Plot_DO_Fentanyl_Cohort2_mapping
Hao He
2023-02-21
Last updated: 2023-02-21
Checks: 7 0
Knit directory: DO_Opioid/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200504) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 2200f95. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/Picture1.png
Untracked files:
Untracked: Rplot_rz.png
Untracked: TIMBR.test.prop.bm.MN.RET.RData
Untracked: TIMBR.test.random.RData
Untracked: TIMBR.test.rz.transformed_TVb_ml.RData
Untracked: analysis/DDO_morphine1_second_set_69k.stdout
Untracked: analysis/DO_Fentanyl.R
Untracked: analysis/DO_Fentanyl.err
Untracked: analysis/DO_Fentanyl.out
Untracked: analysis/DO_Fentanyl.sh
Untracked: analysis/DO_Fentanyl_69k.R
Untracked: analysis/DO_Fentanyl_69k.err
Untracked: analysis/DO_Fentanyl_69k.out
Untracked: analysis/DO_Fentanyl_69k.sh
Untracked: analysis/DO_Fentanyl_Cohort2_GCTA_herit.R
Untracked: analysis/DO_Fentanyl_Cohort2_gemma.R
Untracked: analysis/DO_Fentanyl_Cohort2_mapping.R
Untracked: analysis/DO_Fentanyl_Cohort2_mapping.err
Untracked: analysis/DO_Fentanyl_Cohort2_mapping.out
Untracked: analysis/DO_Fentanyl_Cohort2_mapping.sh
Untracked: analysis/DO_Fentanyl_GCTA_herit.R
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.R
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.err
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.out
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.sh
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.R
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.err
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.out
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.sh
Untracked: analysis/DO_Fentanyl_array.R
Untracked: analysis/DO_Fentanyl_array.err
Untracked: analysis/DO_Fentanyl_array.out
Untracked: analysis/DO_Fentanyl_array.sh
Untracked: analysis/DO_Fentanyl_combining2Cohort_GCTA_herit.R
Untracked: analysis/DO_Fentanyl_combining2Cohort_gemma.R
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping.R
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping.err
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping.out
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping.sh
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping_CoxPH.R
Untracked: analysis/DO_Fentanyl_finalreport_to_plink.sh
Untracked: analysis/DO_Fentanyl_gemma.R
Untracked: analysis/DO_Fentanyl_gemma.err
Untracked: analysis/DO_Fentanyl_gemma.out
Untracked: analysis/DO_Fentanyl_gemma.sh
Untracked: analysis/DO_morphine1.R
Untracked: analysis/DO_morphine1.Rout
Untracked: analysis/DO_morphine1.sh
Untracked: analysis/DO_morphine1.stderr
Untracked: analysis/DO_morphine1.stdout
Untracked: analysis/DO_morphine1_SNP.R
Untracked: analysis/DO_morphine1_SNP.Rout
Untracked: analysis/DO_morphine1_SNP.sh
Untracked: analysis/DO_morphine1_SNP.stderr
Untracked: analysis/DO_morphine1_SNP.stdout
Untracked: analysis/DO_morphine1_combined.R
Untracked: analysis/DO_morphine1_combined.Rout
Untracked: analysis/DO_morphine1_combined.sh
Untracked: analysis/DO_morphine1_combined.stderr
Untracked: analysis/DO_morphine1_combined.stdout
Untracked: analysis/DO_morphine1_combined_69k.R
Untracked: analysis/DO_morphine1_combined_69k.Rout
Untracked: analysis/DO_morphine1_combined_69k.sh
Untracked: analysis/DO_morphine1_combined_69k.stderr
Untracked: analysis/DO_morphine1_combined_69k.stdout
Untracked: analysis/DO_morphine1_combined_69k_m2.R
Untracked: analysis/DO_morphine1_combined_69k_m2.Rout
Untracked: analysis/DO_morphine1_combined_69k_m2.sh
Untracked: analysis/DO_morphine1_combined_69k_m2.stderr
Untracked: analysis/DO_morphine1_combined_69k_m2.stdout
Untracked: analysis/DO_morphine1_combined_weight_DOB.R
Untracked: analysis/DO_morphine1_combined_weight_DOB.Rout
Untracked: analysis/DO_morphine1_combined_weight_DOB.err
Untracked: analysis/DO_morphine1_combined_weight_DOB.out
Untracked: analysis/DO_morphine1_combined_weight_DOB.sh
Untracked: analysis/DO_morphine1_combined_weight_DOB.stderr
Untracked: analysis/DO_morphine1_combined_weight_DOB.stdout
Untracked: analysis/DO_morphine1_combined_weight_age.R
Untracked: analysis/DO_morphine1_combined_weight_age.err
Untracked: analysis/DO_morphine1_combined_weight_age.out
Untracked: analysis/DO_morphine1_combined_weight_age.sh
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.R
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.err
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.out
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.sh
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.R
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.err
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.out
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.sh
Untracked: analysis/DO_morphine1_cph.R
Untracked: analysis/DO_morphine1_cph.Rout
Untracked: analysis/DO_morphine1_cph.sh
Untracked: analysis/DO_morphine1_second_set.R
Untracked: analysis/DO_morphine1_second_set.Rout
Untracked: analysis/DO_morphine1_second_set.sh
Untracked: analysis/DO_morphine1_second_set.stderr
Untracked: analysis/DO_morphine1_second_set.stdout
Untracked: analysis/DO_morphine1_second_set_69k.R
Untracked: analysis/DO_morphine1_second_set_69k.Rout
Untracked: analysis/DO_morphine1_second_set_69k.sh
Untracked: analysis/DO_morphine1_second_set_69k.stderr
Untracked: analysis/DO_morphine1_second_set_SNP.R
Untracked: analysis/DO_morphine1_second_set_SNP.Rout
Untracked: analysis/DO_morphine1_second_set_SNP.sh
Untracked: analysis/DO_morphine1_second_set_SNP.stderr
Untracked: analysis/DO_morphine1_second_set_SNP.stdout
Untracked: analysis/DO_morphine1_second_set_weight_DOB.R
Untracked: analysis/DO_morphine1_second_set_weight_DOB.Rout
Untracked: analysis/DO_morphine1_second_set_weight_DOB.err
Untracked: analysis/DO_morphine1_second_set_weight_DOB.out
Untracked: analysis/DO_morphine1_second_set_weight_DOB.sh
Untracked: analysis/DO_morphine1_second_set_weight_DOB.stderr
Untracked: analysis/DO_morphine1_second_set_weight_DOB.stdout
Untracked: analysis/DO_morphine1_second_set_weight_age.R
Untracked: analysis/DO_morphine1_second_set_weight_age.Rout
Untracked: analysis/DO_morphine1_second_set_weight_age.err
Untracked: analysis/DO_morphine1_second_set_weight_age.out
Untracked: analysis/DO_morphine1_second_set_weight_age.sh
Untracked: analysis/DO_morphine1_second_set_weight_age.stderr
Untracked: analysis/DO_morphine1_second_set_weight_age.stdout
Untracked: analysis/DO_morphine1_weight_DOB.R
Untracked: analysis/DO_morphine1_weight_DOB.sh
Untracked: analysis/DO_morphine1_weight_age.R
Untracked: analysis/DO_morphine1_weight_age.sh
Untracked: analysis/DO_morphine_gemma.R
Untracked: analysis/DO_morphine_gemma.err
Untracked: analysis/DO_morphine_gemma.out
Untracked: analysis/DO_morphine_gemma.sh
Untracked: analysis/DO_morphine_gemma_firstmin.R
Untracked: analysis/DO_morphine_gemma_firstmin.err
Untracked: analysis/DO_morphine_gemma_firstmin.out
Untracked: analysis/DO_morphine_gemma_firstmin.sh
Untracked: analysis/DO_morphine_gemma_withpermu.R
Untracked: analysis/DO_morphine_gemma_withpermu.err
Untracked: analysis/DO_morphine_gemma_withpermu.out
Untracked: analysis/DO_morphine_gemma_withpermu.sh
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.R
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.err
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.out
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.sh
Untracked: analysis/Plot_DO_morphine1_SNP.R
Untracked: analysis/Plot_DO_morphine1_SNP.Rout
Untracked: analysis/Plot_DO_morphine1_SNP.sh
Untracked: analysis/Plot_DO_morphine1_SNP.stderr
Untracked: analysis/Plot_DO_morphine1_SNP.stdout
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.R
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.Rout
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.sh
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.stderr
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.stdout
Untracked: analysis/download_GSE100356_sra.sh
Untracked: analysis/fentanyl_2cohorts_coxph.R
Untracked: analysis/fentanyl_2cohorts_coxph.err
Untracked: analysis/fentanyl_2cohorts_coxph.out
Untracked: analysis/fentanyl_2cohorts_coxph.sh
Untracked: analysis/fentanyl_scanone.cph.R
Untracked: analysis/fentanyl_scanone.cph.err
Untracked: analysis/fentanyl_scanone.cph.out
Untracked: analysis/fentanyl_scanone.cph.sh
Untracked: analysis/geo_rnaseq.R
Untracked: analysis/heritability_first_second_batch.R
Untracked: analysis/nf-rnaseq-b6.R
Untracked: analysis/plot_fentanyl_2cohorts_coxph.R
Untracked: analysis/scripts/
Untracked: analysis/tibmr.R
Untracked: analysis/timbr_demo.R
Untracked: analysis/workflow_proc.R
Untracked: analysis/workflow_proc.sh
Untracked: analysis/workflow_proc.stderr
Untracked: analysis/workflow_proc.stdout
Untracked: analysis/x.R
Untracked: code/cfw/
Untracked: code/gemma_plot.R
Untracked: code/process.sanger.snp.R
Untracked: code/reconst_utils.R
Untracked: data/69k_grid_pgmap.RData
Untracked: data/Composite Post Kevins Program Group 2 Fentanyl Prepped for Hao.xlsx
Untracked: data/DO_WBP_Data_JAB_to_map.xlsx
Untracked: data/Fentanyl_alternate_metrics.xlsx
Untracked: data/FinalReport/
Untracked: data/GM/
Untracked: data/GM_covar.csv
Untracked: data/GM_covar_07092018_morphine.csv
Untracked: data/Jackson_Lab_Bubier_MURGIGV01/
Untracked: data/MPD_Upload_October.csv
Untracked: data/MPD_Upload_October_updated_sex.csv
Untracked: data/Master Fentanyl DO Study Sheet.xlsx
Untracked: data/MasterMorphine Second Set DO w DOB2.xlsx
Untracked: data/MasterMorphine Second Set DO.xlsx
Untracked: data/Morphine CC DO mice Updated with Published inbred strains.csv
Untracked: data/Morphine_CC_DO_mice_Updated_with_Published_inbred_strains.csv
Untracked: data/cc_variants.sqlite
Untracked: data/combined/
Untracked: data/fentanyl/
Untracked: data/fentanyl2/
Untracked: data/fentanyl_1_2/
Untracked: data/fentanyl_2cohorts_coxph_data.Rdata
Untracked: data/first/
Untracked: data/founder_geno.csv
Untracked: data/genetic_map.csv
Untracked: data/gm.json
Untracked: data/gwas.sh
Untracked: data/marker_grid_0.02cM_plus.txt
Untracked: data/mouse_genes_mgi.sqlite
Untracked: data/pheno.csv
Untracked: data/pheno_qtl2.csv
Untracked: data/pheno_qtl2_07092018_morphine.csv
Untracked: data/pheno_qtl2_w_dob.csv
Untracked: data/physical_map.csv
Untracked: data/rnaseq/
Untracked: data/sample_geno.csv
Untracked: data/second/
Untracked: figure/
Untracked: glimma-plots/
Untracked: output/DO_Fentanyl_Cohort2_MinDepressionRR_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_MinDepressionRR_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_RRDepressionRateHrSLOPE_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_RRRecoveryRateHrSLOPE_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_RRRecoveryRateHrSLOPE_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_StartofRecoveryHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_StartofRecoveryHr_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_Statusbin_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_Statusbin_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_SteadyStateDepressionDurationHrINTERVAL_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoDead(Hr)_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoDeadHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoDeadHr_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoProjectedRecoveryHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoProjectedRecoveryHr_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoSteadyRRDepression(Hr)_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoSteadyRRDepressionHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoSteadyRRDepressionHr_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoThresholdRecoveryHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoThresholdRecoveryHr_coefplot_blup.pdf
Untracked: output/DO_morphine_Min.depression.png
Untracked: output/DO_morphine_Min.depression22222_violin_chr5.pdf
Untracked: output/DO_morphine_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_Min.depression_coefplot_blup_chr5.png
Untracked: output/DO_morphine_Min.depression_coefplot_blup_chrX.png
Untracked: output/DO_morphine_Min.depression_coefplot_chr5.png
Untracked: output/DO_morphine_Min.depression_coefplot_chrX.png
Untracked: output/DO_morphine_Min.depression_peak_genes_chr5.png
Untracked: output/DO_morphine_Min.depression_violin_chr5.png
Untracked: output/DO_morphine_Recovery.Time.png
Untracked: output/DO_morphine_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr11.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr4.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr7.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr9.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr11.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr4.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr7.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr9.png
Untracked: output/DO_morphine_Status_bin.png
Untracked: output/DO_morphine_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_Survival.Time.png
Untracked: output/DO_morphine_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_Survival.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_Survival.Time_coefplot_blup_chr17.png
Untracked: output/DO_morphine_Survival.Time_coefplot_blup_chr8.png
Untracked: output/DO_morphine_Survival.Time_coefplot_chr17.png
Untracked: output/DO_morphine_Survival.Time_coefplot_chr8.png
Untracked: output/DO_morphine_combine_batch_peak_violin.pdf
Untracked: output/DO_morphine_combined_69k_m2_Min.depression.png
Untracked: output/DO_morphine_combined_69k_m2_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_combined_69k_m2_Recovery.Time.png
Untracked: output/DO_morphine_combined_69k_m2_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_combined_69k_m2_Status_bin.png
Untracked: output/DO_morphine_combined_69k_m2_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_combined_69k_m2_Survival.Time.png
Untracked: output/DO_morphine_combined_69k_m2_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Survival.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_coxph_24hrs_kinship_QTL.png
Untracked: output/DO_morphine_cphout.RData
Untracked: output/DO_morphine_first_batch_peak_in_second_batch_violin.pdf
Untracked: output/DO_morphine_first_batch_peak_in_second_batch_violin_sidebyside.pdf
Untracked: output/DO_morphine_first_batch_peak_violin.pdf
Untracked: output/DO_morphine_operm.cph.RData
Untracked: output/DO_morphine_second_batch_on_first_batch_peak_violin.pdf
Untracked: output/DO_morphine_second_batch_peak_ch6surv_on_first_batchviolin.pdf
Untracked: output/DO_morphine_second_batch_peak_ch6surv_on_first_batchviolin2.pdf
Untracked: output/DO_morphine_second_batch_peak_in_first_batch_violin.pdf
Untracked: output/DO_morphine_second_batch_peak_in_first_batch_violin_sidebyside.pdf
Untracked: output/DO_morphine_second_batch_peak_violin.pdf
Untracked: output/DO_morphine_secondbatch_69k_Min.depression.png
Untracked: output/DO_morphine_secondbatch_69k_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_69k_Recovery.Time.png
Untracked: output/DO_morphine_secondbatch_69k_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_69k_Status_bin.png
Untracked: output/DO_morphine_secondbatch_69k_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_69k_Survival.Time.png
Untracked: output/DO_morphine_secondbatch_69k_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Survival.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Min.depression.png
Untracked: output/DO_morphine_secondbatch_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Recovery.Time.png
Untracked: output/DO_morphine_secondbatch_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Status_bin.png
Untracked: output/DO_morphine_secondbatch_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Survival.Time.png
Untracked: output/DO_morphine_secondbatch_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Survival.Time_coefplot_blup.pdf
Untracked: output/Fentanyl/
Untracked: output/KPNA3.pdf
Untracked: output/SSC4D.pdf
Untracked: output/TIMBR.test.RData
Untracked: output/apr_69kchr_combined.RData
Untracked: output/apr_69kchr_k_loco_combined.rds
Untracked: output/apr_69kchr_second_set.RData
Untracked: output/combine_batch_variation.RData
Untracked: output/combined_gm.RData
Untracked: output/combined_gm.k_loco.rds
Untracked: output/combined_gm.k_overall.rds
Untracked: output/combined_gm.probs_8state.rds
Untracked: output/coxph/
Untracked: output/do.morphine.RData
Untracked: output/do.morphine.k_loco.rds
Untracked: output/do.morphine.probs_36state.rds
Untracked: output/do.morphine.probs_8state.rds
Untracked: output/do_Fentanyl_combine2cohort_MeanDepressionBR_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_MeanDepressionBR_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_MinDepressionBR_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_MinDepressionBR_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_MinDepressionRR_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_MinDepressionRR_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_RRRecoveryRateHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_RRRecoveryRateHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_StartofRecoveryHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_StartofRecoveryHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_Statusbin_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_Statusbin_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_SteadyStateDepressionDurationHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_SteadyStateDepressionDurationHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_SurvivalTime_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_SurvivalTime_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoDeadHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoDeadHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoMostlyDeadHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoMostlyDeadHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoProjectedRecoveryHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoProjectedRecoveryHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoRecoveryHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoRecoveryHr_coefplot_blup.pdf
Untracked: output/first_batch_variation.RData
Untracked: output/first_second_survival_peak_chr.xlsx
Untracked: output/hsq_1_first_batch_herit_qtl2.RData
Untracked: output/hsq_2_second_batch_herit_qtl2.RData
Untracked: output/old_temp/
Untracked: output/pr_69kchr_combined.RData
Untracked: output/pr_69kchr_second_set.RData
Untracked: output/qtl.morphine.69k.out.combined.RData
Untracked: output/qtl.morphine.69k.out.combined_m2.RData
Untracked: output/qtl.morphine.69k.out.second_set.RData
Untracked: output/qtl.morphine.operm.RData
Untracked: output/qtl.morphine.out.RData
Untracked: output/qtl.morphine.out.combined_gm.RData
Untracked: output/qtl.morphine.out.combined_gm.female.RData
Untracked: output/qtl.morphine.out.combined_gm.male.RData
Untracked: output/qtl.morphine.out.combined_weight_DOB.RData
Untracked: output/qtl.morphine.out.combined_weight_age.RData
Untracked: output/qtl.morphine.out.female.RData
Untracked: output/qtl.morphine.out.male.RData
Untracked: output/qtl.morphine.out.second_set.RData
Untracked: output/qtl.morphine.out.second_set.female.RData
Untracked: output/qtl.morphine.out.second_set.male.RData
Untracked: output/qtl.morphine.out.second_set.weight_DOB.RData
Untracked: output/qtl.morphine.out.second_set.weight_age.RData
Untracked: output/qtl.morphine.out.weight_DOB.RData
Untracked: output/qtl.morphine.out.weight_age.RData
Untracked: output/qtl.morphine1.snpout.RData
Untracked: output/qtl.morphine2.snpout.RData
Untracked: output/second_batch_pheno.csv
Untracked: output/second_batch_variation.RData
Untracked: output/second_set_apr_69kchr_k_loco.rds
Untracked: output/second_set_gm.RData
Untracked: output/second_set_gm.k_loco.rds
Untracked: output/second_set_gm.probs_36state.rds
Untracked: output/second_set_gm.probs_8state.rds
Untracked: output/topSNP_chr5_mindepression.csv
Untracked: output/zoompeak_Min.depression_9.pdf
Untracked: output/zoompeak_Recovery.Time_16.pdf
Untracked: output/zoompeak_Status_bin_11.pdf
Untracked: output/zoompeak_Survival.Time_1.pdf
Untracked: output/zoompeak_fentanyl_Survival.Time_2.pdf
Untracked: sra-tools_v2.10.7.sif
Unstaged changes:
Modified: _workflowr.yml
Modified: analysis/marker_violin.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/Plot_DO_Fentanyl_Cohort2_mapping.Rmd) and HTML (docs/Plot_DO_Fentanyl_Cohort2_mapping.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 2200f95 | xhyuo | 2023-02-21 | update with sex specific Fentanyl plot |
| html | 8611541 | xhyuo | 2023-02-21 | Build site. |
| Rmd | d1a7028 | xhyuo | 2023-02-21 | update with sex specific Fentanyl |
| html | fd4dc88 | xhyuo | 2023-01-11 | Build site. |
| Rmd | aff7cd2 | xhyuo | 2023-01-11 | update with histogram |
| html | 6eeaa6a | xhyuo | 2023-01-06 | Build site. |
| Rmd | 6d89b79 | xhyuo | 2023-01-06 | update with histogram and gcta h2, figure size |
| html | fe30eec | xhyuo | 2023-01-06 | Build site. |
| Rmd | 15a2b6f | xhyuo | 2023-01-06 | update with histogram and gcta h2 |
| html | b77b34e | xhyuo | 2022-06-29 | Build site. |
| Rmd | 3f07442 | xhyuo | 2022-06-29 | Plot_DO_Fentanyl_Cohort2 |
| html | 7faf7f1 | xhyuo | 2022-06-29 | Build site. |
| Rmd | e9fb99f | xhyuo | 2022-06-29 | Fentanyl_Cohort2 |
Last update: 2023-02-21
loading libraries
library(ggplot2)
library(gridExtra)
library(GGally)
library(parallel)
library(qtl2)
library(parallel)
library(survival)
library(regress)
library(abind)
library(openxlsx)
library(tidyverse)
rz.transform <- function(y) {
rankY=rank(y, ties.method="average", na.last="keep")
rzT=qnorm(rankY/(length(na.exclude(rankY))+1))
return(rzT)
}
load("code/do.colors.RData")Read phenotype data
# Read phenotype data -----------------------------------------------------
#batch 20220502
do.pheno <- readxl::read_excel(path = "data/Composite Post Kevins Program Group 2 Fentanyl Prepped for Hao.xlsx",
col_types = c("text", "text", "text", "date", "numeric", "numeric", "date", rep("numeric", 9), "date"))
# Warning in read_fun(path = enc2native(normalizePath(path)), sheet_i = sheet, :
# Expecting date in D263 / R263C4: got 'BLANK'
# Warning in read_fun(path = enc2native(normalizePath(path)), sheet_i = sheet, :
# Expecting numeric in F263 / R263C6: got 'BLANK'
do.pheno <- do.pheno %>%
dplyr::rename_with(., ~str_remove_all(.x, " ")) %>%
dplyr::filter(!is.na(MouseID)) %>%
dplyr::mutate(Status_bin = case_when(
EarTag == "DEAD" ~ 0,
is.na(EarTag) ~ 1
)) %>%
as.data.frame()
# Read genotype data -----------------------------------------------------------
load("data/Jackson_Lab_Bubier_MURGIGV01/gm_DO765_qc_newid.RData")#gm_after_qc
overlap.id = intersect(ind_ids(gm_after_qc), as.character(do.pheno$MouseID))
#subset
gm = gm_after_qc[overlap.id, ]
do.pheno = do.pheno[do.pheno$MouseID %in% overlap.id, ]
rownames(do.pheno) = do.pheno$MouseID
do.pheno = do.pheno[overlap.id,]
all.equal(ind_ids(gm), do.pheno$MouseID)
# [1] TRUE
#covar
do.pheno$Sex <- as.factor(do.pheno$Sex)
#boxplot on the raw data------------
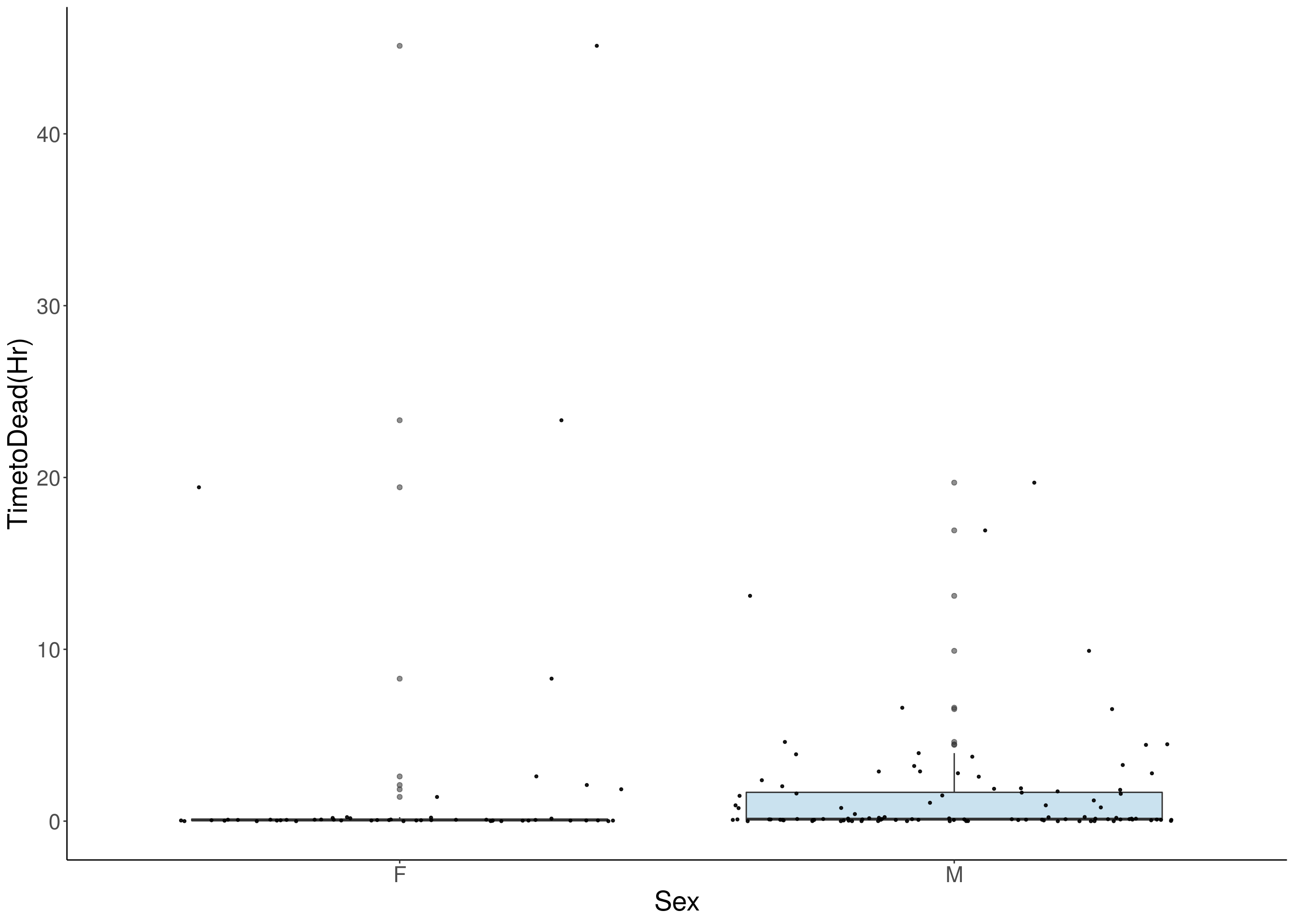
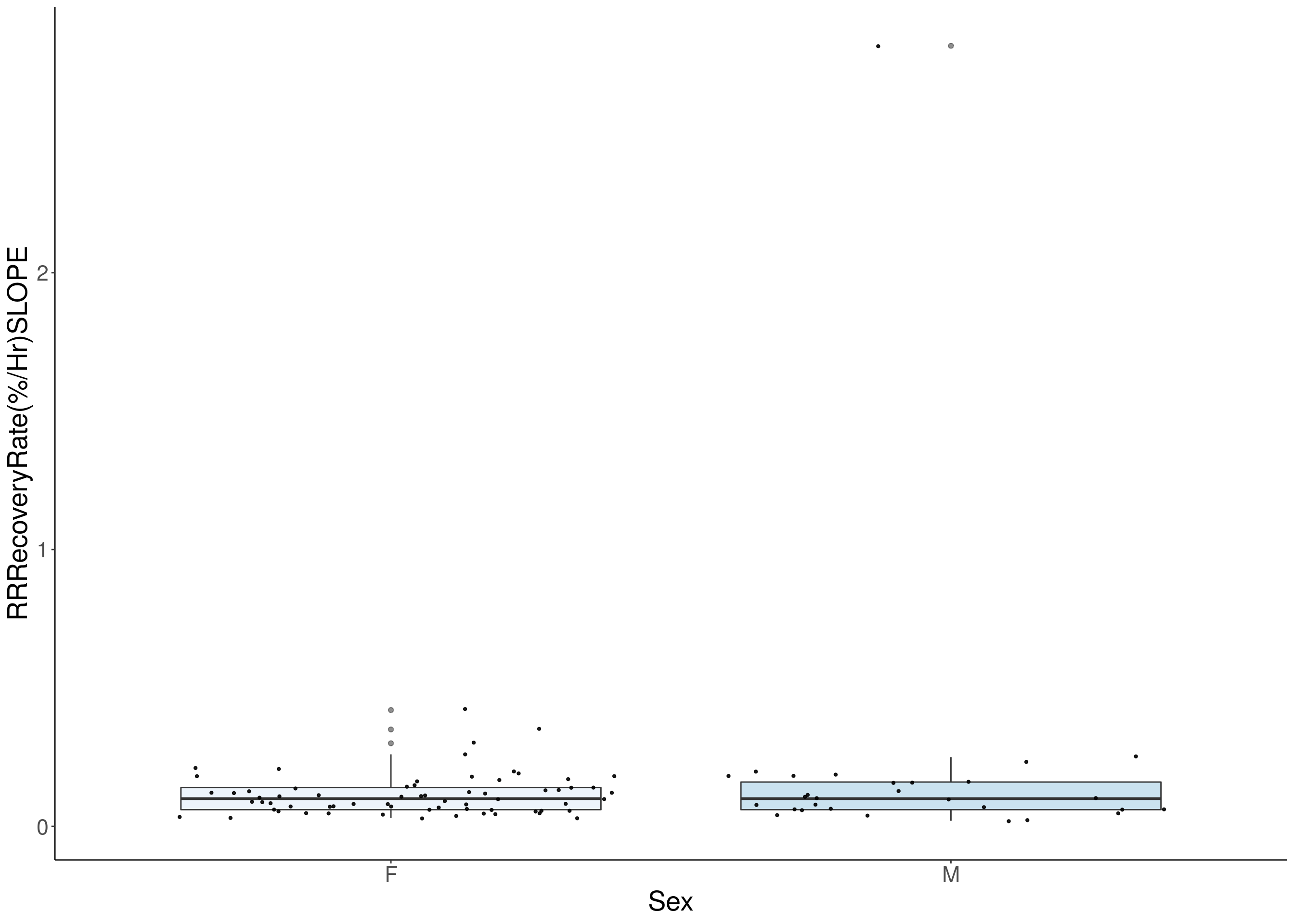
for(i in 8:16){
df <- do.pheno[,c(1, 3, i)]
df <- df[complete.cases(df), ]
p <- ggplot(df, aes(x=Sex, y=df[,3], group = Sex, fill = Sex, alpha = 0.9)) +
geom_boxplot(show.legend = F , outlier.size = 1.5, notchwidth = 0.85) +
geom_jitter(color="black", size=0.8, alpha=0.9) +
scale_fill_brewer(palette="Blues") +
ylab(paste0(colnames(df)[3])) +
xlab("Sex") +
labs(fill = "") +
theme(legend.position = "none",
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
axis.line = element_line(colour = "black"),
text = element_text(size=21),
axis.title=element_text(size=21)) +
guides(shape = guide_legend(override.aes = list(size = 12)))
print(p)
}
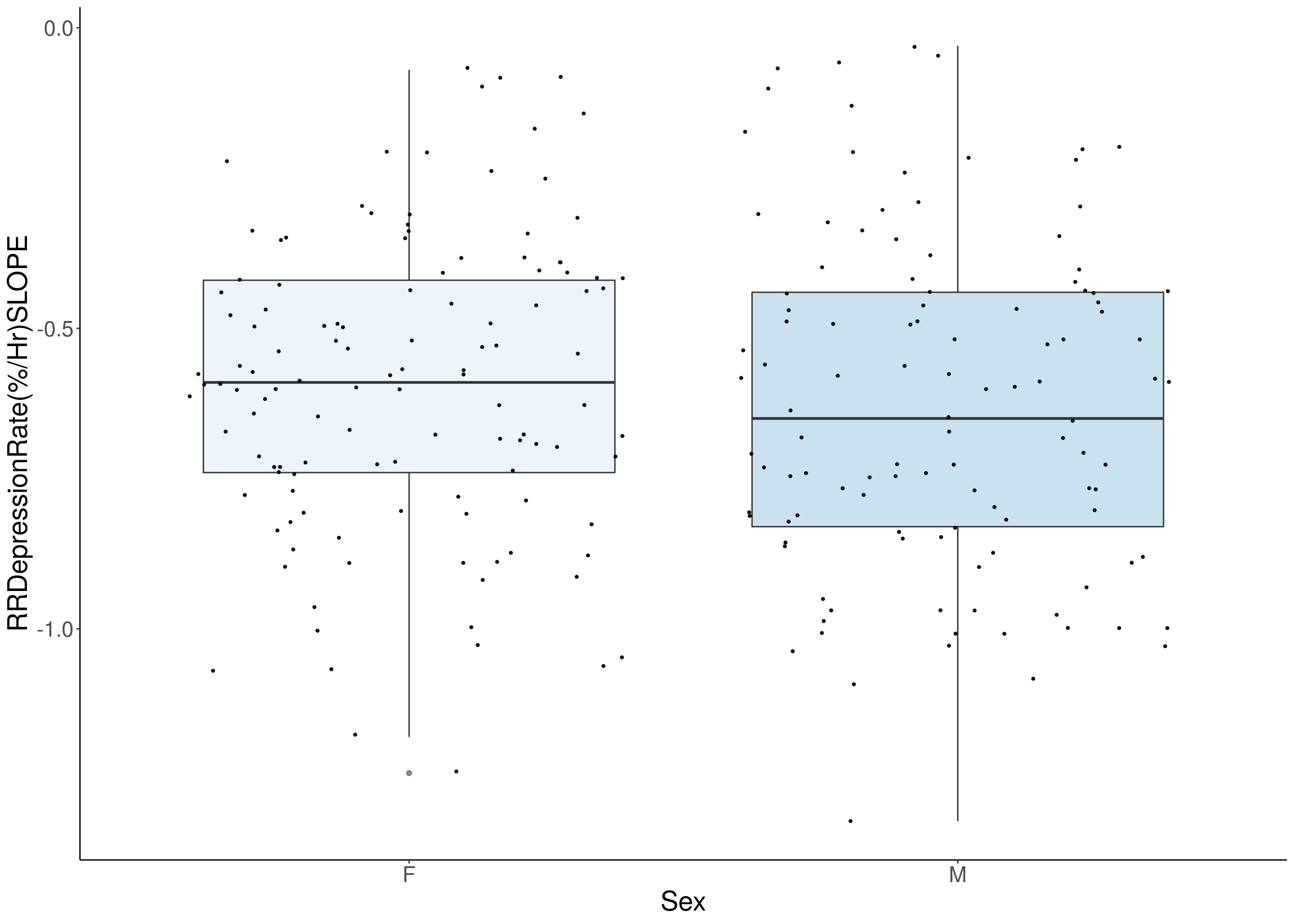
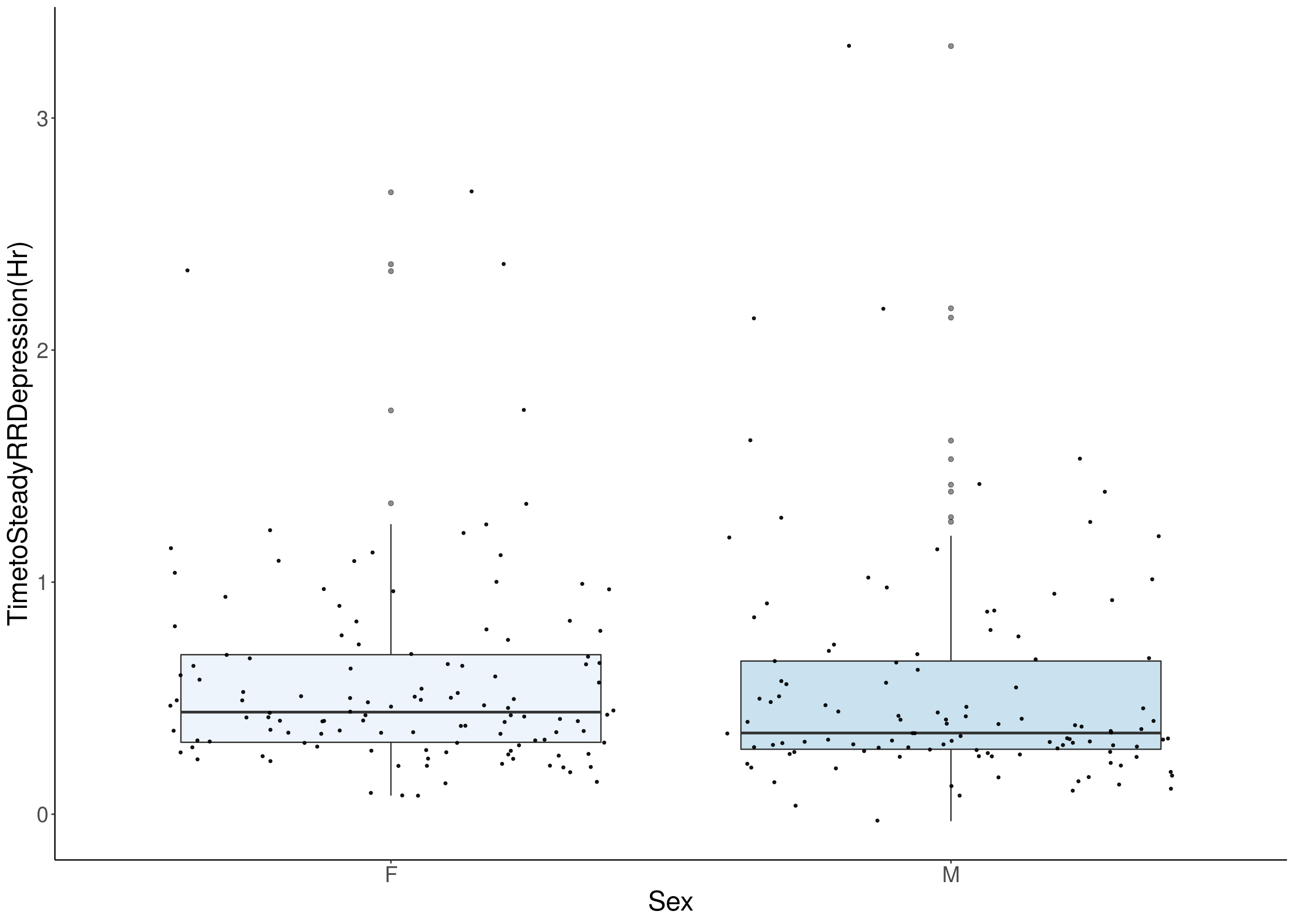
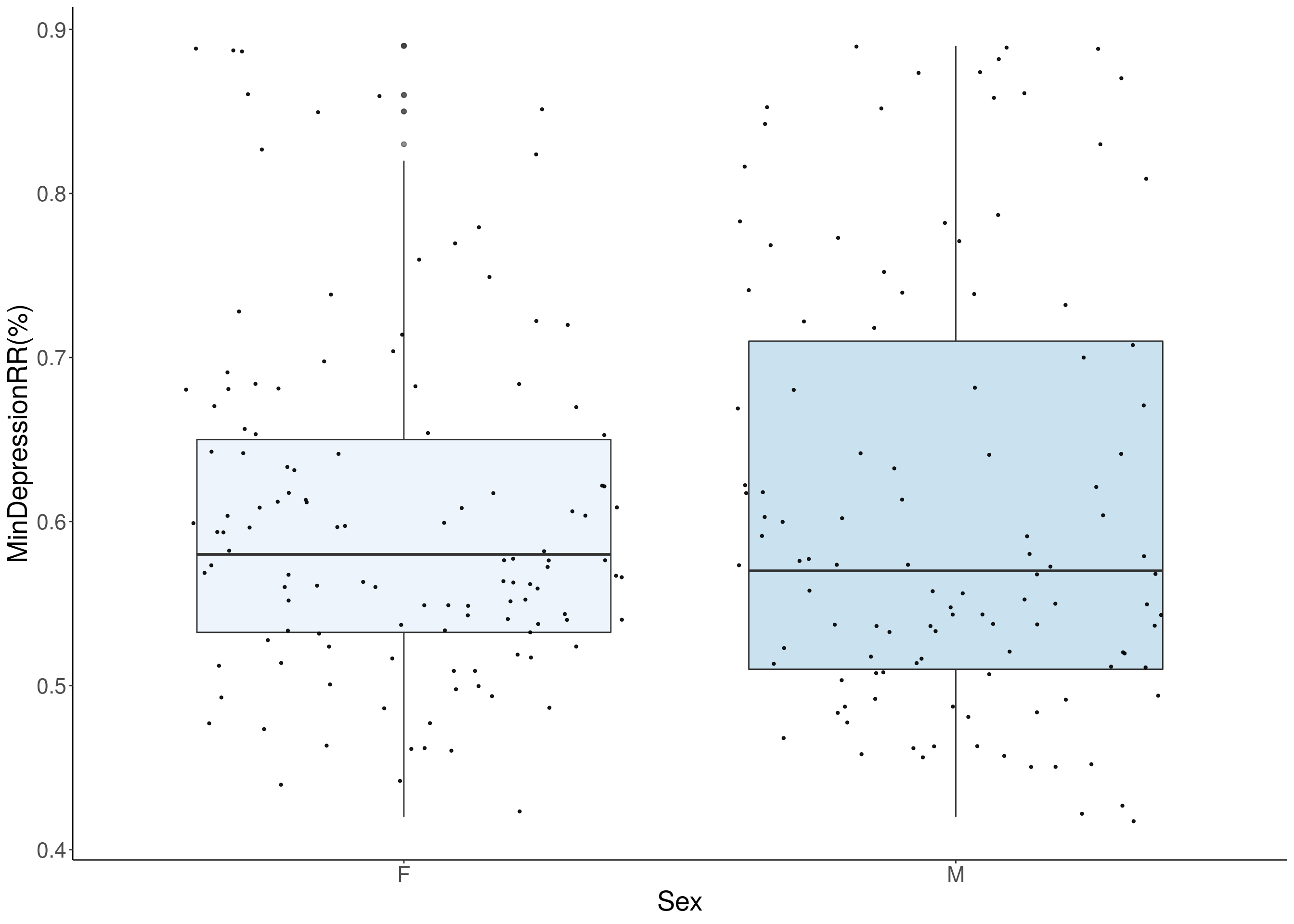
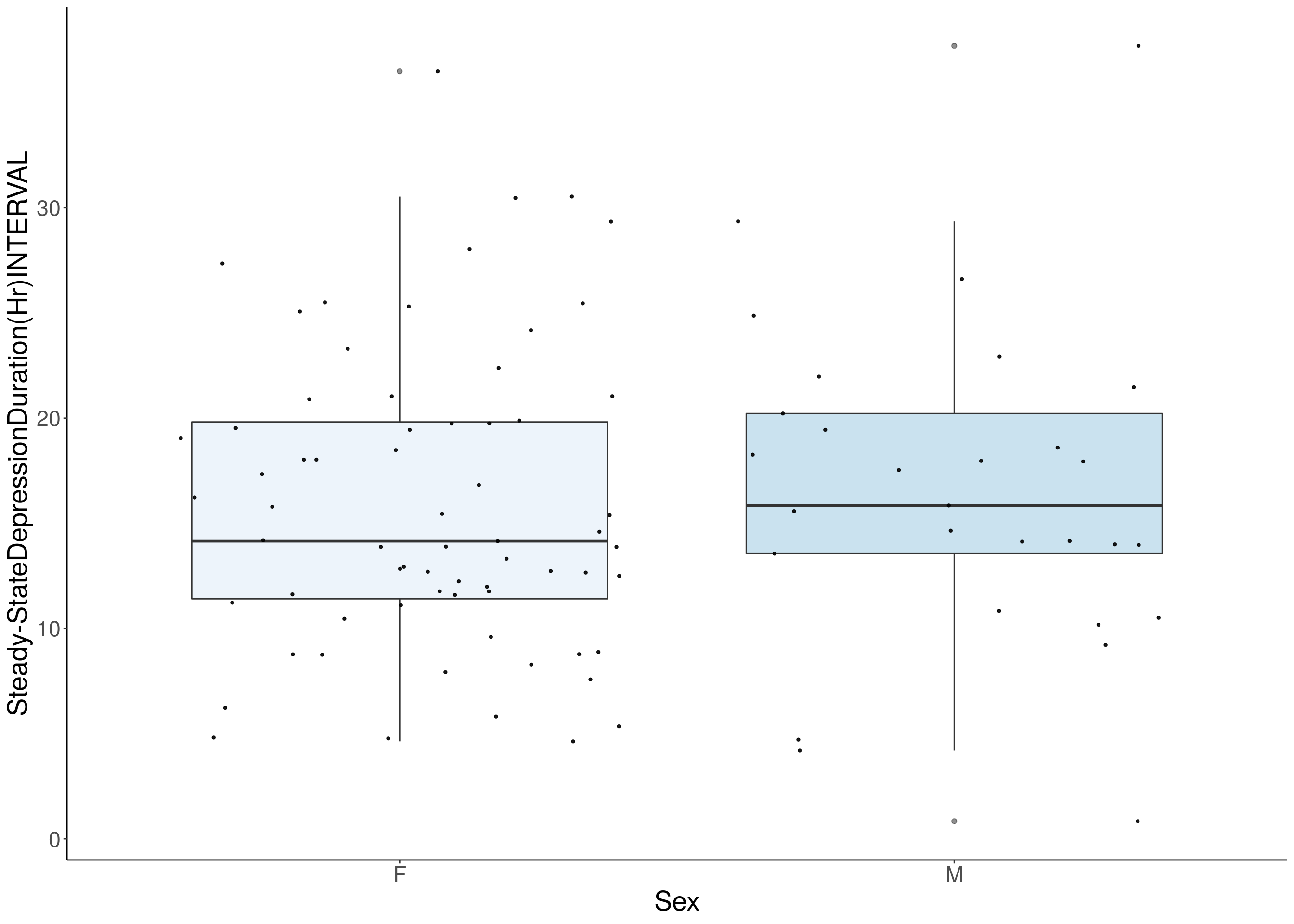
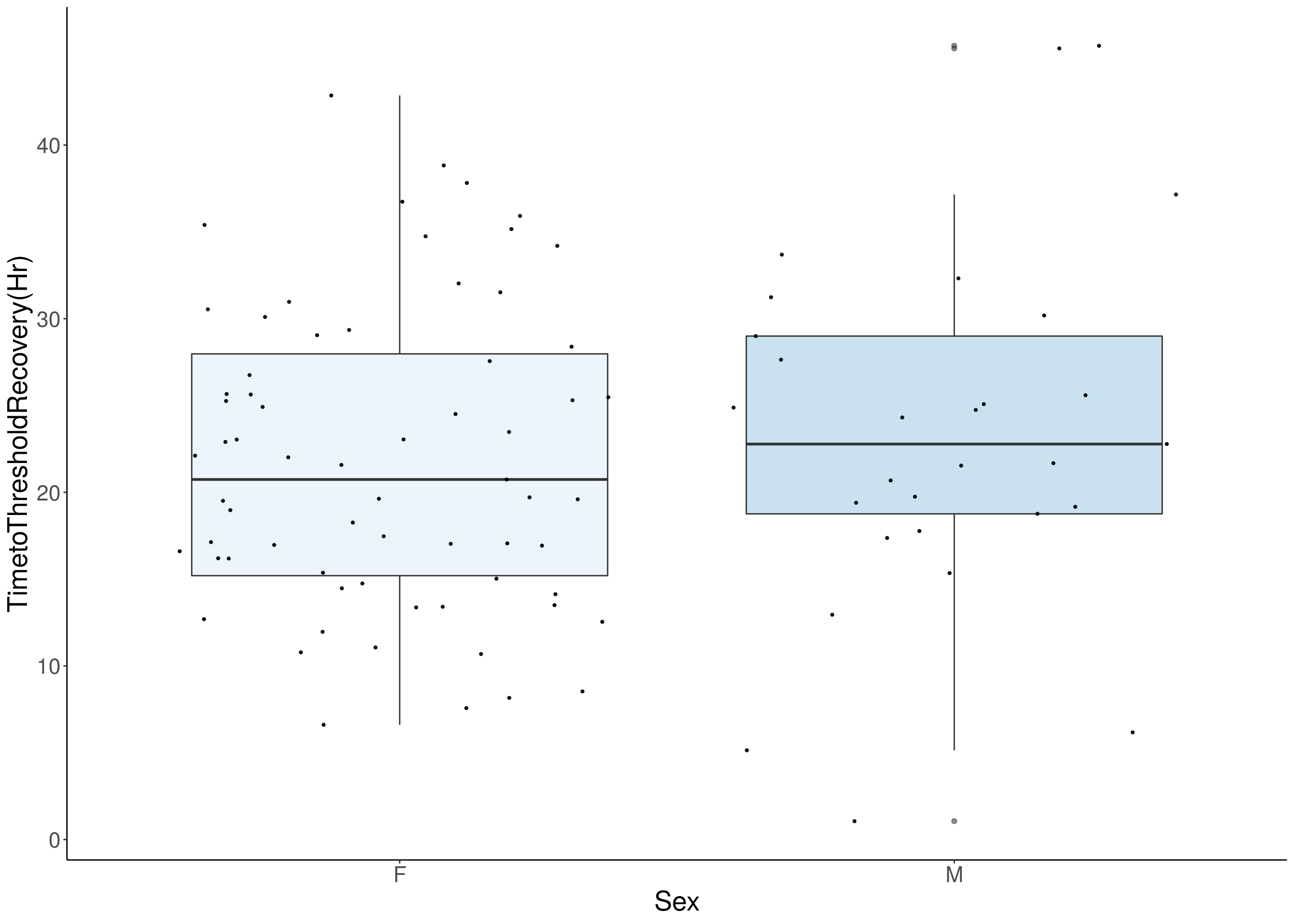
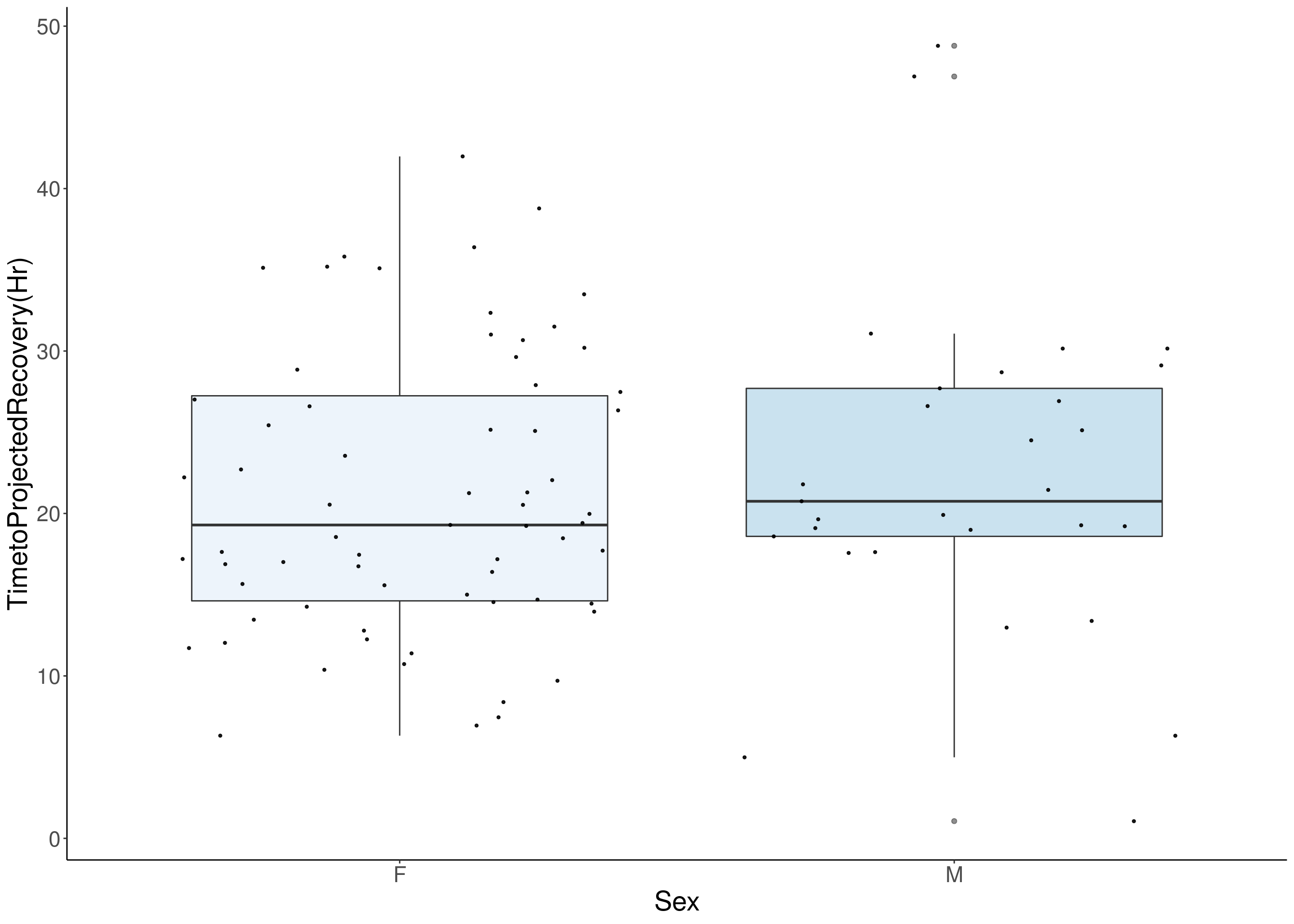
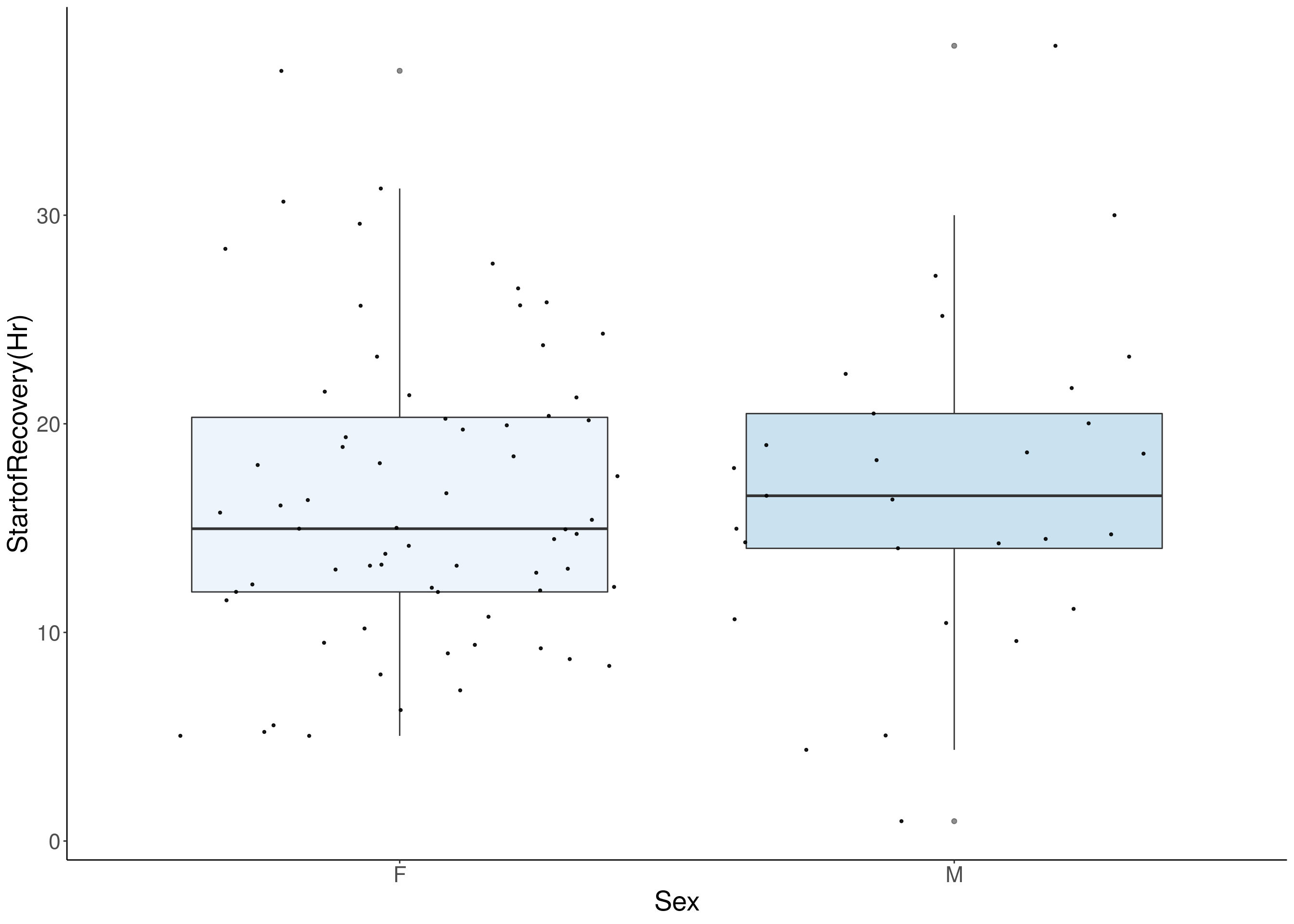
#histogram
for(i in c(8:10, 12:16)){
df <- do.pheno[,c(1, 3, i)]
p <- ggplot(data=df, aes(x=df[,3])) +
geom_histogram() +
ylab("Number of DO mice") +
xlab(paste0(colnames(df)[3])) +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
panel.background = element_blank(), axis.line = element_line(colour = "black"),
text = element_text(size=21))
print(p)
}
# Warning: Removed 132 rows containing non-finite values (stat_bin).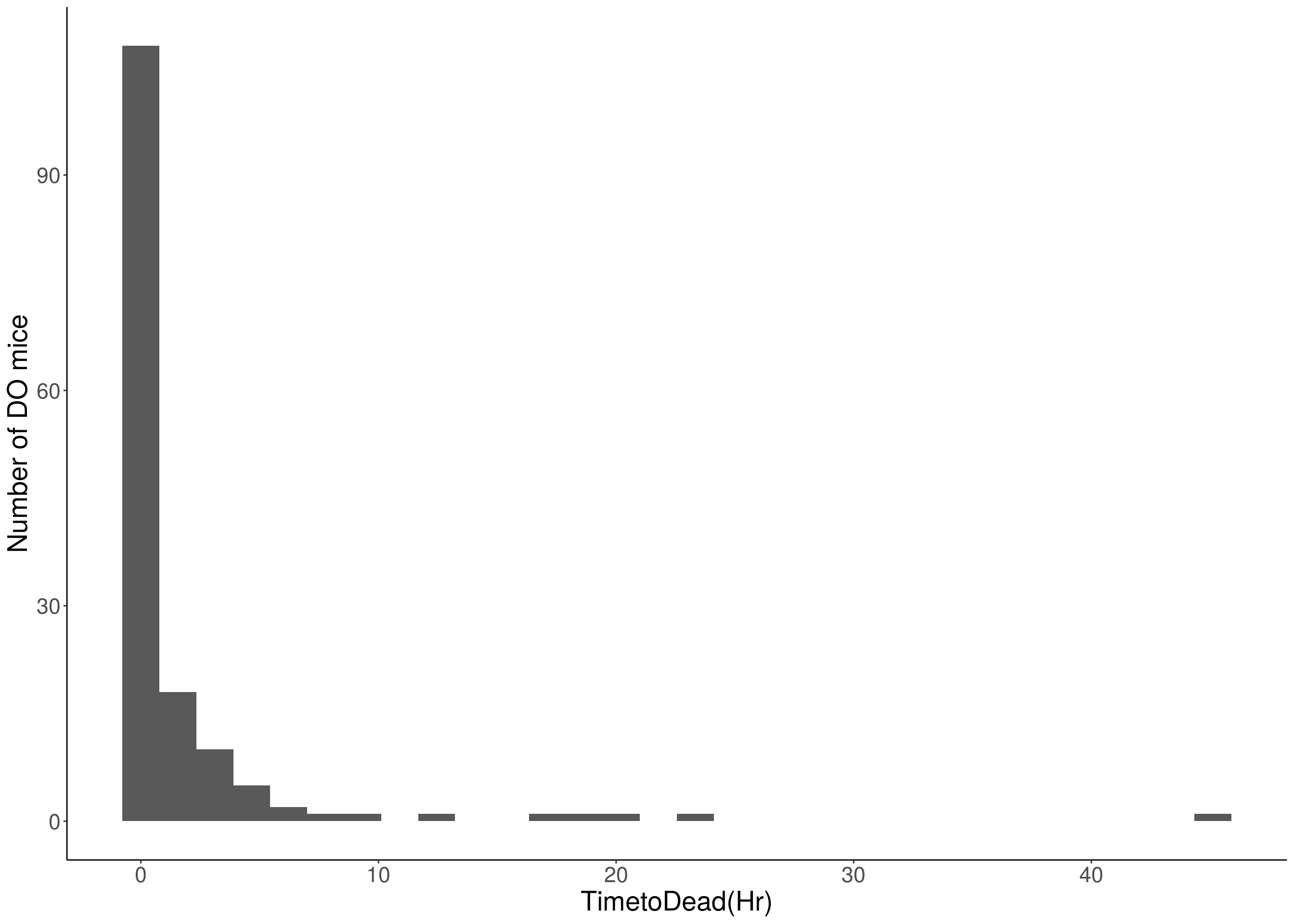
# Warning: Removed 48 rows containing non-finite values (stat_bin).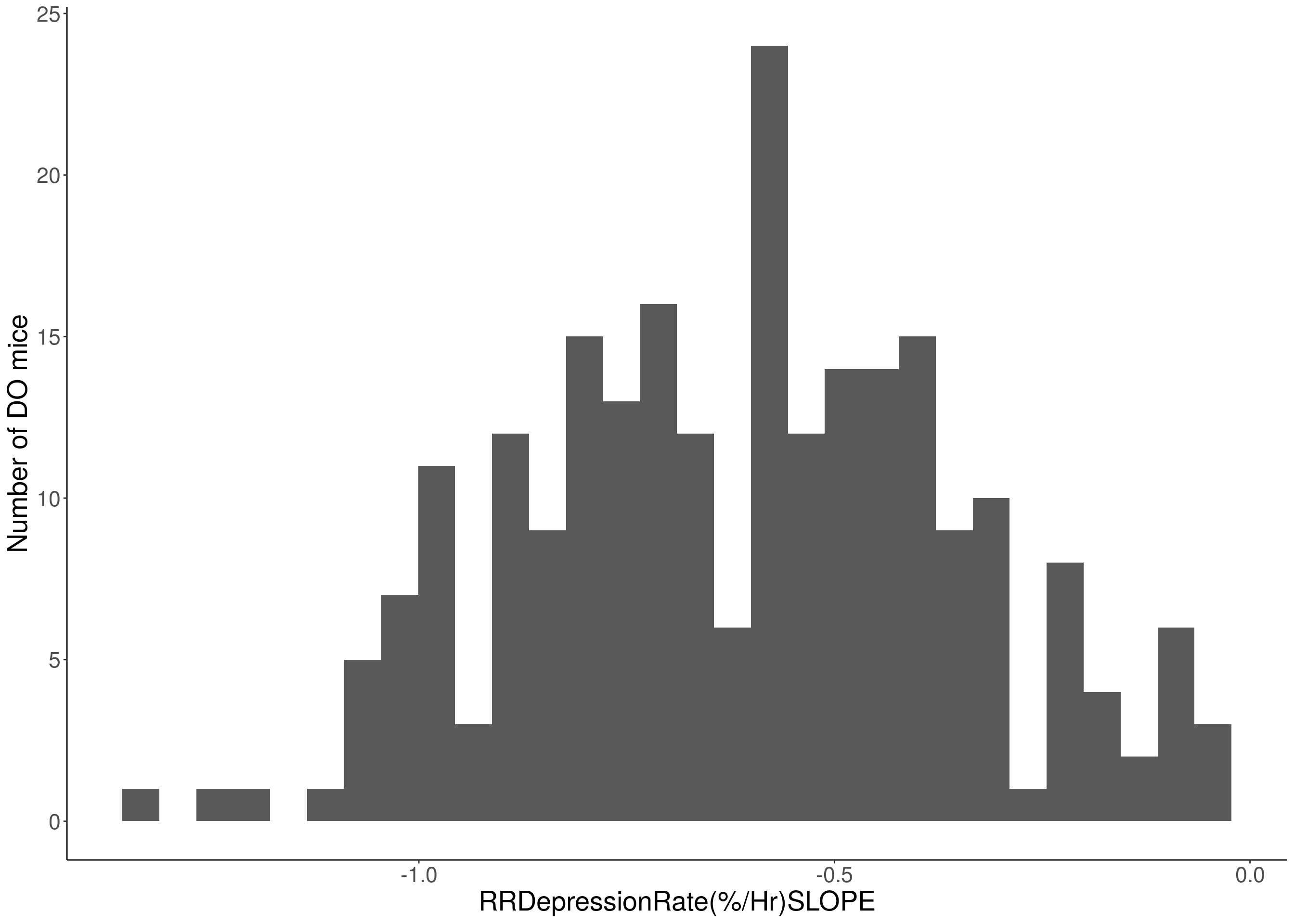
# Warning: Removed 48 rows containing non-finite values (stat_bin).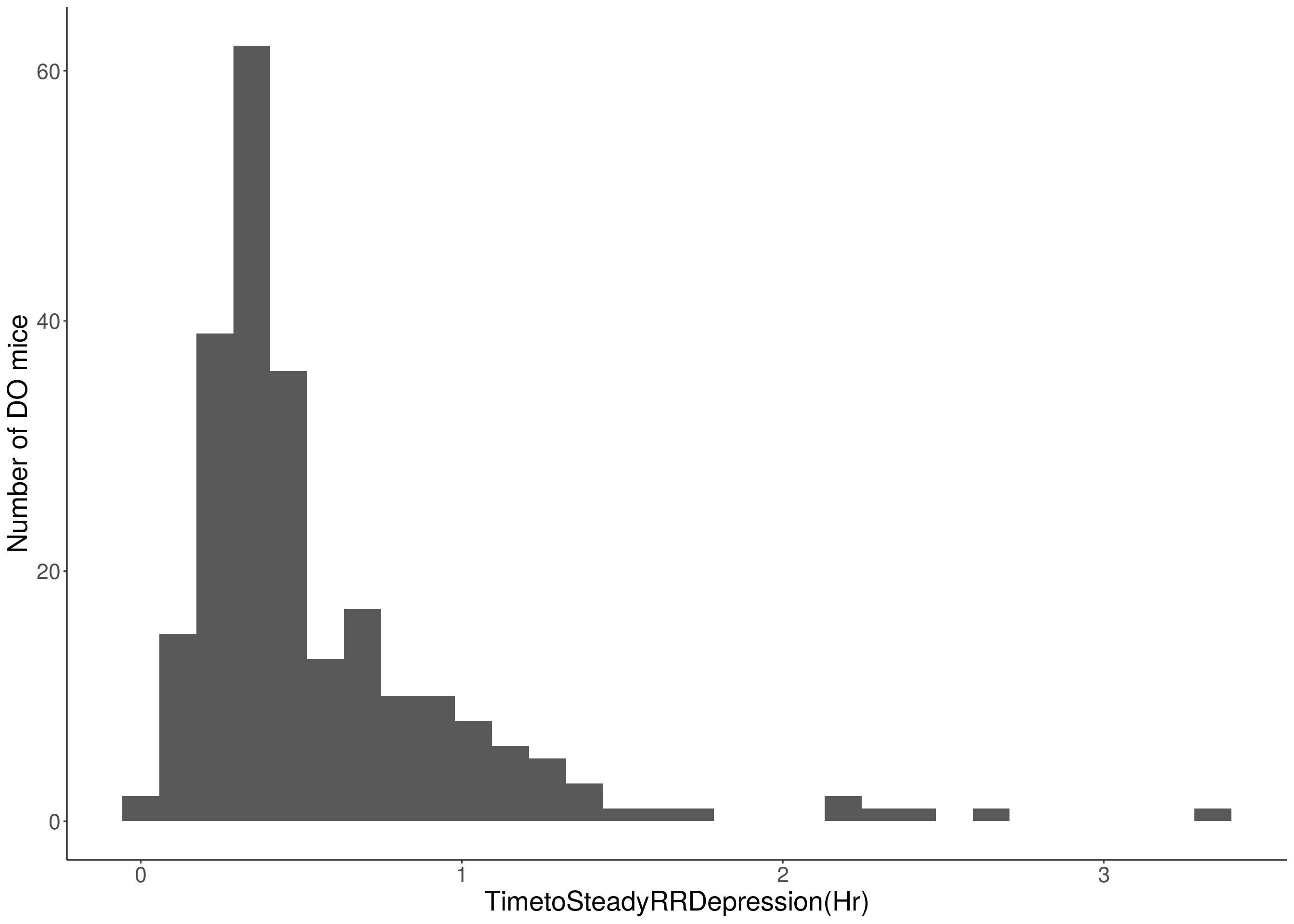
# Warning: Removed 187 rows containing non-finite values (stat_bin).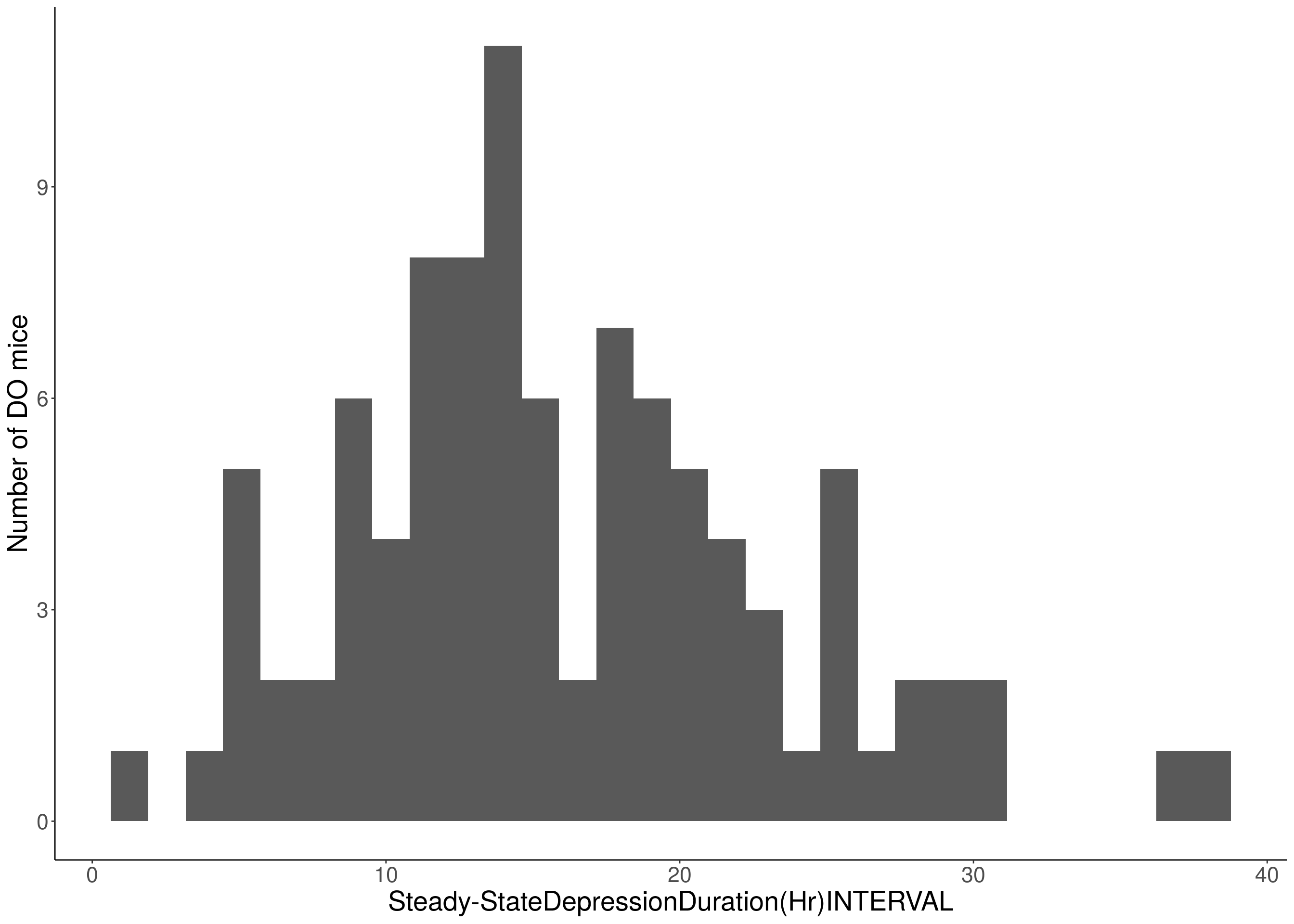
# Warning: Removed 187 rows containing non-finite values (stat_bin).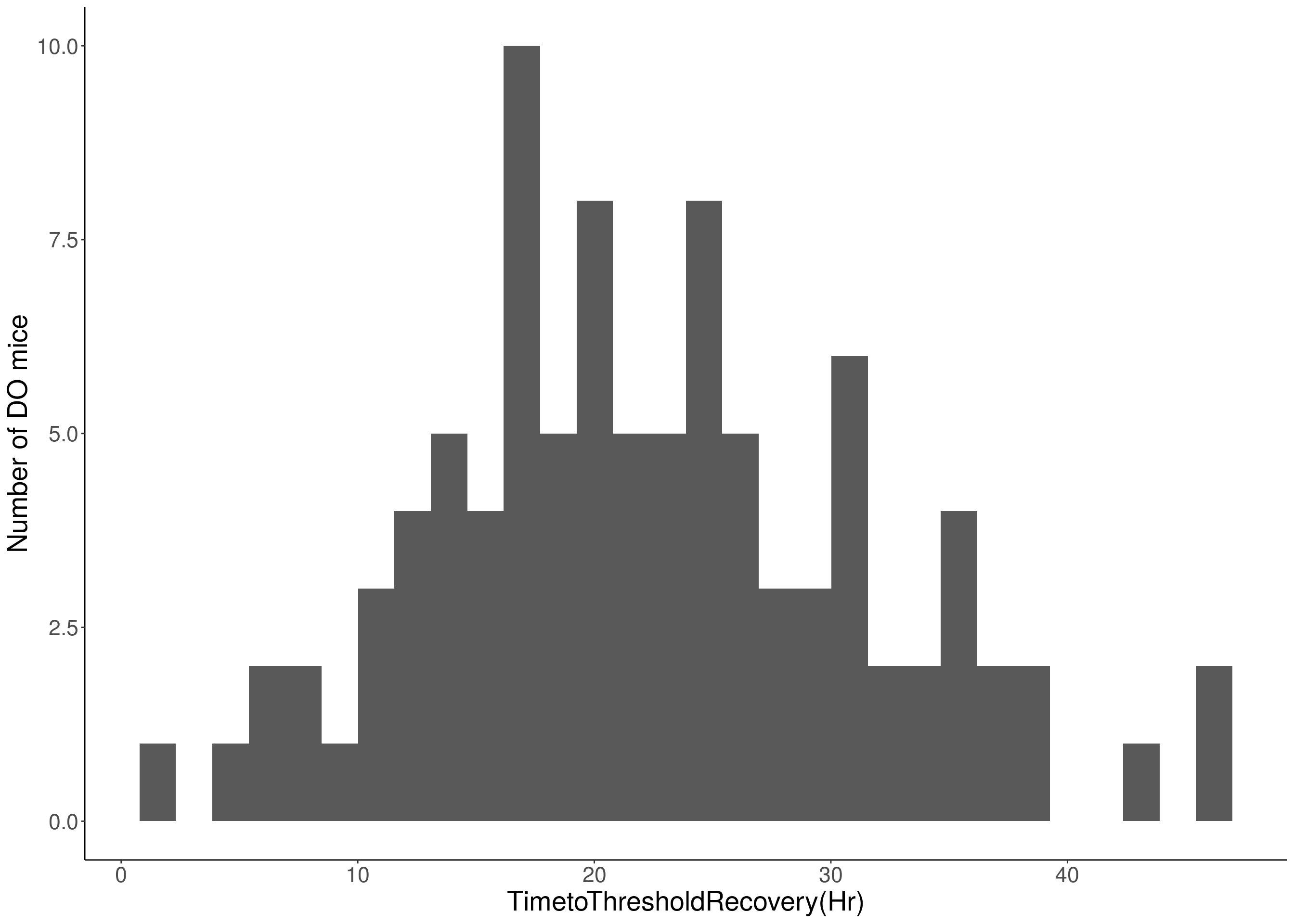
# Warning: Removed 187 rows containing non-finite values (stat_bin).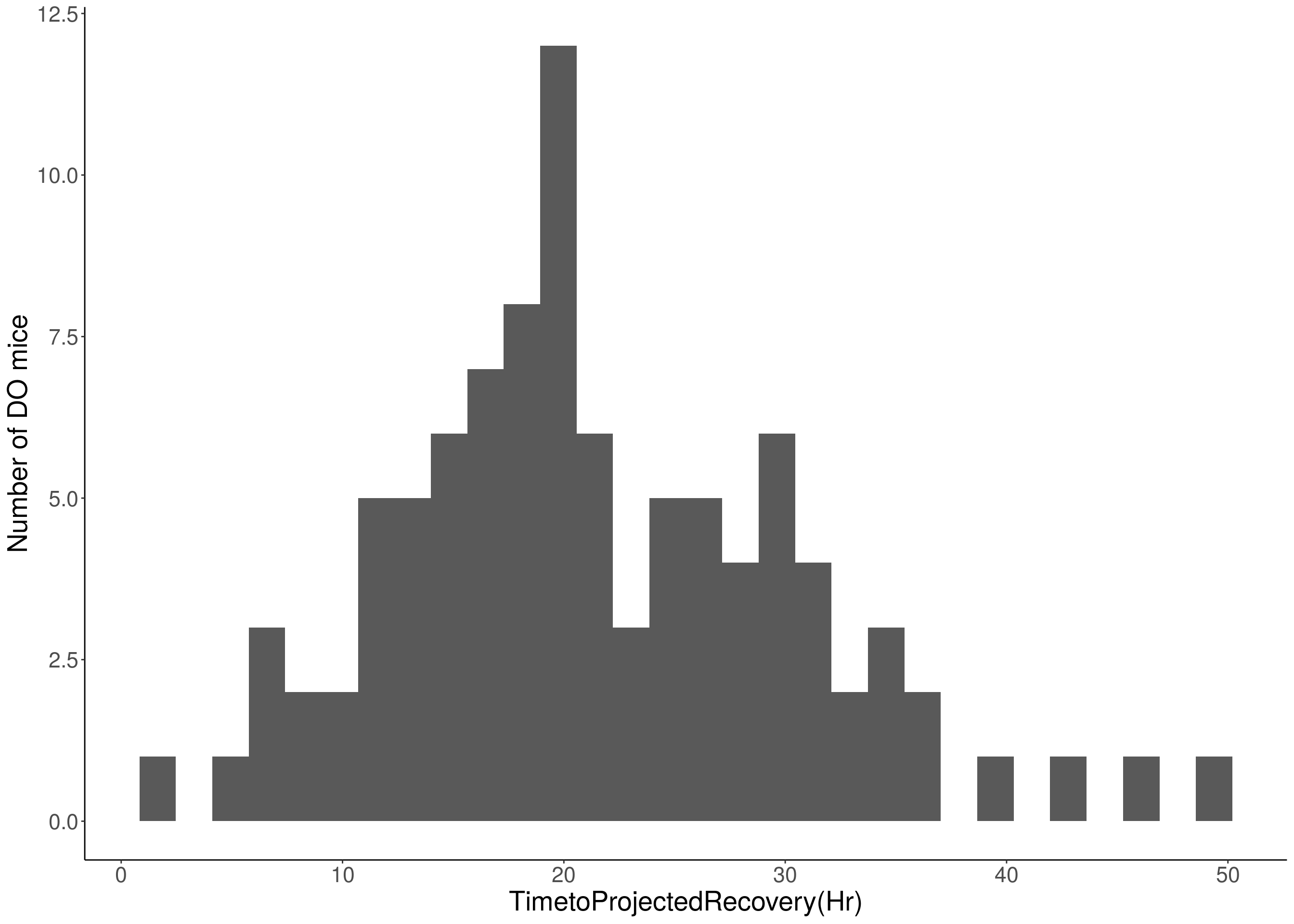
# Warning: Removed 187 rows containing non-finite values (stat_bin).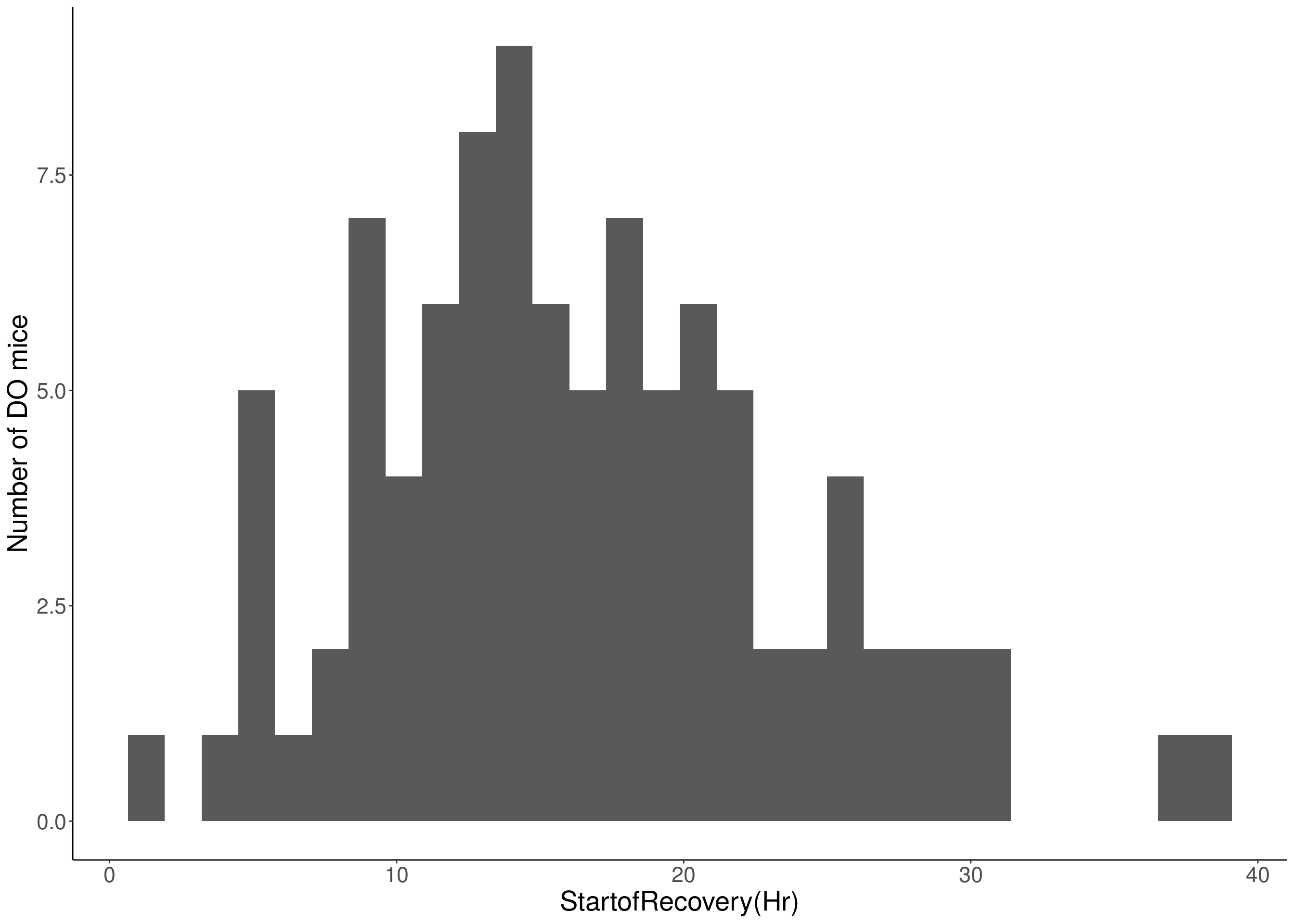
# Warning: Removed 187 rows containing non-finite values (stat_bin).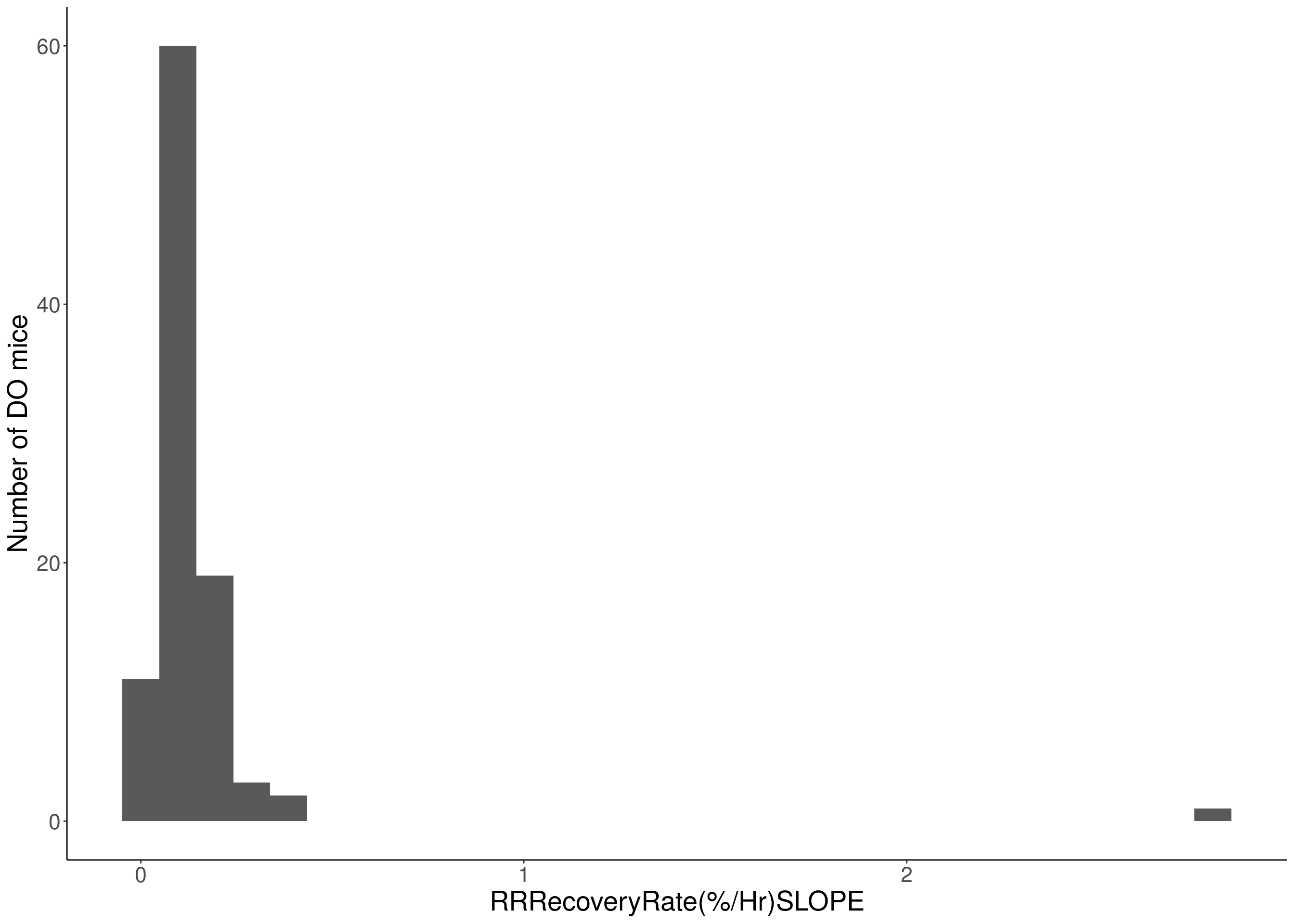
#for "MinDepressionRR(%)"
i = 11
df <- do.pheno[,c(1, 3, i)]
p <- ggplot(data=df, aes(x=df[,3])) +
geom_histogram() +
ylab("Number of DO mice") +
xlab(paste0(colnames(df)[3])) +
xlim(0.4, 1) +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
panel.background = element_blank(), axis.line = element_line(colour = "black"),
text = element_text(size=21))
print(p)
# Warning: Removed 48 rows containing non-finite values (stat_bin).
# Warning: Removed 2 rows containing missing values (geom_bar).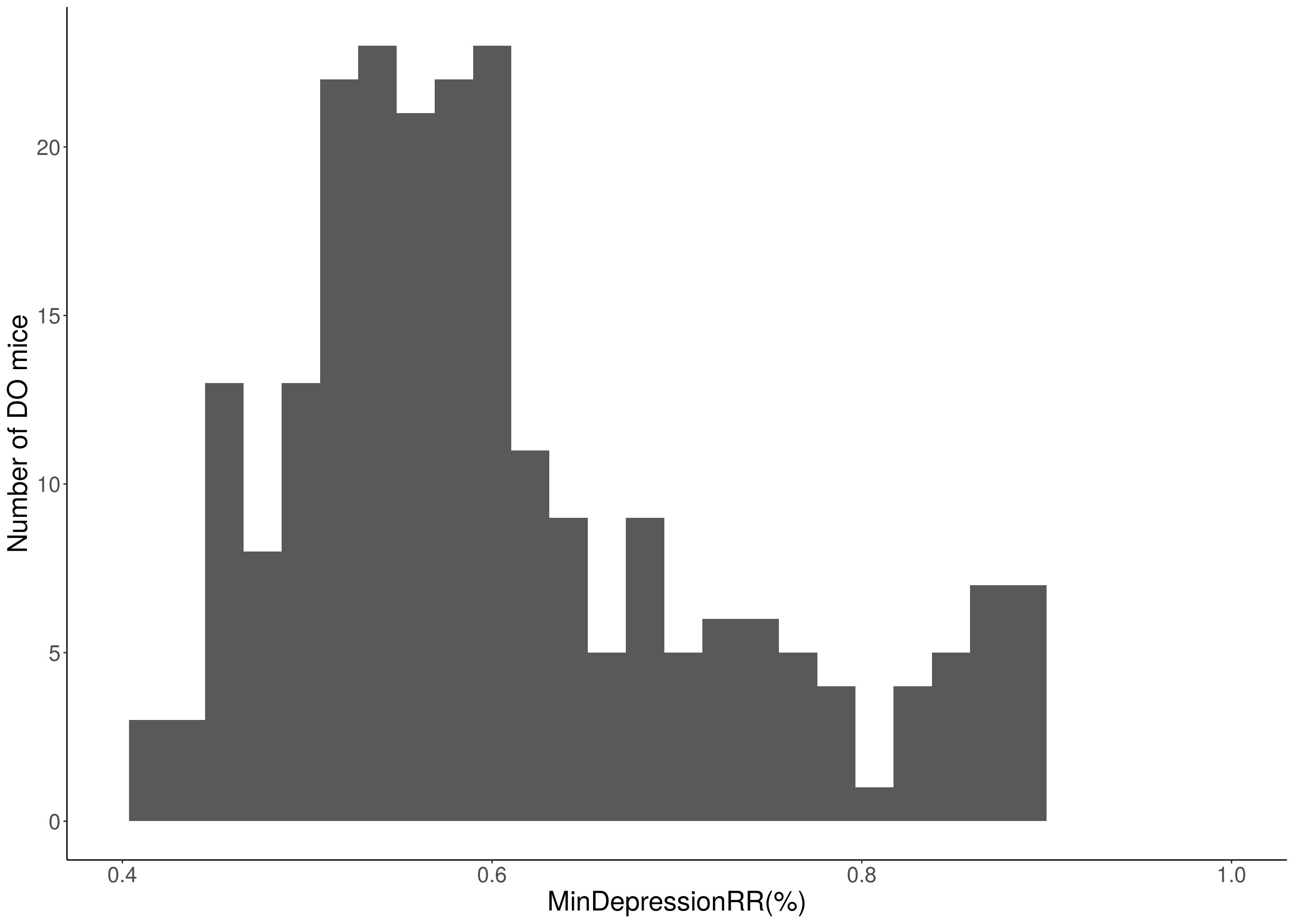
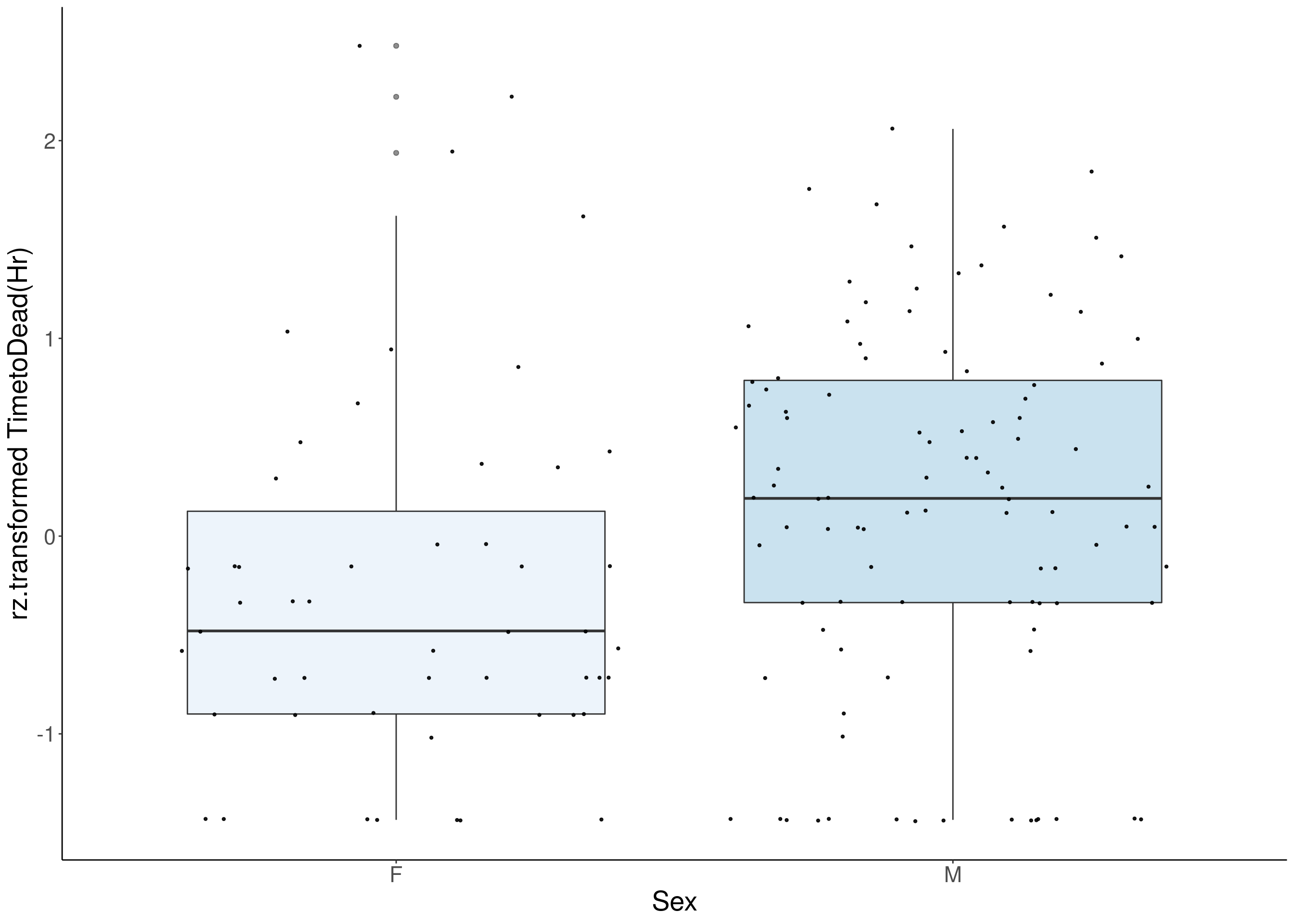
#boxplot on the rankz data------------
for(i in 8:16){
df <- do.pheno[,c(1, 3, i)]
df <- df[complete.cases(df), ]
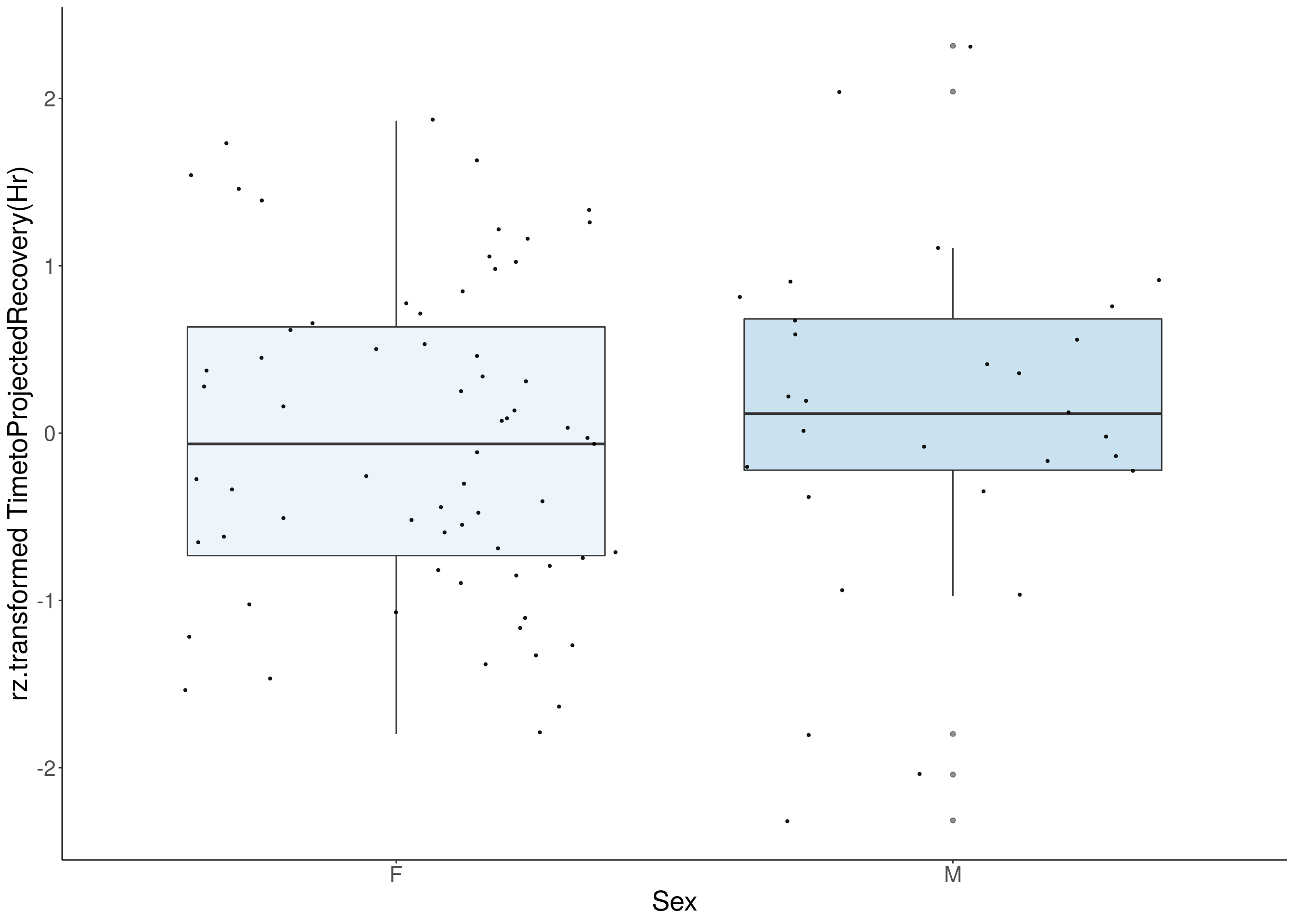
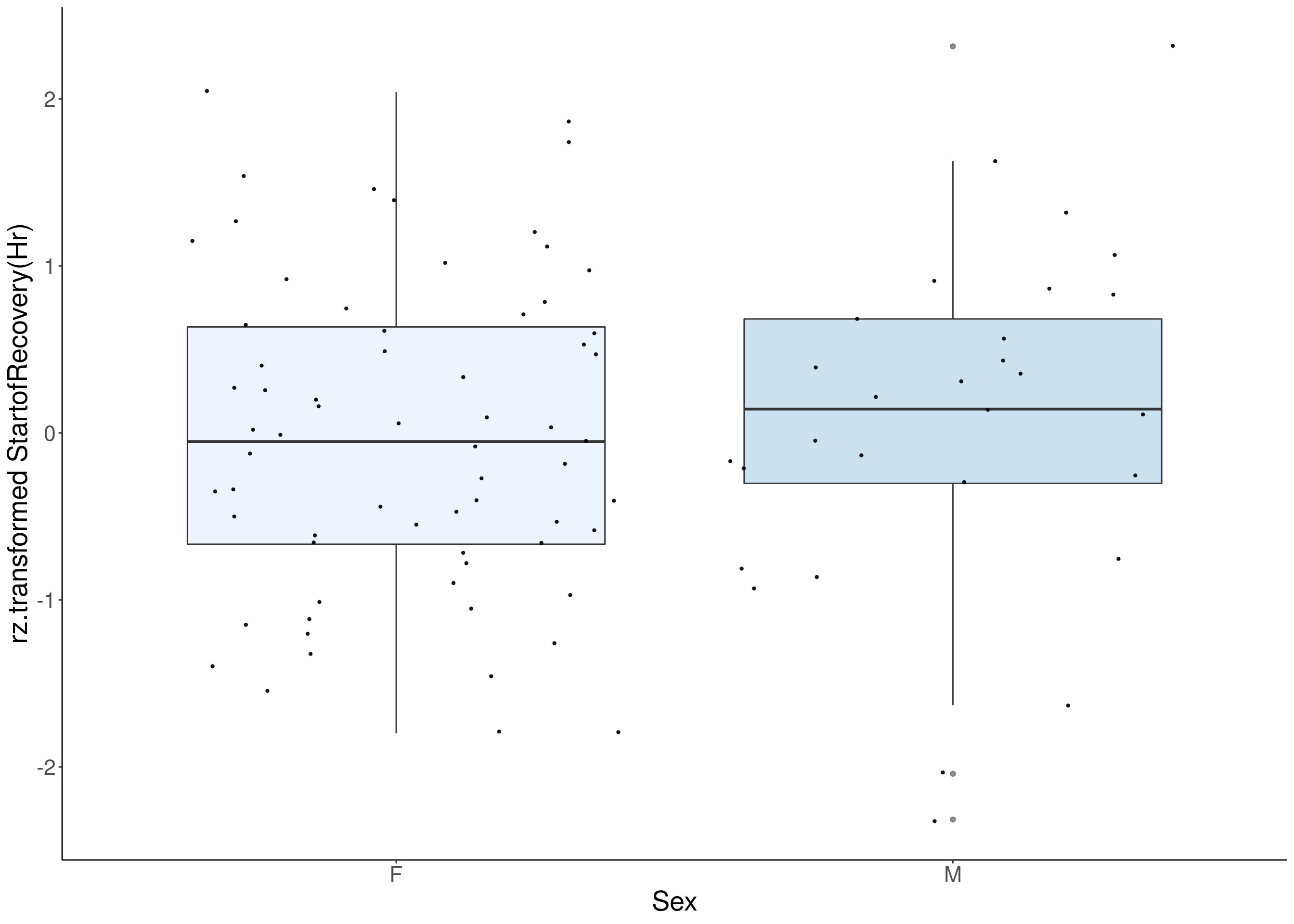
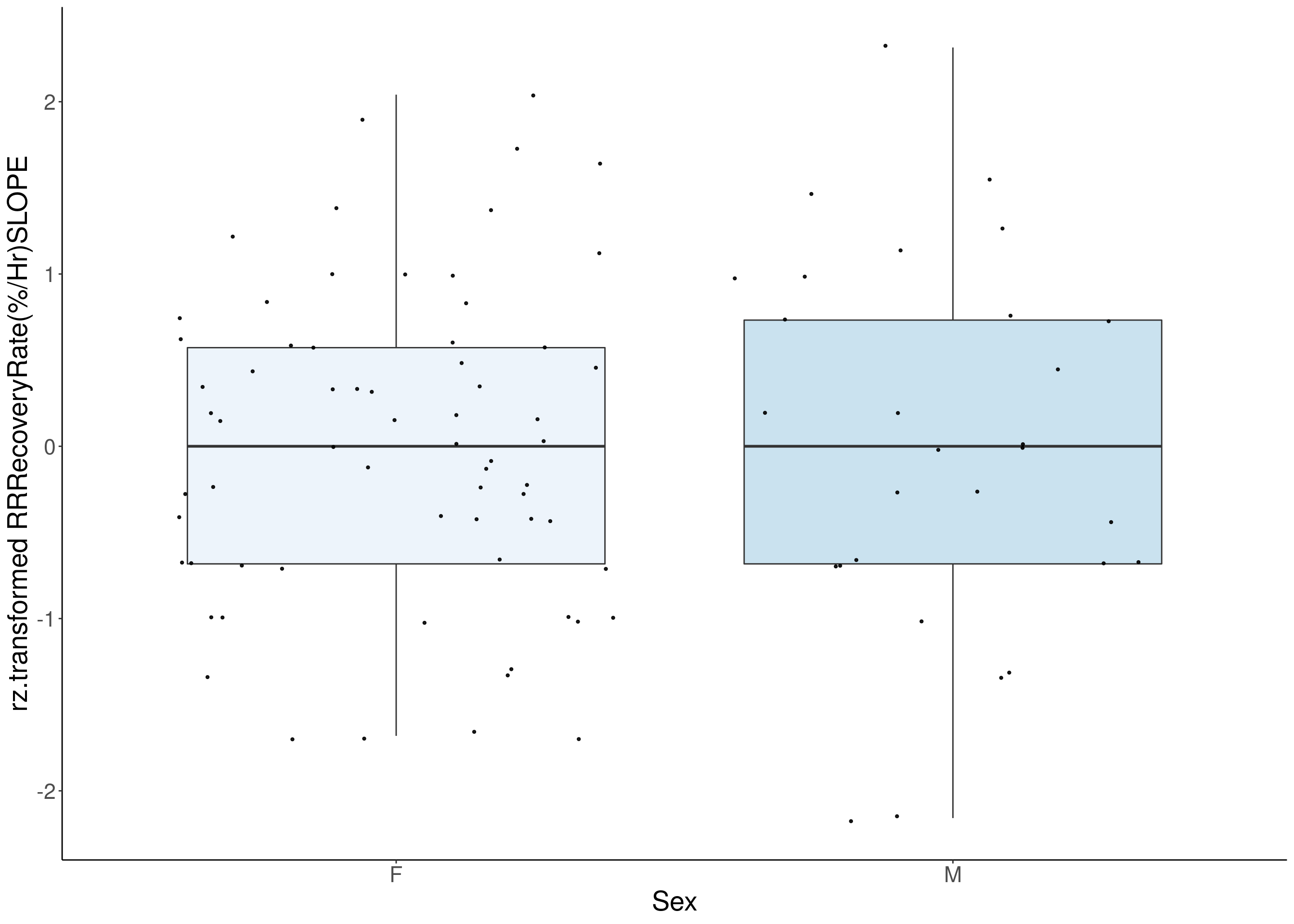
p <- ggplot(df, aes(x=Sex, y= rz.transform(df[,3]), group = Sex, fill = Sex, alpha = 0.9)) +
geom_boxplot(show.legend = F , outlier.size = 1.5, notchwidth = 0.85) +
geom_jitter(color="black", size=0.8, alpha=0.9) +
scale_fill_brewer(palette="Blues") +
ylab(paste0("rz.transformed ", colnames(df)[3])) +
xlab("Sex") +
labs(fill = "") +
theme(legend.position = "none",
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
axis.line = element_line(colour = "black"),
text = element_text(size=21),
axis.title=element_text(size=21)) +
guides(shape = guide_legend(override.aes = list(size = 12)))
print(p)
}
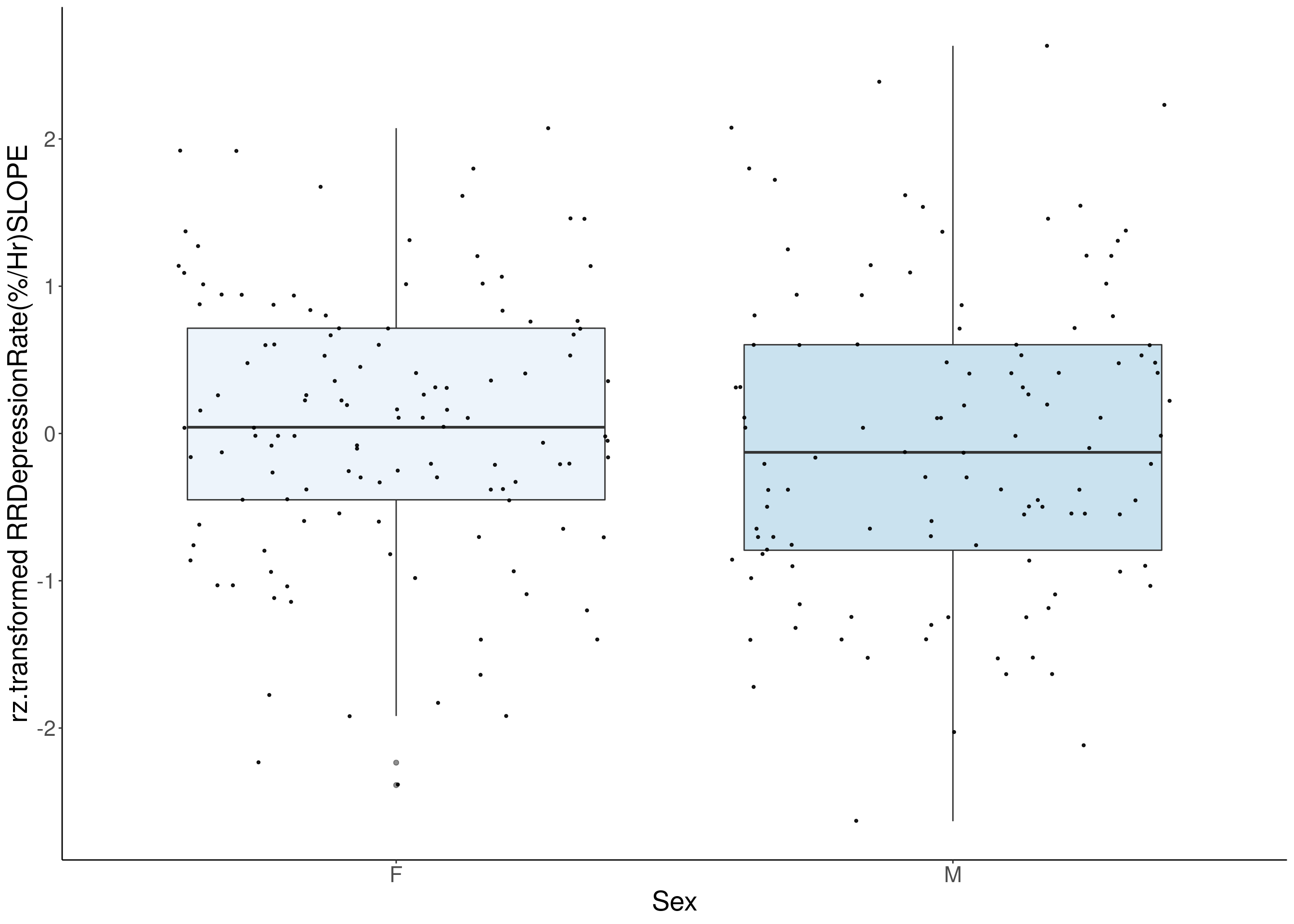
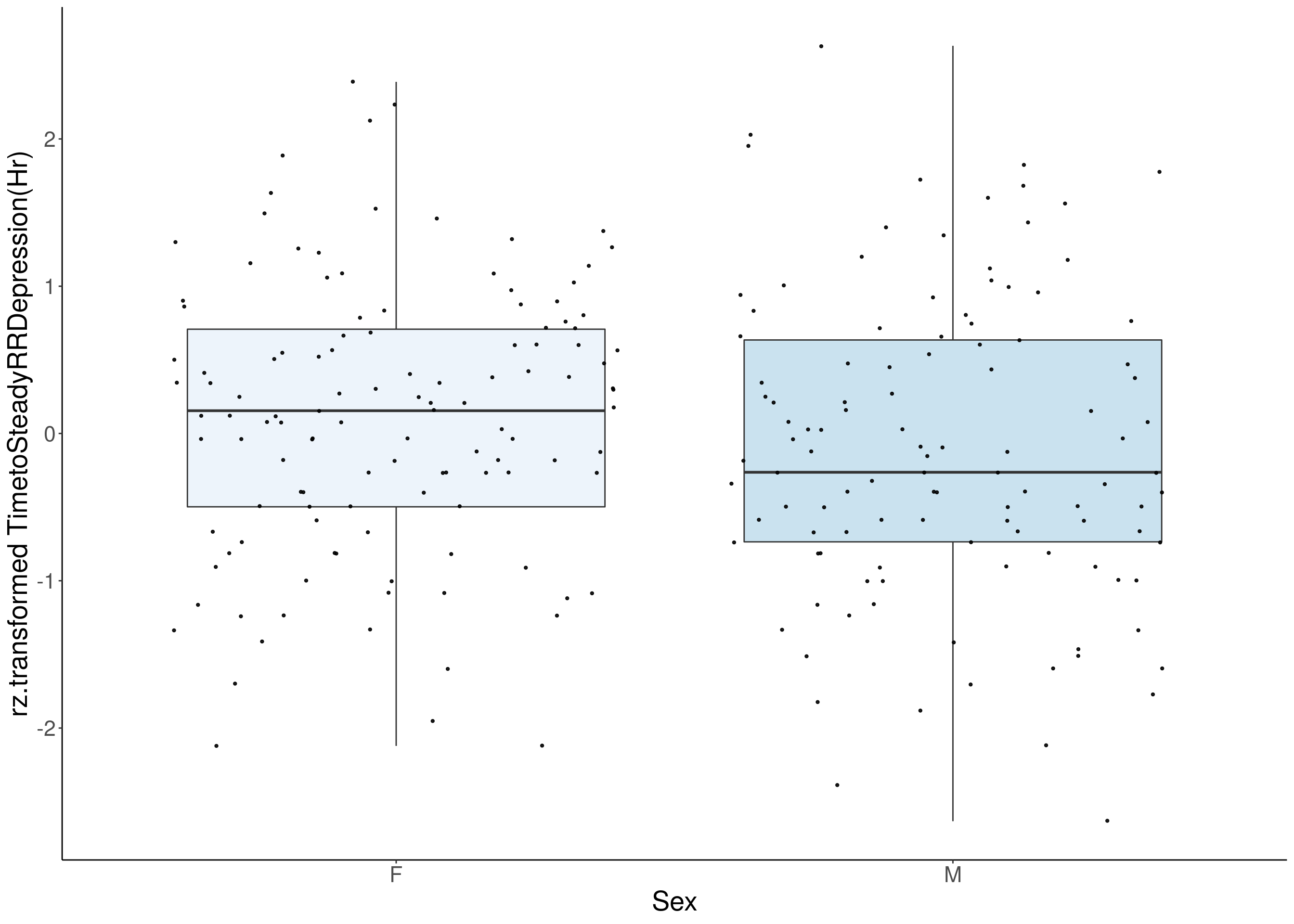
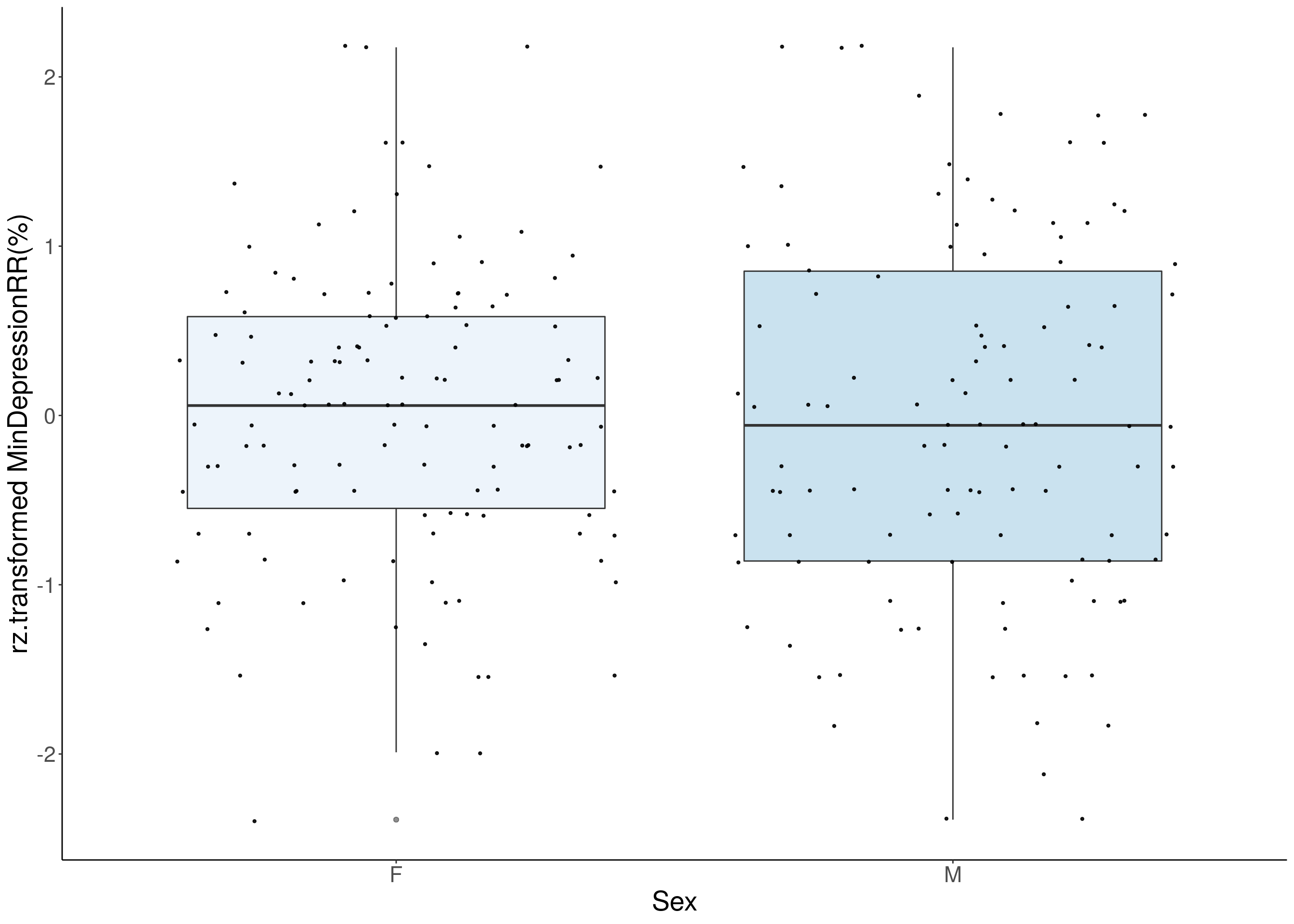
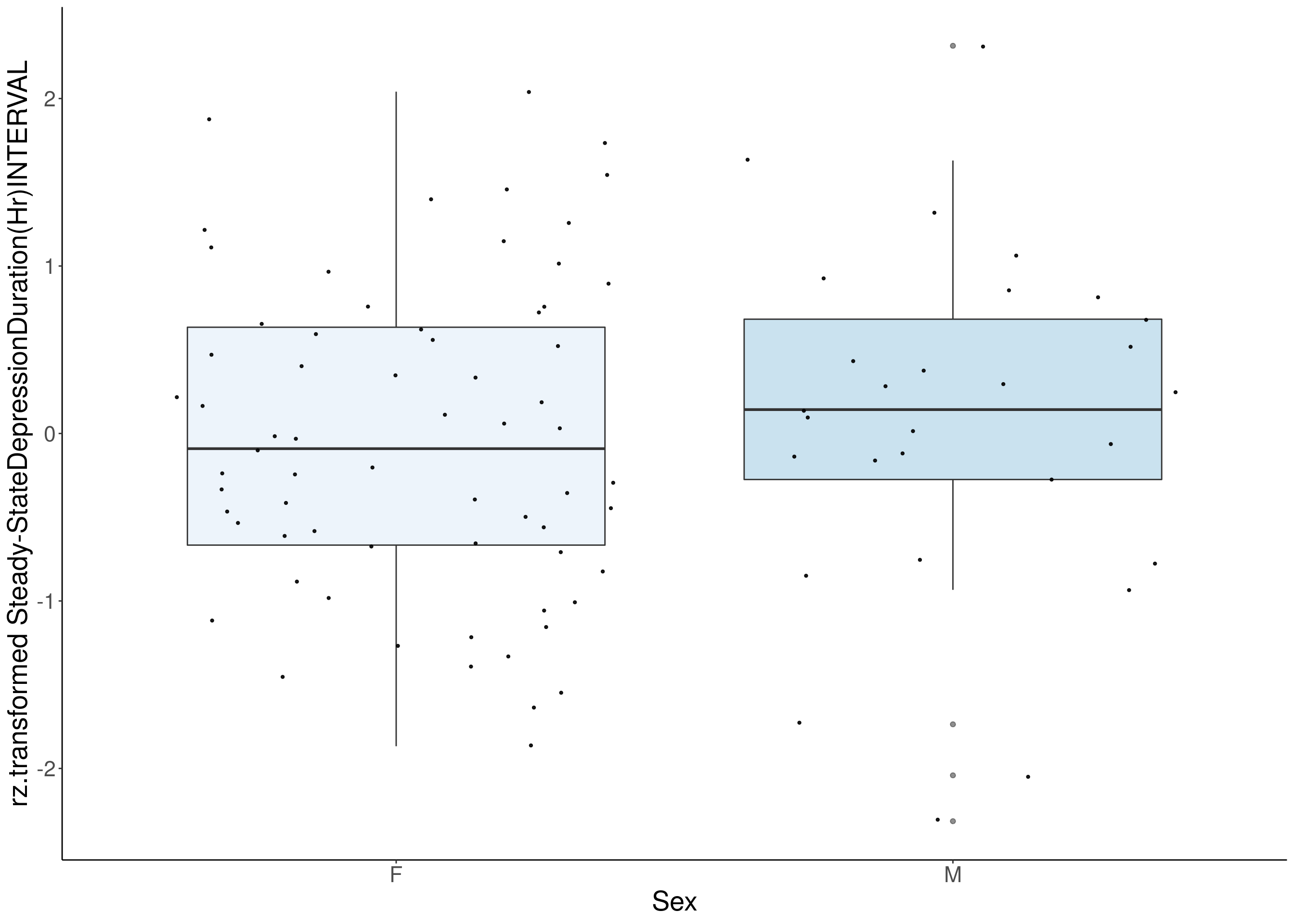
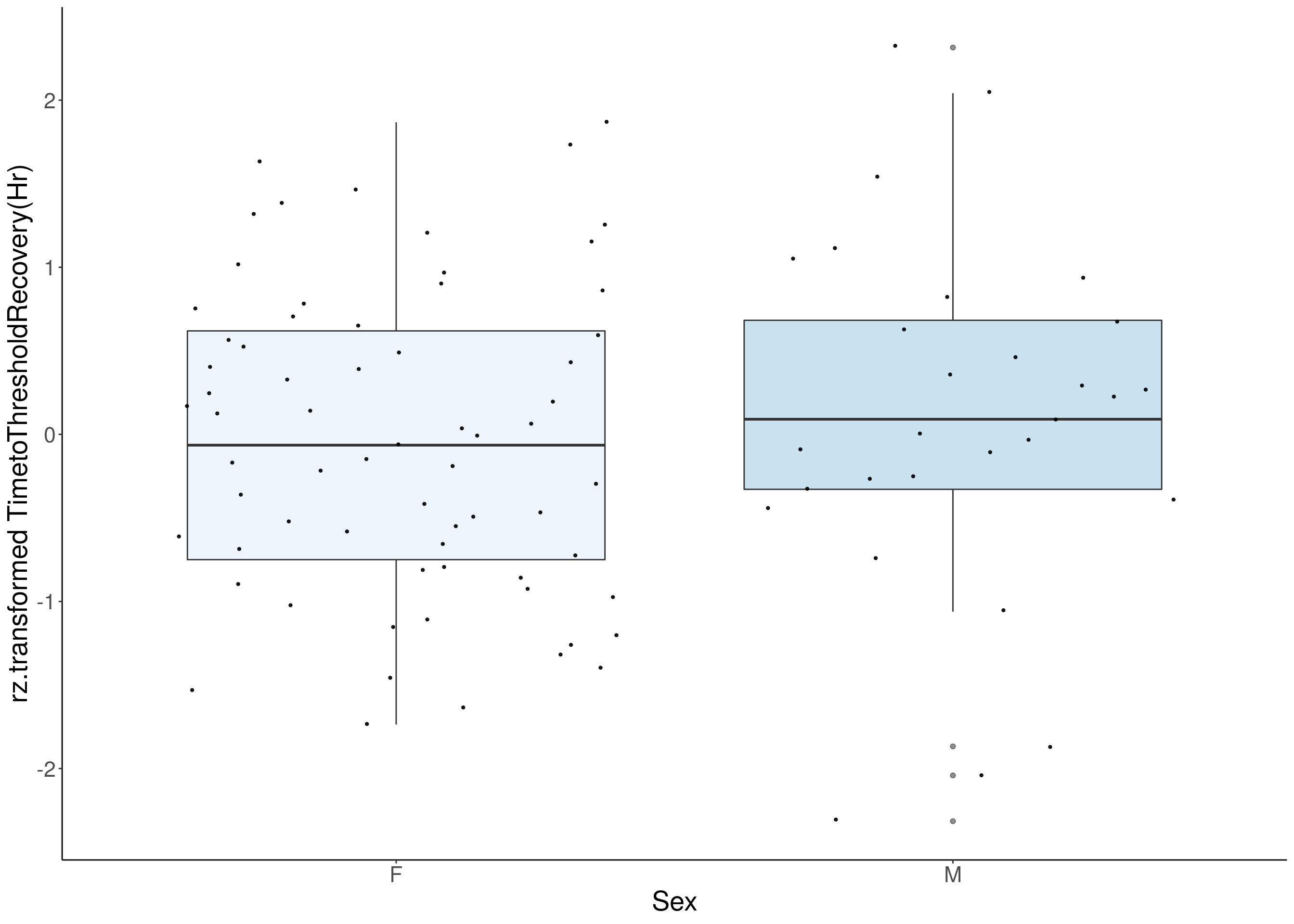
#histogram
for(i in 8:16){
df <- do.pheno[,c(1, 3, i)]
p <- ggplot(data=df, aes(x=rz.transform(df[,3]))) +
geom_histogram() +
ylab("Number of DO mice") +
xlab(paste0("rz.transformed ", colnames(df)[3])) +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
panel.background = element_blank(), axis.line = element_line(colour = "black"),
text = element_text(size=21))
print(p)
}
# Warning: Removed 132 rows containing non-finite values (stat_bin).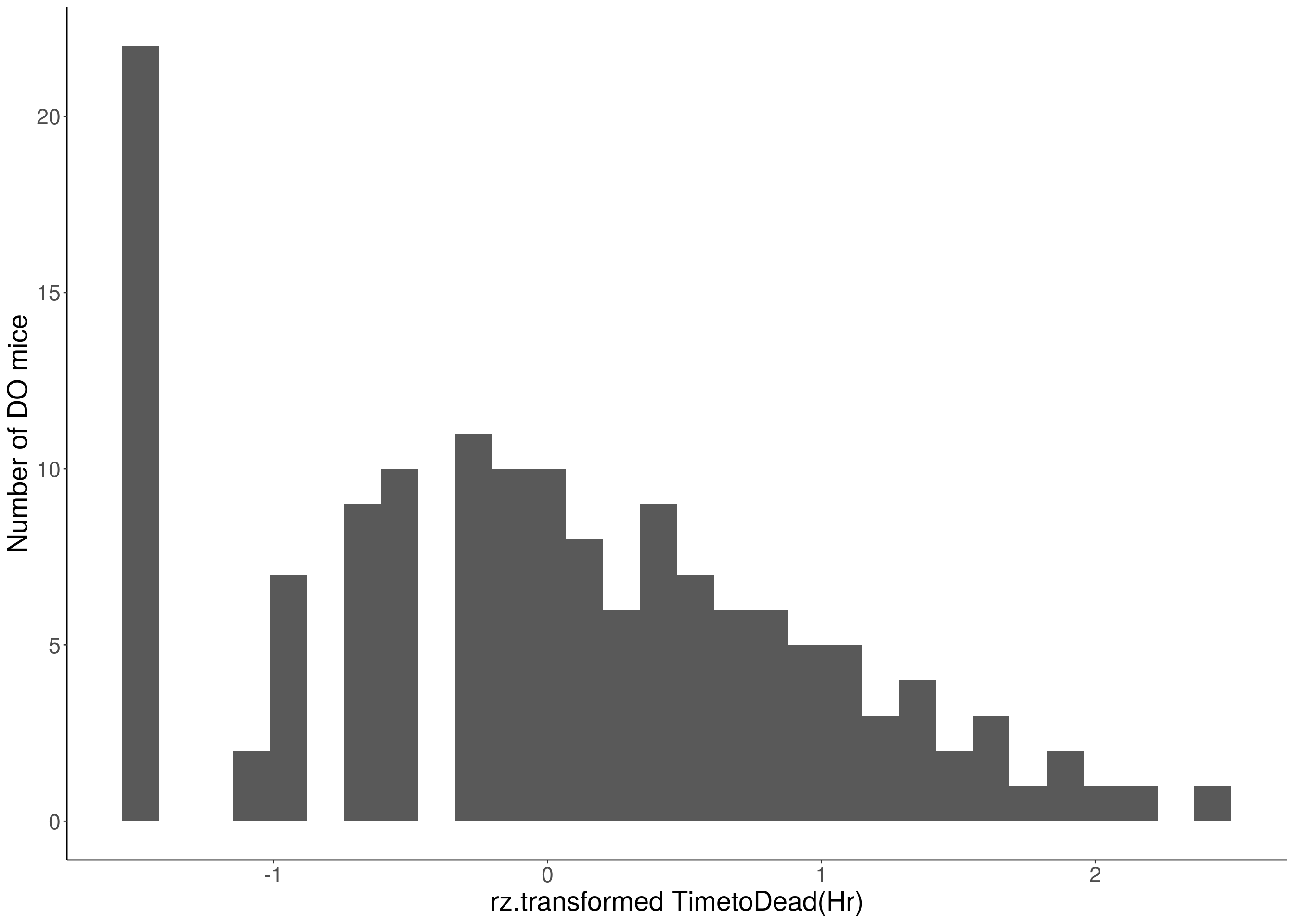
# Warning: Removed 48 rows containing non-finite values (stat_bin).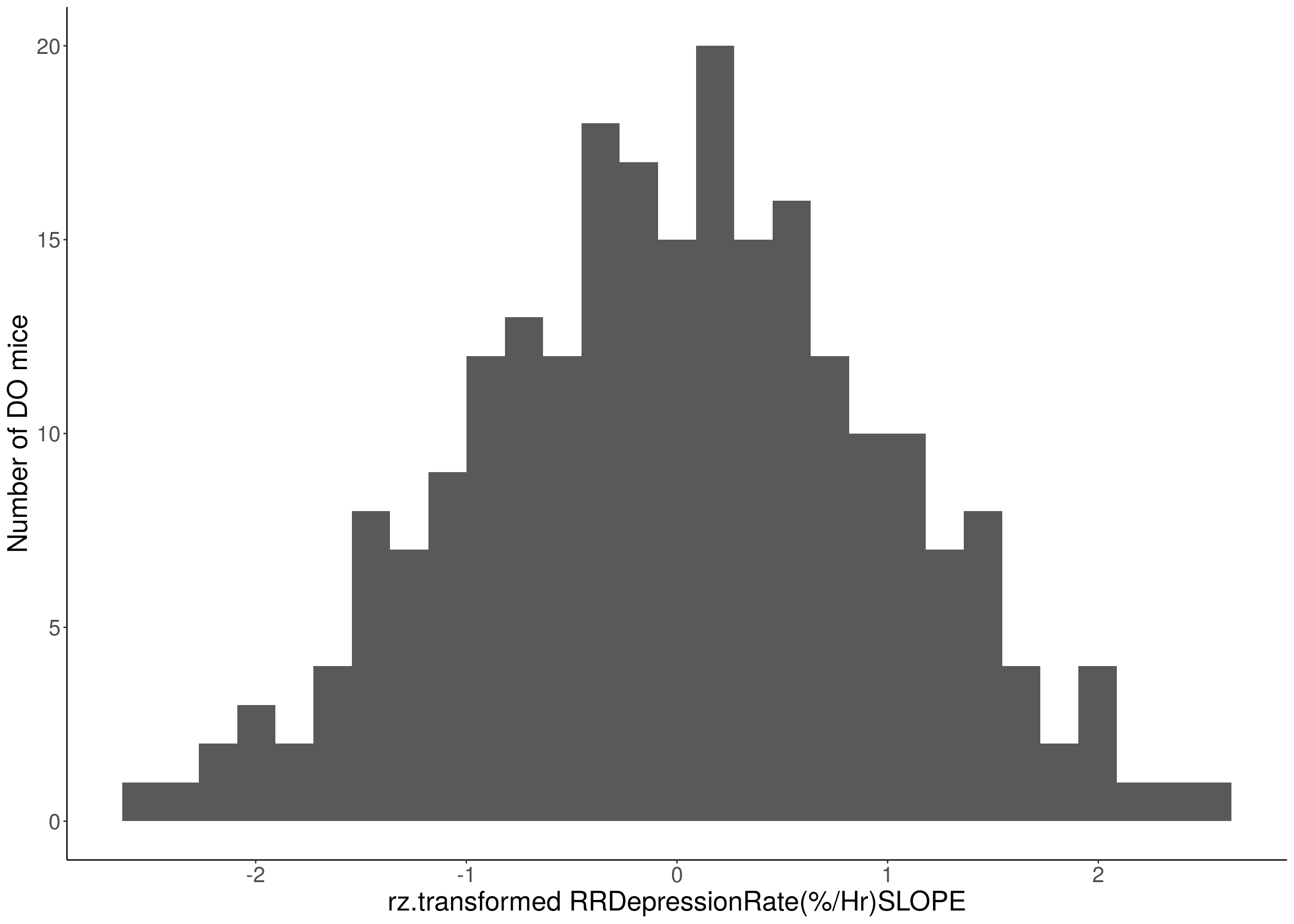
# Warning: Removed 48 rows containing non-finite values (stat_bin).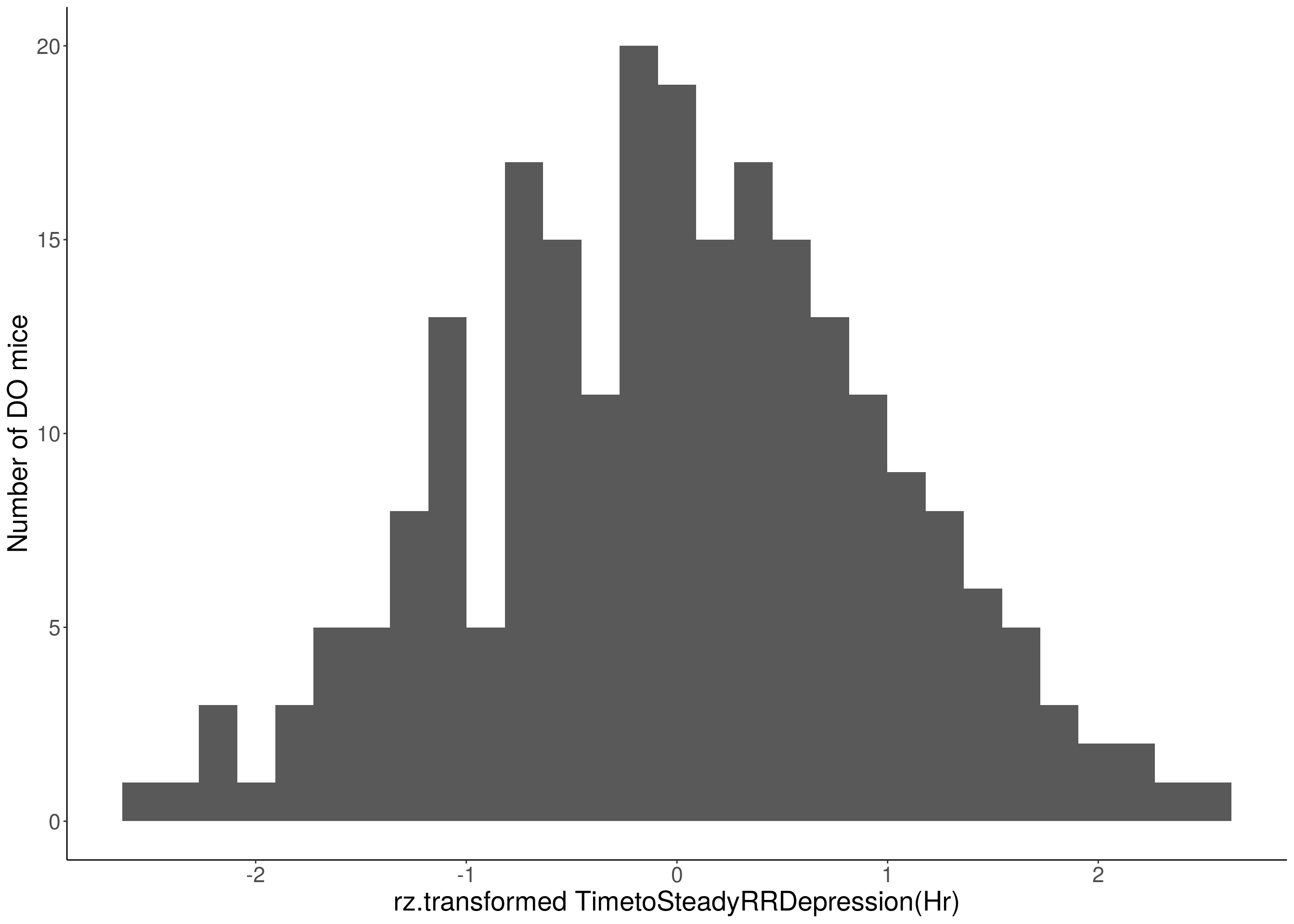
# Warning: Removed 48 rows containing non-finite values (stat_bin).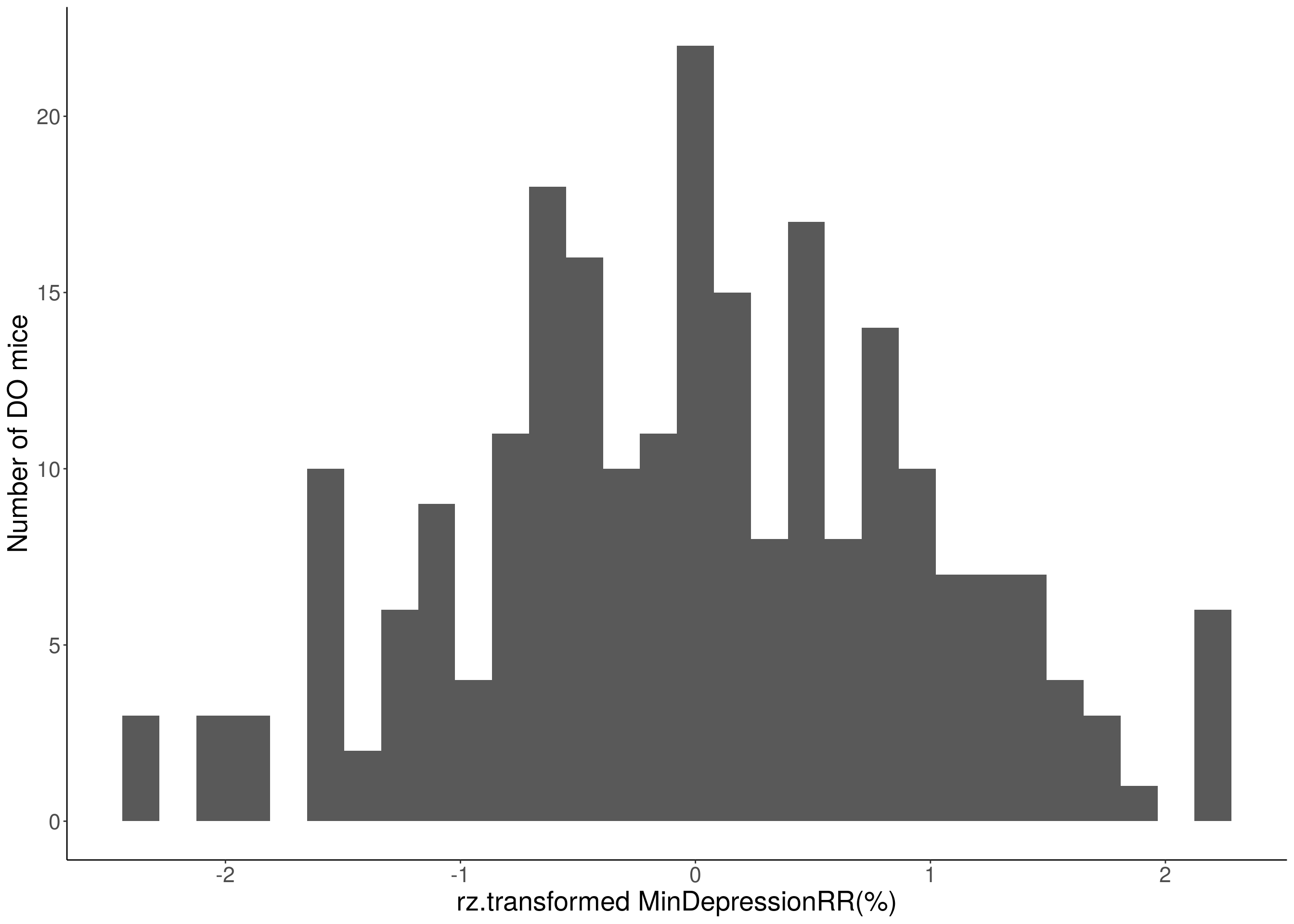
# Warning: Removed 187 rows containing non-finite values (stat_bin).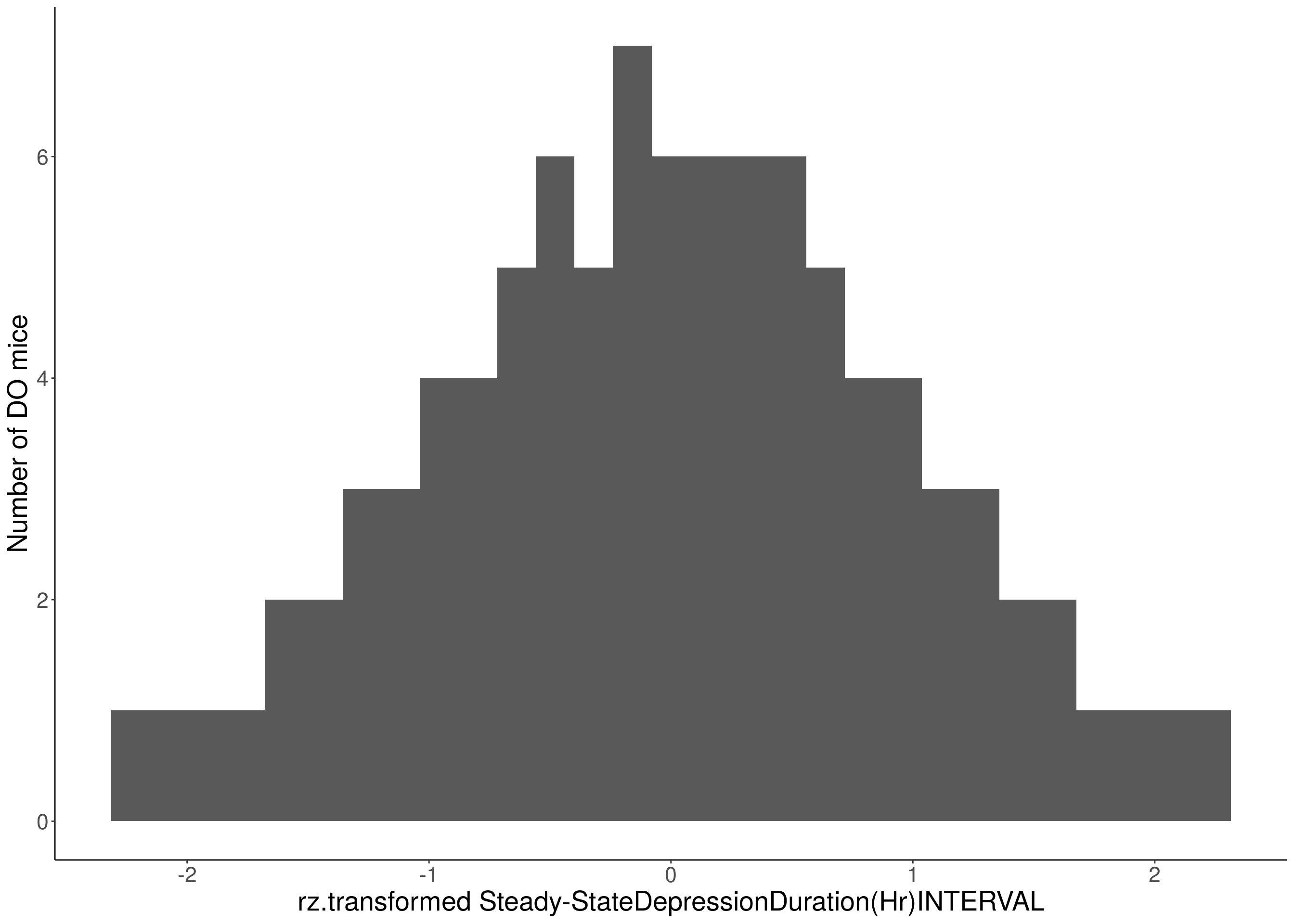
# Warning: Removed 187 rows containing non-finite values (stat_bin).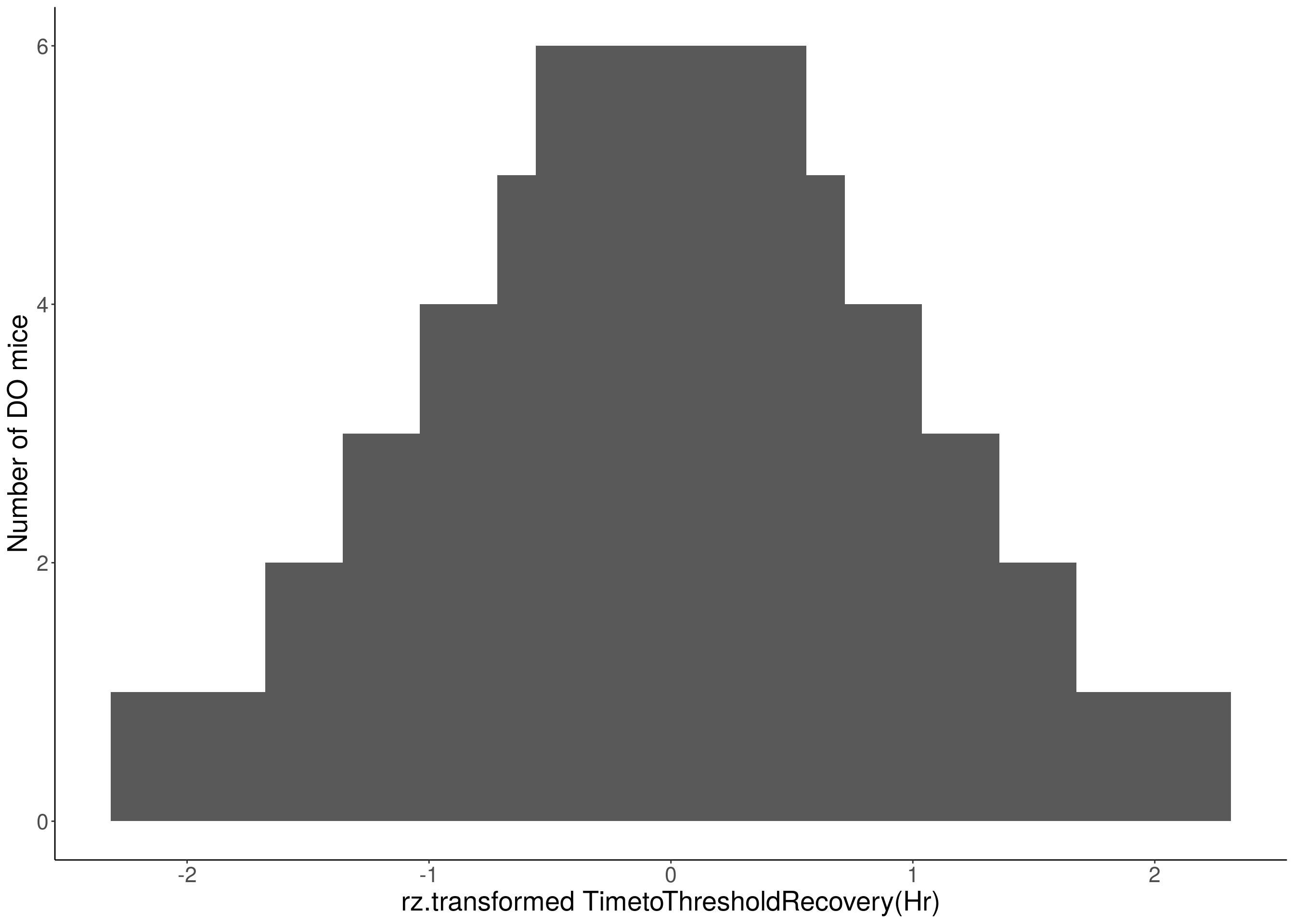
# Warning: Removed 187 rows containing non-finite values (stat_bin).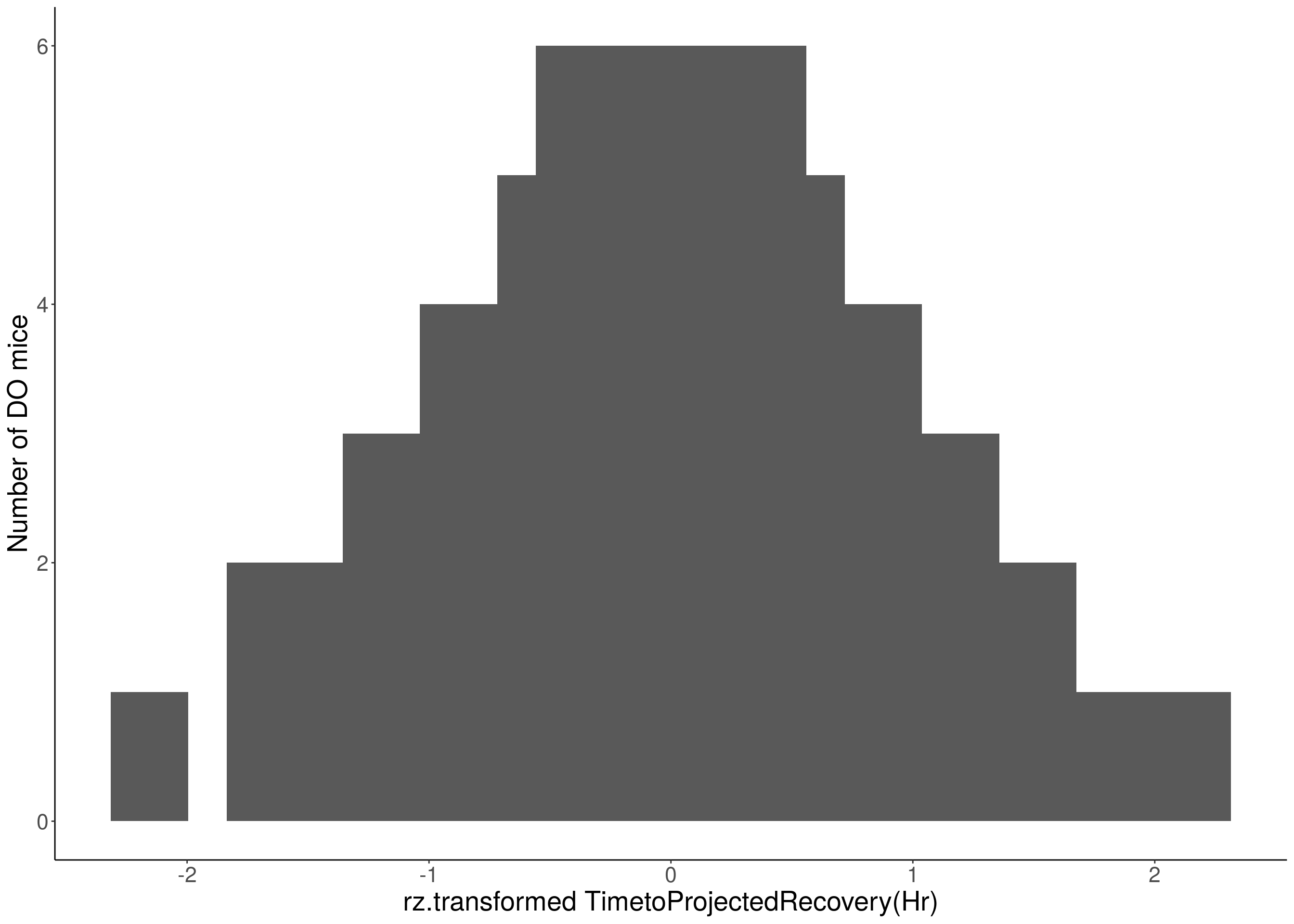
# Warning: Removed 187 rows containing non-finite values (stat_bin).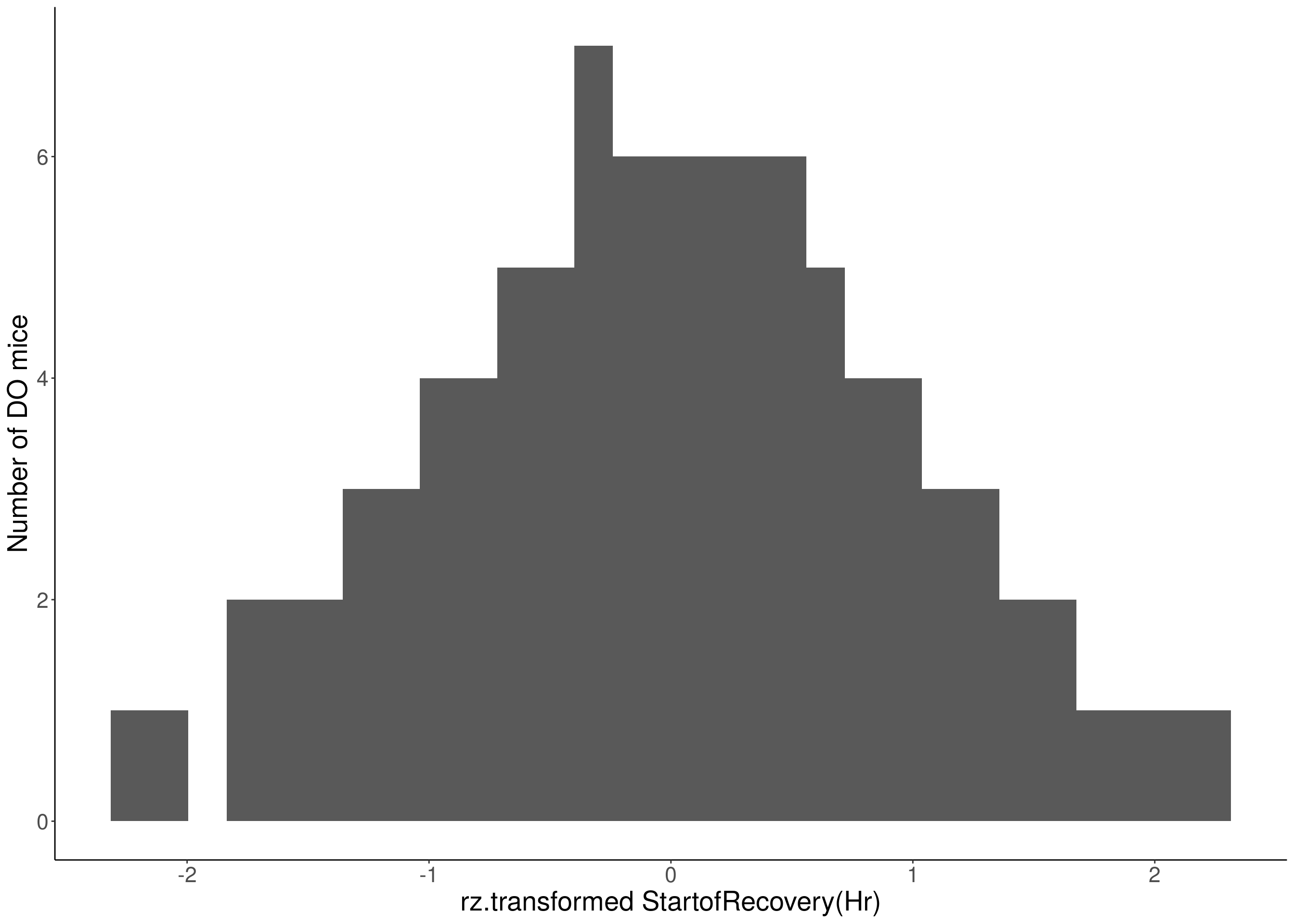
# Warning: Removed 187 rows containing non-finite values (stat_bin).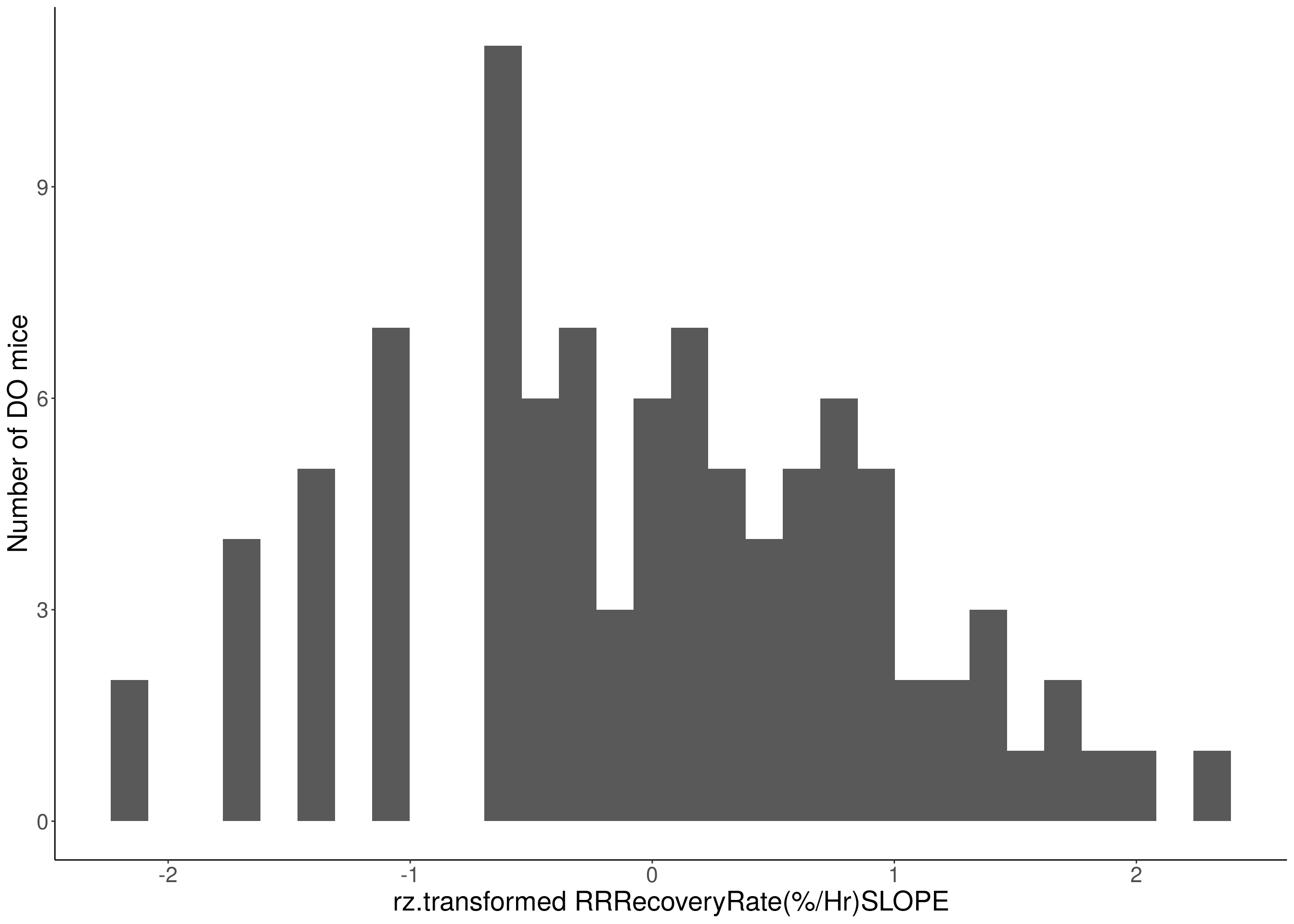
Plot qtl mapping on array
Plot by using its output qtl.do_Fentanyl_Cohort2.out.RData
load("output/Fentanyl/qtl.do_Fentanyl_Cohort2.out.RData")
#pmap and gmap
chr.names <- c("1","2","3","4","5","6","7","8","9","10","11","12","13","14","15","16","17","18","19", "X")
gmap <- list()
pmap <- list()
for (chr in chr.names){
gmap[[chr]] <- gm$gmap[[chr]]
pmap[[chr]] <- gm$pmap[[chr]]
}
attr(gmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
attr(pmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
#genome-wide plot
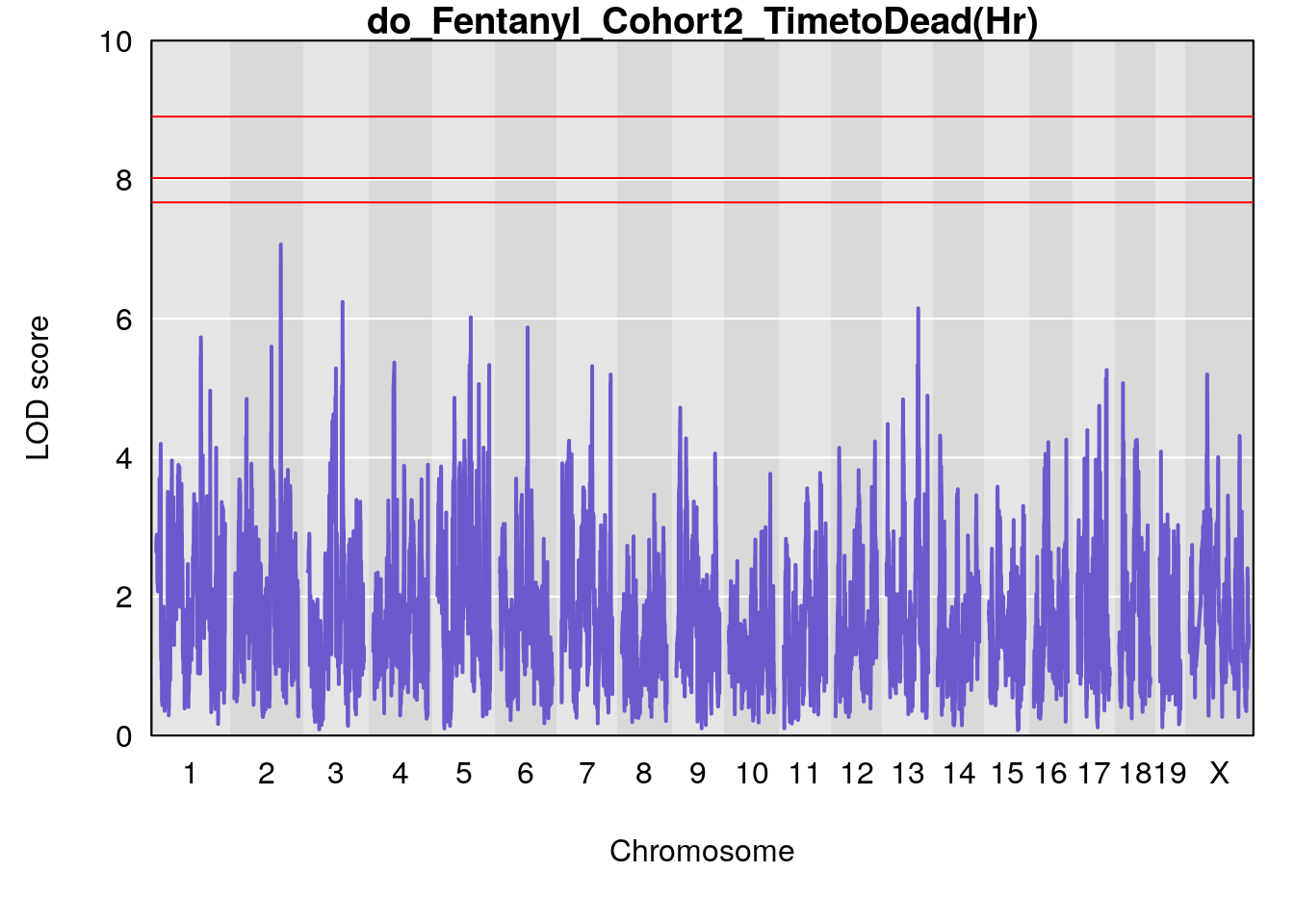
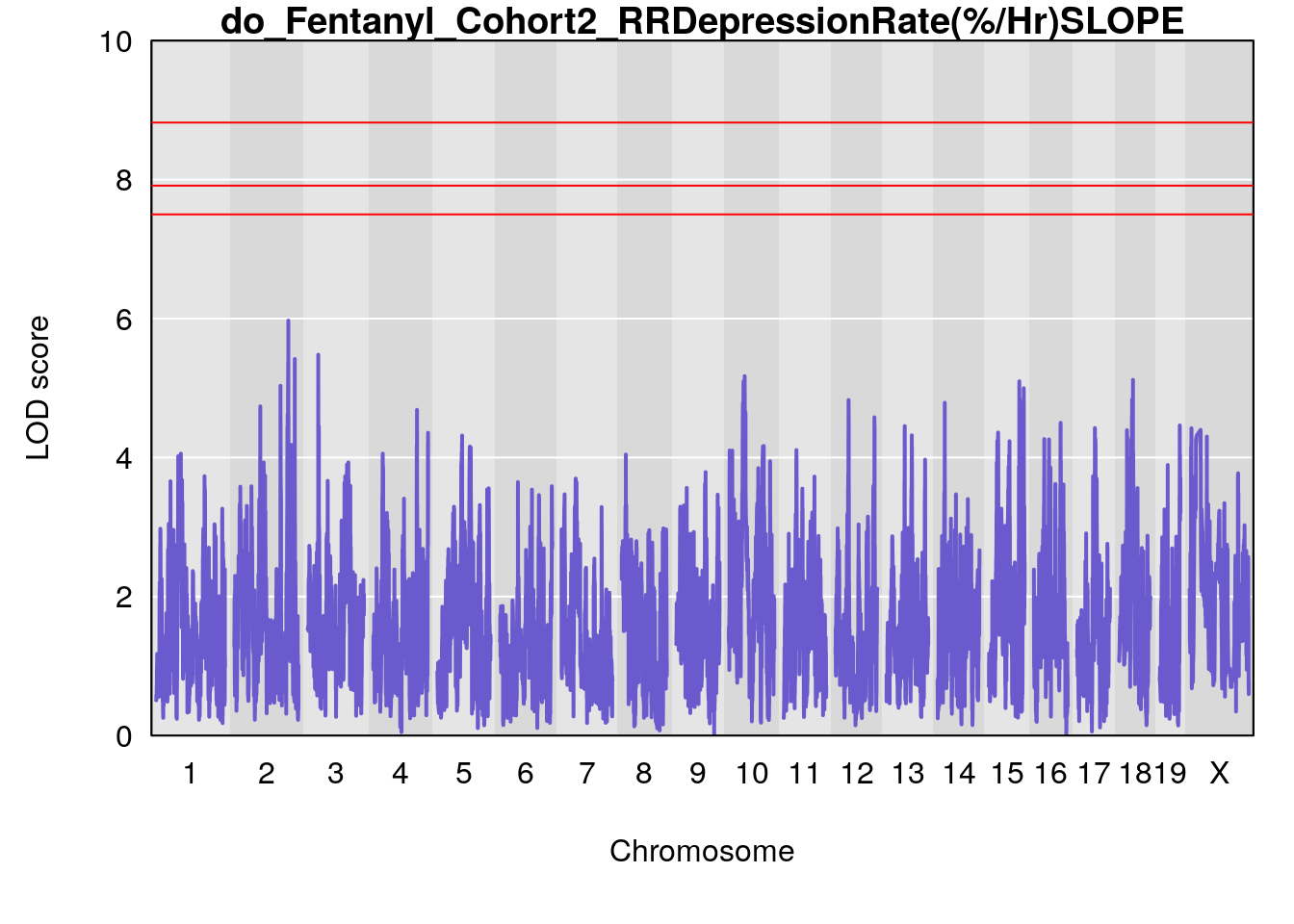
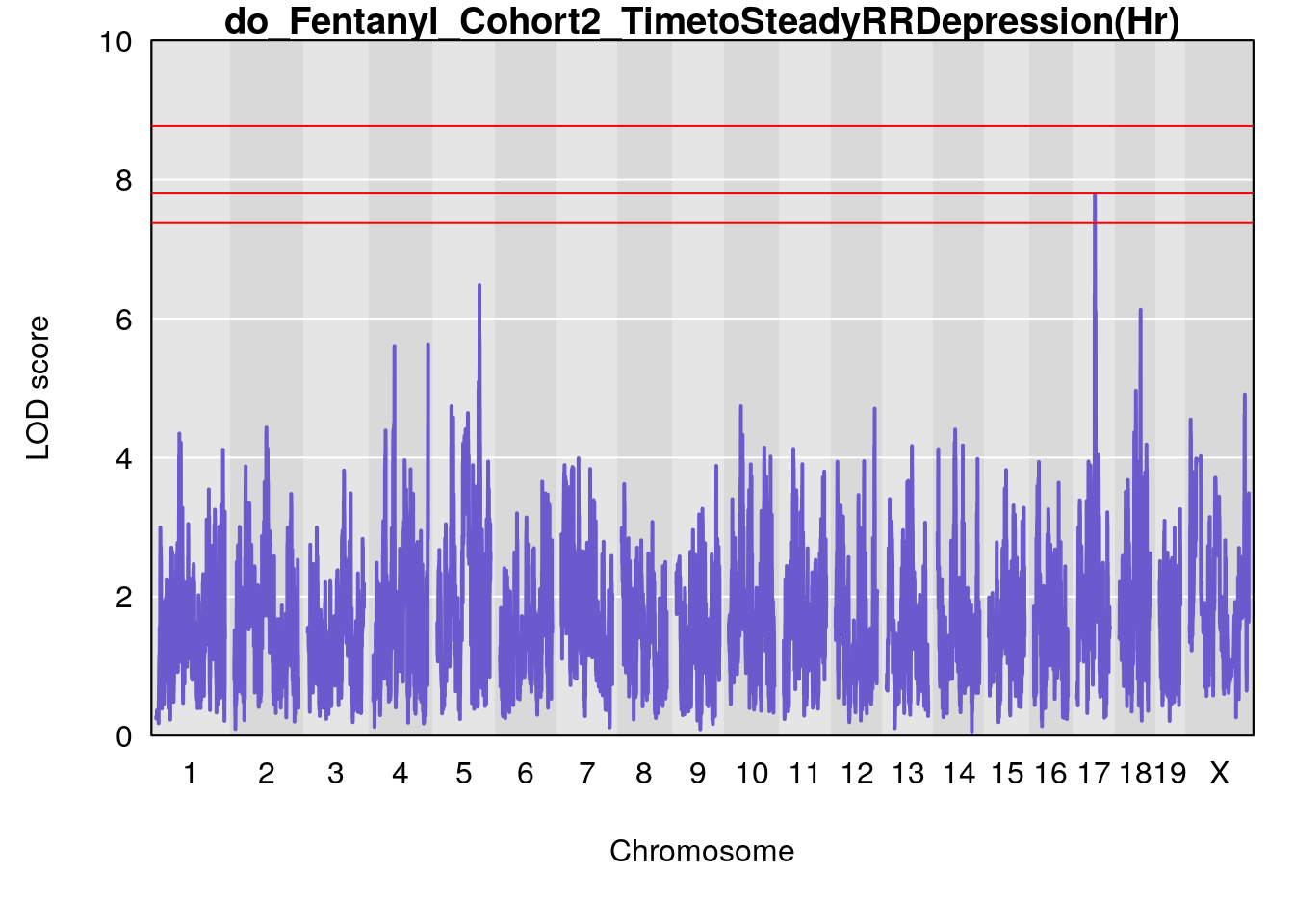
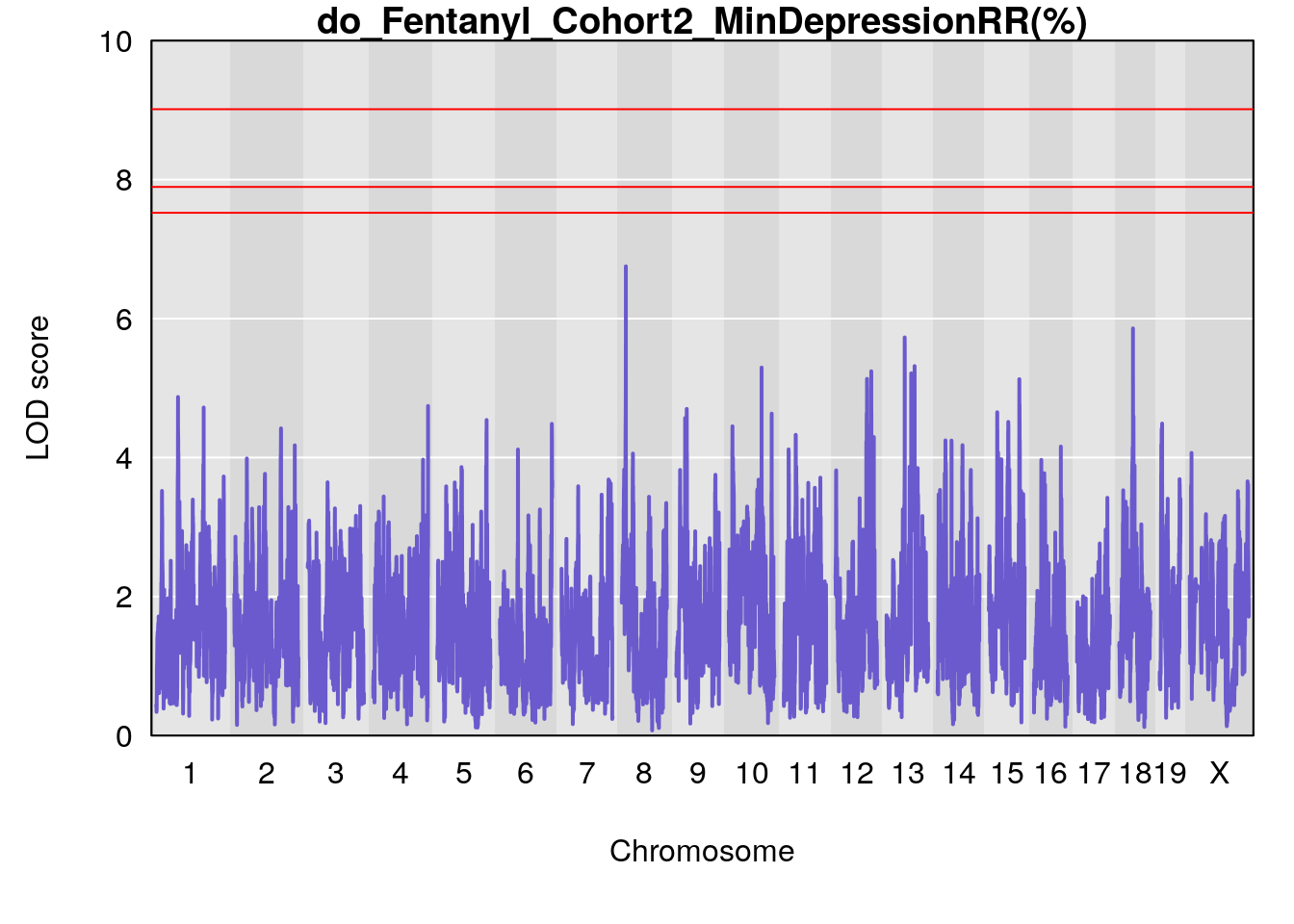
for(i in names(qtl.morphine.out)){
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.morphine.out[[i]]) # overall maximum LOD score
plot(qtl.morphine.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, 10))
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("do_Fentanyl_Cohort2_",i))
}
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |

| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
pdf(file = paste0("output/Fentanyl/do_Fentanyl_Cohort2.pdf"), width = 16, height =8)
#save genome-wide plo
for(i in names(qtl.morphine.out)){
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.morphine.out[[i]]) # overall maximum LOD score
plot(qtl.morphine.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, 10))
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("do_Fentanyl_Cohort2_",i))
}
dev.off()
# png
# 2
#peaks coeff plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
if(nrow(peaks) > 0) {
print(peaks)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
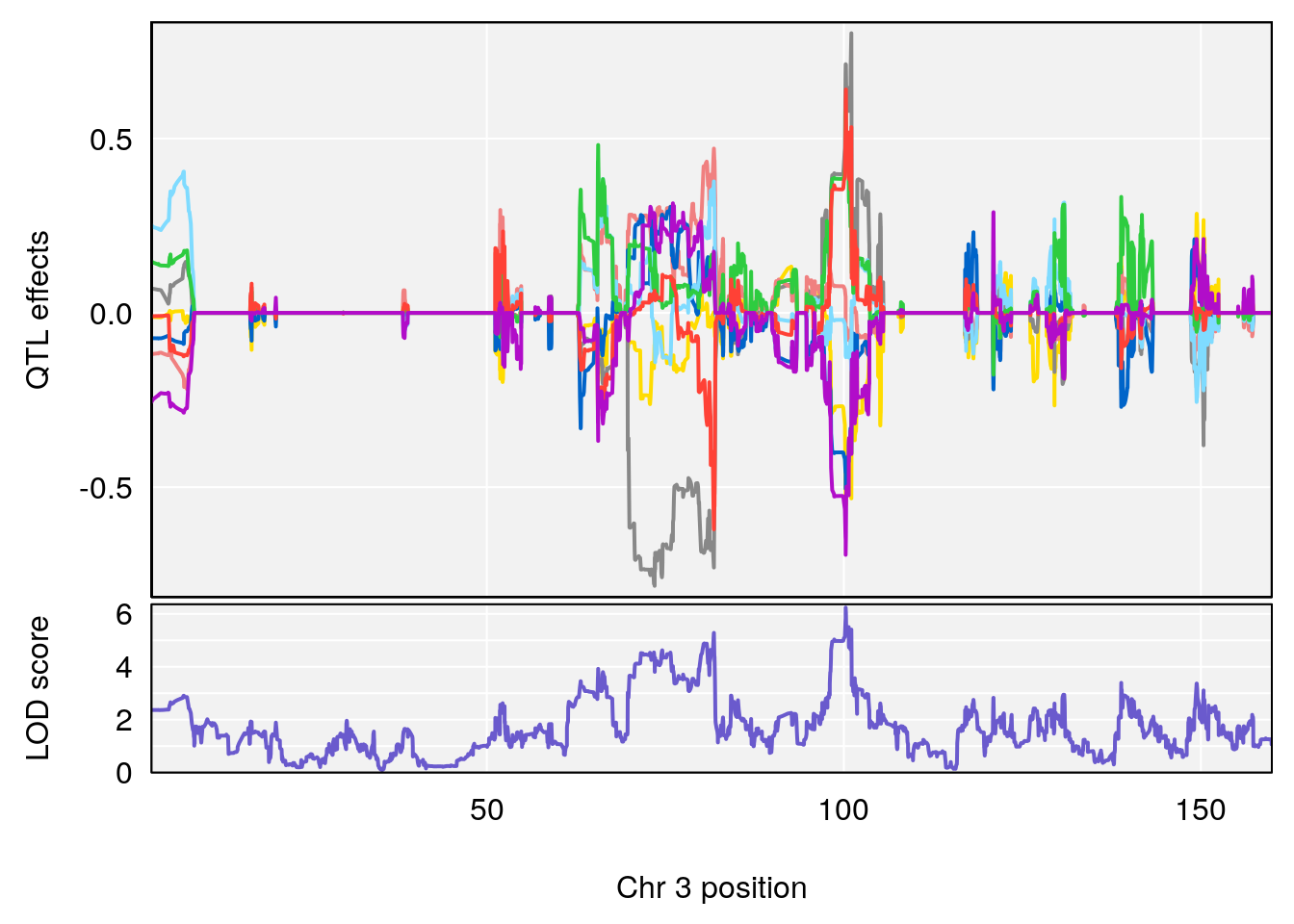
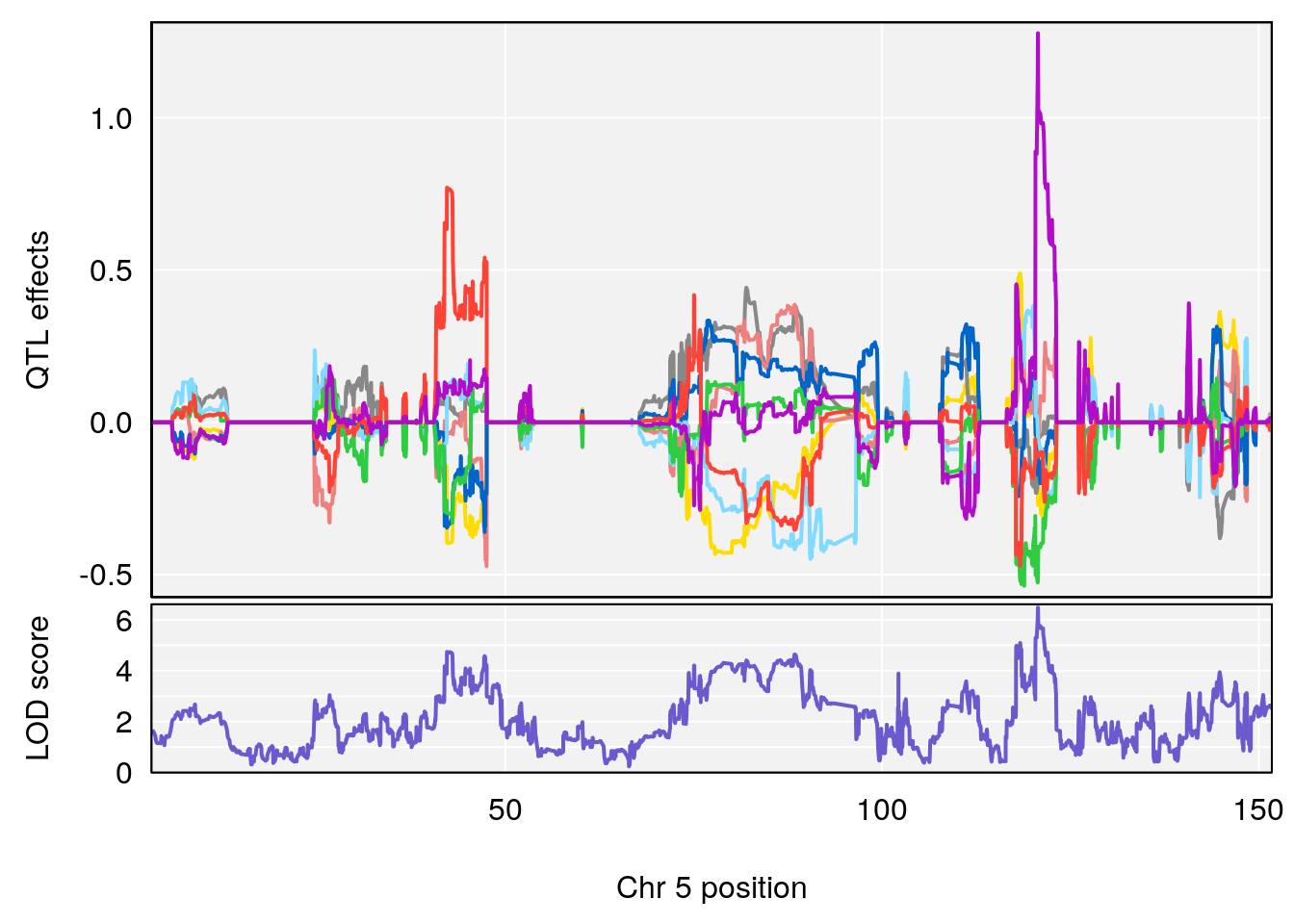
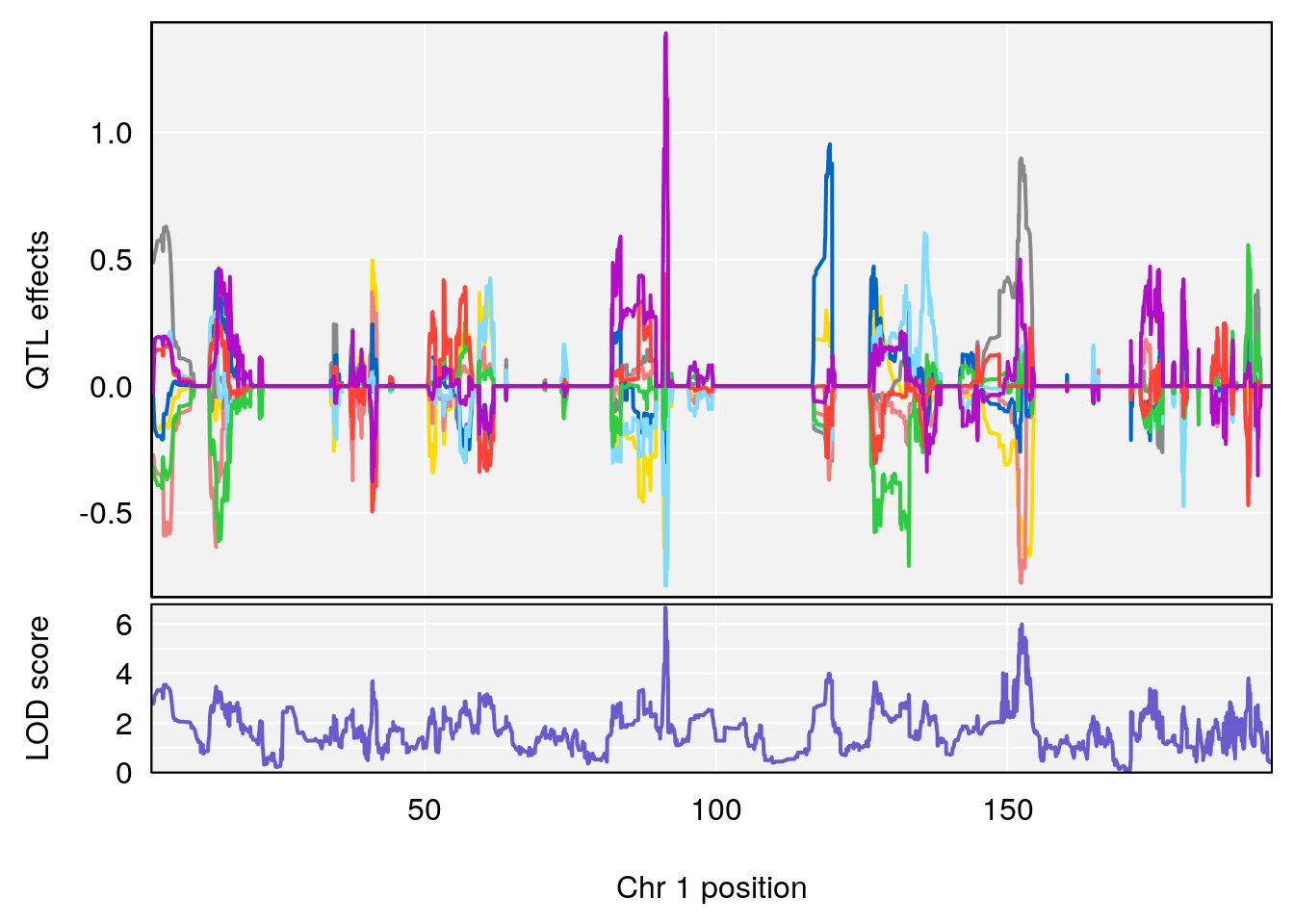
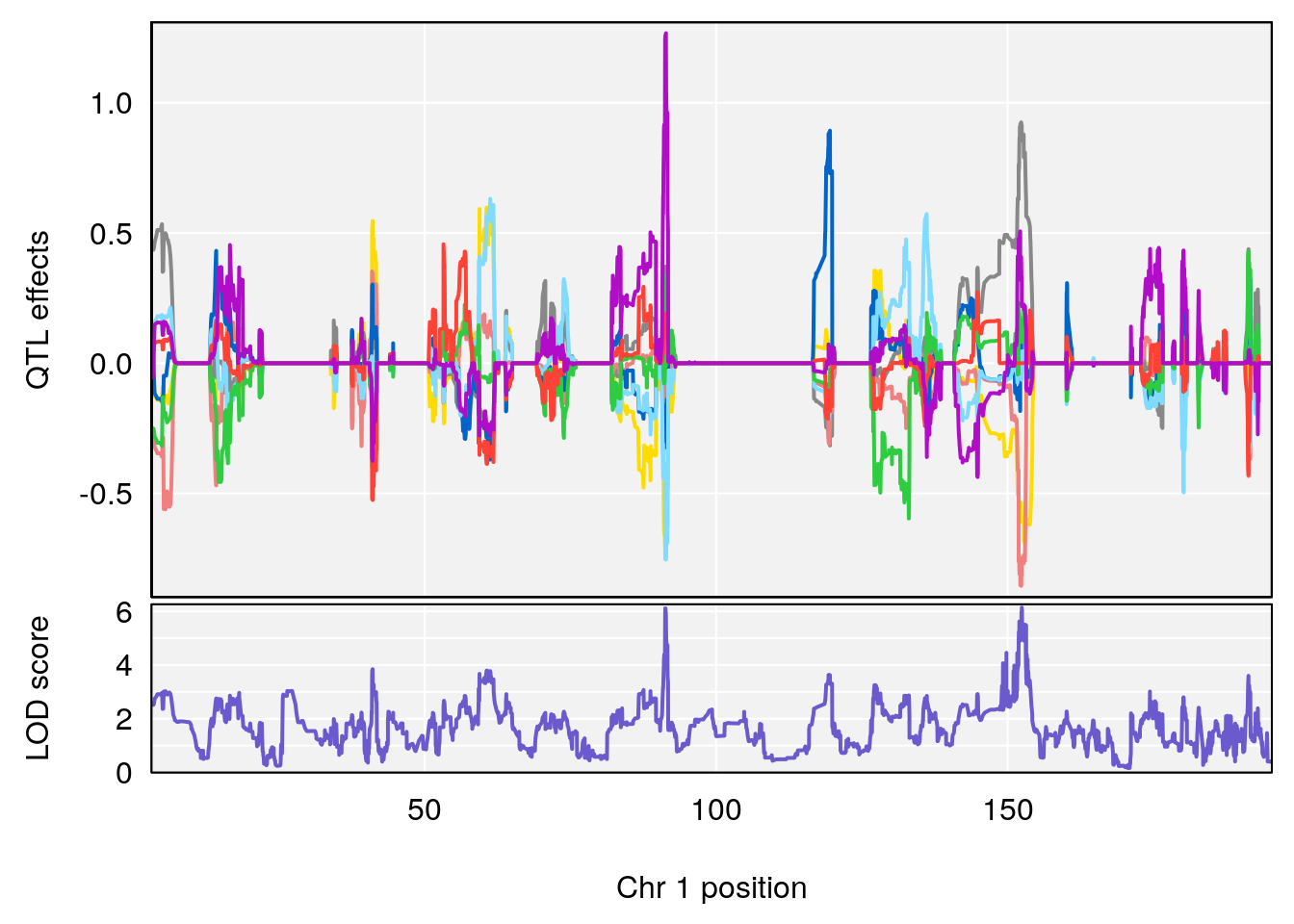
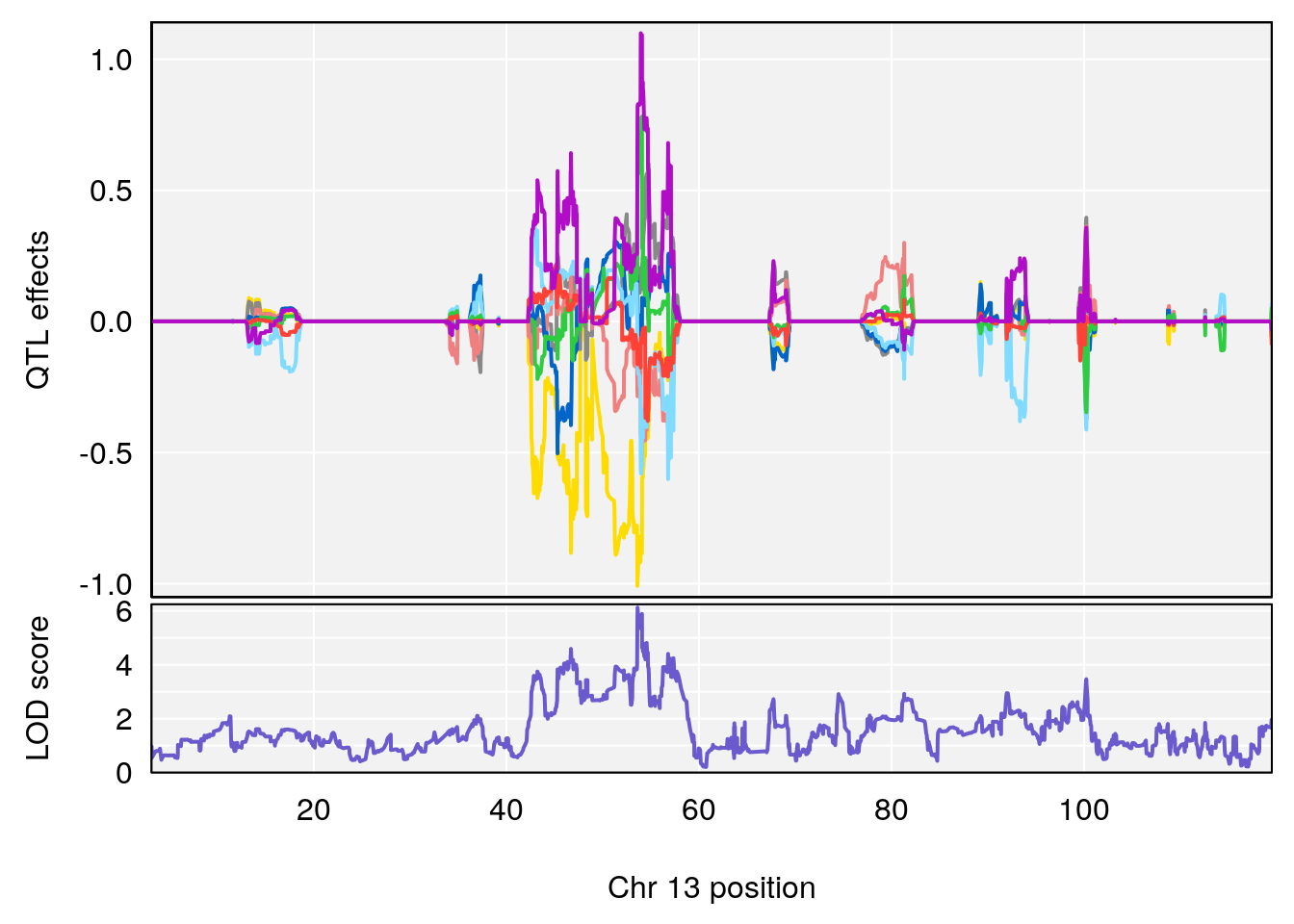
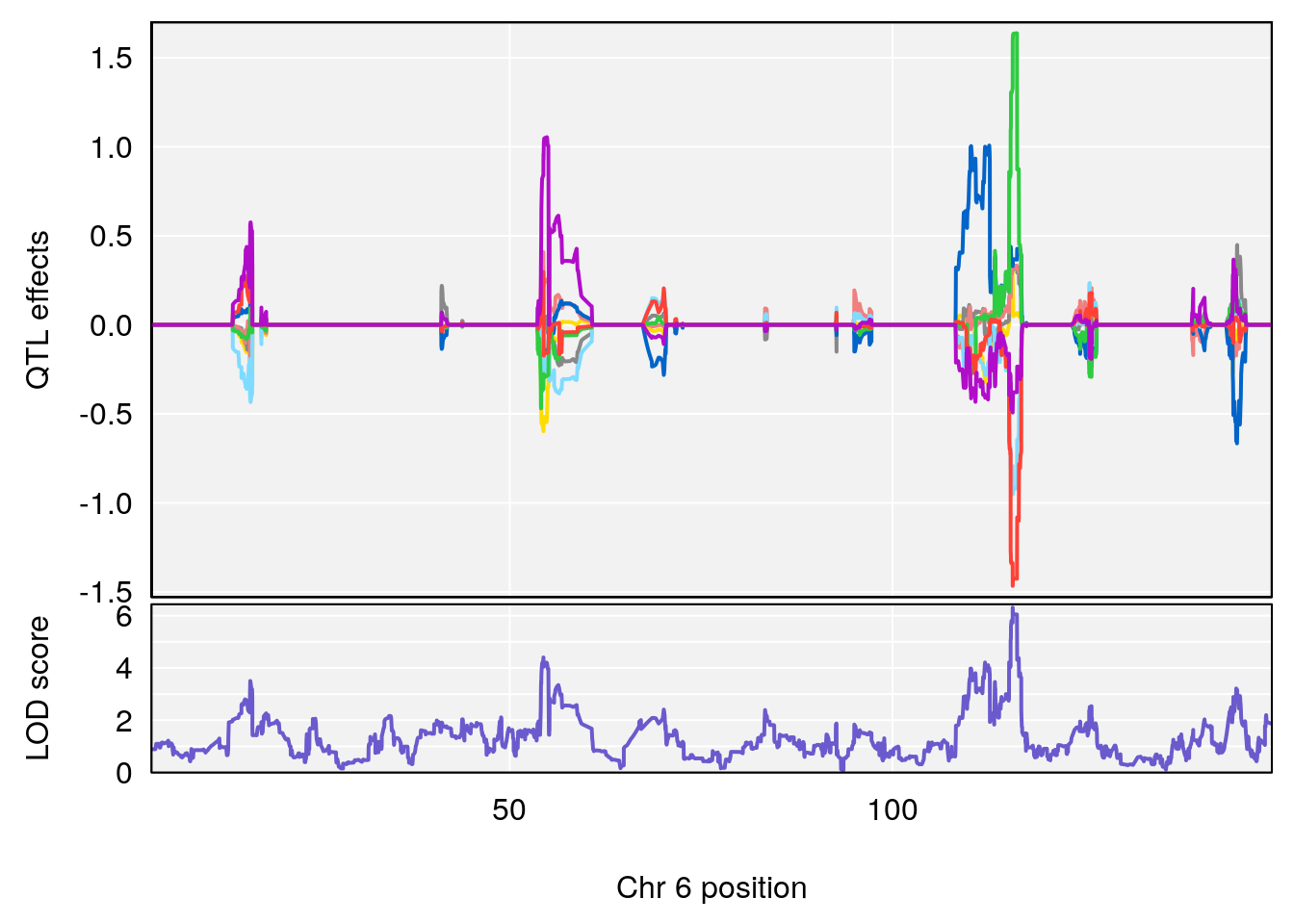
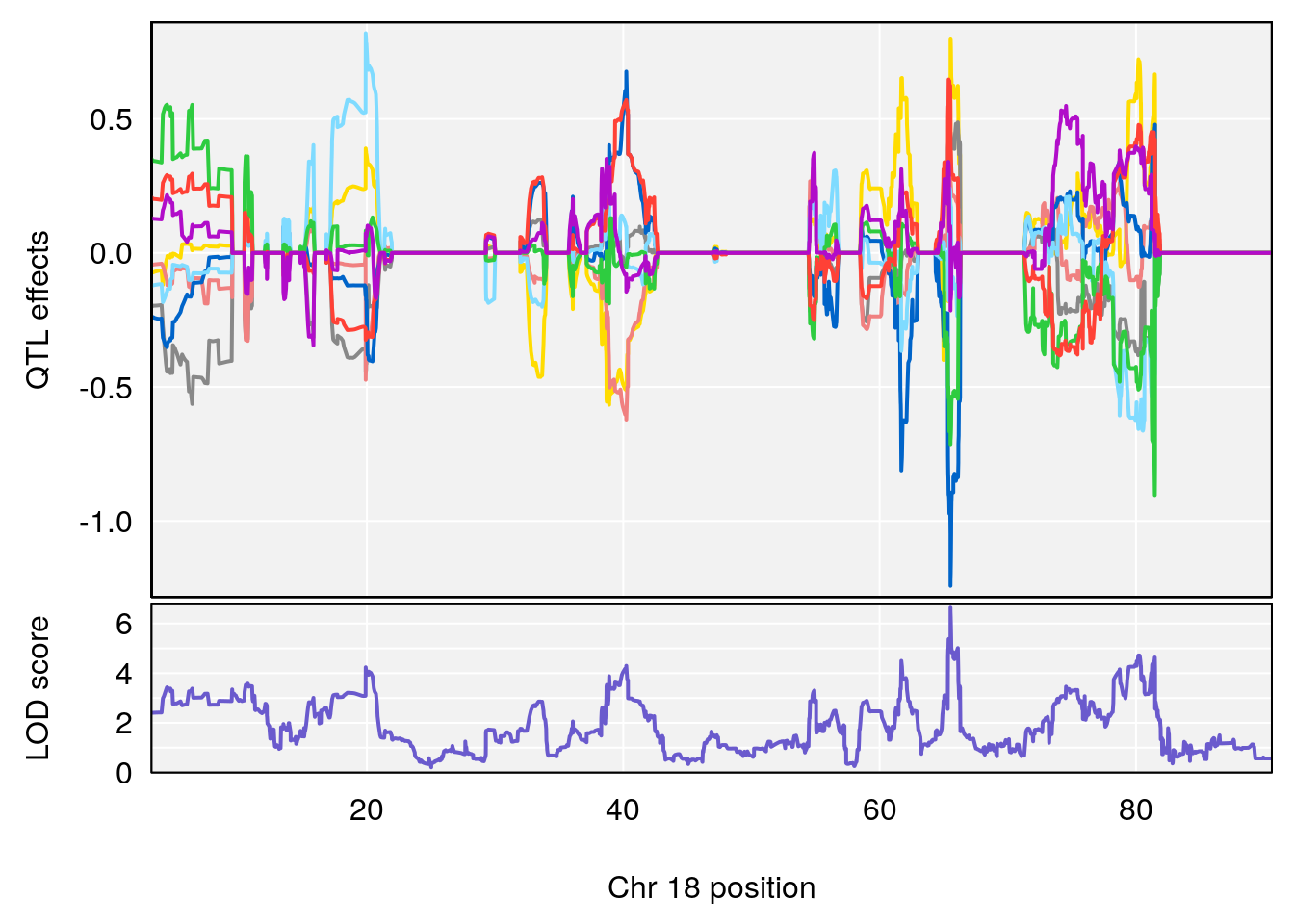
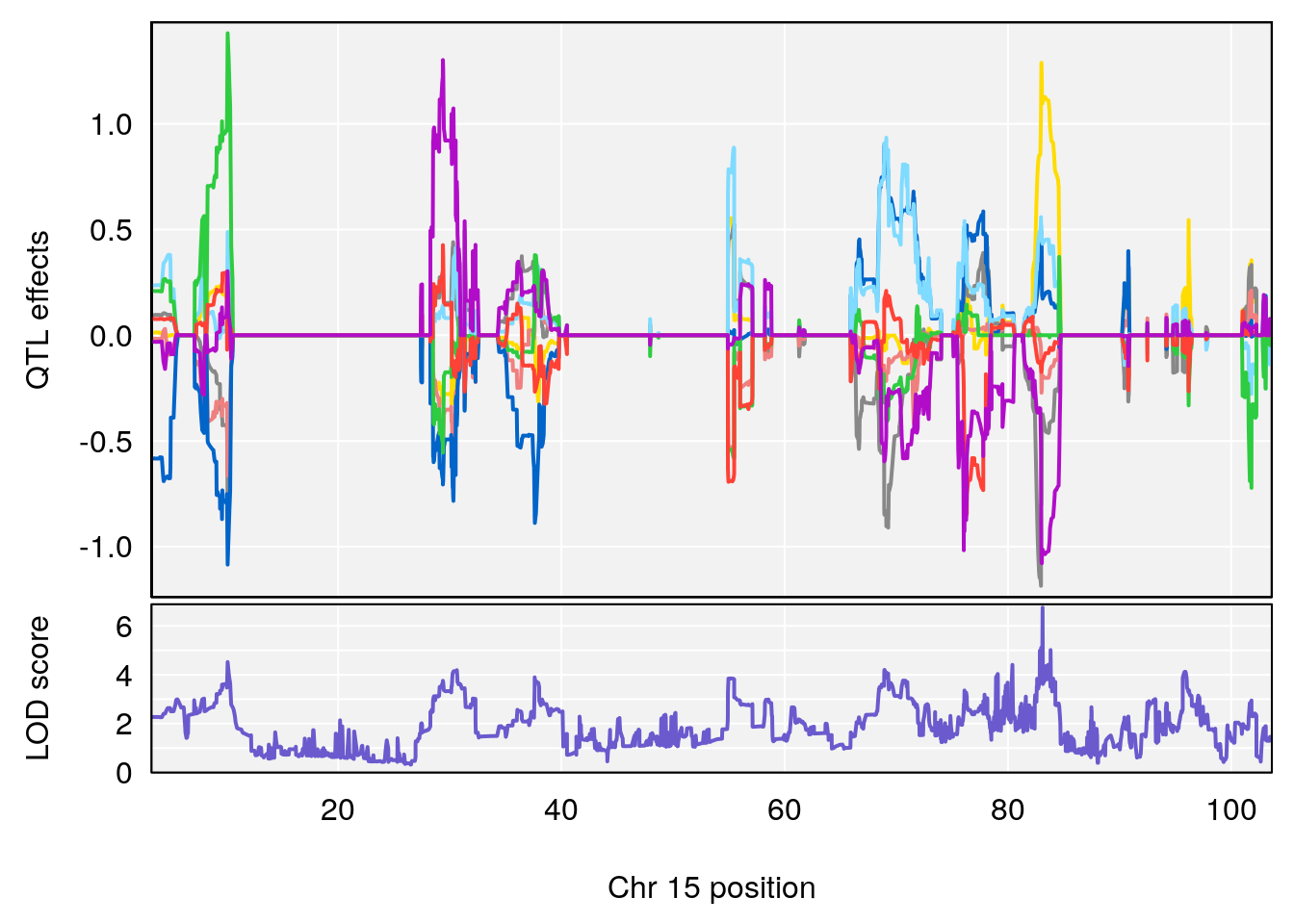
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
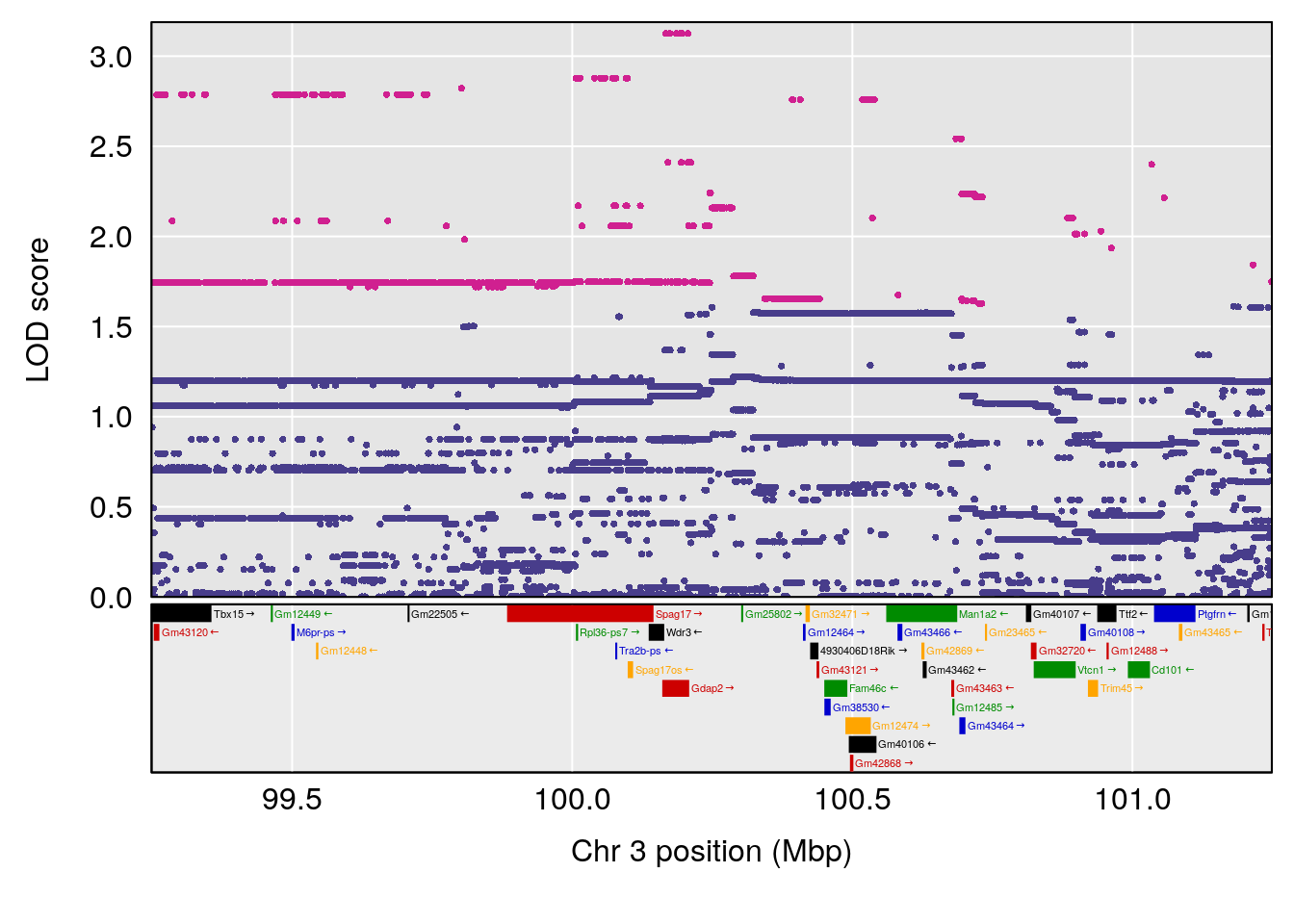
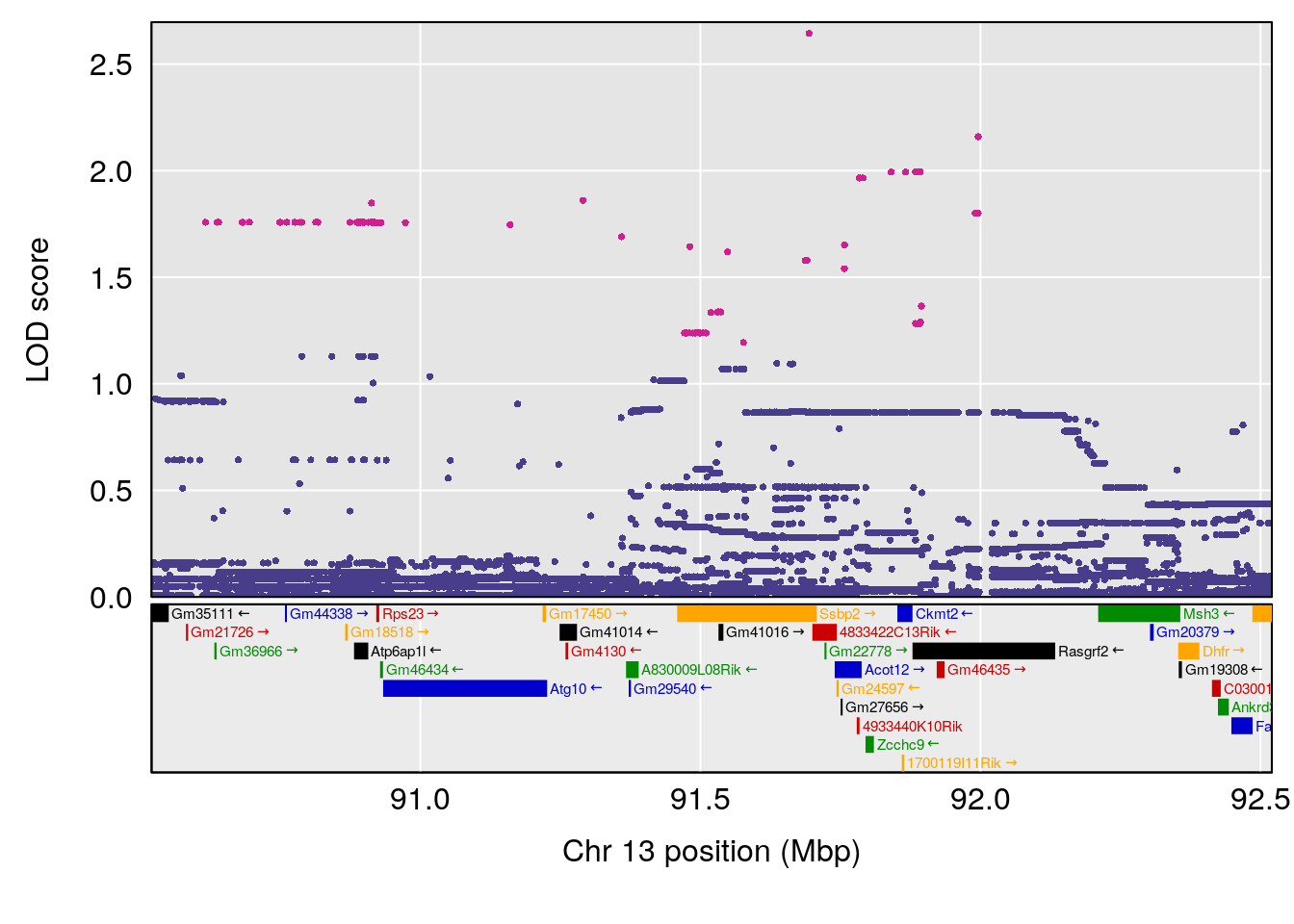
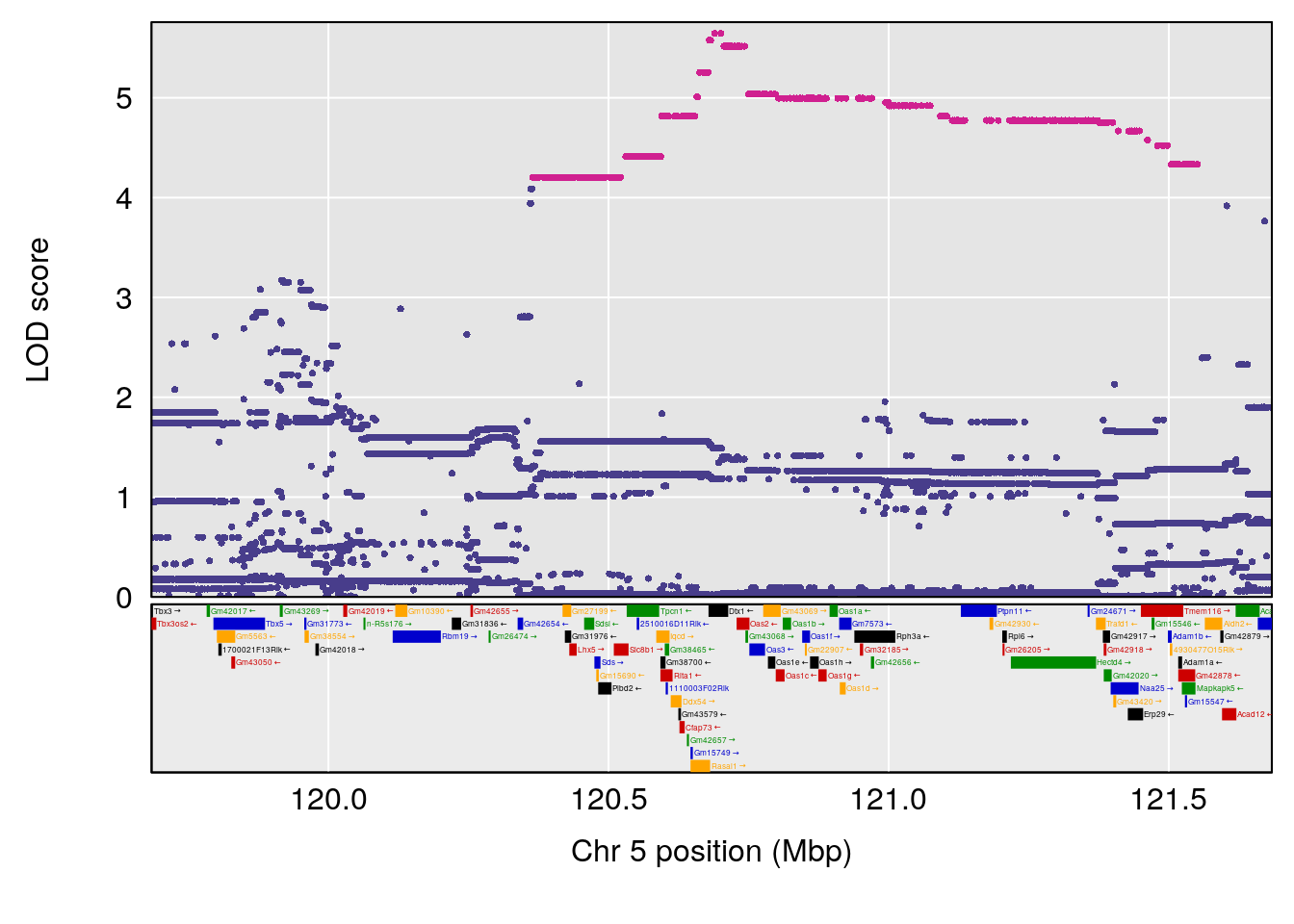
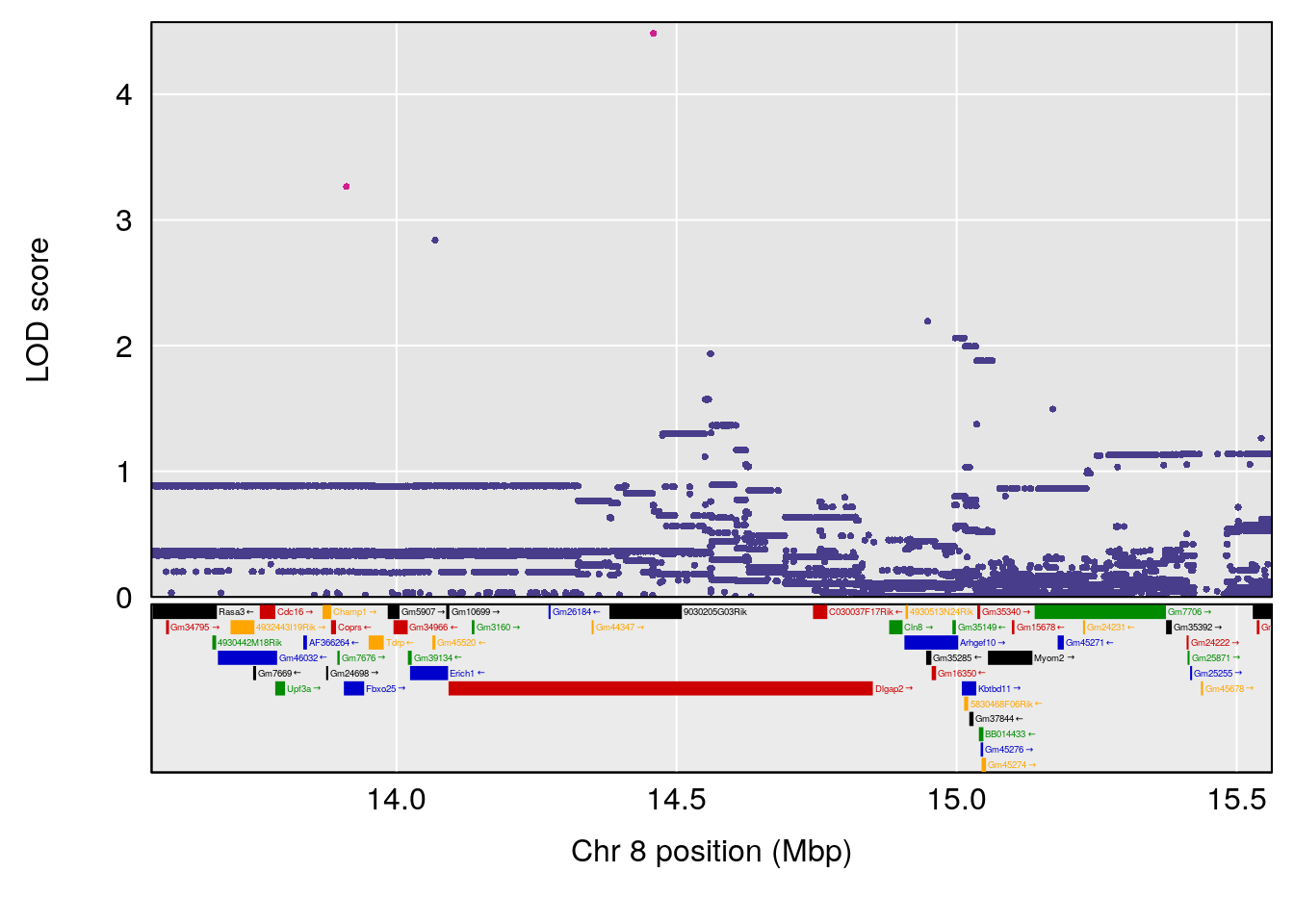
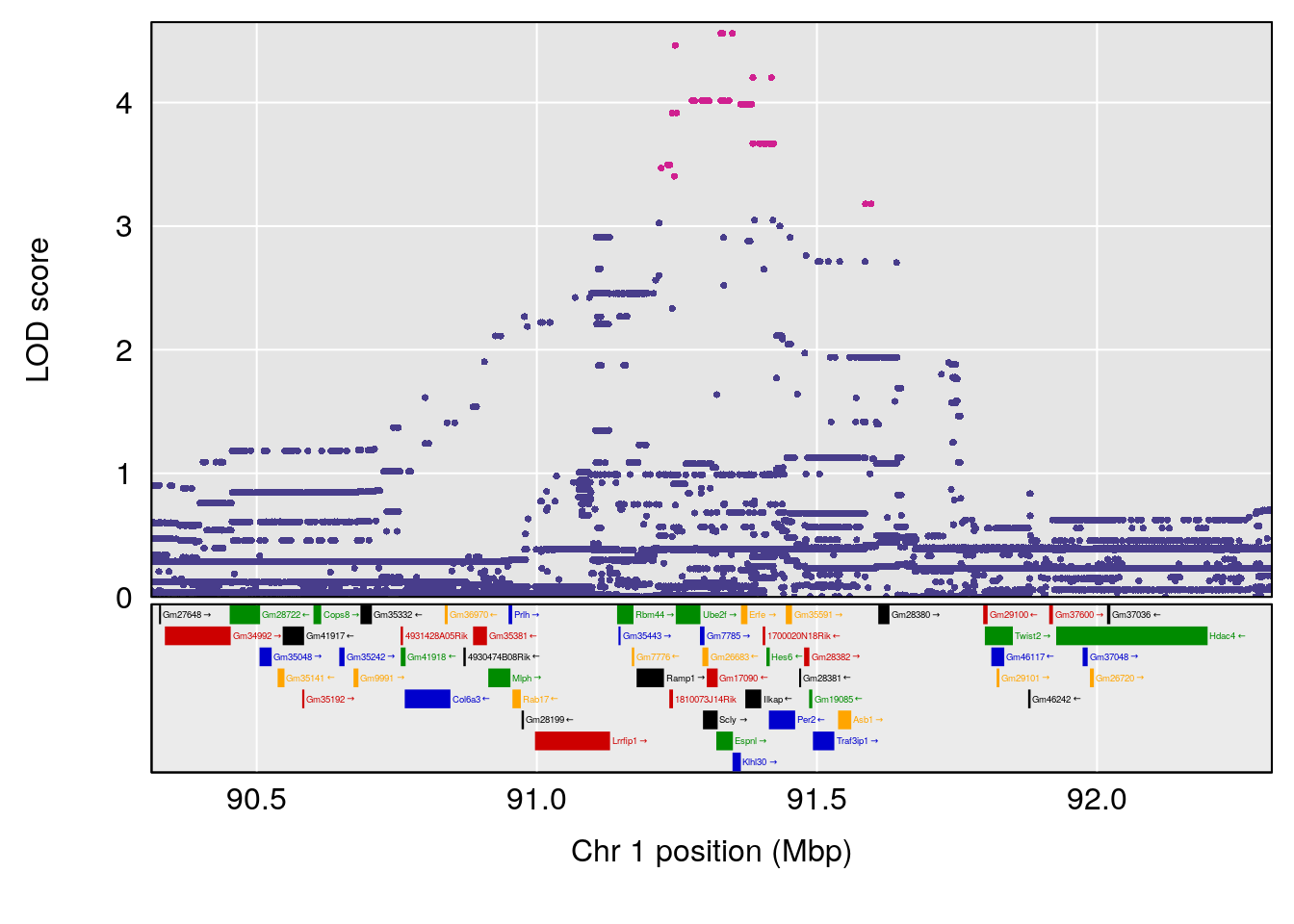
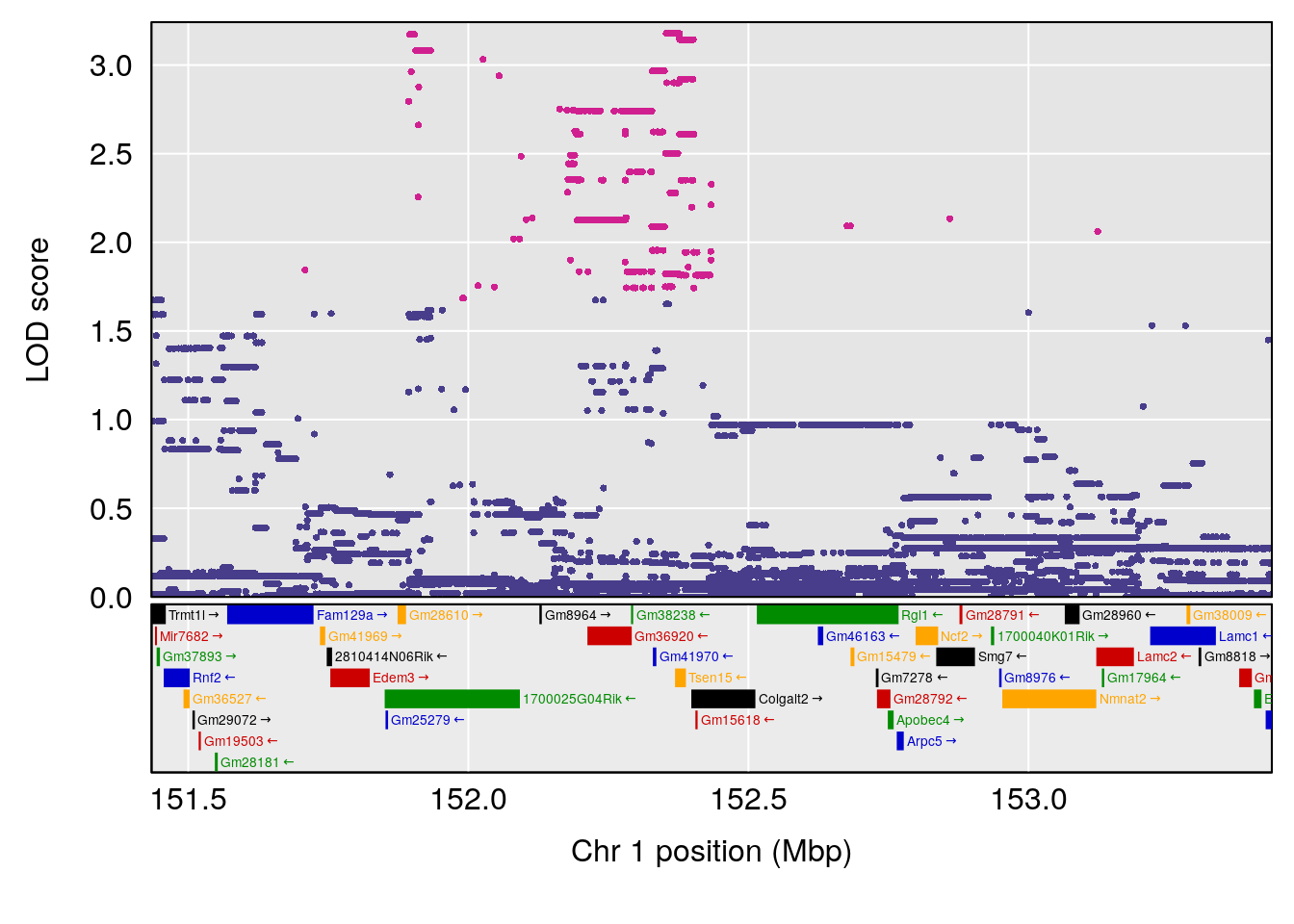
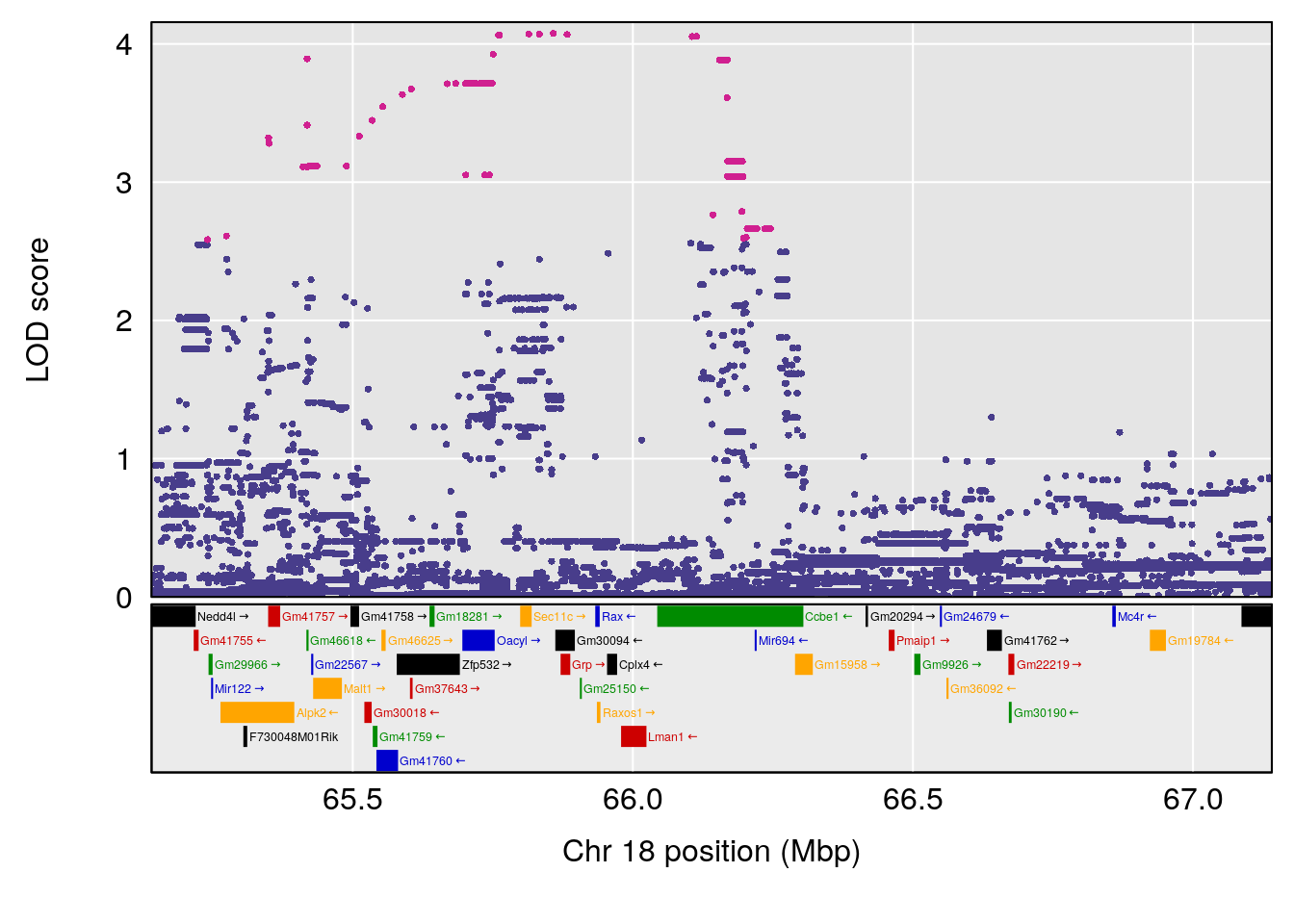
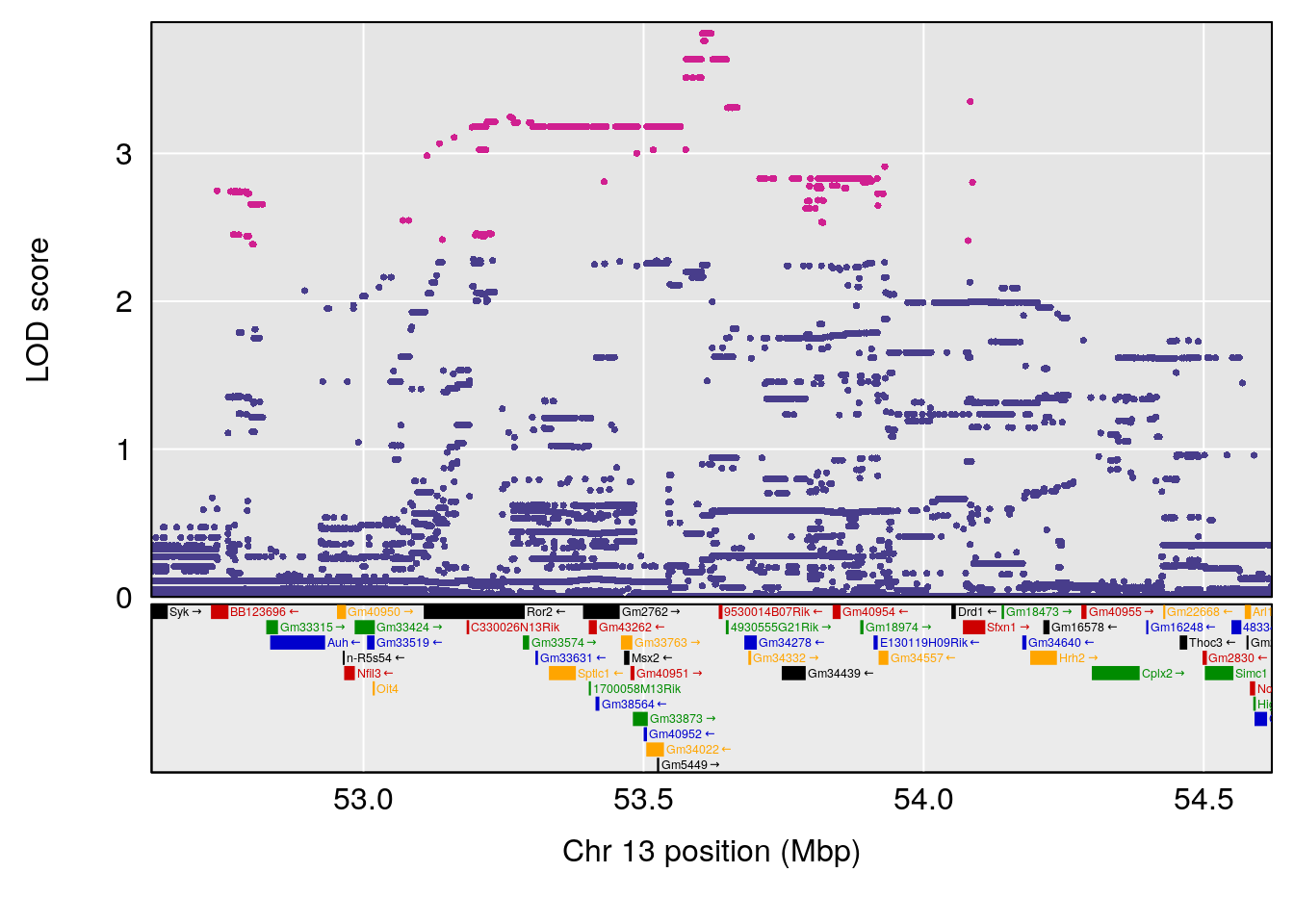
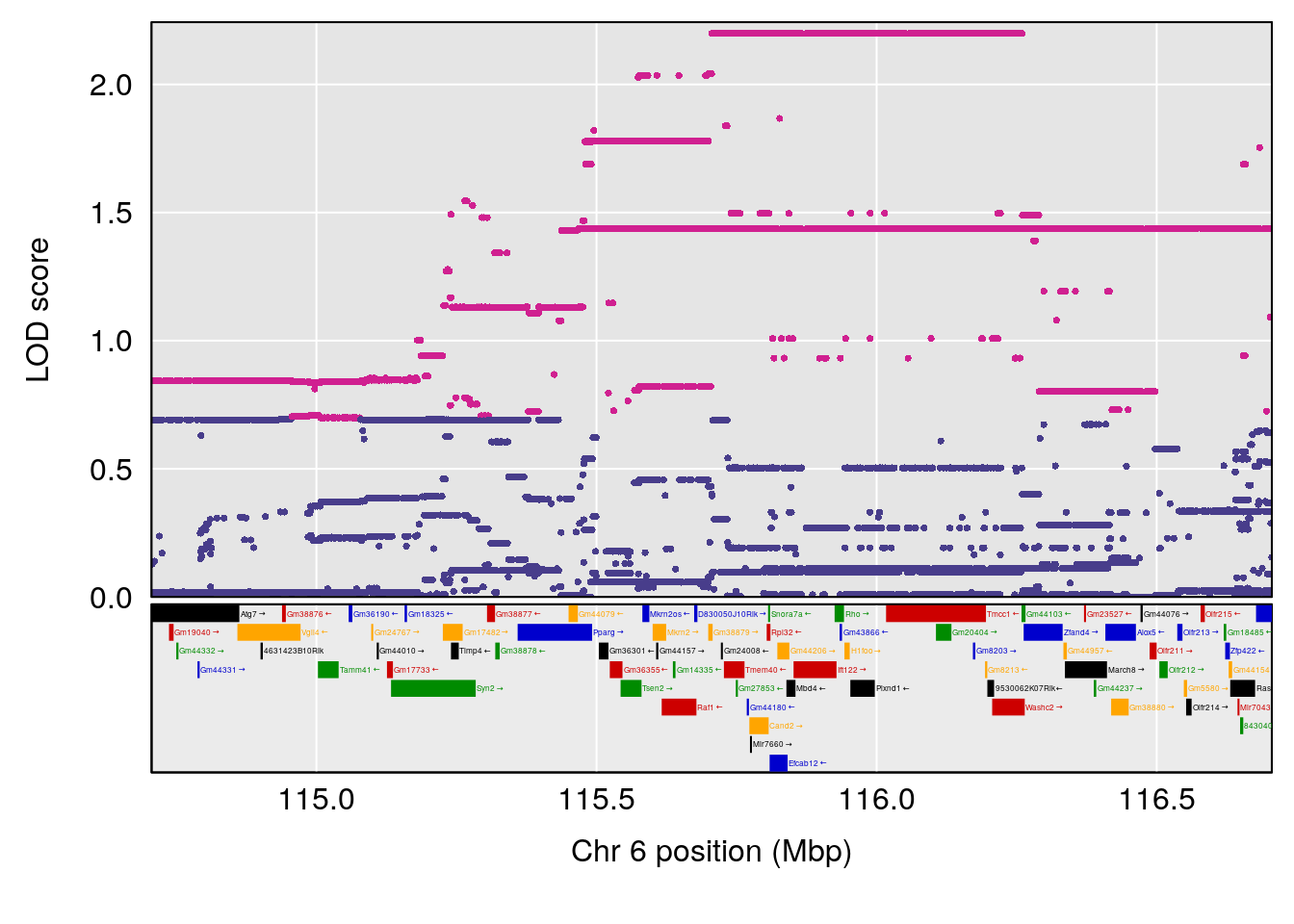
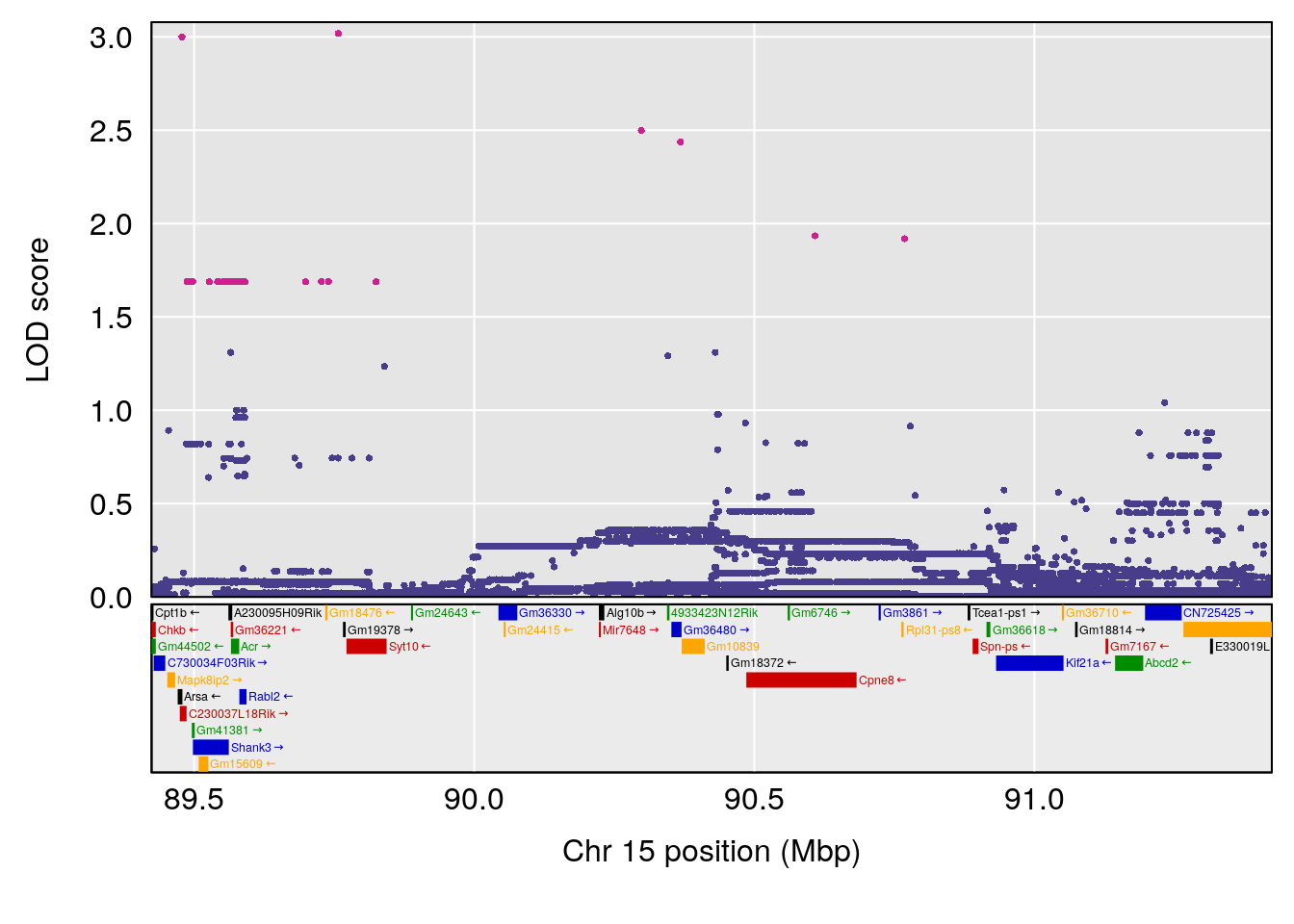
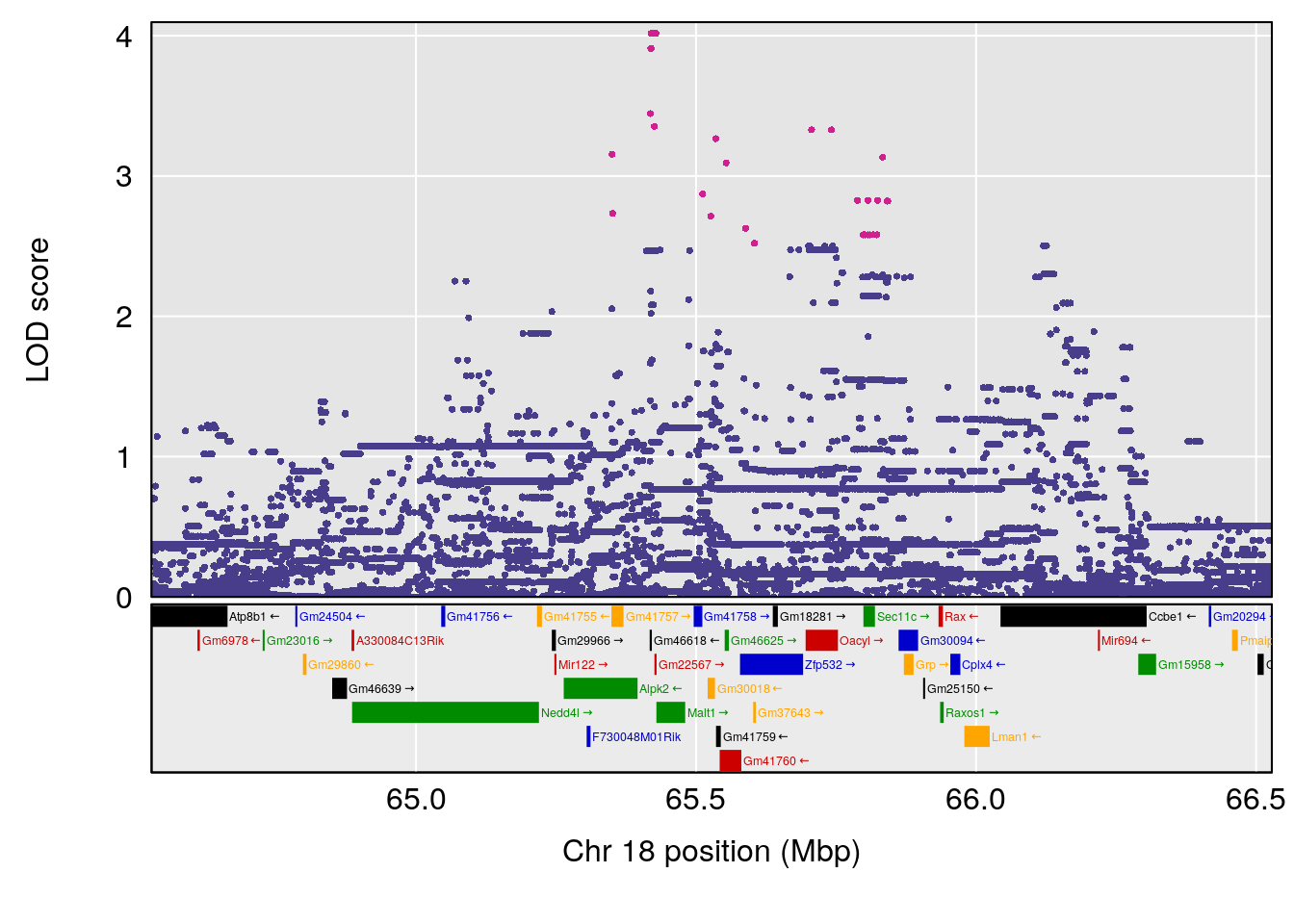
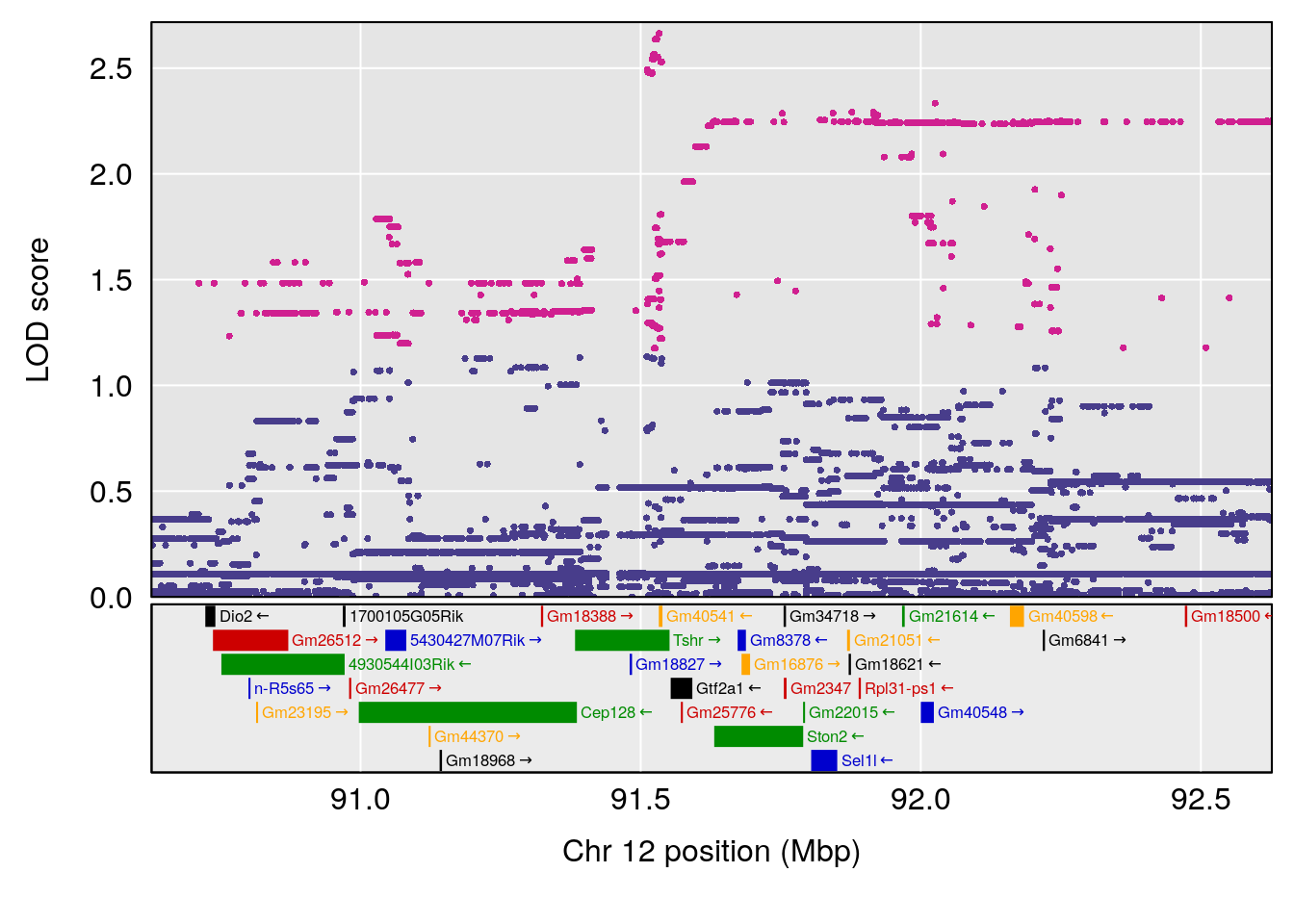
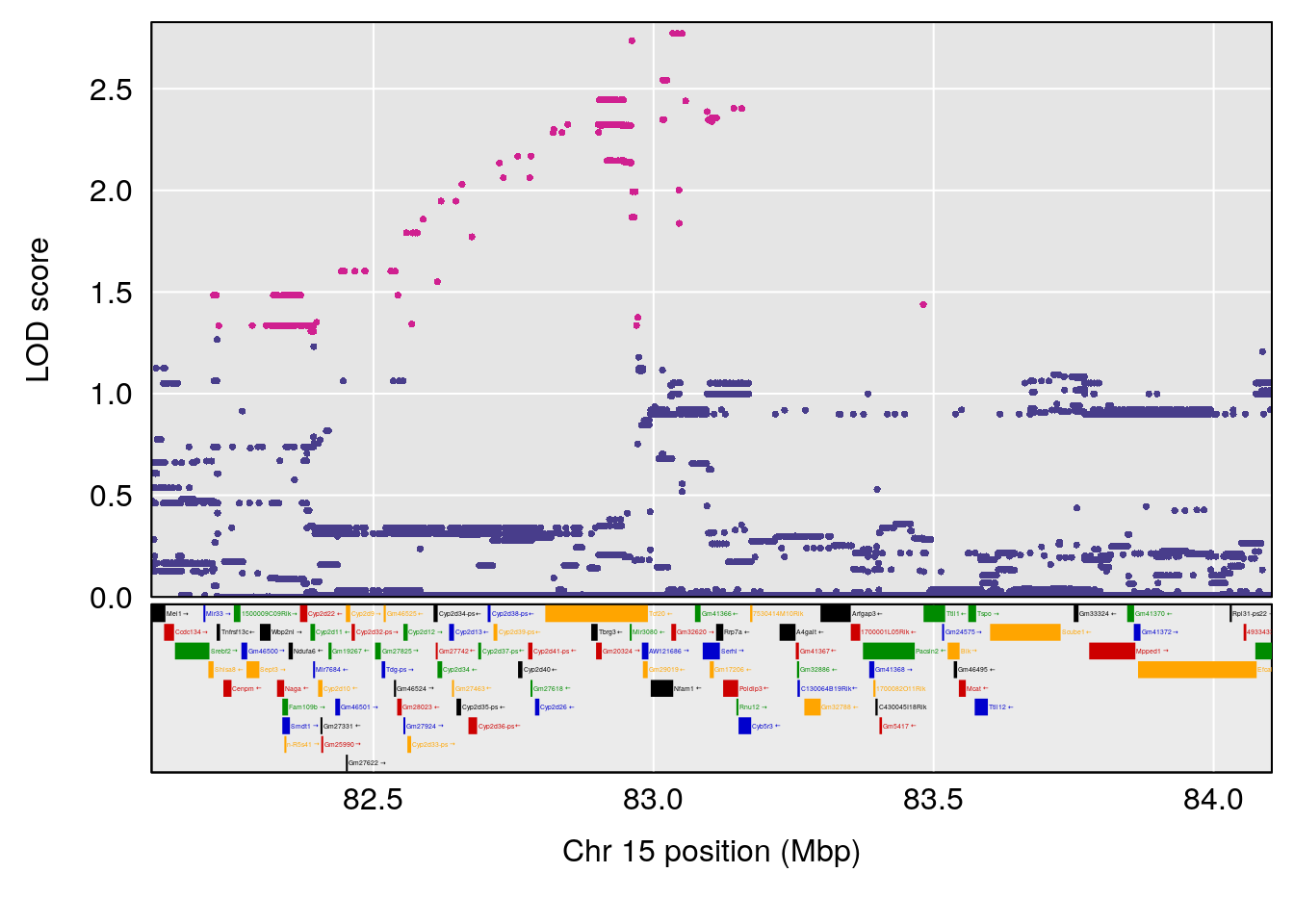
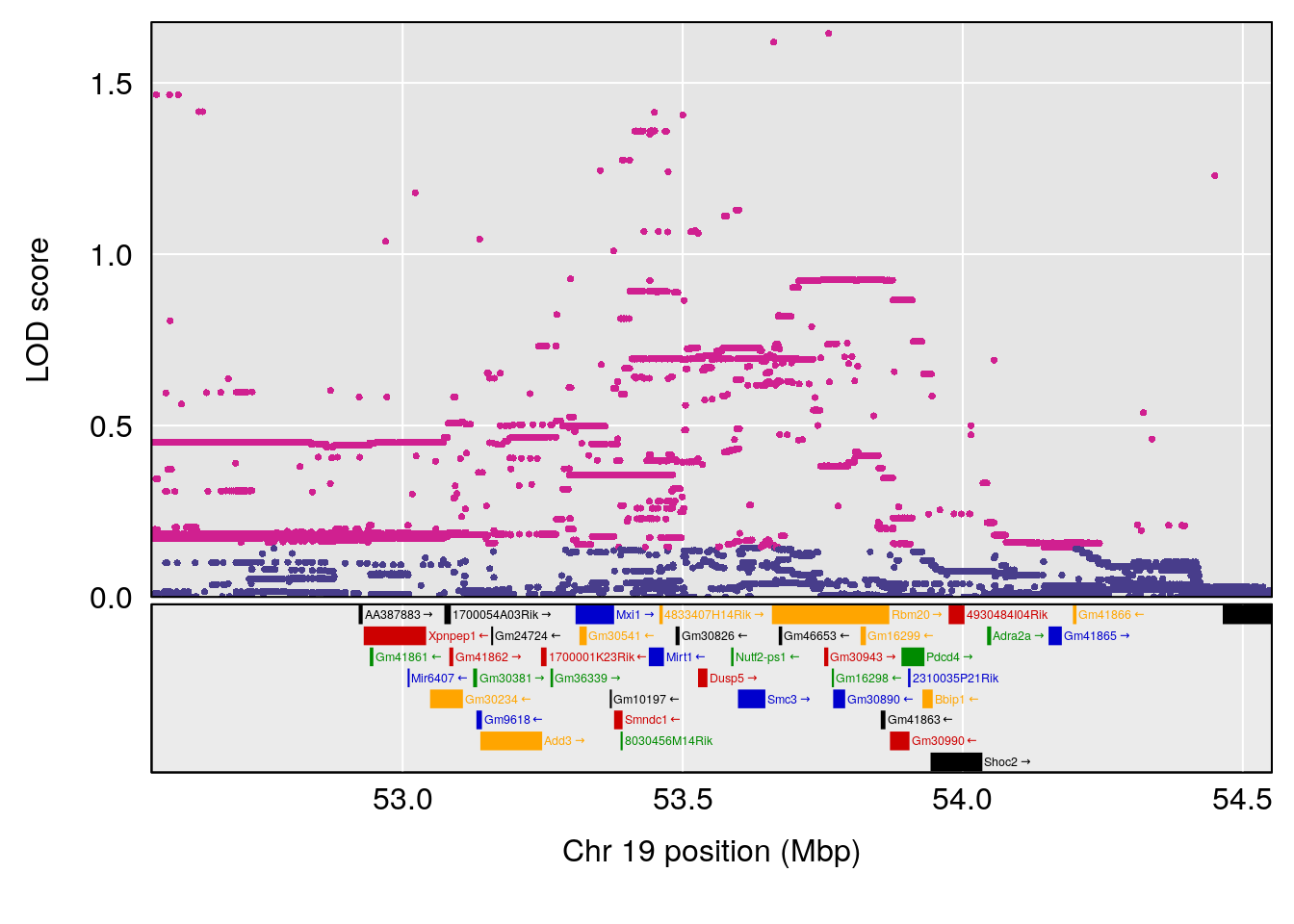
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
}
}
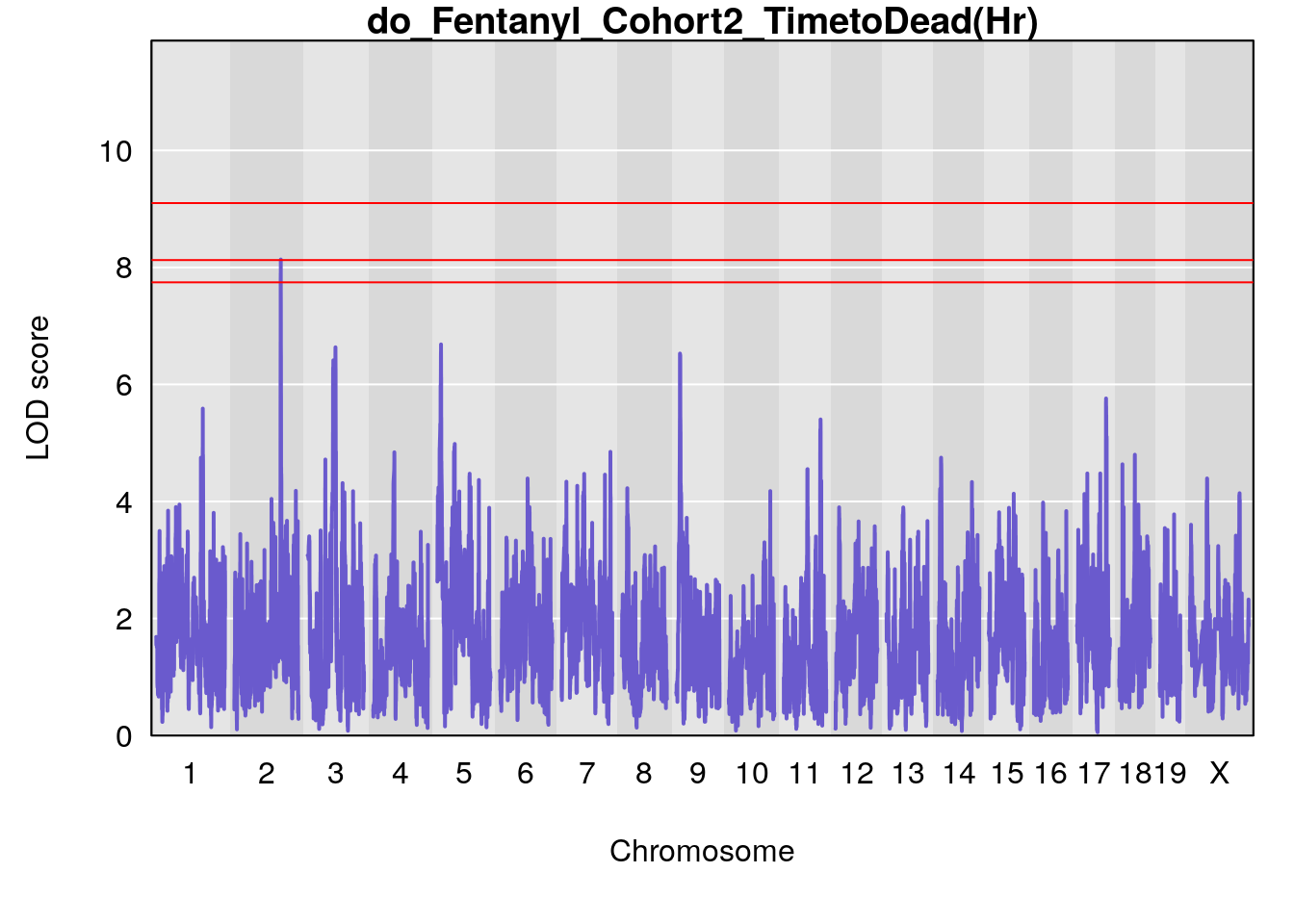
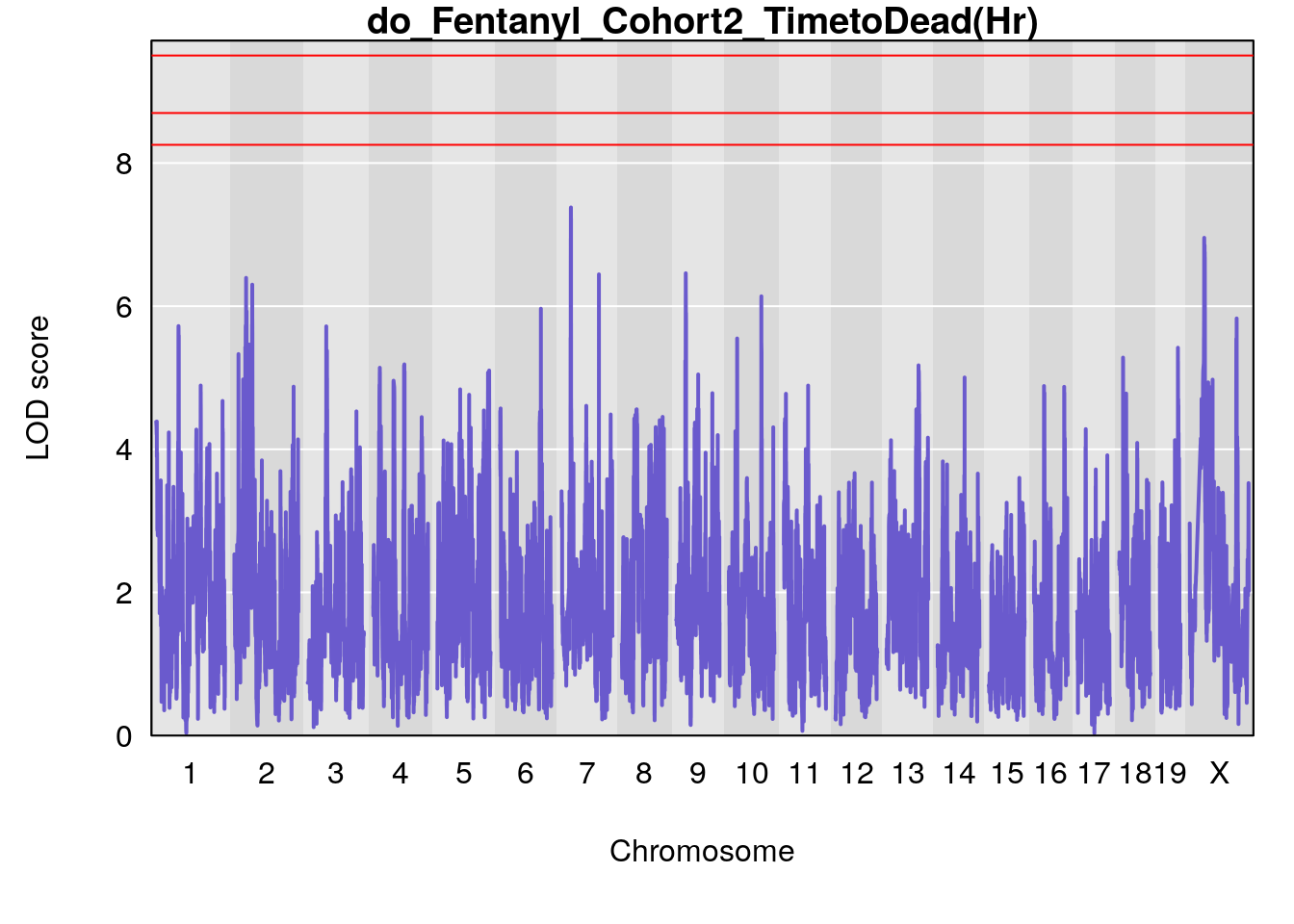
# [1] "TimetoDead(Hr)"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
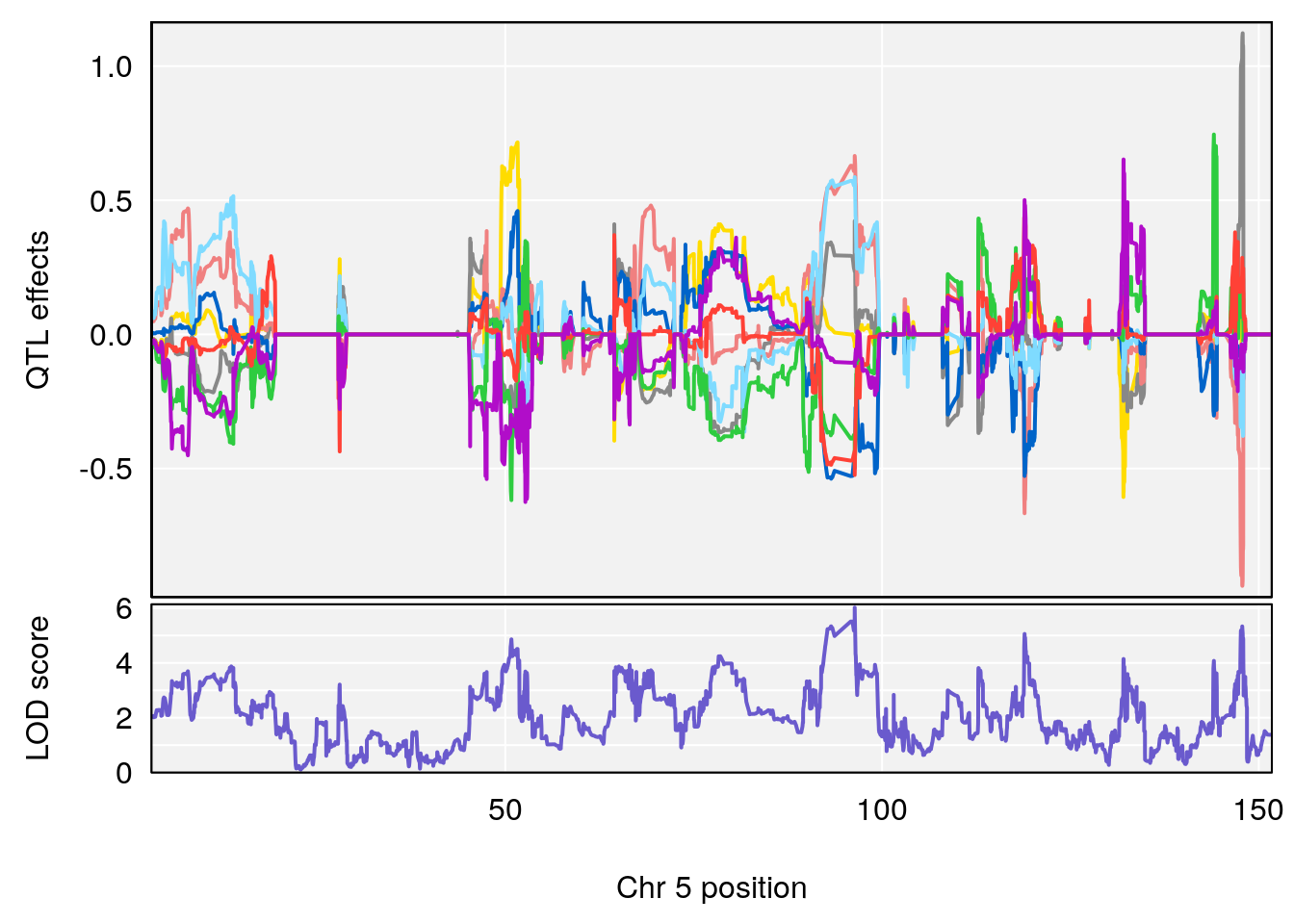
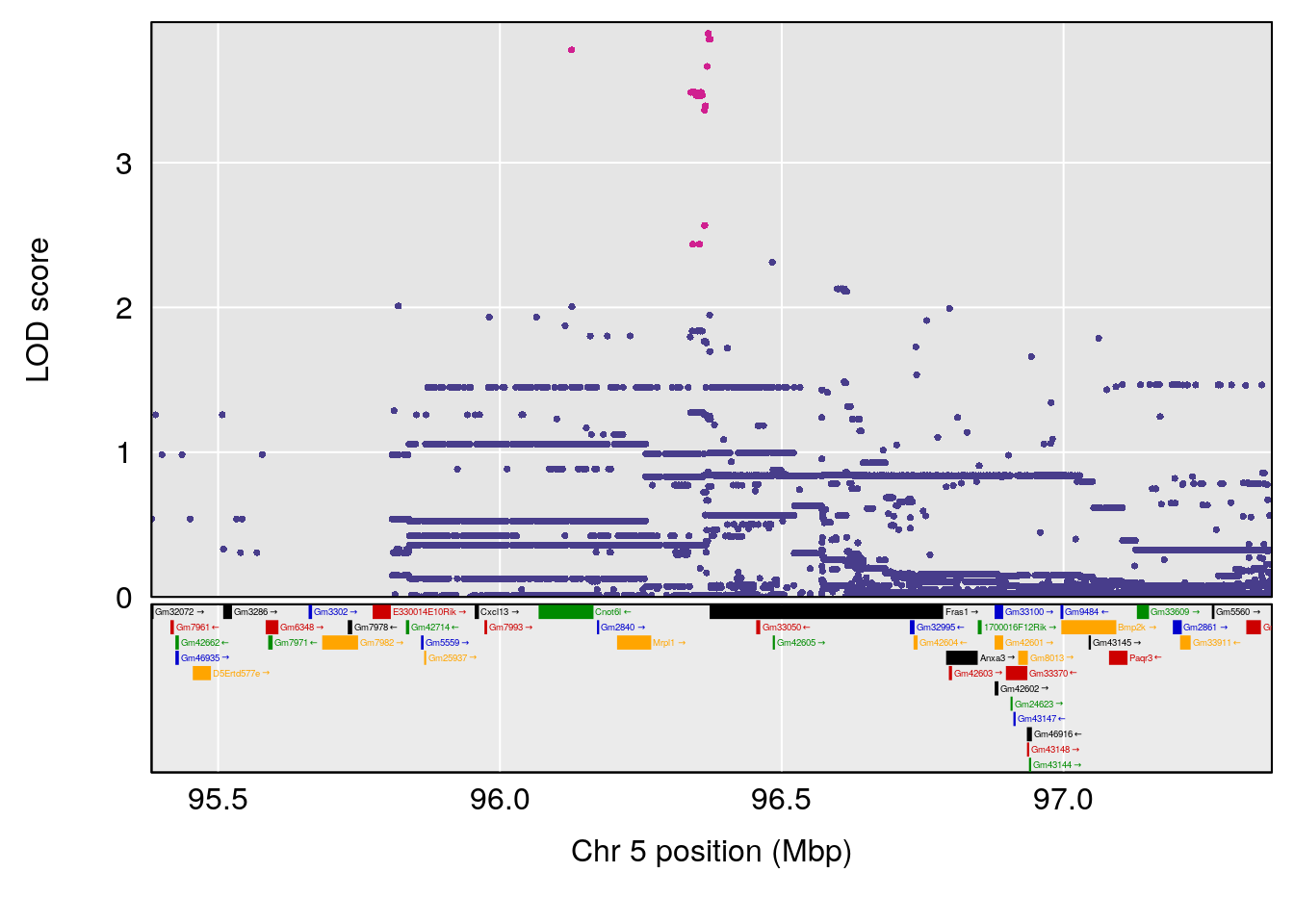
# 1 1 pheno1 2 132.62489 7.067034 106.68530 133.3807
# 2 1 pheno1 3 100.24885 6.239943 80.21006 101.0701
# 3 1 pheno1 5 96.36969 6.019079 50.65575 147.9508
# 4 1 pheno1 13 91.52025 6.147871 48.80519 117.2887
# [1] 1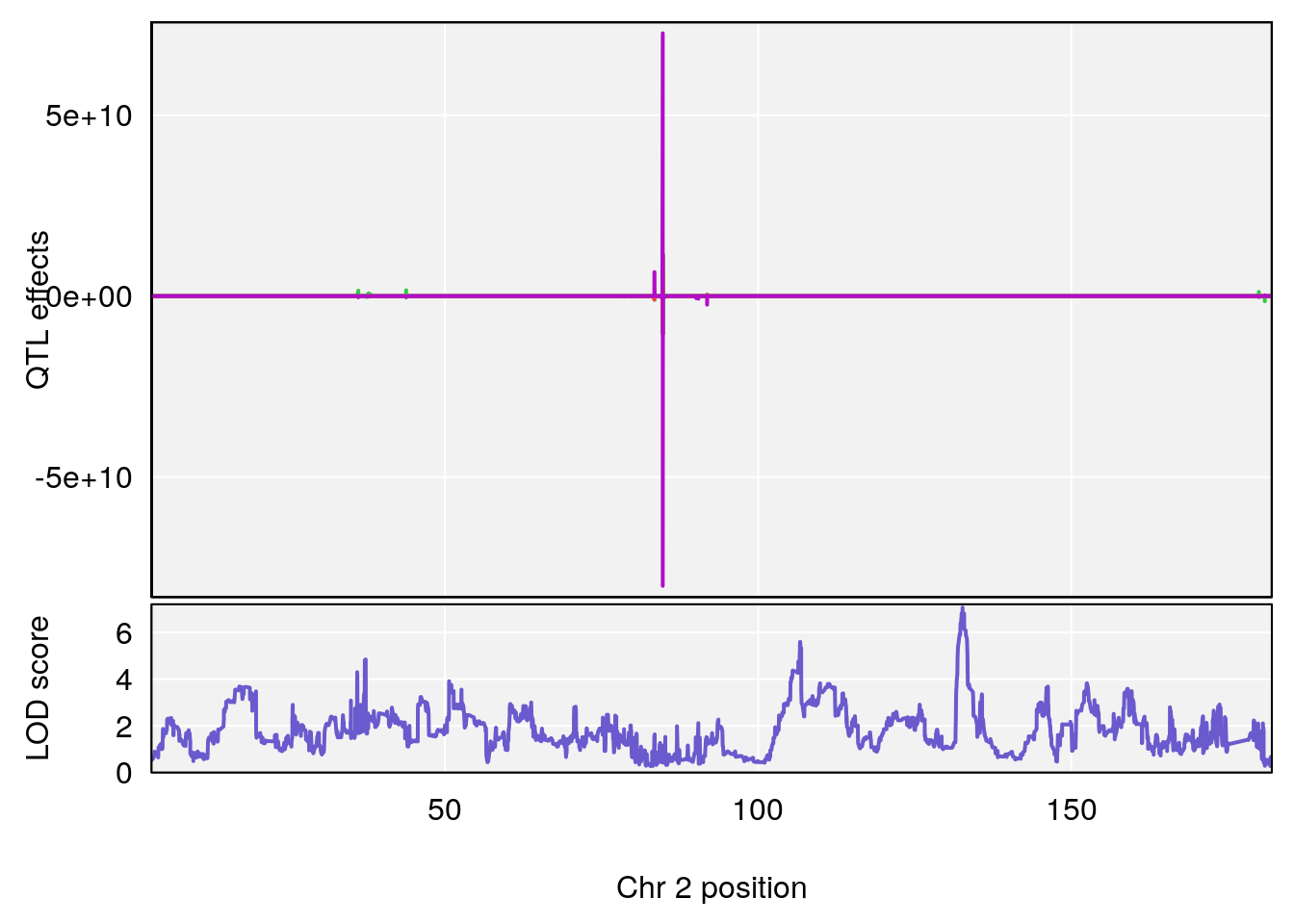
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
# [1] 2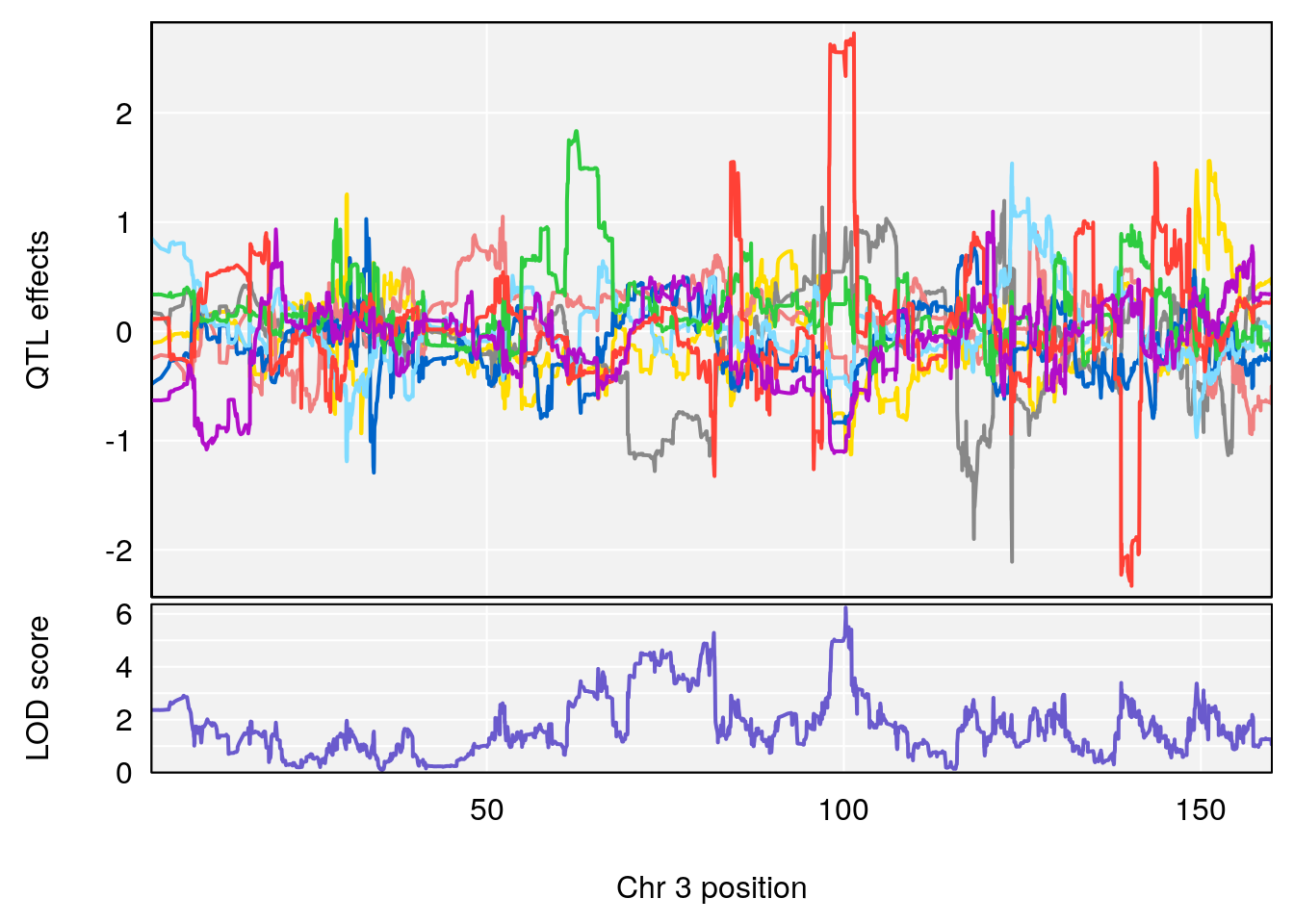
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
# [1] 3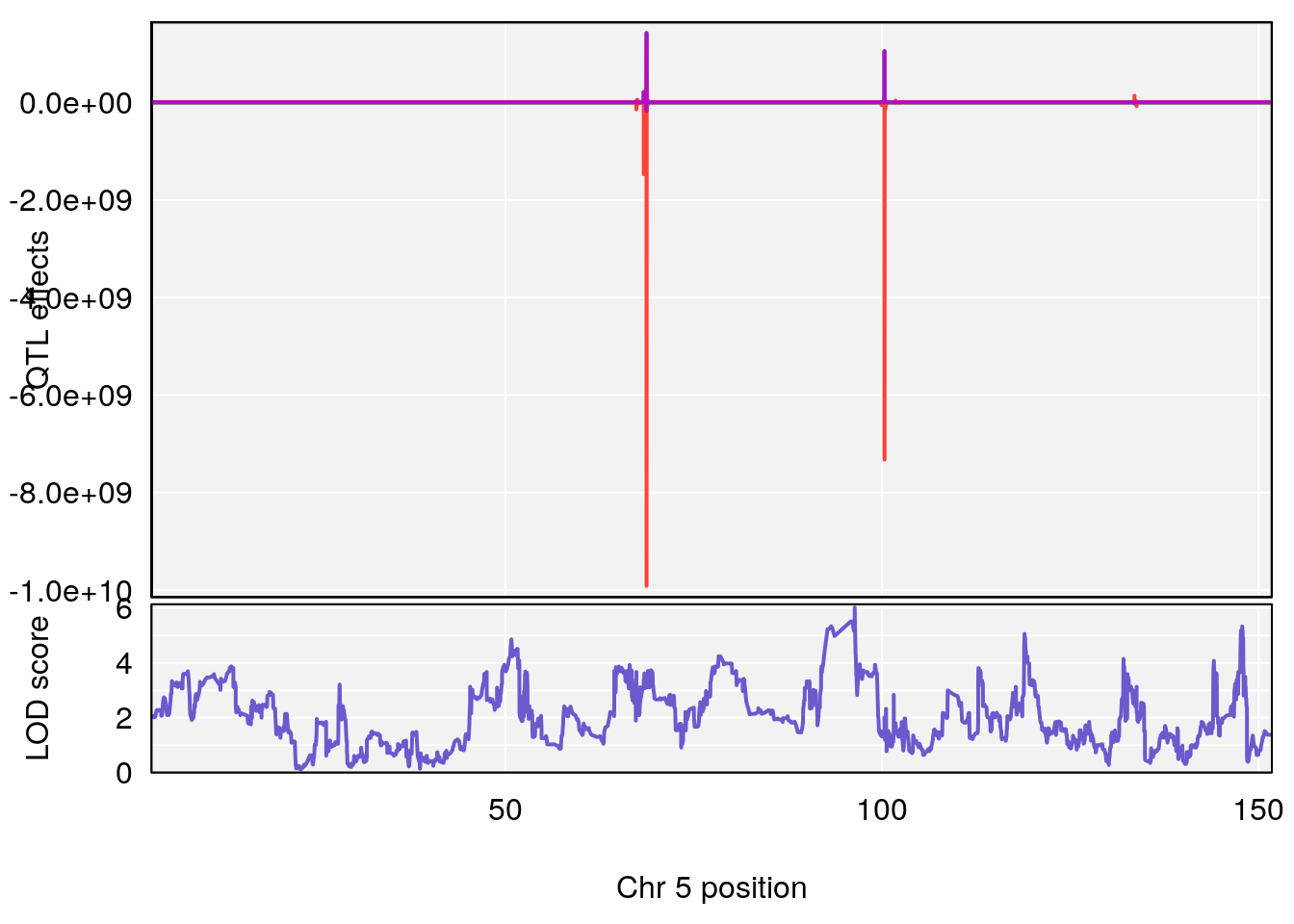
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
# [1] 4
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
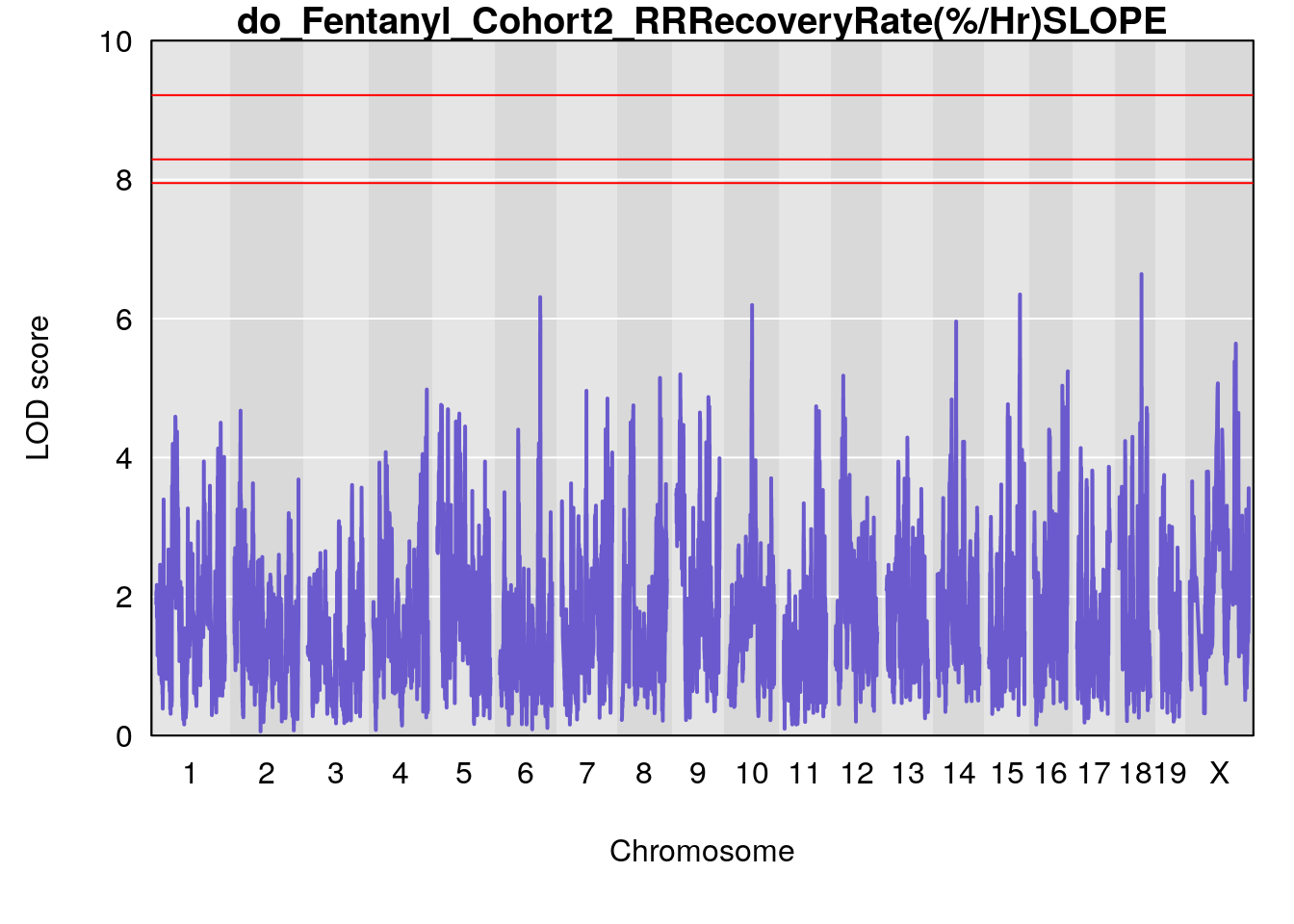
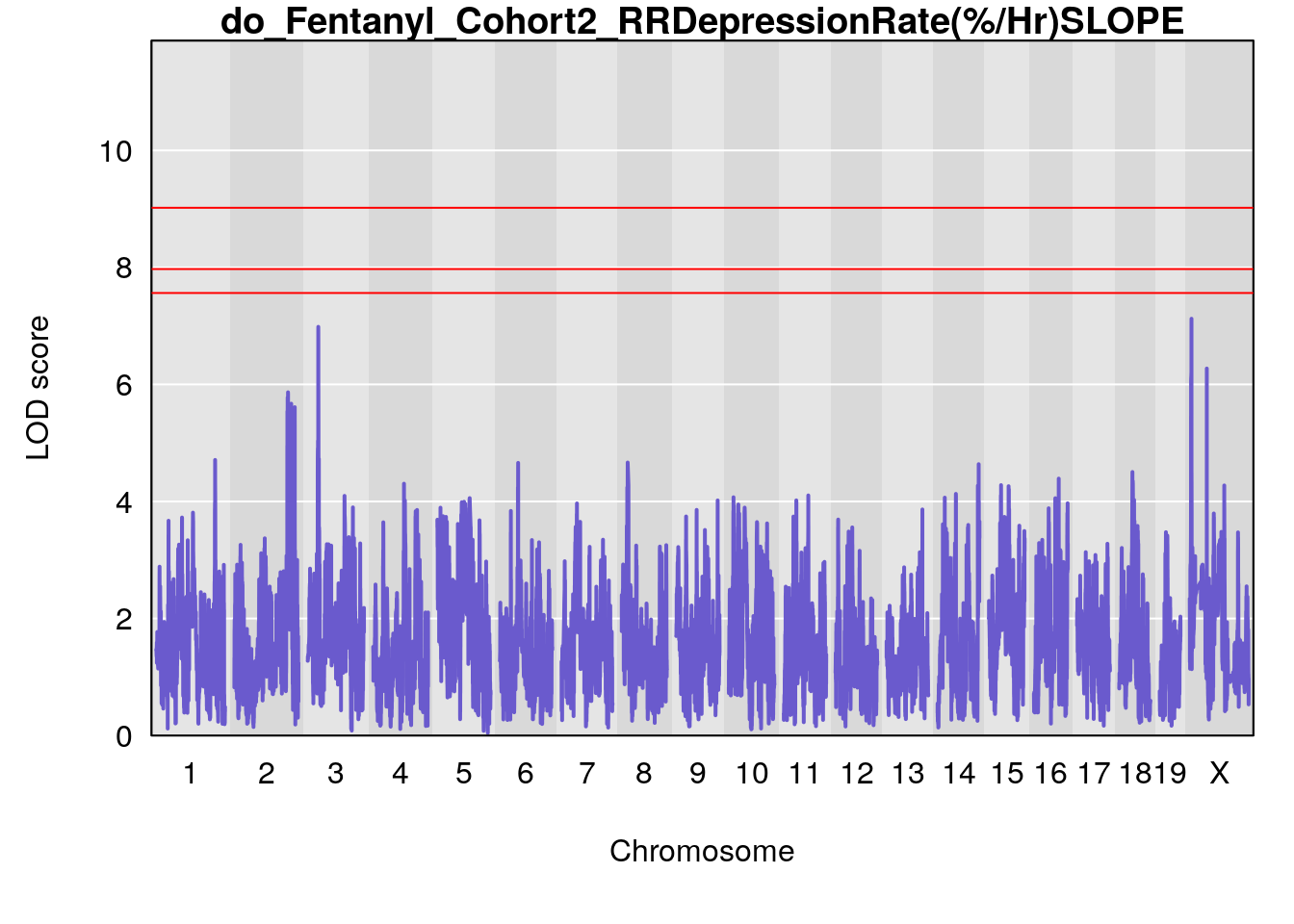
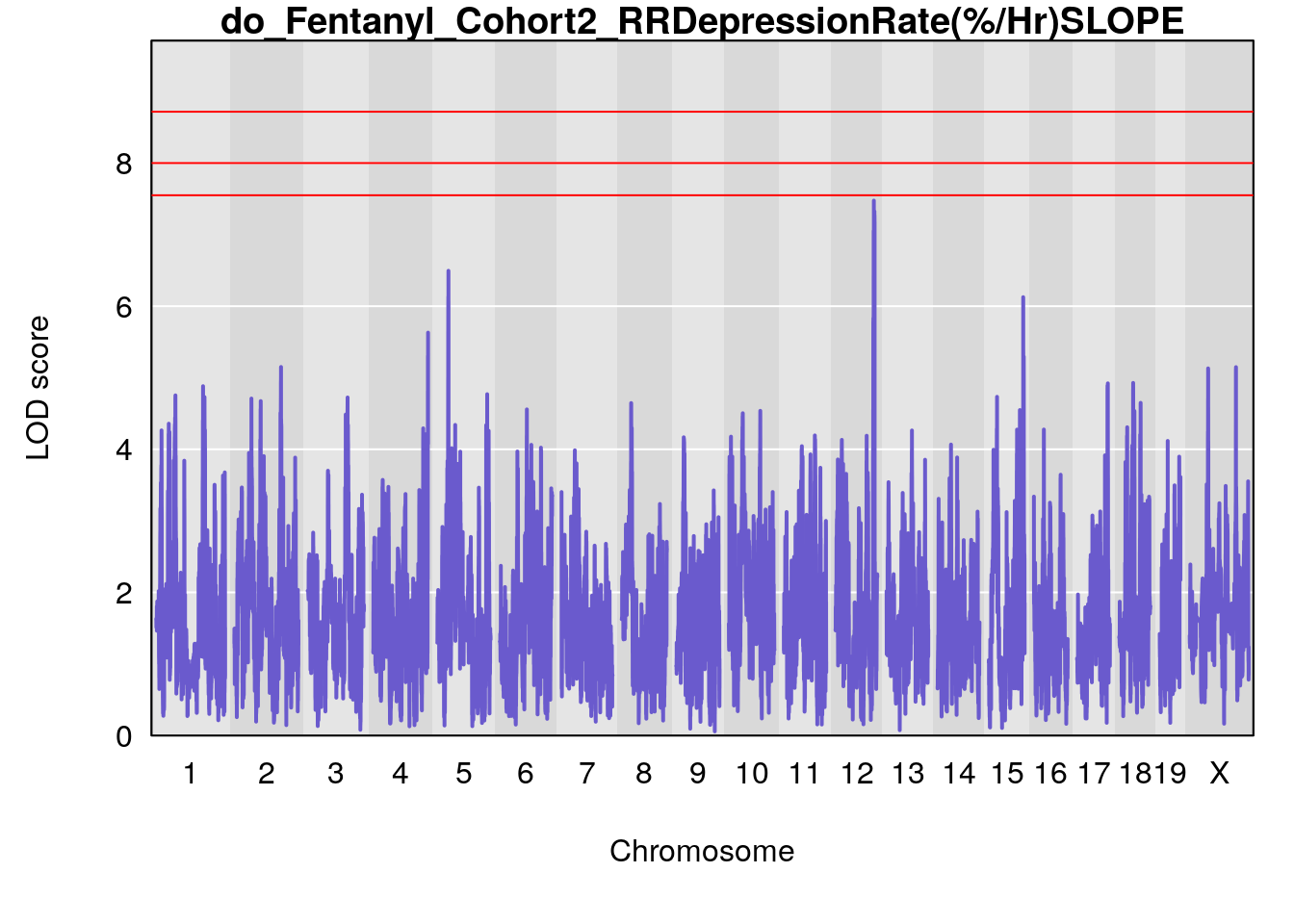
# [1] "RRDepressionRate(%/Hr)SLOPE"
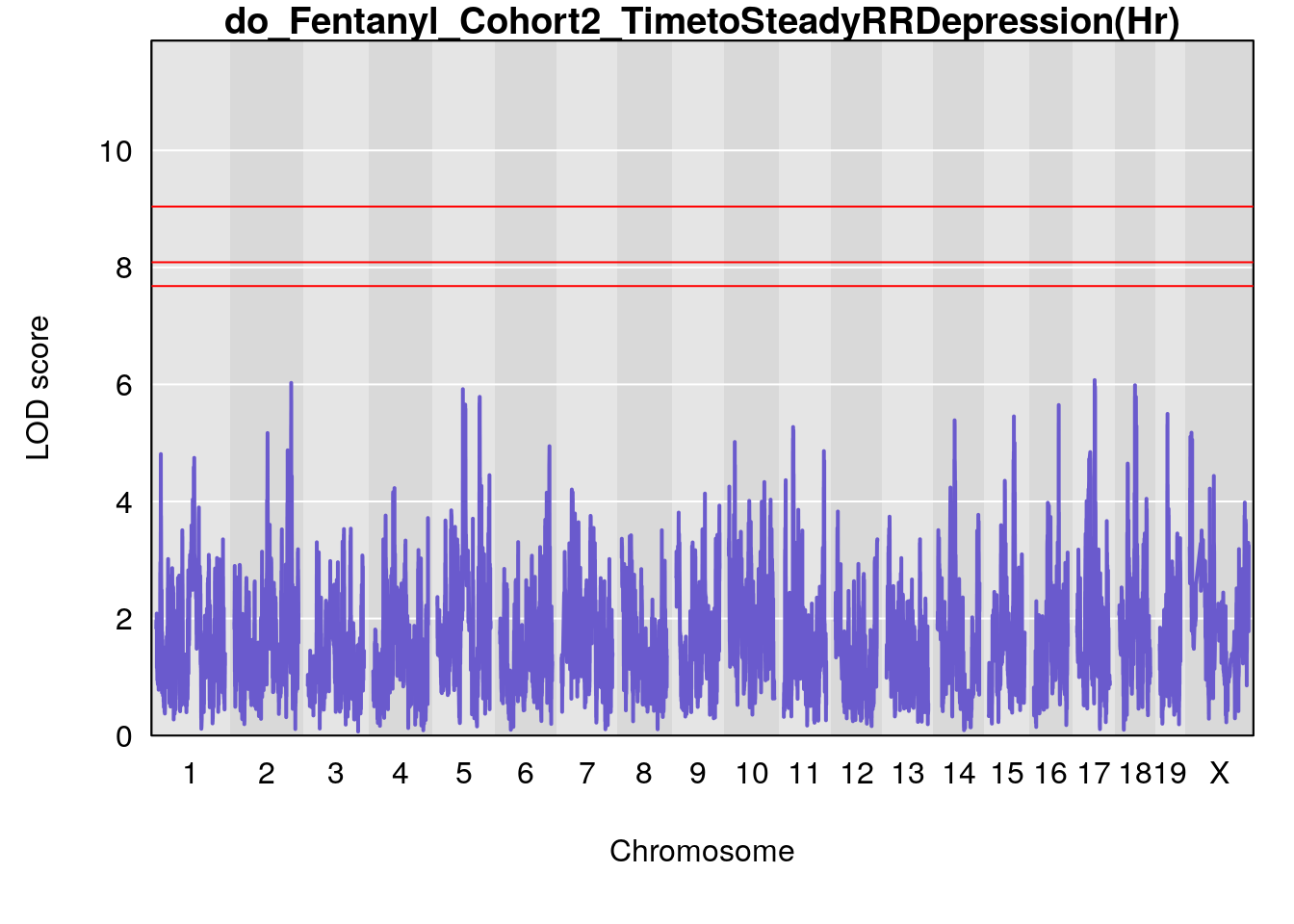
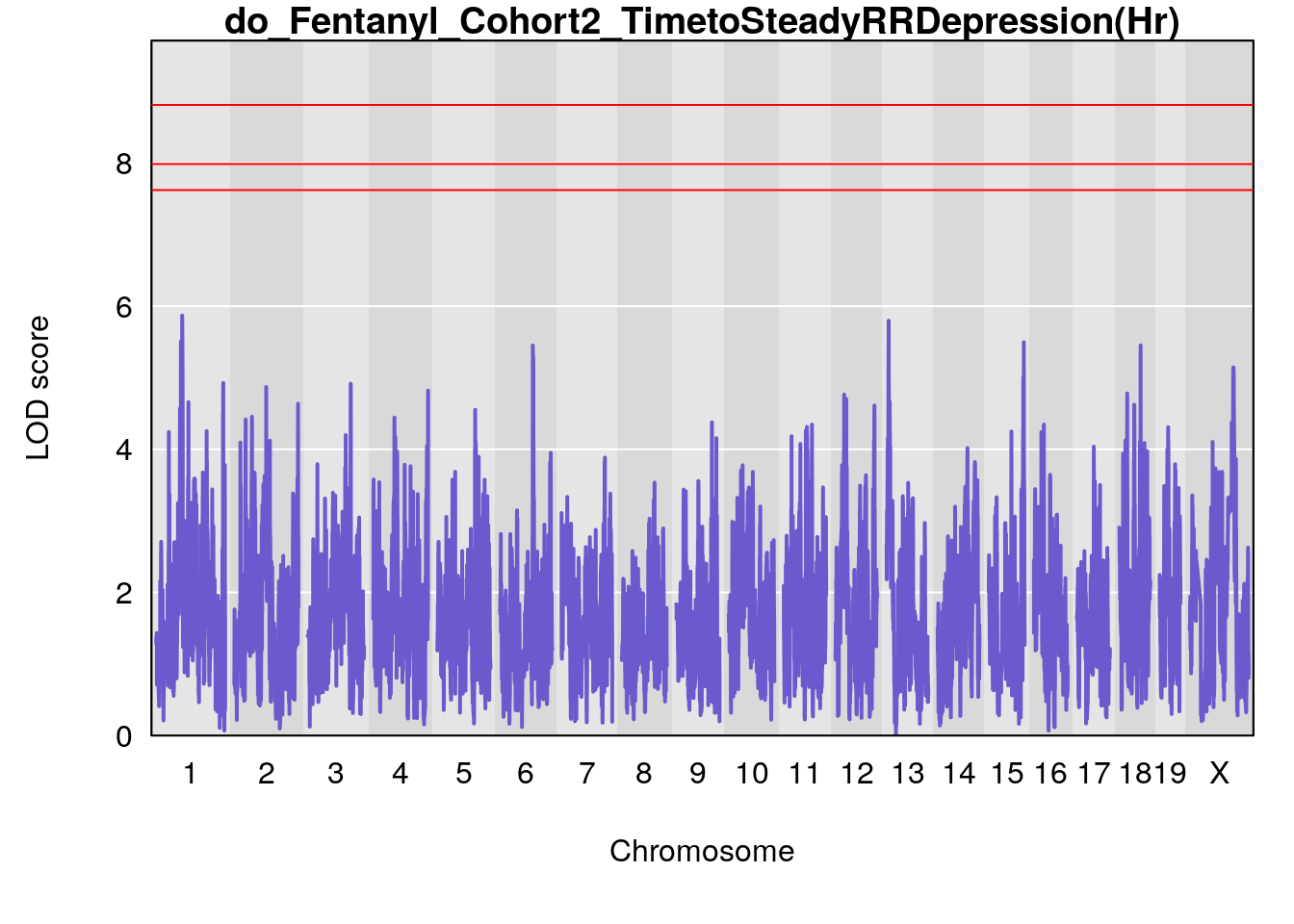
# [1] "TimetoSteadyRRDepression(Hr)"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 5 120.68390 6.483622 118.27549 121.58096
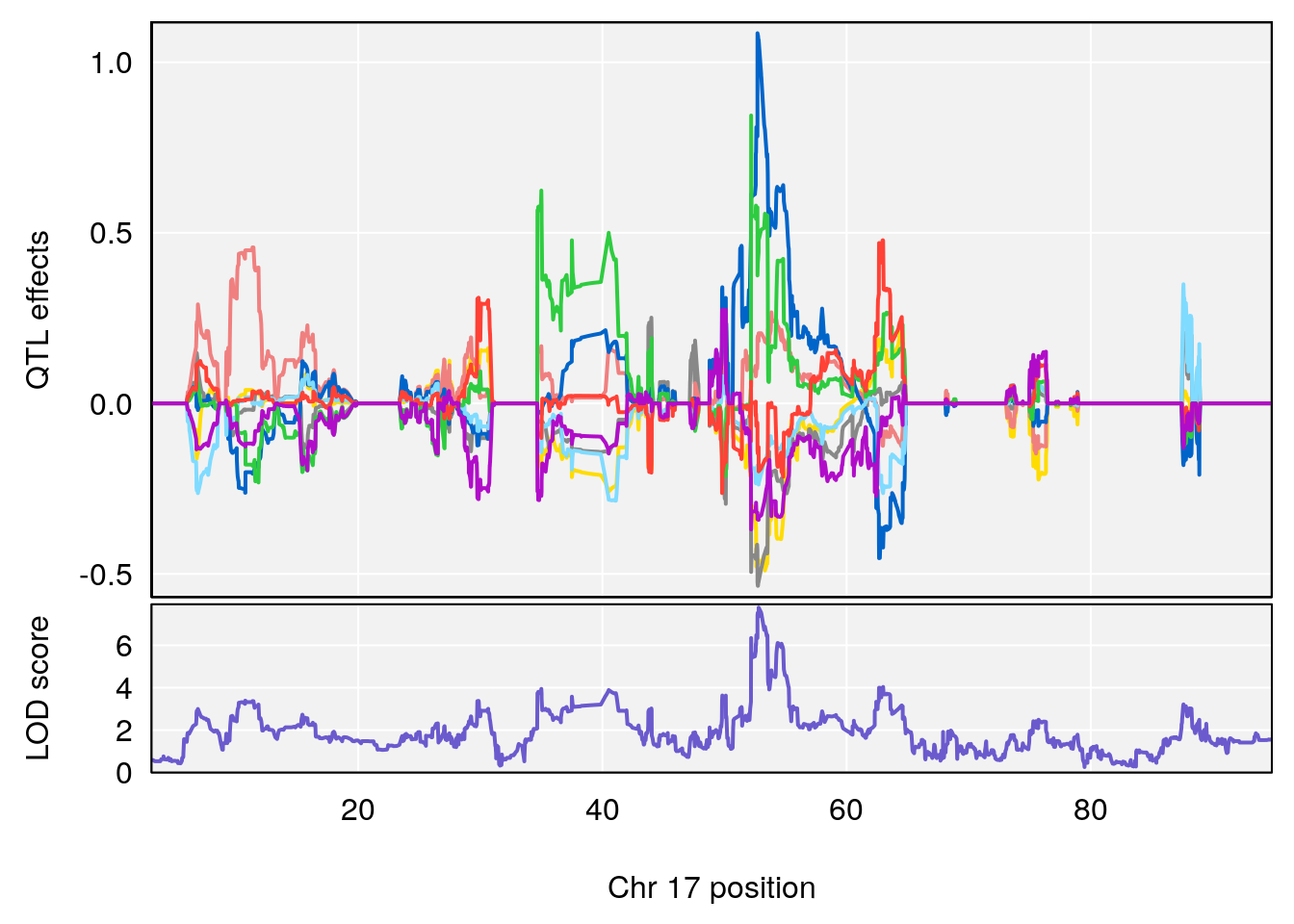
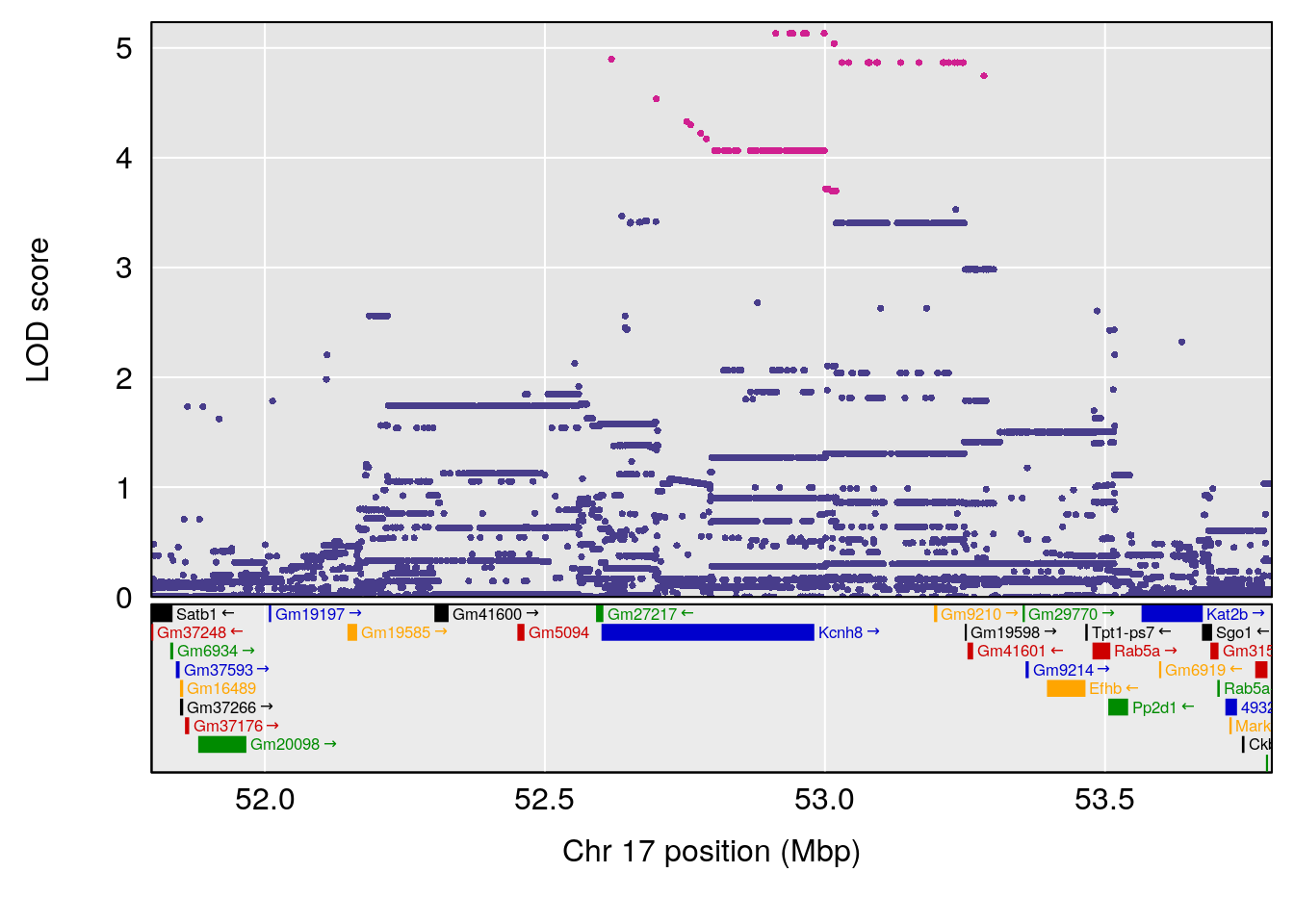
# 2 1 pheno1 17 52.79771 7.779978 52.16947 53.54382
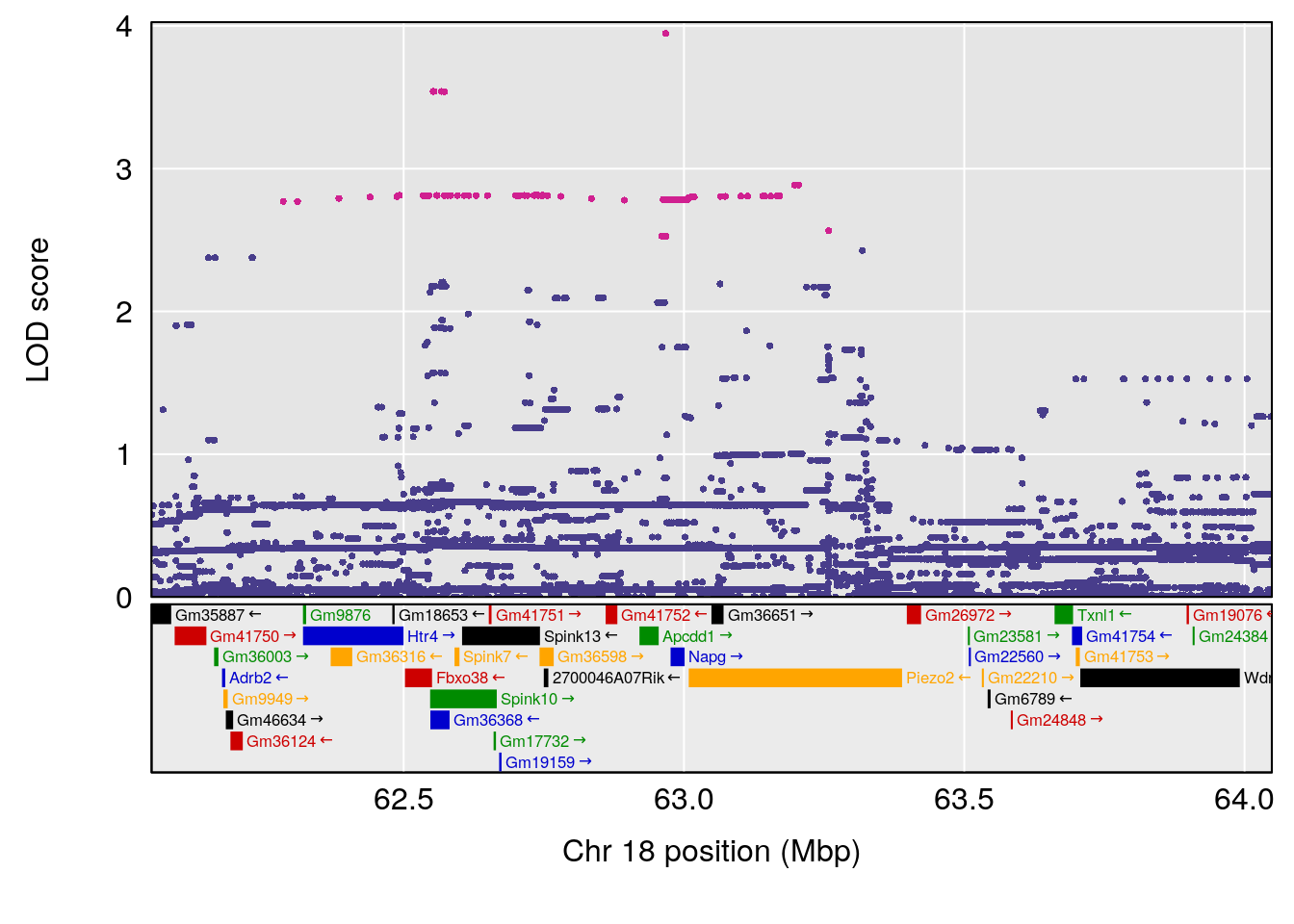
# 3 1 pheno1 18 63.05026 6.127351 49.68747 63.25613
# [1] 1
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
# [1] 2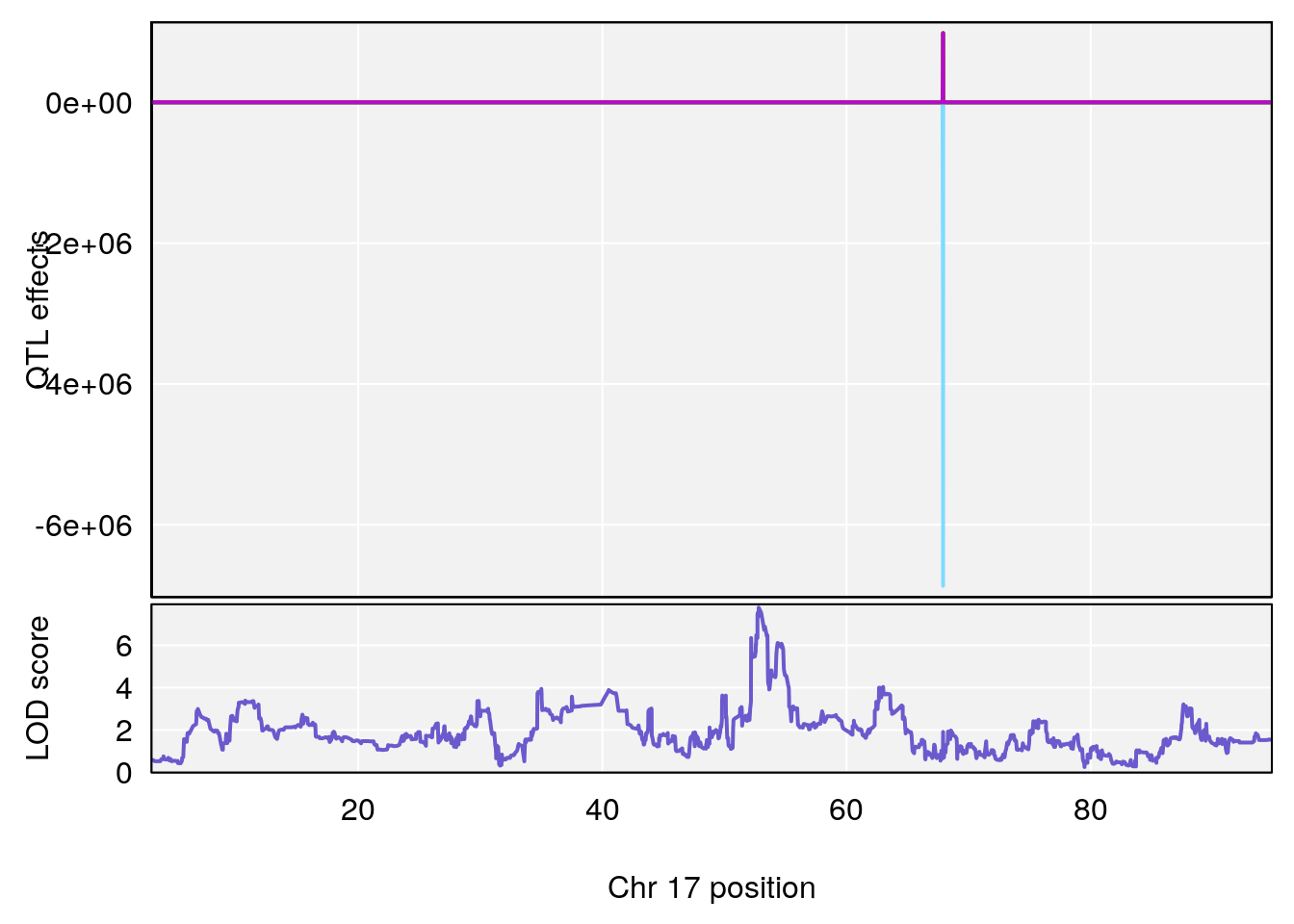
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
# [1] 3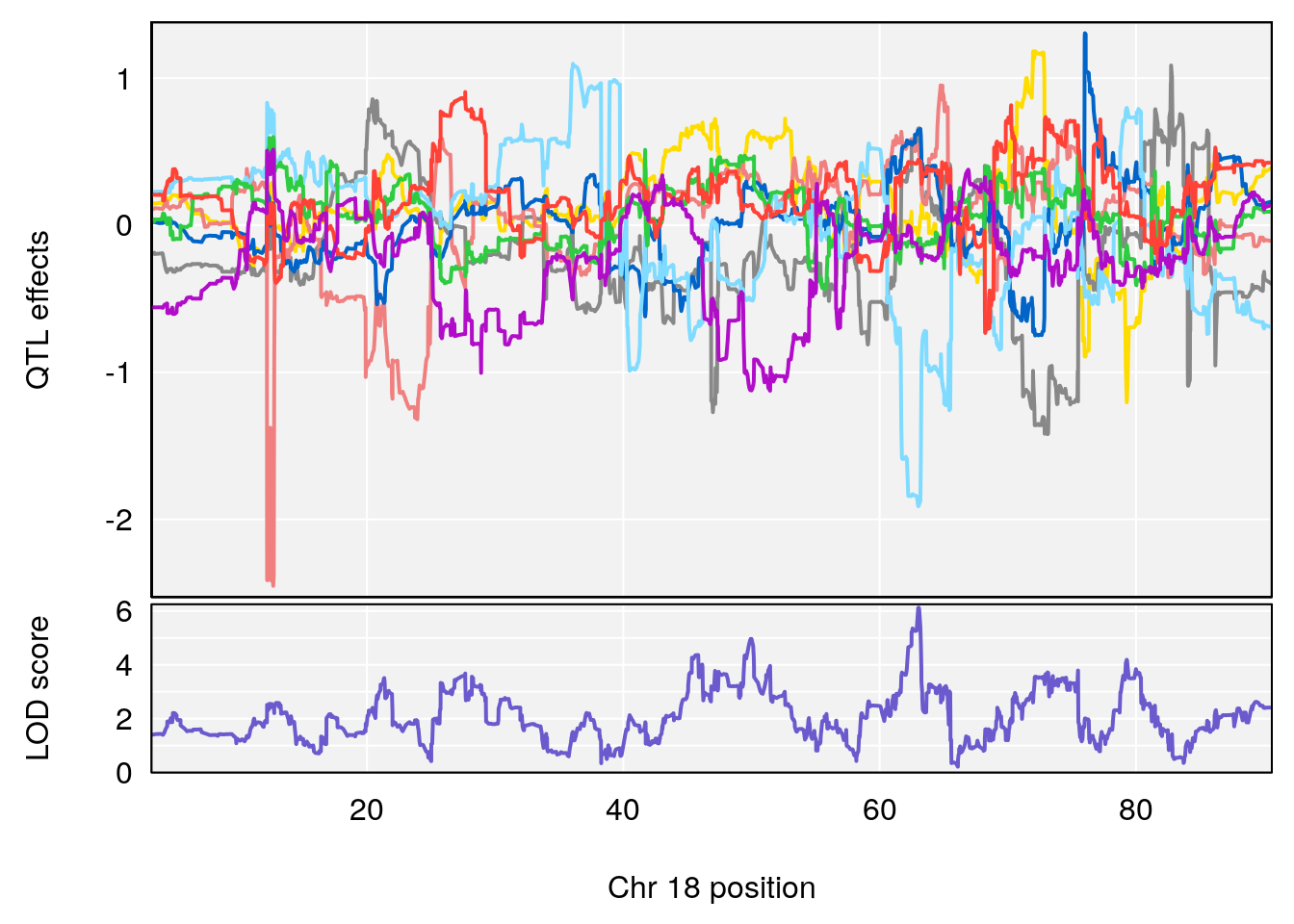
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
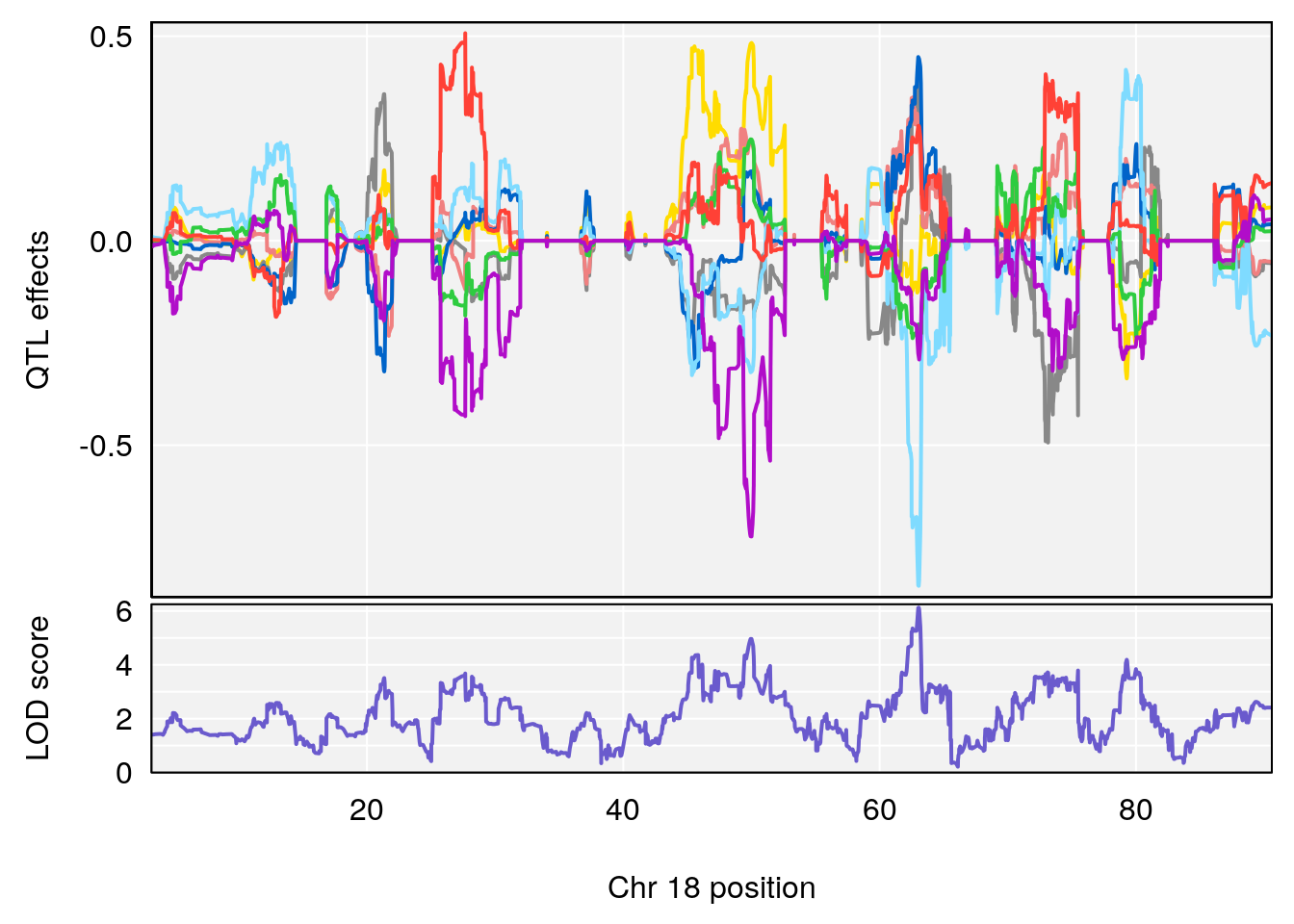
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
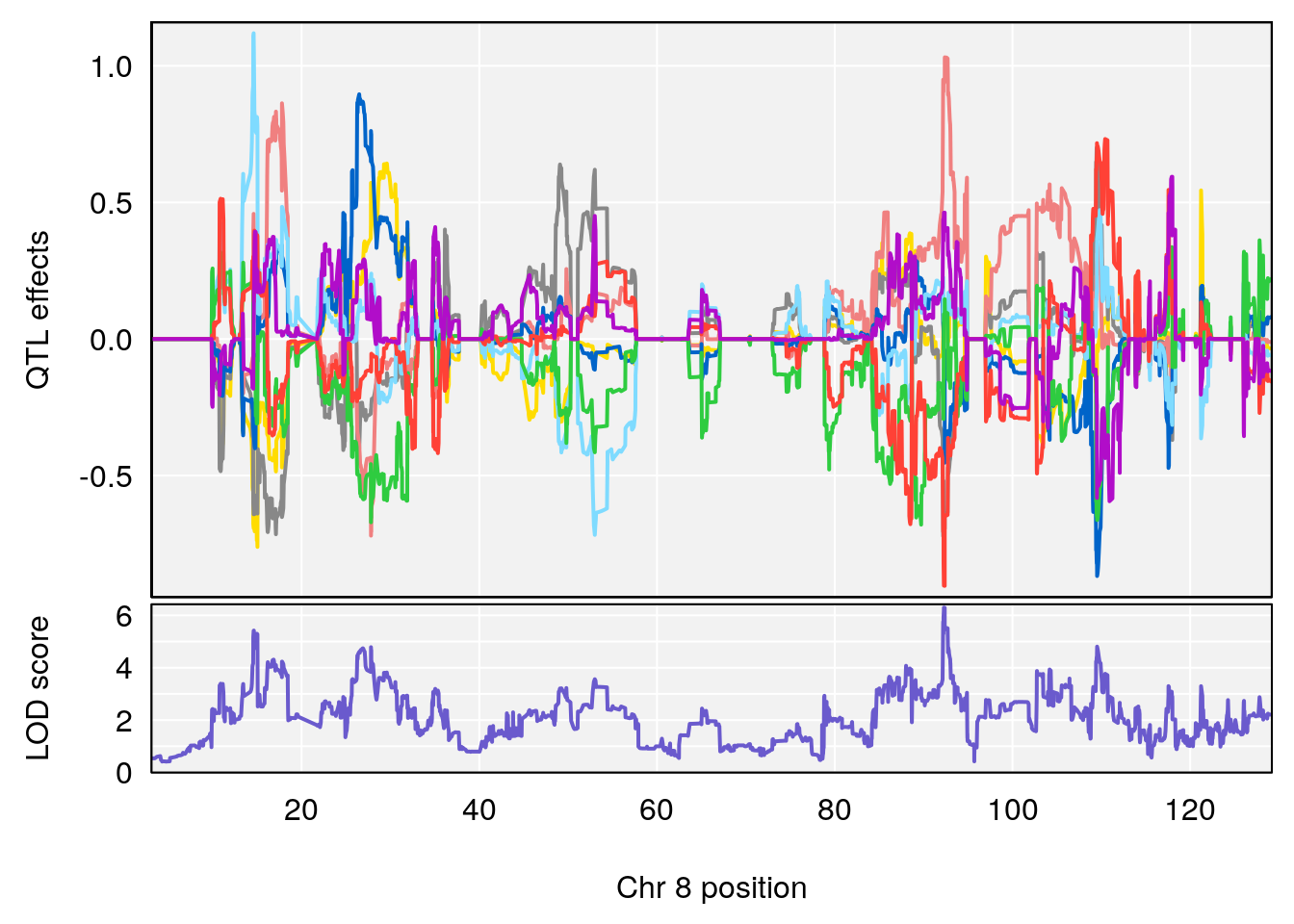
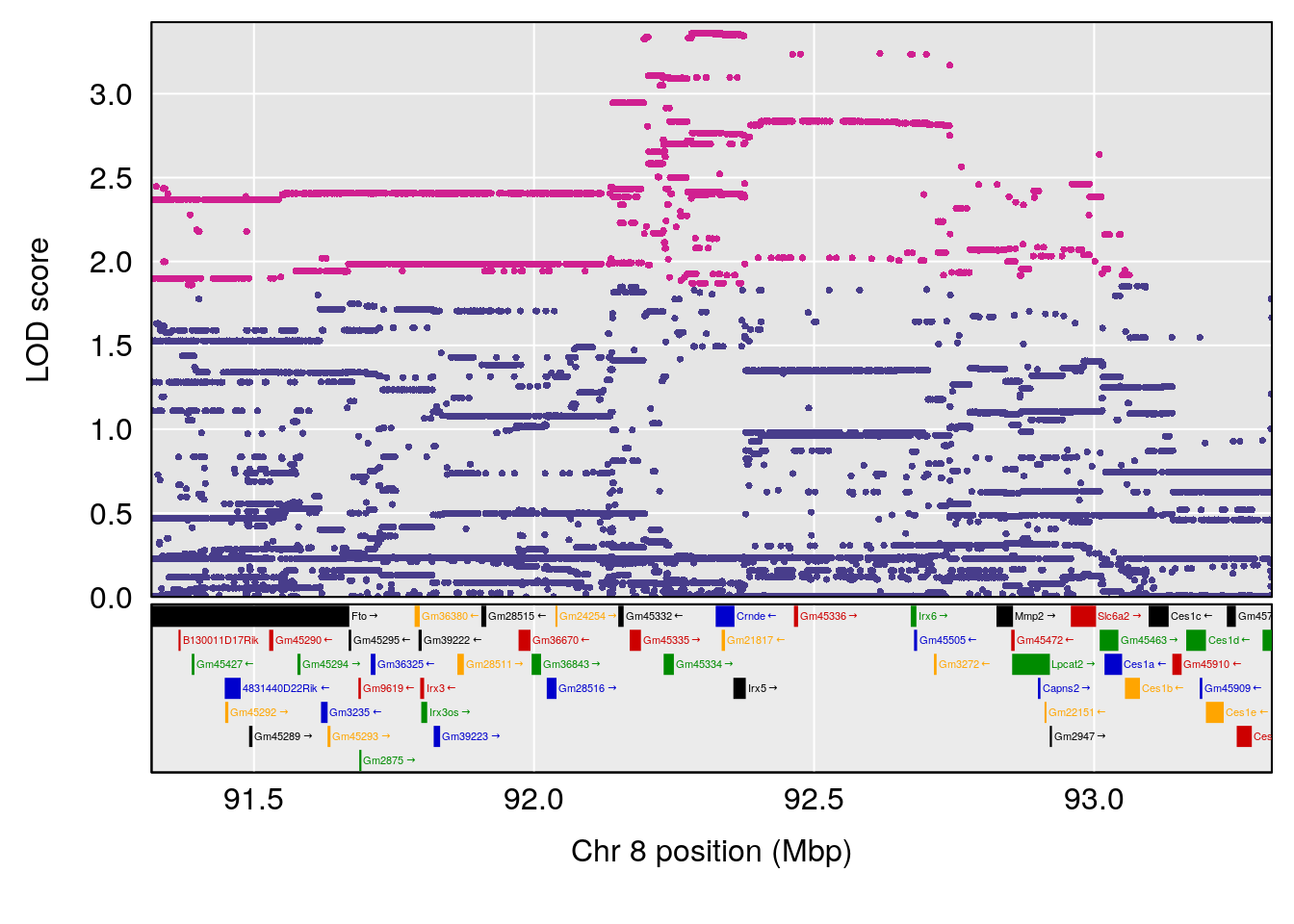
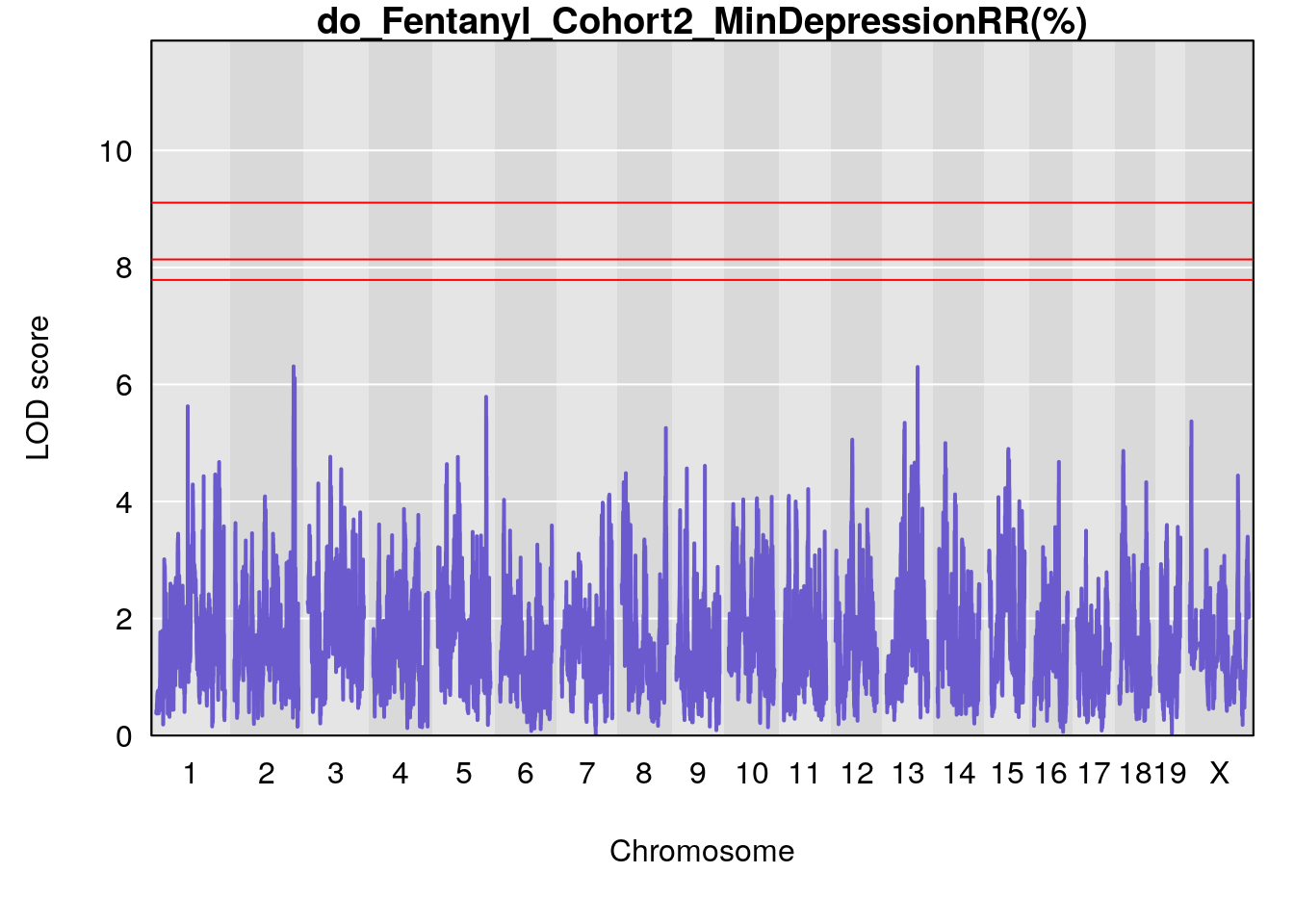
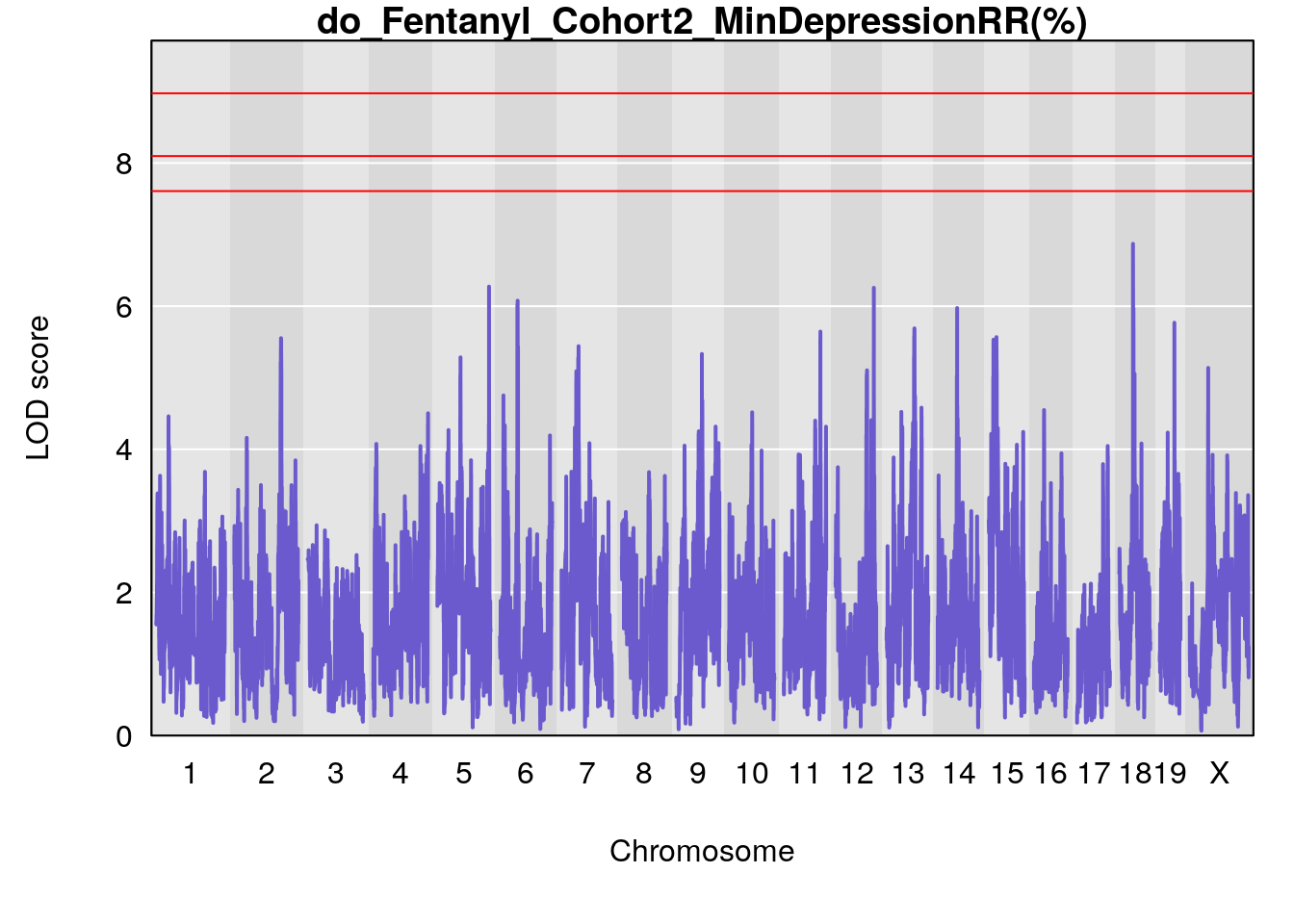
# [1] "MinDepressionRR(%)"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 8 14.56268 6.752336 14.38322 14.60657
# [1] 1
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
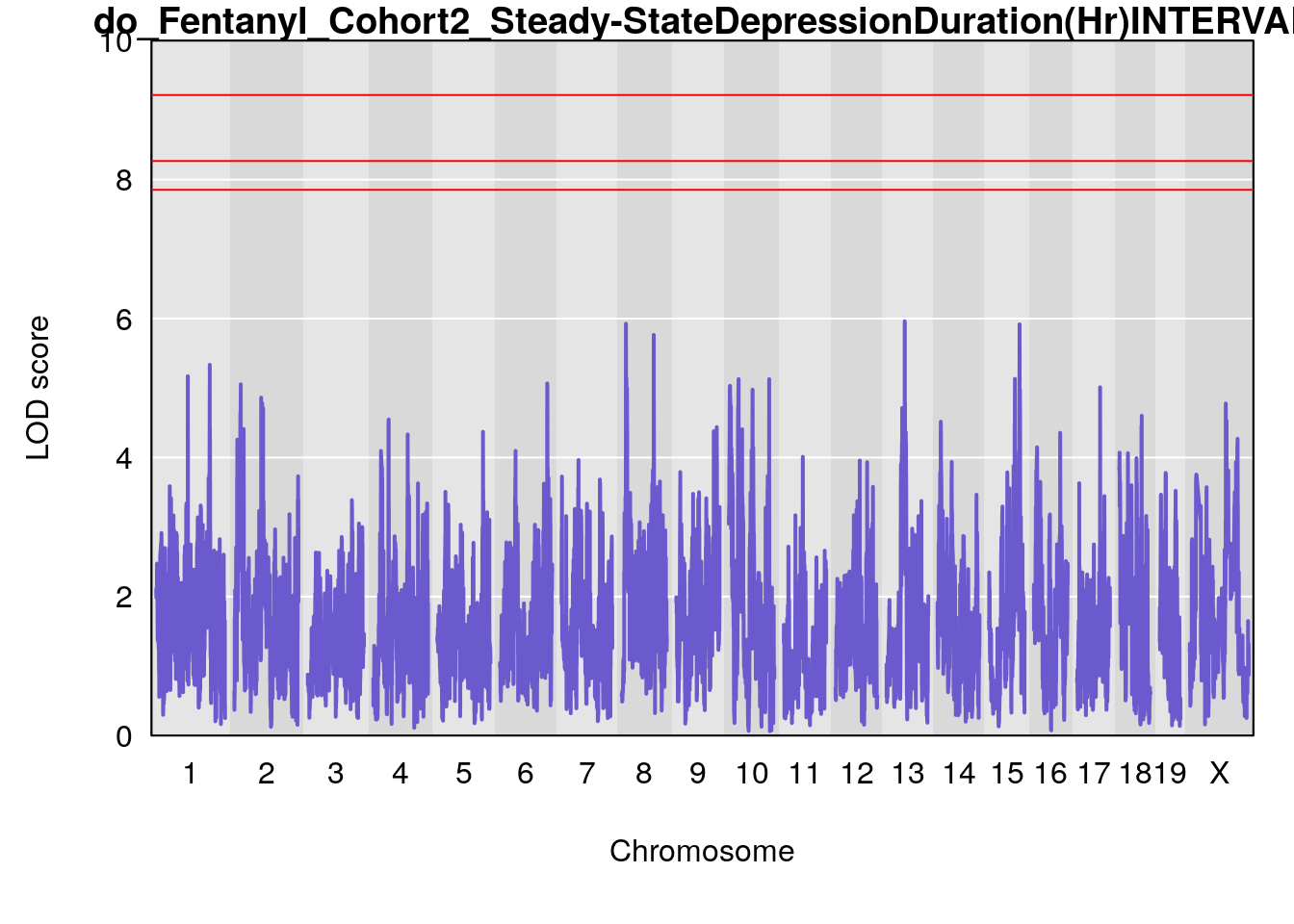
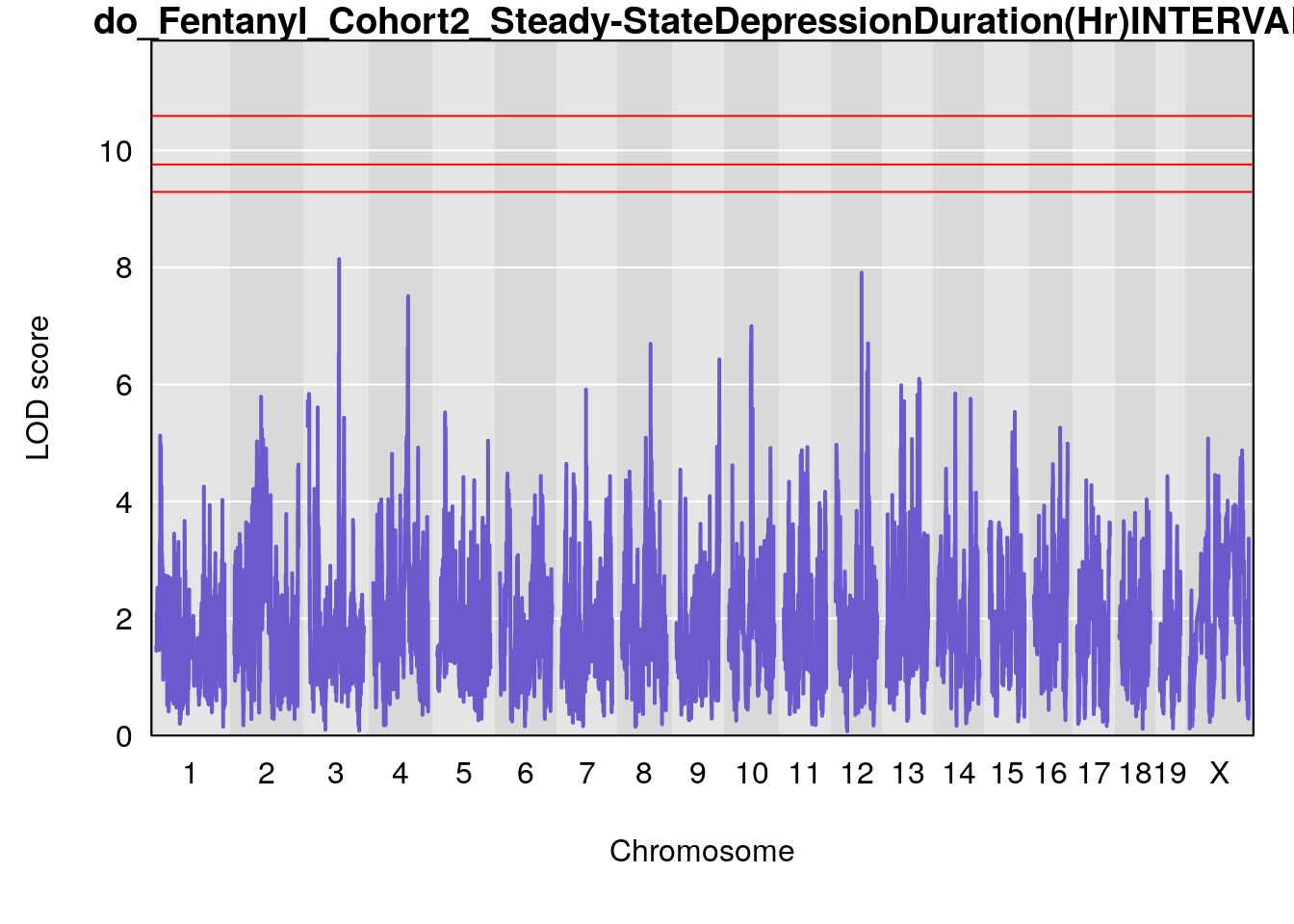
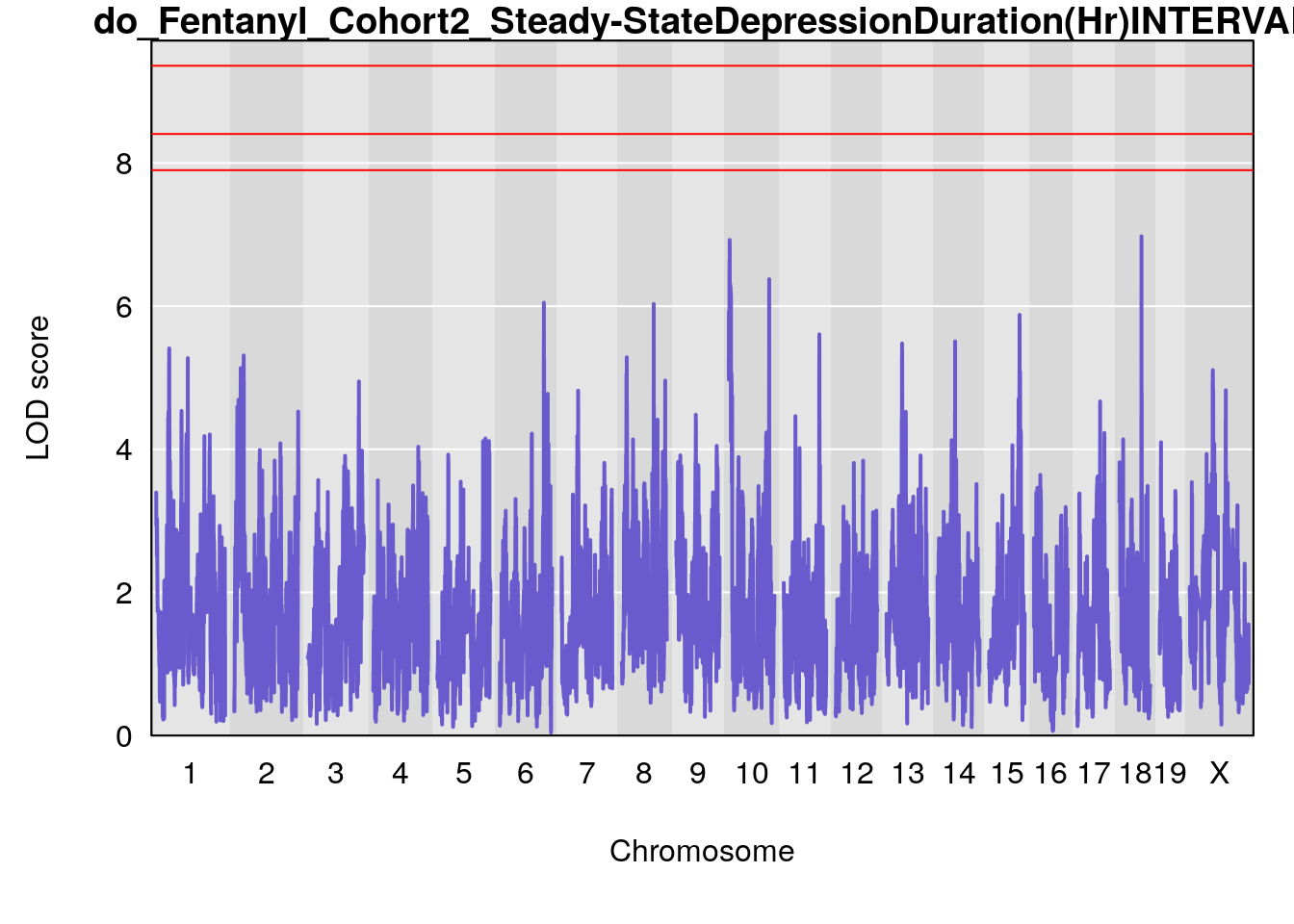
# [1] "Steady-StateDepressionDuration(Hr)INTERVAL"
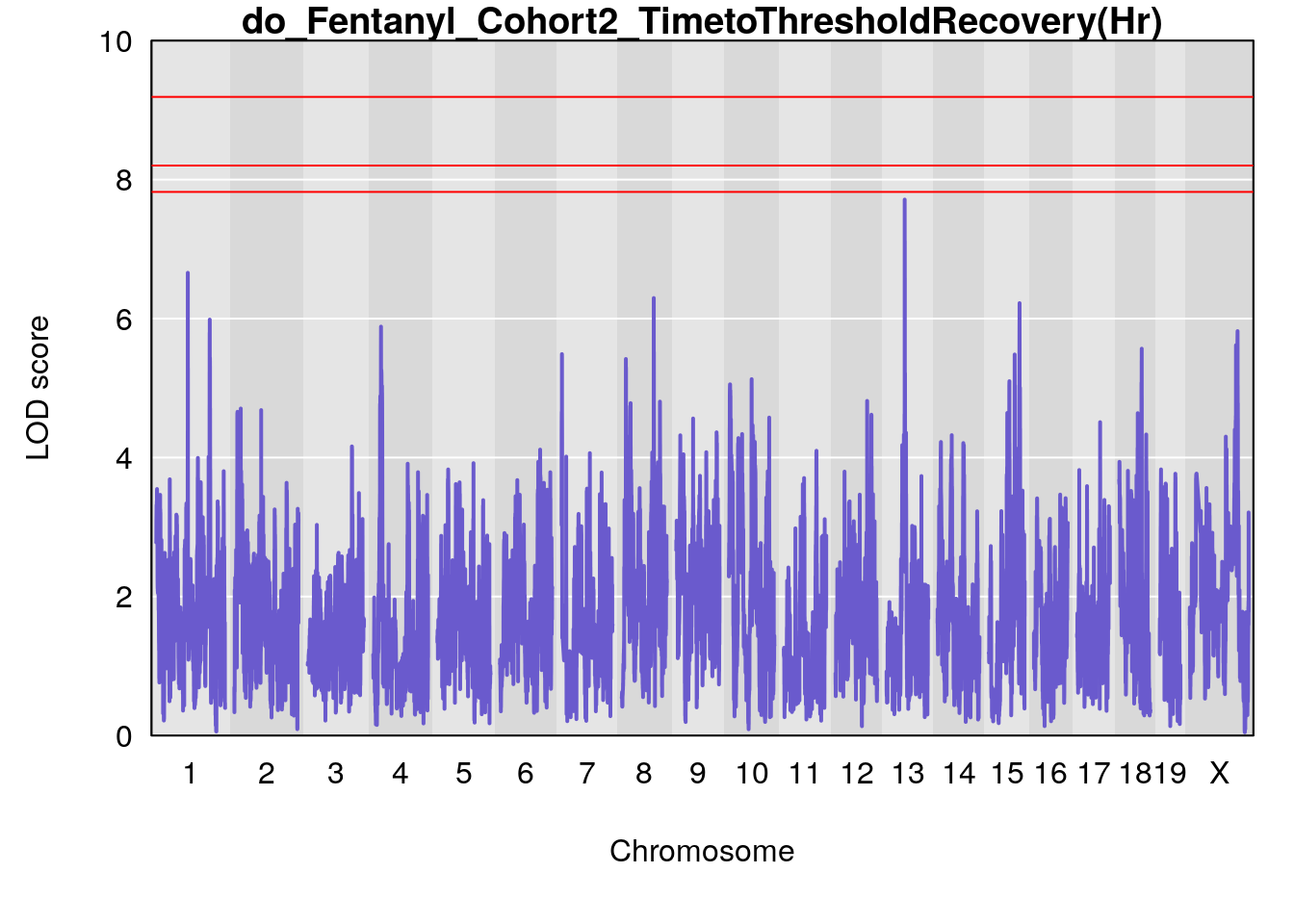
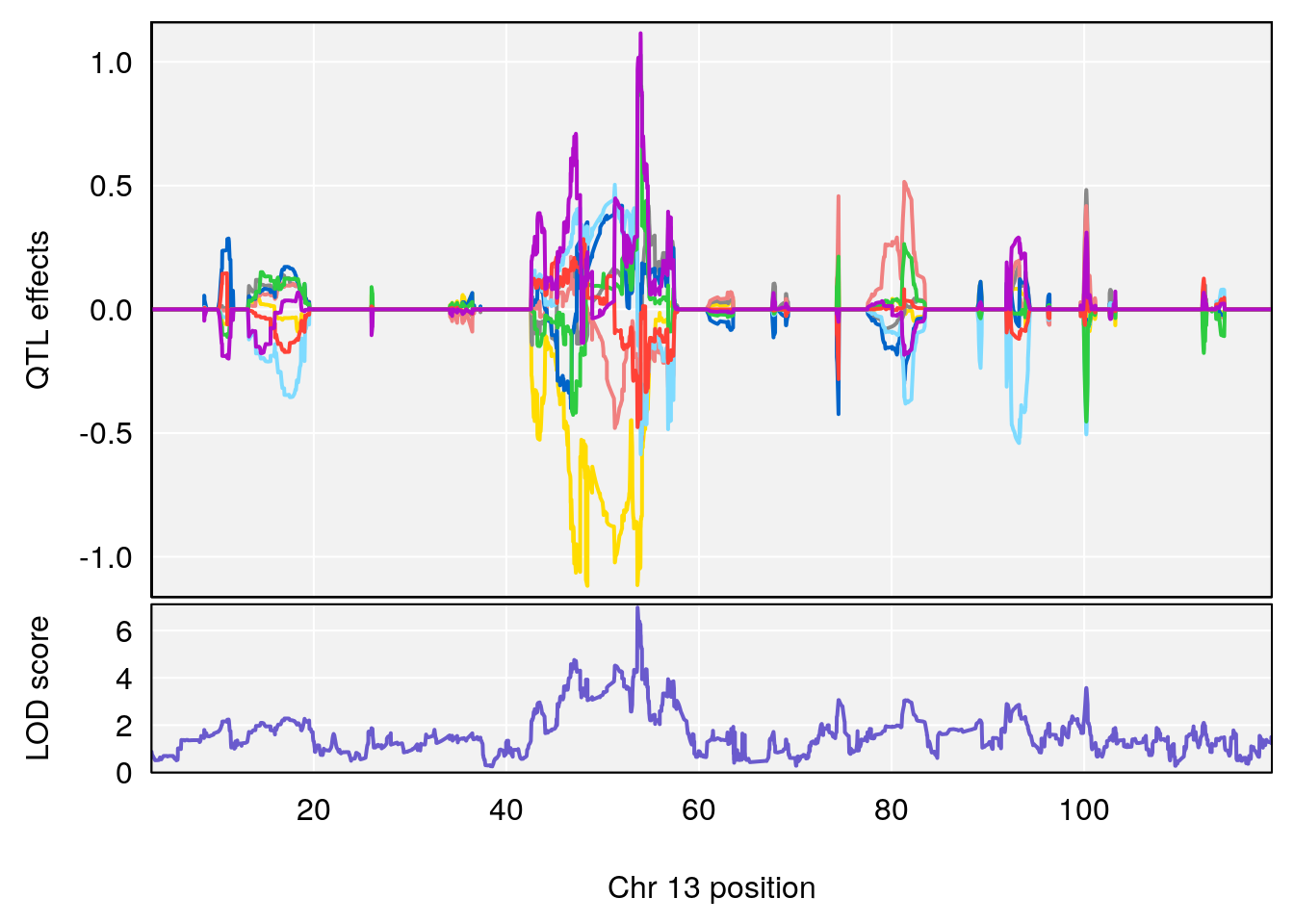
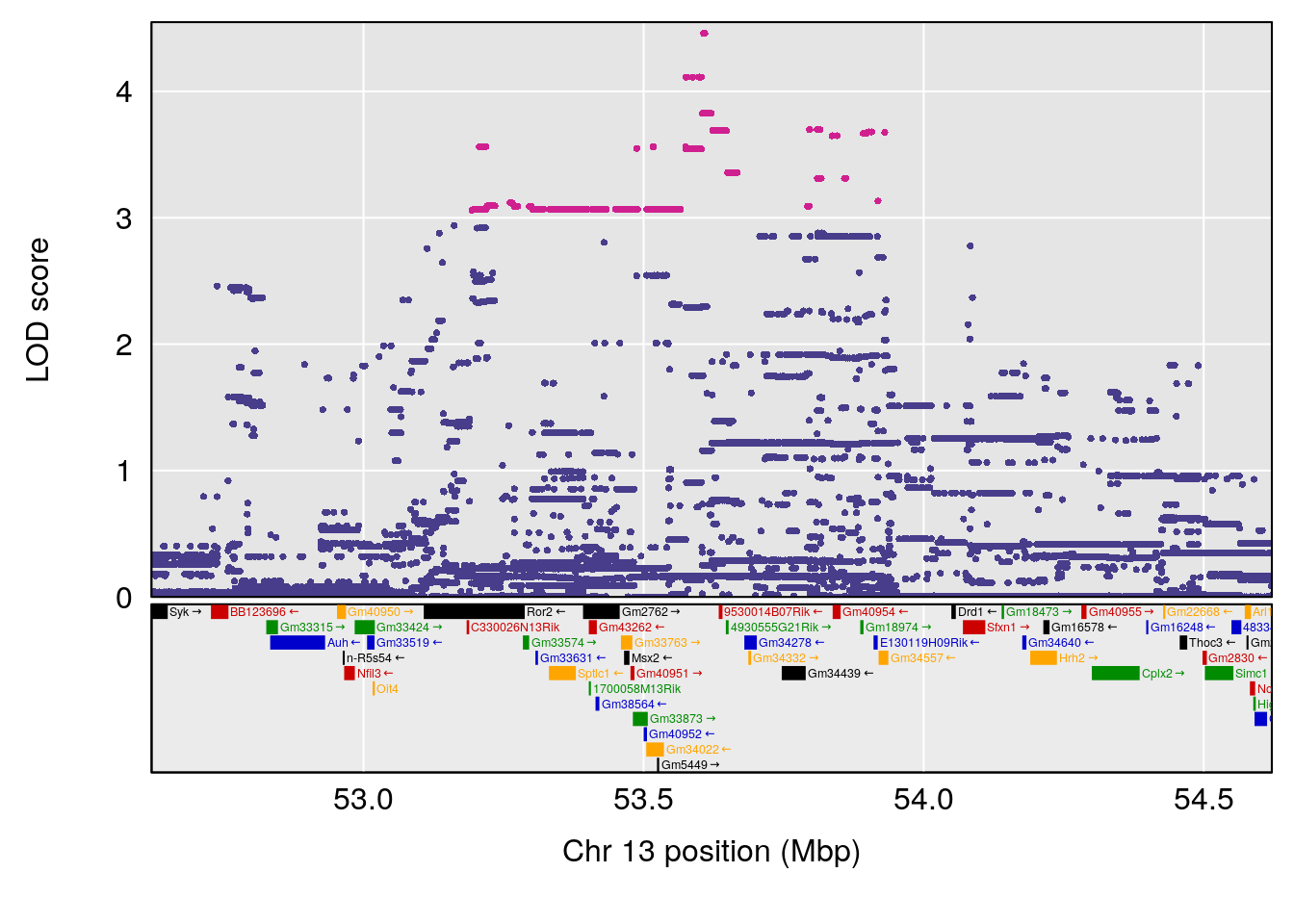
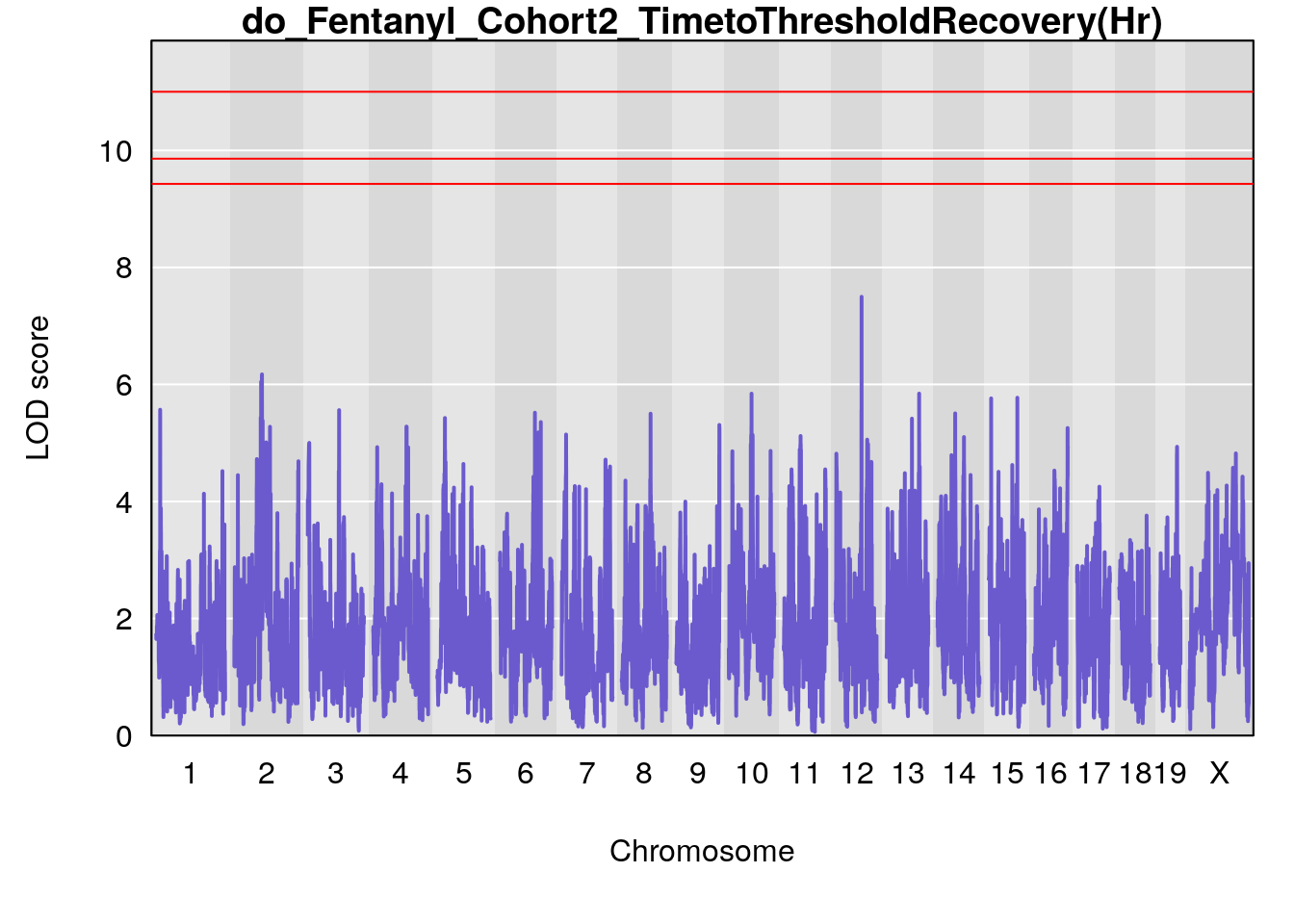
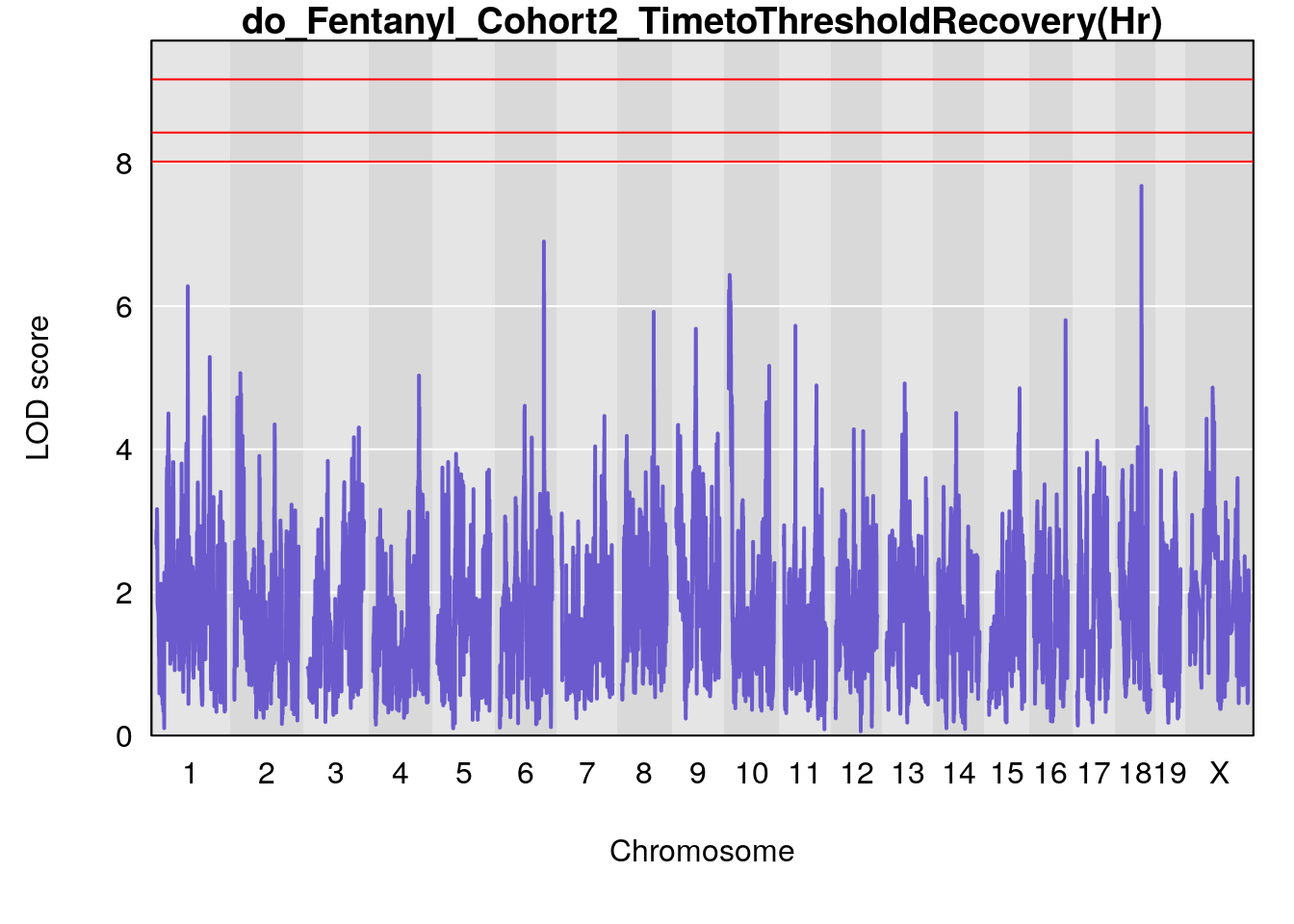
# [1] "TimetoThresholdRecovery(Hr)"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 91.31201 6.658992 91.21626 153.19382
# 2 1 pheno1 8 92.31720 6.295785 14.56005 109.54470
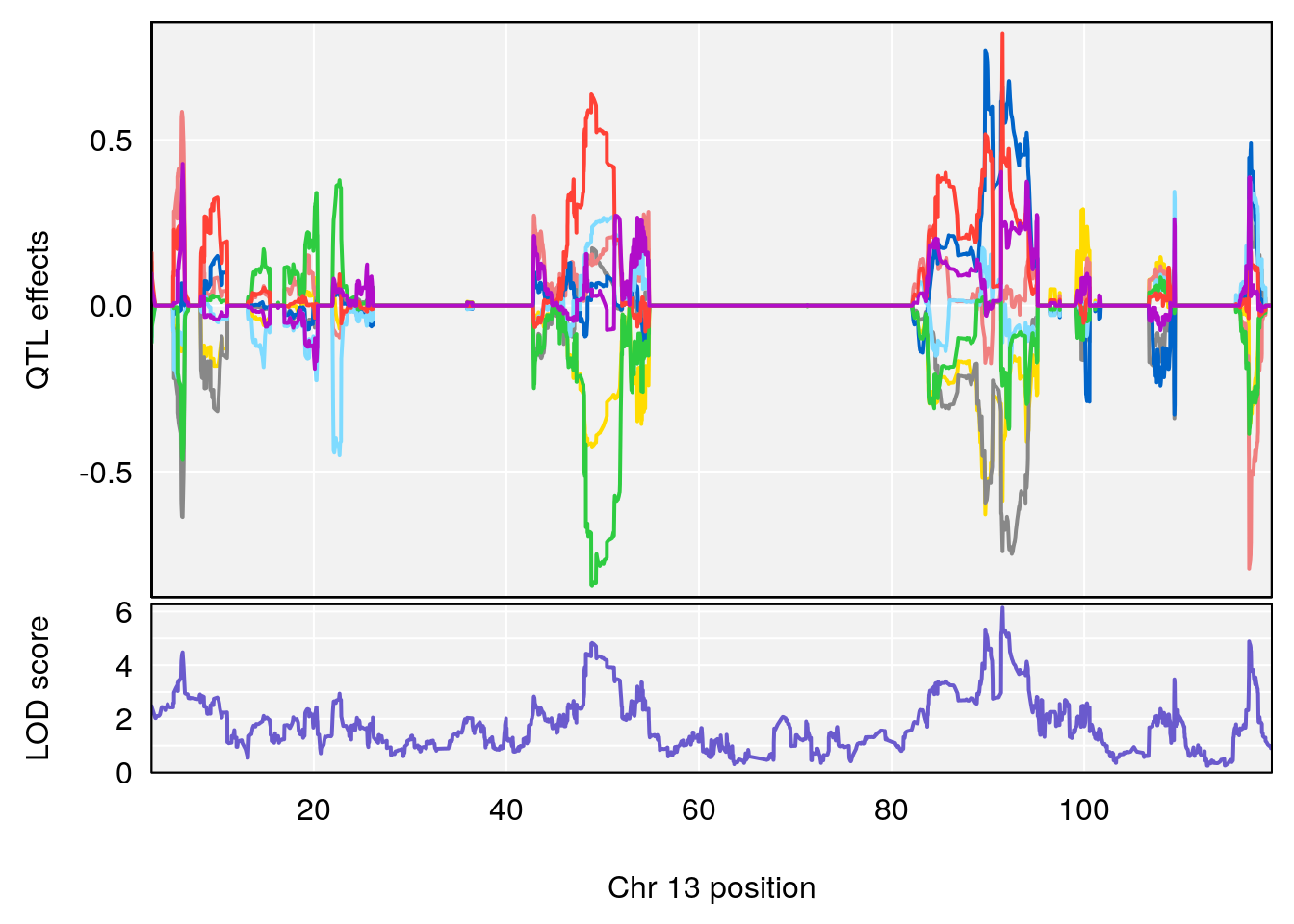
# 3 1 pheno1 13 53.62165 7.712830 53.57538 54.08557
# 4 1 pheno1 15 89.24166 6.222582 60.56487 90.52322
# [1] 1
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
# [1] 2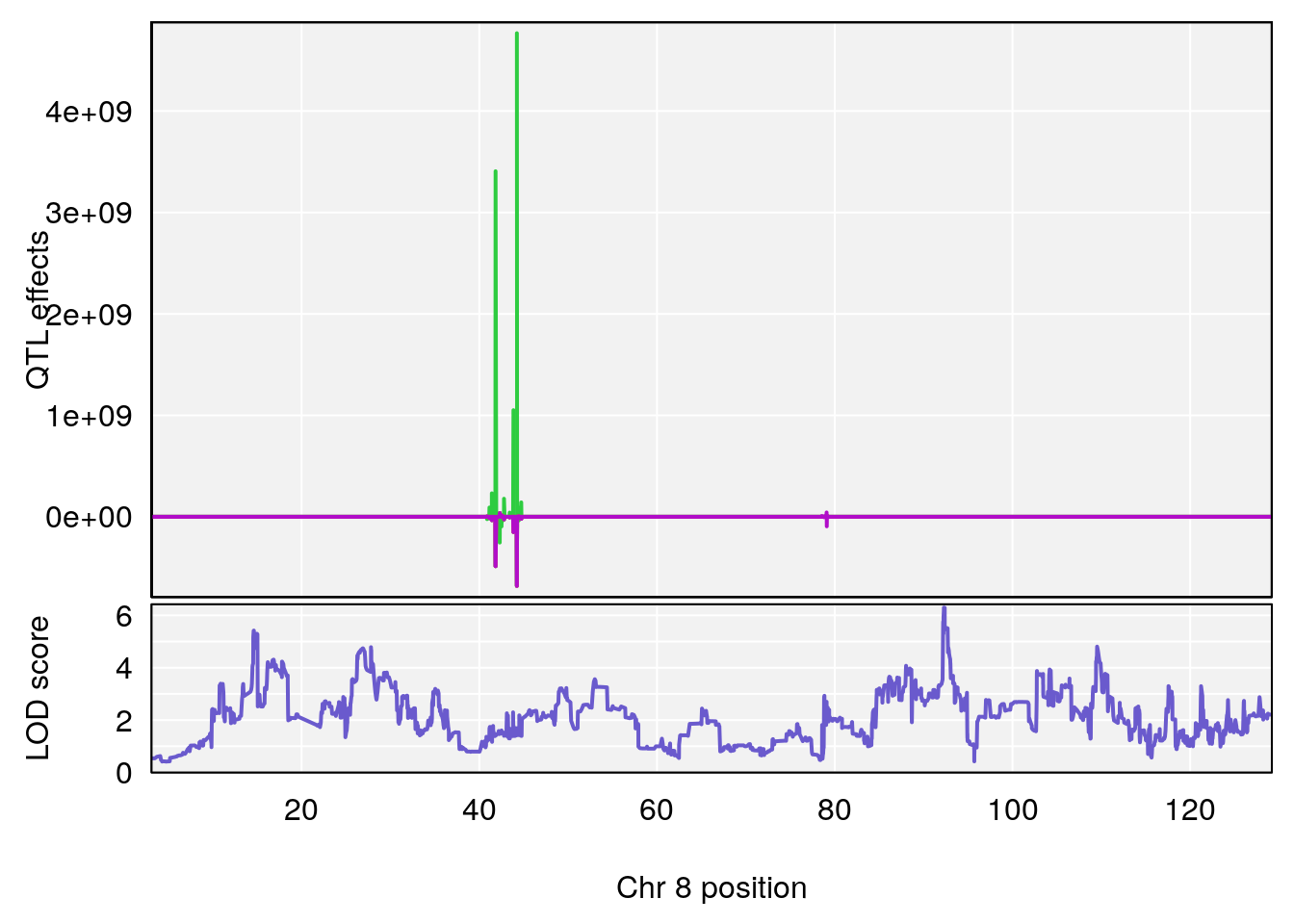
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
# [1] 3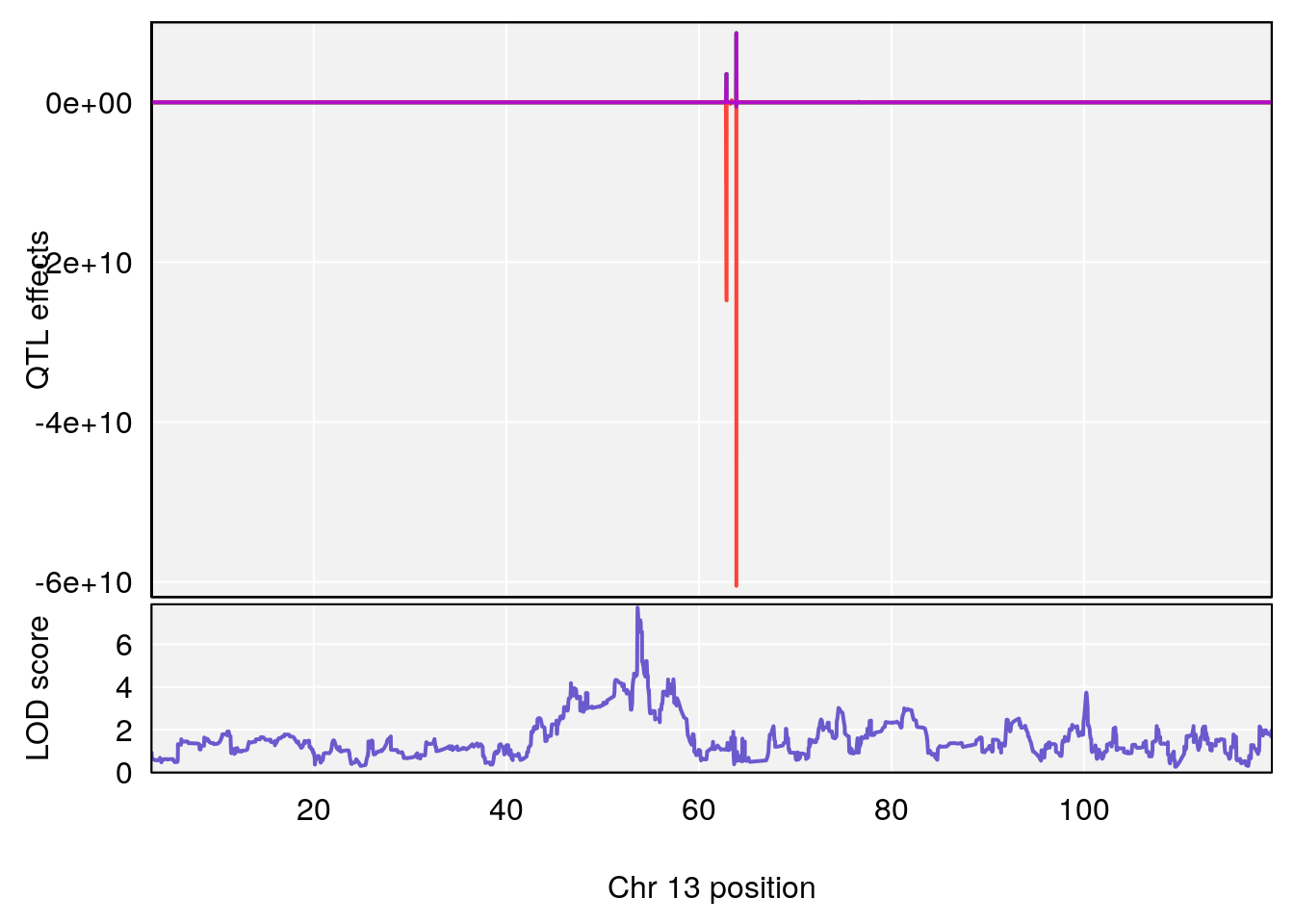
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
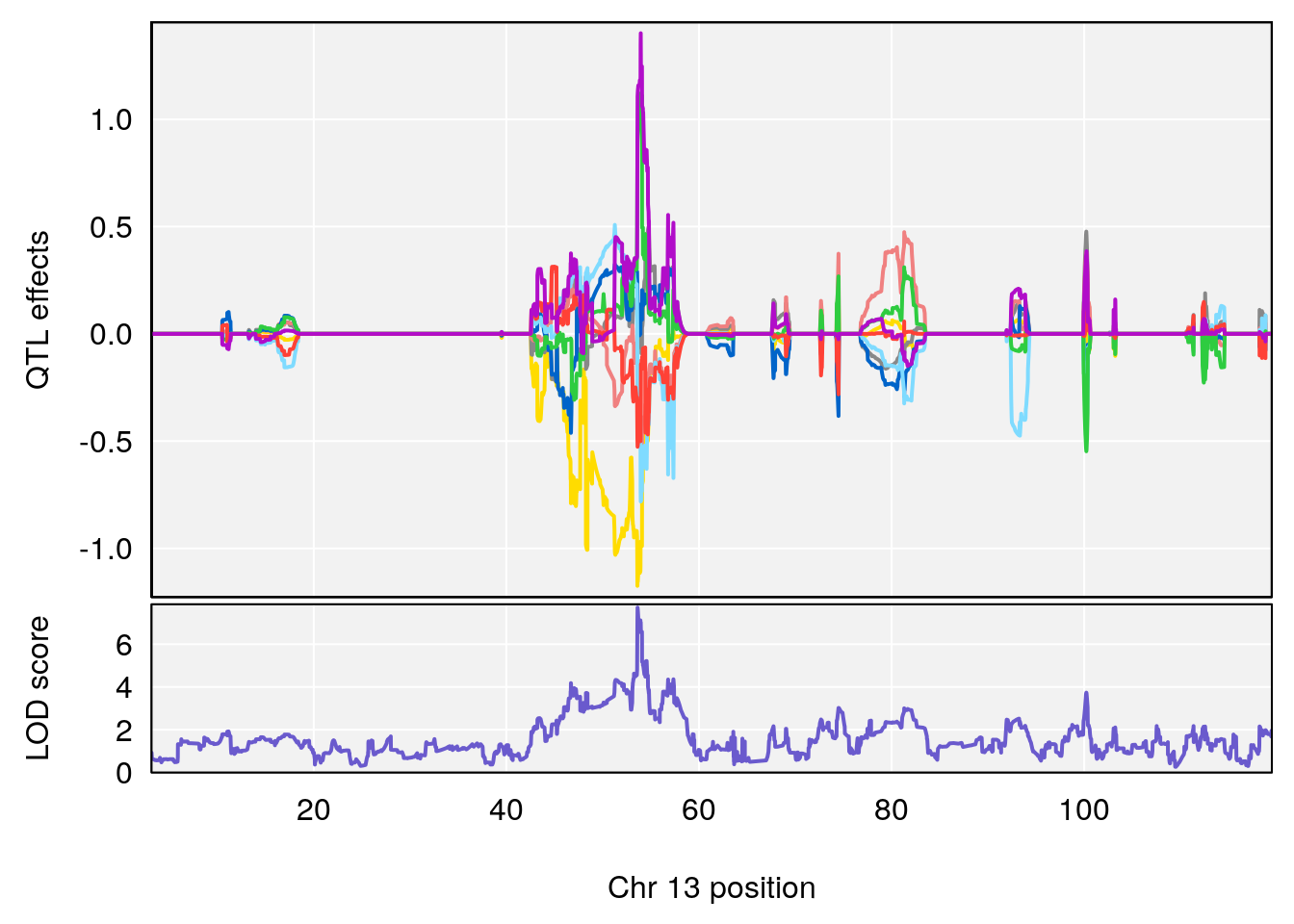
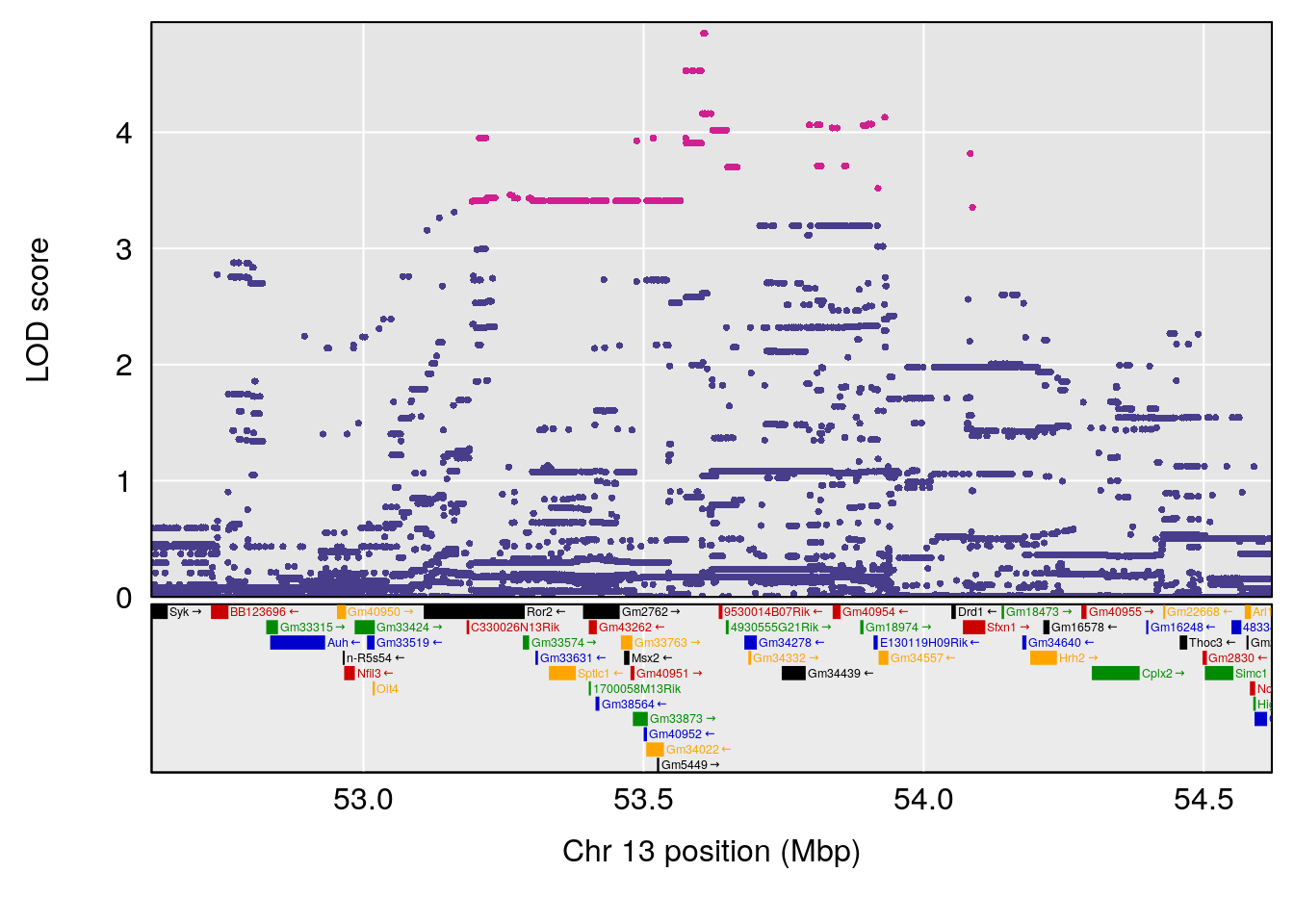
# [1] 4
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
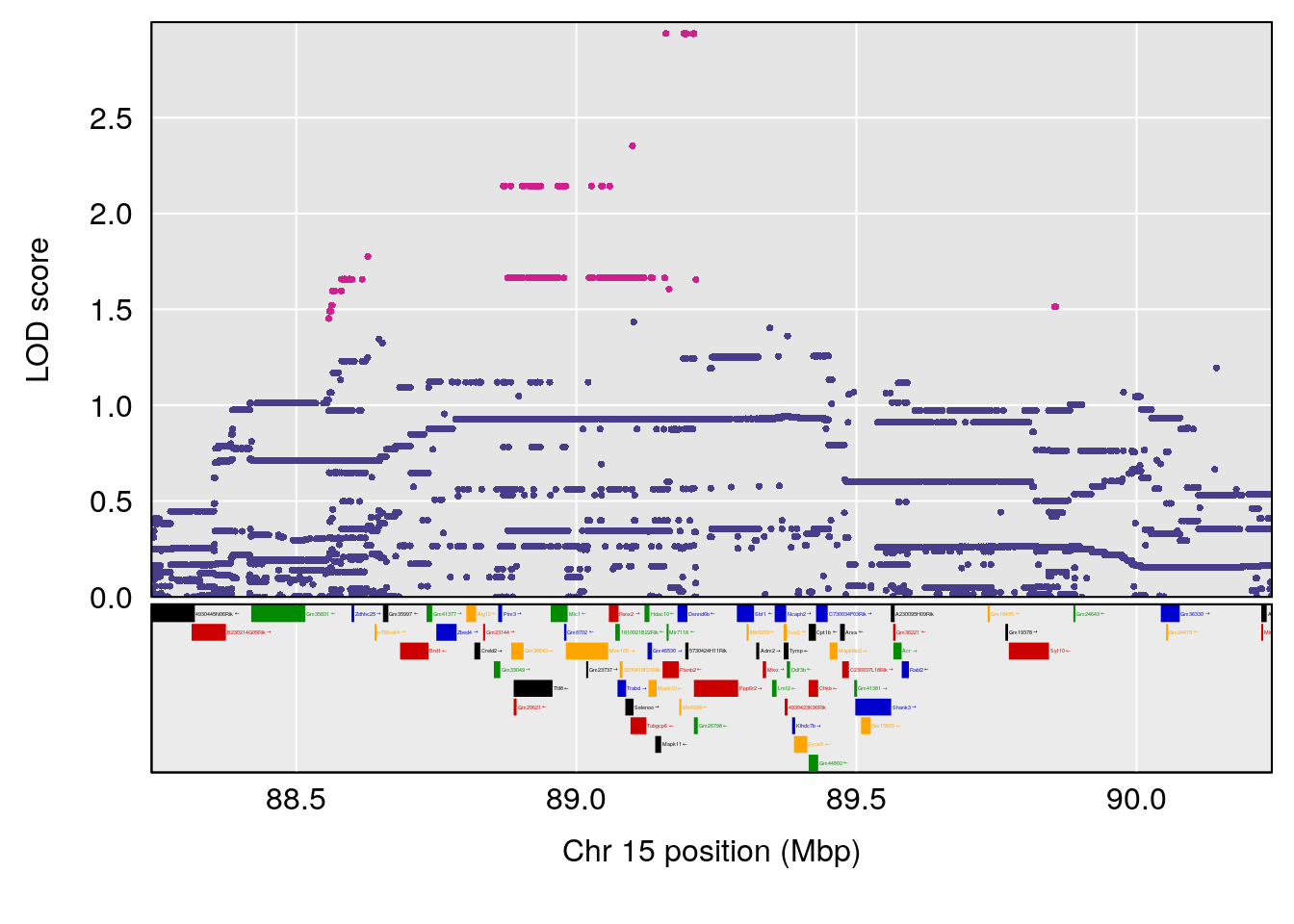
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
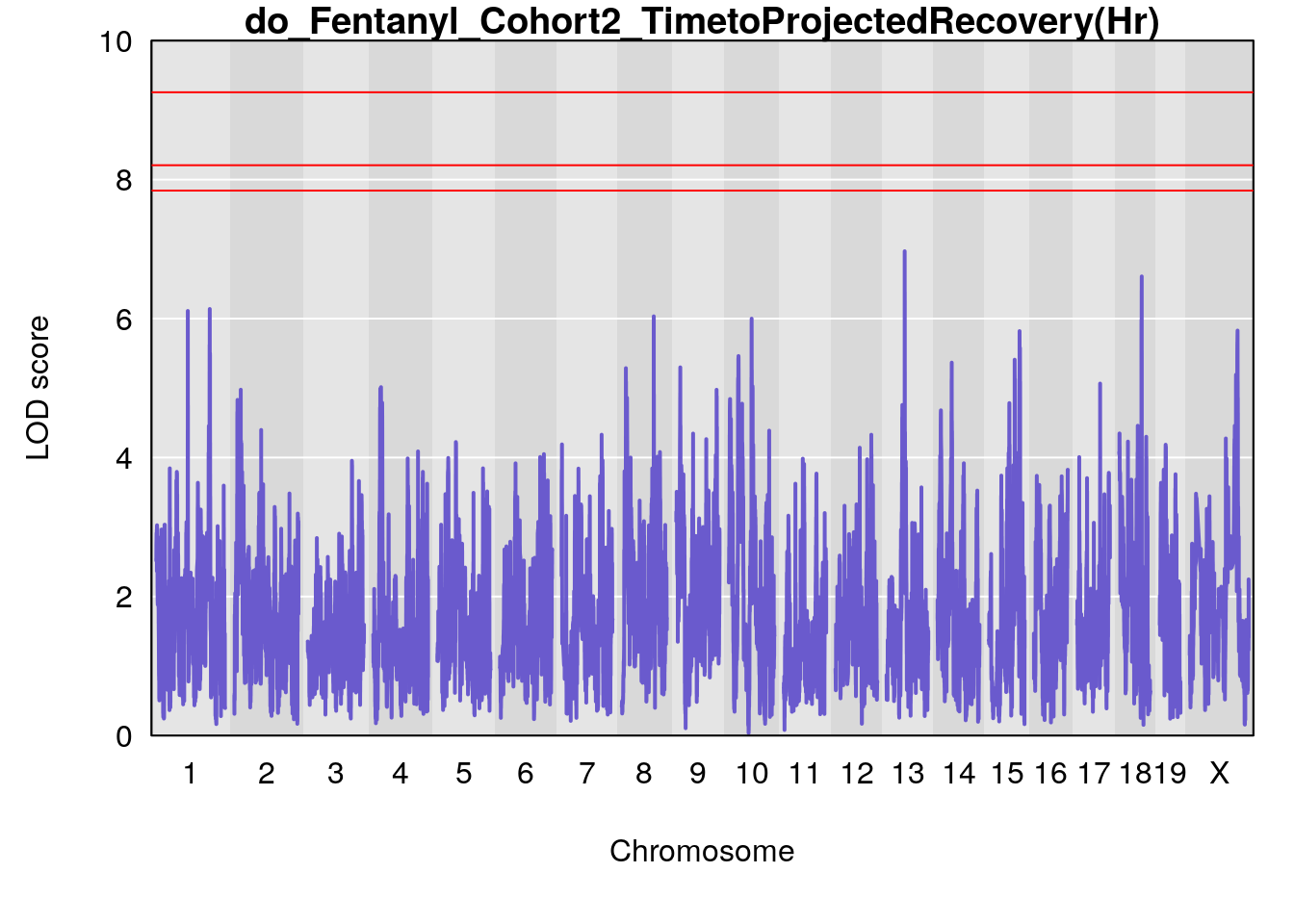
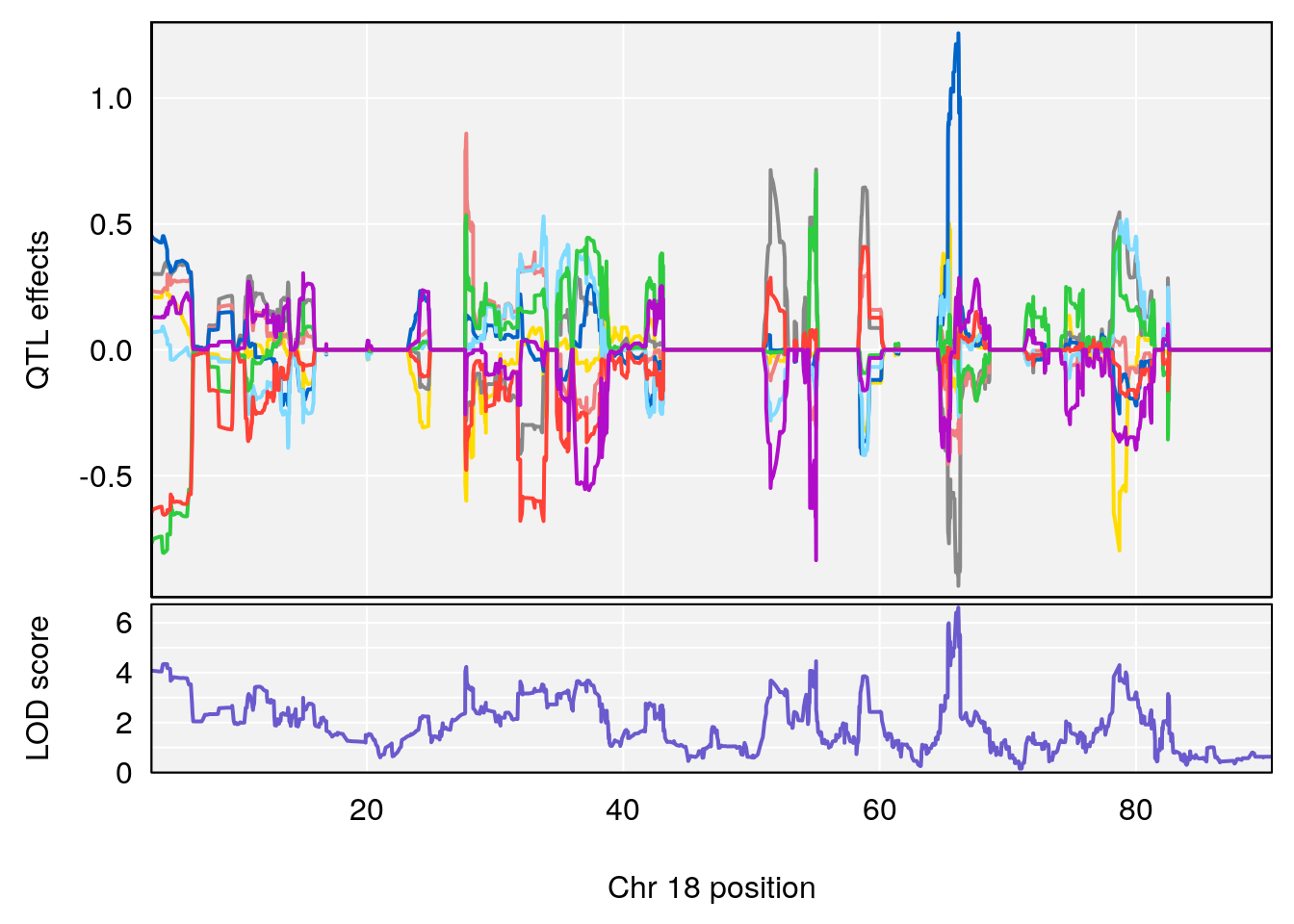
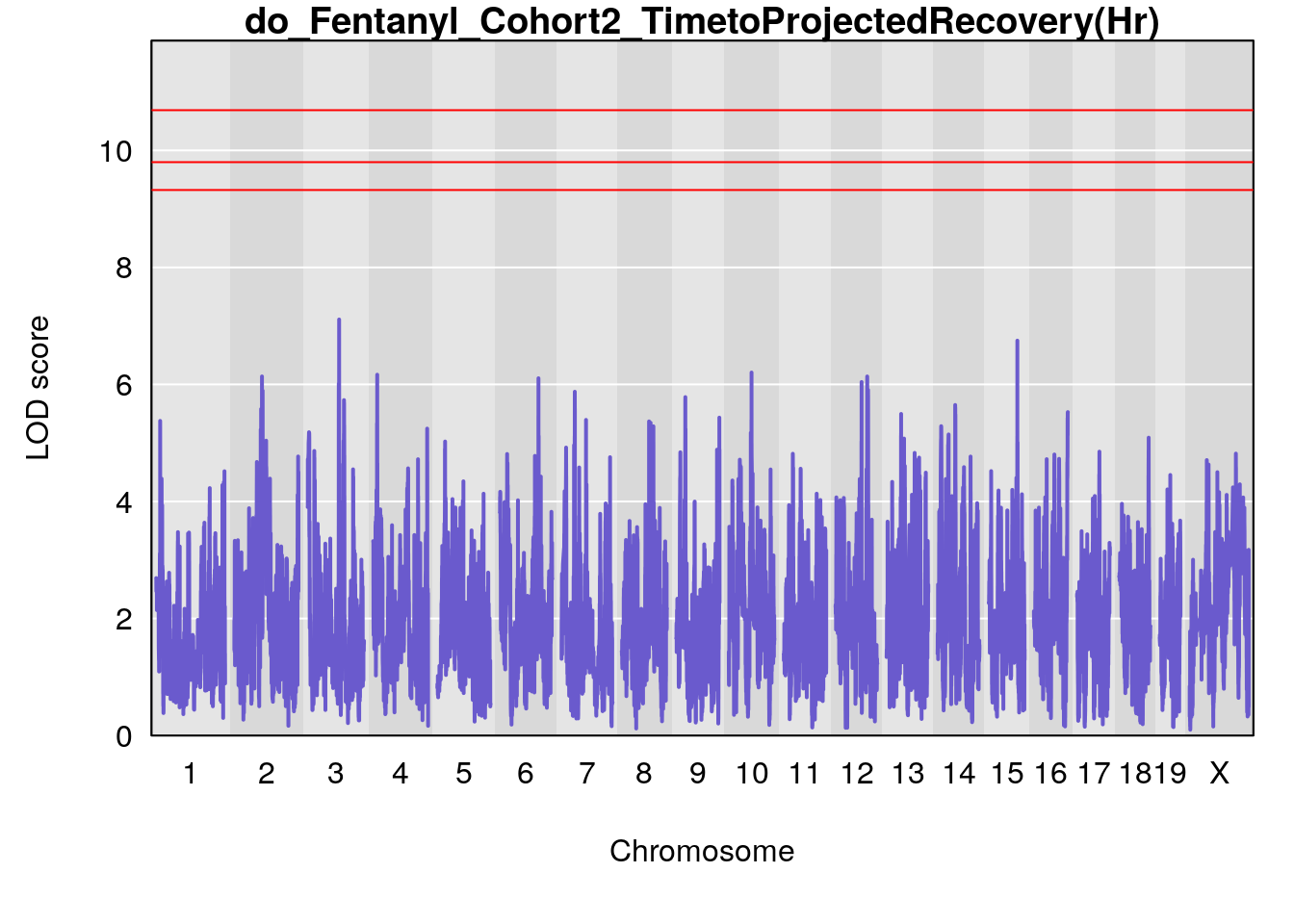
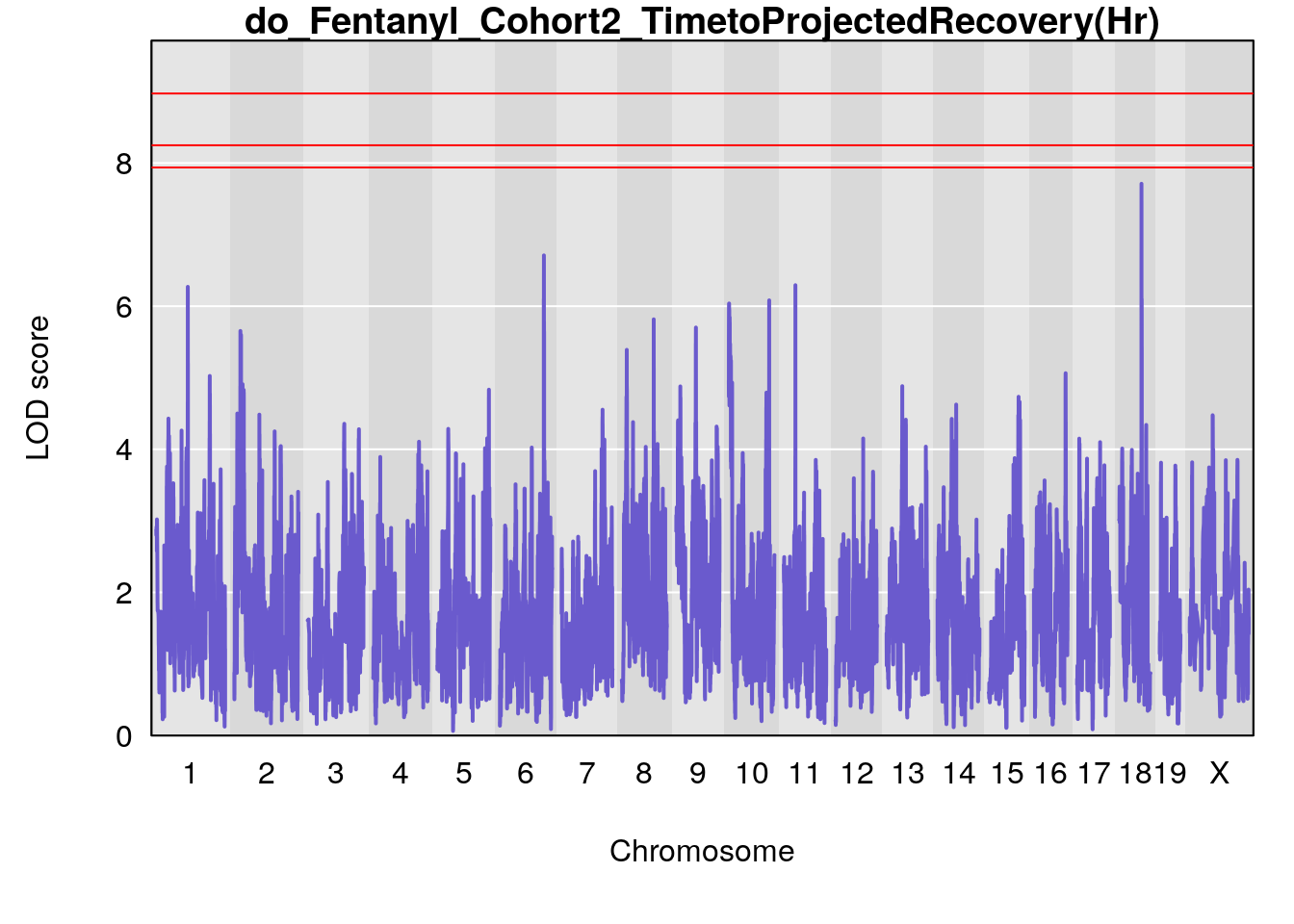
# [1] "TimetoProjectedRecovery(Hr)"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 152.43451 6.138085 91.21626 153.19382
# 2 1 pheno1 8 92.31720 6.032551 14.55032 92.73082
# 3 1 pheno1 13 53.62165 6.969155 53.57538 53.97165
# 4 1 pheno1 18 66.14084 6.606701 65.31382 66.30142
# [1] 1
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
# [1] 2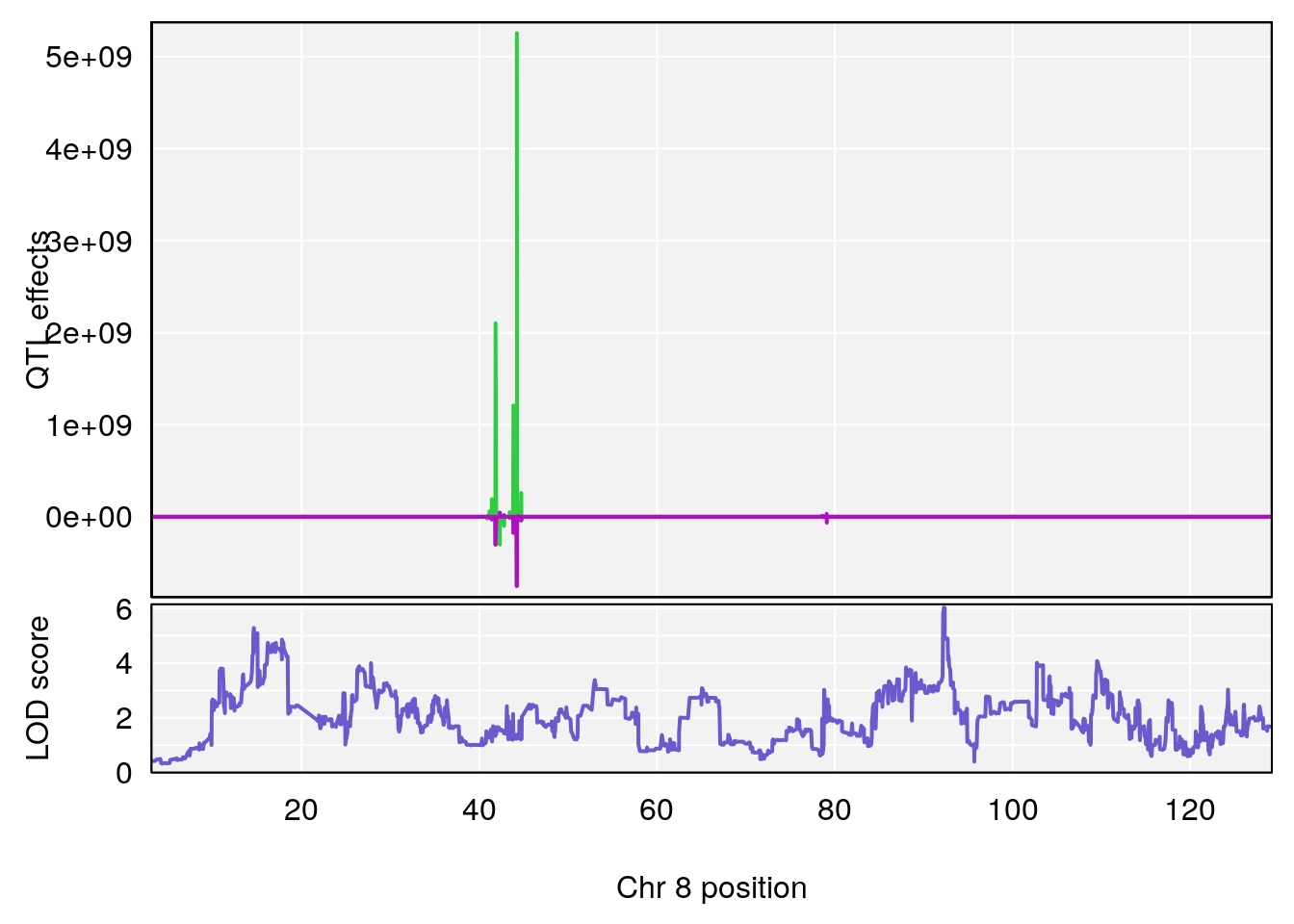
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
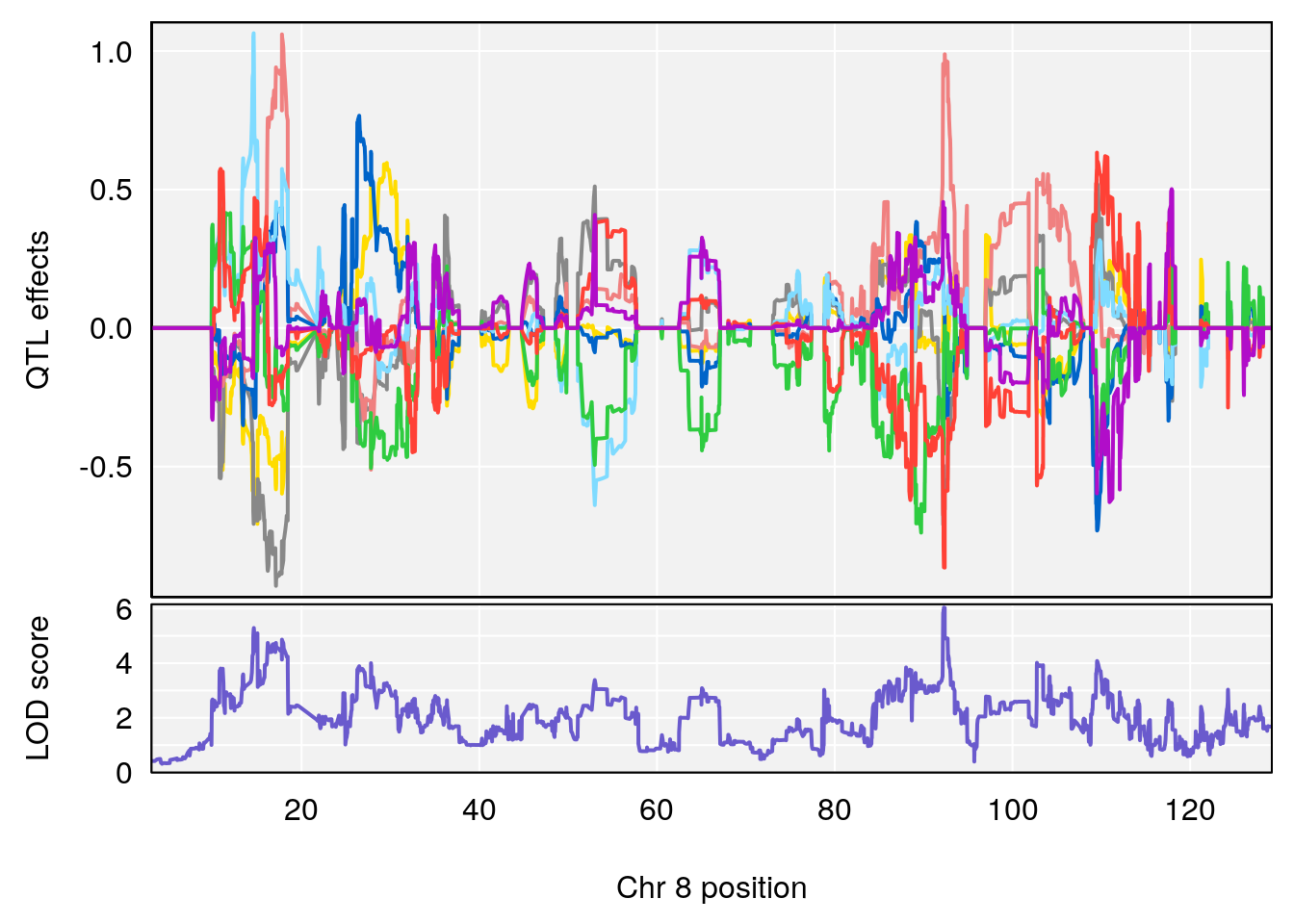
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
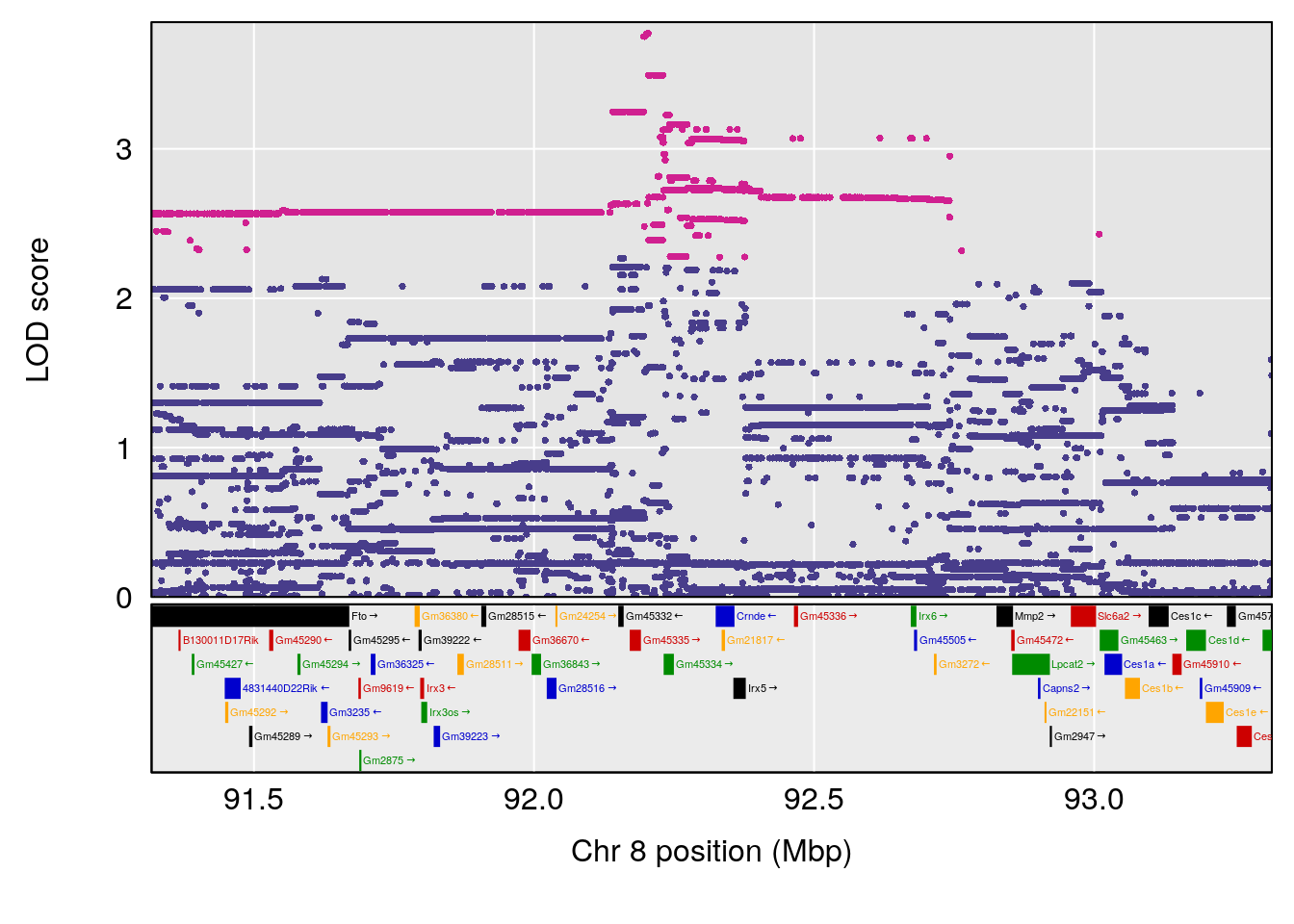
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
# [1] 3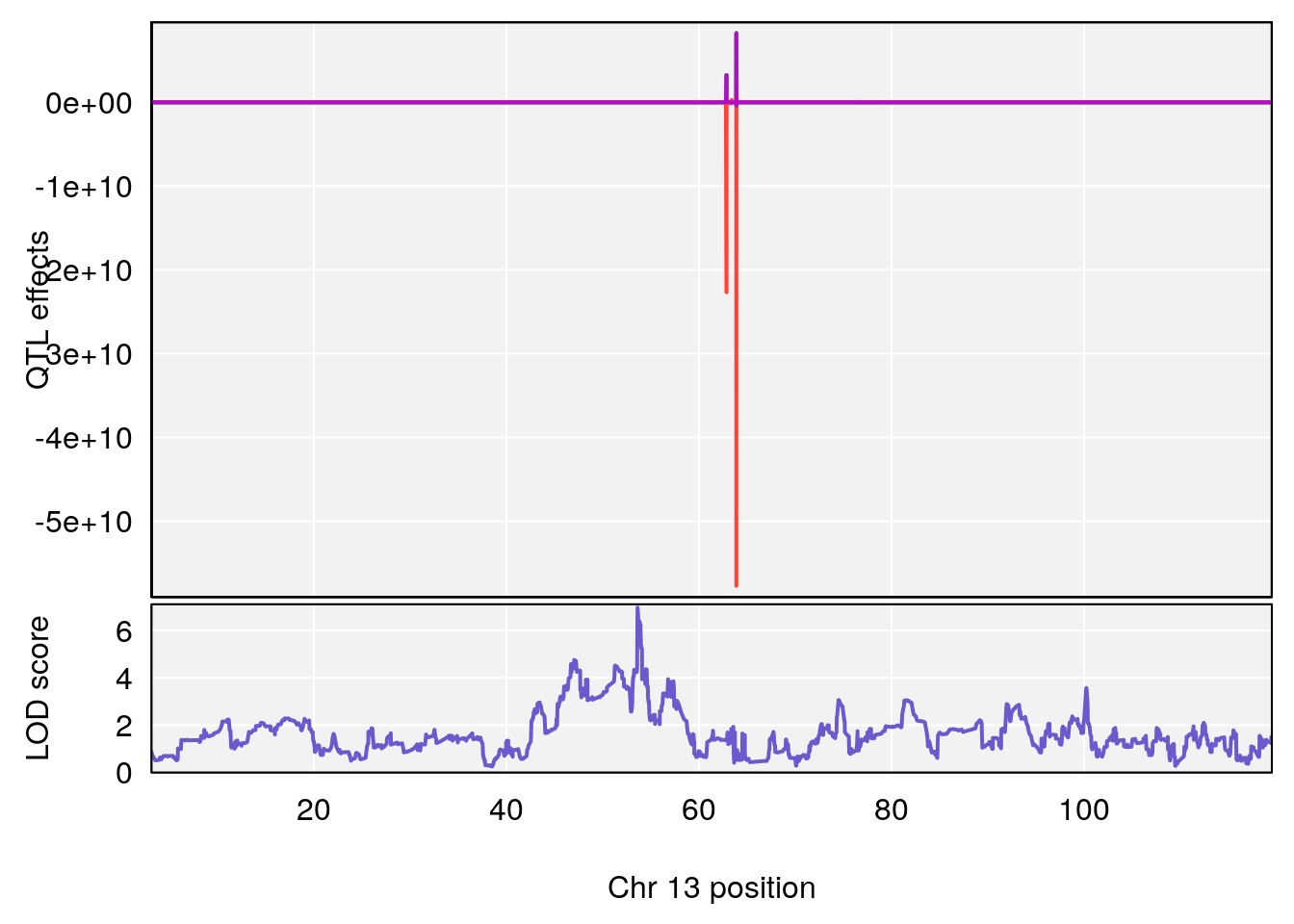
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
# [1] 4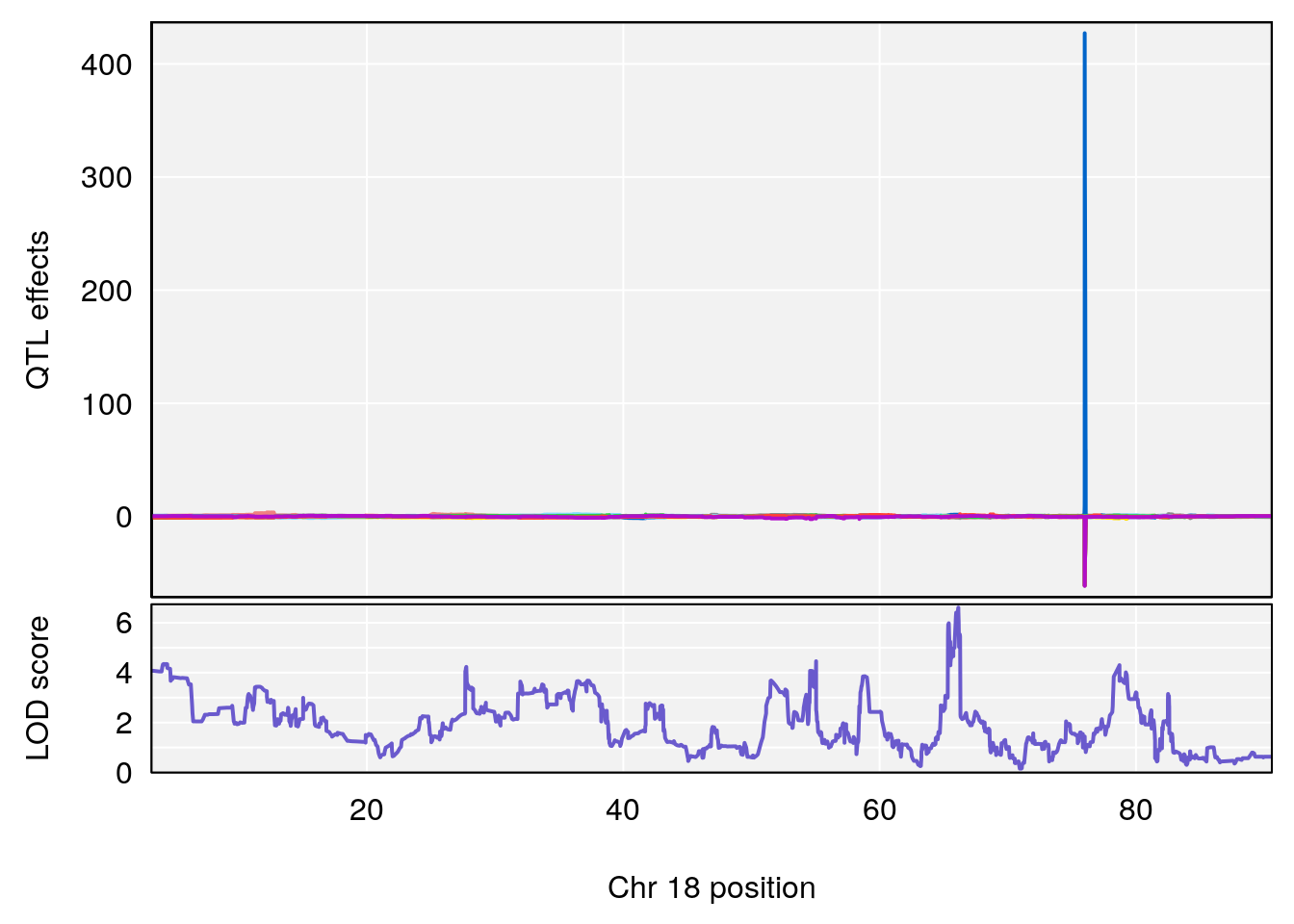
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
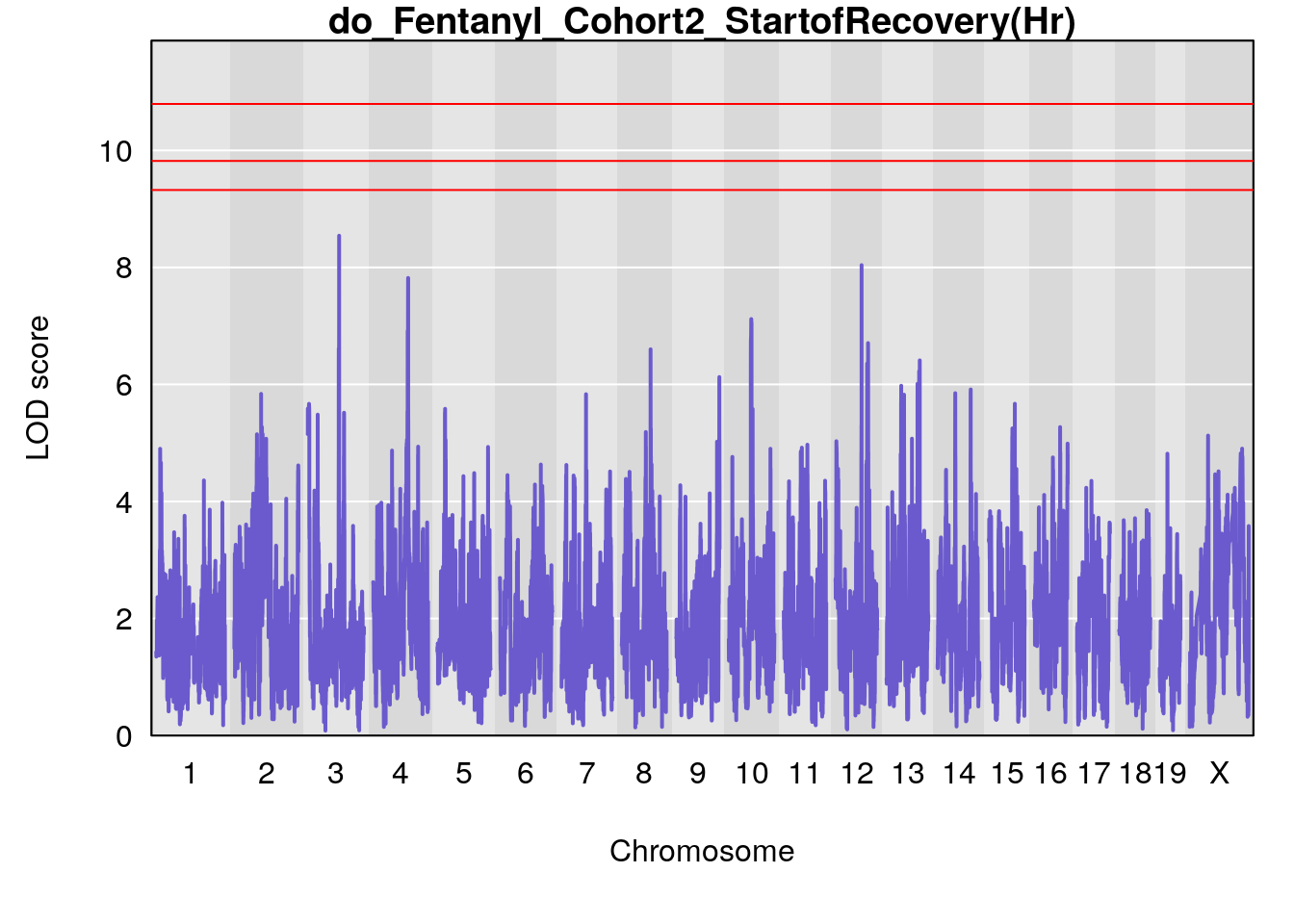
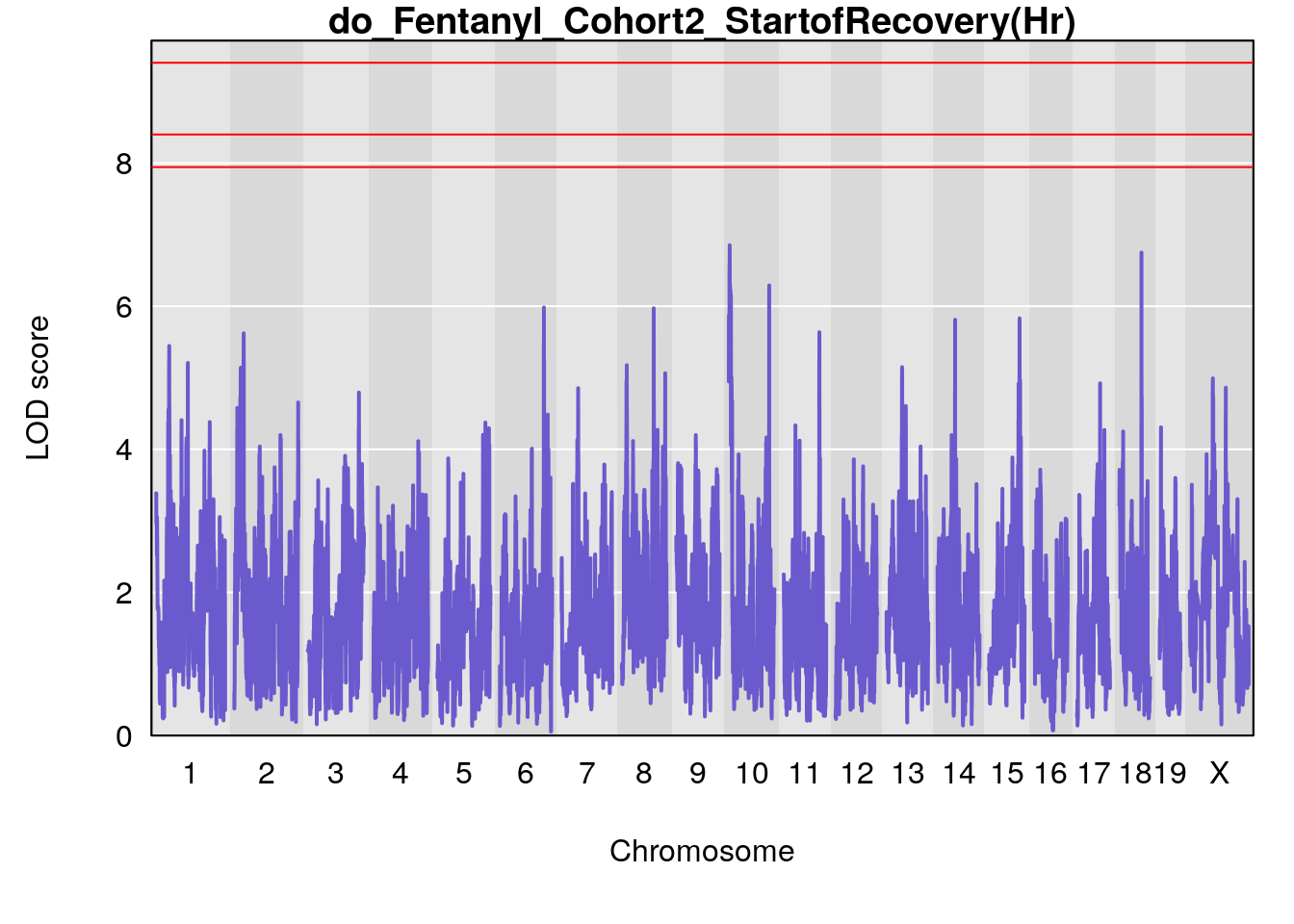
# [1] "StartofRecovery(Hr)"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 13 53.62165 6.12149 53.57538 54.62068
# [1] 1
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
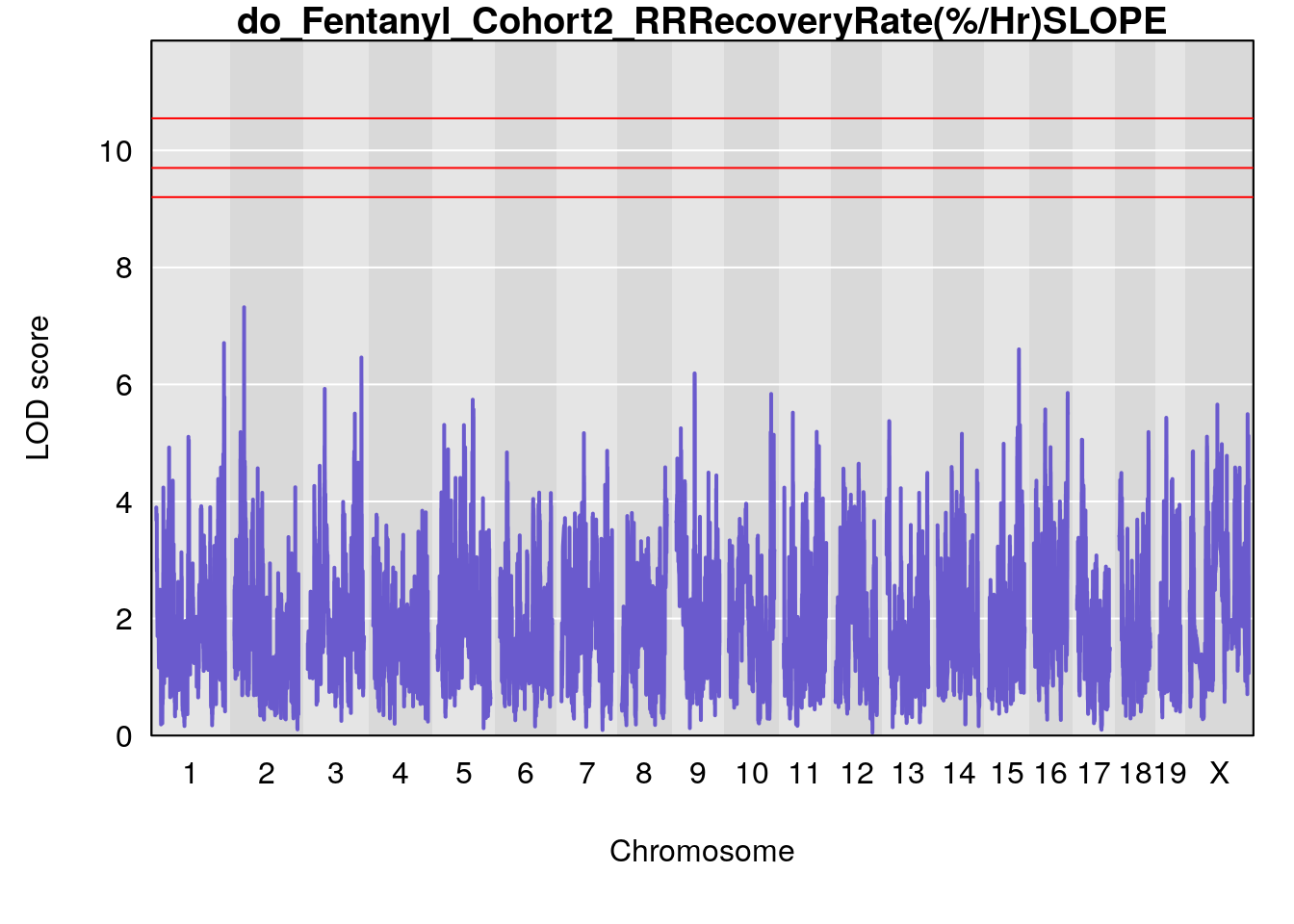
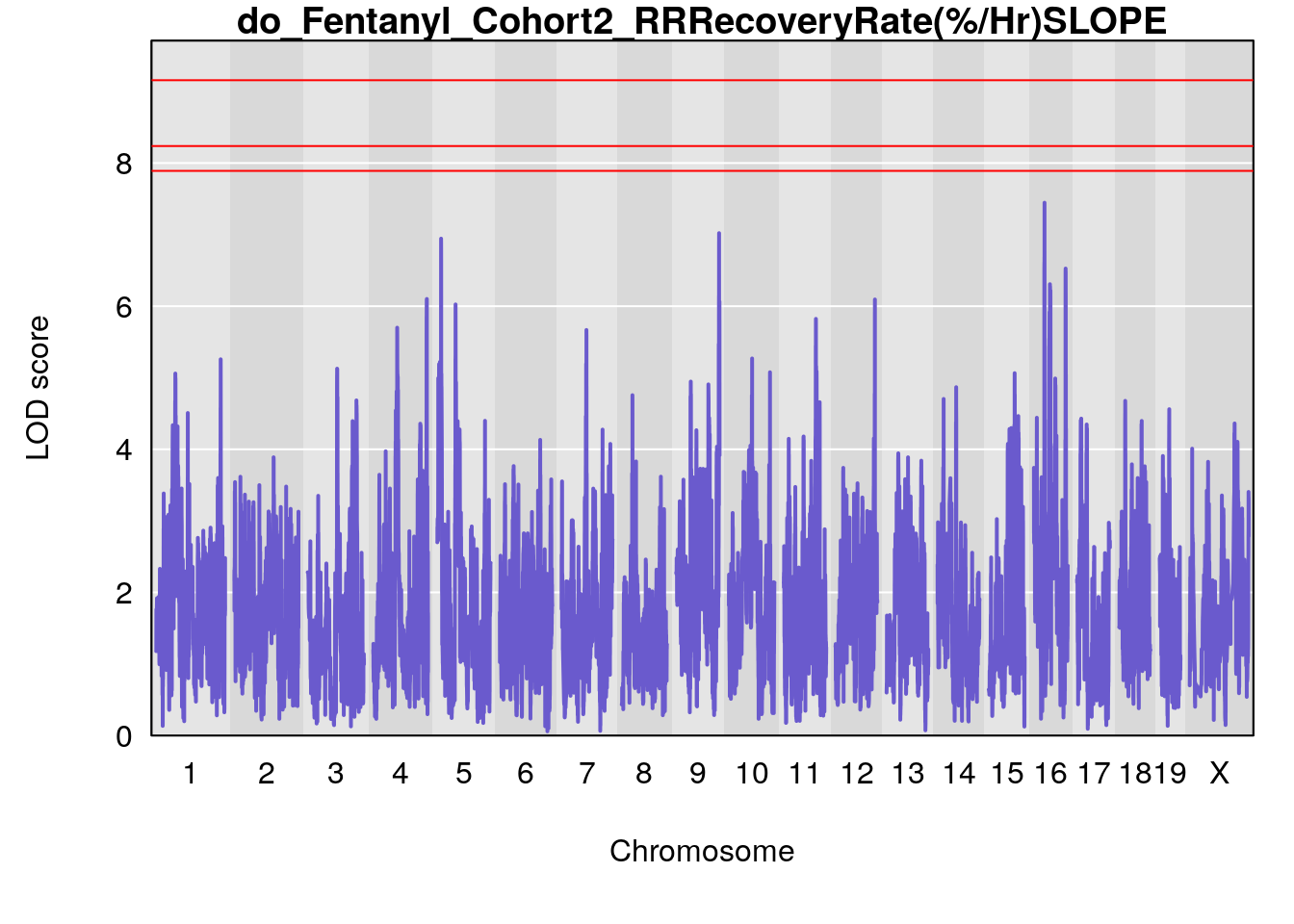
# [1] "RRRecoveryRate(%/Hr)SLOPE"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 6 115.70560 6.308902 115.43247 116.28985
# 2 1 pheno1 10 68.11376 6.195341 66.55149 68.20195
# 3 1 pheno1 15 90.42378 6.347611 88.92565 90.85895
# 4 1 pheno1 18 65.52797 6.640763 65.36123 65.58896
# [1] 1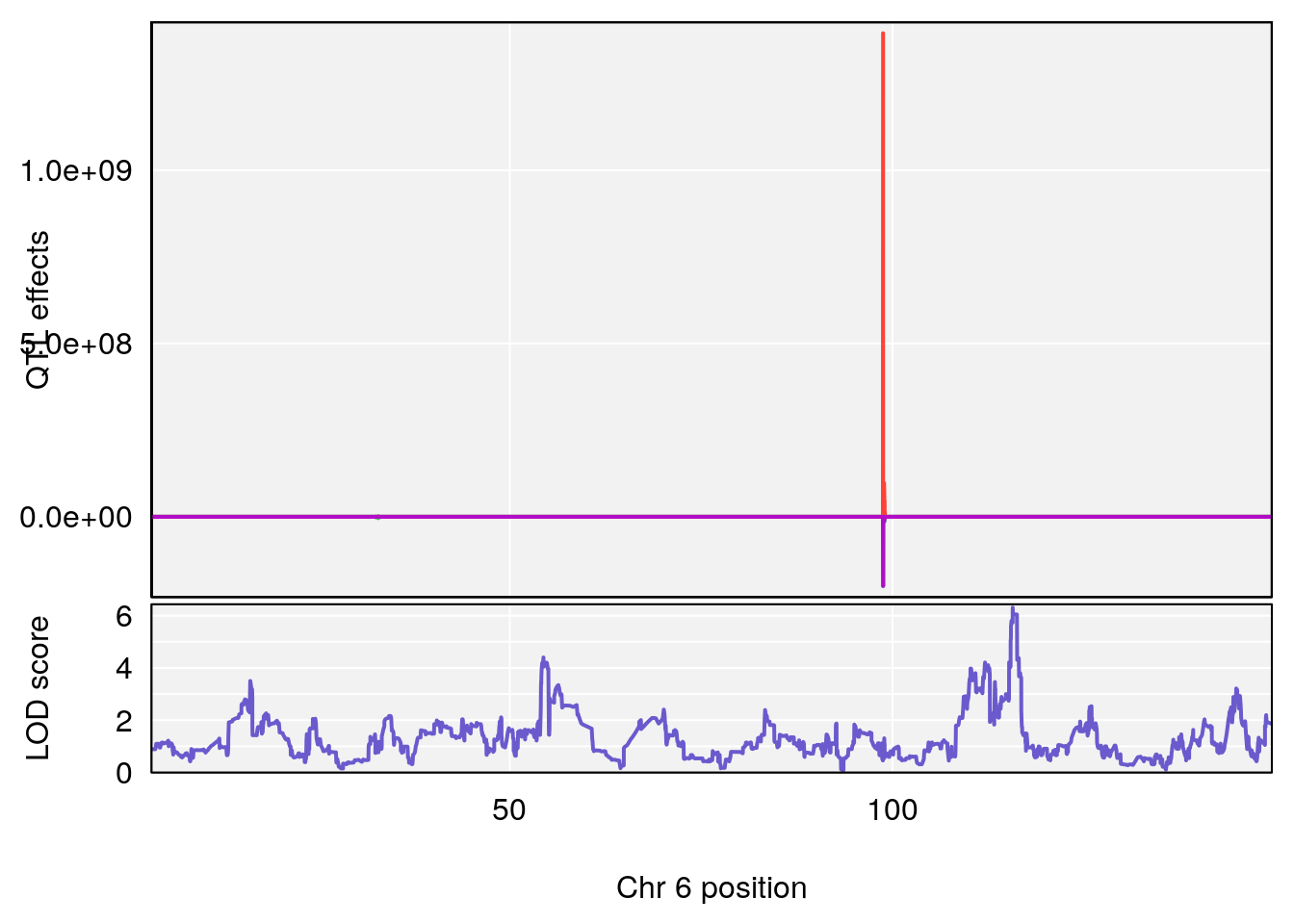
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
# [1] 2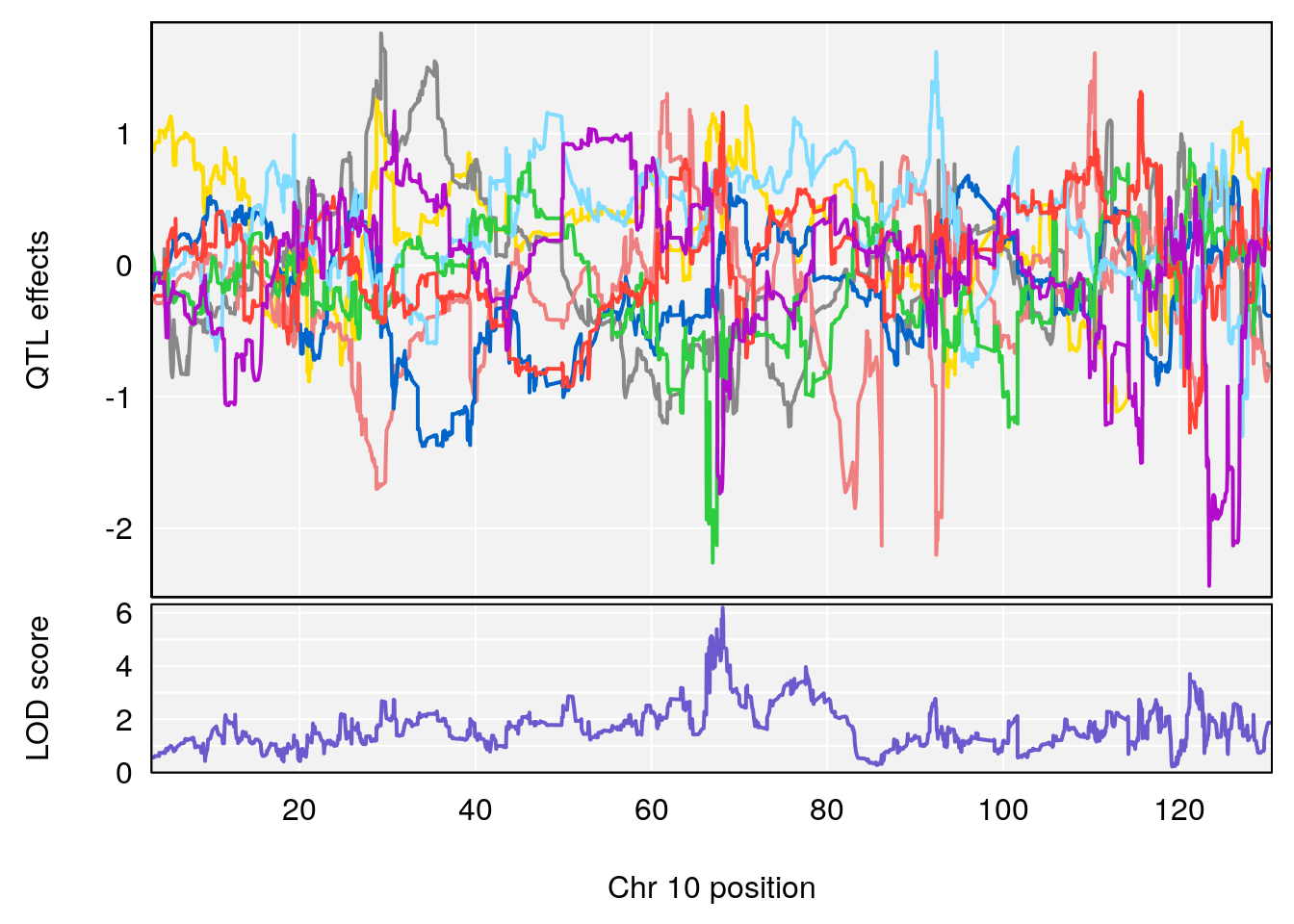
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
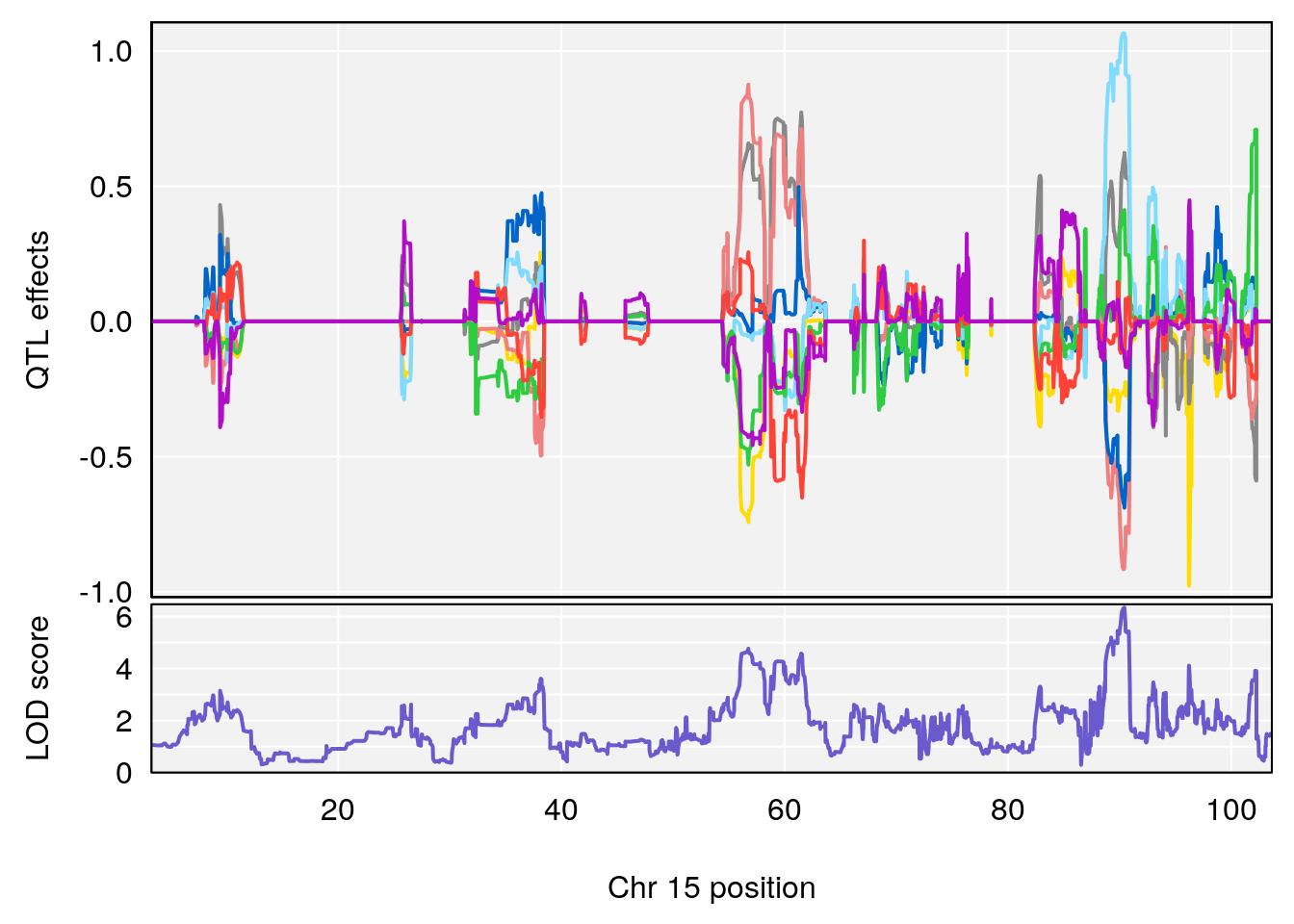
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
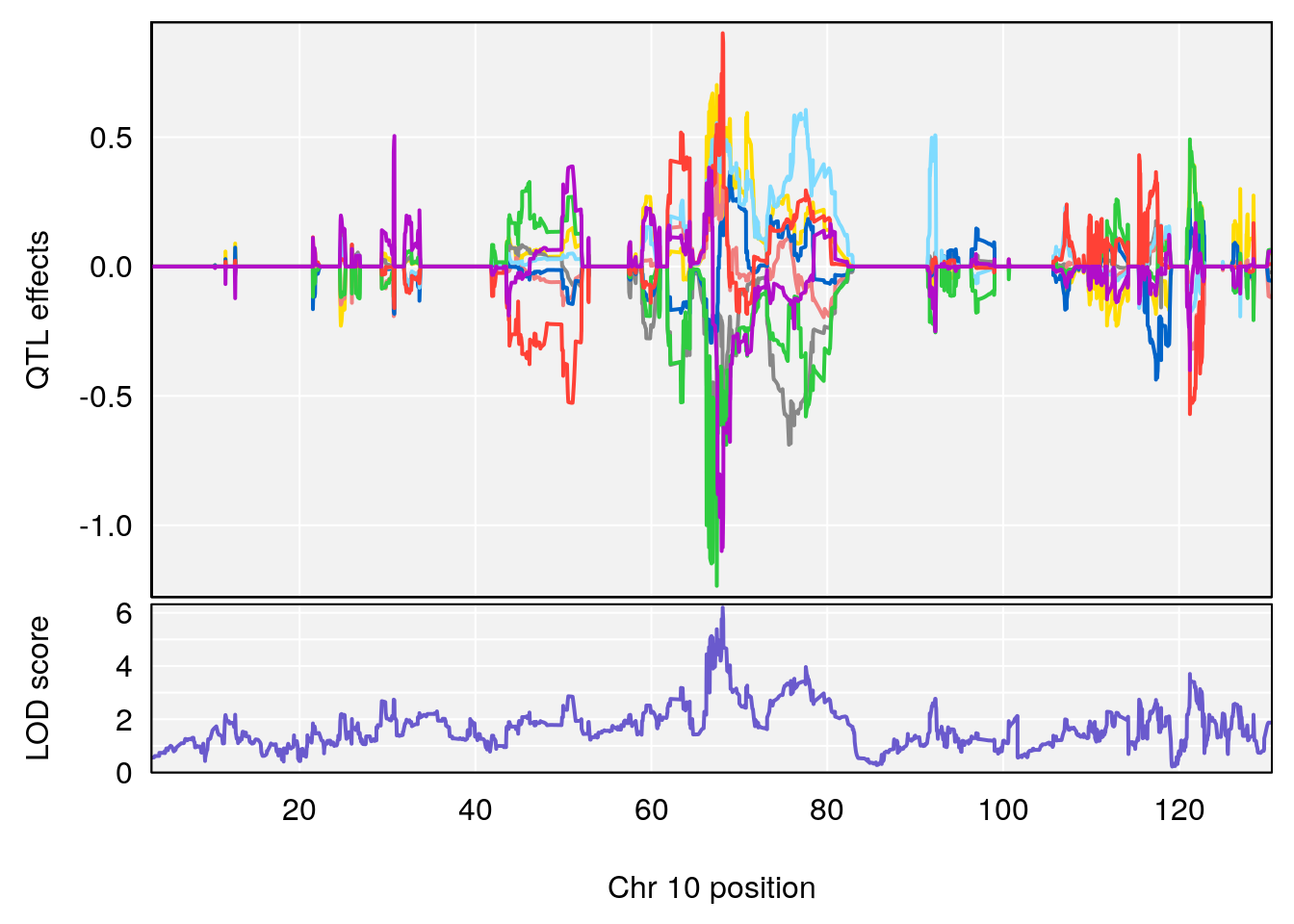
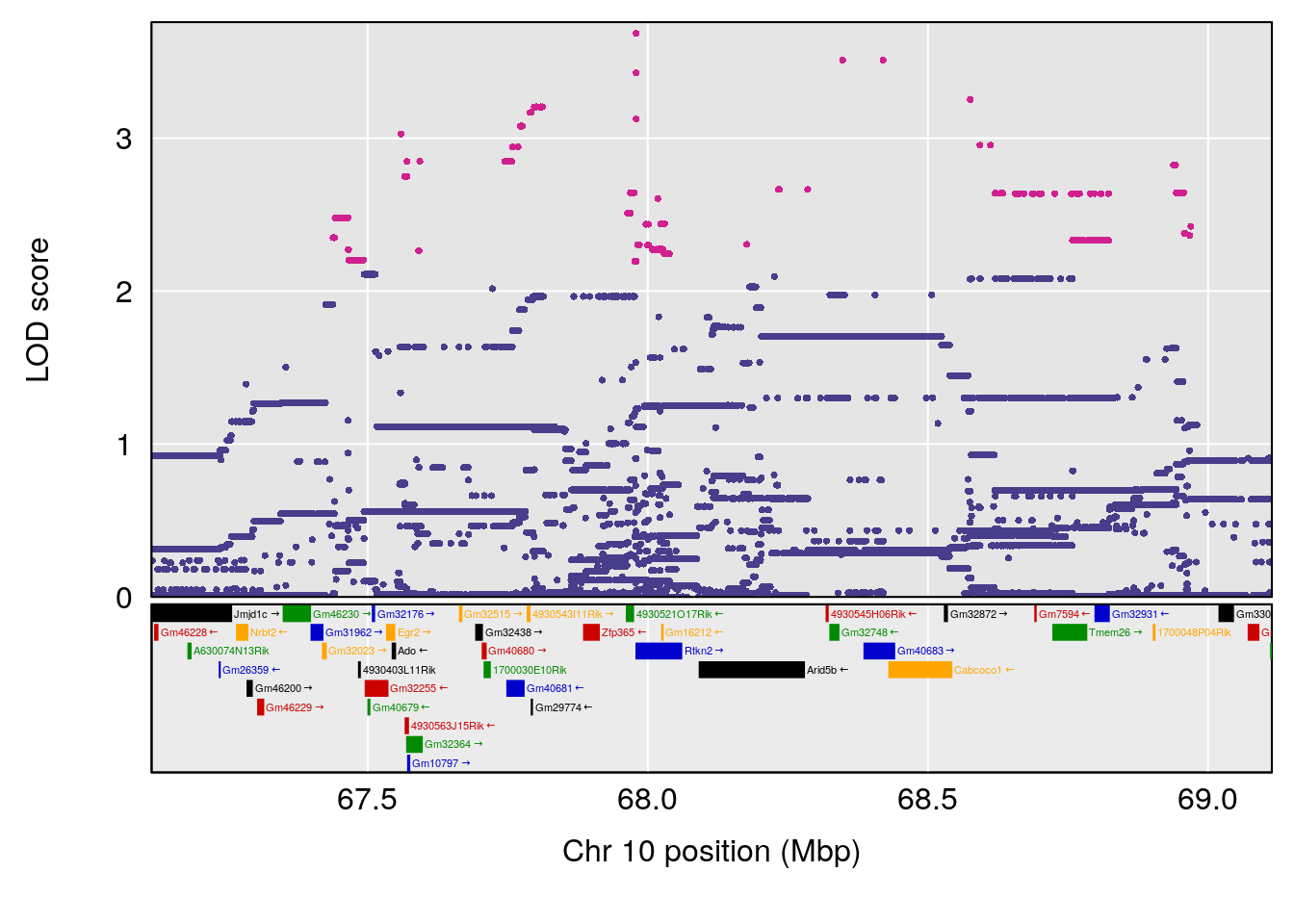
# [1] 3
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
# [1] 4
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
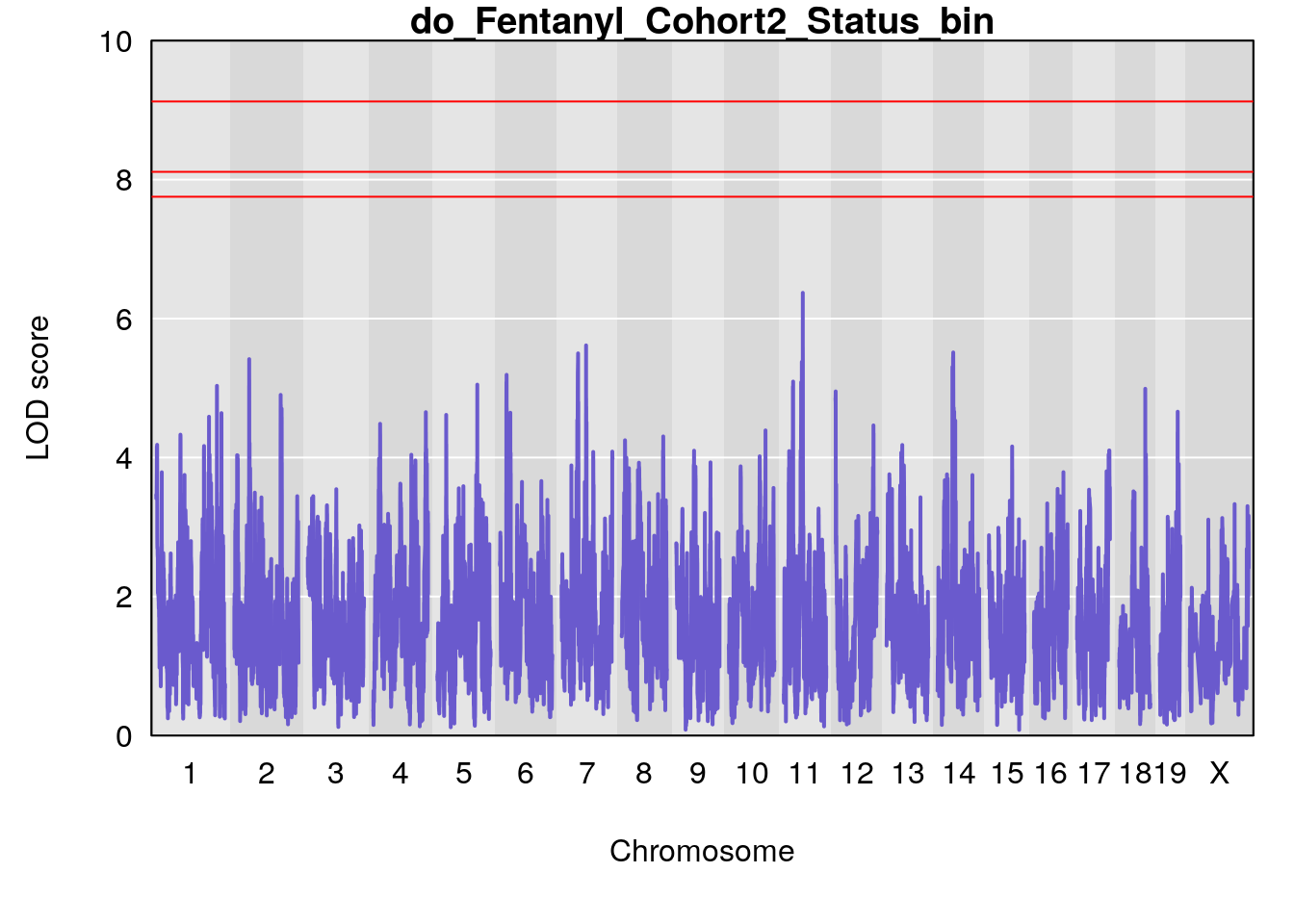
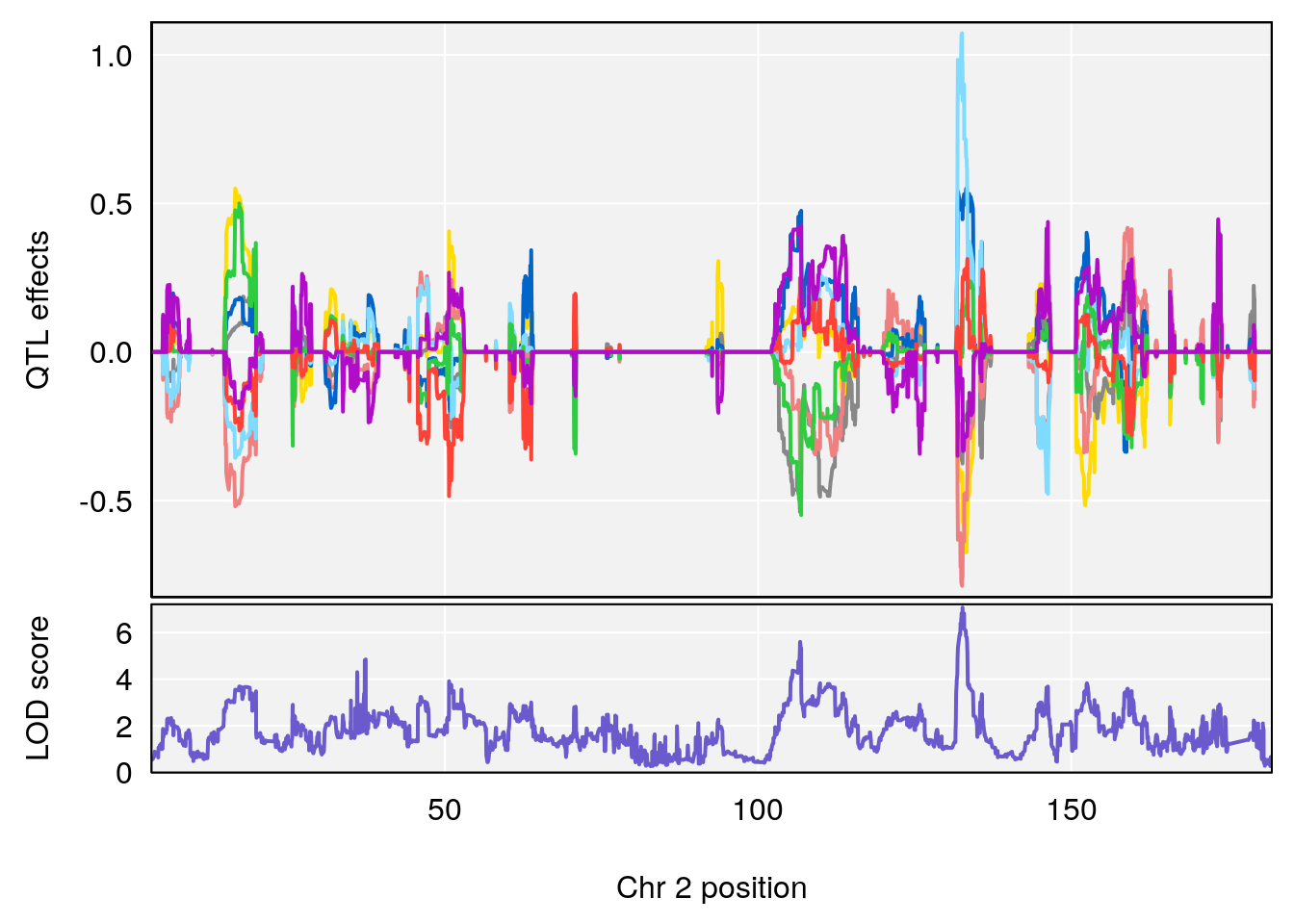
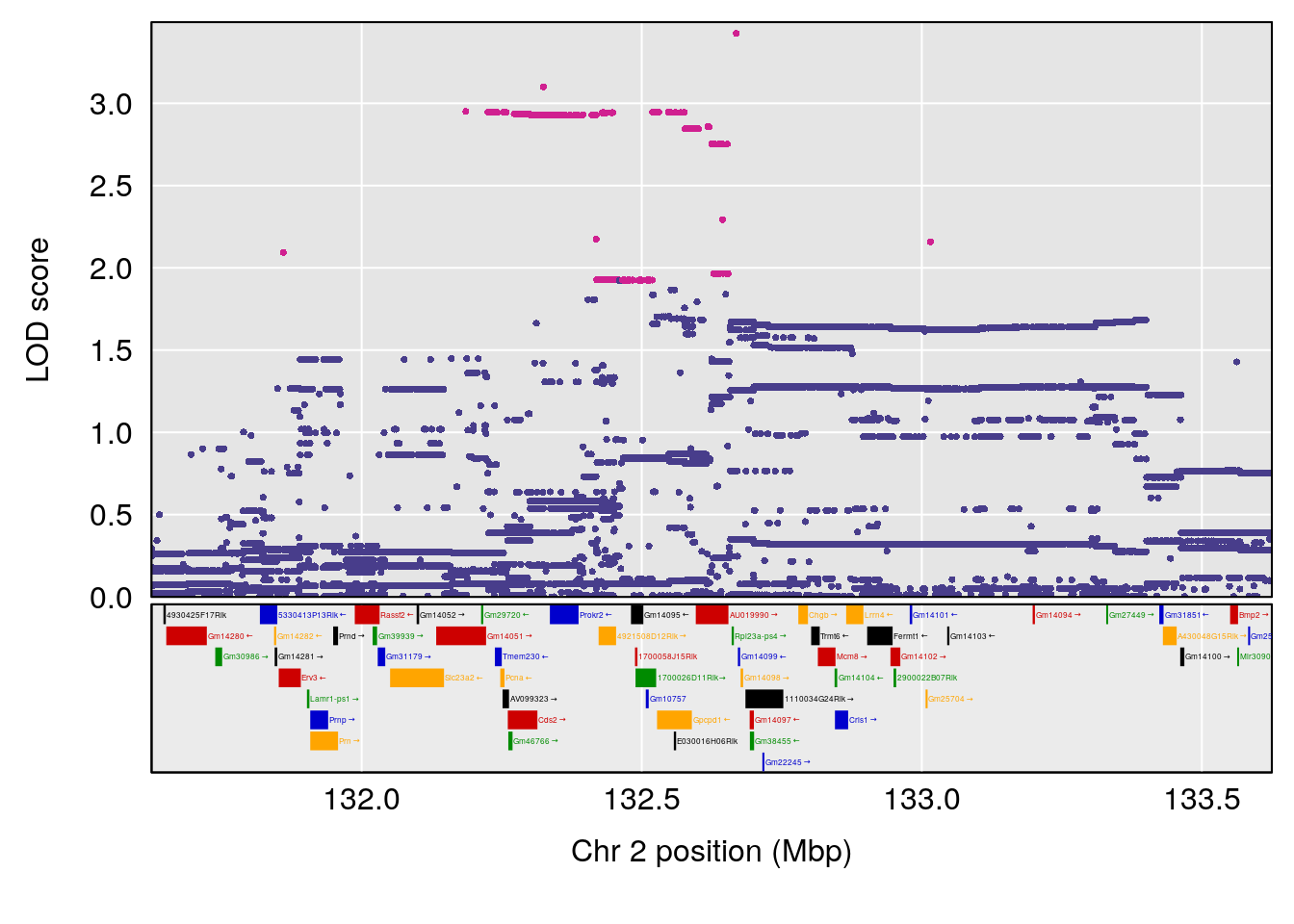
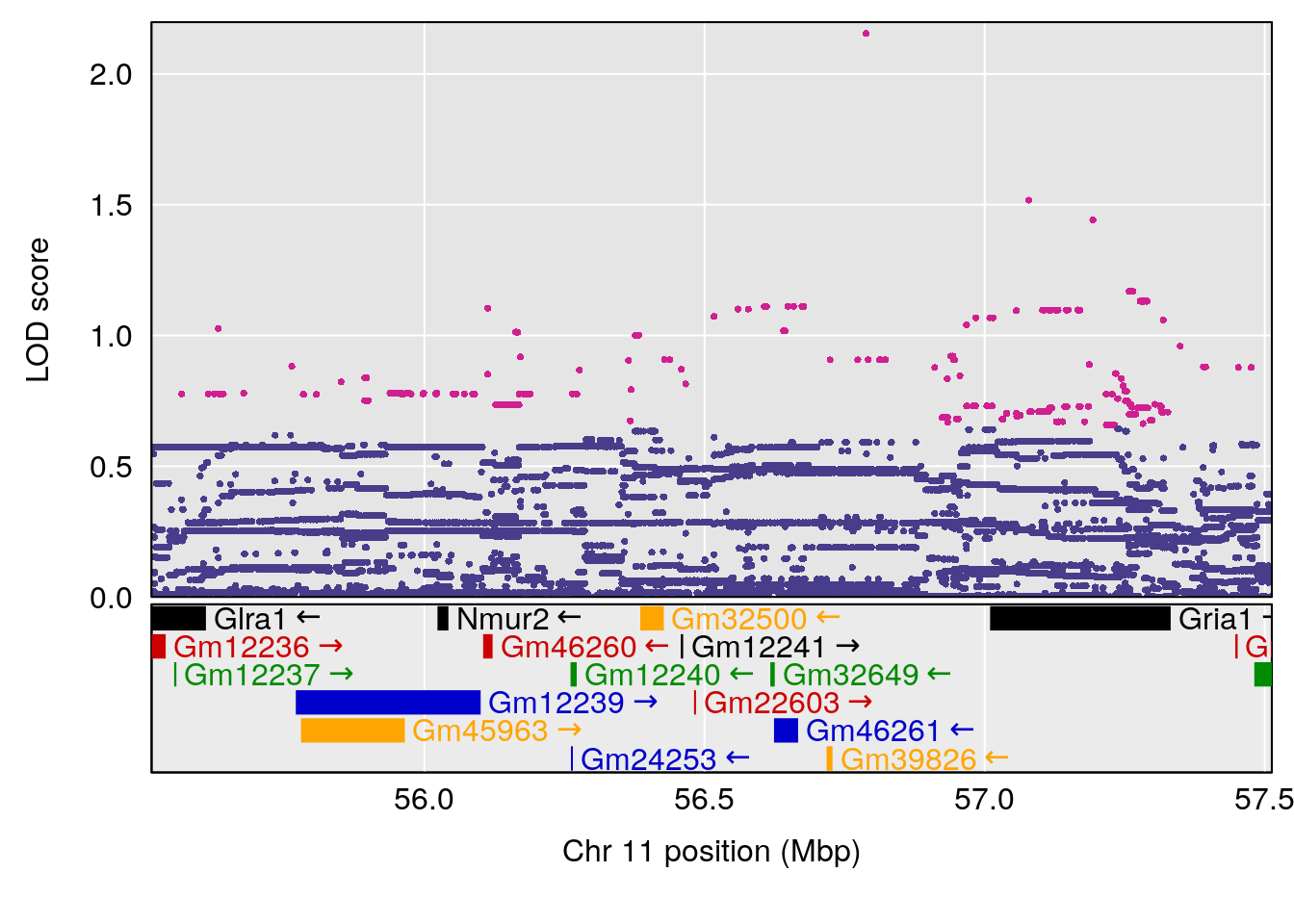
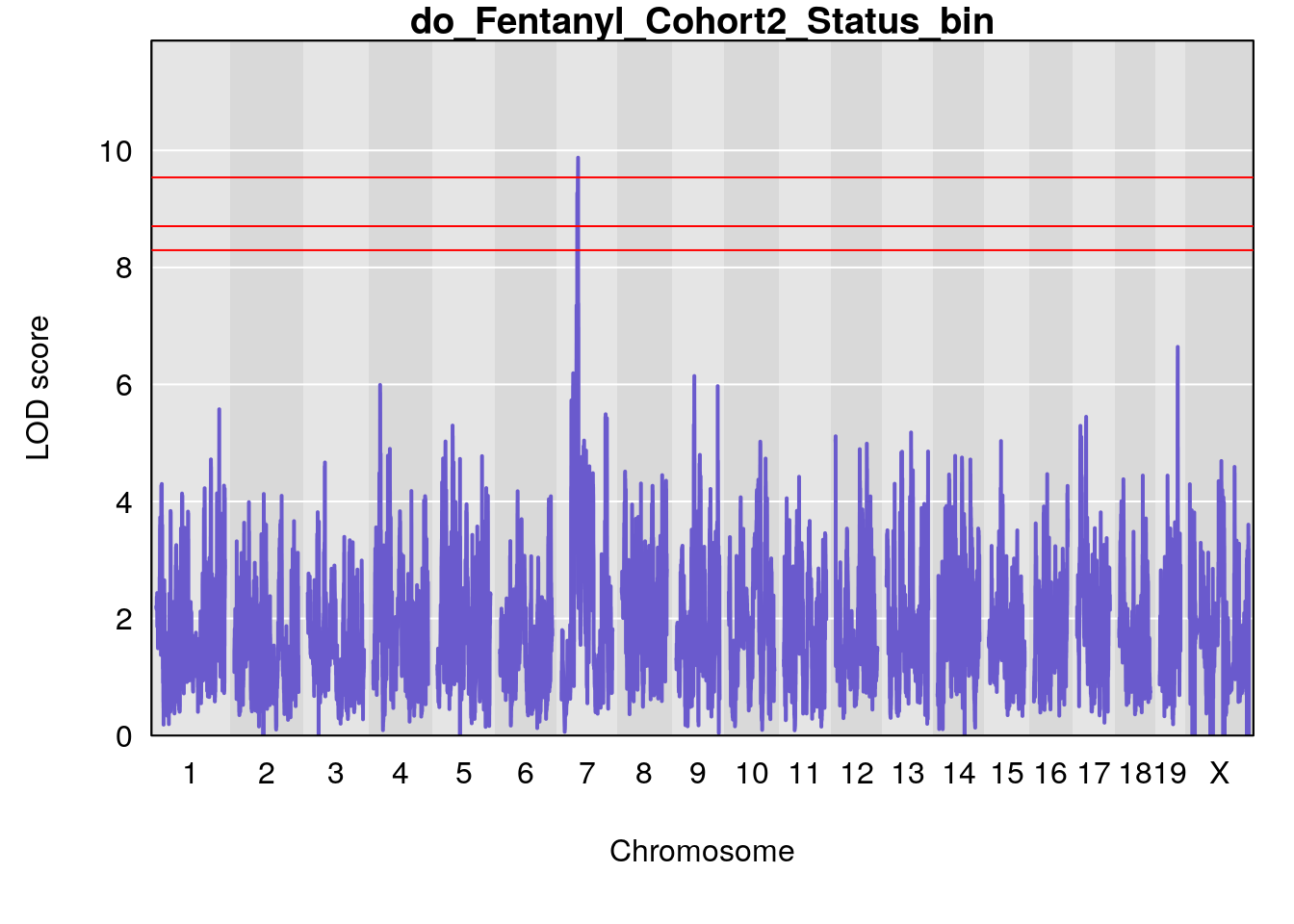
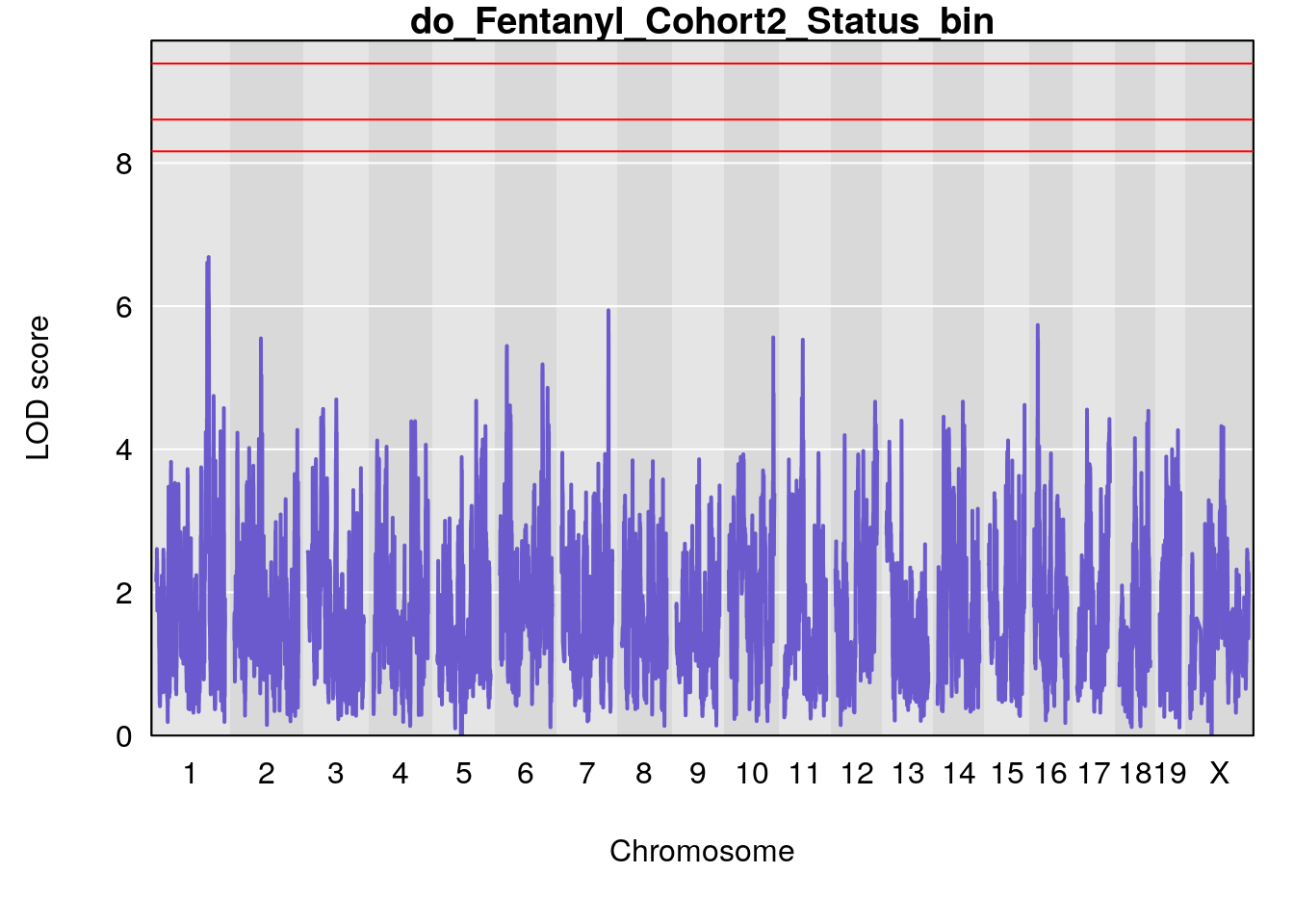
# [1] "Status_bin"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 11 56.51261 6.370706 28.86938 56.80903
# [1] 1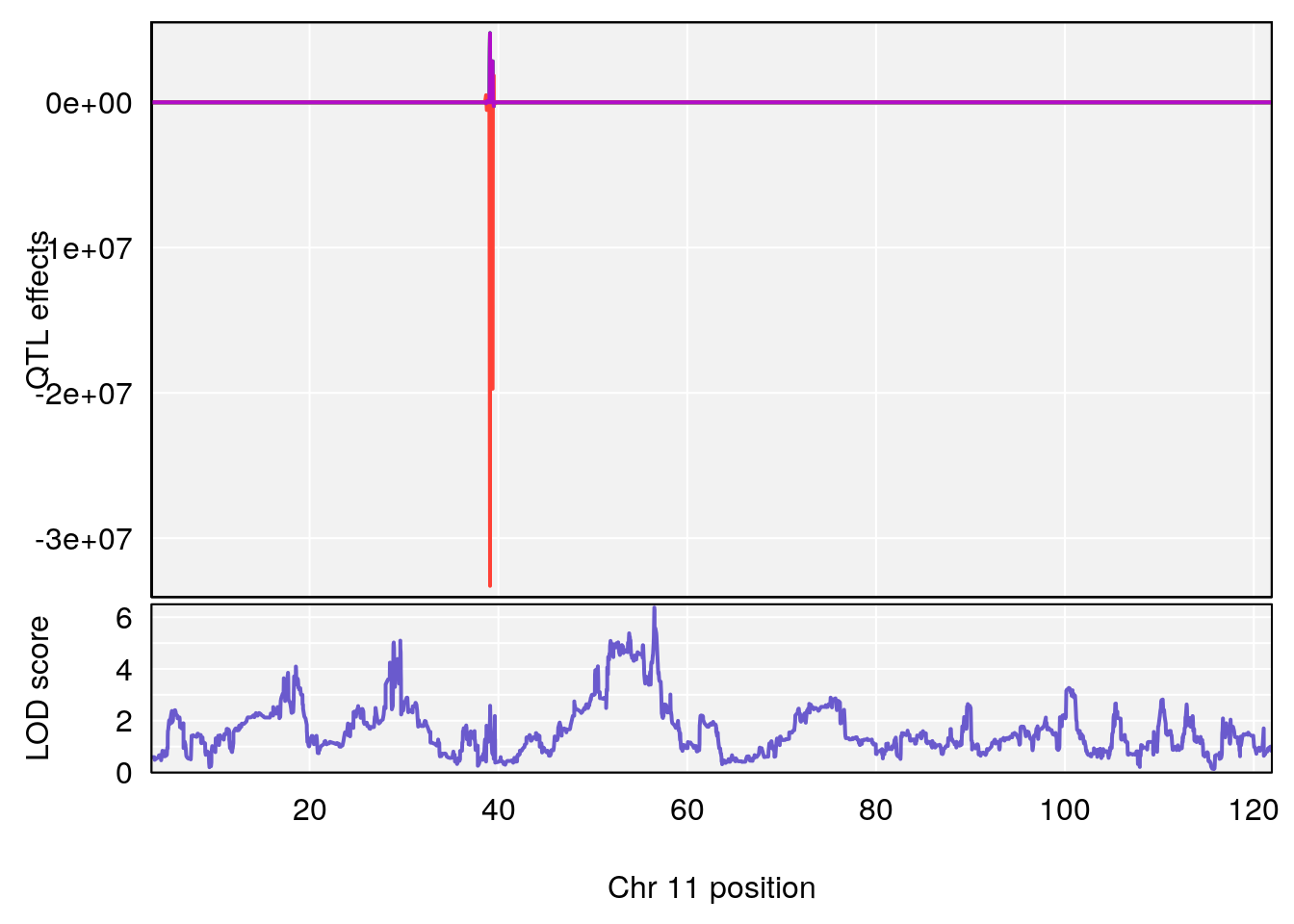
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
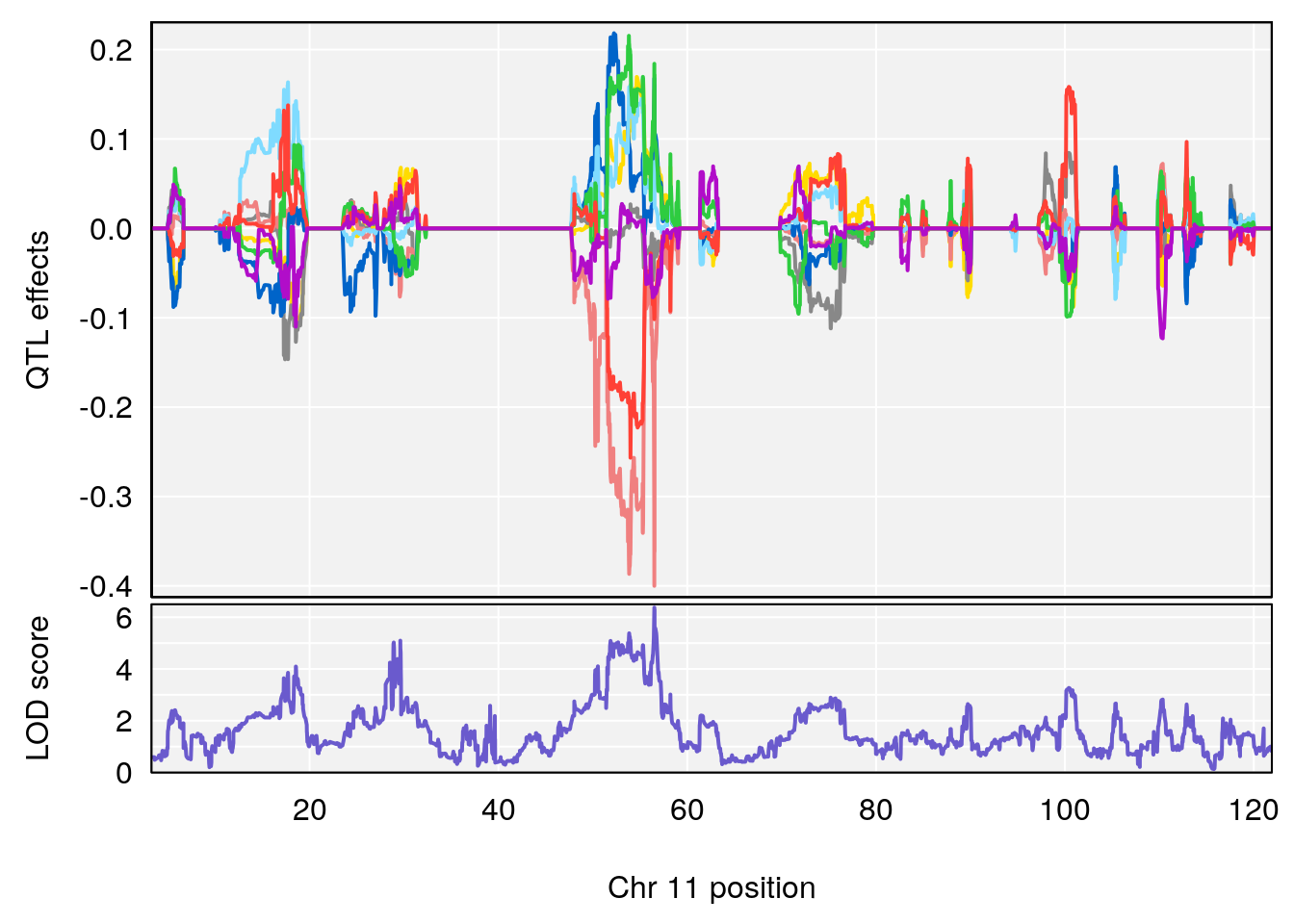
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
| Version | Author | Date |
|---|---|---|
| 7faf7f1 | xhyuo | 2022-06-29 |
#save peaks coeff plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
if(nrow(peaks) > 0){
fname <- paste("output/DO_Fentanyl_Cohort2_", str_replace_all(i, "[[:punct:]]", "") ,"_coefplot.pdf",sep="")
pdf(file = fname, width = 16, height =8)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
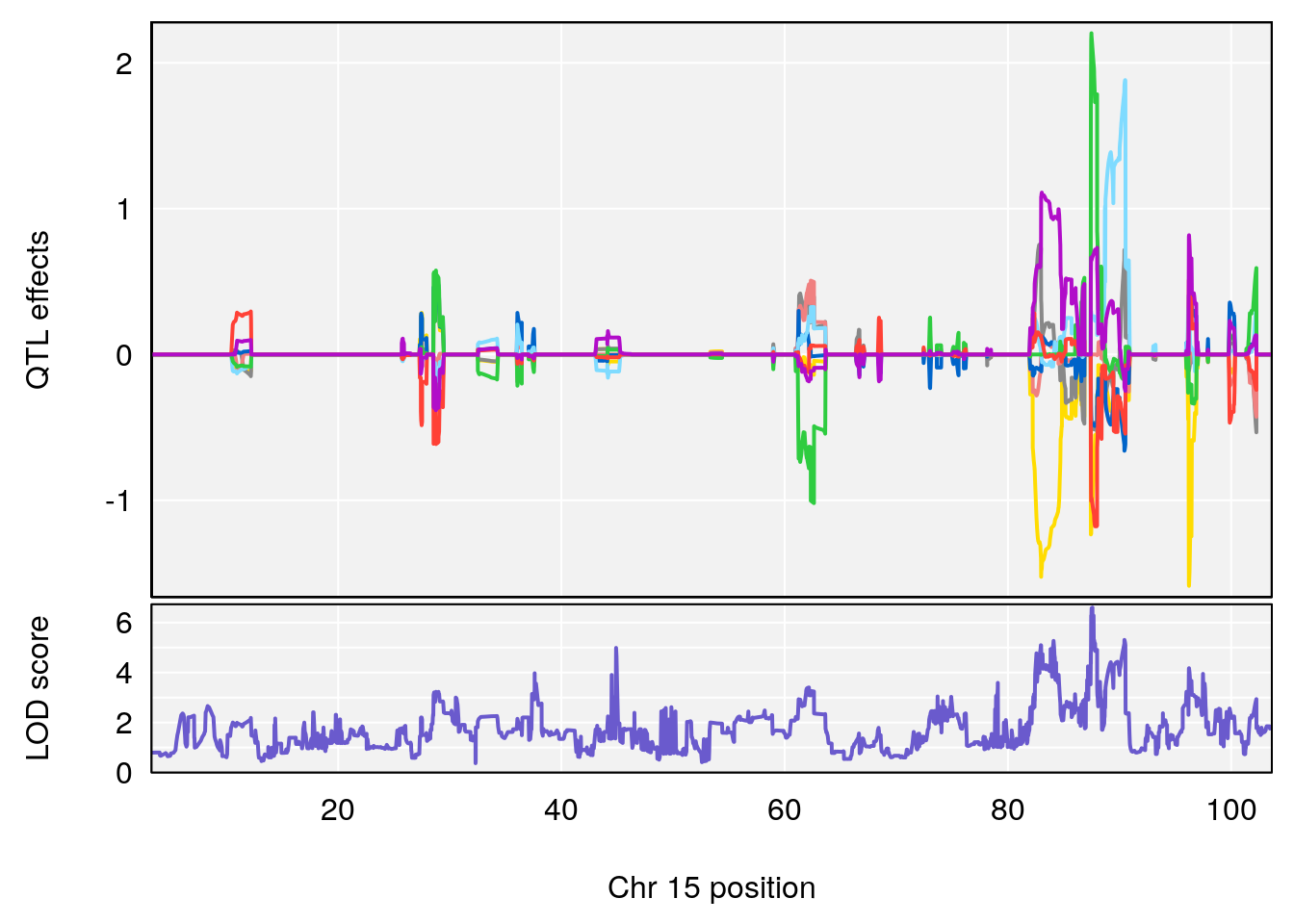
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
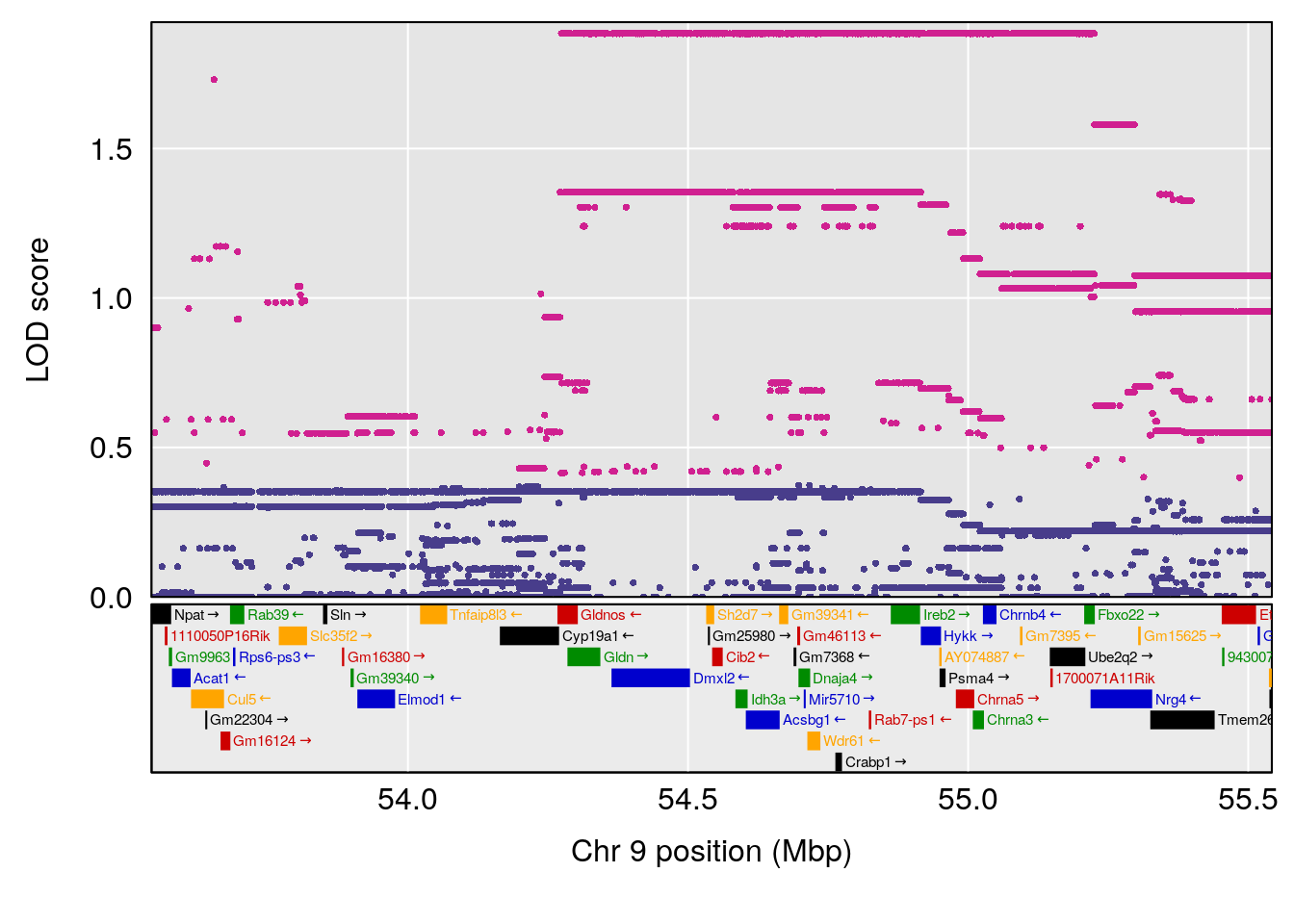
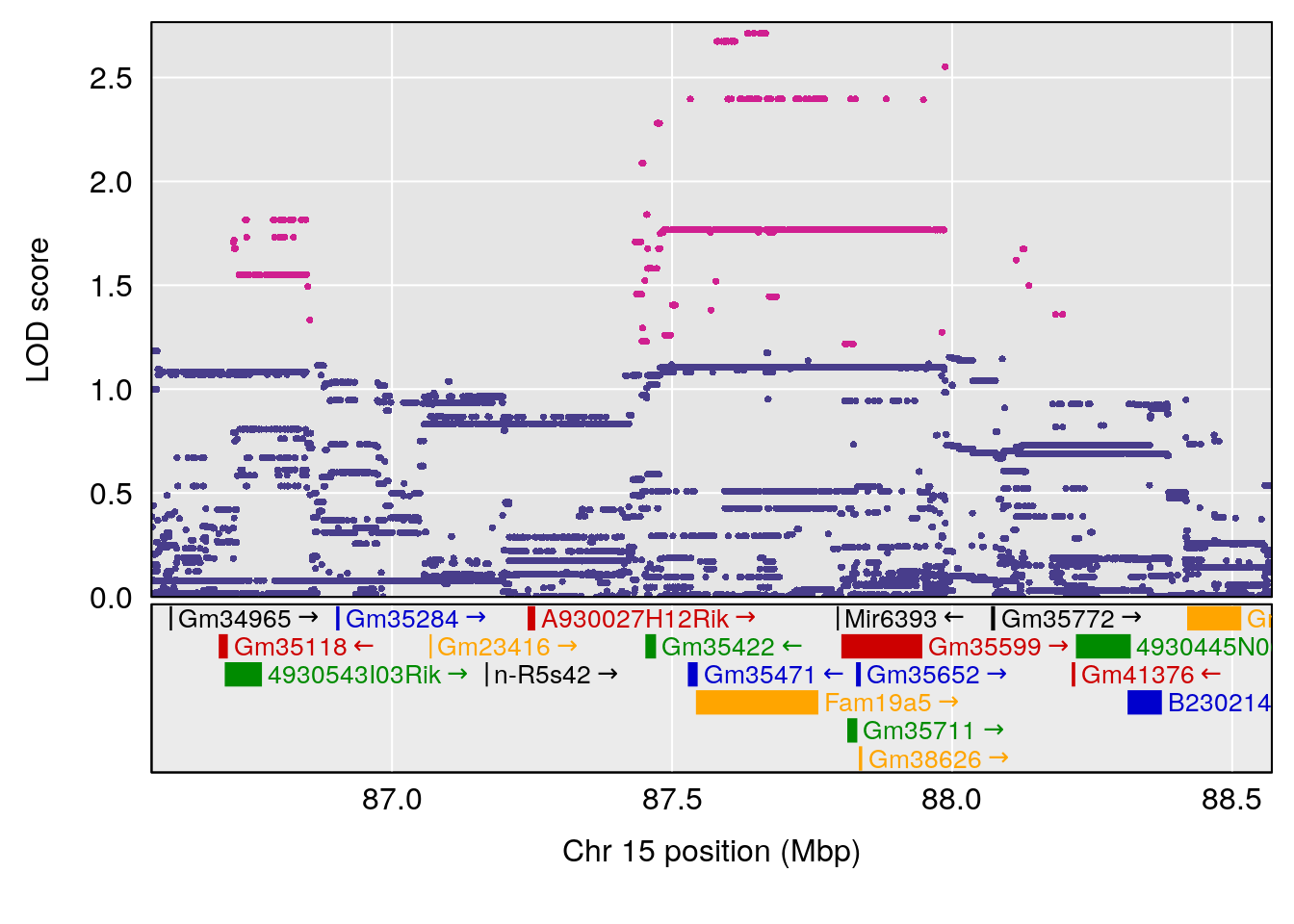
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
dev.off()
}
}
# [1] "TimetoDead(Hr)"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] "RRDepressionRate(%/Hr)SLOPE"
# [1] "TimetoSteadyRRDepression(Hr)"
# [1] 1
# [1] 2
# [1] 3
# [1] "MinDepressionRR(%)"
# [1] 1
# [1] "Steady-StateDepressionDuration(Hr)INTERVAL"
# [1] "TimetoThresholdRecovery(Hr)"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] "TimetoProjectedRecovery(Hr)"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] "StartofRecovery(Hr)"
# [1] 1
# [1] "RRRecoveryRate(%/Hr)SLOPE"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] "Status_bin"
# [1] 1
#save peaks coeff blup plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
if(nrow(peaks) > 0) {
fname <- paste("output/DO_Fentanyl_Cohort2_", str_replace_all(i, "[[:punct:]]", "") ,"_coefplot_blup.pdf",sep="")
pdf(file = fname, width = 16, height =8)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
dev.off()
}
}
# [1] "TimetoDead(Hr)"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] "RRDepressionRate(%/Hr)SLOPE"
# [1] "TimetoSteadyRRDepression(Hr)"
# [1] 1
# [1] 2
# [1] 3
# [1] "MinDepressionRR(%)"
# [1] 1
# [1] "Steady-StateDepressionDuration(Hr)INTERVAL"
# [1] "TimetoThresholdRecovery(Hr)"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] "TimetoProjectedRecovery(Hr)"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] "StartofRecovery(Hr)"
# [1] 1
# [1] "RRRecoveryRate(%/Hr)SLOPE"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] "Status_bin"
# [1] 1heritability
#plot heritability by qtl2 array --------------
herit <- data.frame(Phenotype = names(unlist(qtl.morphine.hsq)),
Heritability = round(unlist(qtl.morphine.hsq),3))
herit <- herit %>%
arrange(desc(Heritability))
herit$Phenotype <- factor(herit$Phenotype, levels = herit$Phenotype)
#histgram
p2 <- ggplot(data=herit, aes(x=Phenotype, y=Heritability)) + #, fill=Domain, color = Domain)) +
geom_bar(stat="identity", fill = "blue", color = "blue", show.legend = FALSE) +
scale_y_continuous(breaks=seq(0.0, 1.0, 0.1)) +
geom_text(aes(label = Heritability, y = Heritability + 0.005), position = position_dodge(0.9),vjust = 0) +
theme(axis.text.x = element_text(angle = 90, hjust = 1)) +
ggtitle("DO Fentanyl_Cohort2 Heritability by qtl2 array")
p2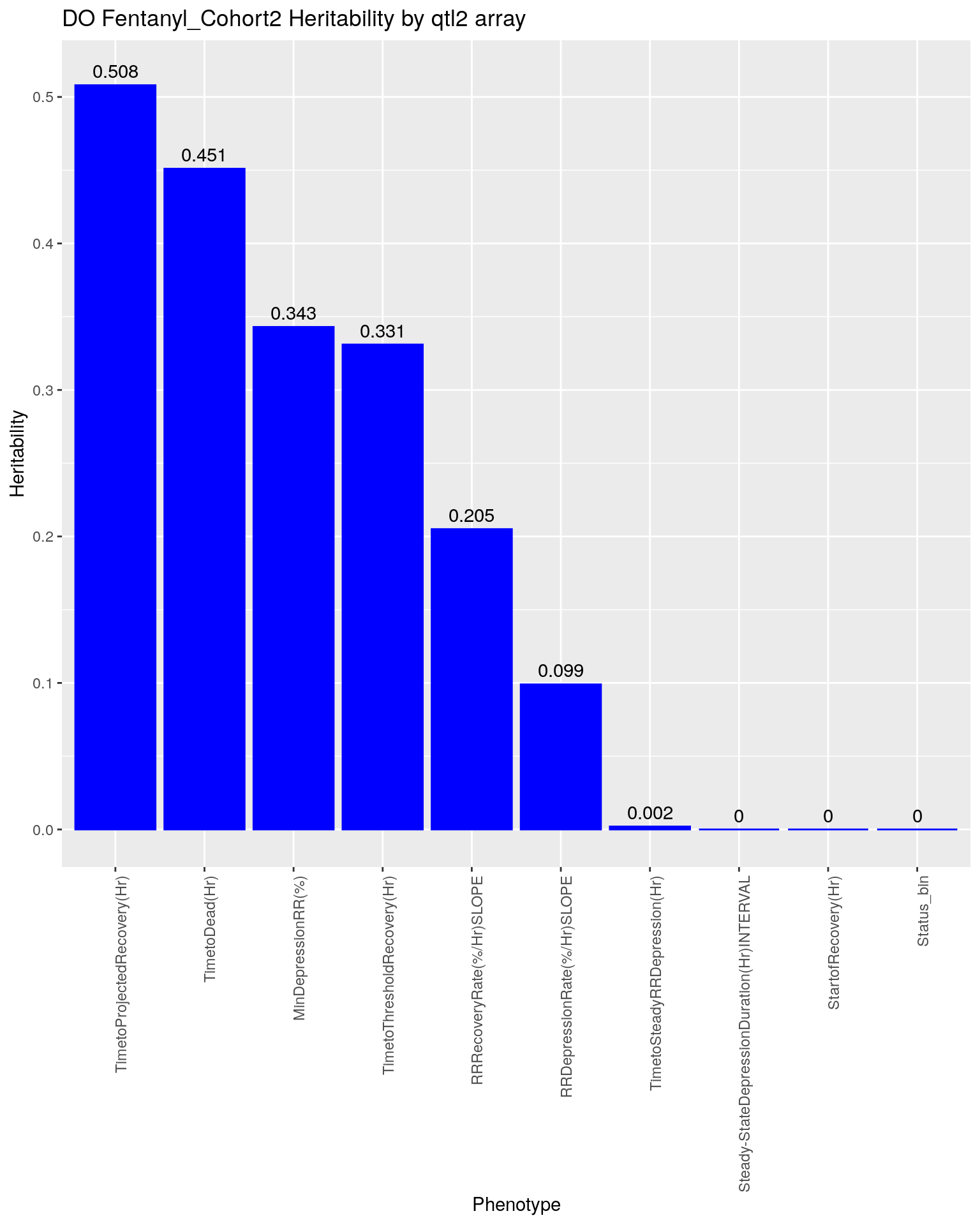
#plot heritability by GCTA
h <- read.csv("data/FinalReport/GCTA/DO_Fentanyl_Cohort2/DO_Fentanyl_Cohort2_heritability_by_GCTA.csv", header = TRUE)
h <- h %>%
arrange(desc(Heritability))
h$Phenotype <- factor(h$Phenotype, levels = h$Phenotype)
h$Heritability <- round(h$Heritability,2)
#histgram
pdf(file = paste0("data/FinalReport/GCTA/DO_Fentanyl_Cohort2/DO_Fentanyl_Cohort2","_heritability_by_GCTA.pdf"), height = 10, width = 10)
p3<-ggplot(data=h, aes(x=Phenotype, y=Heritability)) + #, fill=Domain, color = Domain)) +
geom_bar(stat="identity", fill = "blue", color = "blue", show.legend = FALSE) +
scale_y_continuous(breaks=seq(0.0, 1.0, 0.1)) +
geom_text(aes(label = Heritability, y = Heritability + 0.005), position = position_dodge(0.9),vjust = 0) +
theme(axis.text.x = element_text(angle = 90, hjust = 1)) +
ggtitle("Heritability by GCTA")
p3
dev.off()
# png
# 2
# png
# 2
p3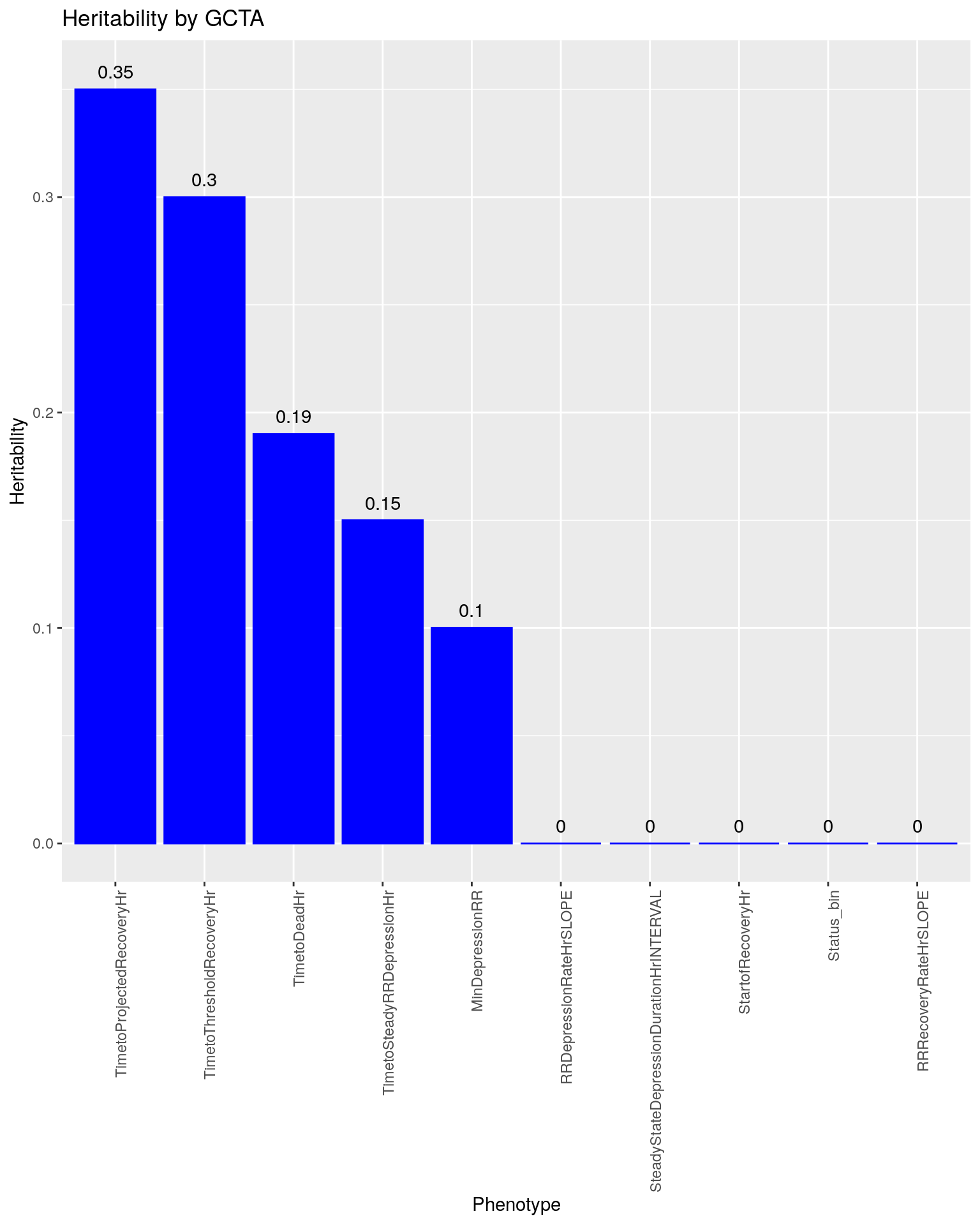
Plot qtl mapping on array male
load("output/Fentanyl/qtl.do_Fentanyl_Cohort2.out.male.RData")
#pmap and gmap
chr.names <- c("1","2","3","4","5","6","7","8","9","10","11","12","13","14","15","16","17","18","19", "X")
gmap <- list()
pmap <- list()
for (chr in chr.names){
gmap[[chr]] <- gm$gmap[[chr]]
pmap[[chr]] <- gm$pmap[[chr]]
}
attr(gmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
attr(pmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
#genome-wide plot
for(i in names(qtl.morphine.out)){
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.morphine.out[[i]]) # overall maximum LOD score
plot(qtl.morphine.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, max(sapply(qtl.morphine.out, maxlod)) +2))
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("do_Fentanyl_Cohort2_",i))
}
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
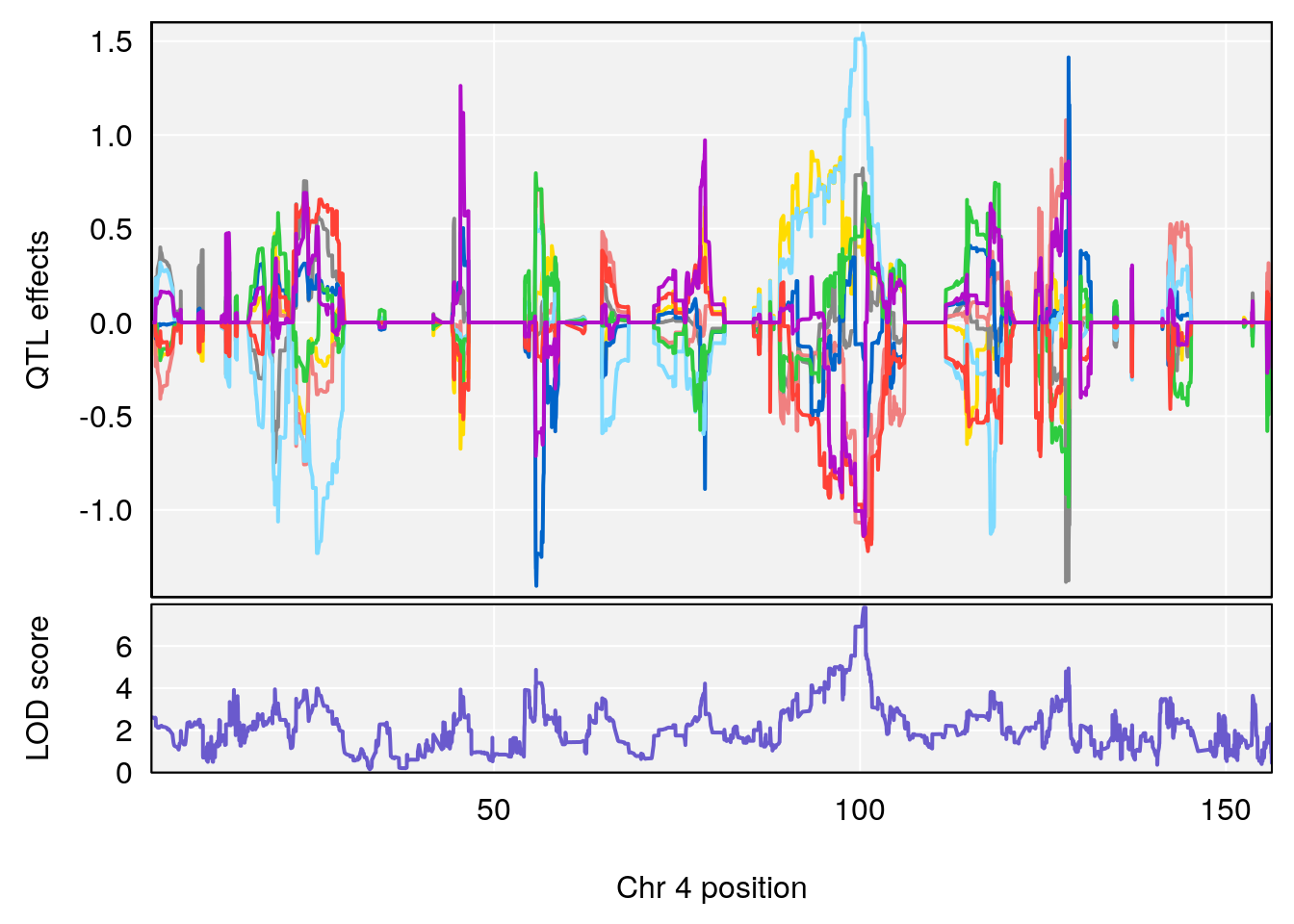
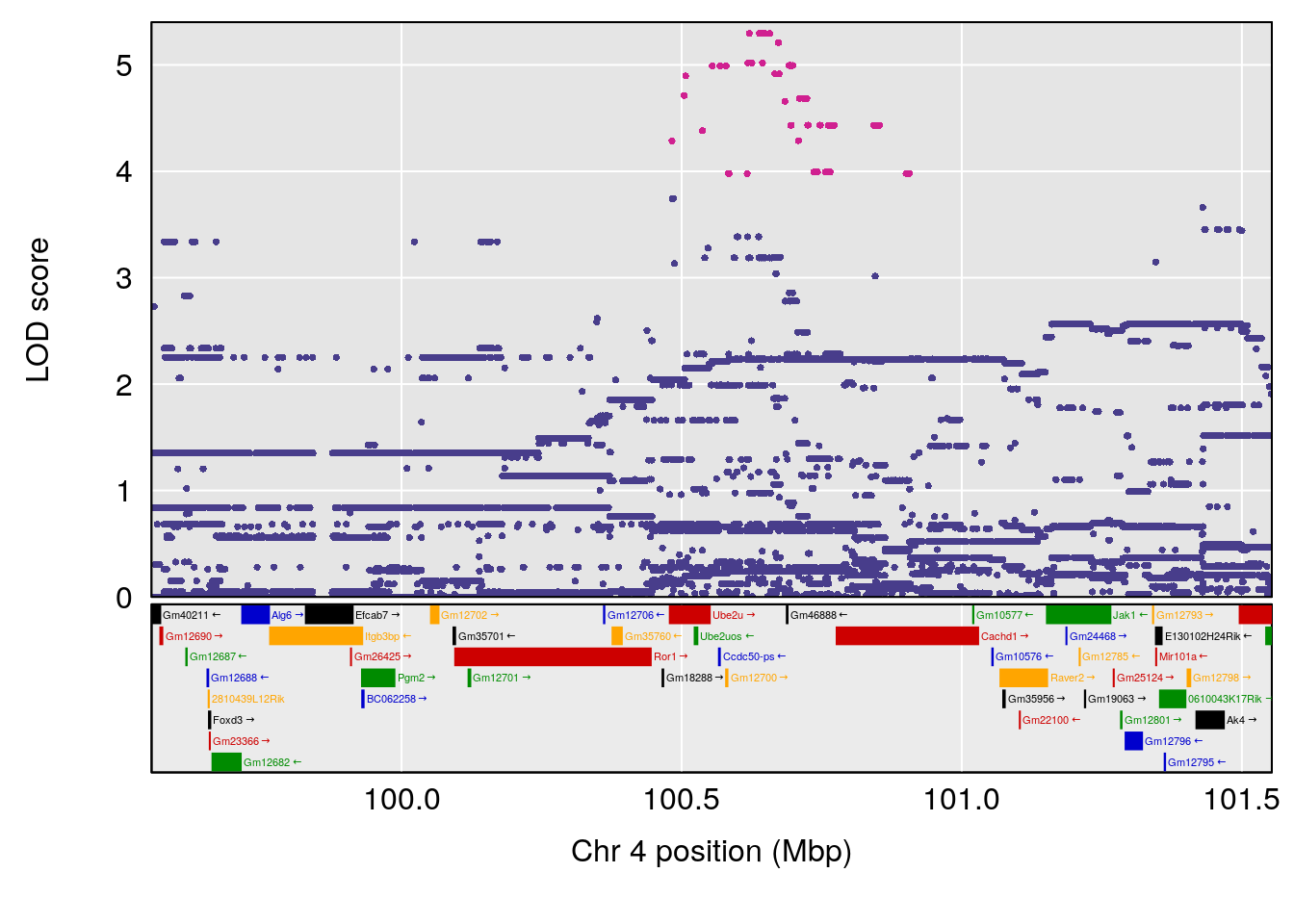
#peaks coeff plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
if(nrow(peaks) > 0) {
print(peaks)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
}
}
# [1] "TimetoDead(Hr)"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
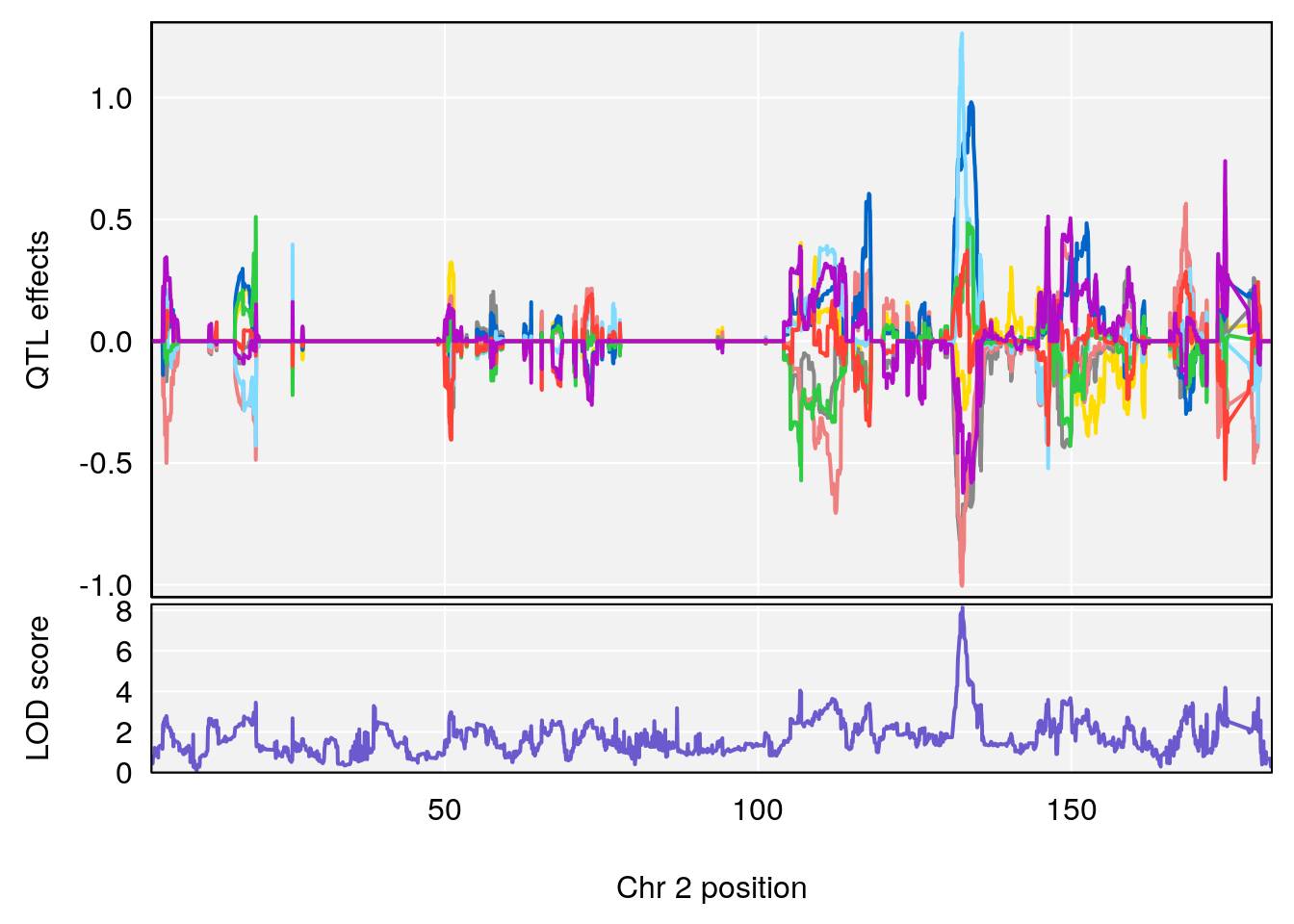
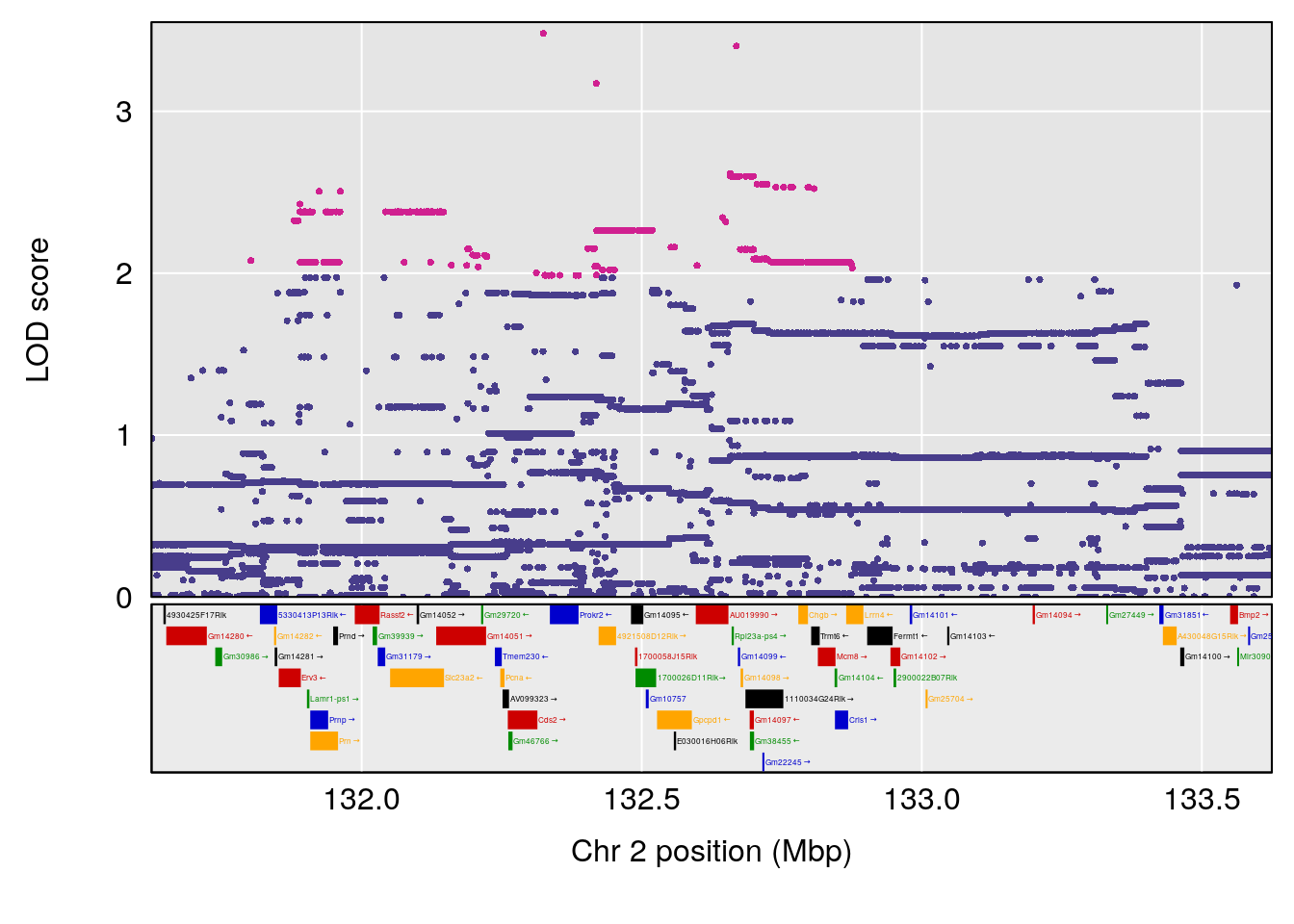
# 1 1 pheno1 2 132.62489 8.135472 131.88985 132.94007
# 2 1 pheno1 3 80.46385 6.635020 73.35207 81.19227
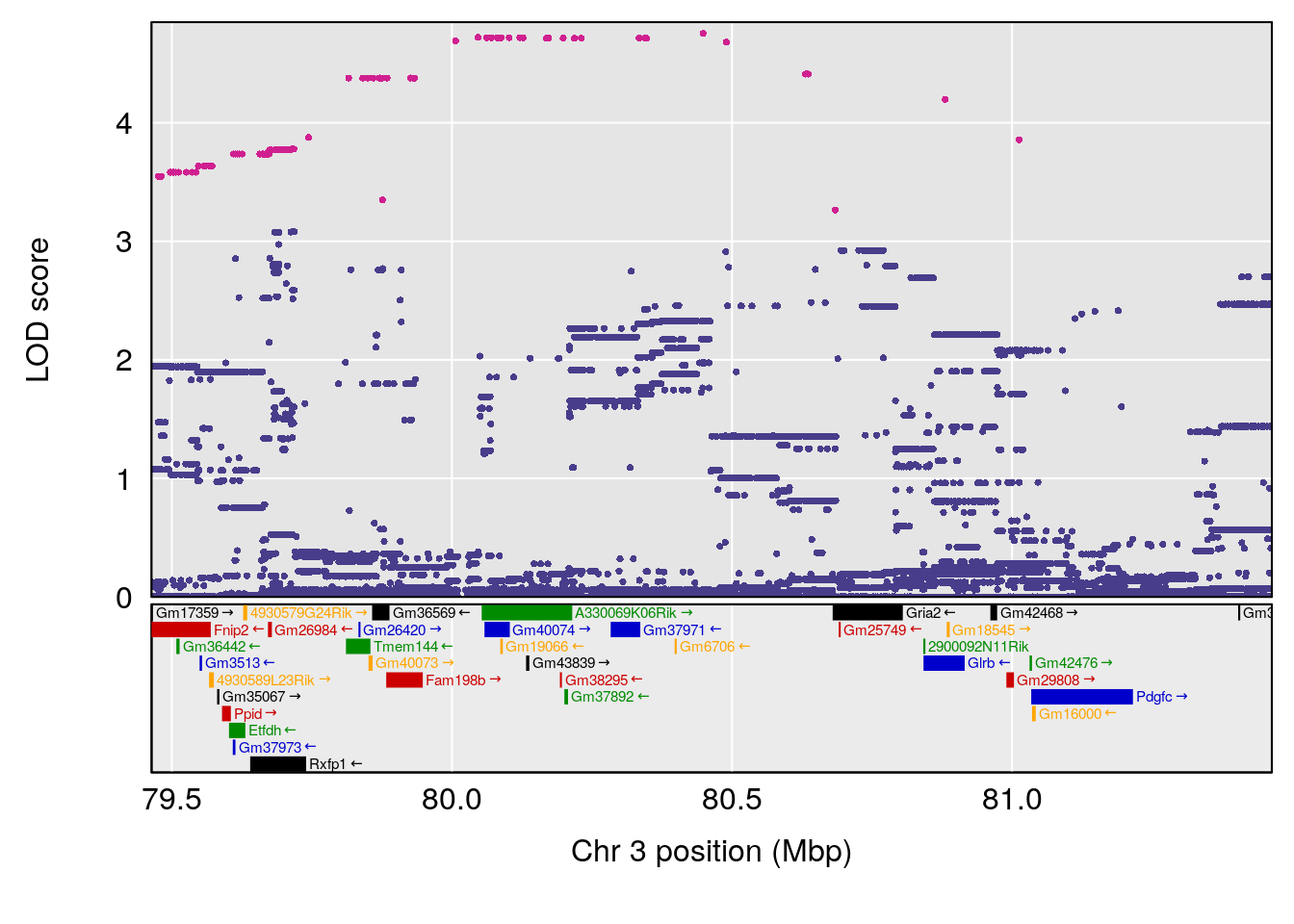
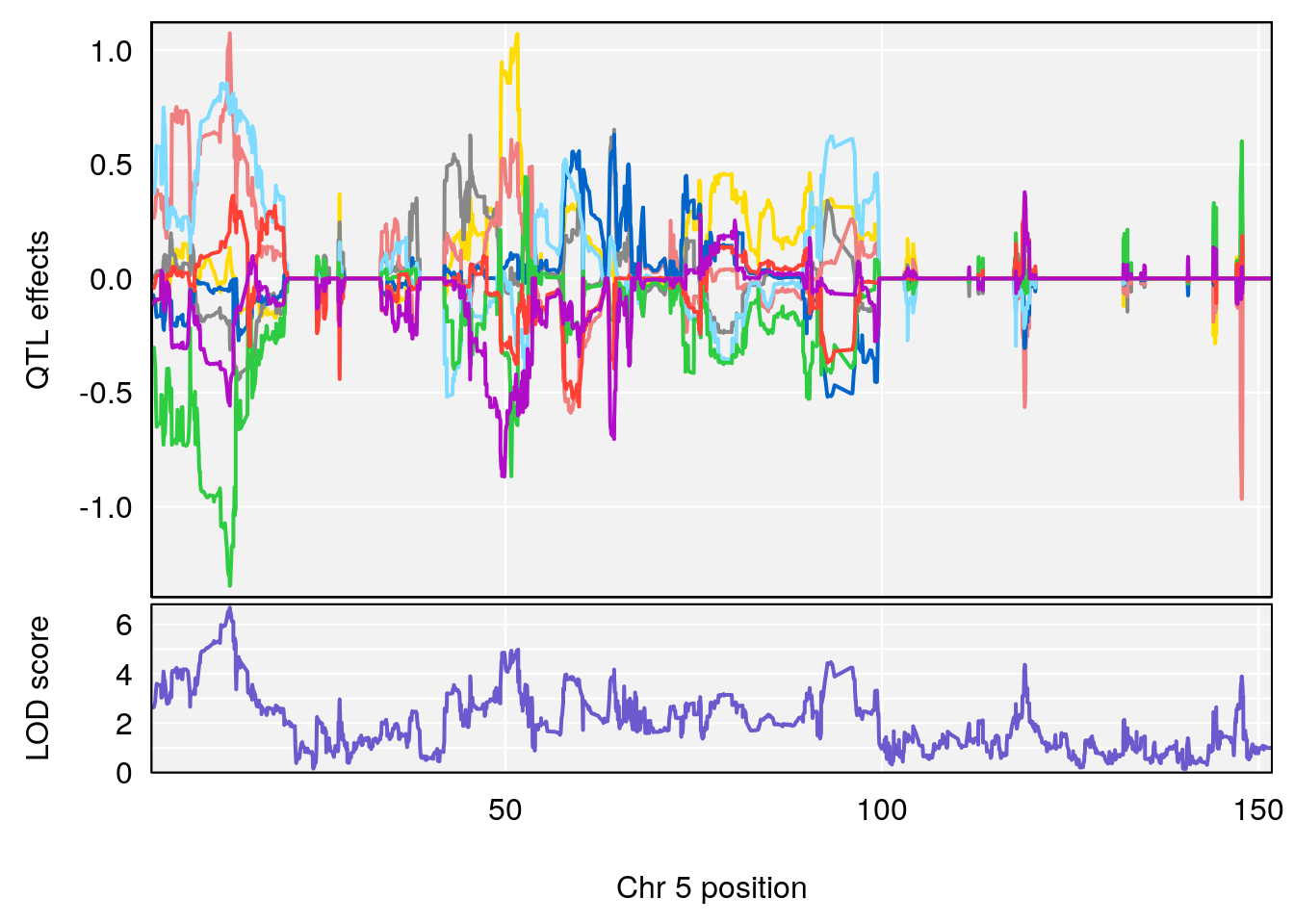
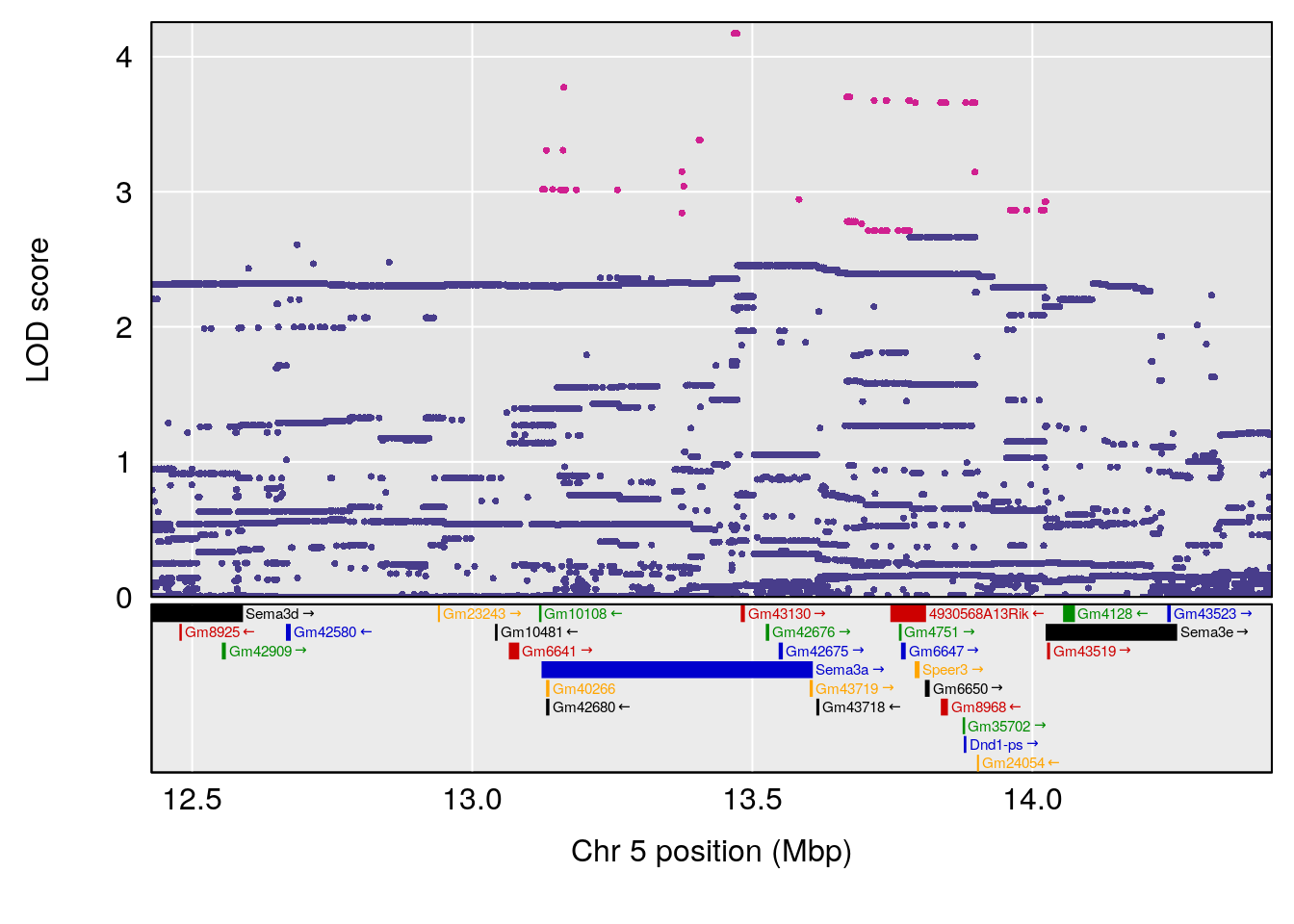
# 3 1 pheno1 5 13.42743 6.684415 10.86410 14.21411
# 4 1 pheno1 9 13.71424 6.527954 13.12685 15.44563
# [1] 1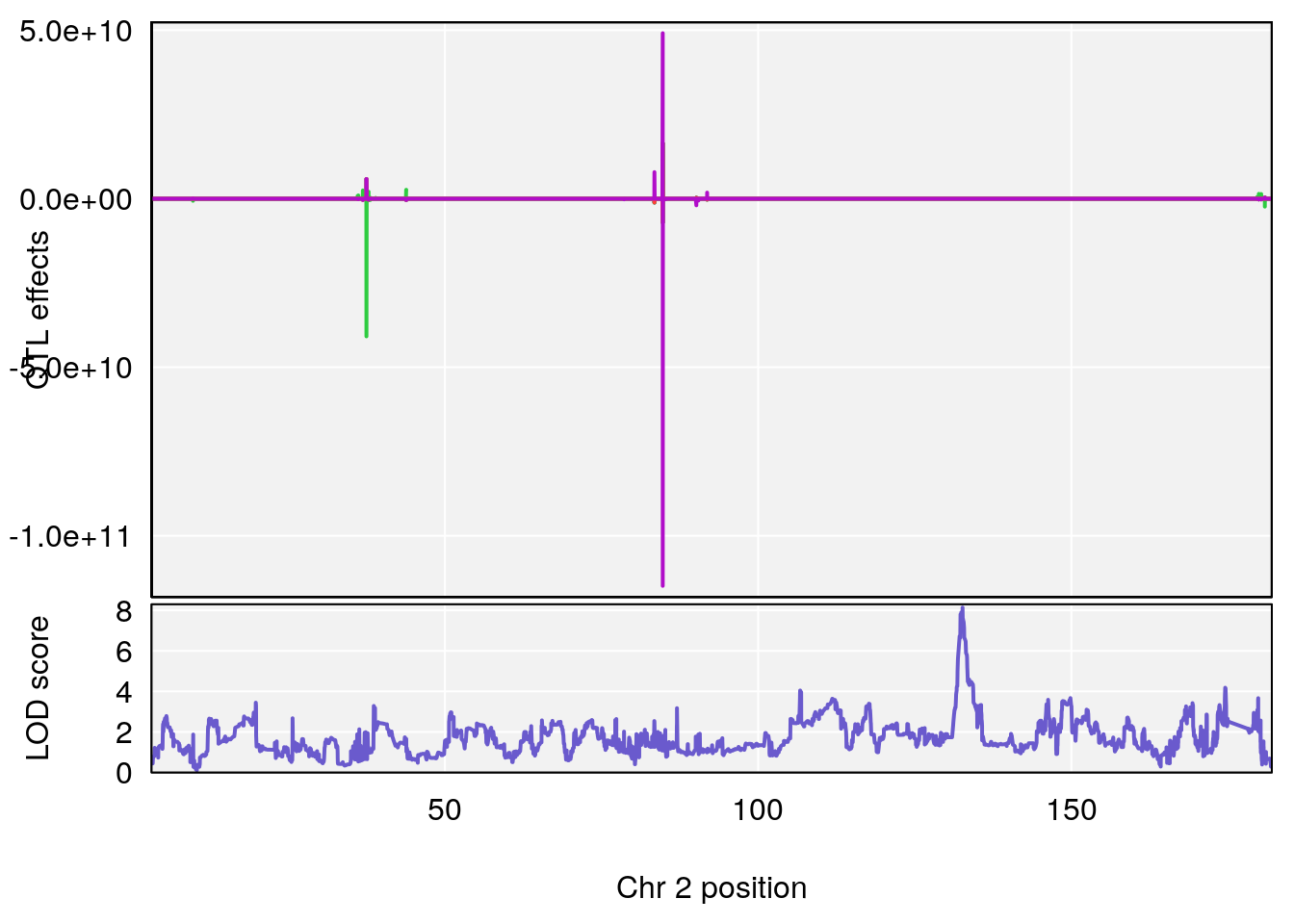
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 3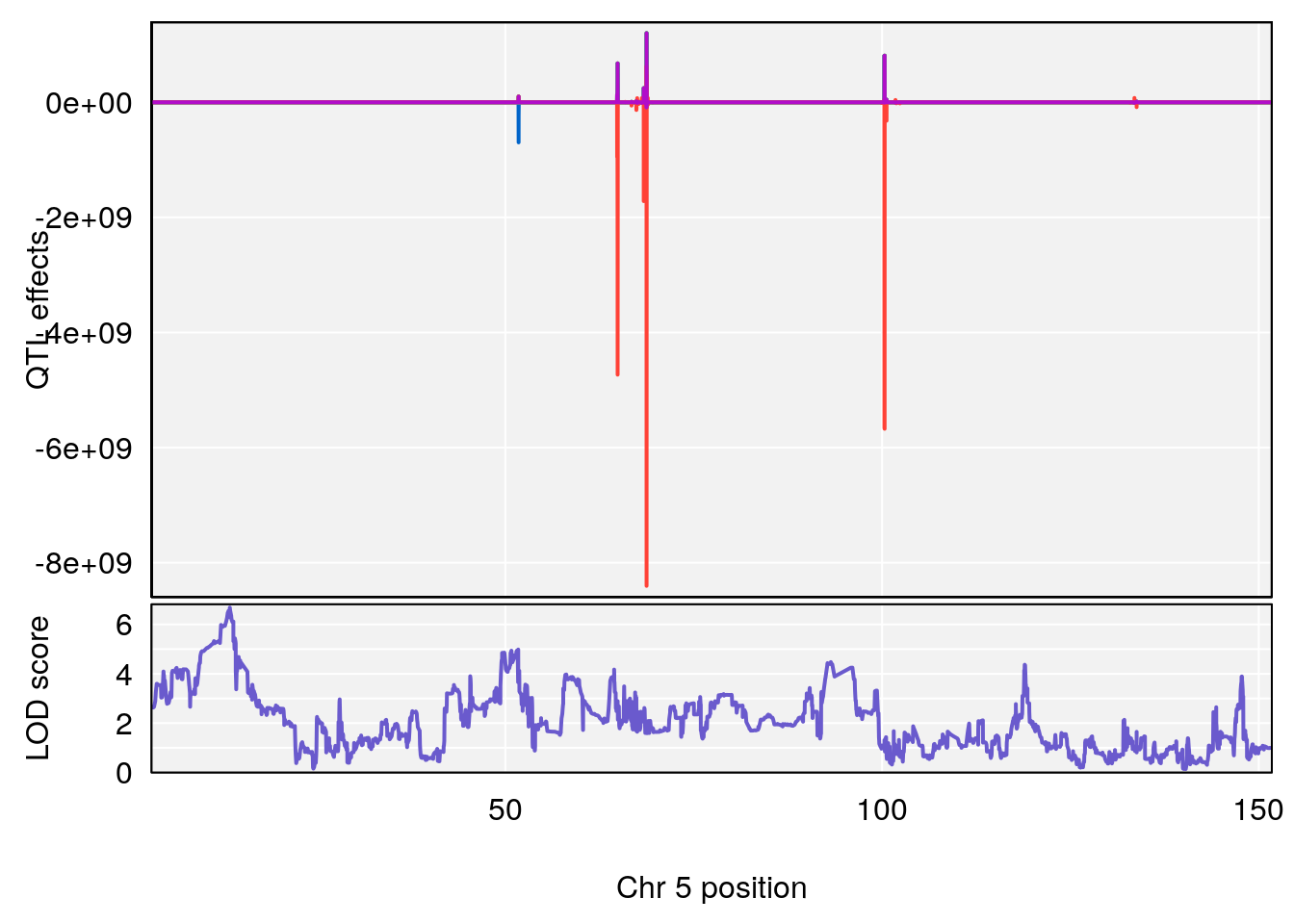
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
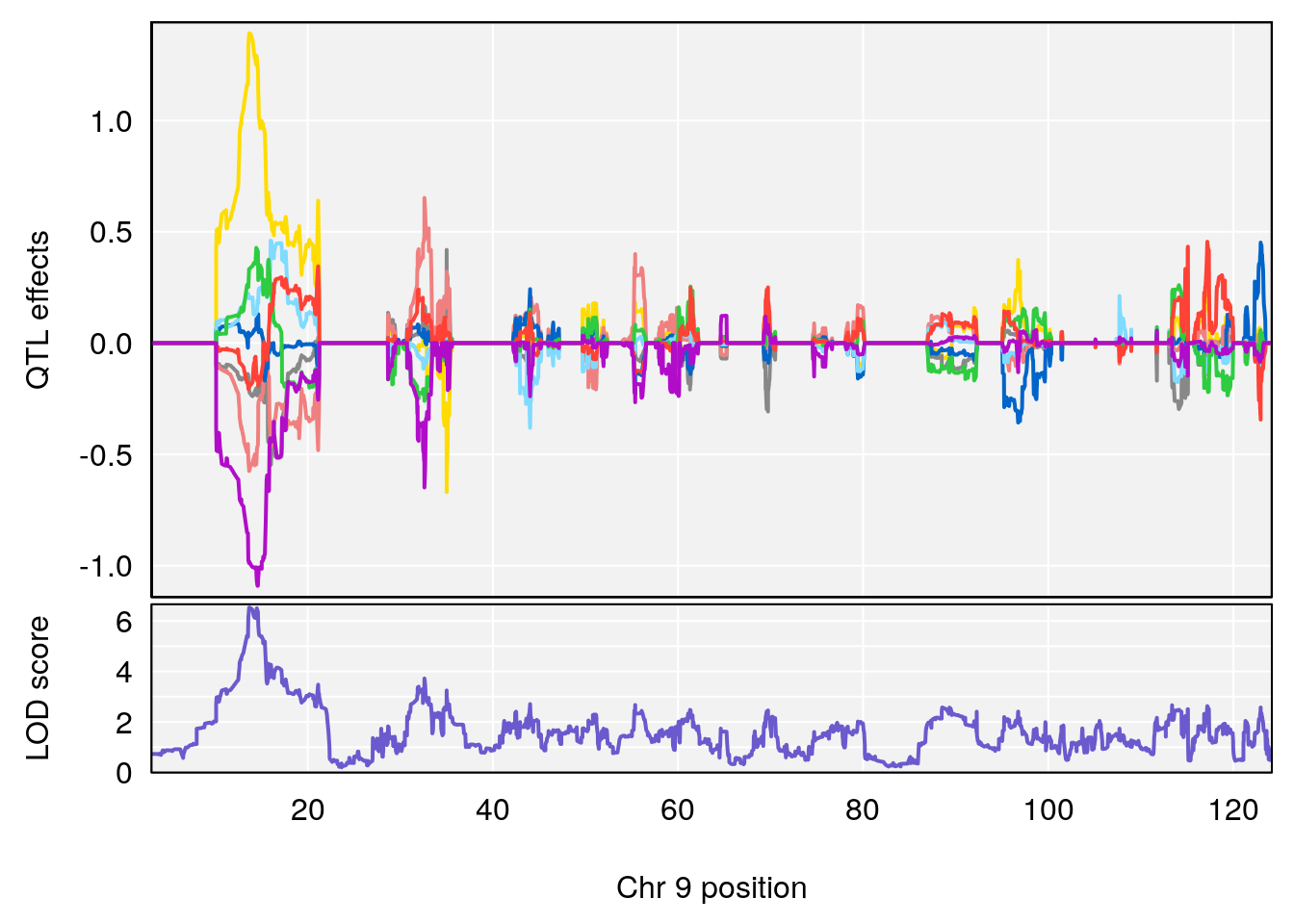
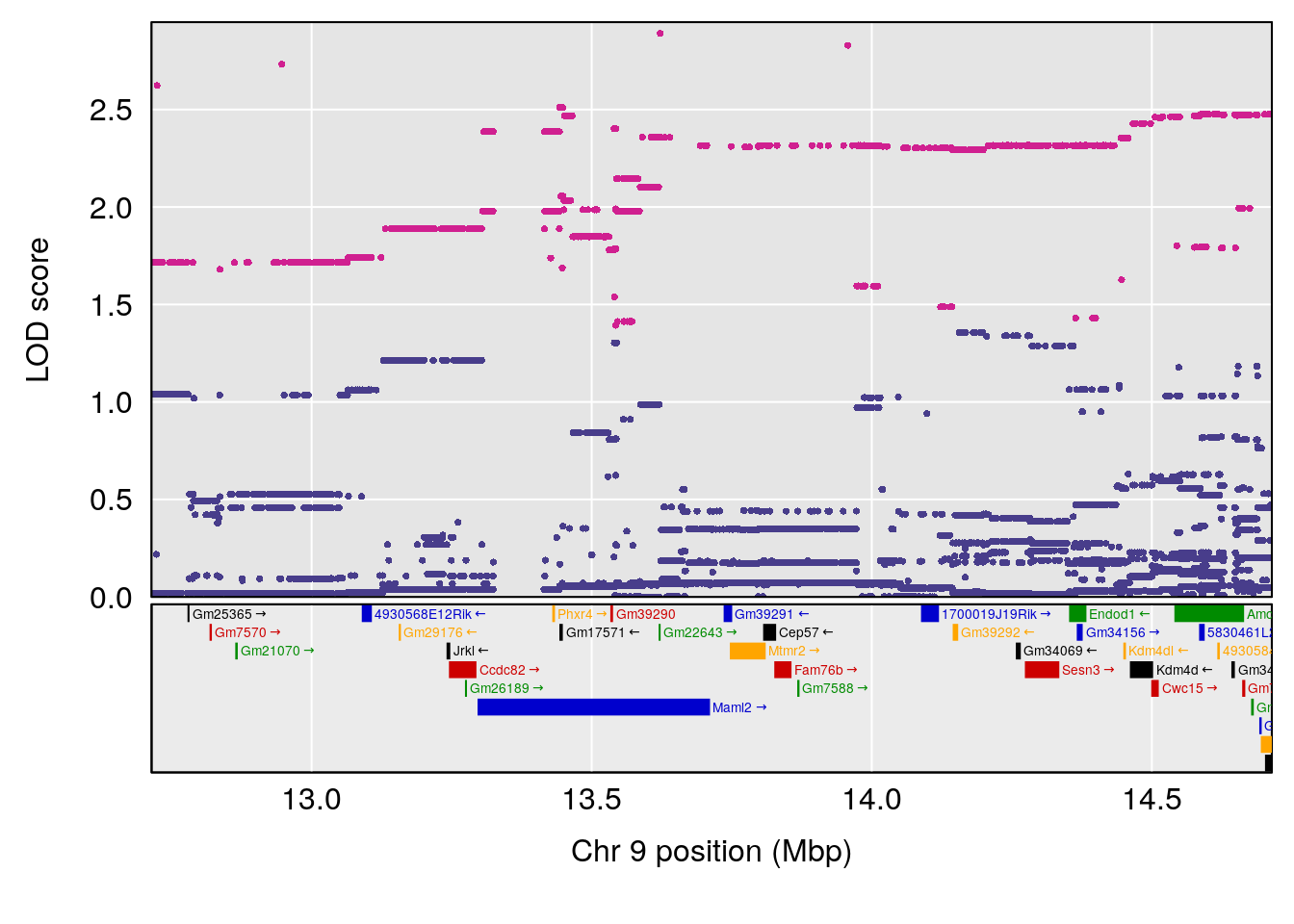
# [1] 4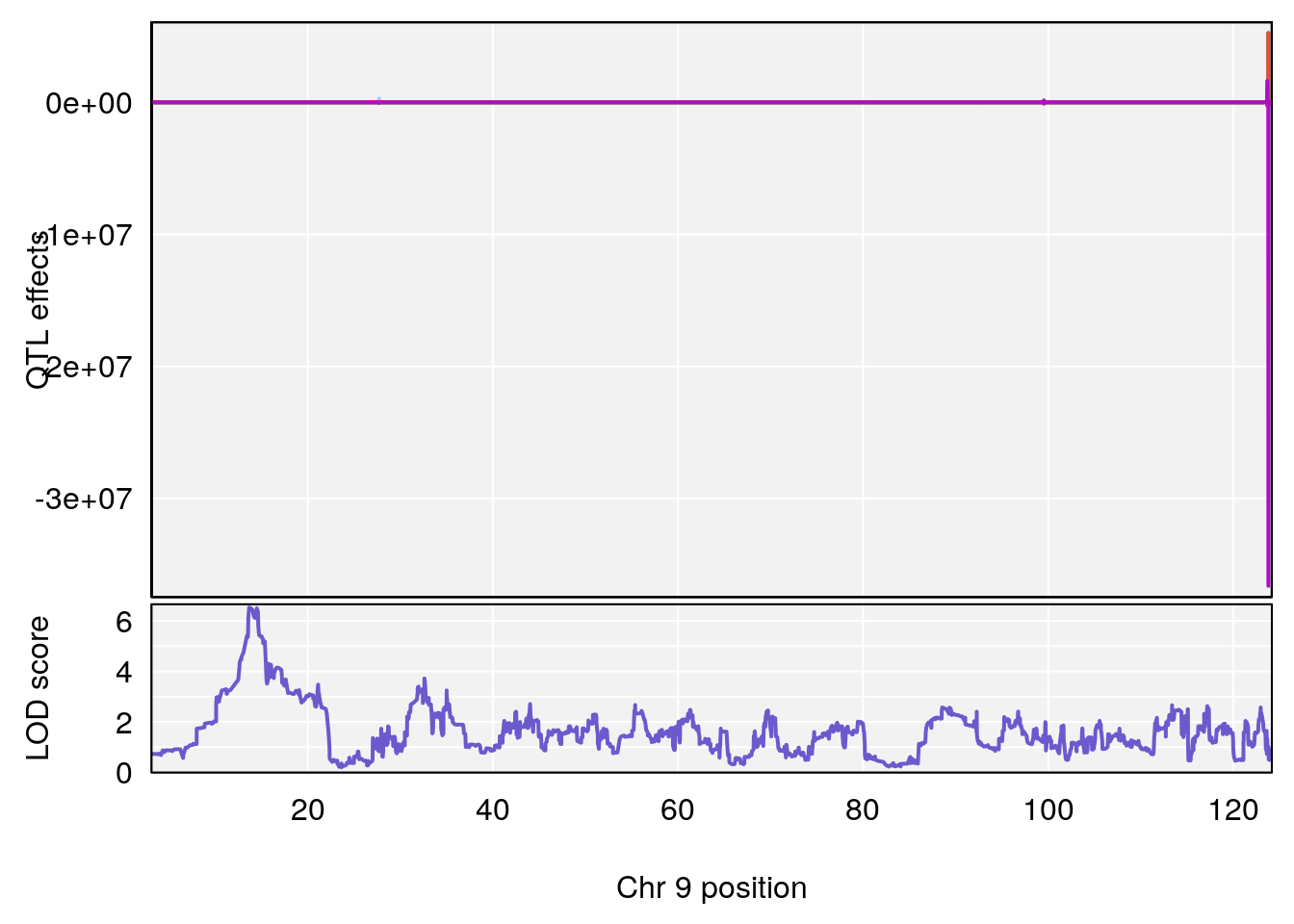
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
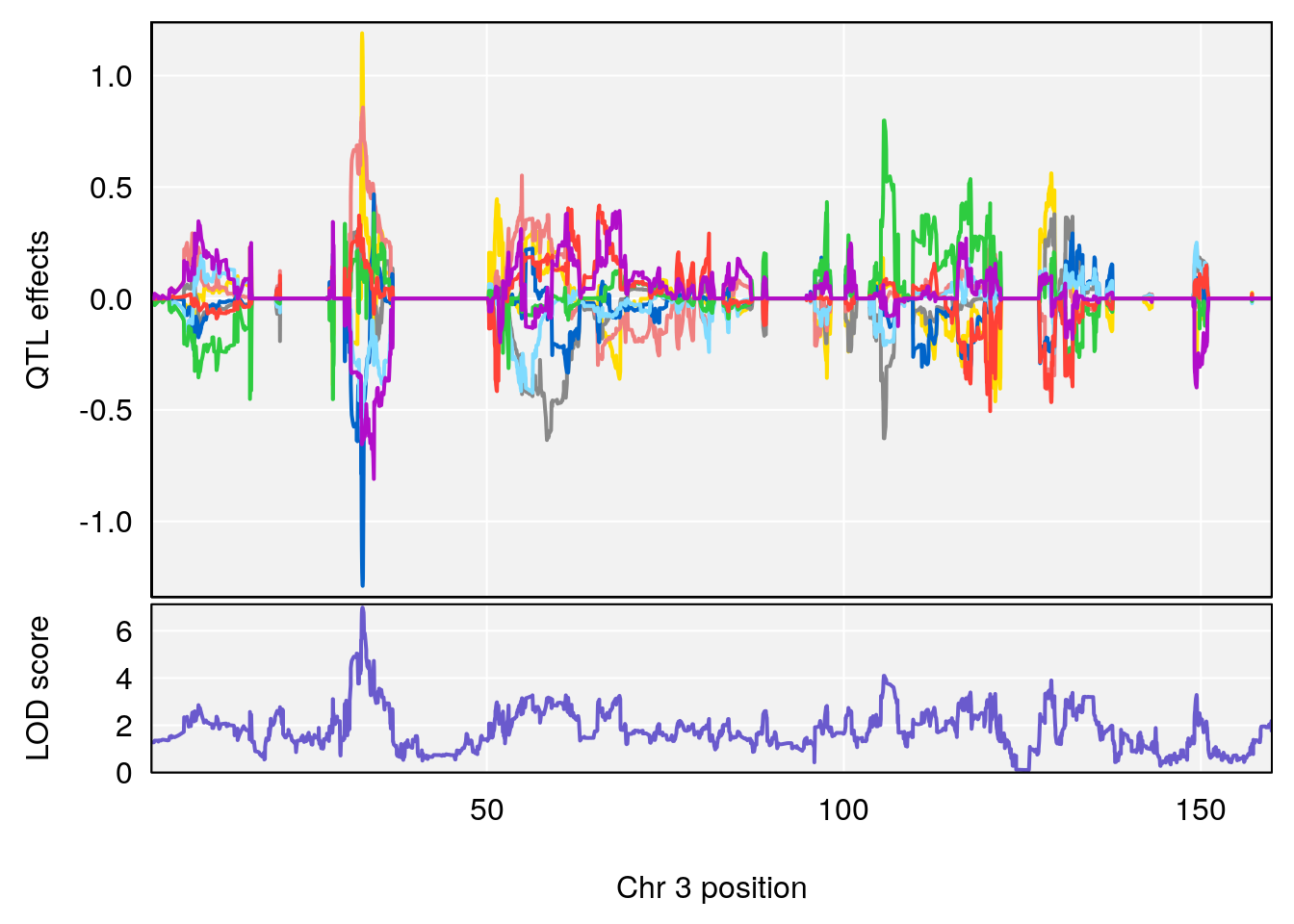
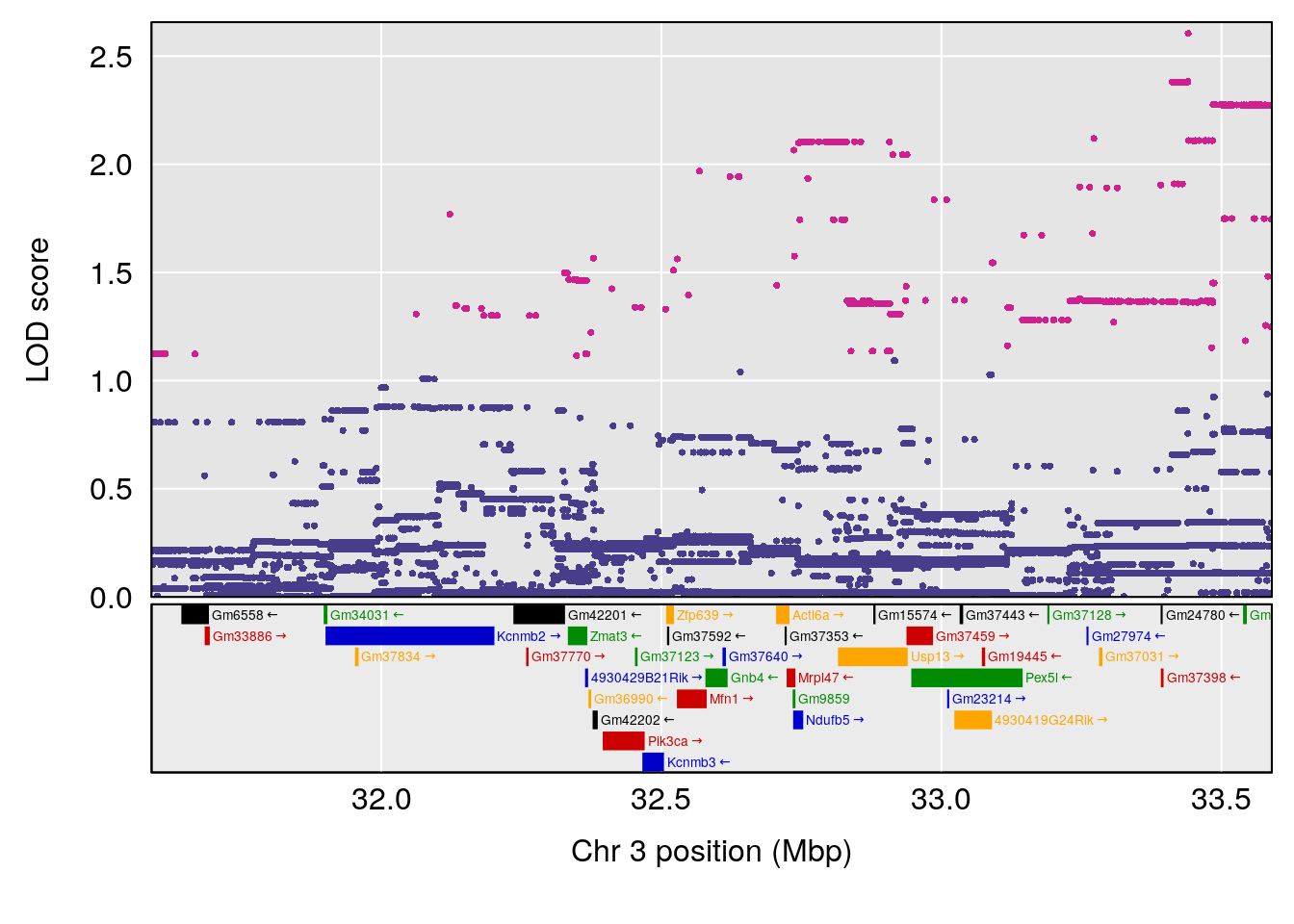
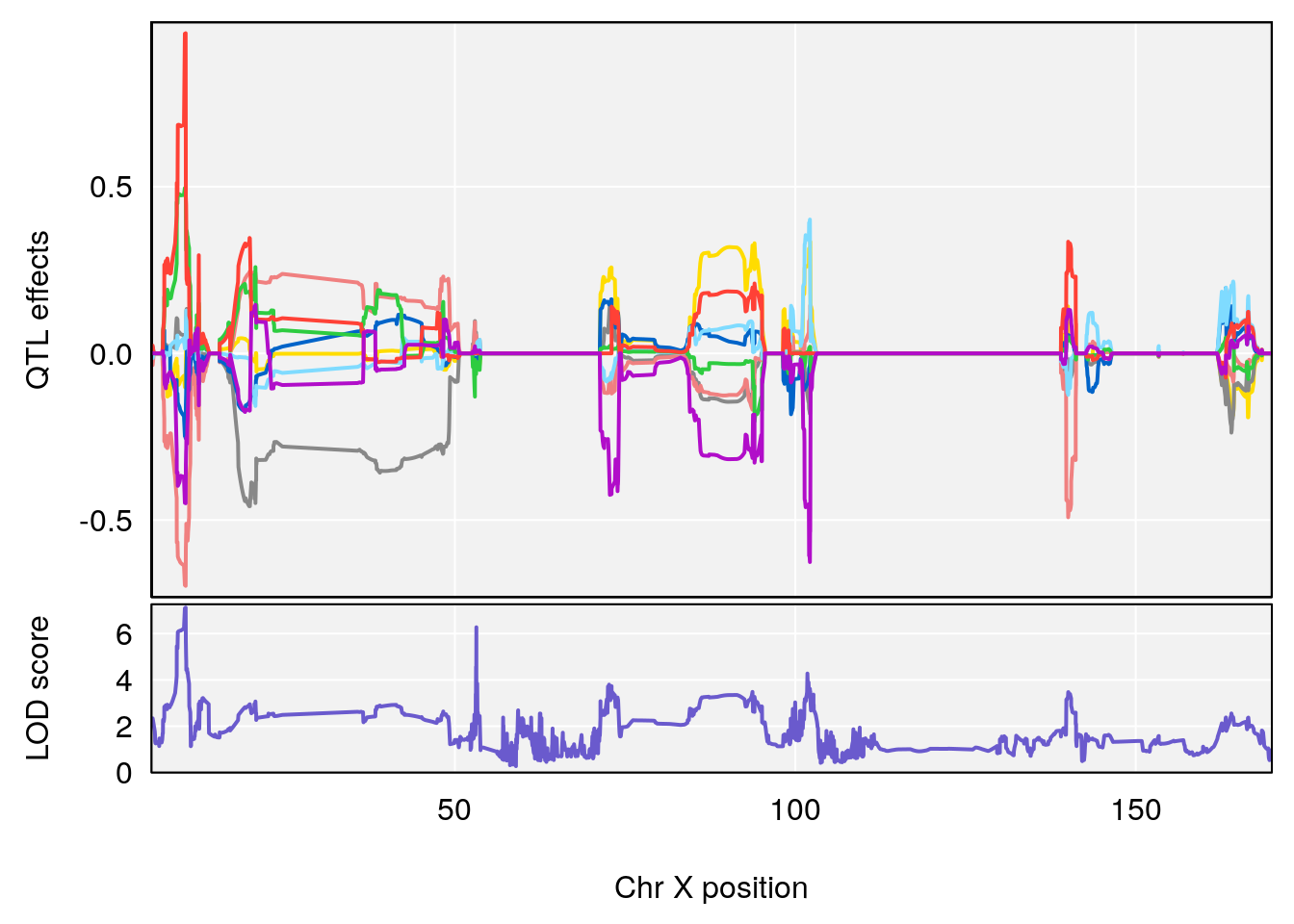
# [1] "RRDepressionRate(%/Hr)SLOPE"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 3 32.58987 6.984733 32.379800 33.07485
# 2 1 pheno1 X 10.46006 7.122931 9.261486 53.15896
# [1] 1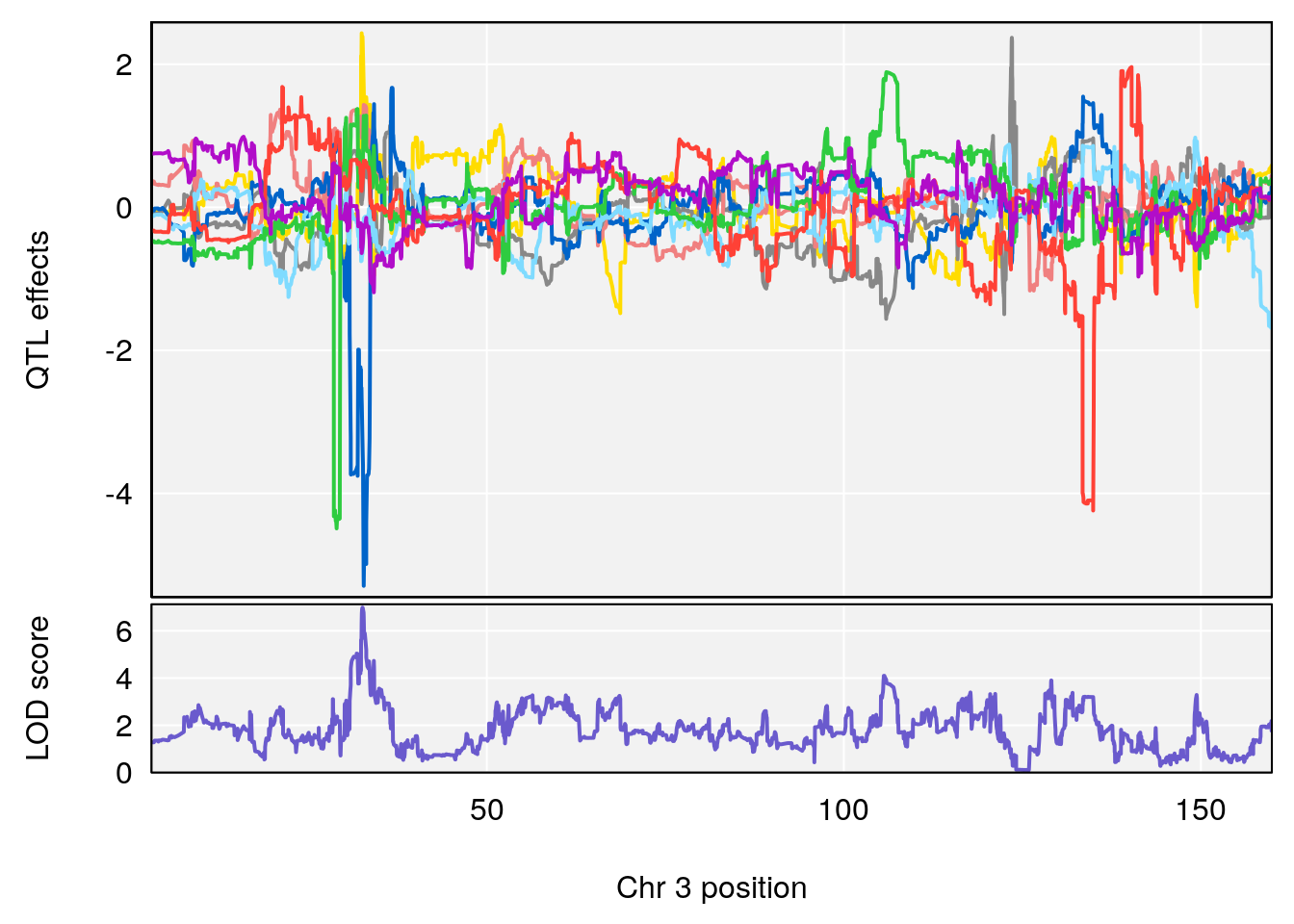
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
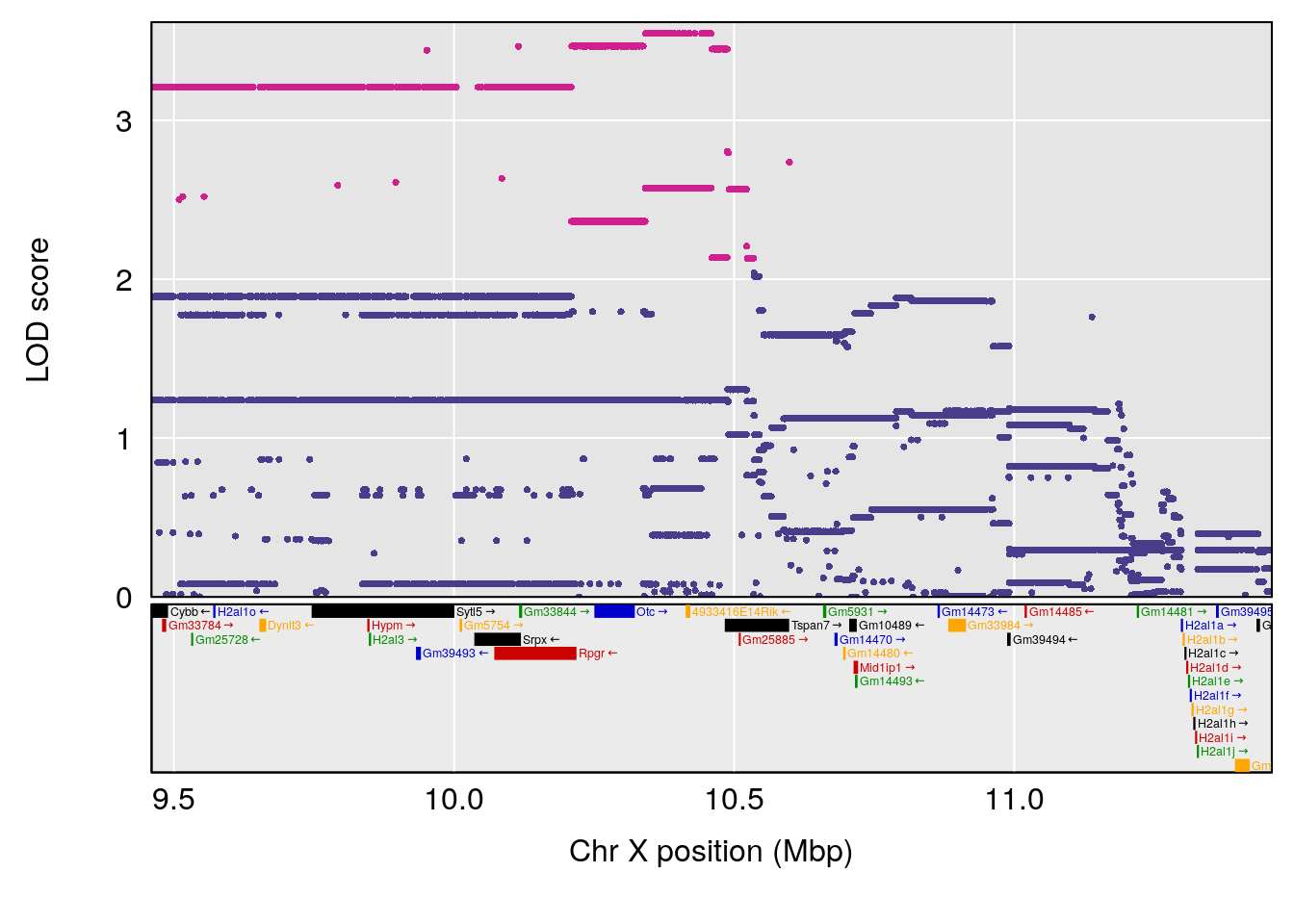
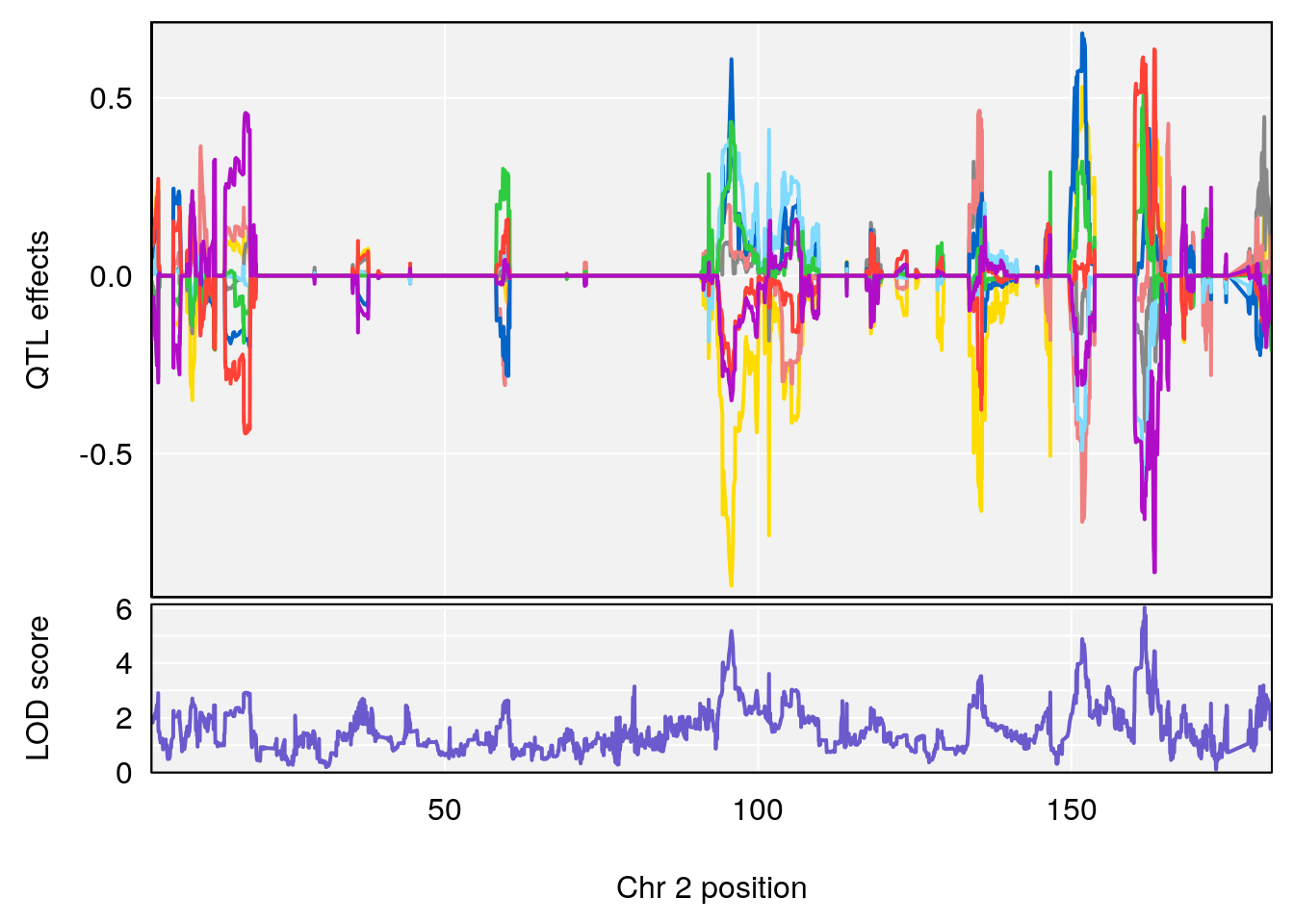
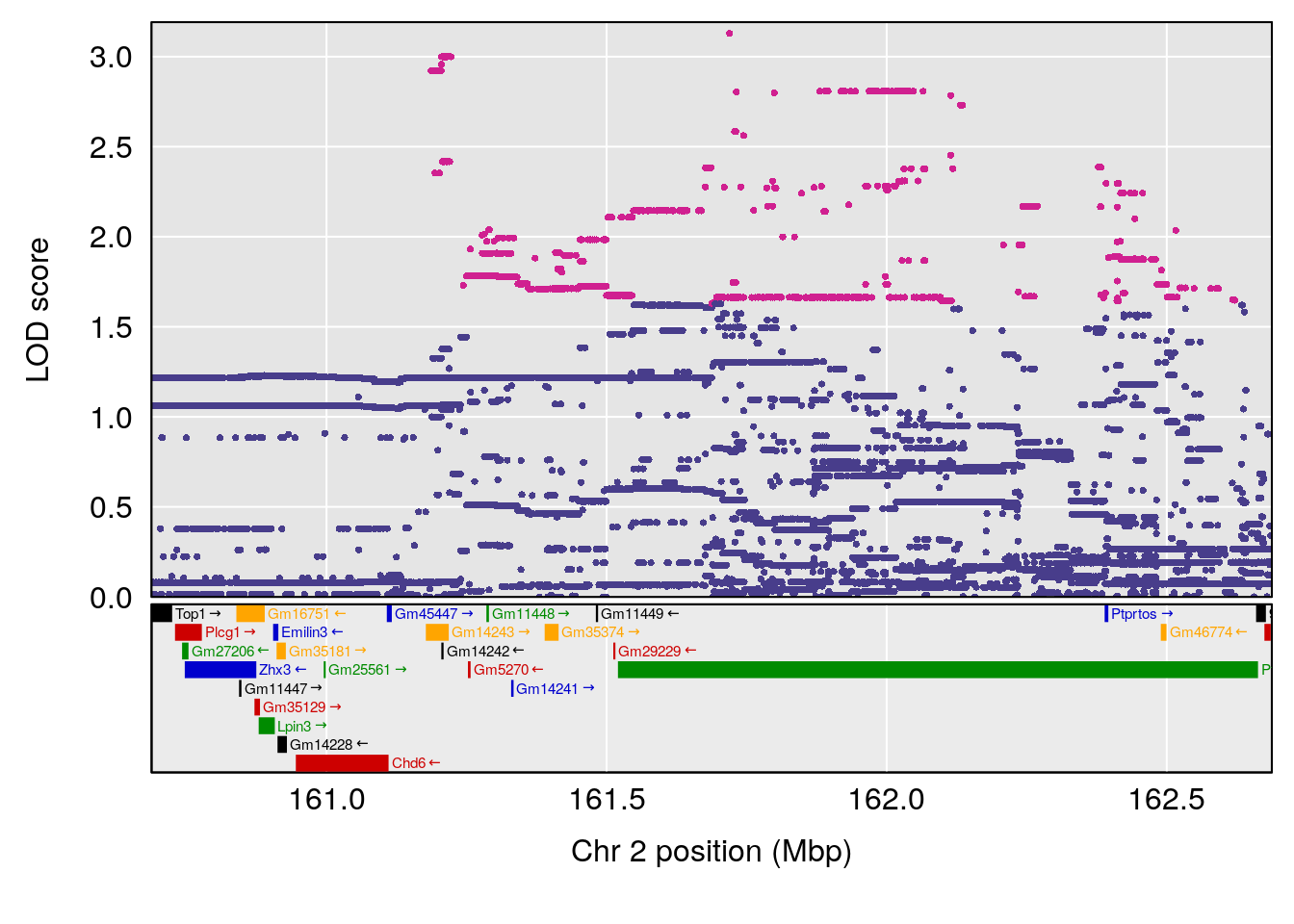
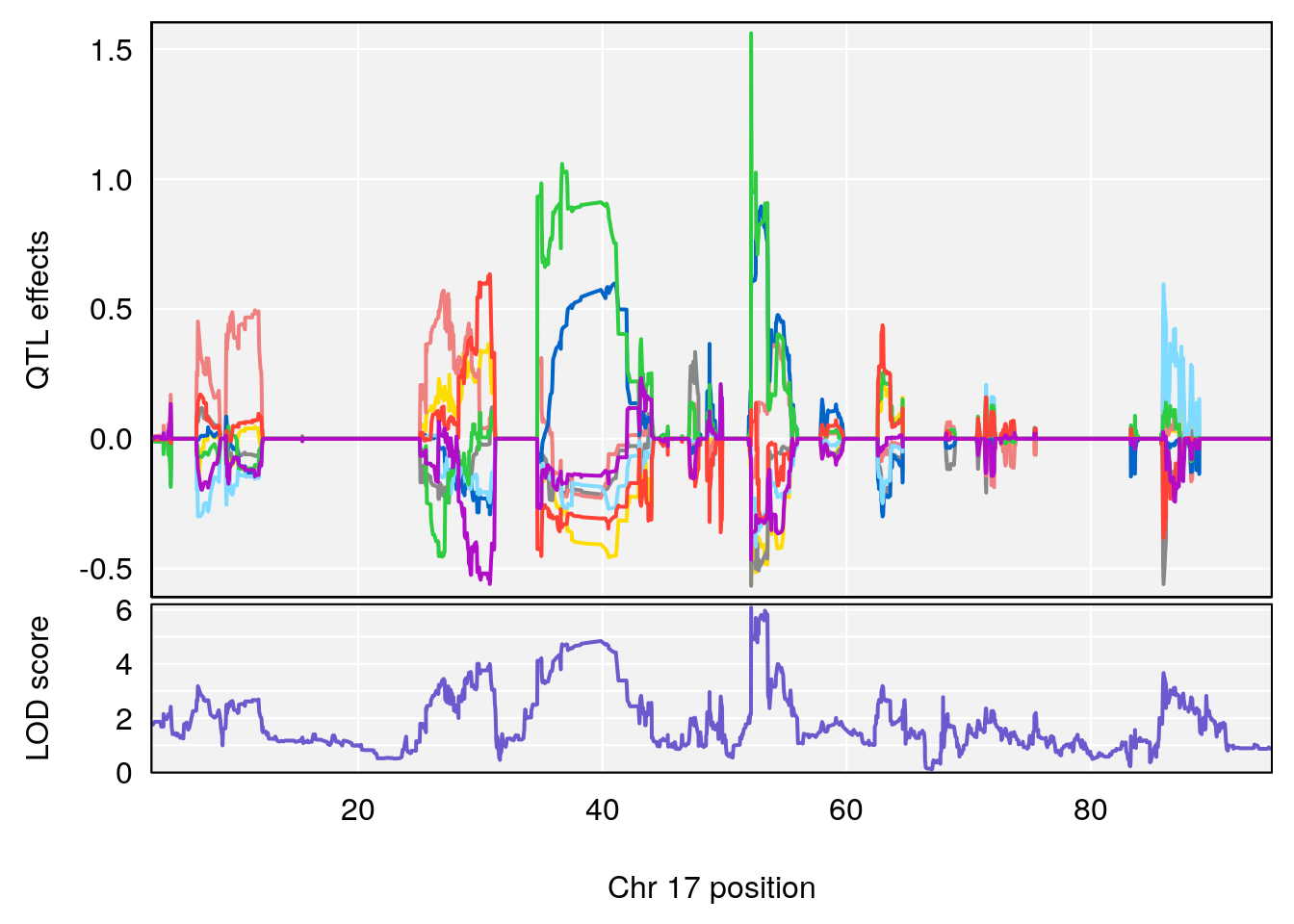
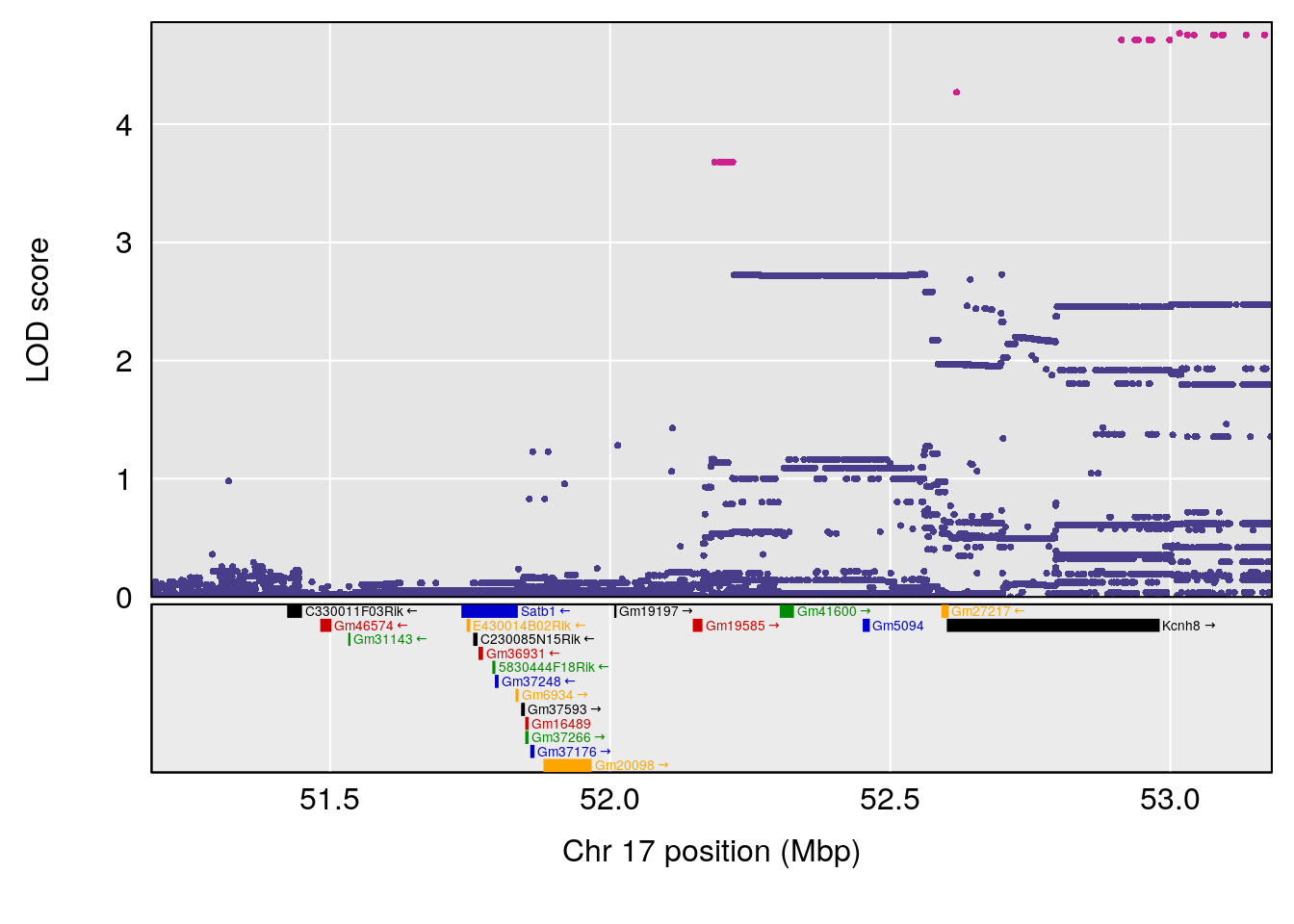
# [1] "TimetoSteadyRRDepression(Hr)"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 2 161.6875 6.026396 95.37519 161.87139
# 2 1 pheno1 17 52.1811 6.074600 36.63825 53.54382
# [1] 1
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2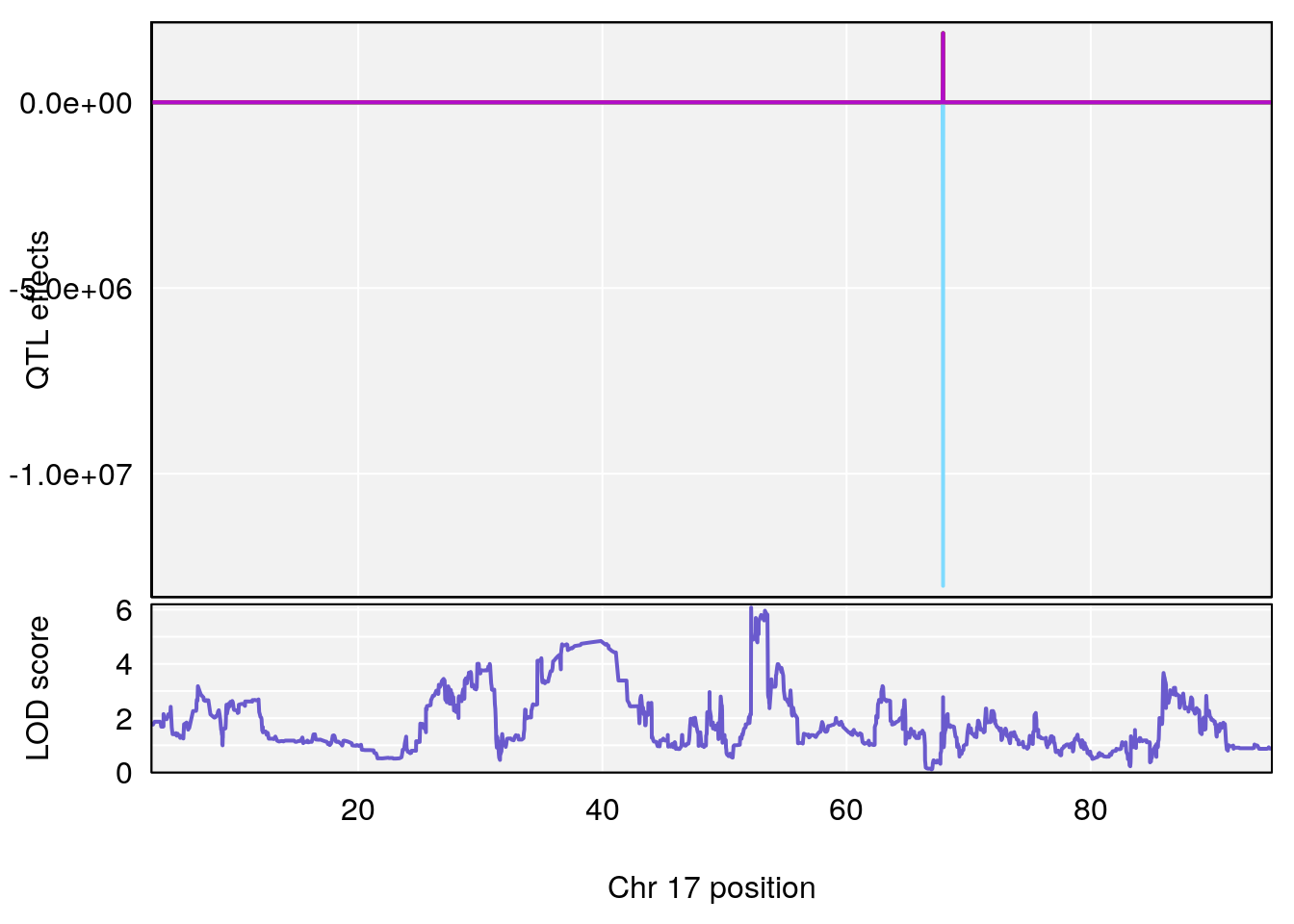
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
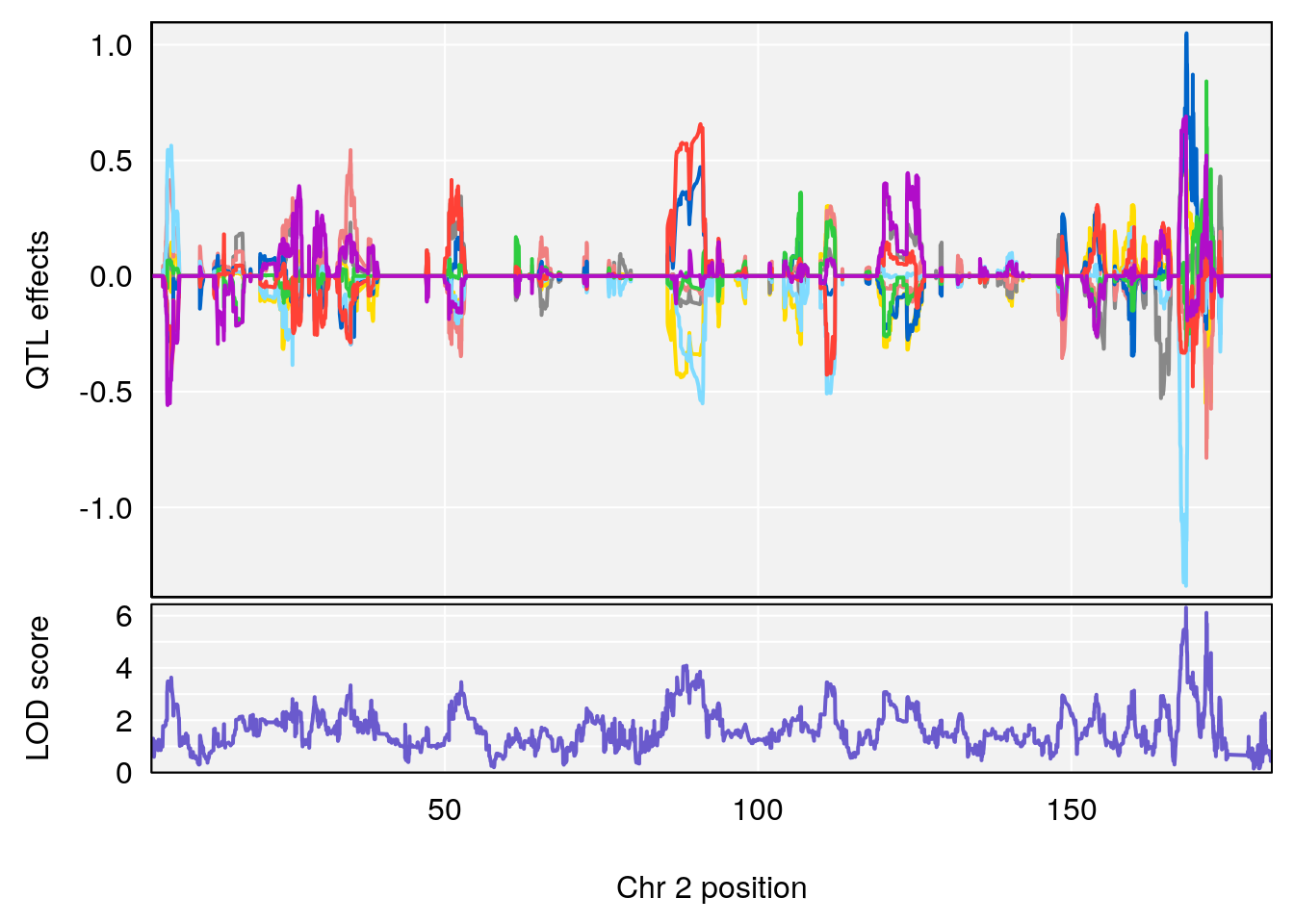
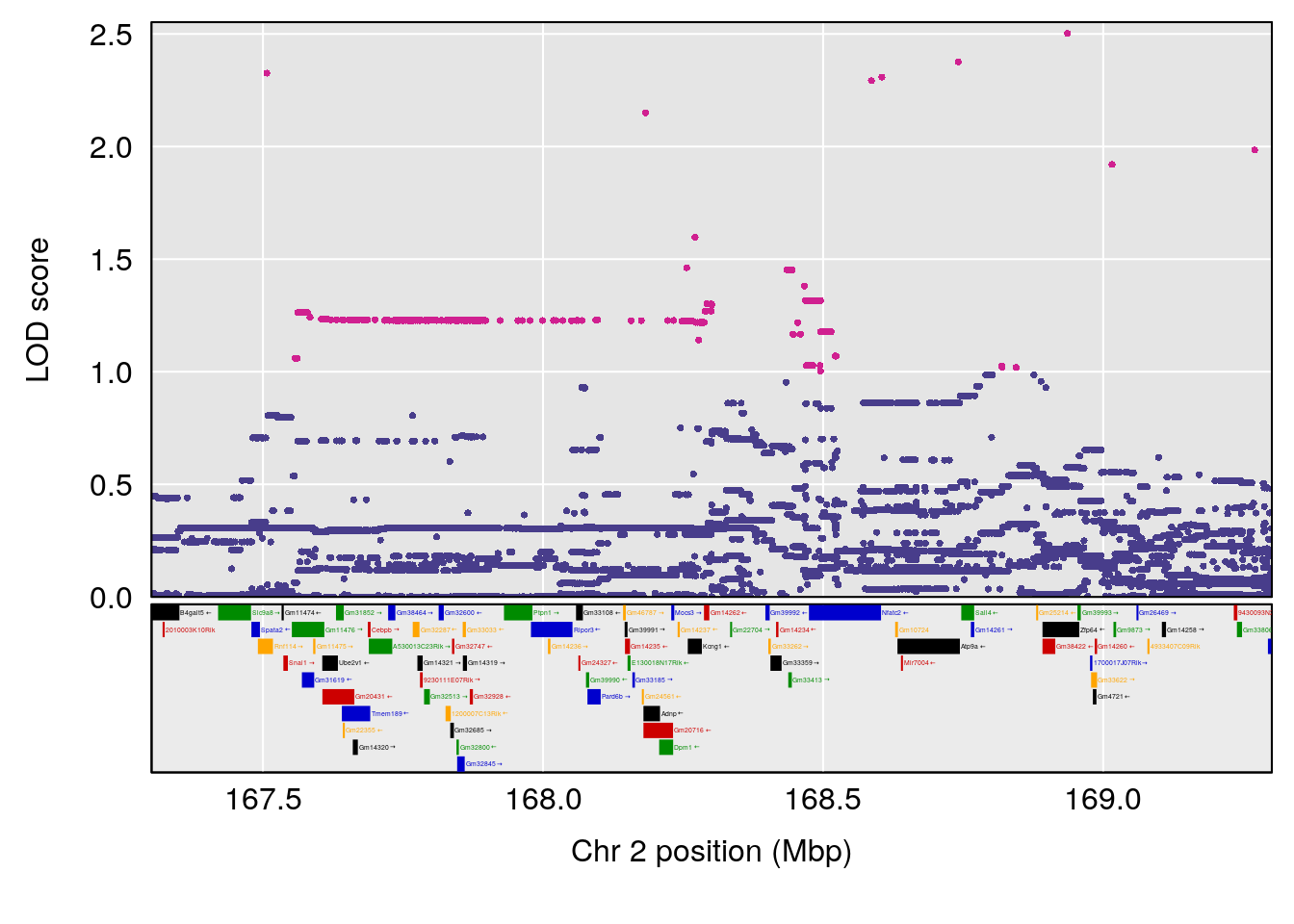
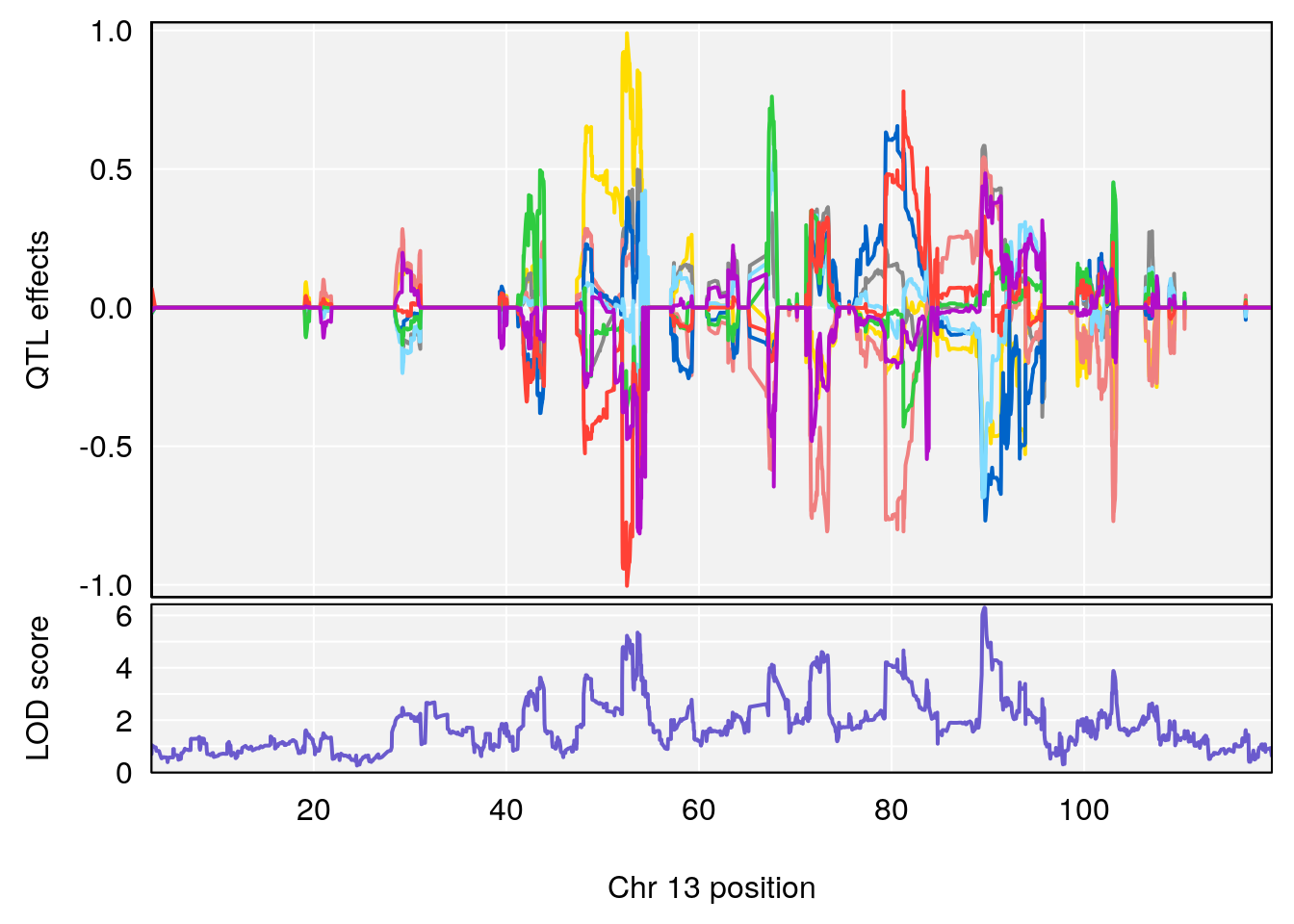
# [1] "MinDepressionRR(%)"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 2 168.30118 6.308975 167.55191 171.64499
# 2 1 pheno1 13 89.64923 6.299108 52.44224 90.31856
# [1] 1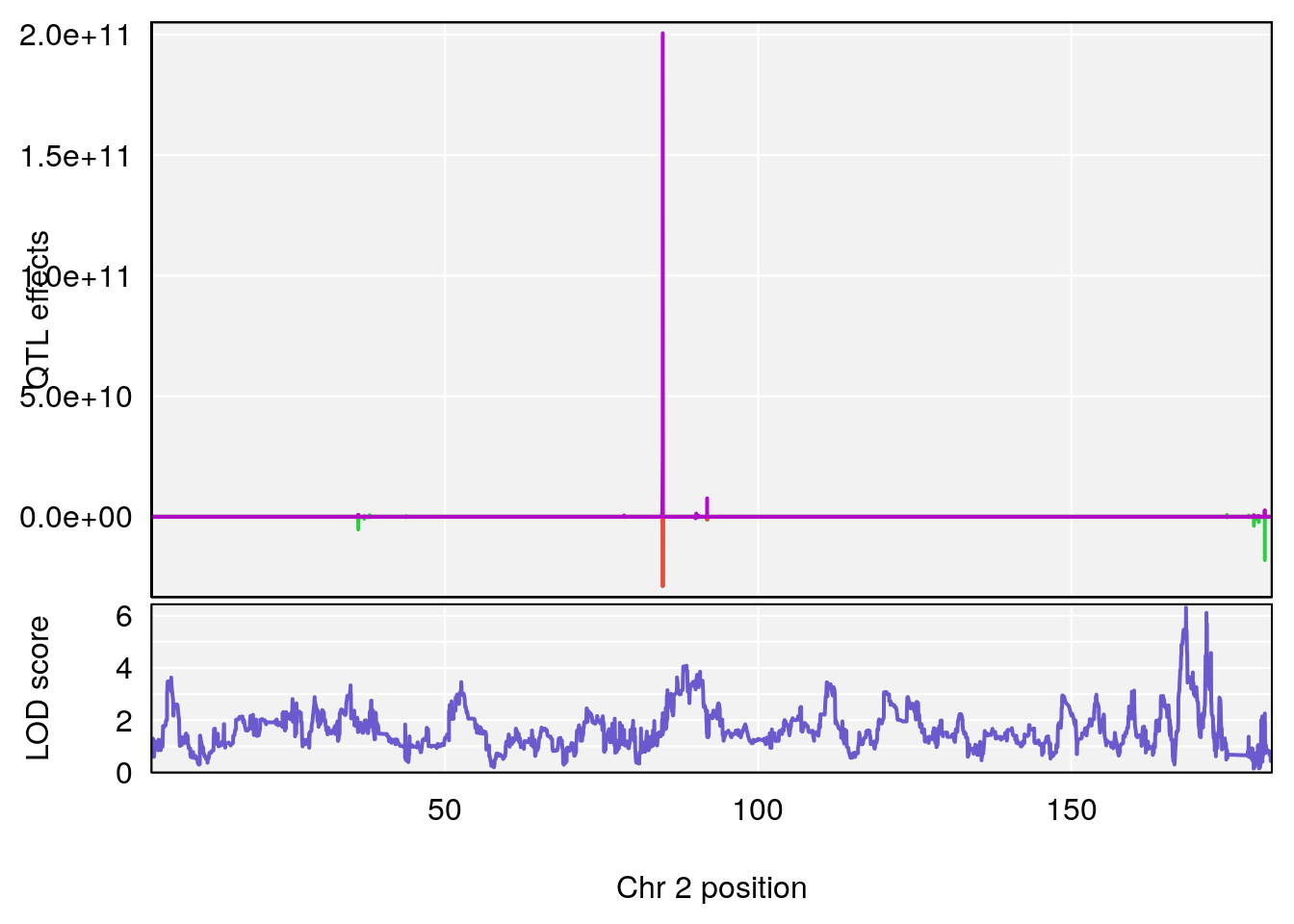
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
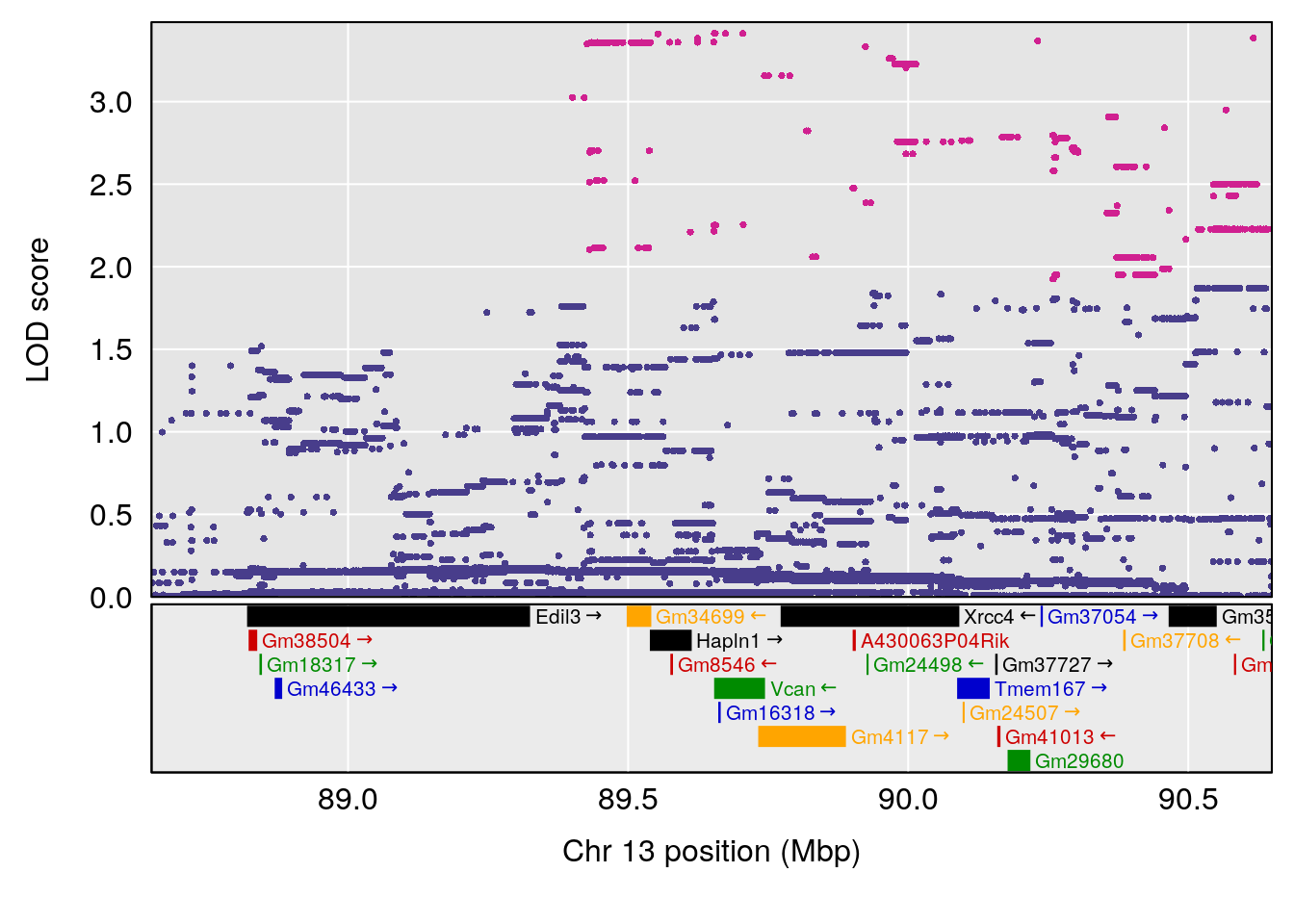
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
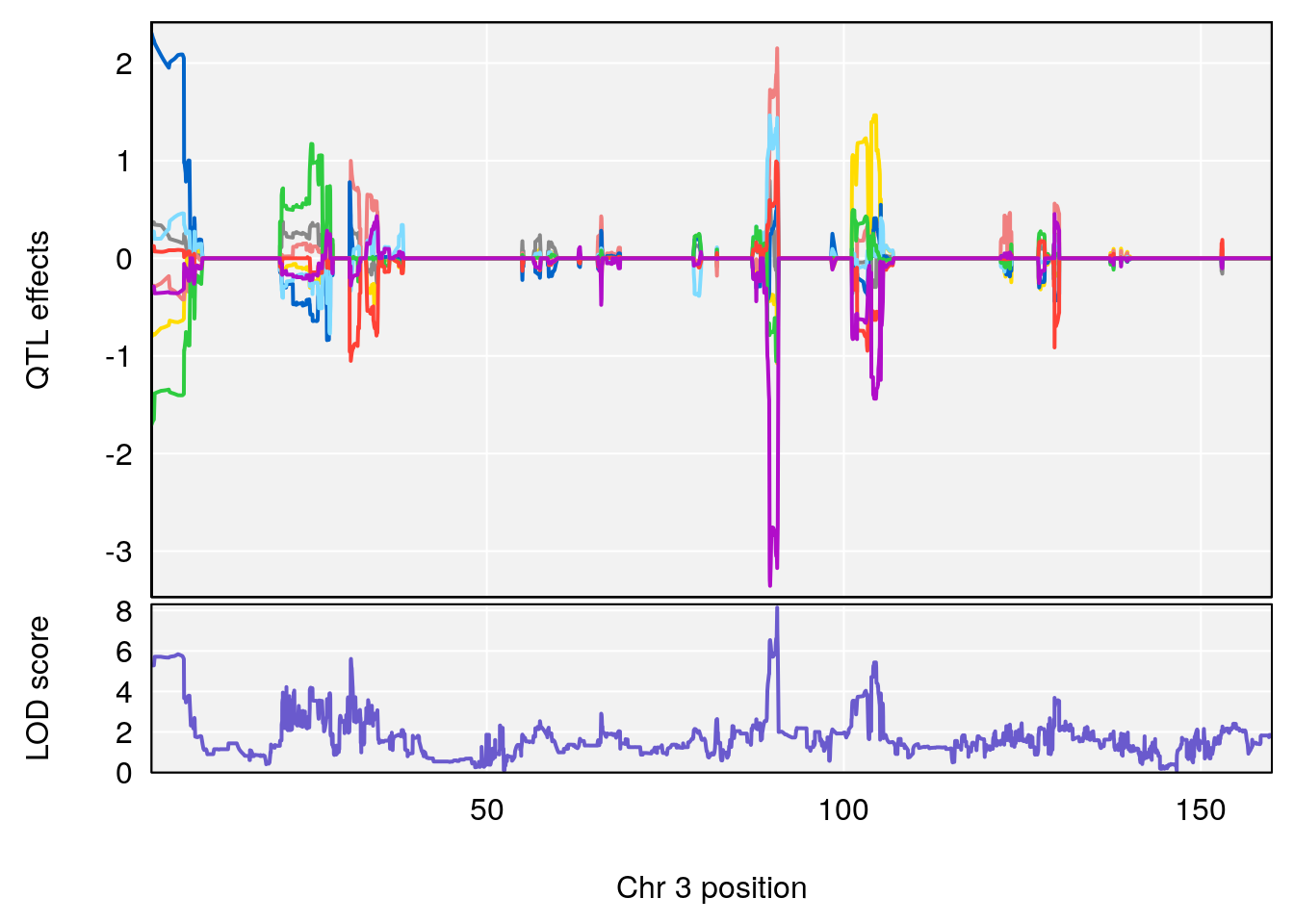
# [1] "Steady-StateDepressionDuration(Hr)INTERVAL"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 3 90.66061 8.142433 90.57609 90.70492
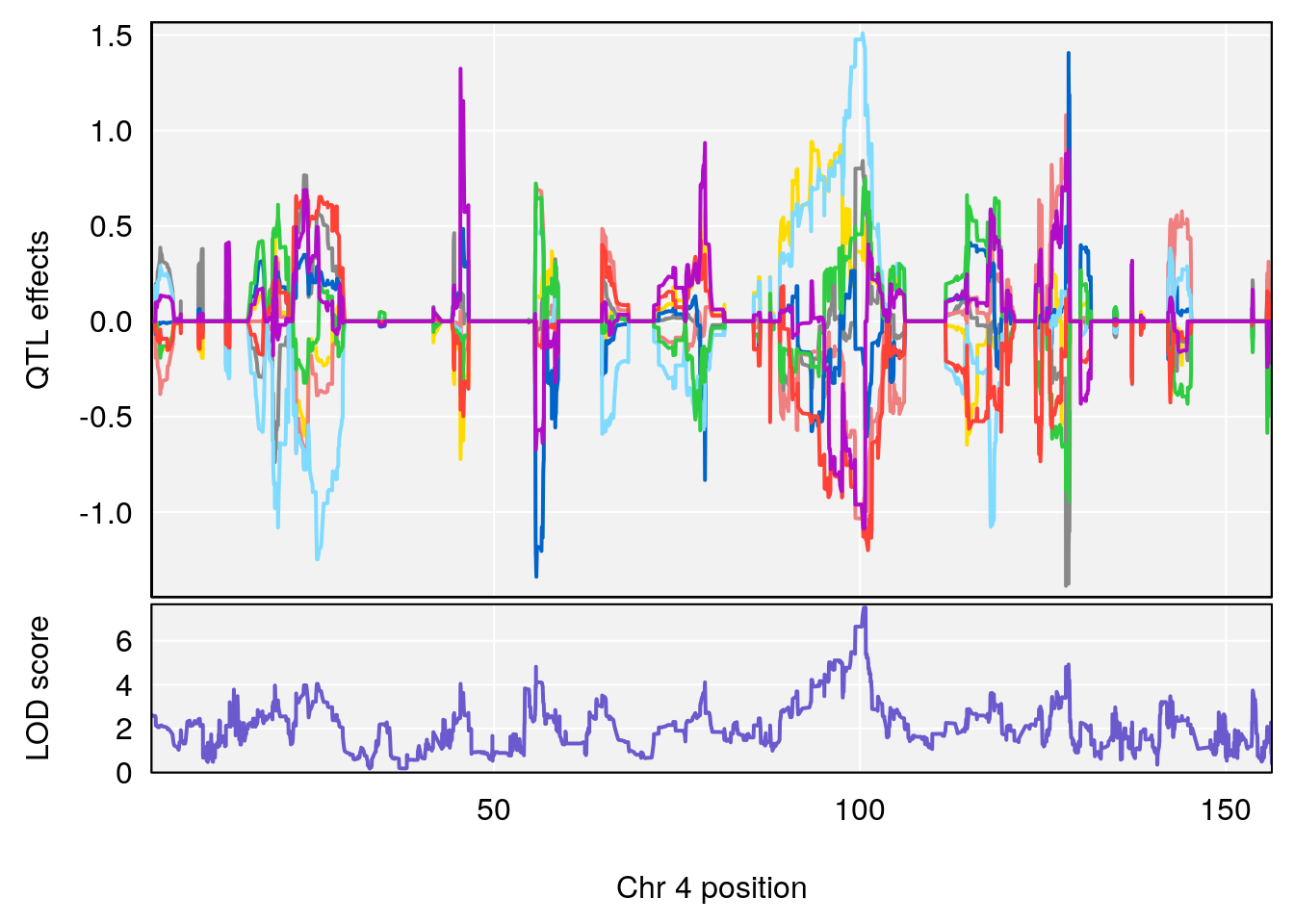
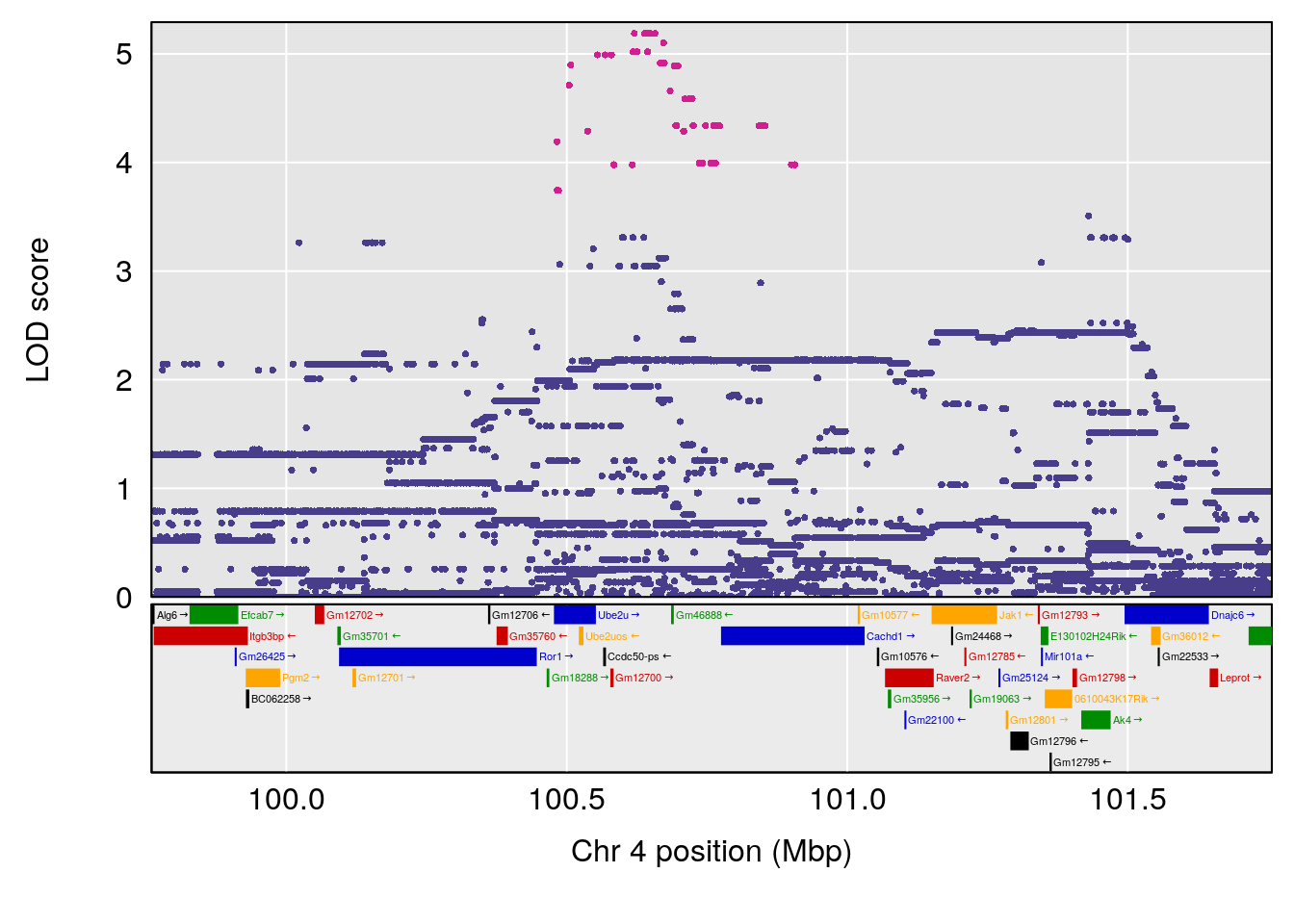
# 2 1 pheno1 4 100.75953 7.510841 99.32926 100.79381
# 3 1 pheno1 8 83.45834 6.695546 83.35490 84.22023
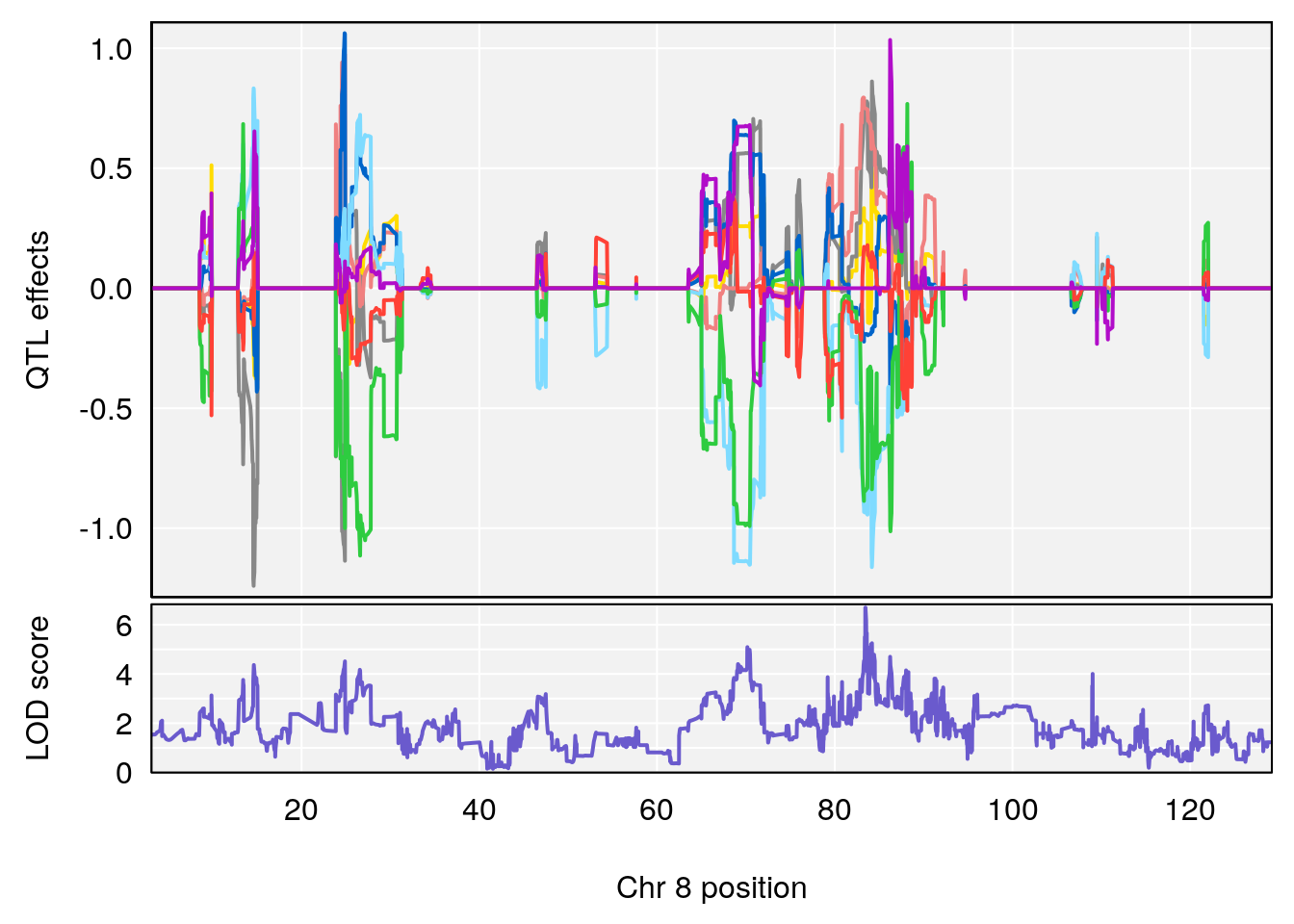
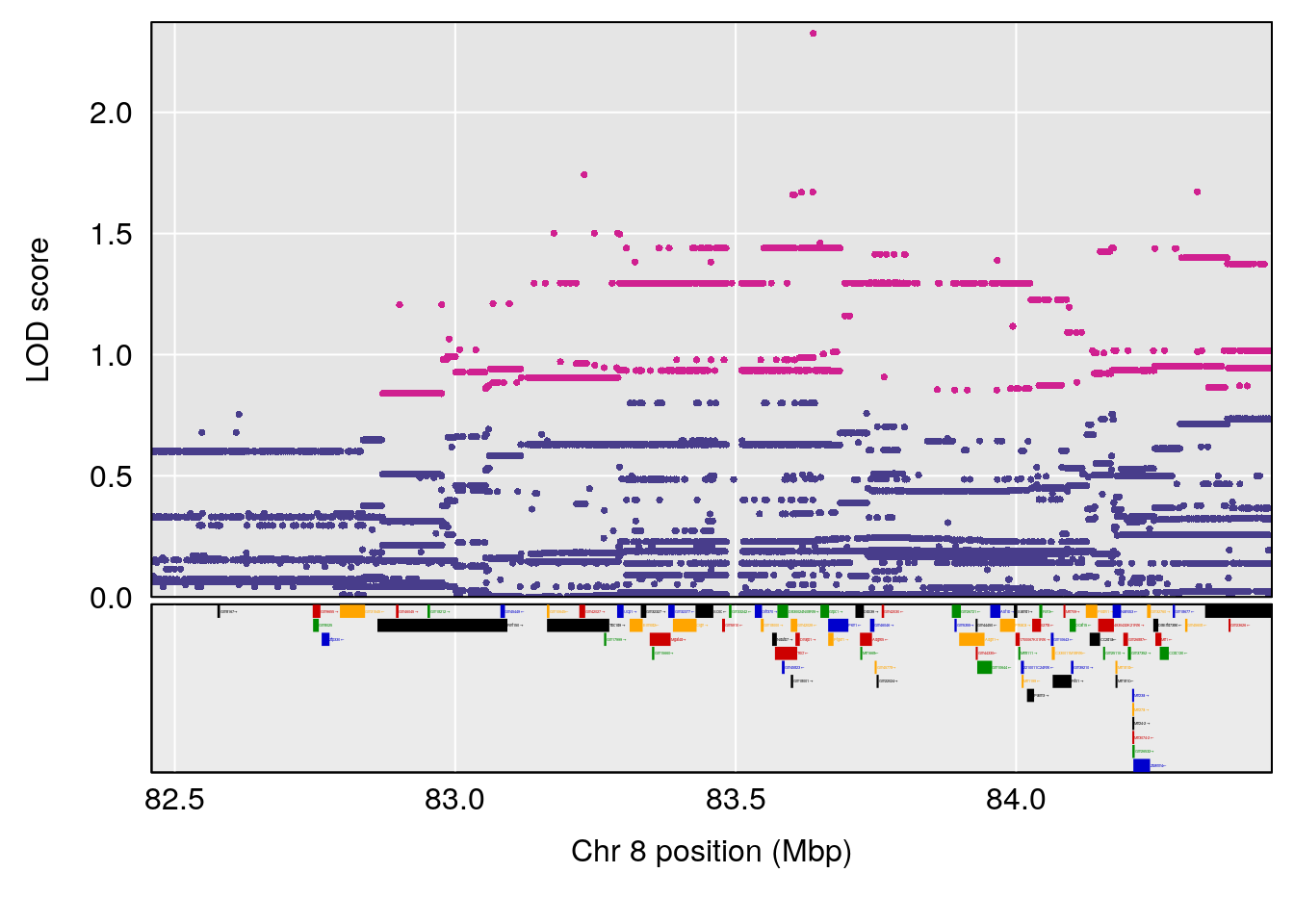
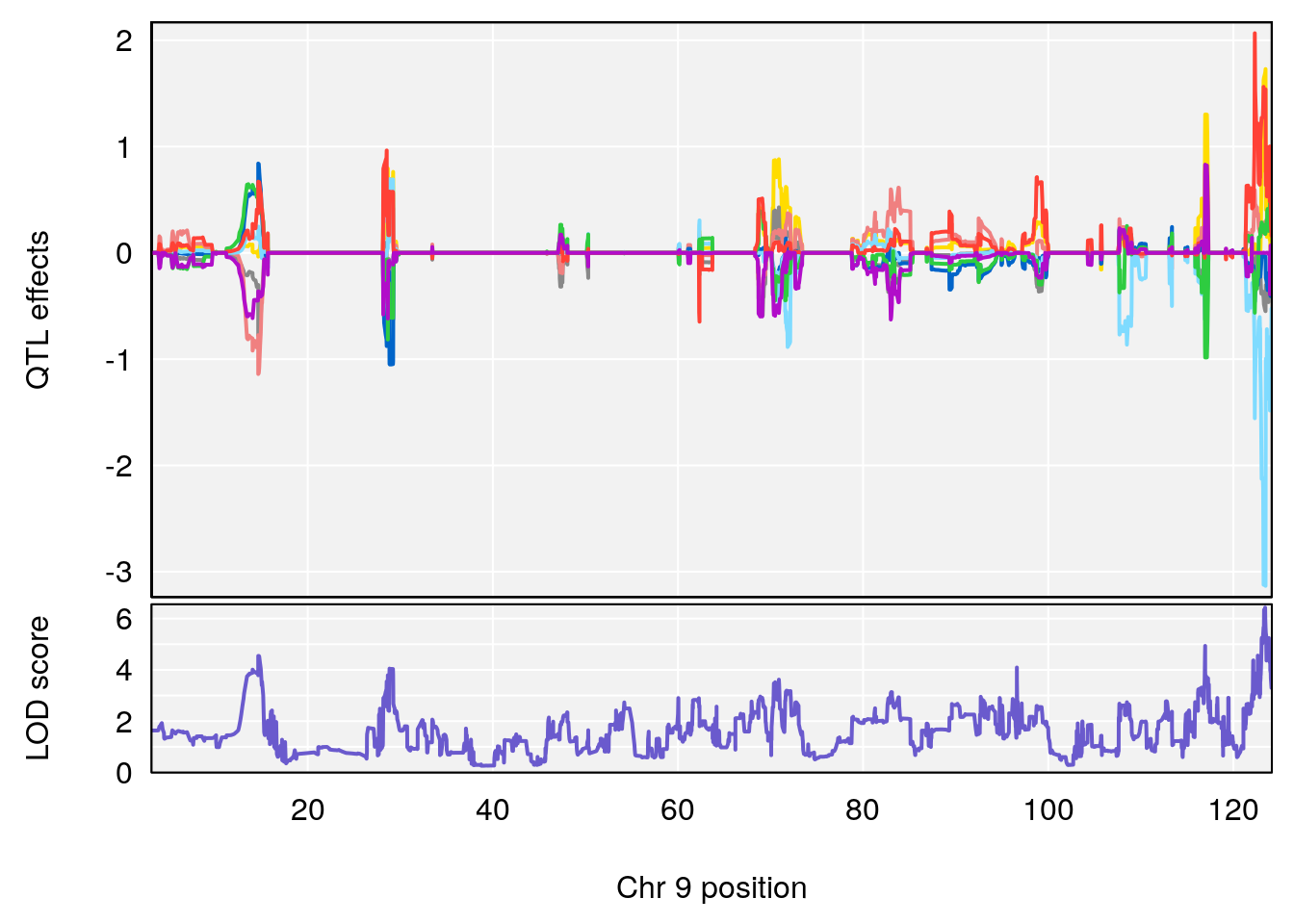
# 4 1 pheno1 9 123.42827 6.428620 116.93394 123.85184
# 5 1 pheno1 10 66.55650 6.997621 64.45014 69.65303
# 6 1 pheno1 12 75.98747 7.912513 75.95610 94.21761
# 7 1 pheno1 13 93.91029 6.097764 42.38805 95.63661
# [1] 1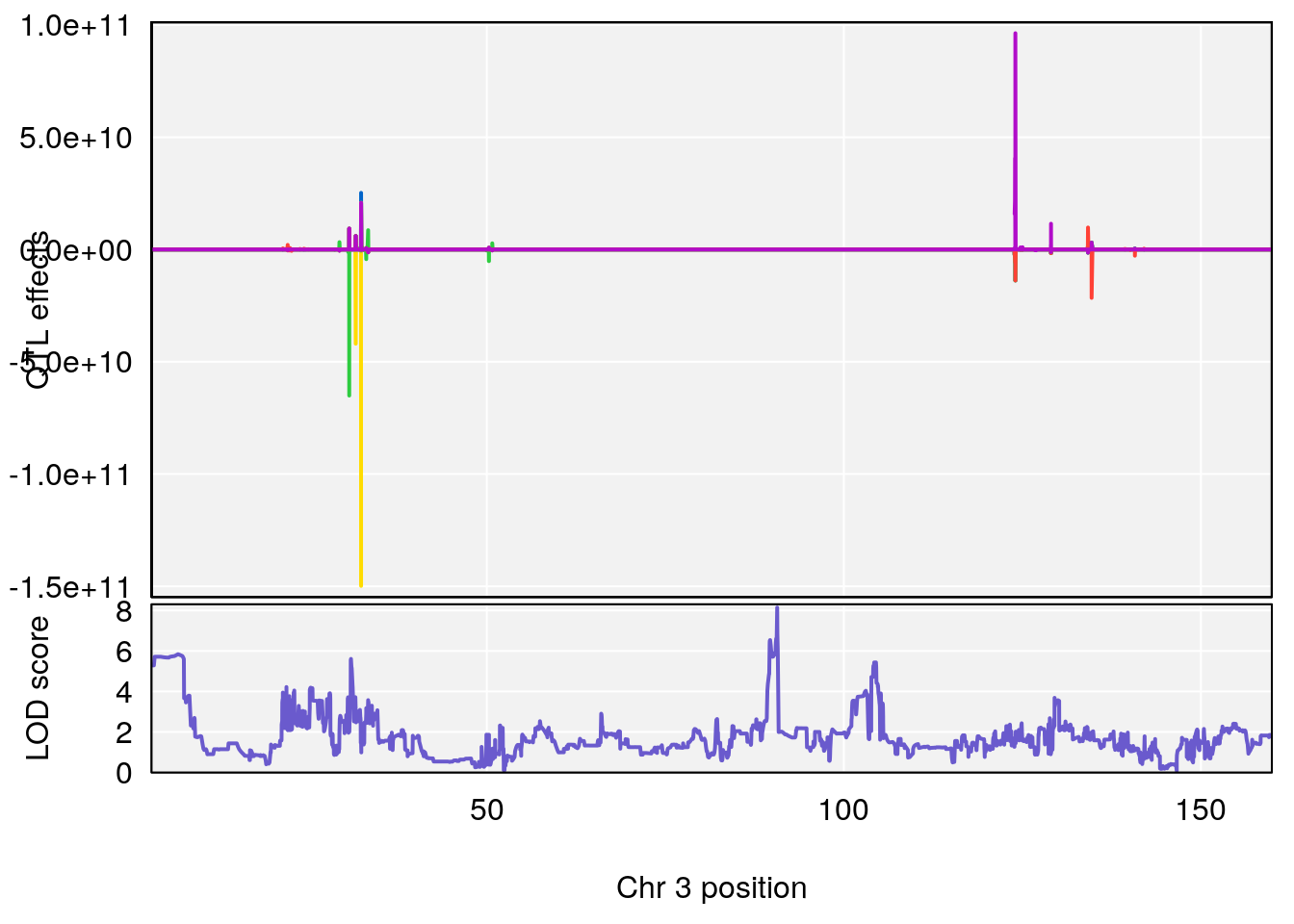
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
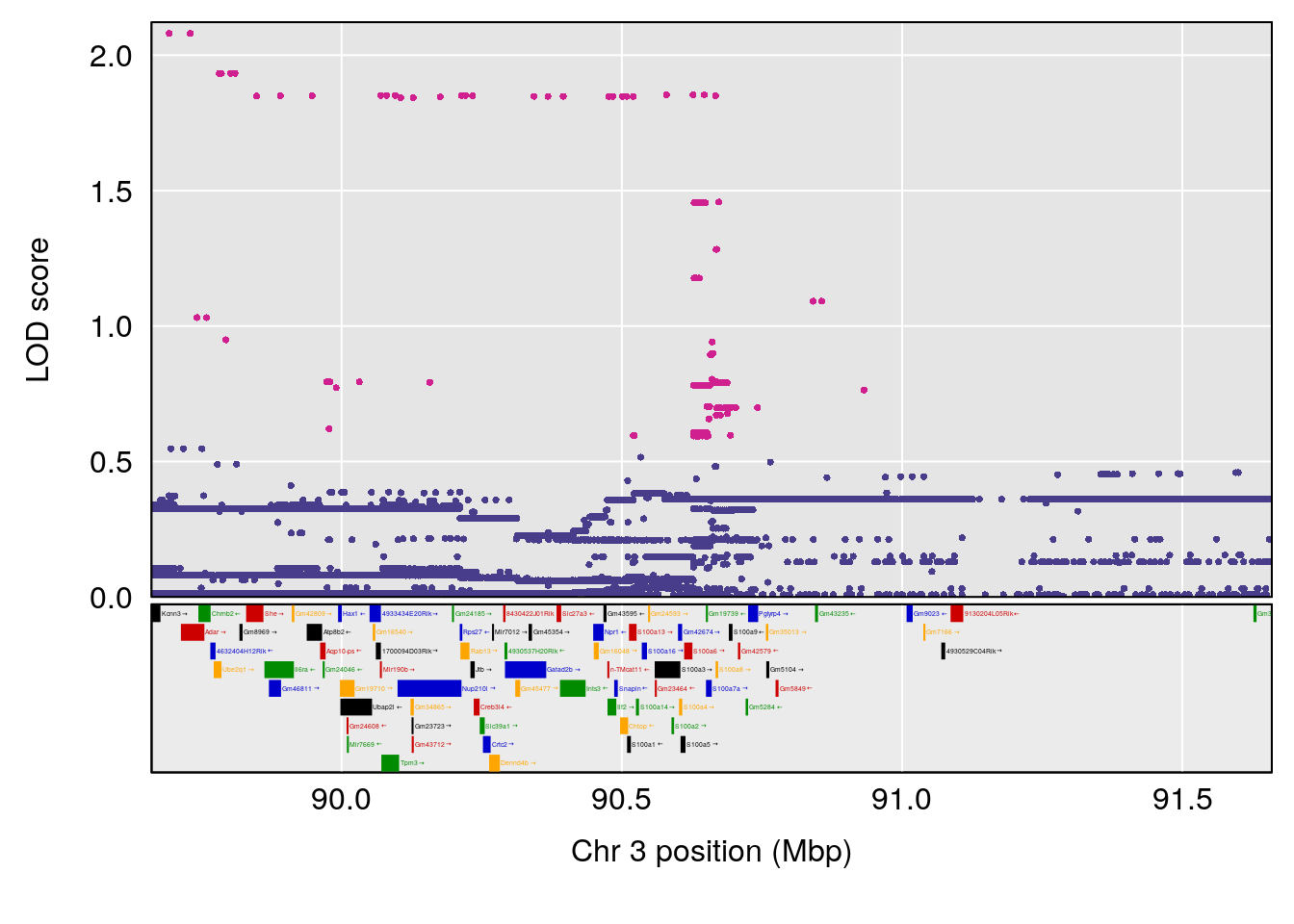
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2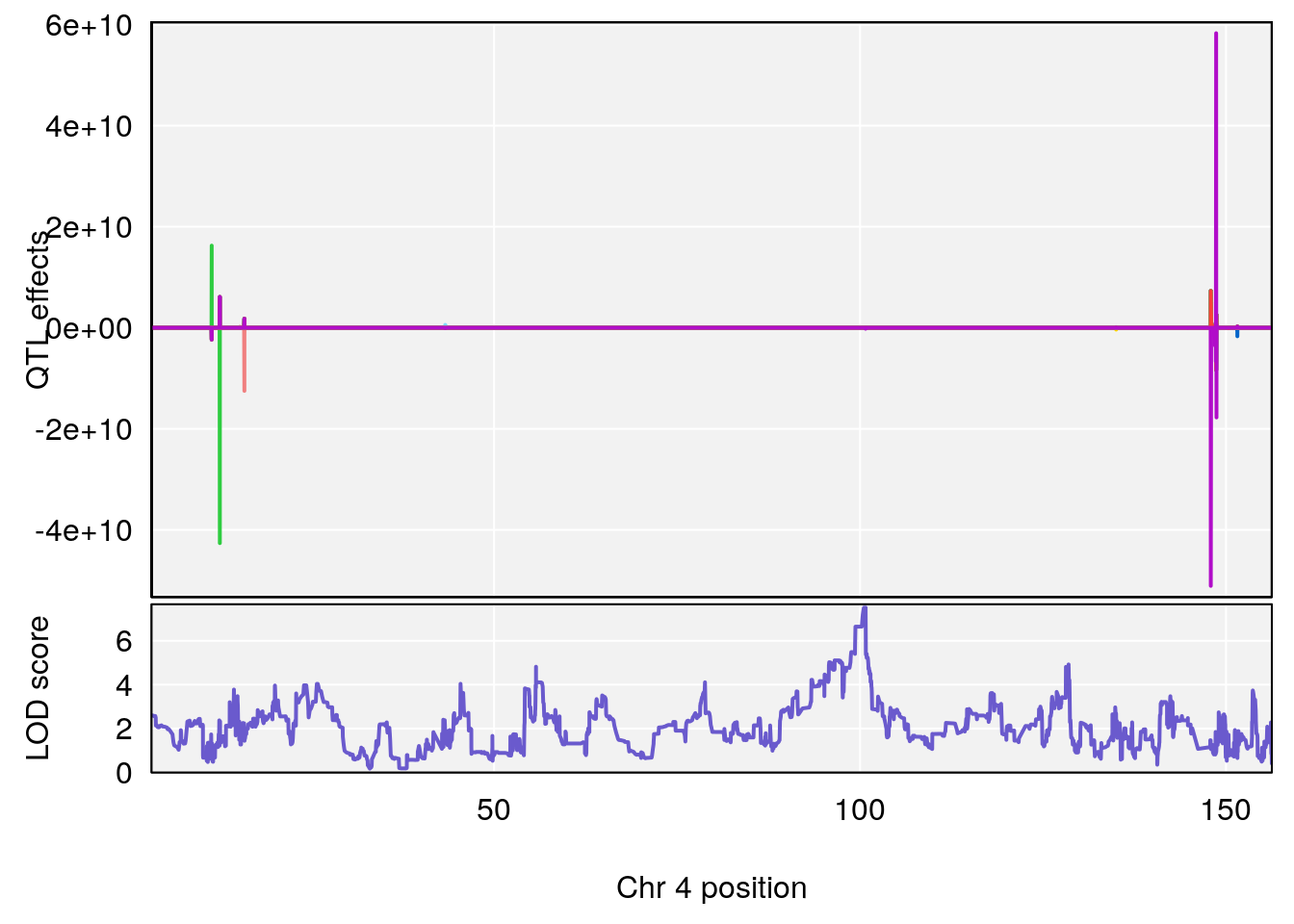
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 3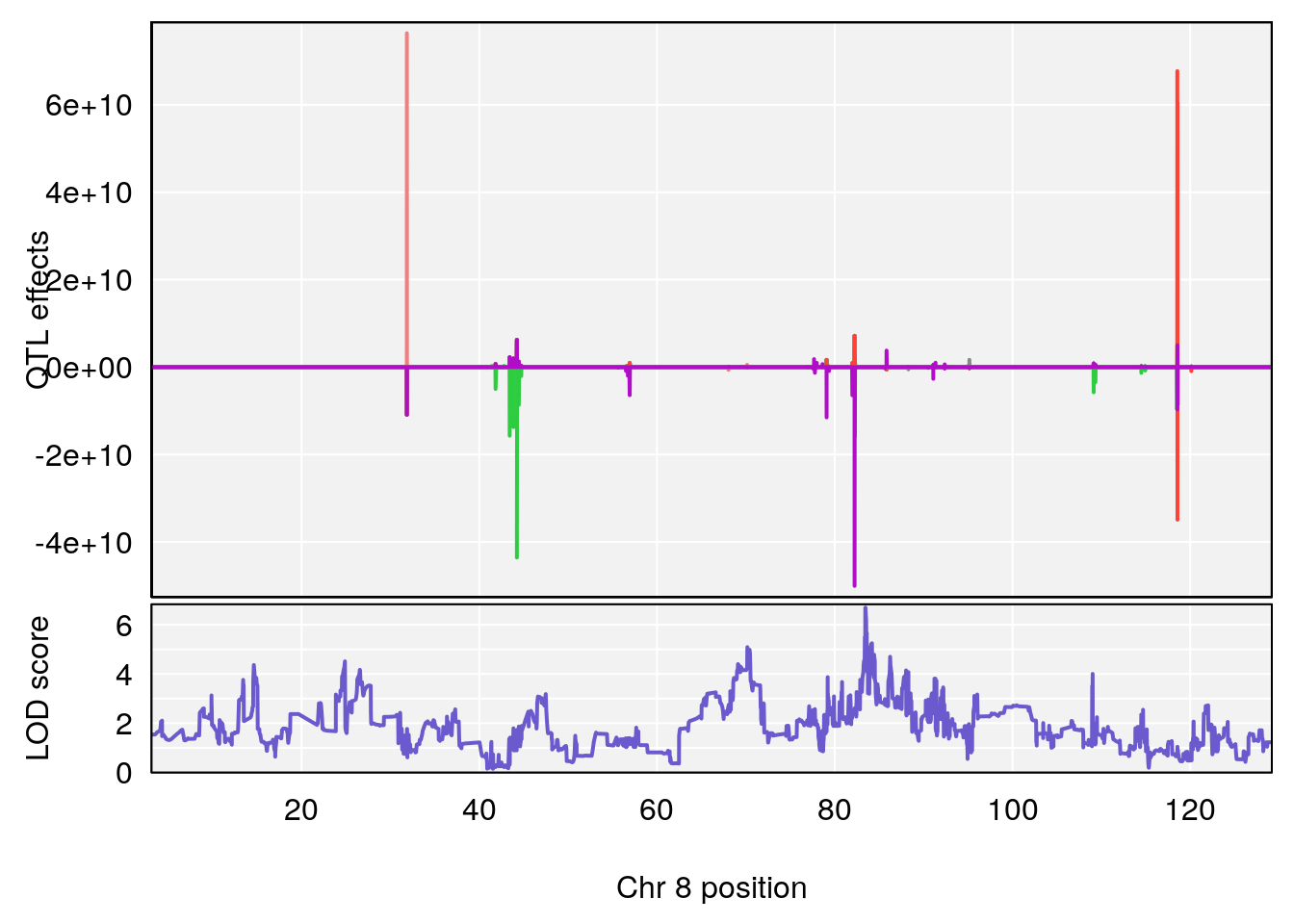
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 4
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
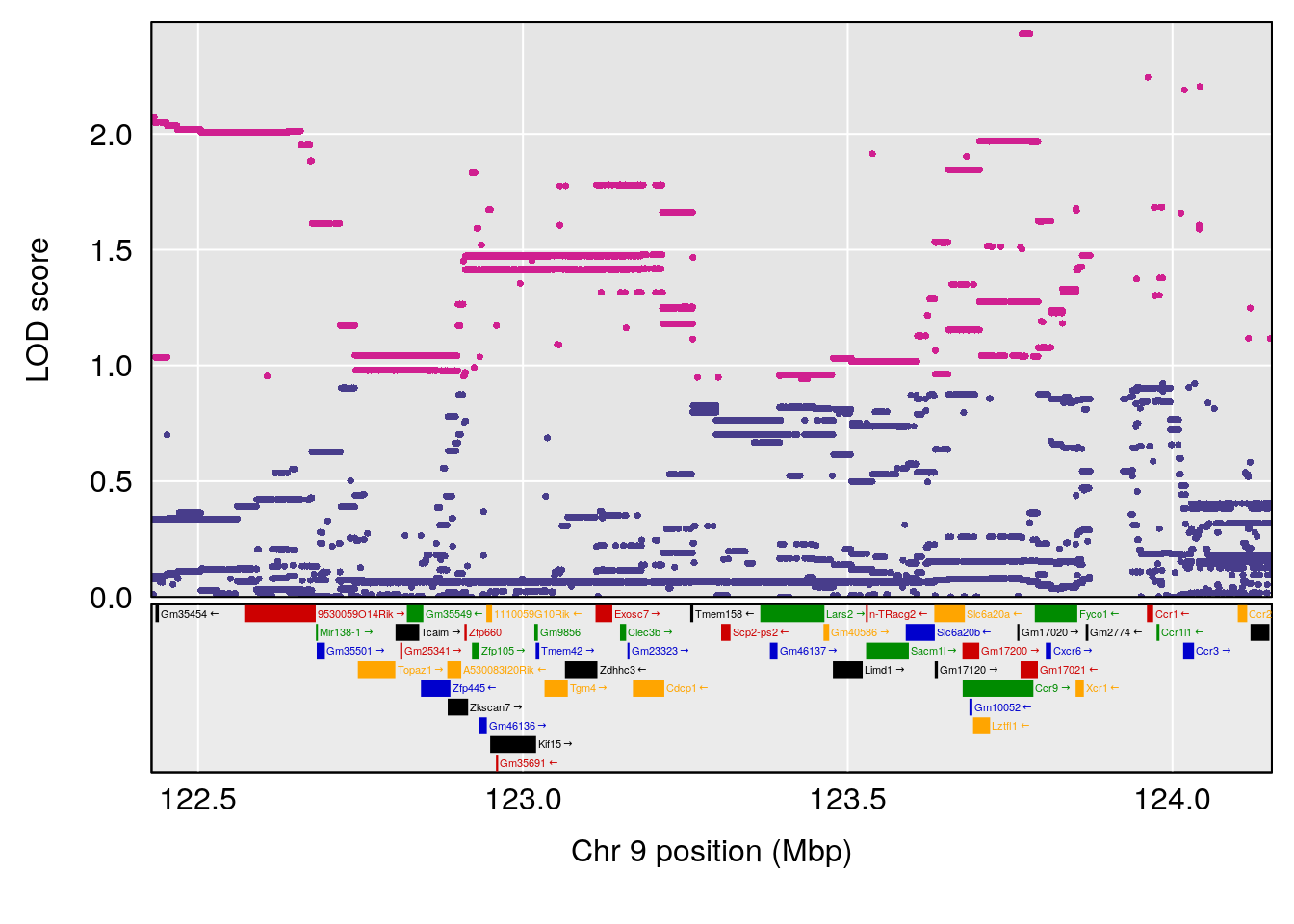
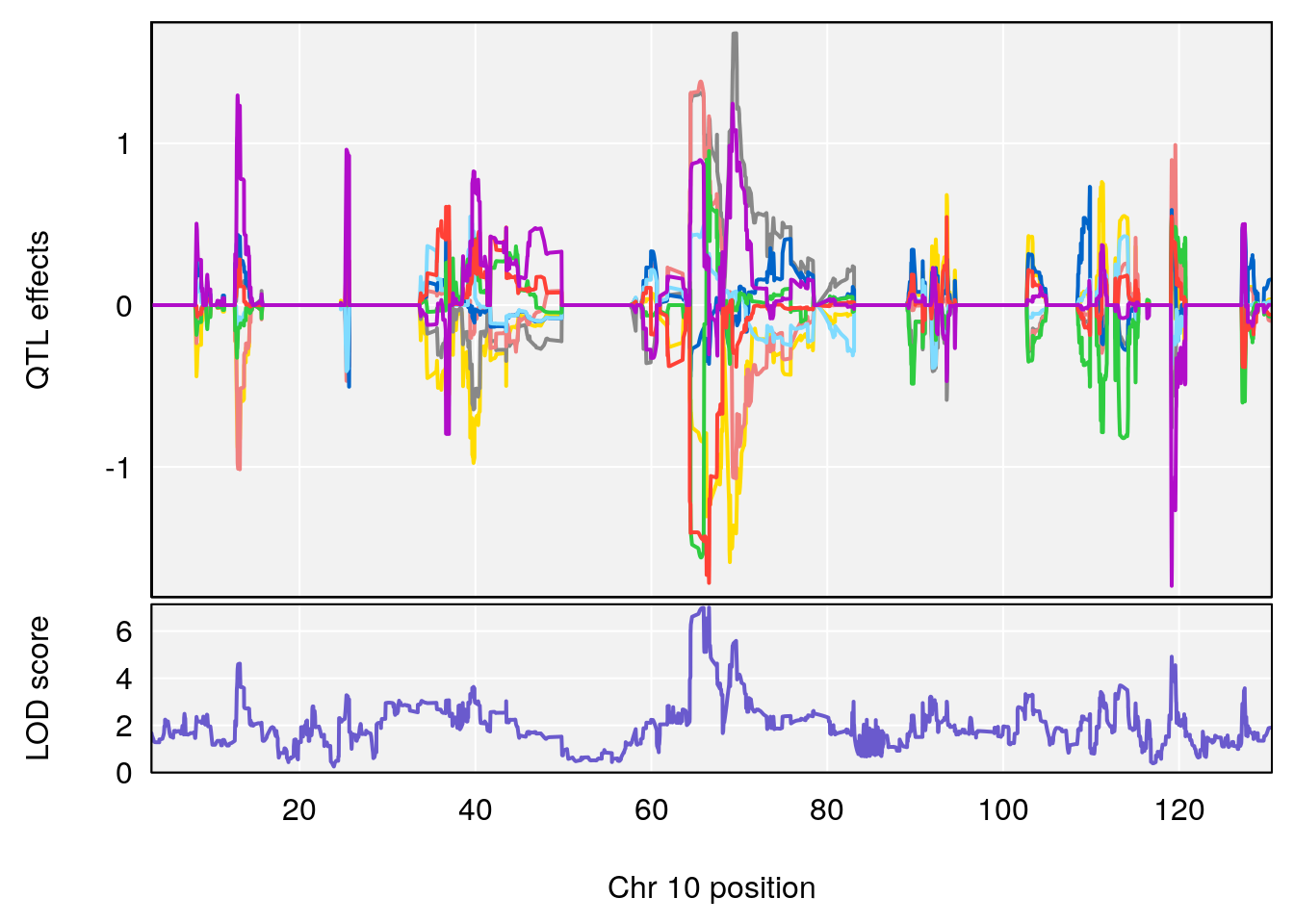
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
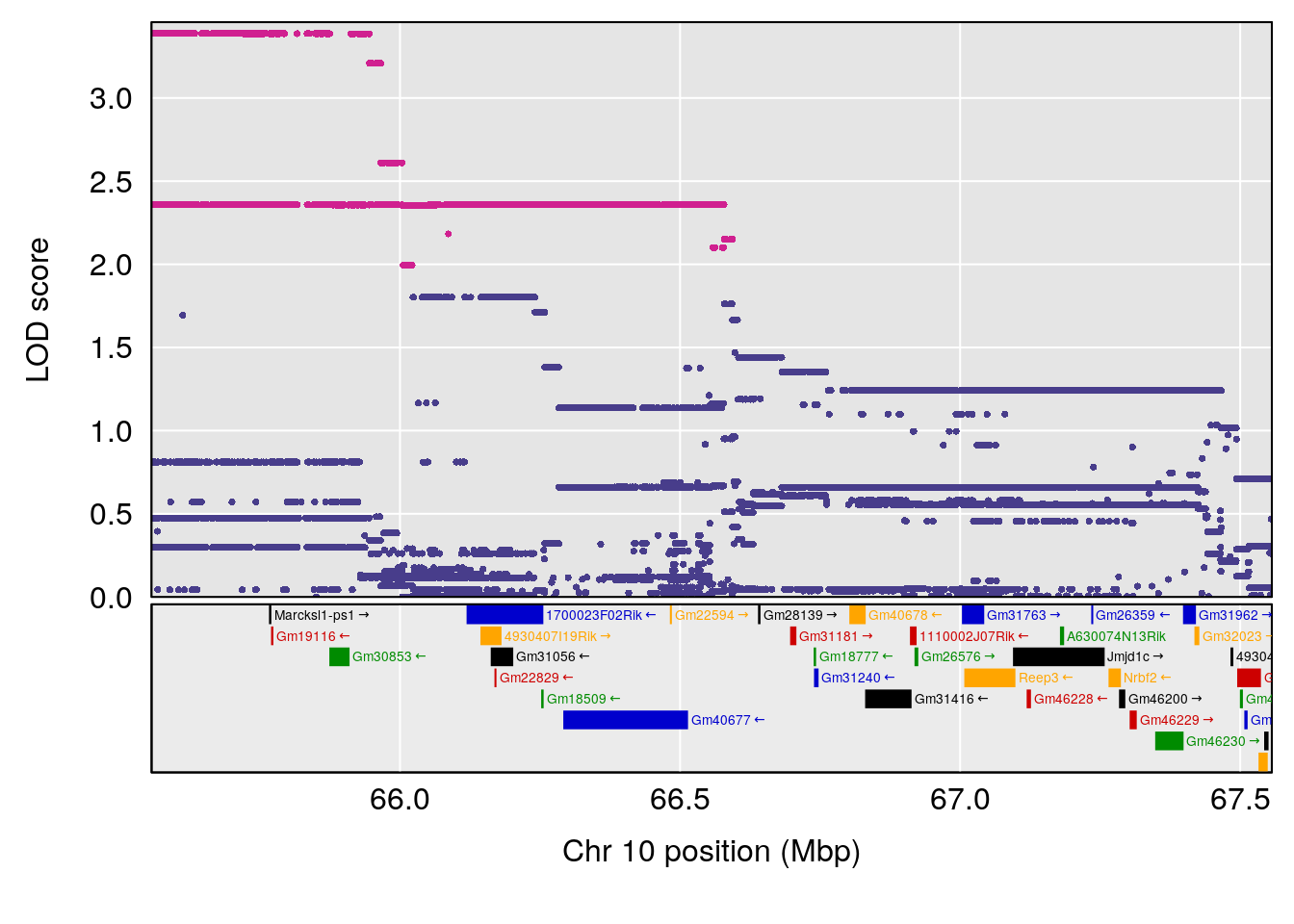
# [1] 5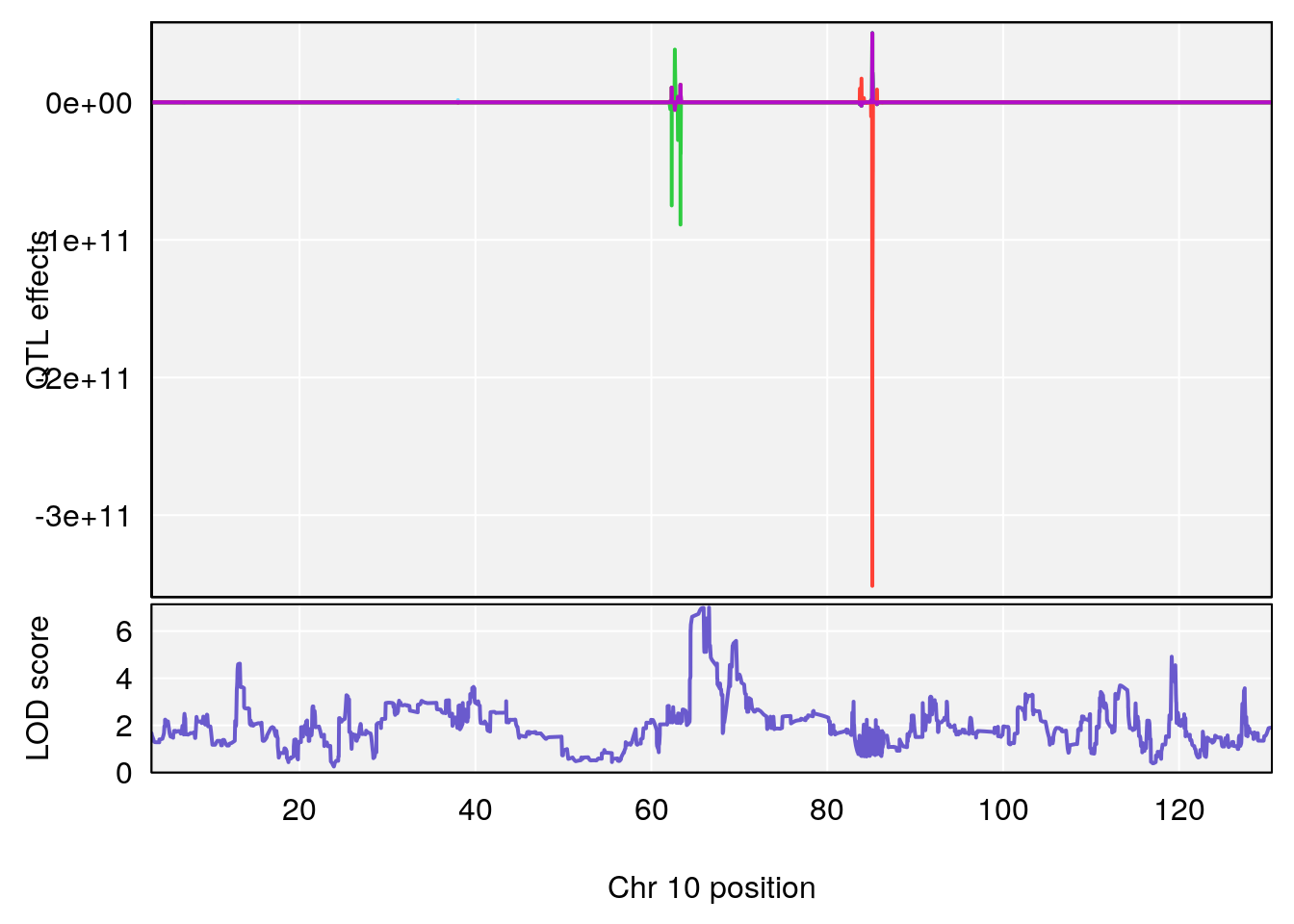
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
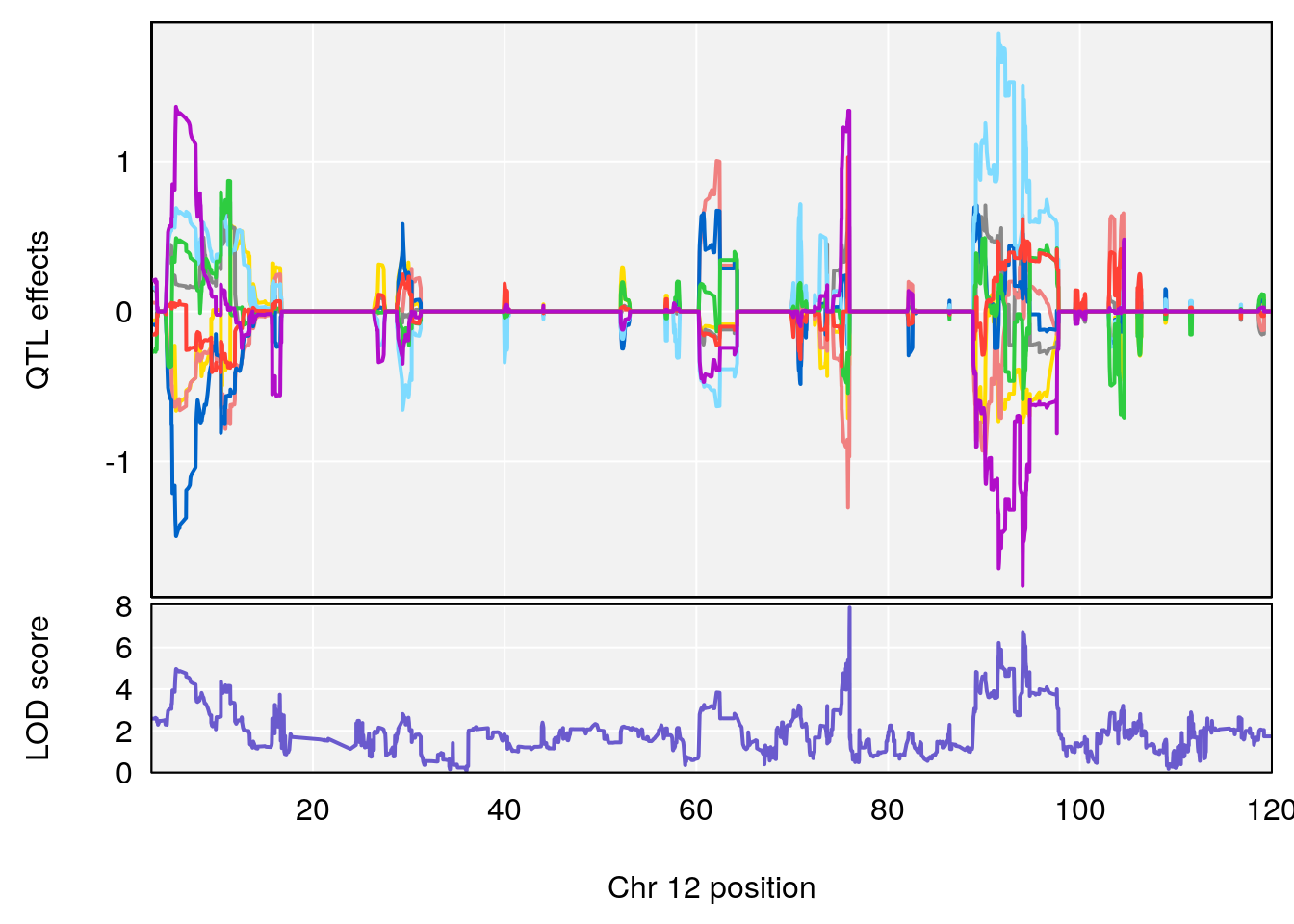
# [1] 6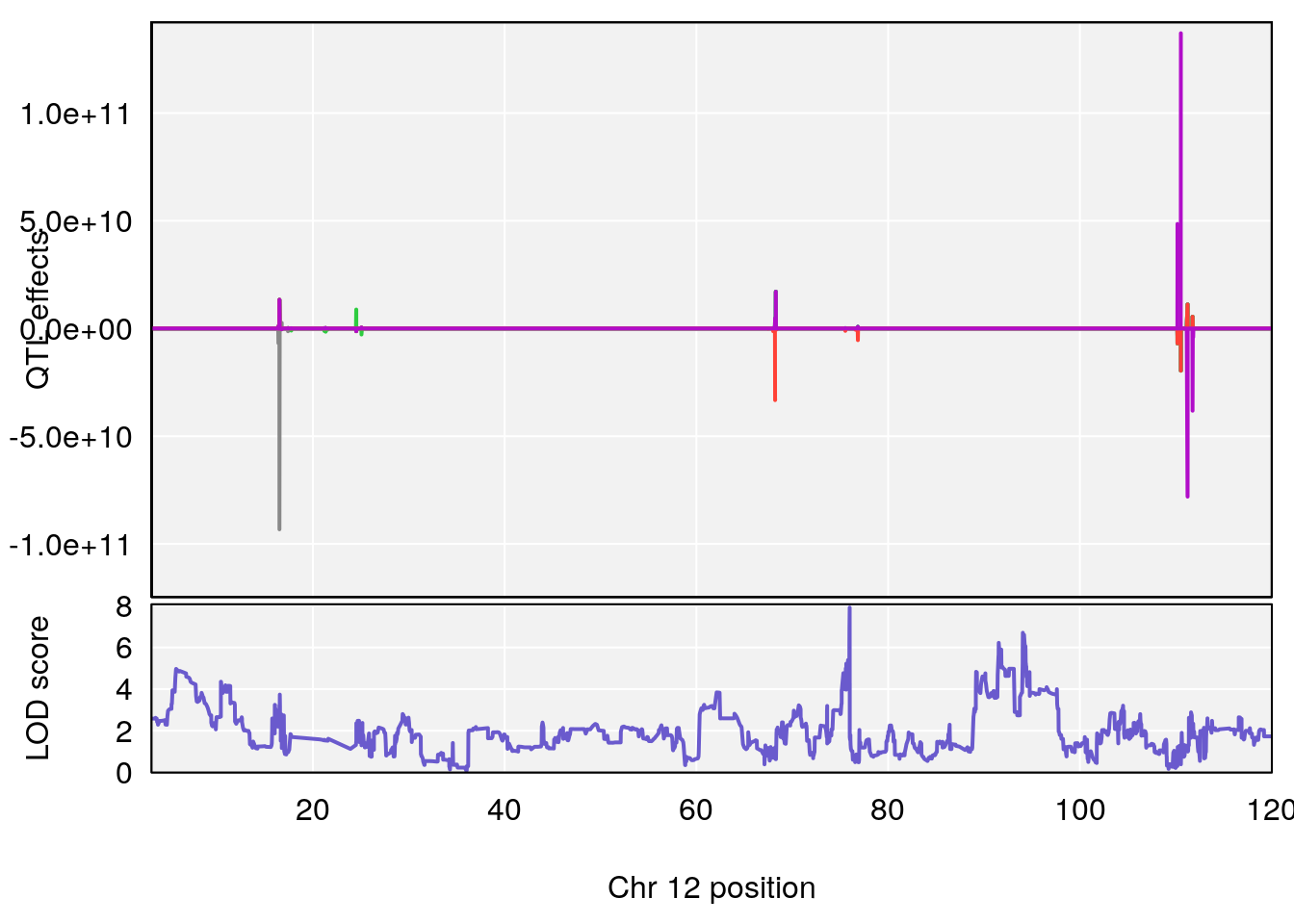
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
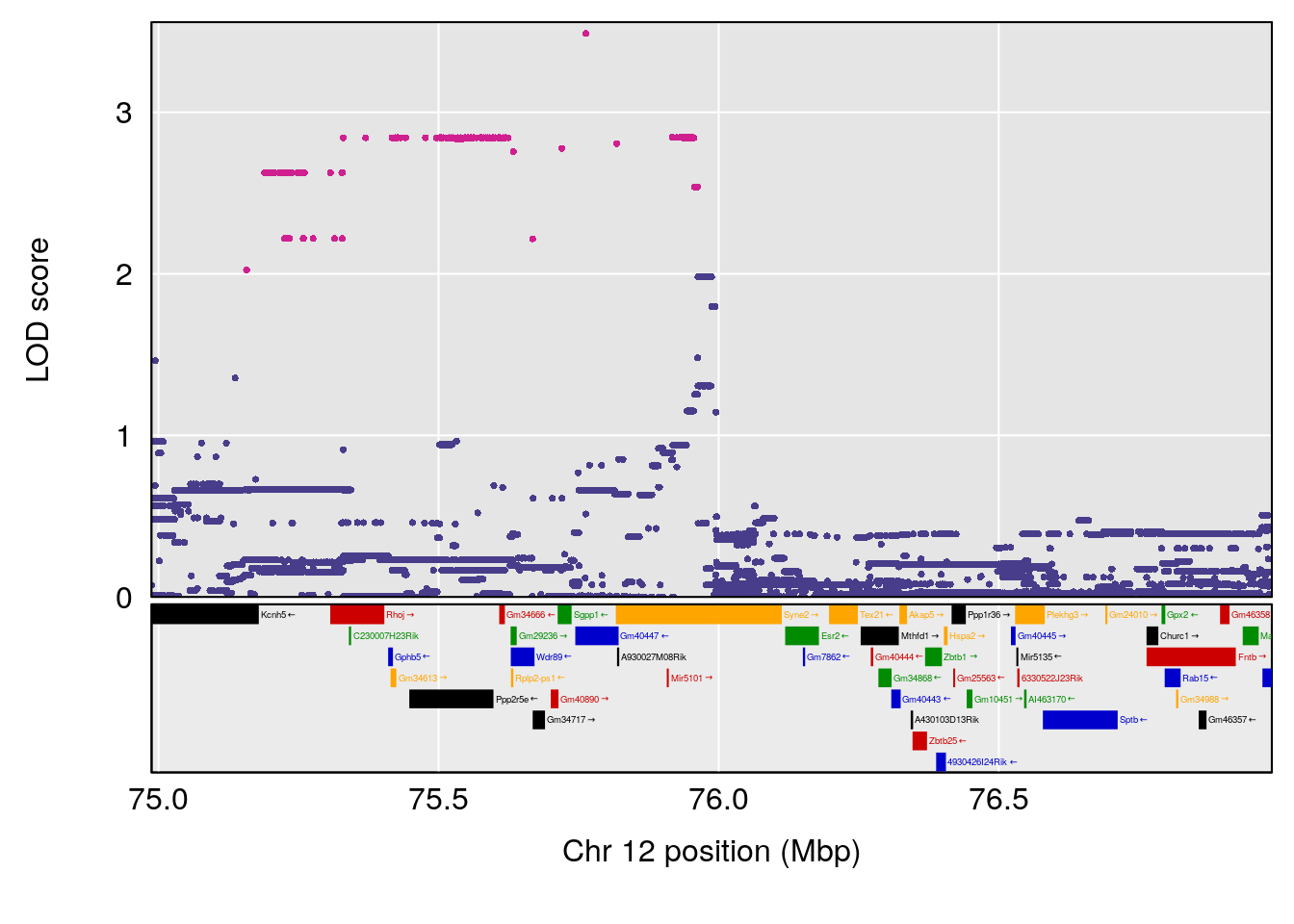
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 7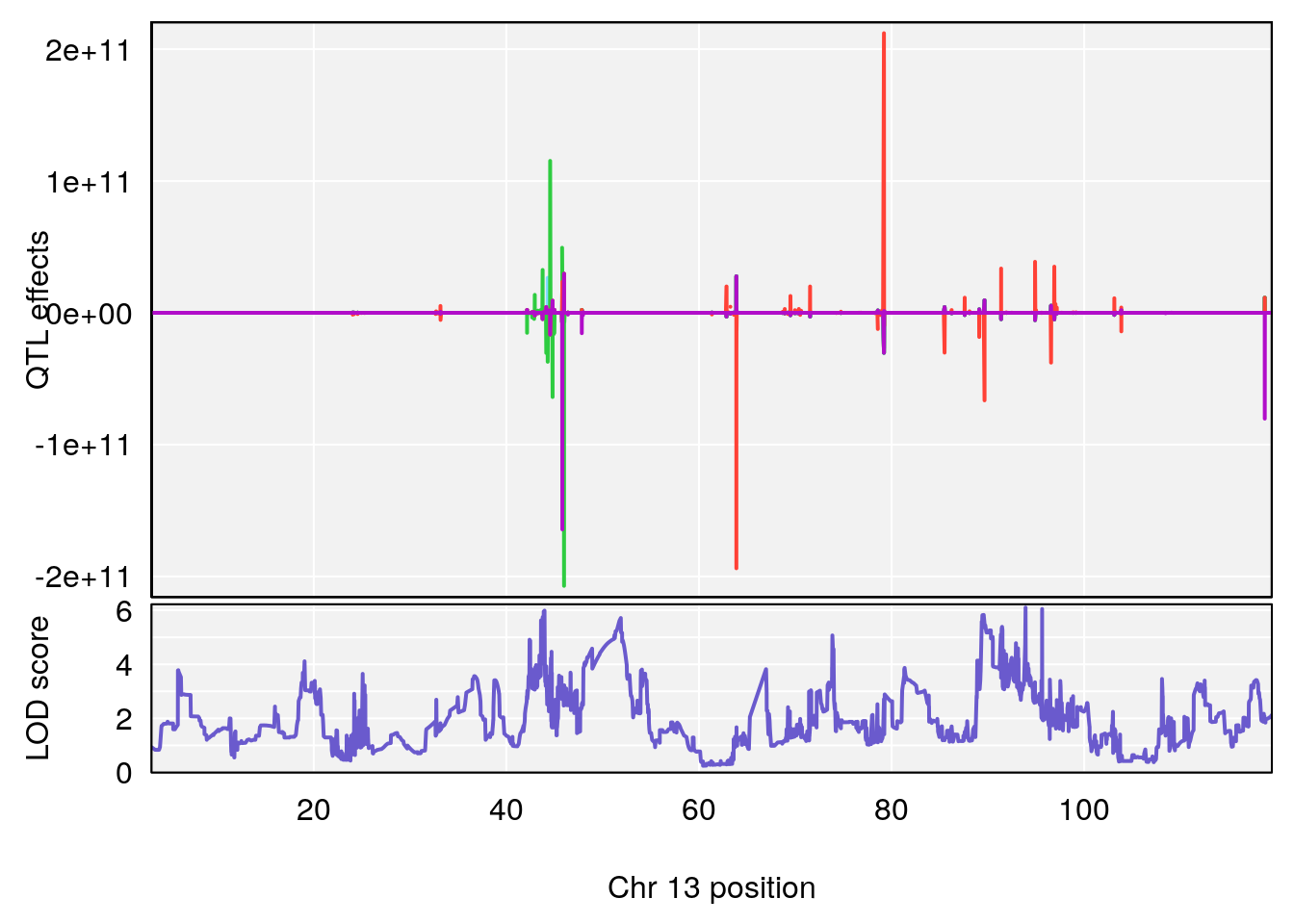
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
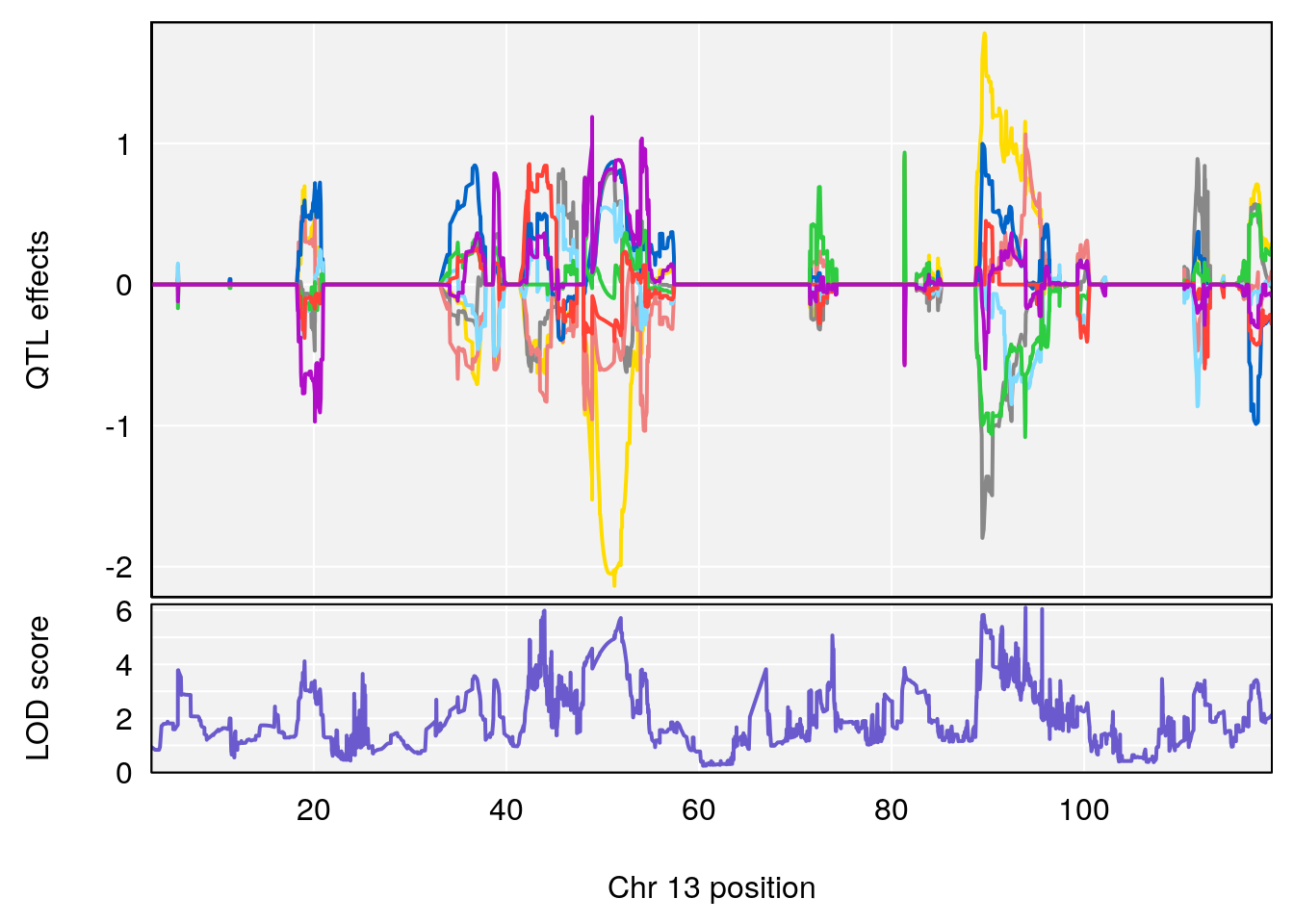
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
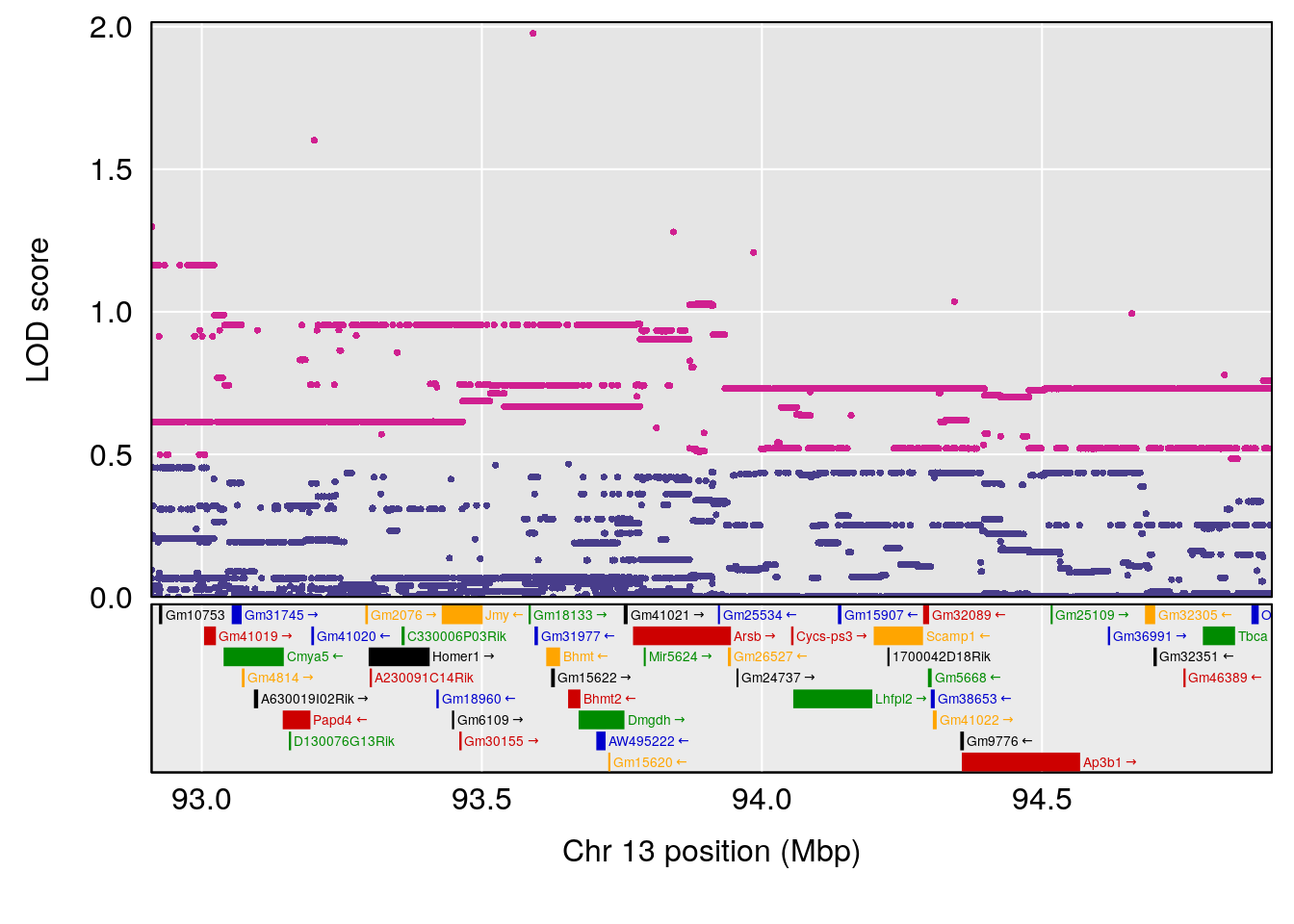
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "TimetoThresholdRecovery(Hr)"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 2 80.18535 6.172261 65.91339 181.81225
# 2 1 pheno1 12 75.98747 7.498411 75.94321 75.99396
# [1] 1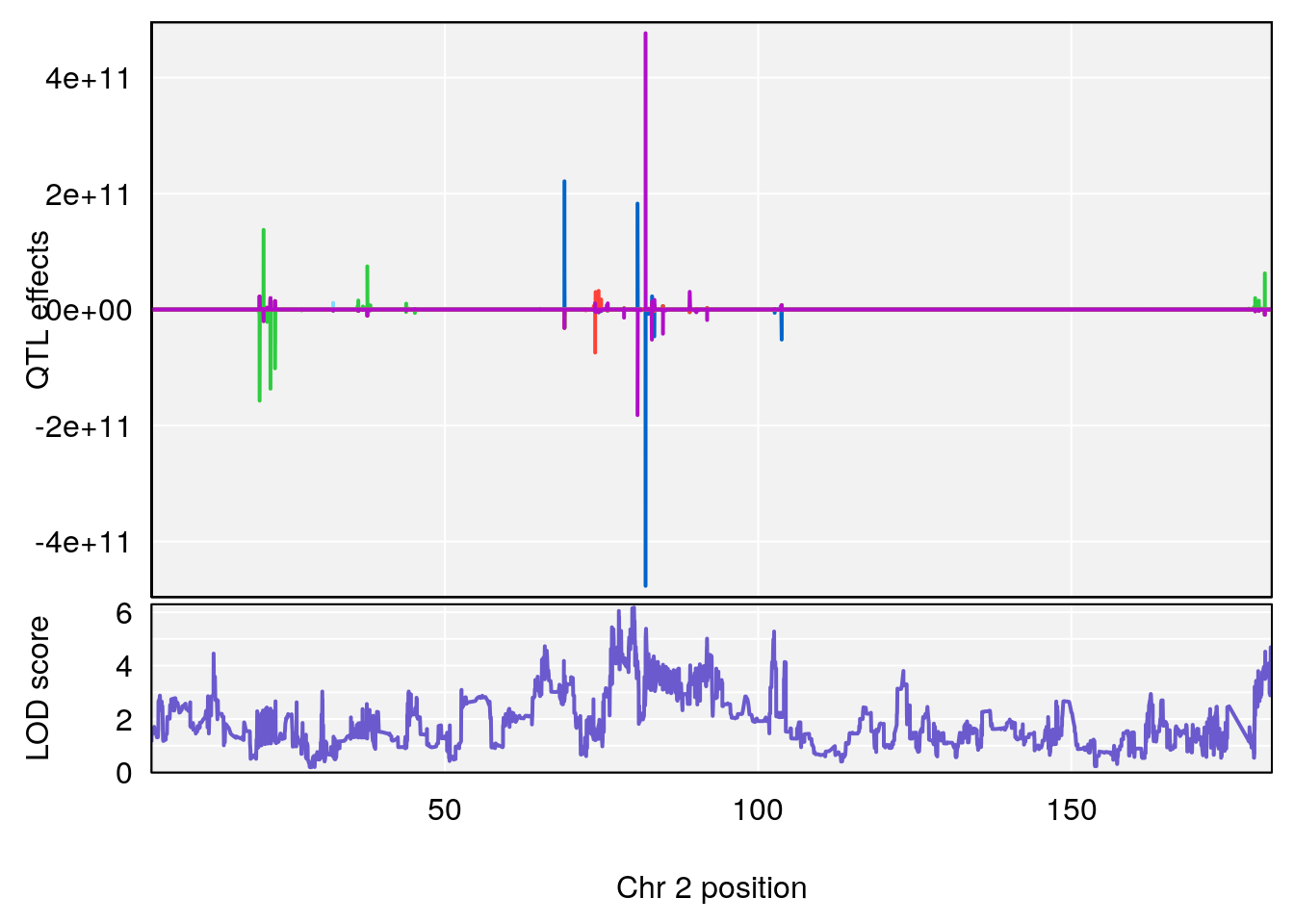
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
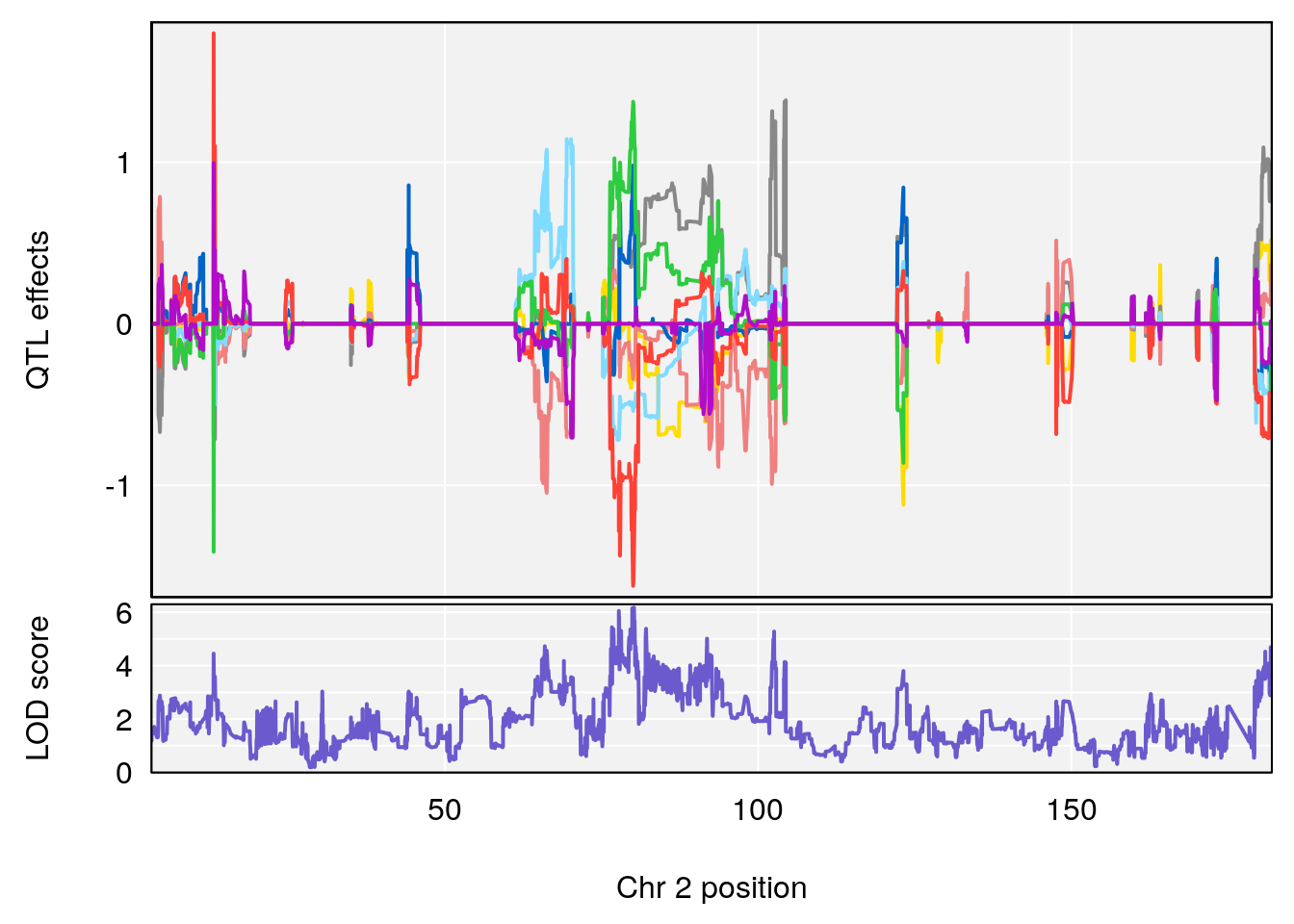
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
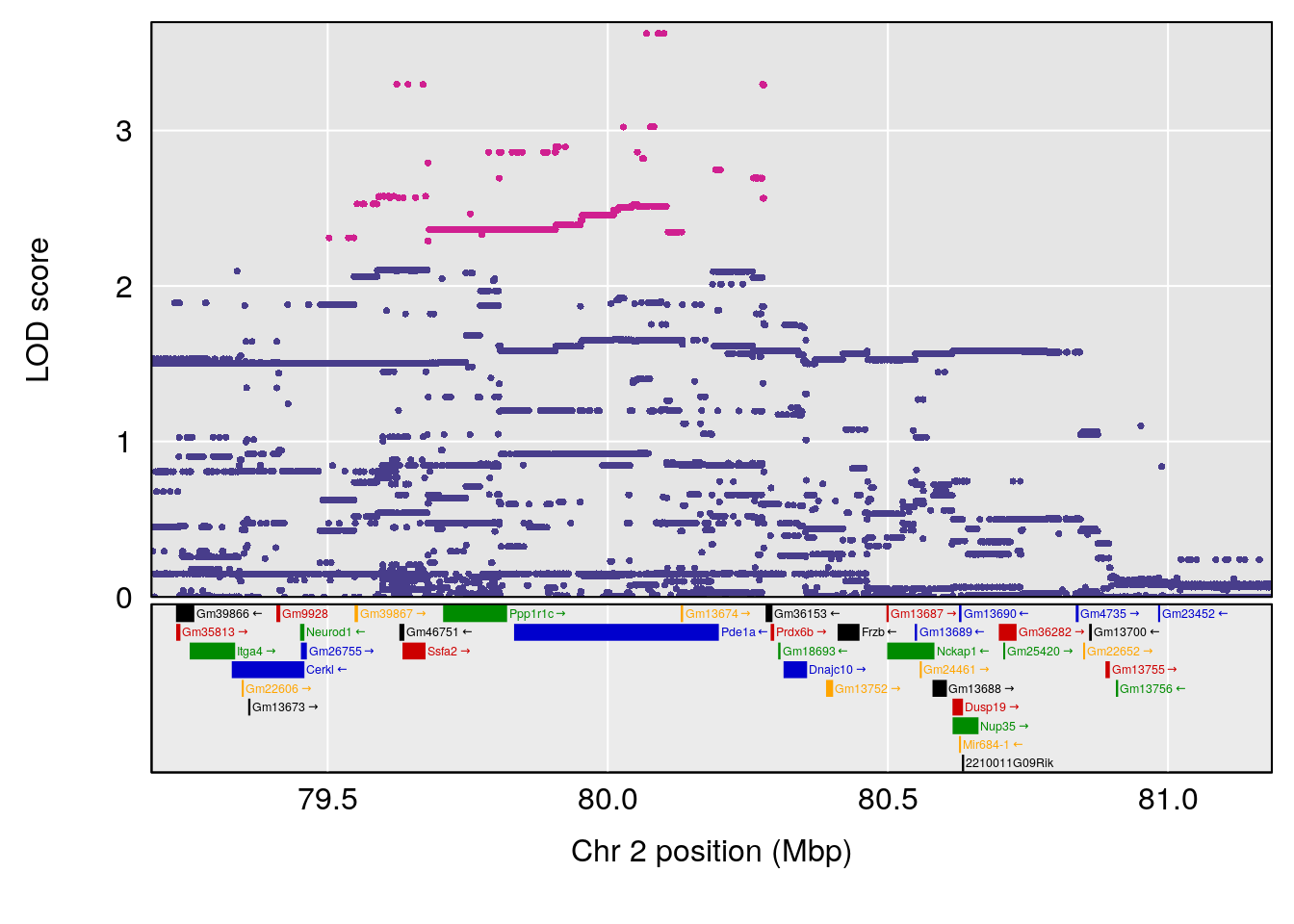
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
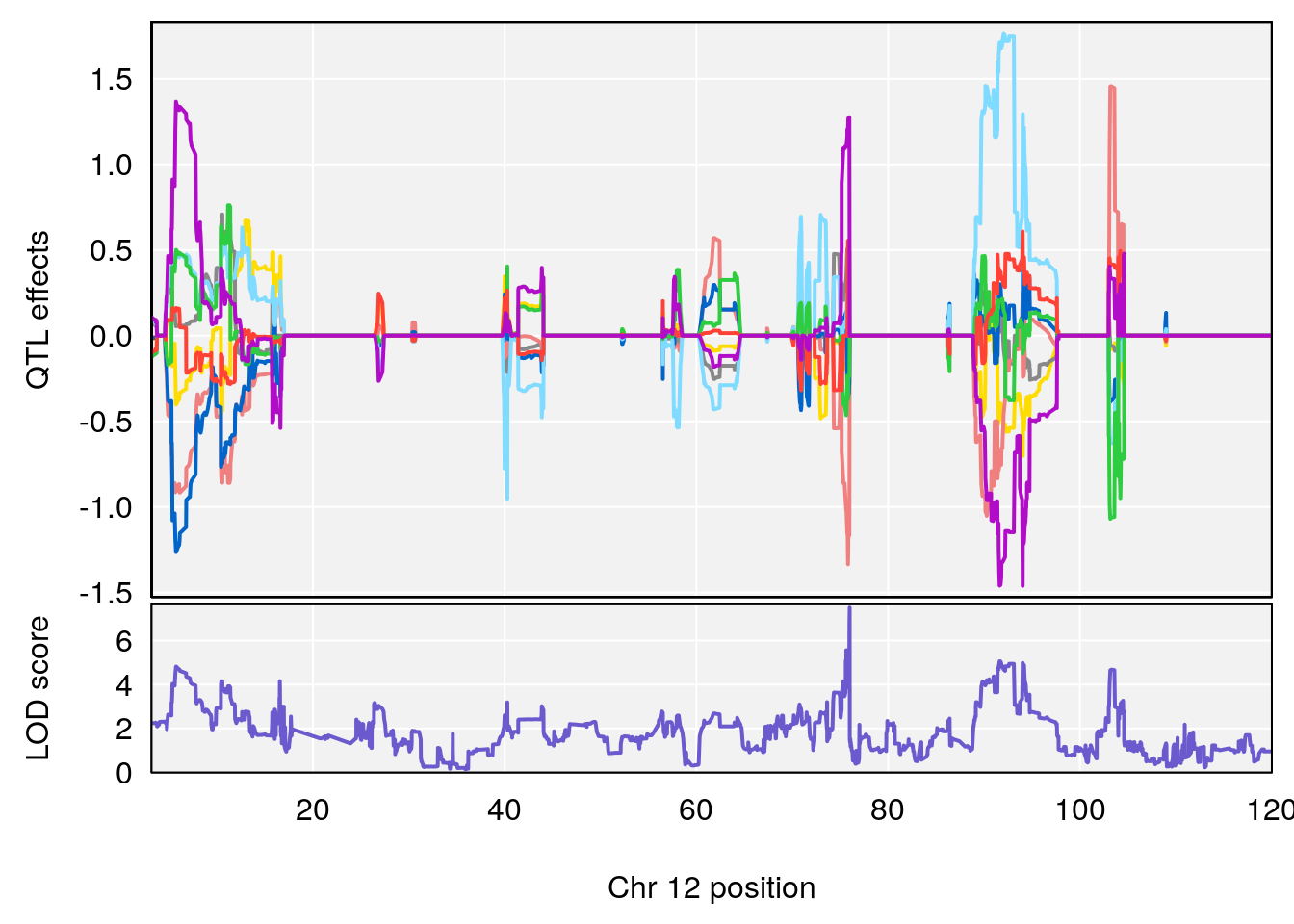
# [1] 2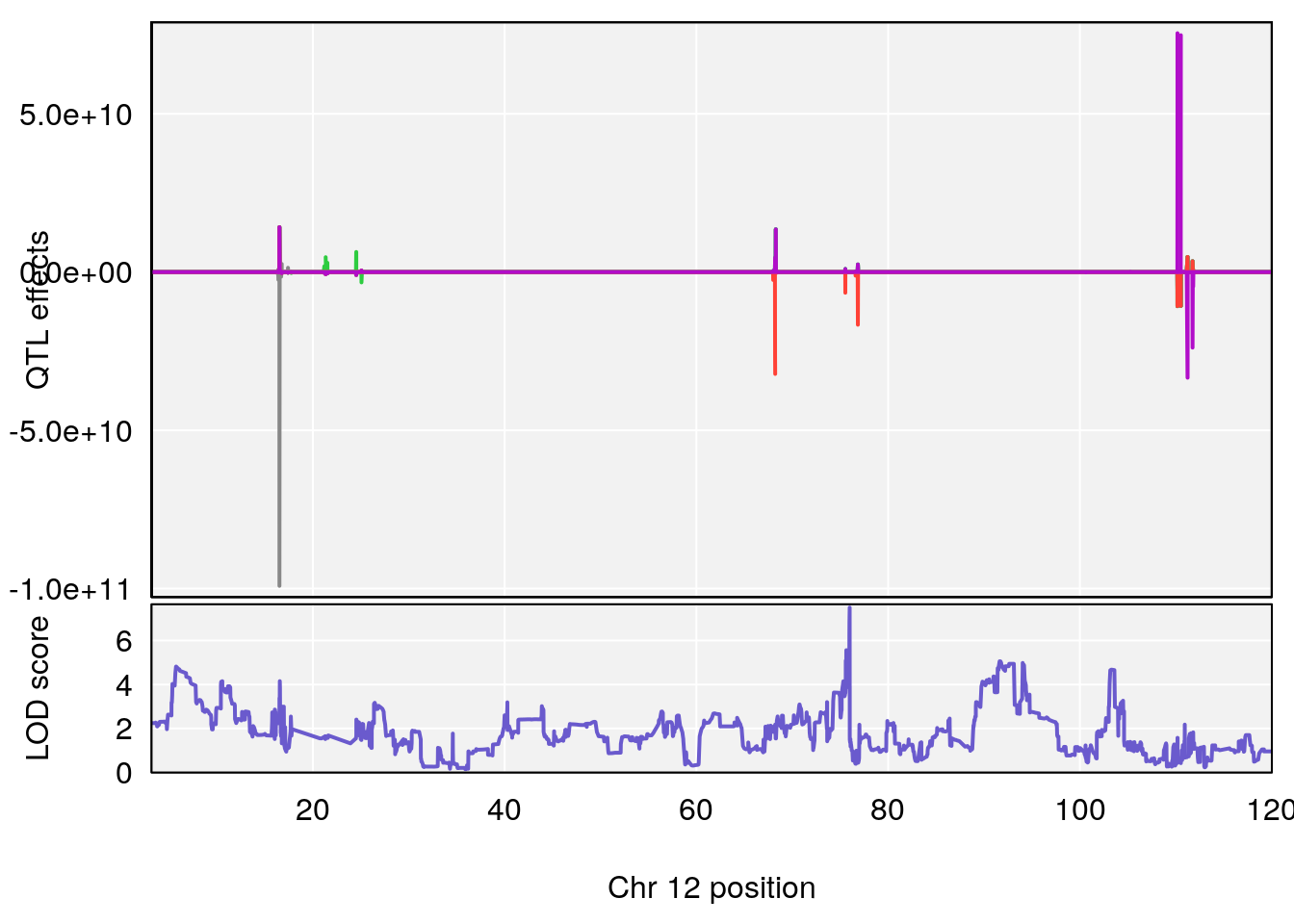
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
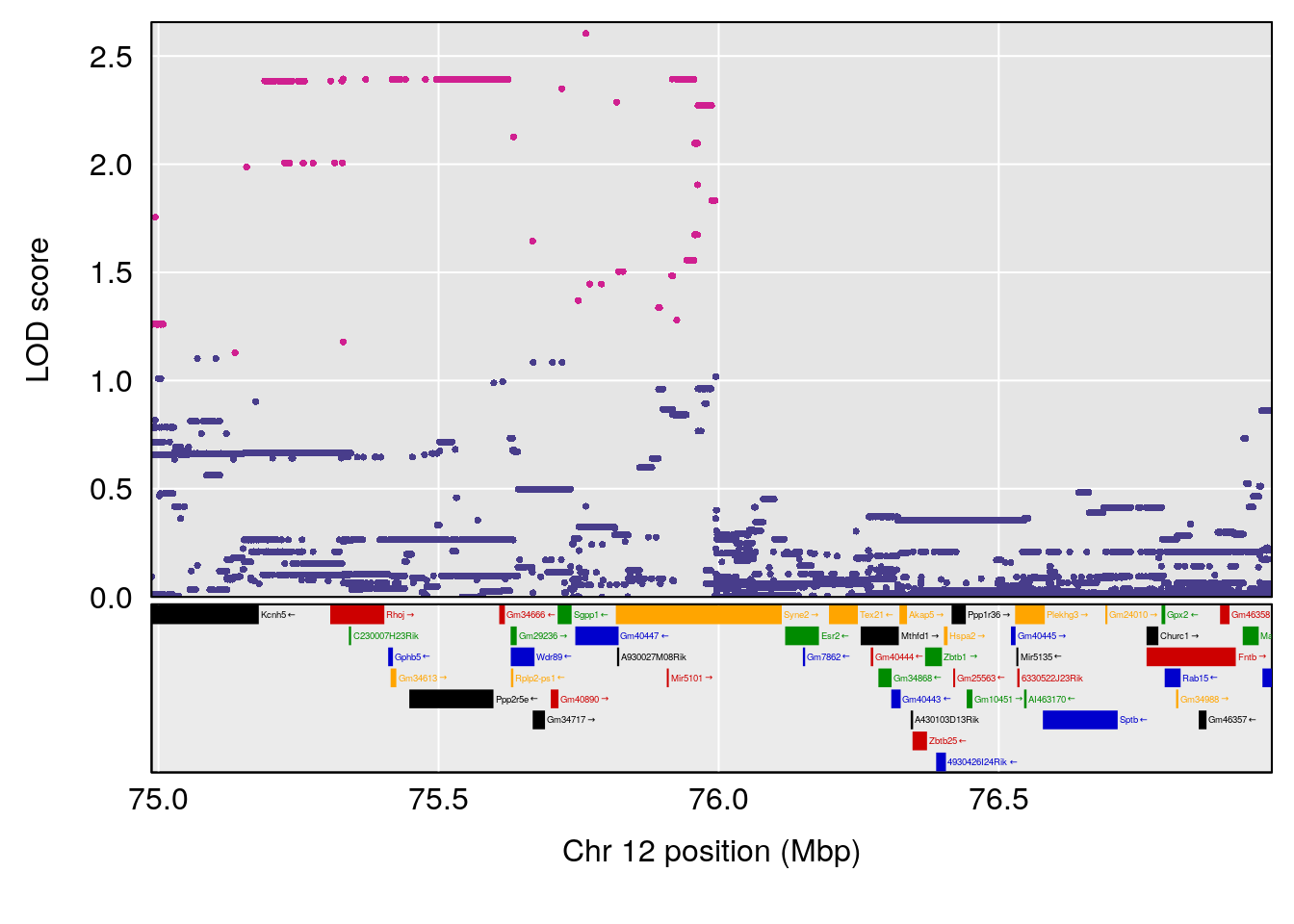
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "TimetoProjectedRecovery(Hr)"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 2 80.18535 6.137005 65.91339 180.94469
# 2 1 pheno1 3 90.66241 7.109982 89.60391 104.52824
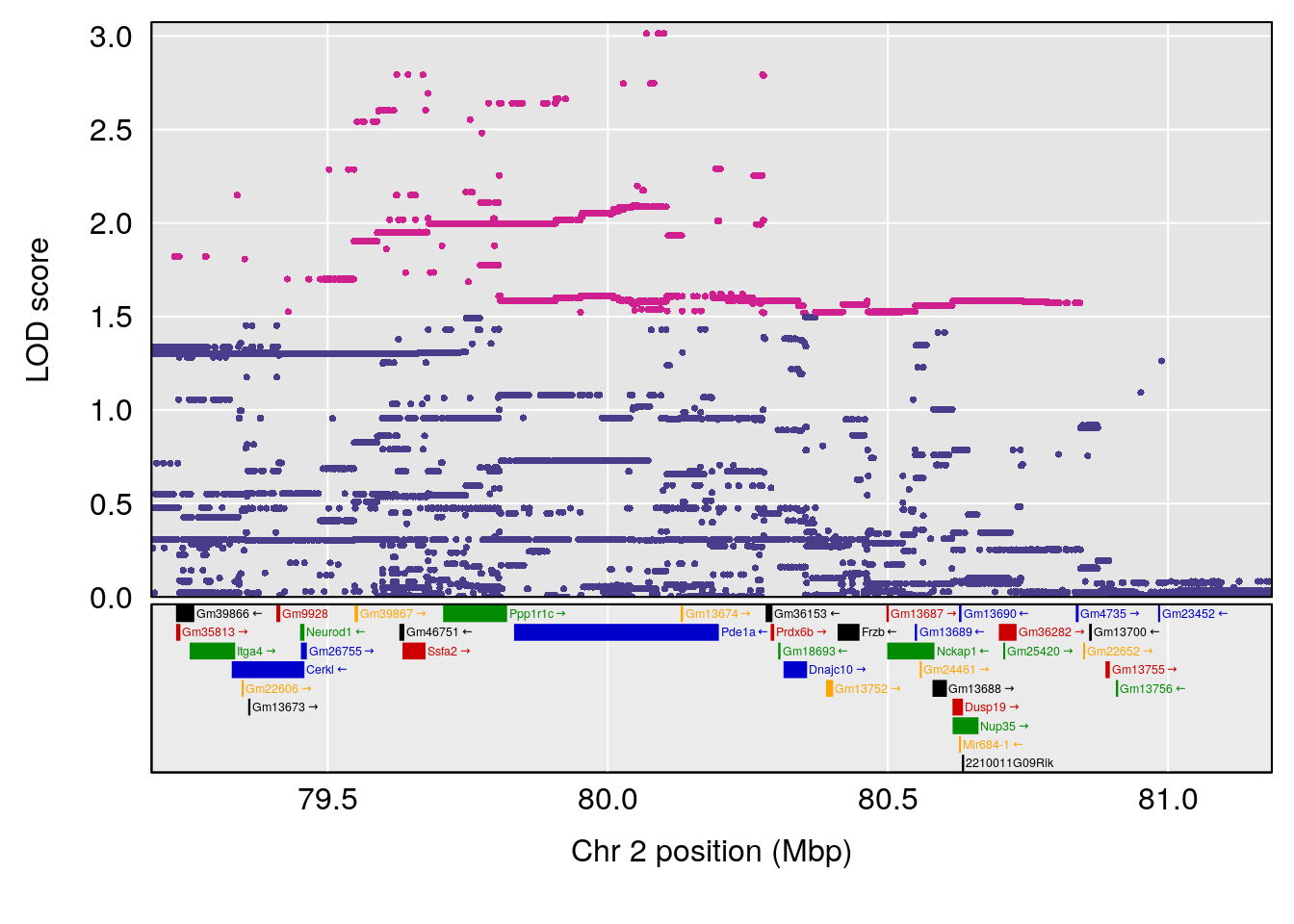
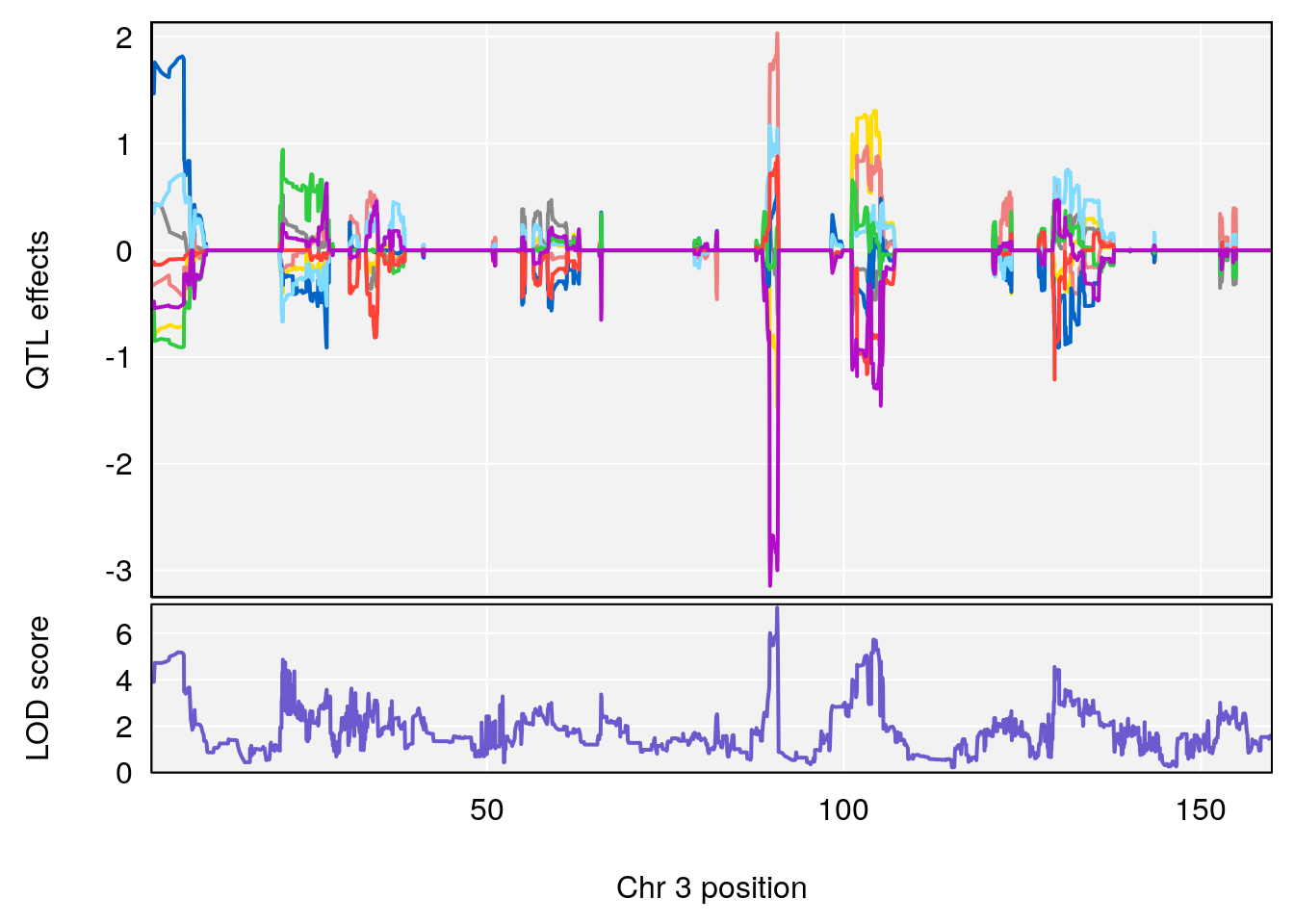
# 3 1 pheno1 4 14.49143 6.166787 13.87348 153.99966
# 4 1 pheno1 6 110.87154 6.105875 23.39815 111.51060
# 5 1 pheno1 10 66.55650 6.203671 33.86040 67.04043
# 6 1 pheno1 12 91.62691 6.136398 75.62742 94.37674
# 7 1 pheno1 15 83.10382 6.748883 83.06880 83.13036
# [1] 1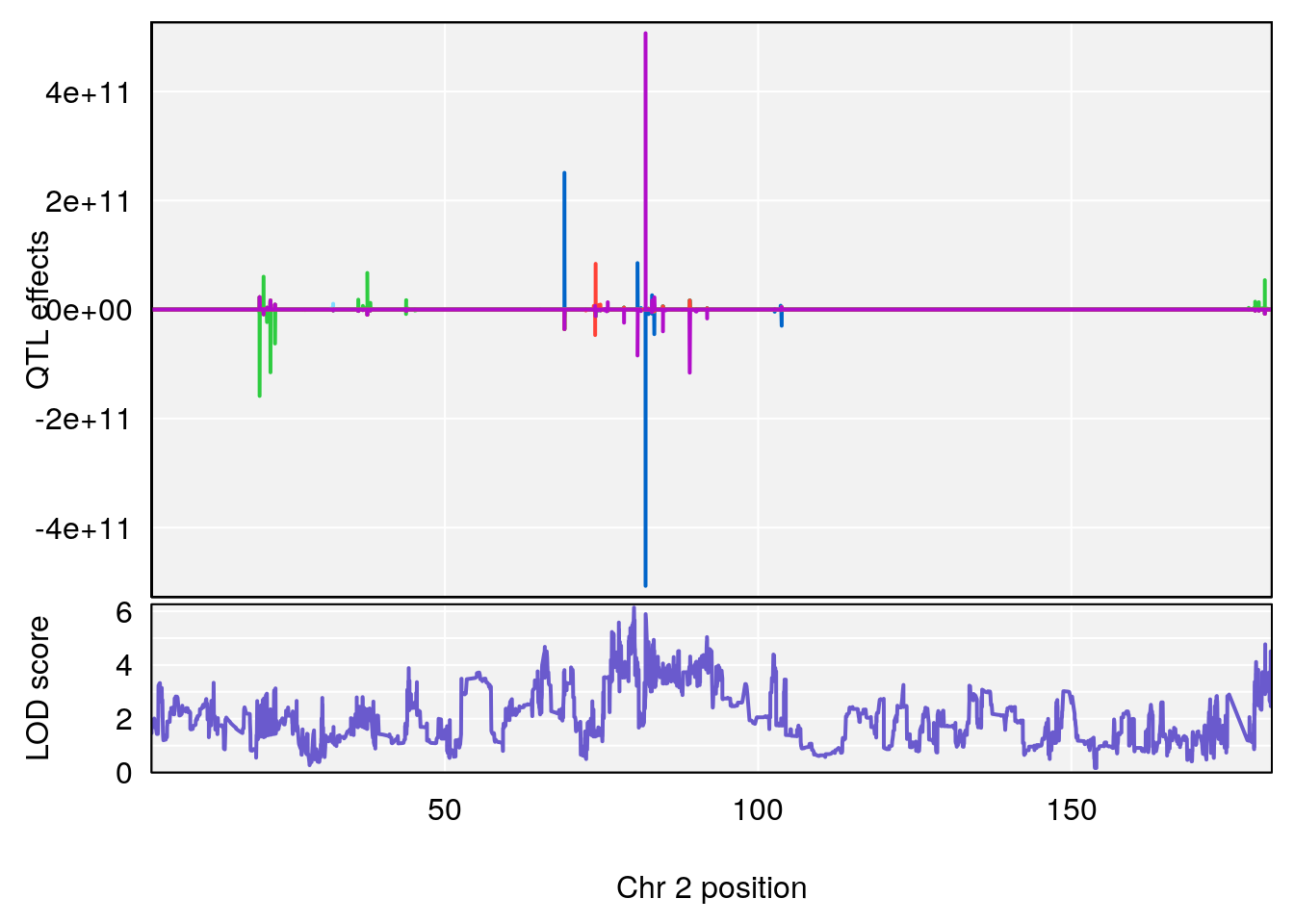
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |

| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2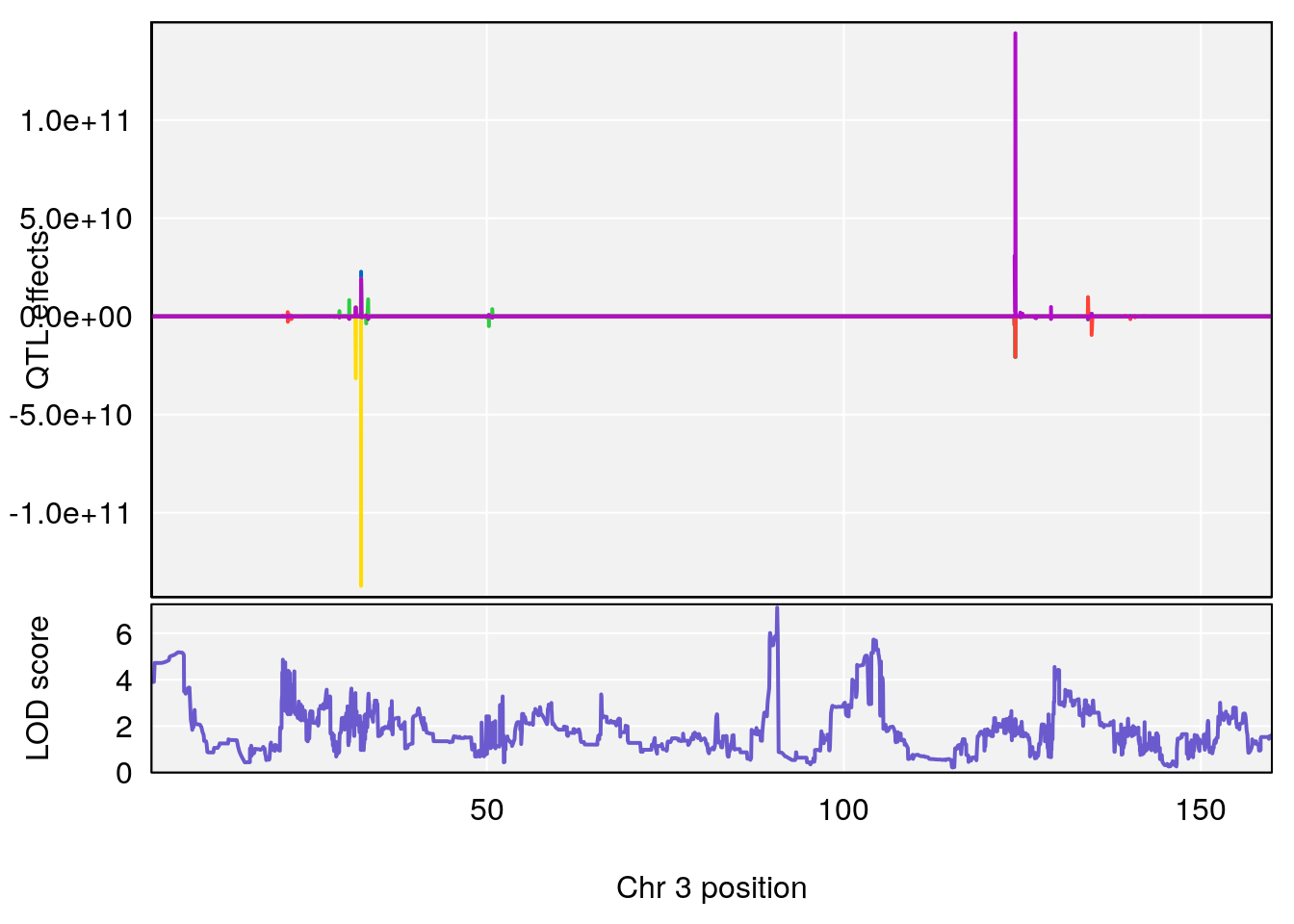
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
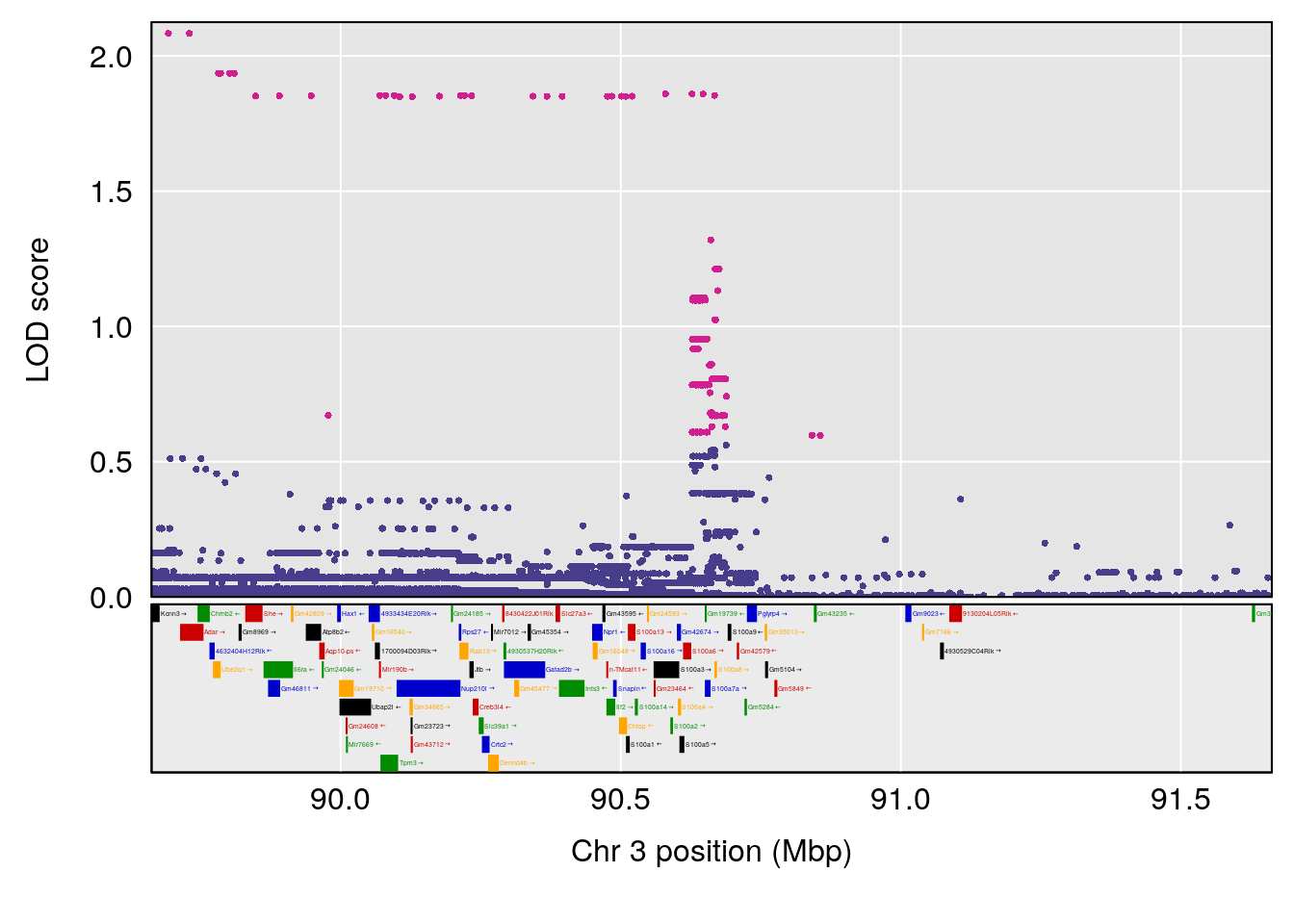
# [1] 3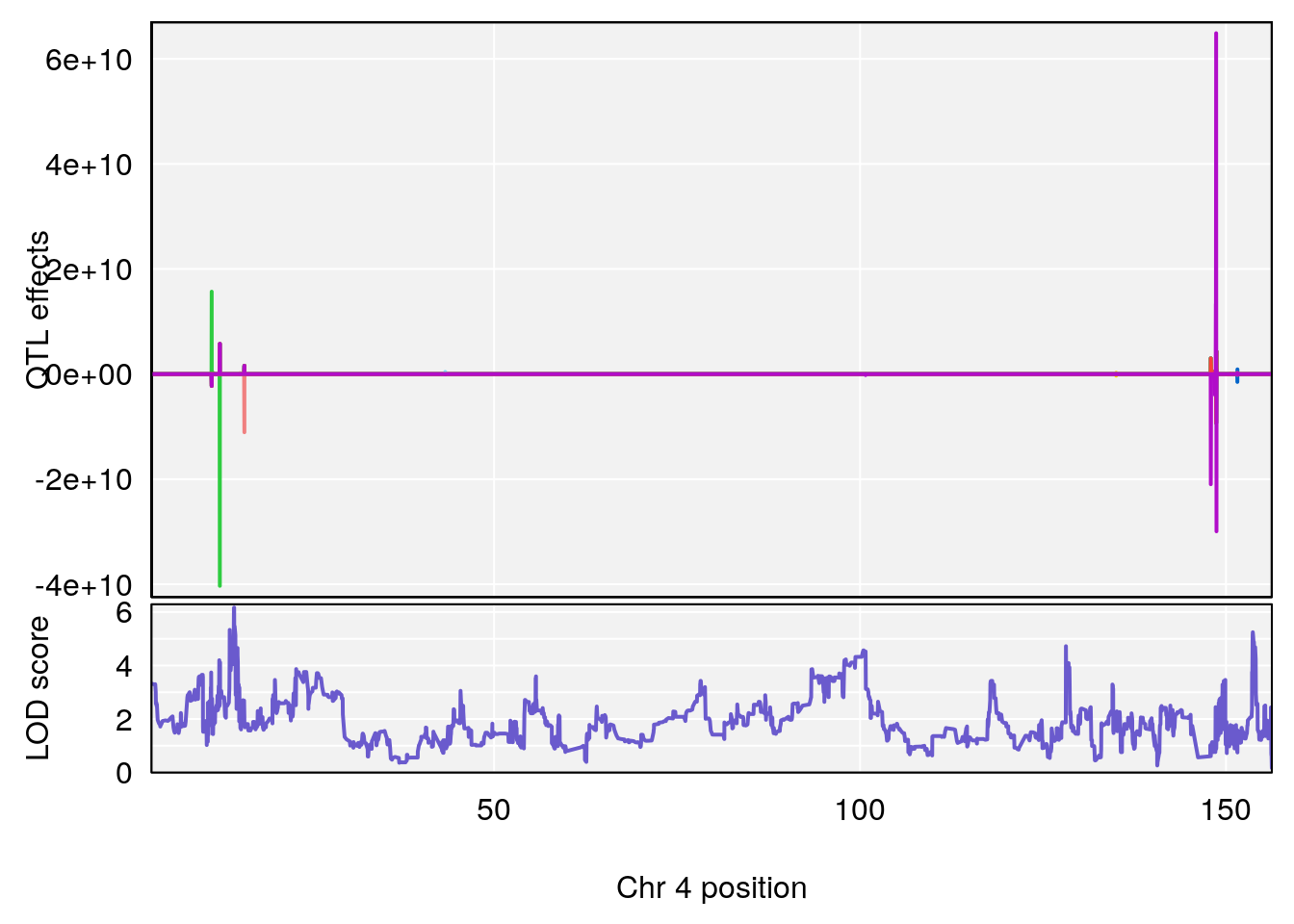
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
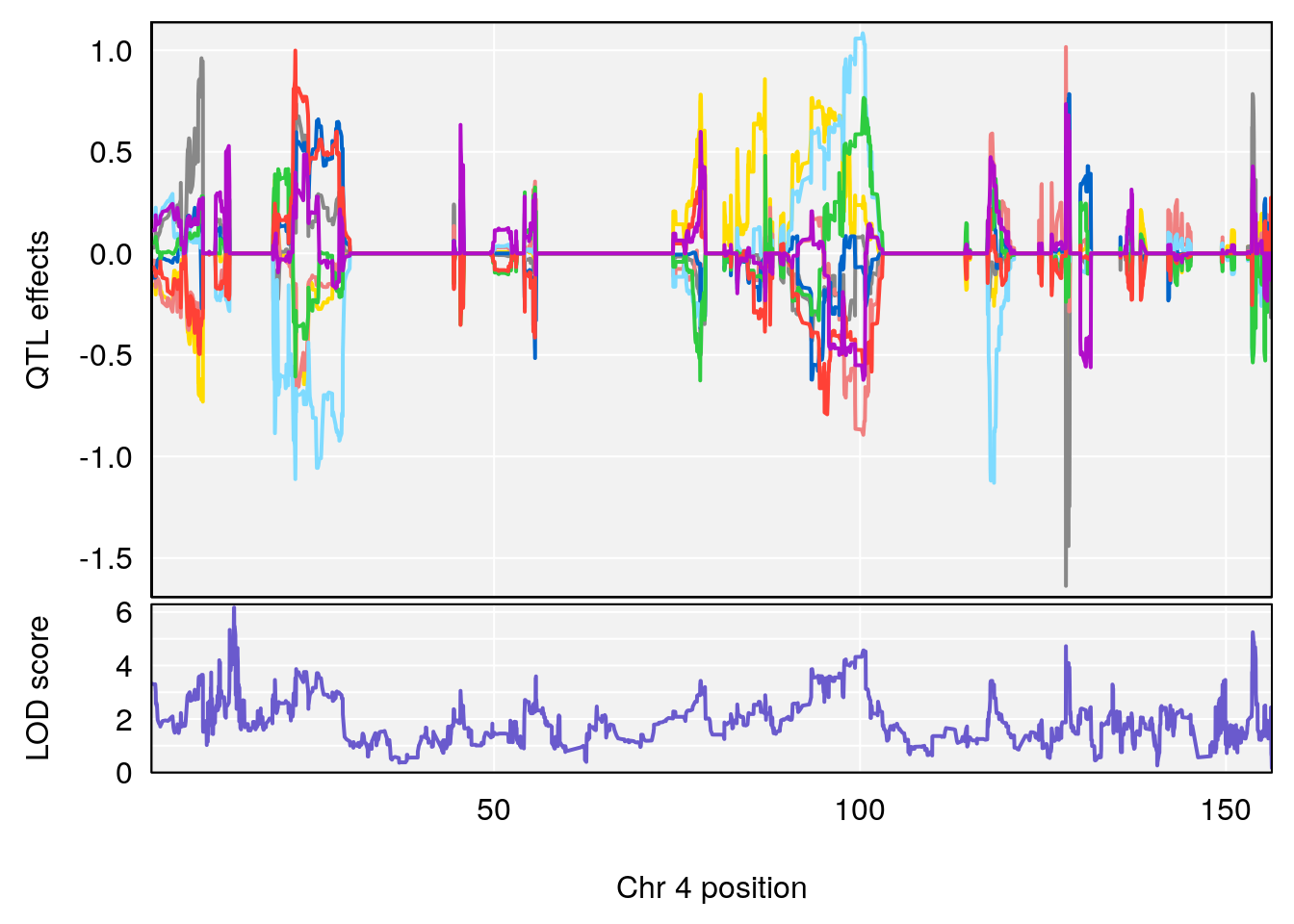
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
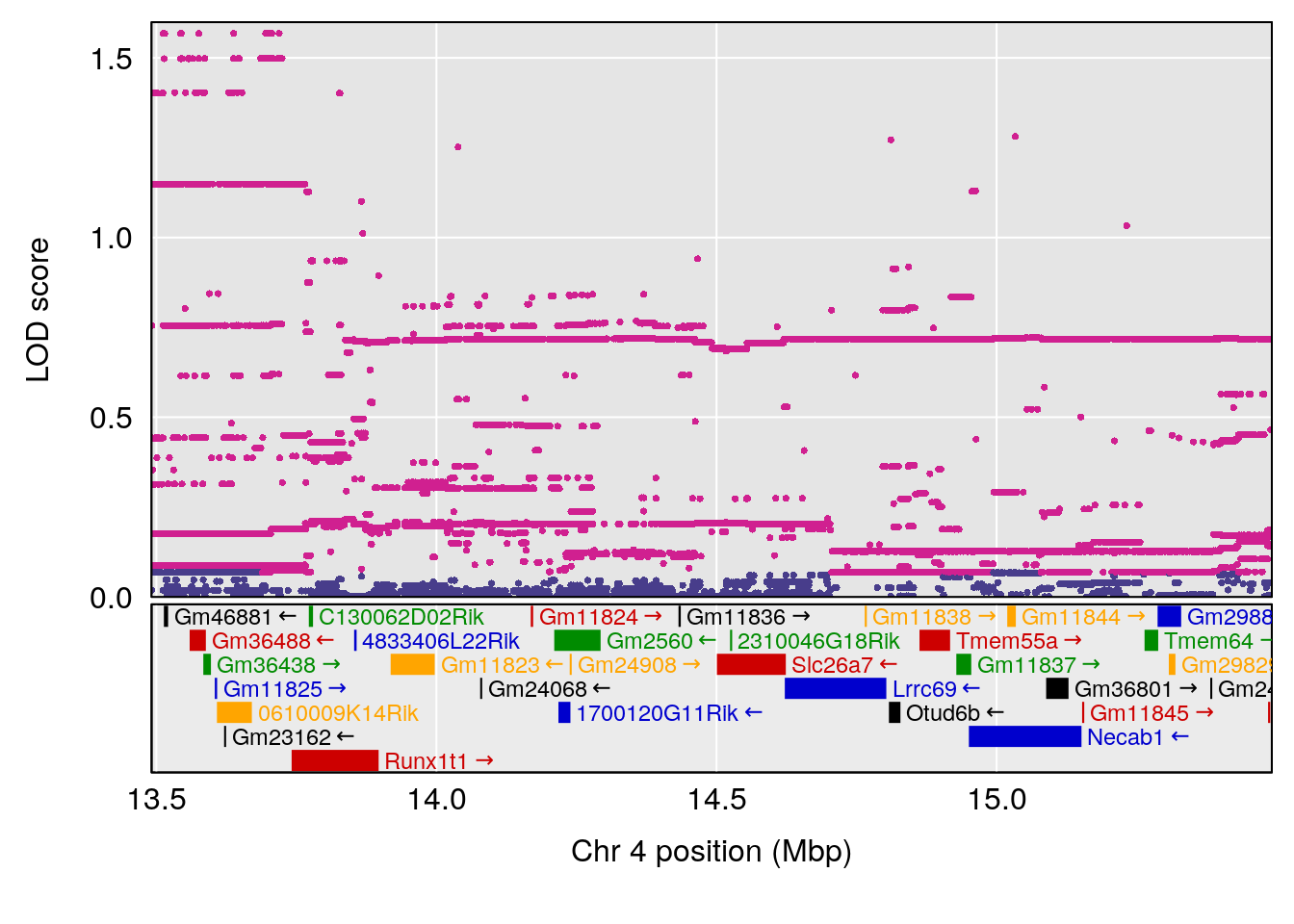
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
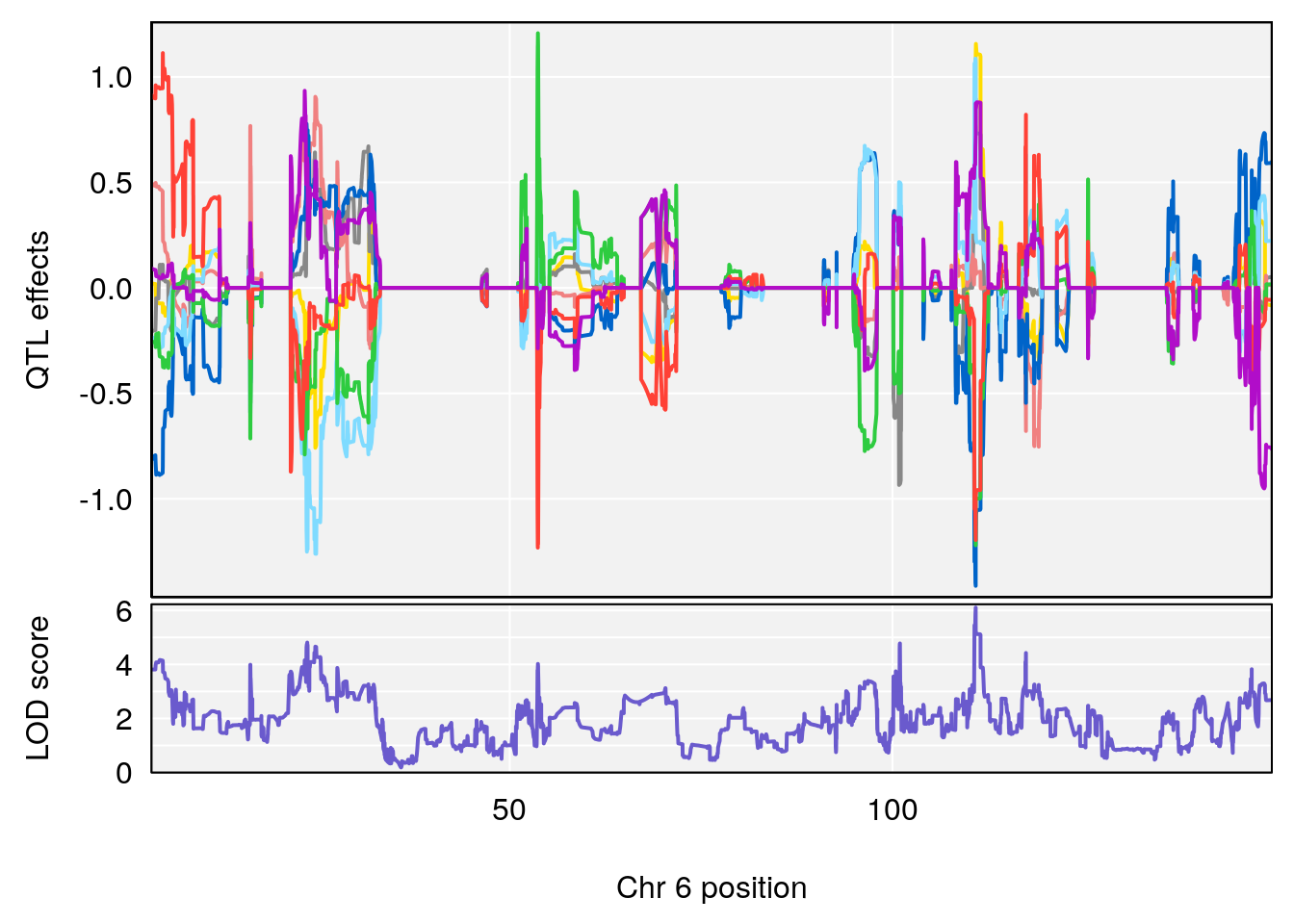
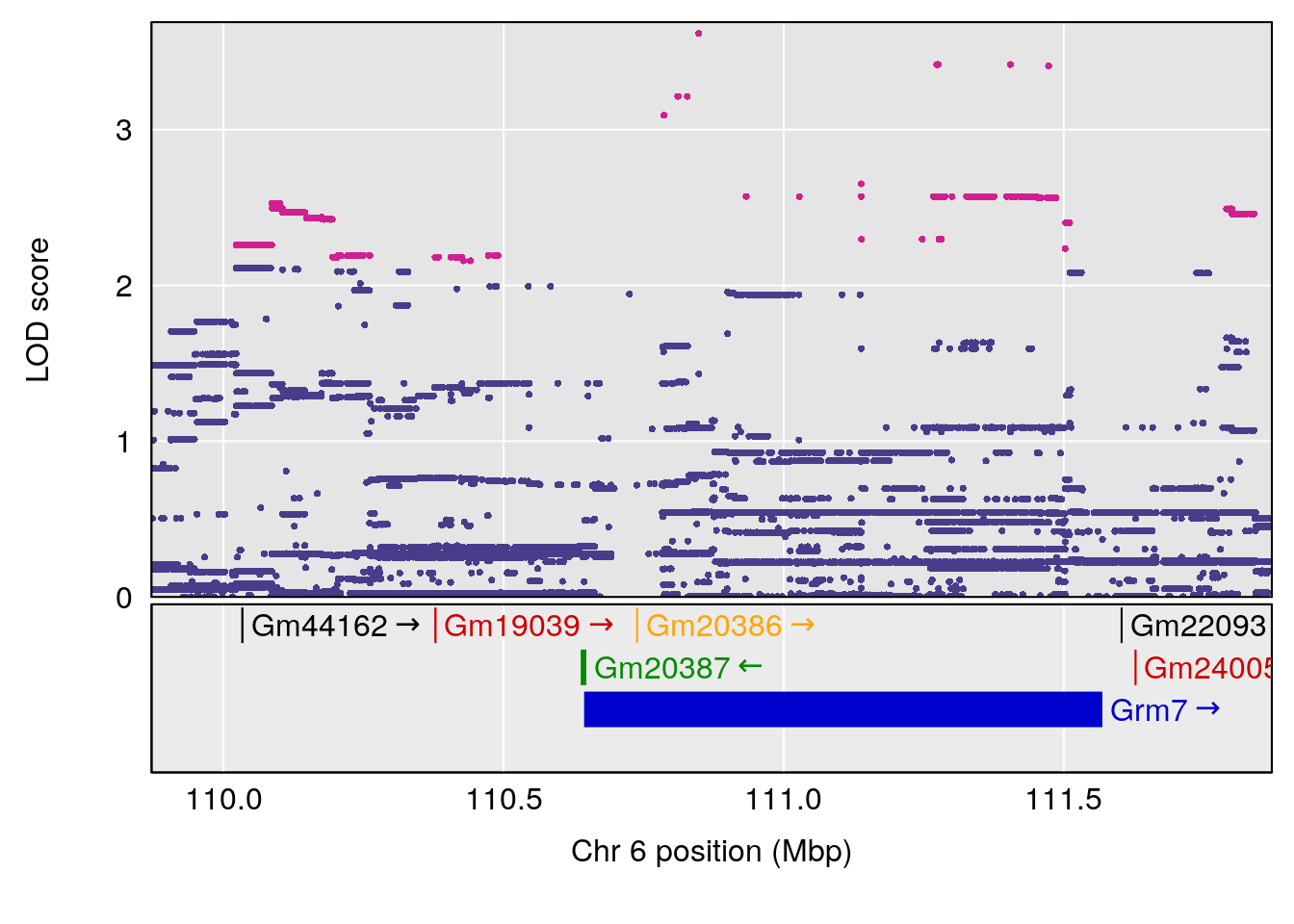
# [1] 4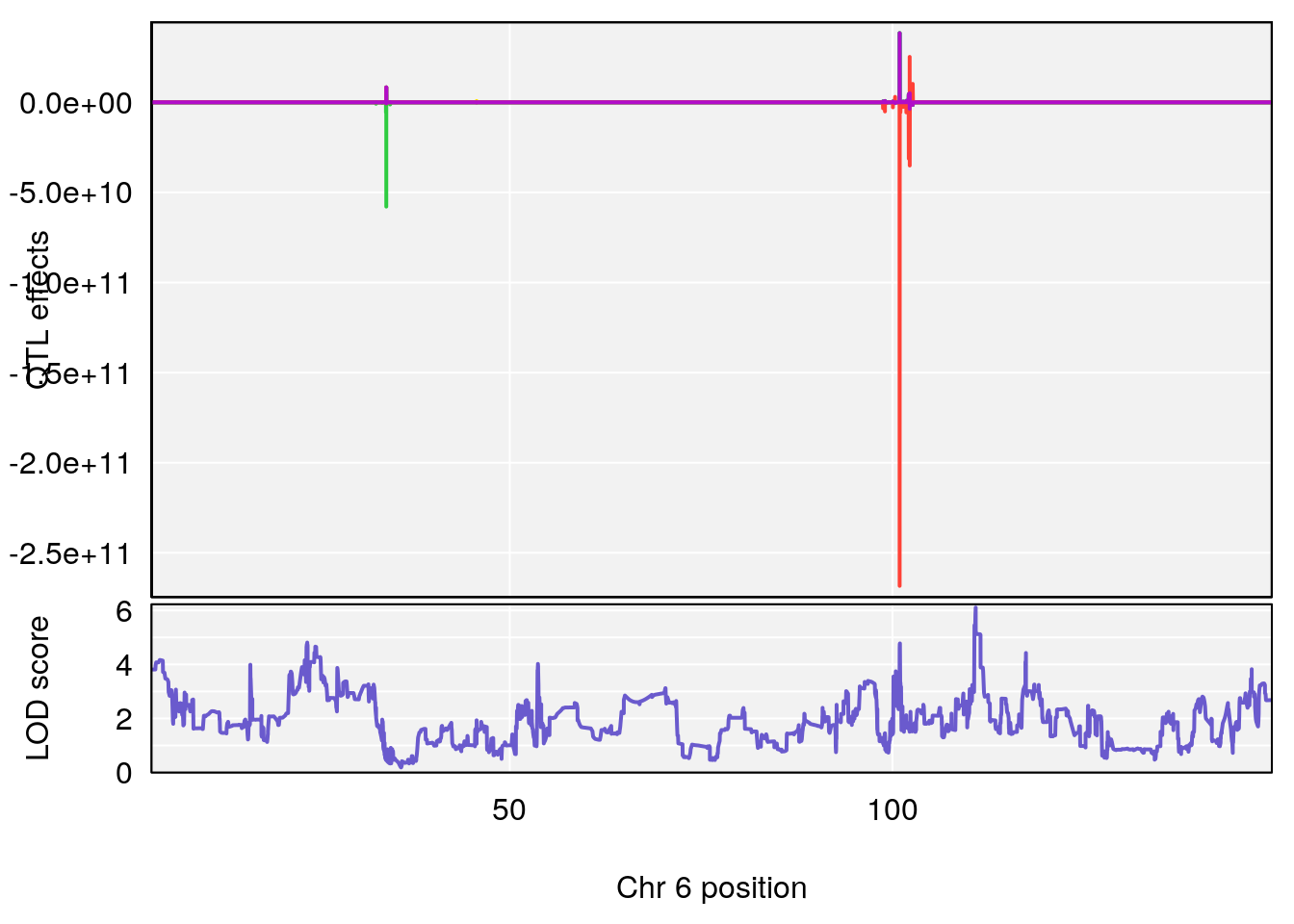
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 5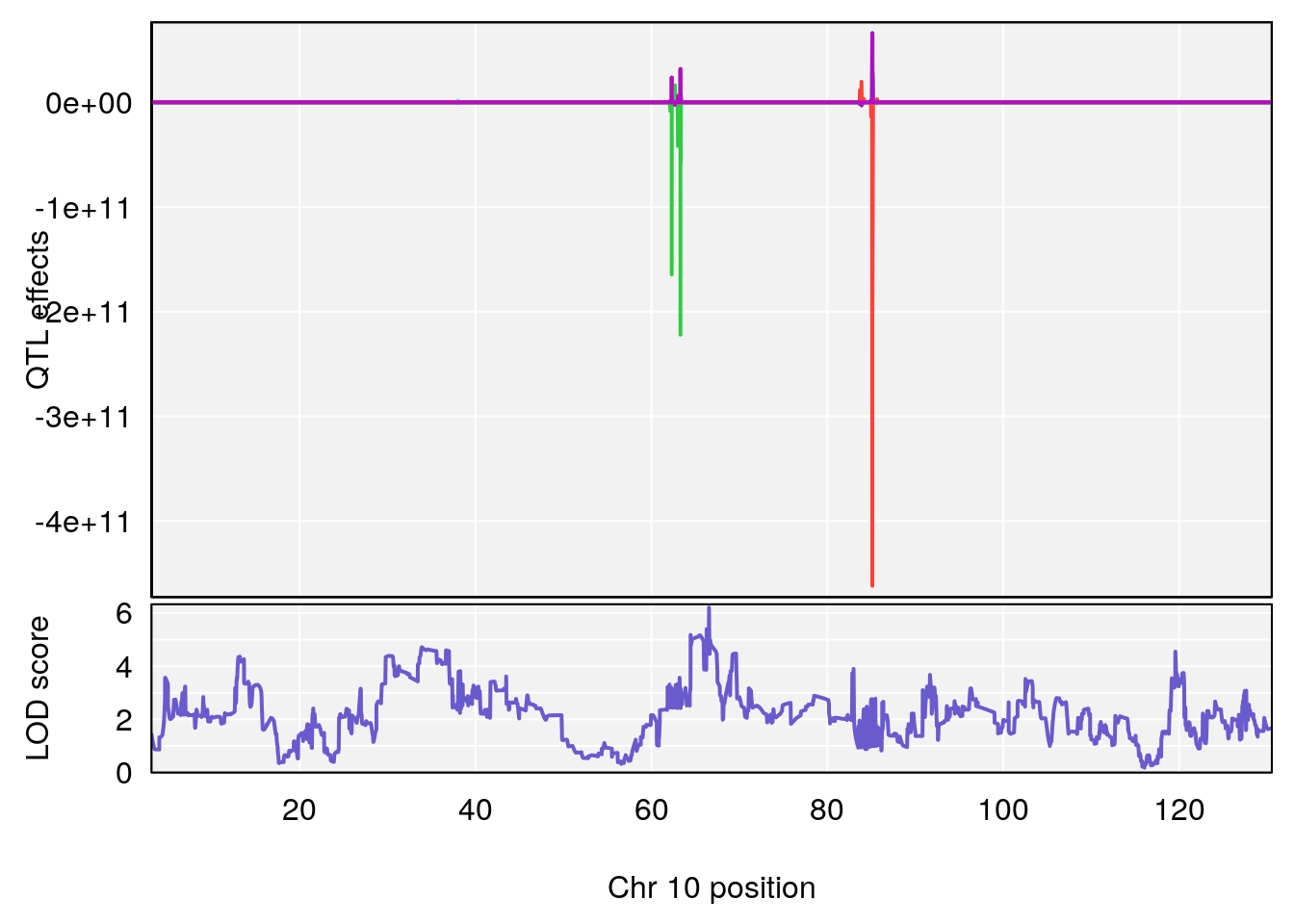
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
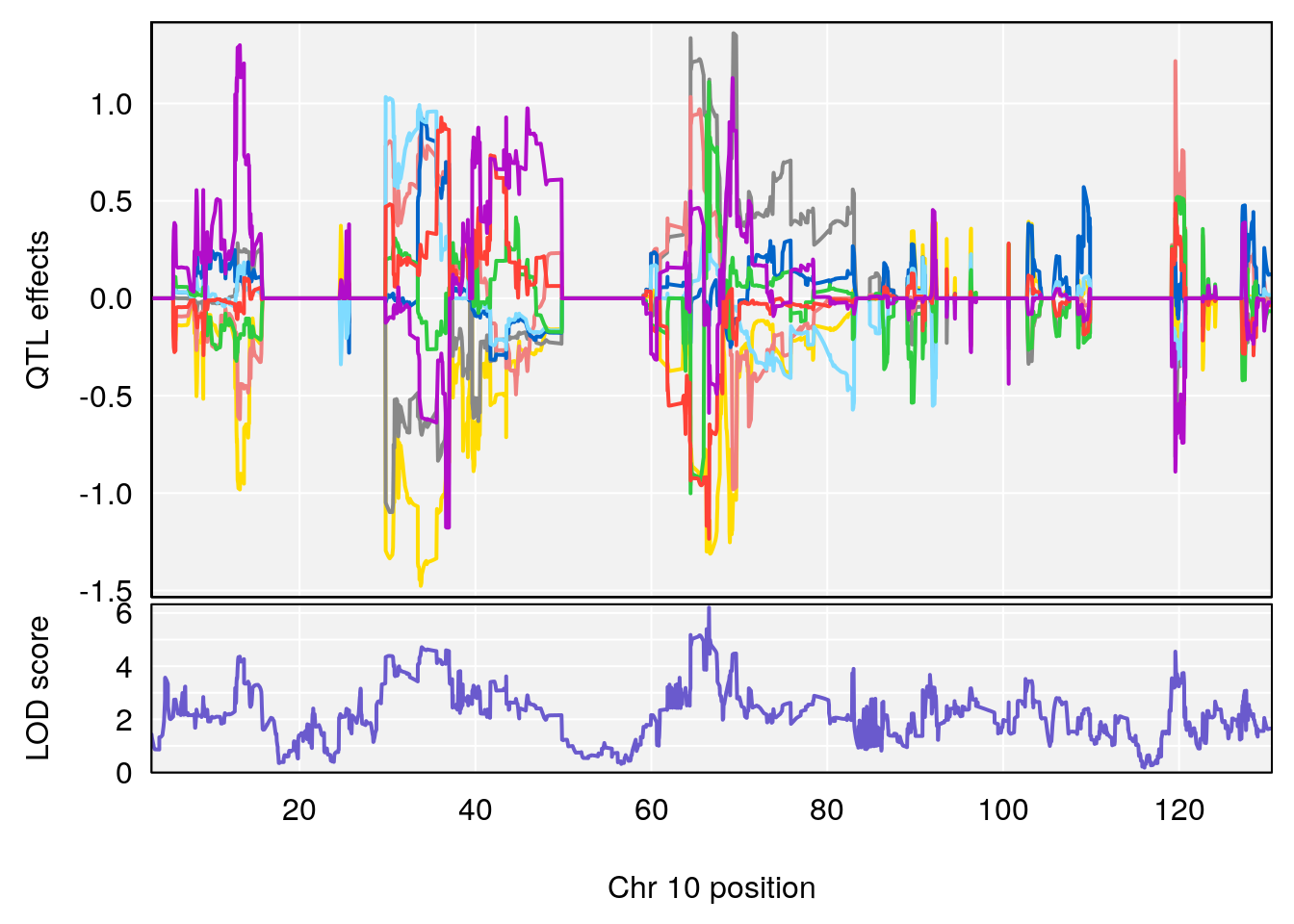
| Version | Author | Date |
|---|---|---|
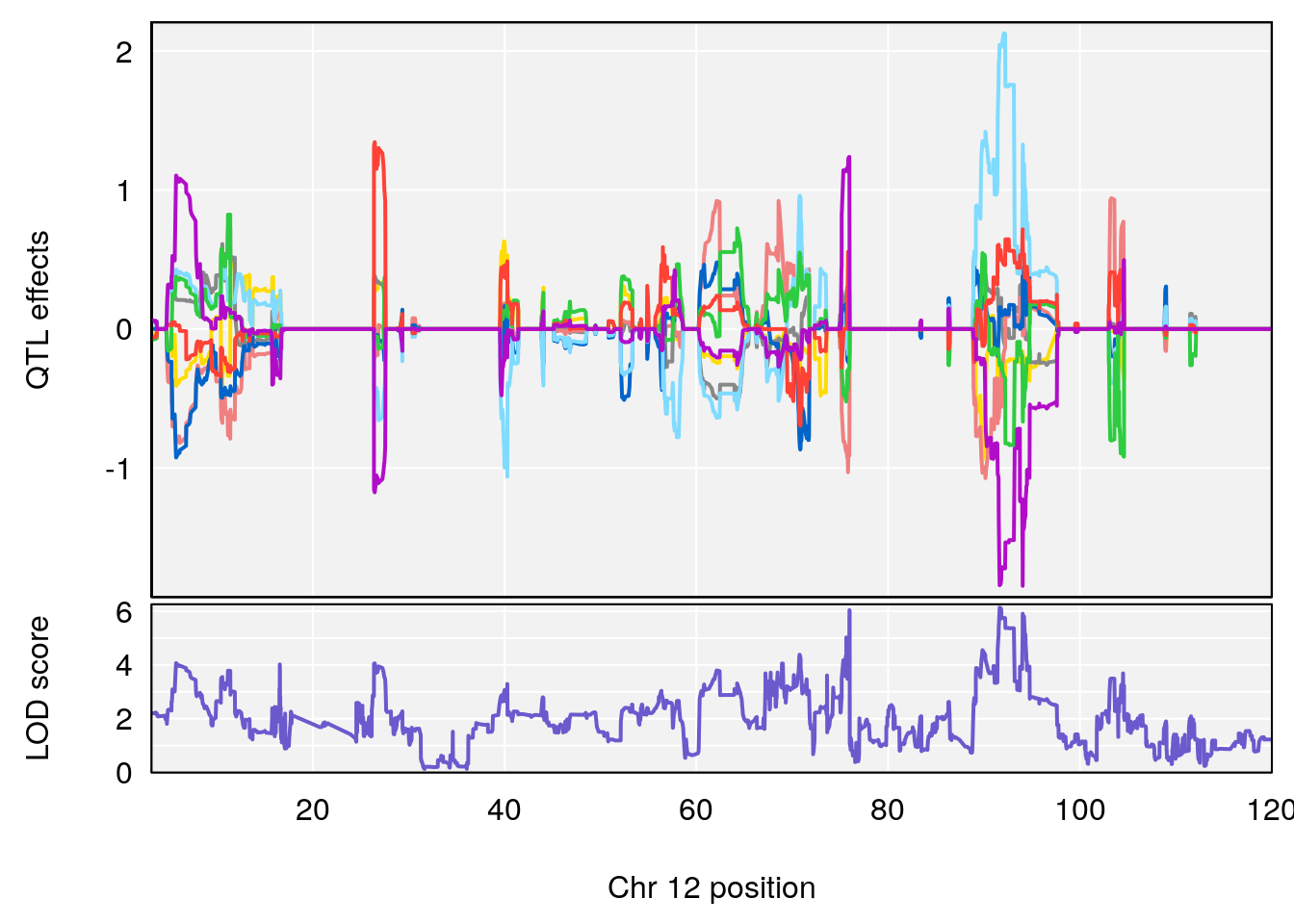
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
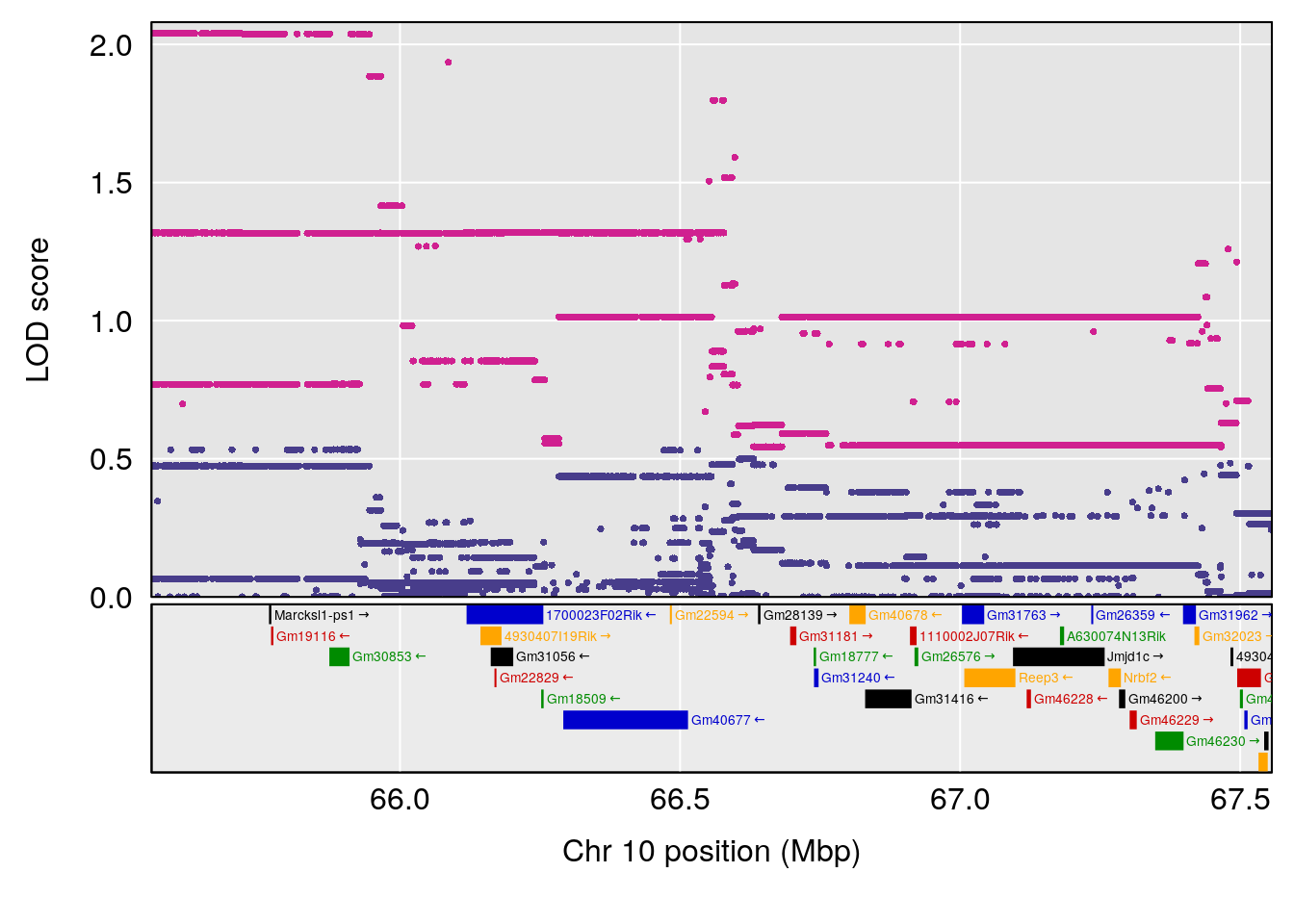
# [1] 6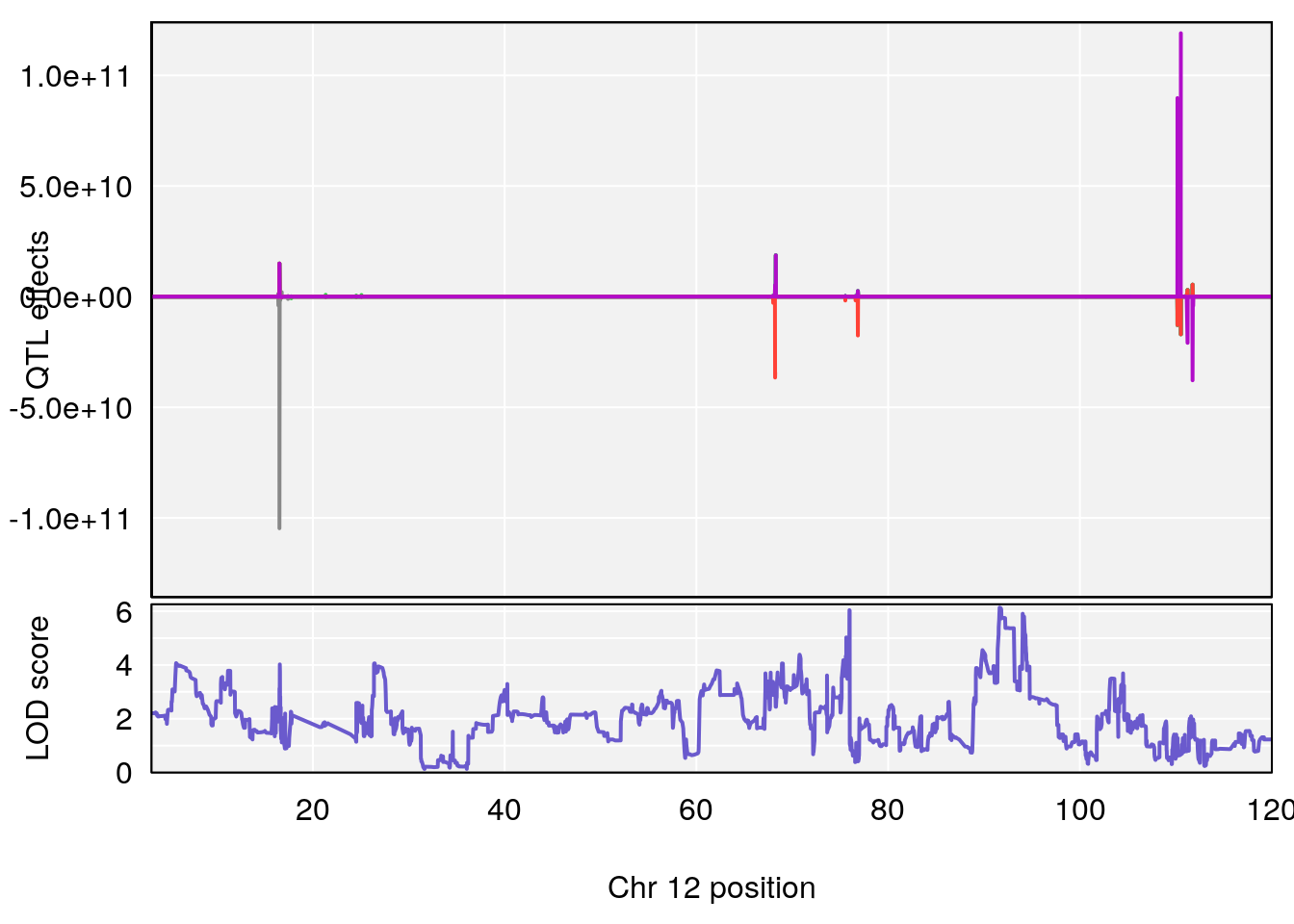
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 7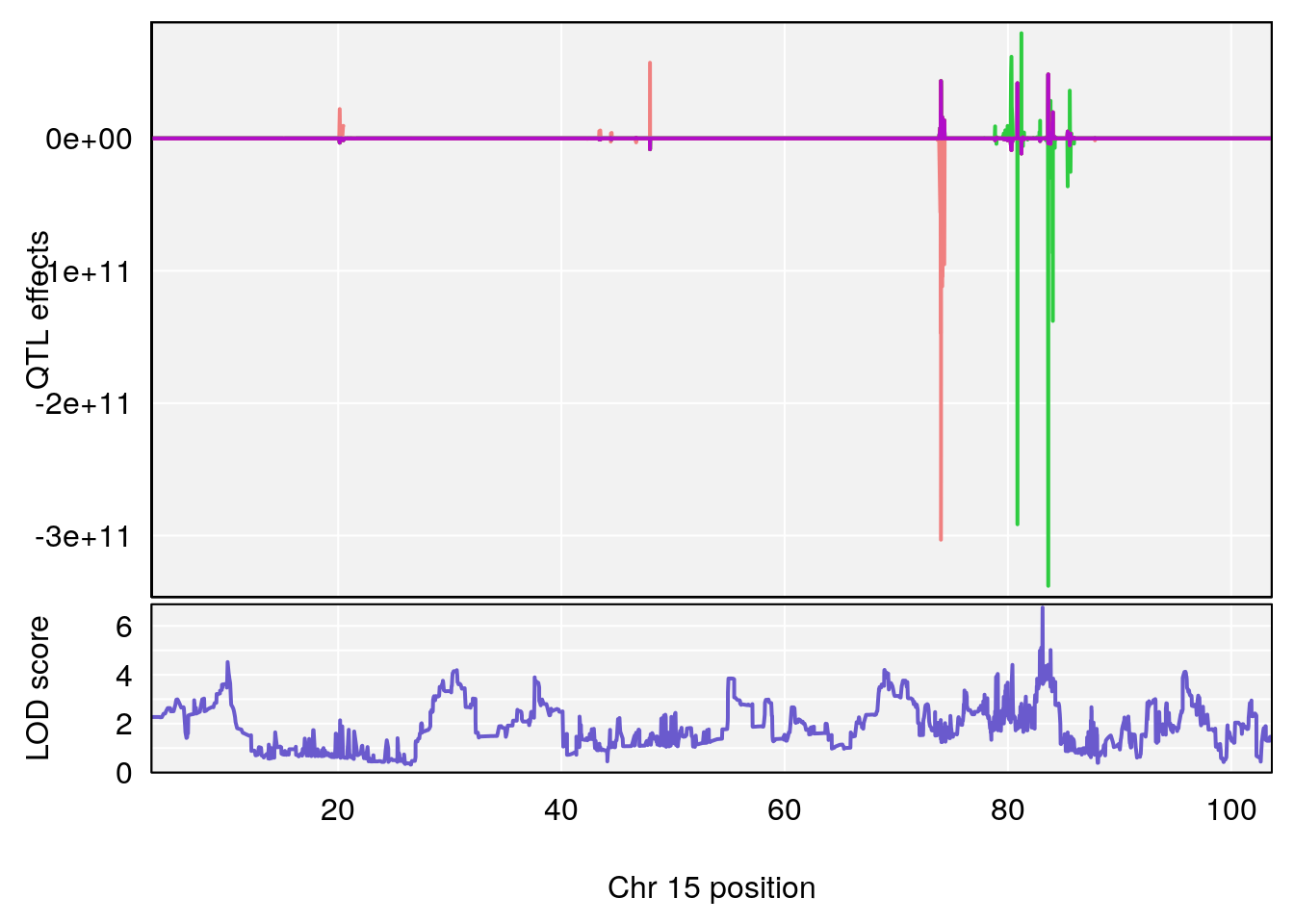
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "StartofRecovery(Hr)"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 3 90.66061 8.542034 90.57609 90.70492
# 2 1 pheno1 4 100.55380 7.820399 99.32926 100.79381
# 3 1 pheno1 8 83.45834 6.600689 70.15607 84.22023
# 4 1 pheno1 9 123.42827 6.126531 116.90024 123.85184
# 5 1 pheno1 10 65.71837 7.117583 64.45014 66.60381
# 6 1 pheno1 12 75.98747 8.039338 75.95610 94.18281
# 7 1 pheno1 13 95.62986 6.411262 43.57533 95.63661
# [1] 1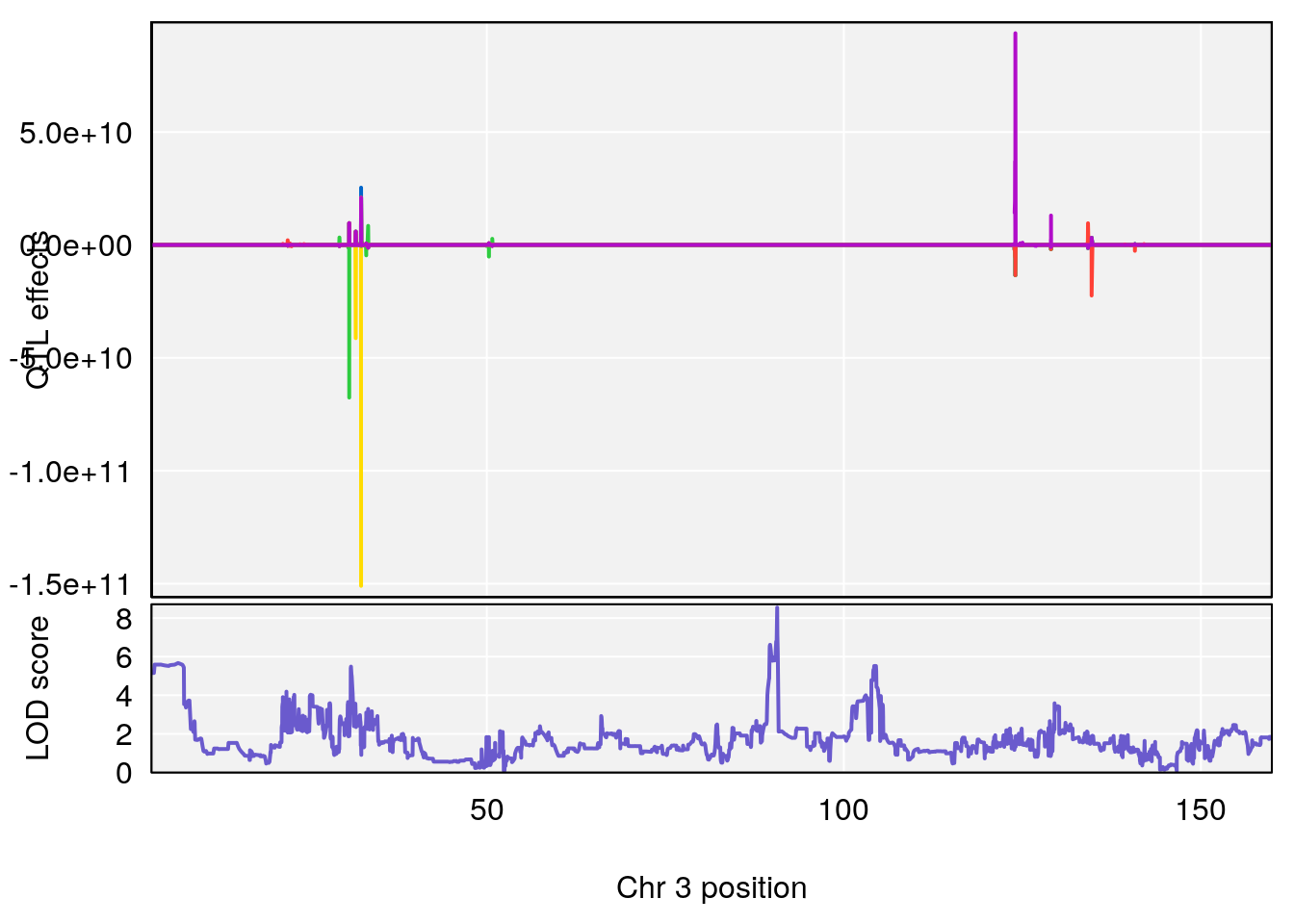
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
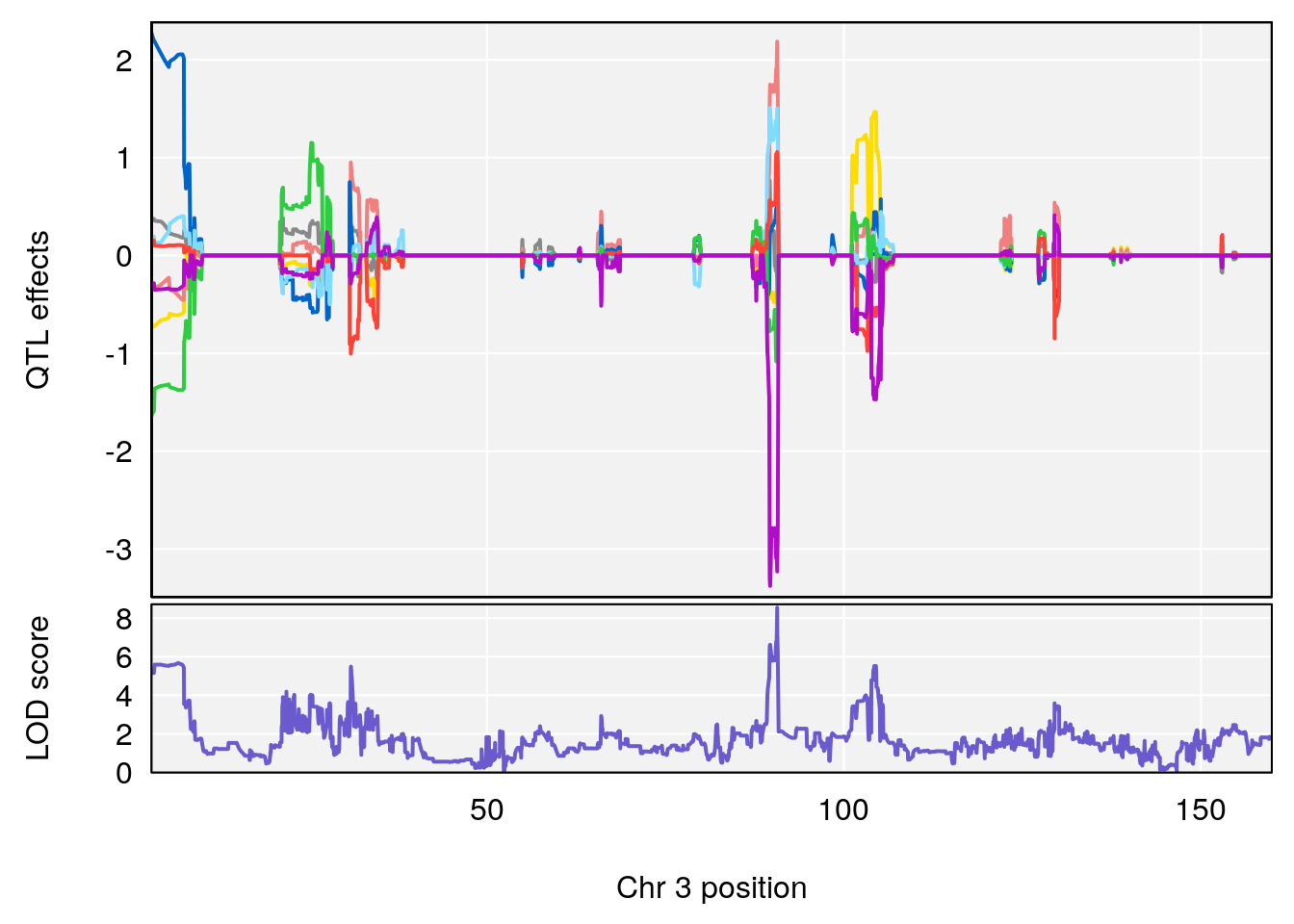
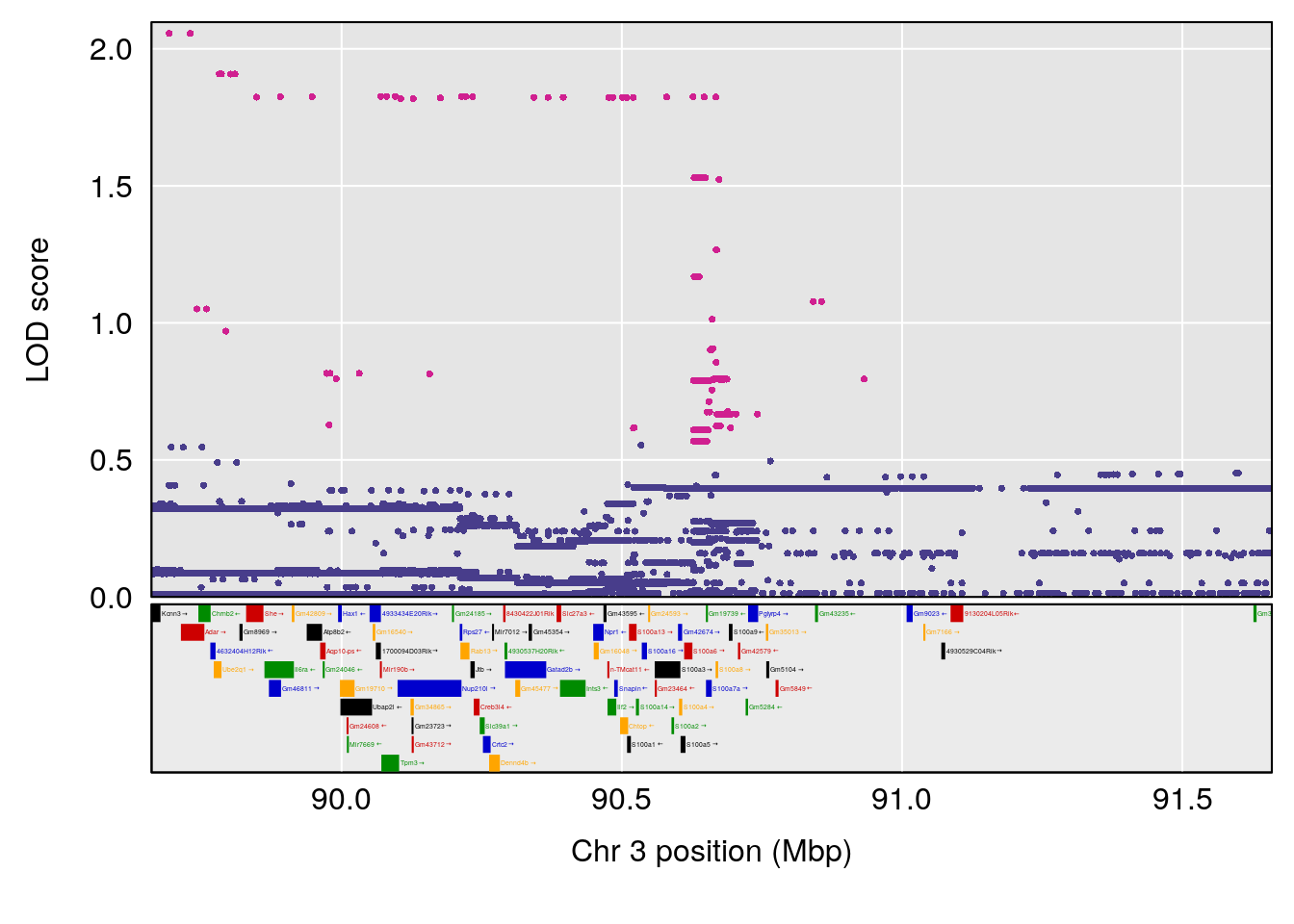
# [1] 2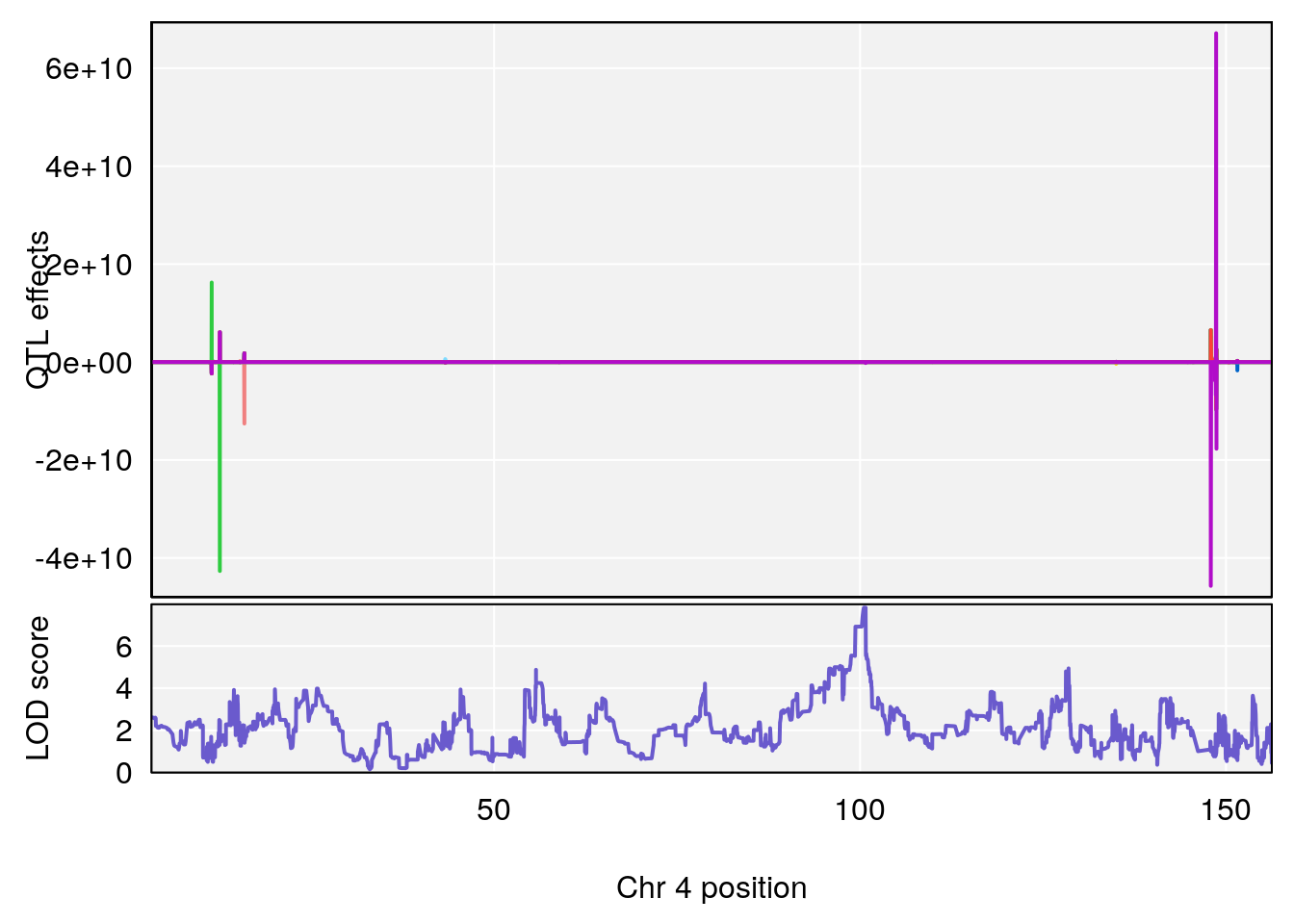
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 3
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
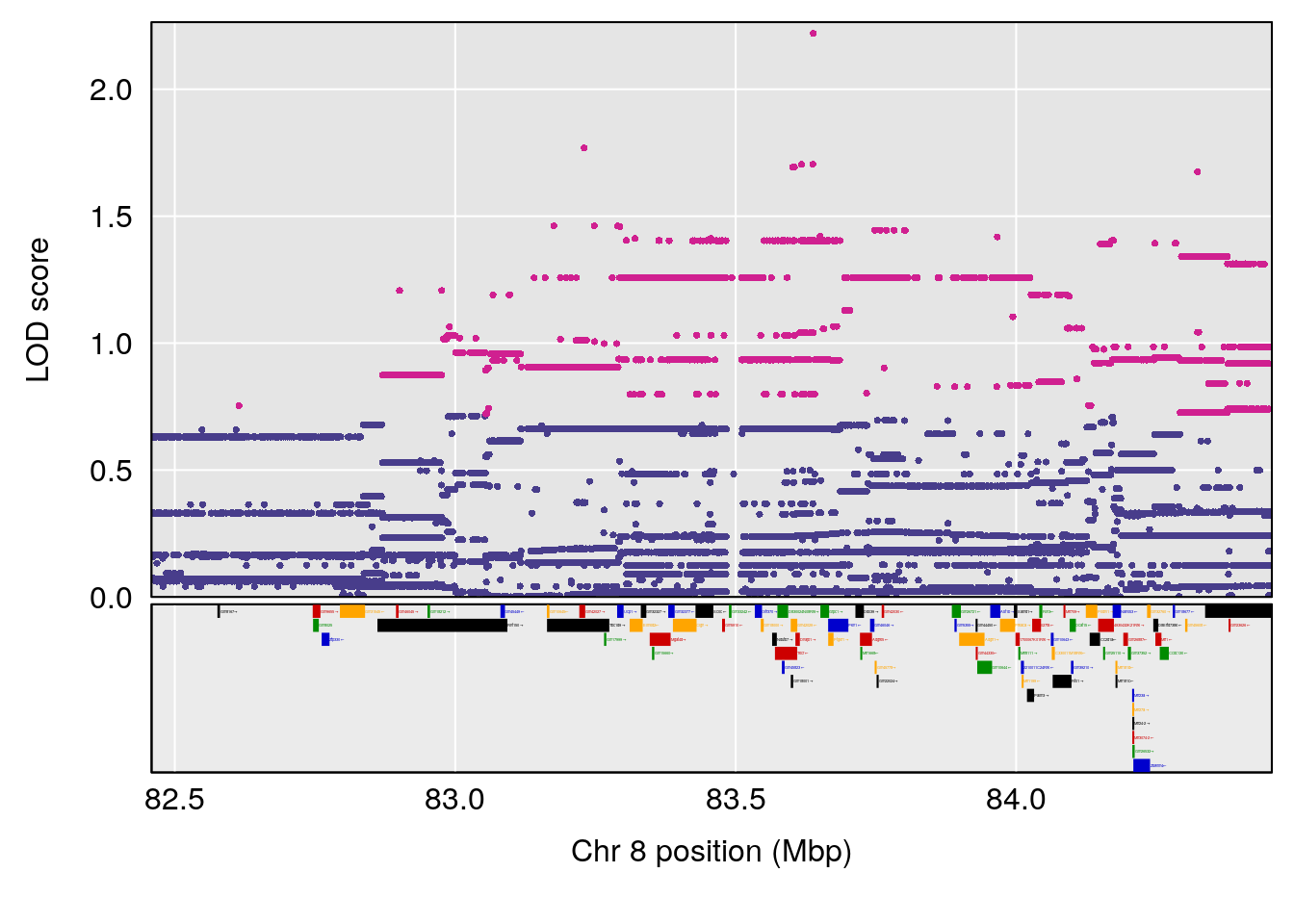
# [1] 4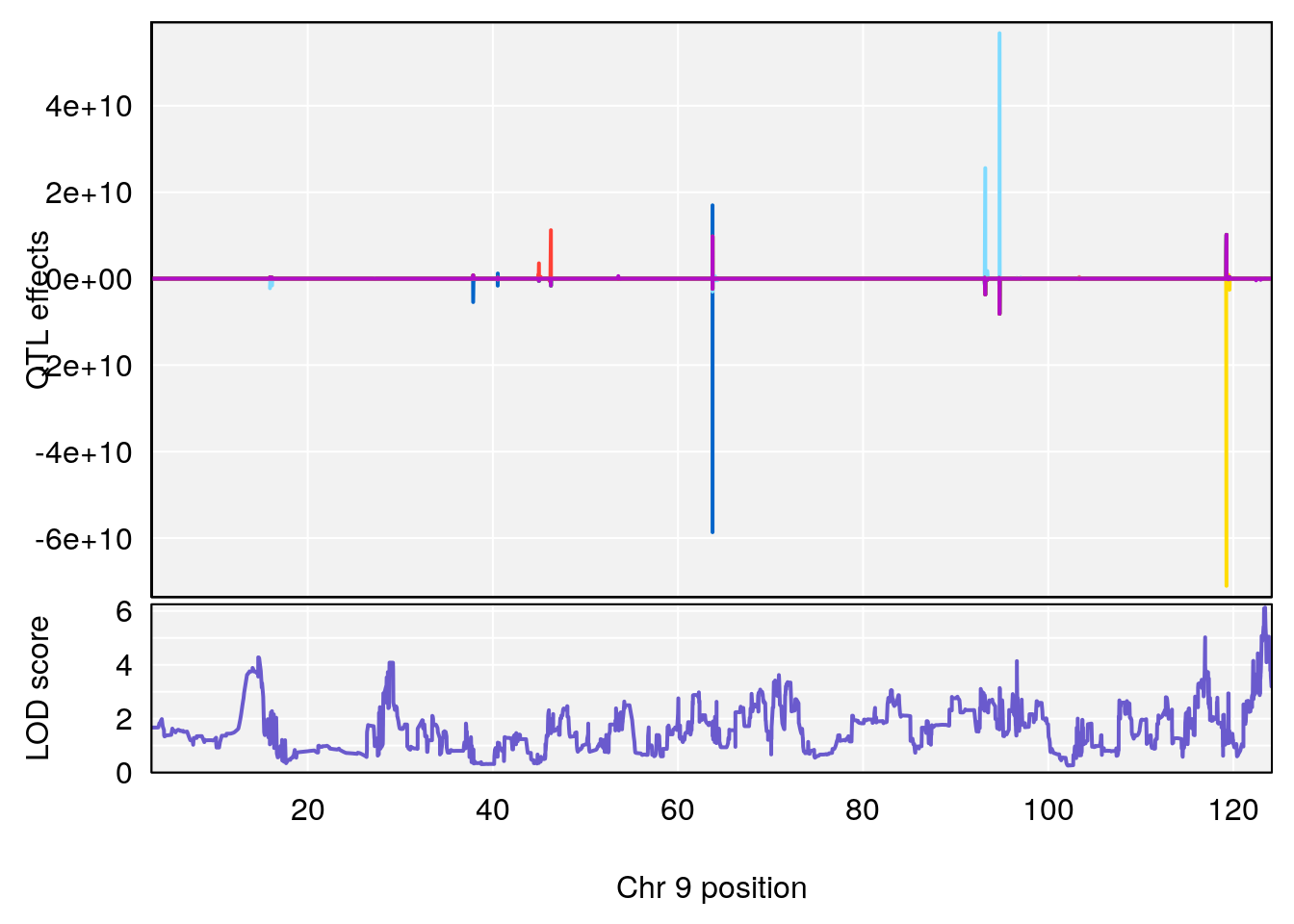
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
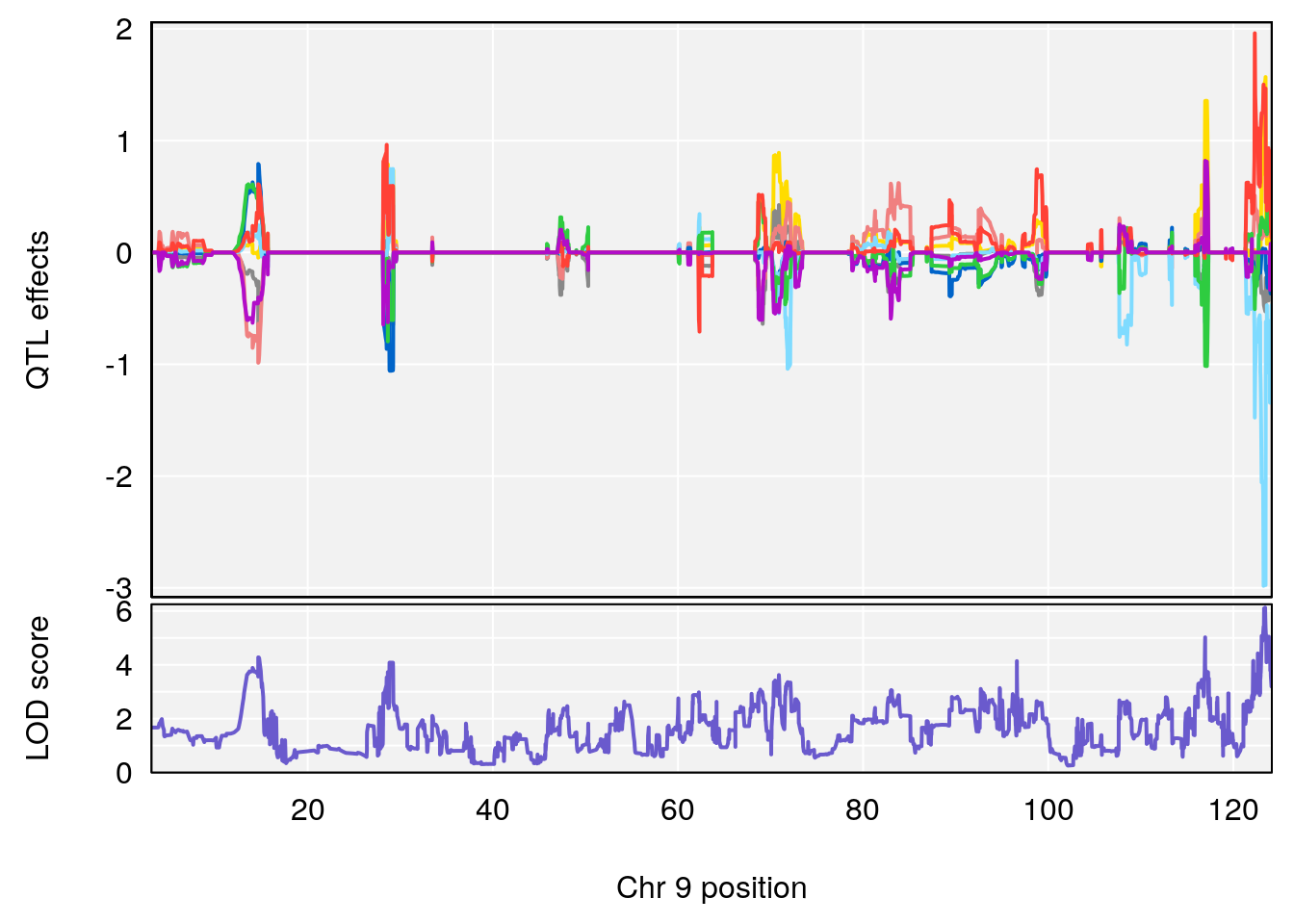
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
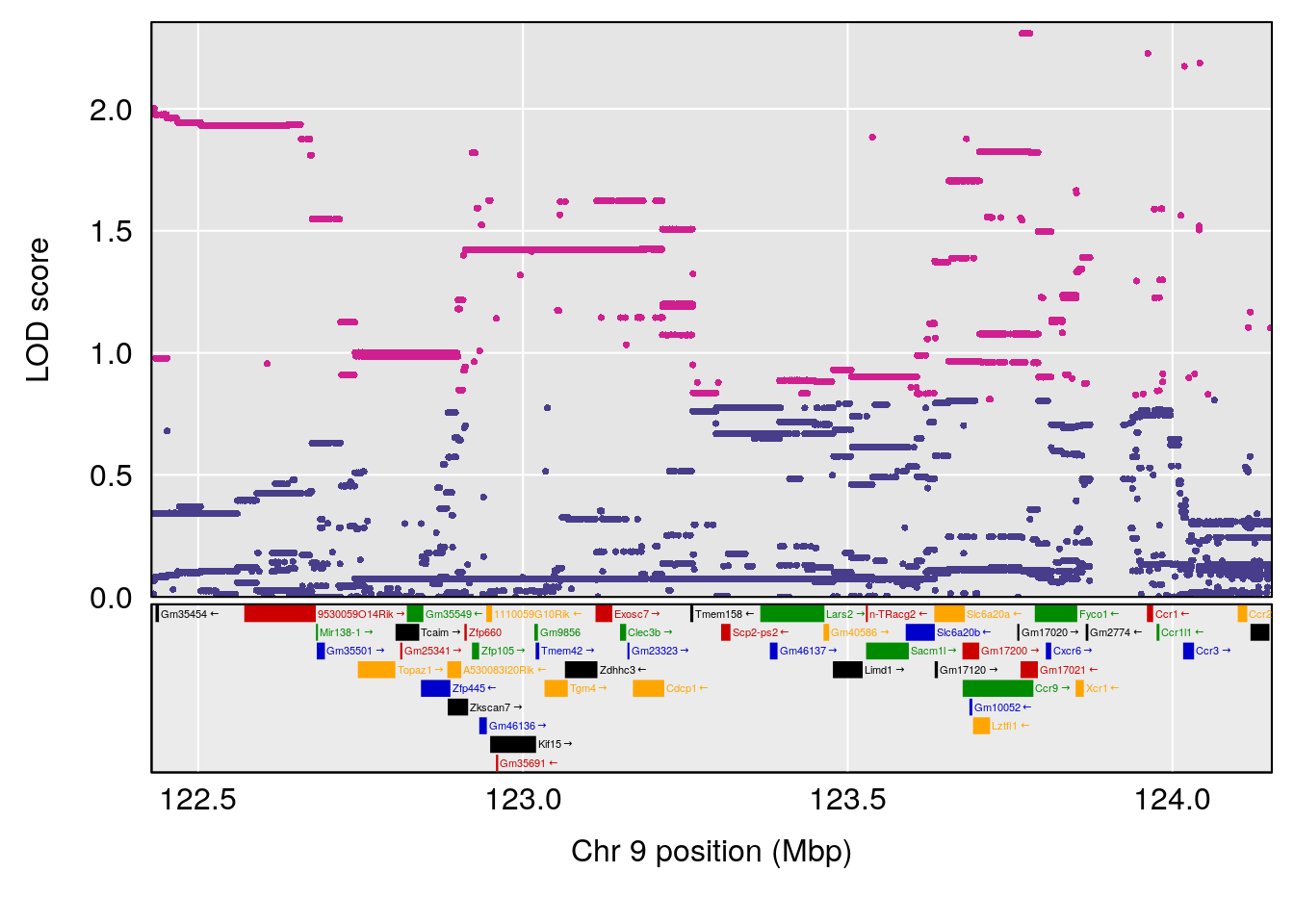
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
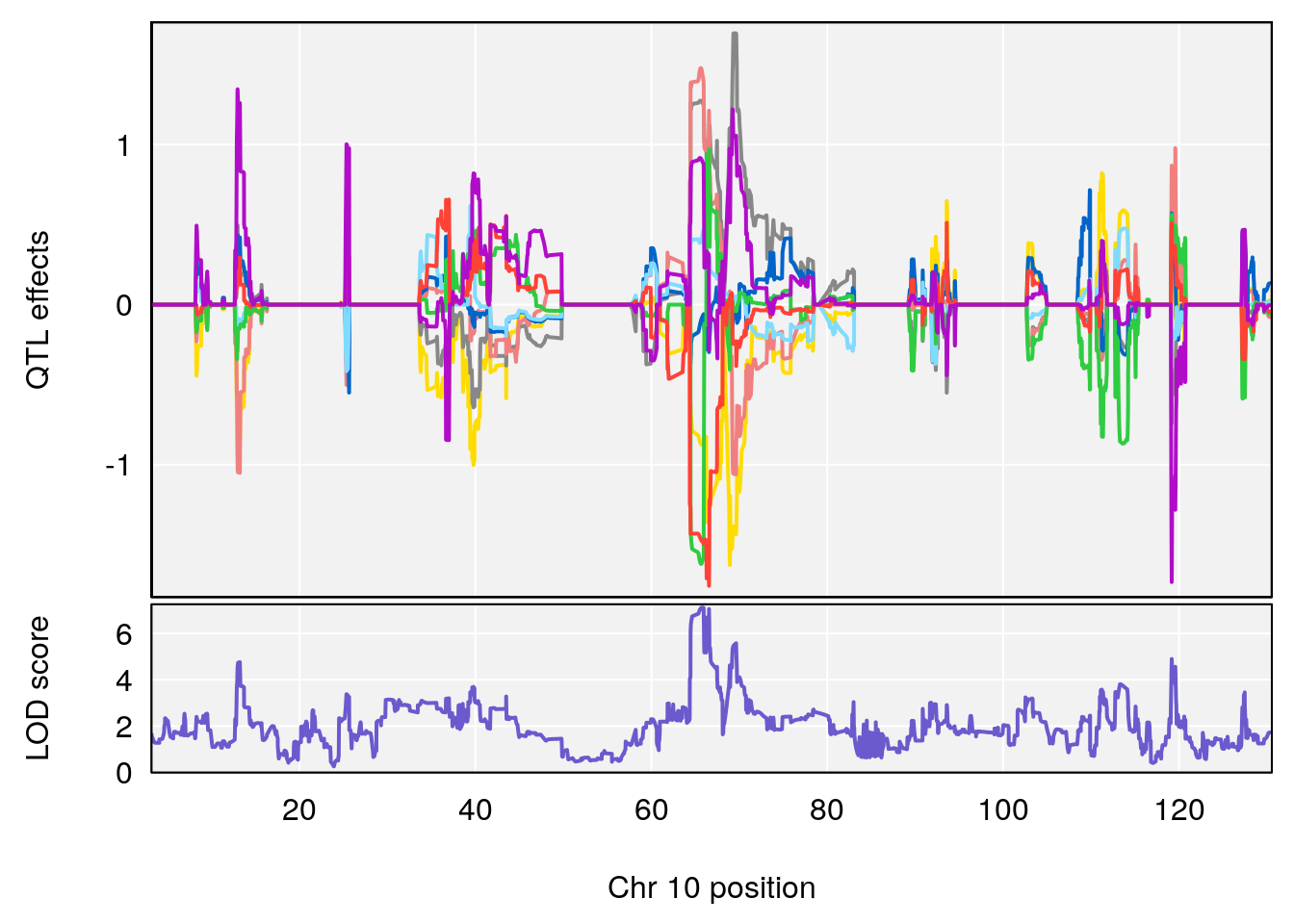
# [1] 5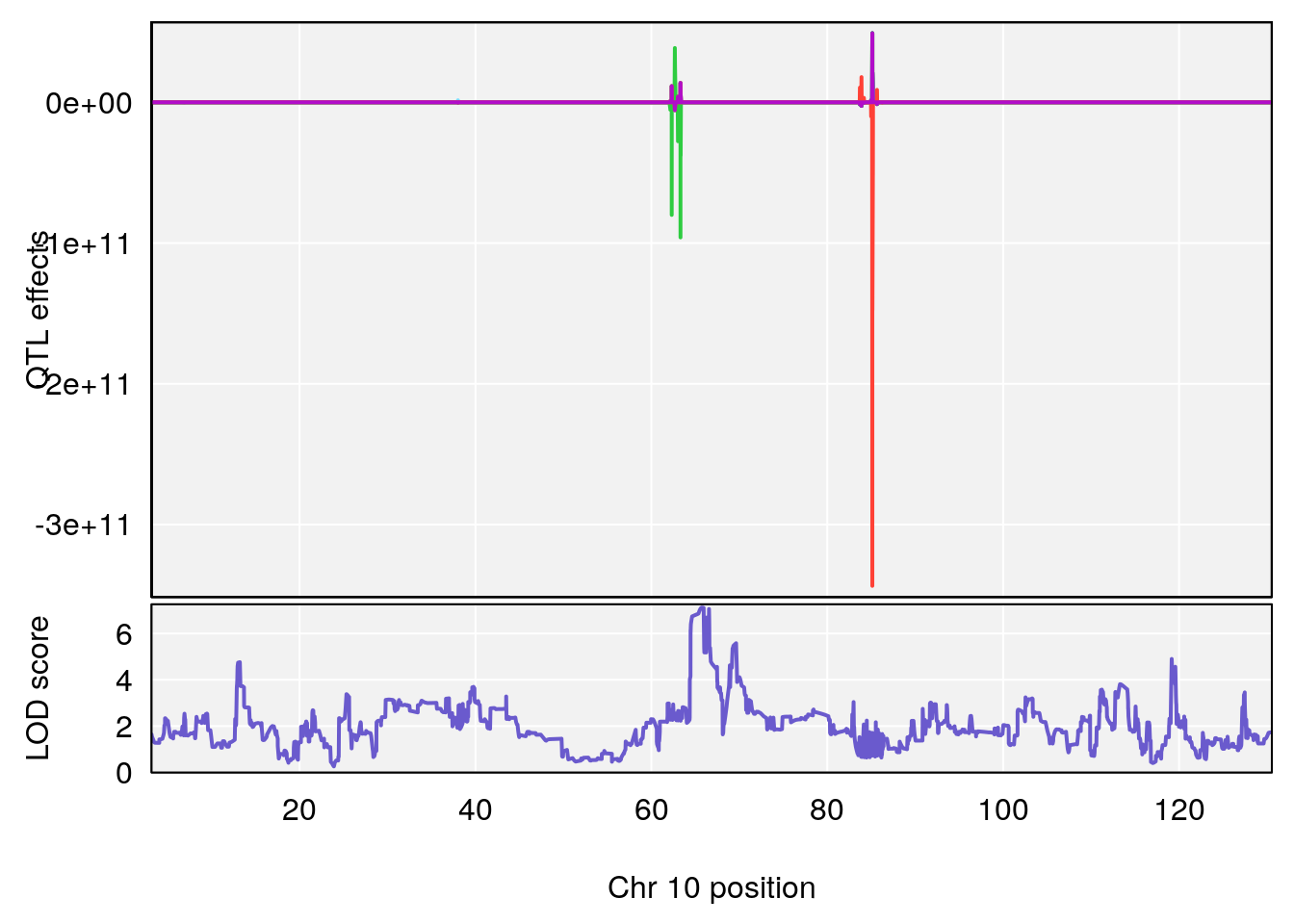
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
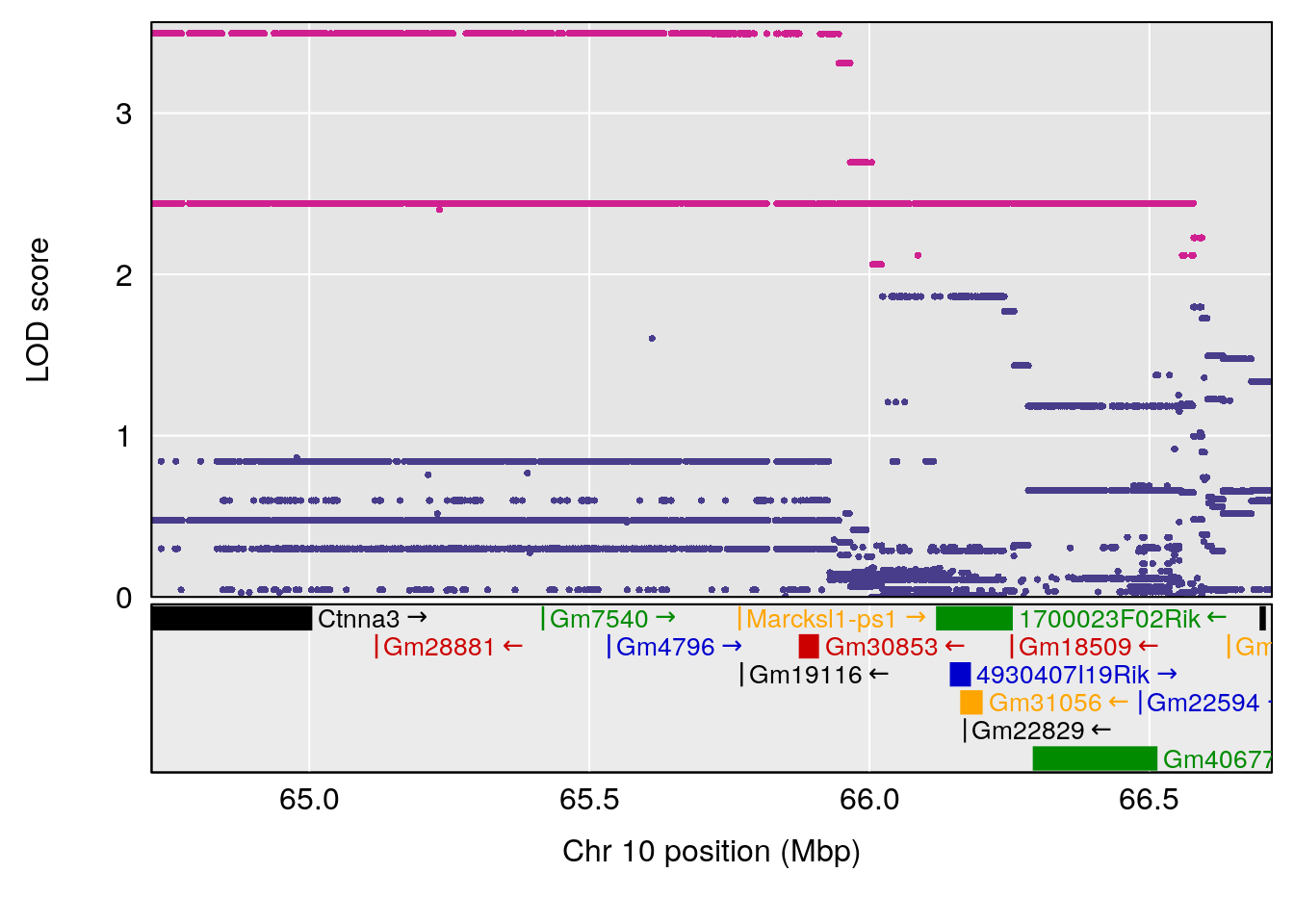
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 6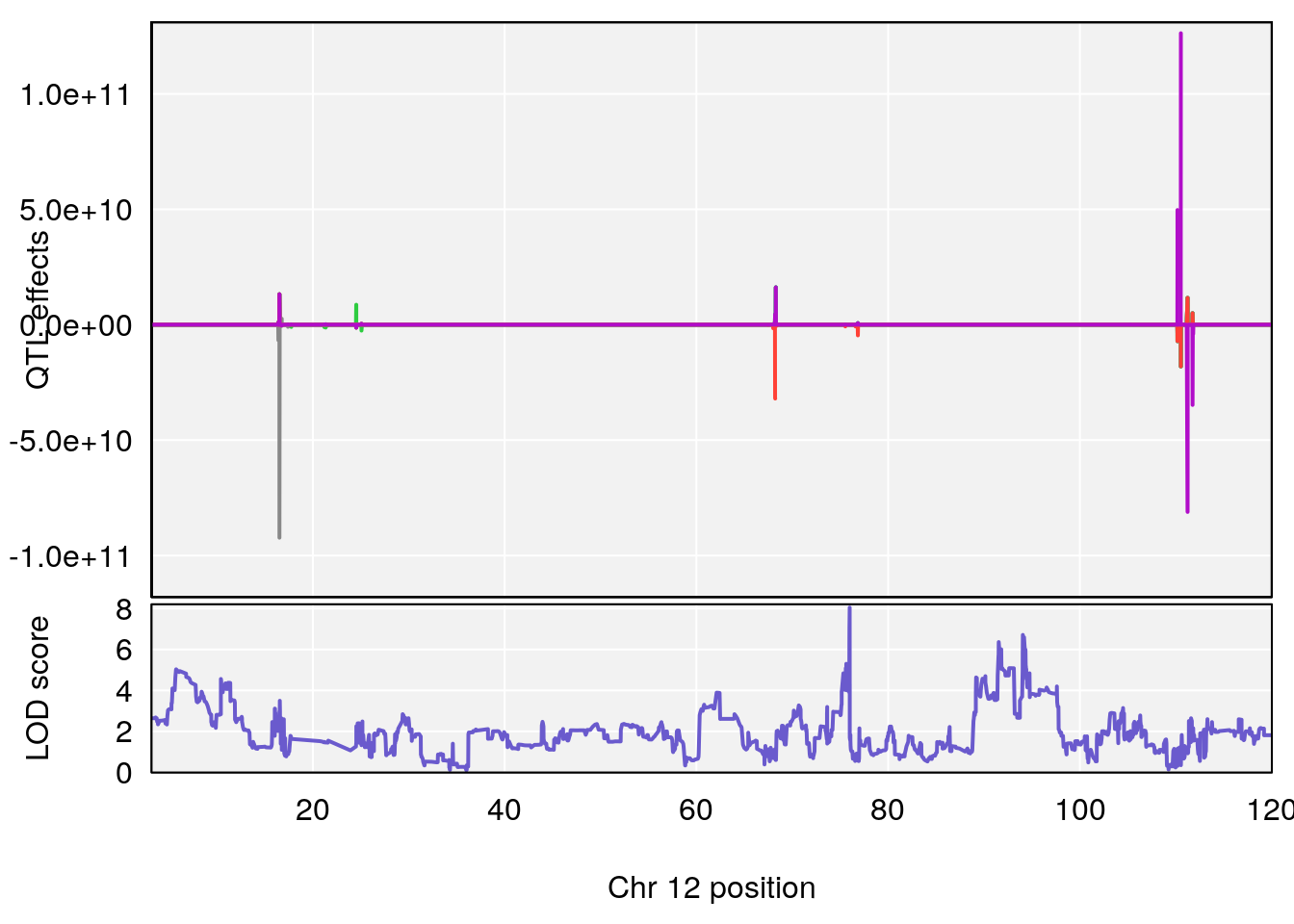
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
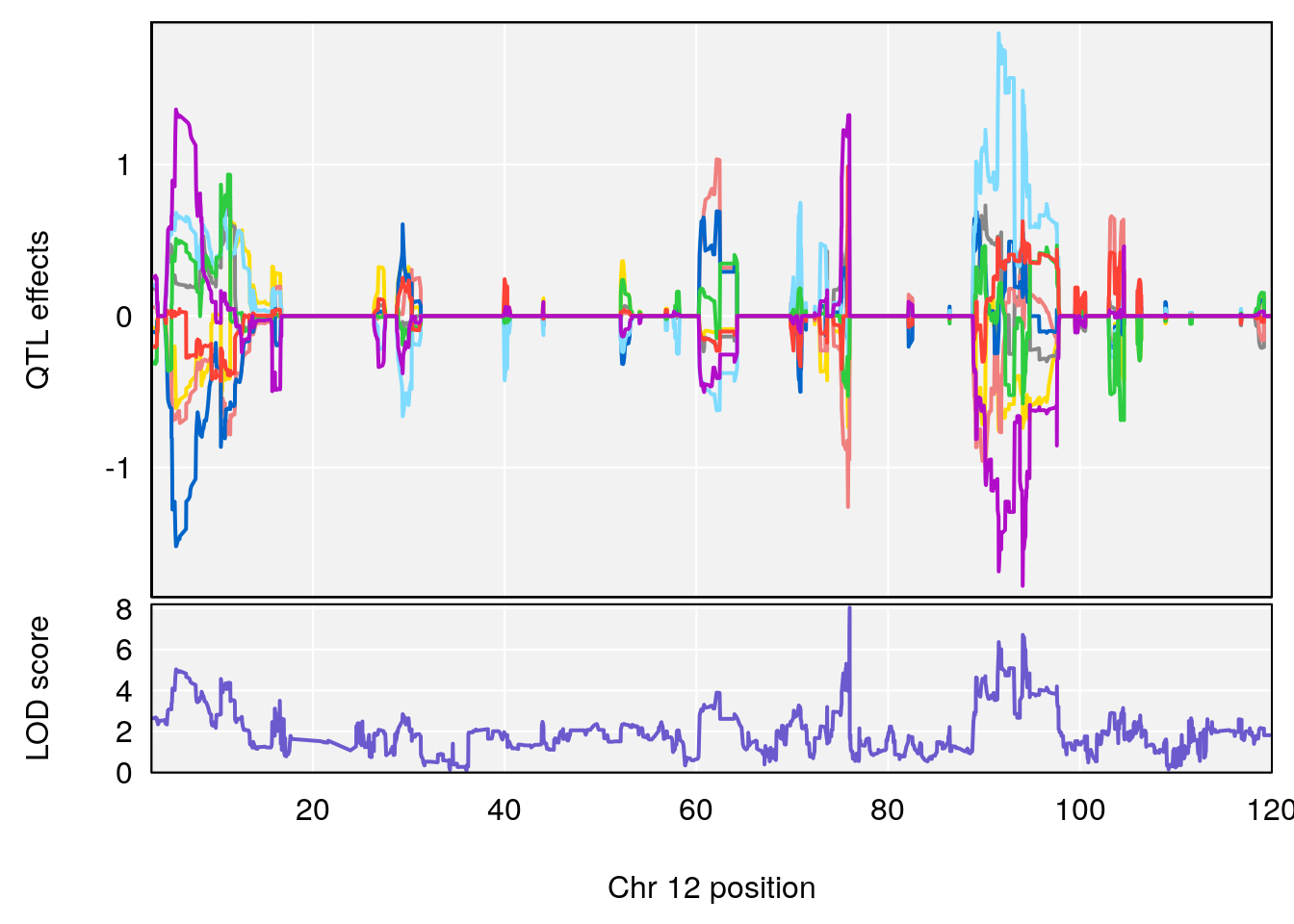
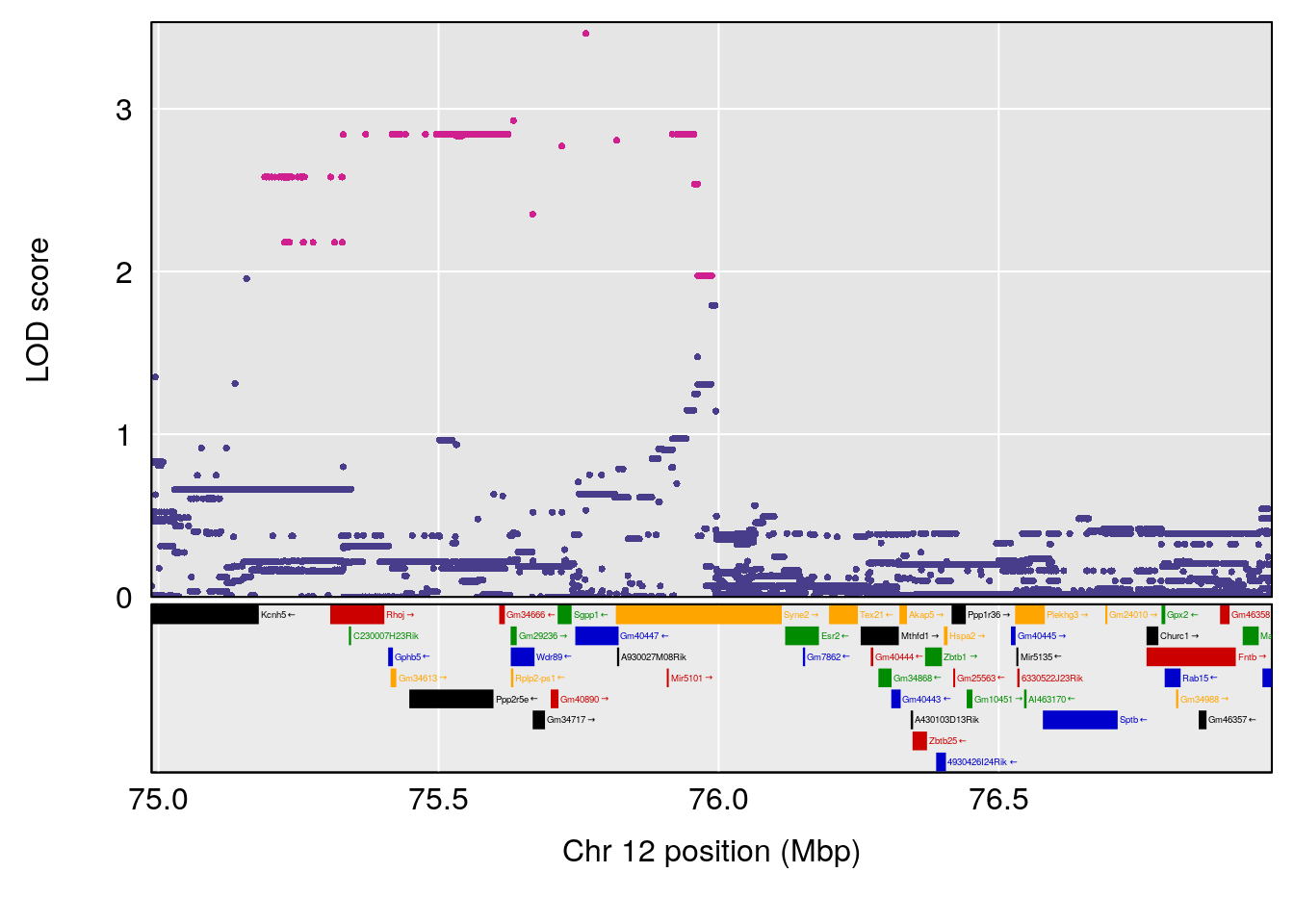
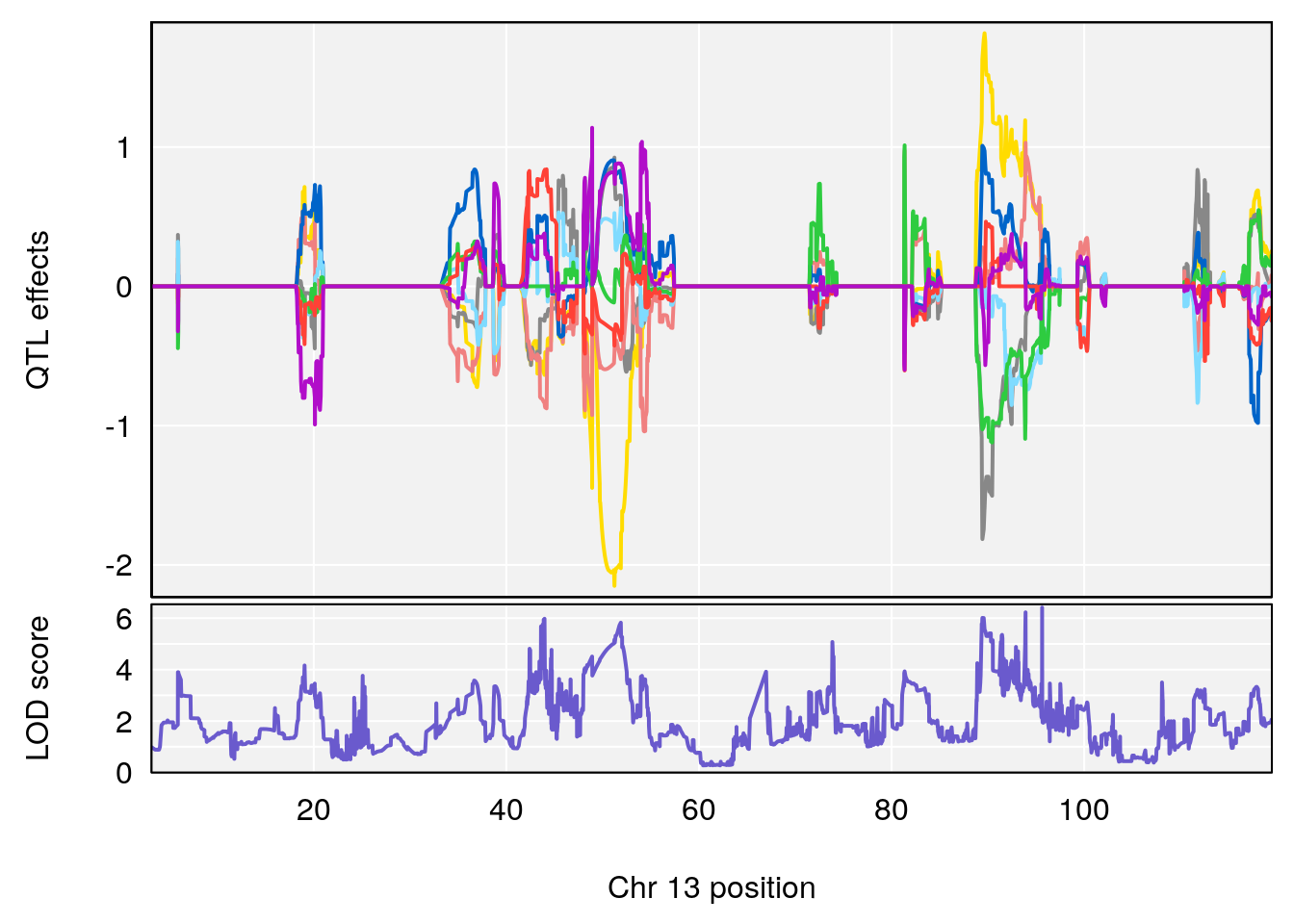
# [1] 7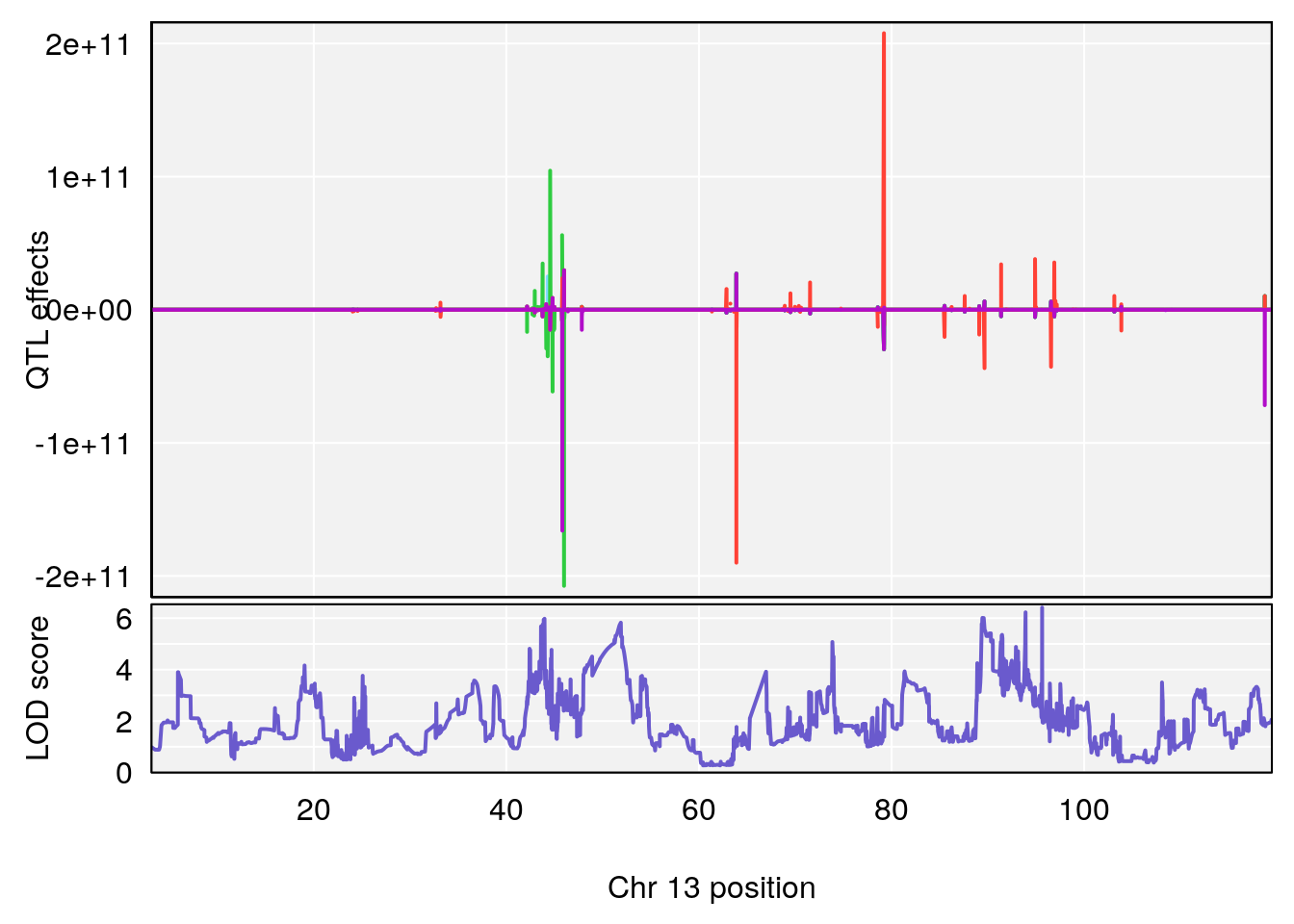
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
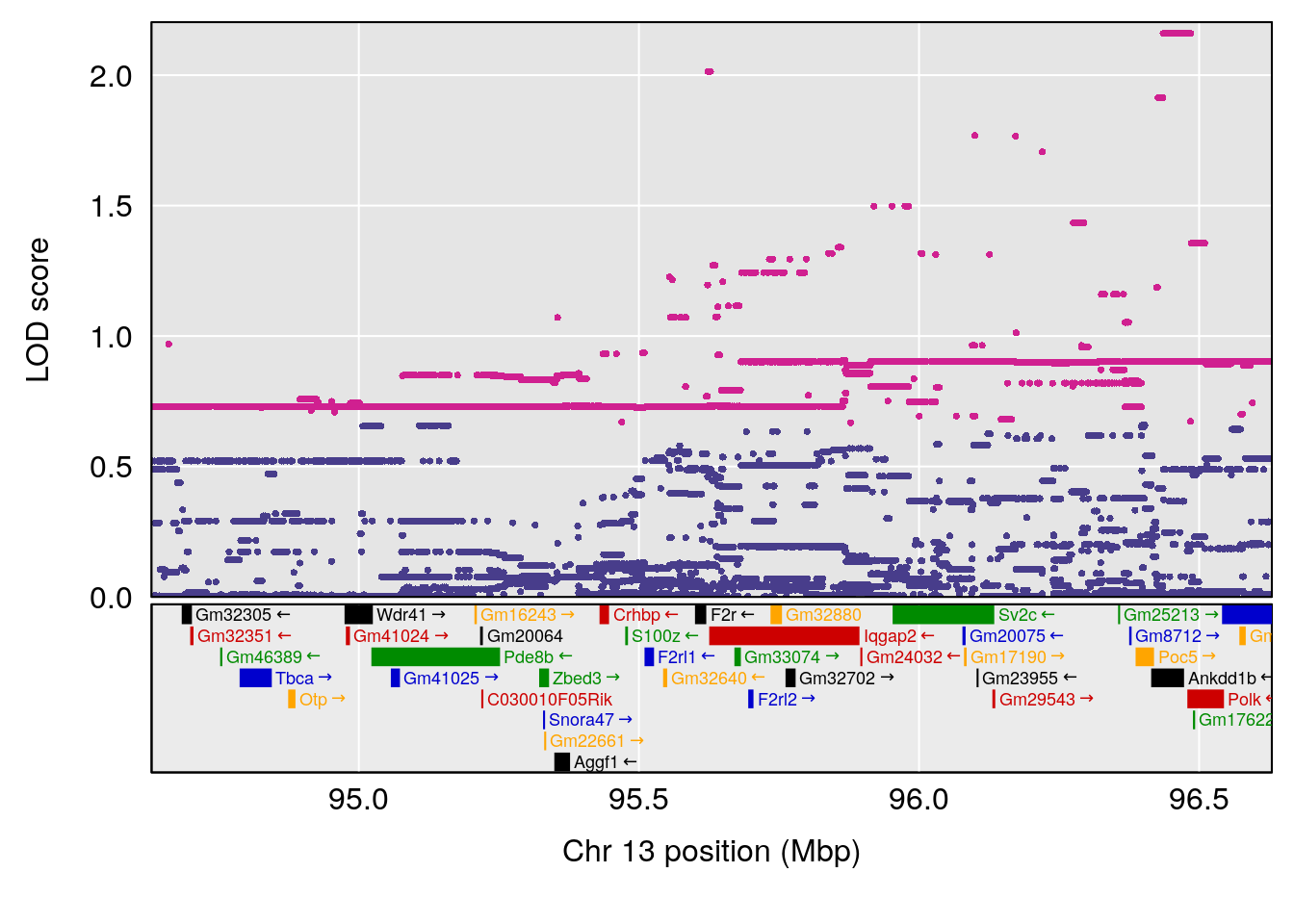
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
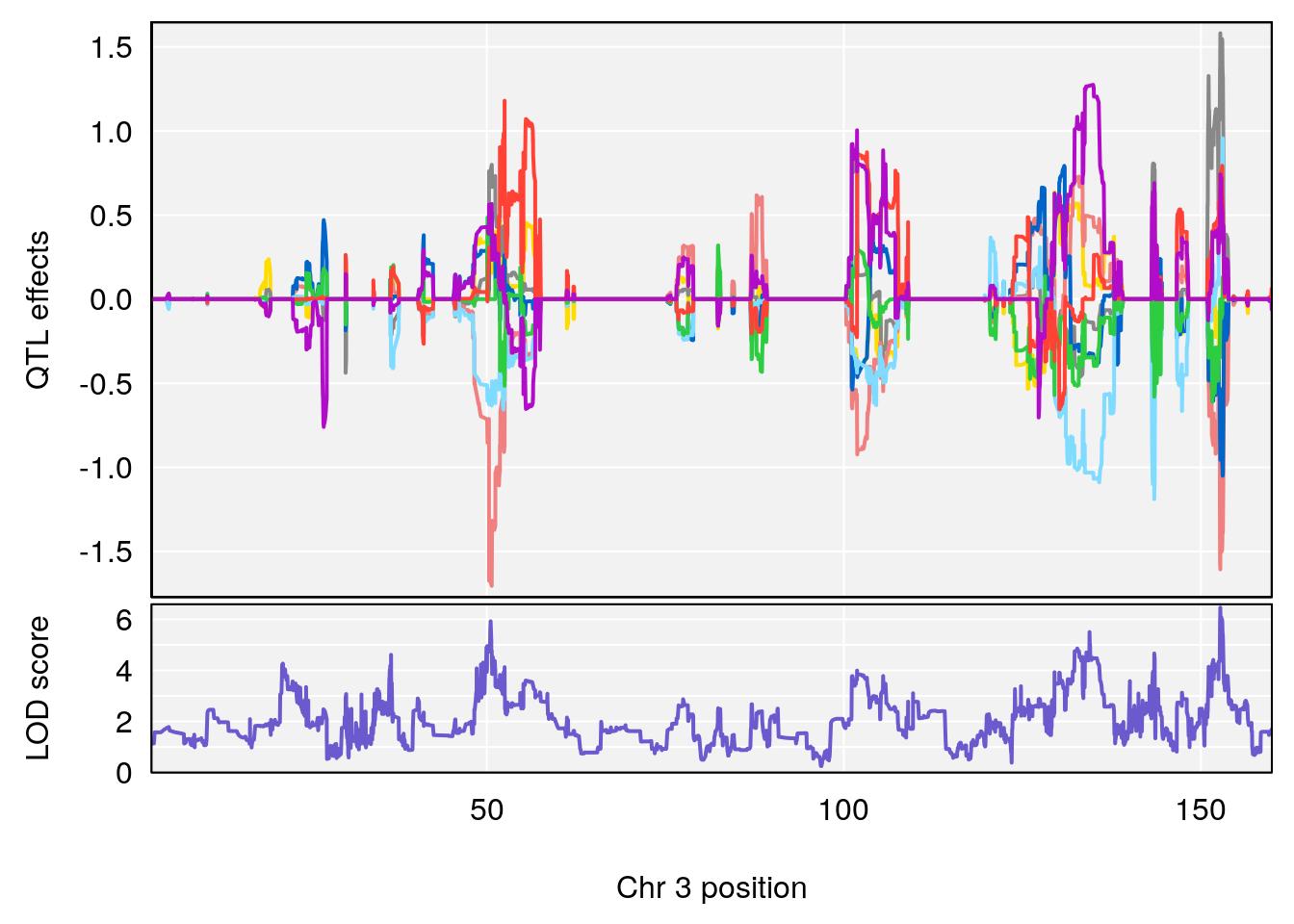
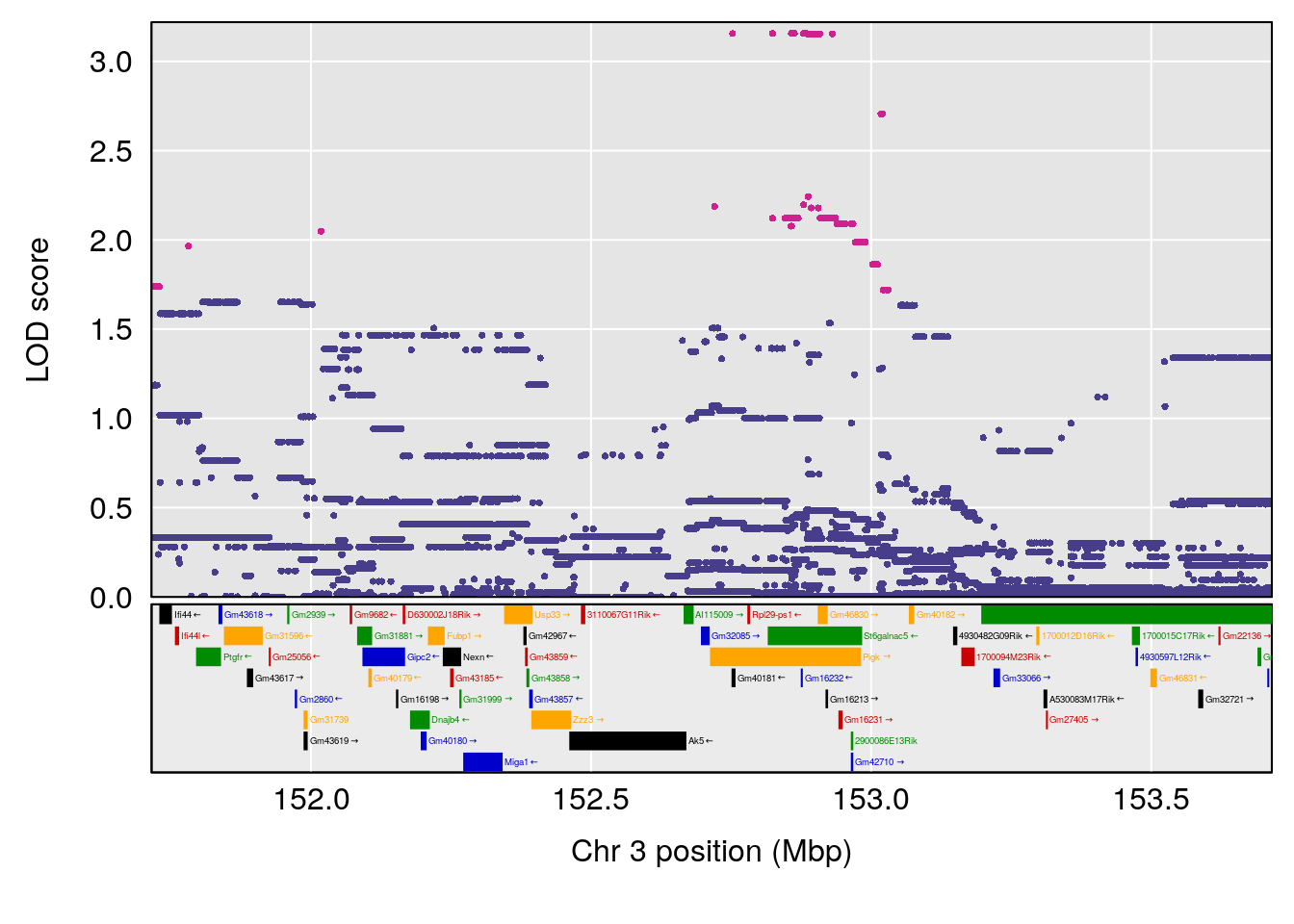
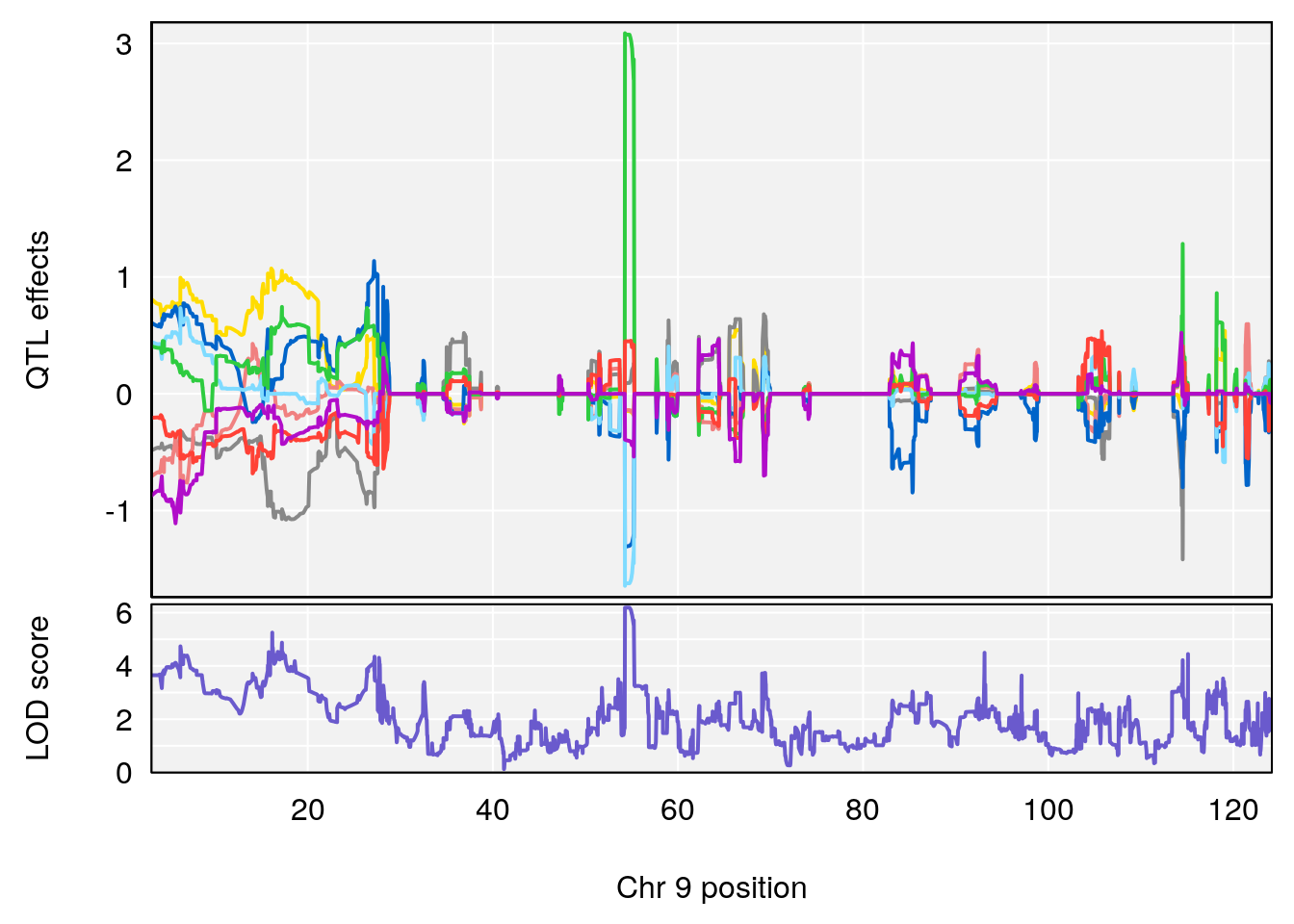
# [1] "RRRecoveryRate(%/Hr)SLOPE"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 192.12423 6.708234 191.931595 193.43806
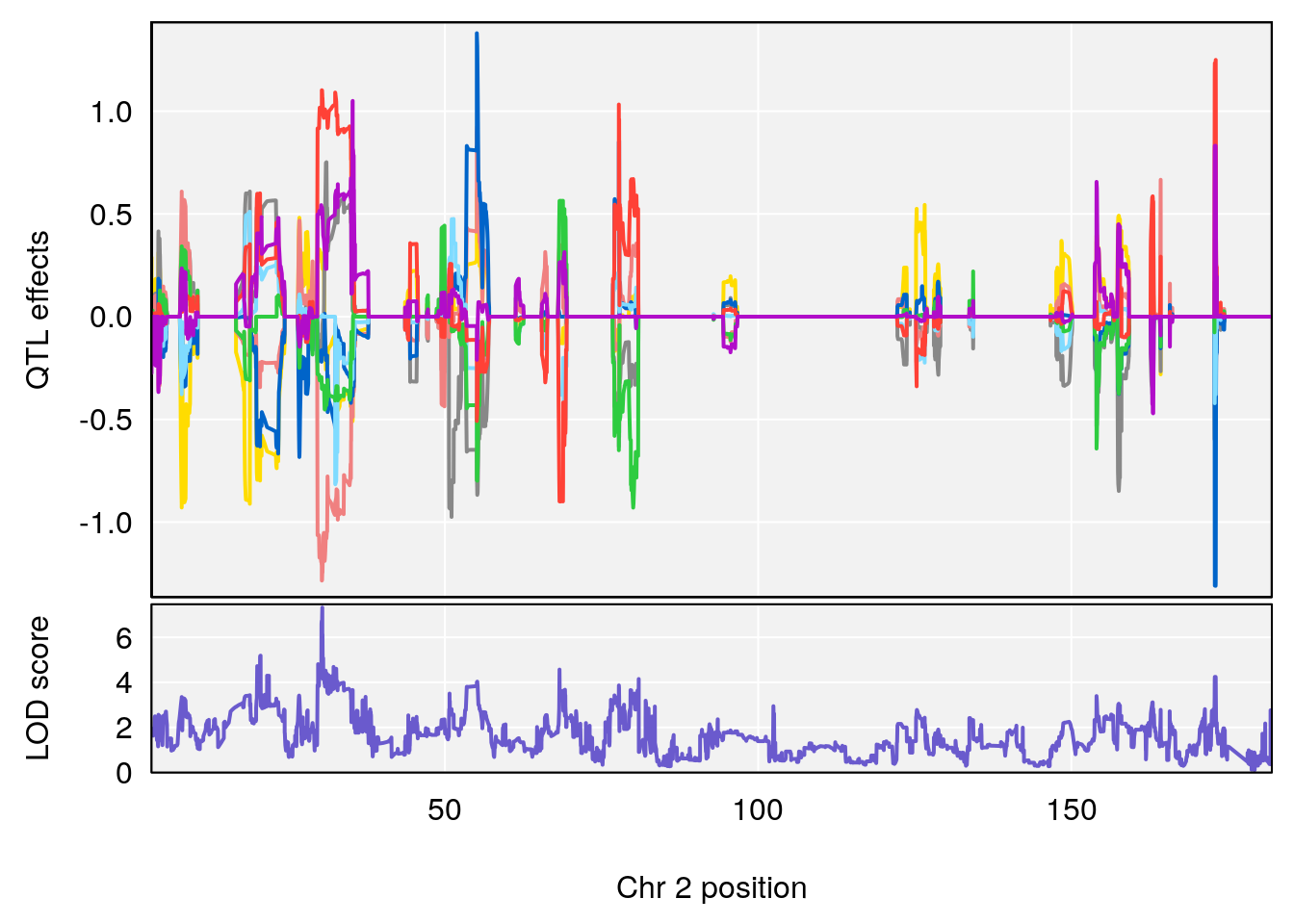
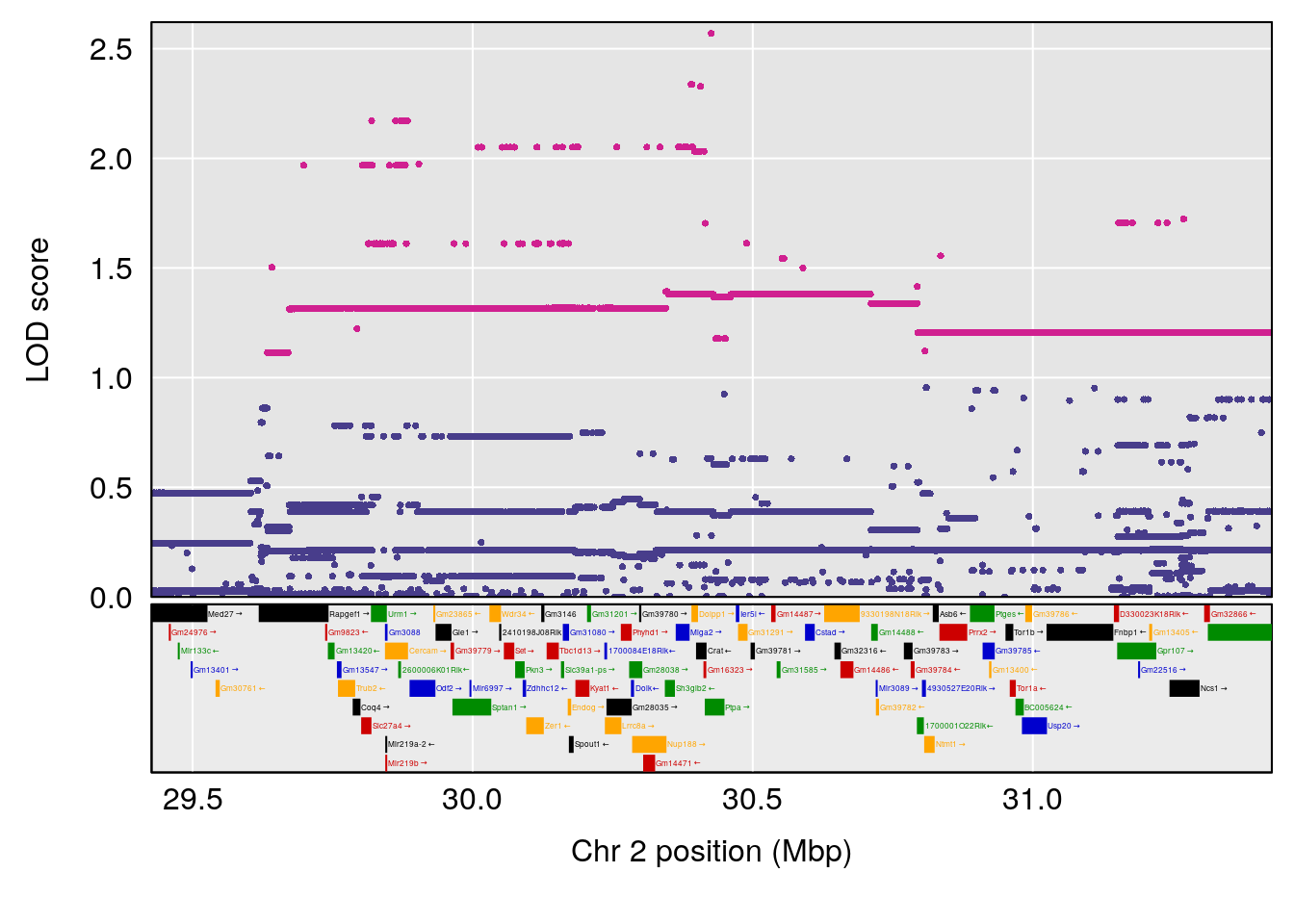
# 2 1 pheno1 2 30.42672 7.319624 30.269862 30.50527
# 3 1 pheno1 3 152.71526 6.460920 50.440226 153.14209
# 4 1 pheno1 9 54.54239 6.189928 6.239837 55.29730
# 5 1 pheno1 15 87.57068 6.600476 84.049458 90.51963
# [1] 1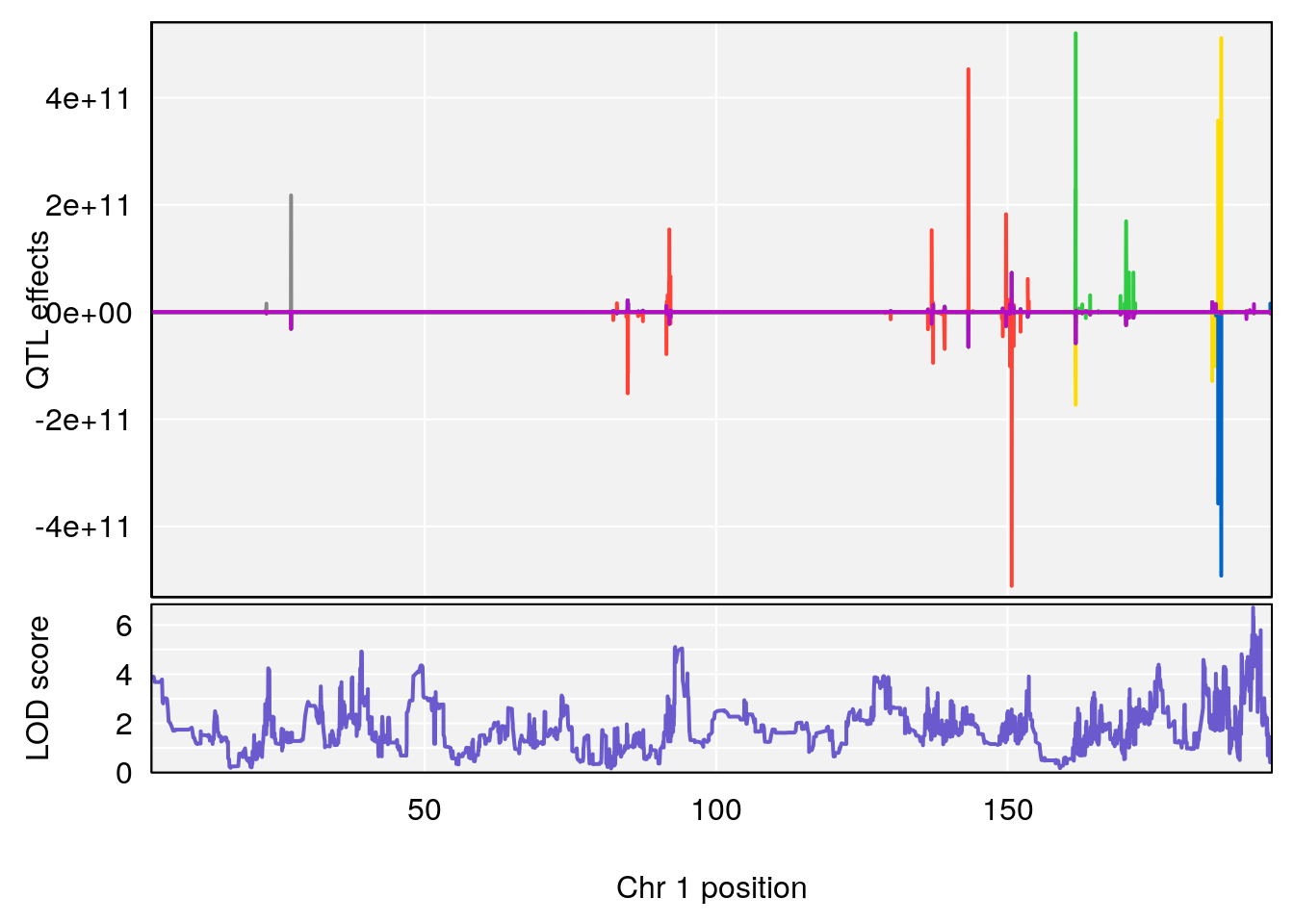
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
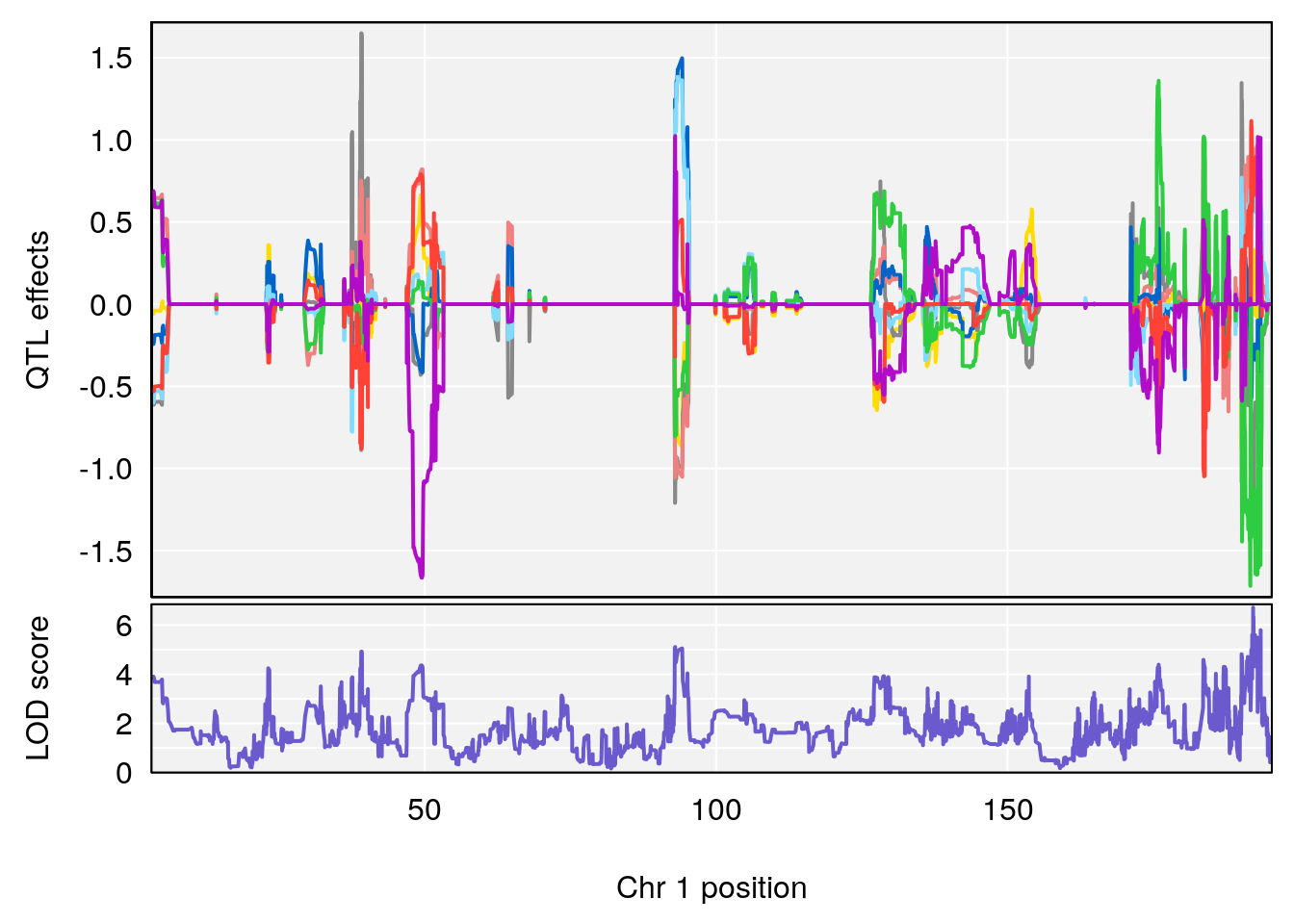
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
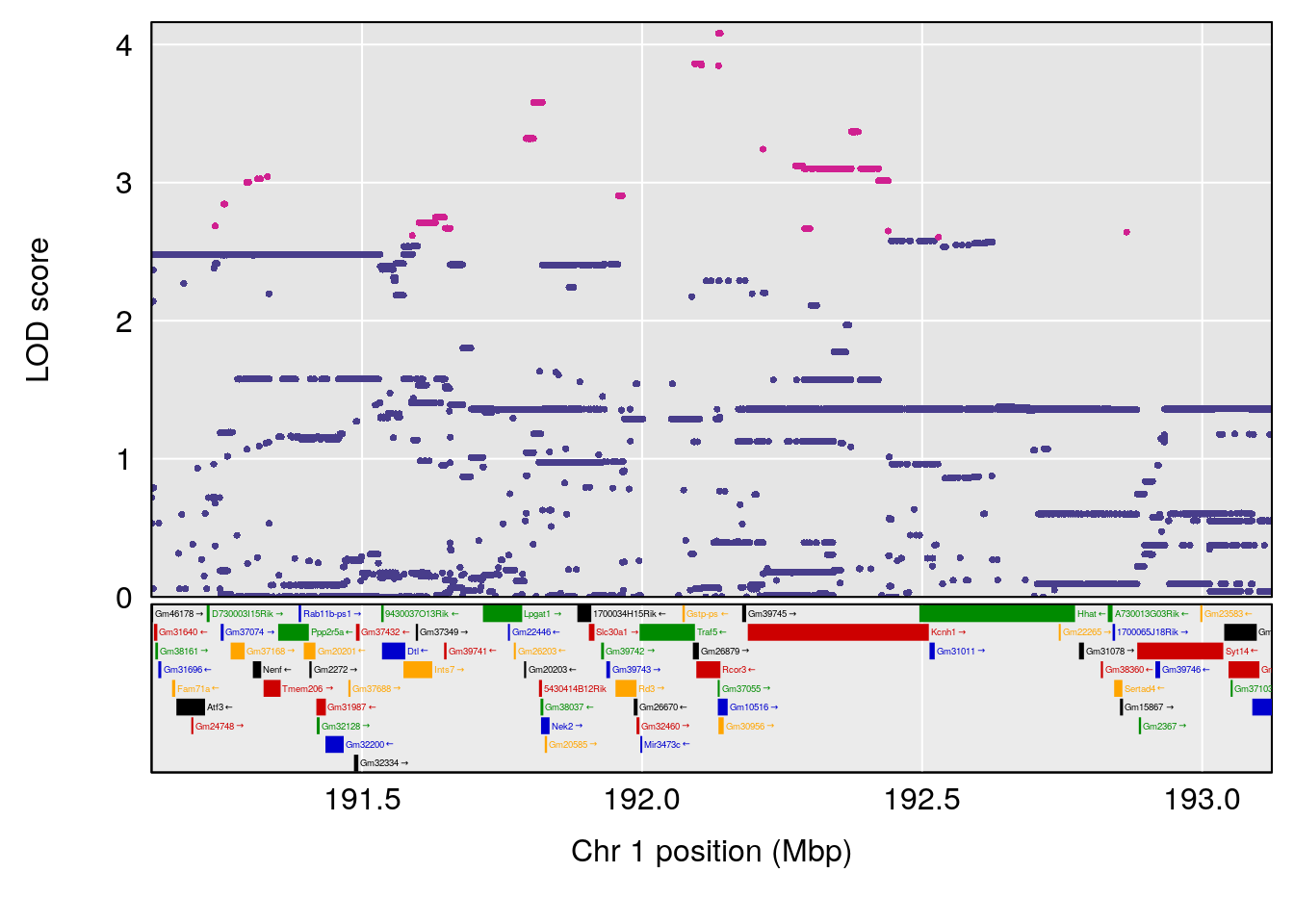
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2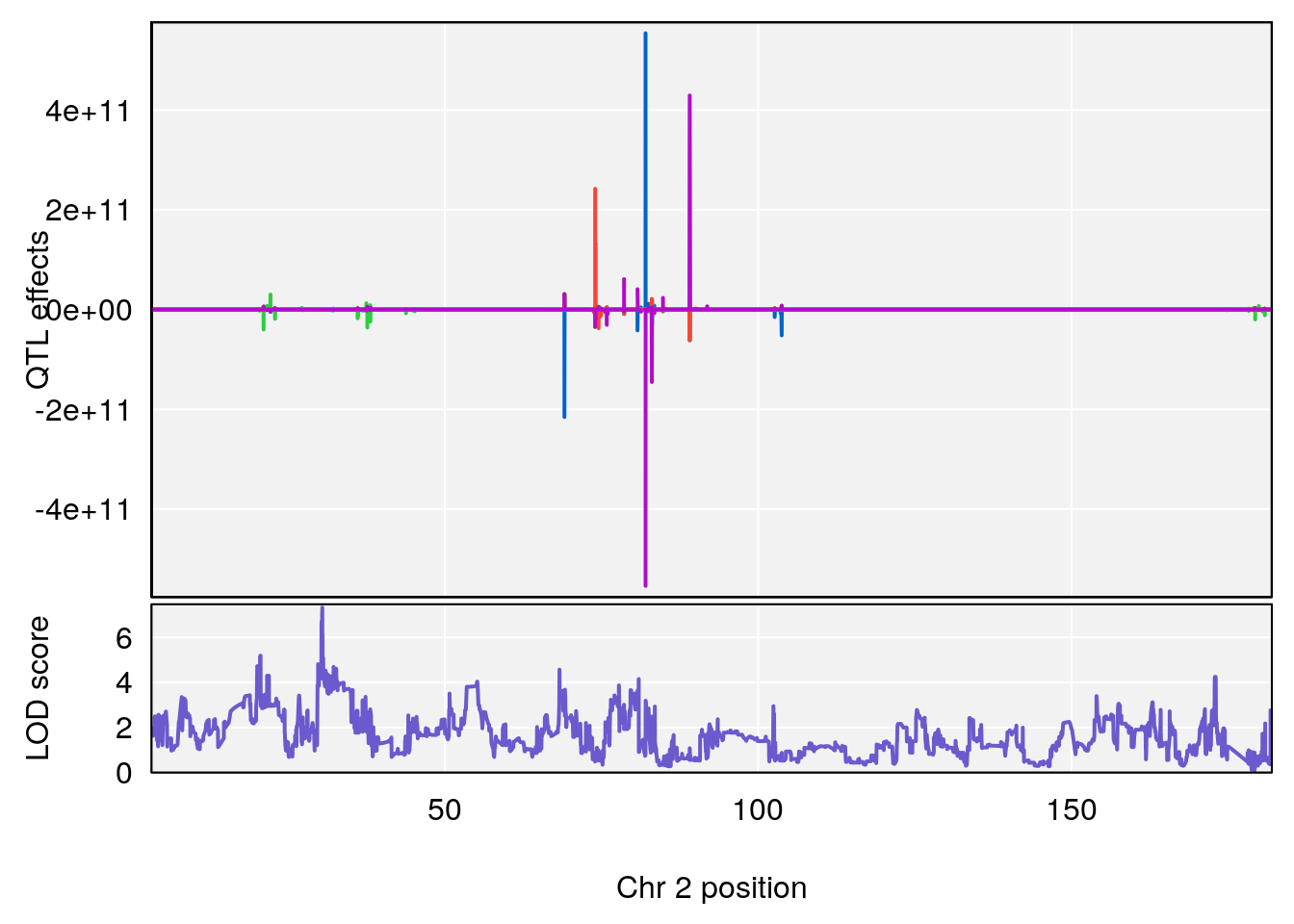
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 3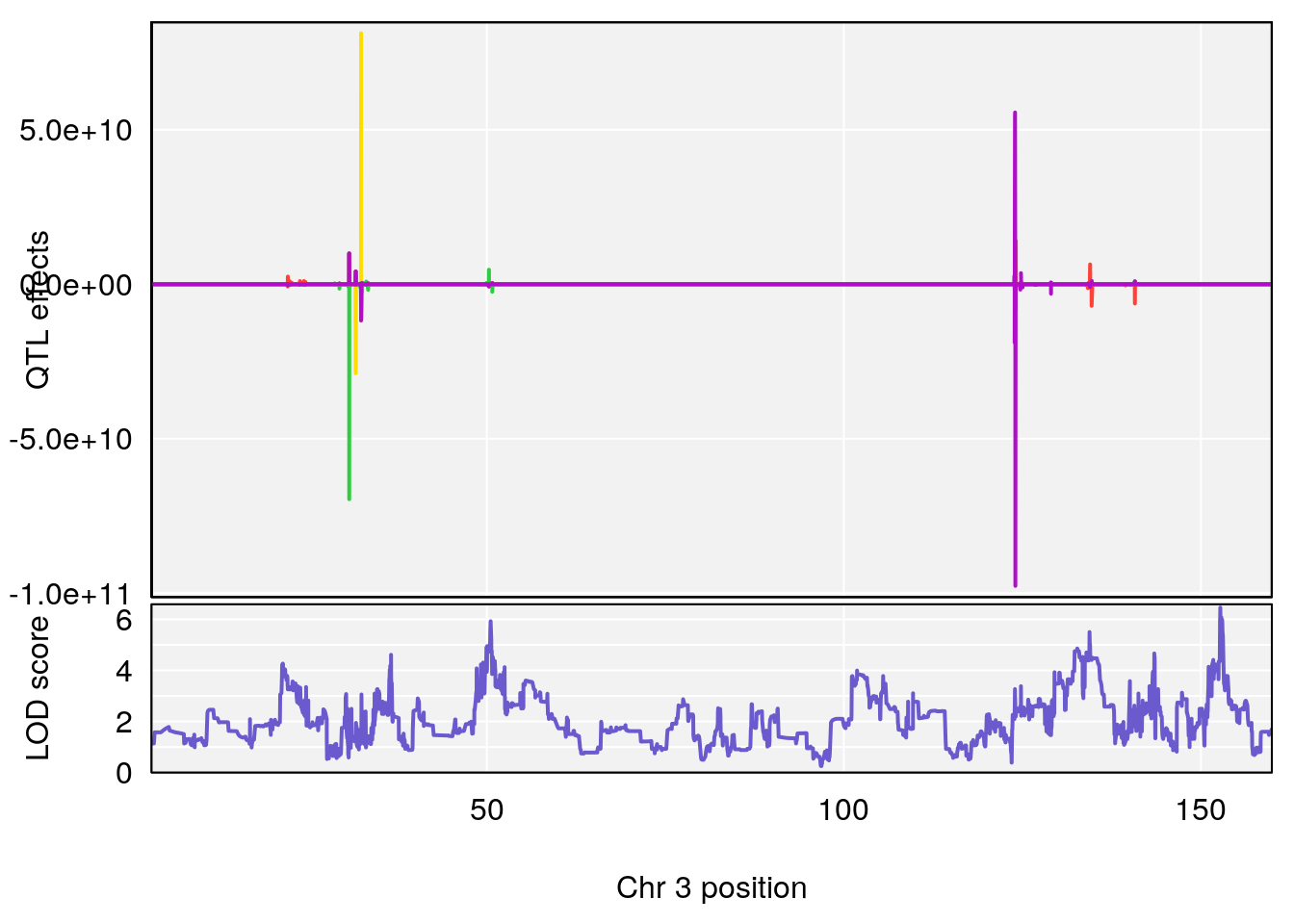
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 4
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 5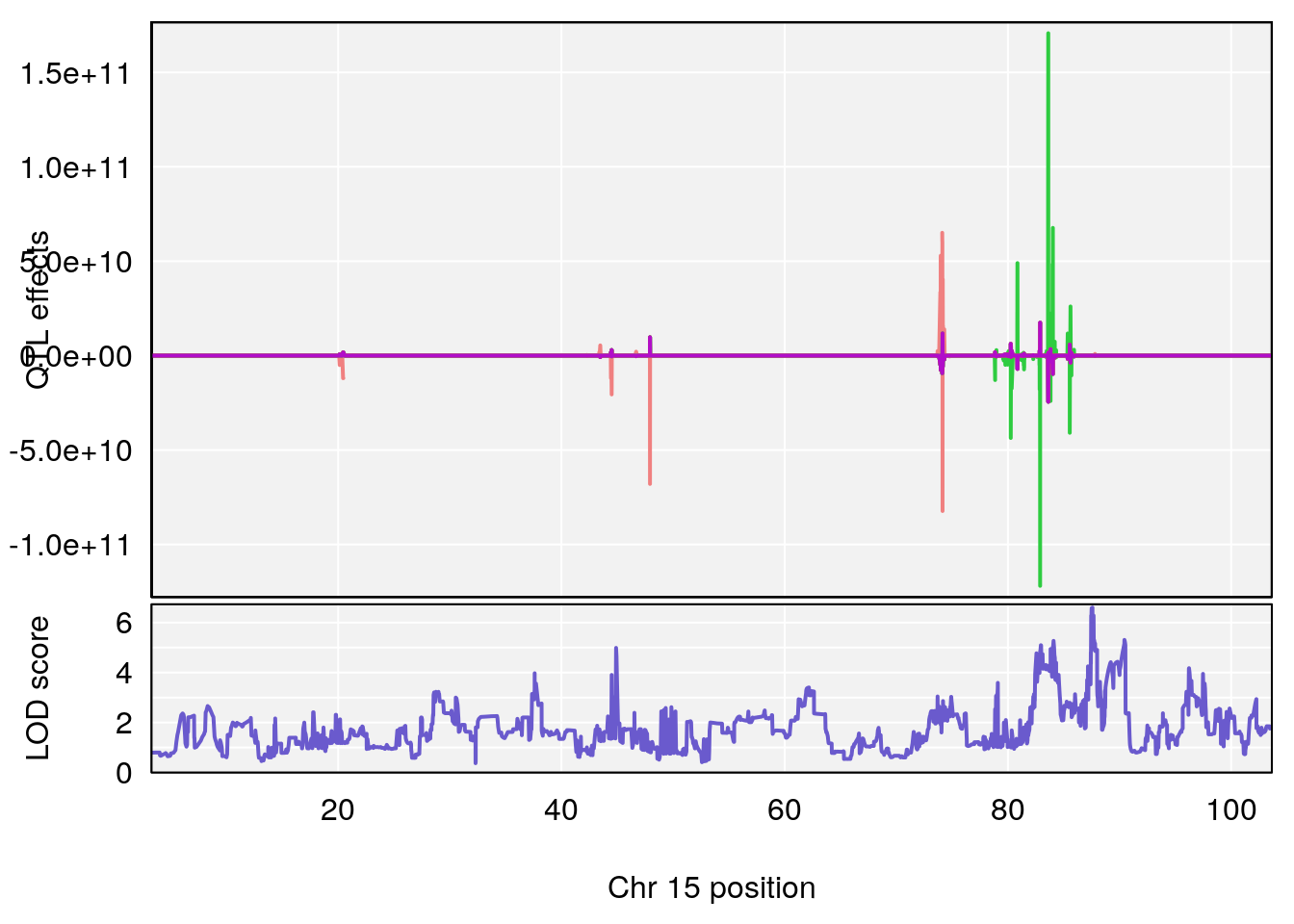
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
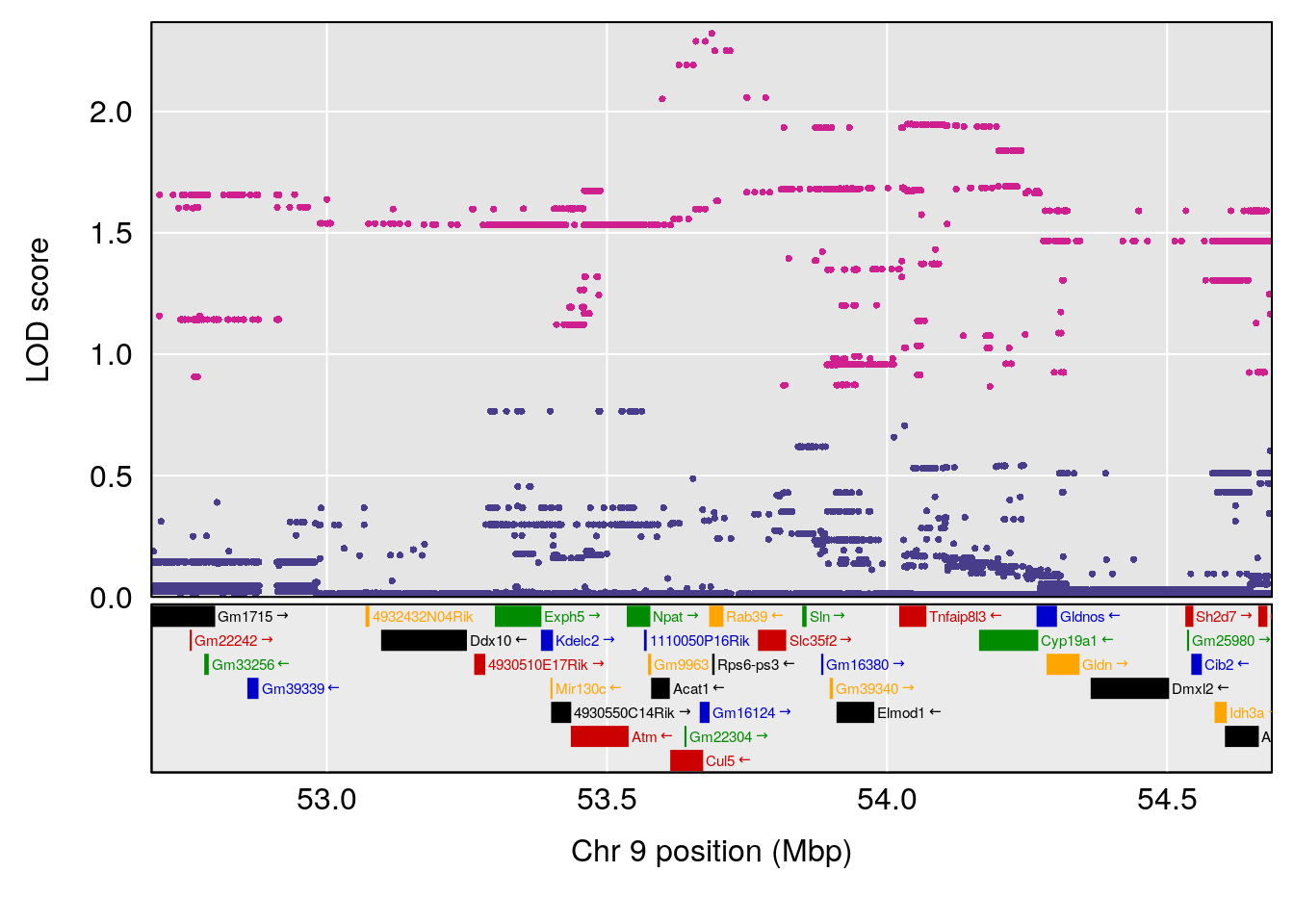
# [1] "Status_bin"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
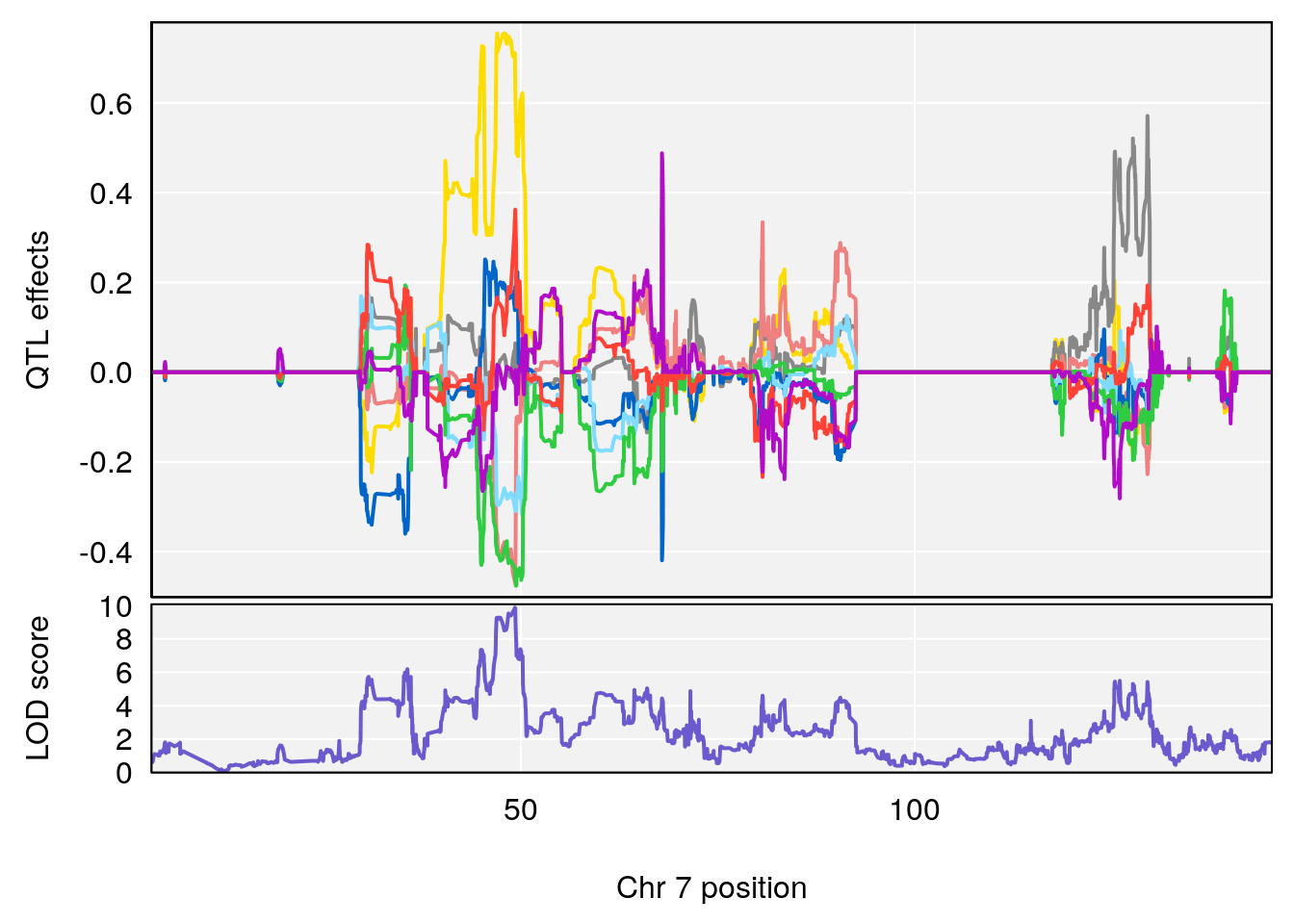
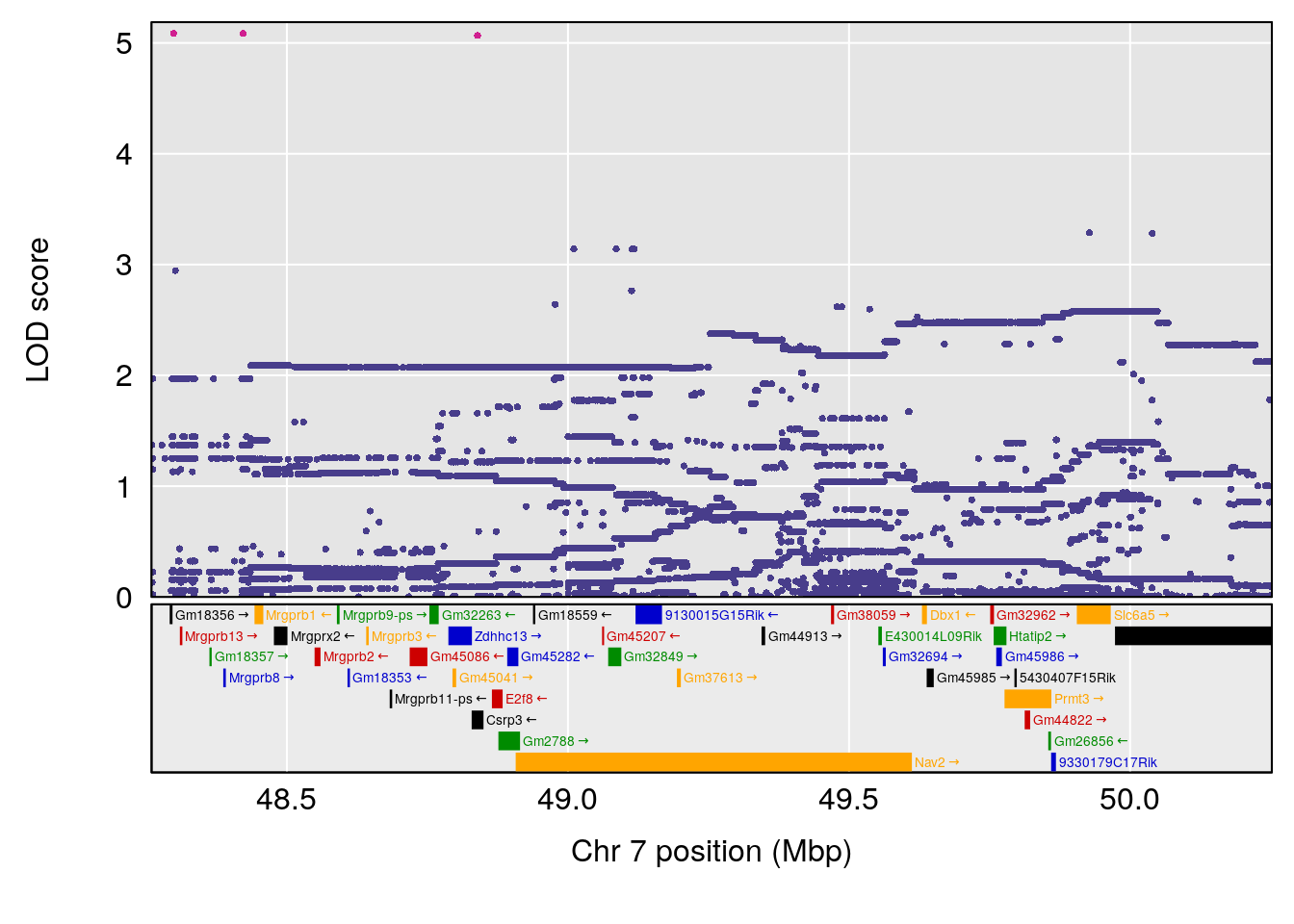
# 1 1 pheno1 7 49.25215 9.875931 46.83042 49.37829
# 2 1 pheno1 9 53.68682 6.144273 52.14281 119.66040
# 3 1 pheno1 19 53.55175 6.642364 53.11032 53.65224
# [1] 1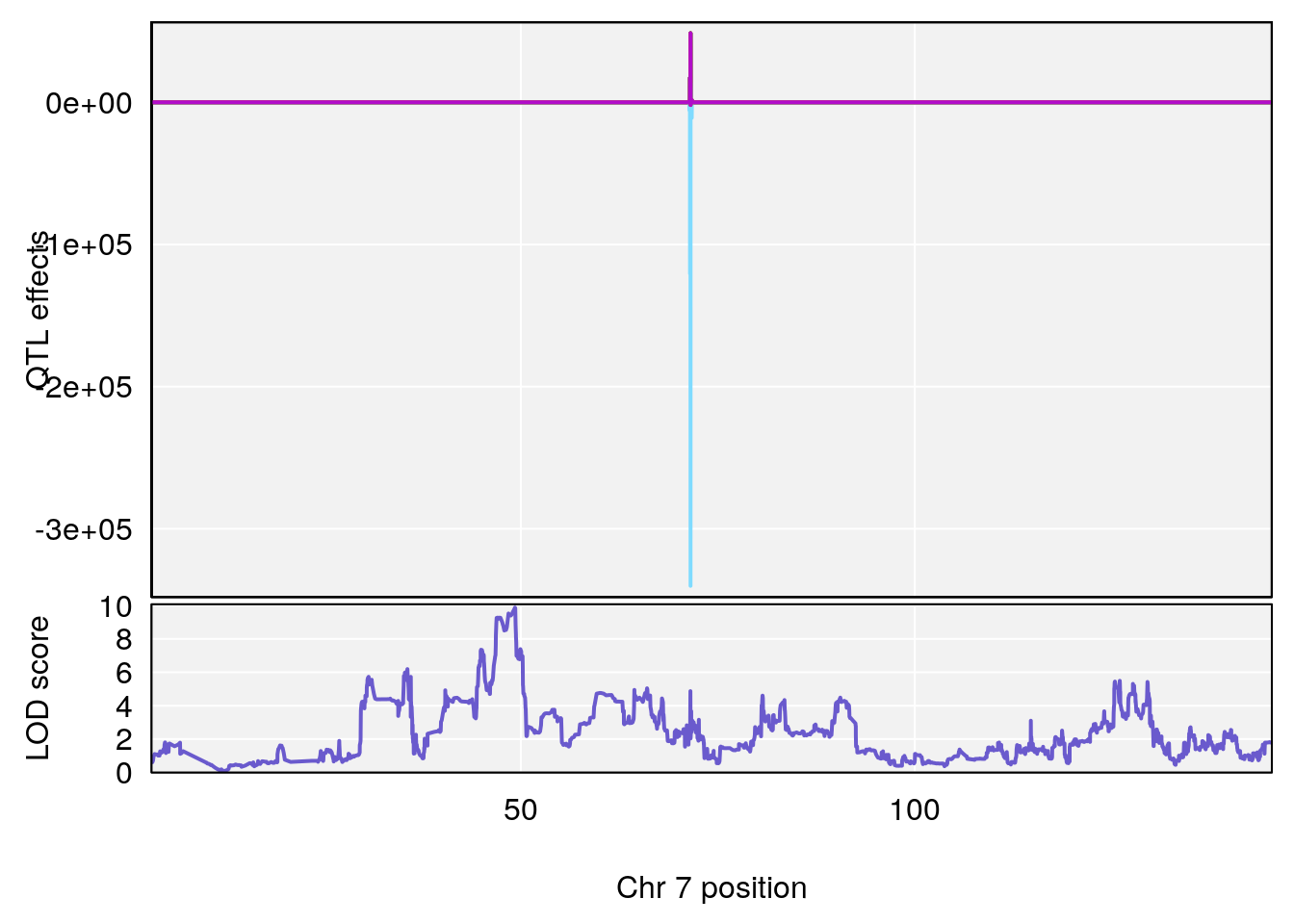
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2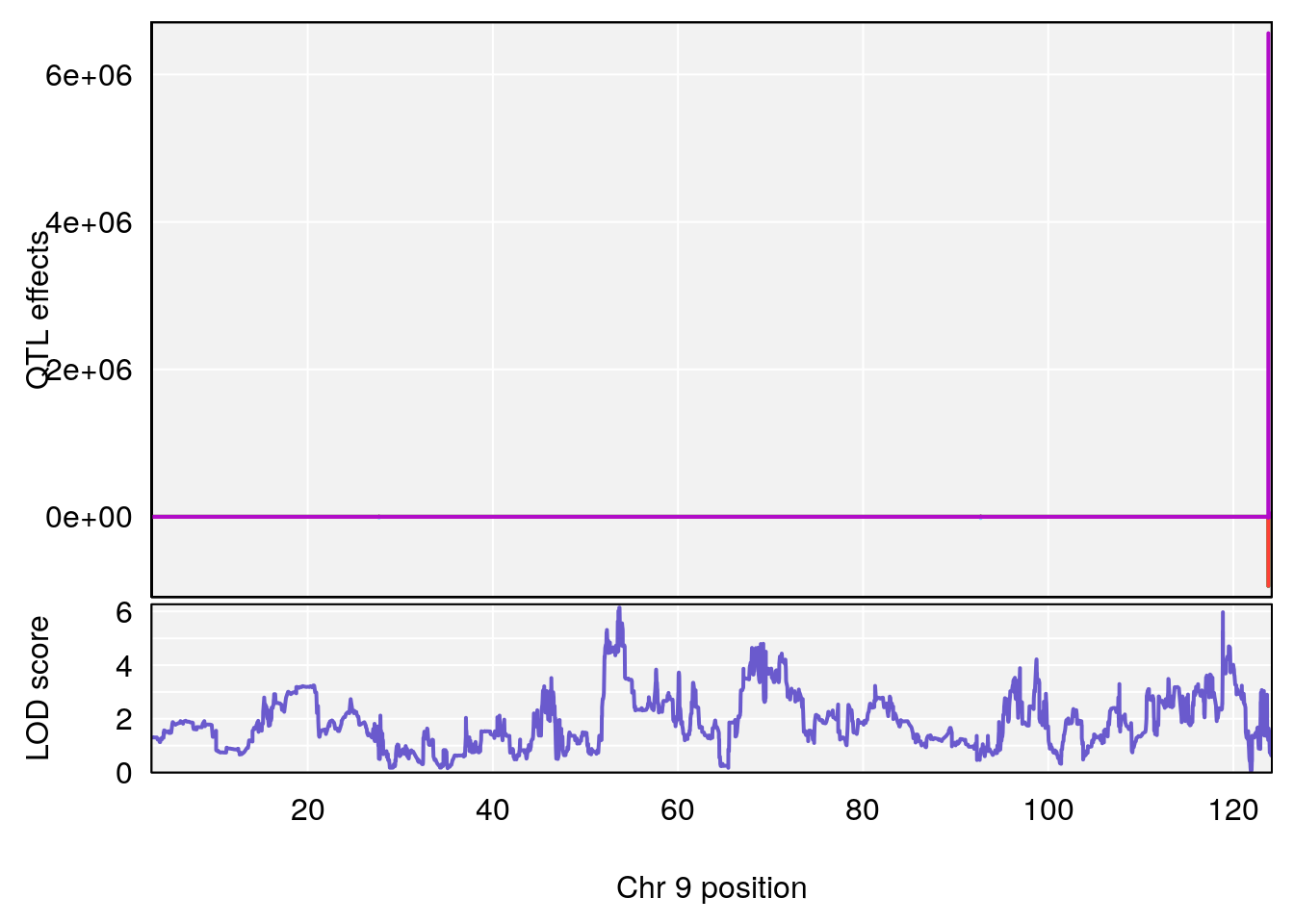
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
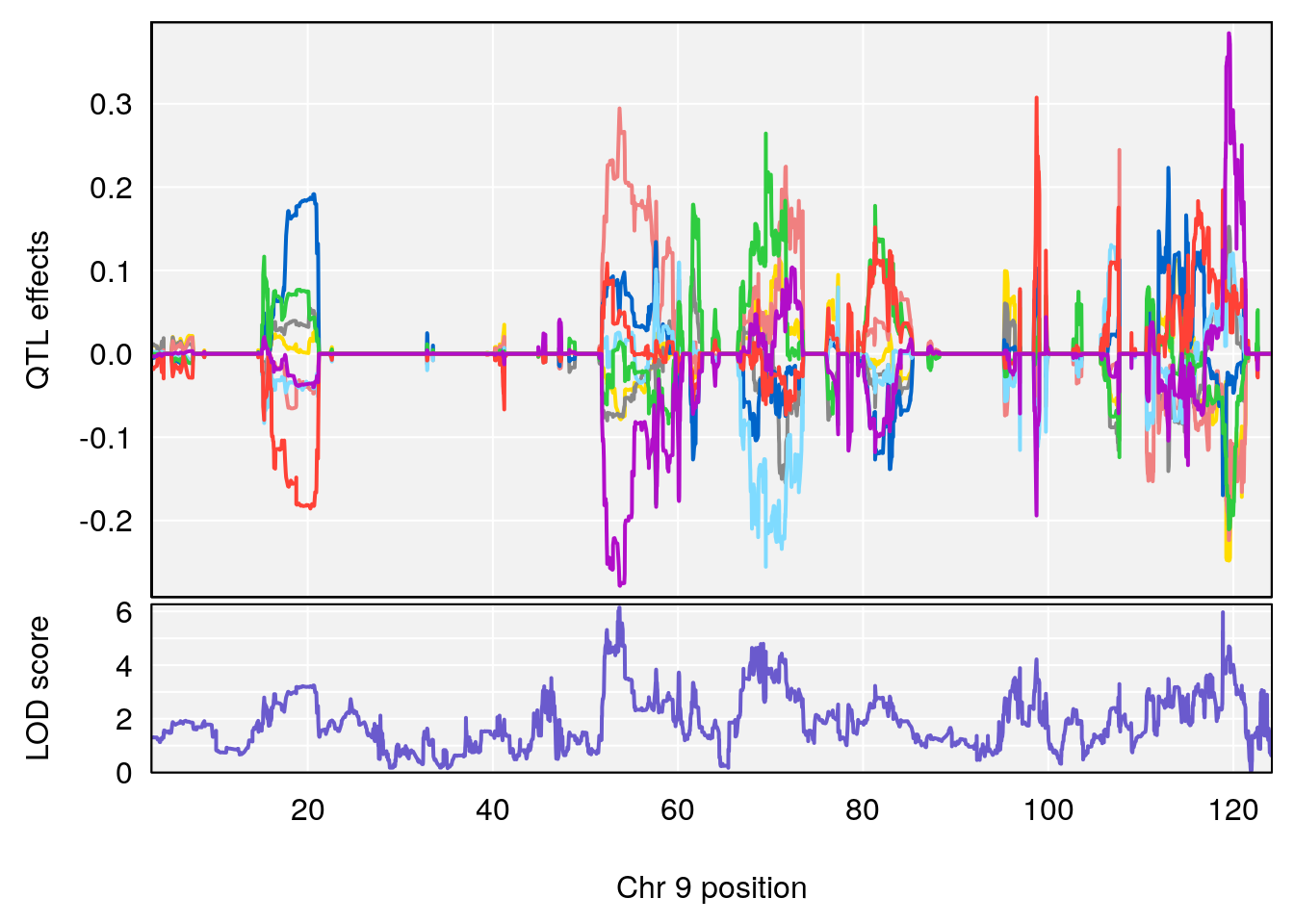
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 3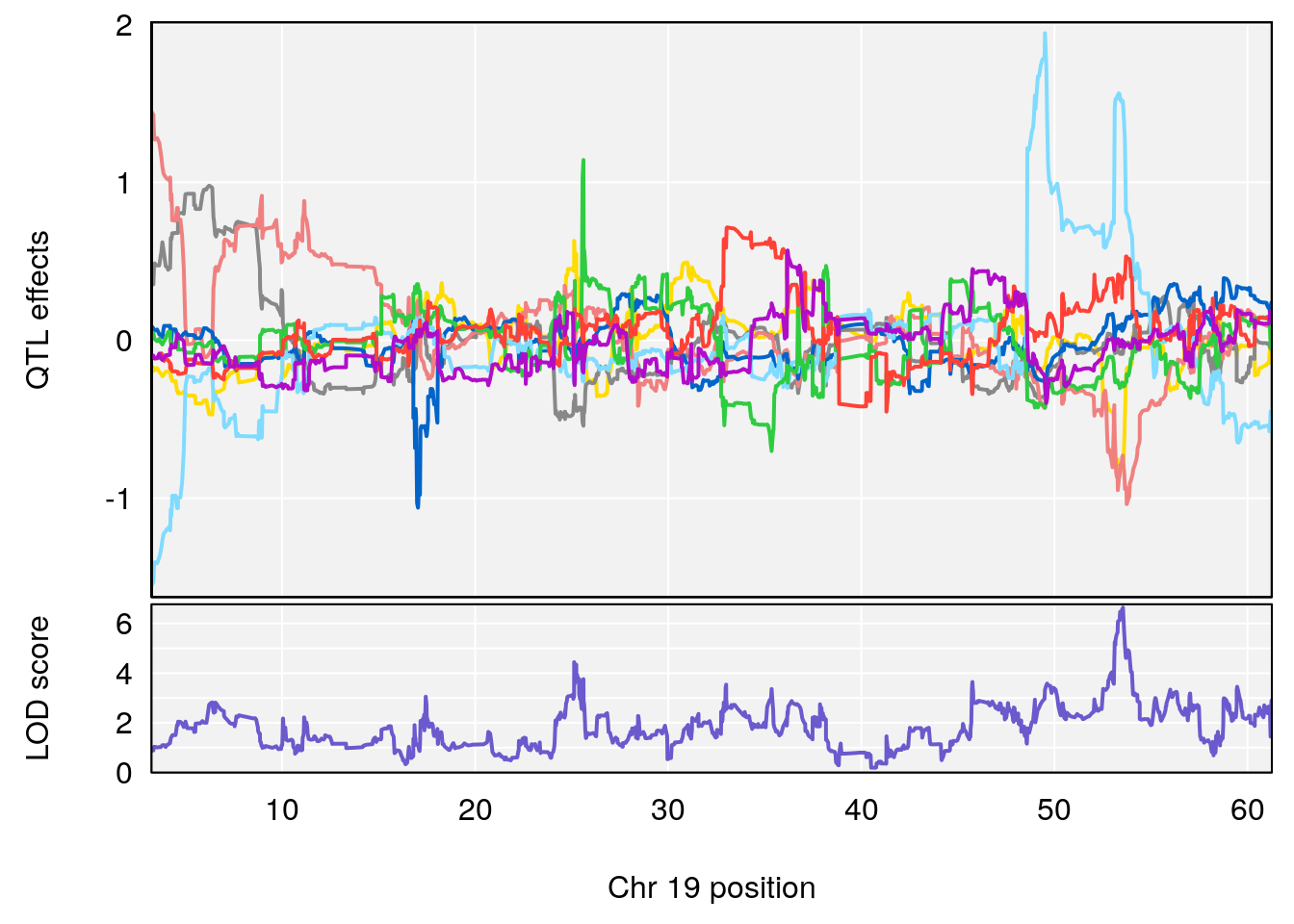
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
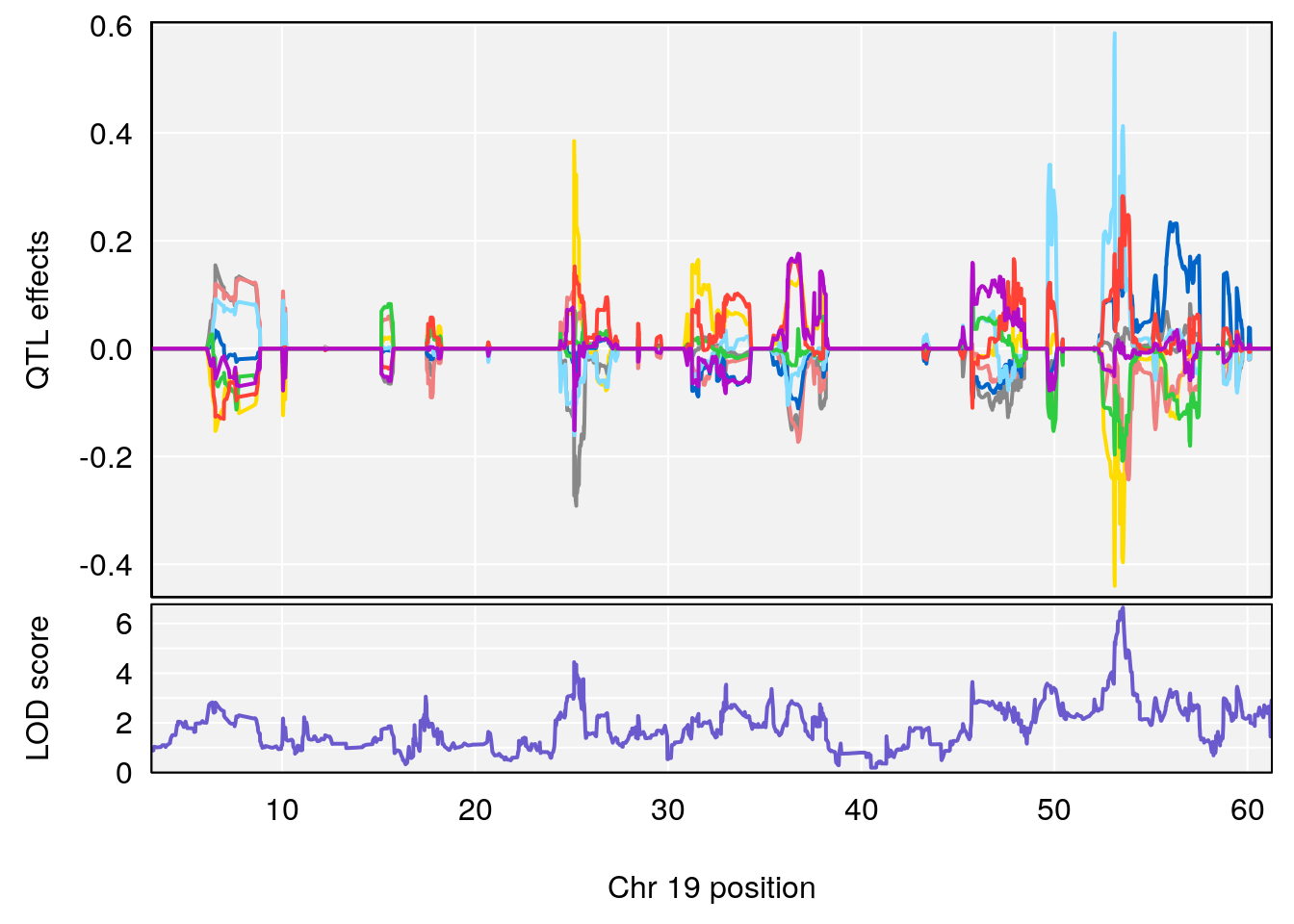
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
Plot qtl mapping on array female
load("output/Fentanyl/qtl.do_Fentanyl_Cohort2.out.female.RData")
#pmap and gmap
chr.names <- c("1","2","3","4","5","6","7","8","9","10","11","12","13","14","15","16","17","18","19", "X")
gmap <- list()
pmap <- list()
for (chr in chr.names){
gmap[[chr]] <- gm$gmap[[chr]]
pmap[[chr]] <- gm$pmap[[chr]]
}
attr(gmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
attr(pmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
#genome-wide plot
for(i in names(qtl.morphine.out)){
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.morphine.out[[i]]) # overall maximum LOD score
plot(qtl.morphine.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, max(sapply(qtl.morphine.out, maxlod)) +2))
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.morphine.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("do_Fentanyl_Cohort2_",i))
}
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
#peaks coeff plot
for(i in names(qtl.morphine.out)){
print(i)
peaks <- find_peaks(qtl.morphine.out[[i]], map=pmap, threshold=6, drop=1.5)
if(nrow(peaks) > 0) {
print(peaks)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.morphine.out[[i]], bgcolor="gray95", legend=NULL)
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
}
}
# [1] "TimetoDead(Hr)"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
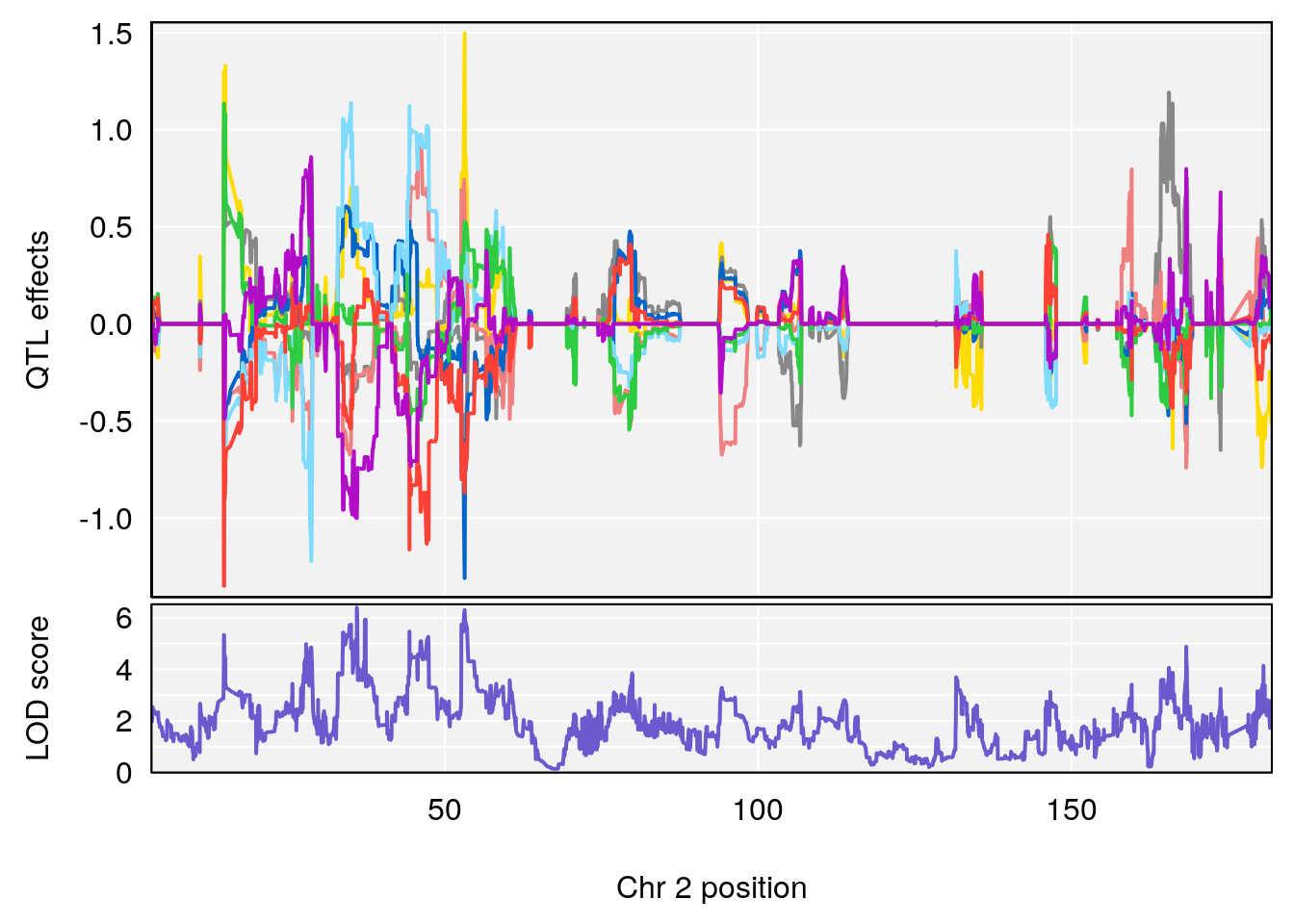
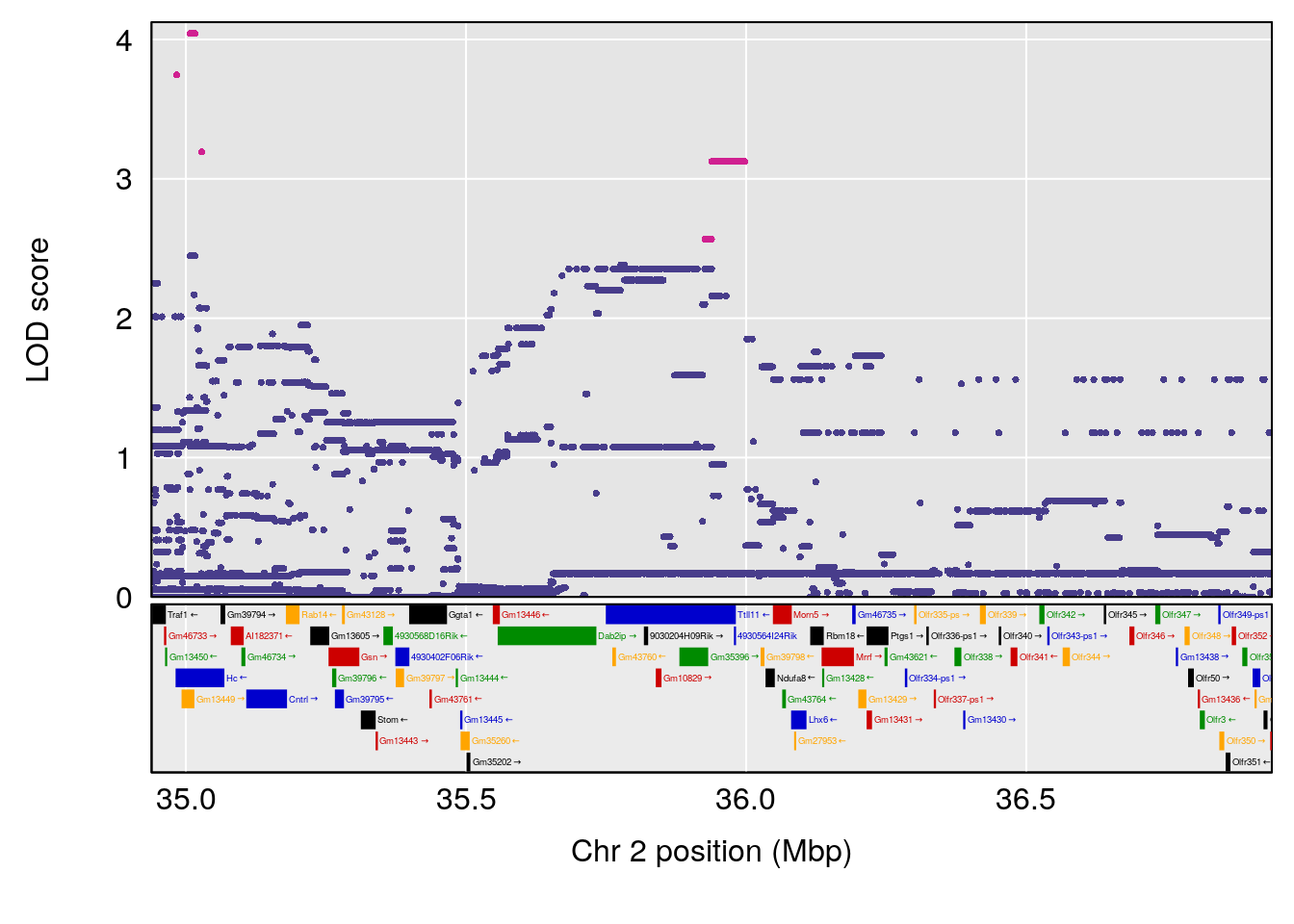
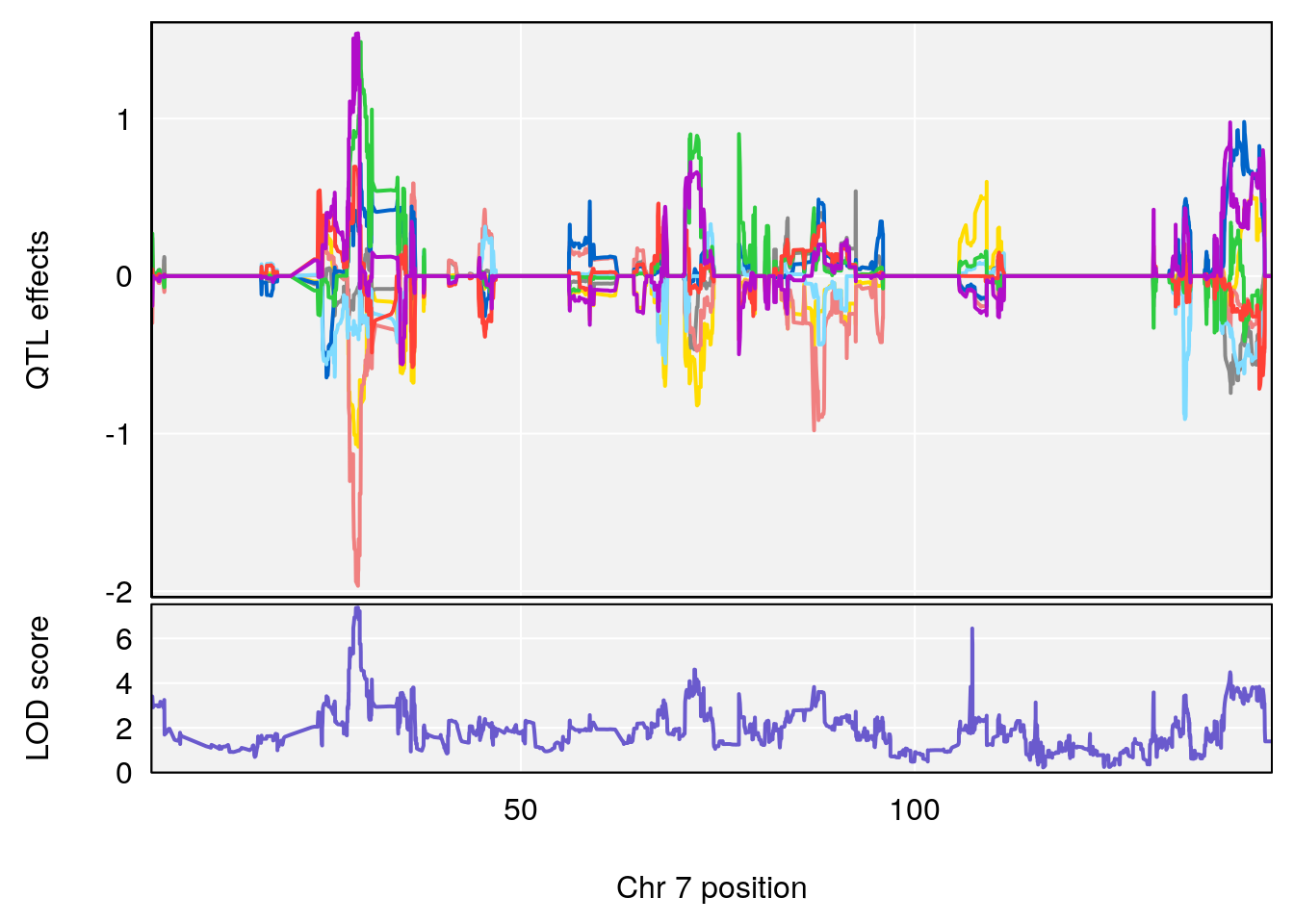
# 1 1 pheno1 2 35.93828 6.395693 14.71230 53.73635
# 2 1 pheno1 7 29.33386 7.380186 28.72162 107.32907
# 3 1 pheno1 9 29.84406 6.460520 28.55111 64.47467
# 4 1 pheno1 10 93.92129 6.138204 26.38250 94.50163
# 5 1 pheno1 X 45.99921 6.955609 44.92816 136.45503
# [1] 1
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
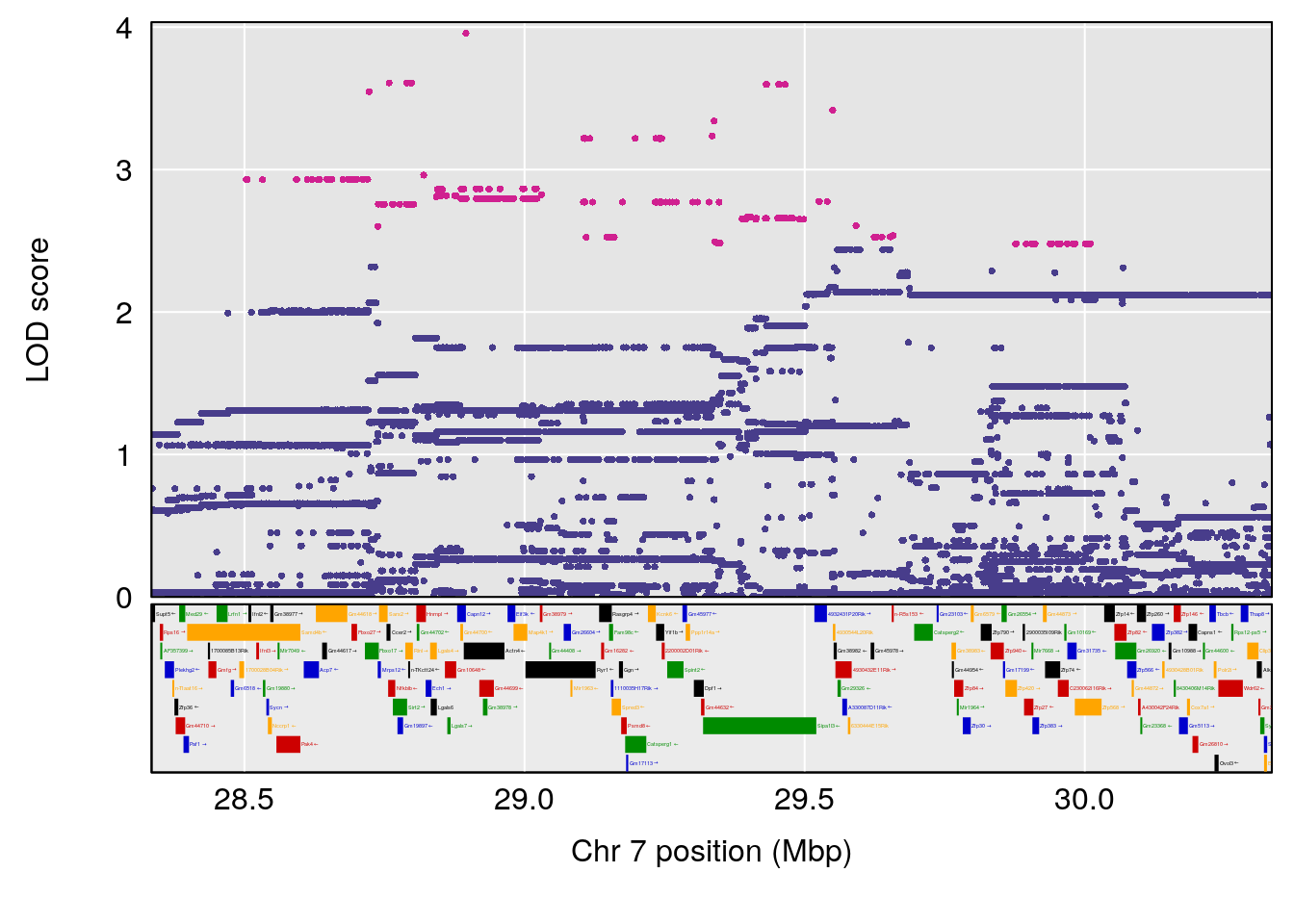
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
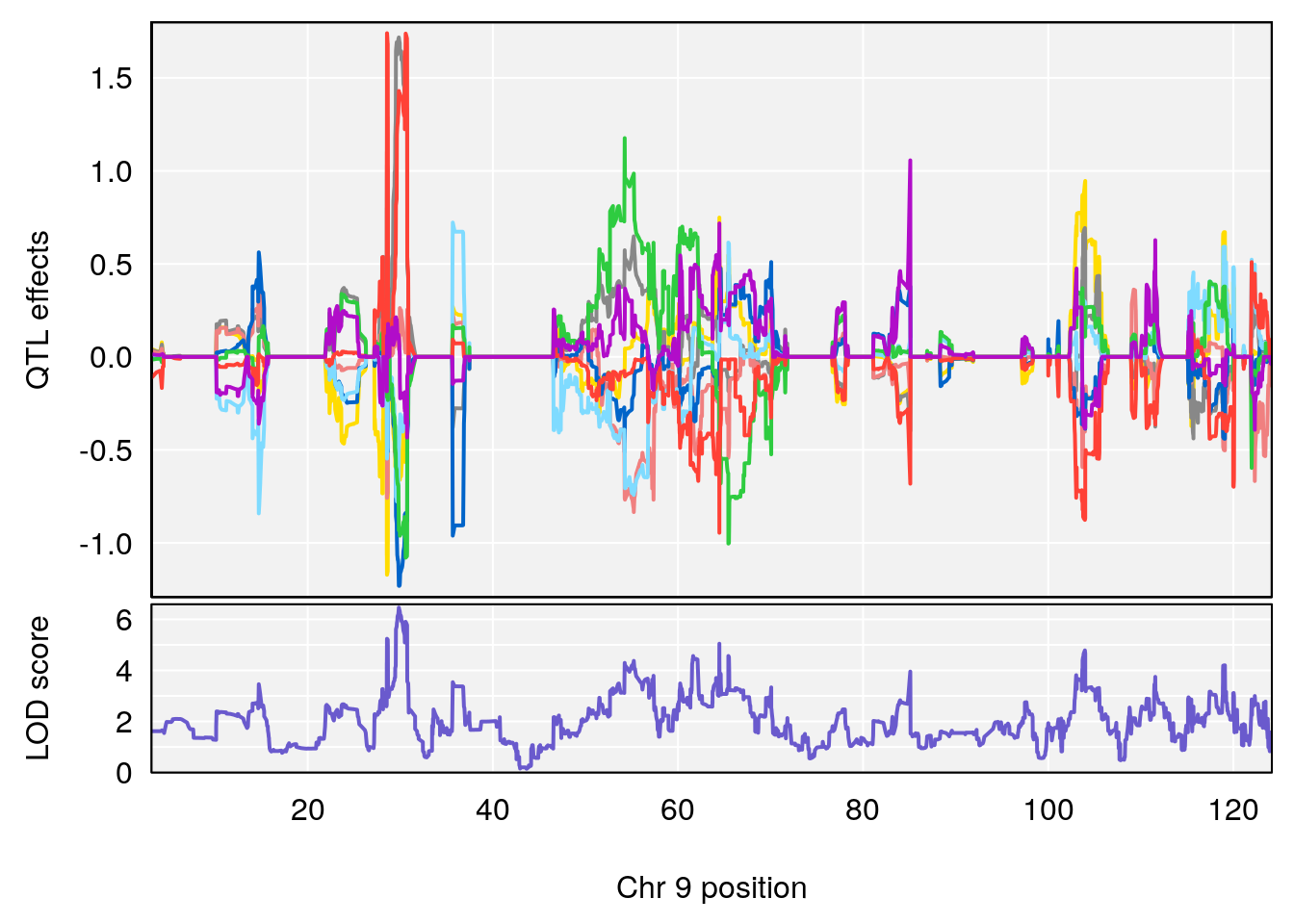
# [1] 3
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
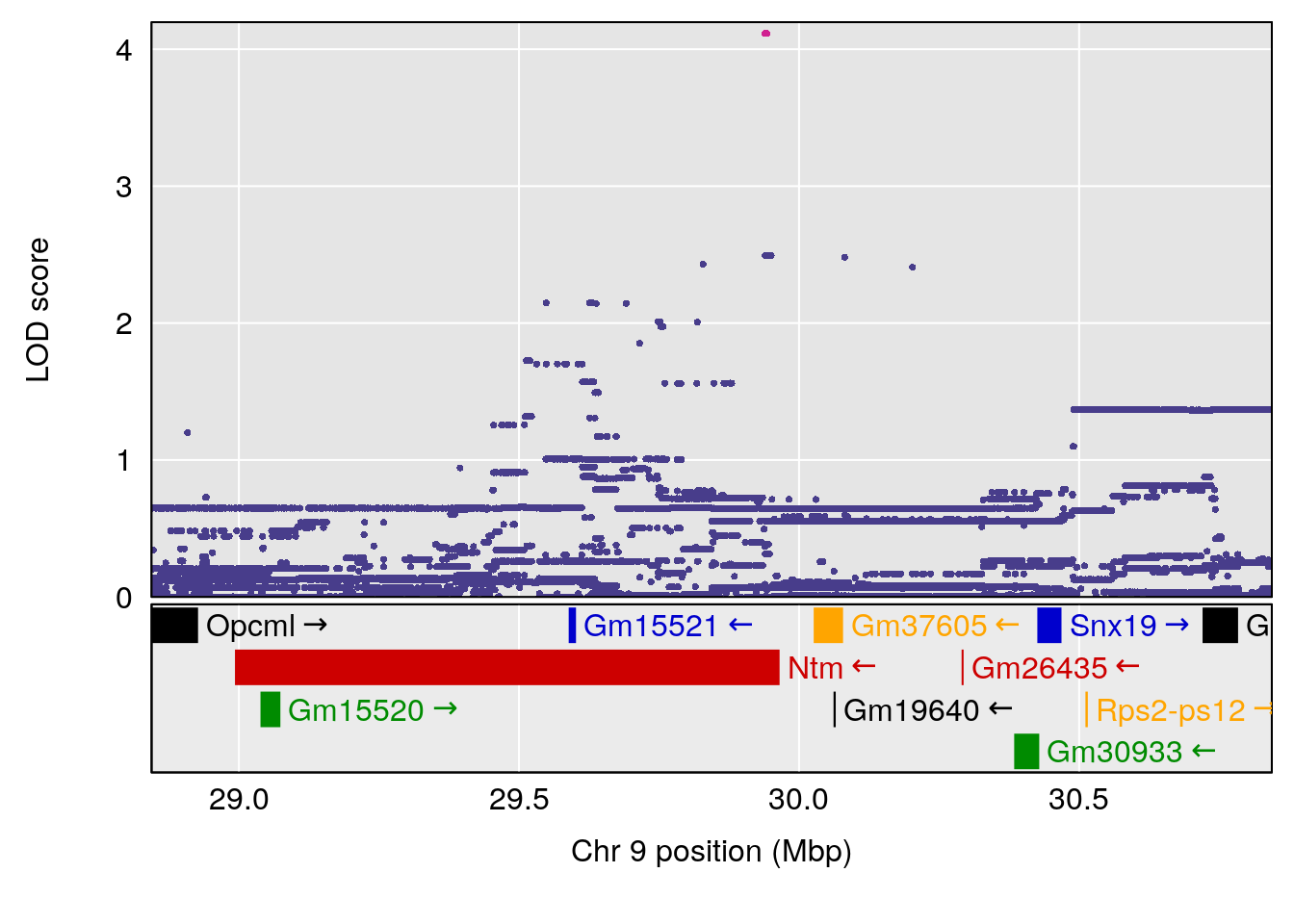
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
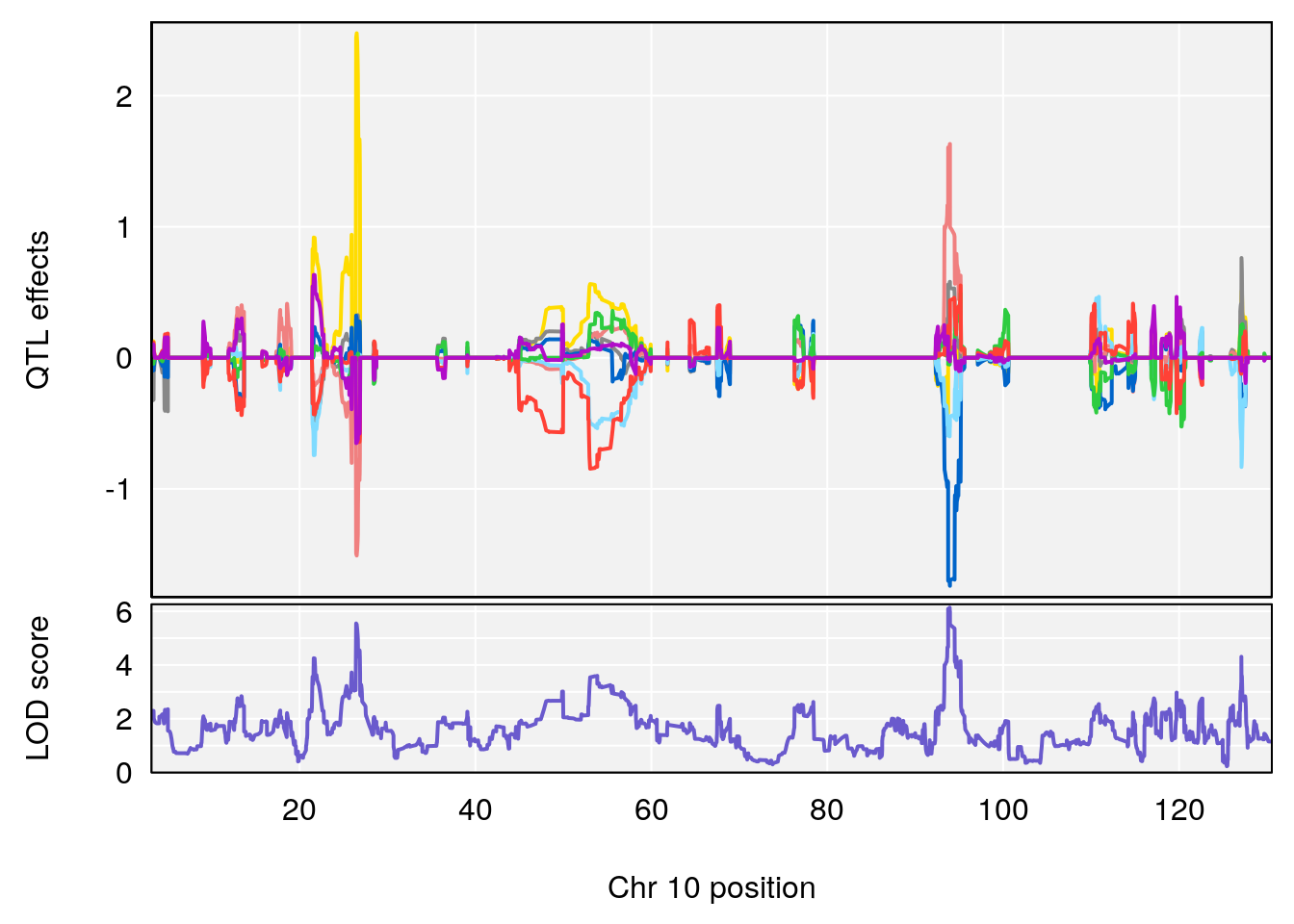
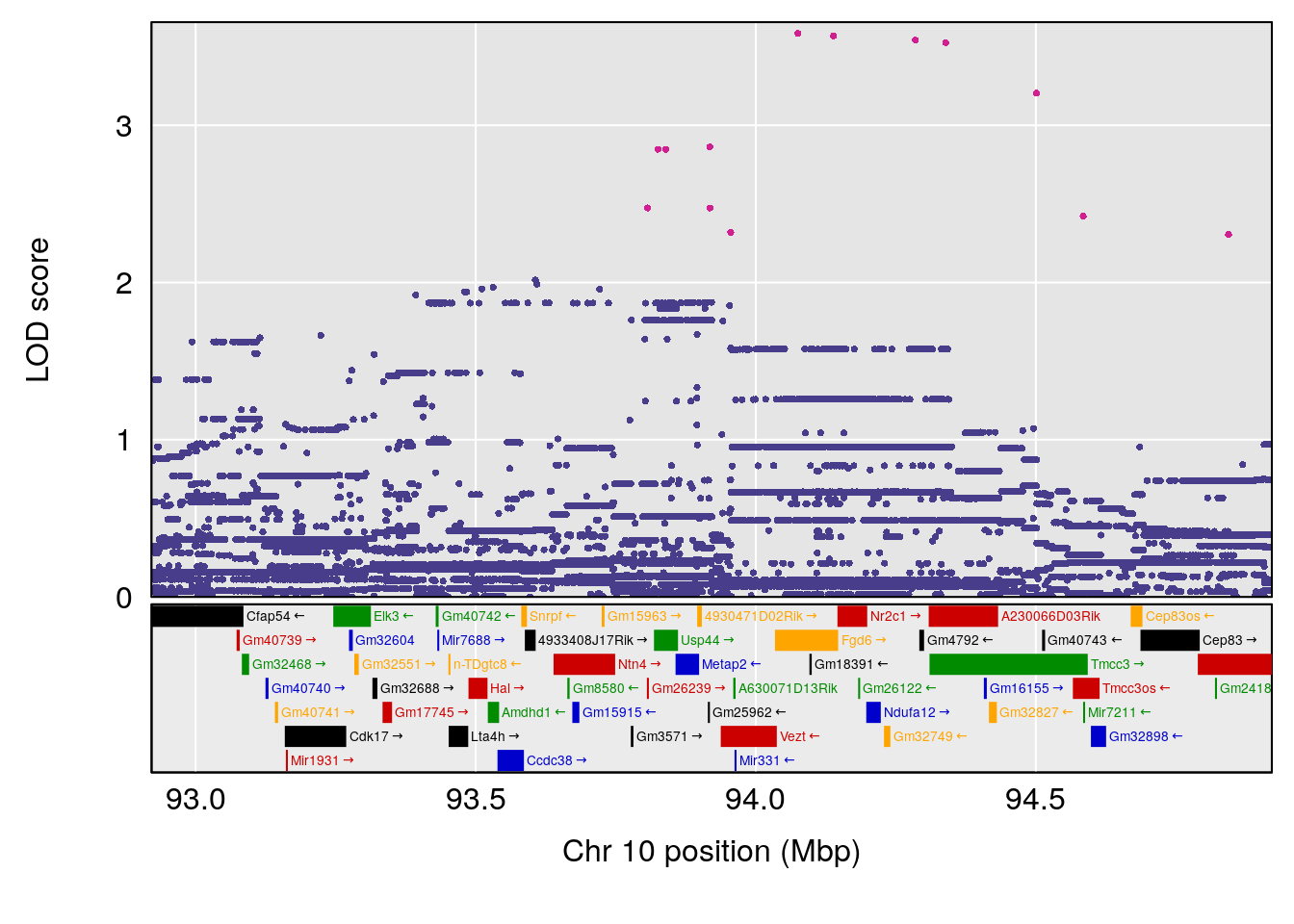
# [1] 4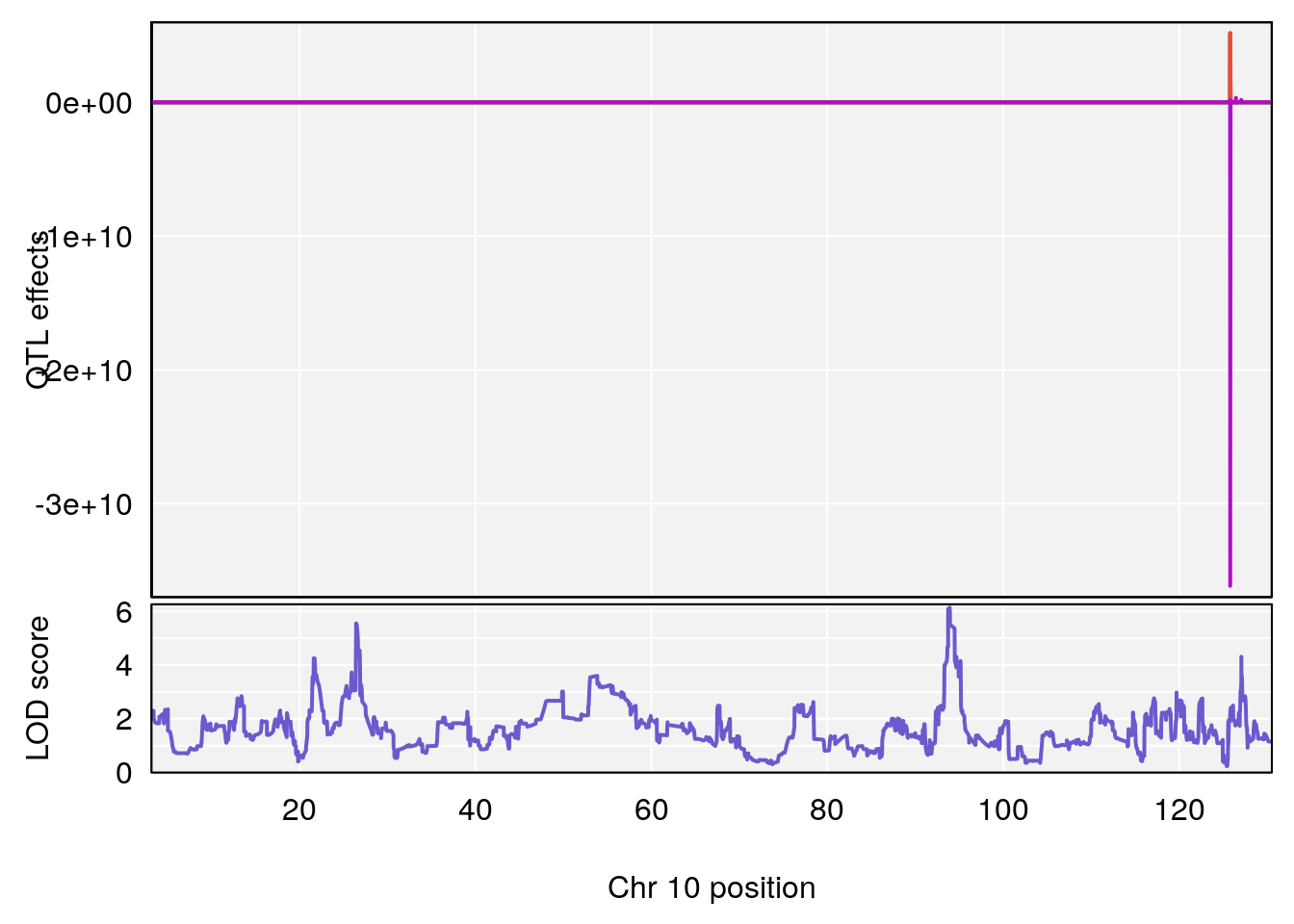
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 5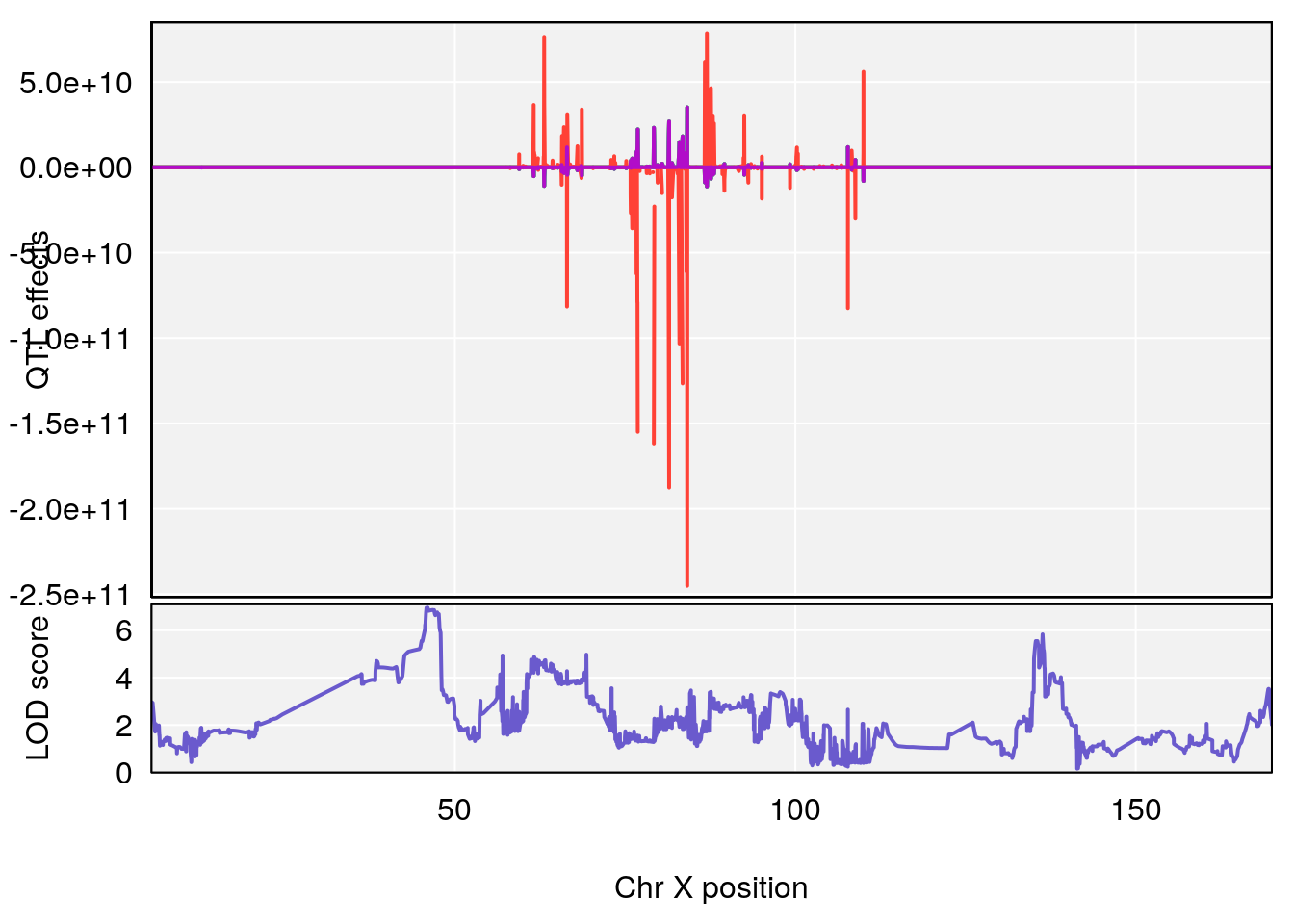
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
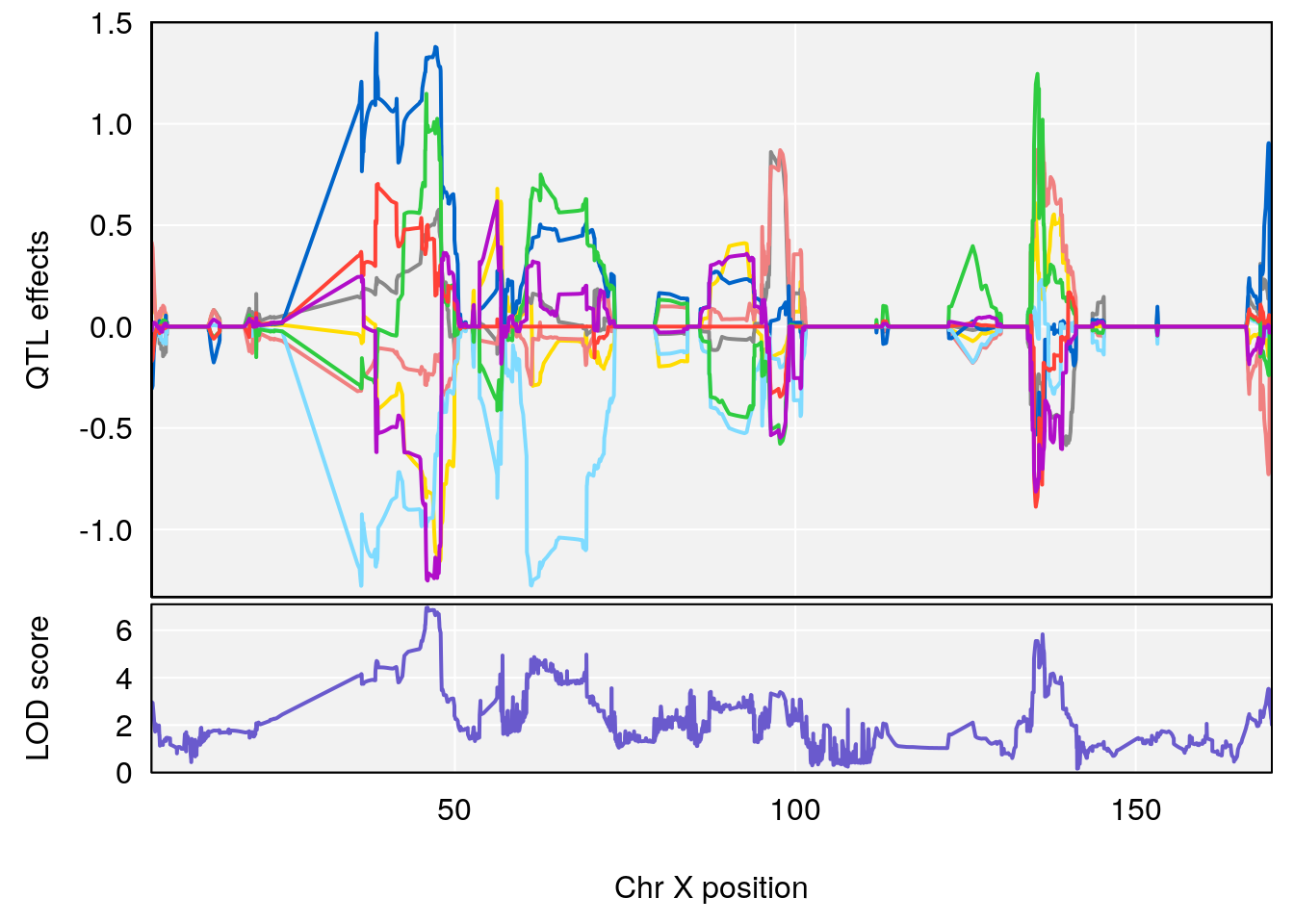
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
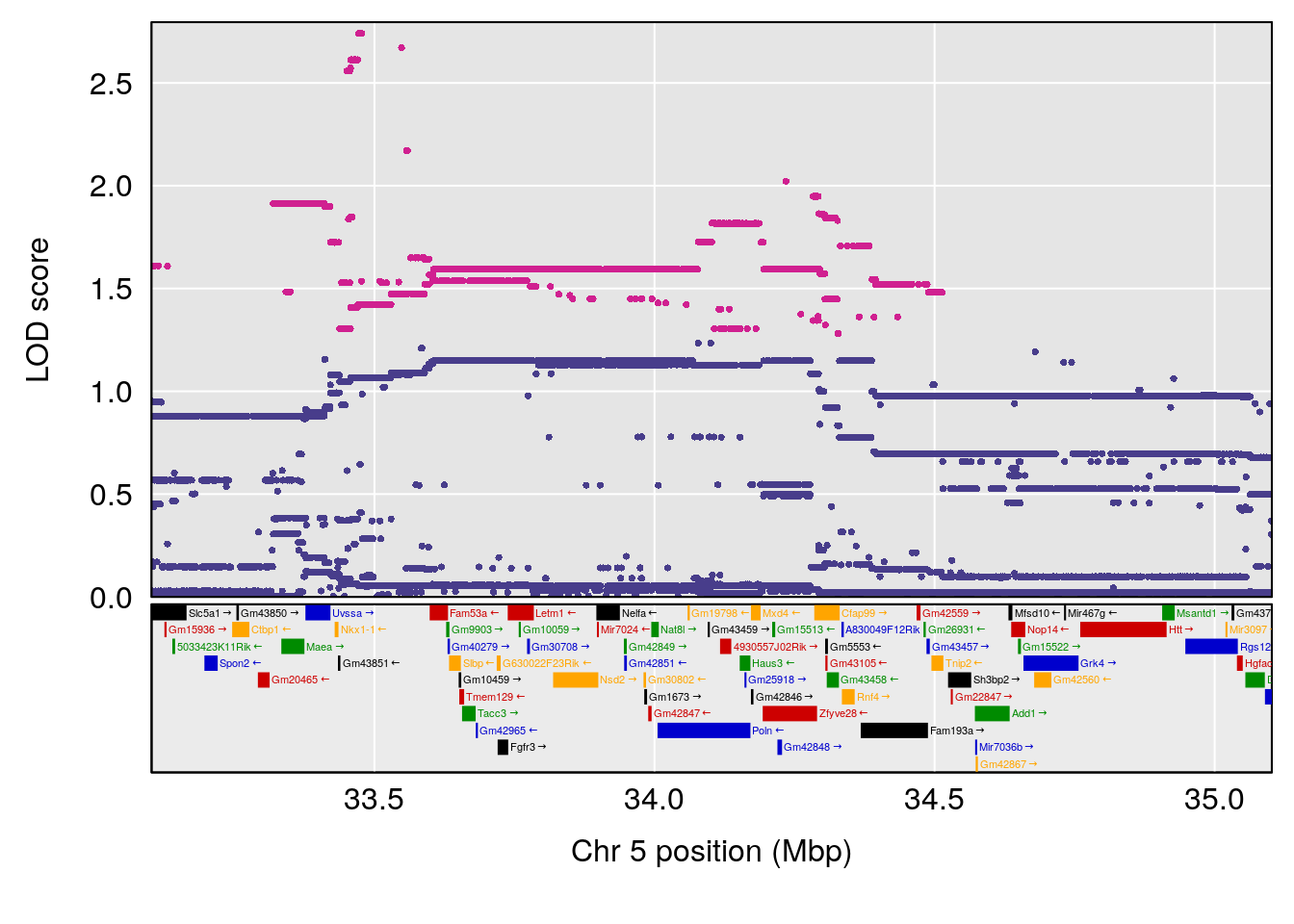
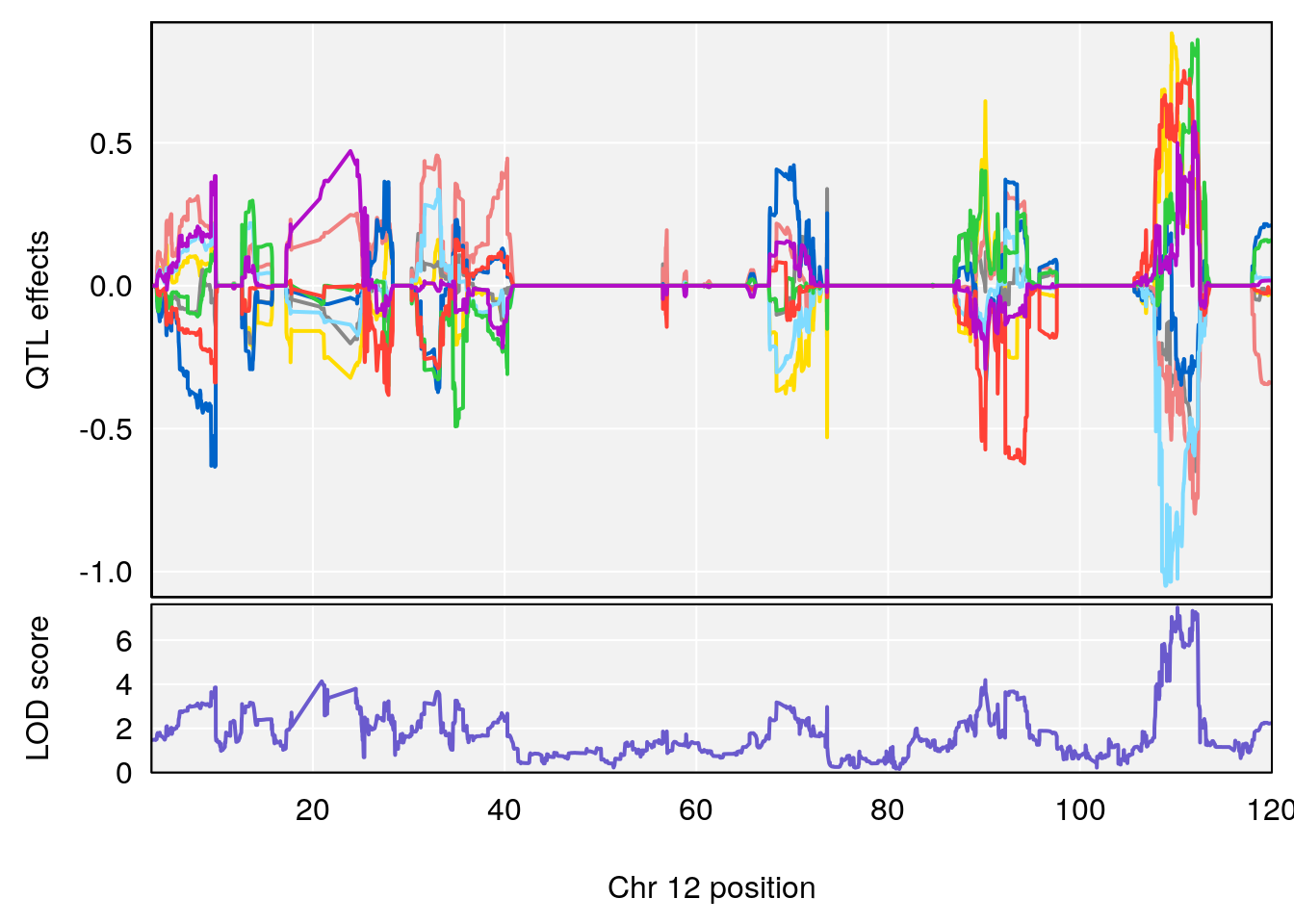
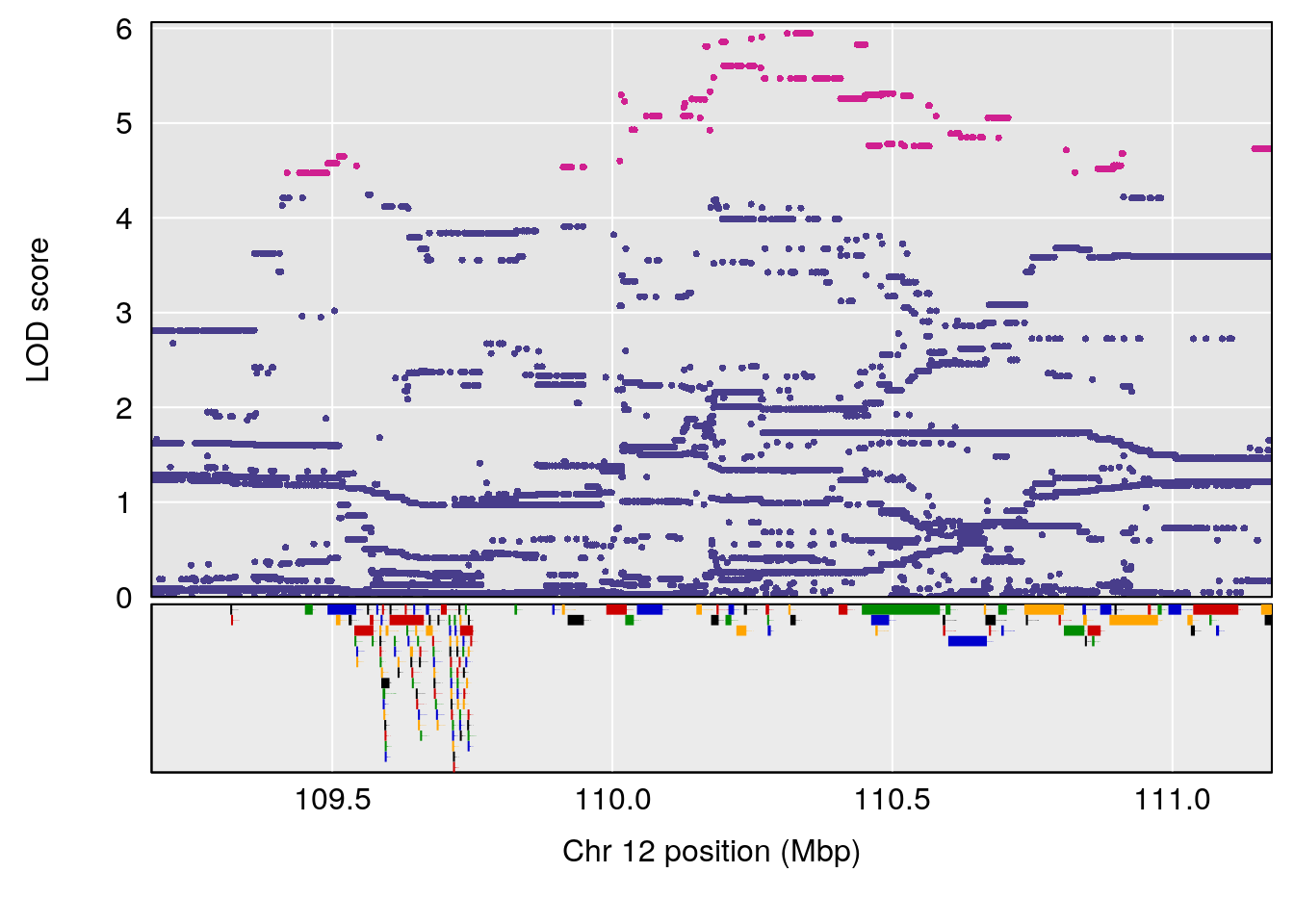
# [1] "RRDepressionRate(%/Hr)SLOPE"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 5 34.10212 6.494372 33.30631 34.38771
# 2 1 pheno1 12 110.17735 7.475173 109.51204 112.34613
# 3 1 pheno1 15 99.39531 6.125827 26.44955 101.12575
# [1] 1
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
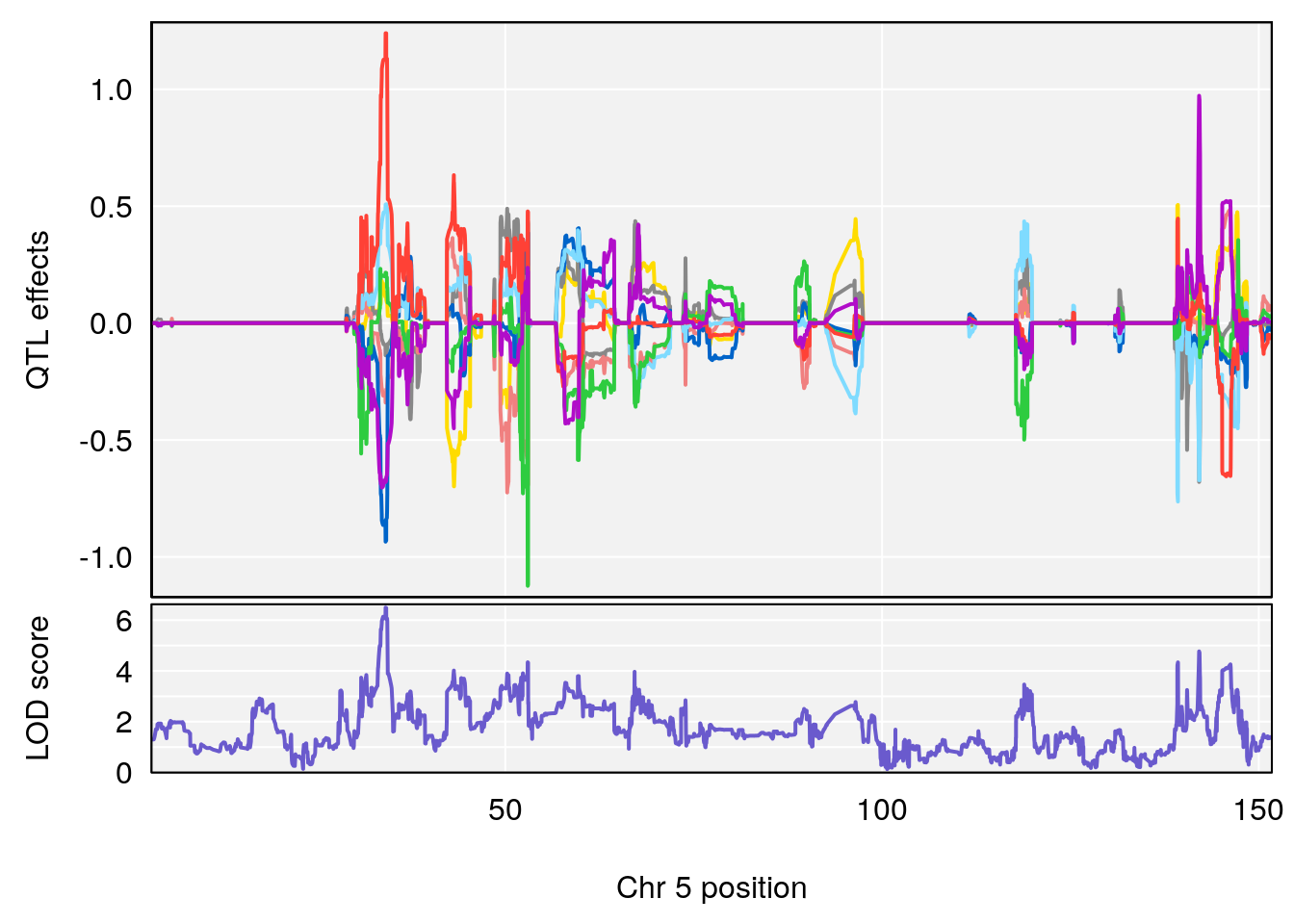
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2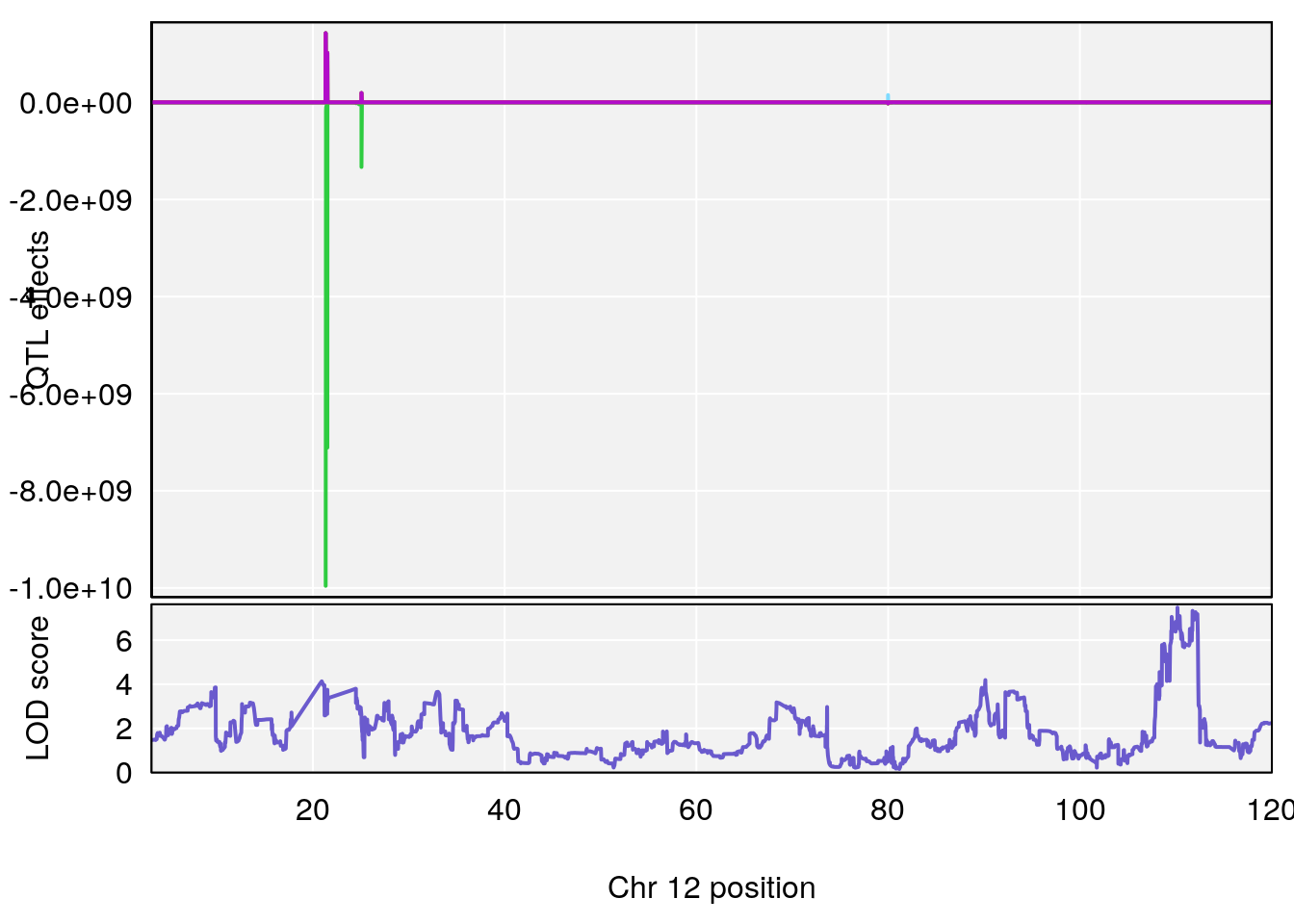
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 3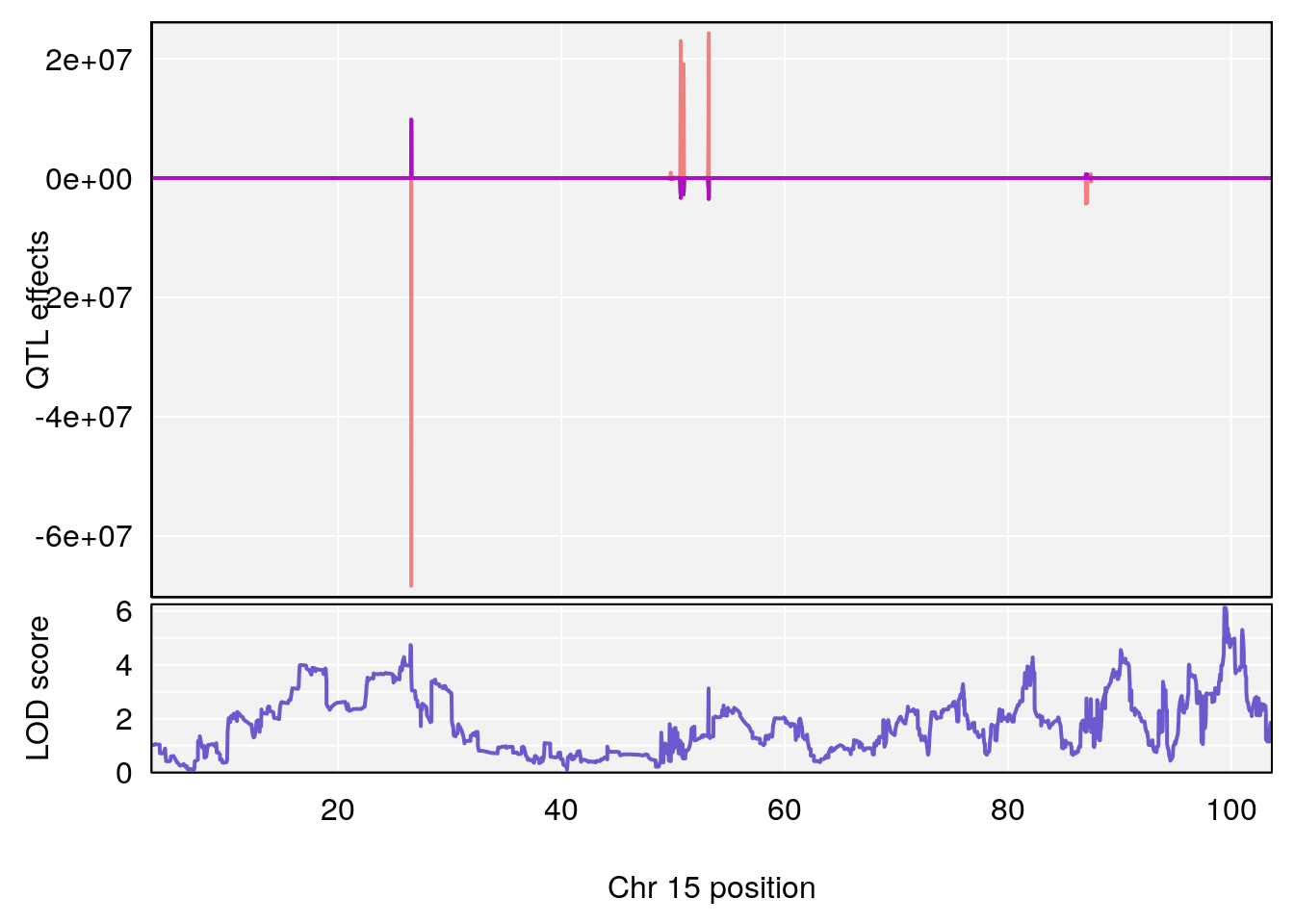
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
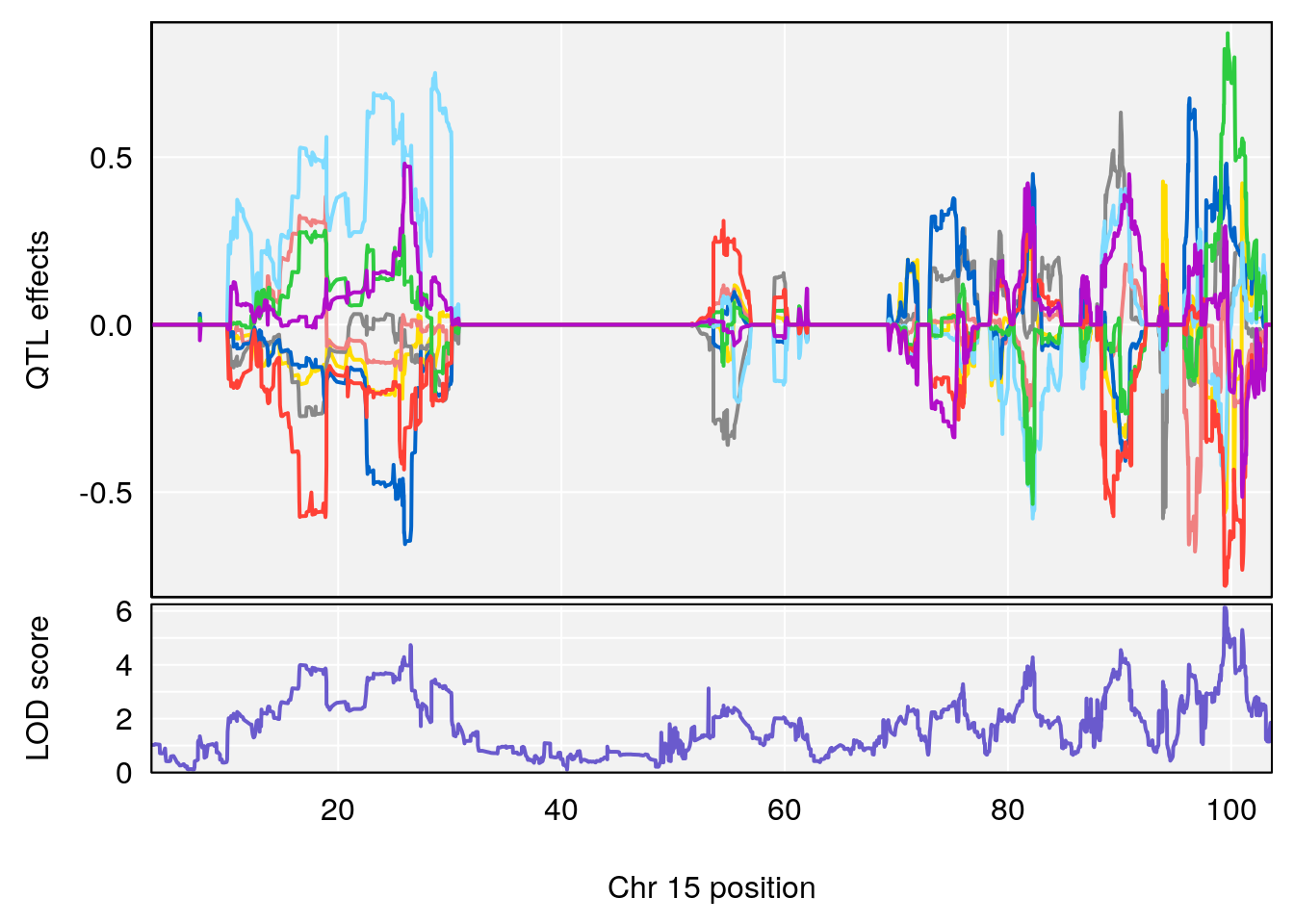
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
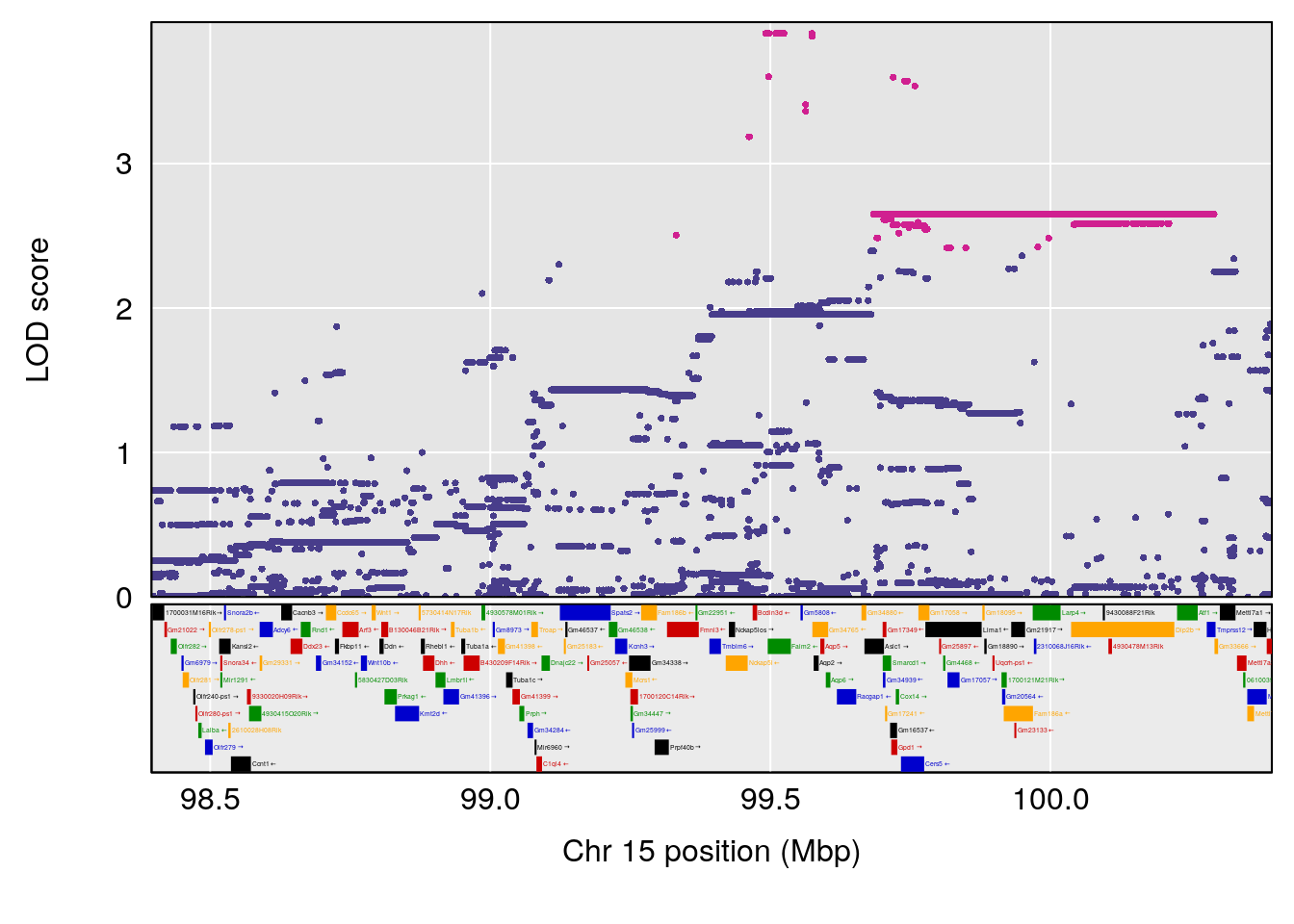
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "TimetoSteadyRRDepression(Hr)"
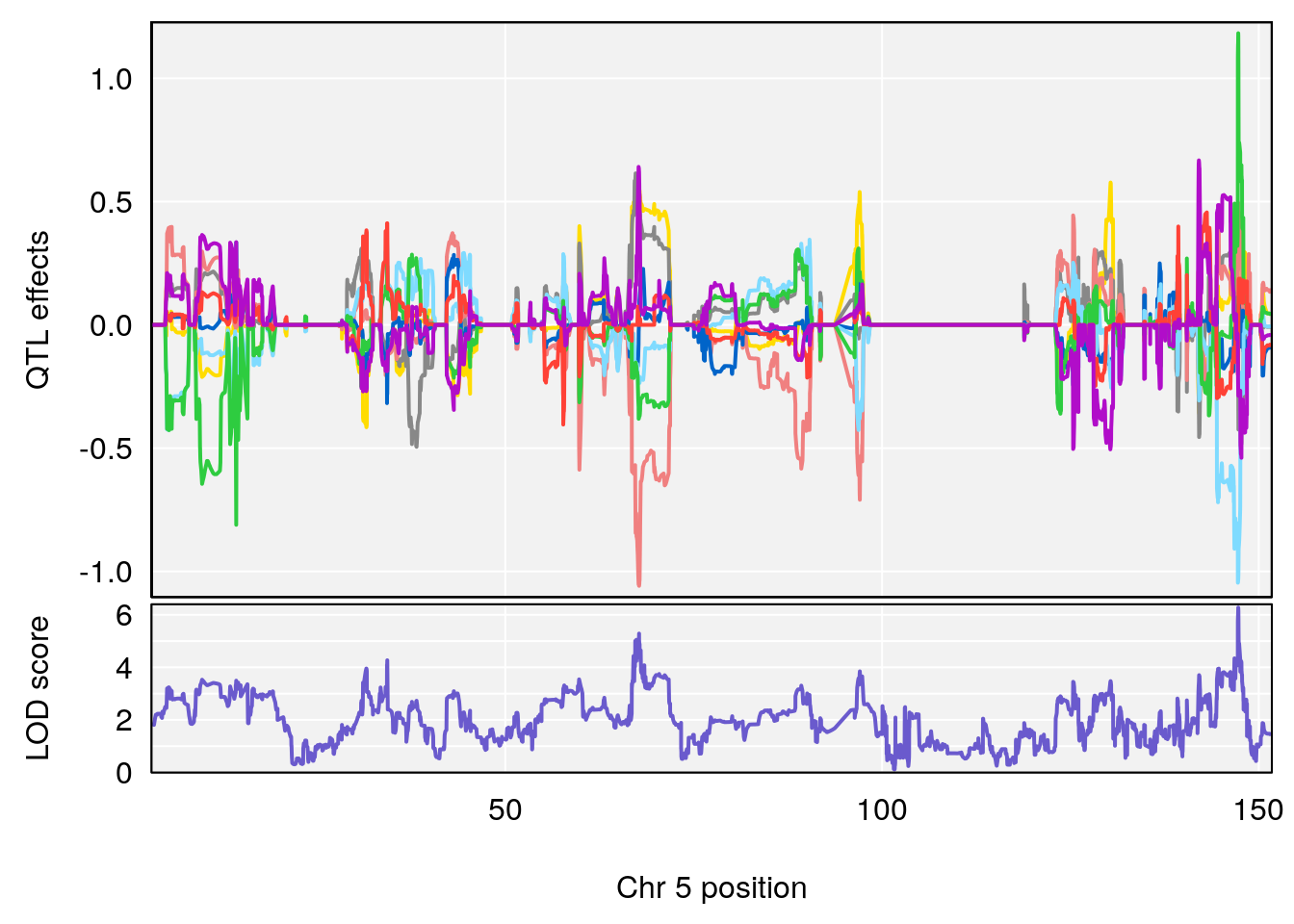
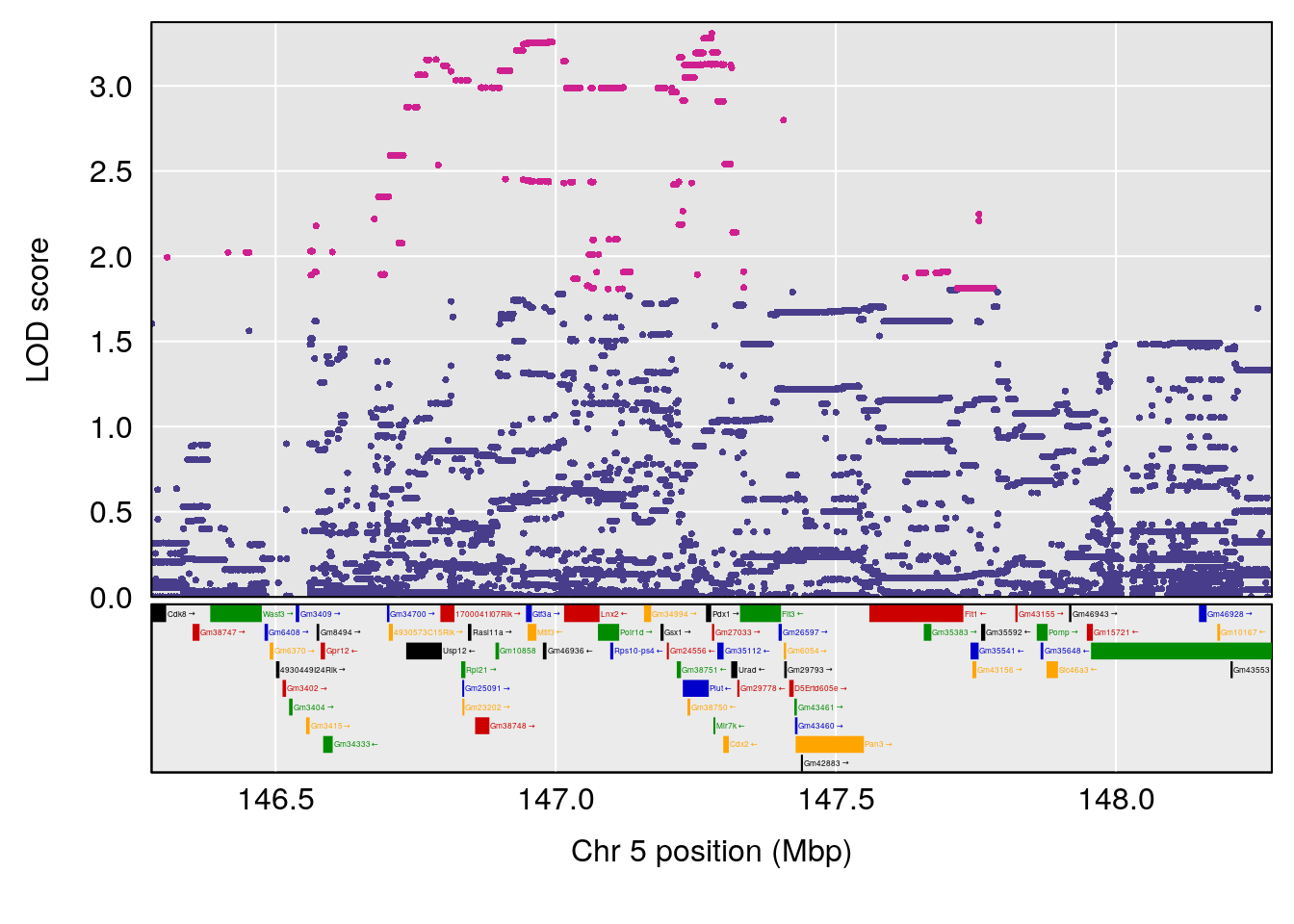
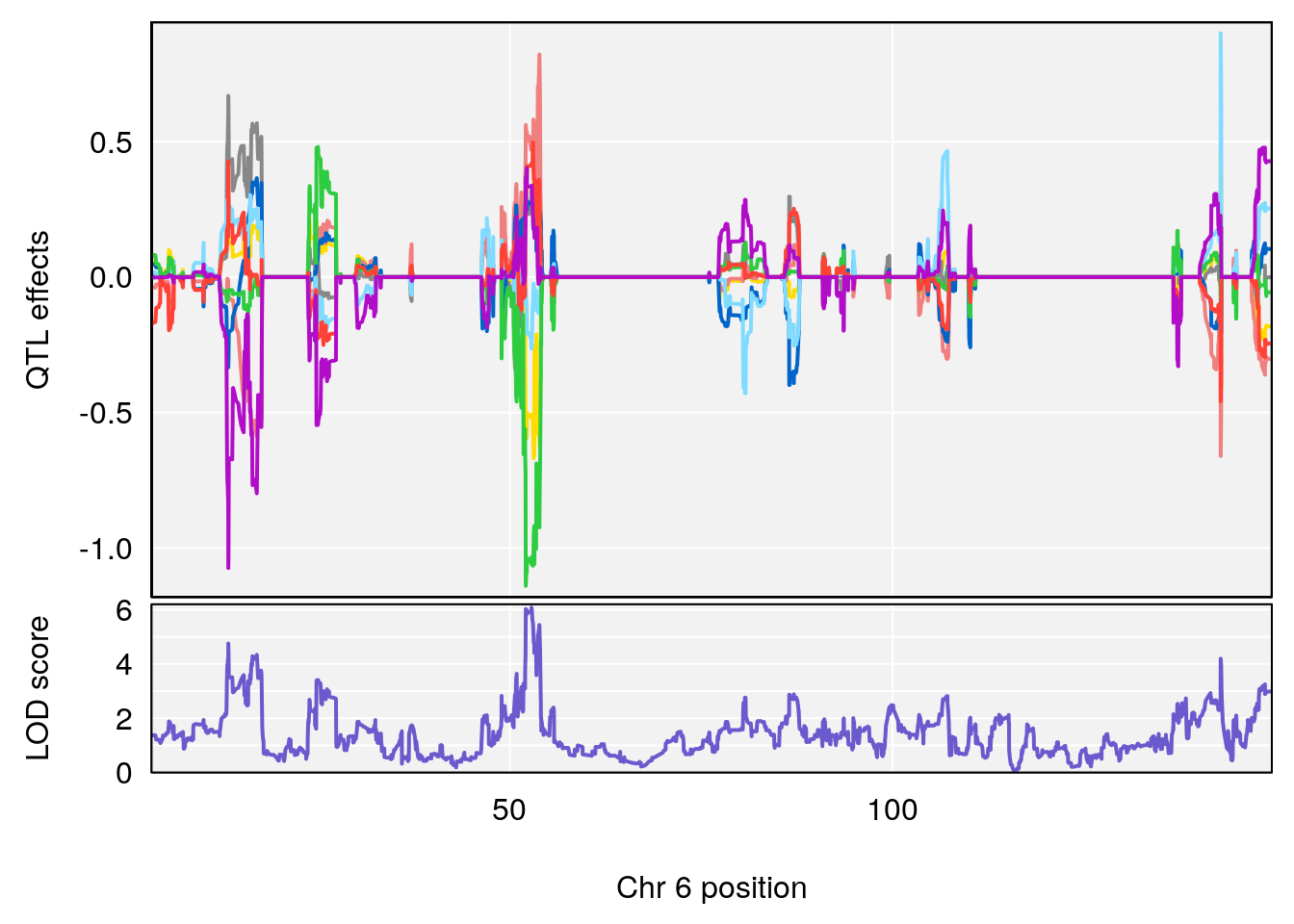
# [1] "MinDepressionRR(%)"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 5 147.27839 6.273736 67.23641 147.43417
# 2 1 pheno1 6 52.83155 6.076547 13.21830 53.95921
# 3 1 pheno1 12 110.14878 6.256748 90.07443 110.17562
# 4 1 pheno1 18 41.80133 6.871462 41.74898 43.29028
# [1] 1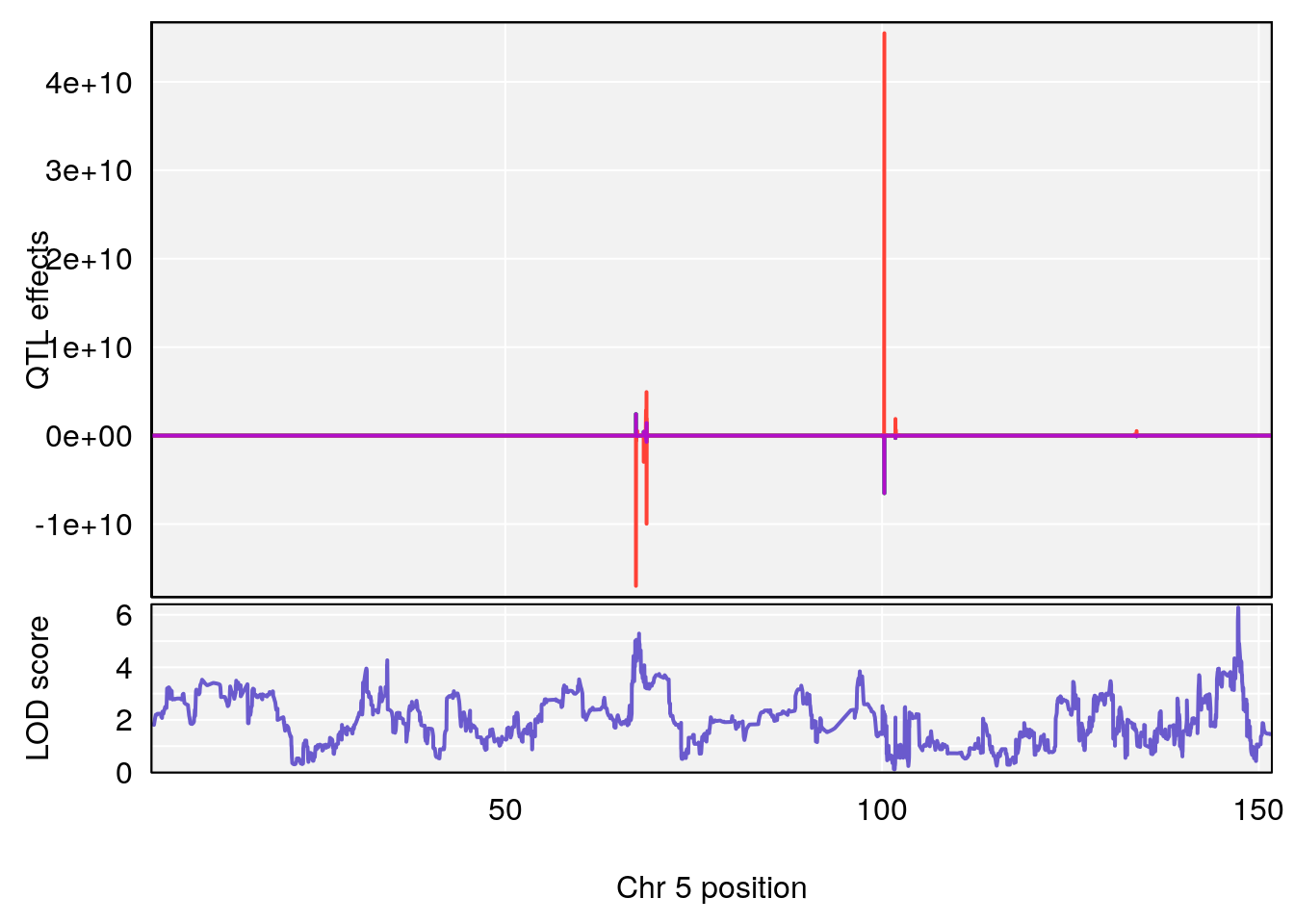
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2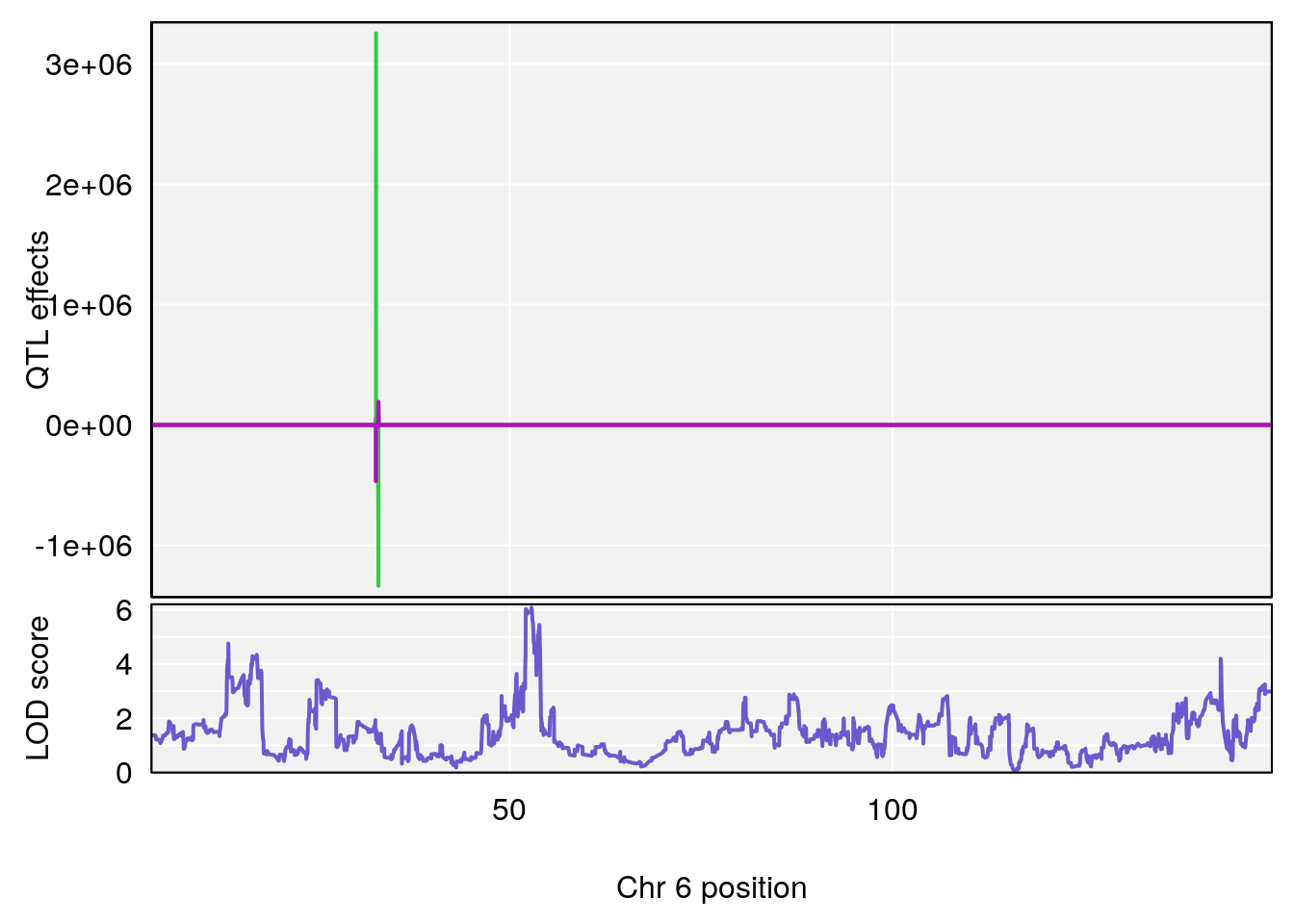
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
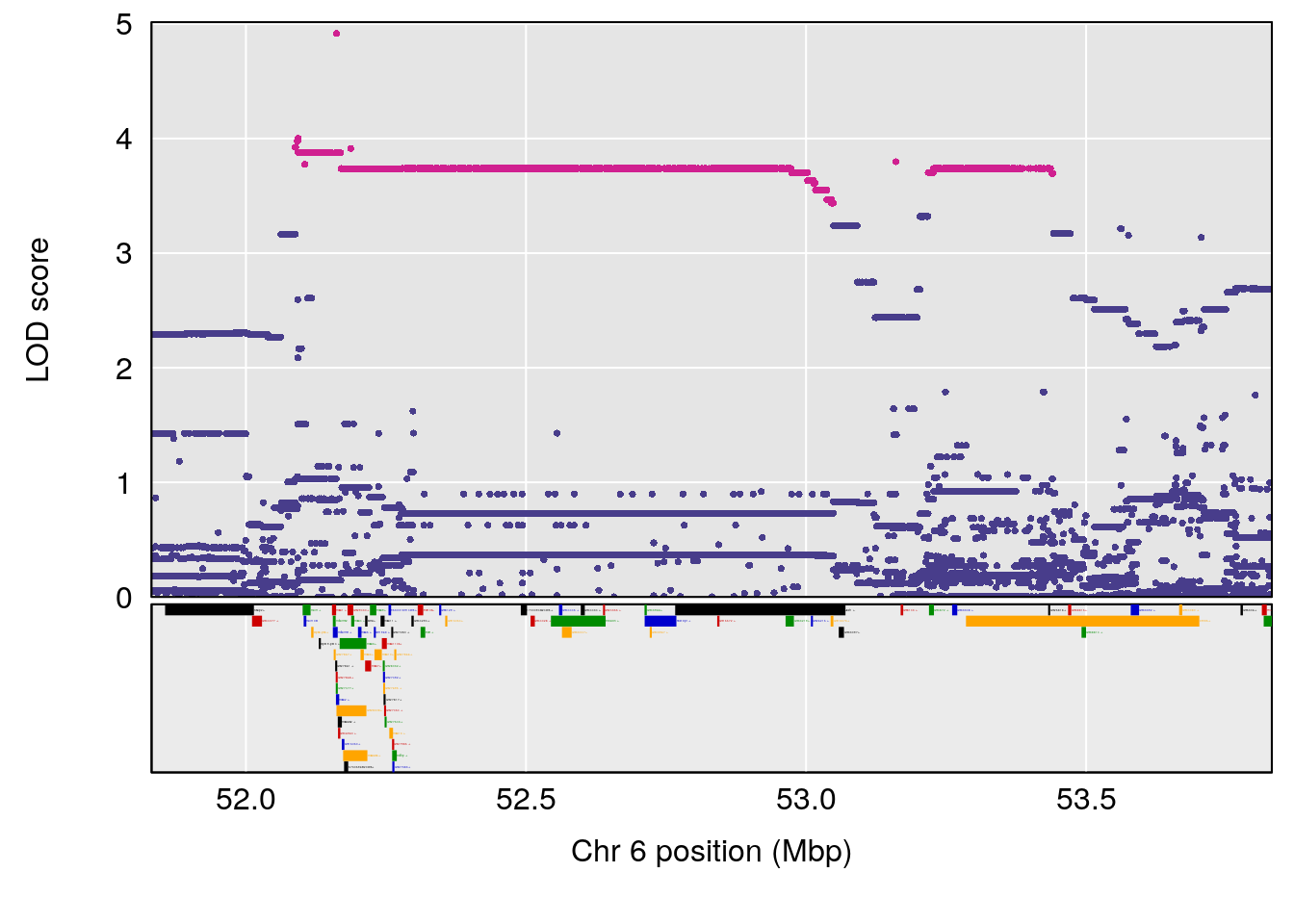
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 3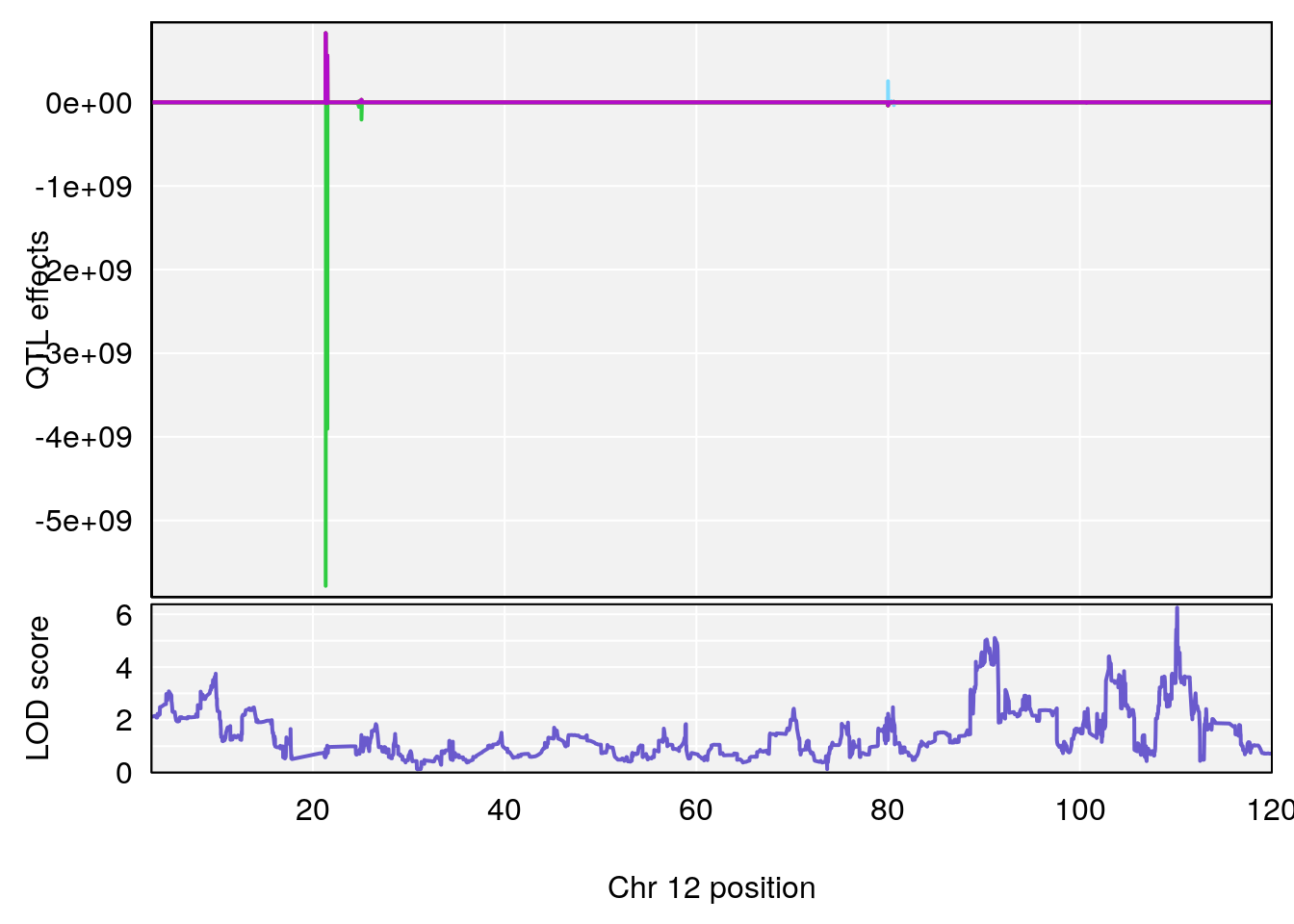
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
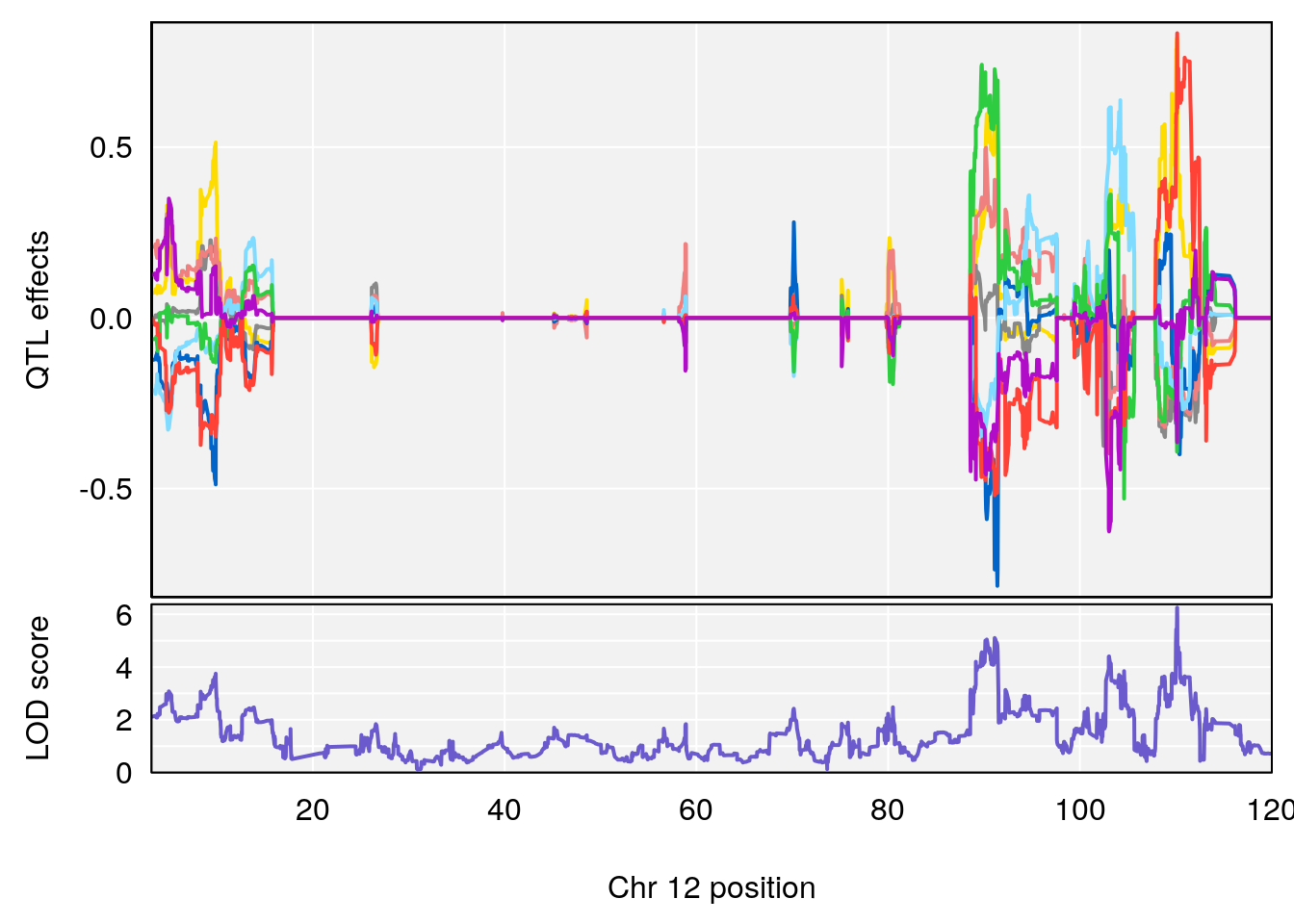
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
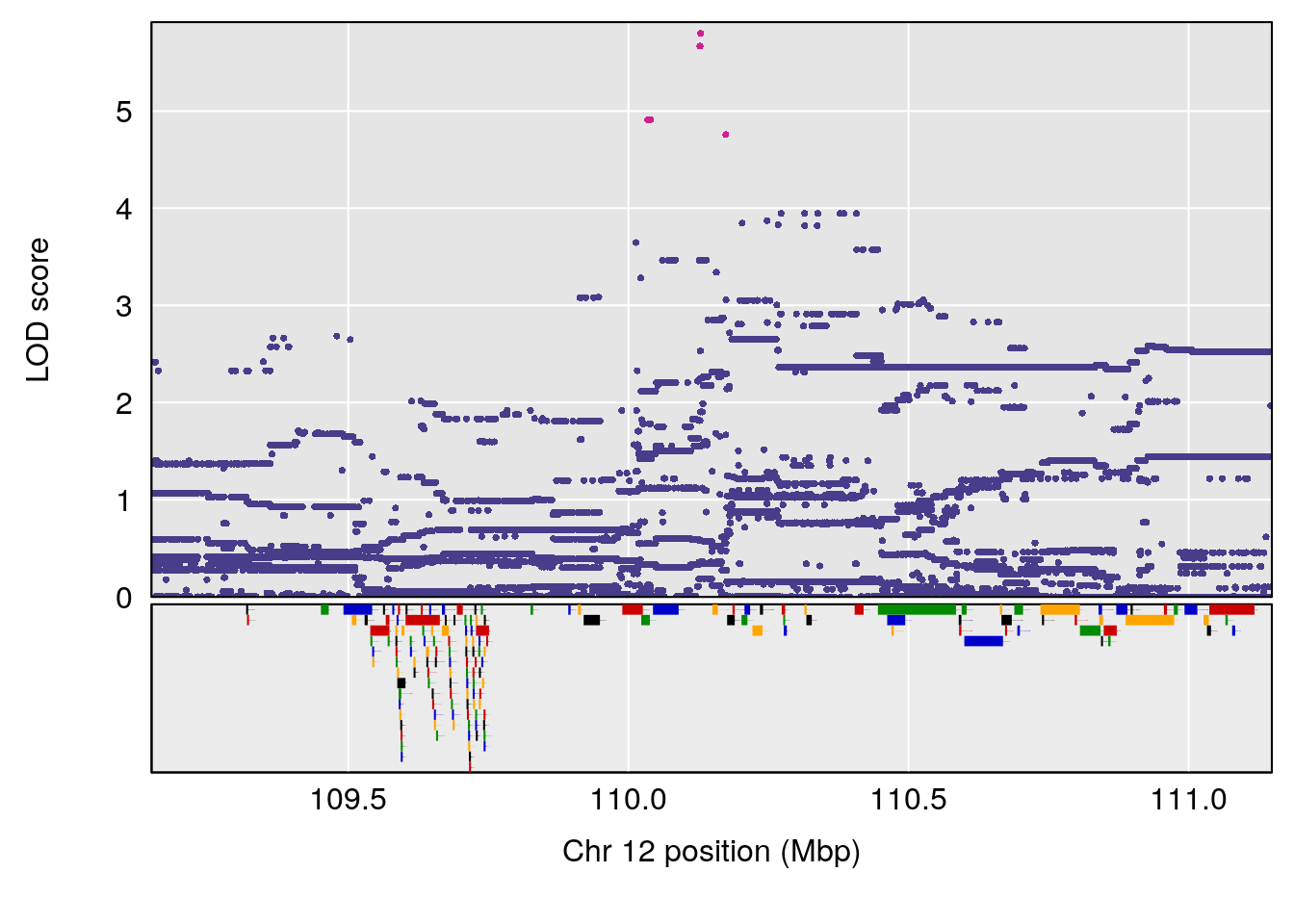
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
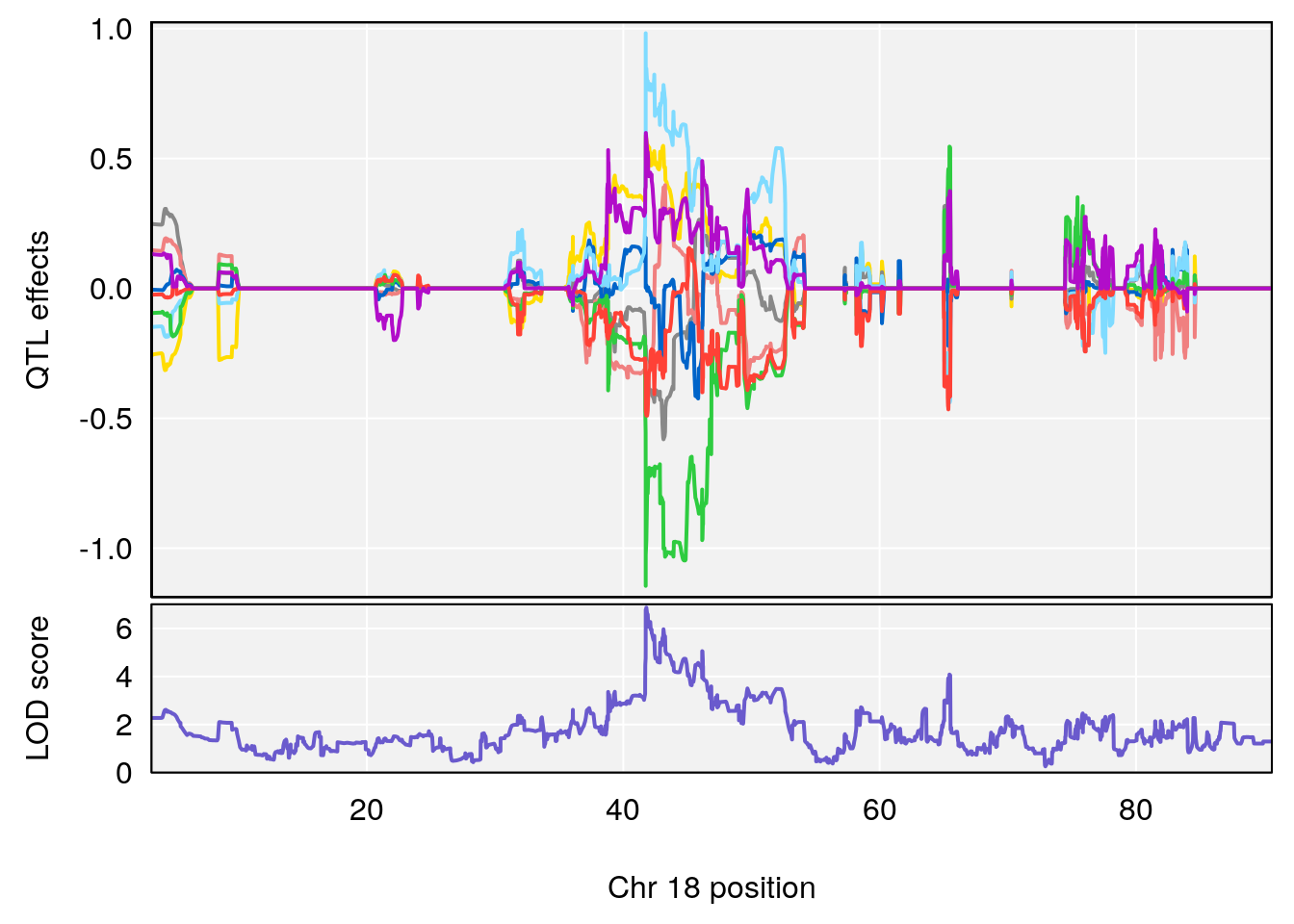
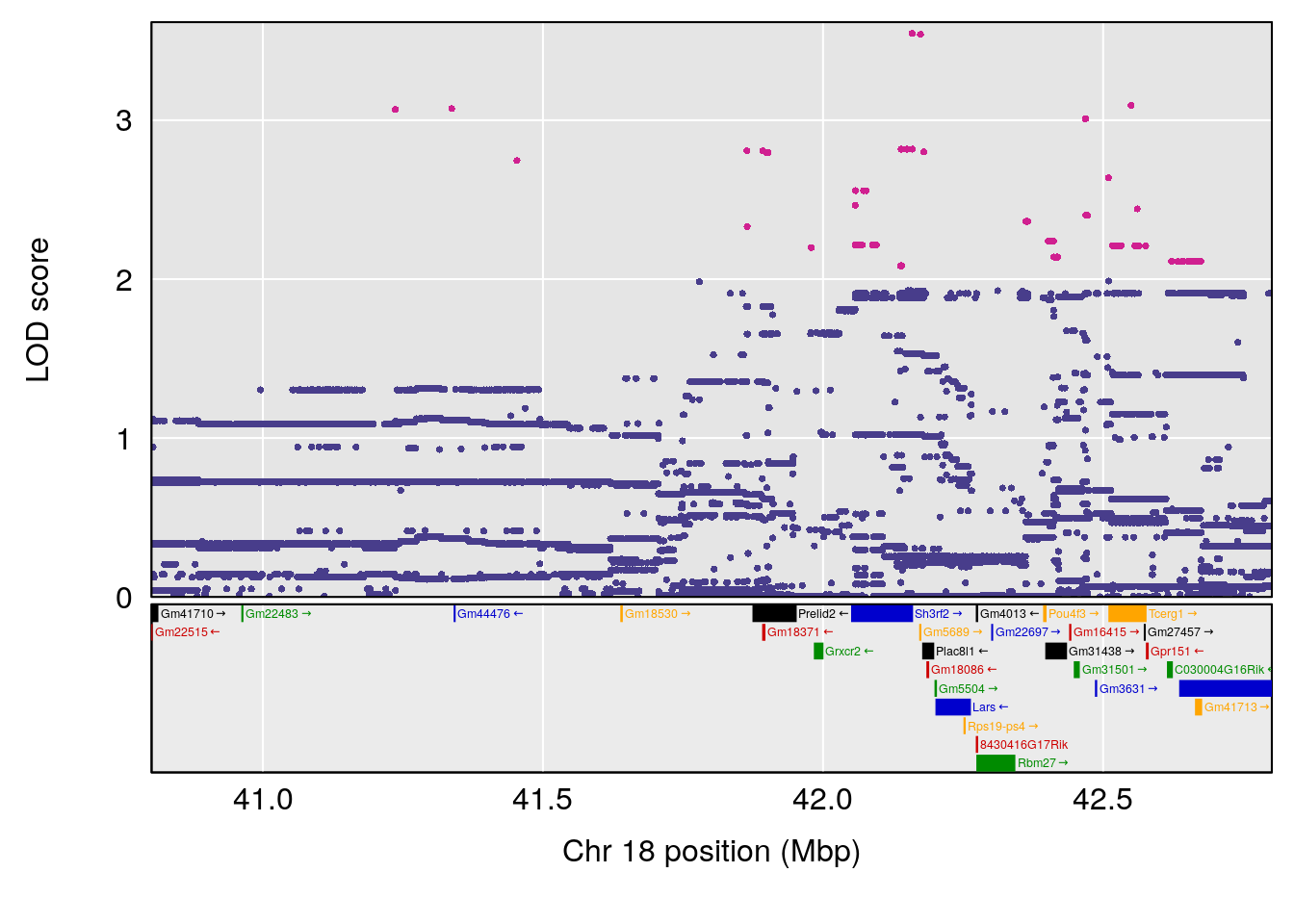
# [1] 4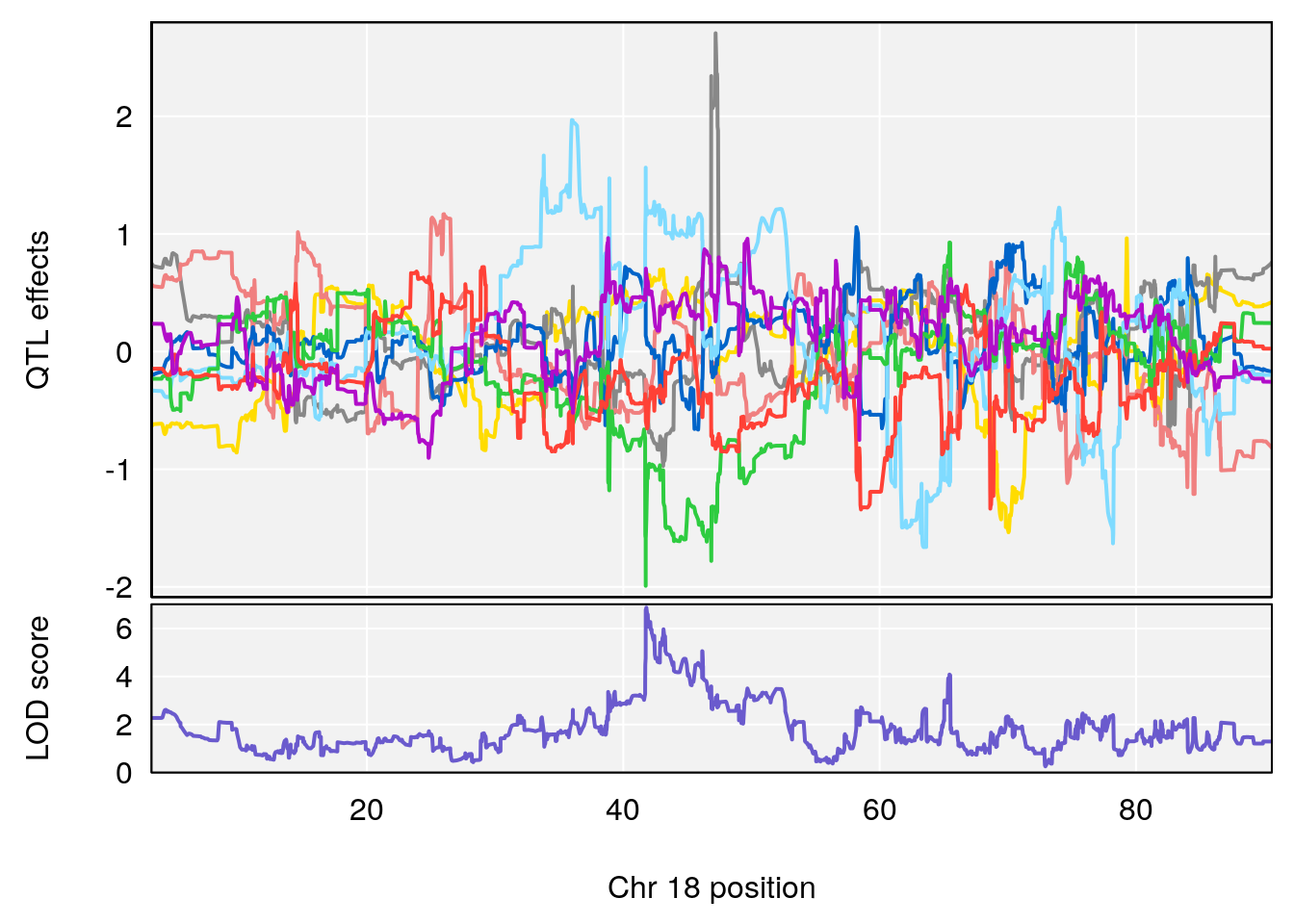
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "Steady-StateDepressionDuration(Hr)INTERVAL"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 6 126.017905 6.048217 125.696341 136.70757
# 2 1 pheno1 8 92.203842 6.030778 16.080653 124.33745
# 3 1 pheno1 10 5.635019 6.925830 3.400722 115.81149
# 4 1 pheno1 18 65.434900 6.977069 65.313821 65.52767
# [1] 1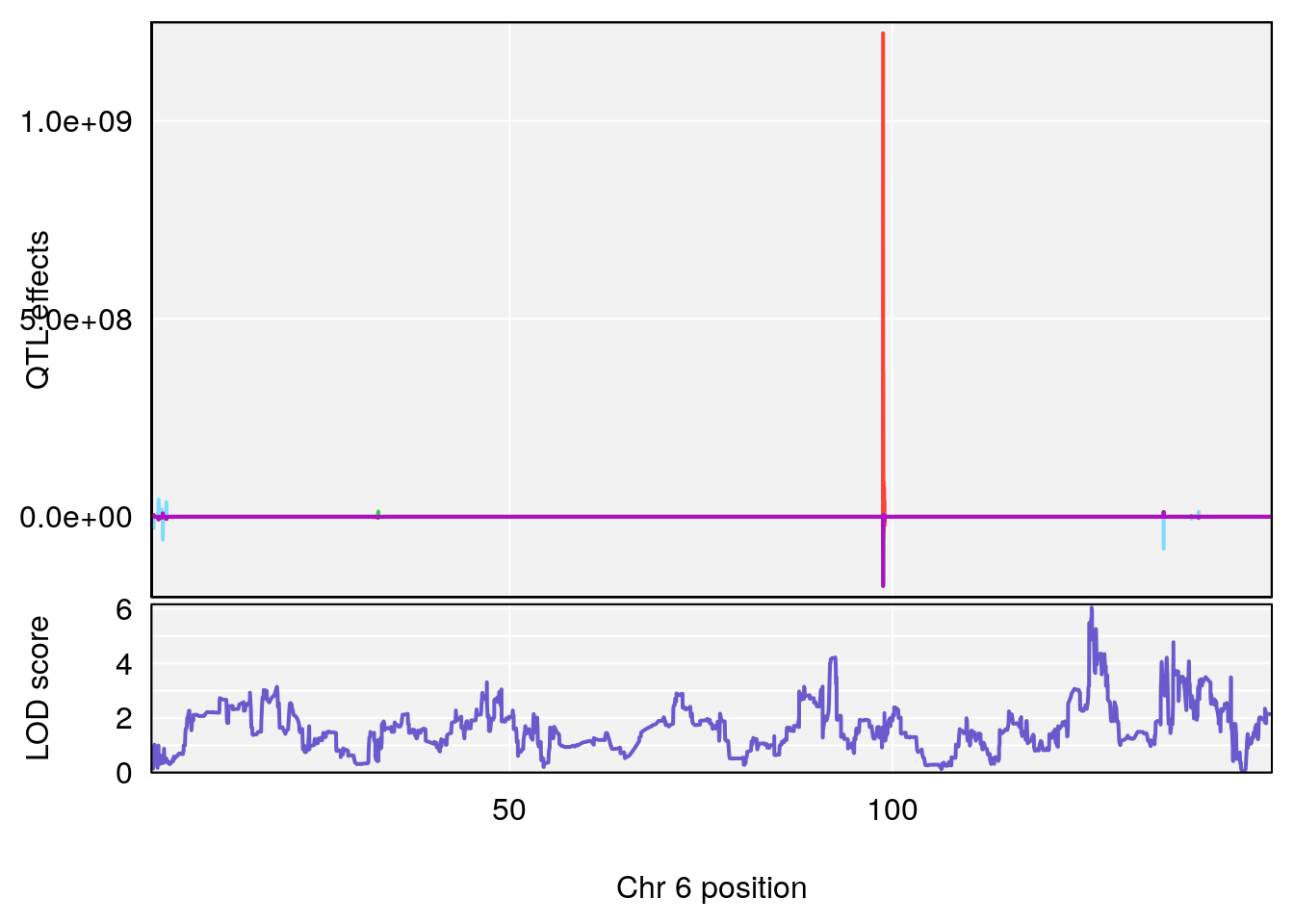
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
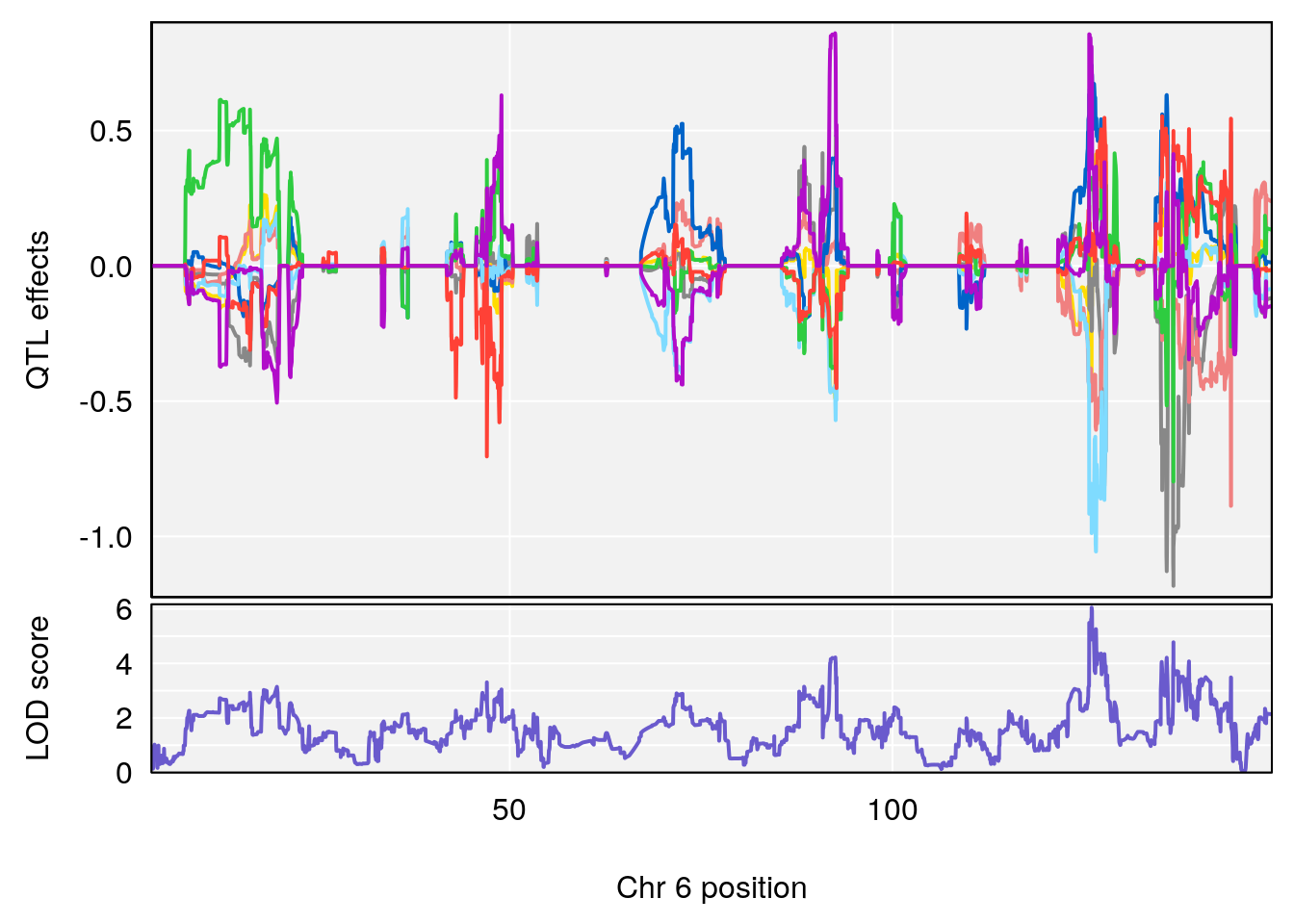
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
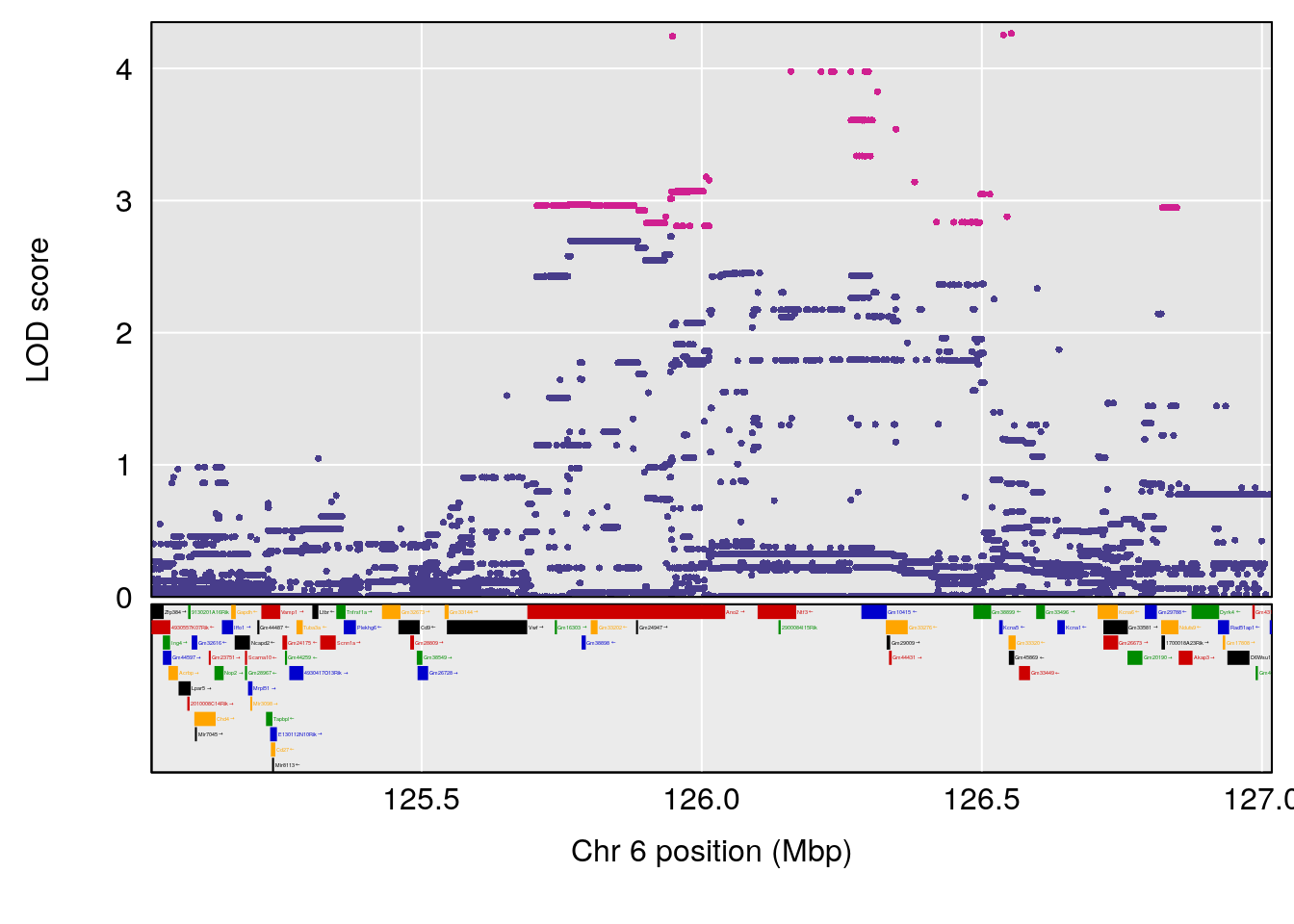
# [1] 2
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
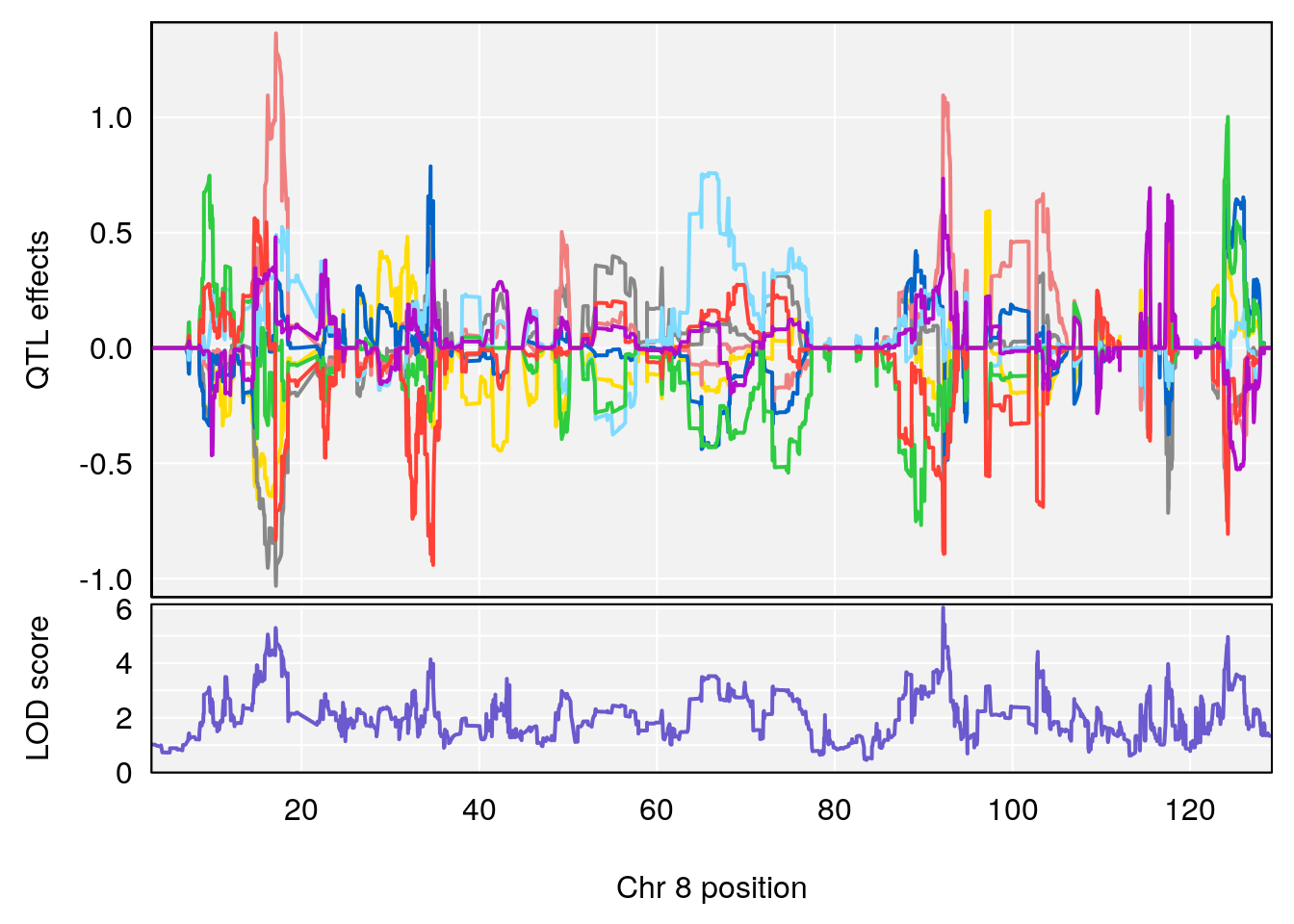
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
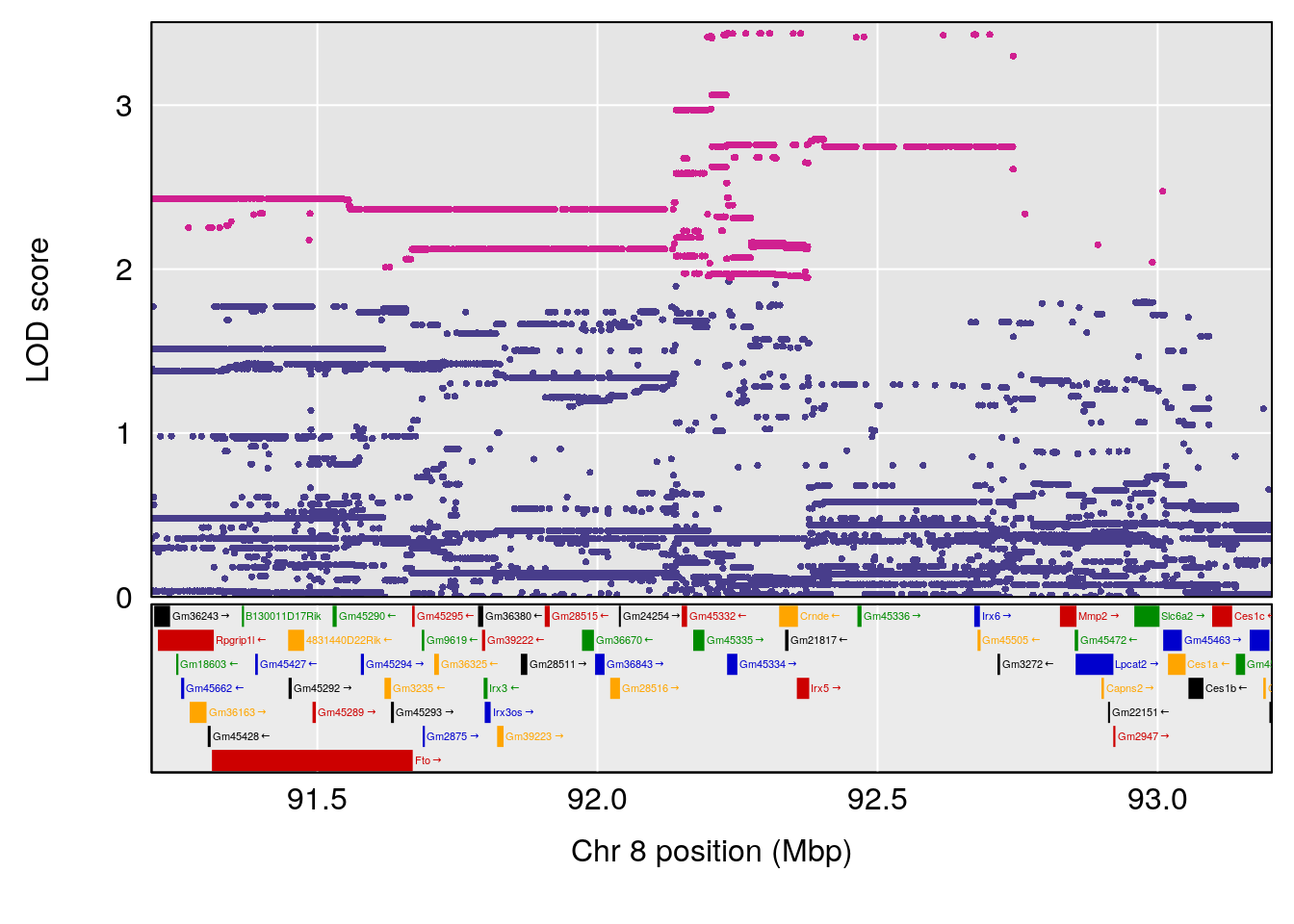
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 3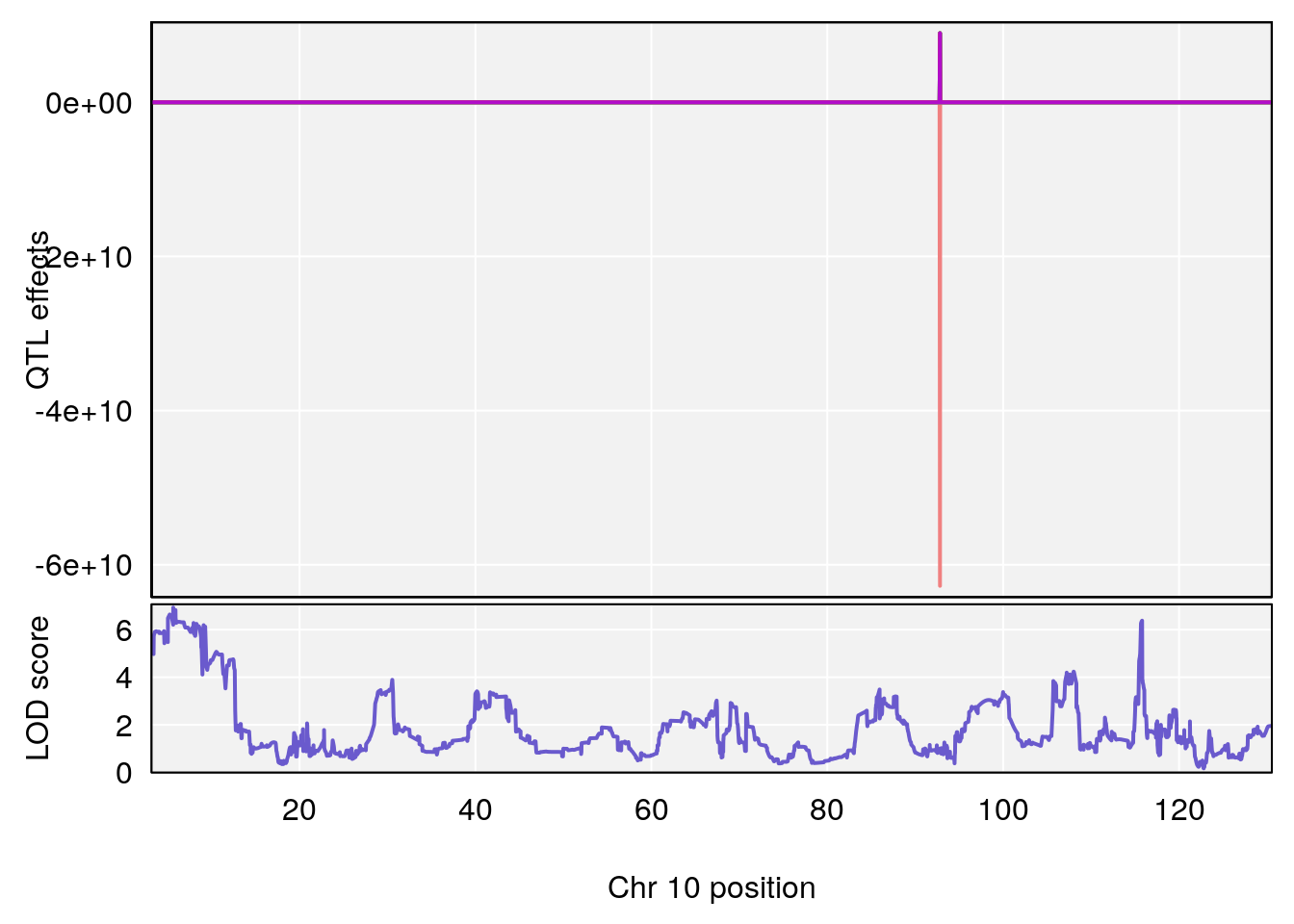
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
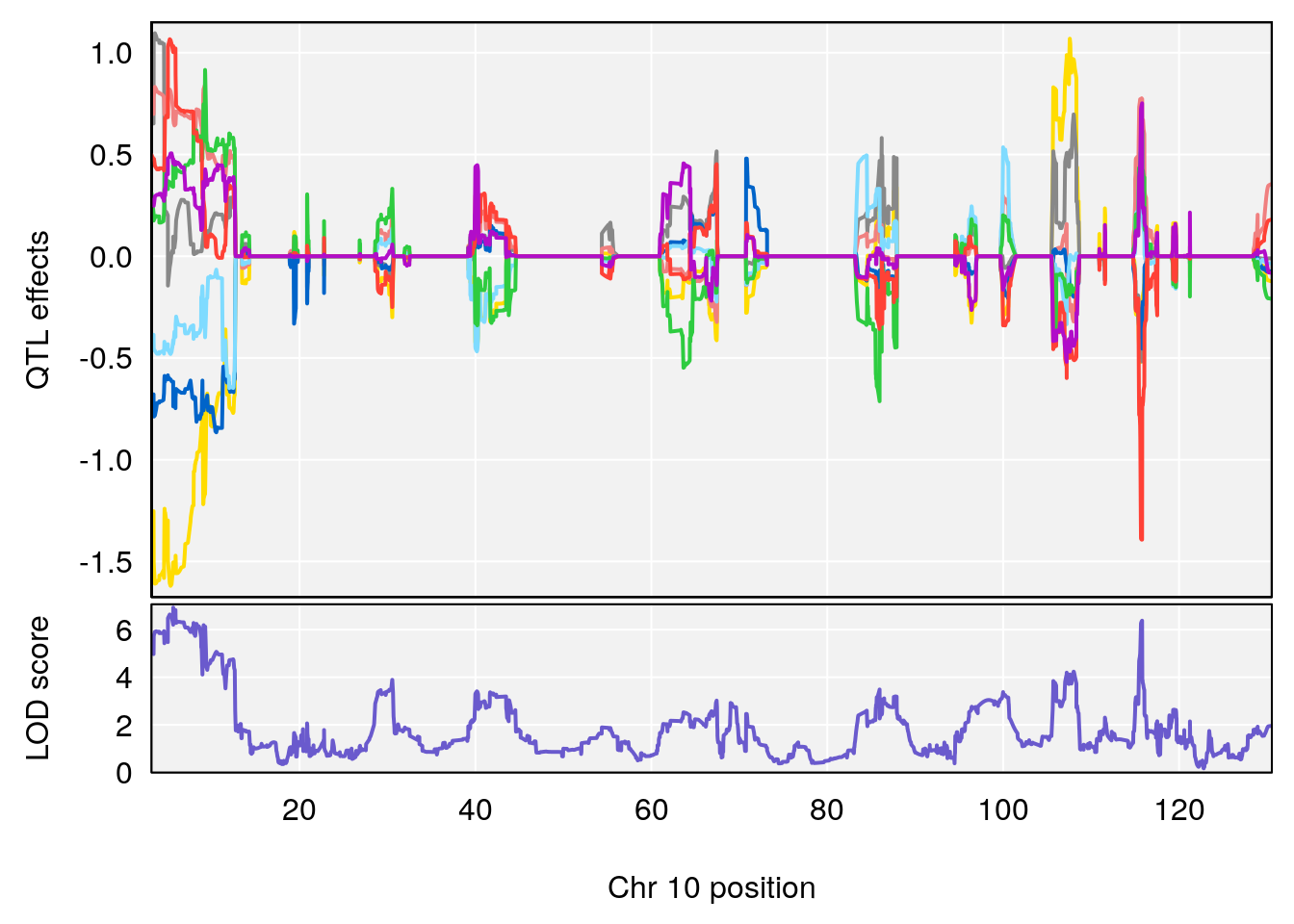
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
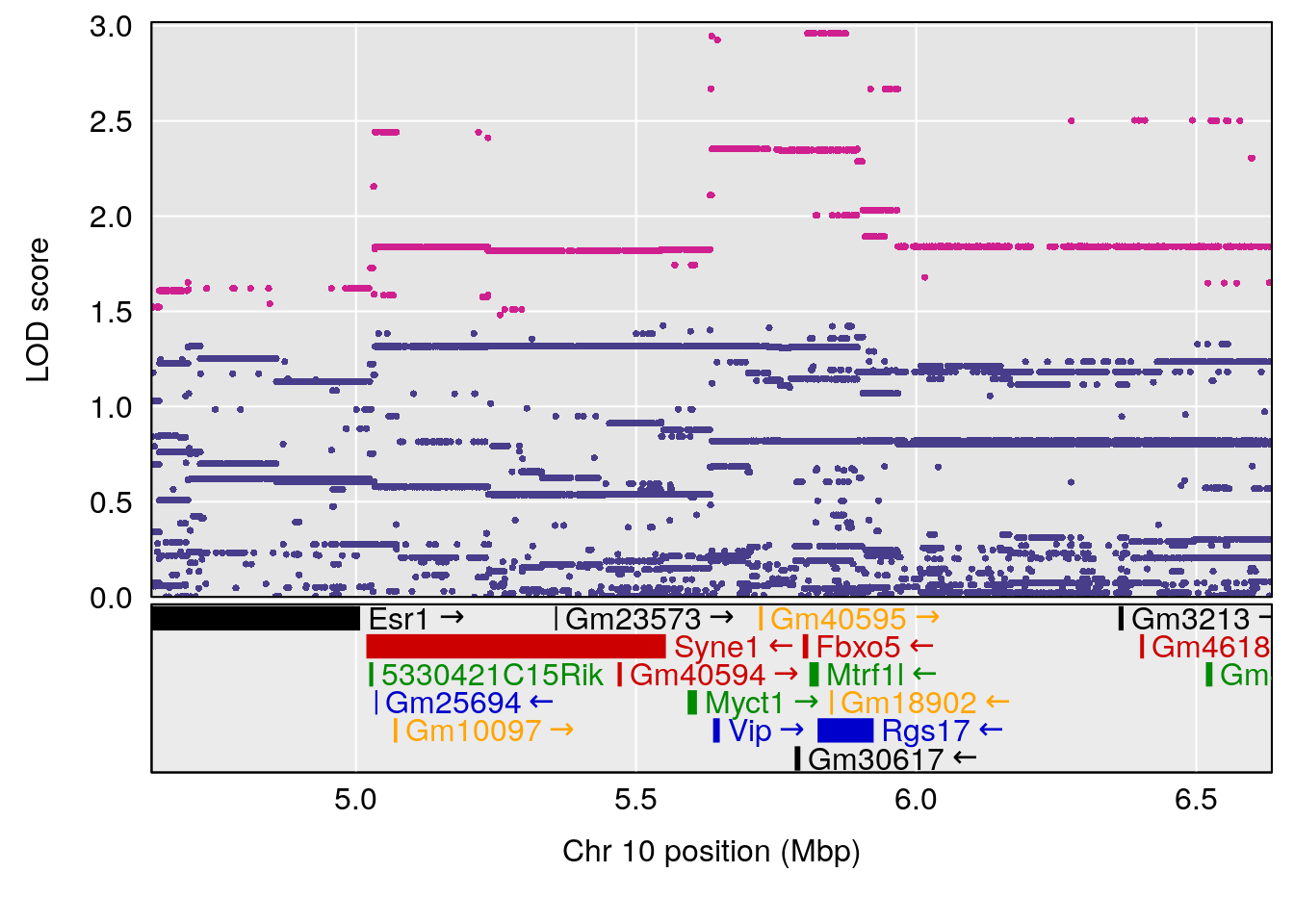
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 4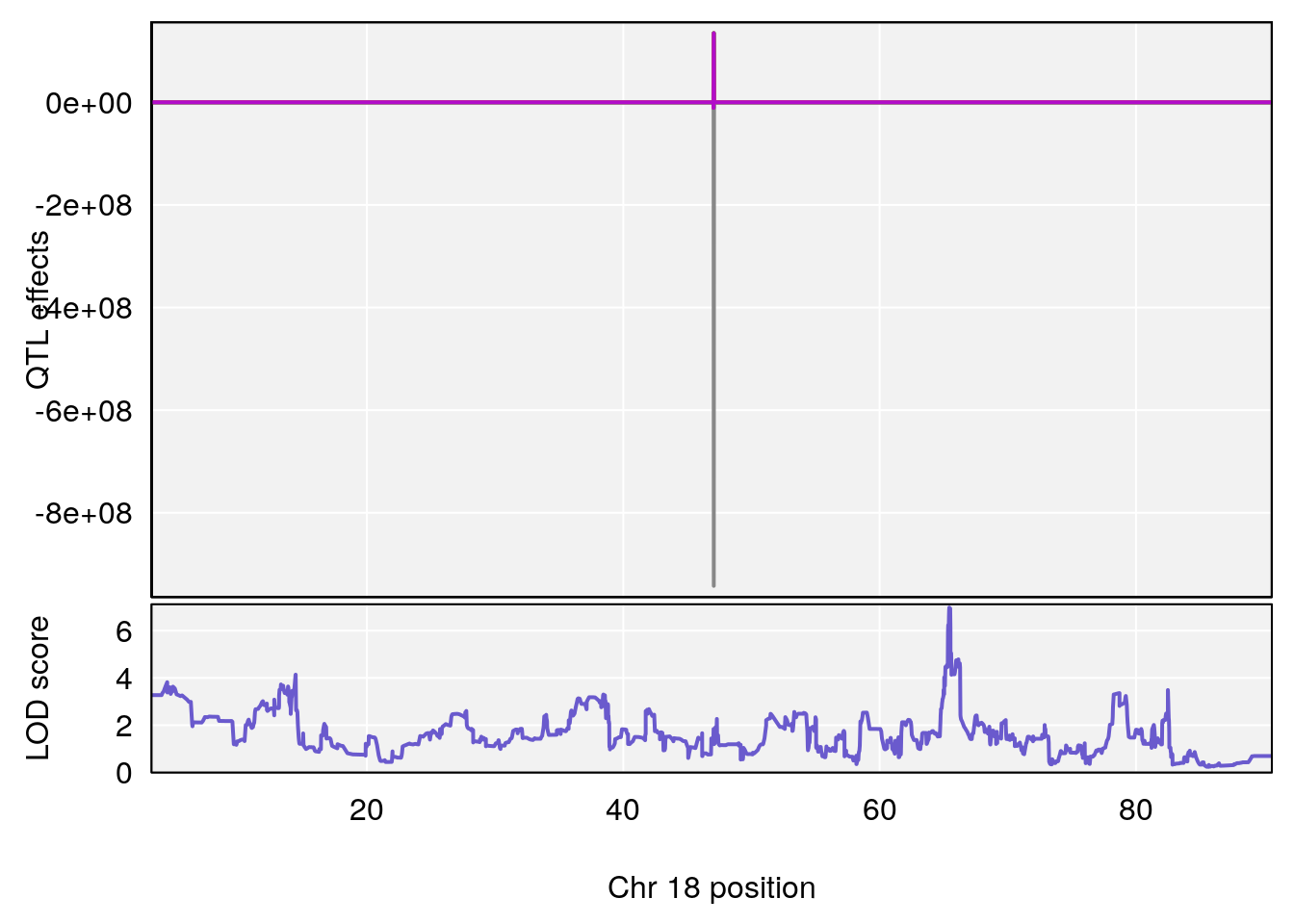
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
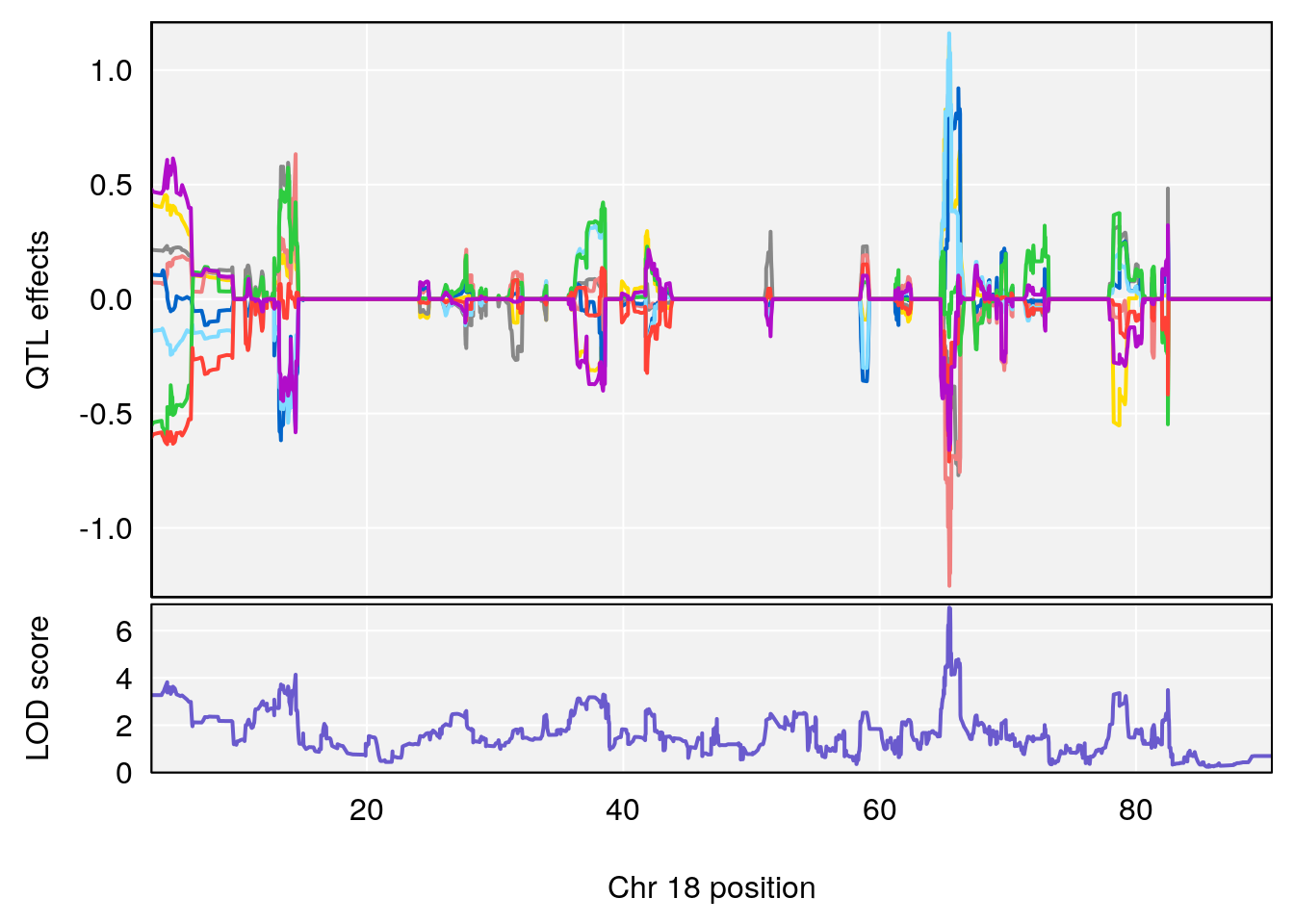
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
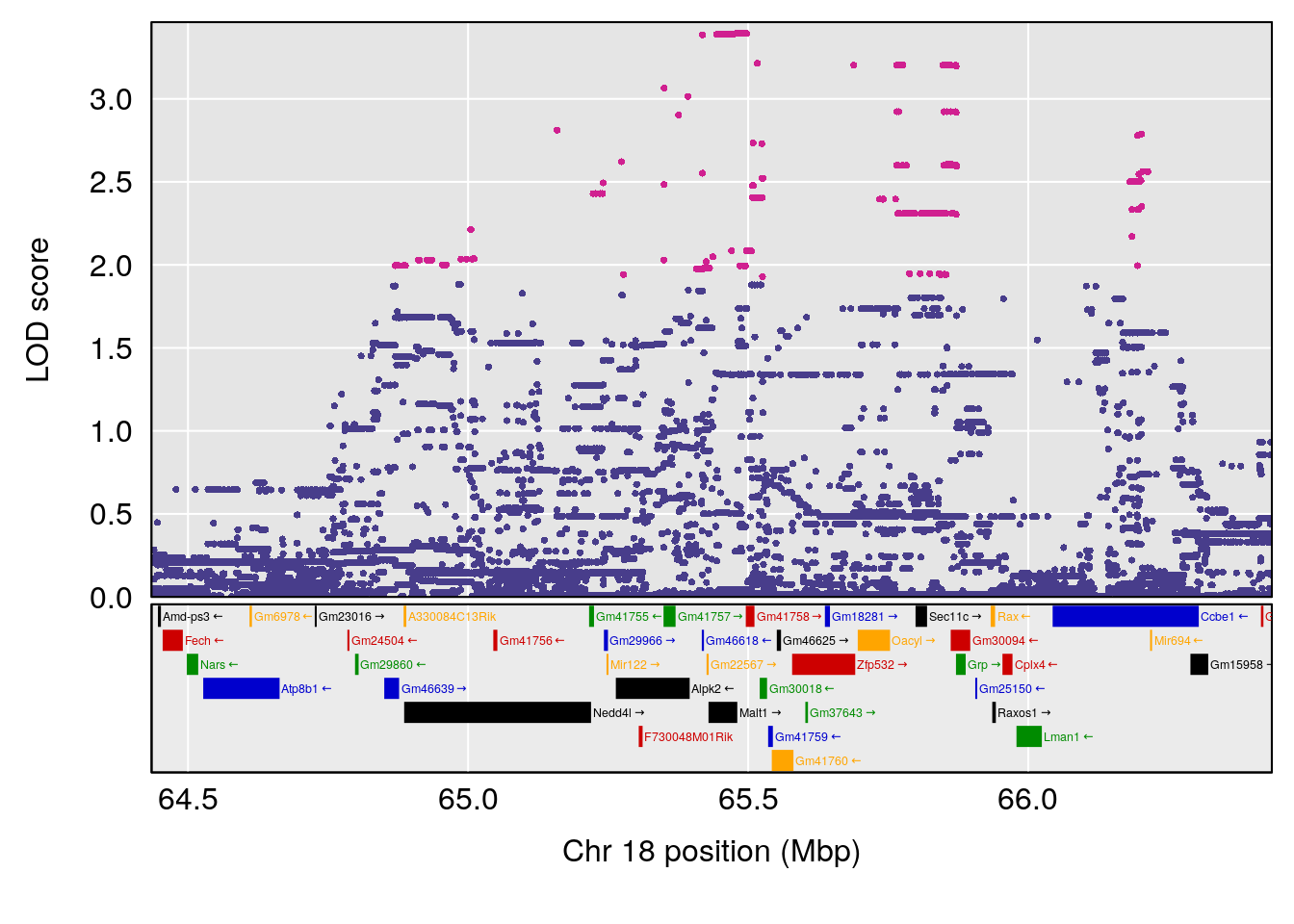
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "TimetoThresholdRecovery(Hr)"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 91.312012 6.278565 91.216258 152.86541
# 2 1 pheno1 6 126.017905 6.903066 125.696341 126.63864
# 3 1 pheno1 10 5.635019 6.436675 3.174515 115.81149
# 4 1 pheno1 18 65.438329 7.680994 65.313821 65.52797
# [1] 1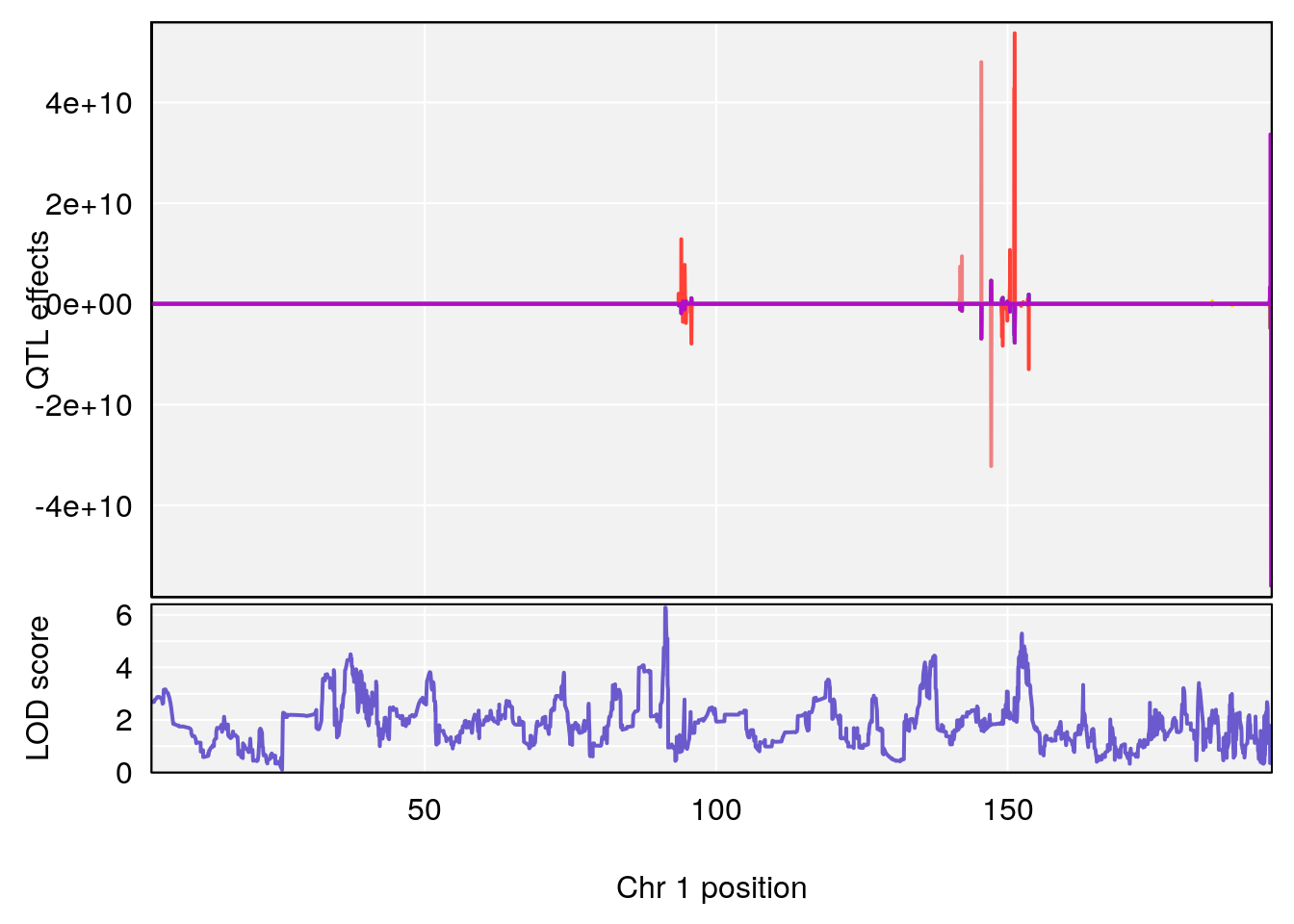
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
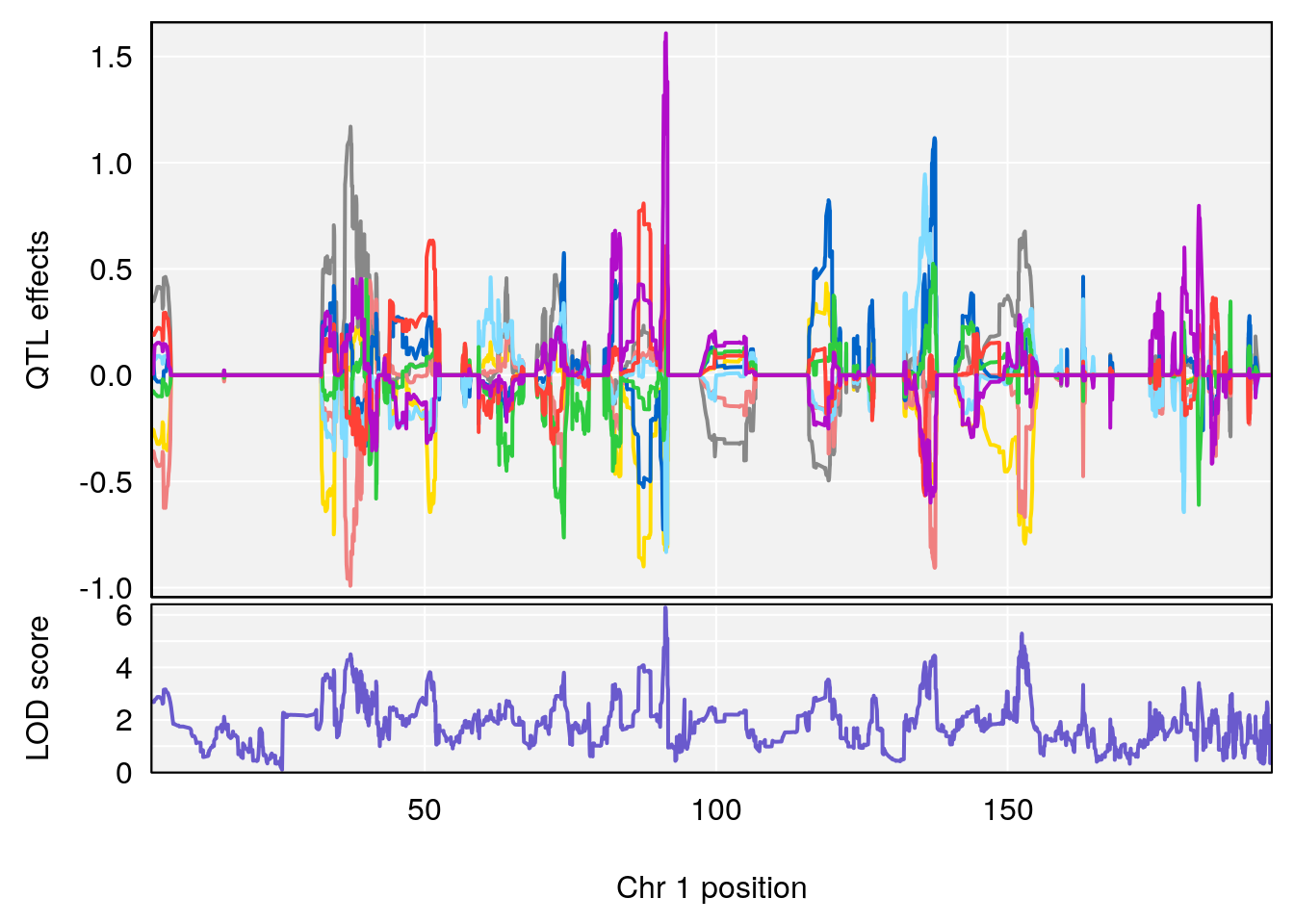
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
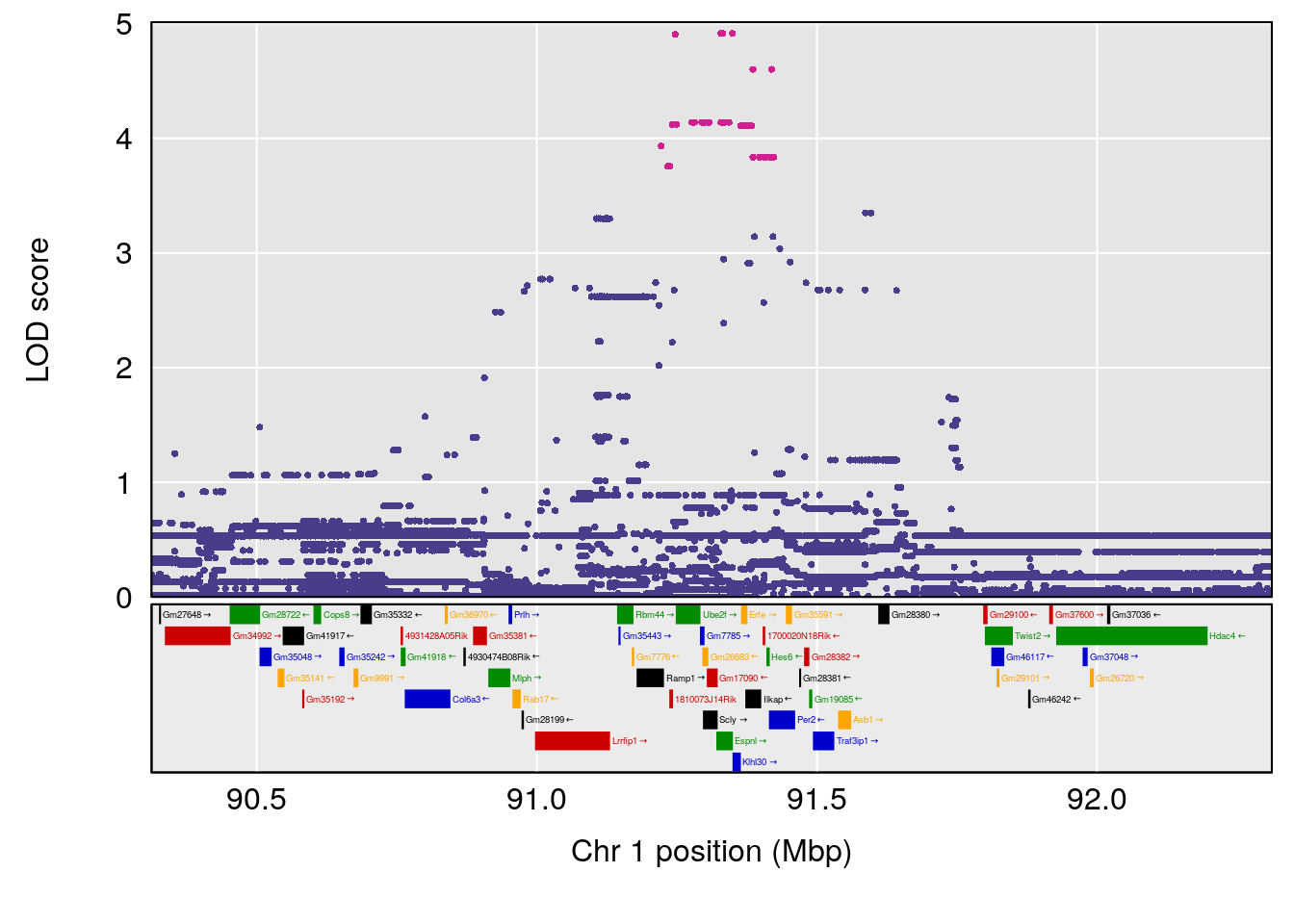
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
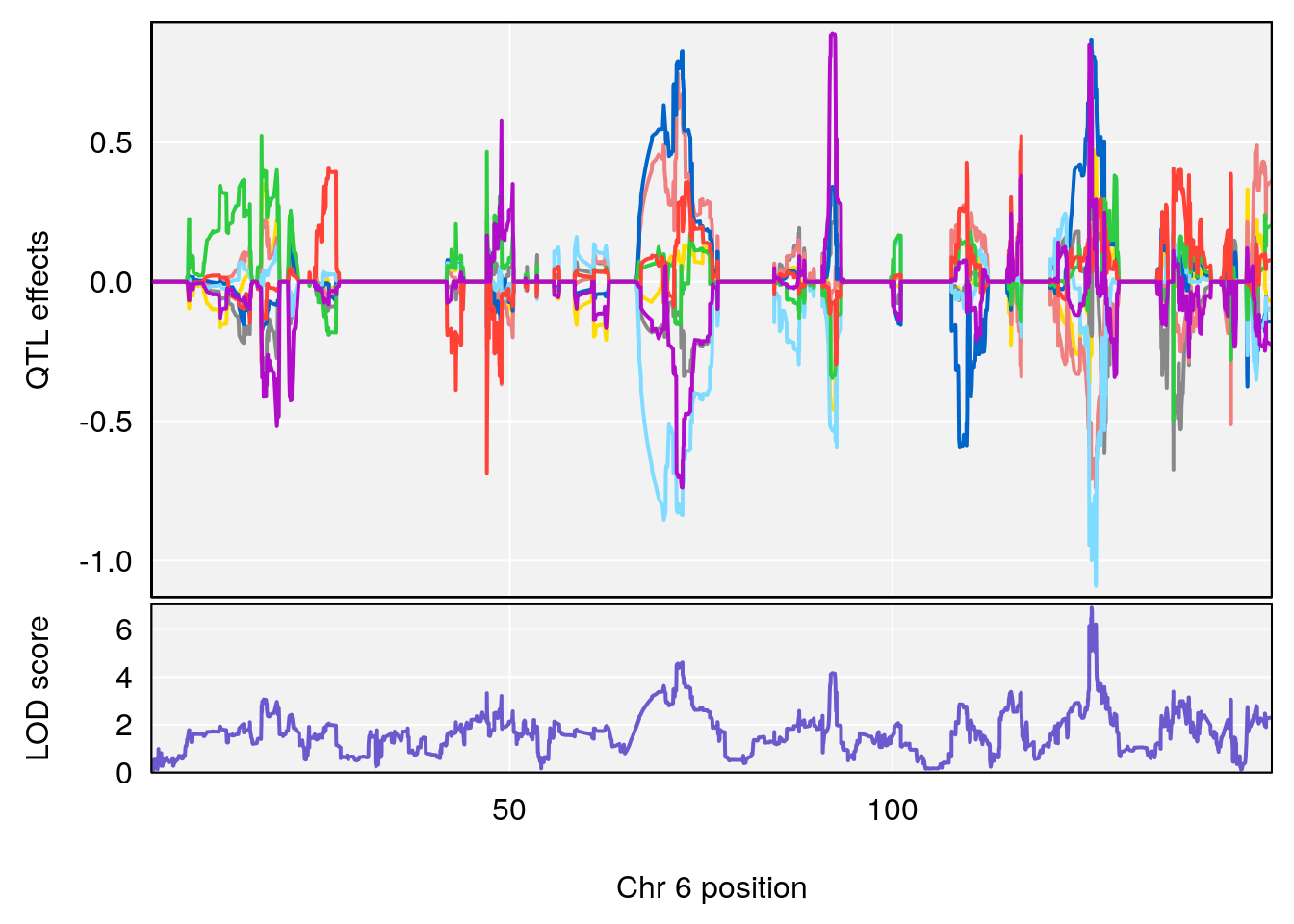
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
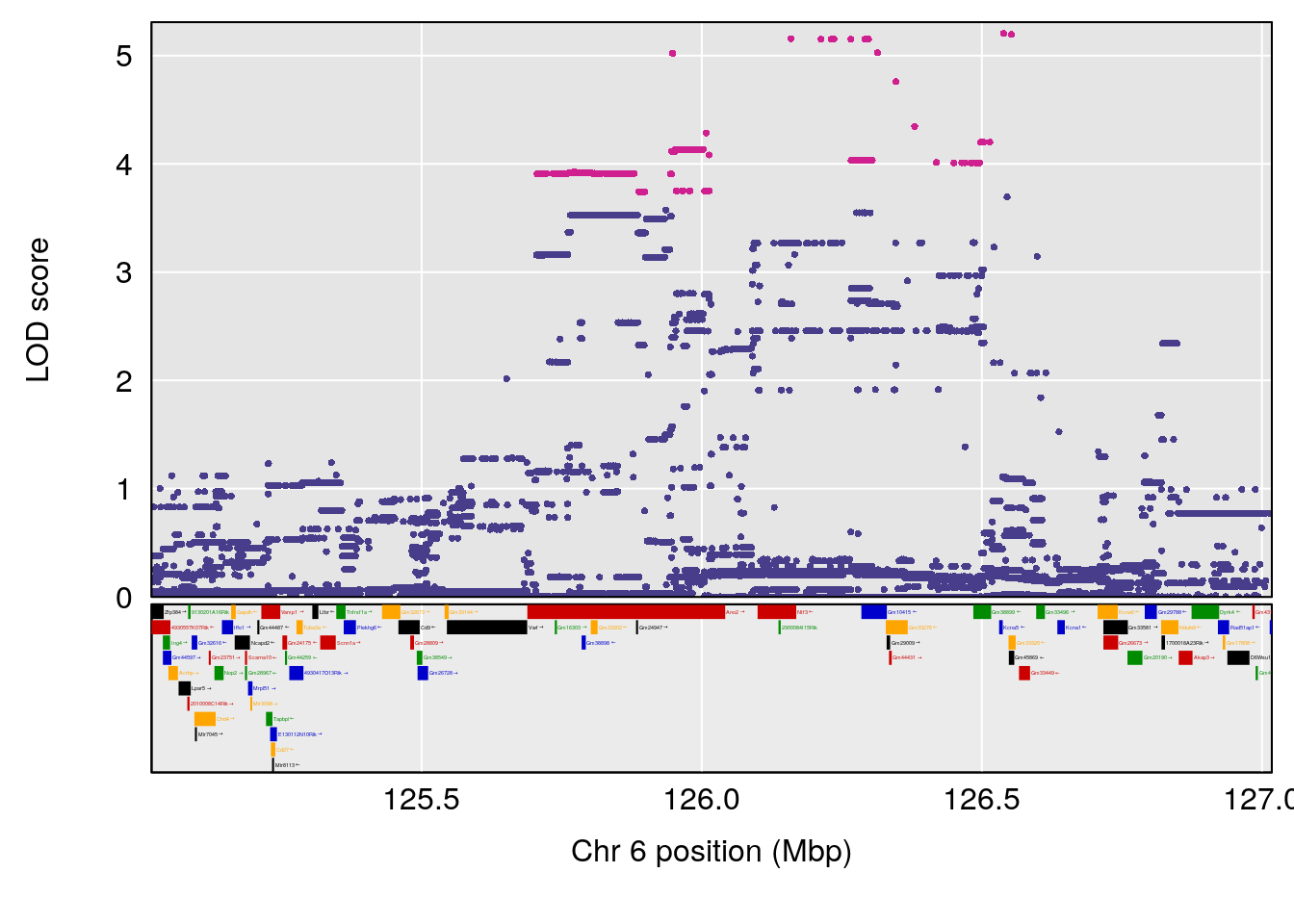
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 3
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
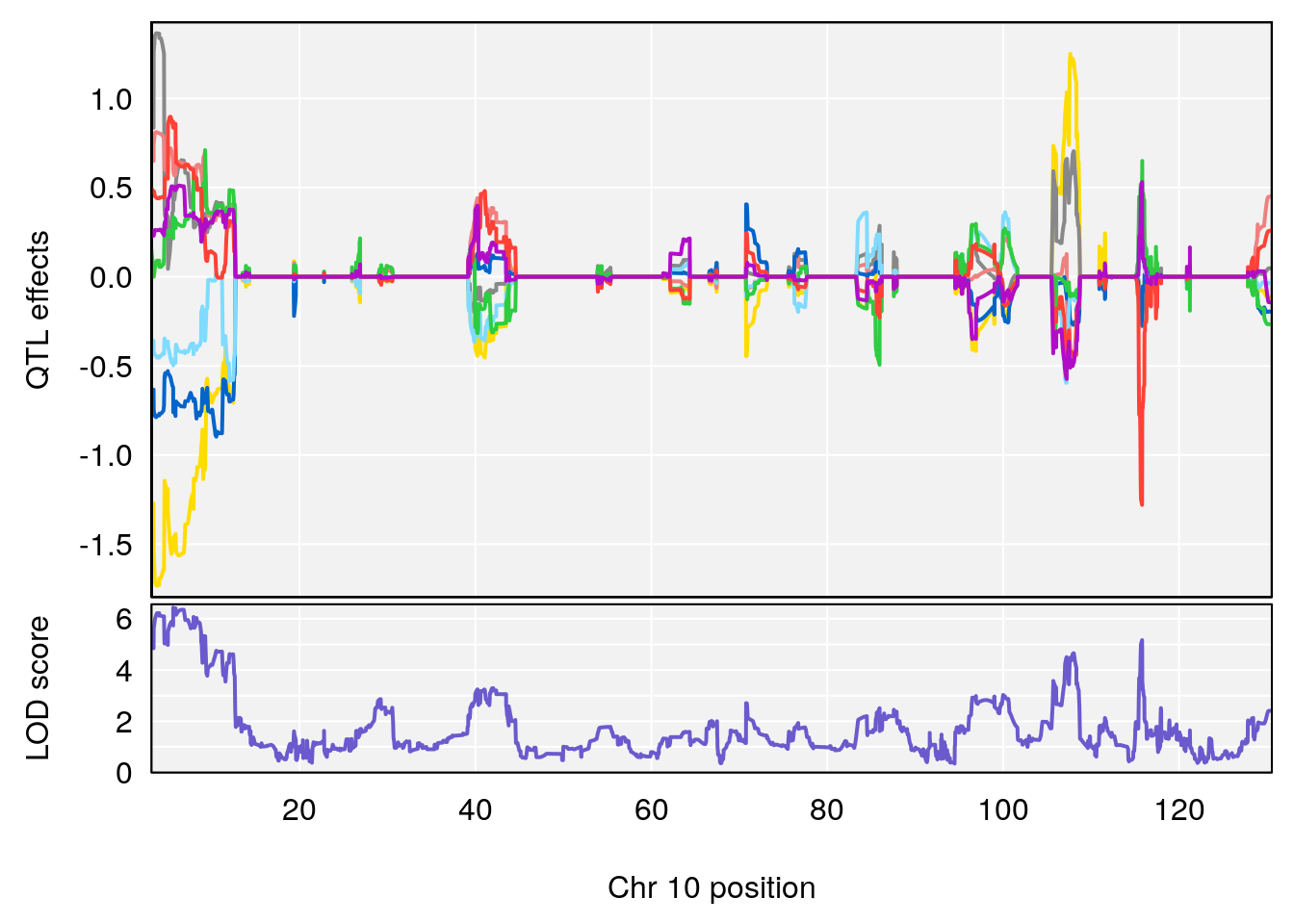
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
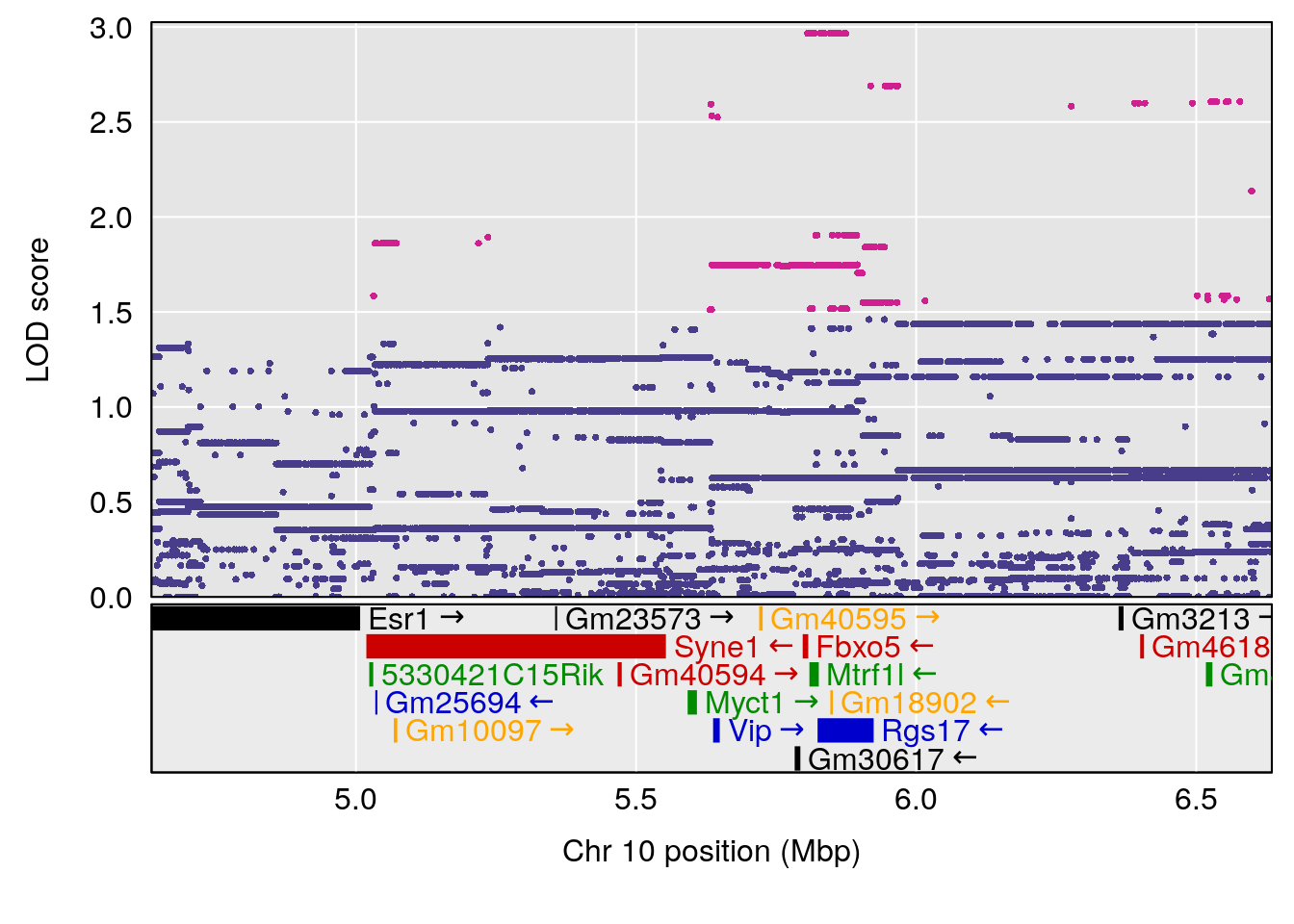
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 4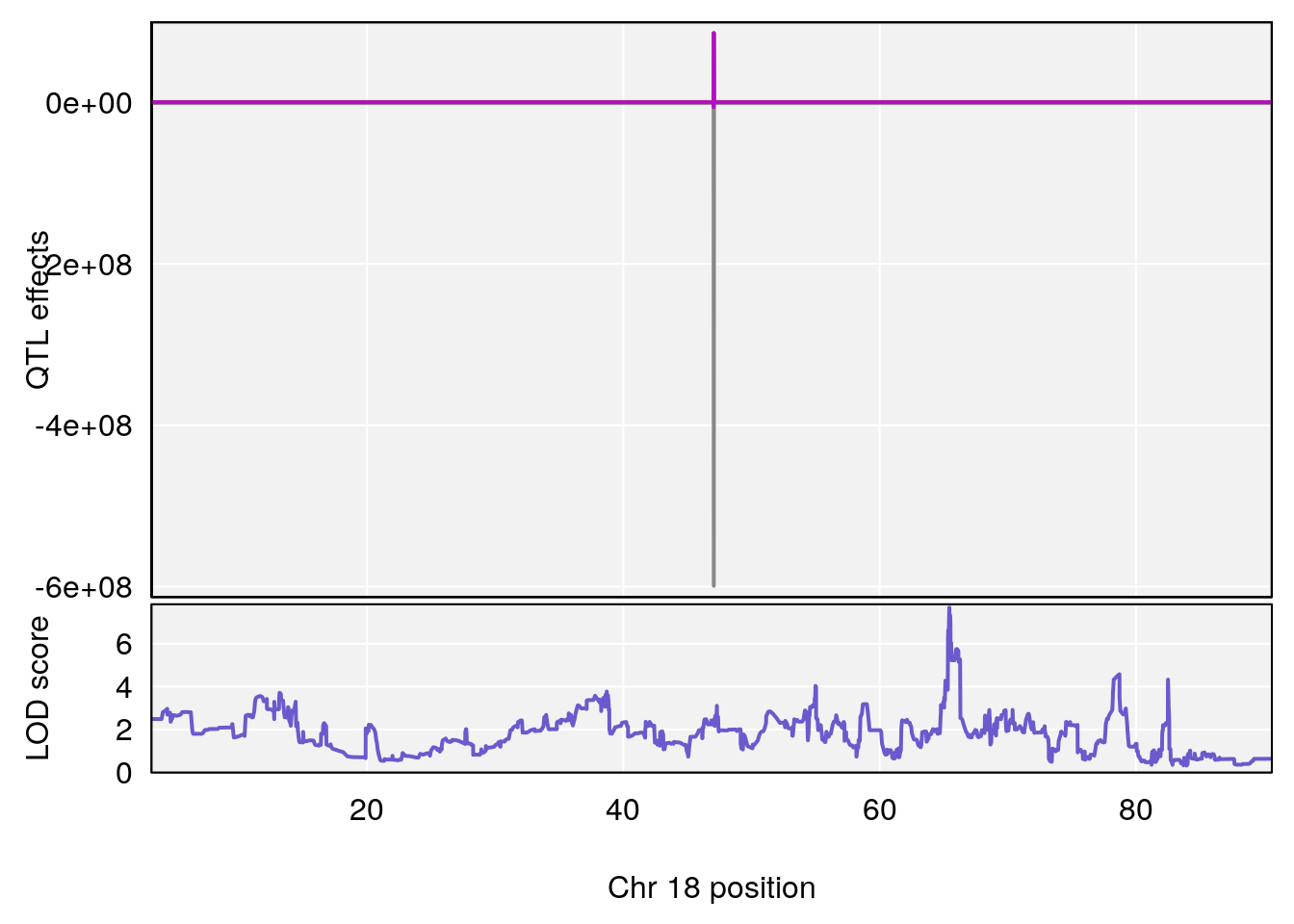
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
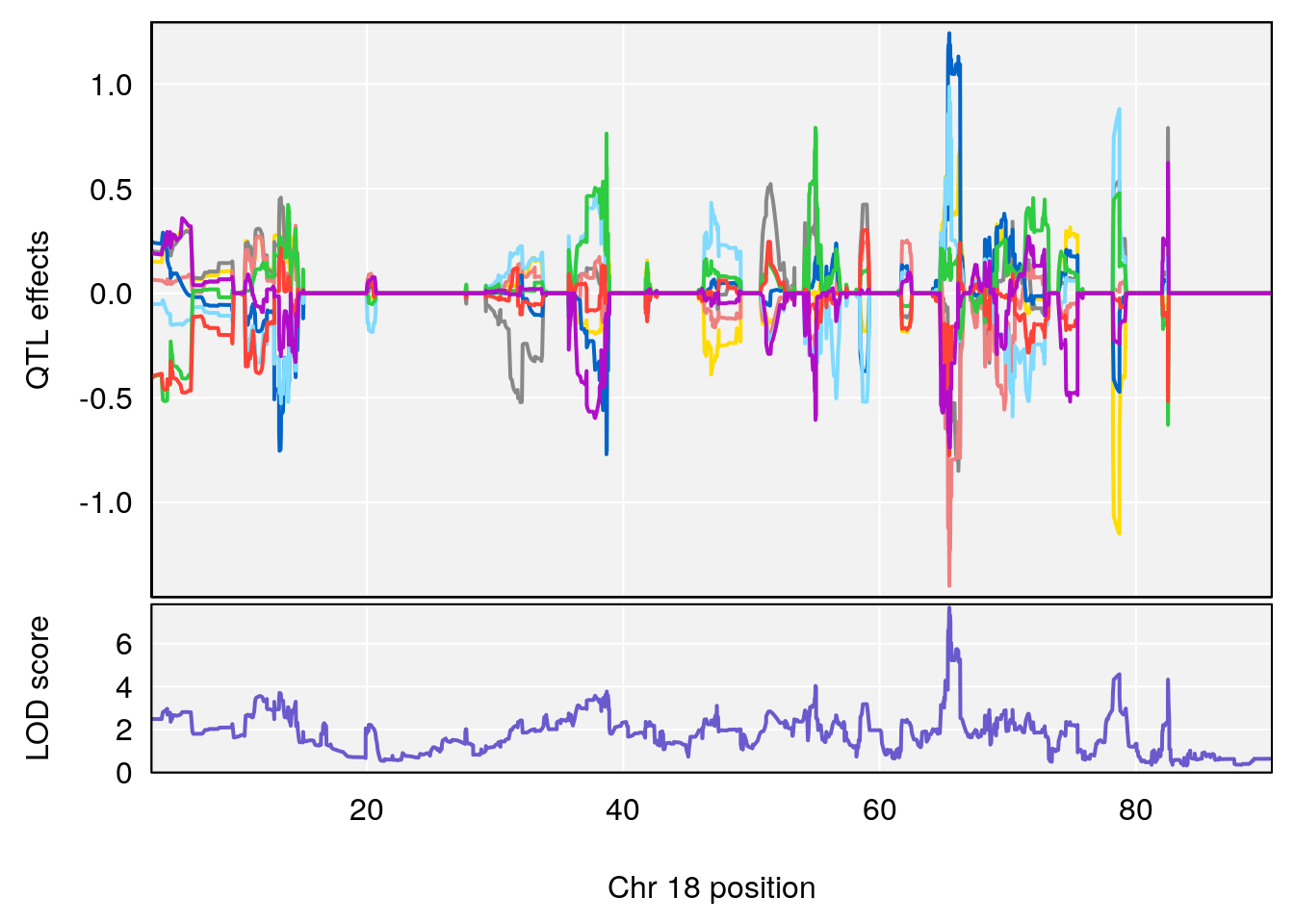
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
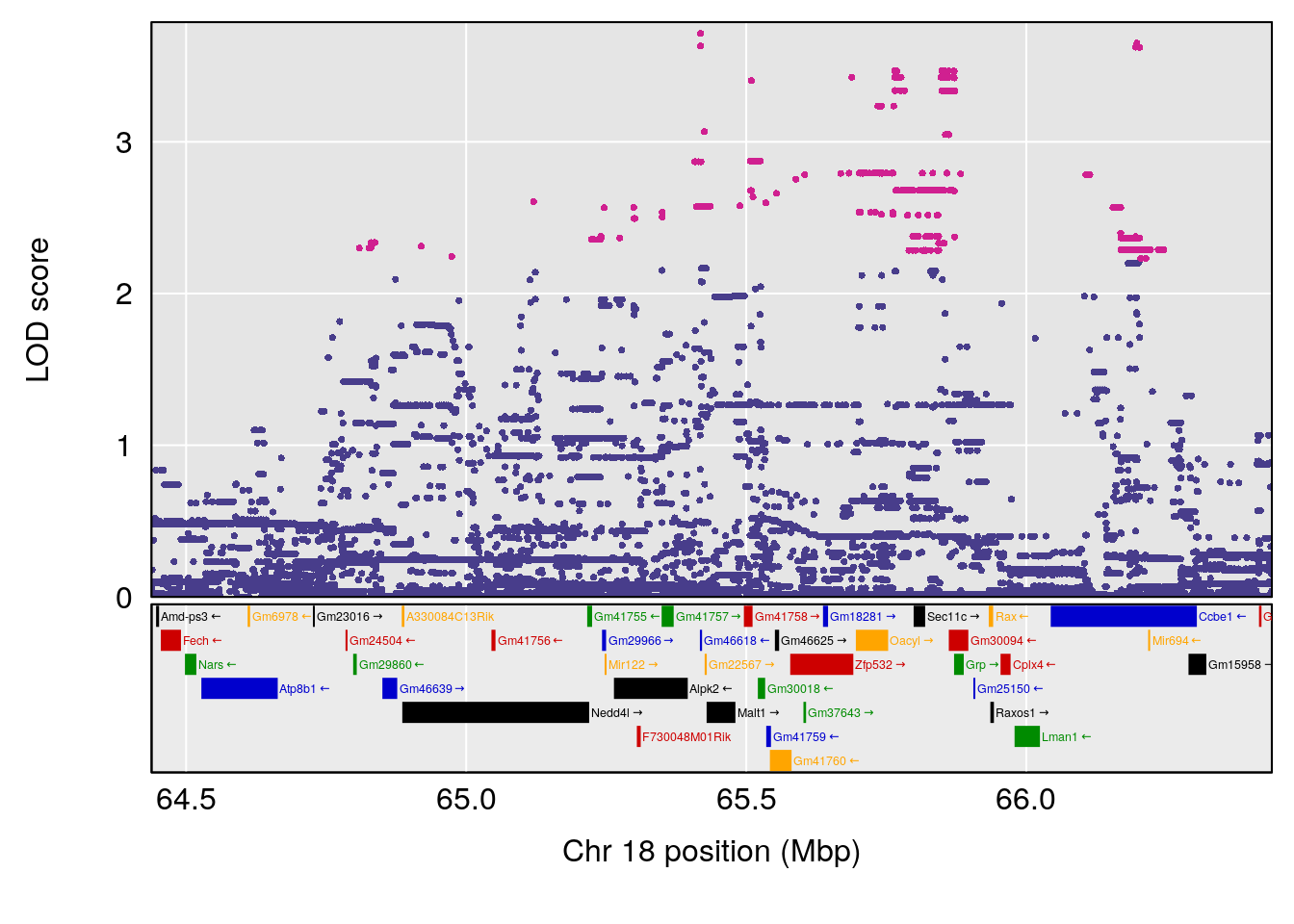
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "TimetoProjectedRecovery(Hr)"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 91.31201 6.270337 91.209902 152.86541
# 2 1 pheno1 6 126.01790 6.709756 125.696341 126.61414
# 3 1 pheno1 10 115.79817 6.081346 3.174515 115.81494
# 4 1 pheno1 11 35.91853 6.293340 35.844802 35.99807
# 5 1 pheno1 18 65.43490 7.710831 65.313821 65.52767
# [1] 1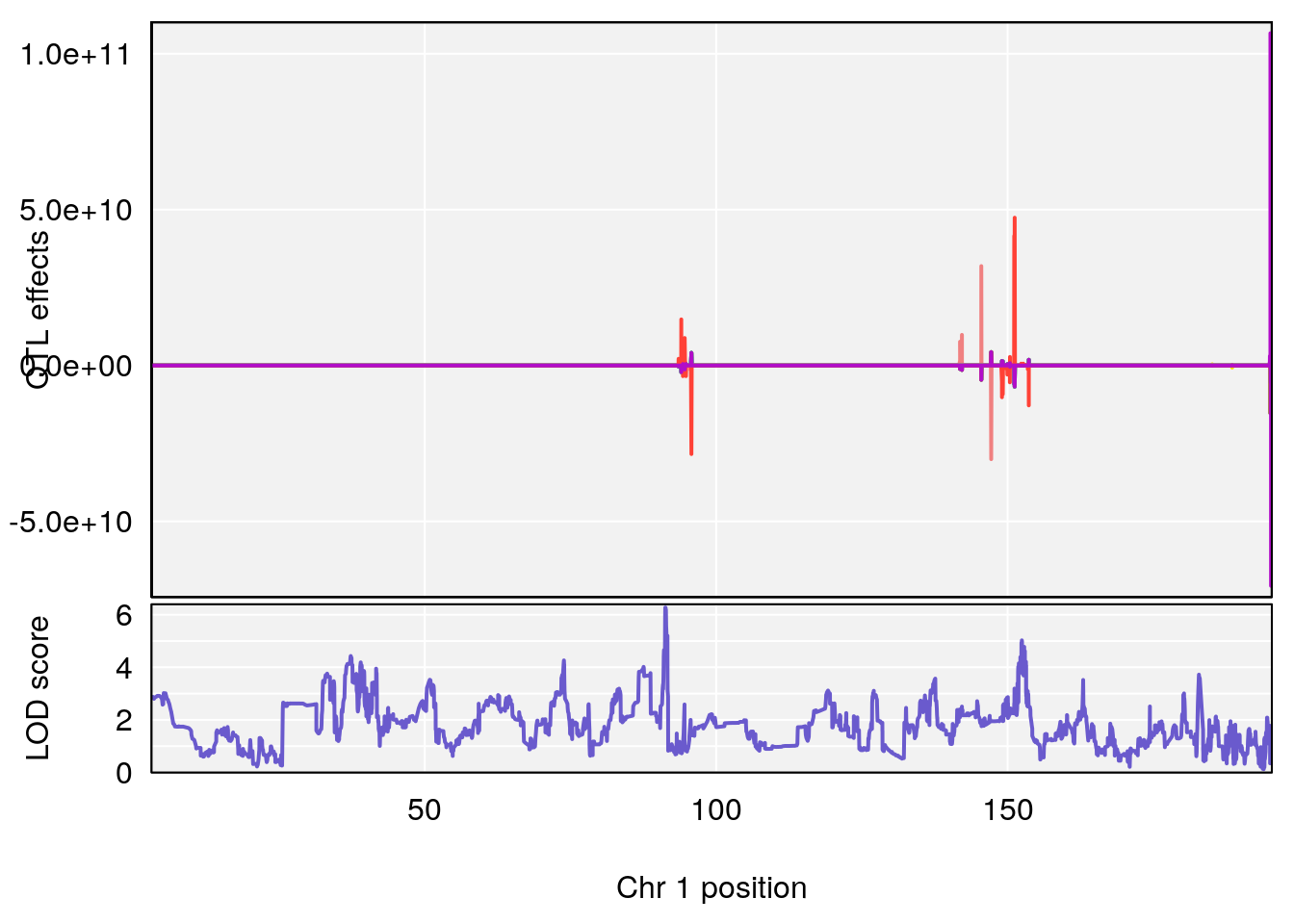
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
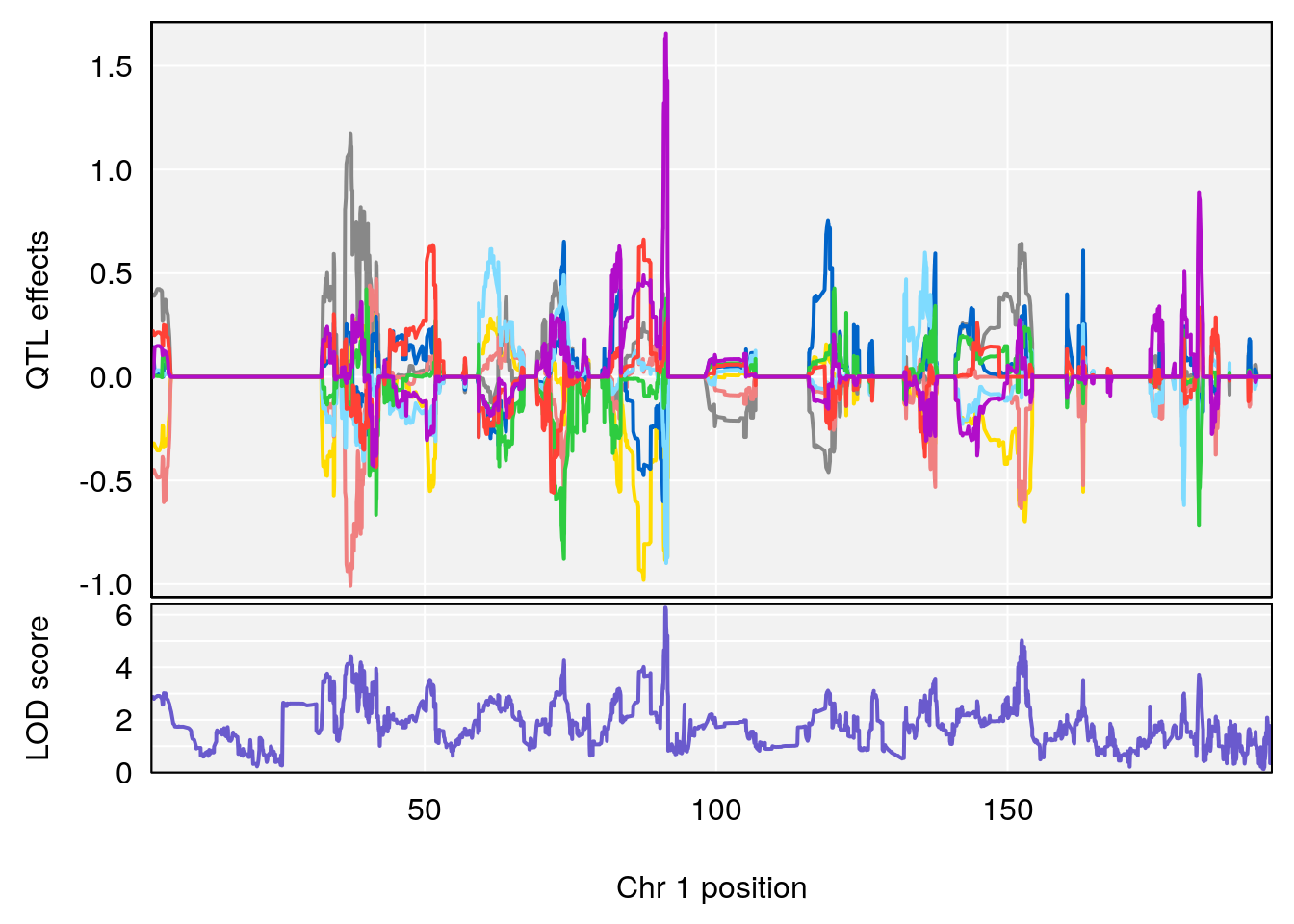
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
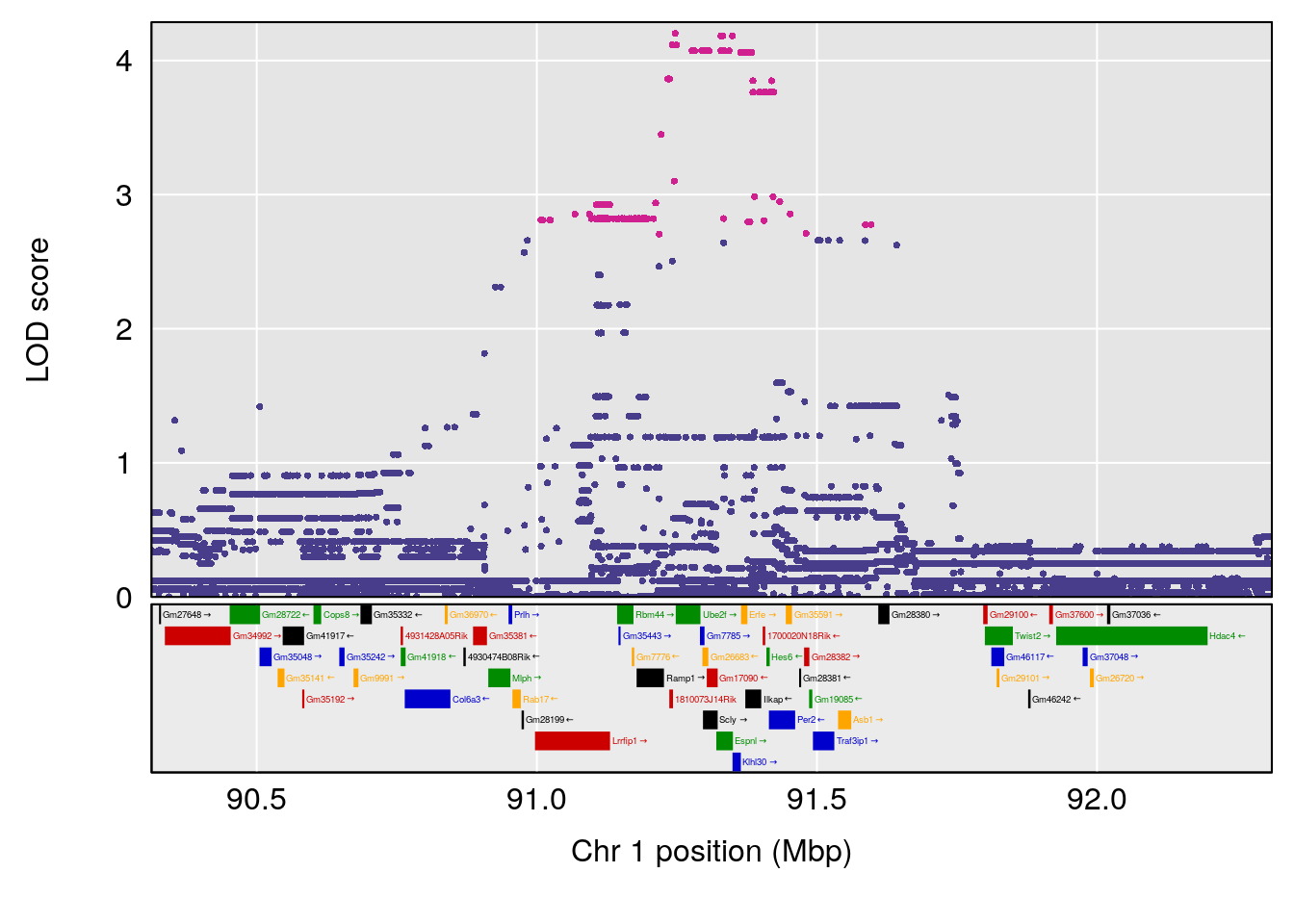
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
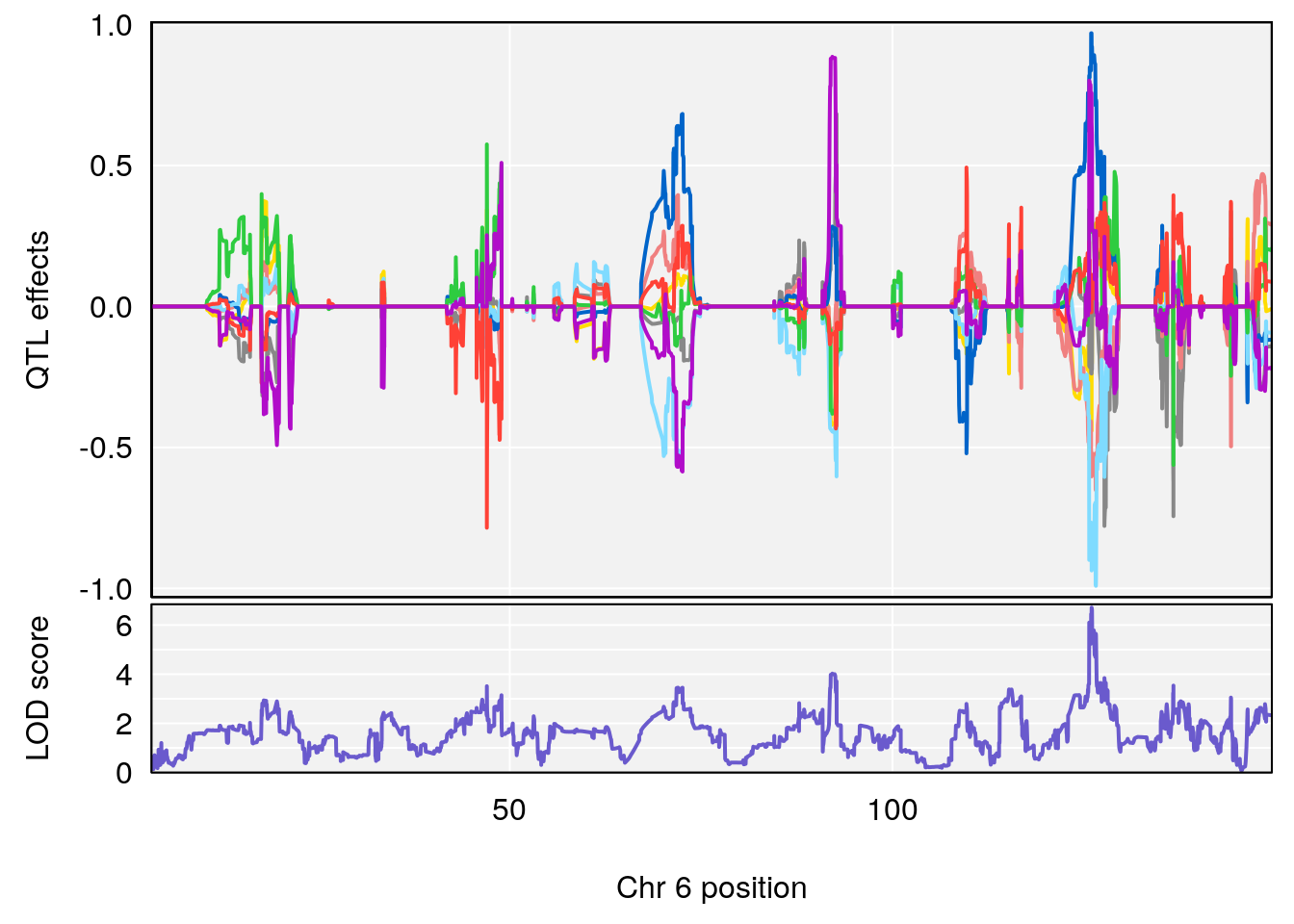
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
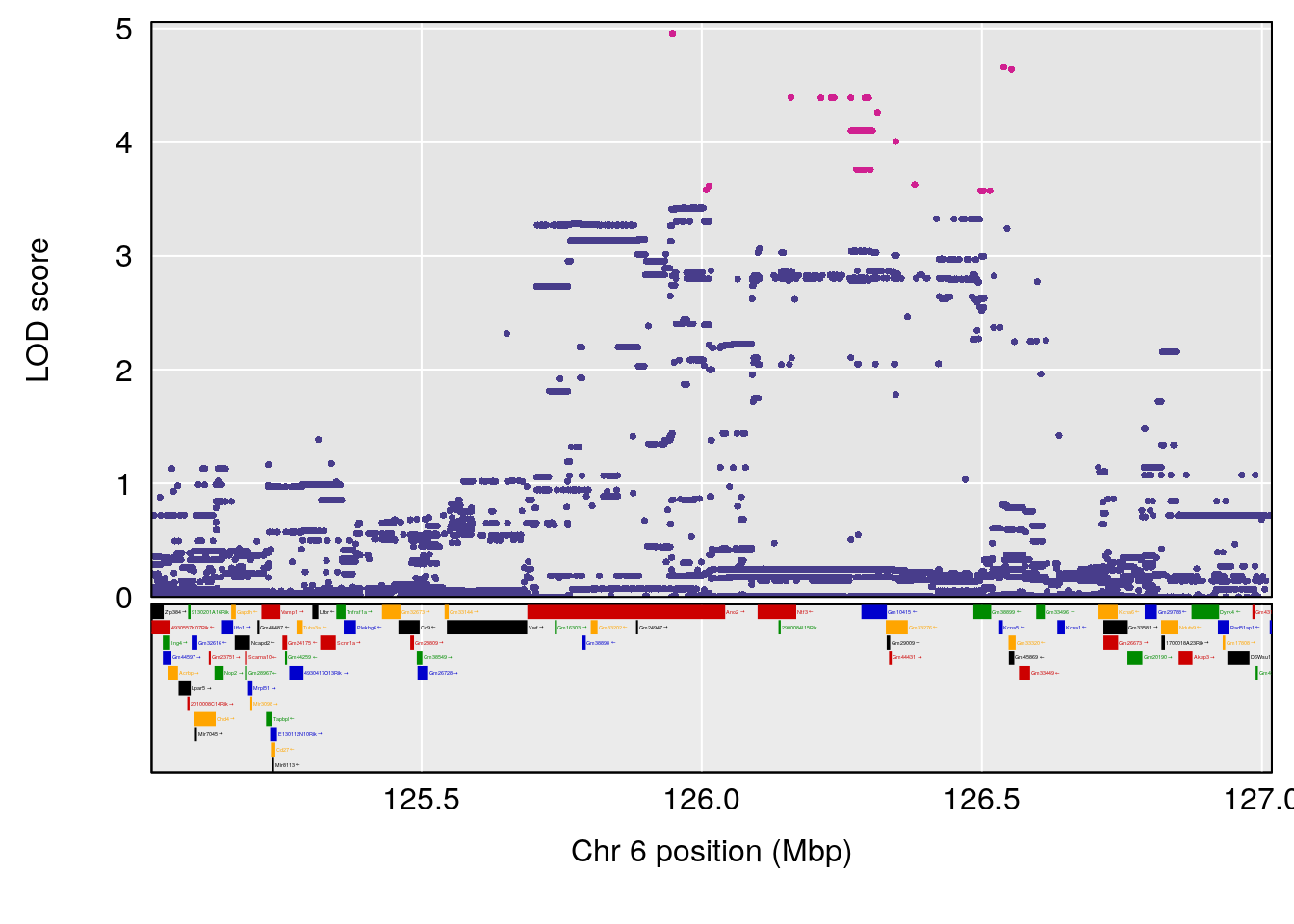
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 3
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
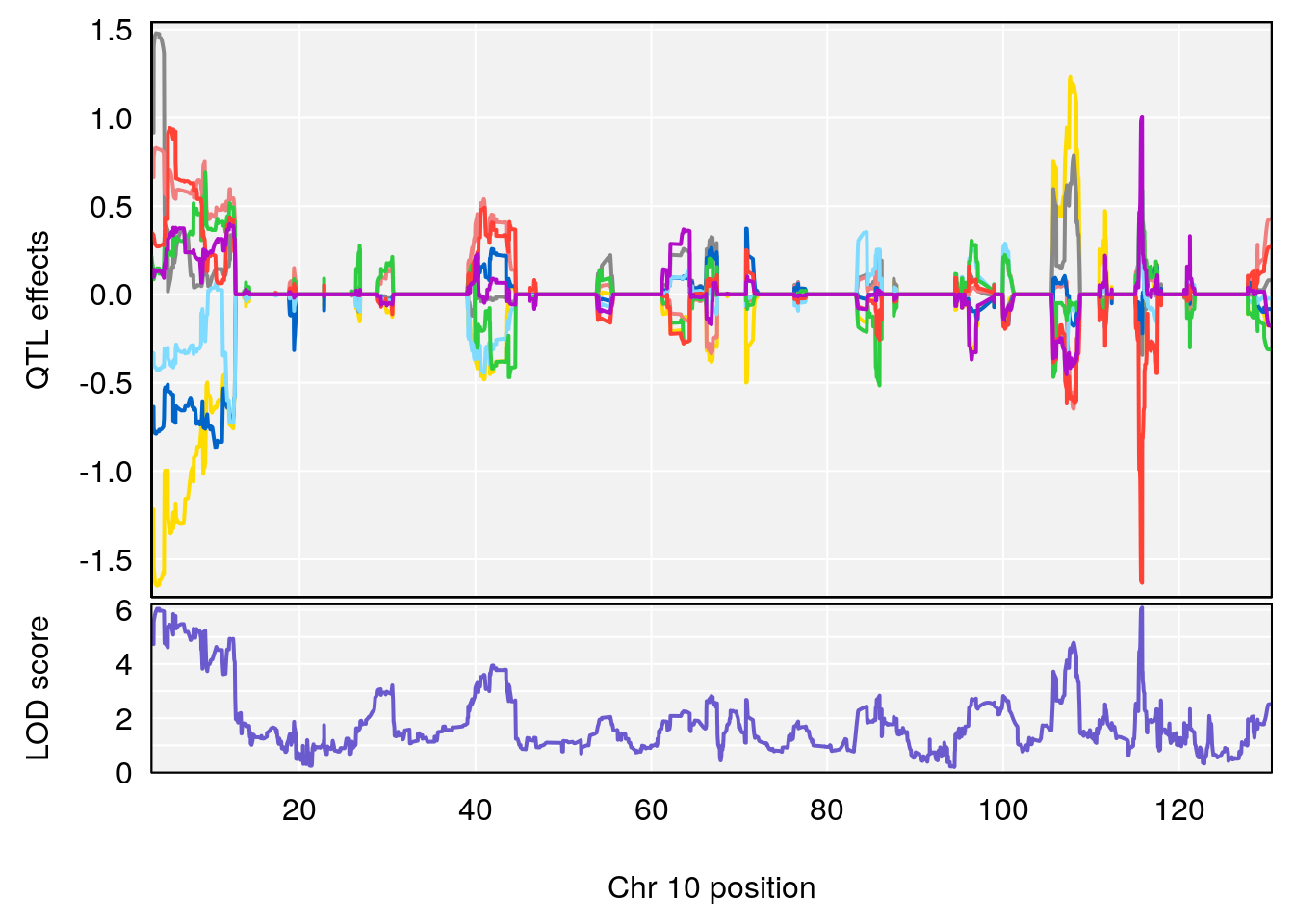
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
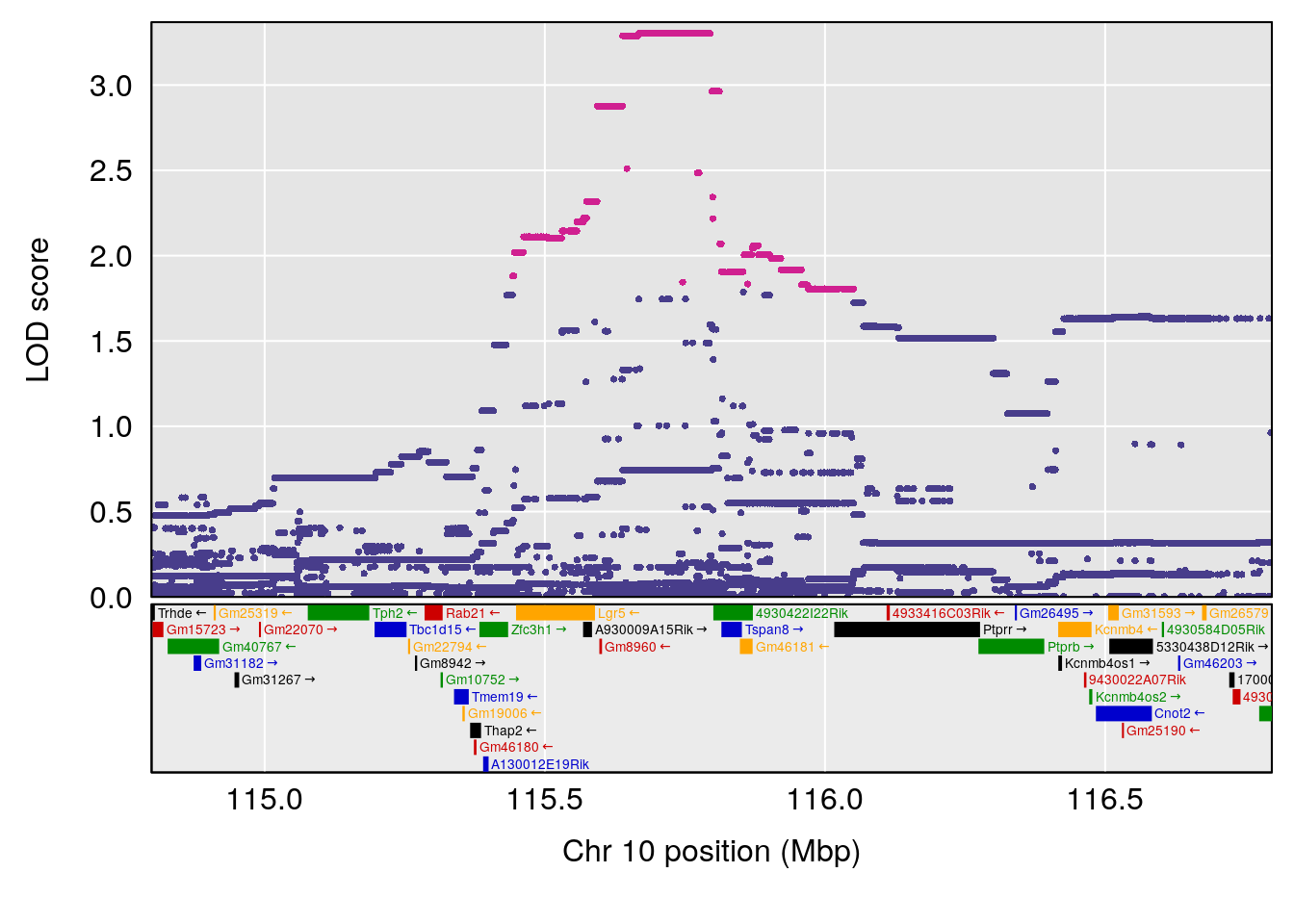
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 4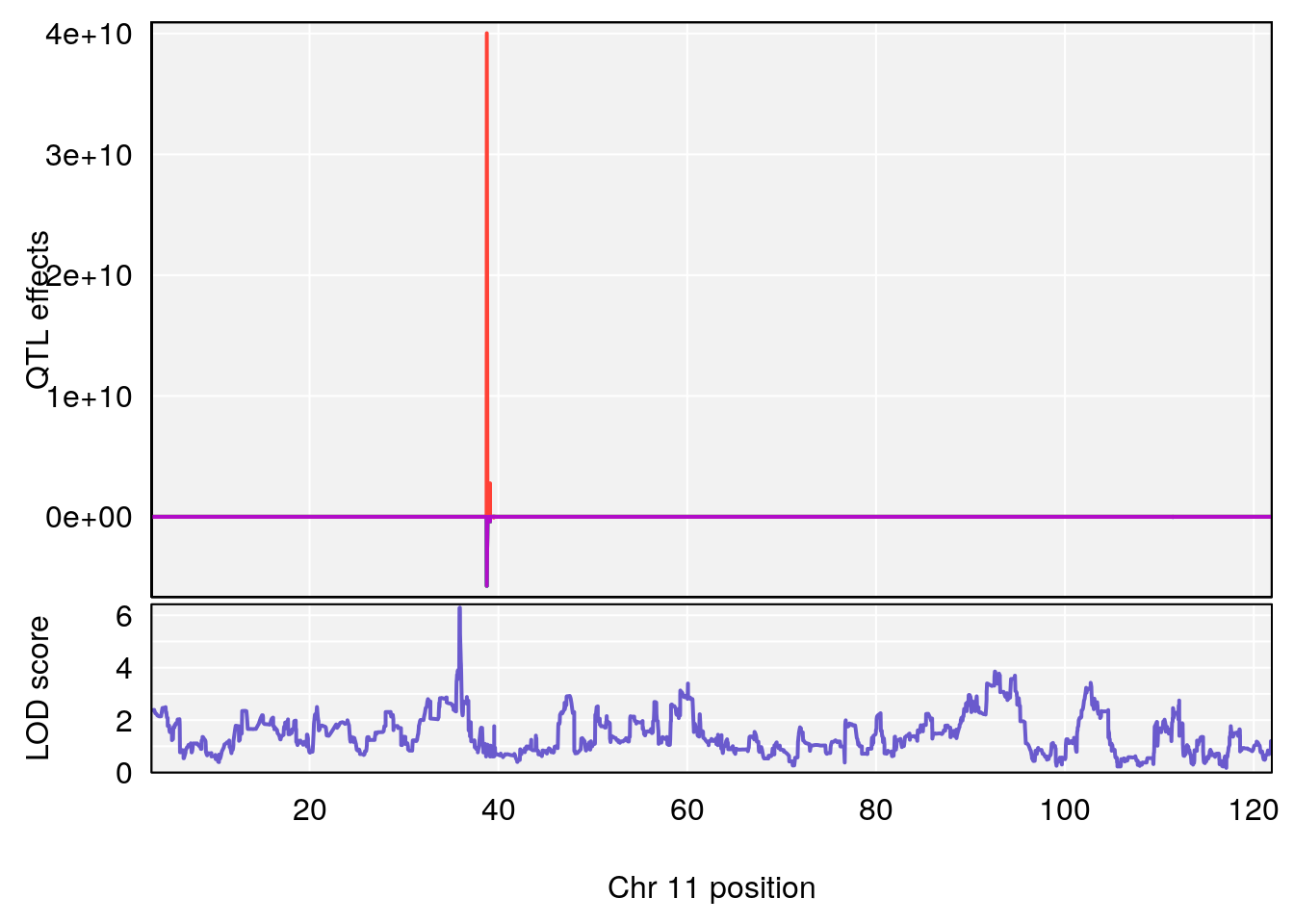
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
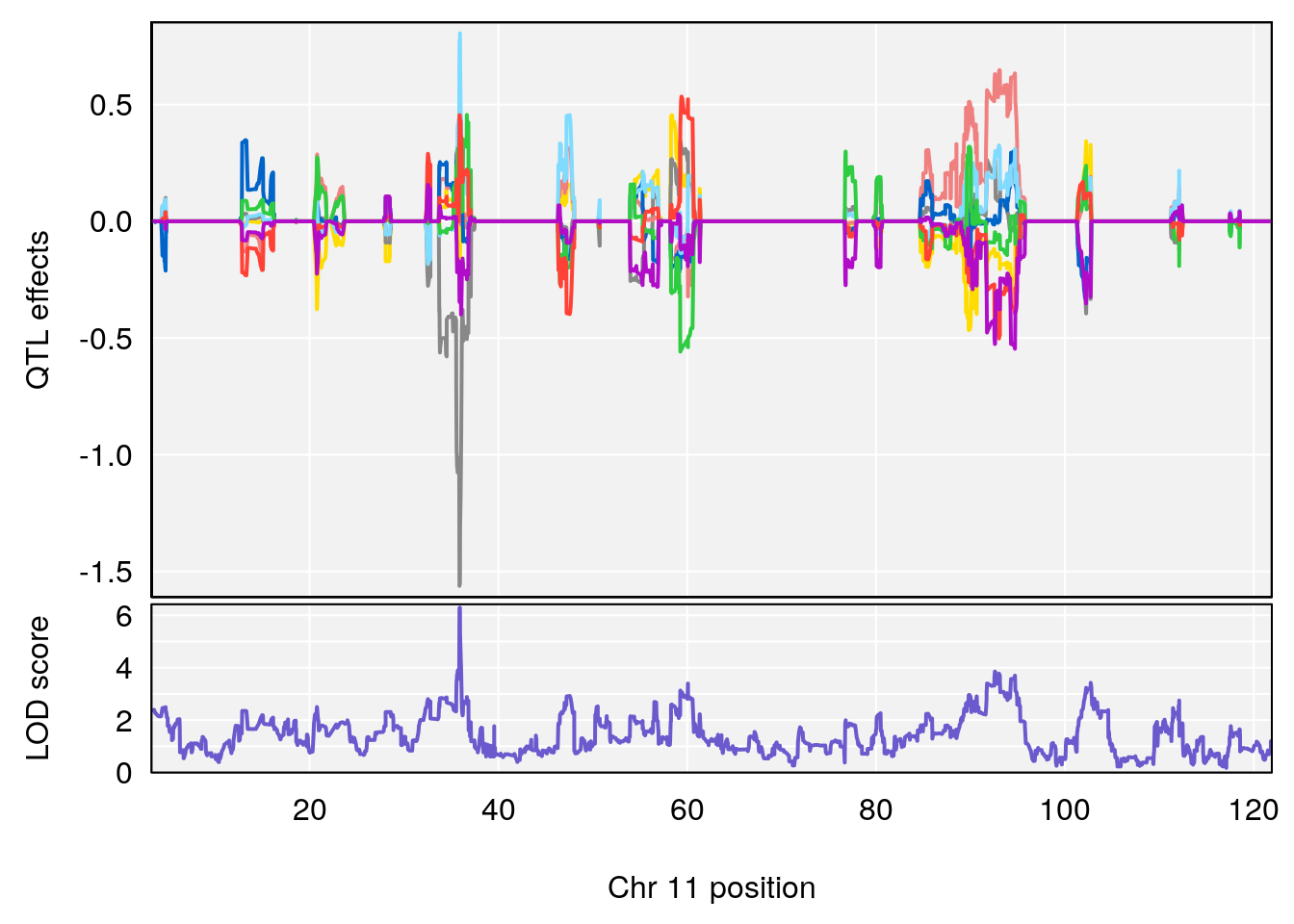
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
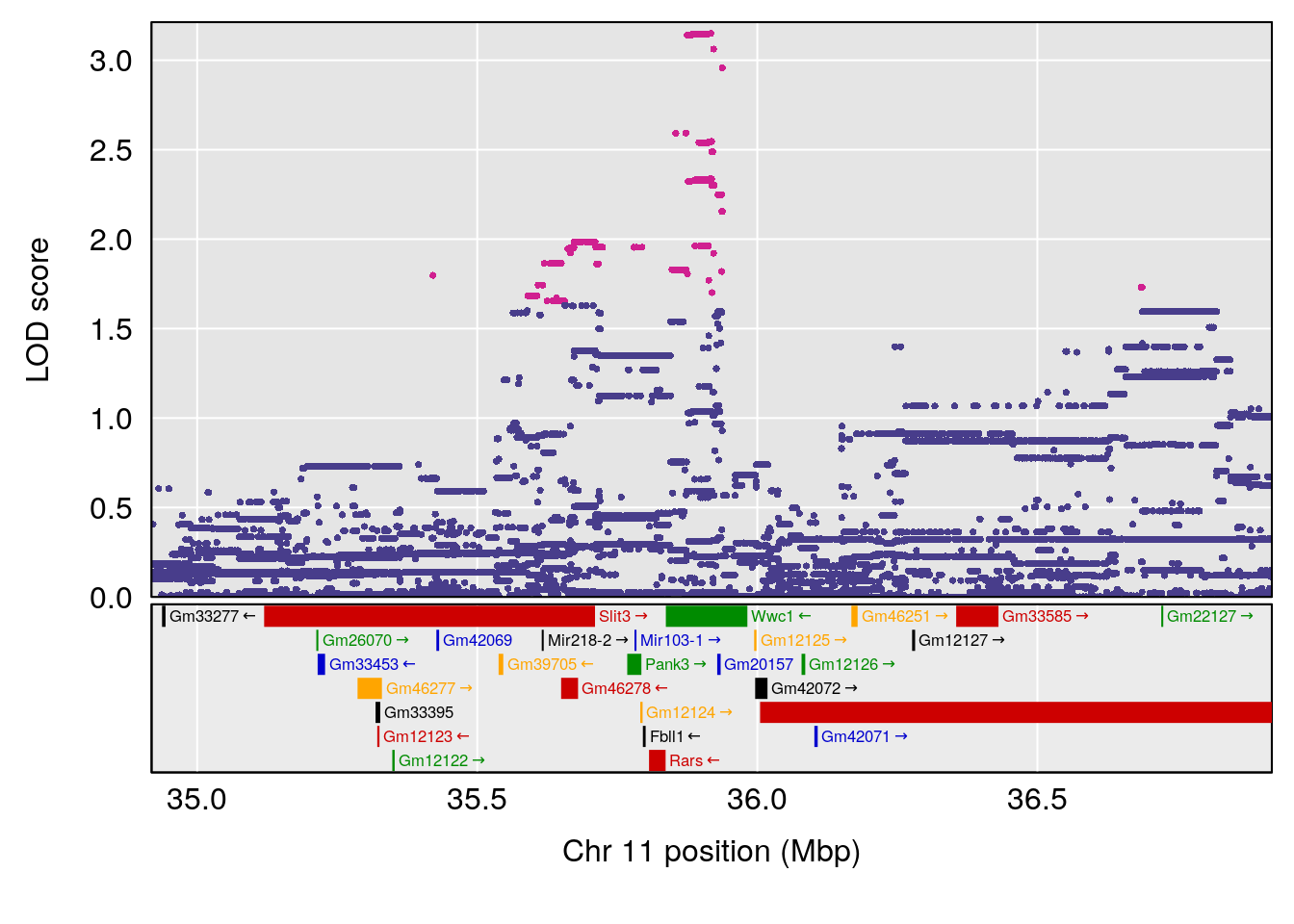
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 5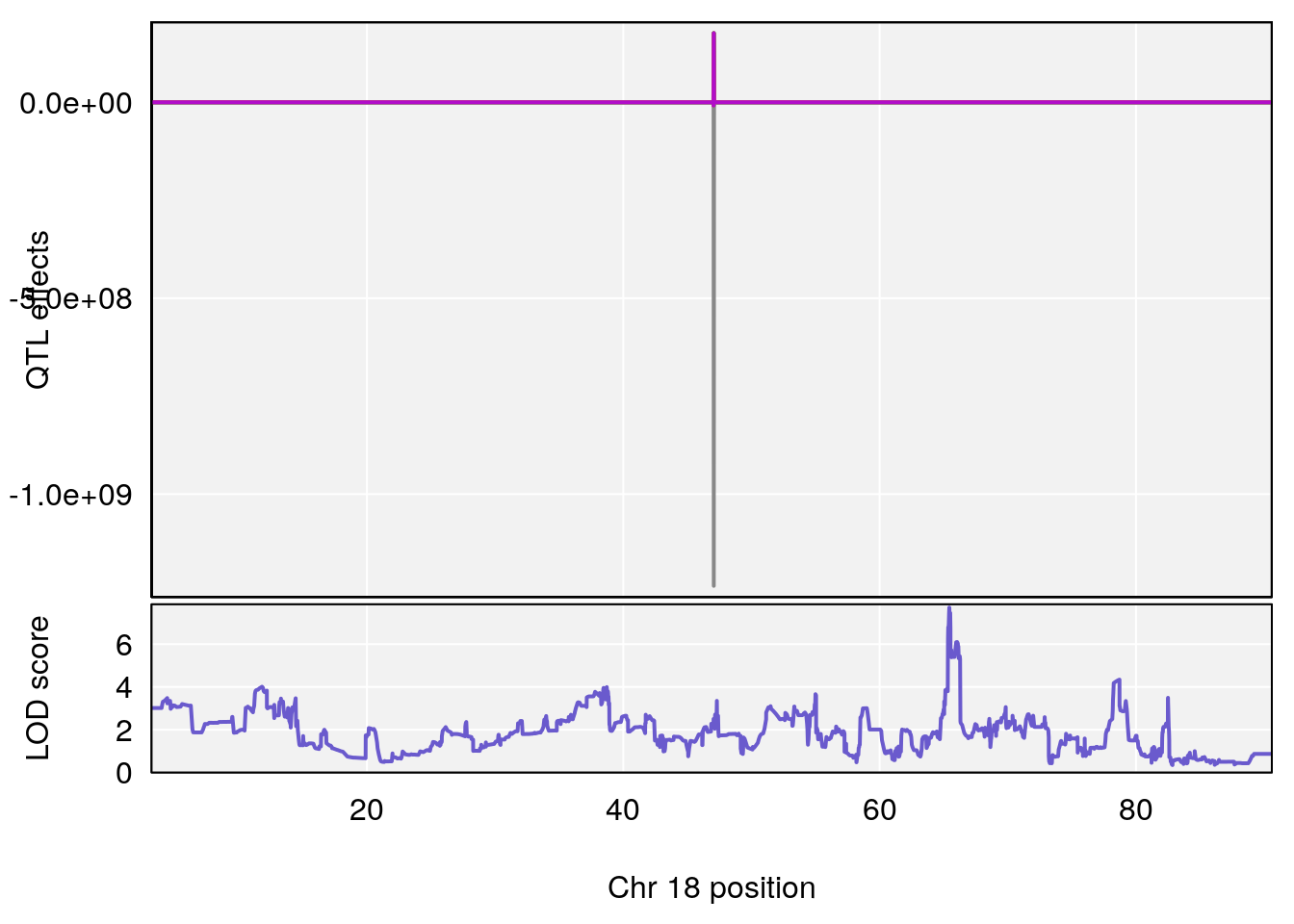
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
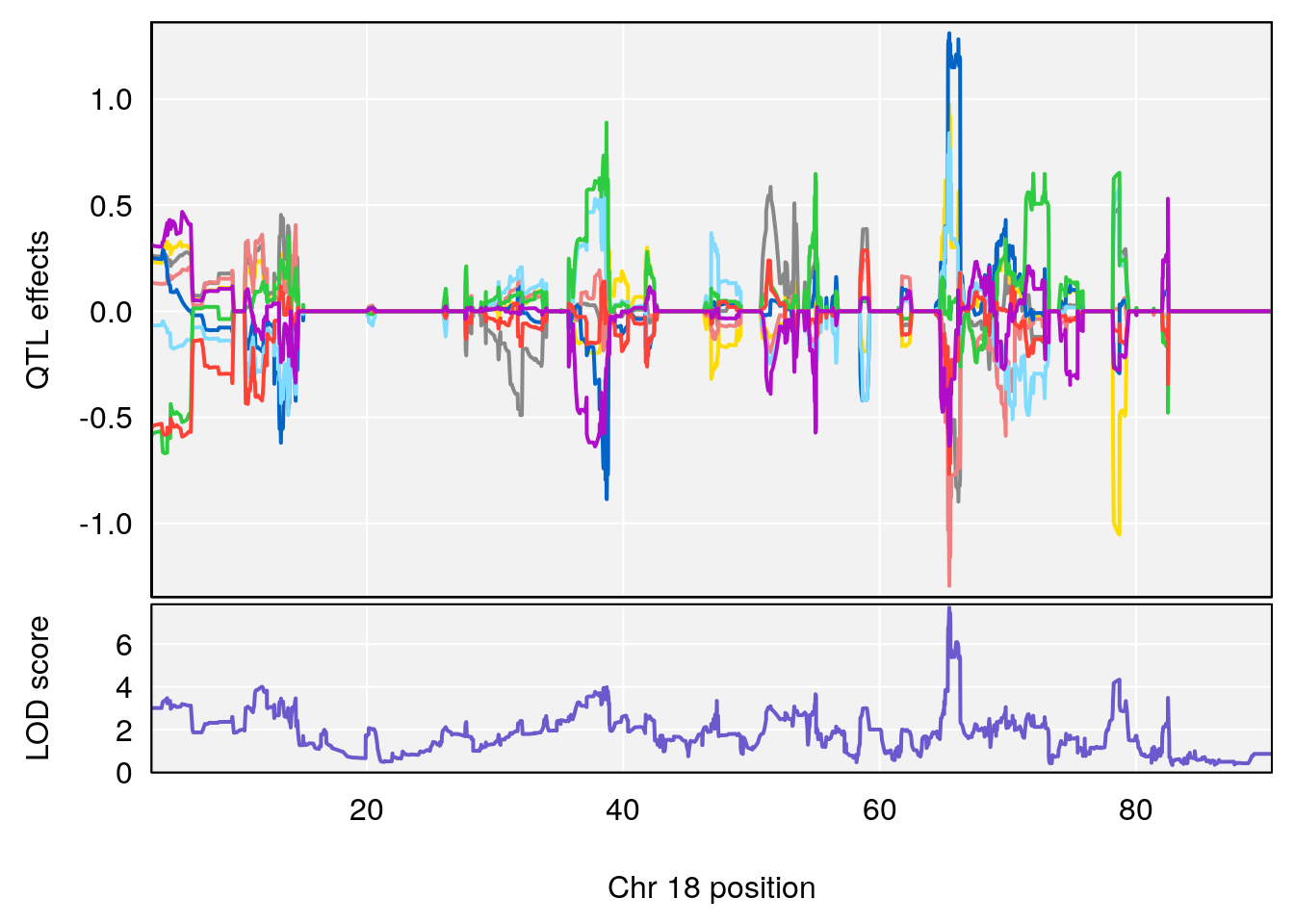
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
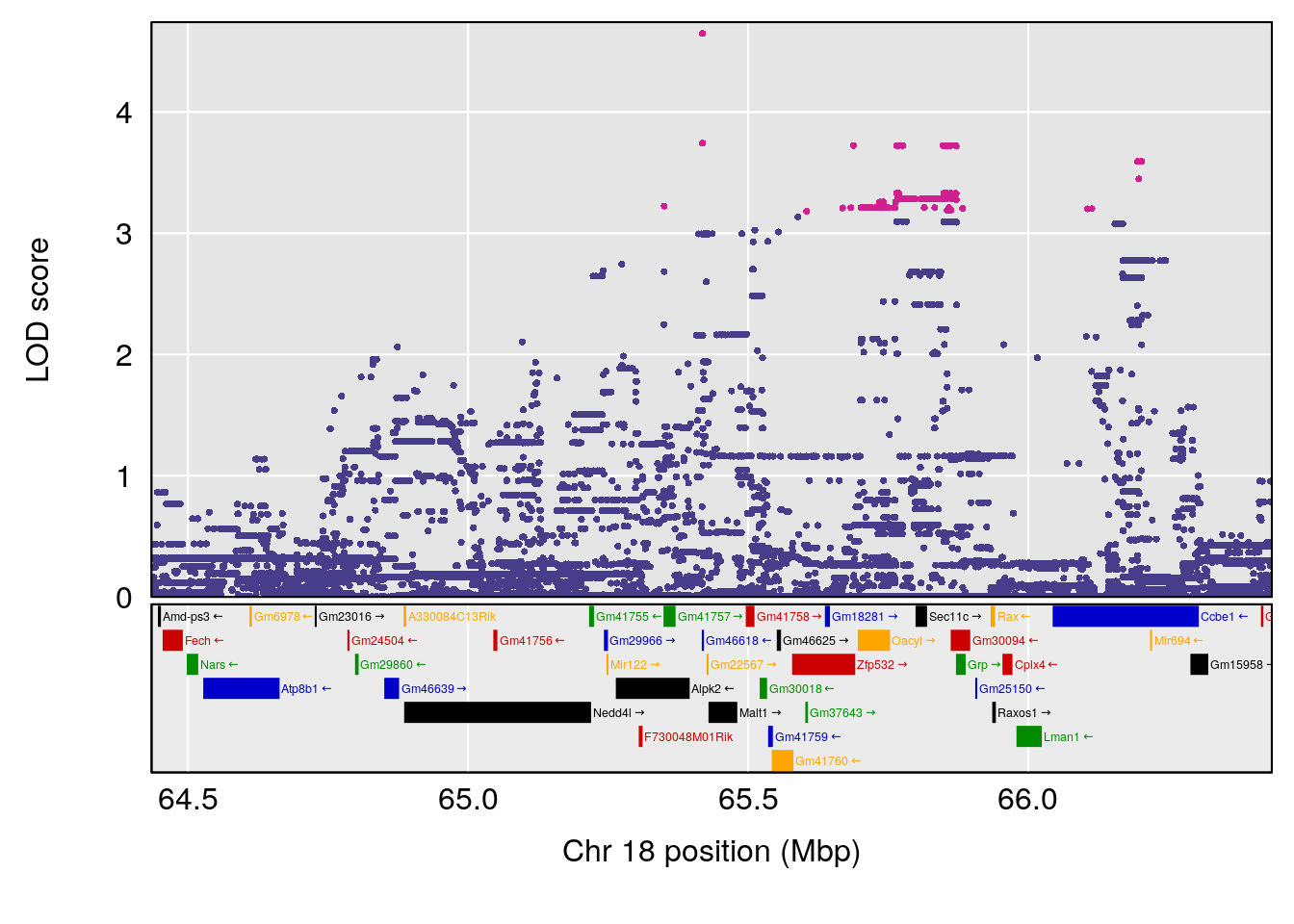
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "StartofRecovery(Hr)"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 10 5.635019 6.852134 3.400722 115.81149
# 2 1 pheno1 18 65.434900 6.749689 65.313821 65.52767
# [1] 1
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
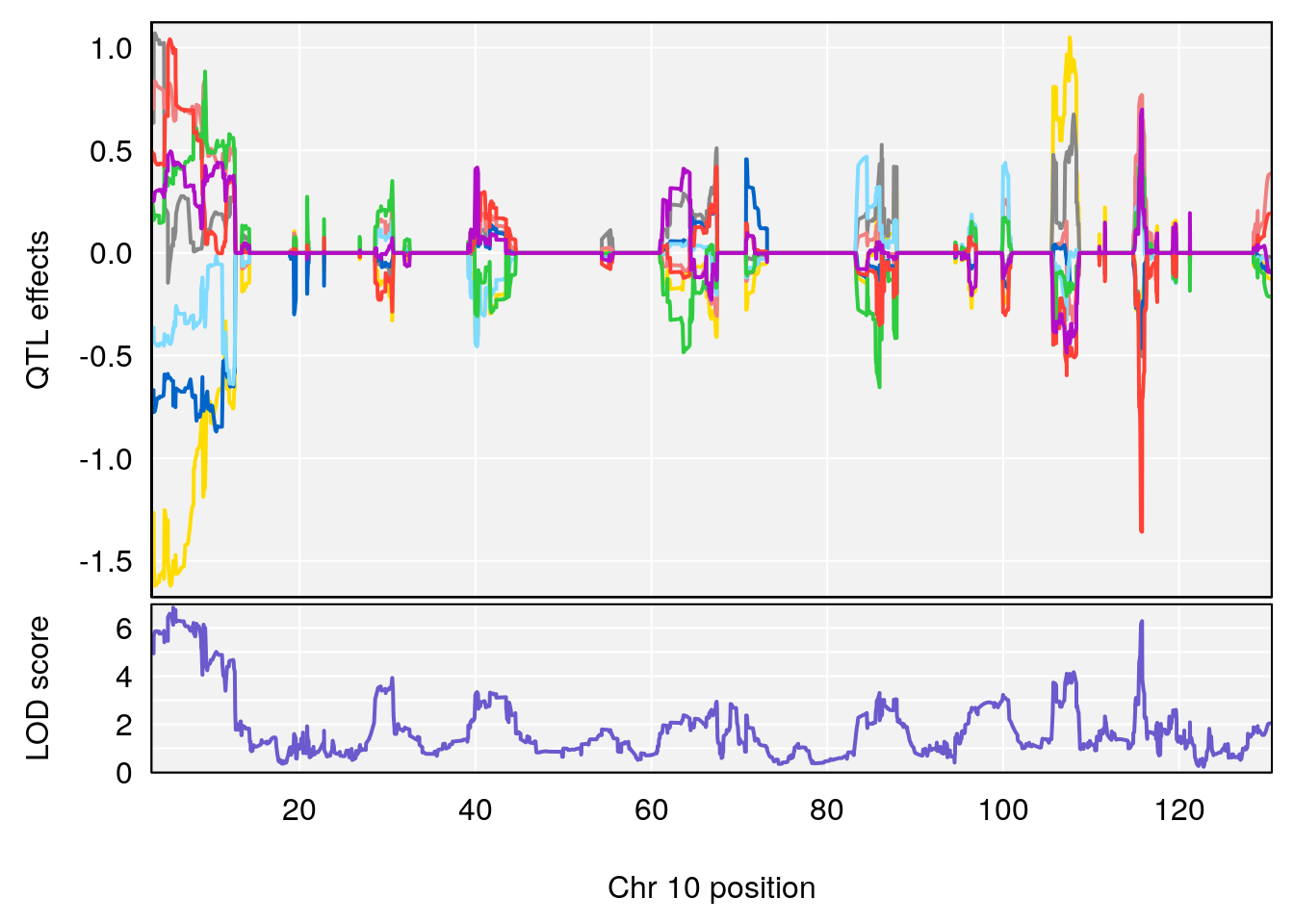
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
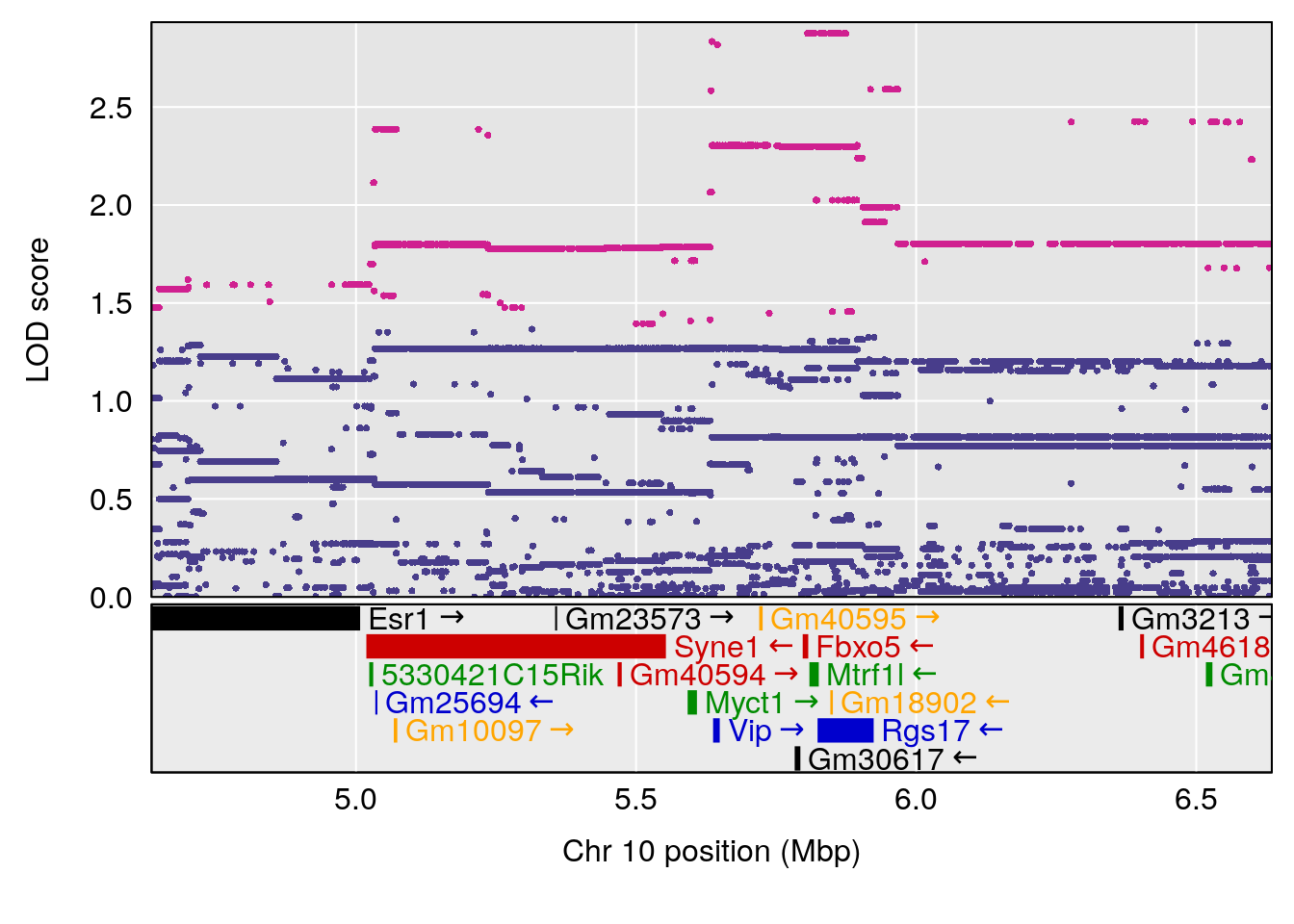
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
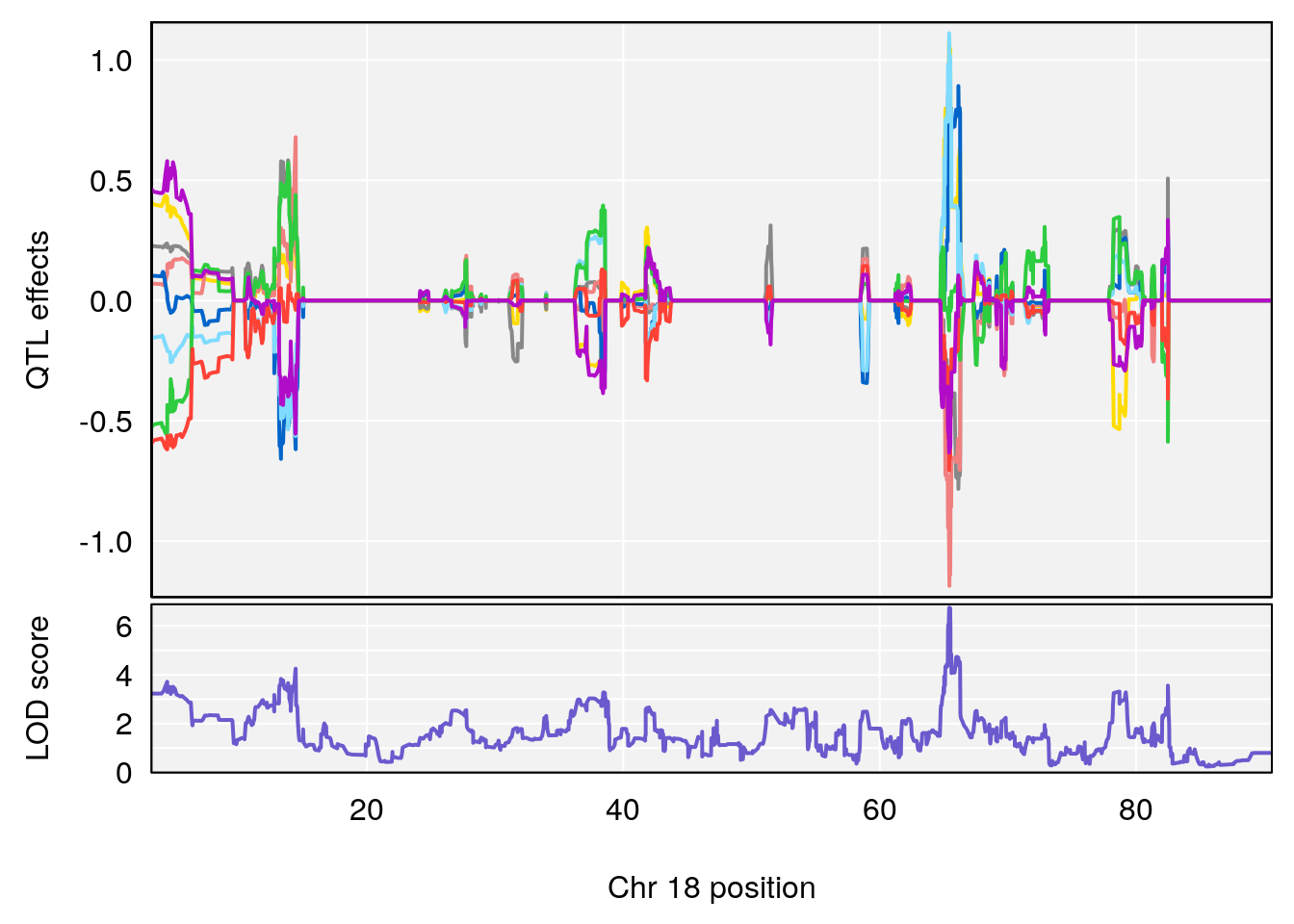
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
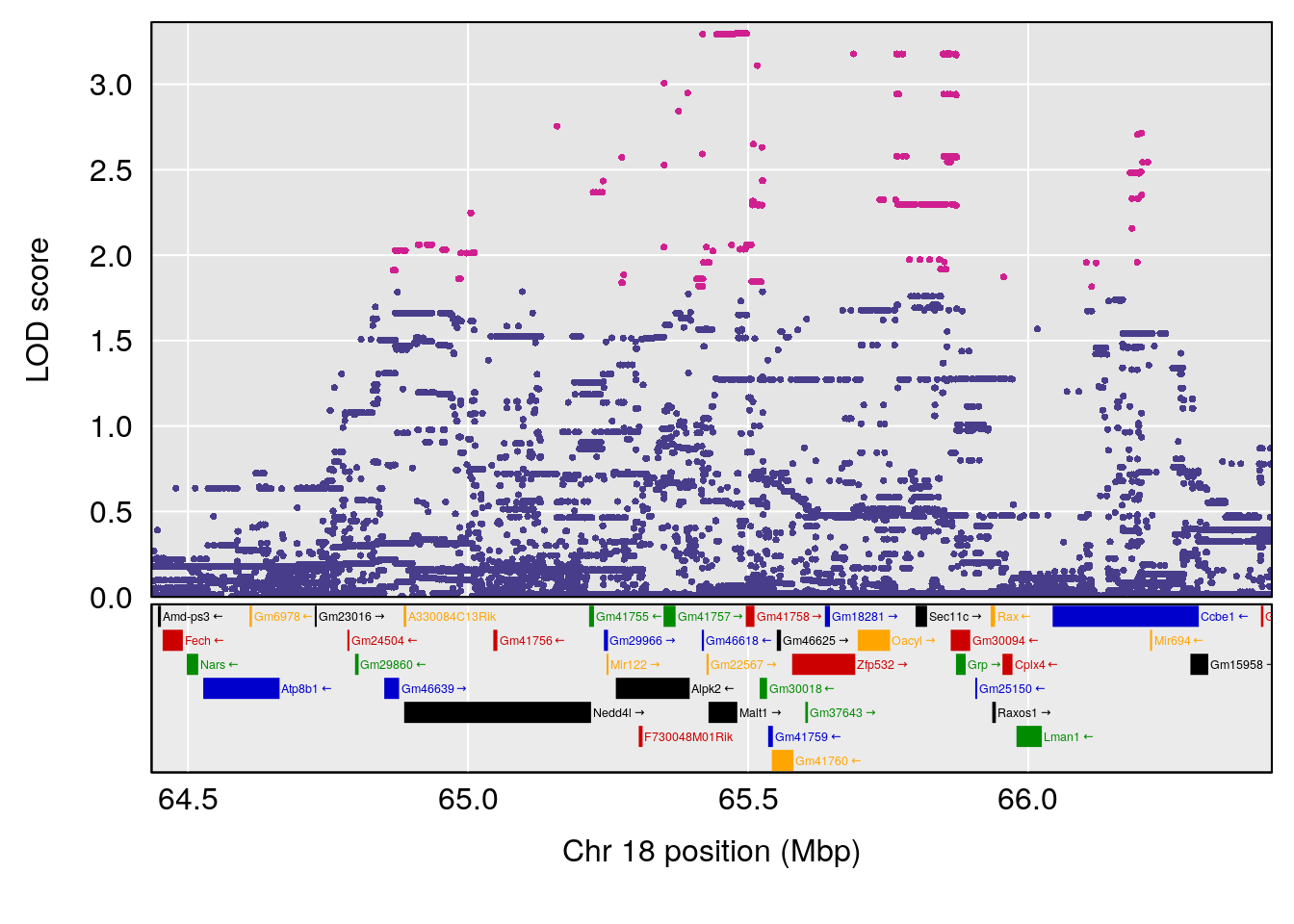
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "RRRecoveryRate(%/Hr)SLOPE"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 4 152.69816 6.100750 68.40926 152.83582
# 2 1 pheno1 5 13.65223 6.943854 13.42743 54.23998
# 3 1 pheno1 9 122.67218 7.020565 122.31448 123.93203
# 4 1 pheno1 12 113.10511 6.093534 112.53083 113.14986
# 5 1 pheno1 16 33.09290 7.445865 32.54616 92.27653
# [1] 1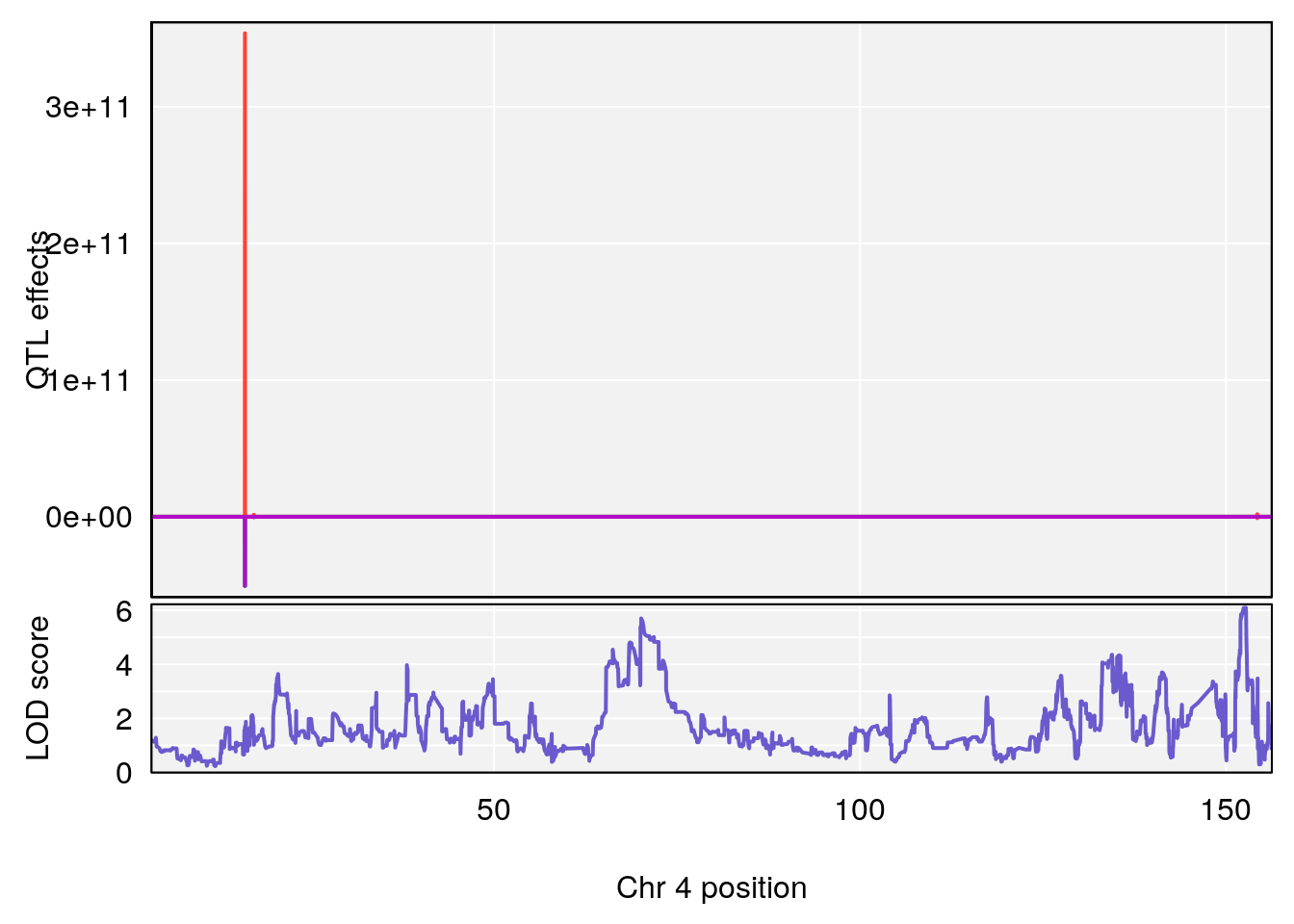
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
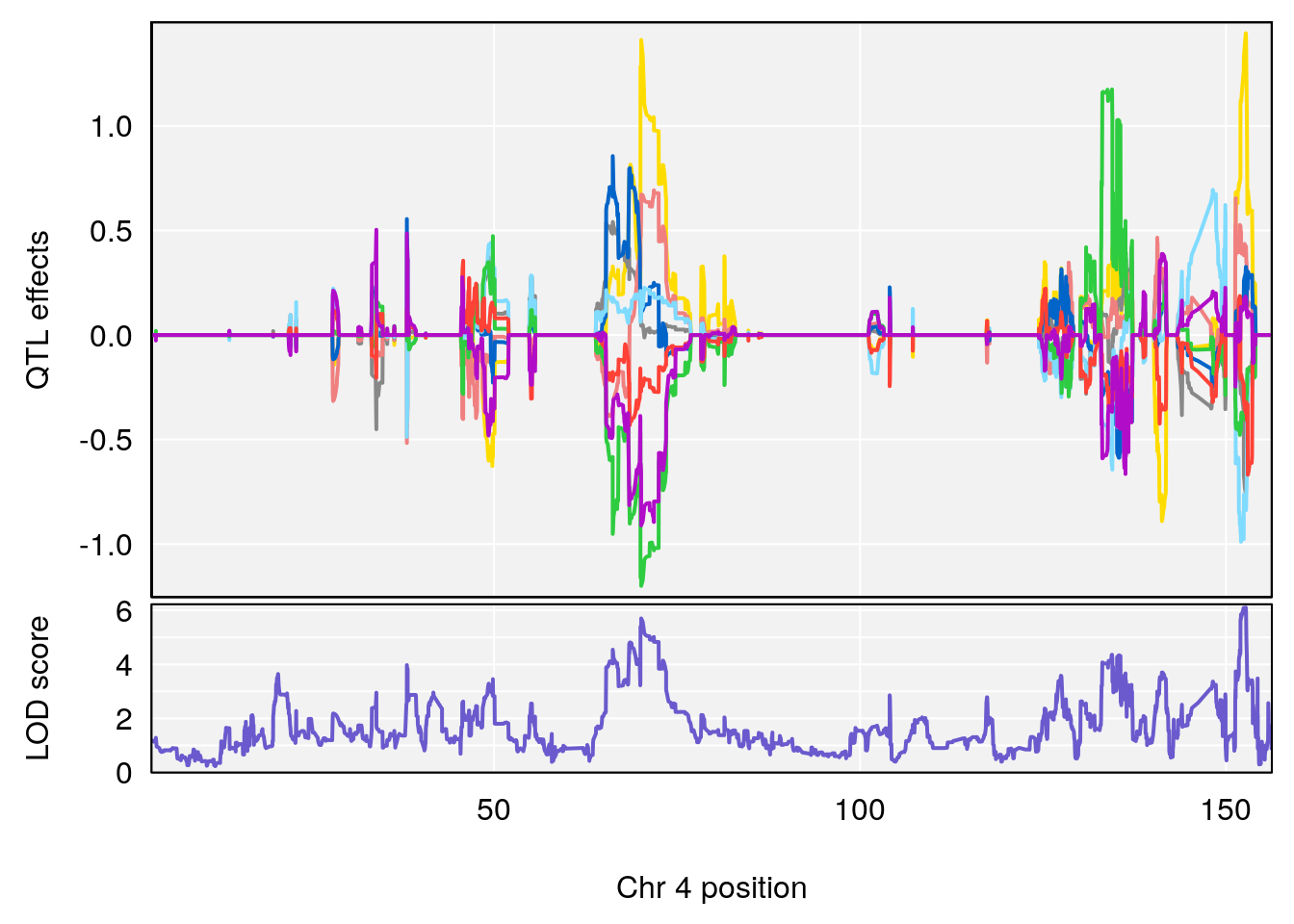
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
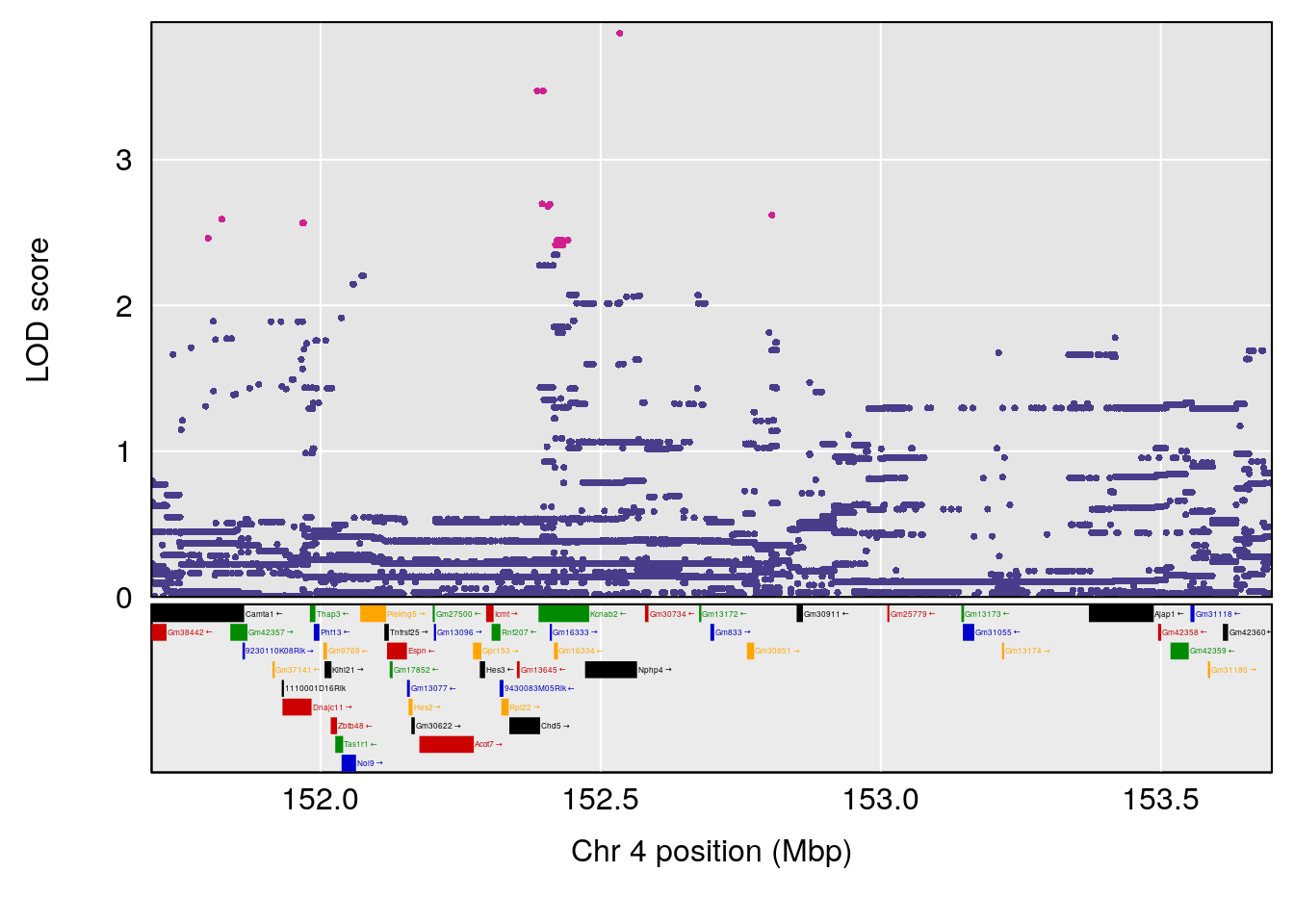
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2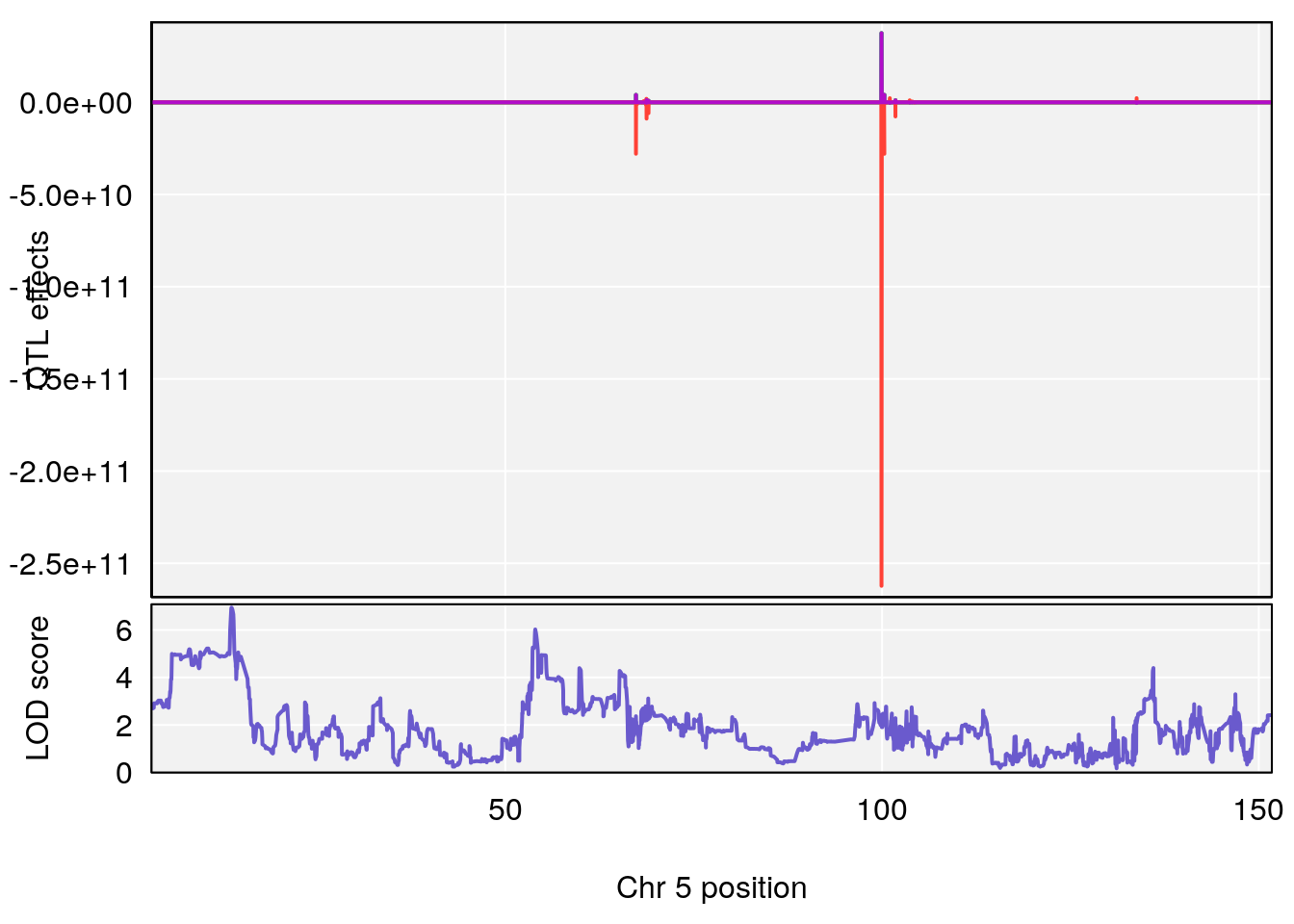
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
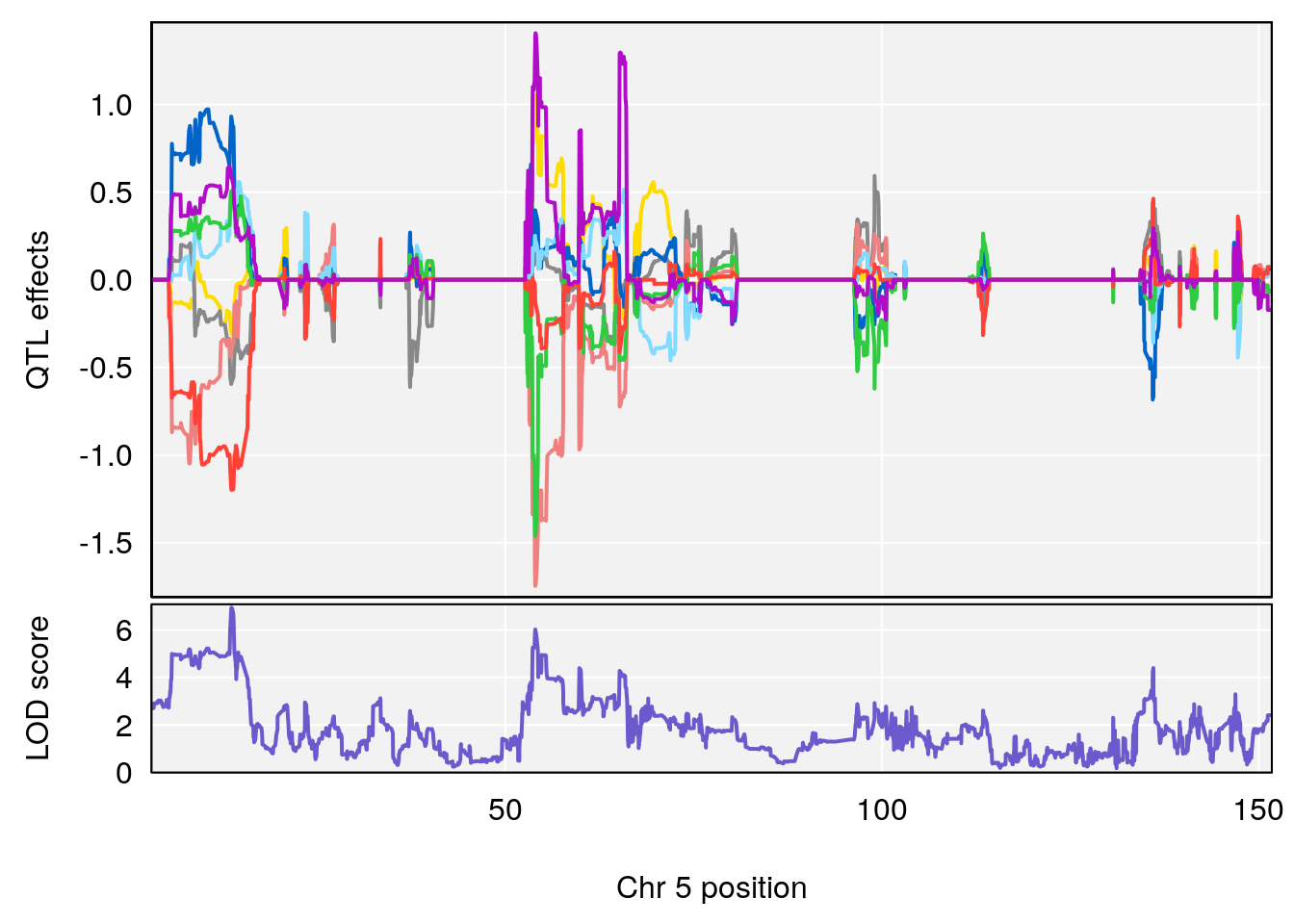
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
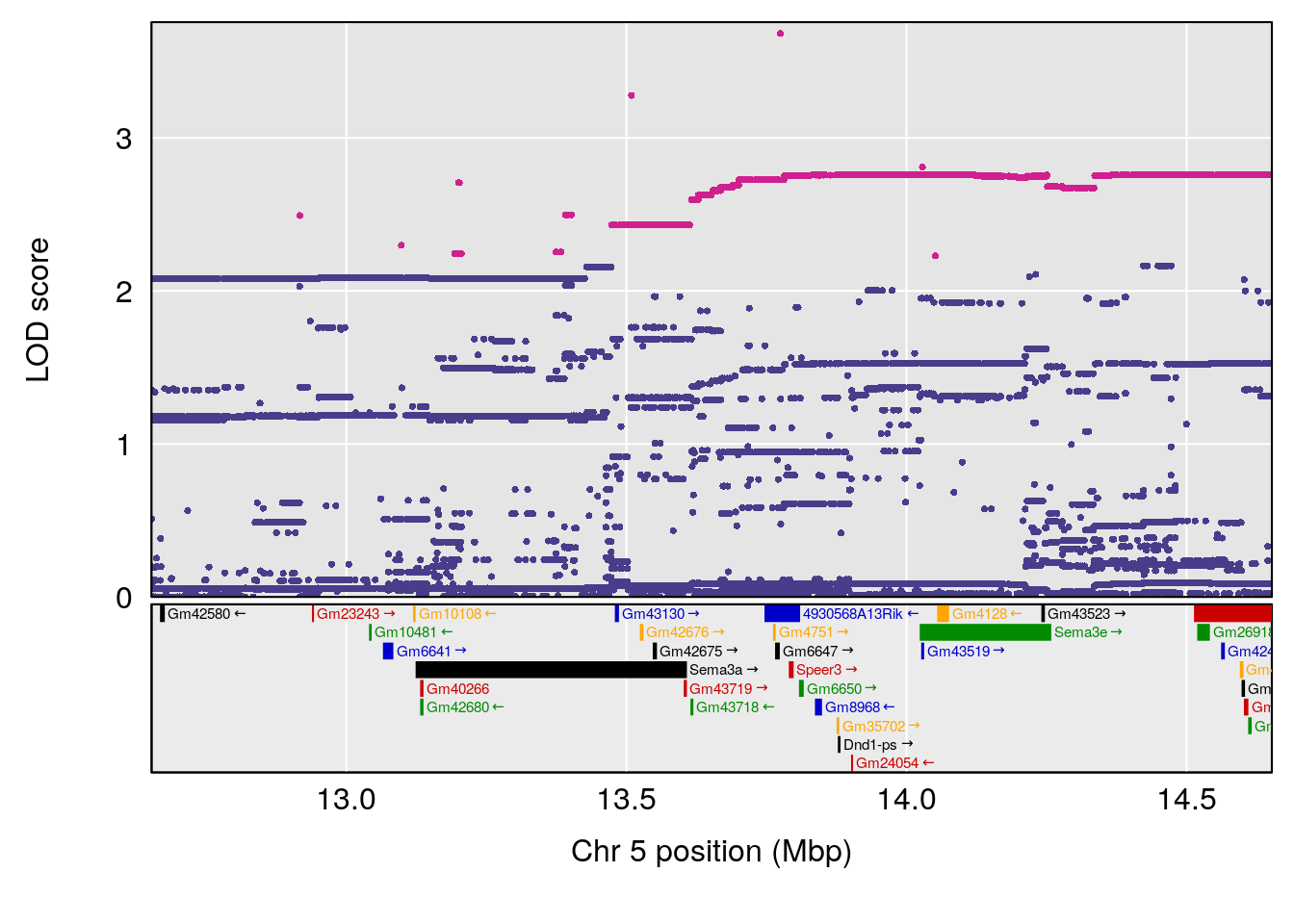
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 3
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
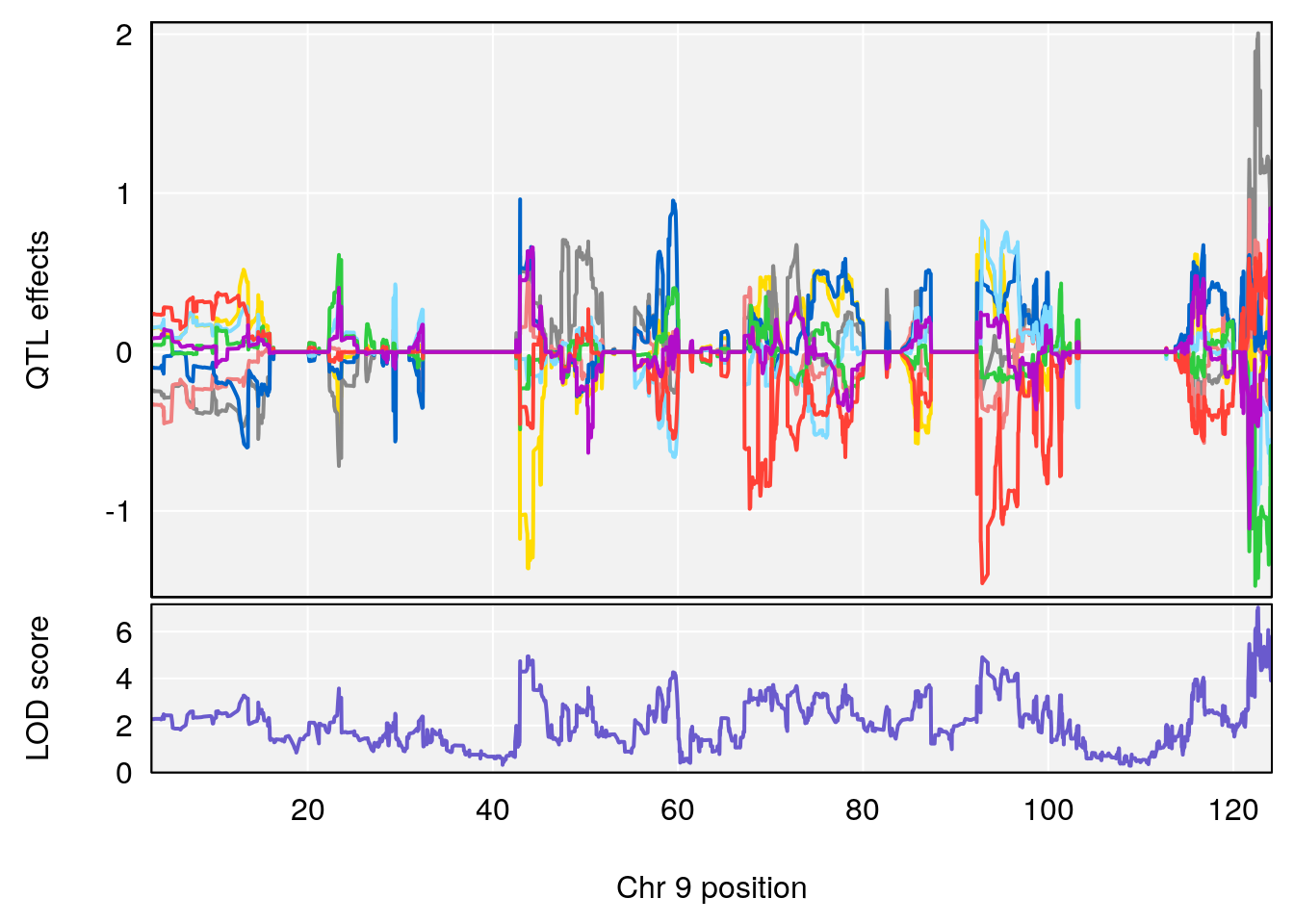
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
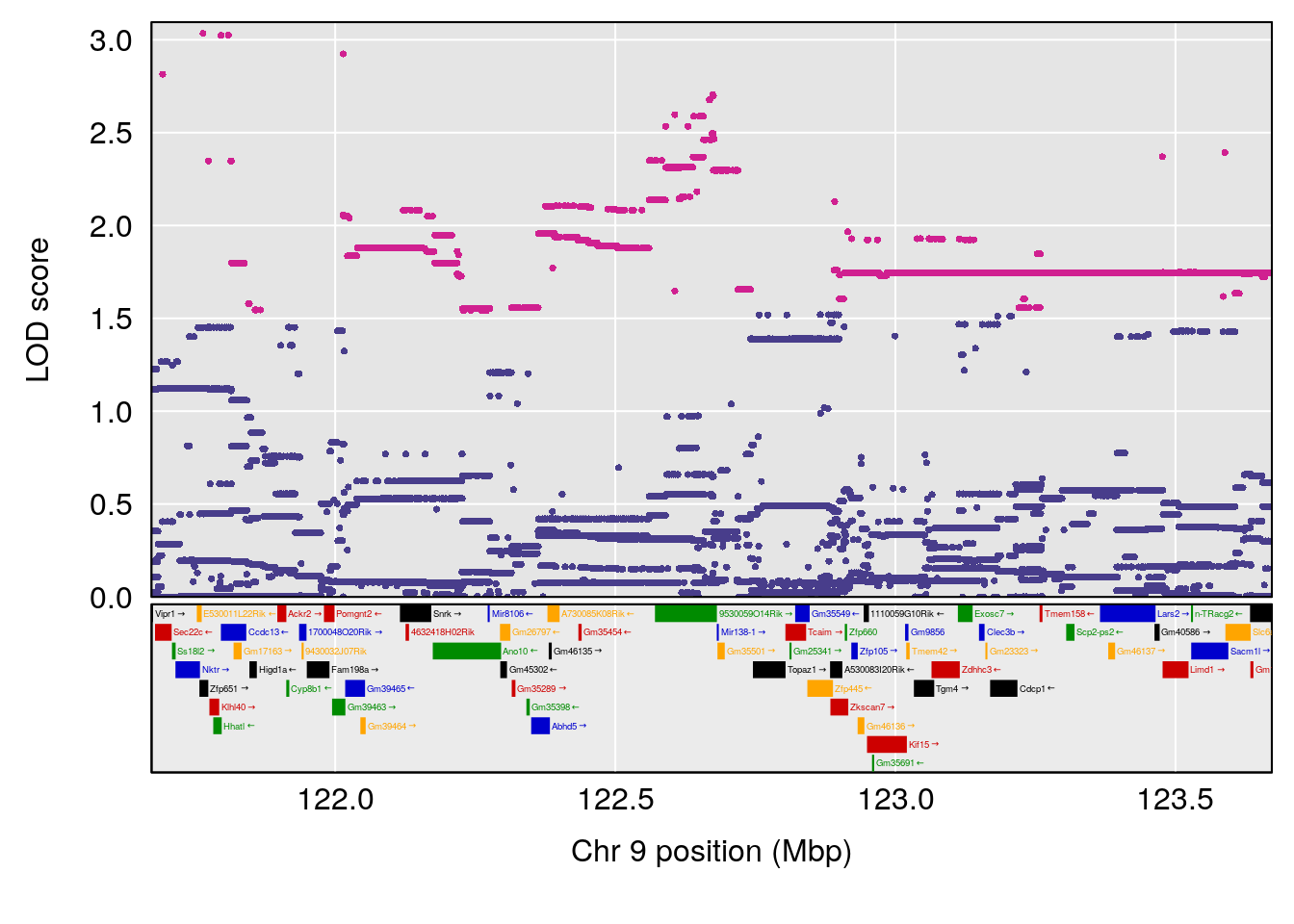
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 4
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
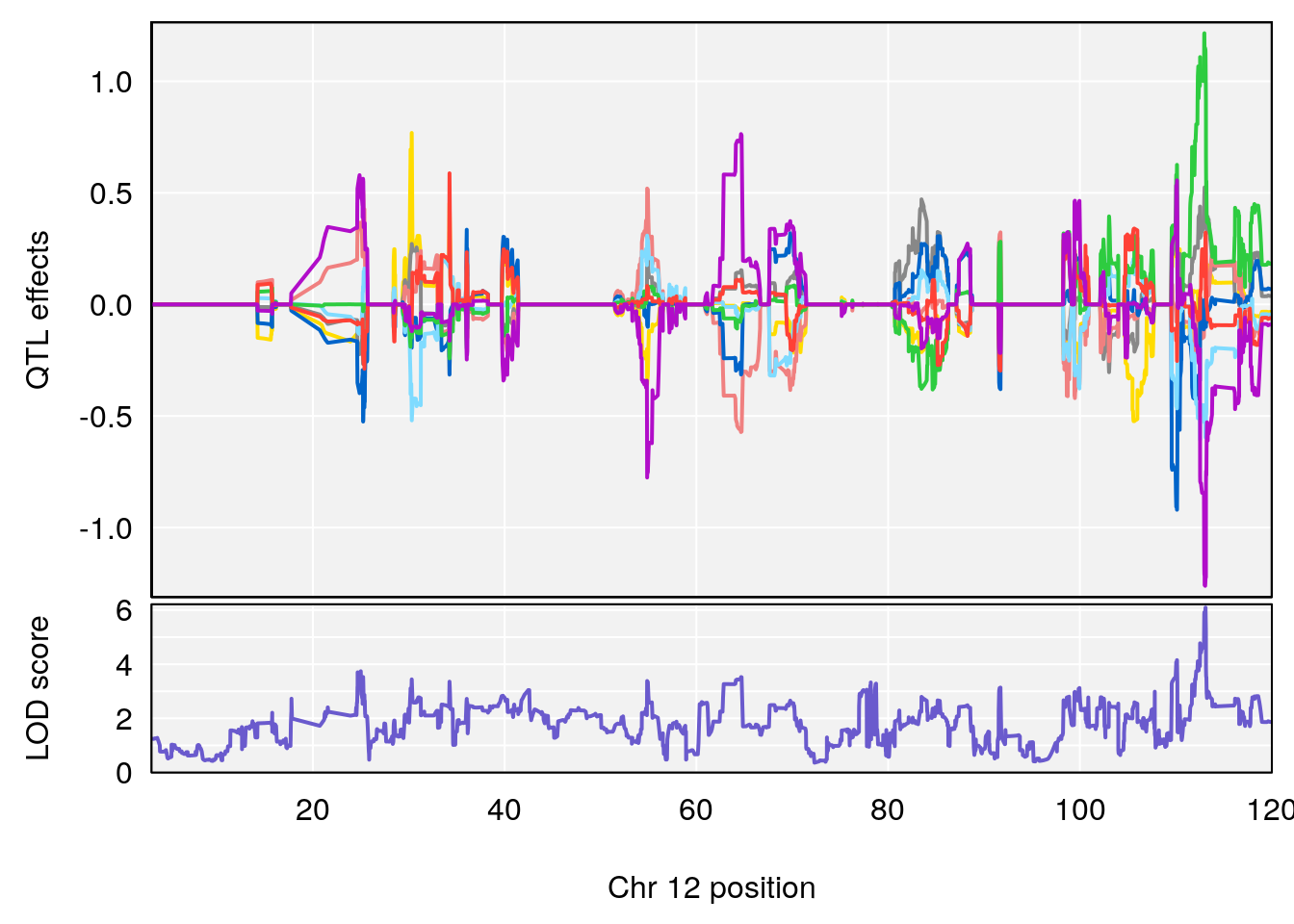
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
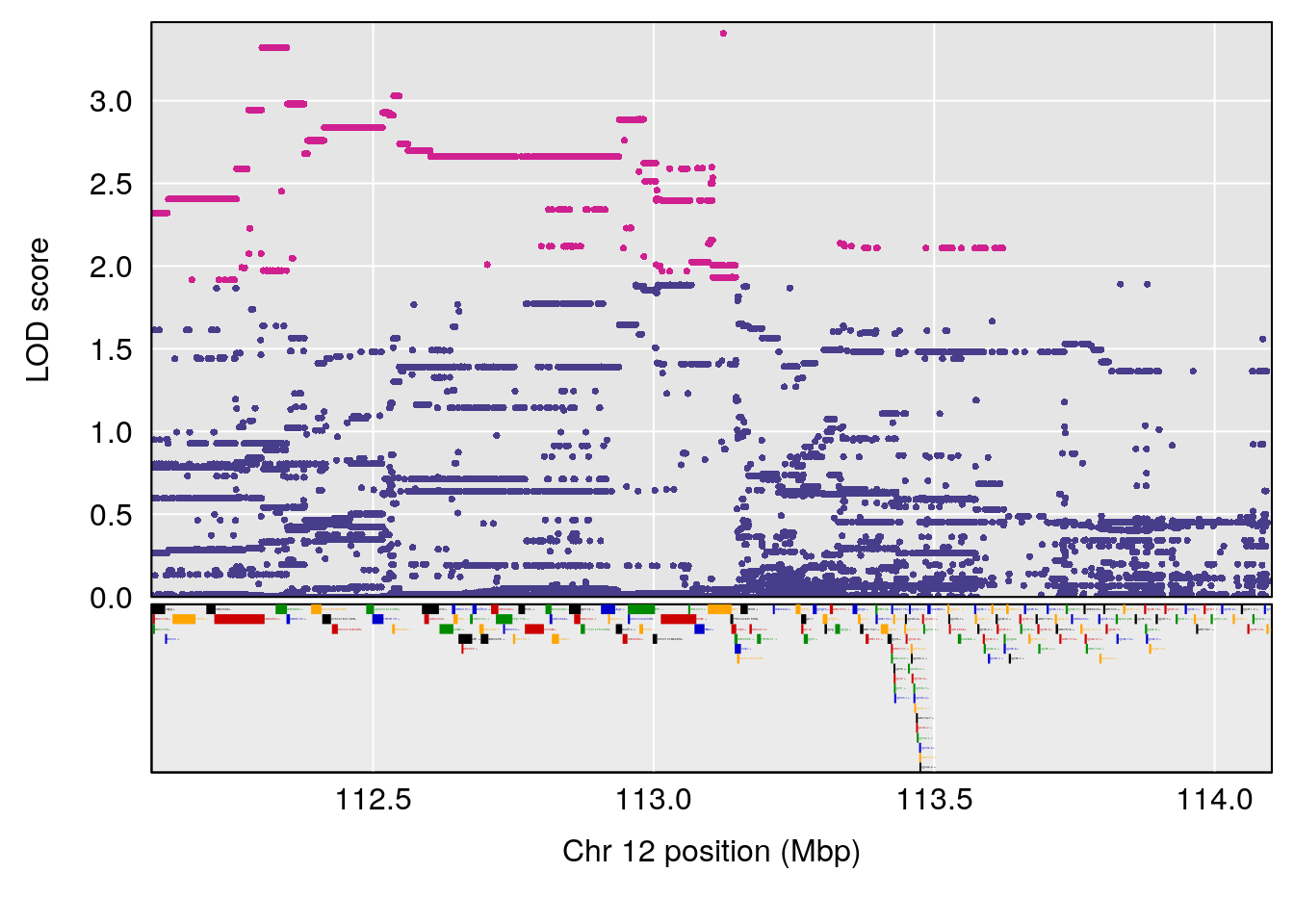
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 5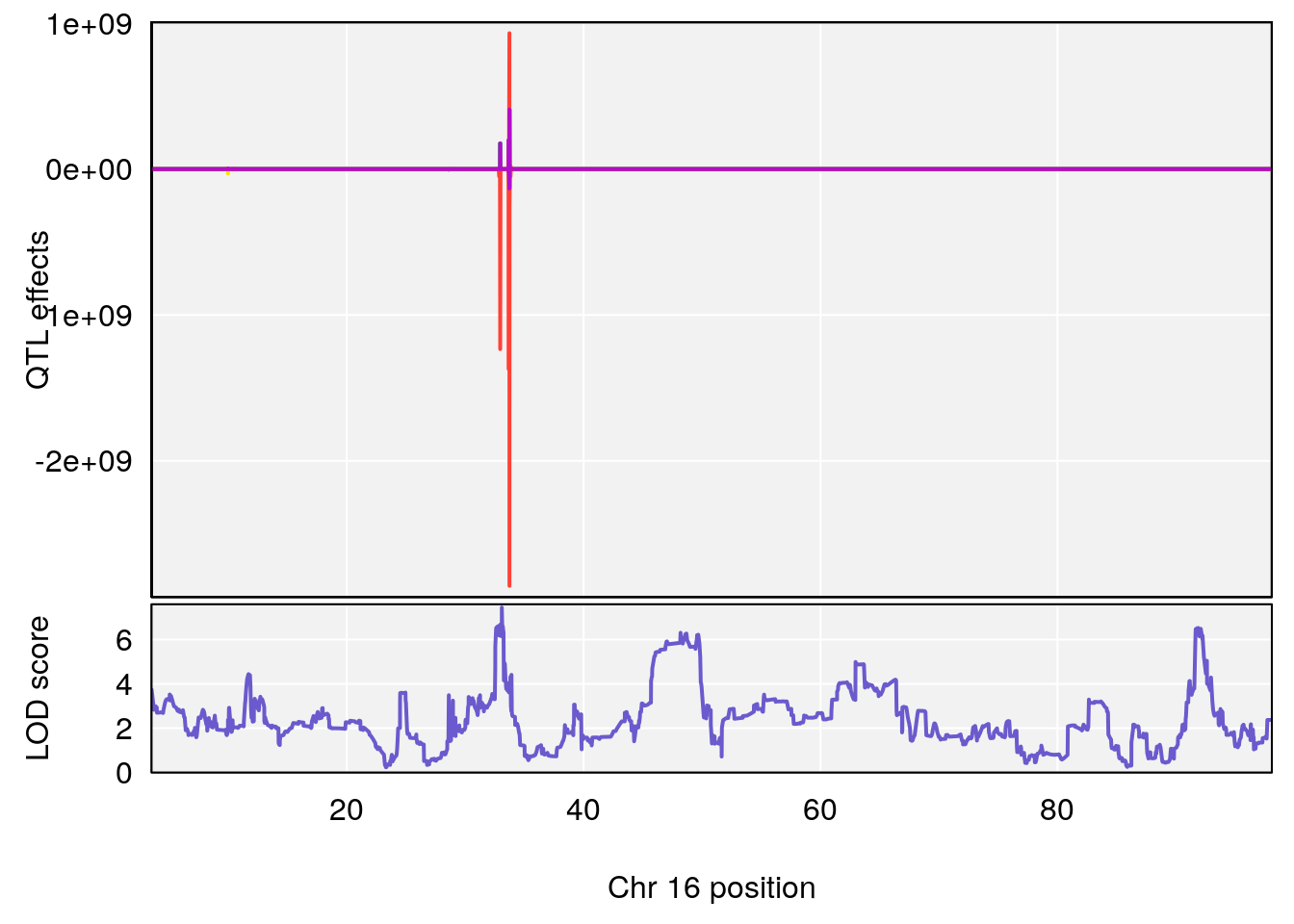
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
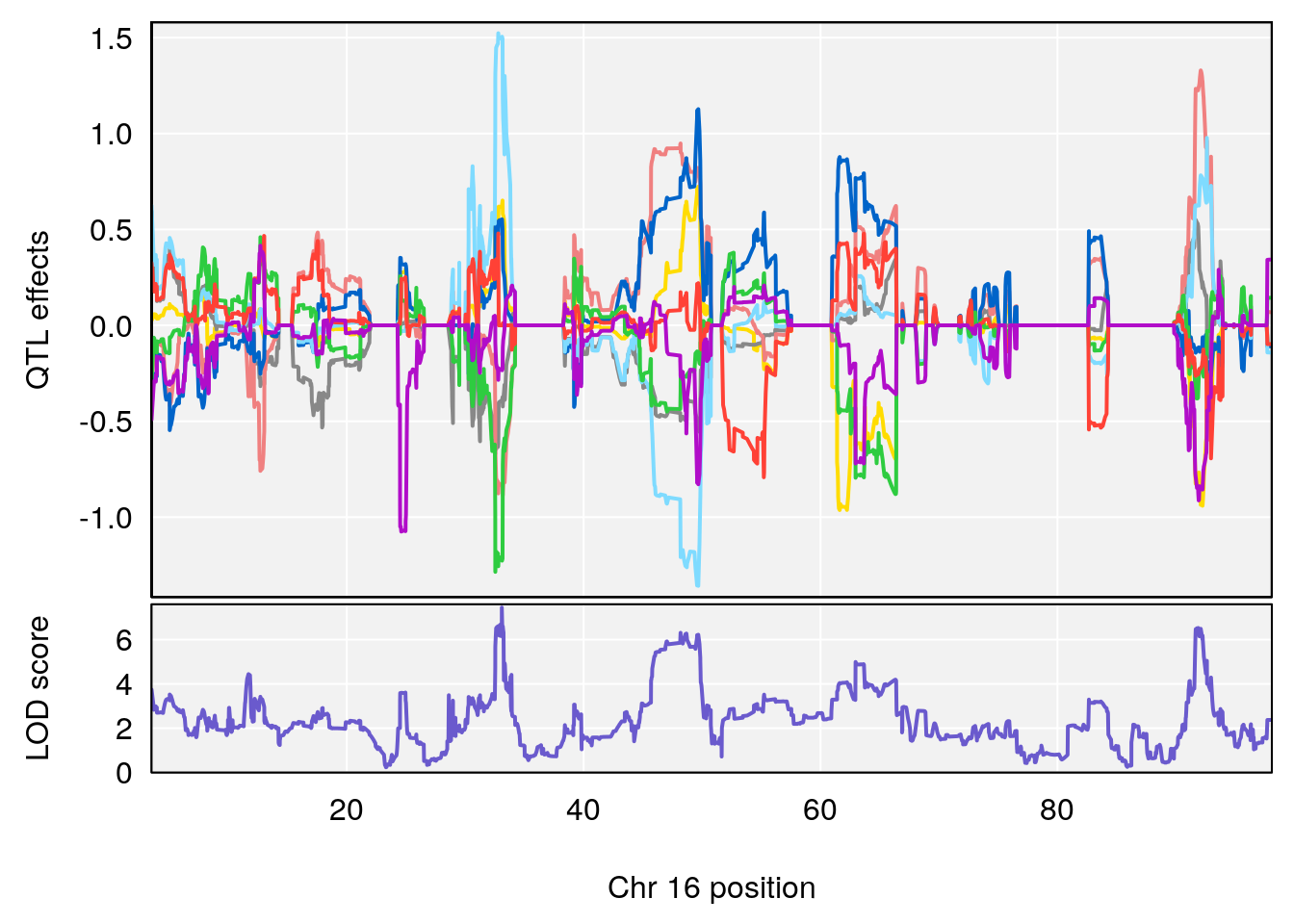
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
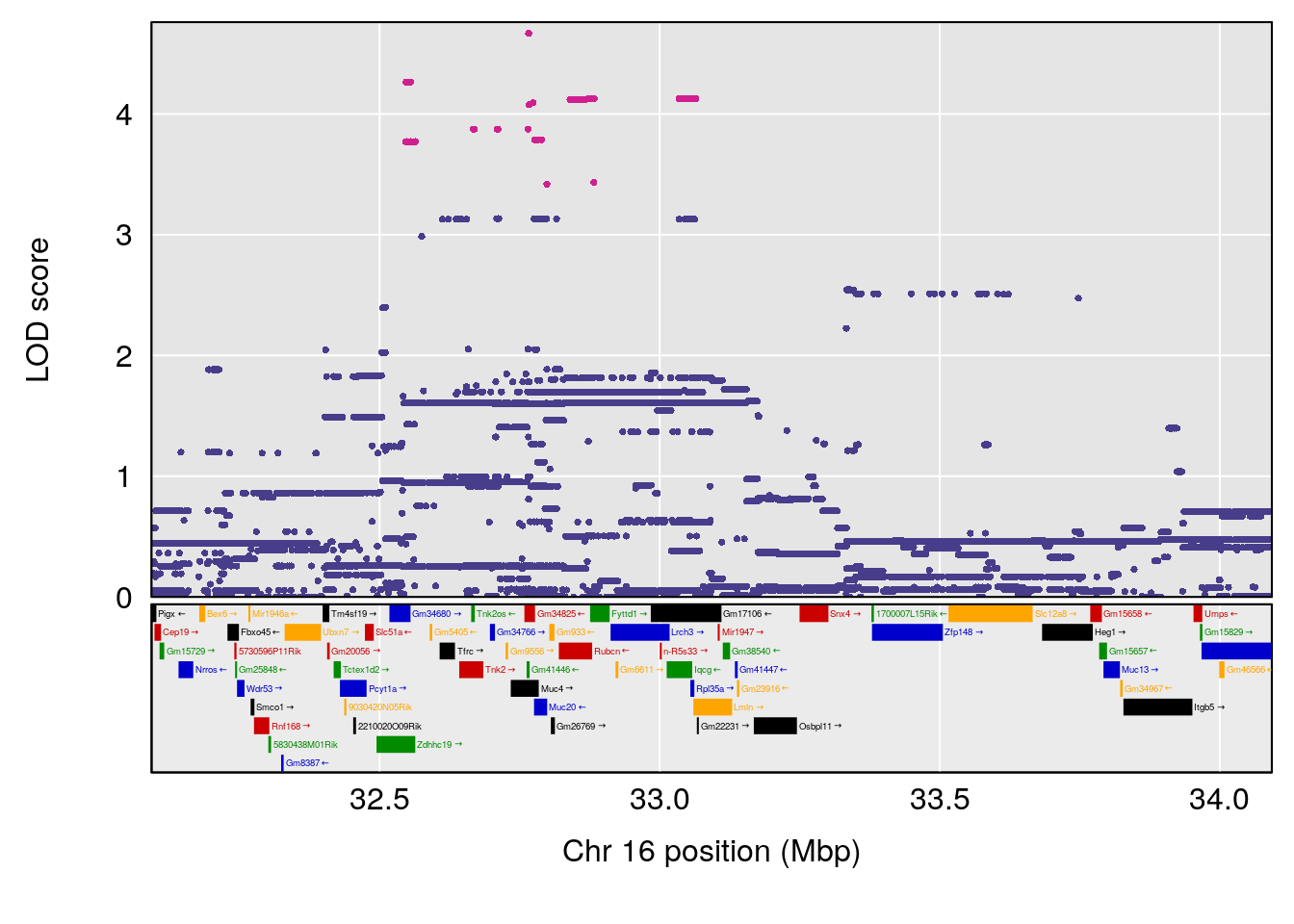
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "Status_bin"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 149.3413 6.686633 145.0415 150.4074
# [1] 1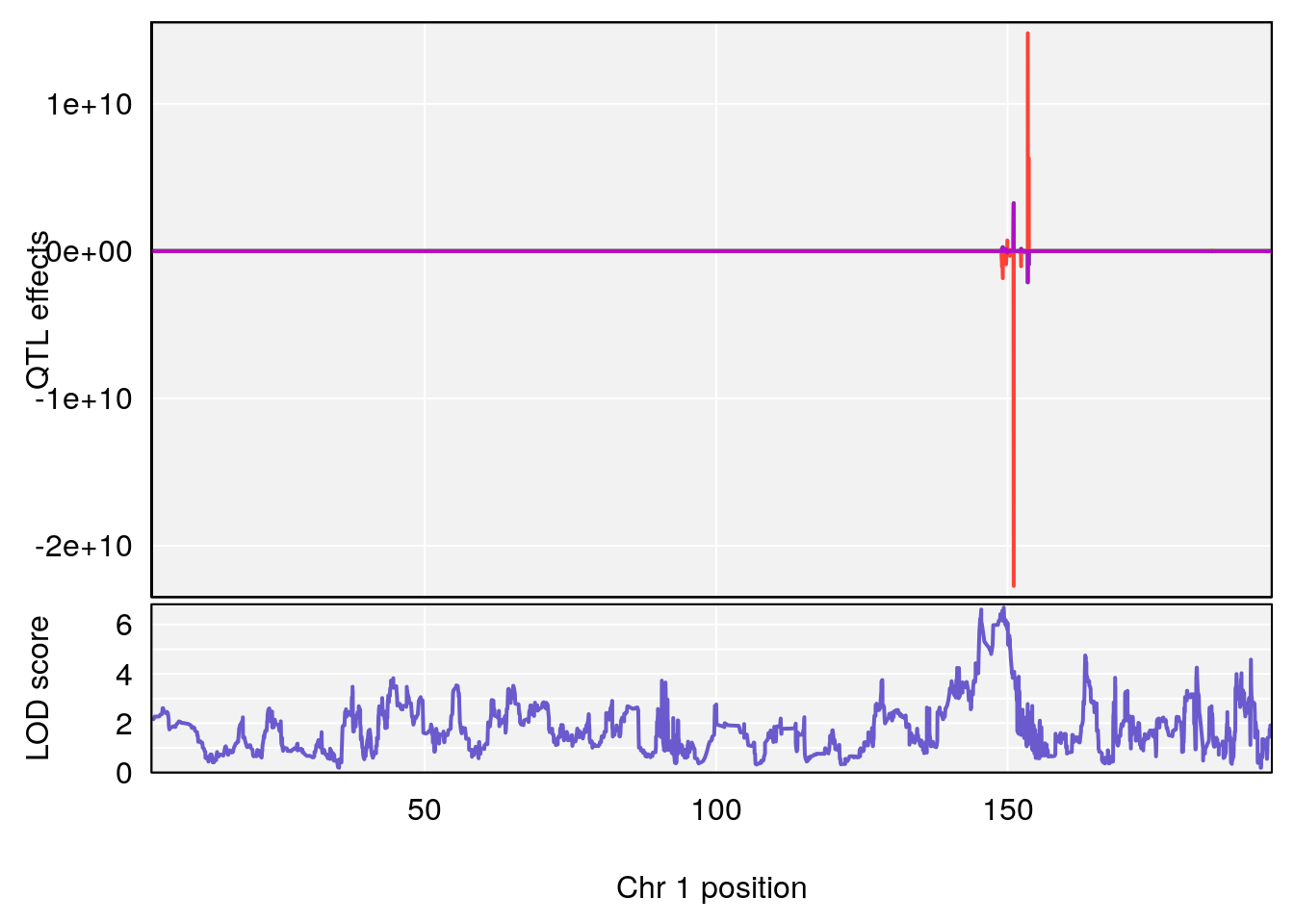
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
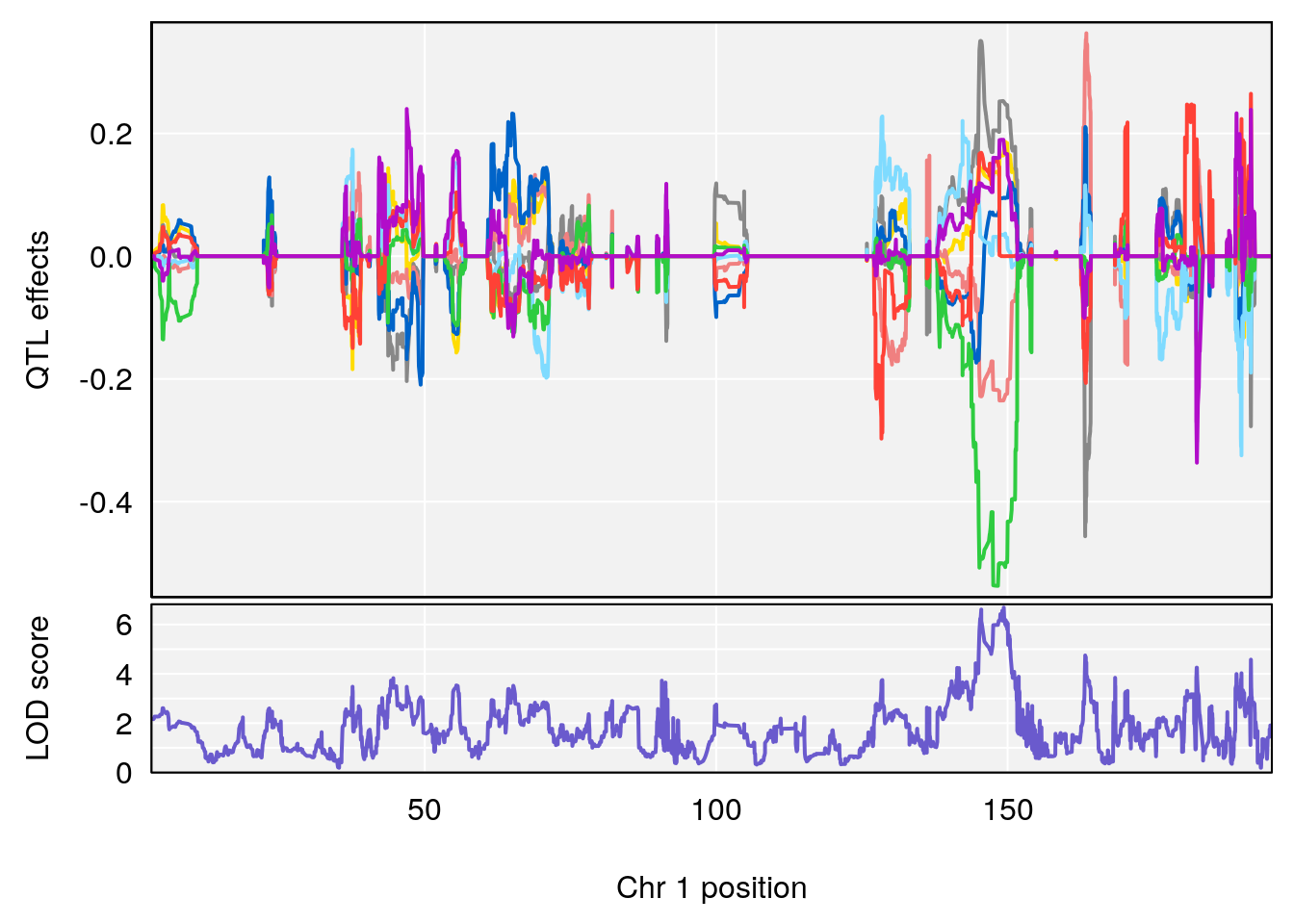
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
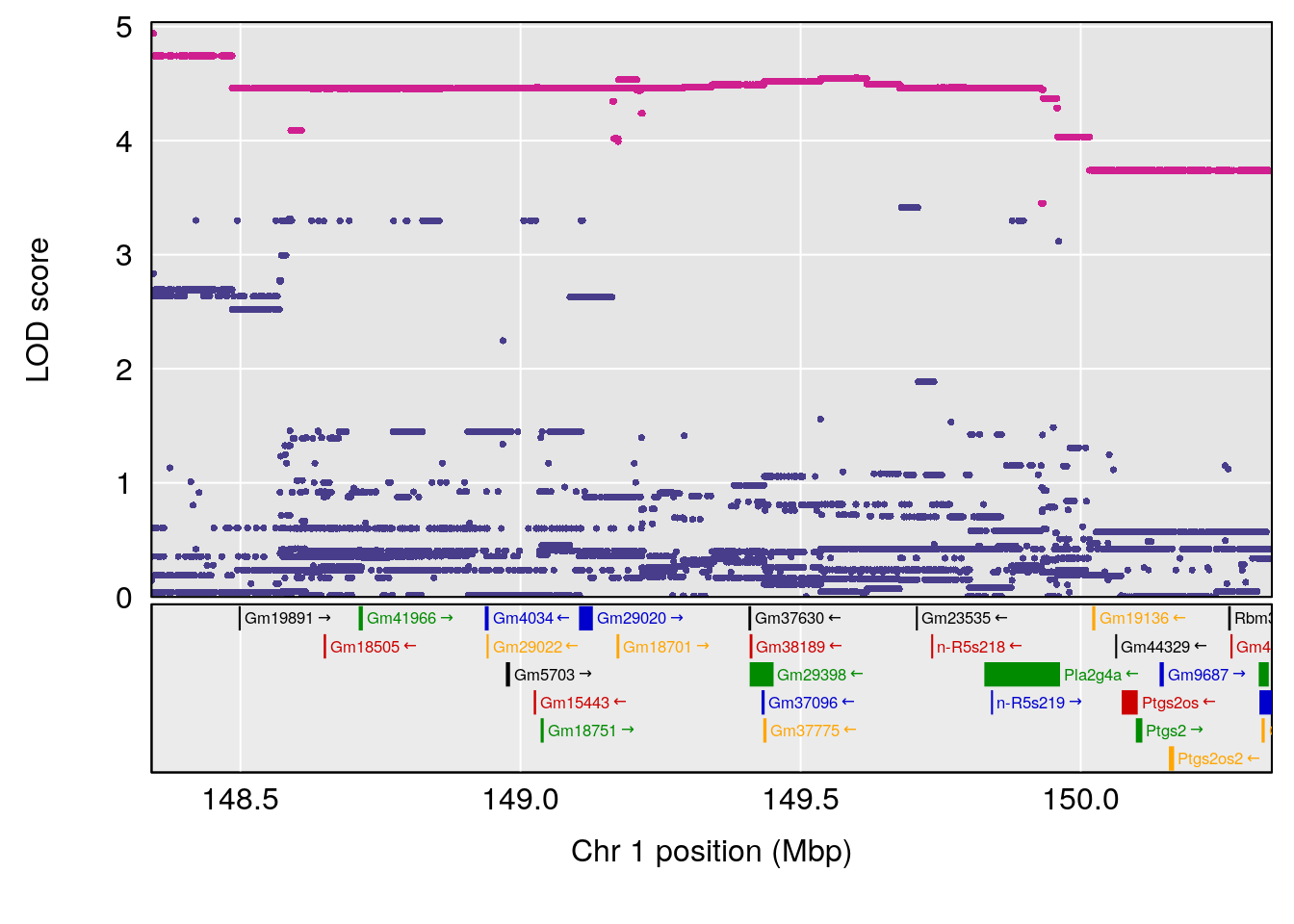
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
sessionInfo()
# R version 4.0.3 (2020-10-10)
# Platform: x86_64-pc-linux-gnu (64-bit)
# Running under: Ubuntu 20.04.2 LTS
#
# Matrix products: default
# BLAS/LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.8.so
#
# locale:
# [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
# [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
# [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=C
# [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
# [9] LC_ADDRESS=C LC_TELEPHONE=C
# [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
#
# attached base packages:
# [1] parallel stats graphics grDevices utils datasets methods
# [8] base
#
# other attached packages:
# [1] forcats_0.5.1 stringr_1.4.0 dplyr_1.0.4 purrr_0.3.4
# [5] readr_1.4.0 tidyr_1.1.2 tibble_3.0.6 tidyverse_1.3.0
# [9] openxlsx_4.2.3 abind_1.4-5 regress_1.3-21 survival_3.2-7
# [13] qtl2_0.24 GGally_2.1.0 gridExtra_2.3 ggplot2_3.3.3
# [17] workflowr_1.6.2
#
# loaded via a namespace (and not attached):
# [1] httr_1.4.2 bit64_4.0.5 jsonlite_1.7.2 splines_4.0.3
# [5] modelr_0.1.8 assertthat_0.2.1 highr_0.8 blob_1.2.1
# [9] cellranger_1.1.0 yaml_2.2.1 pillar_1.4.7 RSQLite_2.2.3
# [13] backports_1.2.1 lattice_0.20-41 glue_1.4.2 digest_0.6.27
# [17] RColorBrewer_1.1-2 promises_1.2.0.1 rvest_0.3.6 colorspace_2.0-0
# [21] htmltools_0.5.1.1 httpuv_1.5.5 Matrix_1.3-2 plyr_1.8.6
# [25] pkgconfig_2.0.3 broom_0.7.4 haven_2.3.1 scales_1.1.1
# [29] whisker_0.4 later_1.1.0.1 git2r_0.28.0 farver_2.0.3
# [33] generics_0.1.0 ellipsis_0.3.1 cachem_1.0.4 withr_2.4.1
# [37] cli_2.3.0 magrittr_2.0.1 crayon_1.4.1 readxl_1.3.1
# [41] memoise_2.0.0 evaluate_0.14 fs_1.5.0 xml2_1.3.2
# [45] tools_4.0.3 data.table_1.13.6 hms_1.0.0 lifecycle_1.0.0
# [49] reprex_1.0.0 munsell_0.5.0 zip_2.1.1 compiler_4.0.3
# [53] rlang_1.0.2 grid_4.0.3 rstudioapi_0.13 labeling_0.4.2
# [57] rmarkdown_2.6 gtable_0.3.0 DBI_1.1.1 reshape_0.8.8
# [61] R6_2.5.0 lubridate_1.7.9.2 knitr_1.31 fastmap_1.1.0
# [65] bit_4.0.4 rprojroot_2.0.2 stringi_1.5.3 Rcpp_1.0.6
# [69] vctrs_0.3.6 dbplyr_2.1.0 tidyselect_1.1.0 xfun_0.21This R Markdown site was created with workflowr