/DO_Fentanyl_array.out
Untracked: analysis/DO_Fentanyl_array.sh
Untracked: analysis/DO_Fentanyl_combining2Cohort_GCTA_herit.R
Untracked: analysis/DO_Fentanyl_combining2Cohort_gemma.R
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping.R
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping.err
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping.out
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping.sh
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping_CoxPH.R
Untracked: analysis/DO_Fentanyl_finalreport_to_plink.sh
Untracked: analysis/DO_Fentanyl_gemma.R
Untracked: analysis/DO_Fentanyl_gemma.err
Untracked: analysis/DO_Fentanyl_gemma.out
Untracked: analysis/DO_Fentanyl_gemma.sh
Untracked: analysis/DO_morphine1.R
Untracked: analysis/DO_morphine1.Rout
Untracked: analysis/DO_morphine1.sh
Untracked: analysis/DO_morphine1.stderr
Untracked: analysis/DO_morphine1.stdout
Untracked: analysis/DO_morphine1_SNP.R
Untracked: analysis/DO_morphine1_SNP.Rout
Untracked: analysis/DO_morphine1_SNP.sh
Untracked: analysis/DO_morphine1_SNP.stderr
Untracked: analysis/DO_morphine1_SNP.stdout
Untracked: analysis/DO_morphine1_combined.R
Untracked: analysis/DO_morphine1_combined.Rout
Untracked: analysis/DO_morphine1_combined.sh
Untracked: analysis/DO_morphine1_combined.stderr
Untracked: analysis/DO_morphine1_combined.stdout
Untracked: analysis/DO_morphine1_combined_69k.R
Untracked: analysis/DO_morphine1_combined_69k.Rout
Untracked: analysis/DO_morphine1_combined_69k.sh
Untracked: analysis/DO_morphine1_combined_69k.stderr
Untracked: analysis/DO_morphine1_combined_69k.stdout
Untracked: analysis/DO_morphine1_combined_69k_m2.R
Untracked: analysis/DO_morphine1_combined_69k_m2.Rout
Untracked: analysis/DO_morphine1_combined_69k_m2.sh
Untracked: analysis/DO_morphine1_combined_69k_m2.stderr
Untracked: analysis/DO_morphine1_combined_69k_m2.stdout
Untracked: analysis/DO_morphine1_combined_weight_DOB.R
Untracked: analysis/DO_morphine1_combined_weight_DOB.Rout
Untracked: analysis/DO_morphine1_combined_weight_DOB.err
Untracked: analysis/DO_morphine1_combined_weight_DOB.out
Untracked: analysis/DO_morphine1_combined_weight_DOB.sh
Untracked: analysis/DO_morphine1_combined_weight_DOB.stderr
Untracked: analysis/DO_morphine1_combined_weight_DOB.stdout
Untracked: analysis/DO_morphine1_combined_weight_age.R
Untracked: analysis/DO_morphine1_combined_weight_age.err
Untracked: analysis/DO_morphine1_combined_weight_age.out
Untracked: analysis/DO_morphine1_combined_weight_age.sh
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.R
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.err
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.out
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.sh
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.R
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.err
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.out
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.sh
Untracked: analysis/DO_morphine1_cph.R
Untracked: analysis/DO_morphine1_cph.Rout
Untracked: analysis/DO_morphine1_cph.sh
Untracked: analysis/DO_morphine1_second_set.R
Untracked: analysis/DO_morphine1_second_set.Rout
Untracked: analysis/DO_morphine1_second_set.sh
Untracked: analysis/DO_morphine1_second_set.stderr
Untracked: analysis/DO_morphine1_second_set.stdout
Untracked: analysis/DO_morphine1_second_set_69k.R
Untracked: analysis/DO_morphine1_second_set_69k.Rout
Untracked: analysis/DO_morphine1_second_set_69k.sh
Untracked: analysis/DO_morphine1_second_set_69k.stderr
Untracked: analysis/DO_morphine1_second_set_SNP.R
Untracked: analysis/DO_morphine1_second_set_SNP.Rout
Untracked: analysis/DO_morphine1_second_set_SNP.sh
Untracked: analysis/DO_morphine1_second_set_SNP.stderr
Untracked: analysis/DO_morphine1_second_set_SNP.stdout
Untracked: analysis/DO_morphine1_second_set_weight_DOB.R
Untracked: analysis/DO_morphine1_second_set_weight_DOB.Rout
Untracked: analysis/DO_morphine1_second_set_weight_DOB.err
Untracked: analysis/DO_morphine1_second_set_weight_DOB.out
Untracked: analysis/DO_morphine1_second_set_weight_DOB.sh
Untracked: analysis/DO_morphine1_second_set_weight_DOB.stderr
Untracked: analysis/DO_morphine1_second_set_weight_DOB.stdout
Untracked: analysis/DO_morphine1_second_set_weight_age.R
Untracked: analysis/DO_morphine1_second_set_weight_age.Rout
Untracked: analysis/DO_morphine1_second_set_weight_age.err
Untracked: analysis/DO_morphine1_second_set_weight_age.out
Untracked: analysis/DO_morphine1_second_set_weight_age.sh
Untracked: analysis/DO_morphine1_second_set_weight_age.stderr
Untracked: analysis/DO_morphine1_second_set_weight_age.stdout
Untracked: analysis/DO_morphine1_weight_DOB.R
Untracked: analysis/DO_morphine1_weight_DOB.sh
Untracked: analysis/DO_morphine1_weight_age.R
Untracked: analysis/DO_morphine1_weight_age.sh
Untracked: analysis/DO_morphine_gemma.R
Untracked: analysis/DO_morphine_gemma.err
Untracked: analysis/DO_morphine_gemma.out
Untracked: analysis/DO_morphine_gemma.sh
Untracked: analysis/DO_morphine_gemma_firstmin.R
Untracked: analysis/DO_morphine_gemma_firstmin.err
Untracked: analysis/DO_morphine_gemma_firstmin.out
Untracked: analysis/DO_morphine_gemma_firstmin.sh
Untracked: analysis/DO_morphine_gemma_withpermu.R
Untracked: analysis/DO_morphine_gemma_withpermu.err
Untracked: analysis/DO_morphine_gemma_withpermu.out
Untracked: analysis/DO_morphine_gemma_withpermu.sh
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.R
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.err
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.out
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.sh
Untracked: analysis/Plot_DO_morphine1_SNP.R
Untracked: analysis/Plot_DO_morphine1_SNP.Rout
Untracked: analysis/Plot_DO_morphine1_SNP.sh
Untracked: analysis/Plot_DO_morphine1_SNP.stderr
Untracked: analysis/Plot_DO_morphine1_SNP.stdout
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.R
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.Rout
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.sh
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.stderr
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.stdout
Untracked: analysis/download_GSE100356_sra.sh
Untracked: analysis/fentanyl_2cohorts_coxph.R
Untracked: analysis/fentanyl_2cohorts_coxph.err
Untracked: analysis/fentanyl_2cohorts_coxph.out
Untracked: analysis/fentanyl_2cohorts_coxph.sh
Untracked: analysis/fentanyl_scanone.cph.R
Untracked: analysis/fentanyl_scanone.cph.err
Untracked: analysis/fentanyl_scanone.cph.out
Untracked: analysis/fentanyl_scanone.cph.sh
Untracked: analysis/geo_rnaseq.R
Untracked: analysis/heritability_first_second_batch.R
Untracked: analysis/nf-rnaseq-b6.R
Untracked: analysis/plot_fentanyl_2cohorts_coxph.R
Untracked: analysis/scripts/
Untracked: analysis/tibmr.R
Untracked: analysis/timbr_demo.R
Untracked: analysis/workflow_proc.R
Untracked: analysis/workflow_proc.sh
Untracked: analysis/workflow_proc.stderr
Untracked: analysis/workflow_proc.stdout
Untracked: analysis/x.R
Untracked: code/cfw/
Untracked: code/gemma_plot.R
Untracked: code/process.sanger.snp.R
Untracked: code/reconst_utils.R
Untracked: data/69k_grid_pgmap.RData
Untracked: data/Composite Post Kevins Program Group 2 Fentanyl Prepped for Hao.xlsx
Untracked: data/DO_WBP_Data_JAB_to_map.xlsx
Untracked: data/Fentanyl_alternate_metrics.xlsx
Untracked: data/FinalReport/
Untracked: data/GM/
Untracked: data/GM_covar.csv
Untracked: data/GM_covar_07092018_morphine.csv
Untracked: data/Jackson_Lab_Bubier_MURGIGV01/
Untracked: data/MPD_Upload_October.csv
Untracked: data/MPD_Upload_October_updated_sex.csv
Untracked: data/Master Fentanyl DO Study Sheet.xlsx
Untracked: data/MasterMorphine Second Set DO w DOB2.xlsx
Untracked: data/MasterMorphine Second Set DO.xlsx
Untracked: data/Morphine CC DO mice Updated with Published inbred strains.csv
Untracked: data/Morphine_CC_DO_mice_Updated_with_Published_inbred_strains.csv
Untracked: data/cc_variants.sqlite
Untracked: data/combined/
Untracked: data/fentanyl/
Untracked: data/fentanyl2/
Untracked: data/fentanyl_1_2/
Untracked: data/fentanyl_2cohorts_coxph_data.Rdata
Untracked: data/first/
Untracked: data/founder_geno.csv
Untracked: data/genetic_map.csv
Untracked: data/gm.json
Untracked: data/gwas.sh
Untracked: data/marker_grid_0.02cM_plus.txt
Untracked: data/mouse_genes_mgi.sqlite
Untracked: data/pheno.csv
Untracked: data/pheno_qtl2.csv
Untracked: data/pheno_qtl2_07092018_morphine.csv
Untracked: data/pheno_qtl2_w_dob.csv
Untracked: data/physical_map.csv
Untracked: data/rnaseq/
Untracked: data/sample_geno.csv
Untracked: data/second/
Untracked: figure/
Untracked: glimma-plots/
Untracked: output/DO_Fentanyl_Cohort2_MinDepressionRR_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_MinDepressionRR_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_RRDepressionRateHrSLOPE_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_RRRecoveryRateHrSLOPE_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_RRRecoveryRateHrSLOPE_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_StartofRecoveryHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_StartofRecoveryHr_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_Statusbin_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_Statusbin_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_SteadyStateDepressionDurationHrINTERVAL_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoDead(Hr)_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoDeadHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoDeadHr_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoProjectedRecoveryHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoProjectedRecoveryHr_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoSteadyRRDepression(Hr)_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoSteadyRRDepressionHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoSteadyRRDepressionHr_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoThresholdRecoveryHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoThresholdRecoveryHr_coefplot_blup.pdf
Untracked: output/DO_morphine_Min.depression.png
Untracked: output/DO_morphine_Min.depression22222_violin_chr5.pdf
Untracked: output/DO_morphine_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_Min.depression_coefplot_blup_chr5.png
Untracked: output/DO_morphine_Min.depression_coefplot_blup_chrX.png
Untracked: output/DO_morphine_Min.depression_coefplot_chr5.png
Untracked: output/DO_morphine_Min.depression_coefplot_chrX.png
Untracked: output/DO_morphine_Min.depression_peak_genes_chr5.png
Untracked: output/DO_morphine_Min.depression_violin_chr5.png
Untracked: output/DO_morphine_Recovery.Time.png
Untracked: output/DO_morphine_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr11.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr4.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr7.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr9.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr11.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr4.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr7.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr9.png
Untracked: output/DO_morphine_Status_bin.png
Untracked: output/DO_morphine_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_Survival.Time.png
Untracked: output/DO_morphine_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_Survival.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_Survival.Time_coefplot_blup_chr17.png
Untracked: output/DO_morphine_Survival.Time_coefplot_blup_chr8.png
Untracked: output/DO_morphine_Survival.Time_coefplot_chr17.png
Untracked: output/DO_morphine_Survival.Time_coefplot_chr8.png
Untracked: output/DO_morphine_combine_batch_peak_violin.pdf
Untracked: output/DO_morphine_combined_69k_m2_Min.depression.png
Untracked: output/DO_morphine_combined_69k_m2_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_combined_69k_m2_Recovery.Time.png
Untracked: output/DO_morphine_combined_69k_m2_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_combined_69k_m2_Status_bin.png
Untracked: output/DO_morphine_combined_69k_m2_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_combined_69k_m2_Survival.Time.png
Untracked: output/DO_morphine_combined_69k_m2_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Survival.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_coxph_24hrs_kinship_QTL.png
Untracked: output/DO_morphine_cphout.RData
Untracked: output/DO_morphine_first_batch_peak_in_second_batch_violin.pdf
Untracked: output/DO_morphine_first_batch_peak_in_second_batch_violin_sidebyside.pdf
Untracked: output/DO_morphine_first_batch_peak_violin.pdf
Untracked: output/DO_morphine_operm.cph.RData
Untracked: output/DO_morphine_second_batch_on_first_batch_peak_violin.pdf
Untracked: output/DO_morphine_second_batch_peak_ch6surv_on_first_batchviolin.pdf
Untracked: output/DO_morphine_second_batch_peak_ch6surv_on_first_batchviolin2.pdf
Untracked: output/DO_morphine_second_batch_peak_in_first_batch_violin.pdf
Untracked: output/DO_morphine_second_batch_peak_in_first_batch_violin_sidebyside.pdf
Untracked: output/DO_morphine_second_batch_peak_violin.pdf
Untracked: output/DO_morphine_secondbatch_69k_Min.depression.png
Untracked: output/DO_morphine_secondbatch_69k_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_69k_Recovery.Time.png
Untracked: output/DO_morphine_secondbatch_69k_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_69k_Status_bin.png
Untracked: output/DO_morphine_secondbatch_69k_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_69k_Survival.Time.png
Untracked: output/DO_morphine_secondbatch_69k_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Survival.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Min.depression.png
Untracked: output/DO_morphine_secondbatch_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Recovery.Time.png
Untracked: output/DO_morphine_secondbatch_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Status_bin.png
Untracked: output/DO_morphine_secondbatch_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Survival.Time.png
Untracked: output/DO_morphine_secondbatch_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Survival.Time_coefplot_blup.pdf
Untracked: output/Fentanyl/
Untracked: output/KPNA3.pdf
Untracked: output/SSC4D.pdf
Untracked: output/TIMBR.test.RData
Untracked: output/apr_69kchr_combined.RData
Untracked: output/apr_69kchr_k_loco_combined.rds
Untracked: output/apr_69kchr_second_set.RData
Untracked: output/combine_batch_variation.RData
Untracked: output/combined_gm.RData
Untracked: output/combined_gm.k_loco.rds
Untracked: output/combined_gm.k_overall.rds
Untracked: output/combined_gm.probs_8state.rds
Untracked: output/coxph/
Untracked: output/do.morphine.RData
Untracked: output/do.morphine.k_loco.rds
Untracked: output/do.morphine.probs_36state.rds
Untracked: output/do.morphine.probs_8state.rds
Untracked: output/do_Fentanyl_combine2cohort_MeanDepressionBR_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_MeanDepressionBR_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_MinDepressionBR_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_MinDepressionBR_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_MinDepressionRR_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_MinDepressionRR_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_RRRecoveryRateHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_RRRecoveryRateHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_StartofRecoveryHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_StartofRecoveryHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_Statusbin_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_Statusbin_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_SteadyStateDepressionDurationHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_SteadyStateDepressionDurationHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_SurvivalTime_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_SurvivalTime_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoDeadHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoDeadHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoMostlyDeadHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoMostlyDeadHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoProjectedRecoveryHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoProjectedRecoveryHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoRecoveryHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoRecoveryHr_coefplot_blup.pdf
Untracked: output/first_batch_variation.RData
Untracked: output/first_second_survival_peak_chr.xlsx
Untracked: output/hsq_1_first_batch_herit_qtl2.RData
Untracked: output/hsq_2_second_batch_herit_qtl2.RData
Untracked: output/old_temp/
Untracked: output/pr_69kchr_combined.RData
Untracked: output/pr_69kchr_second_set.RData
Untracked: output/qtl.morphine.69k.out.combined.RData
Untracked: output/qtl.morphine.69k.out.combined_m2.RData
Untracked: output/qtl.morphine.69k.out.second_set.RData
Untracked: output/qtl.morphine.operm.RData
Untracked: output/qtl.morphine.out.RData
Untracked: output/qtl.morphine.out.combined_gm.RData
Untracked: output/qtl.morphine.out.combined_gm.female.RData
Untracked: output/qtl.morphine.out.combined_gm.male.RData
Untracked: output/qtl.morphine.out.combined_weight_DOB.RData
Untracked: output/qtl.morphine.out.combined_weight_age.RData
Untracked: output/qtl.morphine.out.female.RData
Untracked: output/qtl.morphine.out.male.RData
Untracked: output/qtl.morphine.out.second_set.RData
Untracked: output/qtl.morphine.out.second_set.female.RData
Untracked: output/qtl.morphine.out.second_set.male.RData
Untracked: output/qtl.morphine.out.second_set.weight_DOB.RData
Untracked: output/qtl.morphine.out.second_set.weight_age.RData
Untracked: output/qtl.morphine.out.weight_DOB.RData
Untracked: output/qtl.morphine.out.weight_age.RData
Untracked: output/qtl.morphine1.snpout.RData
Untracked: output/qtl.morphine2.snpout.RData
Untracked: output/second_batch_pheno.csv
Untracked: output/second_batch_variation.RData
Untracked: output/second_set_apr_69kchr_k_loco.rds
Untracked: output/second_set_gm.RData
Untracked: output/second_set_gm.k_loco.rds
Untracked: output/second_set_gm.probs_36state.rds
Untracked: output/second_set_gm.probs_8state.rds
Untracked: output/topSNP_chr5_mindepression.csv
Untracked: output/zoompeak_Min.depression_9.pdf
Untracked: output/zoompeak_Recovery.Time_16.pdf
Untracked: output/zoompeak_Status_bin_11.pdf
Untracked: output/zoompeak_Survival.Time_1.pdf
Untracked: output/zoompeak_fentanyl_Survival.Time_2.pdf
Untracked: sra-tools_v2.10.7.sif
Unstaged changes:
Modified: _workflowr.yml
Modified: analysis/Plot_DO_Fentanyl_Cohort2_mapping.Rmd
Modified: analysis/Plot_DO_Fentanyl_combining2Cohort_mapping.Rmd
Modified: analysis/Plot_DO_fentanyl.Rmd
Modified: analysis/marker_violin.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/Plot_DO_morphine_combined.Rmd) and HTML (docs/Plot_DO_morphine_combined.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | cc3ae27 | xhyuo | 2023-02-21 | update with sex specific morphine plot |
| html | 8fe8543 | xhyuo | 2023-02-20 | Build site. |
| Rmd | b46775c | xhyuo | 2023-02-20 | update with sex specific morphine |
| html | 422da68 | xhyuo | 2020-07-12 | Build site. |
| Rmd | f210d8a | xhyuo | 2020-07-12 | Plot_DO_morphine_combined |
Last update: 2023-02-21
loading libraries
library(ggplot2)
library(gridExtra)
library(GGally)
library(parallel)
library(qtl2)
library(parallel)
library(survival)
library(regress)
library(abind)
library("readxl")
rz.transform <- function(y) {
rankY=rank(y, ties.method="average", na.last="keep")
rzT=qnorm(rankY/(length(na.exclude(rankY))+1))
return(rzT)
}
load("code/do.colors.RData")Read phenotype data
# Read phenotype data for first batch-----------------------------------------------------
#morphine pheno
morphine.pheno <- read.csv("data/pheno_qtl2_07092018_morphine.csv",header = T)
#remove duplicate id
morphine.pheno <- morphine.pheno[!morphine.pheno$X %in% c("morphine_DO_jbubier_13645_C9", "morphine_DO_jbubier_13554_H1"),]
morphine.pheno <- data.frame(ID = morphine.pheno$X,
Sex = NA,
GENERATION = NA,
GROUP = NA,
Date = NA,
Status24 = morphine.pheno$Status,
Survival.Time = morphine.pheno$Survival.Time,
Recovery.Time = morphine.pheno$Recovery.Time,
Min.depression = morphine.pheno$Min.depression,
Status_bin = morphine.pheno$Status_bin,
RL = "NO")
rownames(morphine.pheno) <- gsub("morphine_DO_jbubier_","", morphine.pheno$ID)
rownames(morphine.pheno) <- gsub("\\_.*","", rownames(morphine.pheno))
# Read phenotype data for second batch-----------------------------------------------------
second_set <- read_xlsx(path = "data/MasterMorphine Second Set DO.xlsx",
sheet = 1)
second_set <- as.data.frame(second_set)
colnames(second_set)[2] <- "Sex"
colnames(second_set)[6] <- "Status24"
#character
second_set[,c("ID", "Sex", "GROUP", "Status24")] <- lapply(second_set[, c("ID", "Sex", "GROUP", "Status24")], as.character)
second_set$Status_bin <- ifelse(second_set$Status24 == "ALIVE", 1, 0)
rownames(second_set) <- second_set$ID
#remove outlier
second_set <- second_set[second_set$ID != "empty",]
colnames(second_set) <- c("ID","Sex","GENERATION","GROUP","Date",
"Status24", "Survival.Time","Recovery.Time", "Min.depression",
"Status_bin")
#remove negative survival time subjects
second_set <- second_set[!(!is.na(second_set$Survival.Time) & second_set$Survival.Time < 0),]
second_set$Survival.Time[!is.na(second_set$Survival.Time) & second_set$Survival.Time > 15] <- NA
second_set$Recovery.Time[!is.na(second_set$Recovery.Time) & second_set$Recovery.Time > 24] <- NA
second_set$Min.depression[!is.na(second_set$Min.depression) & second_set$Min.depression > 1] <- NA
second_set$RL = "YES"
#combined pheno
pheno <- rbind(morphine.pheno, second_set)
#load json object
load("/projects/csna/csna_workflow/data/Jackson_Lab_11_batches/gm_DO2816_qc.RData")
#replace id names
old_ids <- paste0(as.character(do.call(rbind.data.frame, strsplit(ind_ids(gm_DO2816_qc), "_"))[,6]))
old_ids <- make.unique(as.character(old_ids), sep = "_")
new_ids <- setNames(old_ids,
ind_ids(gm_DO2816_qc))
combined_gm <- replace_ids(gm_DO2816_qc, new_ids)[intersect(rownames(pheno),old_ids), ]
combined_gm
# Object of class cross2 (crosstype "do")
#
# Total individuals 646
# No. genotyped individuals 646
# No. phenotyped individuals 646
# No. with both geno & pheno 646
#
# No. phenotypes 1
# No. covariates 4
# No. phenotype covariates 0
#
# No. chromosomes 20
# Total markers 112455
#
# No. markers by chr:
# 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
# 8530 8649 6403 6600 6559 6427 6281 5660 5854 5439 6338 5151 5257 5025 4547 4355
# 17 18 19 X
# 4321 3986 3102 3971
#update pheno
pheno <- merge(pheno, combined_gm$covar, by = "row.names", all.y = "TRUE")
rownames(pheno) <- pheno$Row.names
pheno <- pheno[,-1]
#remove generation 32 35
pheno <- pheno[!pheno$ngen %in% c("32", "35"),]
combined_gm <- combined_gm[rownames(pheno),]
combined_gm
# Object of class cross2 (crosstype "do")
#
# Total individuals 637
# No. genotyped individuals 637
# No. phenotyped individuals 637
# No. with both geno & pheno 637
#
# No. phenotypes 1
# No. covariates 4
# No. phenotype covariates 0
#
# No. chromosomes 20
# Total markers 112455
#
# No. markers by chr:
# 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
# 8530 8649 6403 6600 6559 6427 6281 5660 5854 5439 6338 5151 5257 5025 4547 4355
# 17 18 19 X
# 4321 3986 3102 3971
#covar
#first batch
covar1 <- read.csv("data/GM_covar_07092018_morphine.csv",header = T)
#remove duplicate id
covar1 <- covar1[!covar1$X %in% c("morphine_DO_jbubier_13645_C9", "morphine_DO_jbubier_13554_H1"),]
rownames(covar1) <- gsub("morphine_DO_jbubier_","", covar1$id)
rownames(covar1) <- gsub("\\_.*","", rownames(covar1))
covar1 <- covar1[,c(2:5)]
colnames(covar1) <- c("ID", "Sex", "GENERATION", "Date")
#second batch
covar2 <- second_set[,c("ID", "Sex", "GENERATION", "Date")]
covar2$Date <- as.character(covar2$Date)
#combine covar
covar <- rbind(covar1, covar2)
covar <- covar[rownames(pheno),]
covar$Sex <- as.factor(covar$Sex)
covar$Date <- as.factor(covar$Date)
all.equal(rownames(pheno), rownames(covar))
# [1] TRUE
#boxplot for the pheno by generation
data <- cbind(pheno, covar)
#boxplot
#survival time
surv <- data[,c(1, 7, 11, 14, 15, 19)]
surv$ngen <- factor(surv$ngen, levels = as.character(c(29:34)))
p1 <- ggplot(surv, aes(x=ngen, y=Survival.Time, group = ngen, fill = ngen, alpha = 0.9)) +
geom_boxplot(show.legend = F , outlier.size = 1.5, notchwidth = 0.85) +
geom_jitter(color="black", size=0.8, alpha=0.9) +
scale_fill_brewer(palette="Blues") +
ylab("Survival Time") +
xlab("Generation") +
labs(fill = "") +
theme(legend.position = "none",
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
axis.line = element_line(colour = "black"),
text = element_text(size=16),
axis.title=element_text(size=16)) +
guides(shape = guide_legend(override.aes = list(size = 12)))
p1
# Warning: Removed 384 rows containing non-finite values (stat_boxplot).
# Warning: Removed 384 rows containing missing values (geom_point).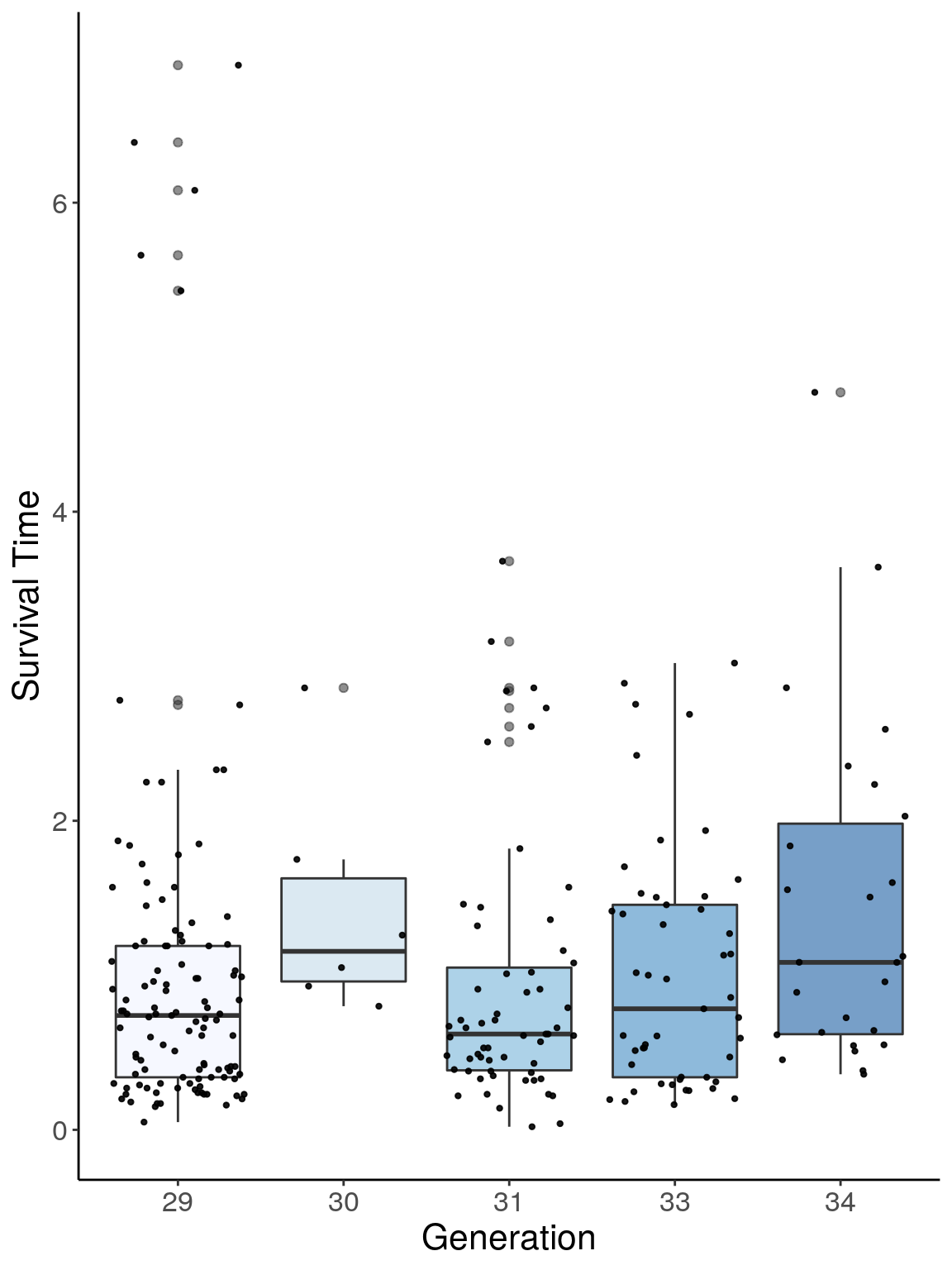
p1.1 <- ggplot(surv, aes(x=sex, y=Survival.Time, group = sex, fill = sex, alpha = 0.9)) +
geom_boxplot(show.legend = F , outlier.size = 1.5, notchwidth = 0.85) +
geom_jitter(color="black", size=0.8, alpha=0.9) +
scale_fill_brewer(palette="Blues") +
ylab("Survival Time") +
xlab("sex") +
labs(fill = "") +
theme(legend.position = "none",
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
axis.line = element_line(colour = "black"),
text = element_text(size=16),
axis.title=element_text(size=16)) +
guides(shape = guide_legend(override.aes = list(size = 12)))
p1.1
# Warning: Removed 384 rows containing non-finite values (stat_boxplot).
# Warning: Removed 384 rows containing missing values (geom_point).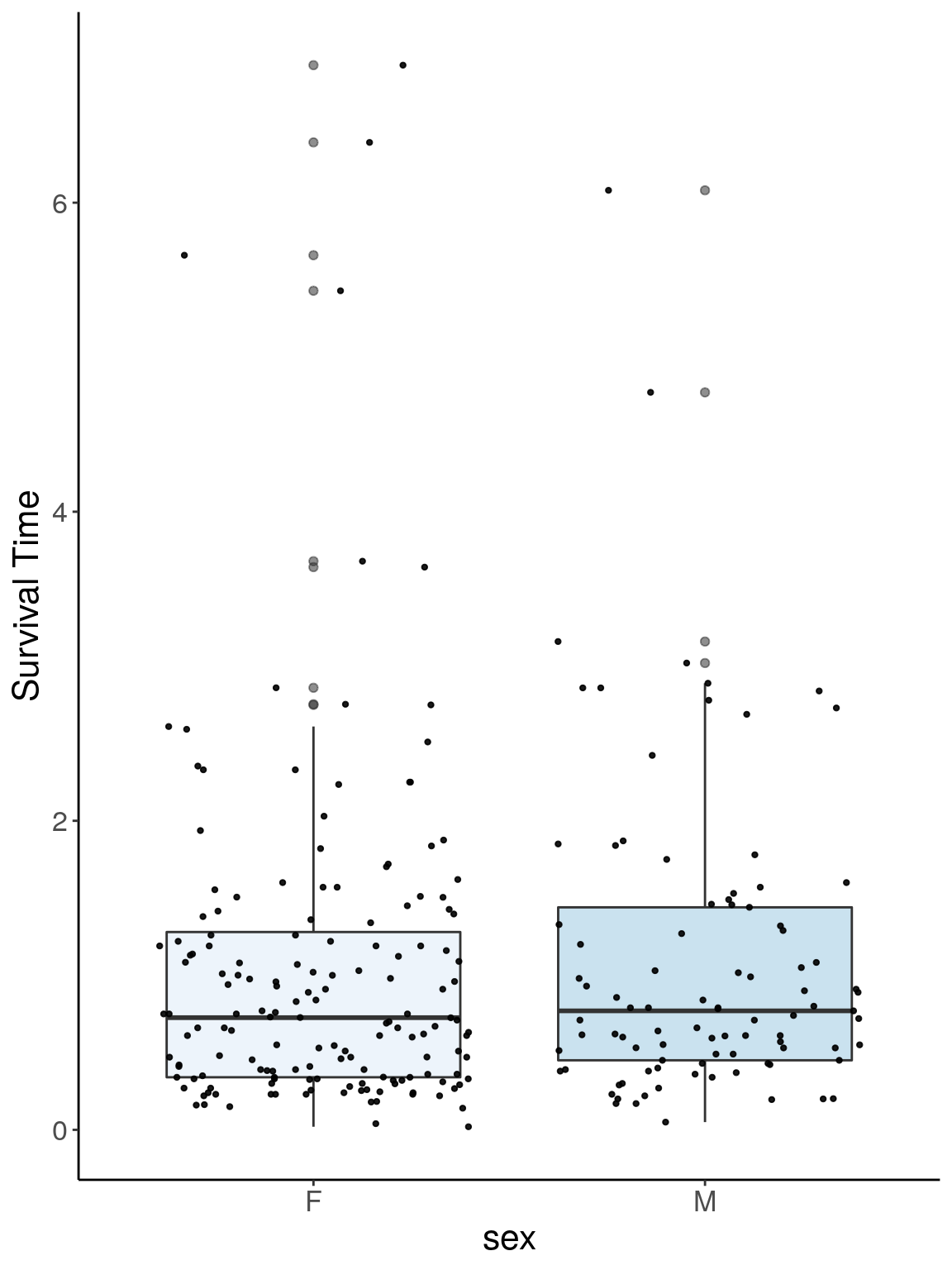
p1.2 <- ggplot(surv, aes(x=RL, y=Survival.Time, group = RL, fill = RL, alpha = 0.9)) +
geom_boxplot(show.legend = F , outlier.size = 1.5, notchwidth = 0.85) +
geom_jitter(color="black", size=0.8, alpha=0.9) +
scale_fill_brewer(palette="Blues") +
ylab("Survival Time") +
xlab("RL") +
labs(fill = "") +
theme(legend.position = "none",
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
axis.line = element_line(colour = "black"),
text = element_text(size=16),
axis.title=element_text(size=16)) +
guides(shape = guide_legend(override.aes = list(size = 12)))
p1.2
# Warning: Removed 384 rows containing non-finite values (stat_boxplot).
# Warning: Removed 384 rows containing missing values (geom_point).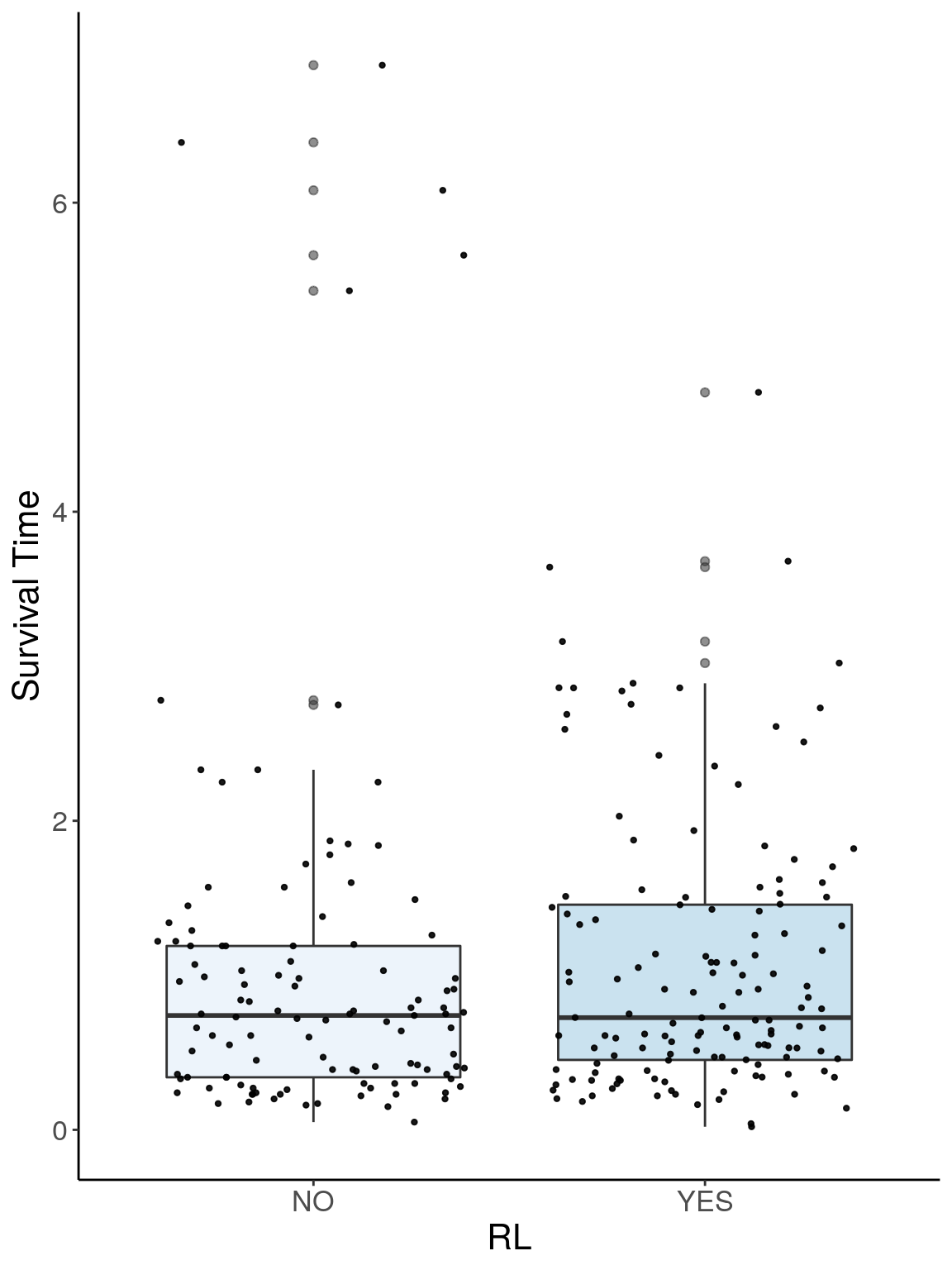
reco <- data[,c(1, 8, 11, 14, 15, 19)]
reco$ngen <- factor(reco$ngen, levels = as.character(c(29:35)))
p2 <- ggplot(reco, aes(x=ngen, y=Recovery.Time, group = ngen, fill = ngen, alpha = 0.9)) +
geom_boxplot(show.legend = F , outlier.size = 1.5, notchwidth = 0.85) +
geom_jitter(color="black", size=0.8, alpha=0.9) +
scale_fill_brewer(palette="Blues") +
ylab("Recovery Time") +
xlab("Generation") +
labs(fill = "") +
theme(legend.position = "none",
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
axis.line = element_line(colour = "black"),
text = element_text(size=16),
axis.title=element_text(size=16)) +
guides(shape = guide_legend(override.aes = list(size = 12)))
p2
# Warning: Removed 266 rows containing non-finite values (stat_boxplot).
# Warning: Removed 266 rows containing missing values (geom_point).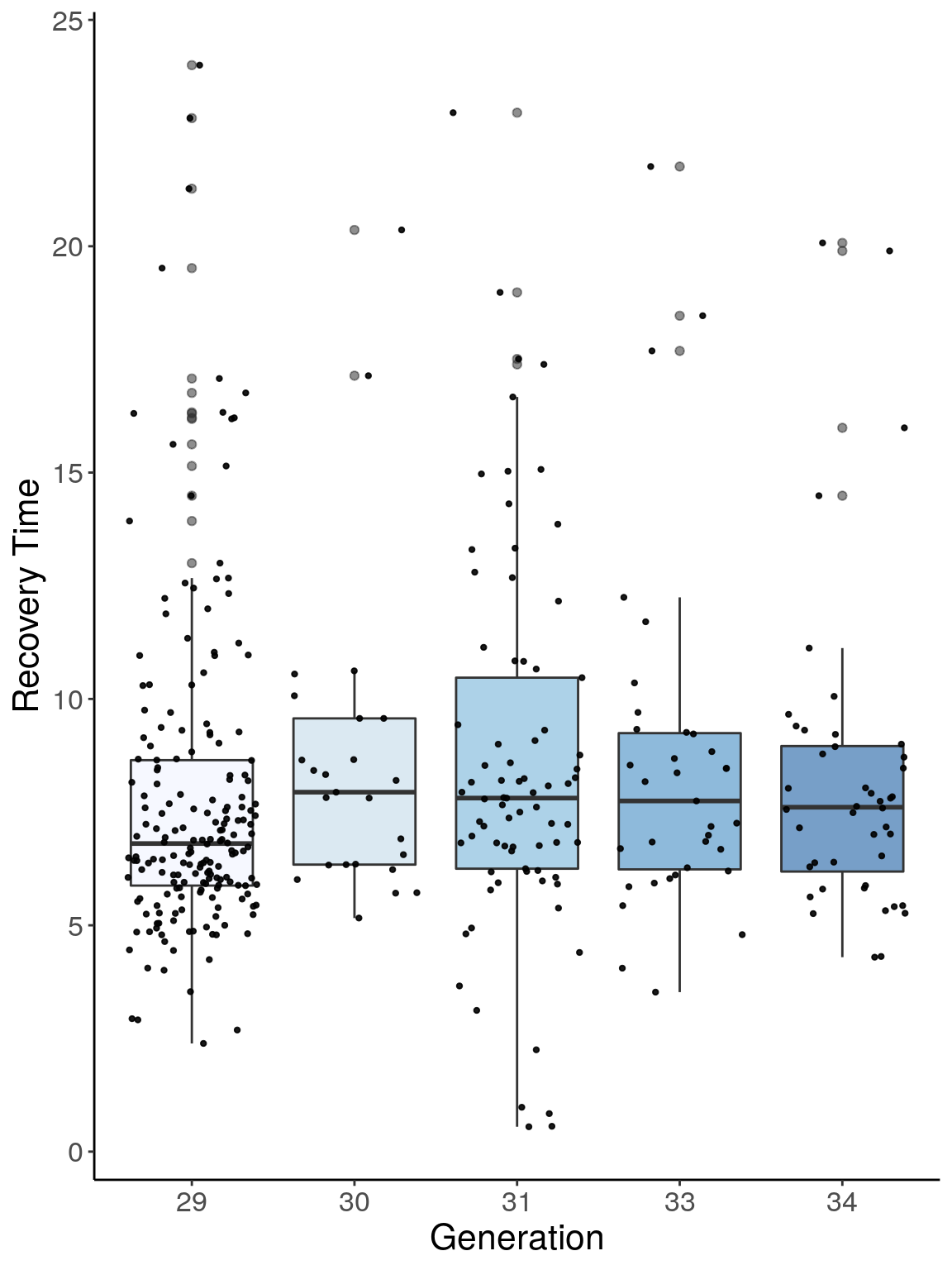
p2.1 <- ggplot(reco, aes(x=sex, y=Recovery.Time, group = sex, fill = sex, alpha = 0.9)) +
geom_boxplot(show.legend = F , outlier.size = 1.5, notchwidth = 0.85) +
geom_jitter(color="black", size=0.8, alpha=0.9) +
scale_fill_brewer(palette="Blues") +
ylab("Recovery Time") +
xlab("sex") +
labs(fill = "") +
theme(legend.position = "none",
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
axis.line = element_line(colour = "black"),
text = element_text(size=16),
axis.title=element_text(size=16)) +
guides(shape = guide_legend(override.aes = list(size = 12)))
p2.1
# Warning: Removed 266 rows containing non-finite values (stat_boxplot).
# Warning: Removed 266 rows containing missing values (geom_point).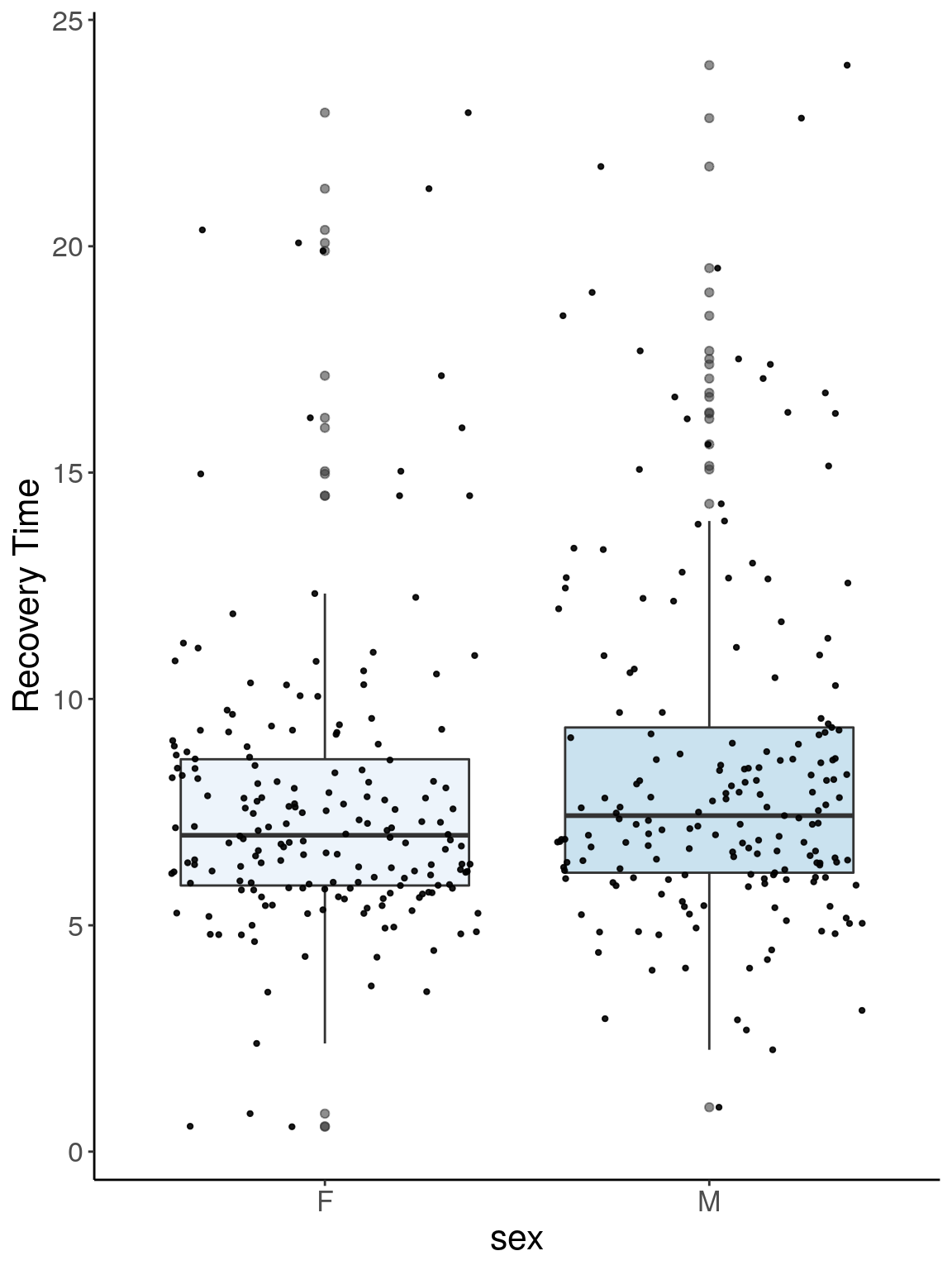
p2.2 <- ggplot(reco, aes(x=RL, y=Recovery.Time, group = RL, fill = RL, alpha = 0.9)) +
geom_boxplot(show.legend = F , outlier.size = 1.5, notchwidth = 0.85) +
geom_jitter(color="black", size=0.8, alpha=0.9) +
scale_fill_brewer(palette="Blues") +
ylab("Recovery Time") +
xlab("RL") +
labs(fill = "") +
theme(legend.position = "none",
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
axis.line = element_line(colour = "black"),
text = element_text(size=16),
axis.title=element_text(size=16)) +
guides(shape = guide_legend(override.aes = list(size = 12)))
p2.2
# Warning: Removed 266 rows containing non-finite values (stat_boxplot).
# Warning: Removed 266 rows containing missing values (geom_point).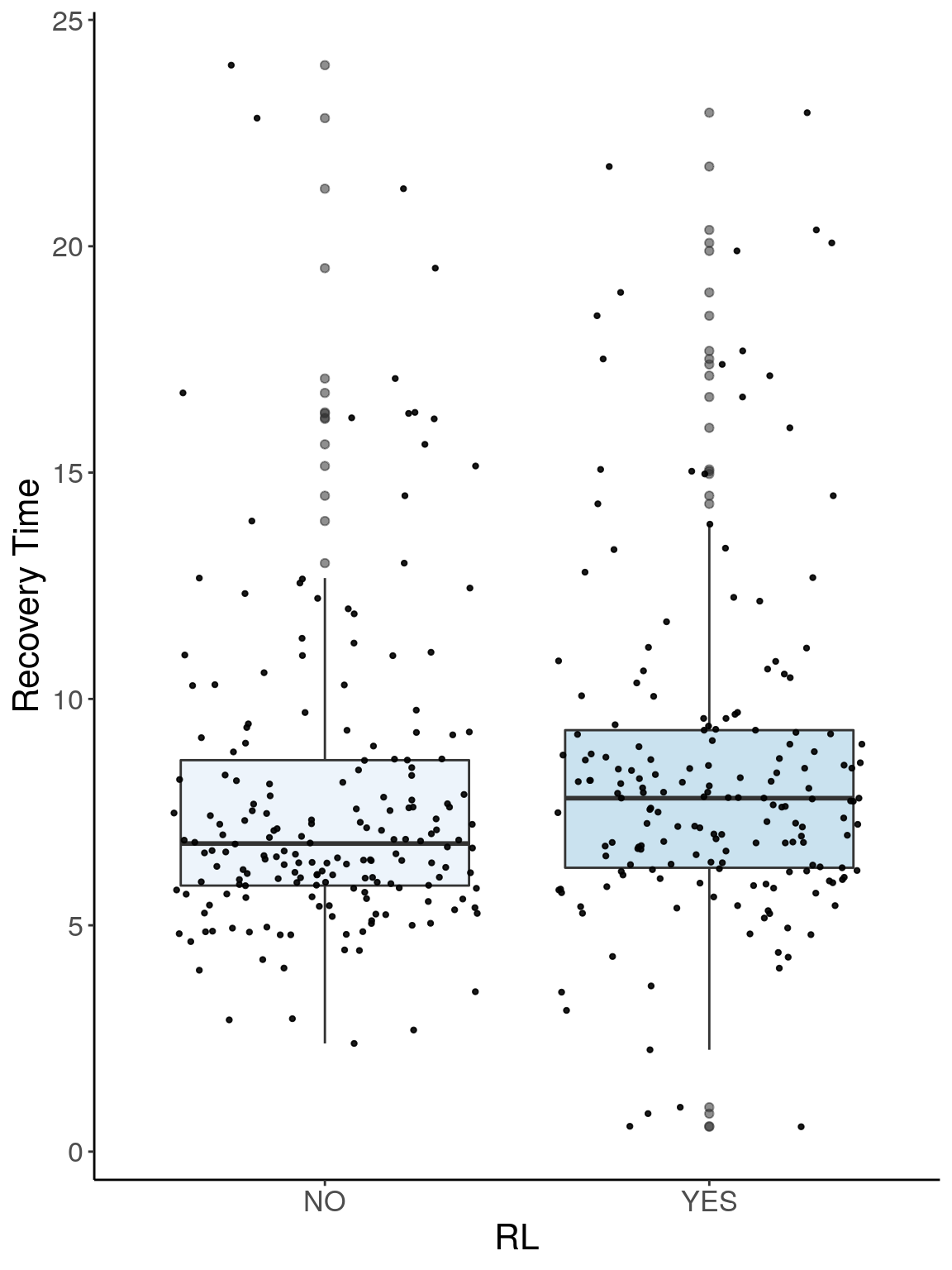
dep <- data[,c(1, 9, 11, 14, 15, 19)]
dep$ngen <- factor(dep$ngen, levels = as.character(c(29:35)))
p3 <- ggplot(dep, aes(x=ngen, y=Min.depression, group = ngen, fill = ngen, alpha = 0.9)) +
geom_boxplot(show.legend = F , outlier.size = 1.5, notchwidth = 0.85) +
geom_jitter(color="black", size=0.8, alpha=0.9) +
scale_fill_brewer(palette="Blues") +
ylab("Min depression") +
xlab("Generation") +
labs(fill = "") +
theme(legend.position = "none",
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
axis.line = element_line(colour = "black"),
text = element_text(size=16),
axis.title=element_text(size=16)) +
guides(shape = guide_legend(override.aes = list(size = 12)))
p3
# Warning: Removed 41 rows containing non-finite values (stat_boxplot).
# Warning: Removed 41 rows containing missing values (geom_point).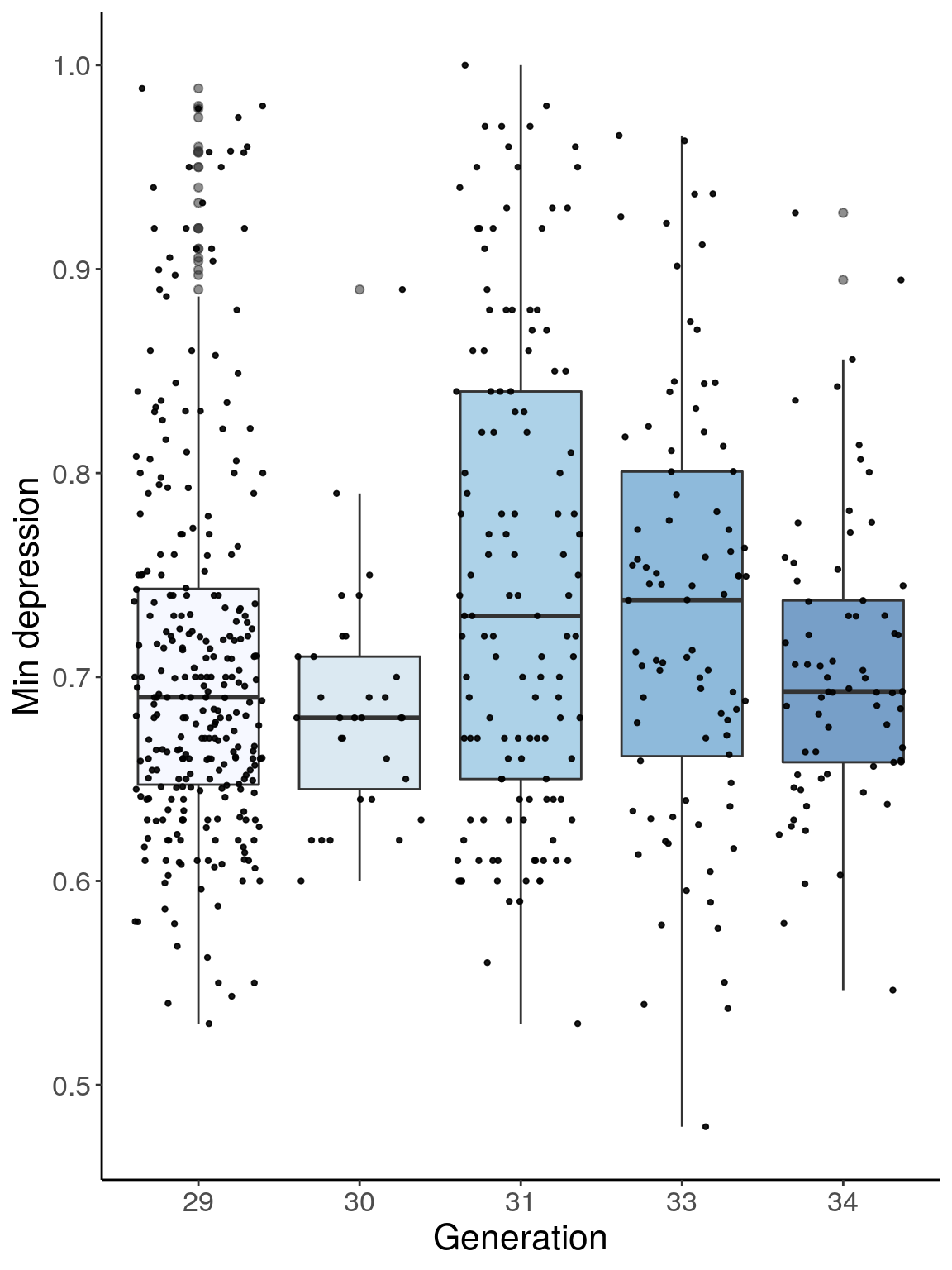
p3.1 <- ggplot(dep, aes(x=sex, y=Min.depression, group = sex, fill = sex, alpha = 0.9)) +
geom_boxplot(show.legend = F , outlier.size = 1.5, notchwidth = 0.85) +
geom_jitter(color="black", size=0.8, alpha=0.9) +
scale_fill_brewer(palette="Blues") +
ylab("Min depression") +
xlab("sex") +
labs(fill = "") +
theme(legend.position = "none",
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
axis.line = element_line(colour = "black"),
text = element_text(size=16),
axis.title=element_text(size=16)) +
guides(shape = guide_legend(override.aes = list(size = 12)))
p3.1
# Warning: Removed 41 rows containing non-finite values (stat_boxplot).
# Warning: Removed 41 rows containing missing values (geom_point).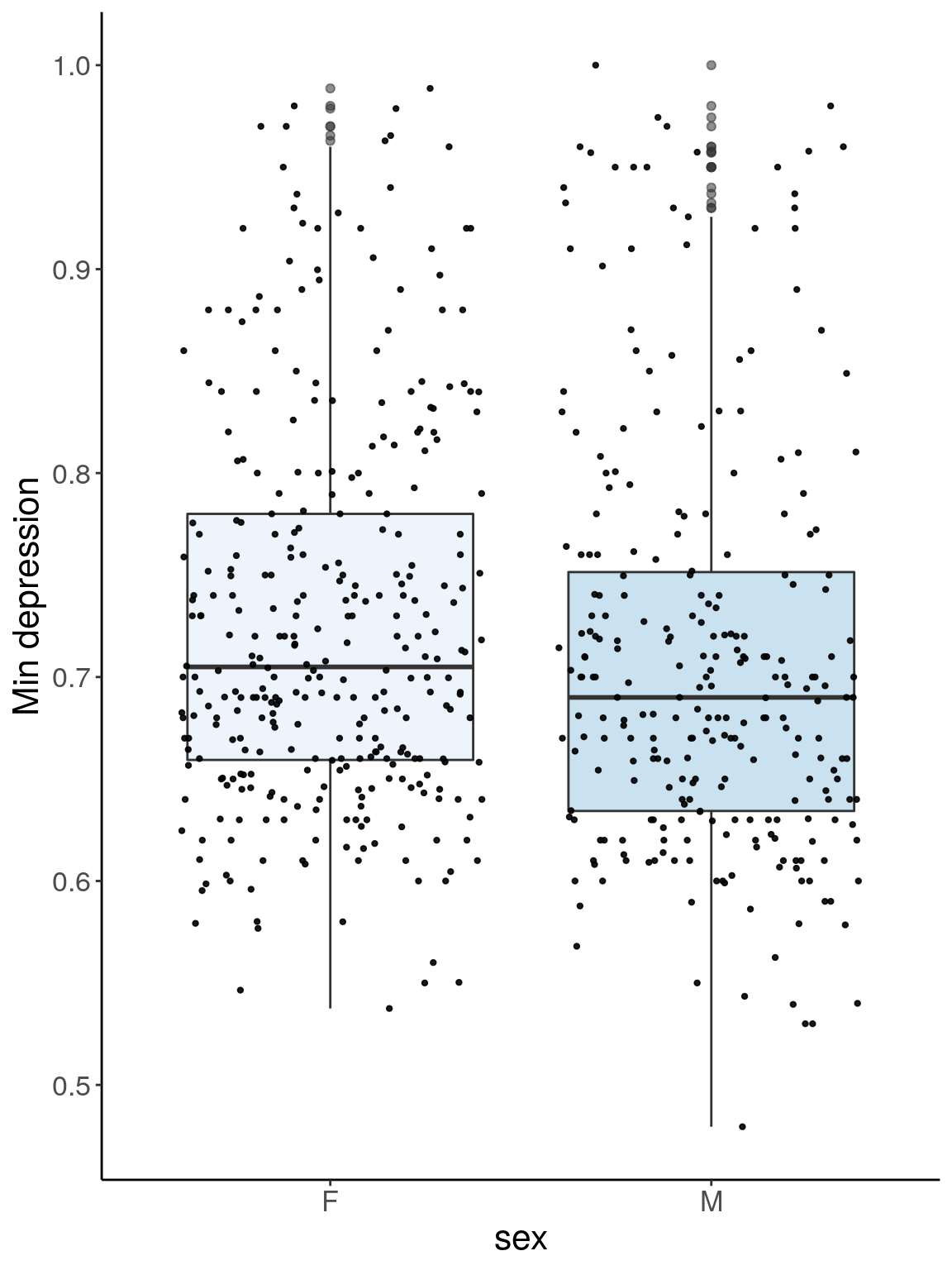
p3.2 <- ggplot(dep, aes(x=RL, y=Min.depression, group = RL, fill = RL, alpha = 0.9)) +
geom_boxplot(show.legend = F , outlier.size = 1.5, notchwidth = 0.85) +
geom_jitter(color="black", size=0.8, alpha=0.9) +
scale_fill_brewer(palette="Blues") +
ylab("Min depression") +
xlab("RL") +
labs(fill = "") +
theme(legend.position = "none",
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
axis.line = element_line(colour = "black"),
text = element_text(size=16),
axis.title=element_text(size=16)) +
guides(shape = guide_legend(override.aes = list(size = 12)))
p3.2
# Warning: Removed 41 rows containing non-finite values (stat_boxplot).
# Warning: Removed 41 rows containing missing values (geom_point).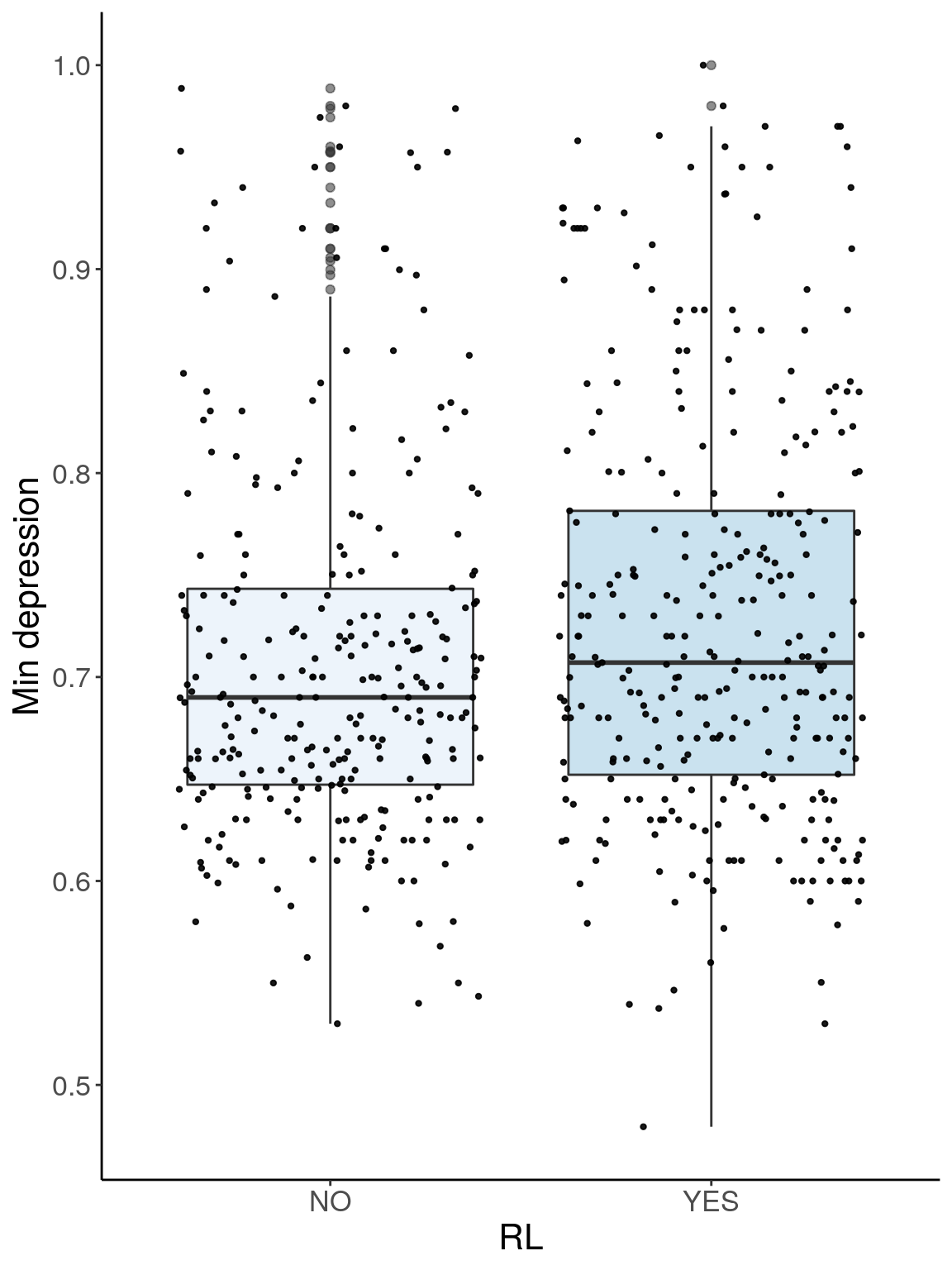
Plot qtl mapping on 69k
Plot by using its output qtl.morphine.69k.out.combined.RData
load("output/qtl.morphine.69k.out.combined.RData")
#pmap and gmap
load("data/69k_grid_pgmap.RData")
#genome-wide plot
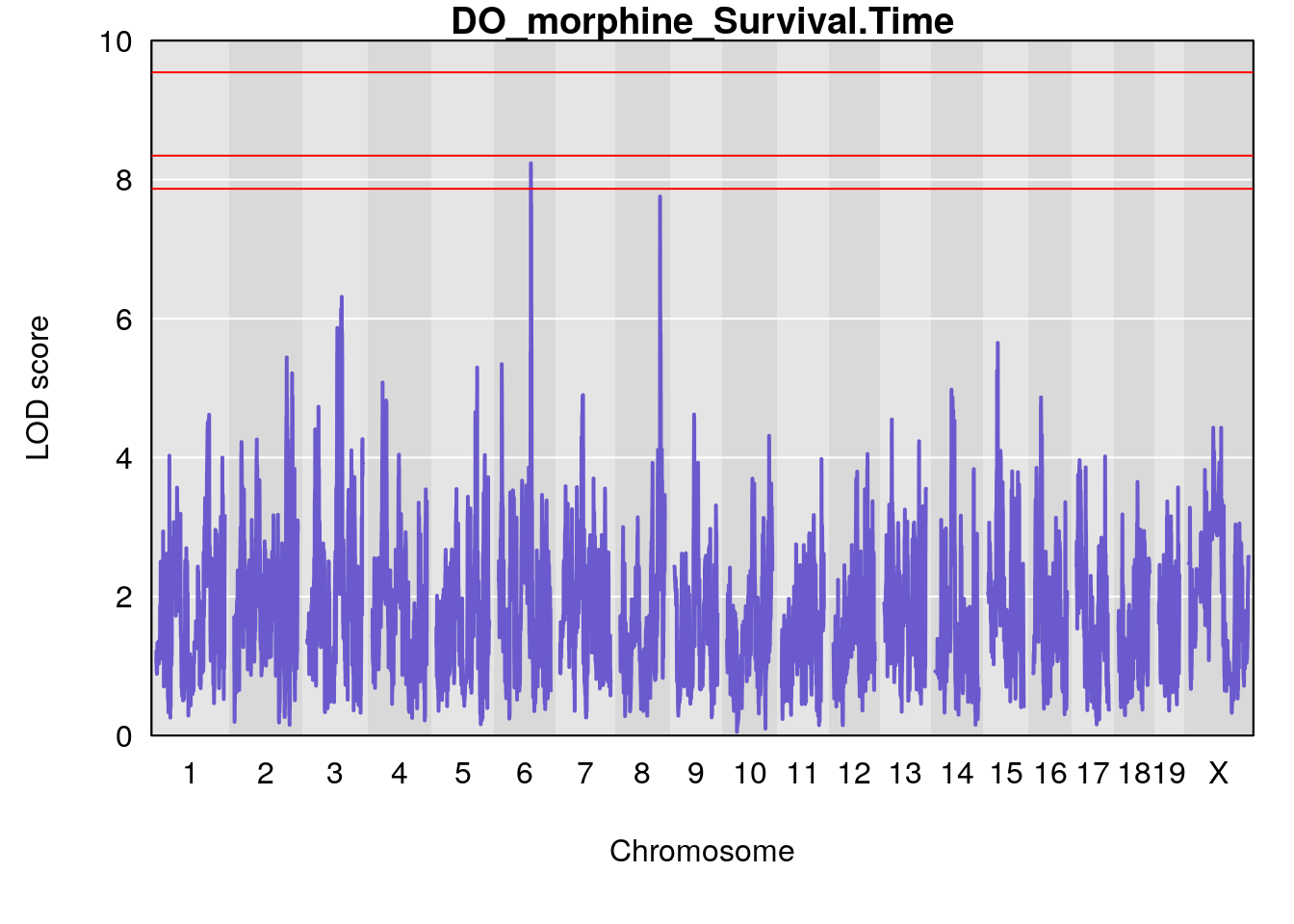
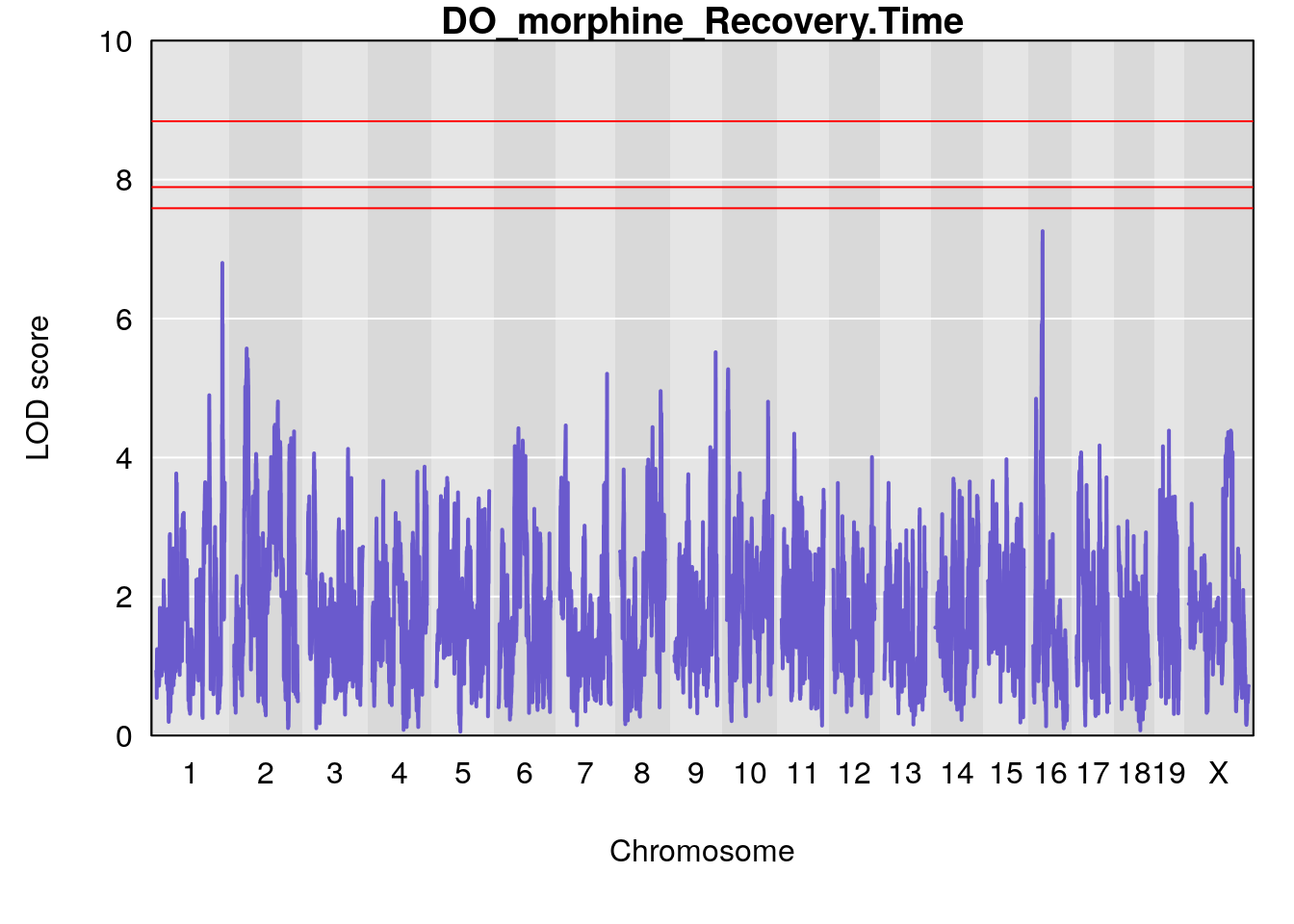
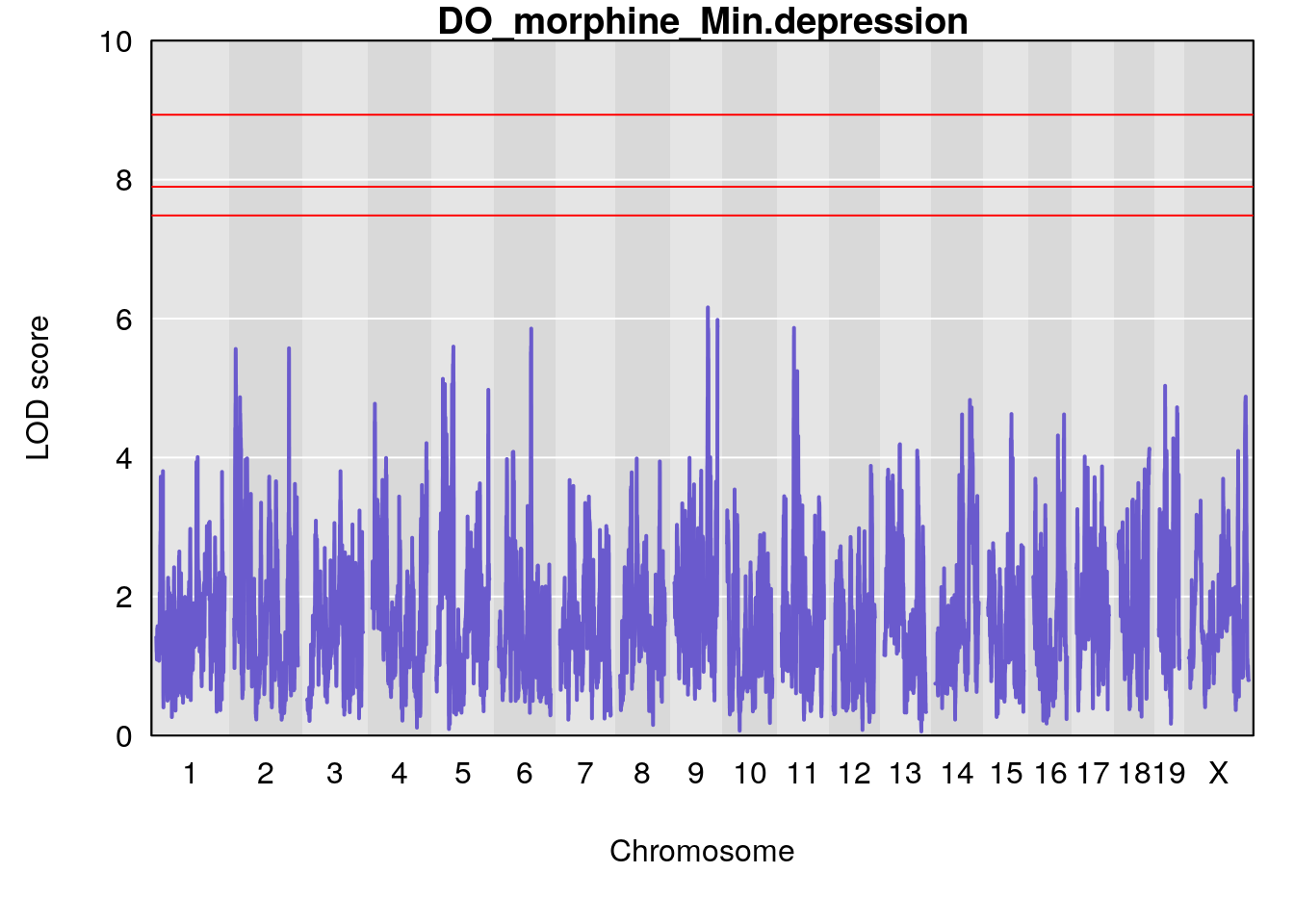
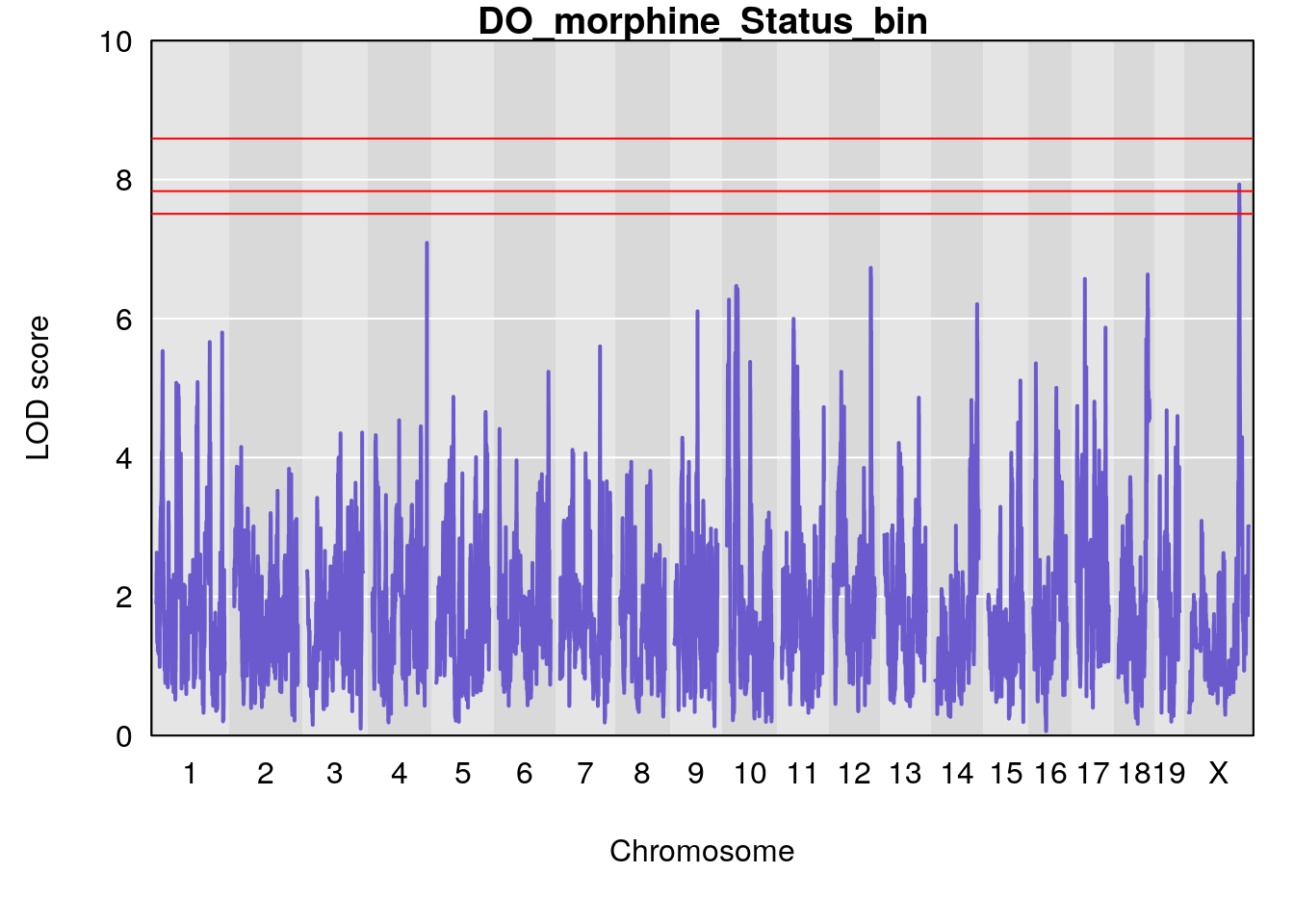
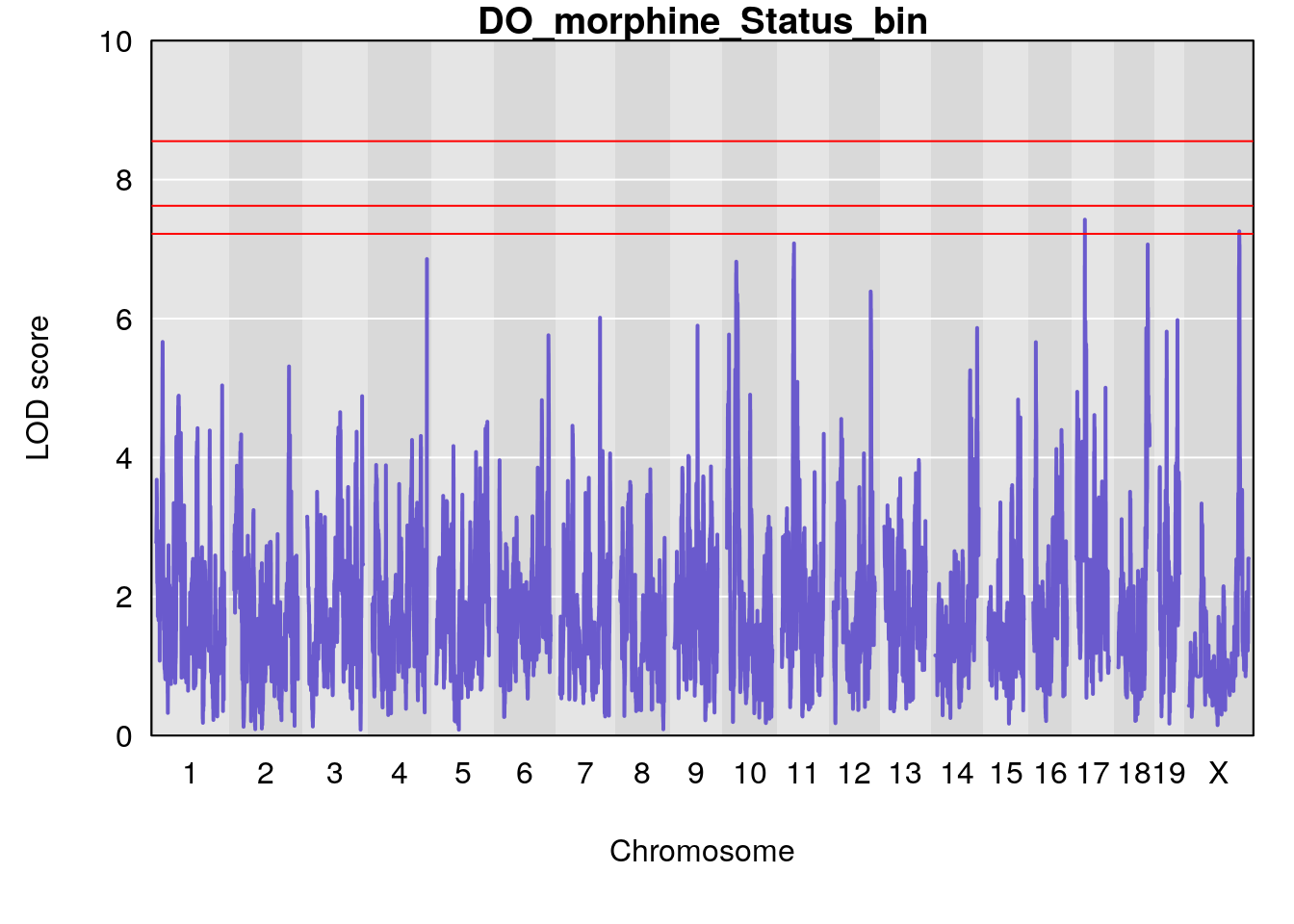
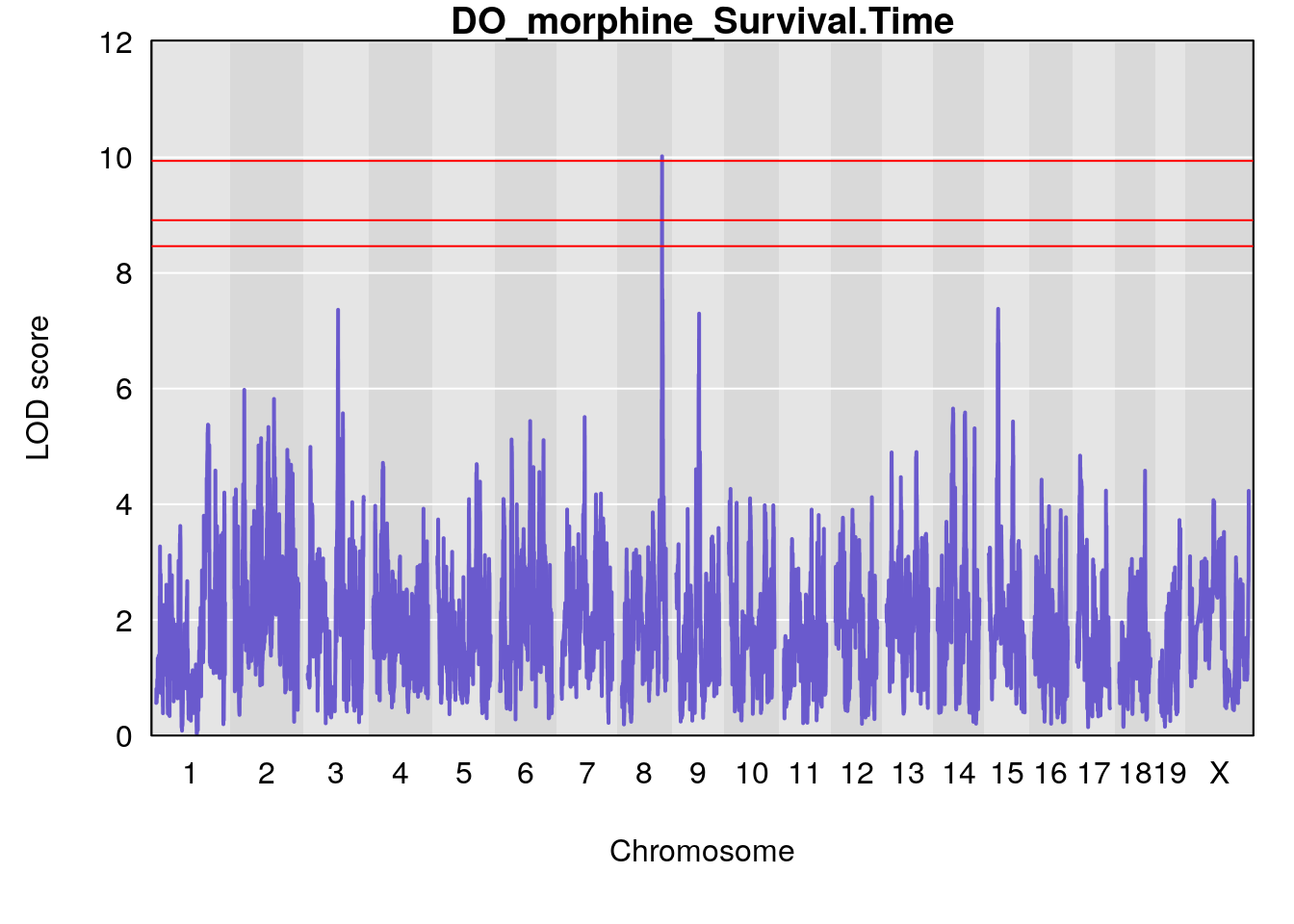
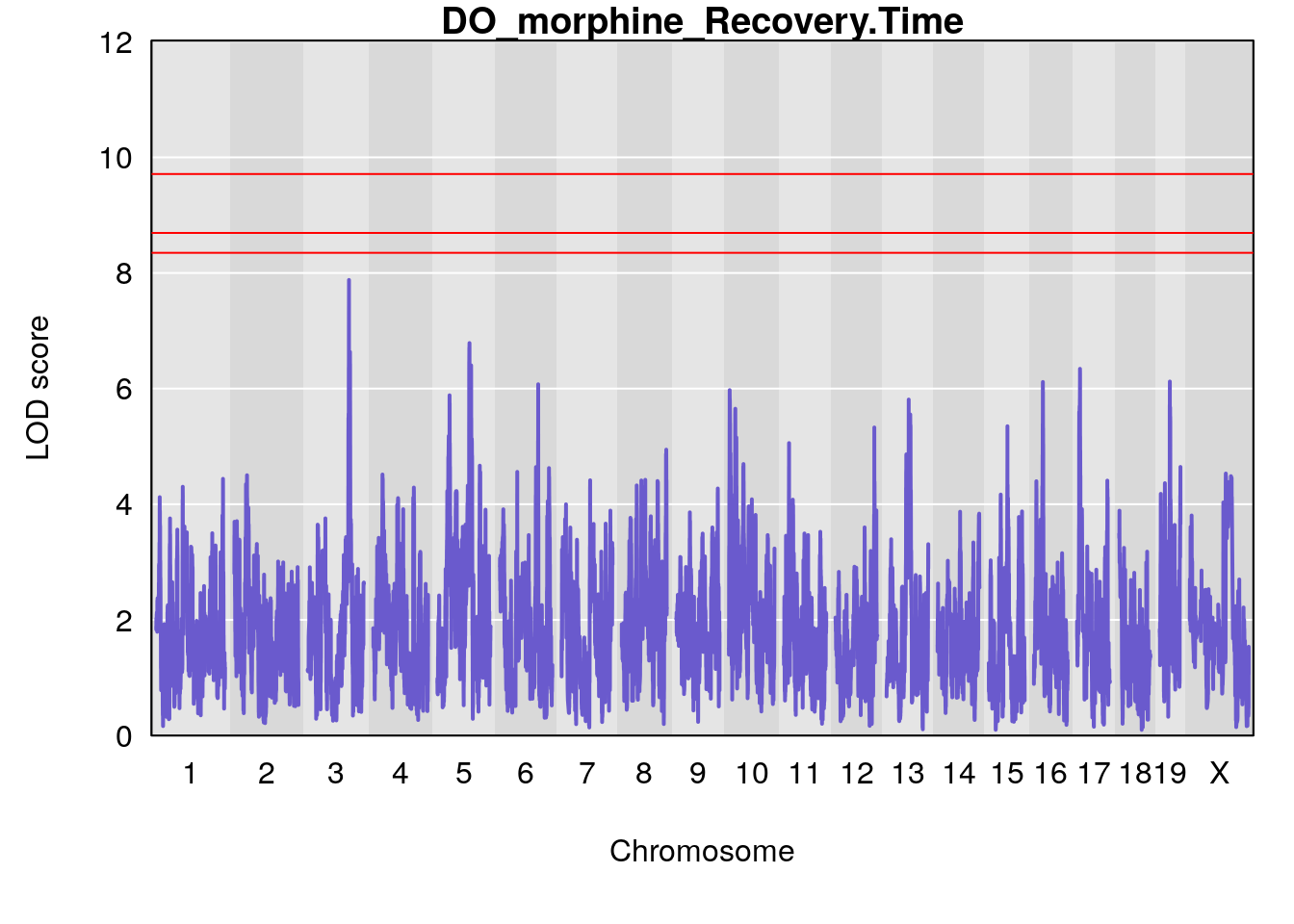
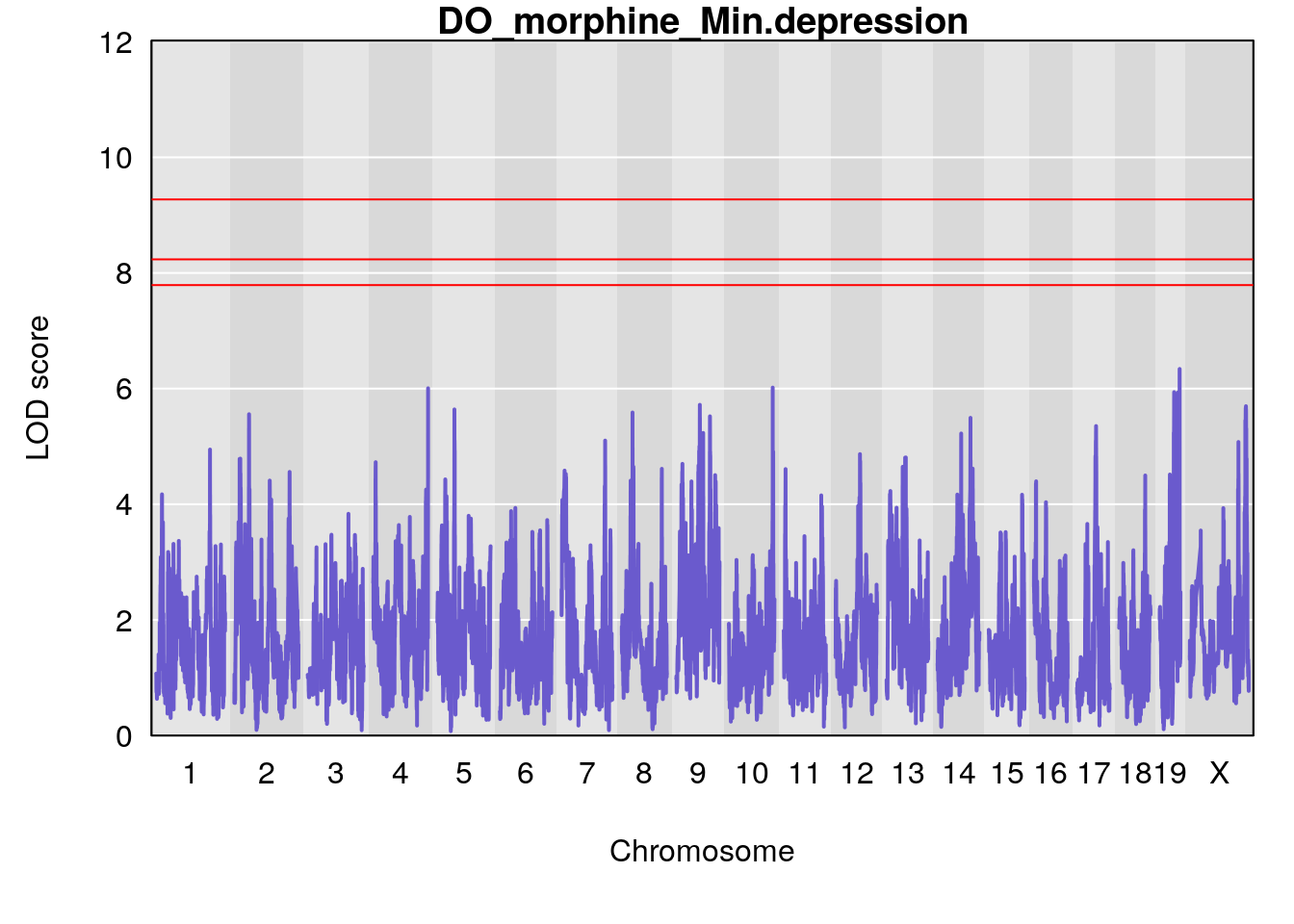
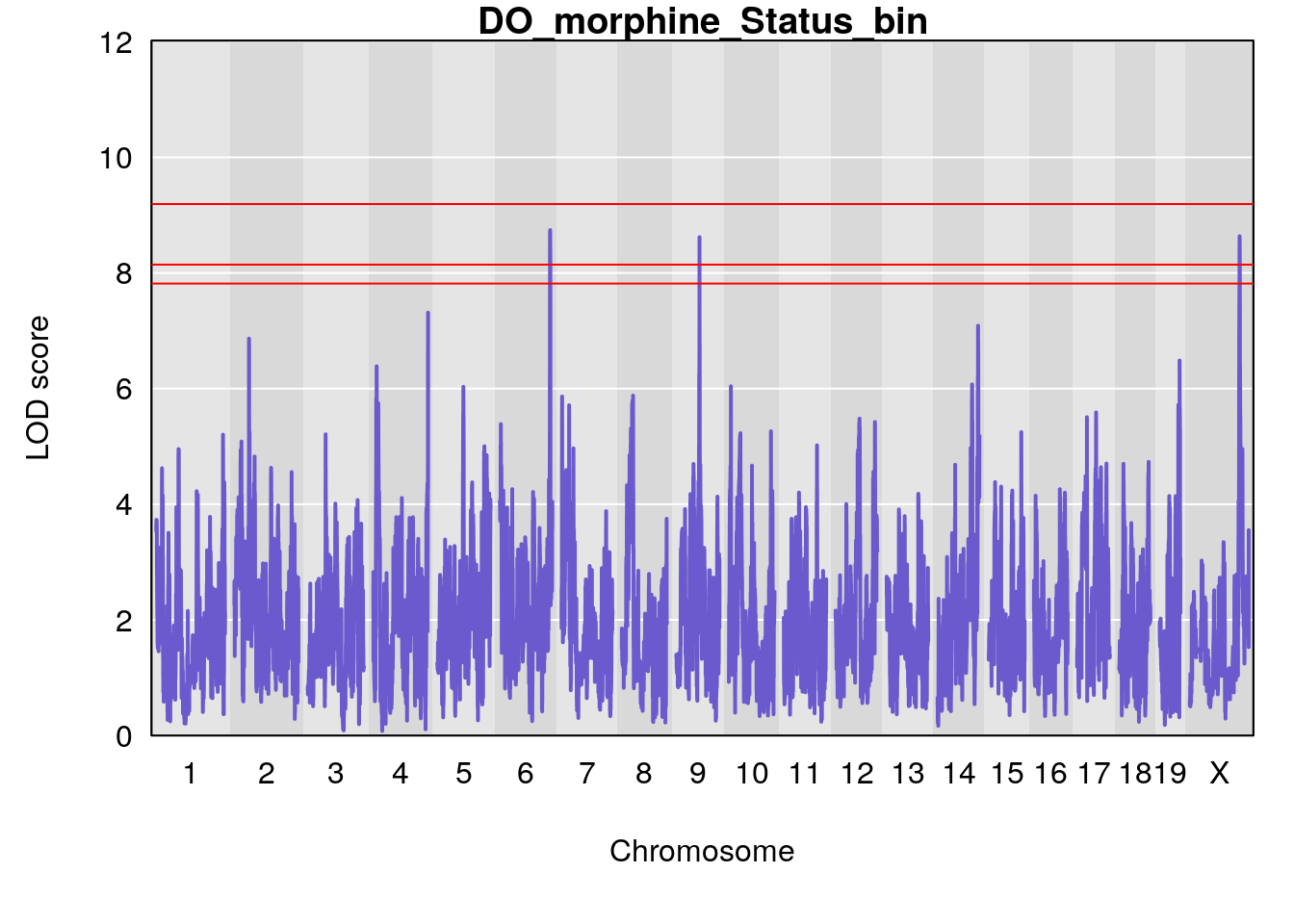
for(i in names(qtl.morphine.out)){
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.morphine.out[[i]]) # overall maximum LOD score
plot(qtl.morphine.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, 10))
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("DO_morphine_",i))
}
#save genome-wide plot
for(i in names(qtl.morphine.out)){
png(file = paste0("output/DO_morphine_secondbatch_", i, ".png"), width = 16, height =8, res=300, units = "in")
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.morphine.out[[i]]) # overall maximum LOD score
plot(qtl.morphine.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, 10))
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("DO_morphine_",i))
dev.off()
}
#peaks coeff plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
print(peaks)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
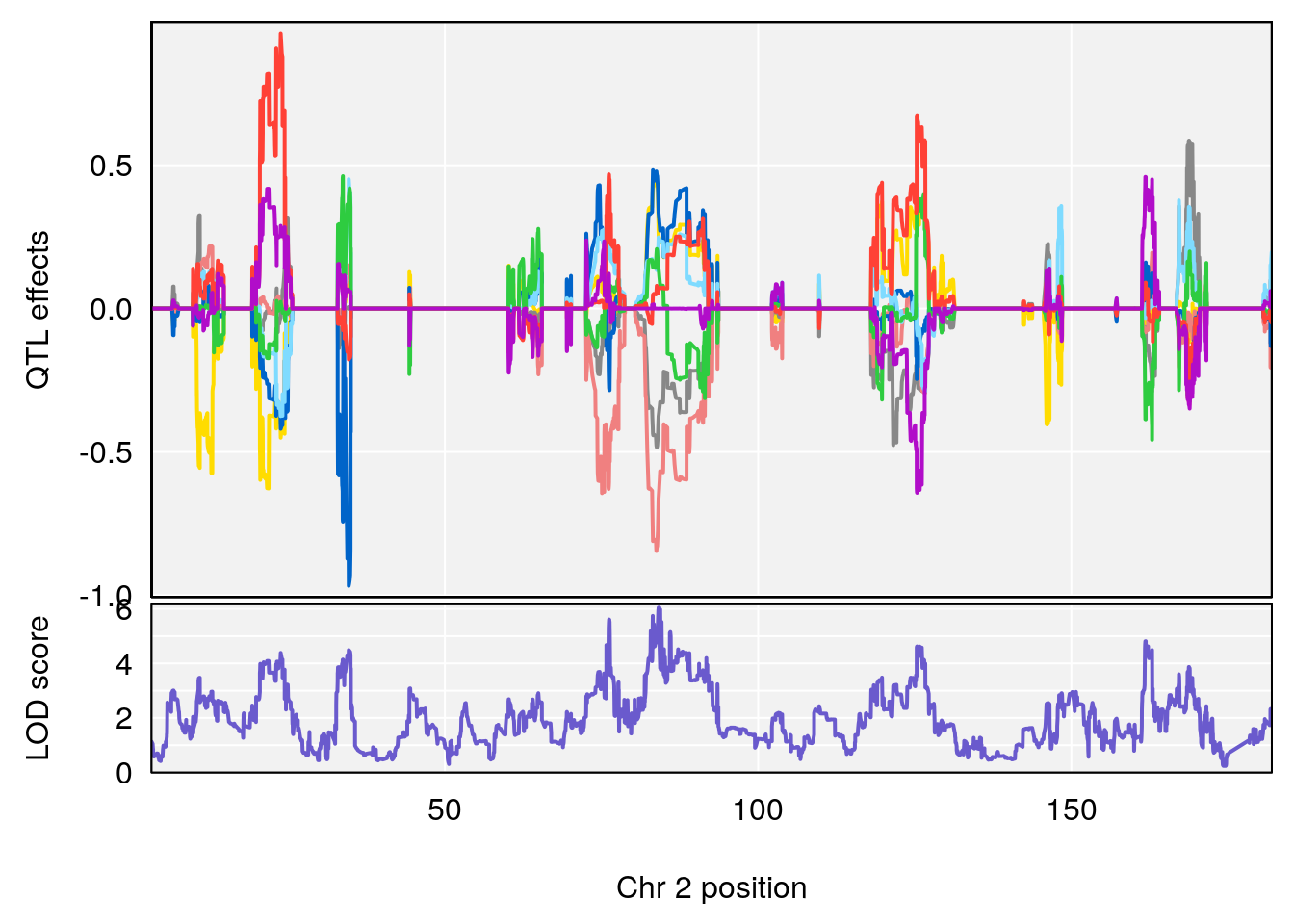
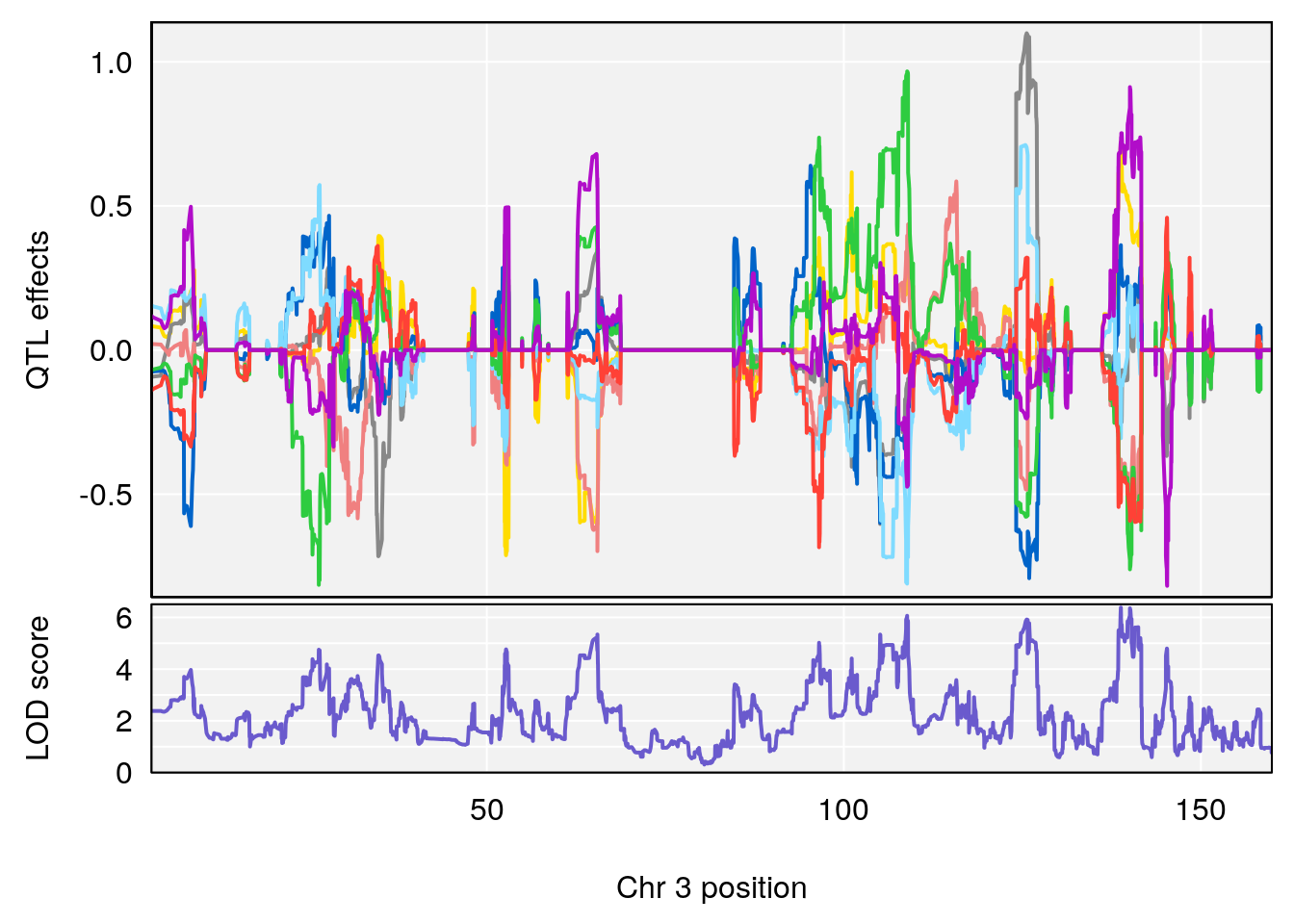
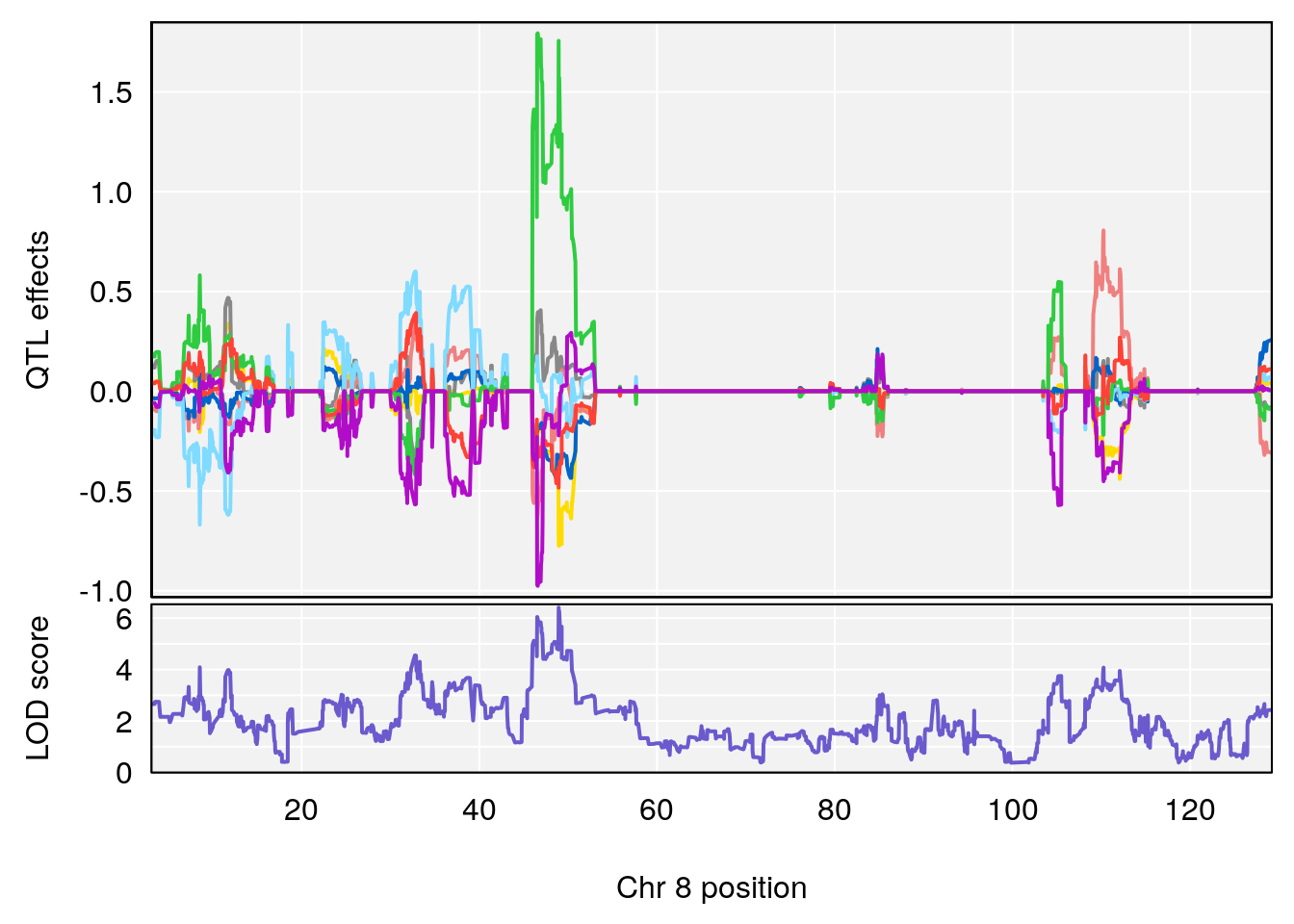
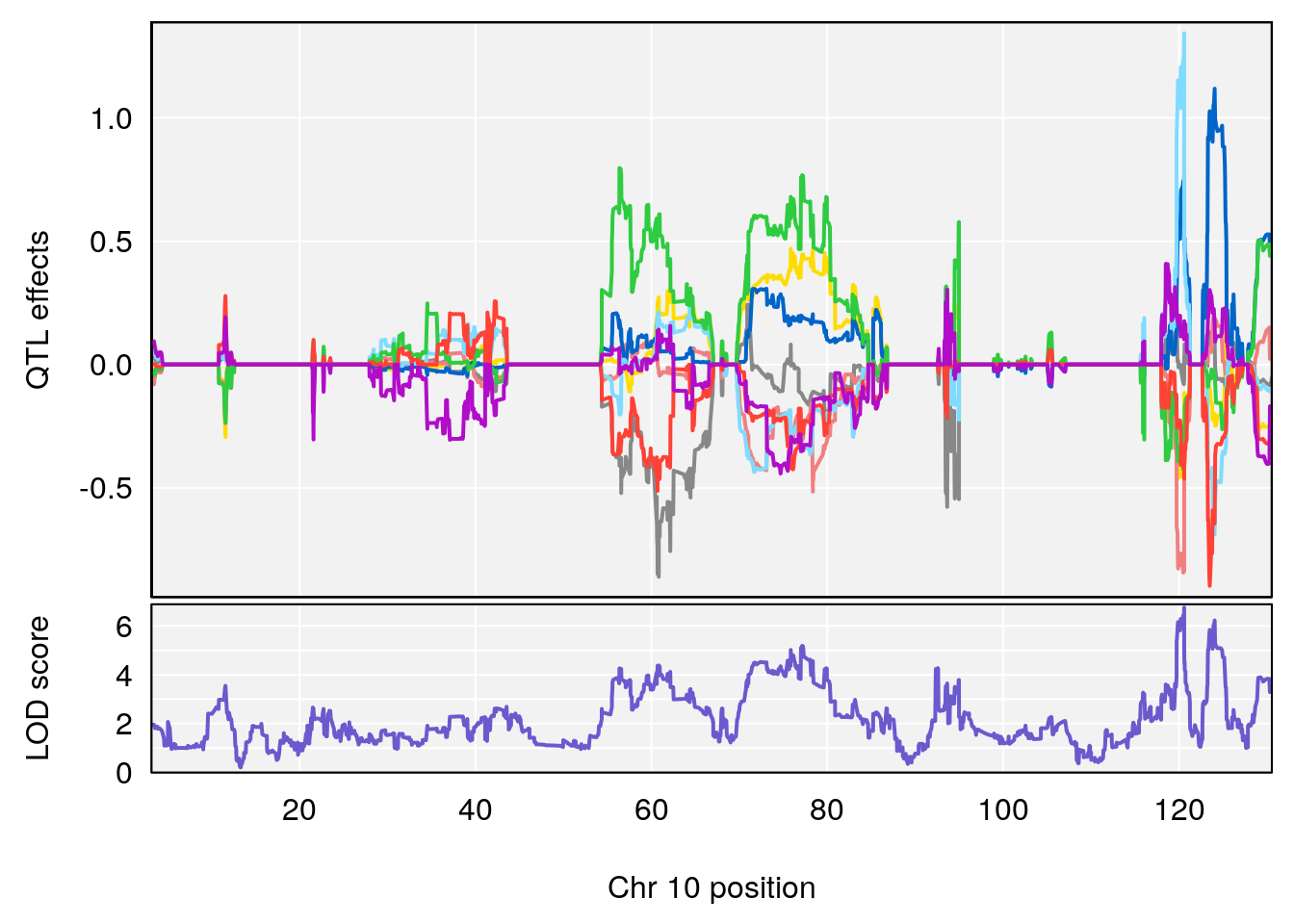
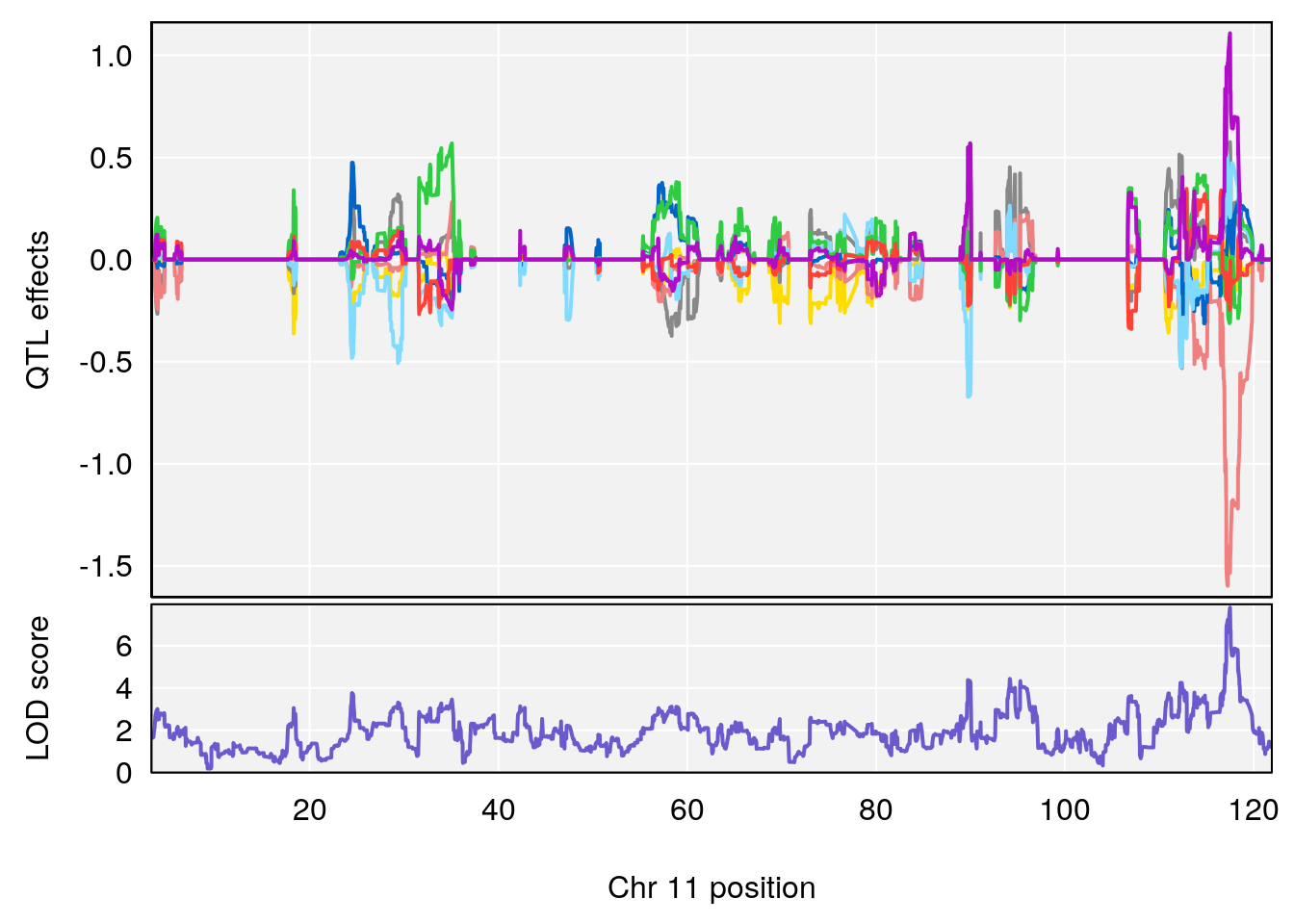
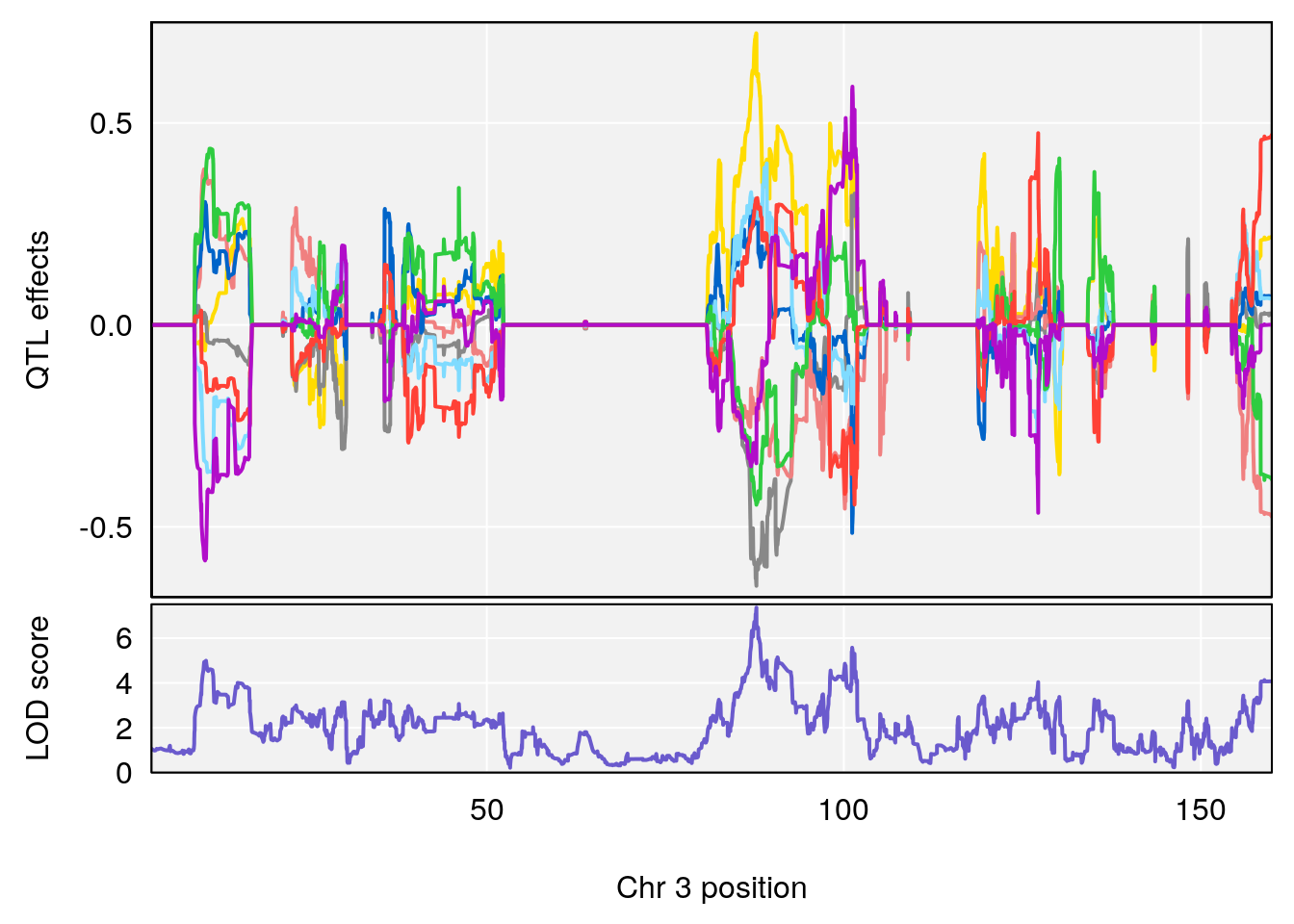
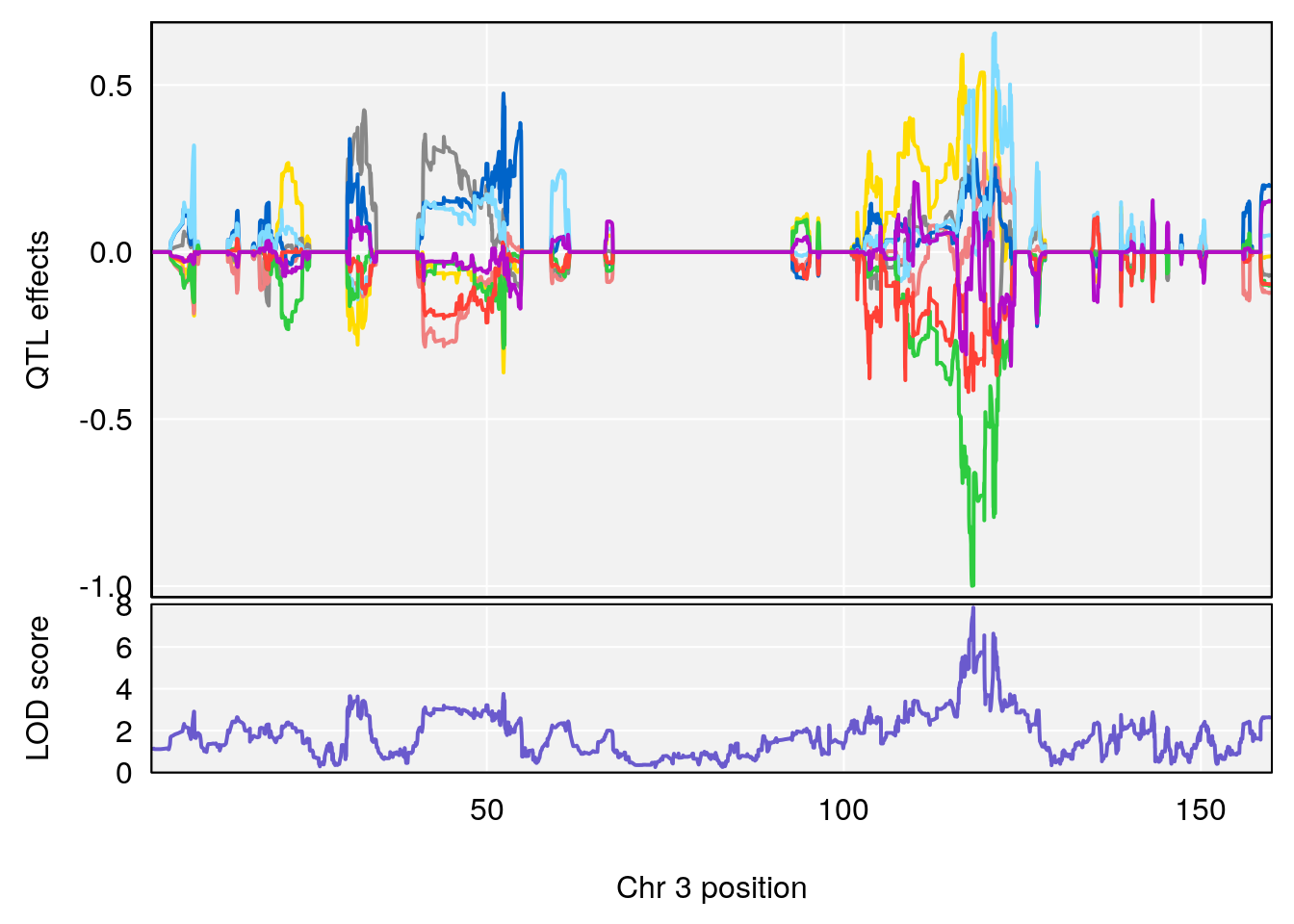
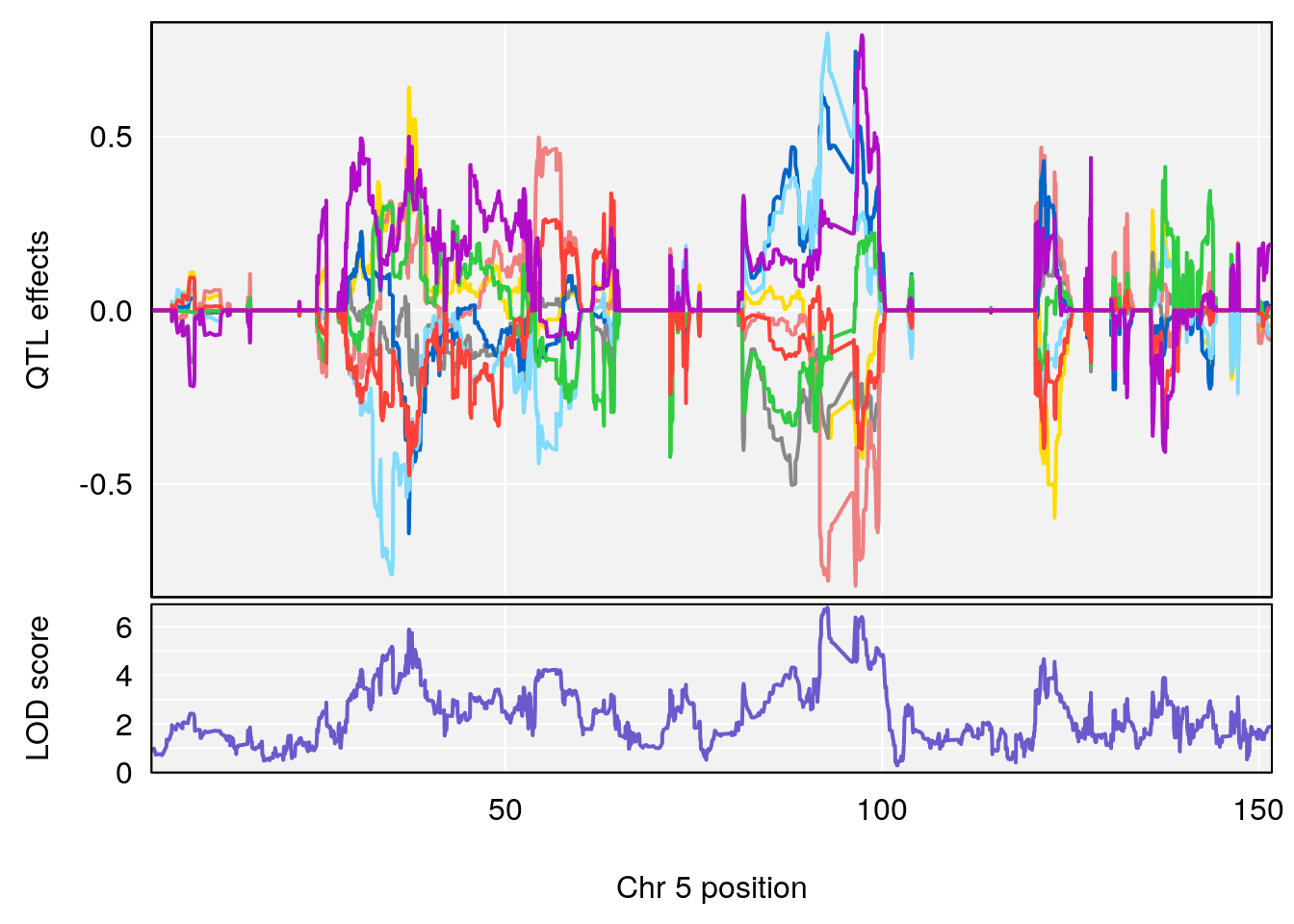
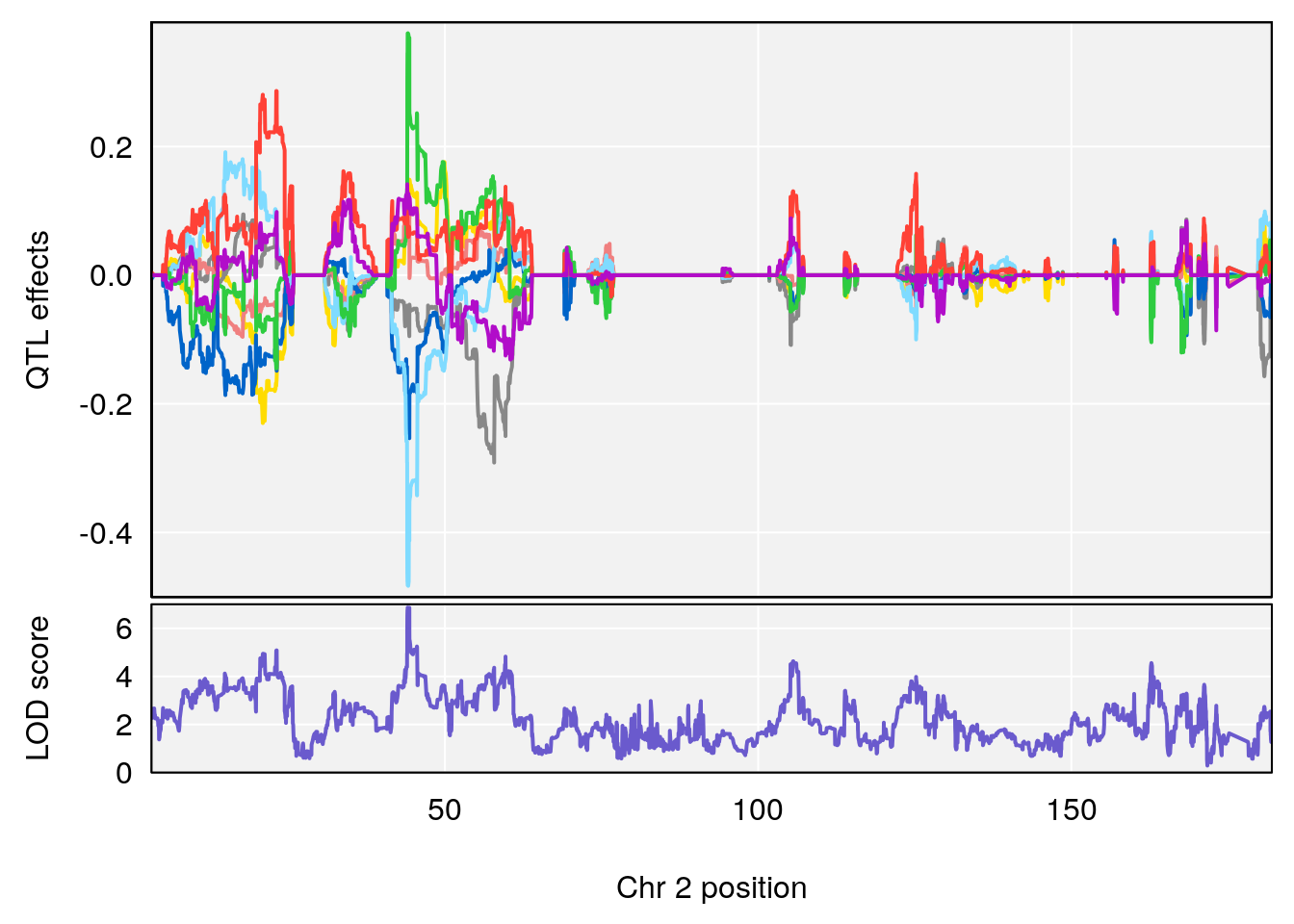
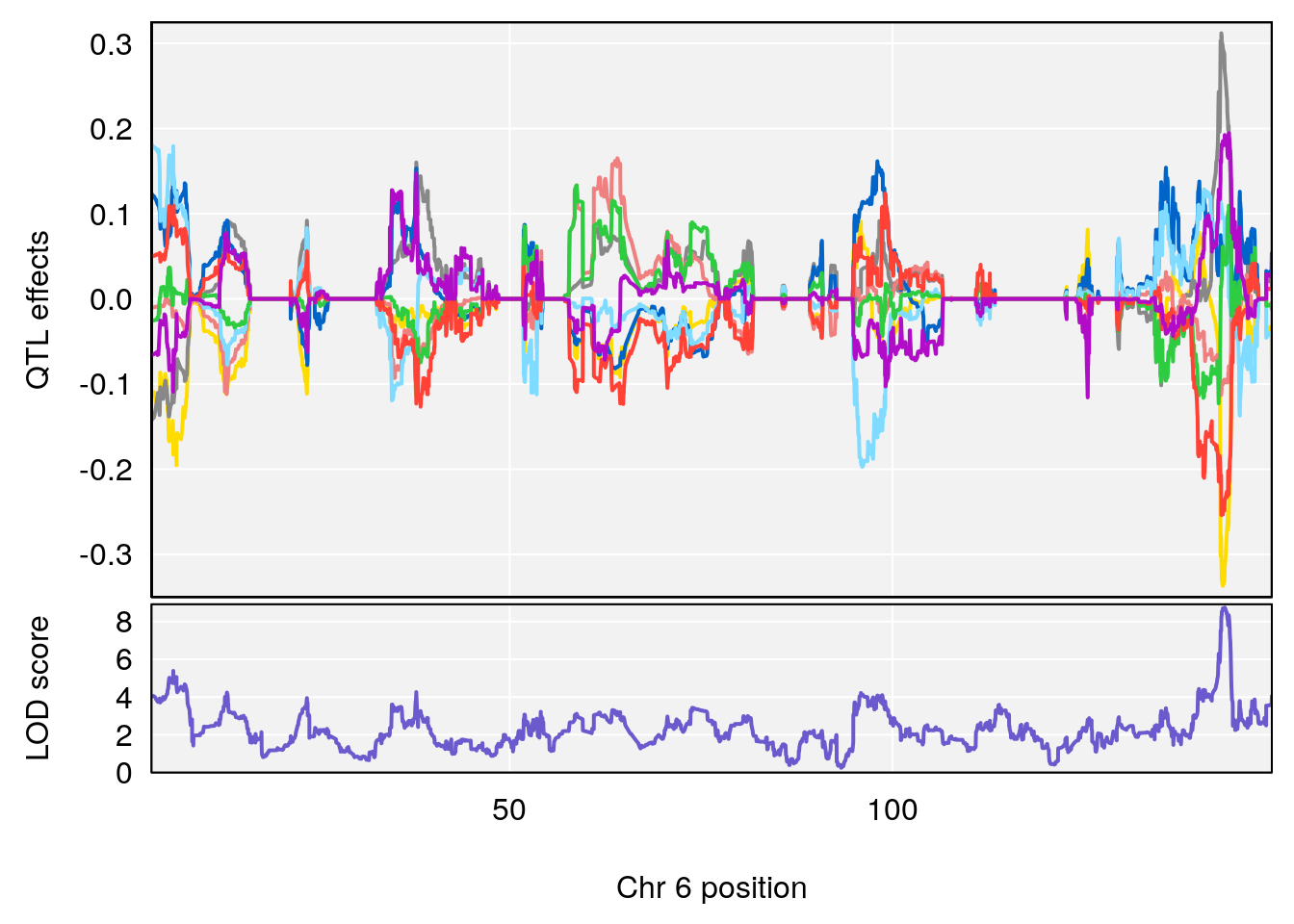
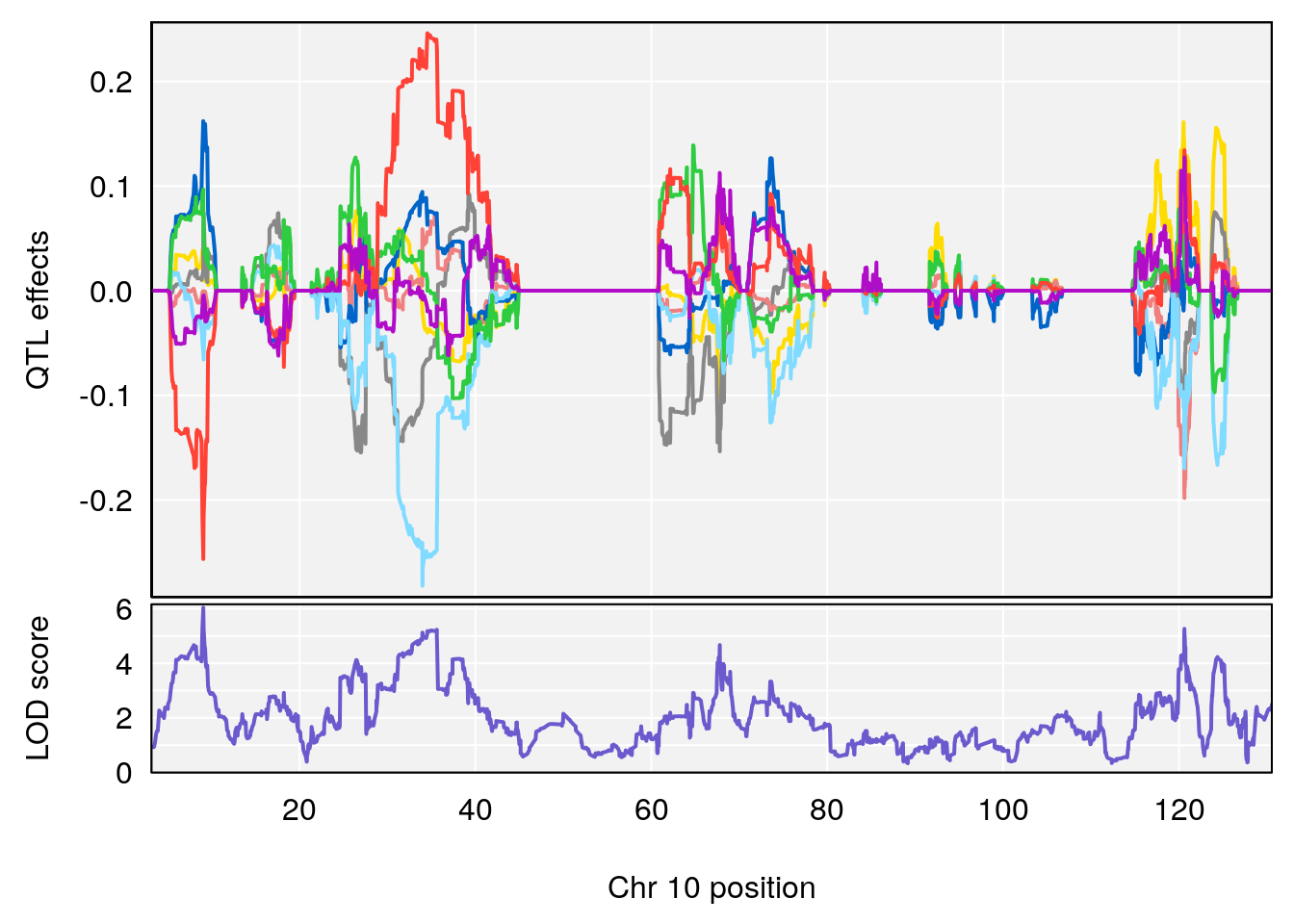
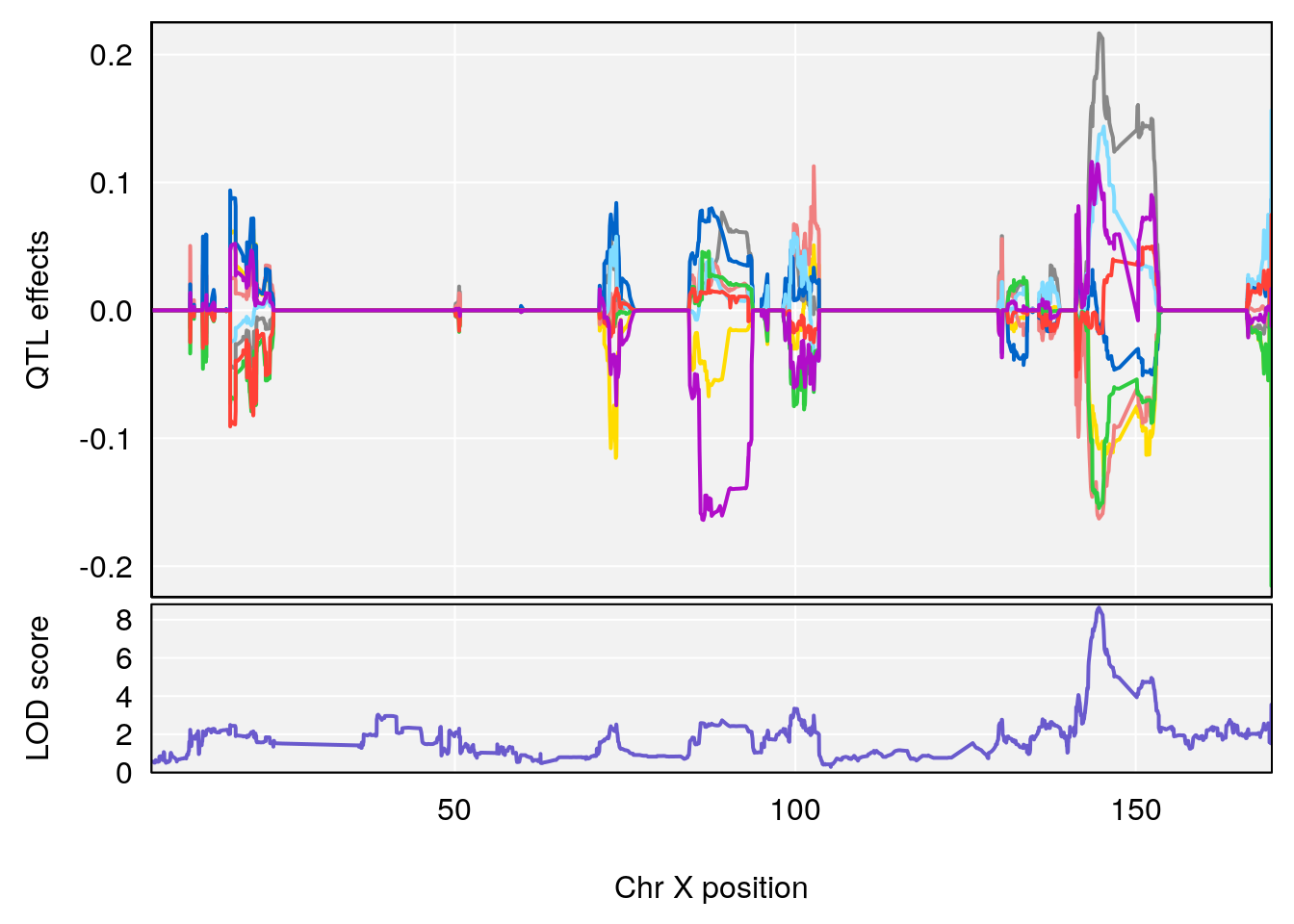
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
}
}
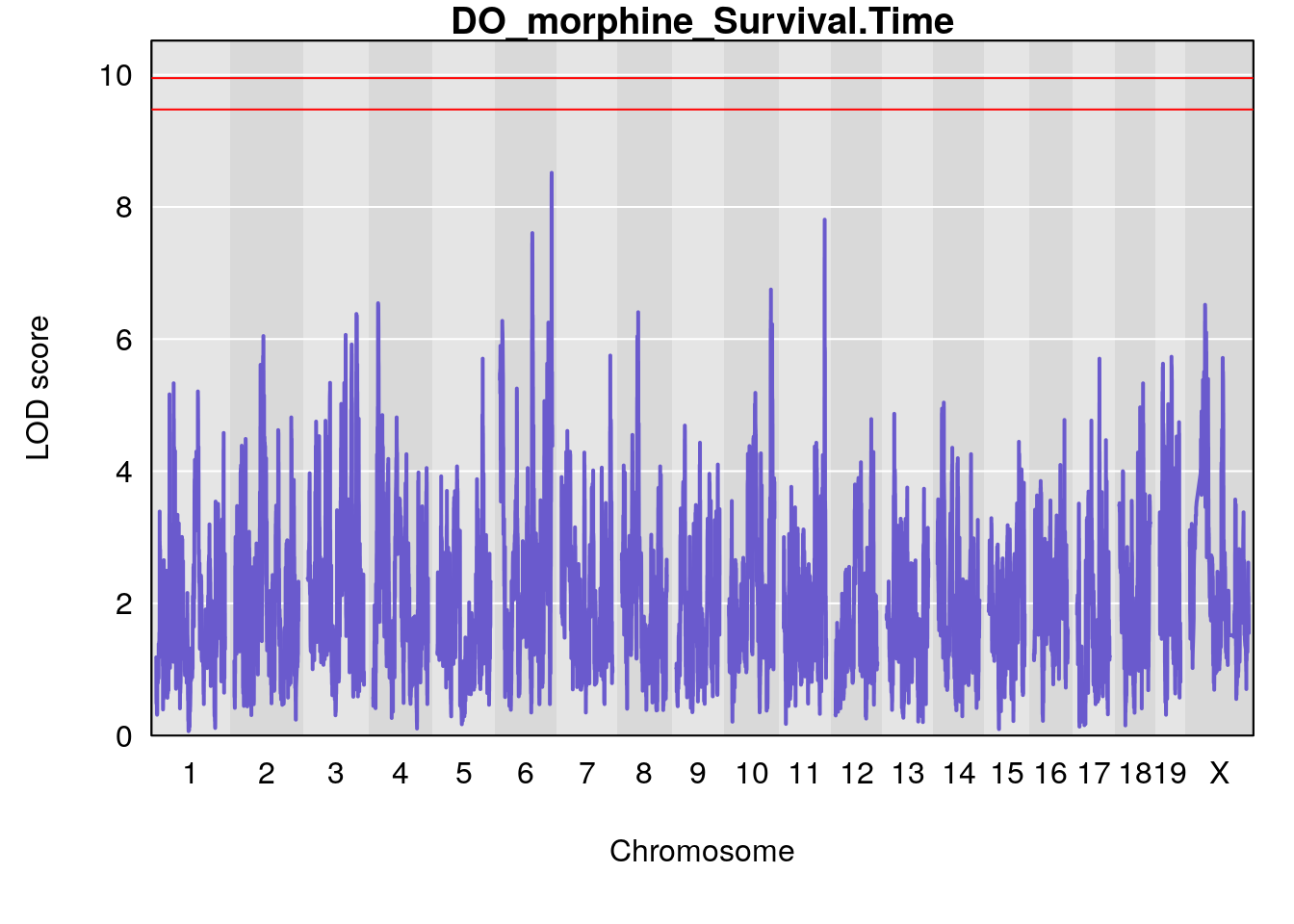
# [1] "Survival.Time"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 3 100.22014 6.314064 86.91920 101.53654
# 2 1 pheno1 6 93.76935 8.235696 93.60032 94.88262
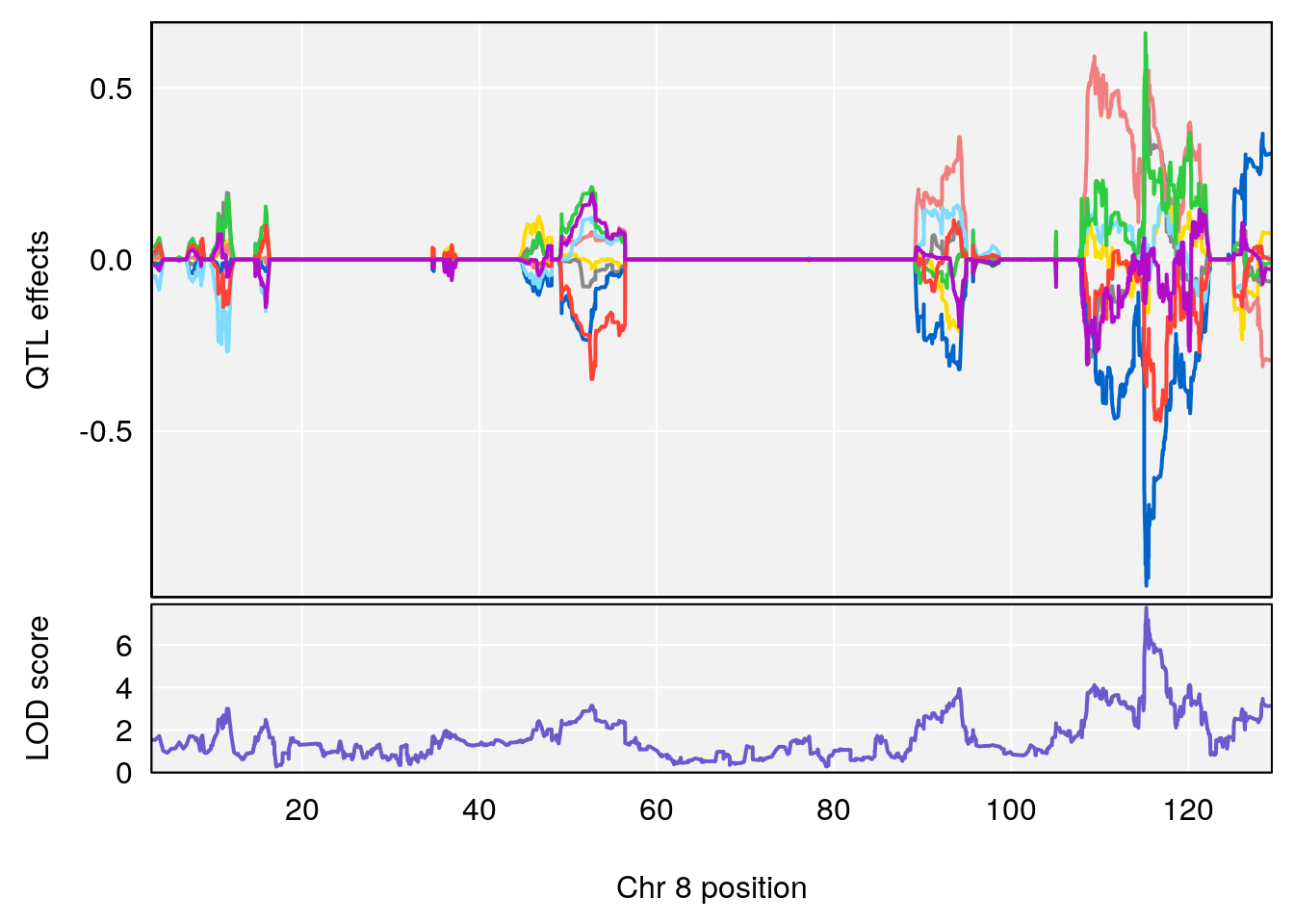
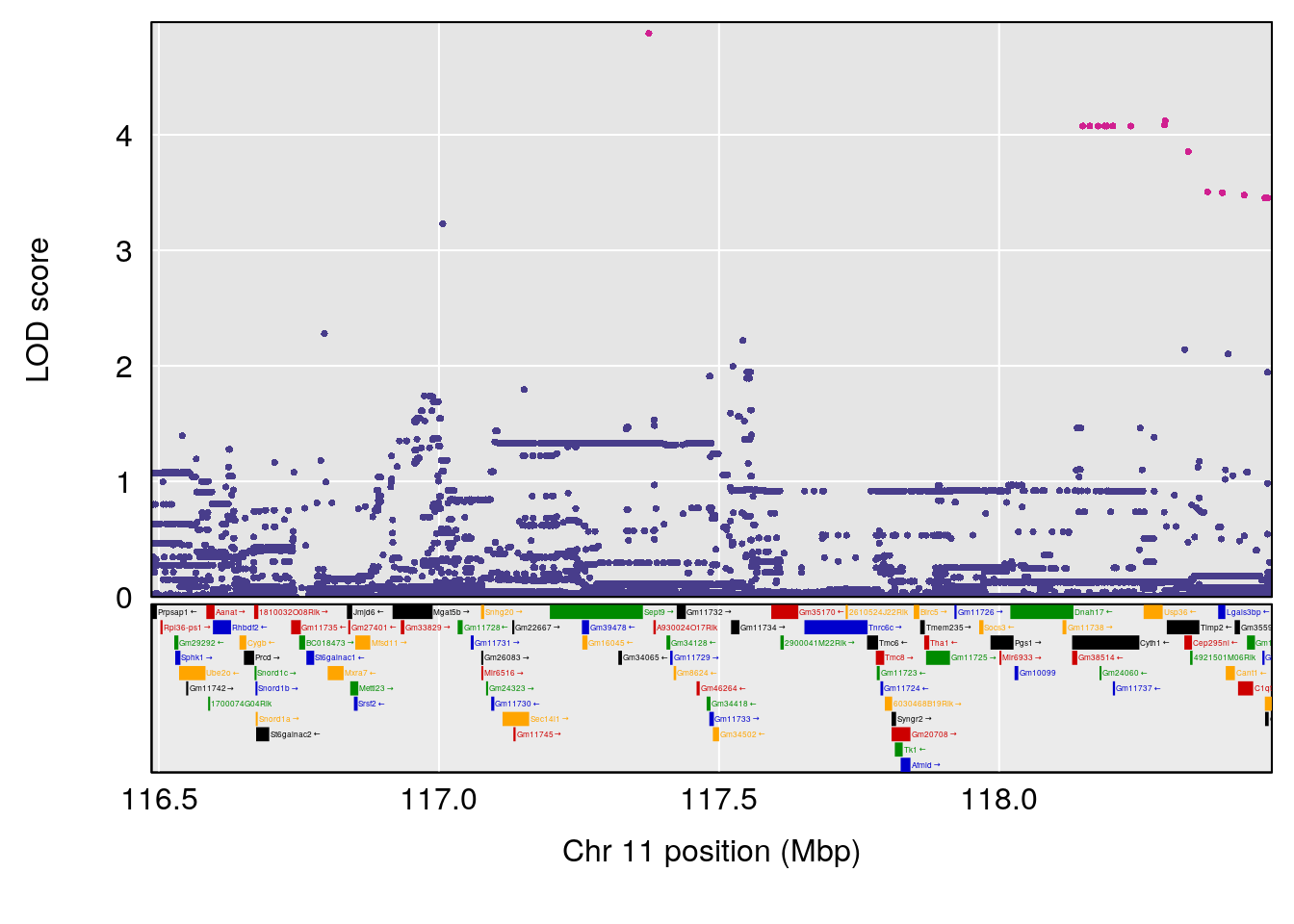
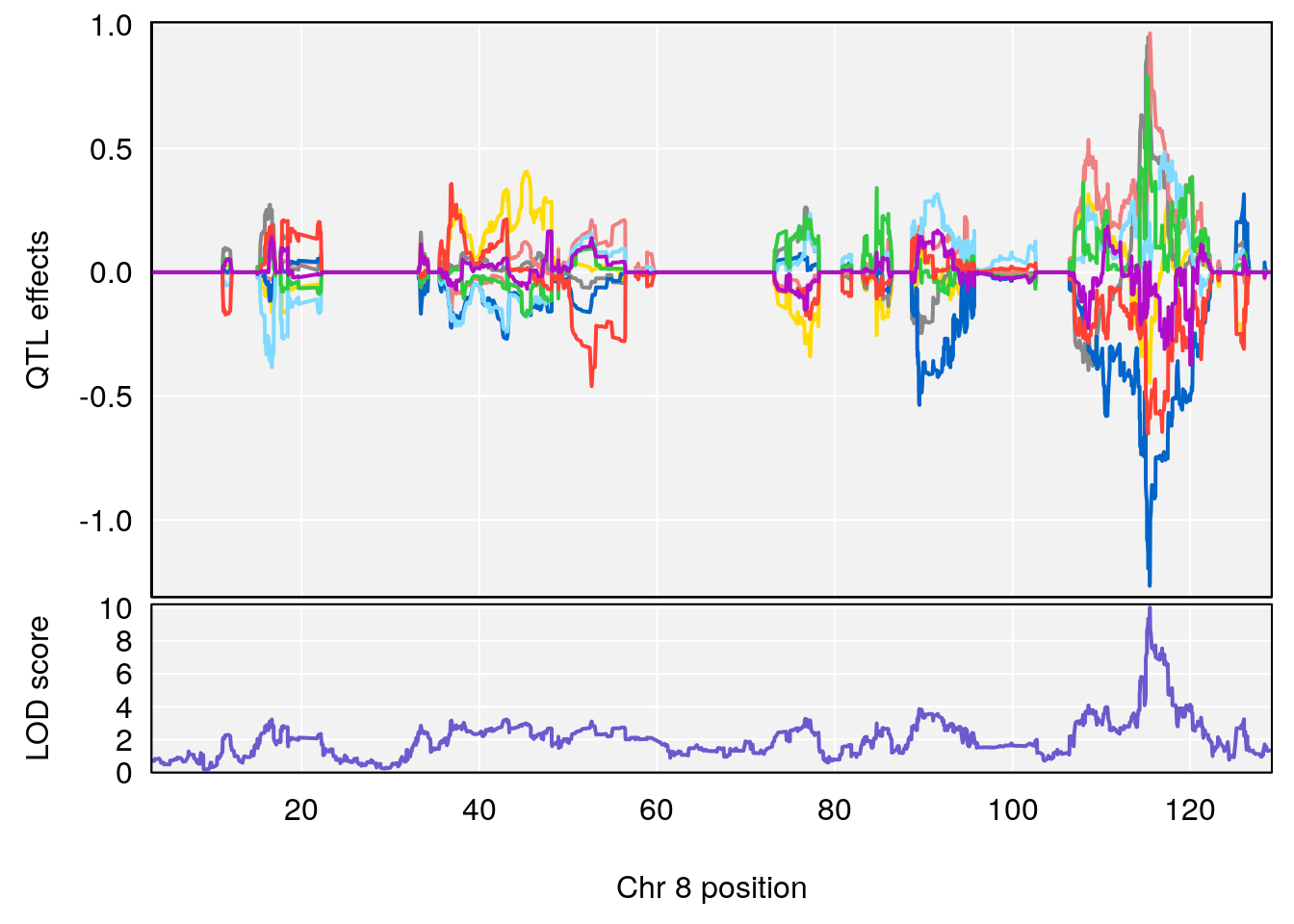
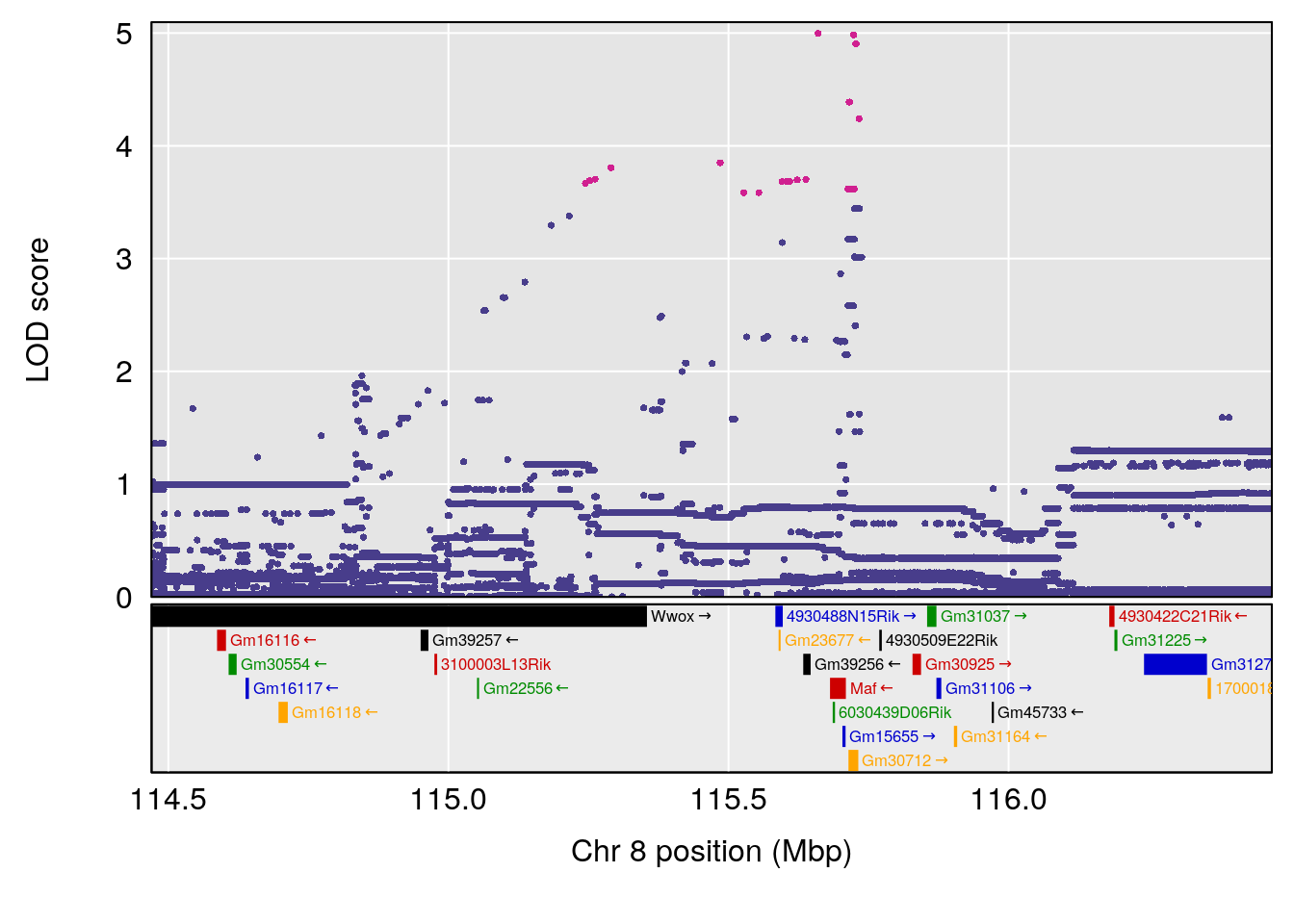
# 3 1 pheno1 8 115.24260 7.756984 115.11279 115.81949
# [1] 1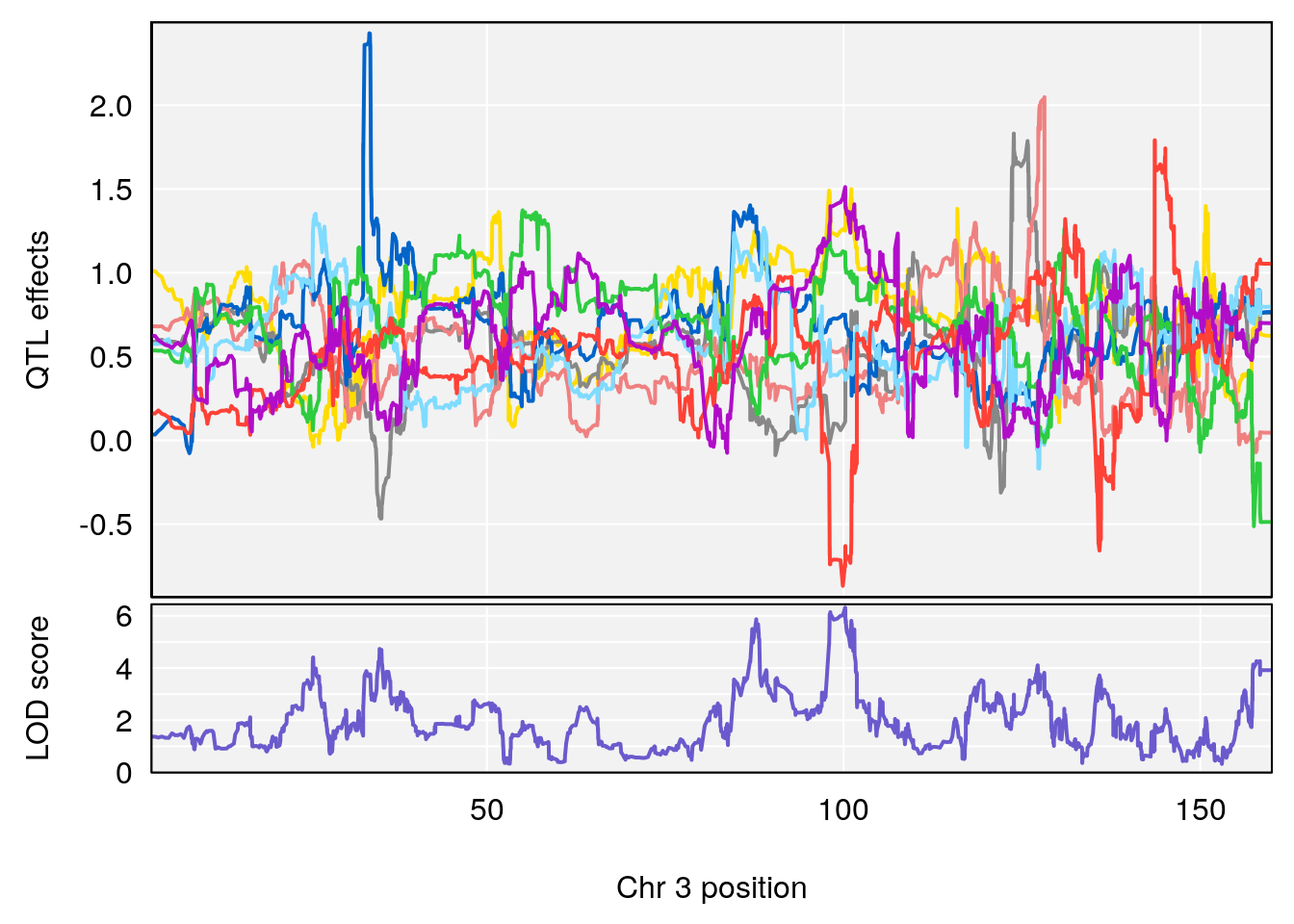
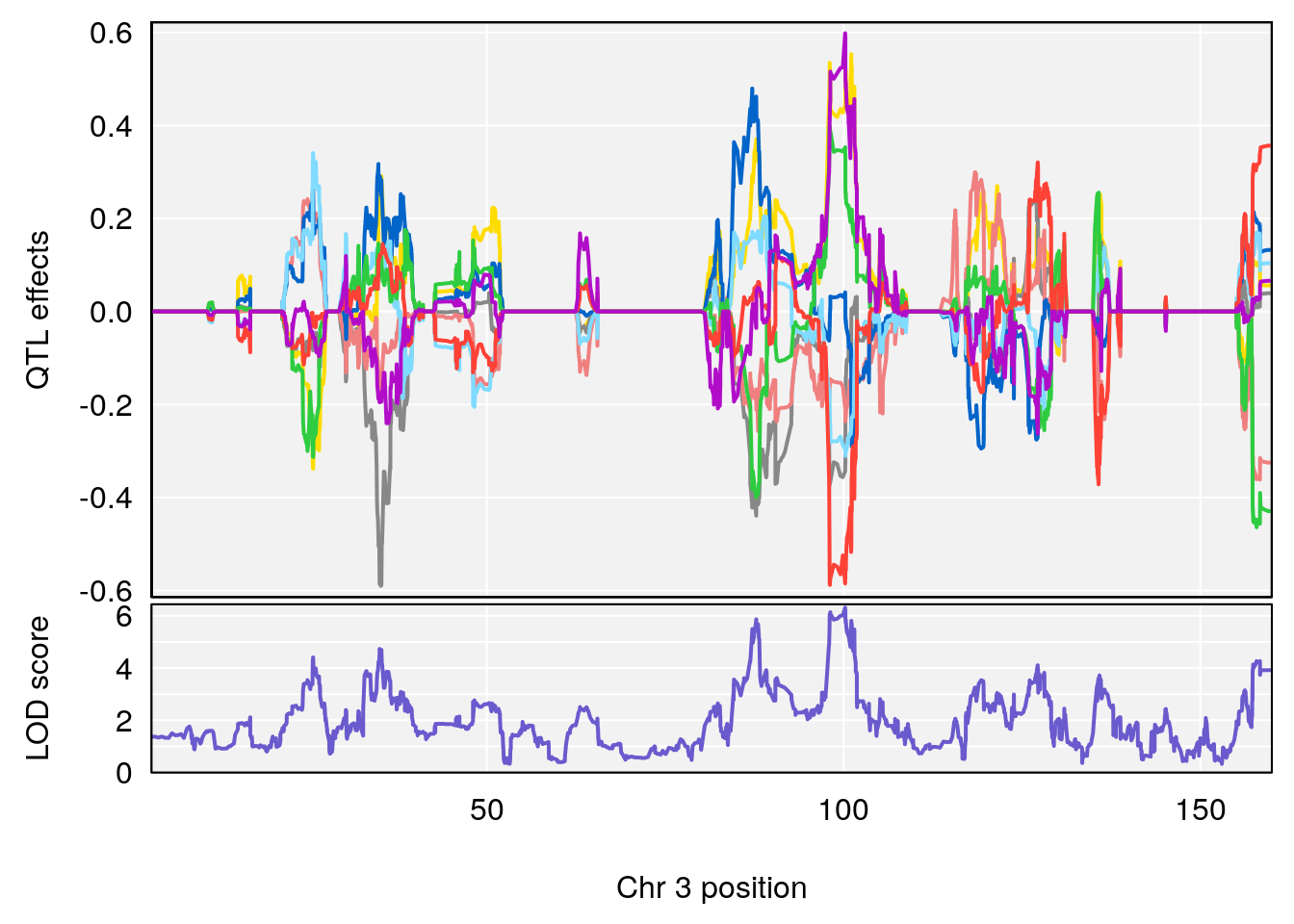
# [1] 2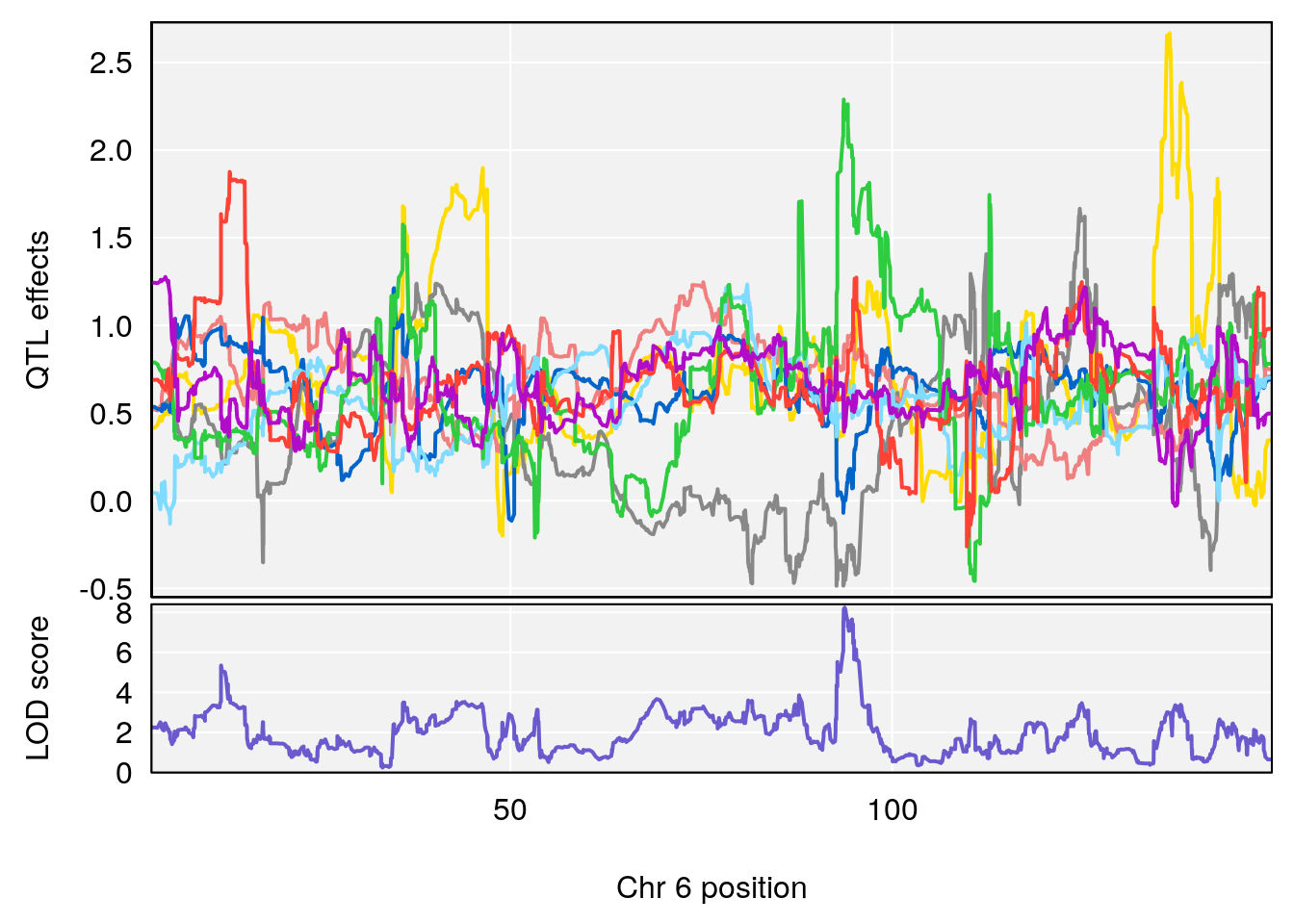
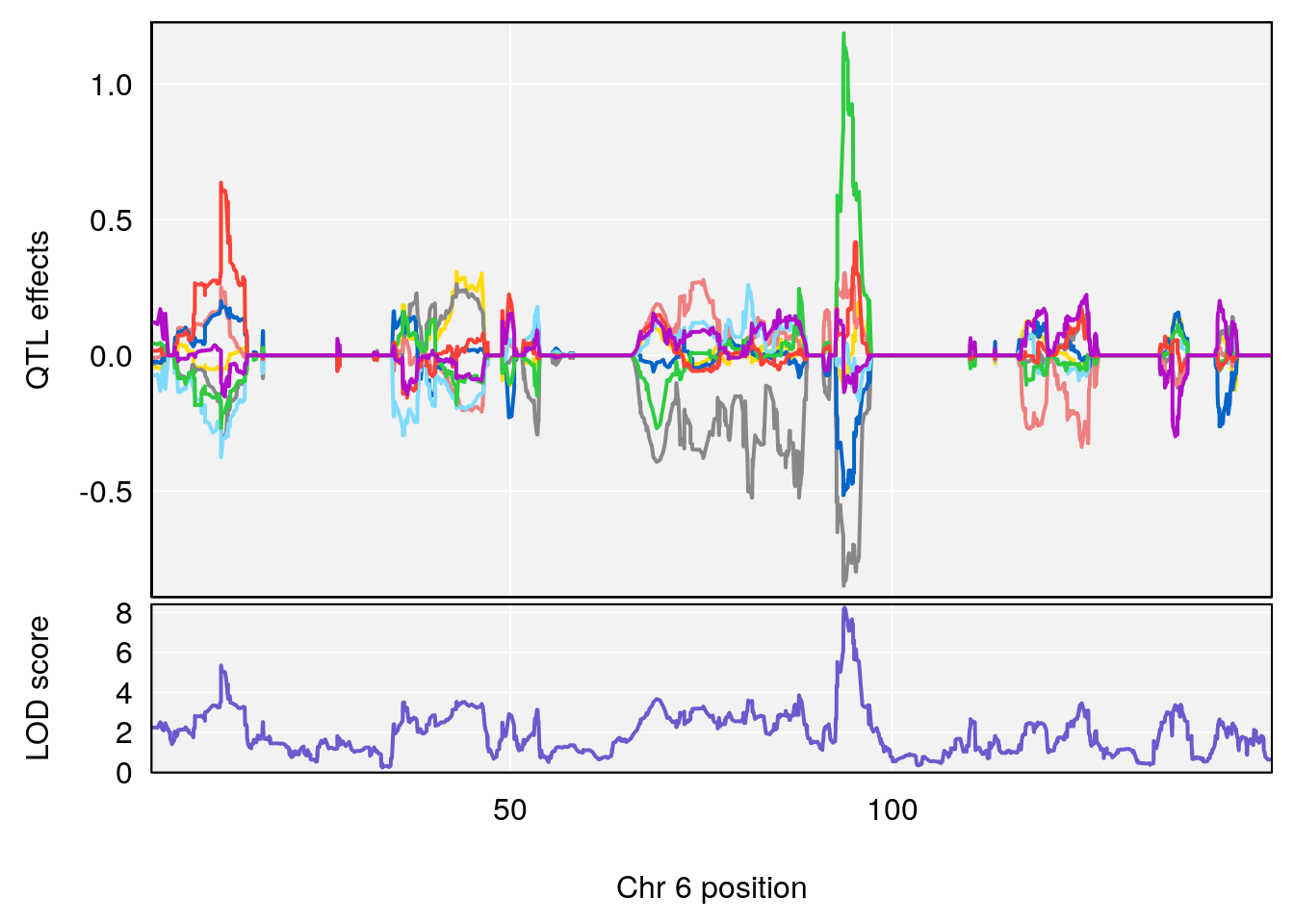
# [1] 3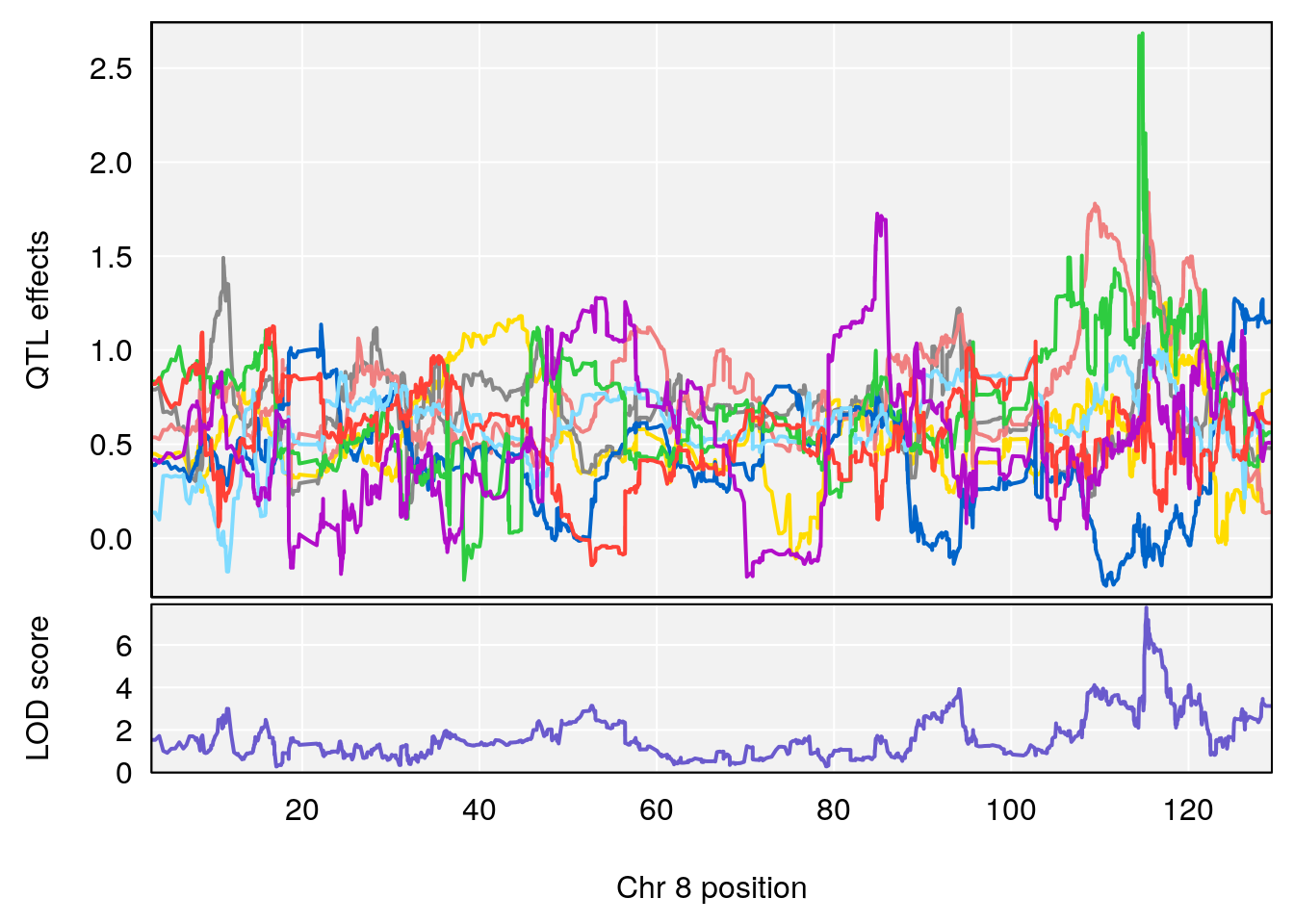
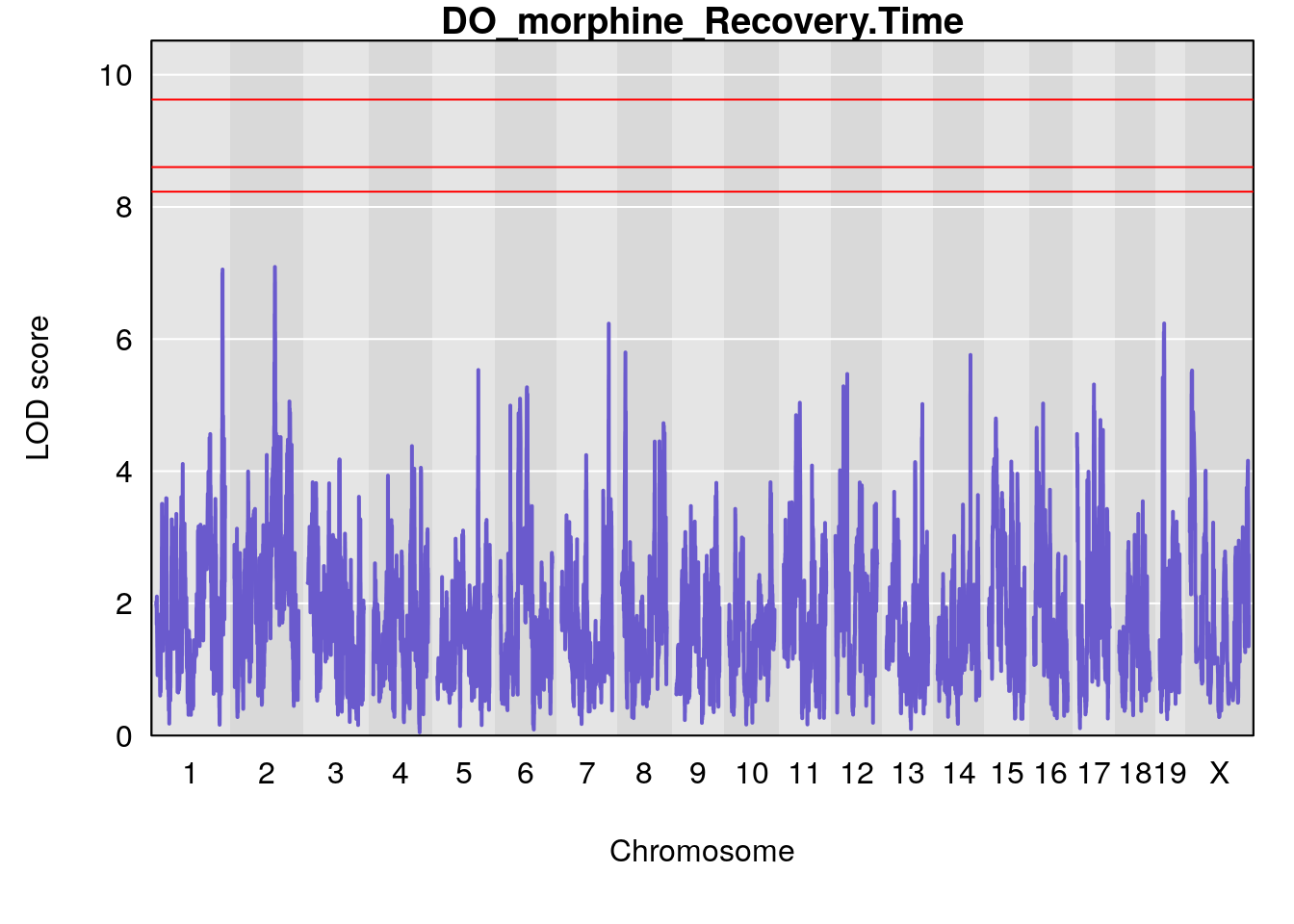
# [1] "Recovery.Time"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
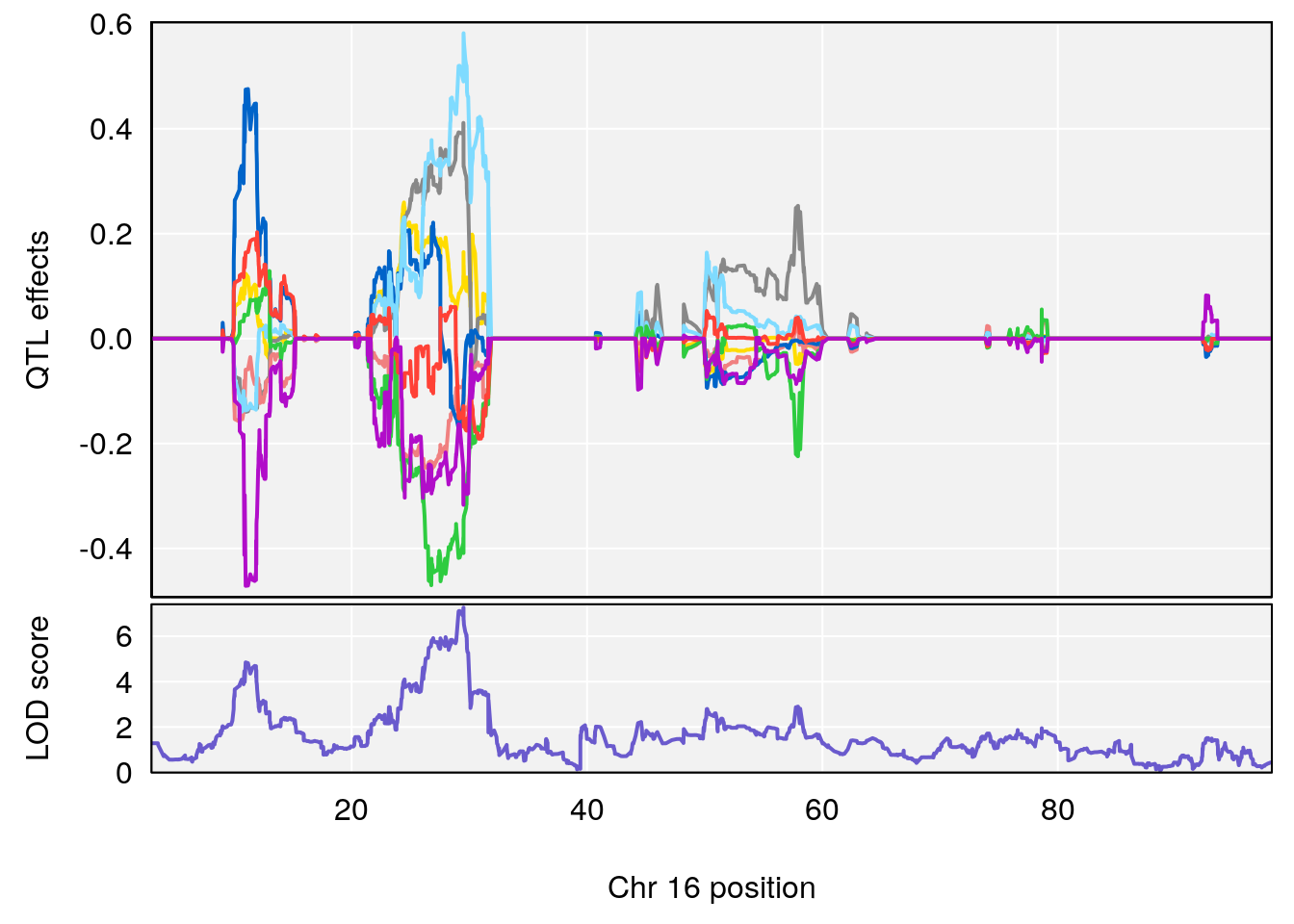
# 1 1 pheno1 1 188.47009 6.800872 188.08317 189.42004
# 2 1 pheno1 16 29.50467 7.258734 26.76501 29.78912
# [1] 1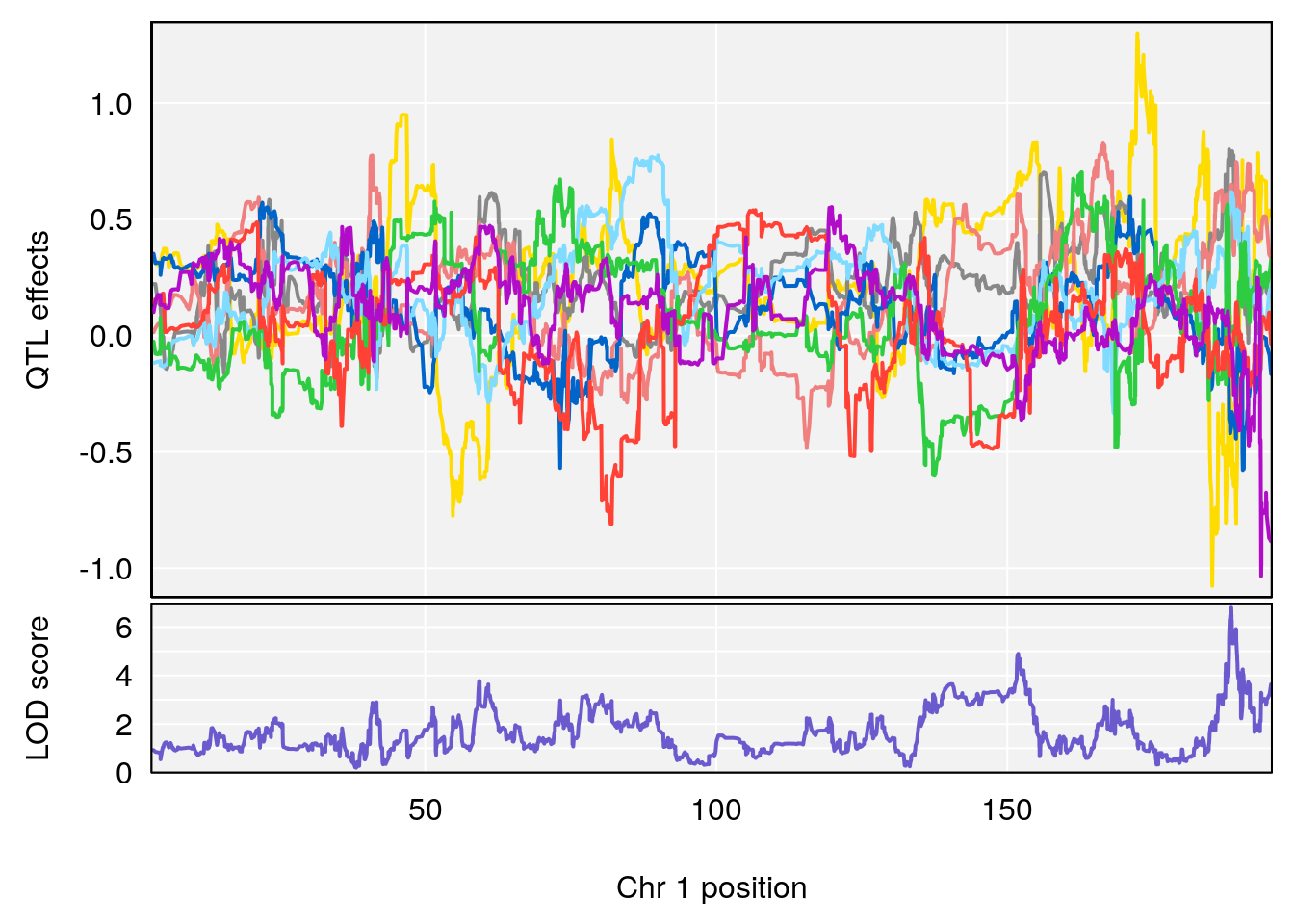
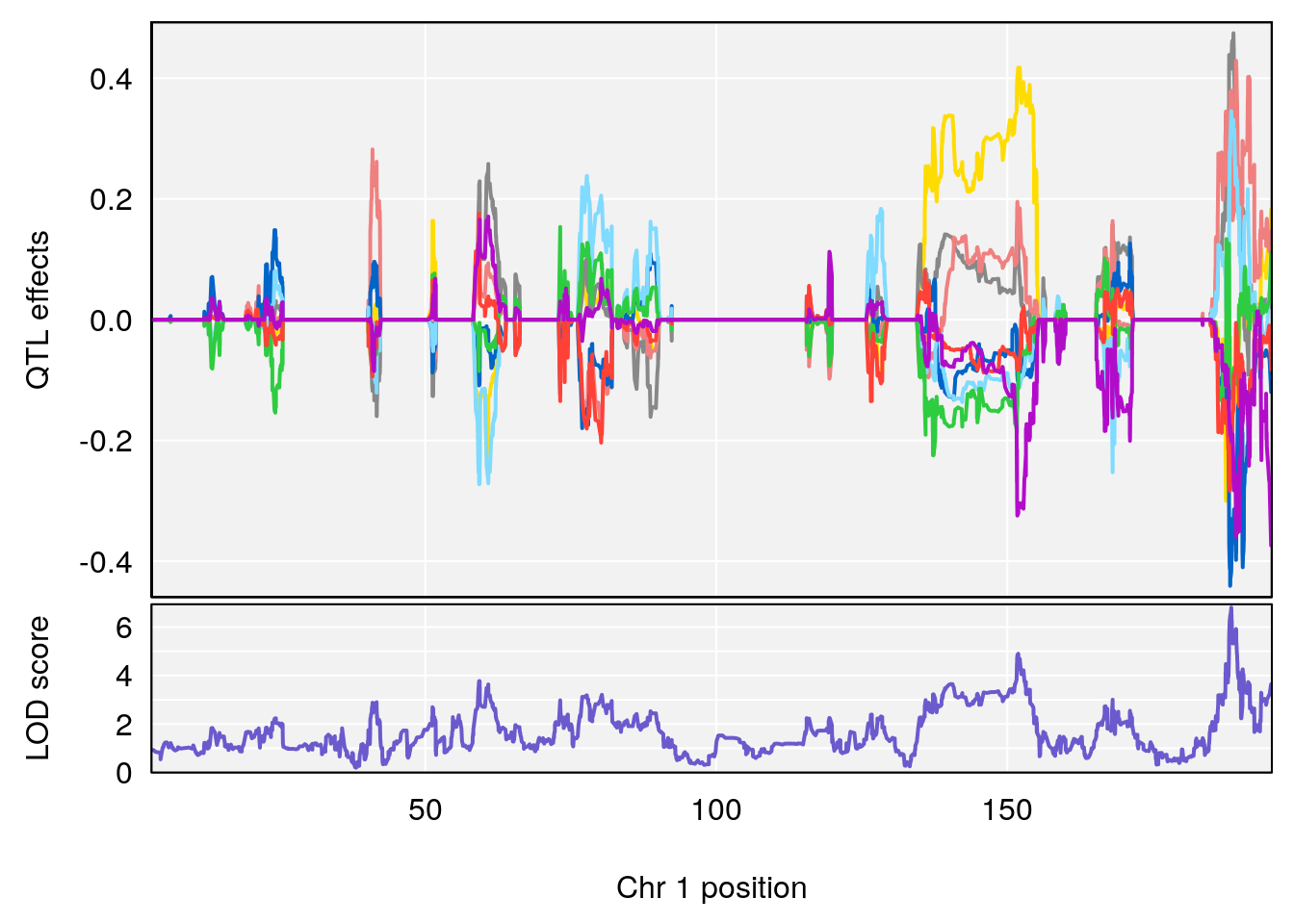
# [1] 2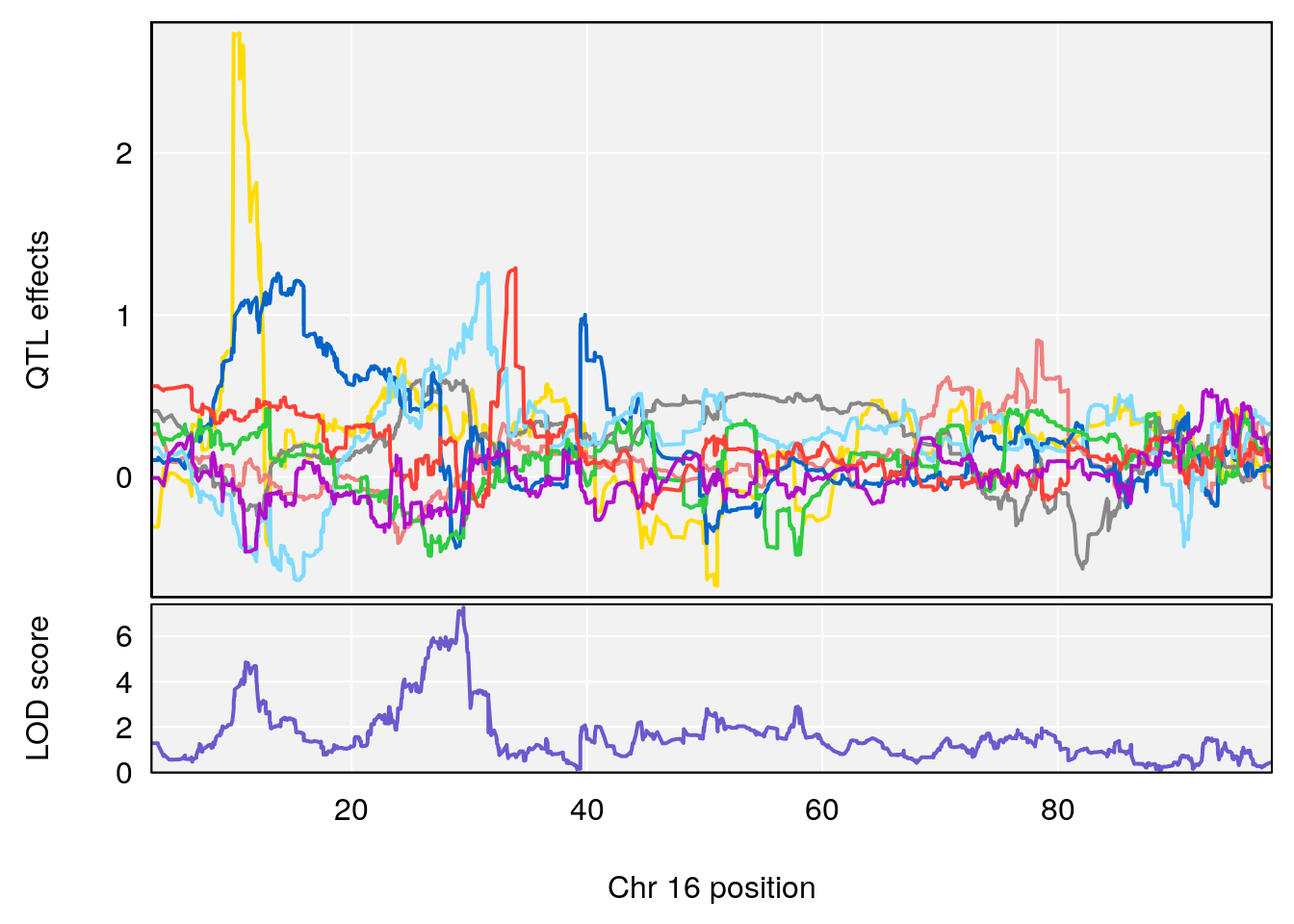
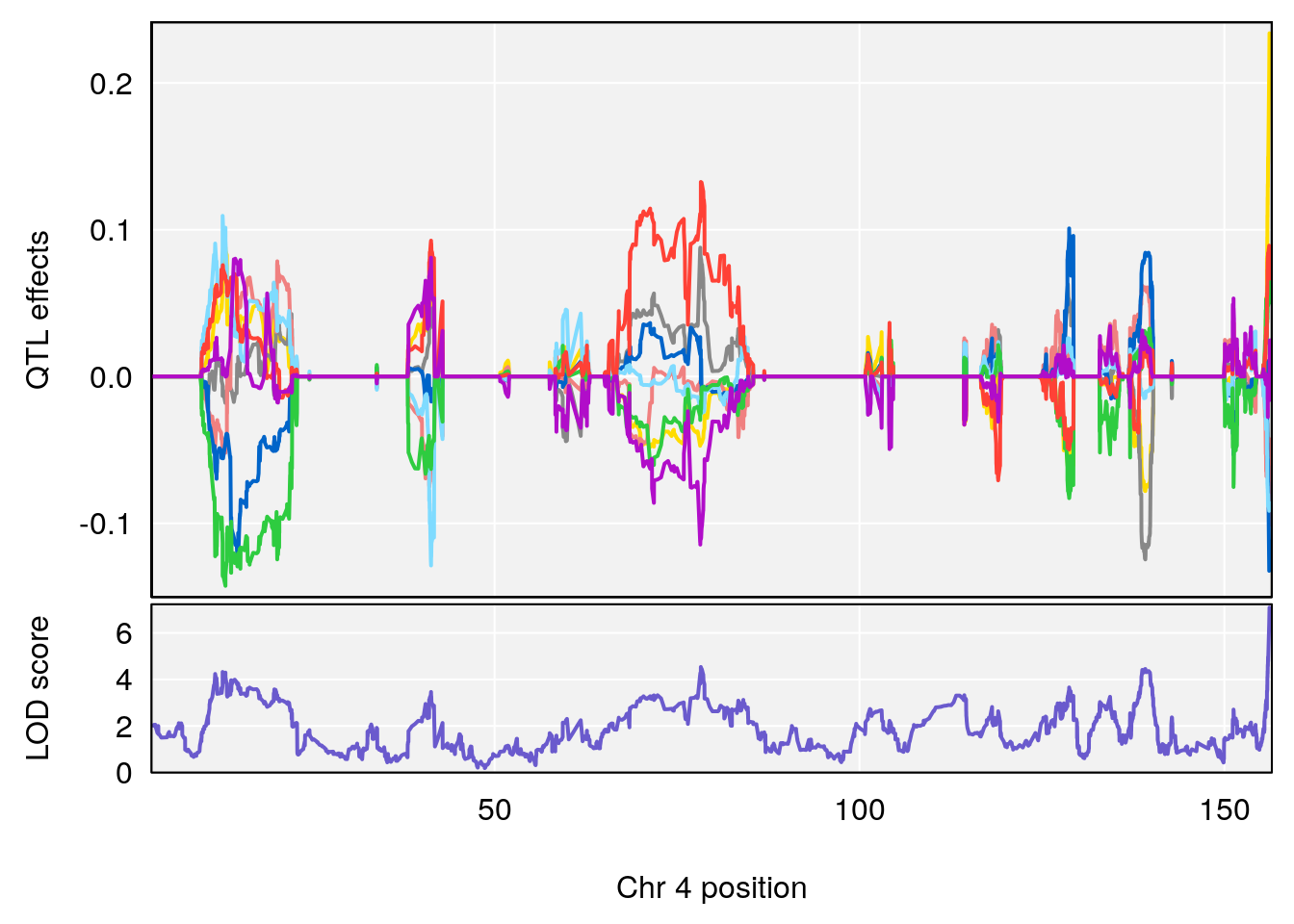
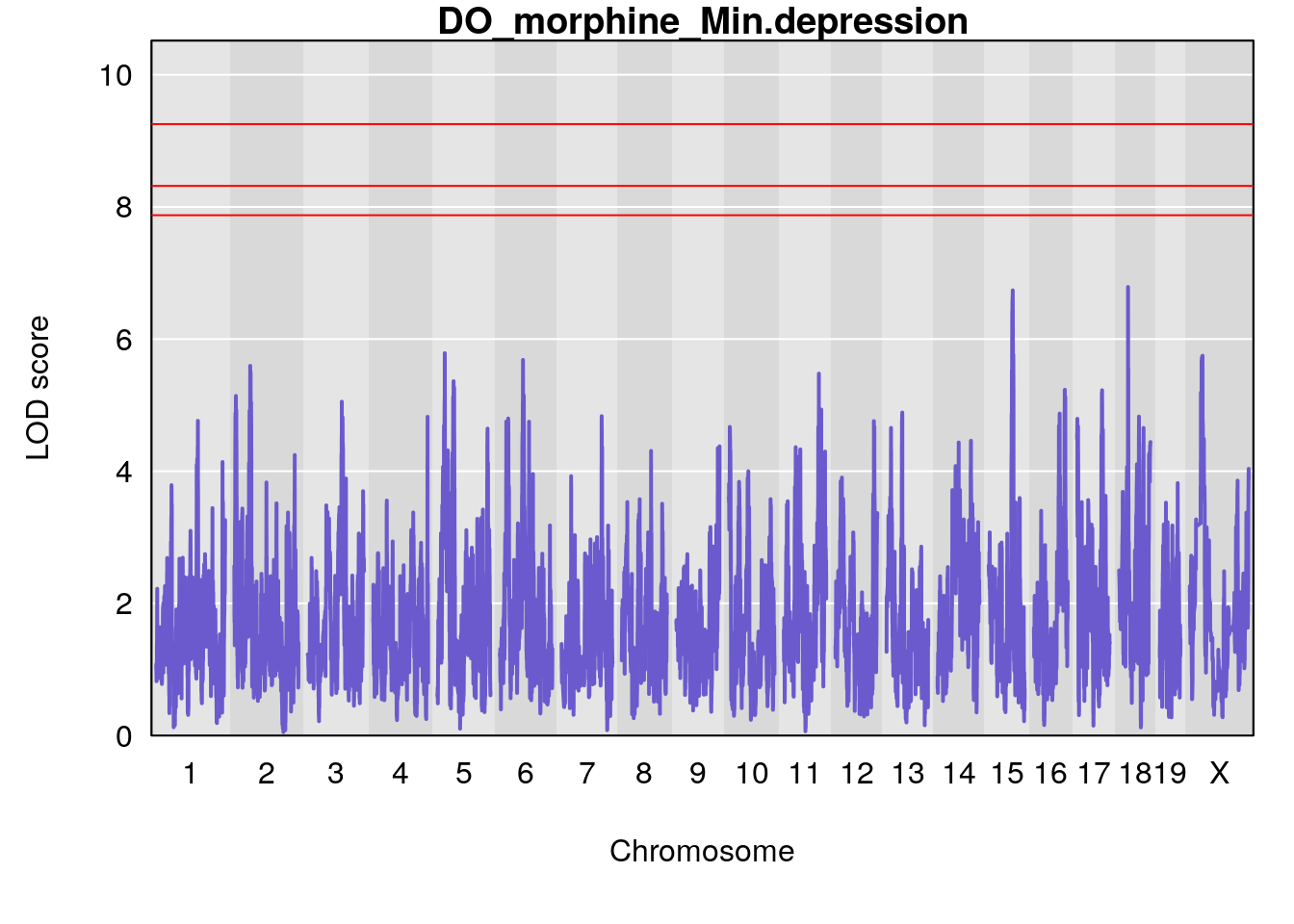
# [1] "Min.depression"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
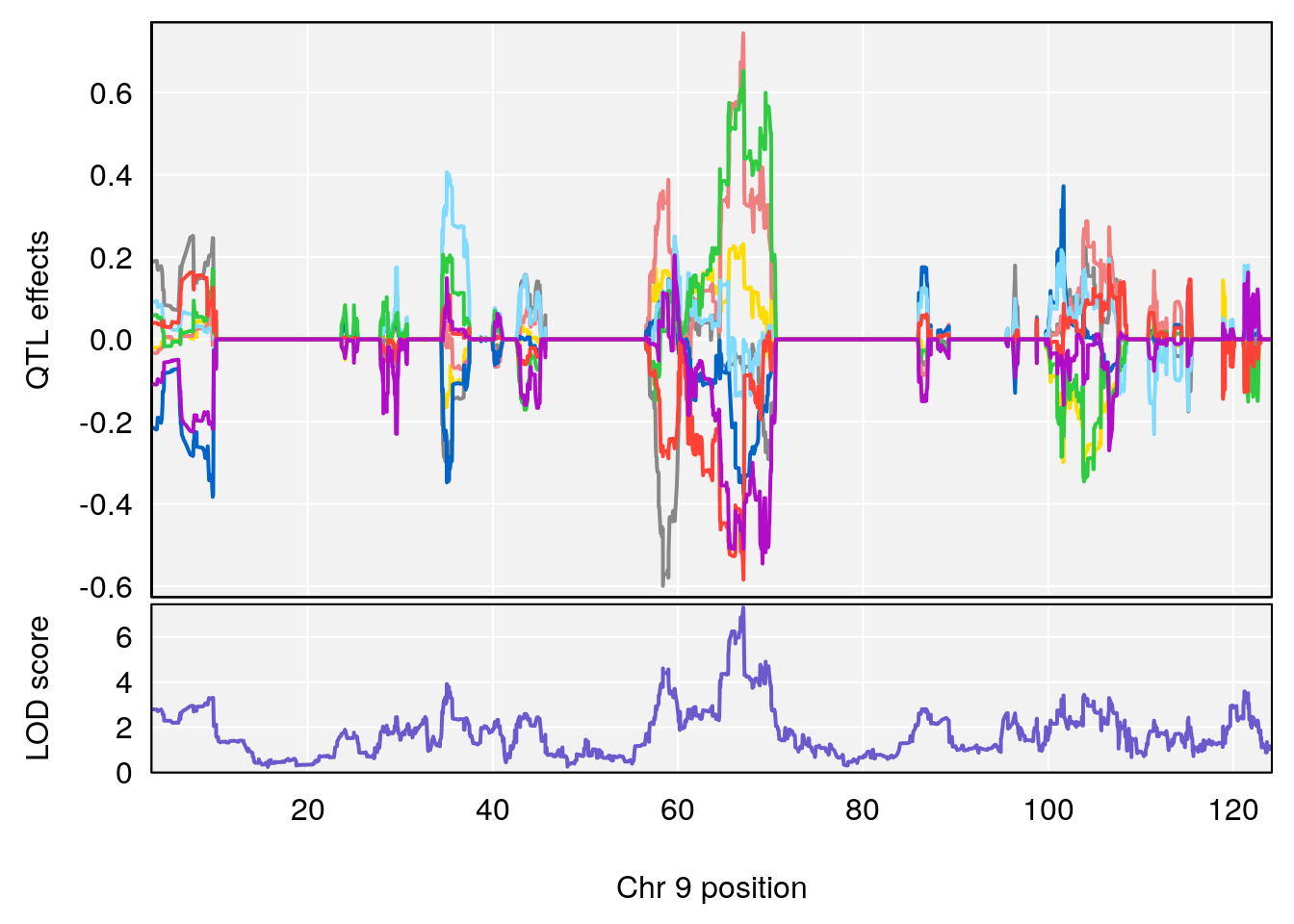
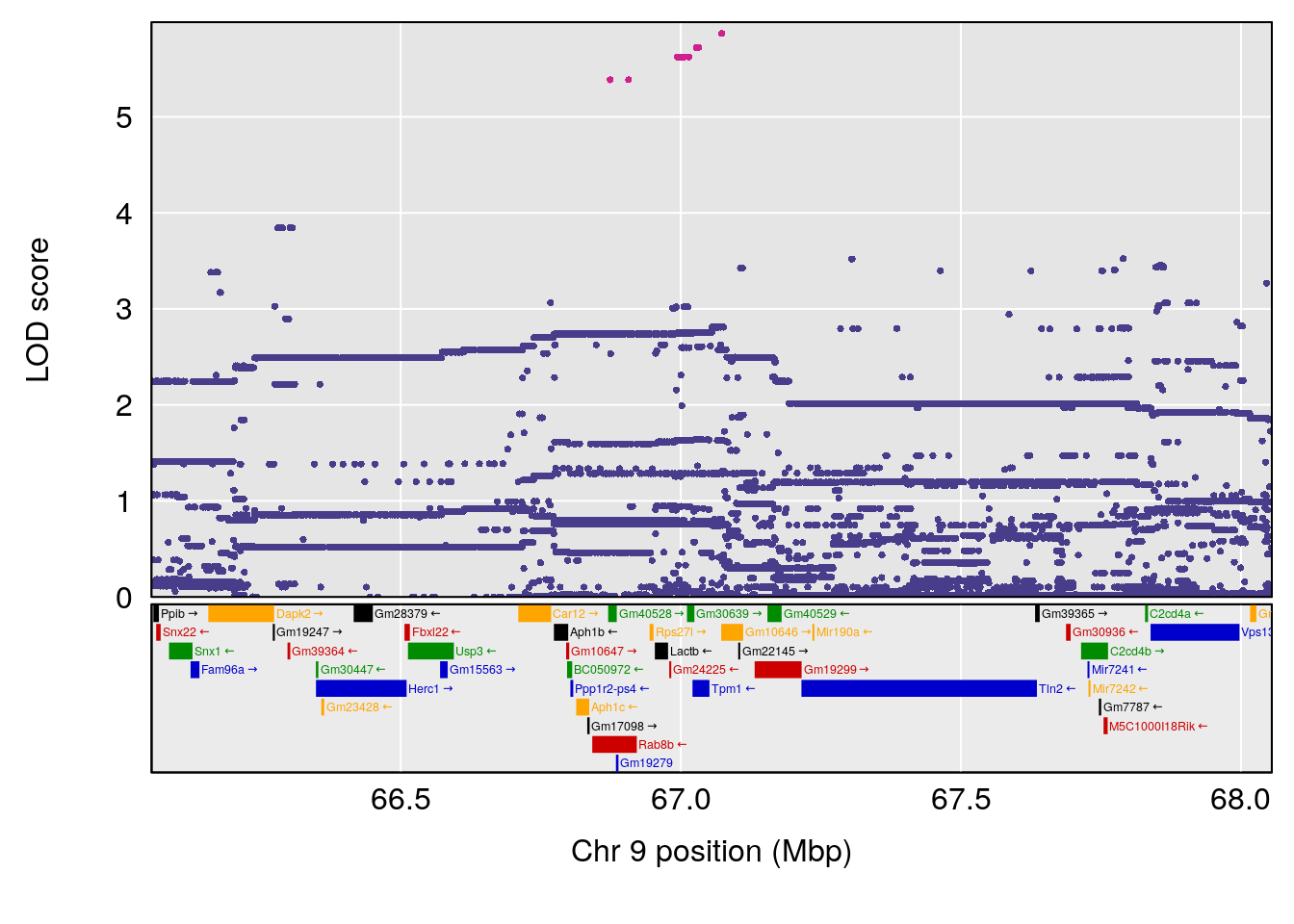
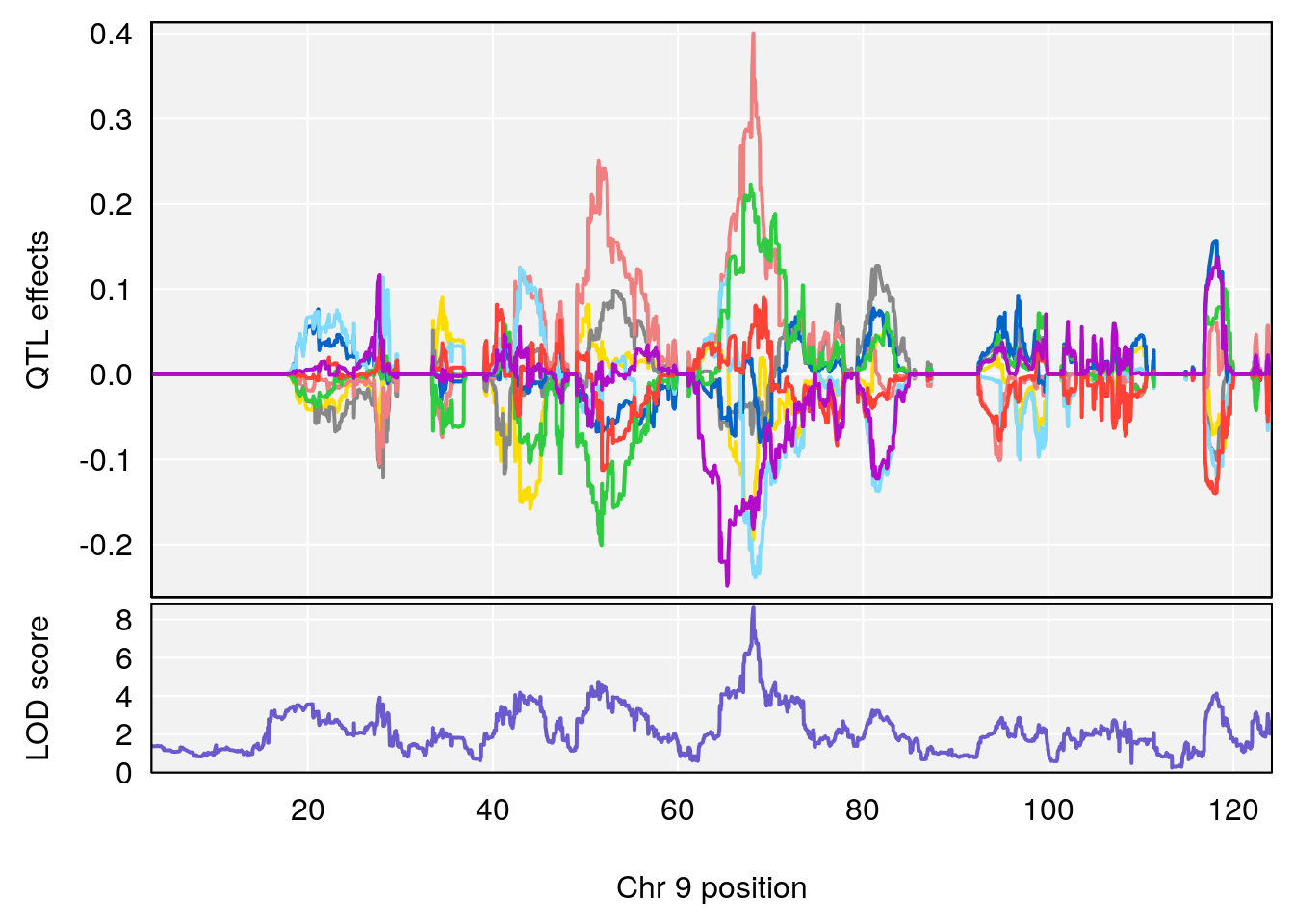
# 1 1 pheno1 9 97.15519 6.161653 96.10171 124.5827
# [1] 1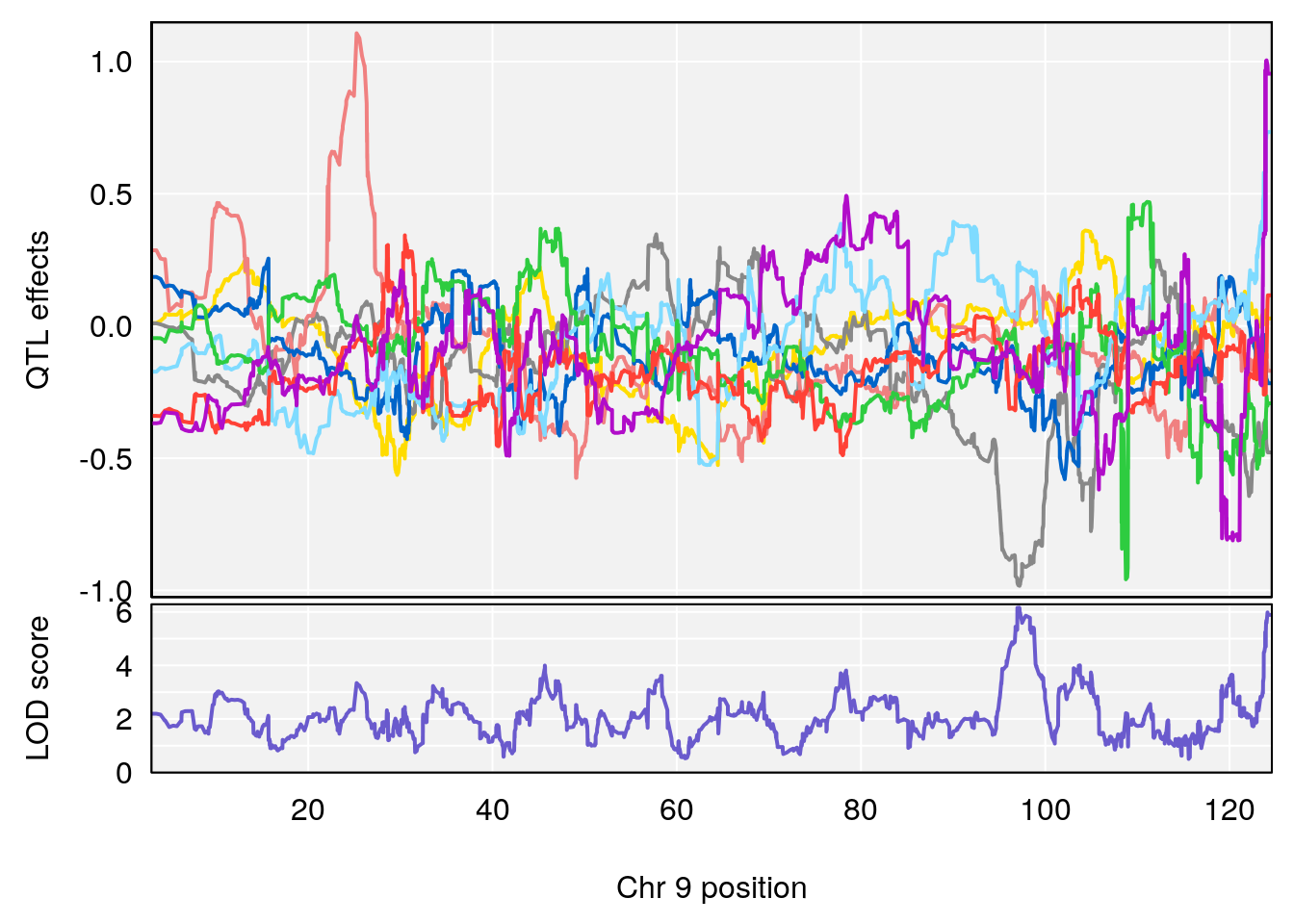
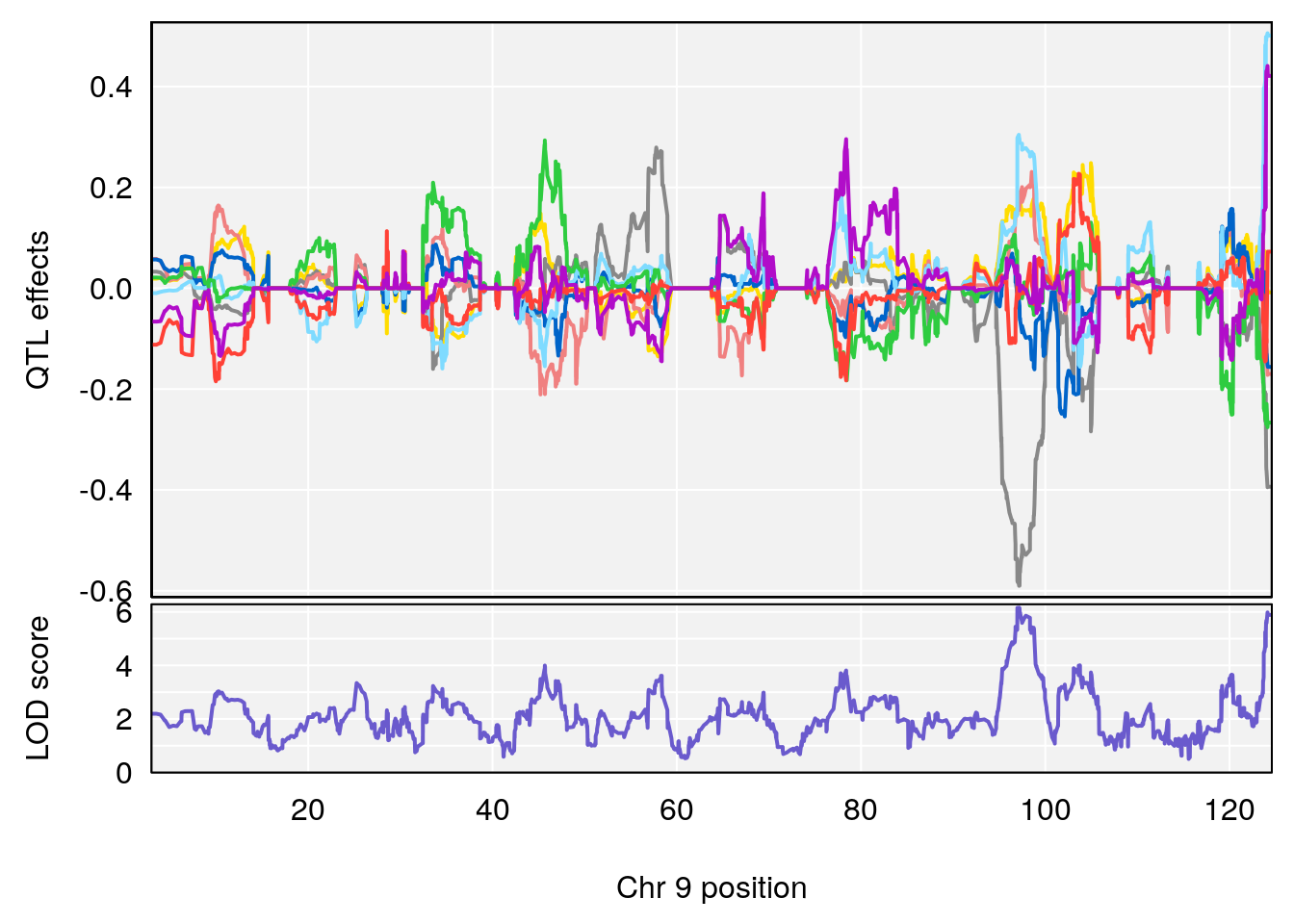
# [1] "Status_bin"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
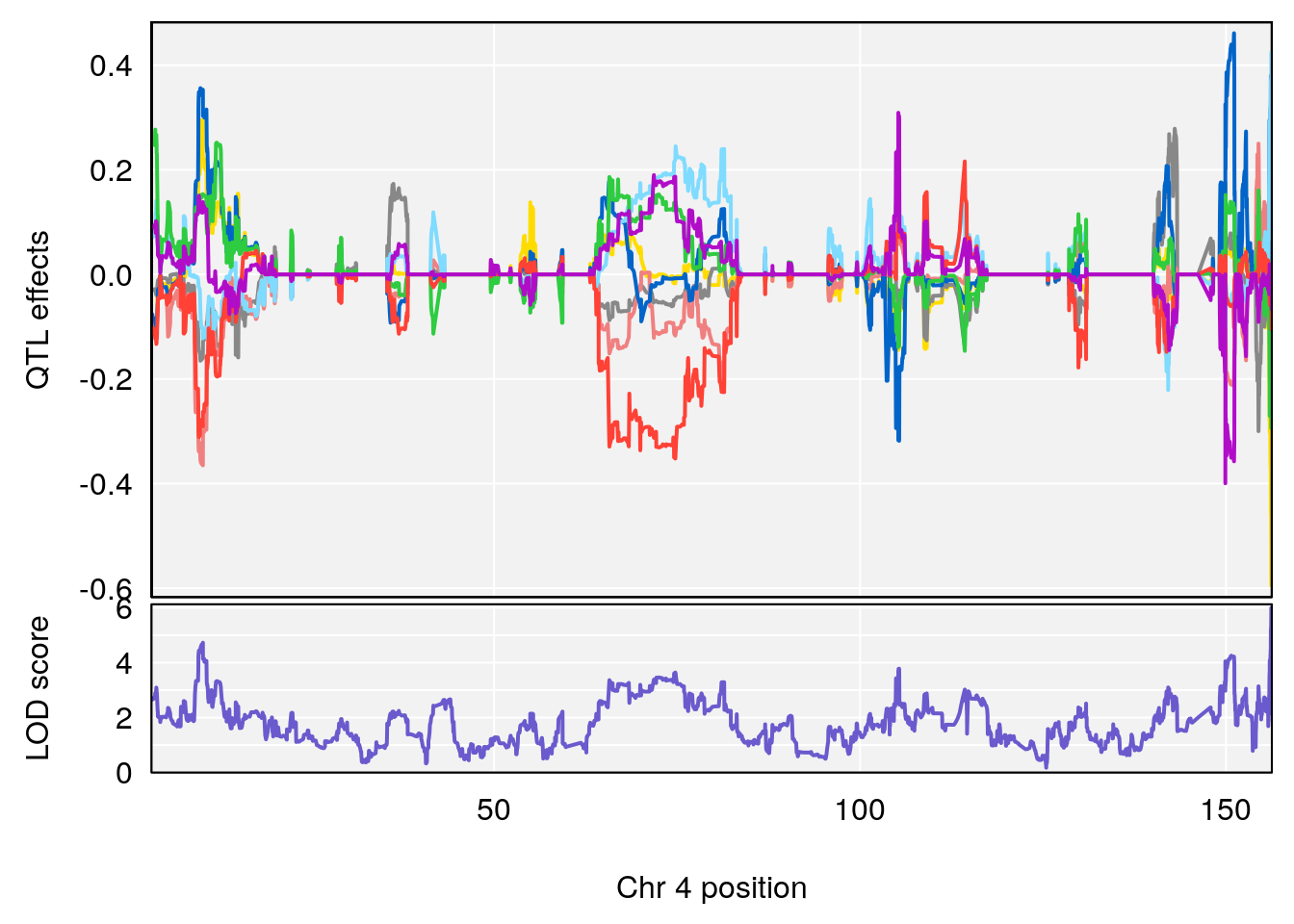
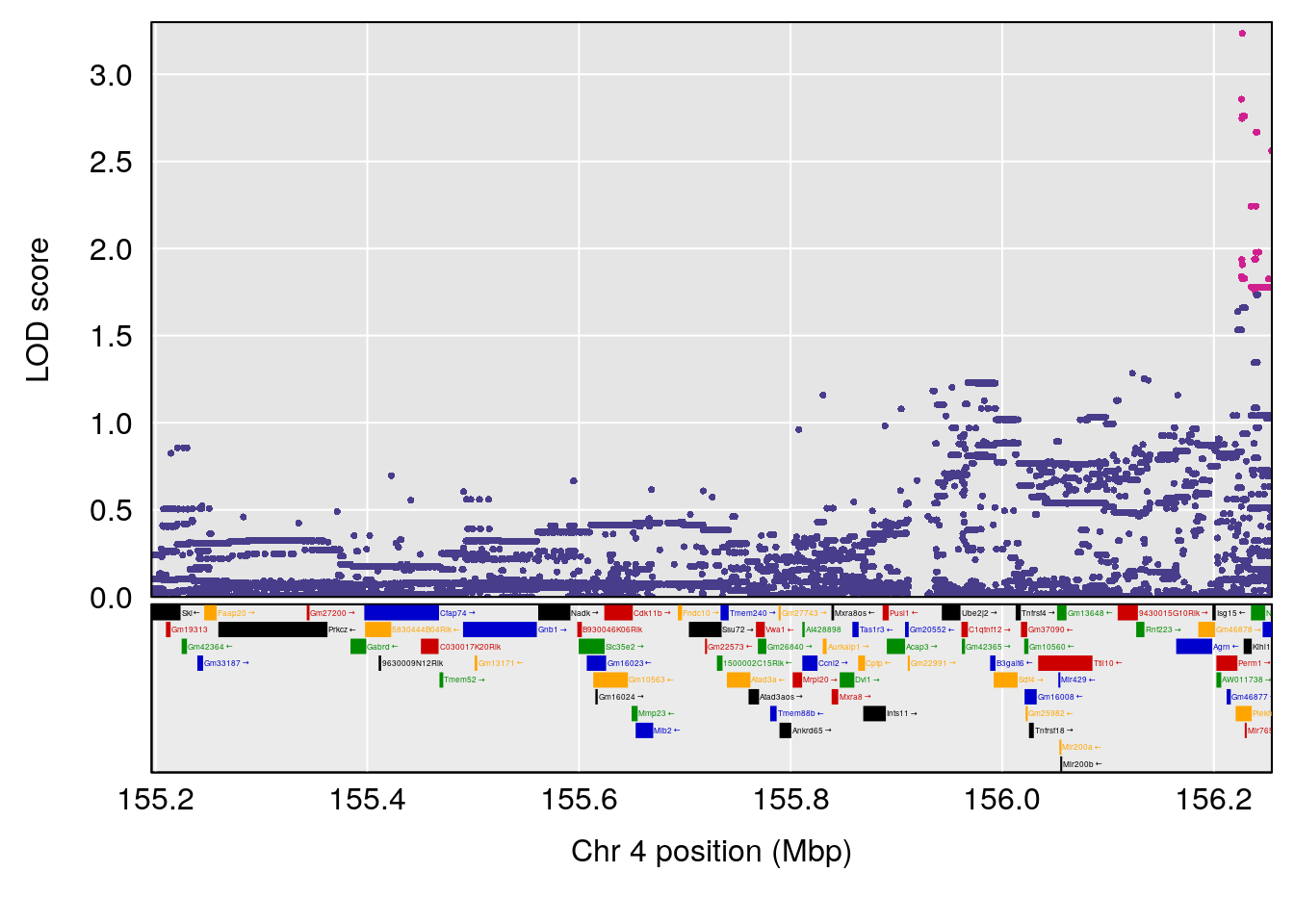
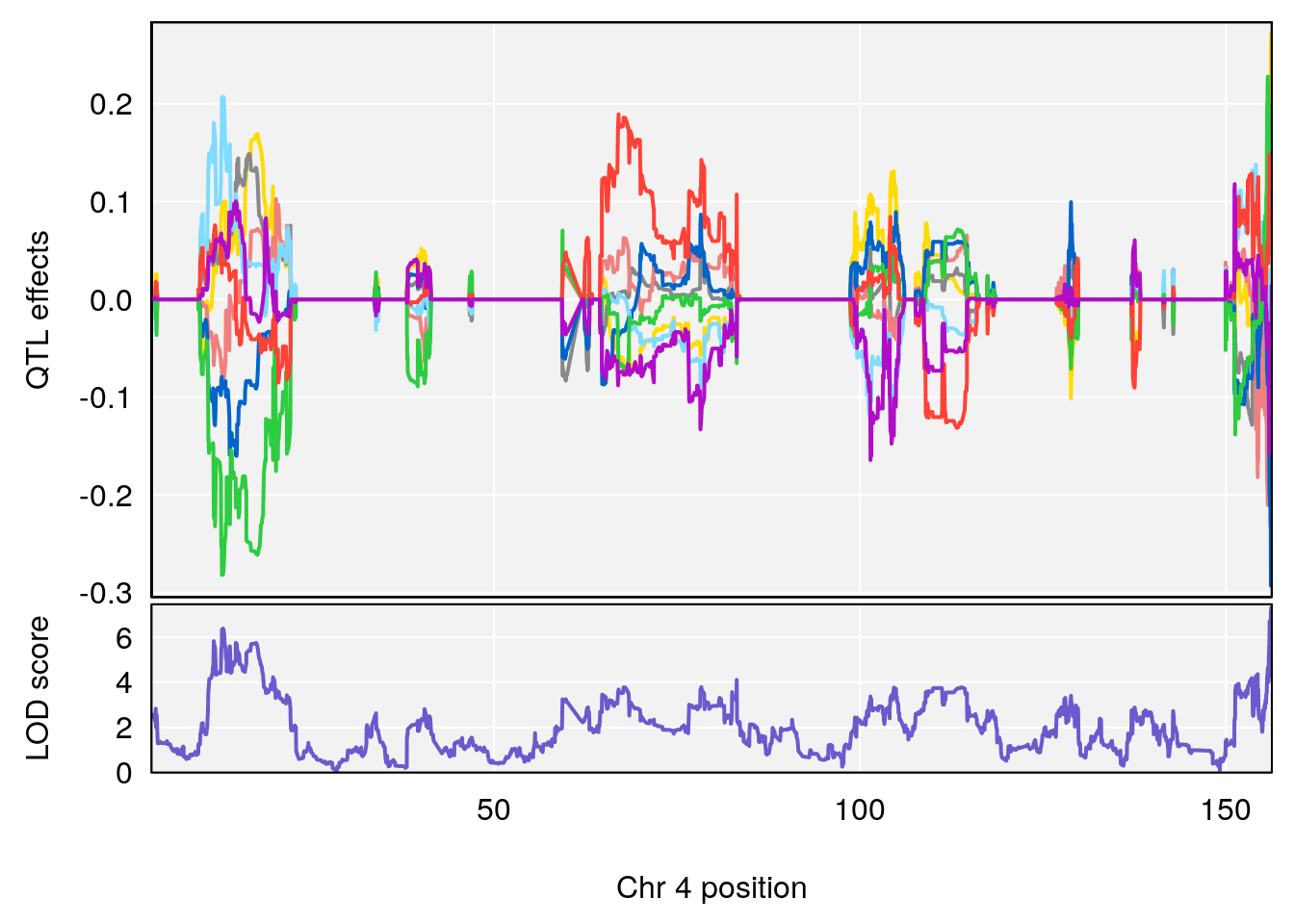
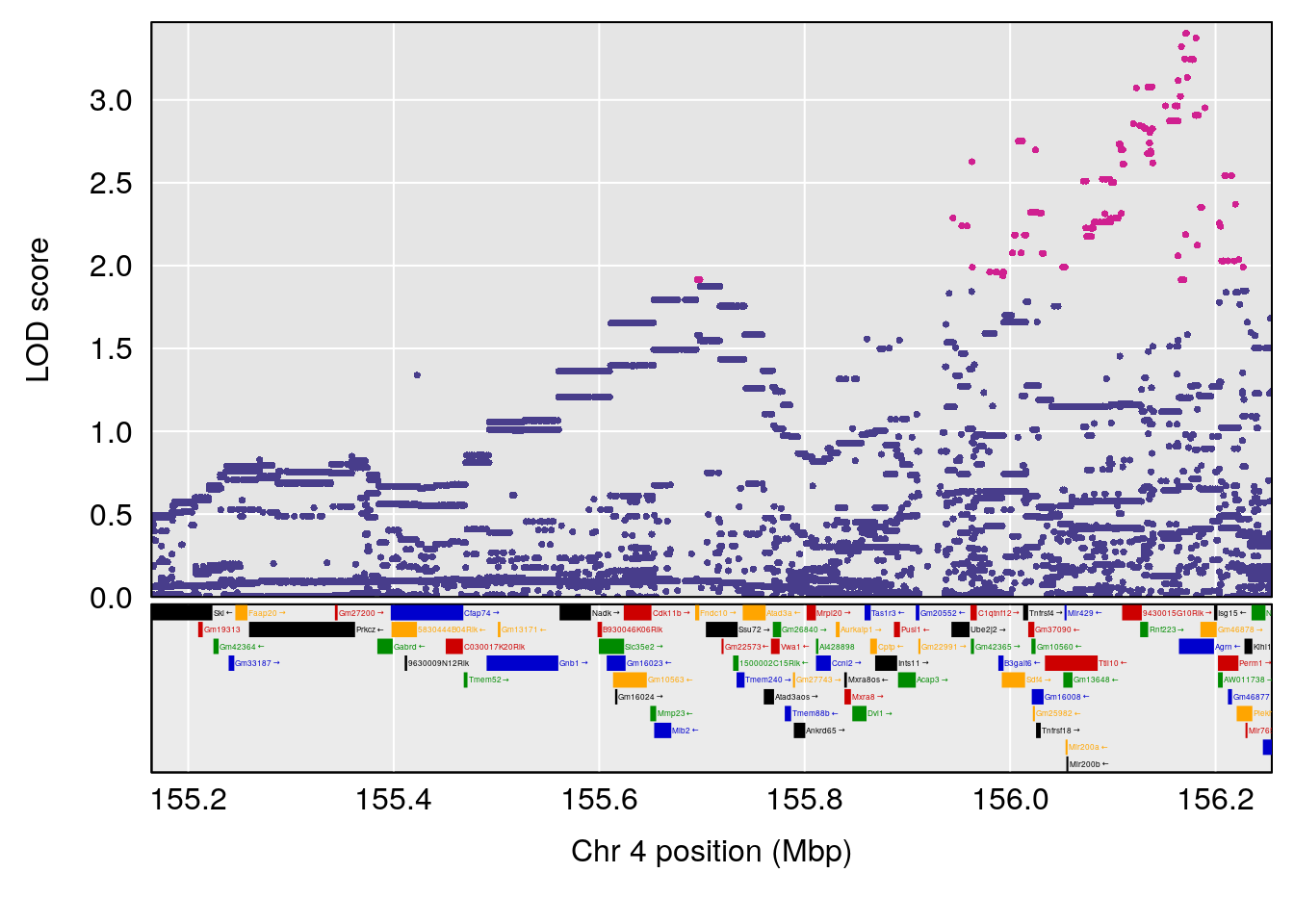
# 1 1 pheno1 4 156.17629 7.090528 156.009474 156.49607
# 2 1 pheno1 9 68.16325 6.104652 67.850302 68.56932
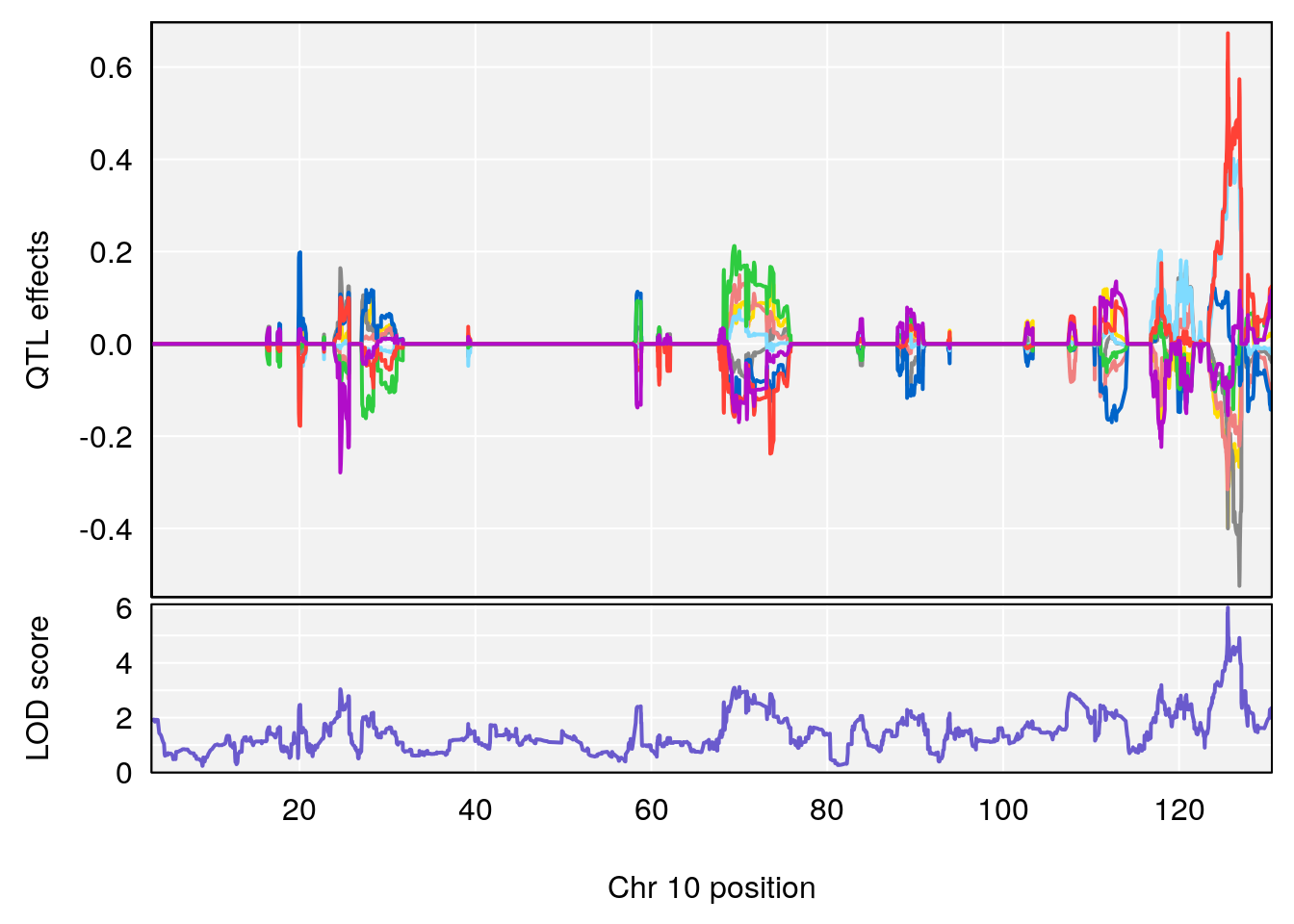
# 3 1 pheno1 10 29.38717 6.470057 5.600518 68.73517
# 4 1 pheno1 12 108.10231 6.730559 25.430755 109.67503
# 5 1 pheno1 14 120.28595 6.208071 104.068913 121.29650
# 6 1 pheno1 17 27.00602 6.571641 26.634222 85.12396
# 7 1 pheno1 18 85.35144 6.635902 81.255418 86.51269
# 8 1 pheno1 X 144.48572 7.929565 143.604522 145.79861
# [1] 1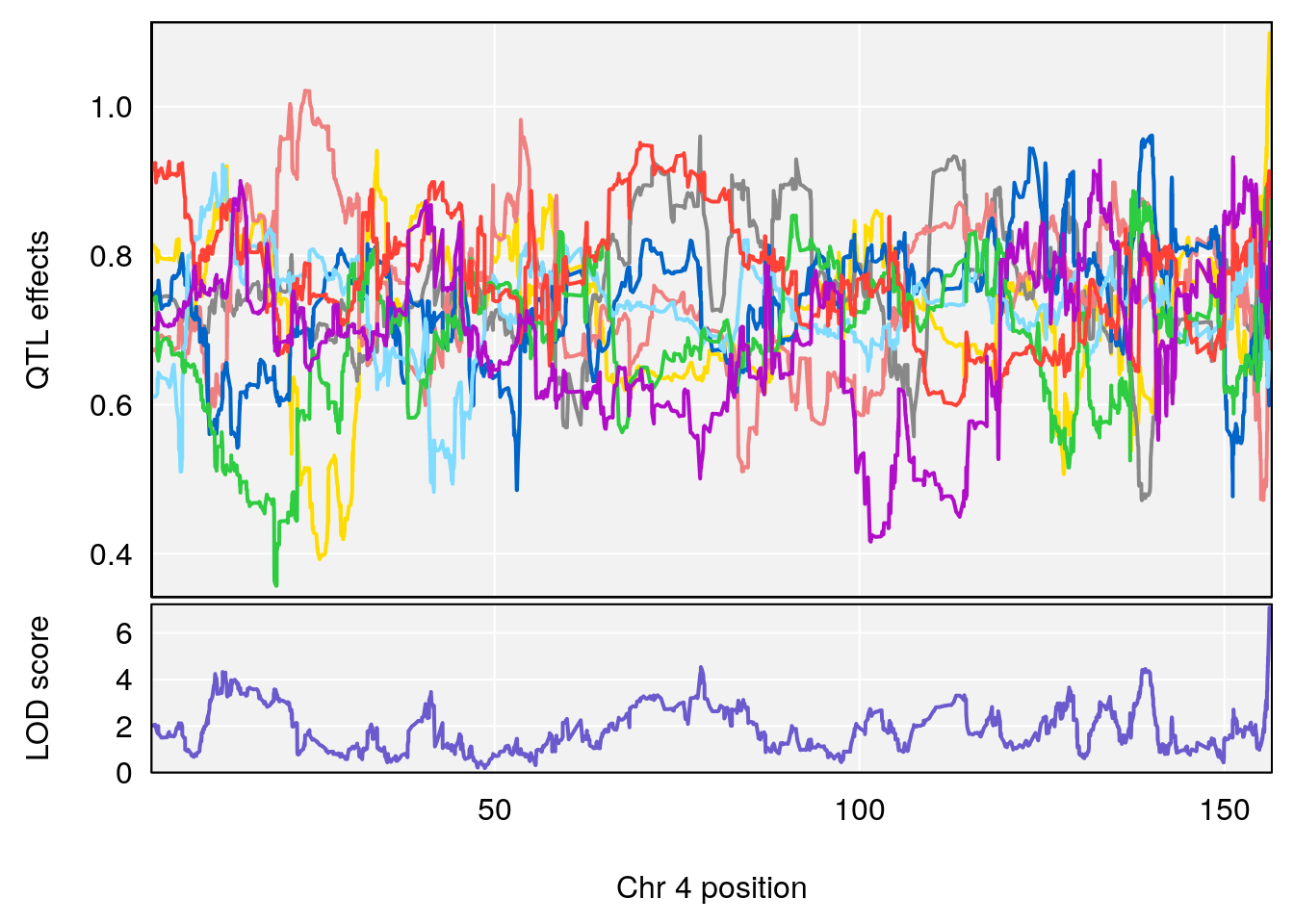
# [1] 2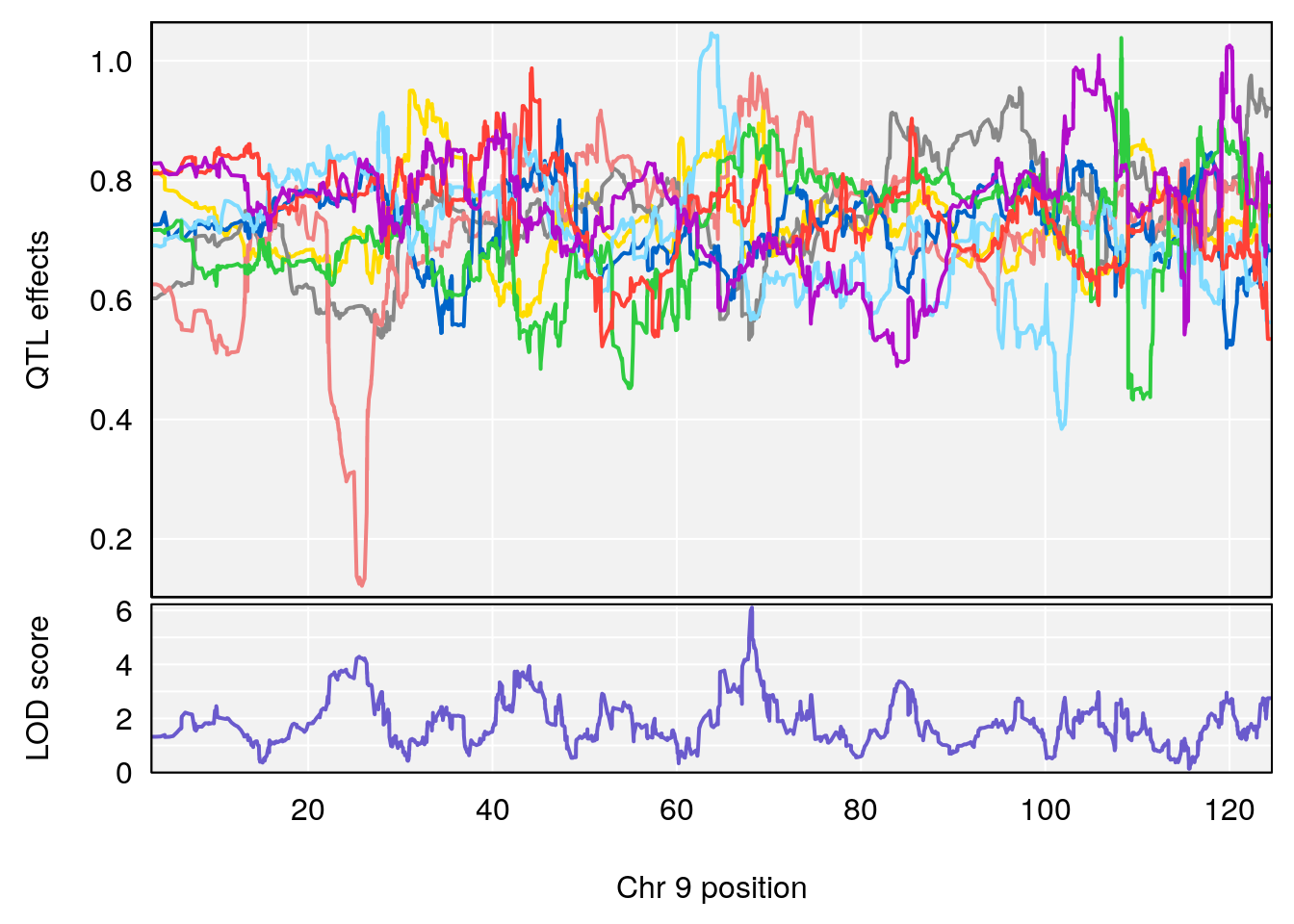
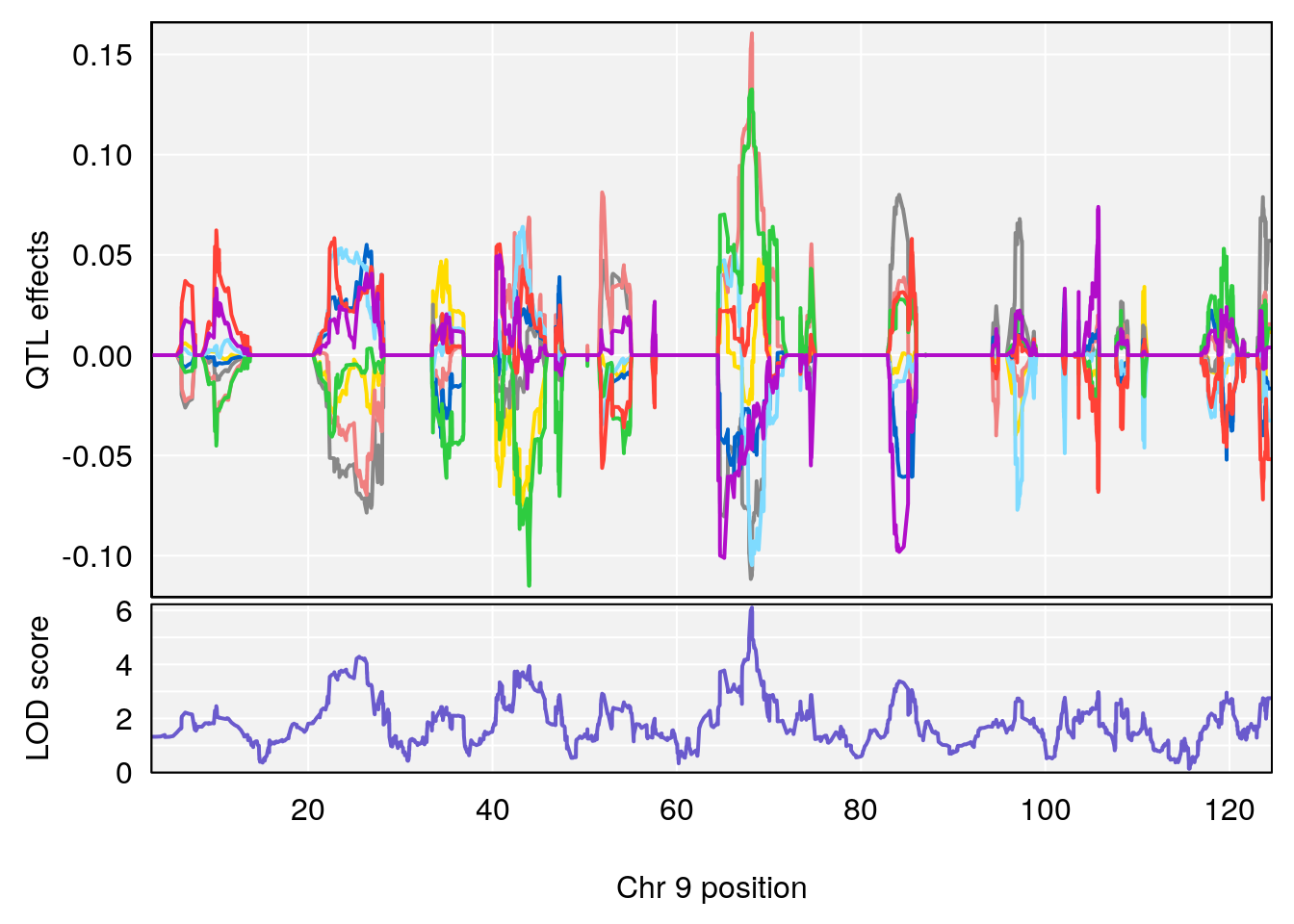
# [1] 3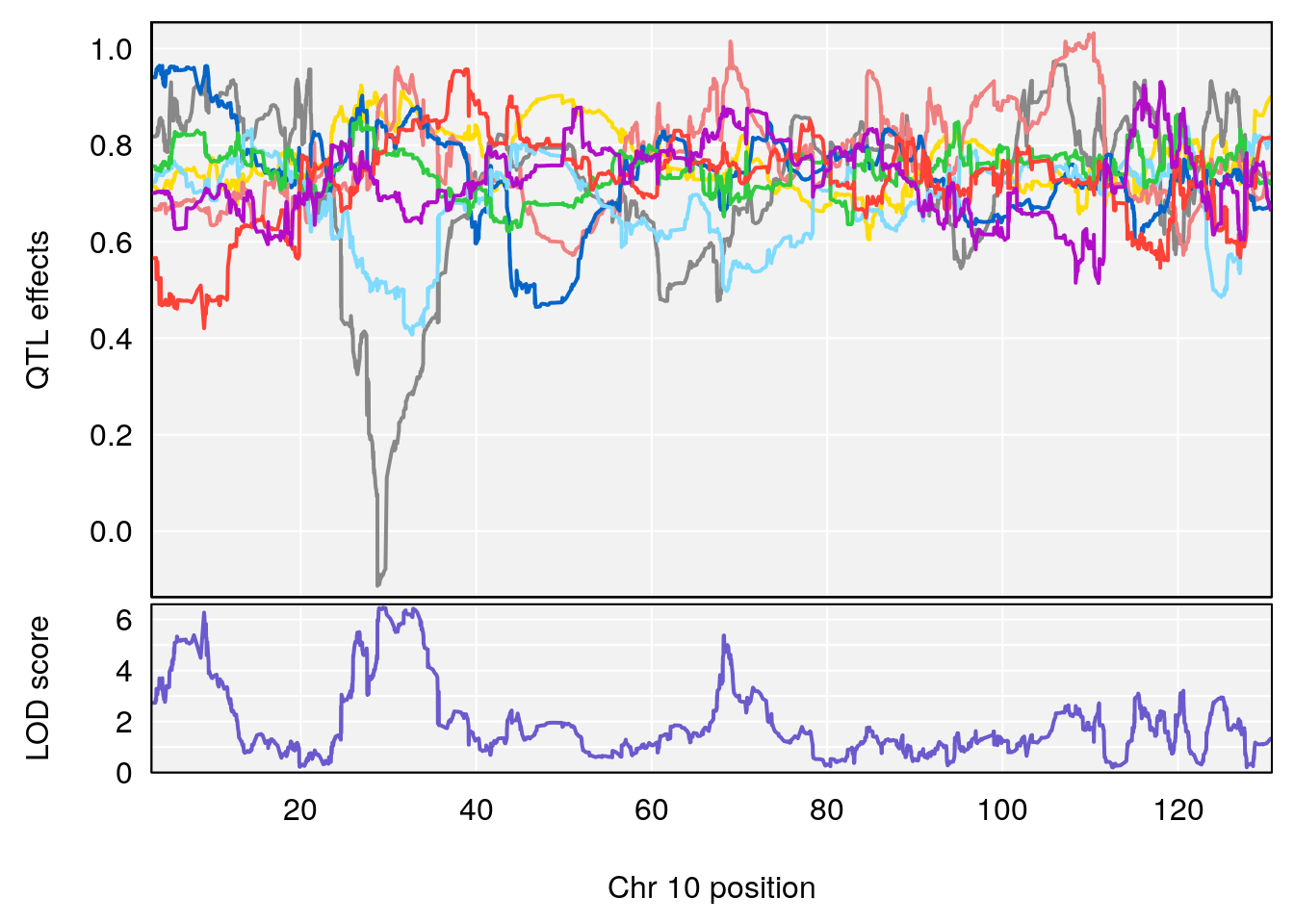
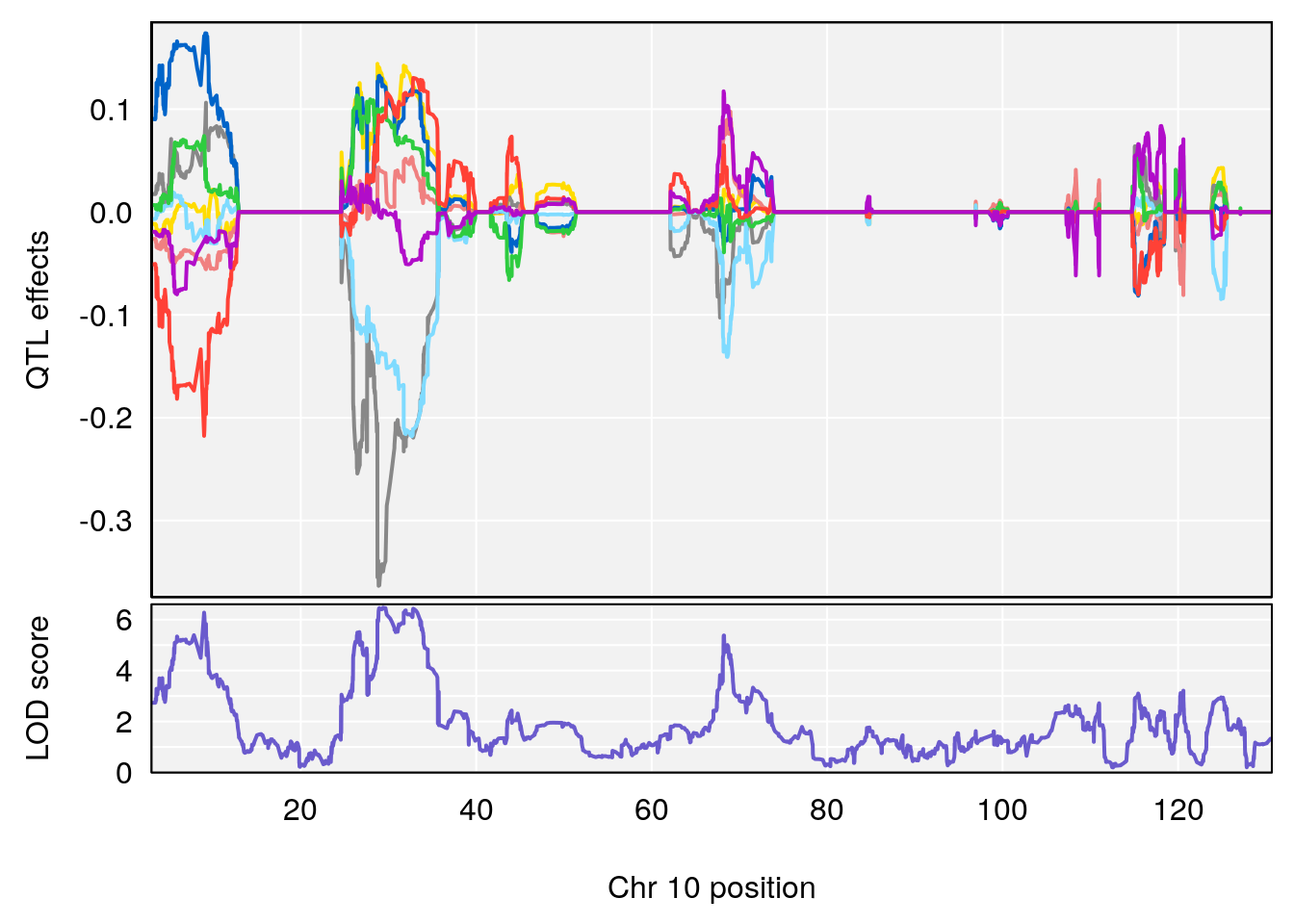
# [1] 4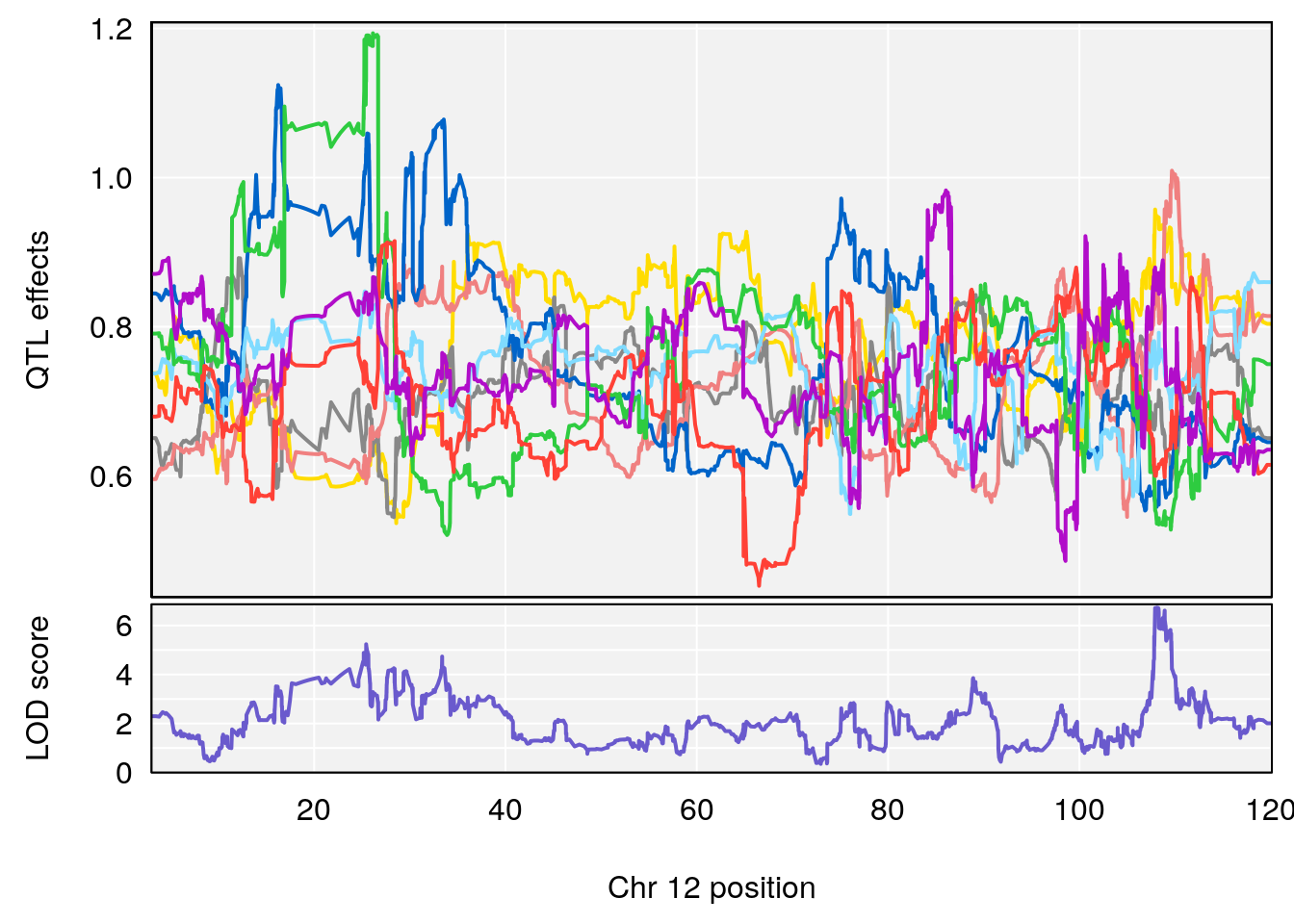
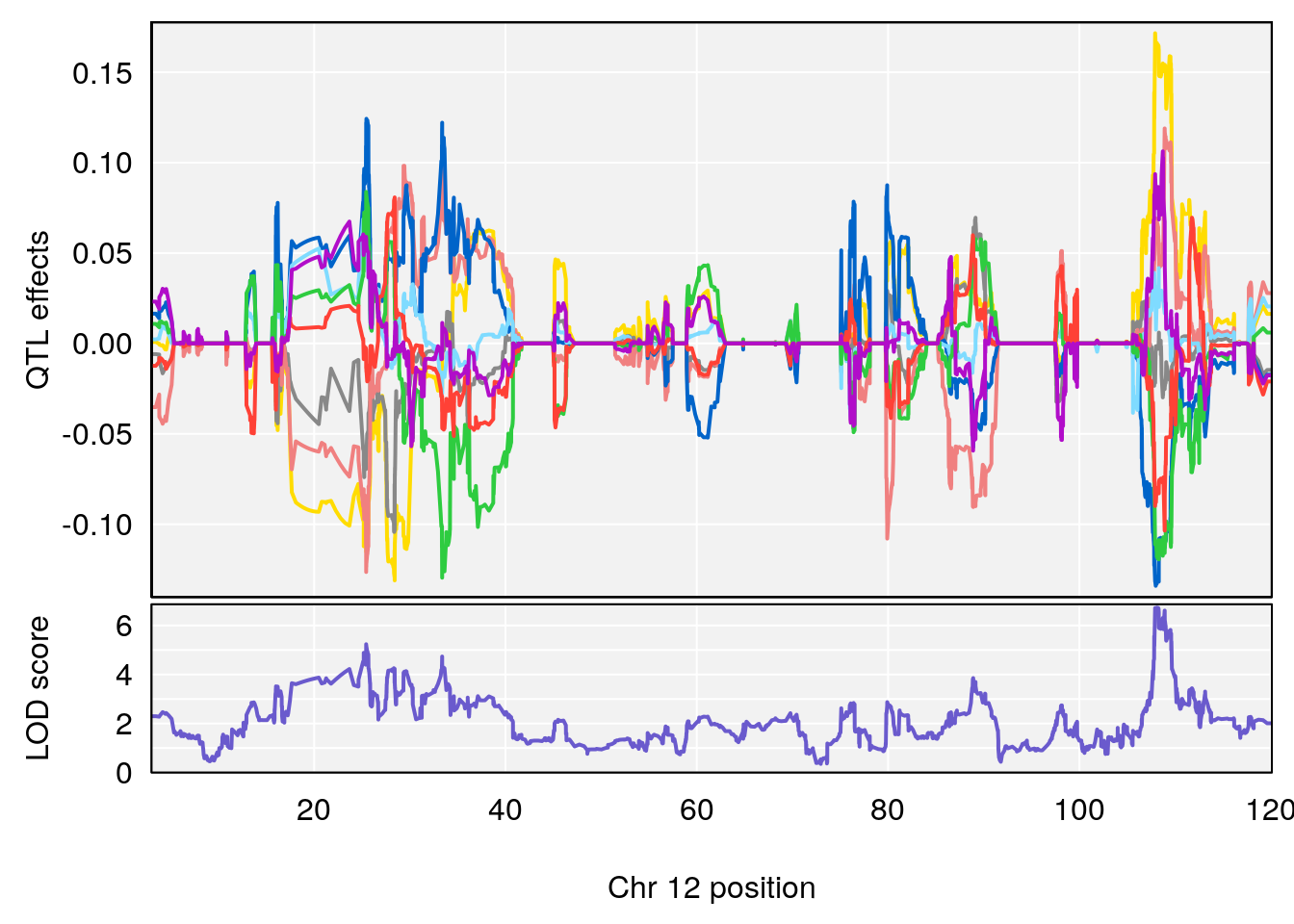
# [1] 5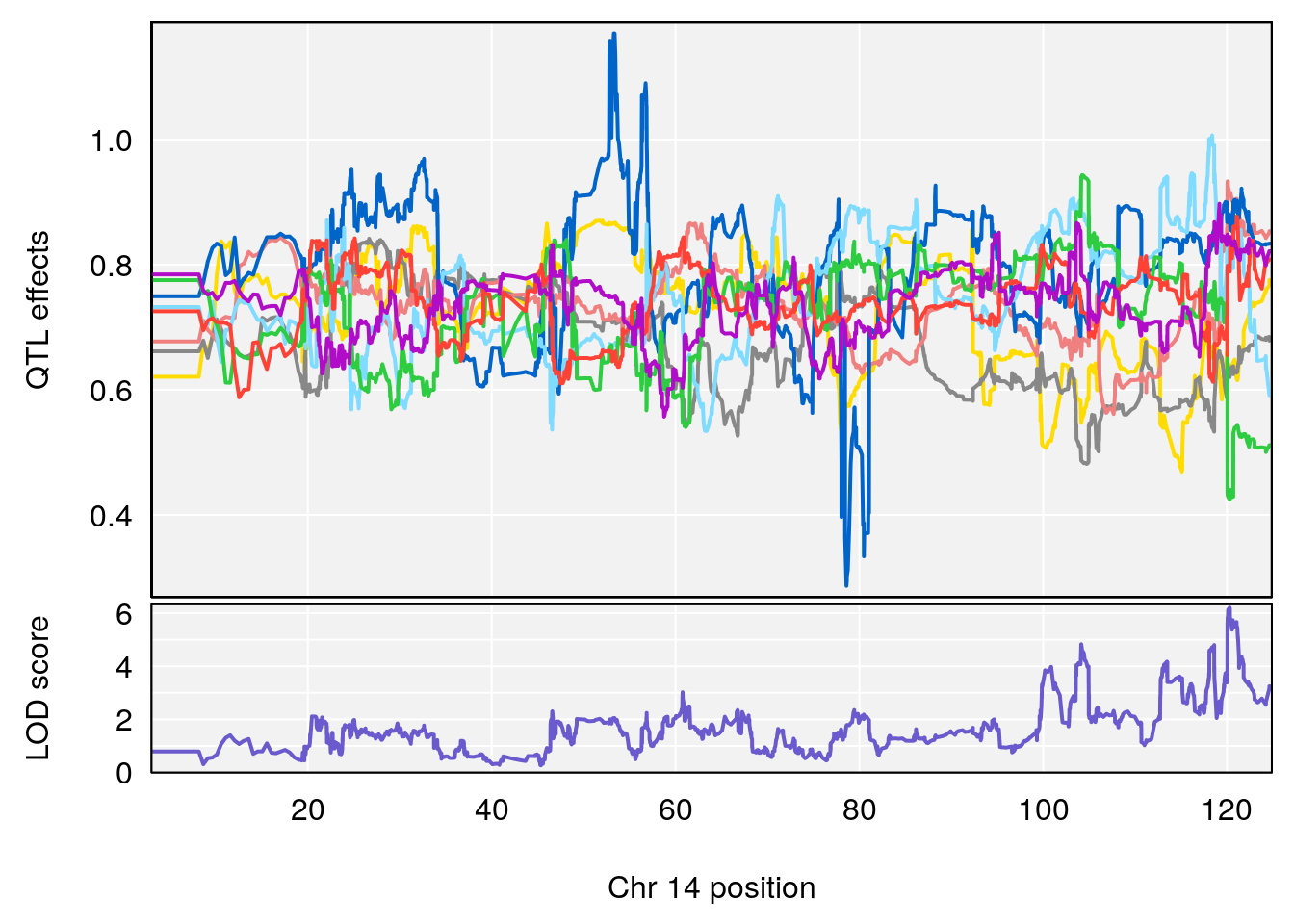
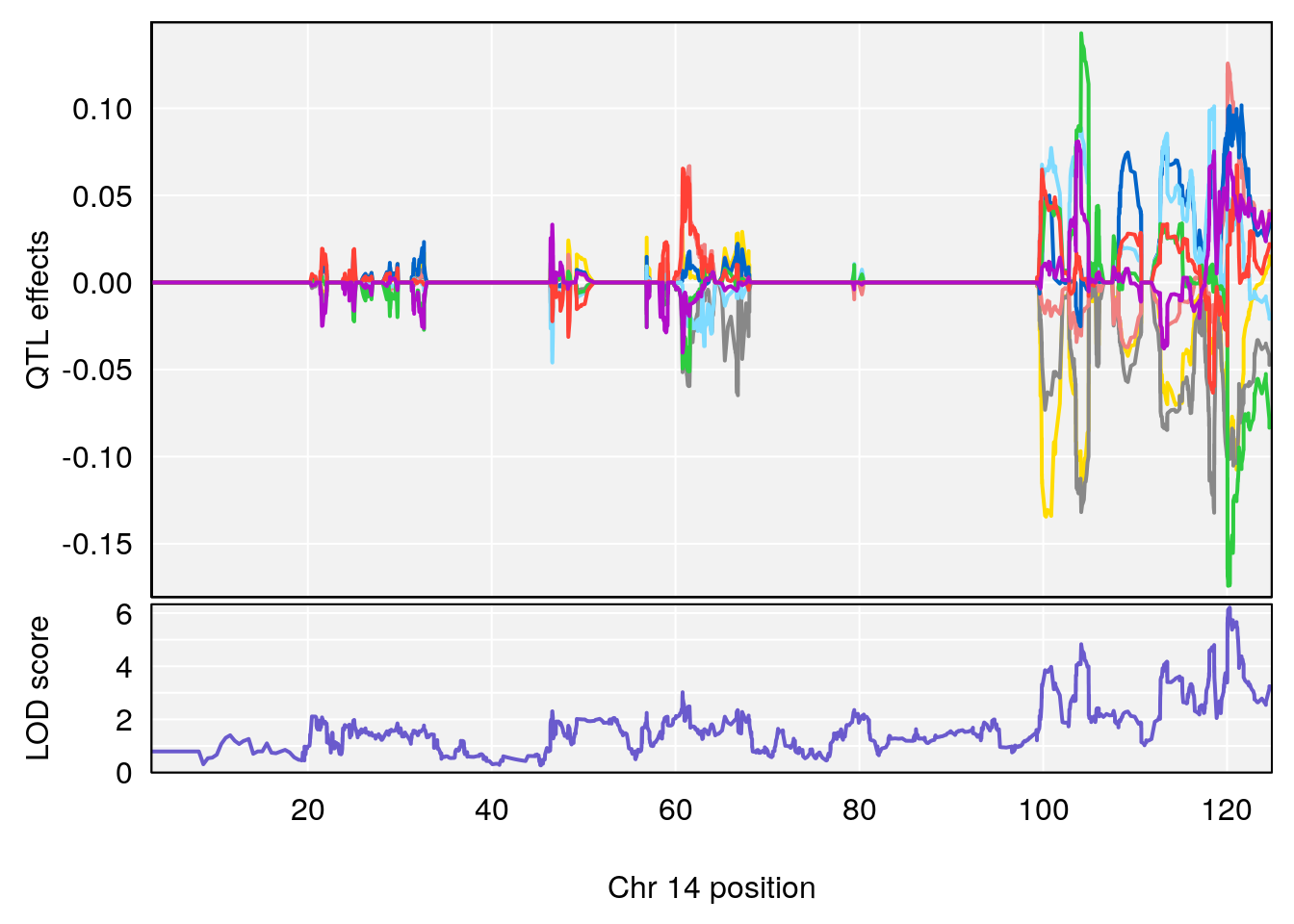
# [1] 6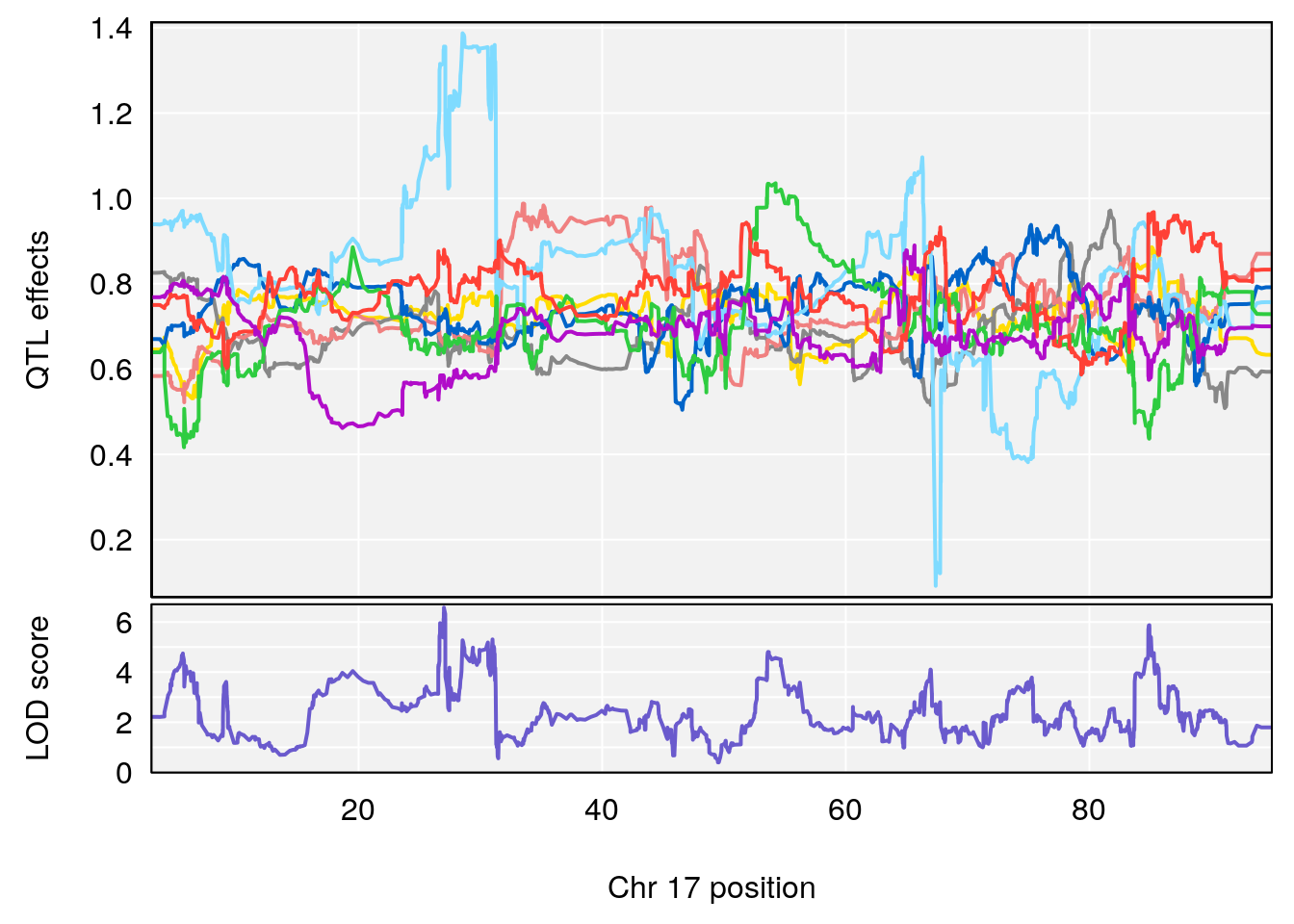
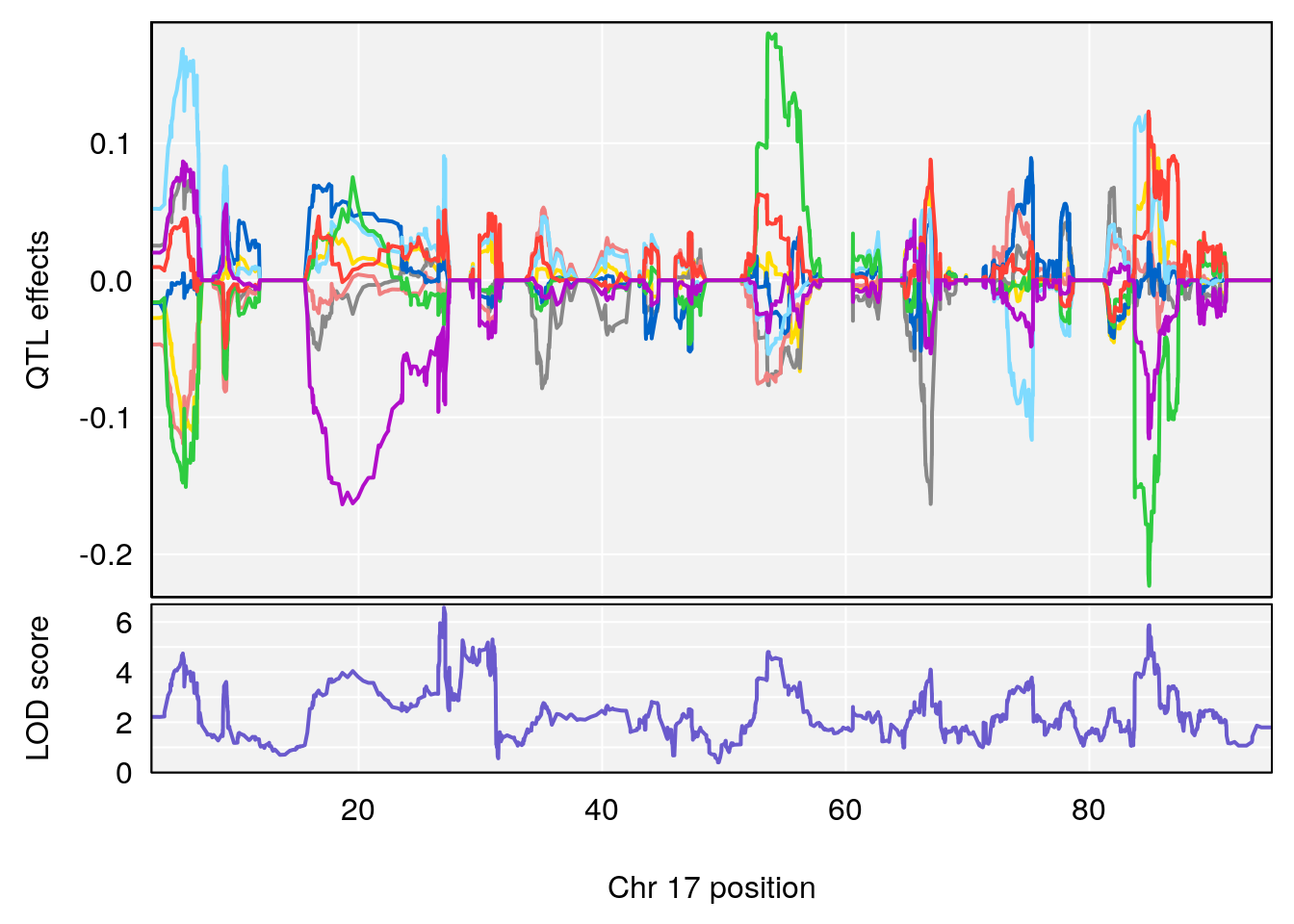
# [1] 7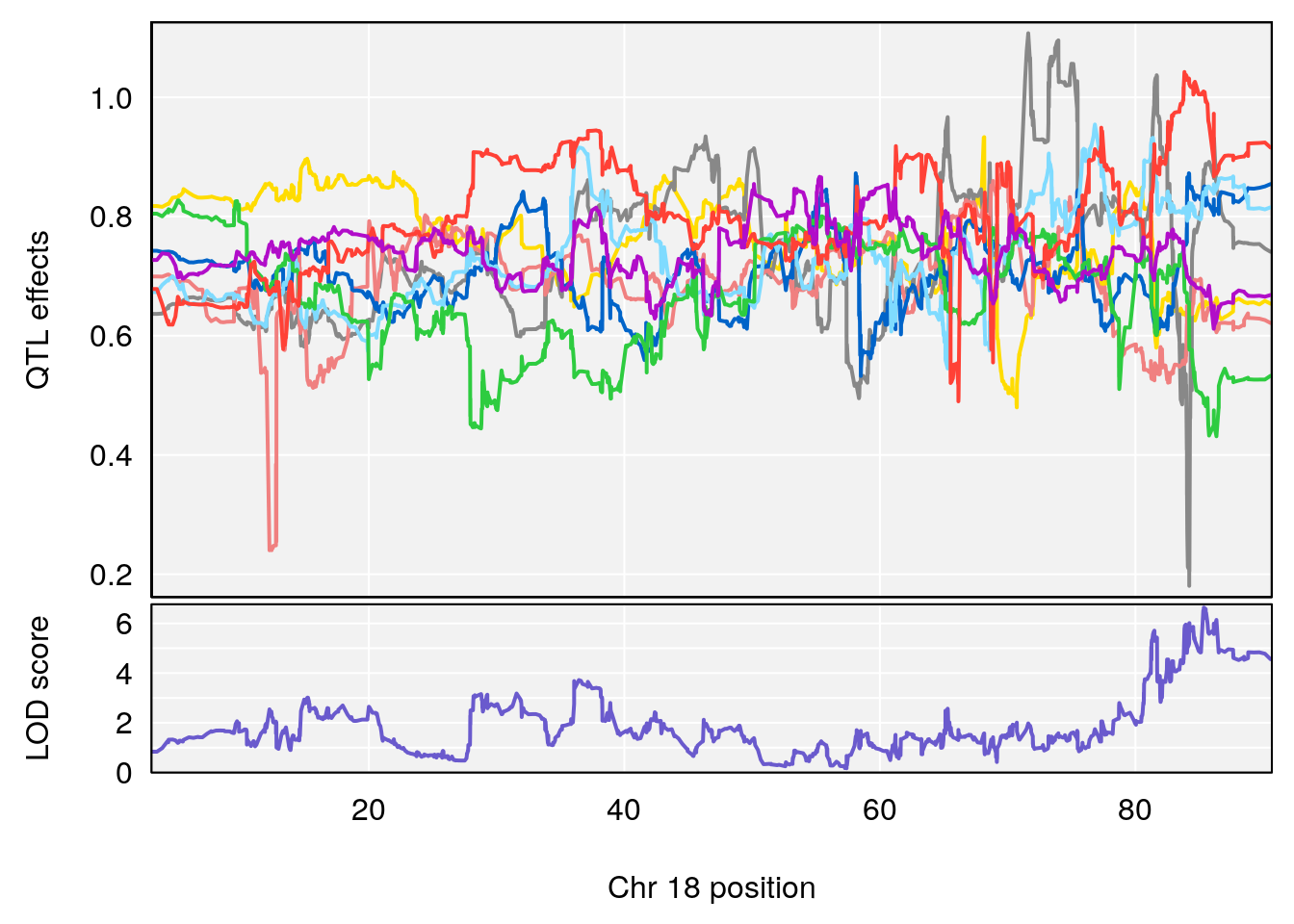
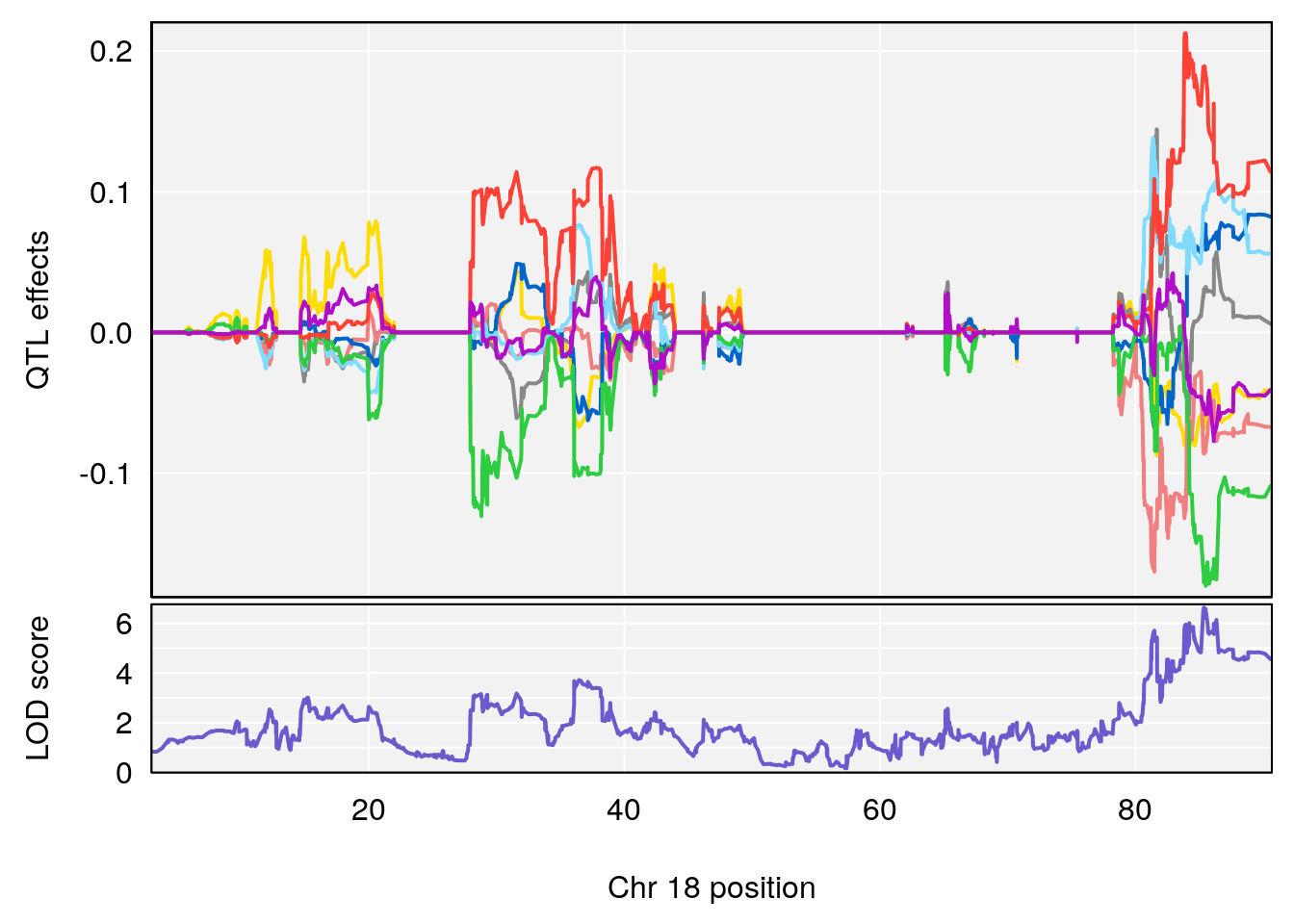
# [1] 8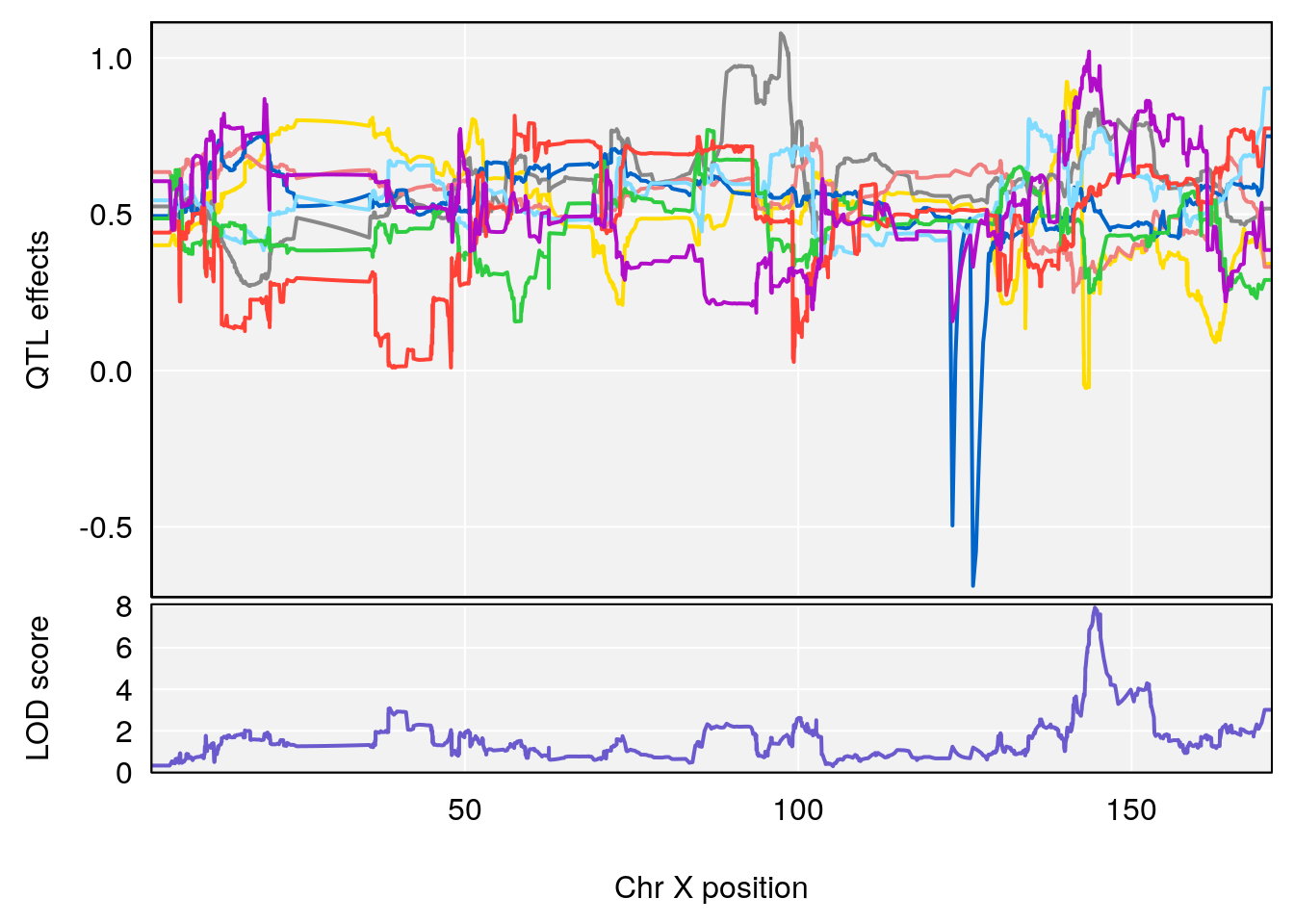
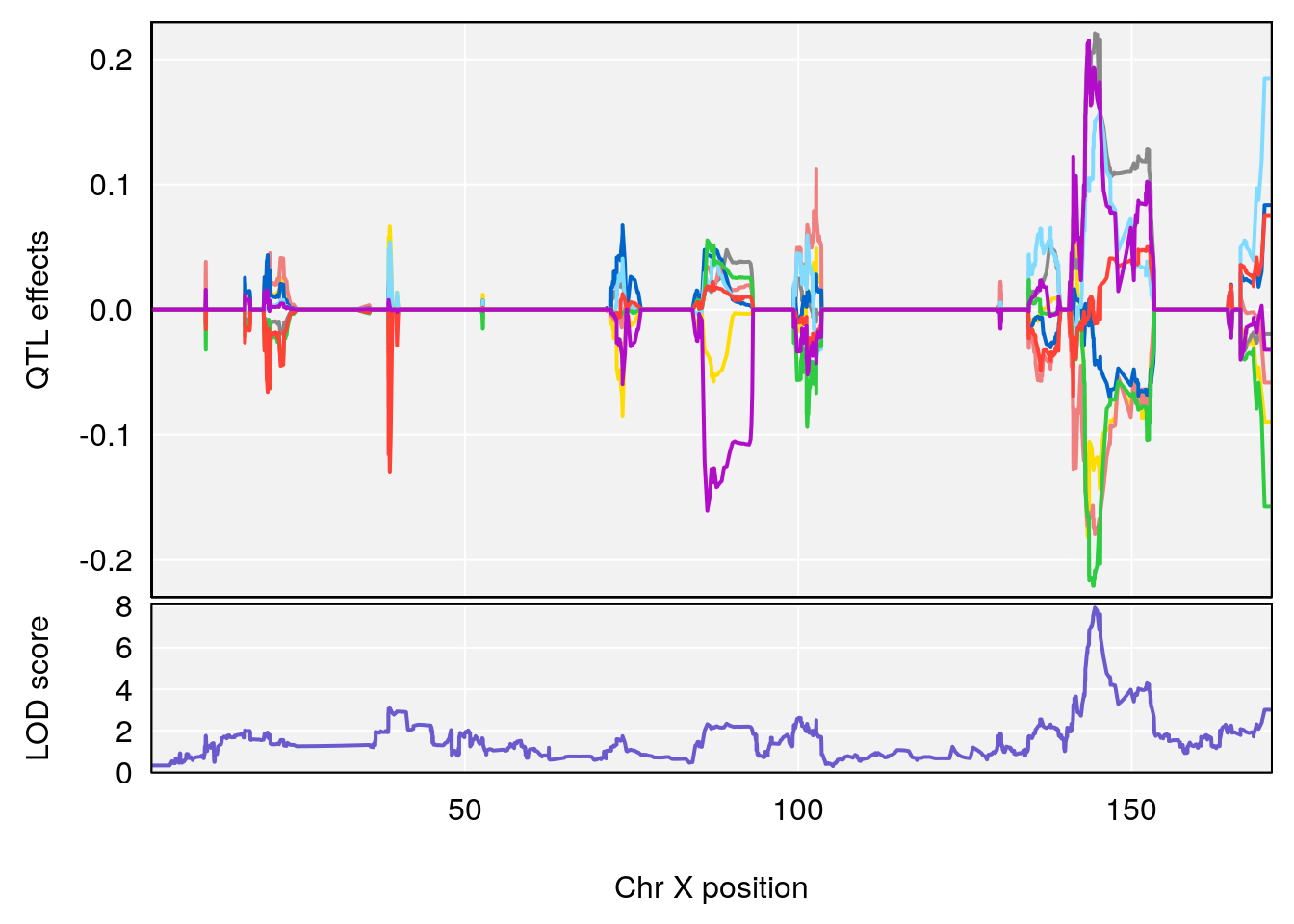
#save peaks coeff plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
fname <- paste("output/DO_morphine_secondbatch_",i,"_coefplot.pdf",sep="")
pdf(file = fname, width = 16, height =8)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
}
dev.off()
}
# [1] "Survival.Time"
# [1] 1
# [1] 2
# [1] 3
# [1] "Recovery.Time"
# [1] 1
# [1] 2
# [1] "Min.depression"
# [1] 1
# [1] "Status_bin"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] 5
# [1] 6
# [1] 7
# [1] 8
#save peaks coeff blup plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
fname <- paste("output/DO_morphine_secondbatch_",i,"_coefplot_blup.pdf",sep="")
pdf(file = fname, width = 16, height =8)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
}
dev.off()
}
# [1] "Survival.Time"
# [1] 1
# [1] 2
# [1] 3
# [1] "Recovery.Time"
# [1] 1
# [1] 2
# [1] "Min.depression"
# [1] 1
# [1] "Status_bin"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] 5
# [1] 6
# [1] 7
# [1] 8Plot qtl mapping on 69K_m2
Plot by using its output qtl.morphine.69k.out.combined_m2.RData
load("output/qtl.morphine.69k.out.combined_m2.RData")
#pmap and gmap
load("data/69k_grid_pgmap.RData")
#genome-wide plot
for(i in names(qtl.morphine.out)){
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.morphine.out[[i]]) # overall maximum LOD score
plot(qtl.morphine.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, 10))
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("DO_morphine_",i))
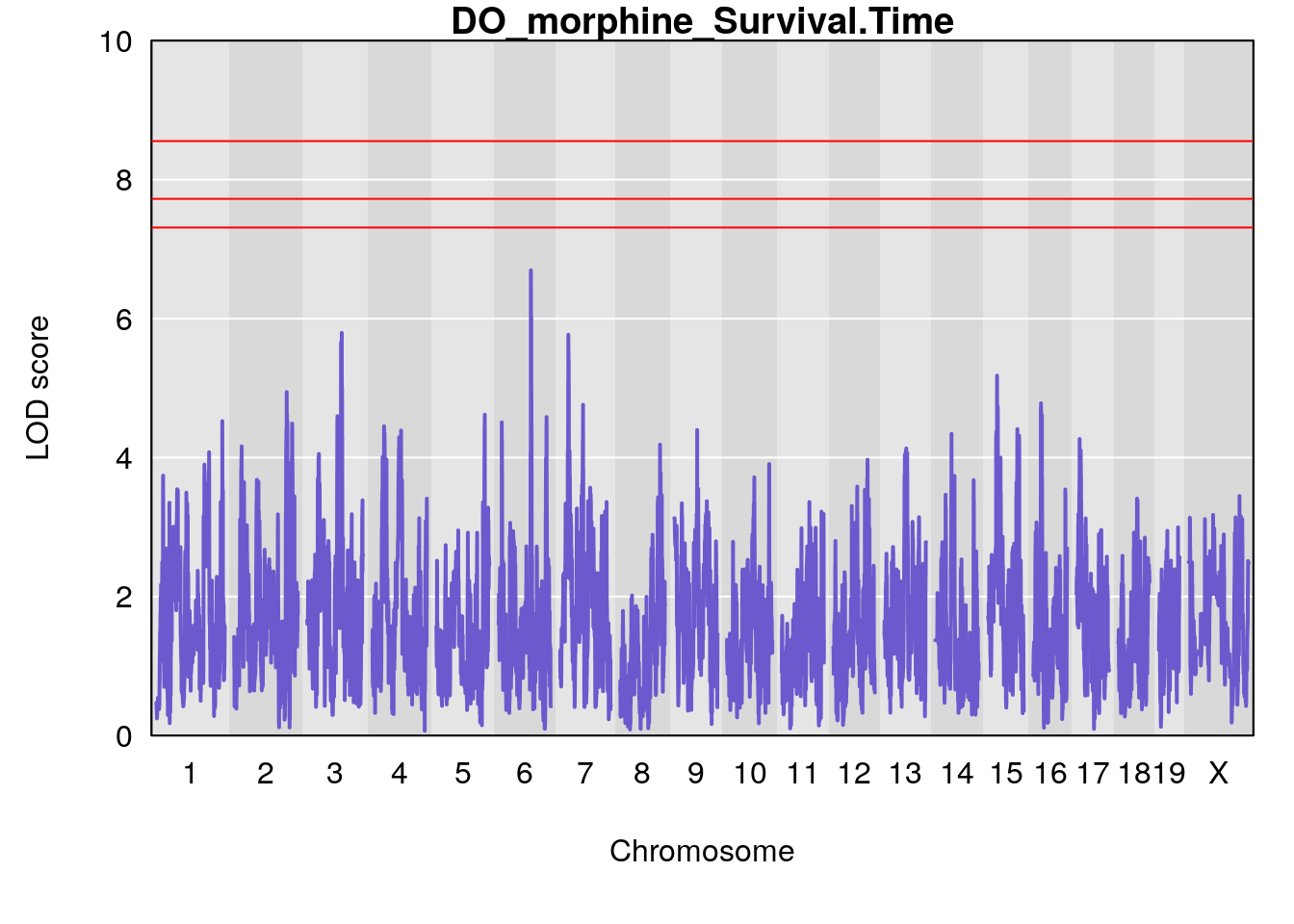
}
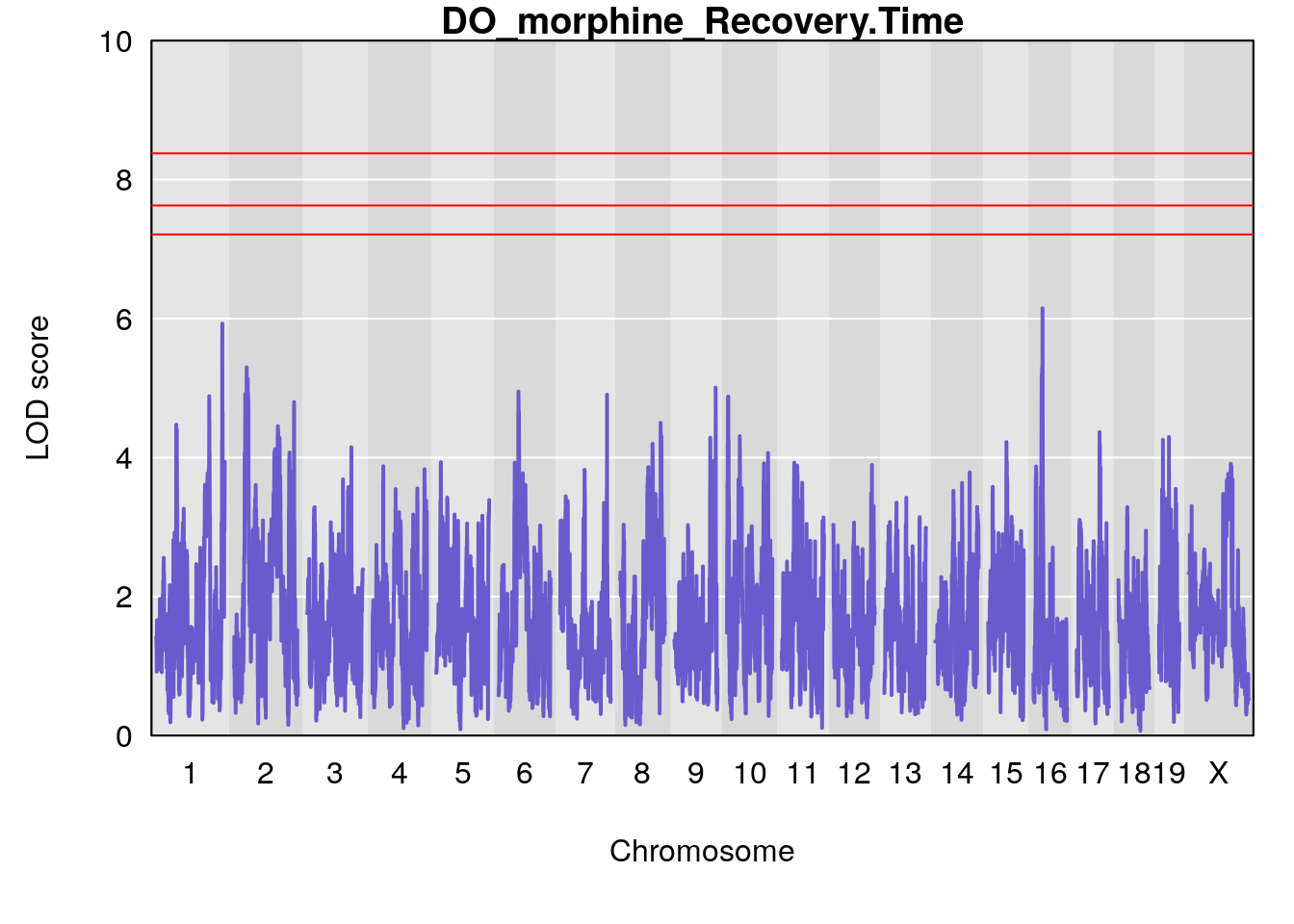
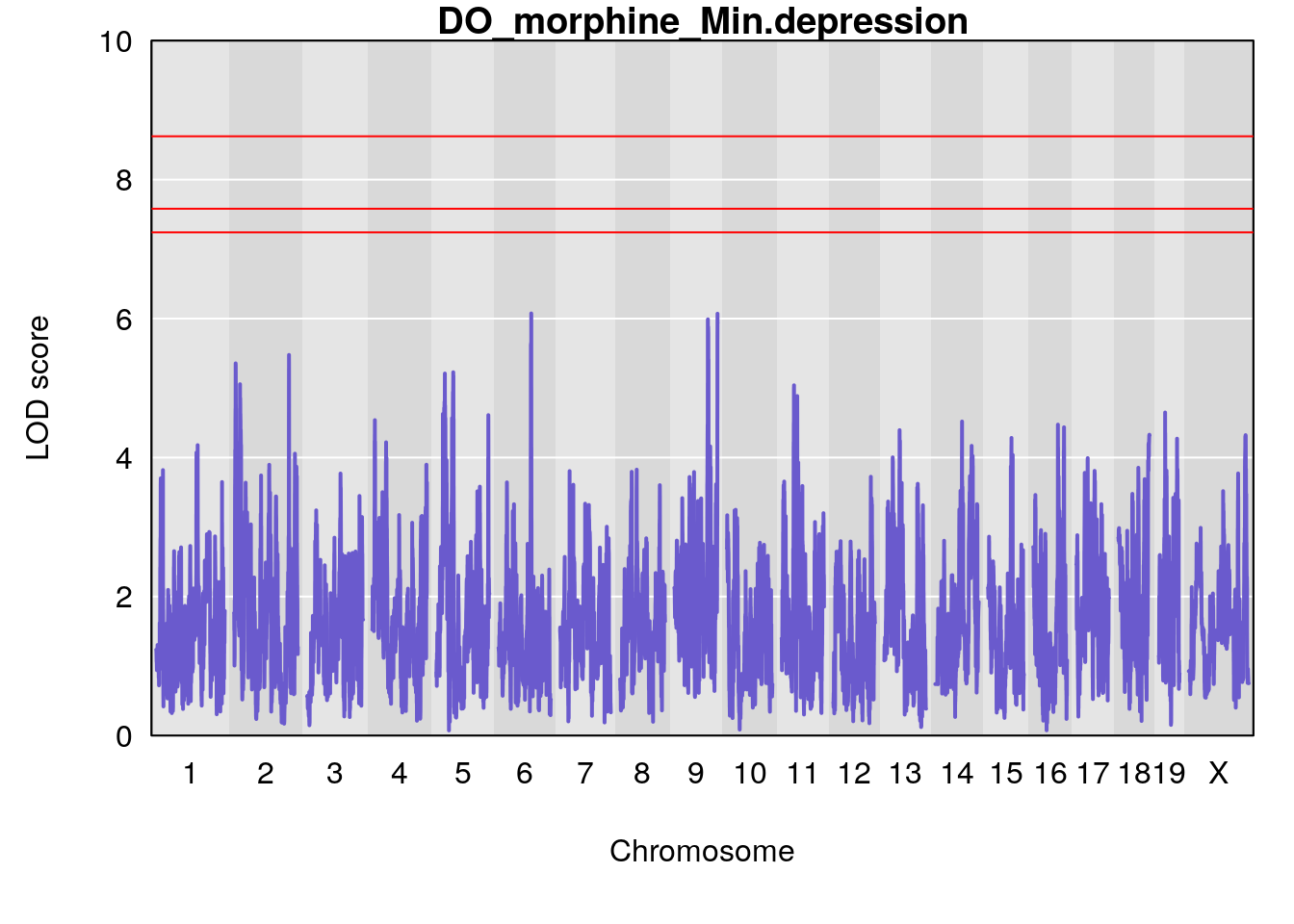
#save genome-wide plot
for(i in names(qtl.morphine.out)){
png(file = paste0("output/DO_morphine_combined_69k_m2_", i, ".png"), width = 16, height =8, res=300, units = "in")
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.morphine.out[[i]]) # overall maximum LOD score
plot(qtl.morphine.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, 10))
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("DO_morphine_",i))
dev.off()
}
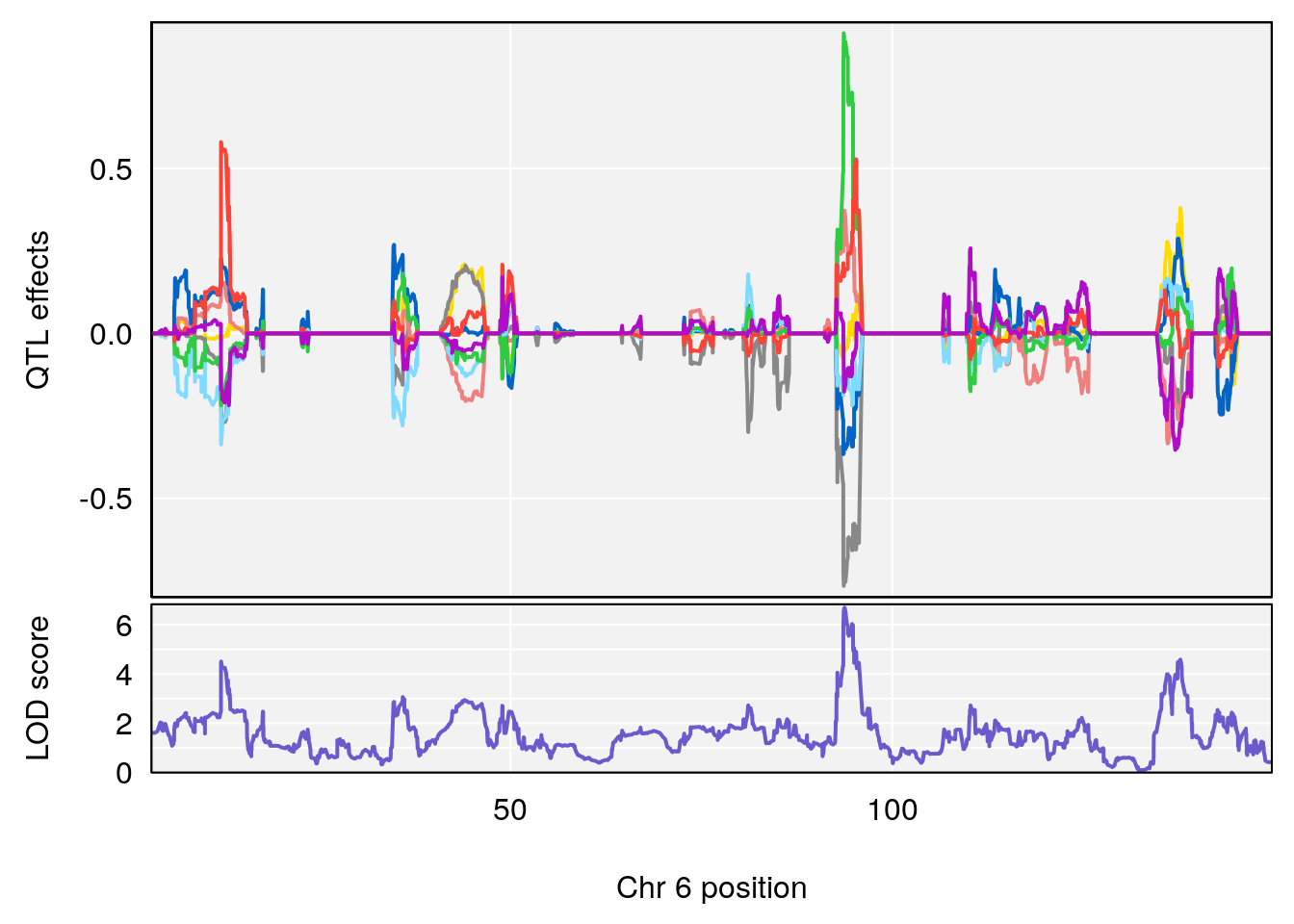
#peaks coeff plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
print(peaks)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
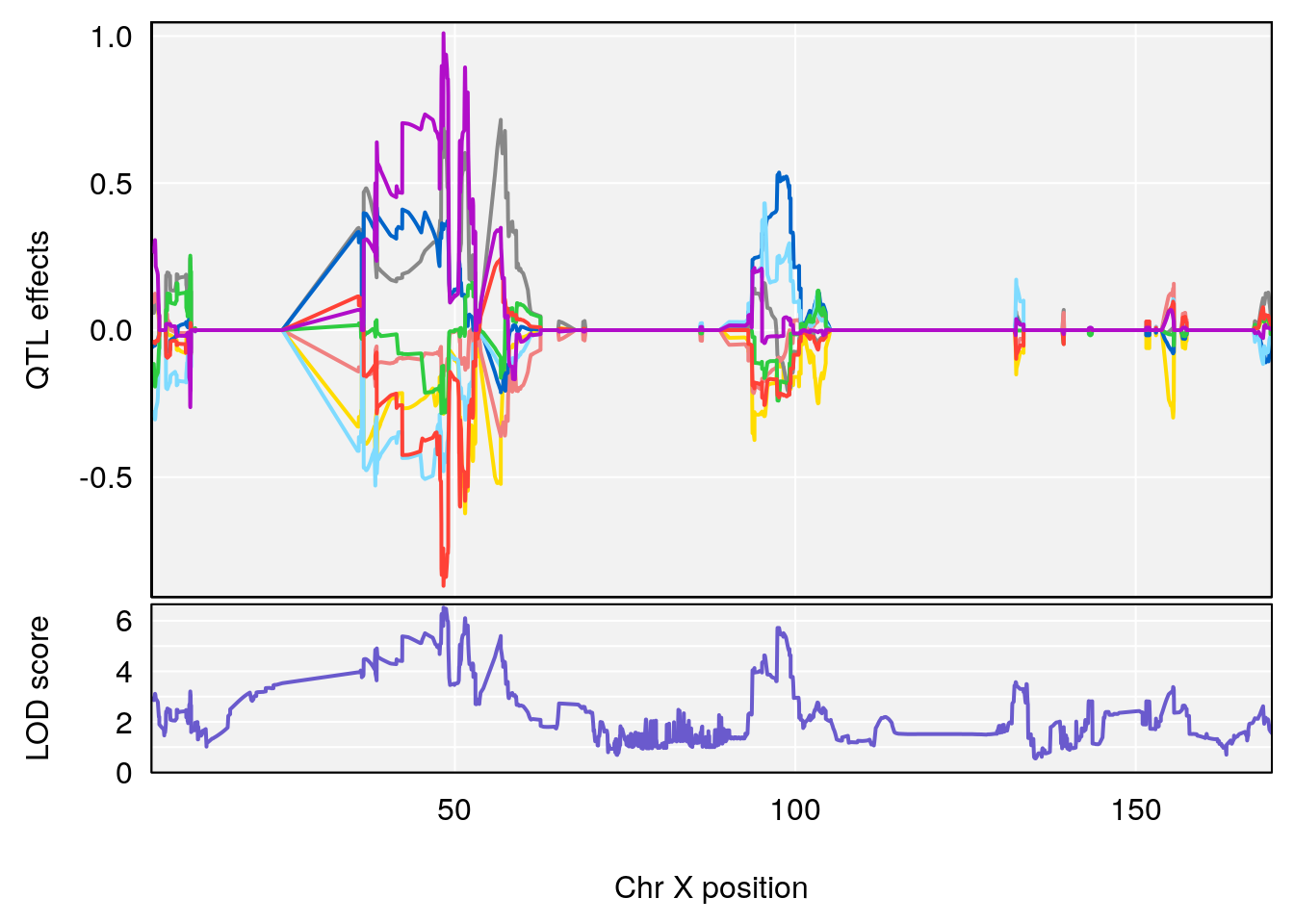
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
}
}
# [1] "Survival.Time"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 6 93.76935 6.696094 93.60032 94.88262
# [1] 1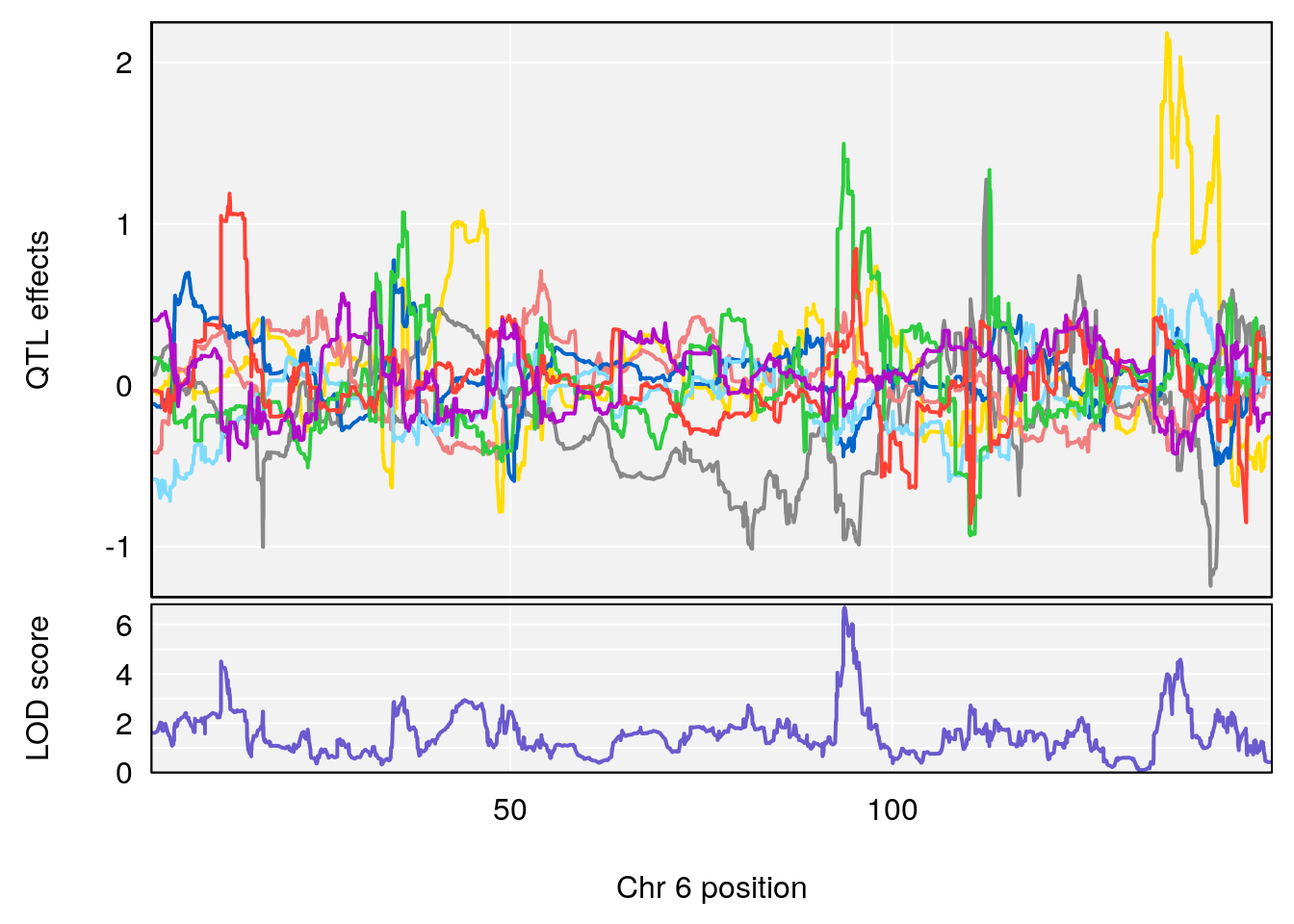
# [1] "Recovery.Time"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 16 29.08447 6.148843 26.48461 29.65254
# [1] 1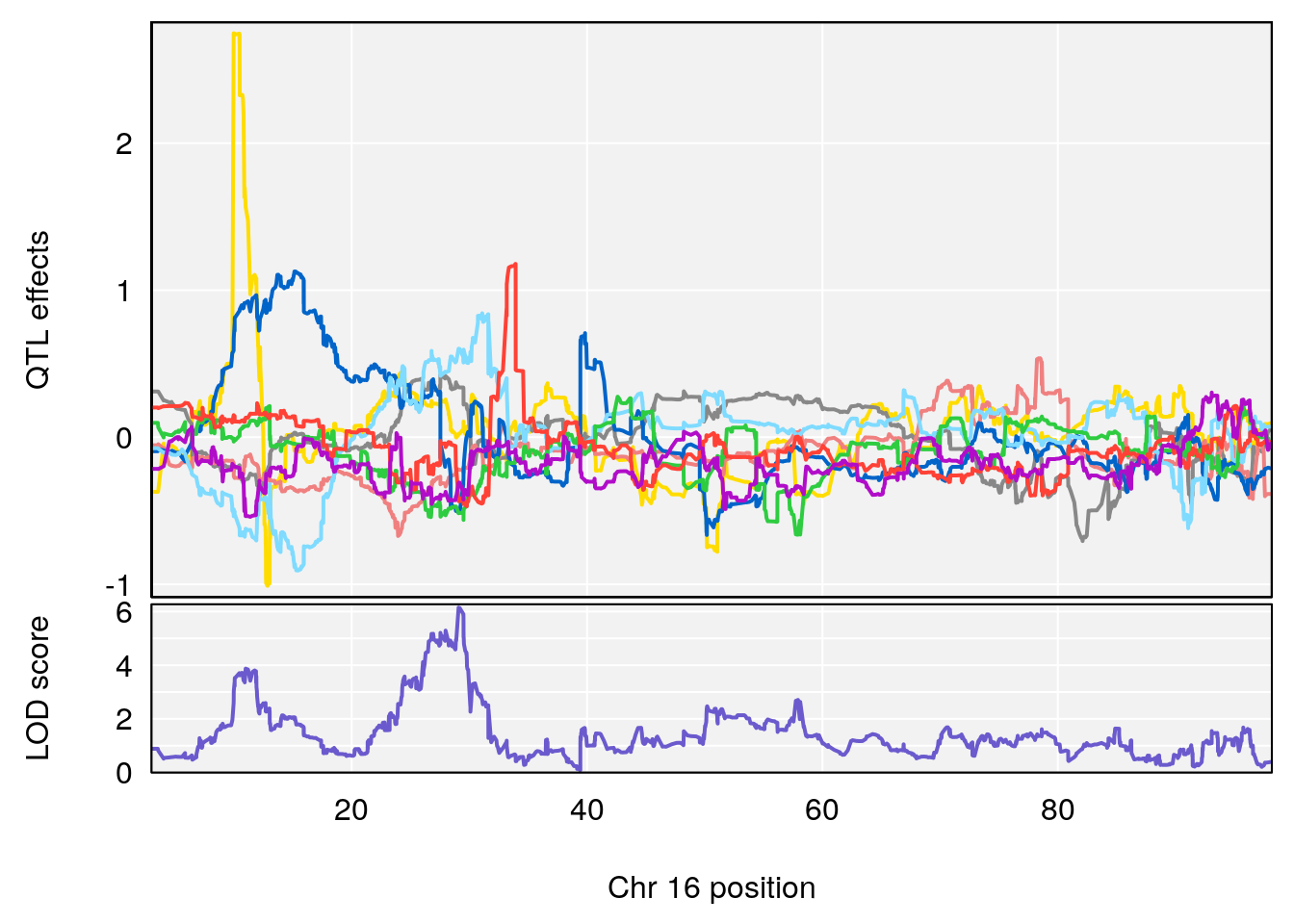
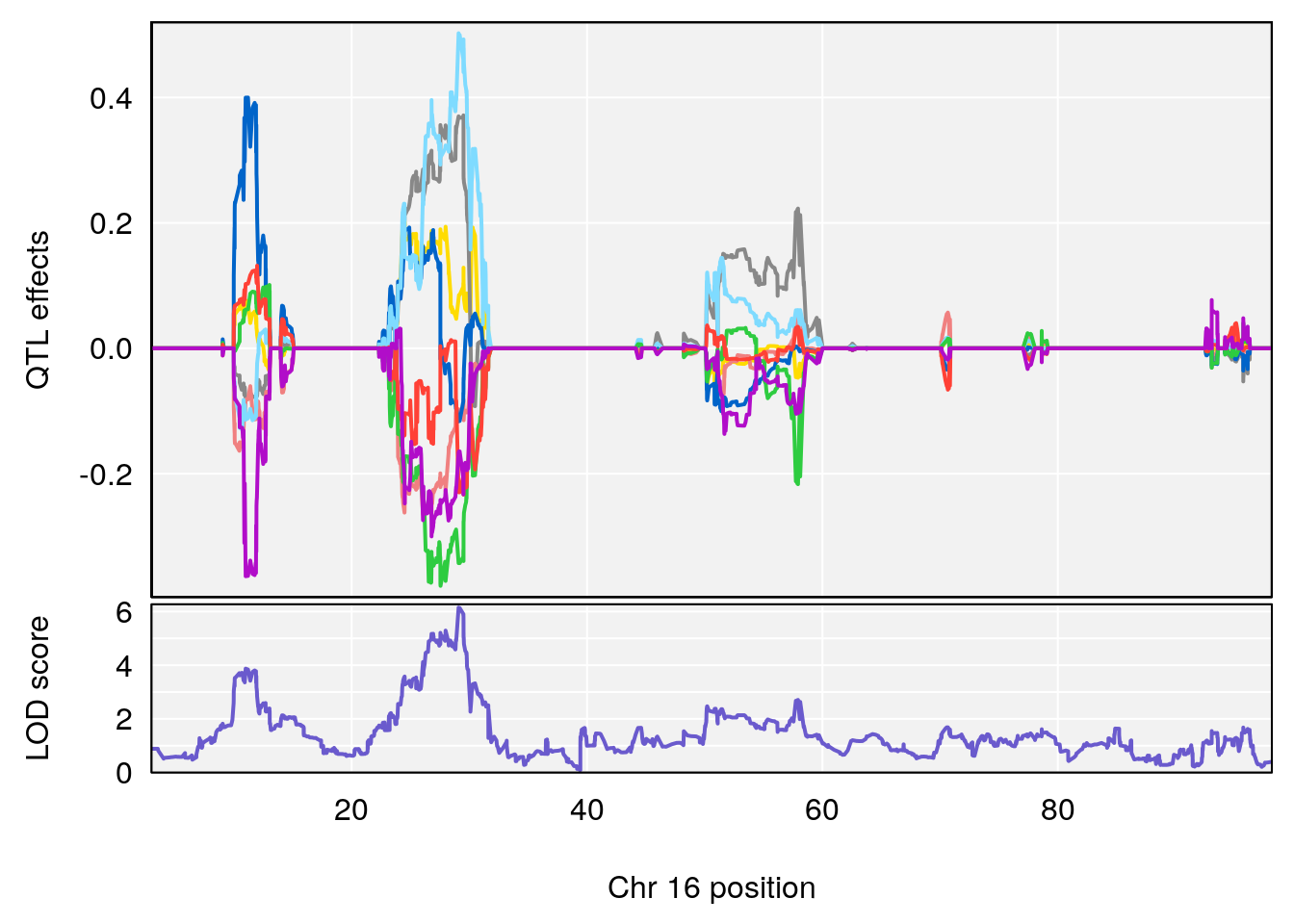
# [1] "Min.depression"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 6 94.75287 6.074533 93.60032 95.01174
# 2 1 pheno1 9 124.11695 6.070446 96.69146 124.58265
# [1] 1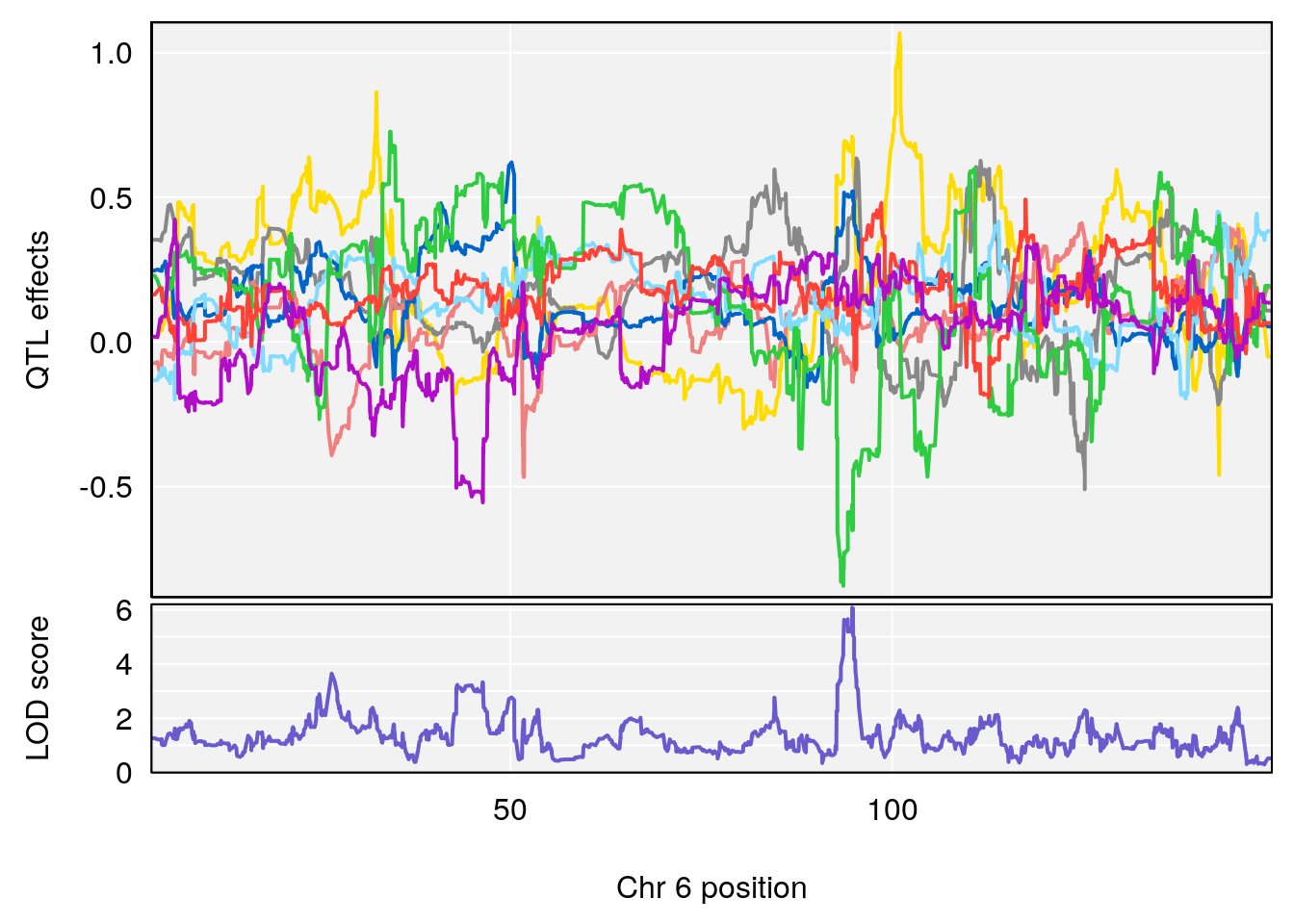
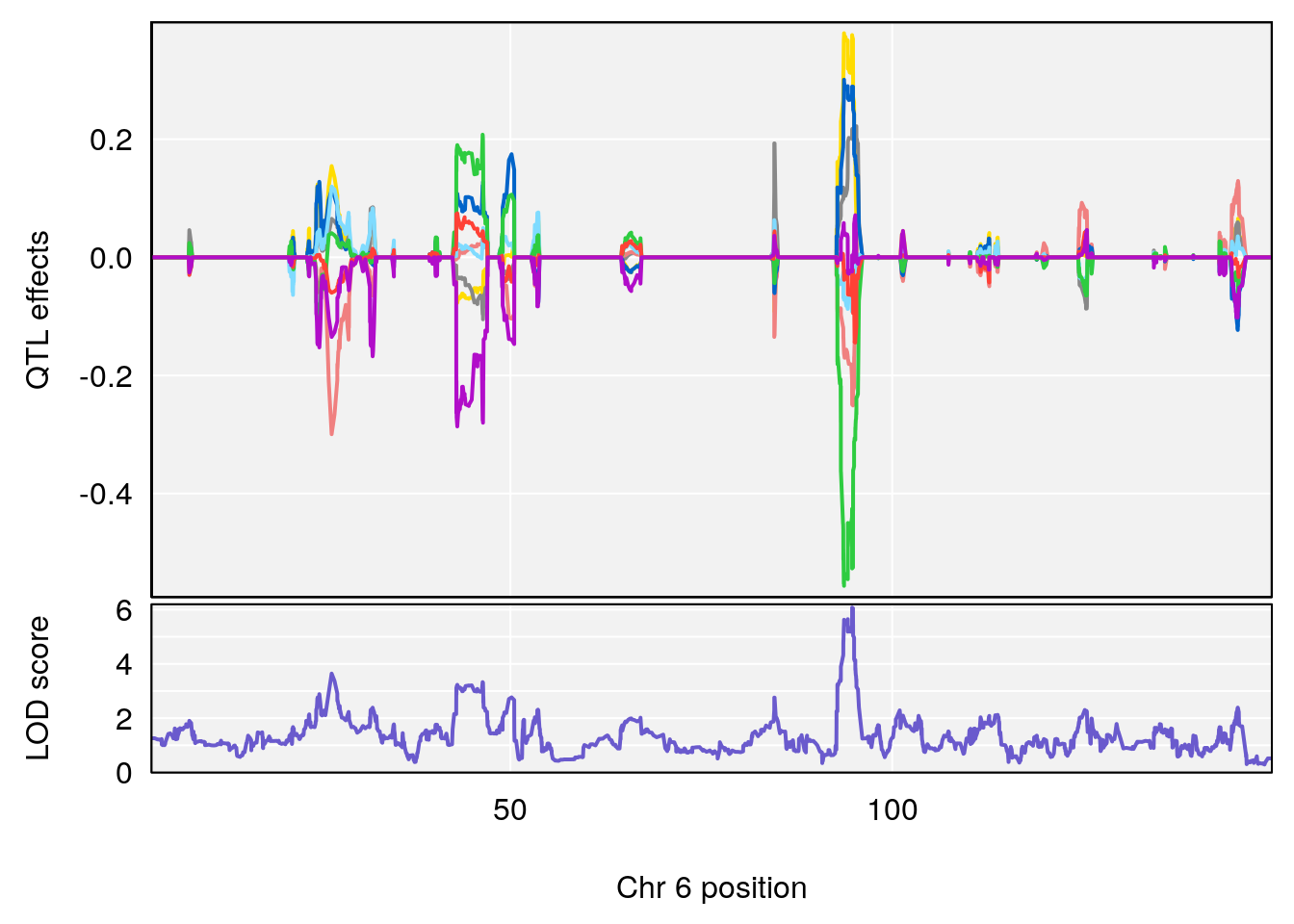
# [1] 2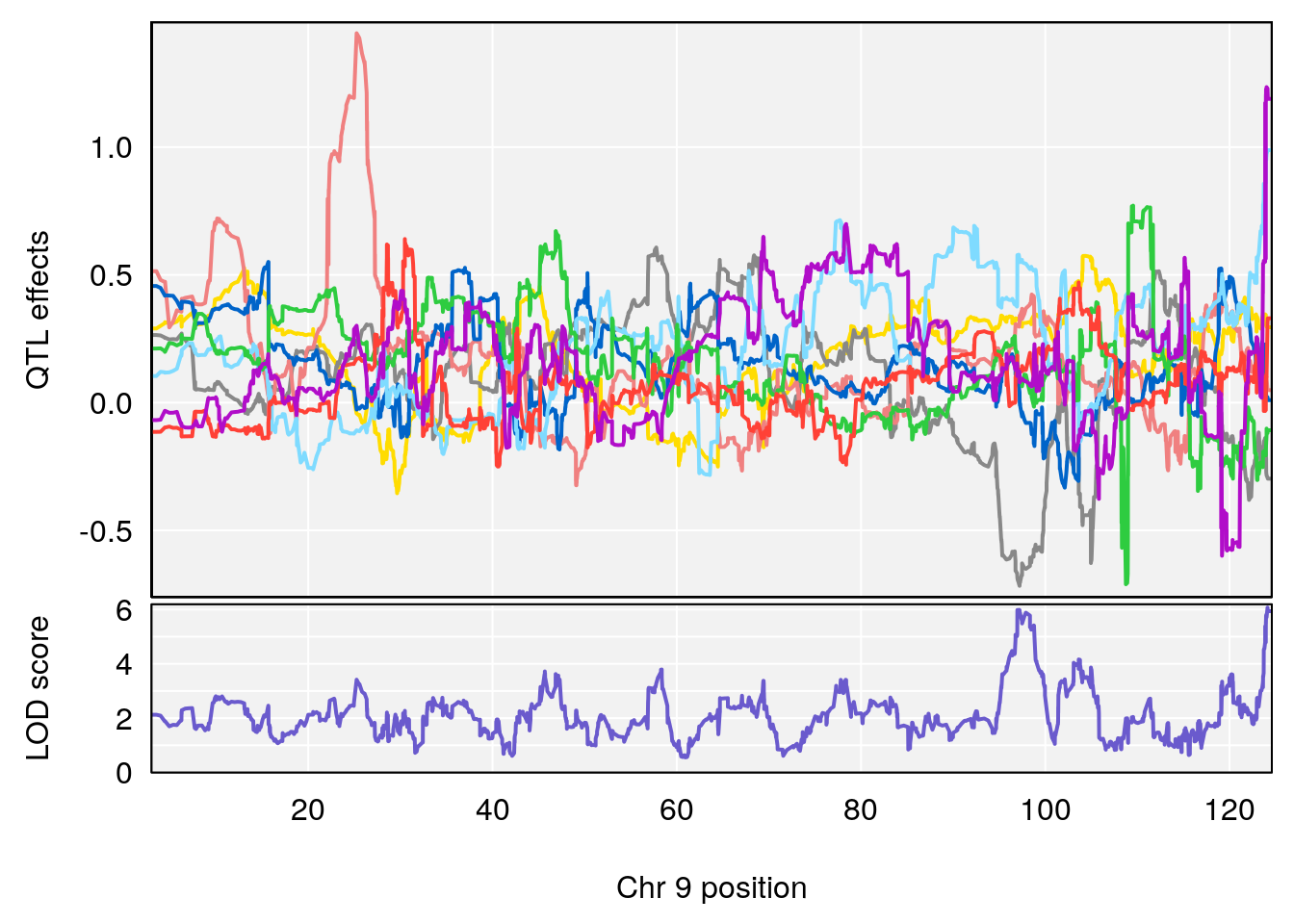
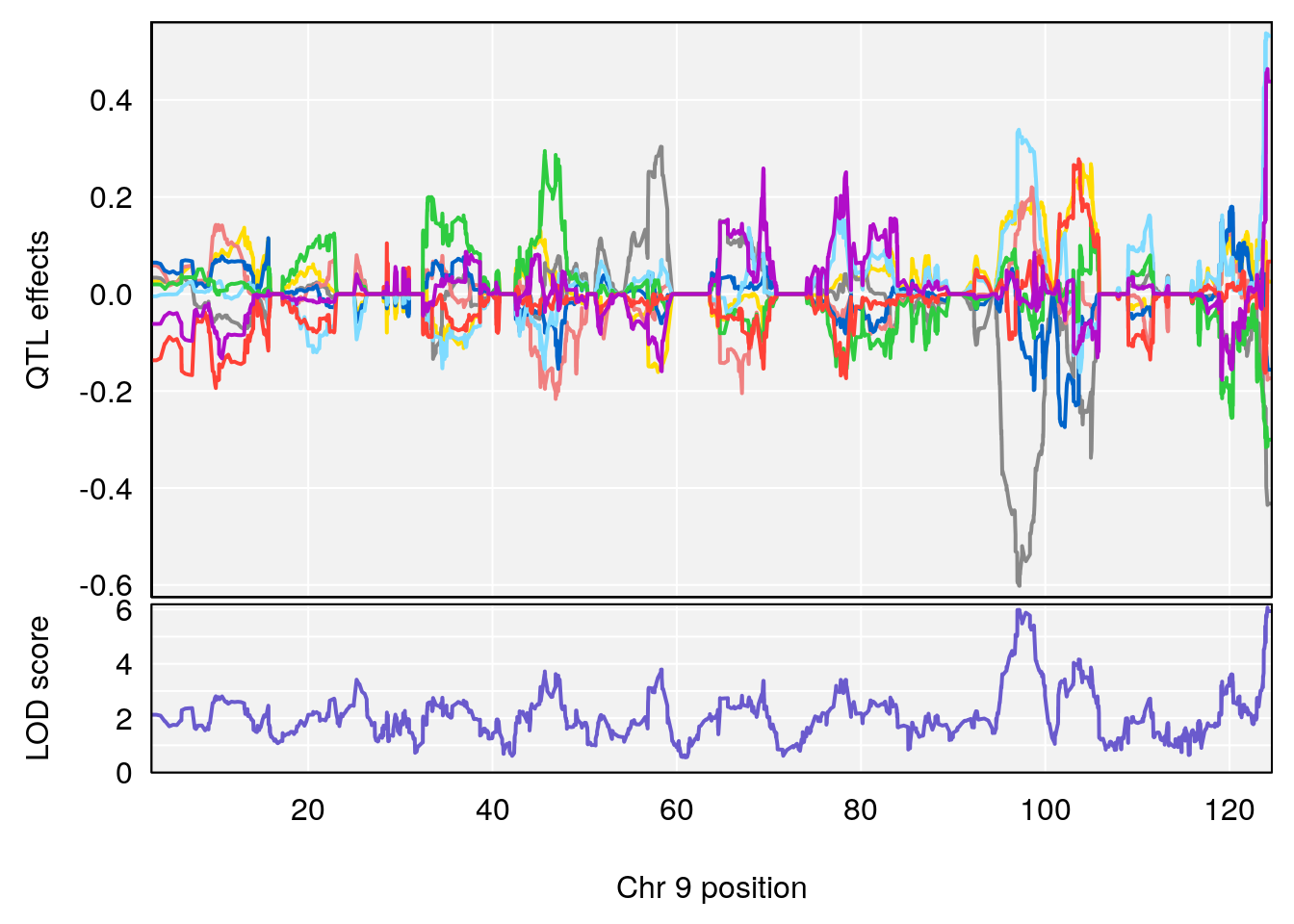
# [1] "Status_bin"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 4 156.17629 6.857142 156.009474 156.49607
# 2 1 pheno1 7 115.16657 6.012871 114.526442 115.46115
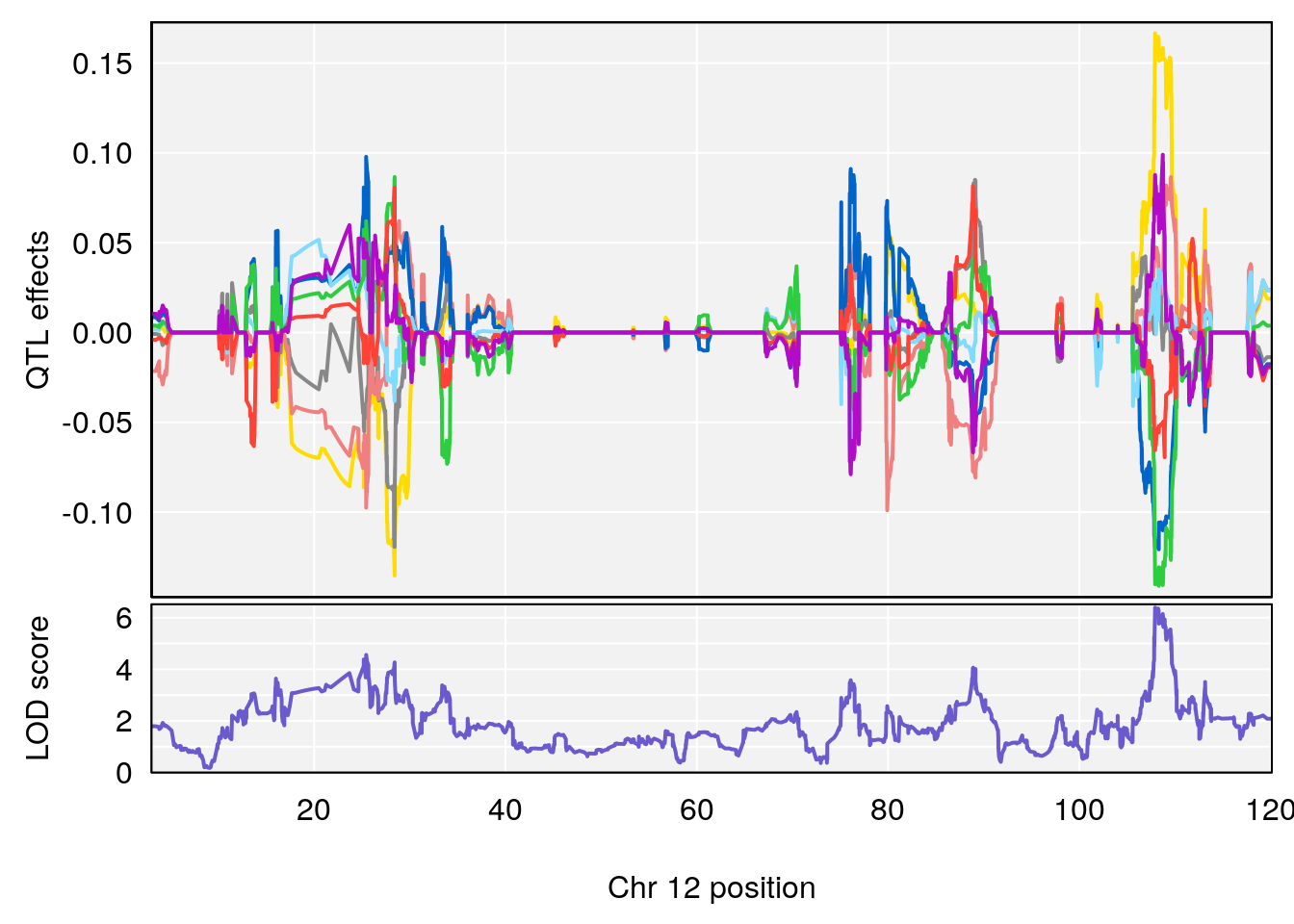
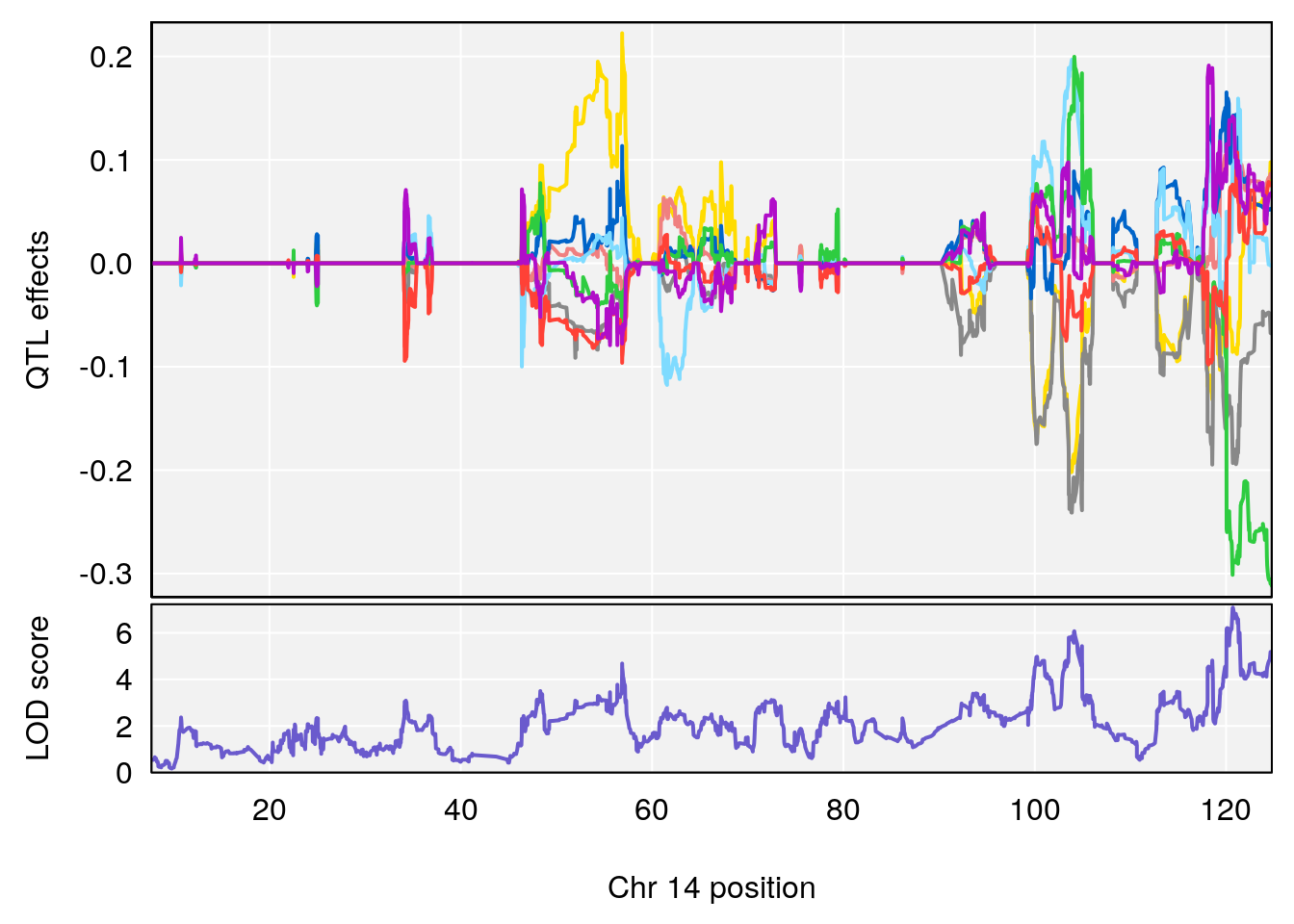
# 3 1 pheno1 10 29.38717 6.819337 8.597626 33.97126
# 4 1 pheno1 11 37.81053 7.082165 35.662586 38.39621
# 5 1 pheno1 12 107.91869 6.389835 107.816680 109.67503
# 6 1 pheno1 17 27.00602 7.424161 26.634222 28.58929
# 7 1 pheno1 18 85.49980 7.067088 81.327205 86.51269
# 8 1 pheno1 X 144.48572 7.256846 143.604522 145.79861
# [1] 1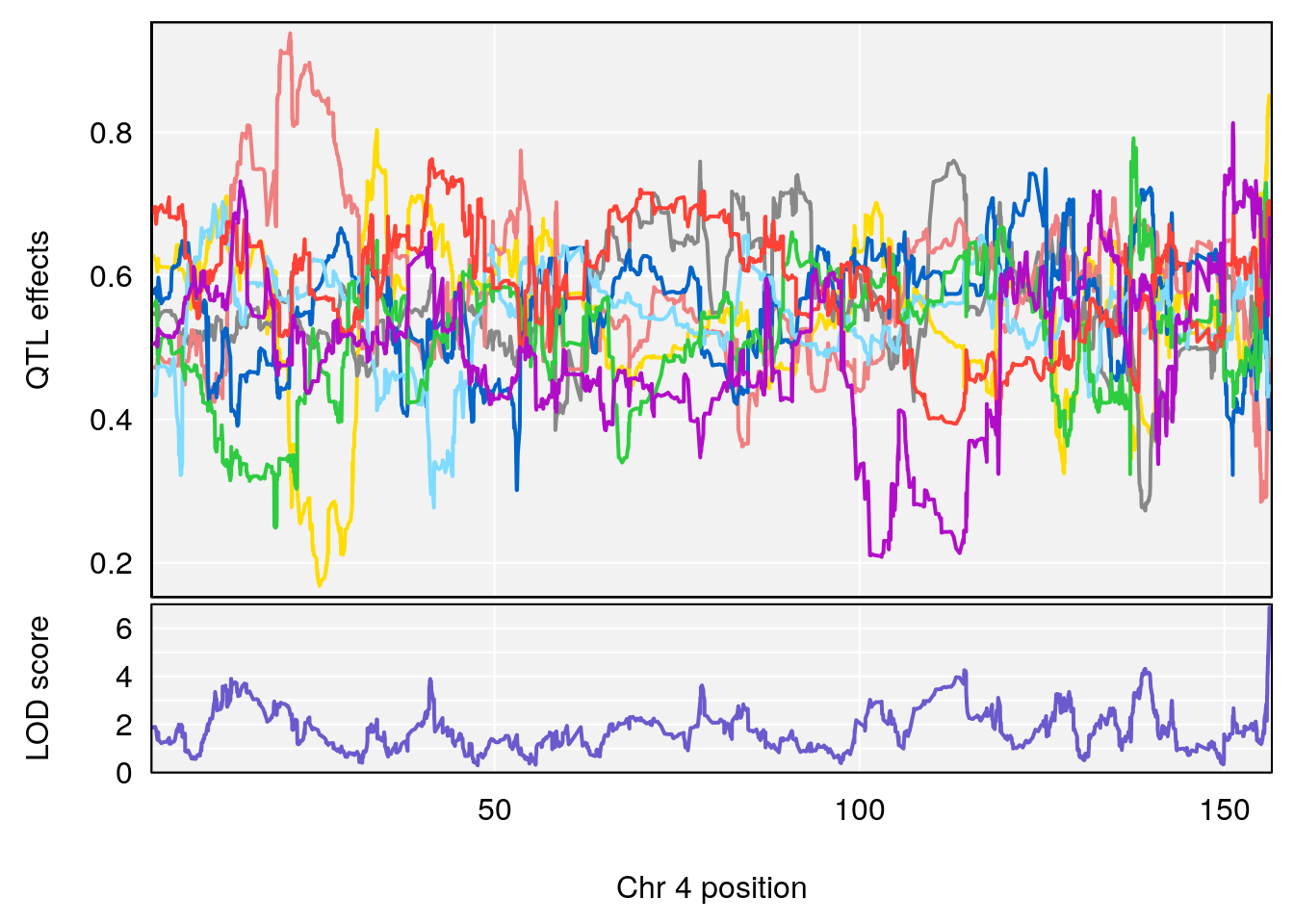
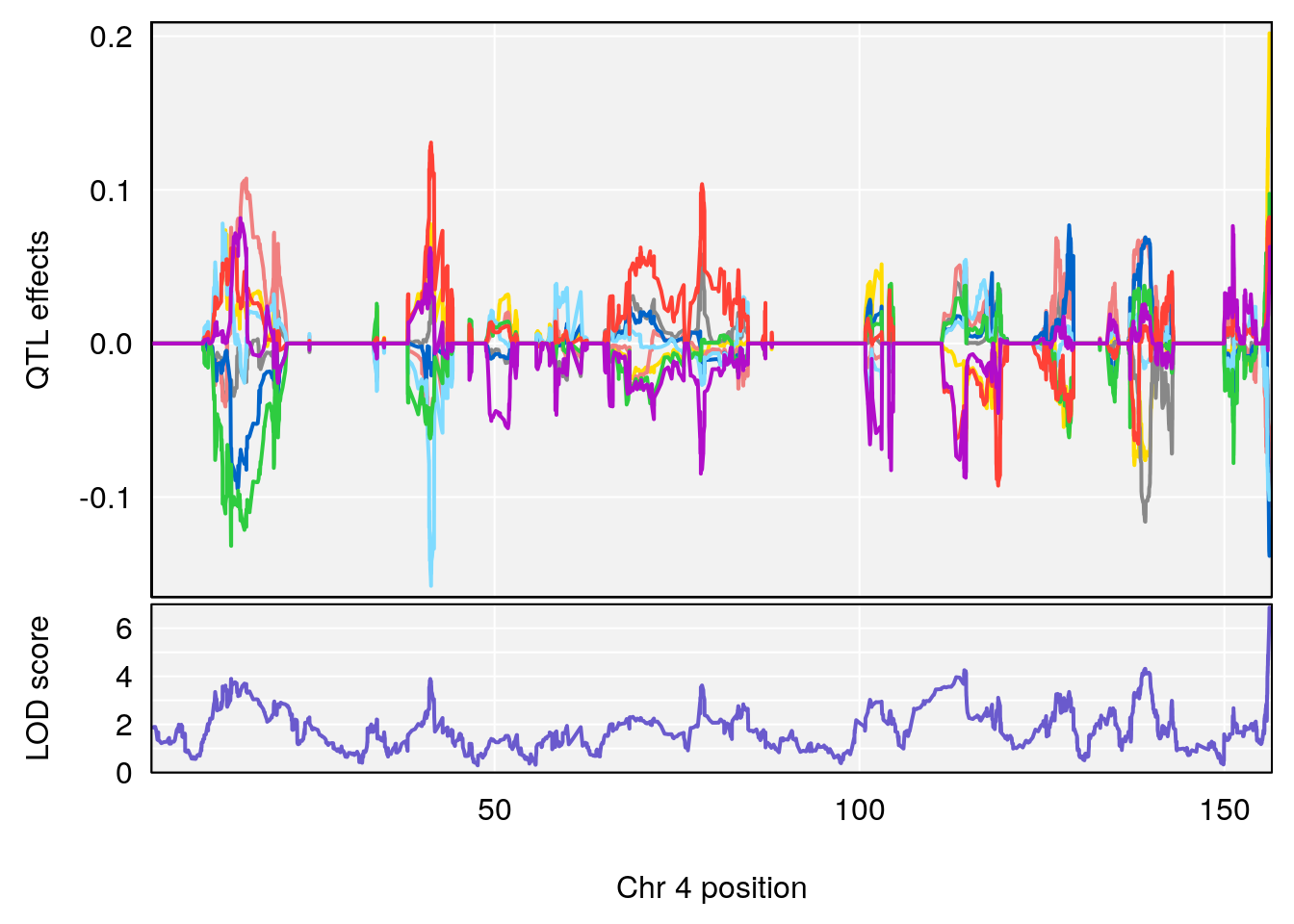
# [1] 2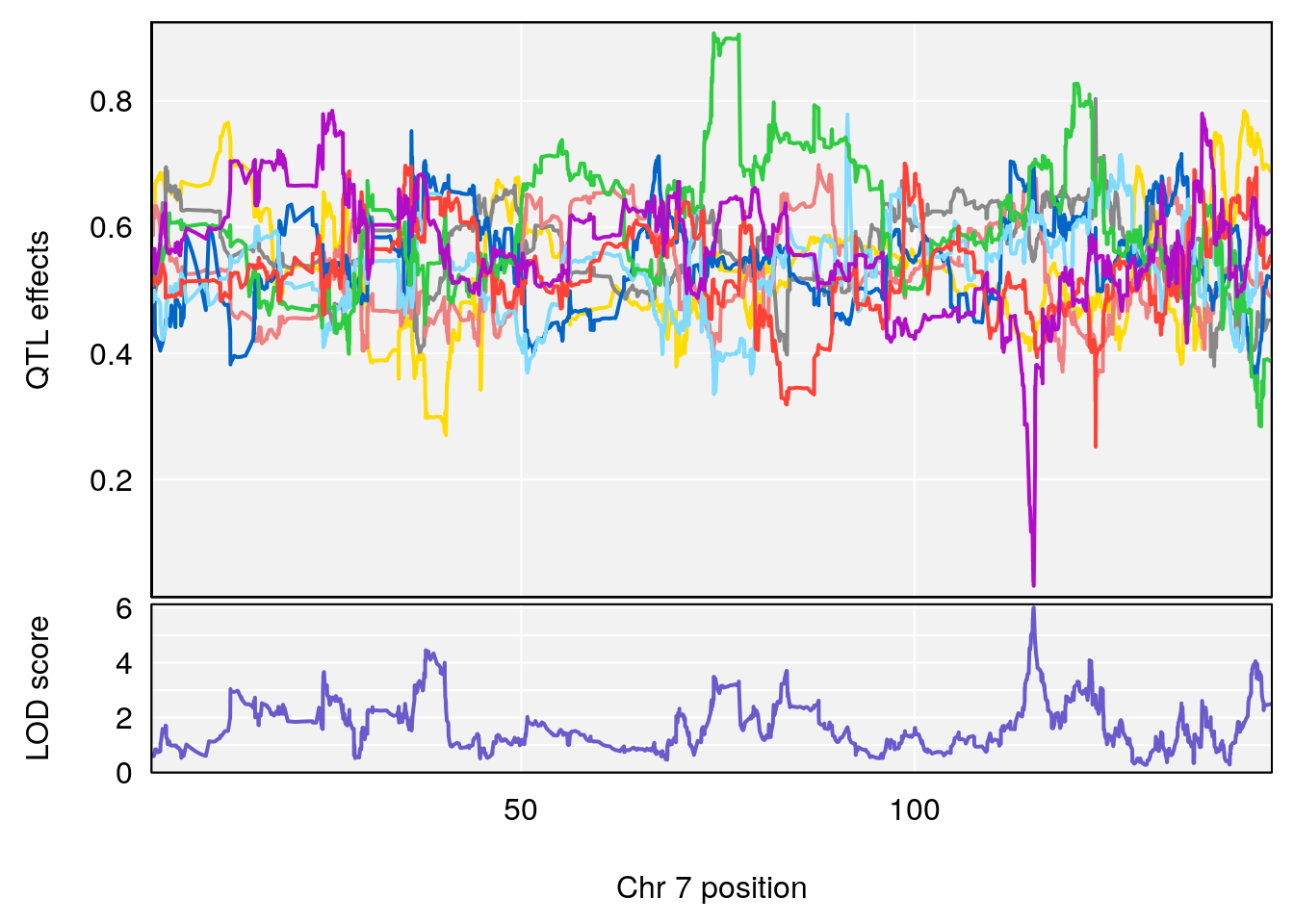
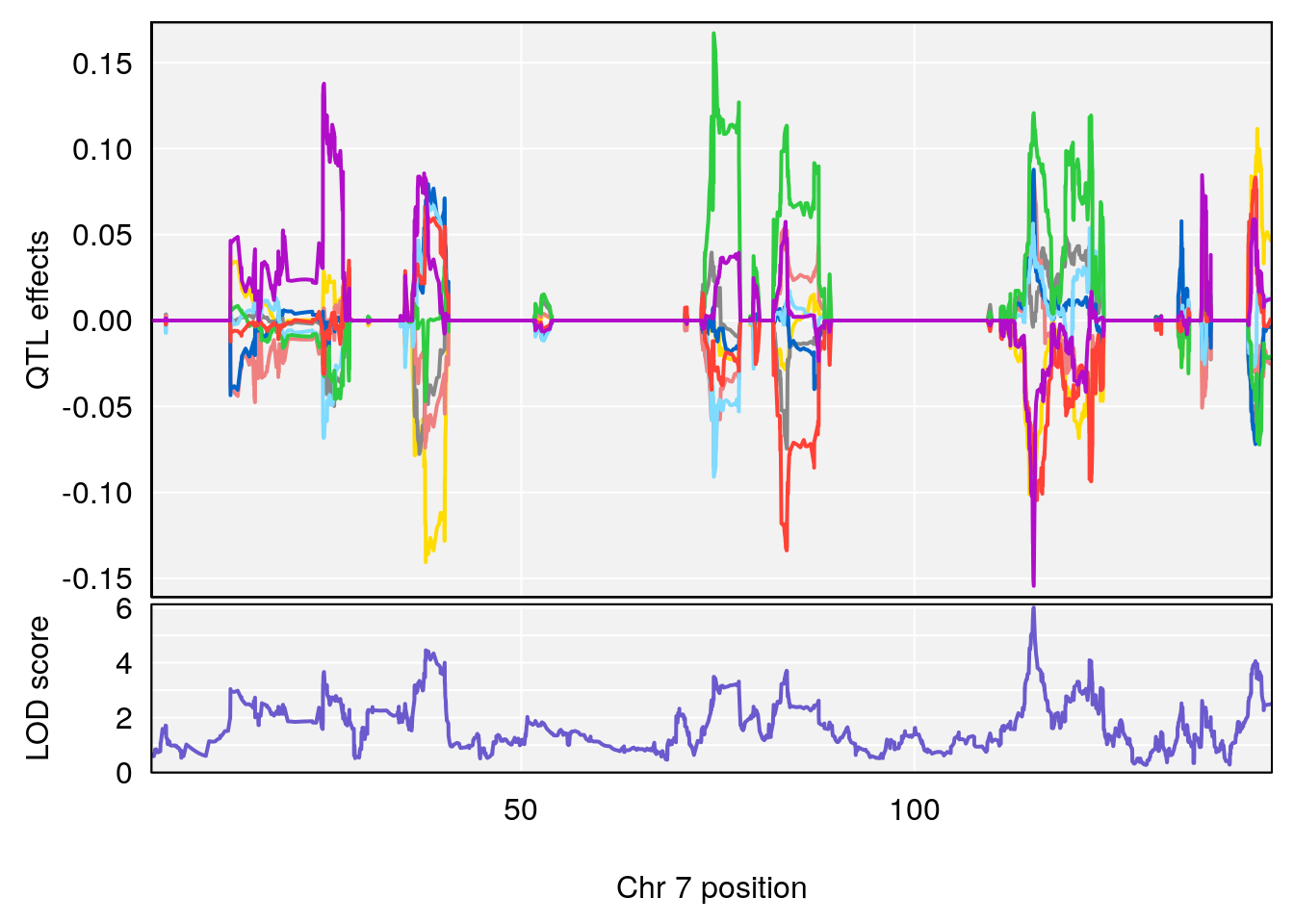
# [1] 3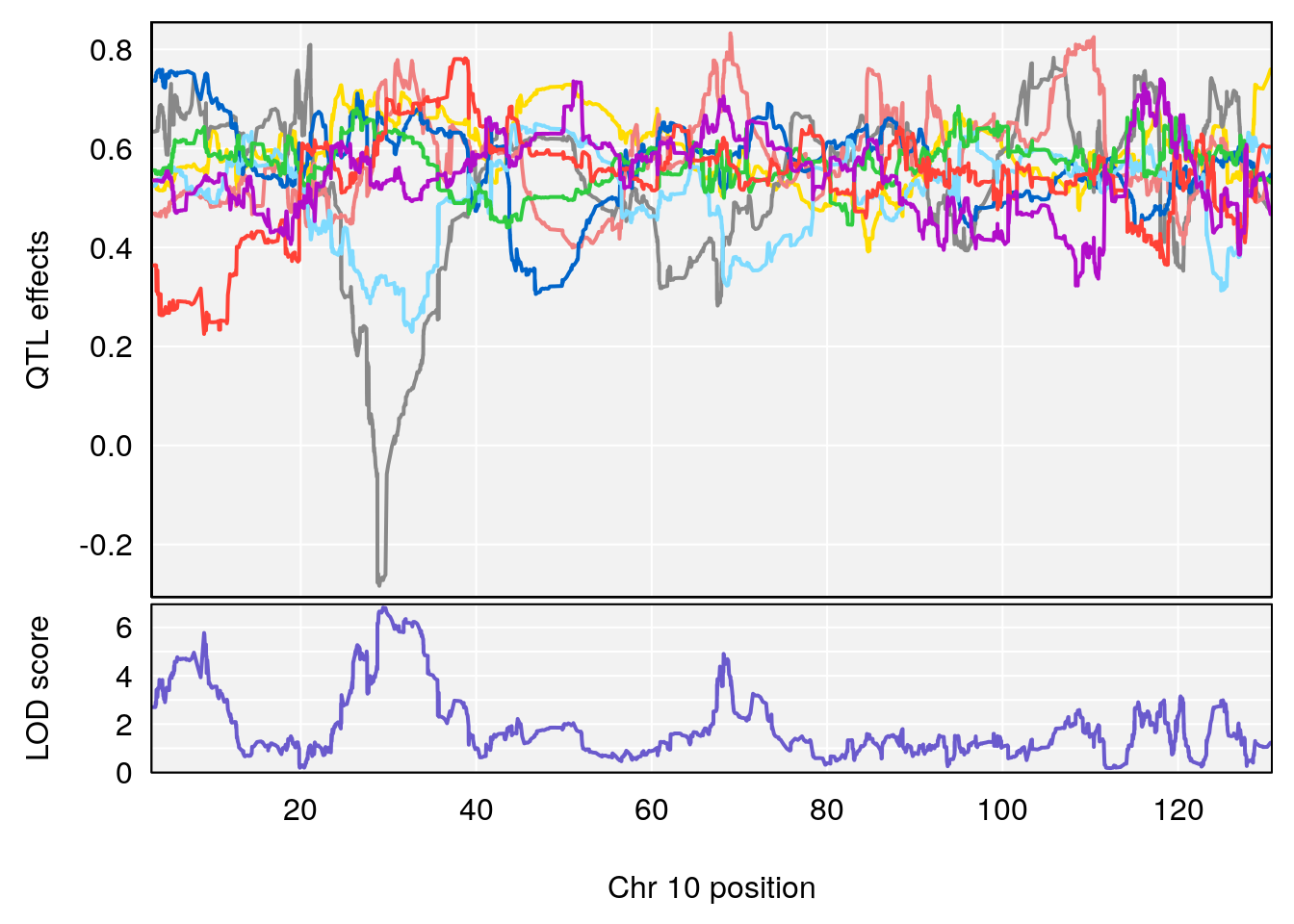
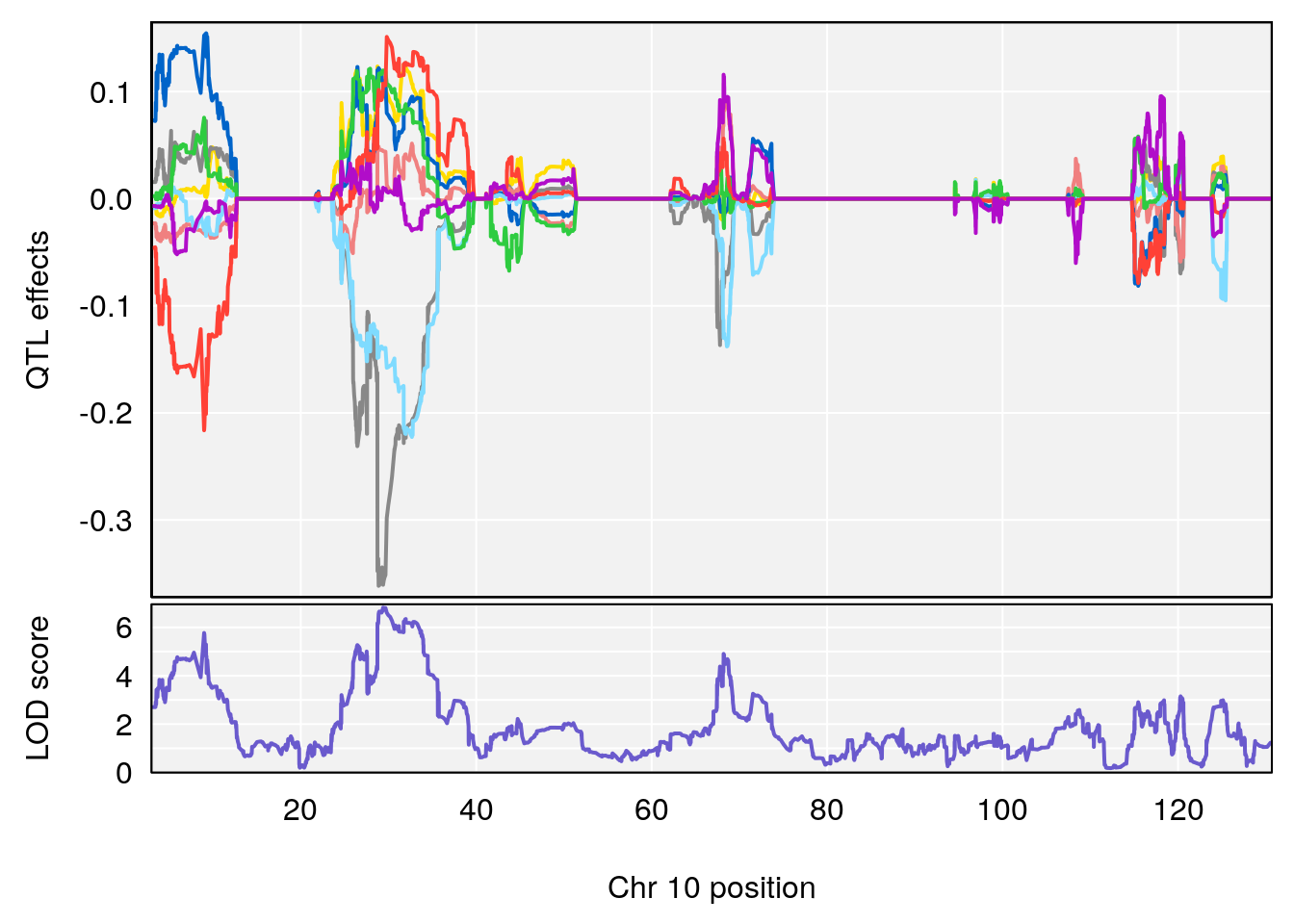
# [1] 4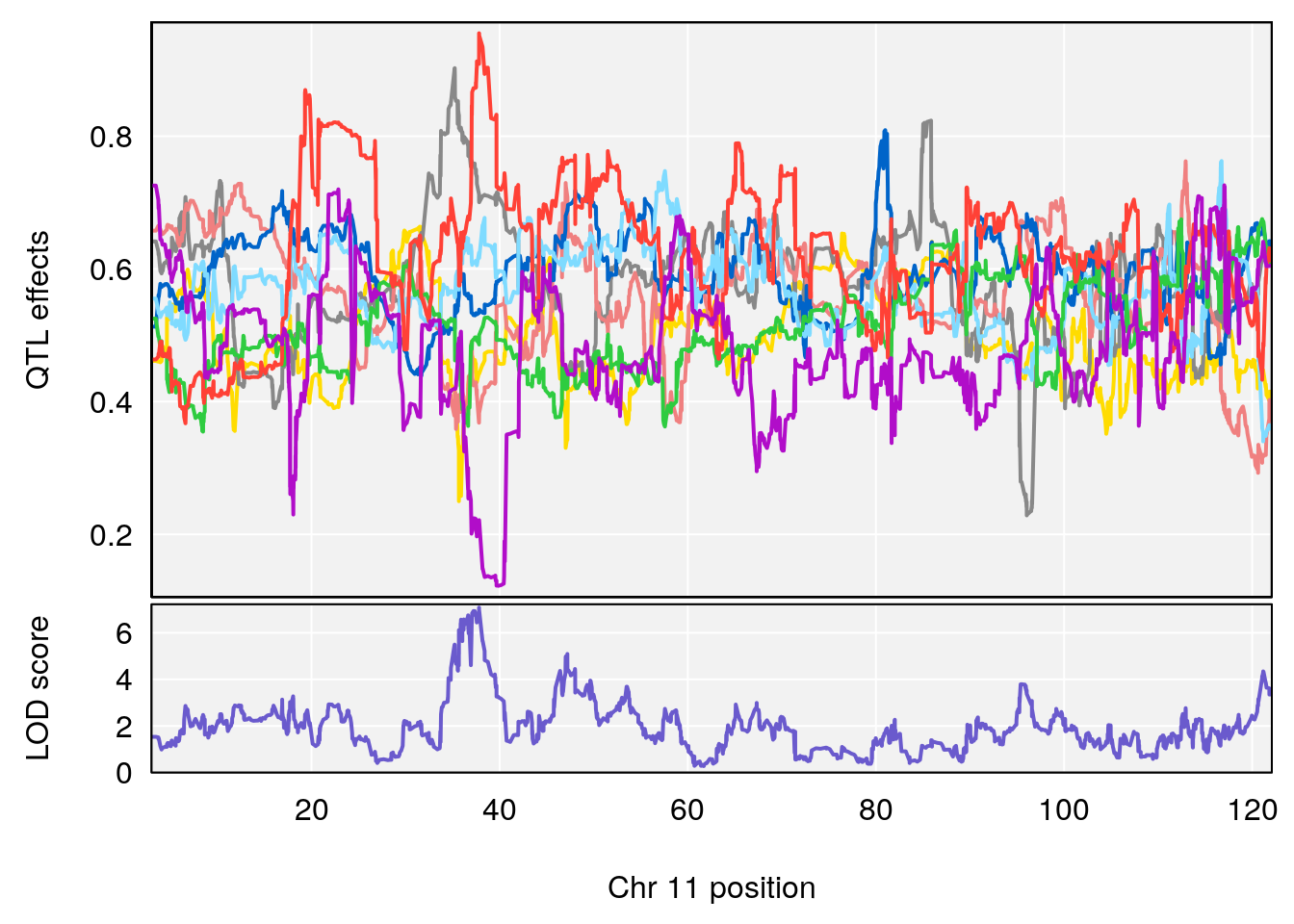
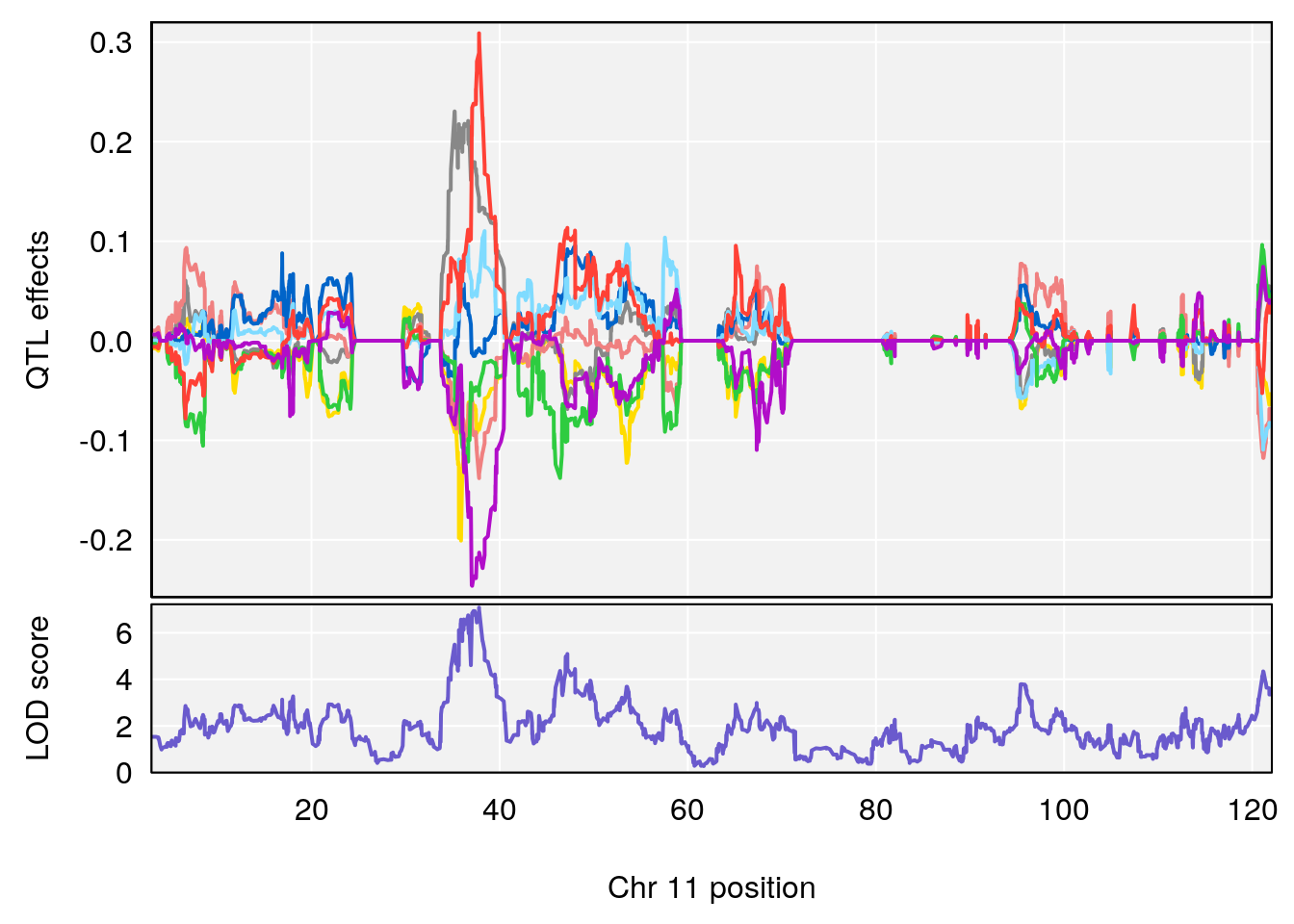
# [1] 5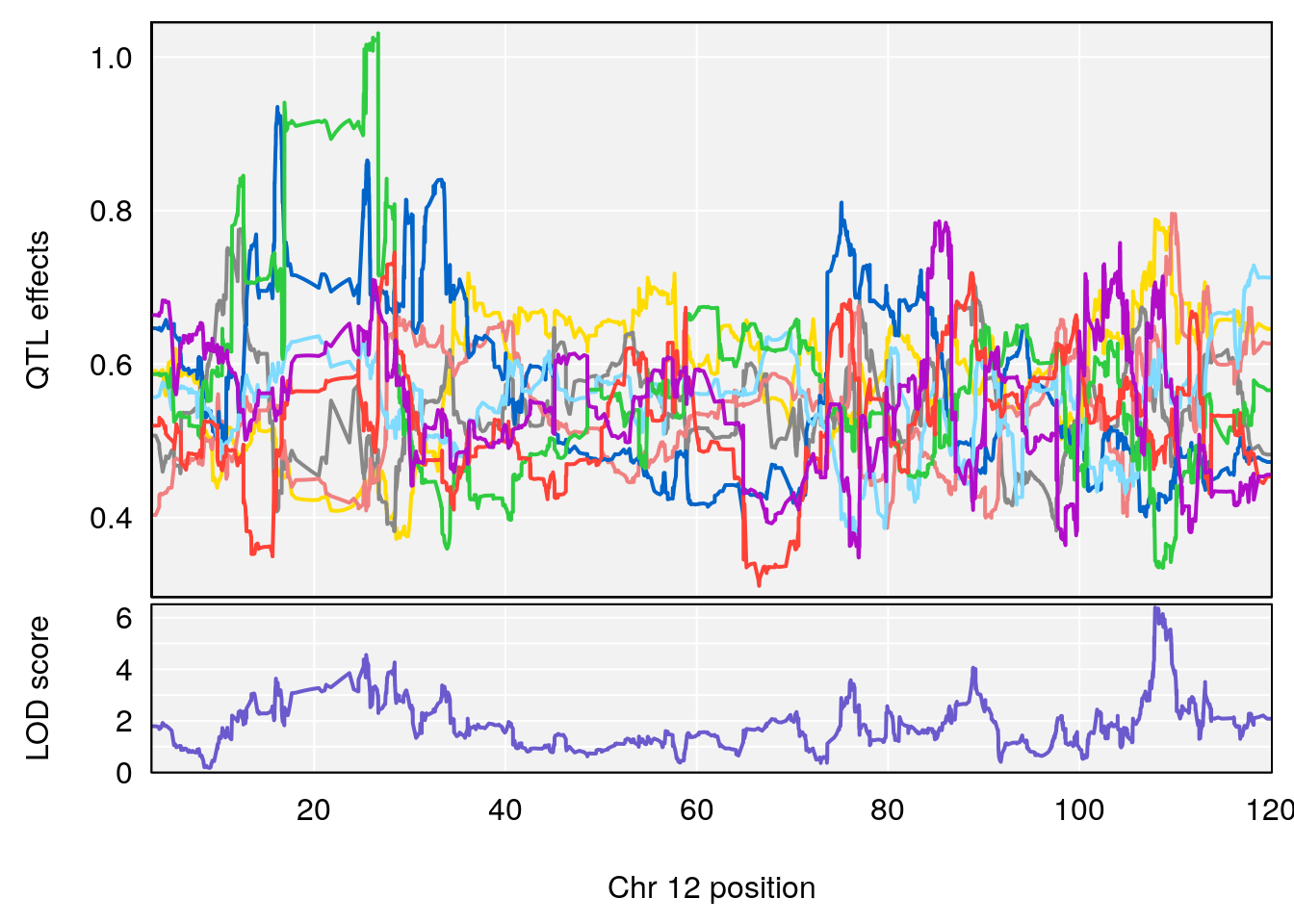
# [1] 6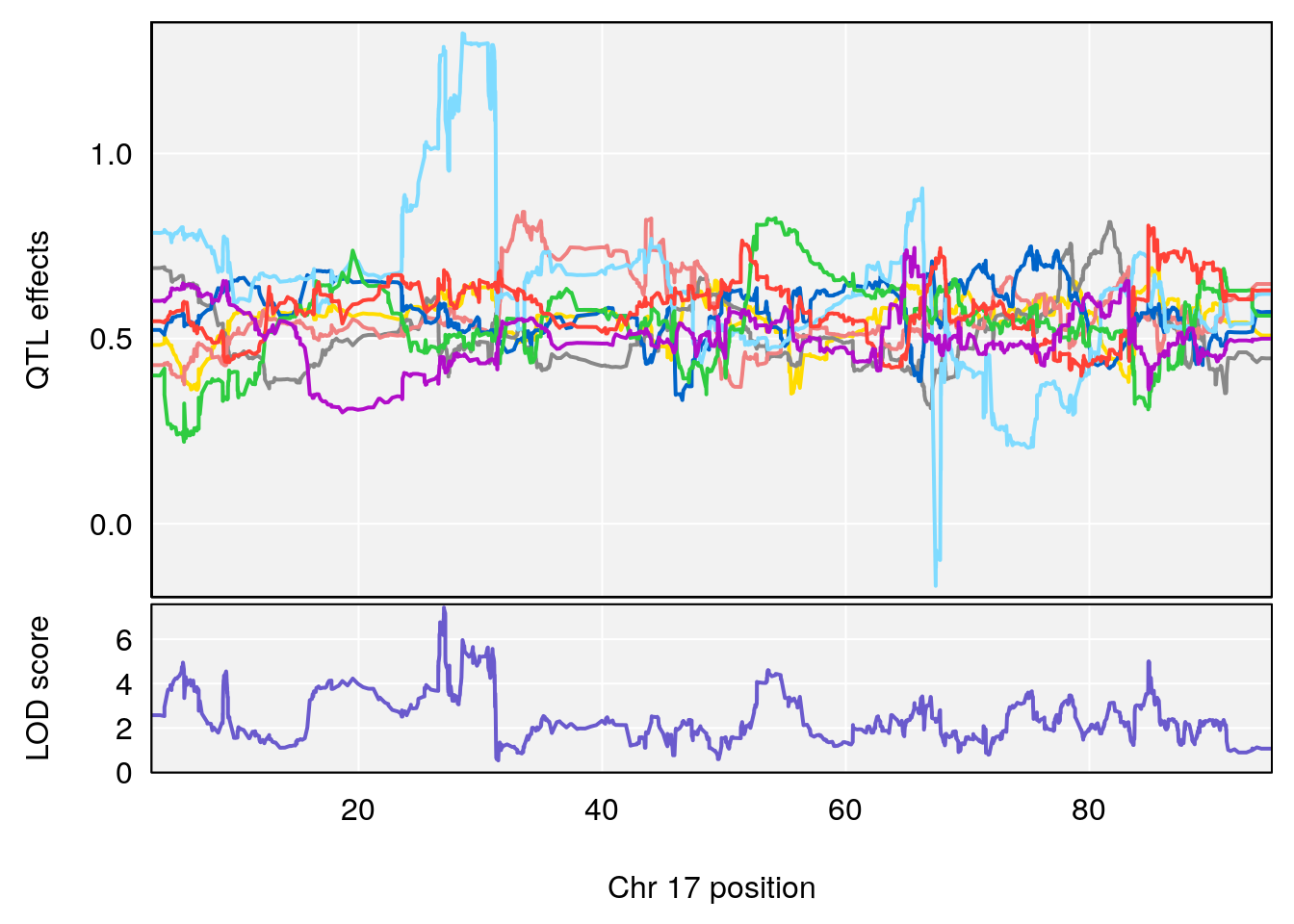
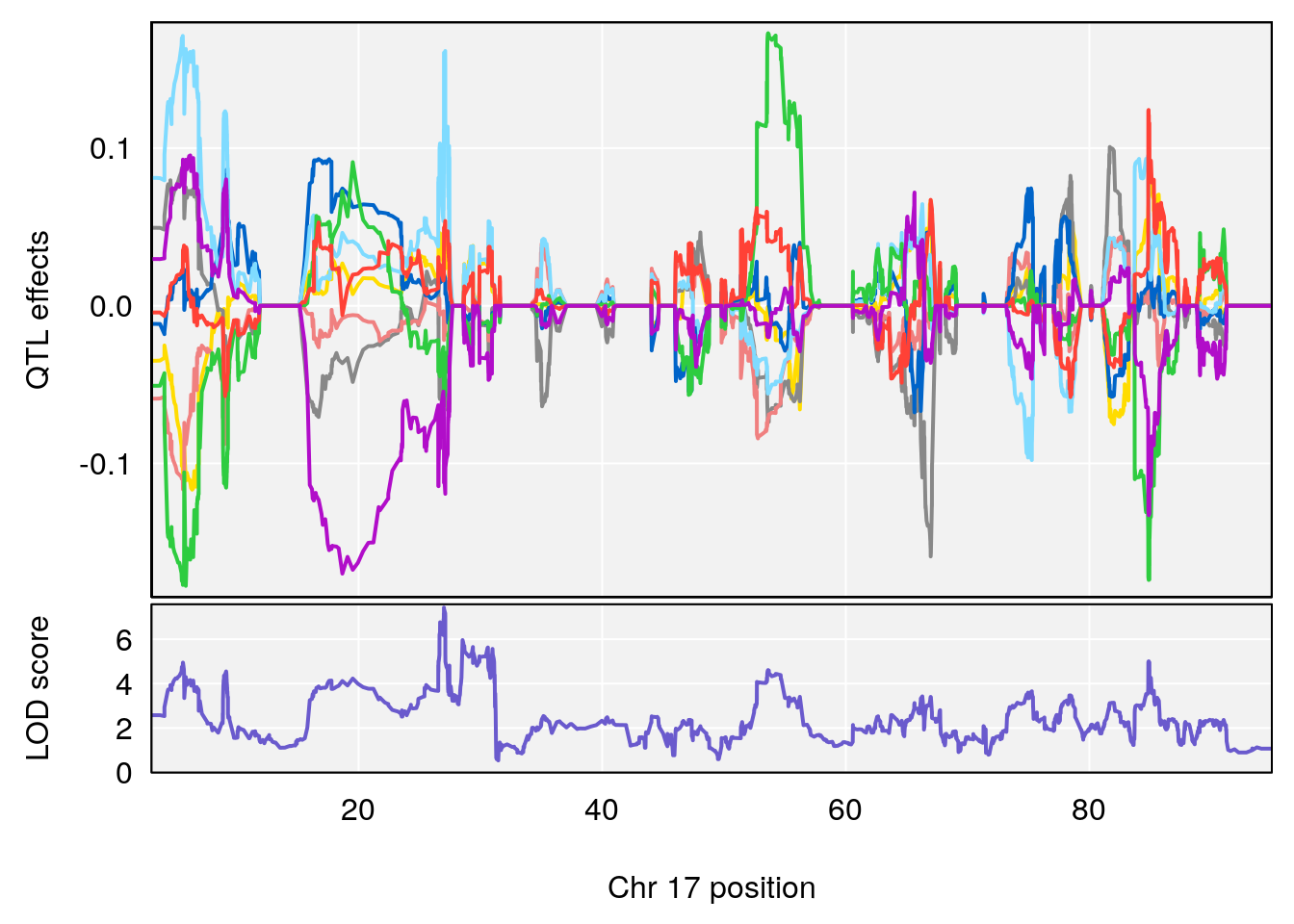
# [1] 7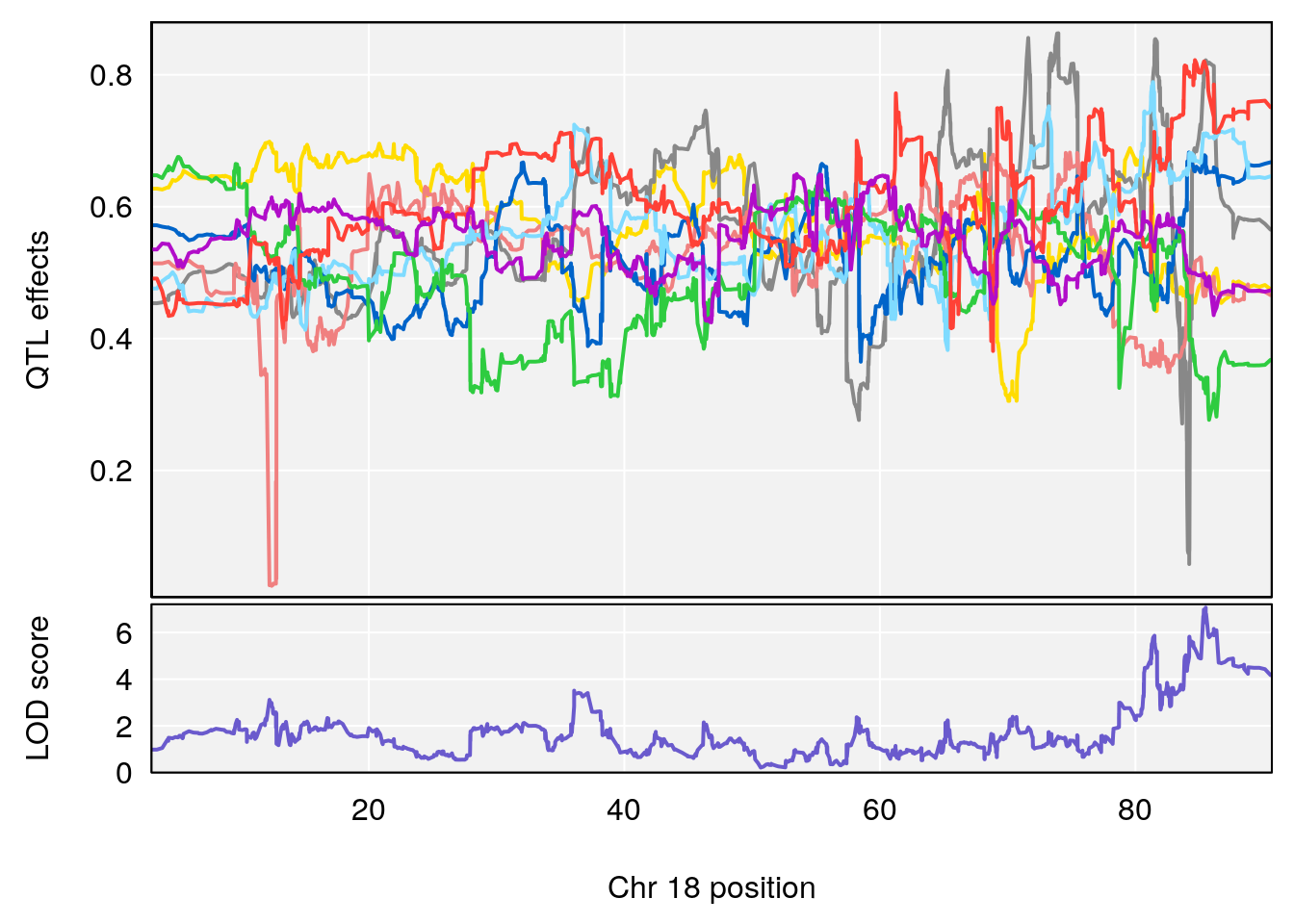
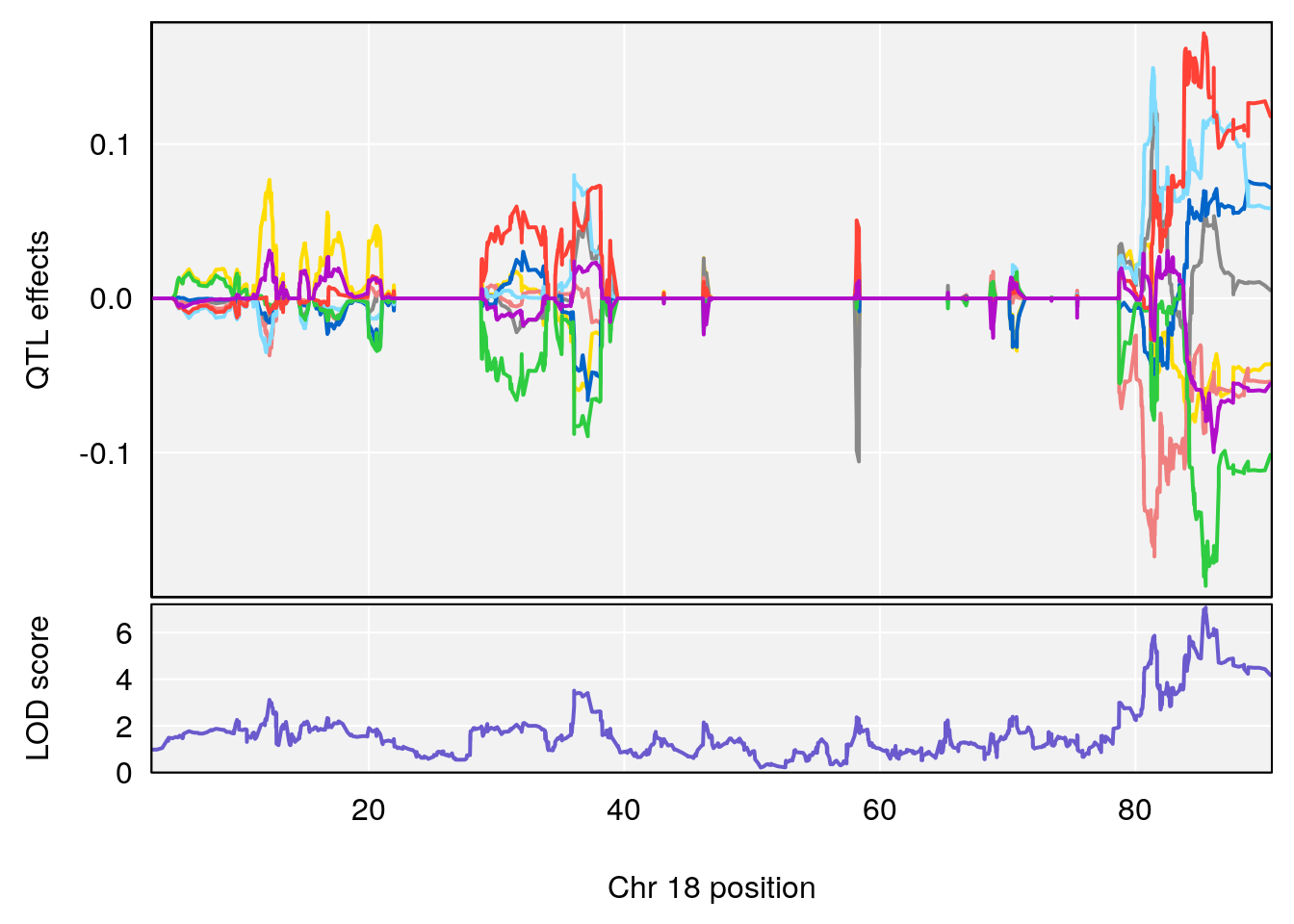
# [1] 8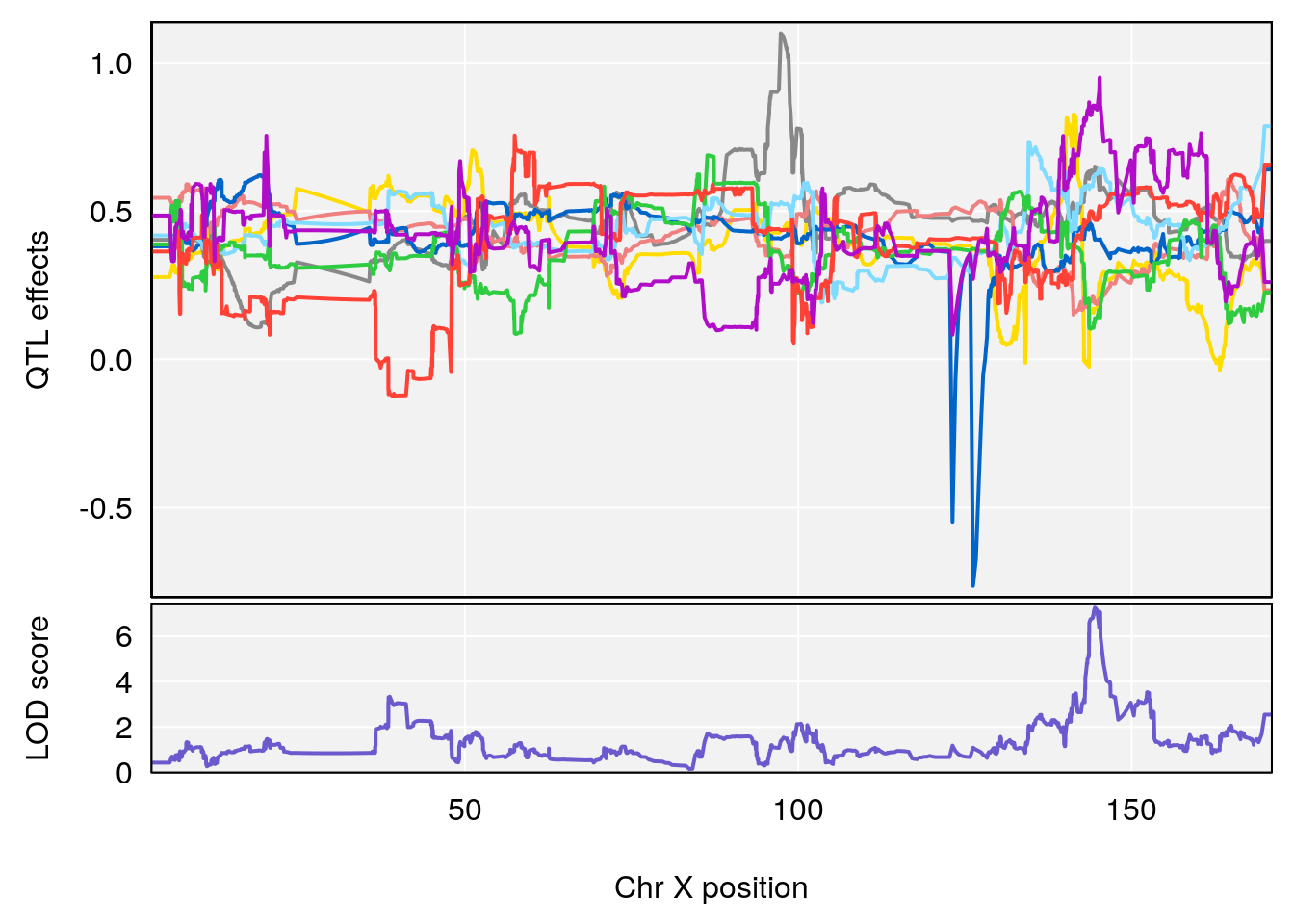
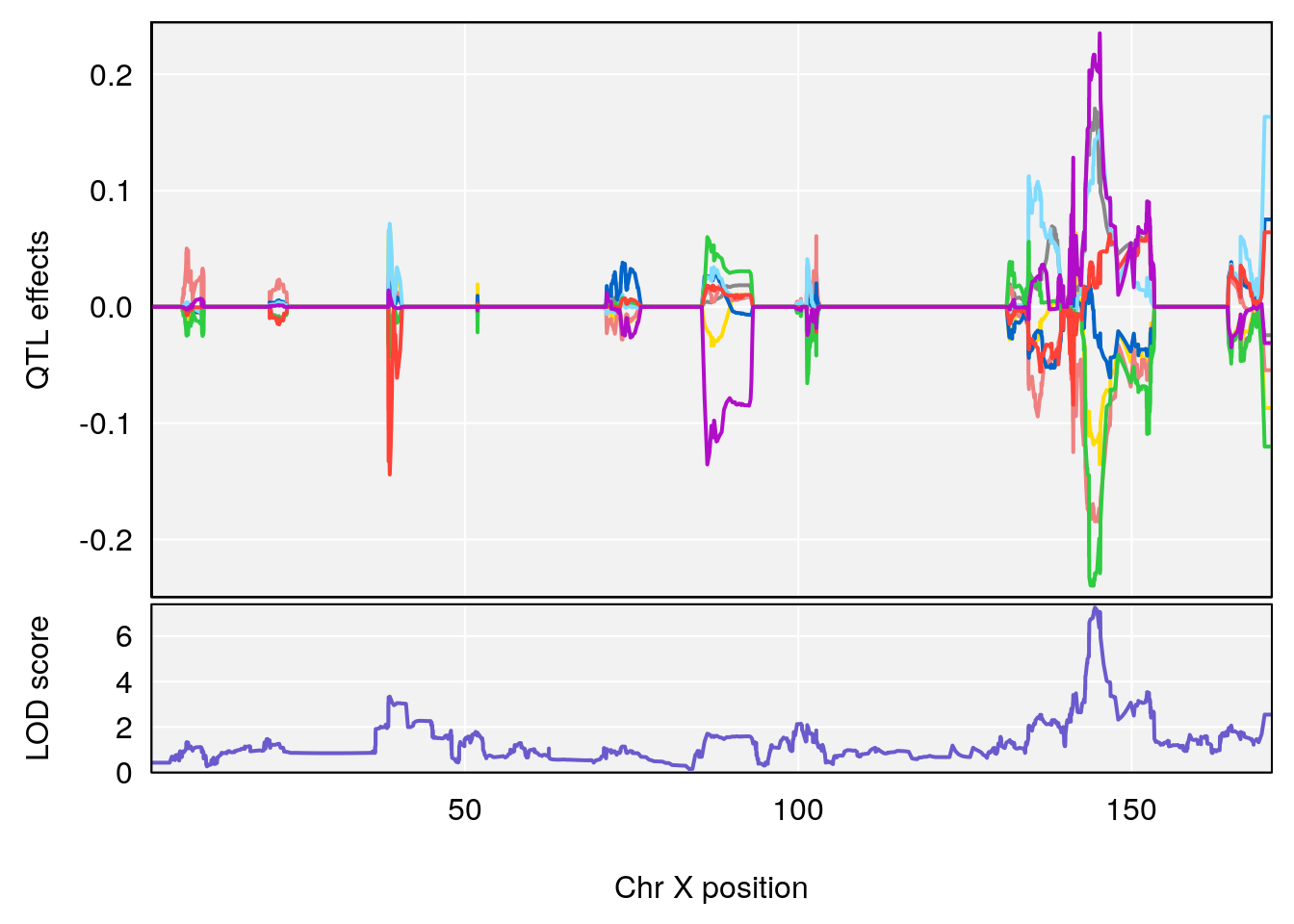
#save peaks coeff plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
fname <- paste("output/DO_morphine_combined_69k_m2_",i,"_coefplot.pdf",sep="")
pdf(file = fname, width = 16, height =8)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
}
dev.off()
}
# [1] "Survival.Time"
# [1] 1
# [1] "Recovery.Time"
# [1] 1
# [1] "Min.depression"
# [1] 1
# [1] 2
# [1] "Status_bin"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] 5
# [1] 6
# [1] 7
# [1] 8
#save peaks coeff blup plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
fname <- paste("output/DO_morphine_combined_69k_m2_",i,"_coefplot_blup.pdf",sep="")
pdf(file = fname, width = 16, height =8)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
}
dev.off()
}
# [1] "Survival.Time"
# [1] 1
# [1] "Recovery.Time"
# [1] 1
# [1] "Min.depression"
# [1] 1
# [1] 2
# [1] "Status_bin"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] 5
# [1] 6
# [1] 7
# [1] 8Plot qtl mapping on array male
load("output/qtl.morphine.out.combined_gm.male.RData")
#pmap and gmap
chr.names <- c("1","2","3","4","5","6","7","8","9","10","11","12","13","14","15","16","17","18","19", "X")
gmap <- list()
pmap <- list()
for (chr in chr.names){
gmap[[chr]] <- combined_gm$gmap[[chr]]
pmap[[chr]] <- combined_gm$pmap[[chr]]
}
attr(gmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
attr(pmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
#genome-wide plot
for(i in names(qtl.morphine.out)){
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.morphine.out[[i]]) # overall maximum LOD score
plot(qtl.morphine.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, max(sapply(qtl.morphine.out, maxlod)) +2))
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("DO_morphine_",i))
}
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
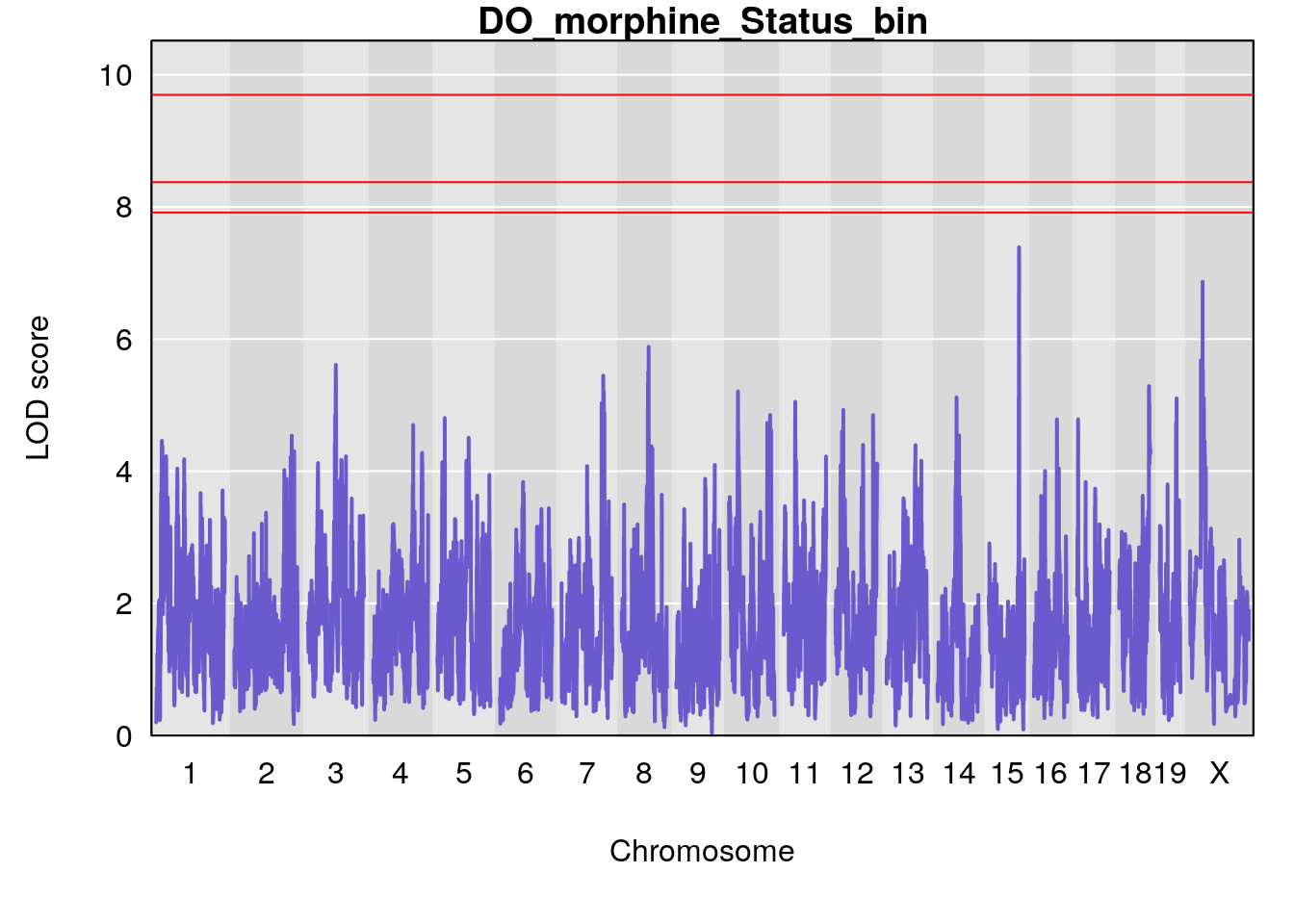
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
#peaks coeff plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
print(peaks)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
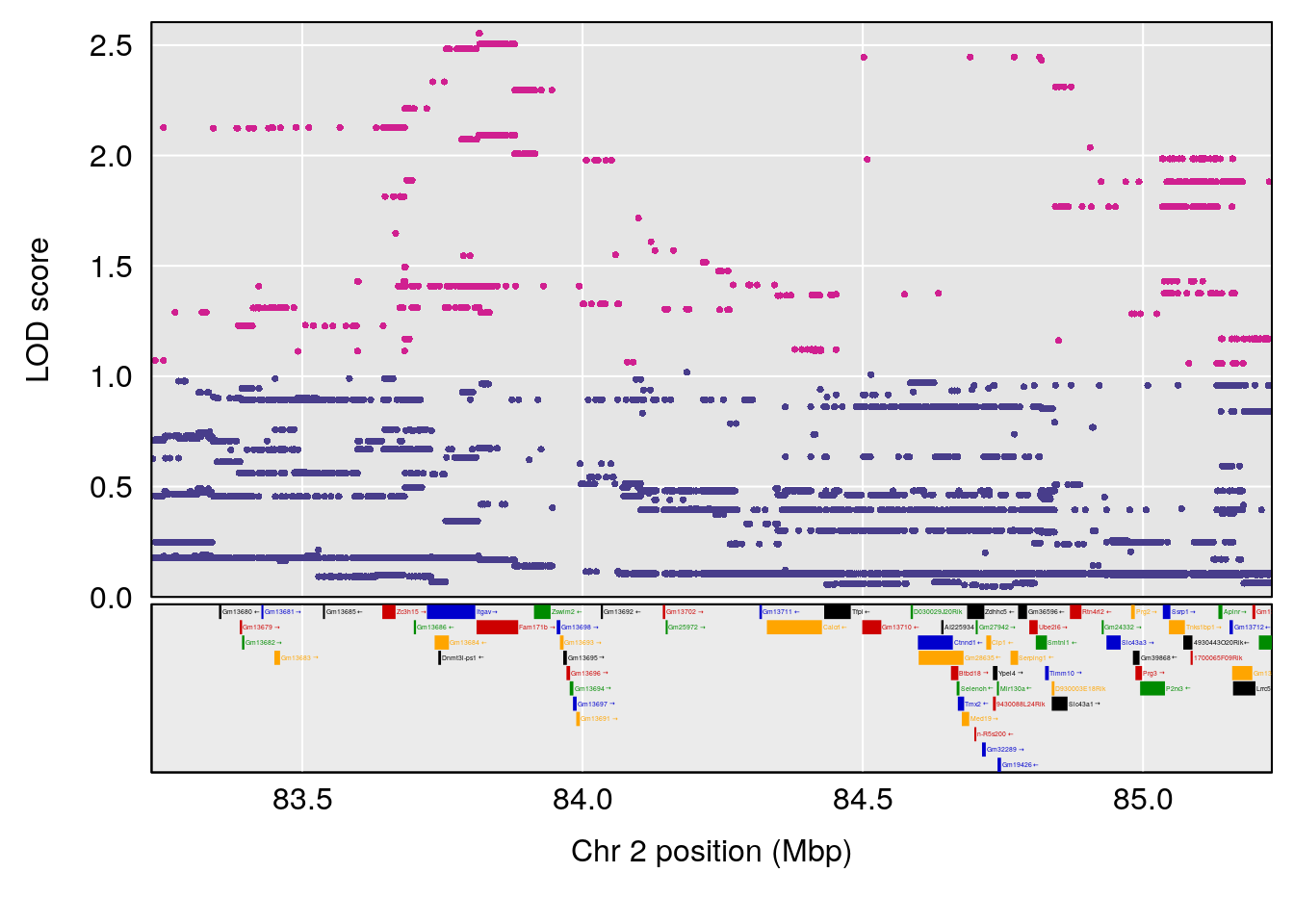
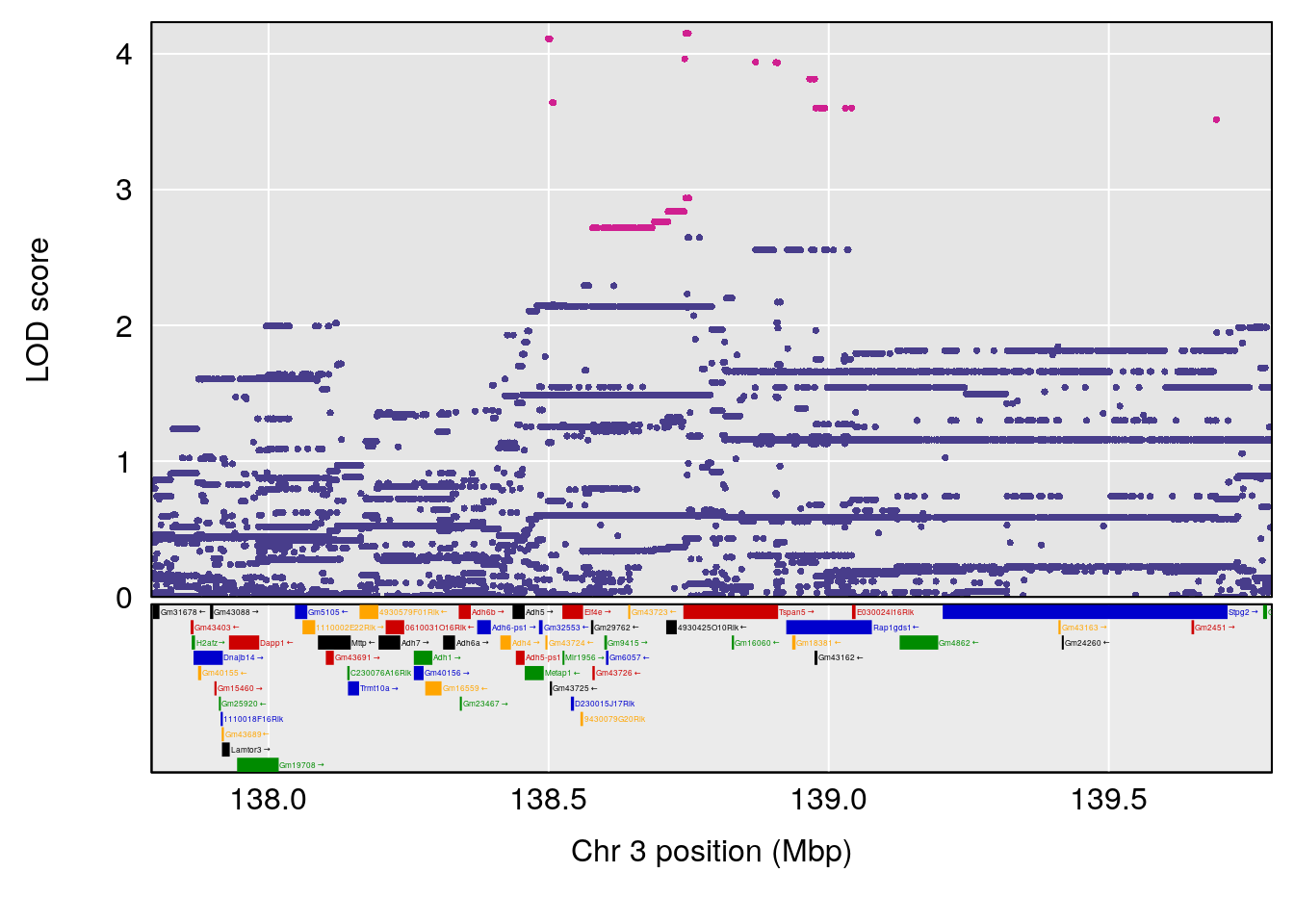
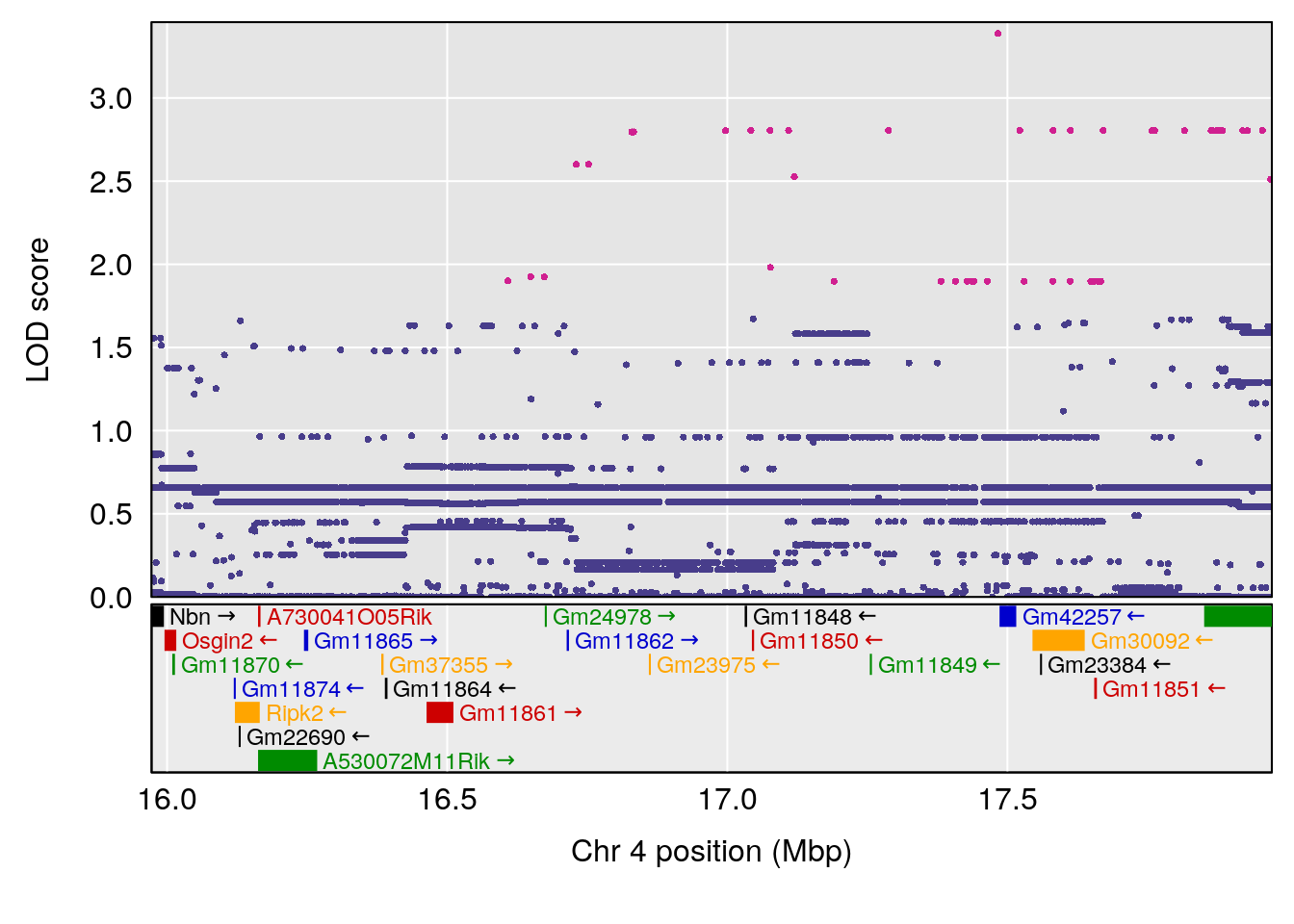
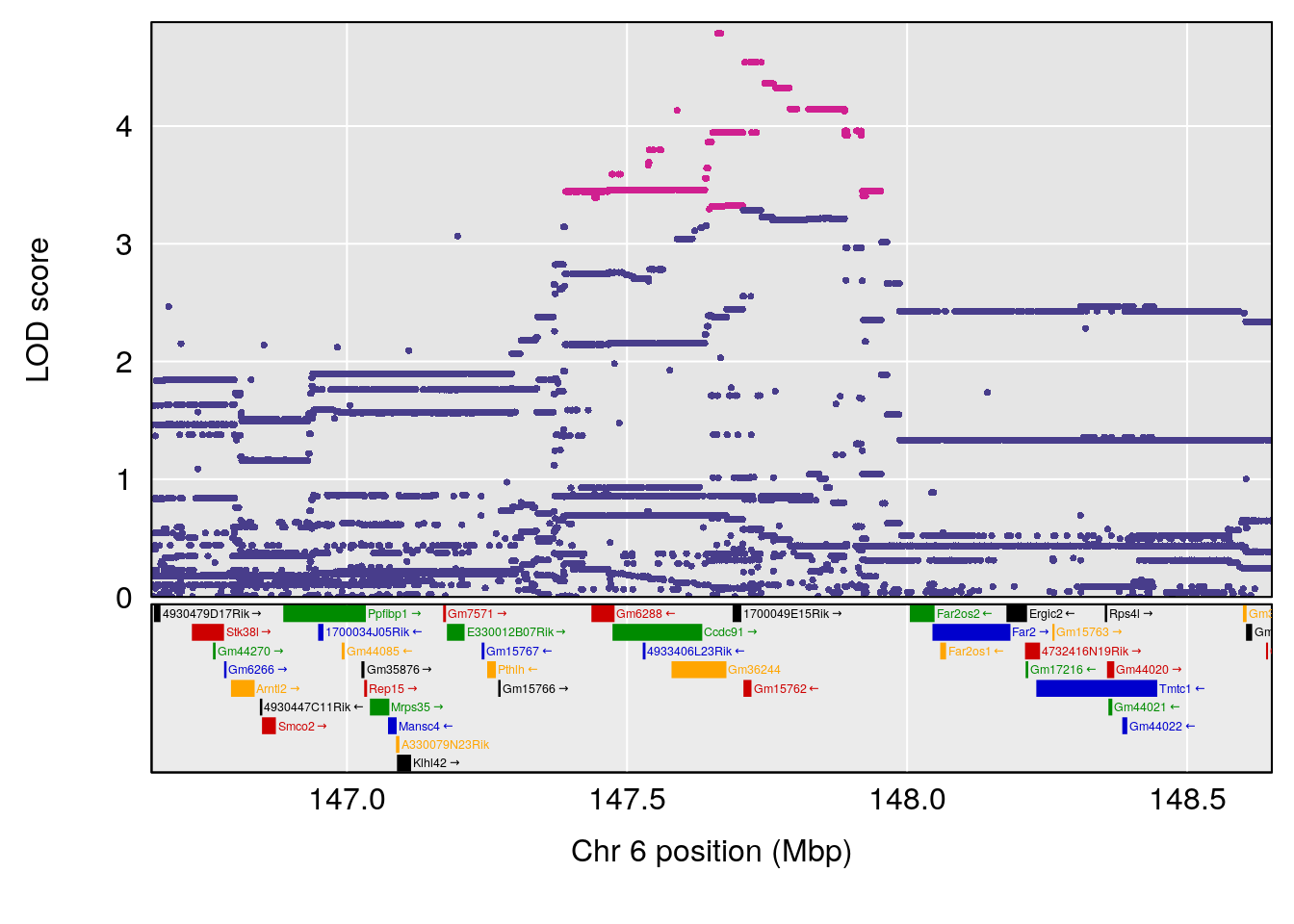
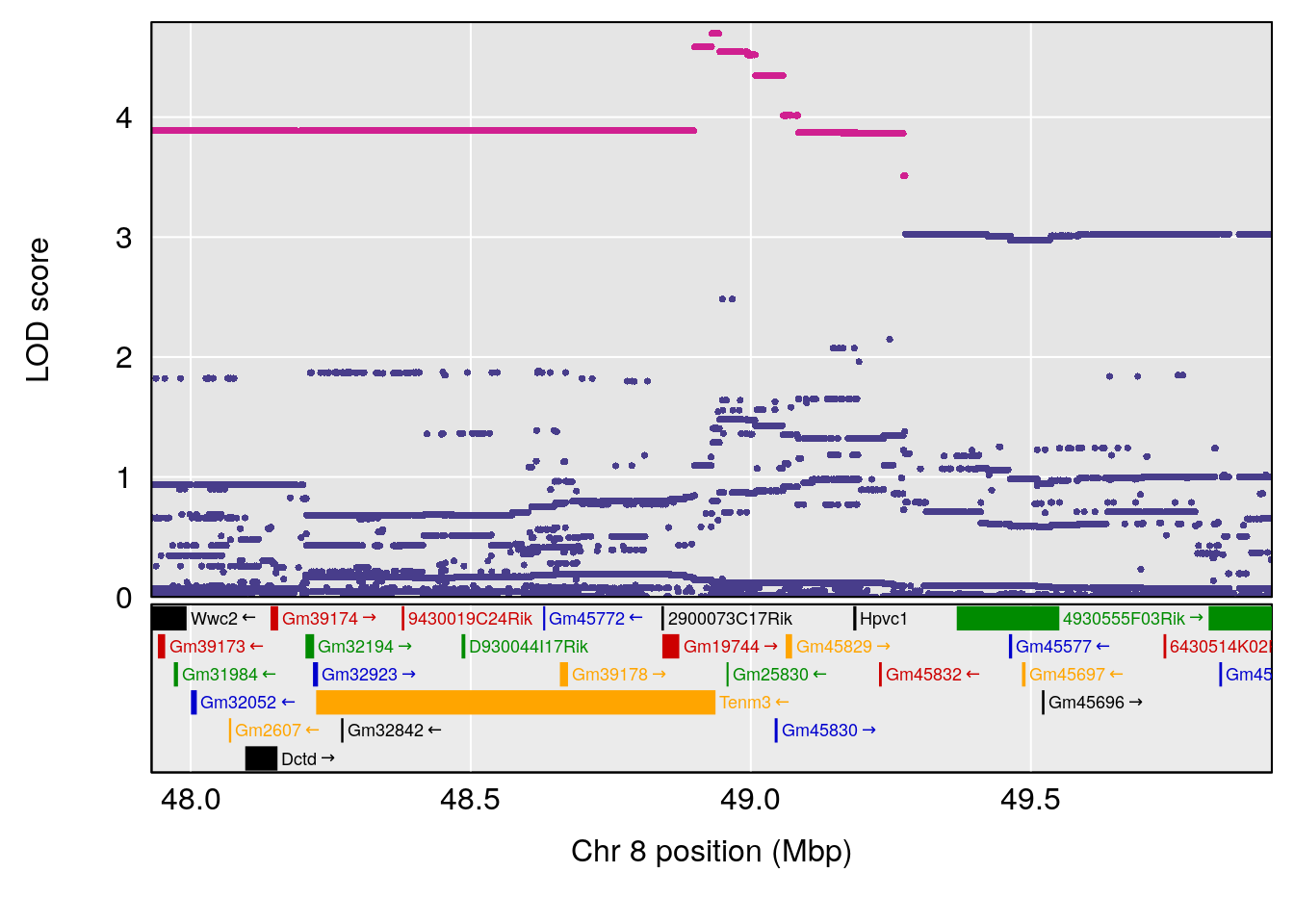
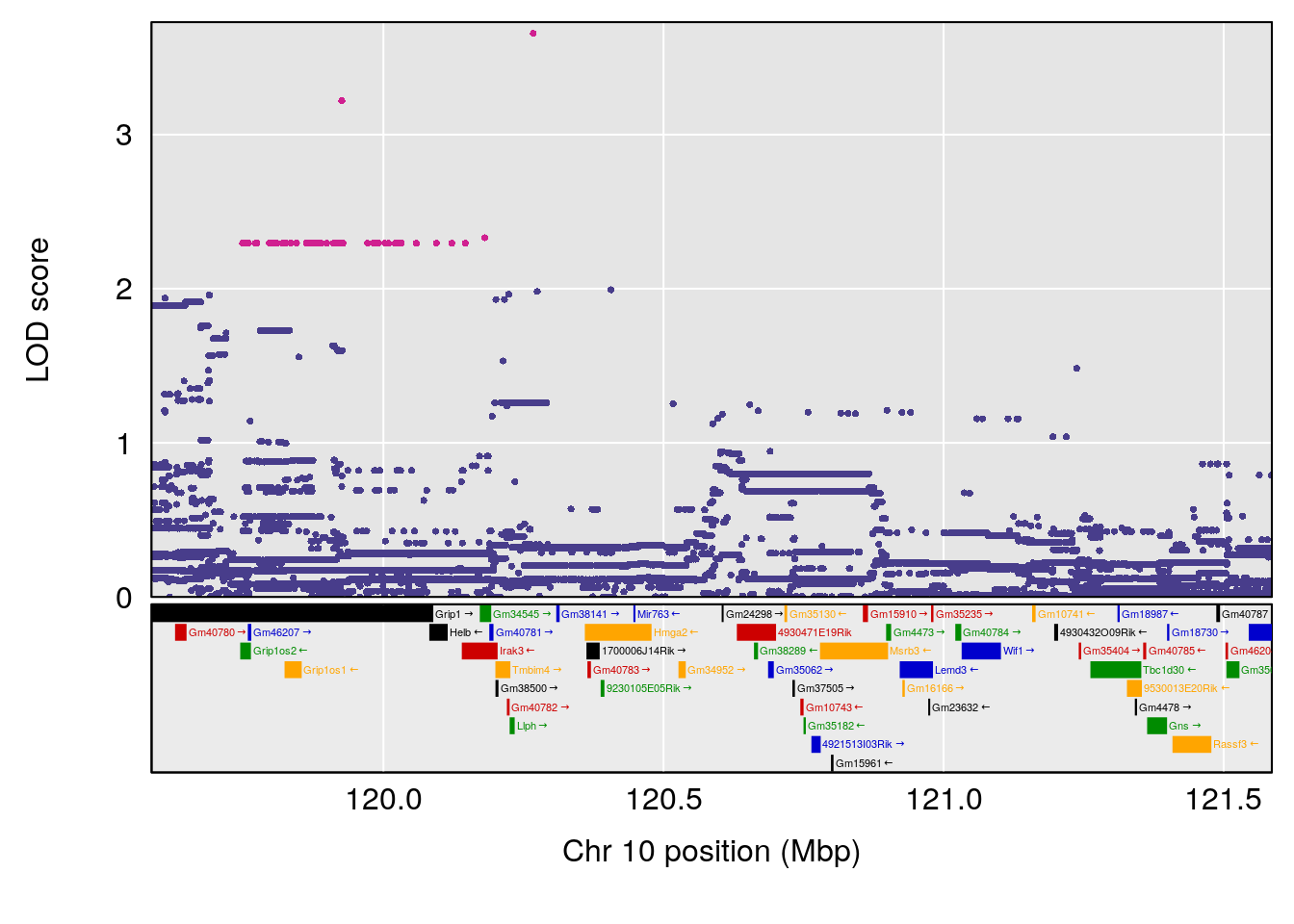
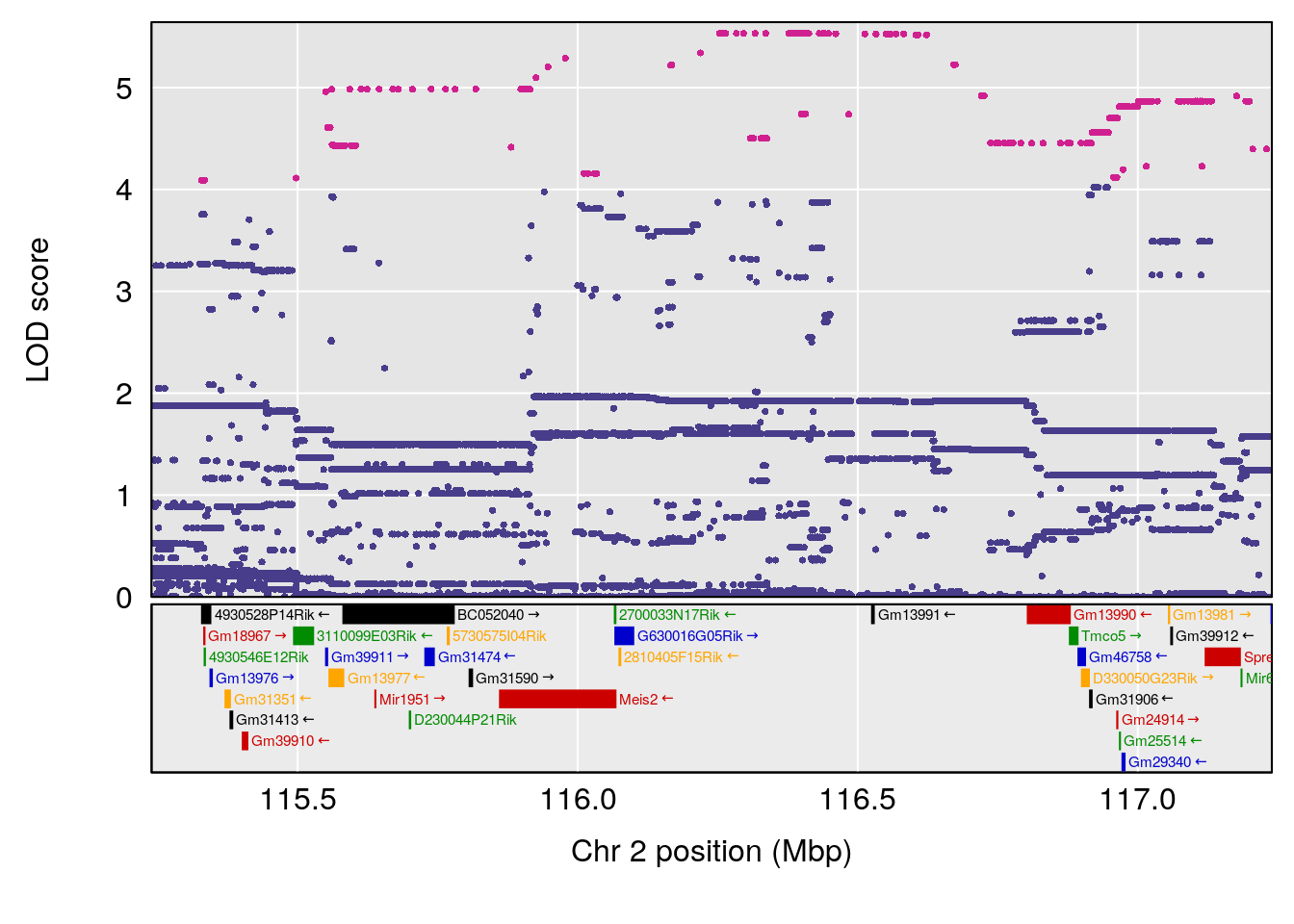
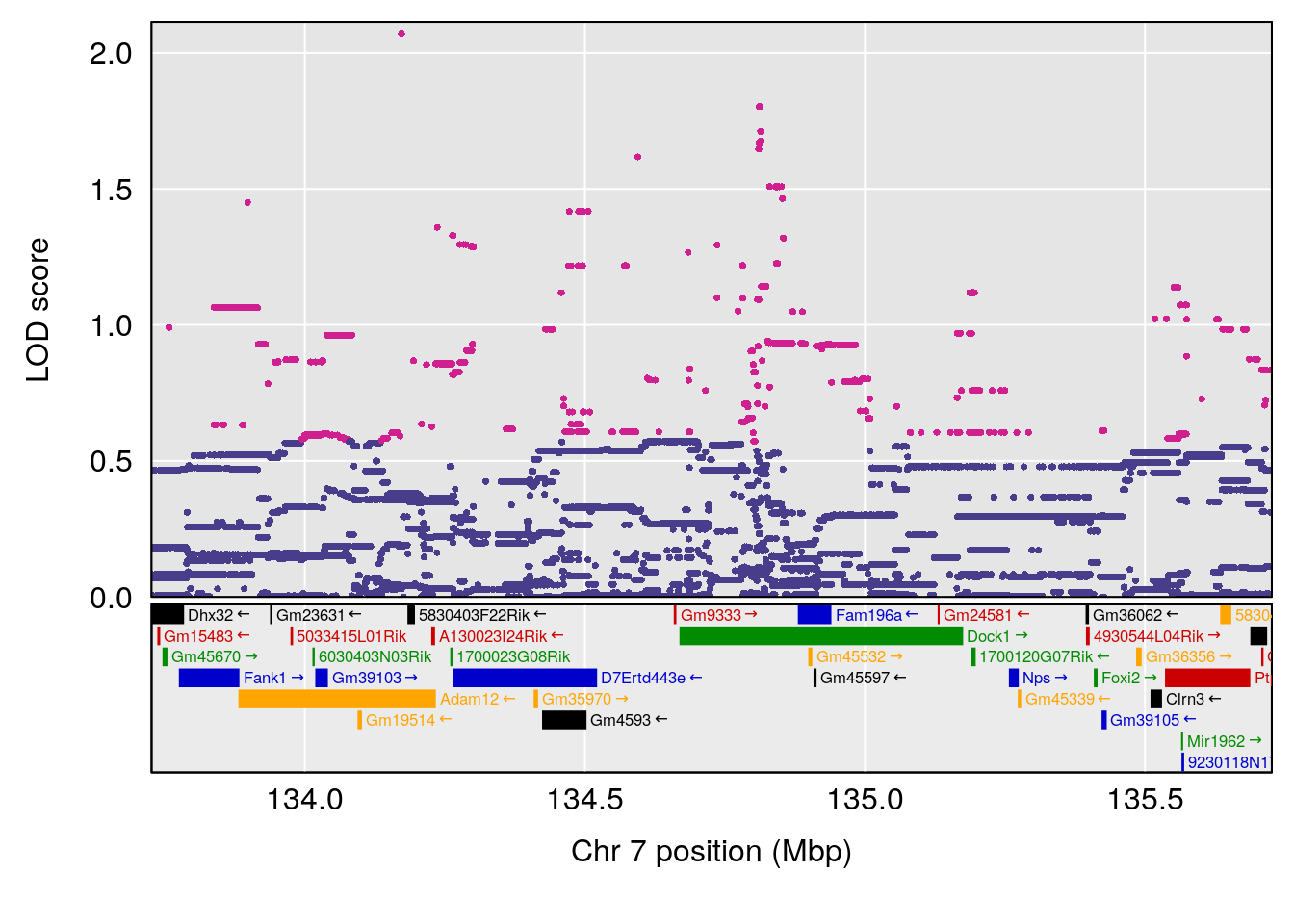
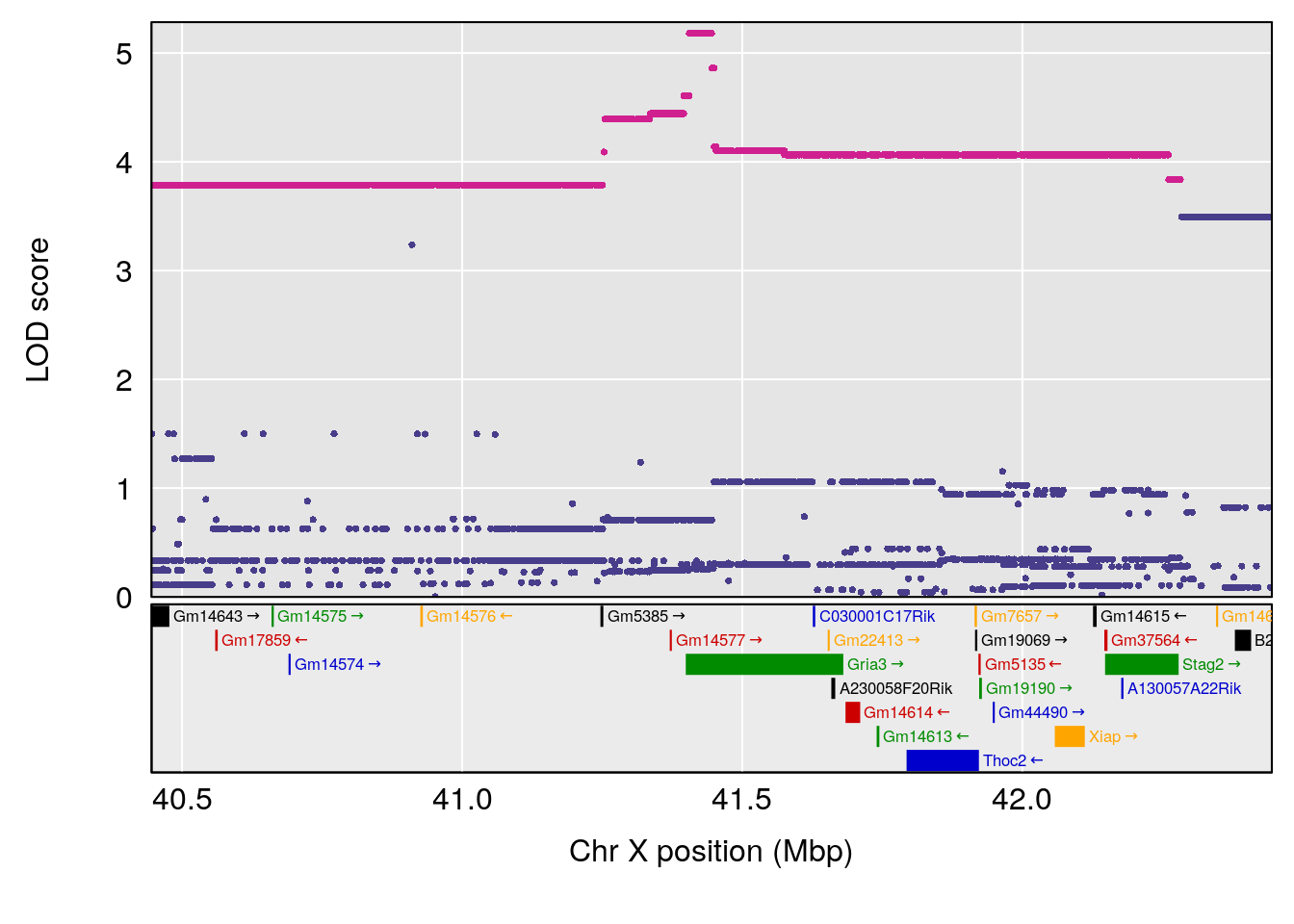
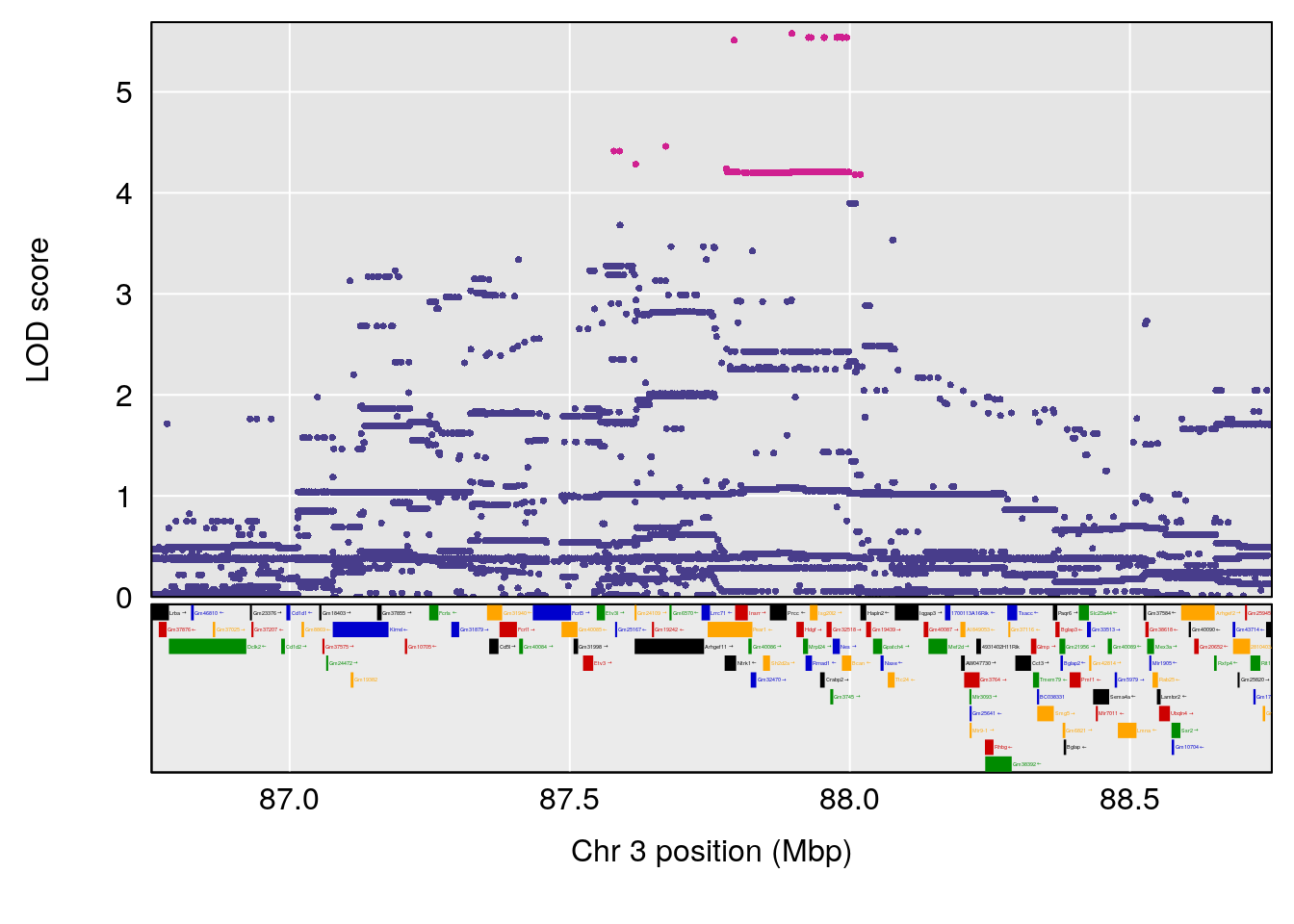
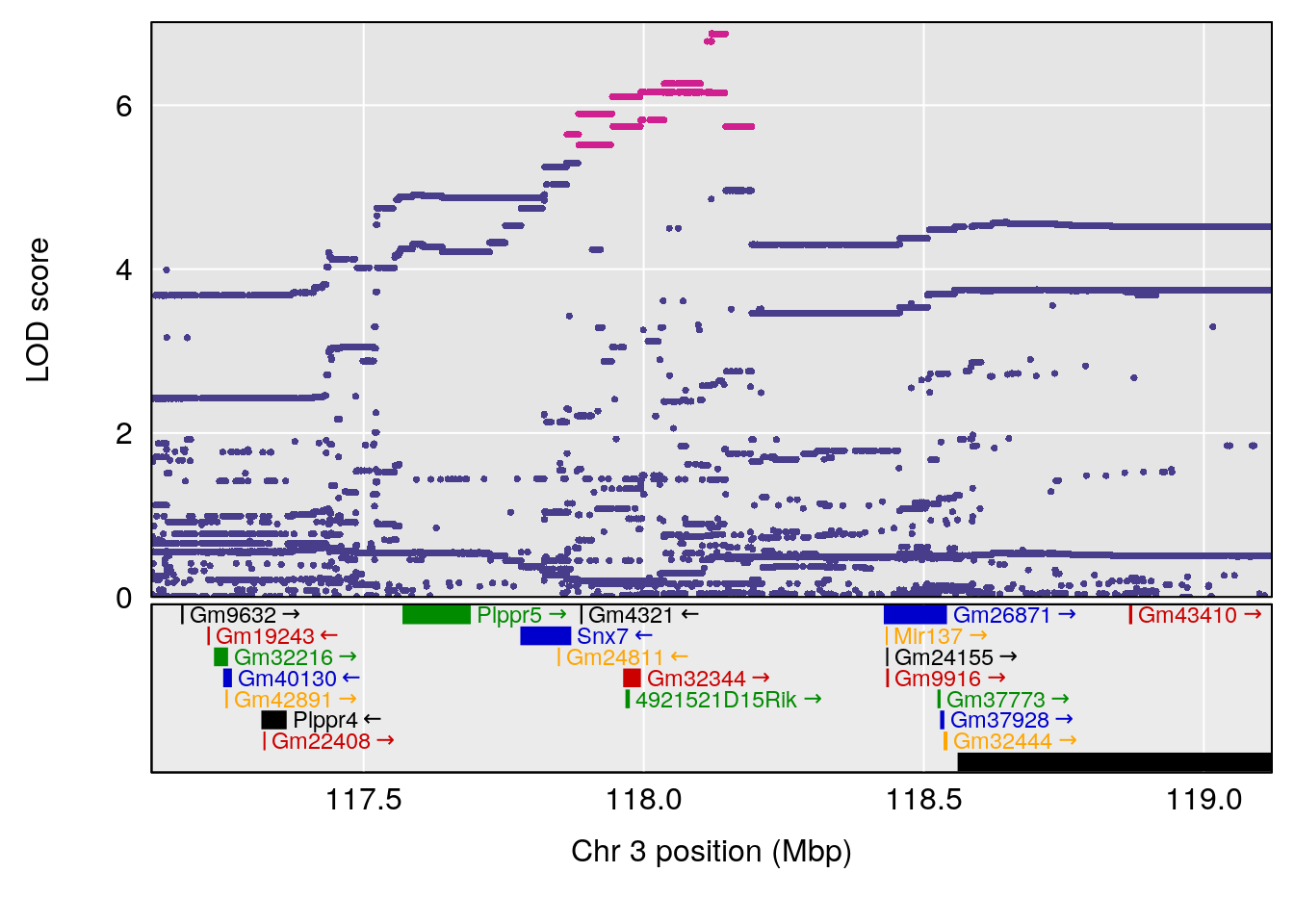
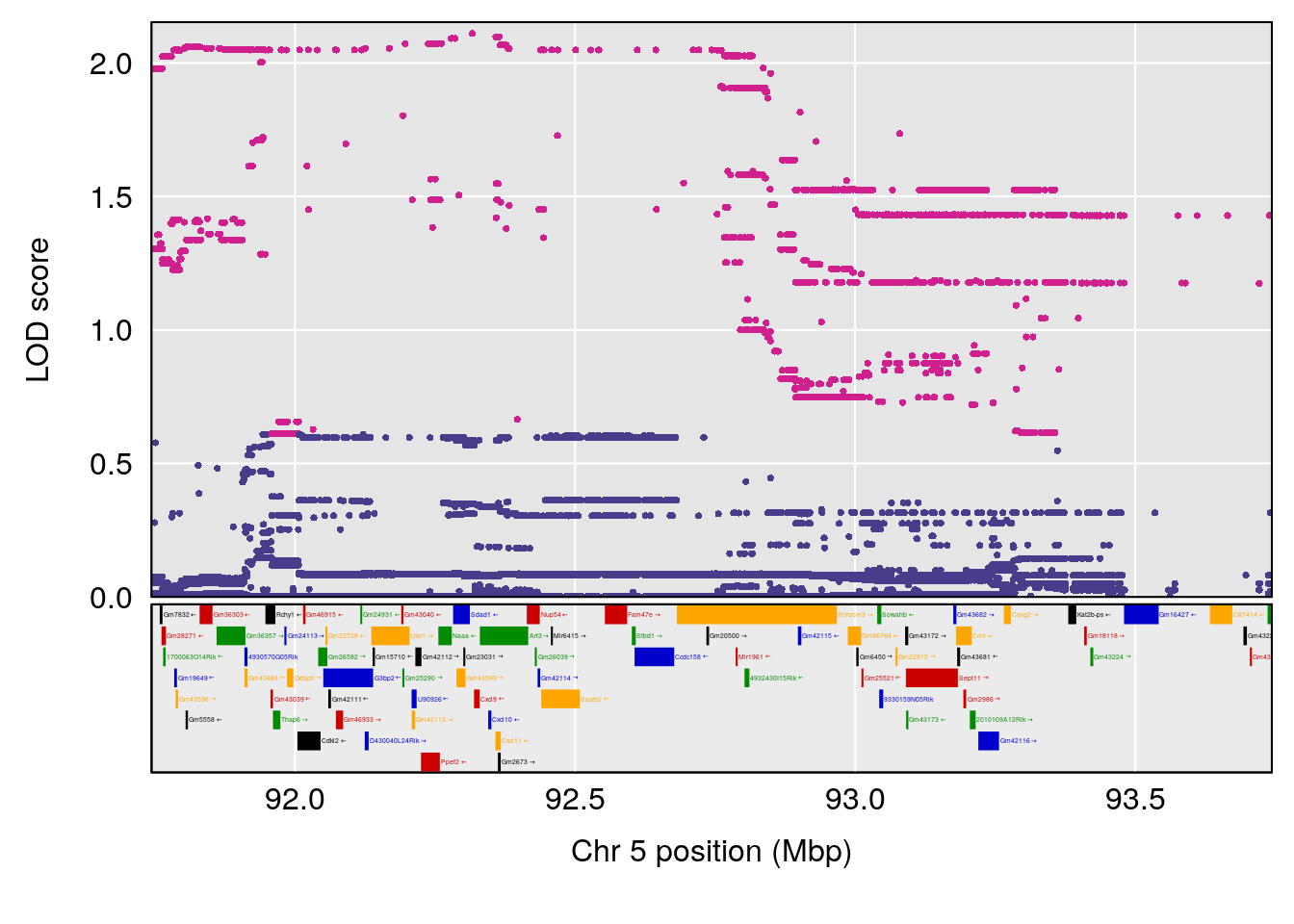
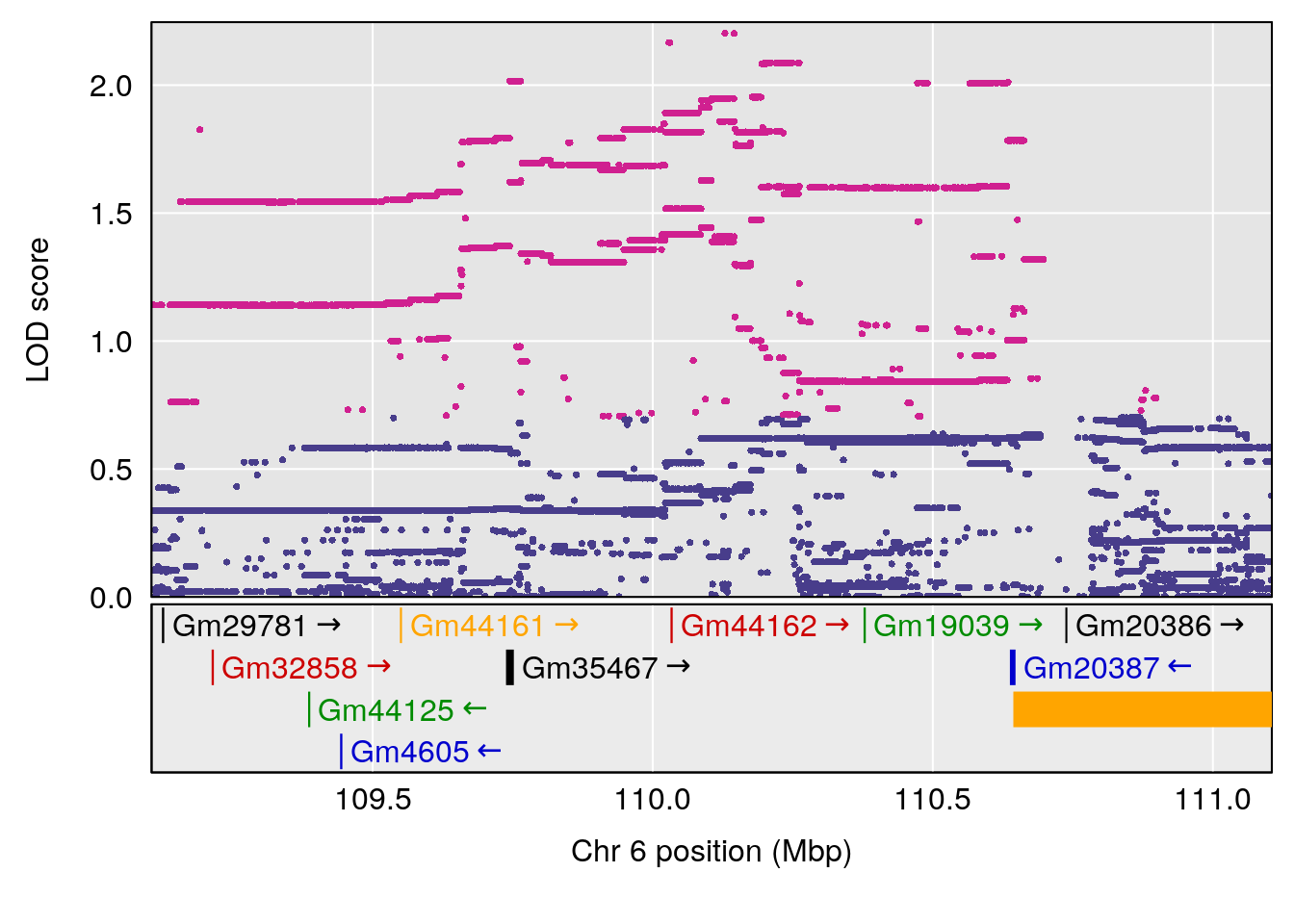
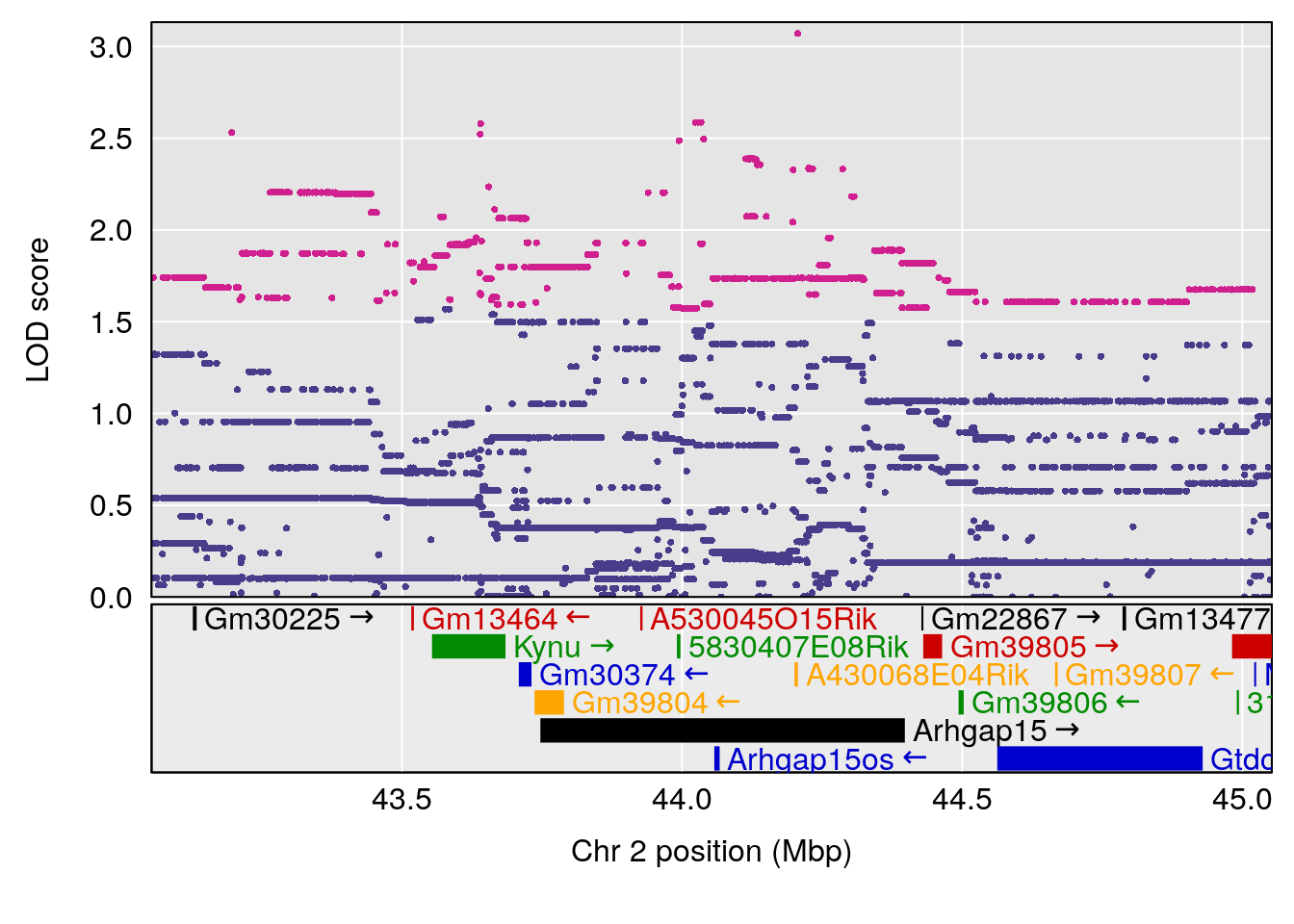
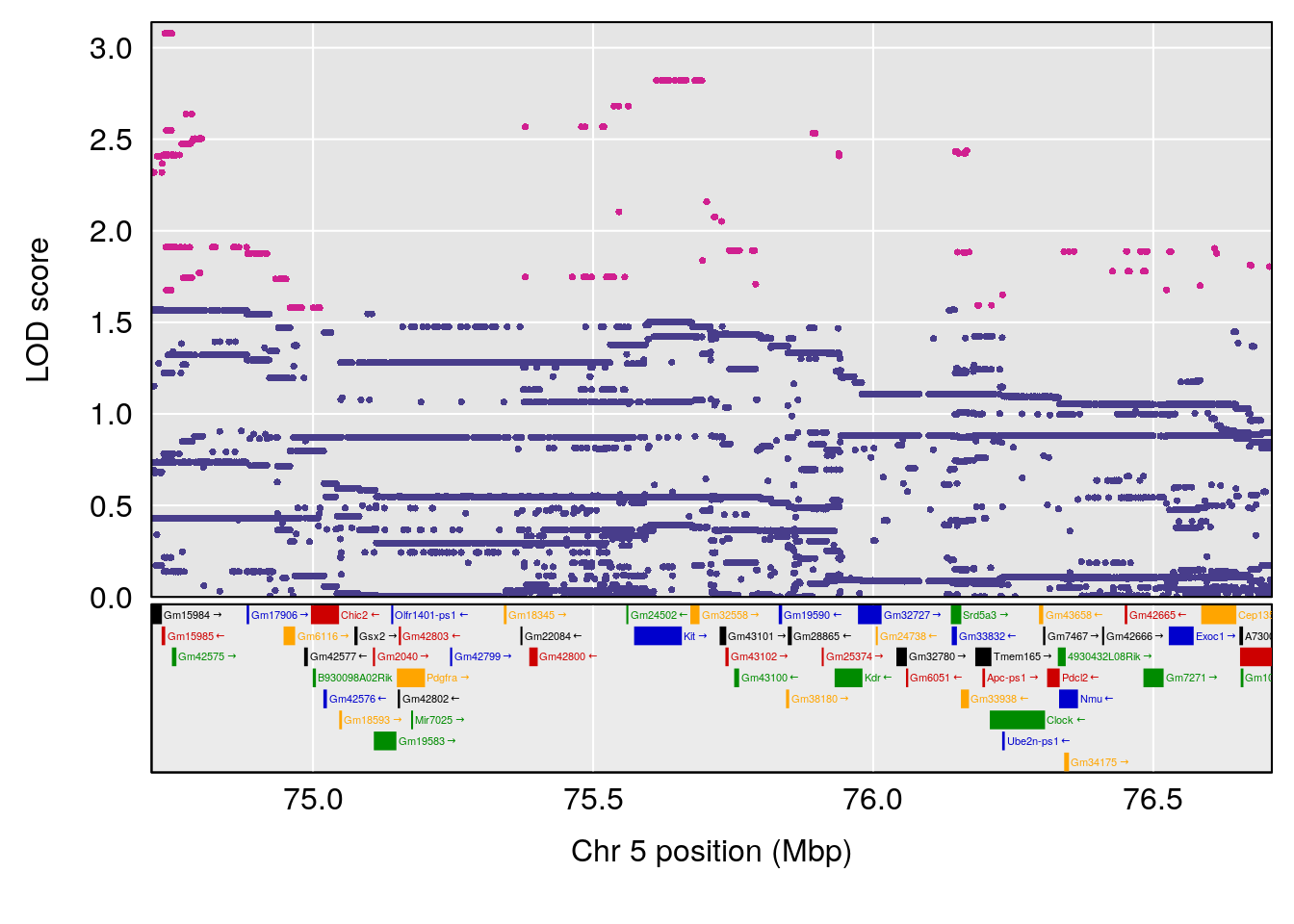
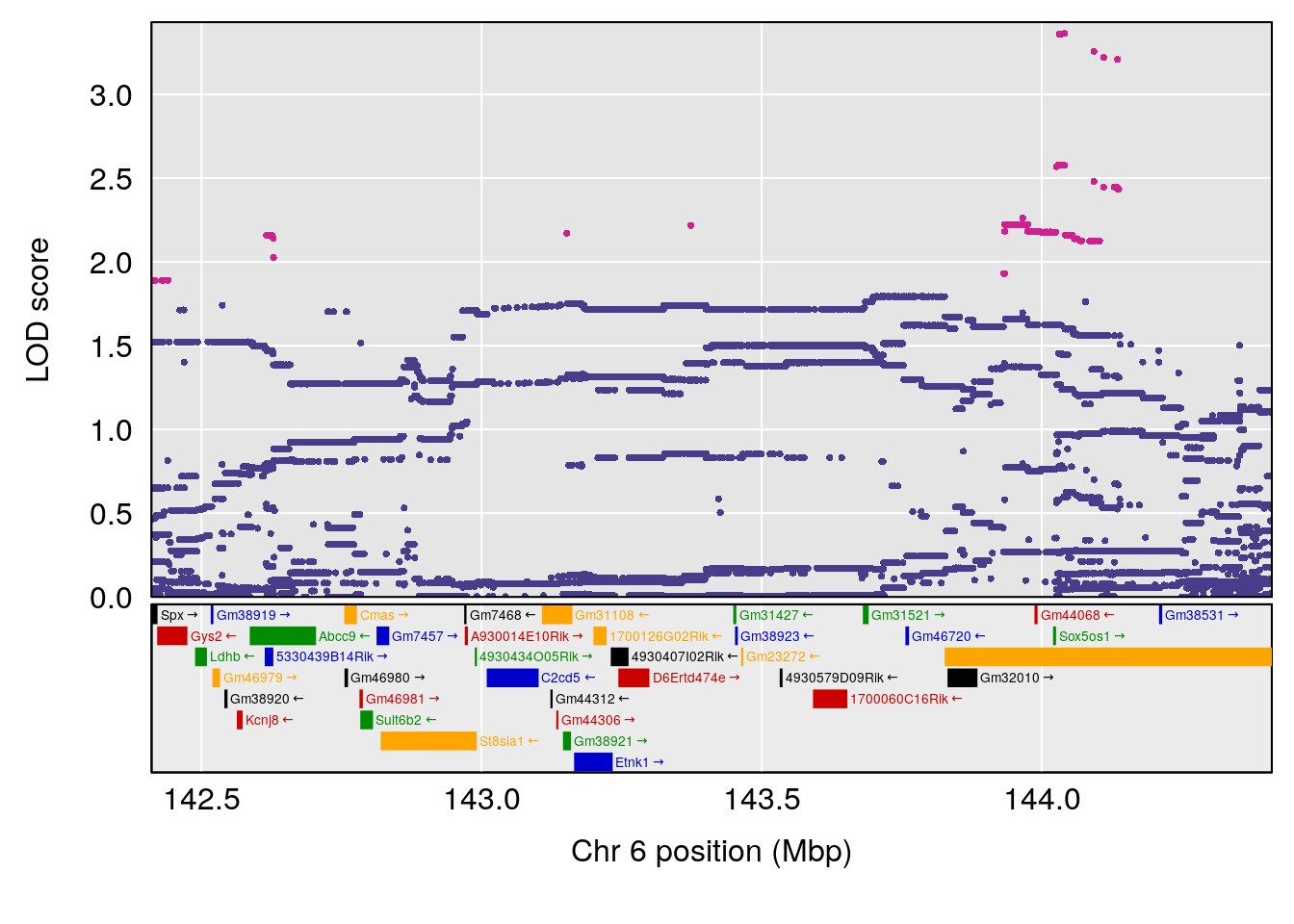
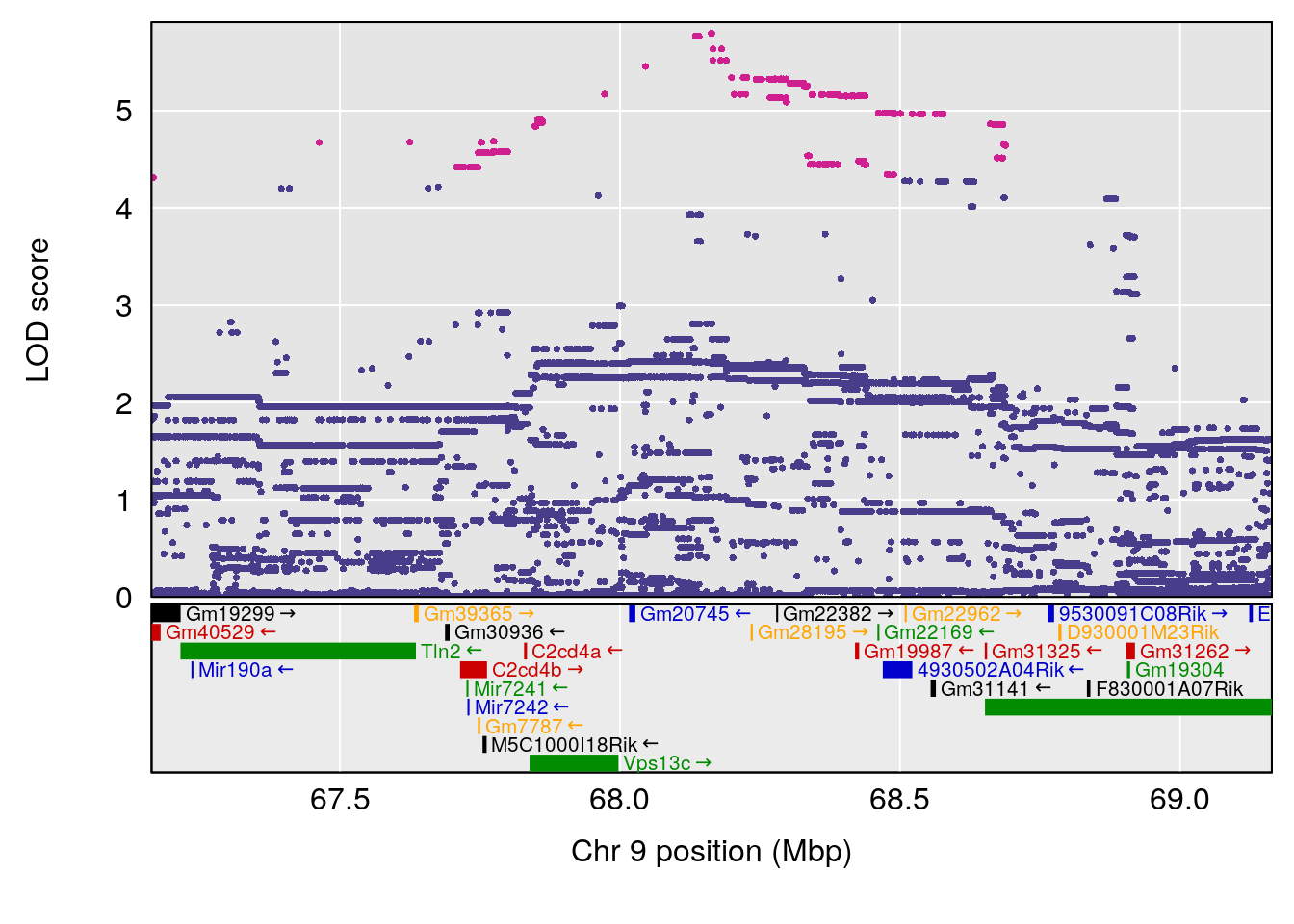
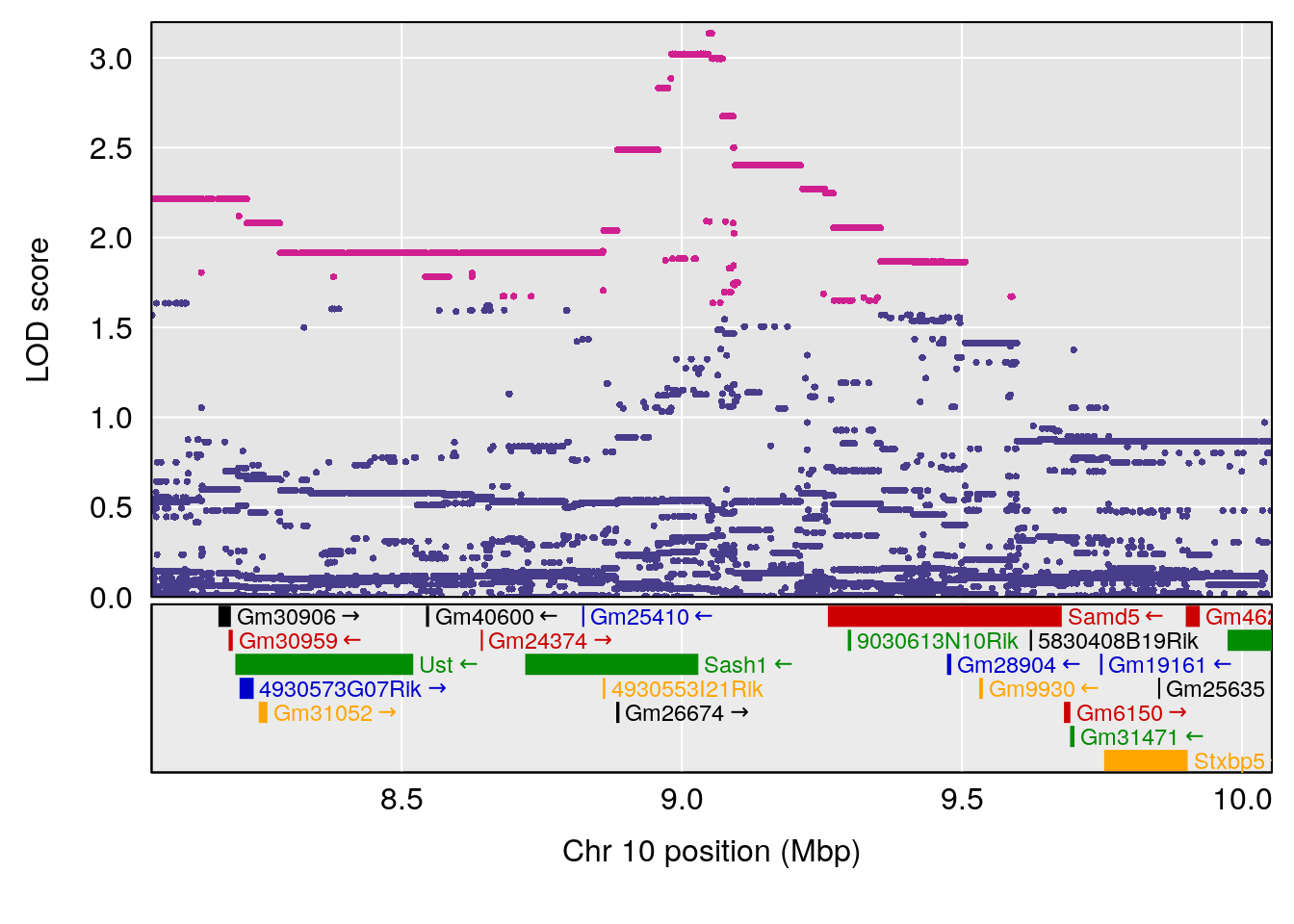
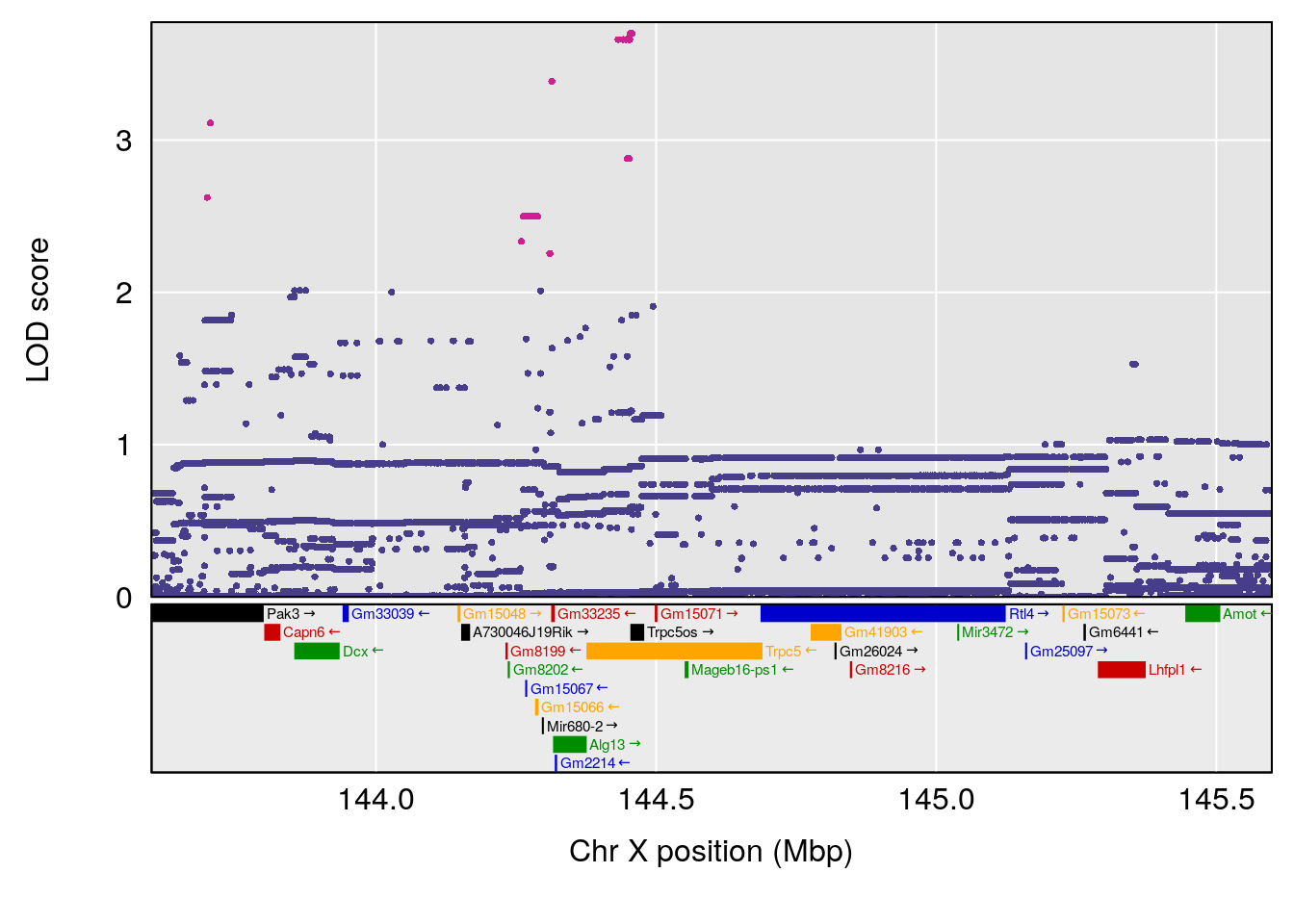
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
}
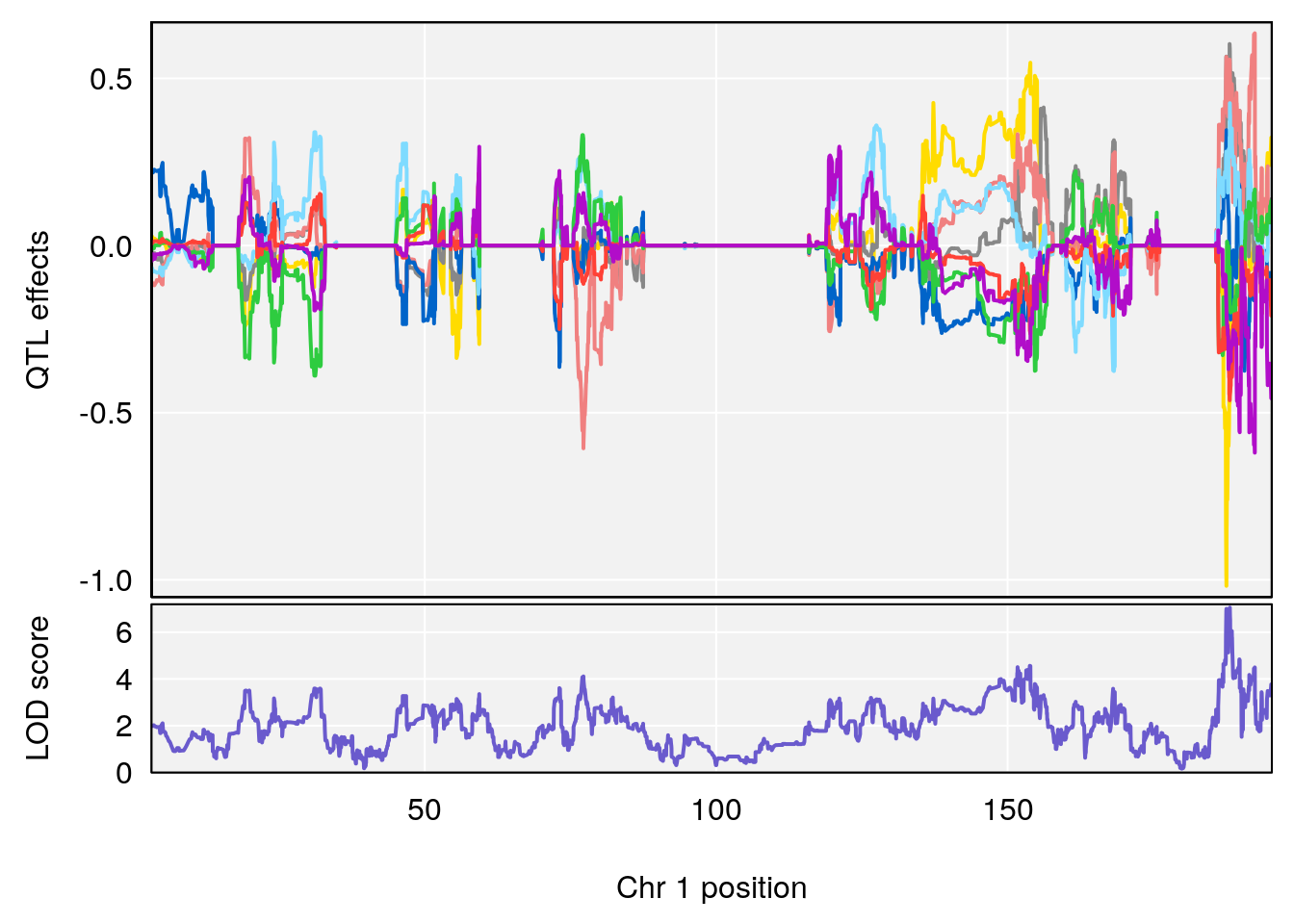
# [1] "Survival.Time"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 2 84.22989 6.046307 75.76507 162.24291
# 2 1 pheno1 3 138.79100 6.380273 64.39701 141.65520
# 3 1 pheno1 4 16.97218 6.542984 16.62465 18.61874
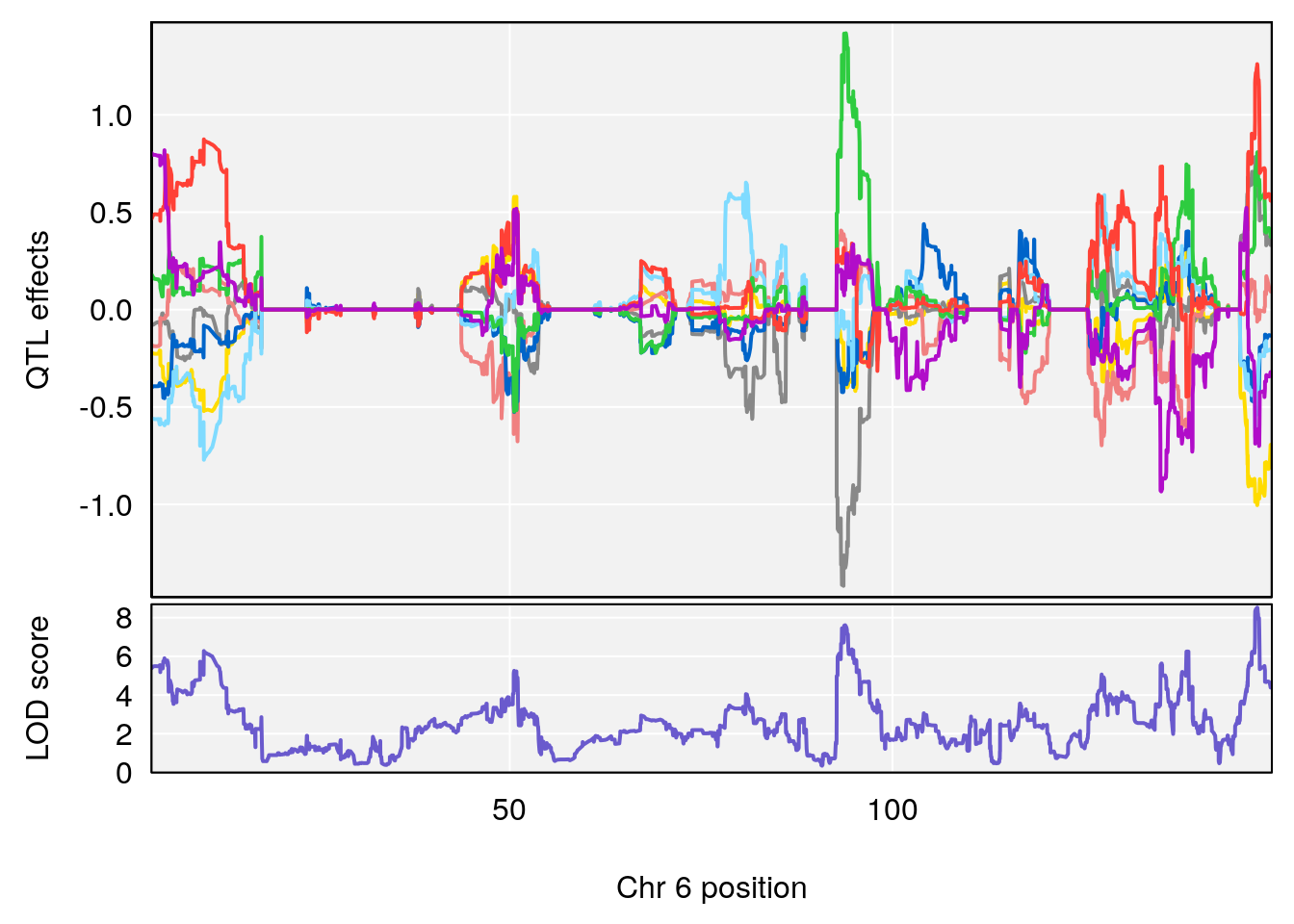
# 4 1 pheno1 6 147.65112 8.516164 93.37250 147.95398
# 5 1 pheno1 8 48.93015 6.406429 45.94523 49.27541
# 6 1 pheno1 10 120.58604 6.749862 119.72620 124.20834
# 7 1 pheno1 11 117.48686 7.807276 117.09262 117.56616
# 8 1 pheno1 X 48.33118 6.518895 42.26096 98.87872
# [1] 1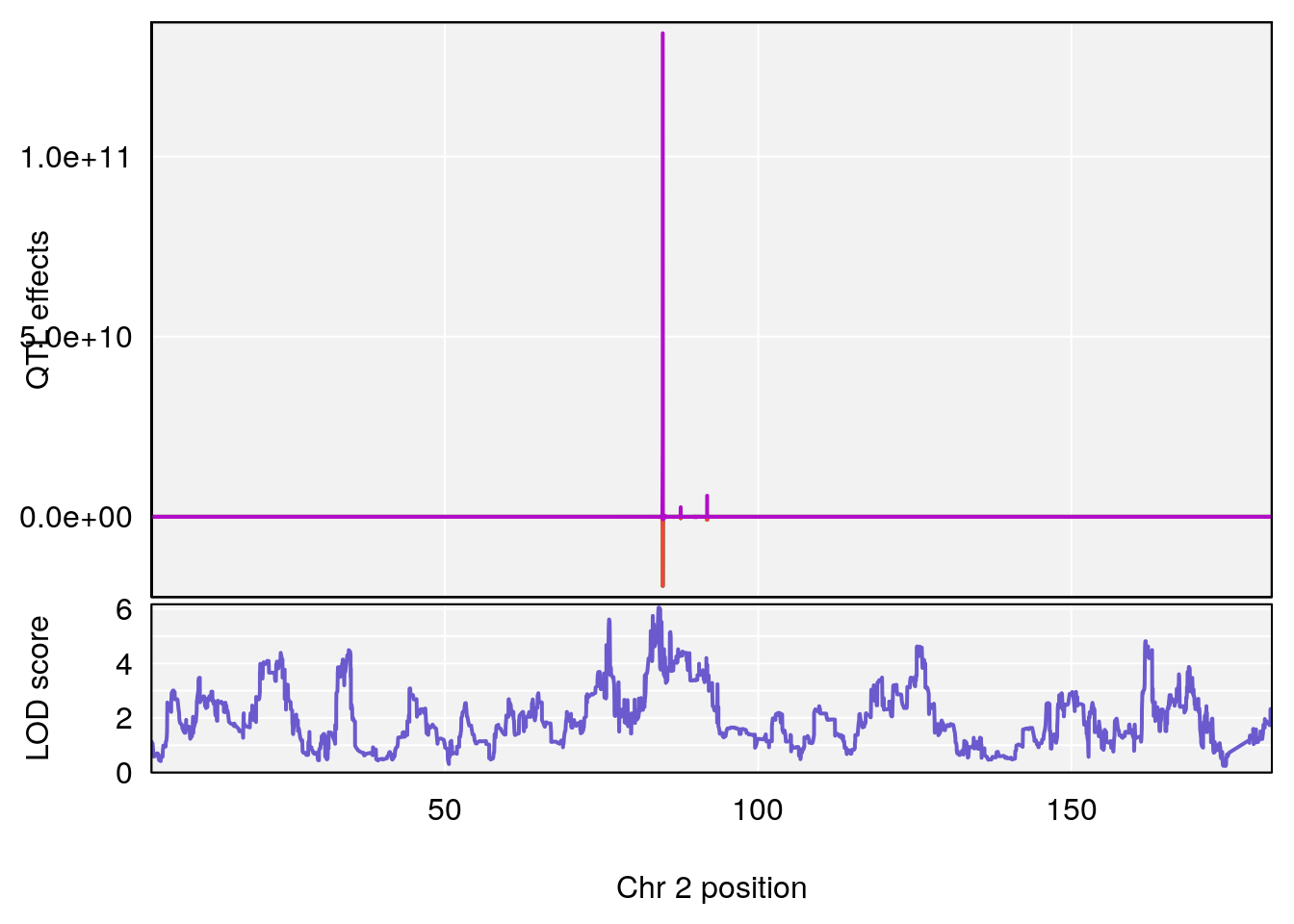
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 2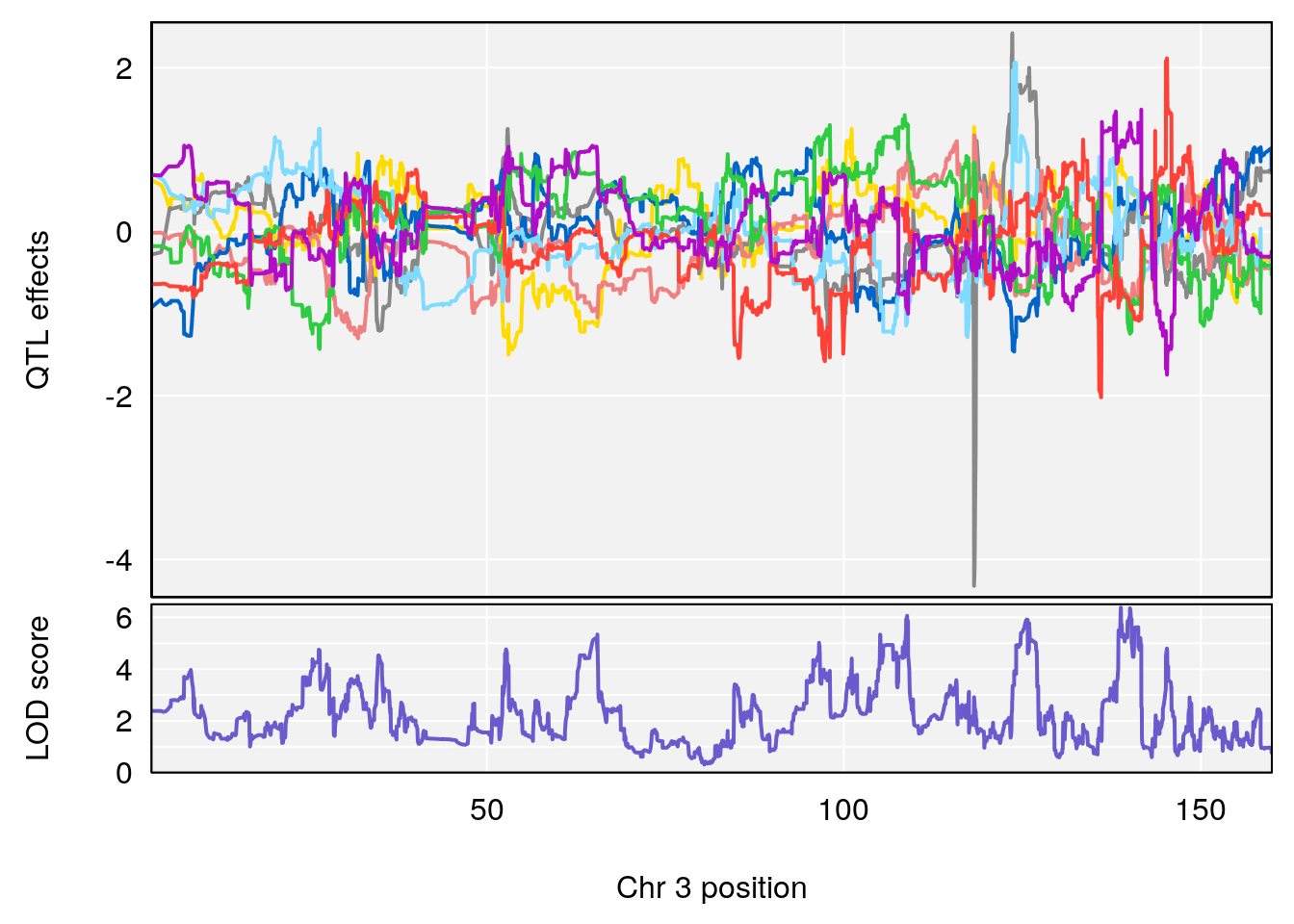
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 3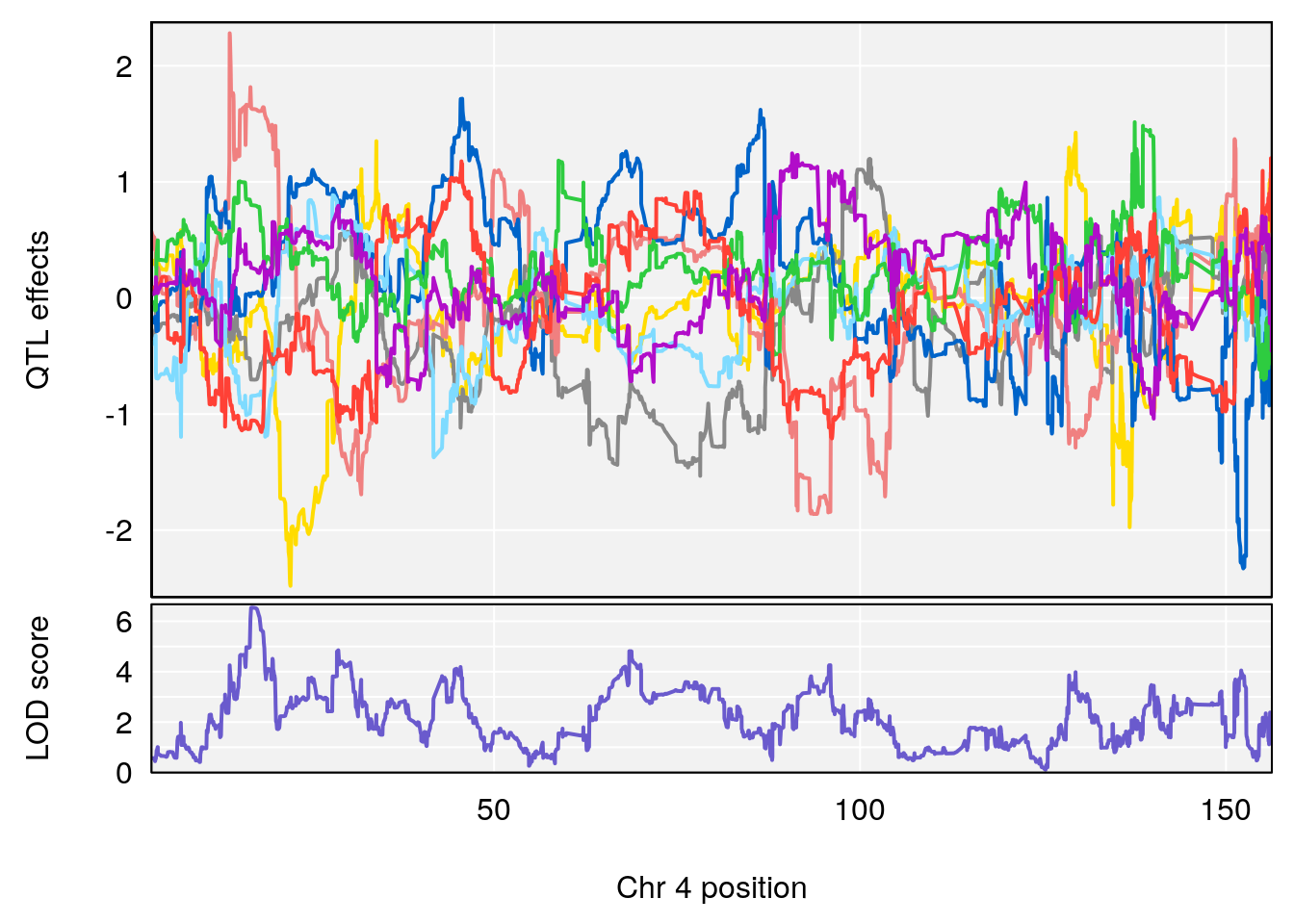
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
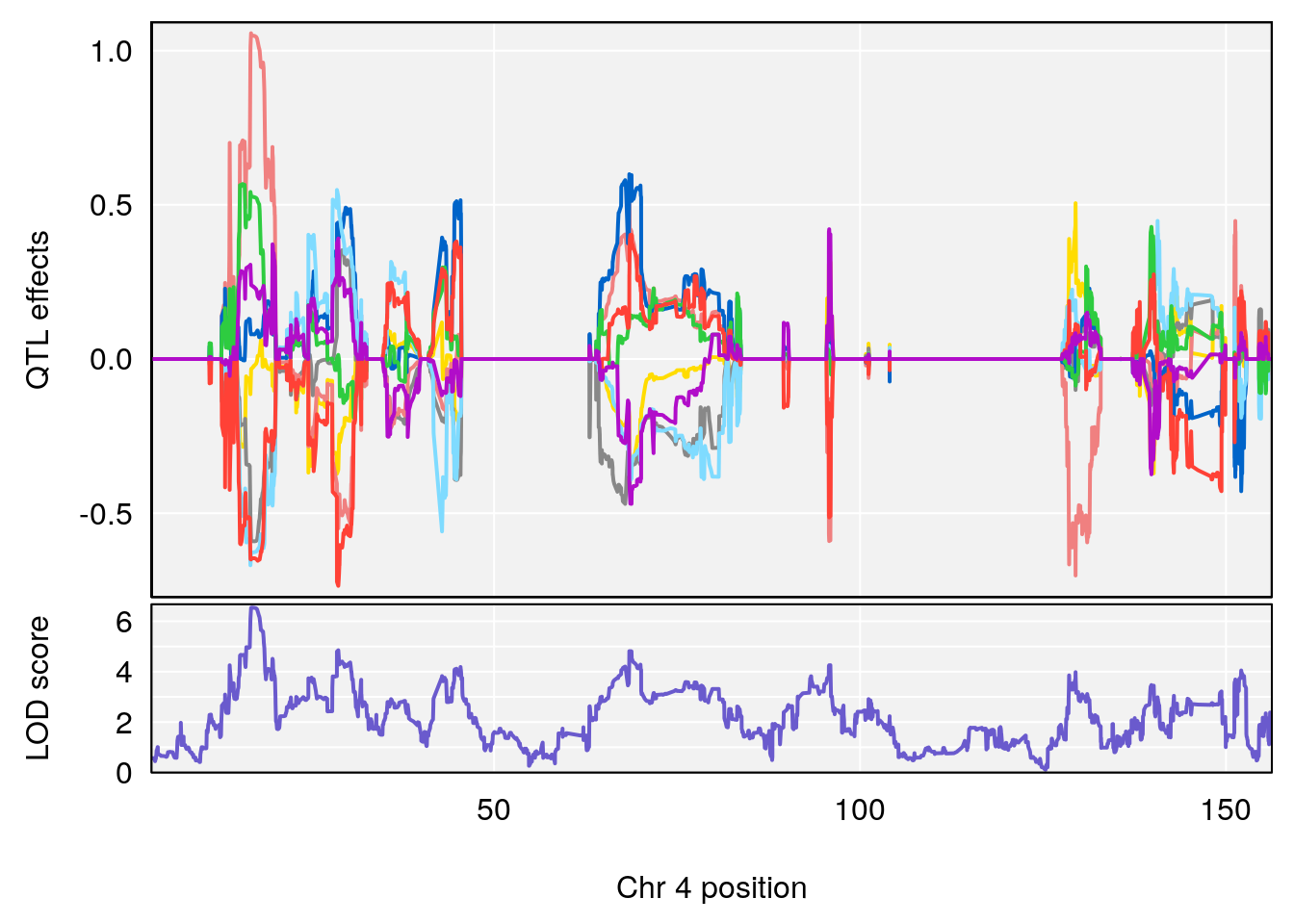
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 4
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 5
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 6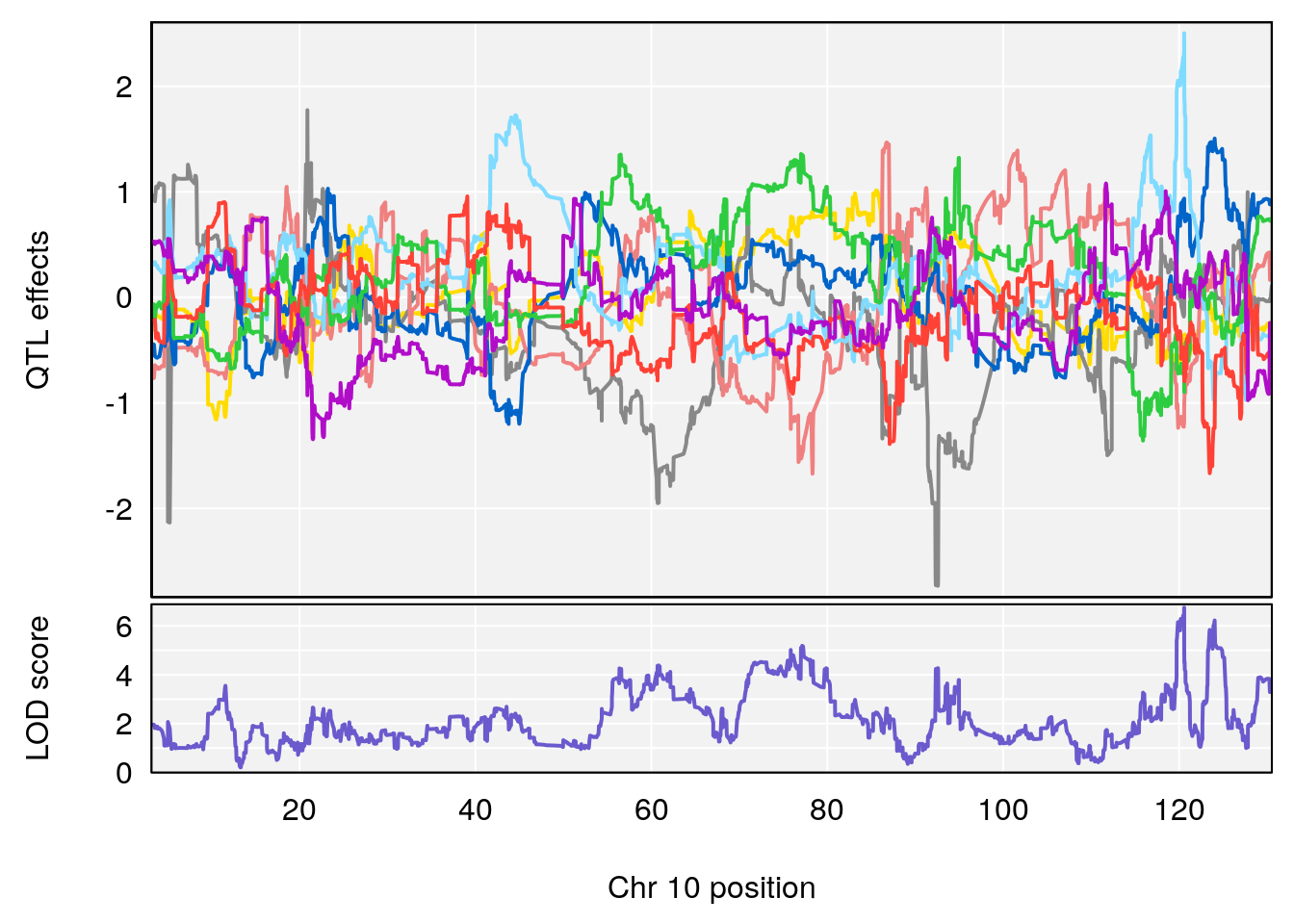
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 7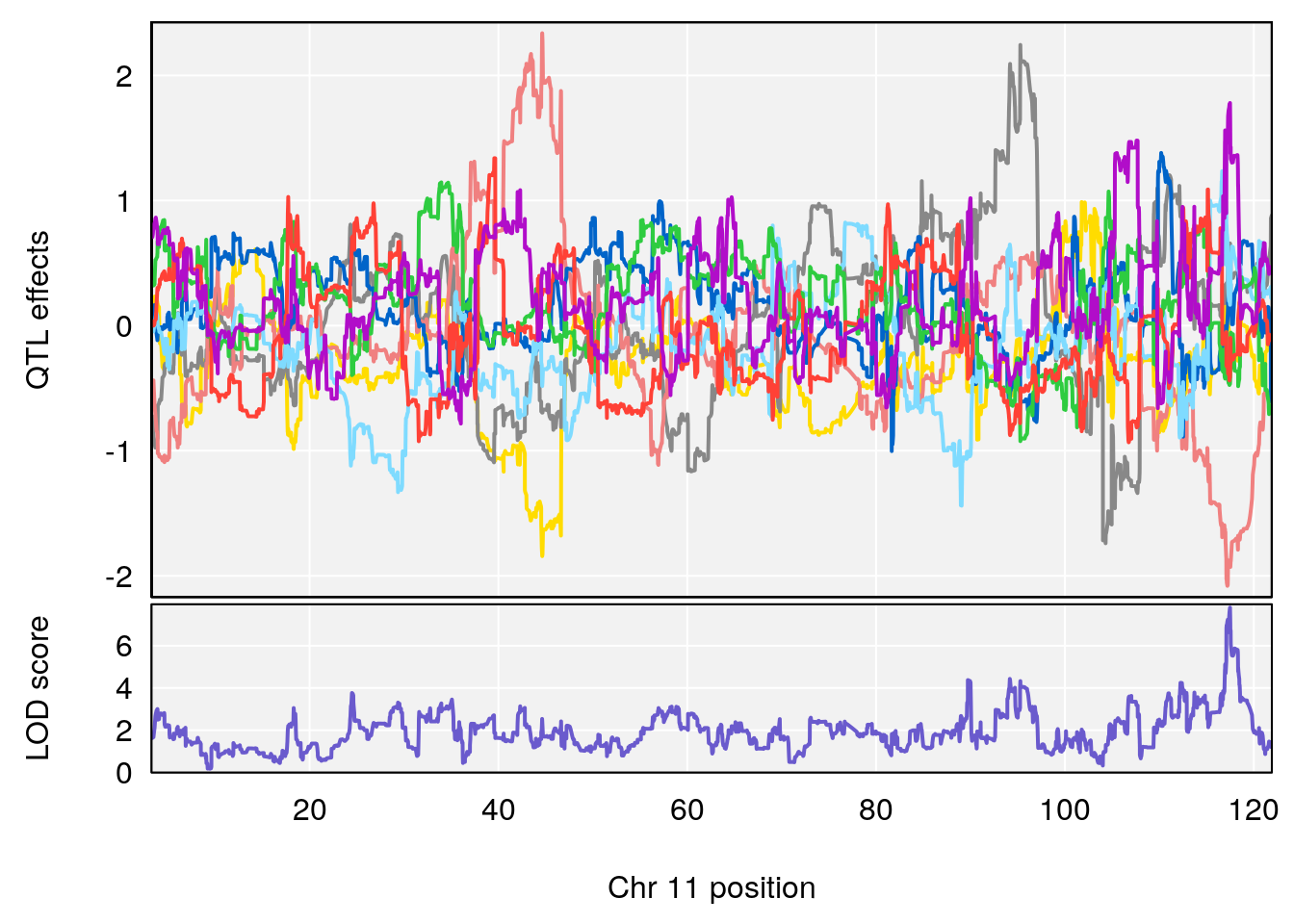
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 8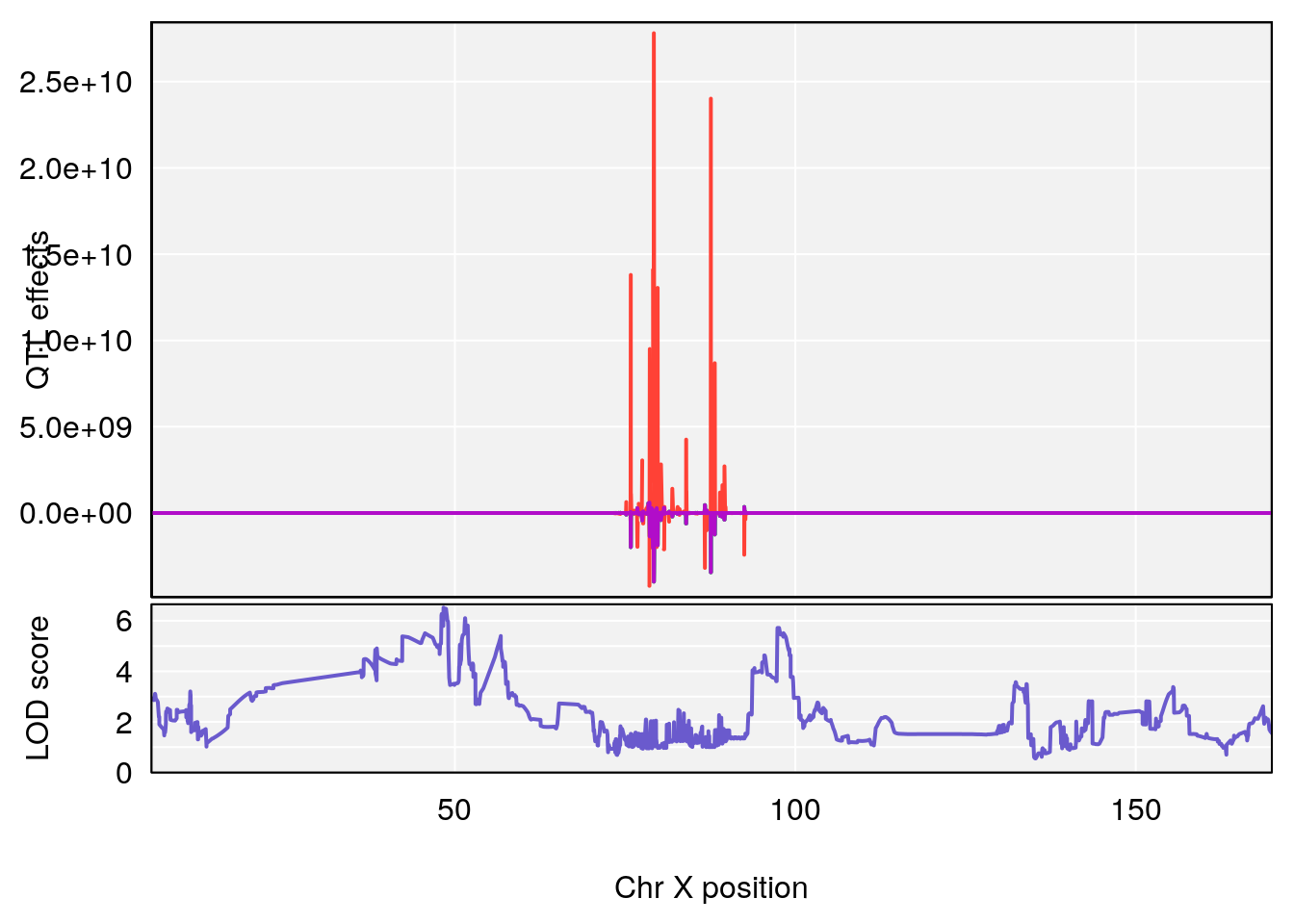
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
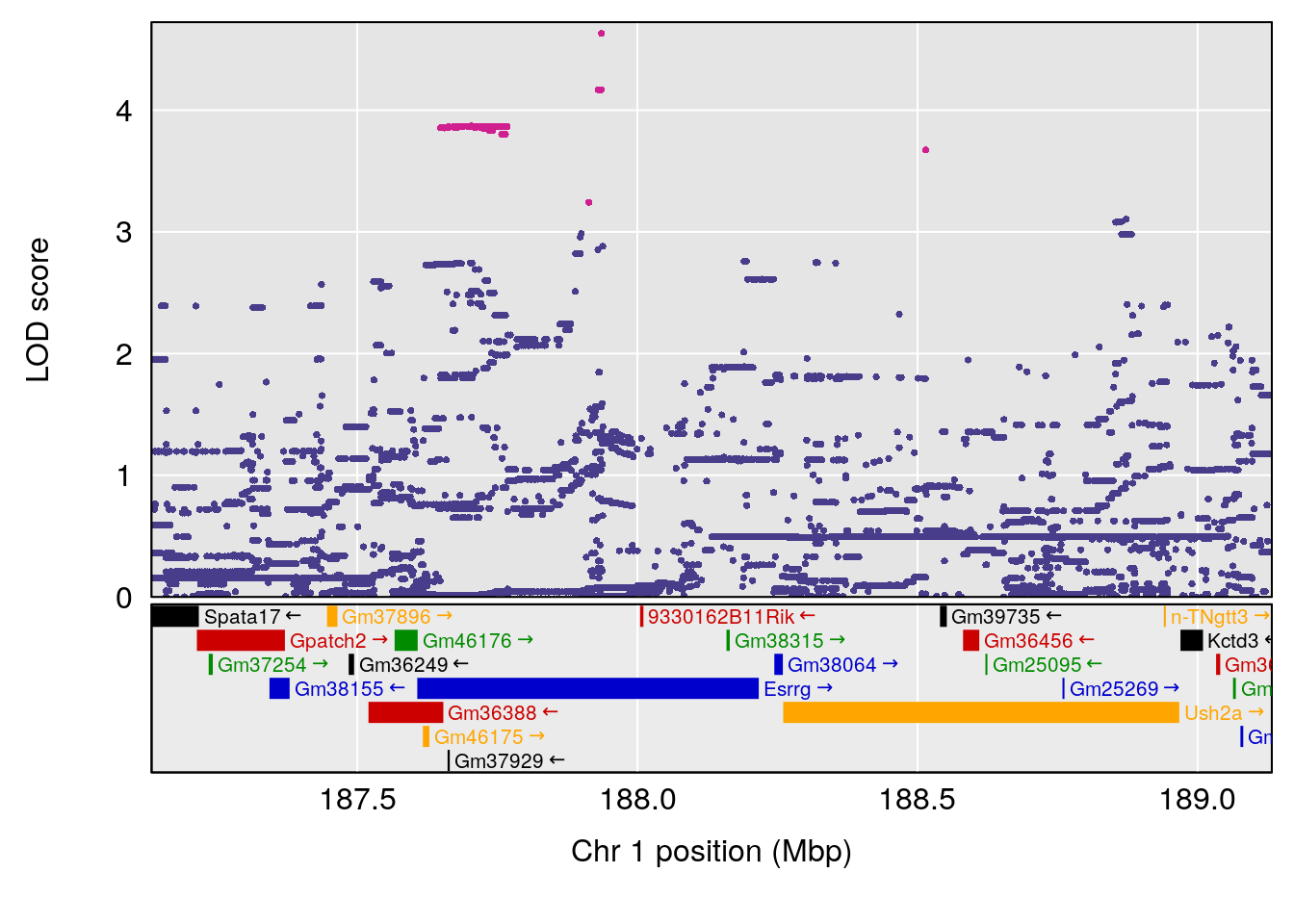
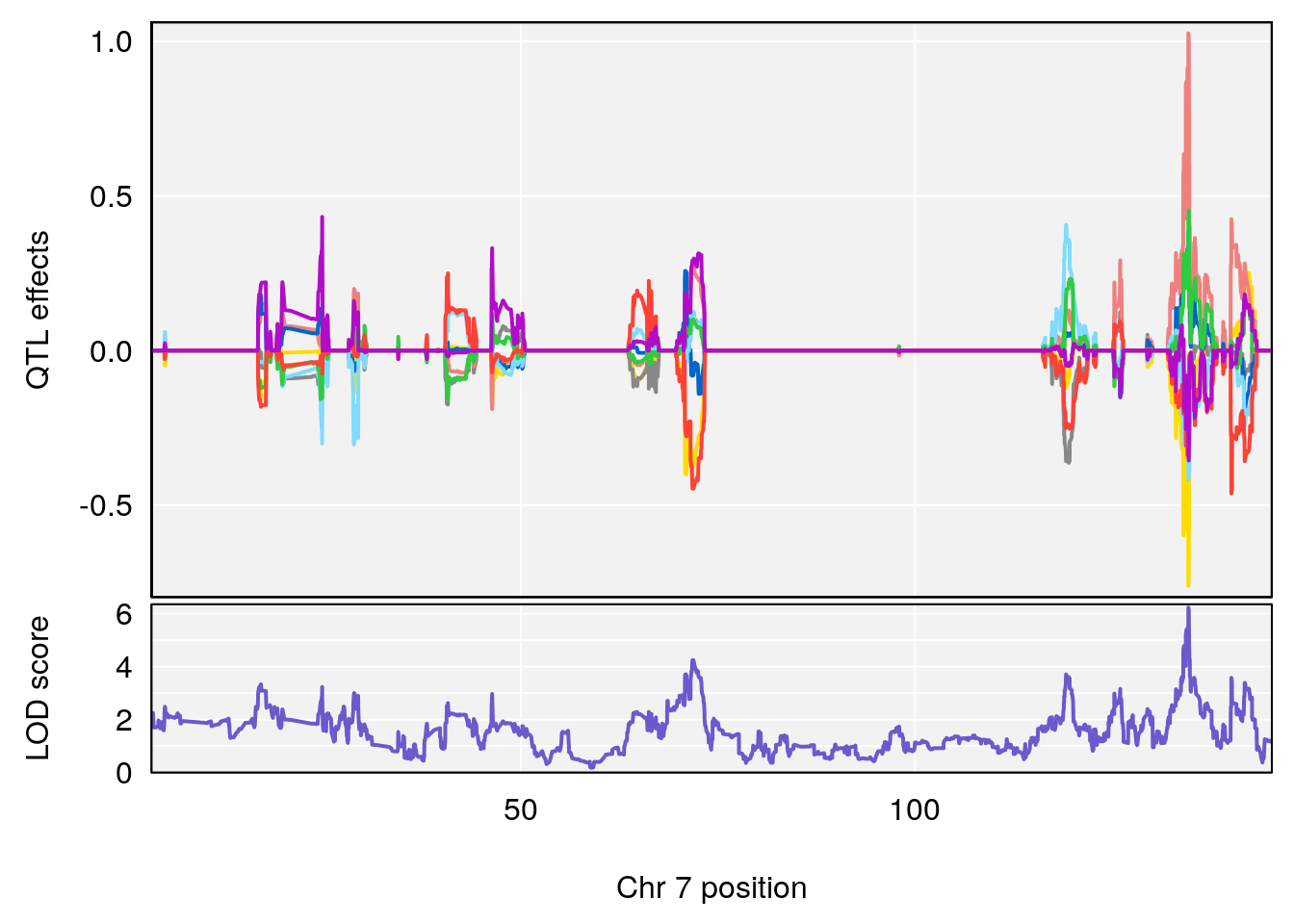
# [1] "Recovery.Time"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 188.13243 7.053141 187.4489 188.56172
# 2 1 pheno1 2 116.23927 7.091868 114.6279 117.24777
# 3 1 pheno1 7 134.72640 6.233802 134.1477 134.83353
# 4 1 pheno1 19 15.78318 6.236367 11.6891 16.37066
# [1] 1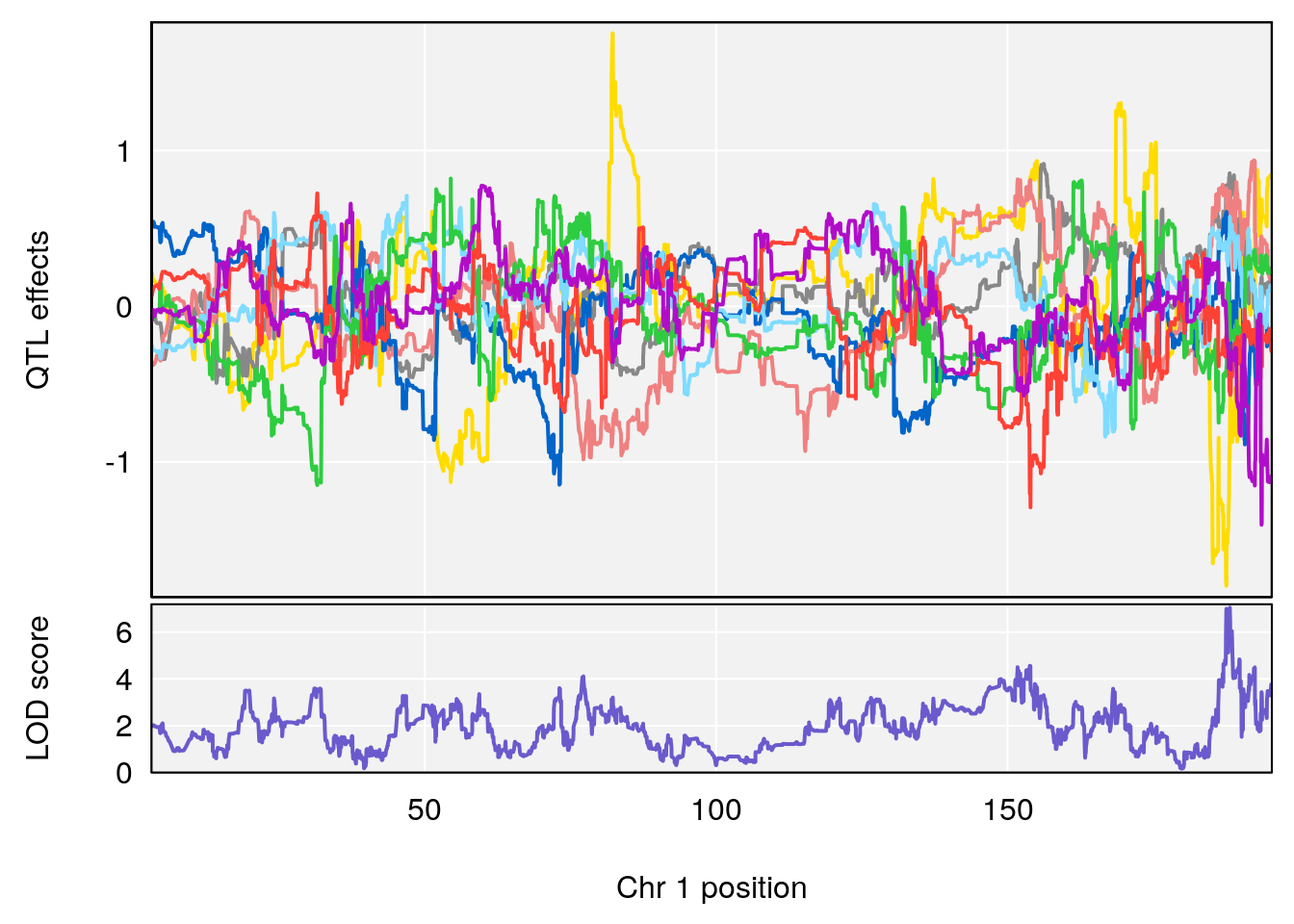
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 2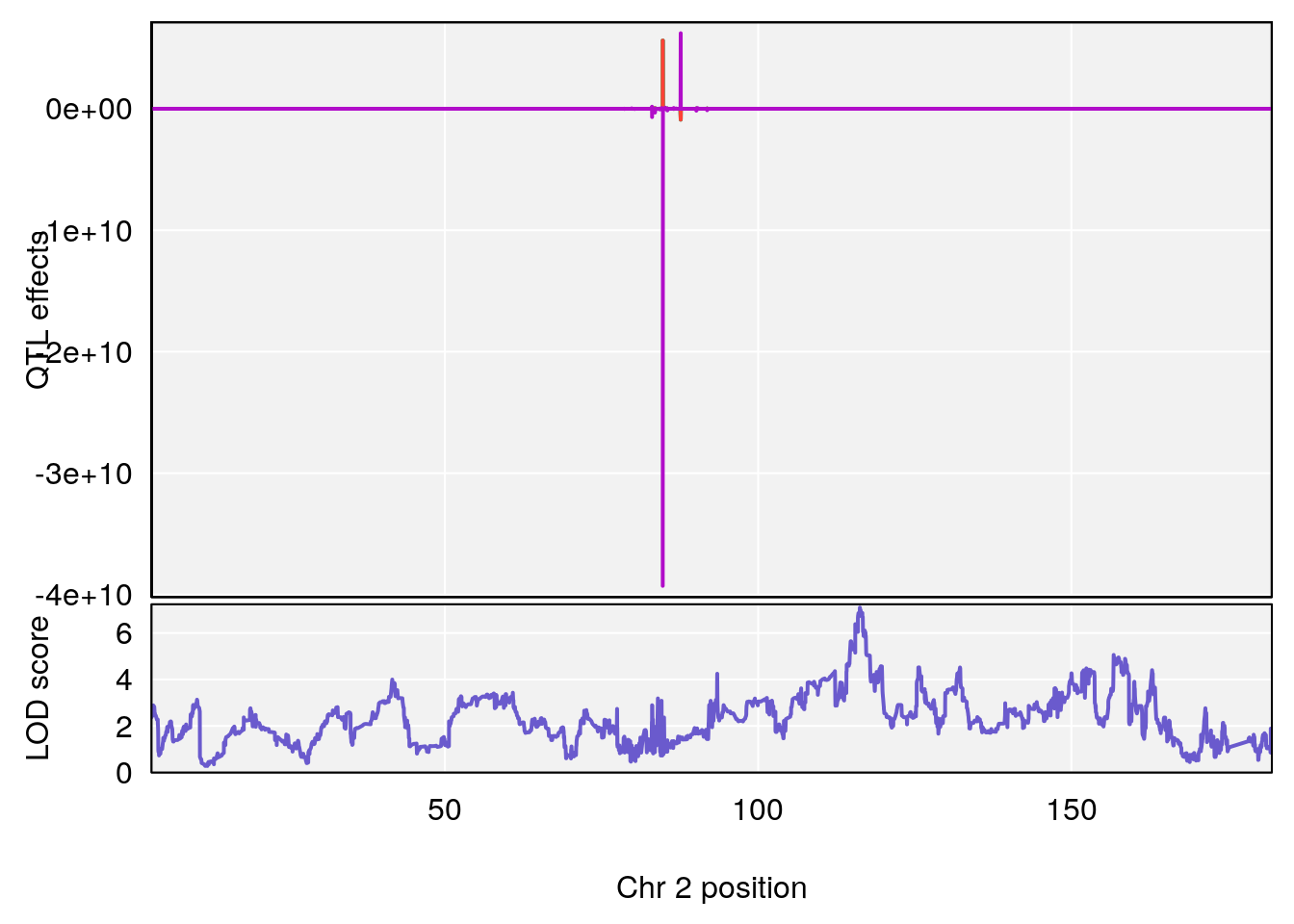
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
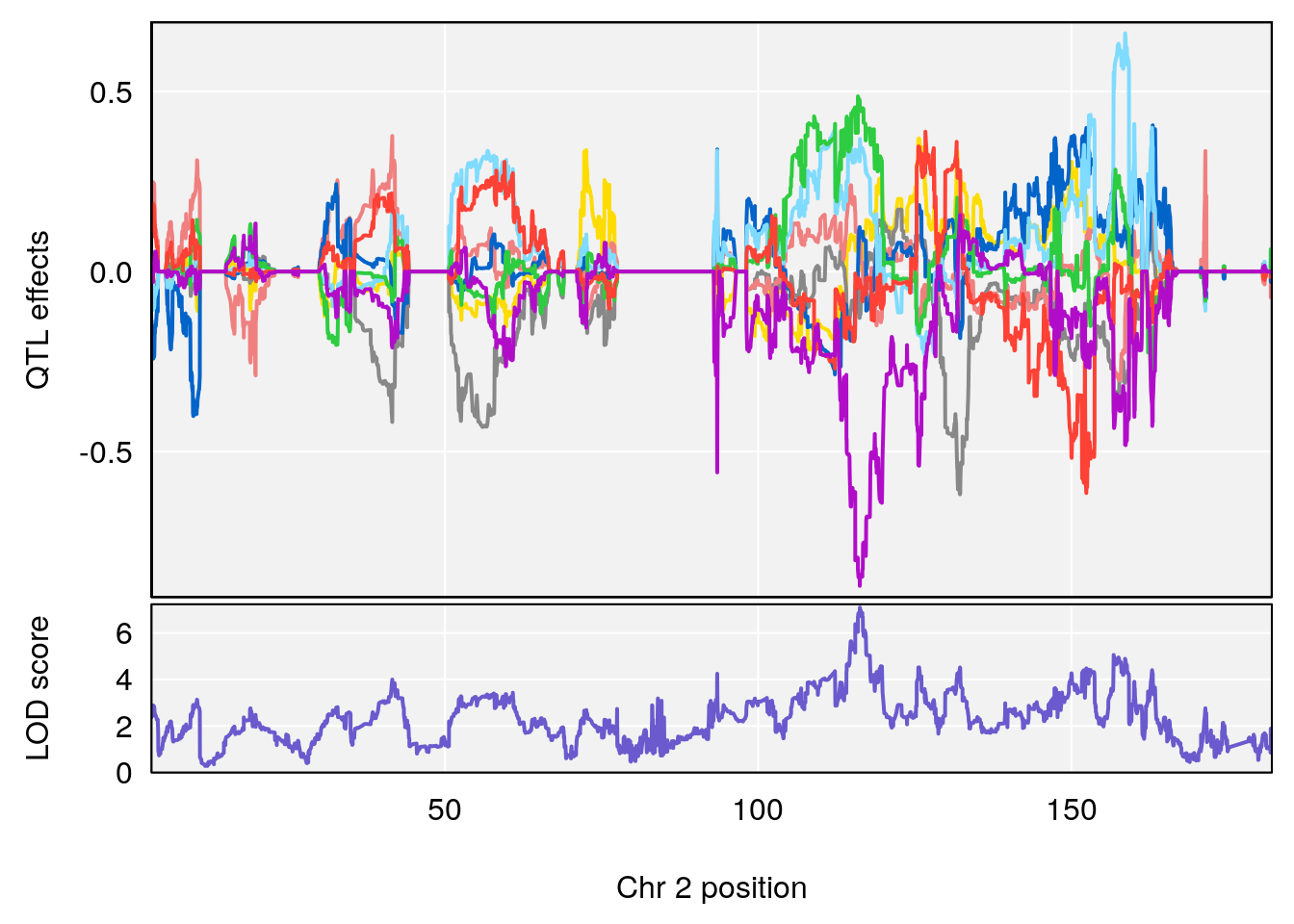
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 3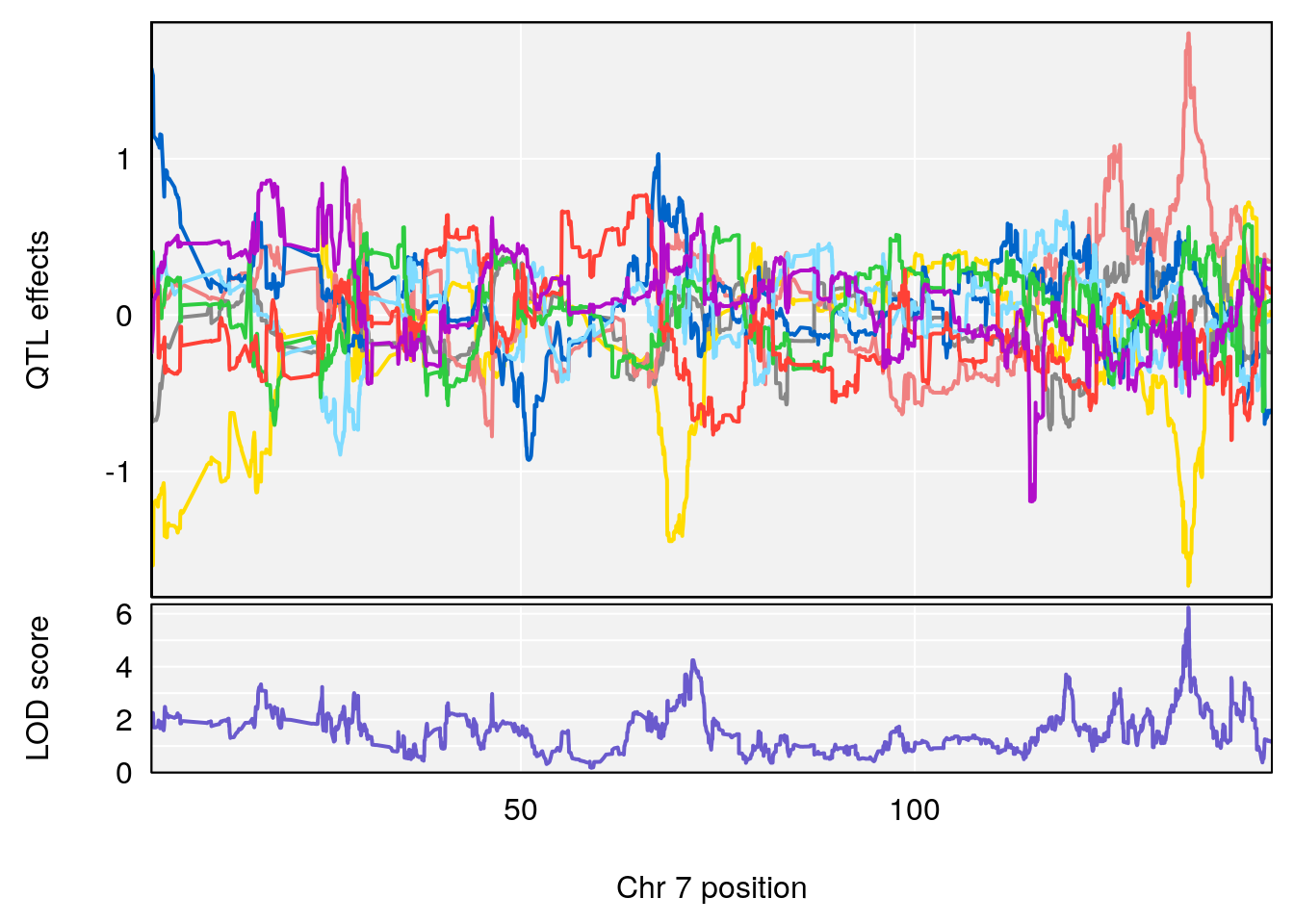
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 4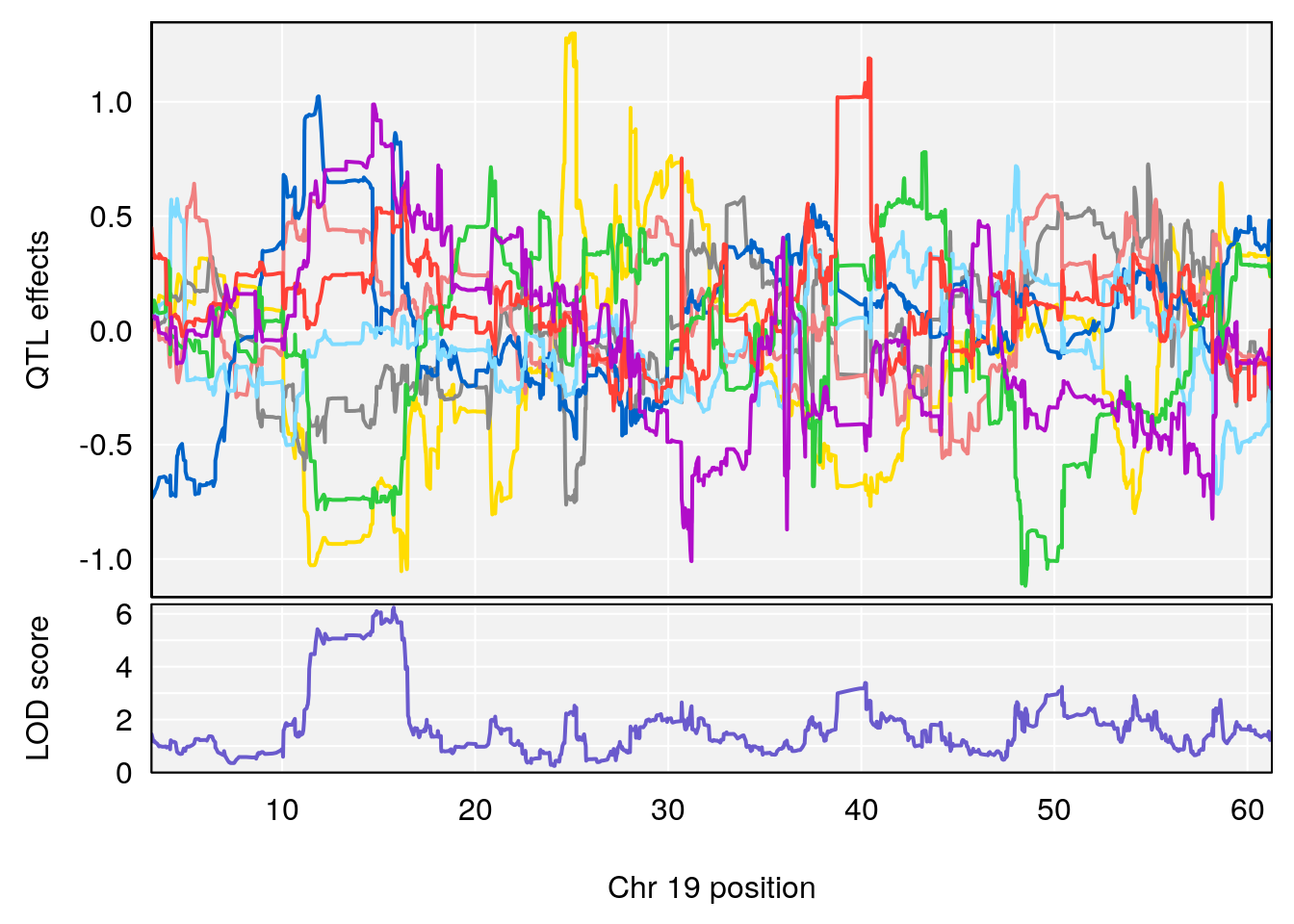
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
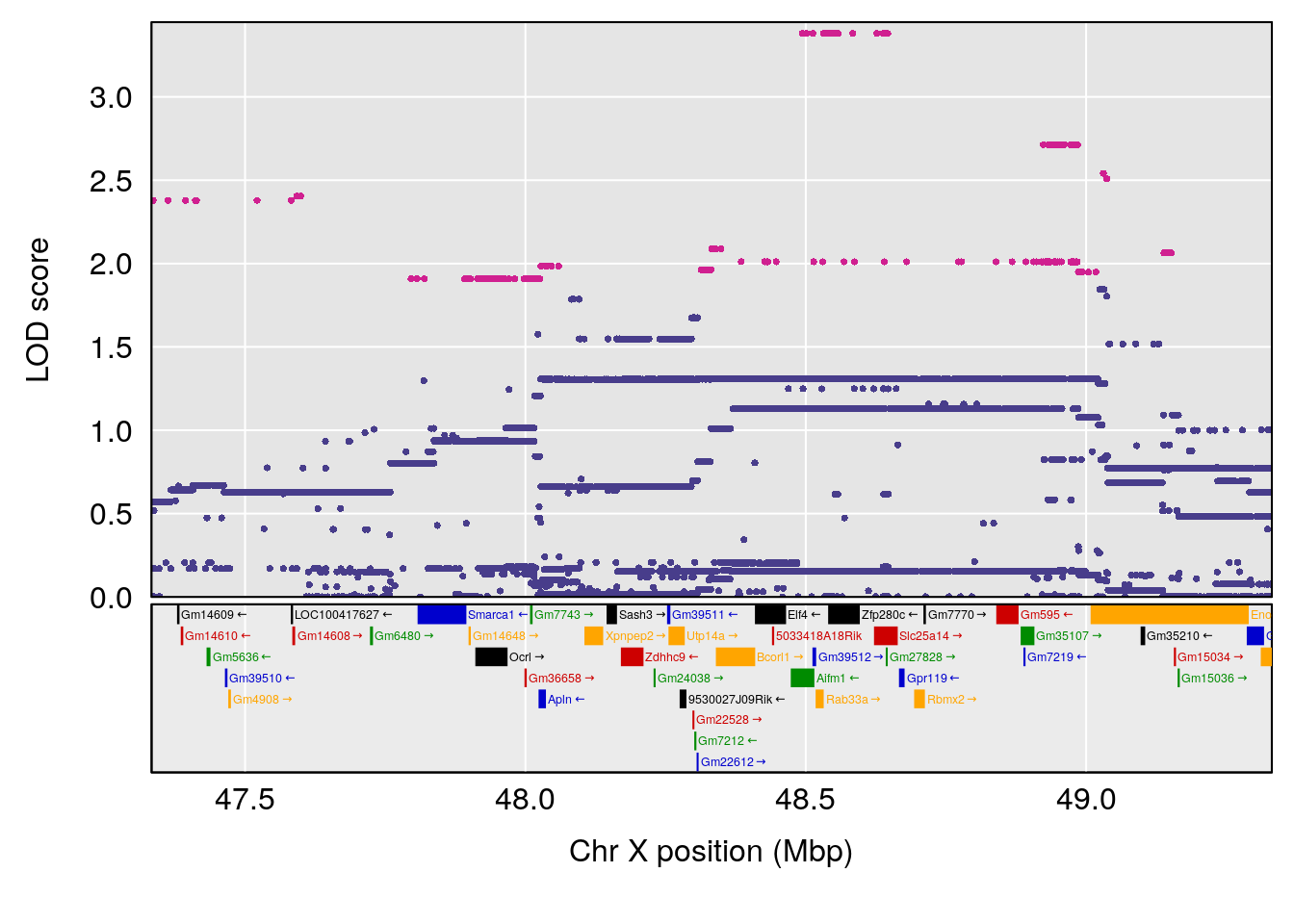
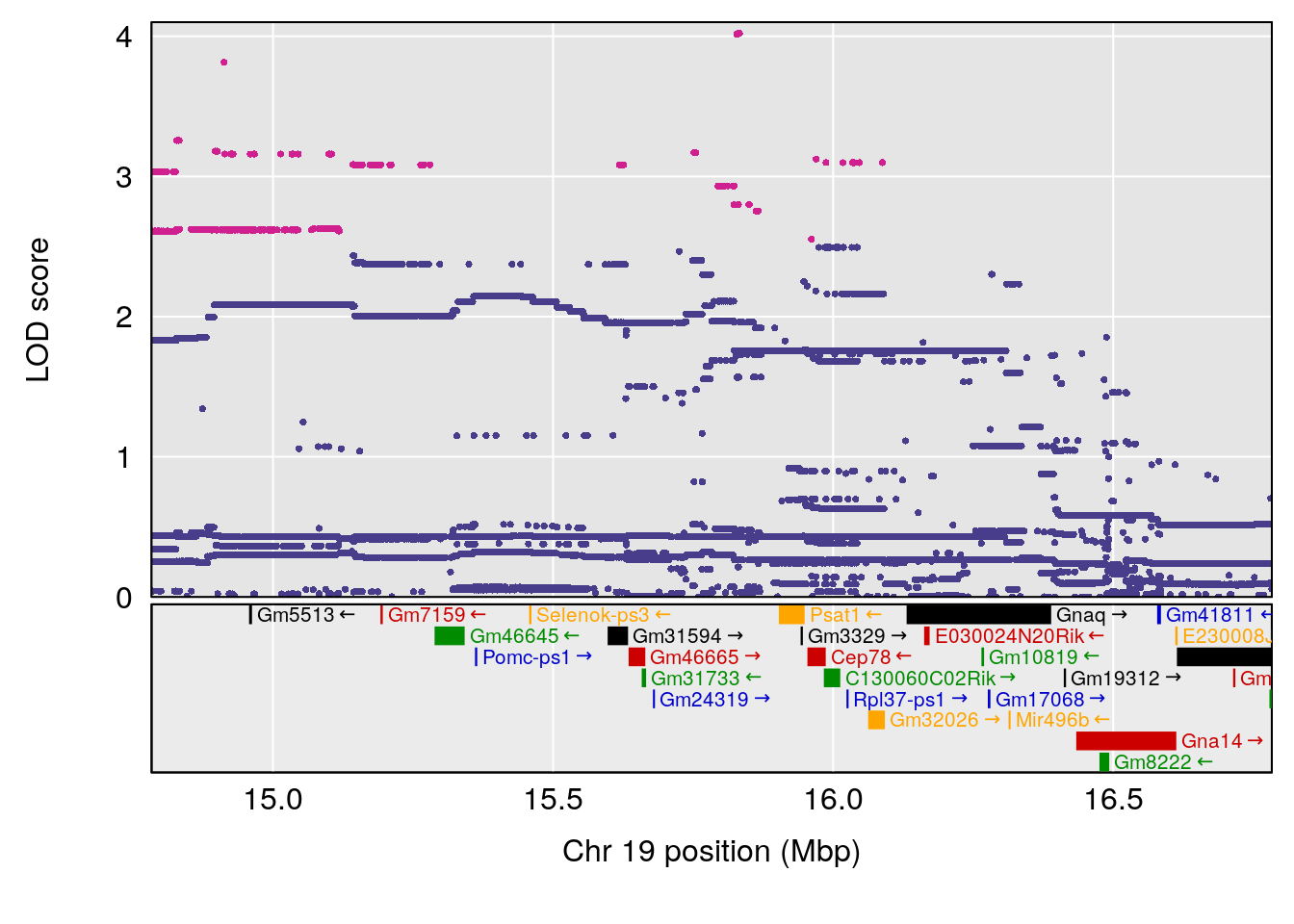
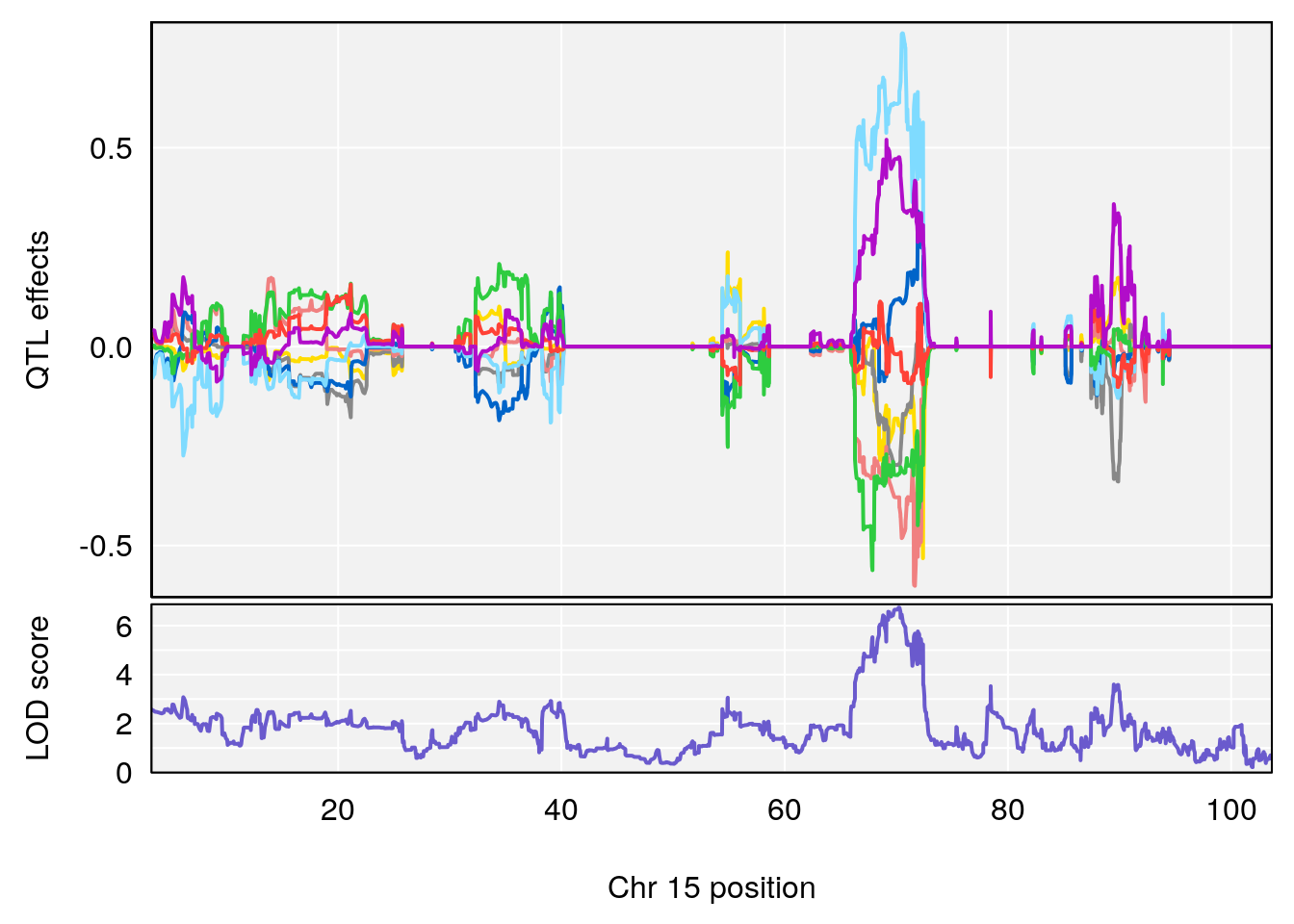
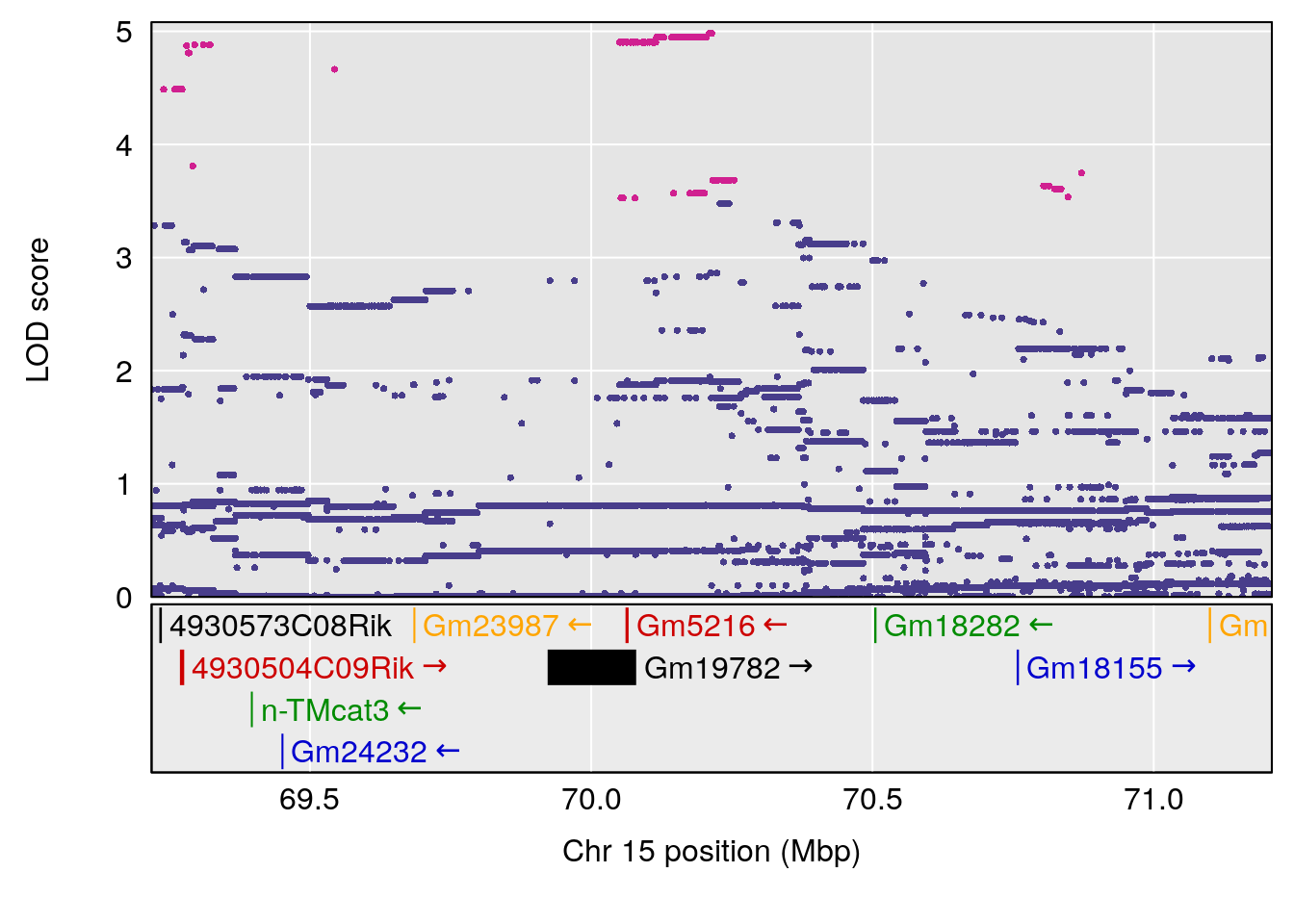
# [1] "Min.depression"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
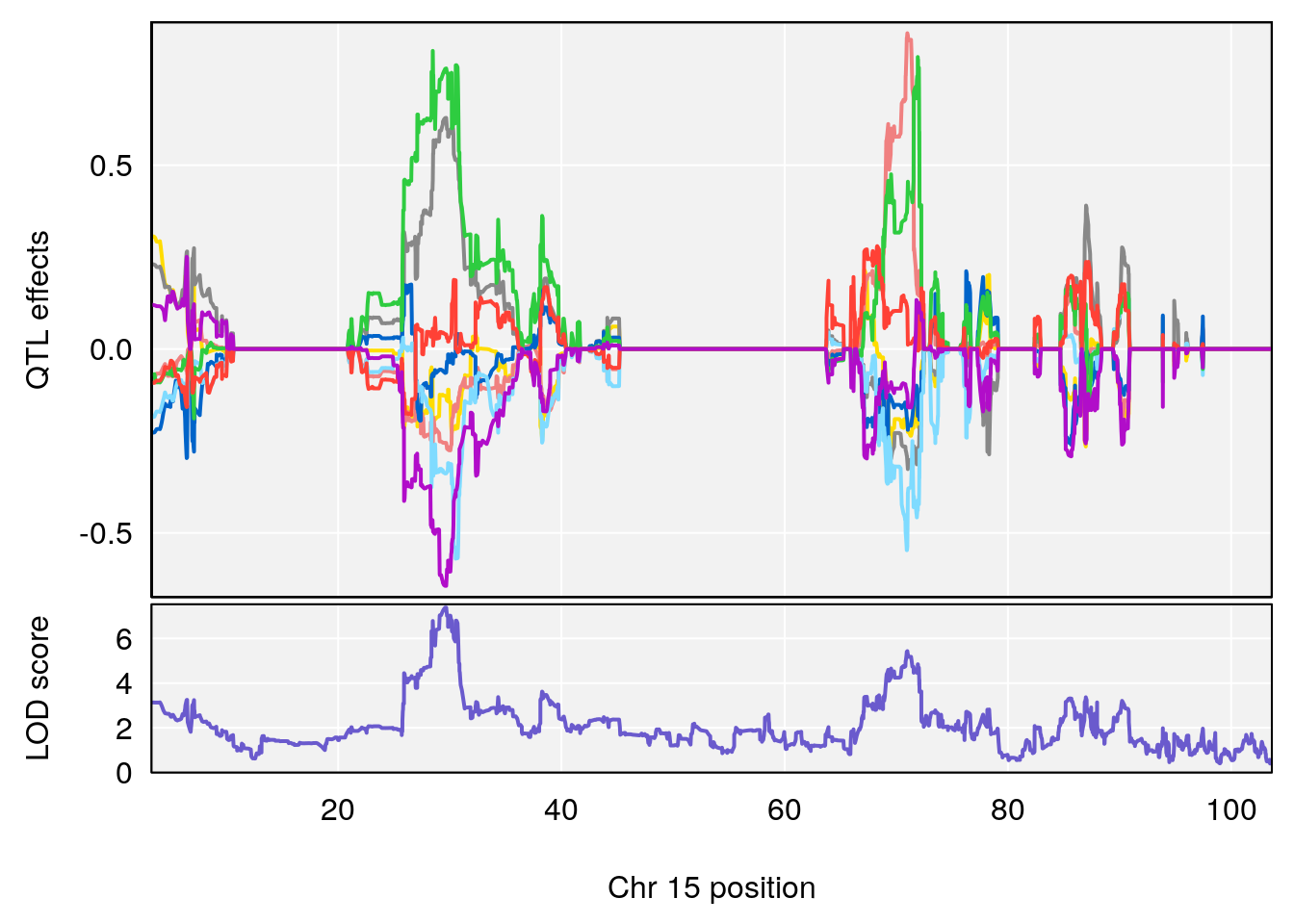
# 1 1 pheno1 15 70.21387 6.737292 67.74024 72.24396
# 2 1 pheno1 18 27.95989 6.788787 27.78975 28.82732
# [1] 1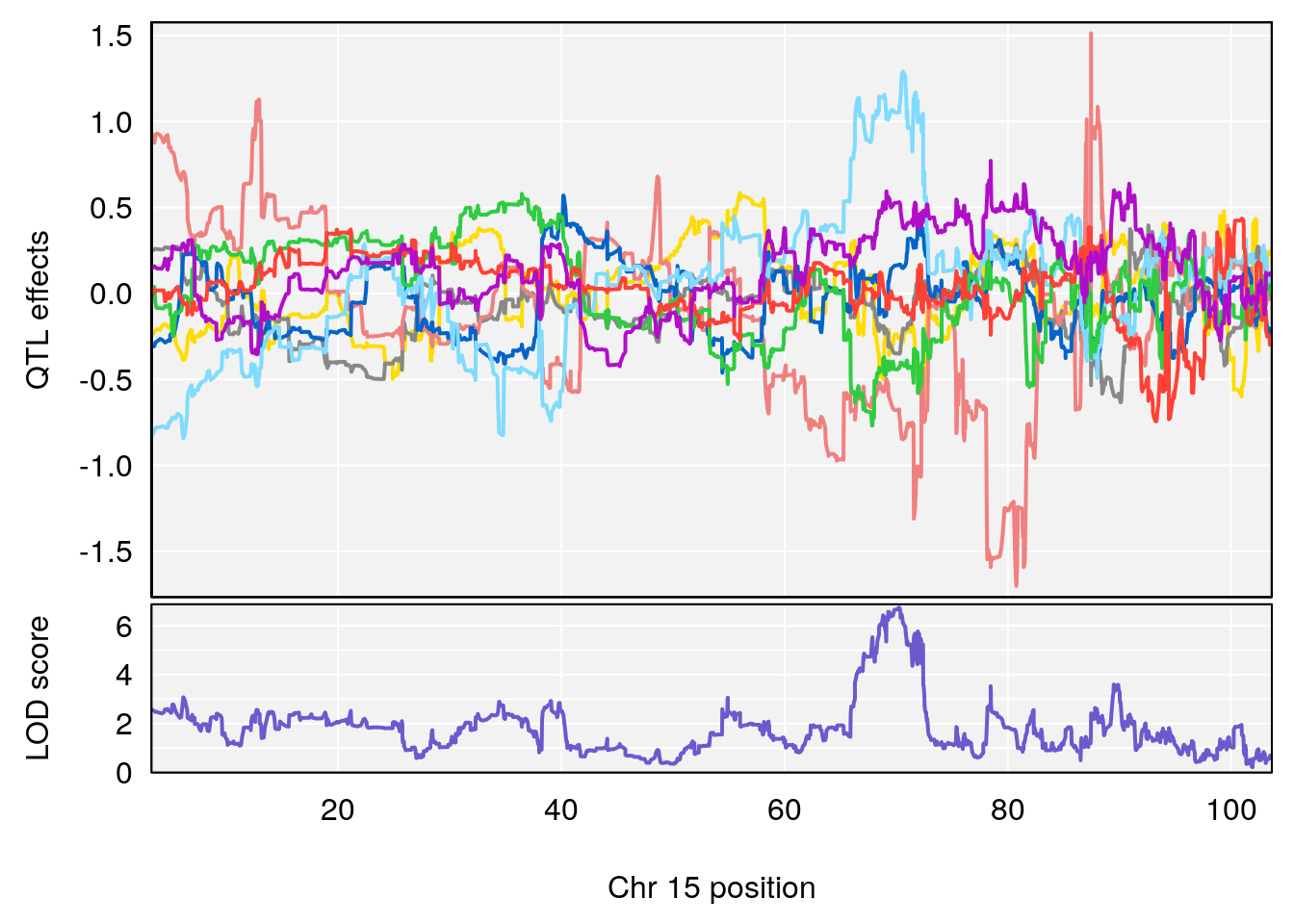
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 2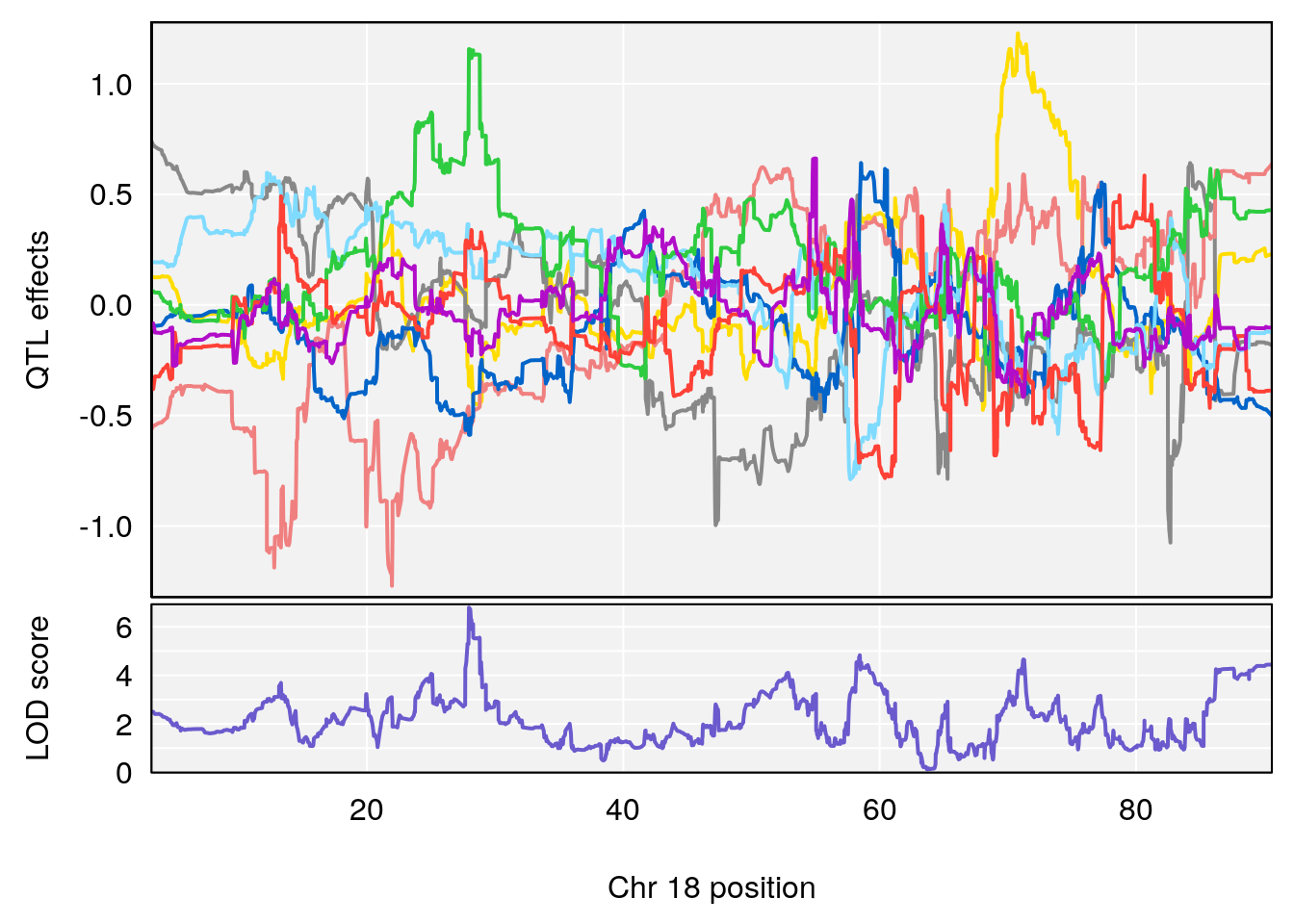
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
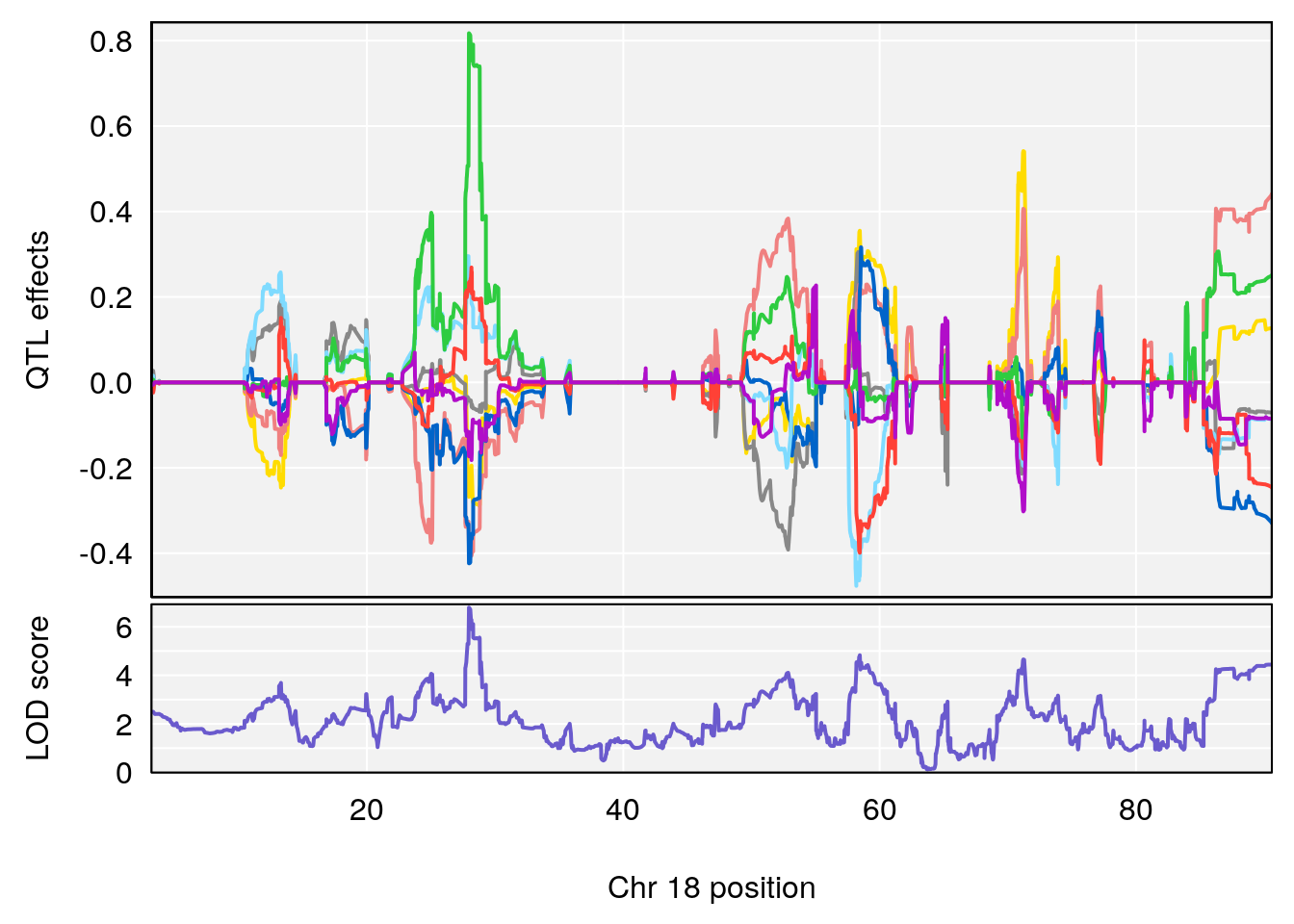
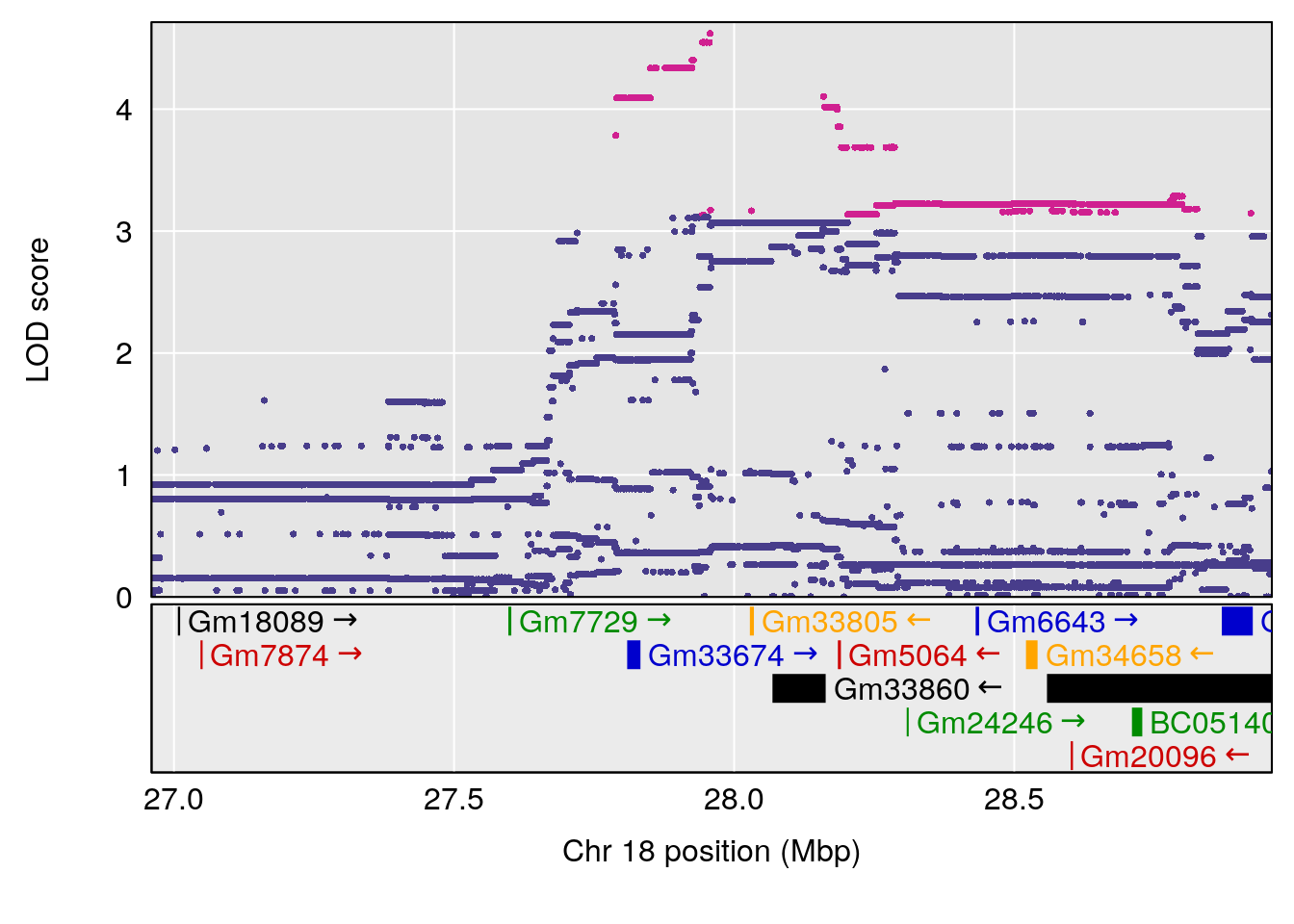
# [1] "Status_bin"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
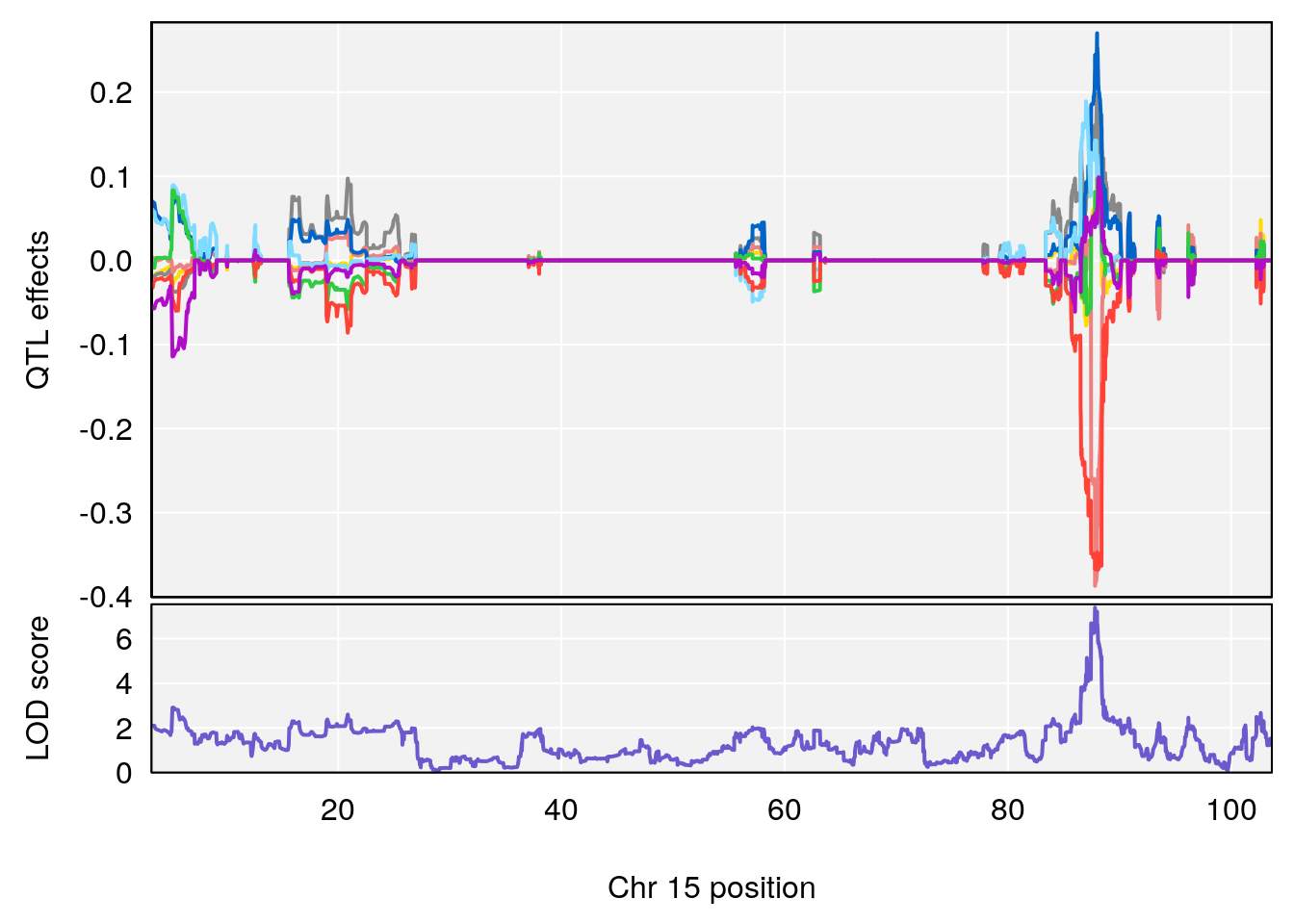
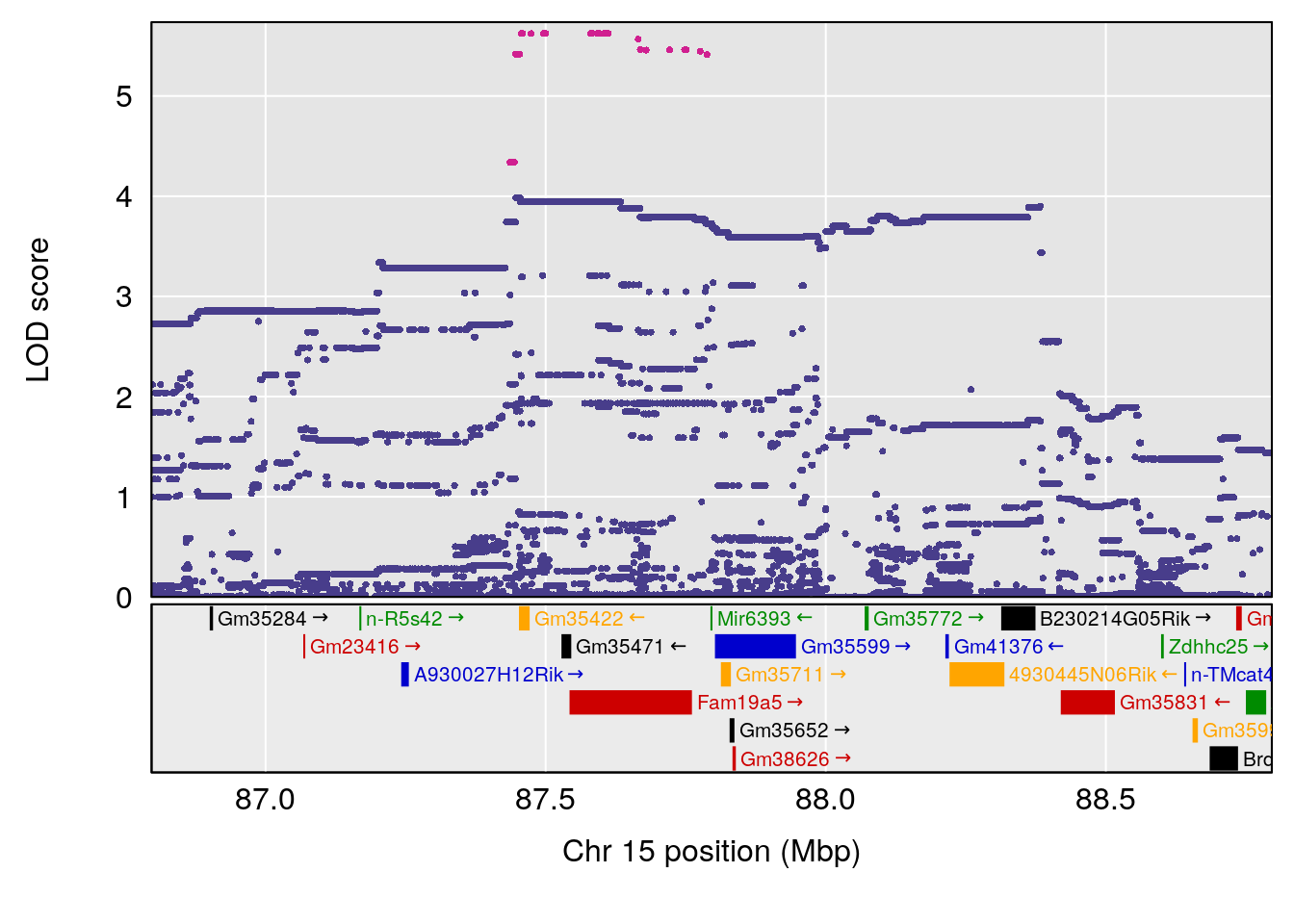
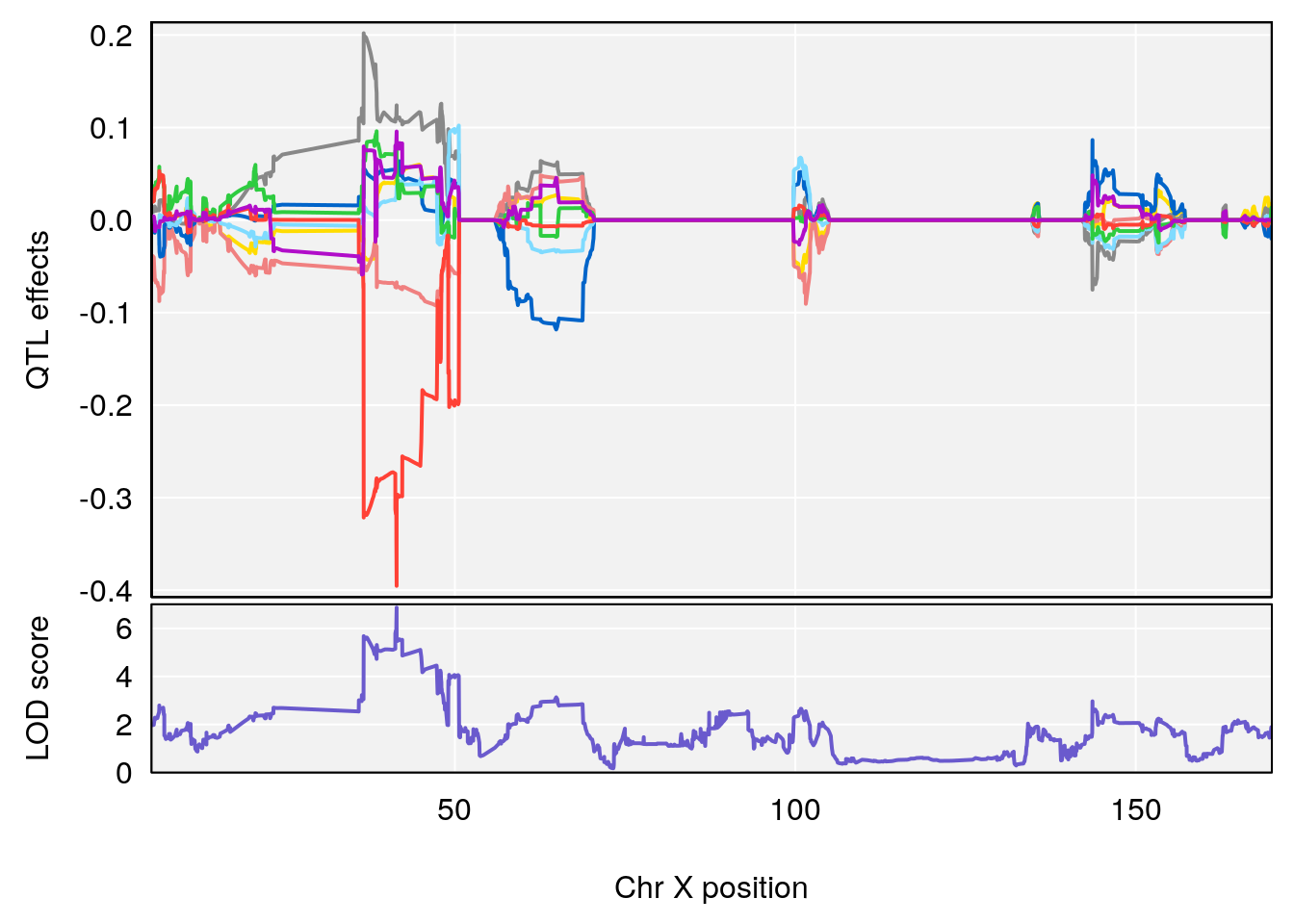
# 1 1 pheno1 15 87.79636 7.389801 87.42813 88.11448
# 2 1 pheno1 X 41.44546 6.867848 36.58281 42.28244
# [1] 1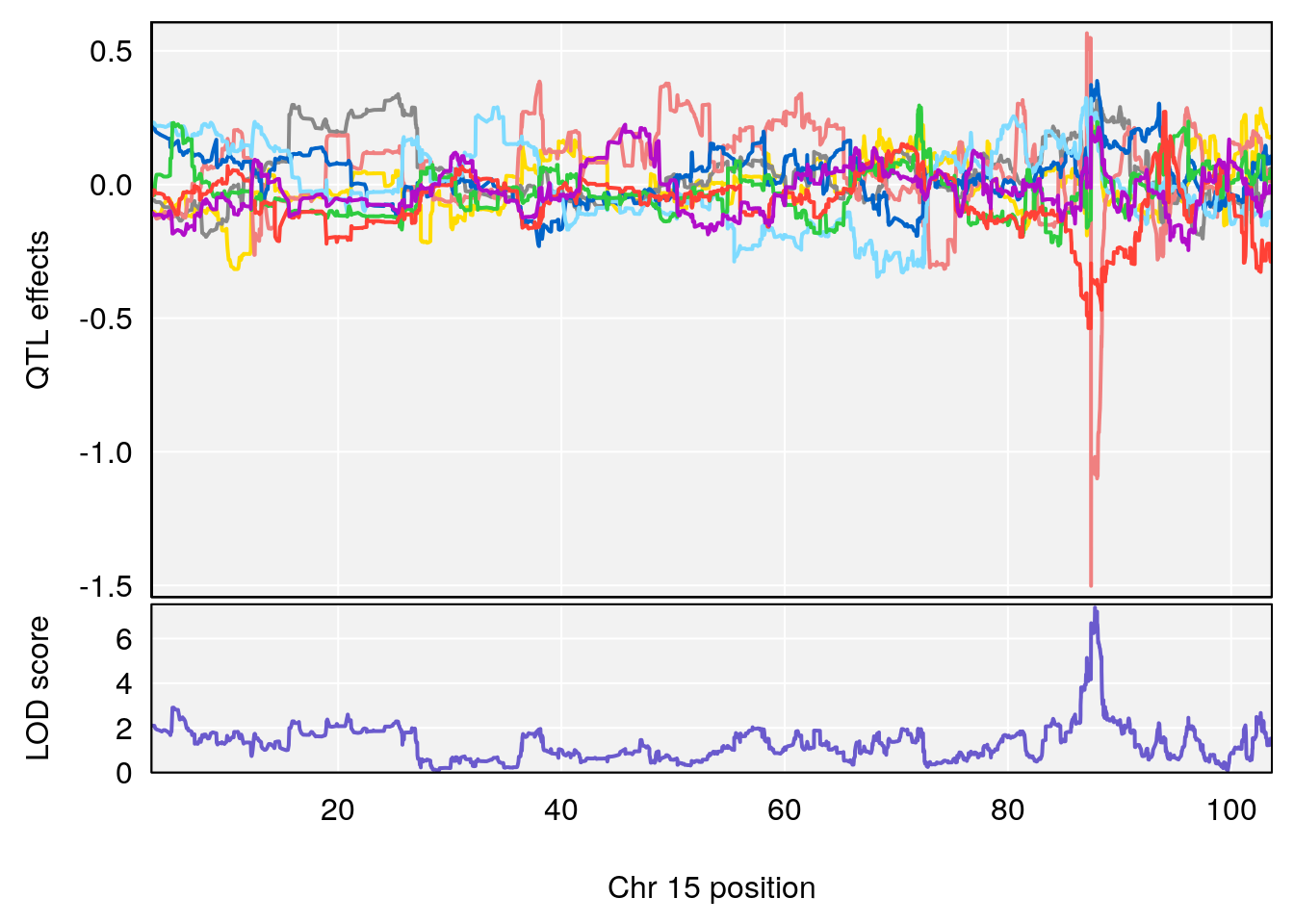
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 2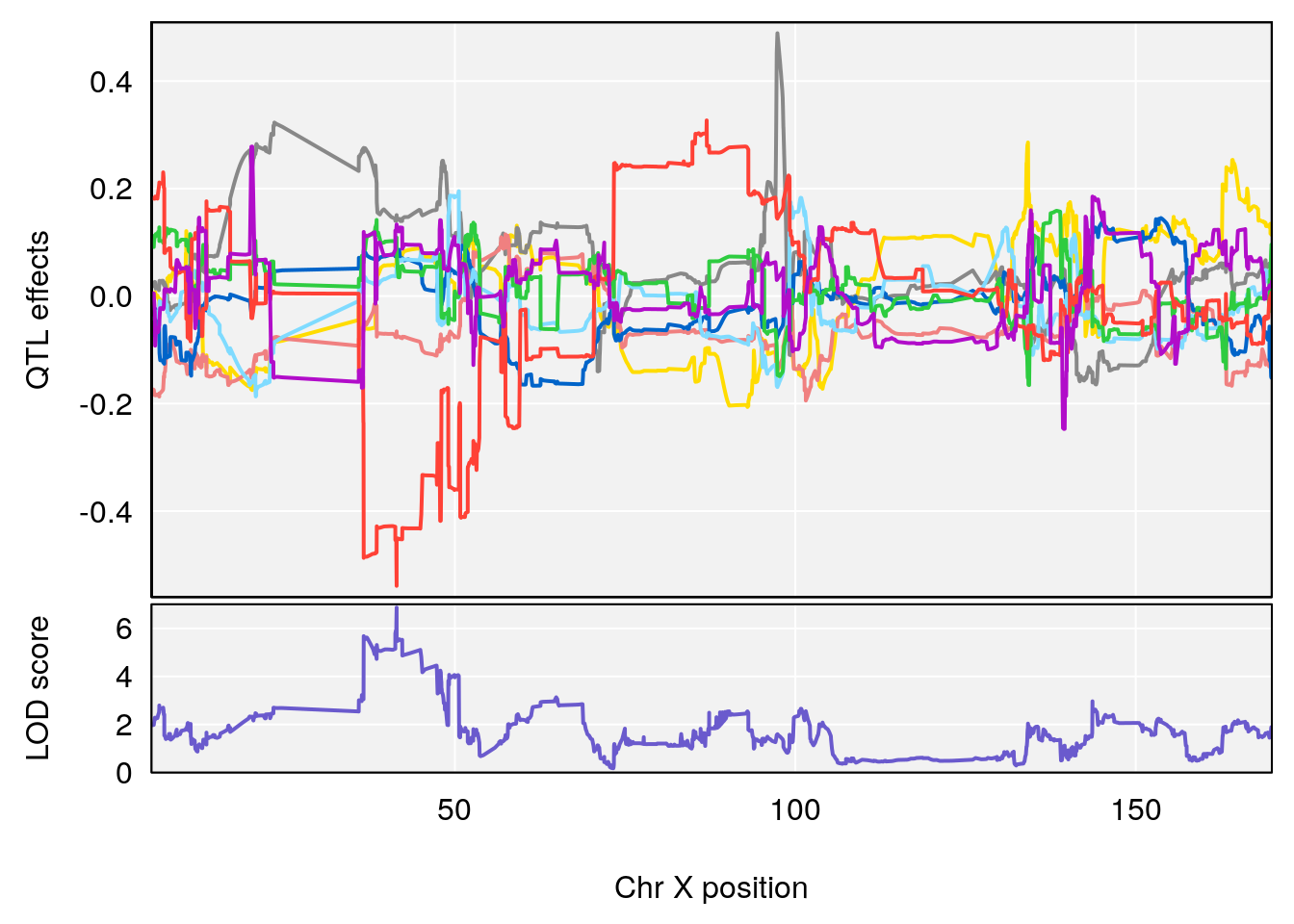
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
Plot qtl mapping on array female
load("output/qtl.morphine.out.combined_gm.female.RData")
#pmap and gmap
chr.names <- c("1","2","3","4","5","6","7","8","9","10","11","12","13","14","15","16","17","18","19", "X")
gmap <- list()
pmap <- list()
for (chr in chr.names){
gmap[[chr]] <- combined_gm$gmap[[chr]]
pmap[[chr]] <- combined_gm$pmap[[chr]]
}
attr(gmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
attr(pmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
#genome-wide plot
for(i in names(qtl.morphine.out)){
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.morphine.out[[i]]) # overall maximum LOD score
plot(qtl.morphine.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, max(sapply(qtl.morphine.out, maxlod)) +2))
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("DO_morphine_",i))
}
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
#peaks coeff plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
print(peaks)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
}
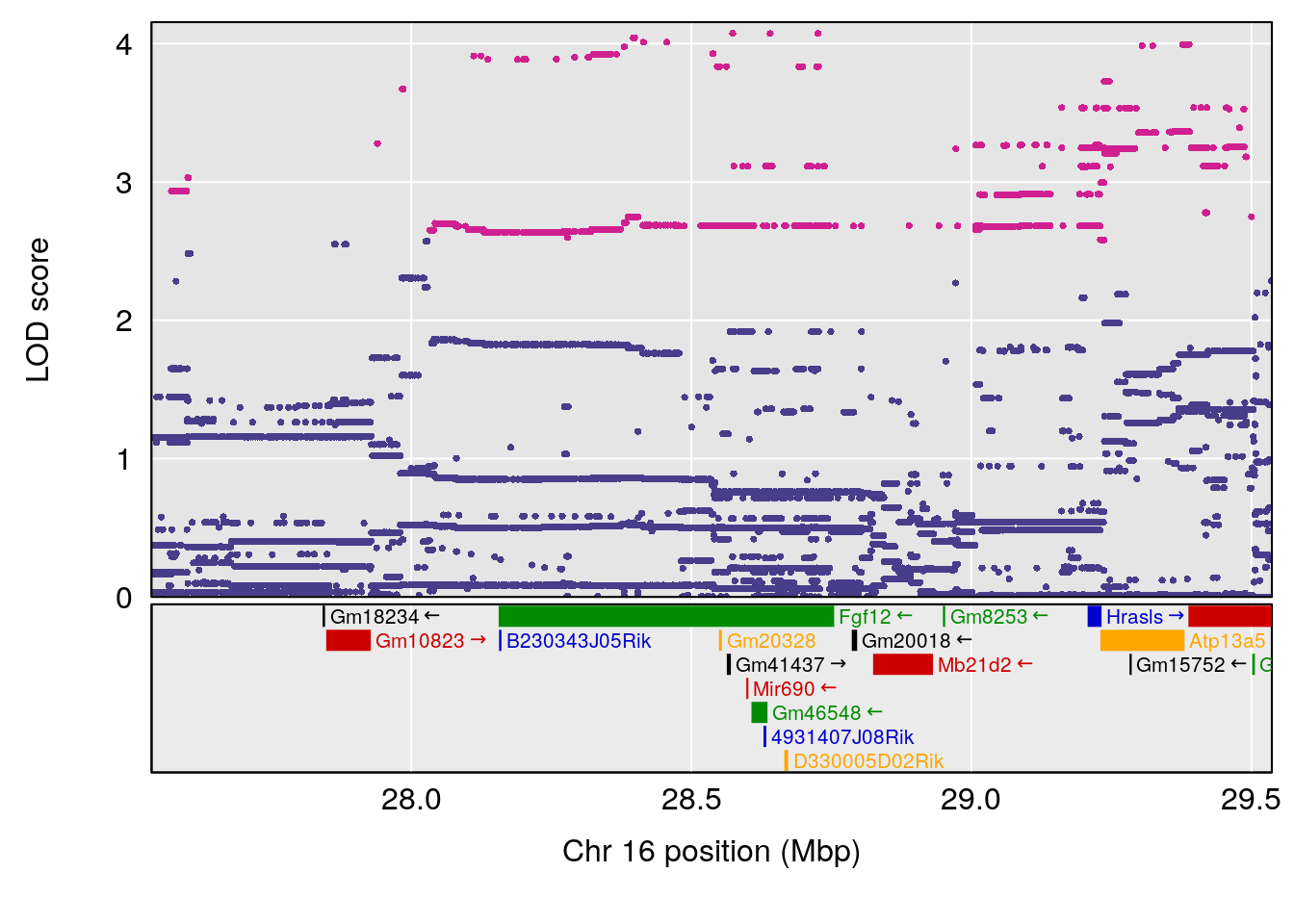
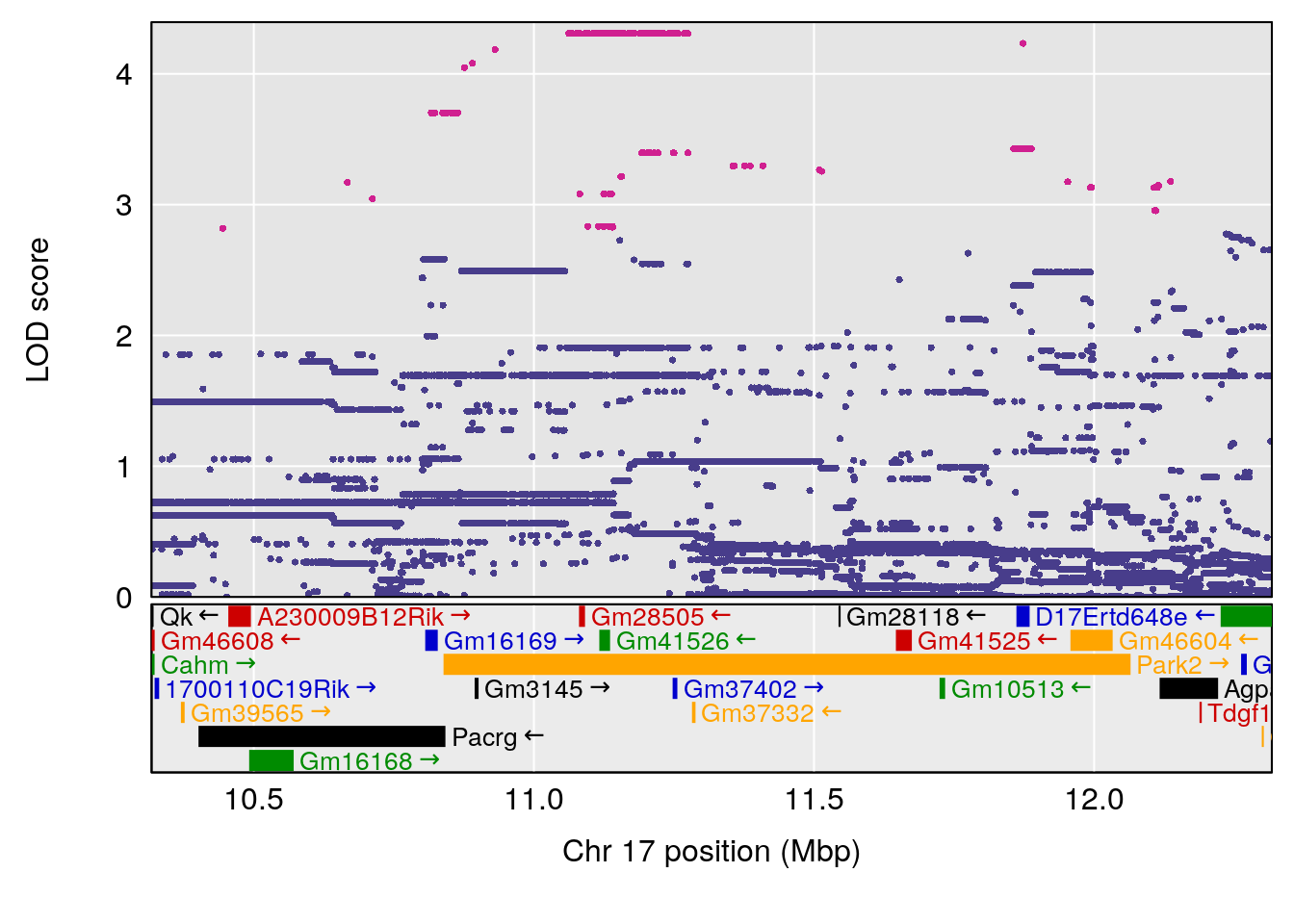
# [1] "Survival.Time"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 3 87.75328 7.363908 87.01462 88.24094
# 2 1 pheno1 8 115.47006 10.018661 115.13712 115.57484
# 3 1 pheno1 9 67.05514 7.297535 65.50554 67.16519
# 4 1 pheno1 15 29.63536 7.380134 28.36166 30.77732
# [1] 1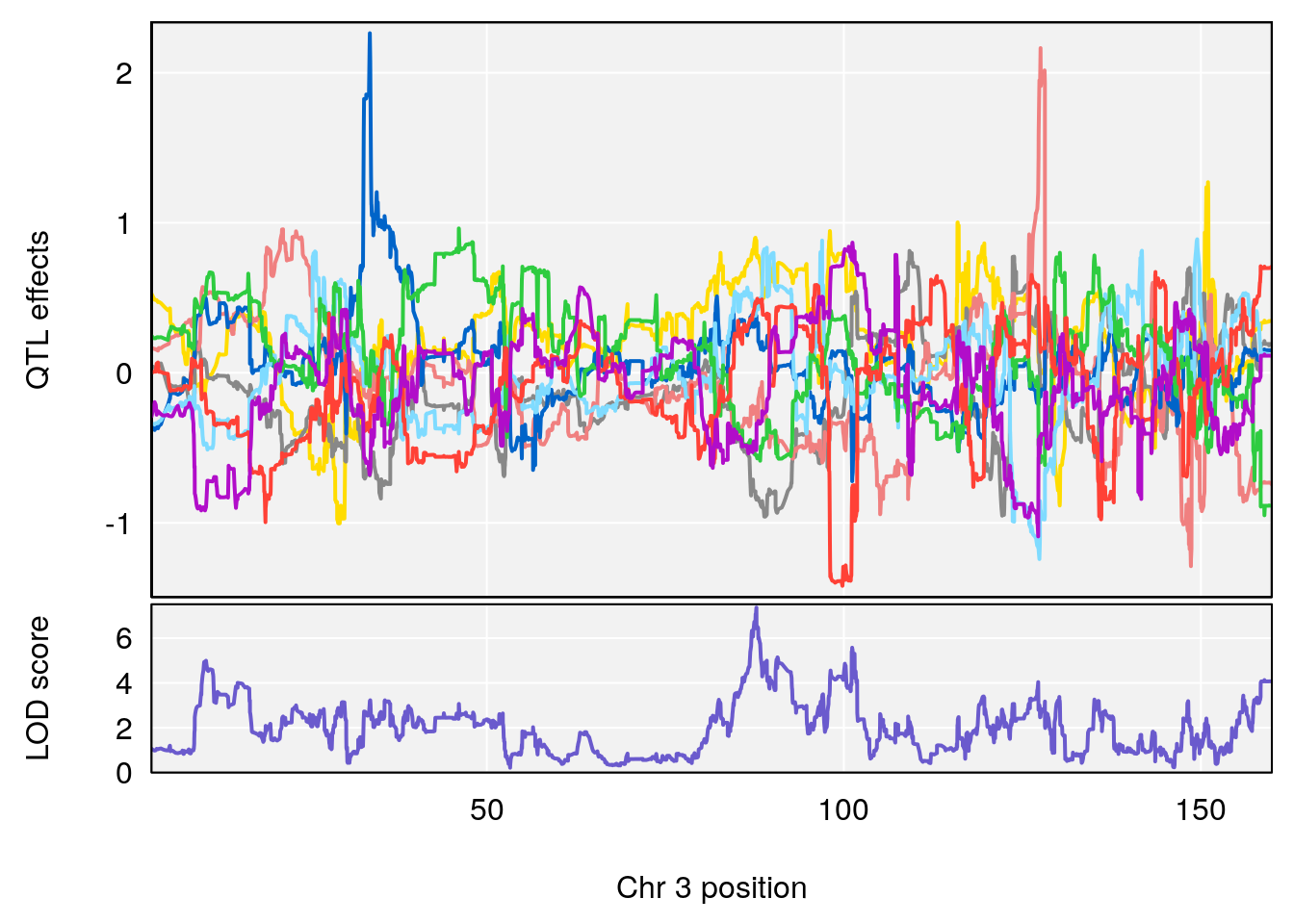
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 2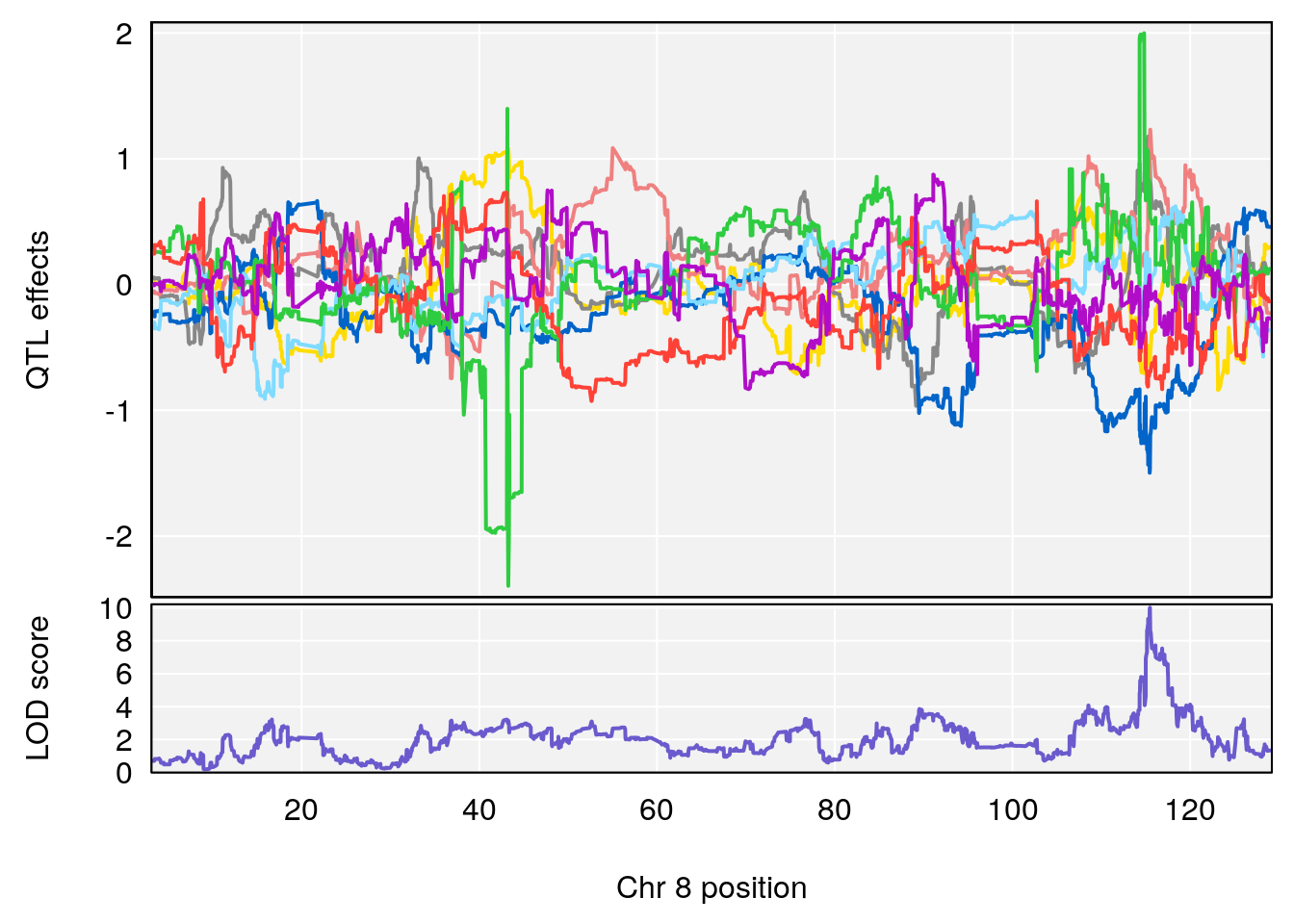
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 3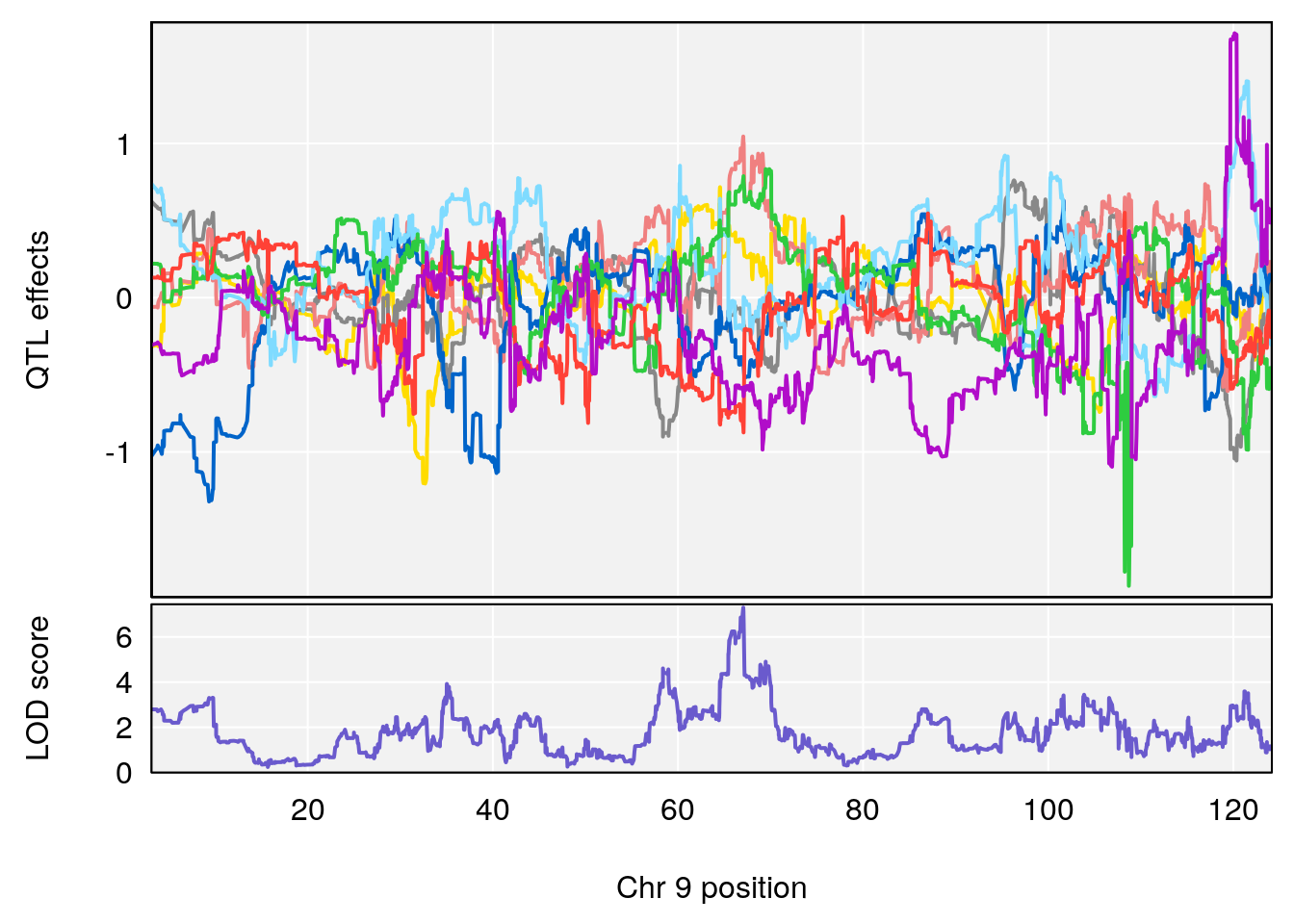
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 4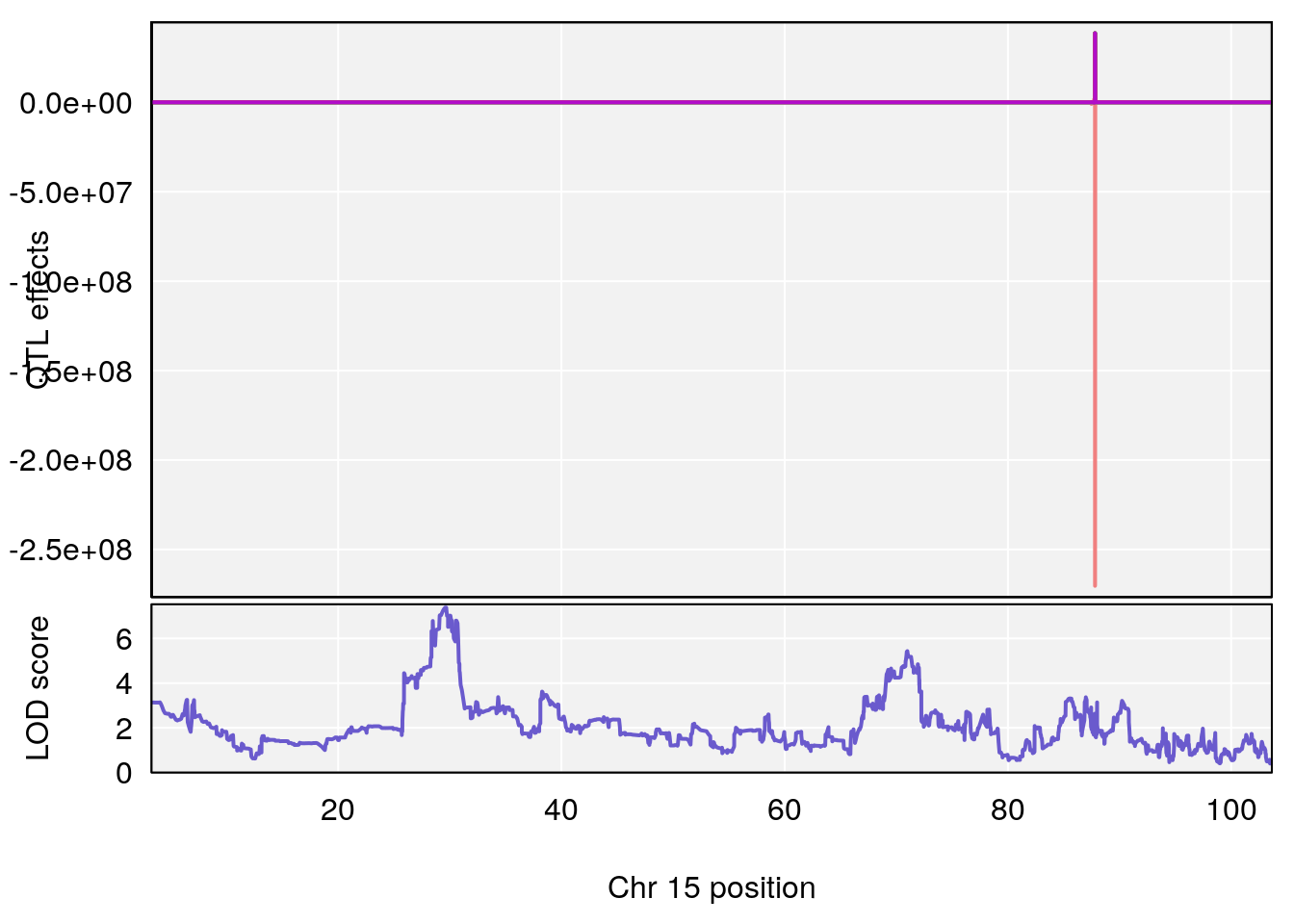
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
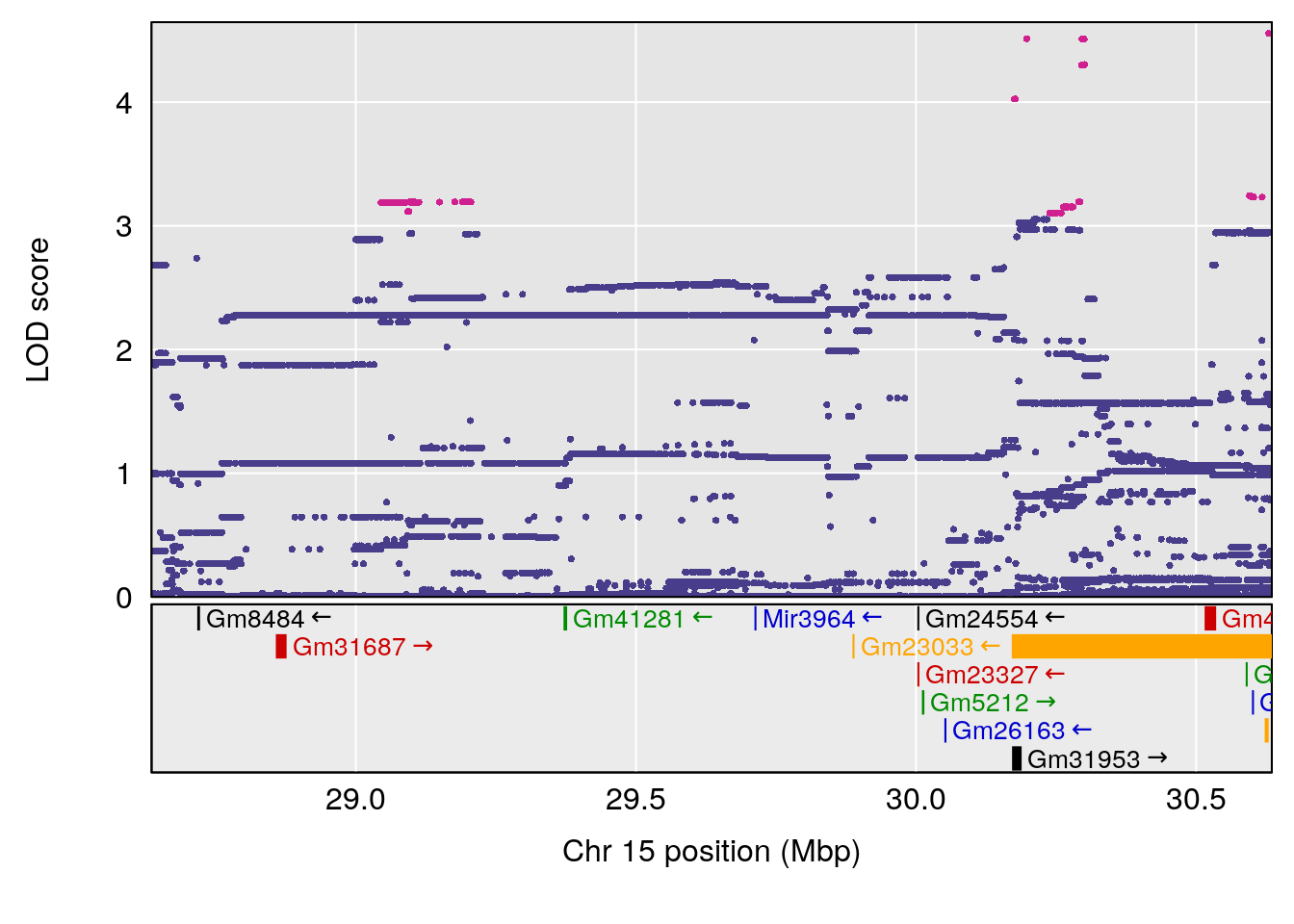
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
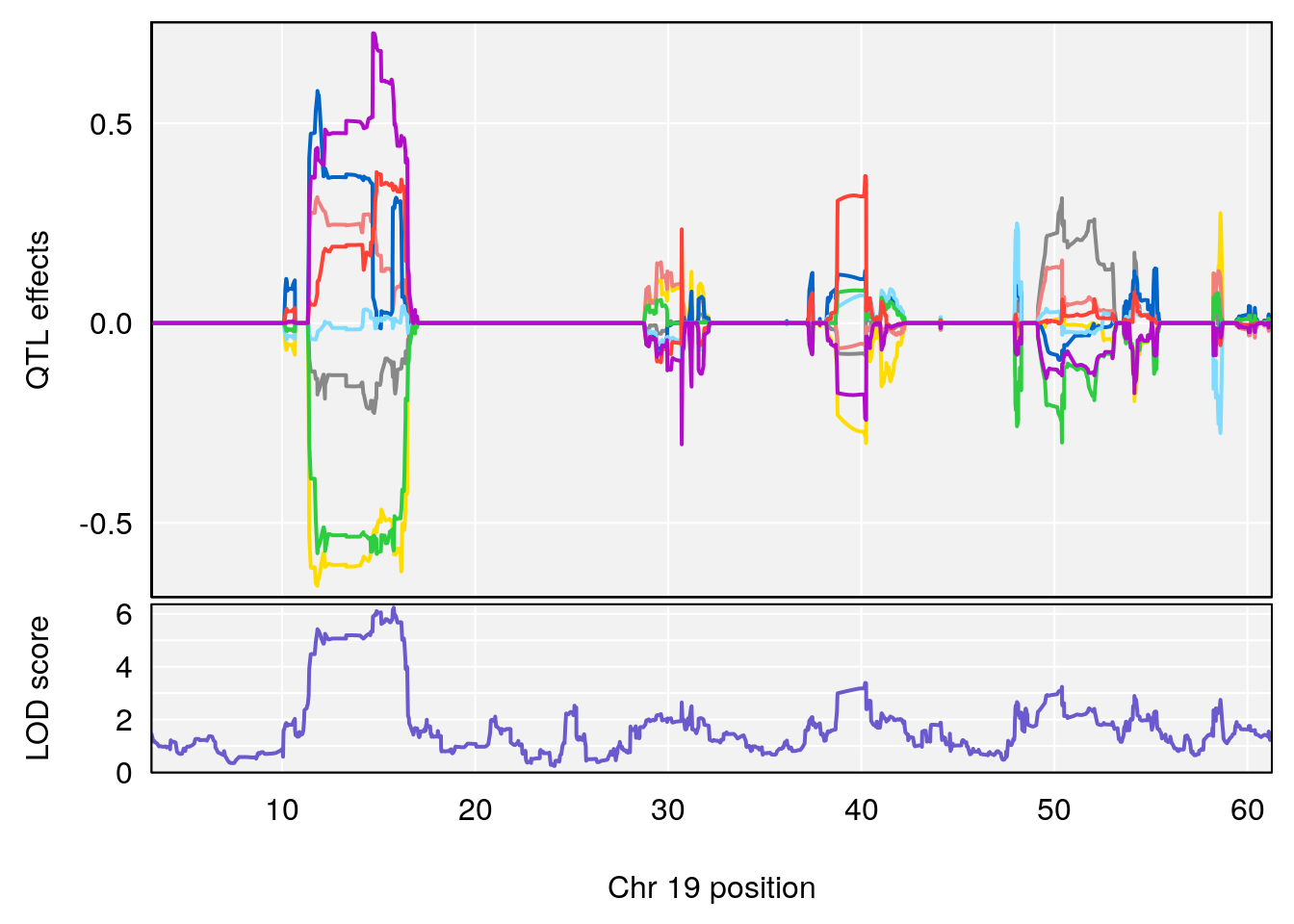
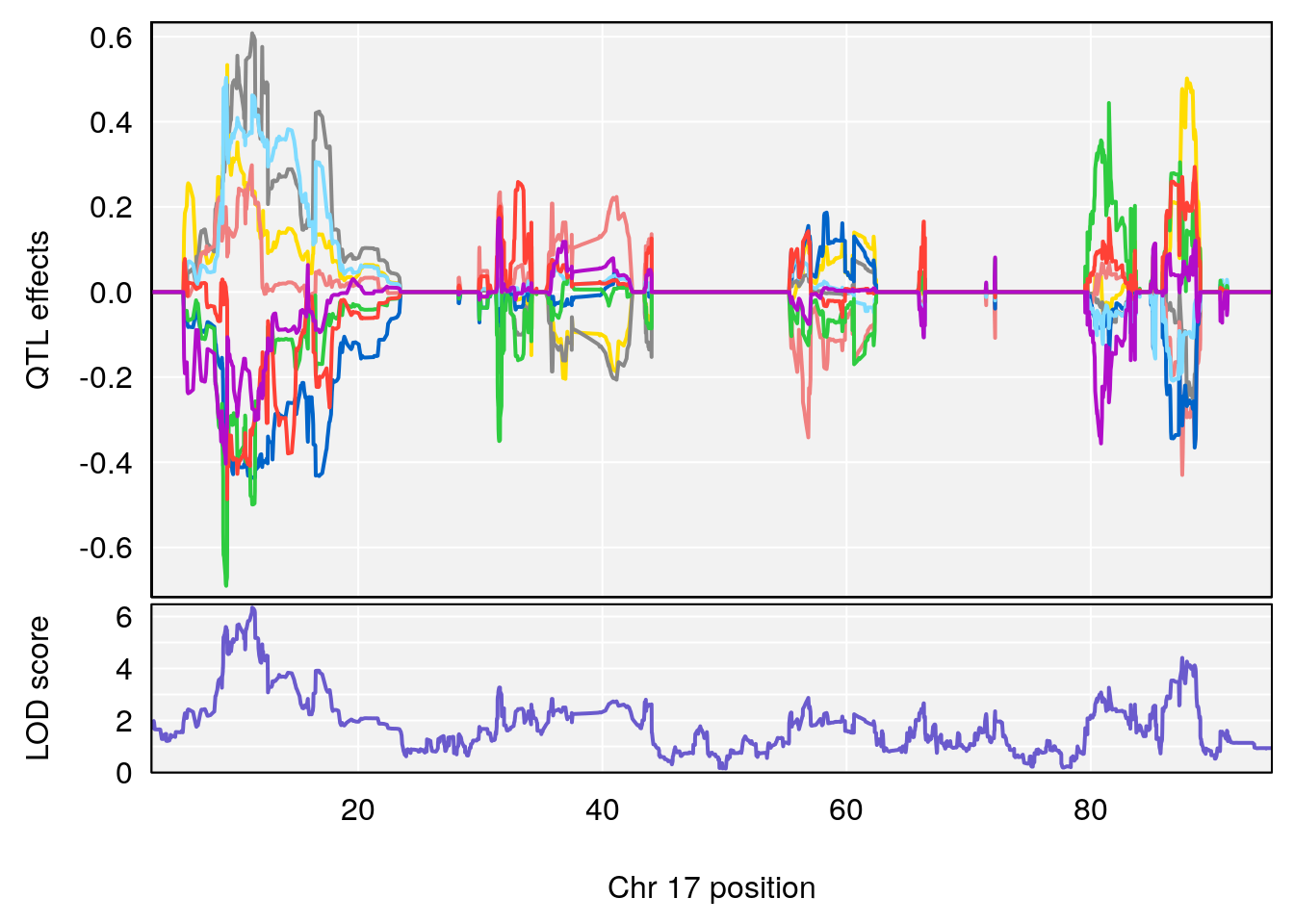
# [1] "Recovery.Time"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 3 118.12137 7.879474 117.724824 121.19240
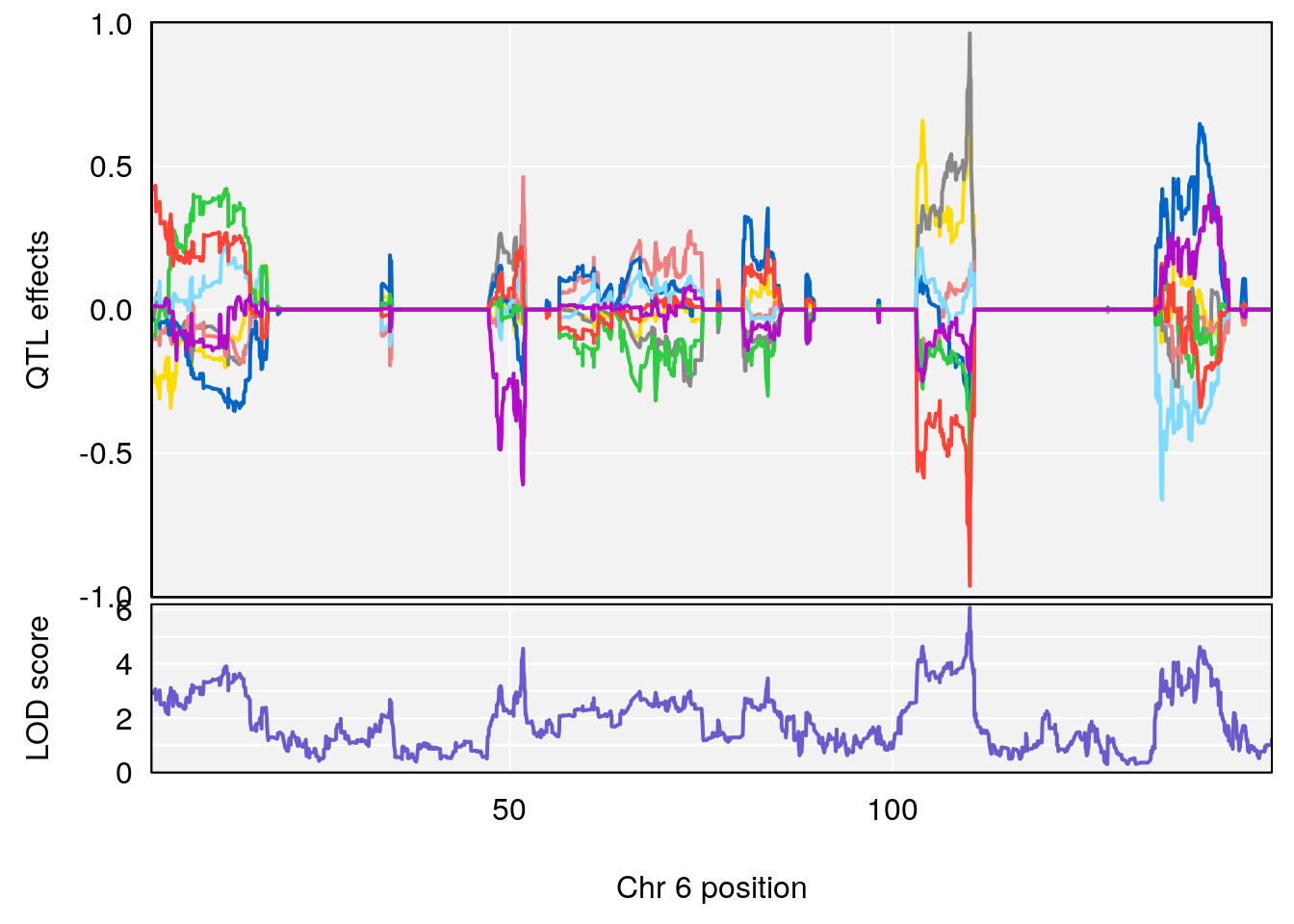
# 2 1 pheno1 5 92.74391 6.788068 37.152061 97.94610
# 3 1 pheno1 6 110.10532 6.075225 103.818523 140.20546
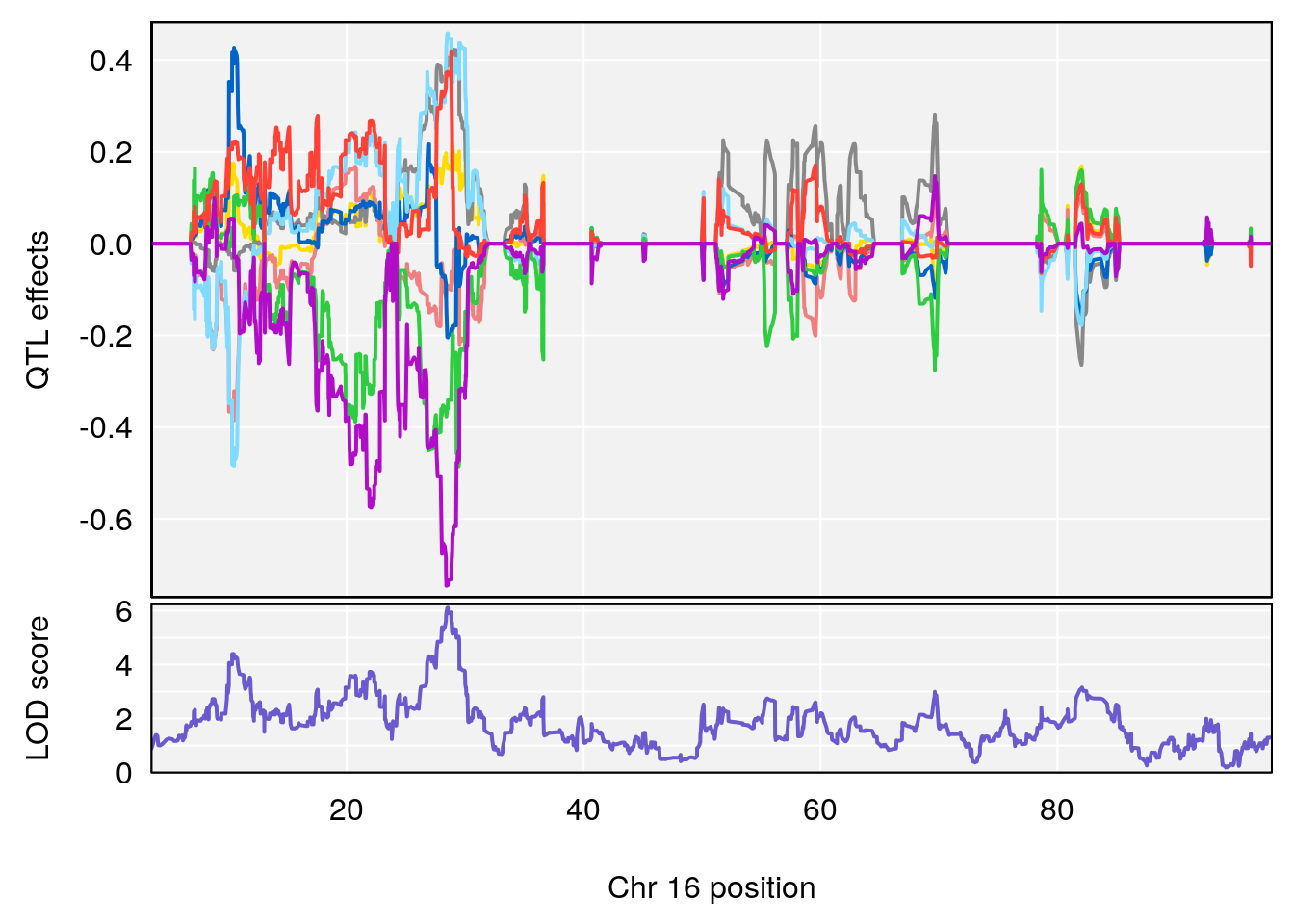
# 4 1 pheno1 16 28.53624 6.114898 27.599995 29.50554
# 5 1 pheno1 17 11.31727 6.342205 8.947284 12.16295
# 6 1 pheno1 19 31.56747 6.122590 31.188745 61.13260
# [1] 1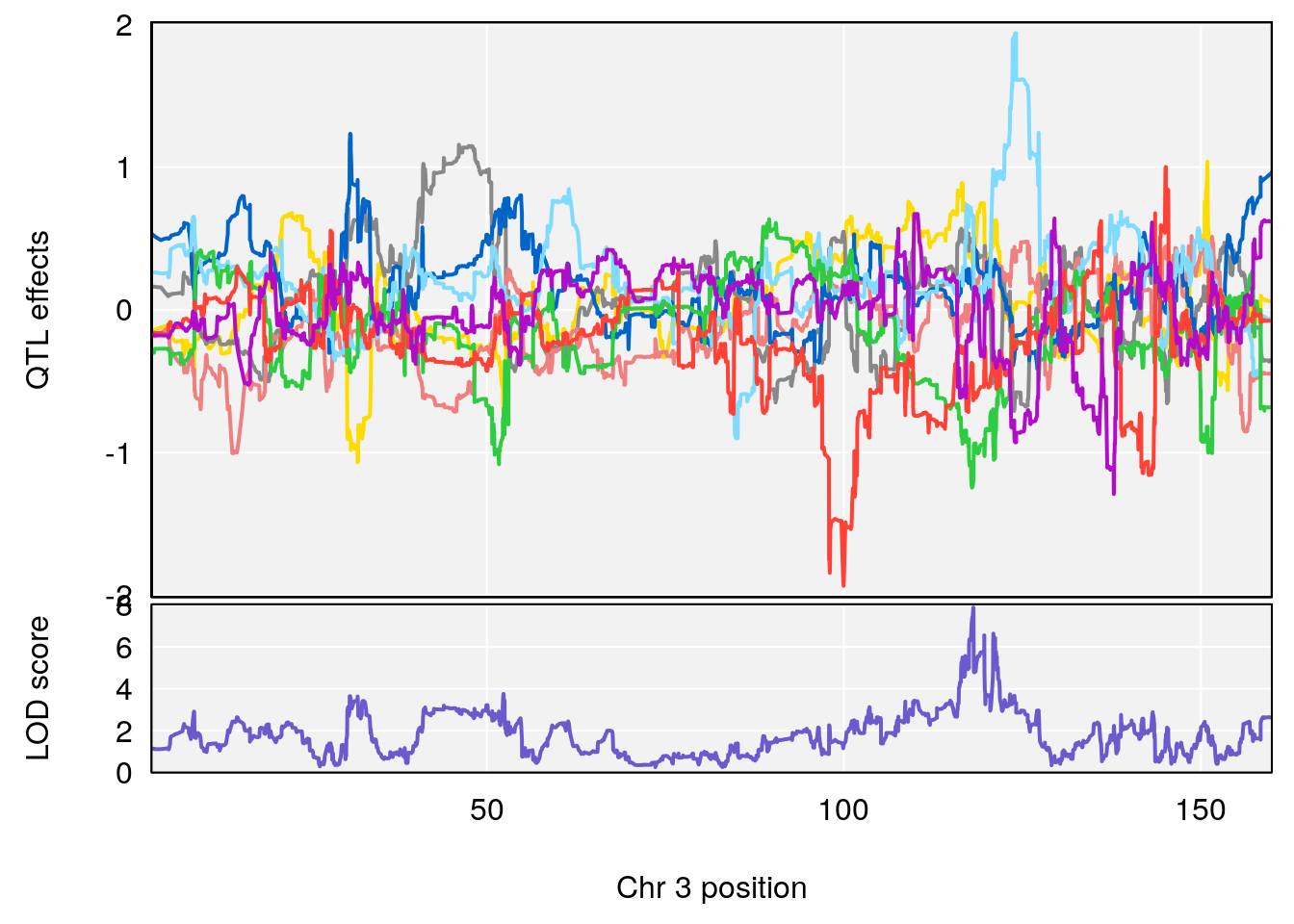
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 2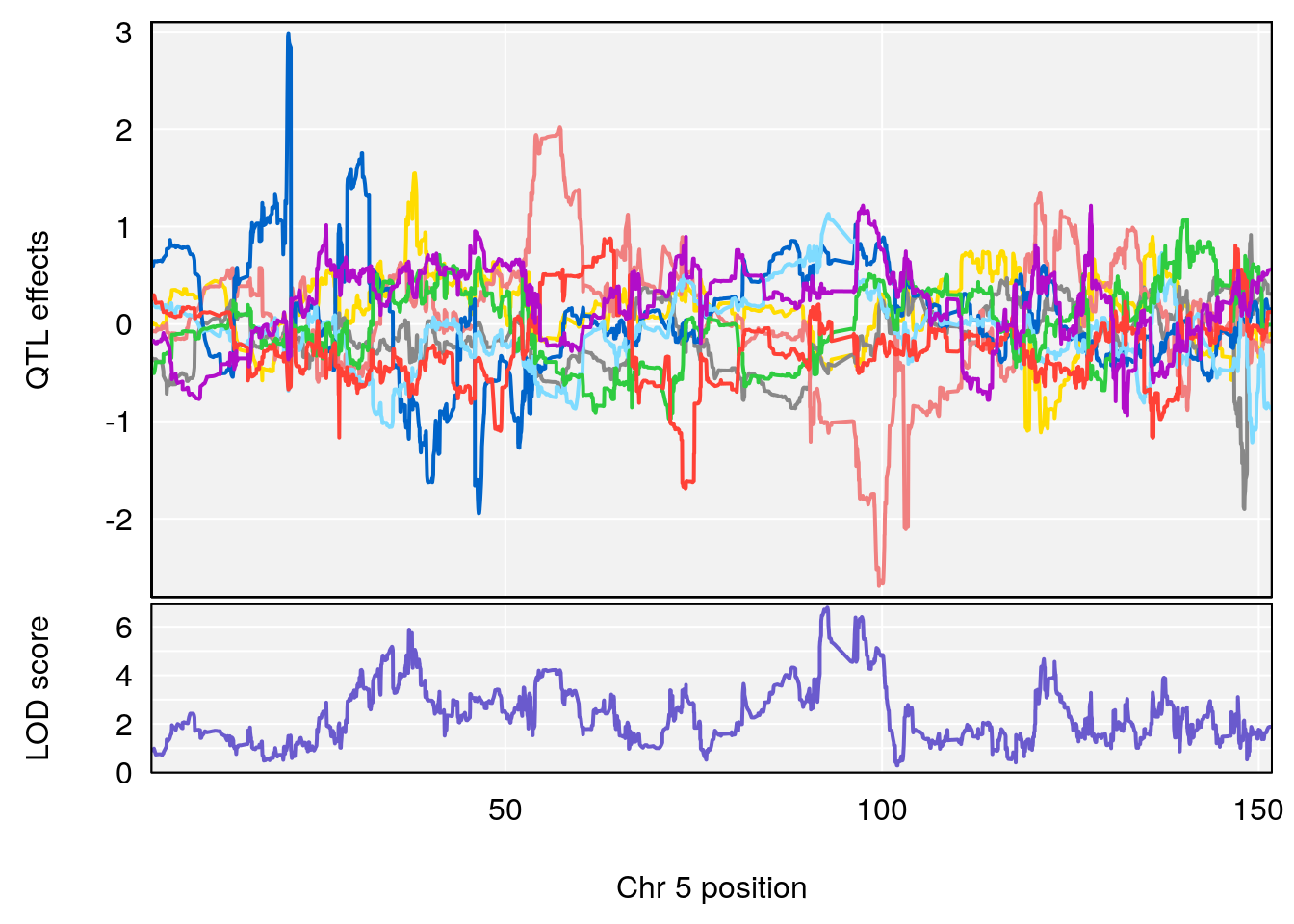
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 3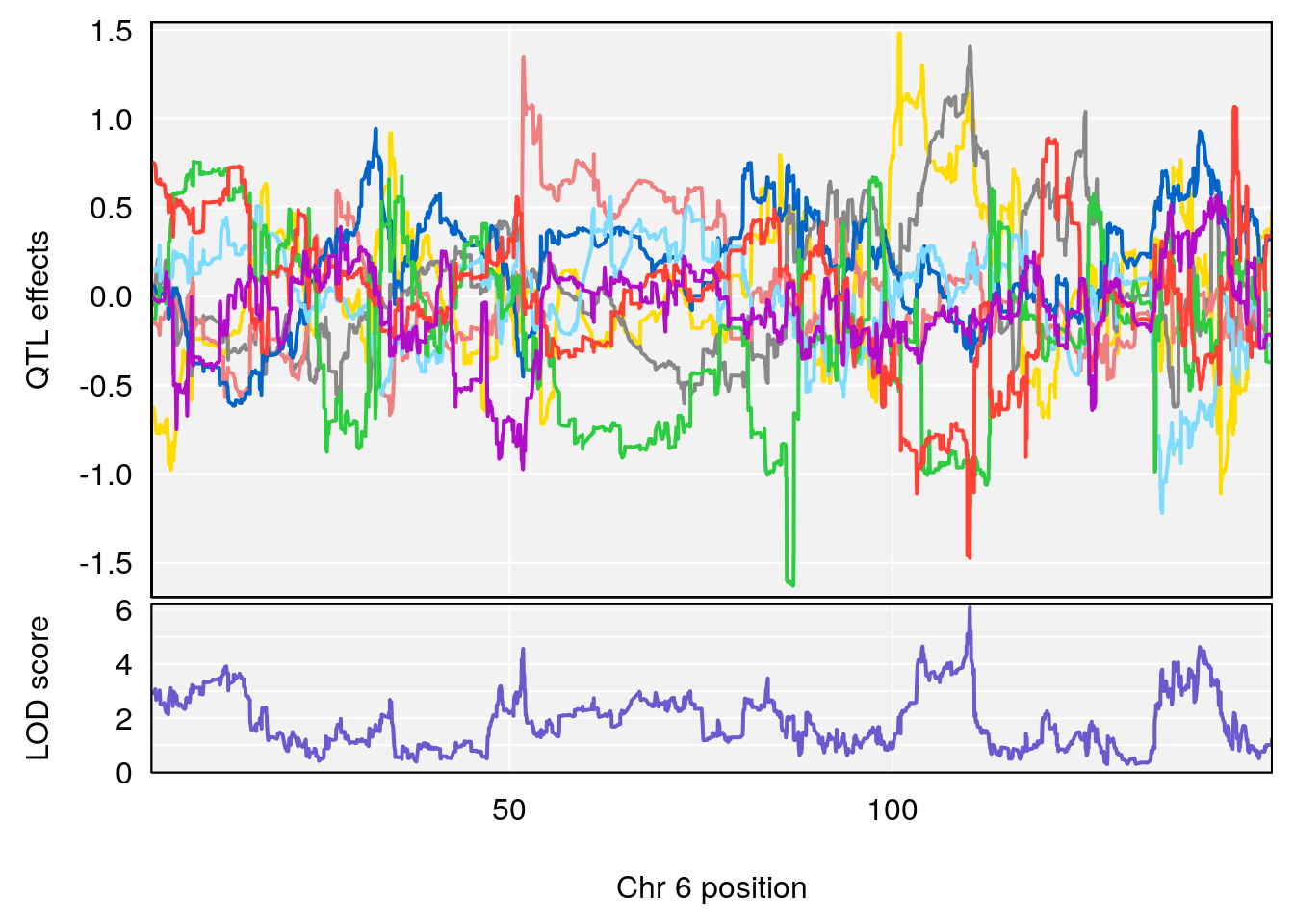
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 4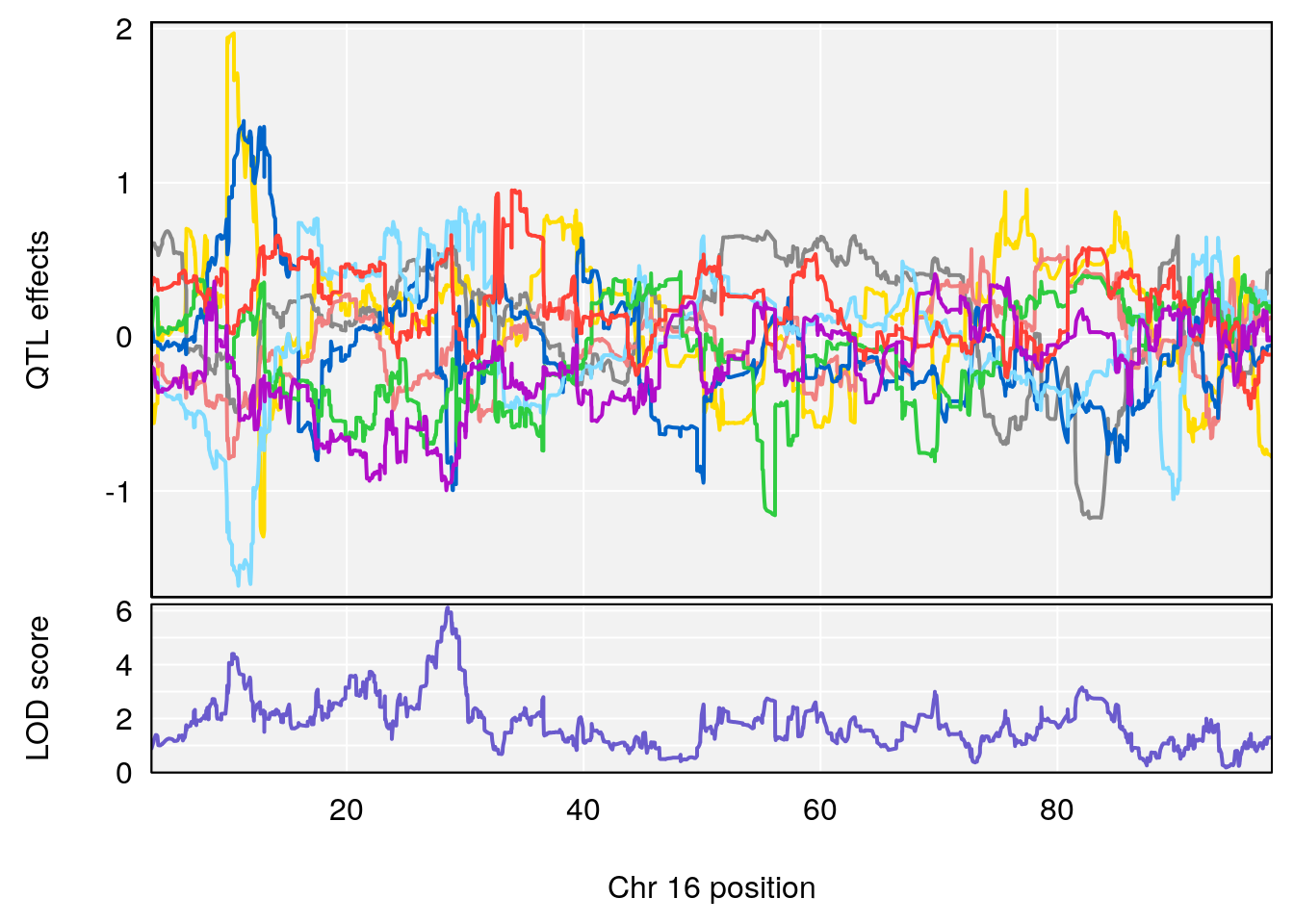
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 5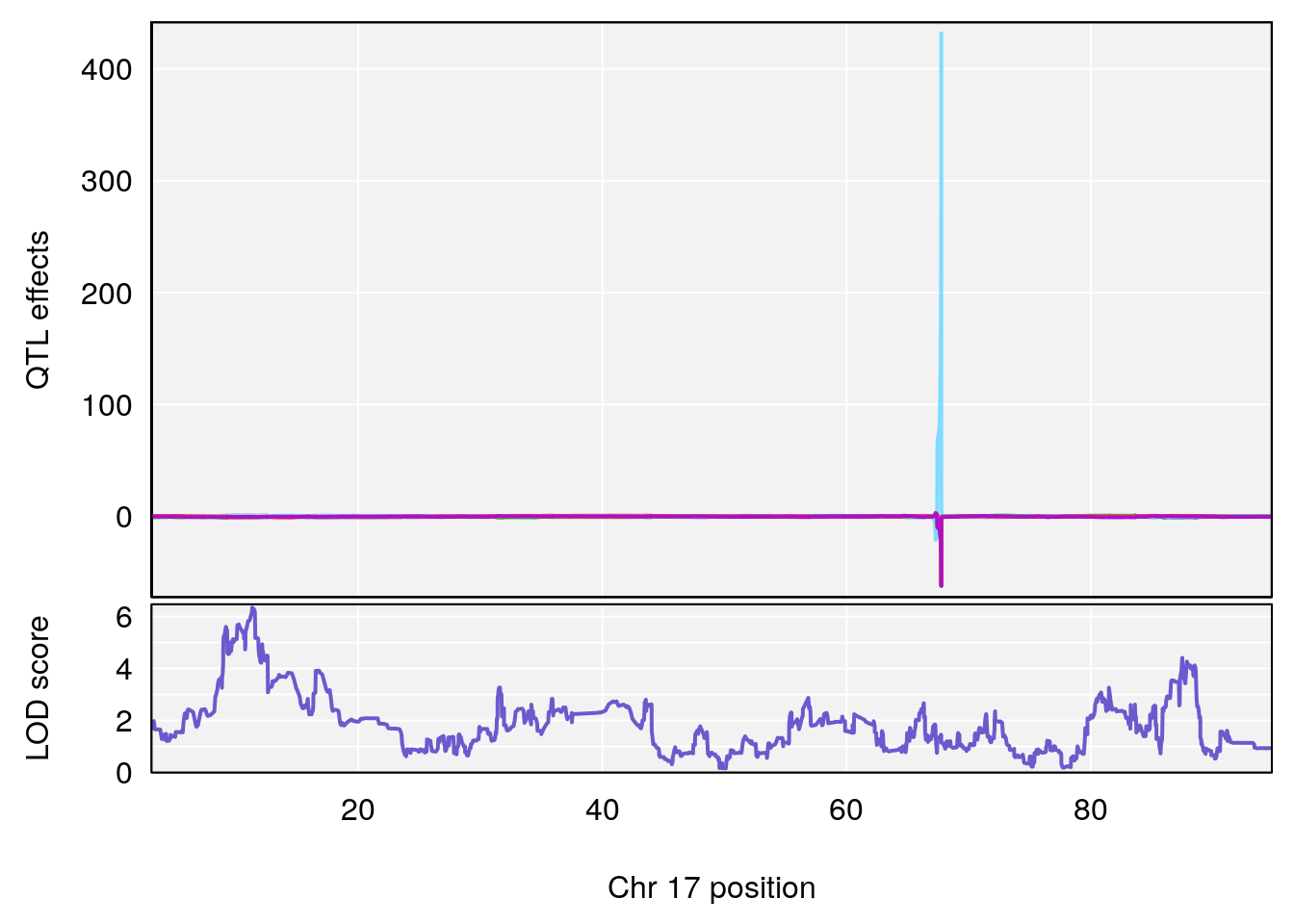
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 6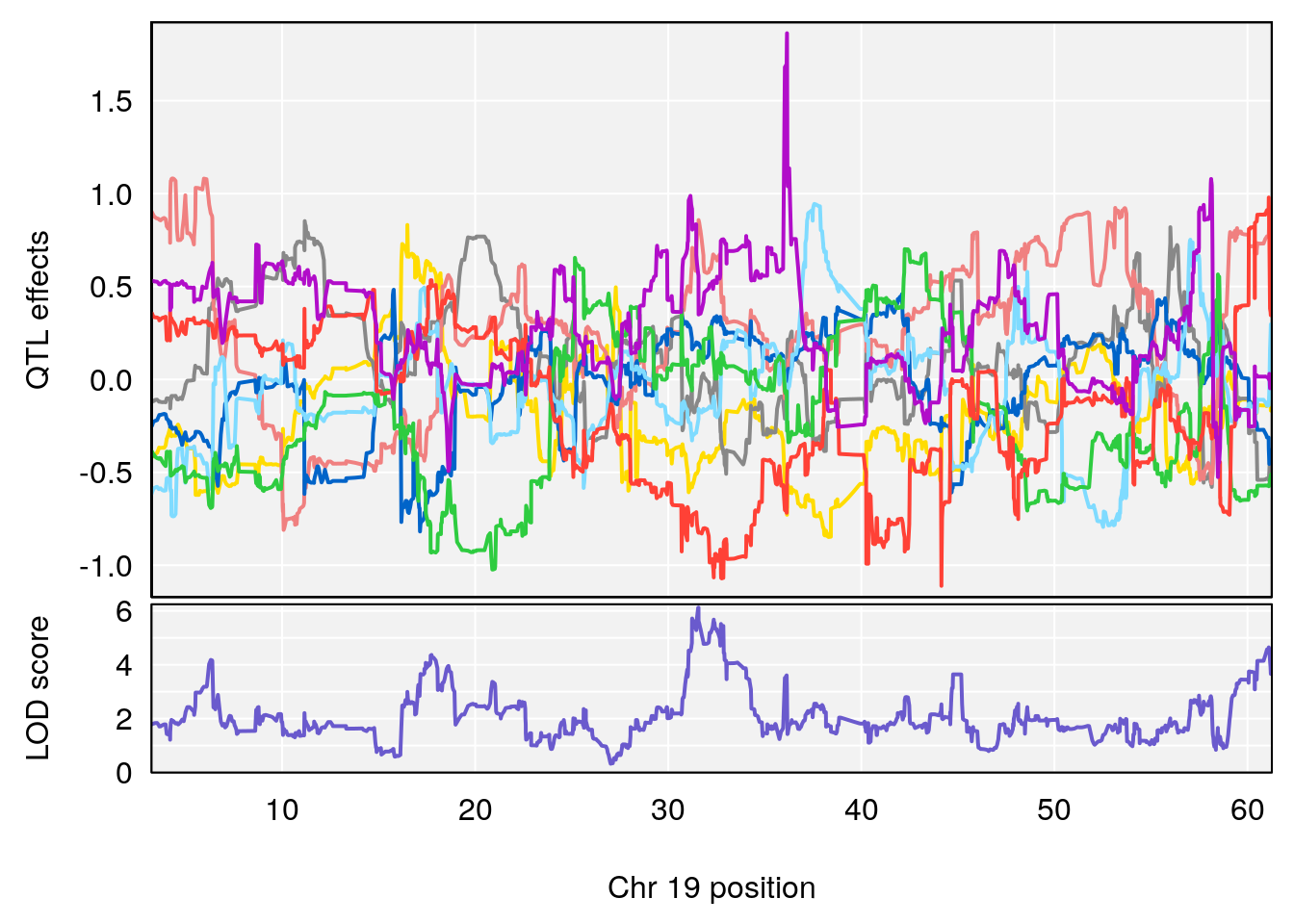
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
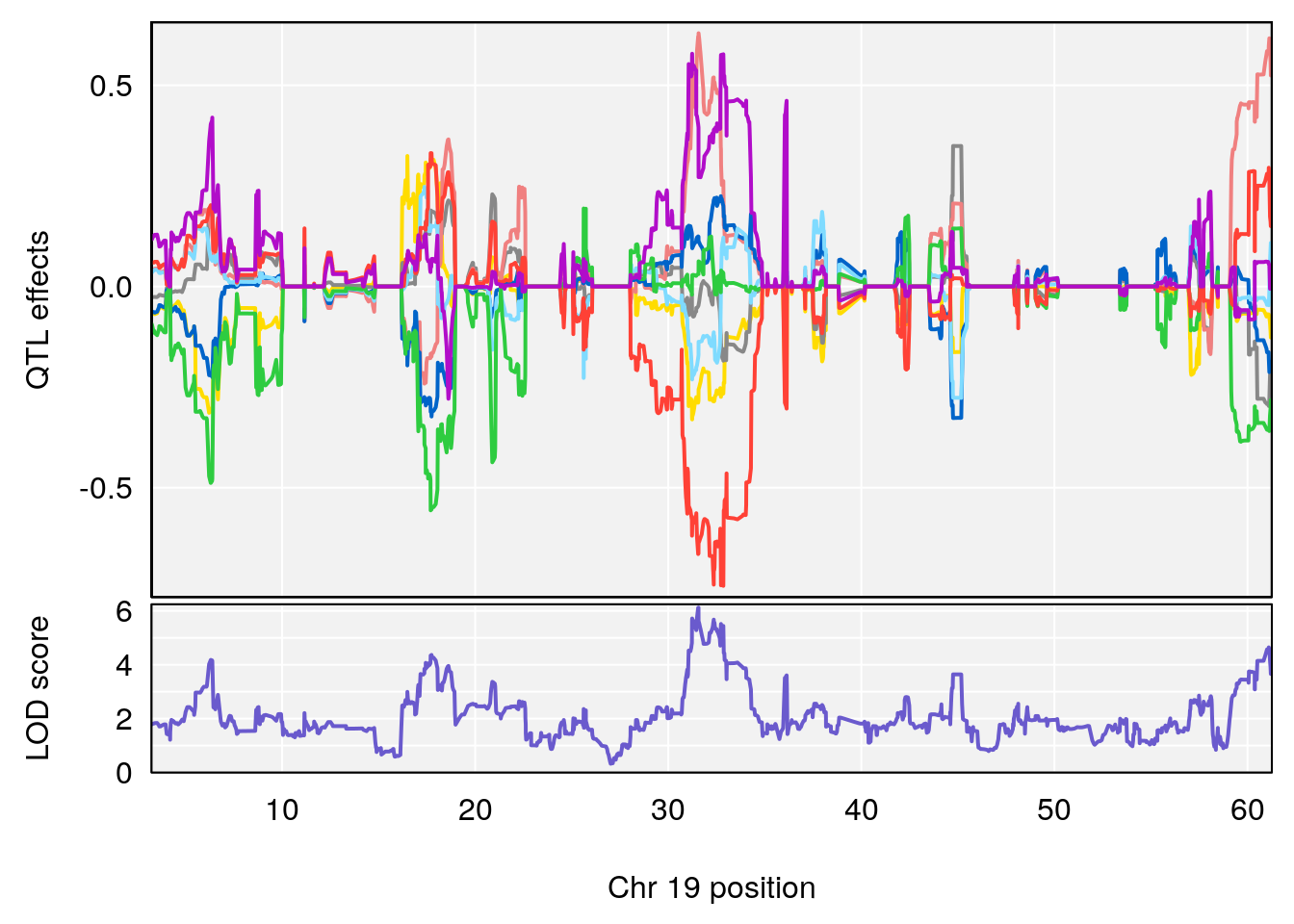
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
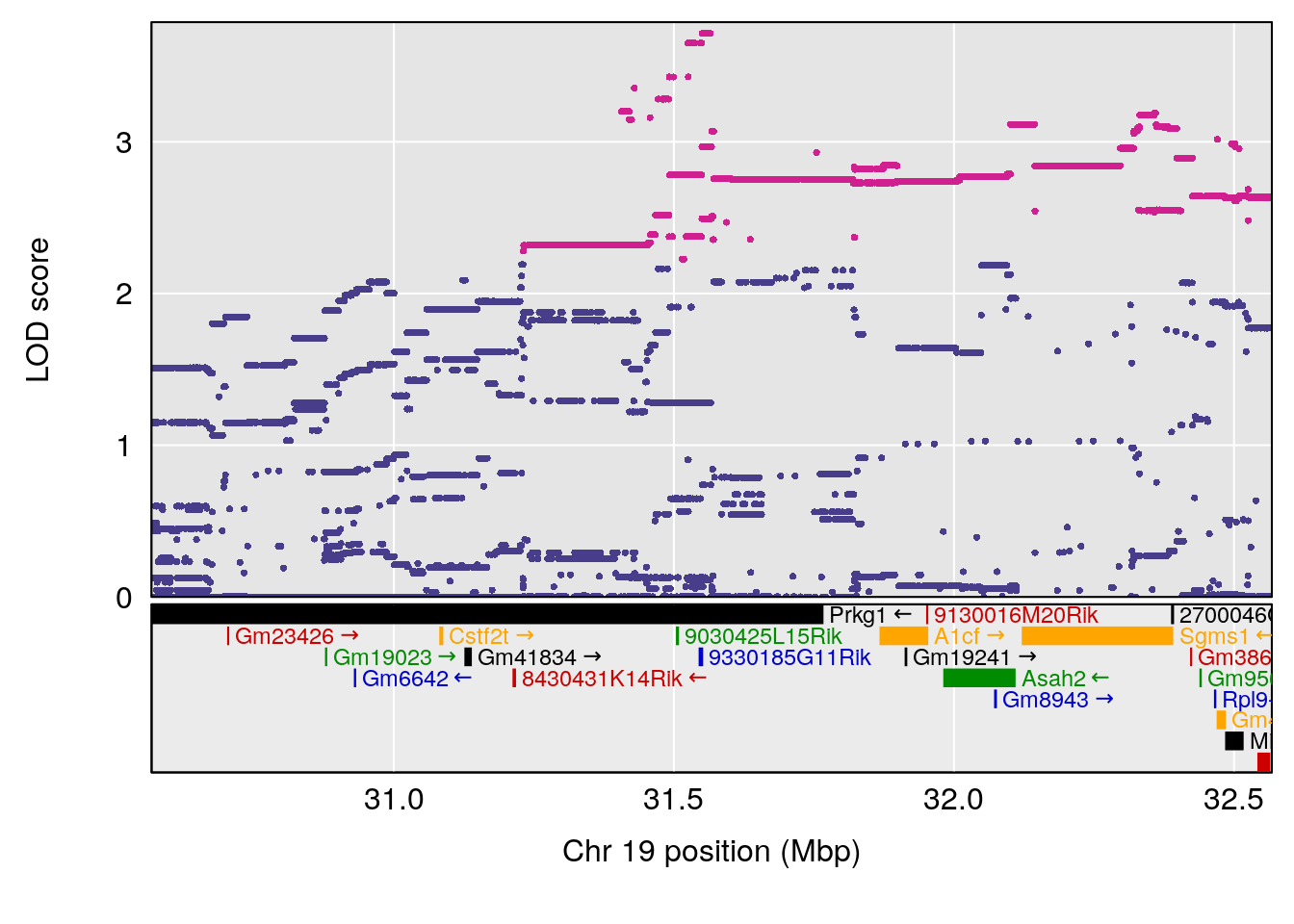
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
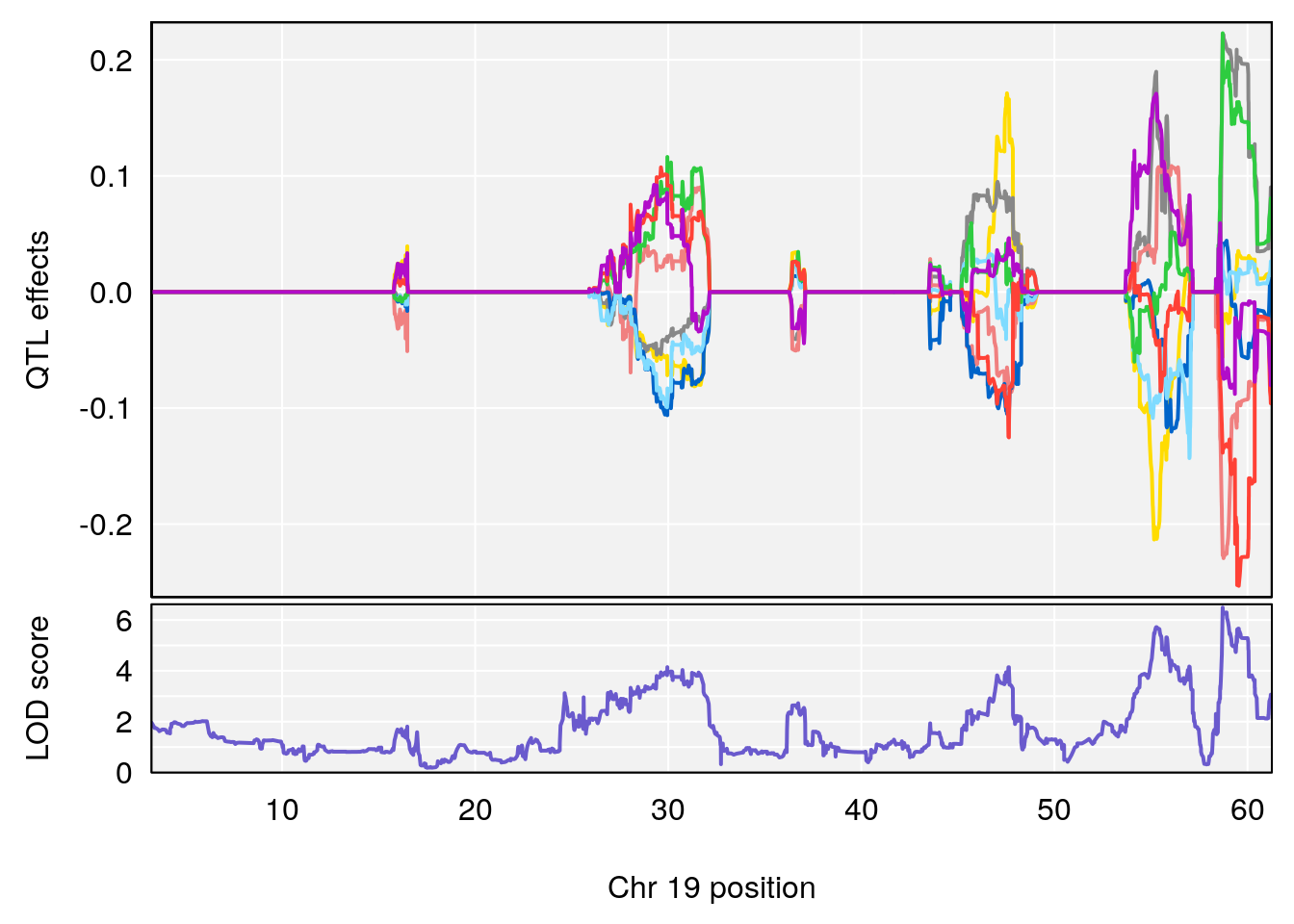
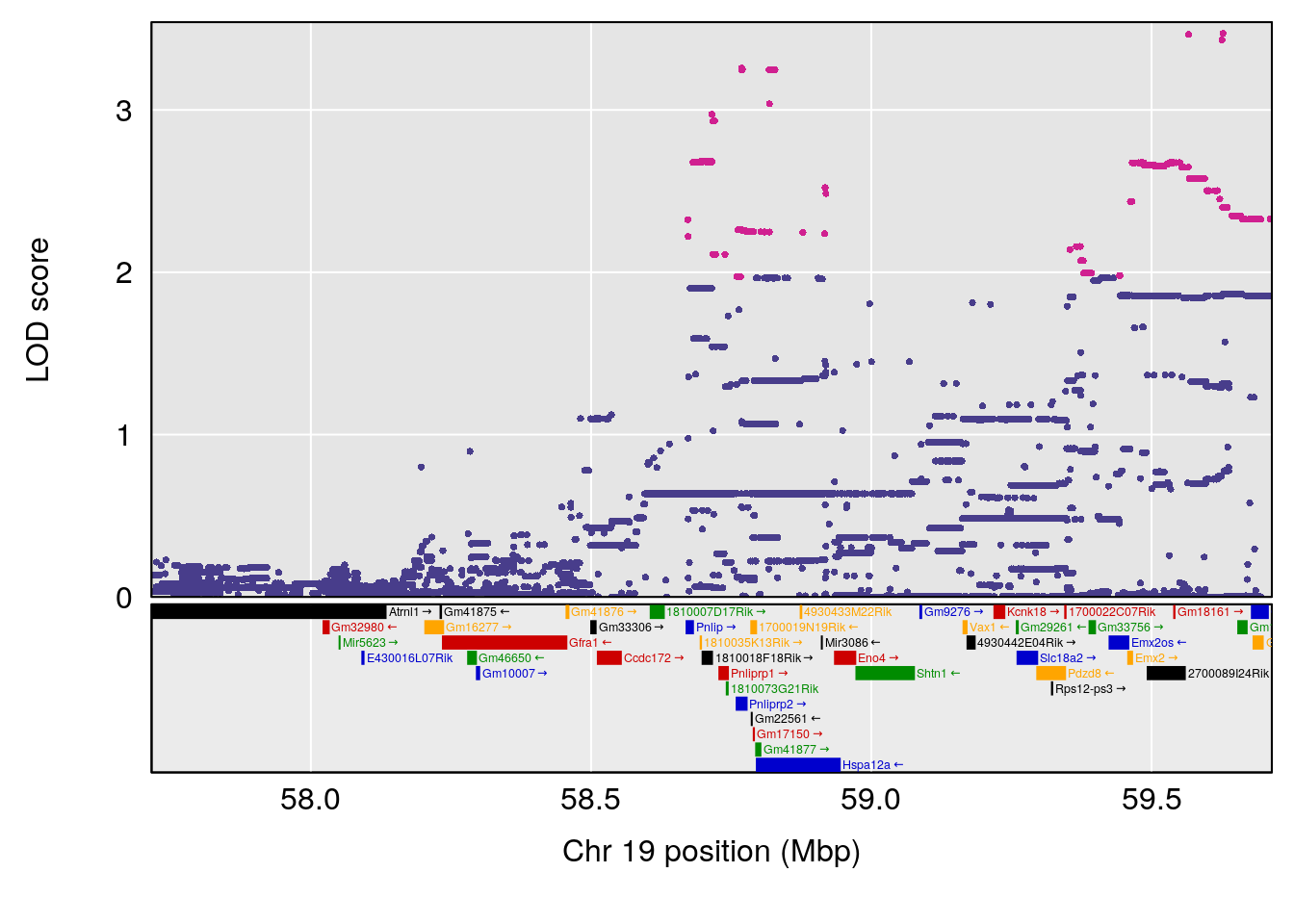
# [1] "Min.depression"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 4 156.19609 6.005957 9.858468 156.25473
# 2 1 pheno1 10 125.55489 6.017892 125.435827 126.91023
# 3 1 pheno1 19 58.87562 6.340088 42.445420 59.03354
# [1] 1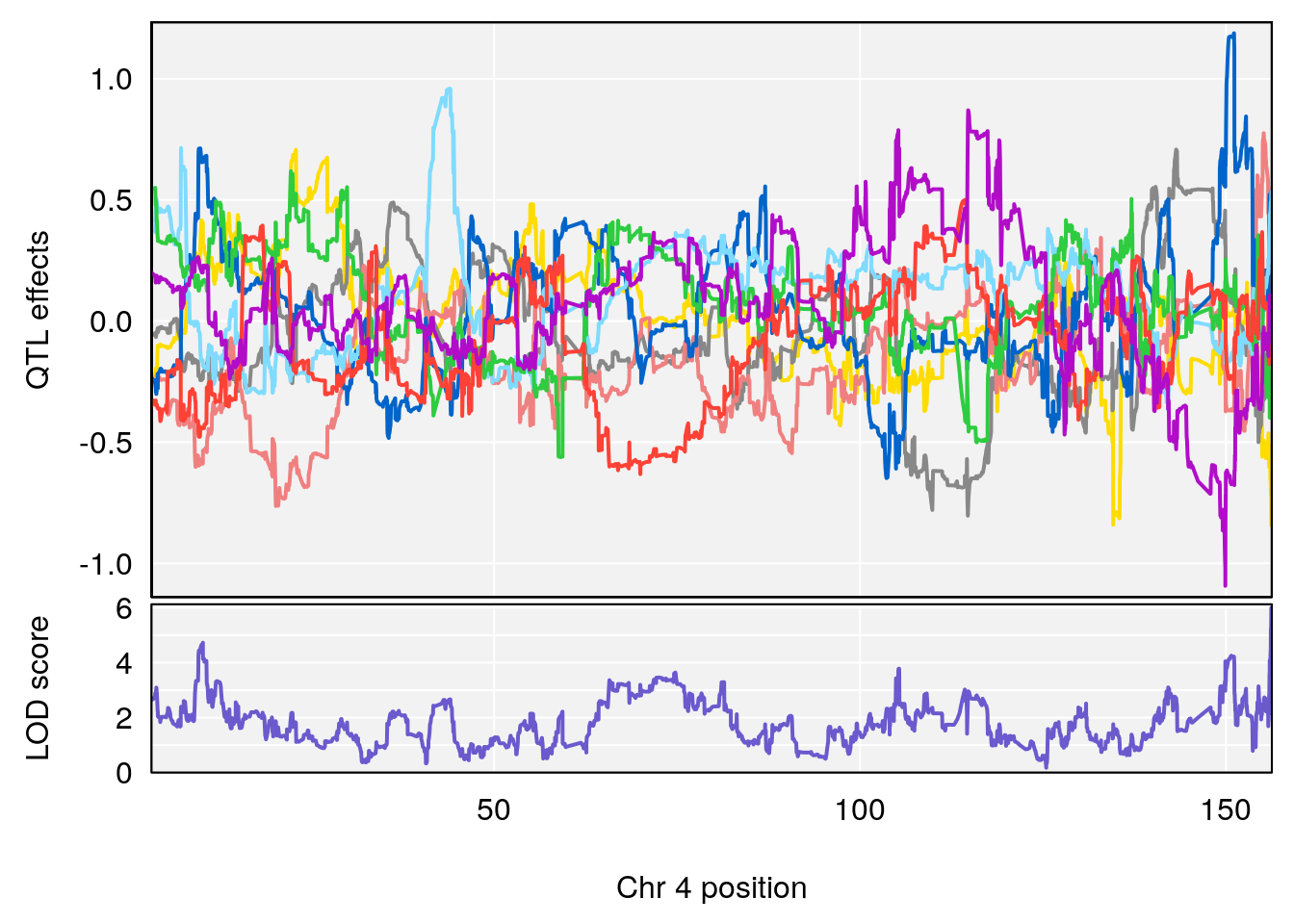
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 2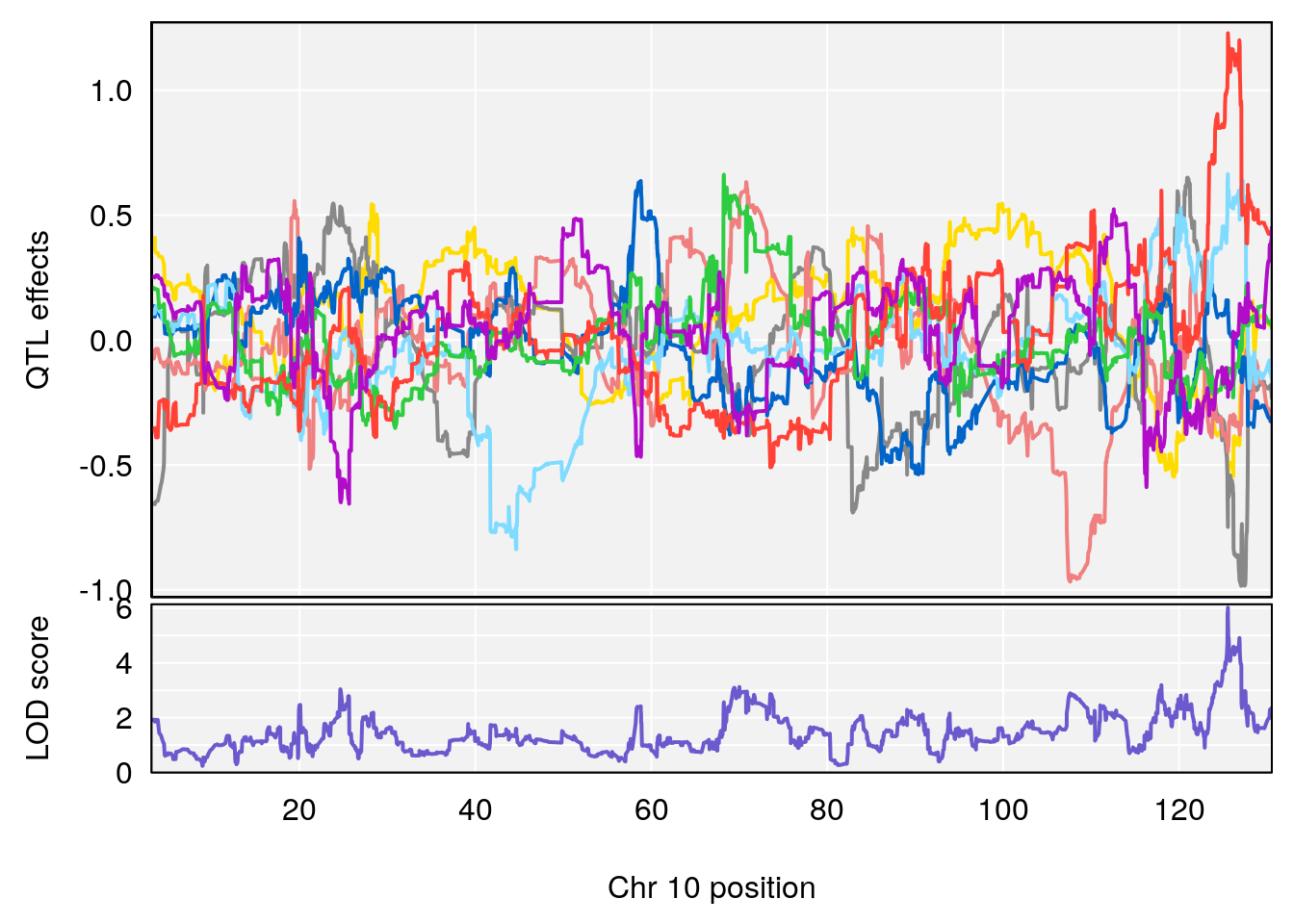
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
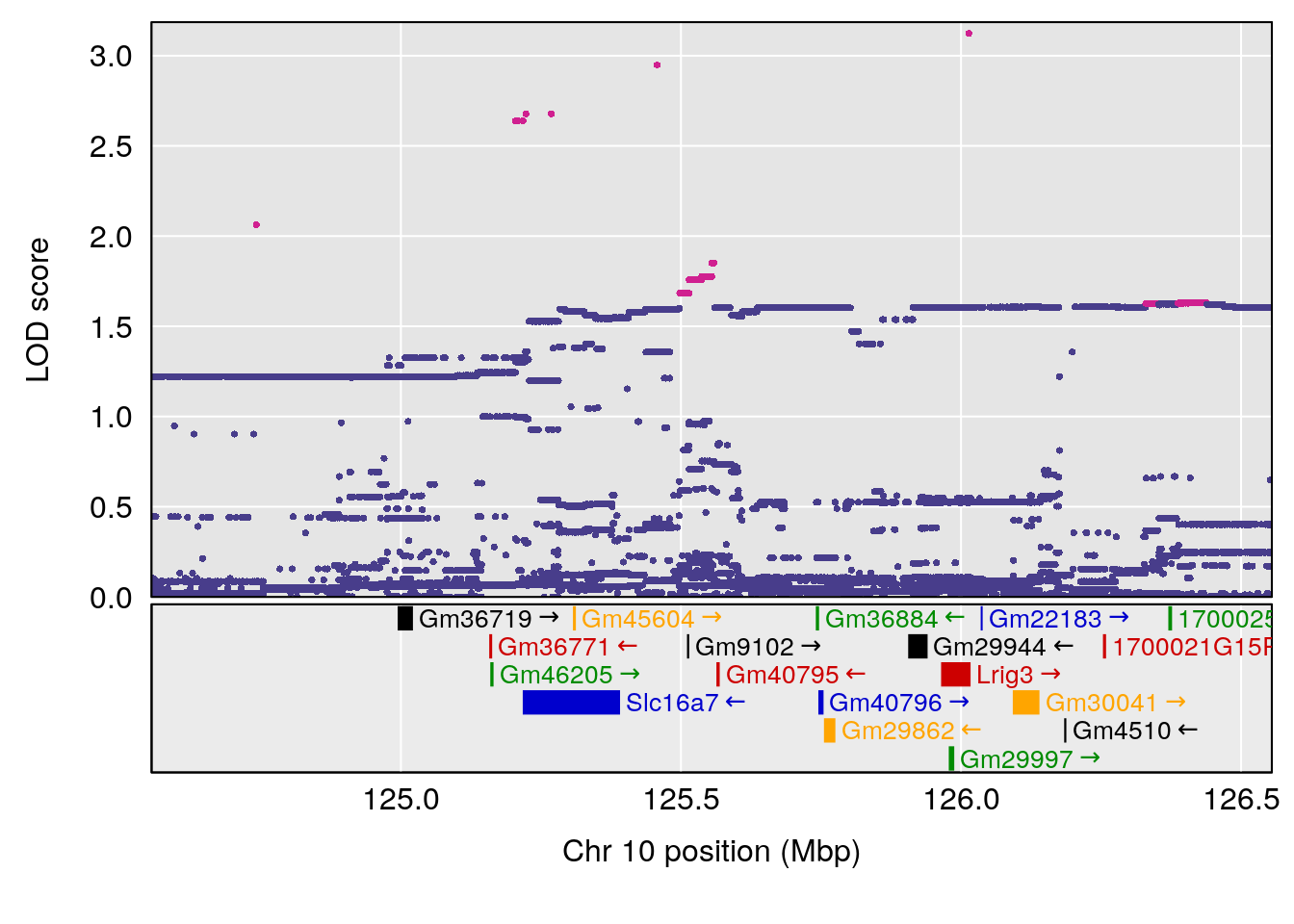
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 3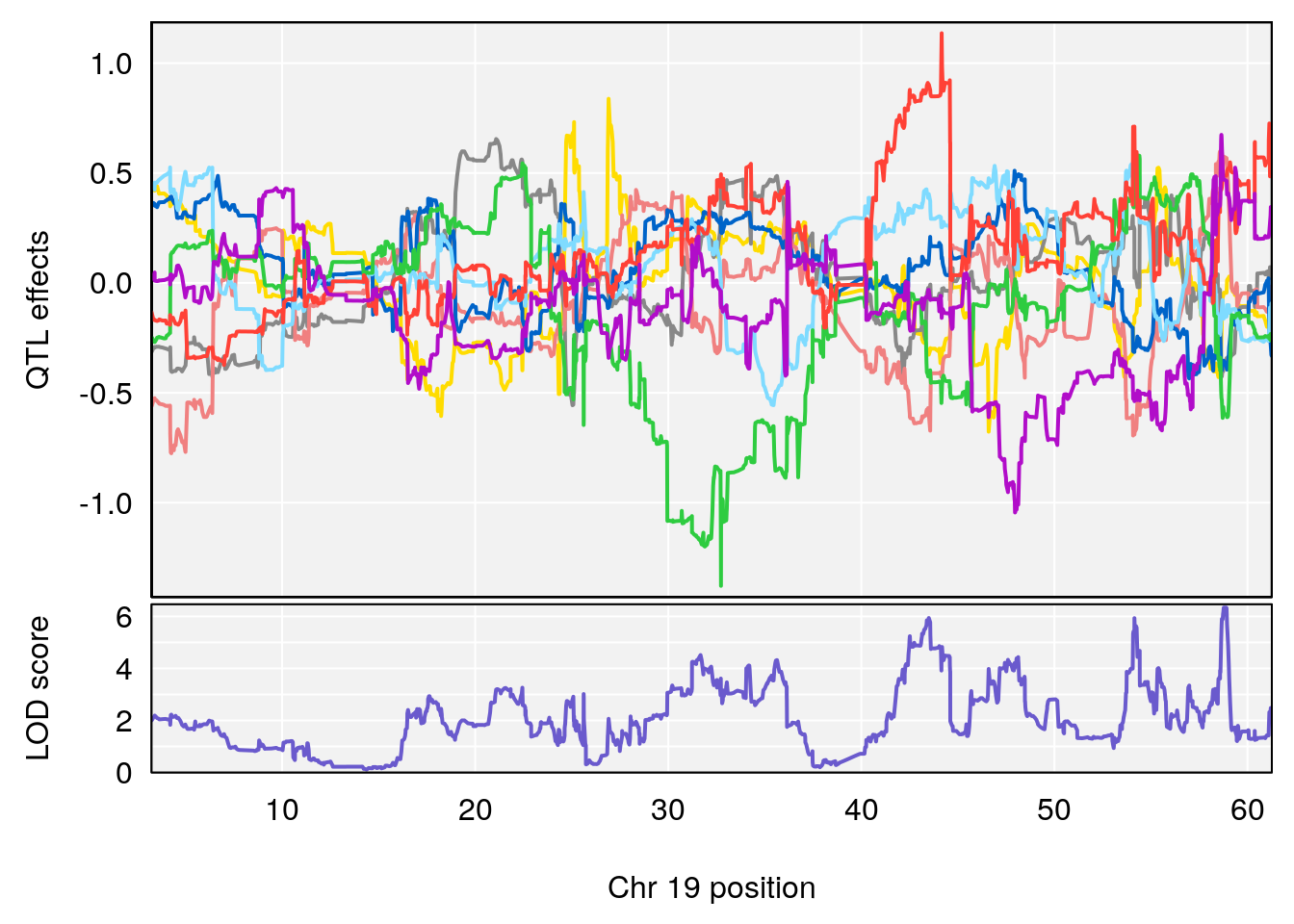
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
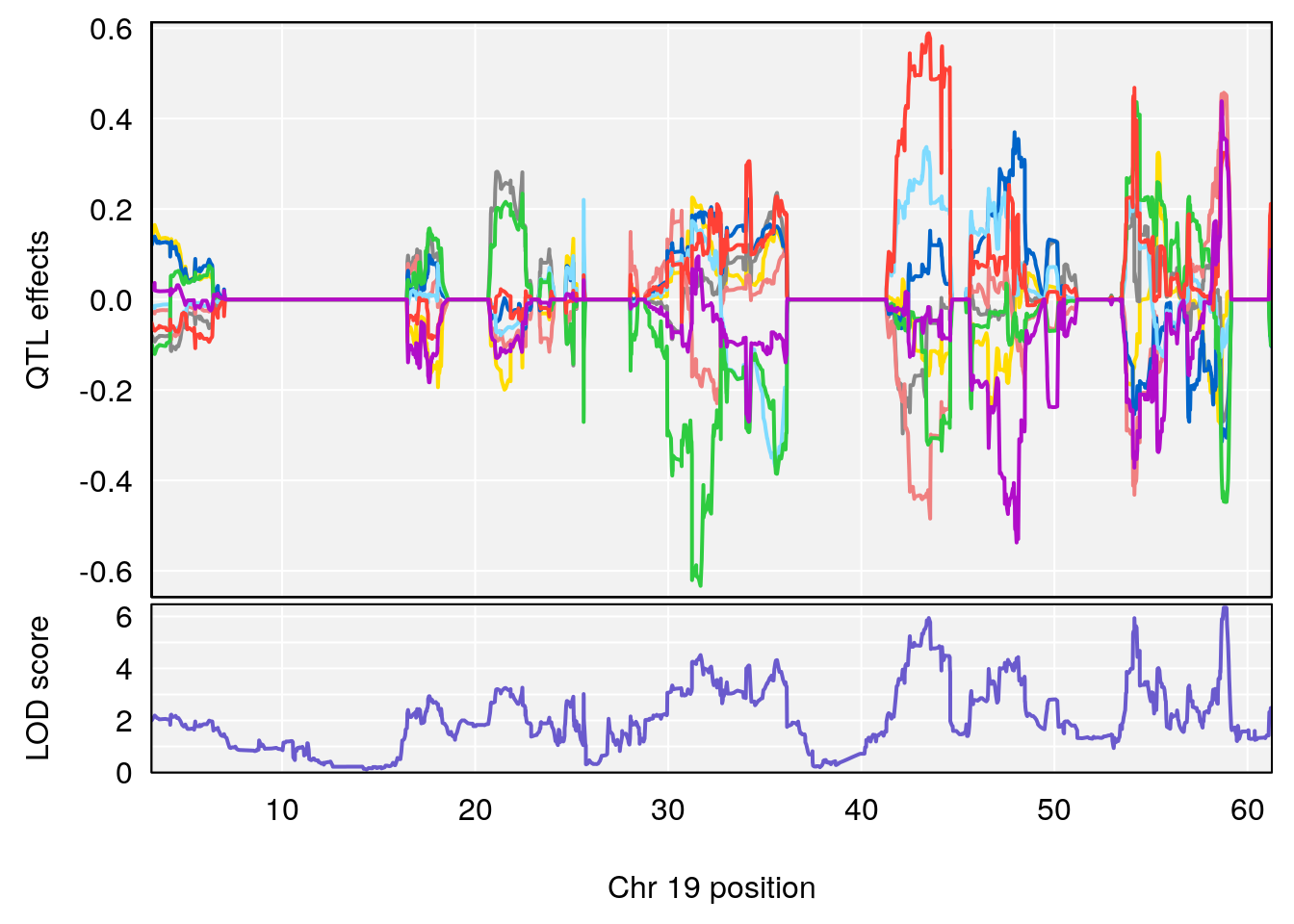
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
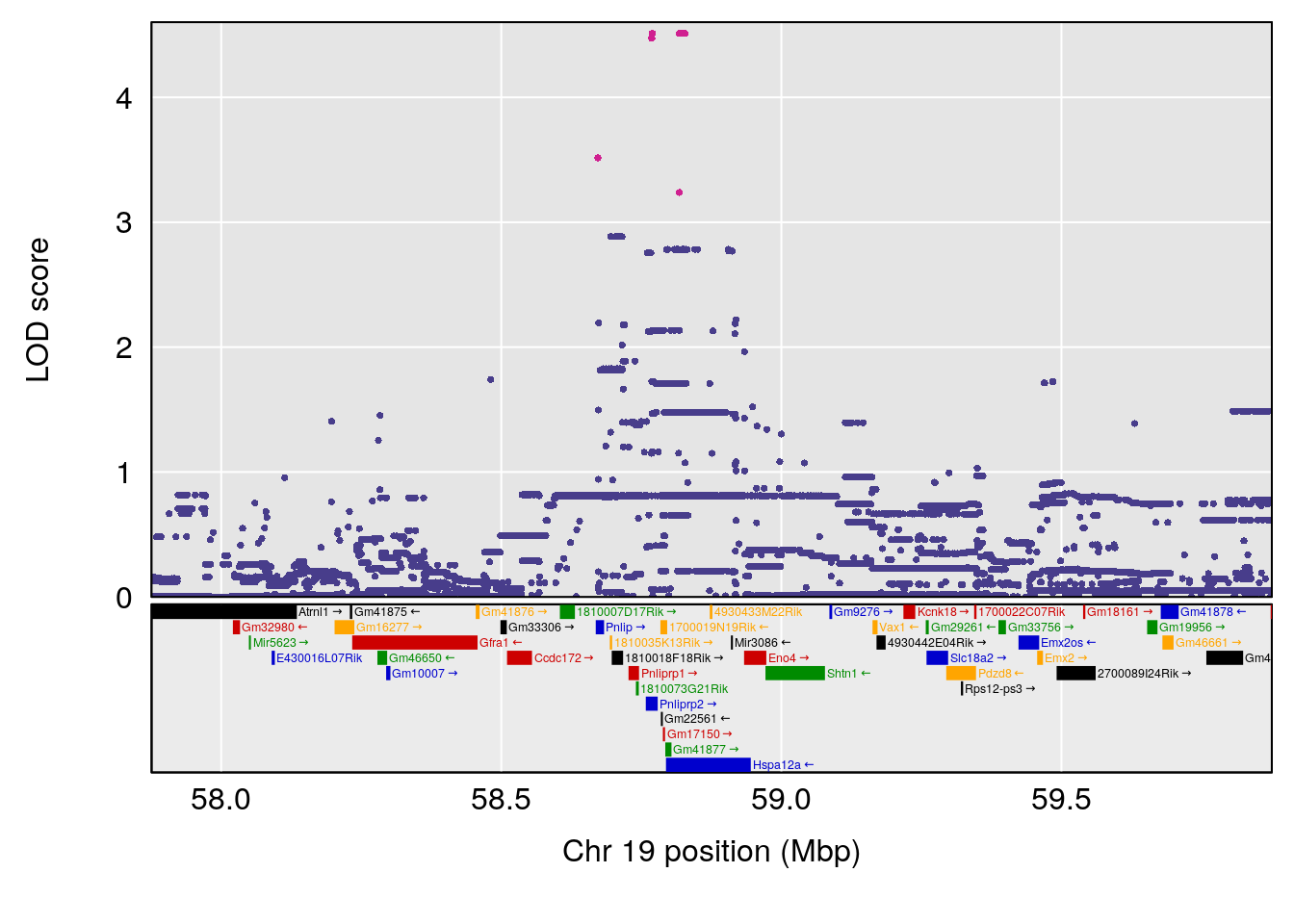
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
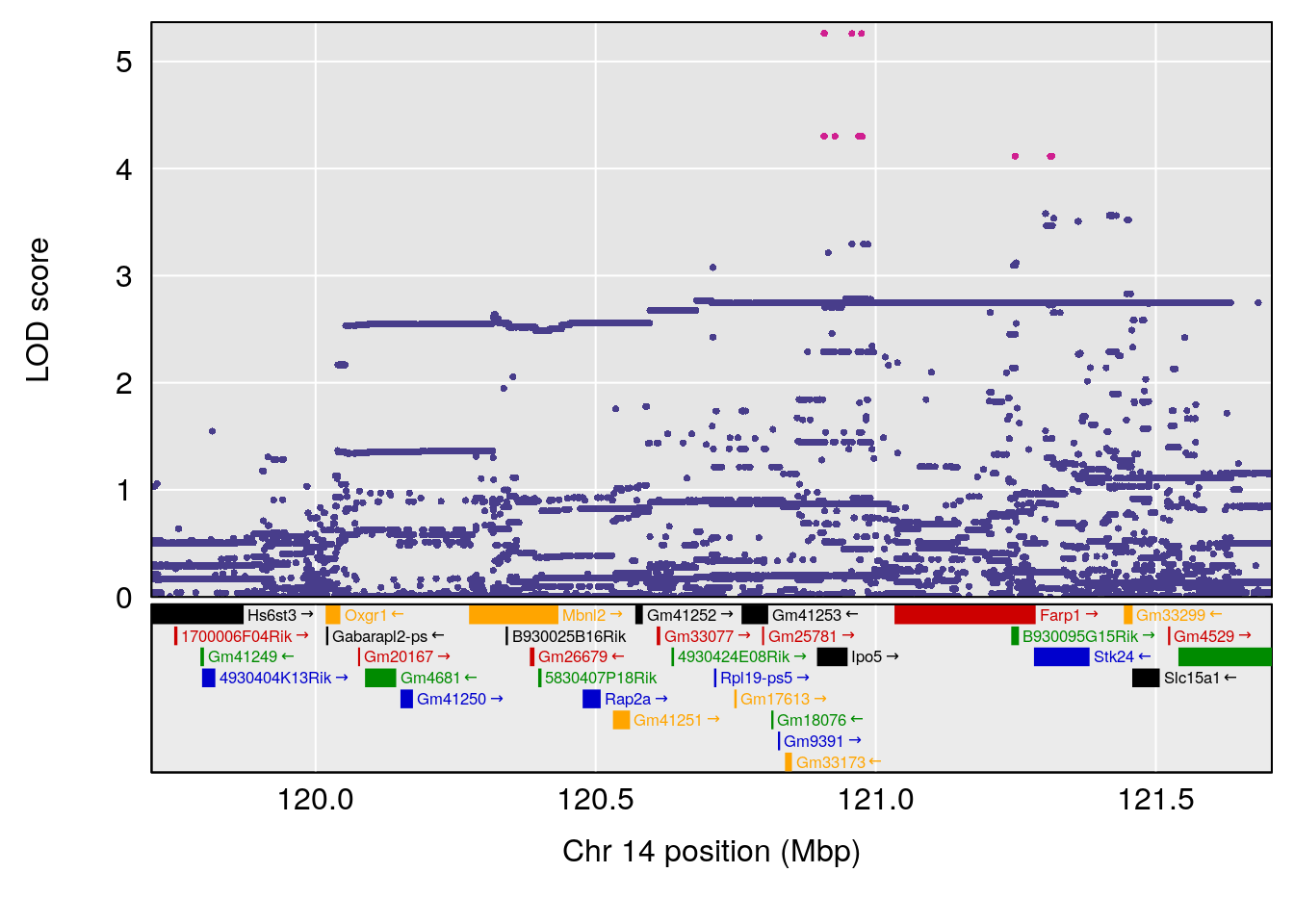
# [1] "Status_bin"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 2 44.052851 6.865421 43.981019 44.45340
# 2 1 pheno1 4 156.164170 7.313472 11.650579 156.22250
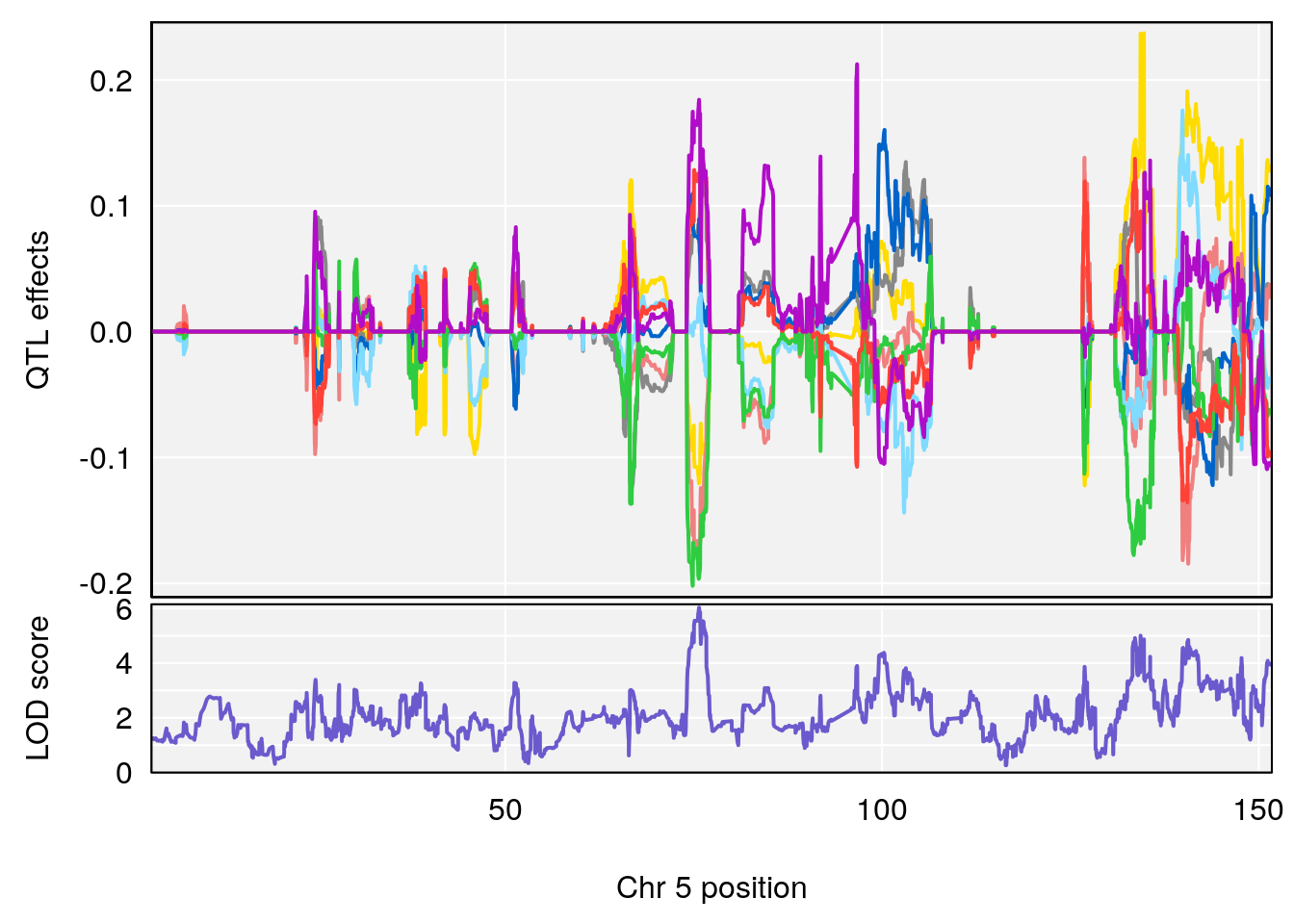
# 3 1 pheno1 5 75.711735 6.031017 74.447060 140.84978
# 4 1 pheno1 6 143.410744 8.741595 142.860829 144.10696
# 5 1 pheno1 9 68.163768 8.619578 67.950946 68.33918
# 6 1 pheno1 10 9.053339 6.040880 7.721133 120.73537
# 7 1 pheno1 14 120.707152 7.088220 103.551070 121.45855
# 8 1 pheno1 19 58.715653 6.486409 55.101359 60.04661
# 9 1 pheno1 X 144.599251 8.636026 143.607480 145.35755
# [1] 1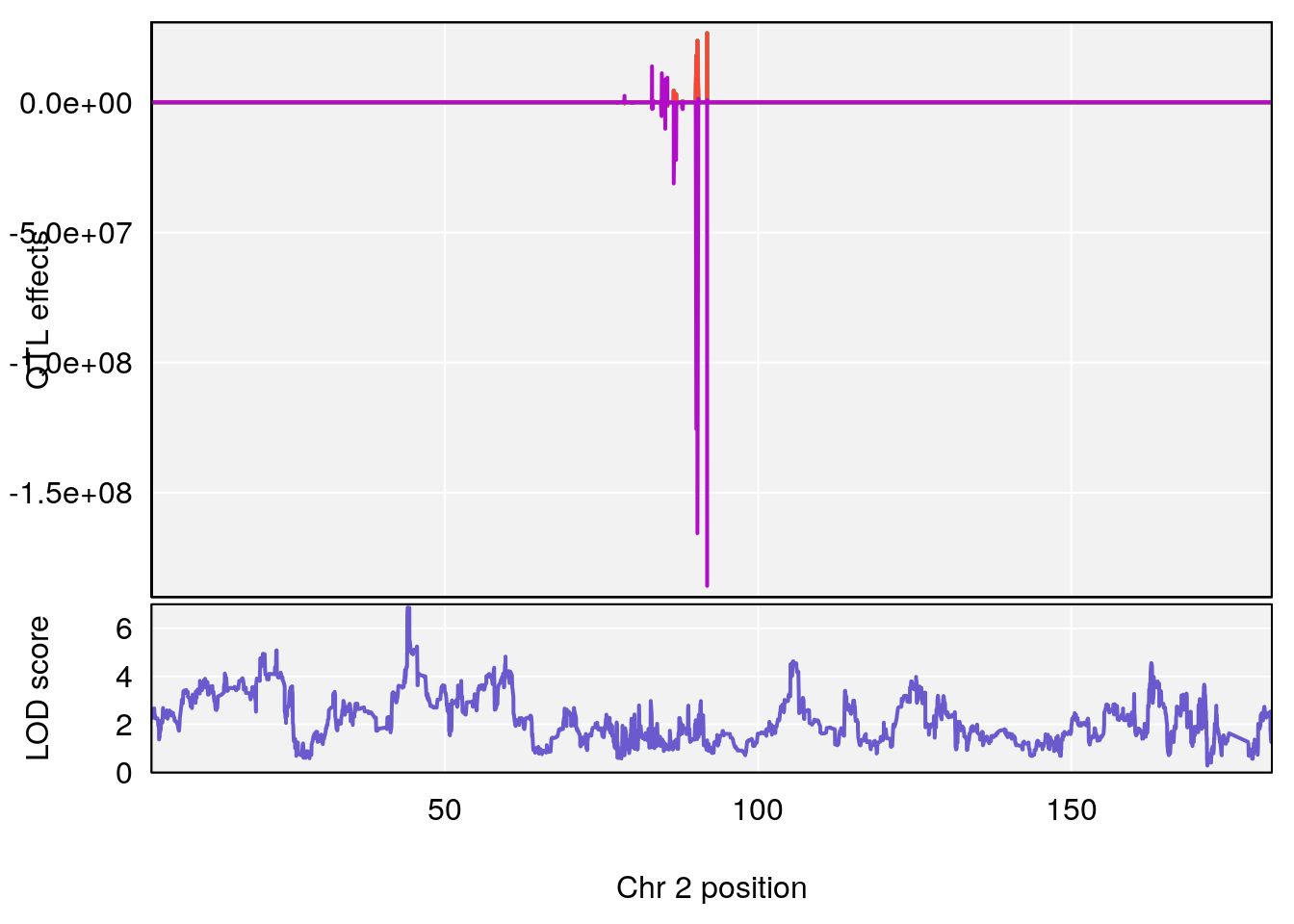
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 2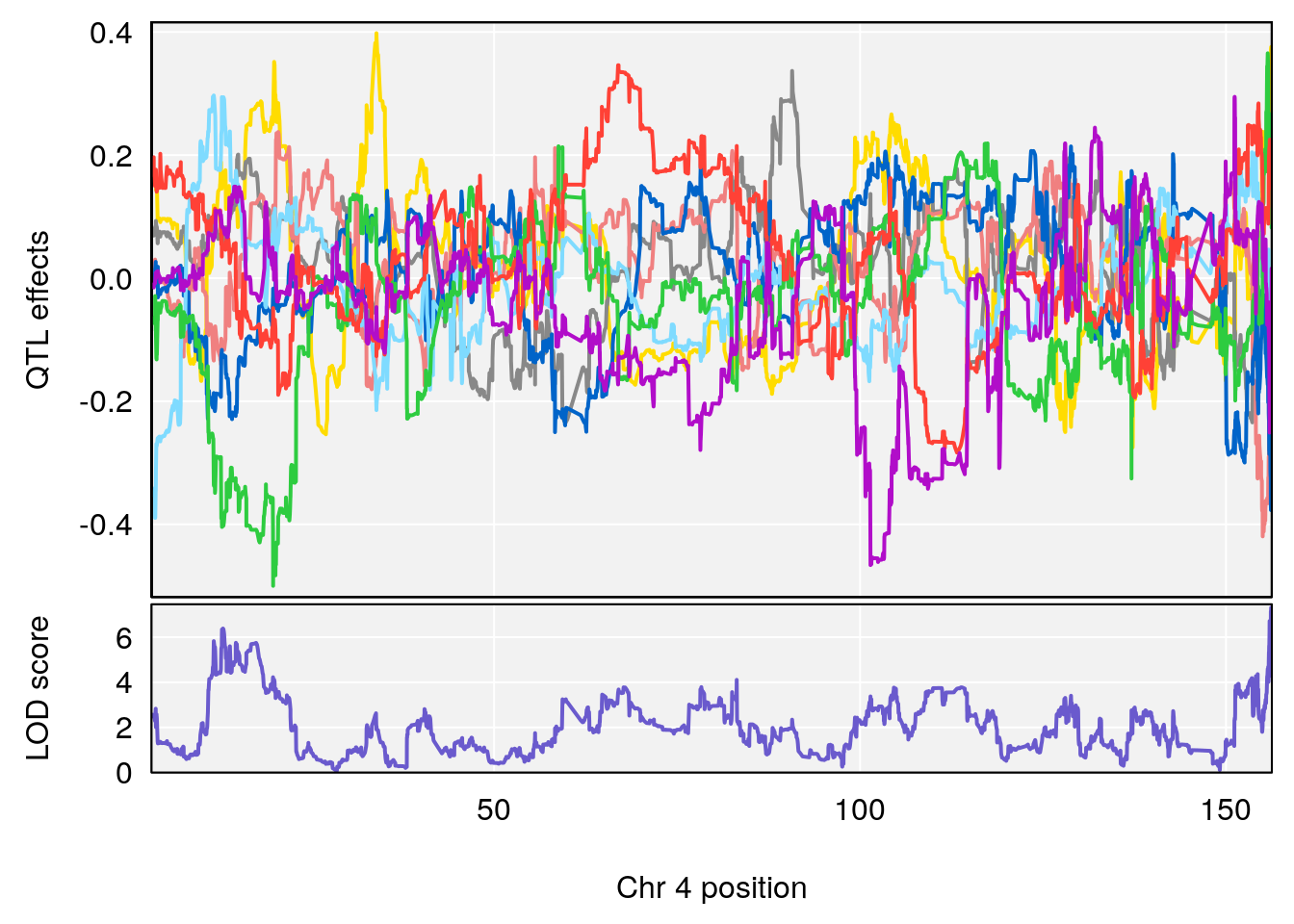
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 3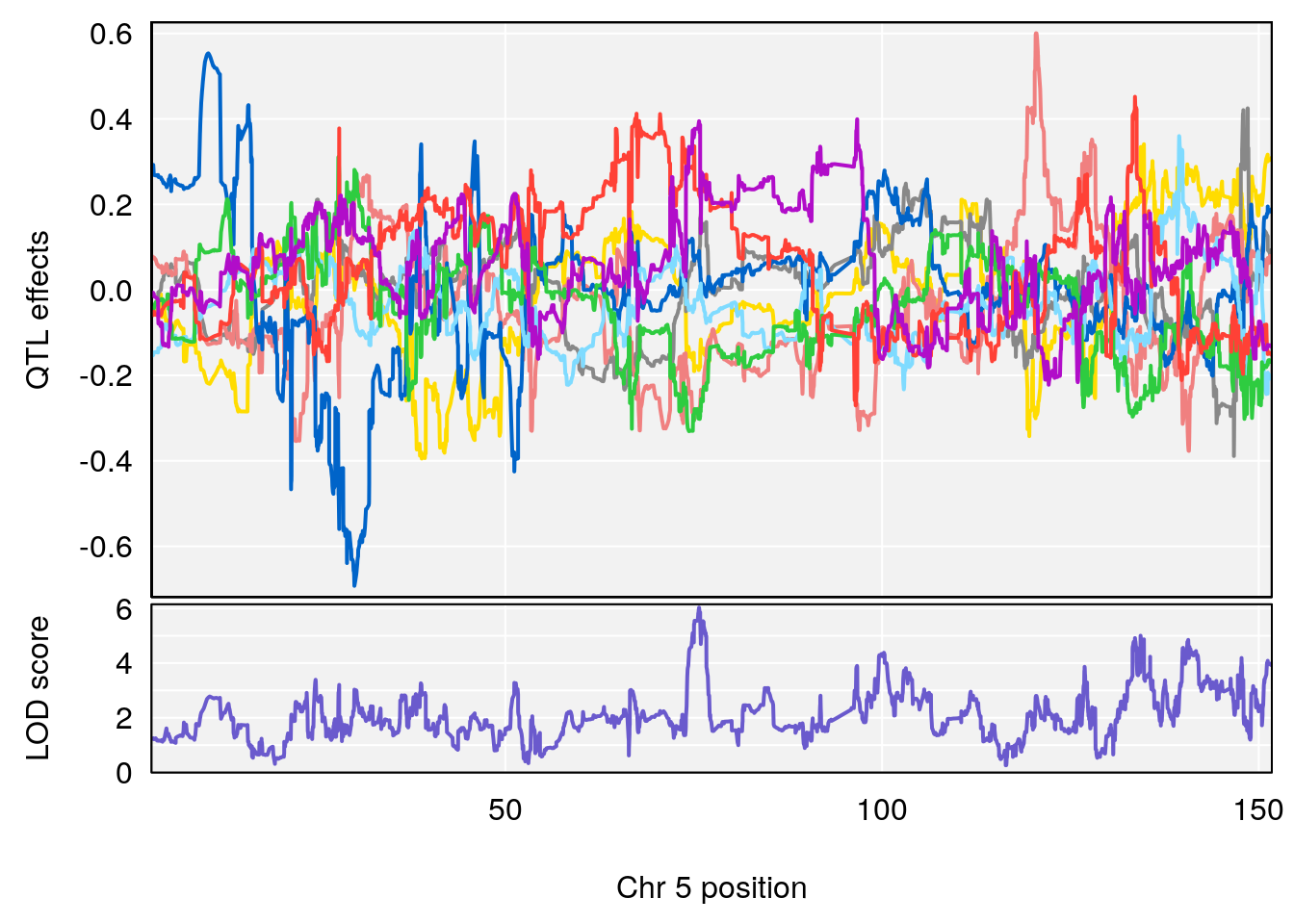
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 4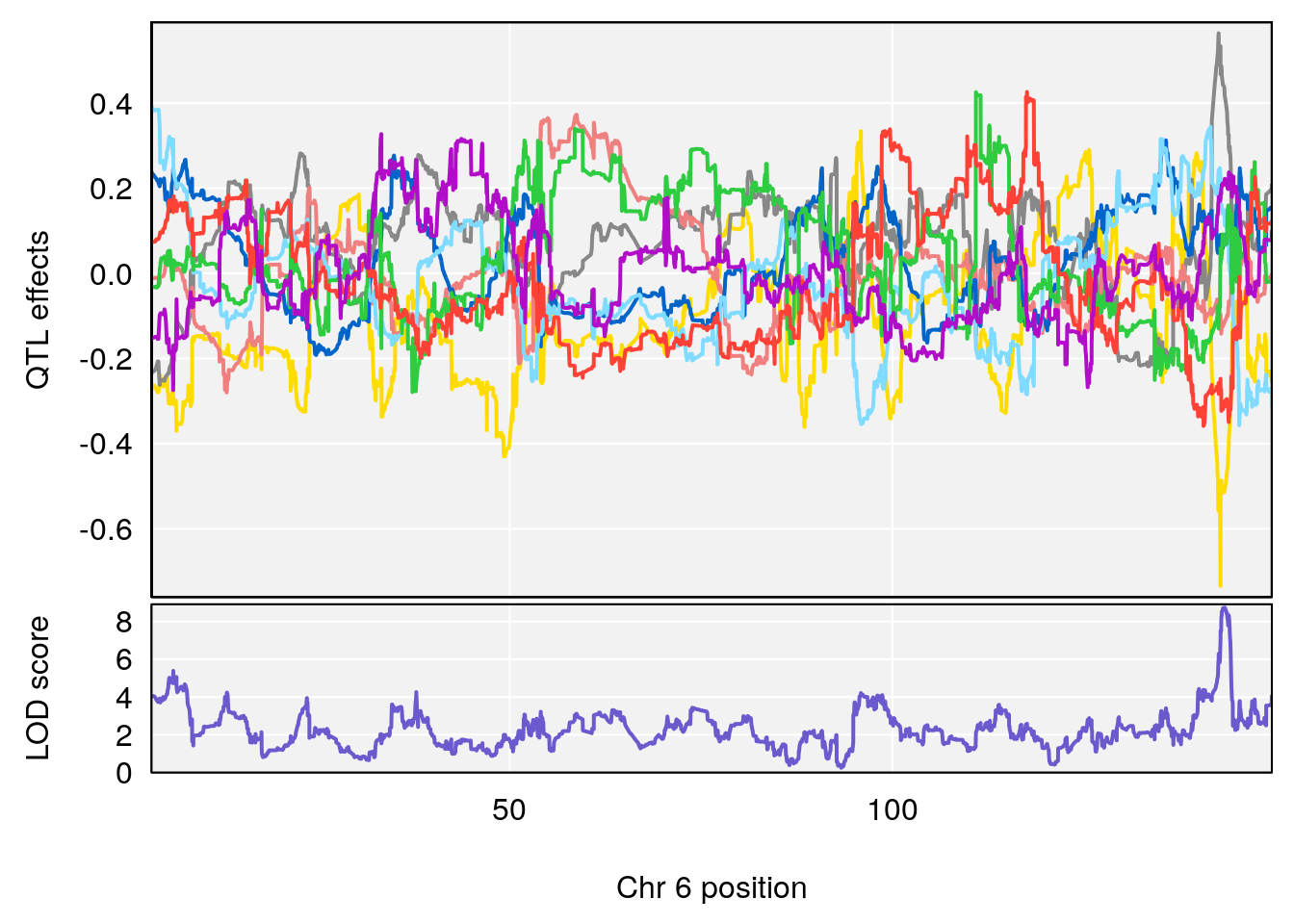
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 5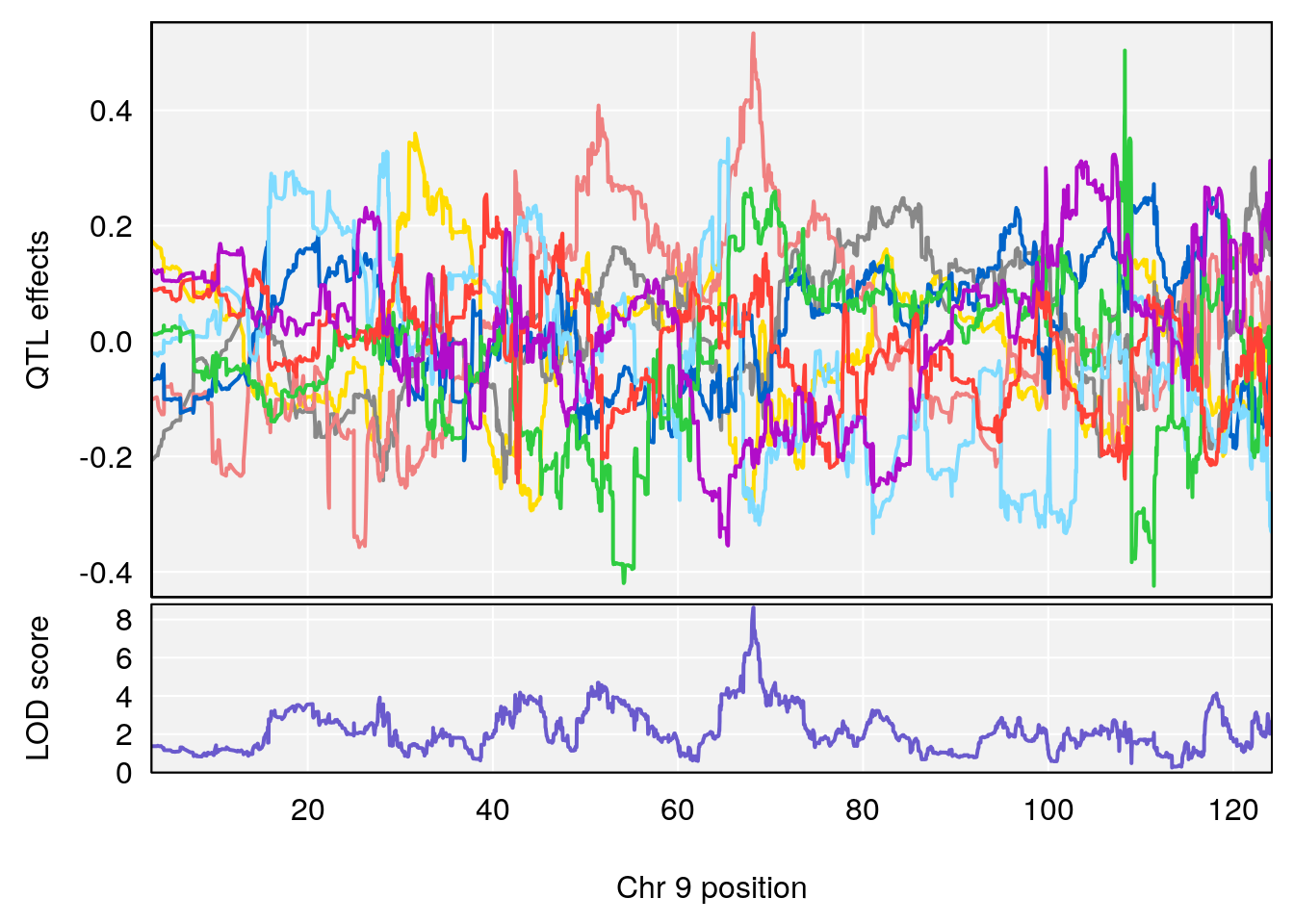
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 6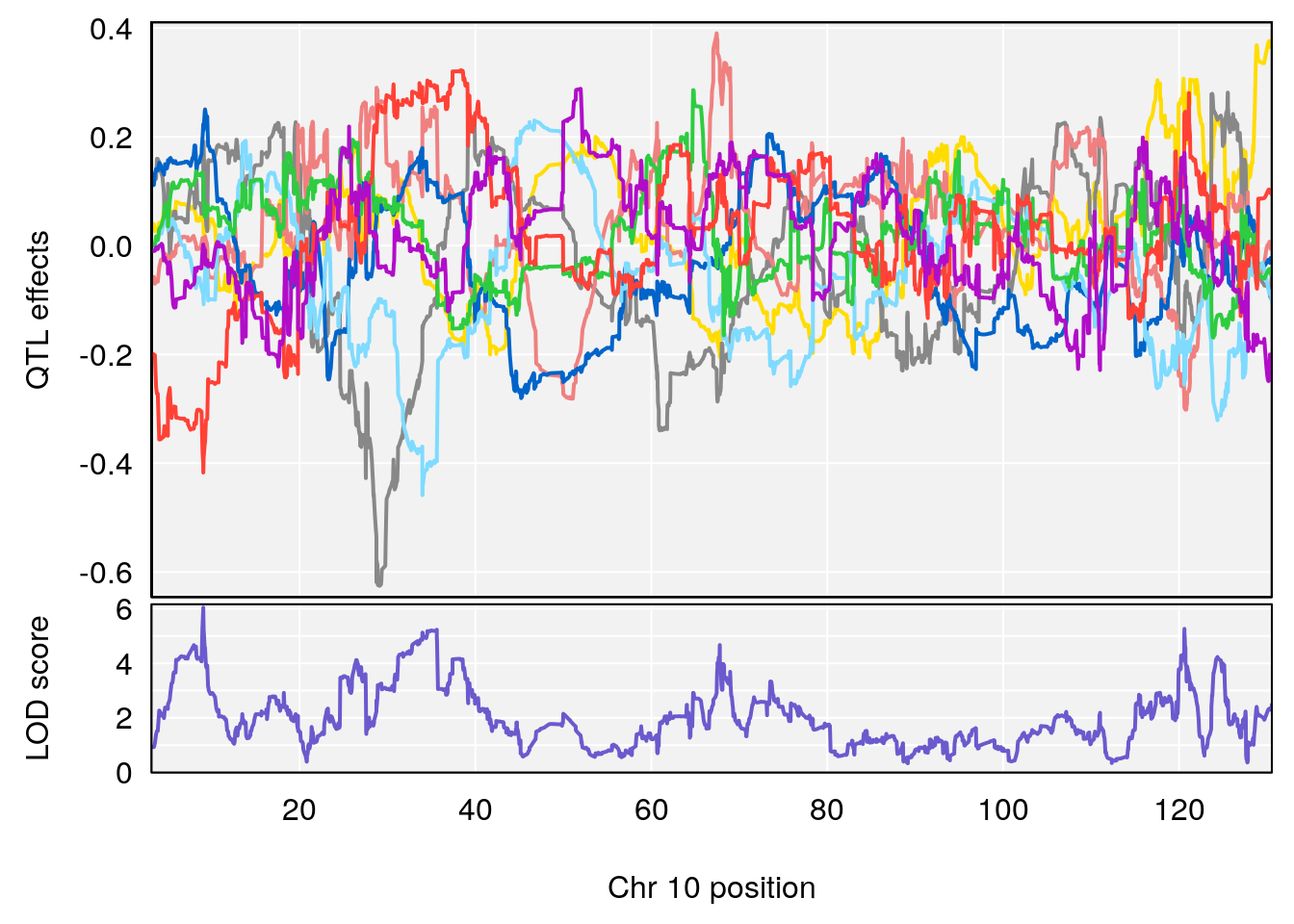
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 7
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 8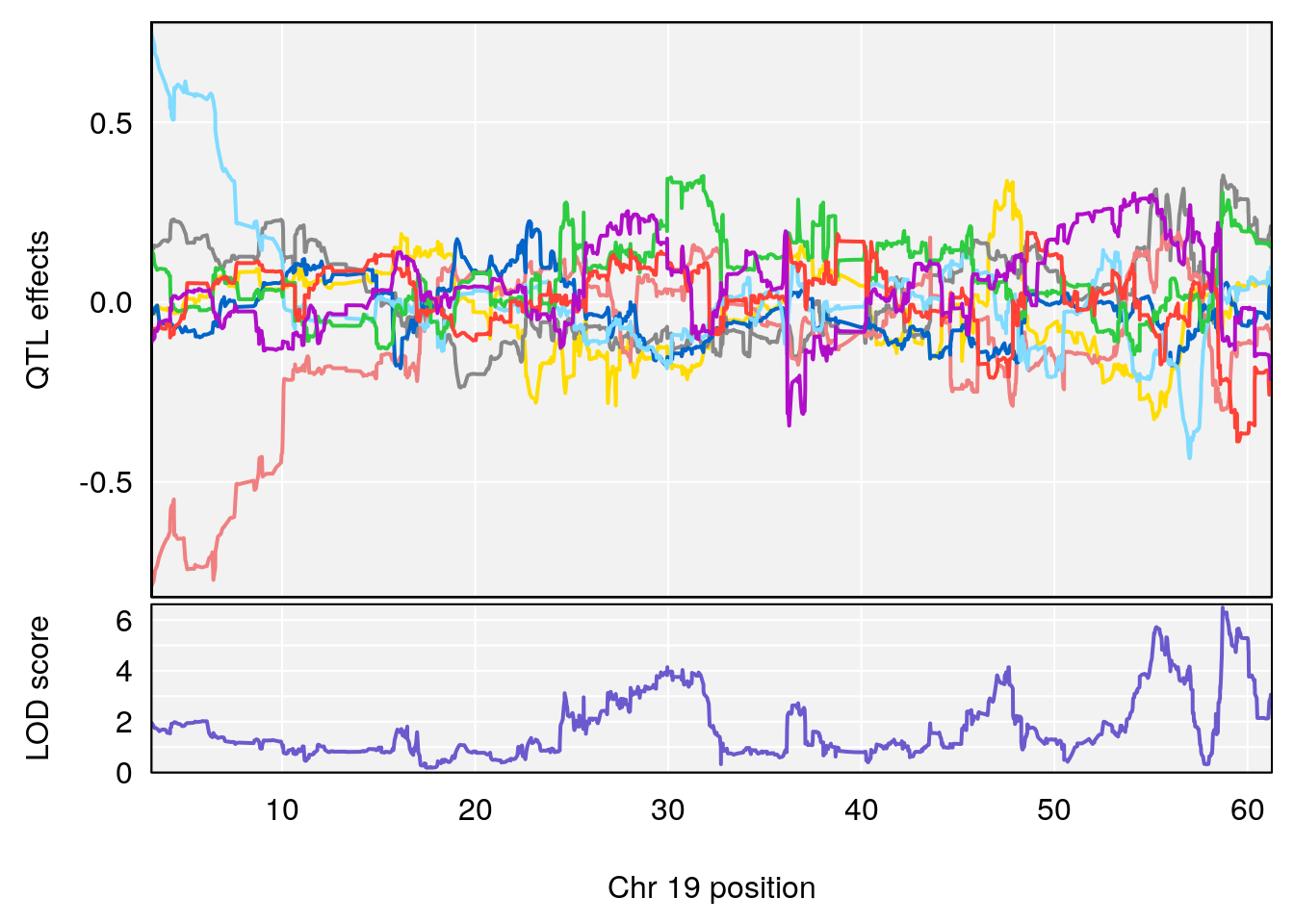
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 9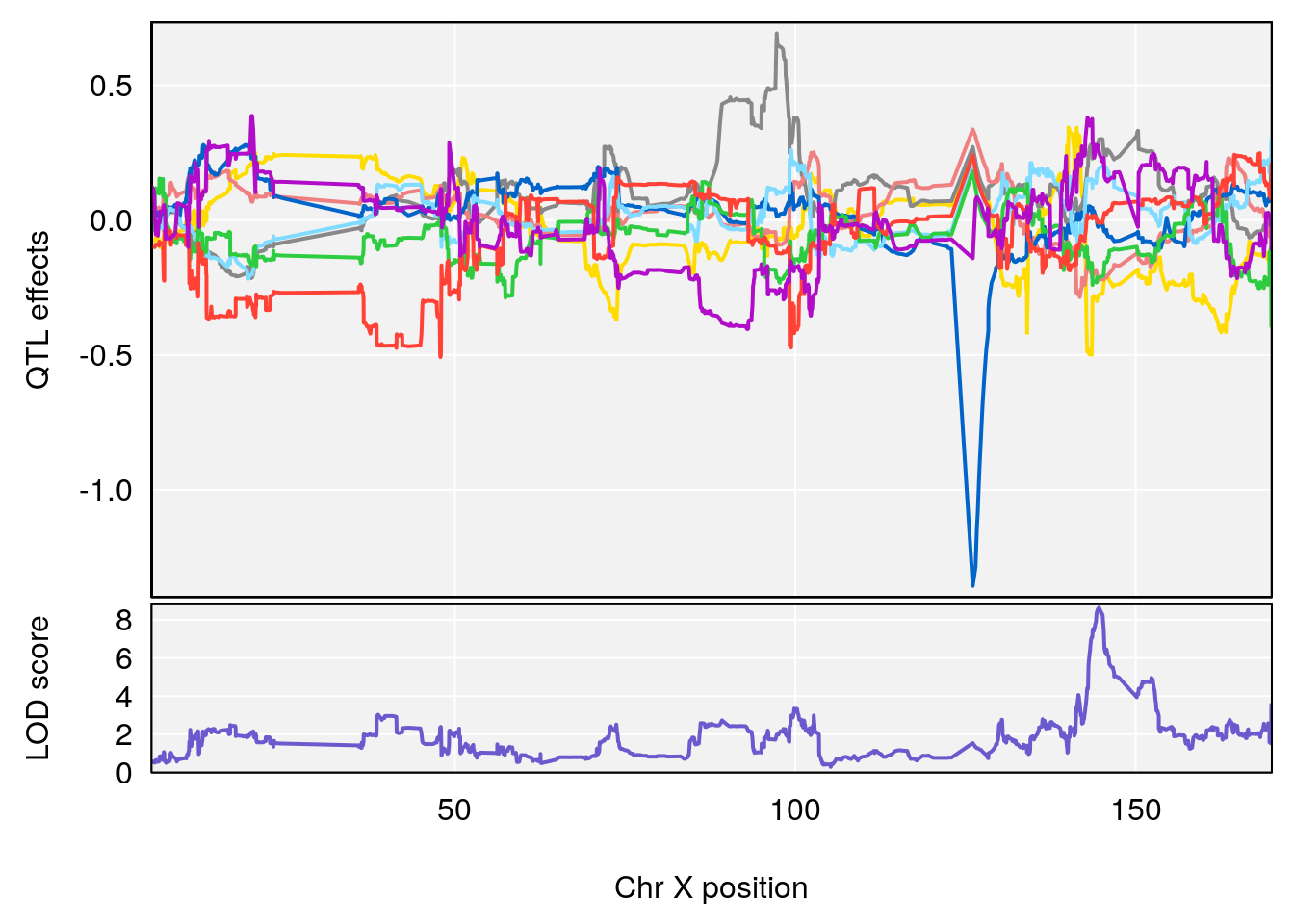
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
sessionInfo()
# R version 4.0.3 (2020-10-10)
# Platform: x86_64-pc-linux-gnu (64-bit)
# Running under: Ubuntu 20.04.2 LTS
#
# Matrix products: default
# BLAS/LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.8.so
#
# locale:
# [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
# [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
# [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=C
# [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
# [9] LC_ADDRESS=C LC_TELEPHONE=C
# [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
#
# attached base packages:
# [1] parallel stats graphics grDevices utils datasets methods
# [8] base
#
# other attached packages:
# [1] readxl_1.3.1 abind_1.4-5 regress_1.3-21 survival_3.2-7
# [5] qtl2_0.24 GGally_2.1.0 gridExtra_2.3 ggplot2_3.3.3
# [9] workflowr_1.6.2
#
# loaded via a namespace (and not attached):
# [1] tidyselect_1.1.0 xfun_0.21 purrr_0.3.4 lattice_0.20-41
# [5] splines_4.0.3 colorspace_2.0-0 vctrs_0.3.6 generics_0.1.0
# [9] htmltools_0.5.1.1 yaml_2.2.1 blob_1.2.1 rlang_1.0.2
# [13] later_1.1.0.1 pillar_1.4.7 glue_1.4.2 withr_2.4.1
# [17] DBI_1.1.1 bit64_4.0.5 RColorBrewer_1.1-2 lifecycle_1.0.0
# [21] plyr_1.8.6 stringr_1.4.0 cellranger_1.1.0 munsell_0.5.0
# [25] gtable_0.3.0 evaluate_0.14 memoise_2.0.0 labeling_0.4.2
# [29] knitr_1.31 fastmap_1.1.0 httpuv_1.5.5 highr_0.8
# [33] Rcpp_1.0.6 promises_1.2.0.1 scales_1.1.1 cachem_1.0.4
# [37] farver_2.0.3 fs_1.5.0 bit_4.0.4 digest_0.6.27
# [41] stringi_1.5.3 dplyr_1.0.4 grid_4.0.3 rprojroot_2.0.2
# [45] cli_2.3.0 tools_4.0.3 magrittr_2.0.1 tibble_3.0.6
# [49] RSQLite_2.2.3 crayon_1.4.1 whisker_0.4 pkgconfig_2.0.3
# [53] Matrix_1.3-2 ellipsis_0.3.1 data.table_1.13.6 assertthat_0.2.1
# [57] rmarkdown_2.6 reshape_0.8.8 R6_2.5.0 git2r_0.28.0
# [61] compiler_4.0.3This R Markdown site was created with workflowr