Plot_DO_morphine_secondbatch
Hao He
2023-02-21
Last updated: 2023-02-21
Checks: 7 0
Knit directory: DO_Opioid/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200504) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version cc3ae27. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/Picture1.png
Untracked files:
Untracked: Rplot_rz.png
Untracked: TIMBR.test.prop.bm.MN.RET.RData
Untracked: TIMBR.test.random.RData
Untracked: TIMBR.test.rz.transformed_TVb_ml.RData
Untracked: analysis/DDO_morphine1_second_set_69k.stdout
Untracked: analysis/DO_Fentanyl.R
Untracked: analysis/DO_Fentanyl.err
Untracked: analysis/DO_Fentanyl.out
Untracked: analysis/DO_Fentanyl.sh
Untracked: analysis/DO_Fentanyl_69k.R
Untracked: analysis/DO_Fentanyl_69k.err
Untracked: analysis/DO_Fentanyl_69k.out
Untracked: analysis/DO_Fentanyl_69k.sh
Untracked: analysis/DO_Fentanyl_Cohort2_GCTA_herit.R
Untracked: analysis/DO_Fentanyl_Cohort2_gemma.R
Untracked: analysis/DO_Fentanyl_Cohort2_mapping.R
Untracked: analysis/DO_Fentanyl_Cohort2_mapping.err
Untracked: analysis/DO_Fentanyl_Cohort2_mapping.out
Untracked: analysis/DO_Fentanyl_Cohort2_mapping.sh
Untracked: analysis/DO_Fentanyl_GCTA_herit.R
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.R
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.err
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.out
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.sh
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.R
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.err
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.out
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.sh
Untracked: analysis/DO_Fentanyl_array.R
Untracked: analysis/DO_Fentanyl_array.err
Untracked: analysis/DO_Fentanyl_array.out
Untracked: analysis/DO_Fentanyl_array.sh
Untracked: analysis/DO_Fentanyl_combining2Cohort_GCTA_herit.R
Untracked: analysis/DO_Fentanyl_combining2Cohort_gemma.R
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping.R
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping.err
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping.out
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping.sh
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping_CoxPH.R
Untracked: analysis/DO_Fentanyl_finalreport_to_plink.sh
Untracked: analysis/DO_Fentanyl_gemma.R
Untracked: analysis/DO_Fentanyl_gemma.err
Untracked: analysis/DO_Fentanyl_gemma.out
Untracked: analysis/DO_Fentanyl_gemma.sh
Untracked: analysis/DO_morphine1.R
Untracked: analysis/DO_morphine1.Rout
Untracked: analysis/DO_morphine1.sh
Untracked: analysis/DO_morphine1.stderr
Untracked: analysis/DO_morphine1.stdout
Untracked: analysis/DO_morphine1_SNP.R
Untracked: analysis/DO_morphine1_SNP.Rout
Untracked: analysis/DO_morphine1_SNP.sh
Untracked: analysis/DO_morphine1_SNP.stderr
Untracked: analysis/DO_morphine1_SNP.stdout
Untracked: analysis/DO_morphine1_combined.R
Untracked: analysis/DO_morphine1_combined.Rout
Untracked: analysis/DO_morphine1_combined.sh
Untracked: analysis/DO_morphine1_combined.stderr
Untracked: analysis/DO_morphine1_combined.stdout
Untracked: analysis/DO_morphine1_combined_69k.R
Untracked: analysis/DO_morphine1_combined_69k.Rout
Untracked: analysis/DO_morphine1_combined_69k.sh
Untracked: analysis/DO_morphine1_combined_69k.stderr
Untracked: analysis/DO_morphine1_combined_69k.stdout
Untracked: analysis/DO_morphine1_combined_69k_m2.R
Untracked: analysis/DO_morphine1_combined_69k_m2.Rout
Untracked: analysis/DO_morphine1_combined_69k_m2.sh
Untracked: analysis/DO_morphine1_combined_69k_m2.stderr
Untracked: analysis/DO_morphine1_combined_69k_m2.stdout
Untracked: analysis/DO_morphine1_combined_weight_DOB.R
Untracked: analysis/DO_morphine1_combined_weight_DOB.Rout
Untracked: analysis/DO_morphine1_combined_weight_DOB.err
Untracked: analysis/DO_morphine1_combined_weight_DOB.out
Untracked: analysis/DO_morphine1_combined_weight_DOB.sh
Untracked: analysis/DO_morphine1_combined_weight_DOB.stderr
Untracked: analysis/DO_morphine1_combined_weight_DOB.stdout
Untracked: analysis/DO_morphine1_combined_weight_age.R
Untracked: analysis/DO_morphine1_combined_weight_age.err
Untracked: analysis/DO_morphine1_combined_weight_age.out
Untracked: analysis/DO_morphine1_combined_weight_age.sh
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.R
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.err
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.out
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.sh
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.R
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.err
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.out
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.sh
Untracked: analysis/DO_morphine1_cph.R
Untracked: analysis/DO_morphine1_cph.Rout
Untracked: analysis/DO_morphine1_cph.sh
Untracked: analysis/DO_morphine1_second_set.R
Untracked: analysis/DO_morphine1_second_set.Rout
Untracked: analysis/DO_morphine1_second_set.sh
Untracked: analysis/DO_morphine1_second_set.stderr
Untracked: analysis/DO_morphine1_second_set.stdout
Untracked: analysis/DO_morphine1_second_set_69k.R
Untracked: analysis/DO_morphine1_second_set_69k.Rout
Untracked: analysis/DO_morphine1_second_set_69k.sh
Untracked: analysis/DO_morphine1_second_set_69k.stderr
Untracked: analysis/DO_morphine1_second_set_SNP.R
Untracked: analysis/DO_morphine1_second_set_SNP.Rout
Untracked: analysis/DO_morphine1_second_set_SNP.sh
Untracked: analysis/DO_morphine1_second_set_SNP.stderr
Untracked: analysis/DO_morphine1_second_set_SNP.stdout
Untracked: analysis/DO_morphine1_second_set_weight_DOB.R
Untracked: analysis/DO_morphine1_second_set_weight_DOB.Rout
Untracked: analysis/DO_morphine1_second_set_weight_DOB.err
Untracked: analysis/DO_morphine1_second_set_weight_DOB.out
Untracked: analysis/DO_morphine1_second_set_weight_DOB.sh
Untracked: analysis/DO_morphine1_second_set_weight_DOB.stderr
Untracked: analysis/DO_morphine1_second_set_weight_DOB.stdout
Untracked: analysis/DO_morphine1_second_set_weight_age.R
Untracked: analysis/DO_morphine1_second_set_weight_age.Rout
Untracked: analysis/DO_morphine1_second_set_weight_age.err
Untracked: analysis/DO_morphine1_second_set_weight_age.out
Untracked: analysis/DO_morphine1_second_set_weight_age.sh
Untracked: analysis/DO_morphine1_second_set_weight_age.stderr
Untracked: analysis/DO_morphine1_second_set_weight_age.stdout
Untracked: analysis/DO_morphine1_weight_DOB.R
Untracked: analysis/DO_morphine1_weight_DOB.sh
Untracked: analysis/DO_morphine1_weight_age.R
Untracked: analysis/DO_morphine1_weight_age.sh
Untracked: analysis/DO_morphine_gemma.R
Untracked: analysis/DO_morphine_gemma.err
Untracked: analysis/DO_morphine_gemma.out
Untracked: analysis/DO_morphine_gemma.sh
Untracked: analysis/DO_morphine_gemma_firstmin.R
Untracked: analysis/DO_morphine_gemma_firstmin.err
Untracked: analysis/DO_morphine_gemma_firstmin.out
Untracked: analysis/DO_morphine_gemma_firstmin.sh
Untracked: analysis/DO_morphine_gemma_withpermu.R
Untracked: analysis/DO_morphine_gemma_withpermu.err
Untracked: analysis/DO_morphine_gemma_withpermu.out
Untracked: analysis/DO_morphine_gemma_withpermu.sh
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.R
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.err
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.out
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.sh
Untracked: analysis/Plot_DO_morphine1_SNP.R
Untracked: analysis/Plot_DO_morphine1_SNP.Rout
Untracked: analysis/Plot_DO_morphine1_SNP.sh
Untracked: analysis/Plot_DO_morphine1_SNP.stderr
Untracked: analysis/Plot_DO_morphine1_SNP.stdout
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.R
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.Rout
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.sh
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.stderr
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.stdout
Untracked: analysis/download_GSE100356_sra.sh
Untracked: analysis/fentanyl_2cohorts_coxph.R
Untracked: analysis/fentanyl_2cohorts_coxph.err
Untracked: analysis/fentanyl_2cohorts_coxph.out
Untracked: analysis/fentanyl_2cohorts_coxph.sh
Untracked: analysis/fentanyl_scanone.cph.R
Untracked: analysis/fentanyl_scanone.cph.err
Untracked: analysis/fentanyl_scanone.cph.out
Untracked: analysis/fentanyl_scanone.cph.sh
Untracked: analysis/geo_rnaseq.R
Untracked: analysis/heritability_first_second_batch.R
Untracked: analysis/nf-rnaseq-b6.R
Untracked: analysis/plot_fentanyl_2cohorts_coxph.R
Untracked: analysis/scripts/
Untracked: analysis/tibmr.R
Untracked: analysis/timbr_demo.R
Untracked: analysis/workflow_proc.R
Untracked: analysis/workflow_proc.sh
Untracked: analysis/workflow_proc.stderr
Untracked: analysis/workflow_proc.stdout
Untracked: analysis/x.R
Untracked: code/cfw/
Untracked: code/gemma_plot.R
Untracked: code/process.sanger.snp.R
Untracked: code/reconst_utils.R
Untracked: data/69k_grid_pgmap.RData
Untracked: data/Composite Post Kevins Program Group 2 Fentanyl Prepped for Hao.xlsx
Untracked: data/DO_WBP_Data_JAB_to_map.xlsx
Untracked: data/Fentanyl_alternate_metrics.xlsx
Untracked: data/FinalReport/
Untracked: data/GM/
Untracked: data/GM_covar.csv
Untracked: data/GM_covar_07092018_morphine.csv
Untracked: data/Jackson_Lab_Bubier_MURGIGV01/
Untracked: data/MPD_Upload_October.csv
Untracked: data/MPD_Upload_October_updated_sex.csv
Untracked: data/Master Fentanyl DO Study Sheet.xlsx
Untracked: data/MasterMorphine Second Set DO w DOB2.xlsx
Untracked: data/MasterMorphine Second Set DO.xlsx
Untracked: data/Morphine CC DO mice Updated with Published inbred strains.csv
Untracked: data/Morphine_CC_DO_mice_Updated_with_Published_inbred_strains.csv
Untracked: data/cc_variants.sqlite
Untracked: data/combined/
Untracked: data/fentanyl/
Untracked: data/fentanyl2/
Untracked: data/fentanyl_1_2/
Untracked: data/fentanyl_2cohorts_coxph_data.Rdata
Untracked: data/first/
Untracked: data/founder_geno.csv
Untracked: data/genetic_map.csv
Untracked: data/gm.json
Untracked: data/gwas.sh
Untracked: data/marker_grid_0.02cM_plus.txt
Untracked: data/mouse_genes_mgi.sqlite
Untracked: data/pheno.csv
Untracked: data/pheno_qtl2.csv
Untracked: data/pheno_qtl2_07092018_morphine.csv
Untracked: data/pheno_qtl2_w_dob.csv
Untracked: data/physical_map.csv
Untracked: data/rnaseq/
Untracked: data/sample_geno.csv
Untracked: data/second/
Untracked: figure/
Untracked: glimma-plots/
Untracked: output/DO_Fentanyl_Cohort2_MinDepressionRR_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_MinDepressionRR_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_RRDepressionRateHrSLOPE_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_RRRecoveryRateHrSLOPE_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_RRRecoveryRateHrSLOPE_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_StartofRecoveryHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_StartofRecoveryHr_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_Statusbin_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_Statusbin_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_SteadyStateDepressionDurationHrINTERVAL_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoDead(Hr)_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoDeadHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoDeadHr_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoProjectedRecoveryHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoProjectedRecoveryHr_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoSteadyRRDepression(Hr)_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoSteadyRRDepressionHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoSteadyRRDepressionHr_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoThresholdRecoveryHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoThresholdRecoveryHr_coefplot_blup.pdf
Untracked: output/DO_morphine_Min.depression.png
Untracked: output/DO_morphine_Min.depression22222_violin_chr5.pdf
Untracked: output/DO_morphine_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_Min.depression_coefplot_blup_chr5.png
Untracked: output/DO_morphine_Min.depression_coefplot_blup_chrX.png
Untracked: output/DO_morphine_Min.depression_coefplot_chr5.png
Untracked: output/DO_morphine_Min.depression_coefplot_chrX.png
Untracked: output/DO_morphine_Min.depression_peak_genes_chr5.png
Untracked: output/DO_morphine_Min.depression_violin_chr5.png
Untracked: output/DO_morphine_Recovery.Time.png
Untracked: output/DO_morphine_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr11.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr4.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr7.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr9.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr11.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr4.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr7.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr9.png
Untracked: output/DO_morphine_Status_bin.png
Untracked: output/DO_morphine_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_Survival.Time.png
Untracked: output/DO_morphine_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_Survival.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_Survival.Time_coefplot_blup_chr17.png
Untracked: output/DO_morphine_Survival.Time_coefplot_blup_chr8.png
Untracked: output/DO_morphine_Survival.Time_coefplot_chr17.png
Untracked: output/DO_morphine_Survival.Time_coefplot_chr8.png
Untracked: output/DO_morphine_combine_batch_peak_violin.pdf
Untracked: output/DO_morphine_combined_69k_m2_Min.depression.png
Untracked: output/DO_morphine_combined_69k_m2_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_combined_69k_m2_Recovery.Time.png
Untracked: output/DO_morphine_combined_69k_m2_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_combined_69k_m2_Status_bin.png
Untracked: output/DO_morphine_combined_69k_m2_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_combined_69k_m2_Survival.Time.png
Untracked: output/DO_morphine_combined_69k_m2_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Survival.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_coxph_24hrs_kinship_QTL.png
Untracked: output/DO_morphine_cphout.RData
Untracked: output/DO_morphine_first_batch_peak_in_second_batch_violin.pdf
Untracked: output/DO_morphine_first_batch_peak_in_second_batch_violin_sidebyside.pdf
Untracked: output/DO_morphine_first_batch_peak_violin.pdf
Untracked: output/DO_morphine_operm.cph.RData
Untracked: output/DO_morphine_second_batch_on_first_batch_peak_violin.pdf
Untracked: output/DO_morphine_second_batch_peak_ch6surv_on_first_batchviolin.pdf
Untracked: output/DO_morphine_second_batch_peak_ch6surv_on_first_batchviolin2.pdf
Untracked: output/DO_morphine_second_batch_peak_in_first_batch_violin.pdf
Untracked: output/DO_morphine_second_batch_peak_in_first_batch_violin_sidebyside.pdf
Untracked: output/DO_morphine_second_batch_peak_violin.pdf
Untracked: output/DO_morphine_secondbatch_69k_Min.depression.png
Untracked: output/DO_morphine_secondbatch_69k_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_69k_Recovery.Time.png
Untracked: output/DO_morphine_secondbatch_69k_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_69k_Status_bin.png
Untracked: output/DO_morphine_secondbatch_69k_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_69k_Survival.Time.png
Untracked: output/DO_morphine_secondbatch_69k_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Survival.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Min.depression.png
Untracked: output/DO_morphine_secondbatch_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Recovery.Time.png
Untracked: output/DO_morphine_secondbatch_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Status_bin.png
Untracked: output/DO_morphine_secondbatch_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Survival.Time.png
Untracked: output/DO_morphine_secondbatch_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Survival.Time_coefplot_blup.pdf
Untracked: output/Fentanyl/
Untracked: output/KPNA3.pdf
Untracked: output/SSC4D.pdf
Untracked: output/TIMBR.test.RData
Untracked: output/apr_69kchr_combined.RData
Untracked: output/apr_69kchr_k_loco_combined.rds
Untracked: output/apr_69kchr_second_set.RData
Untracked: output/combine_batch_variation.RData
Untracked: output/combined_gm.RData
Untracked: output/combined_gm.k_loco.rds
Untracked: output/combined_gm.k_overall.rds
Untracked: output/combined_gm.probs_8state.rds
Untracked: output/coxph/
Untracked: output/do.morphine.RData
Untracked: output/do.morphine.k_loco.rds
Untracked: output/do.morphine.probs_36state.rds
Untracked: output/do.morphine.probs_8state.rds
Untracked: output/do_Fentanyl_combine2cohort_MeanDepressionBR_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_MeanDepressionBR_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_MinDepressionBR_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_MinDepressionBR_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_MinDepressionRR_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_MinDepressionRR_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_RRRecoveryRateHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_RRRecoveryRateHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_StartofRecoveryHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_StartofRecoveryHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_Statusbin_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_Statusbin_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_SteadyStateDepressionDurationHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_SteadyStateDepressionDurationHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_SurvivalTime_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_SurvivalTime_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoDeadHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoDeadHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoMostlyDeadHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoMostlyDeadHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoProjectedRecoveryHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoProjectedRecoveryHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoRecoveryHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoRecoveryHr_coefplot_blup.pdf
Untracked: output/first_batch_variation.RData
Untracked: output/first_second_survival_peak_chr.xlsx
Untracked: output/hsq_1_first_batch_herit_qtl2.RData
Untracked: output/hsq_2_second_batch_herit_qtl2.RData
Untracked: output/old_temp/
Untracked: output/pr_69kchr_combined.RData
Untracked: output/pr_69kchr_second_set.RData
Untracked: output/qtl.morphine.69k.out.combined.RData
Untracked: output/qtl.morphine.69k.out.combined_m2.RData
Untracked: output/qtl.morphine.69k.out.second_set.RData
Untracked: output/qtl.morphine.operm.RData
Untracked: output/qtl.morphine.out.RData
Untracked: output/qtl.morphine.out.combined_gm.RData
Untracked: output/qtl.morphine.out.combined_gm.female.RData
Untracked: output/qtl.morphine.out.combined_gm.male.RData
Untracked: output/qtl.morphine.out.combined_weight_DOB.RData
Untracked: output/qtl.morphine.out.combined_weight_age.RData
Untracked: output/qtl.morphine.out.female.RData
Untracked: output/qtl.morphine.out.male.RData
Untracked: output/qtl.morphine.out.second_set.RData
Untracked: output/qtl.morphine.out.second_set.female.RData
Untracked: output/qtl.morphine.out.second_set.male.RData
Untracked: output/qtl.morphine.out.second_set.weight_DOB.RData
Untracked: output/qtl.morphine.out.second_set.weight_age.RData
Untracked: output/qtl.morphine.out.weight_DOB.RData
Untracked: output/qtl.morphine.out.weight_age.RData
Untracked: output/qtl.morphine1.snpout.RData
Untracked: output/qtl.morphine2.snpout.RData
Untracked: output/second_batch_pheno.csv
Untracked: output/second_batch_variation.RData
Untracked: output/second_set_apr_69kchr_k_loco.rds
Untracked: output/second_set_gm.RData
Untracked: output/second_set_gm.k_loco.rds
Untracked: output/second_set_gm.probs_36state.rds
Untracked: output/second_set_gm.probs_8state.rds
Untracked: output/topSNP_chr5_mindepression.csv
Untracked: output/zoompeak_Min.depression_9.pdf
Untracked: output/zoompeak_Recovery.Time_16.pdf
Untracked: output/zoompeak_Status_bin_11.pdf
Untracked: output/zoompeak_Survival.Time_1.pdf
Untracked: output/zoompeak_fentanyl_Survival.Time_2.pdf
Untracked: sra-tools_v2.10.7.sif
Unstaged changes:
Modified: _workflowr.yml
Modified: analysis/Plot_DO_Fentanyl_Cohort2_mapping.Rmd
Modified: analysis/Plot_DO_Fentanyl_combining2Cohort_mapping.Rmd
Modified: analysis/Plot_DO_fentanyl.Rmd
Modified: analysis/marker_violin.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/Plot_DO_morphine_secondbatch.Rmd) and HTML (docs/Plot_DO_morphine_secondbatch.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | cc3ae27 | xhyuo | 2023-02-21 | update with sex specific morphine plot |
| html | 8fe8543 | xhyuo | 2023-02-20 | Build site. |
| Rmd | b46775c | xhyuo | 2023-02-20 | update with sex specific morphine |
| html | 9e4e3be | xhyuo | 2021-08-24 | Build site. |
| Rmd | 8b6594f | xhyuo | 2021-08-24 | add snp/gene plot |
| html | c191146 | xhyuo | 2020-06-16 | Build site. |
| Rmd | 45c4bc8 | xhyuo | 2020-06-16 | Plot_DO_morphine_secondbatch |
| html | 5cbc78e | xhyuo | 2020-06-01 | Build site. |
| Rmd | 60e46d6 | xhyuo | 2020-06-01 | Plot_DO_morphine_secondbatch |
| html | b830cde | xhyuo | 2020-06-01 | Build site. |
| Rmd | 8ecb9e6 | xhyuo | 2020-06-01 | Plot_DO_morphine_secondbatch |
Last update: 2023-02-21
loading libraries
library(ggplot2)
library(gridExtra)
library(GGally)
library(parallel)
library(qtl2)
library(parallel)
library(survival)
library(regress)
library(abind)
library(openxlsx)
rz.transform <- function(y) {
rankY=rank(y, ties.method="average", na.last="keep")
rzT=qnorm(rankY/(length(na.exclude(rankY))+1))
return(rzT)
}
load("code/do.colors.RData")Read phenotype data
# Read phenotype data -----------------------------------------------------
second_set <- read.xlsx(xlsxFile = "data/MasterMorphine Second Set DO.xlsx",
sheet = 1)
colnames(second_set)[2] <- "Sex"
colnames(second_set)[6] <- "Status24"
#character
second_set[,c("ID", "Sex", "GROUP", "Status24")] <- lapply(second_set[, c("ID", "Sex", "GROUP", "Status24")], as.character)
second_set$Status_bin <- ifelse(second_set$Status24 == "ALIVE", 1, 0)
rownames(second_set) <- second_set$ID
#remove outlier
second_set <- second_set[second_set$ID != "empty",]
#remove negative survival time subjects
second_set <- second_set[!(!is.na(second_set$Survival.Time) & second_set$Survival.Time < 0),]
second_set$Survival.Time[!is.na(second_set$Survival.Time) & second_set$Survival.Time > 15] <- NA
second_set$Recovery.Time[!is.na(second_set$Recovery.Time) & second_set$Recovery.Time > 24] <- NA
second_set$Min.depression[!is.na(second_set$Min.depression) & second_set$Min.depression > 1] <- NA
#do
do.morphine <- get(load("output/second_set_gm.RData"))
do.pheno <- merge(second_set, do.morphine$covar, by.x = "row.names", by.y = "row.names", all.x = TRUE)
#first batch
do.pheno1 <- read.csv("data/pheno_qtl2.csv")
colnames(do.pheno1)[1] <- "ID"
do.pheno1$Survival.Time <- as.numeric(do.pheno1$Survival.Time)
do.pheno1$Recovery.Time <- as.numeric(do.pheno1$Recovery.Time)
do.pheno1$Min.depression <- as.numeric(do.pheno1$Min.depression)
do.pheno1$ngen <- "29"
#boxplot
#survival time
surv <- do.pheno[,c(2,8,15)]
#surv <- rbind(surv, do.pheno1[,c(1,2,5)])
surv <- surv[complete.cases(surv), ]
surv$ngen <- factor(surv$ngen, levels = as.character(c(30:34)))
p1 <- ggplot(surv, aes(x=ngen, y=Survival.Time, group = ngen, fill = ngen, alpha = 0.9)) +
geom_boxplot(show.legend = F , outlier.size = 1.5, notchwidth = 0.85) +
geom_jitter(color="black", size=0.8, alpha=0.9) +
scale_fill_brewer(palette="Blues") +
ylab("Survival Time") +
xlab("Generation") +
labs(fill = "") +
theme(legend.position = "none",
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
axis.line = element_line(colour = "black"),
text = element_text(size=16),
axis.title=element_text(size=16)) +
guides(shape = guide_legend(override.aes = list(size = 12)))
p1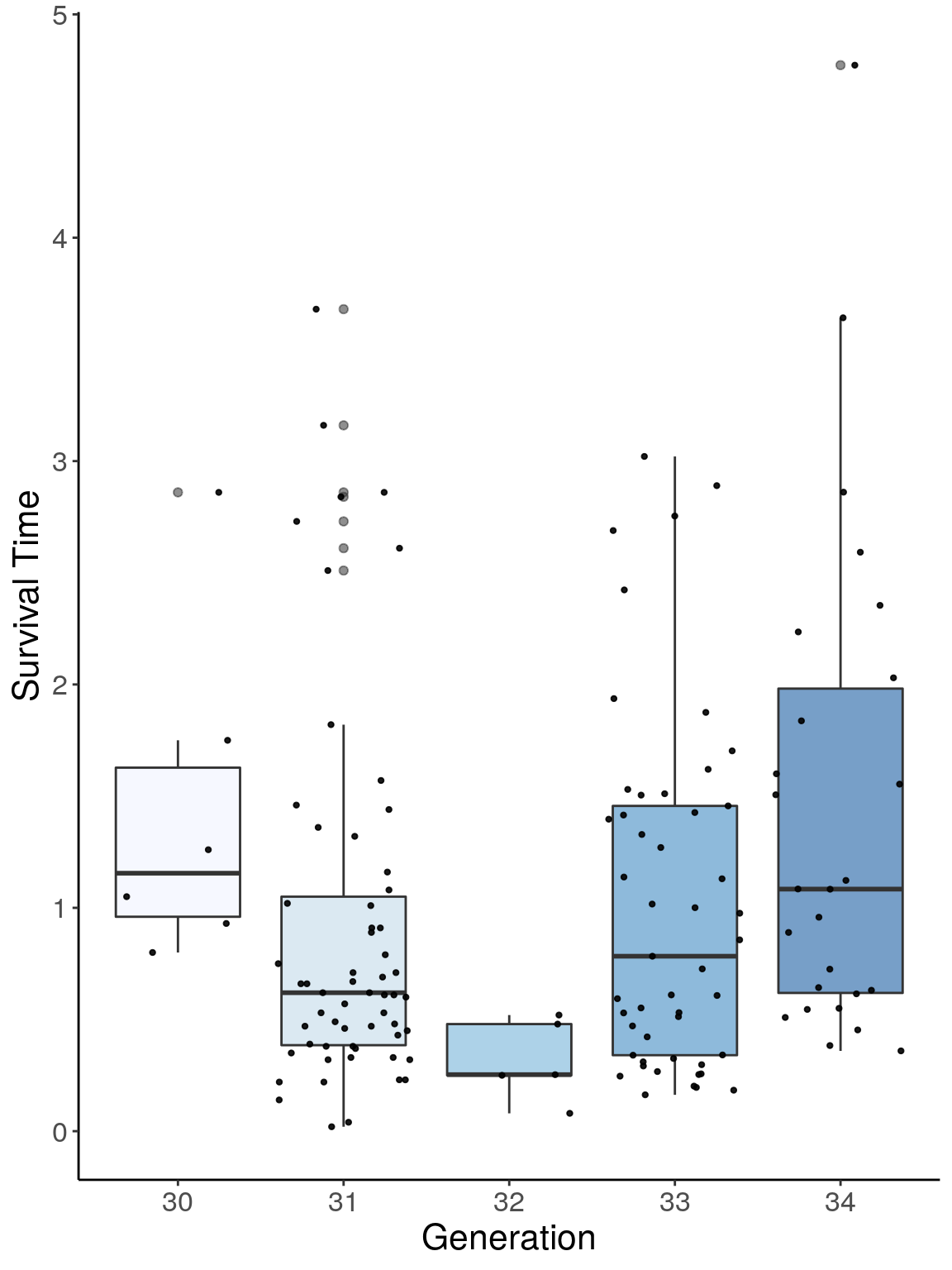
reco <- do.pheno[,c(2,9,15)]
#reco <- rbind(reco, do.pheno1[,c(1,3,5)])
reco <- reco[complete.cases(reco), ]
reco$ngen <- factor(reco$ngen, levels = as.character(c(30:35)))
p2 <- ggplot(reco, aes(x=ngen, y=Recovery.Time, group = ngen, fill = ngen, alpha = 0.9)) +
geom_boxplot(show.legend = F , outlier.size = 1.5, notchwidth = 0.85) +
geom_jitter(color="black", size=0.8, alpha=0.9) +
scale_fill_brewer(palette="Blues") +
ylab("Recovery Time") +
xlab("Generation") +
labs(fill = "") +
theme(legend.position = "none",
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
axis.line = element_line(colour = "black"),
text = element_text(size=16),
axis.title=element_text(size=16)) +
guides(shape = guide_legend(override.aes = list(size = 12)))
p2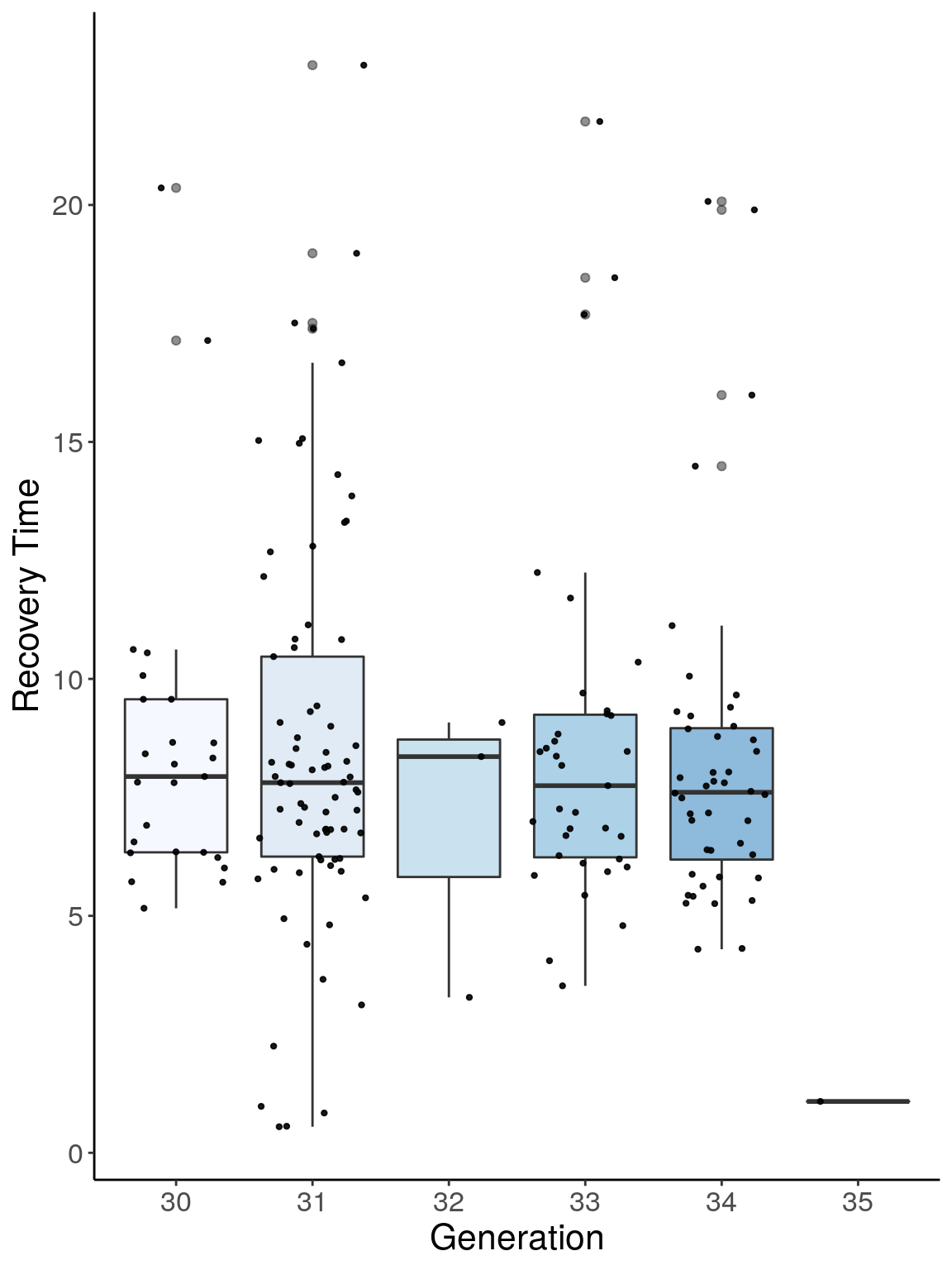
dep <- do.pheno[,c(2,10,15)]
#dep <- rbind(dep, do.pheno1[,c(1,4,5)])
dep <- dep[complete.cases(dep), ]
dep$ngen <- factor(dep$ngen, levels = as.character(c(30:35)))
p3 <- ggplot(dep, aes(x=ngen, y=Min.depression, group = ngen, fill = ngen, alpha = 0.9)) +
geom_boxplot(show.legend = F , outlier.size = 1.5, notchwidth = 0.85) +
geom_jitter(color="black", size=0.8, alpha=0.9) +
scale_fill_brewer(palette="Blues") +
ylab("Min depression") +
xlab("Generation") +
labs(fill = "") +
theme(legend.position = "none",
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
axis.line = element_line(colour = "black"),
text = element_text(size=16),
axis.title=element_text(size=16)) +
guides(shape = guide_legend(override.aes = list(size = 12)))
p3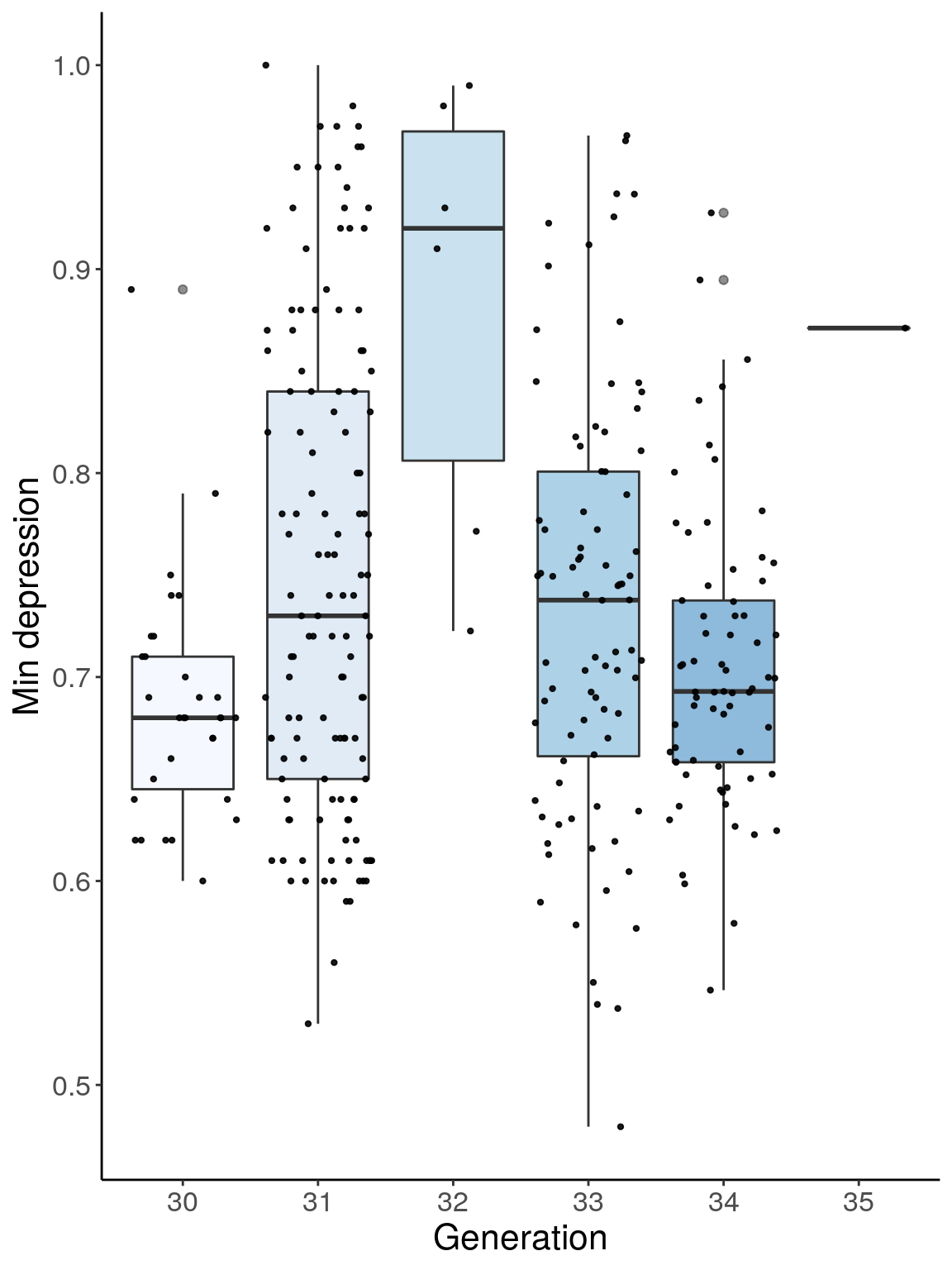
#histogram
a <- ggplot(data=do.pheno, aes(do.pheno$Survival.Time)) +
geom_histogram() +
ylab("Number of DO mice") + xlab("Survival Time (h)")
b <- ggplot(data=do.pheno, aes(do.pheno$Recovery.Time)) +
geom_histogram() +
ylab("Number of DO mice") + xlab("Recovery Time (h)")
c <- ggplot(data=do.pheno, aes(do.pheno$Min.depression)) +
geom_histogram() +
ylab("Number of DO mice") + xlab("Respiratory Depression (% of baseline)")
grid.arrange(a,b,c)
# Warning: Use of `do.pheno$Survival.Time` is discouraged. Use `Survival.Time`
# instead.
# Warning: Removed 211 rows containing non-finite values (stat_bin).
# Warning: Use of `do.pheno$Recovery.Time` is discouraged. Use `Recovery.Time`
# instead.
# Warning: Removed 166 rows containing non-finite values (stat_bin).
# Warning: Use of `do.pheno$Min.depression` is discouraged. Use `Min.depression`
# instead.
# Warning: Removed 17 rows containing non-finite values (stat_bin).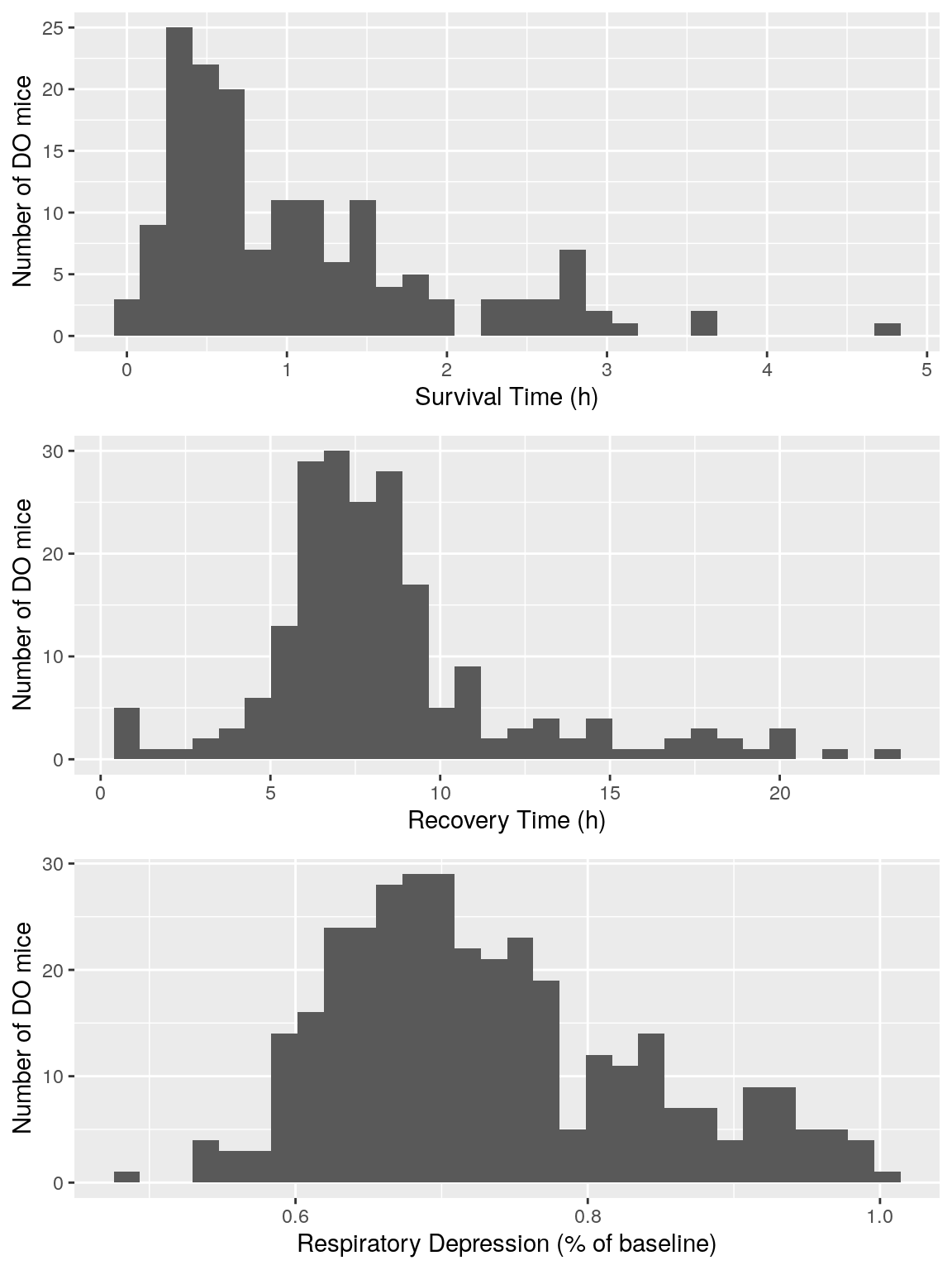
Plot qtl mapping on array
Plot by using its output qtl.morphine.out.second_set.RData
load("output/qtl.morphine.out.second_set.RData")
#pmap and gmap
chr.names <- c("1","2","3","4","5","6","7","8","9","10","11","12","13","14","15","16","17","18","19", "X")
gmap <- list()
pmap <- list()
for (chr in chr.names){
gmap[[chr]] <- do.morphine$gmap[[chr]]
pmap[[chr]] <- do.morphine$pmap[[chr]]
}
attr(gmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
attr(pmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
#genome-wide plot
for(i in names(qtl.morphine.out)){
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.morphine.out[[i]]) # overall maximum LOD score
plot(qtl.morphine.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, 10))
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("DO_morphine_",i))
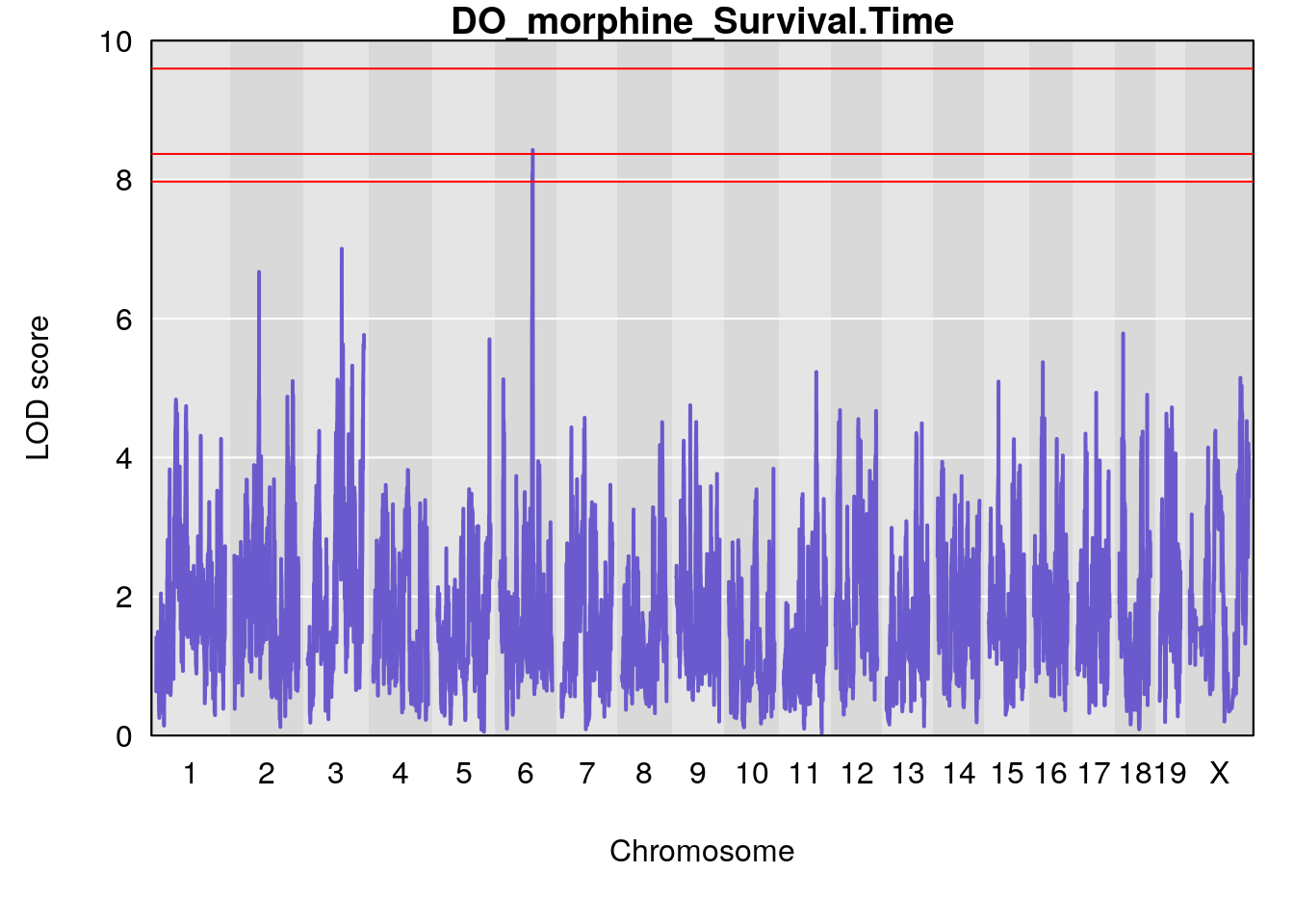
}
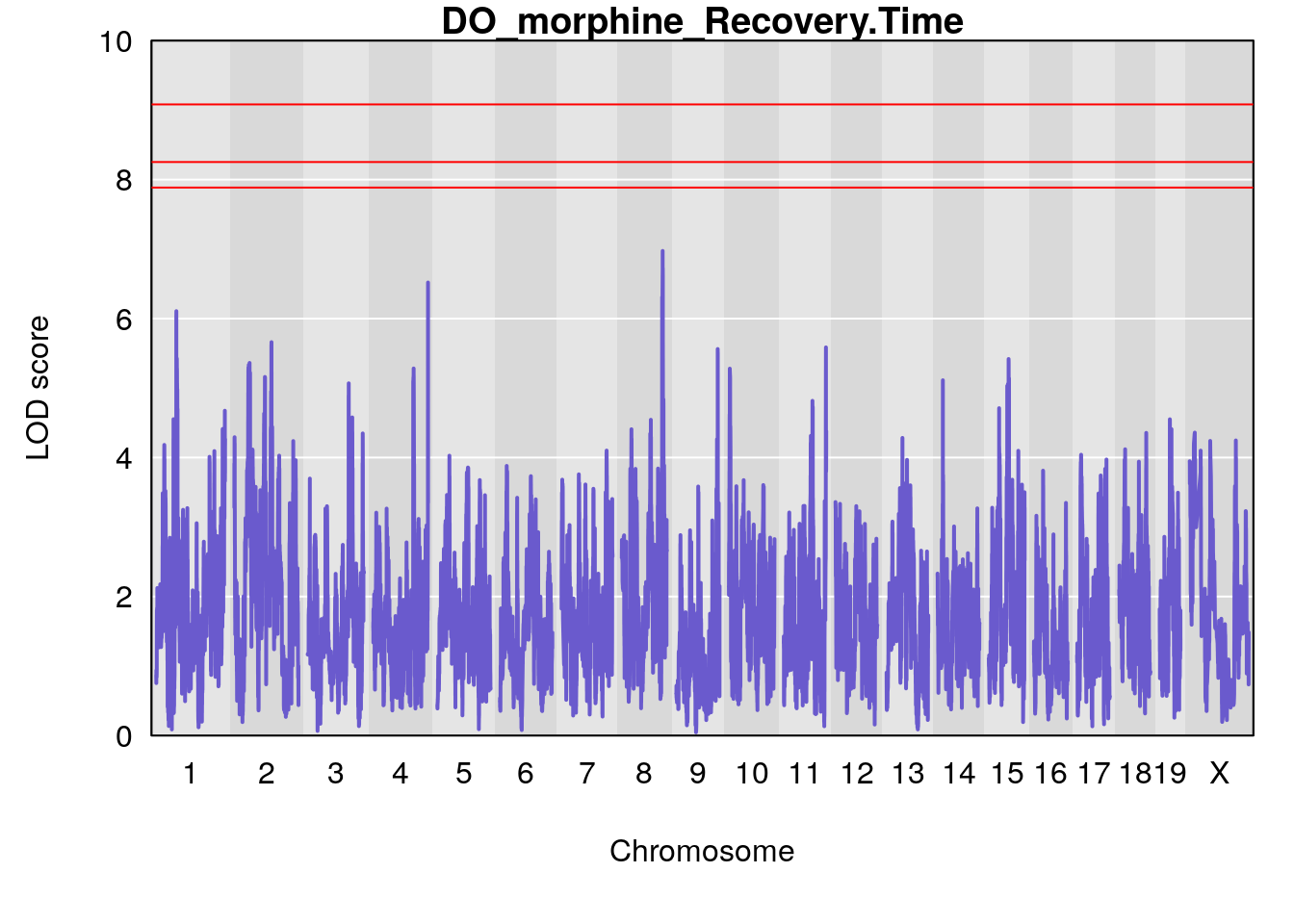
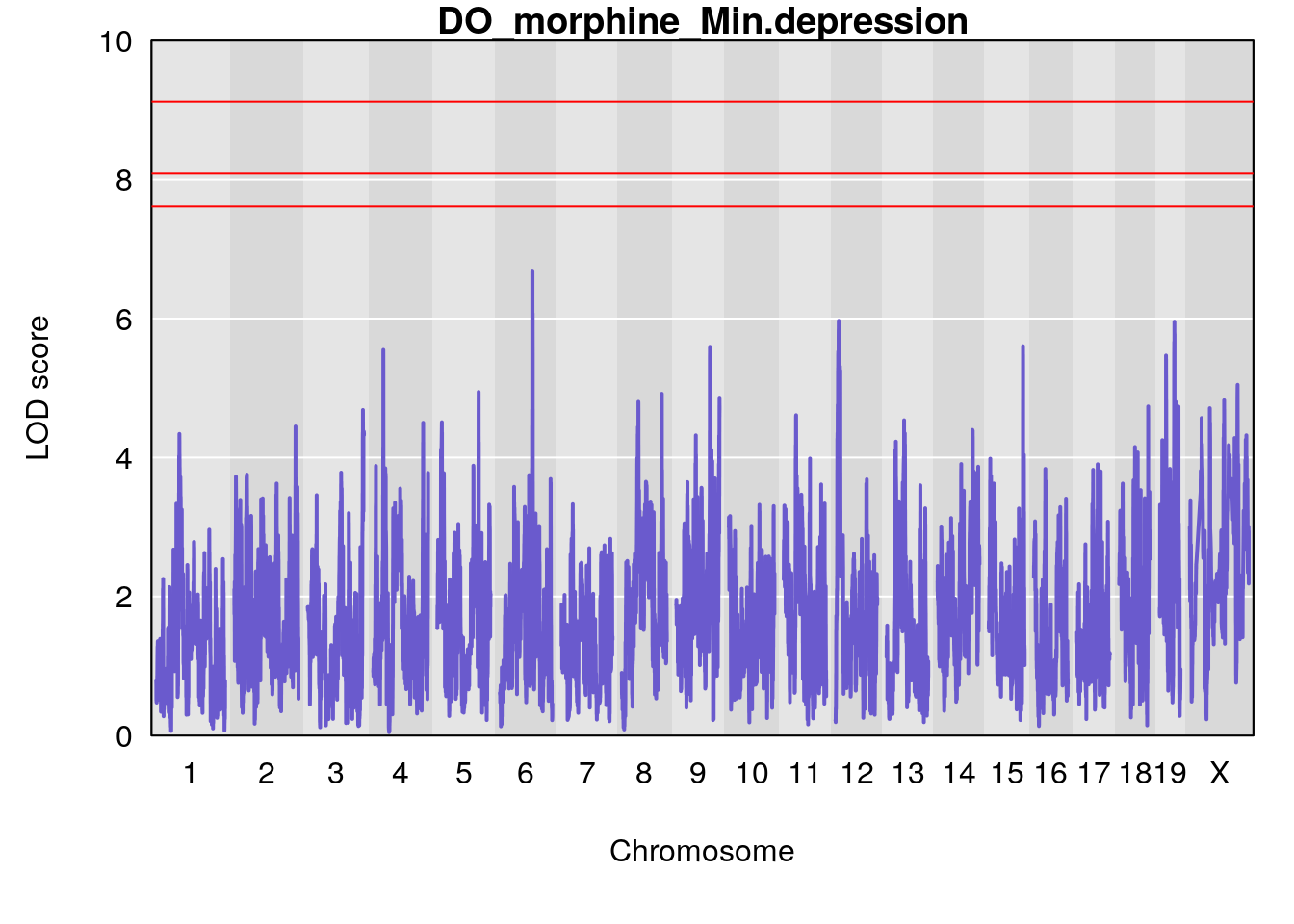
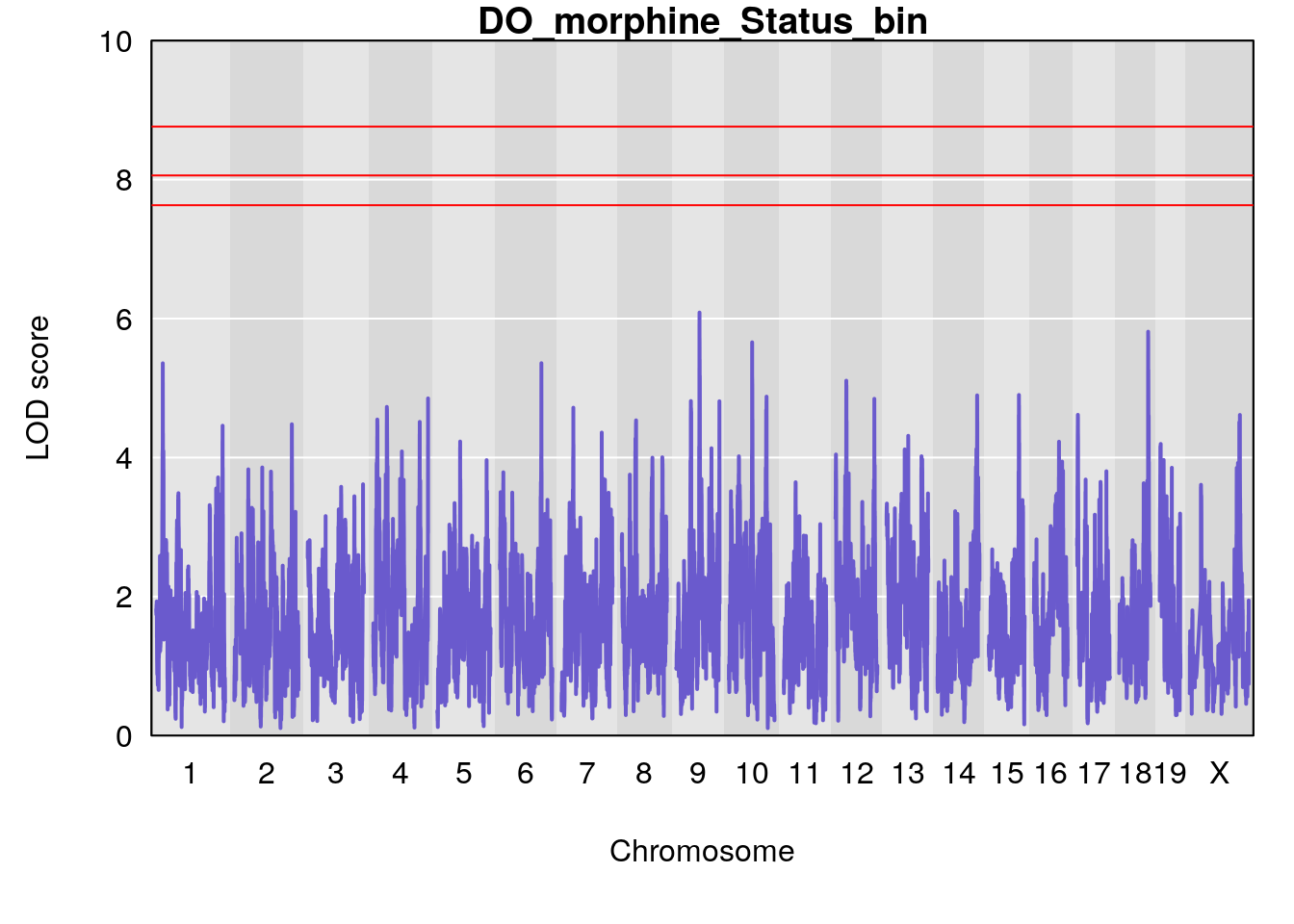
#save genome-wide plot
for(i in names(qtl.morphine.out)){
png(file = paste0("output/DO_morphine_secondbatch_", i, ".png"), width = 16, height =8, res=300, units = "in")
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.morphine.out[[i]]) # overall maximum LOD score
plot(qtl.morphine.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, 10))
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("DO_morphine_",i))
dev.off()
}
#peaks coeff plot
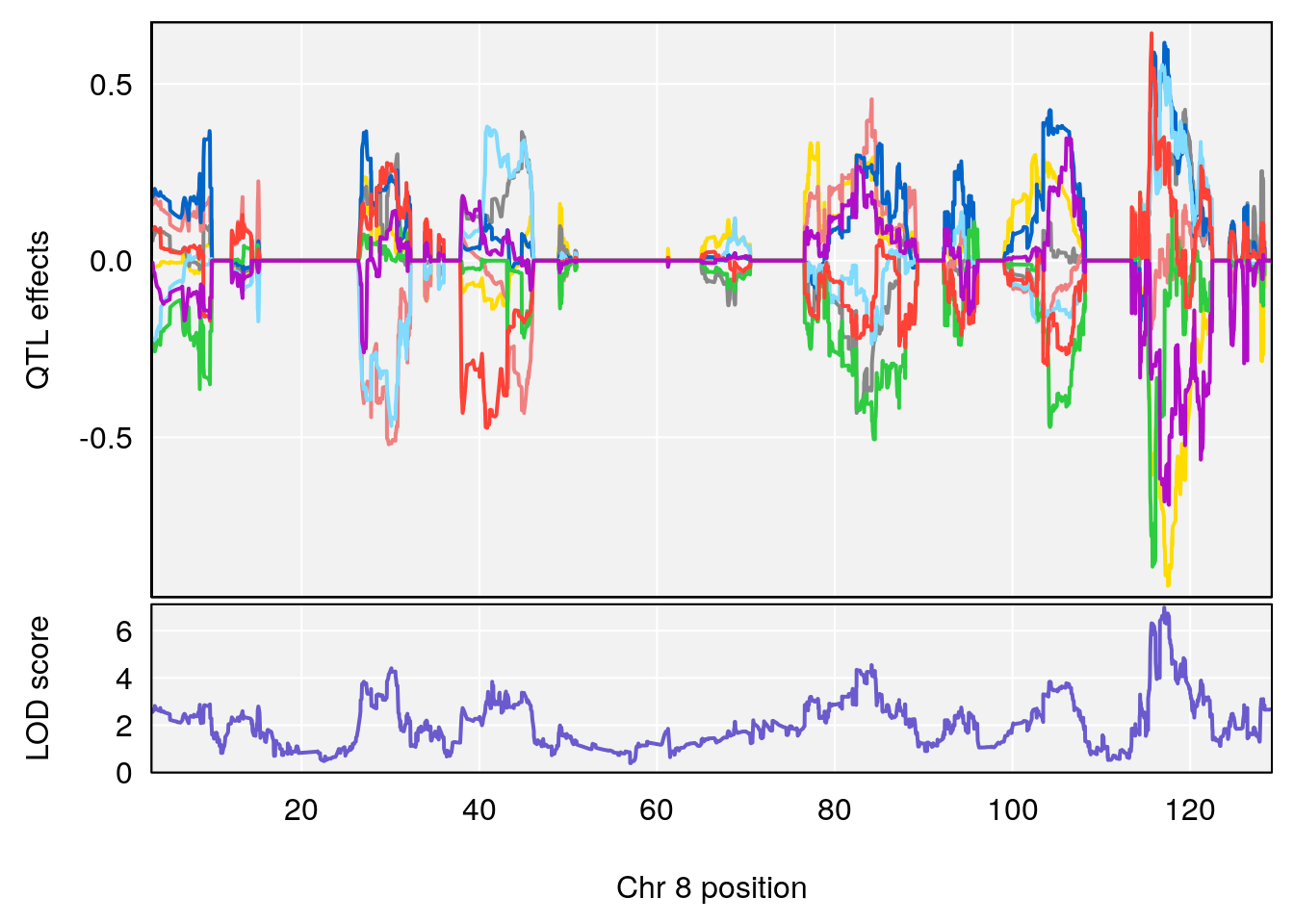
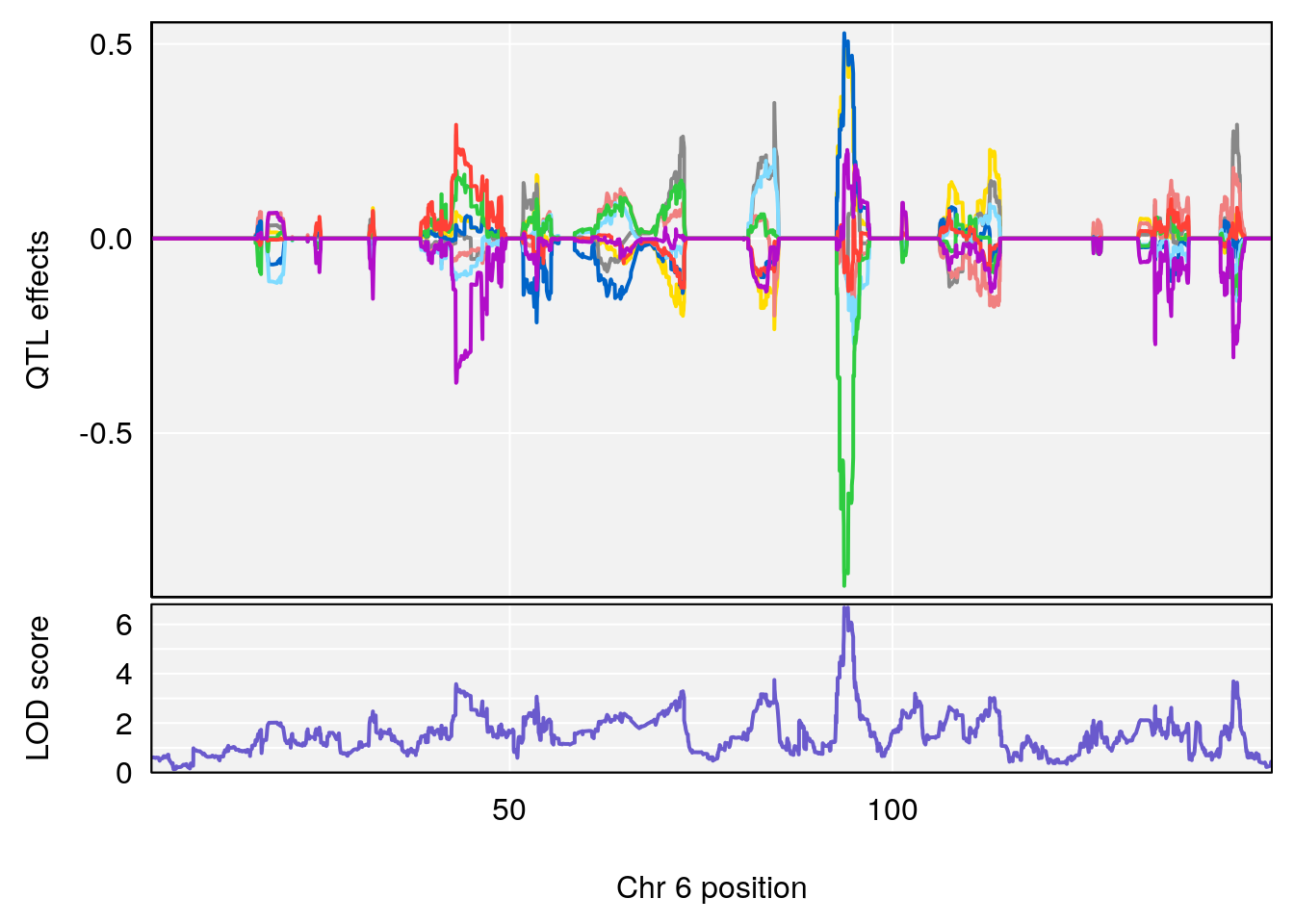
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
print(peaks)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
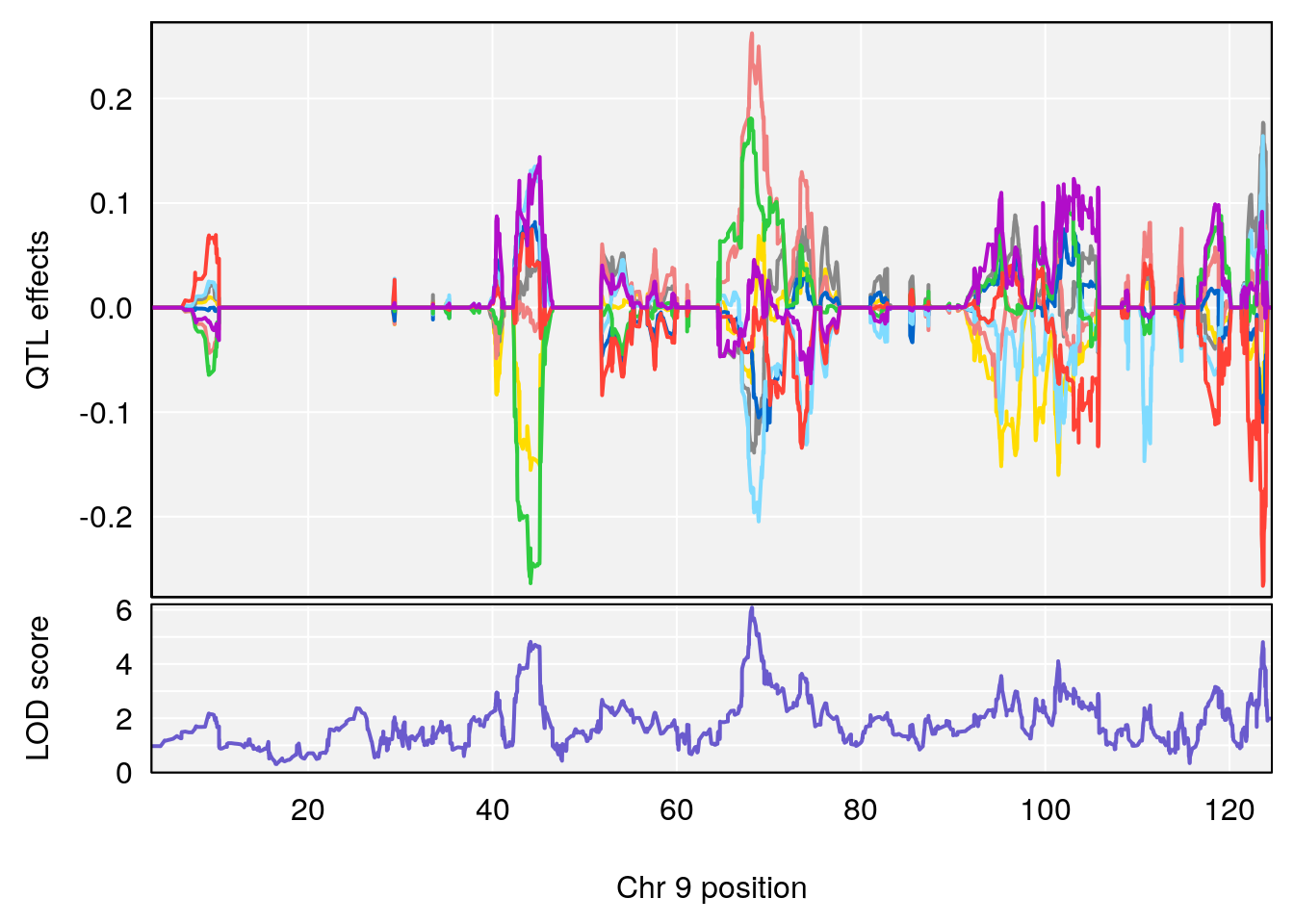
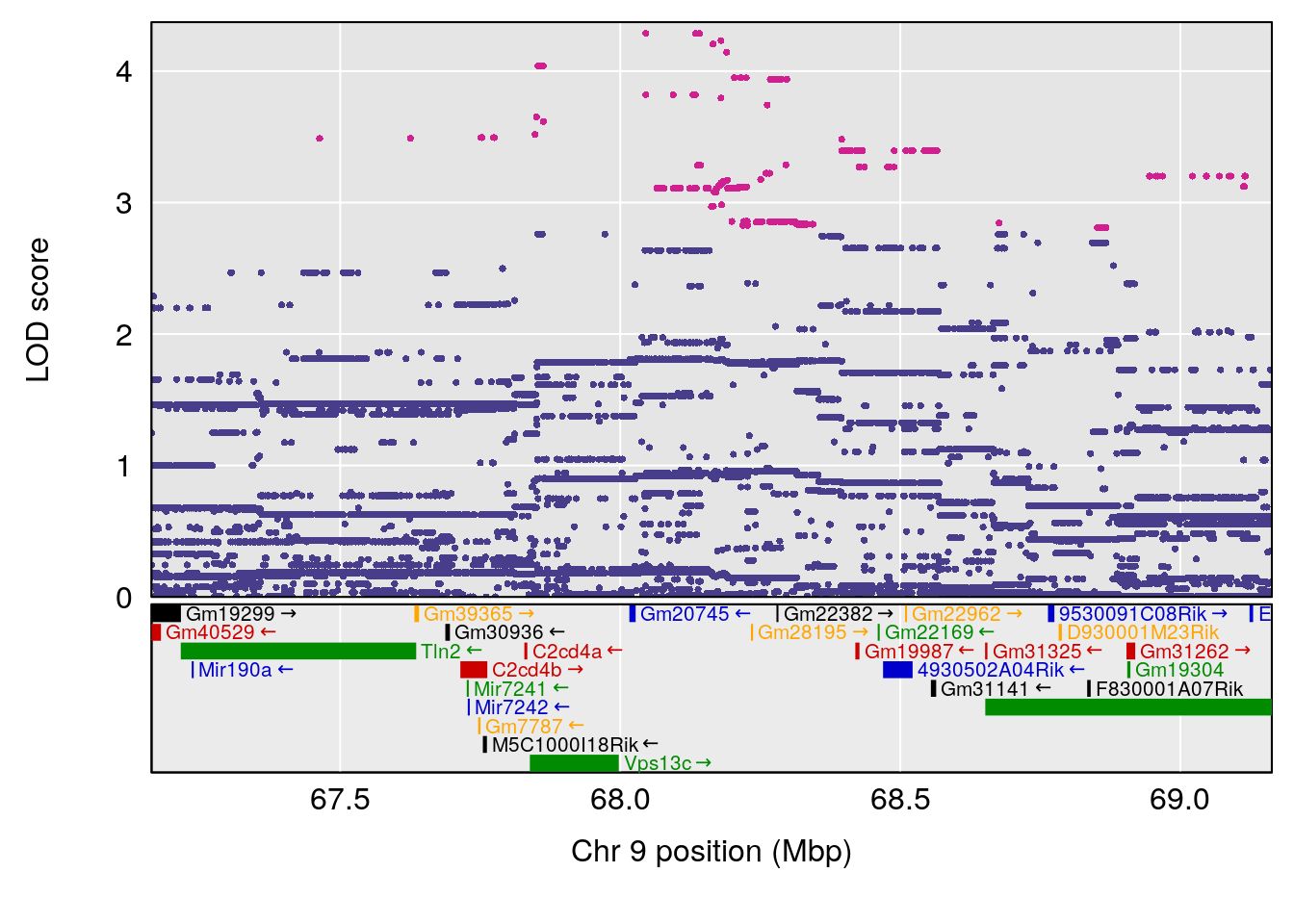
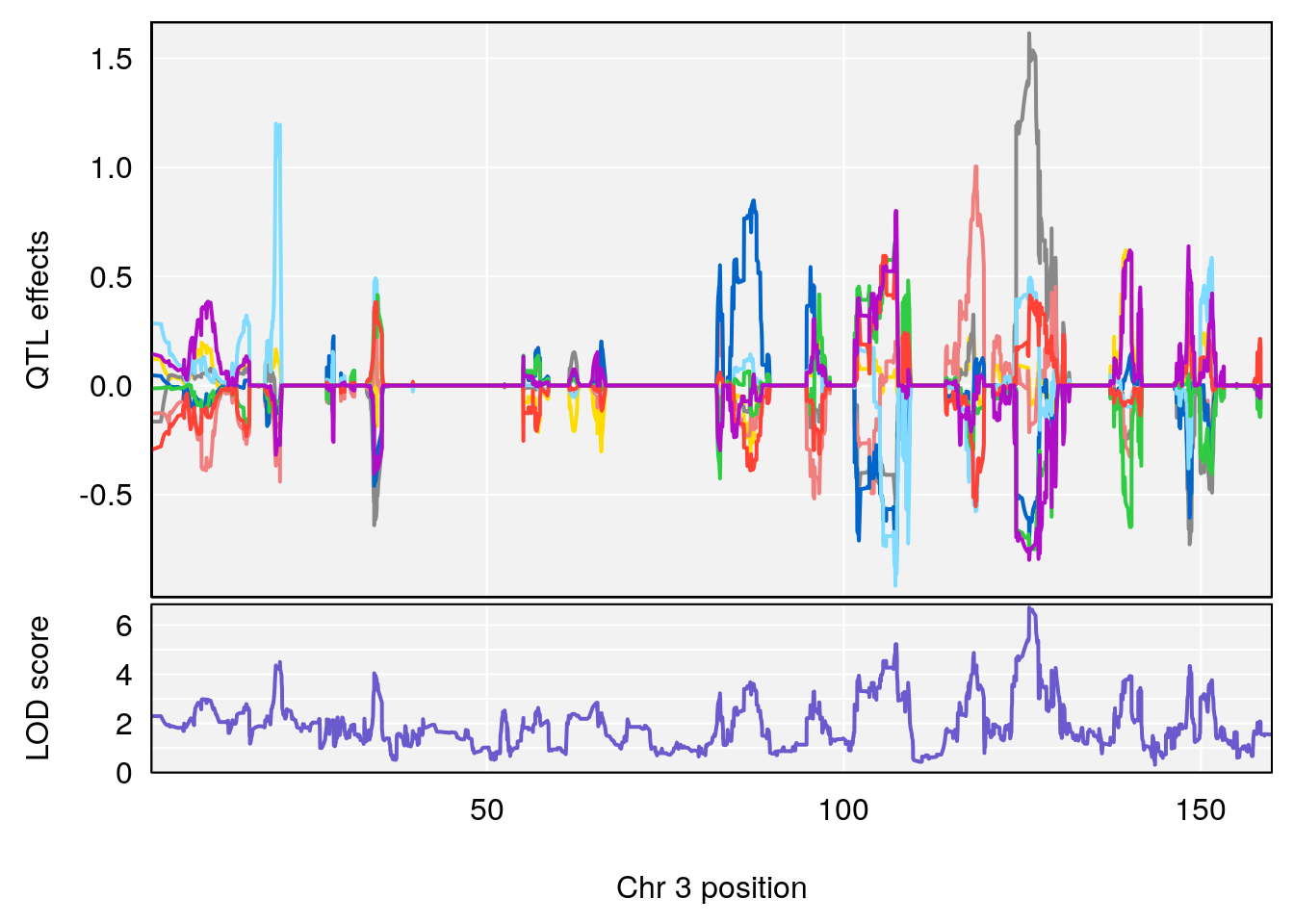
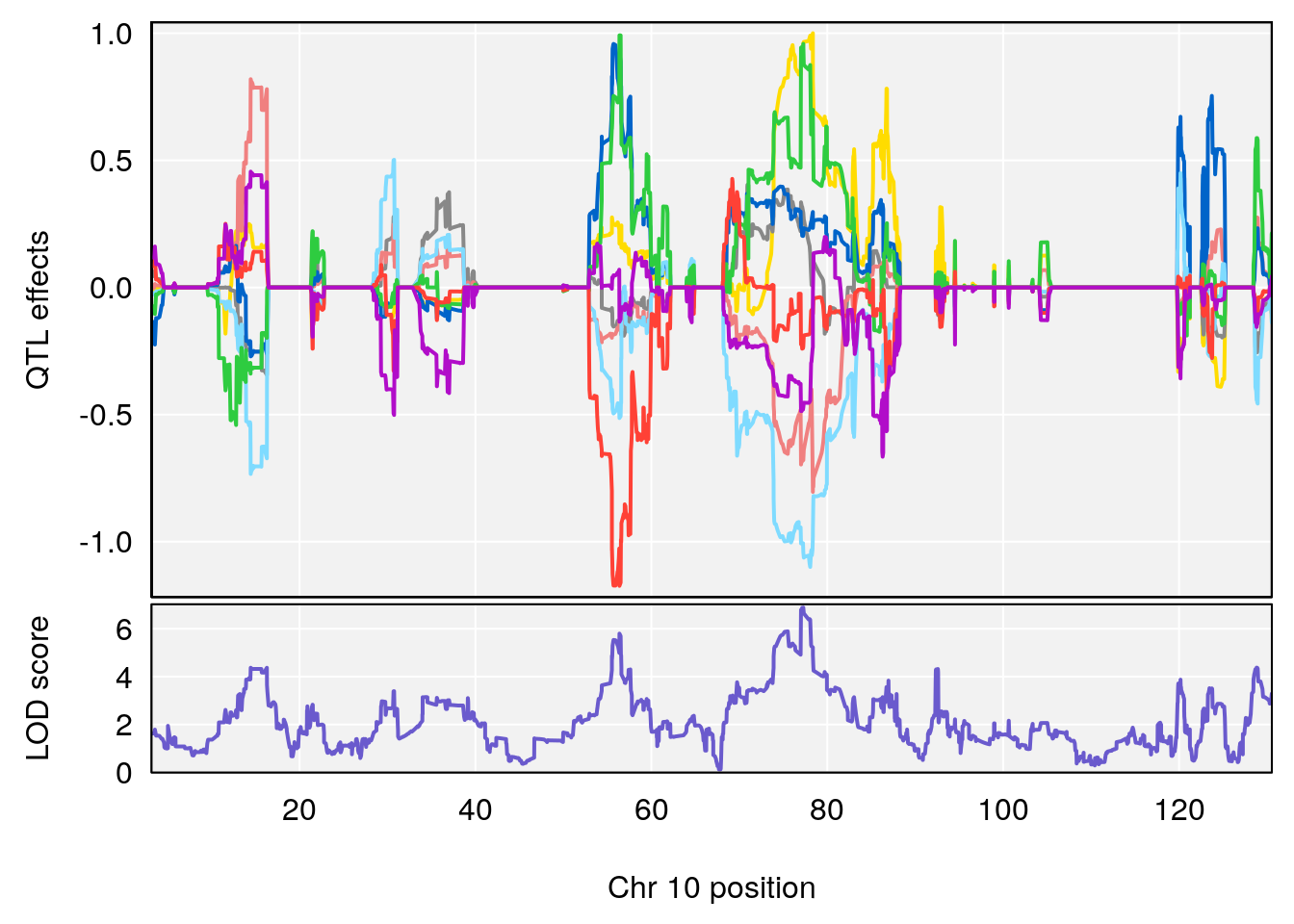
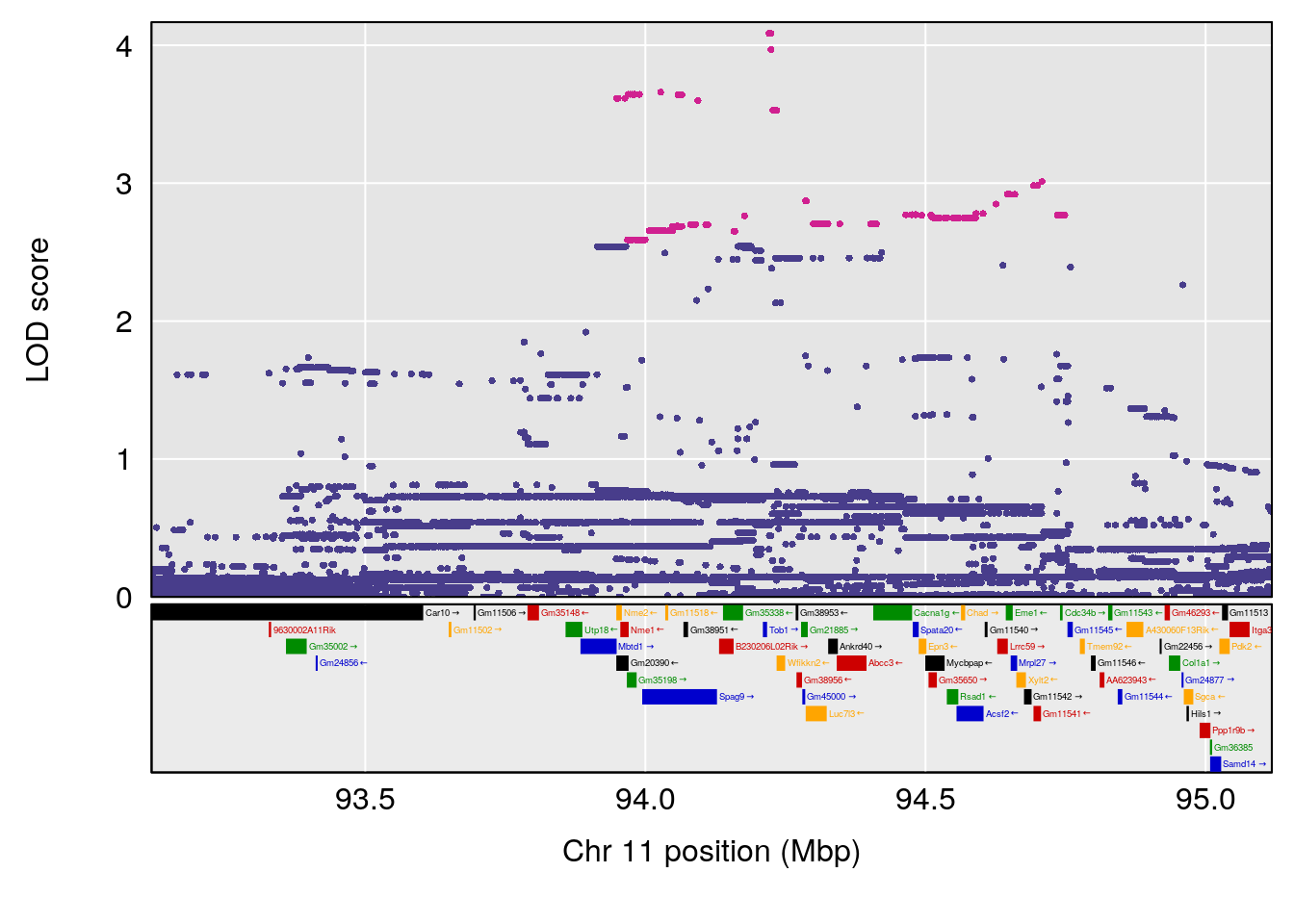
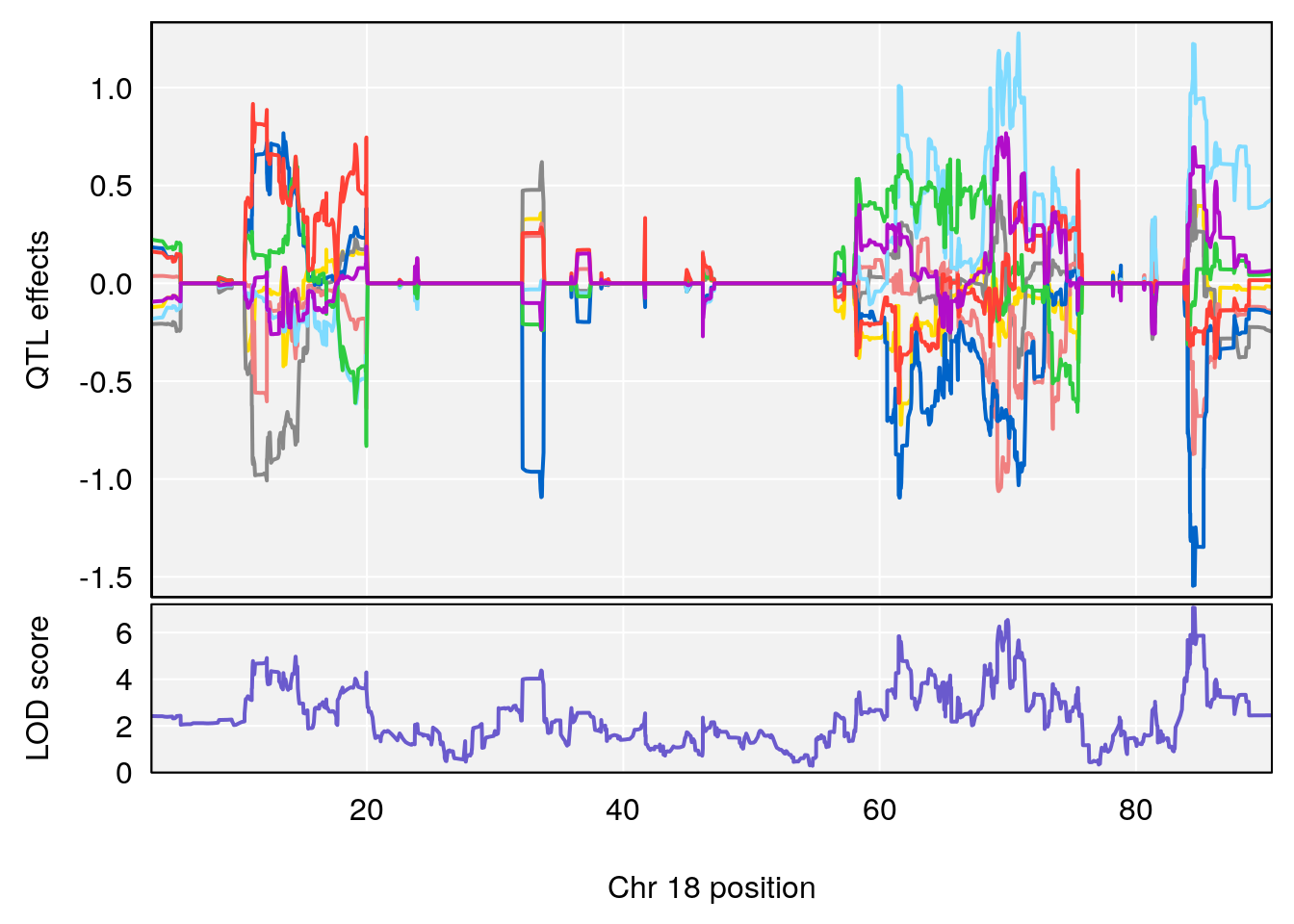
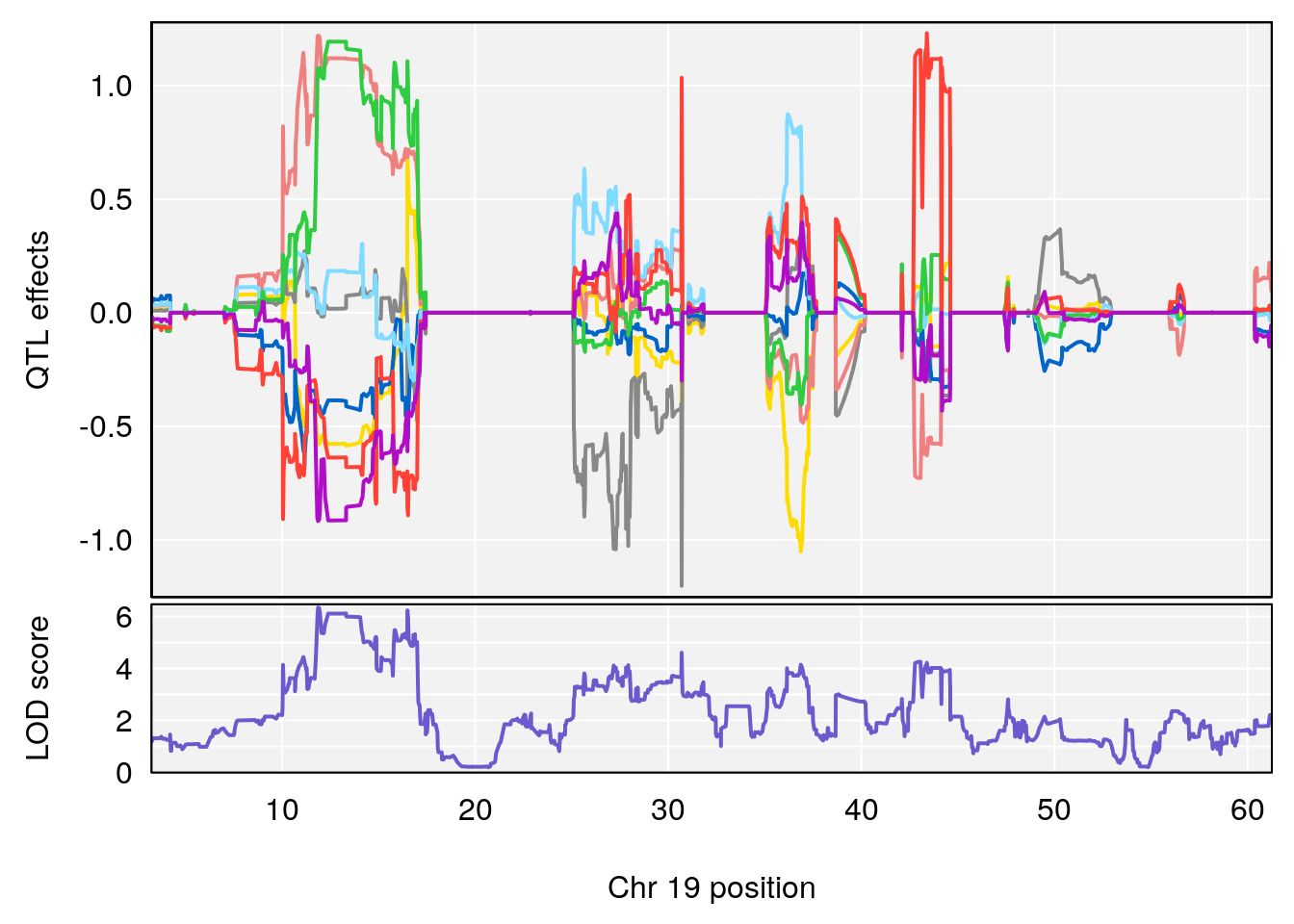
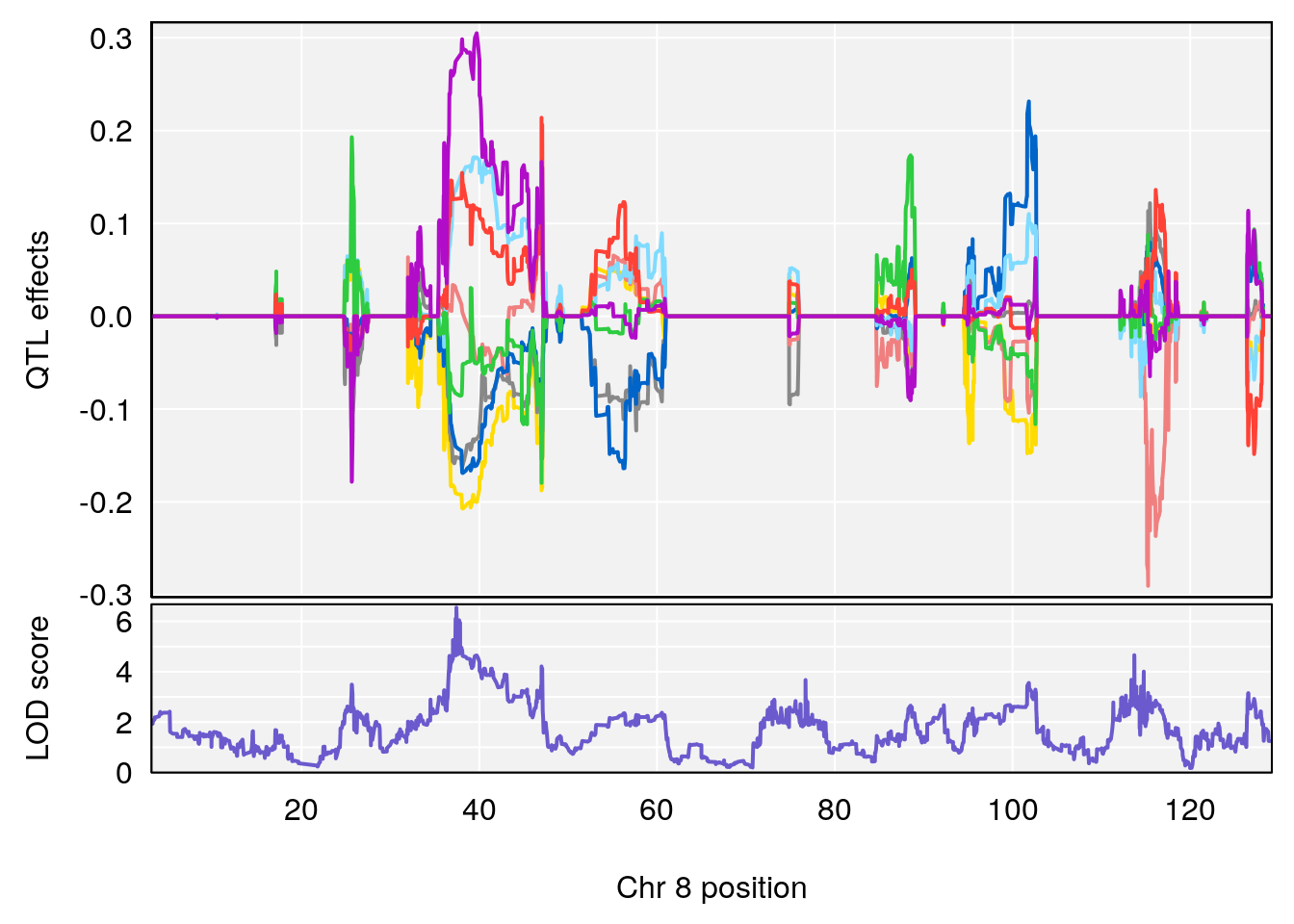
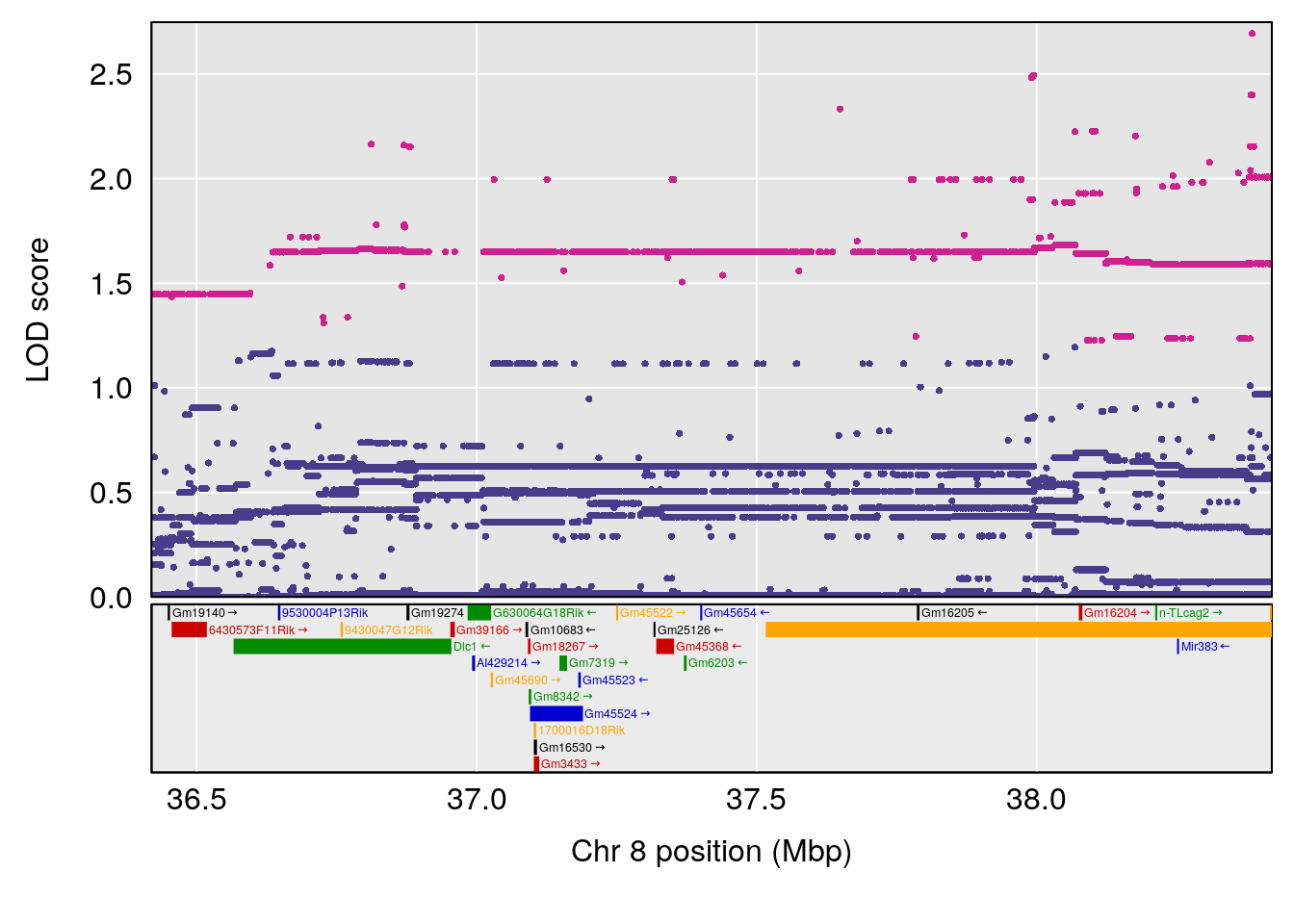
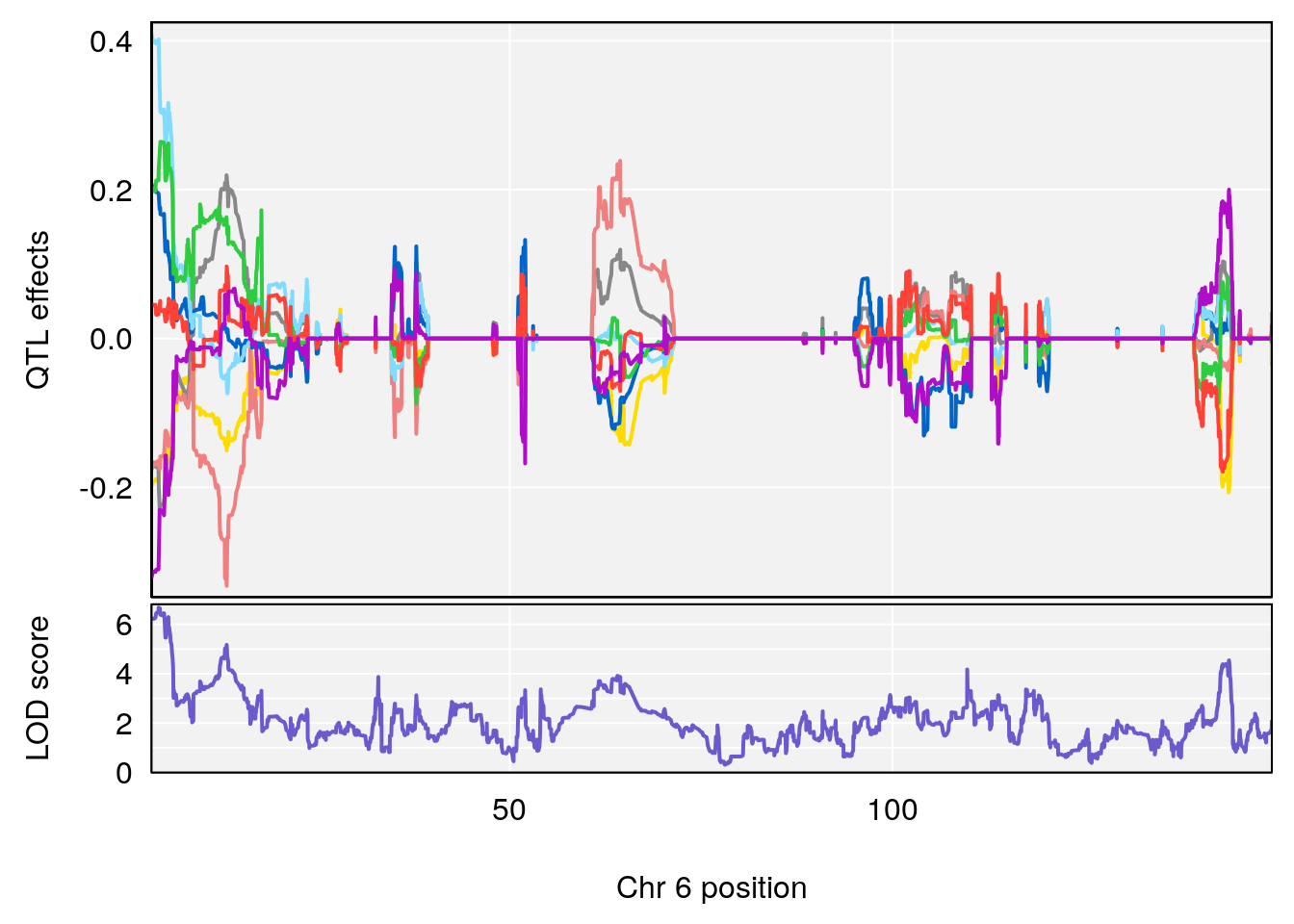
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
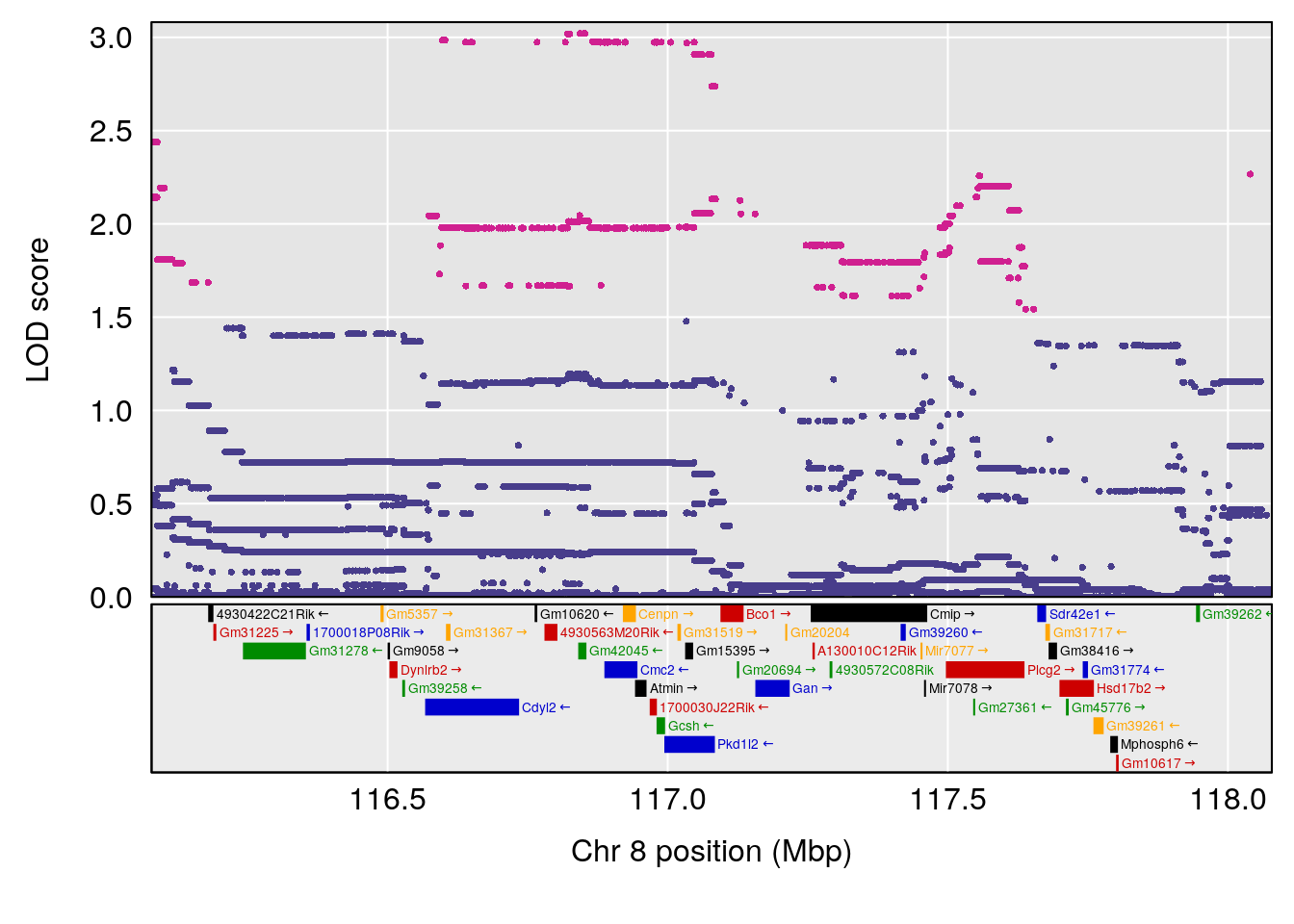
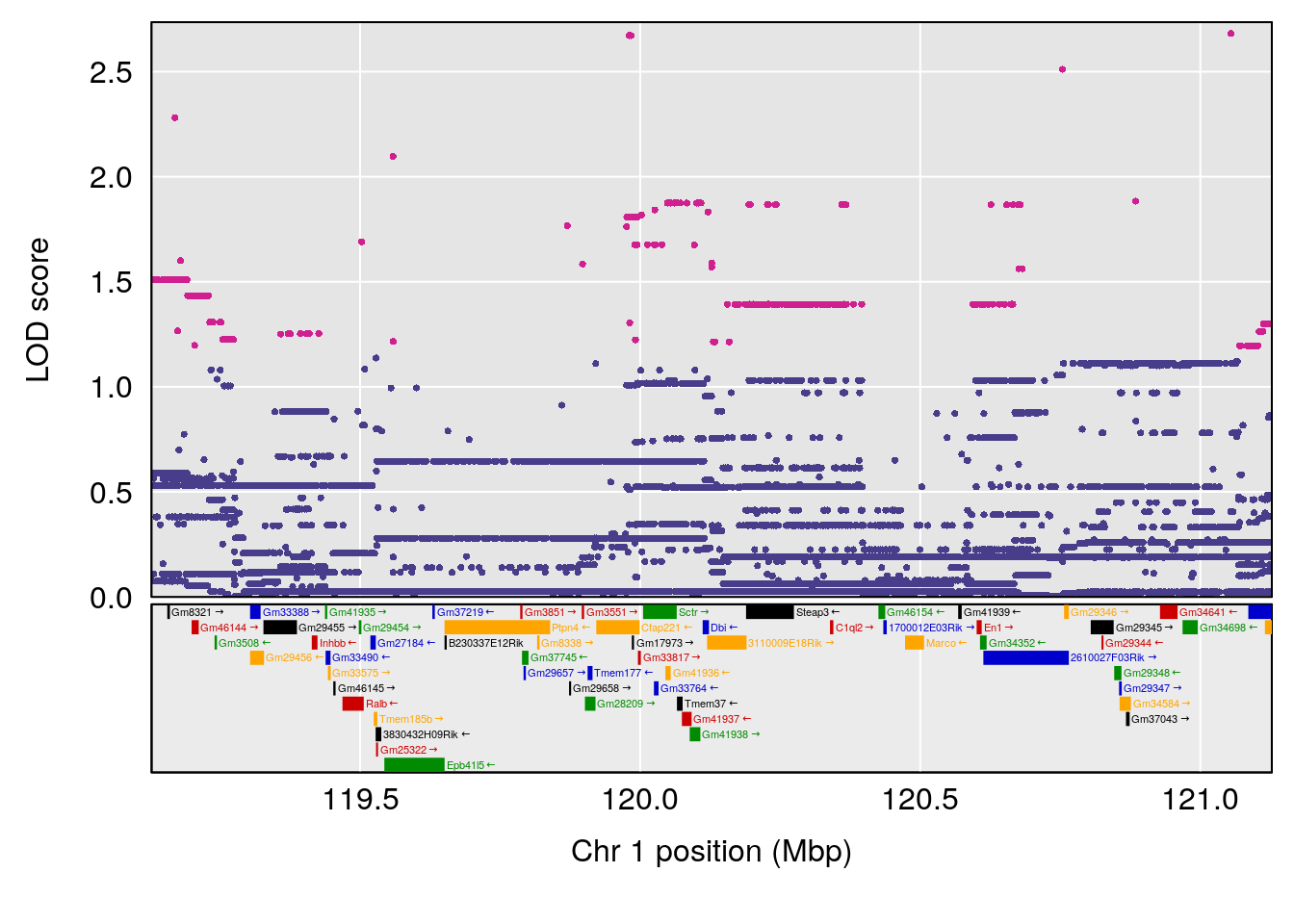
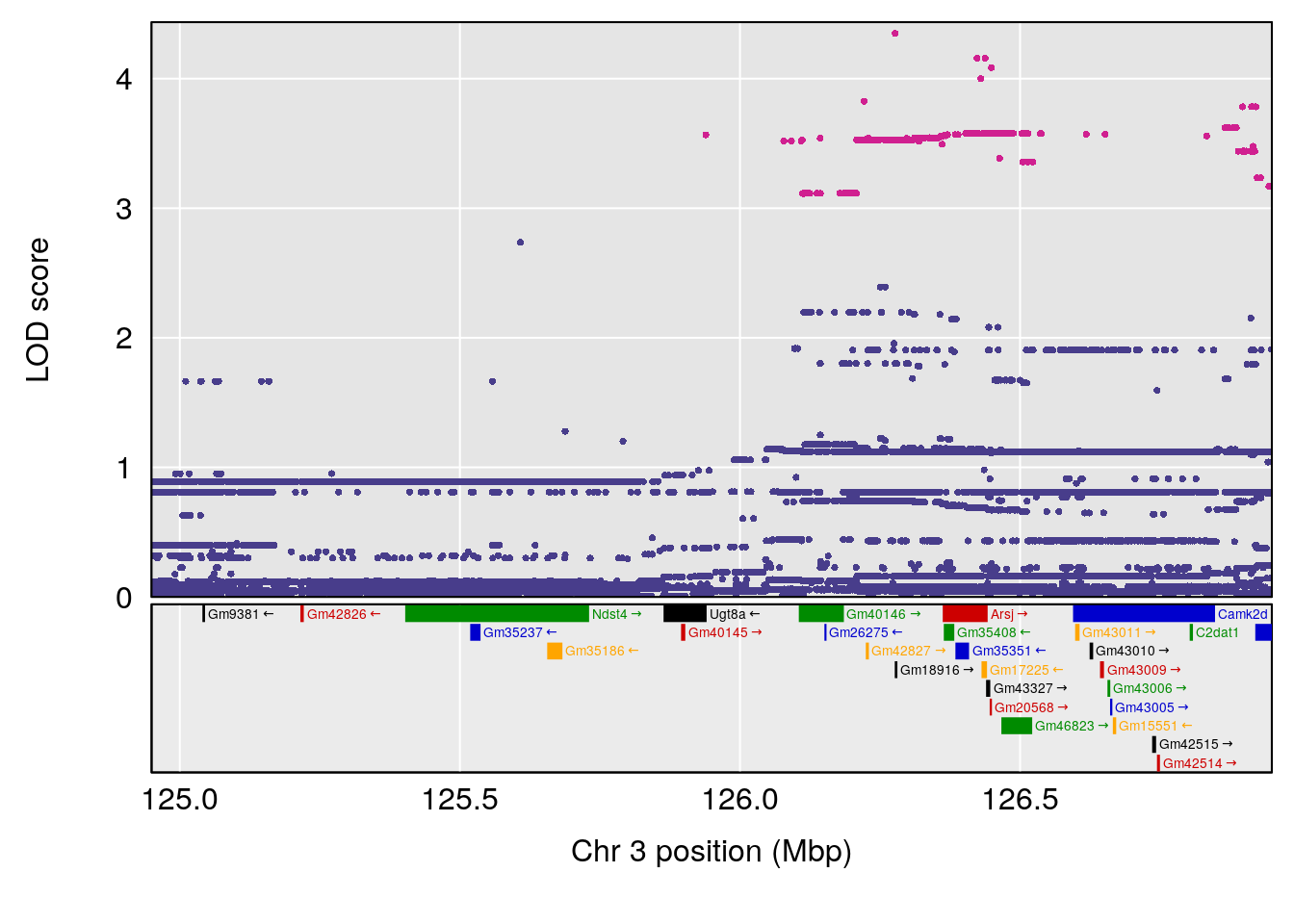
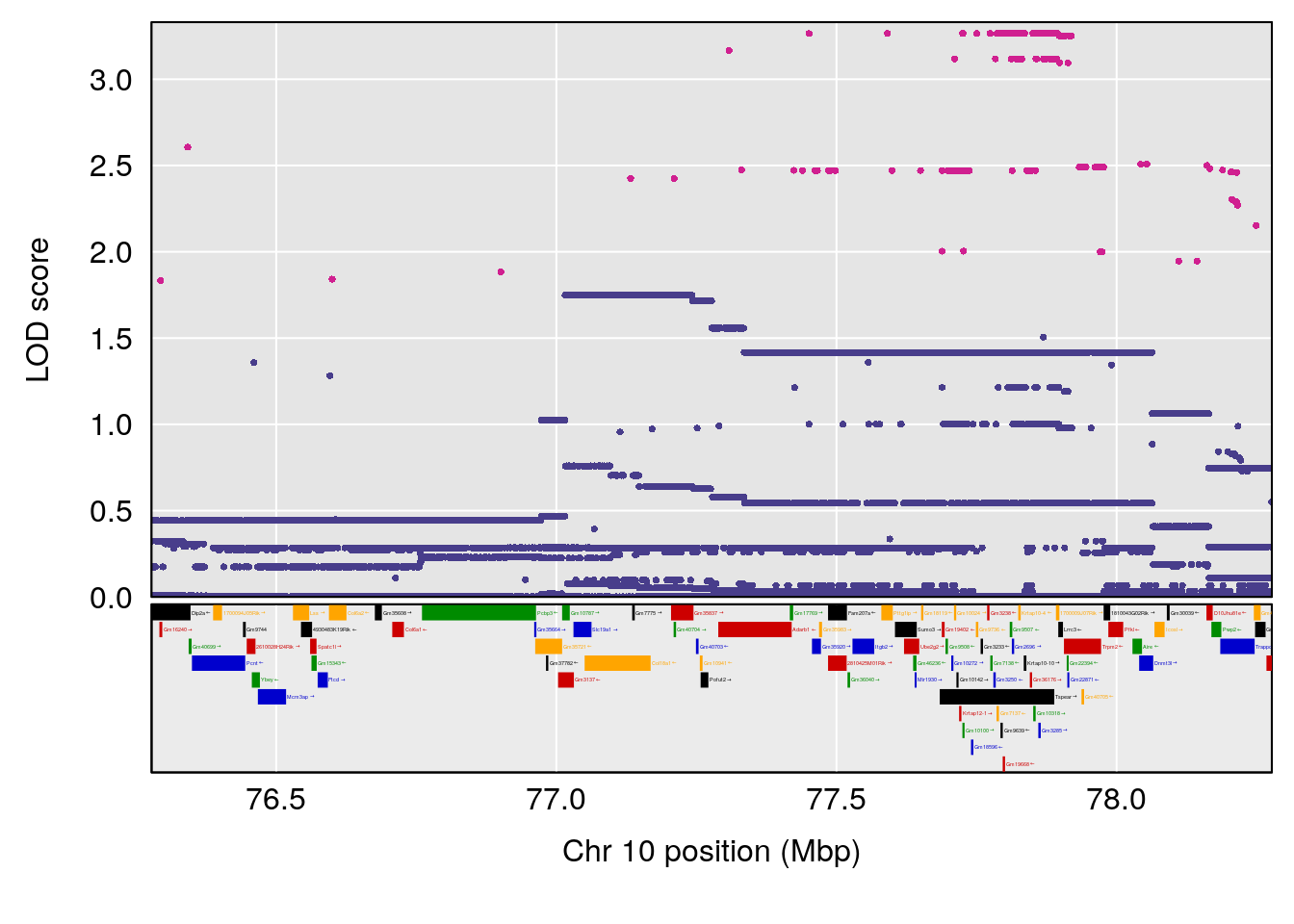
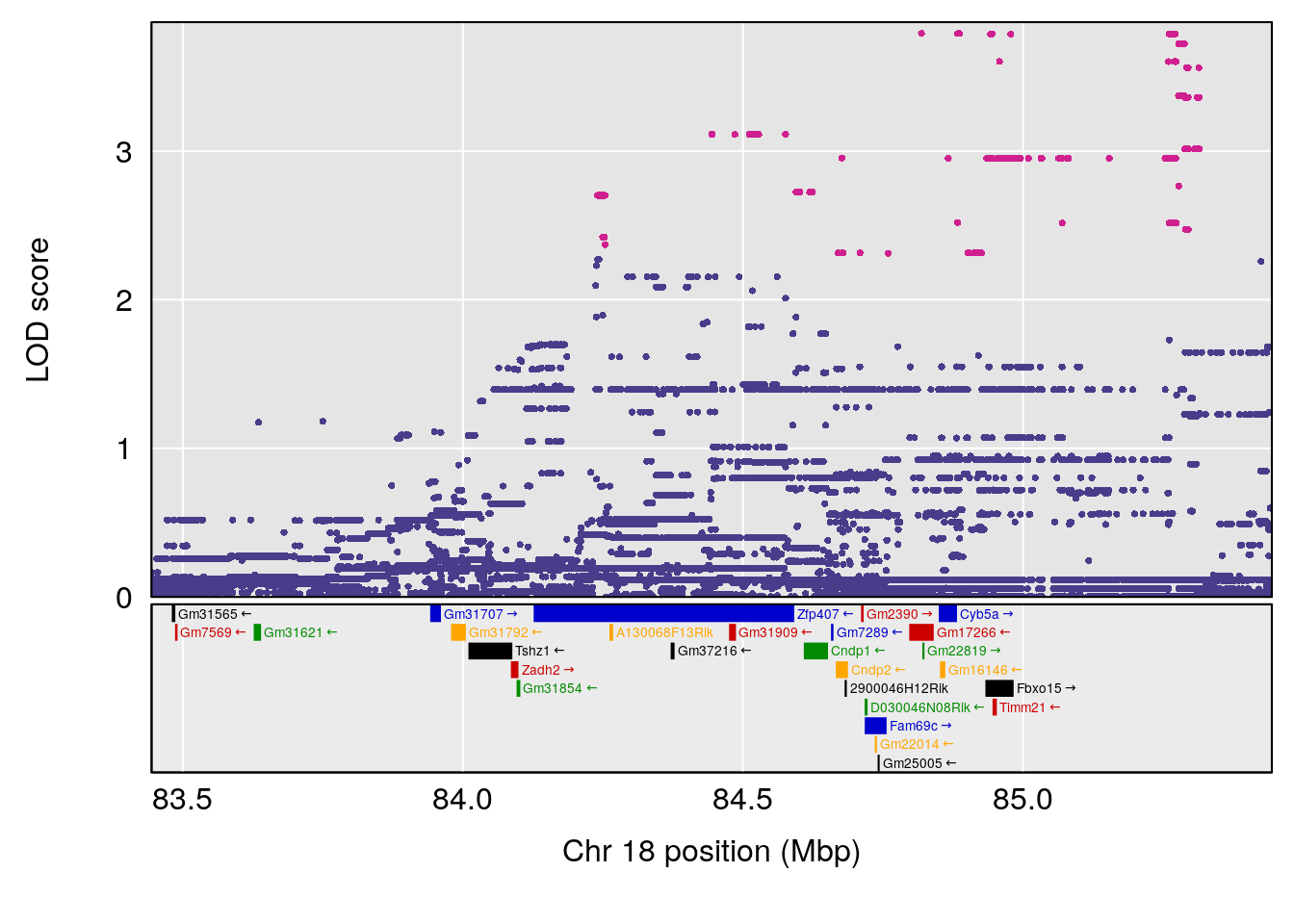
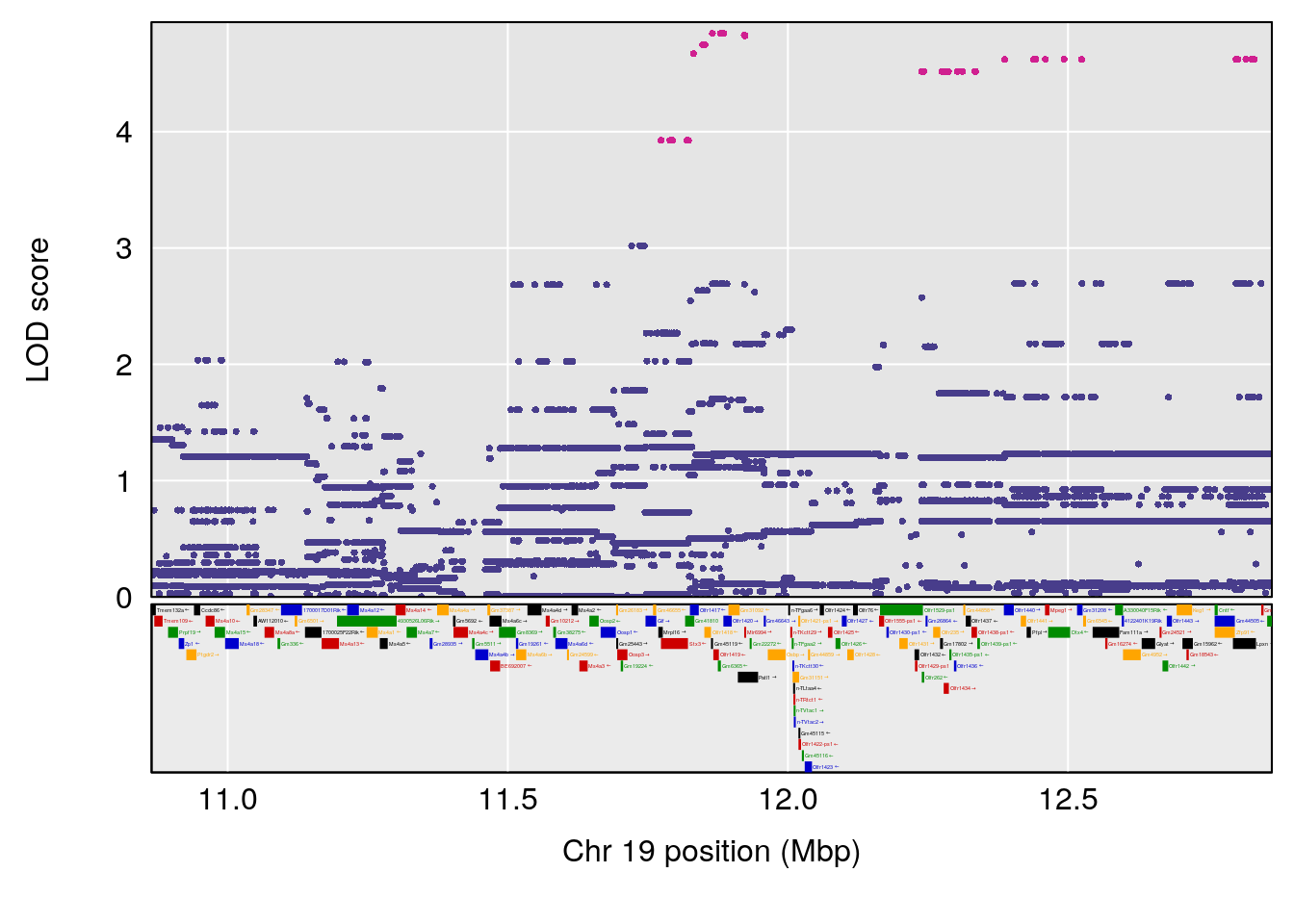
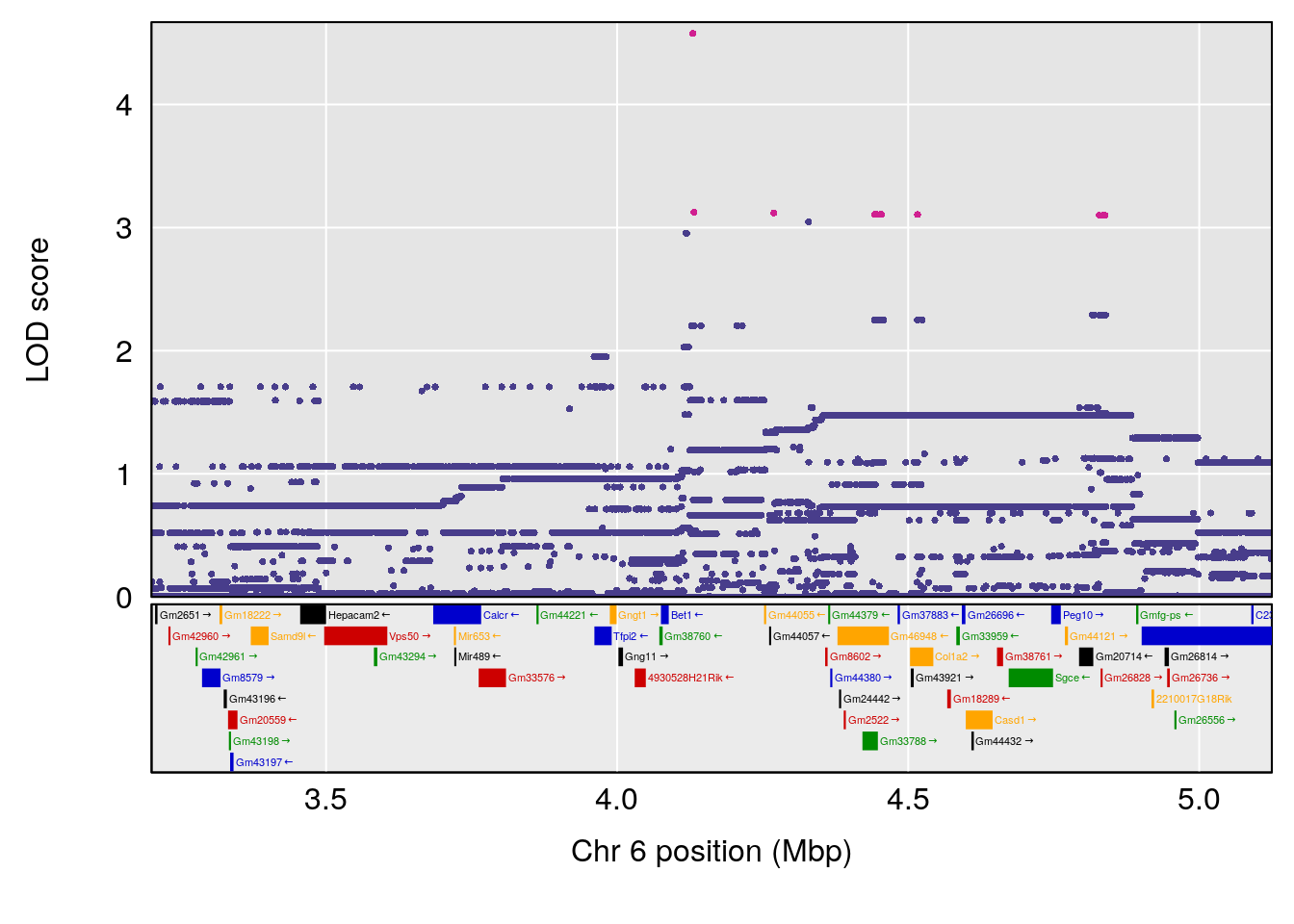
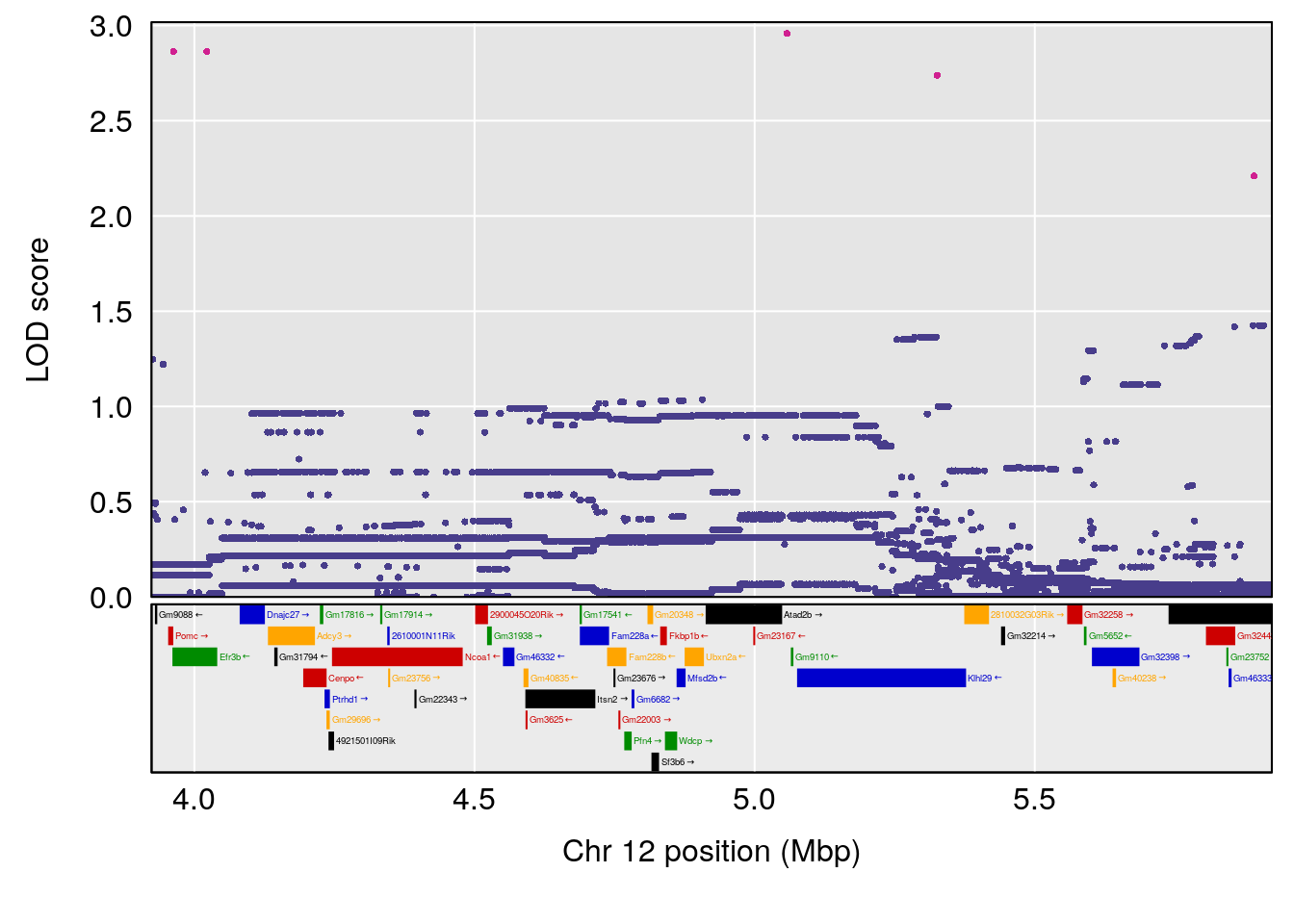
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
}
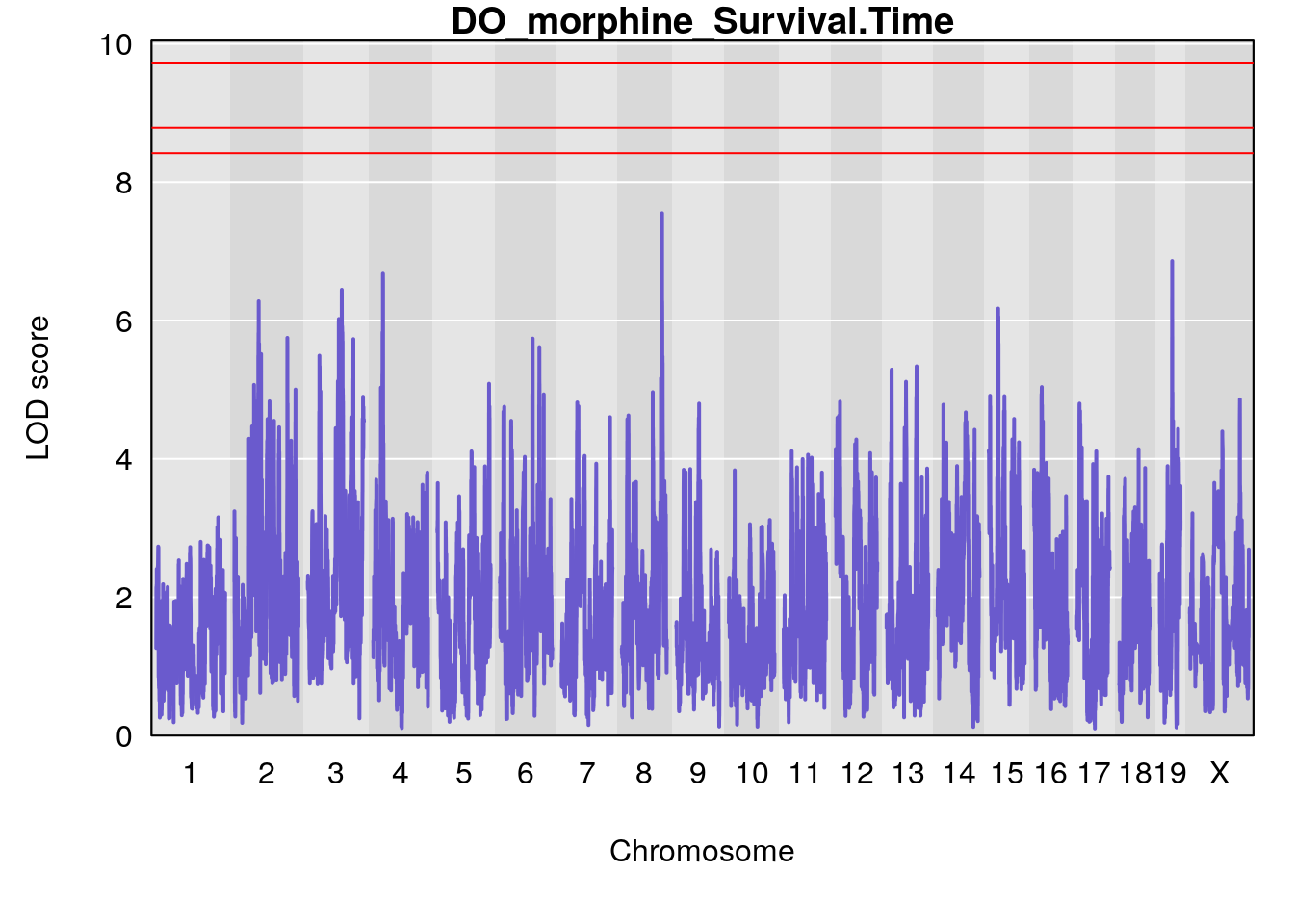
# [1] "Survival.Time"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
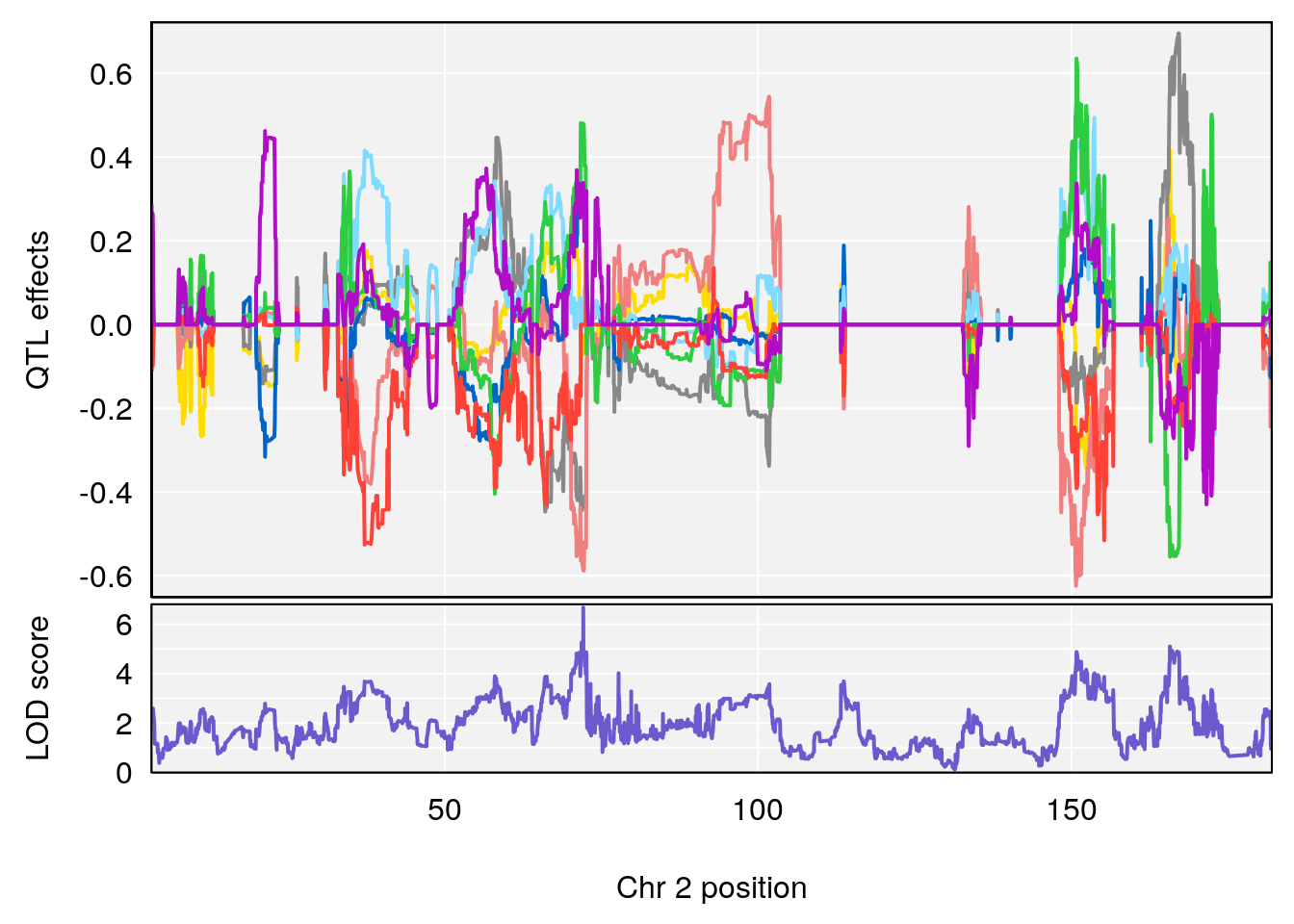
# 1 1 pheno1 2 72.08345 6.672136 71.80655 72.11478
# 2 1 pheno1 3 98.06064 7.005262 97.69551 159.92550
# 3 1 pheno1 6 94.86810 8.427626 93.64635 95.01090
# [1] 1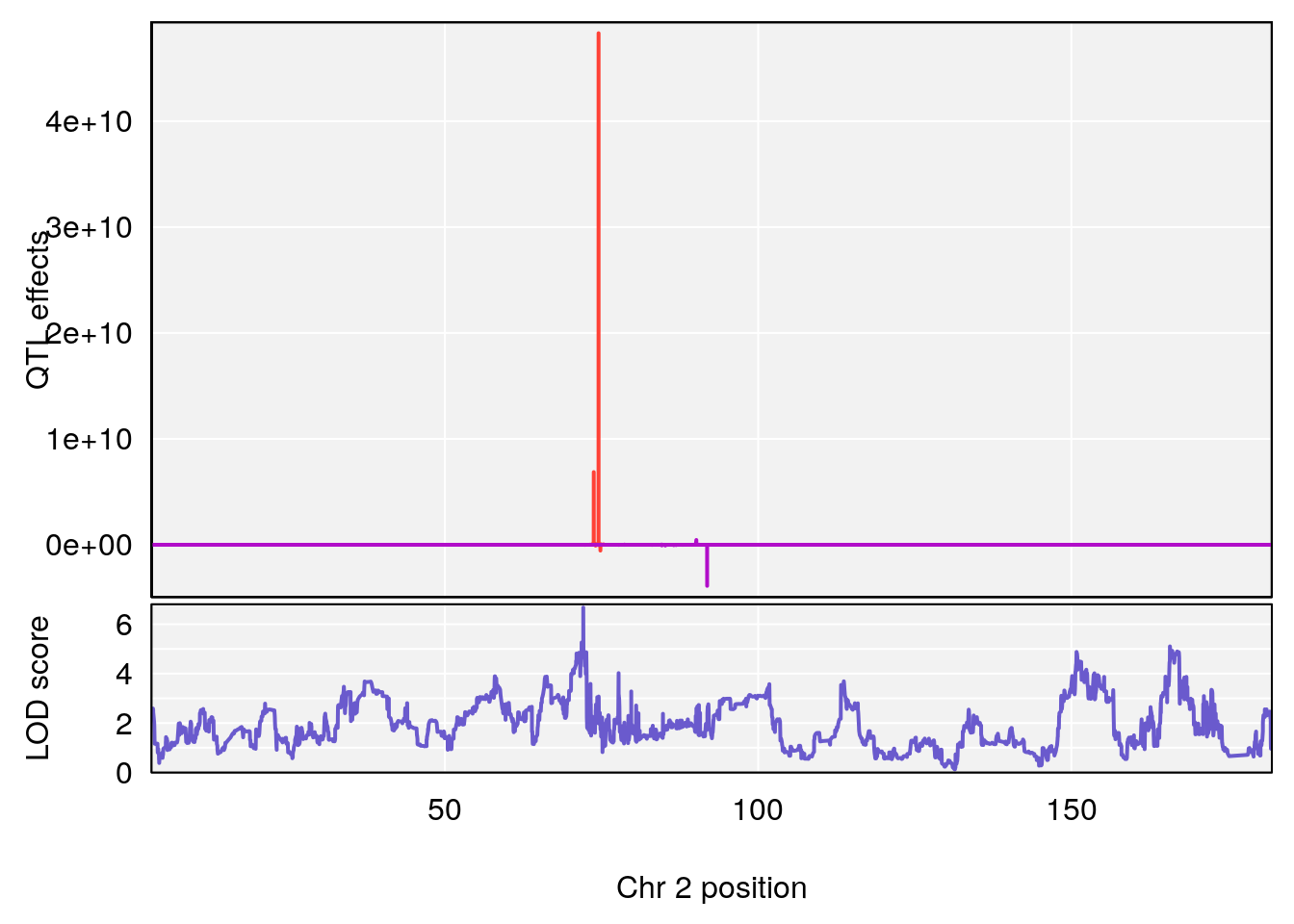
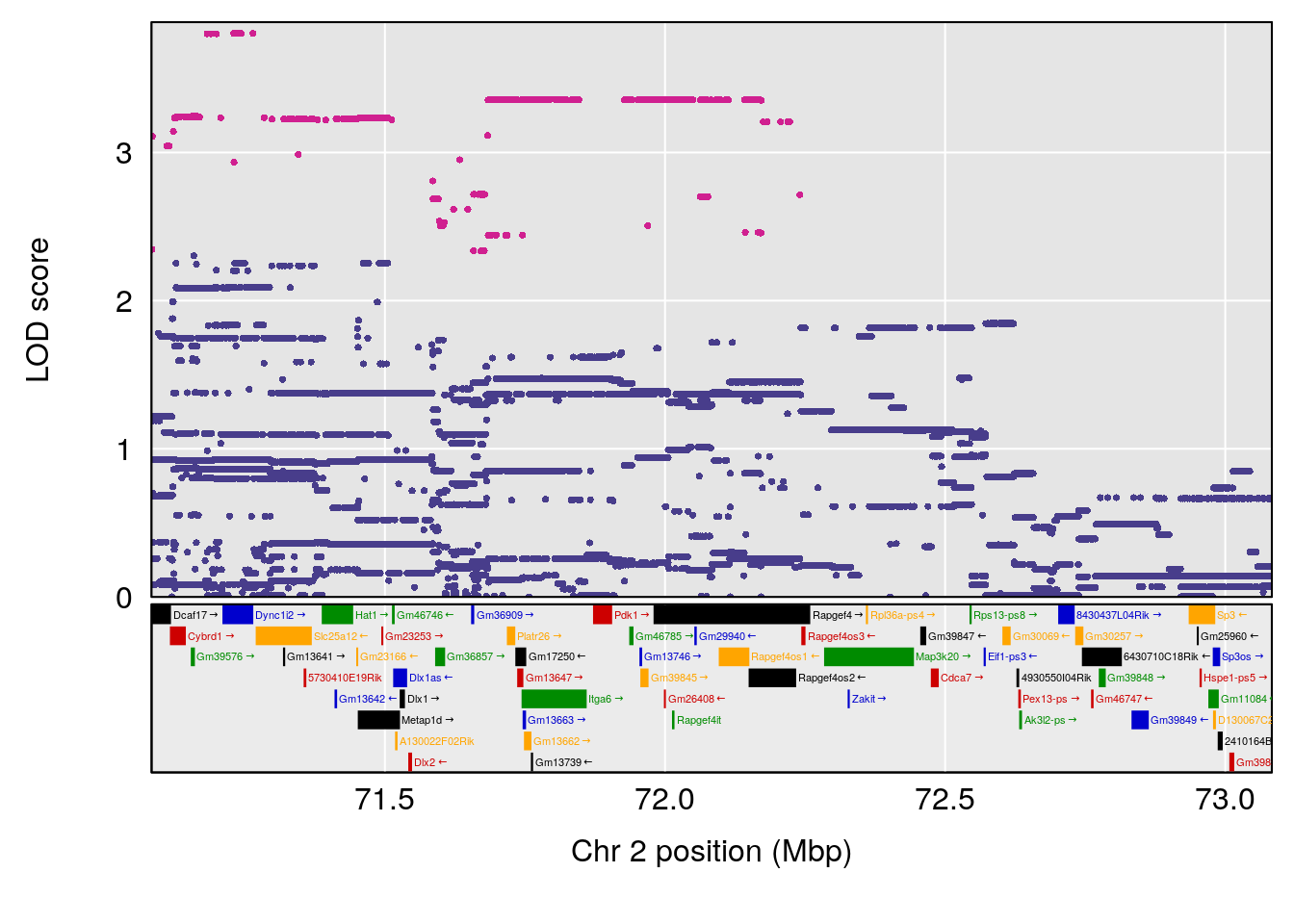
# [1] 2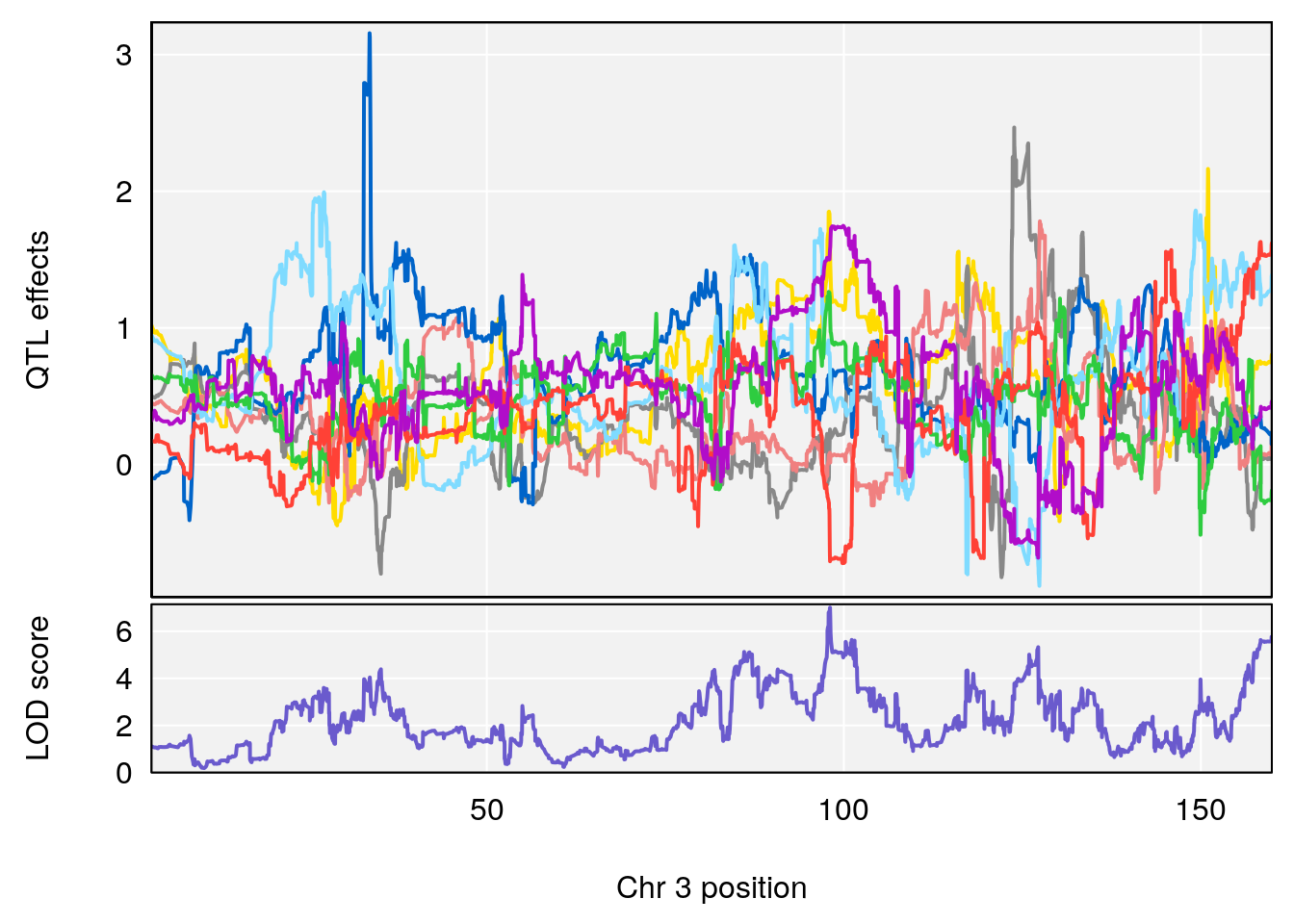
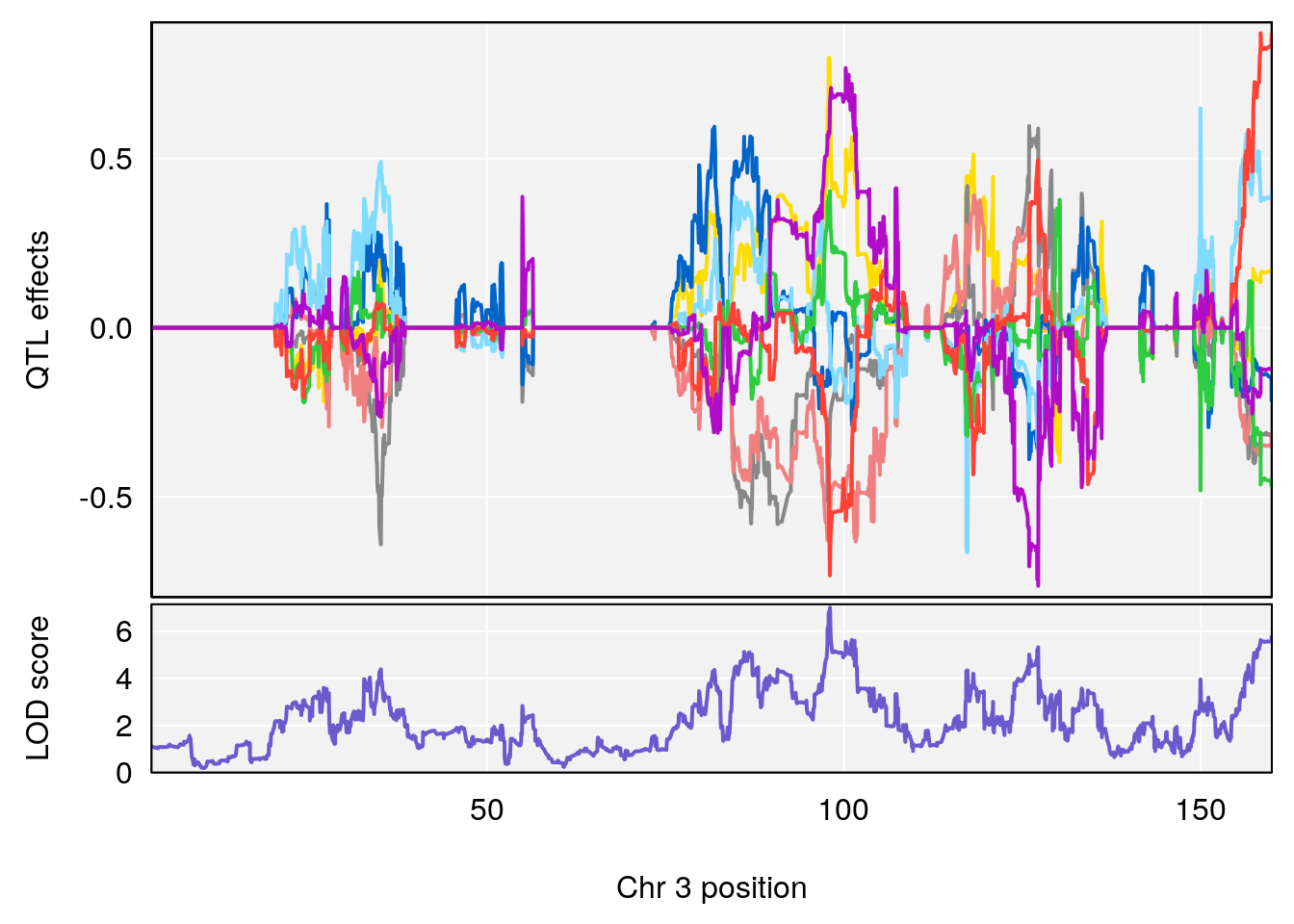
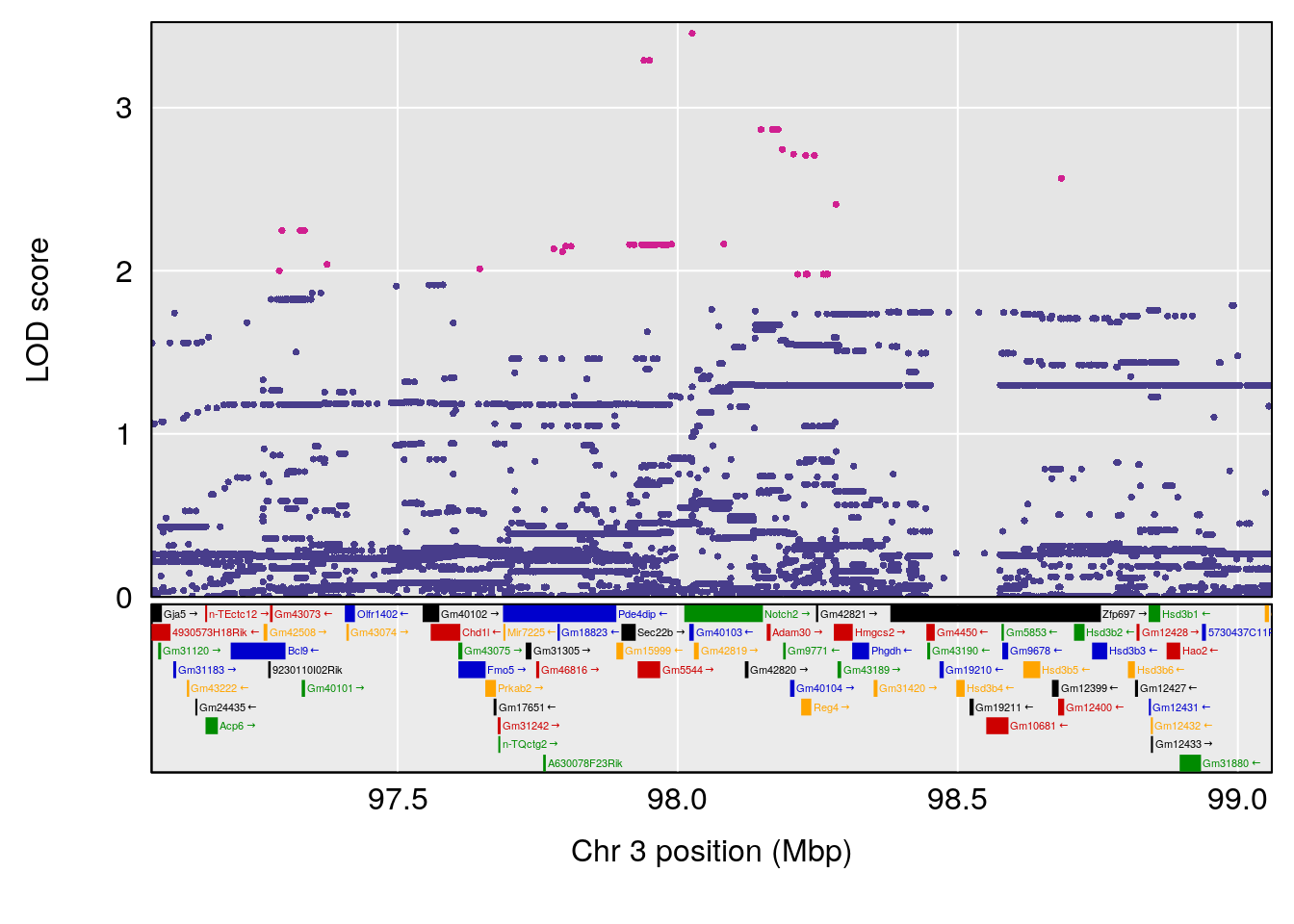
# [1] 3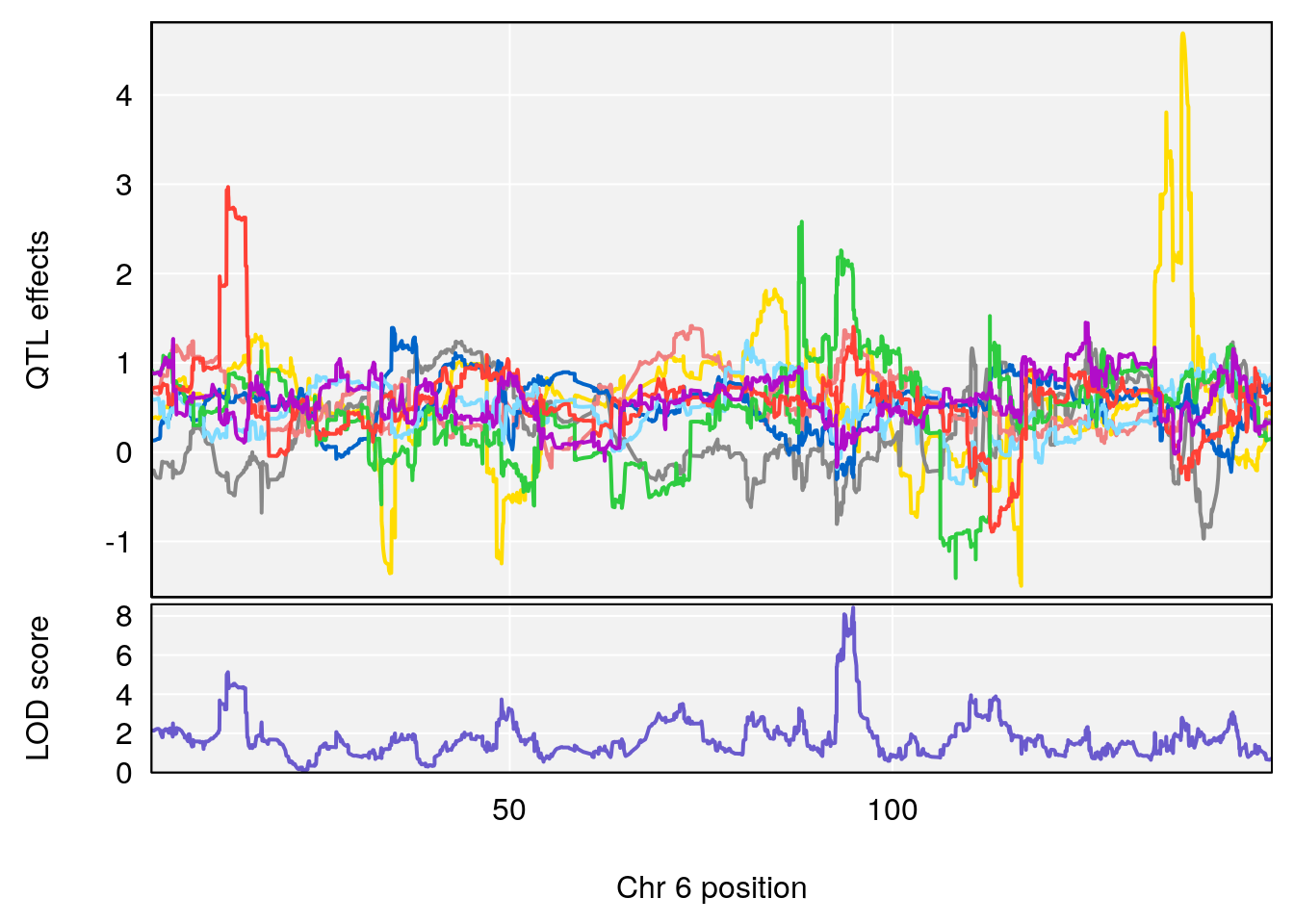
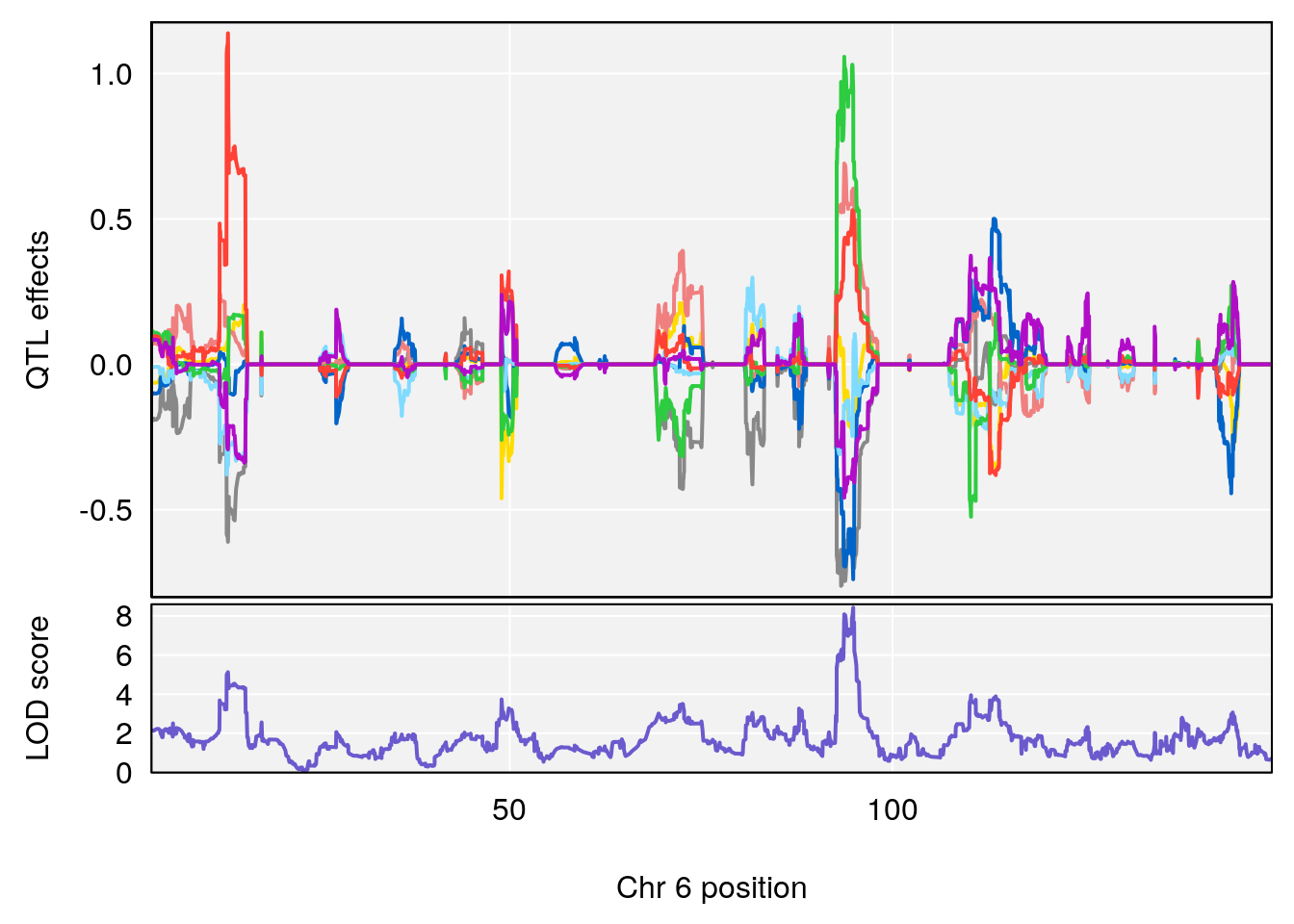
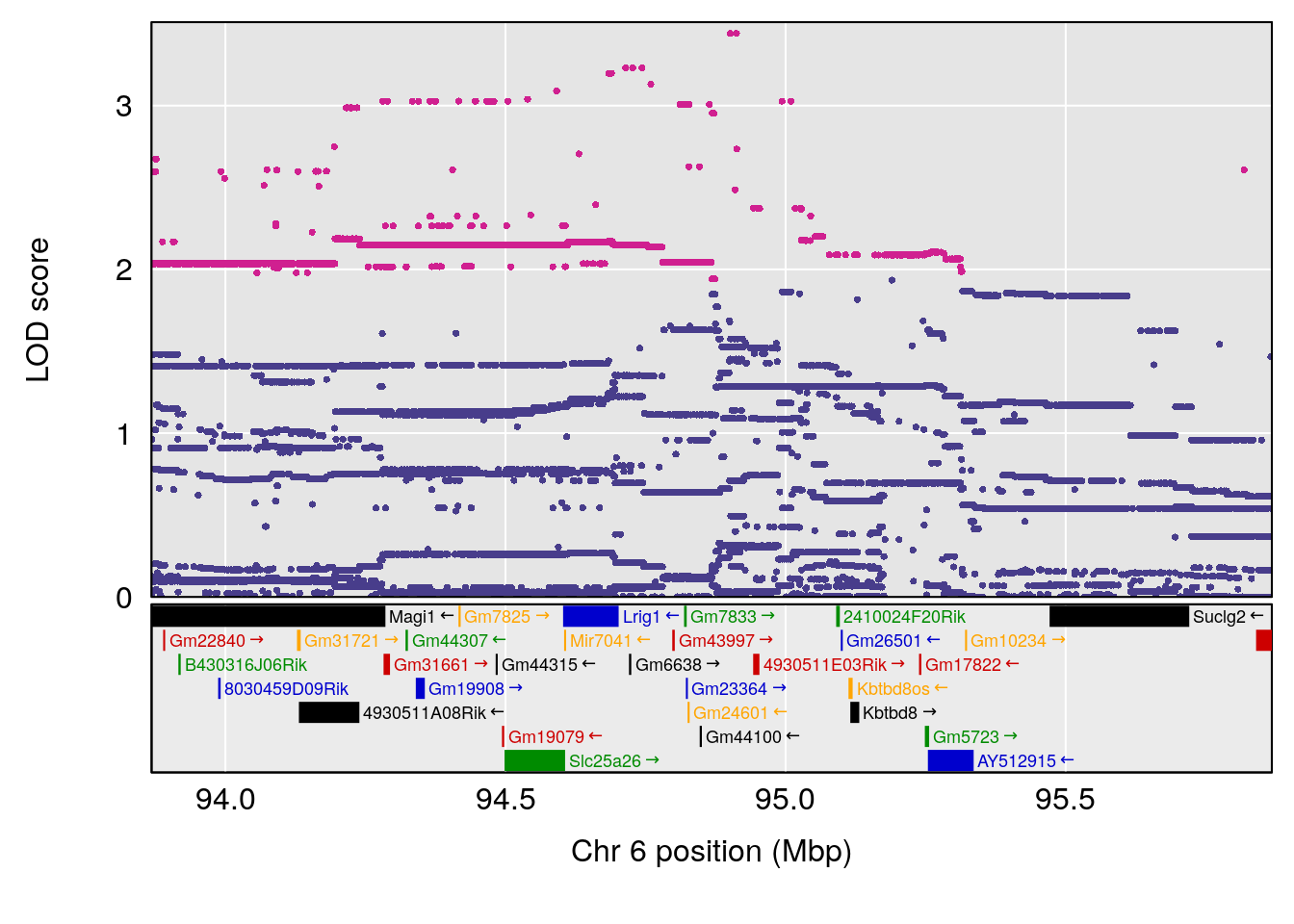
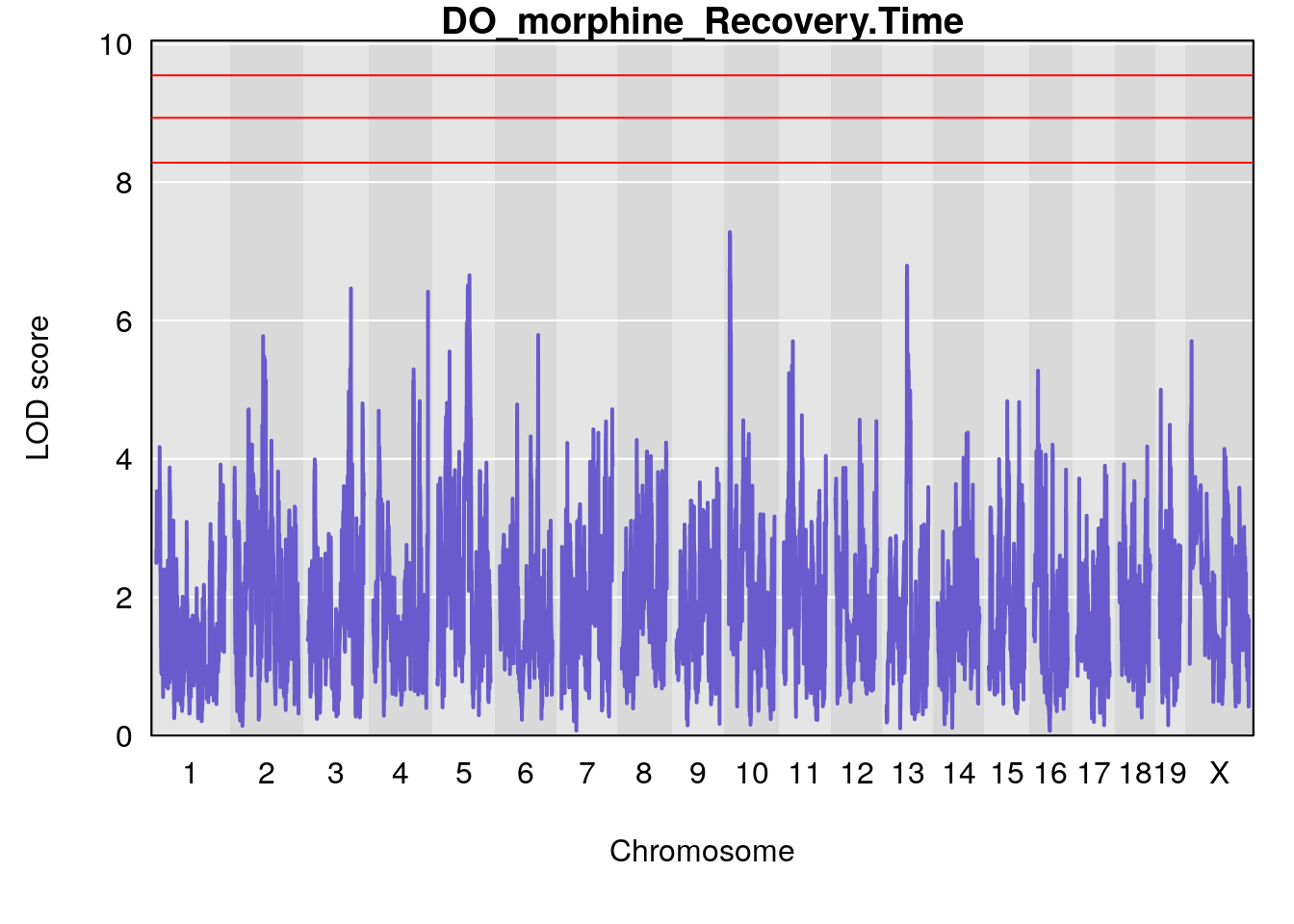
# [1] "Recovery.Time"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 59.35852 6.107005 59.07336 194.5045
# 2 1 pheno1 4 156.04818 6.519153 114.73873 156.1199
# 3 1 pheno1 8 117.07865 6.973714 115.52733 117.7515
# [1] 1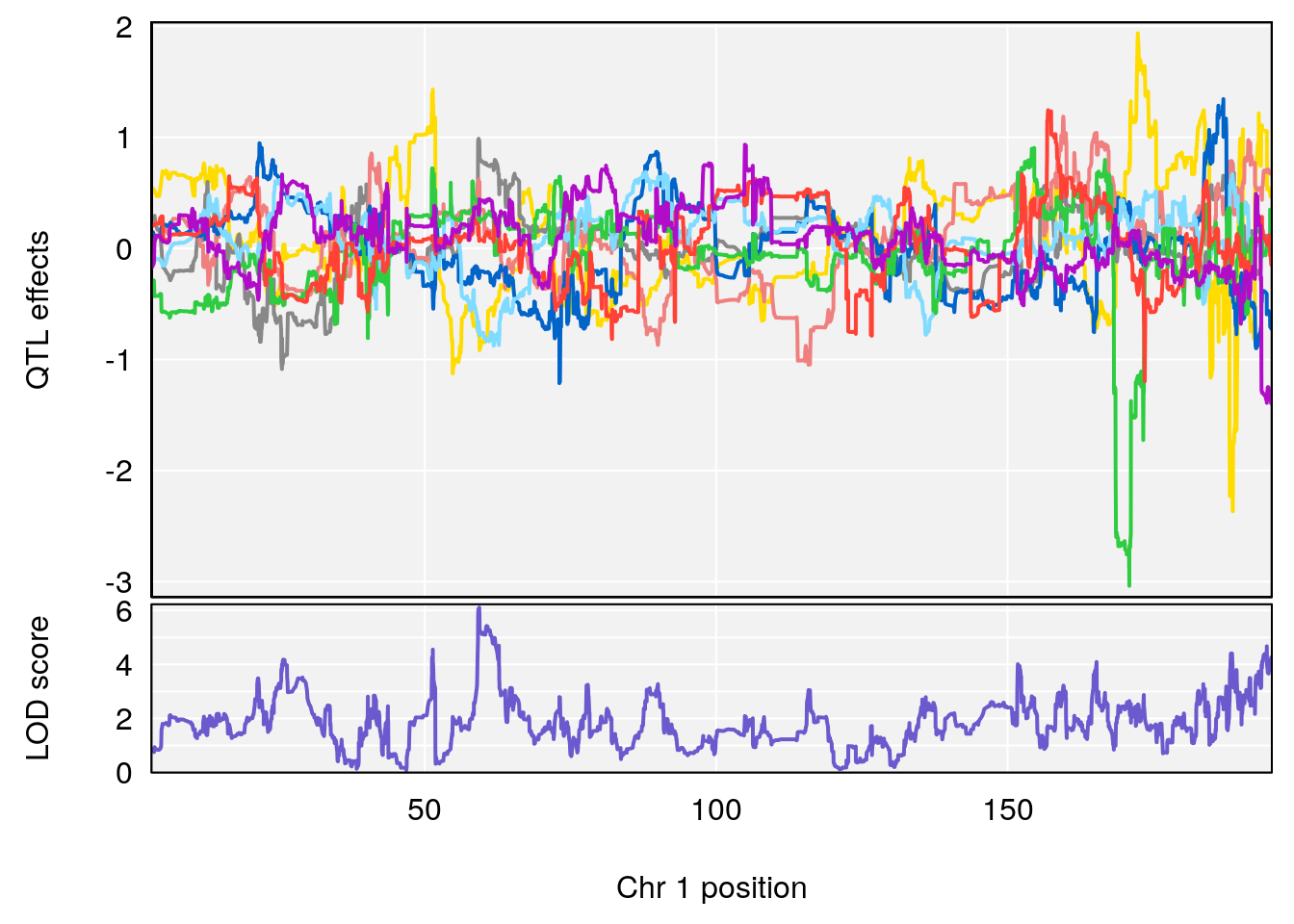
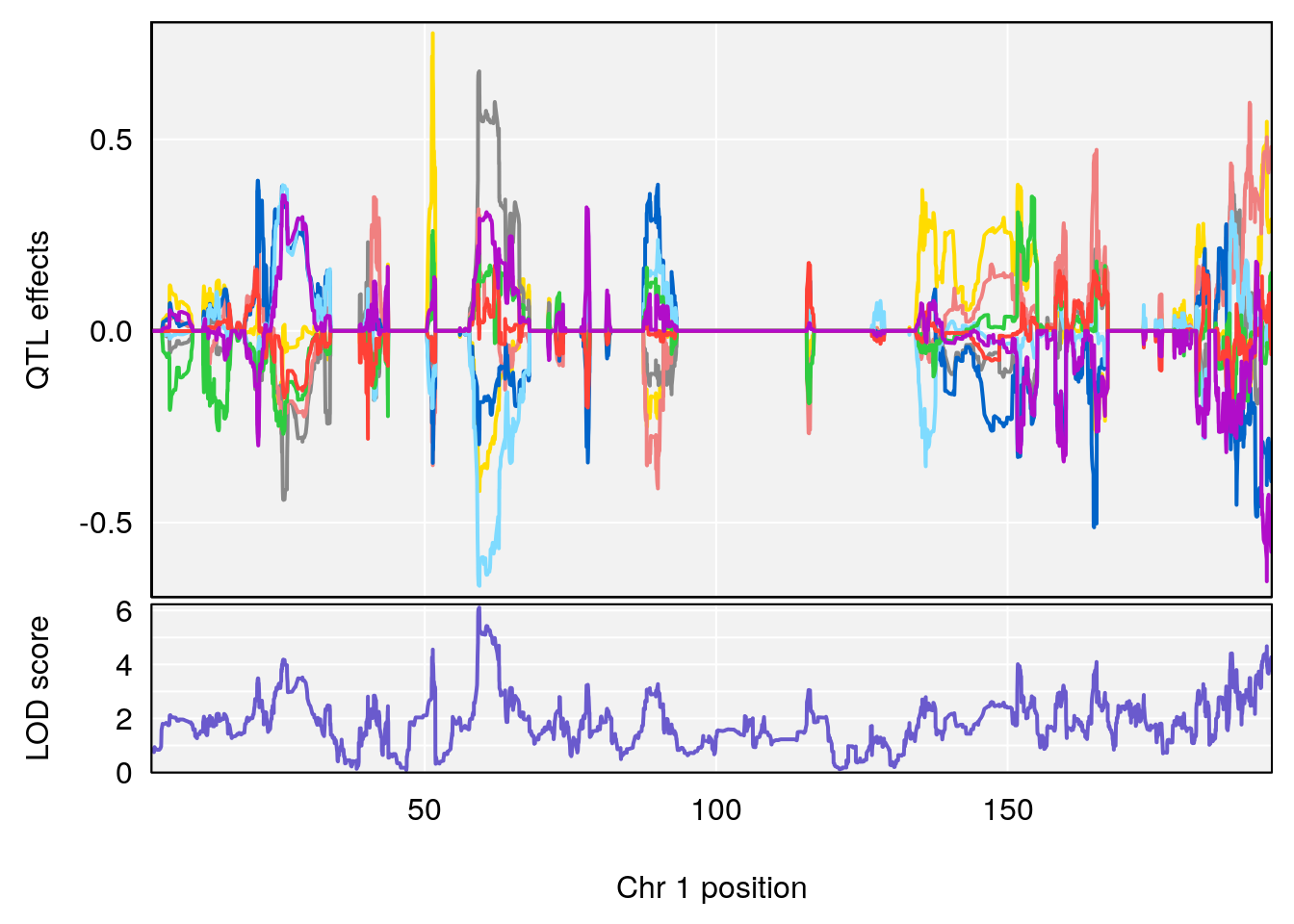
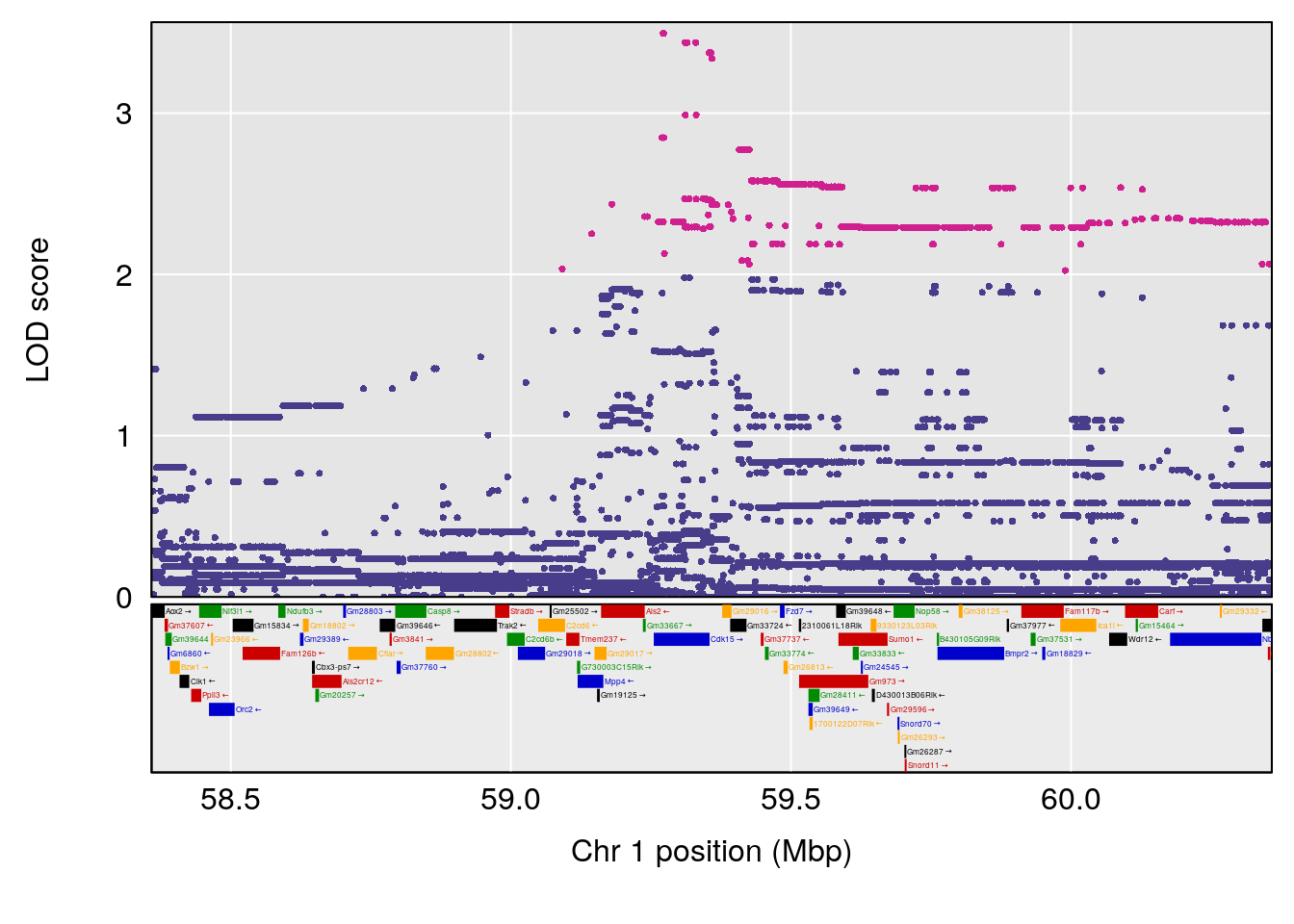
# [1] 2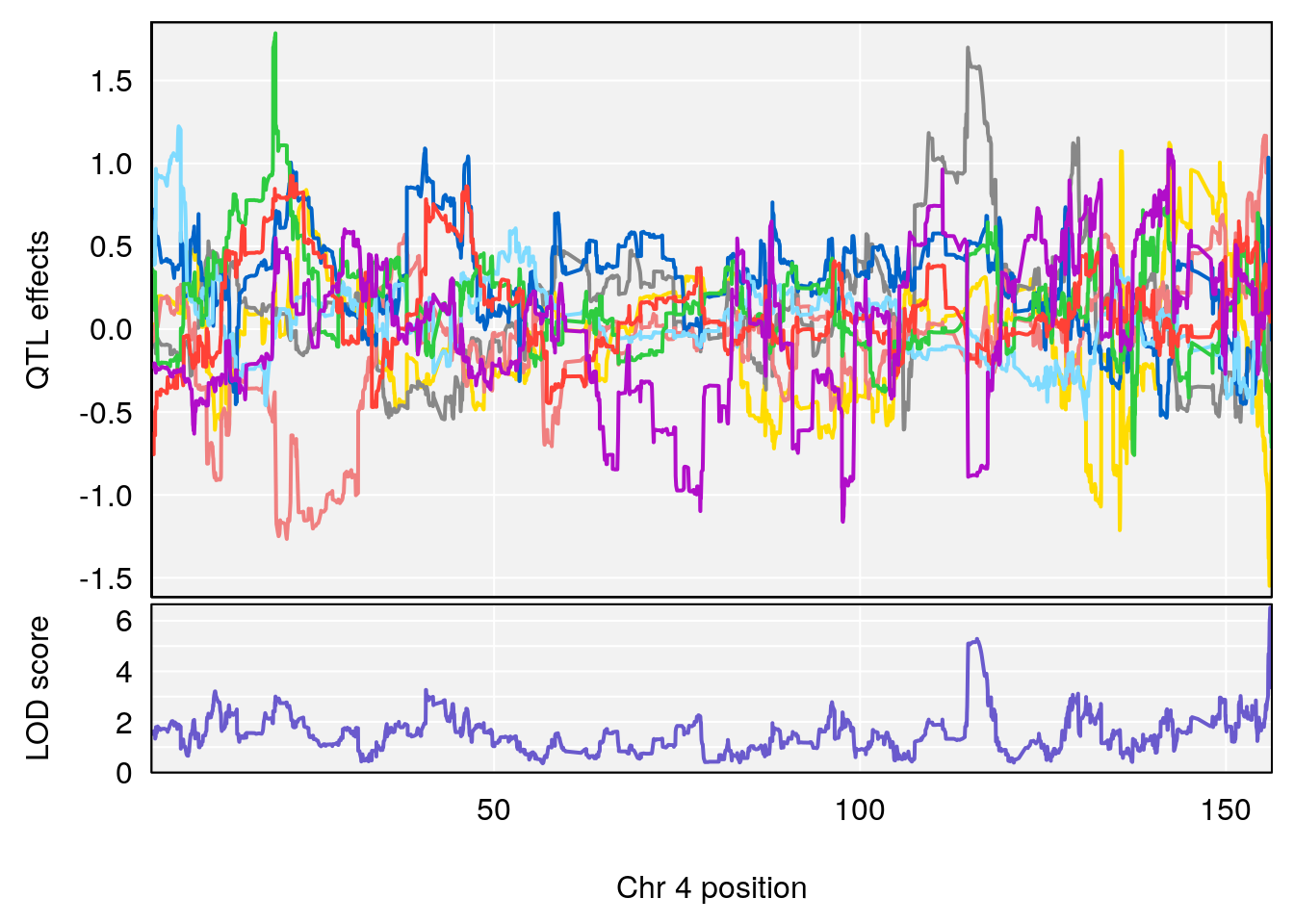
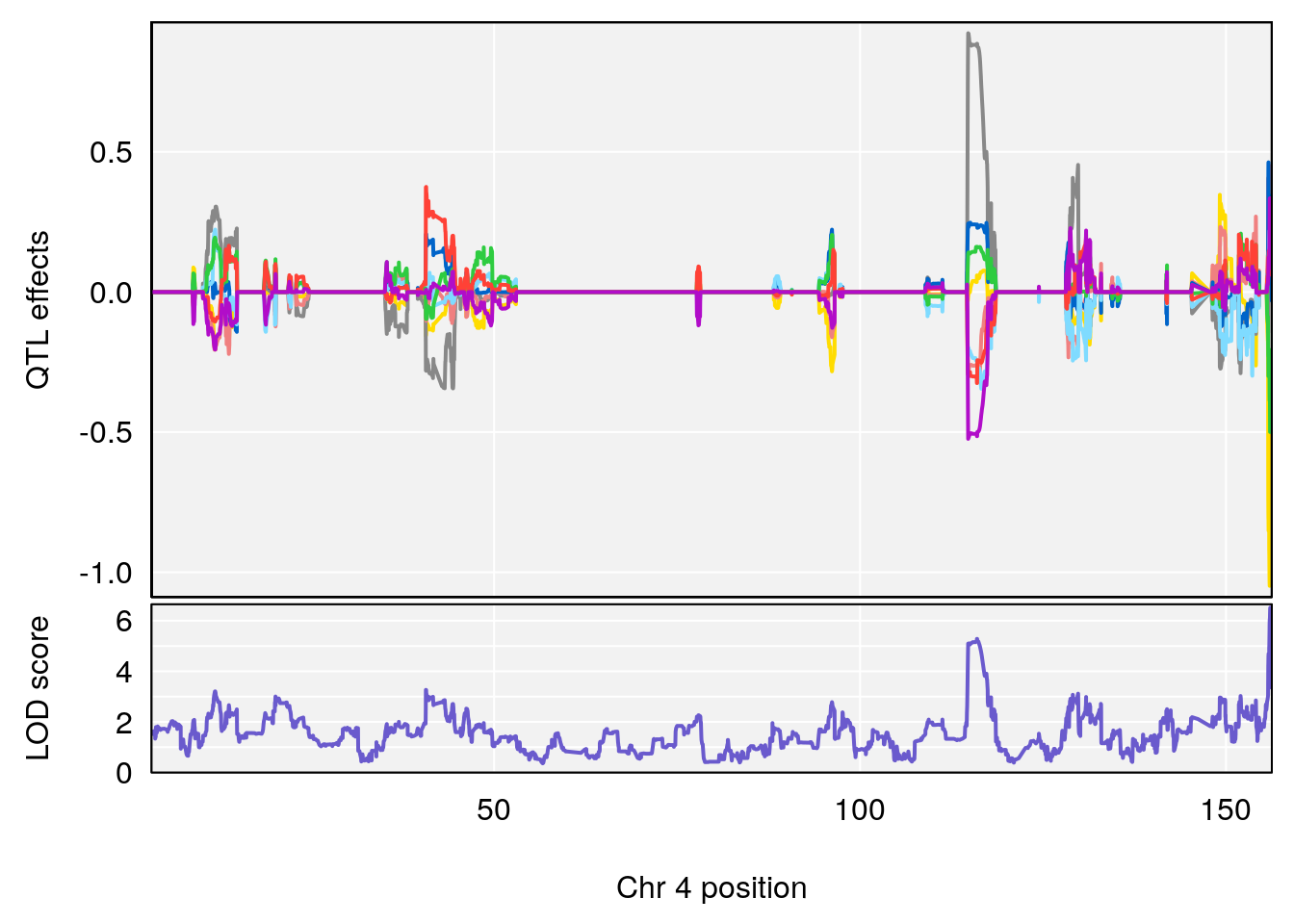
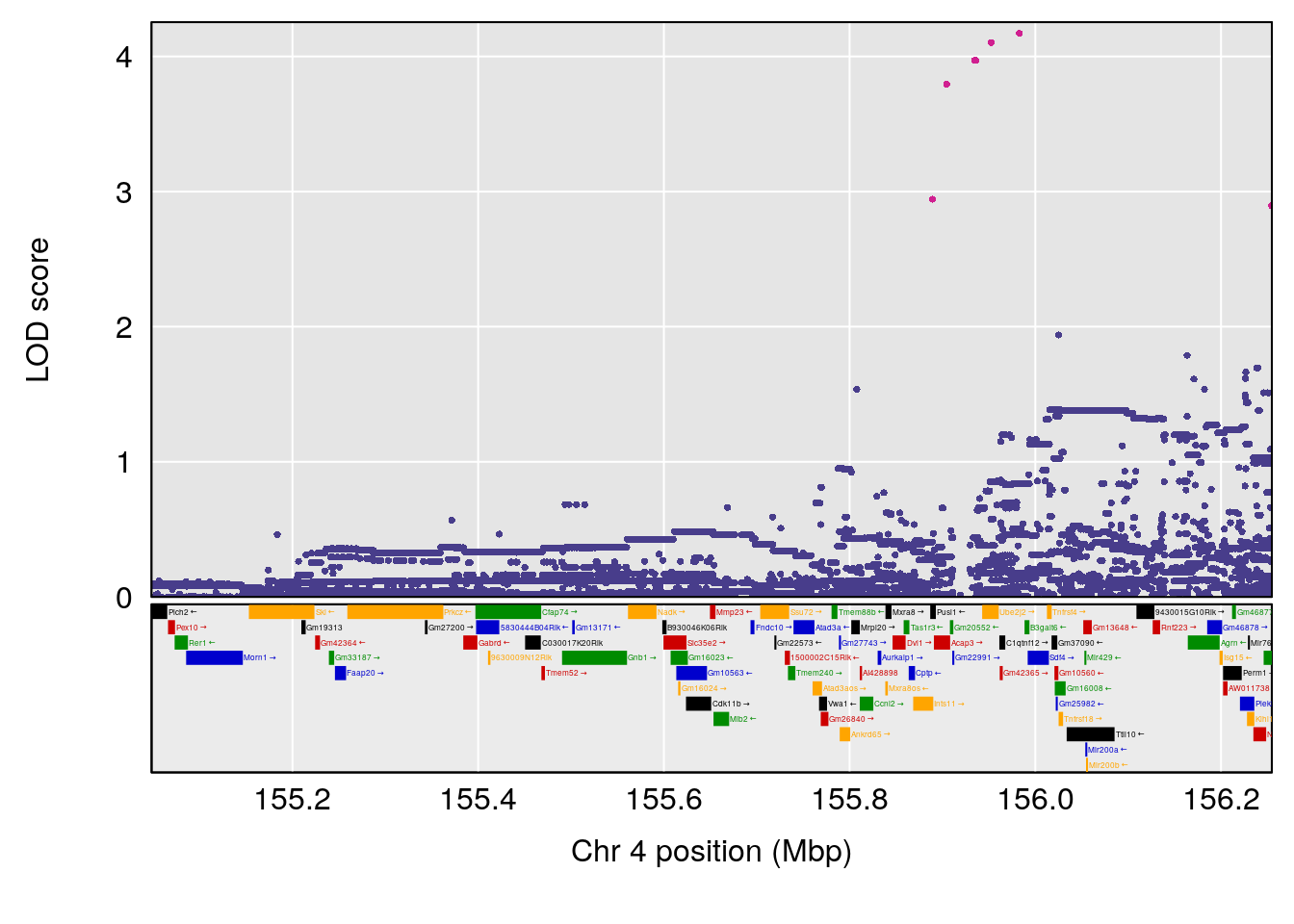
# [1] 3
| Version | Author | Date |
|---|---|---|
| 9e4e3be | xhyuo | 2021-08-24 |
| Version | Author | Date |
|---|---|---|
| 9e4e3be | xhyuo | 2021-08-24 |
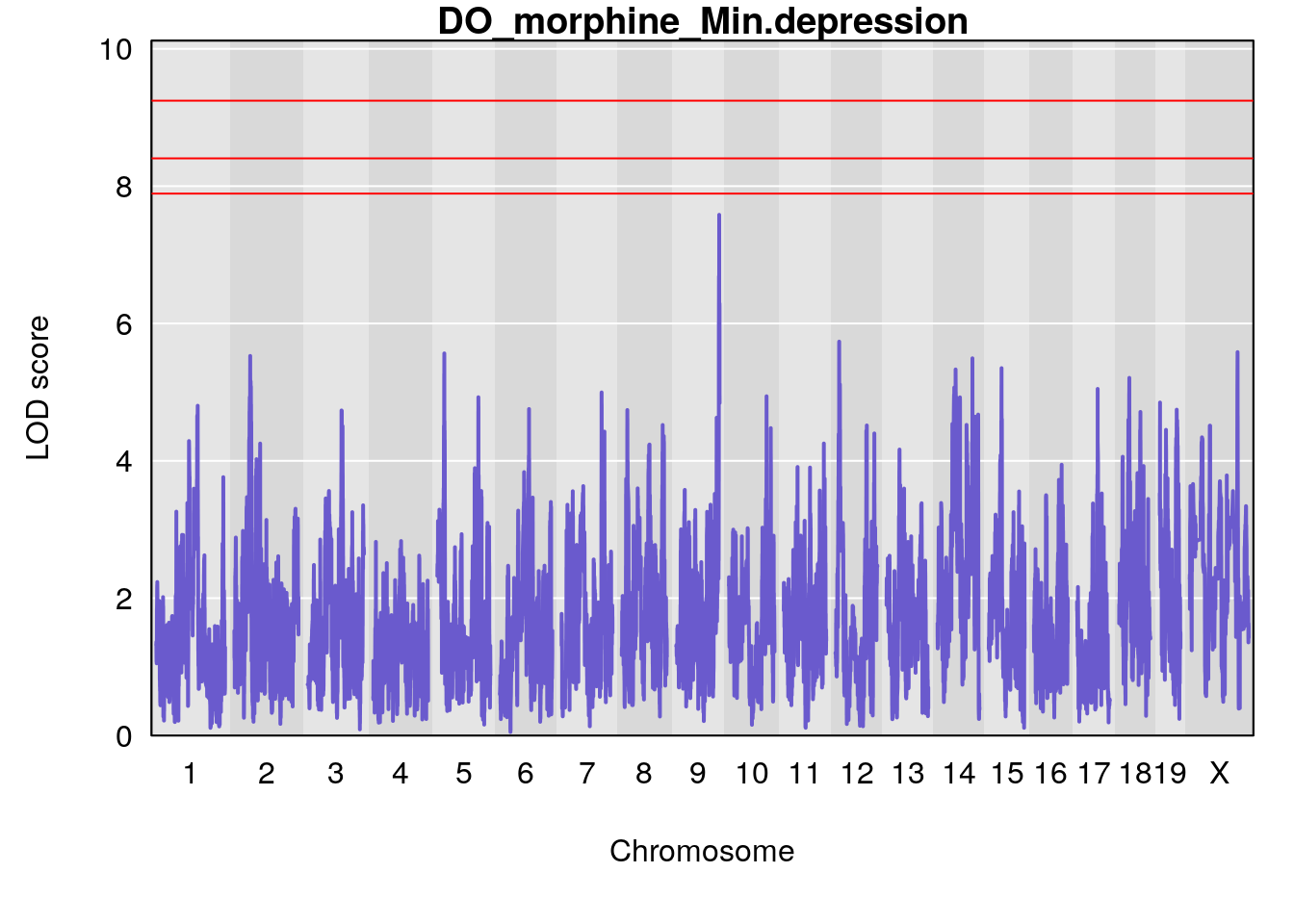
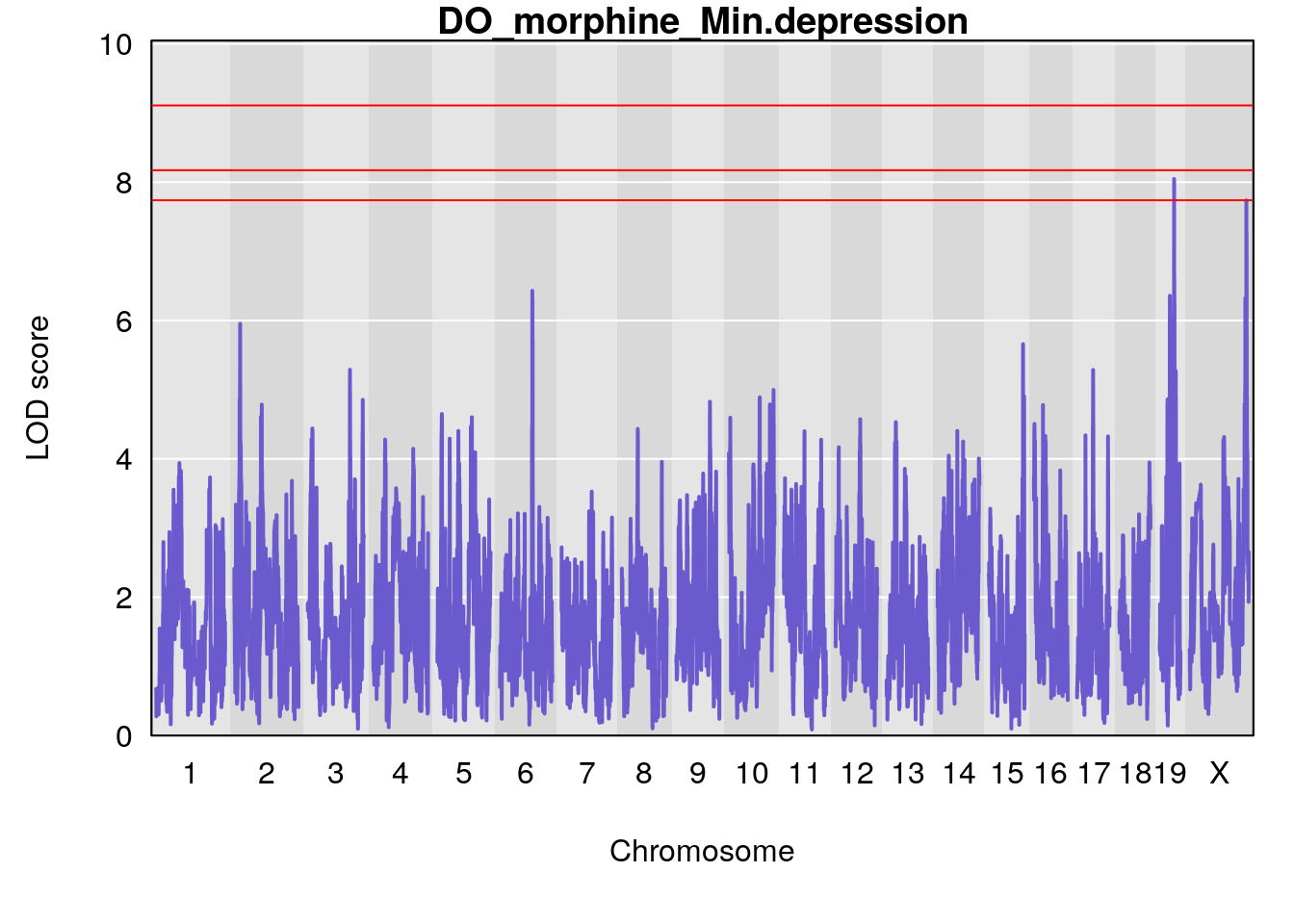
# [1] "Min.depression"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 6 93.69909 6.677445 93.60388 94.87509
# [1] 1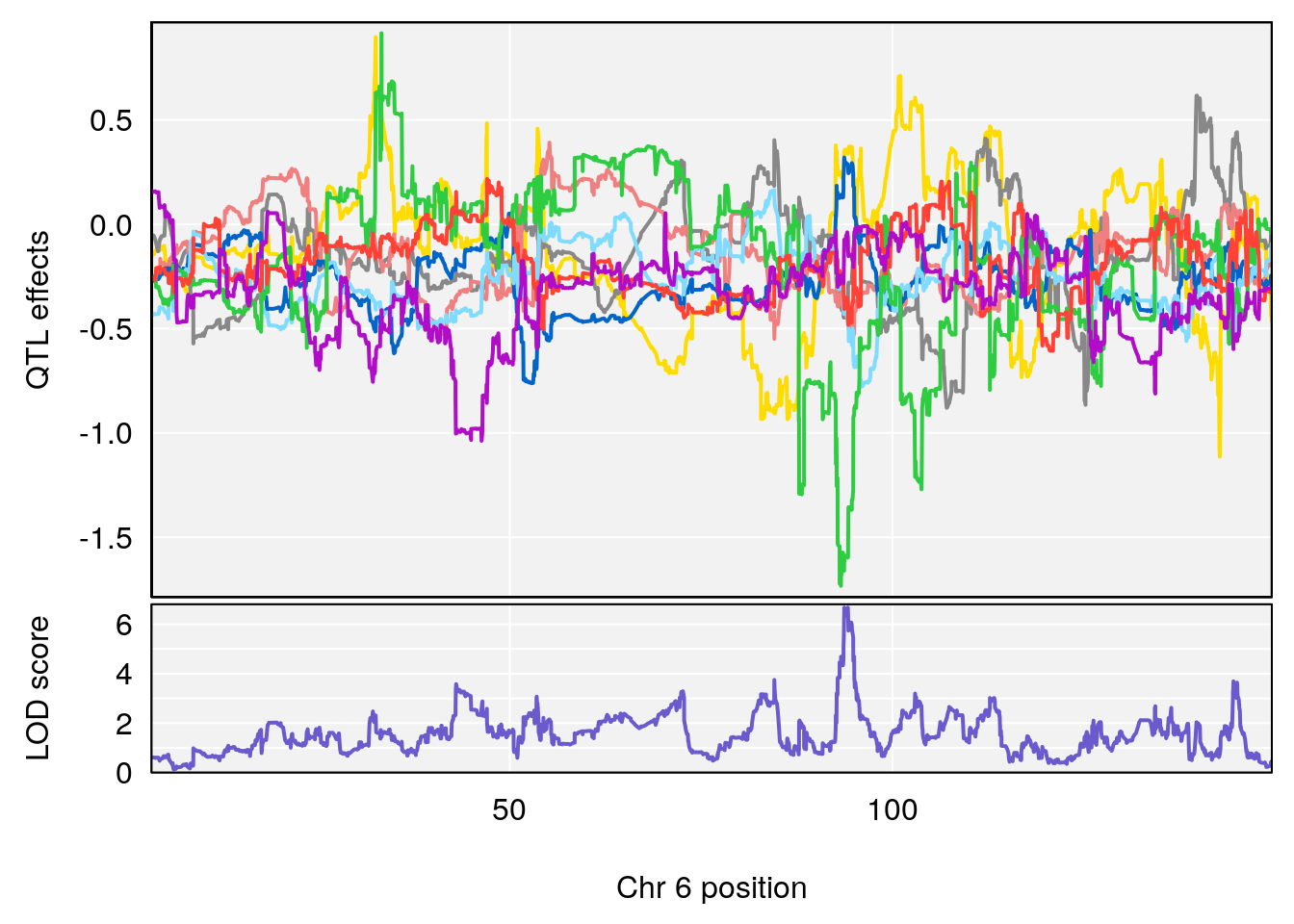
| Version | Author | Date |
|---|---|---|
| 9e4e3be | xhyuo | 2021-08-24 |
| Version | Author | Date |
|---|---|---|
| 9e4e3be | xhyuo | 2021-08-24 |
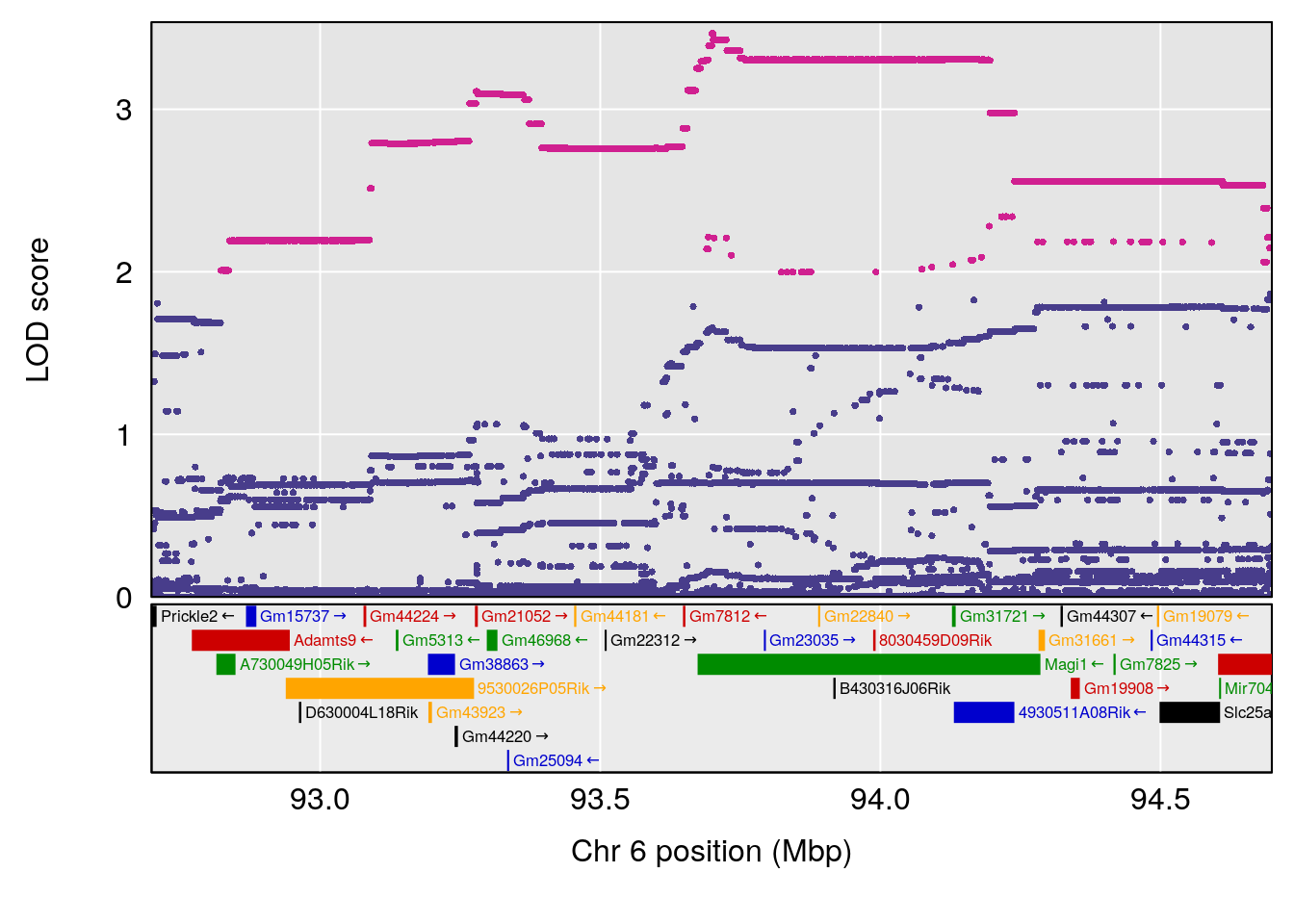
| Version | Author | Date |
|---|---|---|
| 9e4e3be | xhyuo | 2021-08-24 |
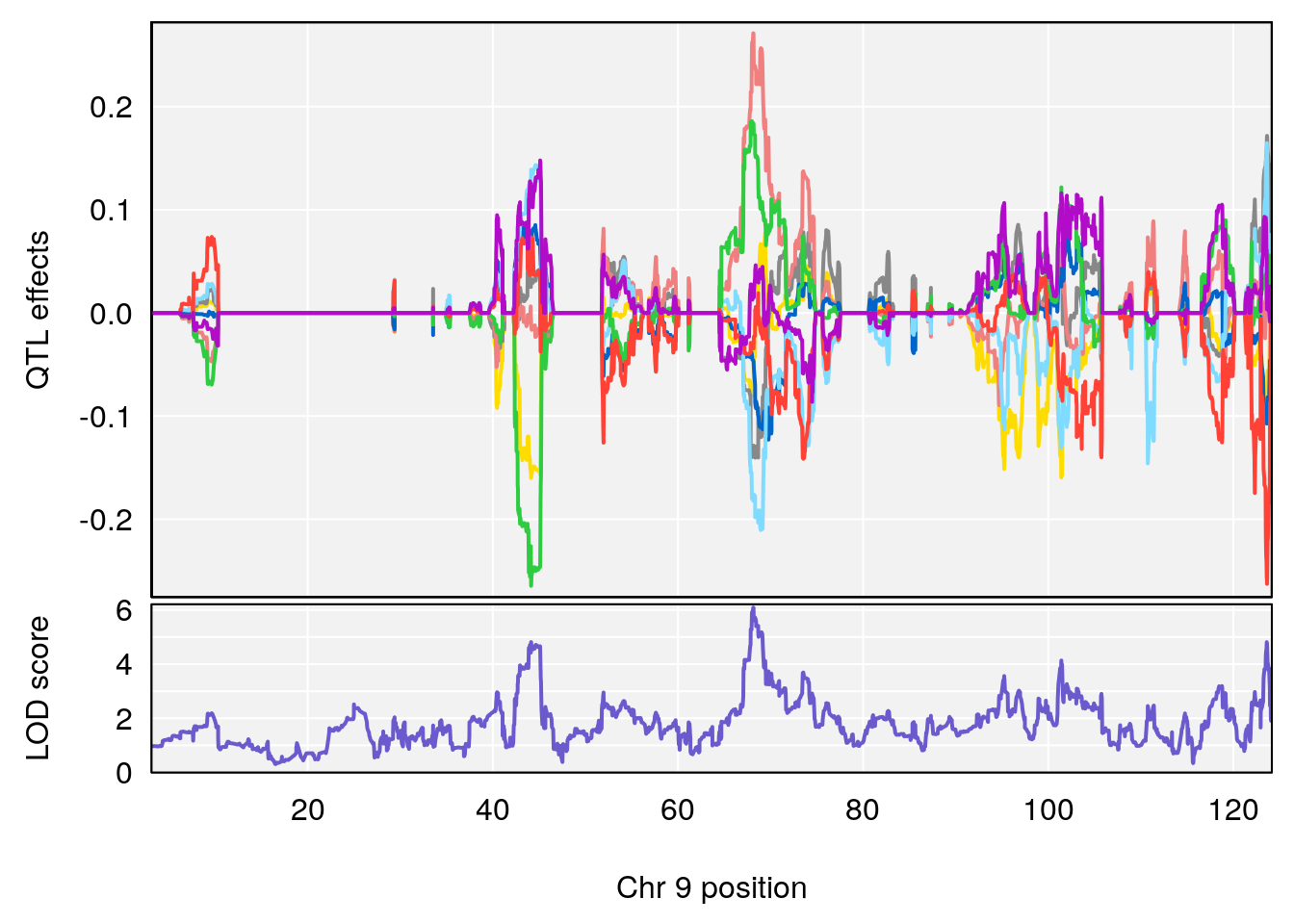
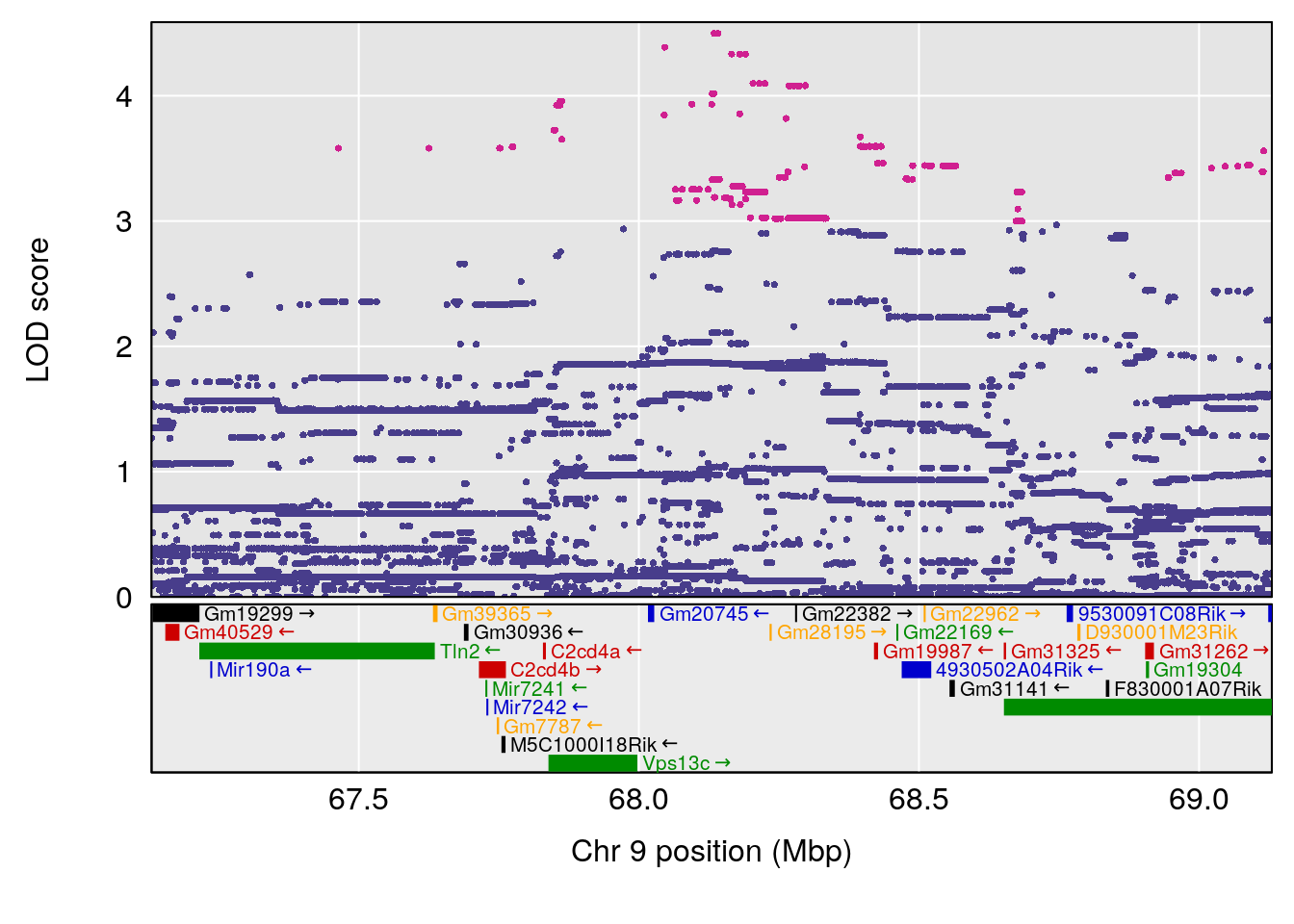
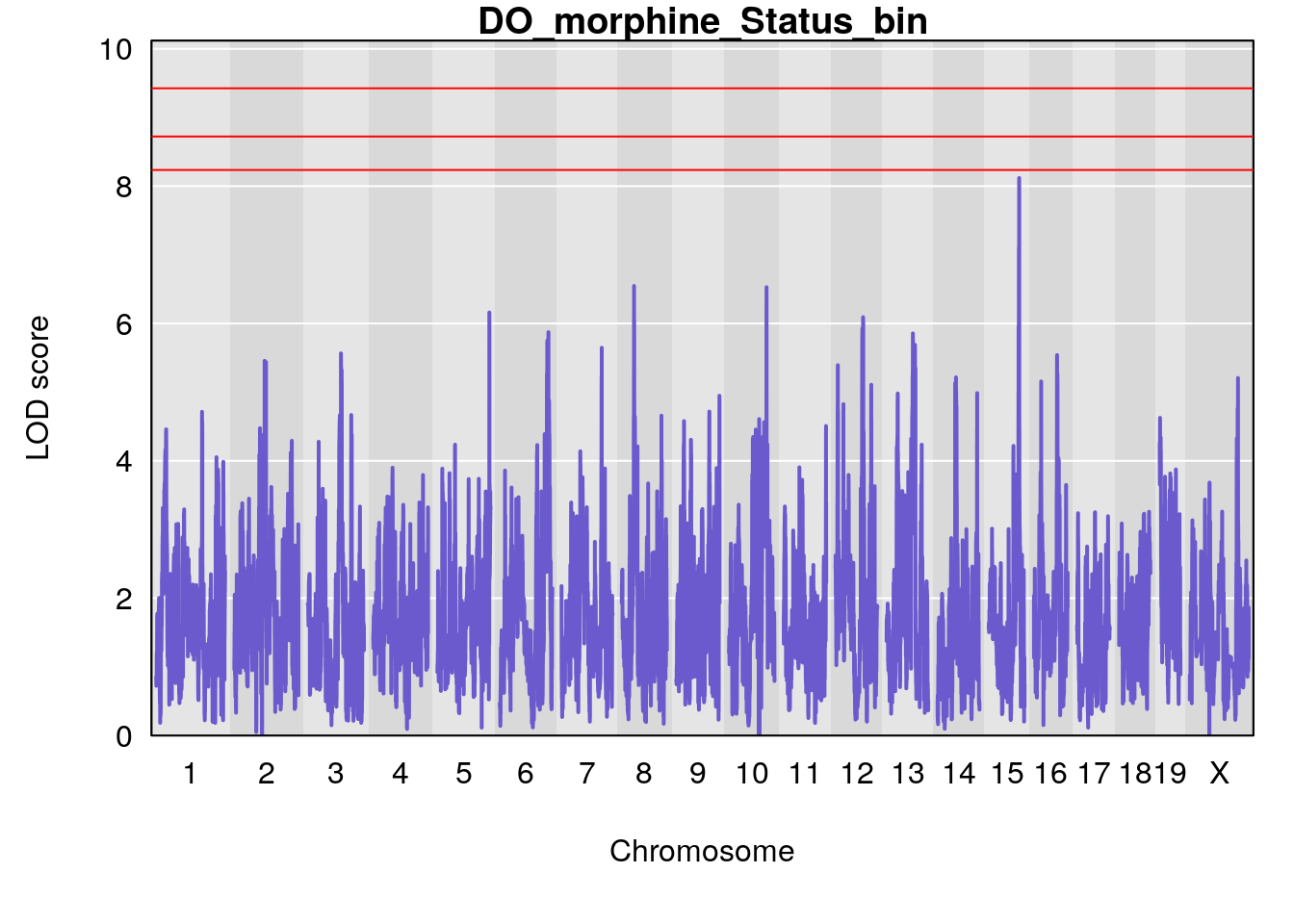
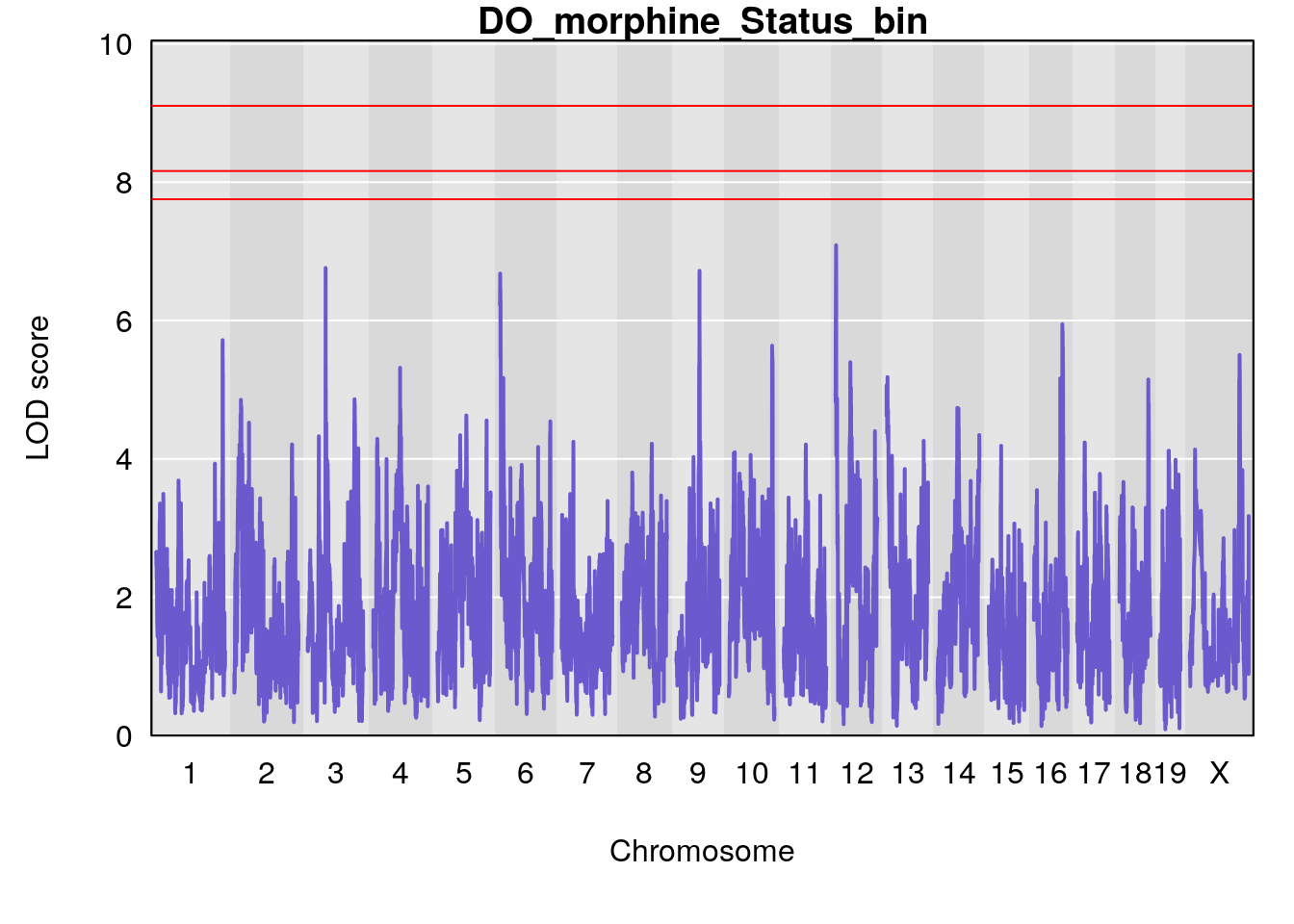
# [1] "Status_bin"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
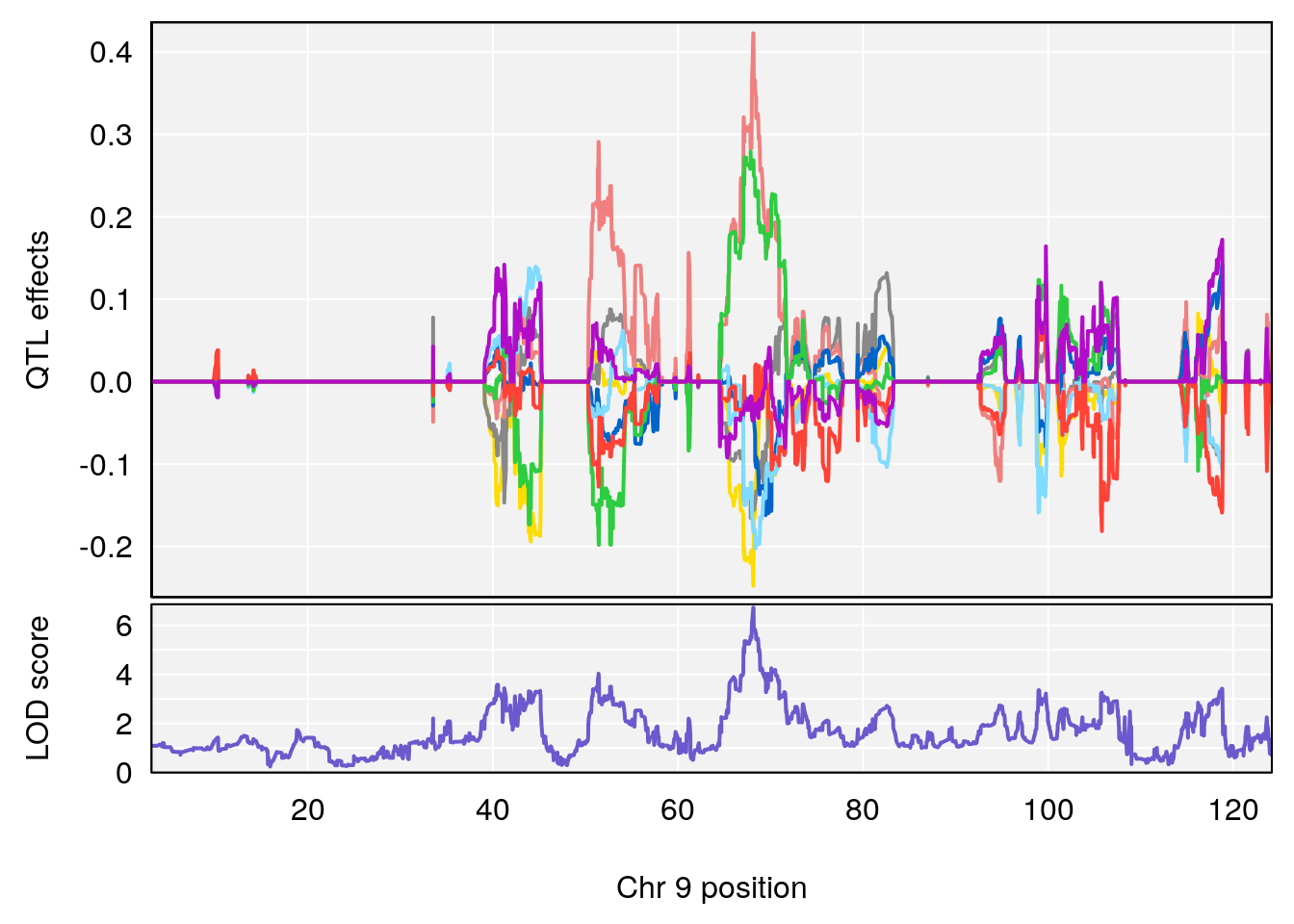
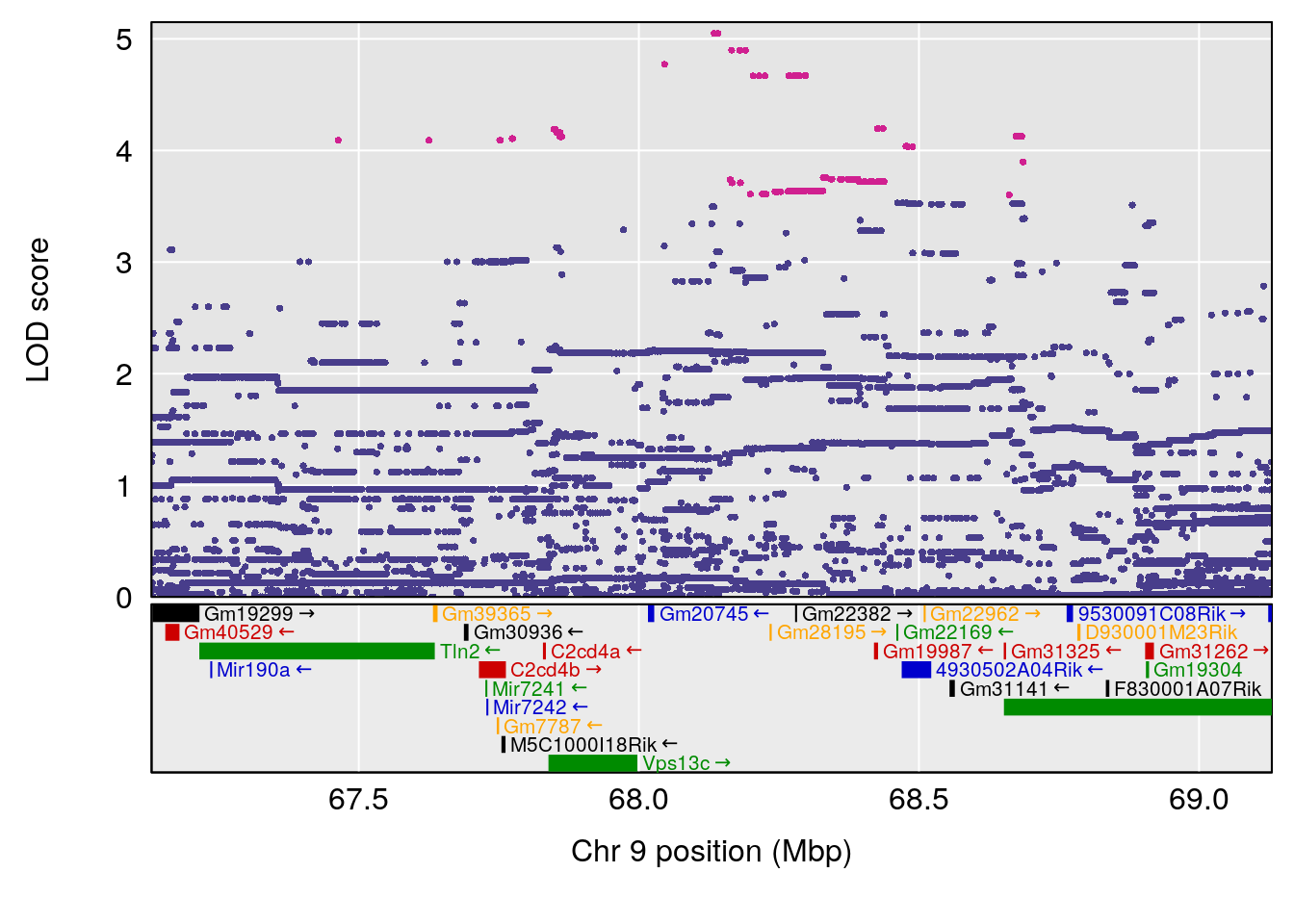
# 1 1 pheno1 9 68.12998 6.087016 43.84234 123.7027
# [1] 1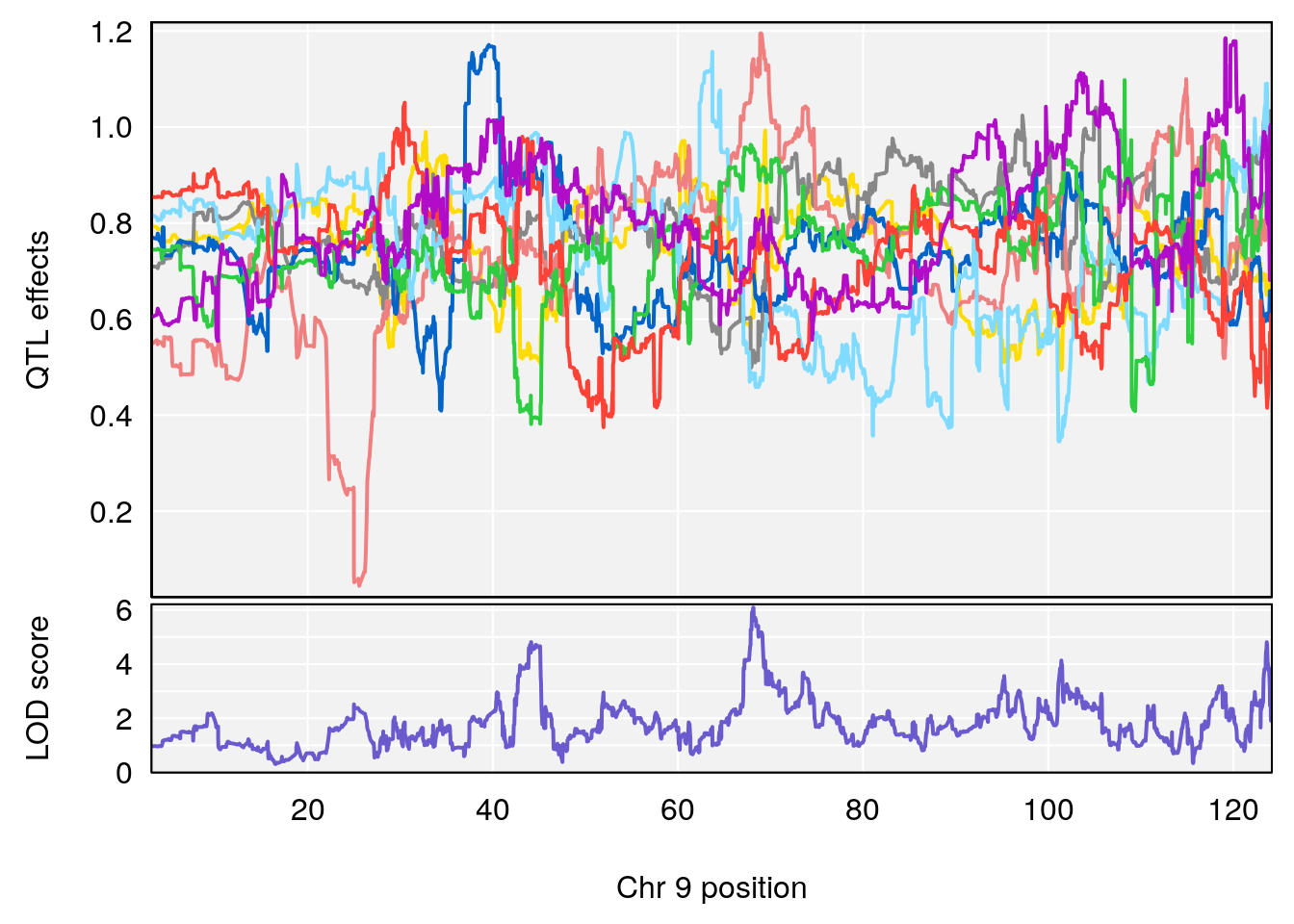
| Version | Author | Date |
|---|---|---|
| 9e4e3be | xhyuo | 2021-08-24 |
| Version | Author | Date |
|---|---|---|
| 9e4e3be | xhyuo | 2021-08-24 |
| Version | Author | Date |
|---|---|---|
| 9e4e3be | xhyuo | 2021-08-24 |
#save peaks coeff plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
fname <- paste("output/DO_morphine_secondbatch_",i,"_coefplot.pdf",sep="")
pdf(file = fname, width = 16, height =8)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
dev.off()
}
# [1] "Survival.Time"
# [1] 1
# [1] 2
# [1] 3
# [1] "Recovery.Time"
# [1] 1
# [1] 2
# [1] 3
# [1] "Min.depression"
# [1] 1
# [1] "Status_bin"
# [1] 1
#save peaks coeff blup plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
fname <- paste("output/DO_morphine_secondbatch_",i,"_coefplot_blup.pdf",sep="")
pdf(file = fname, width = 16, height =8)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
dev.off()
}
# [1] "Survival.Time"
# [1] 1
# [1] 2
# [1] 3
# [1] "Recovery.Time"
# [1] 1
# [1] 2
# [1] 3
# [1] "Min.depression"
# [1] 1
# [1] "Status_bin"
# [1] 1Plot qtl mapping on 69K
Plot by using its output qtl.morphine.69k.out.second_set.RData
load("output/qtl.morphine.69k.out.second_set.RData")
#pmap and gmap
load("data/69k_grid_pgmap.RData")
#genome-wide plot
for(i in names(qtl.morphine.out)){
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.morphine.out[[i]]) # overall maximum LOD score
plot(qtl.morphine.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, 10))
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("DO_morphine_",i))
}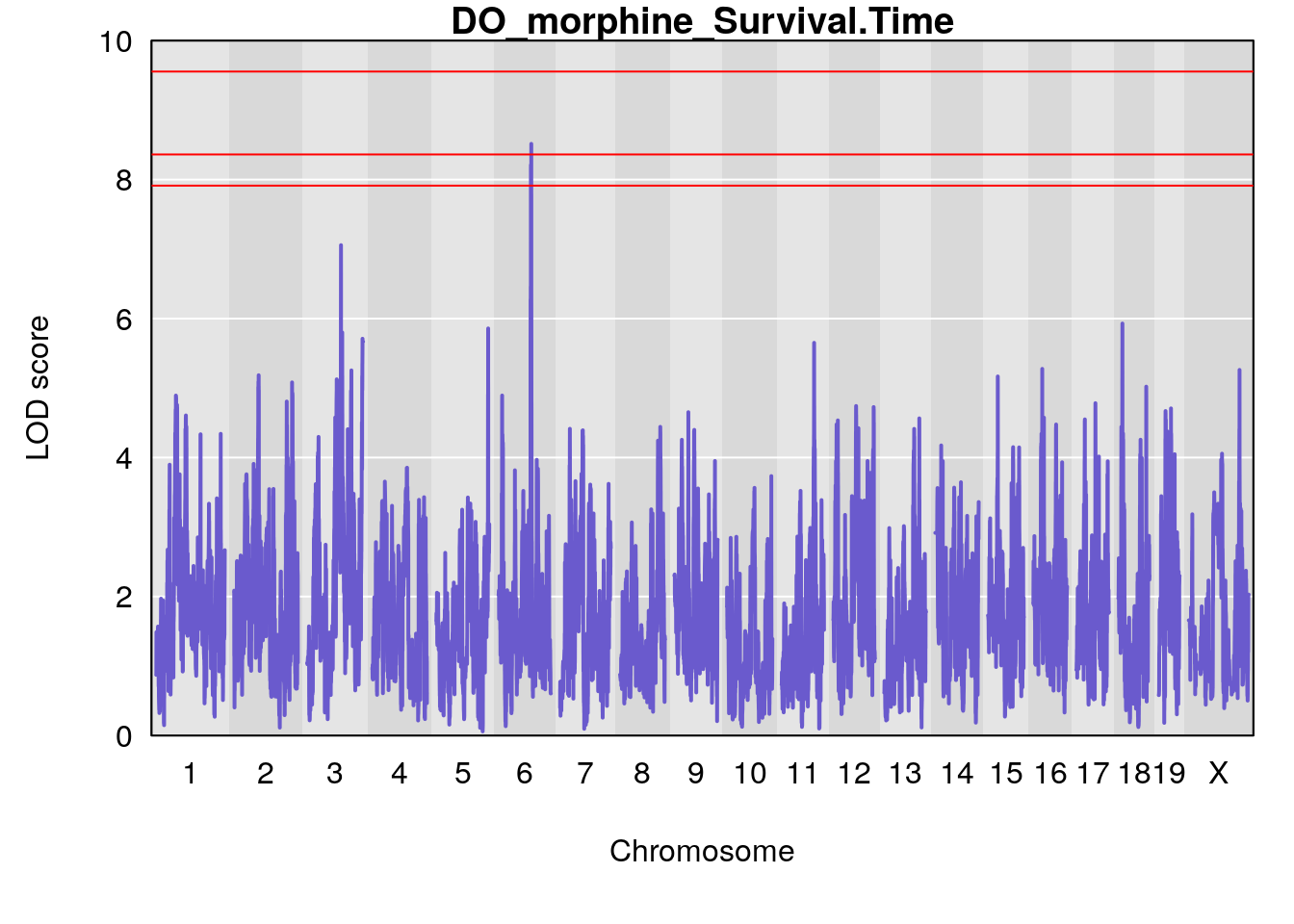
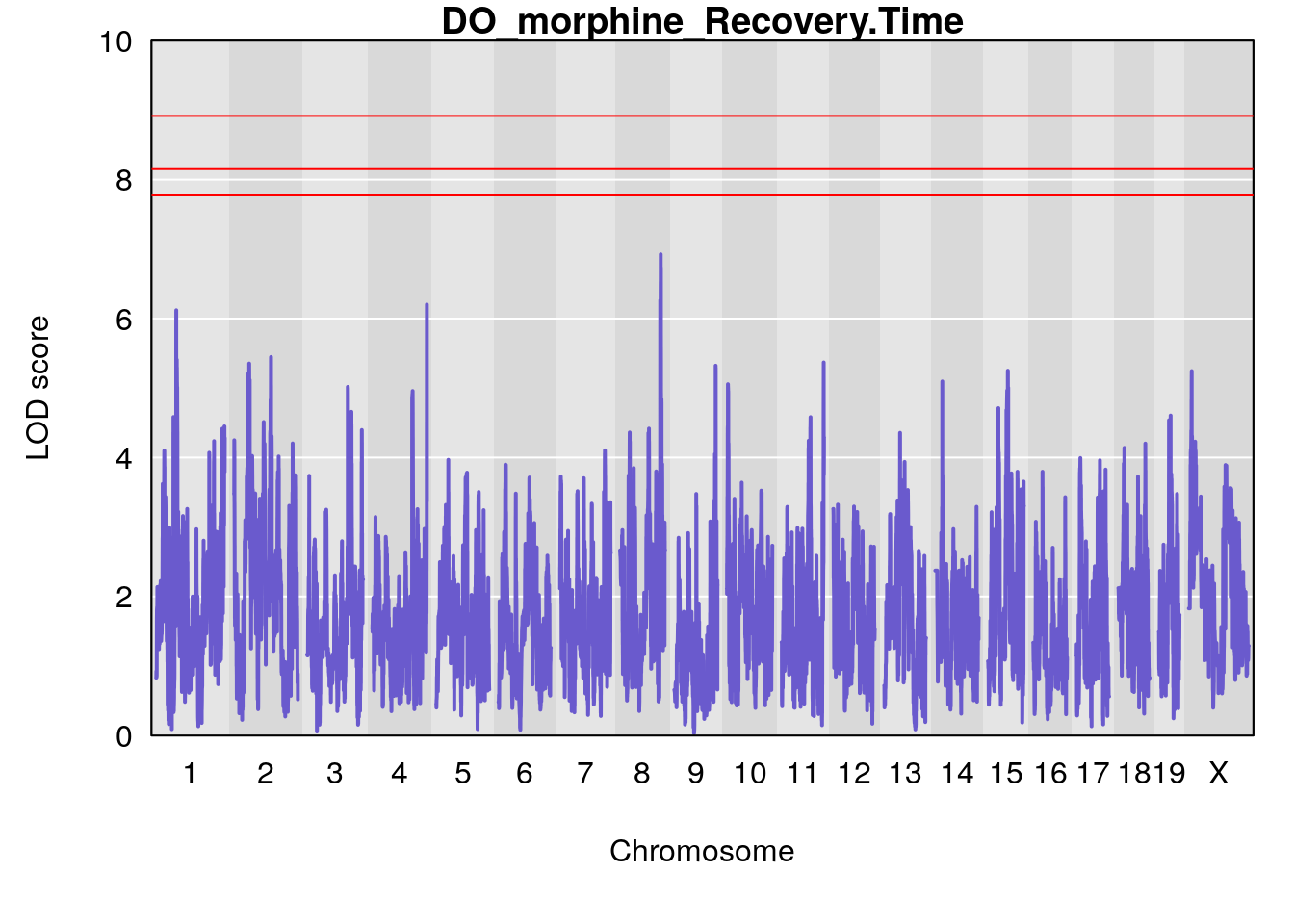
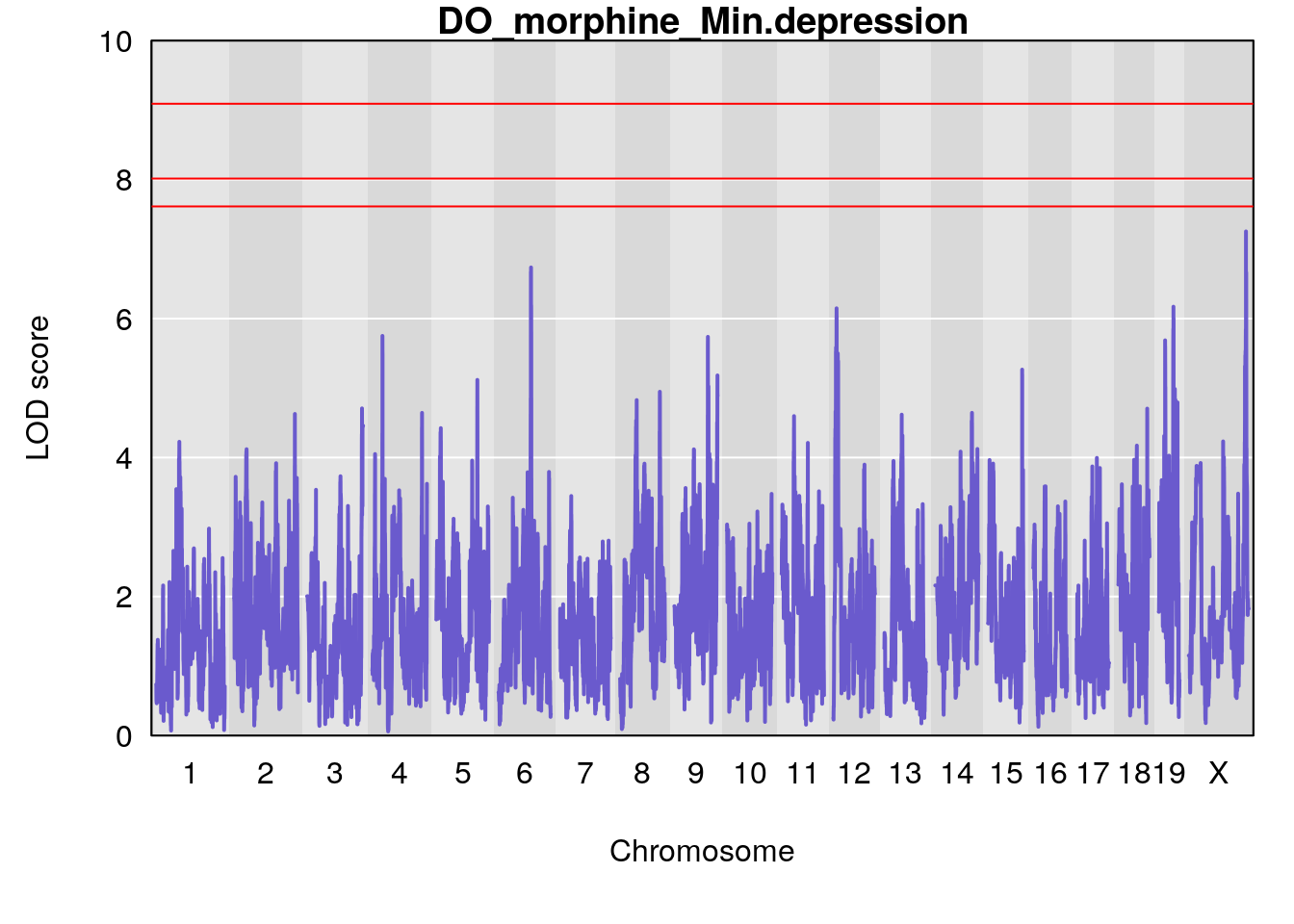
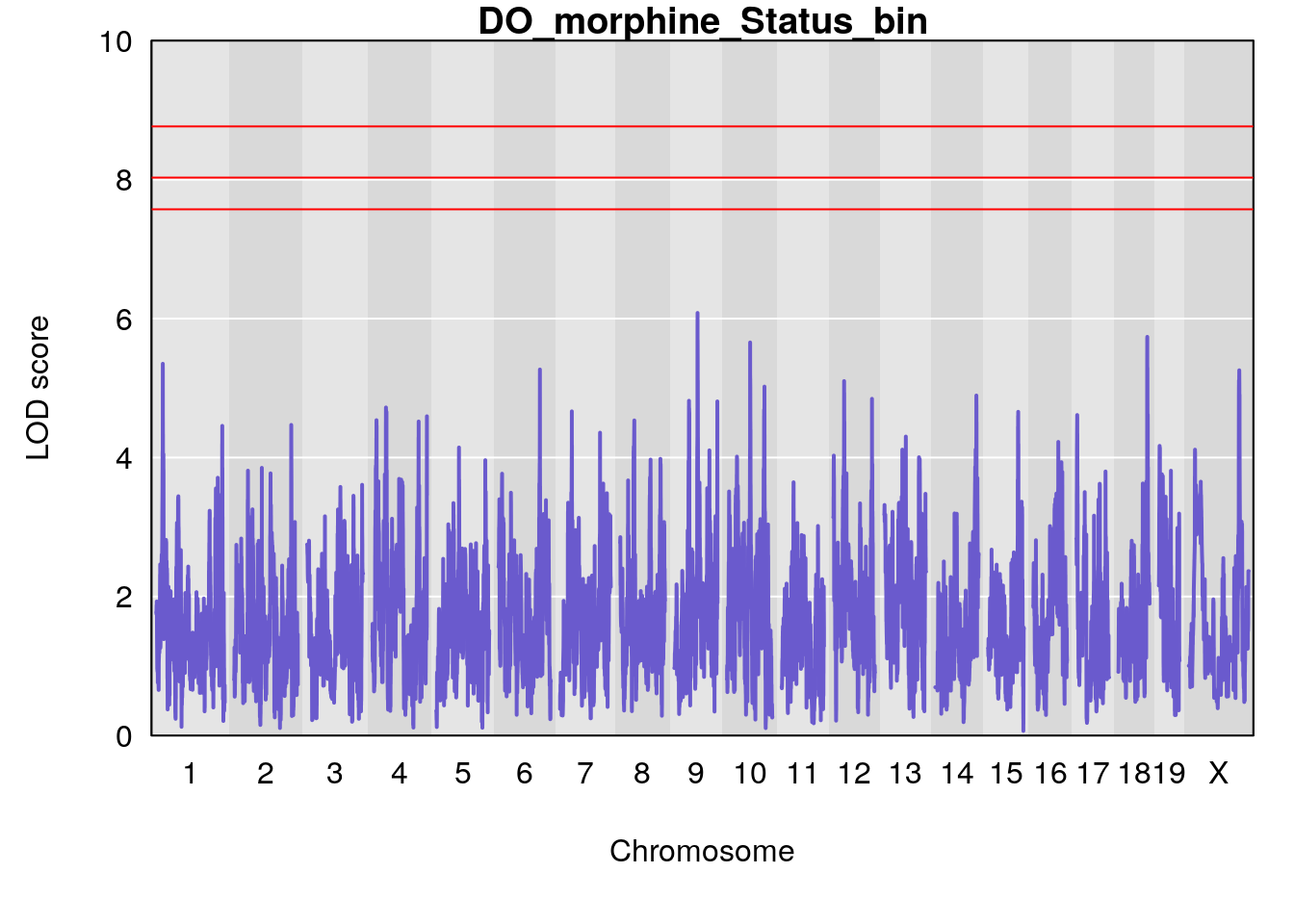
#save genome-wide plot
for(i in names(qtl.morphine.out)){
png(file = paste0("output/DO_morphine_secondbatch_69k_", i, ".png"), width = 16, height =8, res=300, units = "in")
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.morphine.out[[i]]) # overall maximum LOD score
plot(qtl.morphine.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, 10))
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("DO_morphine_",i))
dev.off()
}
#peaks coeff plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
print(peaks)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
}
# [1] "Survival.Time"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
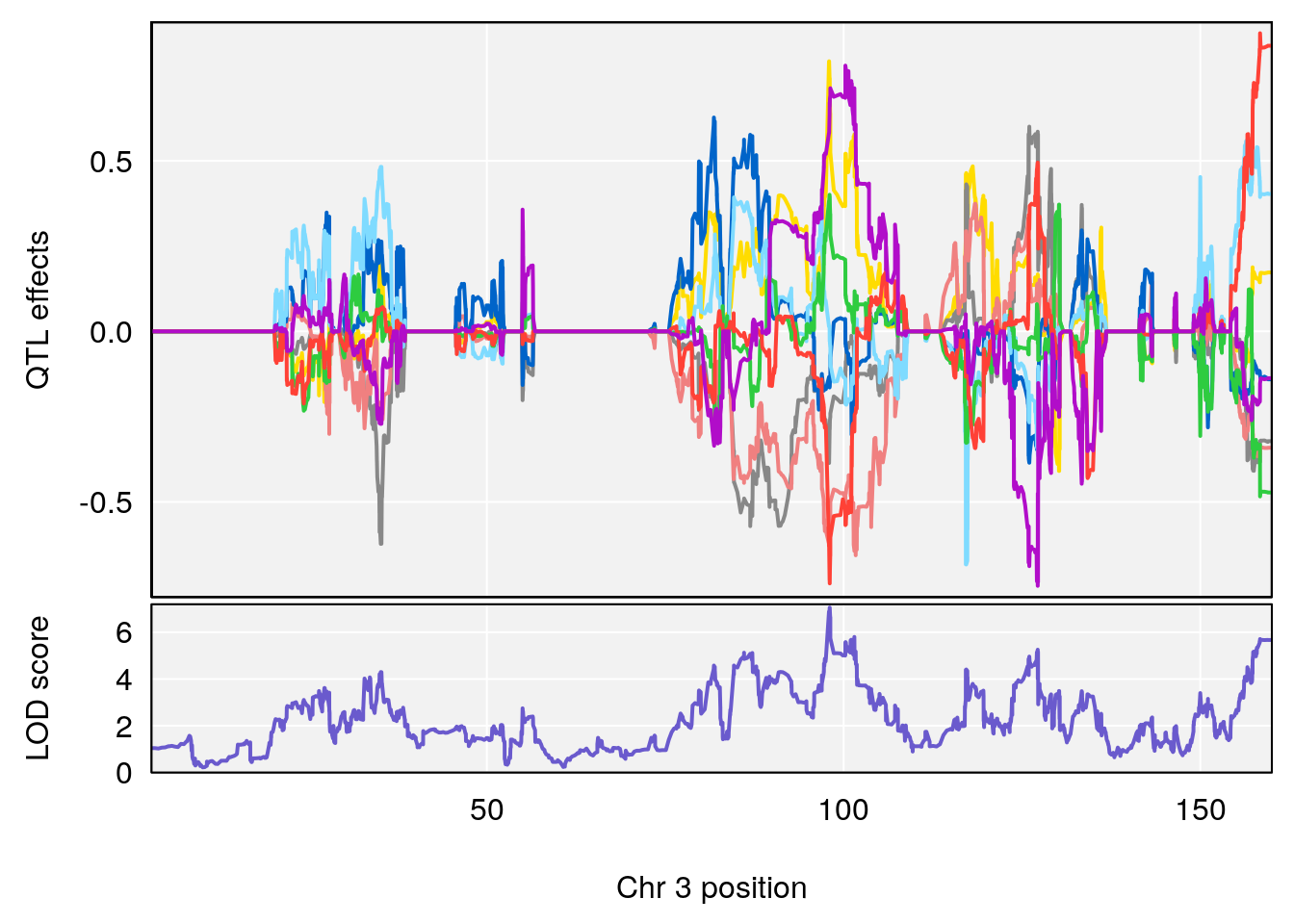
# 1 1 pheno1 3 98.04809 7.056873 97.51576 160.01710
# 2 1 pheno1 6 94.88068 8.514711 93.61447 95.01174
# [1] 1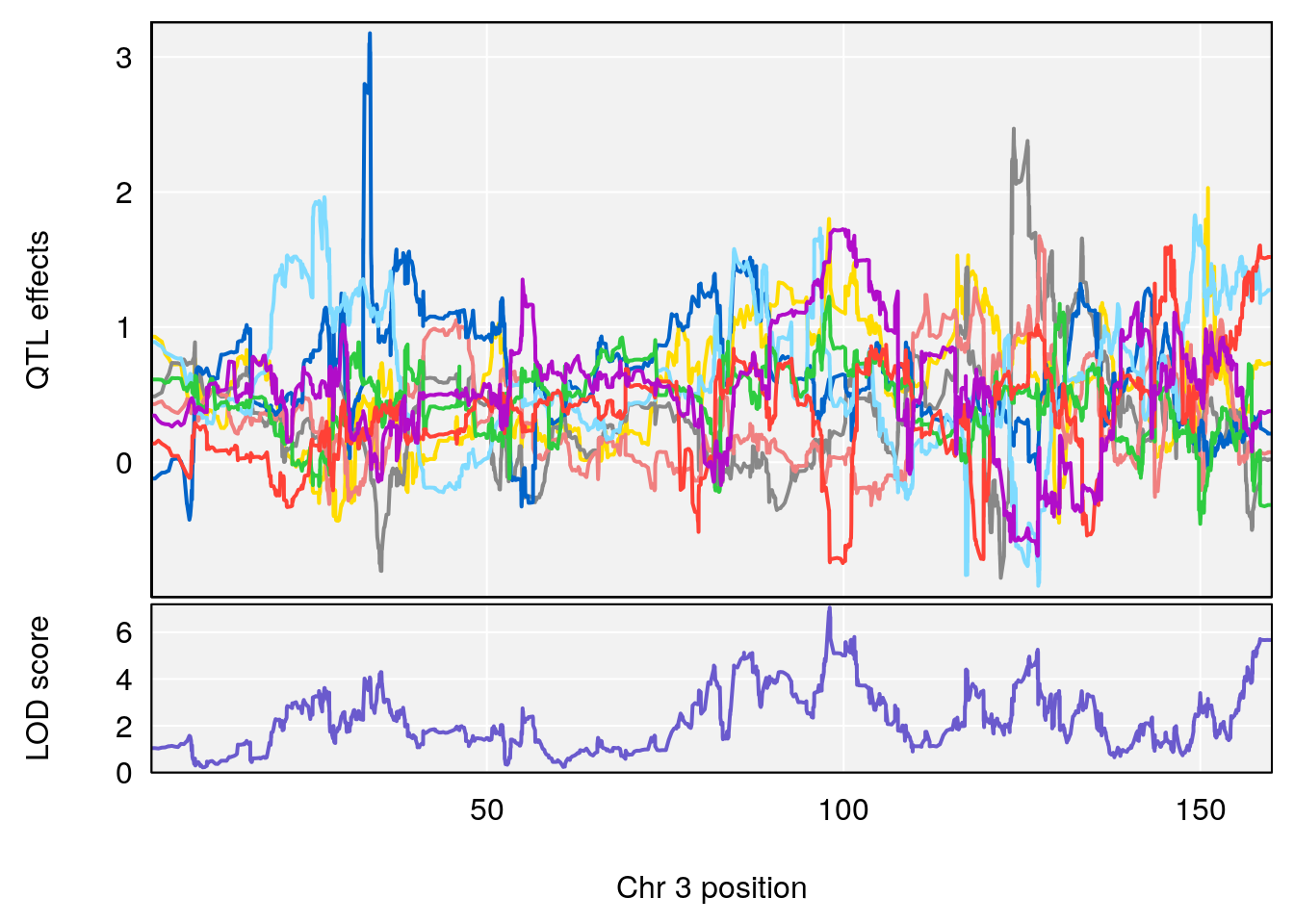
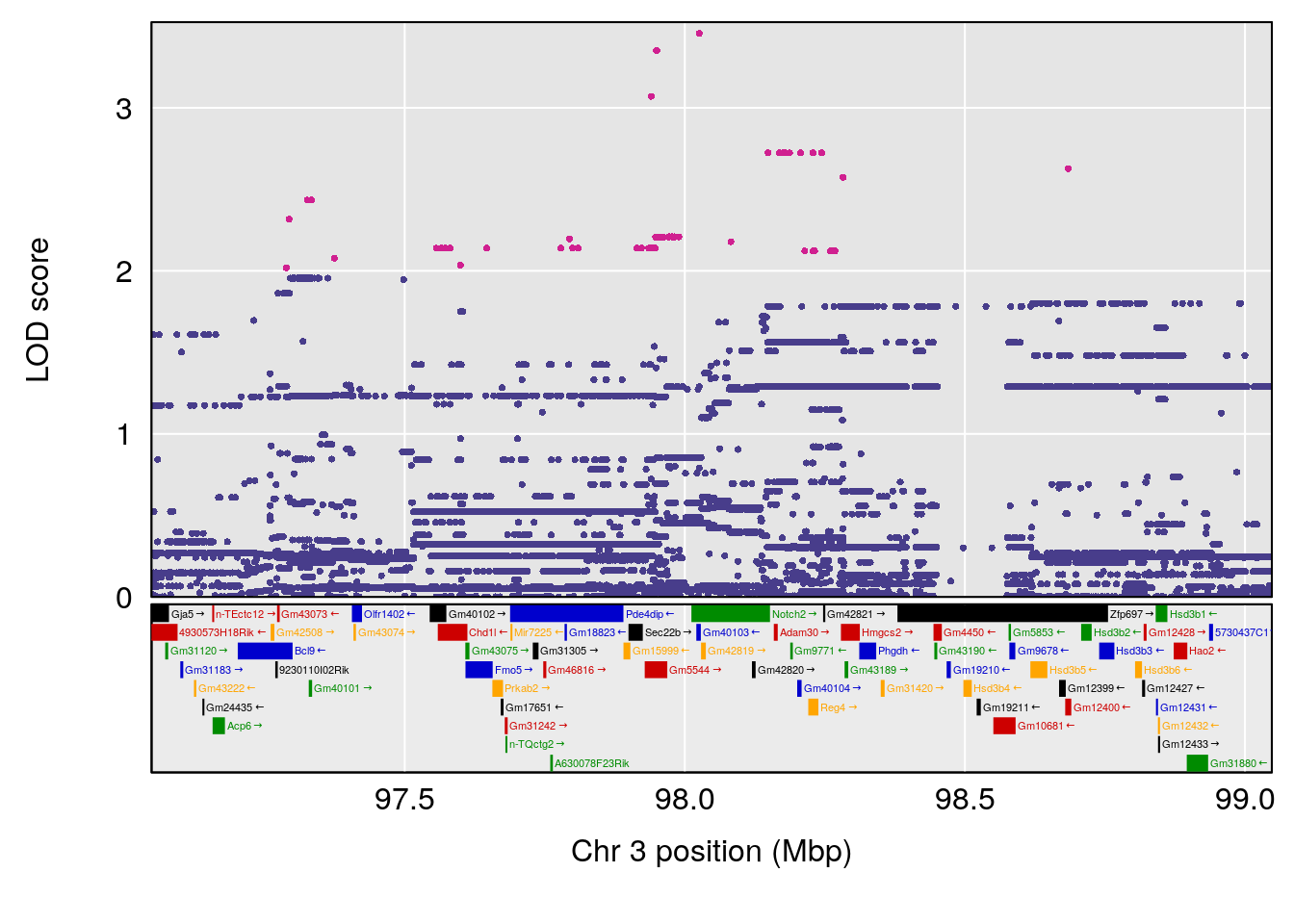
# [1] 2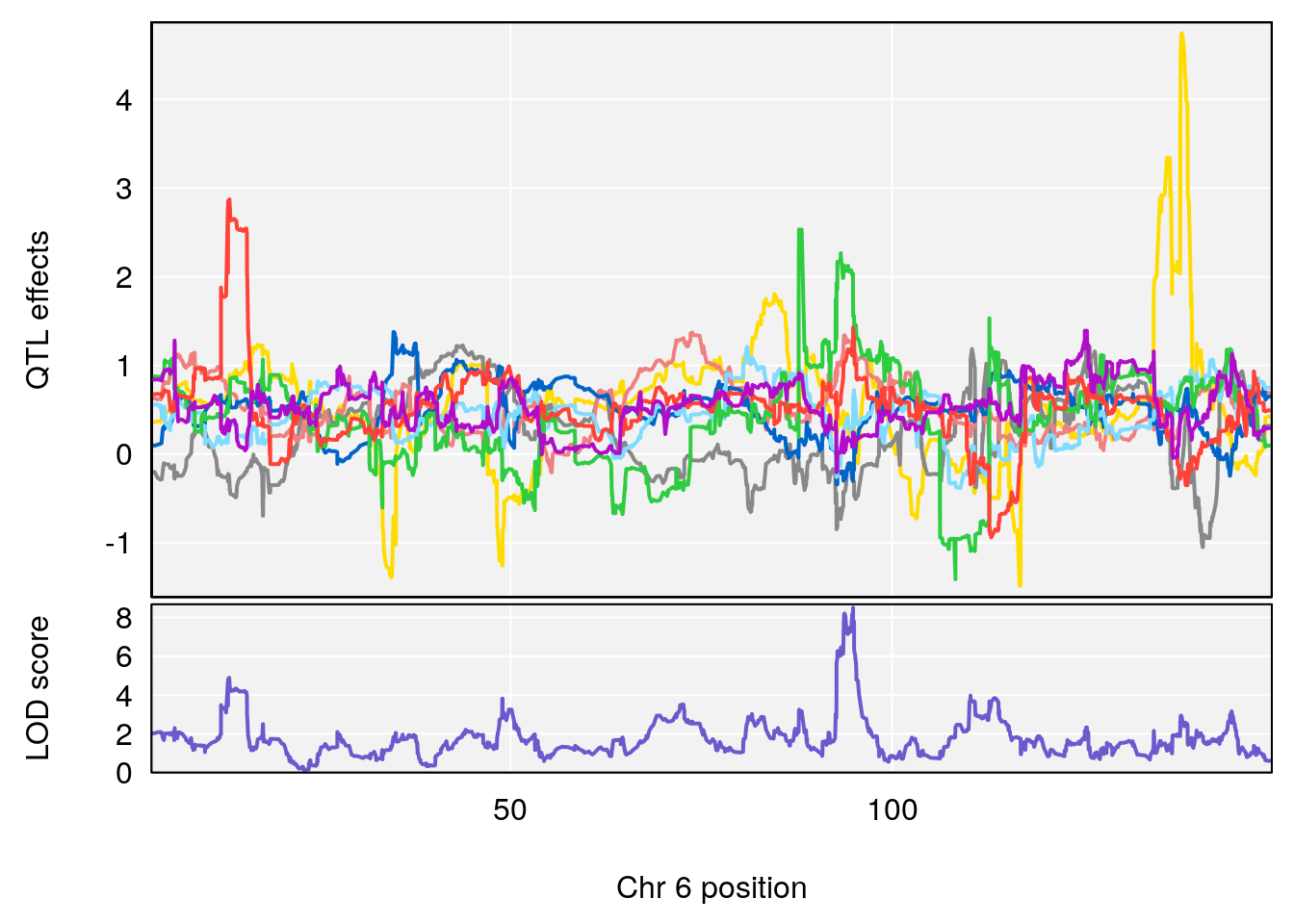
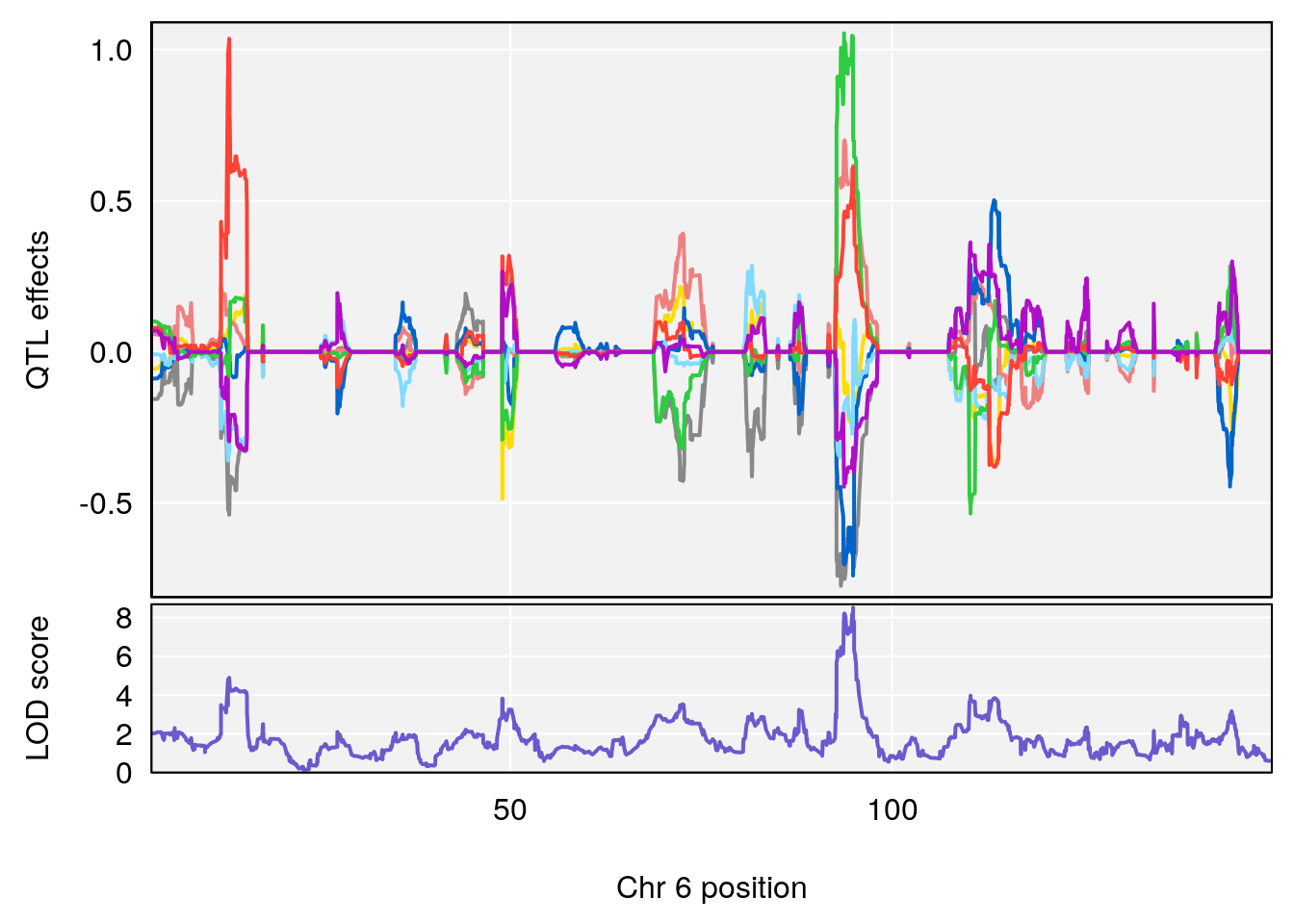
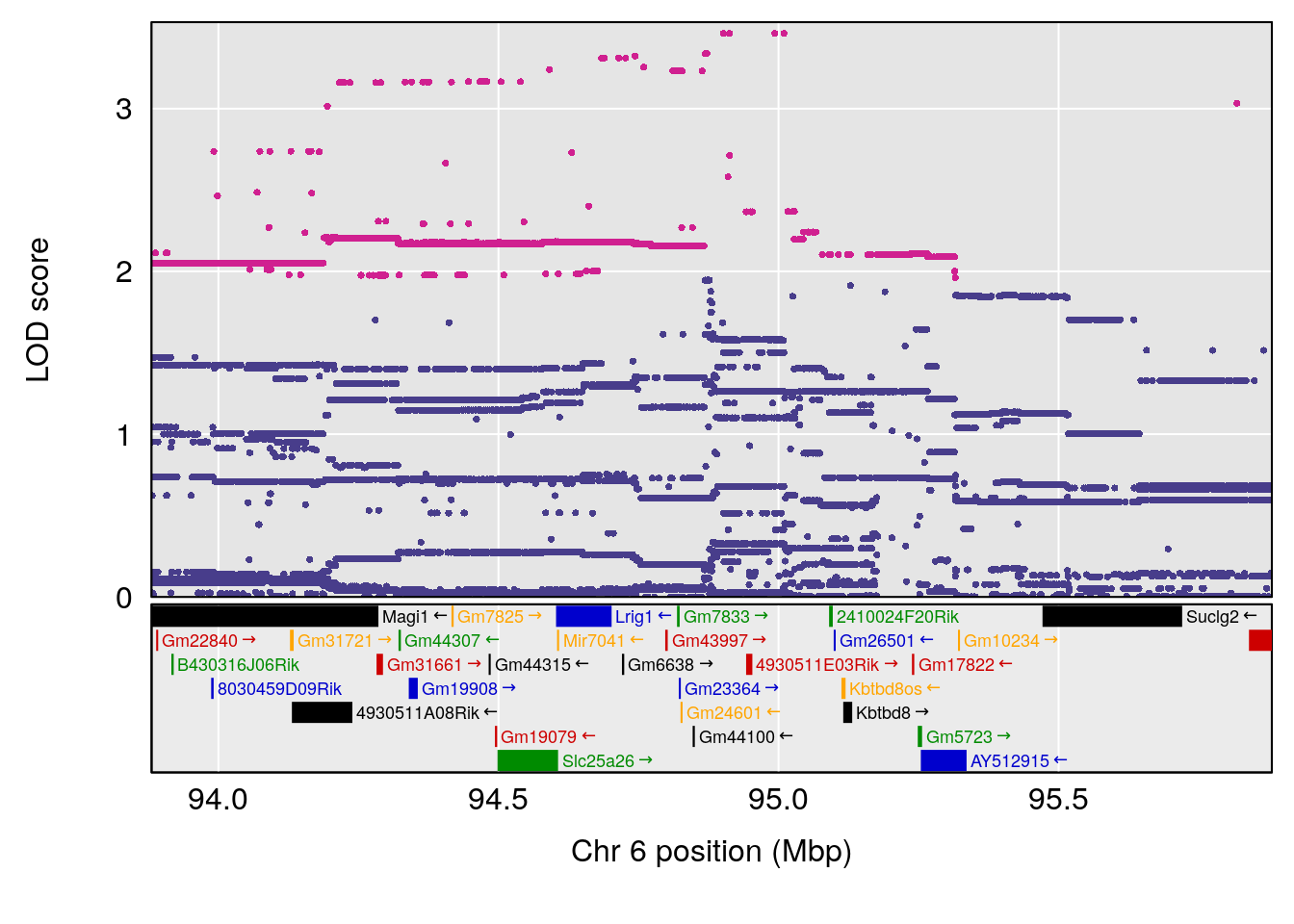
# [1] "Recovery.Time"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 59.37548 6.120300 59.01588 62.32984
# 2 1 pheno1 4 155.99563 6.203463 114.75702 156.13631
# 3 1 pheno1 8 117.09888 6.925422 115.52682 117.86788
# [1] 1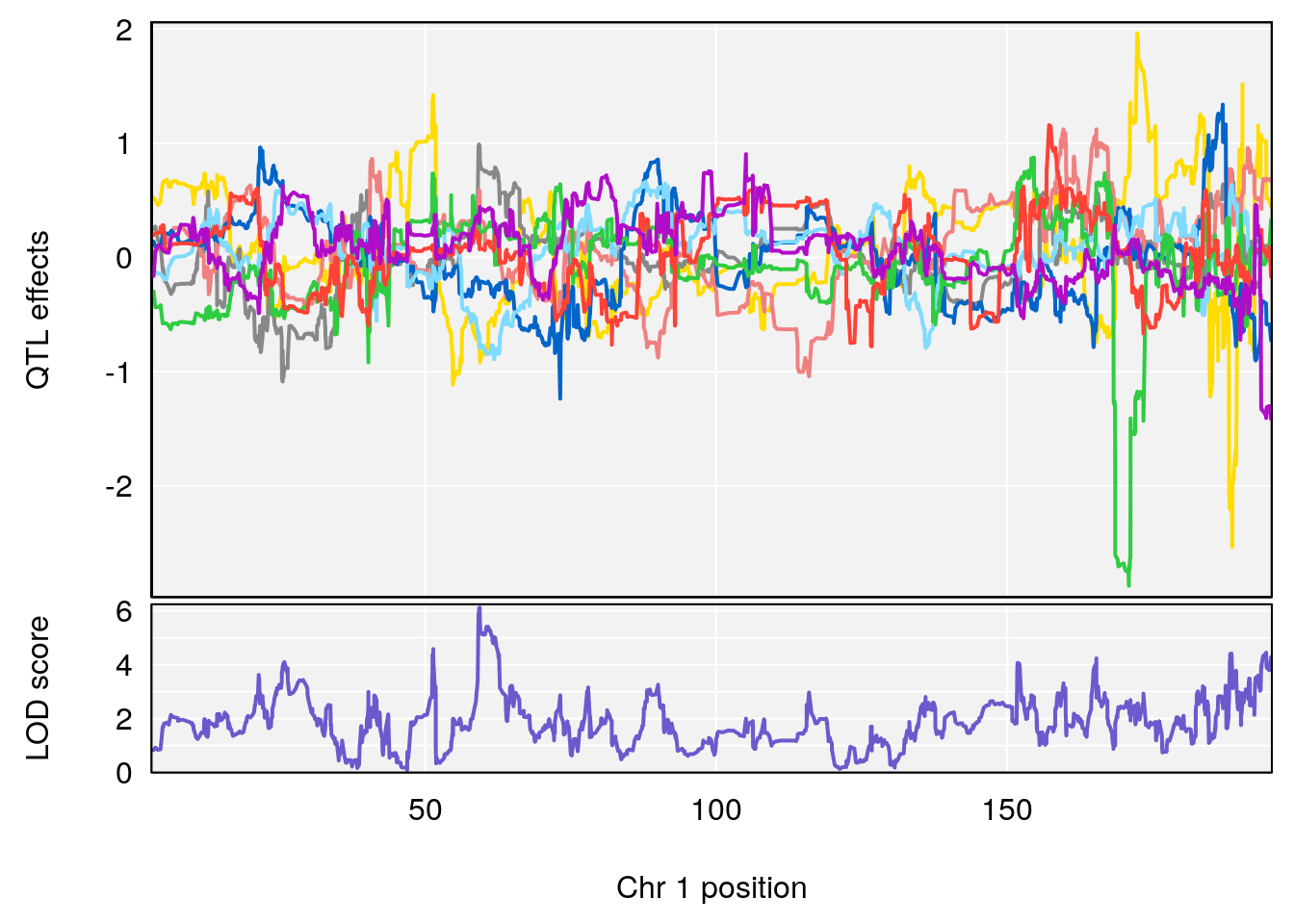
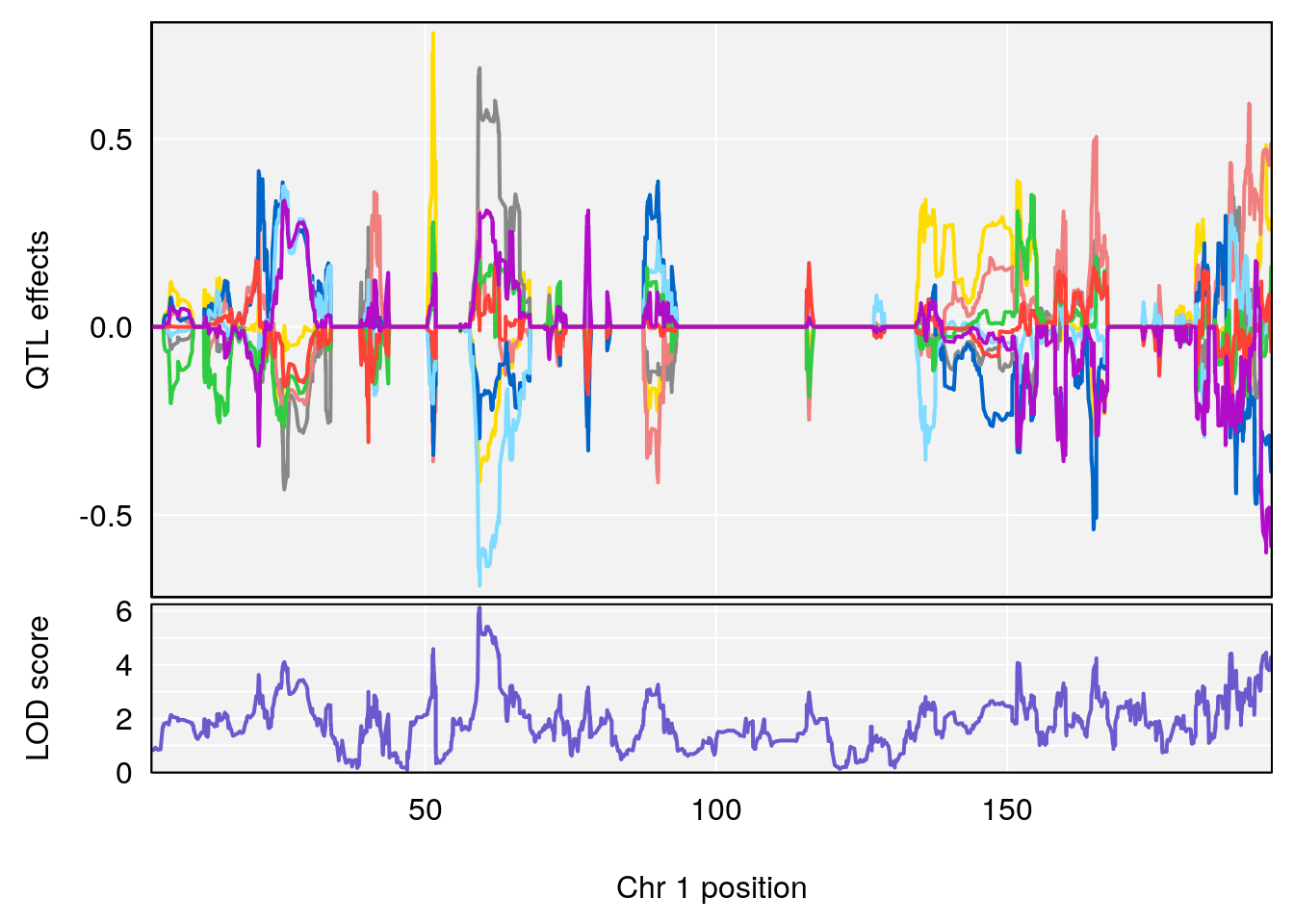
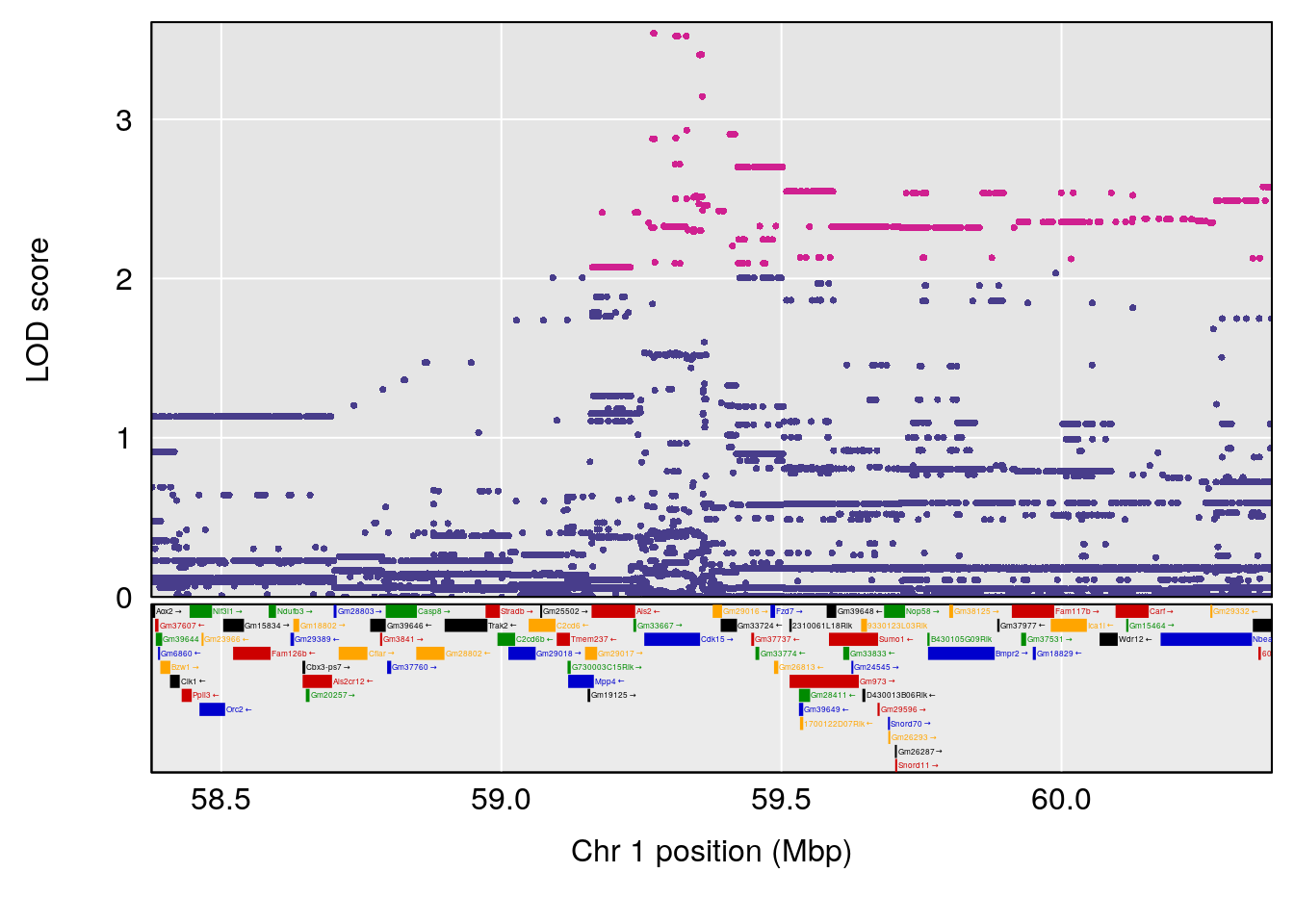
# [1] 2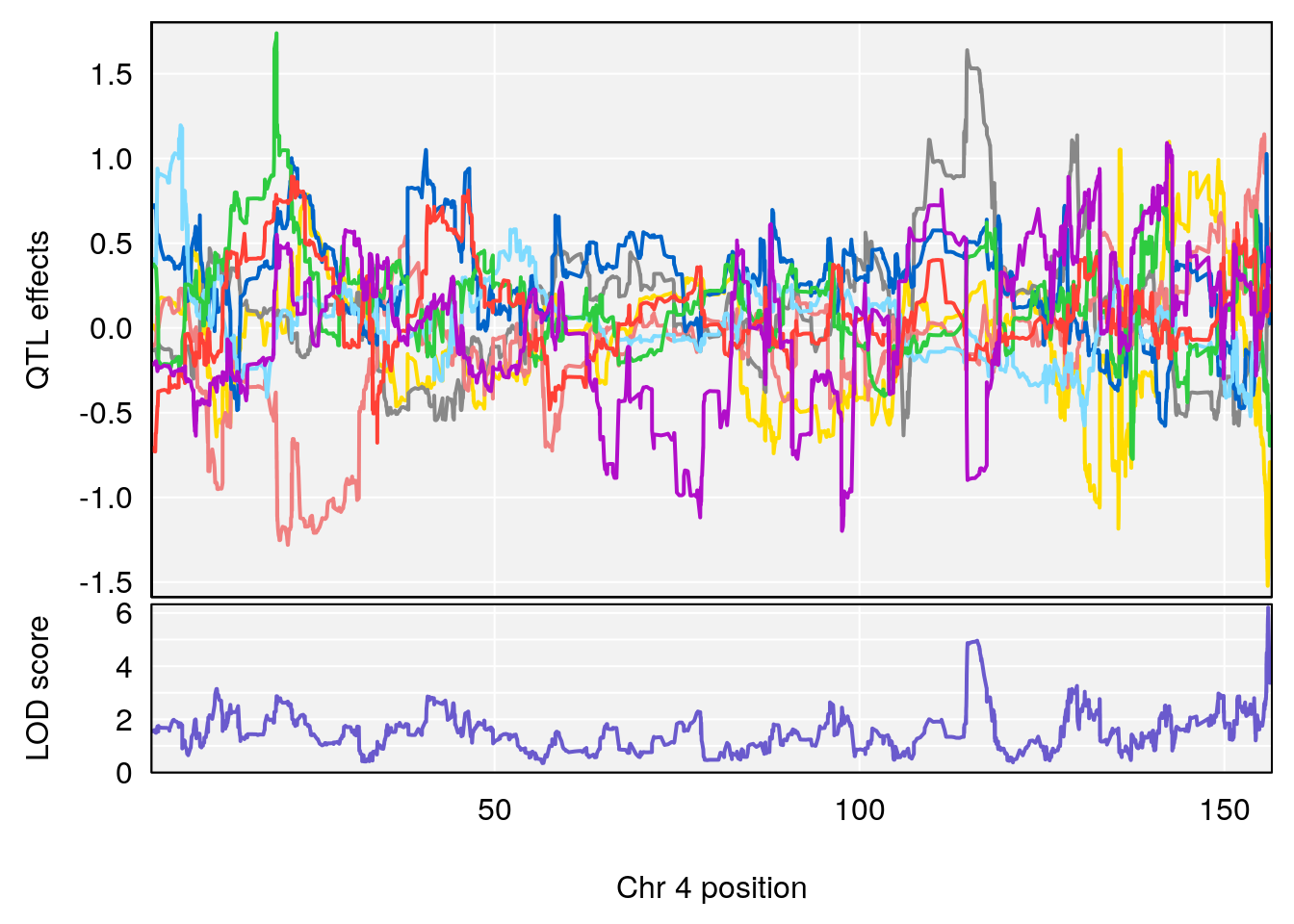
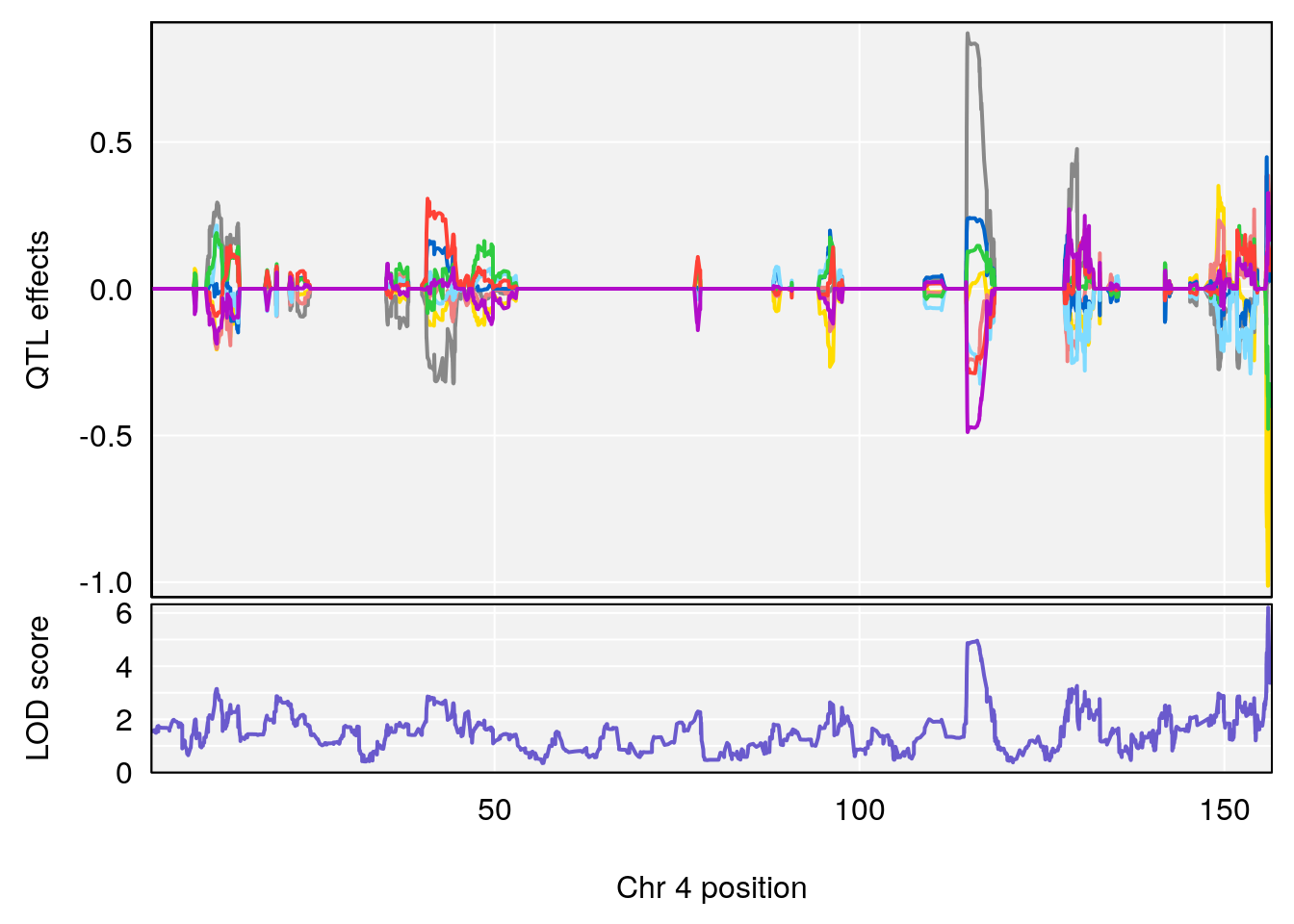
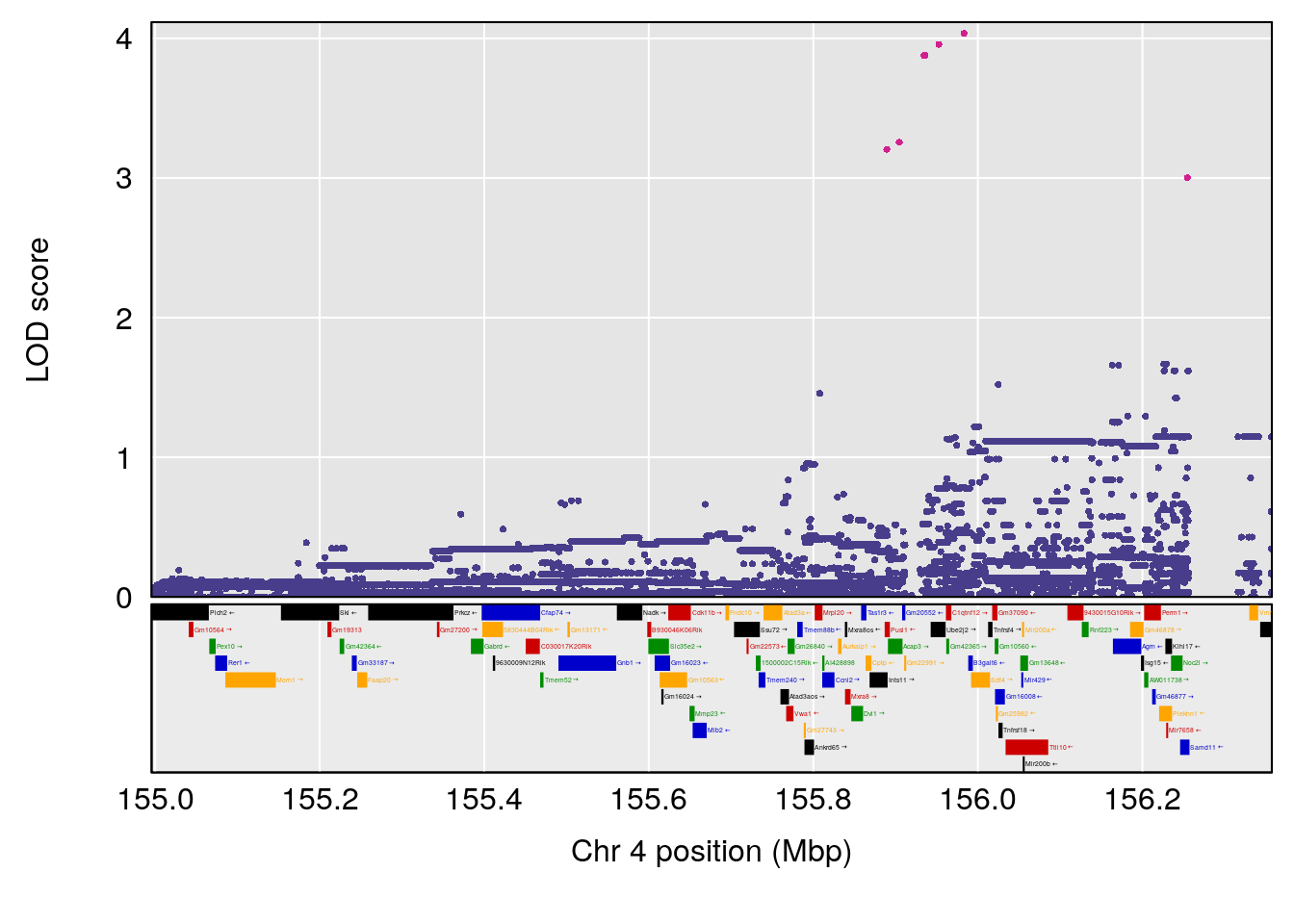
# [1] 3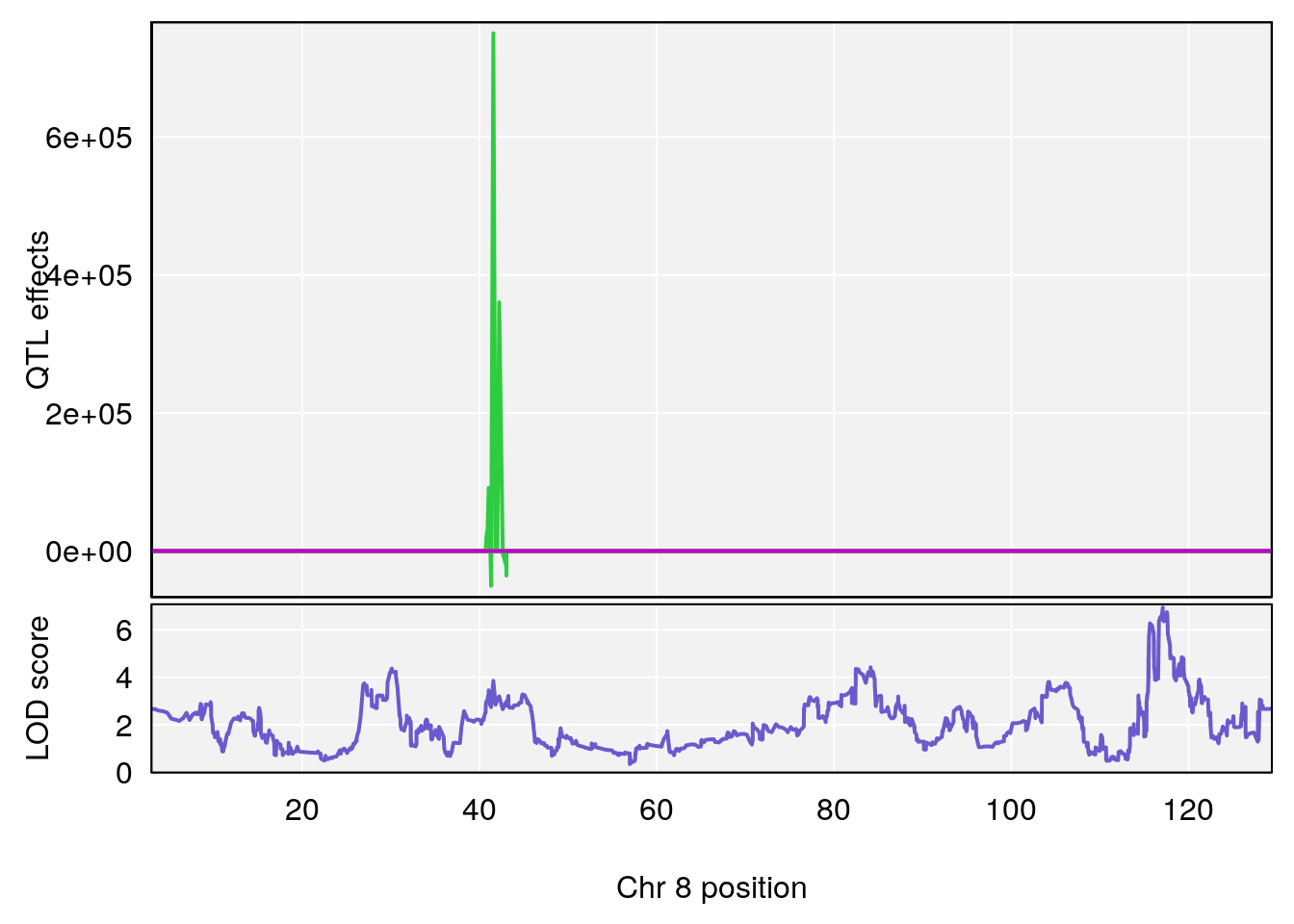
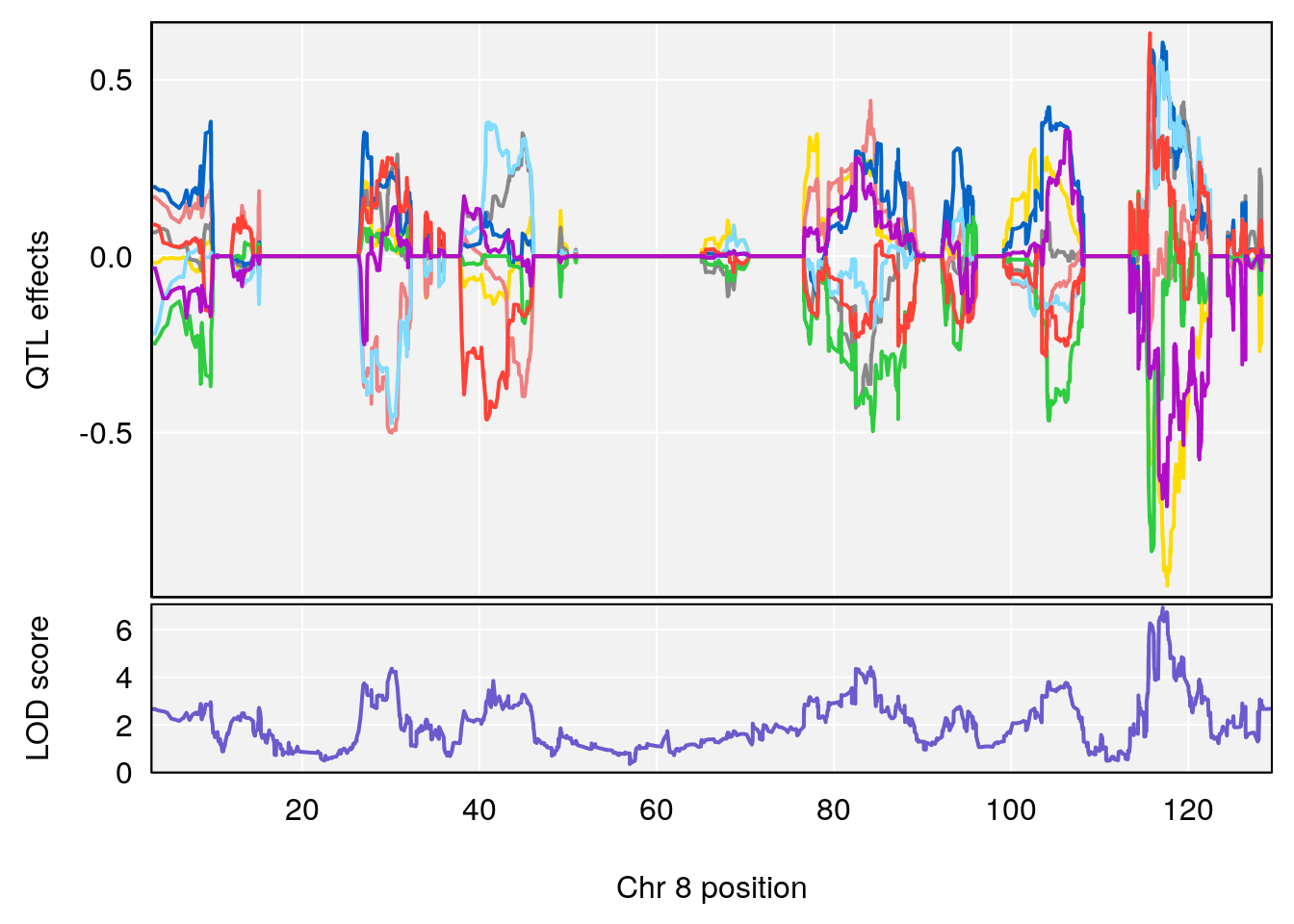
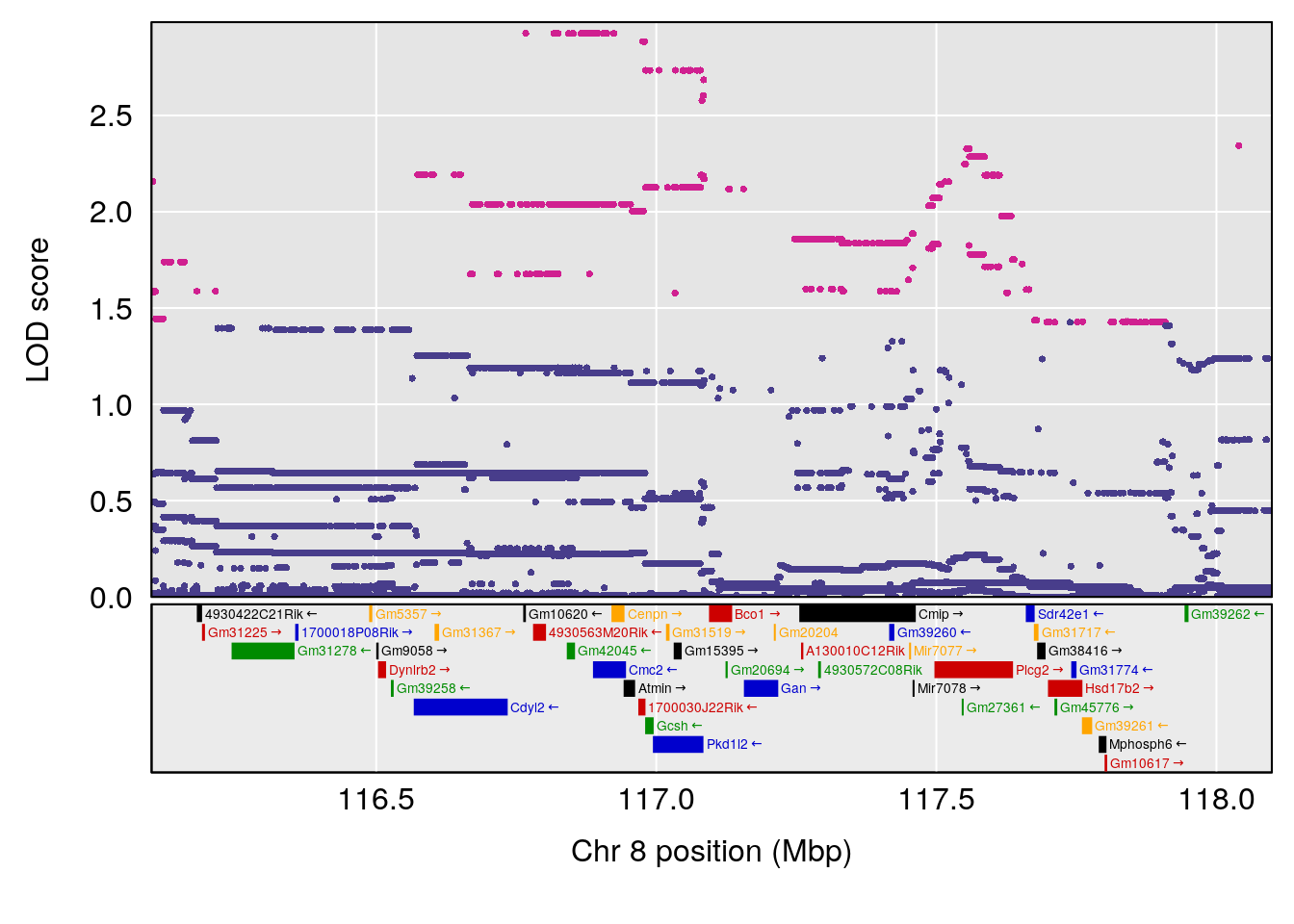
# [1] "Min.depression"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 6 94.17733 6.736136 93.600320 94.87681
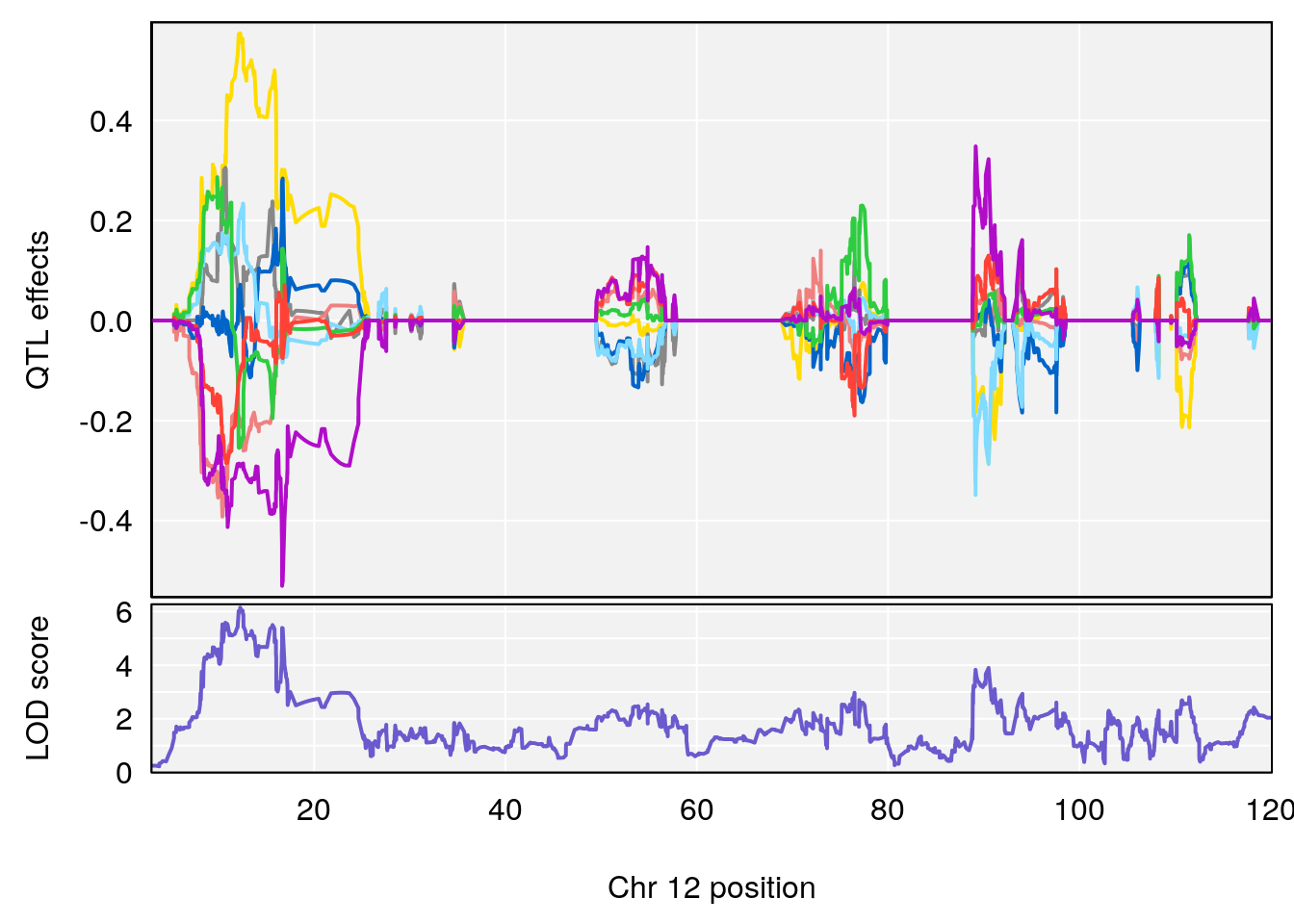
# 2 1 pheno1 12 12.26505 6.147825 9.396249 16.88098
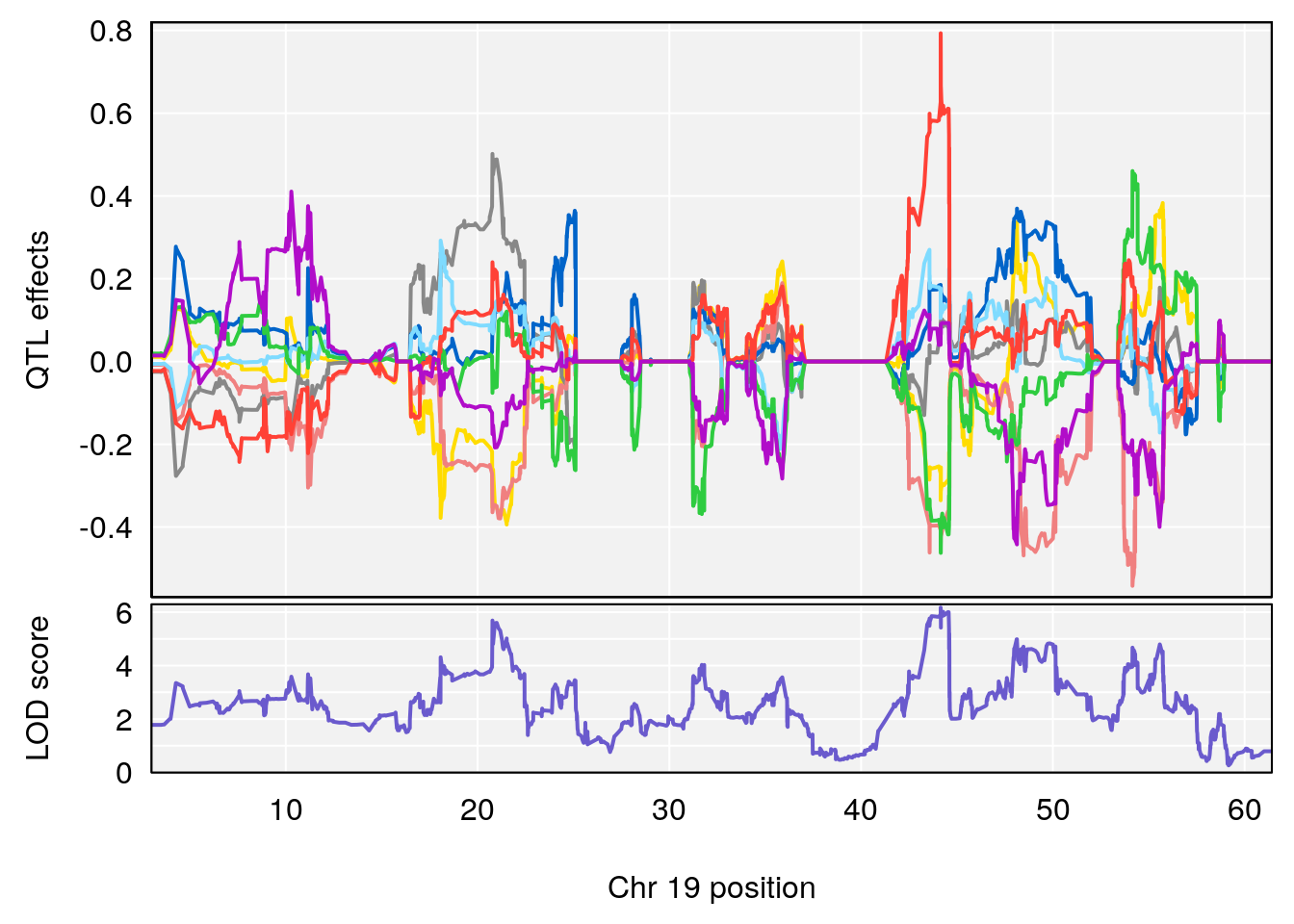
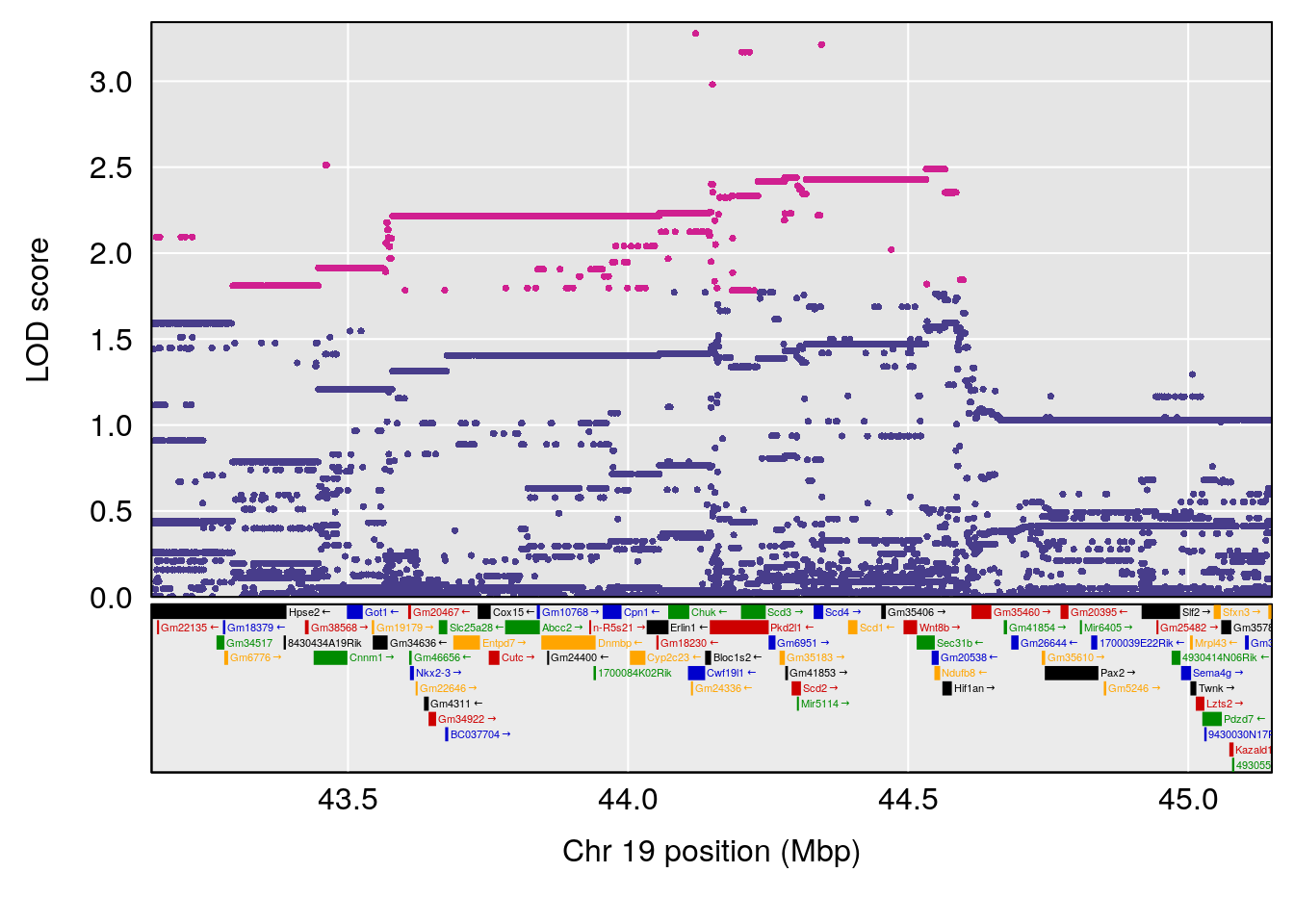
# 3 1 pheno1 19 44.14928 6.171079 20.756178 55.72002
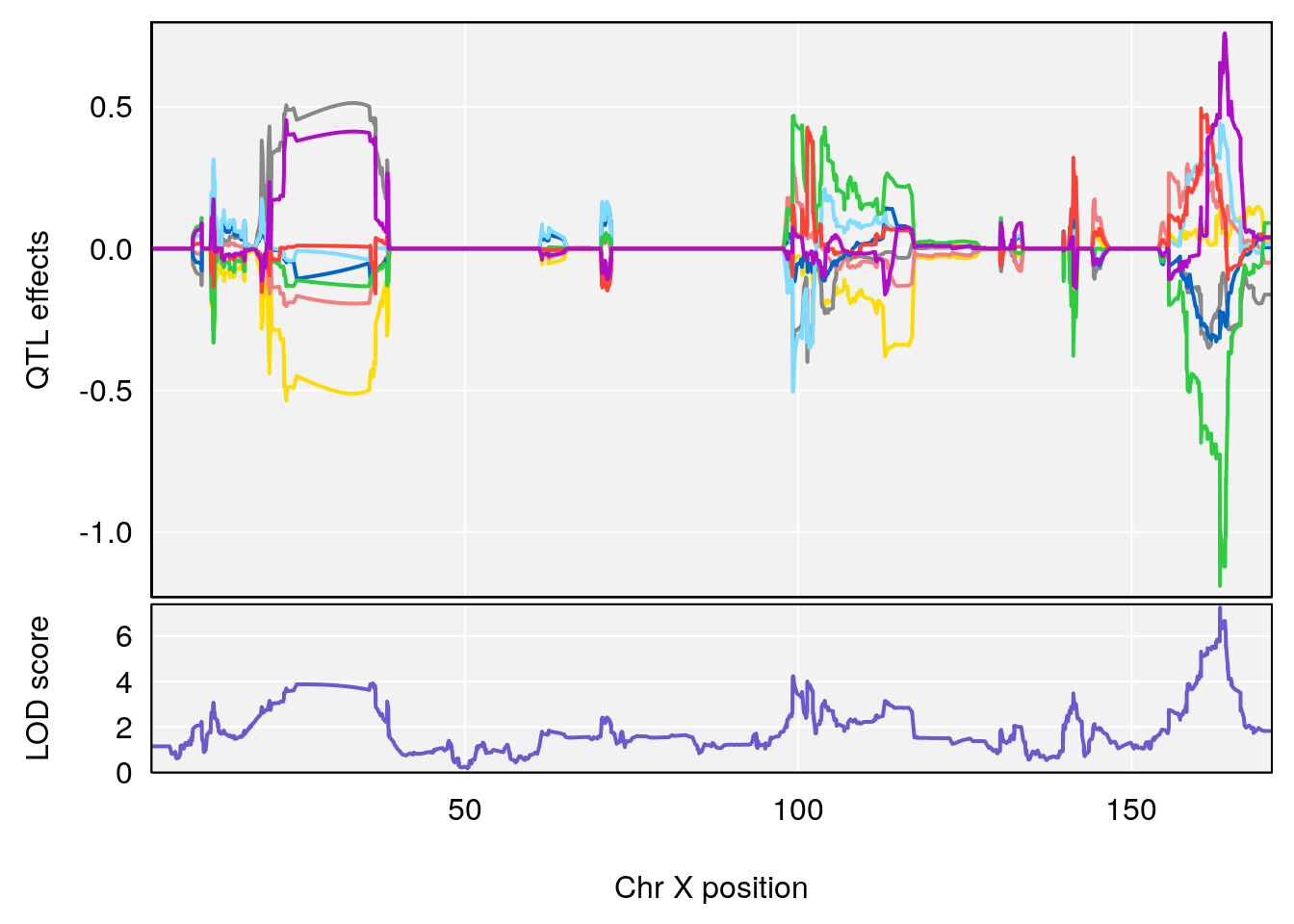
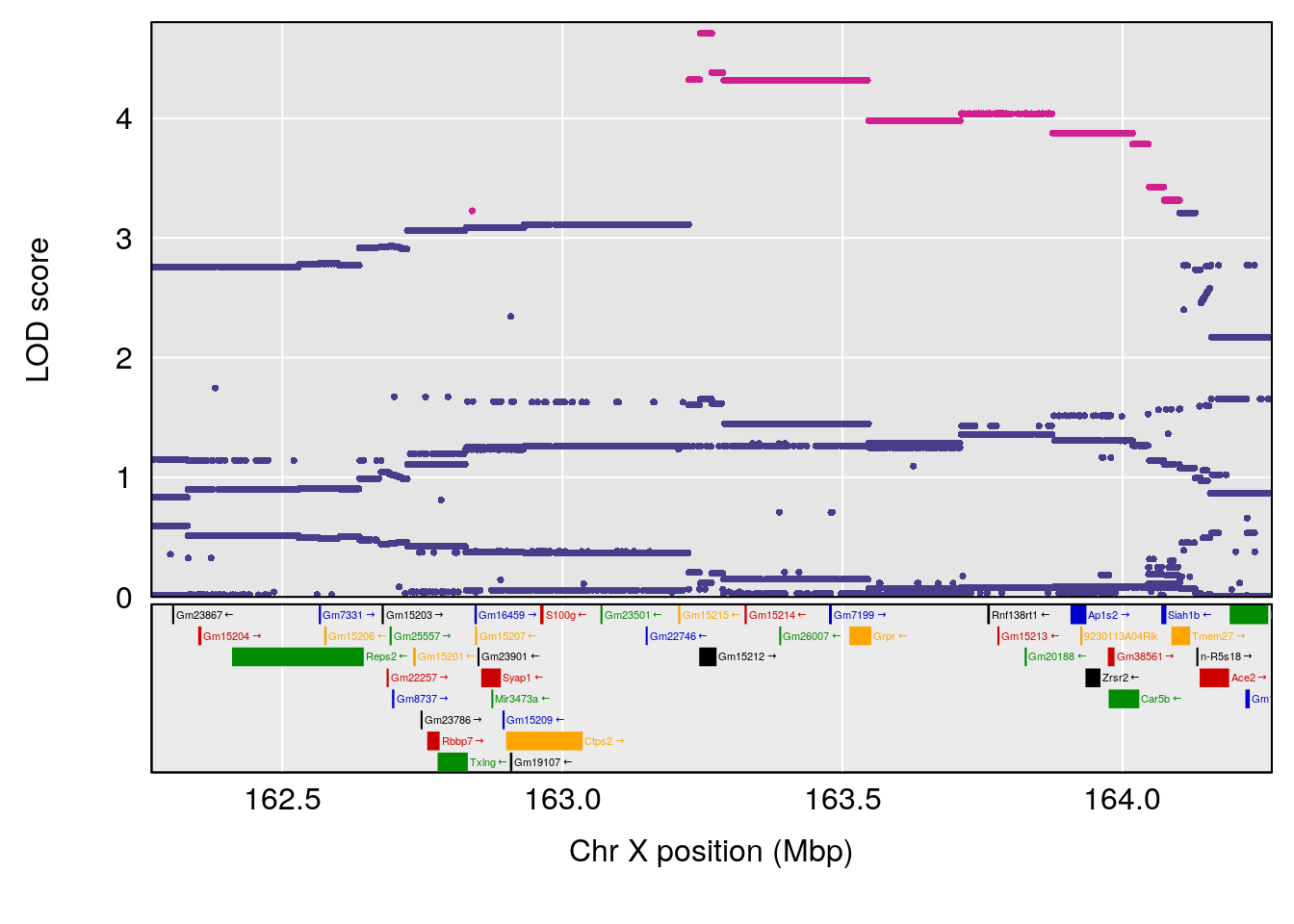
# 4 1 pheno1 X 163.26639 7.254464 162.637334 164.13916
# [1] 1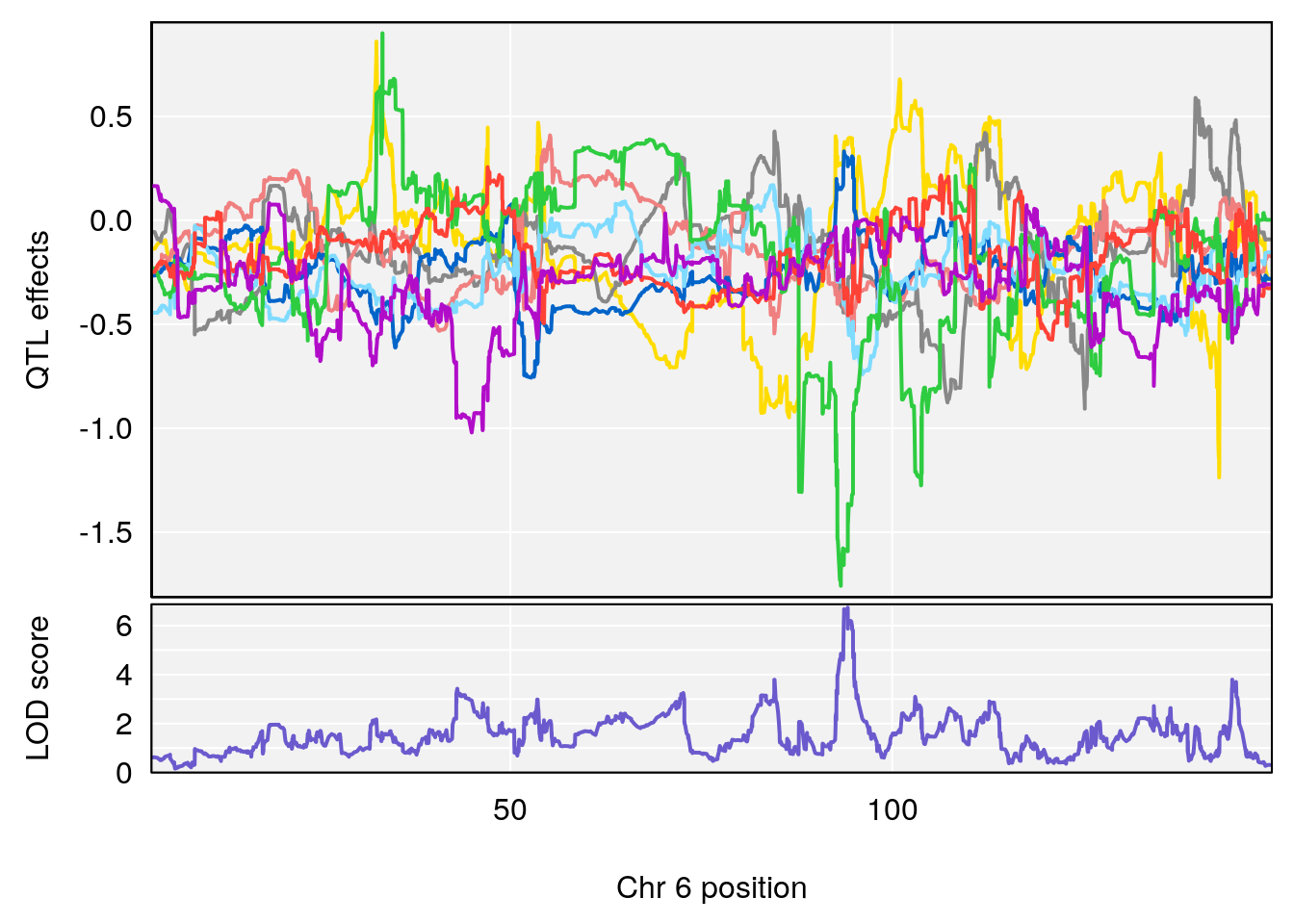
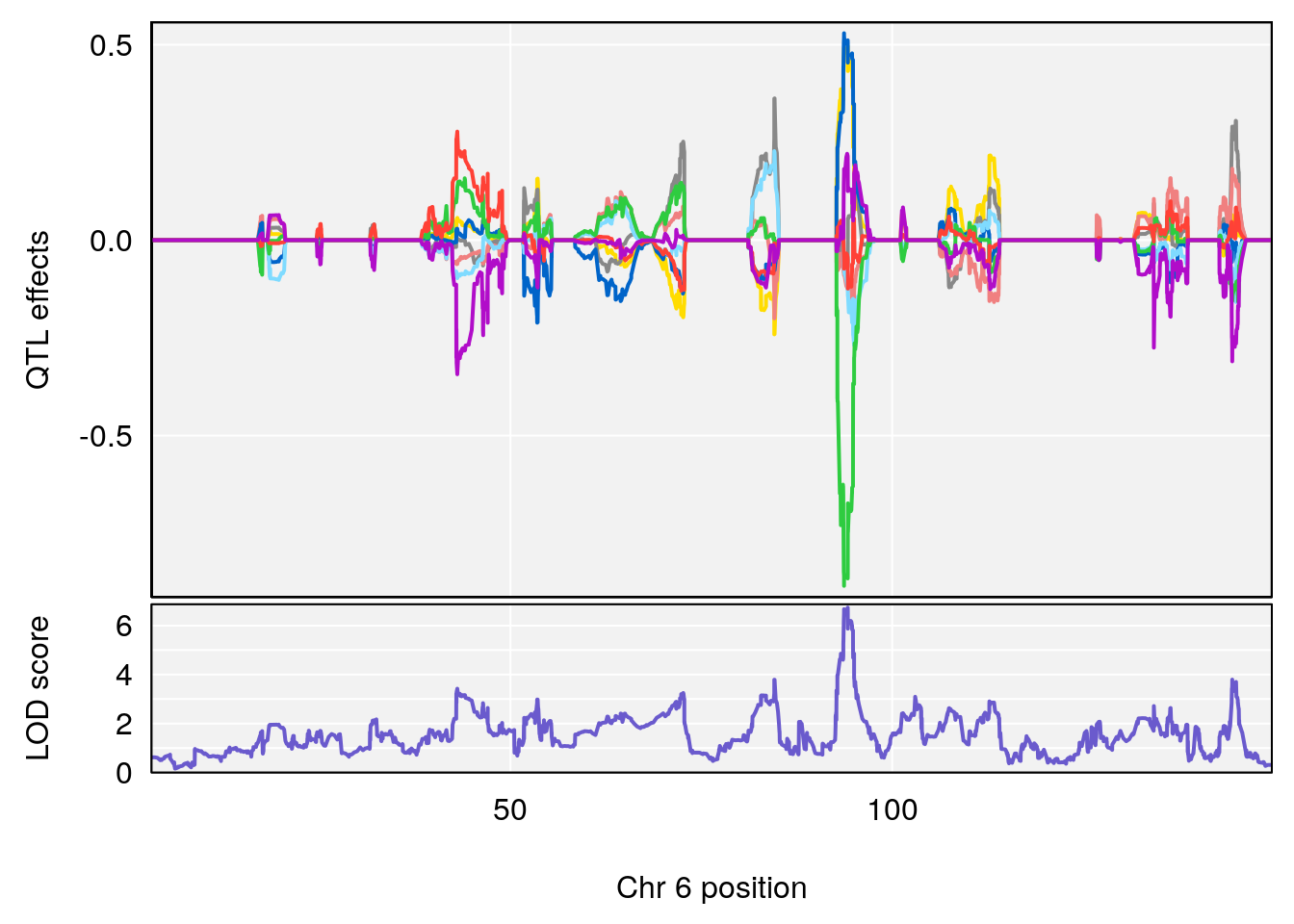
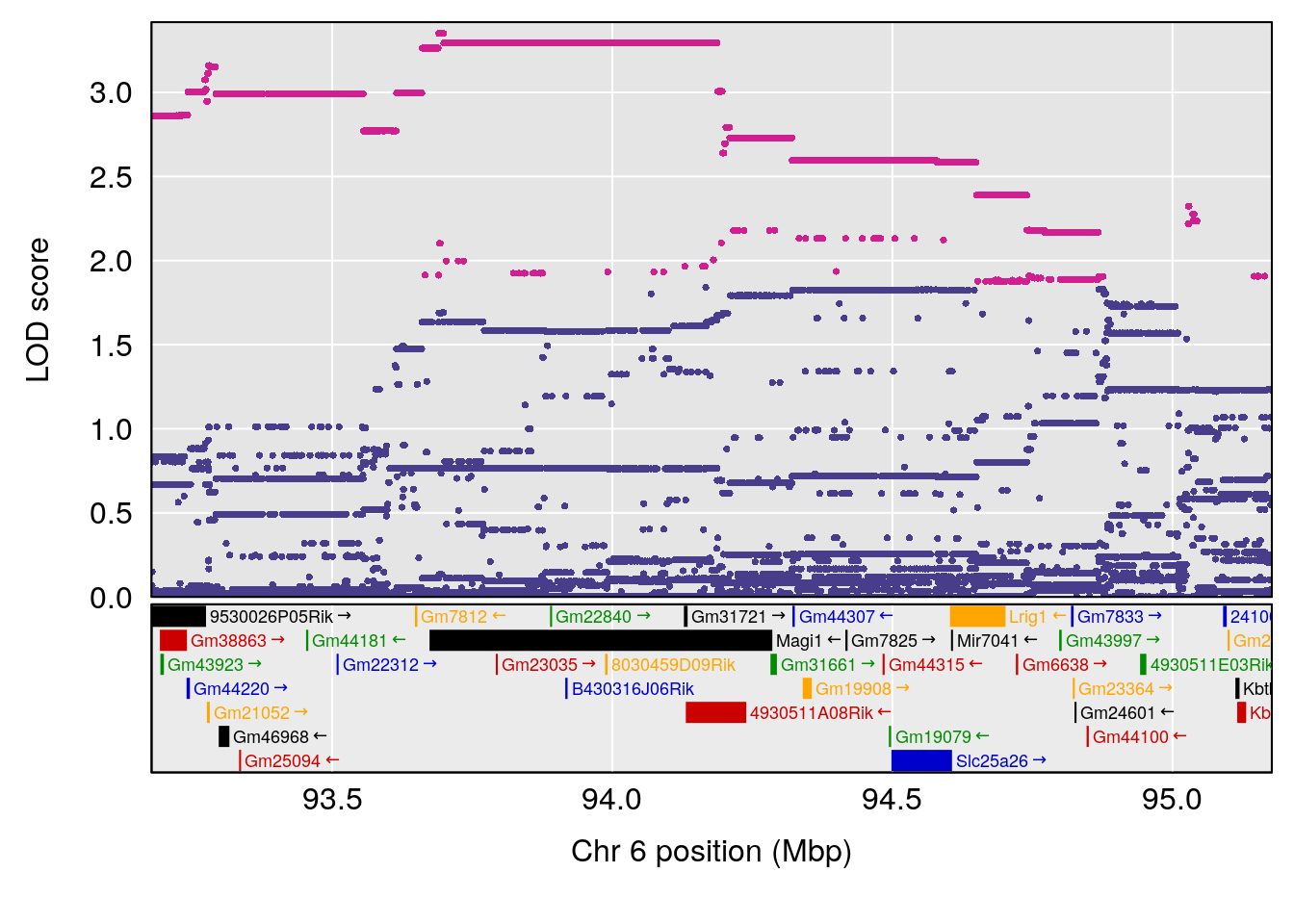
# [1] 2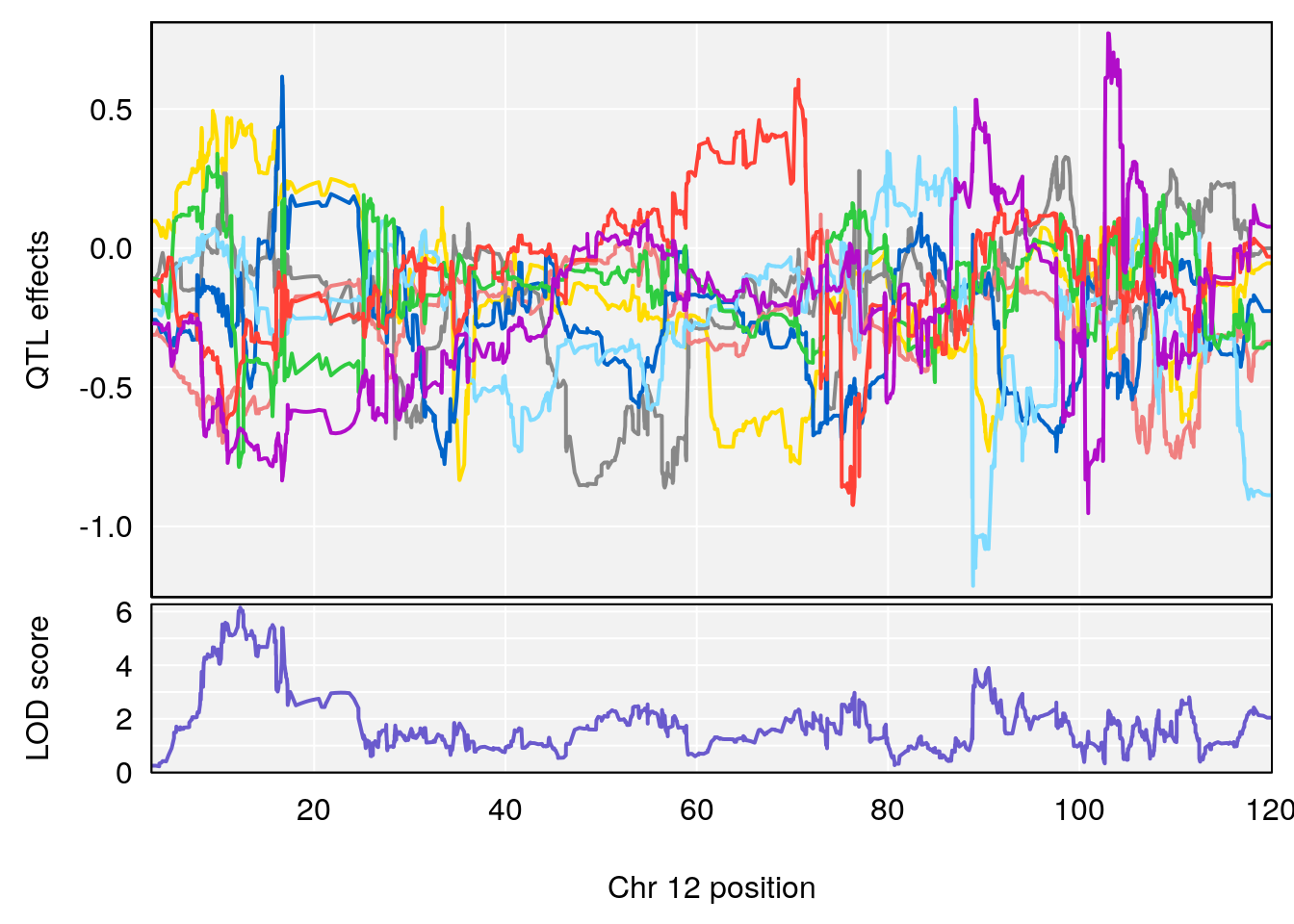
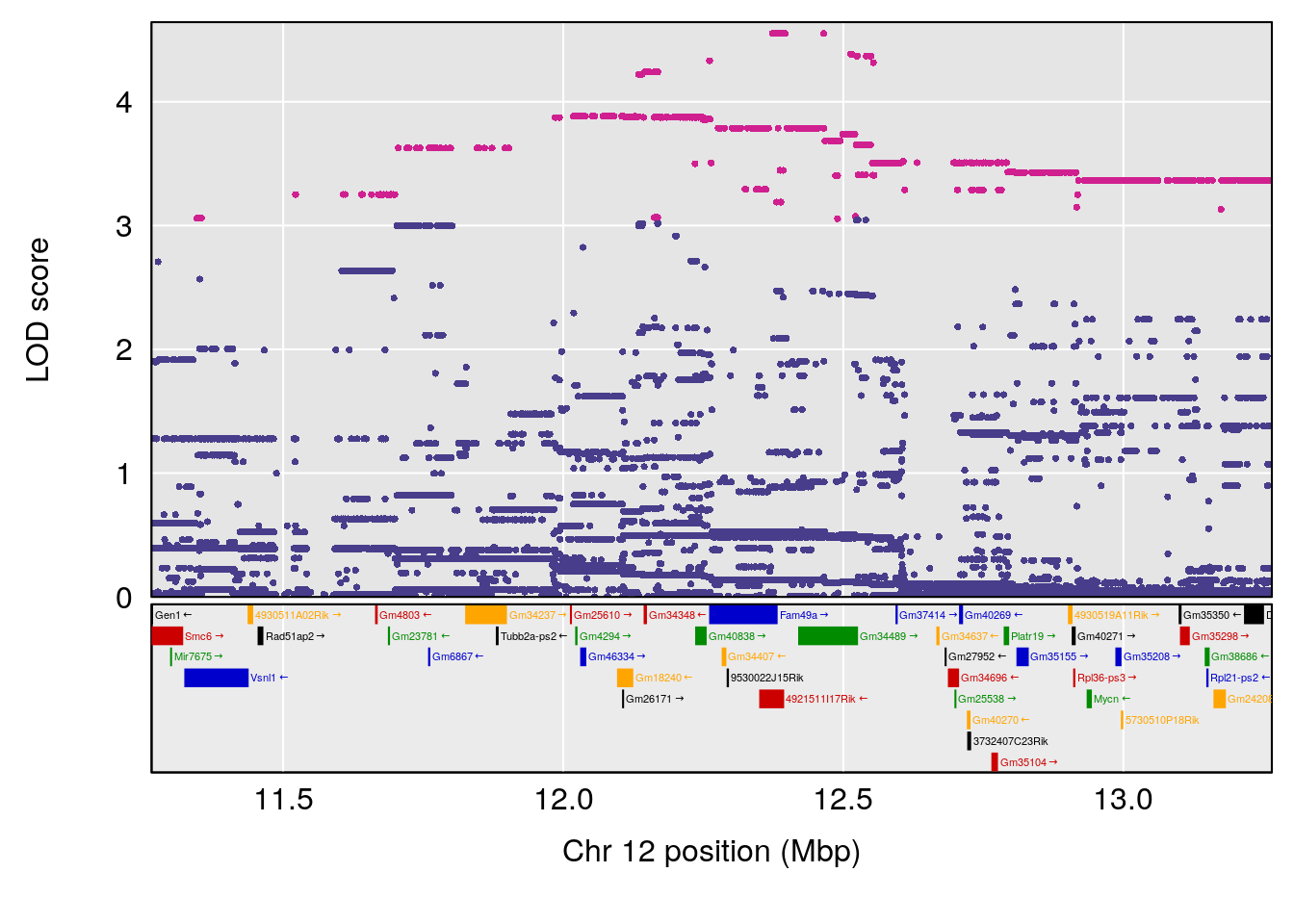
| Version | Author | Date |
|---|---|---|
| 9e4e3be | xhyuo | 2021-08-24 |
# [1] 3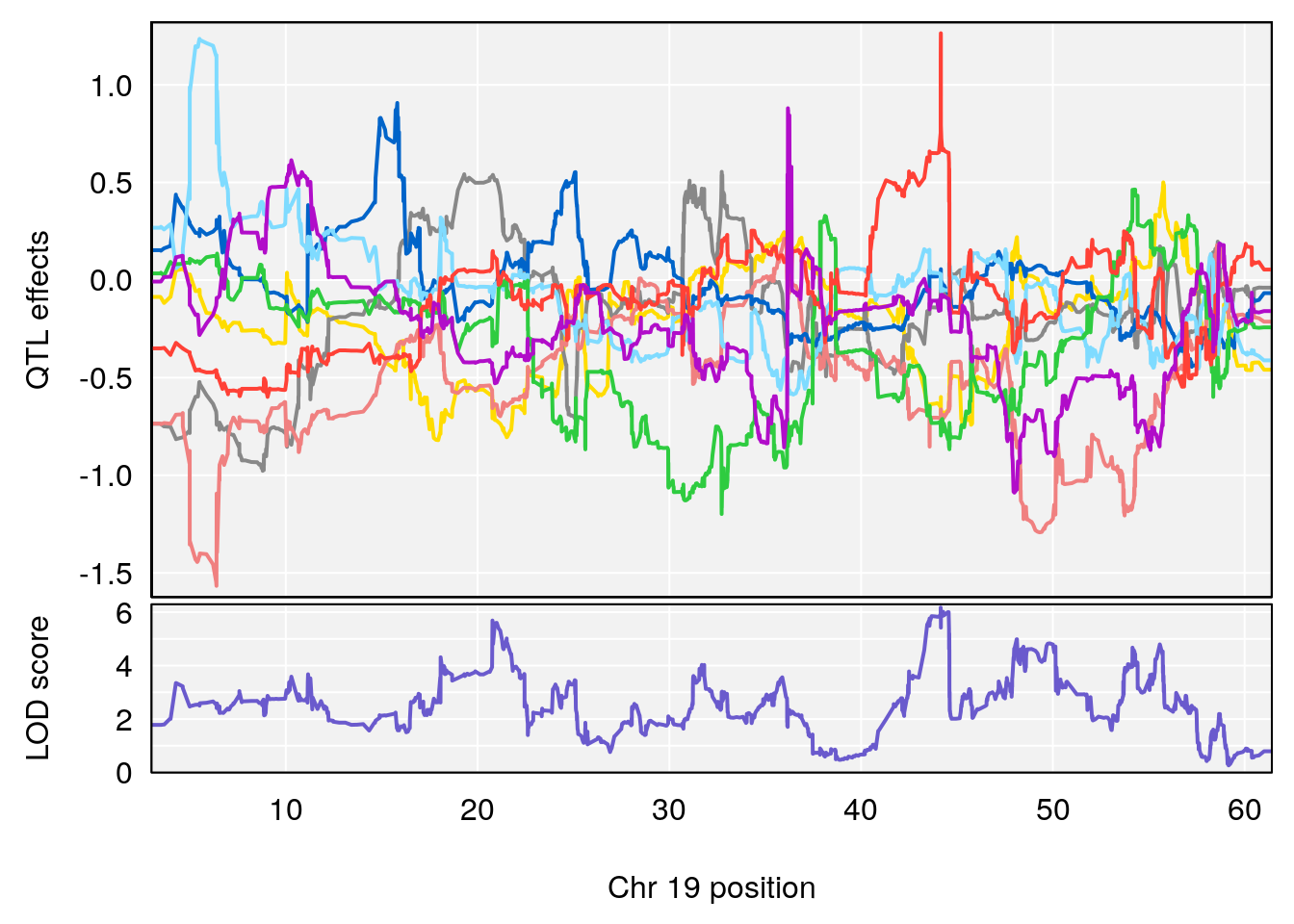
| Version | Author | Date |
|---|---|---|
| 9e4e3be | xhyuo | 2021-08-24 |
| Version | Author | Date |
|---|---|---|
| 9e4e3be | xhyuo | 2021-08-24 |
| Version | Author | Date |
|---|---|---|
| 9e4e3be | xhyuo | 2021-08-24 |
# [1] 4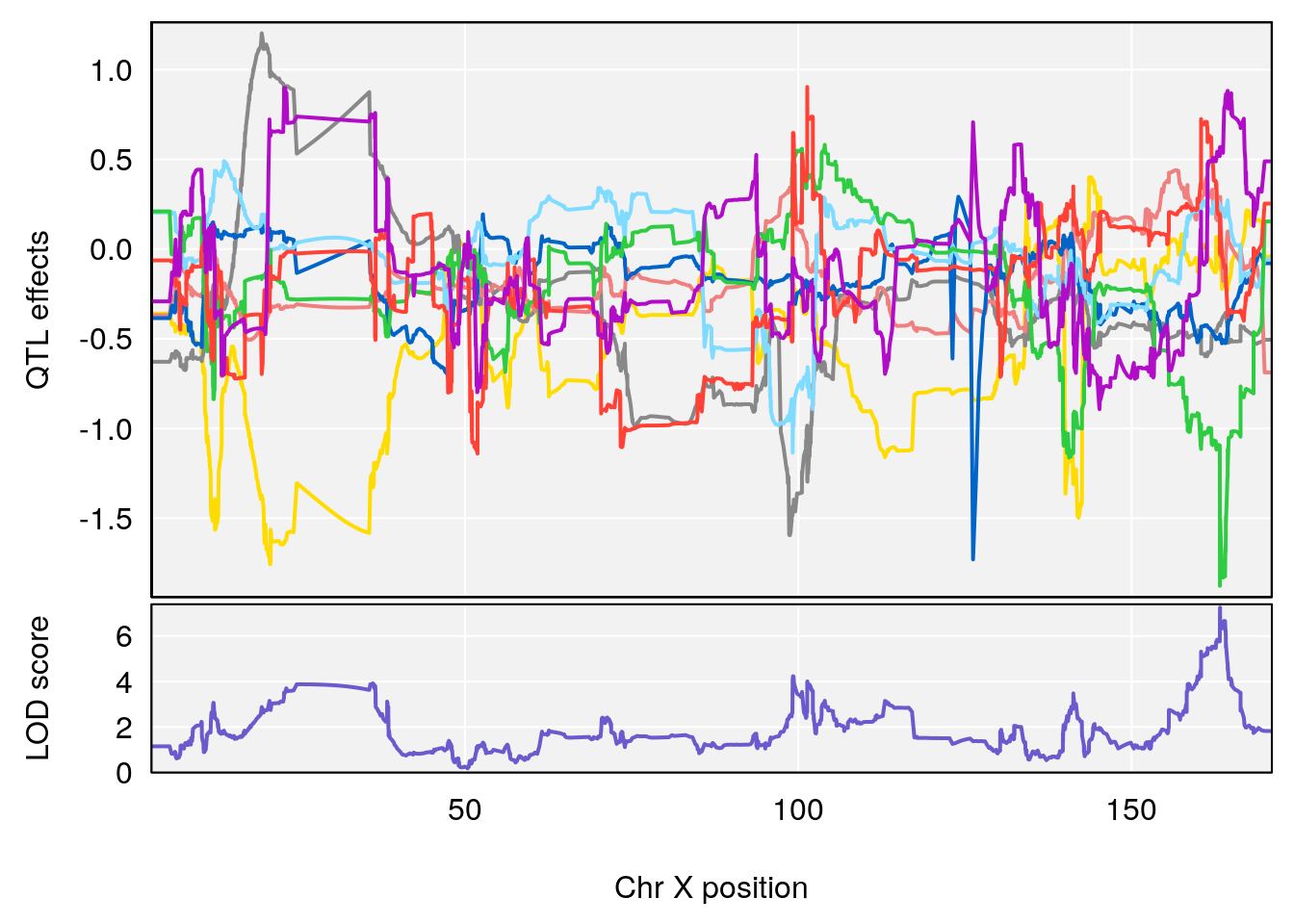
| Version | Author | Date |
|---|---|---|
| 9e4e3be | xhyuo | 2021-08-24 |
| Version | Author | Date |
|---|---|---|
| 9e4e3be | xhyuo | 2021-08-24 |
| Version | Author | Date |
|---|---|---|
| 9e4e3be | xhyuo | 2021-08-24 |
# [1] "Status_bin"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 9 68.16325 6.081293 43.77453 123.6993
# [1] 1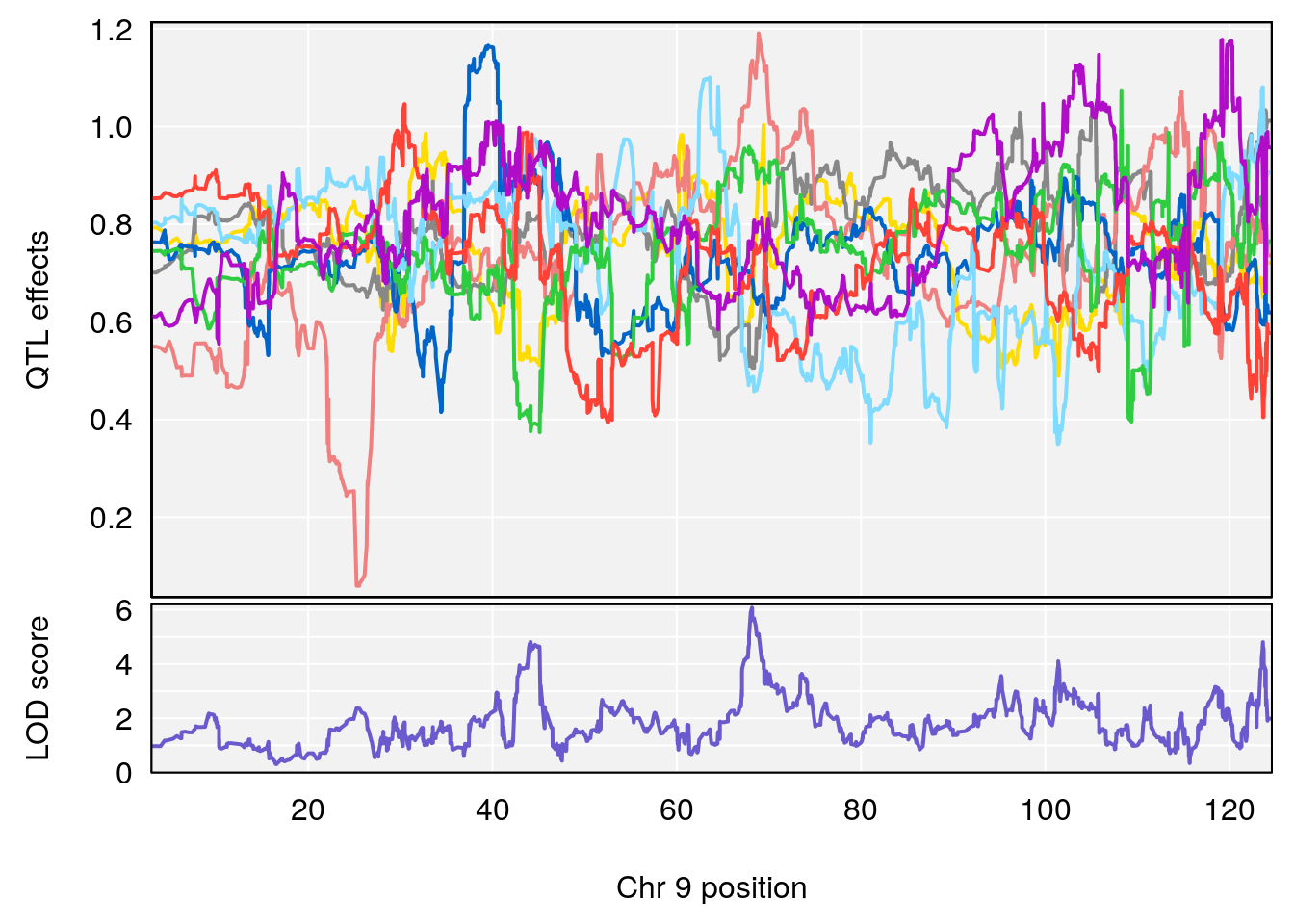
| Version | Author | Date |
|---|---|---|
| 9e4e3be | xhyuo | 2021-08-24 |
| Version | Author | Date |
|---|---|---|
| 9e4e3be | xhyuo | 2021-08-24 |
| Version | Author | Date |
|---|---|---|
| 9e4e3be | xhyuo | 2021-08-24 |
#save peaks coeff plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
fname <- paste("output/DO_morphine_secondbatch_69k_",i,"_coefplot.pdf",sep="")
pdf(file = fname, width = 16, height =8)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
dev.off()
}
# [1] "Survival.Time"
# [1] 1
# [1] 2
# [1] "Recovery.Time"
# [1] 1
# [1] 2
# [1] 3
# [1] "Min.depression"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] "Status_bin"
# [1] 1
#save peaks coeff blup plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
fname <- paste("output/DO_morphine_secondbatch_69k_",i,"_coefplot_blup.pdf",sep="")
pdf(file = fname, width = 16, height =8)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
dev.off()
}
# [1] "Survival.Time"
# [1] 1
# [1] 2
# [1] "Recovery.Time"
# [1] 1
# [1] 2
# [1] 3
# [1] "Min.depression"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] "Status_bin"
# [1] 1Plot qtl mapping on array male
load("output/qtl.morphine.out.second_set.male.RData")
#pmap and gmap
chr.names <- c("1","2","3","4","5","6","7","8","9","10","11","12","13","14","15","16","17","18","19", "X")
gmap <- list()
pmap <- list()
for (chr in chr.names){
gmap[[chr]] <- do.morphine$gmap[[chr]]
pmap[[chr]] <- do.morphine$pmap[[chr]]
}
attr(gmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
attr(pmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
#genome-wide plot
for(i in names(qtl.morphine.out)){
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.morphine.out[[i]]) # overall maximum LOD score
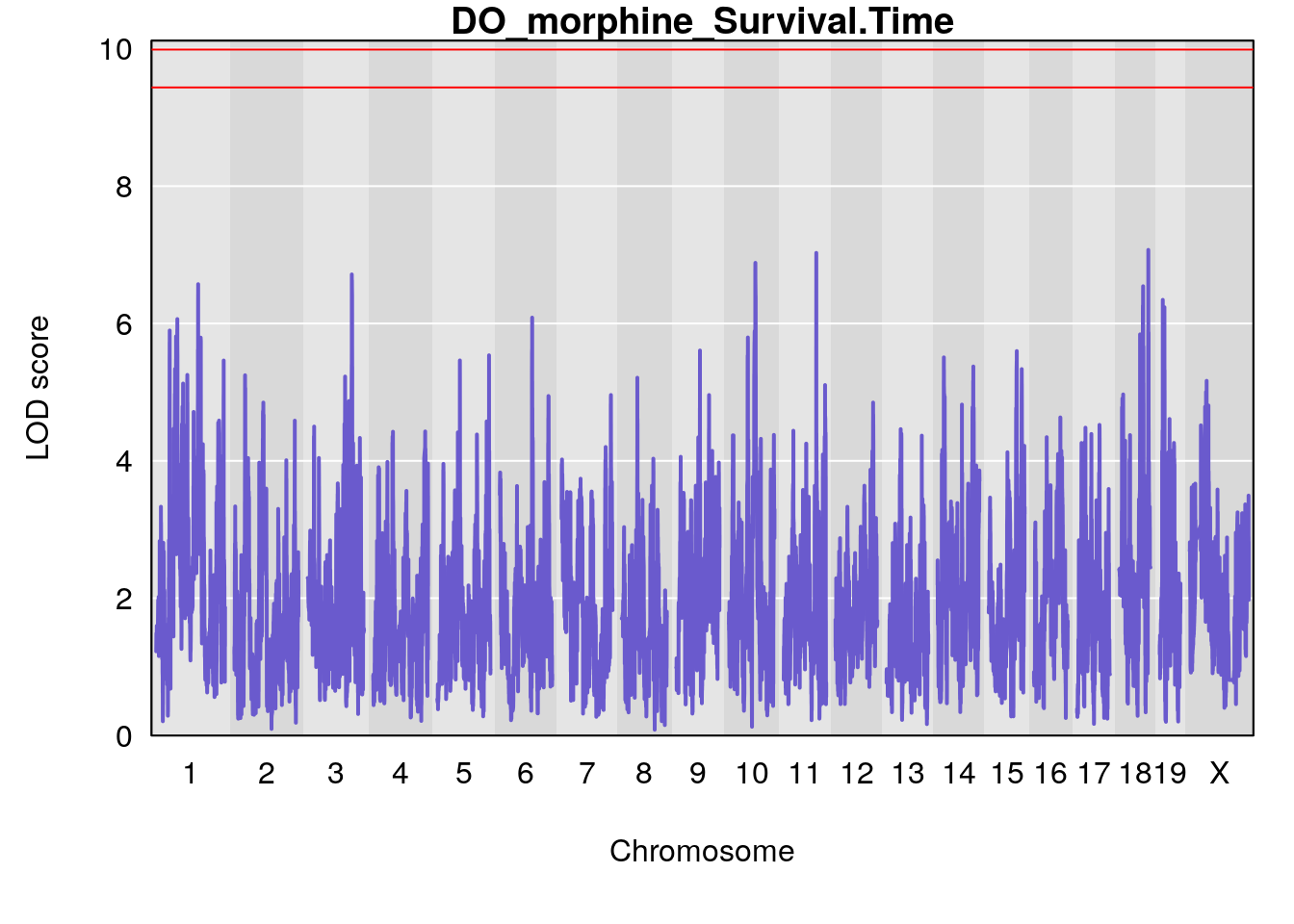
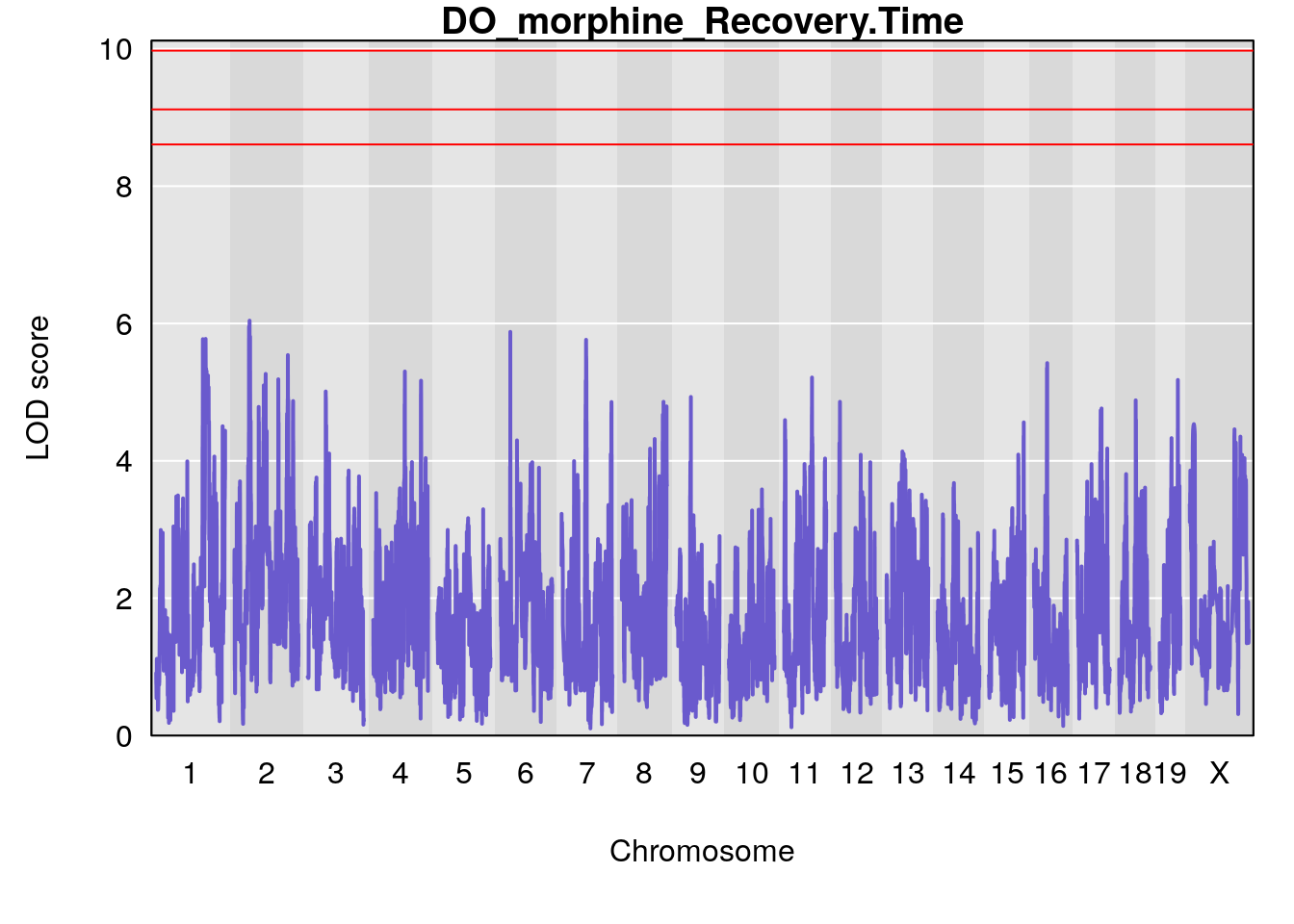
plot(qtl.morphine.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, max(sapply(qtl.morphine.out, maxlod)) +2))
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("DO_morphine_",i))
}
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
#peaks coeff plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
print(peaks)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
}
# [1] "Survival.Time"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 120.12762 6.572499 40.49547 191.33153
# 2 1 pheno1 3 125.94945 6.714517 107.22292 127.25264
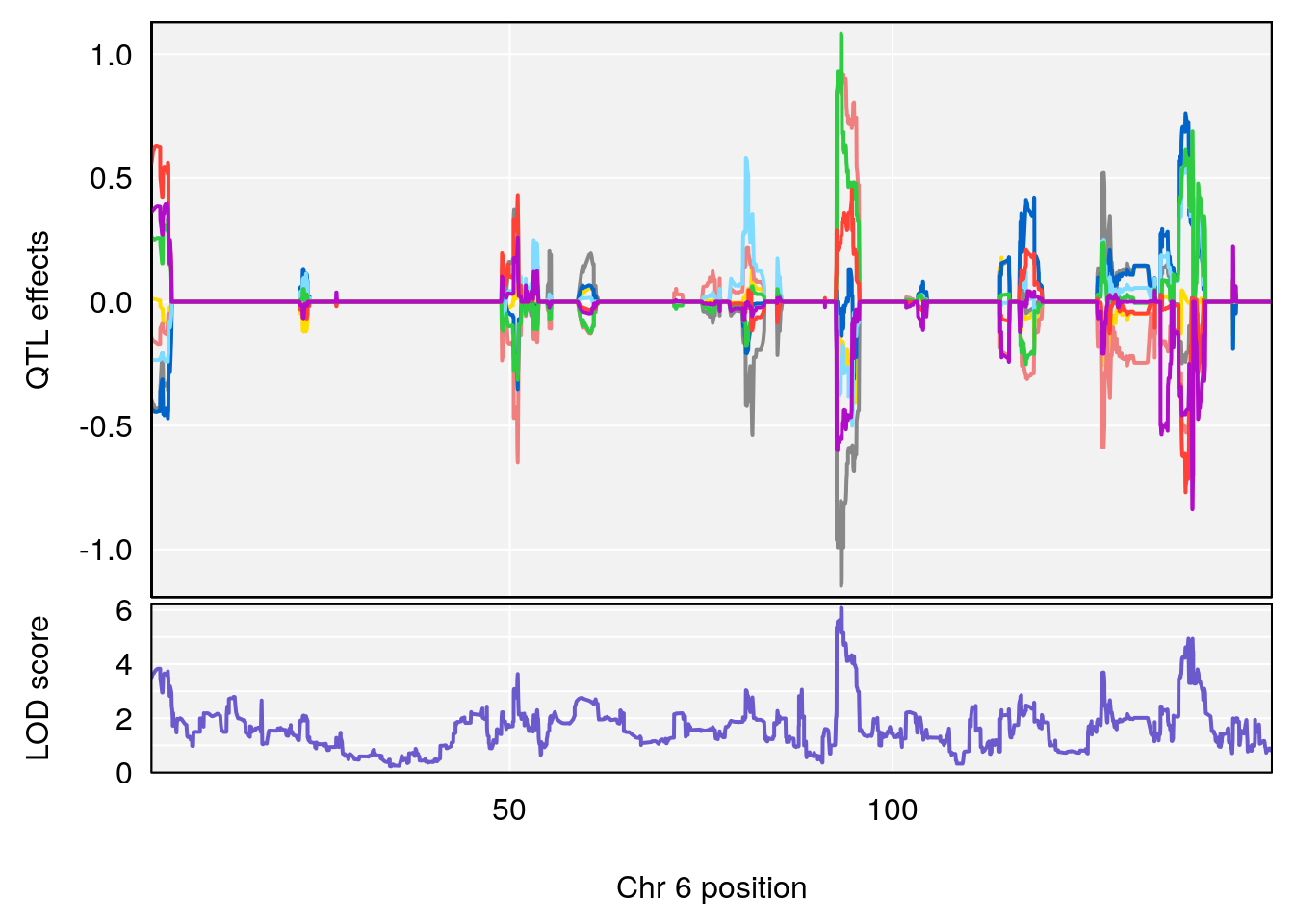
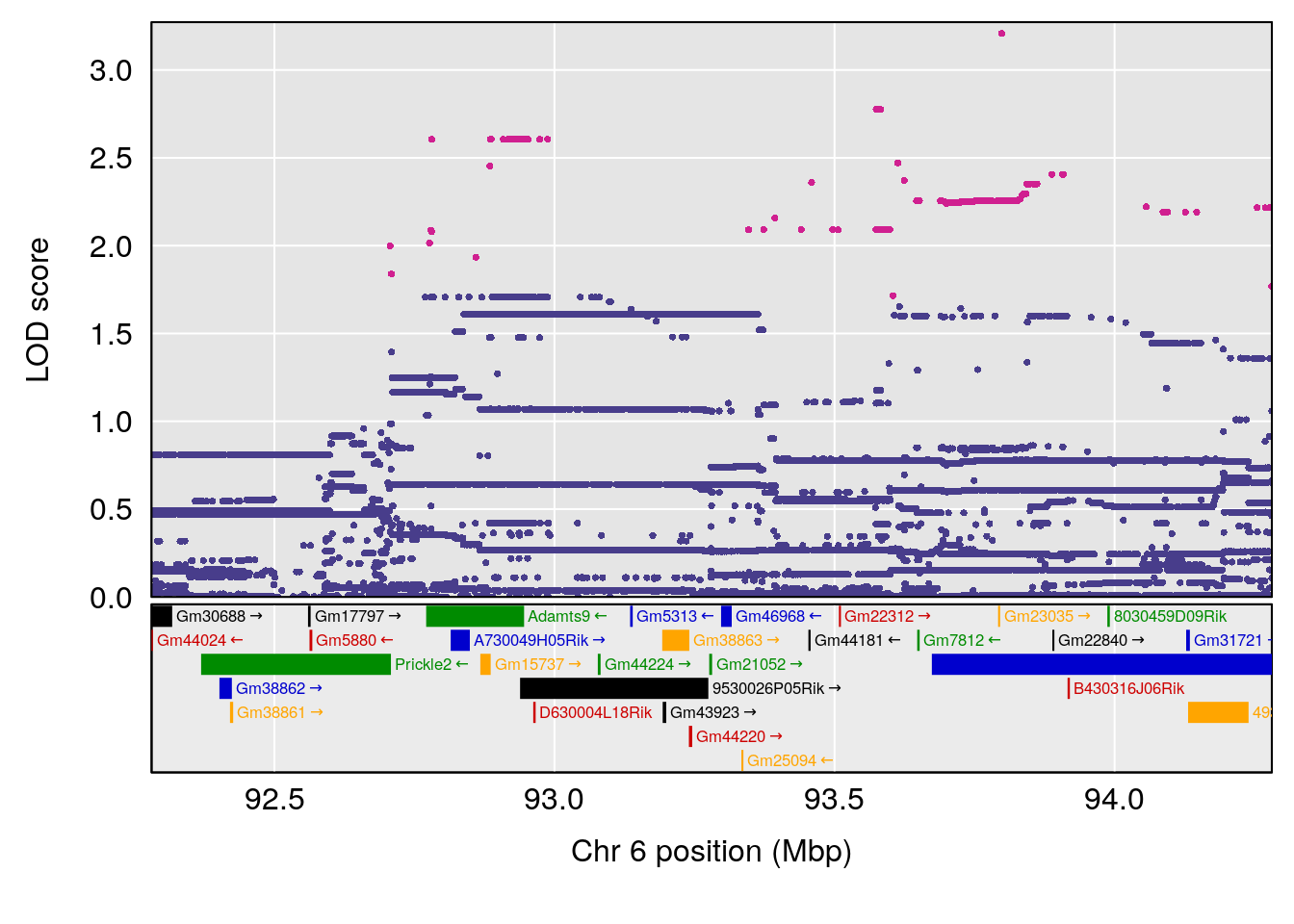
# 3 1 pheno1 6 93.28072 6.085423 92.70653 139.26401
# 4 1 pheno1 10 77.27719 6.883233 55.59002 78.16454
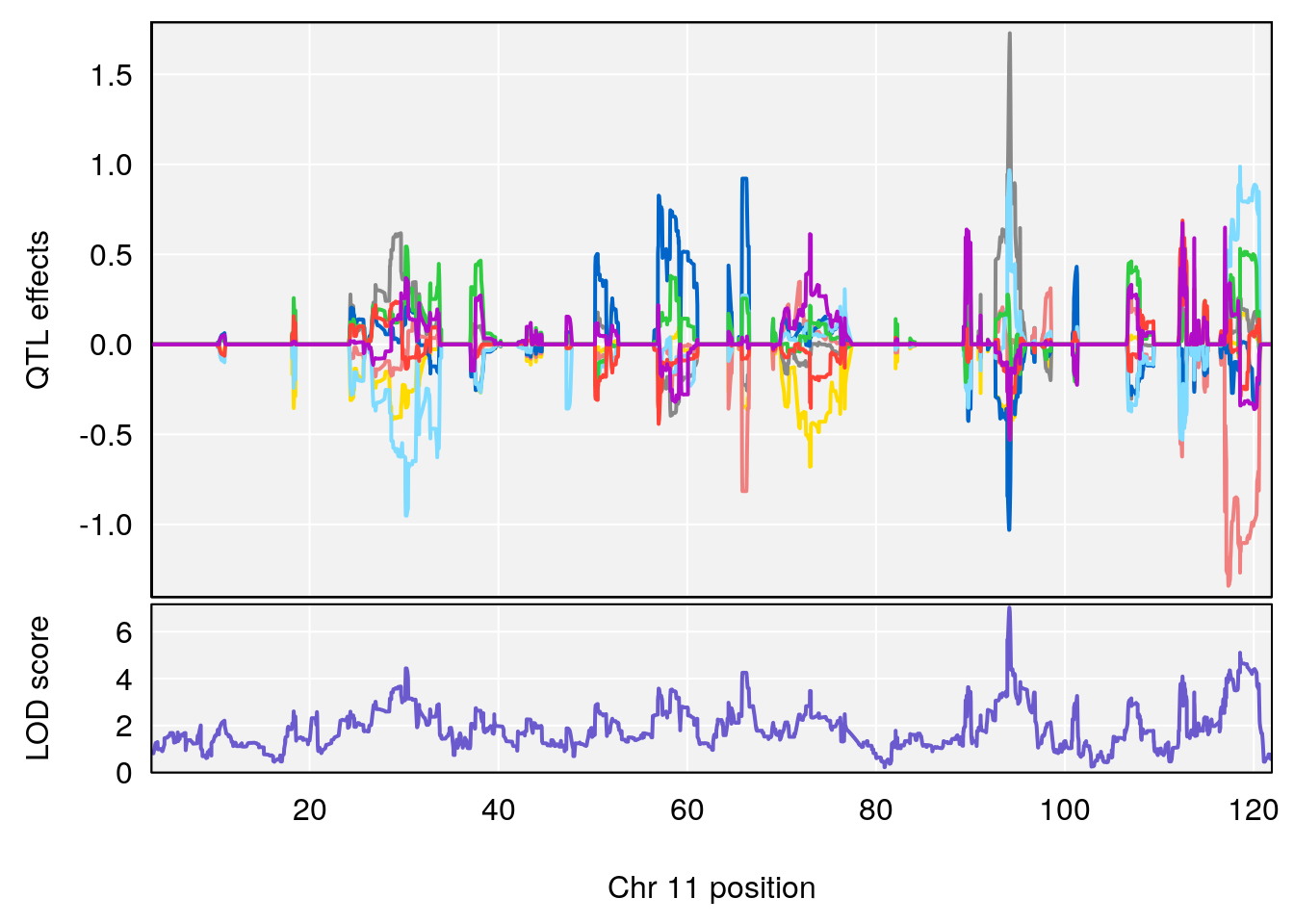
# 5 1 pheno1 11 94.11858 7.028132 93.89976 94.27434
# 6 1 pheno1 18 84.44421 7.071786 61.47853 85.27749
# 7 1 pheno1 19 11.86392 6.344164 11.74479 17.03980
# [1] 1
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 2
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 3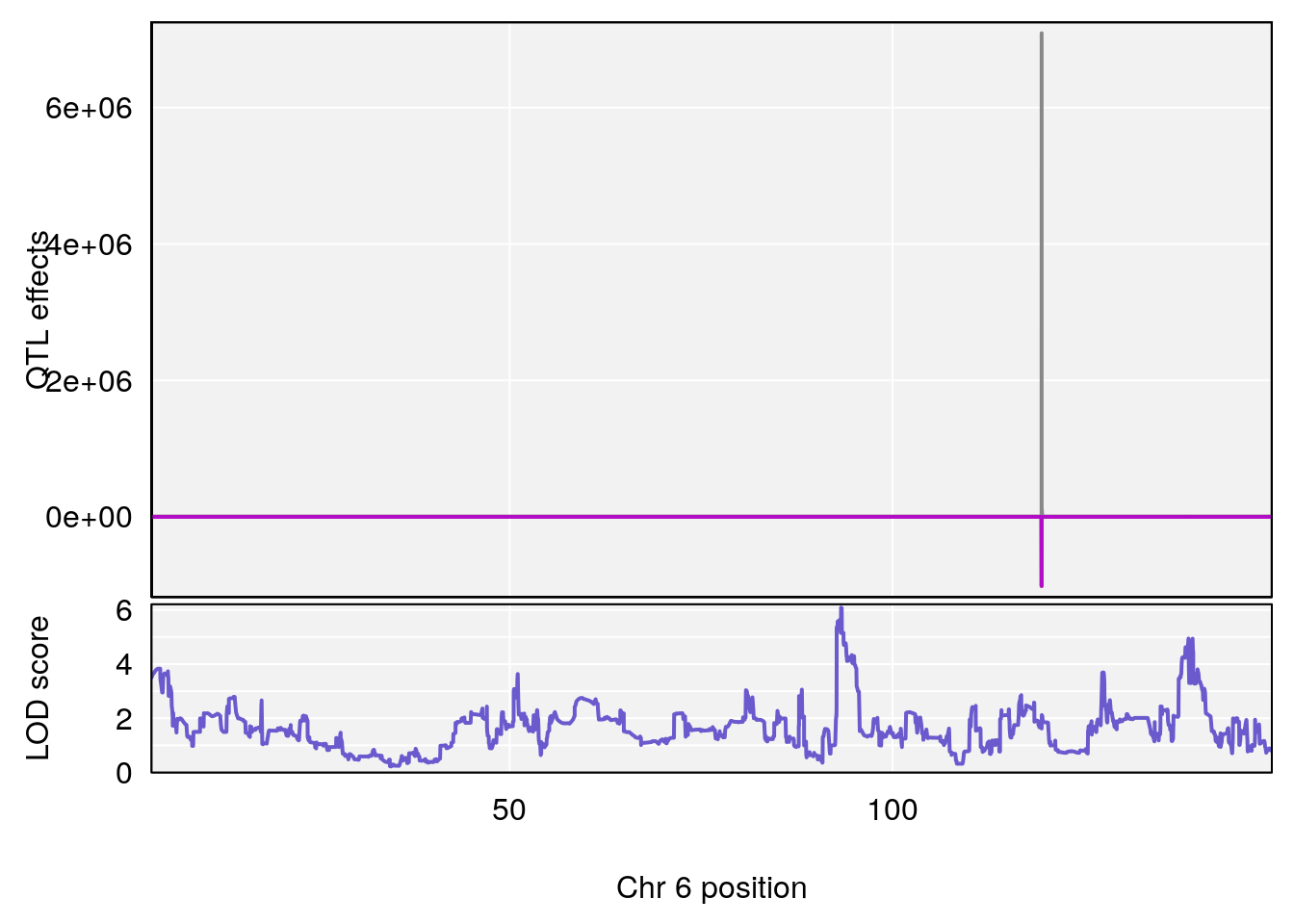
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 4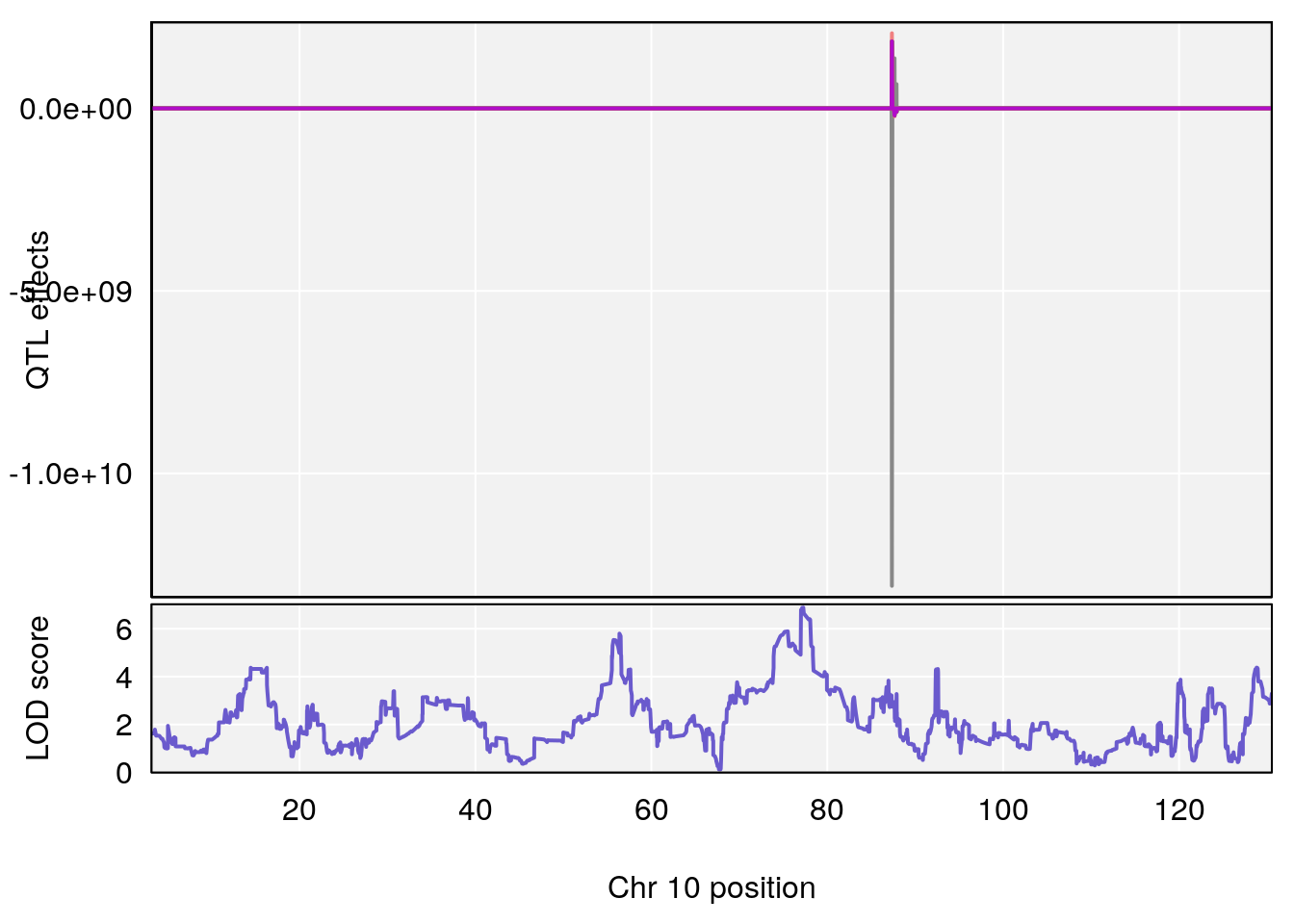
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 5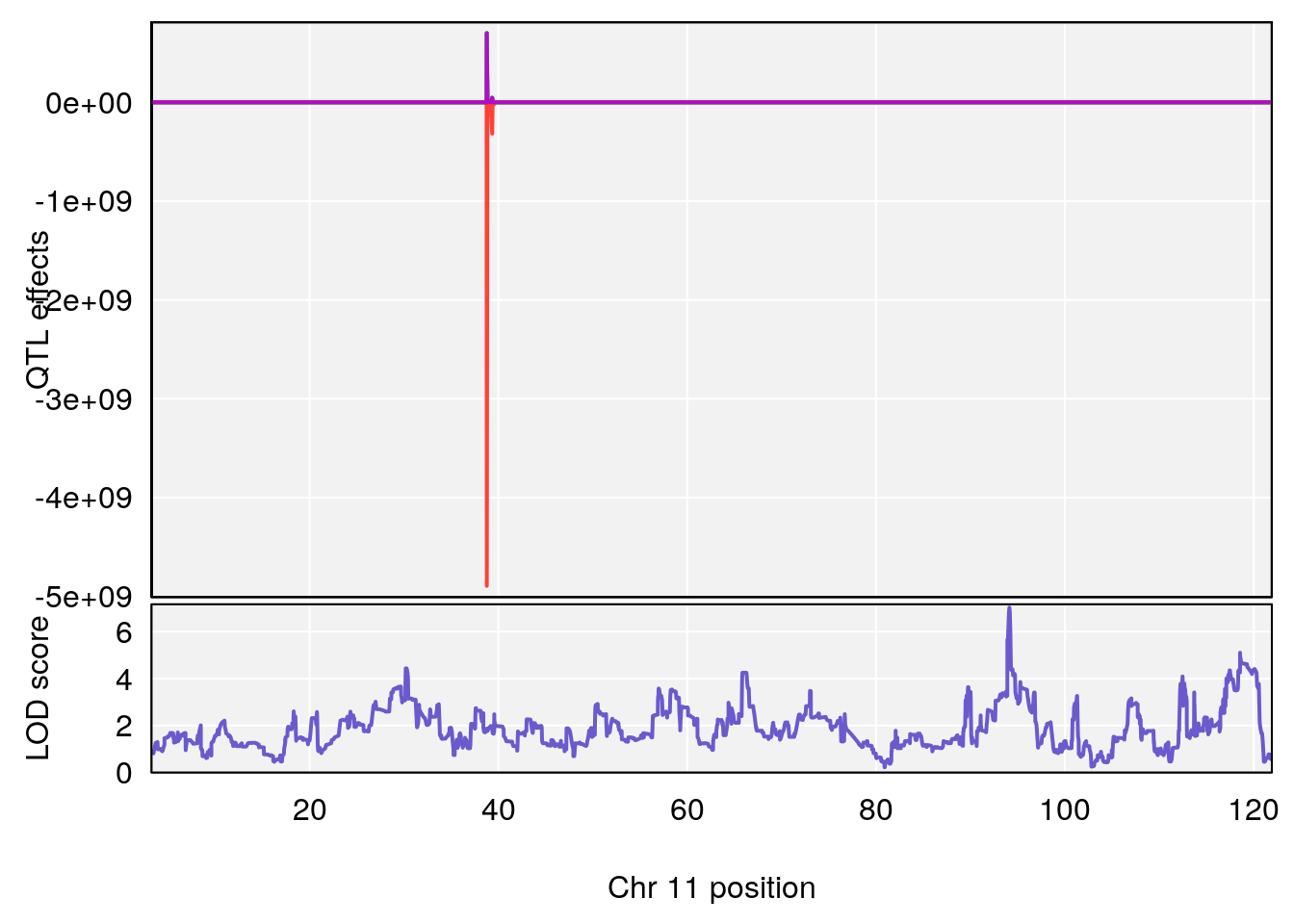
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 6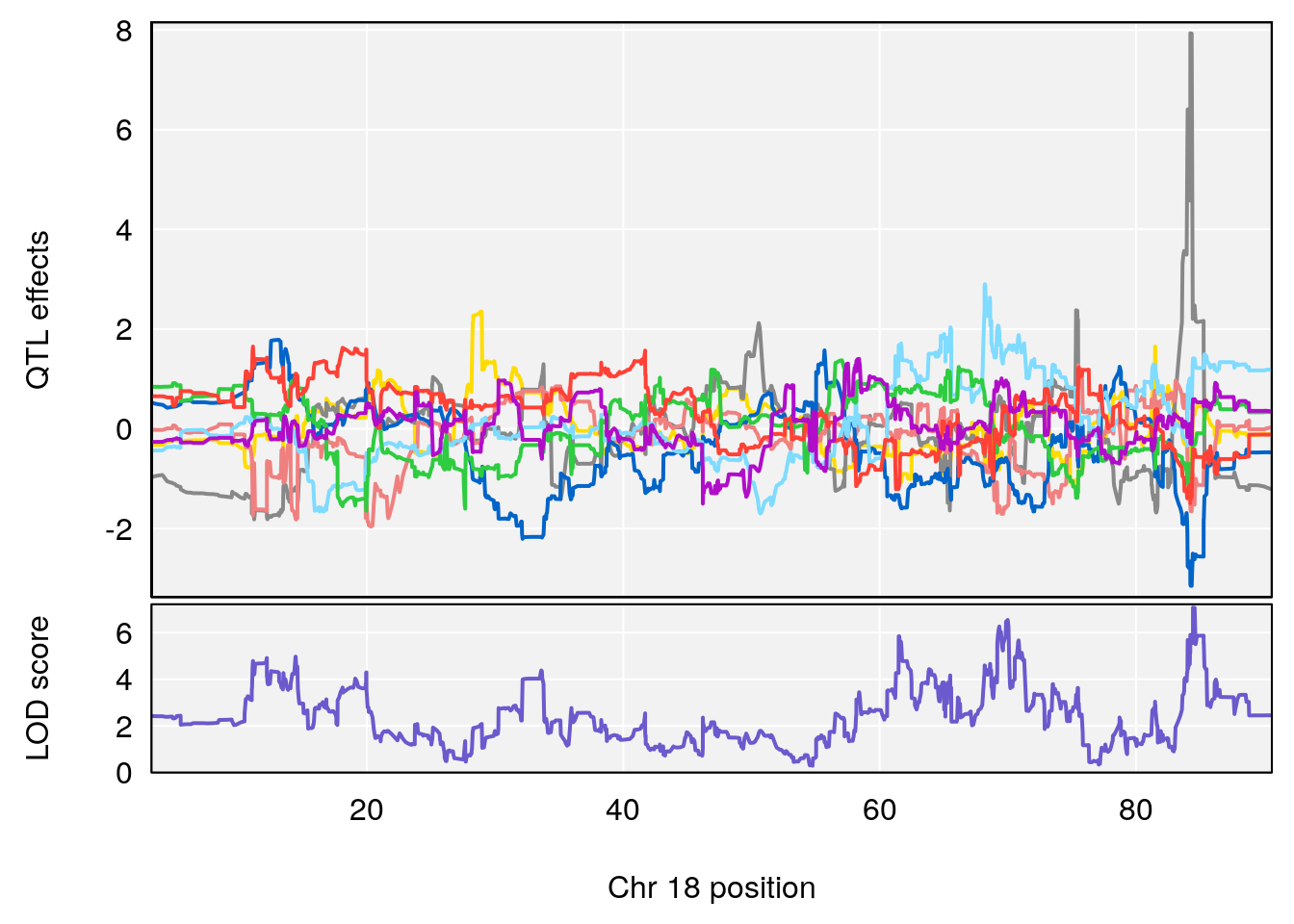
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 7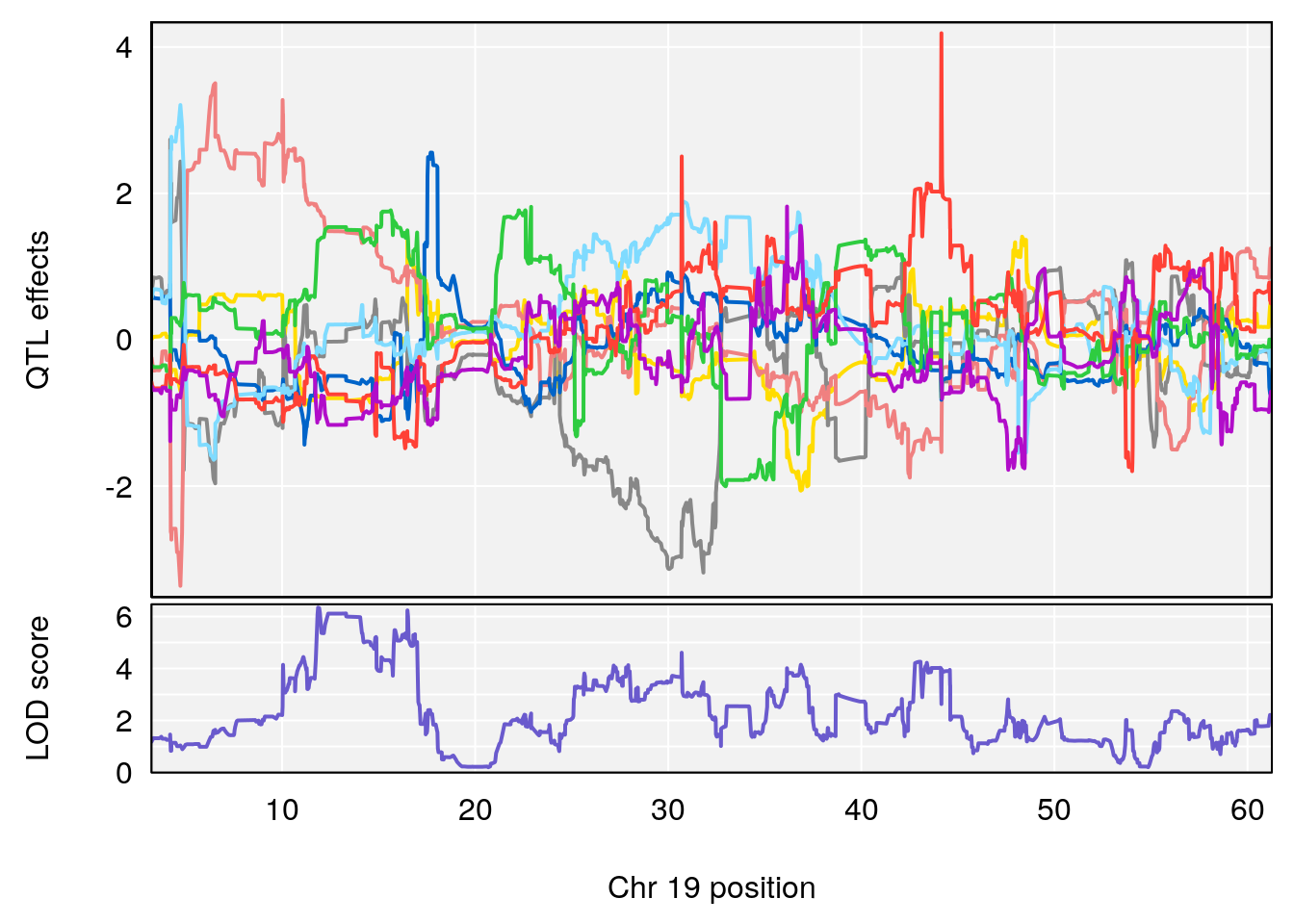
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] "Recovery.Time"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 2 45.6708 6.041384 43.98518 167.0778
# [1] 1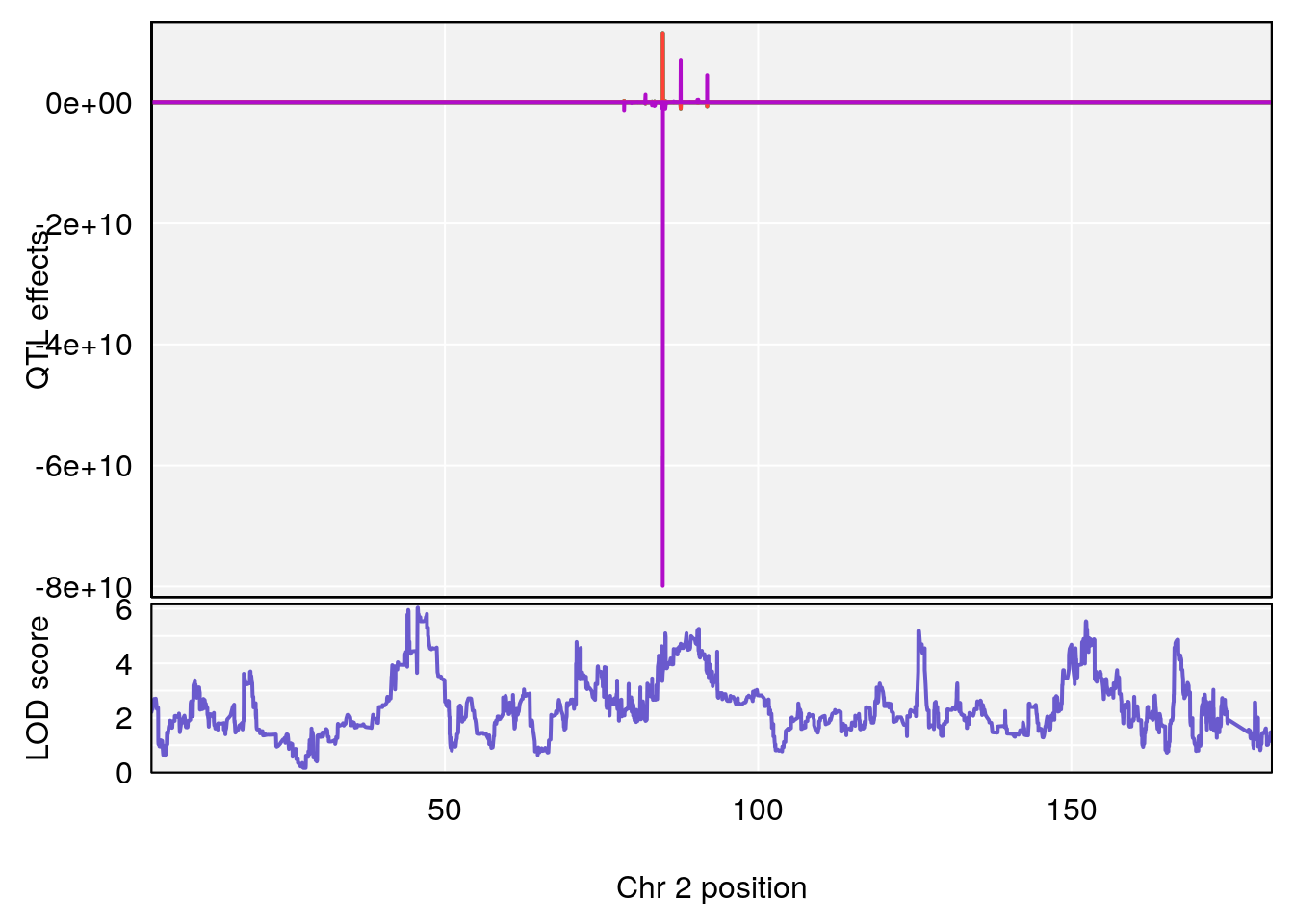
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
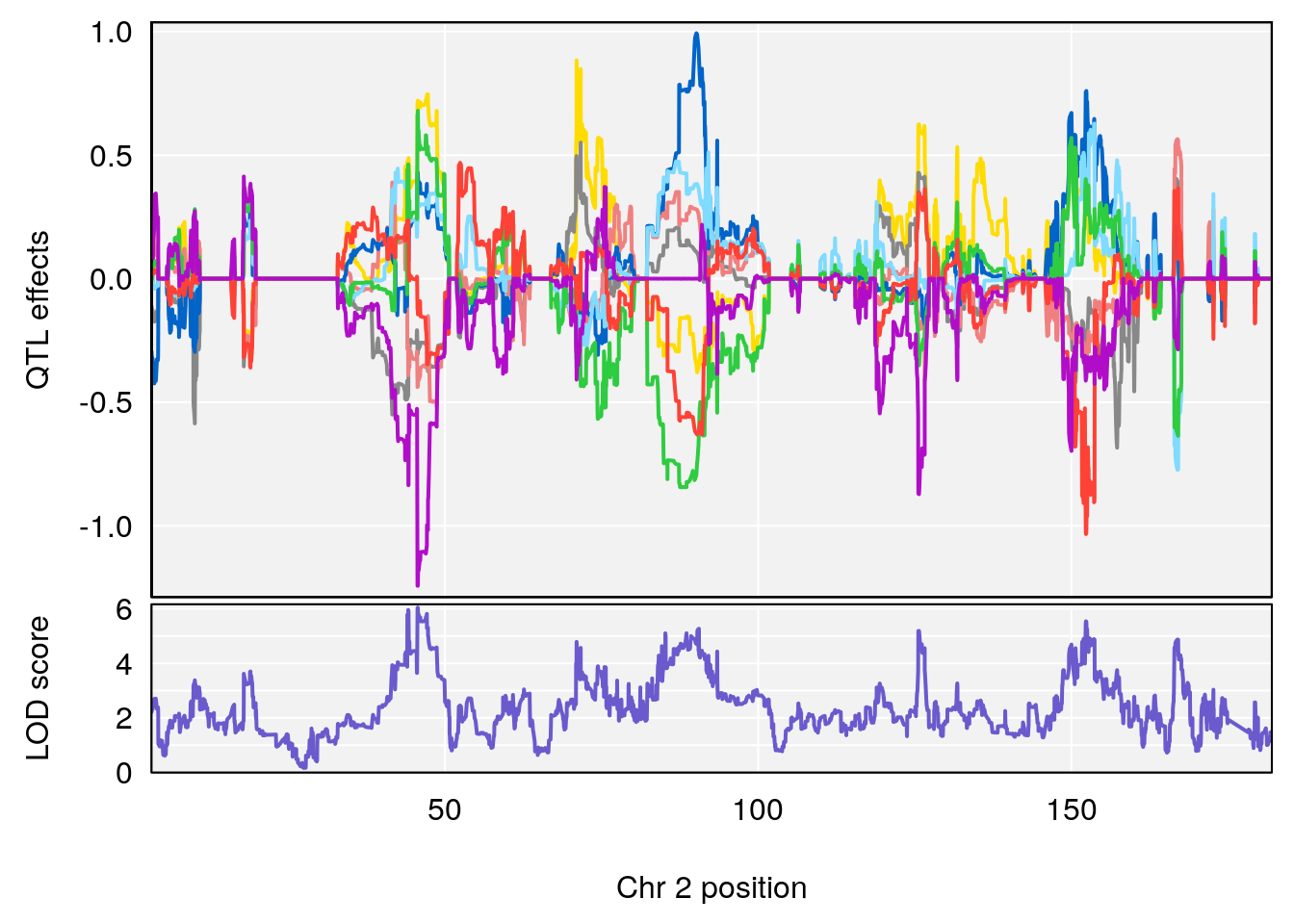
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
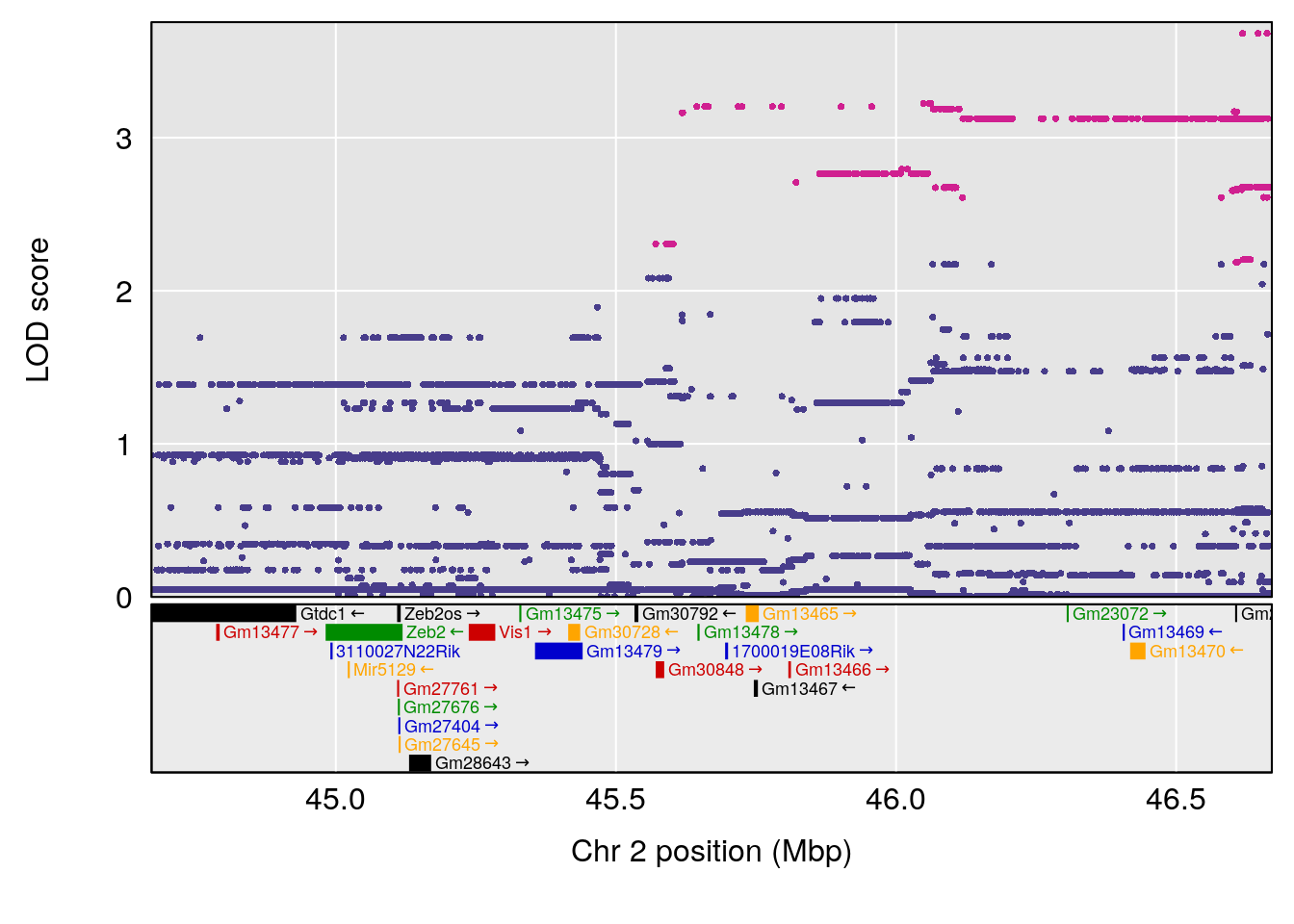
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
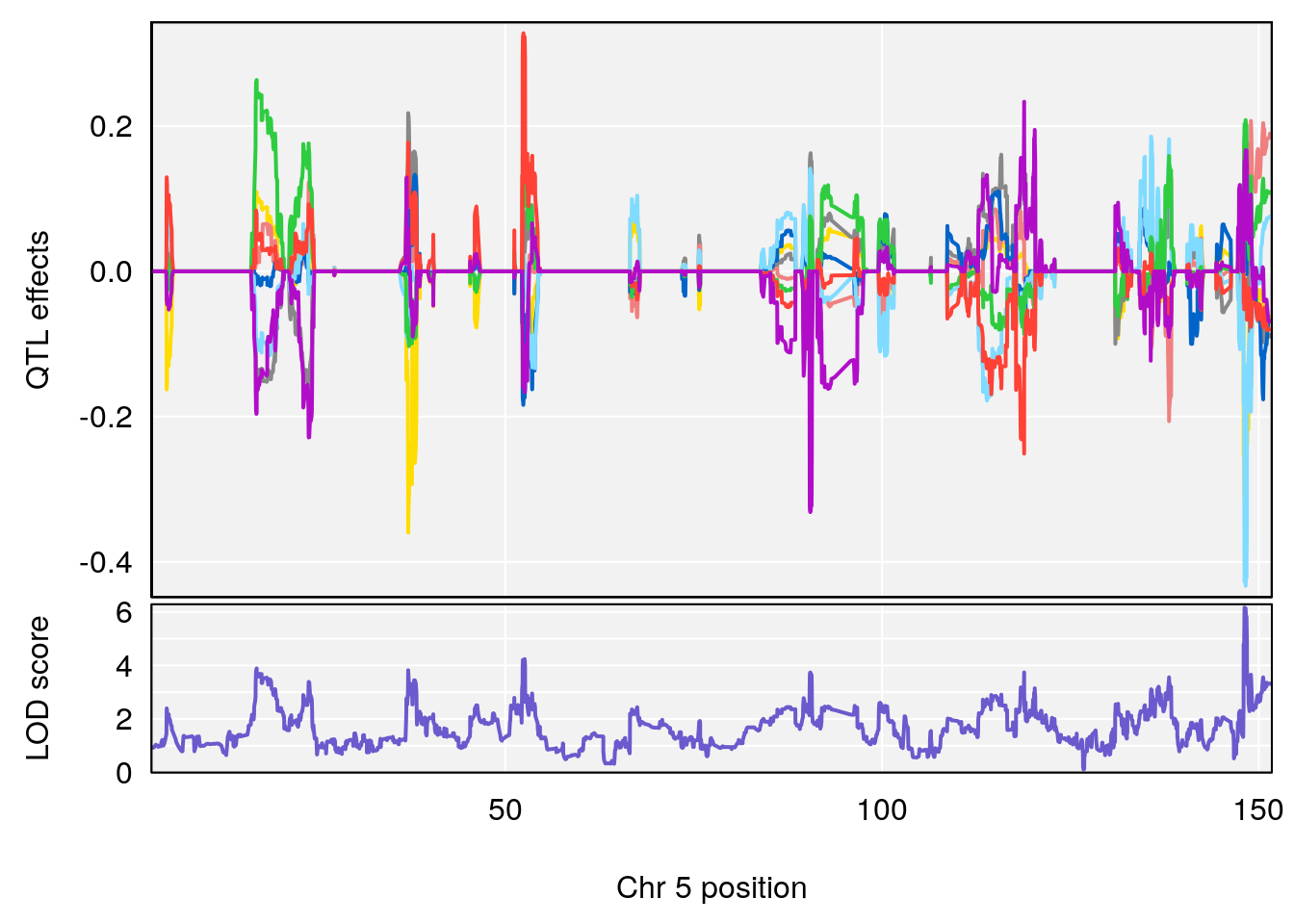
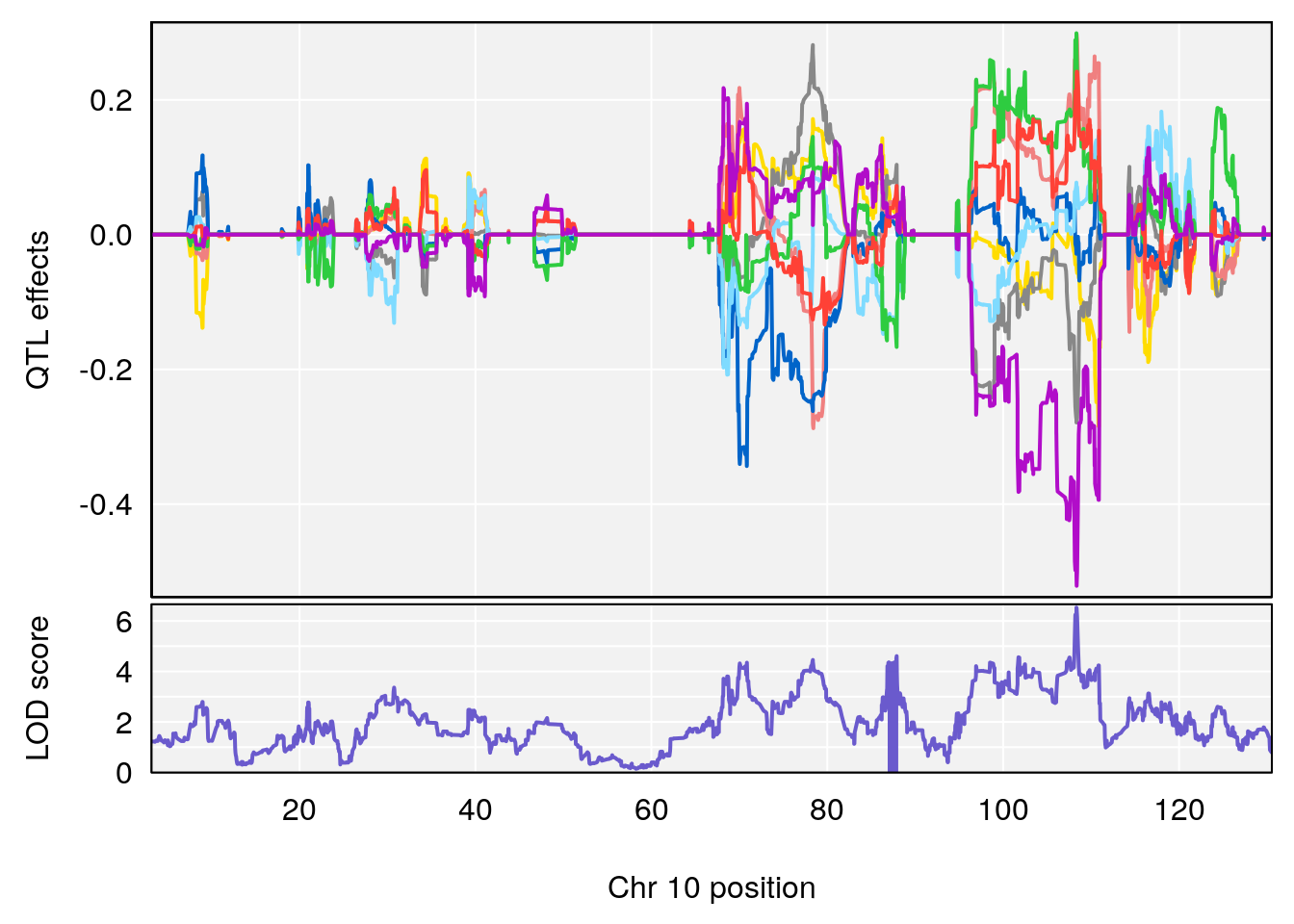
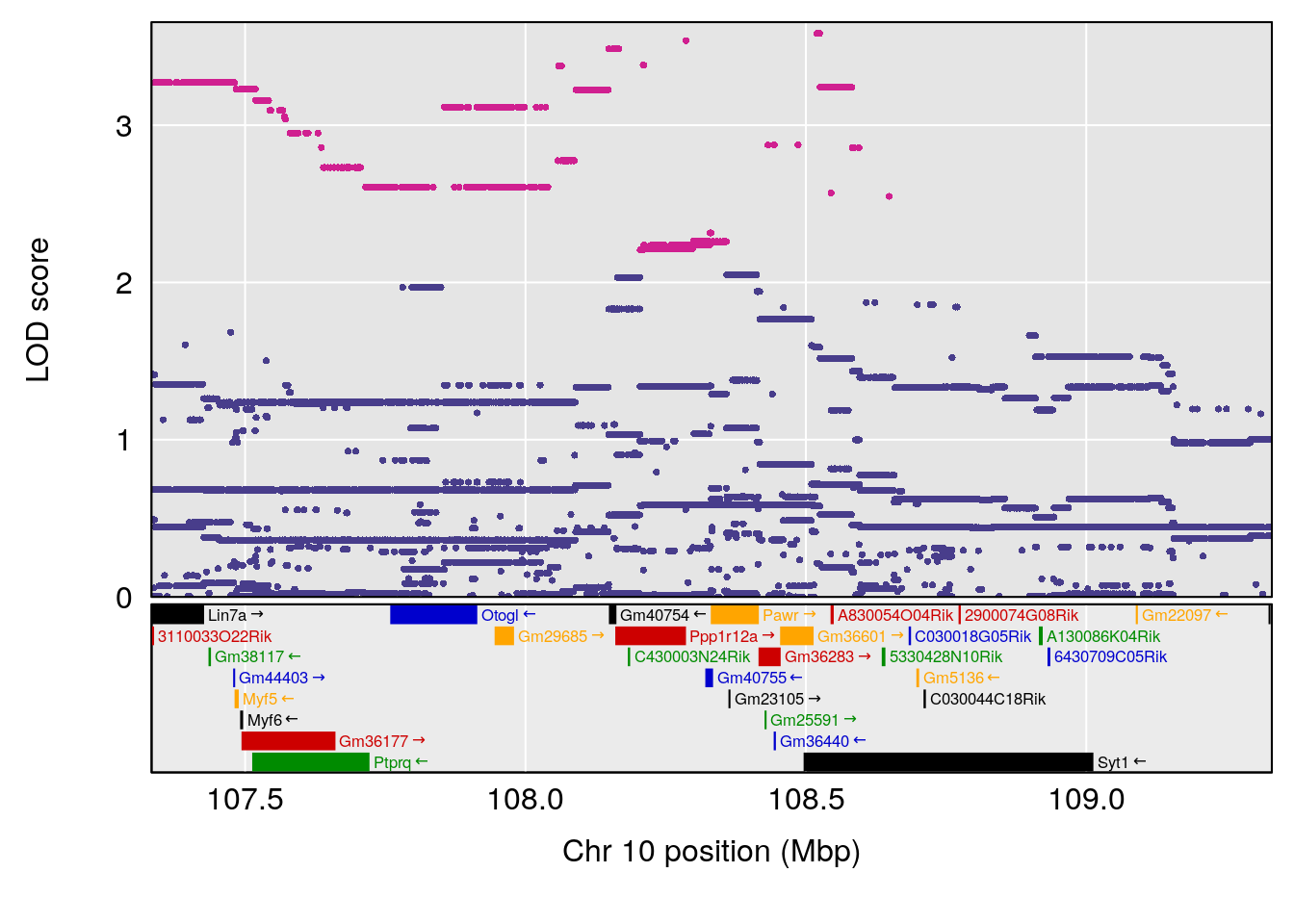
# [1] "Min.depression"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 9 122.9964 7.582778 122.2267 124.0099
# [1] 1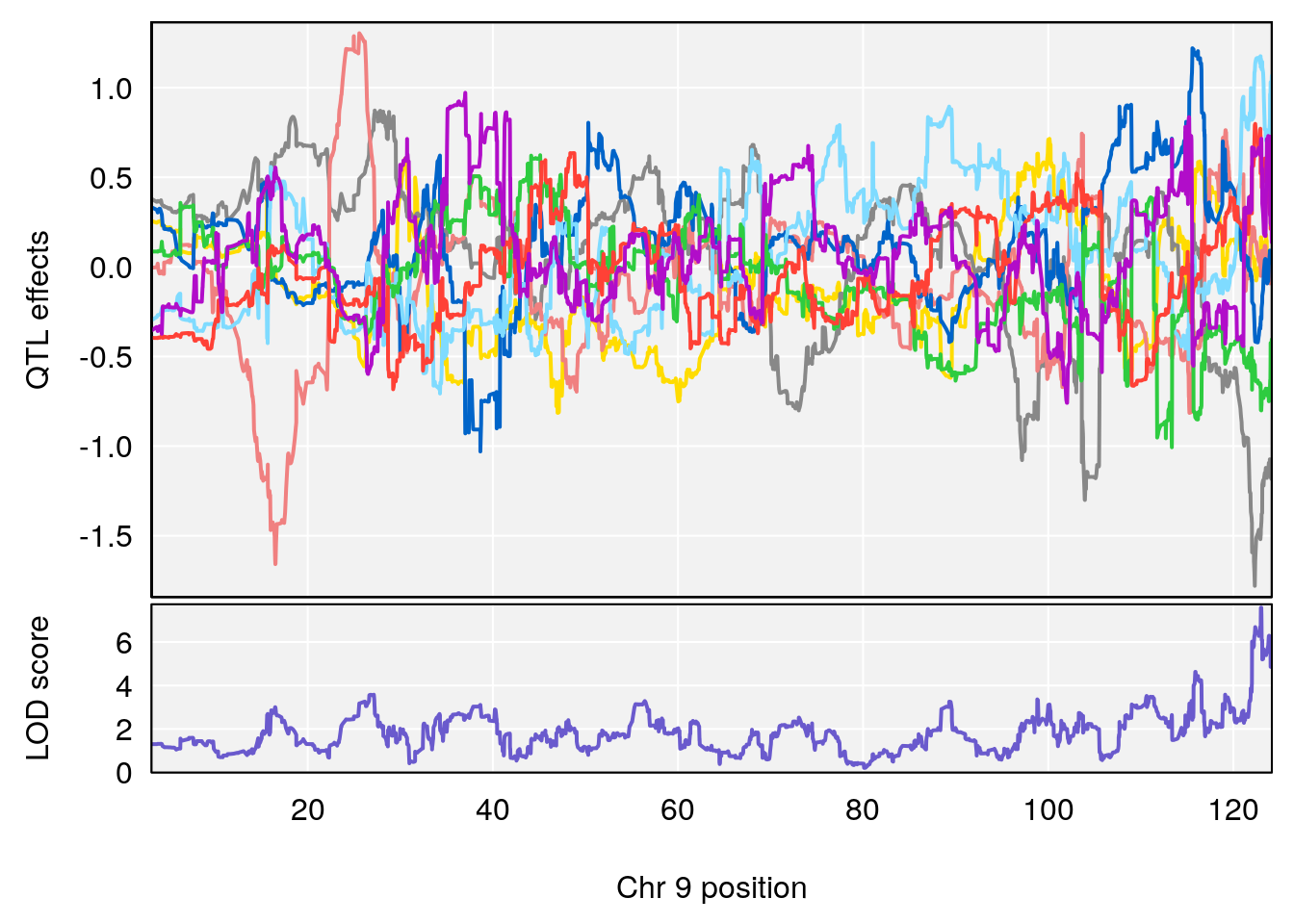
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
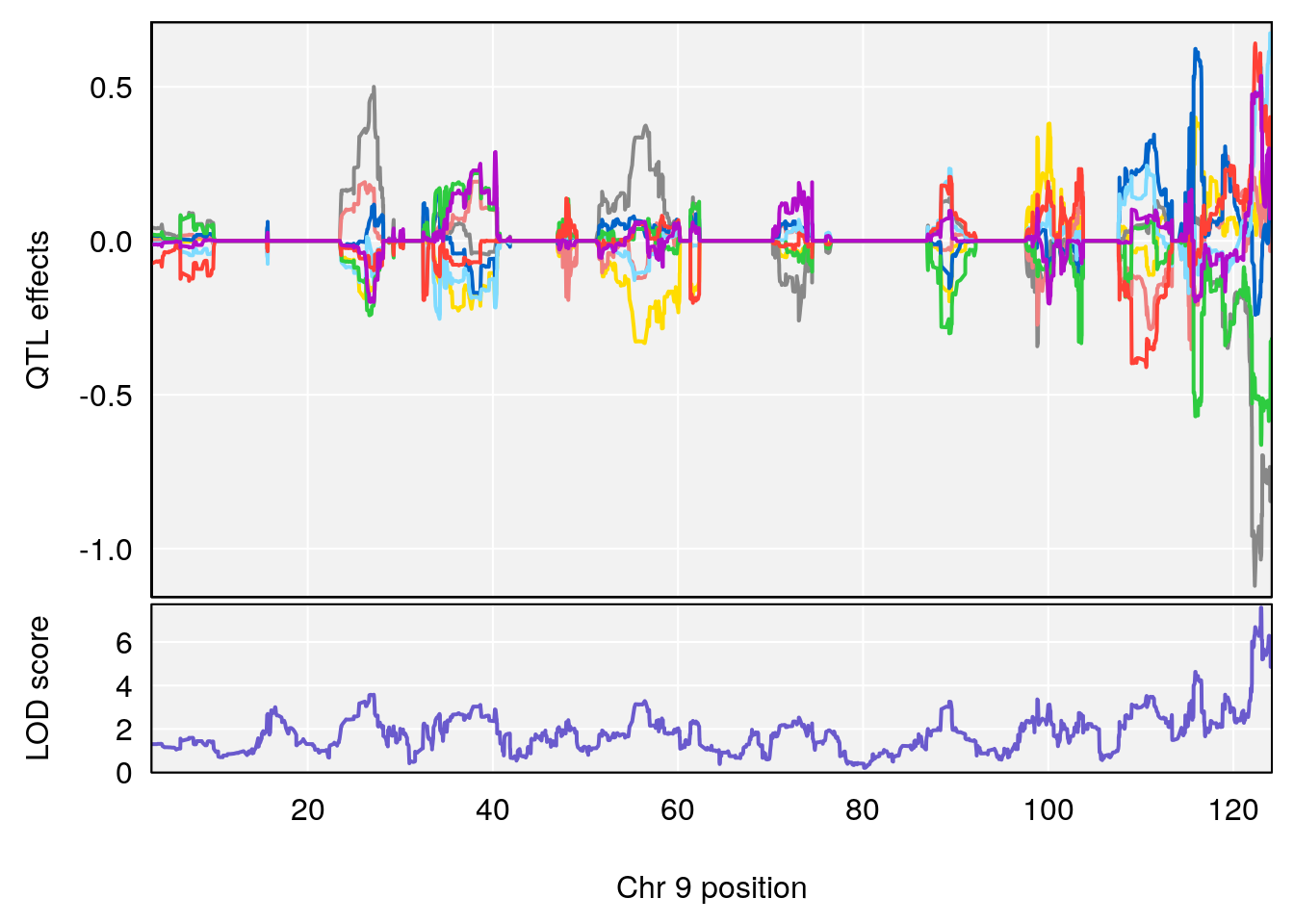
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] "Status_bin"
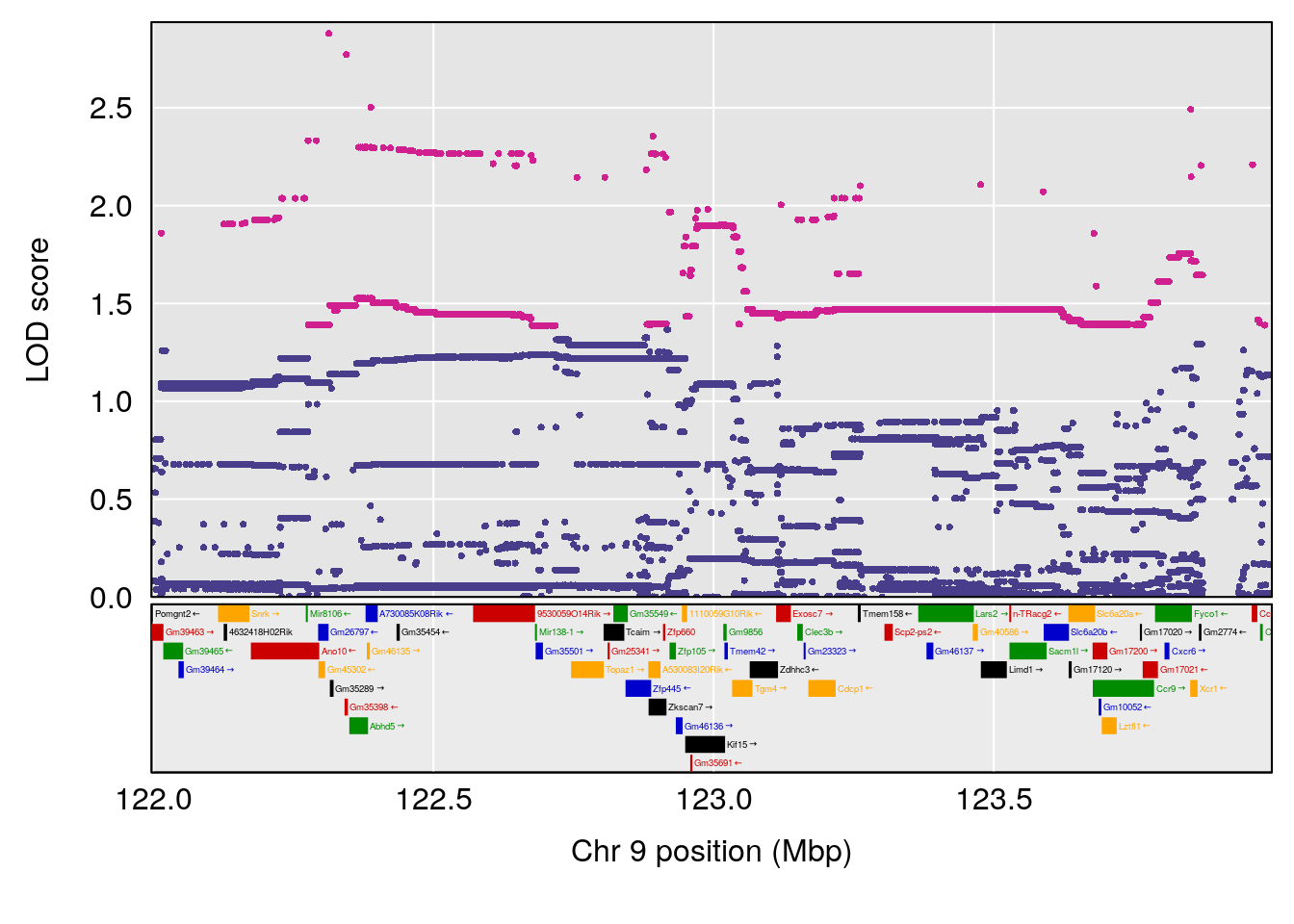
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 5 148.11200 6.160116 147.986160 148.43807
# 2 1 pheno1 8 37.41968 6.546587 37.029877 37.85219
# 3 1 pheno1 10 108.33120 6.527988 108.088576 108.51289
# 4 1 pheno1 12 80.14229 6.091948 9.247594 103.28398
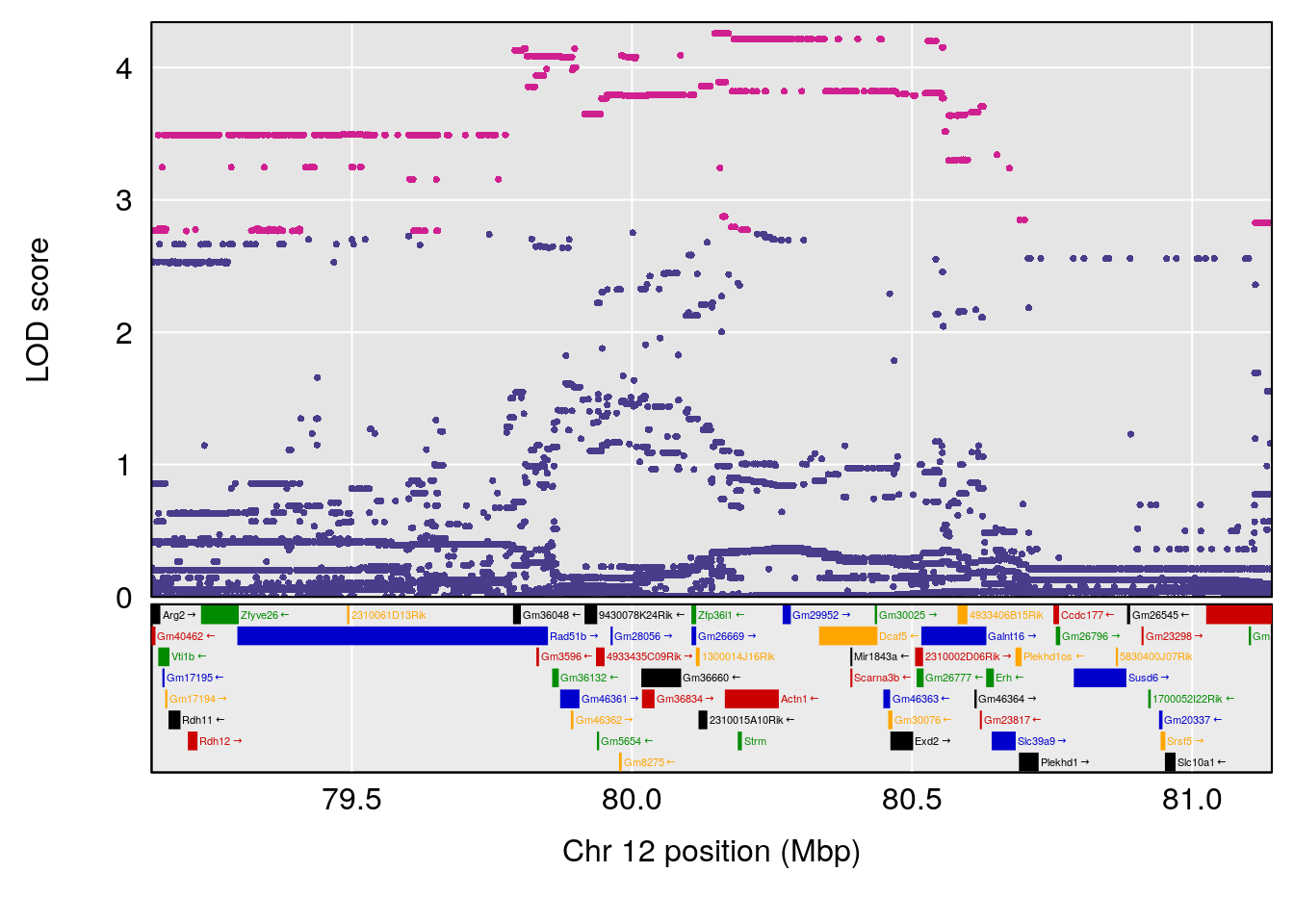
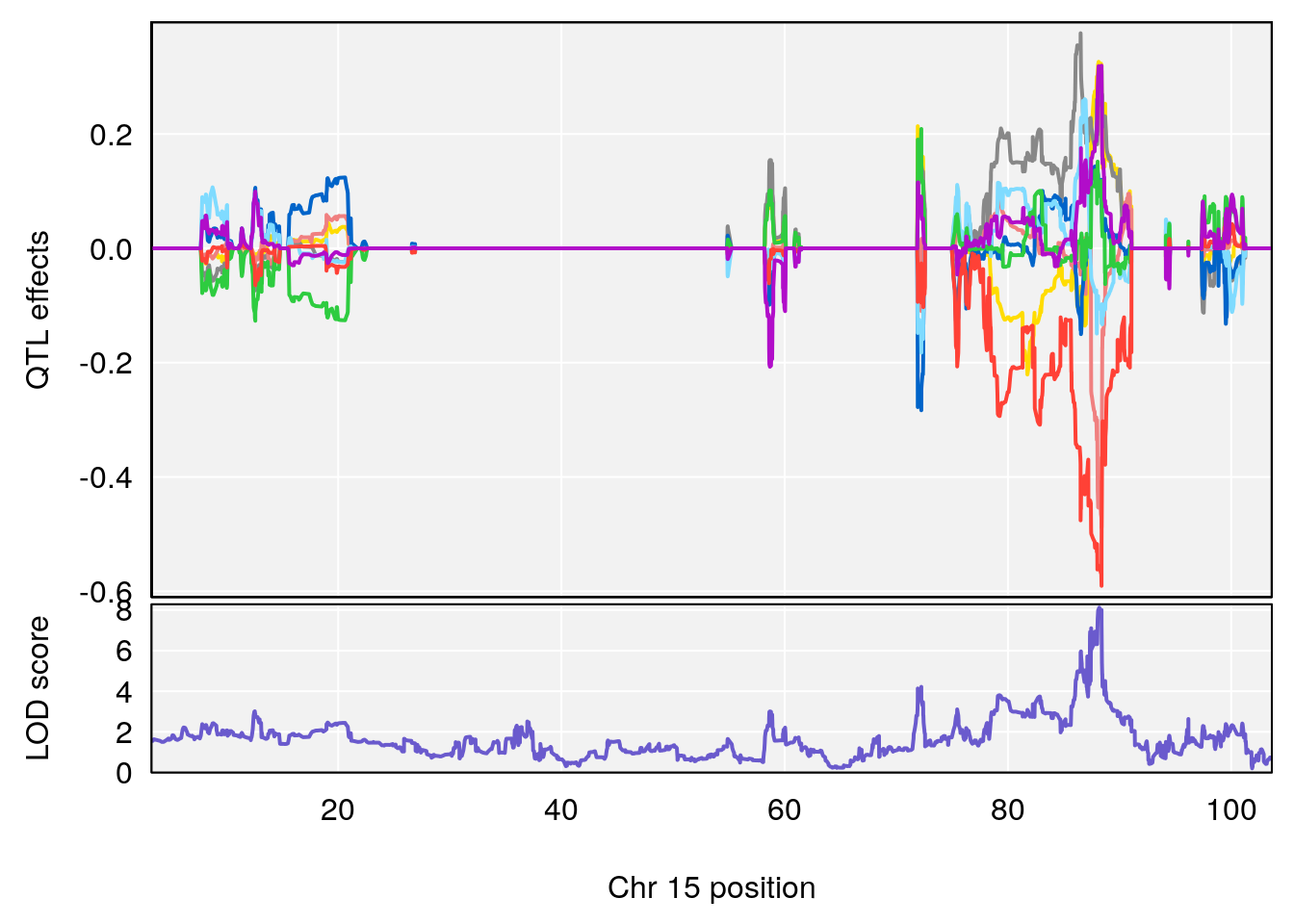
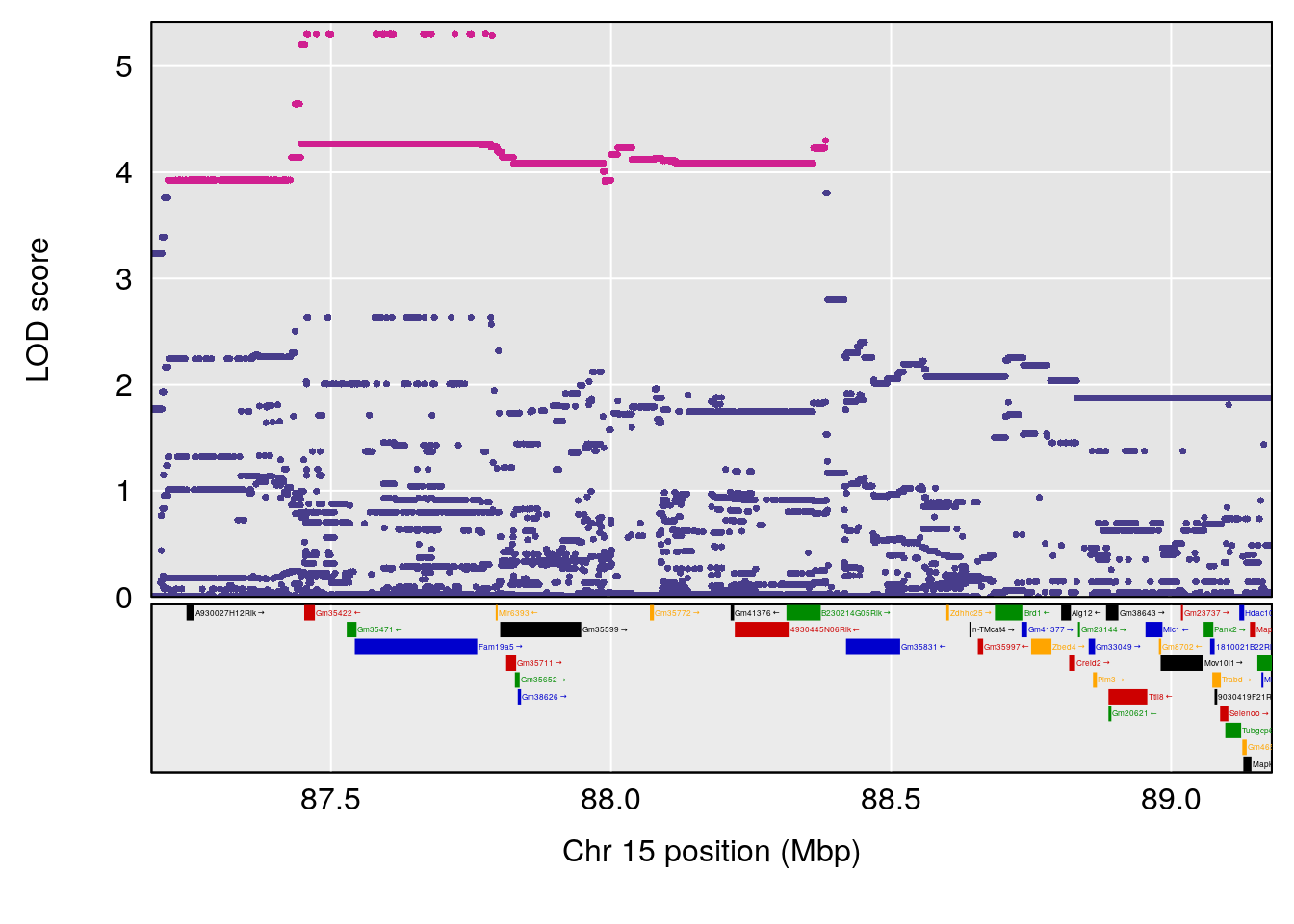
# 5 1 pheno1 15 88.17969 8.118696 87.323190 88.41707
# [1] 1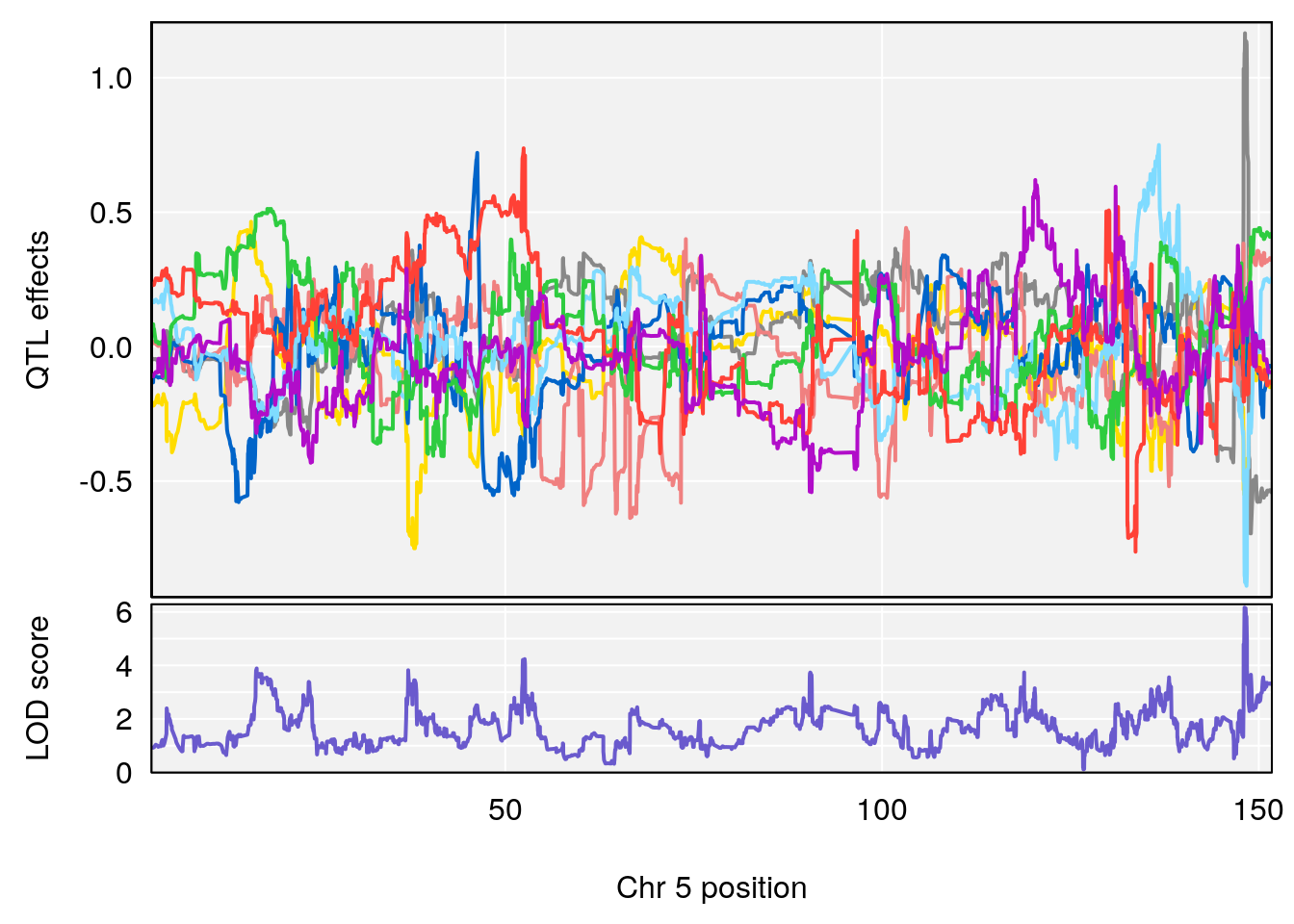
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 2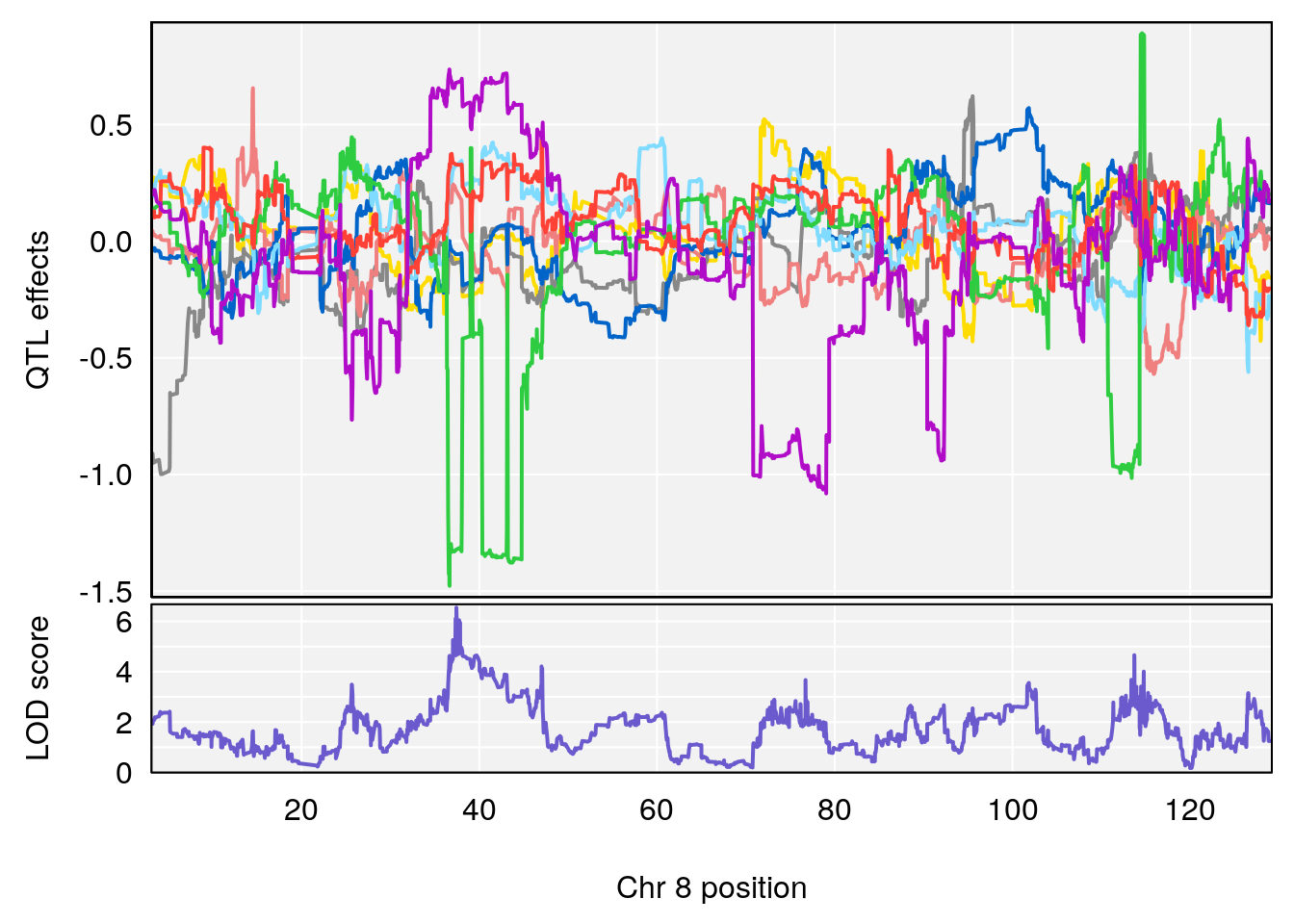
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 3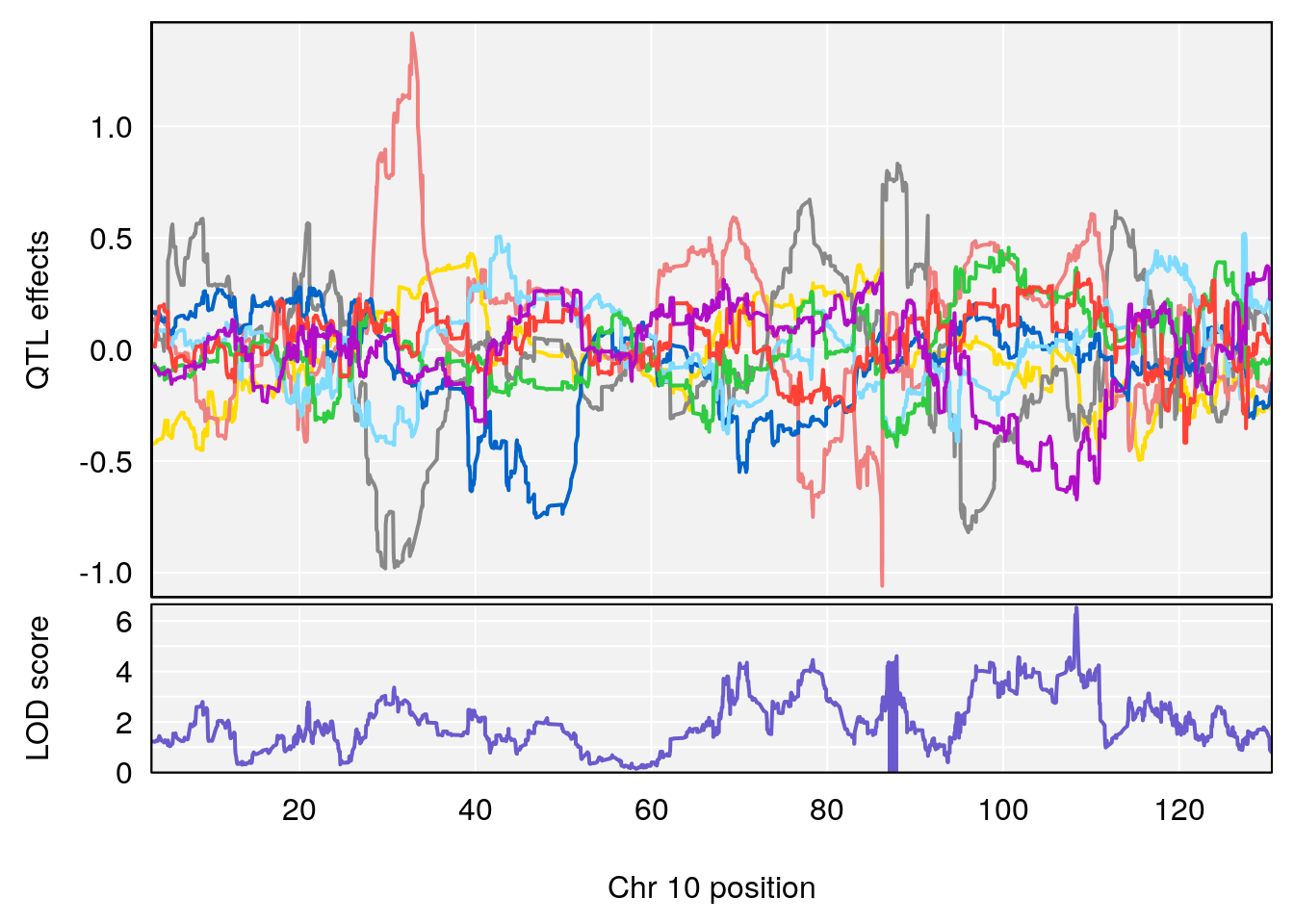
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 4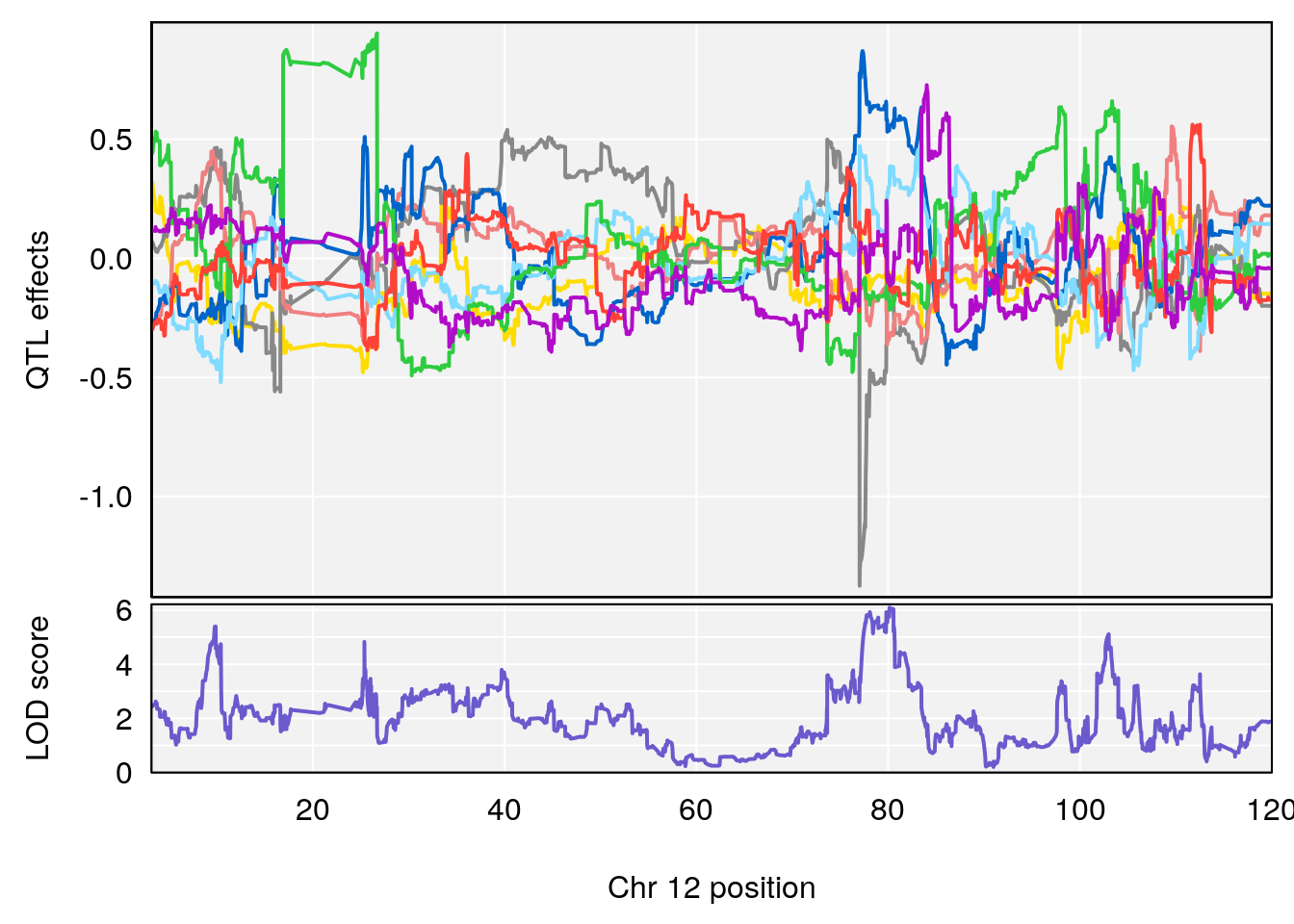
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 5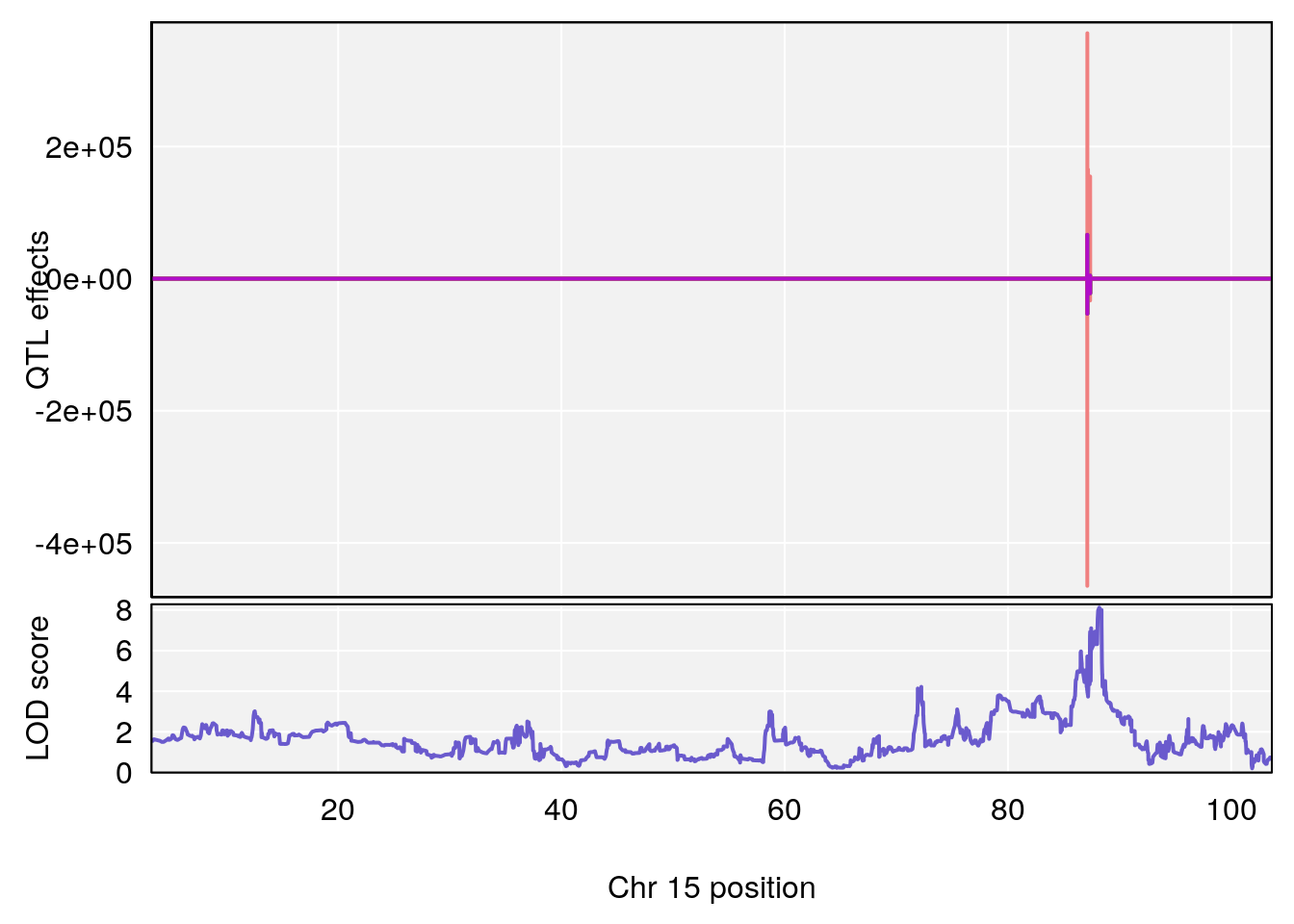
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
Plot qtl mapping on array female
load("output/qtl.morphine.out.second_set.female.RData")
#pmap and gmap
chr.names <- c("1","2","3","4","5","6","7","8","9","10","11","12","13","14","15","16","17","18","19", "X")
gmap <- list()
pmap <- list()
for (chr in chr.names){
gmap[[chr]] <- do.morphine$gmap[[chr]]
pmap[[chr]] <- do.morphine$pmap[[chr]]
}
attr(gmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
attr(pmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
#genome-wide plot
for(i in names(qtl.morphine.out)){
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.morphine.out[[i]]) # overall maximum LOD score
plot(qtl.morphine.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, max(sapply(qtl.morphine.out, maxlod)) +2))
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("DO_morphine_",i))
}
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
#peaks coeff plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
print(peaks)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
}
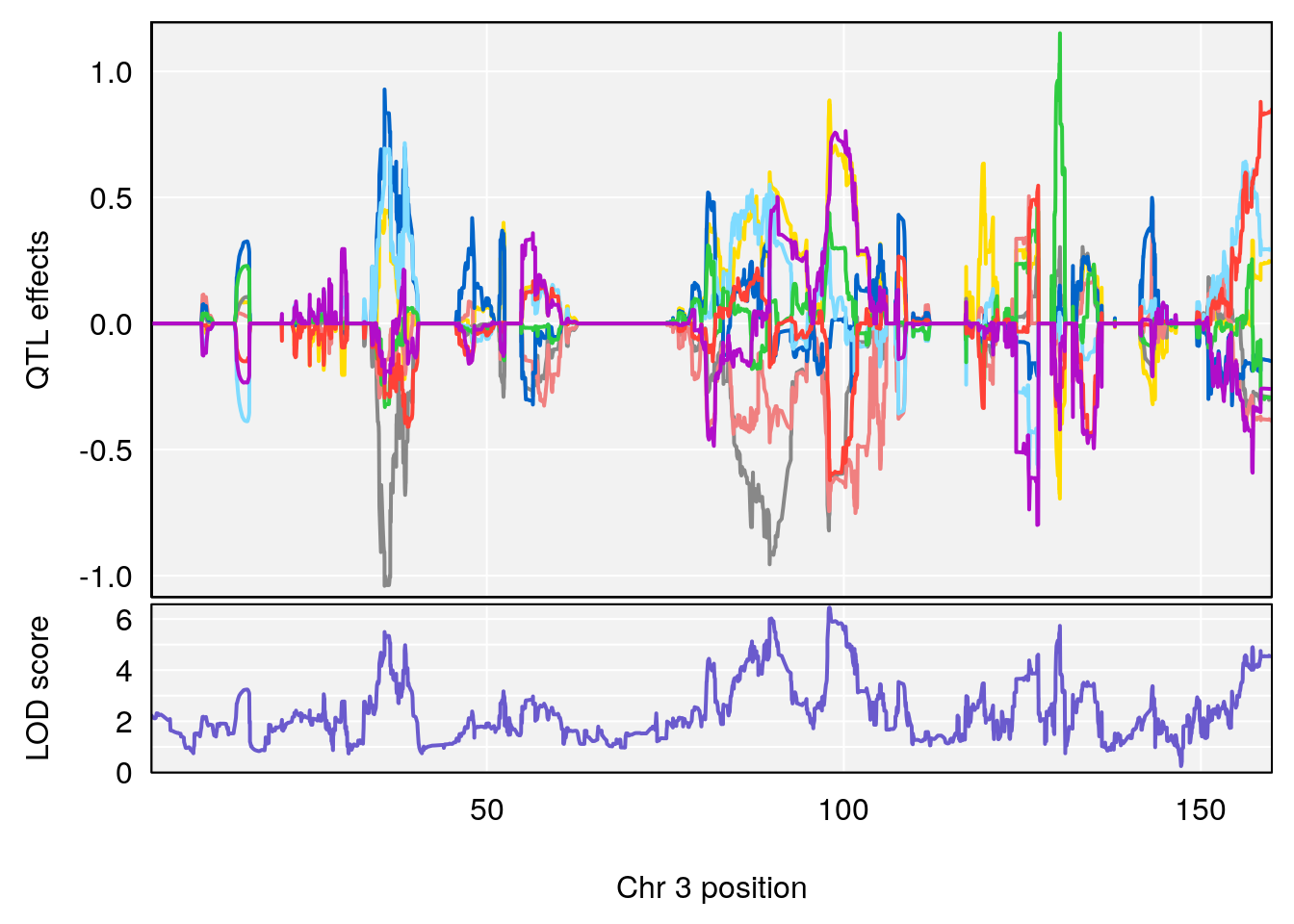
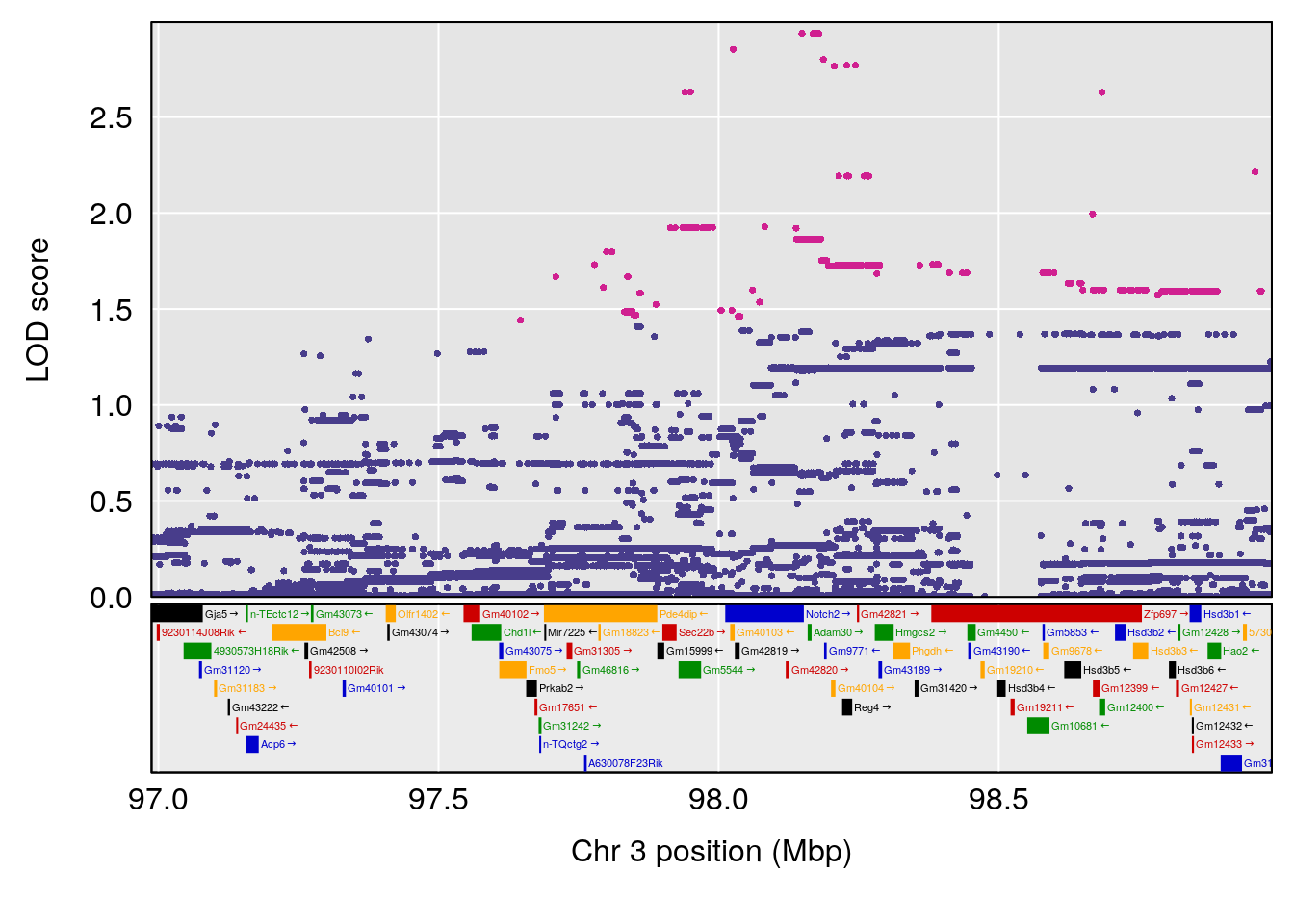
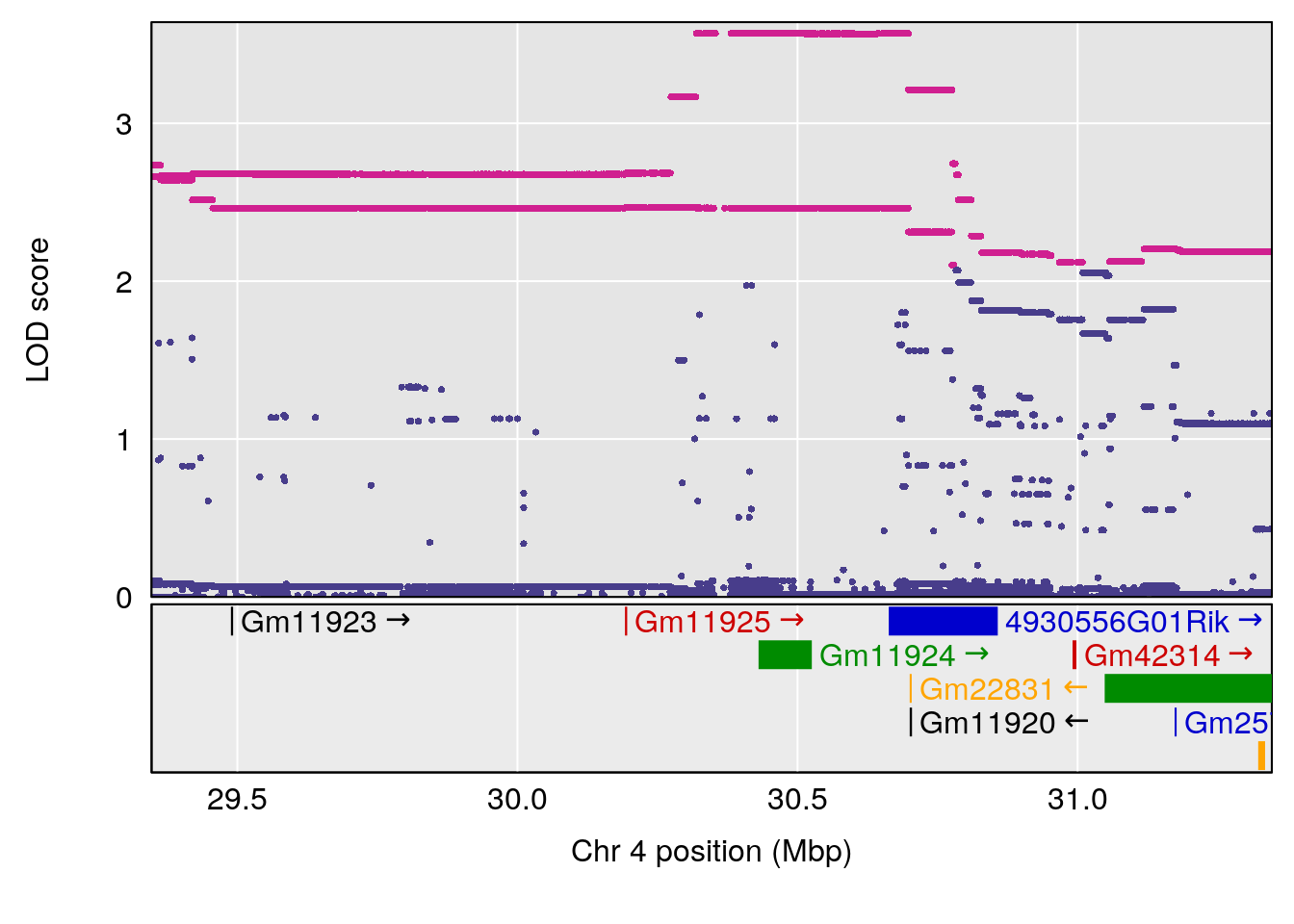
# [1] "Survival.Time"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
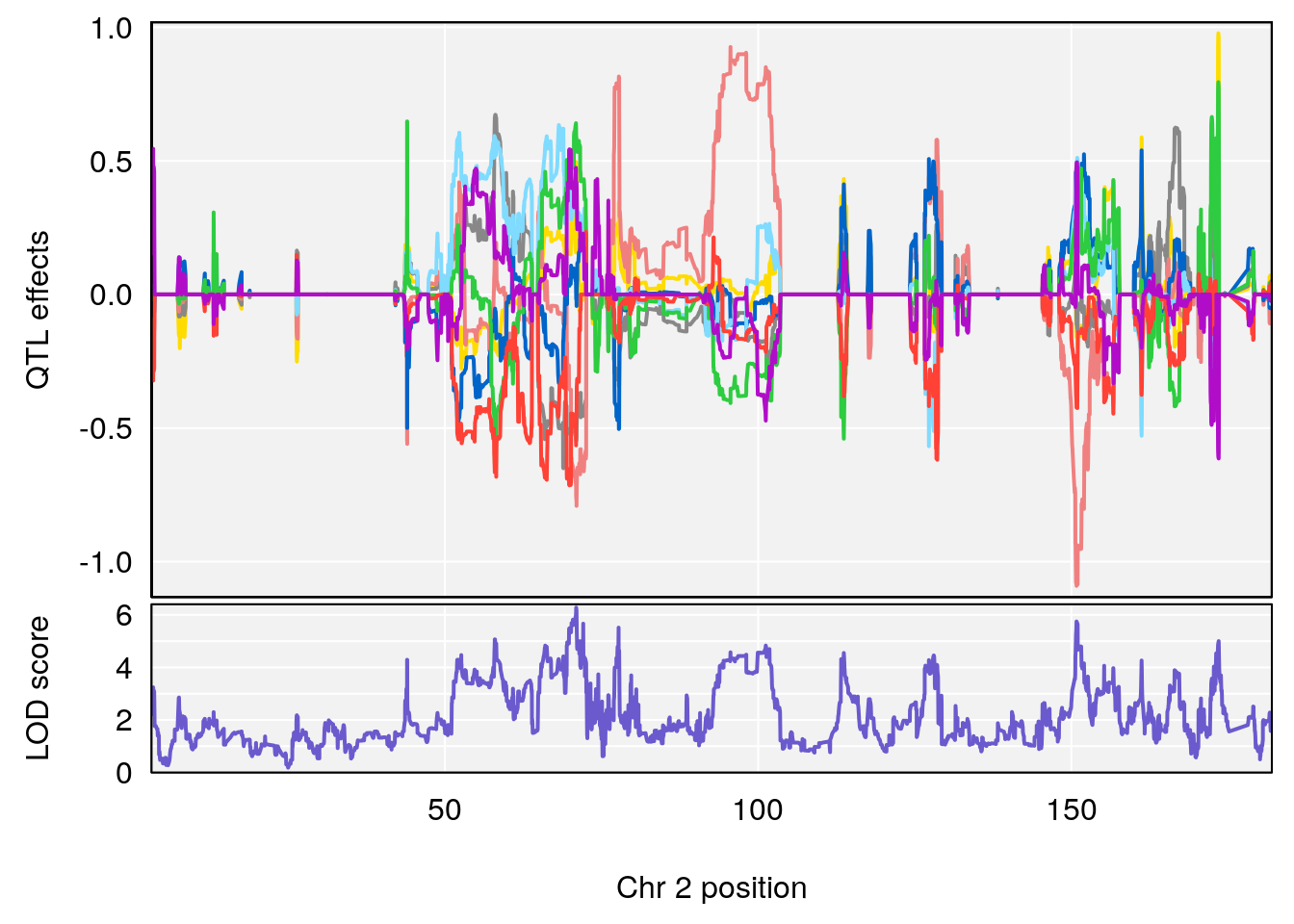
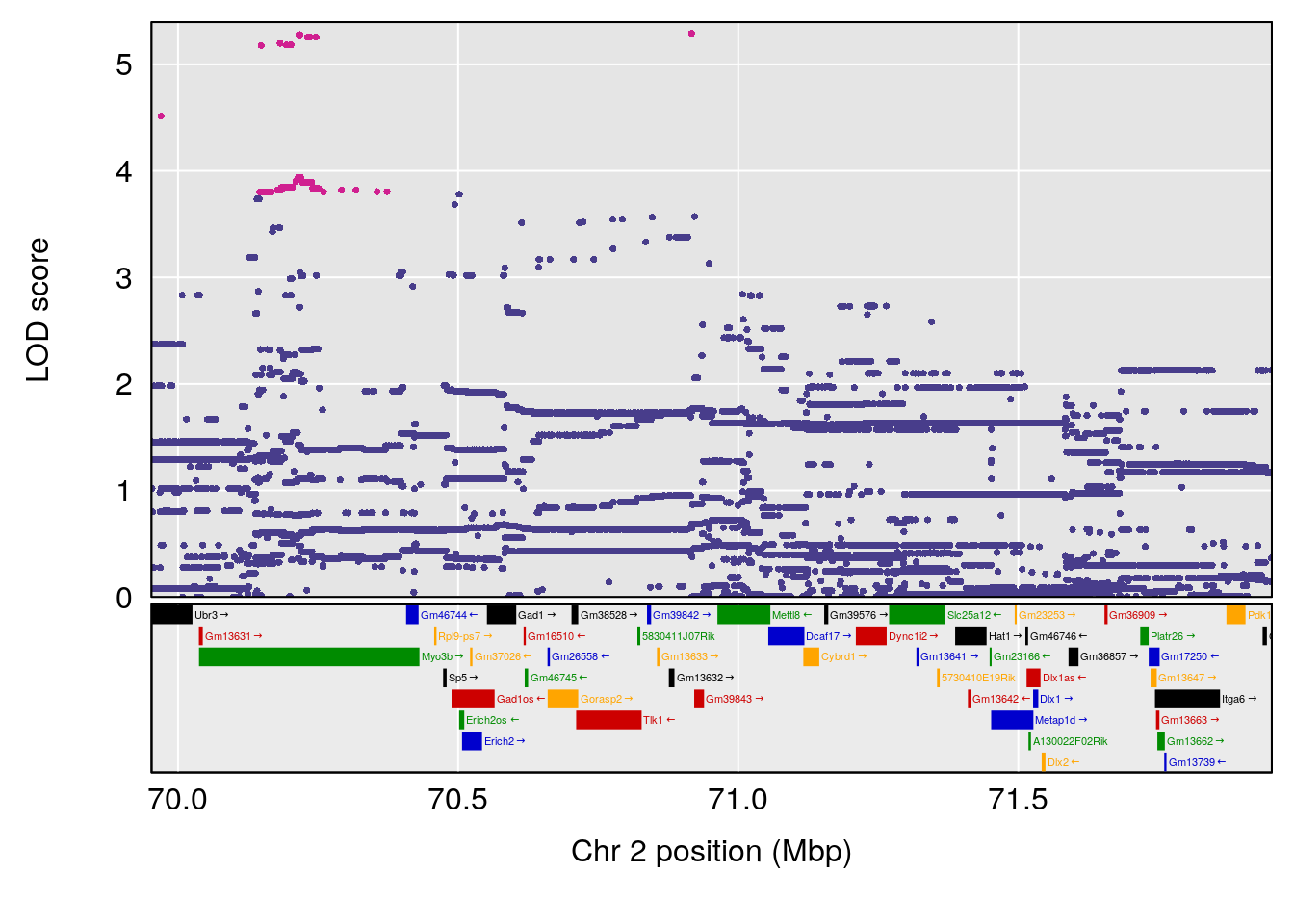
# 1 1 pheno1 2 70.95263 6.278855 57.899382 173.54300
# 2 1 pheno1 3 97.98775 6.445479 35.675687 130.29440
# 3 1 pheno1 4 30.34673 6.677948 28.777007 30.81005
# 4 1 pheno1 8 115.44116 7.553628 115.137119 115.98481
# 5 1 pheno1 15 29.67993 6.173329 7.017907 47.72534
# 6 1 pheno1 19 37.85897 6.861602 37.488231 38.17204
# [1] 1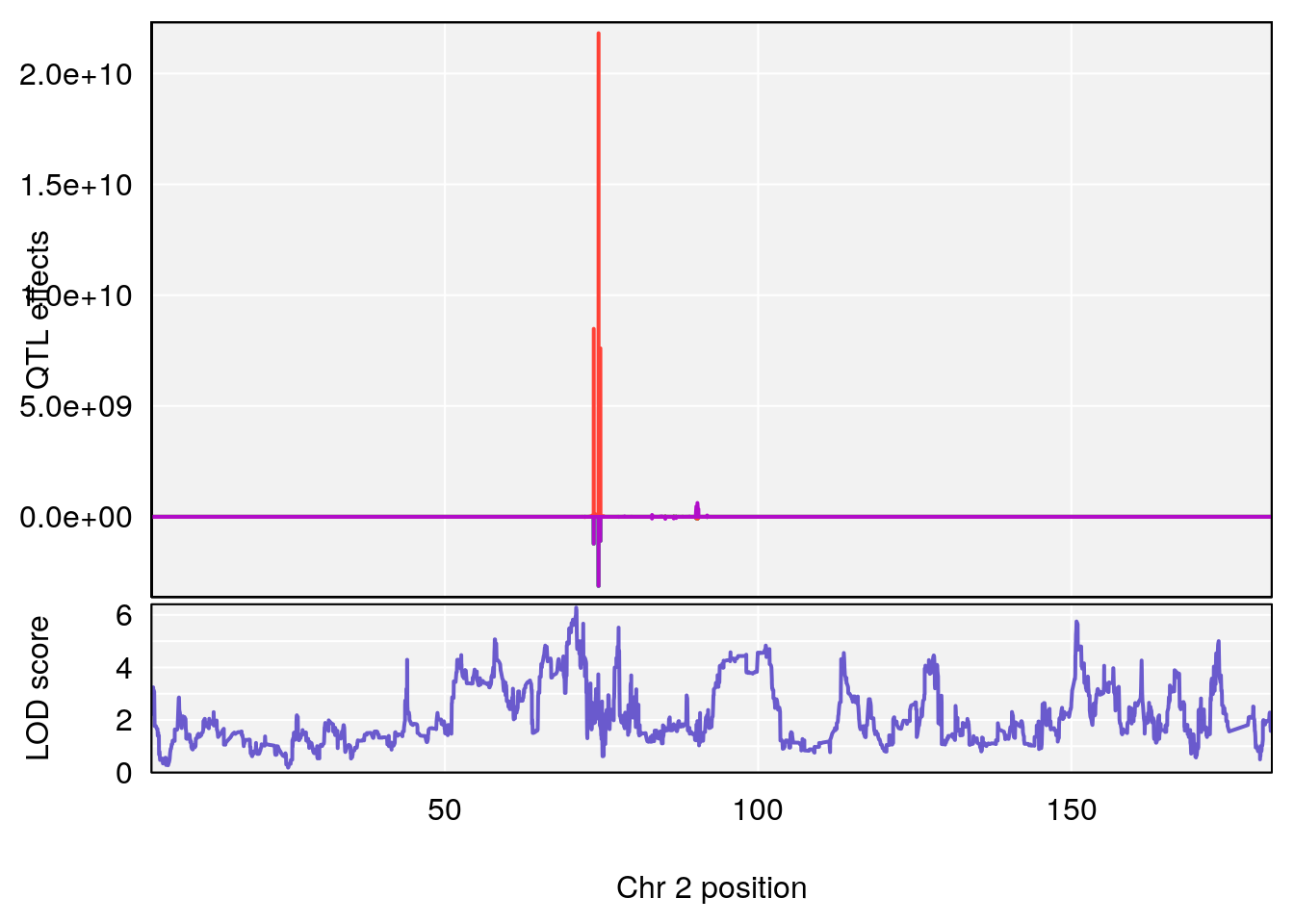
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 2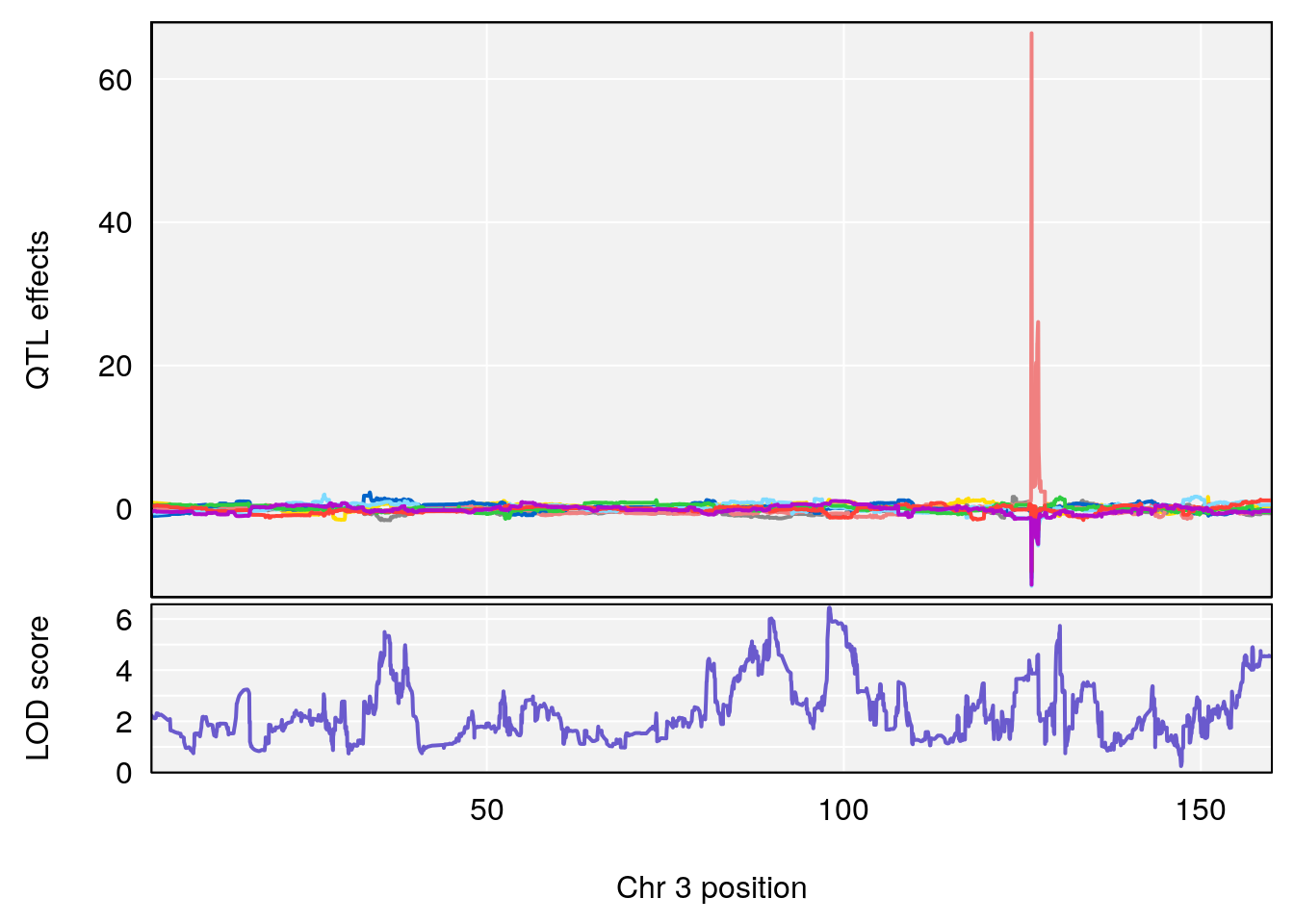
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 3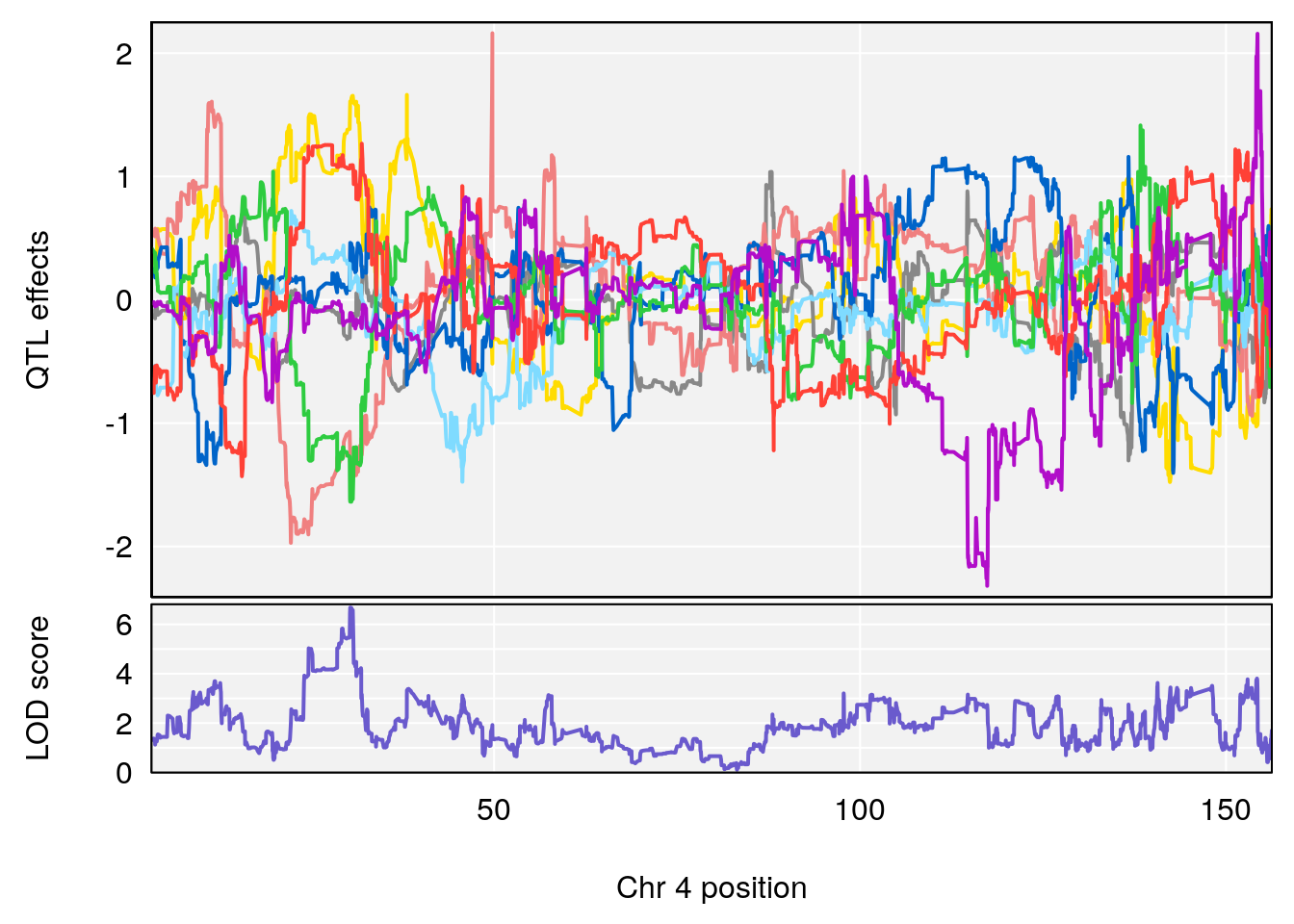
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 4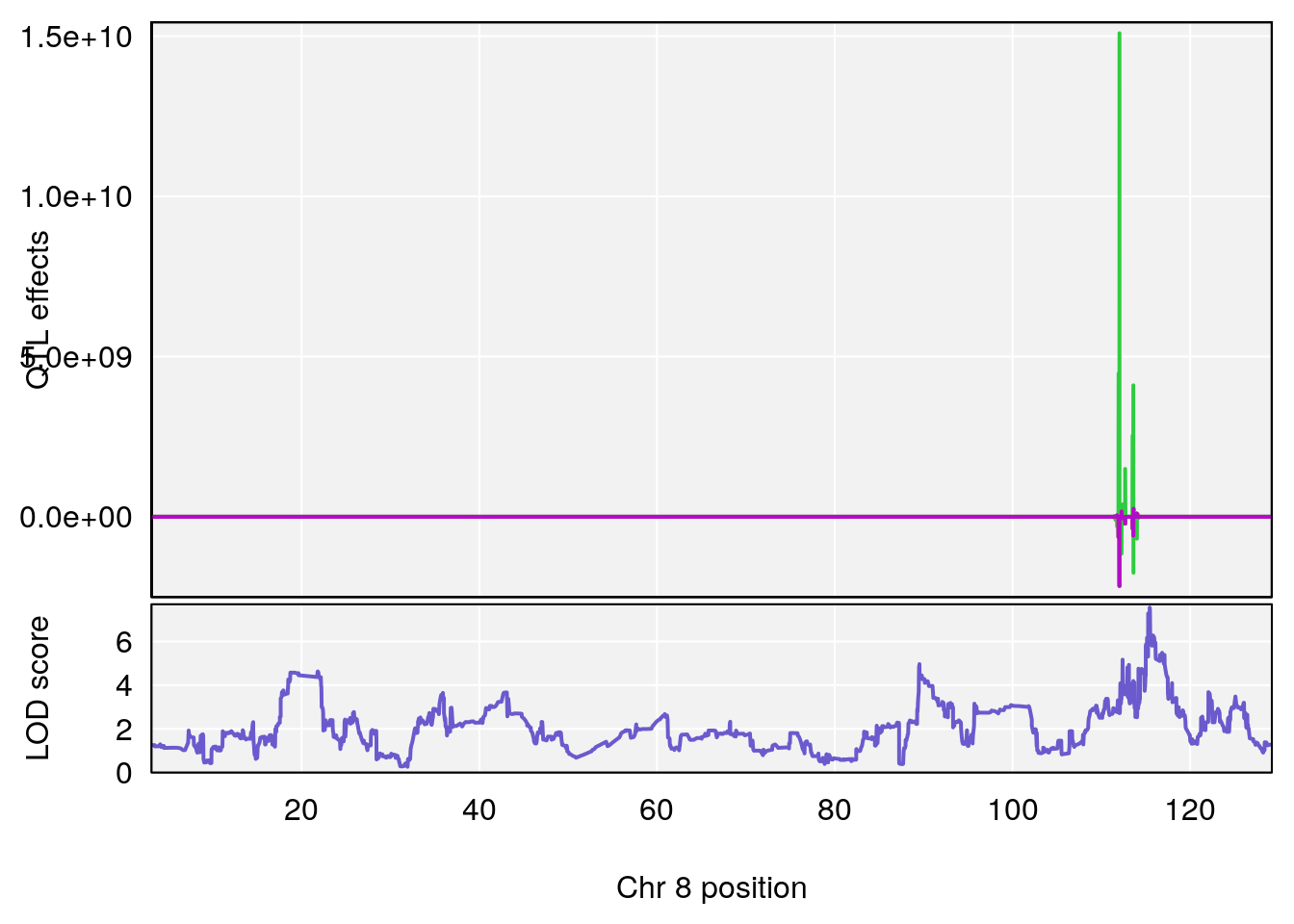
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 5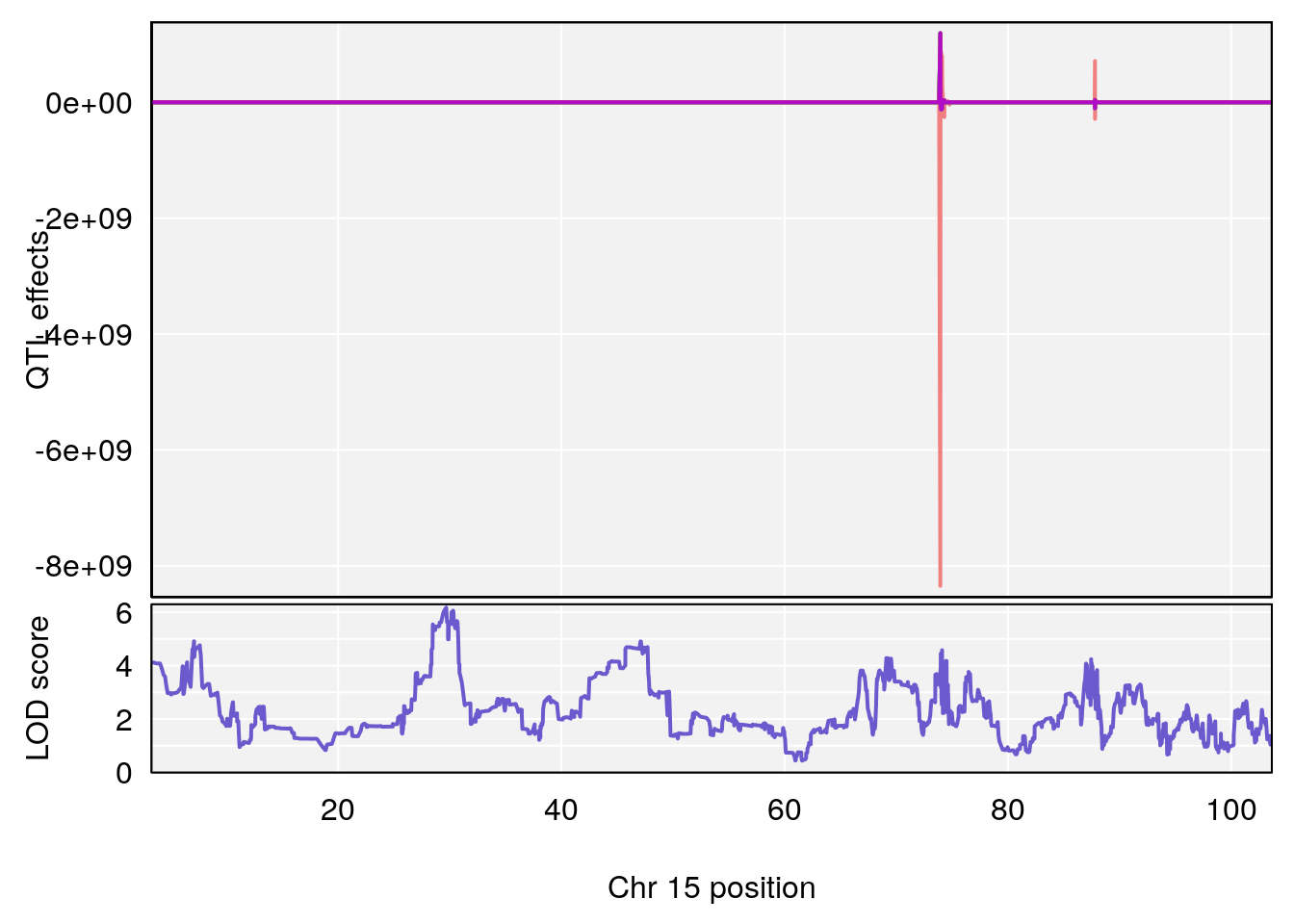
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
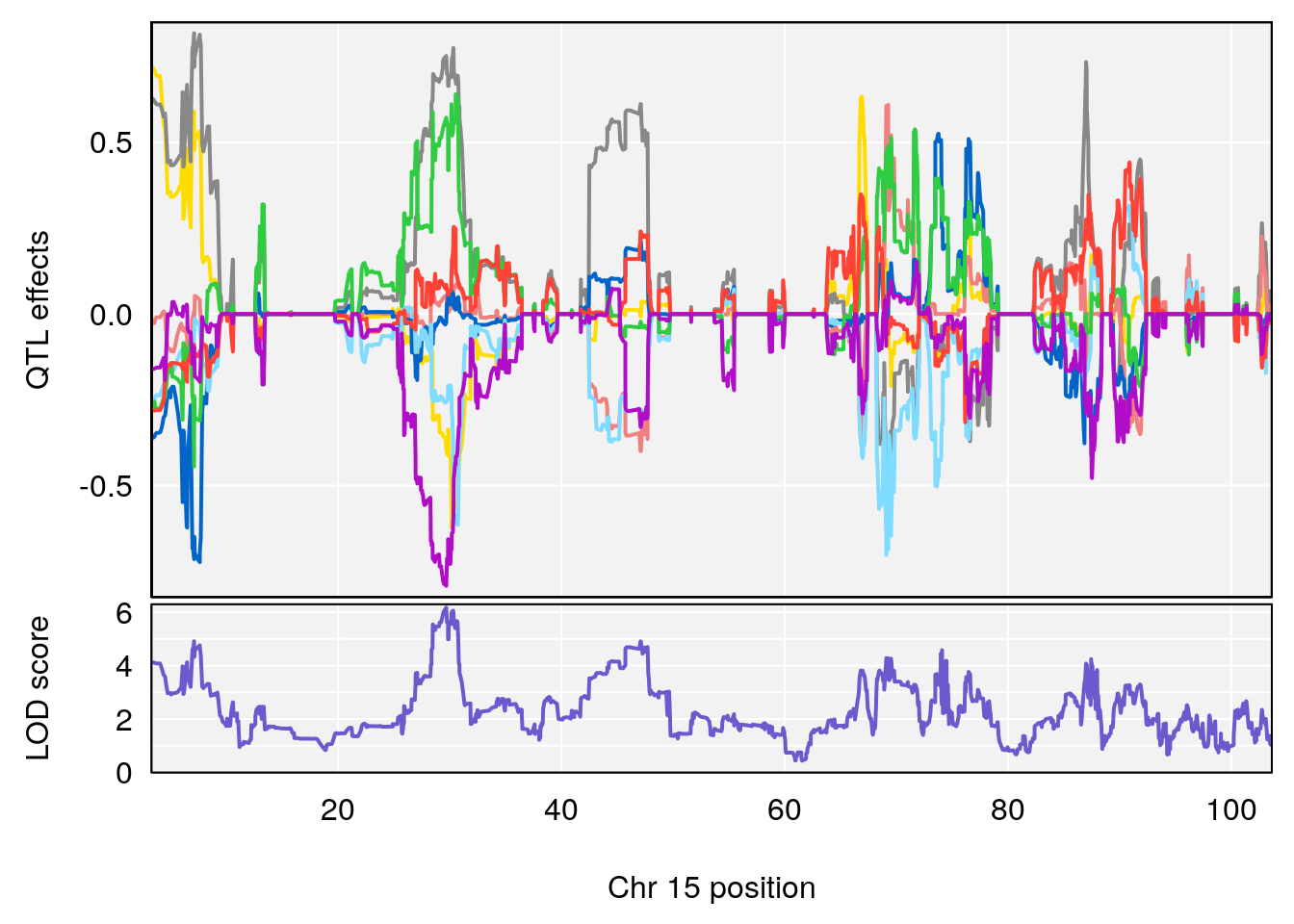
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
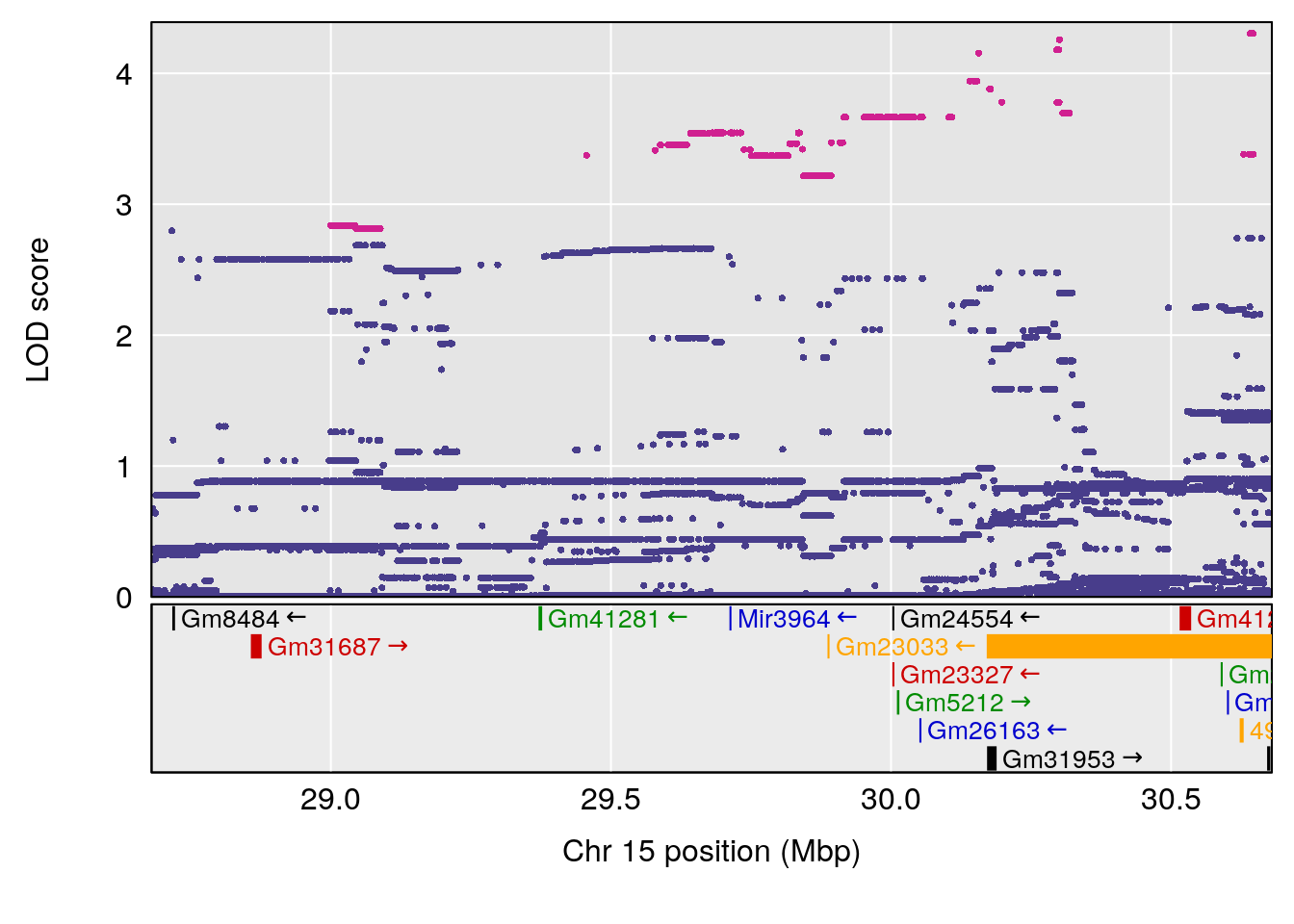
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 6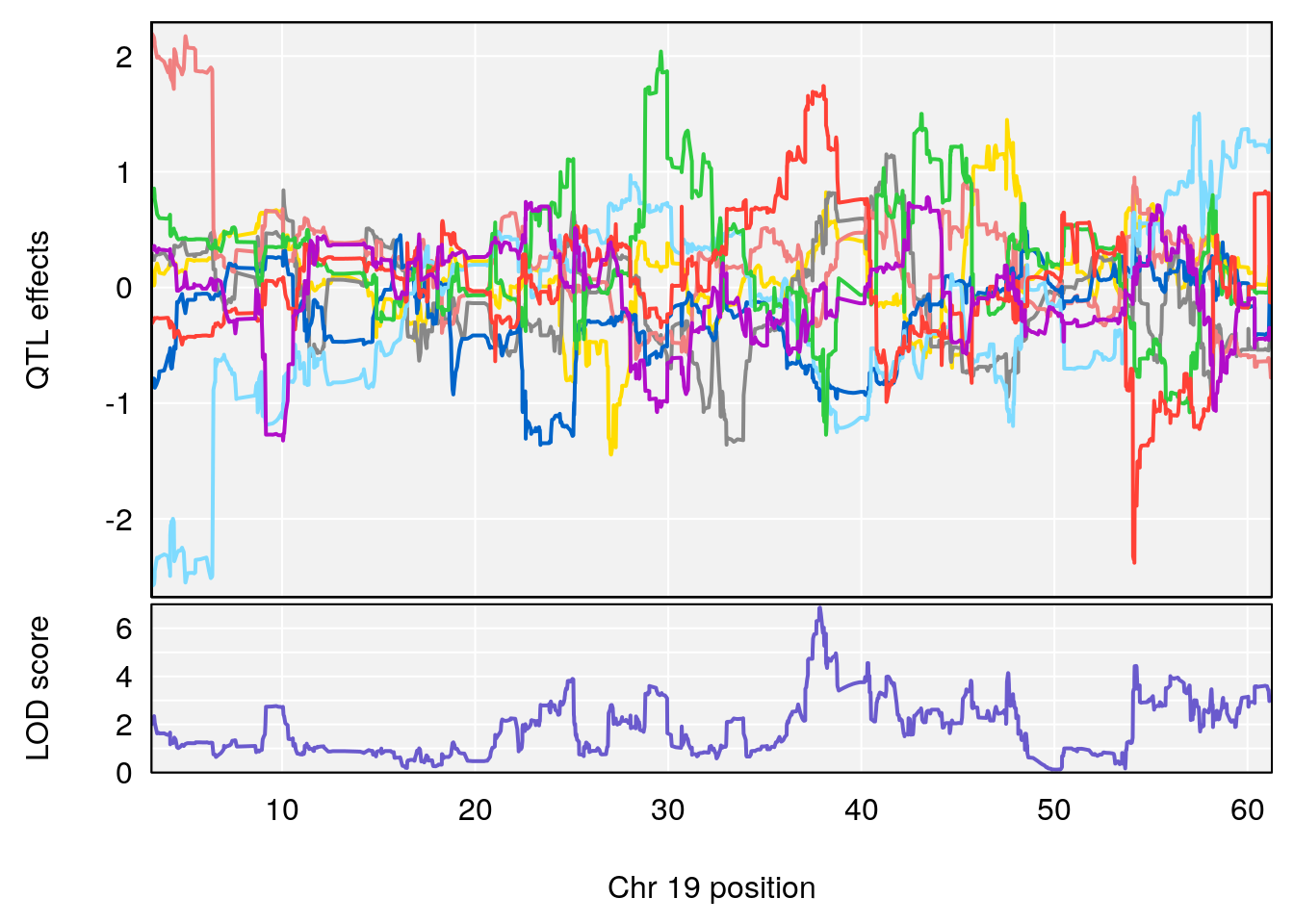
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
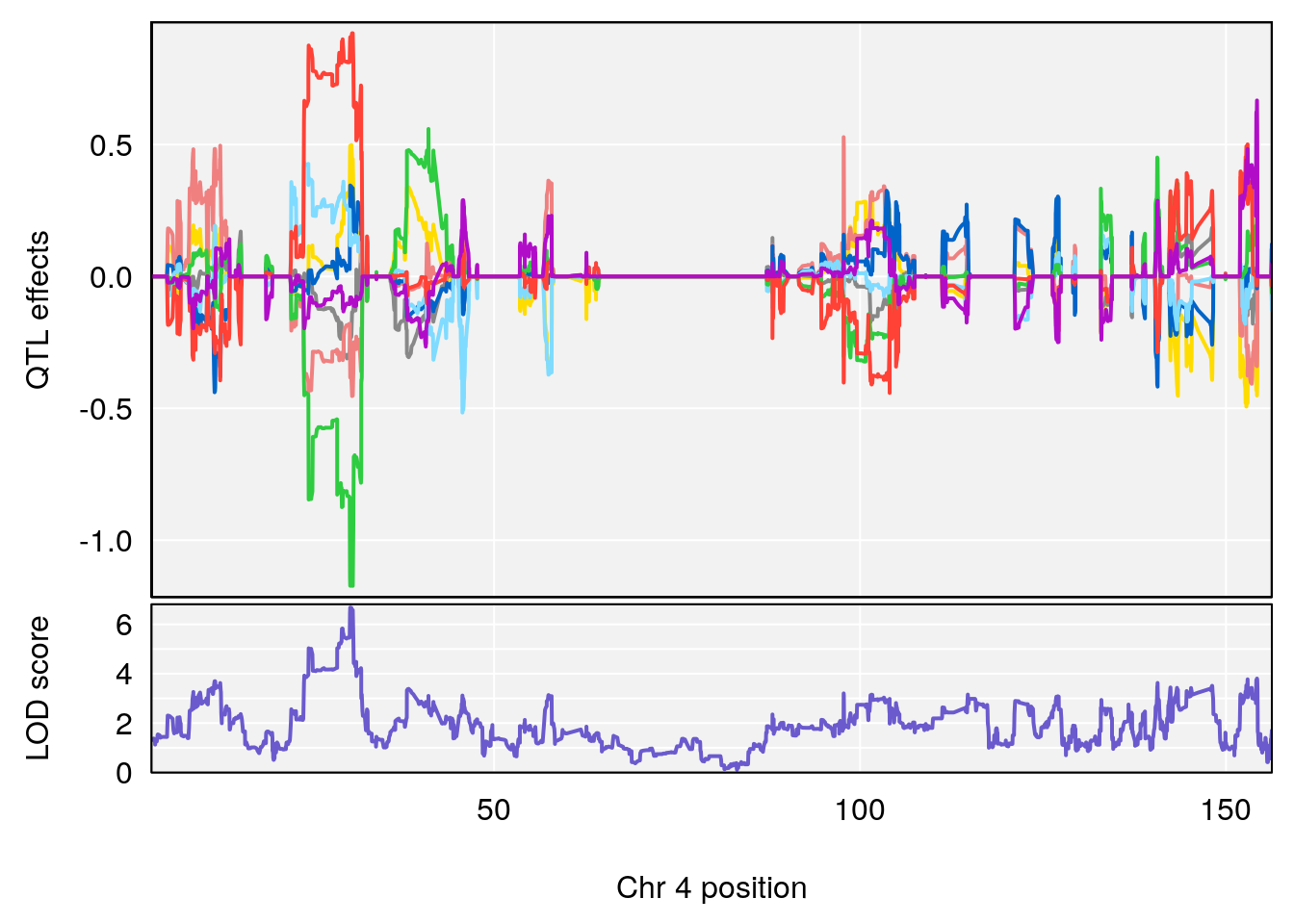
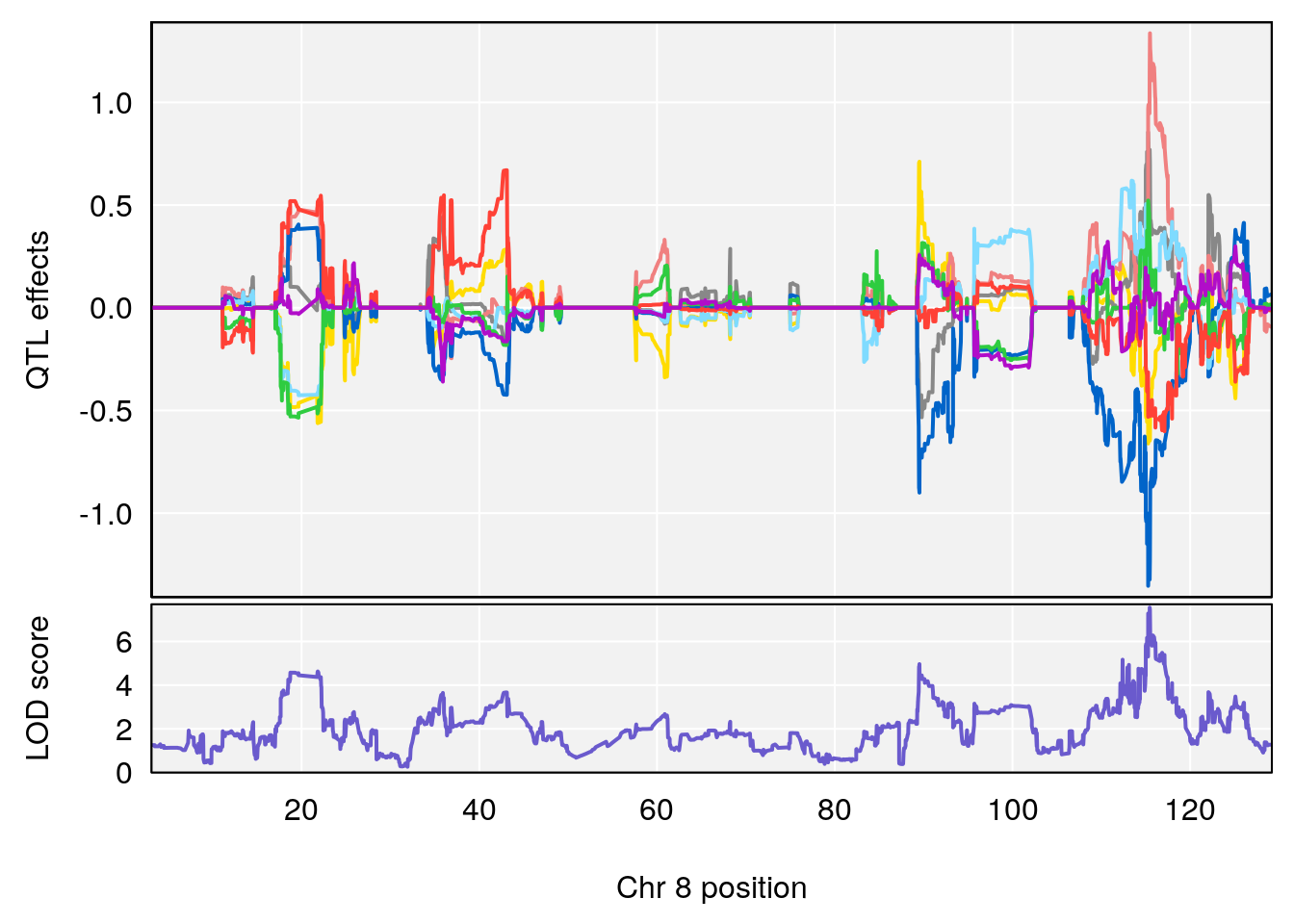
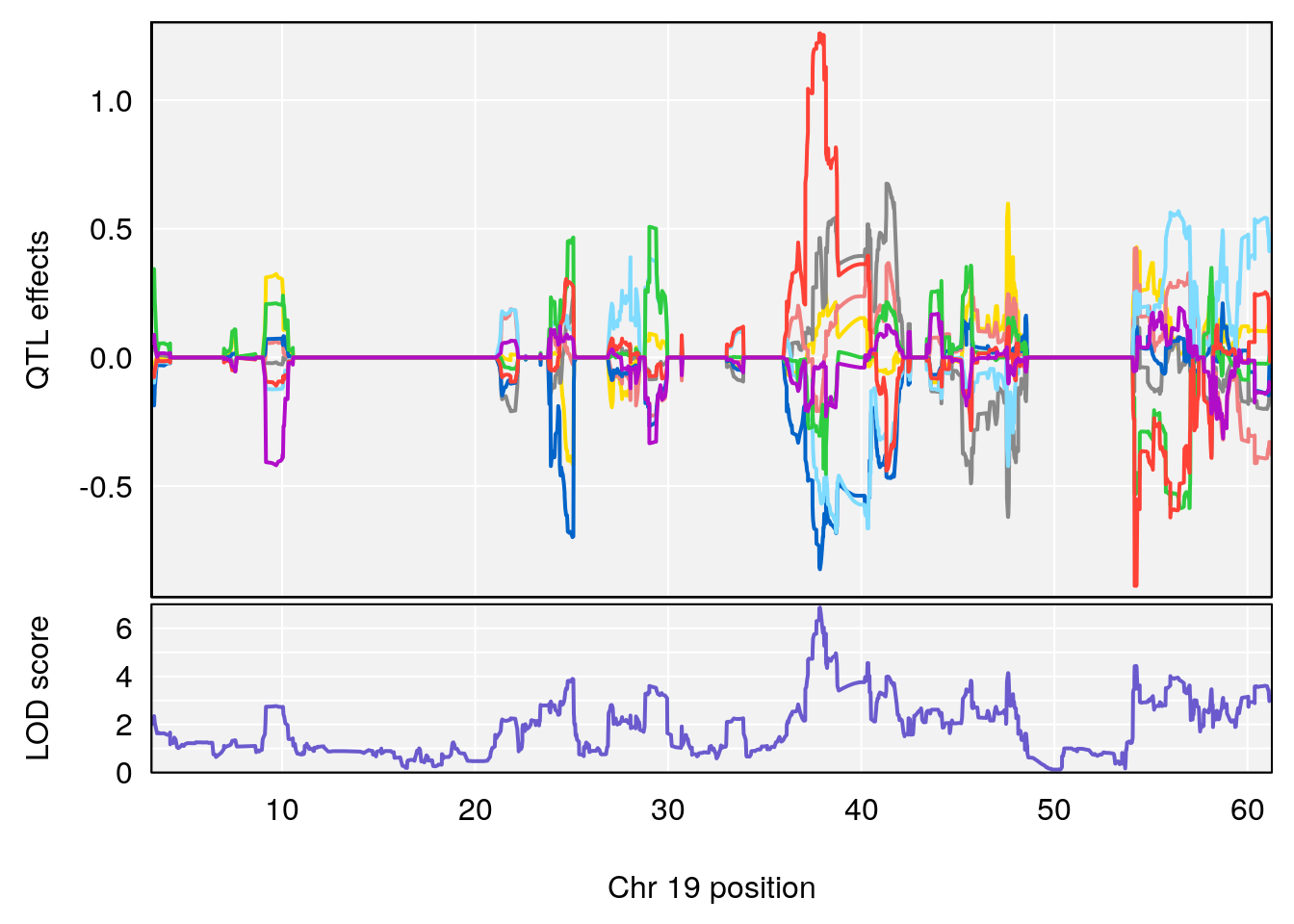
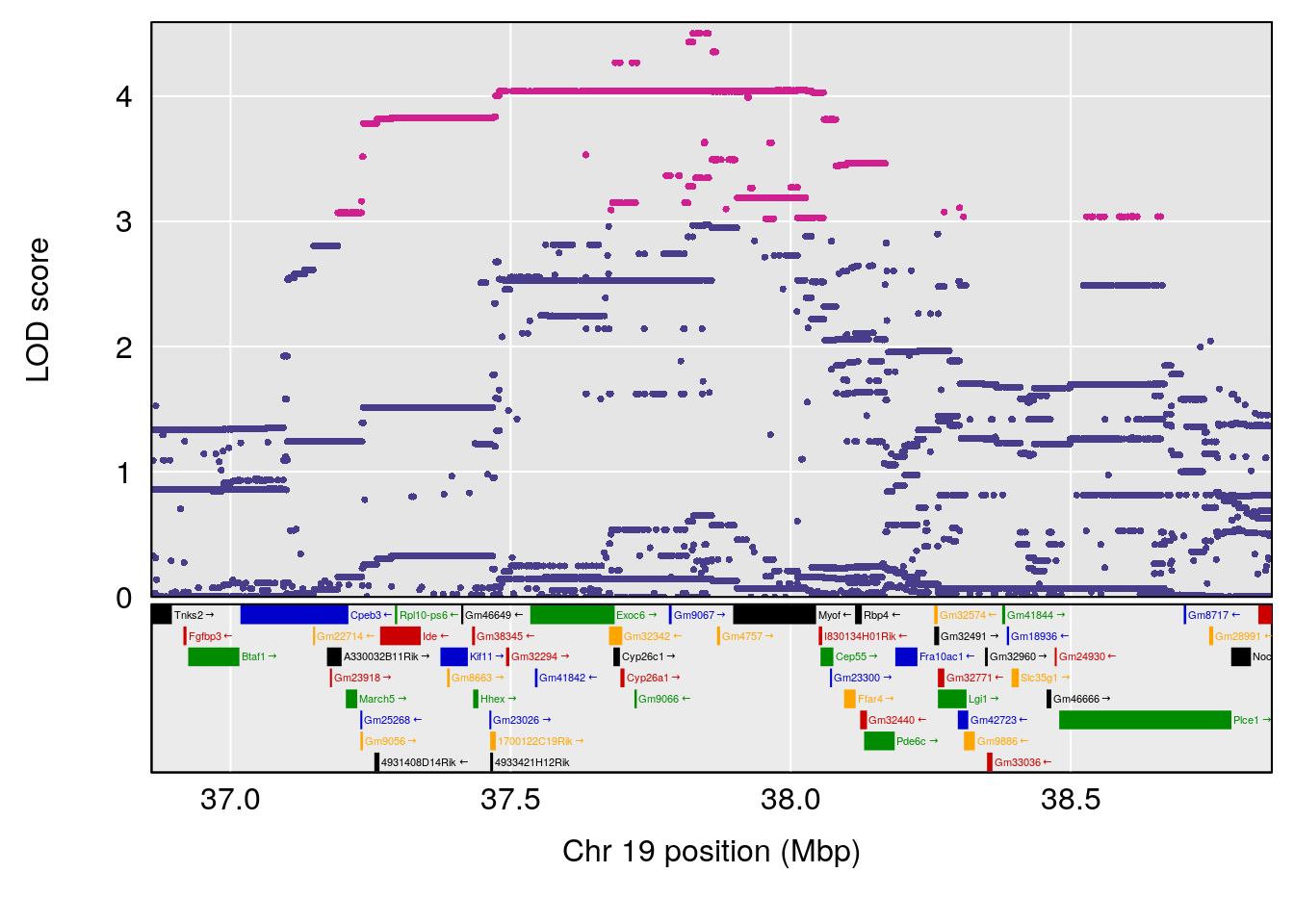
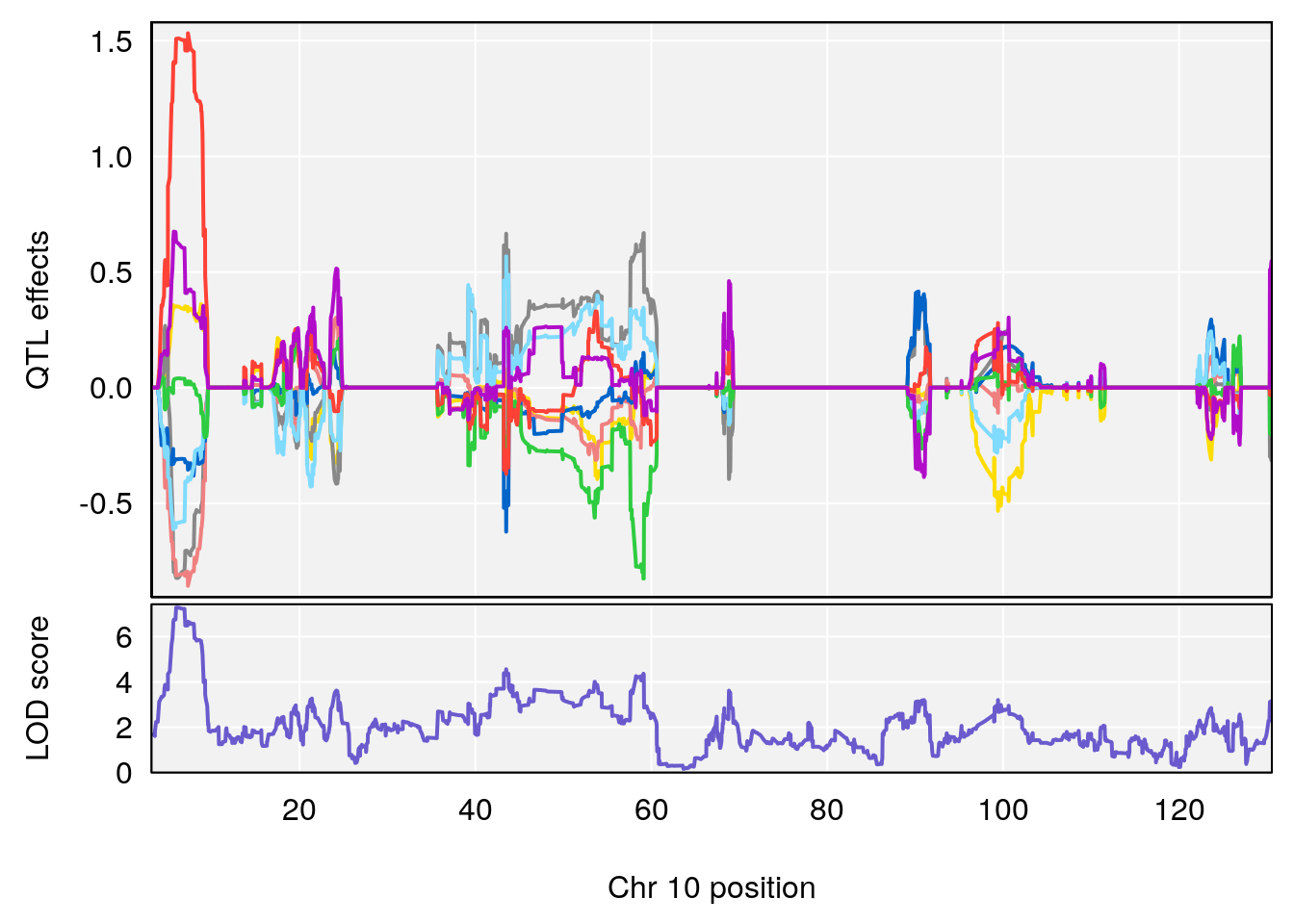
# [1] "Recovery.Time"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 3 123.593760 6.464420 117.457785 123.638818
# 2 1 pheno1 4 156.048178 6.417408 114.723317 156.119928
# 3 1 pheno1 5 92.743909 6.654942 37.152061 95.838678
# 4 1 pheno1 10 5.988497 7.279613 5.445056 8.819529
# 5 1 pheno1 13 60.185807 6.793807 59.610678 63.542749
# [1] 1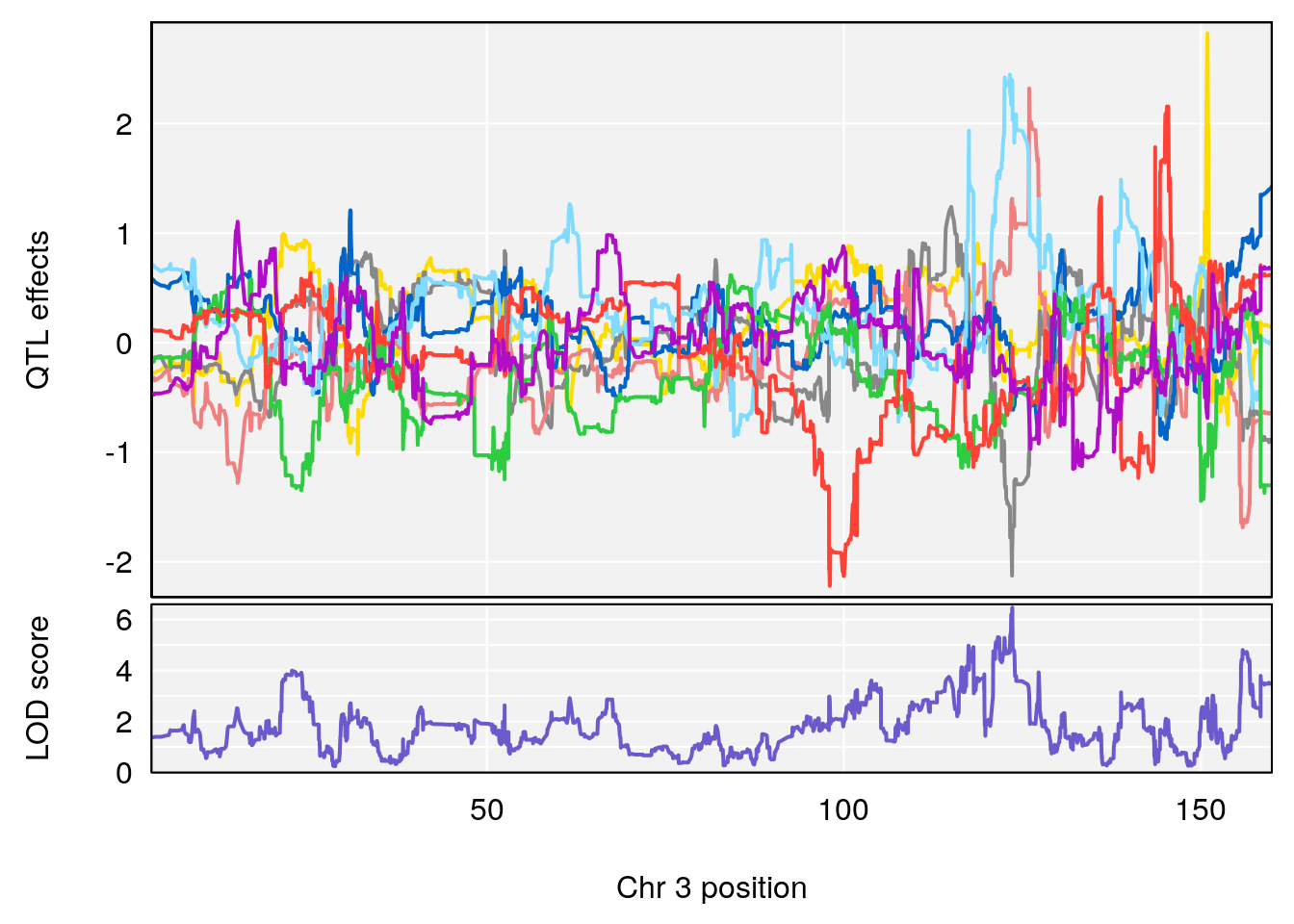
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
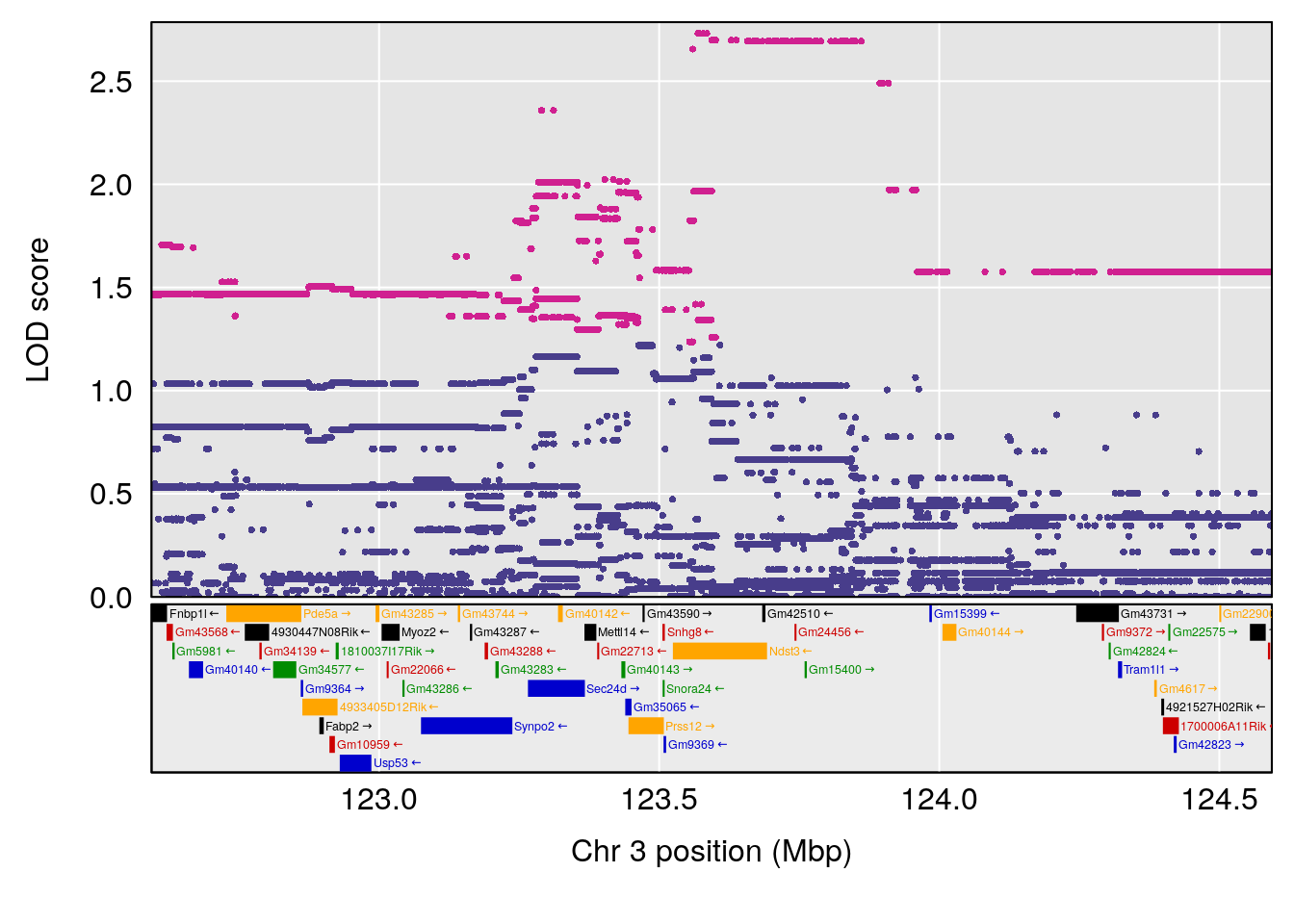
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 2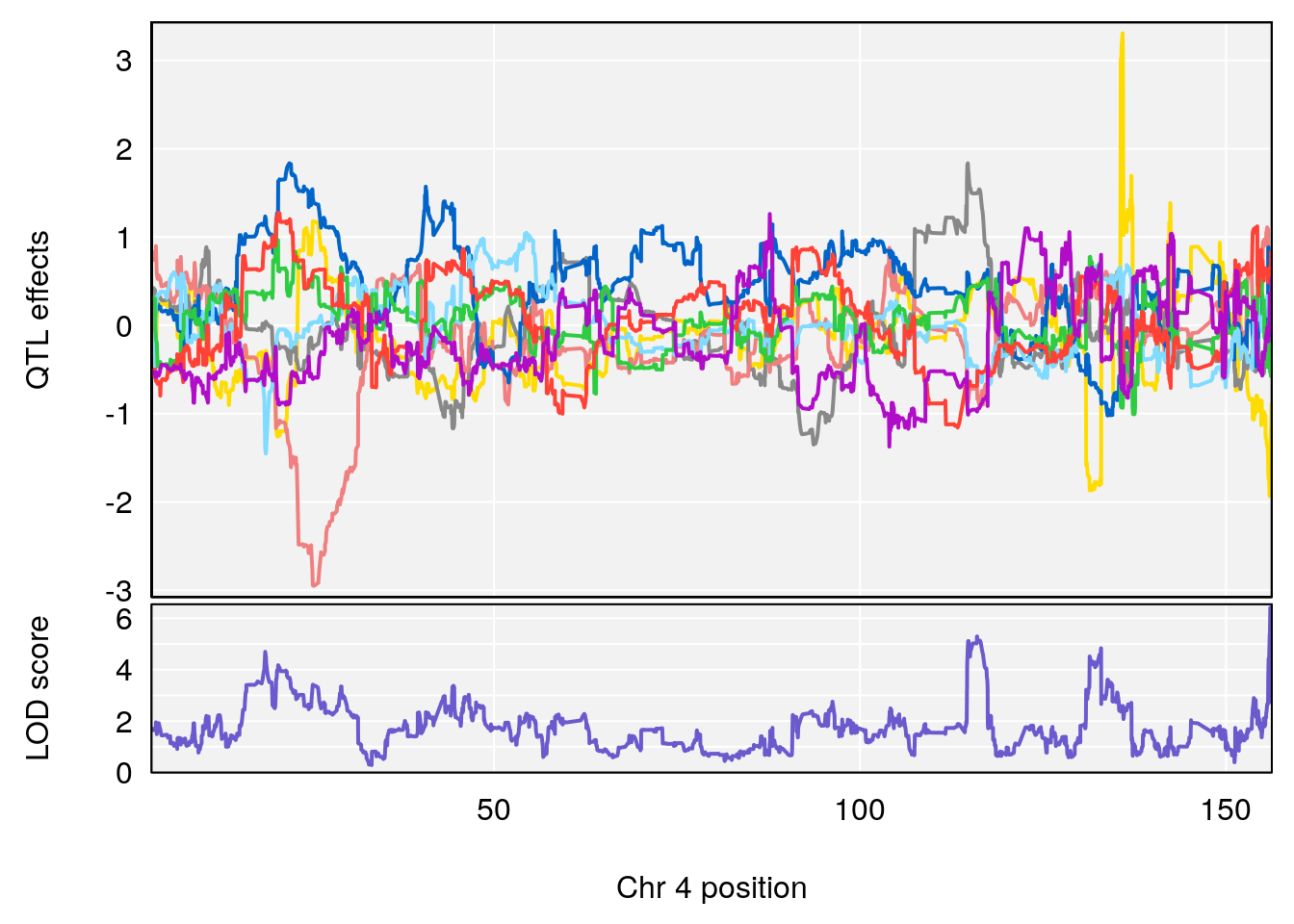
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
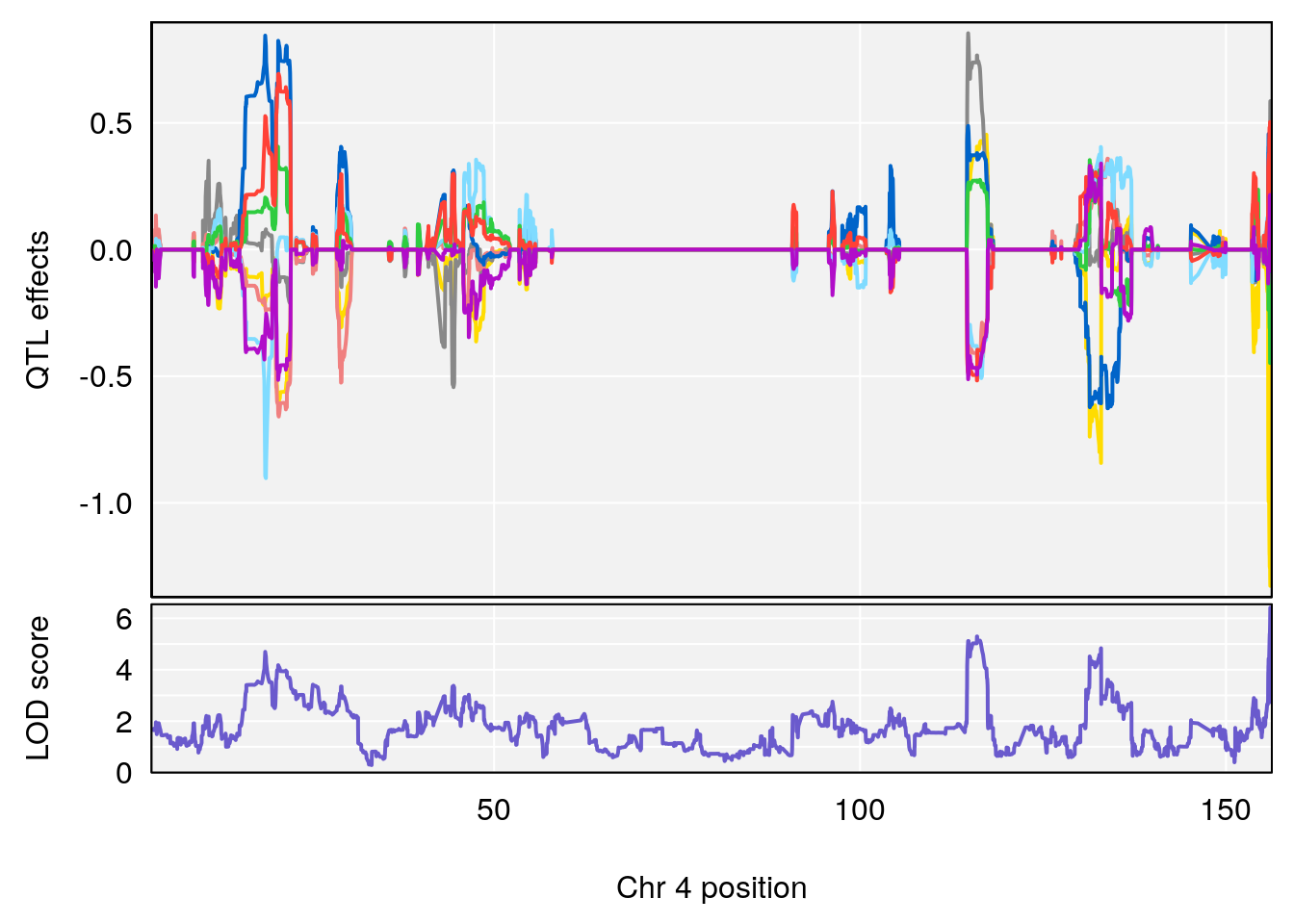
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |

| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 3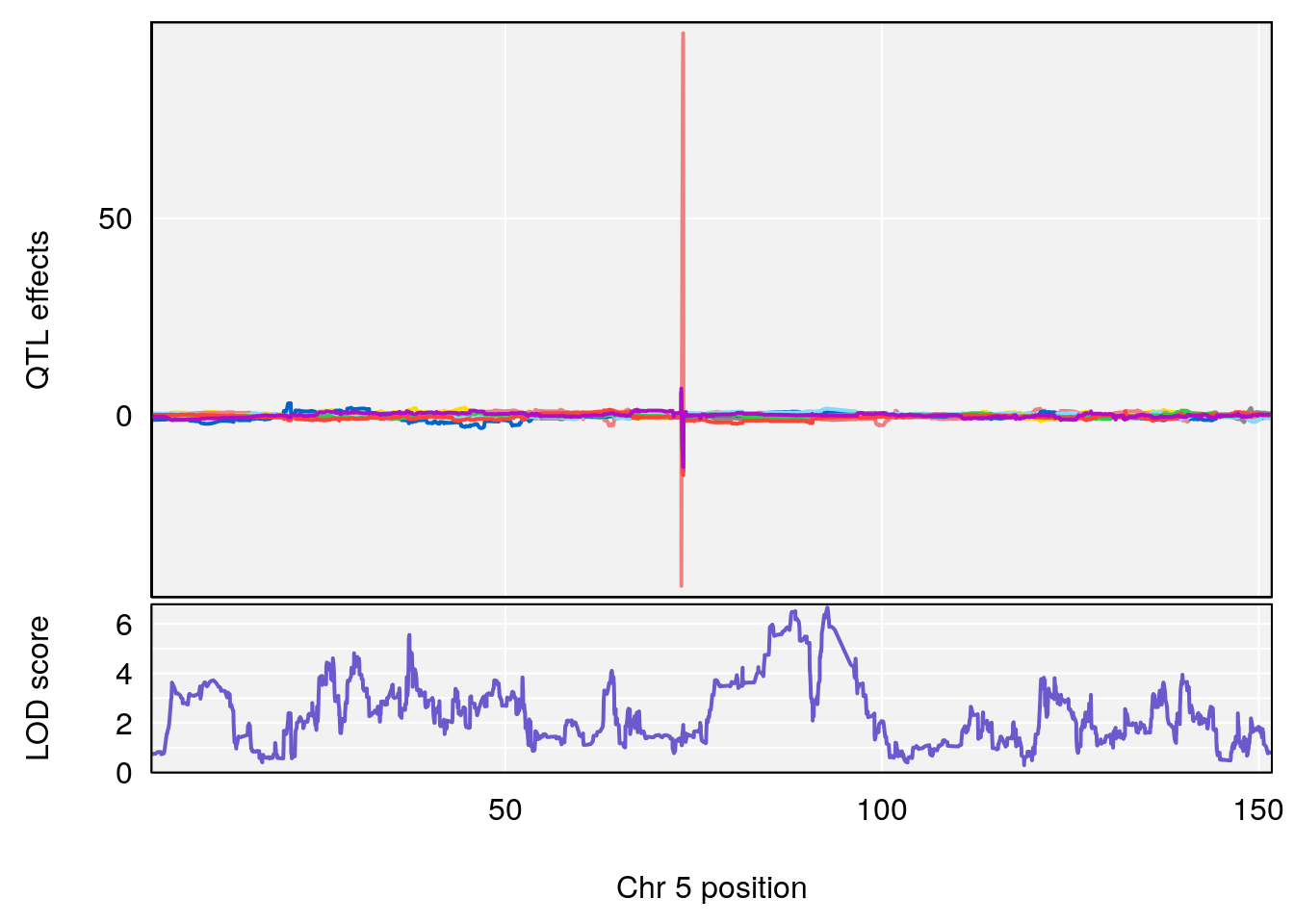
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 4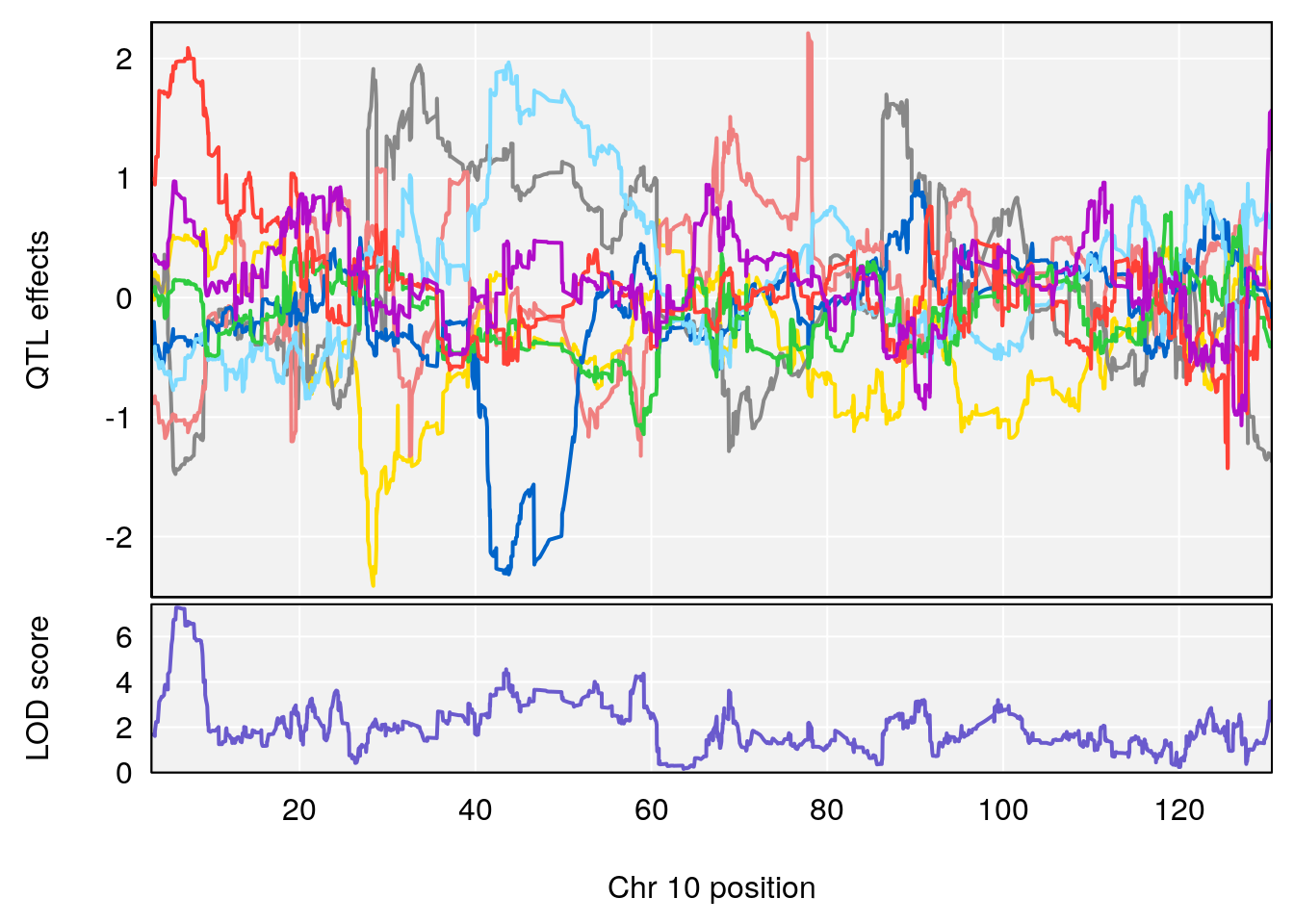
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
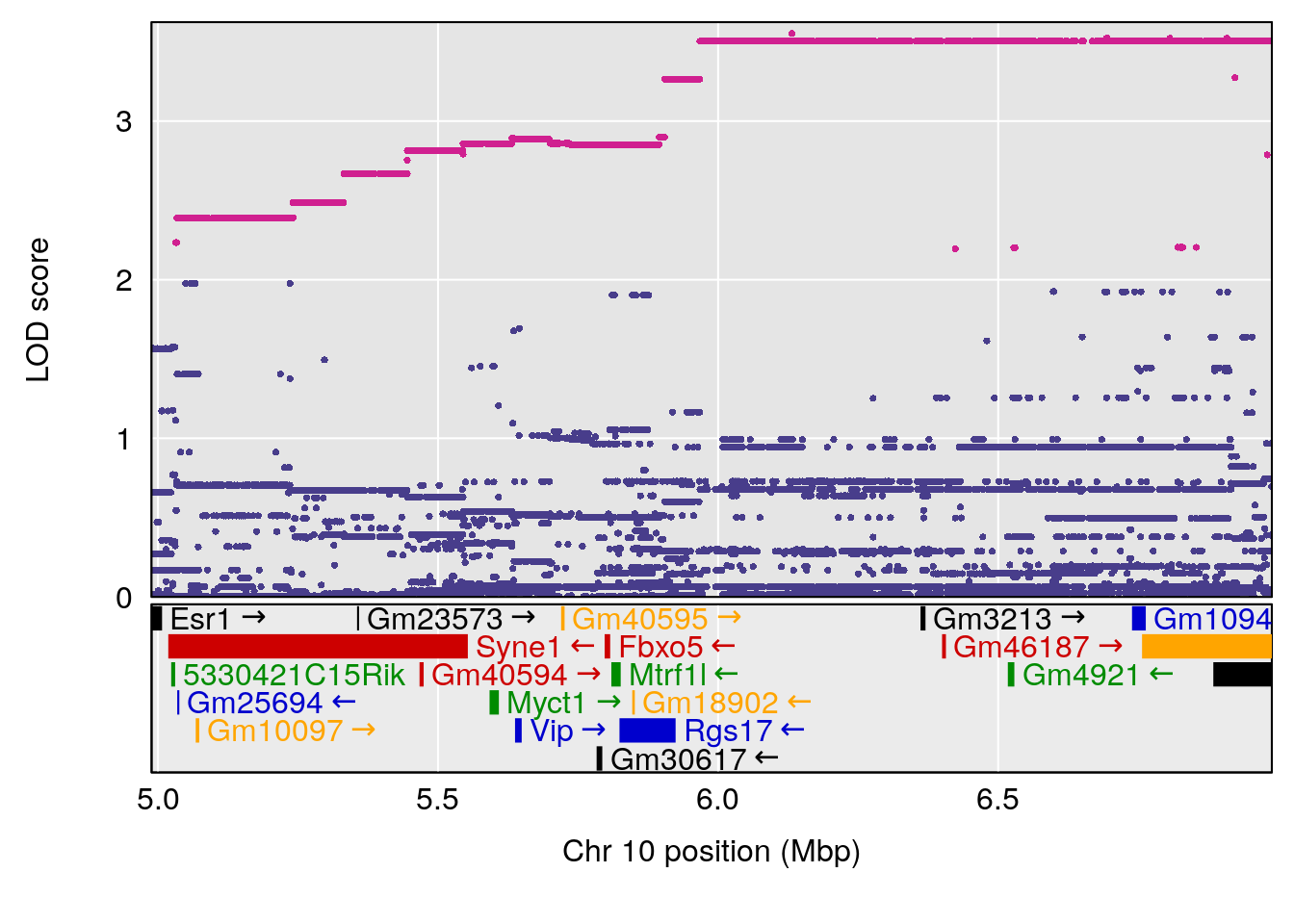
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 5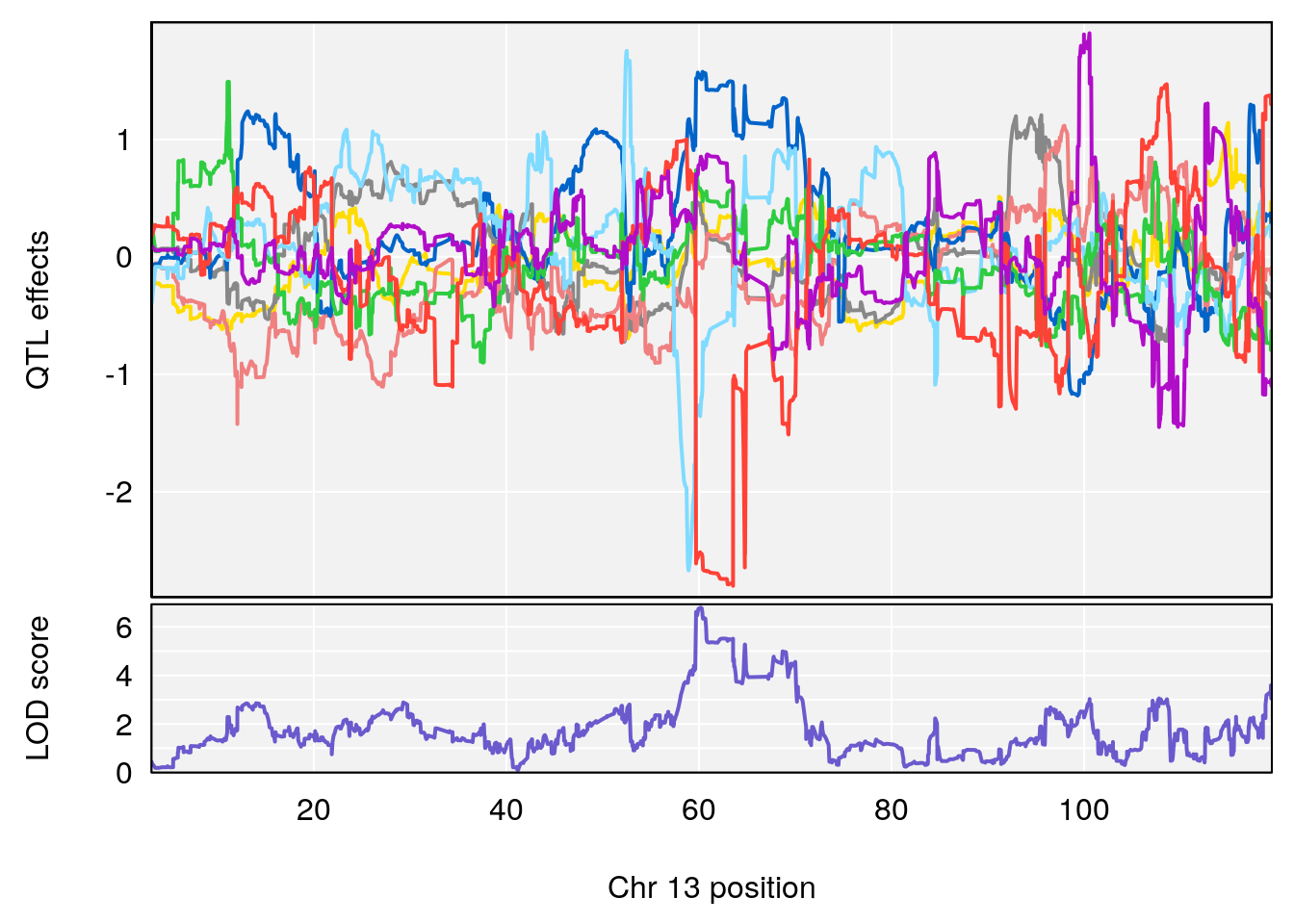
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
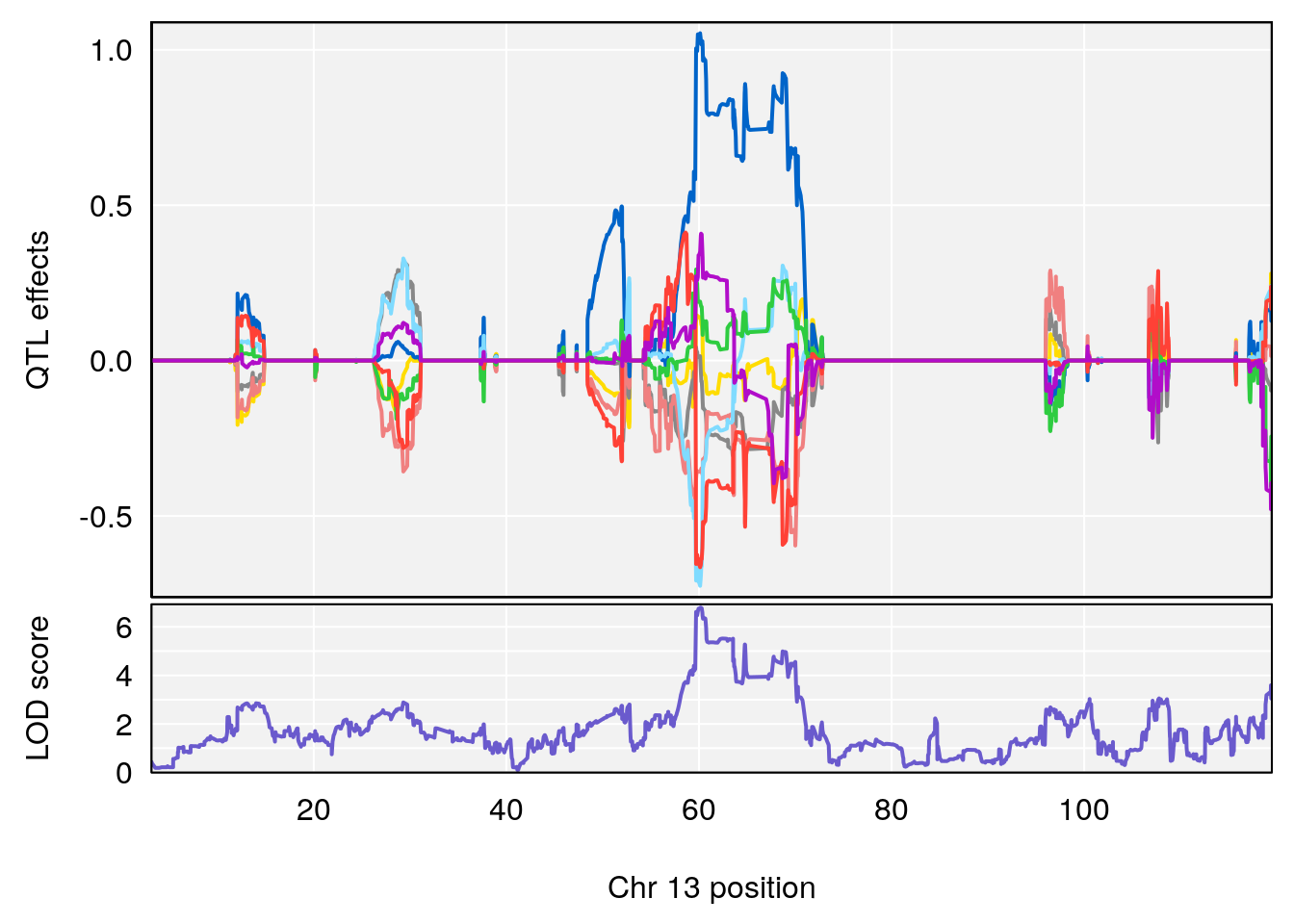
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
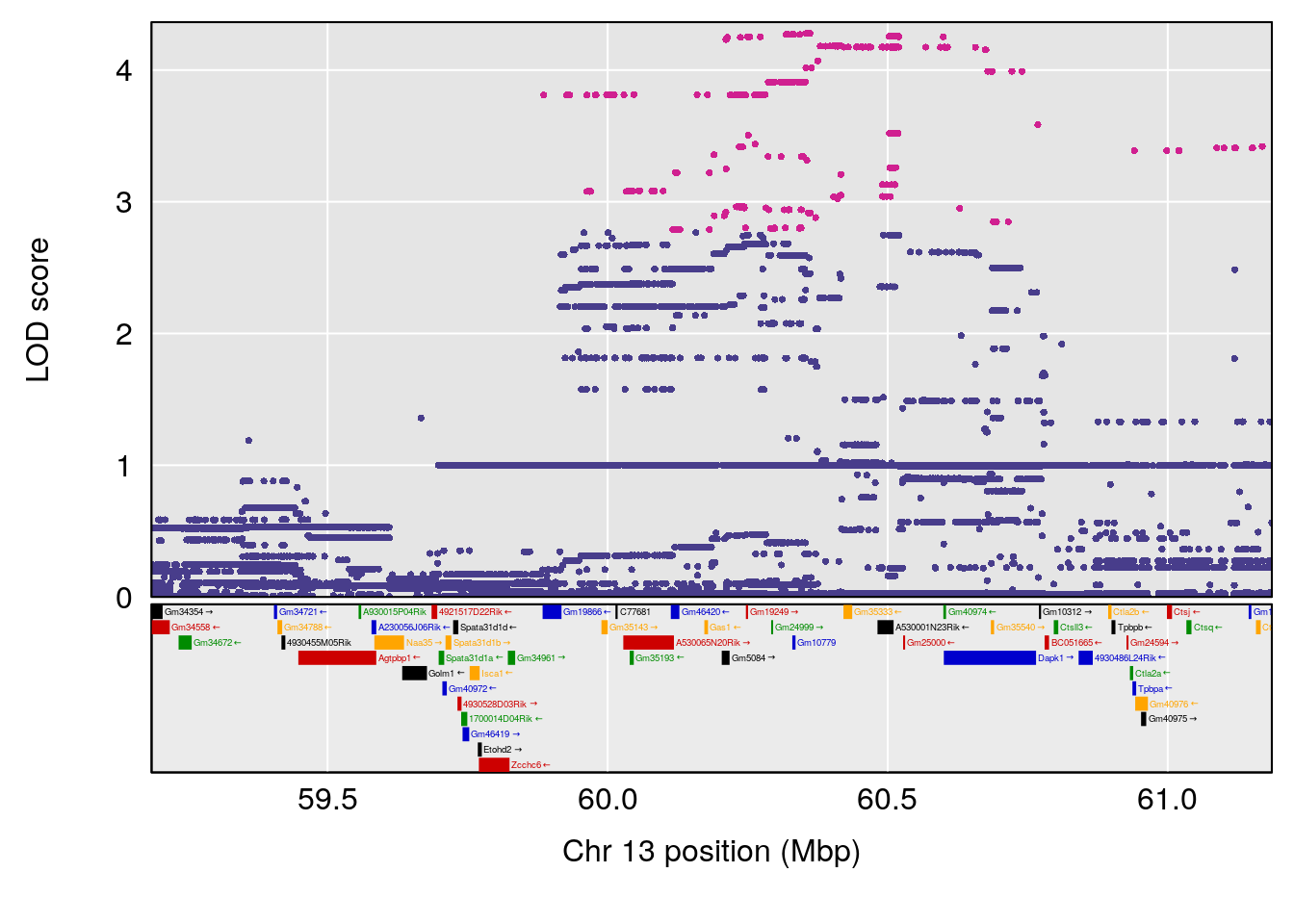
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
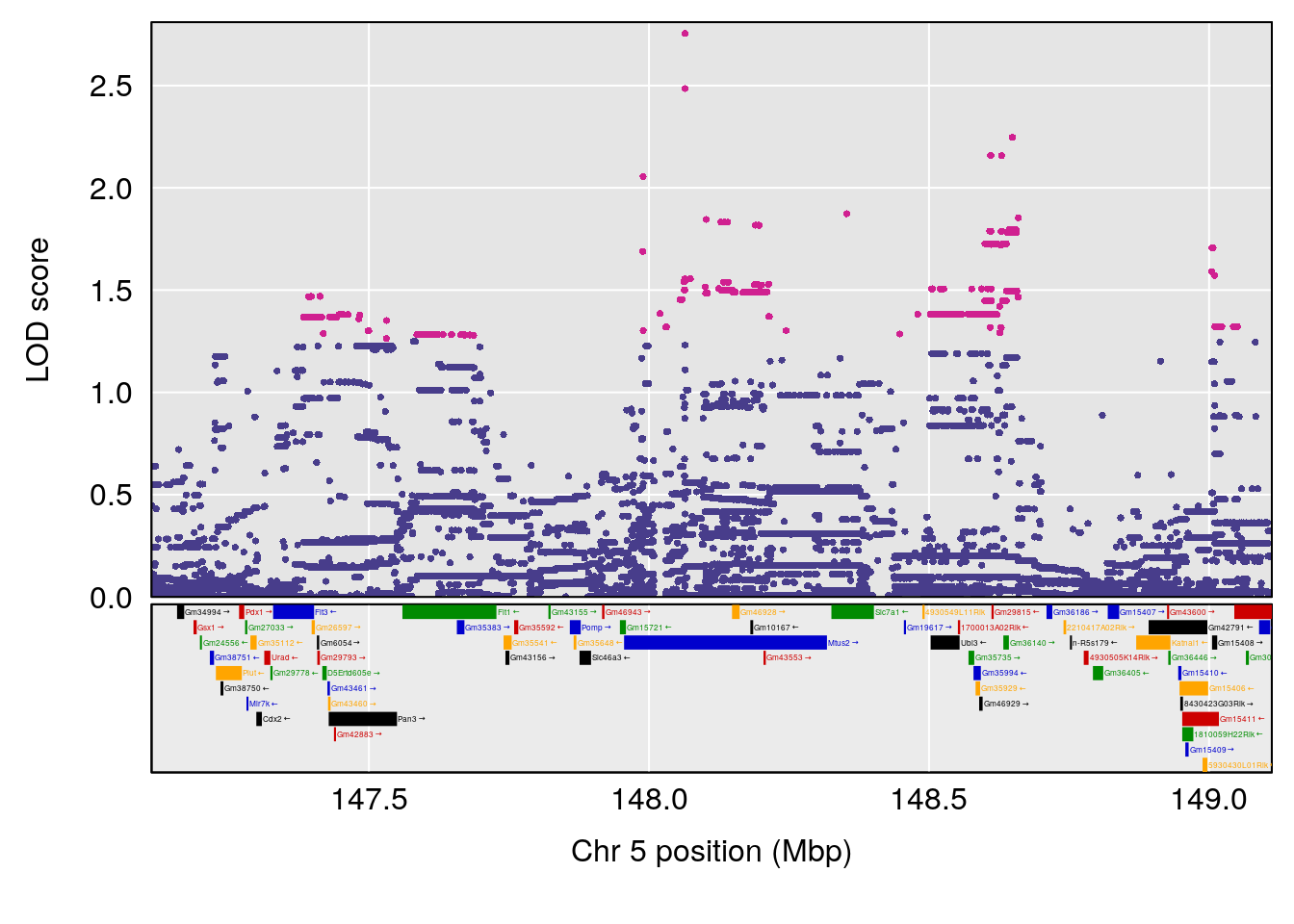
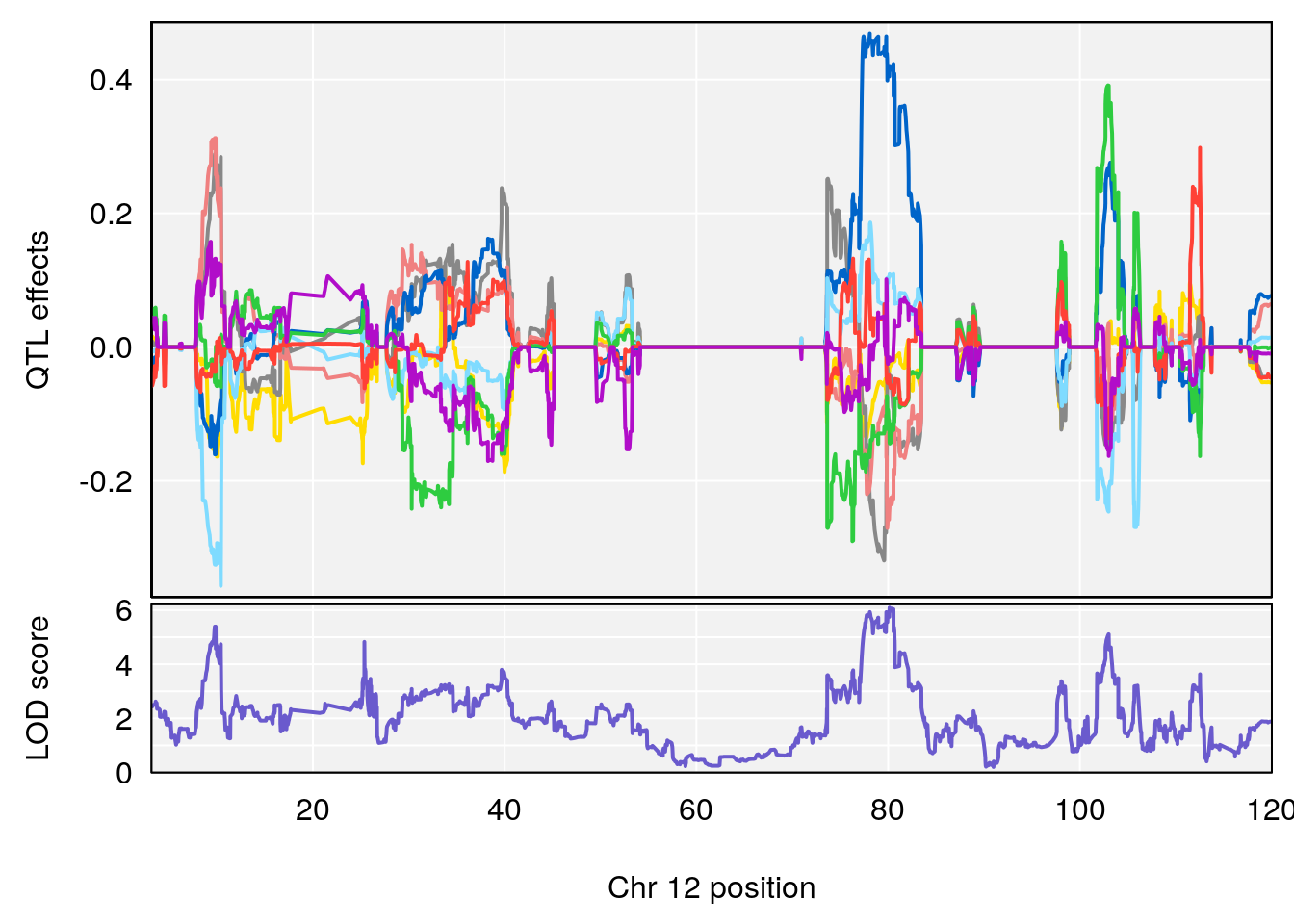
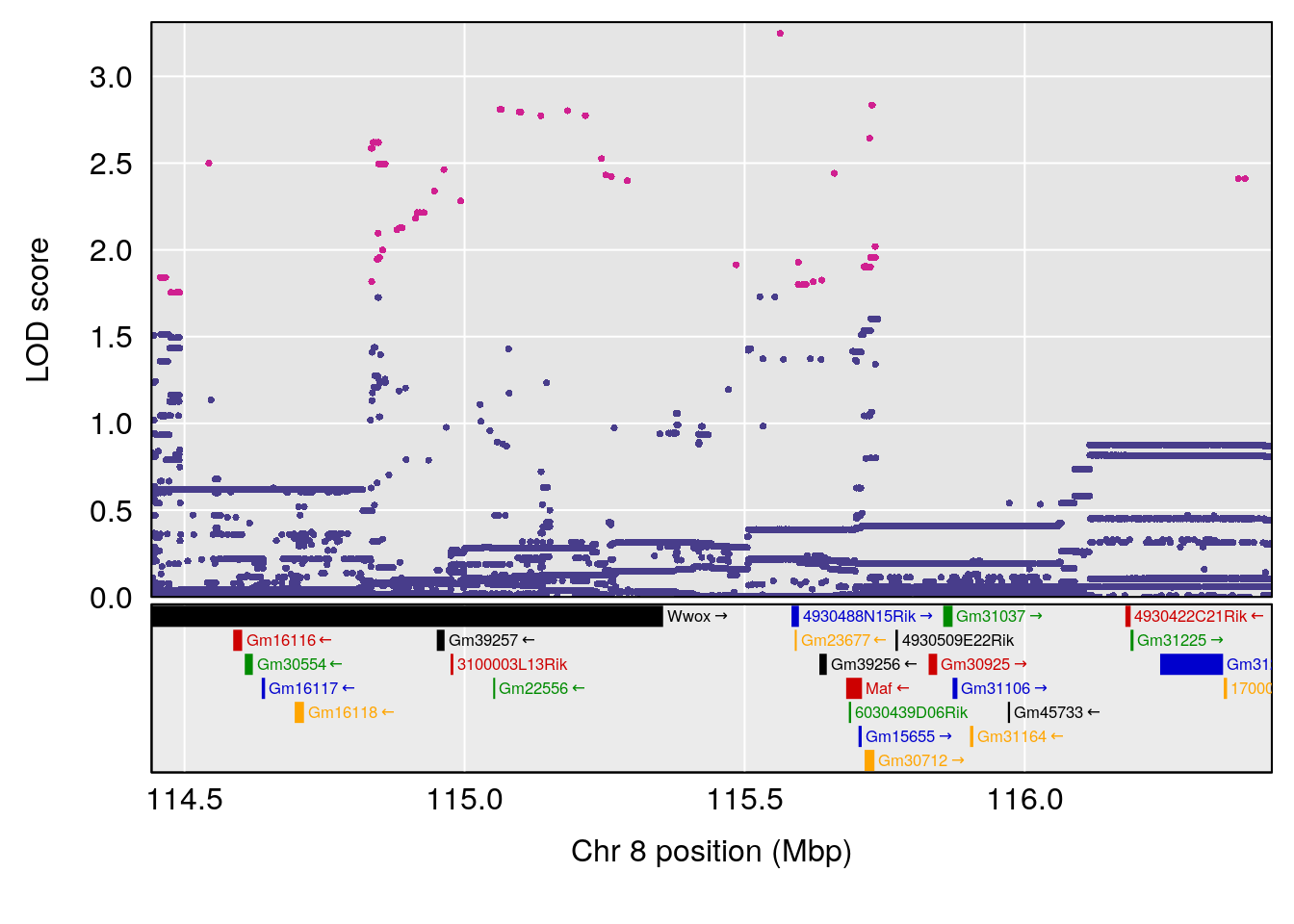
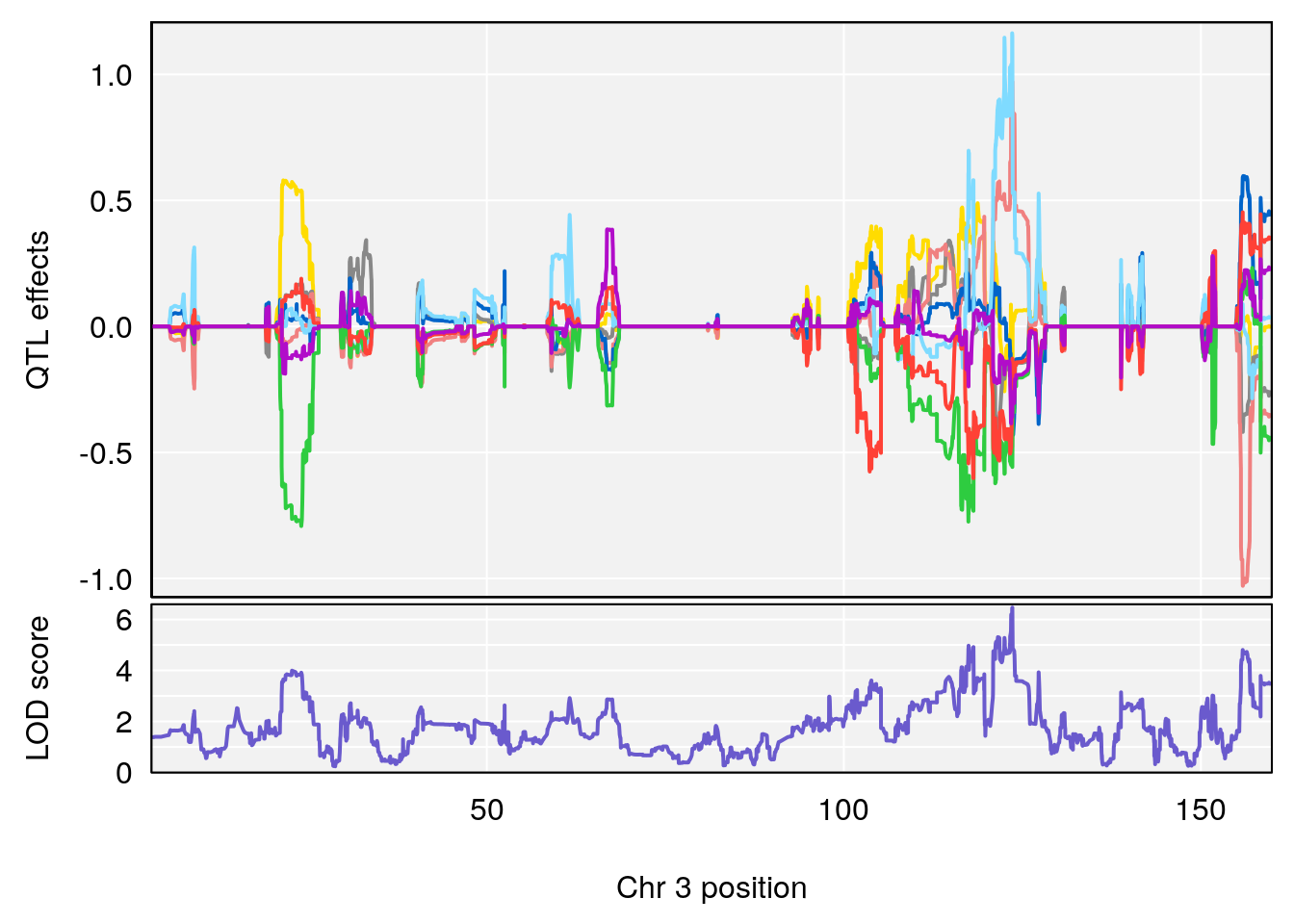
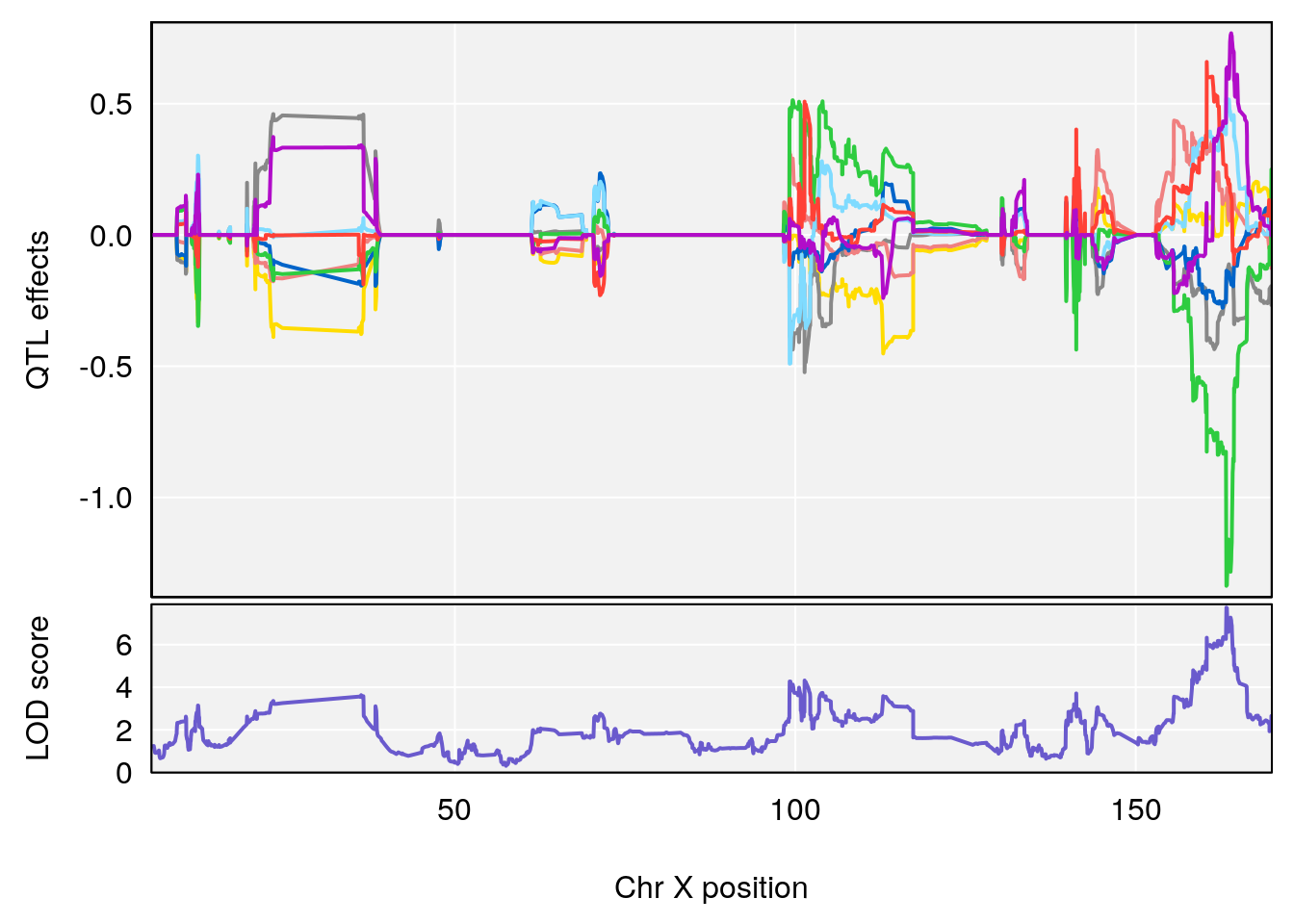
# [1] "Min.depression"
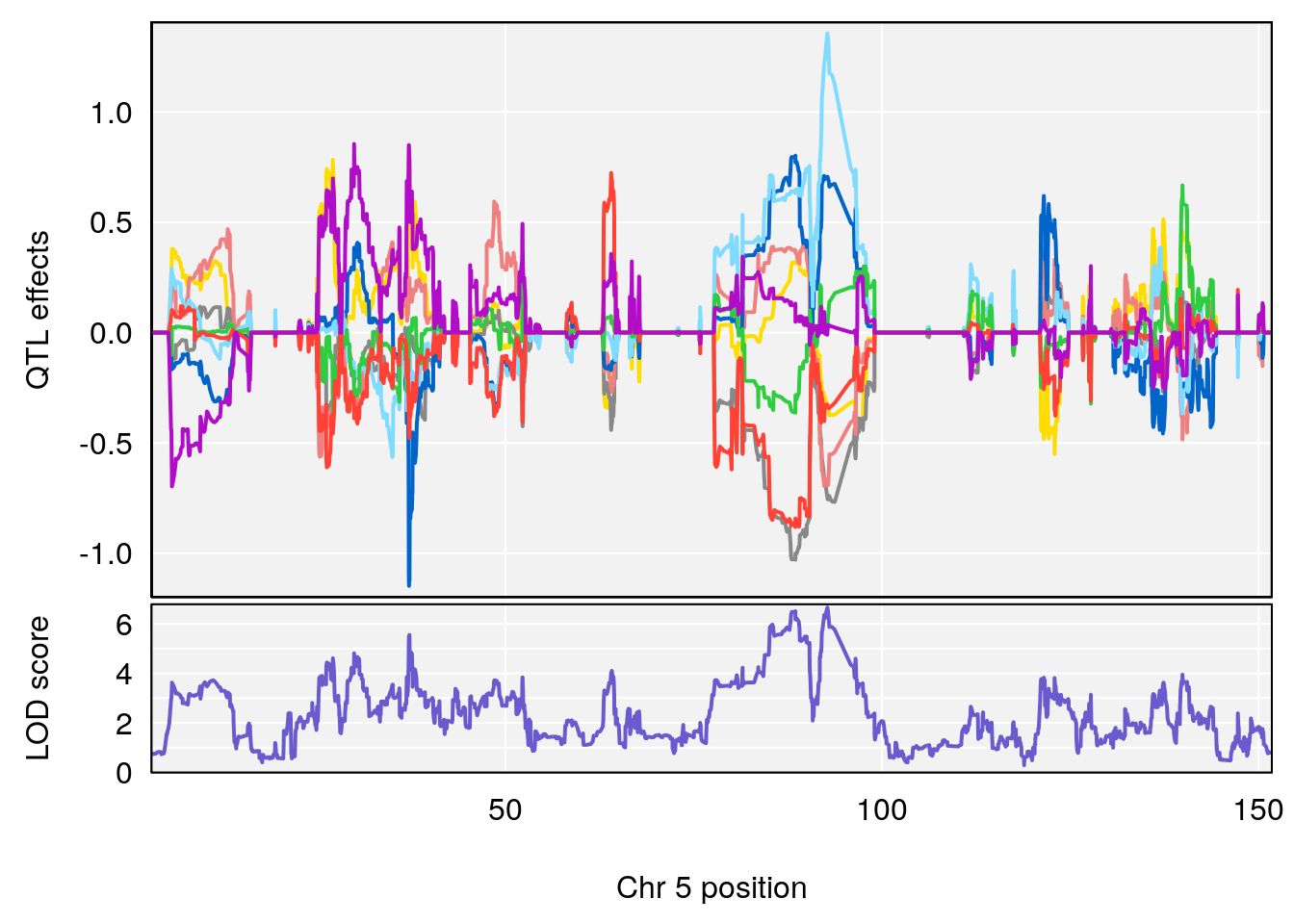
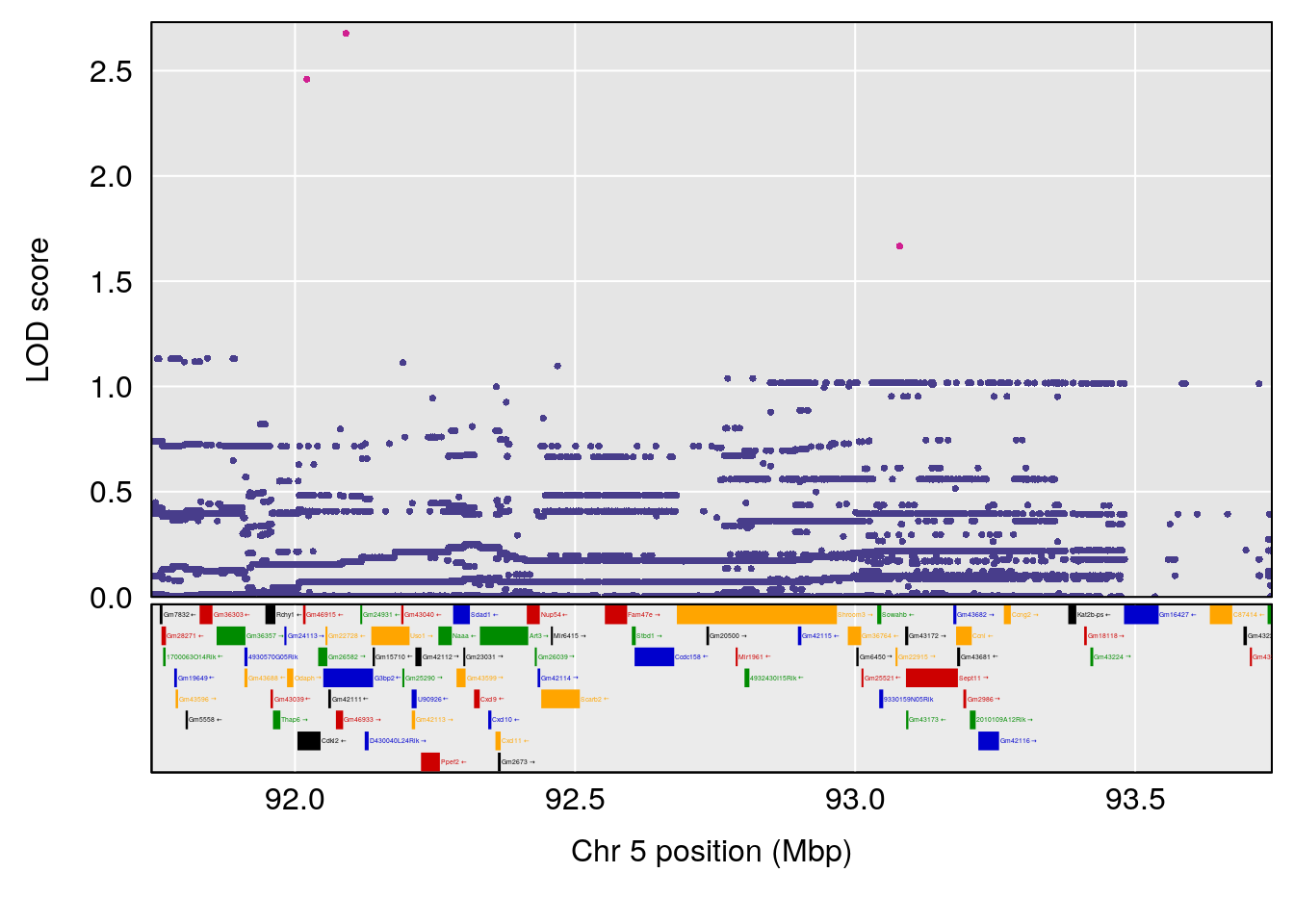
# lodindex lodcolumn chr pos lod ci_lo ci_hi
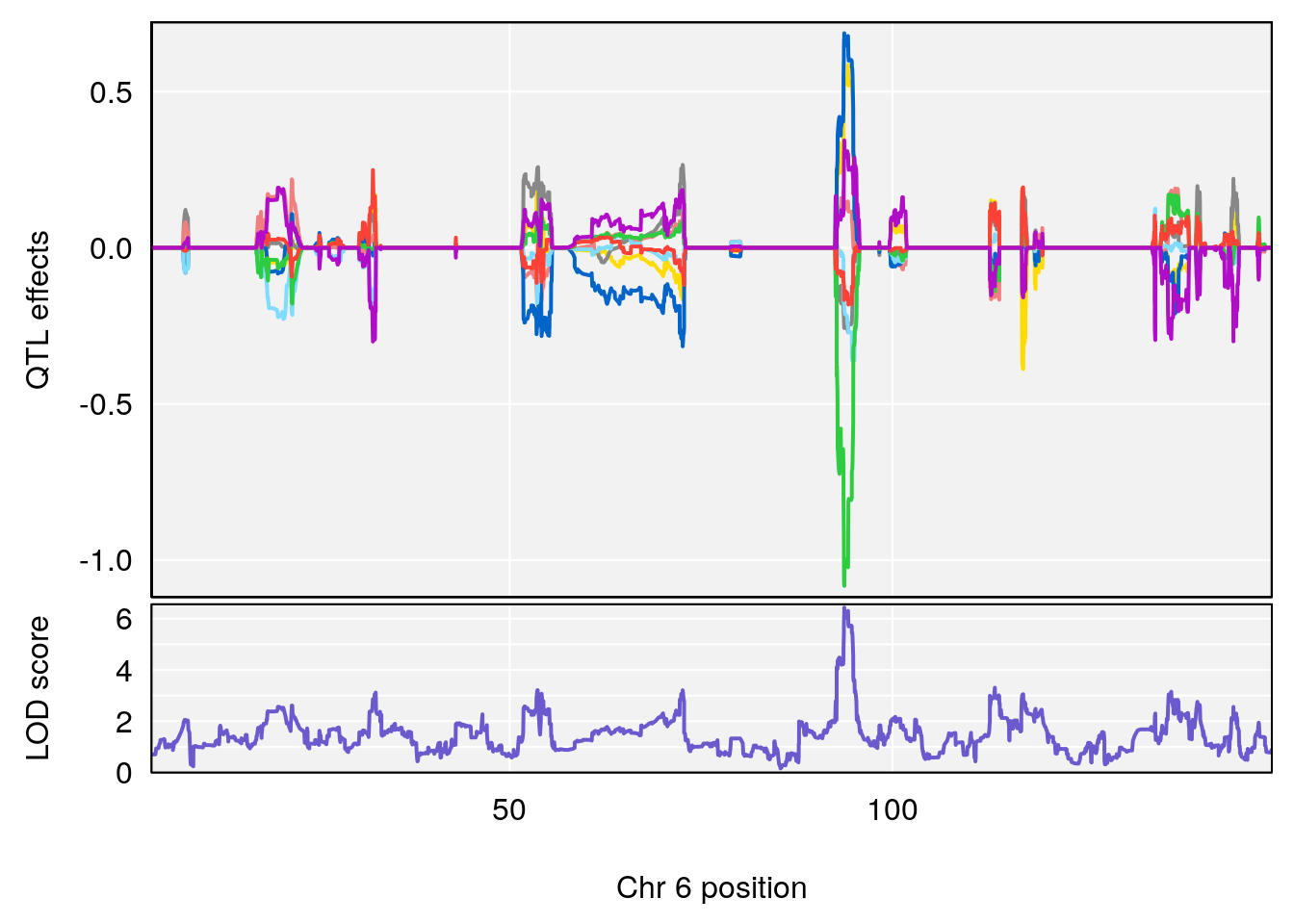
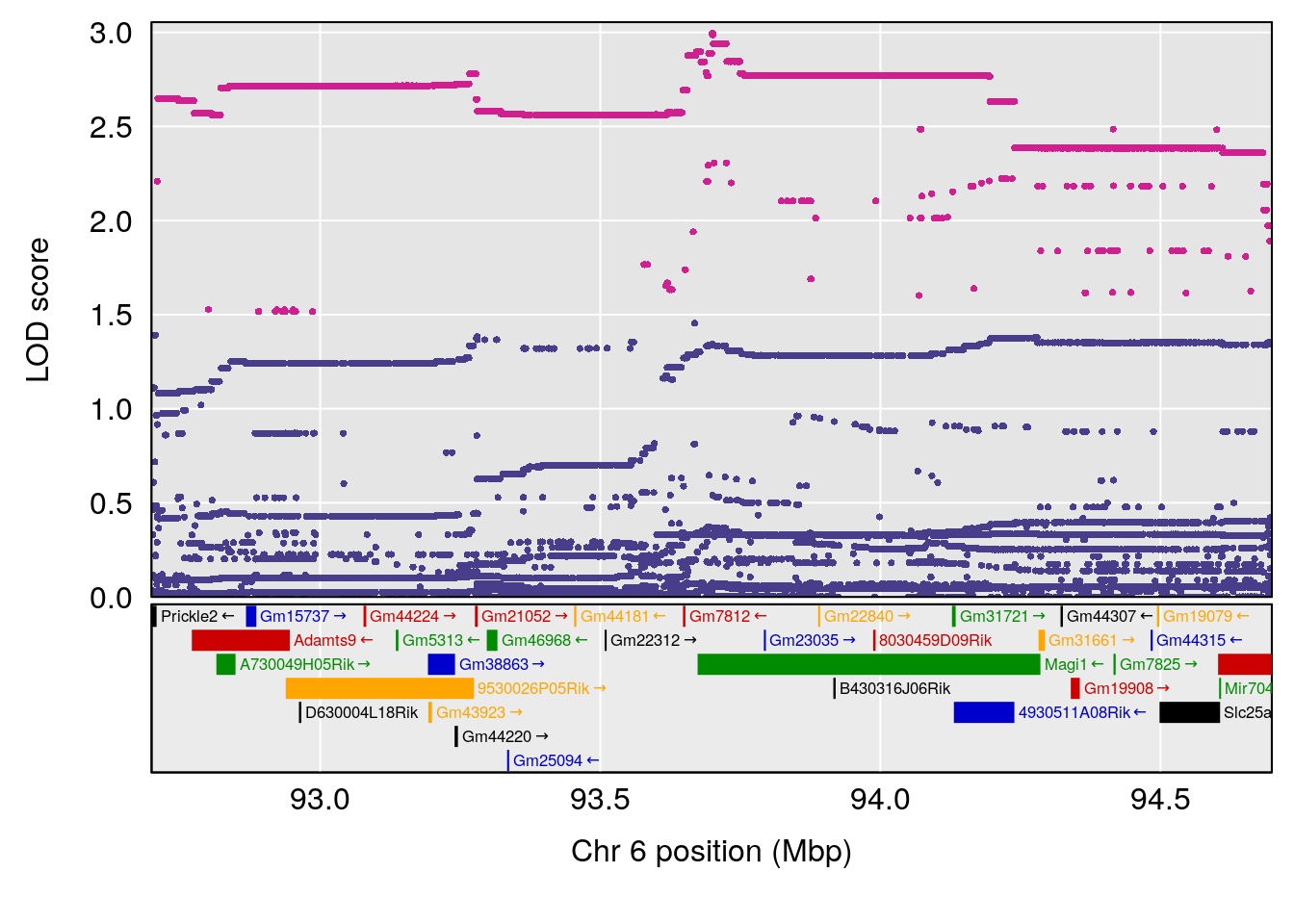
# 1 1 pheno1 6 93.69909 6.430461 93.60934 94.86810
# 2 1 pheno1 19 43.49508 8.046388 43.20074 44.03063
# 3 1 pheno1 X 163.31238 7.736391 160.41596 164.23444
# [1] 1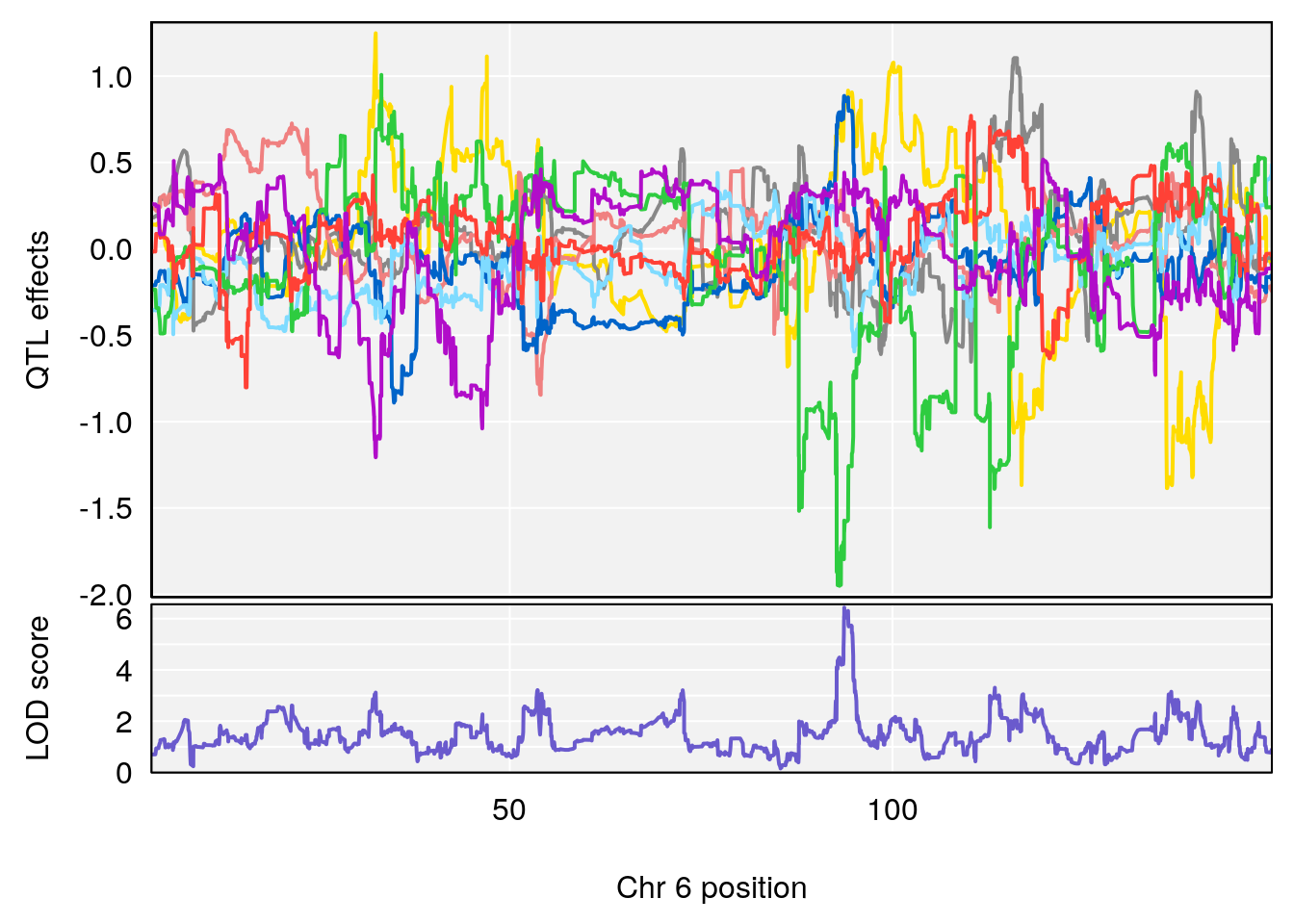
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 2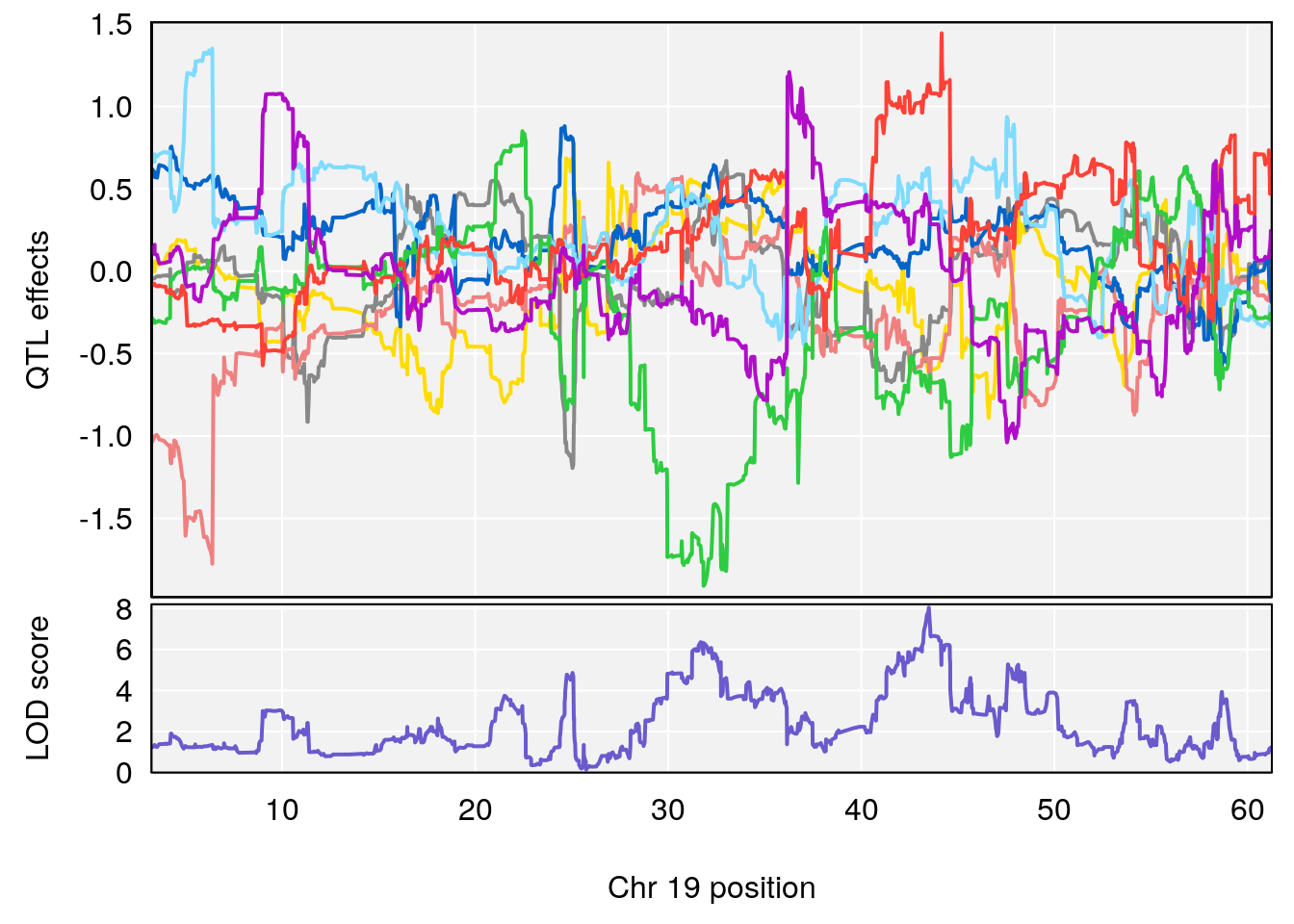
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
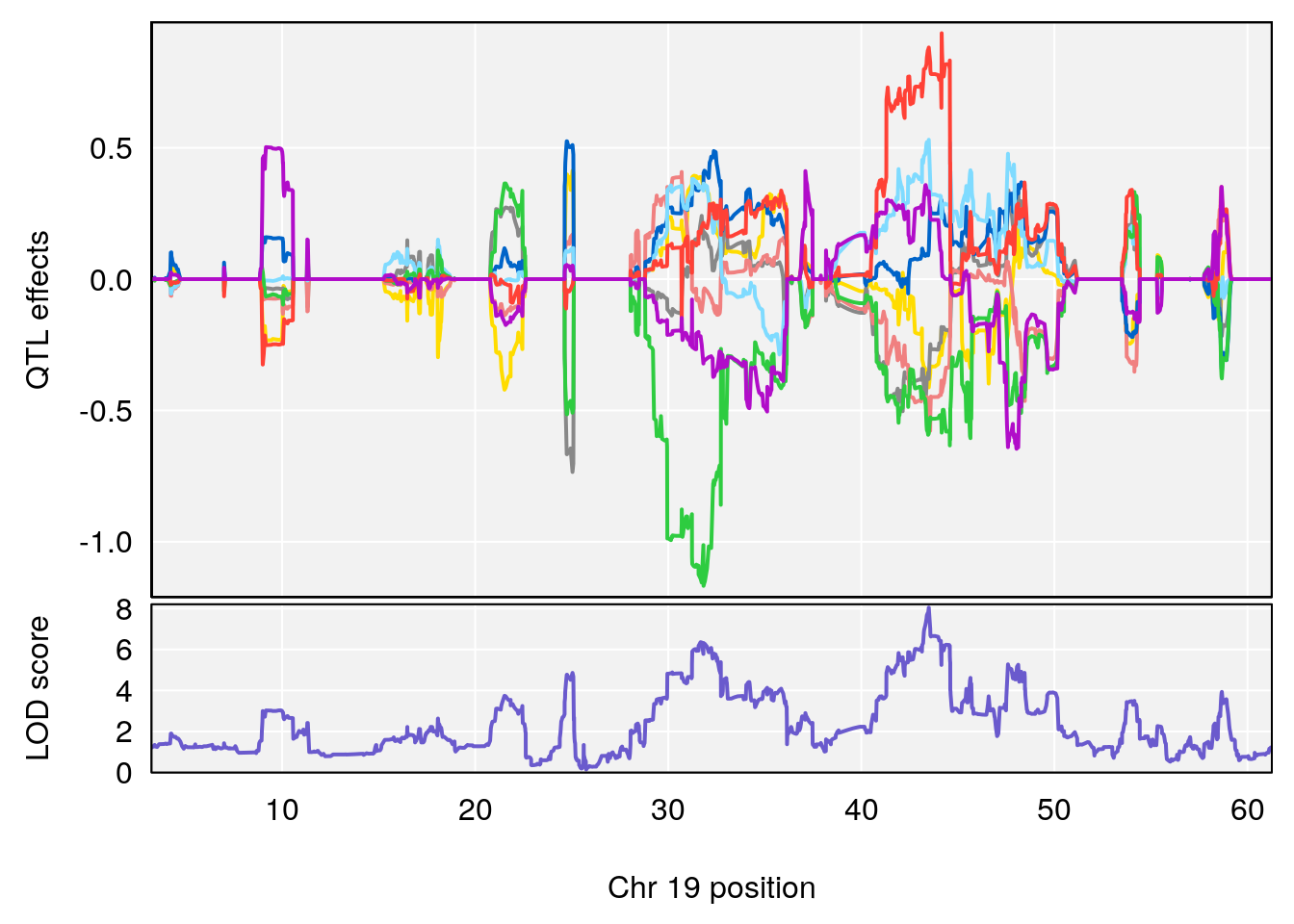
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
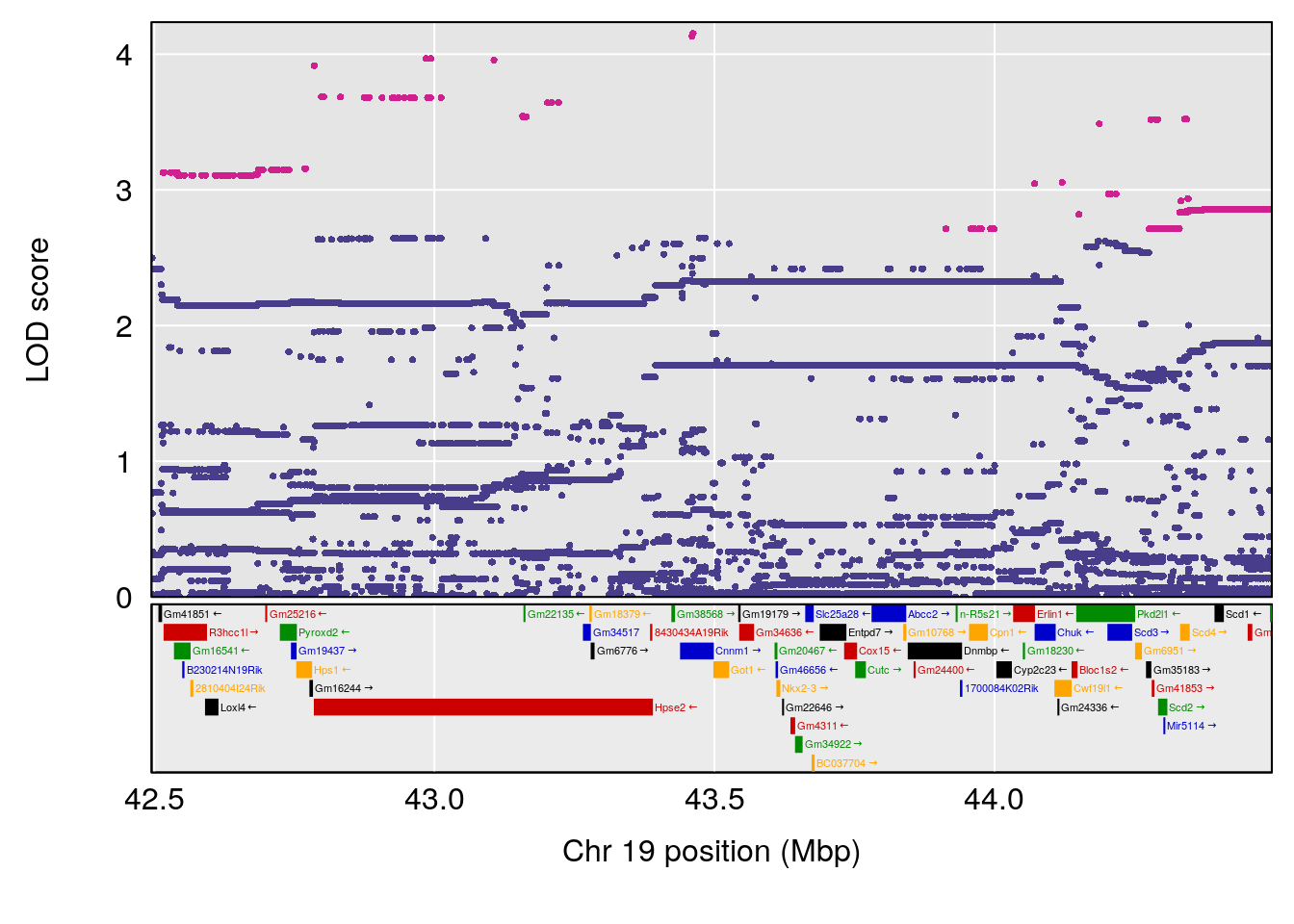
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 3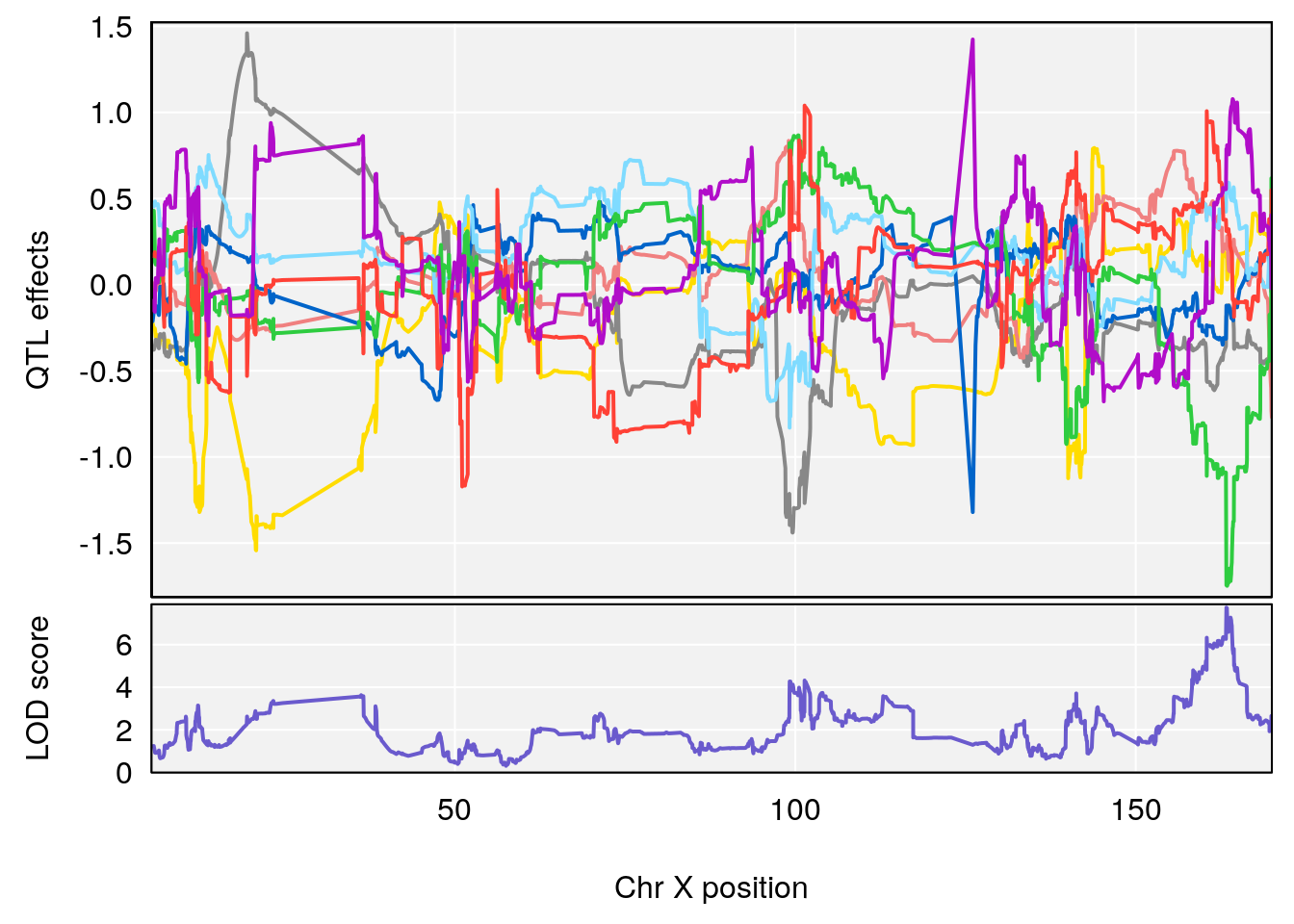
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
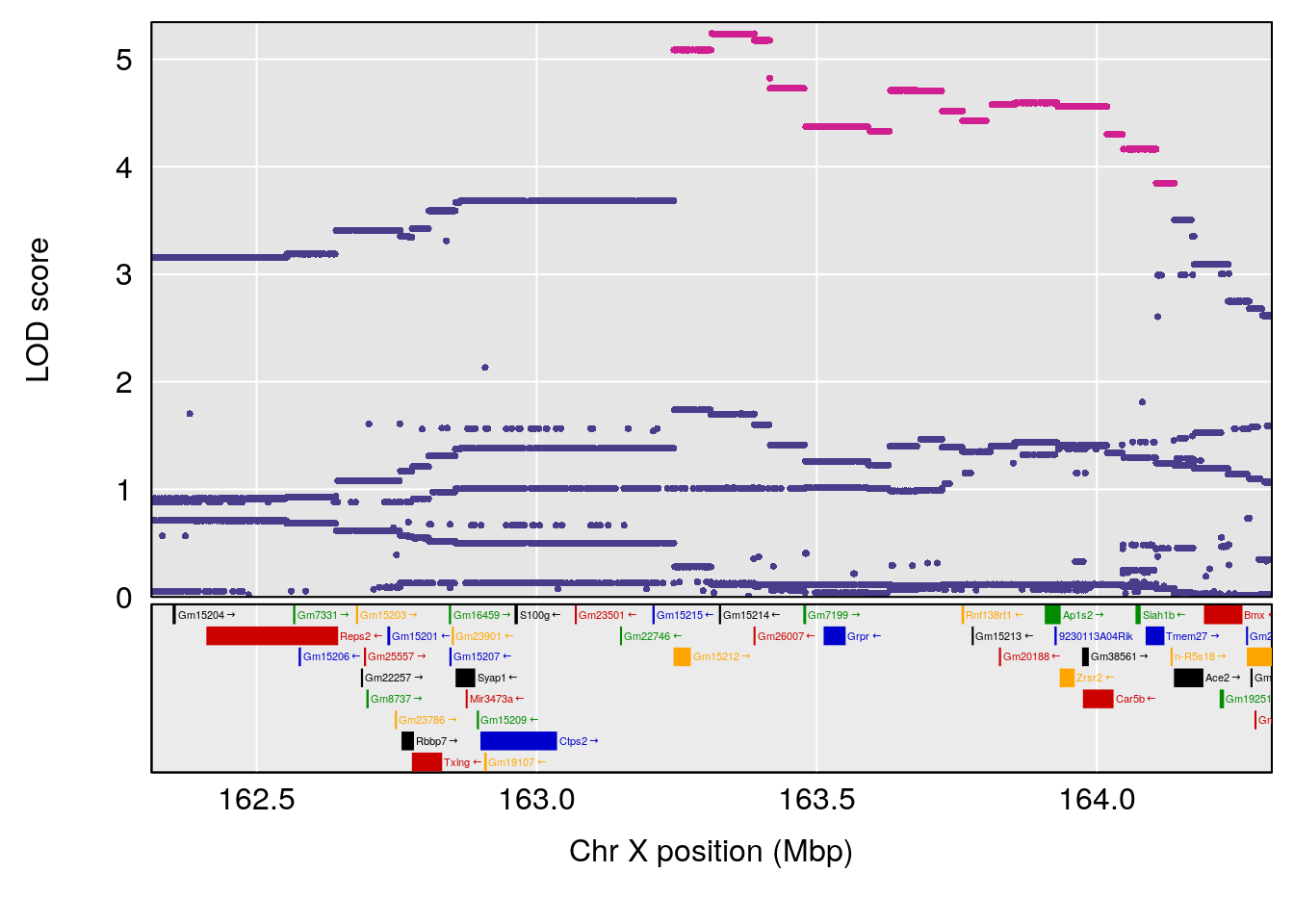
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
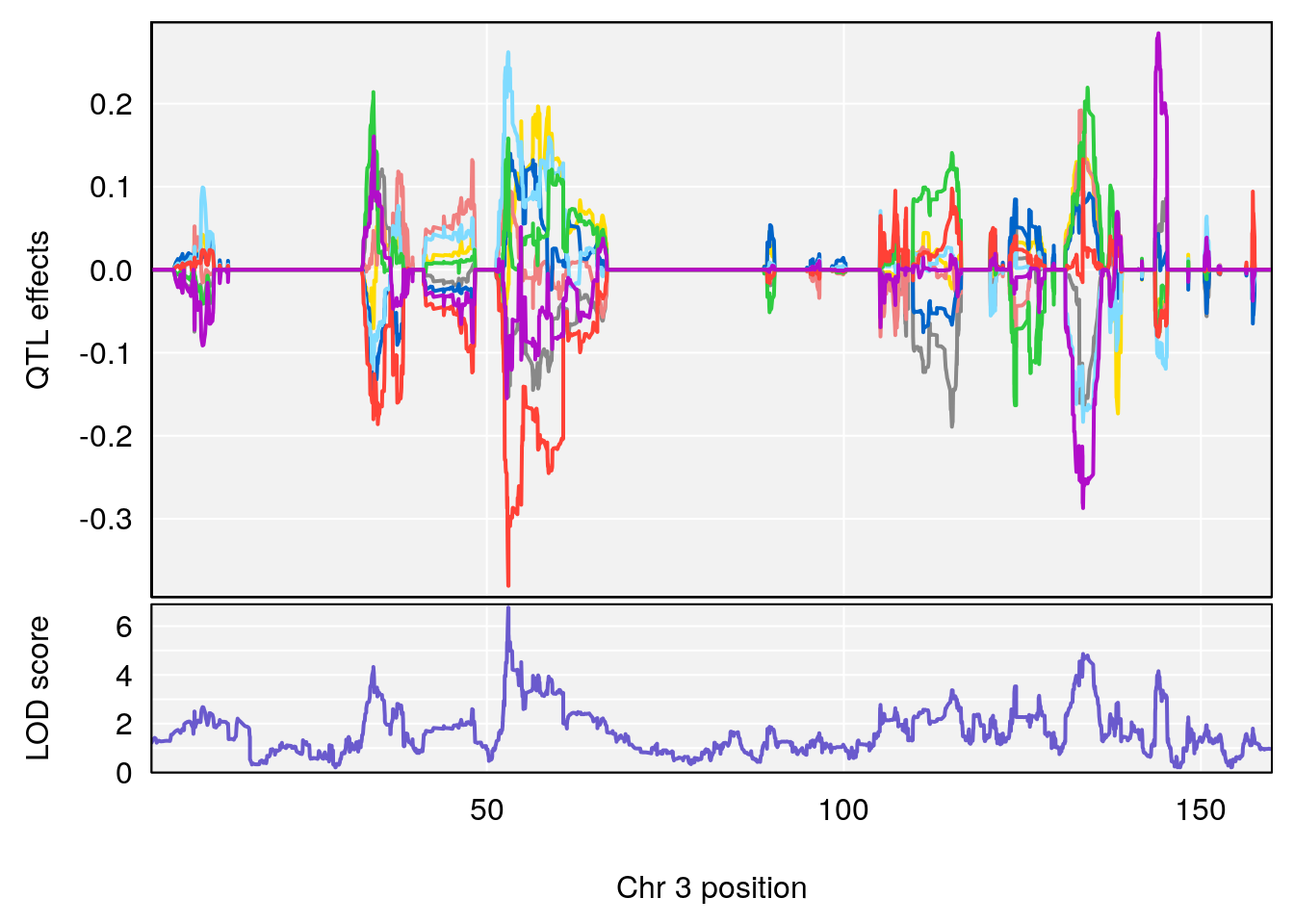
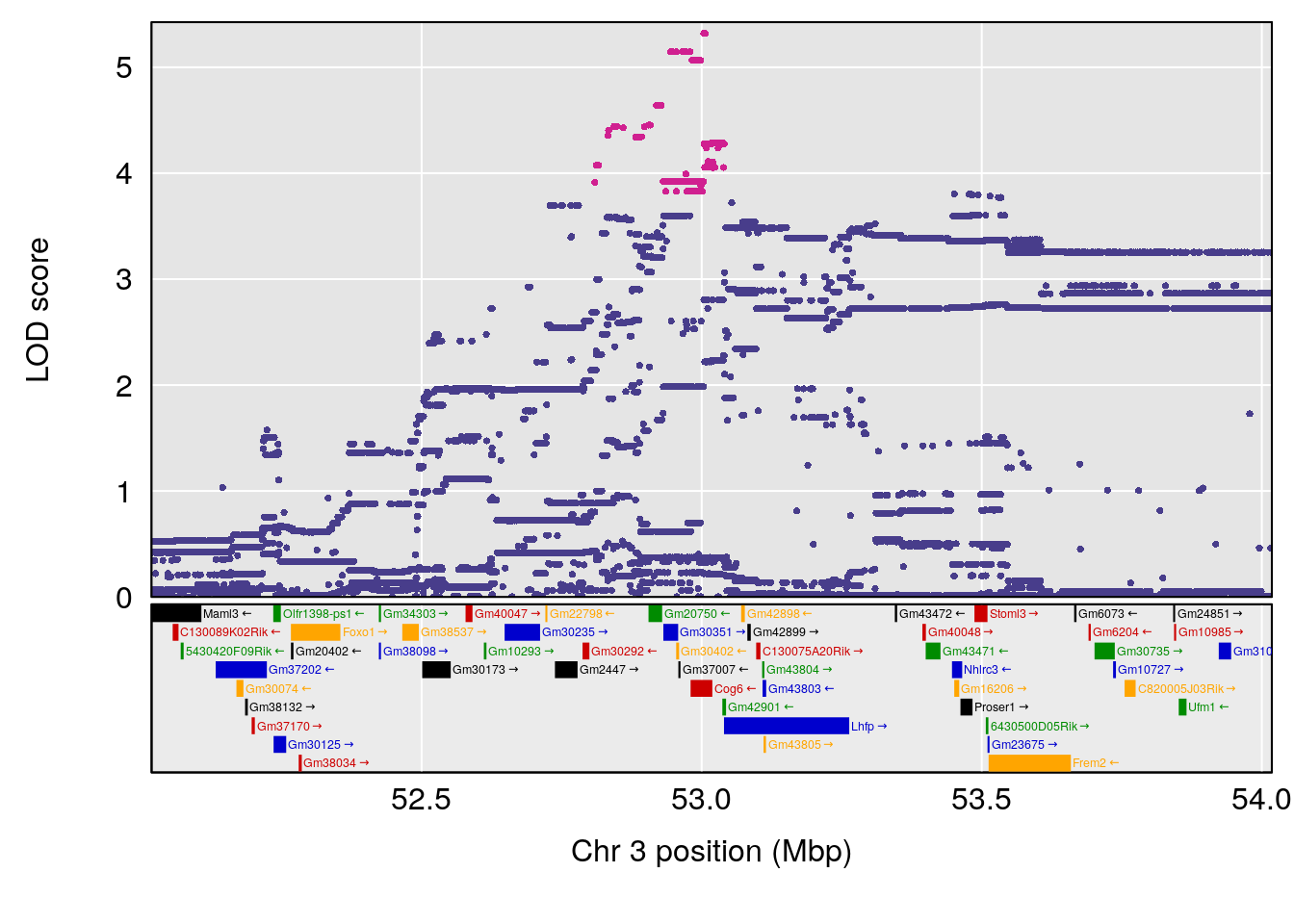
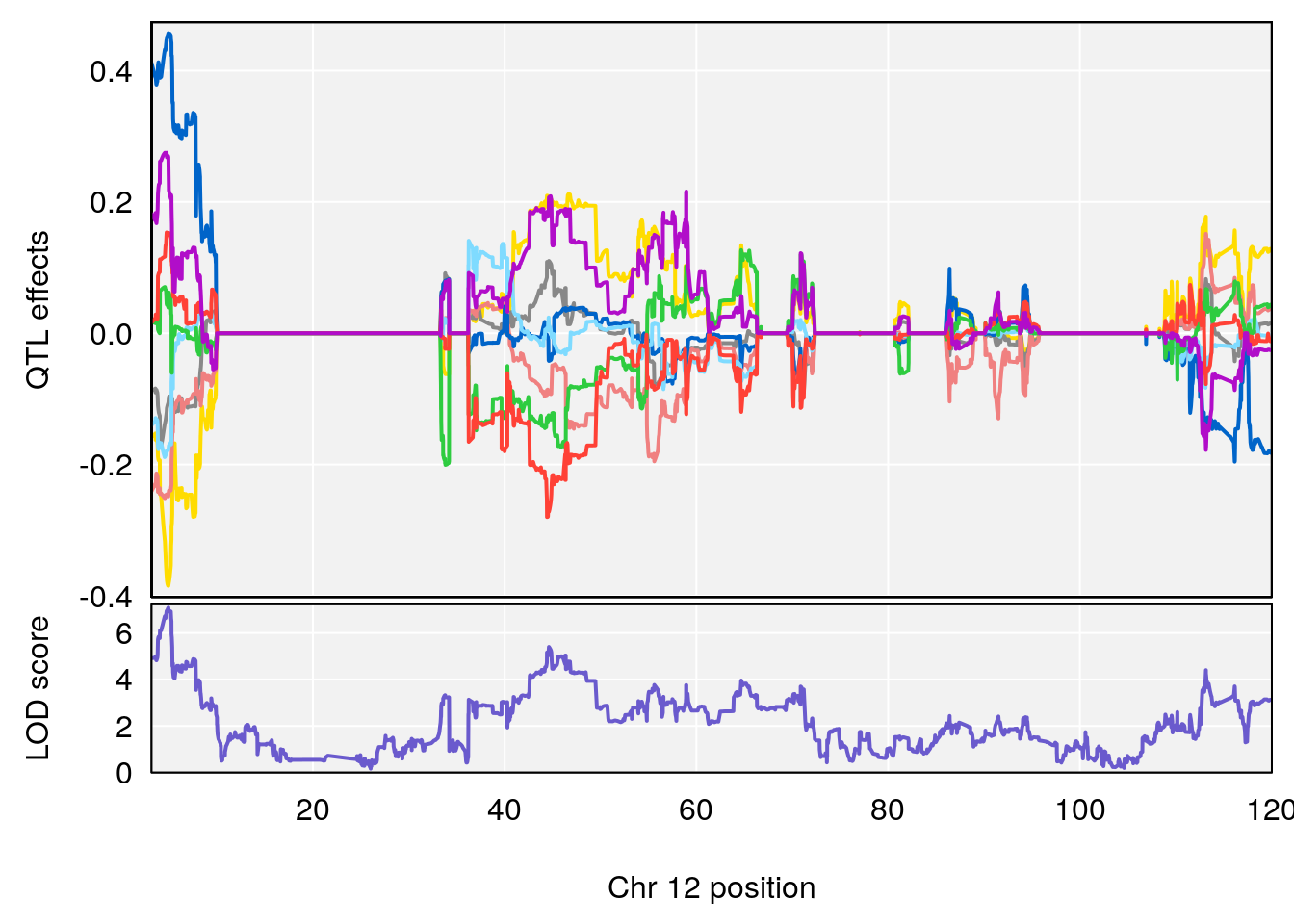
# [1] "Status_bin"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 3 53.017859 6.760367 52.833289 53.290619
# 2 1 pheno1 6 4.124793 6.678702 3.200188 5.832646
# 3 1 pheno1 9 68.129978 6.719334 67.169326 68.687010
# 4 1 pheno1 12 4.923560 7.088994 3.804905 5.324666
# [1] 1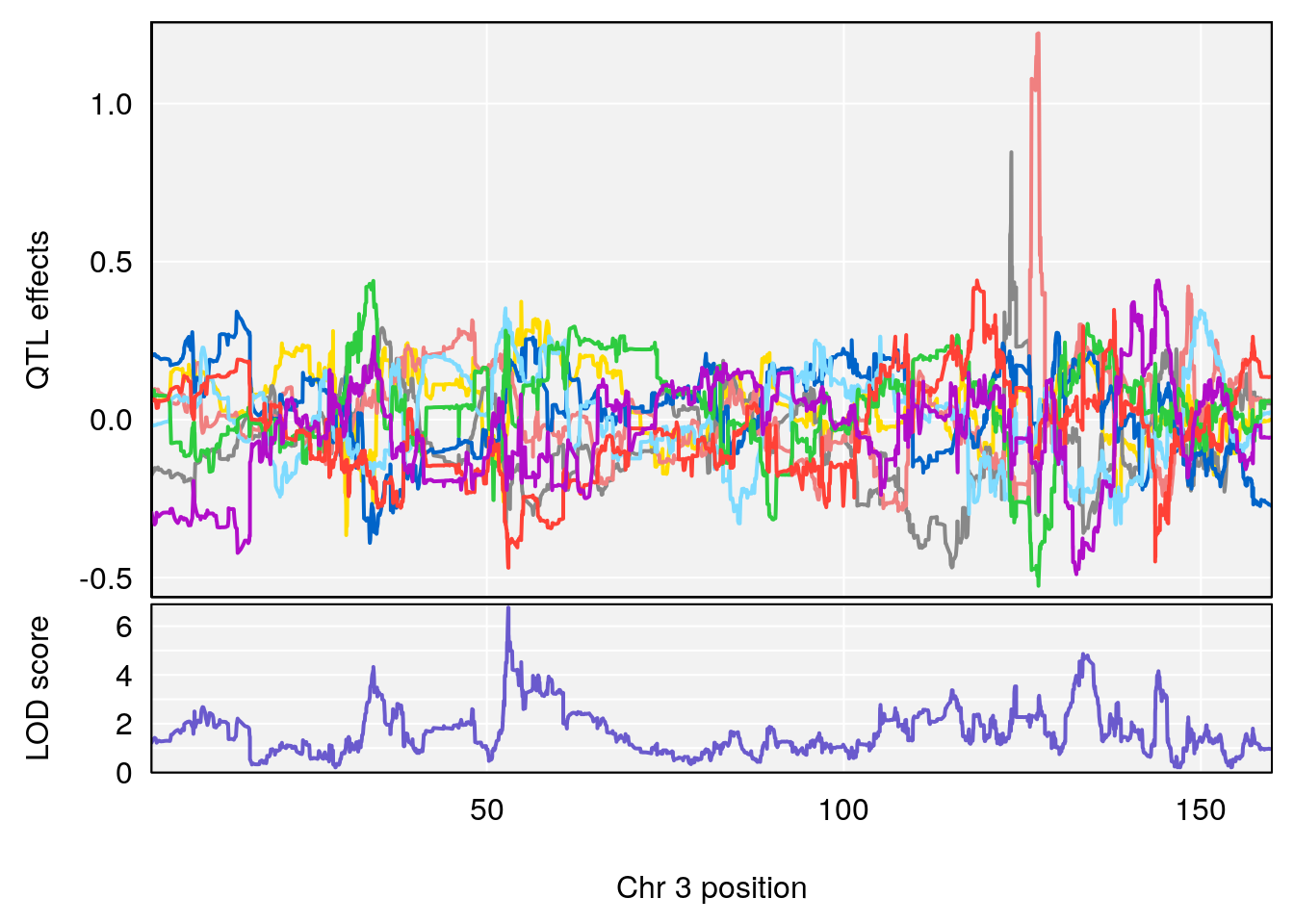
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 2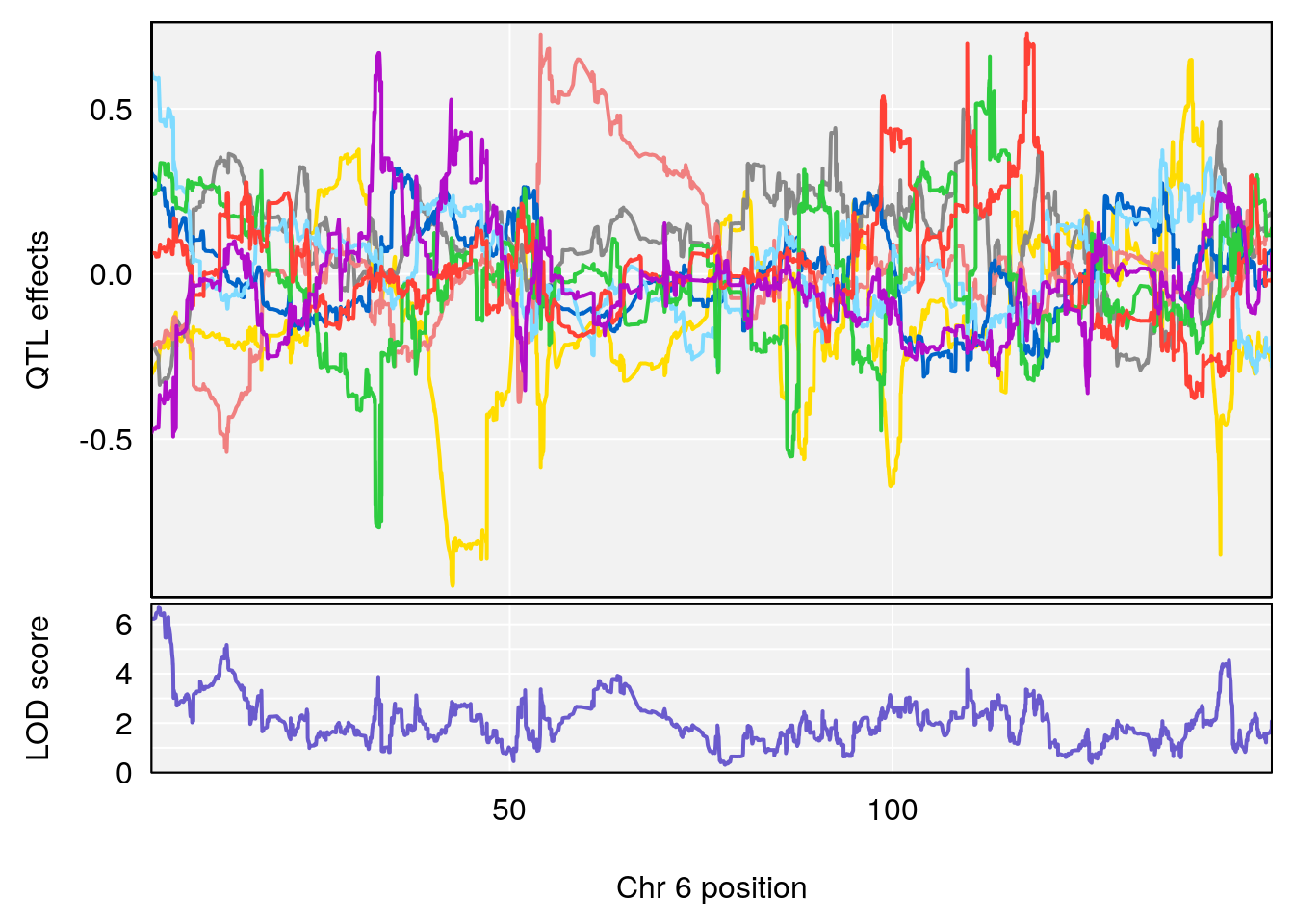
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 3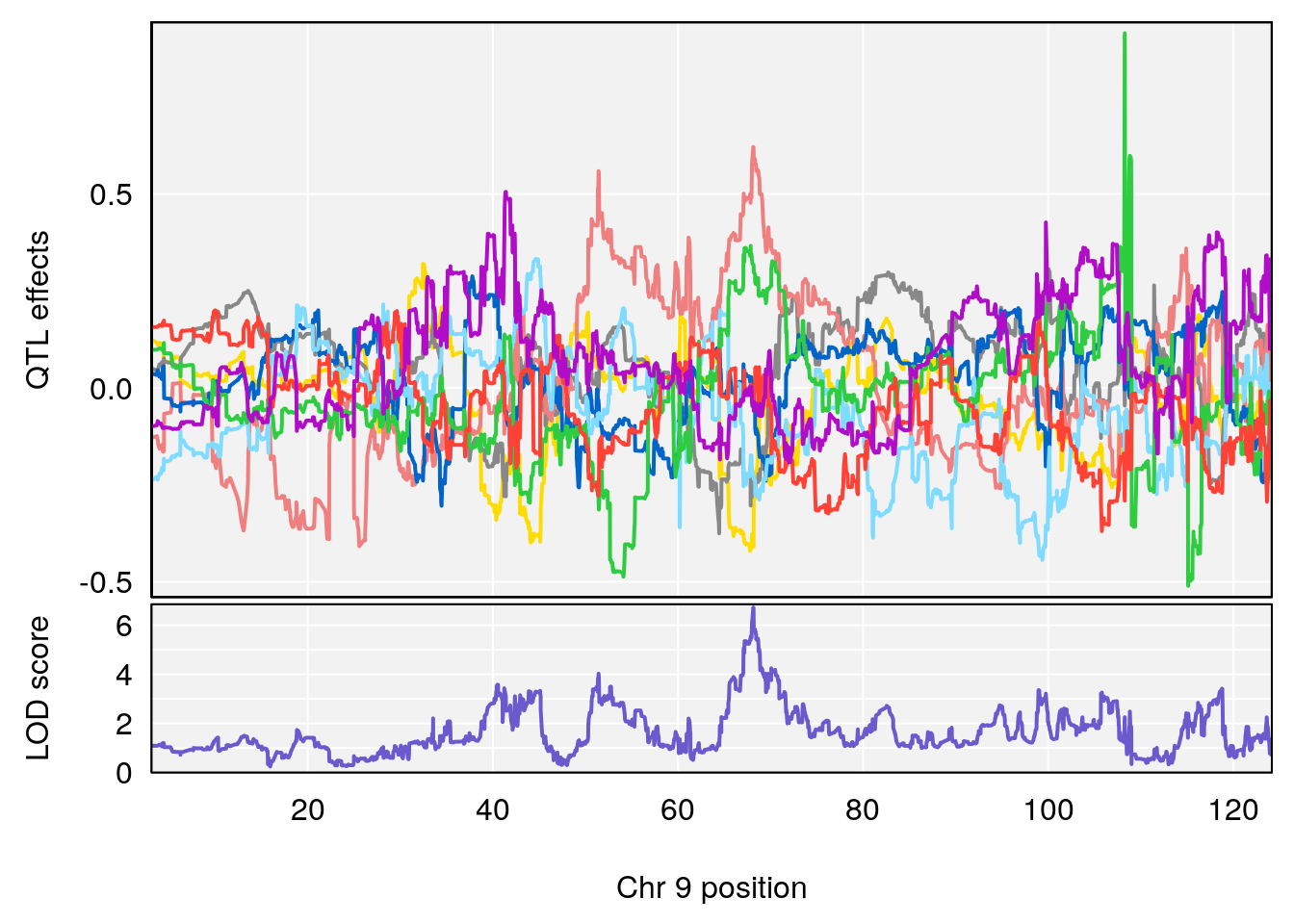
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
# [1] 4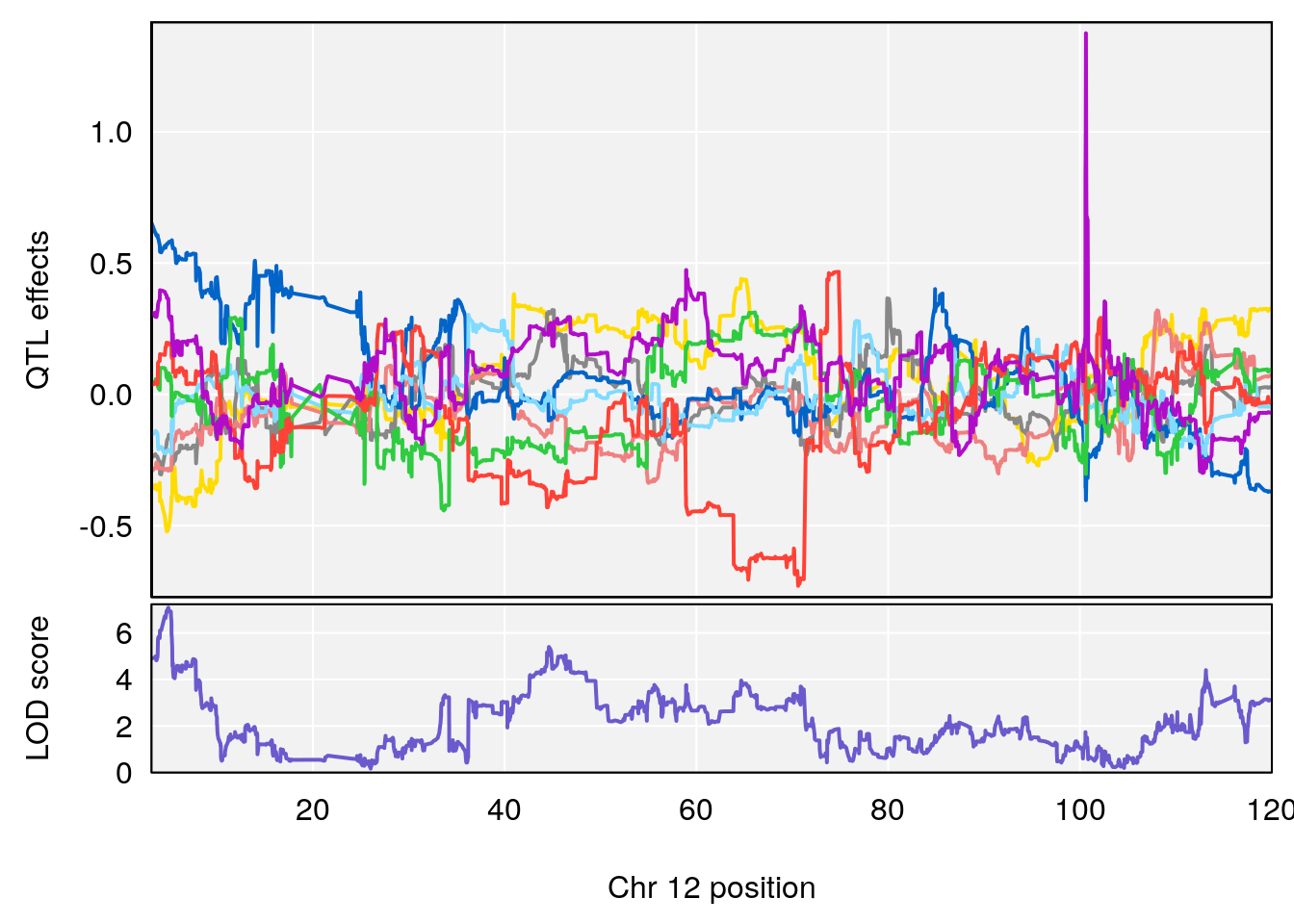
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
| Version | Author | Date |
|---|---|---|
| 8fe8543 | xhyuo | 2023-02-20 |
sessionInfo()
# R version 4.0.3 (2020-10-10)
# Platform: x86_64-pc-linux-gnu (64-bit)
# Running under: Ubuntu 20.04.2 LTS
#
# Matrix products: default
# BLAS/LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.8.so
#
# locale:
# [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
# [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
# [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=C
# [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
# [9] LC_ADDRESS=C LC_TELEPHONE=C
# [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
#
# attached base packages:
# [1] parallel stats graphics grDevices utils datasets methods
# [8] base
#
# other attached packages:
# [1] openxlsx_4.2.3 abind_1.4-5 regress_1.3-21 survival_3.2-7
# [5] qtl2_0.24 GGally_2.1.0 gridExtra_2.3 ggplot2_3.3.3
# [9] workflowr_1.6.2
#
# loaded via a namespace (and not attached):
# [1] tidyselect_1.1.0 xfun_0.21 purrr_0.3.4 lattice_0.20-41
# [5] splines_4.0.3 colorspace_2.0-0 vctrs_0.3.6 generics_0.1.0
# [9] htmltools_0.5.1.1 yaml_2.2.1 blob_1.2.1 rlang_1.0.2
# [13] later_1.1.0.1 pillar_1.4.7 glue_1.4.2 withr_2.4.1
# [17] DBI_1.1.1 bit64_4.0.5 RColorBrewer_1.1-2 lifecycle_1.0.0
# [21] plyr_1.8.6 stringr_1.4.0 munsell_0.5.0 gtable_0.3.0
# [25] zip_2.1.1 evaluate_0.14 memoise_2.0.0 labeling_0.4.2
# [29] knitr_1.31 fastmap_1.1.0 httpuv_1.5.5 highr_0.8
# [33] Rcpp_1.0.6 promises_1.2.0.1 scales_1.1.1 cachem_1.0.4
# [37] farver_2.0.3 fs_1.5.0 bit_4.0.4 digest_0.6.27
# [41] stringi_1.5.3 dplyr_1.0.4 grid_4.0.3 rprojroot_2.0.2
# [45] cli_2.3.0 tools_4.0.3 magrittr_2.0.1 tibble_3.0.6
# [49] RSQLite_2.2.3 crayon_1.4.1 whisker_0.4 pkgconfig_2.0.3
# [53] Matrix_1.3-2 ellipsis_0.3.1 data.table_1.13.6 assertthat_0.2.1
# [57] rmarkdown_2.6 reshape_0.8.8 R6_2.5.0 git2r_0.28.0
# [61] compiler_4.0.3This R Markdown site was created with workflowr