Plot_DO_fentanyl_alternate_metrics
Hao He
2021-12-20
Last updated: 2021-12-20
Checks: 7 0
Knit directory: DO_Opioid/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200504) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 344b6bc. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/Picture1.png
Untracked files:
Untracked: analysis/DDO_morphine1_second_set_69k.stdout
Untracked: analysis/DO_Fentanyl.R
Untracked: analysis/DO_Fentanyl.err
Untracked: analysis/DO_Fentanyl.out
Untracked: analysis/DO_Fentanyl.sh
Untracked: analysis/DO_Fentanyl_69k.R
Untracked: analysis/DO_Fentanyl_69k.err
Untracked: analysis/DO_Fentanyl_69k.out
Untracked: analysis/DO_Fentanyl_69k.sh
Untracked: analysis/DO_Fentanyl_GCTA_herit.R
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.R
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.err
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.out
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.sh
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.R
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.err
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.out
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.sh
Untracked: analysis/DO_Fentanyl_array.R
Untracked: analysis/DO_Fentanyl_array.err
Untracked: analysis/DO_Fentanyl_array.out
Untracked: analysis/DO_Fentanyl_array.sh
Untracked: analysis/DO_Fentanyl_finalreport_to_plink.sh
Untracked: analysis/DO_Fentanyl_gemma.R
Untracked: analysis/DO_Fentanyl_gemma.err
Untracked: analysis/DO_Fentanyl_gemma.out
Untracked: analysis/DO_Fentanyl_gemma.sh
Untracked: analysis/DO_morphine1.R
Untracked: analysis/DO_morphine1.Rout
Untracked: analysis/DO_morphine1.sh
Untracked: analysis/DO_morphine1.stderr
Untracked: analysis/DO_morphine1.stdout
Untracked: analysis/DO_morphine1_SNP.R
Untracked: analysis/DO_morphine1_SNP.Rout
Untracked: analysis/DO_morphine1_SNP.sh
Untracked: analysis/DO_morphine1_SNP.stderr
Untracked: analysis/DO_morphine1_SNP.stdout
Untracked: analysis/DO_morphine1_combined.R
Untracked: analysis/DO_morphine1_combined.Rout
Untracked: analysis/DO_morphine1_combined.sh
Untracked: analysis/DO_morphine1_combined.stderr
Untracked: analysis/DO_morphine1_combined.stdout
Untracked: analysis/DO_morphine1_combined_69k.R
Untracked: analysis/DO_morphine1_combined_69k.Rout
Untracked: analysis/DO_morphine1_combined_69k.sh
Untracked: analysis/DO_morphine1_combined_69k.stderr
Untracked: analysis/DO_morphine1_combined_69k.stdout
Untracked: analysis/DO_morphine1_combined_69k_m2.R
Untracked: analysis/DO_morphine1_combined_69k_m2.Rout
Untracked: analysis/DO_morphine1_combined_69k_m2.sh
Untracked: analysis/DO_morphine1_combined_69k_m2.stderr
Untracked: analysis/DO_morphine1_combined_69k_m2.stdout
Untracked: analysis/DO_morphine1_combined_weight_DOB.R
Untracked: analysis/DO_morphine1_combined_weight_DOB.Rout
Untracked: analysis/DO_morphine1_combined_weight_DOB.err
Untracked: analysis/DO_morphine1_combined_weight_DOB.out
Untracked: analysis/DO_morphine1_combined_weight_DOB.sh
Untracked: analysis/DO_morphine1_combined_weight_DOB.stderr
Untracked: analysis/DO_morphine1_combined_weight_DOB.stdout
Untracked: analysis/DO_morphine1_combined_weight_age.R
Untracked: analysis/DO_morphine1_combined_weight_age.err
Untracked: analysis/DO_morphine1_combined_weight_age.out
Untracked: analysis/DO_morphine1_combined_weight_age.sh
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.R
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.err
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.out
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.sh
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.R
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.err
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.out
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.sh
Untracked: analysis/DO_morphine1_cph.R
Untracked: analysis/DO_morphine1_cph.Rout
Untracked: analysis/DO_morphine1_cph.sh
Untracked: analysis/DO_morphine1_second_set.R
Untracked: analysis/DO_morphine1_second_set.Rout
Untracked: analysis/DO_morphine1_second_set.sh
Untracked: analysis/DO_morphine1_second_set.stderr
Untracked: analysis/DO_morphine1_second_set.stdout
Untracked: analysis/DO_morphine1_second_set_69k.R
Untracked: analysis/DO_morphine1_second_set_69k.Rout
Untracked: analysis/DO_morphine1_second_set_69k.sh
Untracked: analysis/DO_morphine1_second_set_69k.stderr
Untracked: analysis/DO_morphine1_second_set_SNP.R
Untracked: analysis/DO_morphine1_second_set_SNP.Rout
Untracked: analysis/DO_morphine1_second_set_SNP.sh
Untracked: analysis/DO_morphine1_second_set_SNP.stderr
Untracked: analysis/DO_morphine1_second_set_SNP.stdout
Untracked: analysis/DO_morphine1_second_set_weight_DOB.R
Untracked: analysis/DO_morphine1_second_set_weight_DOB.Rout
Untracked: analysis/DO_morphine1_second_set_weight_DOB.err
Untracked: analysis/DO_morphine1_second_set_weight_DOB.out
Untracked: analysis/DO_morphine1_second_set_weight_DOB.sh
Untracked: analysis/DO_morphine1_second_set_weight_DOB.stderr
Untracked: analysis/DO_morphine1_second_set_weight_DOB.stdout
Untracked: analysis/DO_morphine1_second_set_weight_age.R
Untracked: analysis/DO_morphine1_second_set_weight_age.Rout
Untracked: analysis/DO_morphine1_second_set_weight_age.err
Untracked: analysis/DO_morphine1_second_set_weight_age.out
Untracked: analysis/DO_morphine1_second_set_weight_age.sh
Untracked: analysis/DO_morphine1_second_set_weight_age.stderr
Untracked: analysis/DO_morphine1_second_set_weight_age.stdout
Untracked: analysis/DO_morphine1_weight_DOB.R
Untracked: analysis/DO_morphine1_weight_DOB.sh
Untracked: analysis/DO_morphine1_weight_age.R
Untracked: analysis/DO_morphine1_weight_age.sh
Untracked: analysis/DO_morphine_gemma.R
Untracked: analysis/DO_morphine_gemma.err
Untracked: analysis/DO_morphine_gemma.out
Untracked: analysis/DO_morphine_gemma.sh
Untracked: analysis/DO_morphine_gemma_firstmin.R
Untracked: analysis/DO_morphine_gemma_firstmin.err
Untracked: analysis/DO_morphine_gemma_firstmin.out
Untracked: analysis/DO_morphine_gemma_firstmin.sh
Untracked: analysis/DO_morphine_gemma_withpermu.R
Untracked: analysis/DO_morphine_gemma_withpermu.err
Untracked: analysis/DO_morphine_gemma_withpermu.out
Untracked: analysis/DO_morphine_gemma_withpermu.sh
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.R
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.err
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.out
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.sh
Untracked: analysis/Plot_DO_morphine1_SNP.R
Untracked: analysis/Plot_DO_morphine1_SNP.Rout
Untracked: analysis/Plot_DO_morphine1_SNP.sh
Untracked: analysis/Plot_DO_morphine1_SNP.stderr
Untracked: analysis/Plot_DO_morphine1_SNP.stdout
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.R
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.Rout
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.sh
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.stderr
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.stdout
Untracked: analysis/heritability_first_second_batch.R
Untracked: analysis/scripts/
Untracked: analysis/tibmr.R
Untracked: analysis/workflow_proc.R
Untracked: analysis/workflow_proc.sh
Untracked: analysis/workflow_proc.stderr
Untracked: analysis/workflow_proc.stdout
Untracked: analysis/x.R
Untracked: code/cfw/
Untracked: code/gemma_plot.R
Untracked: code/reconst_utils.R
Untracked: data/69k_grid_pgmap.RData
Untracked: data/DO_WBP_Data_JAB_to_map.xlsx
Untracked: data/Fentanyl_alternate_metrics.xlsx
Untracked: data/FinalReport/
Untracked: data/GM/
Untracked: data/GM_covar.csv
Untracked: data/GM_covar_07092018_morphine.csv
Untracked: data/Jackson_Lab_Bubier_MURGIGV01/
Untracked: data/MPD_Upload_October.csv
Untracked: data/MPD_Upload_October_updated_sex.csv
Untracked: data/Master Fentanyl DO Study Sheet.xlsx
Untracked: data/MasterMorphine Second Set DO w DOB2.xlsx
Untracked: data/MasterMorphine Second Set DO.xlsx
Untracked: data/Morphine CC DO mice Updated with Published inbred strains.csv
Untracked: data/Morphine_CC_DO_mice_Updated_with_Published_inbred_strains.csv
Untracked: data/cc_variants.sqlite
Untracked: data/combined/
Untracked: data/fentanyl/
Untracked: data/first/
Untracked: data/founder_geno.csv
Untracked: data/genetic_map.csv
Untracked: data/gm.json
Untracked: data/gwas.sh
Untracked: data/marker_grid_0.02cM_plus.txt
Untracked: data/mouse_genes_mgi.sqlite
Untracked: data/pheno.csv
Untracked: data/pheno_qtl2.csv
Untracked: data/pheno_qtl2_07092018_morphine.csv
Untracked: data/pheno_qtl2_w_dob.csv
Untracked: data/physical_map.csv
Untracked: data/rnaseq/
Untracked: data/sample_geno.csv
Untracked: data/second/
Untracked: figure/
Untracked: glimma-plots/
Untracked: output/DO_morphine_Min.depression.png
Untracked: output/DO_morphine_Min.depression22222_violin_chr5.pdf
Untracked: output/DO_morphine_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_Min.depression_coefplot_blup_chr5.png
Untracked: output/DO_morphine_Min.depression_coefplot_blup_chrX.png
Untracked: output/DO_morphine_Min.depression_coefplot_chr5.png
Untracked: output/DO_morphine_Min.depression_coefplot_chrX.png
Untracked: output/DO_morphine_Min.depression_peak_genes_chr5.png
Untracked: output/DO_morphine_Min.depression_violin_chr5.png
Untracked: output/DO_morphine_Recovery.Time.png
Untracked: output/DO_morphine_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr11.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr4.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr7.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr9.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr11.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr4.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr7.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr9.png
Untracked: output/DO_morphine_Status_bin.png
Untracked: output/DO_morphine_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_Survival.Time.png
Untracked: output/DO_morphine_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_Survival.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_Survival.Time_coefplot_blup_chr17.png
Untracked: output/DO_morphine_Survival.Time_coefplot_blup_chr8.png
Untracked: output/DO_morphine_Survival.Time_coefplot_chr17.png
Untracked: output/DO_morphine_Survival.Time_coefplot_chr8.png
Untracked: output/DO_morphine_combine_batch_peak_violin.pdf
Untracked: output/DO_morphine_combined_69k_m2_Min.depression.png
Untracked: output/DO_morphine_combined_69k_m2_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_combined_69k_m2_Recovery.Time.png
Untracked: output/DO_morphine_combined_69k_m2_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_combined_69k_m2_Status_bin.png
Untracked: output/DO_morphine_combined_69k_m2_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_combined_69k_m2_Survival.Time.png
Untracked: output/DO_morphine_combined_69k_m2_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Survival.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_coxph_24hrs_kinship_QTL.png
Untracked: output/DO_morphine_cphout.RData
Untracked: output/DO_morphine_first_batch_peak_in_second_batch_violin.pdf
Untracked: output/DO_morphine_first_batch_peak_in_second_batch_violin_sidebyside.pdf
Untracked: output/DO_morphine_first_batch_peak_violin.pdf
Untracked: output/DO_morphine_operm.cph.RData
Untracked: output/DO_morphine_second_batch_on_first_batch_peak_violin.pdf
Untracked: output/DO_morphine_second_batch_peak_ch6surv_on_first_batchviolin.pdf
Untracked: output/DO_morphine_second_batch_peak_ch6surv_on_first_batchviolin2.pdf
Untracked: output/DO_morphine_second_batch_peak_in_first_batch_violin.pdf
Untracked: output/DO_morphine_second_batch_peak_in_first_batch_violin_sidebyside.pdf
Untracked: output/DO_morphine_second_batch_peak_violin.pdf
Untracked: output/DO_morphine_secondbatch_69k_Min.depression.png
Untracked: output/DO_morphine_secondbatch_69k_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_69k_Recovery.Time.png
Untracked: output/DO_morphine_secondbatch_69k_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_69k_Status_bin.png
Untracked: output/DO_morphine_secondbatch_69k_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_69k_Survival.Time.png
Untracked: output/DO_morphine_secondbatch_69k_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Survival.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Min.depression.png
Untracked: output/DO_morphine_secondbatch_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Recovery.Time.png
Untracked: output/DO_morphine_secondbatch_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Status_bin.png
Untracked: output/DO_morphine_secondbatch_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Survival.Time.png
Untracked: output/DO_morphine_secondbatch_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Survival.Time_coefplot_blup.pdf
Untracked: output/Fentanyl/
Untracked: output/TIMBR.test.RData
Untracked: output/apr_69kchr_combined.RData
Untracked: output/apr_69kchr_k_loco_combined.rds
Untracked: output/apr_69kchr_second_set.RData
Untracked: output/combine_batch_variation.RData
Untracked: output/combined_gm.RData
Untracked: output/combined_gm.k_loco.rds
Untracked: output/combined_gm.k_overall.rds
Untracked: output/combined_gm.probs_8state.rds
Untracked: output/coxph/
Untracked: output/do.morphine.RData
Untracked: output/do.morphine.k_loco.rds
Untracked: output/do.morphine.probs_36state.rds
Untracked: output/do.morphine.probs_8state.rds
Untracked: output/first_batch_variation.RData
Untracked: output/first_second_survival_peak_chr.xlsx
Untracked: output/hsq_1_first_batch_herit_qtl2.RData
Untracked: output/hsq_2_second_batch_herit_qtl2.RData
Untracked: output/old_temp/
Untracked: output/pr_69kchr_combined.RData
Untracked: output/pr_69kchr_second_set.RData
Untracked: output/qtl.morphine.69k.out.combined.RData
Untracked: output/qtl.morphine.69k.out.combined_m2.RData
Untracked: output/qtl.morphine.69k.out.second_set.RData
Untracked: output/qtl.morphine.operm.RData
Untracked: output/qtl.morphine.out.RData
Untracked: output/qtl.morphine.out.combined_gm.RData
Untracked: output/qtl.morphine.out.combined_weight_DOB.RData
Untracked: output/qtl.morphine.out.combined_weight_age.RData
Untracked: output/qtl.morphine.out.second_set.RData
Untracked: output/qtl.morphine.out.second_set.weight_DOB.RData
Untracked: output/qtl.morphine.out.second_set.weight_age.RData
Untracked: output/qtl.morphine.out.weight_DOB.RData
Untracked: output/qtl.morphine.out.weight_age.RData
Untracked: output/qtl.morphine1.snpout.RData
Untracked: output/qtl.morphine2.snpout.RData
Untracked: output/second_batch_variation.RData
Untracked: output/second_set_apr_69kchr_k_loco.rds
Untracked: output/second_set_gm.RData
Untracked: output/second_set_gm.k_loco.rds
Untracked: output/second_set_gm.probs_36state.rds
Untracked: output/second_set_gm.probs_8state.rds
Untracked: output/topSNP_chr5_mindepression.csv
Untracked: output/zoompeak_Min.depression_9.pdf
Untracked: output/zoompeak_Recovery.Time_16.pdf
Untracked: output/zoompeak_Status_bin_11.pdf
Untracked: output/zoompeak_Survival.Time_1.pdf
Untracked: output/zoompeak_fentanyl_Survival.Time_2.pdf
Unstaged changes:
Modified: _workflowr.yml
Modified: analysis/marker_violin.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/Plot_DO_fentanyl_alternate_metrics.Rmd) and HTML (docs/Plot_DO_fentanyl_alternate_metrics.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 344b6bc | xhyuo | 2021-12-20 | Plot_DO_fentanyl_alternate_metrics |
| html | f412738 | xhyuo | 2021-12-20 | Build site. |
| Rmd | a90242b | xhyuo | 2021-12-20 | Plot_DO_fentanyl_alternate_metrics.Rmd |
Last update: 2021-12-20
##loading libraries
library(ggplot2)
library(gridExtra)
library(GGally)
library(parallel)
library(qtl2)
library(parallel)
library(survival)
library(regress)
library(abind)
library(openxlsx)
library(tidyverse)
library(cowplot)
library(furrr)
library(patchwork)
library(qtl)
theme_set(theme_cowplot())
rz.transform <- function(y) {
rankY=rank(y, ties.method="average", na.last="keep")
rzT=qnorm(rankY/(length(na.exclude(rankY))+1))
return(rzT)
}
Pvaltolod <- function(p){
if(p == 0) p = 1e-10
y = ifelse(p >= 0.5, 0, qchisq(1-2*p, df=1)/(2*log(10)))
y
}
load("code/do.colors.RData")###Read phenotype data
do.pheno <- read.xlsx("data/Master Fentanyl DO Study Sheet.xlsx", sheet = 1,
rows = 1:399, na.strings = "NaN", detectDates = TRUE)
#second 31009 is really 31109
do.pheno[do.pheno$ID == 31009 & do.pheno$File == 20210519, "ID"] = 31109
#remove zombie mice
do.pheno <- do.pheno %>%
dplyr::filter(is.na(Censor)) %>%
dplyr::rename(.,
Min.depression = 9,
Survival.Time = 10,
Recovery.Time = 11) %>%
dplyr::mutate(Status_bin = ifelse(Survival == "ALIVE", 1, 0)) %>%
dplyr::mutate(ID = as.character(ID)) %>%
dplyr::arrange(ID, desc(File)) %>%
dplyr::distinct(ID, .keep_all = TRUE)
#Fentanyl_alternate_metrics.xlsx
do.pheno.alt <- read.xlsx("data/Fentanyl_alternate_metrics.xlsx", sheet = 1,
na.strings = "NaN") %>%
mutate(ID = as.character(Mouse.ID)) %>%
filter(!(if_all(2:11, is.na))) %>%
distinct(ID, .keep_all = TRUE)
colnames(do.pheno.alt) <- str_replace(colnames(do.pheno.alt), "\\(", "")
colnames(do.pheno.alt) <- str_replace(colnames(do.pheno.alt), "\\)", "")
colnames(do.pheno.alt) <- str_replace(colnames(do.pheno.alt), "\\%/", "")
colnames(do.pheno.alt) <- str_replace(colnames(do.pheno.alt), "\\.%", "")
#get sex and dob
do.pheno <- left_join(do.pheno.alt, do.pheno[, 2:4])
# Read genotype data -----------------------------------------------------------
load("data/Jackson_Lab_Bubier_MURGIGV01/gm_DO395_qc_newid.RData")#gm_after_qc
overlap.id = intersect(ind_ids(gm_after_qc), do.pheno$ID)
#subset
gm = gm_after_qc[overlap.id, ]
do.pheno = do.pheno[do.pheno$ID %in% overlap.id, ]
rownames(do.pheno) = do.pheno$ID
do.pheno = do.pheno[overlap.id,]
all.equal(ind_ids(gm), do.pheno$ID)
# [1] TRUE
#covar
do.pheno$Sex <- as.factor(do.pheno$Sex)
do.pheno$dob <- as.factor(do.pheno$dob)
#addcovar
options(na.action='na.pass')
addcovar = model.matrix(~Sex + dob, data = do.pheno[, c("Sex", "dob")])[,-1]
colnames(addcovar) <- c("sex", "dob")
#histgram for raw phenotypes------------
do.pheno %>%
select(2:11) %>%
map2(.,.y = colnames(.), ~ ggplot(do.pheno, aes(x = .x)) +
geom_histogram() +
xlab(.y)
) %>%
wrap_plots( ncol = 3)
# Warning: Removed 204 rows containing non-finite values (stat_bin).
# Warning: Removed 37 rows containing non-finite values (stat_bin).
# Warning: Removed 37 rows containing non-finite values (stat_bin).
# Warning: Removed 37 rows containing non-finite values (stat_bin).
# Warning: Removed 72 rows containing non-finite values (stat_bin).
# Warning: Removed 282 rows containing non-finite values (stat_bin).
# Warning: Removed 282 rows containing non-finite values (stat_bin).
# Warning: Removed 282 rows containing non-finite values (stat_bin).
# Warning: Removed 282 rows containing non-finite values (stat_bin).
# Warning: Removed 282 rows containing non-finite values (stat_bin).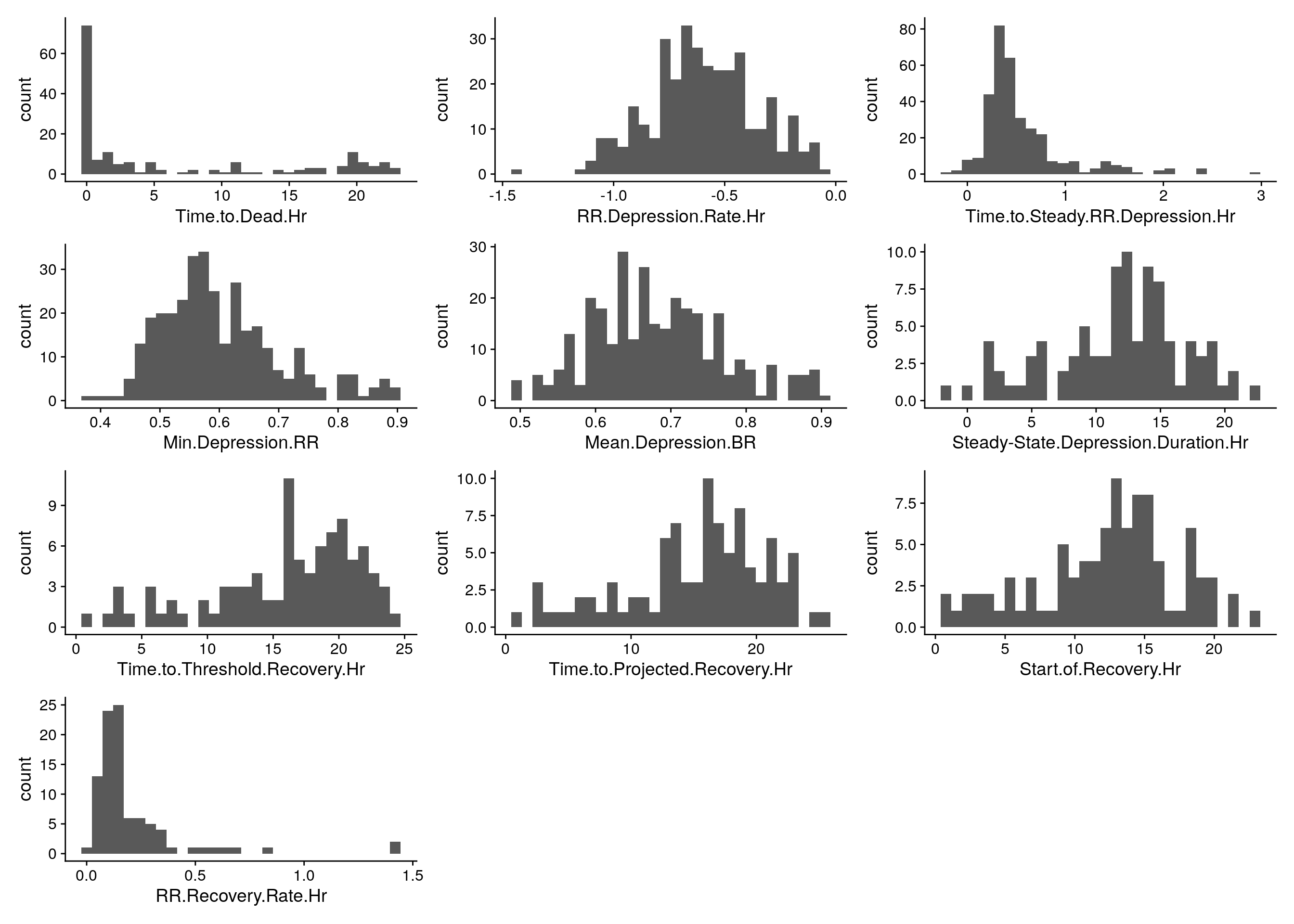
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
#histgram for rankz phenotypes------------
do.pheno %>%
select(2:11) %>%
map2(.,.y = colnames(.), ~ ggplot(do.pheno, aes(x = rz.transform(.x))) +
geom_histogram() +
xlab(.y)
) %>%
wrap_plots( ncol = 3)
# Warning: Removed 204 rows containing non-finite values (stat_bin).
# Warning: Removed 37 rows containing non-finite values (stat_bin).
# Warning: Removed 37 rows containing non-finite values (stat_bin).
# Warning: Removed 37 rows containing non-finite values (stat_bin).
# Warning: Removed 72 rows containing non-finite values (stat_bin).
# Warning: Removed 282 rows containing non-finite values (stat_bin).
# Warning: Removed 282 rows containing non-finite values (stat_bin).
# Warning: Removed 282 rows containing non-finite values (stat_bin).
# Warning: Removed 282 rows containing non-finite values (stat_bin).
# Warning: Removed 282 rows containing non-finite values (stat_bin).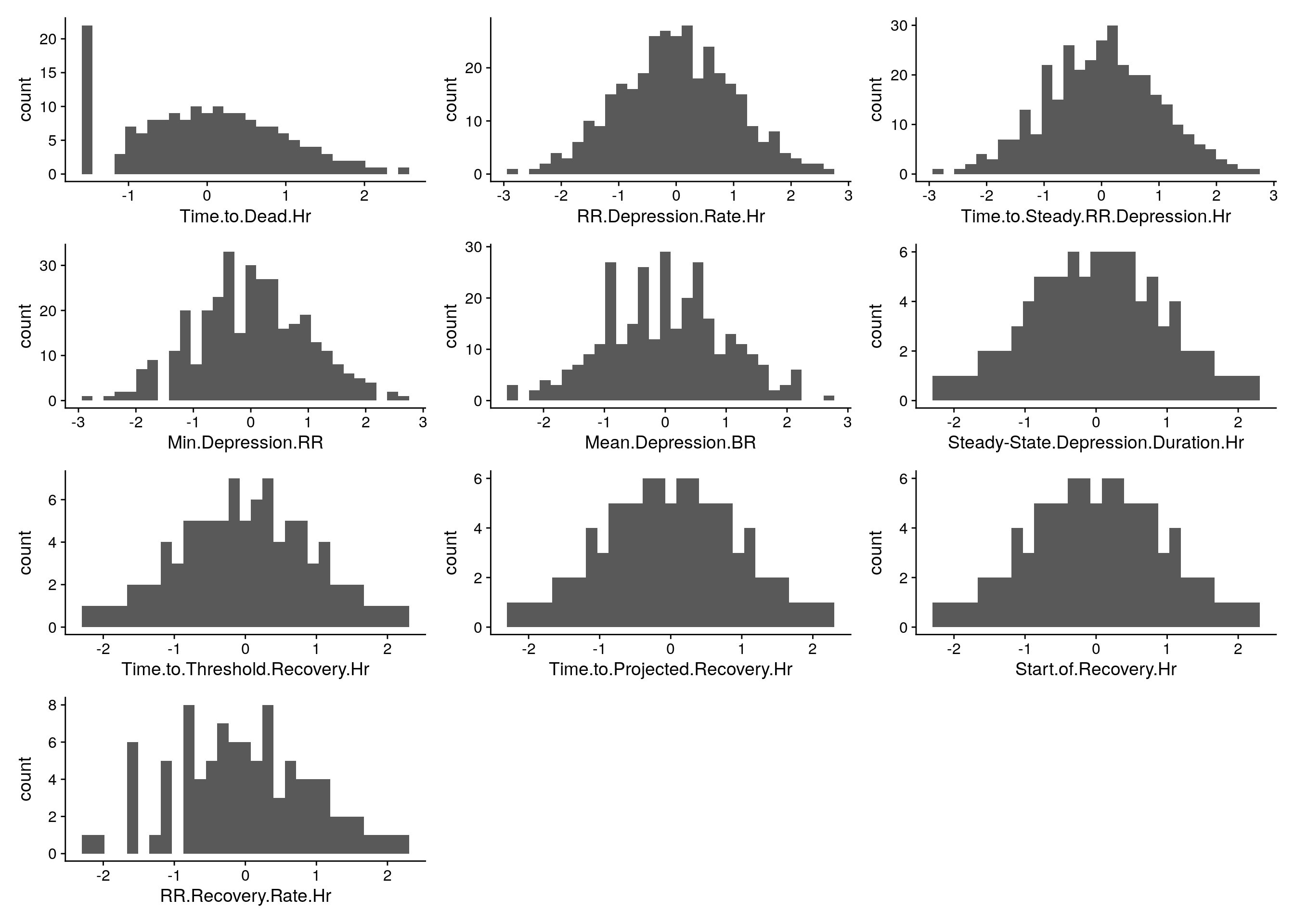
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
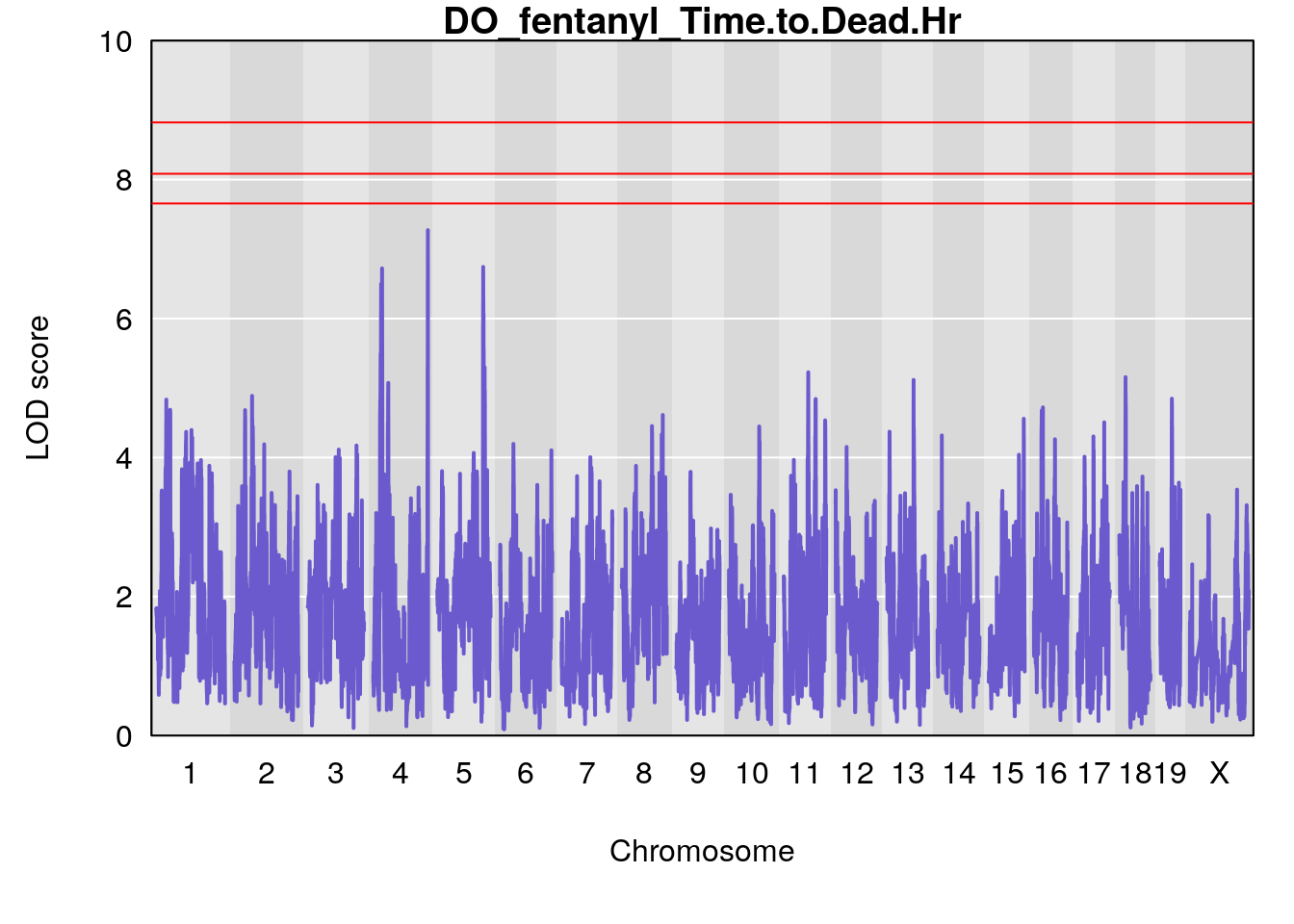
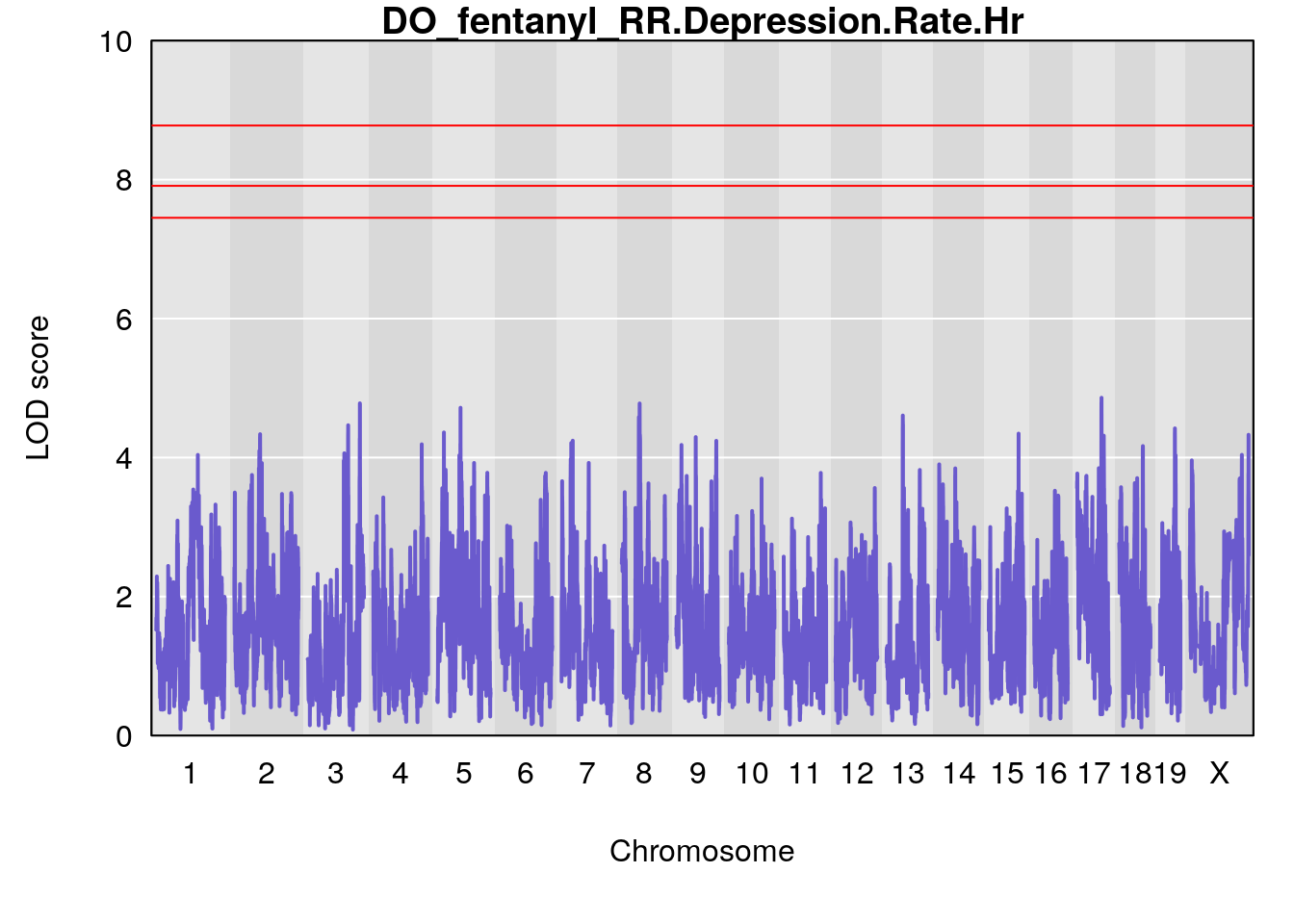
###Plot qtl mapping on array
load("output/Fentanyl/qtl.fentanyl.array.alternate.metrics.out.RData")
#genome-wide plot
for(i in names(qtl.morphine.out)){
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.morphine.out[[i]]) # overall maximum LOD score
plot(qtl.morphine.out[[i]], map=gm$pmap, lodcolumn=1, col="slateblue", ylim=c(0, 10))
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("DO_fentanyl_",i))
}
#save genome-wide plot
for(i in names(qtl.morphine.out)){
png(file = paste0("output/Fentanyl/DO_fentanyl_array_alternate_metrics_", i, ".png"), width = 16, height =8, res=300, units = "in")
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.morphine.out[[i]]) # overall maximum LOD score
plot(qtl.morphine.out[[i]], map=gm$pmap, lodcolumn=1, col="slateblue", ylim=c(0, 10))
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("DO_fentanyl_",i))
dev.off()
}
#peaks coeff plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=gm$pmap, threshold=6, drop=1.5)
print(peaks)
if(dim(peaks)[1] != 0){
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
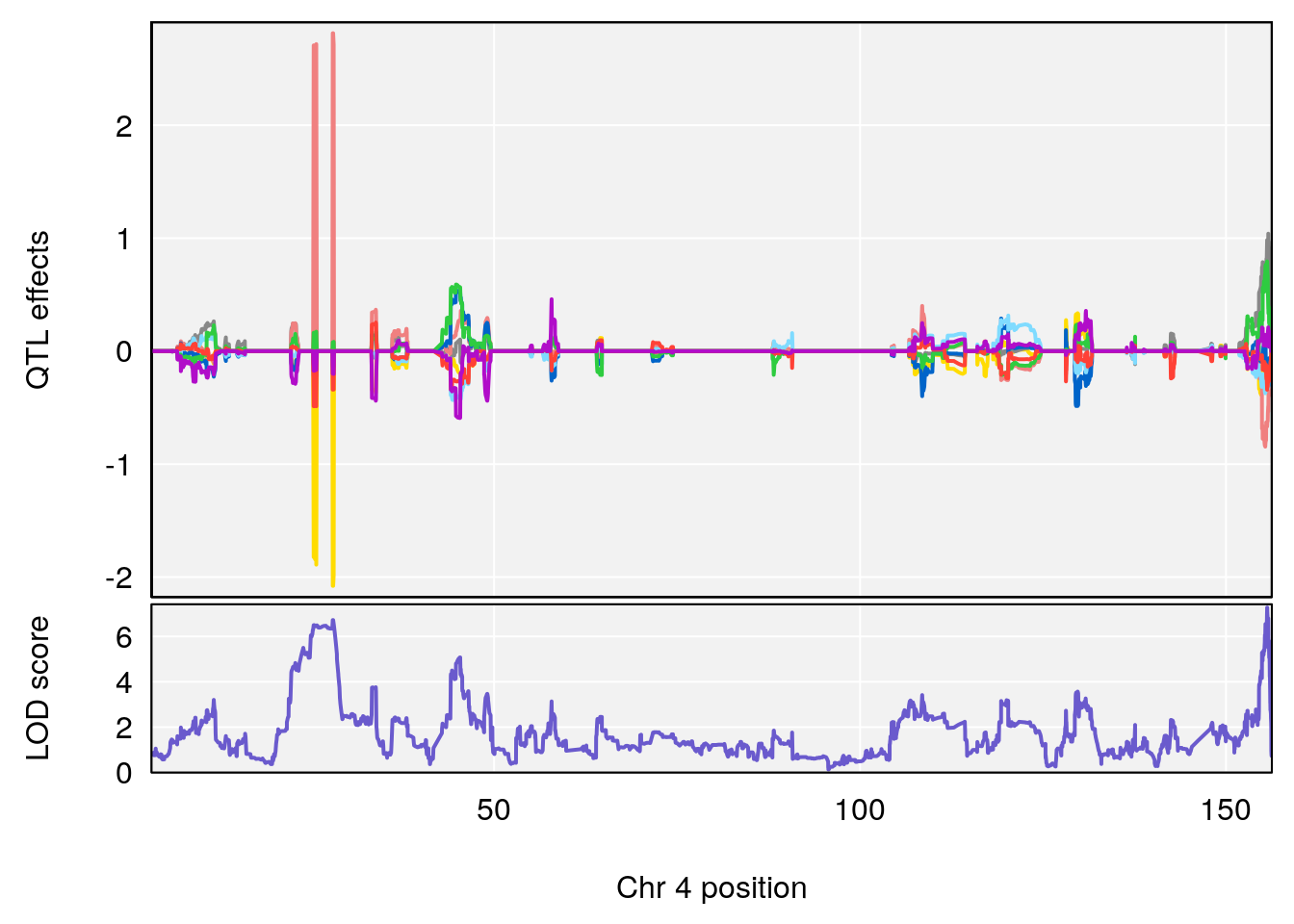
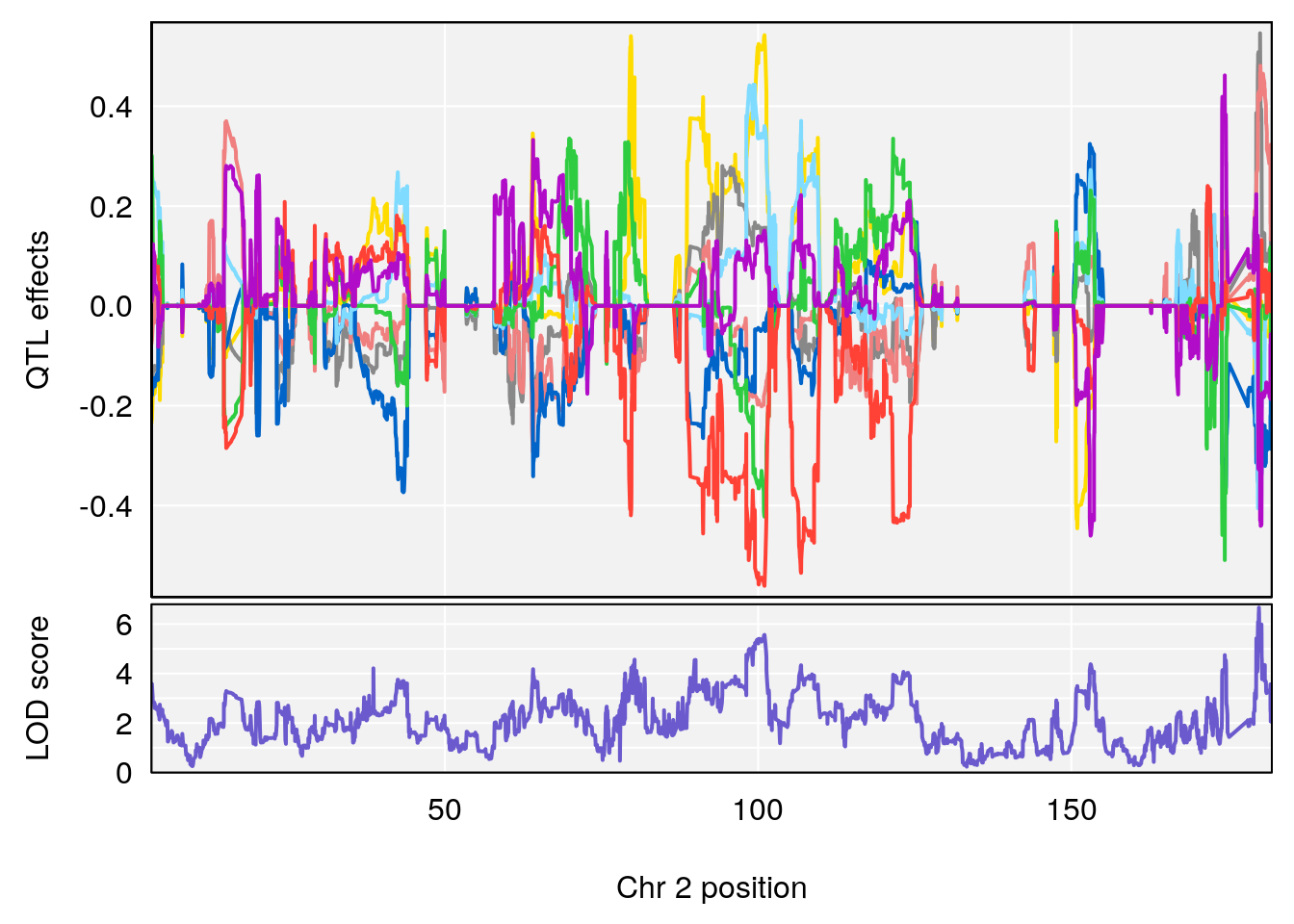
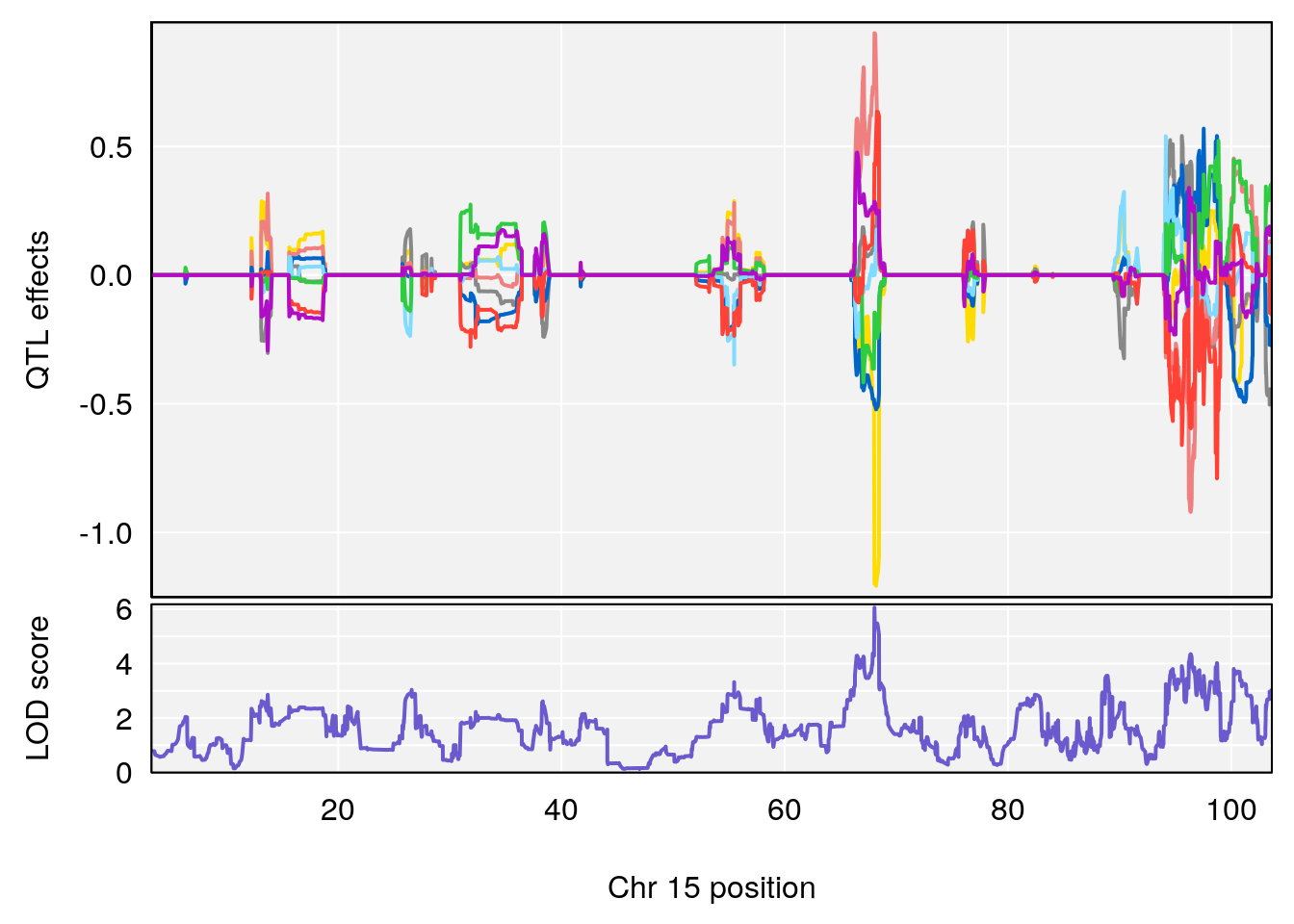
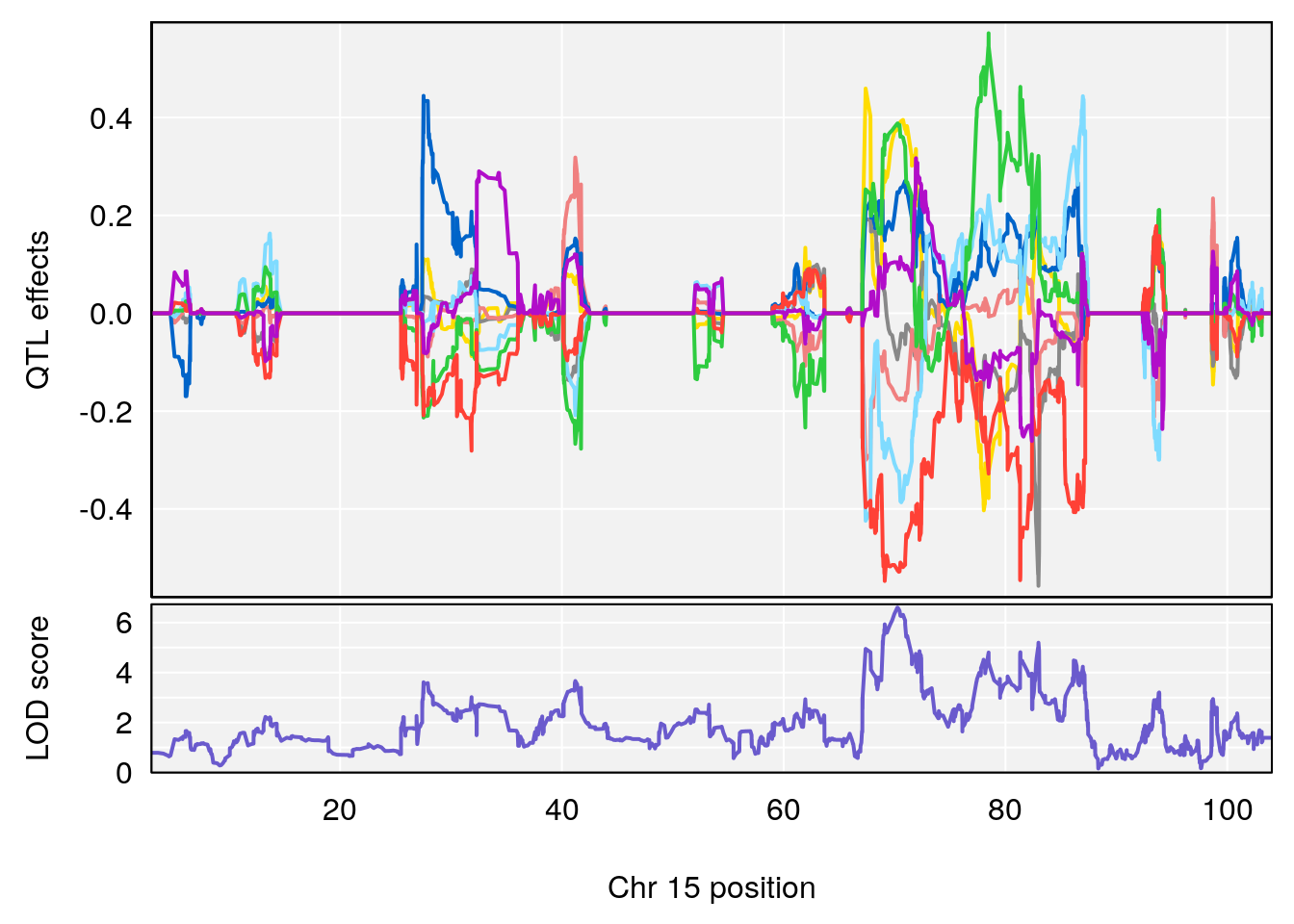
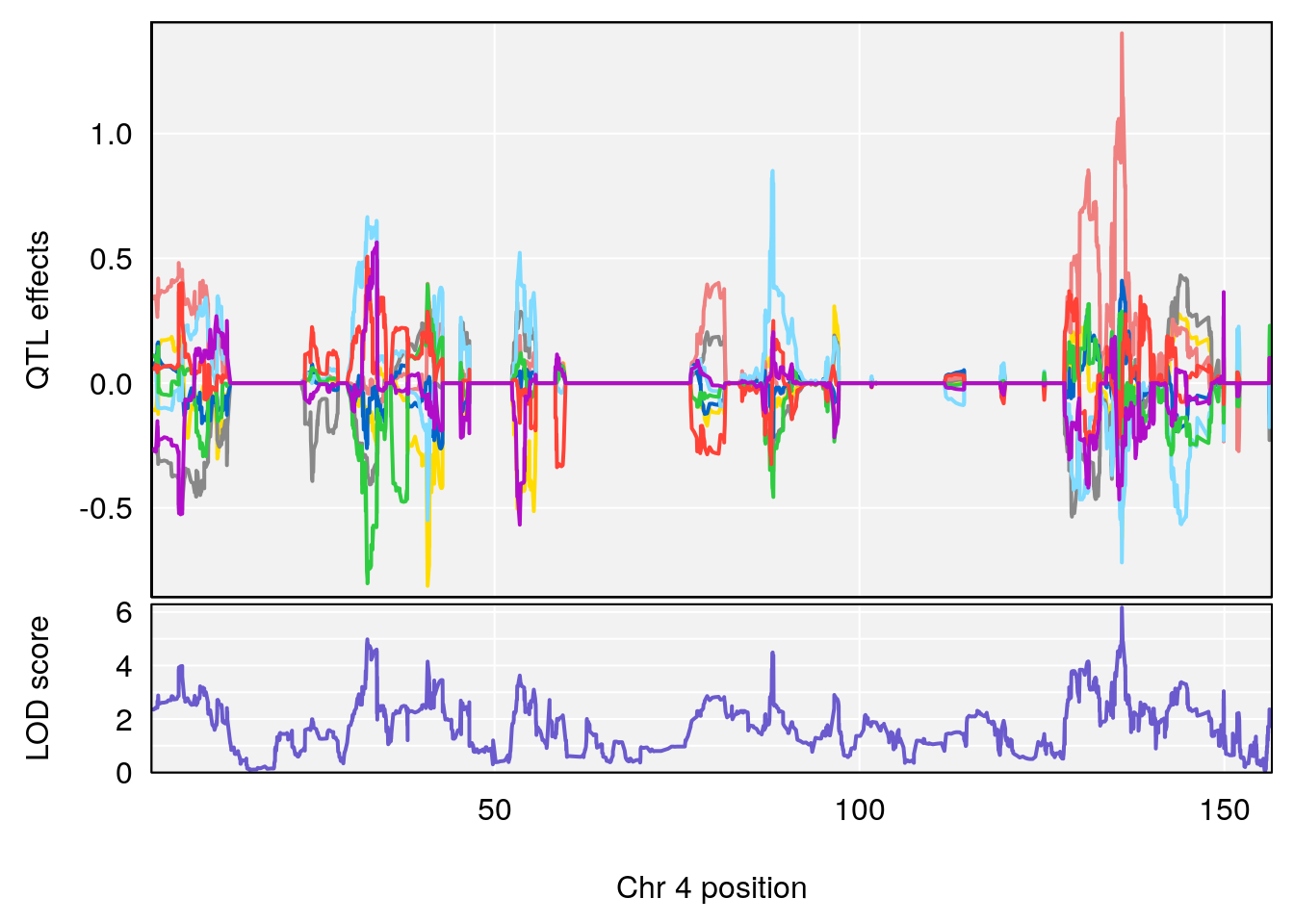
plot_coefCC(coef_c1[[i]][[p]], gm$pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot_coefCC(coef_c2[[i]][[p]], gm$pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
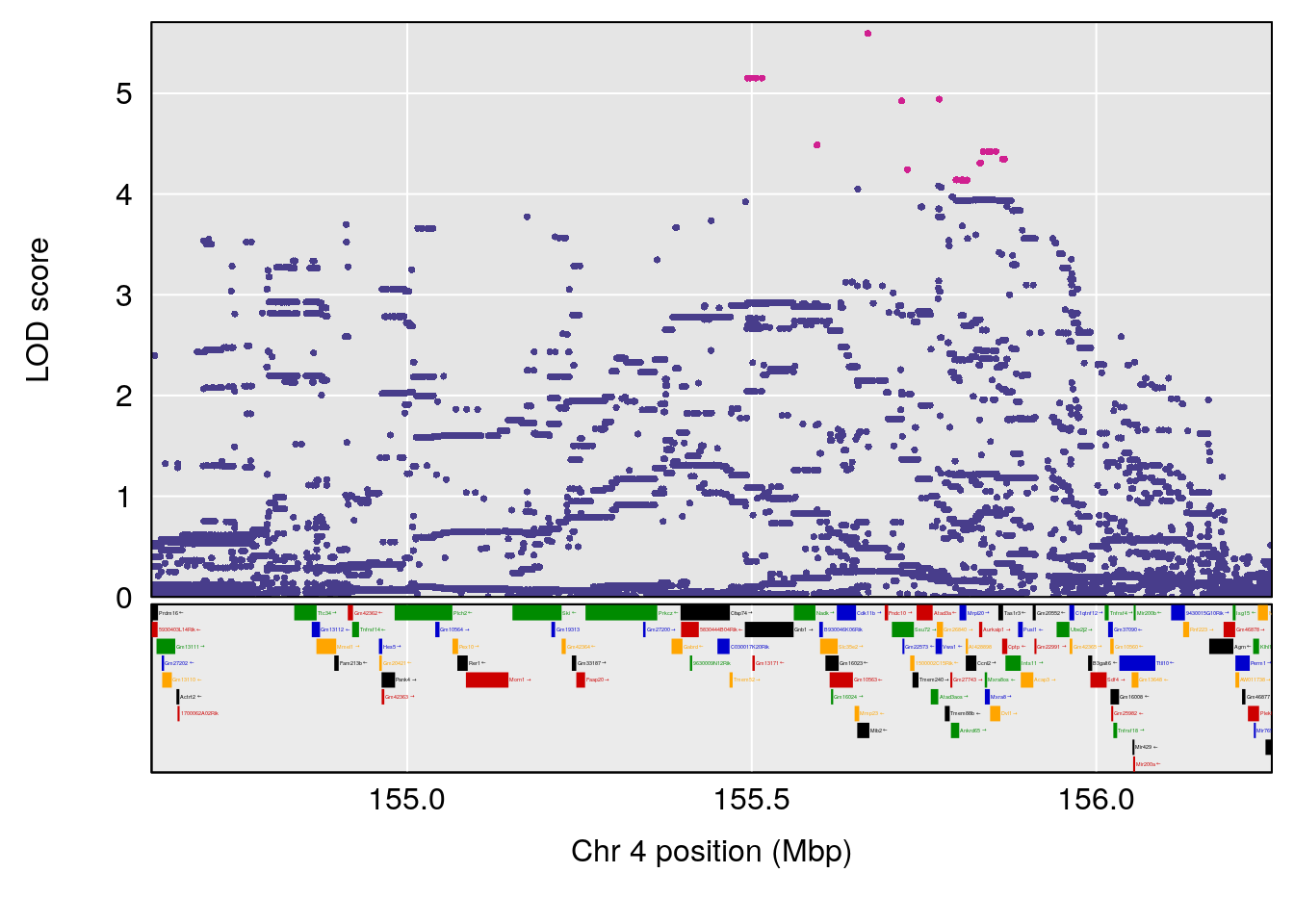
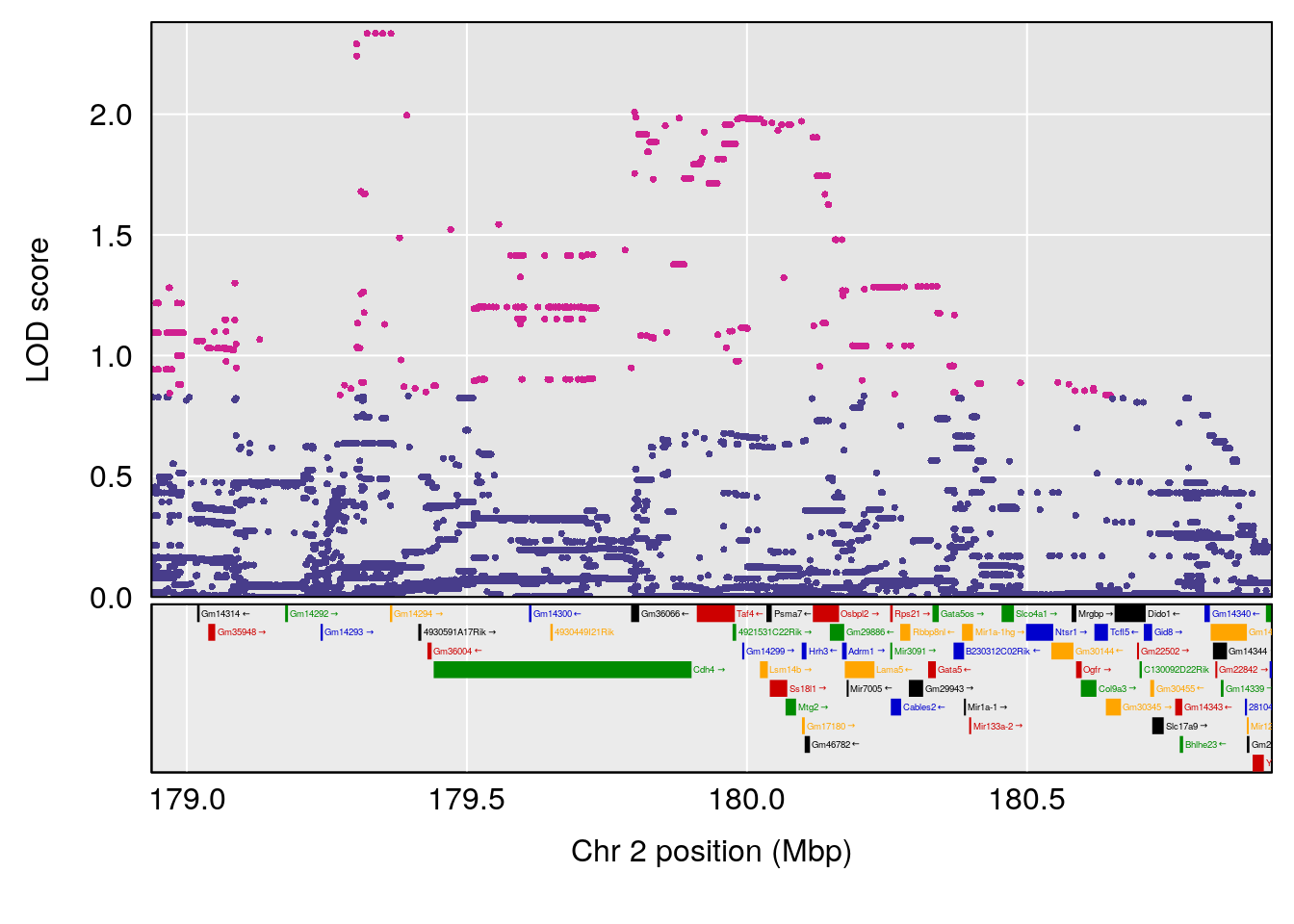
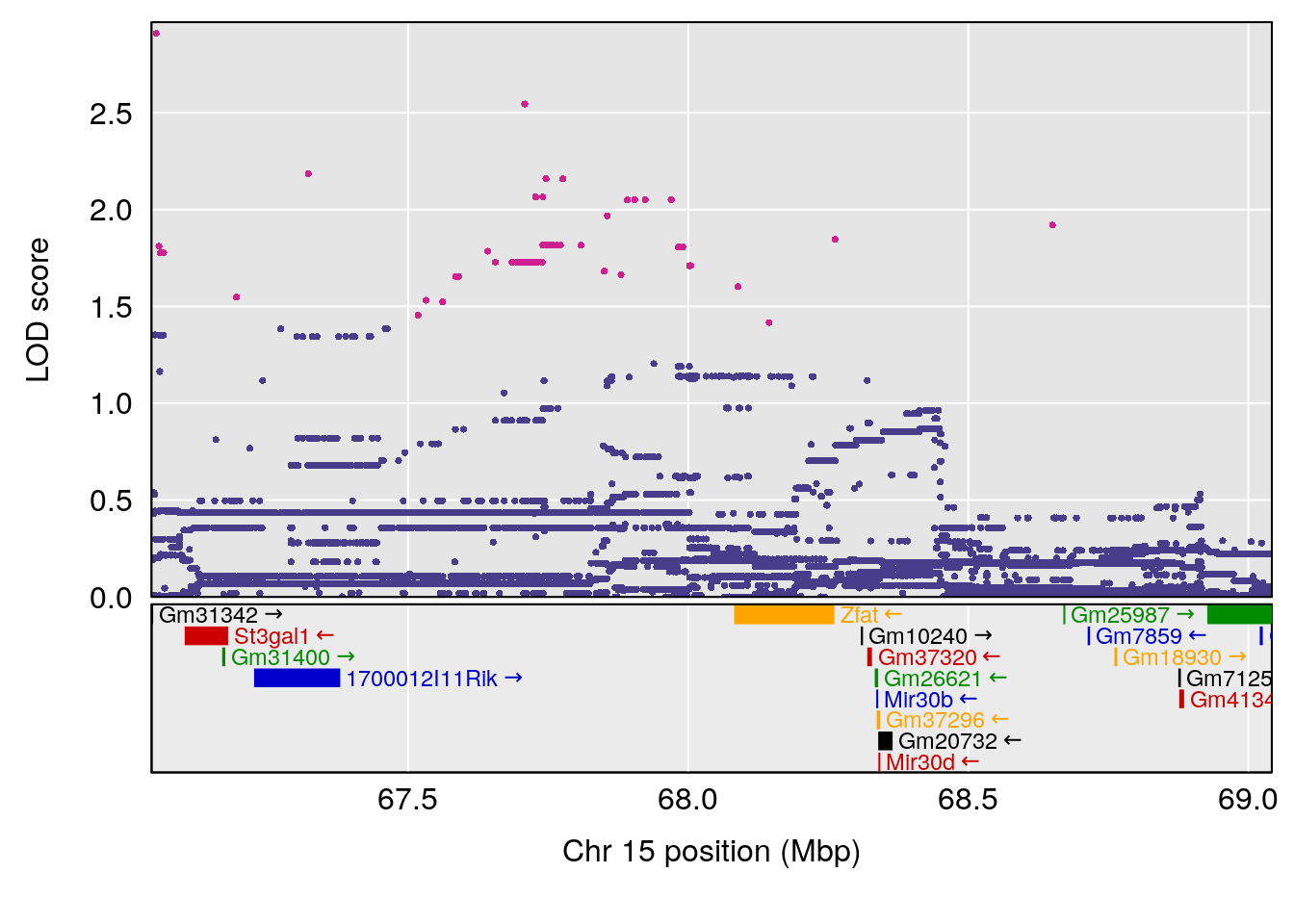
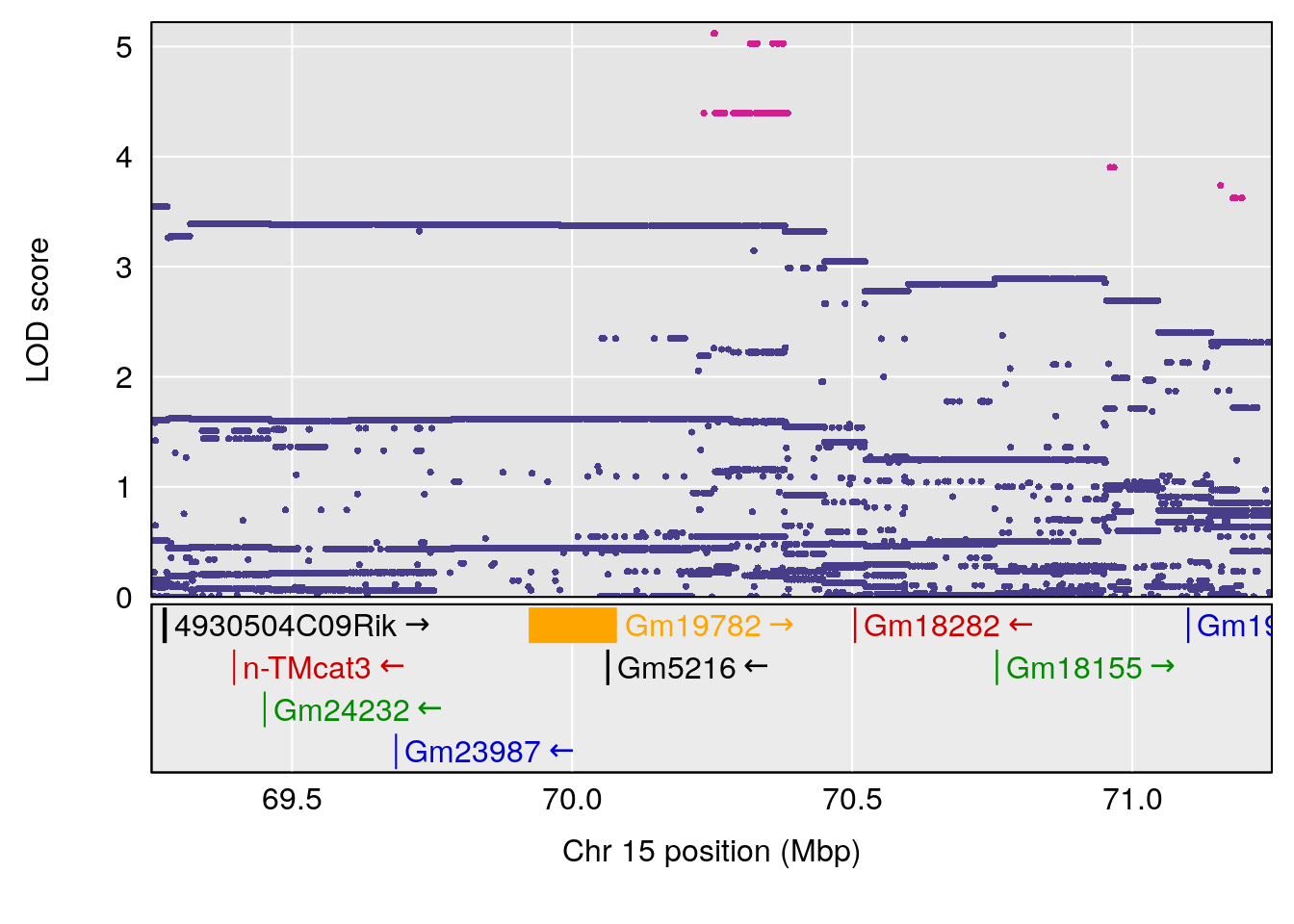
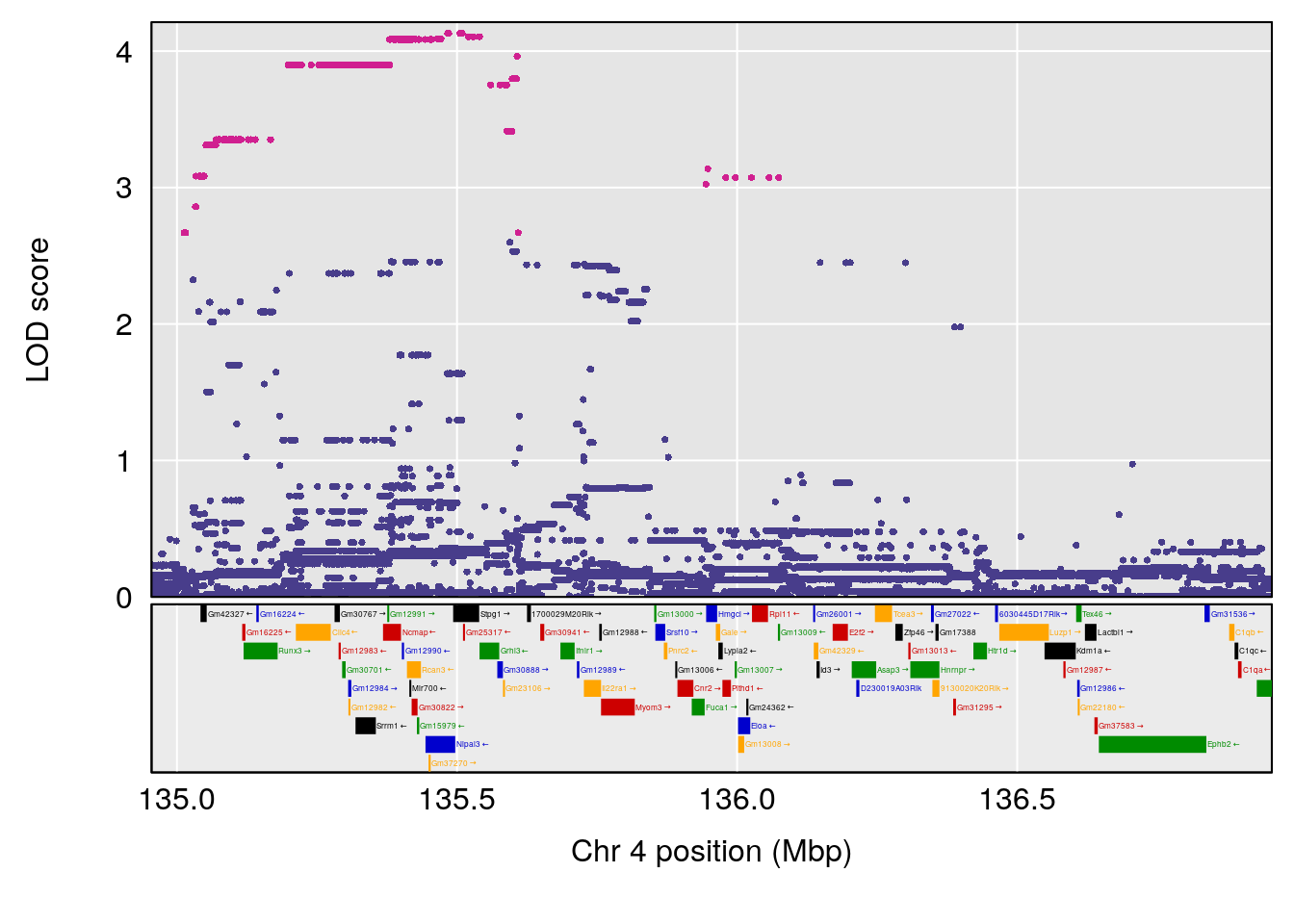
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
}
}
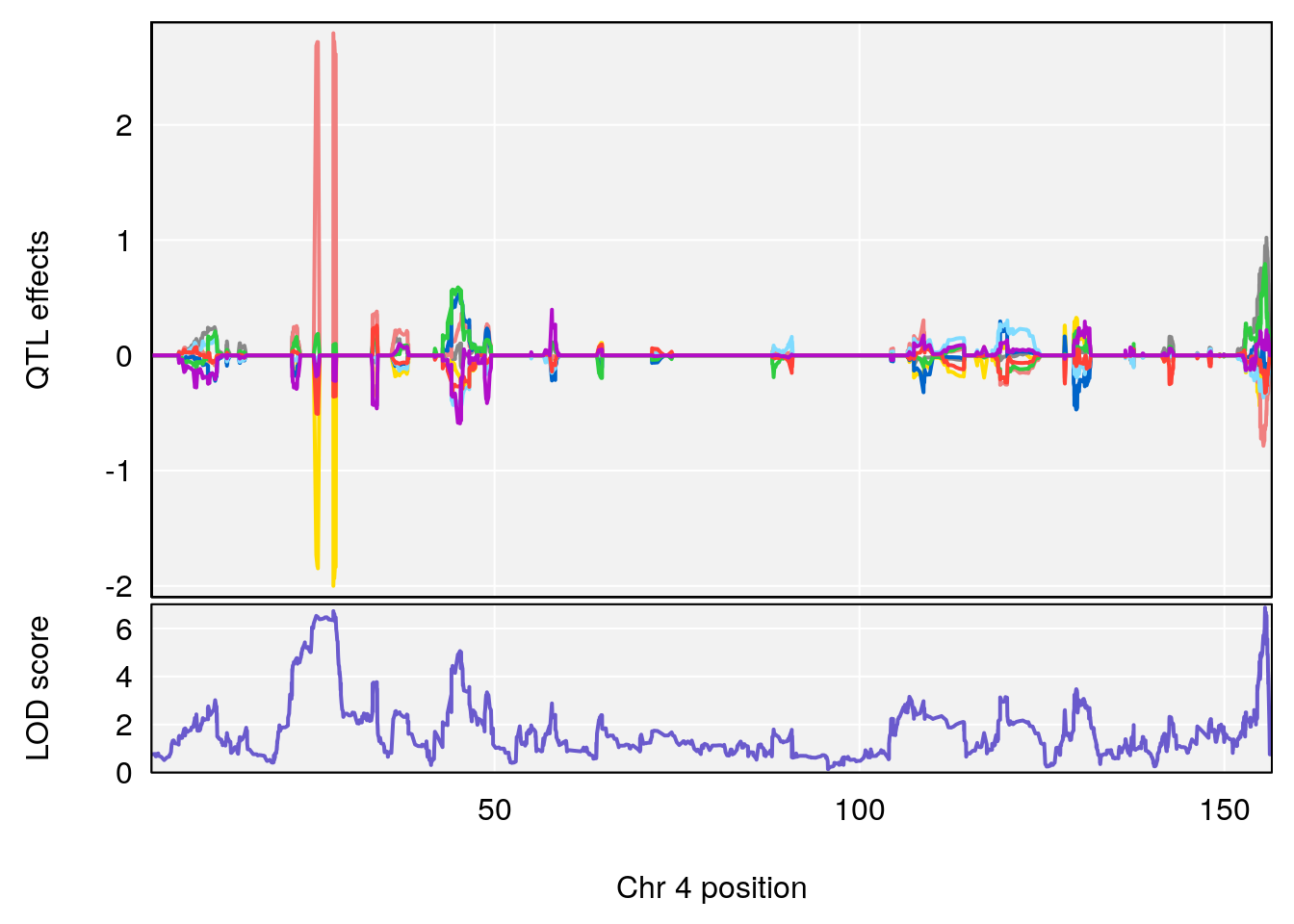
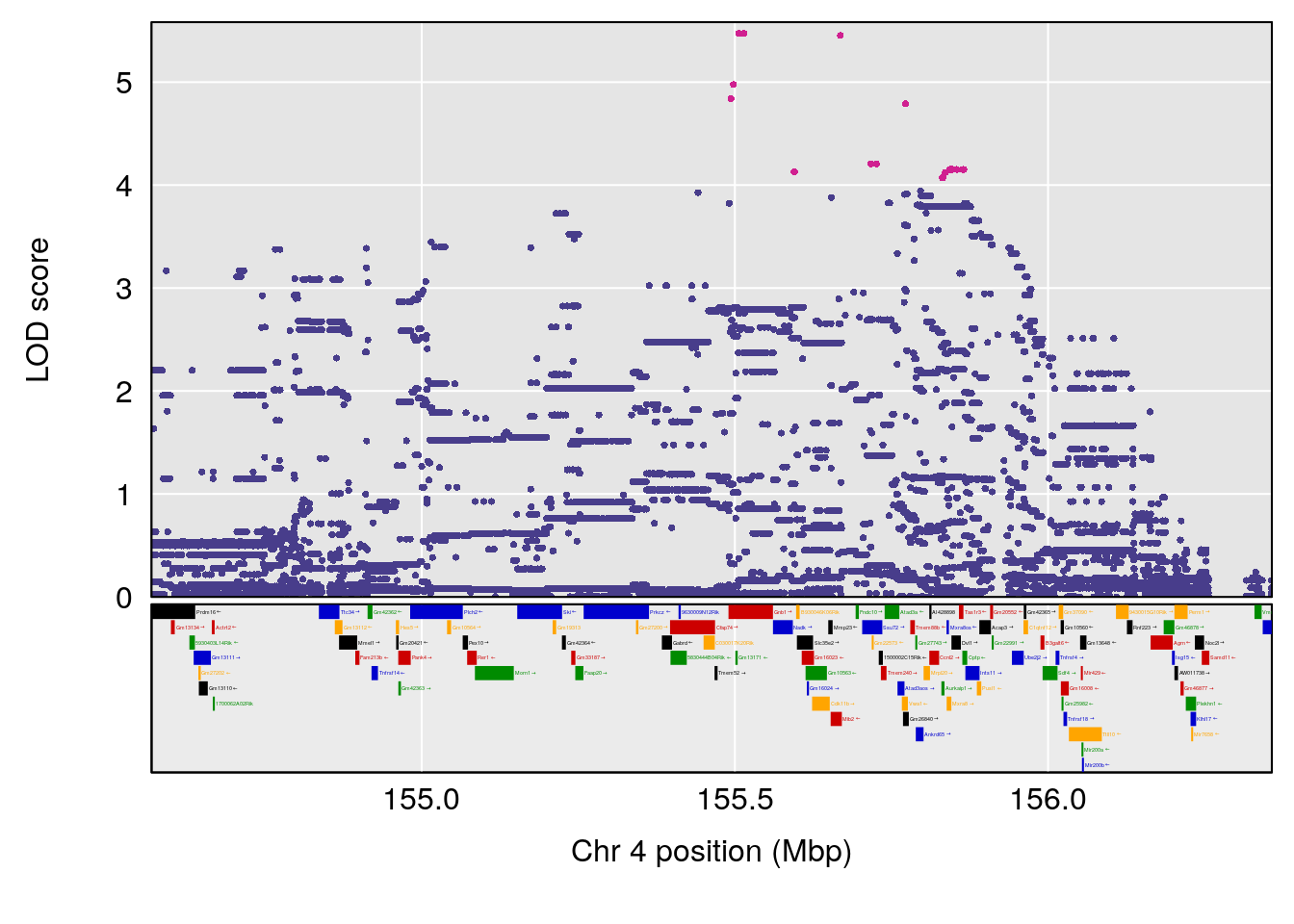
# [1] "Time.to.Dead.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
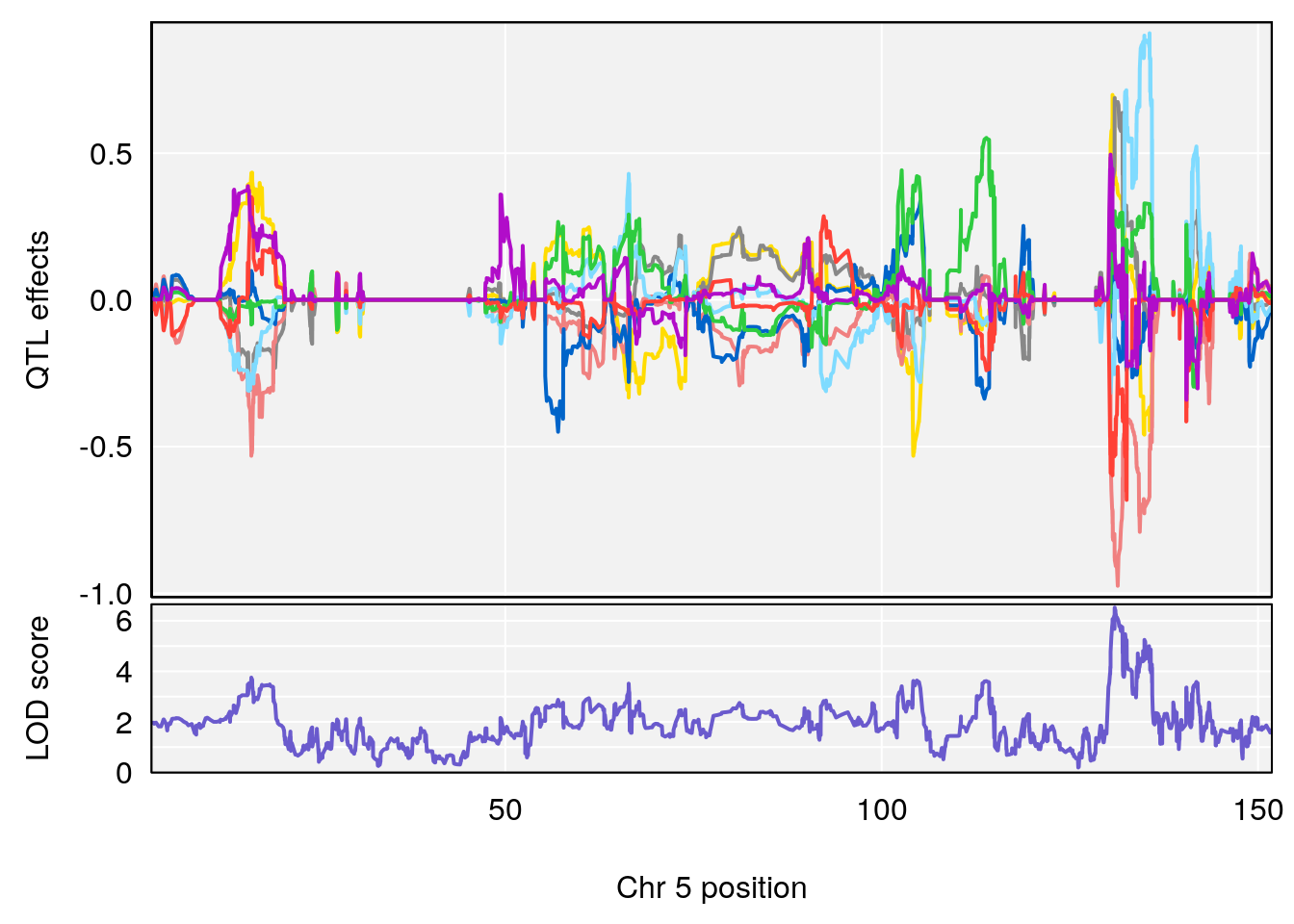
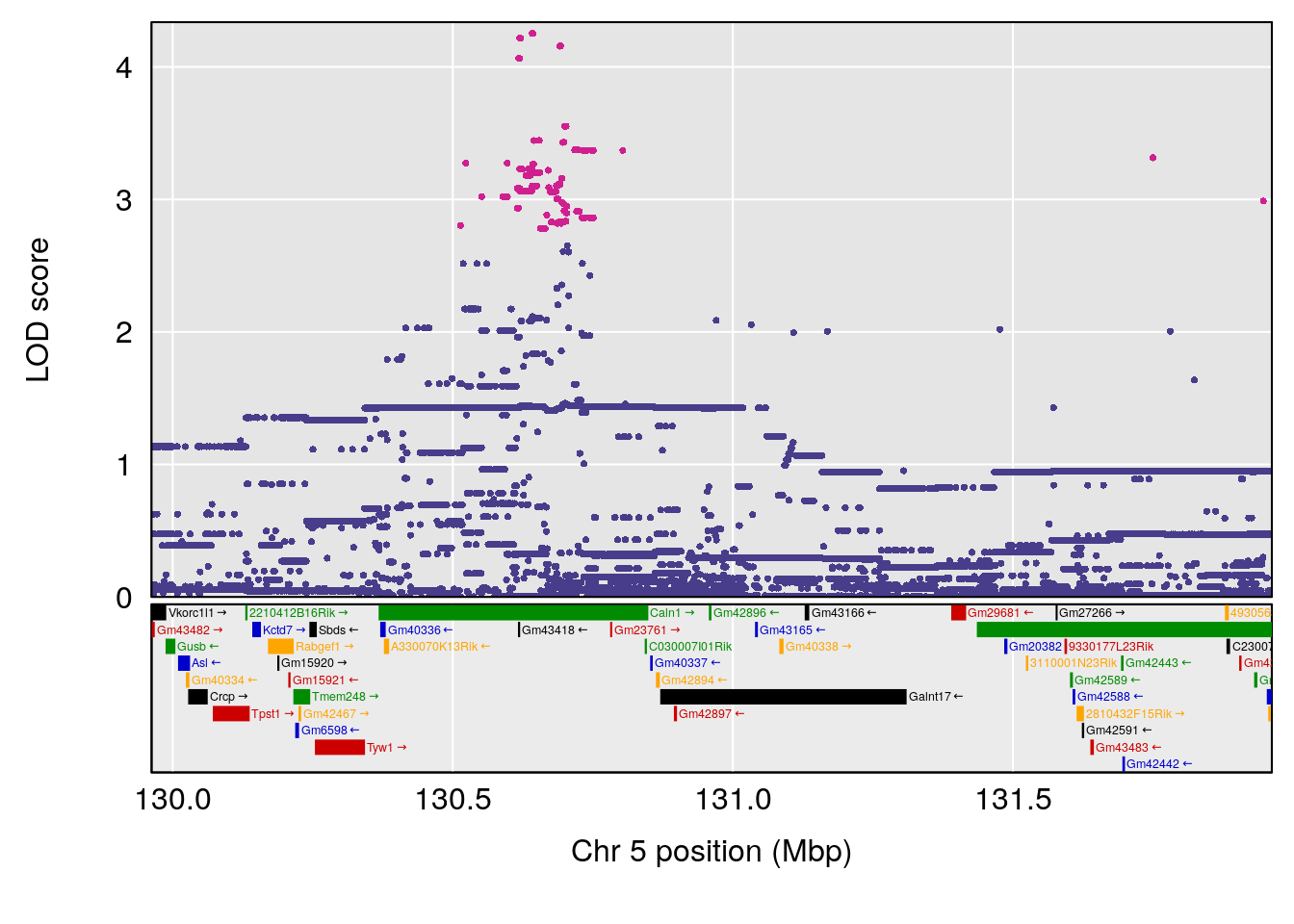
# 1 1 pheno1 4 155.6289 7.273047 24.82559 155.8786
# 2 1 pheno1 5 130.9439 6.743946 130.42272 135.0373
# [1] 1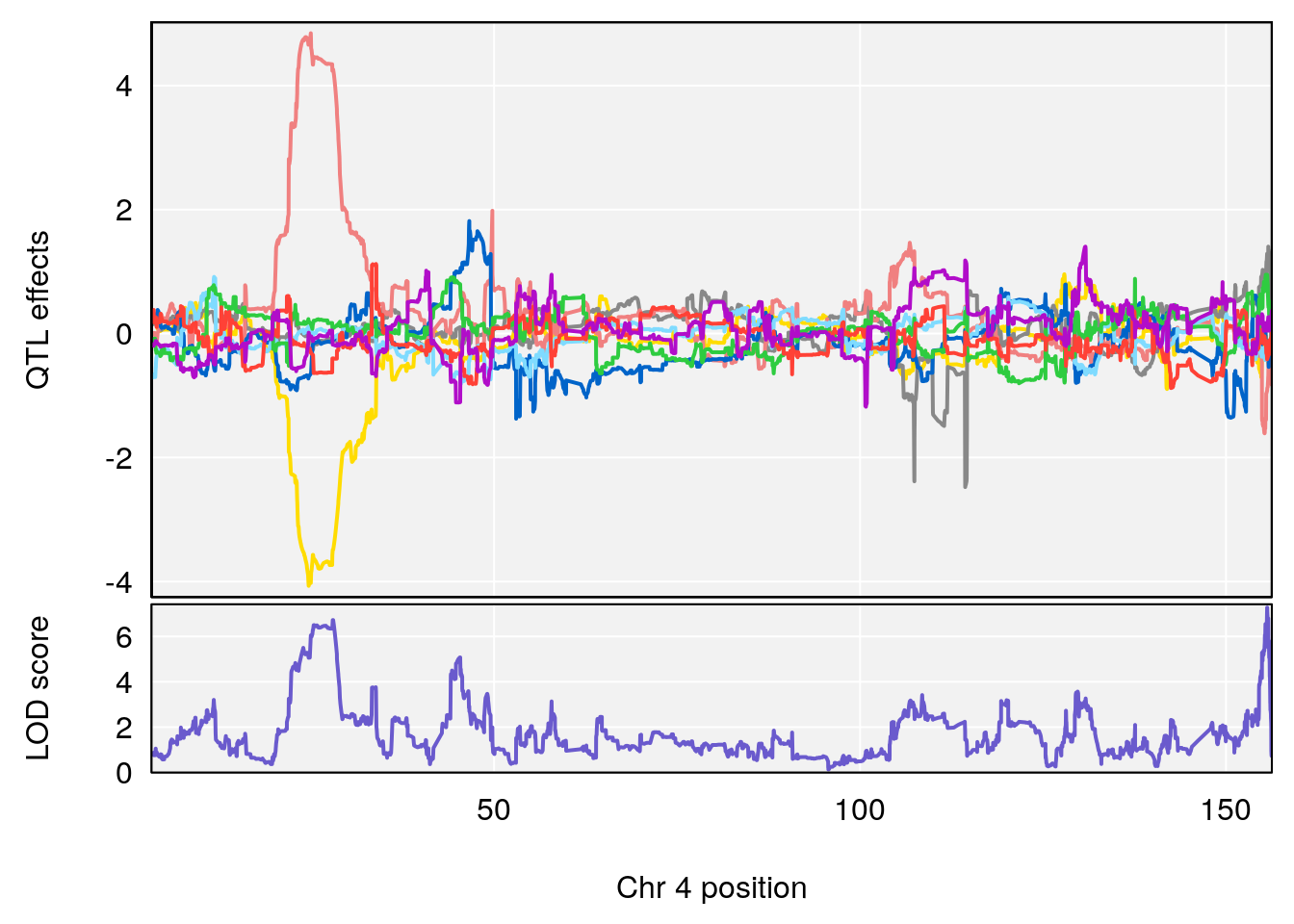
# [1] 2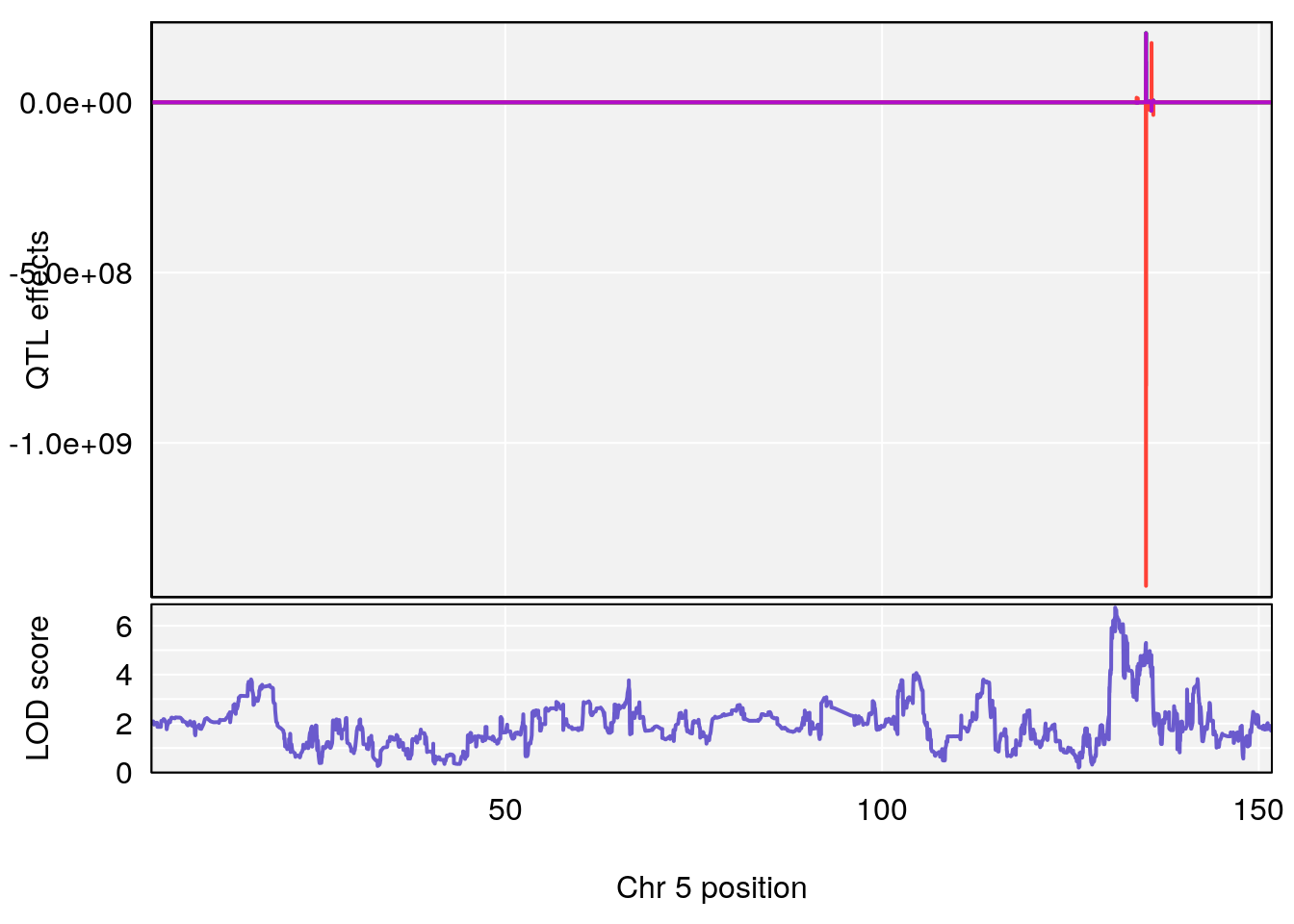
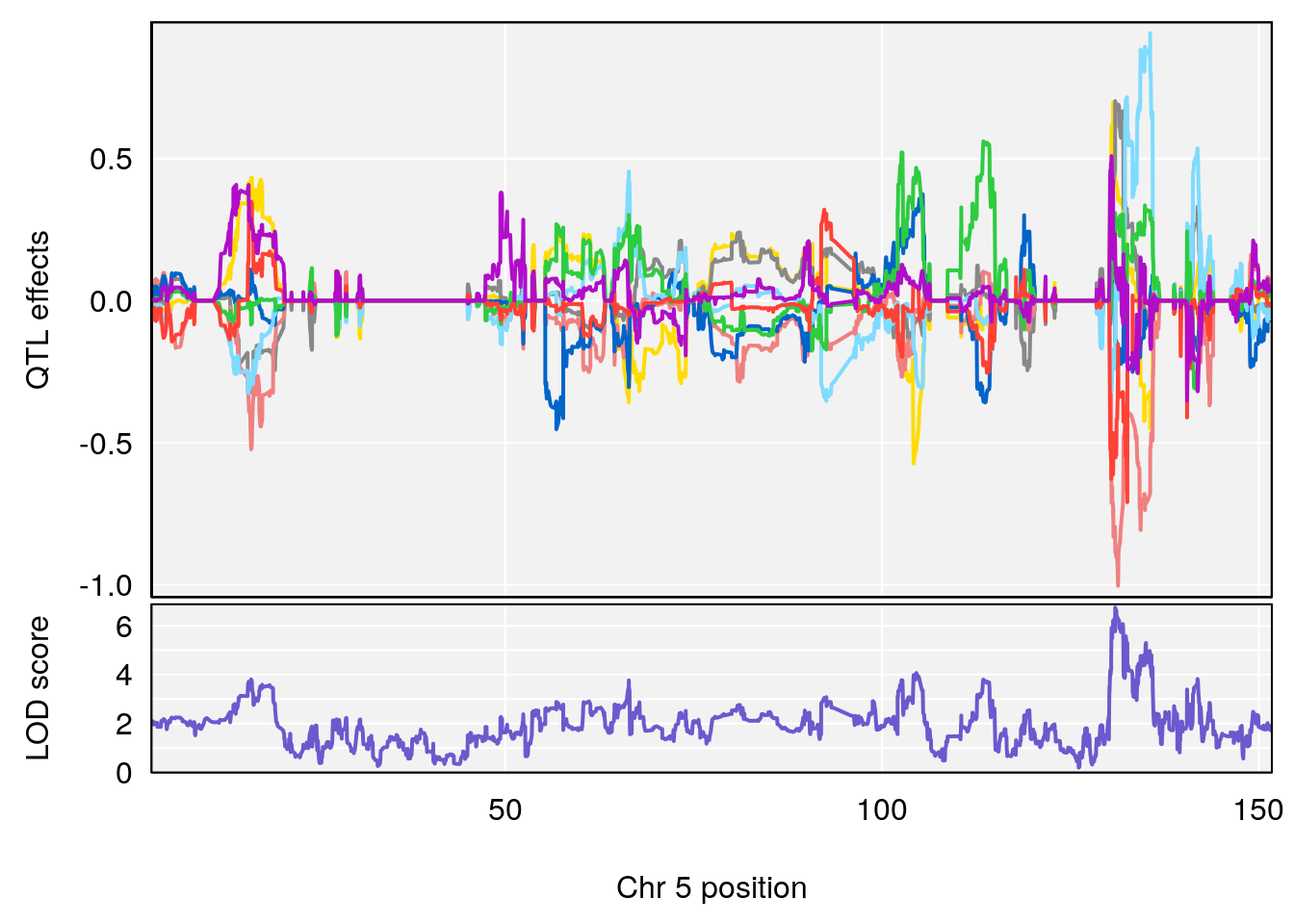
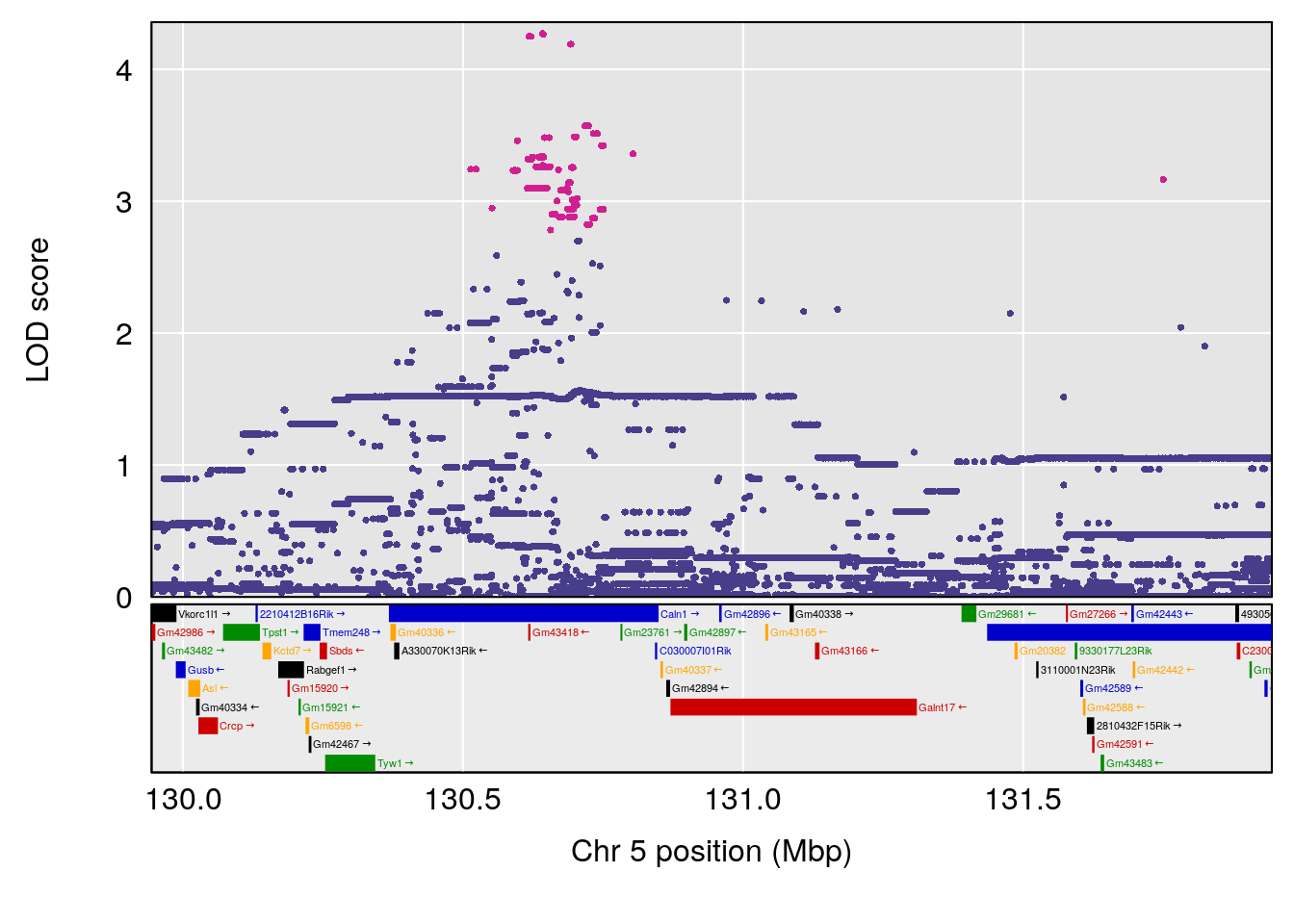
# [1] "RR.Depression.Rate.Hr"
# [1] lodindex lodcolumn chr pos lod ci_lo ci_hi
# <0 rows> (or 0-length row.names)
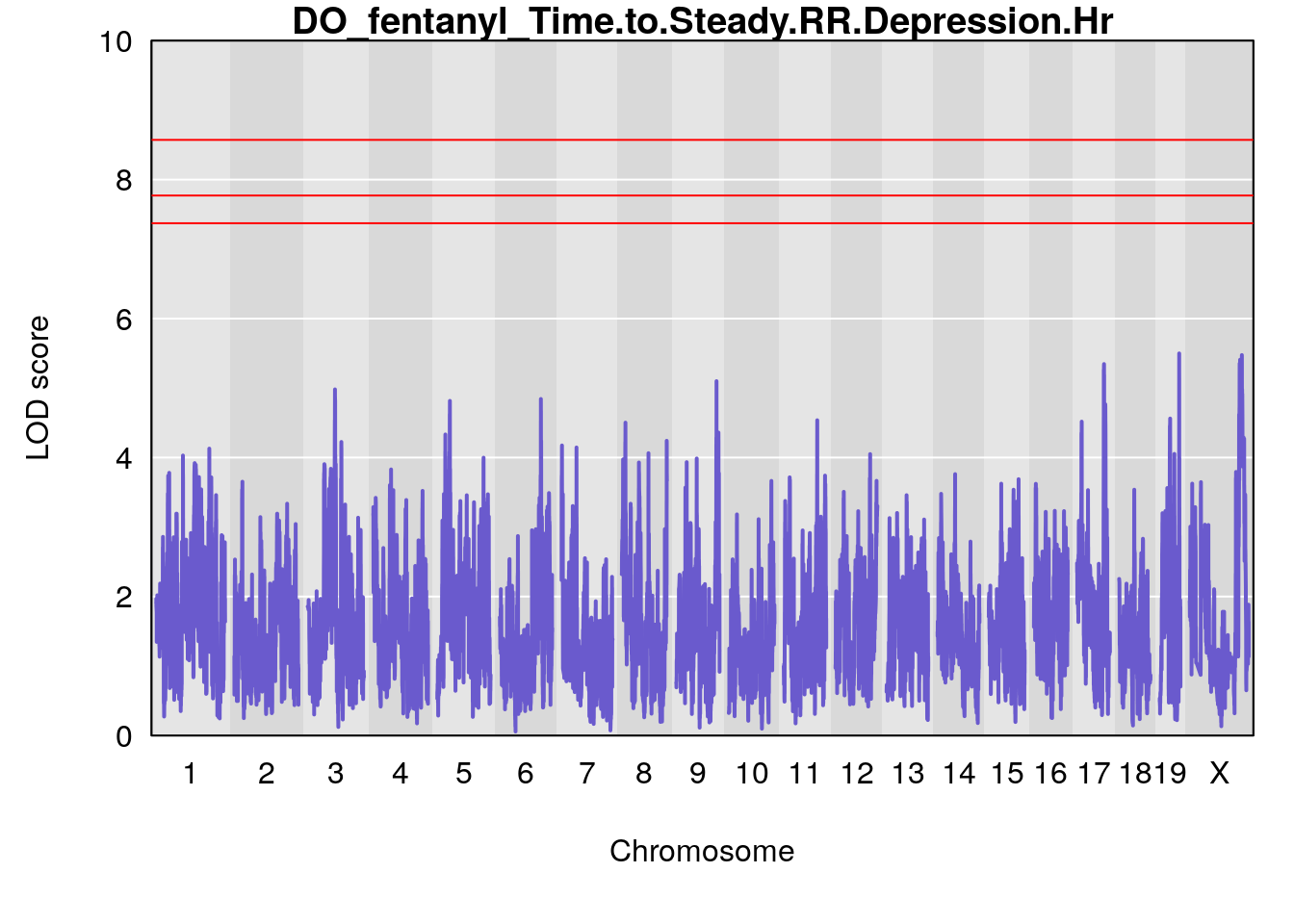
# [1] "Time.to.Steady.RR.Depression.Hr"
# [1] lodindex lodcolumn chr pos lod ci_lo ci_hi
# <0 rows> (or 0-length row.names)
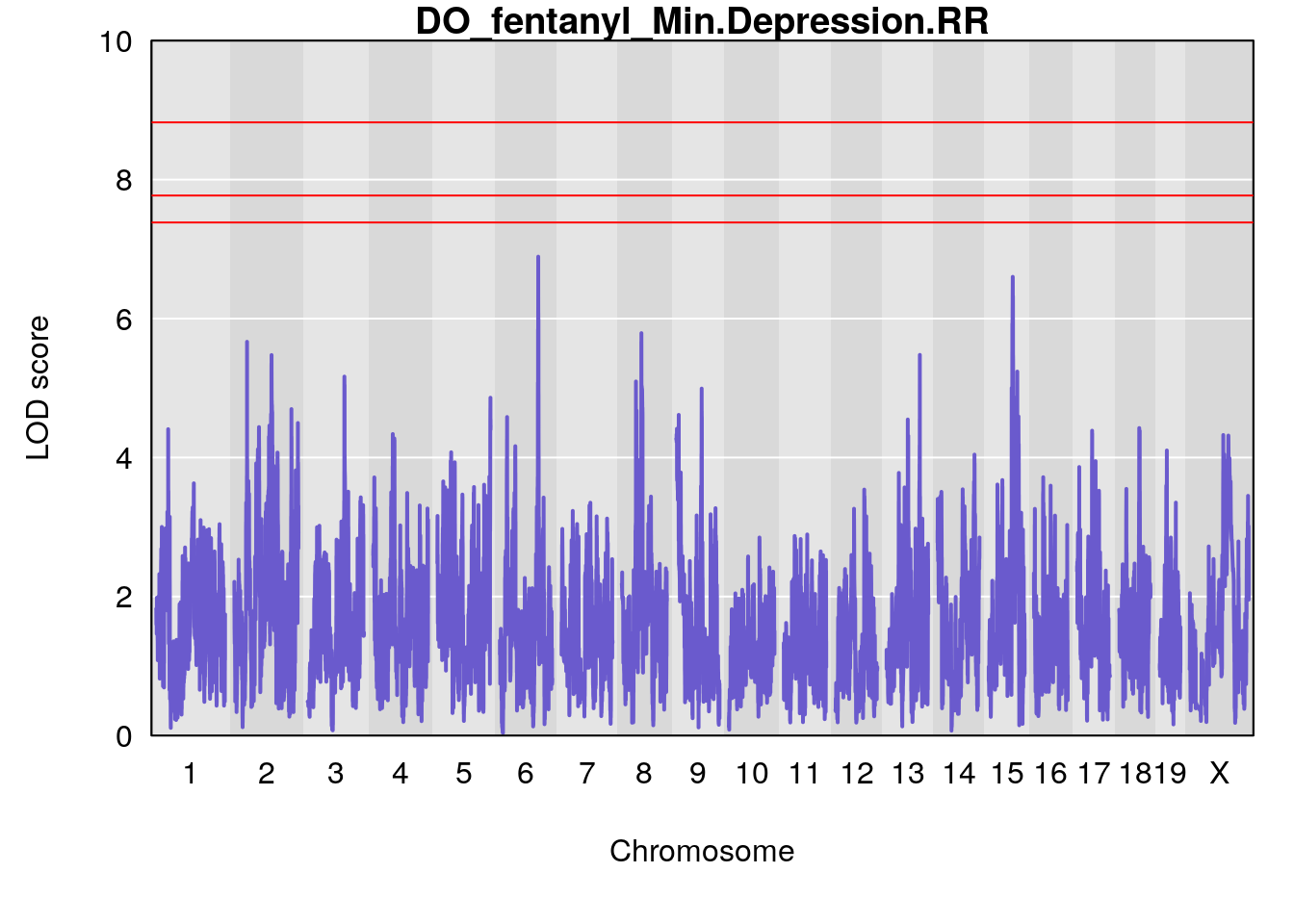
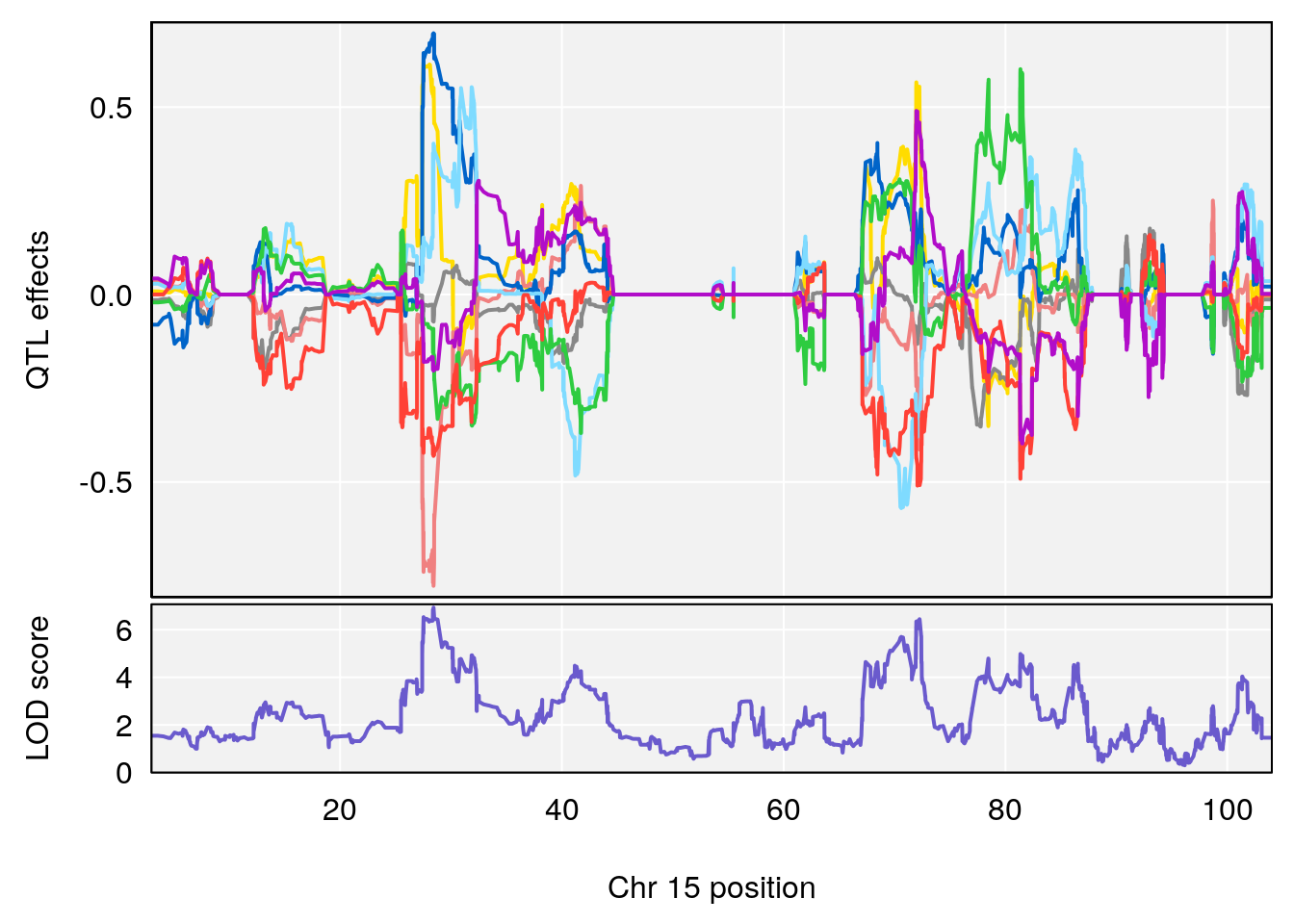
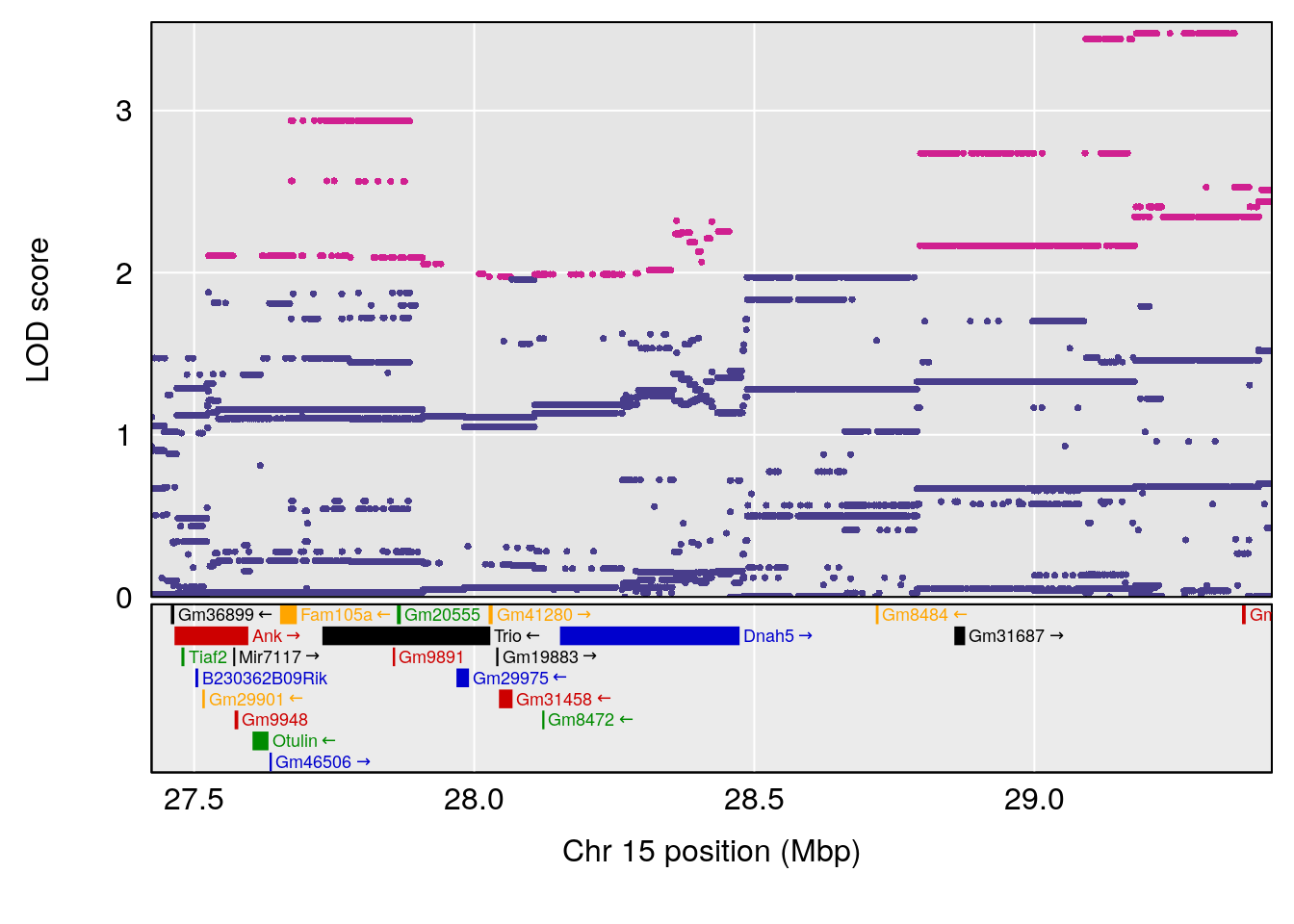
# [1] "Min.Depression.RR"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
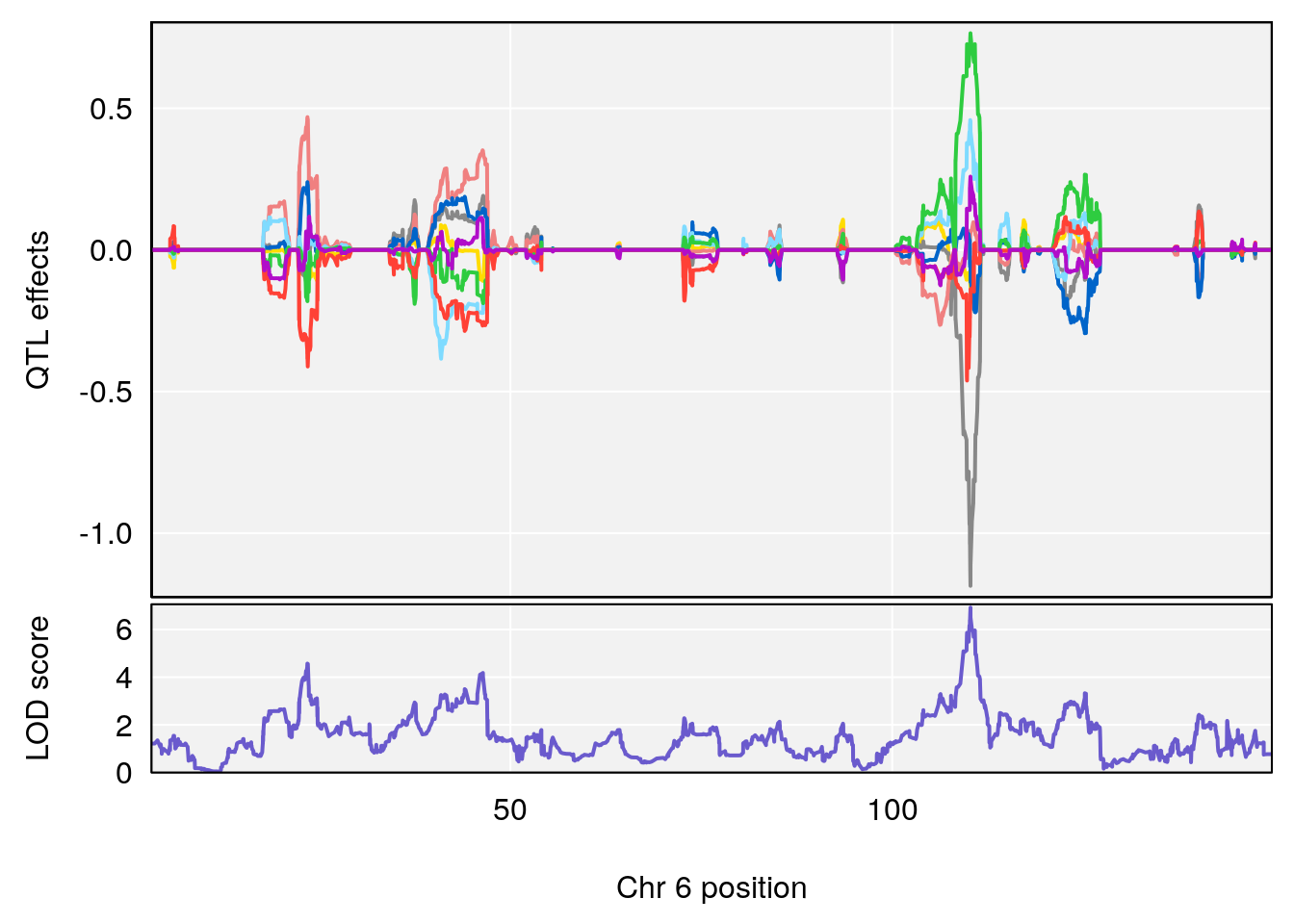
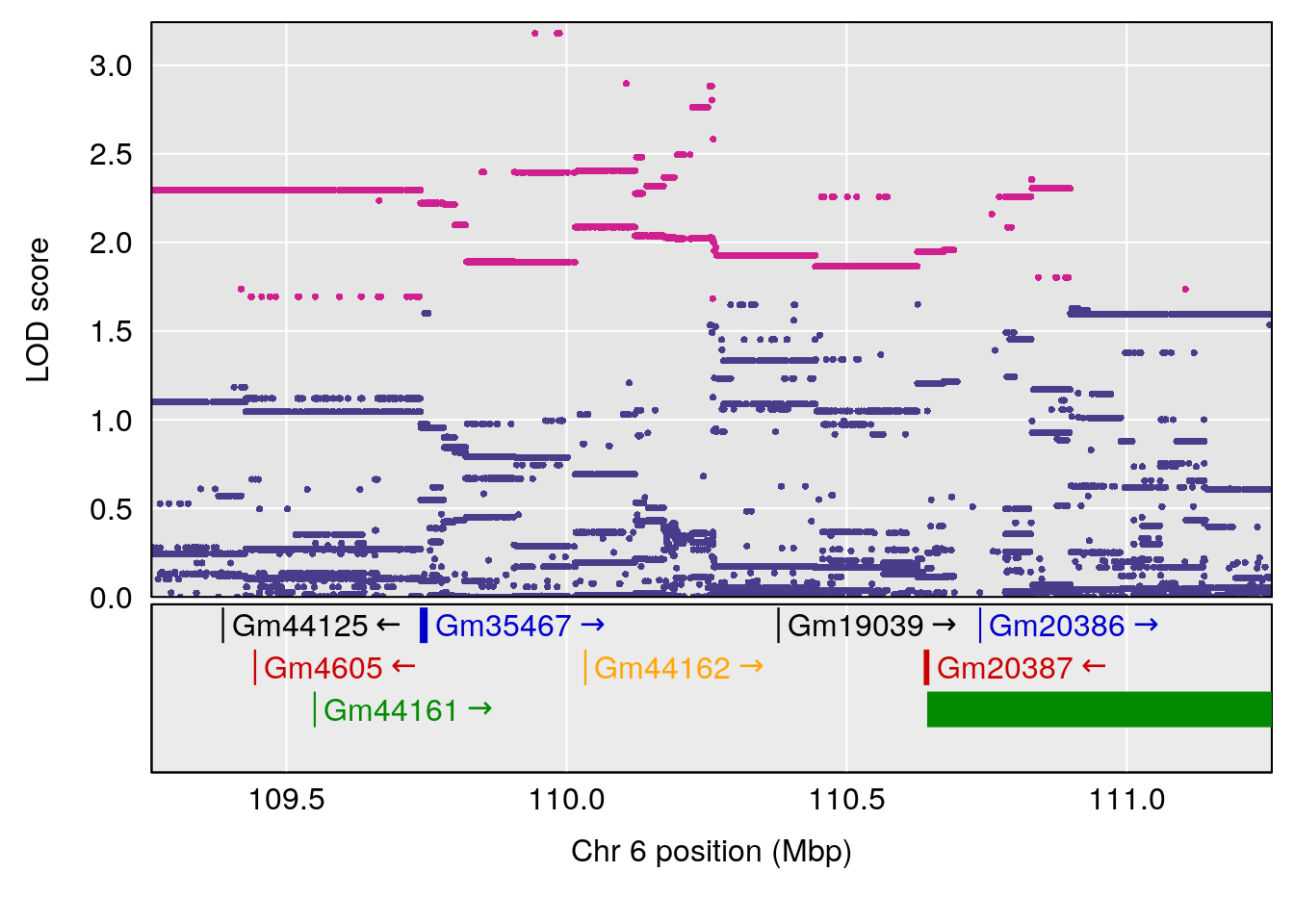
# 1 1 pheno1 6 110.23309 6.891327 109.74468 110.89903
# 2 1 pheno1 15 70.26574 6.600987 68.92851 82.99412
# [1] 1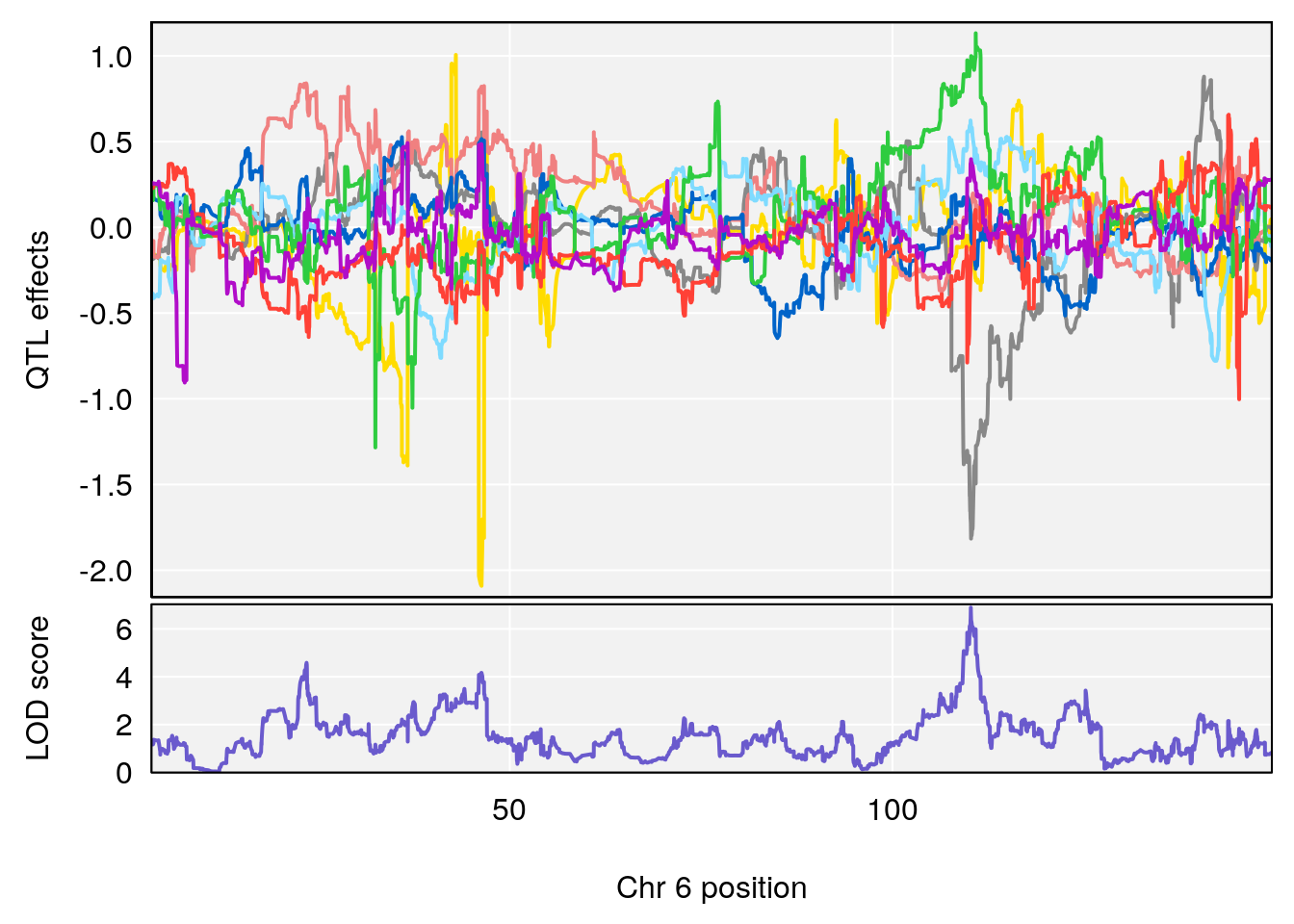
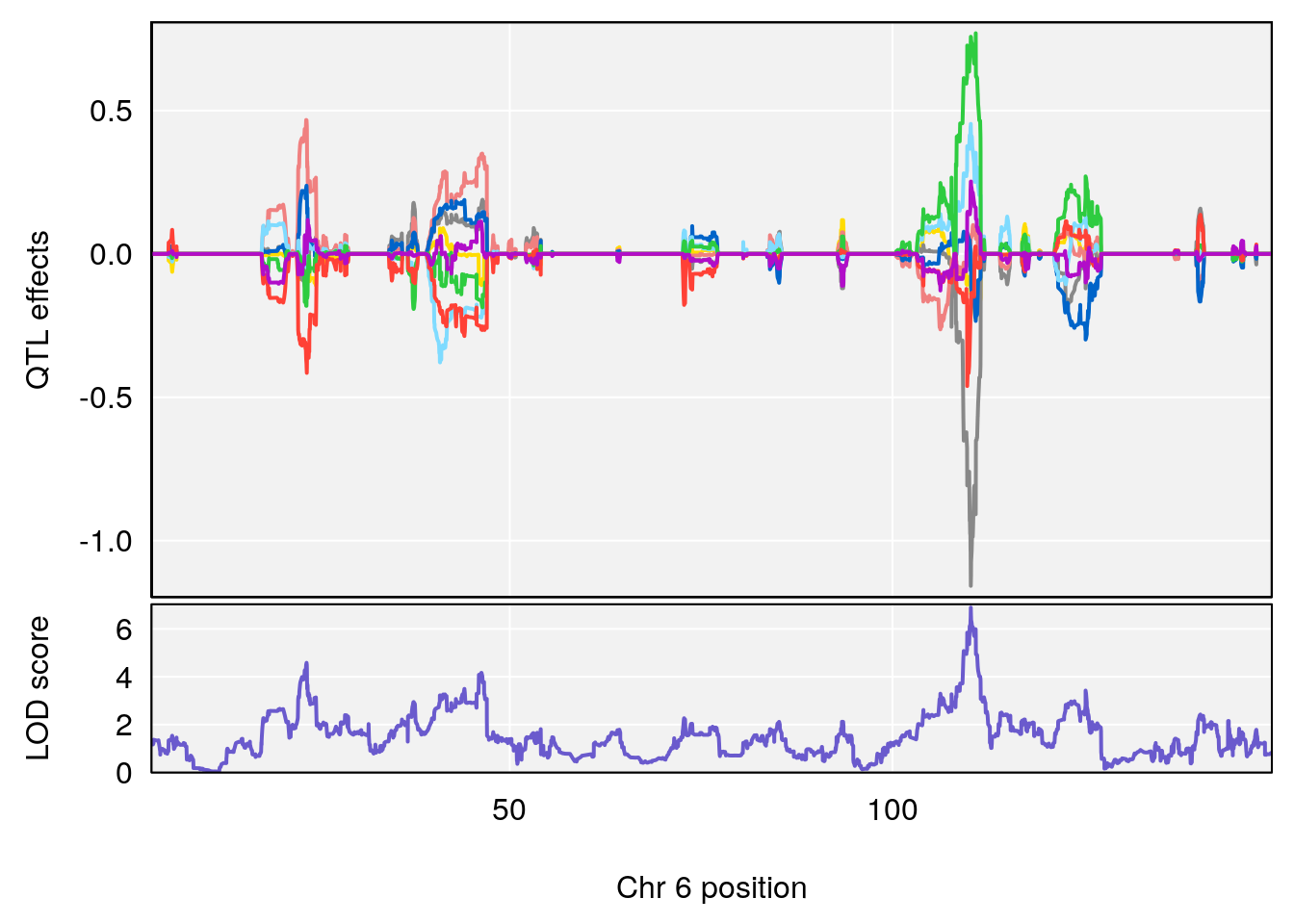
# [1] 2
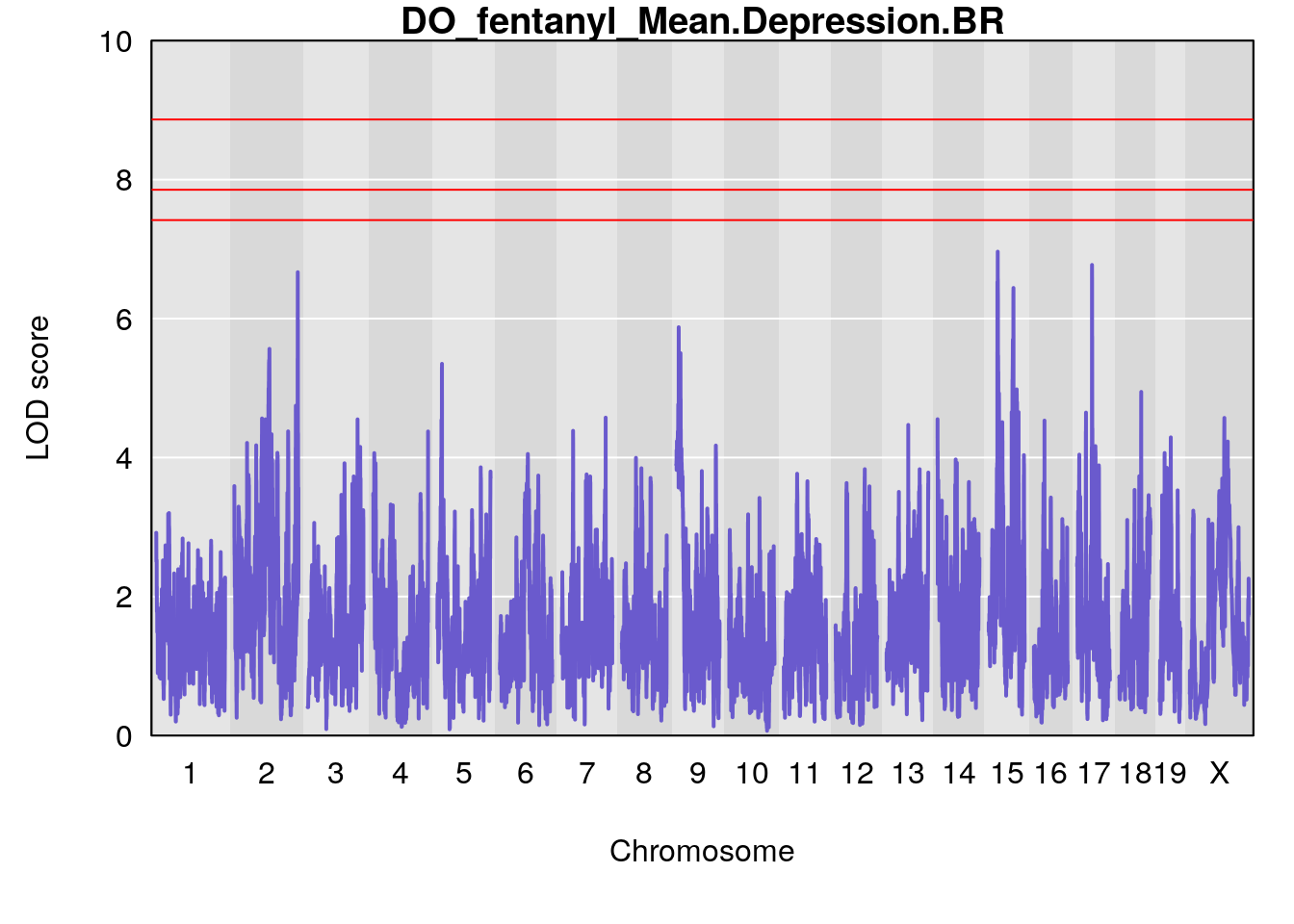
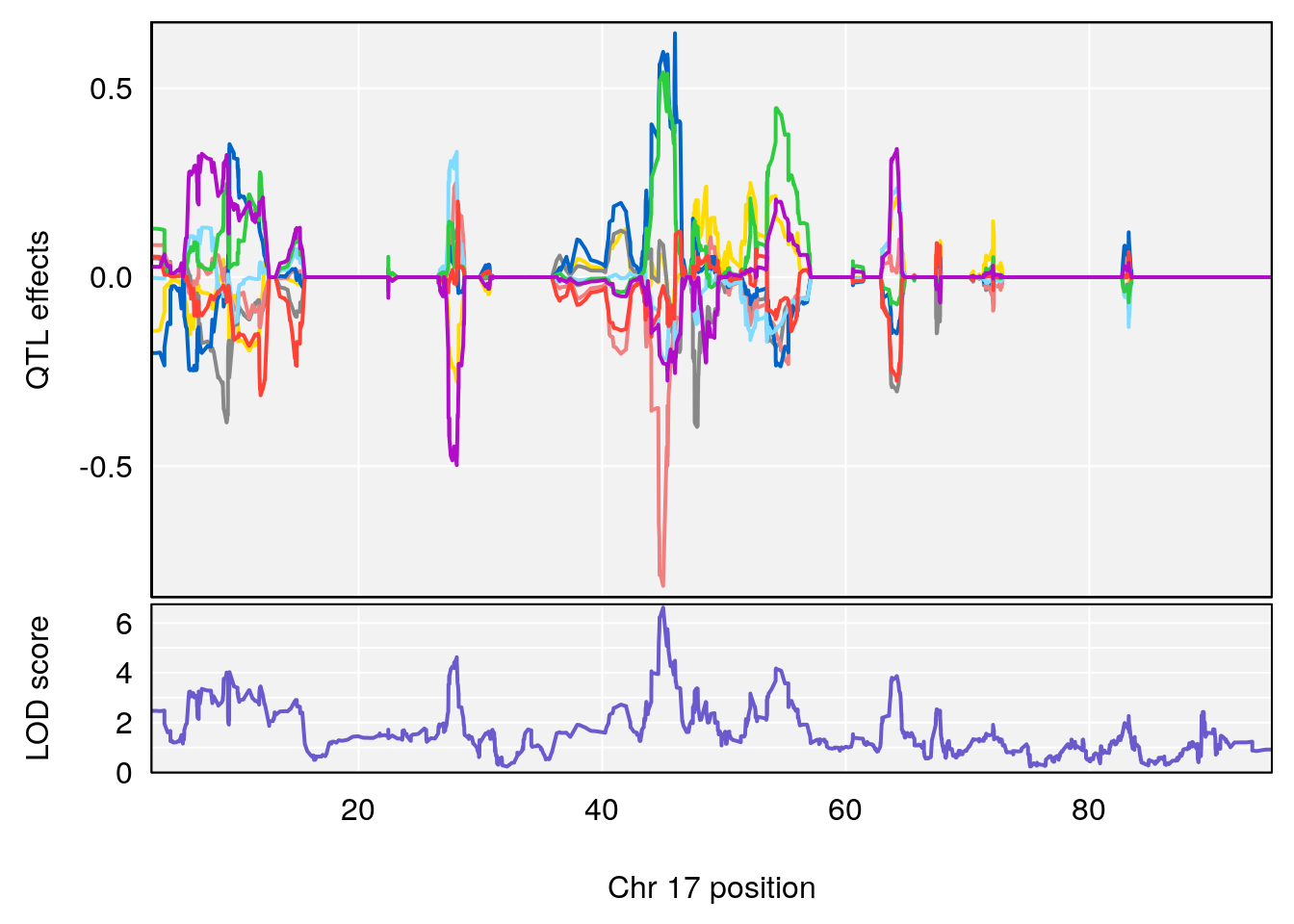
# [1] "Mean.Depression.BR"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 2 179.93697 6.667862 99.48509 180.36898
# 2 1 pheno1 15 28.43474 6.963138 27.40518 72.41165
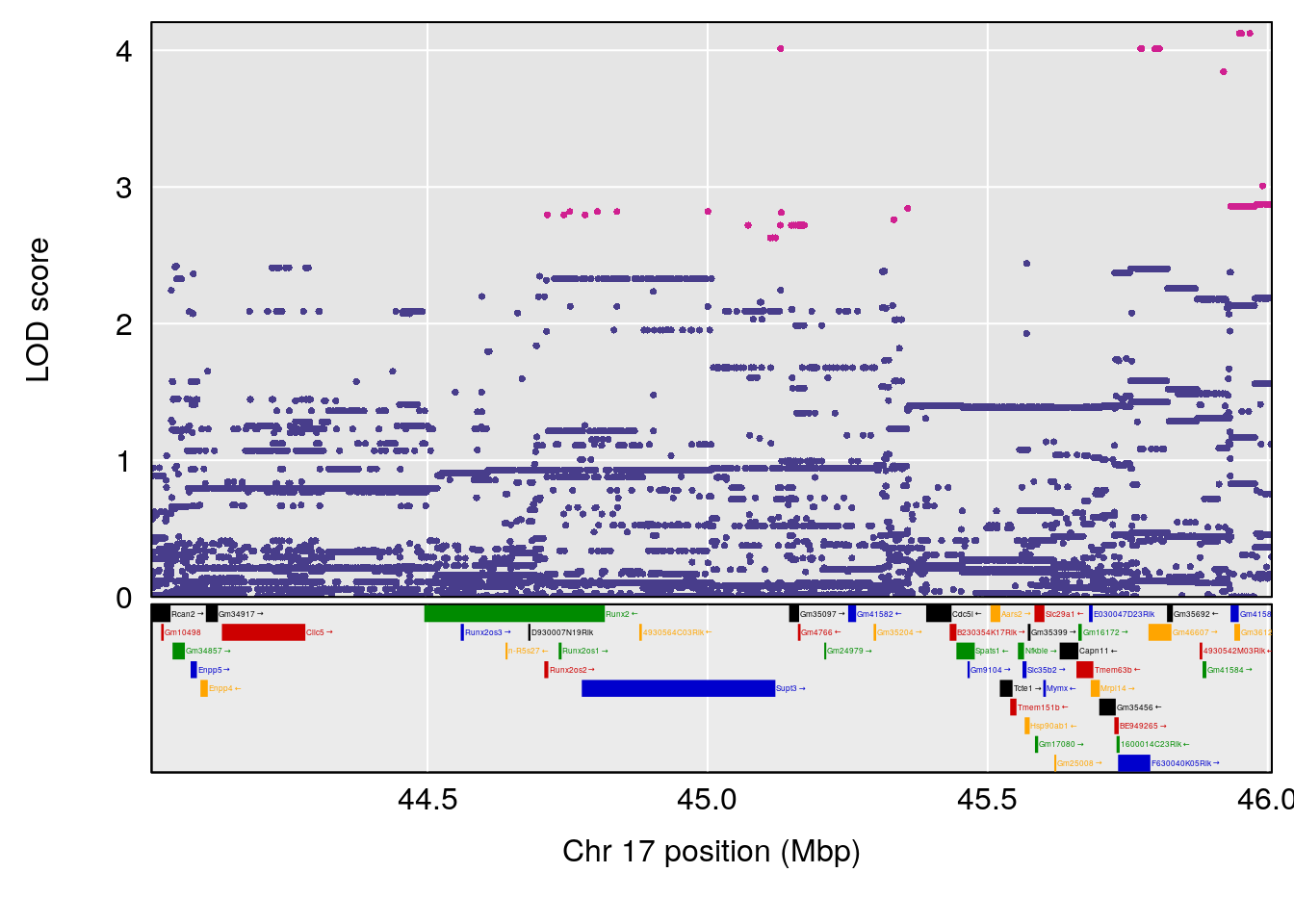
# 3 1 pheno1 17 44.82475 6.772433 44.68776 45.38912
# [1] 1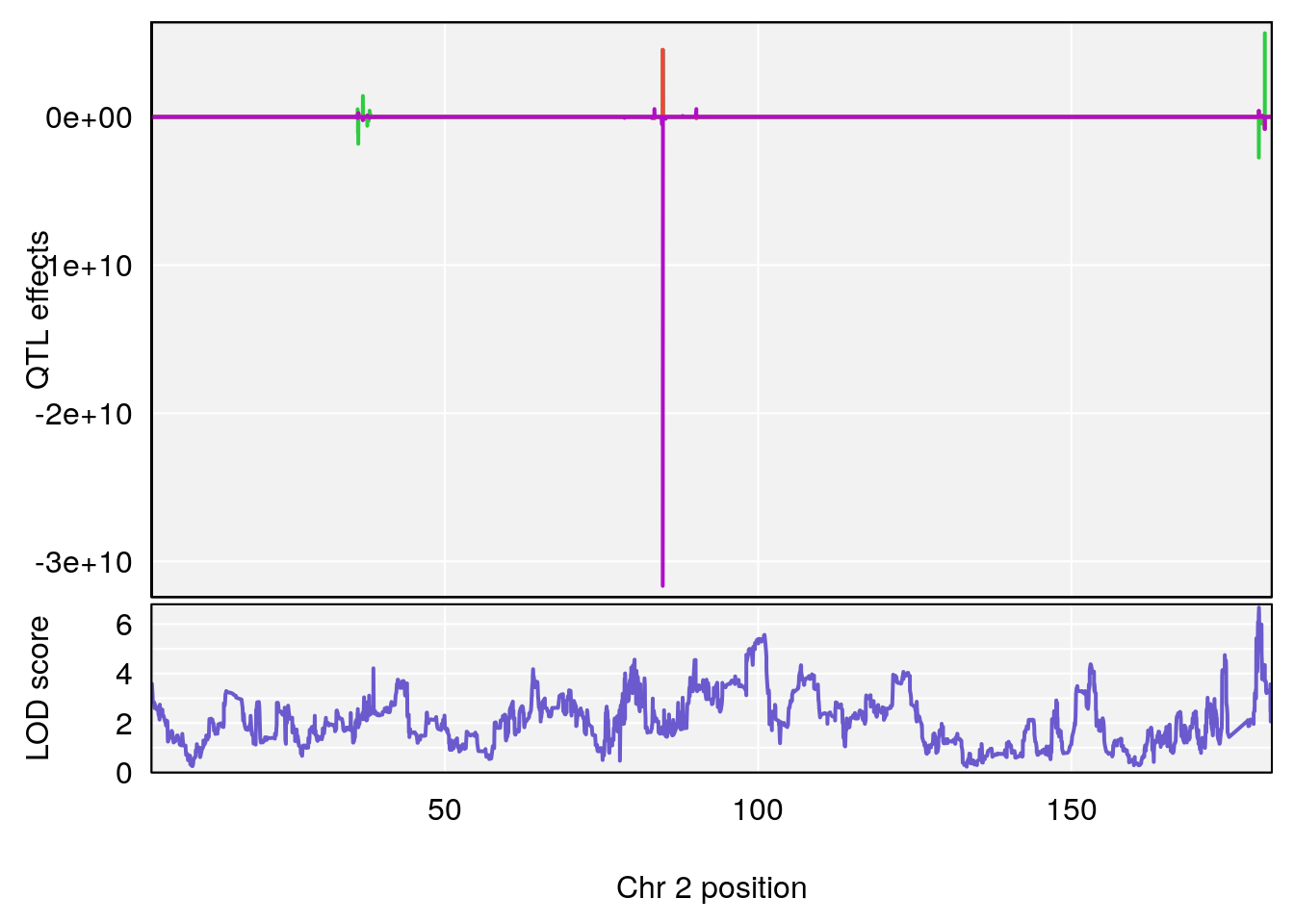
# [1] 2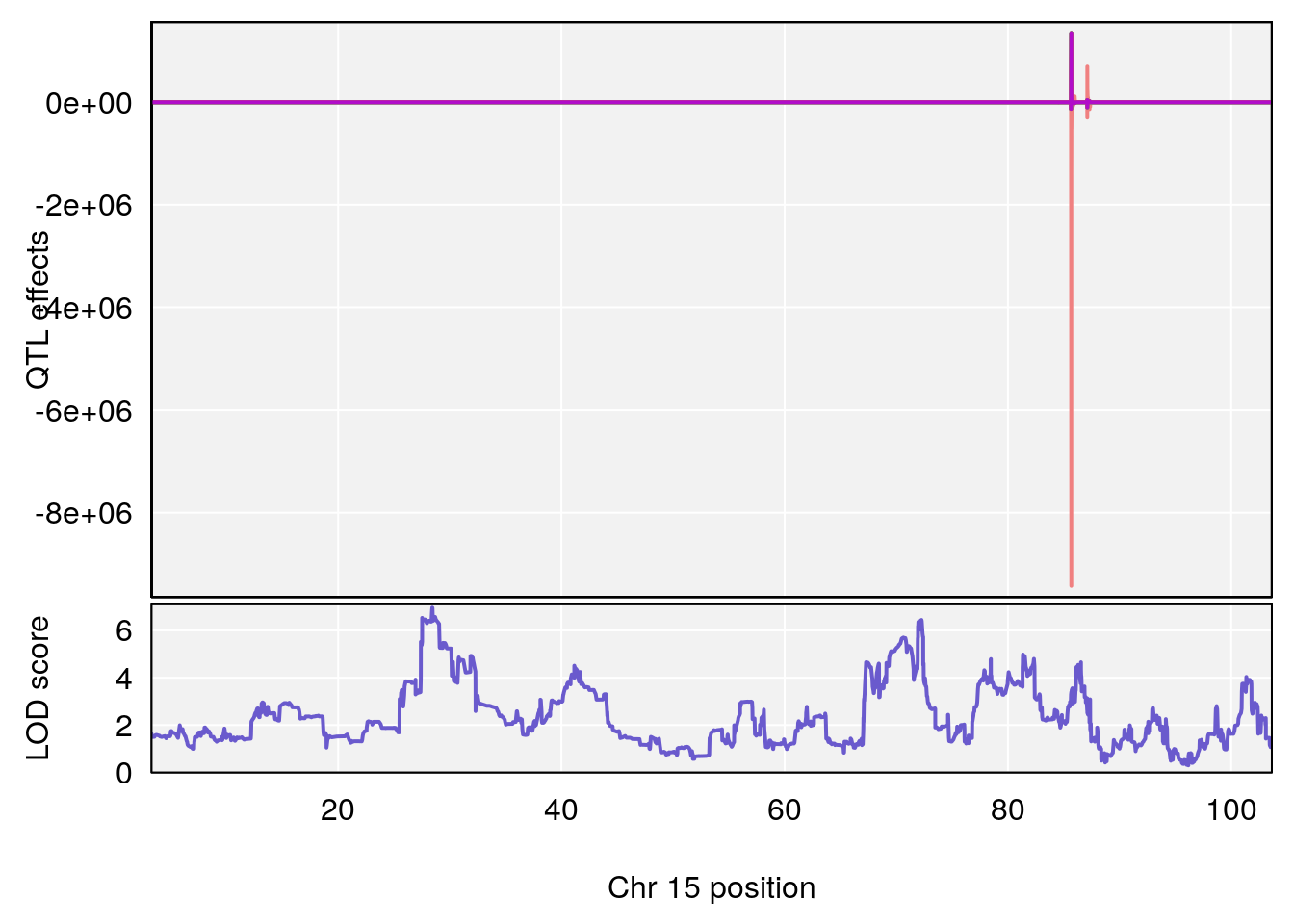
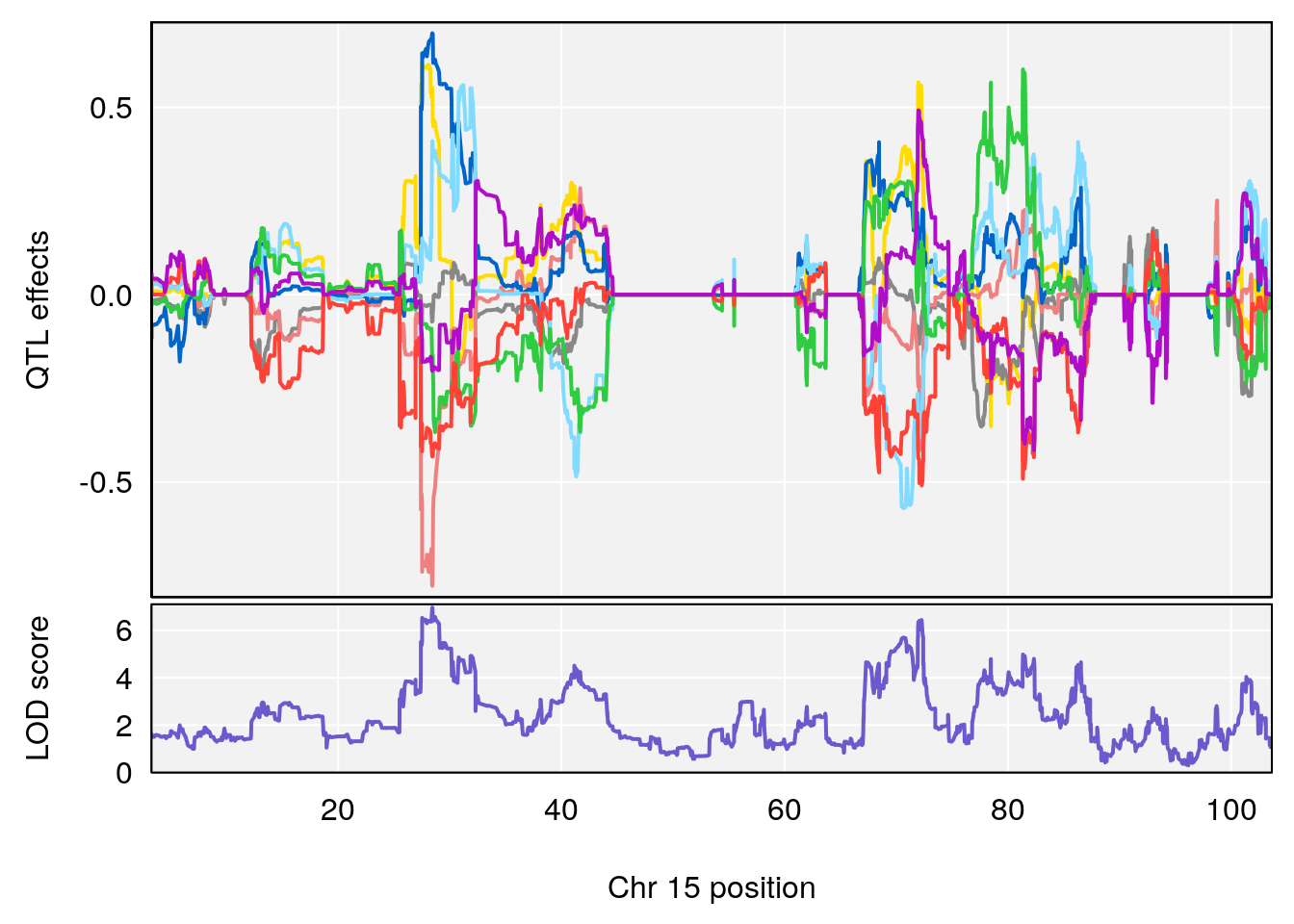
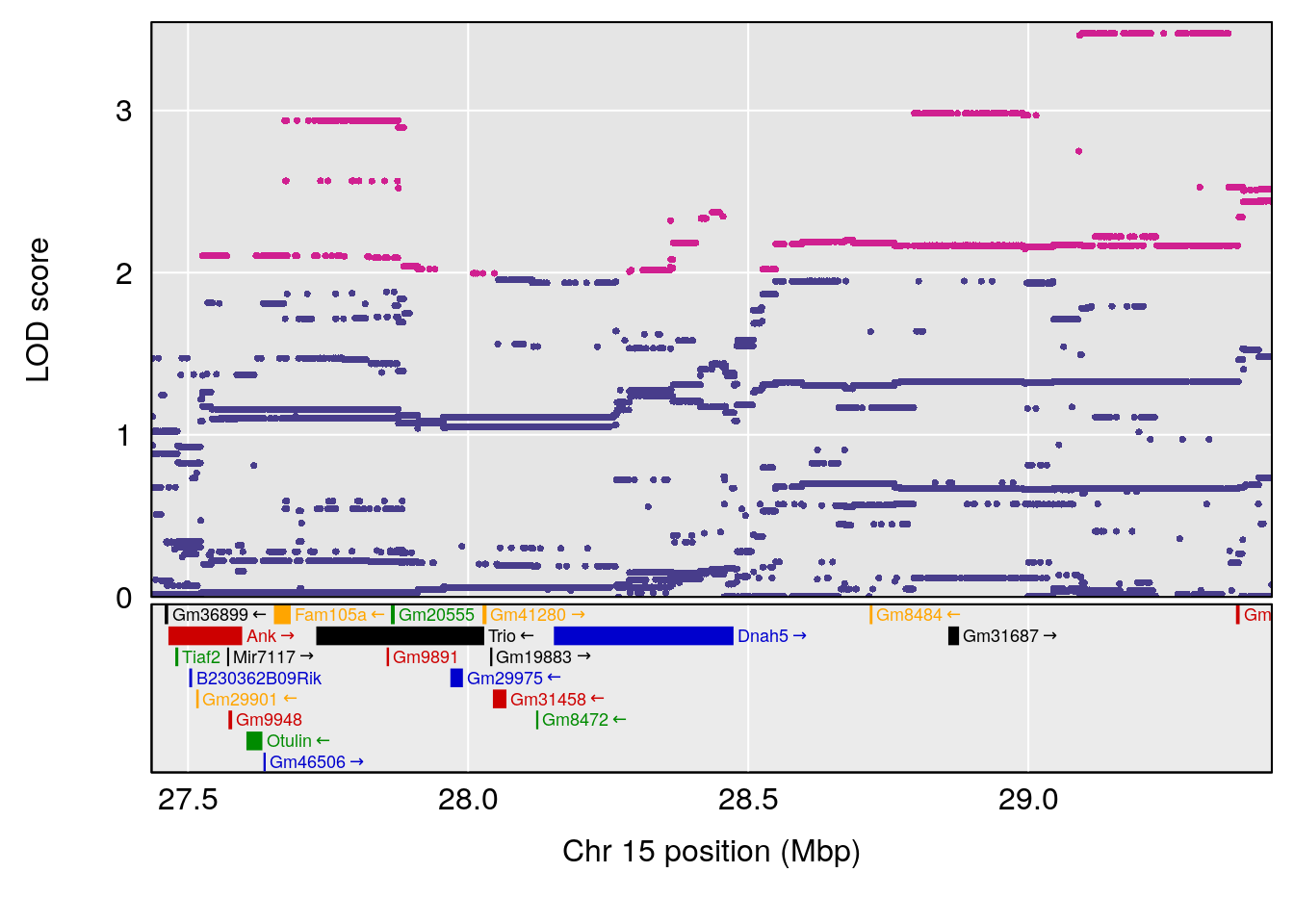
# [1] 3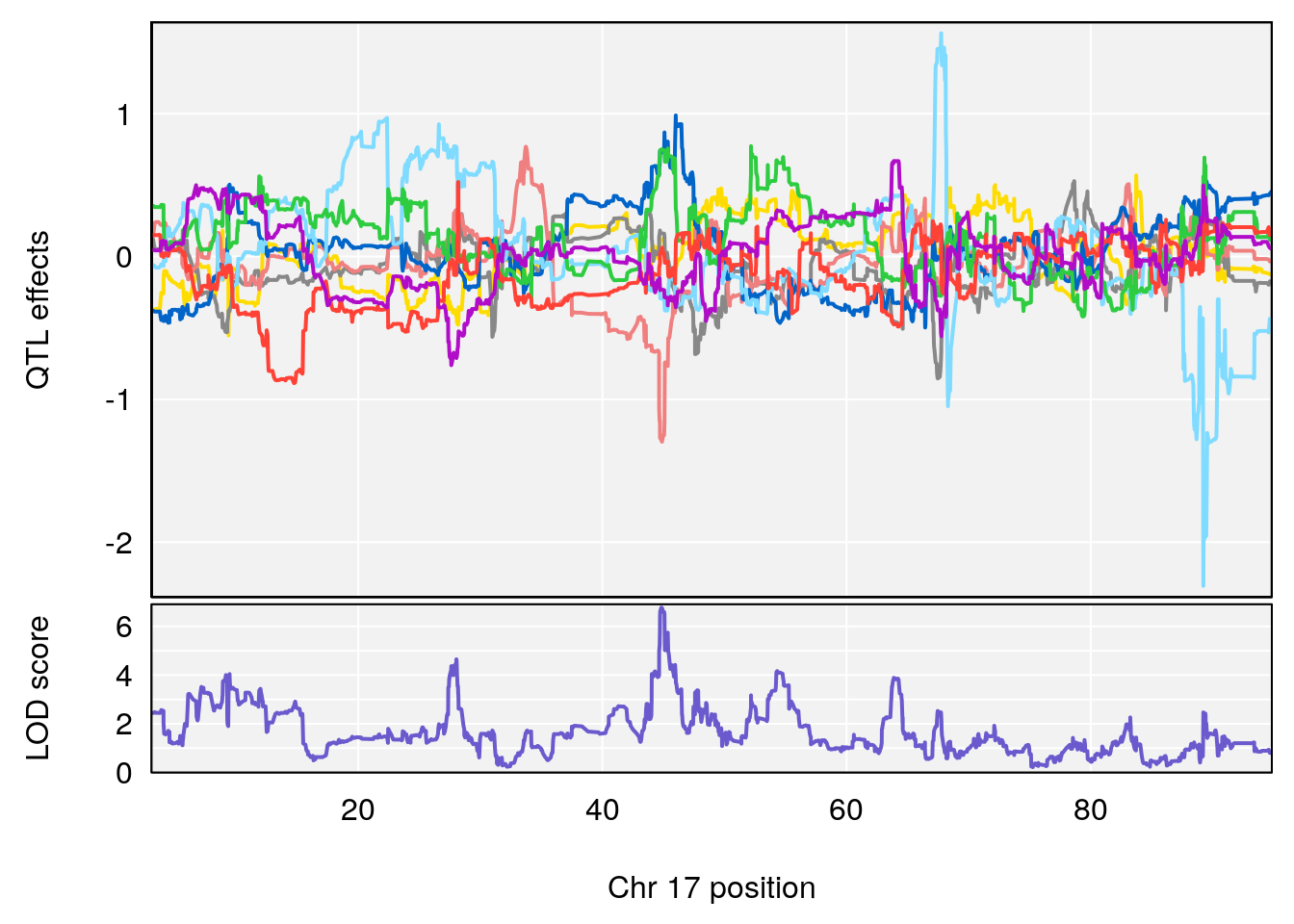
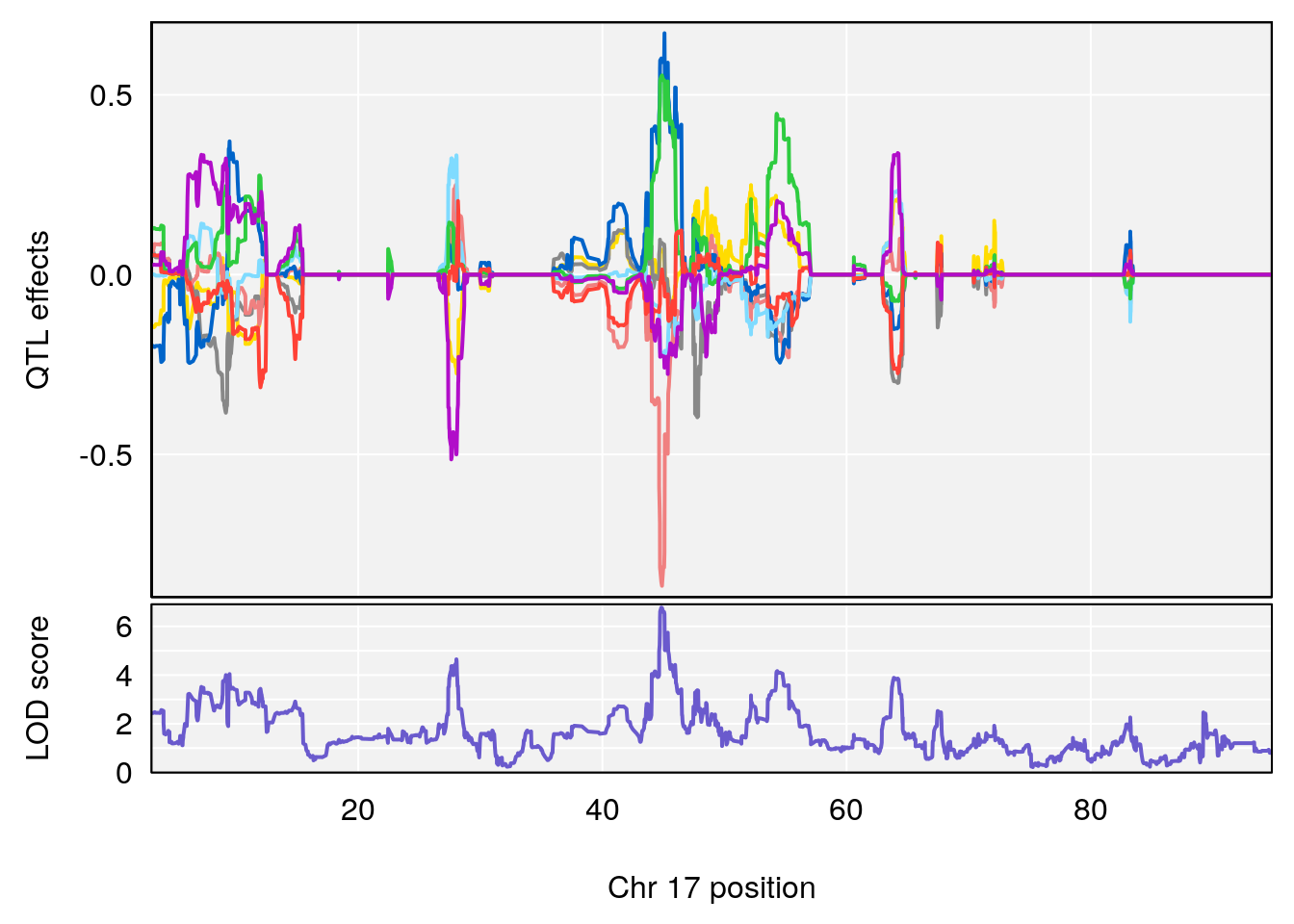
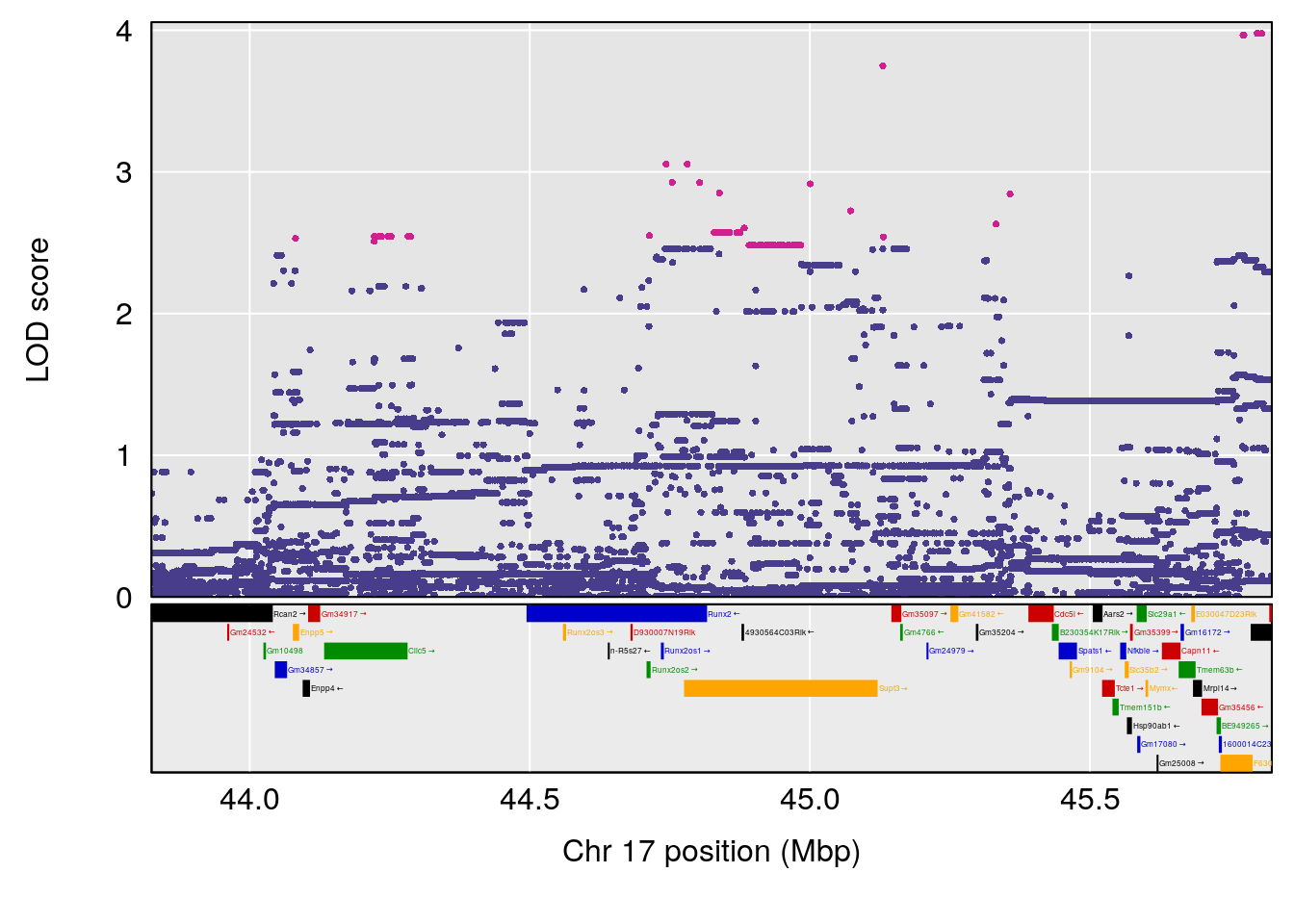
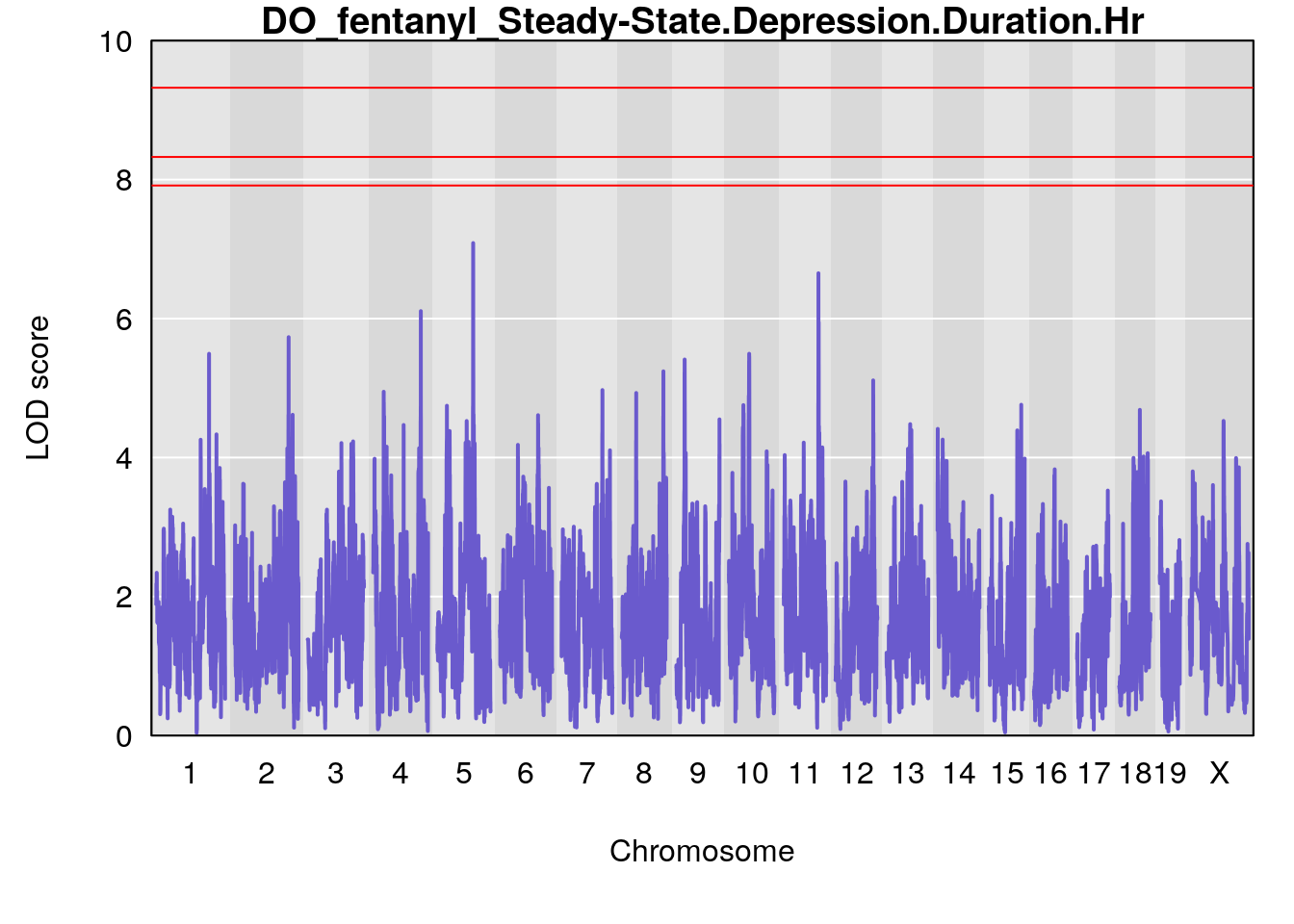
# [1] "Steady-State.Depression.Duration.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 4 135.9927 6.107796 32.37530 136.1999
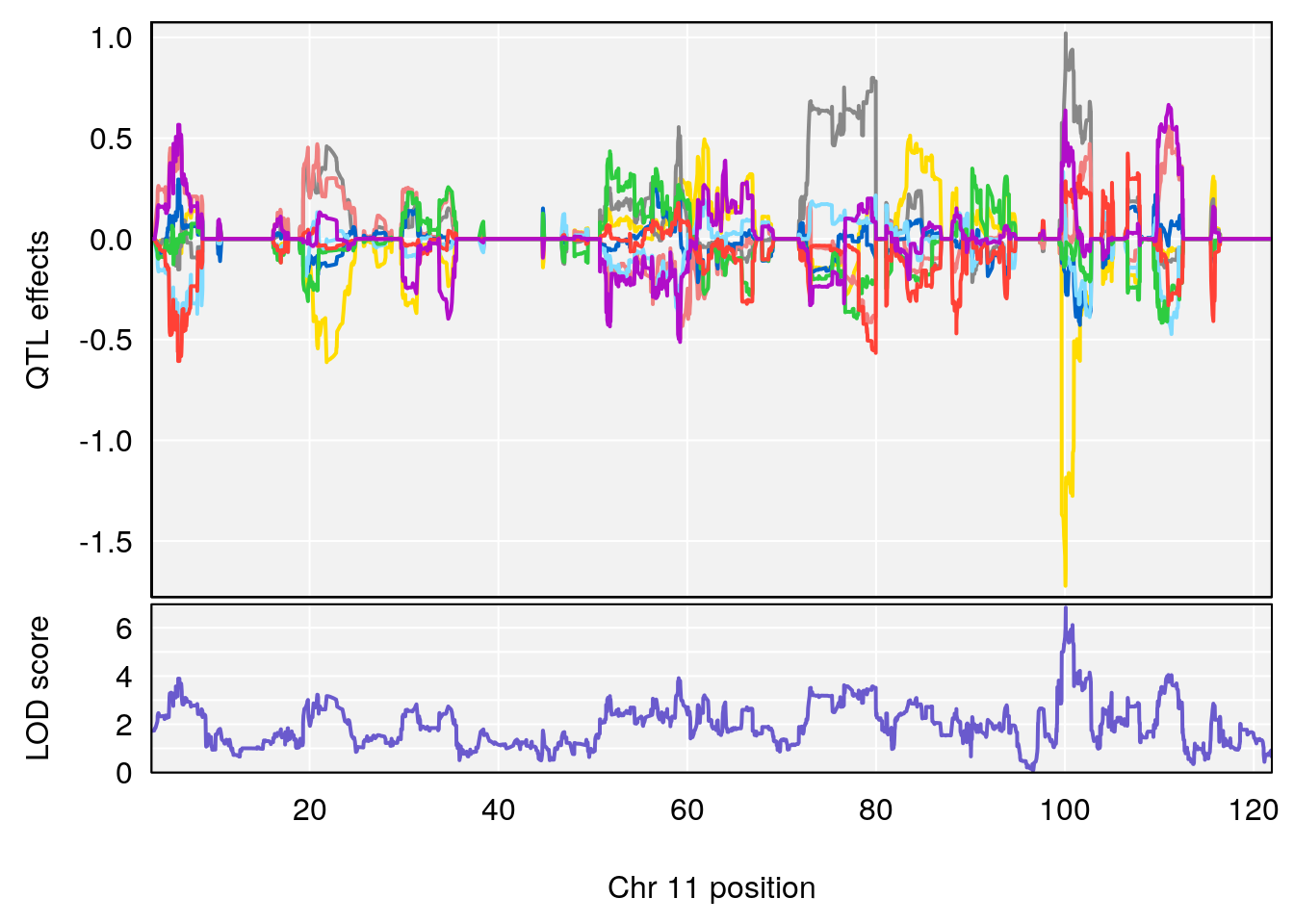
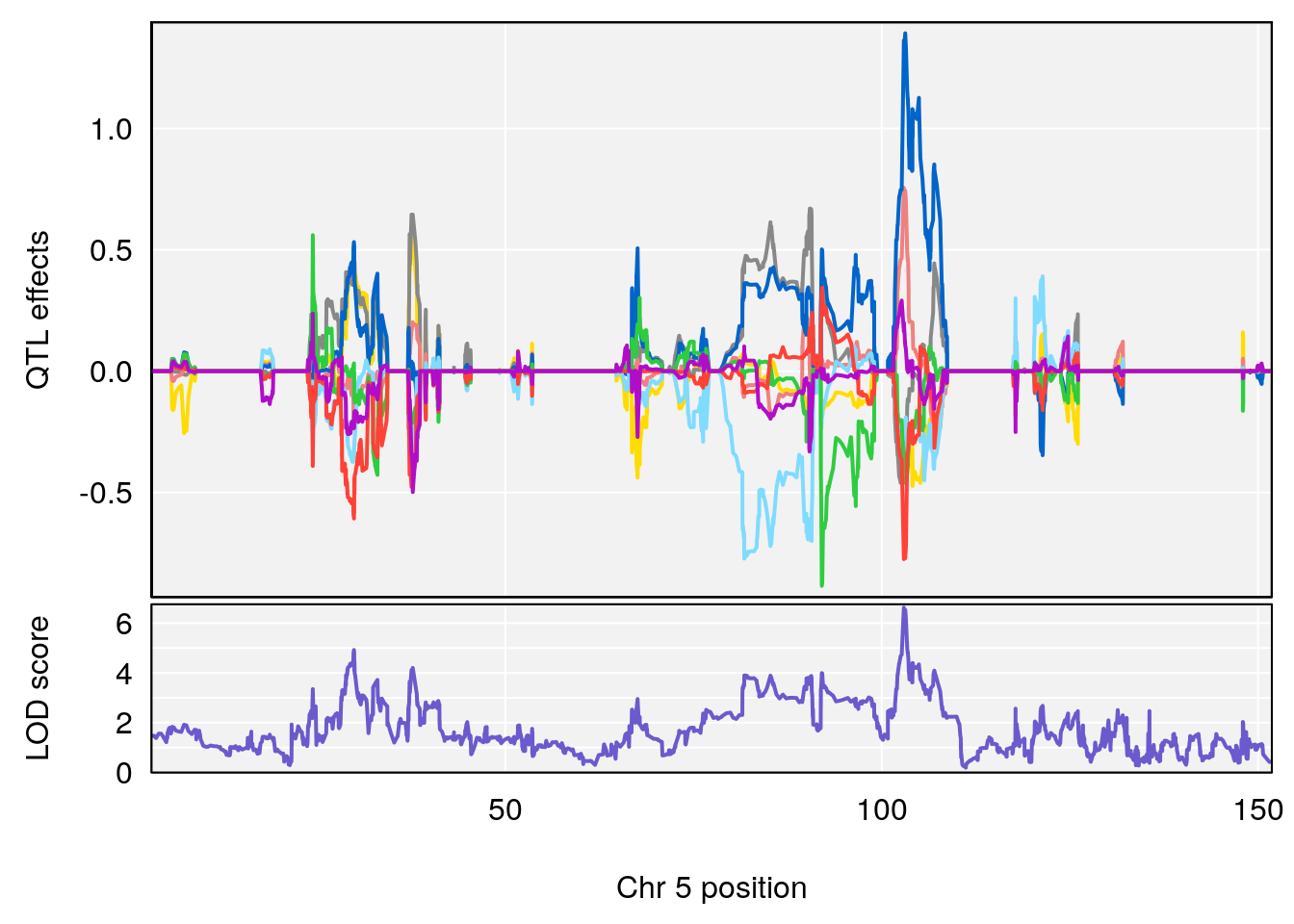
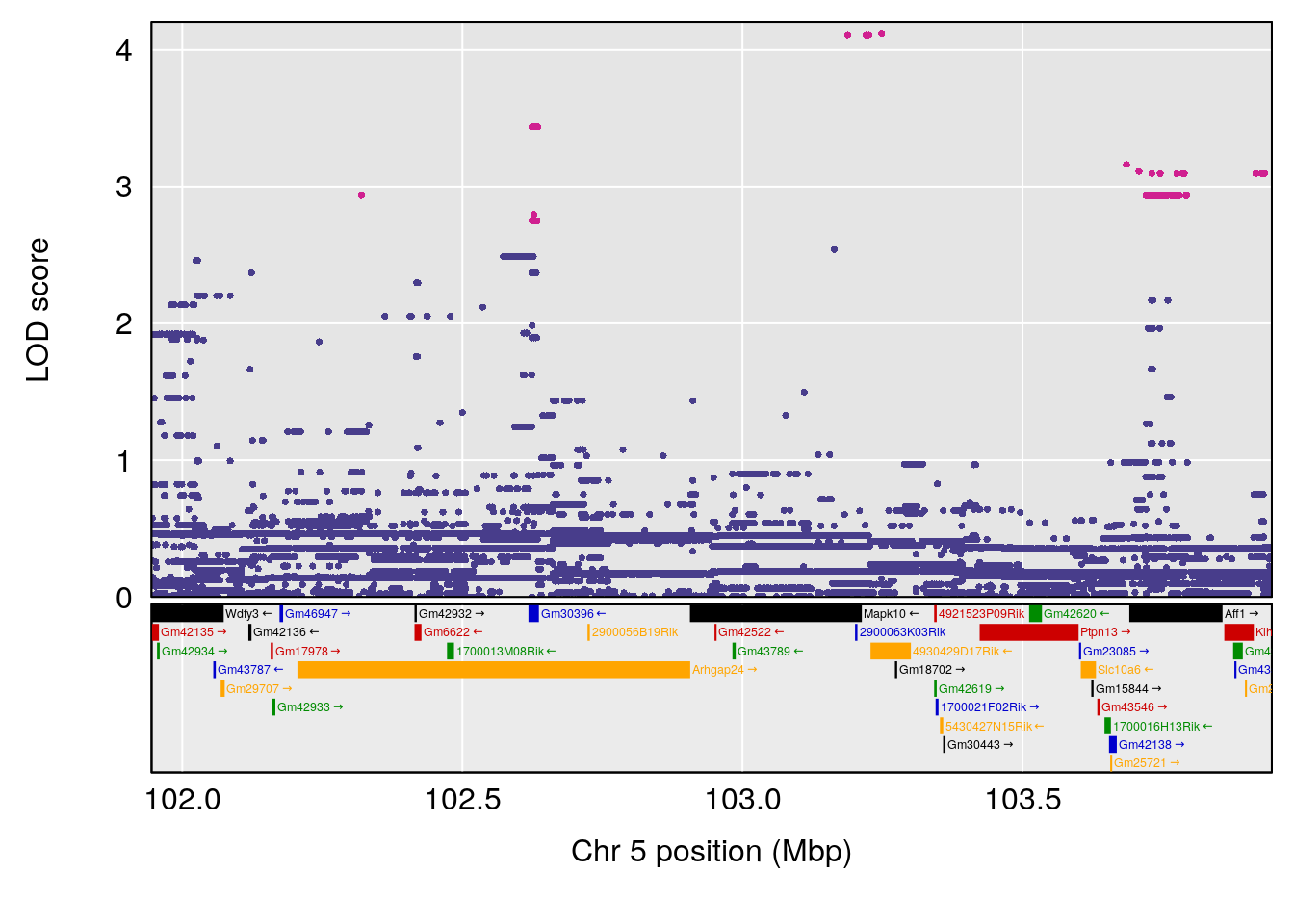
# 2 1 pheno1 5 102.9928 7.087202 102.81969 103.3132
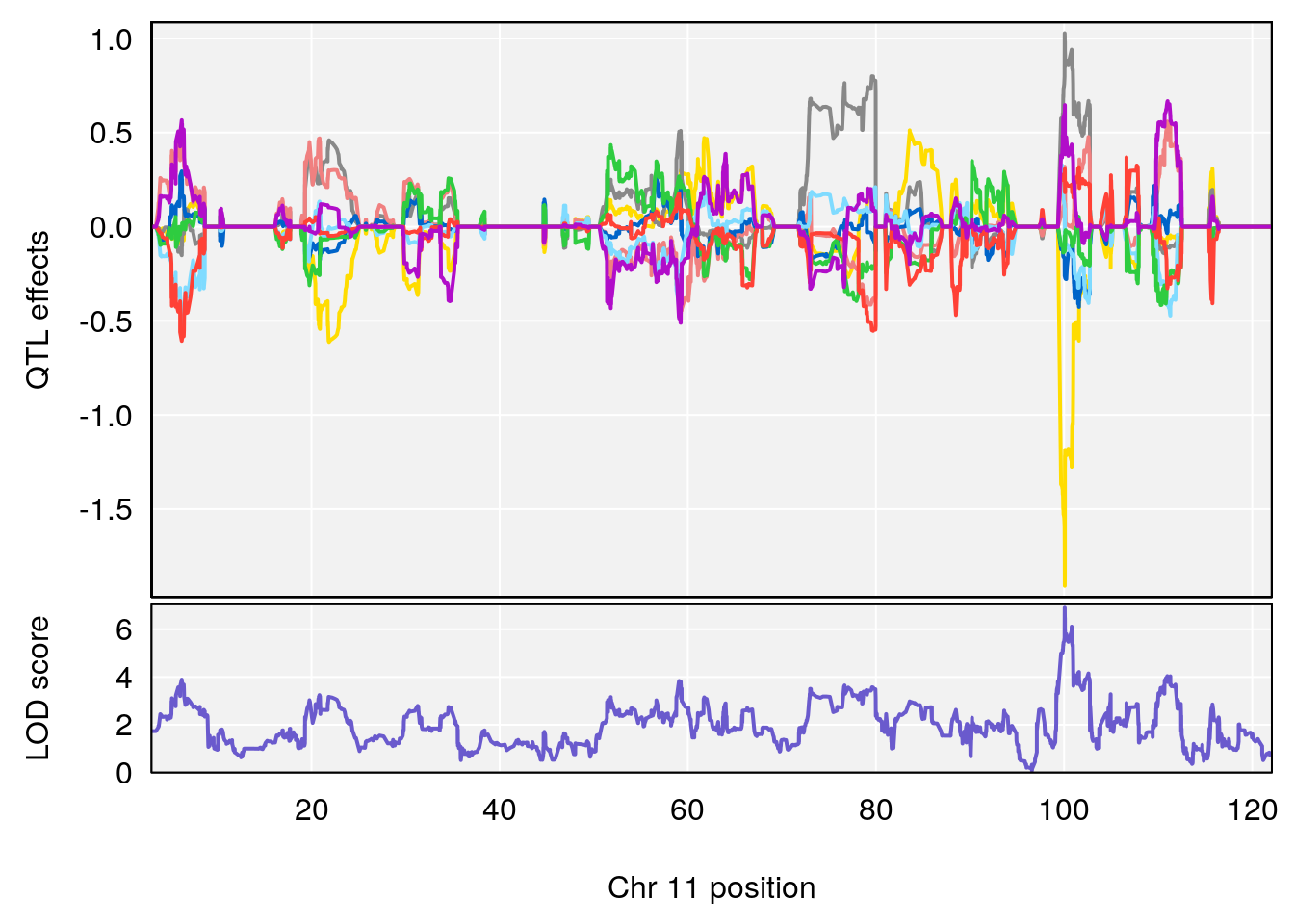
# 3 1 pheno1 11 100.0882 6.652753 99.89319 100.9355
# [1] 1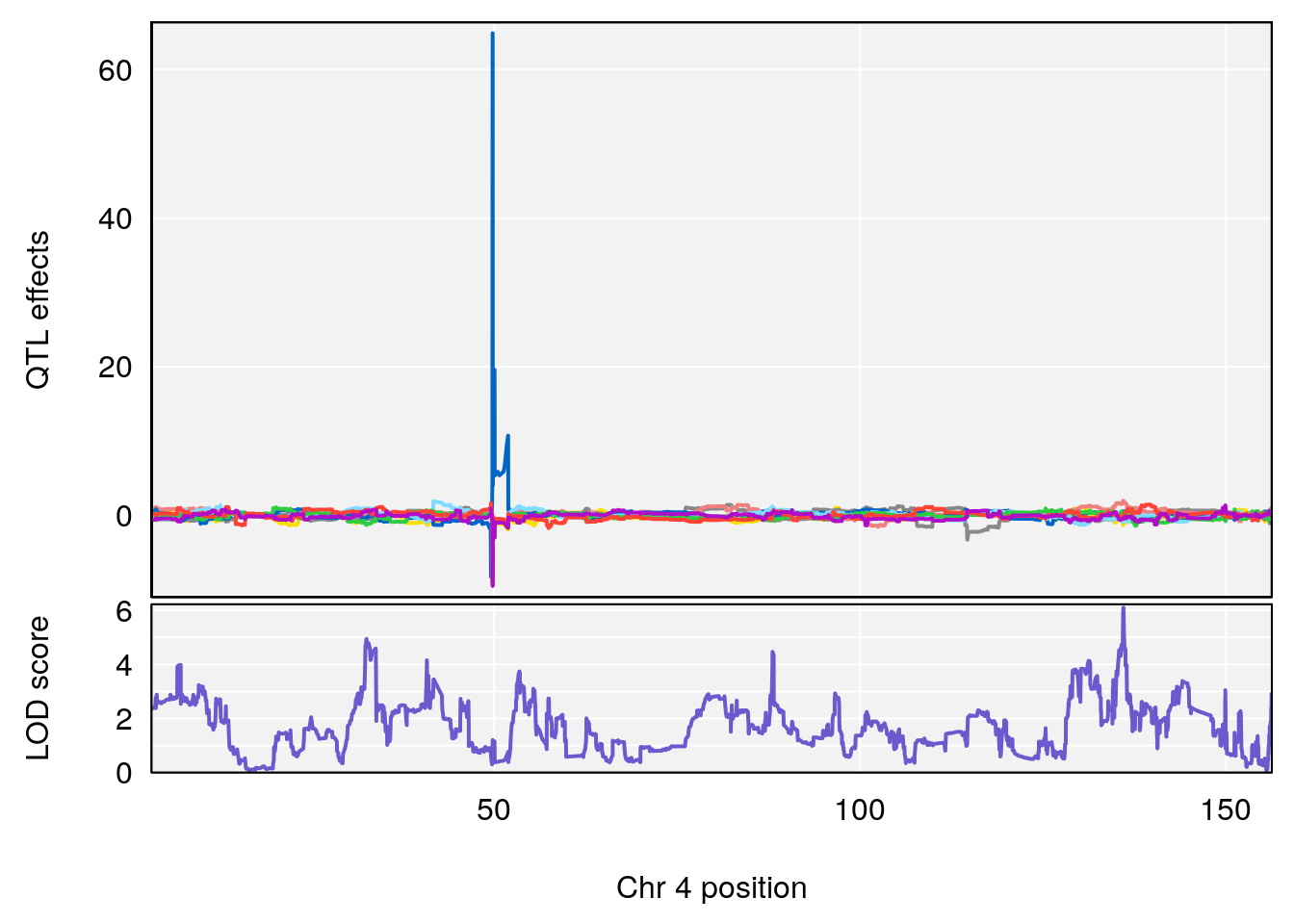
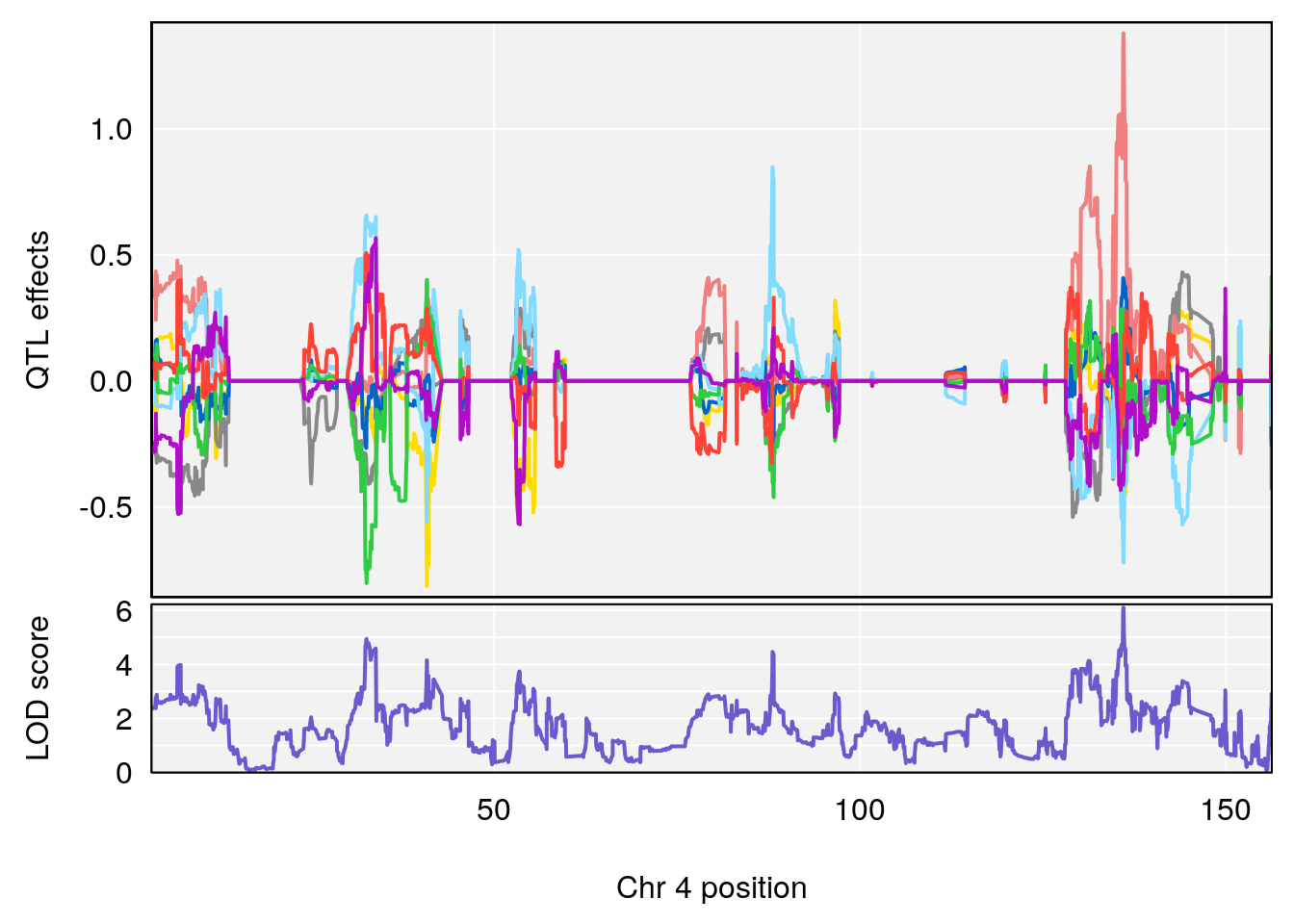
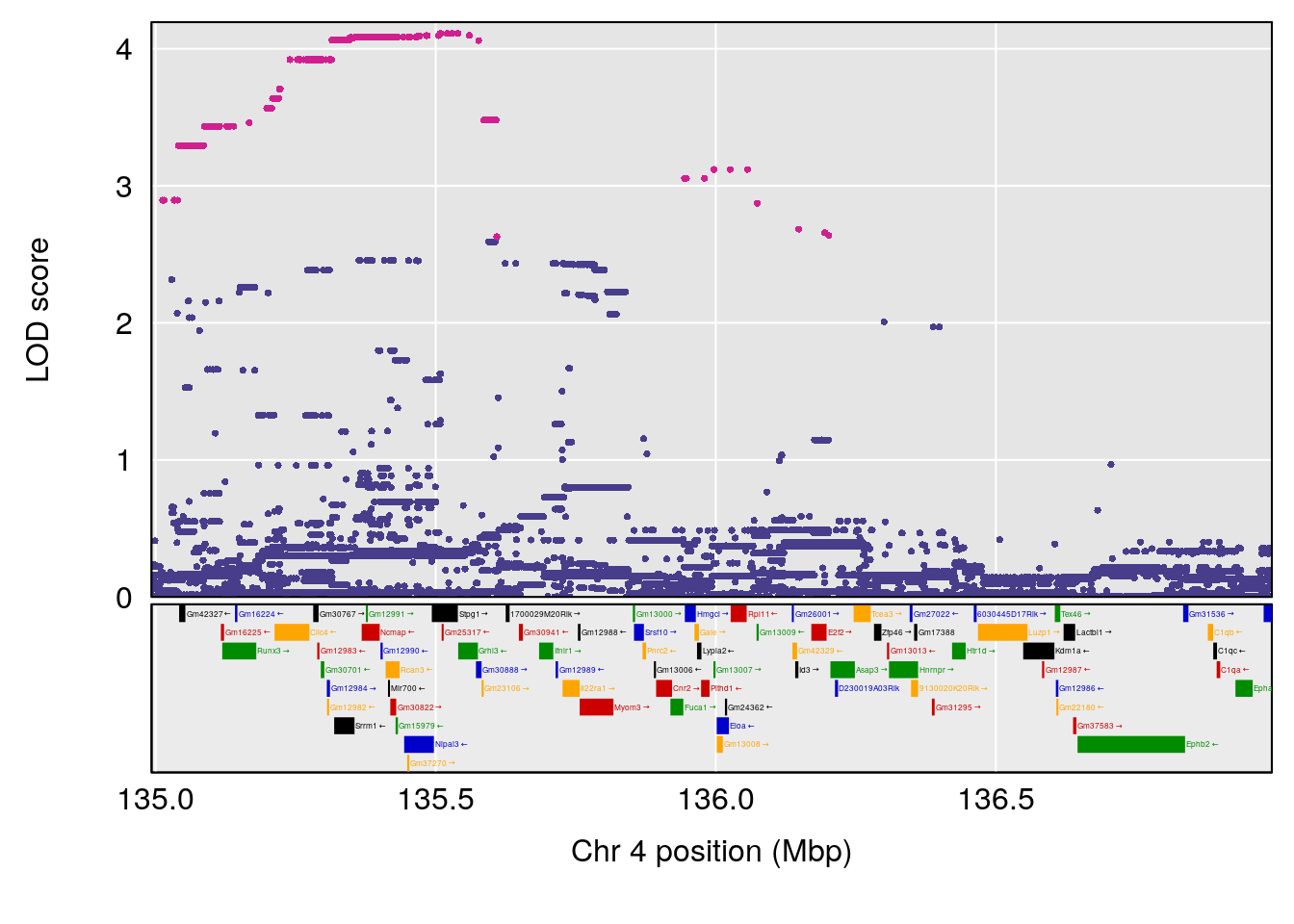
# [1] 2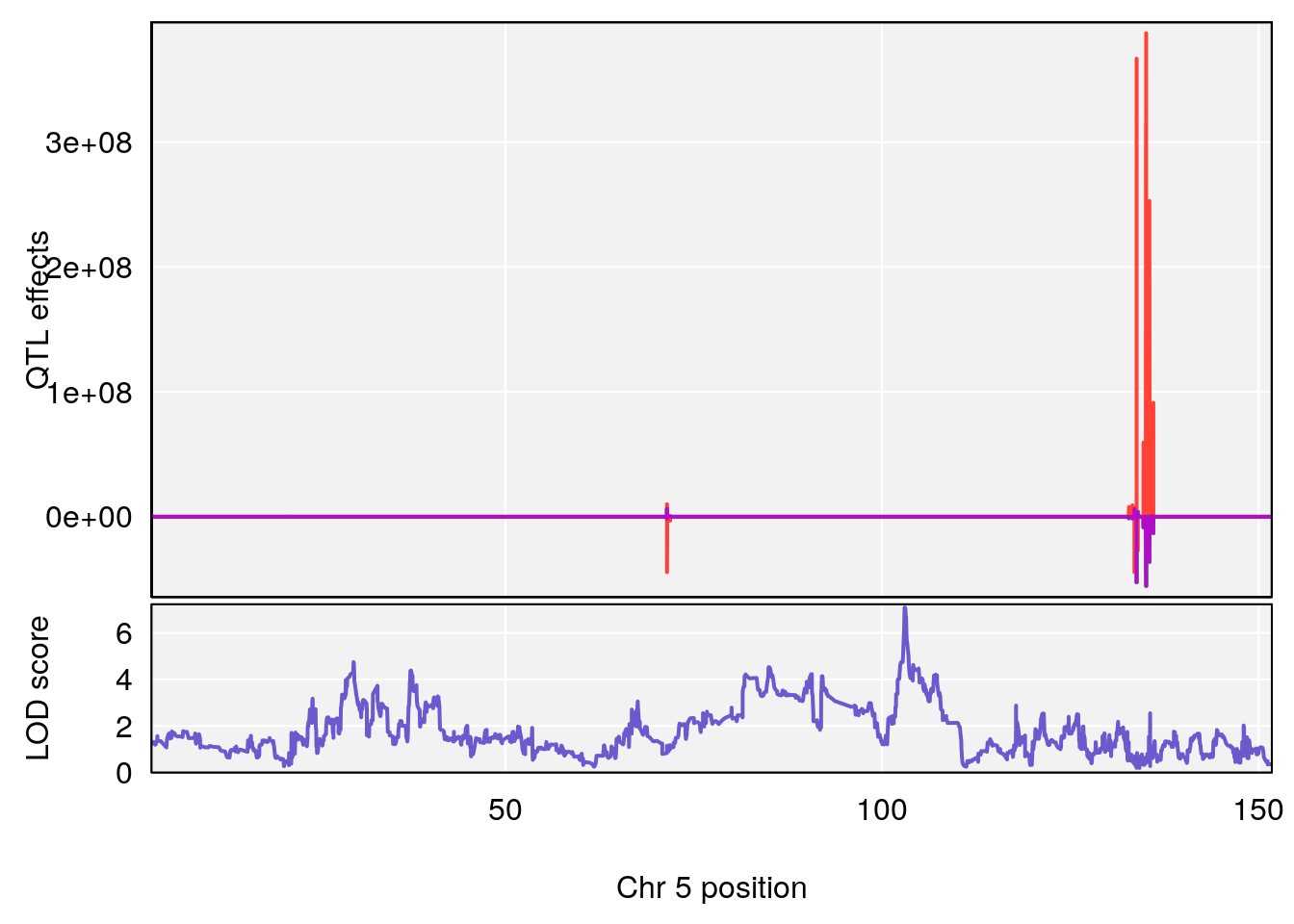
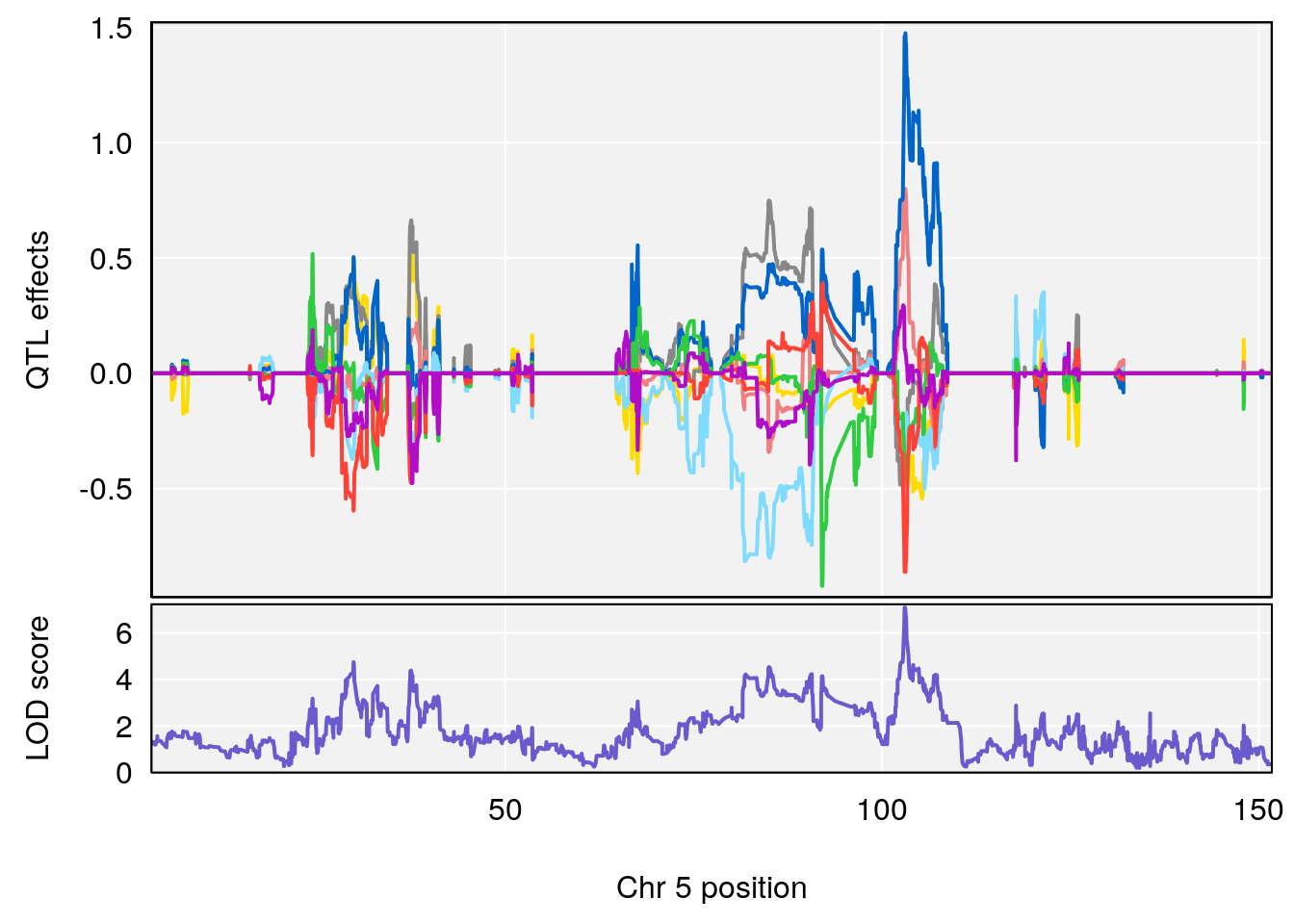
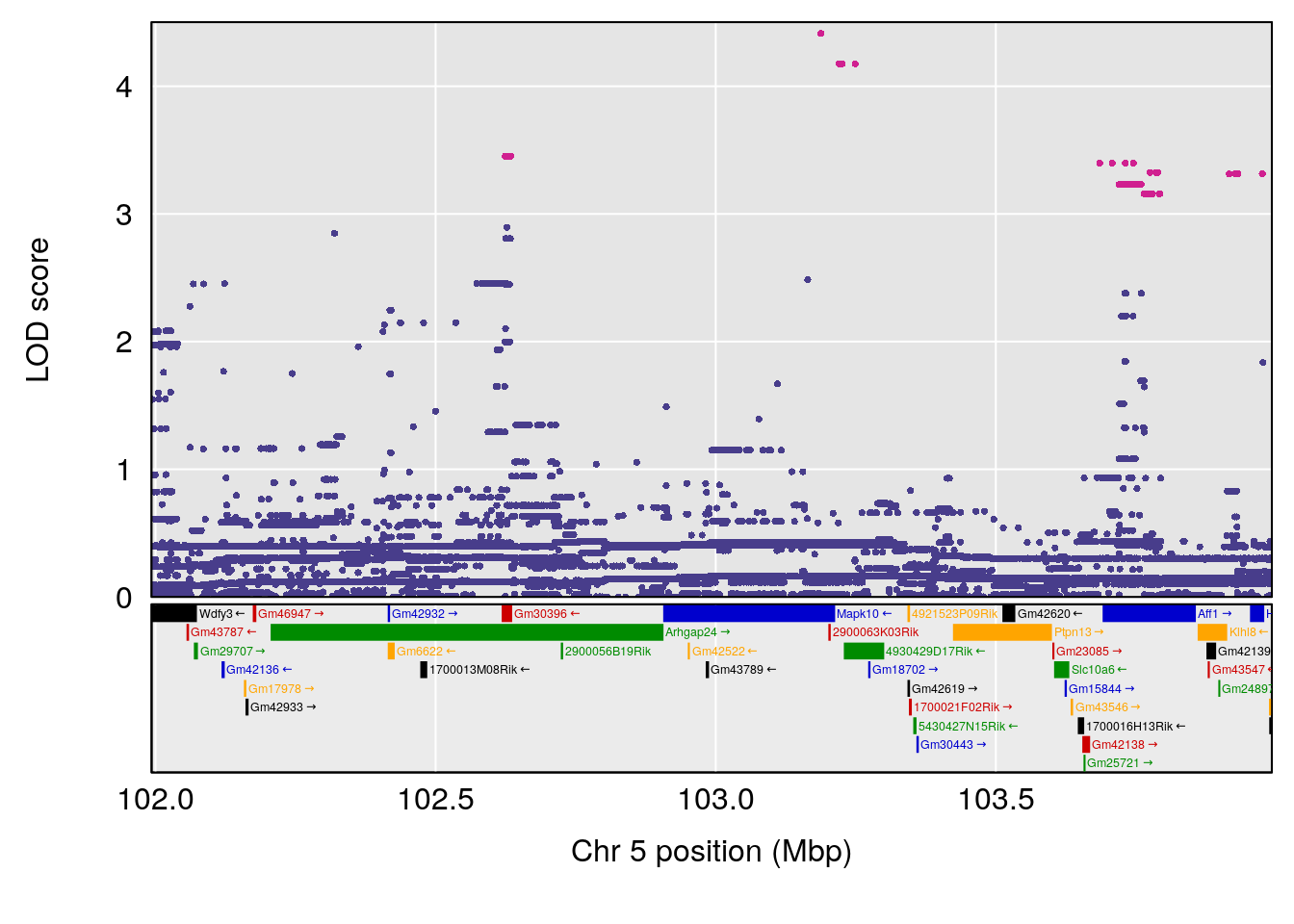
# [1] 3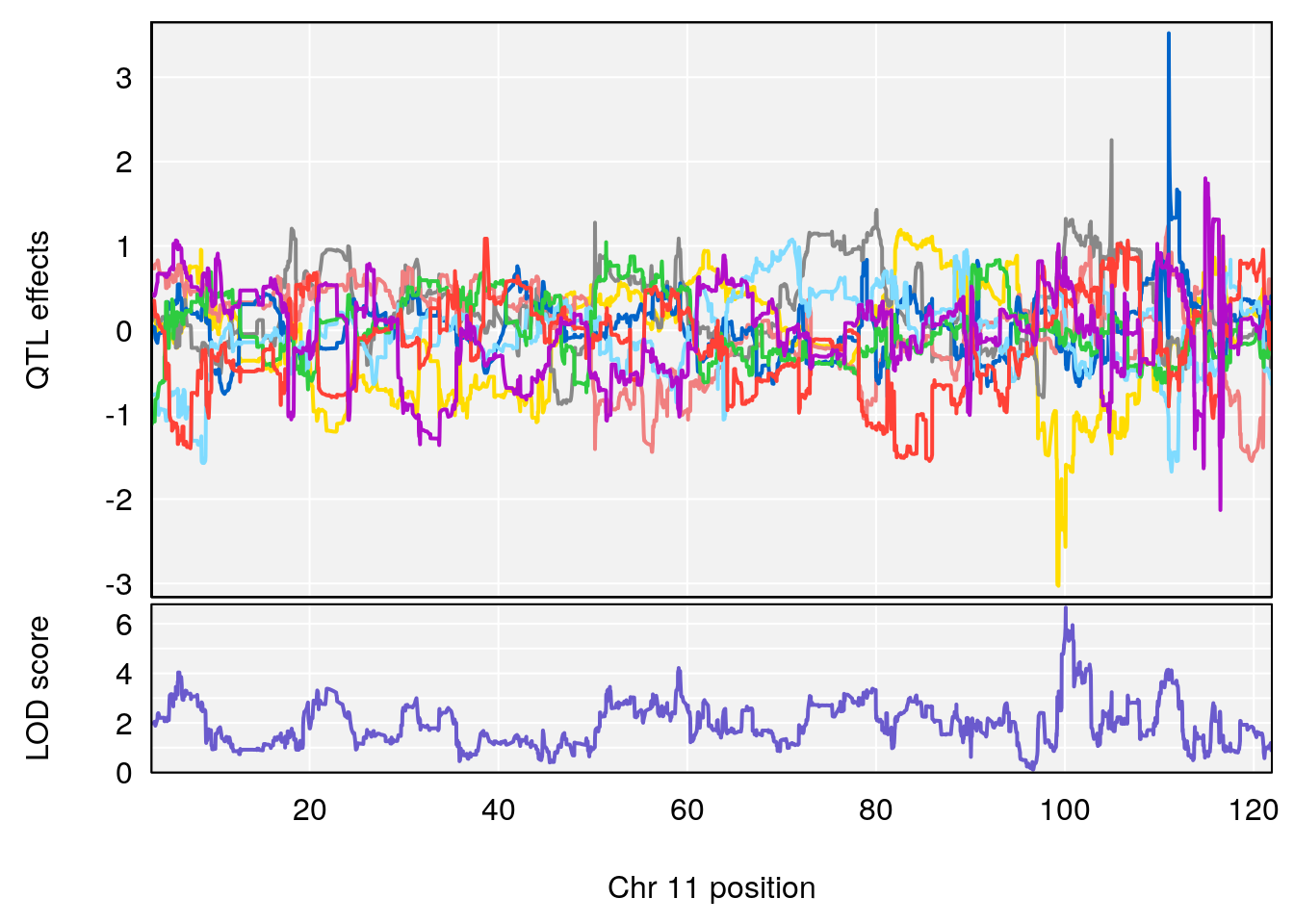
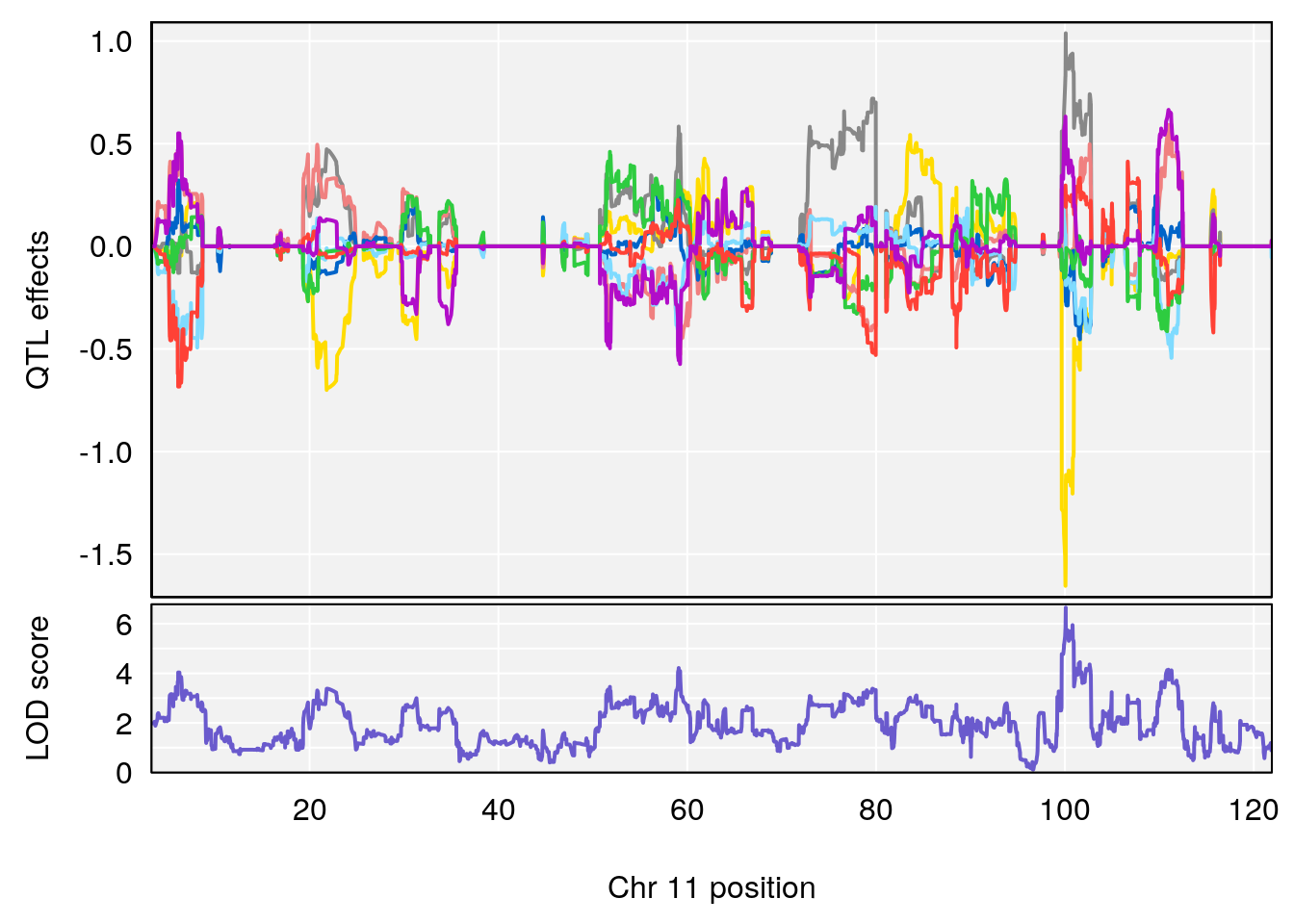
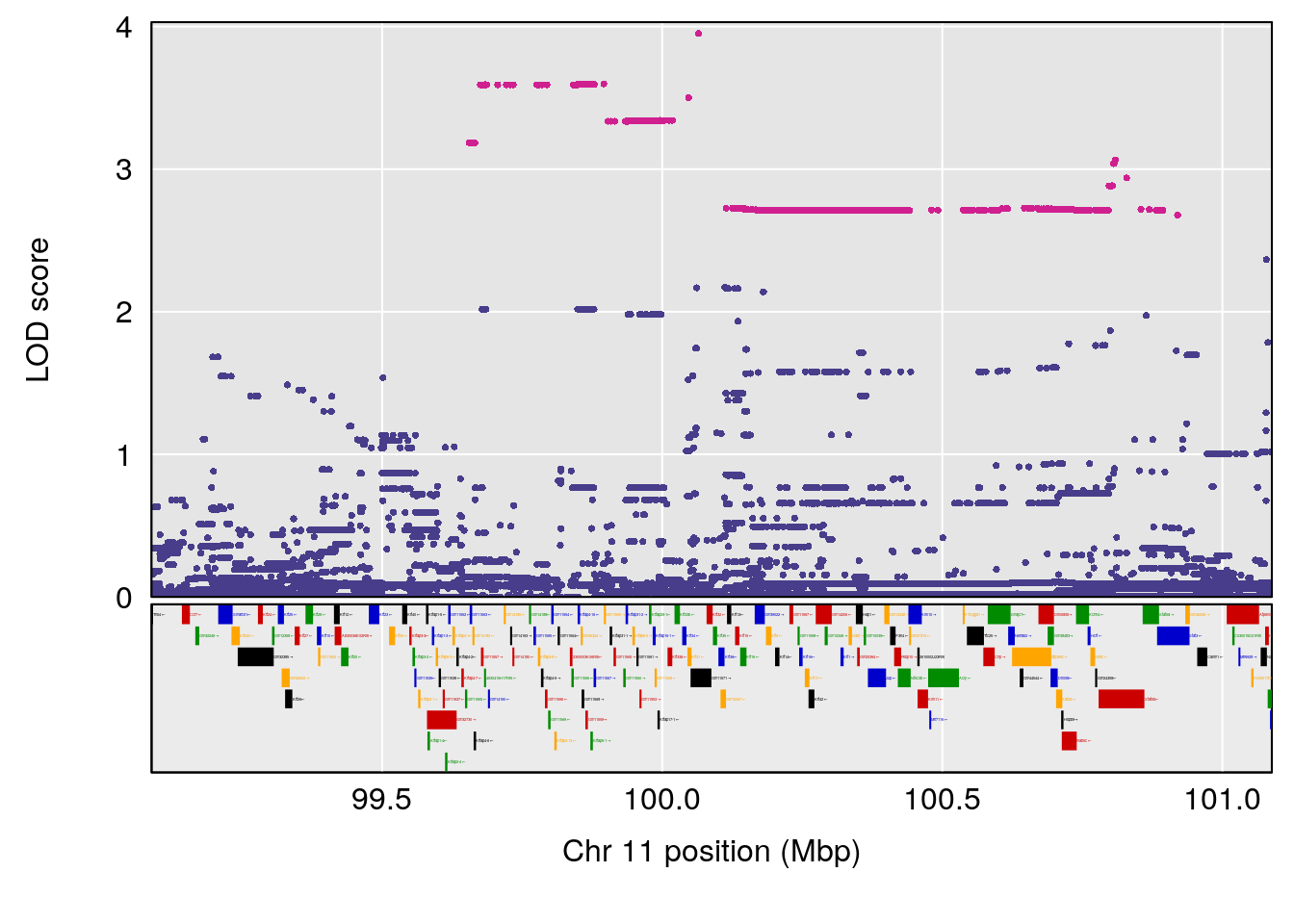
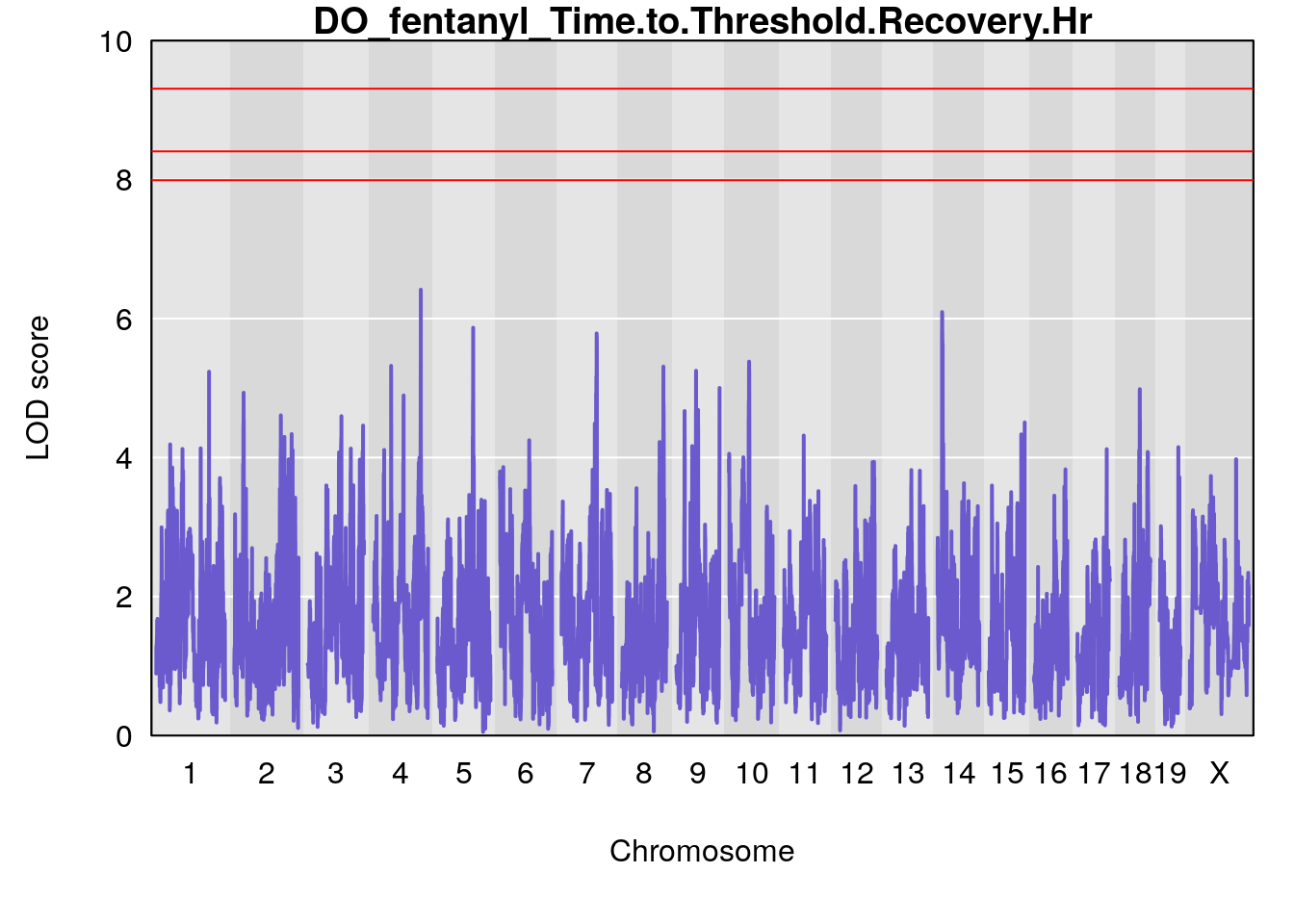
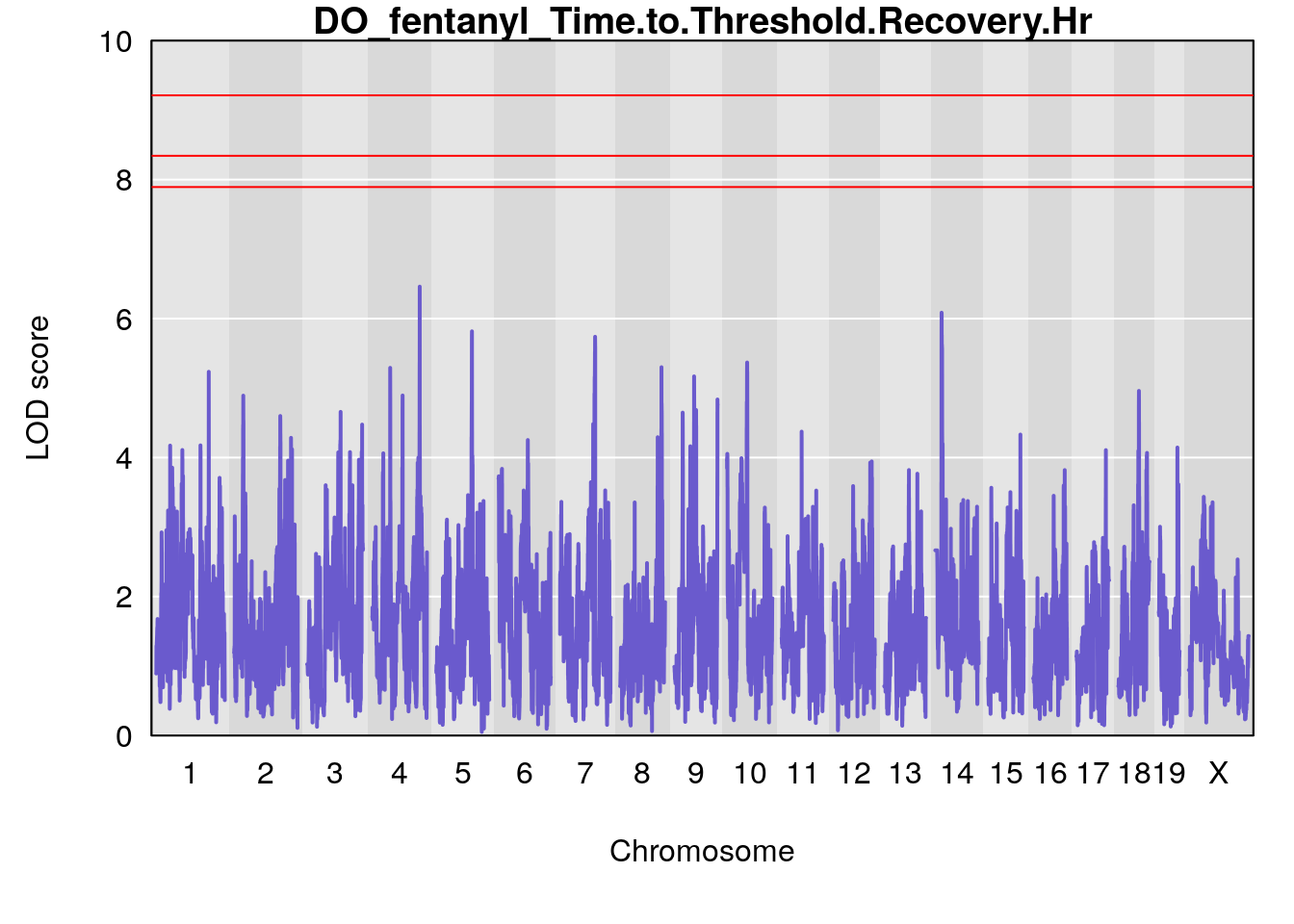
# [1] "Time.to.Threshold.Recovery.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 4 135.94204 6.415931 53.10764 136.06715
# 2 1 pheno1 14 20.50752 6.094258 20.26372 22.25158
# [1] 1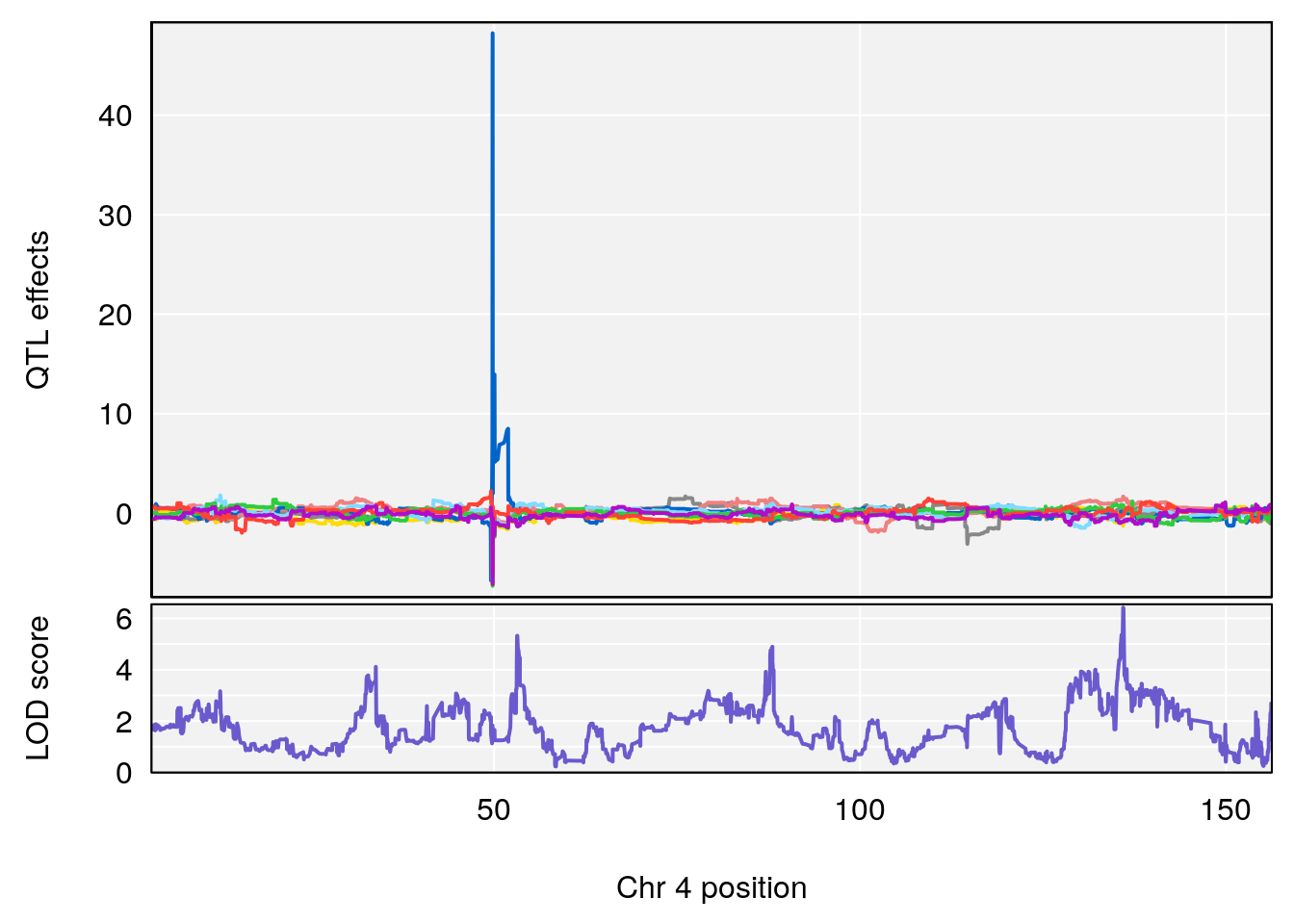
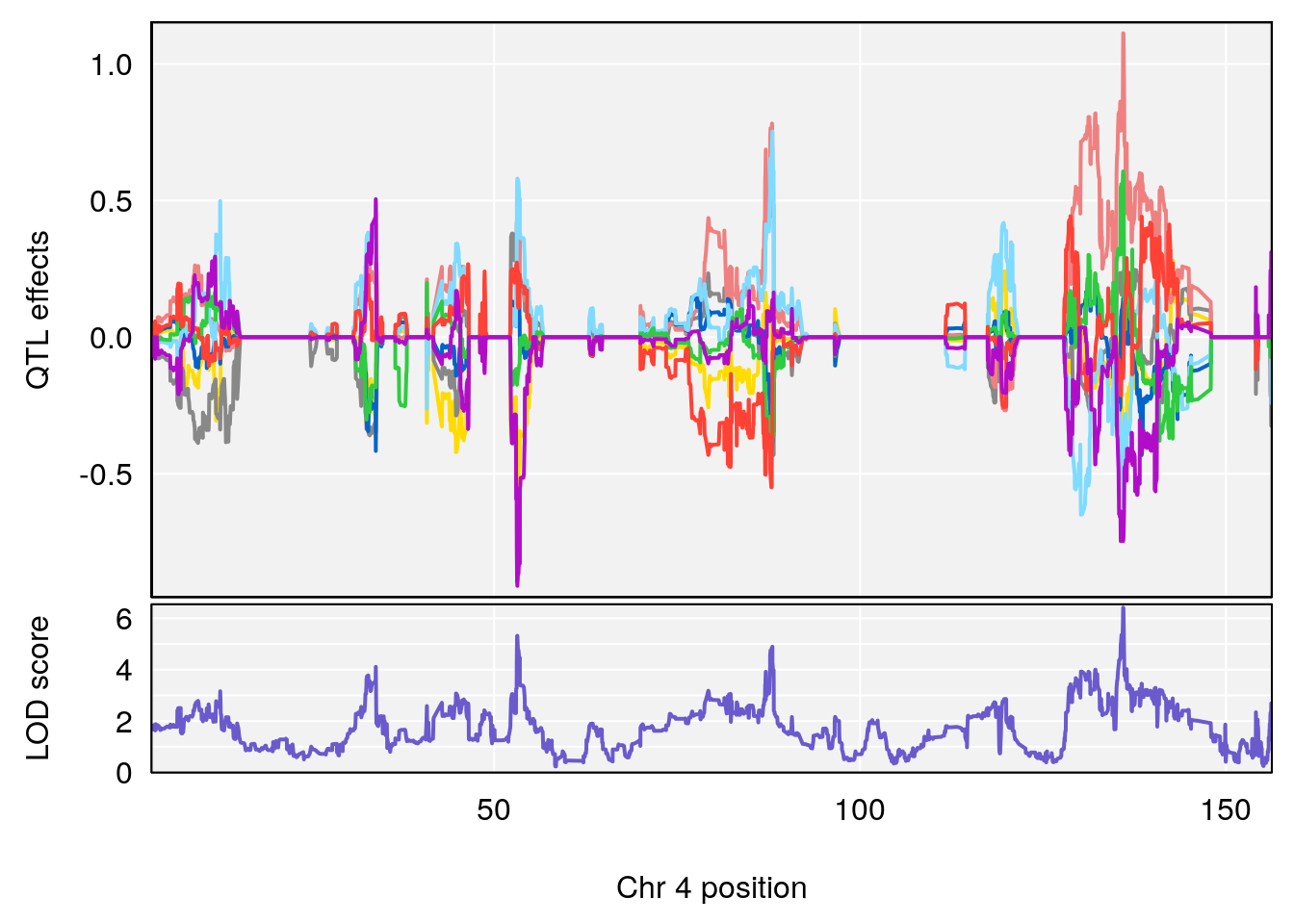
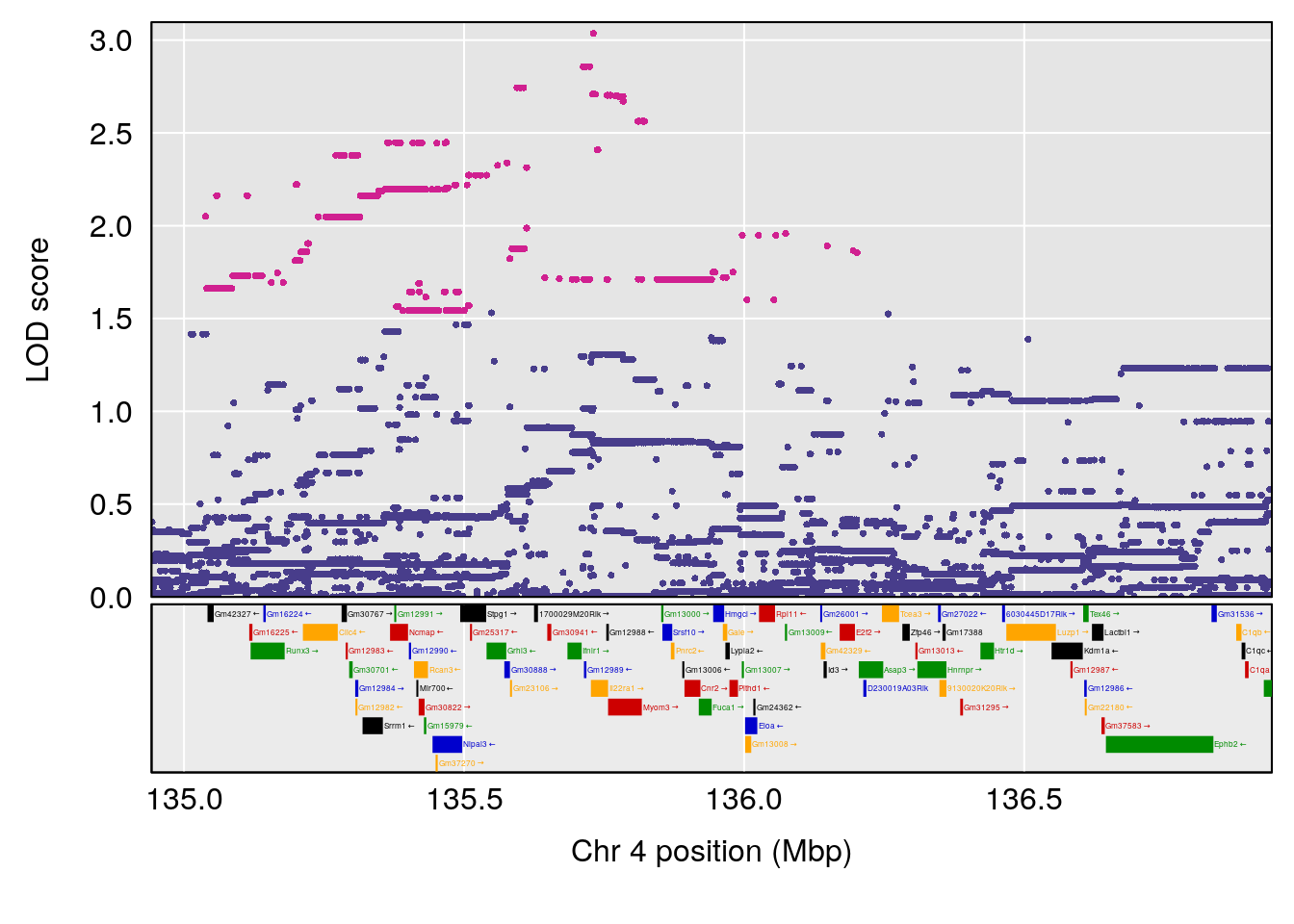
# [1] 2
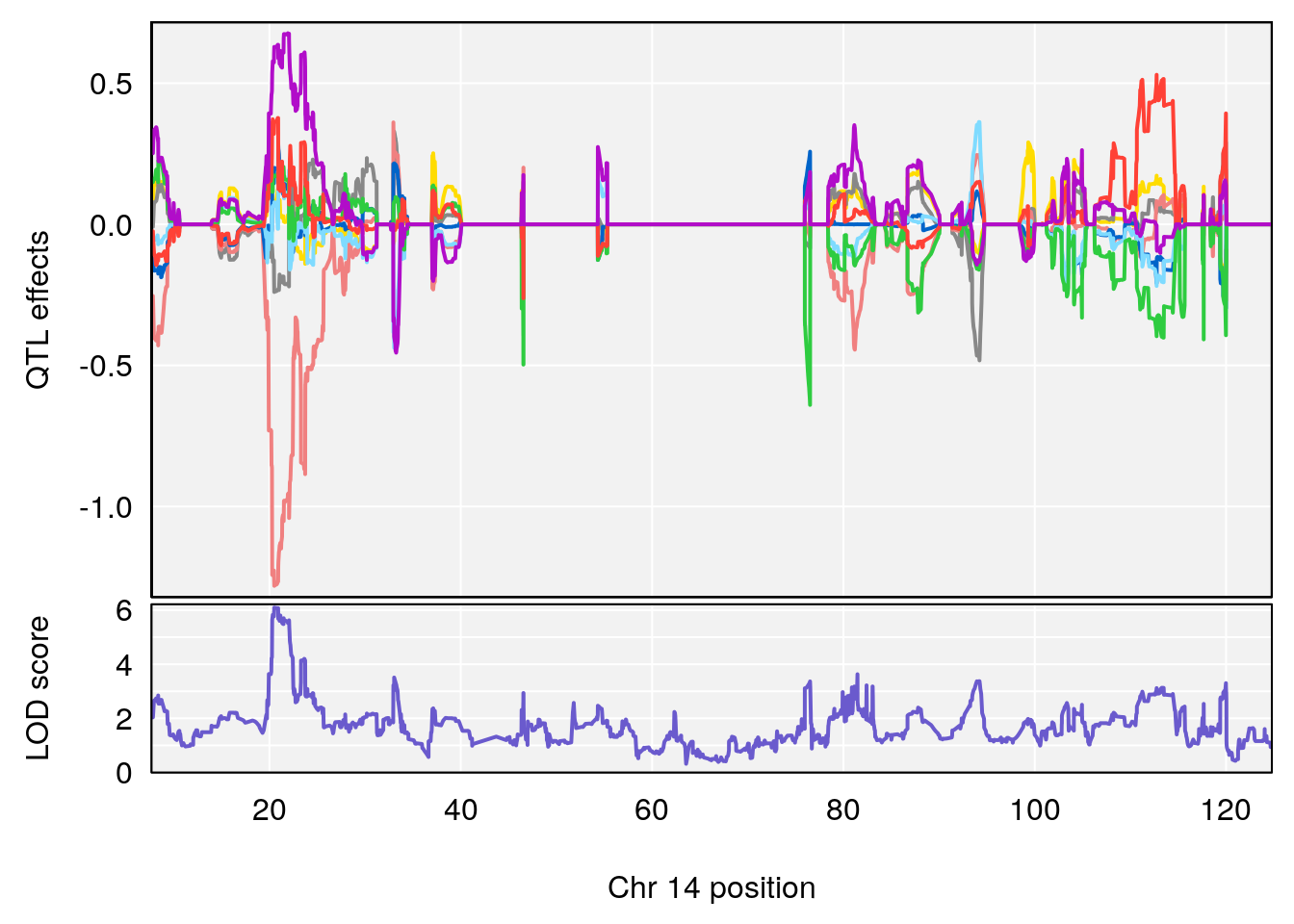
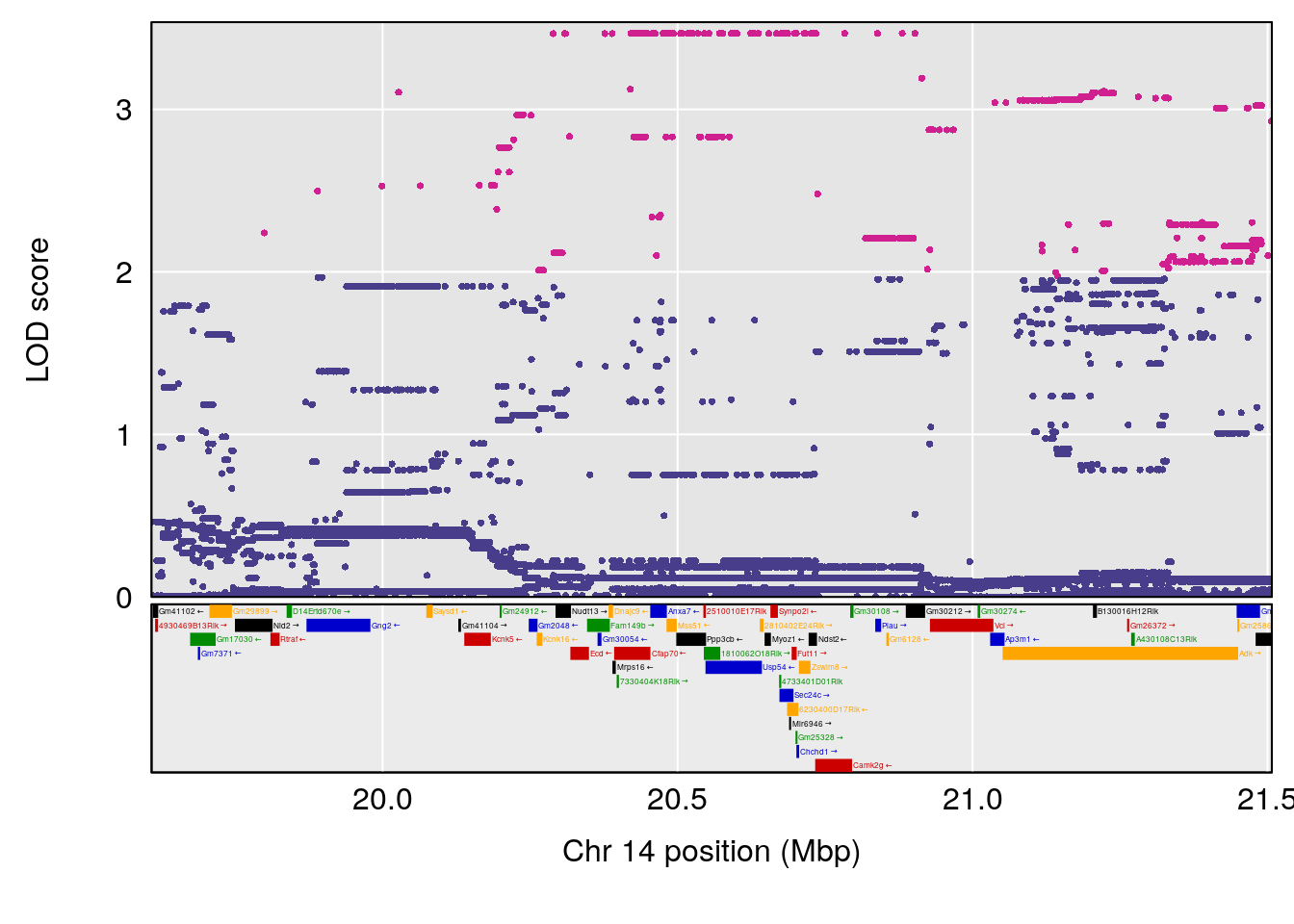
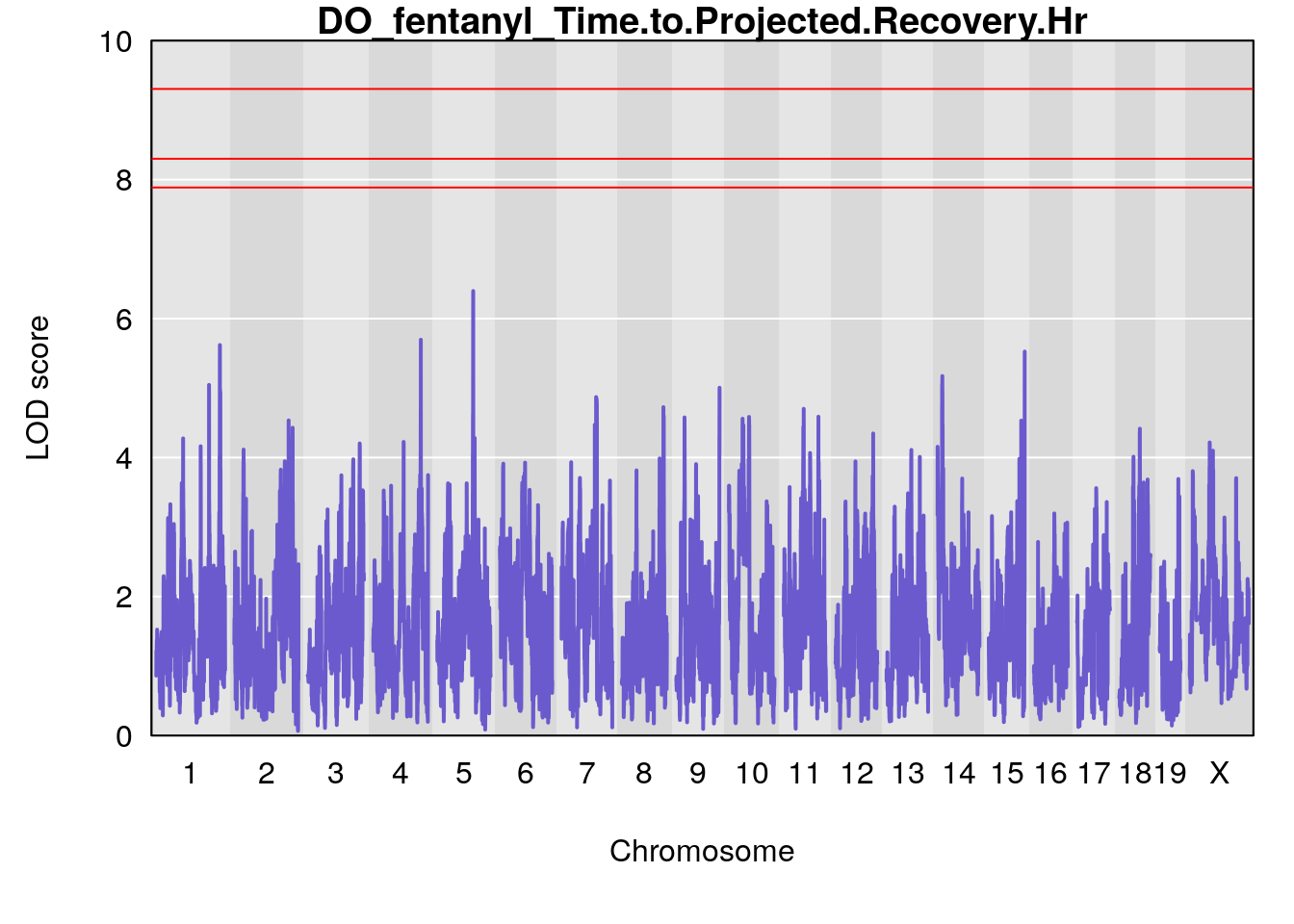
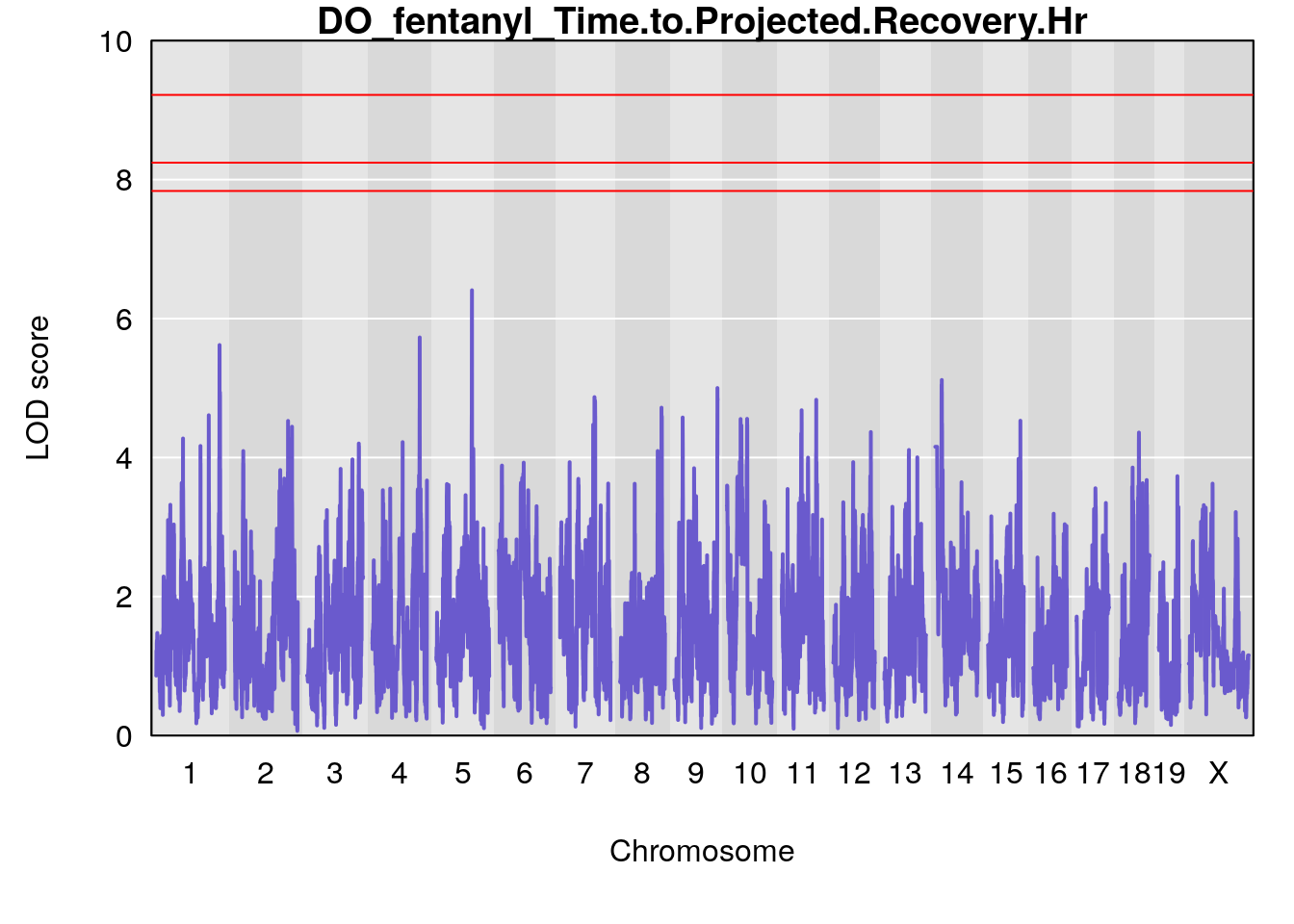
# [1] "Time.to.Projected.Recovery.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 5 103.1582 6.396816 102.756 103.3132
# [1] 1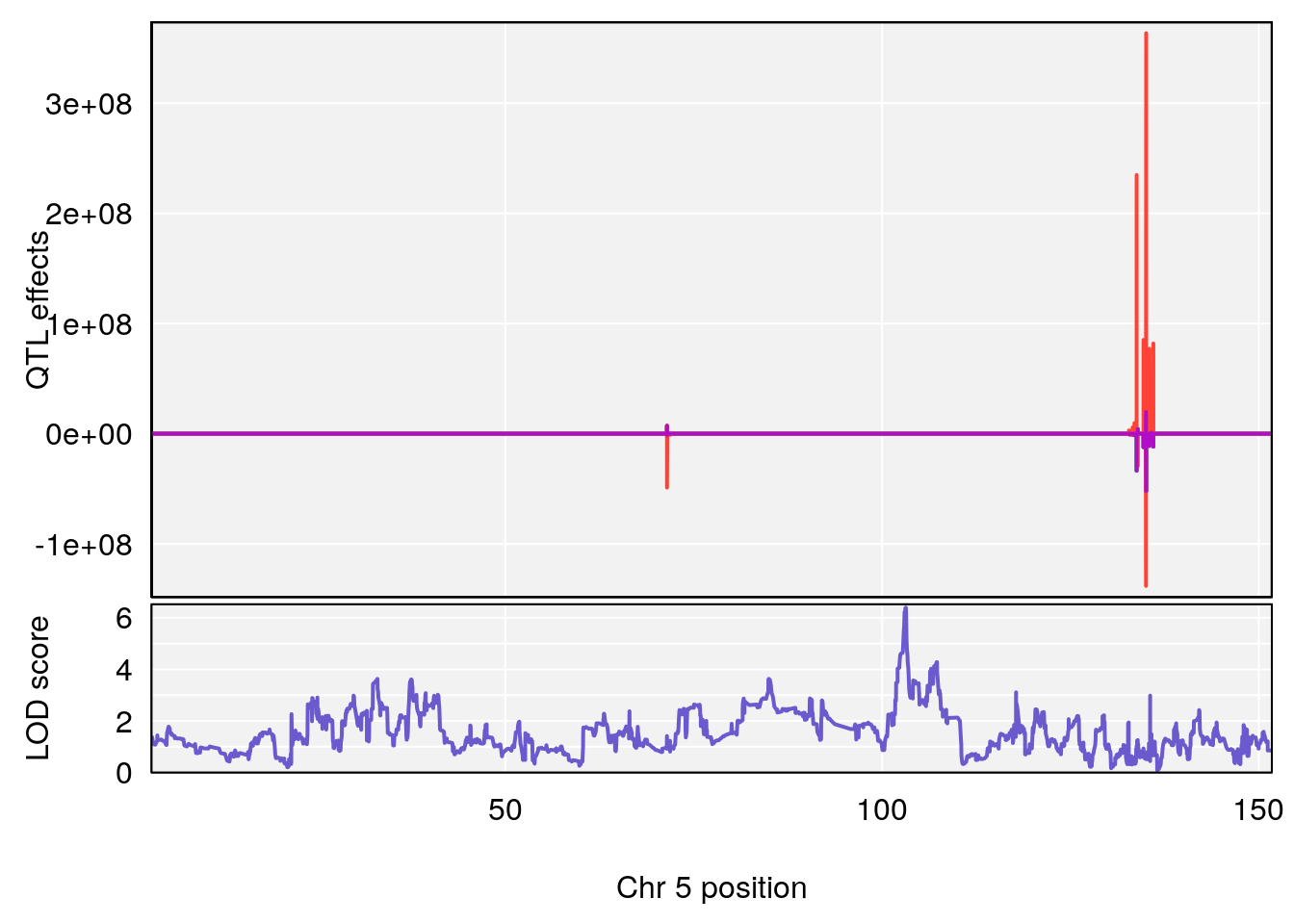
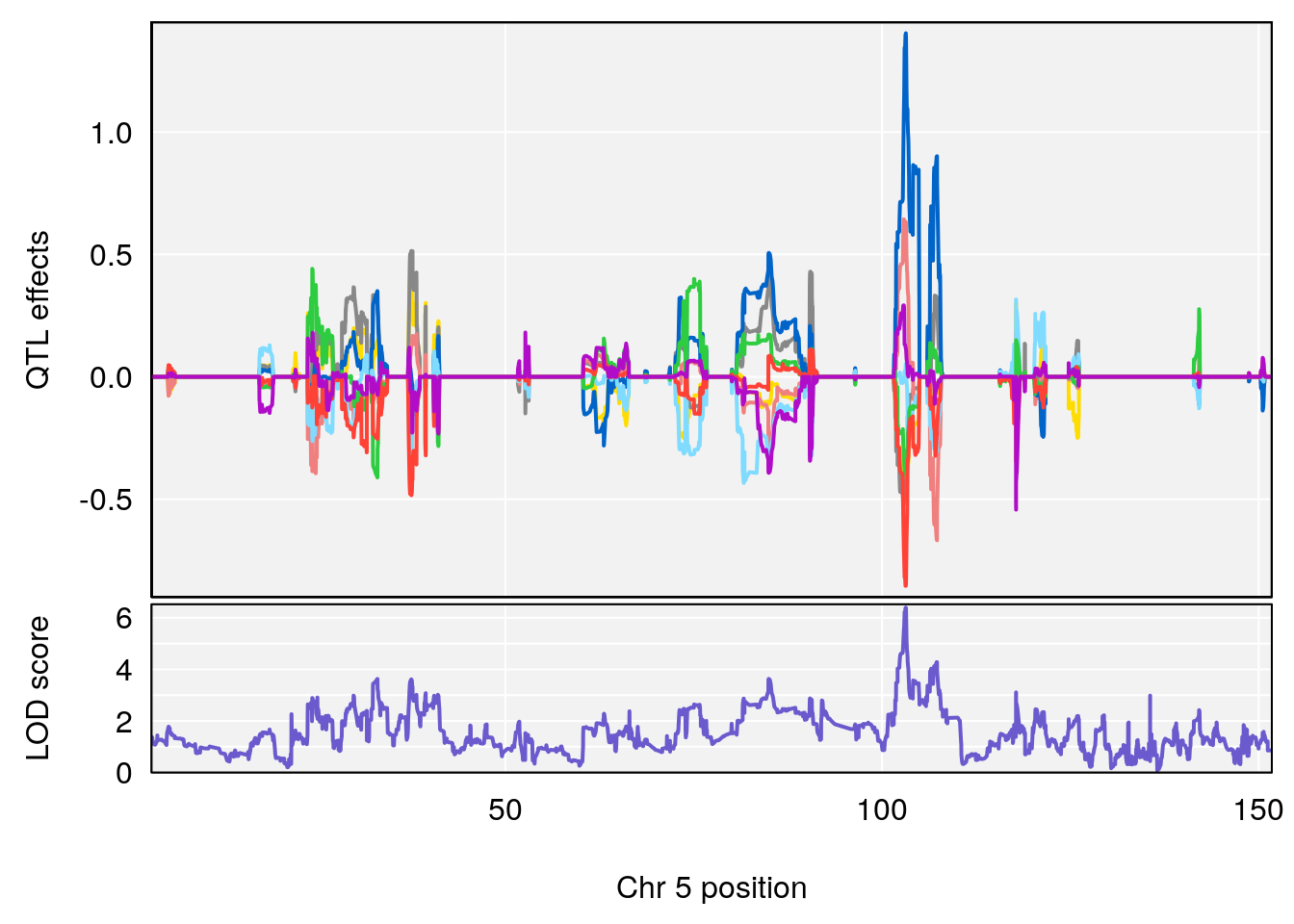
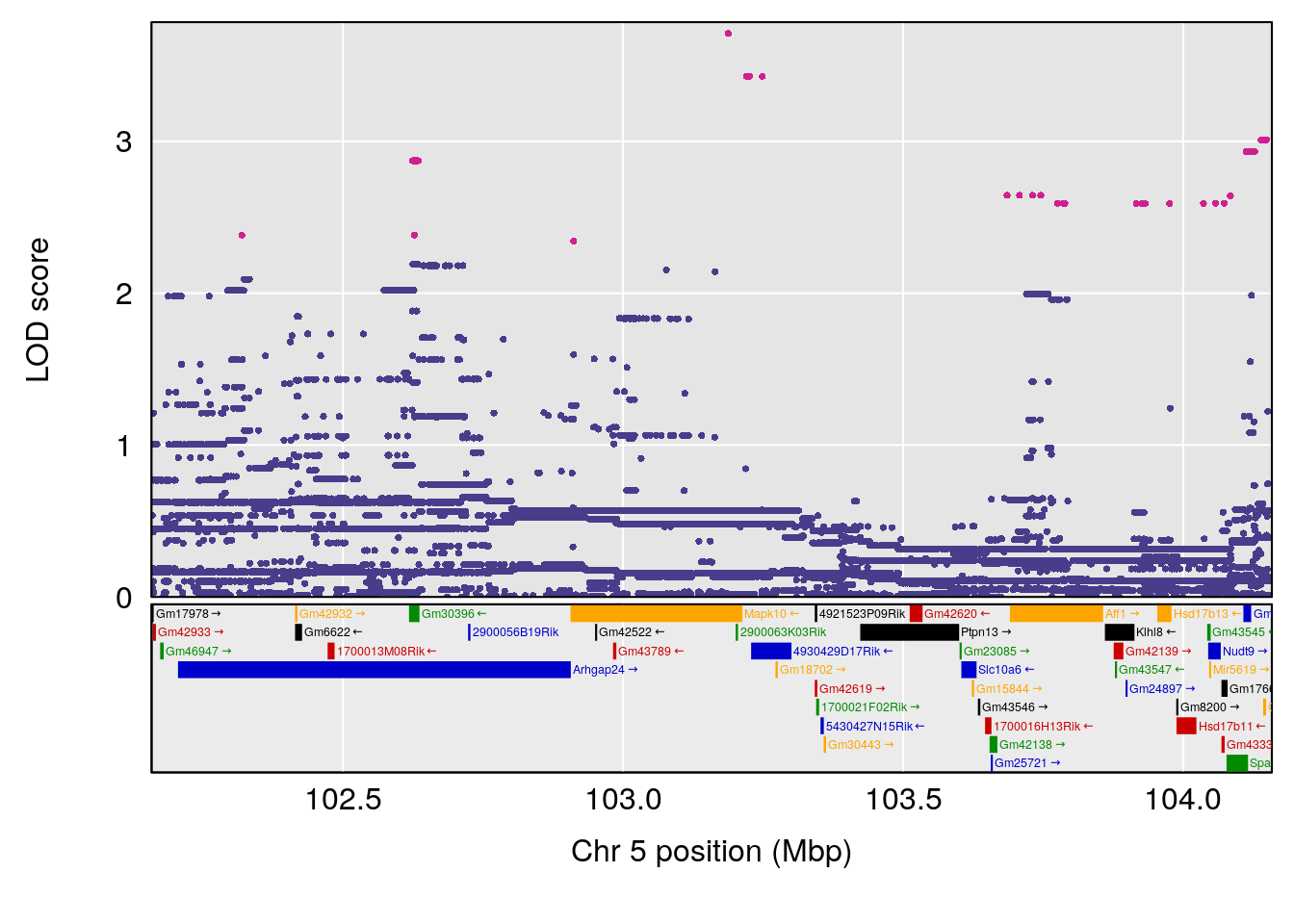
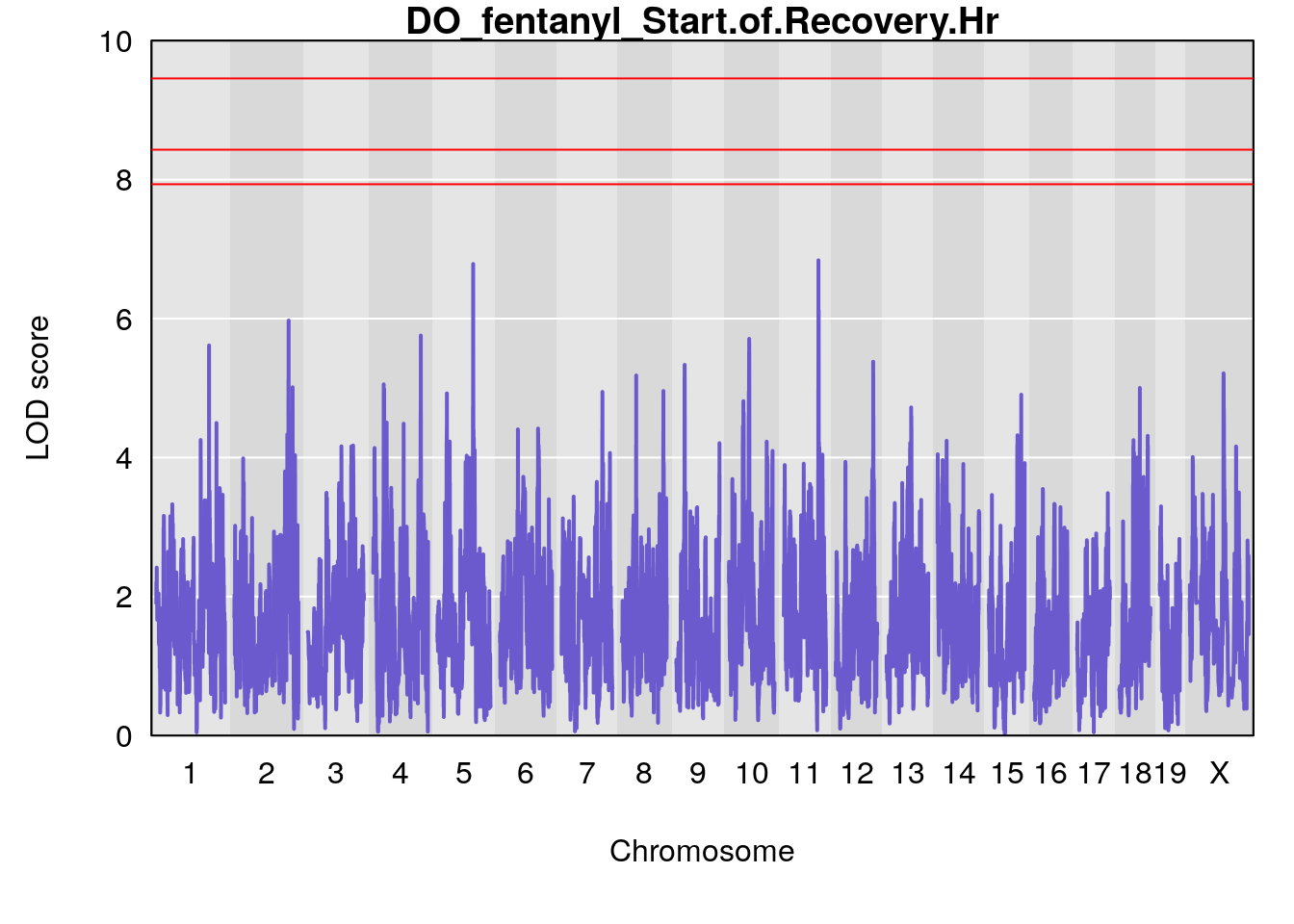
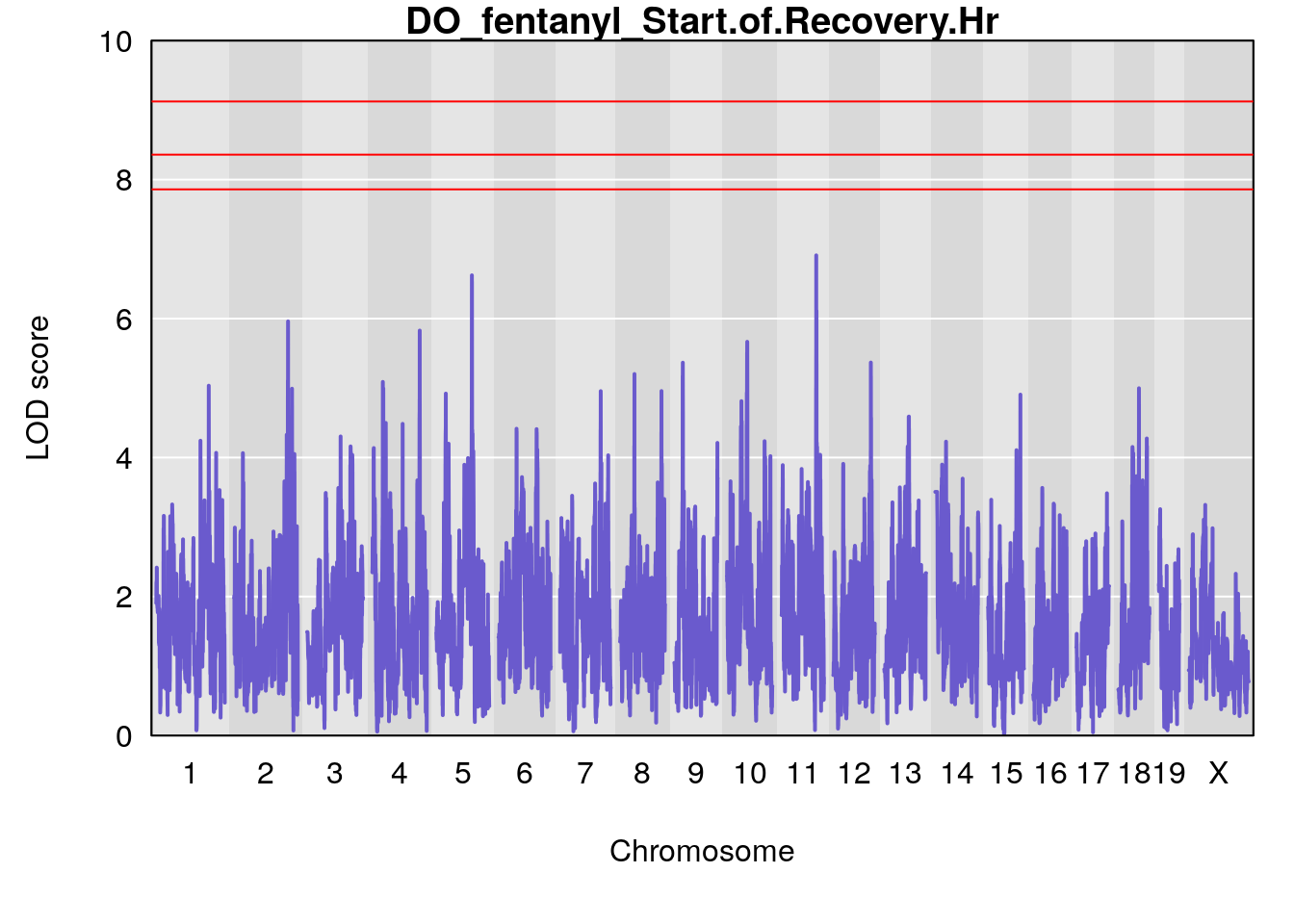
# [1] "Start.of.Recovery.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 5 102.9928 6.785795 102.81969 103.3132
# 2 1 pheno1 11 100.0882 6.838855 99.91068 100.9290
# [1] 1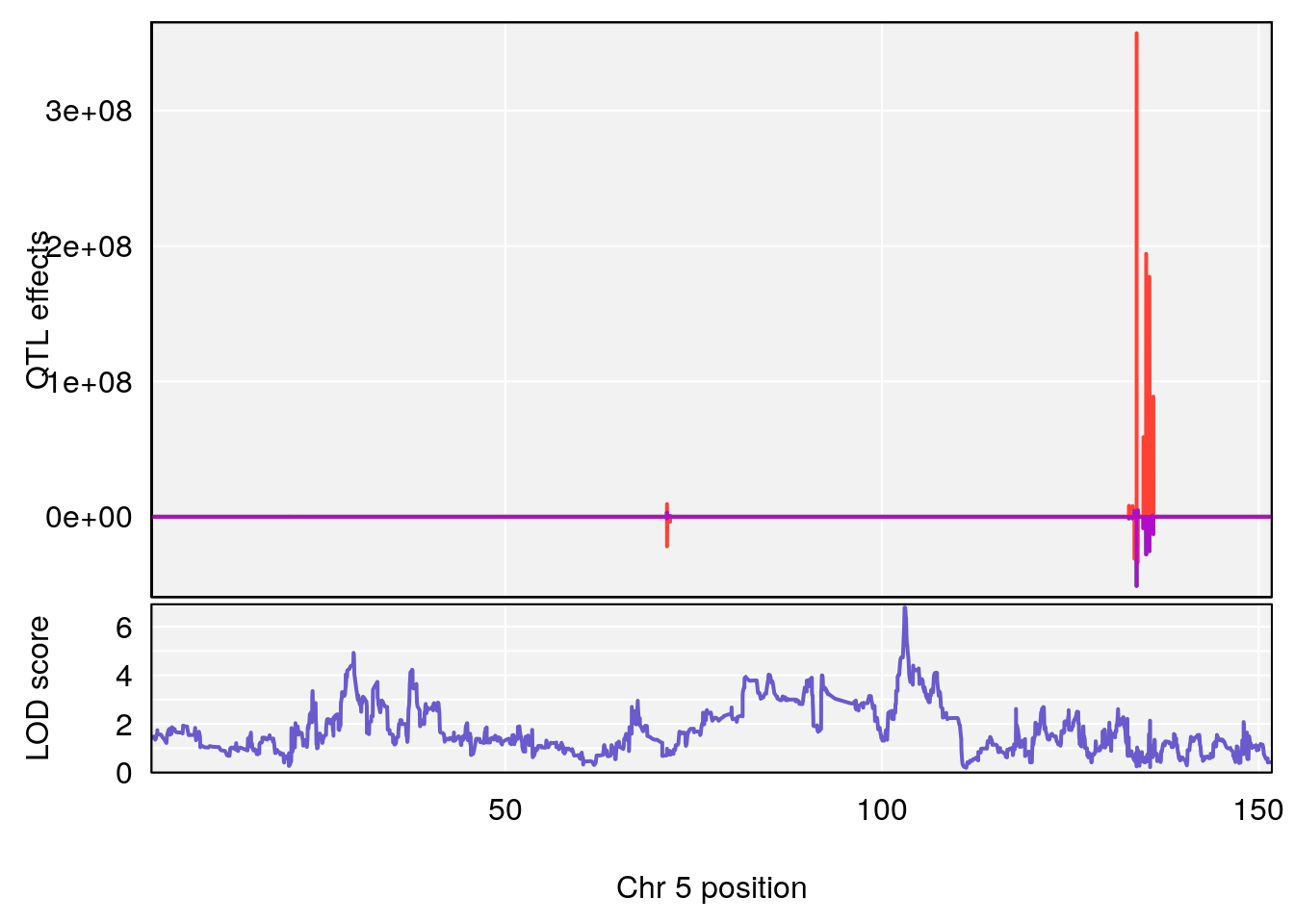
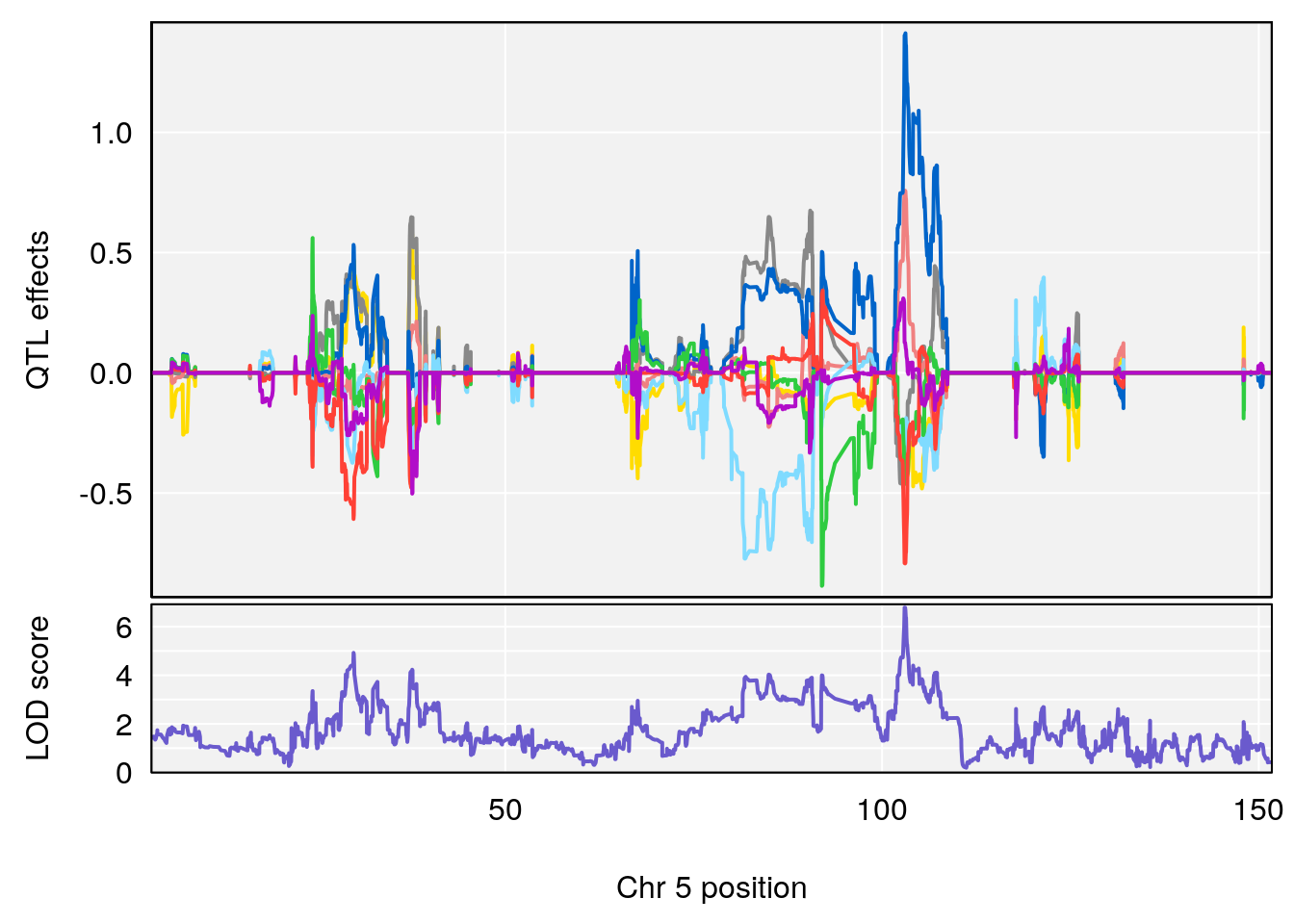
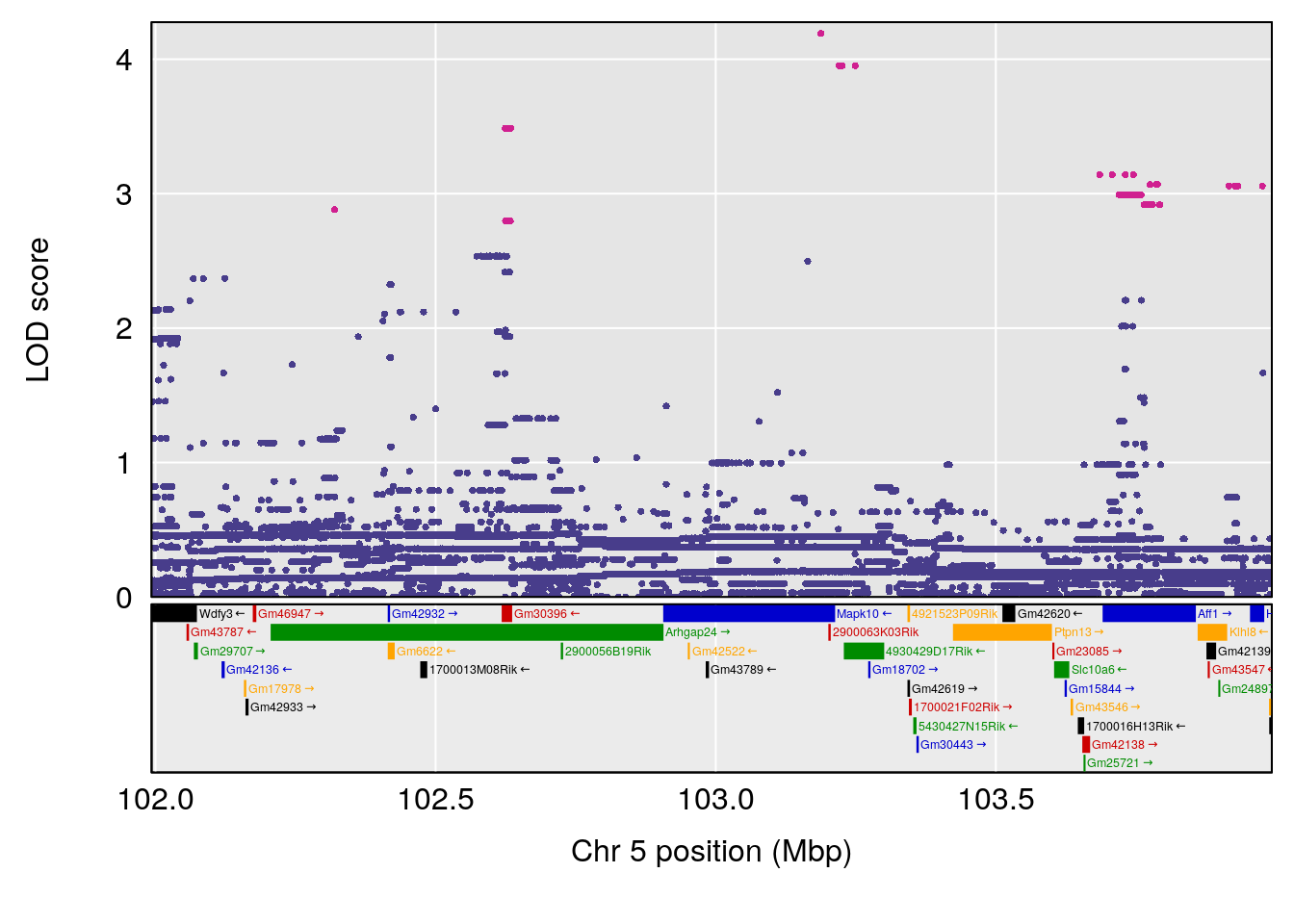
# [1] 2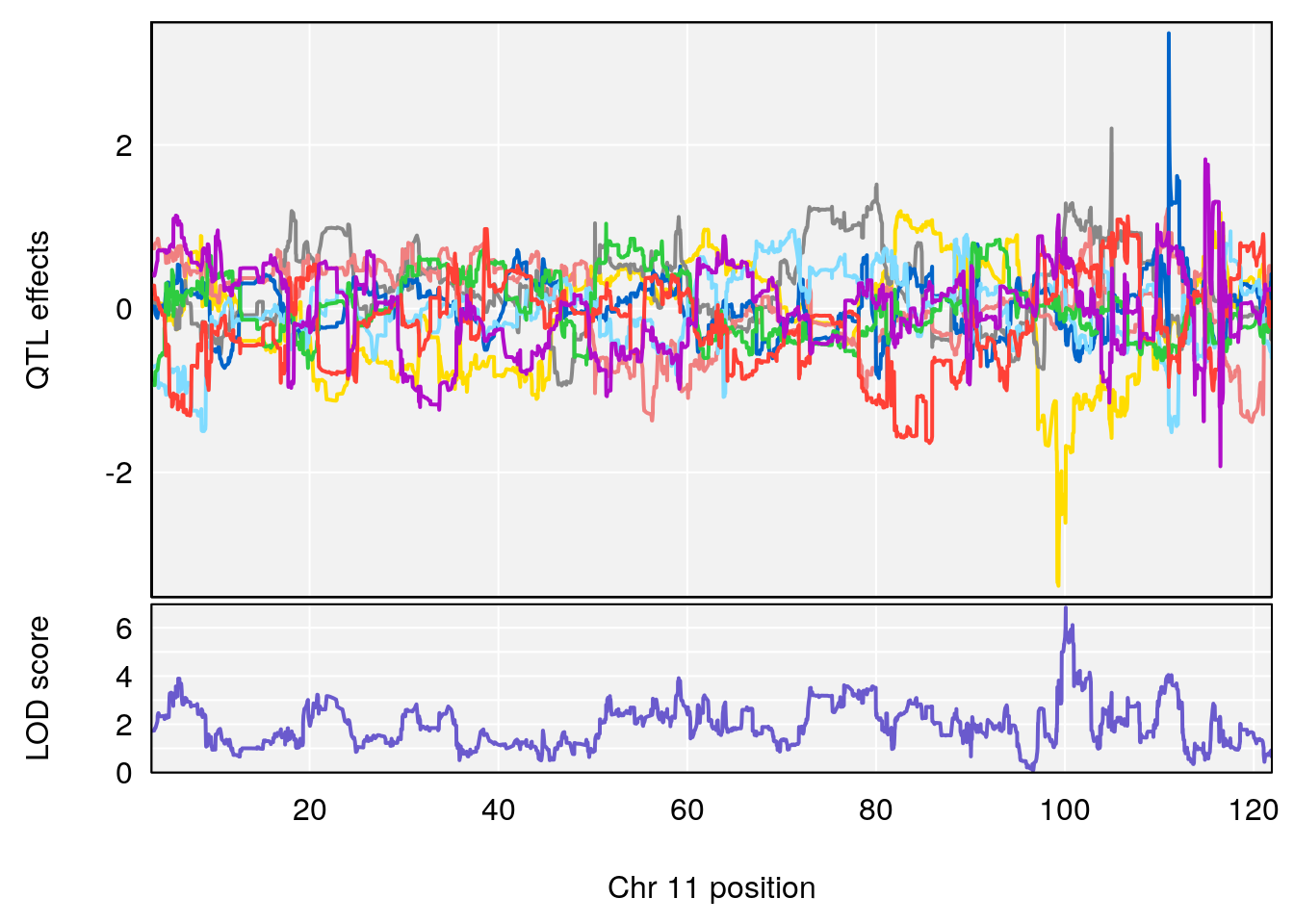
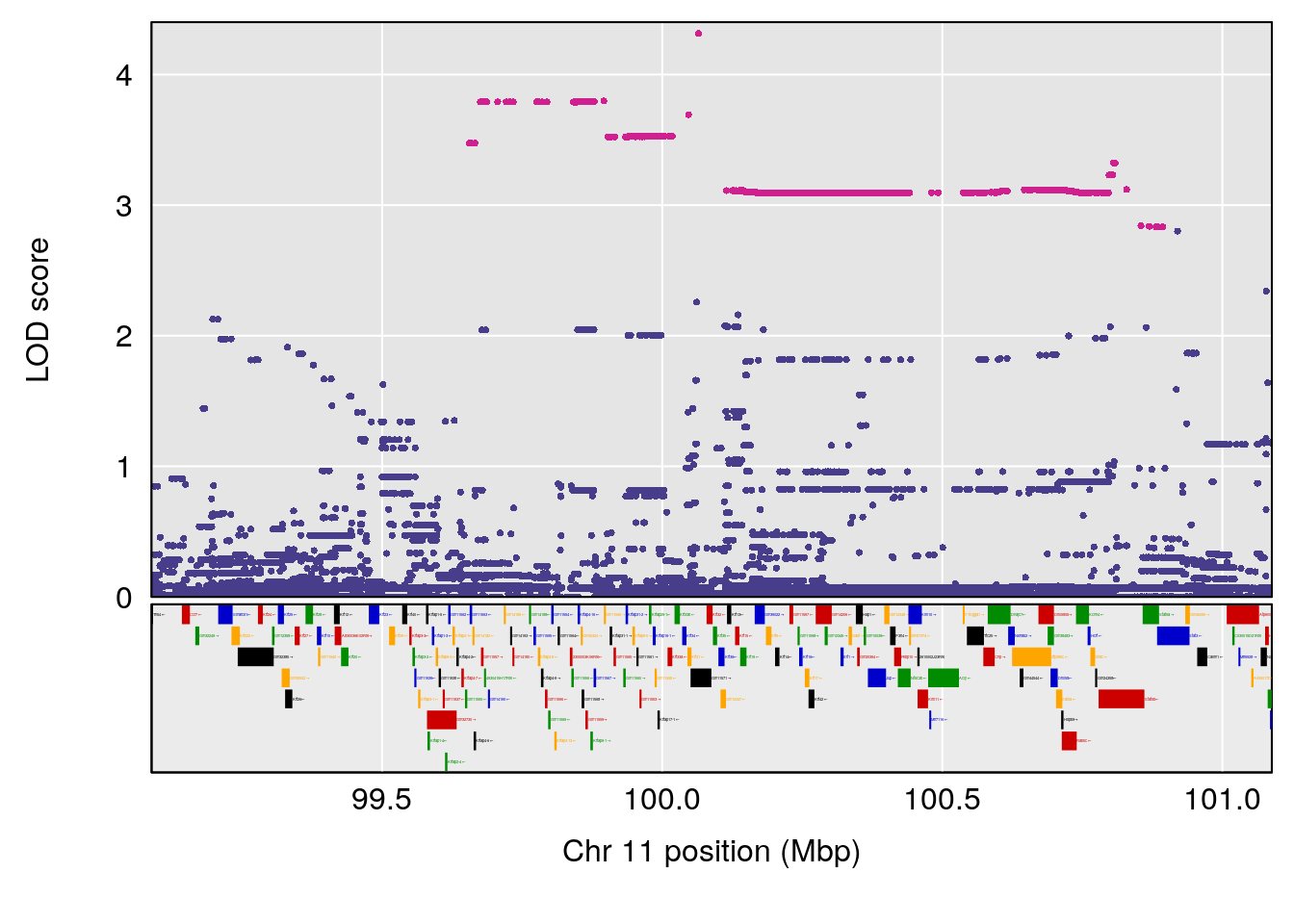
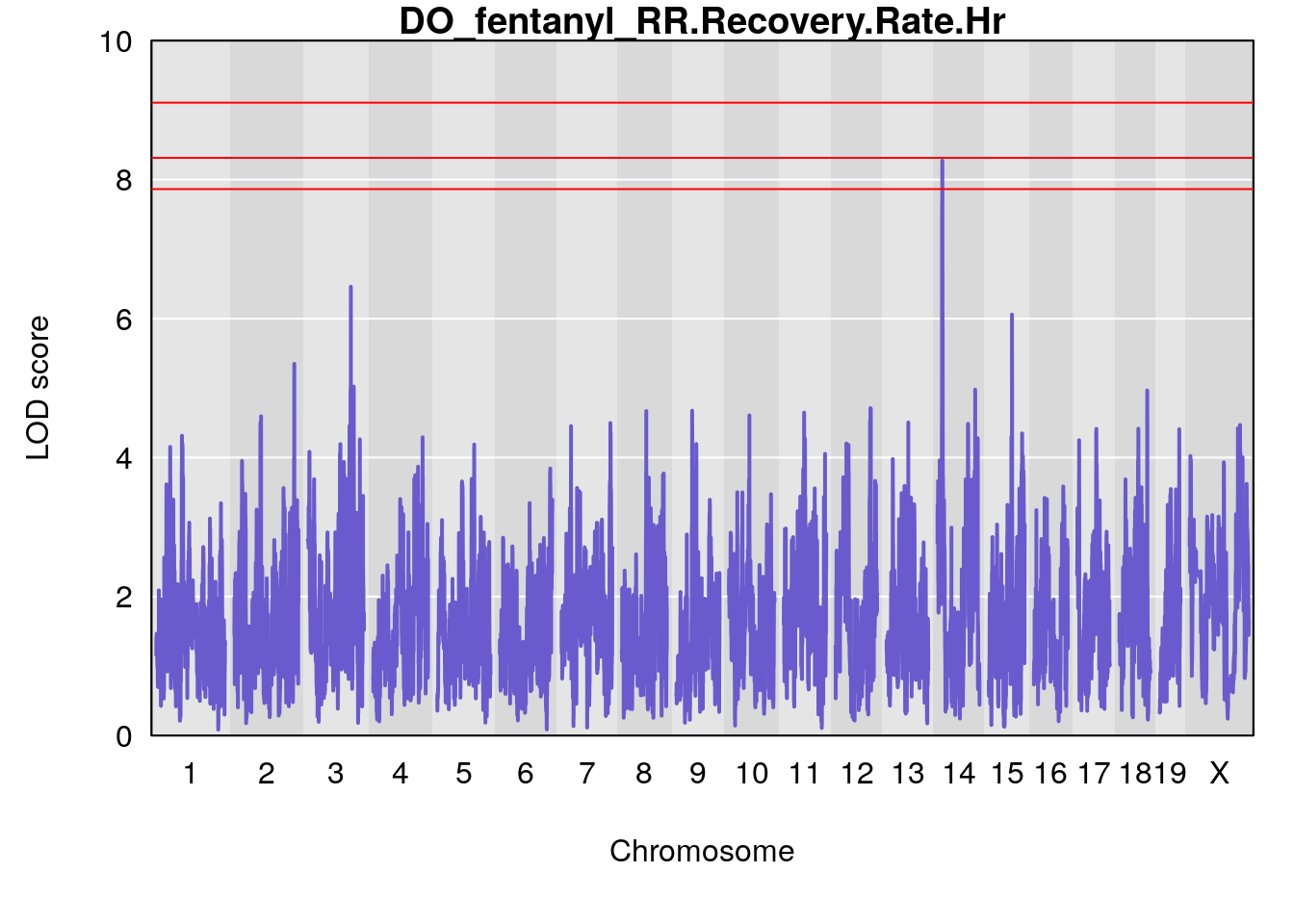
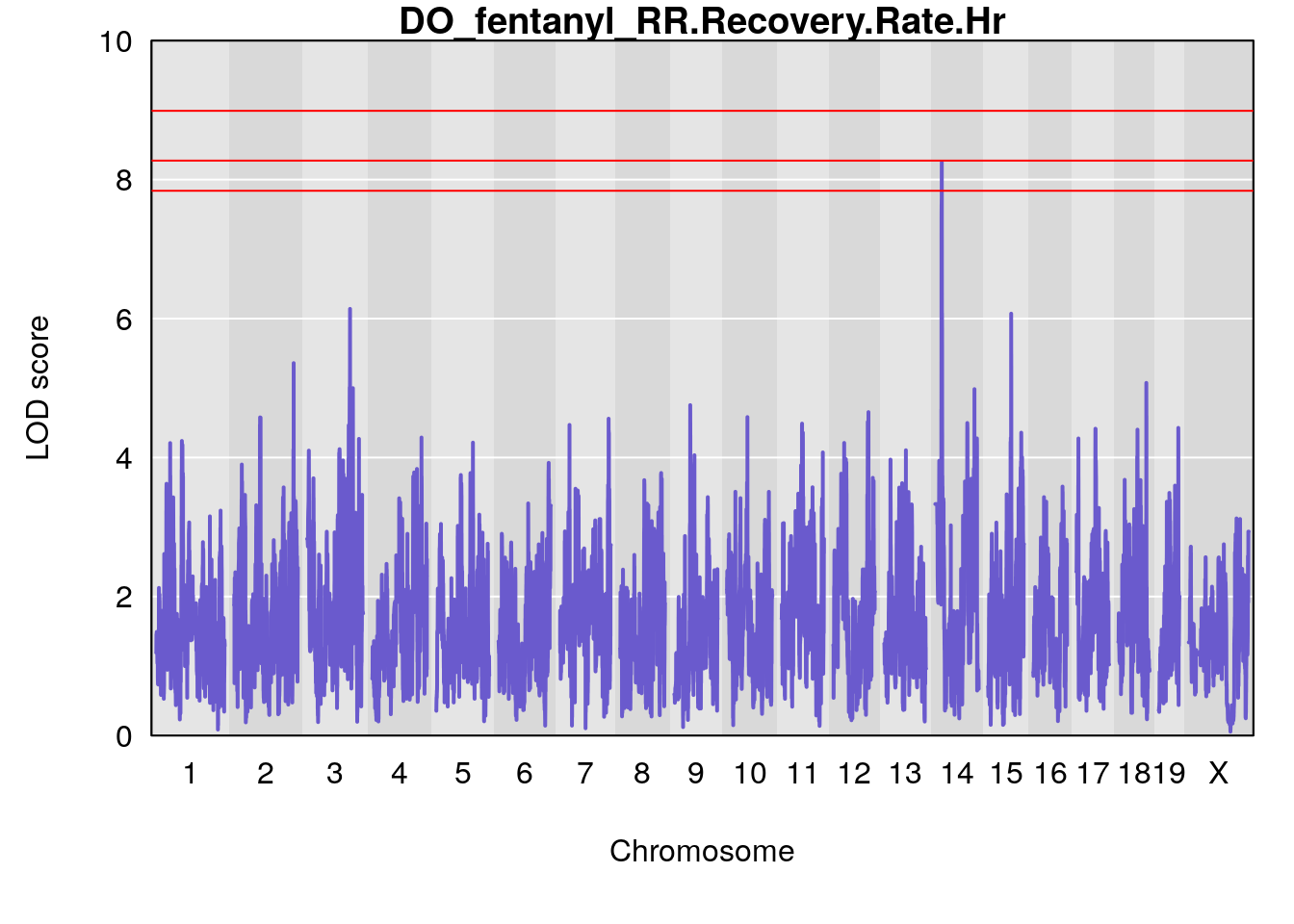
# [1] "RR.Recovery.Rate.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 3 123.39203 6.459037 122.42195 131.46627
# 2 1 pheno1 14 21.13855 8.271971 20.26372 21.51874
# 3 1 pheno1 15 68.04193 6.058226 68.00208 68.45217
# [1] 1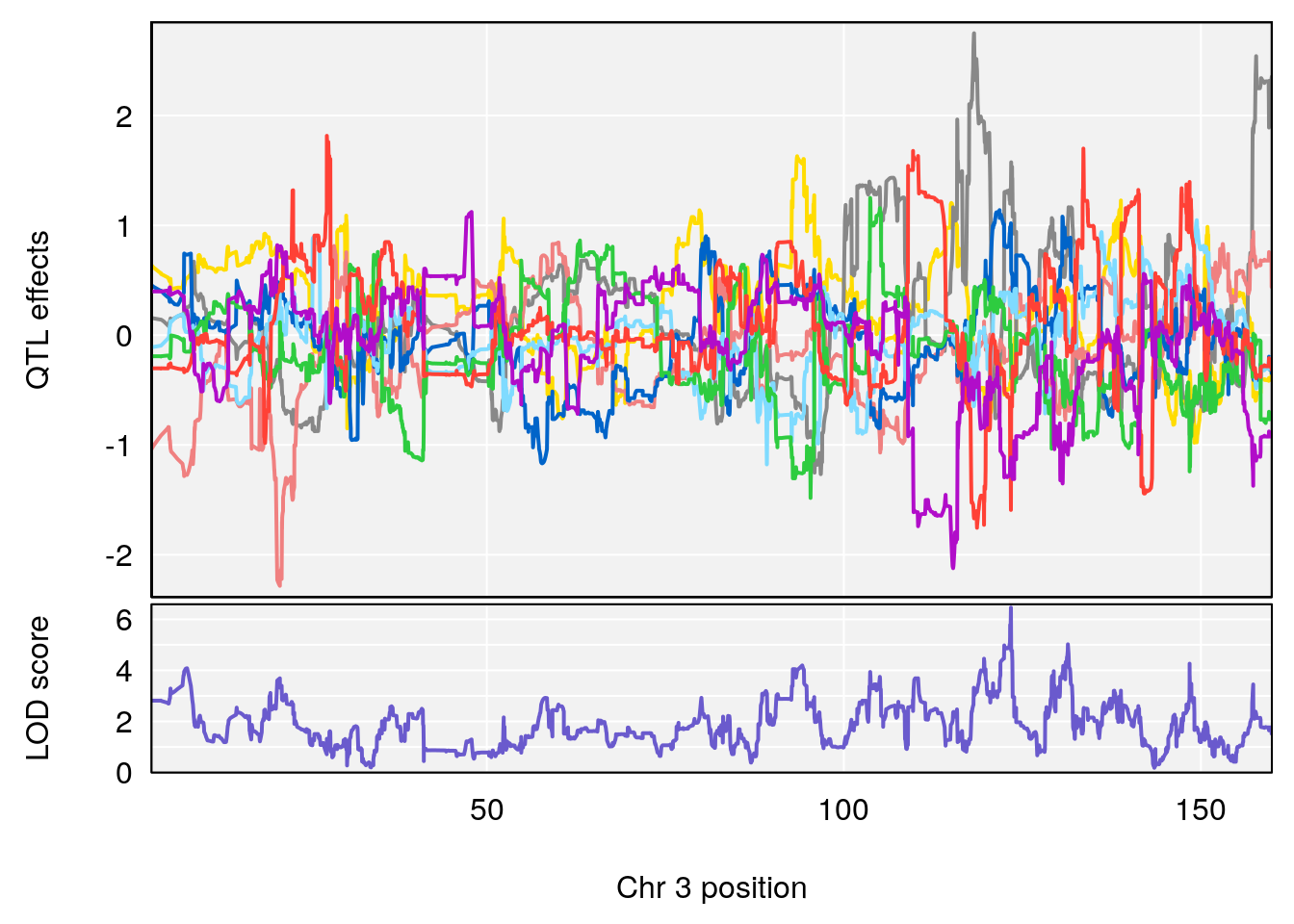
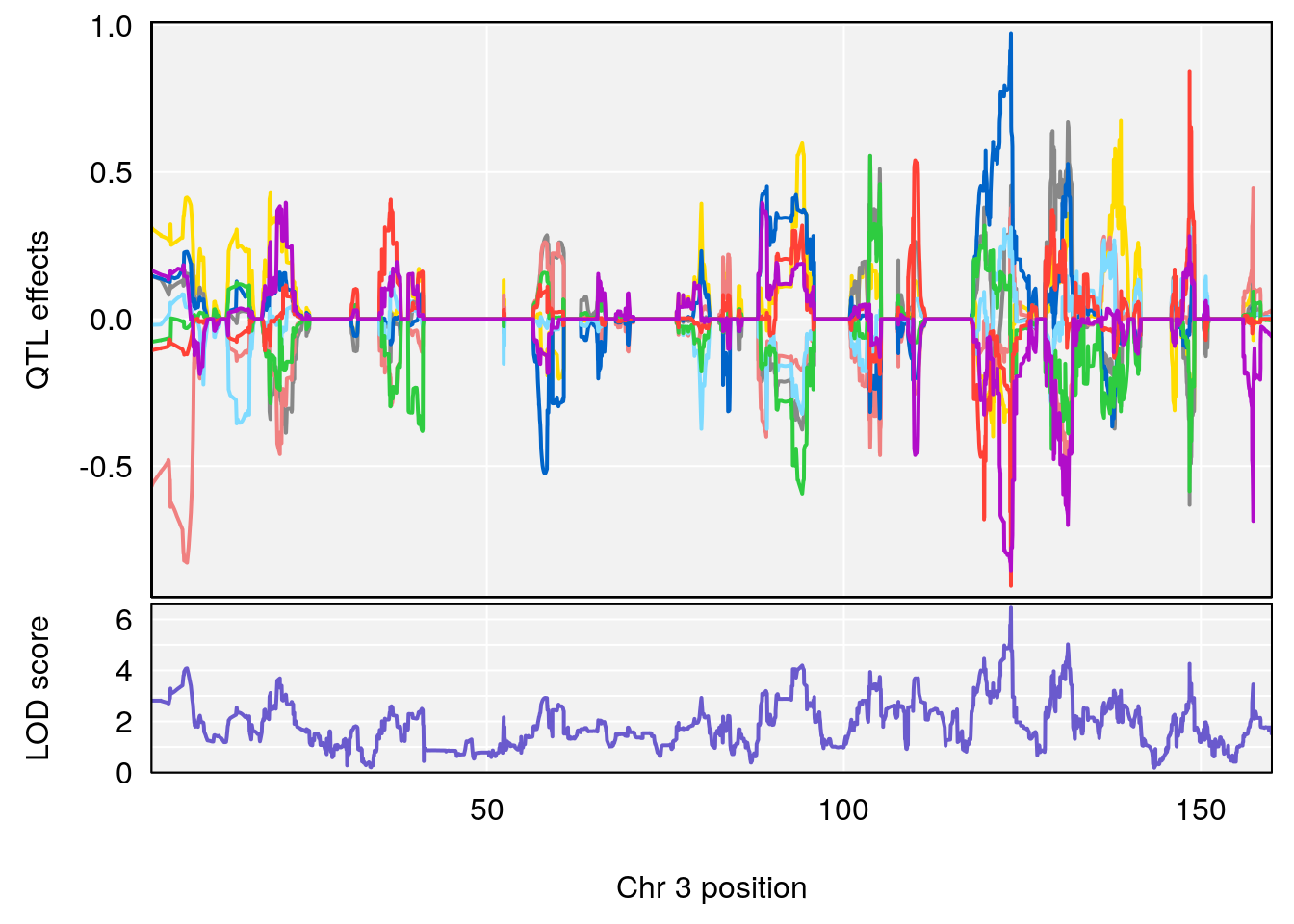
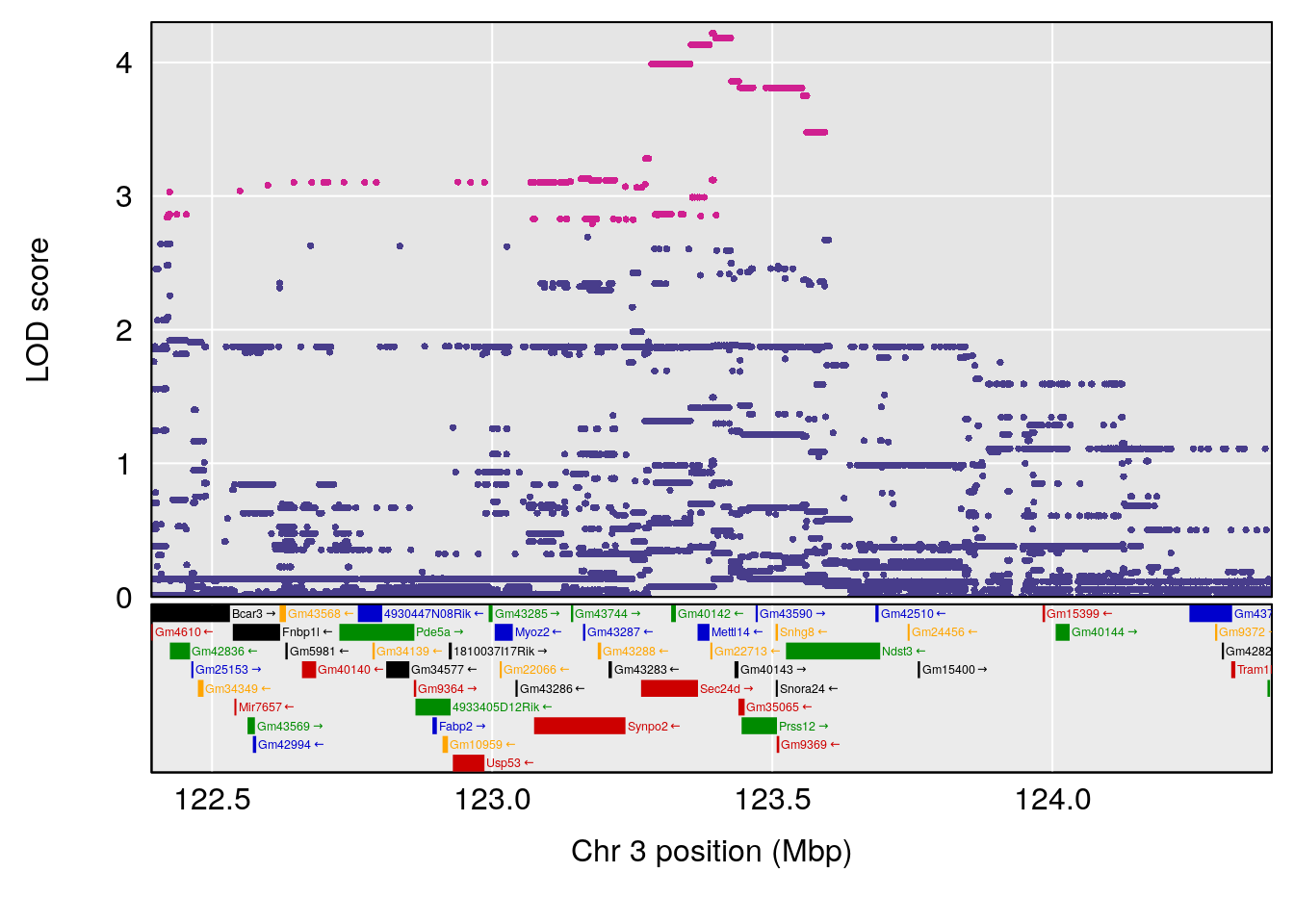
# [1] 2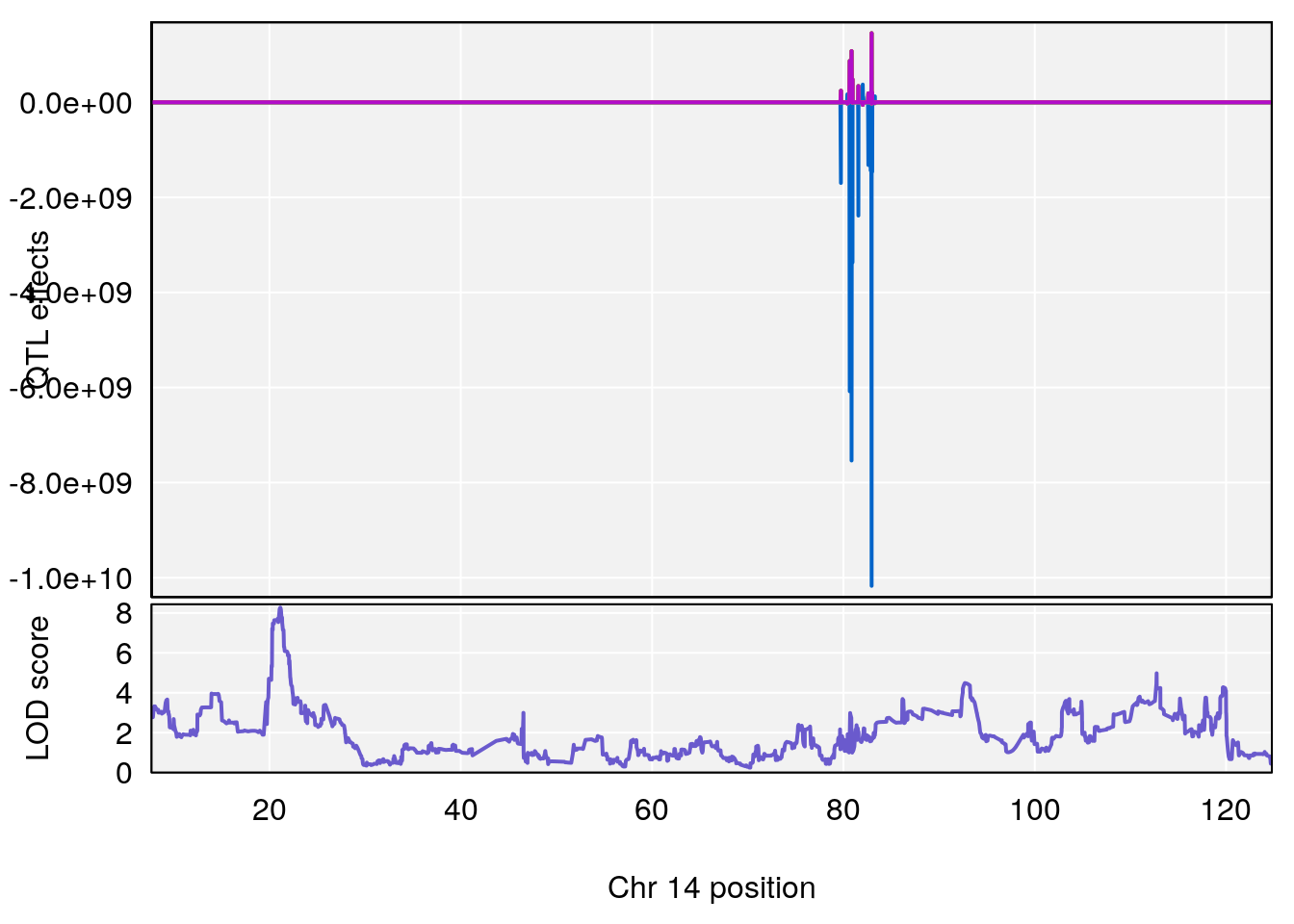
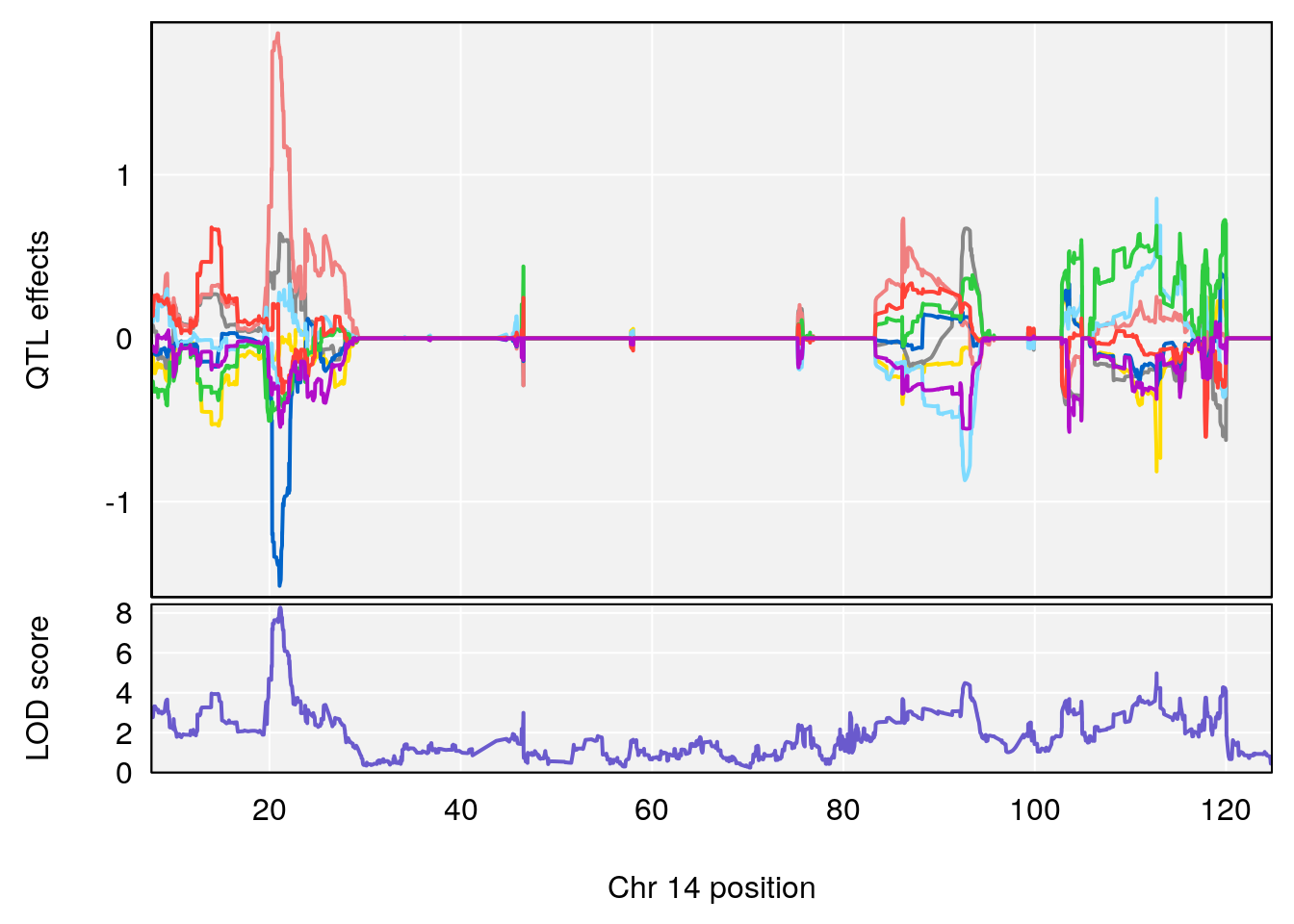
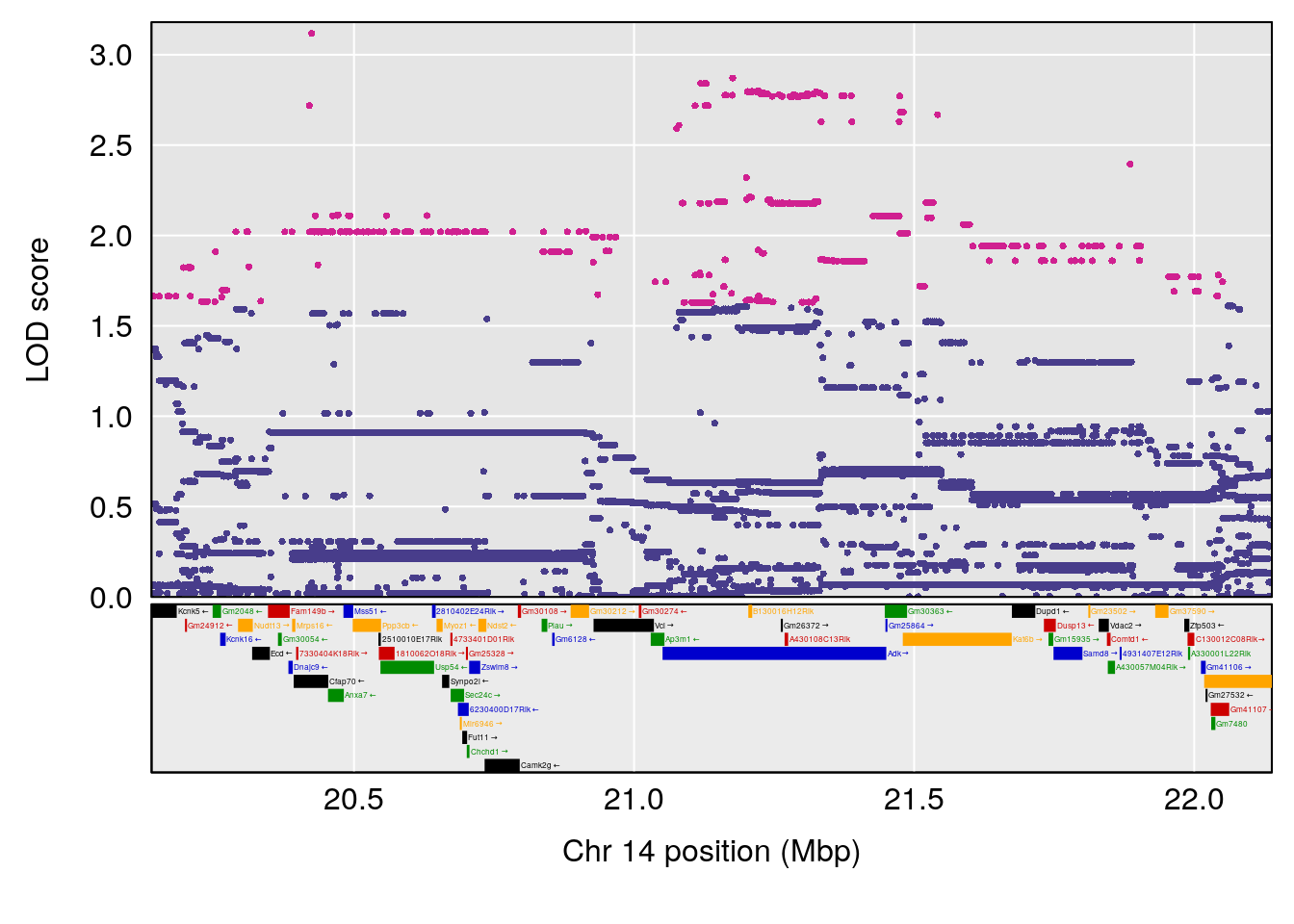
# [1] 3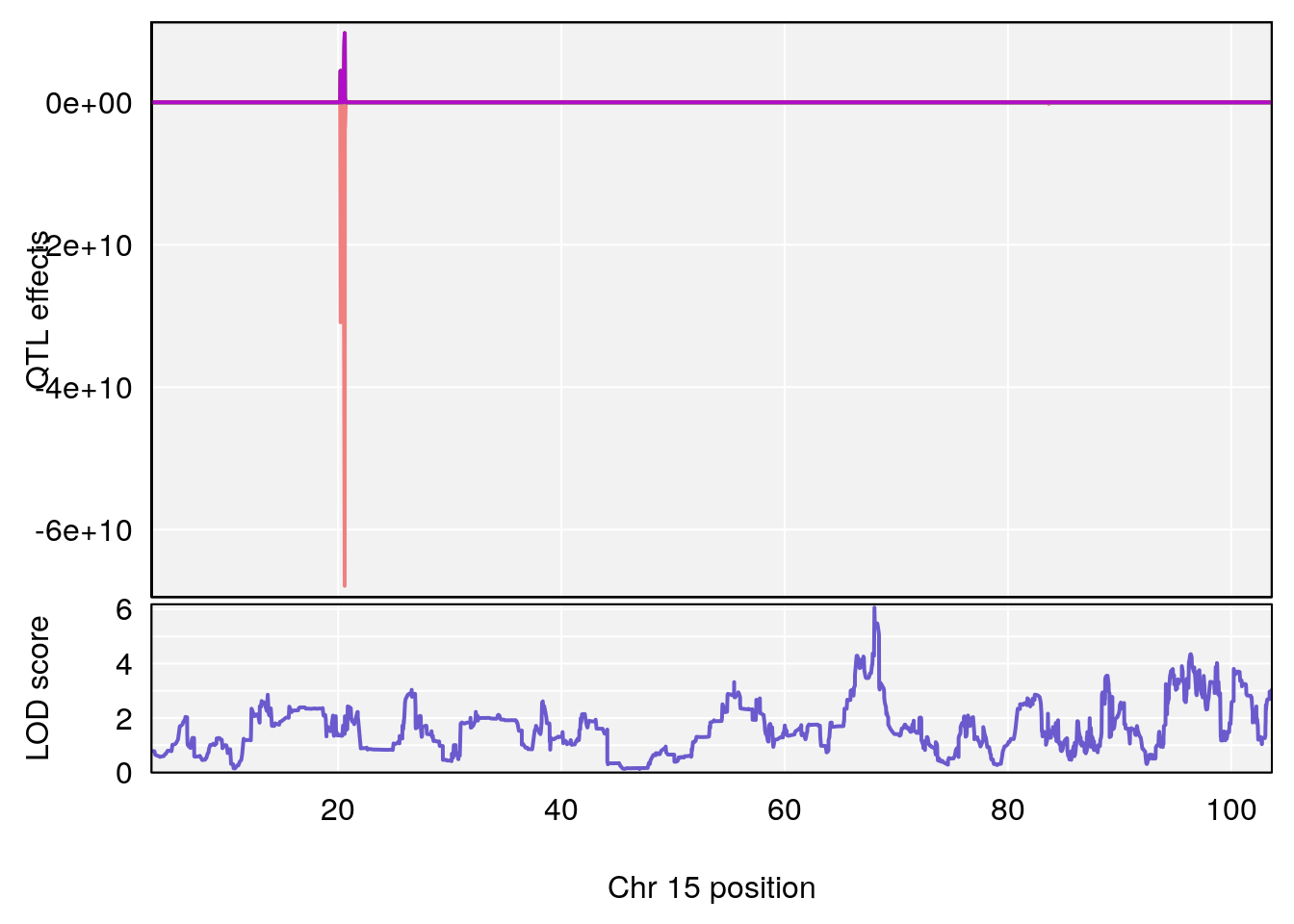
#save peaks coeff plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=gm$pmap, threshold=6, drop=1.5)
fname <- paste("output/Fentanyl/DO_fentanyl_array_alternate_metrics_",i,"_coefplot.pdf",sep="")
pdf(file = fname, width = 16, height =8)
if(dim(peaks)[1] != 0){
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c1[[i]][[p]], gm$pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot_coefCC(coef_c2[[i]][[p]], gm$pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
}
dev.off()
}
# [1] "Time.to.Dead.Hr"
# [1] 1
# [1] 2
# [1] "RR.Depression.Rate.Hr"
# [1] "Time.to.Steady.RR.Depression.Hr"
# [1] "Min.Depression.RR"
# [1] 1
# [1] 2
# [1] "Mean.Depression.BR"
# [1] 1
# [1] 2
# [1] 3
# [1] "Steady-State.Depression.Duration.Hr"
# [1] 1
# [1] 2
# [1] 3
# [1] "Time.to.Threshold.Recovery.Hr"
# [1] 1
# [1] 2
# [1] "Time.to.Projected.Recovery.Hr"
# [1] 1
# [1] "Start.of.Recovery.Hr"
# [1] 1
# [1] 2
# [1] "RR.Recovery.Rate.Hr"
# [1] 1
# [1] 2
# [1] 3
#save peaks coeff blup plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=gm$pmap, threshold=6, drop=1.5)
fname <- paste("output/Fentanyl/DO_fentanyl_array_alternate_metrics_",i,"_coefplot_blup.pdf",sep="")
pdf(file = fname, width = 16, height =8)
if(dim(peaks)[1] != 0){
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c1[[i]][[p]], gm$pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot_coefCC(coef_c2[[i]][[p]], gm$pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
}
dev.off()
}
# [1] "Time.to.Dead.Hr"
# [1] 1
# [1] 2
# [1] "RR.Depression.Rate.Hr"
# [1] "Time.to.Steady.RR.Depression.Hr"
# [1] "Min.Depression.RR"
# [1] 1
# [1] 2
# [1] "Mean.Depression.BR"
# [1] 1
# [1] 2
# [1] 3
# [1] "Steady-State.Depression.Duration.Hr"
# [1] 1
# [1] 2
# [1] 3
# [1] "Time.to.Threshold.Recovery.Hr"
# [1] 1
# [1] 2
# [1] "Time.to.Projected.Recovery.Hr"
# [1] 1
# [1] "Start.of.Recovery.Hr"
# [1] 1
# [1] 2
# [1] "RR.Recovery.Rate.Hr"
# [1] 1
# [1] 2
# [1] 3###Plot qtl mapping on 69K
load("output/Fentanyl/qtl.fentanyl.69k.alternate.metrics.out.RData")
#pmap and gmap
load("data/69k_grid_pgmap.RData")
#genome-wide plot
for(i in names(qtl.morphine.out)){
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.morphine.out[[i]]) # overall maximum LOD score
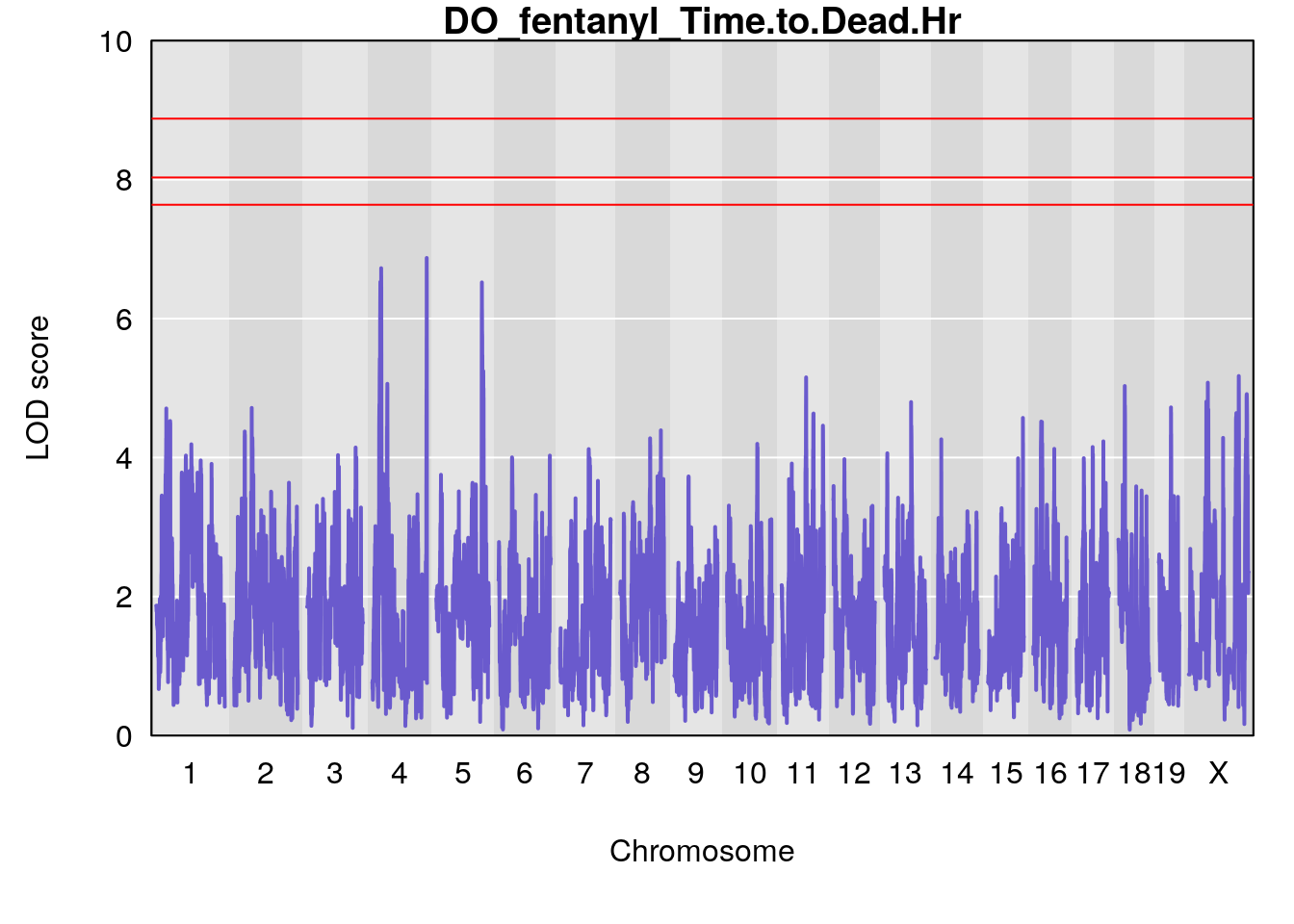
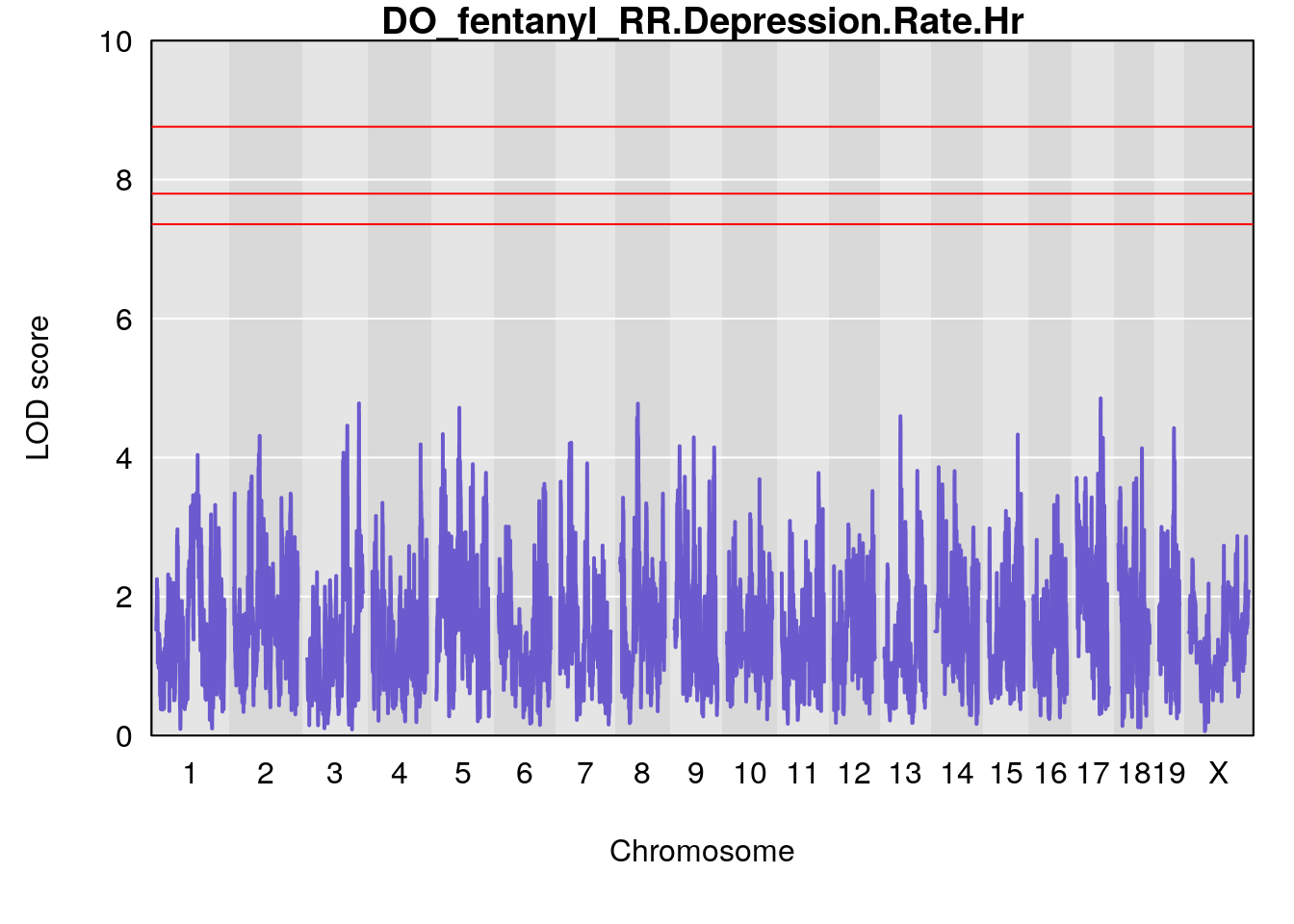
plot(qtl.morphine.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, 10))
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("DO_fentanyl_",i))
}
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
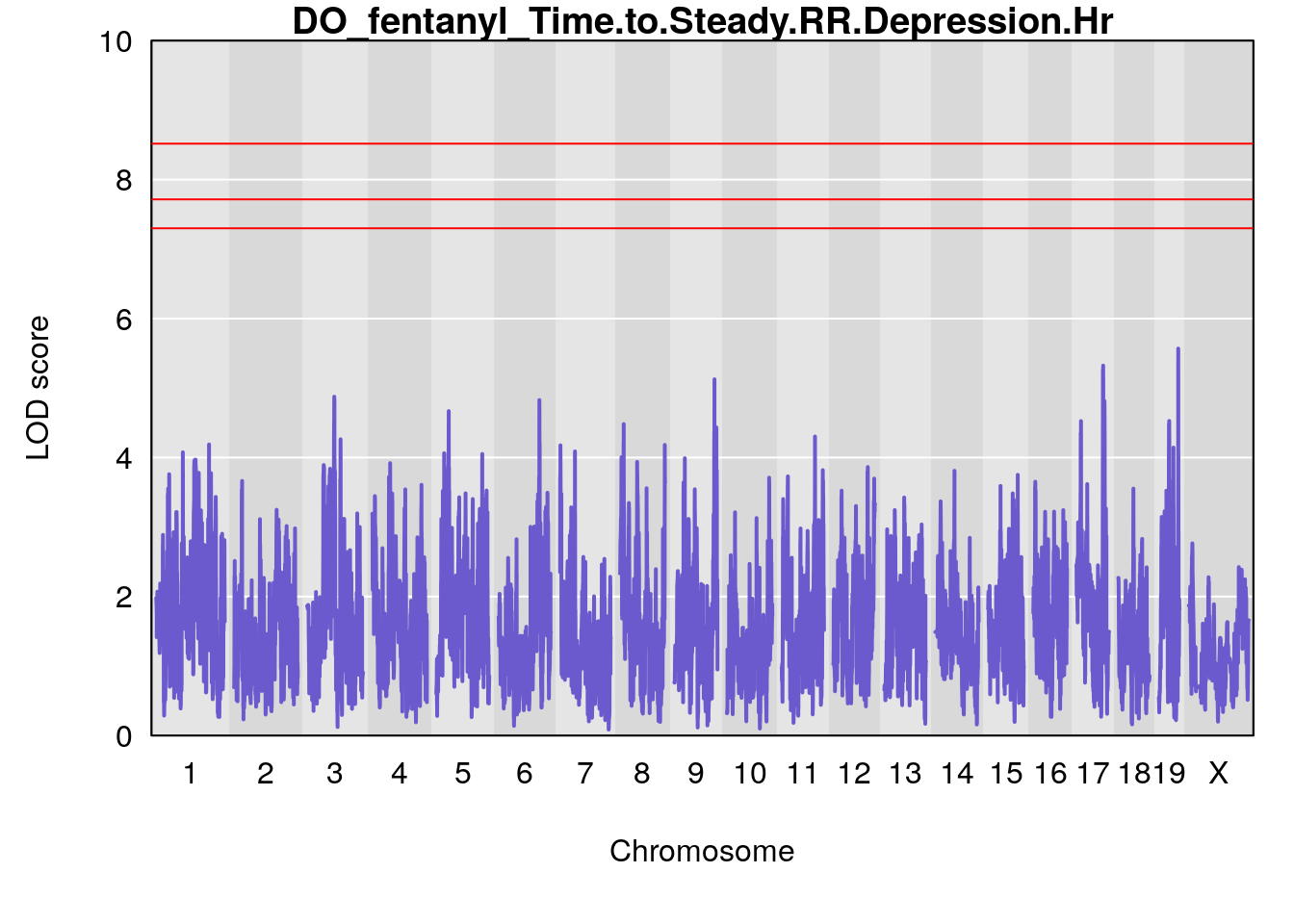
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
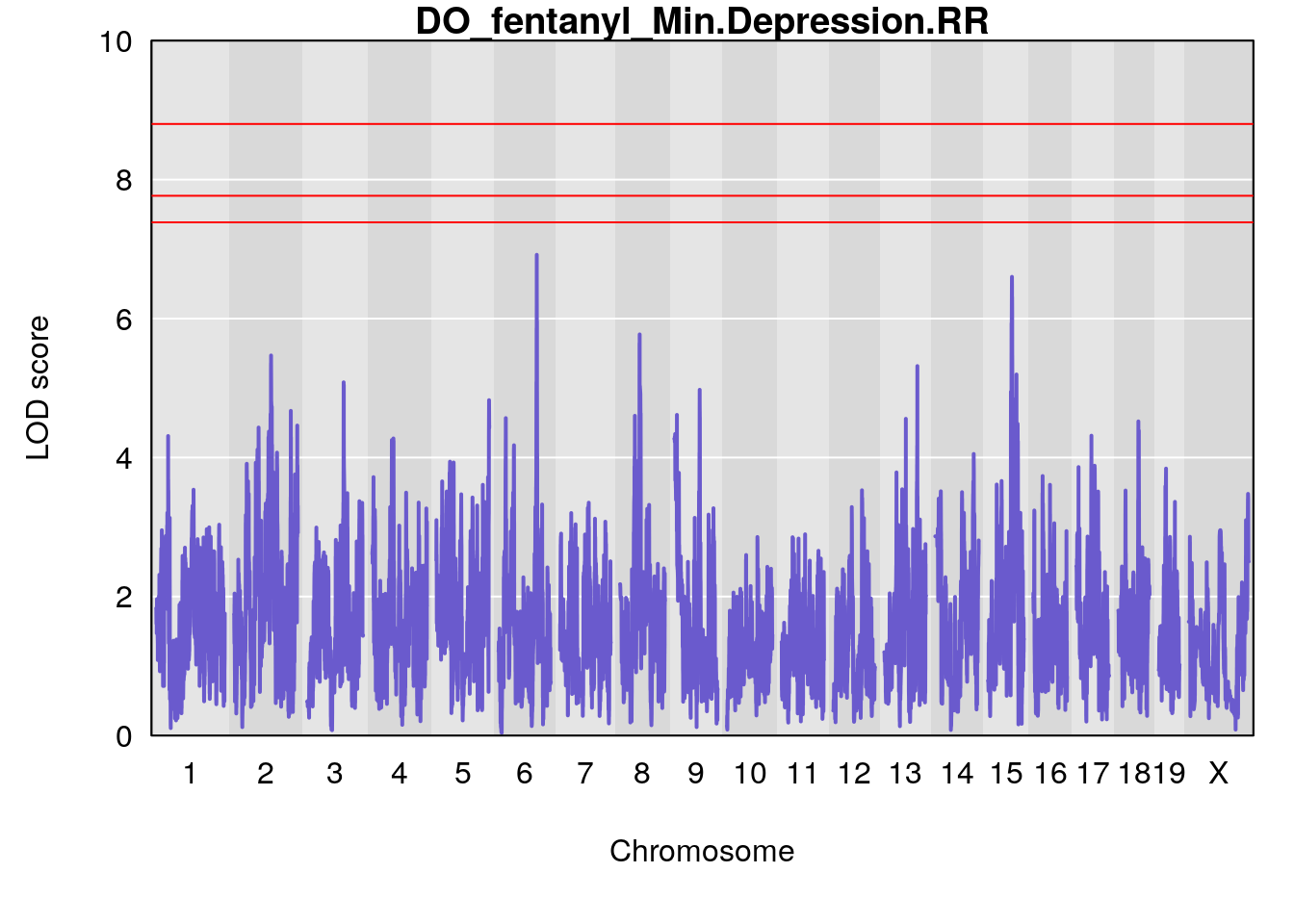
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
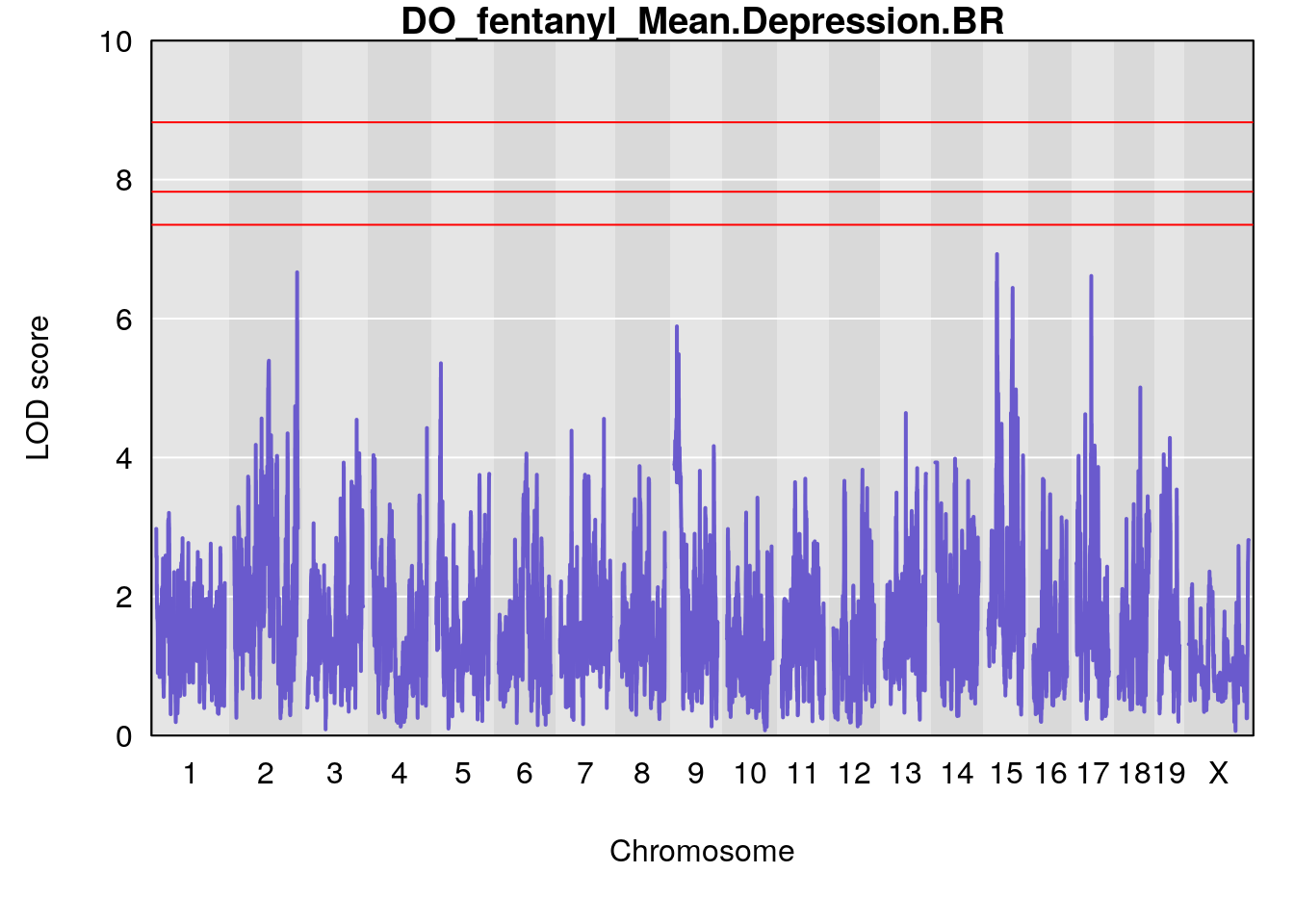
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
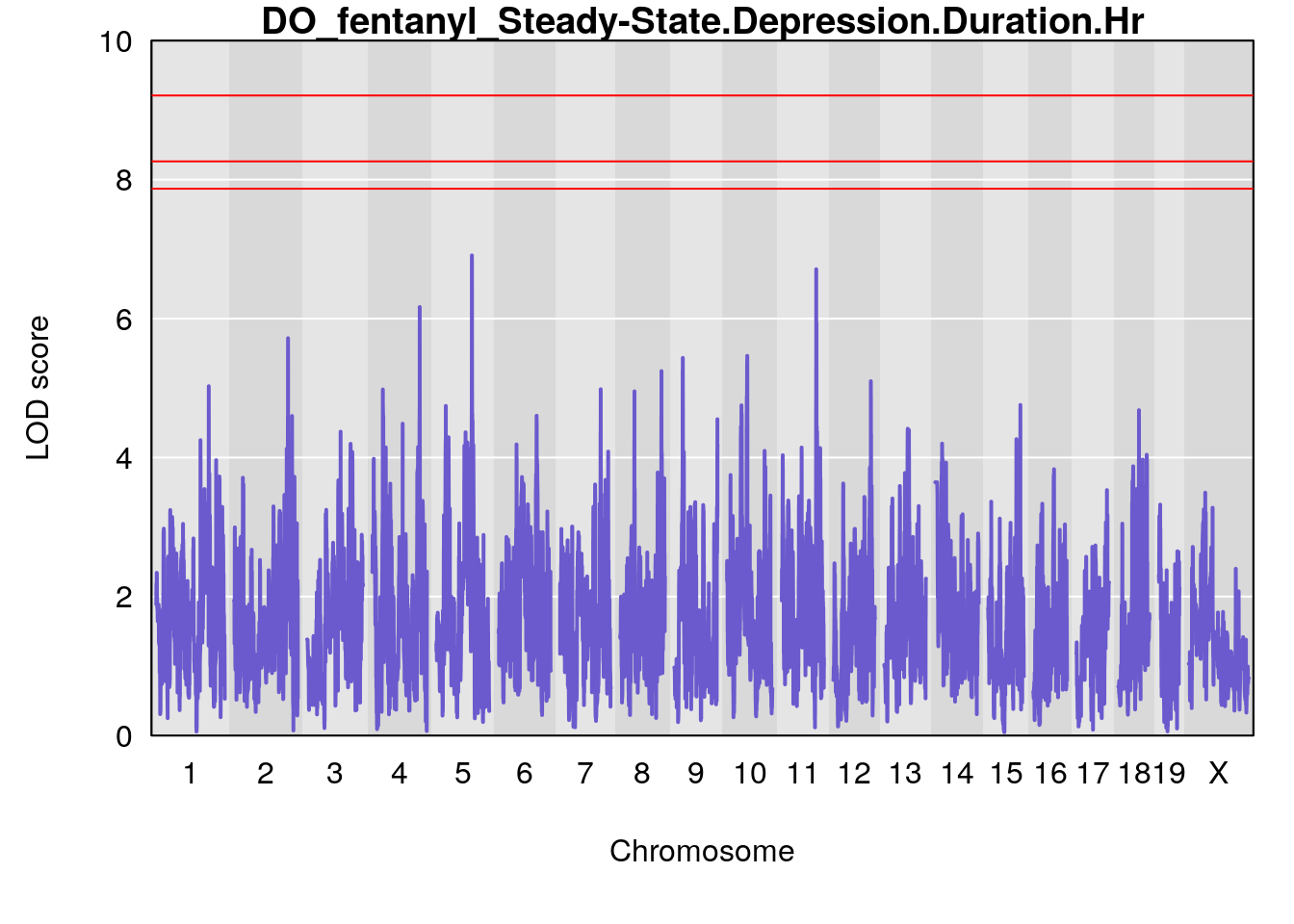
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
#save genome-wide plot
for(i in names(qtl.morphine.out)){
png(file = paste0("output/Fentanyl/DO_fentanyl_69k_alternate_metrics_", i, ".png"), width = 16, height =8, res=300, units = "in")
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.morphine.out[[i]]) # overall maximum LOD score
plot(qtl.morphine.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, 10))
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("DO_fentanyl_",i))
dev.off()
}
#peaks coeff plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
print(peaks)
if(dim(peaks)[1] != 0){
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
}
}
# [1] "Time.to.Dead.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 4 155.5686 6.872808 23.78901 155.8905
# 2 1 pheno1 5 130.9623 6.521364 130.41268 135.0559
# [1] 1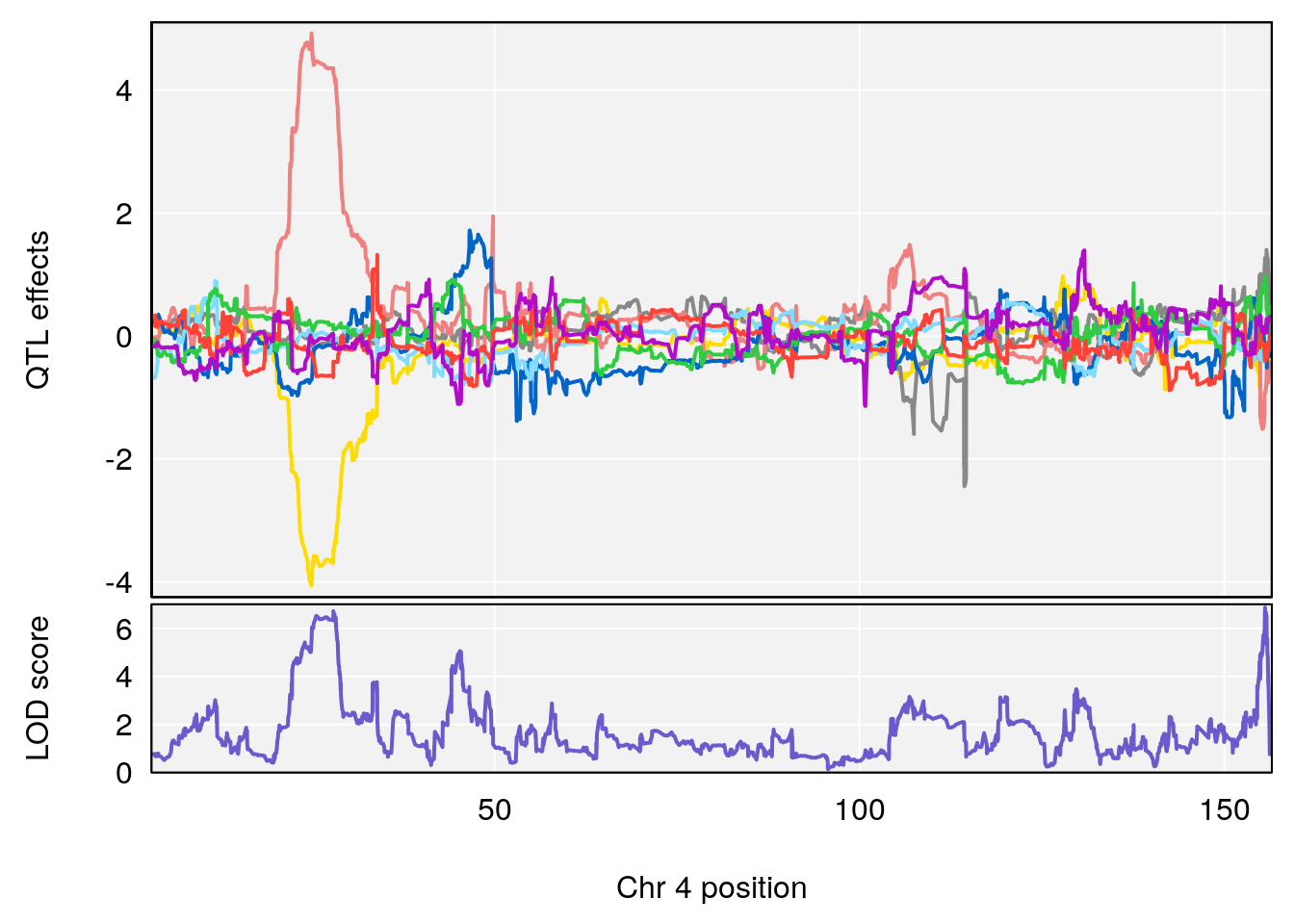
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
# [1] 2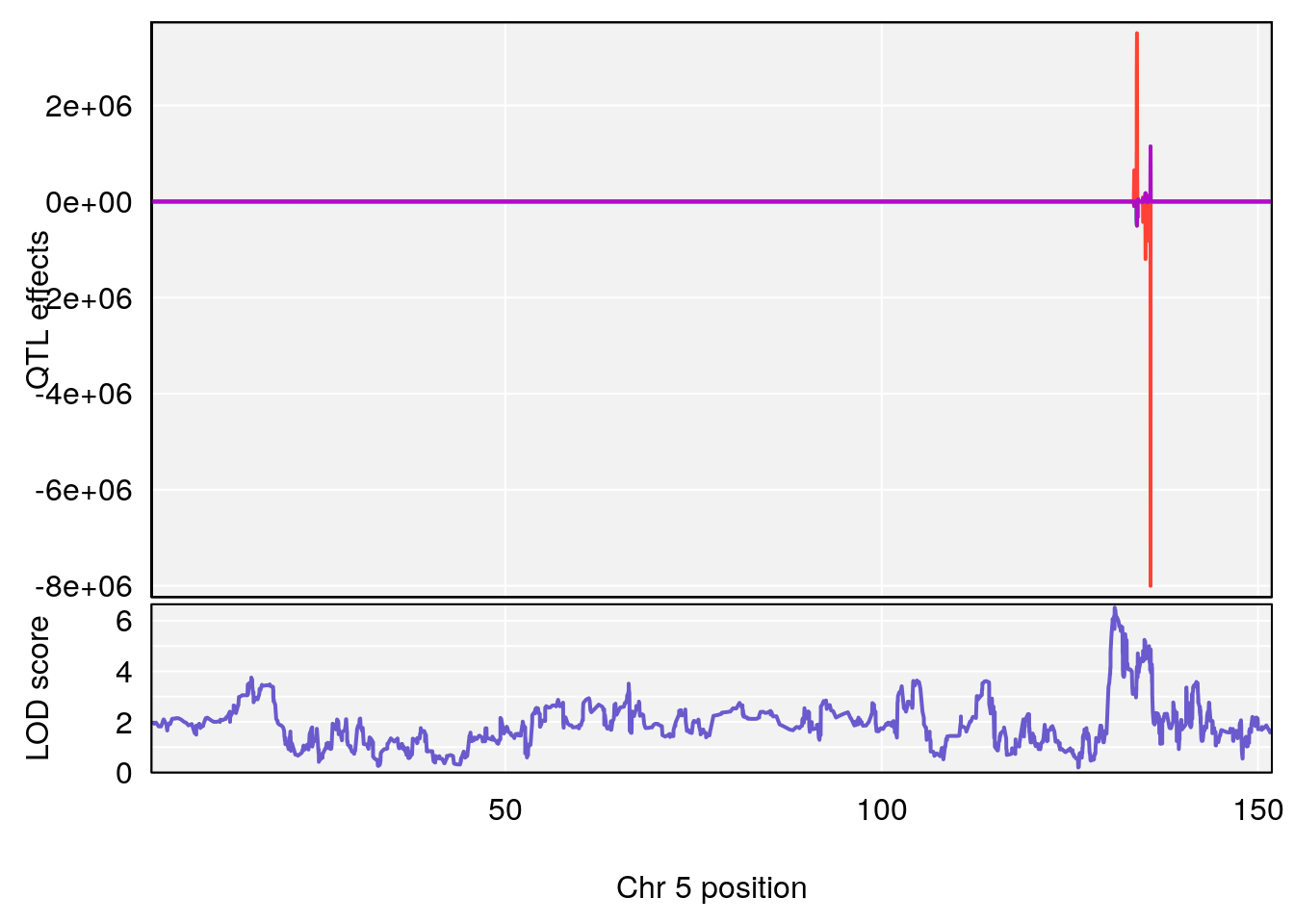
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
# [1] "RR.Depression.Rate.Hr"
# [1] lodindex lodcolumn chr pos lod ci_lo ci_hi
# <0 rows> (or 0-length row.names)
# [1] "Time.to.Steady.RR.Depression.Hr"
# [1] lodindex lodcolumn chr pos lod ci_lo ci_hi
# <0 rows> (or 0-length row.names)
# [1] "Min.Depression.RR"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 6 110.25926 6.917845 109.73966 110.89937
# 2 1 pheno1 15 70.24917 6.600987 68.92043 83.02844
# [1] 1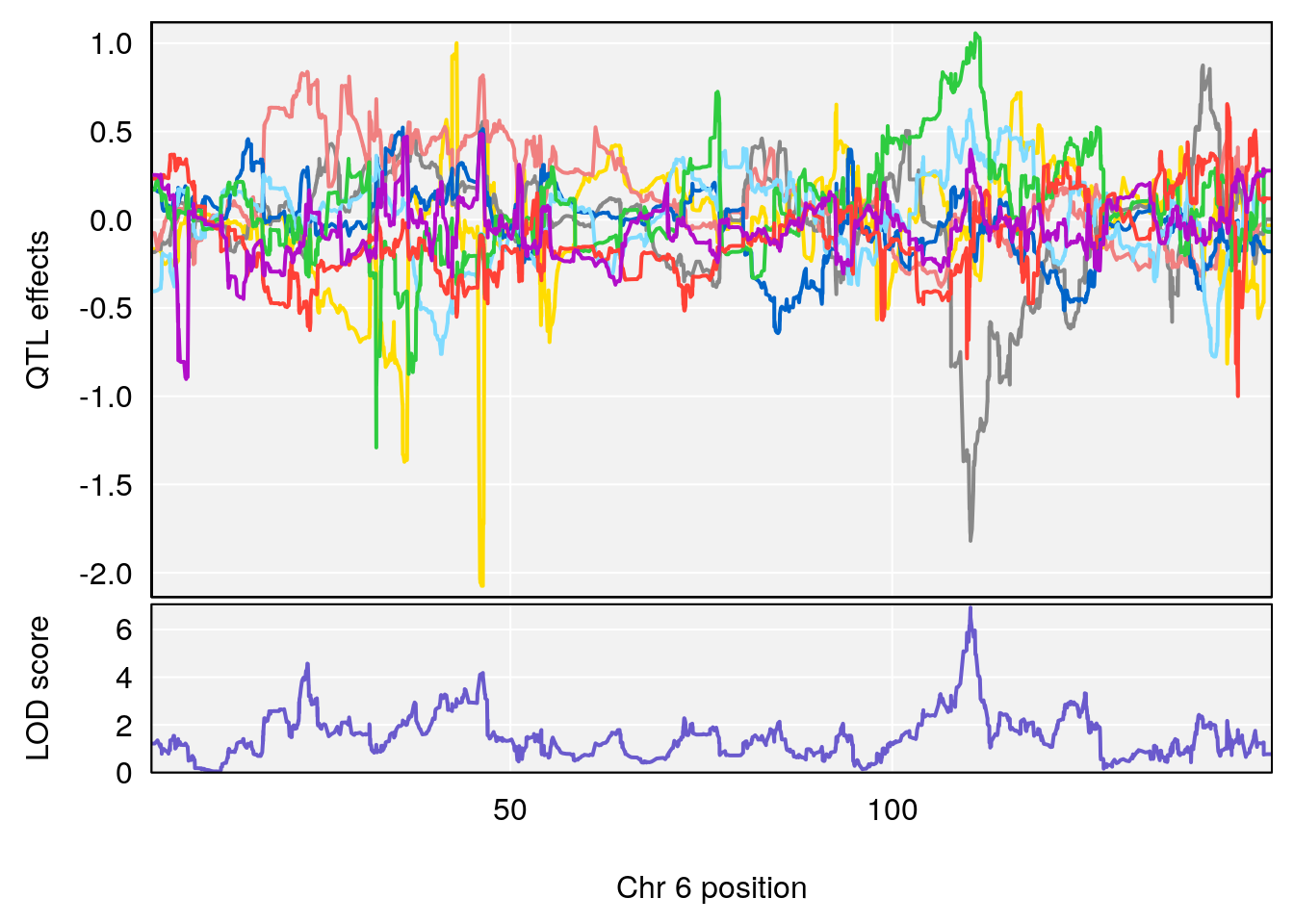
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
# [1] 2
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
# [1] "Mean.Depression.BR"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
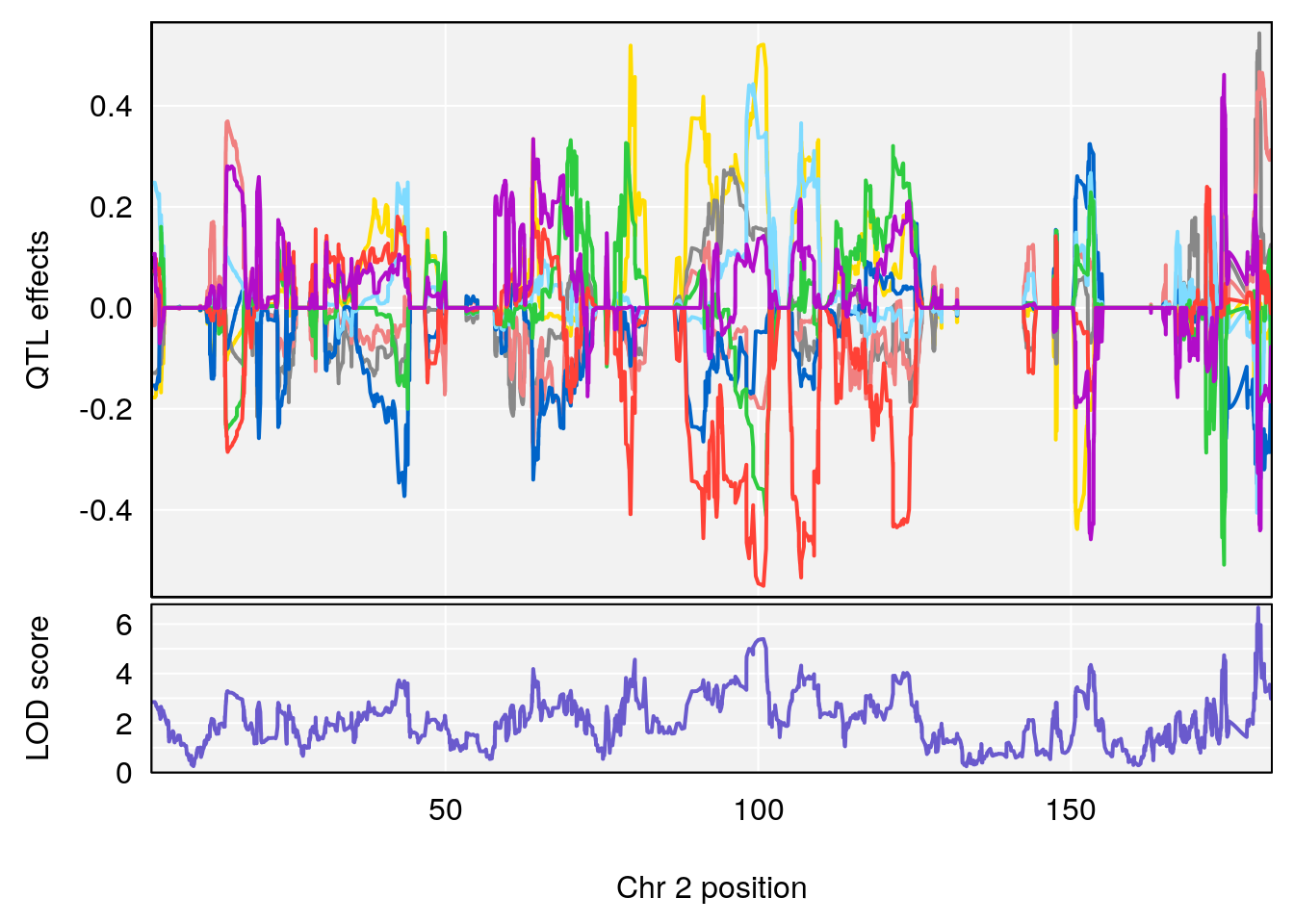
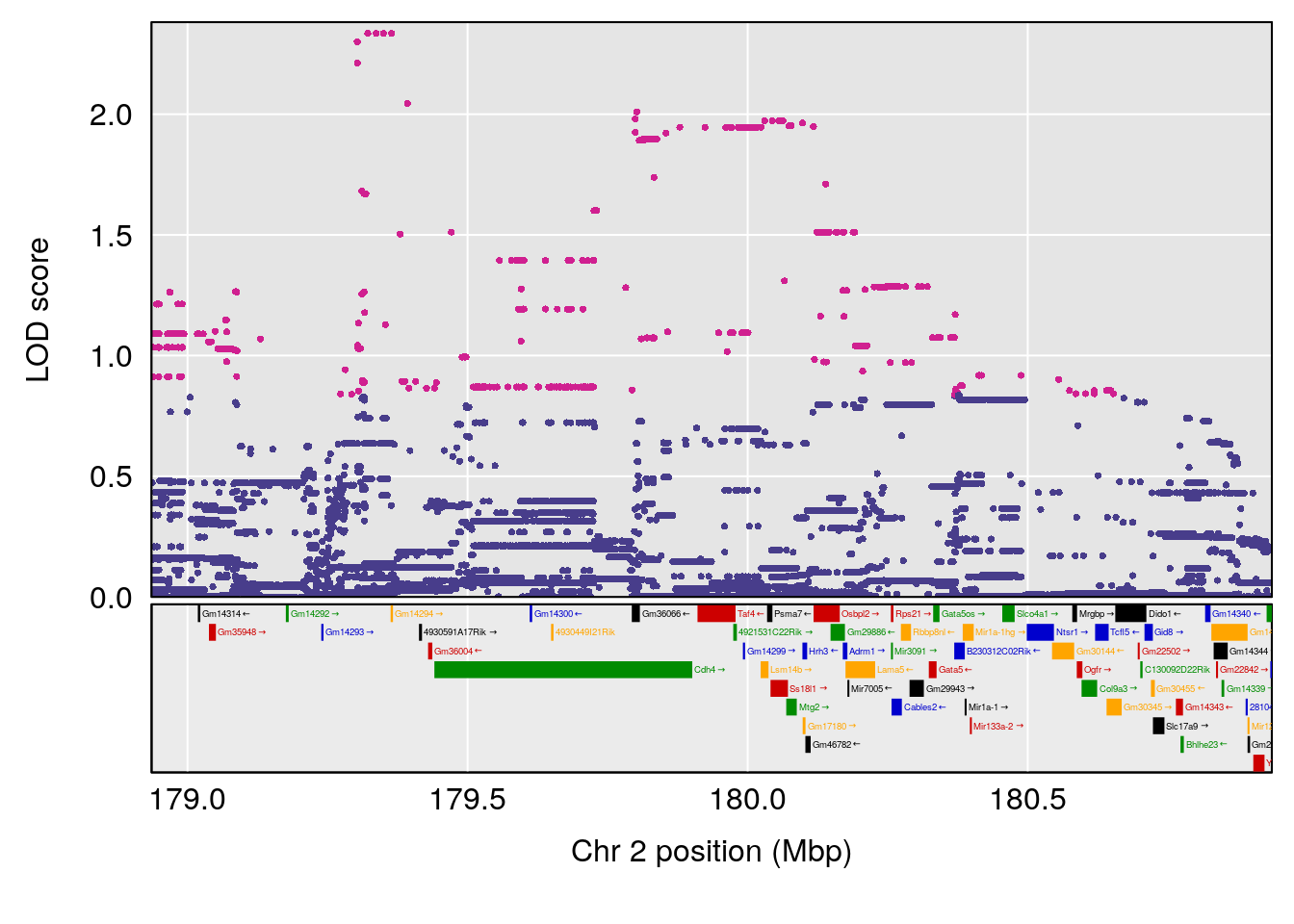
# 1 1 pheno1 2 179.93579 6.666801 99.18788 180.36934
# 2 1 pheno1 15 28.42395 6.928838 27.40287 72.41888
# 3 1 pheno1 17 45.00725 6.614011 44.60730 45.45418
# [1] 1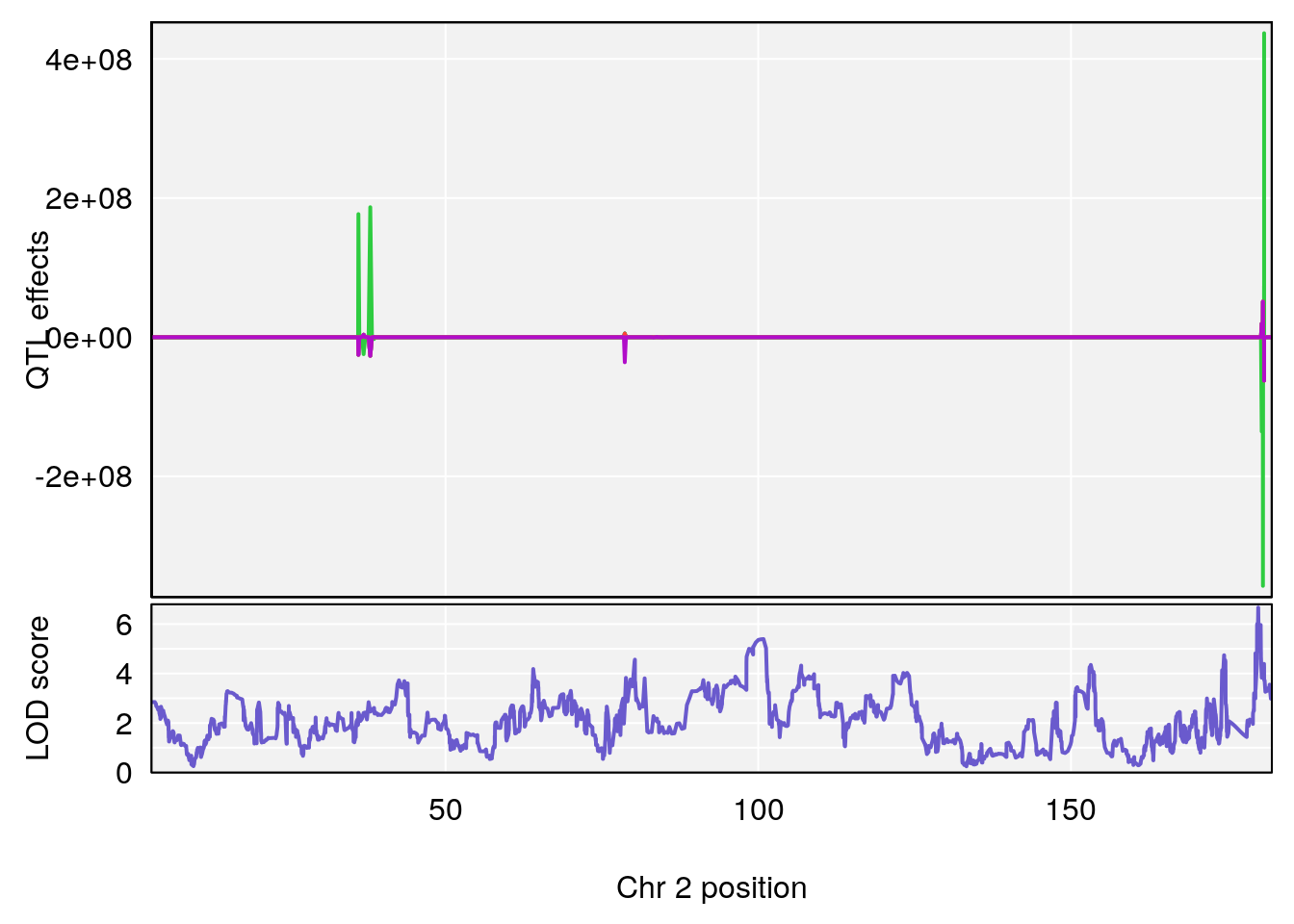
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
# [1] 2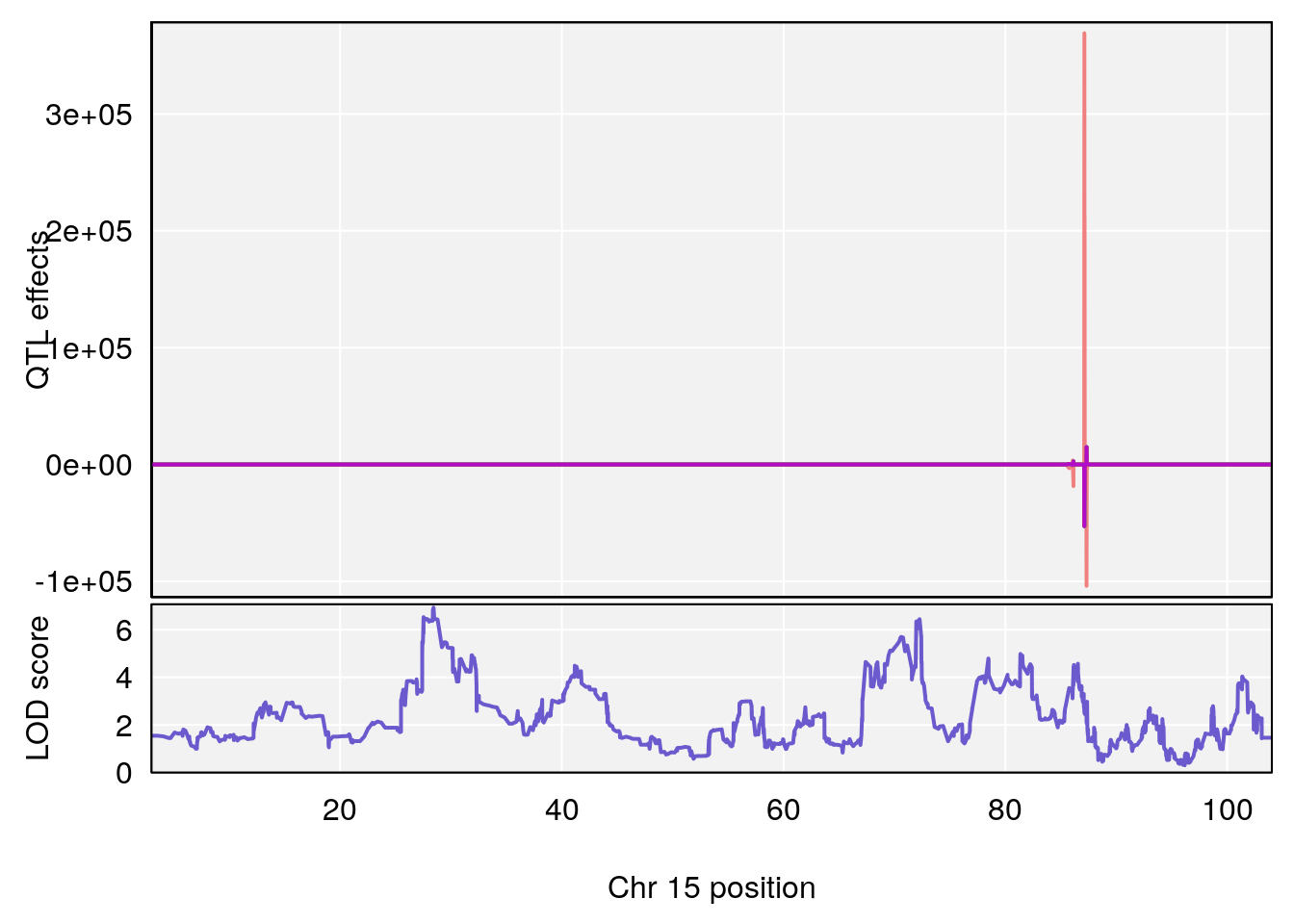
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
# [1] 3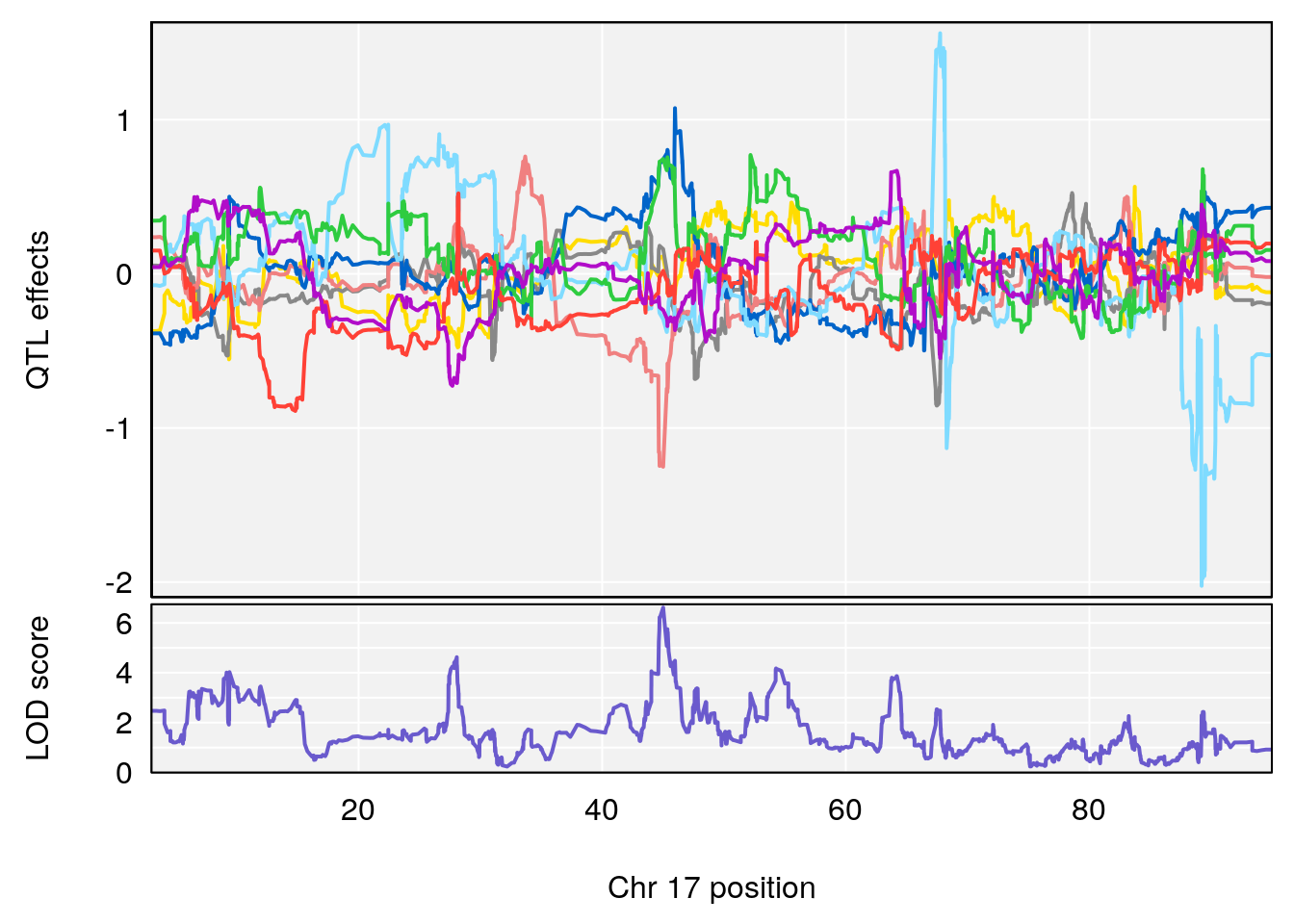
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
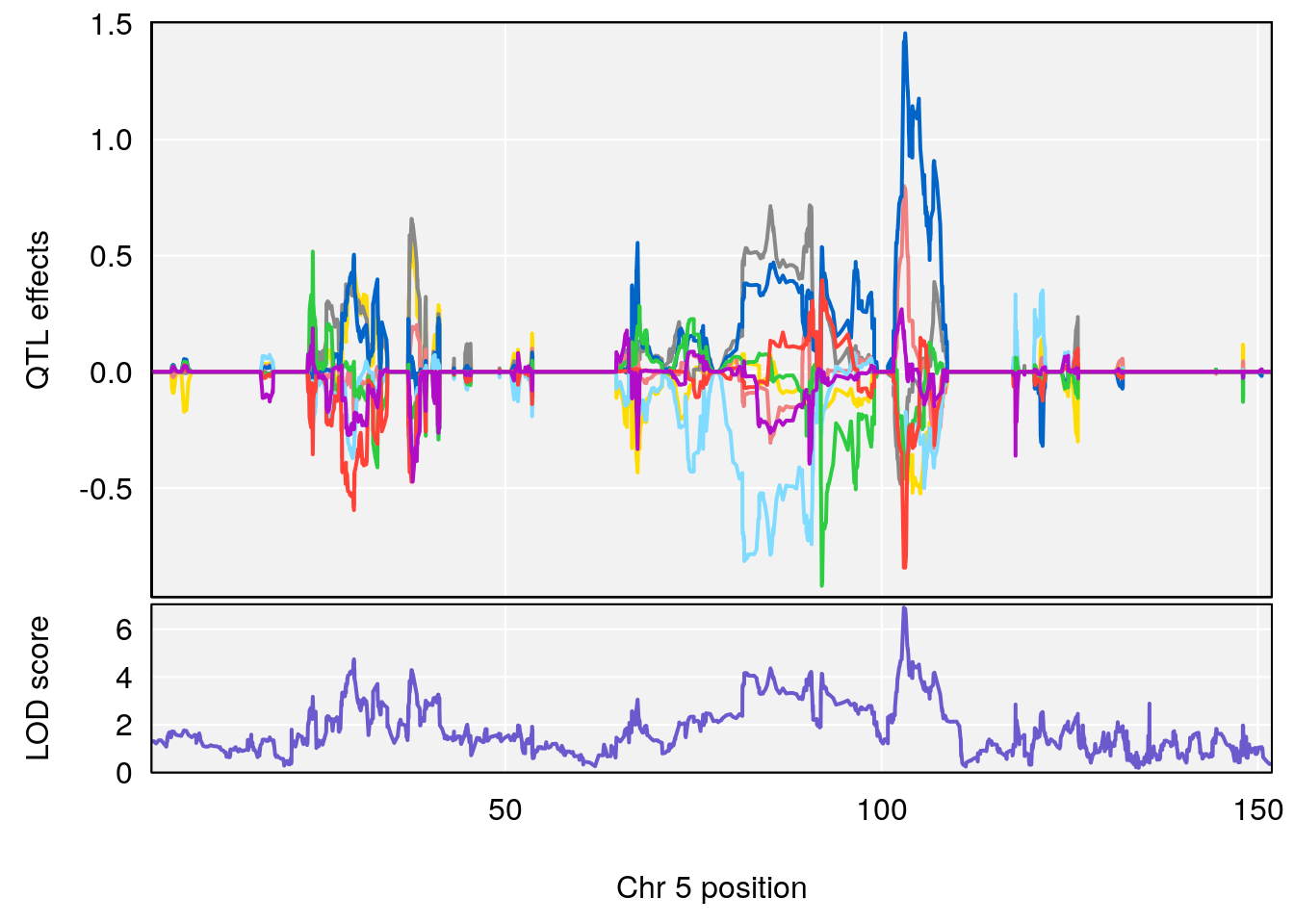
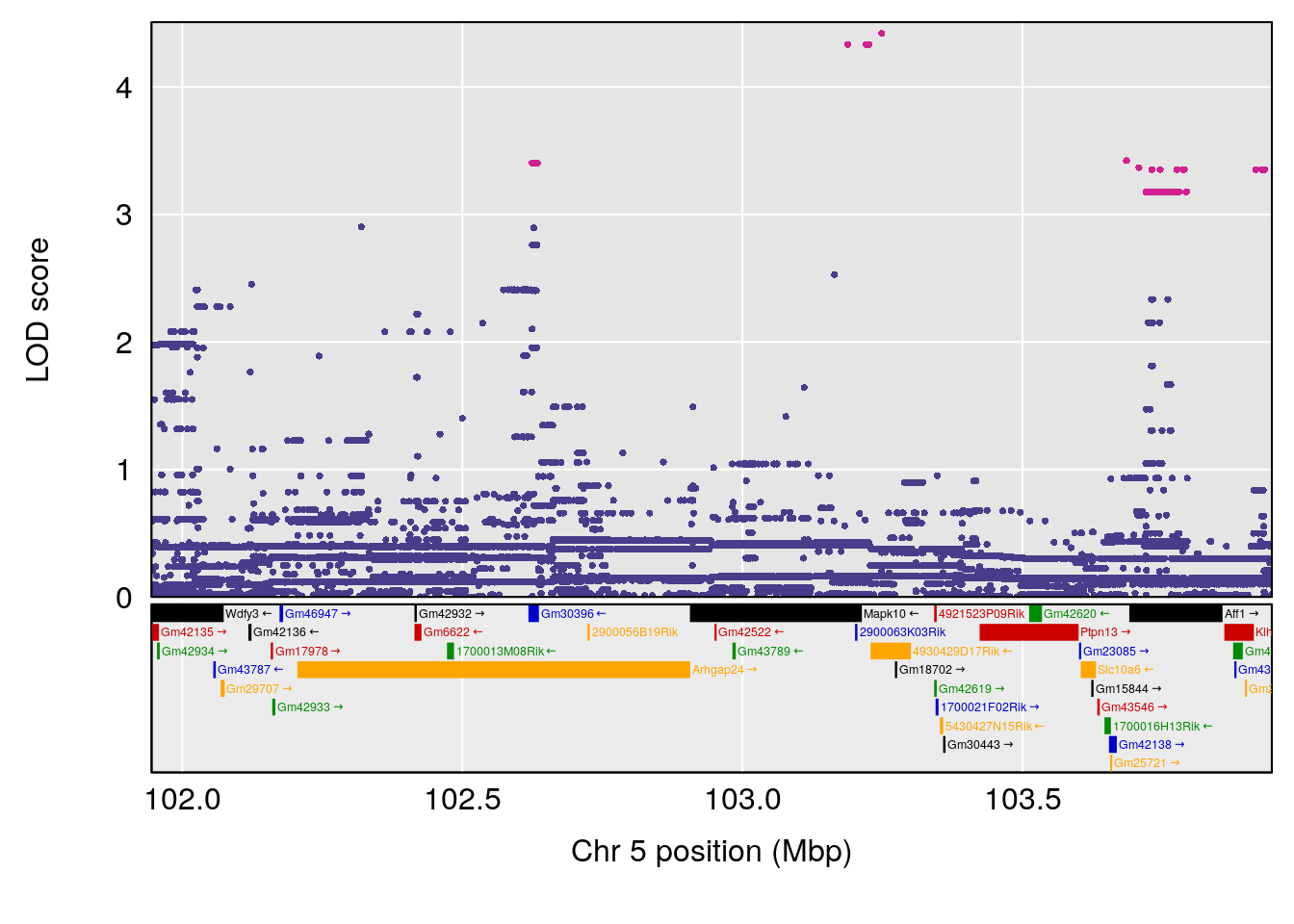
# [1] "Steady-State.Depression.Duration.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 4 135.9546 6.166319 32.37278 136.3792
# 2 1 pheno1 5 102.9451 6.909370 102.66155 103.3928
# 3 1 pheno1 11 100.0623 6.710383 99.90022 100.9337
# [1] 1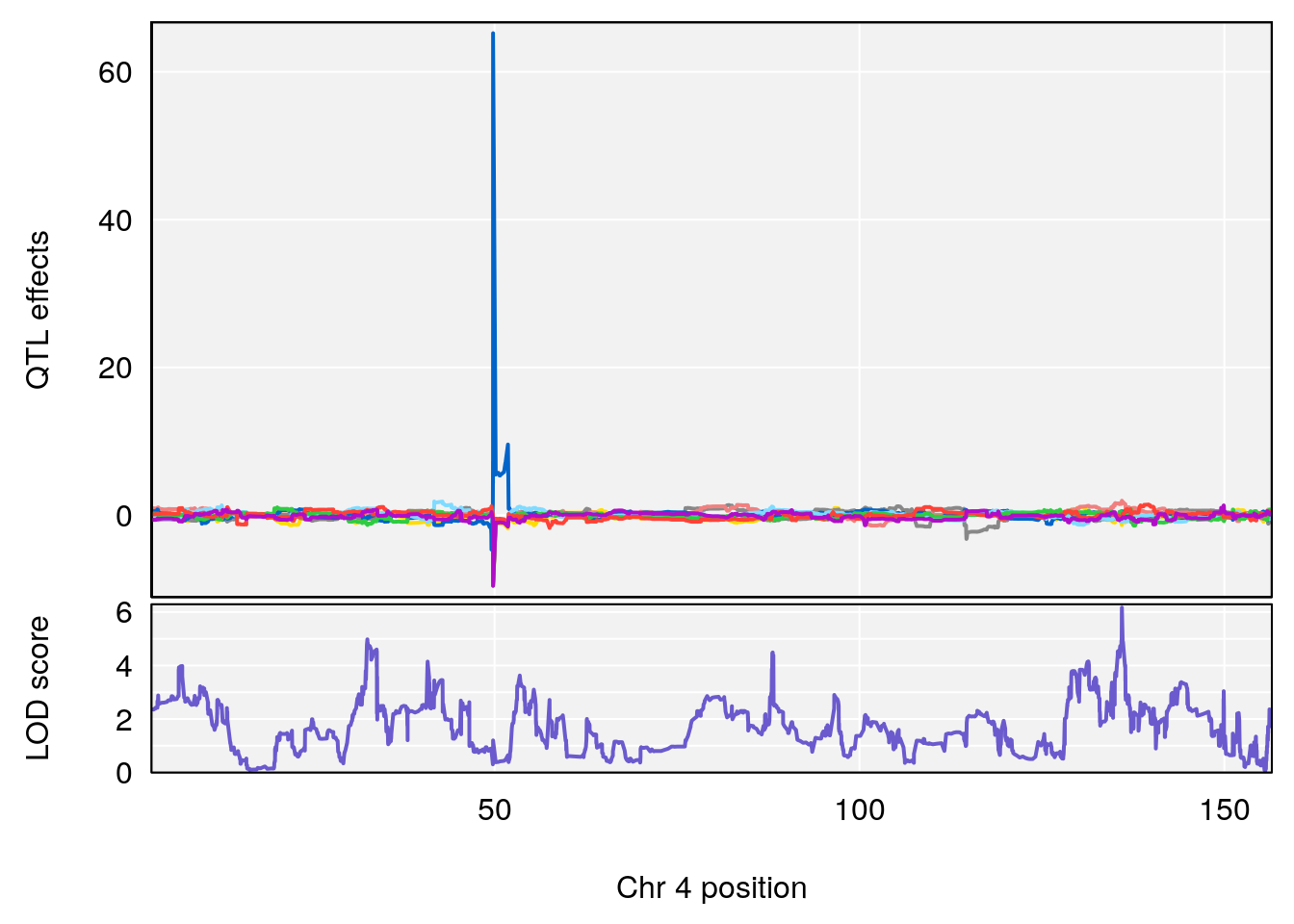
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
# [1] 2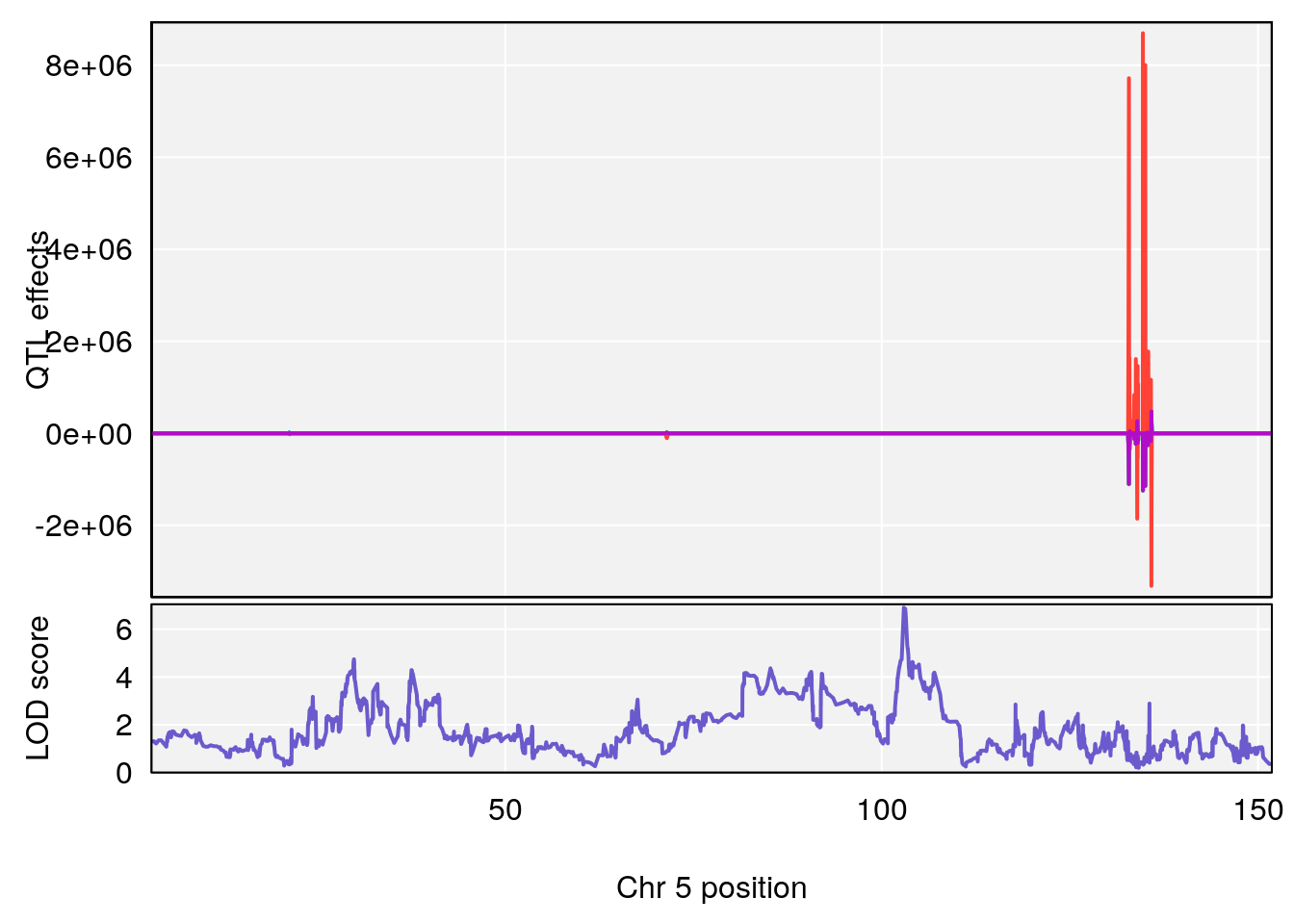
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
# [1] 3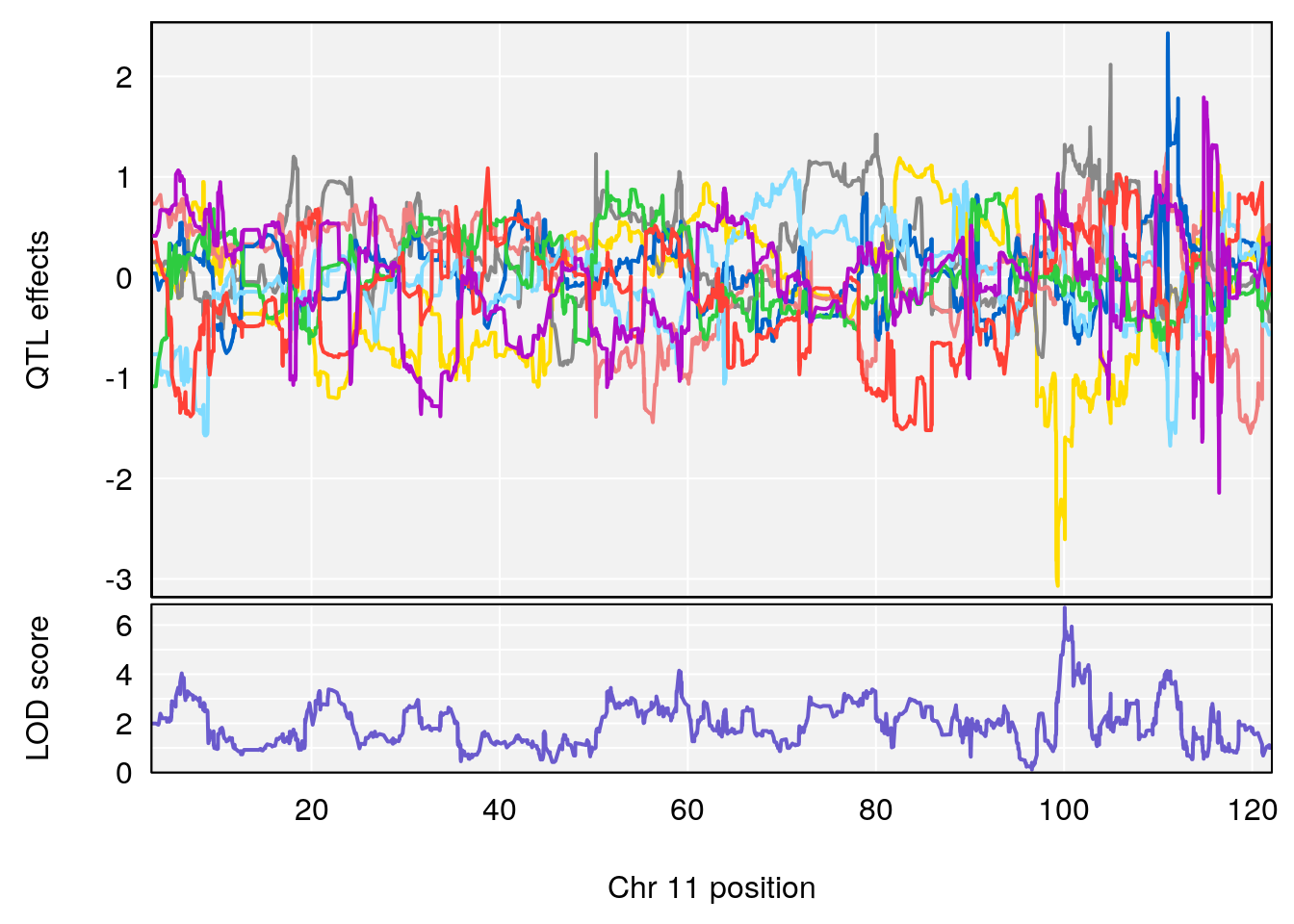
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
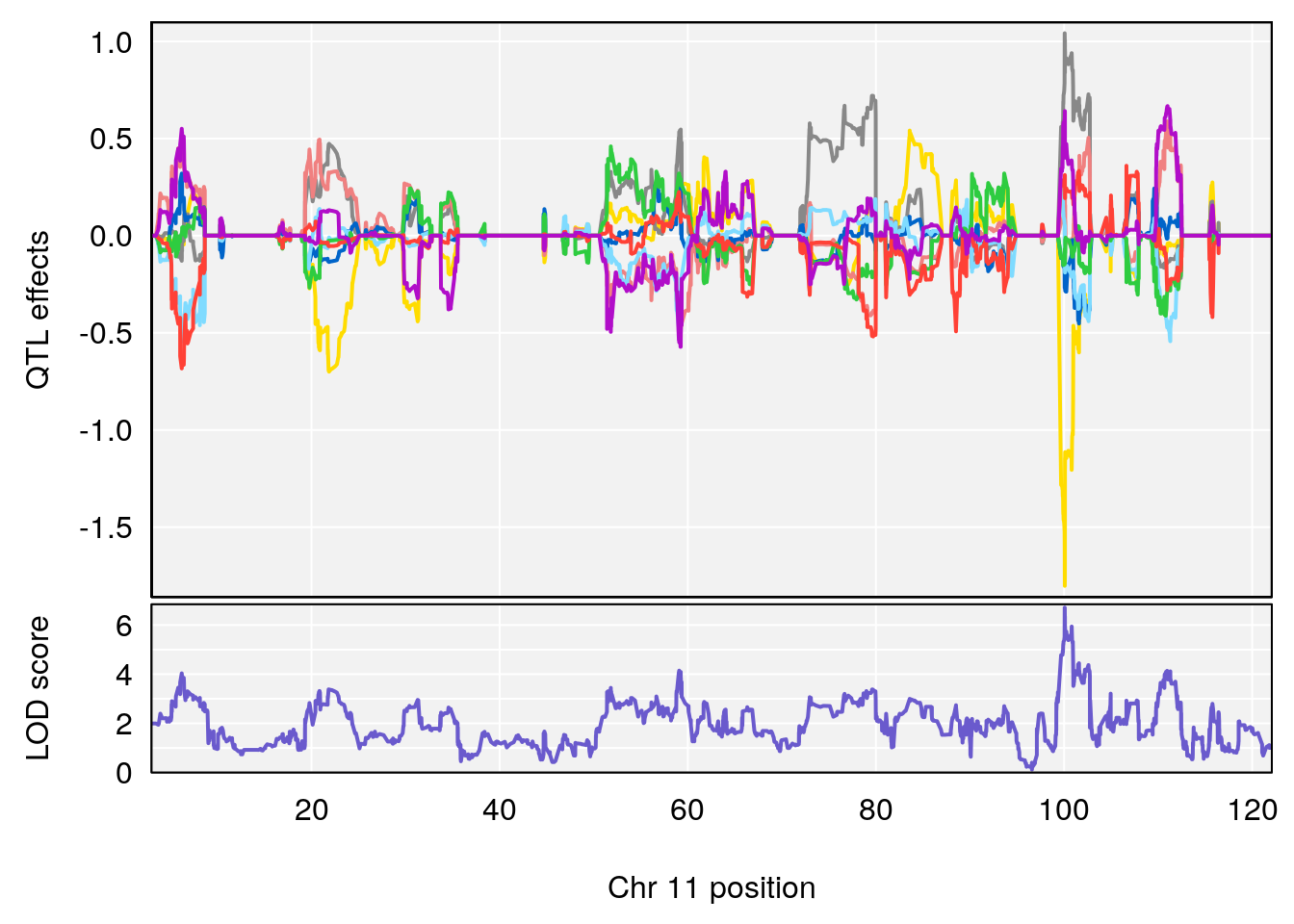
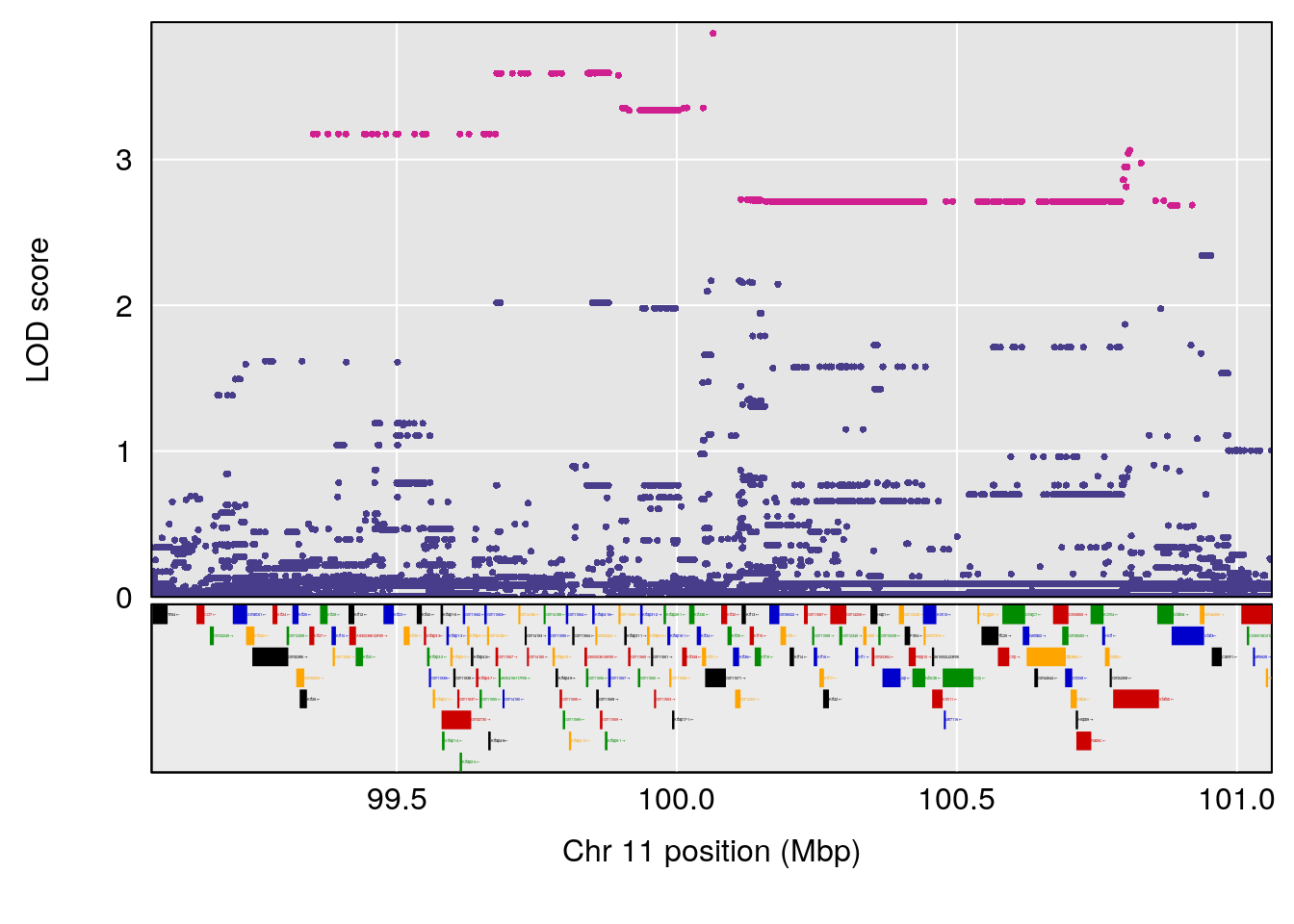
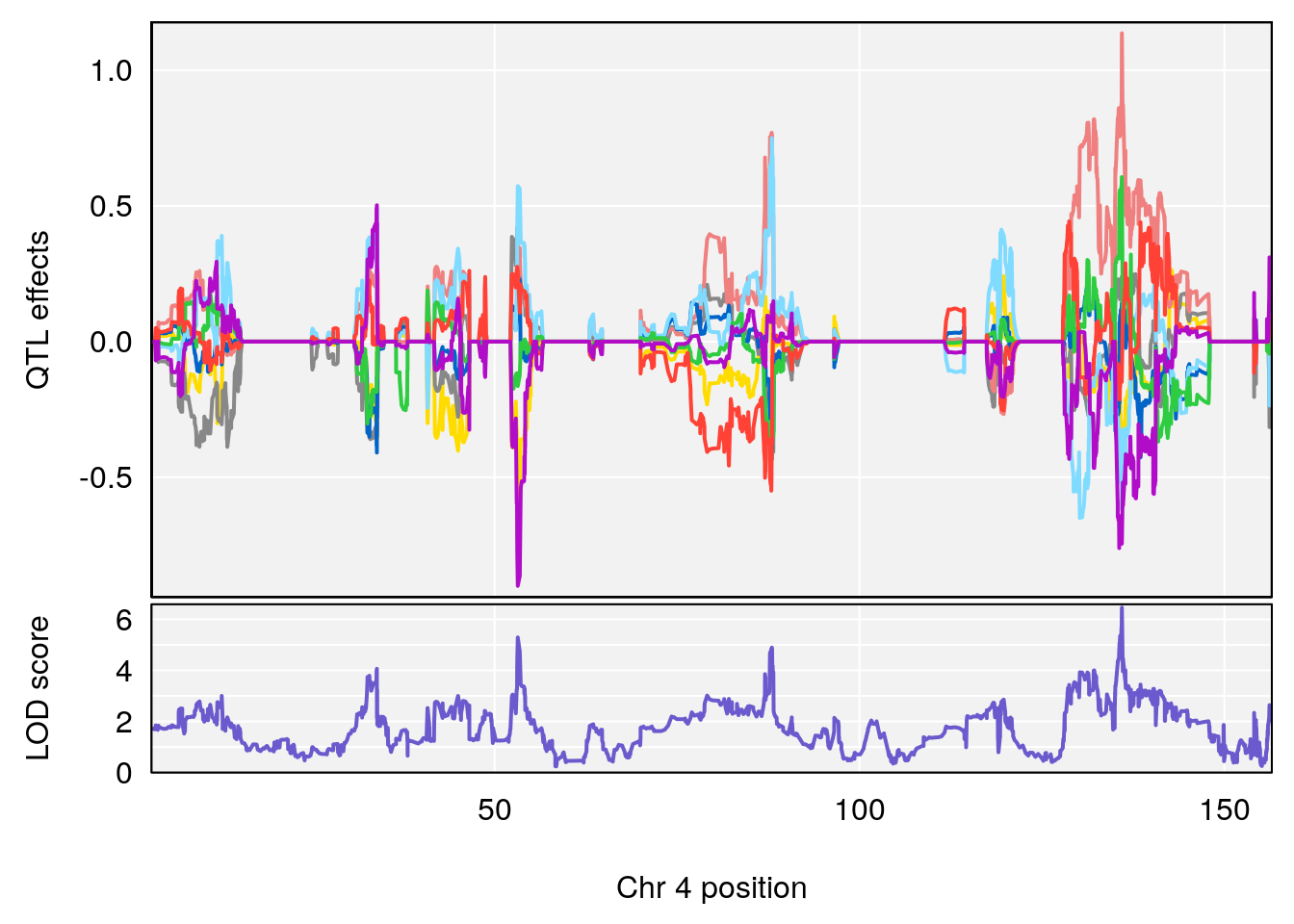
# [1] "Time.to.Threshold.Recovery.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 4 135.9546 6.459910 53.10543 136.07892
# 2 1 pheno1 14 20.6426 6.085256 20.25692 22.26143
# [1] 1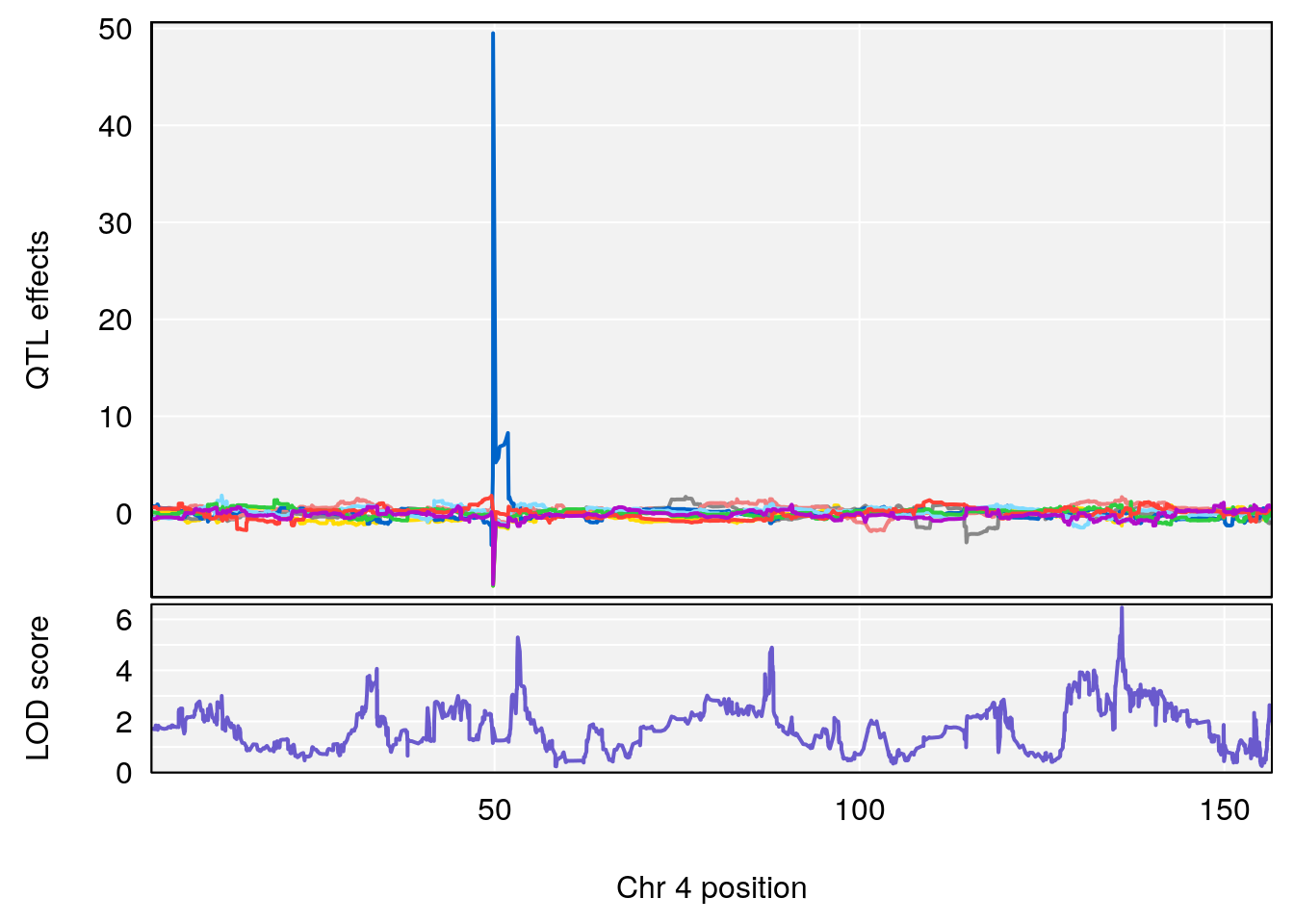
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
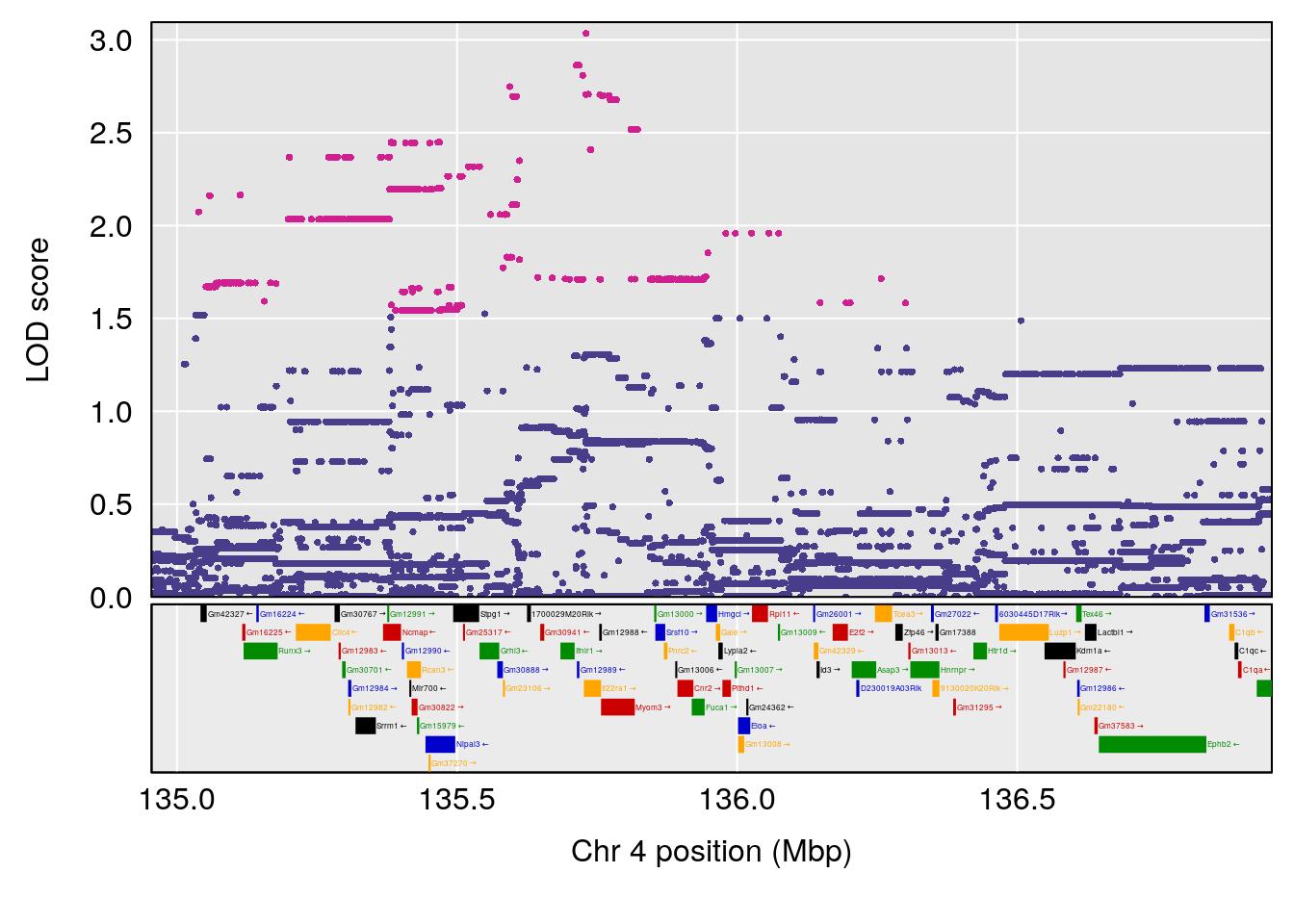
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
# [1] 2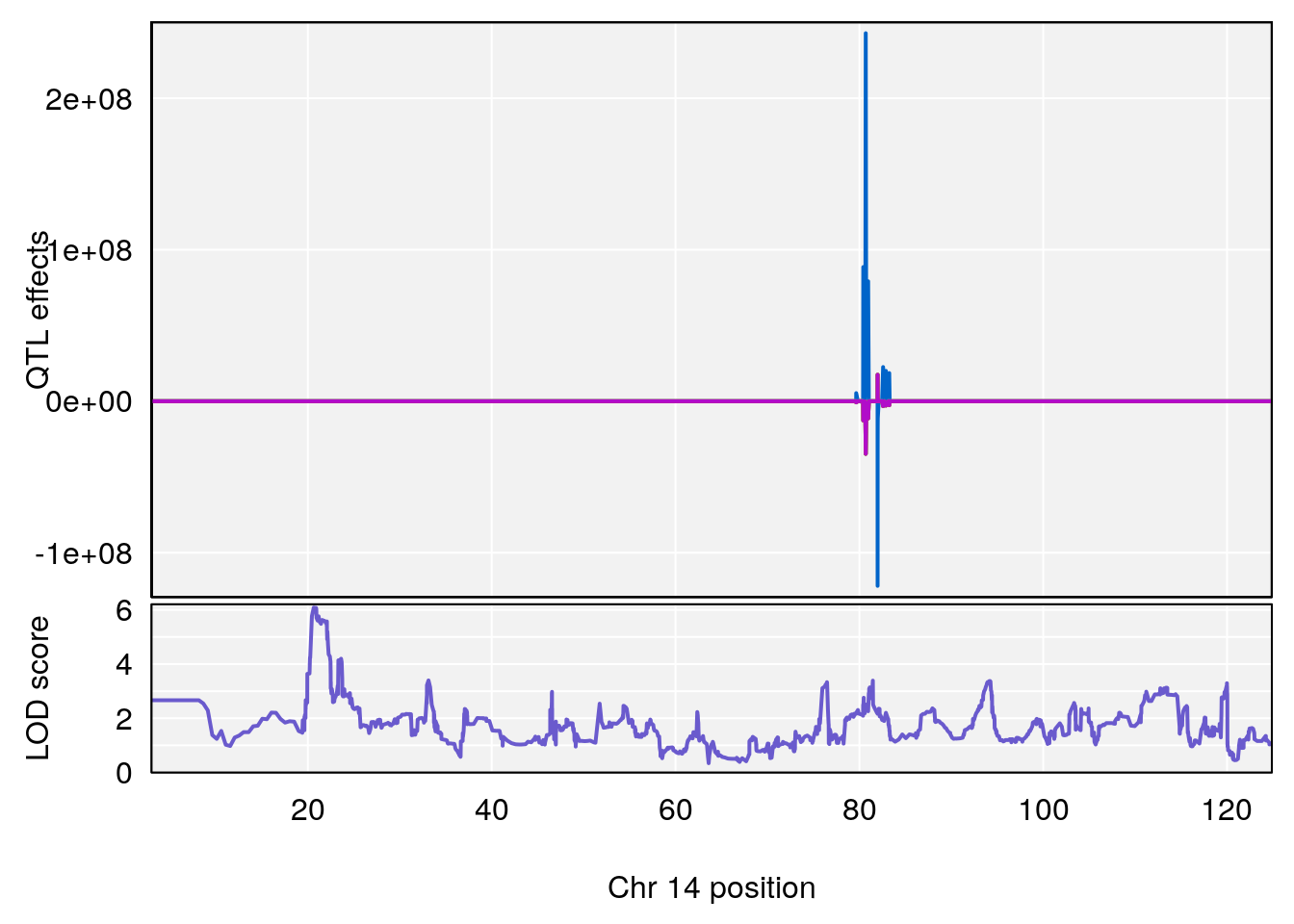
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
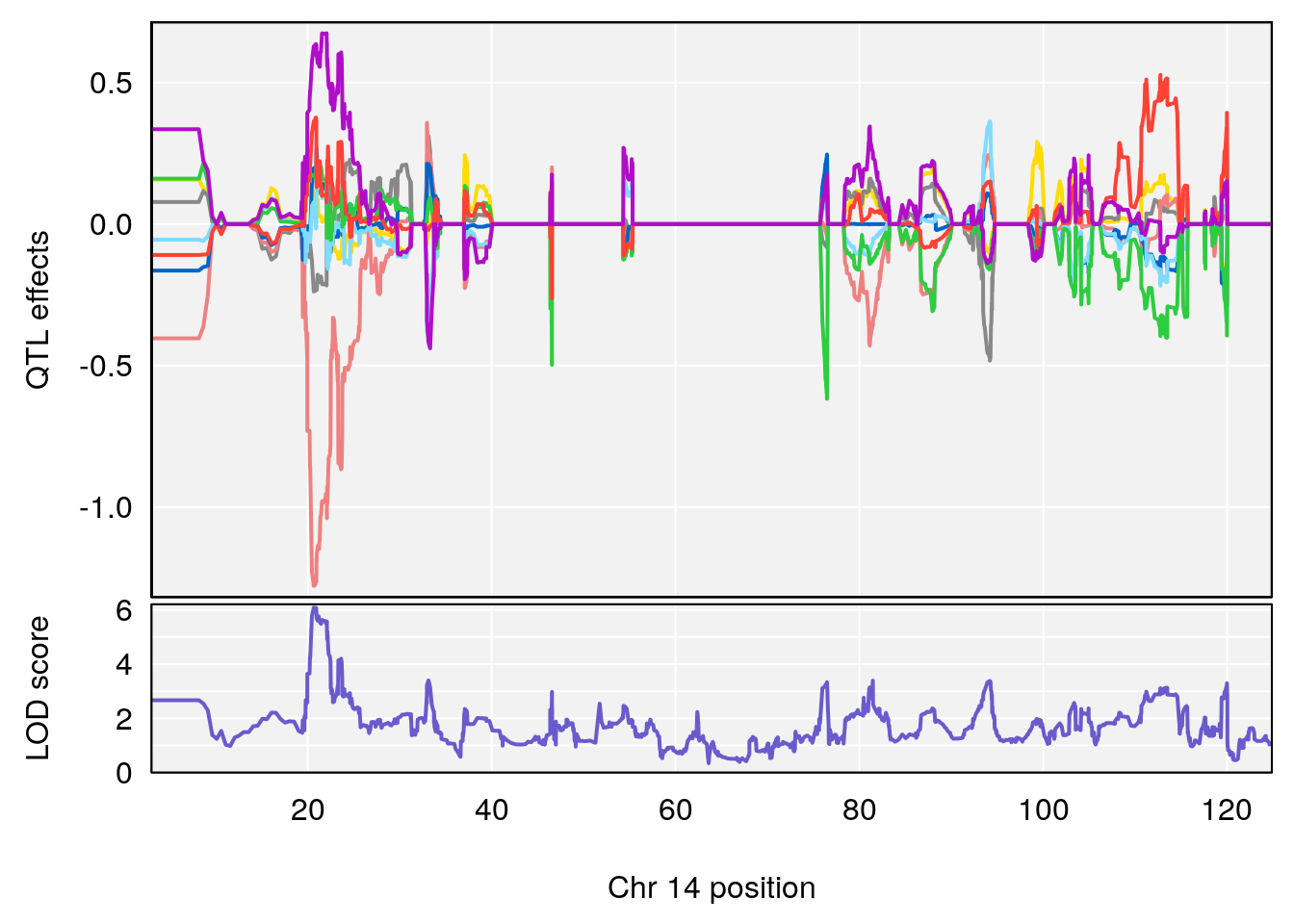
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
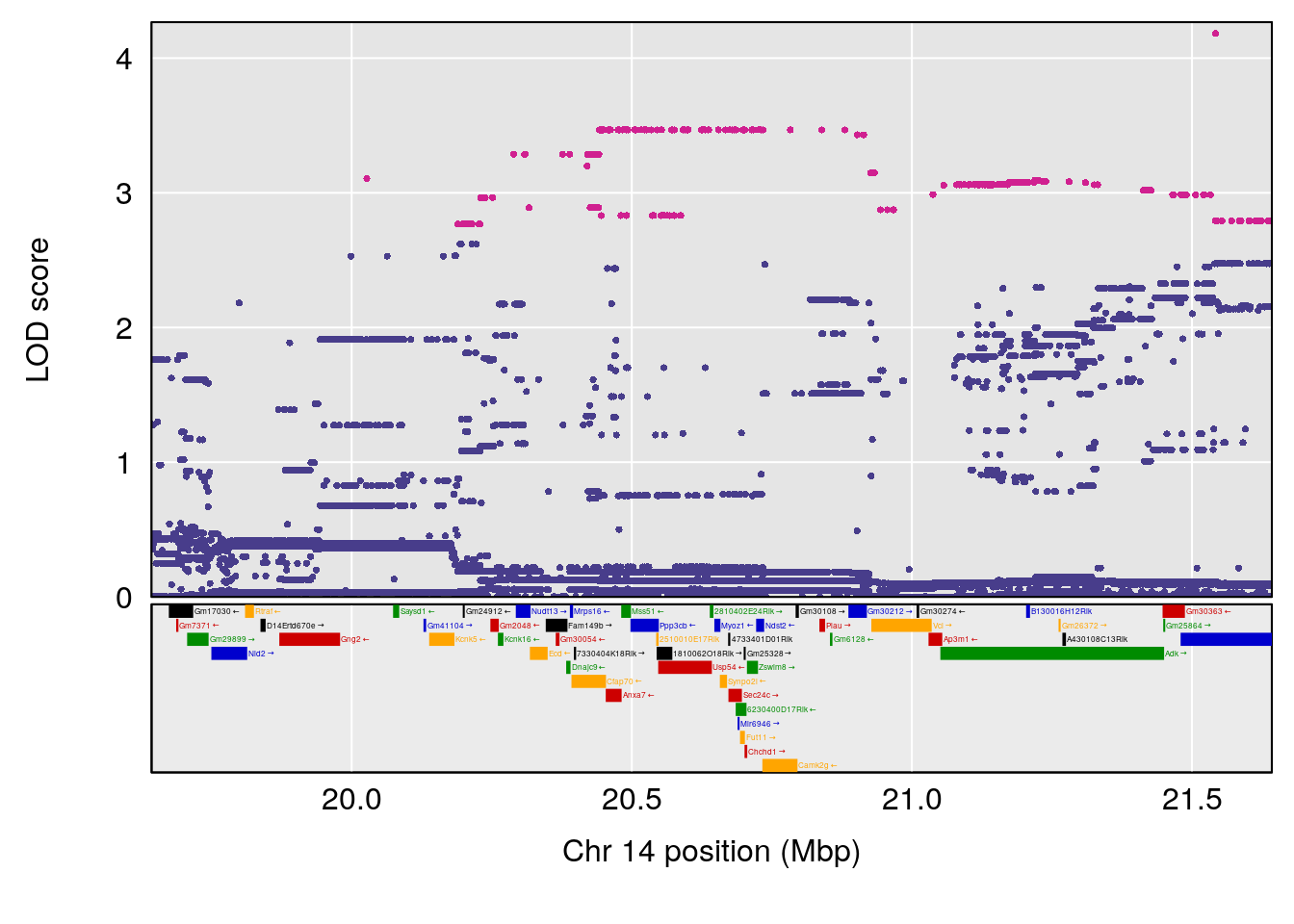
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
# [1] "Time.to.Projected.Recovery.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 5 103.1294 6.407867 102.6616 103.3928
# [1] 1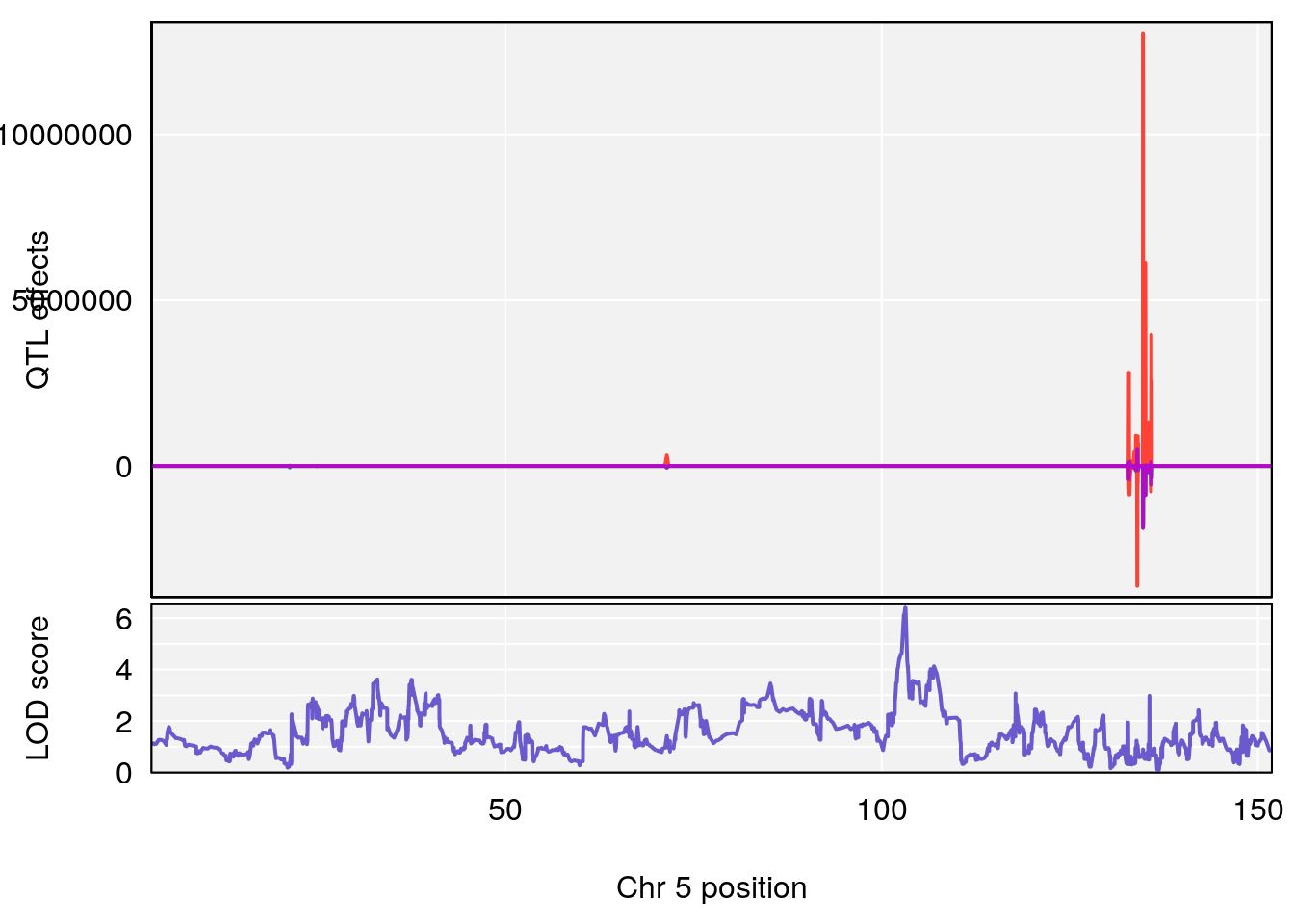
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
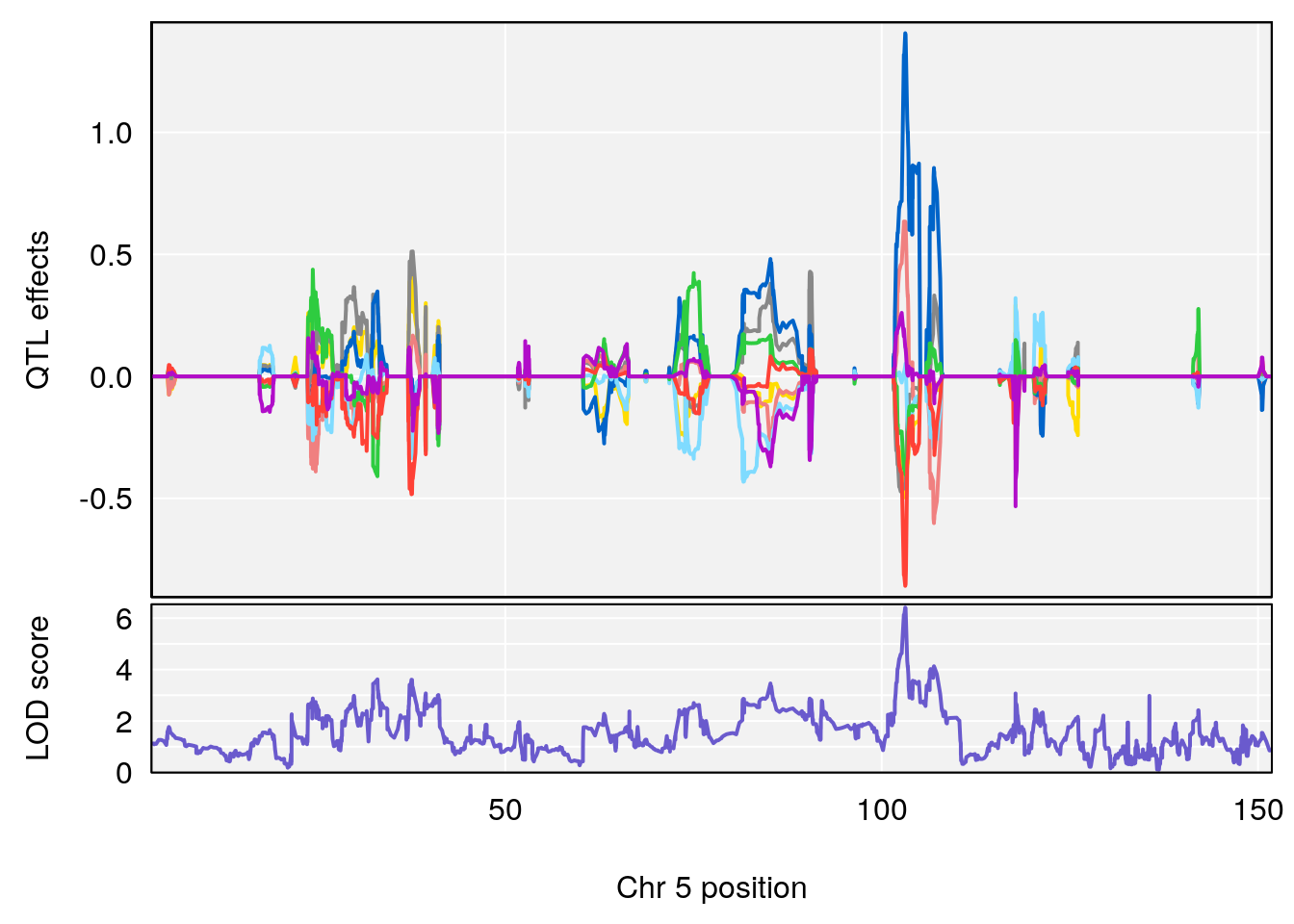
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
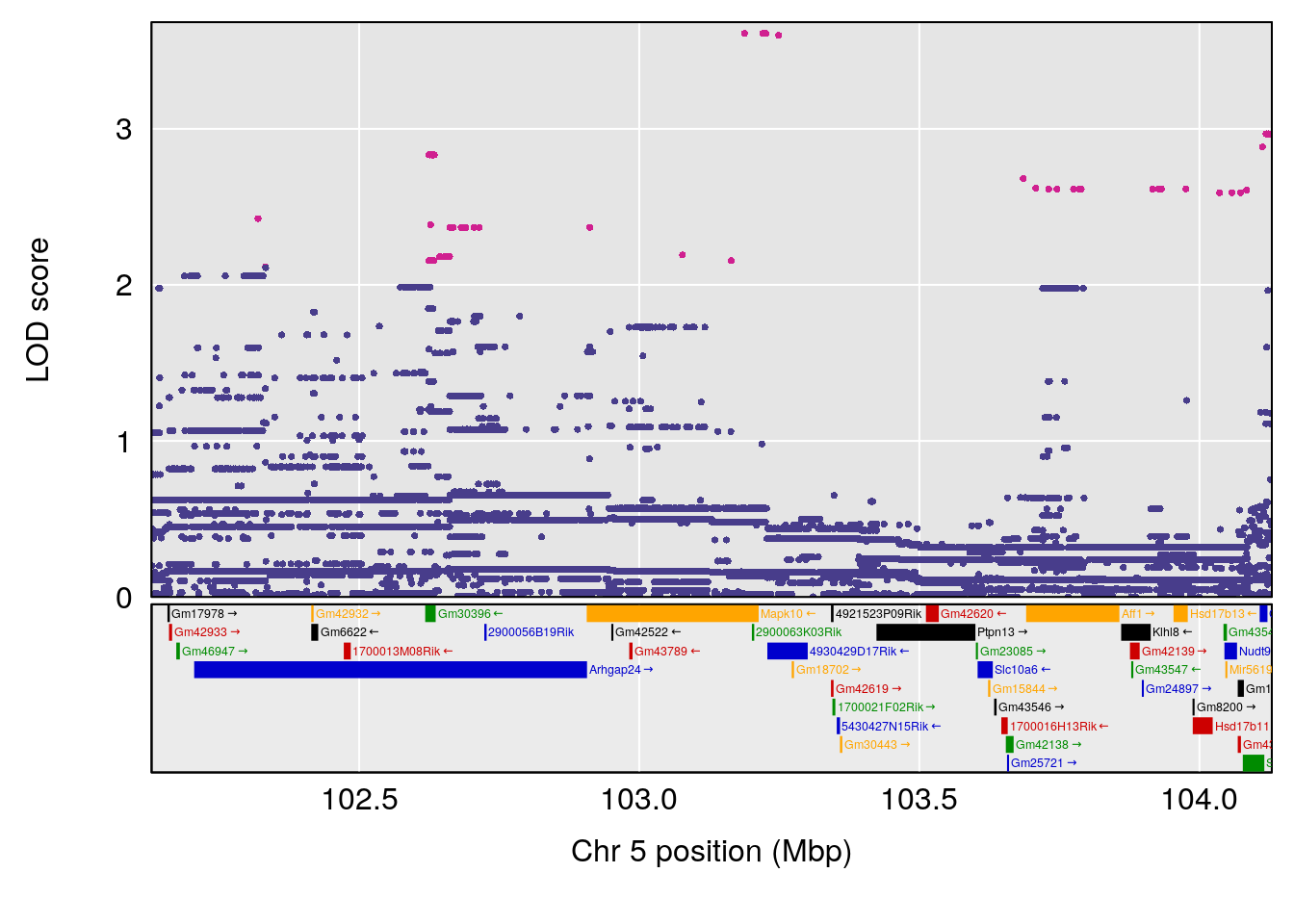
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
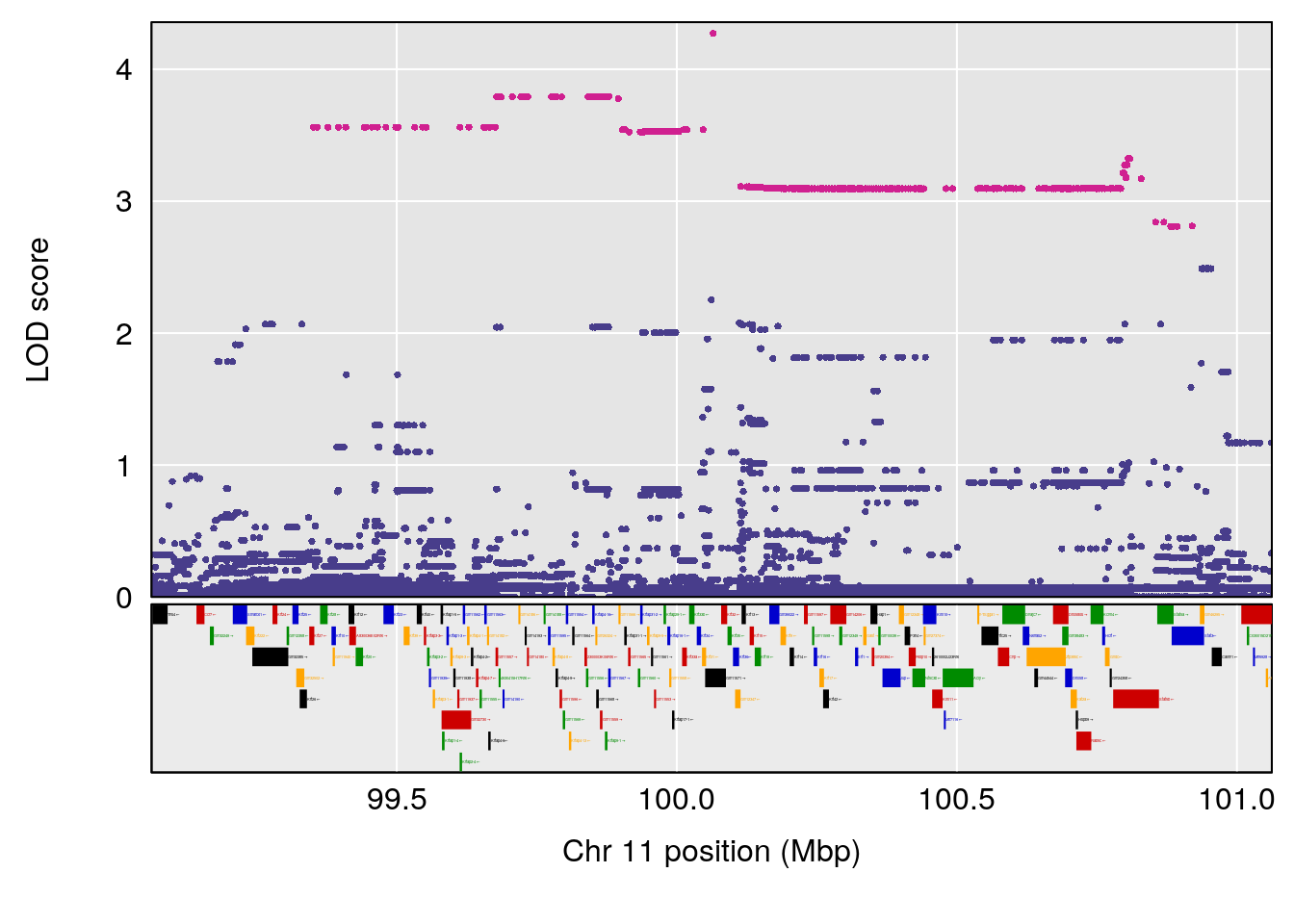
# [1] "Start.of.Recovery.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 5 102.9451 6.622891 102.66155 103.3928
# 2 1 pheno1 11 100.0623 6.910074 99.92203 100.8526
# [1] 1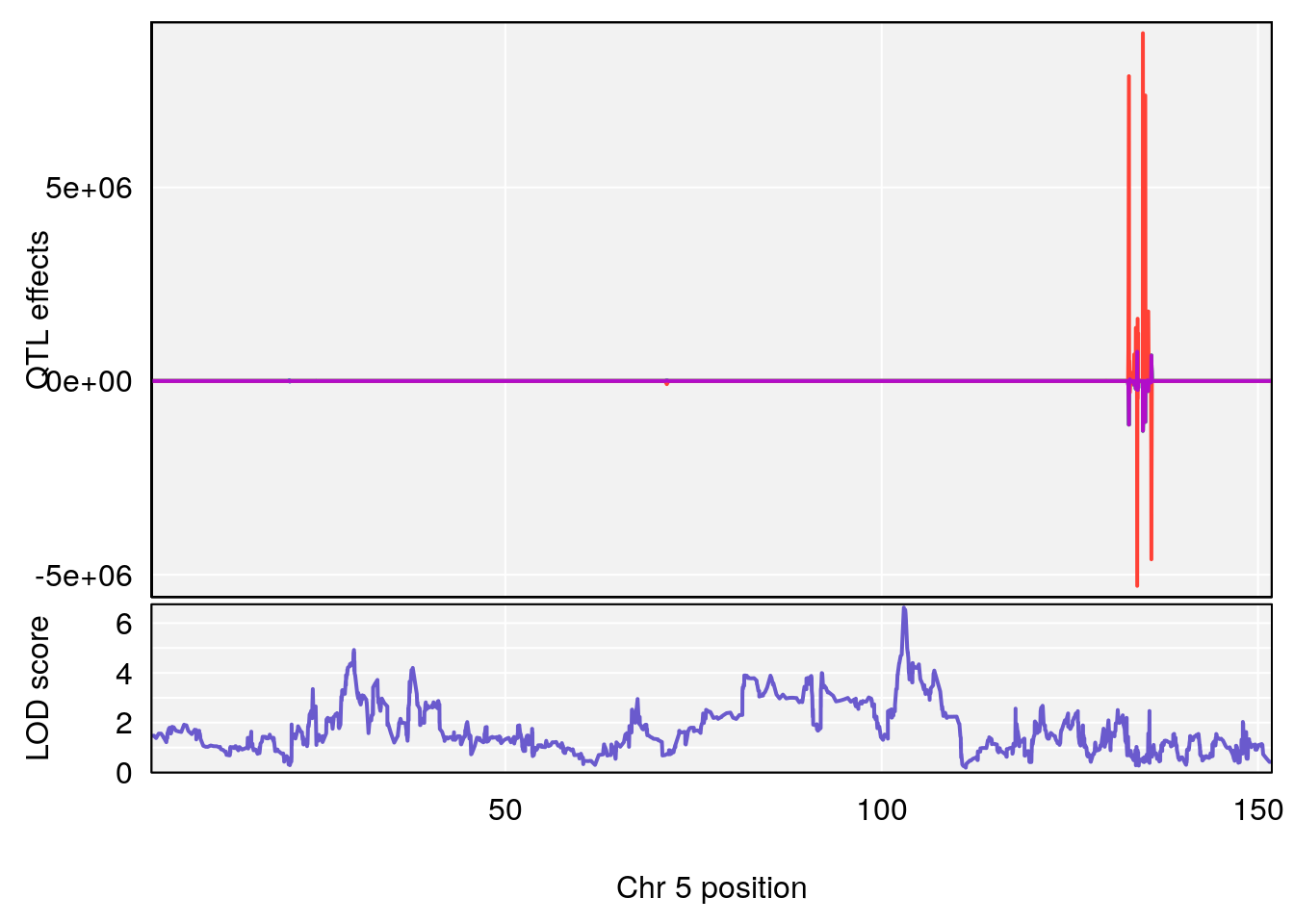
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
# [1] 2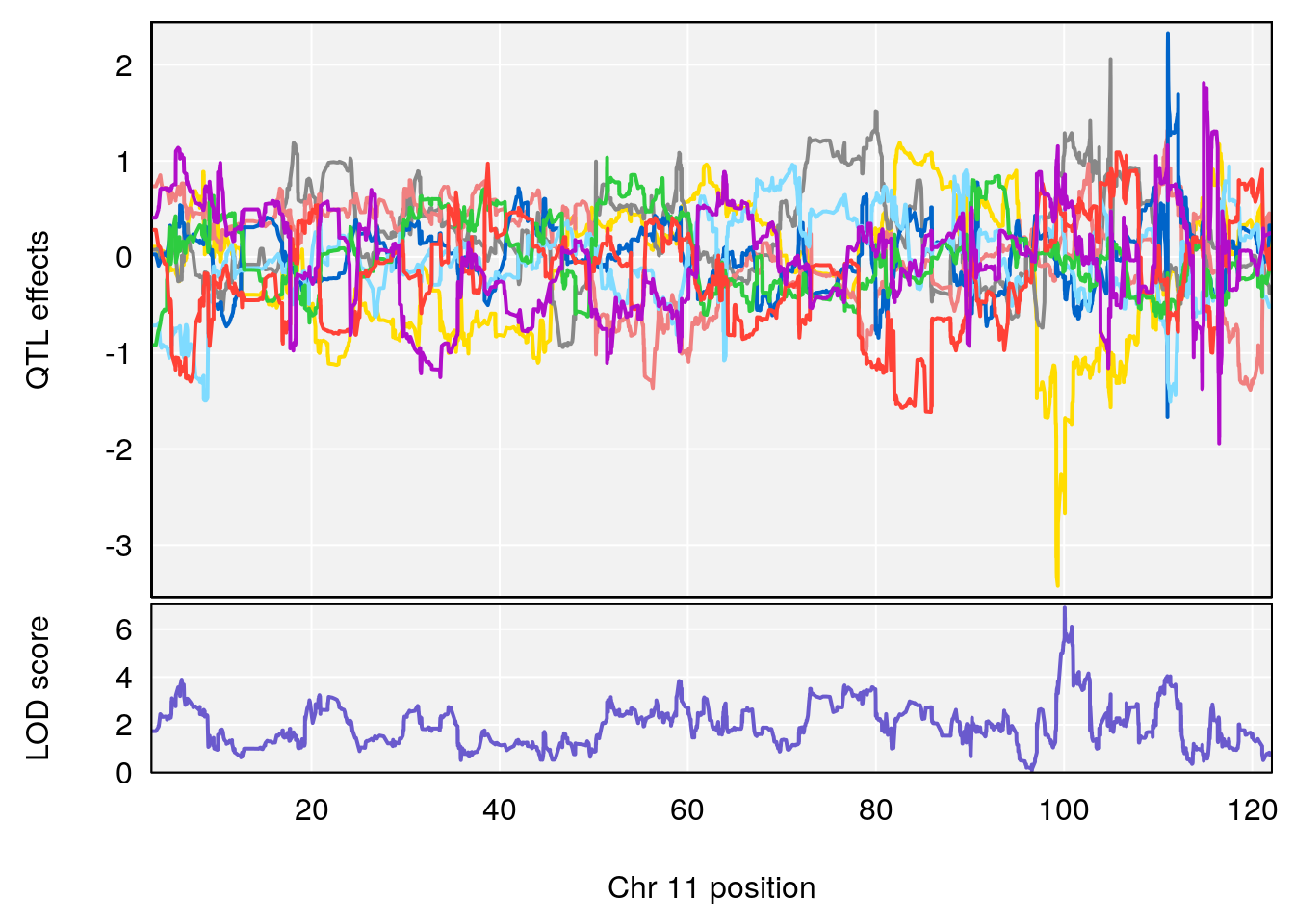
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
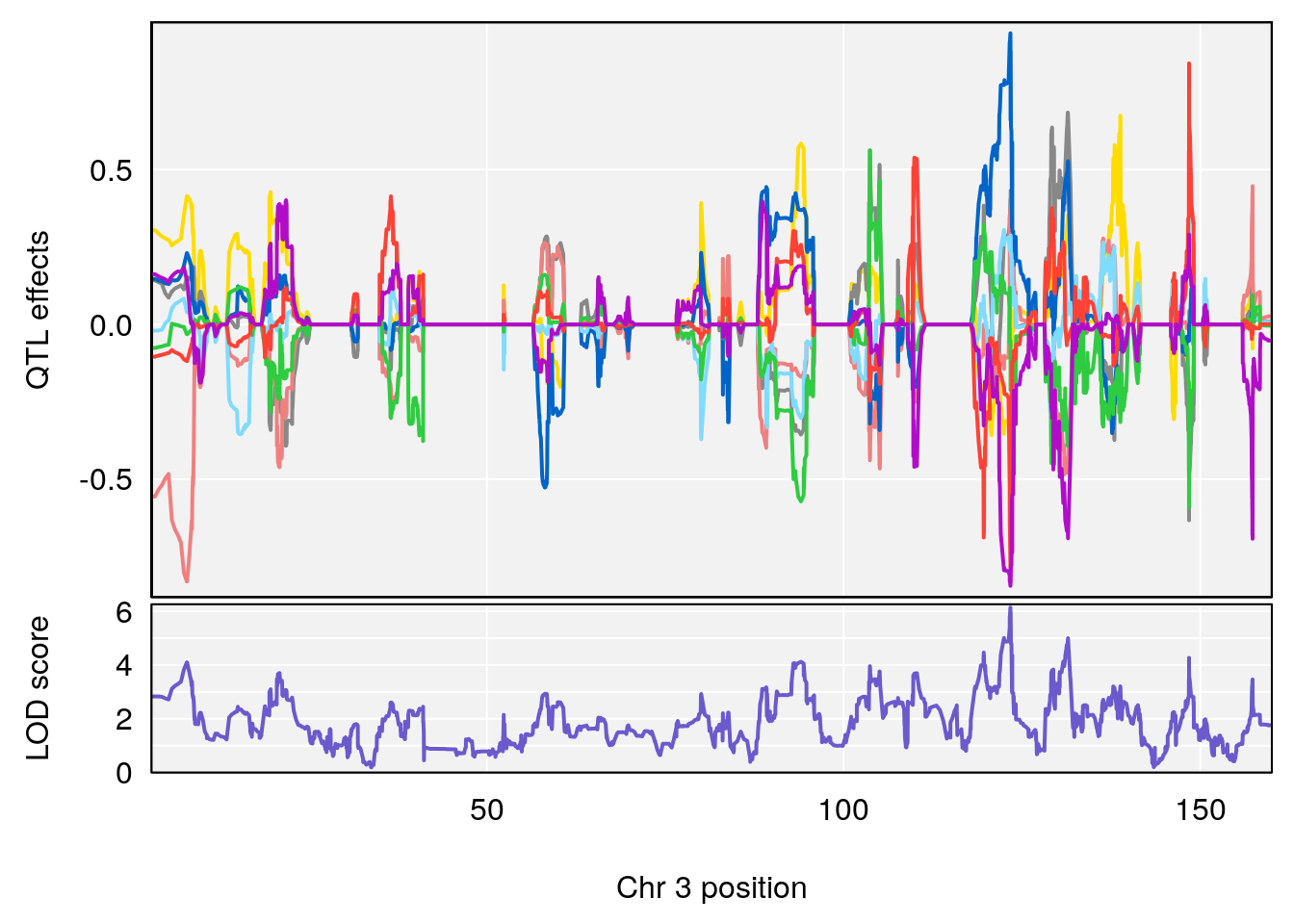
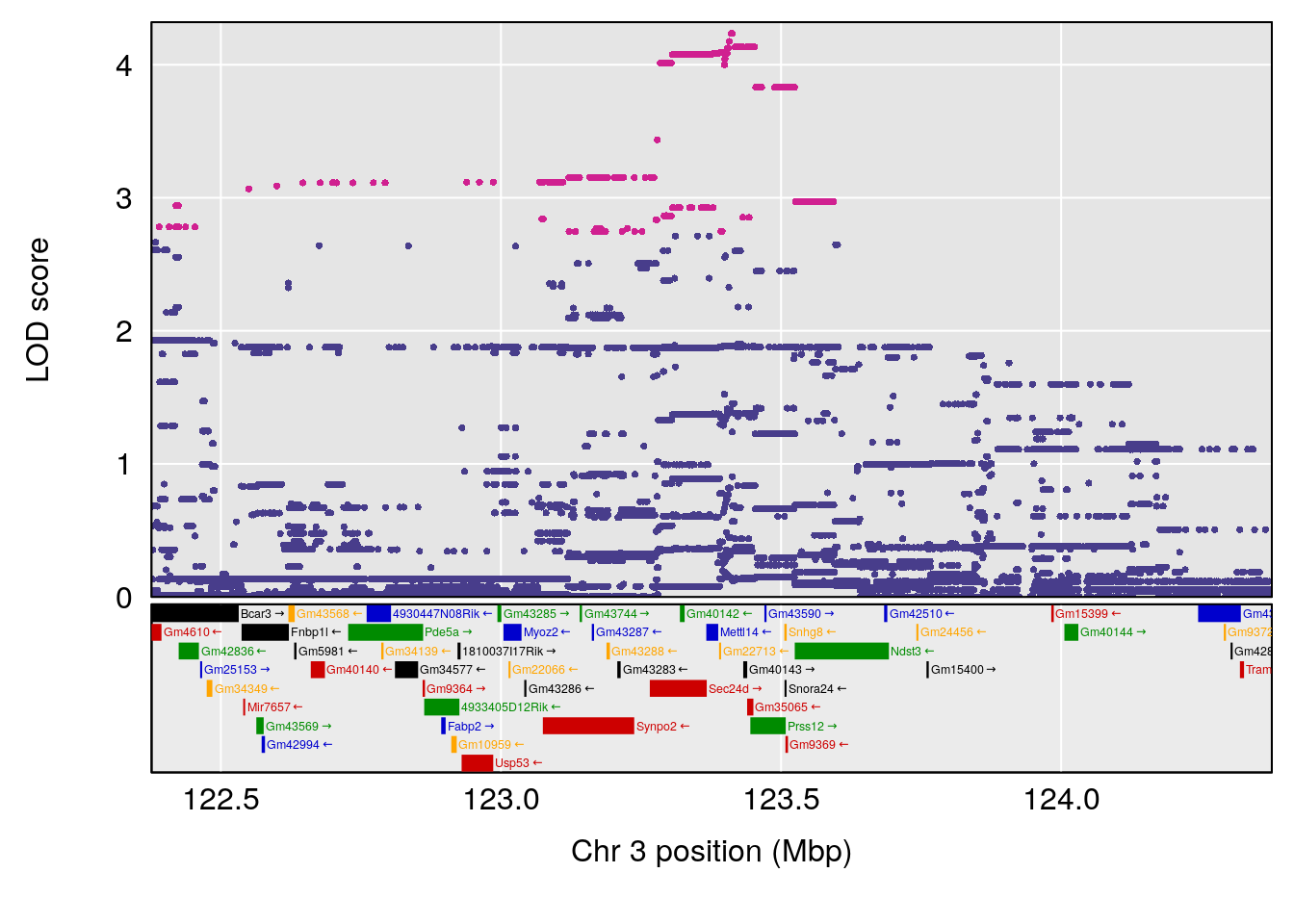
# [1] "RR.Recovery.Rate.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 3 123.37631 6.139661 122.02872 131.78356
# 2 1 pheno1 14 21.17214 8.250728 20.25692 21.53839
# 3 1 pheno1 15 68.07208 6.071145 67.89315 68.61110
# [1] 1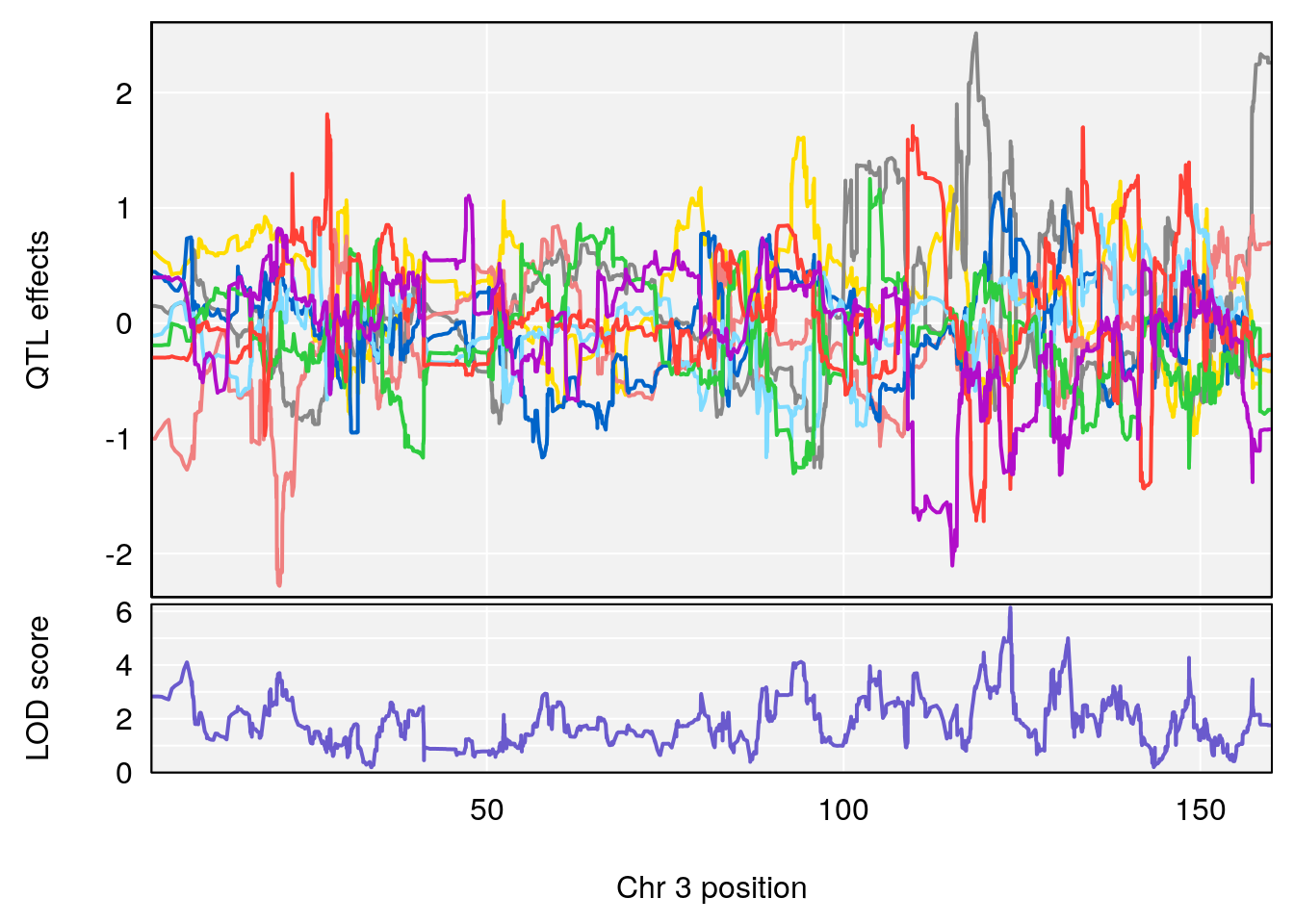
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
# [1] 2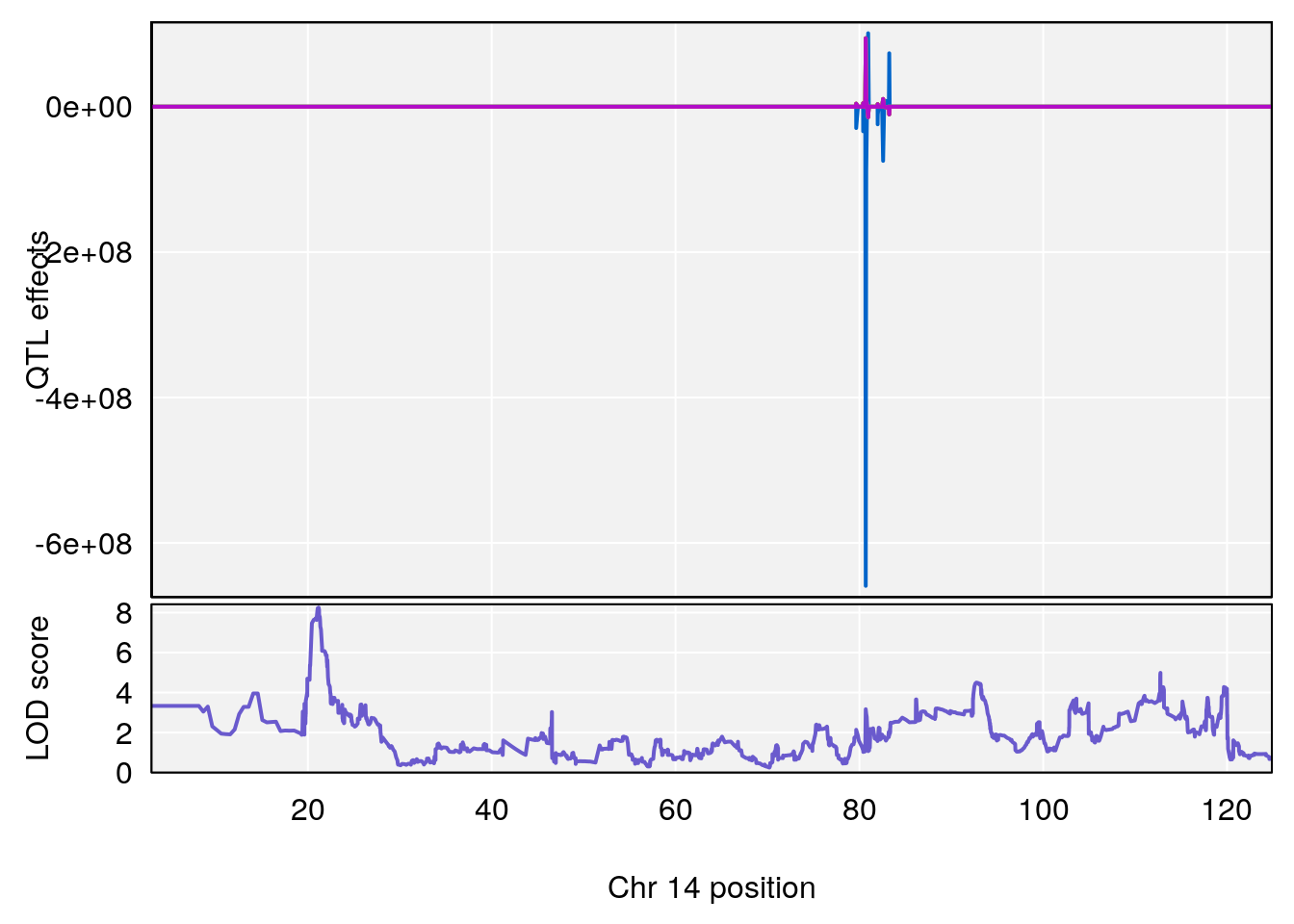
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
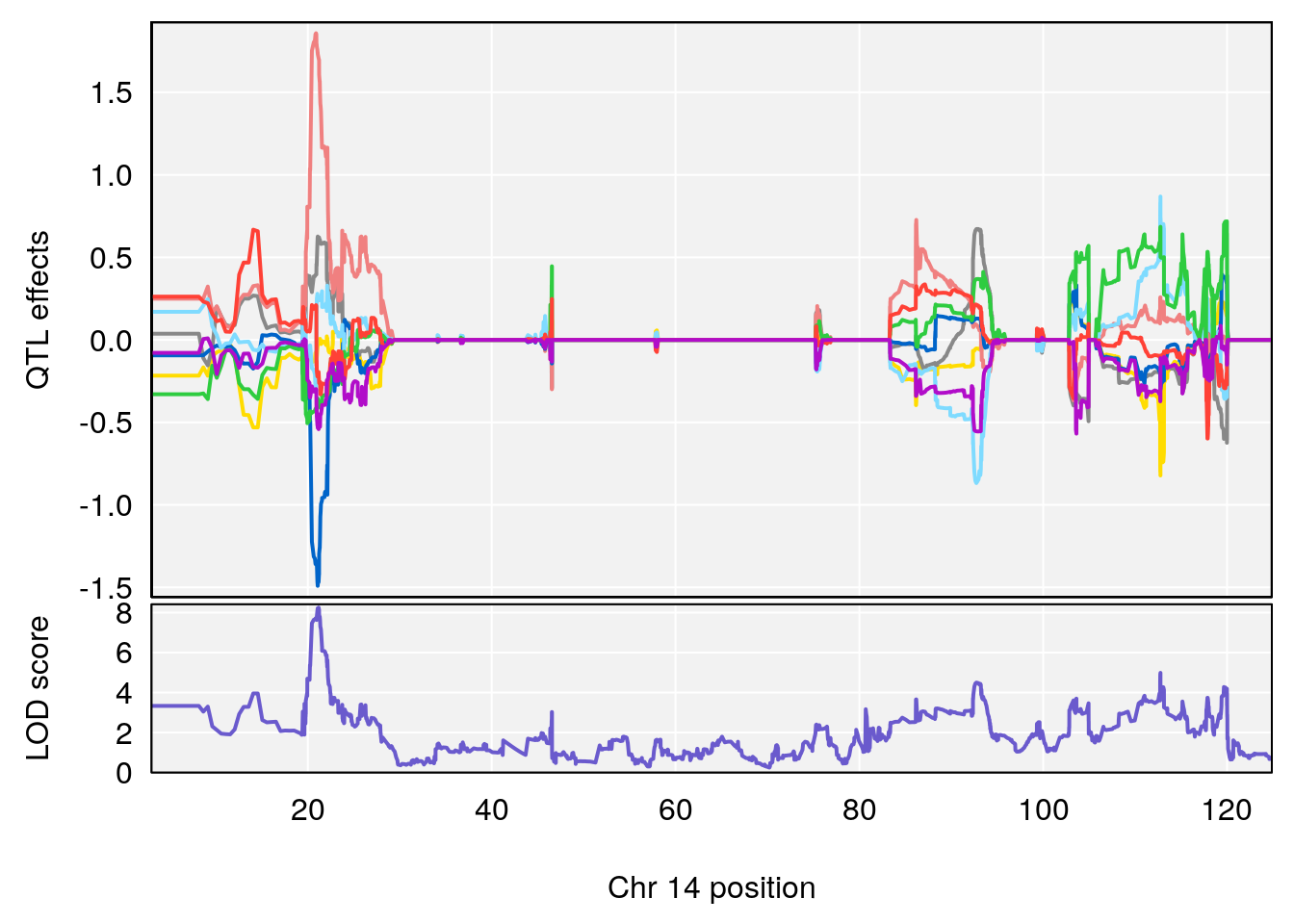
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
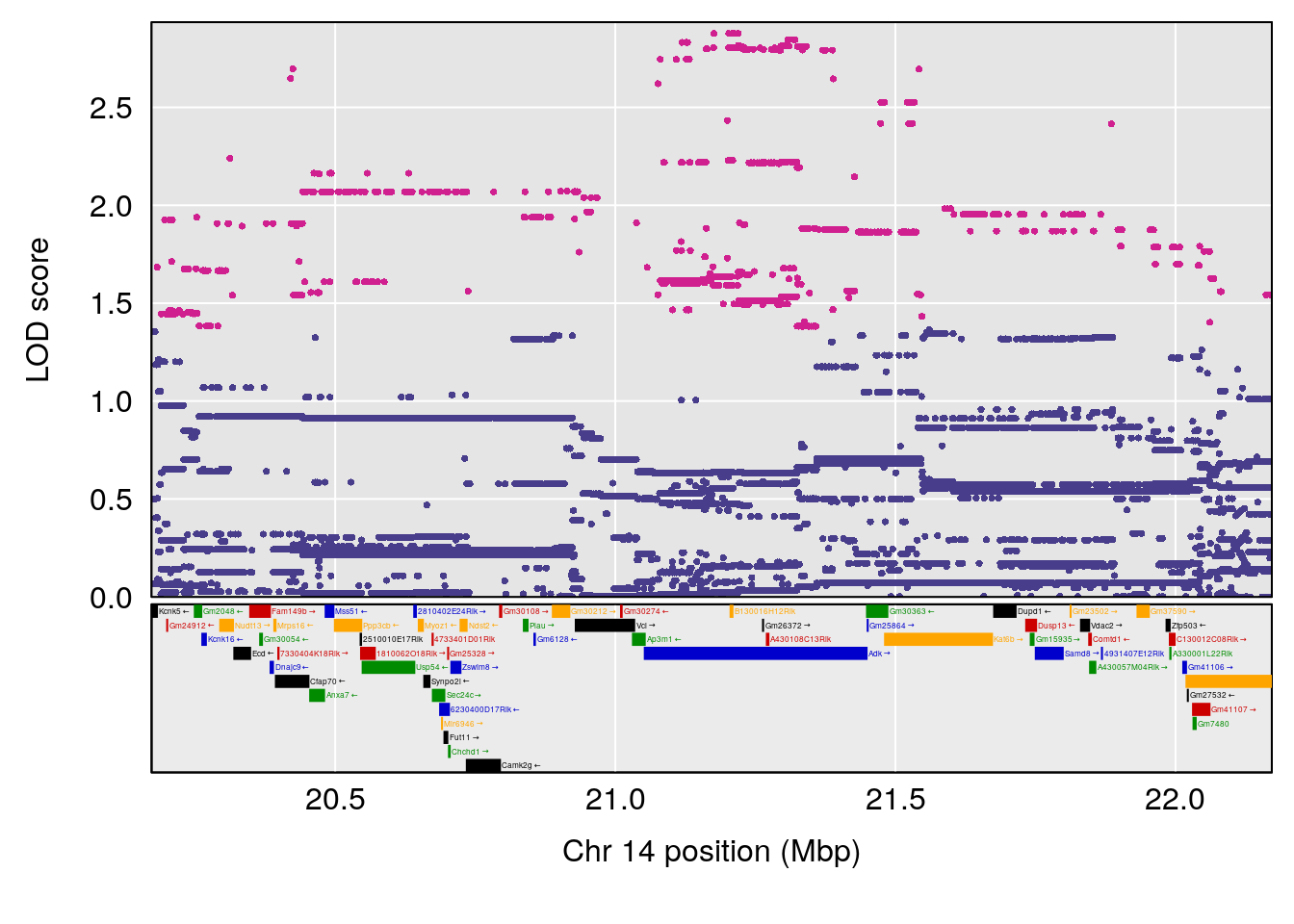
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
# [1] 3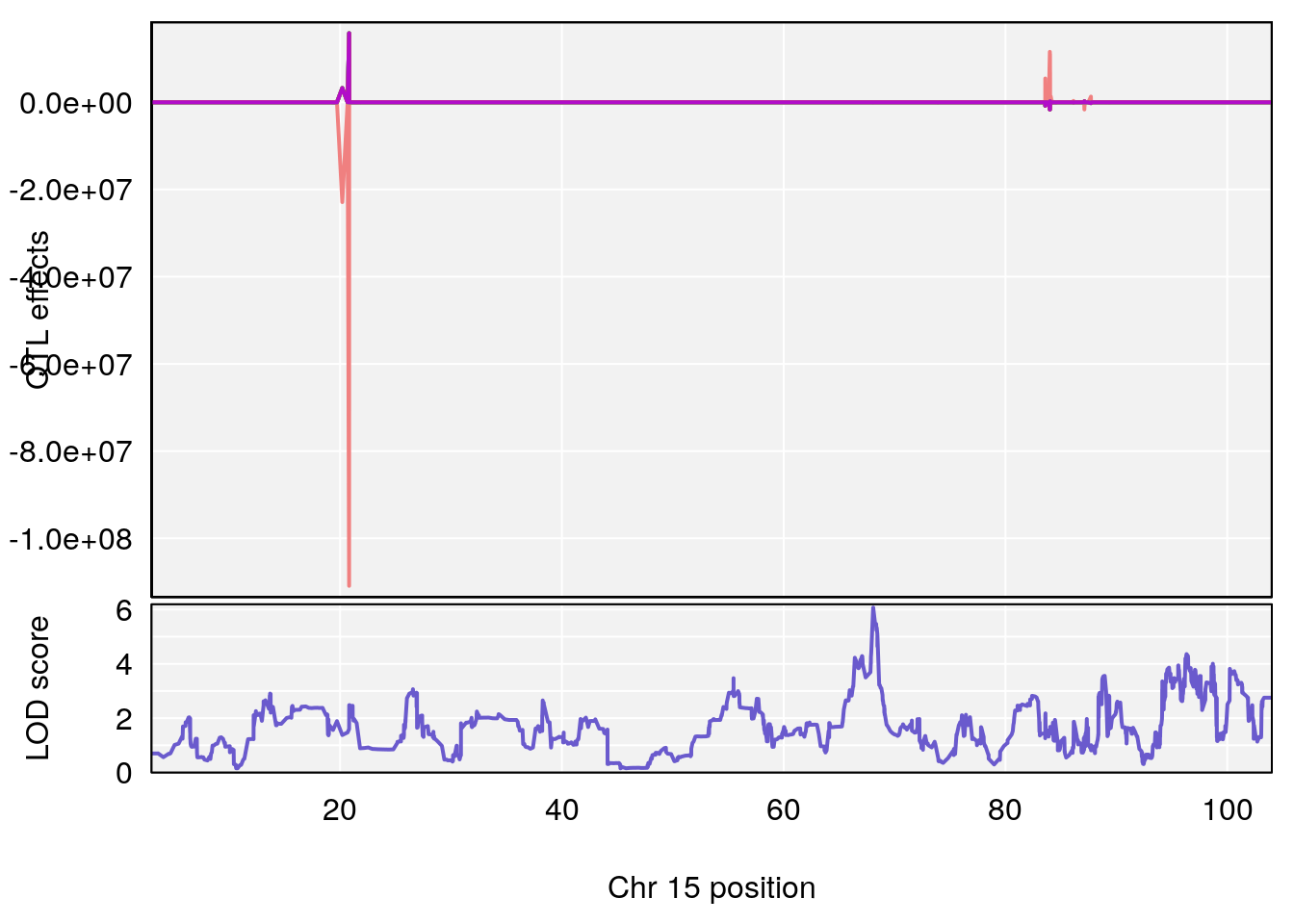
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
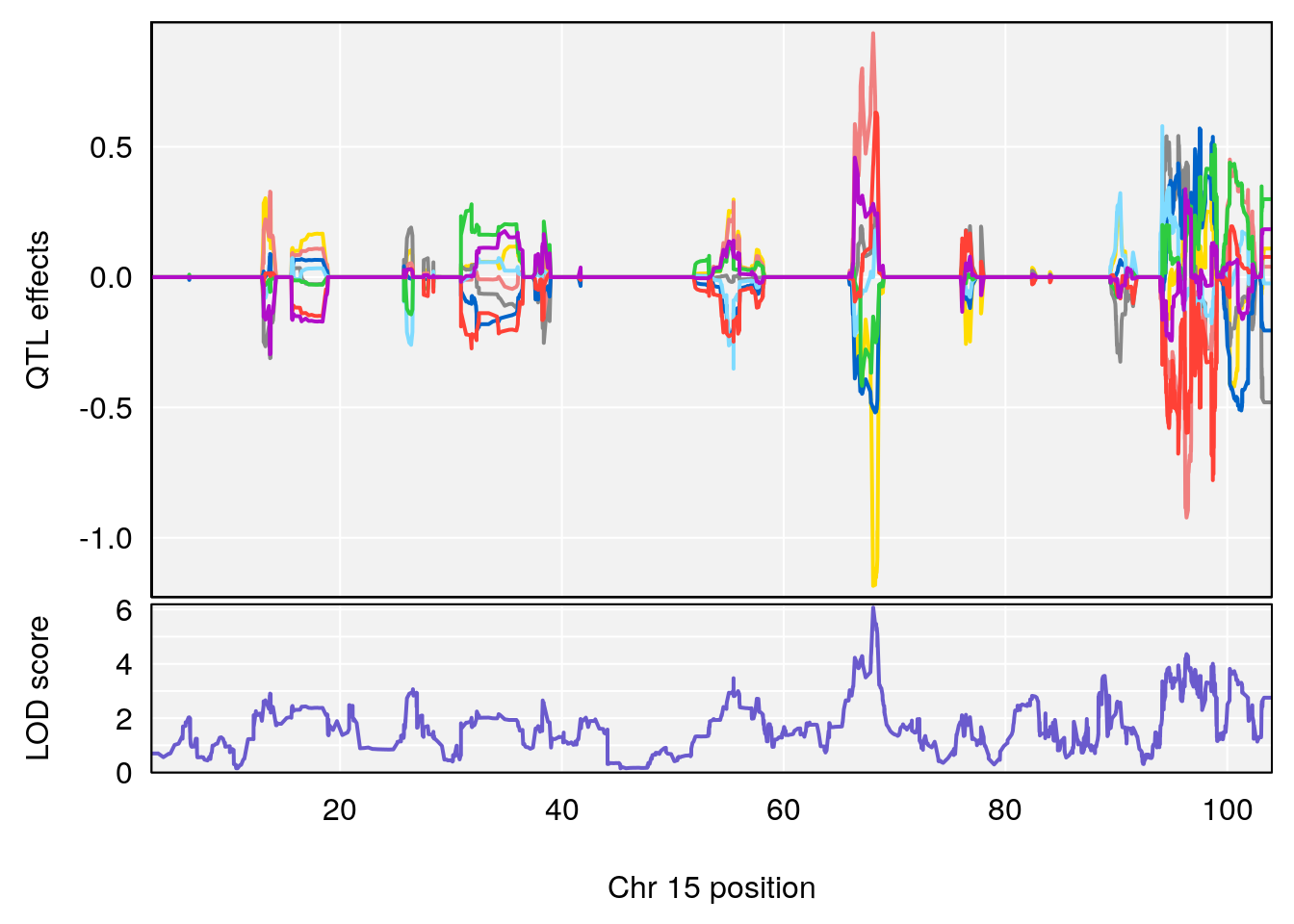
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
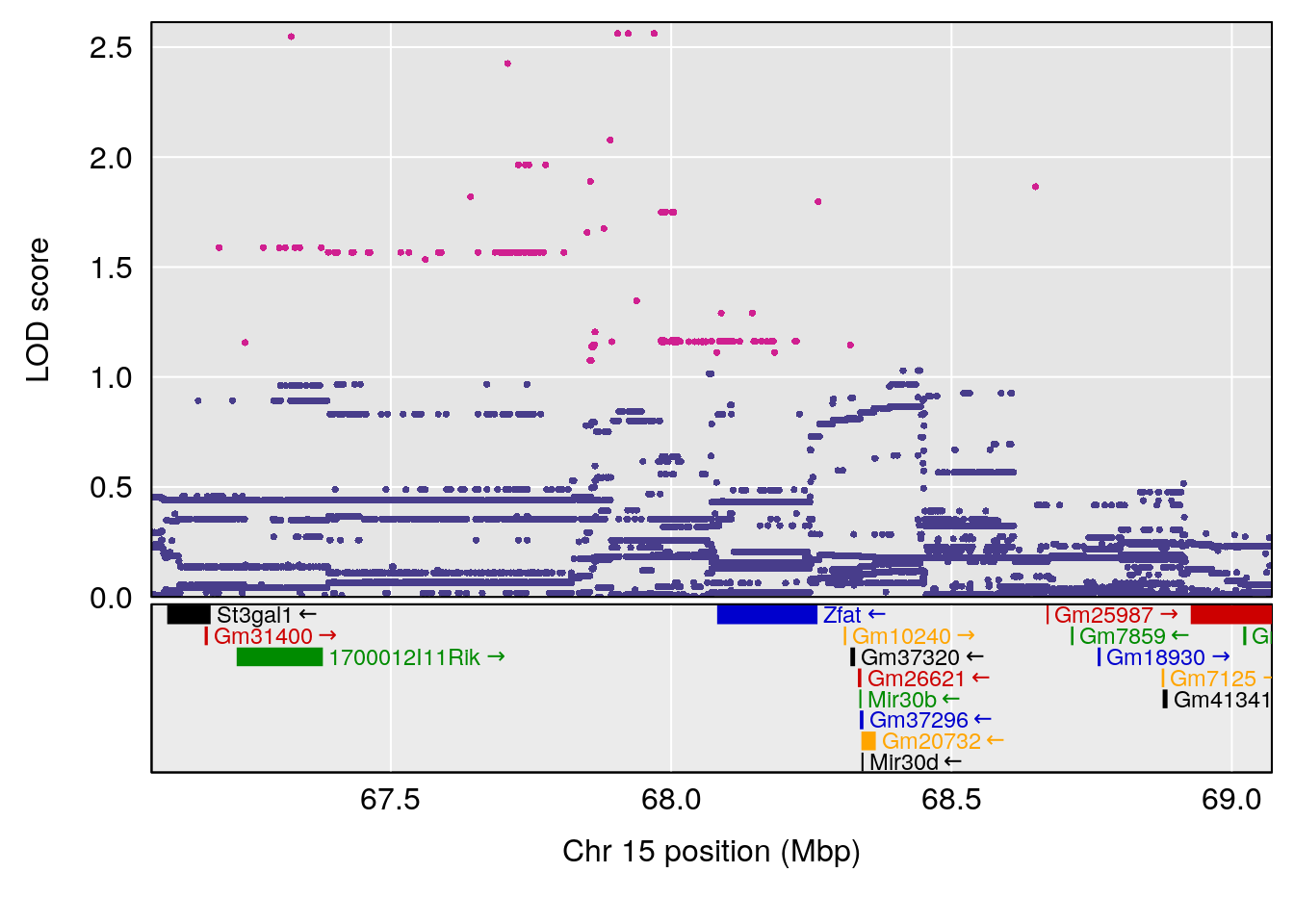
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
#save peaks coeff plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
fname <- paste("output/Fentanyl/DO_fentanyl_69k_alternate_metrics_",i,"_coefplot.pdf",sep="")
pdf(file = fname, width = 16, height =8)
if(dim(peaks)[1] != 0){
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
}
dev.off()
}
# [1] "Time.to.Dead.Hr"
# [1] 1
# [1] 2
# [1] "RR.Depression.Rate.Hr"
# [1] "Time.to.Steady.RR.Depression.Hr"
# [1] "Min.Depression.RR"
# [1] 1
# [1] 2
# [1] "Mean.Depression.BR"
# [1] 1
# [1] 2
# [1] 3
# [1] "Steady-State.Depression.Duration.Hr"
# [1] 1
# [1] 2
# [1] 3
# [1] "Time.to.Threshold.Recovery.Hr"
# [1] 1
# [1] 2
# [1] "Time.to.Projected.Recovery.Hr"
# [1] 1
# [1] "Start.of.Recovery.Hr"
# [1] 1
# [1] 2
# [1] "RR.Recovery.Rate.Hr"
# [1] 1
# [1] 2
# [1] 3
#save peaks coeff blup plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
fname <- paste("output/Fentanyl/DO_fentanyl_69k_alternate_metrics_",i,"_coefplot_blup.pdf",sep="")
pdf(file = fname, width = 16, height =8)
if(dim(peaks)[1] != 0){
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
}
dev.off()
}
# [1] "Time.to.Dead.Hr"
# [1] 1
# [1] 2
# [1] "RR.Depression.Rate.Hr"
# [1] "Time.to.Steady.RR.Depression.Hr"
# [1] "Min.Depression.RR"
# [1] 1
# [1] 2
# [1] "Mean.Depression.BR"
# [1] 1
# [1] 2
# [1] 3
# [1] "Steady-State.Depression.Duration.Hr"
# [1] 1
# [1] 2
# [1] 3
# [1] "Time.to.Threshold.Recovery.Hr"
# [1] 1
# [1] 2
# [1] "Time.to.Projected.Recovery.Hr"
# [1] 1
# [1] "Start.of.Recovery.Hr"
# [1] 1
# [1] 2
# [1] "RR.Recovery.Rate.Hr"
# [1] 1
# [1] 2
# [1] 3###heritability
#plot heritability by qtl2 array
load("output/Fentanyl/qtl.fentanyl.array.alternate.metrics.out.RData")
herit <- data.frame(Phenotype = names(unlist(qtl.morphine.hsq)),
Heritability = round(unlist(qtl.morphine.hsq),2))
herit <- herit %>%
arrange(desc(Heritability))
herit$Phenotype <- factor(herit$Phenotype, levels = herit$Phenotype)
#histgram
pdf(file = paste0("data/FinalReport/GCTA/DO_Fentanyl_alternate_metrics_heritability_by_qtl2_array.pdf"), height = 10, width = 10)
p1<-ggplot(data=herit, aes(x=Phenotype, y=Heritability)) + #, fill=Domain, color = Domain)) +
geom_bar(stat="identity", fill = "blue", color = "blue", show.legend = FALSE) +
scale_y_continuous(breaks=seq(0.0, 1.0, 0.1)) +
geom_text(aes(label = Heritability, y = Heritability + 0.005), position = position_dodge(0.9),vjust = 0) +
theme(axis.text.x = element_text(angle = 90, hjust = 1)) +
ggtitle("Heritability by qtl2 array")
p1
dev.off()
# png
# 2
p1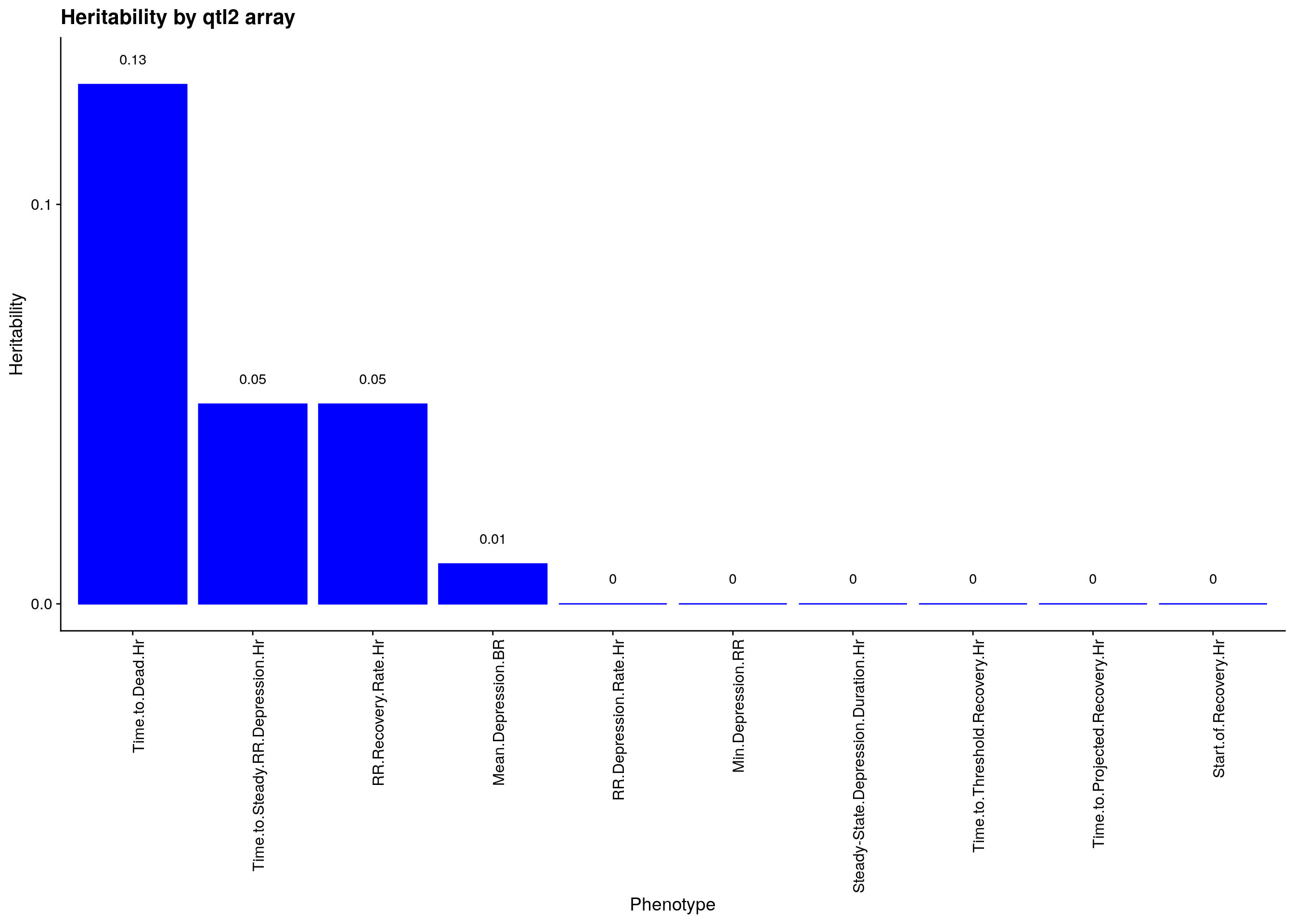
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
#plot heritability by qtl2 69k
load("output/Fentanyl/qtl.fentanyl.69k.alternate.metrics.out.RData")
herit <- data.frame(Phenotype = names(unlist(qtl.morphine.hsq)),
Heritability = round(unlist(qtl.morphine.hsq),2))
herit <- herit %>%
arrange(desc(Heritability))
herit$Phenotype <- factor(herit$Phenotype, levels = herit$Phenotype)
#histgram
pdf(file = paste0("data/FinalReport/GCTA/DO_Fentanyl_alternate_metrics_heritability_by_qtl2_69k.pdf"), height = 10, width = 10)
p2<-ggplot(data=herit, aes(x=Phenotype, y=Heritability)) + #, fill=Domain, color = Domain)) +
geom_bar(stat="identity", fill = "blue", color = "blue", show.legend = FALSE) +
scale_y_continuous(breaks=seq(0.0, 1.0, 0.1)) +
geom_text(aes(label = Heritability, y = Heritability + 0.005), position = position_dodge(0.9),vjust = 0) +
theme(axis.text.x = element_text(angle = 90, hjust = 1)) +
ggtitle("Heritability by qtl2 69k")
p2
dev.off()
# png
# 2
p2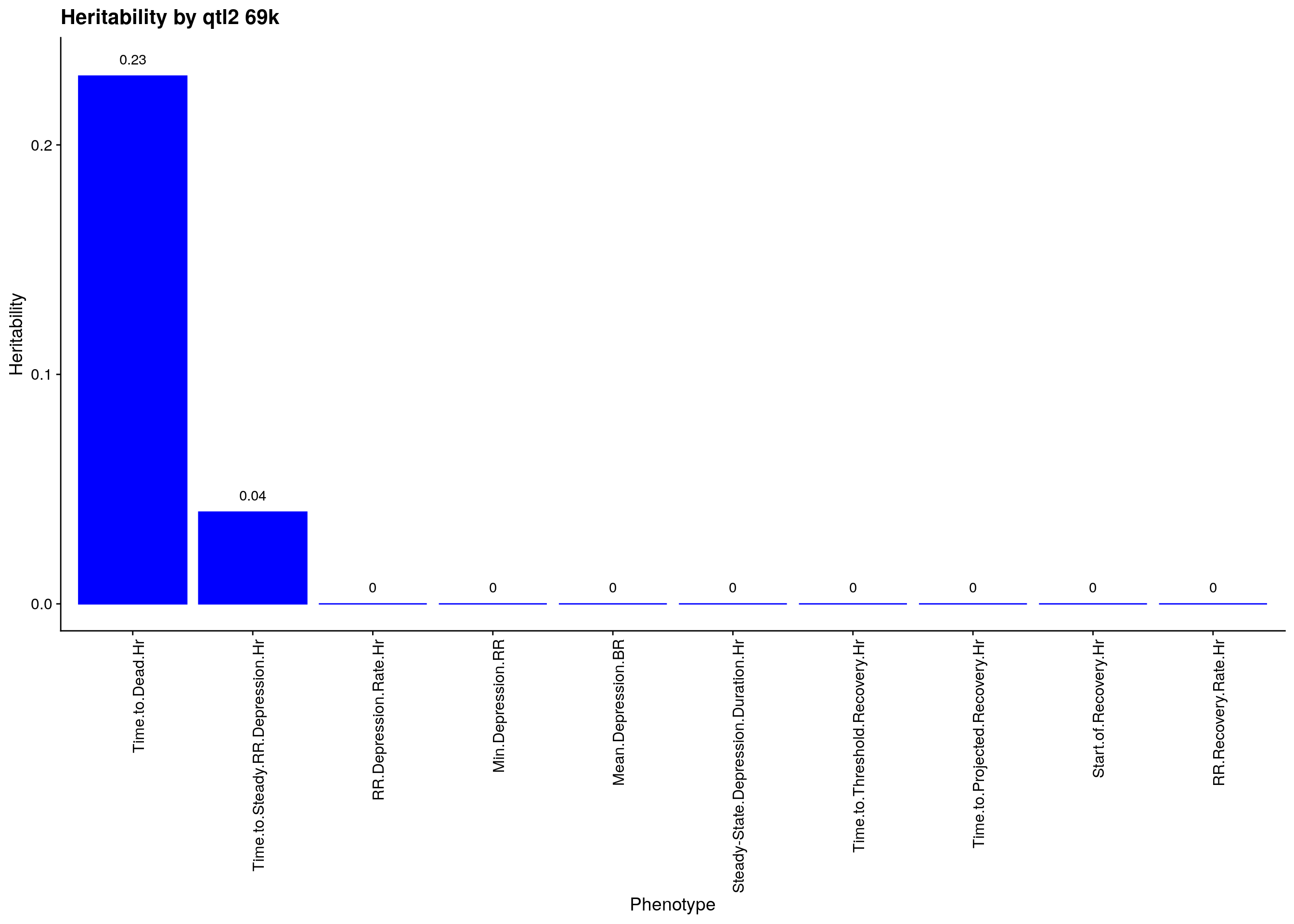
| Version | Author | Date |
|---|---|---|
| f412738 | xhyuo | 2021-12-20 |
sessionInfo()
# R version 4.0.3 (2020-10-10)
# Platform: x86_64-pc-linux-gnu (64-bit)
# Running under: Ubuntu 20.04.2 LTS
#
# Matrix products: default
# BLAS/LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.8.so
#
# locale:
# [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
# [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
# [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=C
# [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
# [9] LC_ADDRESS=C LC_TELEPHONE=C
# [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
#
# attached base packages:
# [1] parallel stats graphics grDevices utils datasets methods
# [8] base
#
# other attached packages:
# [1] qtl_1.47-9 patchwork_1.1.1 furrr_0.2.2 future_1.21.0
# [5] cowplot_1.1.1 forcats_0.5.1 stringr_1.4.0 dplyr_1.0.4
# [9] purrr_0.3.4 readr_1.4.0 tidyr_1.1.2 tibble_3.0.6
# [13] tidyverse_1.3.0 openxlsx_4.2.3 abind_1.4-5 regress_1.3-21
# [17] survival_3.2-7 qtl2_0.24 GGally_2.1.0 gridExtra_2.3
# [21] ggplot2_3.3.3 workflowr_1.6.2
#
# loaded via a namespace (and not attached):
# [1] fs_1.5.0 lubridate_1.7.9.2 bit64_4.0.5 RColorBrewer_1.1-2
# [5] httr_1.4.2 rprojroot_2.0.2 tools_4.0.3 backports_1.2.1
# [9] R6_2.5.0 DBI_1.1.1 colorspace_2.0-0 withr_2.4.1
# [13] tidyselect_1.1.0 bit_4.0.4 compiler_4.0.3 git2r_0.28.0
# [17] cli_2.3.0 rvest_0.3.6 xml2_1.3.2 labeling_0.4.2
# [21] scales_1.1.1 digest_0.6.27 rmarkdown_2.6 pkgconfig_2.0.3
# [25] htmltools_0.5.1.1 parallelly_1.23.0 highr_0.8 dbplyr_2.1.0
# [29] fastmap_1.1.0 rlang_0.4.10 readxl_1.3.1 rstudioapi_0.13
# [33] RSQLite_2.2.3 farver_2.0.3 generics_0.1.0 jsonlite_1.7.2
# [37] zip_2.1.1 magrittr_2.0.1 Matrix_1.2-18 Rcpp_1.0.6
# [41] munsell_0.5.0 lifecycle_1.0.0 stringi_1.5.3 whisker_0.4
# [45] yaml_2.2.1 plyr_1.8.6 grid_4.0.3 blob_1.2.1
# [49] listenv_0.8.0 promises_1.2.0.1 crayon_1.4.1 lattice_0.20-41
# [53] haven_2.3.1 splines_4.0.3 hms_1.0.0 knitr_1.31
# [57] pillar_1.4.7 codetools_0.2-18 reprex_1.0.0 glue_1.4.2
# [61] evaluate_0.14 data.table_1.13.6 modelr_0.1.8 vctrs_0.3.6
# [65] httpuv_1.5.5 cellranger_1.1.0 gtable_0.3.0 reshape_0.8.8
# [69] assertthat_0.2.1 cachem_1.0.4 xfun_0.21 broom_0.7.4
# [73] later_1.1.0.1 memoise_2.0.0 globals_0.14.0 ellipsis_0.3.1This R Markdown site was created with workflowr