DO_Fentanyl_combining2Cohort_mapping
Hao He
2023-02-21
Last updated: 2023-02-21
Checks: 7 0
Knit directory: DO_Opioid/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200504) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 2200f95. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/Picture1.png
Untracked files:
Untracked: Rplot_rz.png
Untracked: TIMBR.test.prop.bm.MN.RET.RData
Untracked: TIMBR.test.random.RData
Untracked: TIMBR.test.rz.transformed_TVb_ml.RData
Untracked: analysis/DDO_morphine1_second_set_69k.stdout
Untracked: analysis/DO_Fentanyl.R
Untracked: analysis/DO_Fentanyl.err
Untracked: analysis/DO_Fentanyl.out
Untracked: analysis/DO_Fentanyl.sh
Untracked: analysis/DO_Fentanyl_69k.R
Untracked: analysis/DO_Fentanyl_69k.err
Untracked: analysis/DO_Fentanyl_69k.out
Untracked: analysis/DO_Fentanyl_69k.sh
Untracked: analysis/DO_Fentanyl_Cohort2_GCTA_herit.R
Untracked: analysis/DO_Fentanyl_Cohort2_gemma.R
Untracked: analysis/DO_Fentanyl_Cohort2_mapping.R
Untracked: analysis/DO_Fentanyl_Cohort2_mapping.err
Untracked: analysis/DO_Fentanyl_Cohort2_mapping.out
Untracked: analysis/DO_Fentanyl_Cohort2_mapping.sh
Untracked: analysis/DO_Fentanyl_GCTA_herit.R
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.R
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.err
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.out
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.sh
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.R
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.err
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.out
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.sh
Untracked: analysis/DO_Fentanyl_array.R
Untracked: analysis/DO_Fentanyl_array.err
Untracked: analysis/DO_Fentanyl_array.out
Untracked: analysis/DO_Fentanyl_array.sh
Untracked: analysis/DO_Fentanyl_combining2Cohort_GCTA_herit.R
Untracked: analysis/DO_Fentanyl_combining2Cohort_gemma.R
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping.R
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping.err
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping.out
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping.sh
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping_CoxPH.R
Untracked: analysis/DO_Fentanyl_finalreport_to_plink.sh
Untracked: analysis/DO_Fentanyl_gemma.R
Untracked: analysis/DO_Fentanyl_gemma.err
Untracked: analysis/DO_Fentanyl_gemma.out
Untracked: analysis/DO_Fentanyl_gemma.sh
Untracked: analysis/DO_morphine1.R
Untracked: analysis/DO_morphine1.Rout
Untracked: analysis/DO_morphine1.sh
Untracked: analysis/DO_morphine1.stderr
Untracked: analysis/DO_morphine1.stdout
Untracked: analysis/DO_morphine1_SNP.R
Untracked: analysis/DO_morphine1_SNP.Rout
Untracked: analysis/DO_morphine1_SNP.sh
Untracked: analysis/DO_morphine1_SNP.stderr
Untracked: analysis/DO_morphine1_SNP.stdout
Untracked: analysis/DO_morphine1_combined.R
Untracked: analysis/DO_morphine1_combined.Rout
Untracked: analysis/DO_morphine1_combined.sh
Untracked: analysis/DO_morphine1_combined.stderr
Untracked: analysis/DO_morphine1_combined.stdout
Untracked: analysis/DO_morphine1_combined_69k.R
Untracked: analysis/DO_morphine1_combined_69k.Rout
Untracked: analysis/DO_morphine1_combined_69k.sh
Untracked: analysis/DO_morphine1_combined_69k.stderr
Untracked: analysis/DO_morphine1_combined_69k.stdout
Untracked: analysis/DO_morphine1_combined_69k_m2.R
Untracked: analysis/DO_morphine1_combined_69k_m2.Rout
Untracked: analysis/DO_morphine1_combined_69k_m2.sh
Untracked: analysis/DO_morphine1_combined_69k_m2.stderr
Untracked: analysis/DO_morphine1_combined_69k_m2.stdout
Untracked: analysis/DO_morphine1_combined_weight_DOB.R
Untracked: analysis/DO_morphine1_combined_weight_DOB.Rout
Untracked: analysis/DO_morphine1_combined_weight_DOB.err
Untracked: analysis/DO_morphine1_combined_weight_DOB.out
Untracked: analysis/DO_morphine1_combined_weight_DOB.sh
Untracked: analysis/DO_morphine1_combined_weight_DOB.stderr
Untracked: analysis/DO_morphine1_combined_weight_DOB.stdout
Untracked: analysis/DO_morphine1_combined_weight_age.R
Untracked: analysis/DO_morphine1_combined_weight_age.err
Untracked: analysis/DO_morphine1_combined_weight_age.out
Untracked: analysis/DO_morphine1_combined_weight_age.sh
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.R
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.err
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.out
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.sh
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.R
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.err
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.out
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.sh
Untracked: analysis/DO_morphine1_cph.R
Untracked: analysis/DO_morphine1_cph.Rout
Untracked: analysis/DO_morphine1_cph.sh
Untracked: analysis/DO_morphine1_second_set.R
Untracked: analysis/DO_morphine1_second_set.Rout
Untracked: analysis/DO_morphine1_second_set.sh
Untracked: analysis/DO_morphine1_second_set.stderr
Untracked: analysis/DO_morphine1_second_set.stdout
Untracked: analysis/DO_morphine1_second_set_69k.R
Untracked: analysis/DO_morphine1_second_set_69k.Rout
Untracked: analysis/DO_morphine1_second_set_69k.sh
Untracked: analysis/DO_morphine1_second_set_69k.stderr
Untracked: analysis/DO_morphine1_second_set_SNP.R
Untracked: analysis/DO_morphine1_second_set_SNP.Rout
Untracked: analysis/DO_morphine1_second_set_SNP.sh
Untracked: analysis/DO_morphine1_second_set_SNP.stderr
Untracked: analysis/DO_morphine1_second_set_SNP.stdout
Untracked: analysis/DO_morphine1_second_set_weight_DOB.R
Untracked: analysis/DO_morphine1_second_set_weight_DOB.Rout
Untracked: analysis/DO_morphine1_second_set_weight_DOB.err
Untracked: analysis/DO_morphine1_second_set_weight_DOB.out
Untracked: analysis/DO_morphine1_second_set_weight_DOB.sh
Untracked: analysis/DO_morphine1_second_set_weight_DOB.stderr
Untracked: analysis/DO_morphine1_second_set_weight_DOB.stdout
Untracked: analysis/DO_morphine1_second_set_weight_age.R
Untracked: analysis/DO_morphine1_second_set_weight_age.Rout
Untracked: analysis/DO_morphine1_second_set_weight_age.err
Untracked: analysis/DO_morphine1_second_set_weight_age.out
Untracked: analysis/DO_morphine1_second_set_weight_age.sh
Untracked: analysis/DO_morphine1_second_set_weight_age.stderr
Untracked: analysis/DO_morphine1_second_set_weight_age.stdout
Untracked: analysis/DO_morphine1_weight_DOB.R
Untracked: analysis/DO_morphine1_weight_DOB.sh
Untracked: analysis/DO_morphine1_weight_age.R
Untracked: analysis/DO_morphine1_weight_age.sh
Untracked: analysis/DO_morphine_gemma.R
Untracked: analysis/DO_morphine_gemma.err
Untracked: analysis/DO_morphine_gemma.out
Untracked: analysis/DO_morphine_gemma.sh
Untracked: analysis/DO_morphine_gemma_firstmin.R
Untracked: analysis/DO_morphine_gemma_firstmin.err
Untracked: analysis/DO_morphine_gemma_firstmin.out
Untracked: analysis/DO_morphine_gemma_firstmin.sh
Untracked: analysis/DO_morphine_gemma_withpermu.R
Untracked: analysis/DO_morphine_gemma_withpermu.err
Untracked: analysis/DO_morphine_gemma_withpermu.out
Untracked: analysis/DO_morphine_gemma_withpermu.sh
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.R
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.err
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.out
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.sh
Untracked: analysis/Plot_DO_morphine1_SNP.R
Untracked: analysis/Plot_DO_morphine1_SNP.Rout
Untracked: analysis/Plot_DO_morphine1_SNP.sh
Untracked: analysis/Plot_DO_morphine1_SNP.stderr
Untracked: analysis/Plot_DO_morphine1_SNP.stdout
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.R
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.Rout
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.sh
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.stderr
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.stdout
Untracked: analysis/download_GSE100356_sra.sh
Untracked: analysis/fentanyl_2cohorts_coxph.R
Untracked: analysis/fentanyl_2cohorts_coxph.err
Untracked: analysis/fentanyl_2cohorts_coxph.out
Untracked: analysis/fentanyl_2cohorts_coxph.sh
Untracked: analysis/fentanyl_scanone.cph.R
Untracked: analysis/fentanyl_scanone.cph.err
Untracked: analysis/fentanyl_scanone.cph.out
Untracked: analysis/fentanyl_scanone.cph.sh
Untracked: analysis/geo_rnaseq.R
Untracked: analysis/heritability_first_second_batch.R
Untracked: analysis/nf-rnaseq-b6.R
Untracked: analysis/plot_fentanyl_2cohorts_coxph.R
Untracked: analysis/scripts/
Untracked: analysis/tibmr.R
Untracked: analysis/timbr_demo.R
Untracked: analysis/workflow_proc.R
Untracked: analysis/workflow_proc.sh
Untracked: analysis/workflow_proc.stderr
Untracked: analysis/workflow_proc.stdout
Untracked: analysis/x.R
Untracked: code/cfw/
Untracked: code/gemma_plot.R
Untracked: code/process.sanger.snp.R
Untracked: code/reconst_utils.R
Untracked: data/69k_grid_pgmap.RData
Untracked: data/Composite Post Kevins Program Group 2 Fentanyl Prepped for Hao.xlsx
Untracked: data/DO_WBP_Data_JAB_to_map.xlsx
Untracked: data/Fentanyl_alternate_metrics.xlsx
Untracked: data/FinalReport/
Untracked: data/GM/
Untracked: data/GM_covar.csv
Untracked: data/GM_covar_07092018_morphine.csv
Untracked: data/Jackson_Lab_Bubier_MURGIGV01/
Untracked: data/MPD_Upload_October.csv
Untracked: data/MPD_Upload_October_updated_sex.csv
Untracked: data/Master Fentanyl DO Study Sheet.xlsx
Untracked: data/MasterMorphine Second Set DO w DOB2.xlsx
Untracked: data/MasterMorphine Second Set DO.xlsx
Untracked: data/Morphine CC DO mice Updated with Published inbred strains.csv
Untracked: data/Morphine_CC_DO_mice_Updated_with_Published_inbred_strains.csv
Untracked: data/cc_variants.sqlite
Untracked: data/combined/
Untracked: data/fentanyl/
Untracked: data/fentanyl2/
Untracked: data/fentanyl_1_2/
Untracked: data/fentanyl_2cohorts_coxph_data.Rdata
Untracked: data/first/
Untracked: data/founder_geno.csv
Untracked: data/genetic_map.csv
Untracked: data/gm.json
Untracked: data/gwas.sh
Untracked: data/marker_grid_0.02cM_plus.txt
Untracked: data/mouse_genes_mgi.sqlite
Untracked: data/pheno.csv
Untracked: data/pheno_qtl2.csv
Untracked: data/pheno_qtl2_07092018_morphine.csv
Untracked: data/pheno_qtl2_w_dob.csv
Untracked: data/physical_map.csv
Untracked: data/rnaseq/
Untracked: data/sample_geno.csv
Untracked: data/second/
Untracked: figure/
Untracked: glimma-plots/
Untracked: output/DO_Fentanyl_Cohort2_MinDepressionRR_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_MinDepressionRR_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_RRDepressionRateHrSLOPE_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_RRRecoveryRateHrSLOPE_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_RRRecoveryRateHrSLOPE_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_StartofRecoveryHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_StartofRecoveryHr_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_Statusbin_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_Statusbin_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_SteadyStateDepressionDurationHrINTERVAL_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoDead(Hr)_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoDeadHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoDeadHr_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoProjectedRecoveryHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoProjectedRecoveryHr_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoSteadyRRDepression(Hr)_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoSteadyRRDepressionHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoSteadyRRDepressionHr_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoThresholdRecoveryHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoThresholdRecoveryHr_coefplot_blup.pdf
Untracked: output/DO_morphine_Min.depression.png
Untracked: output/DO_morphine_Min.depression22222_violin_chr5.pdf
Untracked: output/DO_morphine_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_Min.depression_coefplot_blup_chr5.png
Untracked: output/DO_morphine_Min.depression_coefplot_blup_chrX.png
Untracked: output/DO_morphine_Min.depression_coefplot_chr5.png
Untracked: output/DO_morphine_Min.depression_coefplot_chrX.png
Untracked: output/DO_morphine_Min.depression_peak_genes_chr5.png
Untracked: output/DO_morphine_Min.depression_violin_chr5.png
Untracked: output/DO_morphine_Recovery.Time.png
Untracked: output/DO_morphine_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr11.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr4.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr7.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr9.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr11.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr4.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr7.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr9.png
Untracked: output/DO_morphine_Status_bin.png
Untracked: output/DO_morphine_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_Survival.Time.png
Untracked: output/DO_morphine_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_Survival.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_Survival.Time_coefplot_blup_chr17.png
Untracked: output/DO_morphine_Survival.Time_coefplot_blup_chr8.png
Untracked: output/DO_morphine_Survival.Time_coefplot_chr17.png
Untracked: output/DO_morphine_Survival.Time_coefplot_chr8.png
Untracked: output/DO_morphine_combine_batch_peak_violin.pdf
Untracked: output/DO_morphine_combined_69k_m2_Min.depression.png
Untracked: output/DO_morphine_combined_69k_m2_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_combined_69k_m2_Recovery.Time.png
Untracked: output/DO_morphine_combined_69k_m2_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_combined_69k_m2_Status_bin.png
Untracked: output/DO_morphine_combined_69k_m2_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_combined_69k_m2_Survival.Time.png
Untracked: output/DO_morphine_combined_69k_m2_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Survival.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_coxph_24hrs_kinship_QTL.png
Untracked: output/DO_morphine_cphout.RData
Untracked: output/DO_morphine_first_batch_peak_in_second_batch_violin.pdf
Untracked: output/DO_morphine_first_batch_peak_in_second_batch_violin_sidebyside.pdf
Untracked: output/DO_morphine_first_batch_peak_violin.pdf
Untracked: output/DO_morphine_operm.cph.RData
Untracked: output/DO_morphine_second_batch_on_first_batch_peak_violin.pdf
Untracked: output/DO_morphine_second_batch_peak_ch6surv_on_first_batchviolin.pdf
Untracked: output/DO_morphine_second_batch_peak_ch6surv_on_first_batchviolin2.pdf
Untracked: output/DO_morphine_second_batch_peak_in_first_batch_violin.pdf
Untracked: output/DO_morphine_second_batch_peak_in_first_batch_violin_sidebyside.pdf
Untracked: output/DO_morphine_second_batch_peak_violin.pdf
Untracked: output/DO_morphine_secondbatch_69k_Min.depression.png
Untracked: output/DO_morphine_secondbatch_69k_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_69k_Recovery.Time.png
Untracked: output/DO_morphine_secondbatch_69k_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_69k_Status_bin.png
Untracked: output/DO_morphine_secondbatch_69k_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_69k_Survival.Time.png
Untracked: output/DO_morphine_secondbatch_69k_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Survival.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Min.depression.png
Untracked: output/DO_morphine_secondbatch_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Recovery.Time.png
Untracked: output/DO_morphine_secondbatch_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Status_bin.png
Untracked: output/DO_morphine_secondbatch_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Survival.Time.png
Untracked: output/DO_morphine_secondbatch_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Survival.Time_coefplot_blup.pdf
Untracked: output/Fentanyl/
Untracked: output/KPNA3.pdf
Untracked: output/SSC4D.pdf
Untracked: output/TIMBR.test.RData
Untracked: output/apr_69kchr_combined.RData
Untracked: output/apr_69kchr_k_loco_combined.rds
Untracked: output/apr_69kchr_second_set.RData
Untracked: output/combine_batch_variation.RData
Untracked: output/combined_gm.RData
Untracked: output/combined_gm.k_loco.rds
Untracked: output/combined_gm.k_overall.rds
Untracked: output/combined_gm.probs_8state.rds
Untracked: output/coxph/
Untracked: output/do.morphine.RData
Untracked: output/do.morphine.k_loco.rds
Untracked: output/do.morphine.probs_36state.rds
Untracked: output/do.morphine.probs_8state.rds
Untracked: output/do_Fentanyl_combine2cohort_MeanDepressionBR_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_MeanDepressionBR_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_MinDepressionBR_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_MinDepressionBR_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_MinDepressionRR_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_MinDepressionRR_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_RRRecoveryRateHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_RRRecoveryRateHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_StartofRecoveryHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_StartofRecoveryHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_Statusbin_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_Statusbin_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_SteadyStateDepressionDurationHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_SteadyStateDepressionDurationHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_SurvivalTime_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_SurvivalTime_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoDeadHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoDeadHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoMostlyDeadHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoMostlyDeadHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoProjectedRecoveryHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoProjectedRecoveryHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoRecoveryHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoRecoveryHr_coefplot_blup.pdf
Untracked: output/first_batch_variation.RData
Untracked: output/first_second_survival_peak_chr.xlsx
Untracked: output/hsq_1_first_batch_herit_qtl2.RData
Untracked: output/hsq_2_second_batch_herit_qtl2.RData
Untracked: output/old_temp/
Untracked: output/pr_69kchr_combined.RData
Untracked: output/pr_69kchr_second_set.RData
Untracked: output/qtl.morphine.69k.out.combined.RData
Untracked: output/qtl.morphine.69k.out.combined_m2.RData
Untracked: output/qtl.morphine.69k.out.second_set.RData
Untracked: output/qtl.morphine.operm.RData
Untracked: output/qtl.morphine.out.RData
Untracked: output/qtl.morphine.out.combined_gm.RData
Untracked: output/qtl.morphine.out.combined_gm.female.RData
Untracked: output/qtl.morphine.out.combined_gm.male.RData
Untracked: output/qtl.morphine.out.combined_weight_DOB.RData
Untracked: output/qtl.morphine.out.combined_weight_age.RData
Untracked: output/qtl.morphine.out.female.RData
Untracked: output/qtl.morphine.out.male.RData
Untracked: output/qtl.morphine.out.second_set.RData
Untracked: output/qtl.morphine.out.second_set.female.RData
Untracked: output/qtl.morphine.out.second_set.male.RData
Untracked: output/qtl.morphine.out.second_set.weight_DOB.RData
Untracked: output/qtl.morphine.out.second_set.weight_age.RData
Untracked: output/qtl.morphine.out.weight_DOB.RData
Untracked: output/qtl.morphine.out.weight_age.RData
Untracked: output/qtl.morphine1.snpout.RData
Untracked: output/qtl.morphine2.snpout.RData
Untracked: output/second_batch_pheno.csv
Untracked: output/second_batch_variation.RData
Untracked: output/second_set_apr_69kchr_k_loco.rds
Untracked: output/second_set_gm.RData
Untracked: output/second_set_gm.k_loco.rds
Untracked: output/second_set_gm.probs_36state.rds
Untracked: output/second_set_gm.probs_8state.rds
Untracked: output/topSNP_chr5_mindepression.csv
Untracked: output/zoompeak_Min.depression_9.pdf
Untracked: output/zoompeak_Recovery.Time_16.pdf
Untracked: output/zoompeak_Status_bin_11.pdf
Untracked: output/zoompeak_Survival.Time_1.pdf
Untracked: output/zoompeak_fentanyl_Survival.Time_2.pdf
Untracked: sra-tools_v2.10.7.sif
Unstaged changes:
Modified: _workflowr.yml
Modified: analysis/marker_violin.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/Plot_DO_Fentanyl_combining2Cohort_mapping.Rmd) and HTML (docs/Plot_DO_Fentanyl_combining2Cohort_mapping.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 2200f95 | xhyuo | 2023-02-21 | update with sex specific Fentanyl plot |
| html | 8611541 | xhyuo | 2023-02-21 | Build site. |
| Rmd | d1a7028 | xhyuo | 2023-02-21 | update with sex specific Fentanyl |
| html | fd4dc88 | xhyuo | 2023-01-11 | Build site. |
| Rmd | aff7cd2 | xhyuo | 2023-01-11 | update with histogram |
| html | 6eeaa6a | xhyuo | 2023-01-06 | Build site. |
| Rmd | 6d89b79 | xhyuo | 2023-01-06 | update with histogram and gcta h2, figure size |
| html | fe30eec | xhyuo | 2023-01-06 | Build site. |
| Rmd | 15a2b6f | xhyuo | 2023-01-06 | update with histogram and gcta h2 |
| html | 0971ef2 | xhyuo | 2022-07-05 | Build site. |
| Rmd | 385dbb4 | xhyuo | 2022-07-05 | update Plot_DO_Fentanyl_combining2Cohort |
| html | d014111 | xhyuo | 2022-06-29 | Build site. |
| html | 773a9fa | xhyuo | 2022-06-29 | Build site. |
| Rmd | e97e172 | xhyuo | 2022-06-29 | Plot_DO_Fentanyl_combining2Cohort |
Last update: 2023-02-21
loading libraries
library(ggplot2)
library(gridExtra)
library(GGally)
library(parallel)
library(qtl2)
library(parallel)
library(survival)
library(regress)
library(abind)
library(openxlsx)
library(tidyverse)
rz.transform <- function(y) {
rankY=rank(y, ties.method="average", na.last="keep")
rzT=qnorm(rankY/(length(na.exclude(rankY))+1))
return(rzT)
}
load("code/do.colors.RData")Read phenotype data
# Read phenotype data Master Fentanyl DO Study Sheet.xlsx-----------------------------------------------------
do.pheno1 <- read.xlsx("data/Master Fentanyl DO Study Sheet.xlsx", sheet = 1,
rows = 1:399, na.strings = "NaN", detectDates = TRUE)
#remove zombie mice
do.pheno1 <- do.pheno1 %>%
dplyr::filter(is.na(Censor)) %>%
dplyr::mutate(ID = replace(ID, (ID == 31009 & File == 20210519), 31109)) %>% #second 31009 is really 31109
dplyr::mutate(Survival = replace(Survival, (ID %in% c(30931, 31084)), "ALIVE")) %>% #30931, 31084 should be ALIVE
dplyr::mutate(Survival = replace(Survival, (ID %in% c(31049,30901)), "DEAD")) %>% #31049,30901 should be DEAD
dplyr::mutate(Status_bin = ifelse(Survival == "ALIVE", 1, 0)) %>%
dplyr::mutate(ID = as.character(ID)) %>%
dplyr::arrange(ID, desc(File)) %>%
dplyr::distinct(ID, .keep_all = TRUE) %>%
dplyr::rename(., MouseID = ID)
#Fentanyl_alternate_metrics.xlsx
do.pheno.alt <- read.xlsx("data/Fentanyl_alternate_metrics.xlsx", sheet = 1,
na.strings = "NaN") %>%
dplyr::rename(., MouseID = Mouse.ID) %>%
dplyr::mutate(MouseID = as.character(MouseID)) %>%
dplyr::rename_with(., ~ str_replace(.x, "\\(", "")) %>%
dplyr::rename_with(., ~ str_replace(.x, "\\)", "")) %>%
dplyr::rename_with(., ~ str_replace(.x, "\\%/", "")) %>%
dplyr::rename_with(., ~ str_replace(.x, "\\.%", "")) %>%
dplyr::filter(!(if_all(2:11, is.na))) %>%
dplyr::distinct(MouseID, .keep_all = TRUE)
#get sex and dob
do.pheno1 <- left_join(do.pheno.alt, do.pheno1) %>%
dplyr::rename_with(., ~ str_replace(.x, "\\(", "")) %>%
dplyr::rename_with(., ~ str_replace(.x, "\\)", "")) %>%
dplyr::rename_with(., ~ str_replace(.x, "\\.%", "")) %>%
dplyr::rename_with(., ~ str_replace(.x, "\\-", "")) %>%
dplyr::select(MouseID, Sex, dob, Generation,
Time.to.Dead.Hr:RR.Recovery.Rate.Hr,
Min.Depression.BR, Time.to.Mostly.Dead.Hr, Time.to.Recovery.Hr,
Status_bin, Survival) %>%
dplyr::mutate(Survival = replace(Survival, (!is.na(Time.to.Dead.Hr) & is.na(Survival)), "DEAD")) %>%
dplyr::mutate(event = ifelse(Survival == "DEAD", 1, 0)) %>% #event 0=alive, 1=dead.
dplyr::mutate(Survival.Time48 = replace(Time.to.Dead.Hr, (Survival == "ALIVE" & is.na(Time.to.Dead.Hr)), 24.00)) %>%
dplyr::mutate(cohort = "cohort1")
# Read phenotype data Composite Post Kevins Program Group 2 Fentanyl Prepped for Hao.xlsx-----------------------------------------------------
#batch 20220502
do.pheno2 <- readxl::read_excel(path = "data/Composite Post Kevins Program Group 2 Fentanyl Prepped for Hao.xlsx",
col_types = c("text", "text", "text", "date", "numeric", "numeric", "date", rep("numeric", 9), "date"))
# Warning in read_fun(path = enc2native(normalizePath(path)), sheet_i = sheet, :
# Expecting date in D263 / R263C4: got 'BLANK'
# Warning in read_fun(path = enc2native(normalizePath(path)), sheet_i = sheet, :
# Expecting numeric in F263 / R263C6: got 'BLANK'
do.pheno2 <- do.pheno2 %>%
dplyr::rename_with(., ~str_remove_all(.x, " ")) %>%
dplyr::filter(!is.na(MouseID)) %>%
dplyr::filter(MouseID != "BLANK") %>%
dplyr::mutate(EarTag = replace(EarTag, (MouseID == 35151), "ALIVE")) %>% #35151 should be ALIVE
dplyr::mutate(EarTag = replace(EarTag, (MouseID %in% c(35263, 35108)), "DEAD")) %>% # 35263 35108 should be DEAD
dplyr::mutate(Status_bin = case_when(
EarTag == "DEAD" ~ 0,
(EarTag == "ALIVE" | is.na(EarTag)) ~ 1
)) %>%
dplyr::rename_with(., ~ str_replace(.x, "\\(", "")) %>%
dplyr::rename_with(., ~ str_replace(.x, "\\)", "")) %>%
dplyr::rename_with(., ~ str_replace(.x, "\\%/", "")) %>%
dplyr::rename_with(., ~ str_replace(.x, "\\.%", "")) %>%
dplyr::rename_with(., ~ str_replace(.x, "\\%", "")) %>%
dplyr::rename_with(., ~ str_replace(.x, "\\-", "")) %>%
dplyr::select(MouseID,
Survival = EarTag,
Sex,
dob = DOB,
Time.to.Dead.Hr = TimetoDeadHr,
RR.Depression.Rate.Hr = RRDepressionRateHrSLOPE,
Time.to.Steady.RR.Depression.Hr = TimetoSteadyRRDepressionHr,
Min.Depression.RR = MinDepressionRR,
SteadyState.Depression.Duration.Hr = SteadyStateDepressionDurationHrINTERVAL,
Time.to.Threshold.Recovery.Hr = TimetoThresholdRecoveryHr,
Time.to.Projected.Recovery.Hr = TimetoProjectedRecoveryHr,
Start.of.Recovery.Hr = StartofRecoveryHr,
RR.Recovery.Rate.Hr = RRRecoveryRateHrSLOPE,
Status_bin) %>%
dplyr::mutate(Survival = replace(Survival, is.na(Survival), "ALIVE")) %>%
dplyr::mutate(event = ifelse(Survival == "DEAD", 1, 0)) %>% #event 0=alive, 1=dead.
dplyr::mutate(Survival.Time48 = replace(Time.to.Dead.Hr, (Survival == "ALIVE" & is.na(Time.to.Dead.Hr)), 48.00)) %>%
dplyr::mutate(cohort = "cohort2")
#truncated greater than 24 hrs as missing
do.pheno2 <- do.pheno2 %>%
dplyr::mutate(Time.to.Dead.Hr = ifelse(Time.to.Dead.Hr > 24, NA_real_, Time.to.Dead.Hr)) %>%
dplyr::mutate(SteadyState.Depression.Duration.Hr = ifelse(SteadyState.Depression.Duration.Hr > 24, NA_real_, SteadyState.Depression.Duration.Hr)) %>%
dplyr::mutate(Time.to.Threshold.Recovery.Hr = ifelse(Time.to.Threshold.Recovery.Hr > 24, NA_real_, Time.to.Threshold.Recovery.Hr)) %>%
dplyr::mutate(Time.to.Projected.Recovery.Hr = ifelse(Time.to.Projected.Recovery.Hr > 24, NA_real_, Time.to.Projected.Recovery.Hr)) %>%
dplyr::mutate(Start.of.Recovery.Hr = ifelse(Start.of.Recovery.Hr > 24, NA_real_, Start.of.Recovery.Hr))
#merge with do.pheno
do.pheno <- bind_rows(do.pheno1, do.pheno2)
#data point
table(do.pheno[is.na(do.pheno$Time.to.Dead.Hr), ]$cohort)
#
# cohort1 cohort2
# 212 138
# Read genotype data -----------------------------------------------------------
load("data/Jackson_Lab_Bubier_MURGIGV01/gm_DO765_qc_newid.RData")#gm_after_qc
overlap.id = intersect(ind_ids(gm_after_qc), as.character(do.pheno$MouseID))
#subset
gm = gm_after_qc[overlap.id, ]
do.pheno = do.pheno[do.pheno$MouseID %in% overlap.id, ]
do.pheno = do.pheno[match(ind_ids(gm), do.pheno$MouseID) , ]
rownames(do.pheno) = do.pheno$MouseID
all.equal(ind_ids(gm), do.pheno$MouseID)
# [1] TRUE
#covar
do.pheno$Sex <- as.factor(do.pheno$Sex)
#addcovar
options(na.action='na.pass')
addcovar = model.matrix(~Sex, data = do.pheno[, c("Sex"), drop=F])[,-1, drop=F]
colnames(addcovar) <- c("sex")
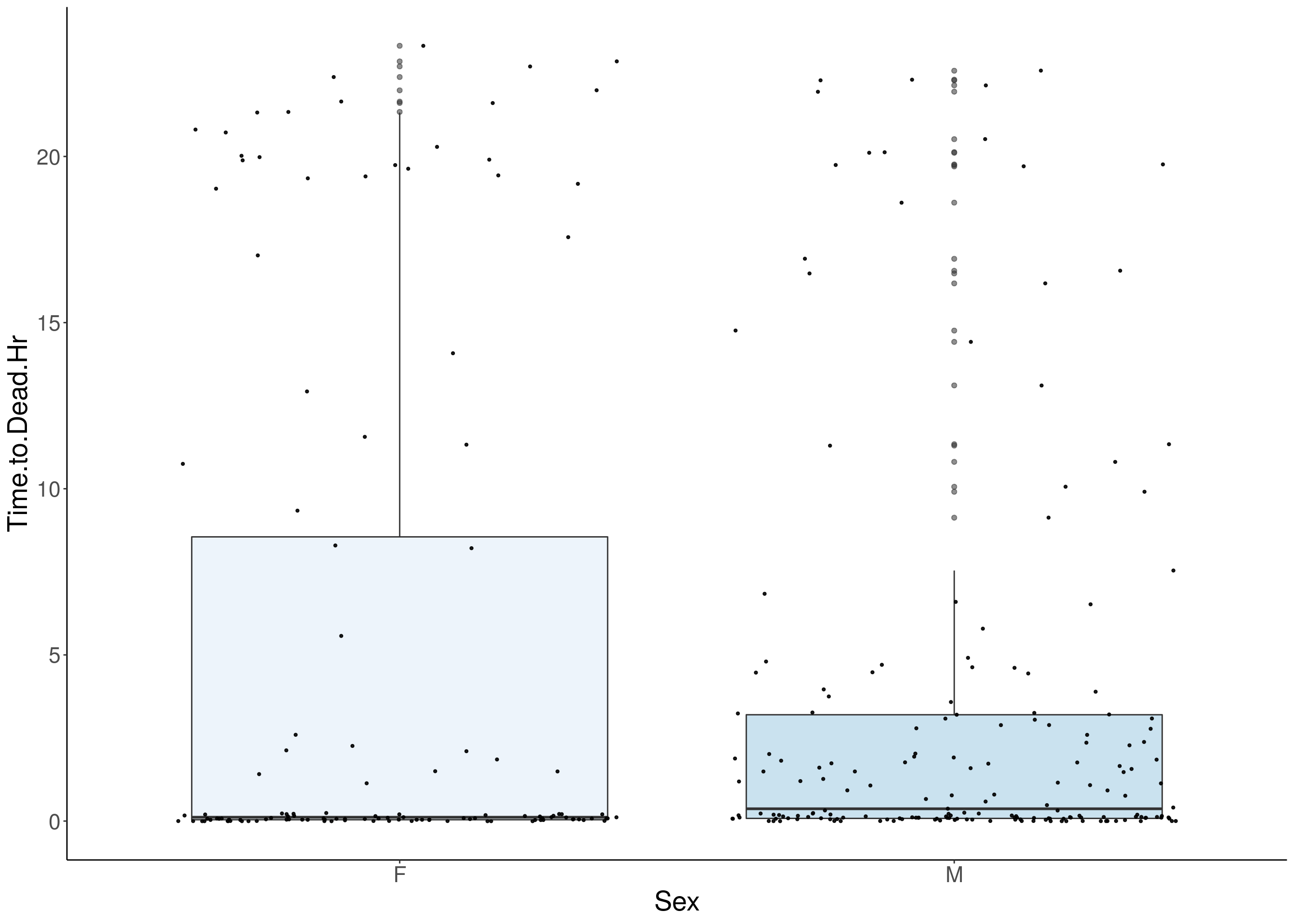
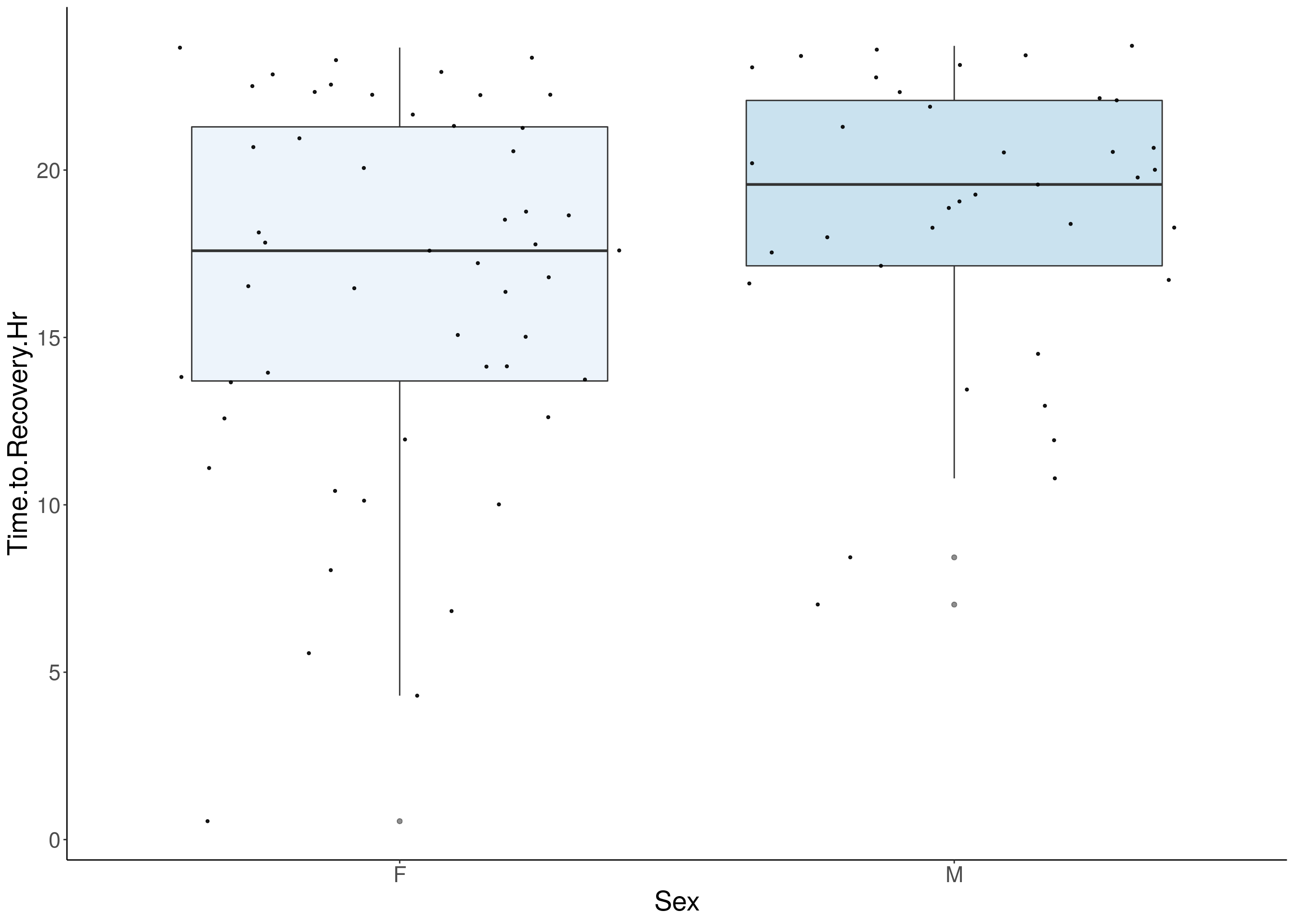
#boxplot on the raw data------------
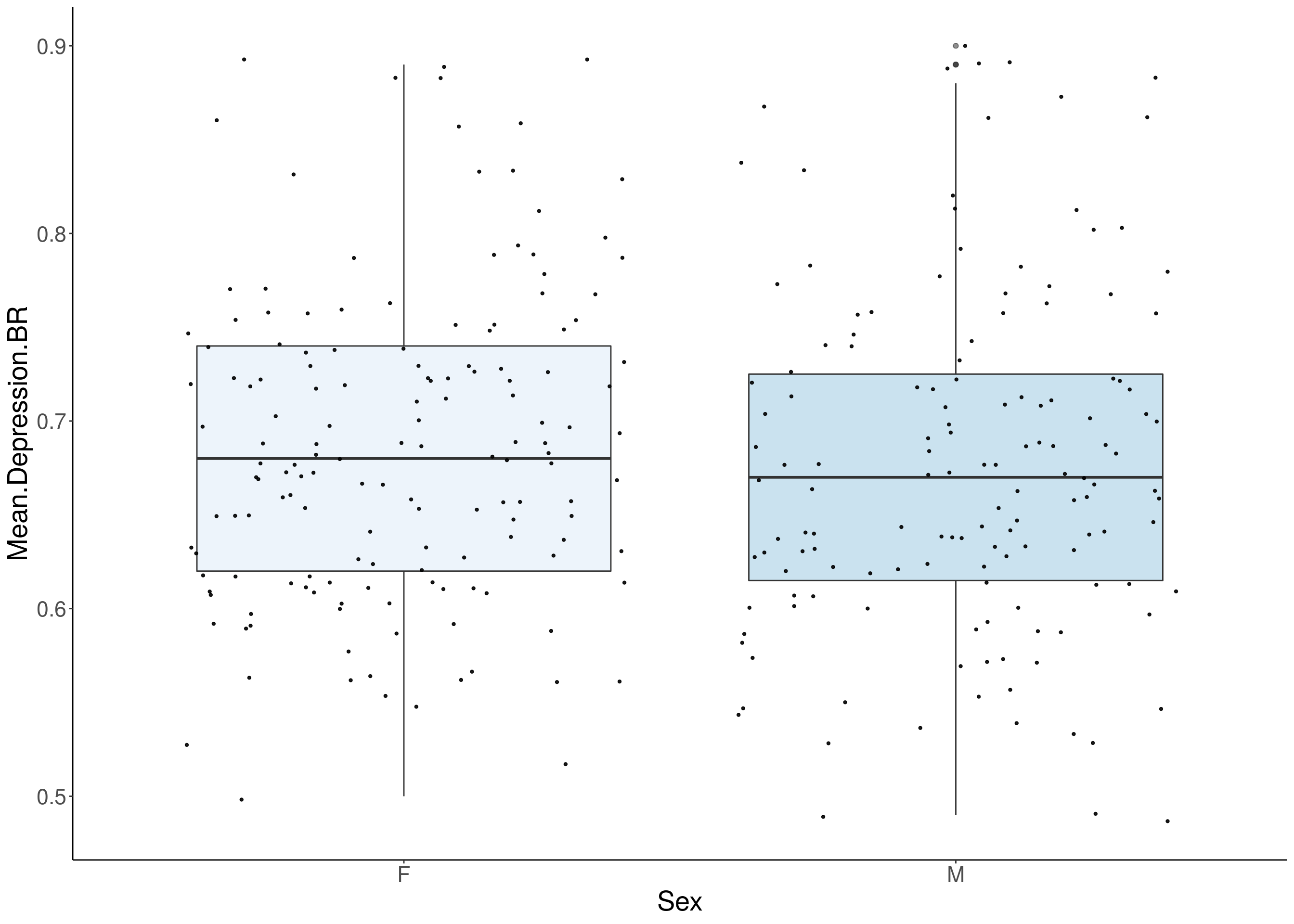
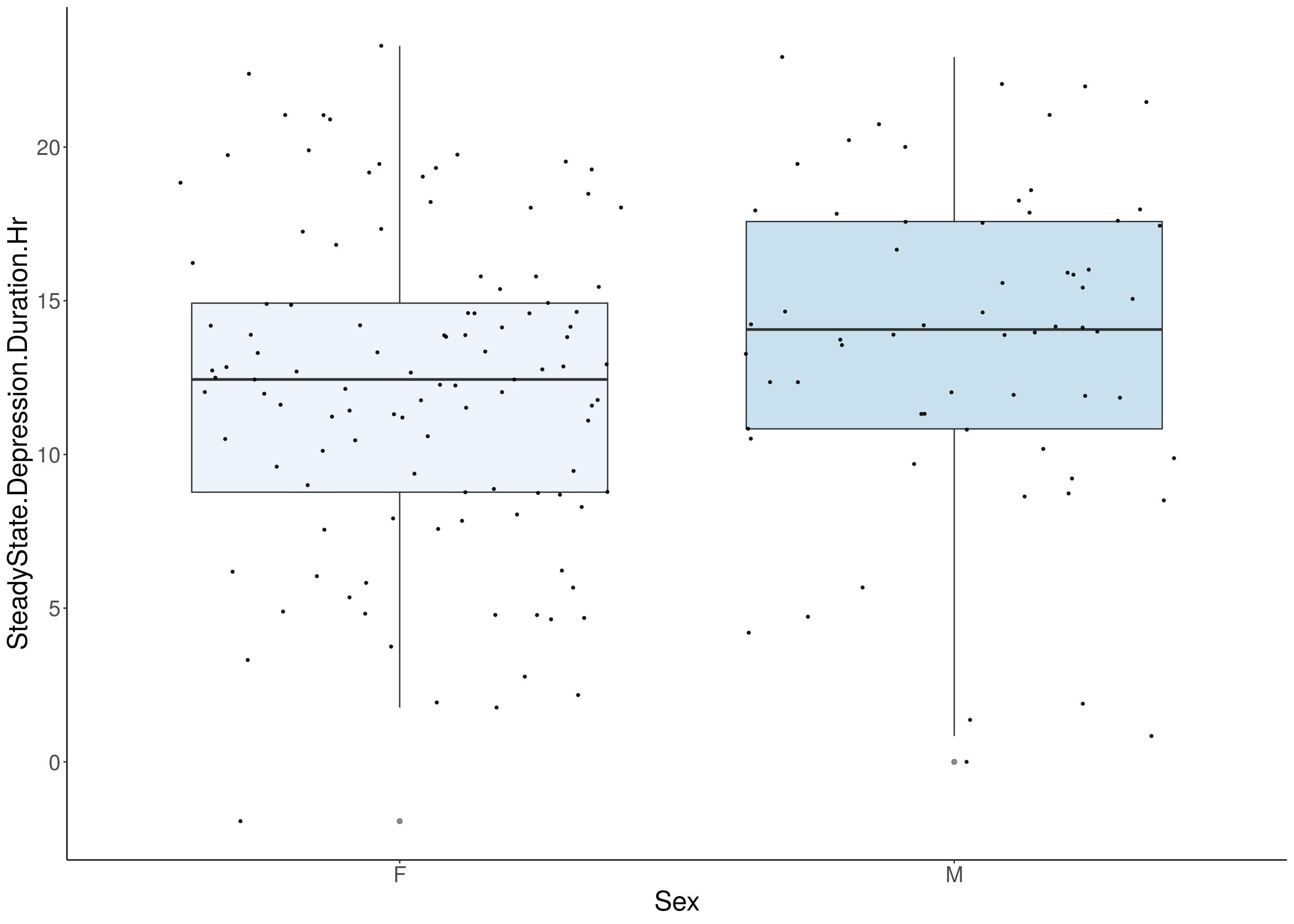
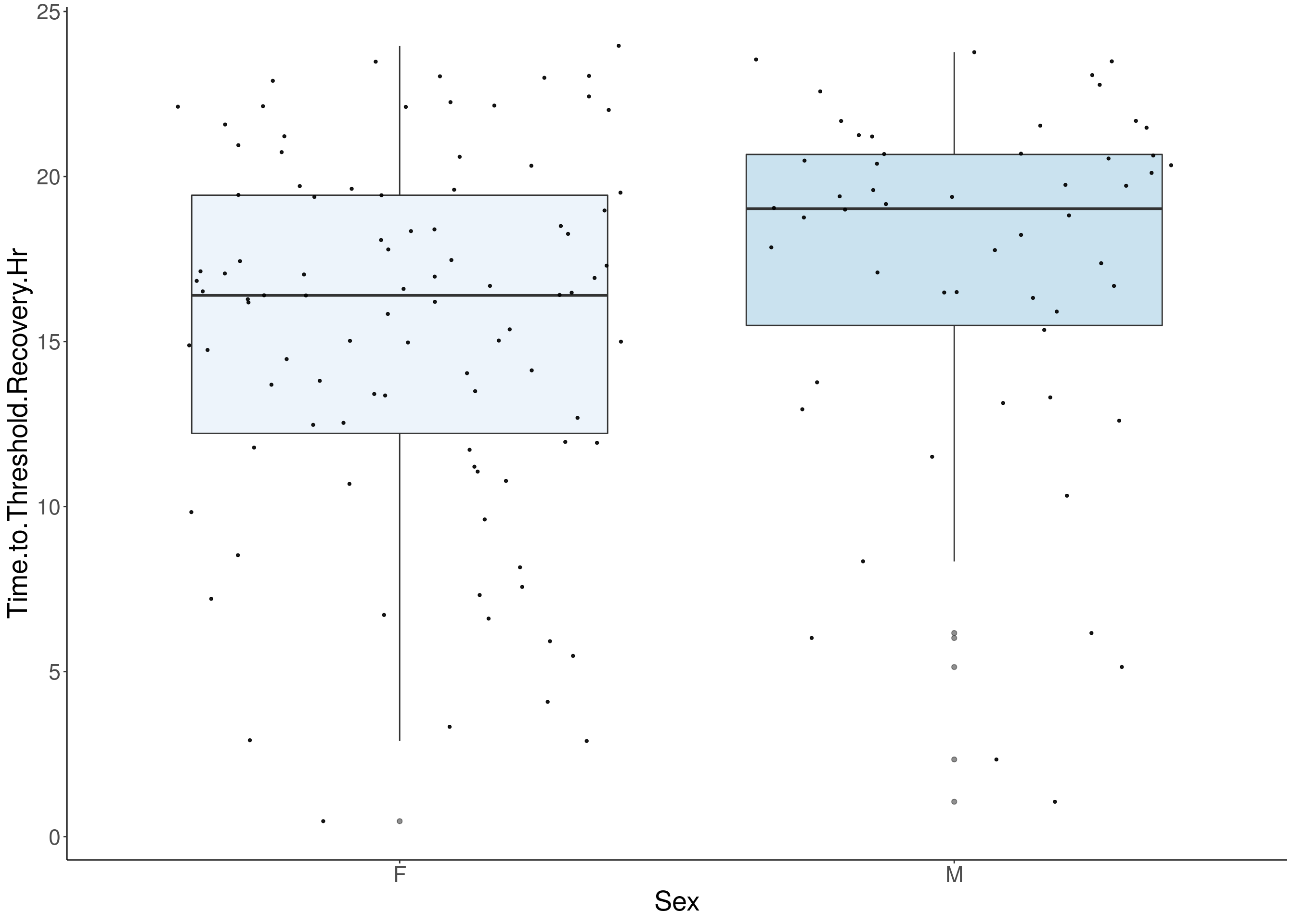
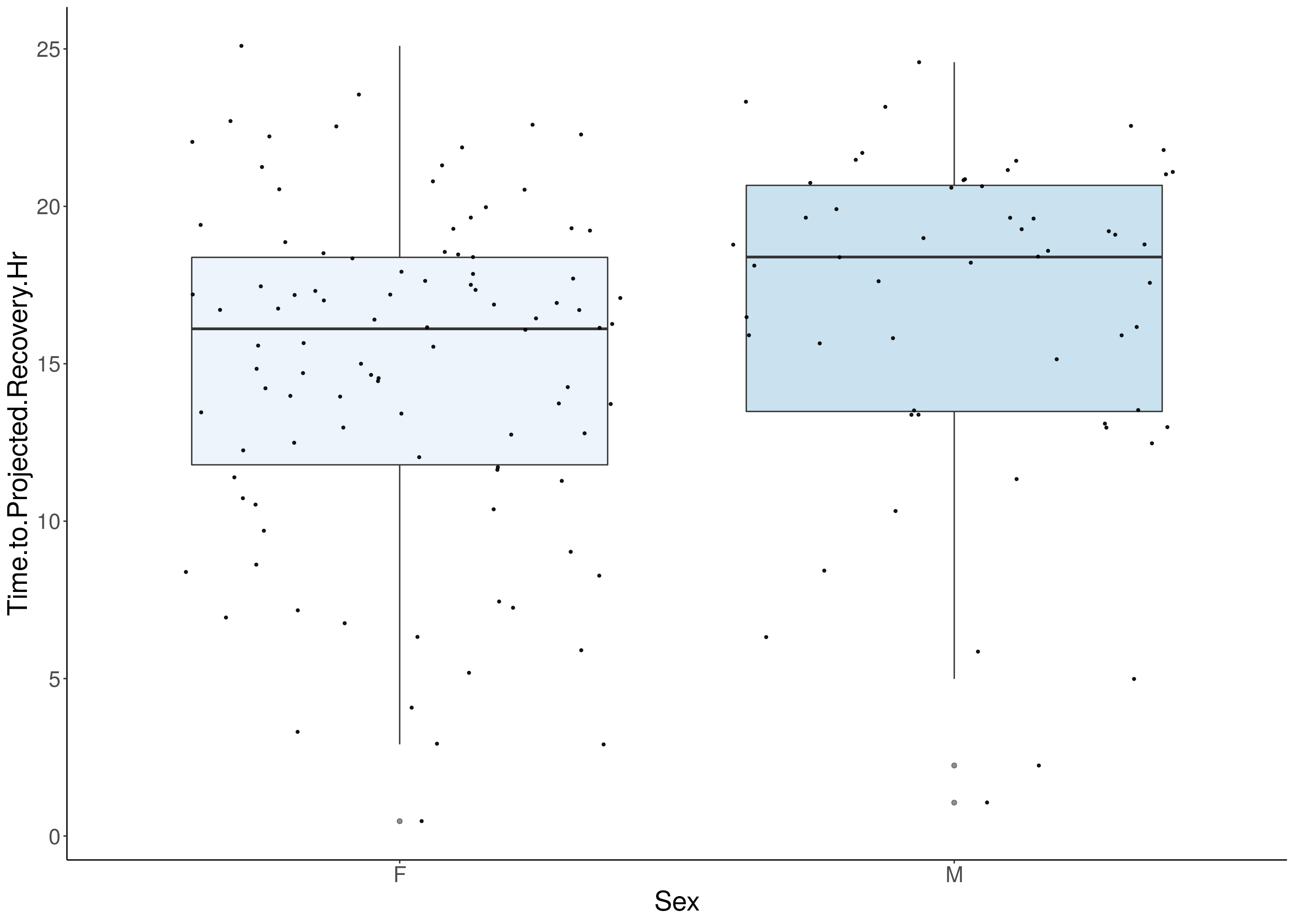
for(i in 5:17){
df <- do.pheno[,c(1,2,i)]
df <- df[complete.cases(df), ]
p <- ggplot(df, aes(x=Sex, y=df[,3], group = Sex, fill = Sex, alpha = 0.9)) +
geom_boxplot(show.legend = F , outlier.size = 1.5, notchwidth = 0.85) +
geom_jitter(color="black", size=0.8, alpha=0.9) +
scale_fill_brewer(palette="Blues") +
ylab(paste0(colnames(df)[3])) +
xlab("Sex") +
labs(fill = "") +
theme(legend.position = "none",
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
axis.line = element_line(colour = "black"),
text = element_text(size=21),
axis.title=element_text(size=21)) +
guides(shape = guide_legend(override.aes = list(size = 12)))
print(p)
}
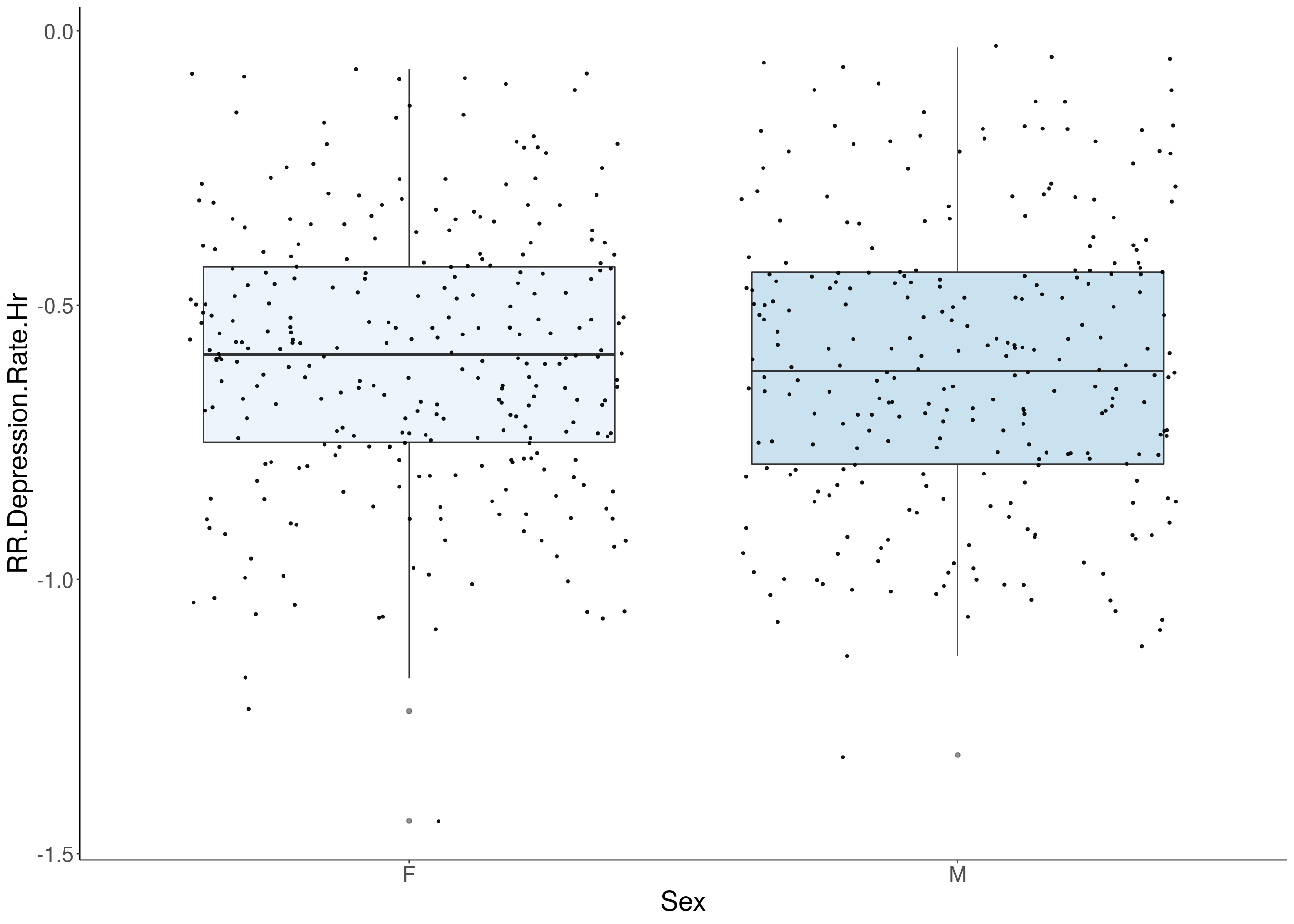
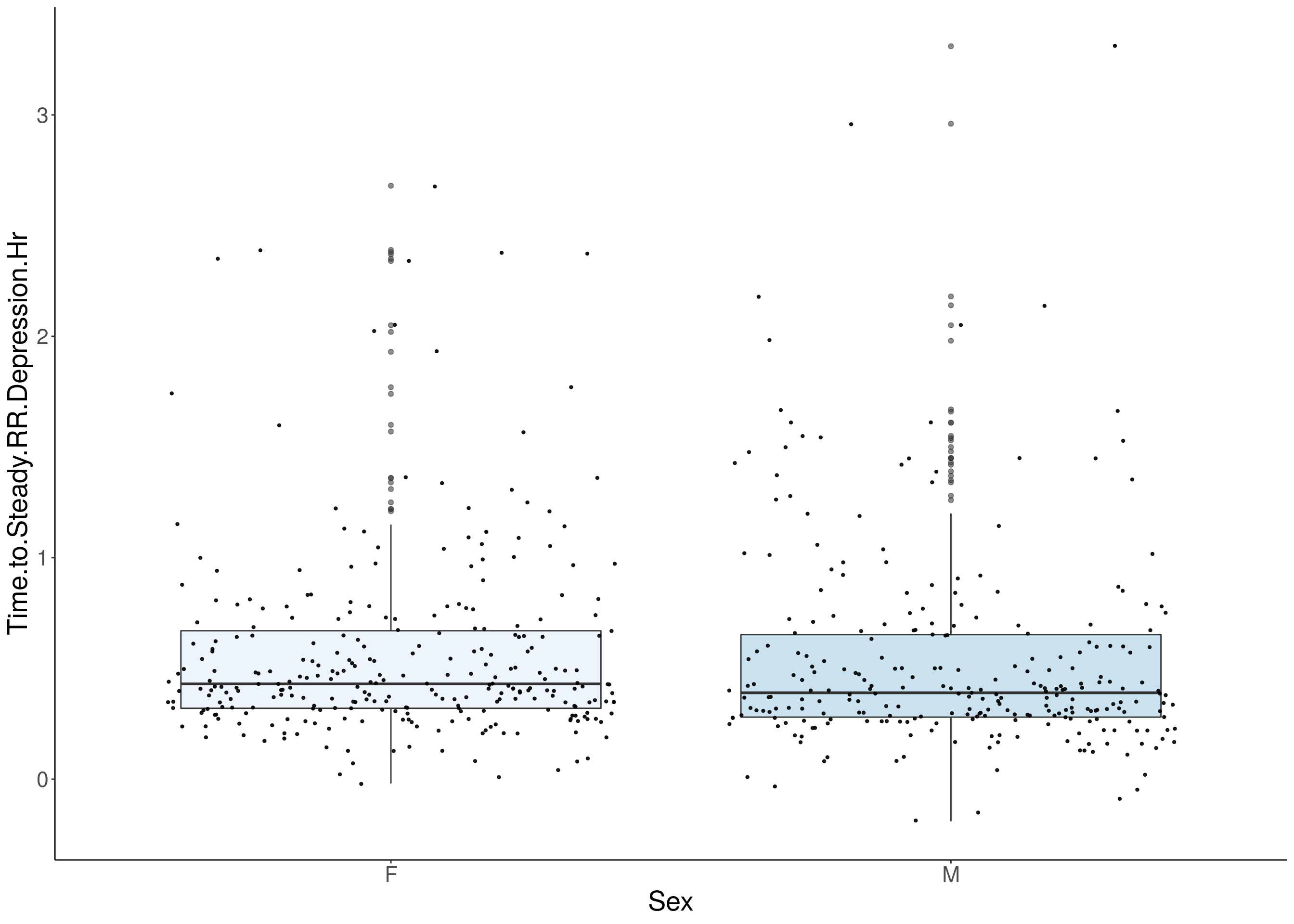
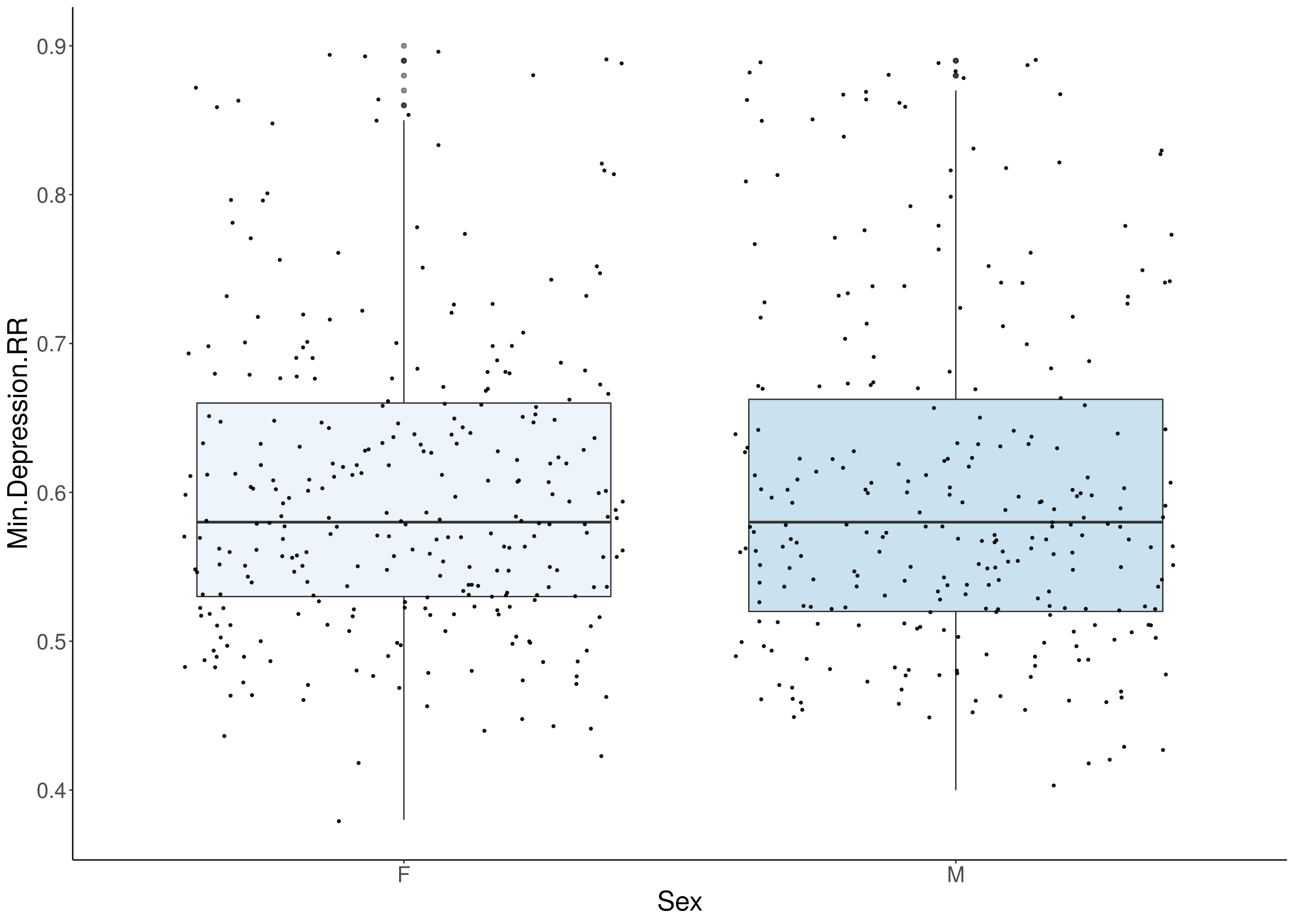
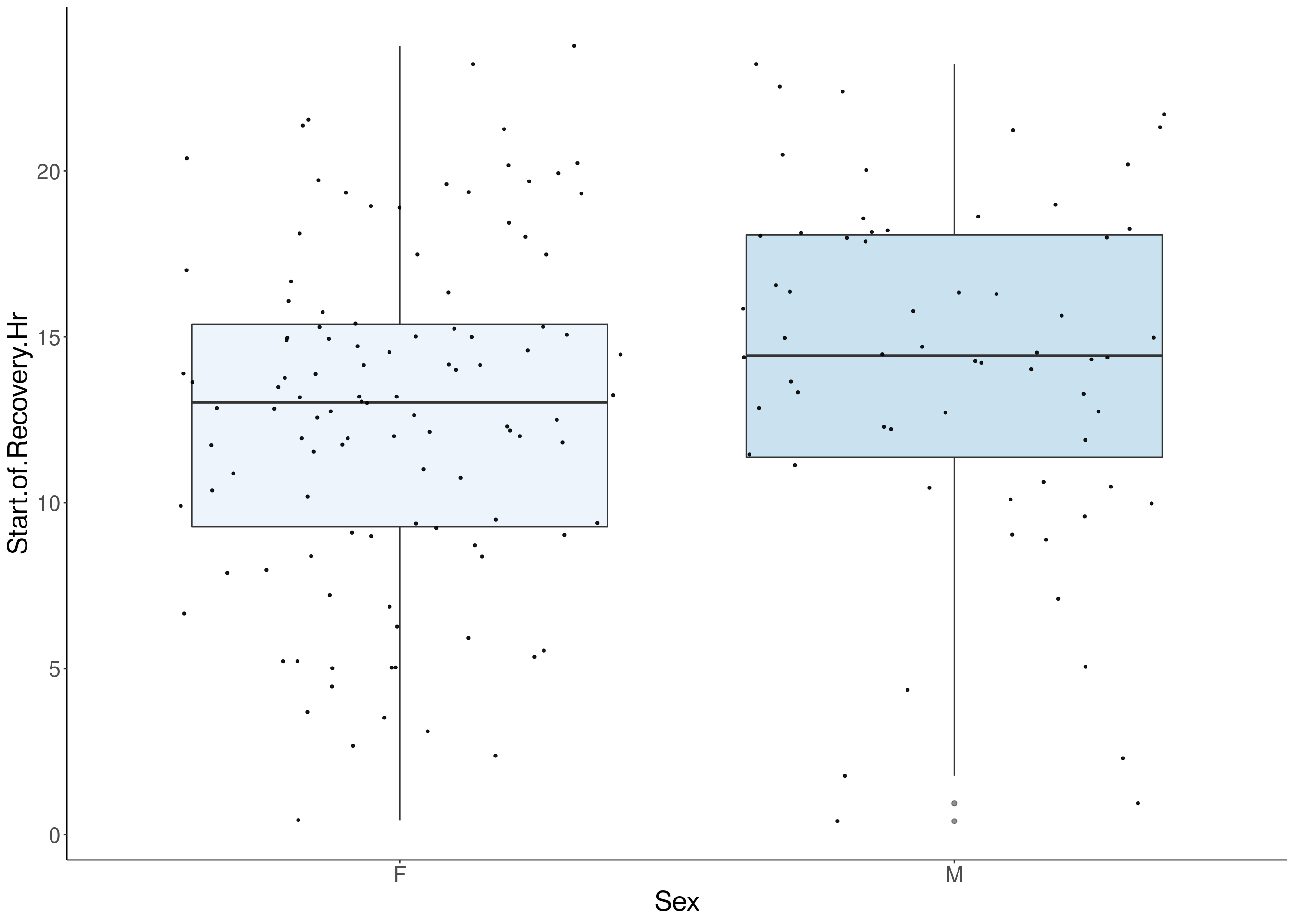
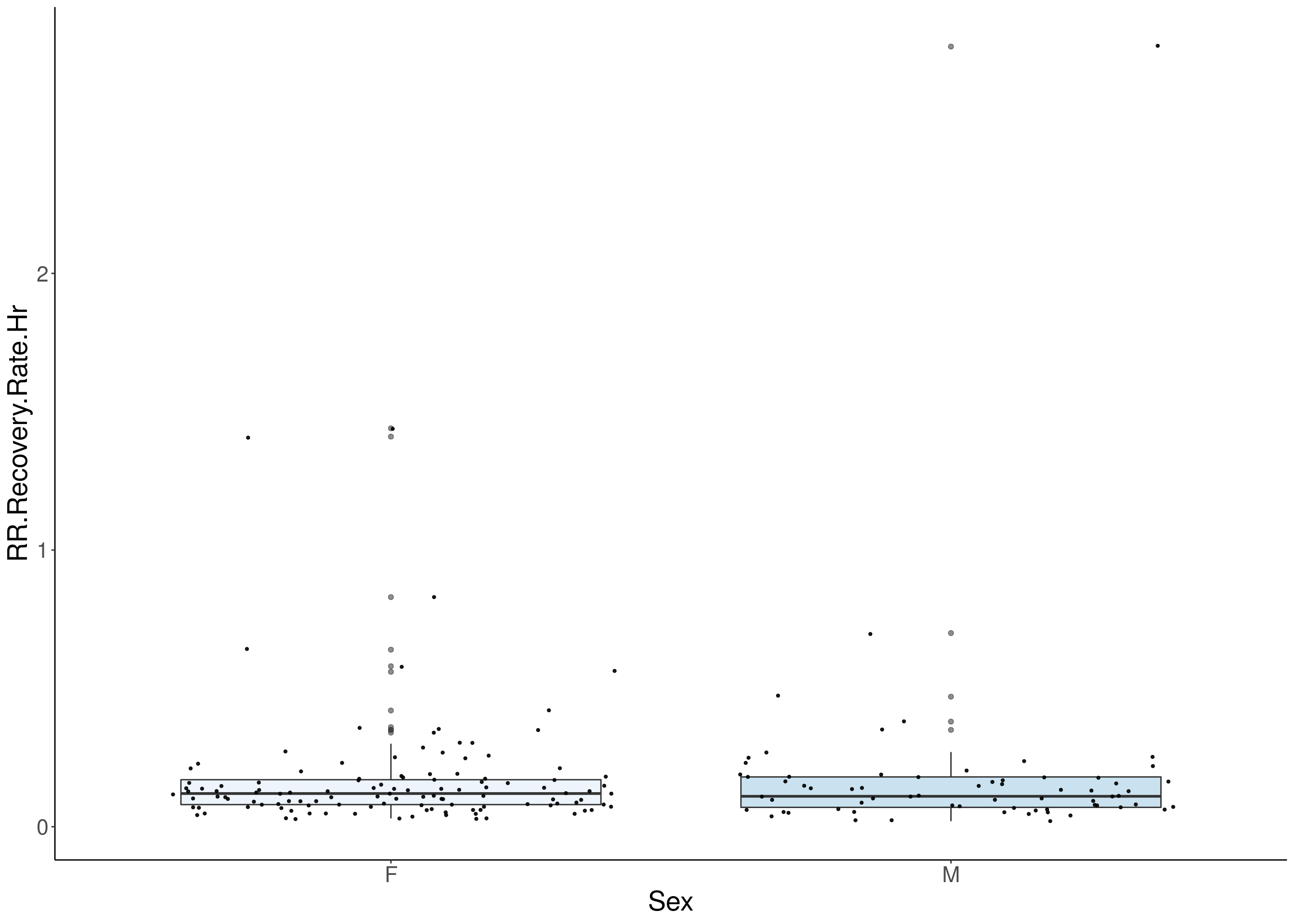
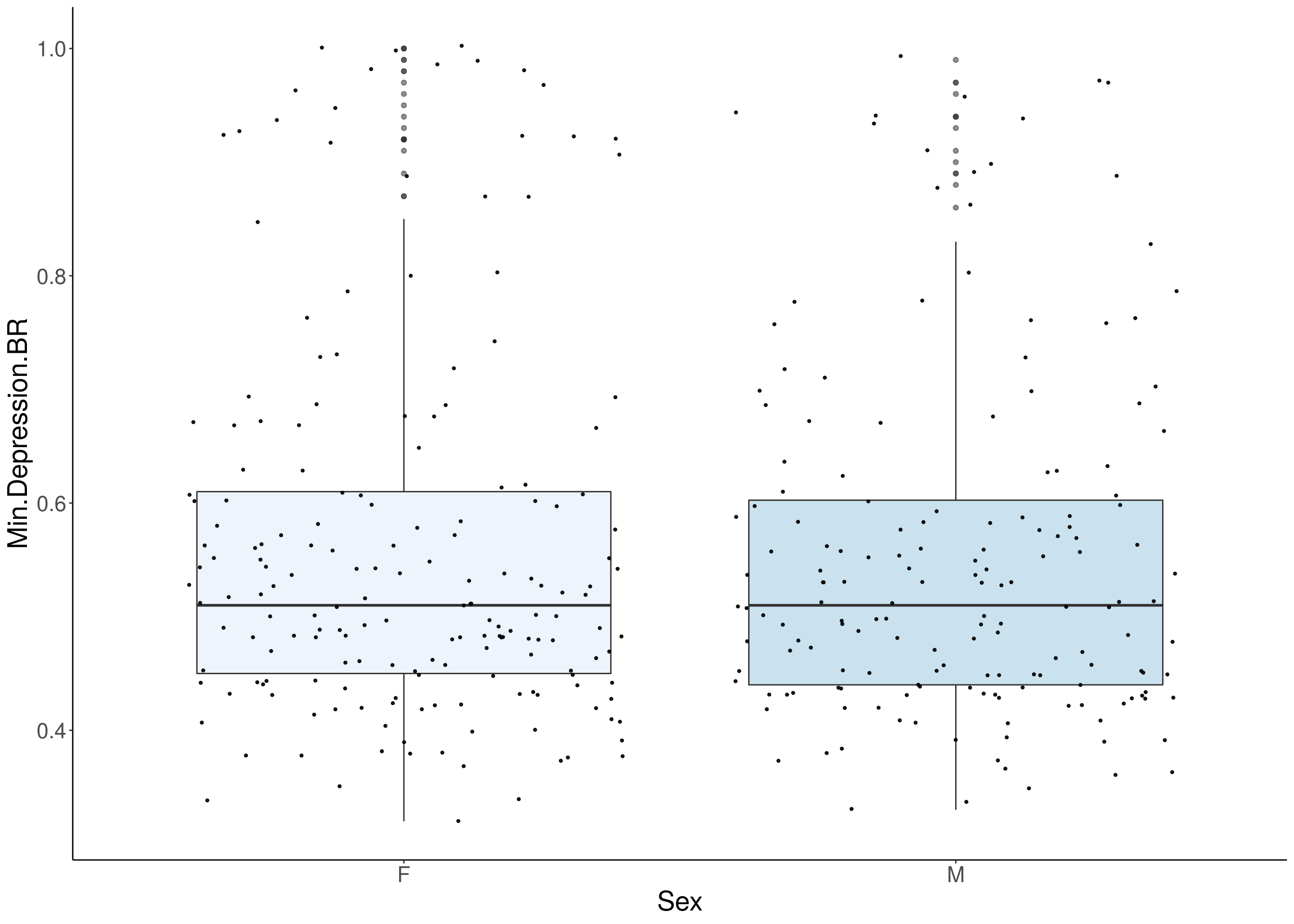
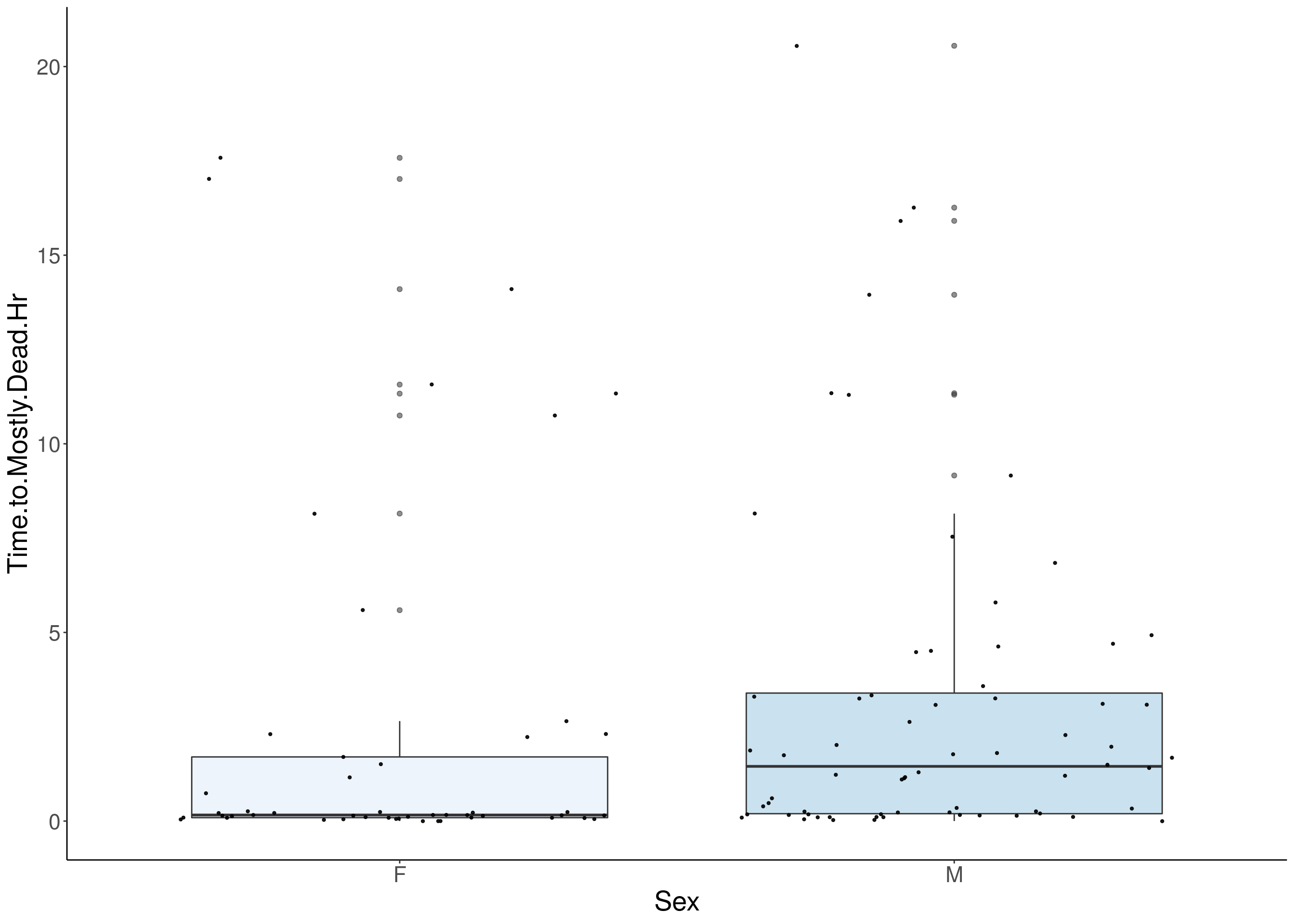
#histogram
for(i in 5:17){
df <- do.pheno[,c(1,2,i)]
p <- ggplot(data=df, aes(x=df[,3])) +
geom_histogram() +
ylab("Number of DO mice") +
xlab(paste0(colnames(df)[3])) +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
panel.background = element_blank(), axis.line = element_line(colour = "black"),
text = element_text(size=21))
print(p)
}
# Warning: Removed 337 rows containing non-finite values (stat_bin).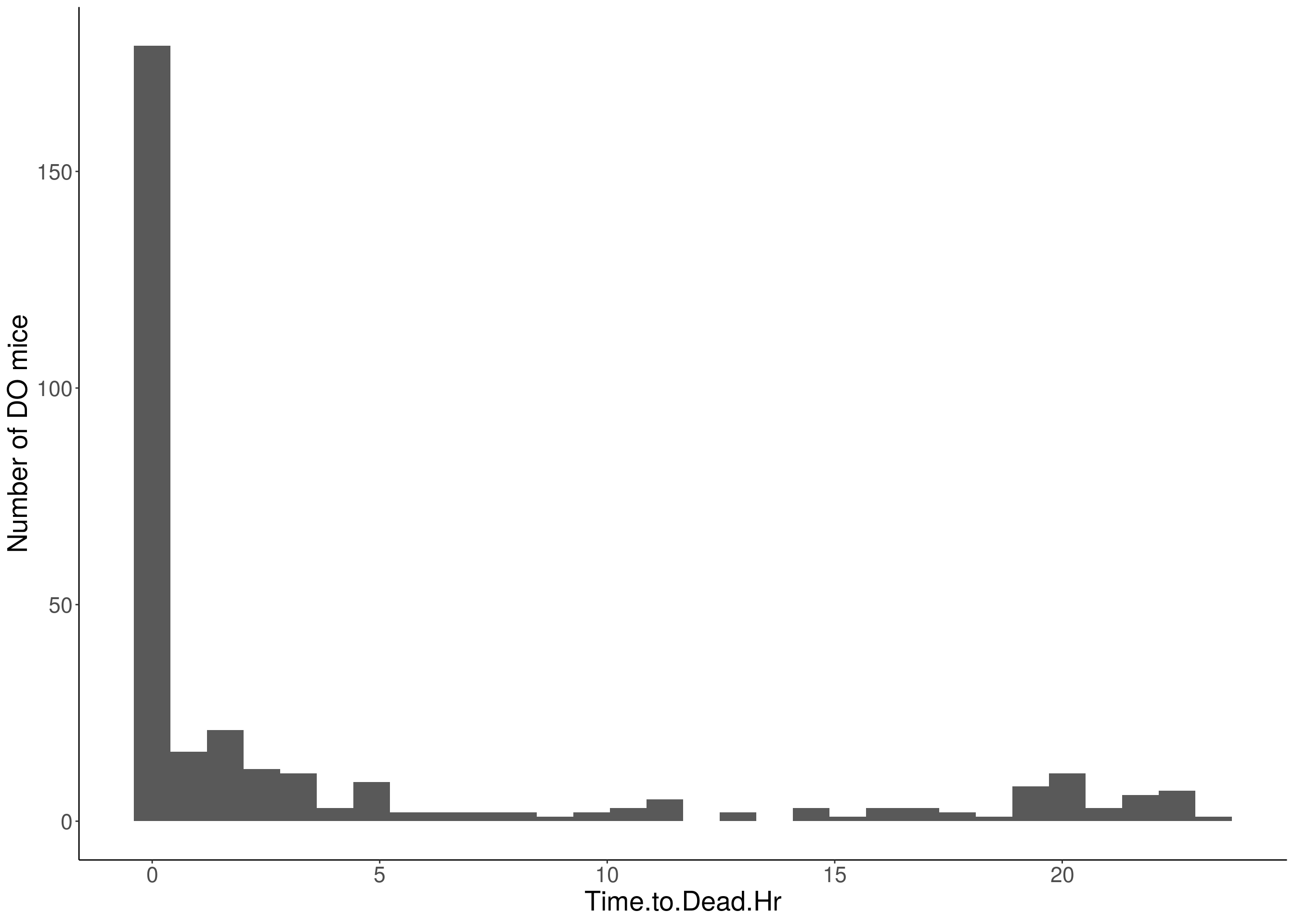
# Warning: Removed 85 rows containing non-finite values (stat_bin).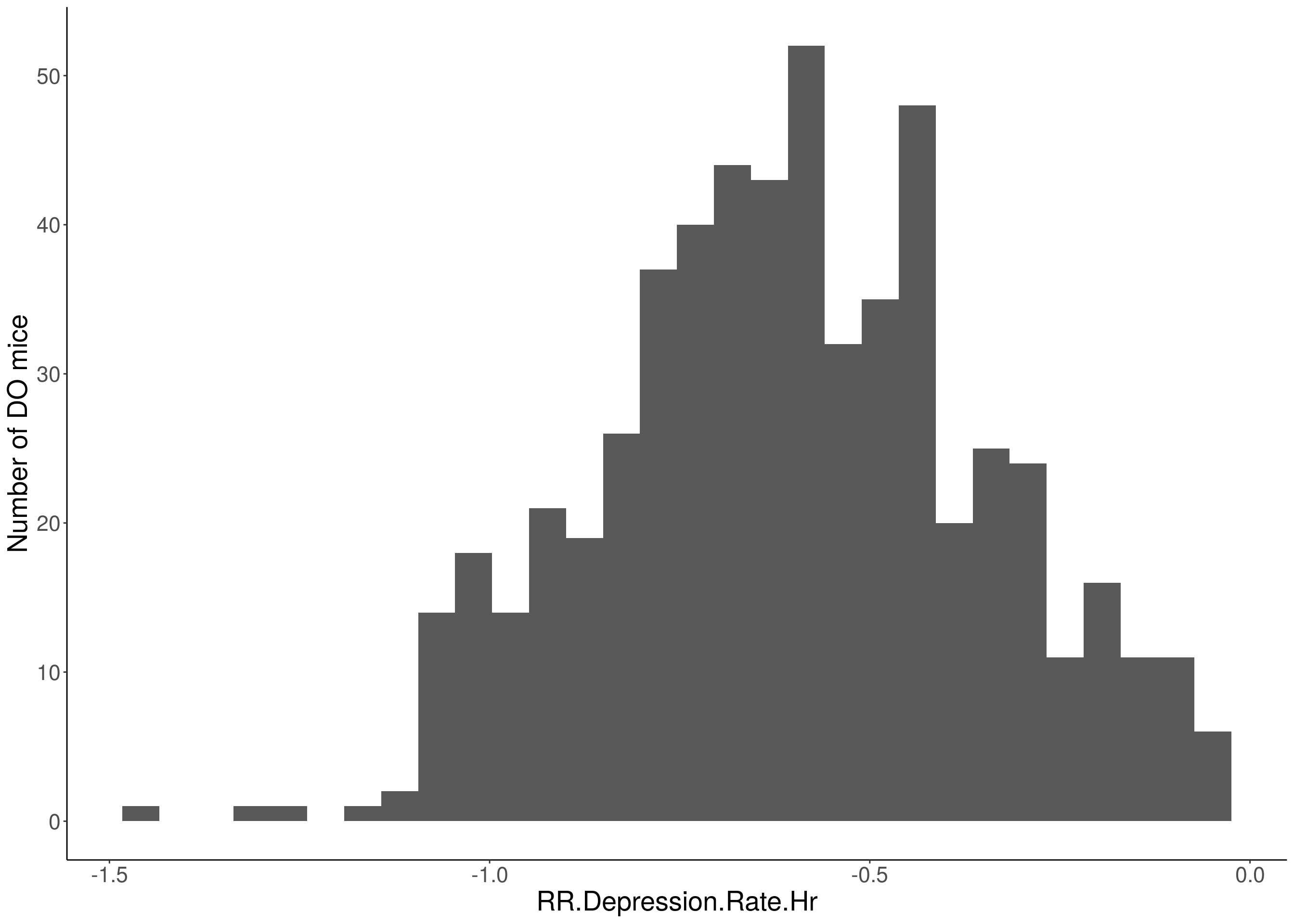
# Warning: Removed 85 rows containing non-finite values (stat_bin).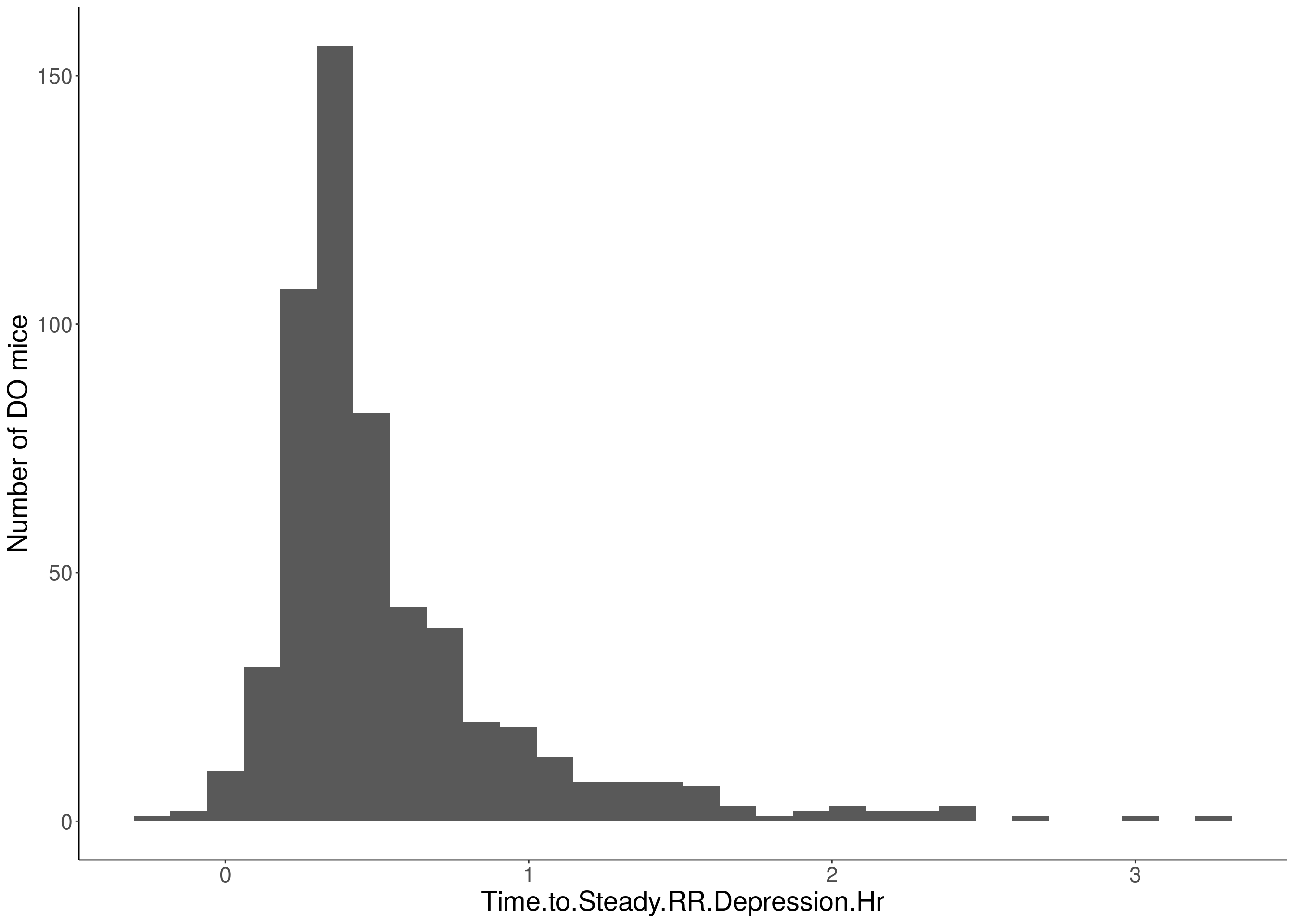
# Warning: Removed 85 rows containing non-finite values (stat_bin).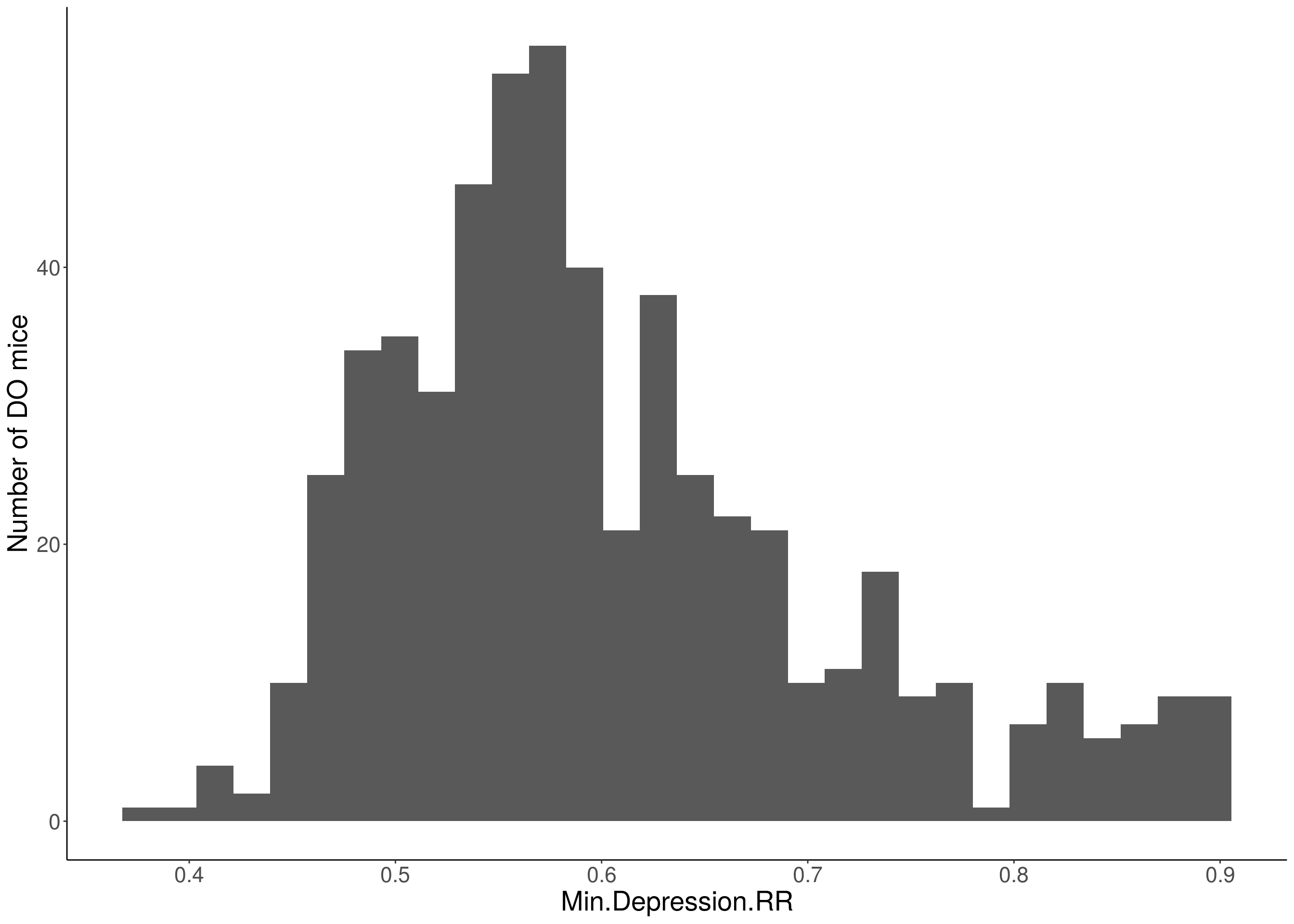
# Warning: Removed 355 rows containing non-finite values (stat_bin).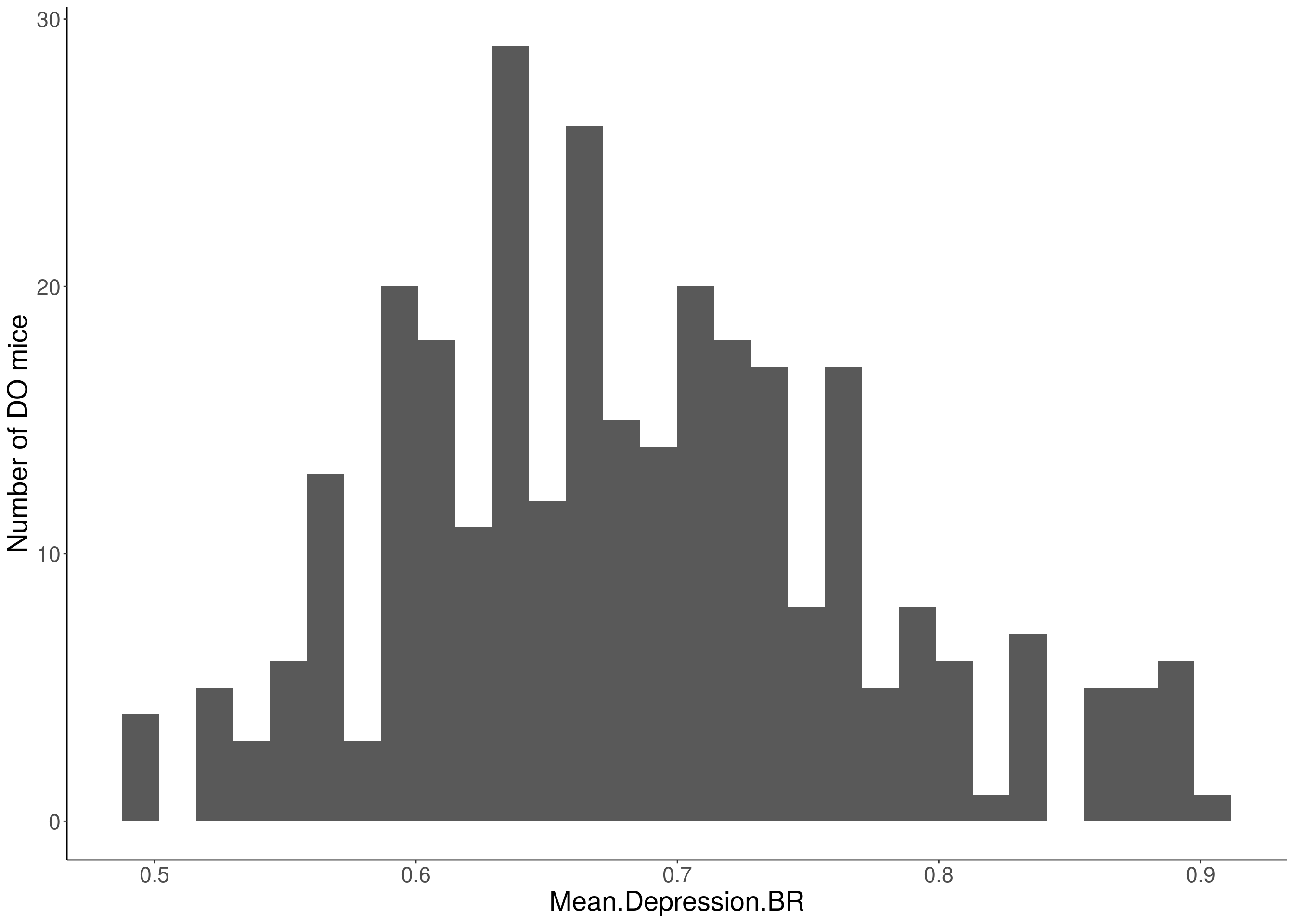
# Warning: Removed 484 rows containing non-finite values (stat_bin).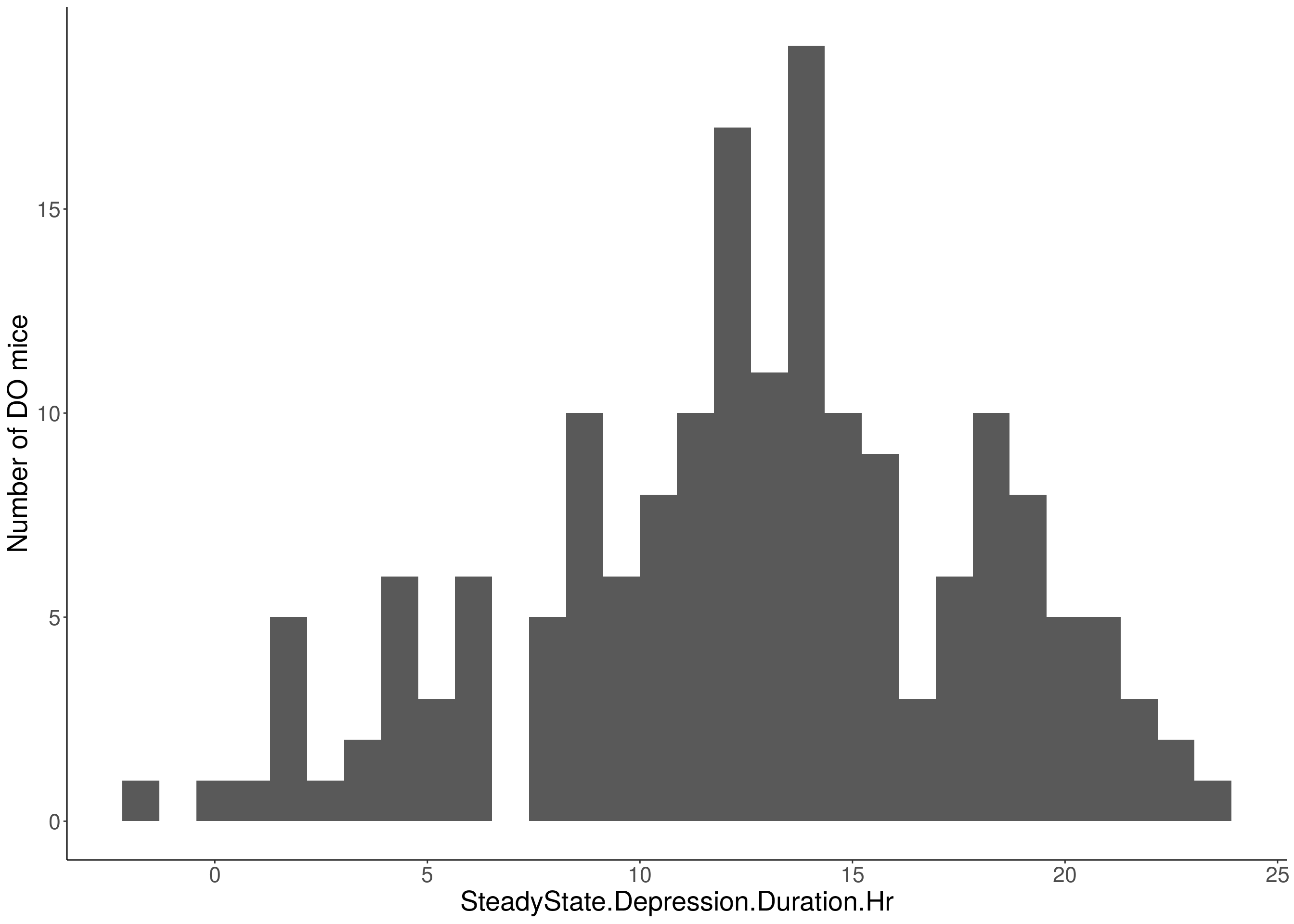
# Warning: Removed 509 rows containing non-finite values (stat_bin).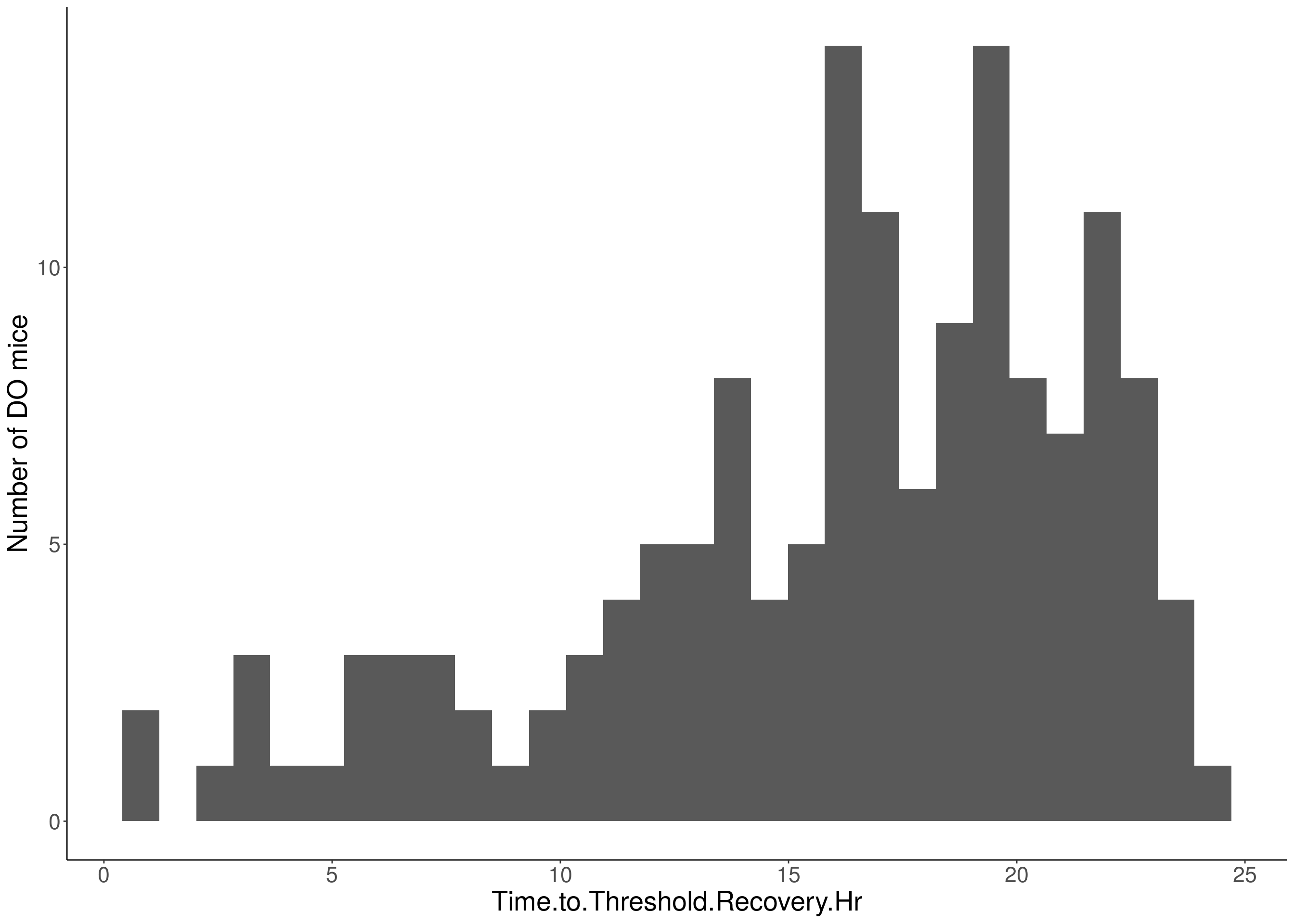
# Warning: Removed 504 rows containing non-finite values (stat_bin).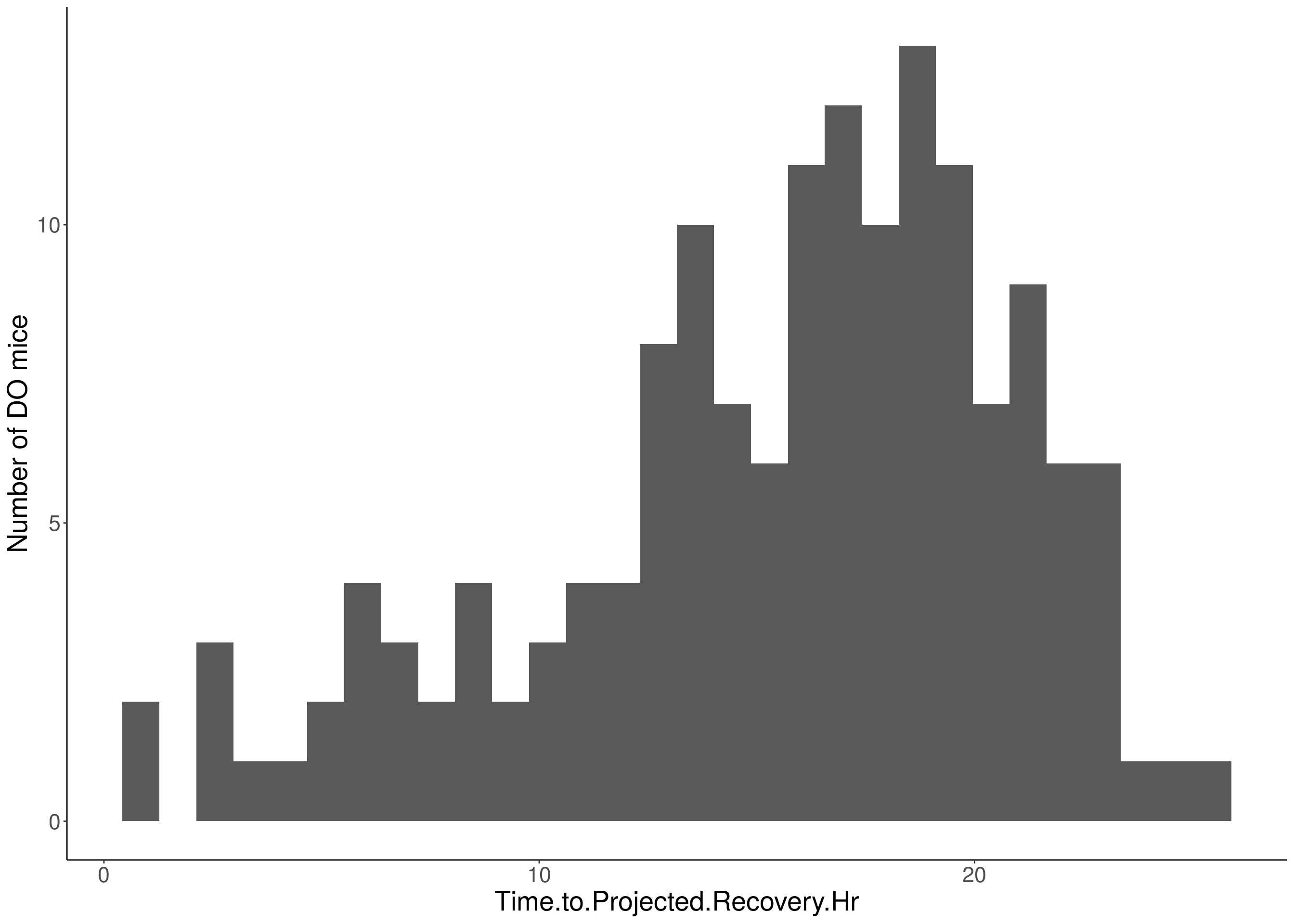
# Warning: Removed 484 rows containing non-finite values (stat_bin).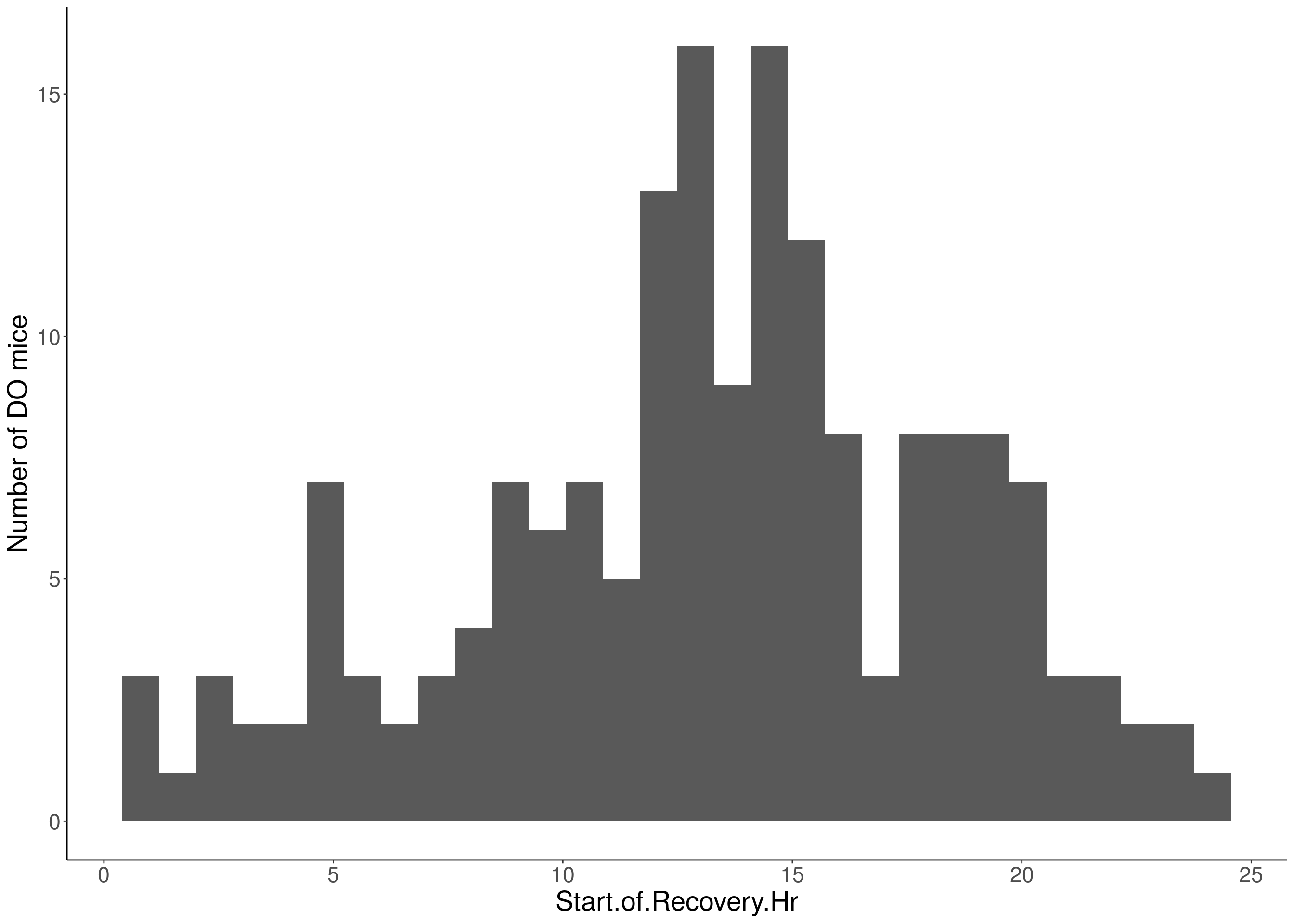
# Warning: Removed 469 rows containing non-finite values (stat_bin).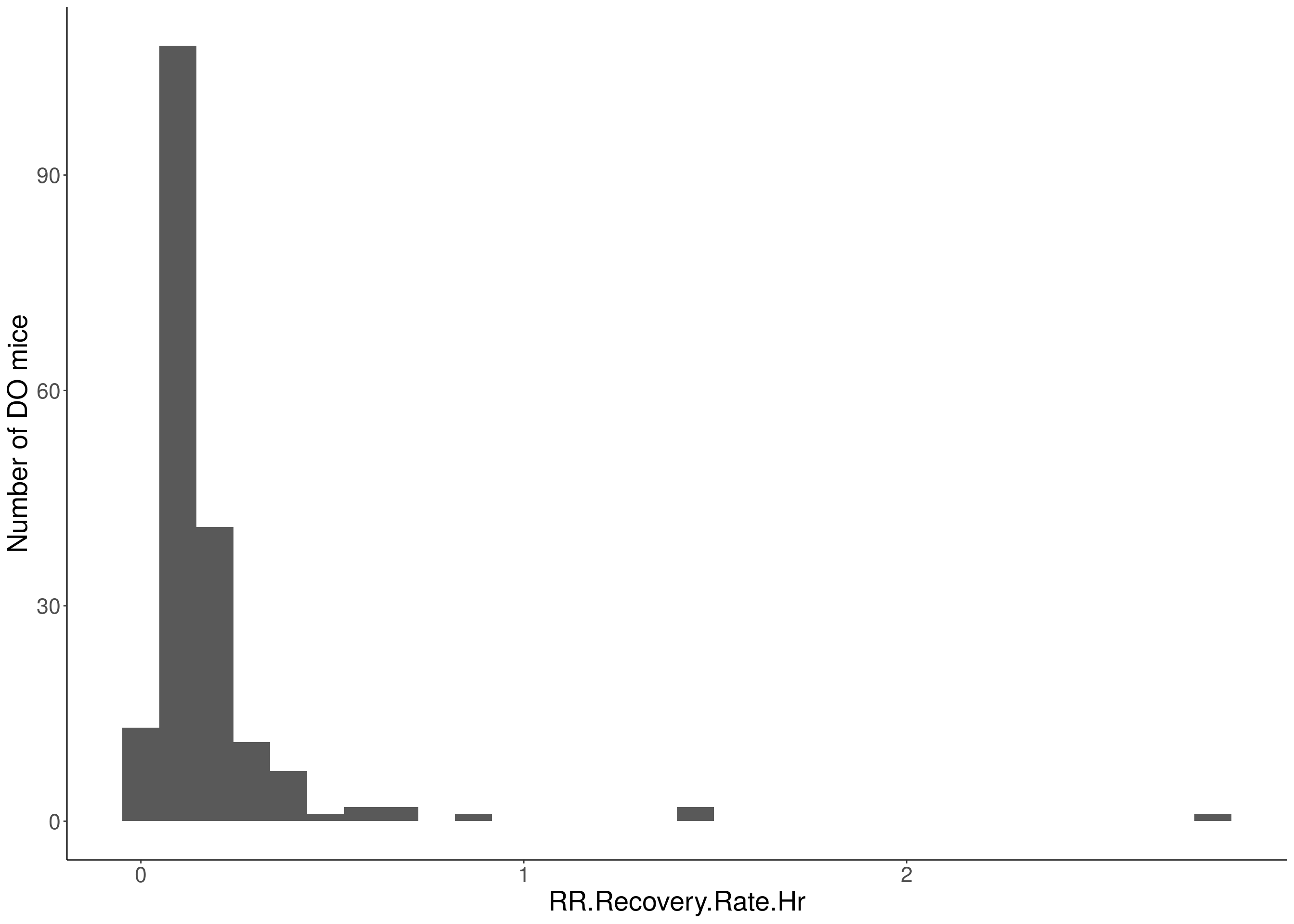
# Warning: Removed 307 rows containing non-finite values (stat_bin).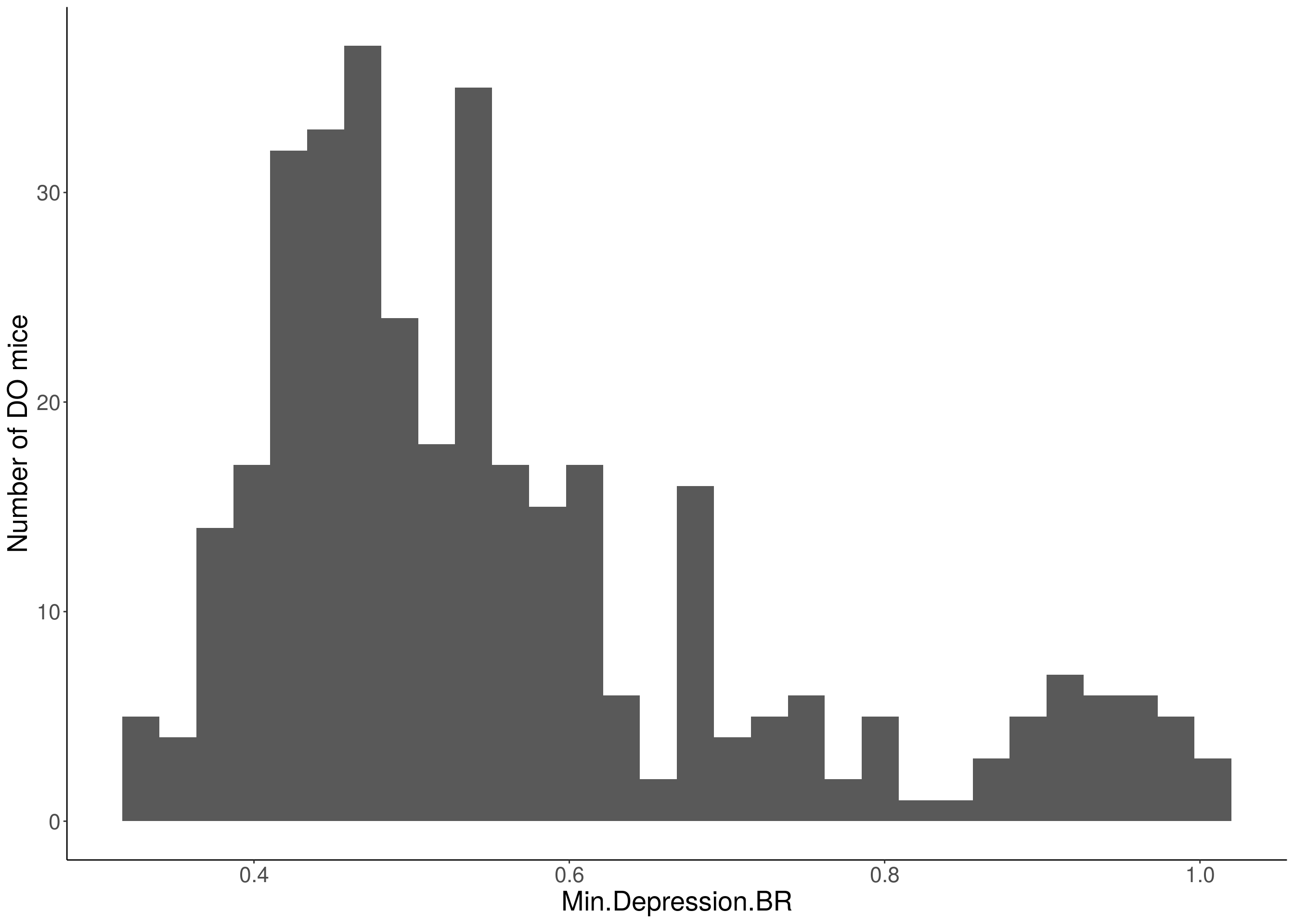
# Warning: Removed 541 rows containing non-finite values (stat_bin).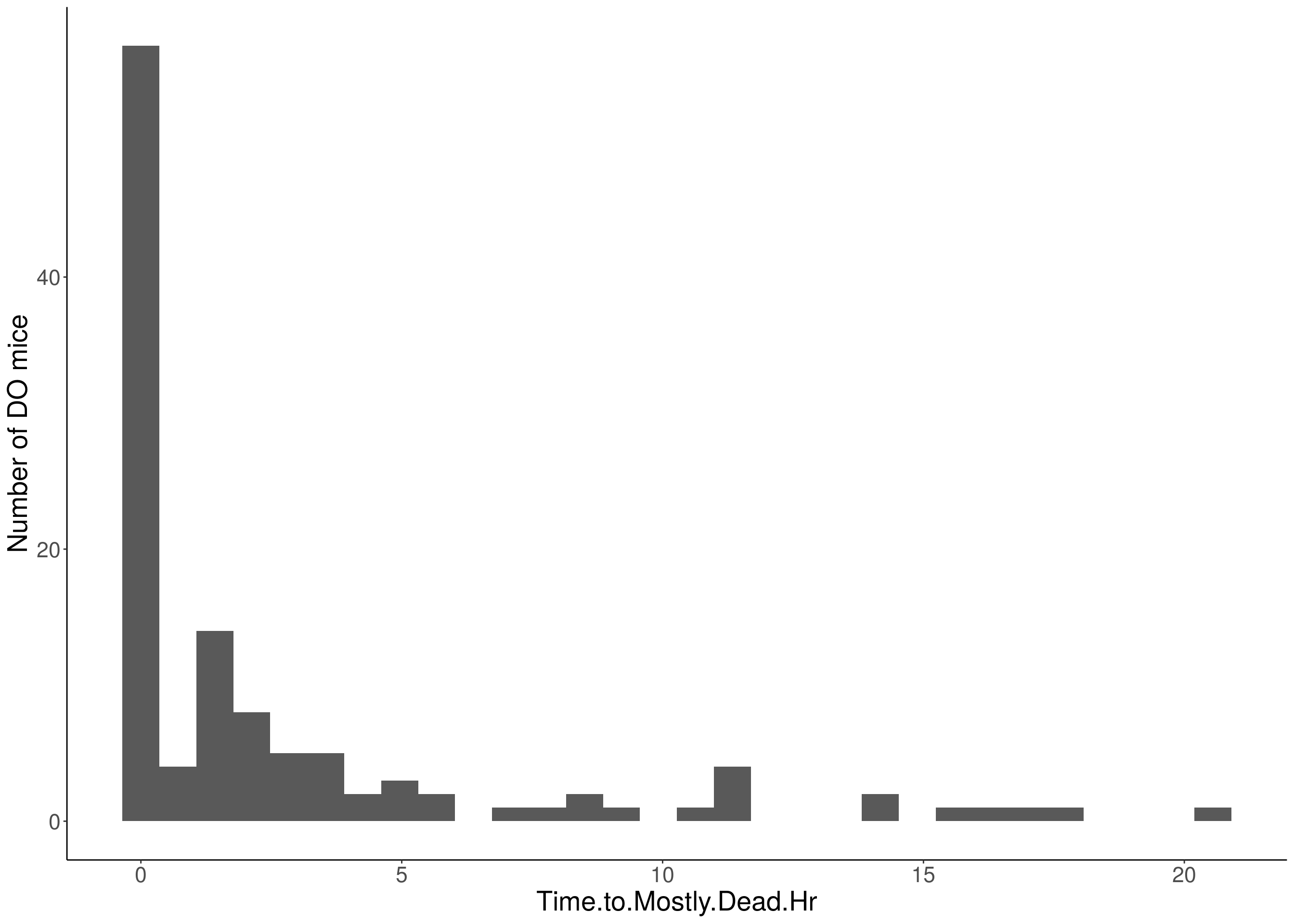
# Warning: Removed 570 rows containing non-finite values (stat_bin).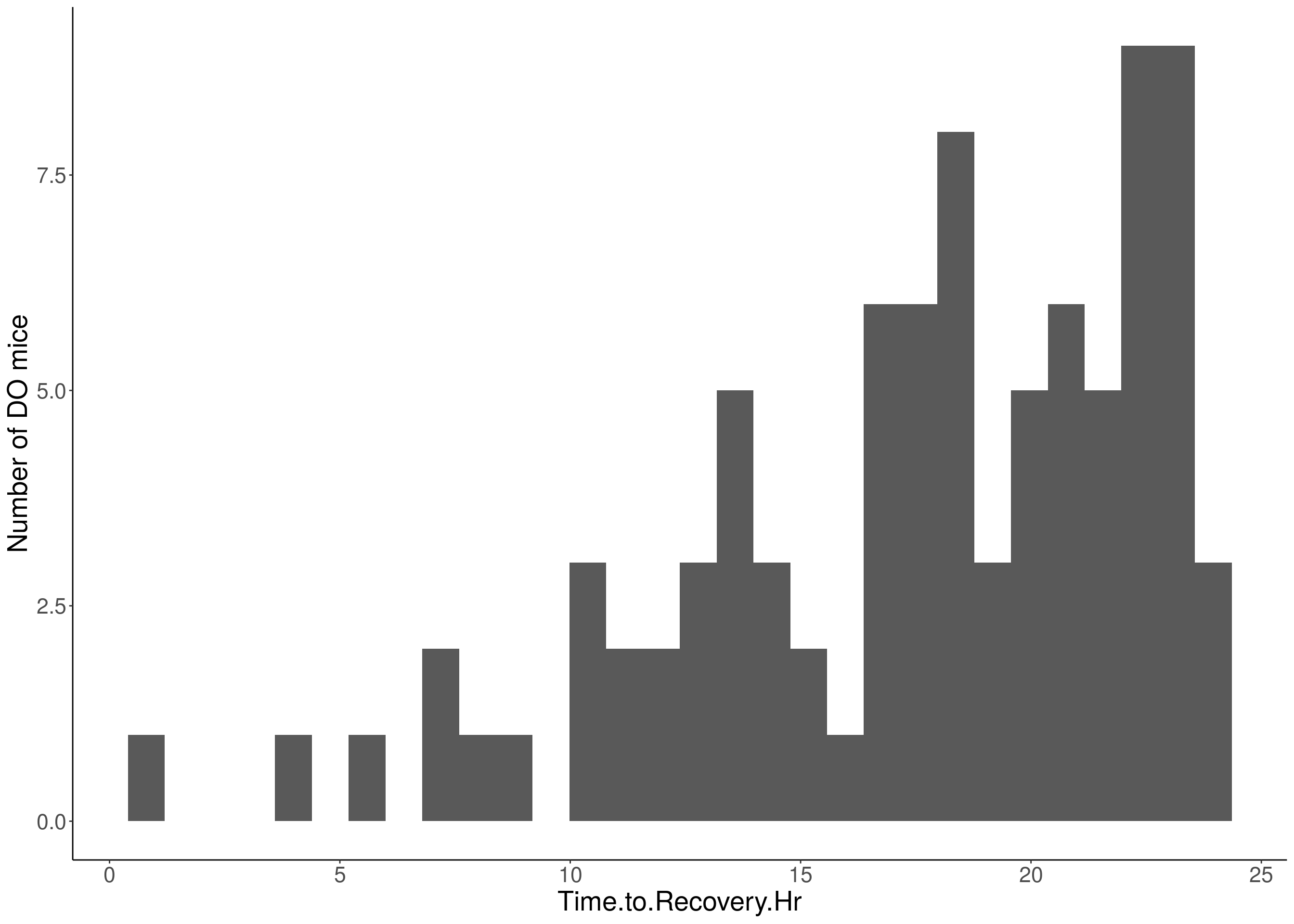
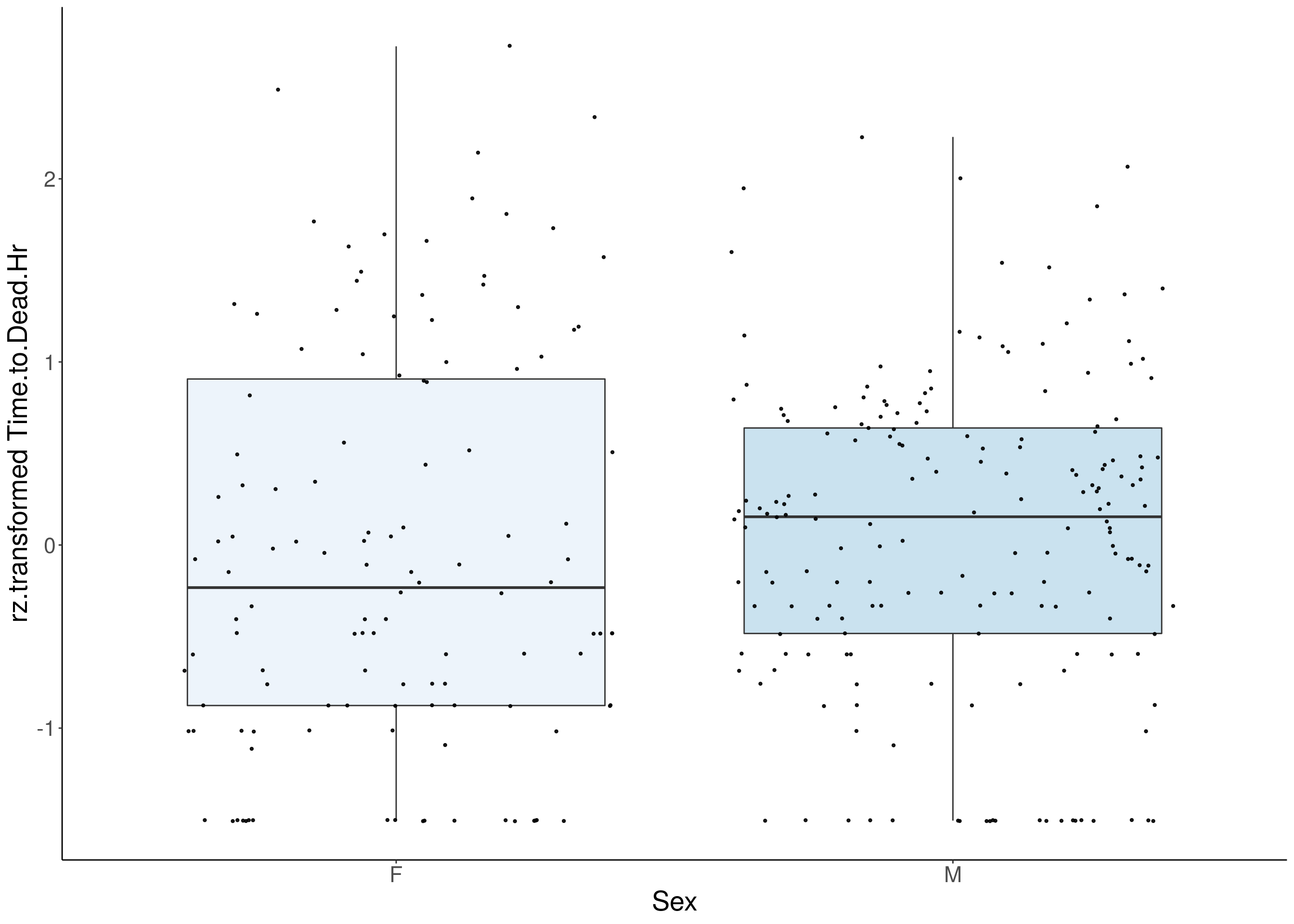
#boxplot on the rankz data------------
for(i in 5:17){
df <- do.pheno[,c(1,2,i)]
df <- df[complete.cases(df), ]
p <- ggplot(df, aes(x=Sex, y= rz.transform(df[,3]), group = Sex, fill = Sex, alpha = 0.9)) +
geom_boxplot(show.legend = F , outlier.size = 1.5, notchwidth = 0.85) +
geom_jitter(color="black", size=0.8, alpha=0.9) +
scale_fill_brewer(palette="Blues") +
ylab(paste0("rz.transformed ", colnames(df)[3])) +
xlab("Sex") +
labs(fill = "") +
theme(legend.position = "none",
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
axis.line = element_line(colour = "black"),
text = element_text(size=21),
axis.title=element_text(size=21)) +
guides(shape = guide_legend(override.aes = list(size = 12)))
print(p)
}
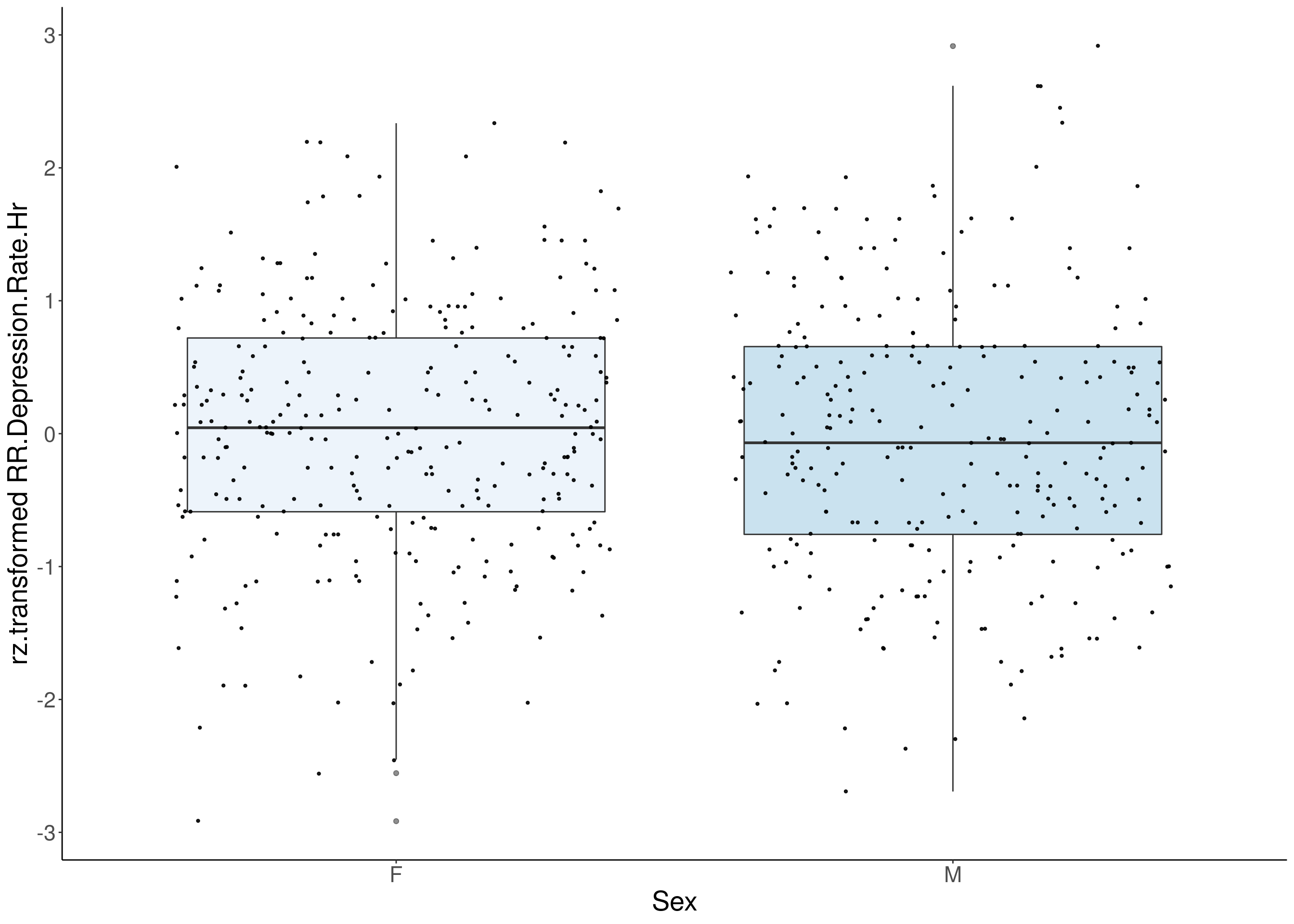
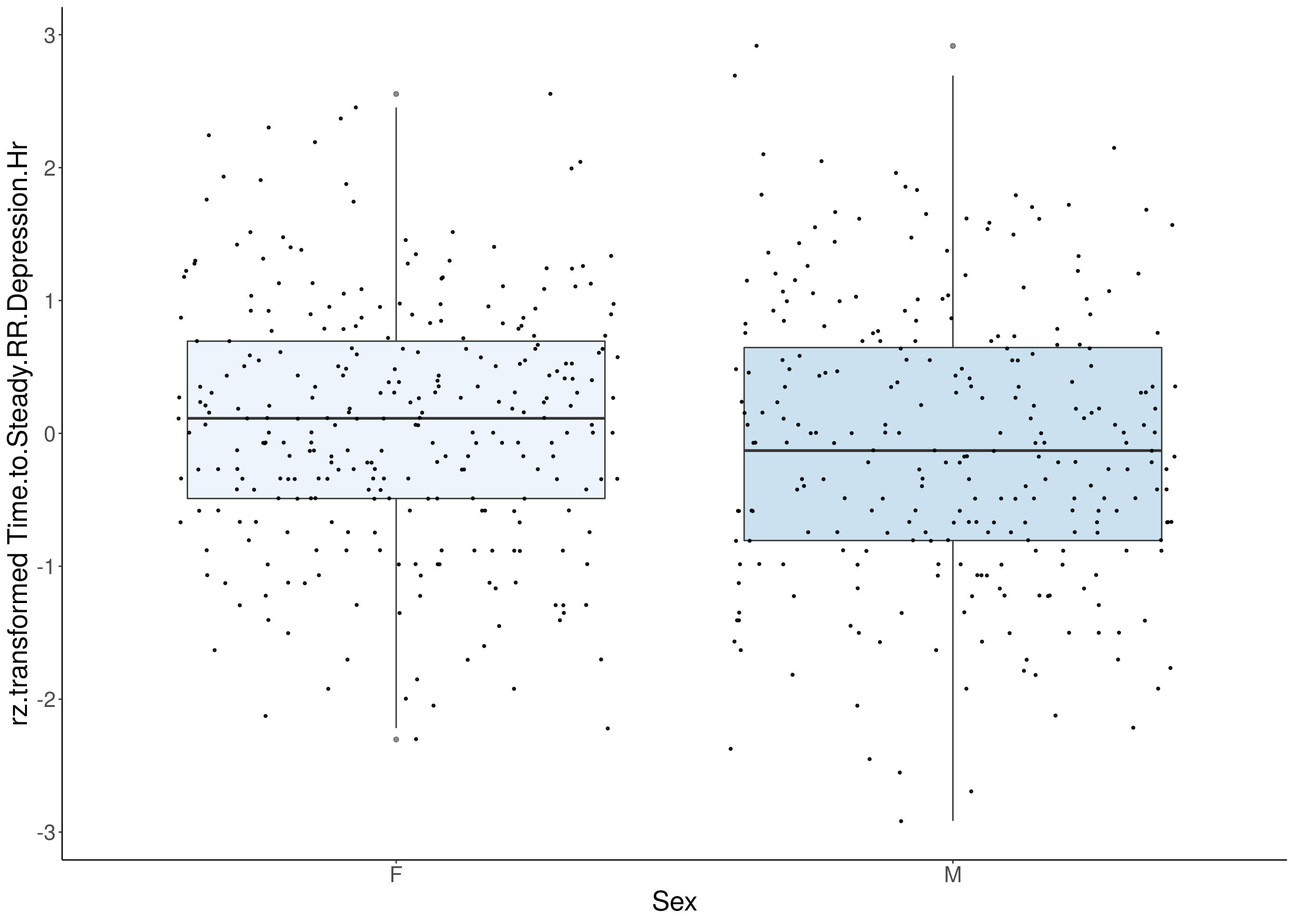
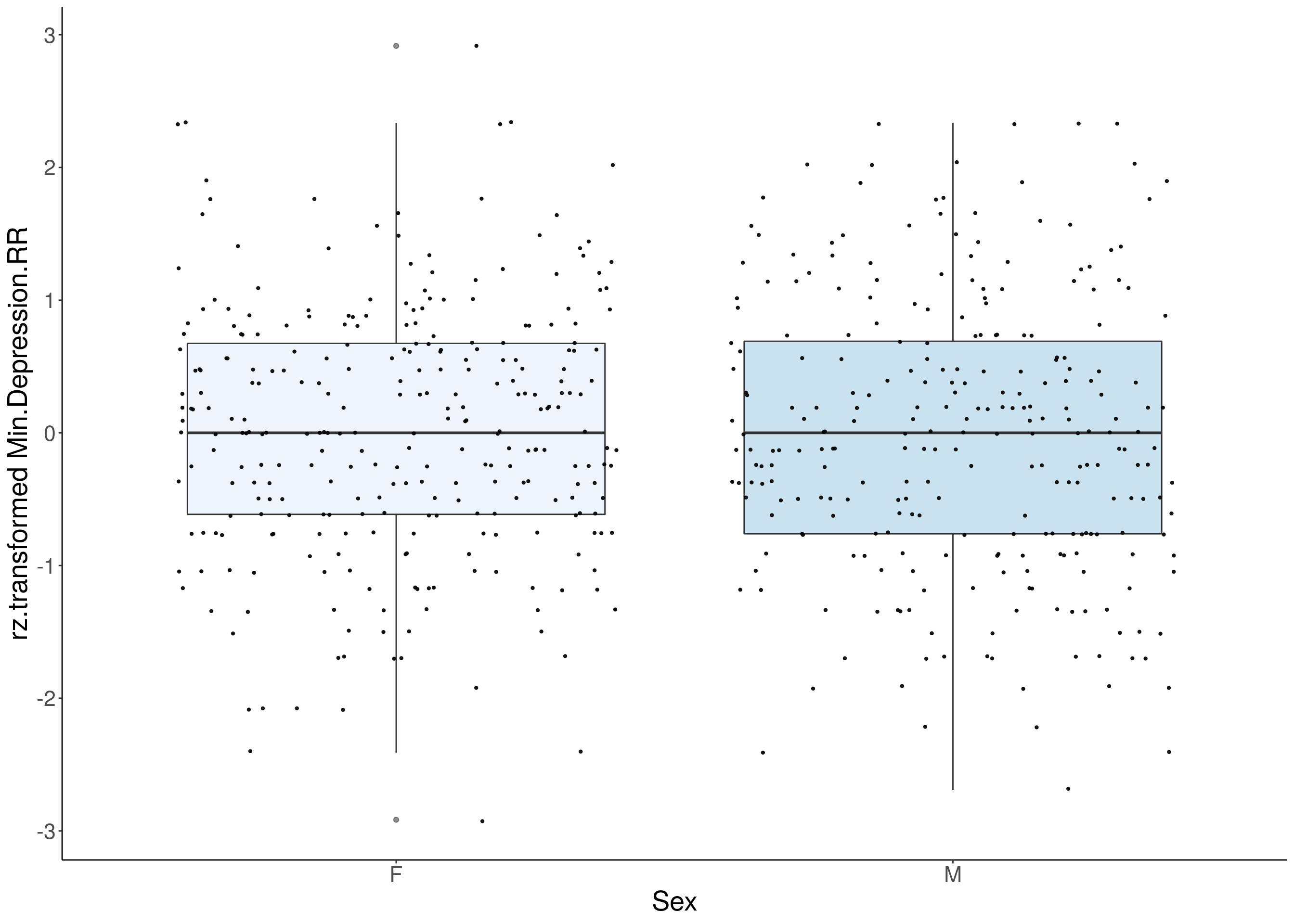
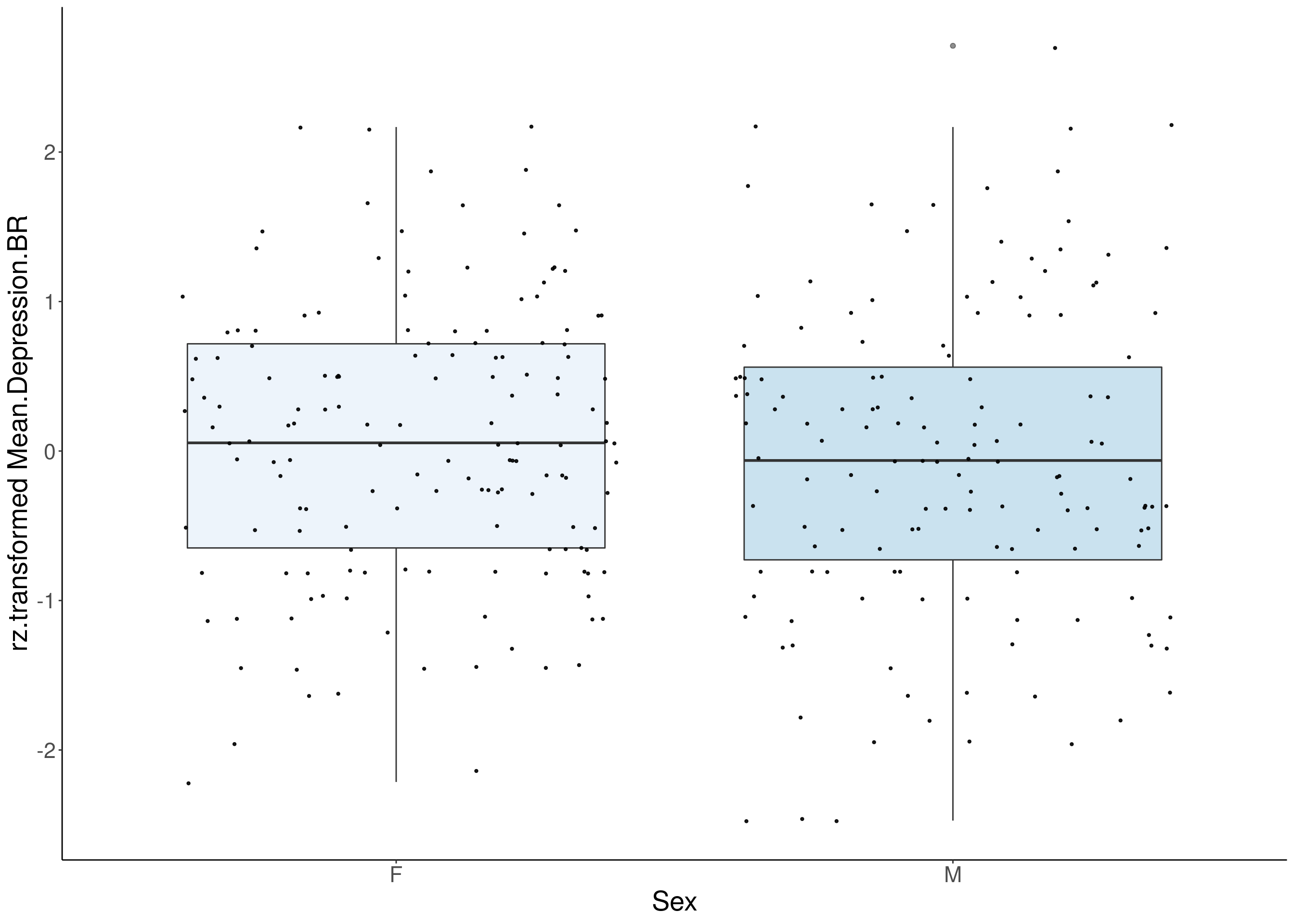
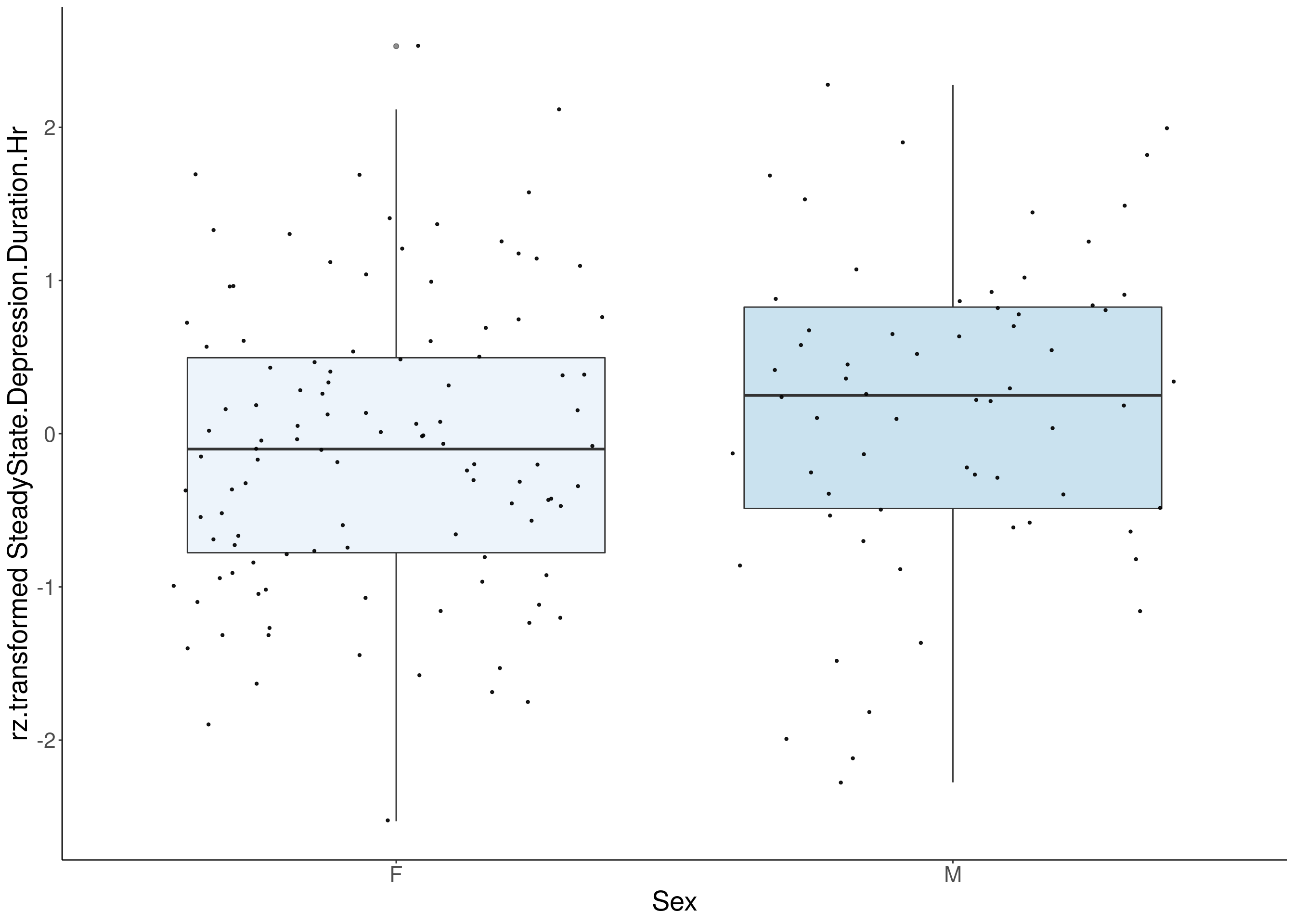
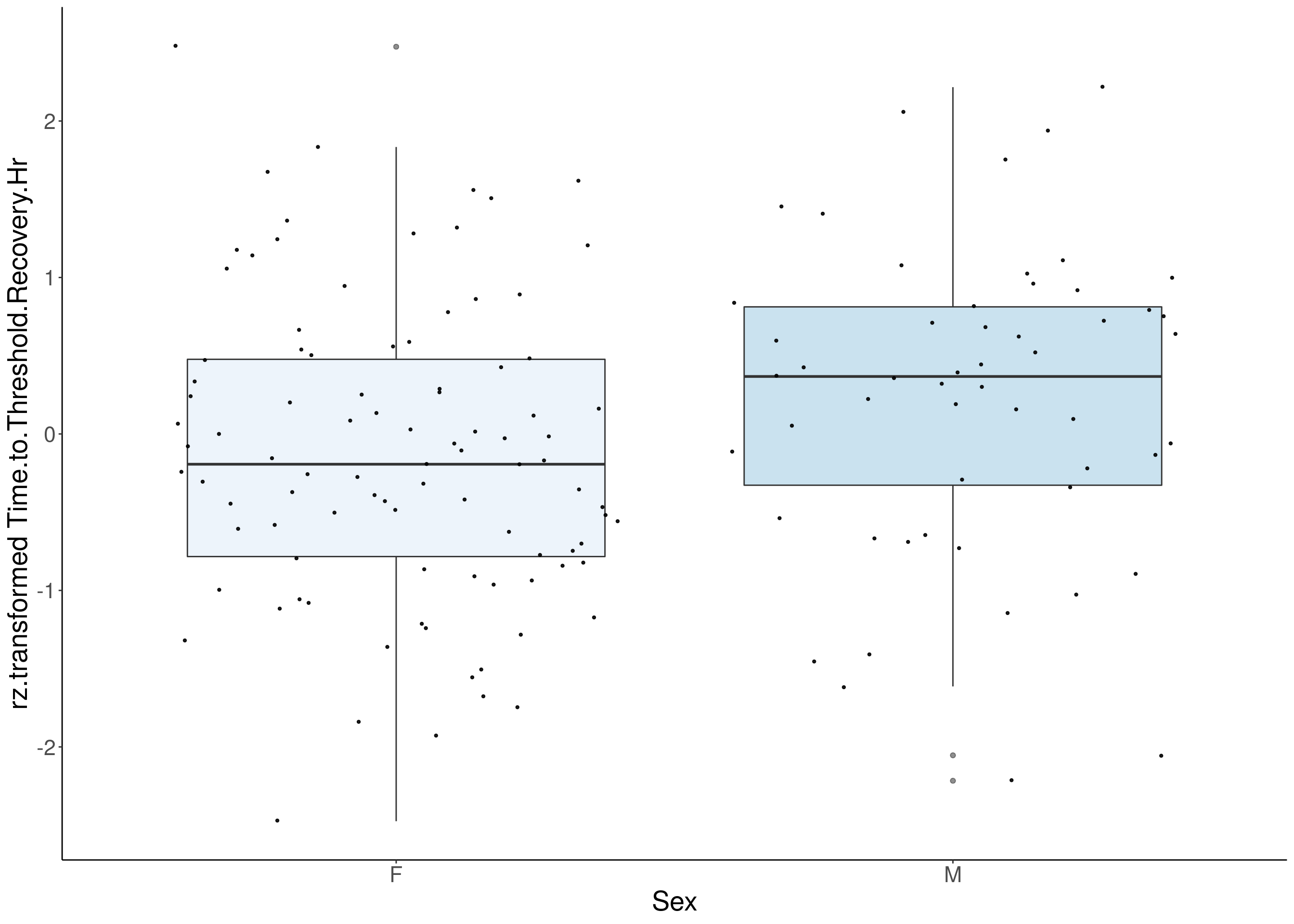
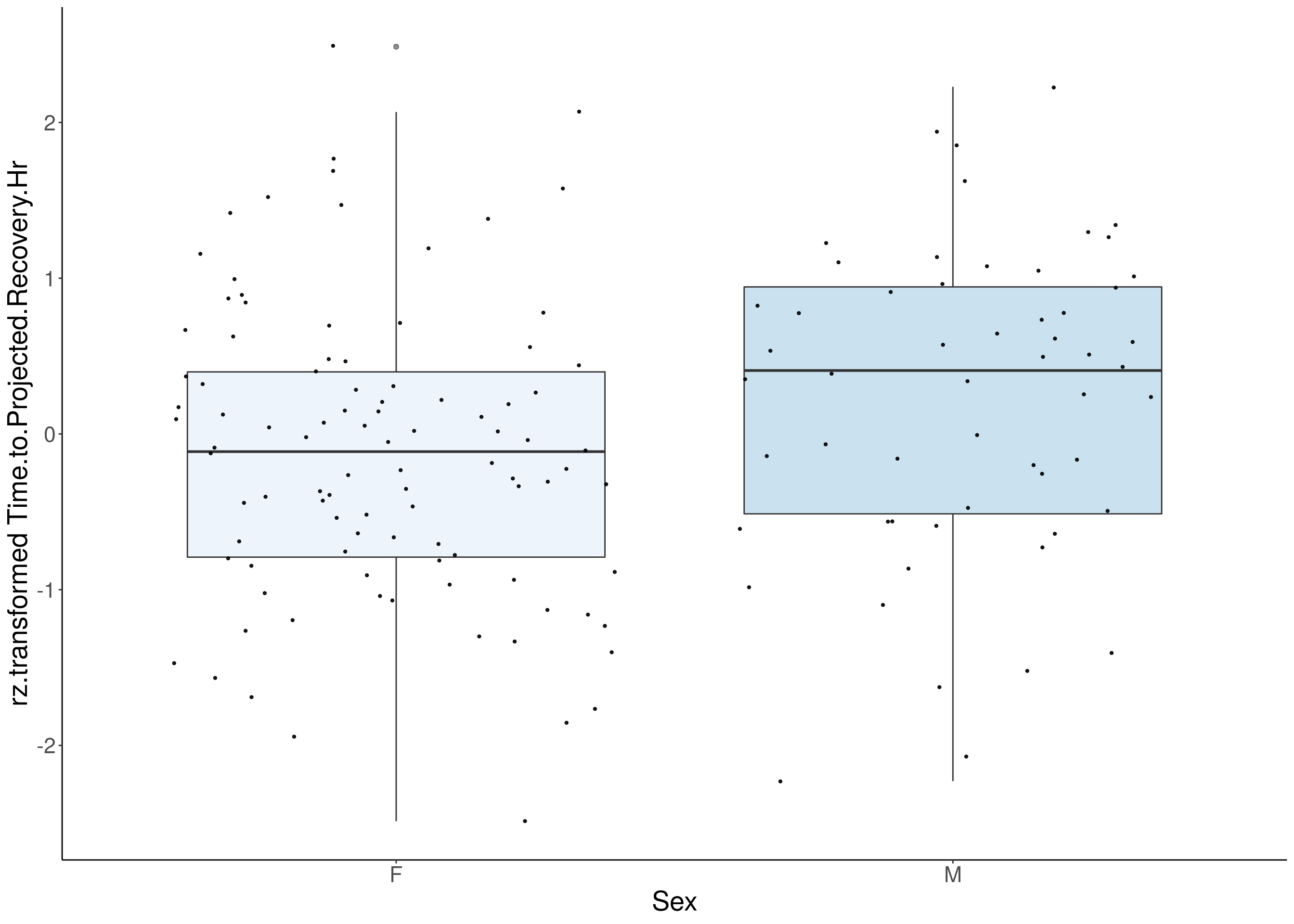
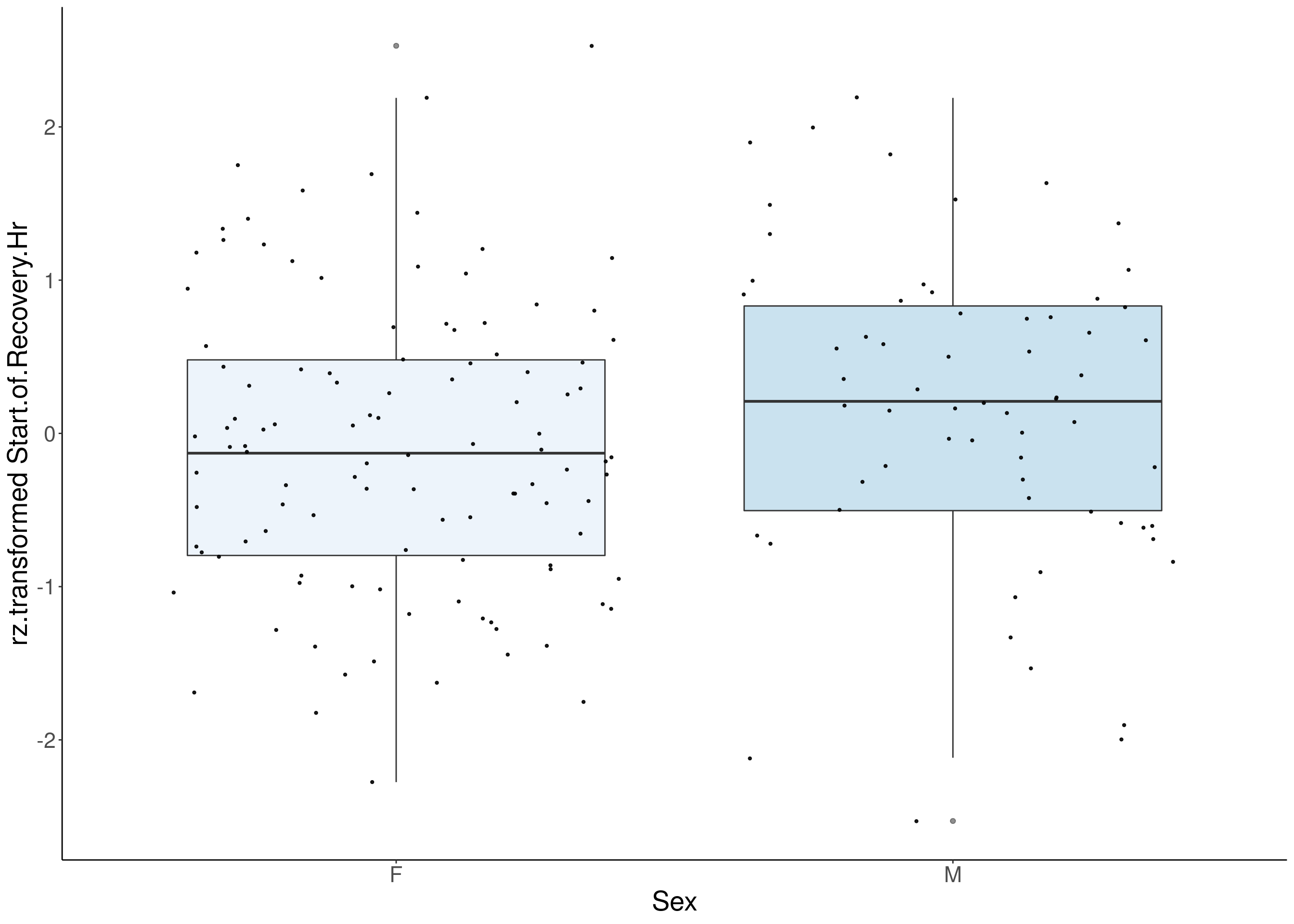
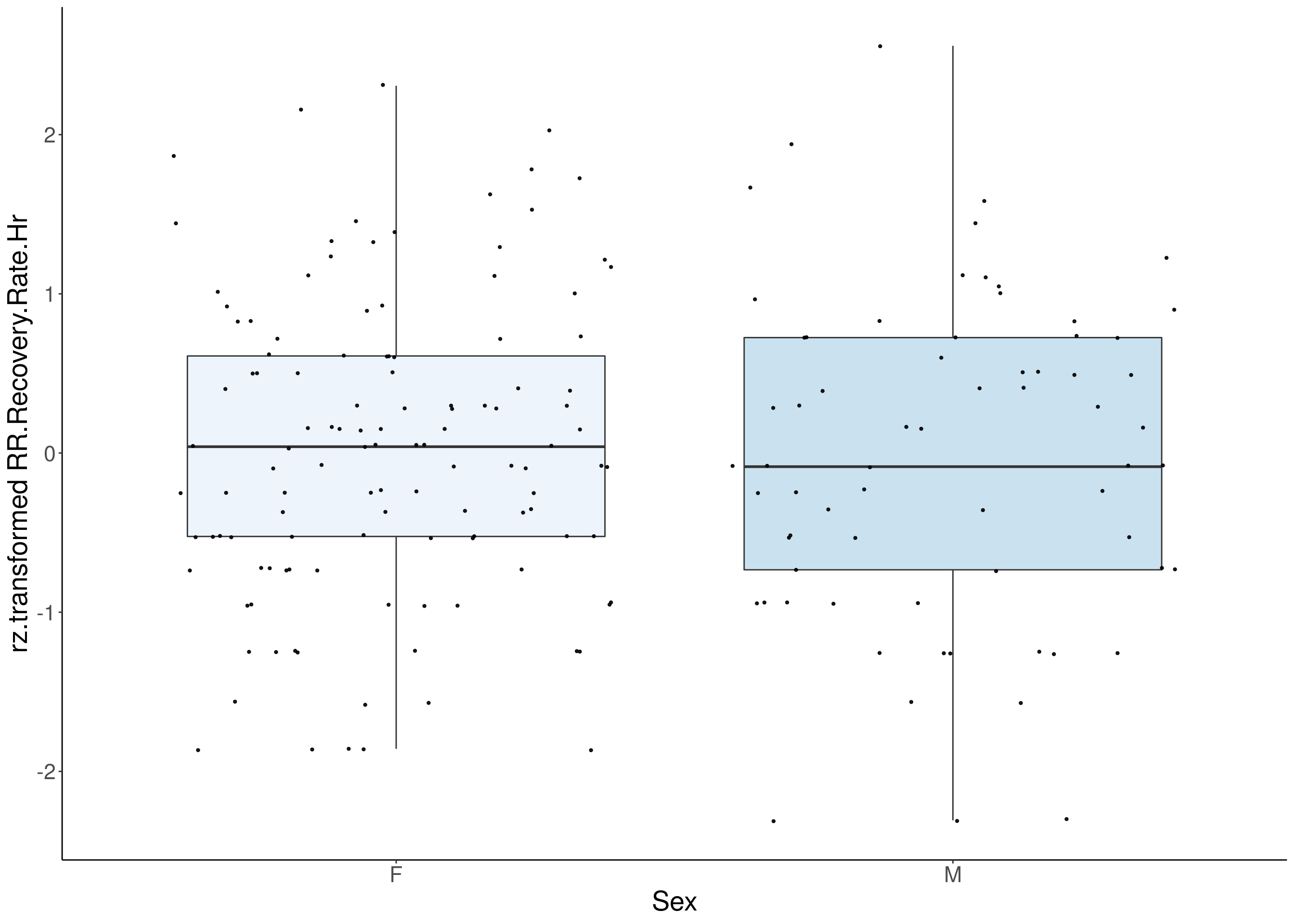
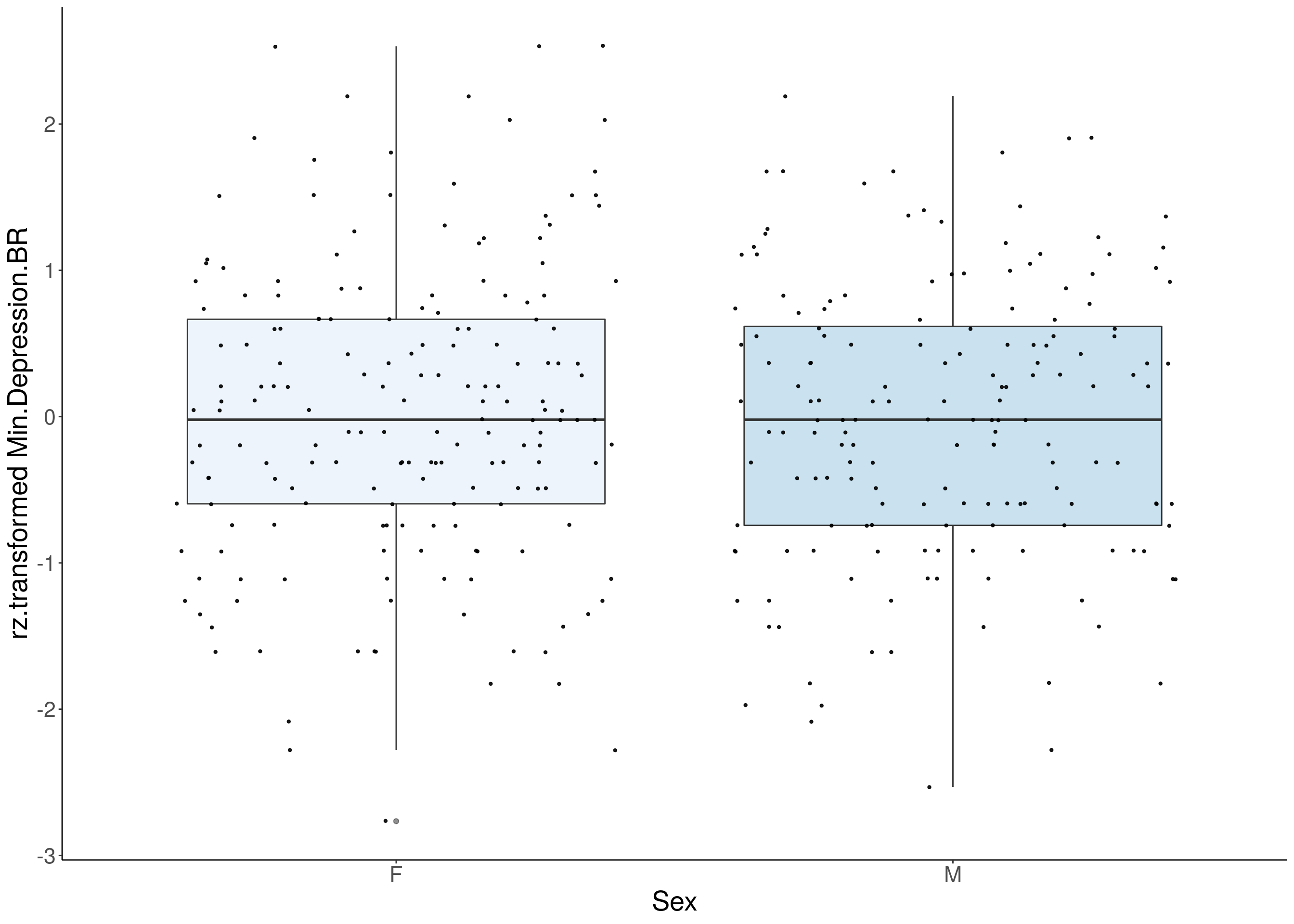
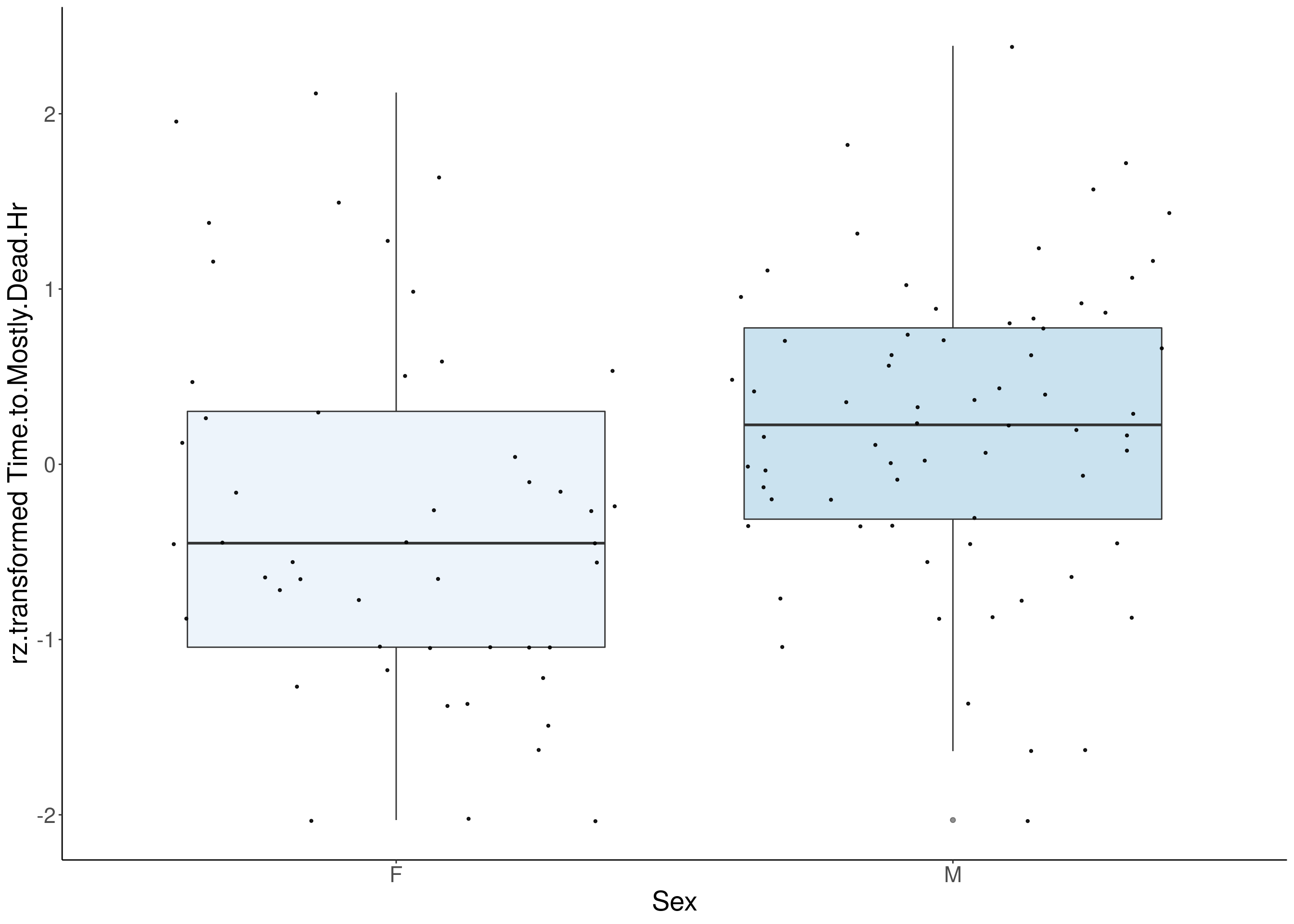
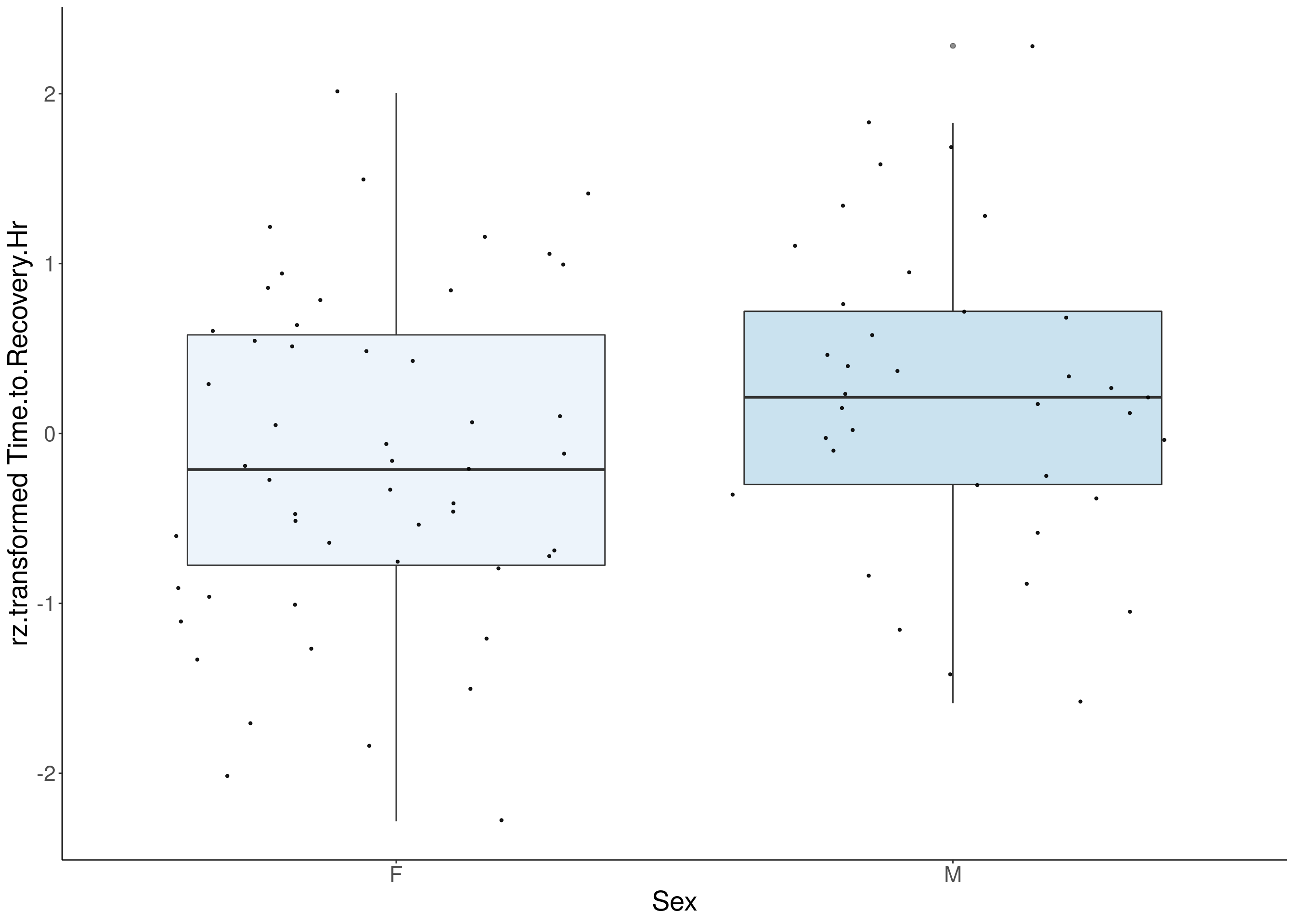
#histogram
for(i in 5:17){
df <- do.pheno[,c(1,2,i)]
p <- ggplot(data=df, aes(x=rz.transform(df[,3]))) +
geom_histogram() +
ylab("Number of DO mice") +
xlab(paste0("rz.transformed ", colnames(df)[3])) +
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
panel.background = element_blank(), axis.line = element_line(colour = "black"),
text = element_text(size=21))
print(p)
}
# Warning: Removed 337 rows containing non-finite values (stat_bin).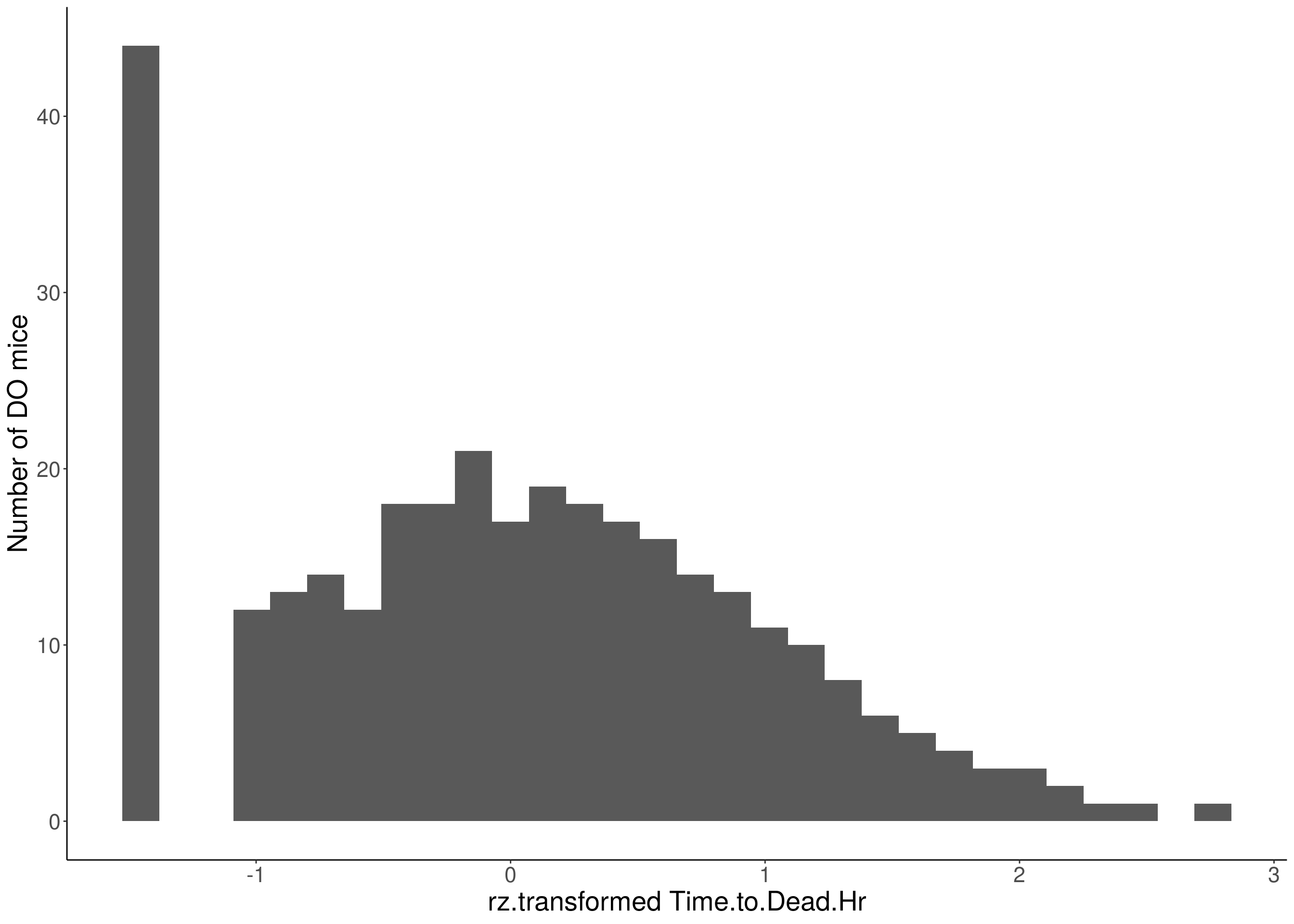
# Warning: Removed 85 rows containing non-finite values (stat_bin).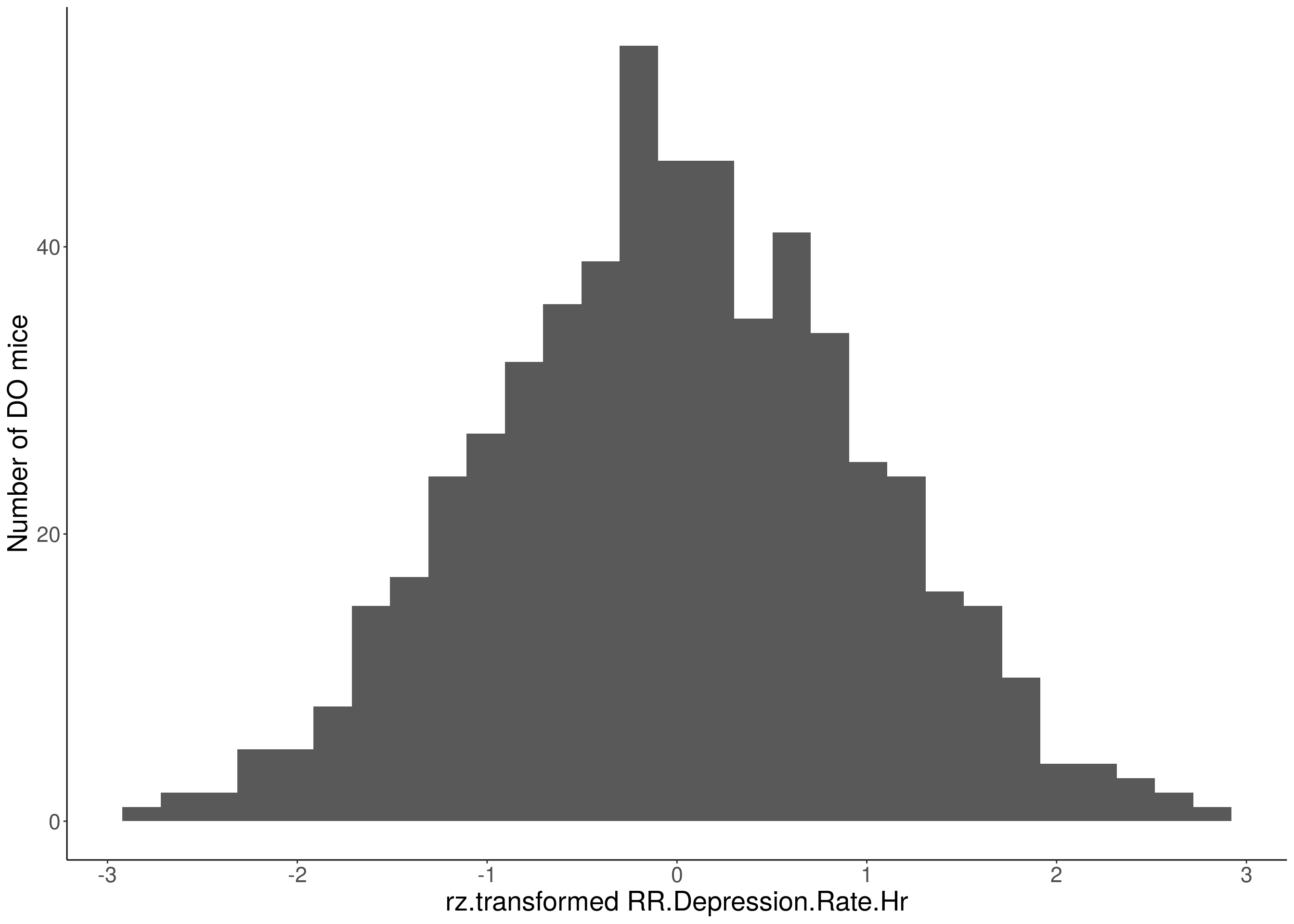
# Warning: Removed 85 rows containing non-finite values (stat_bin).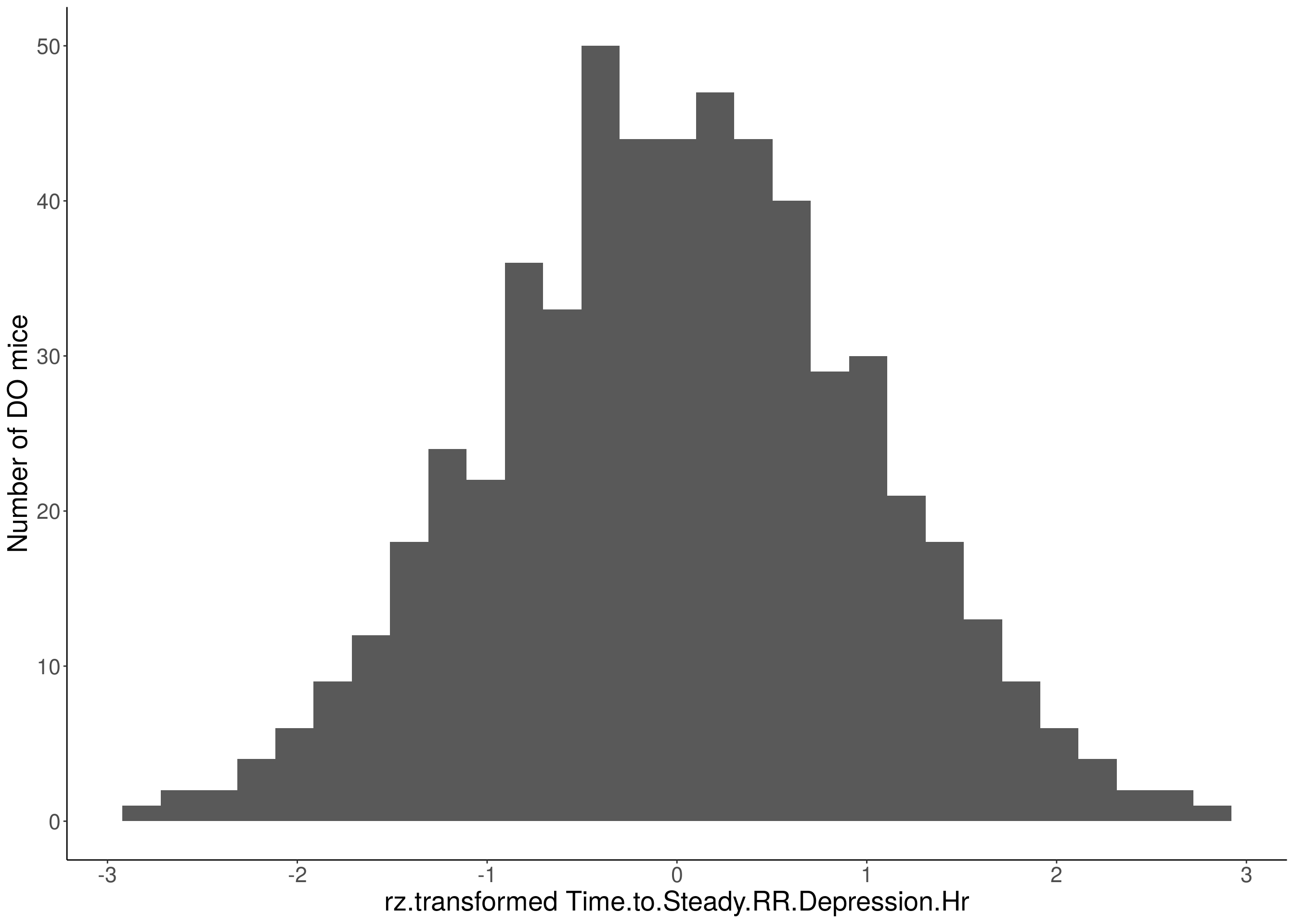
# Warning: Removed 85 rows containing non-finite values (stat_bin).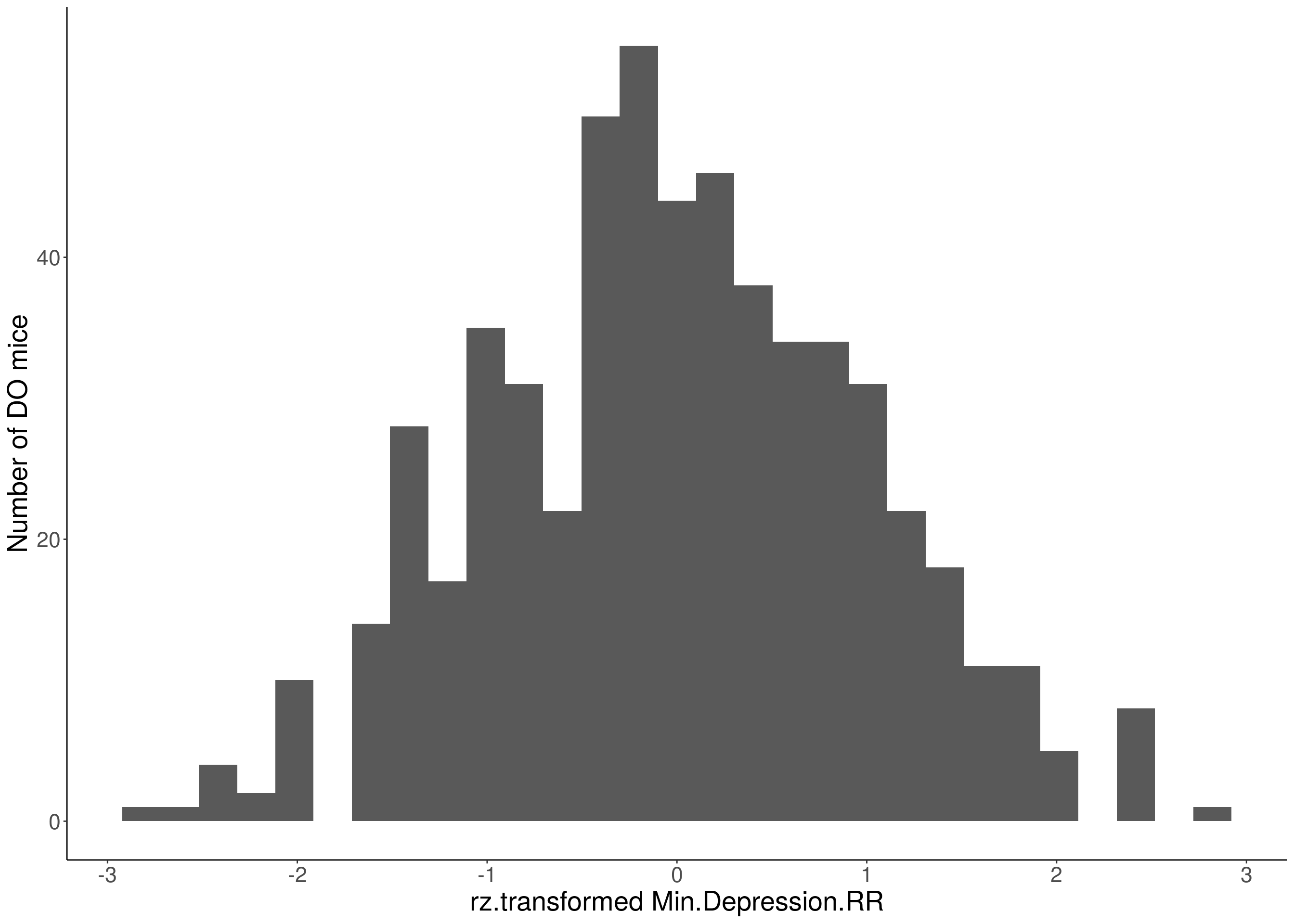
# Warning: Removed 355 rows containing non-finite values (stat_bin).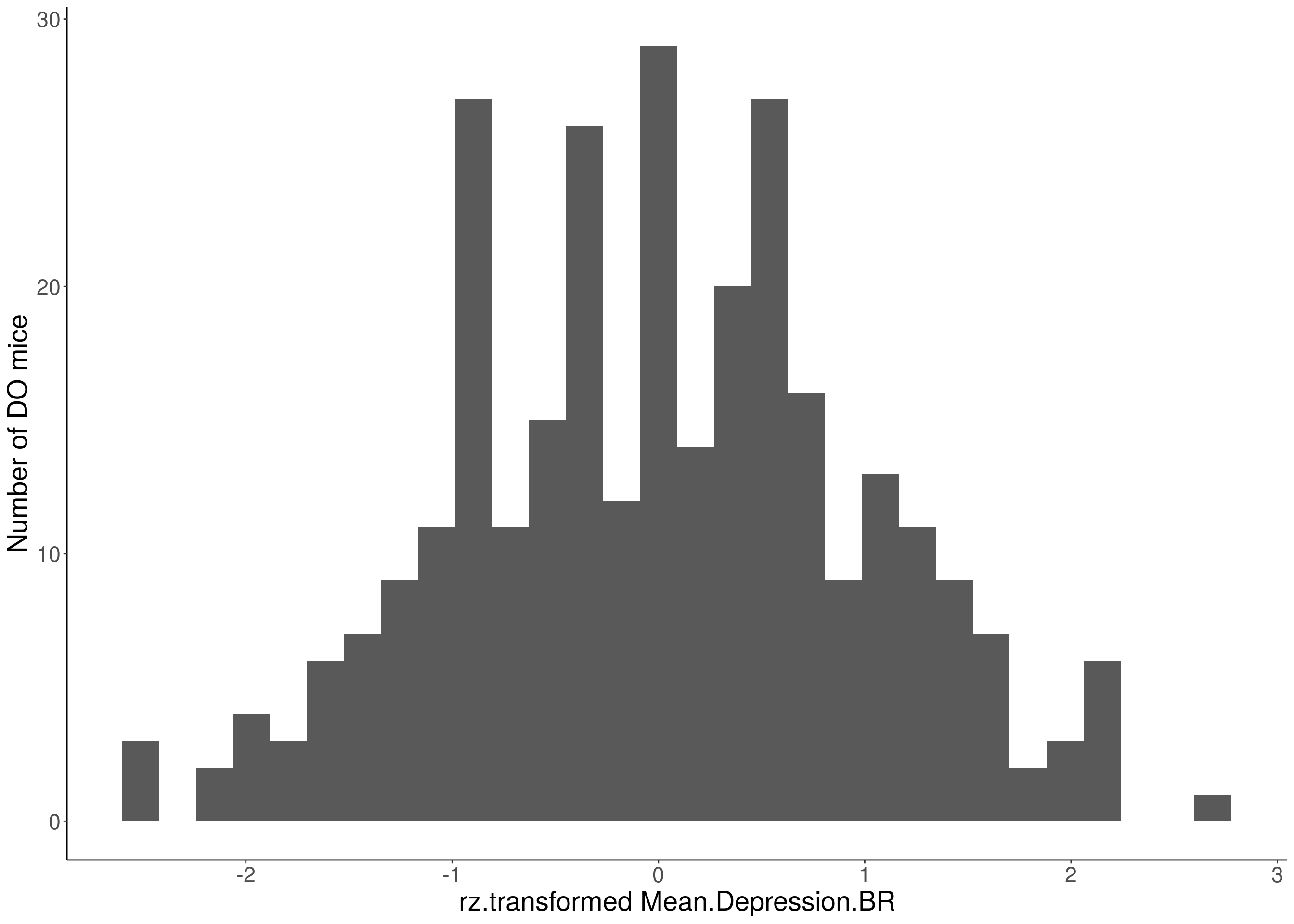
# Warning: Removed 484 rows containing non-finite values (stat_bin).
# Warning: Removed 509 rows containing non-finite values (stat_bin).
# Warning: Removed 504 rows containing non-finite values (stat_bin).
# Warning: Removed 484 rows containing non-finite values (stat_bin).
# Warning: Removed 469 rows containing non-finite values (stat_bin).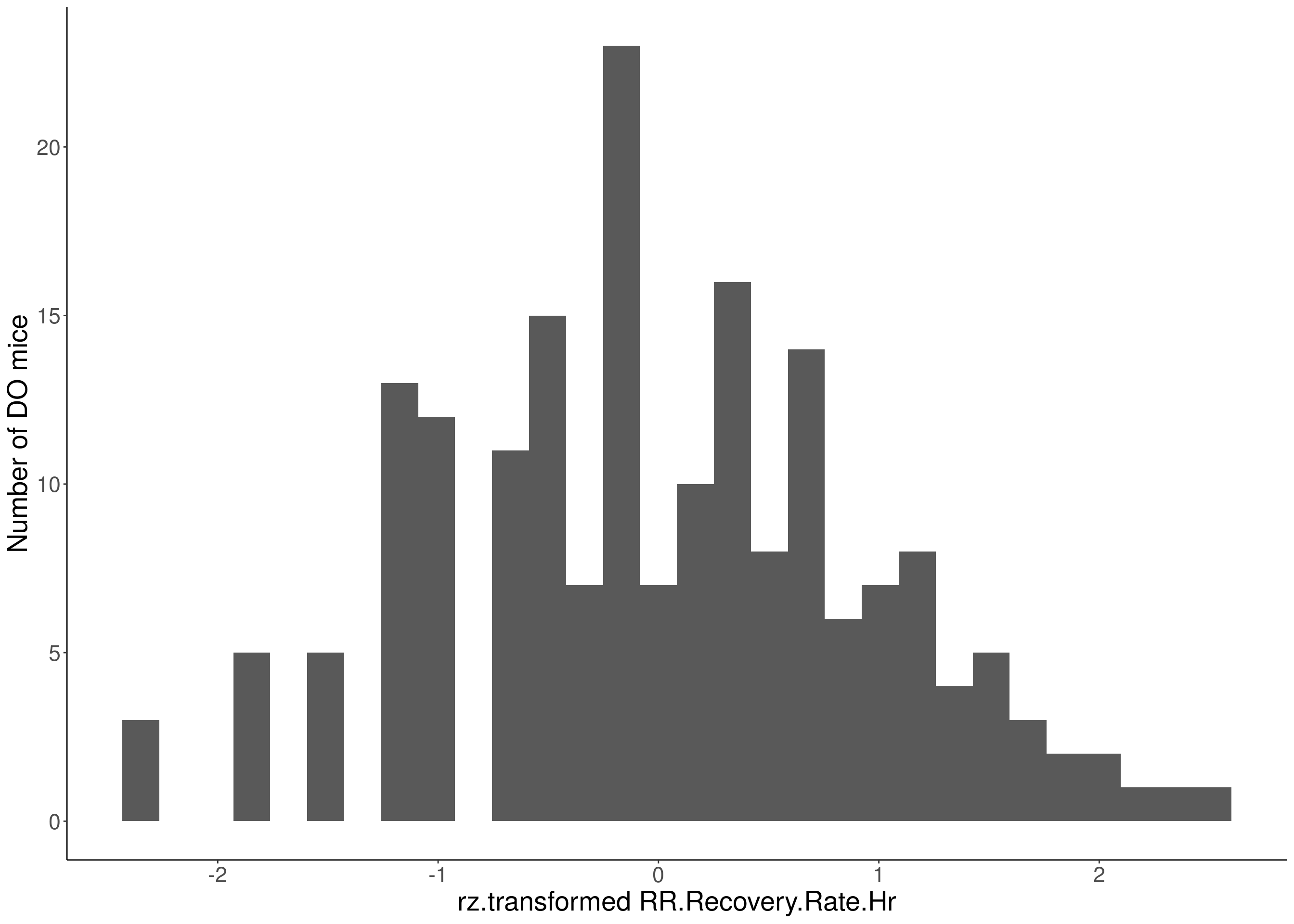
# Warning: Removed 307 rows containing non-finite values (stat_bin).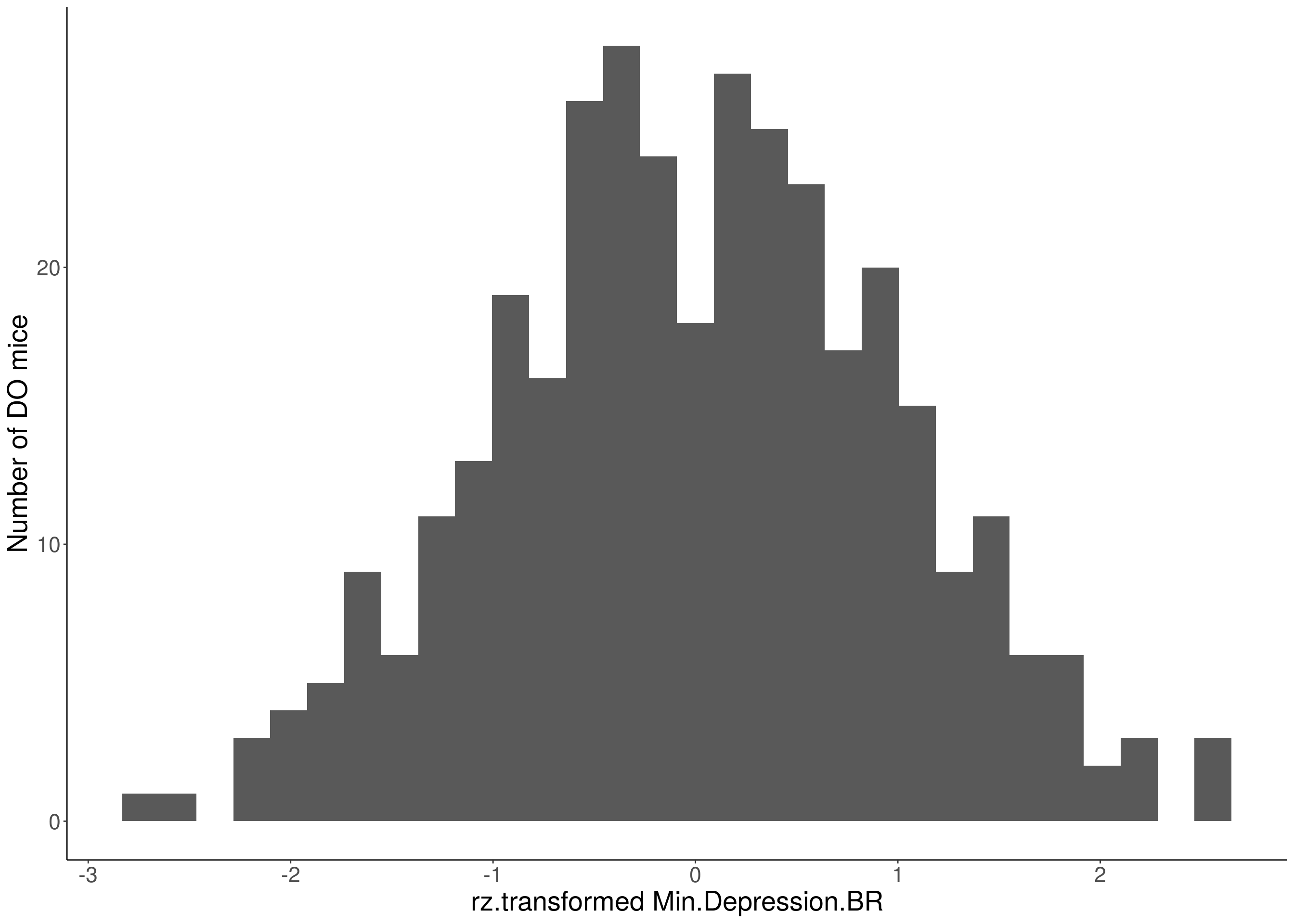
# Warning: Removed 541 rows containing non-finite values (stat_bin).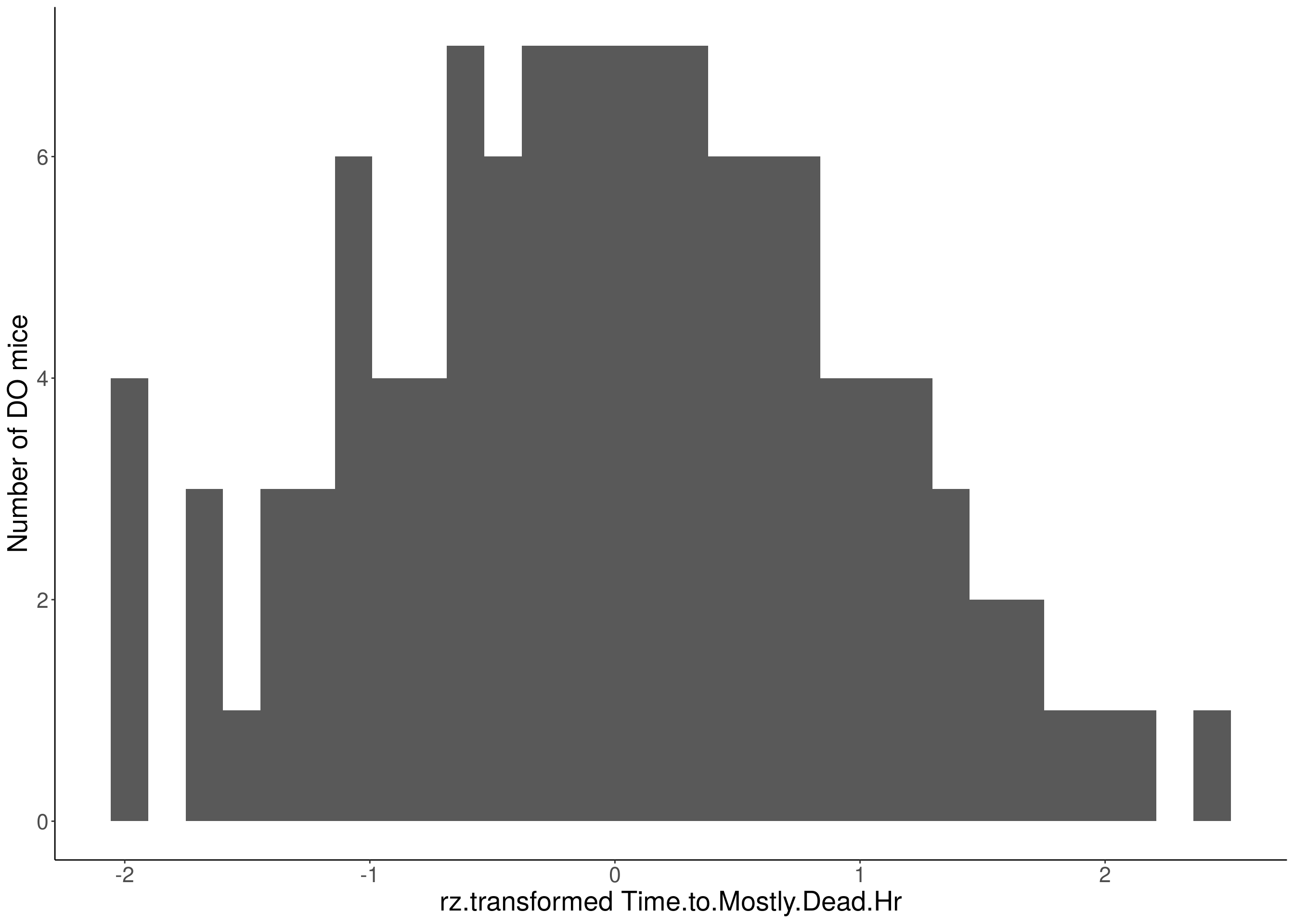
# Warning: Removed 570 rows containing non-finite values (stat_bin).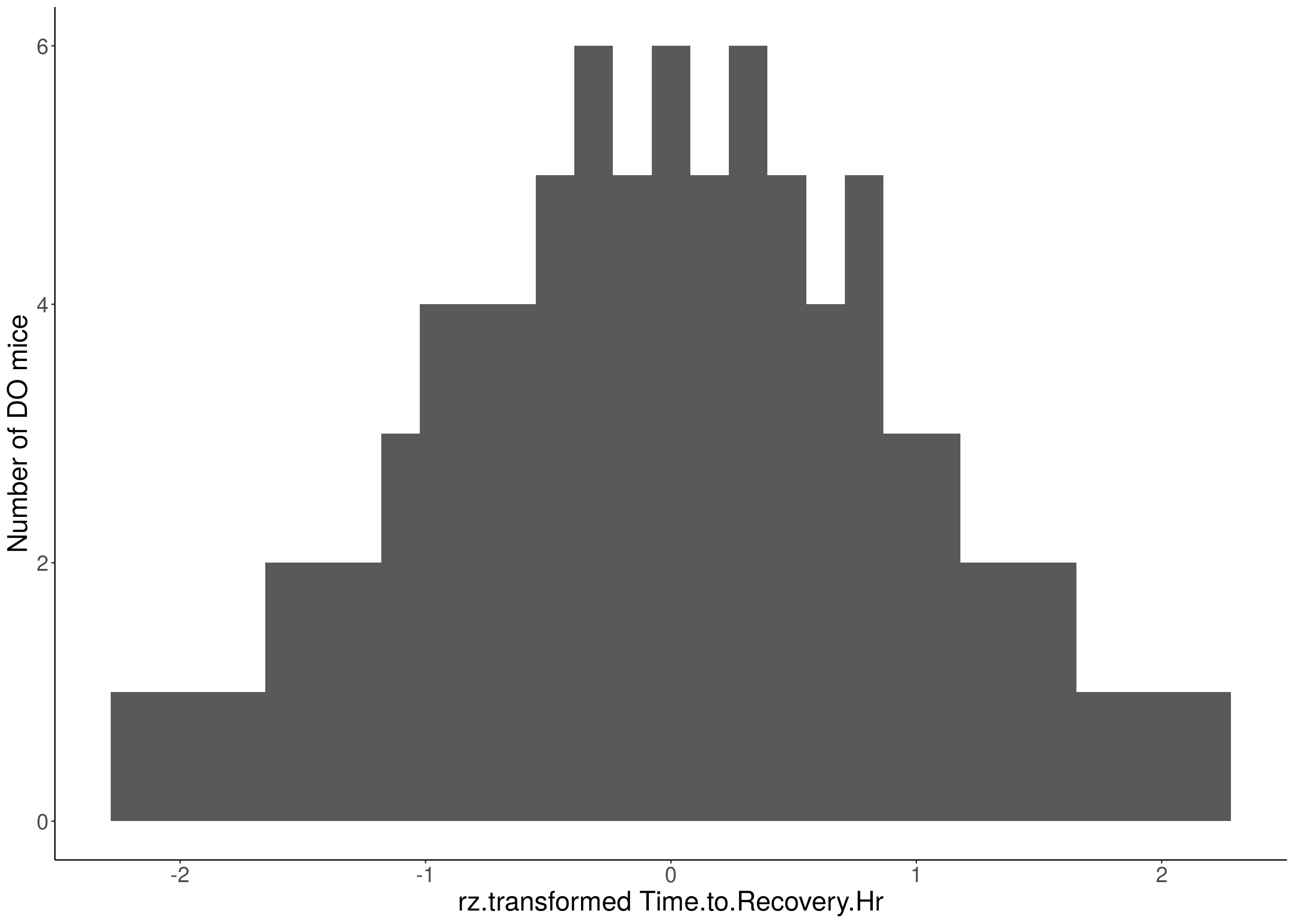
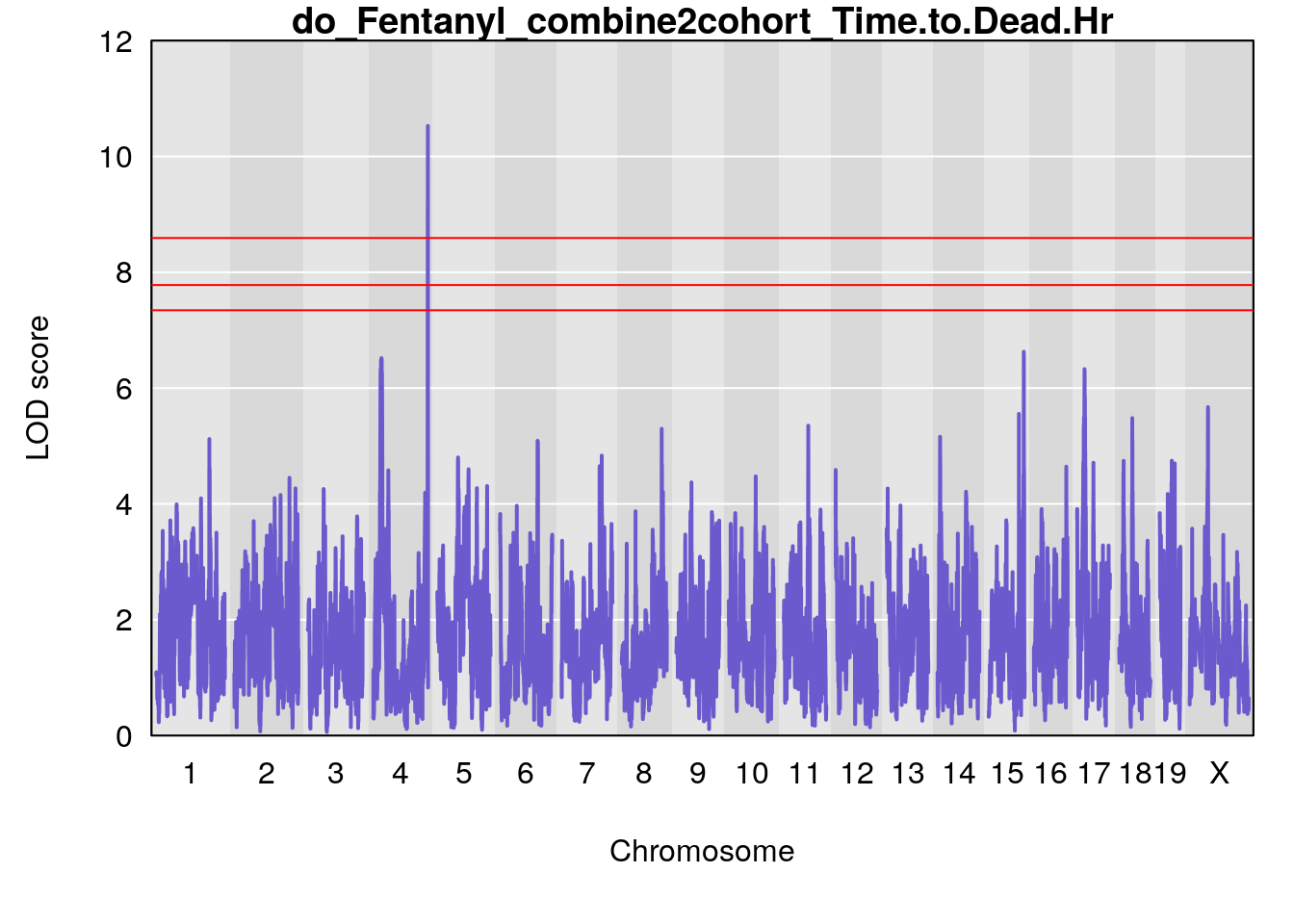
Plot qtl mapping on array
Plot by using its output qtl.fentanyl.combine2cohort.out.RData
load("output/Fentanyl/qtl.fentanyl.combine2cohort.out.RData")
#pmap and gmap
chr.names <- c("1","2","3","4","5","6","7","8","9","10","11","12","13","14","15","16","17","18","19", "X")
gmap <- list()
pmap <- list()
for (chr in chr.names){
gmap[[chr]] <- gm$gmap[[chr]]
pmap[[chr]] <- gm$pmap[[chr]]
}
attr(gmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
attr(pmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
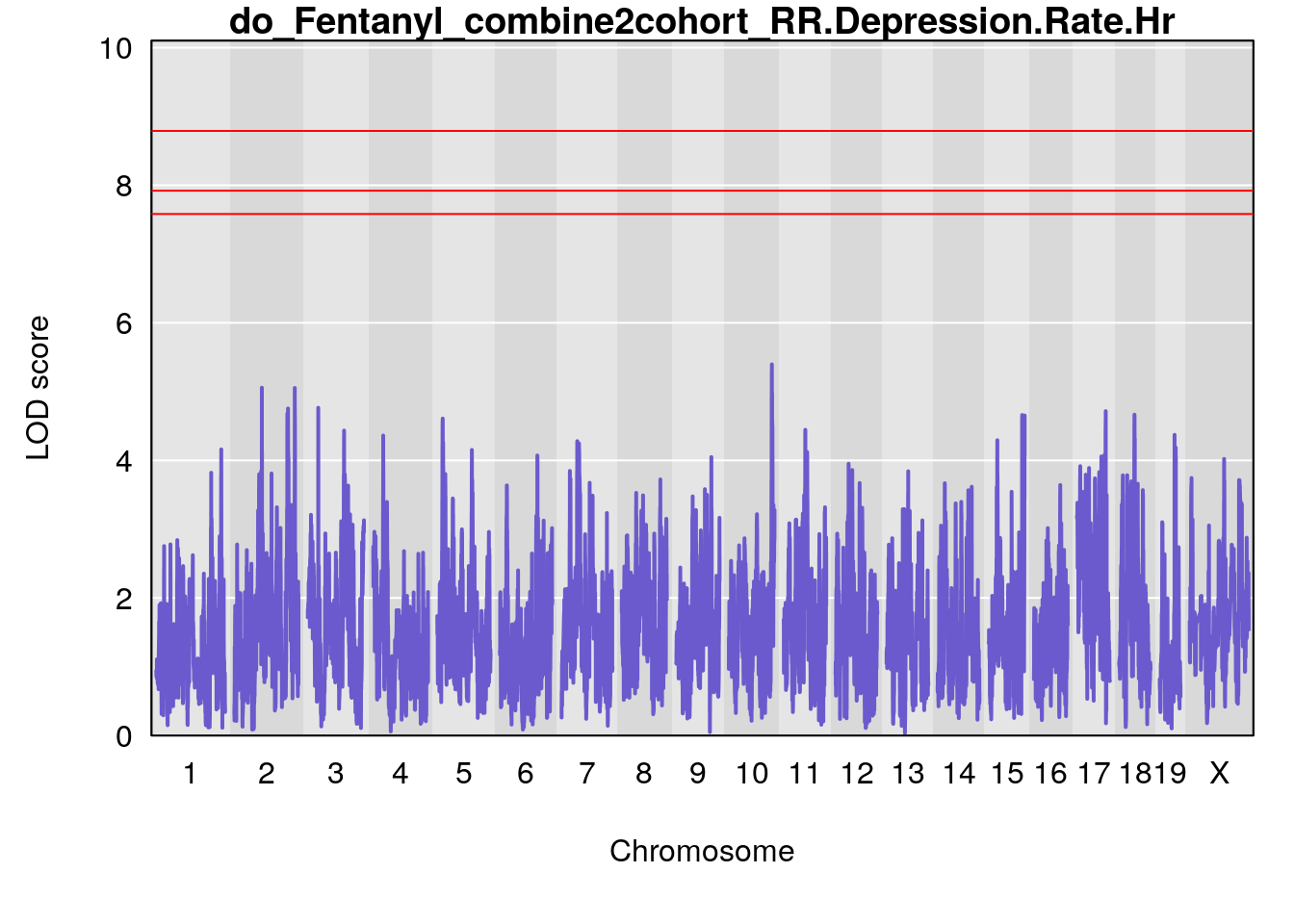
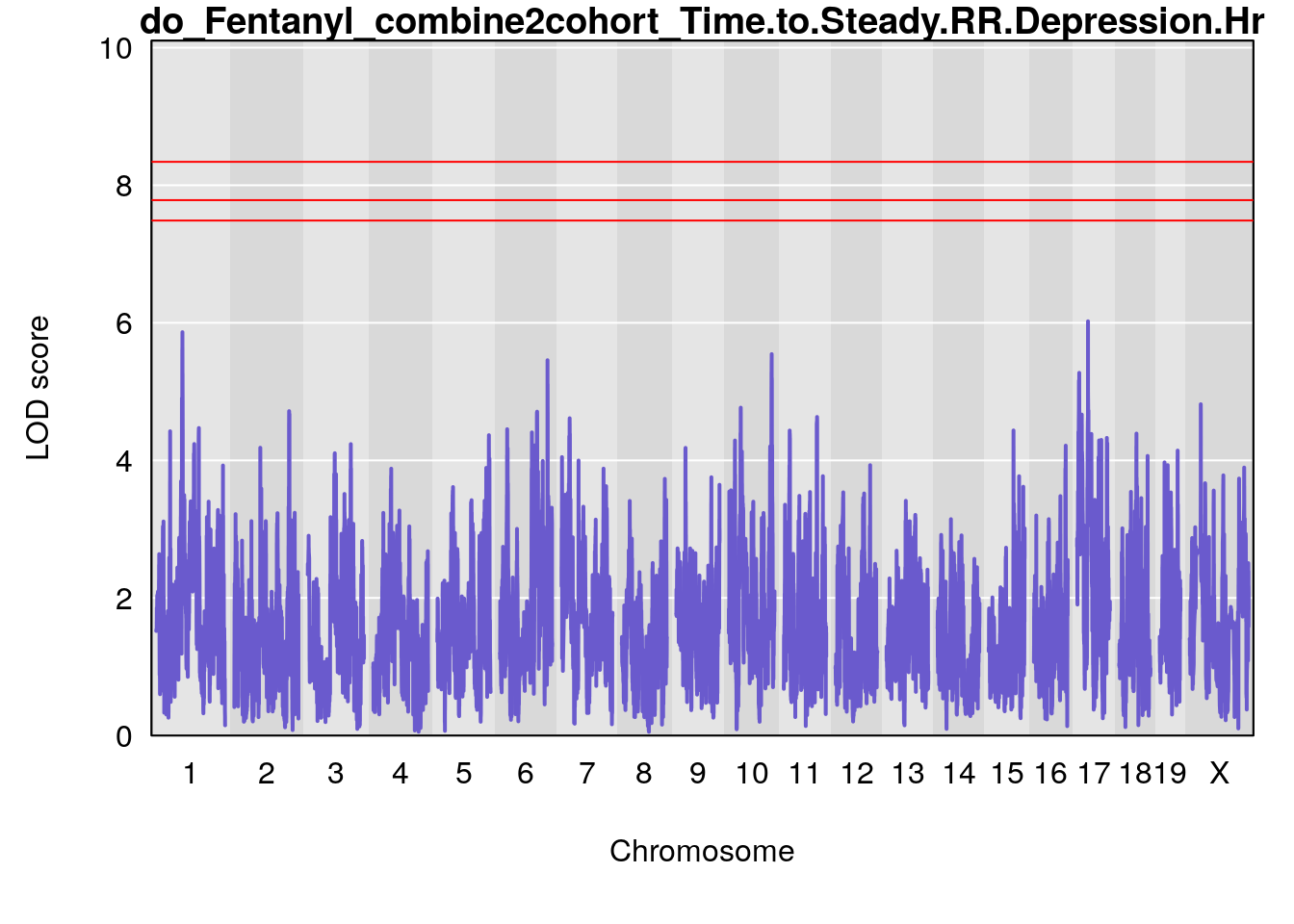
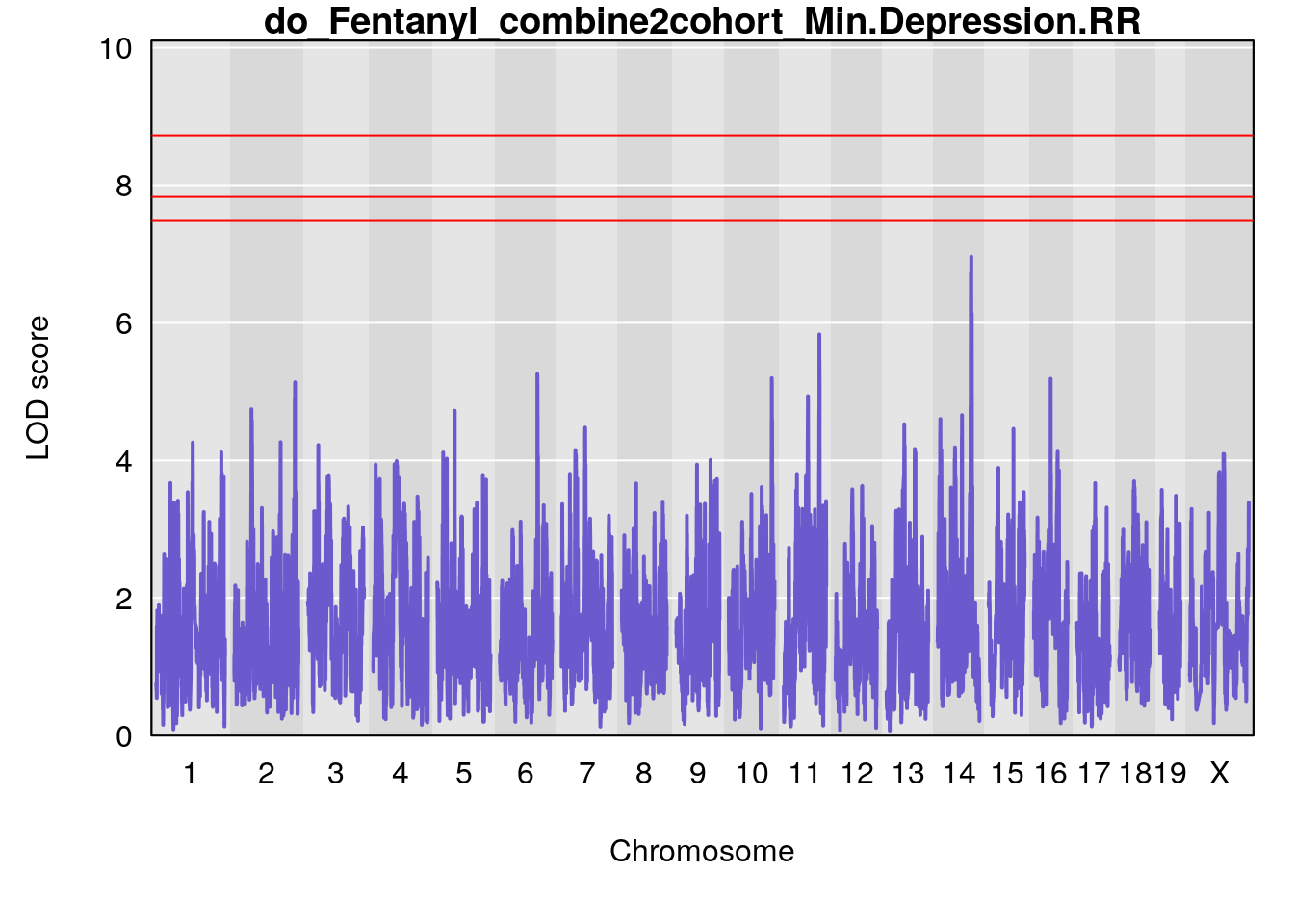
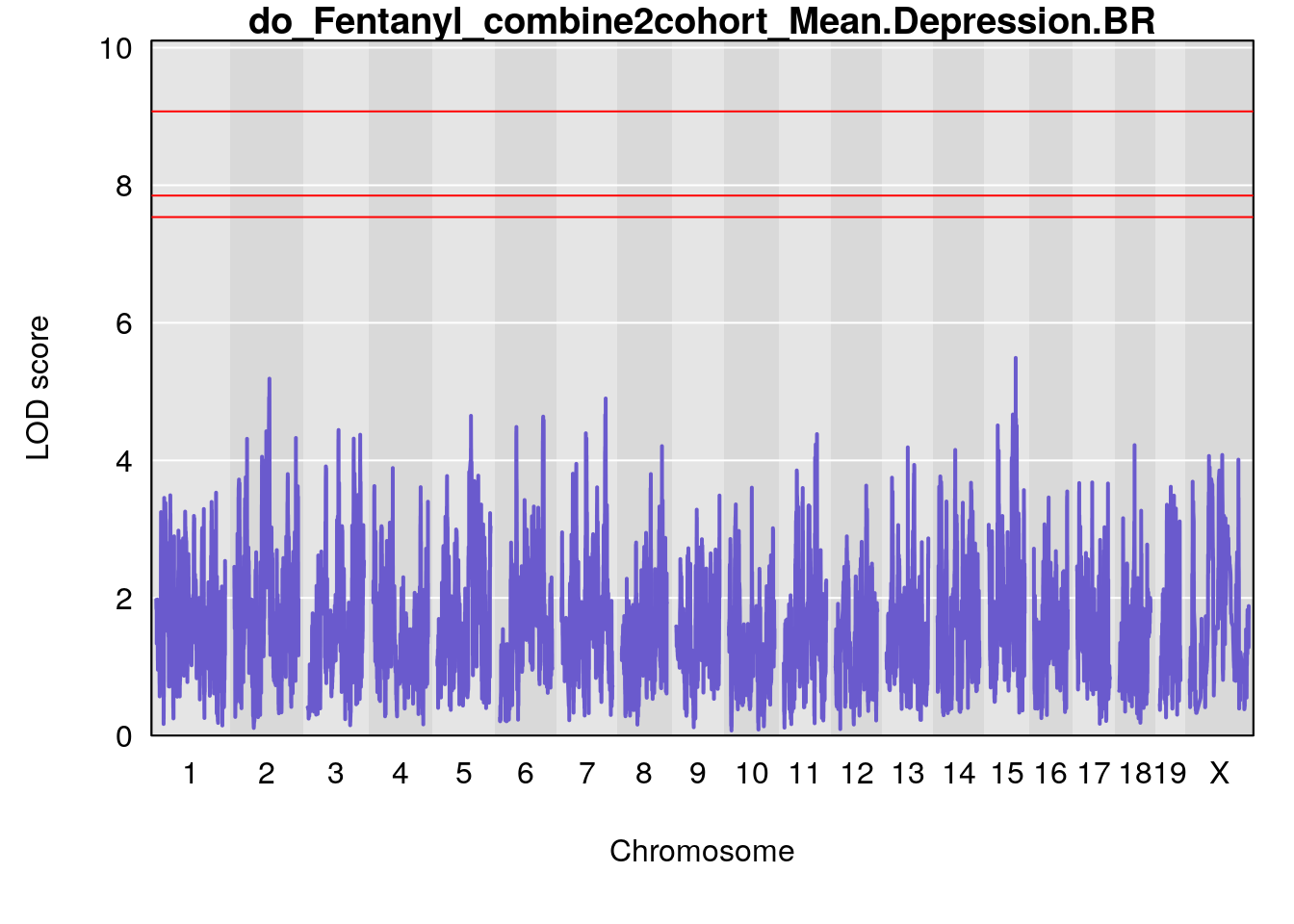
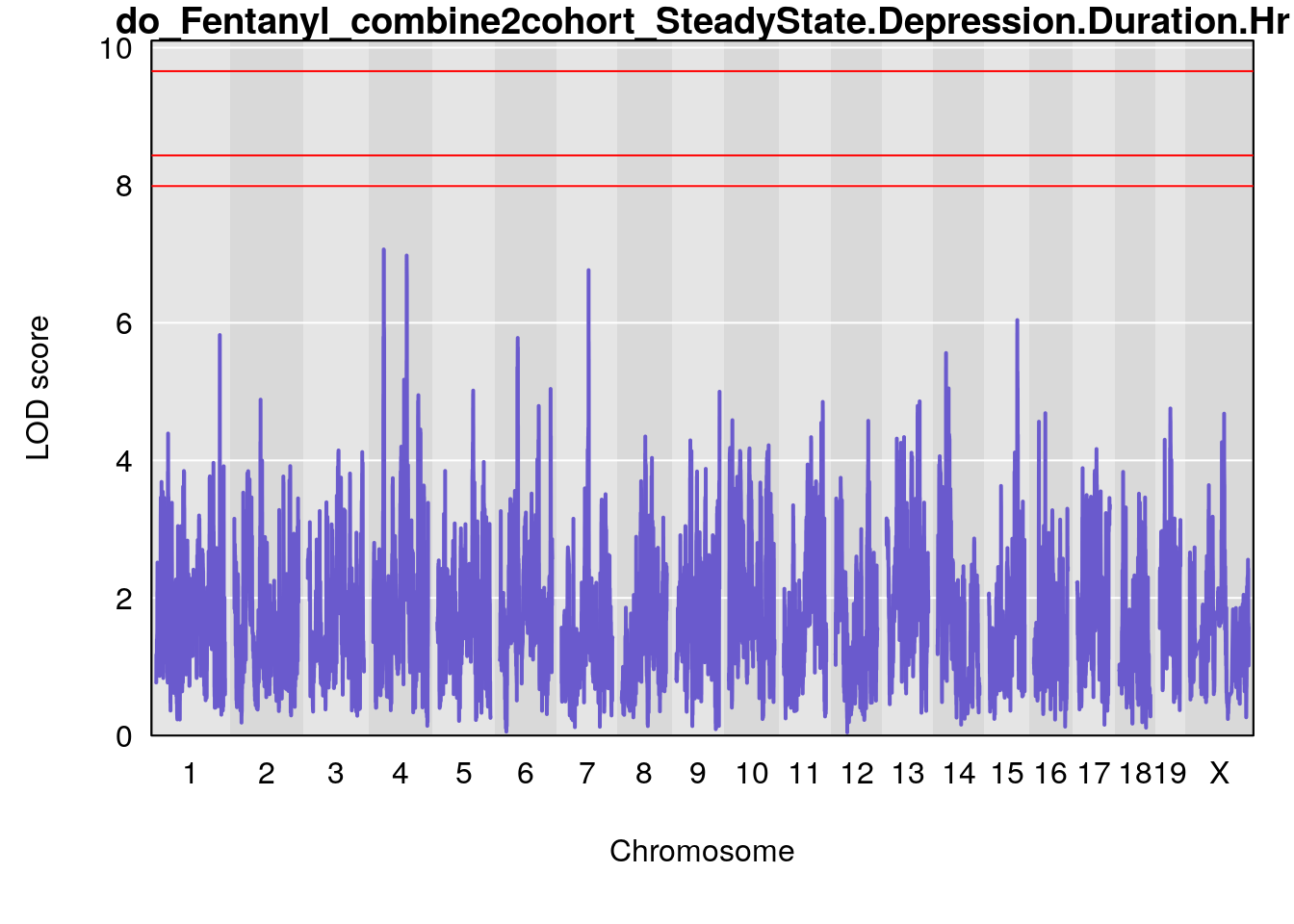
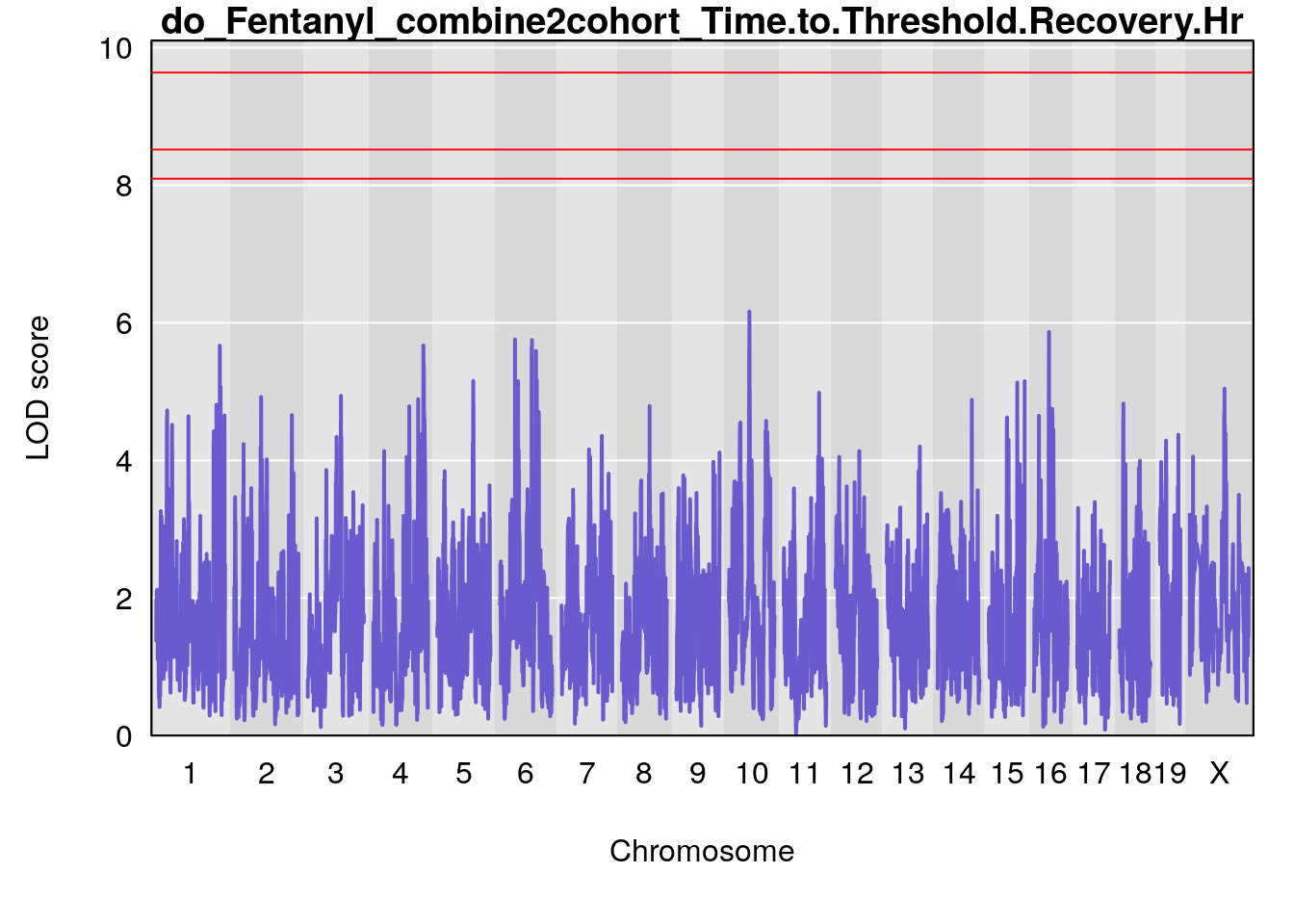
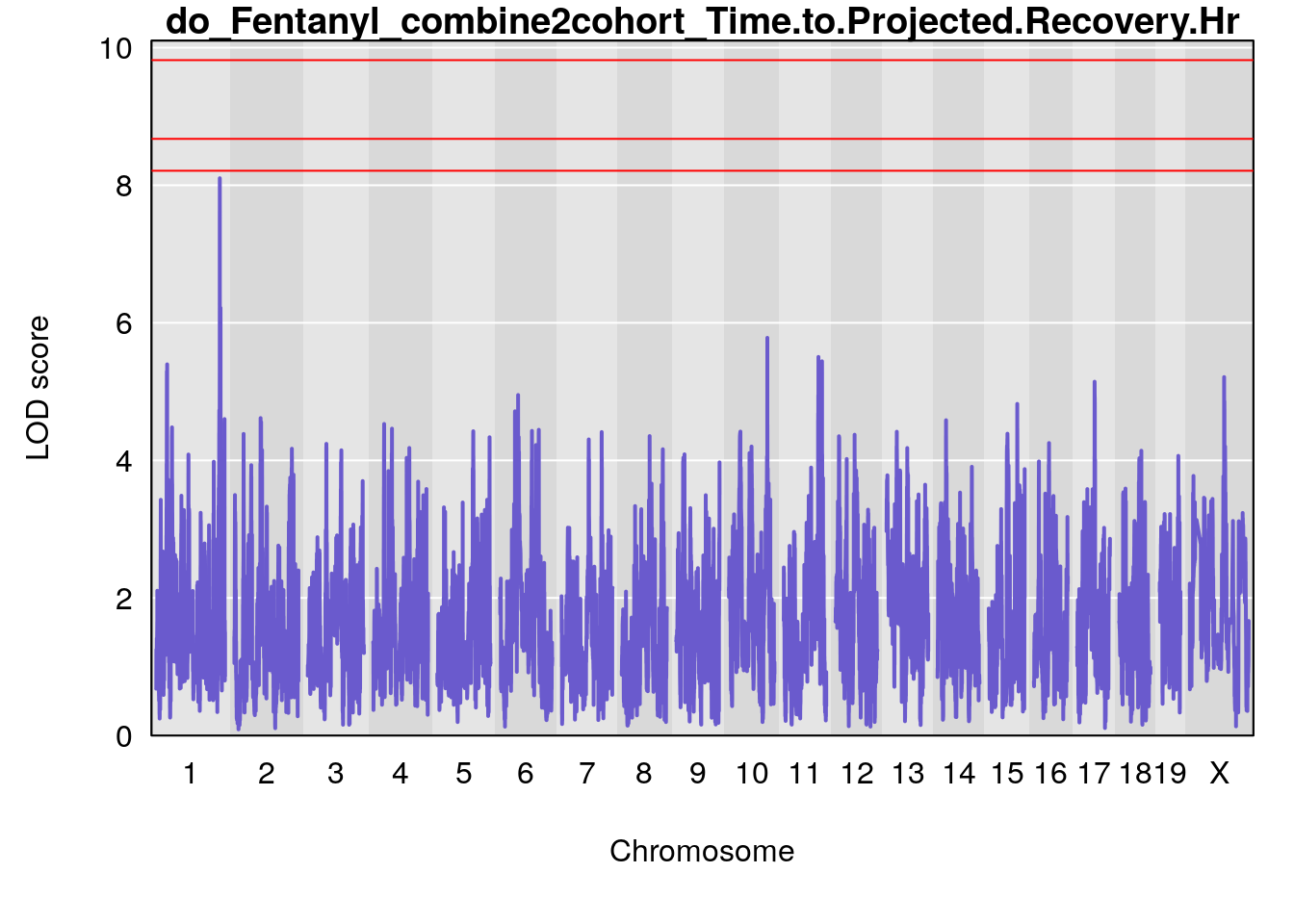
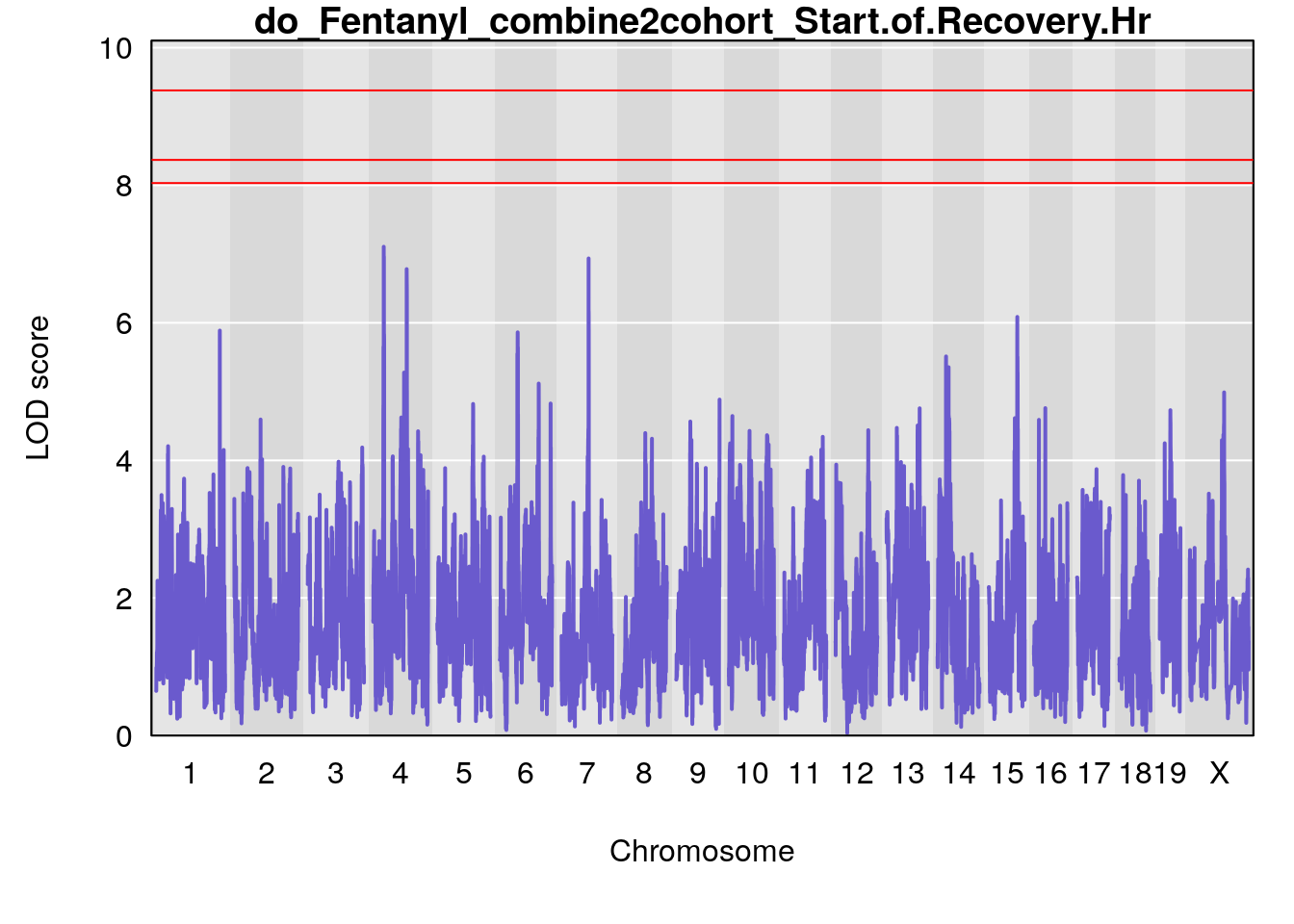
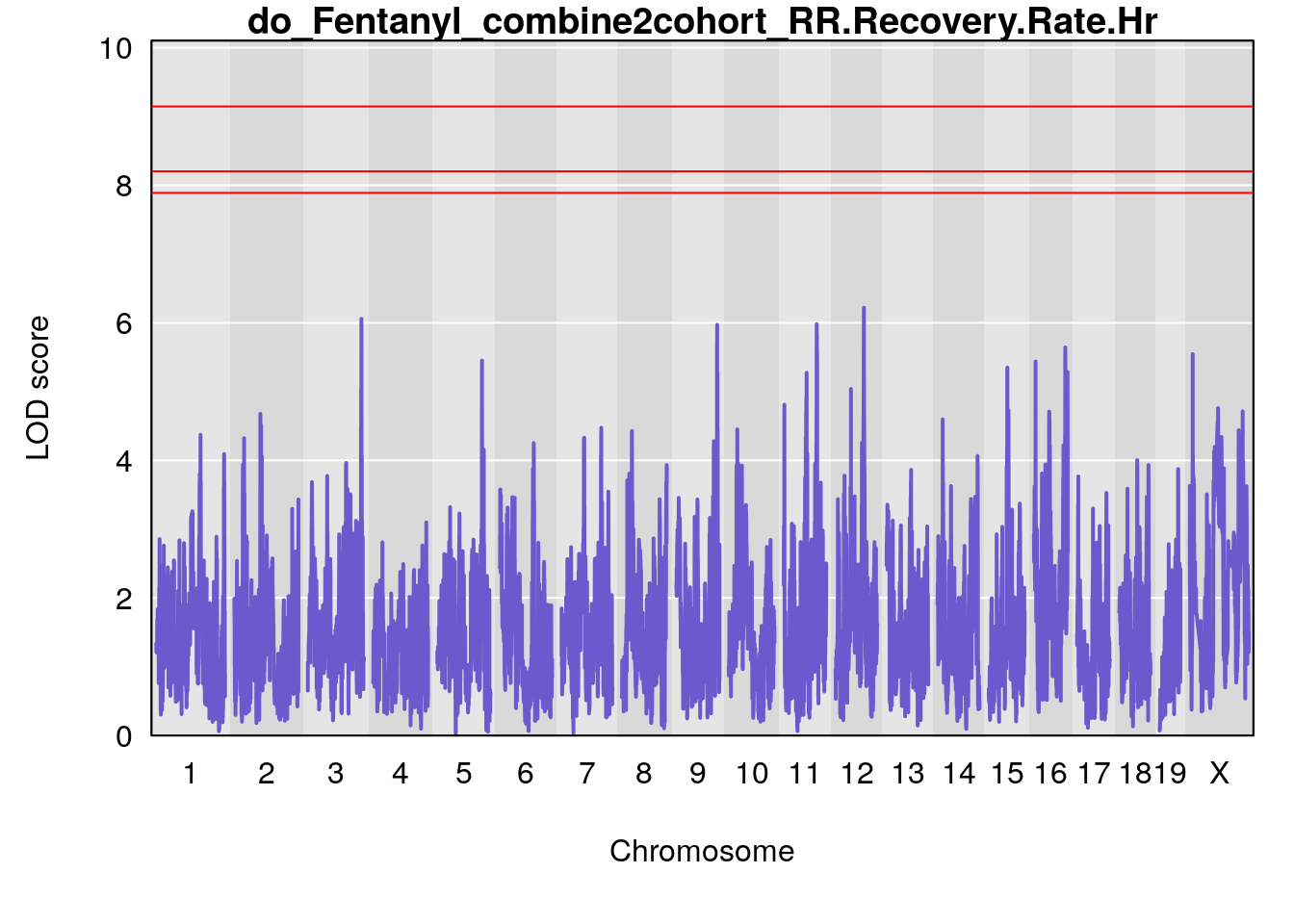
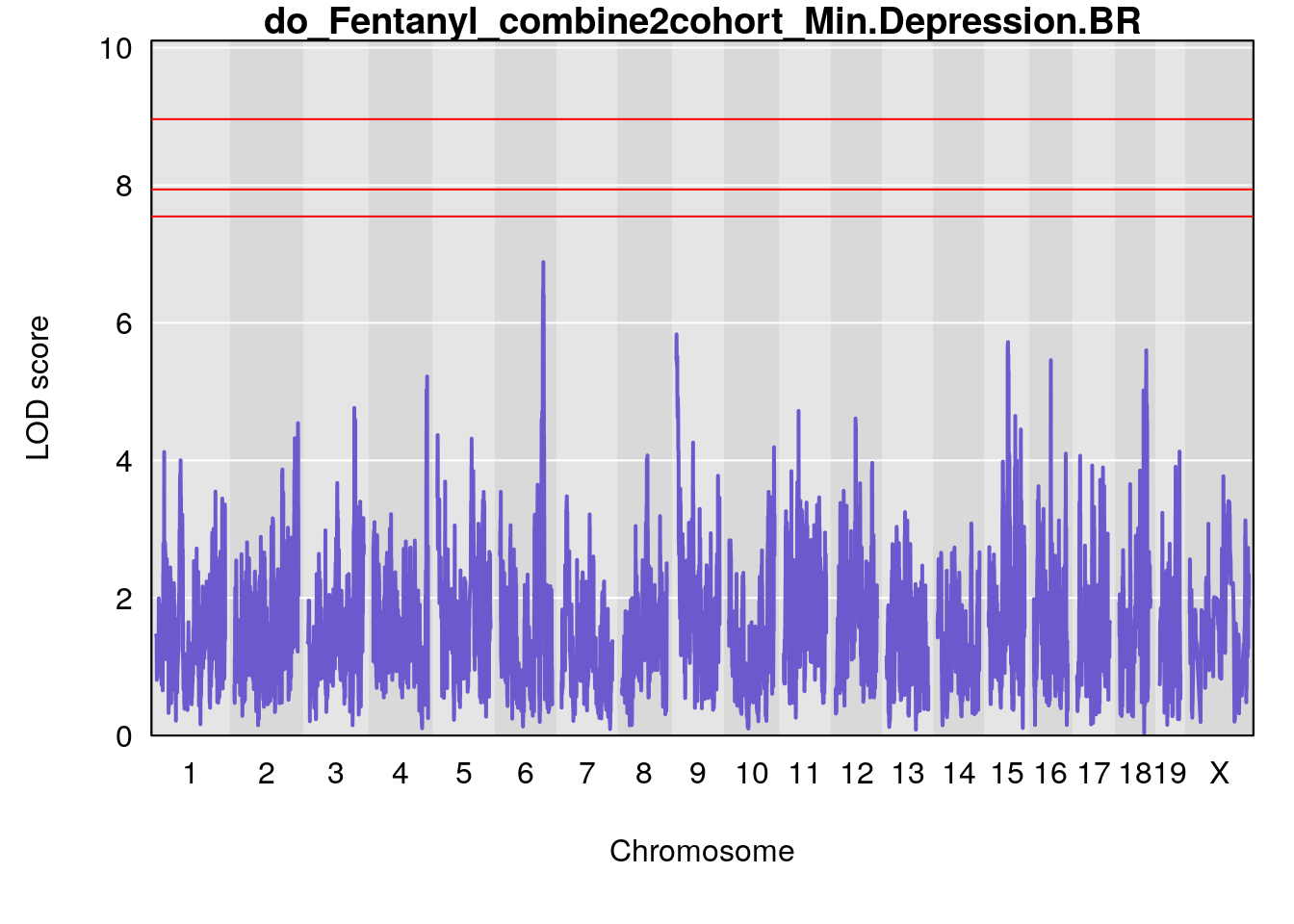
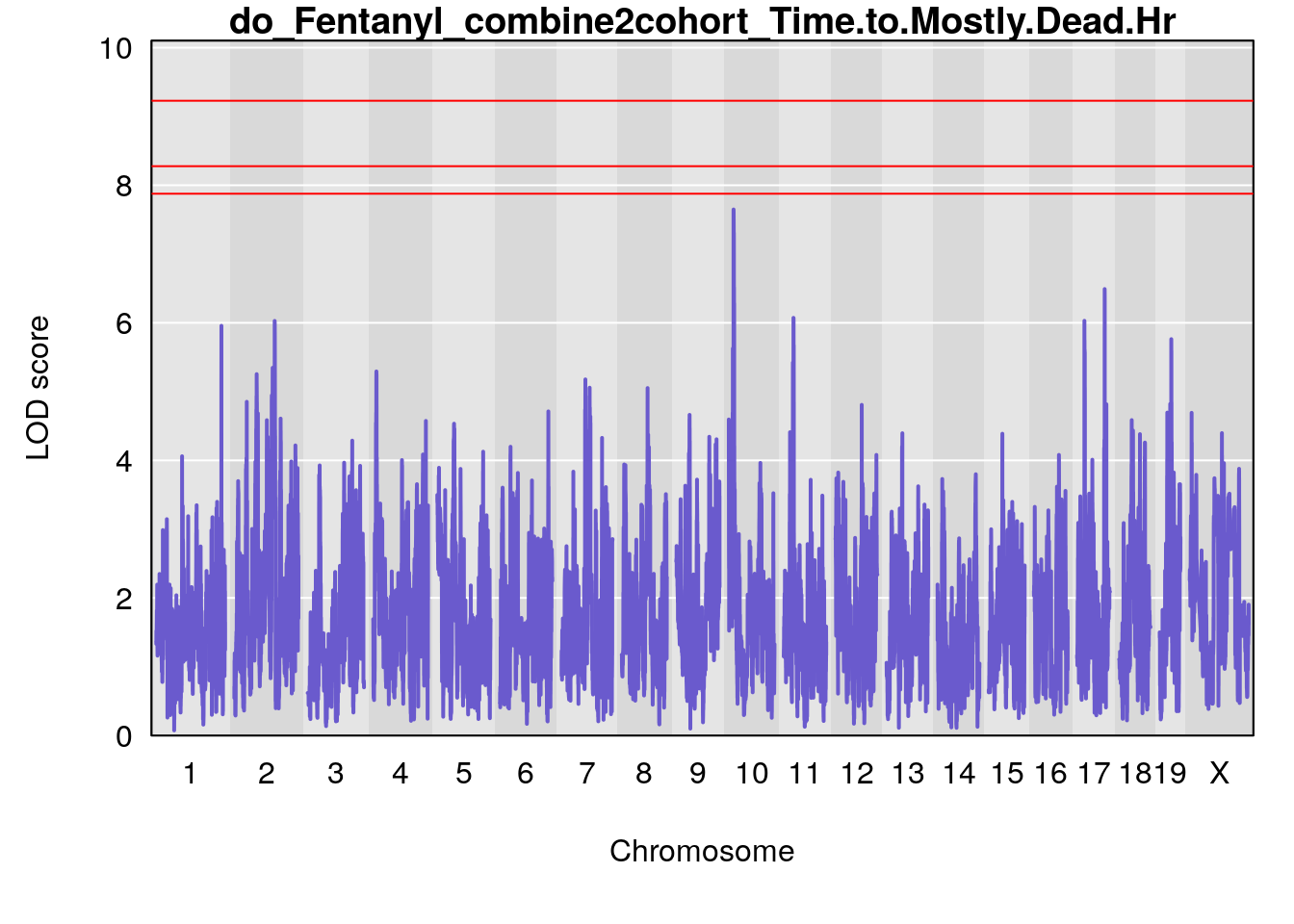
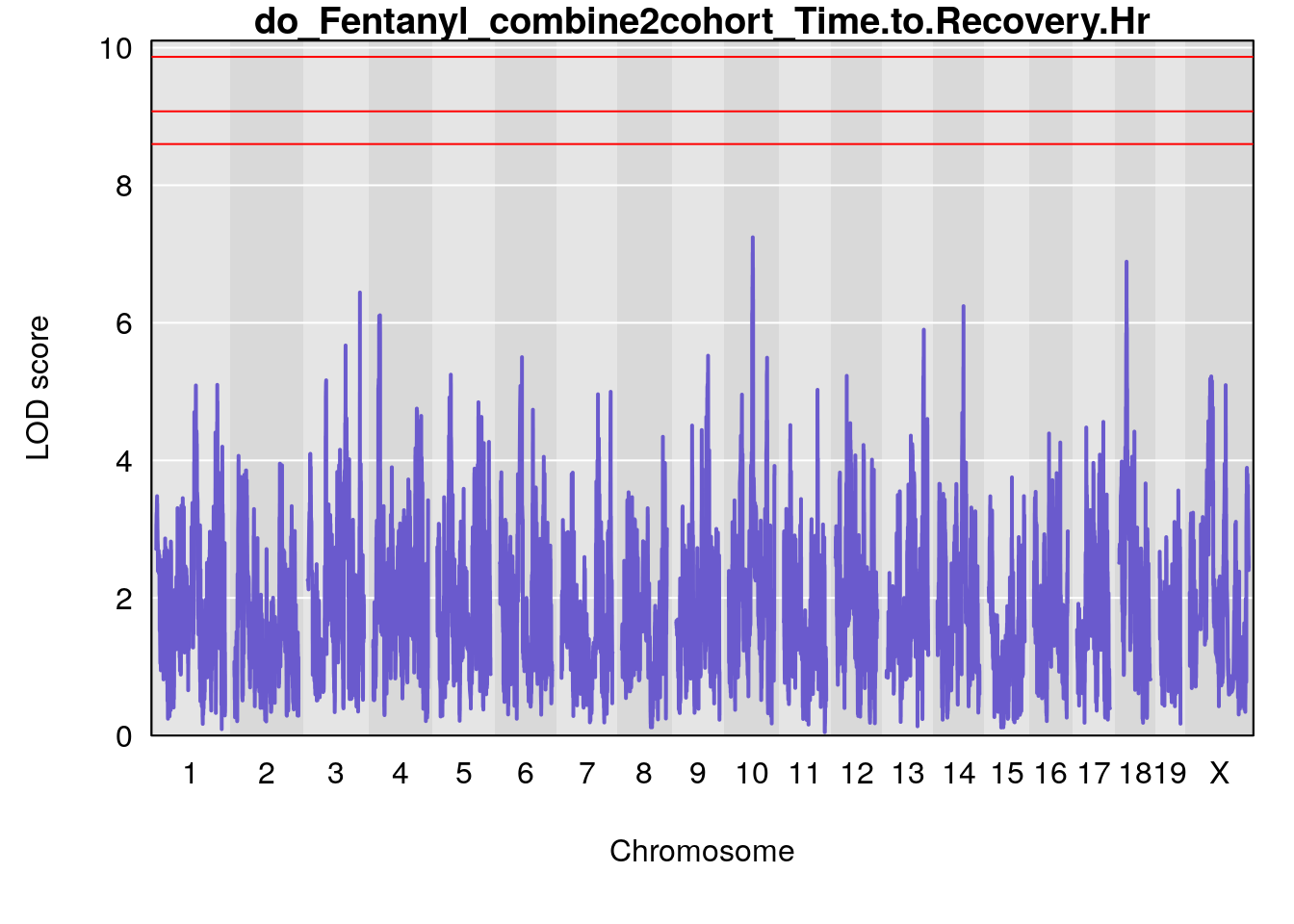
#genome-wide plot
for(i in names(qtl.out)){
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.out[[i]]) # overall maximum LOD score
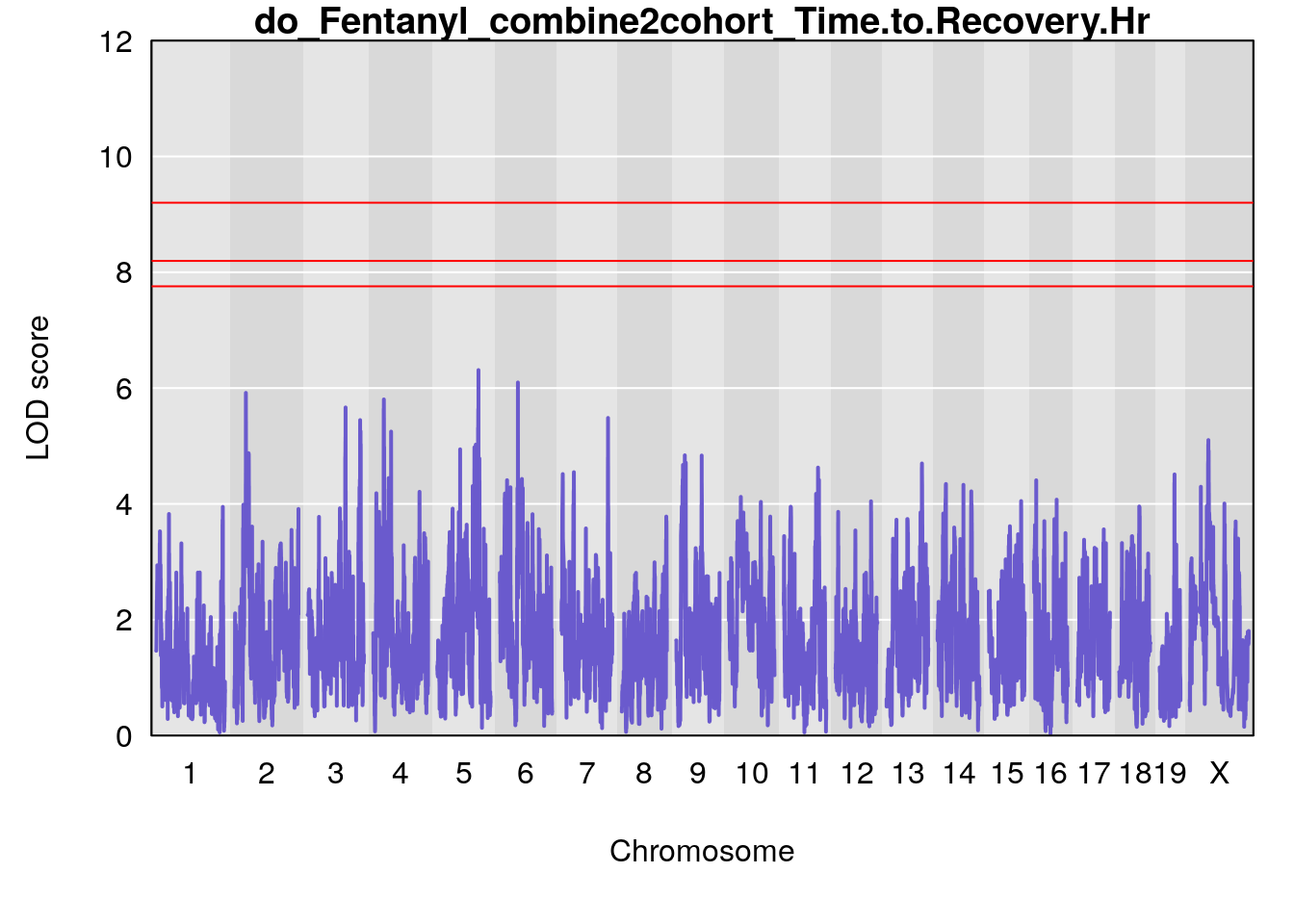
plot(qtl.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, 12))
abline(h=summary(qtl.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("do_Fentanyl_combine2cohort_",i))
}
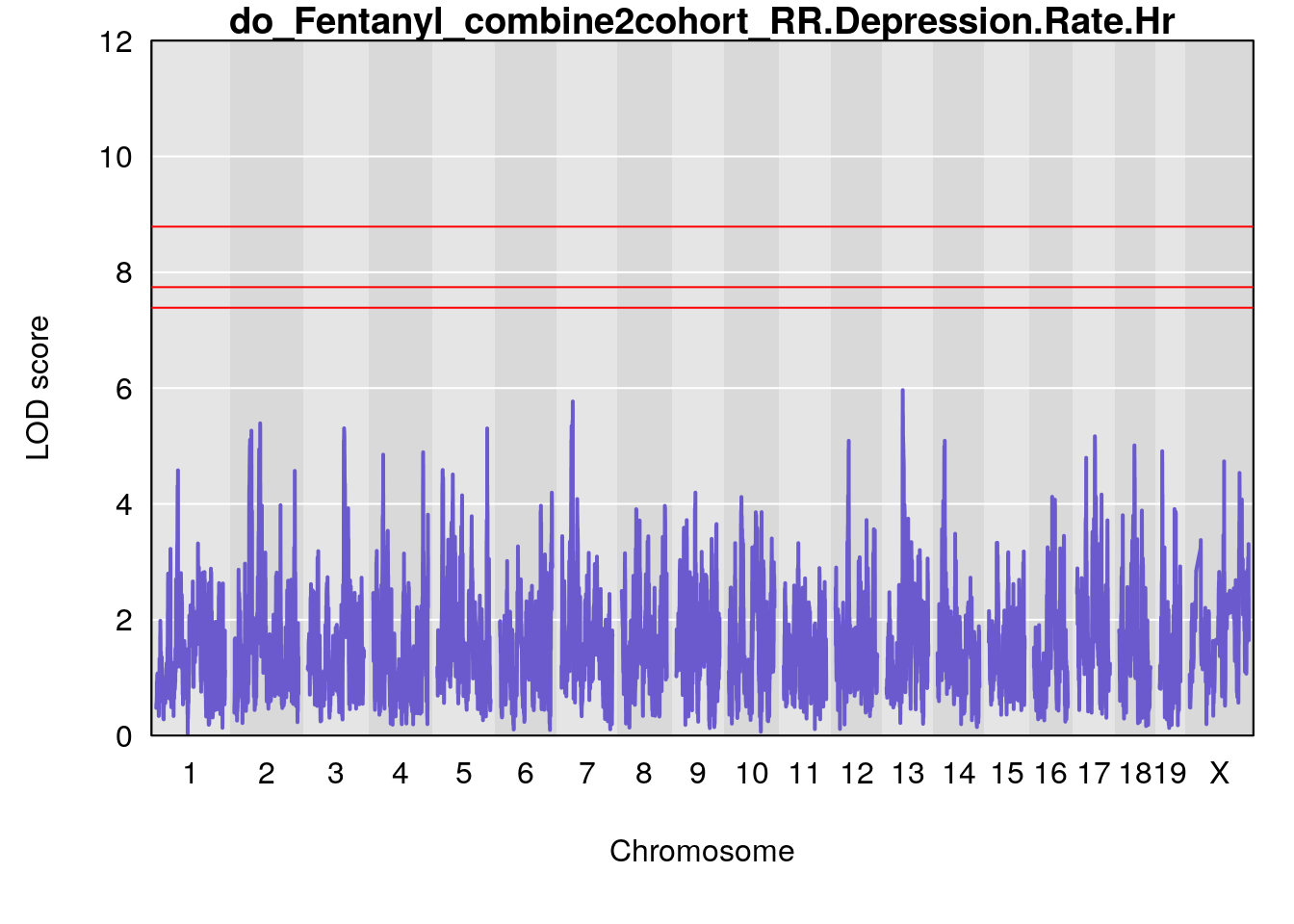
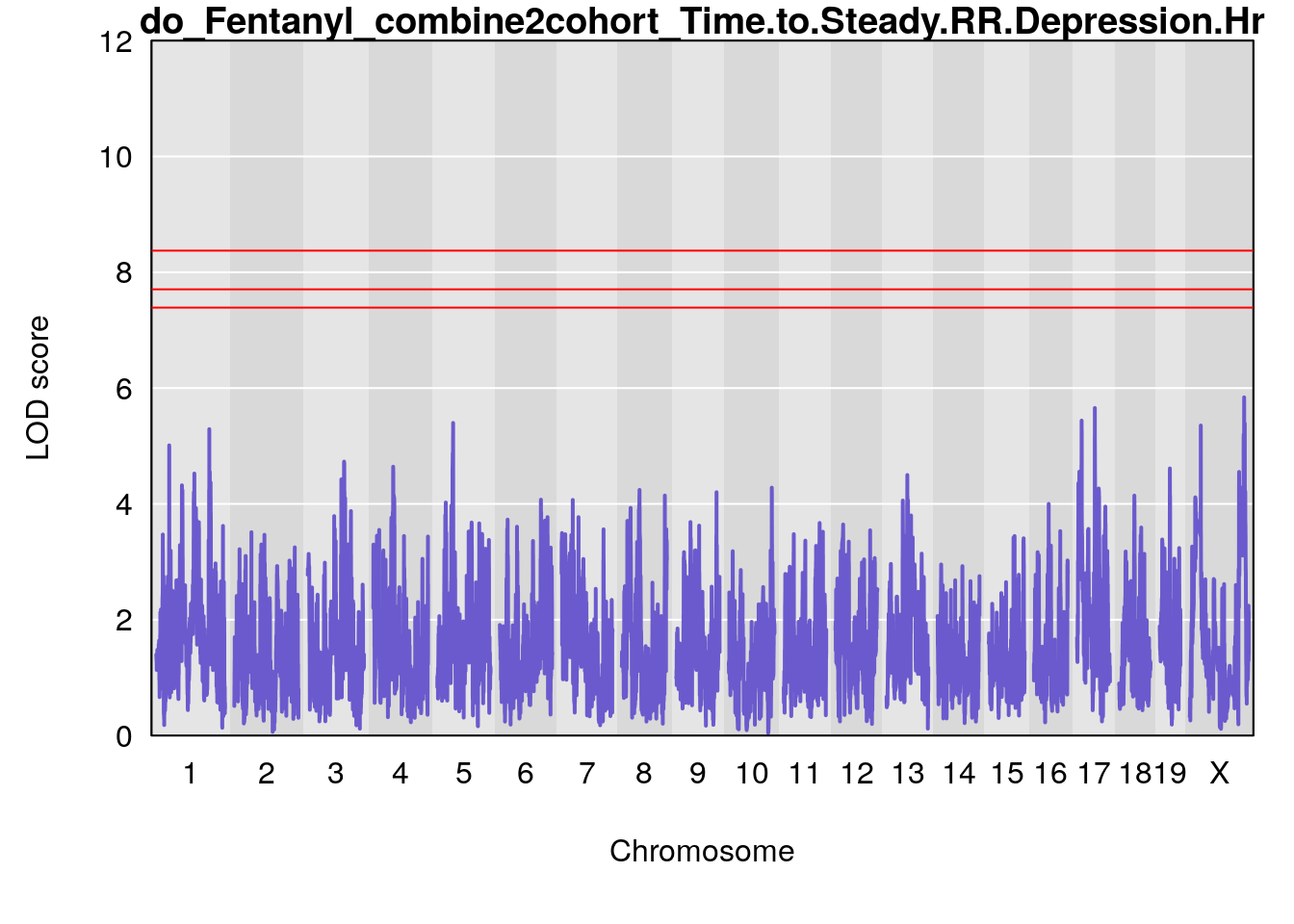
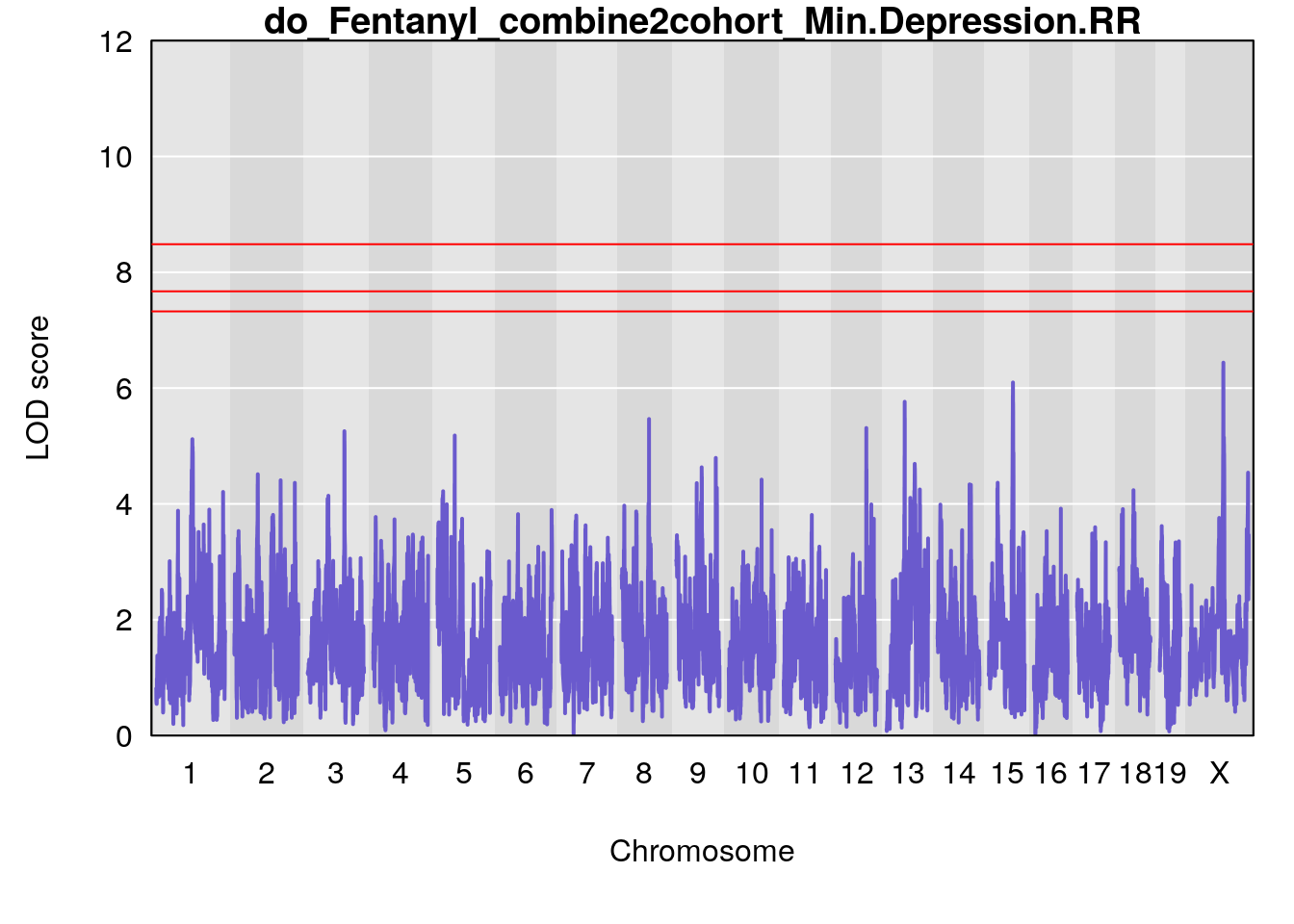
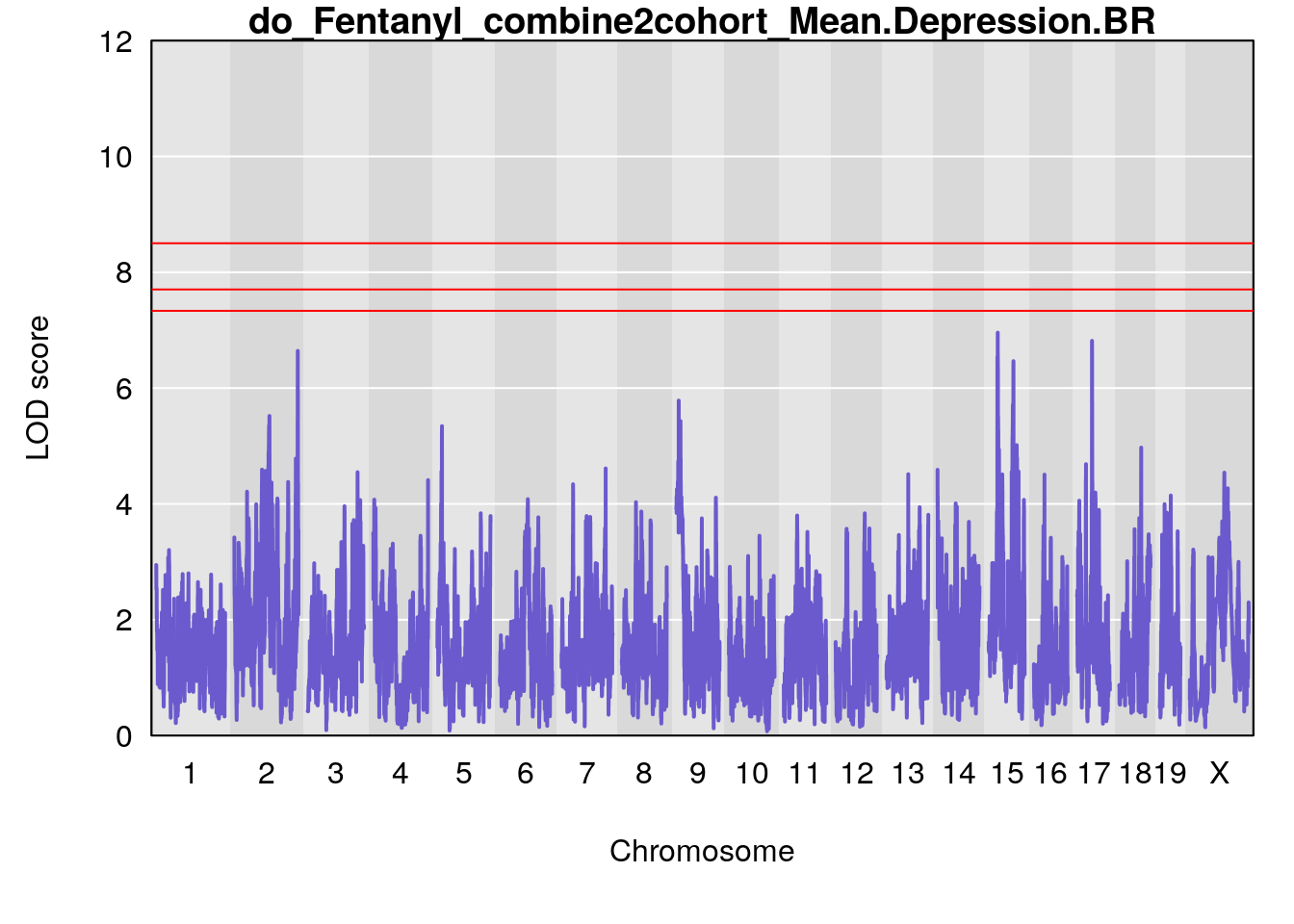
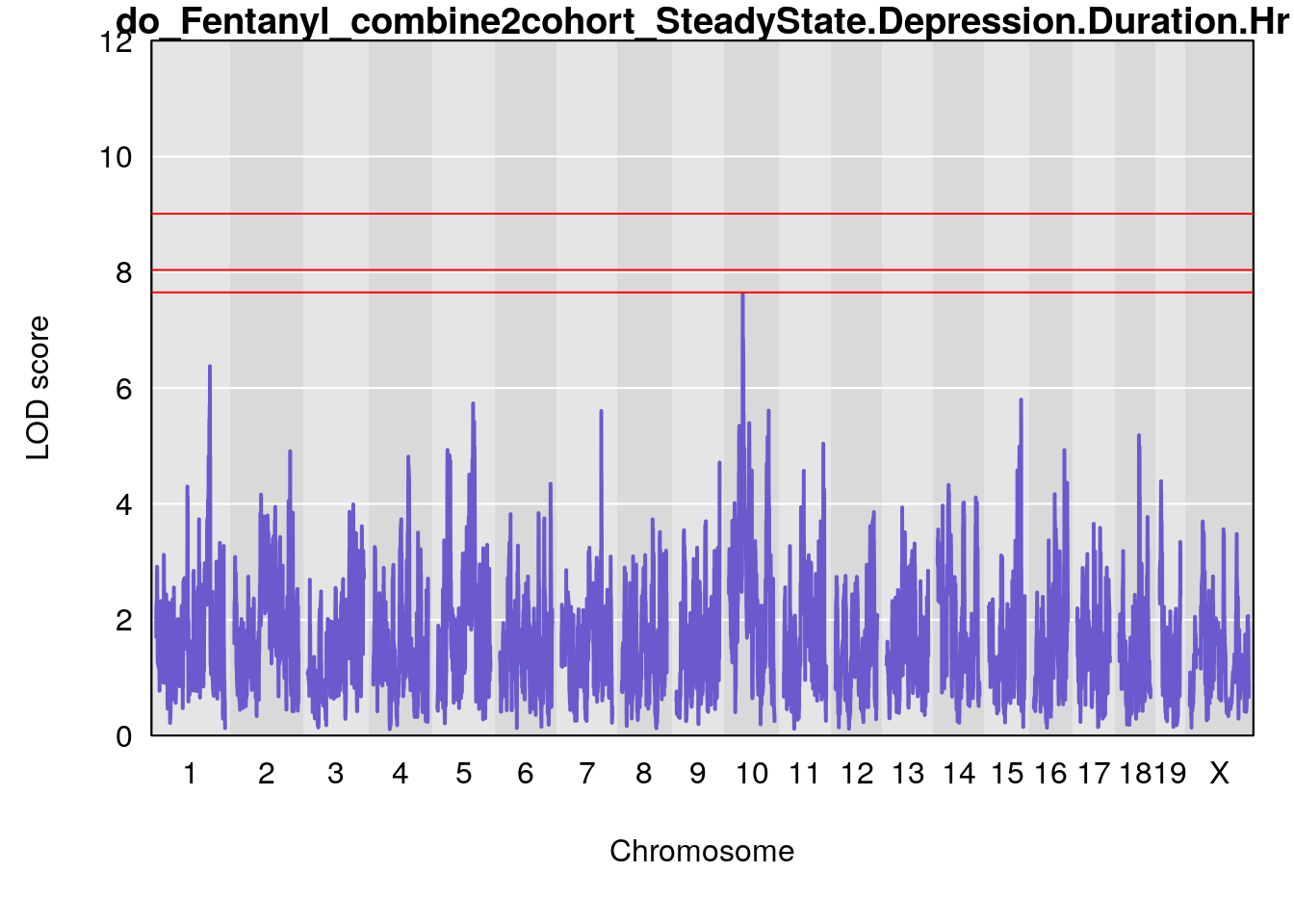
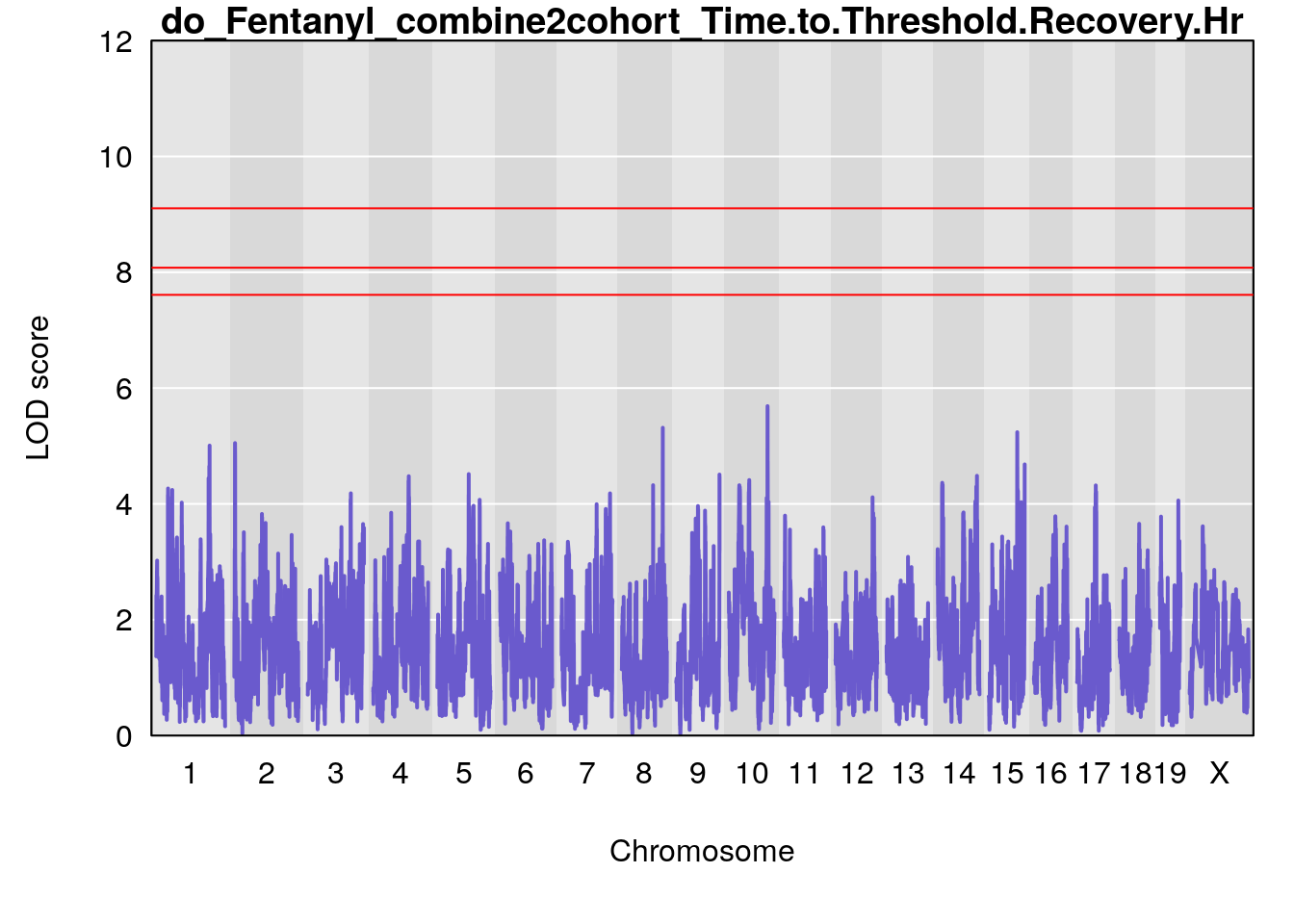
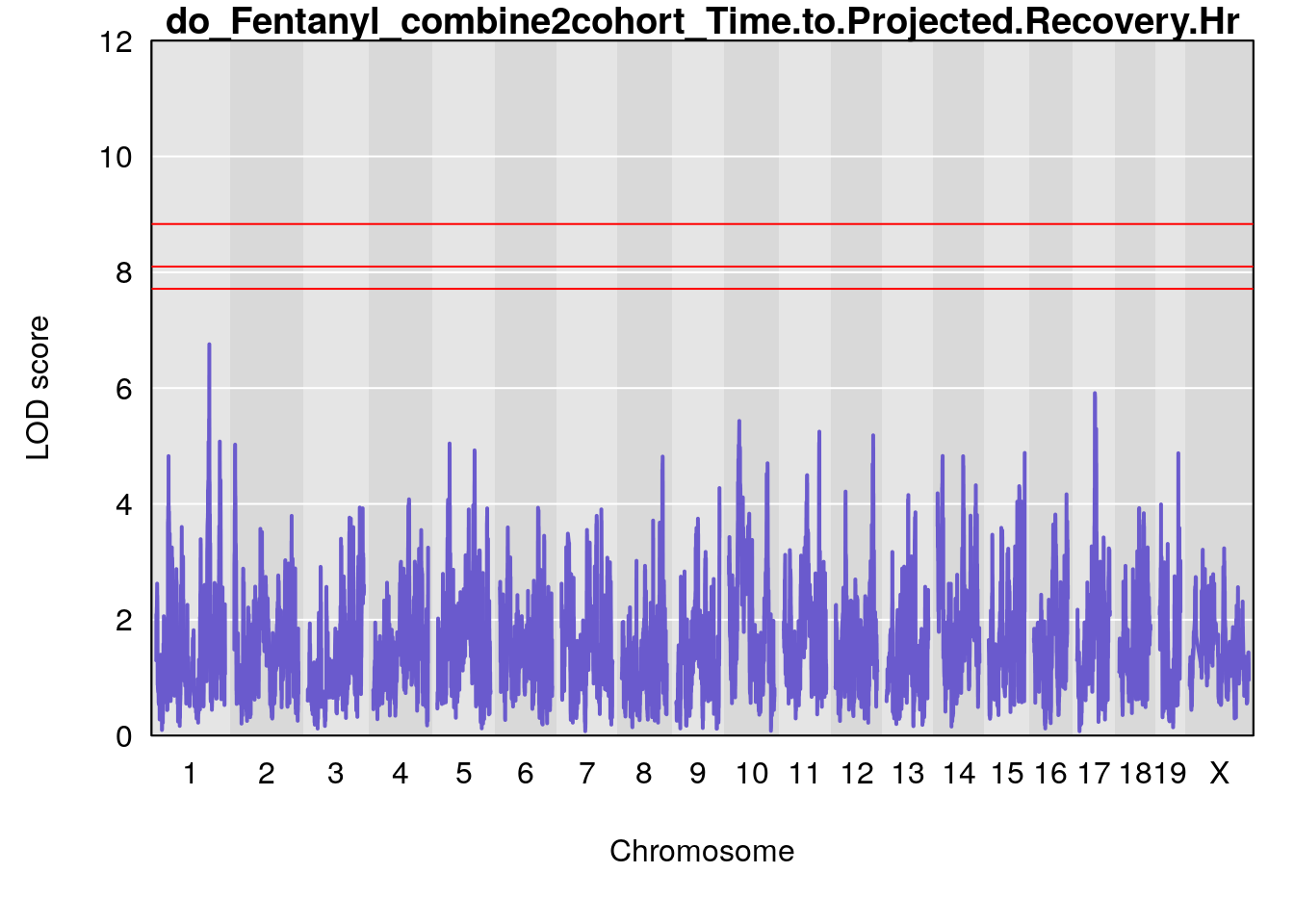
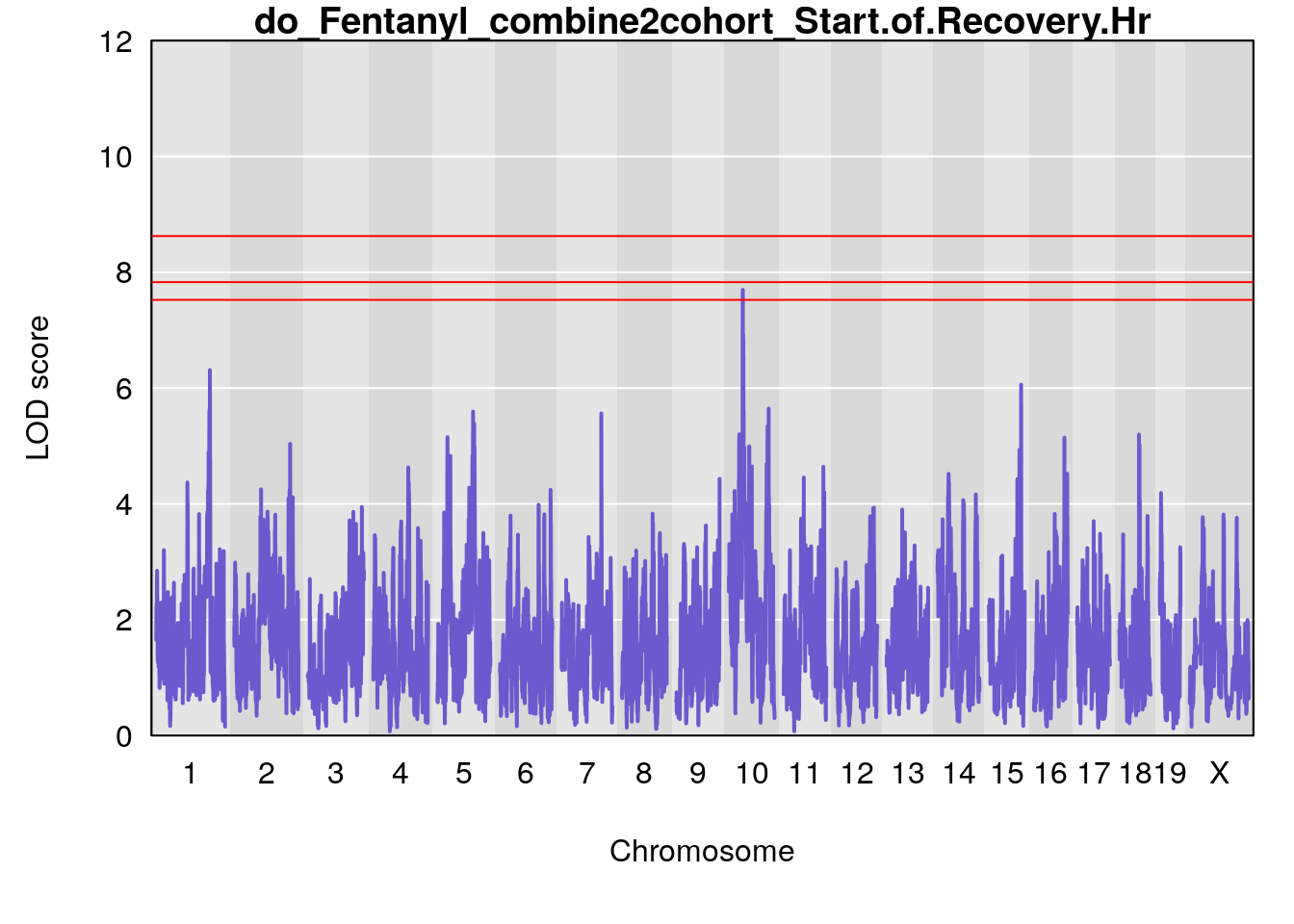
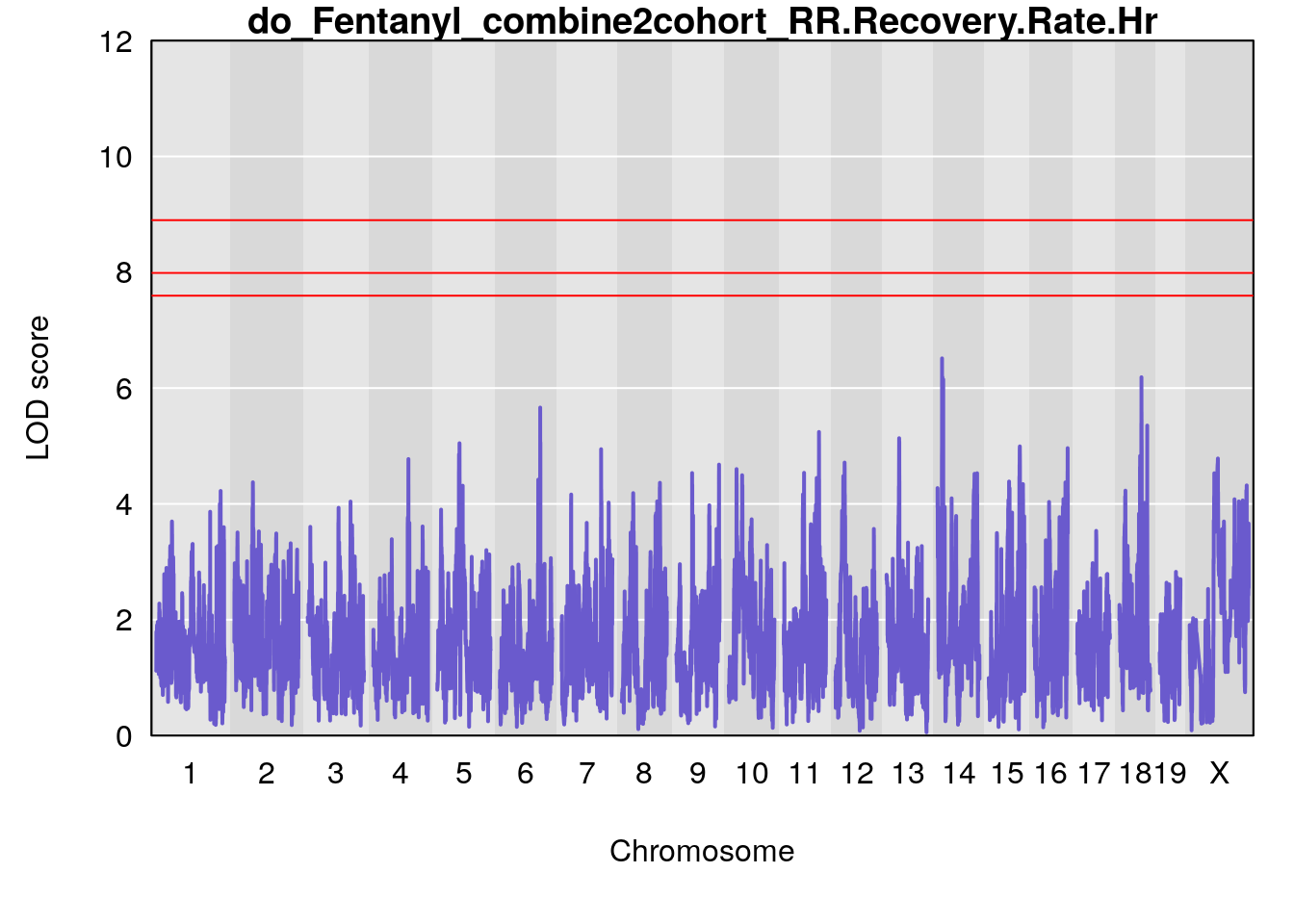
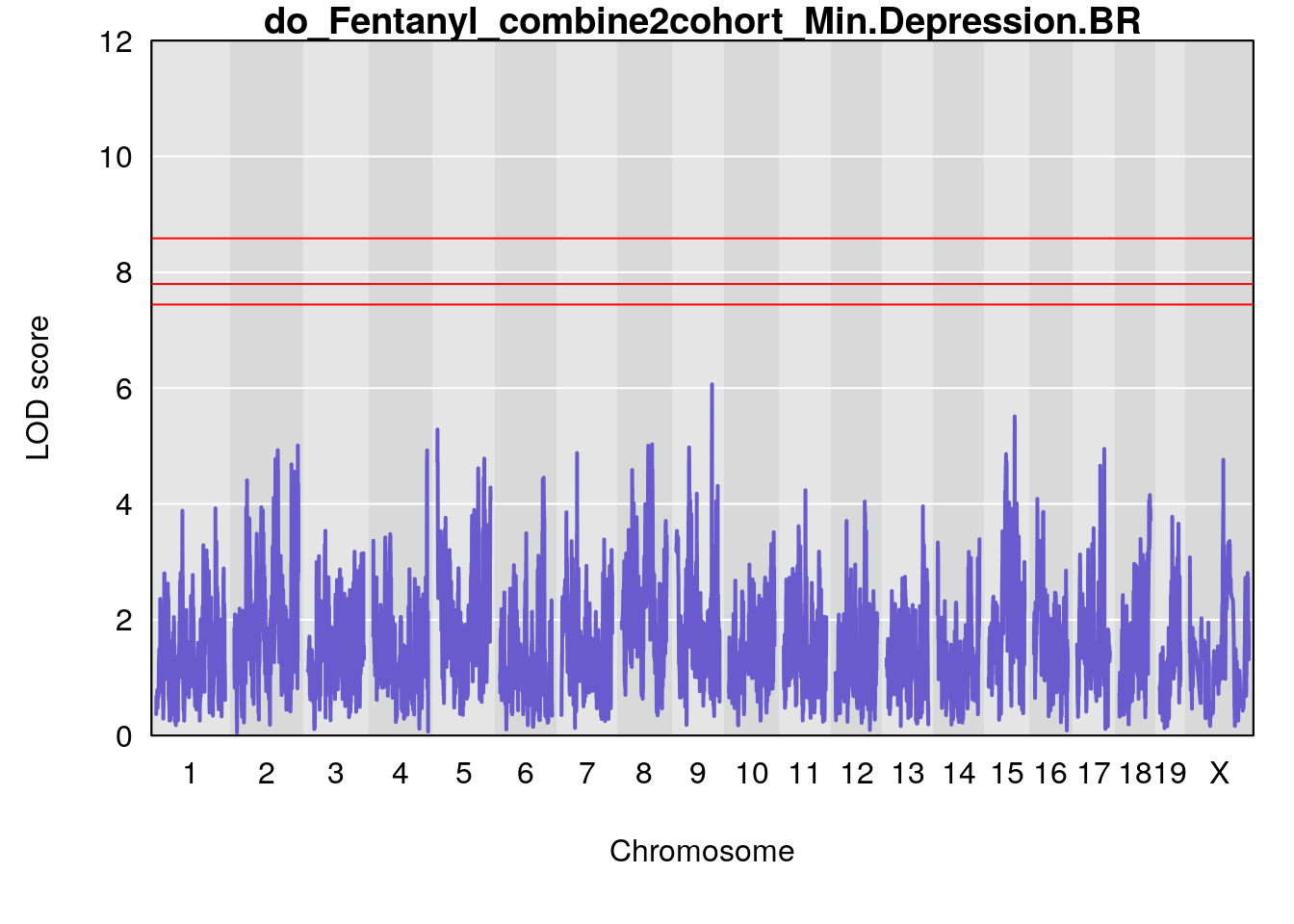
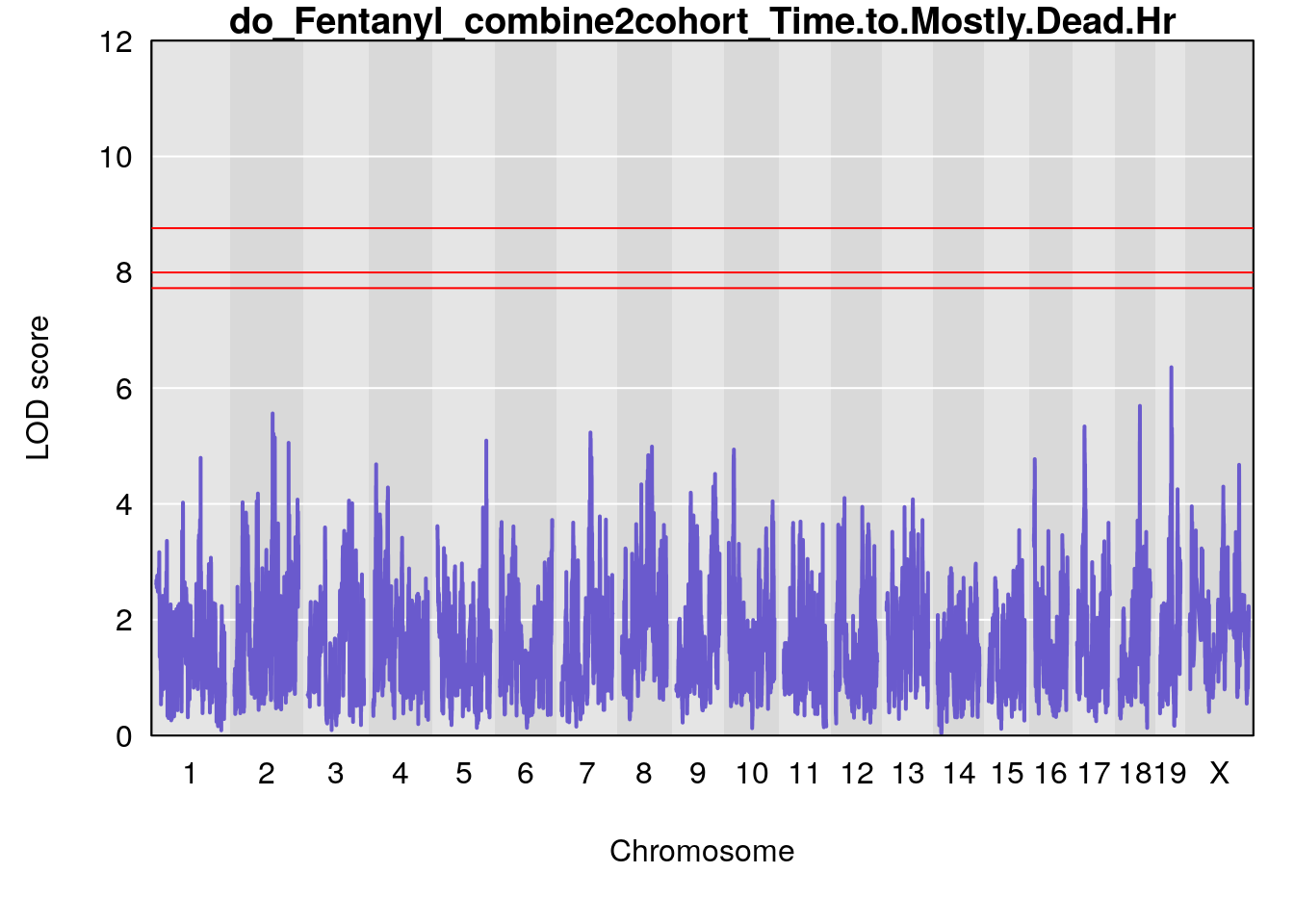
pdf(file = paste0("output/Fentanyl/do_Fentanyl_combine2cohort.pdf"), width = 16, height =8)
#save genome-wide plo
for(i in names(qtl.out)){
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.out[[i]]) # overall maximum LOD score
plot(qtl.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, 10))
abline(h=summary(qtl.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("do_Fentanyl_combine2cohort_",i))
}
dev.off()
# png
# 2
#peaks coeff plot
for(i in names(qtl.out)){
print(i)
peaks <- find_peaks(qtl.out[[i]], map=pmap, threshold=6, drop=1.5)
if(nrow(peaks) > 0) {
print(peaks)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
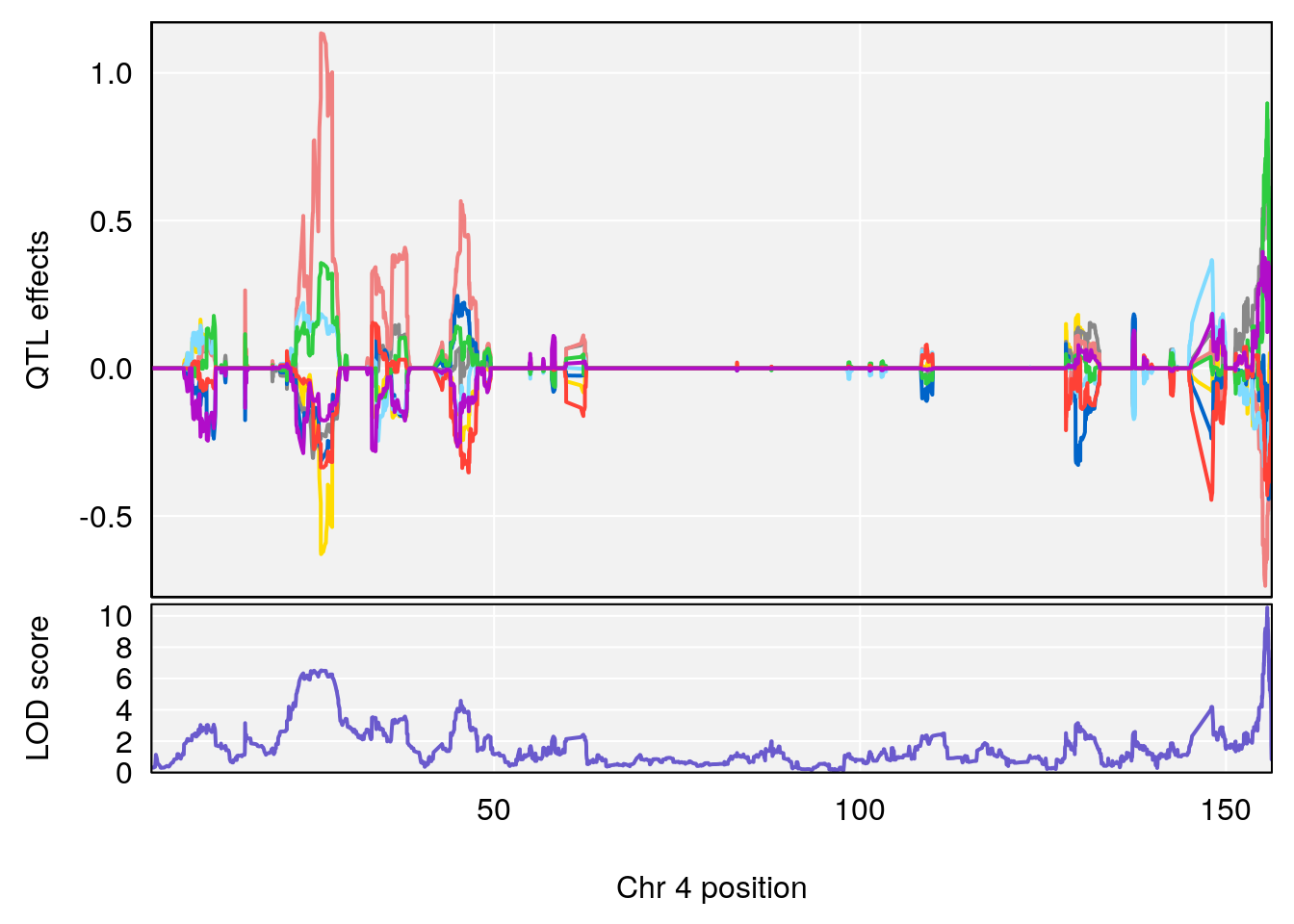
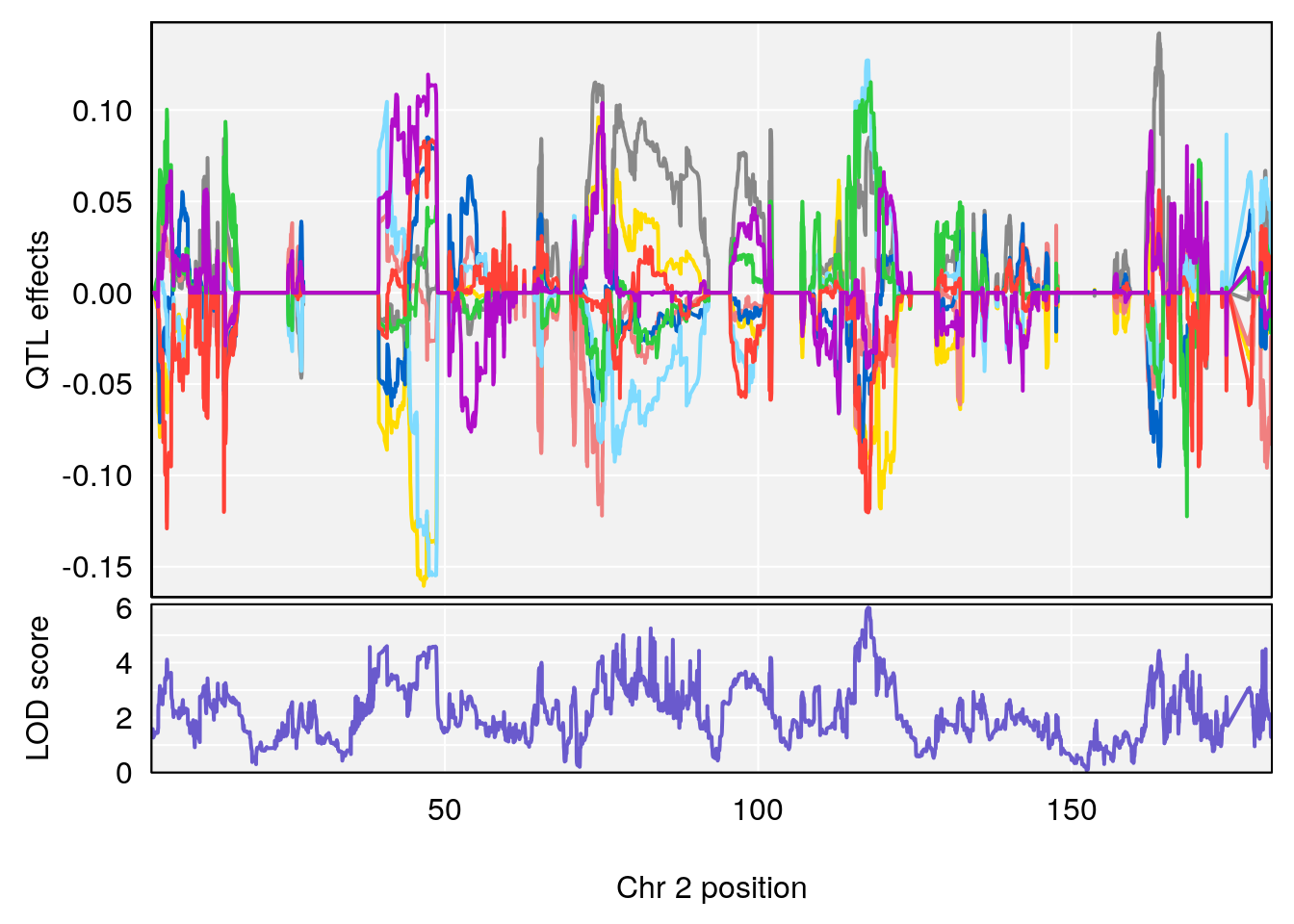
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.out[[i]], bgcolor="gray95", legend=NULL)
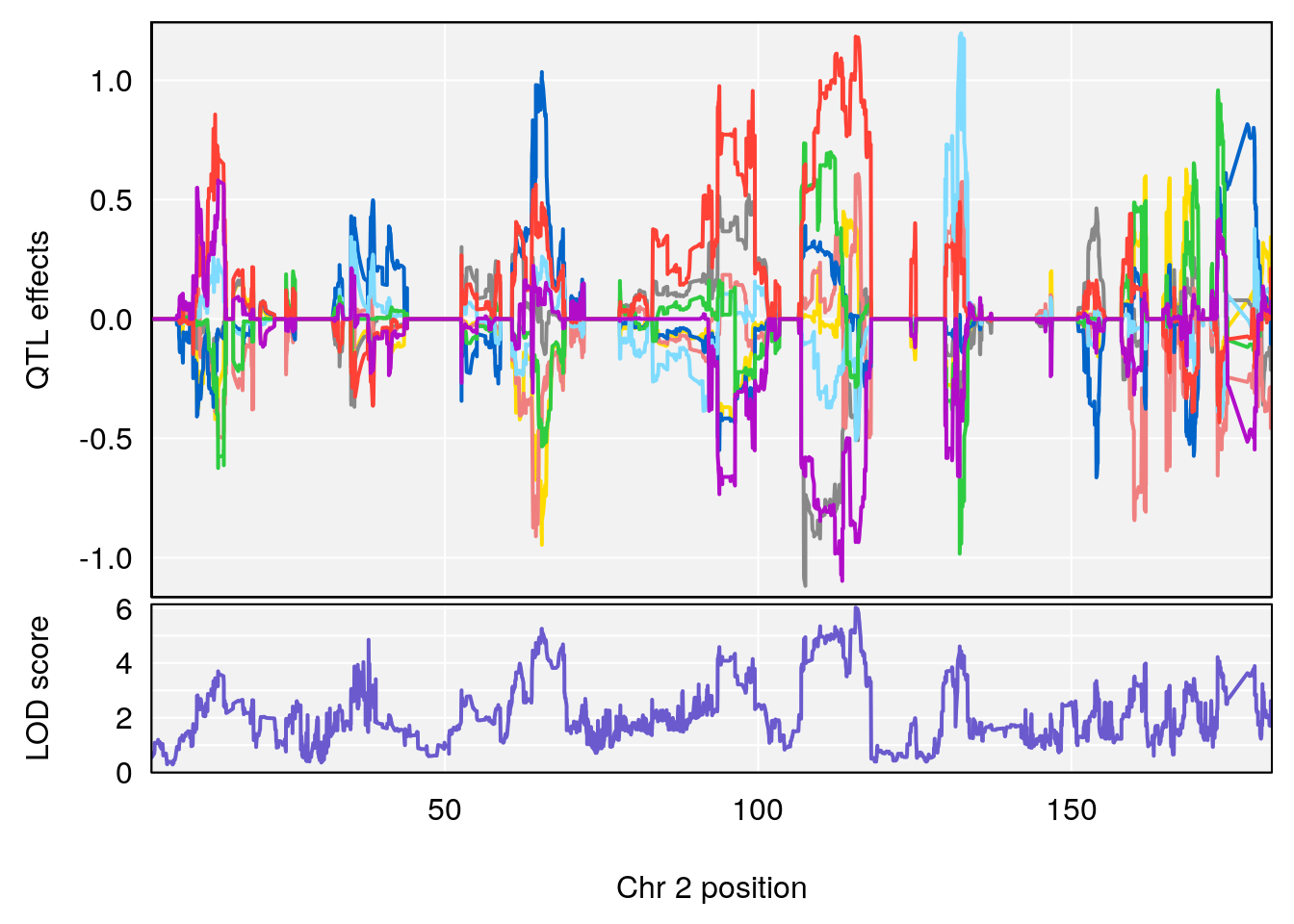
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.out[[i]], bgcolor="gray95", legend=NULL)
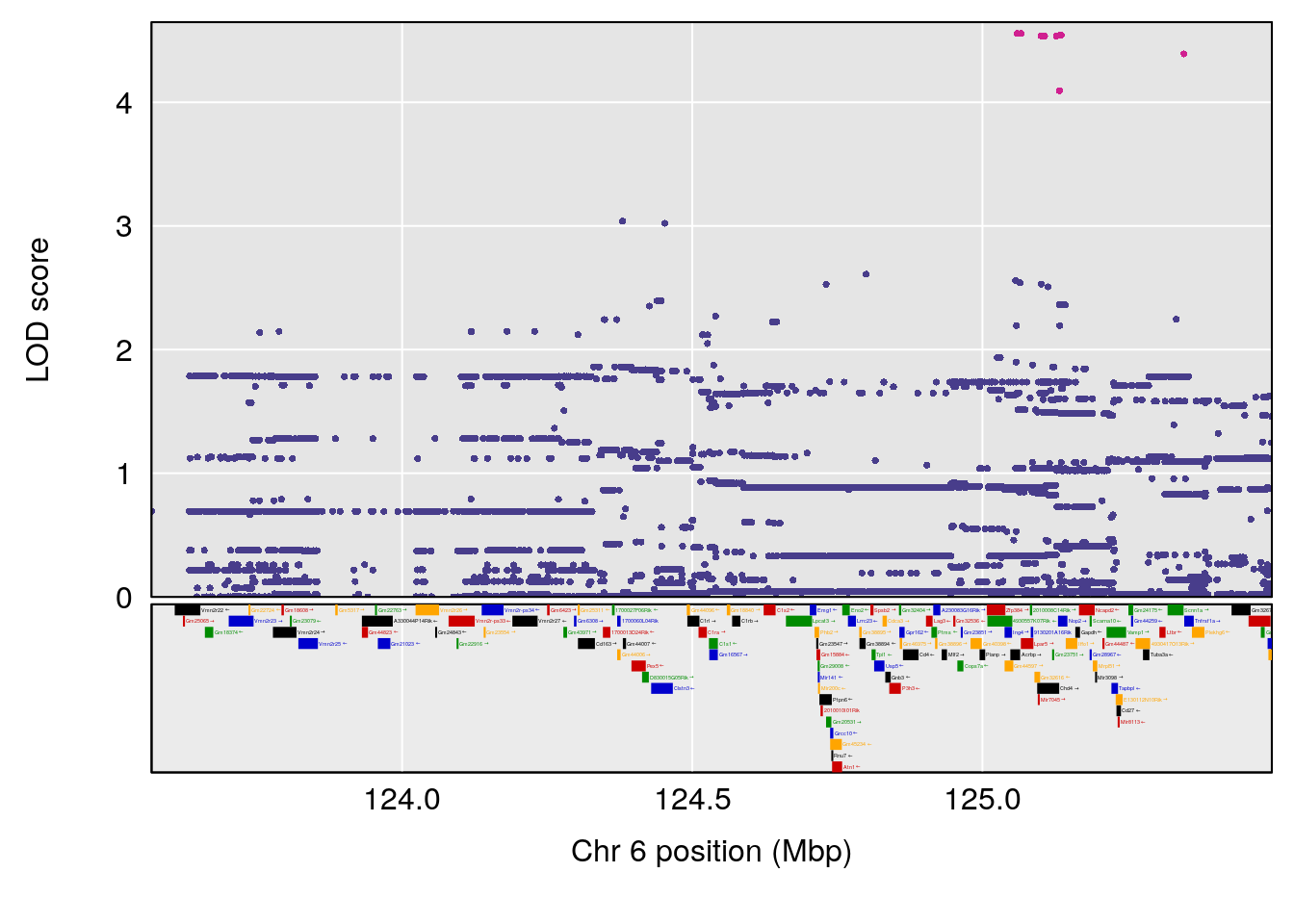
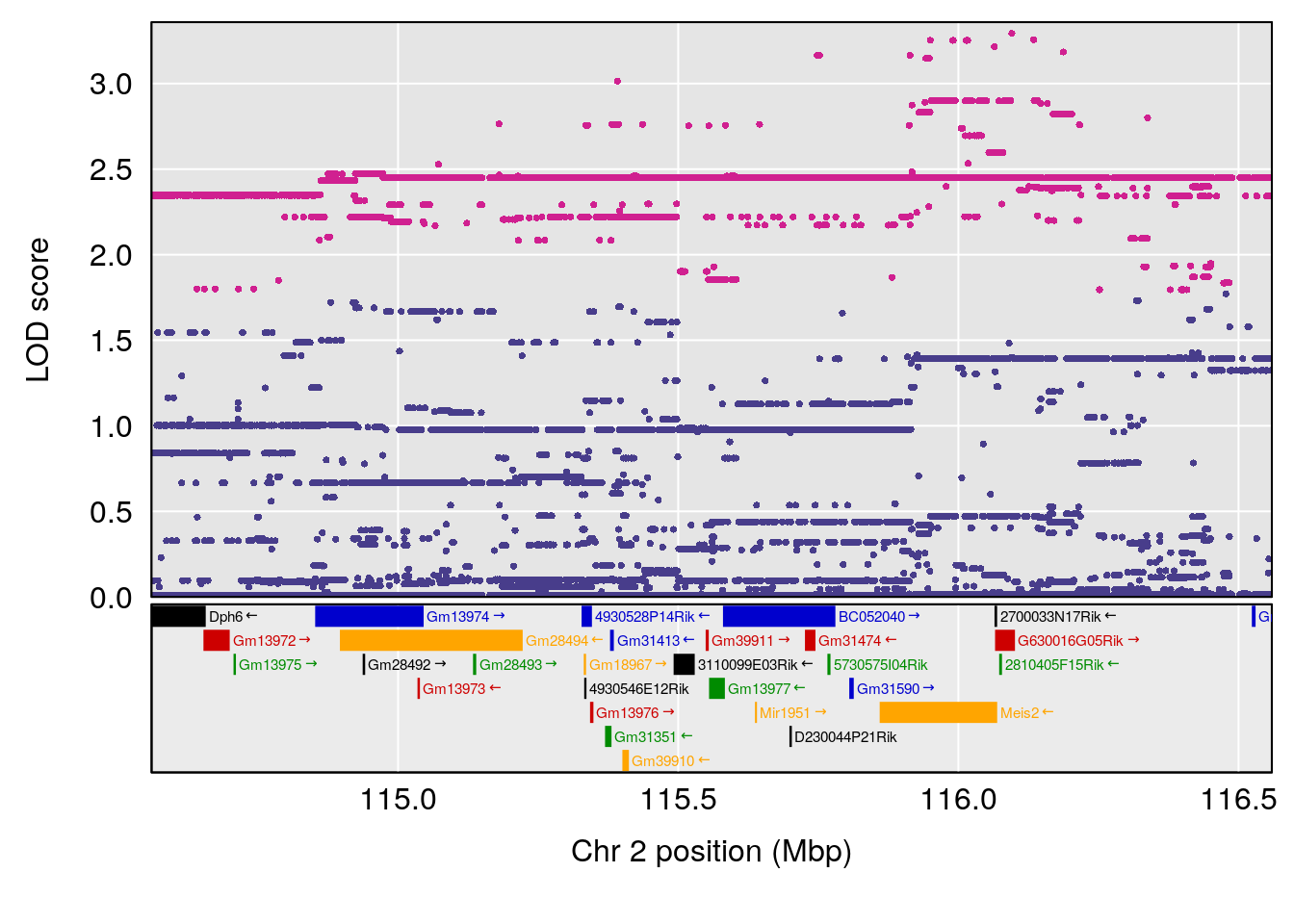
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
}
}
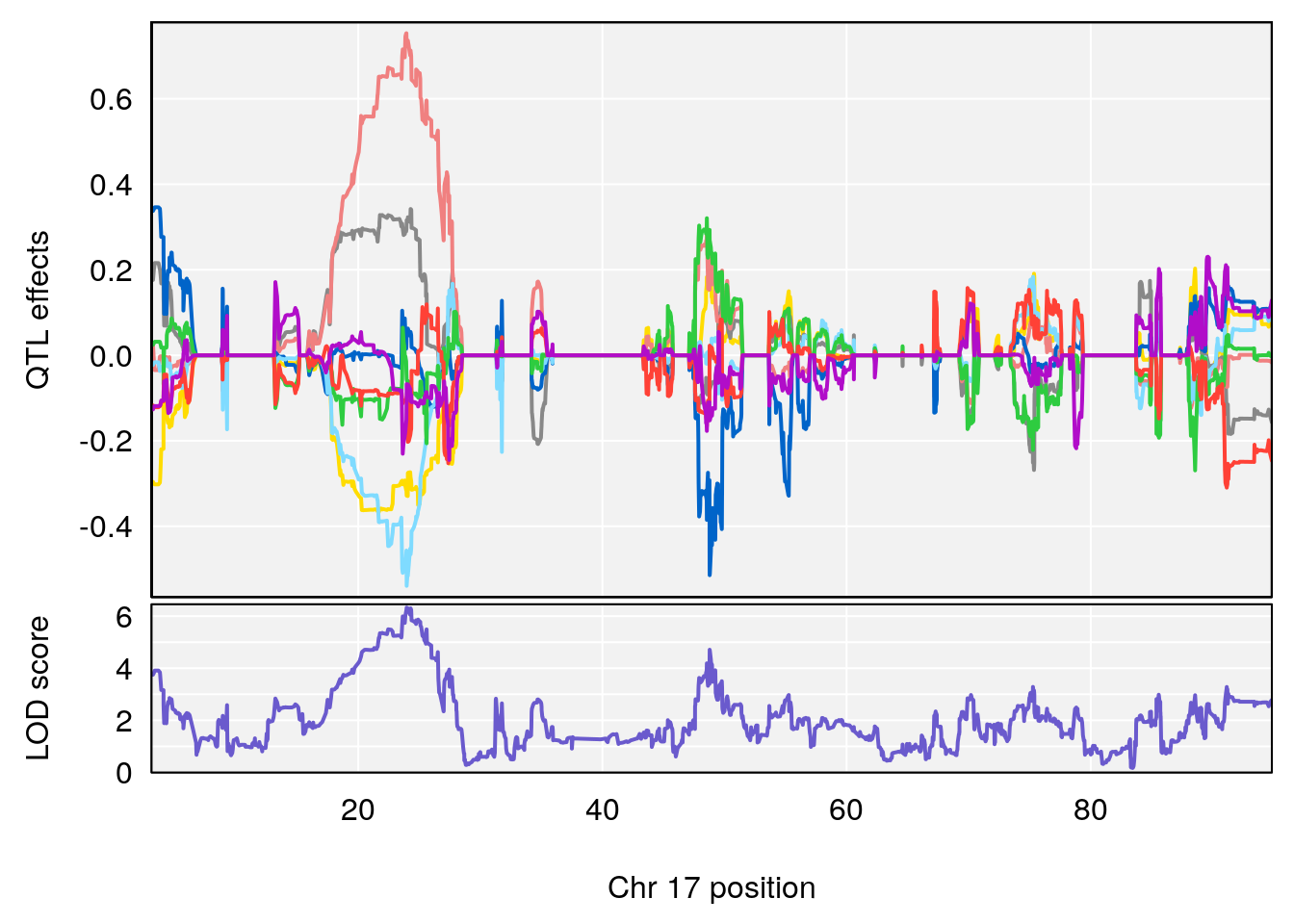
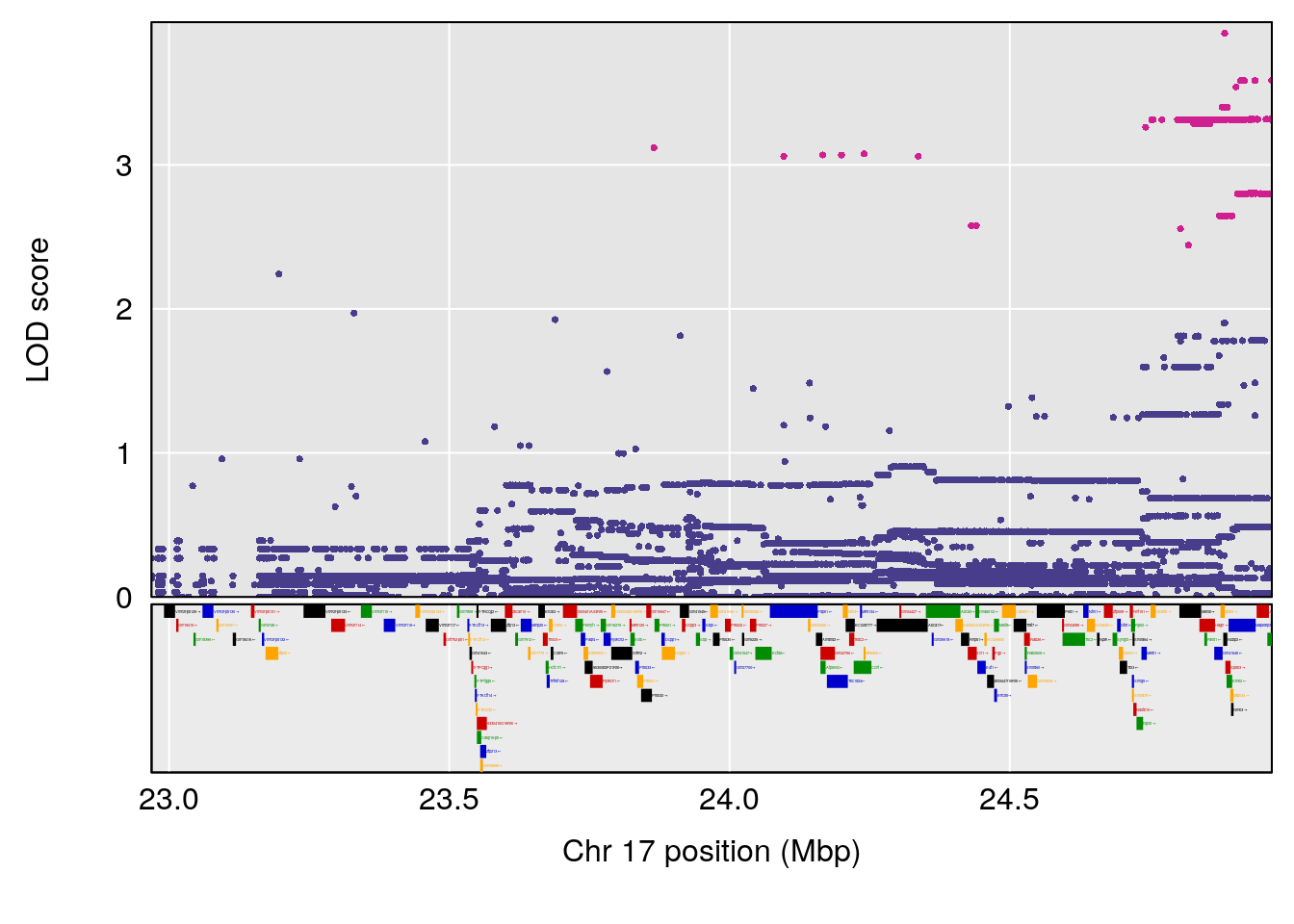
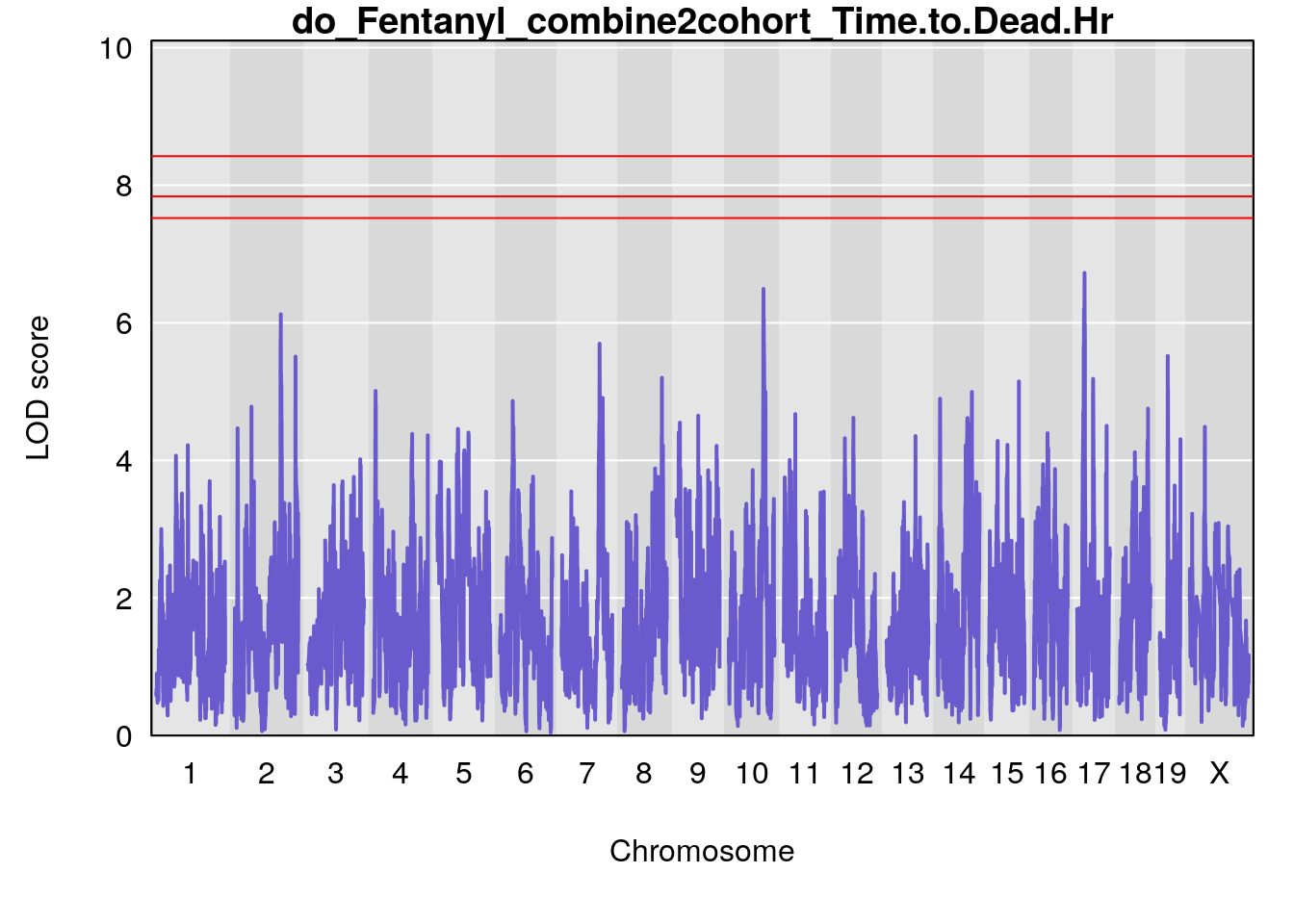
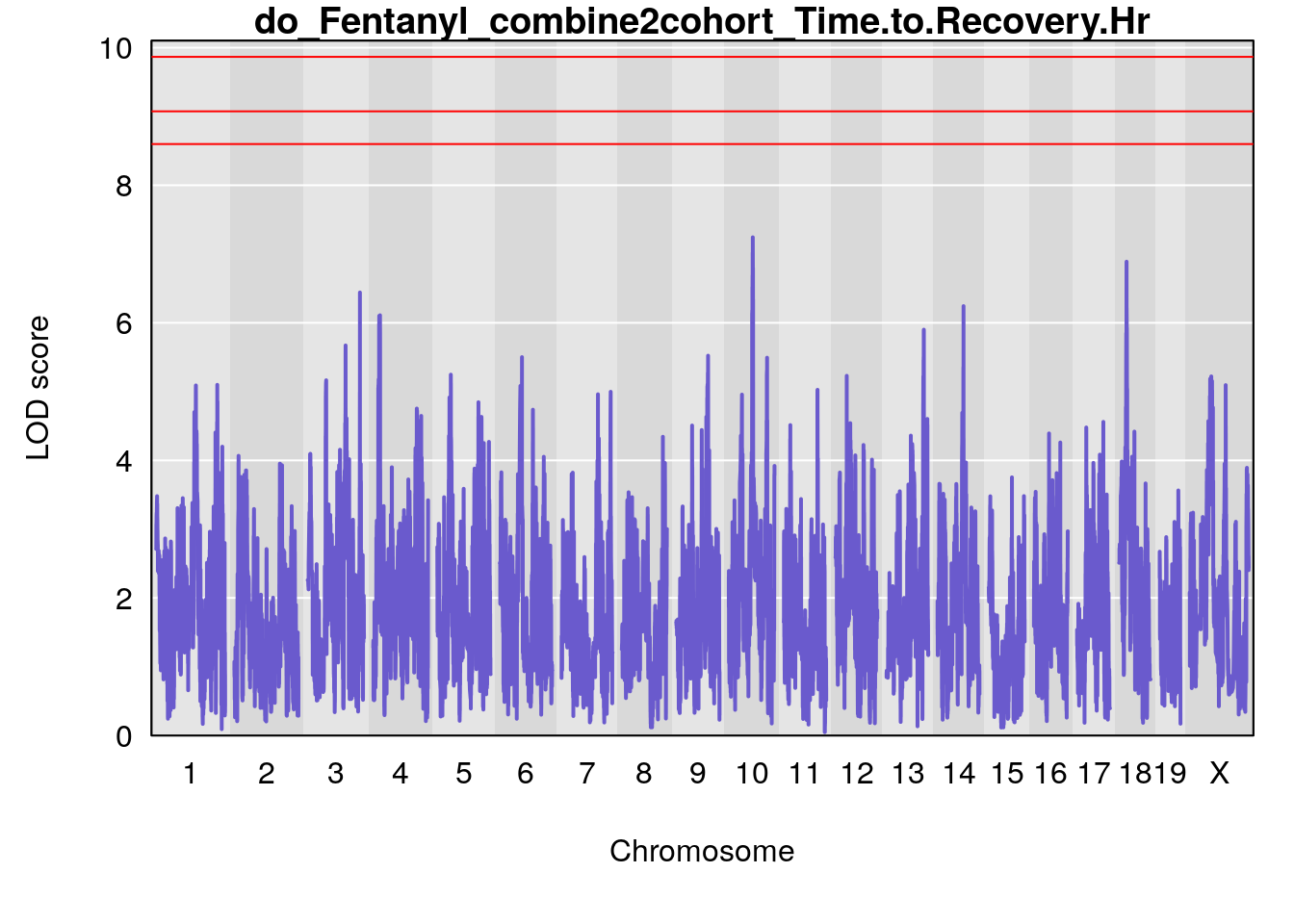
# [1] "Time.to.Dead.Hr"
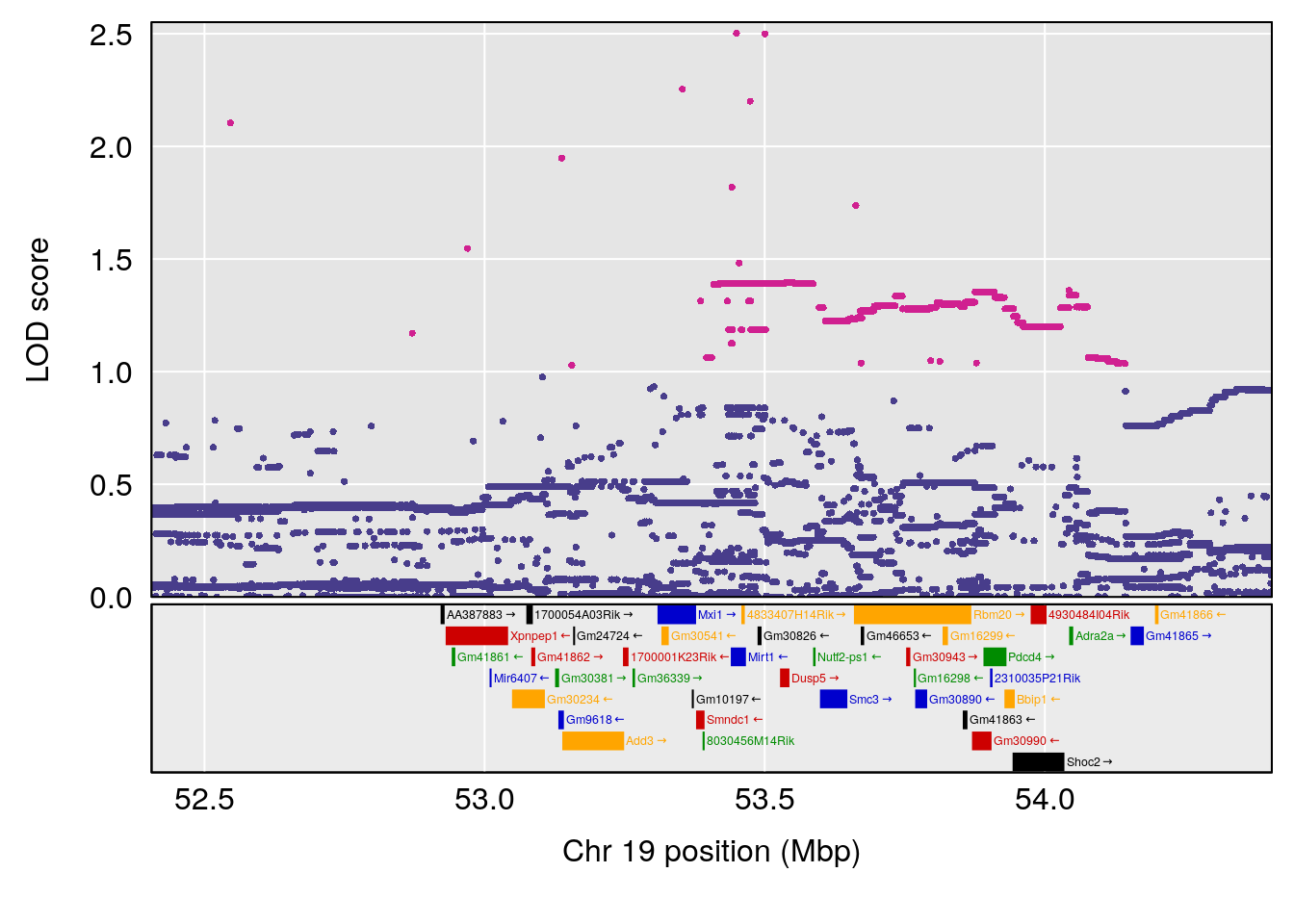
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 4 155.62894 10.526649 155.33847 155.75940
# 2 1 pheno1 15 100.98983 6.626767 87.42813 101.41826
# 3 1 pheno1 17 23.96788 6.327276 21.57341 26.00445
# [1] 1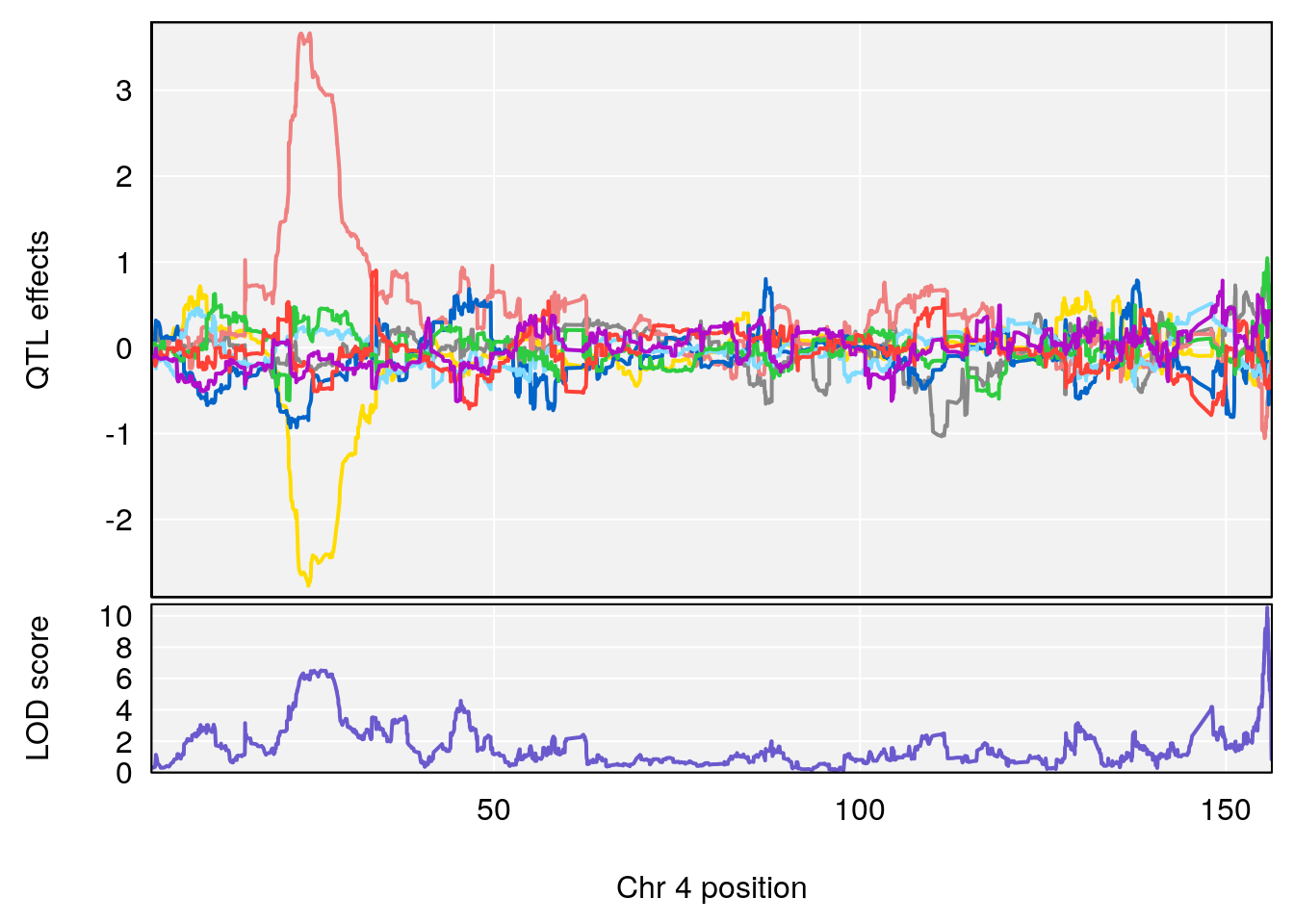
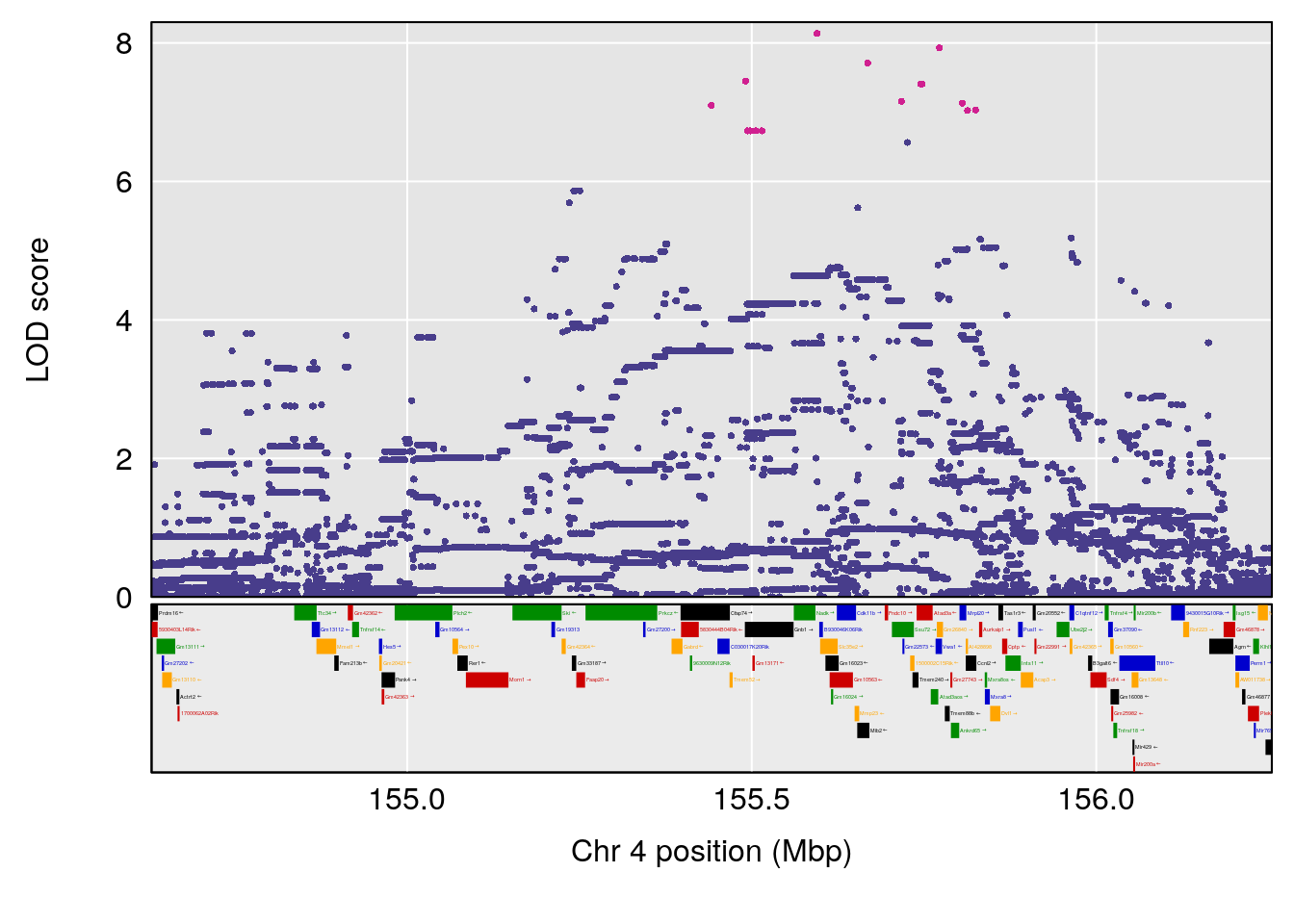
# [1] 2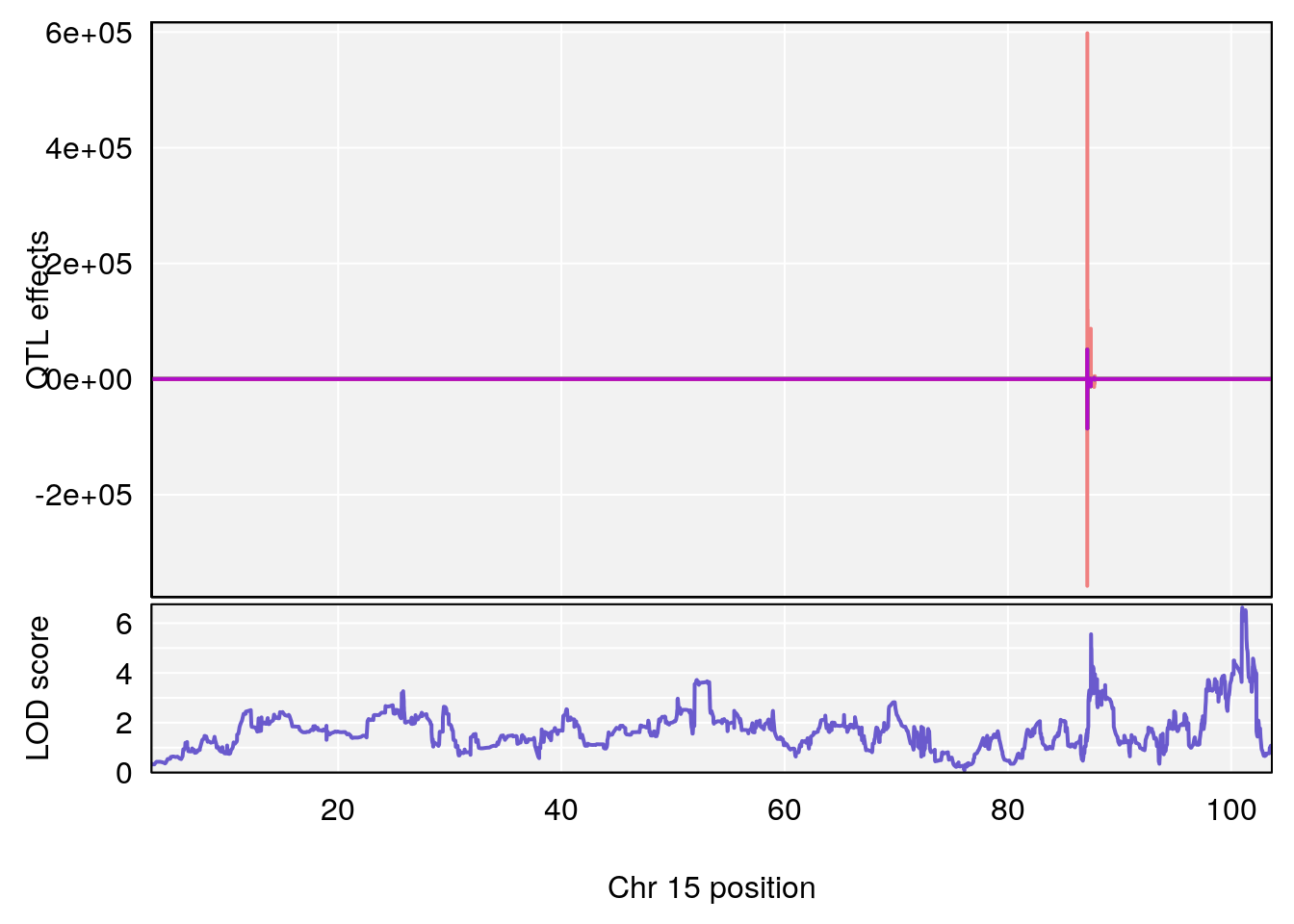
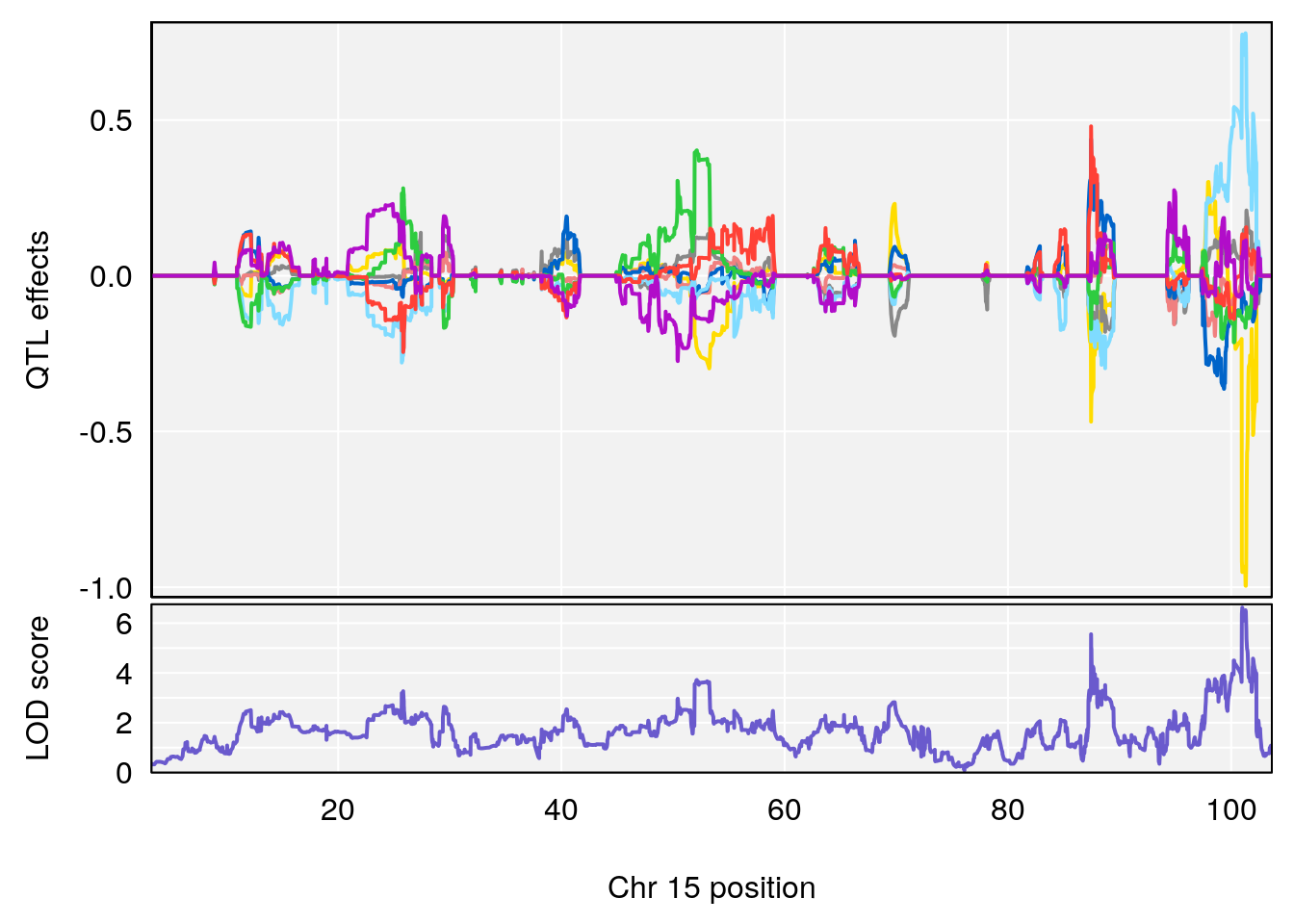
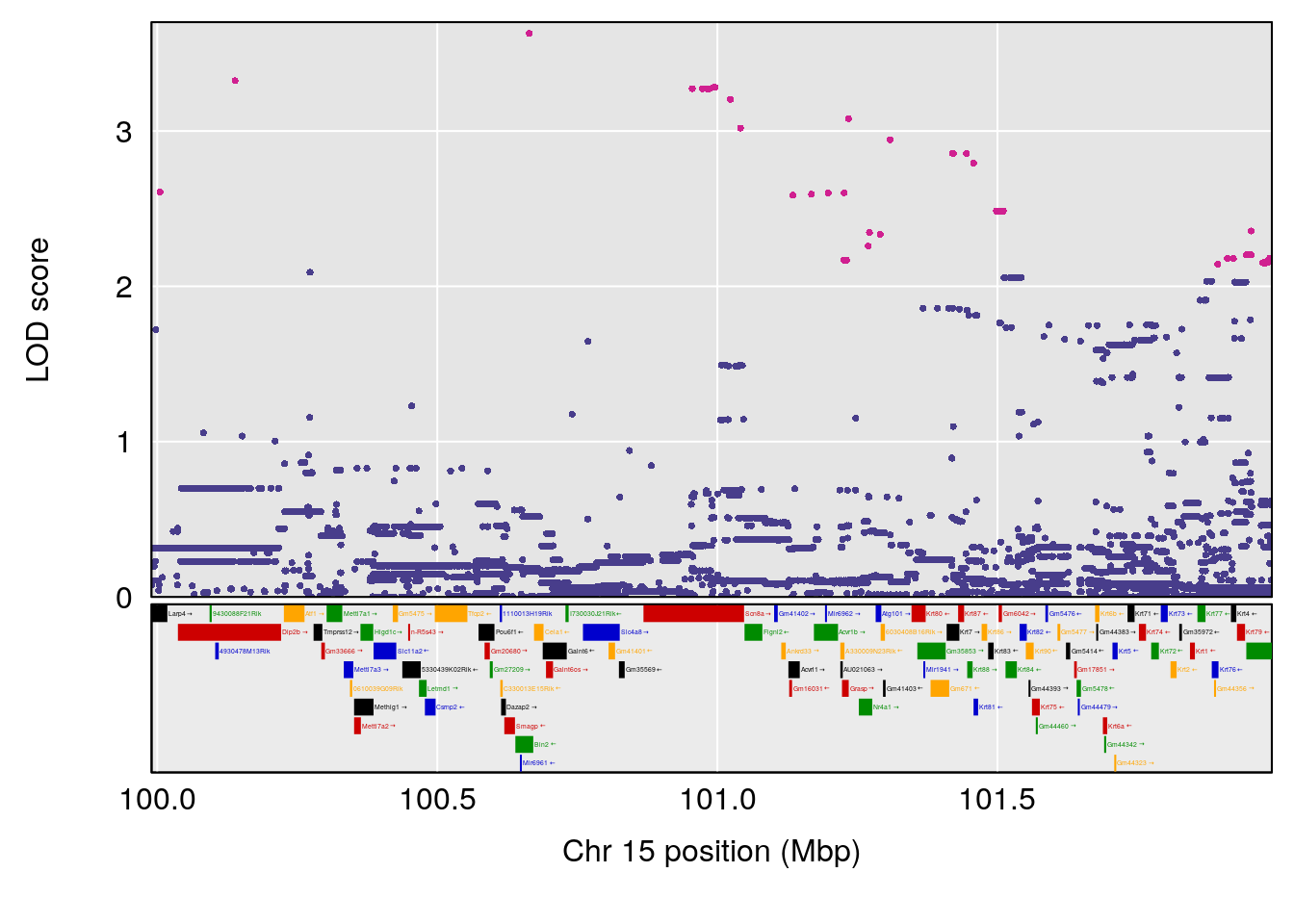
# [1] 3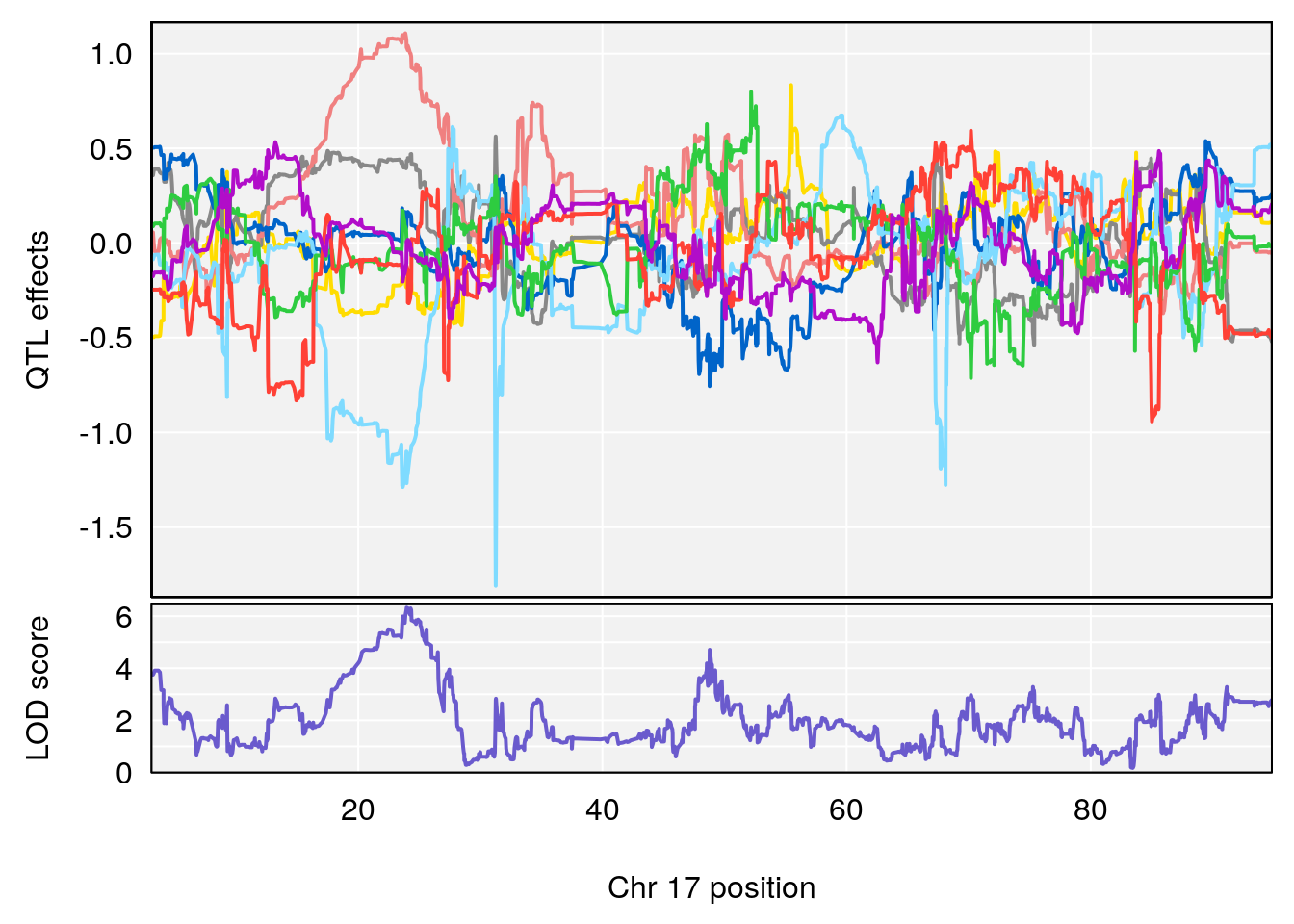
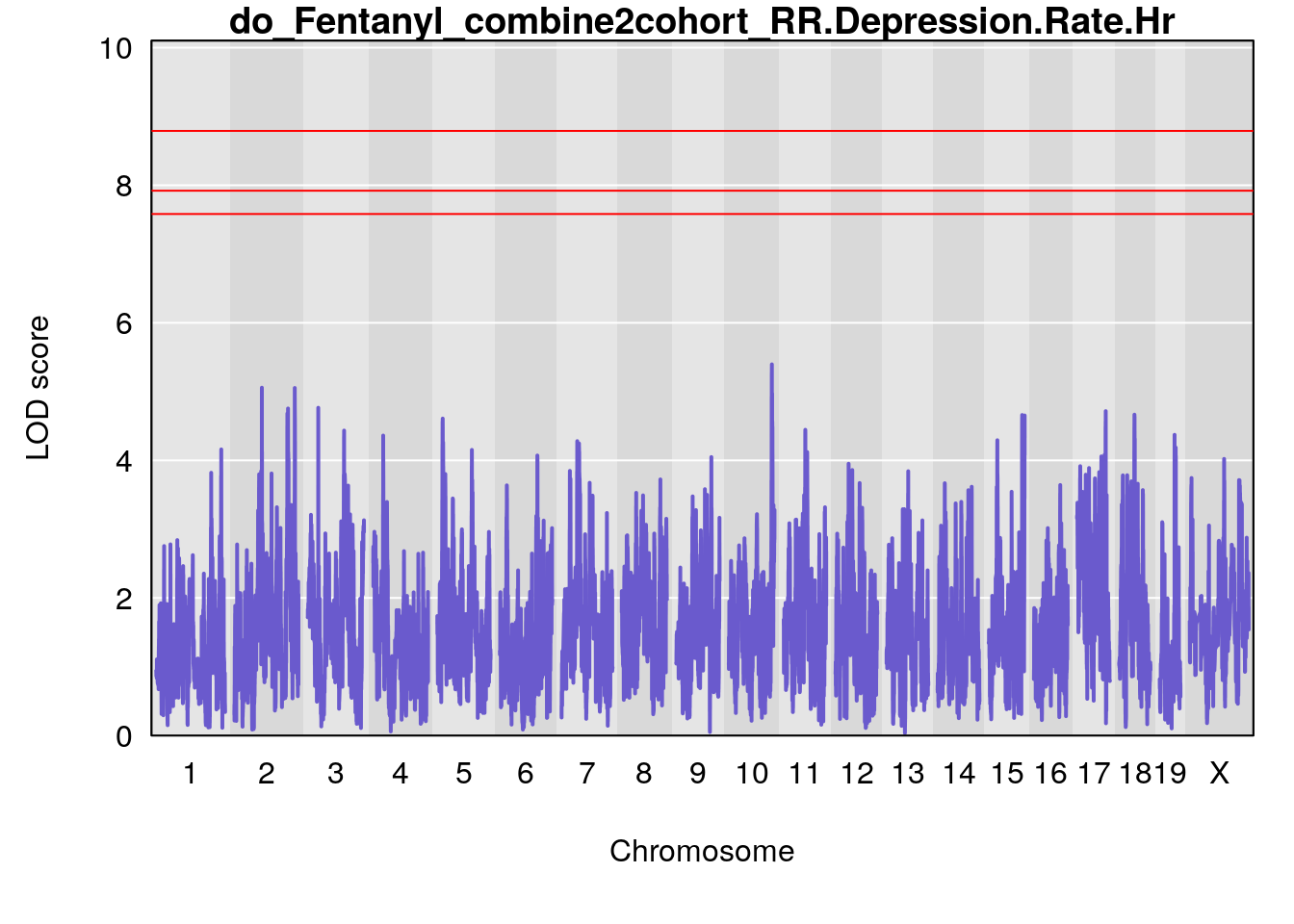
# [1] "RR.Depression.Rate.Hr"
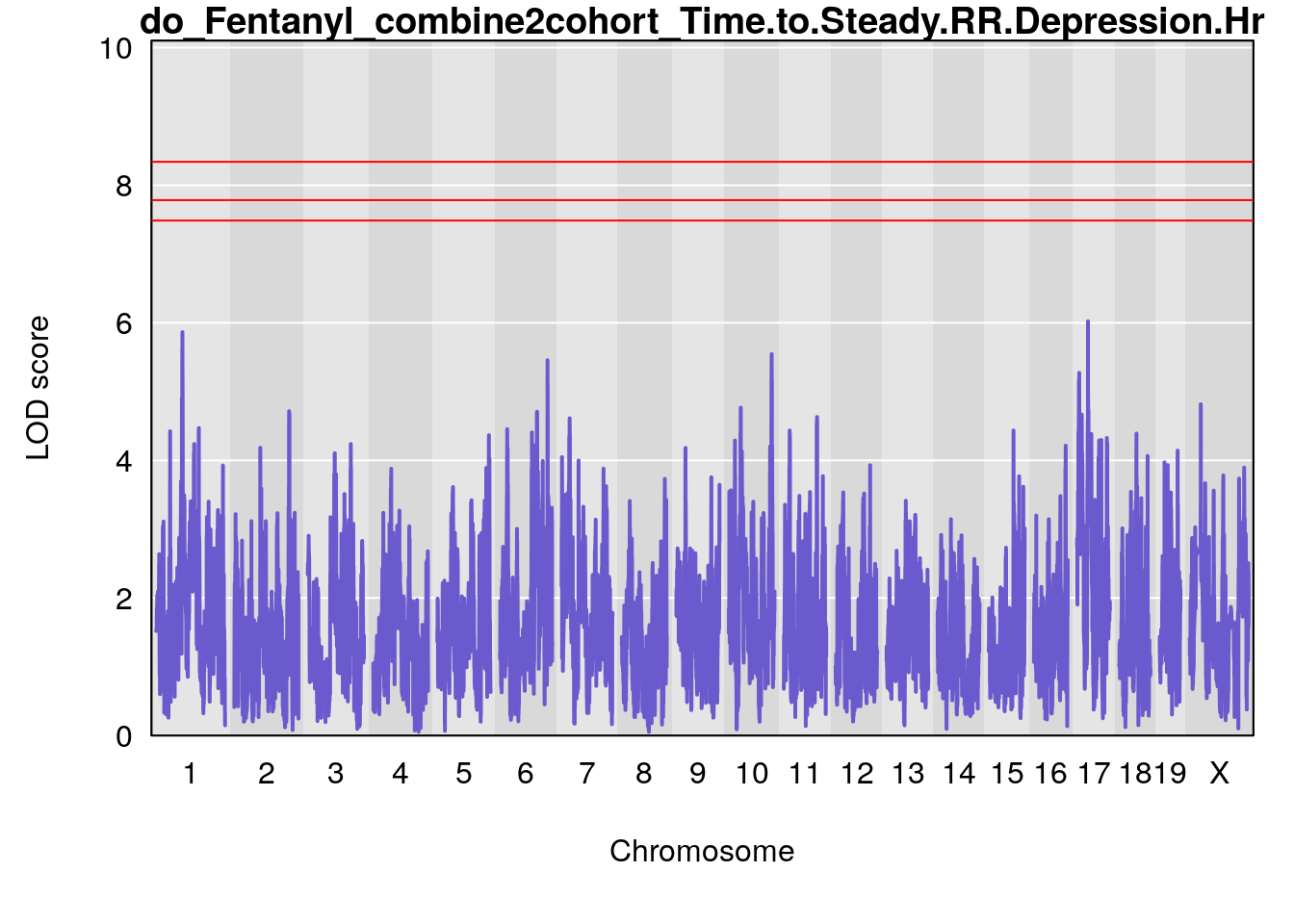
# [1] "Time.to.Steady.RR.Depression.Hr"
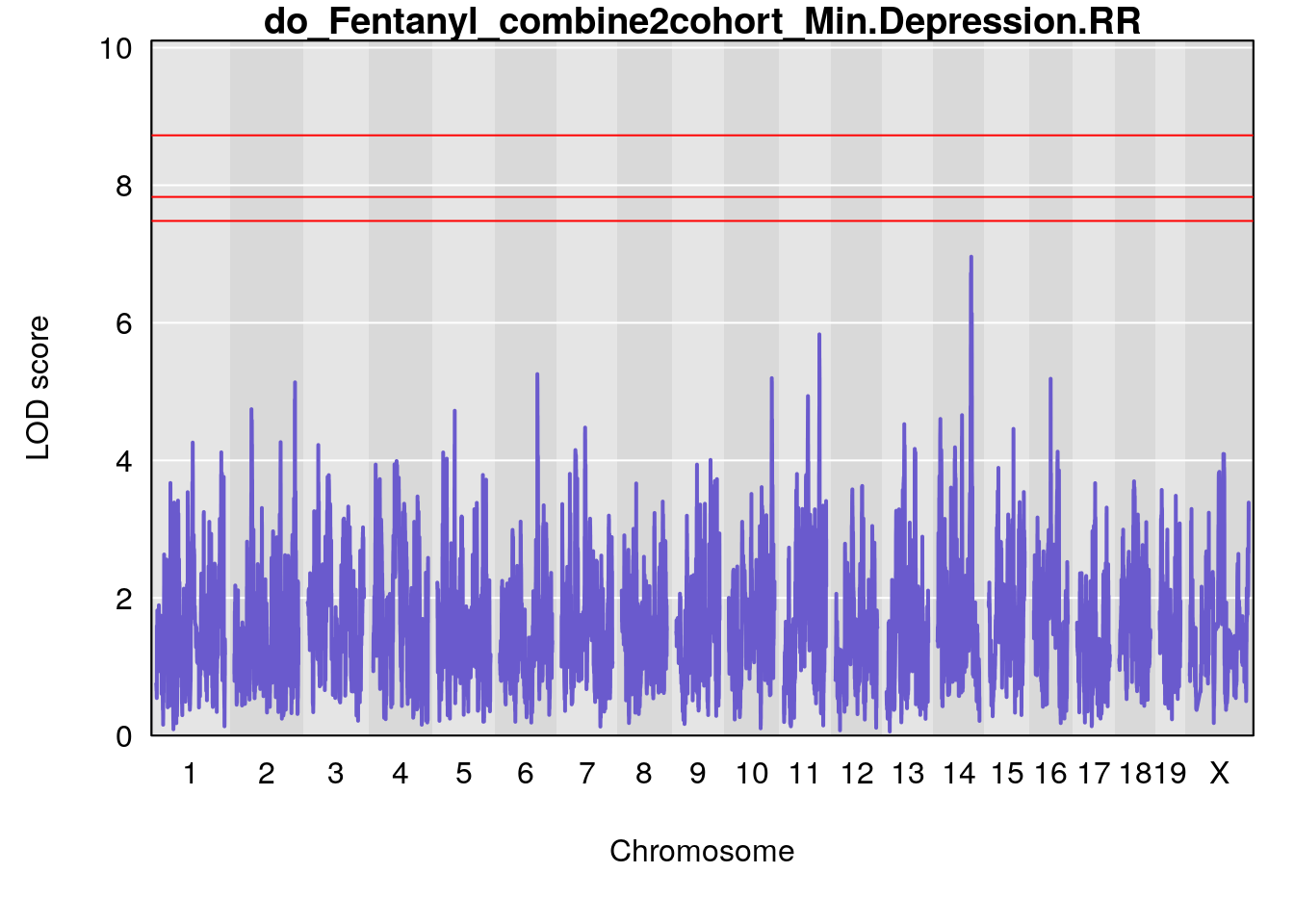
# [1] "Min.Depression.RR"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
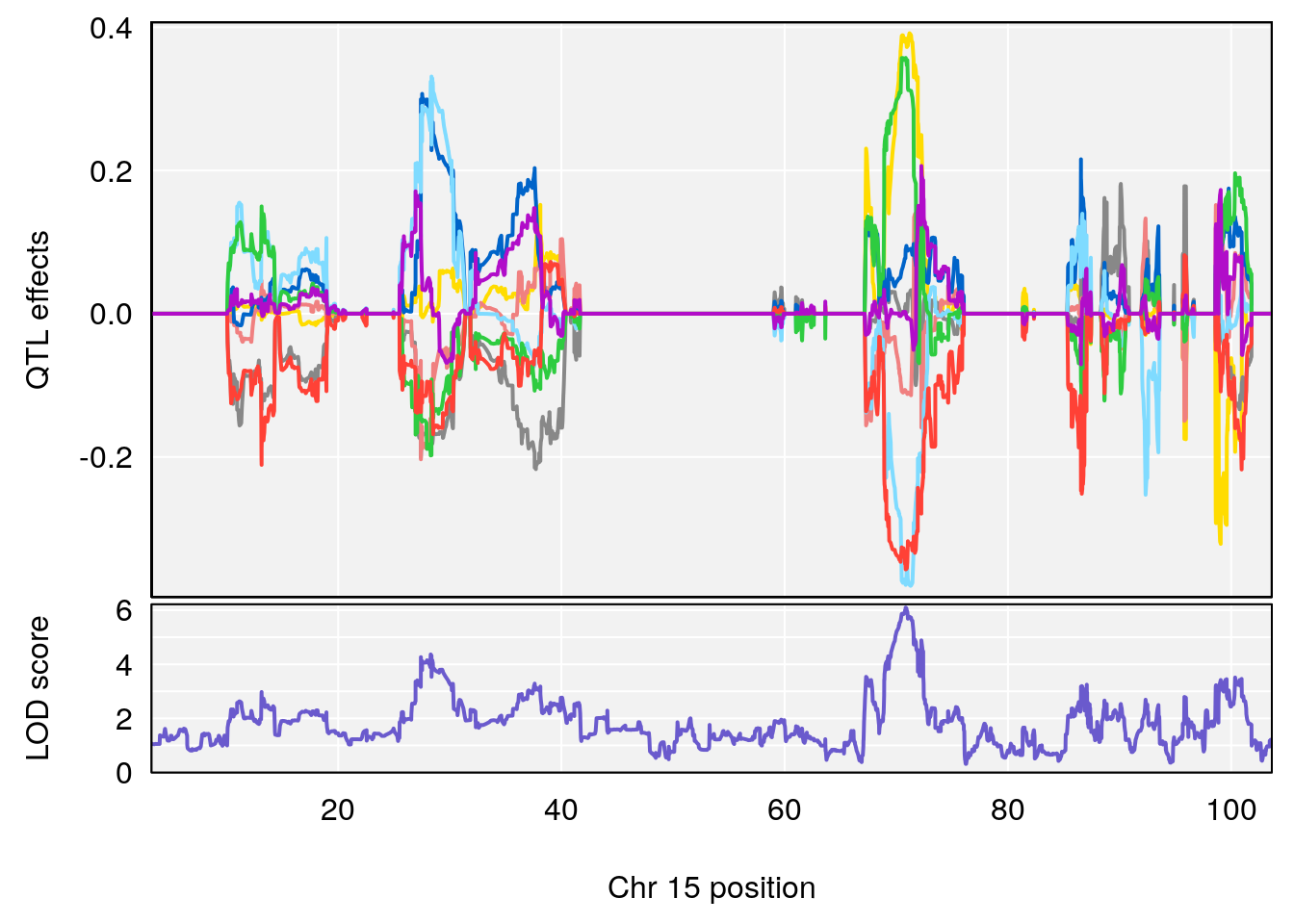
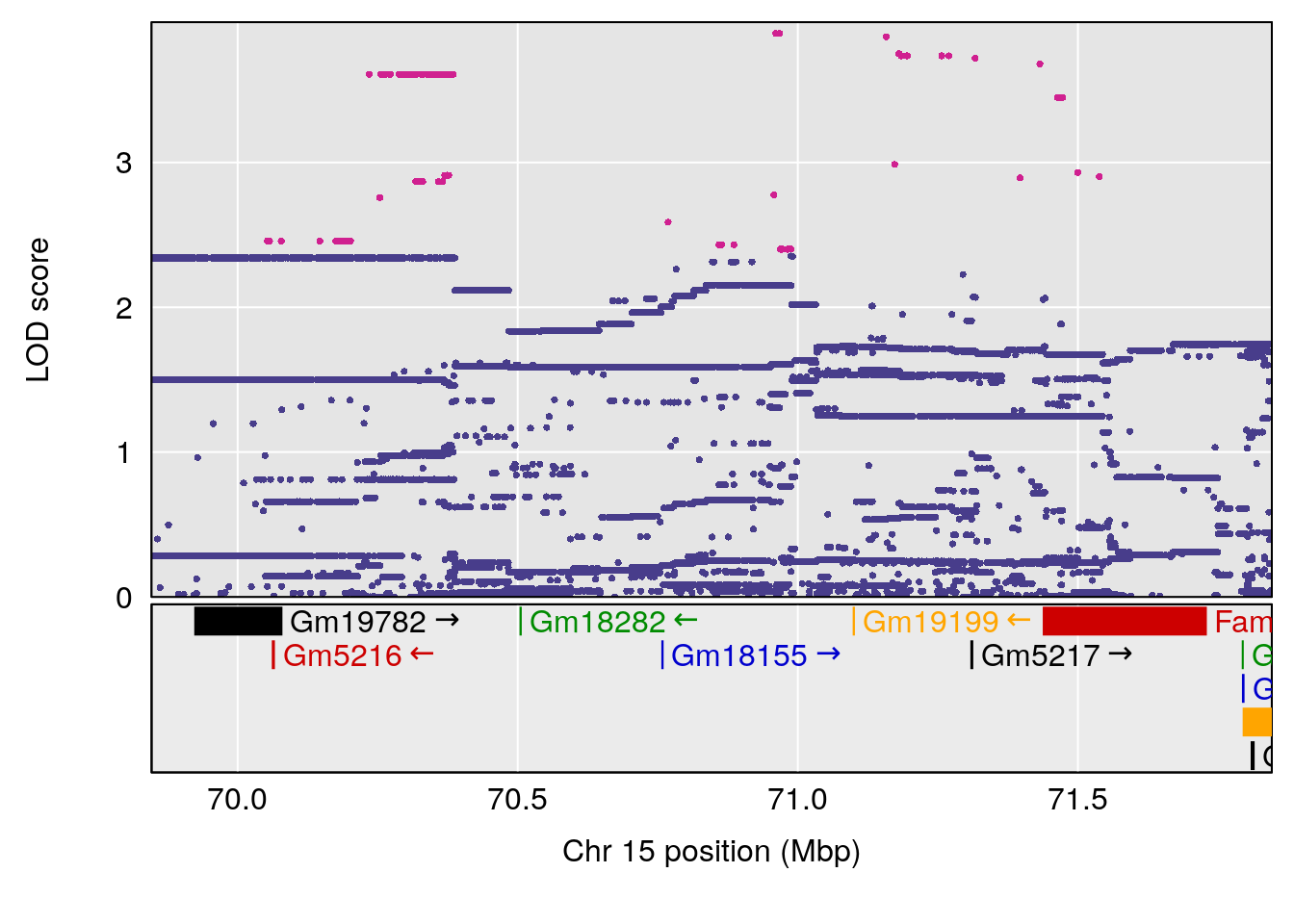
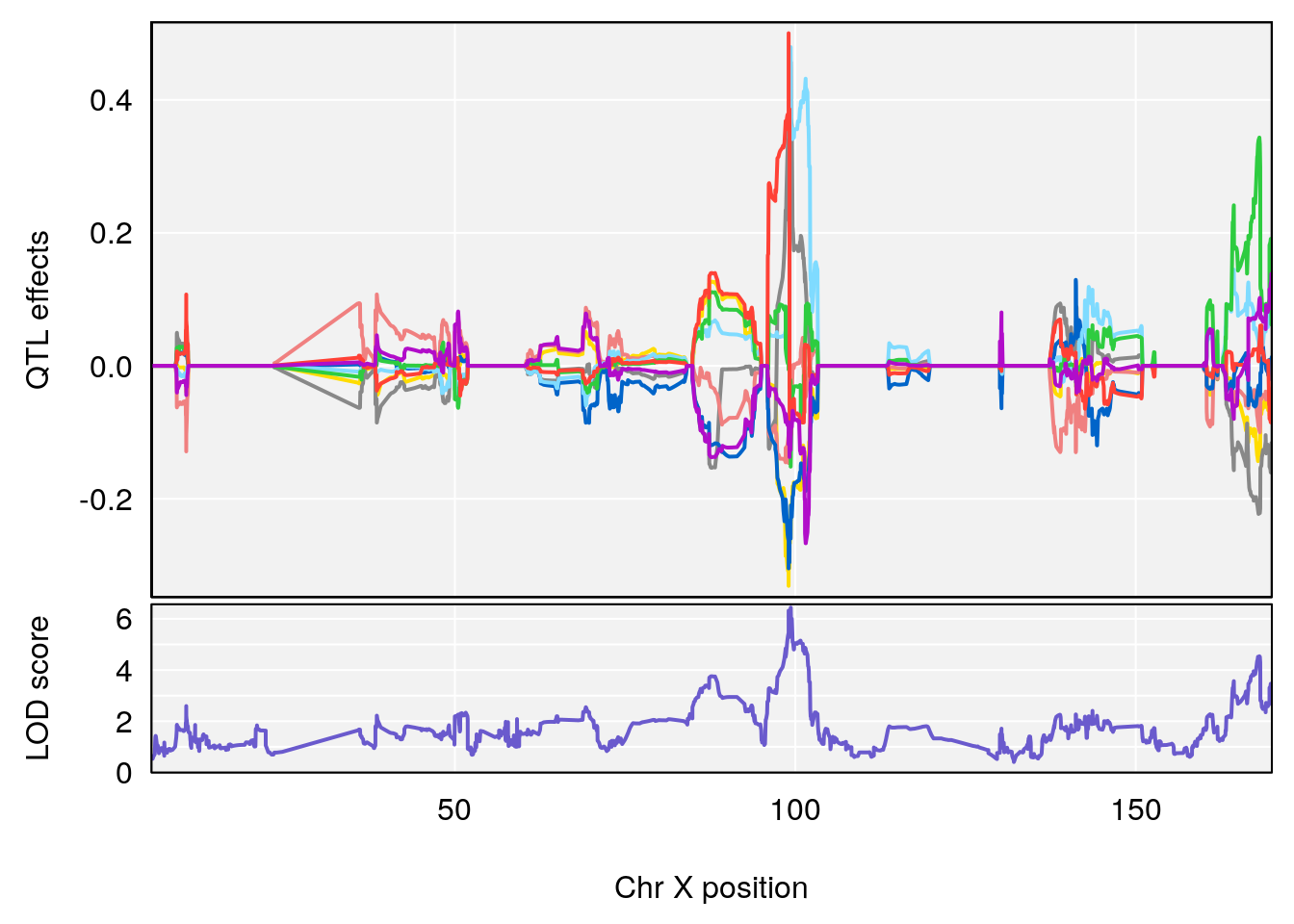
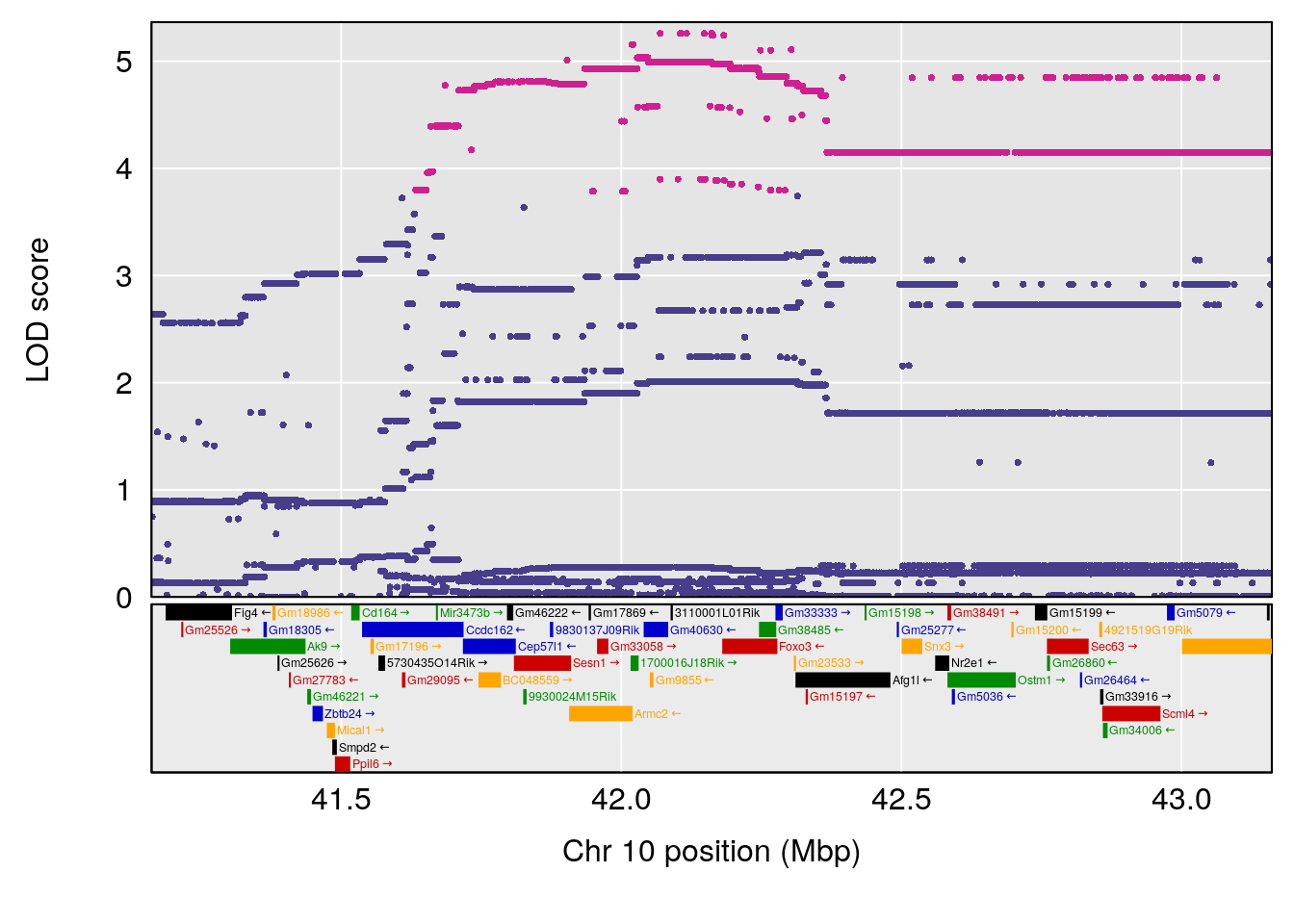
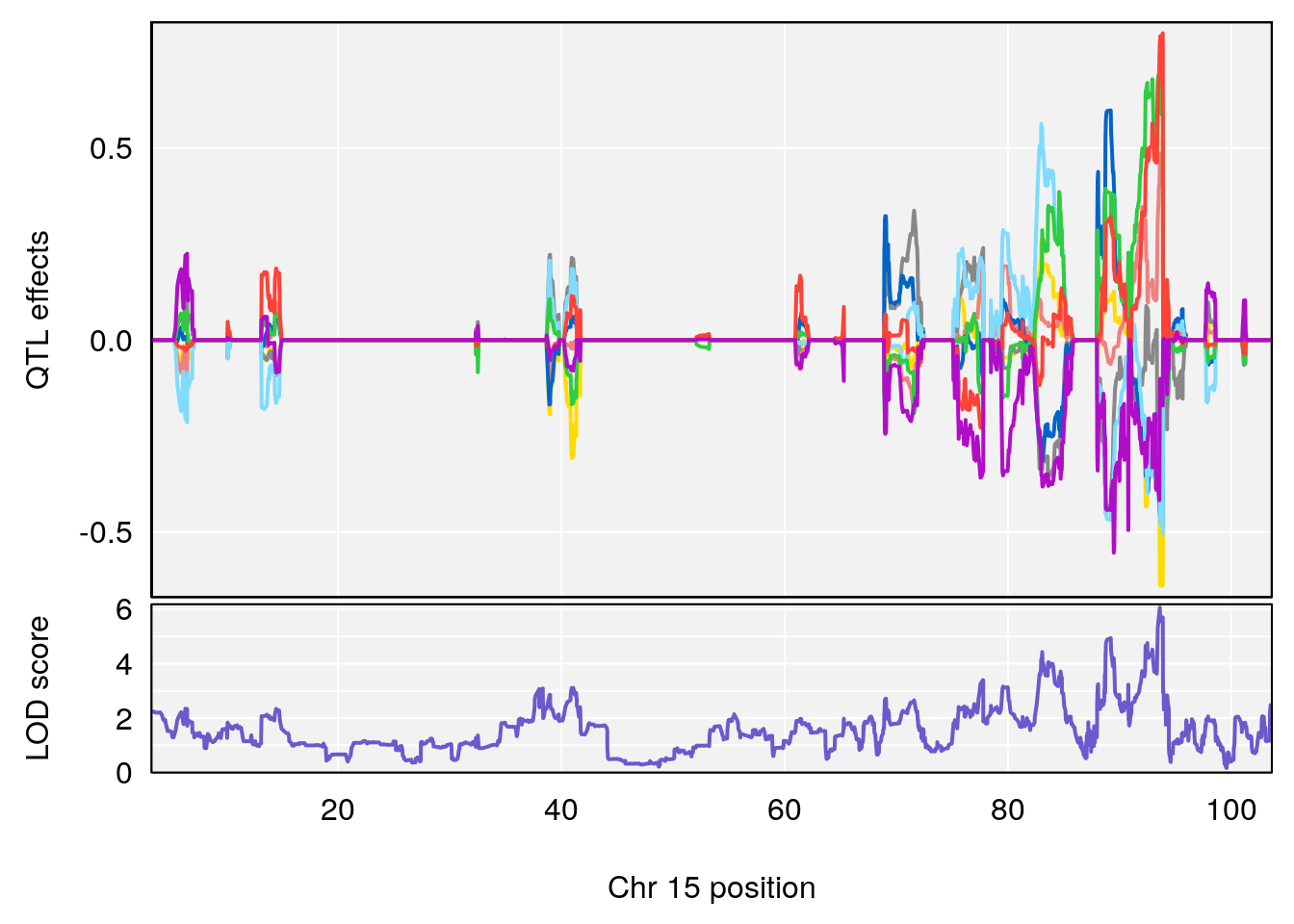
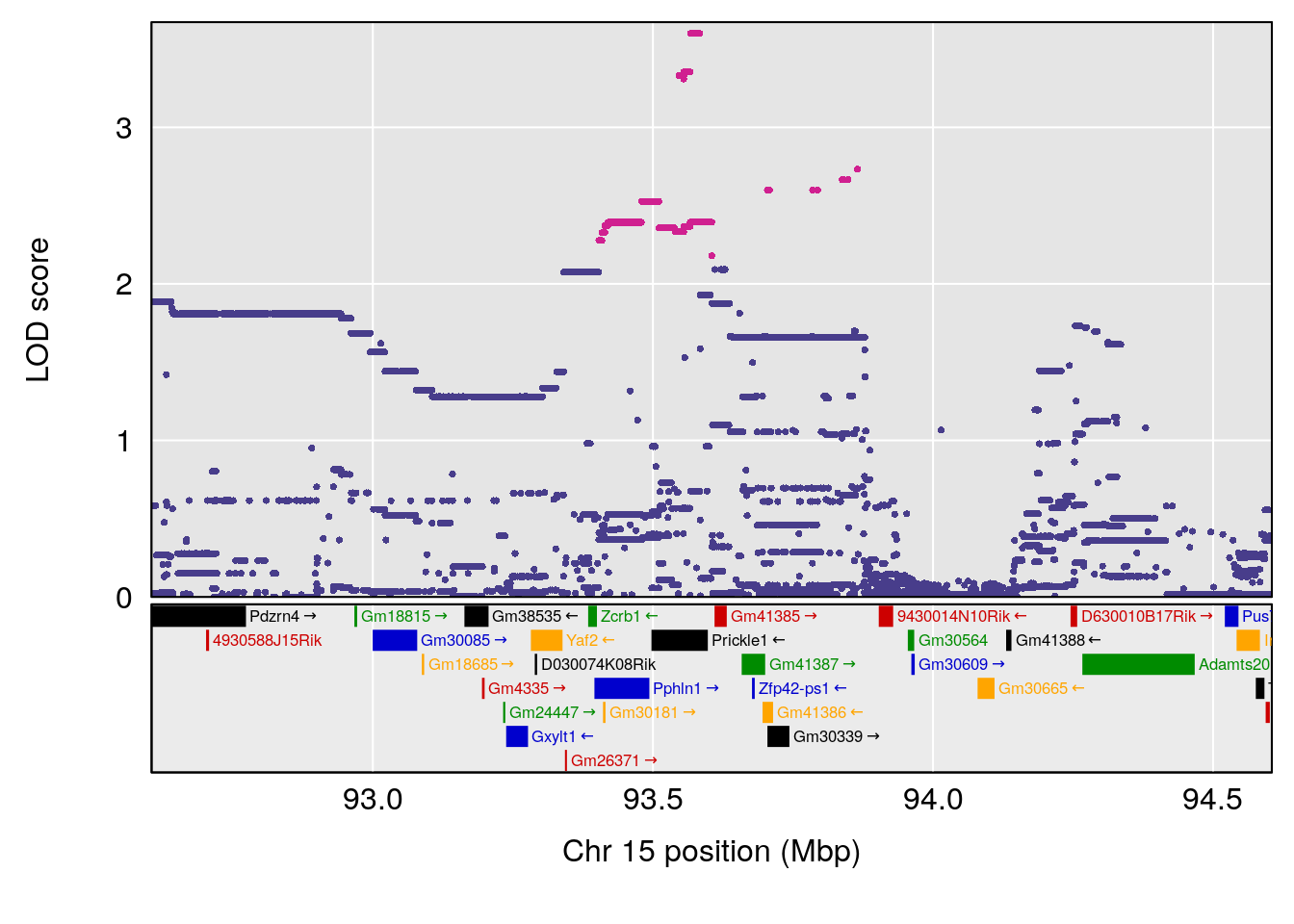
# 1 1 pheno1 15 70.84631 6.097157 69.53075 72.33644
# 2 1 pheno1 X 99.32996 6.439452 98.70699 101.15048
# [1] 1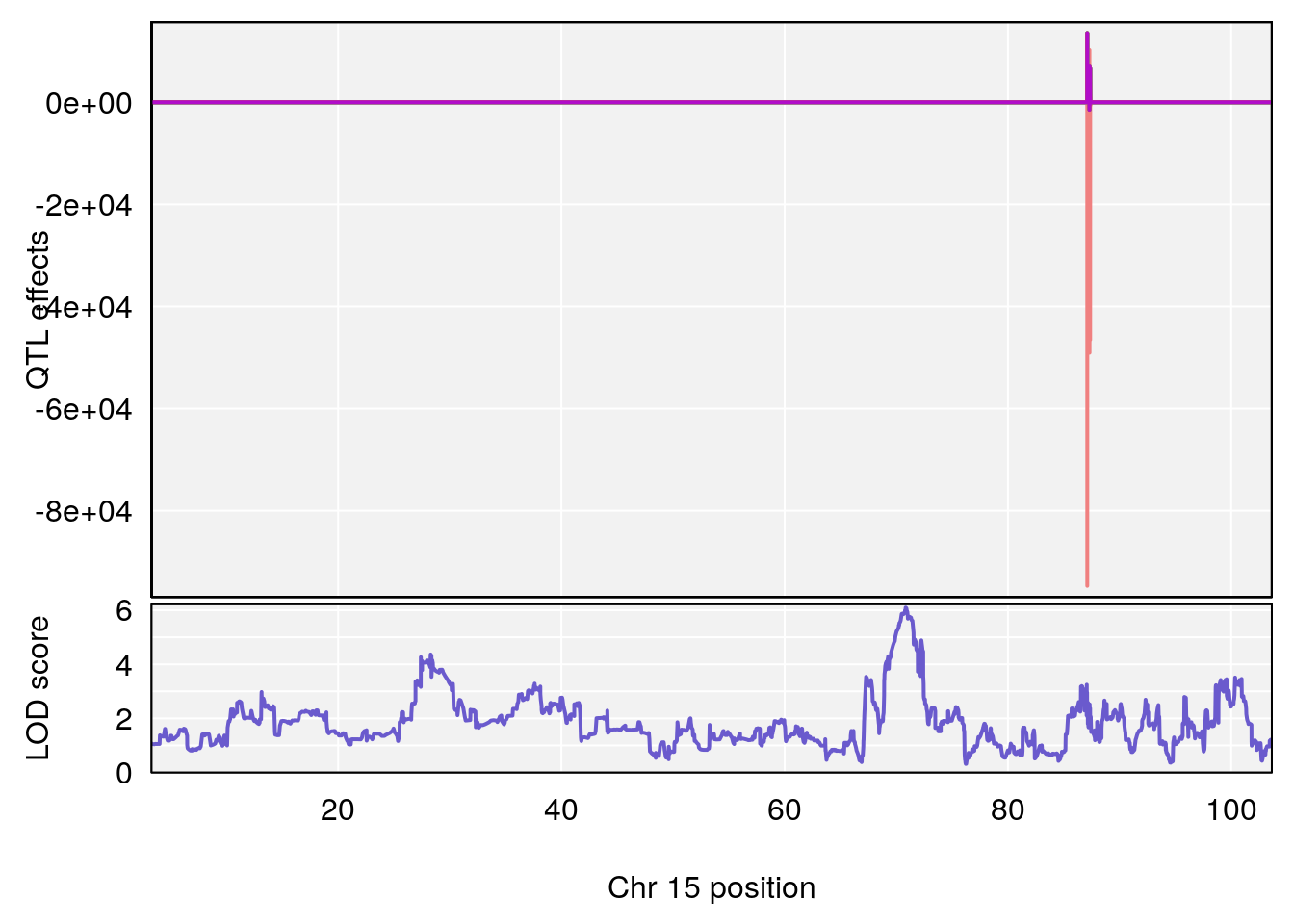
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
# [1] 2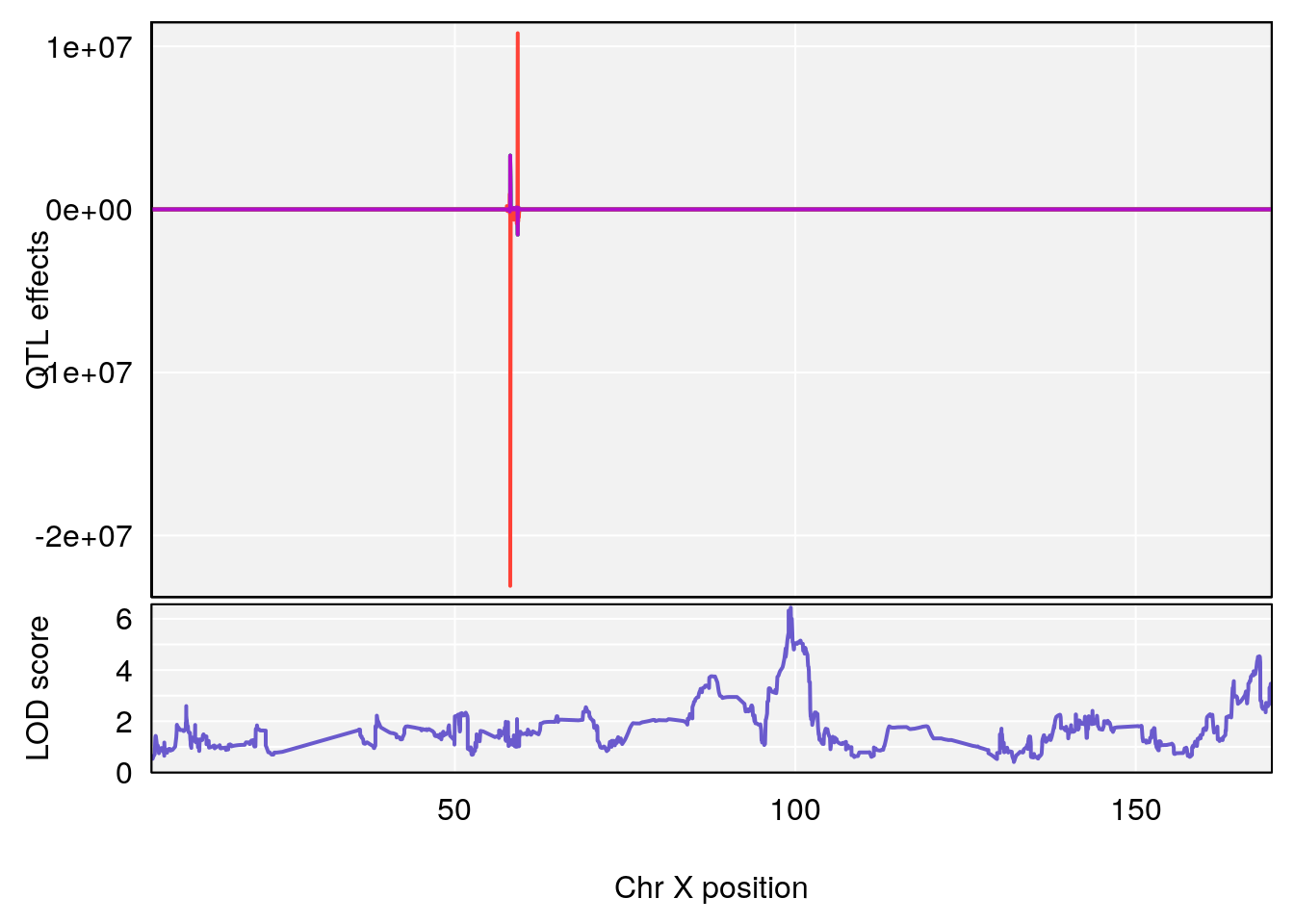
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
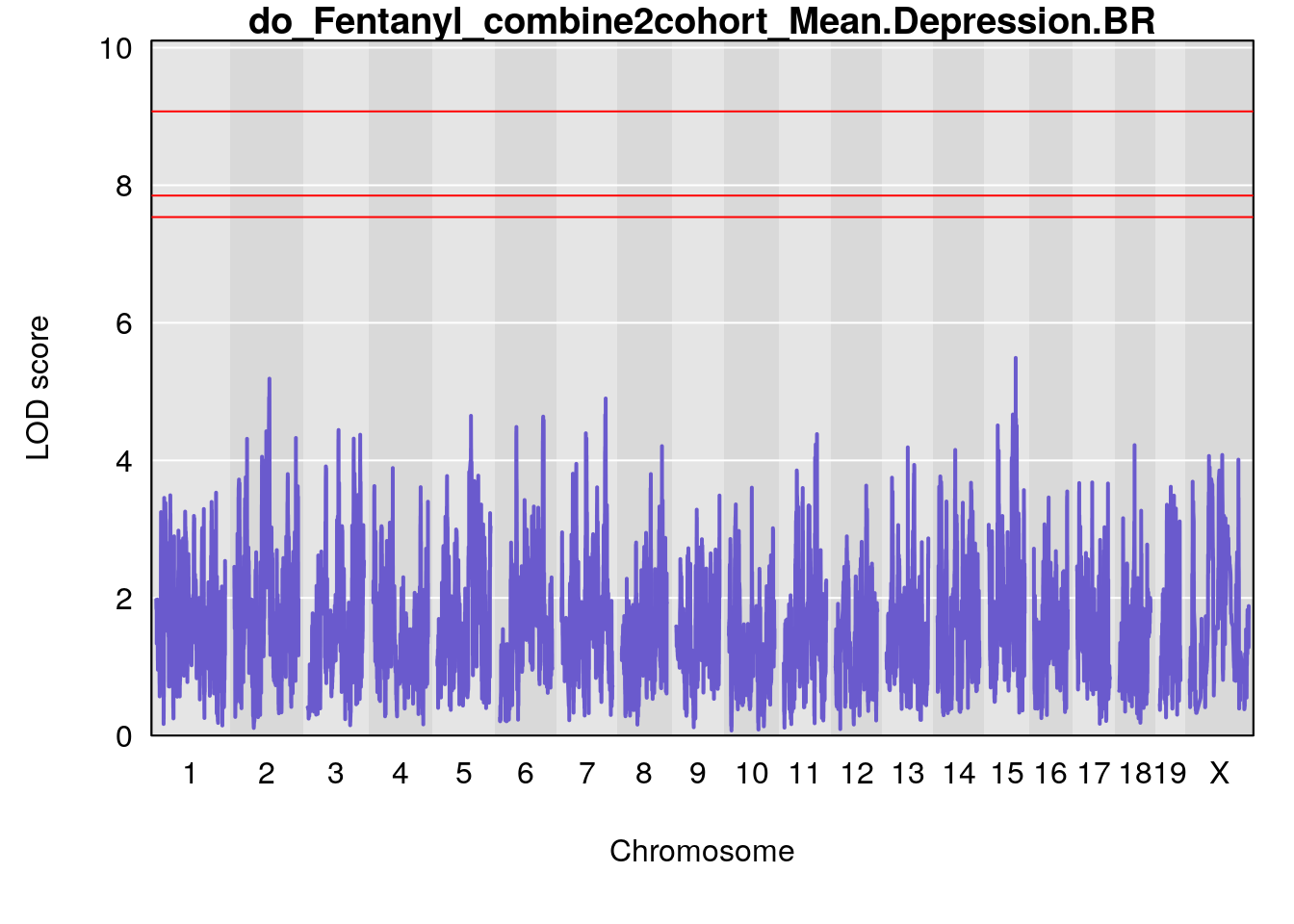
# [1] "Mean.Depression.BR"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
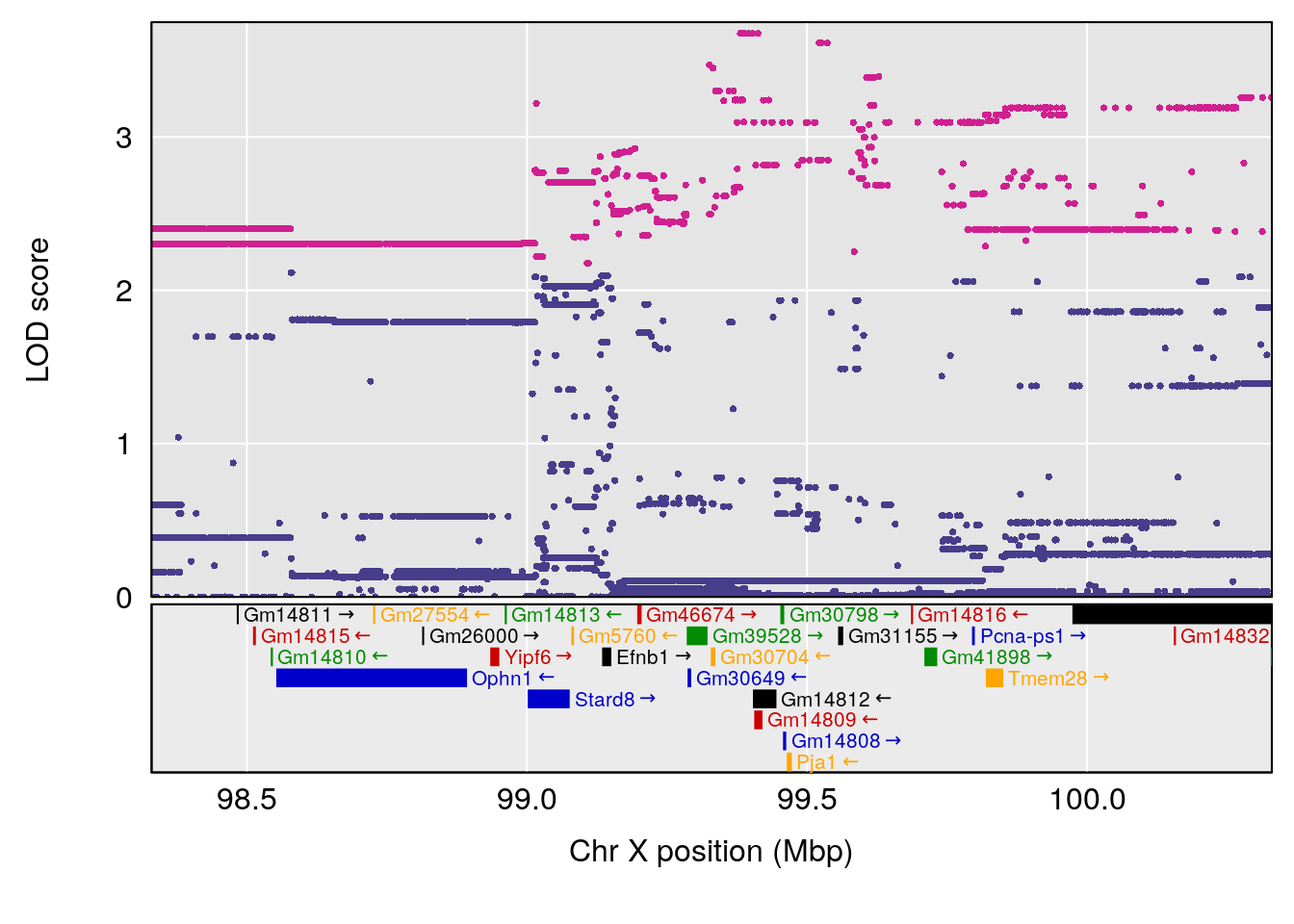
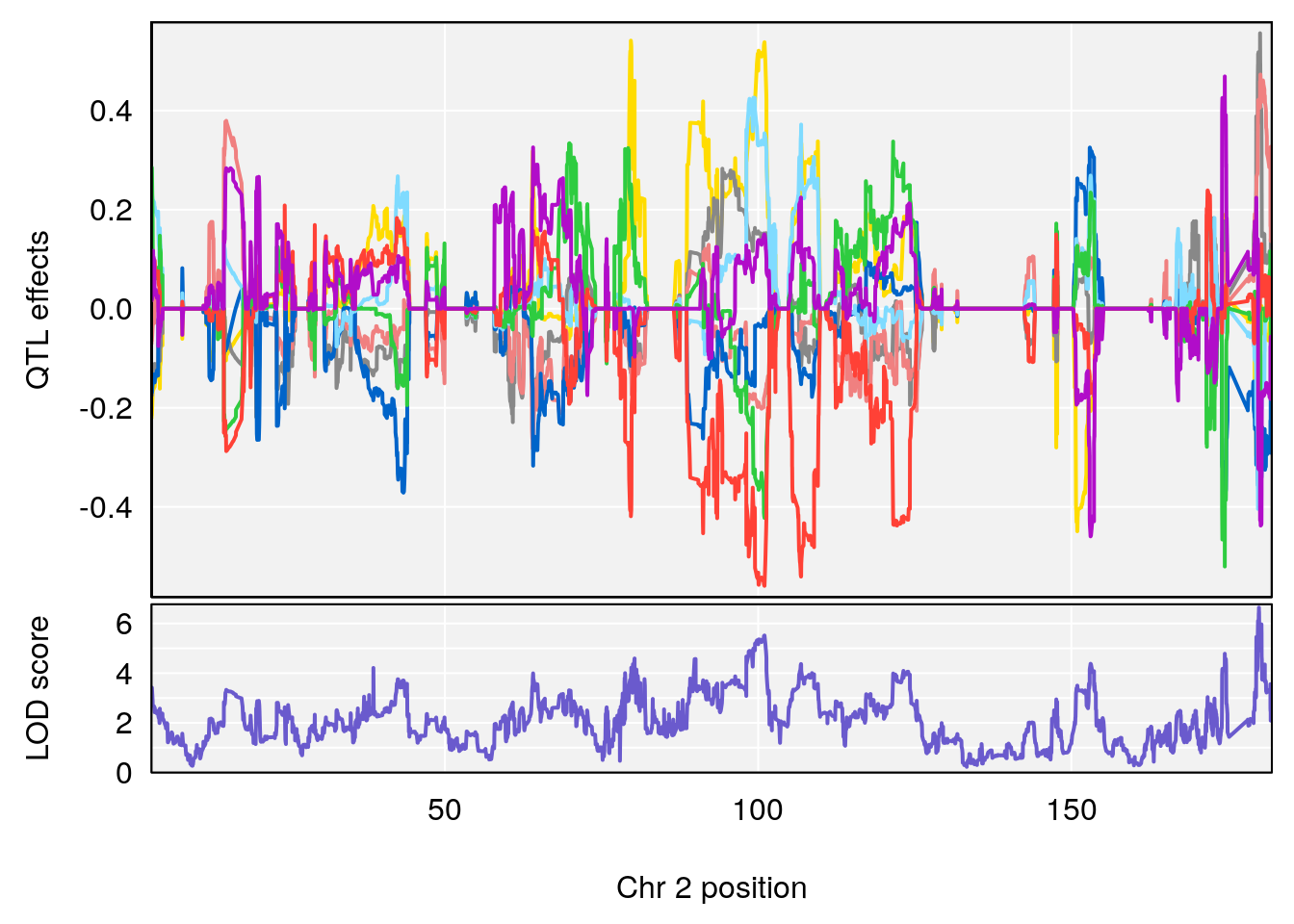
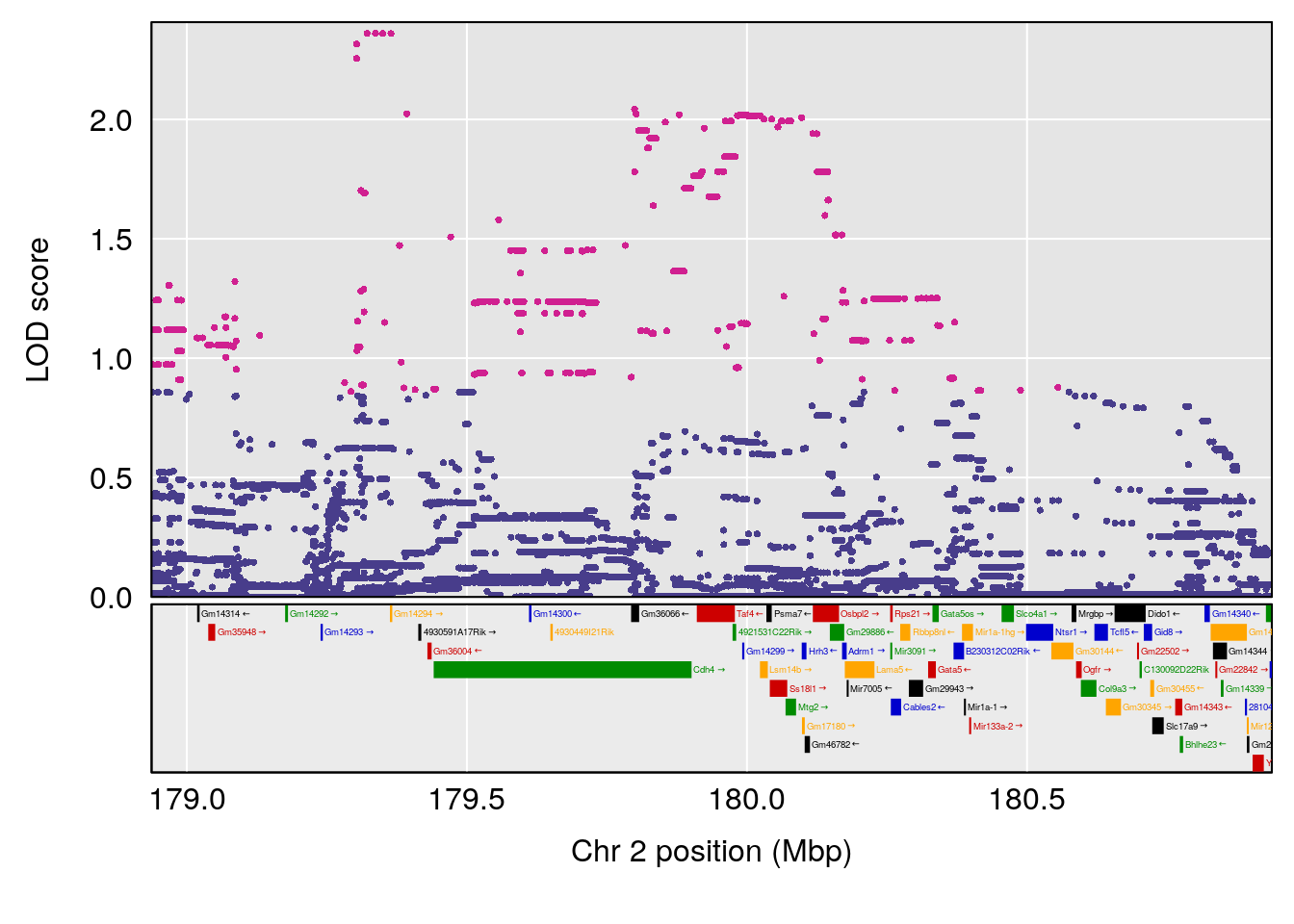
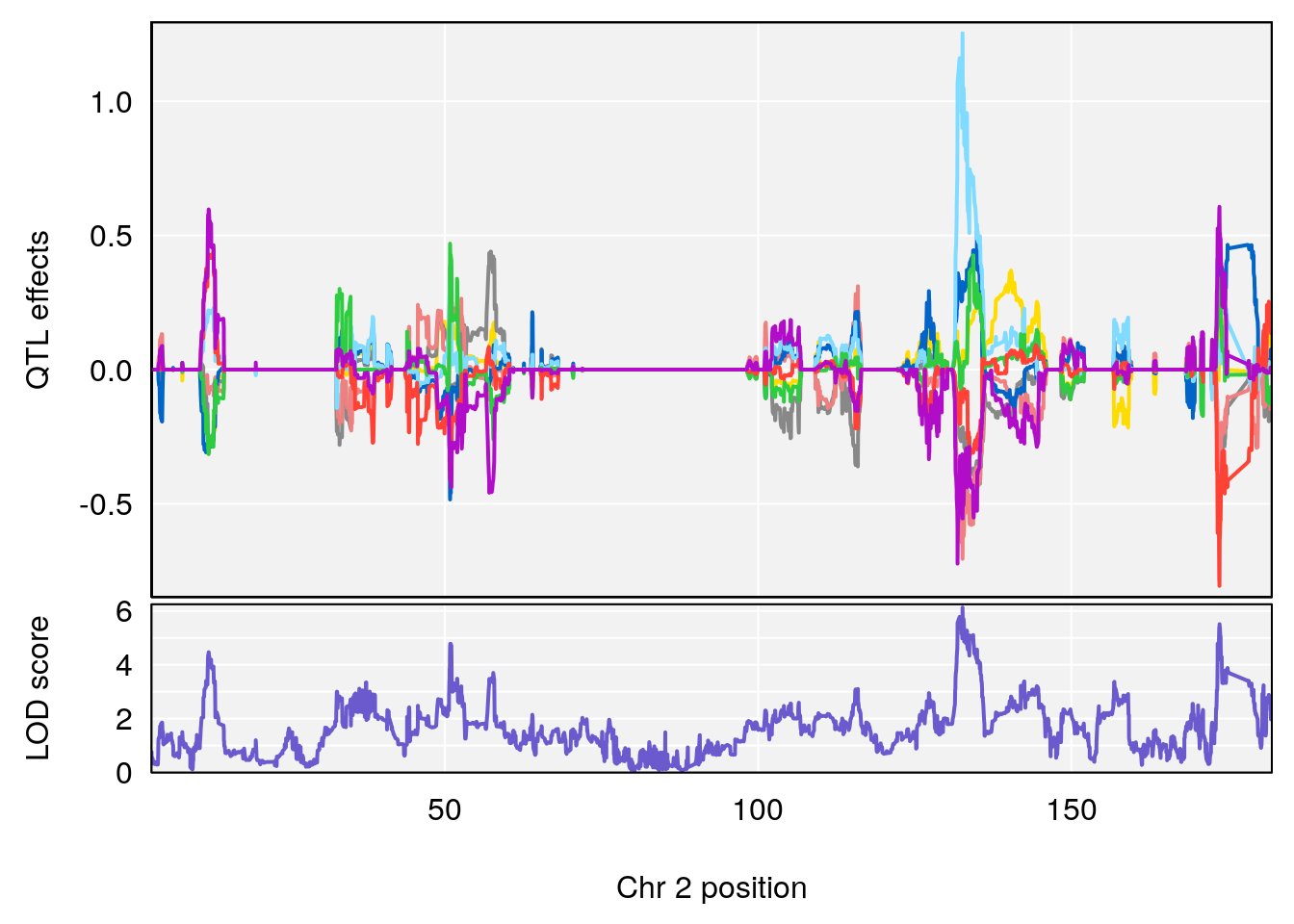
# 1 1 pheno1 2 179.93697 6.642010 99.48509 180.36898
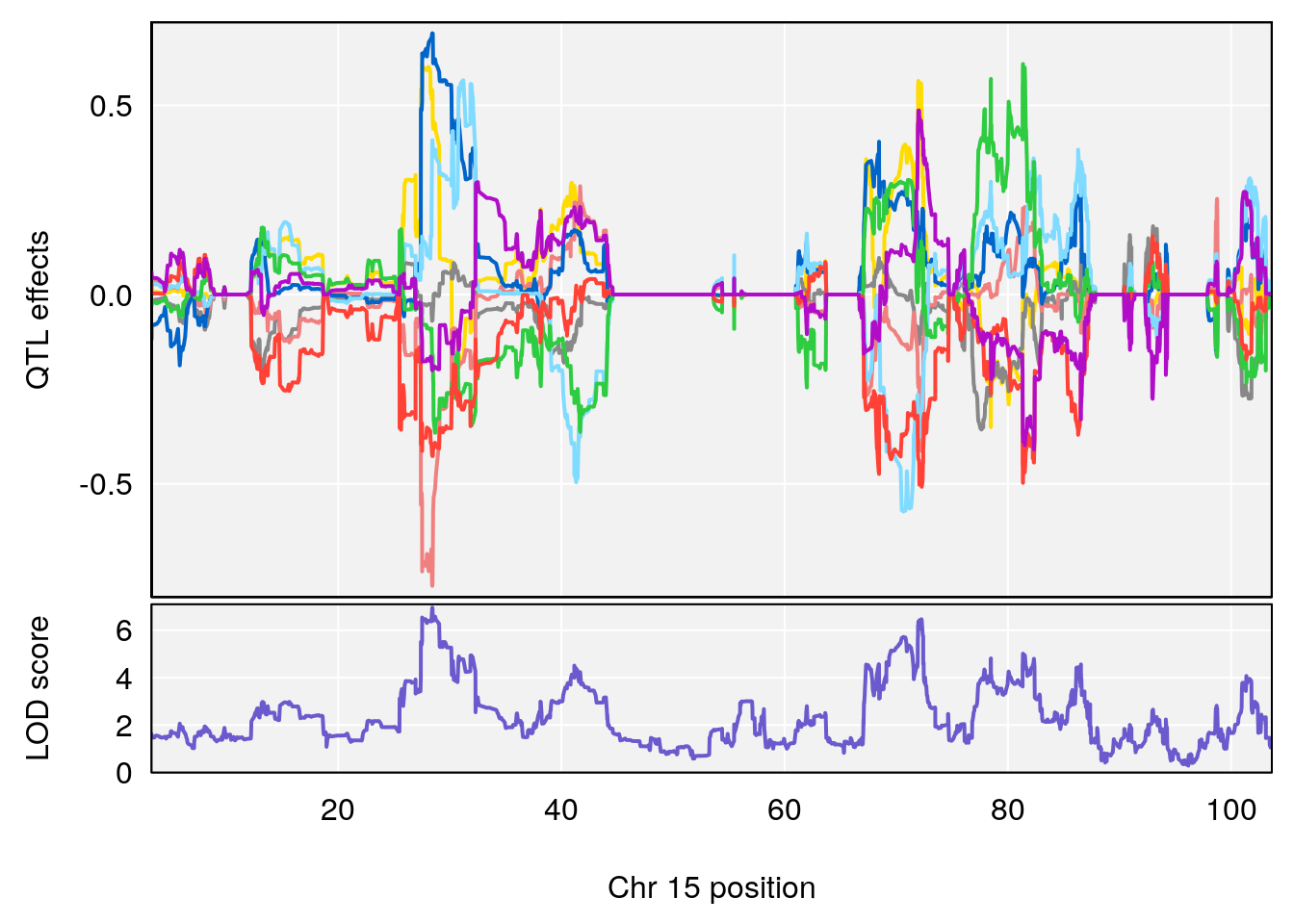
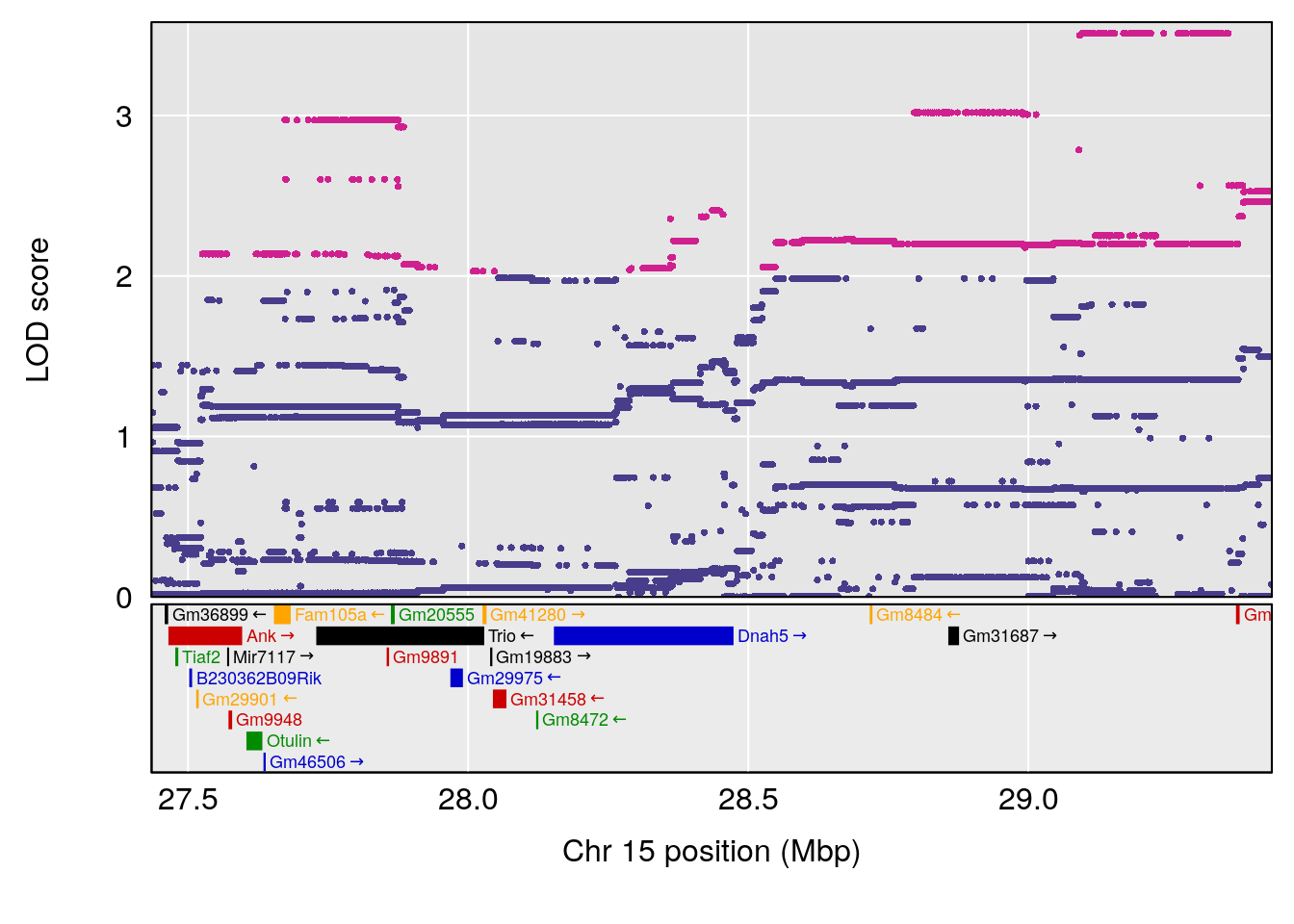
# 2 1 pheno1 15 28.43474 6.957455 27.40518 72.41165
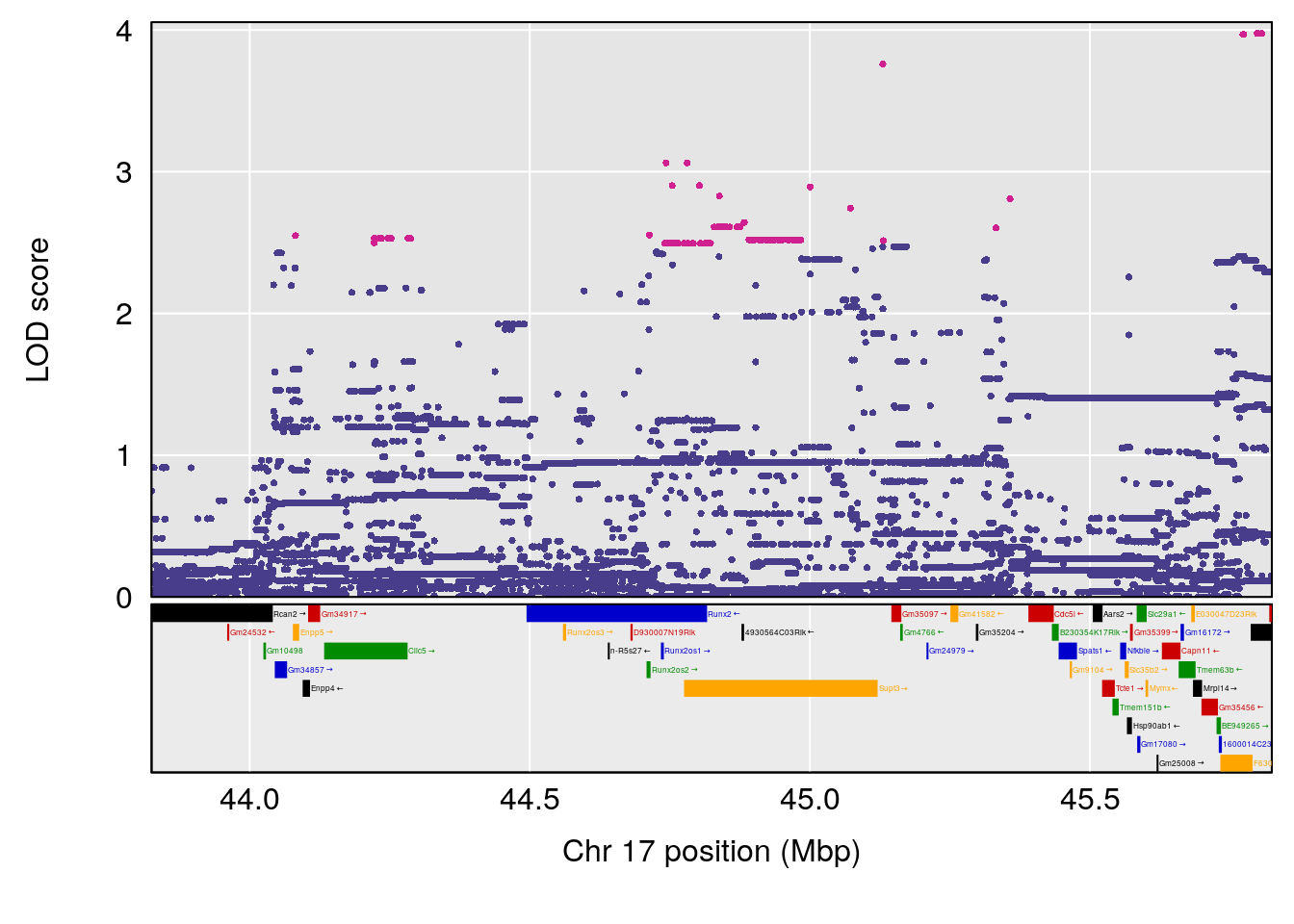
# 3 1 pheno1 17 44.82475 6.816106 44.68776 45.38912
# [1] 1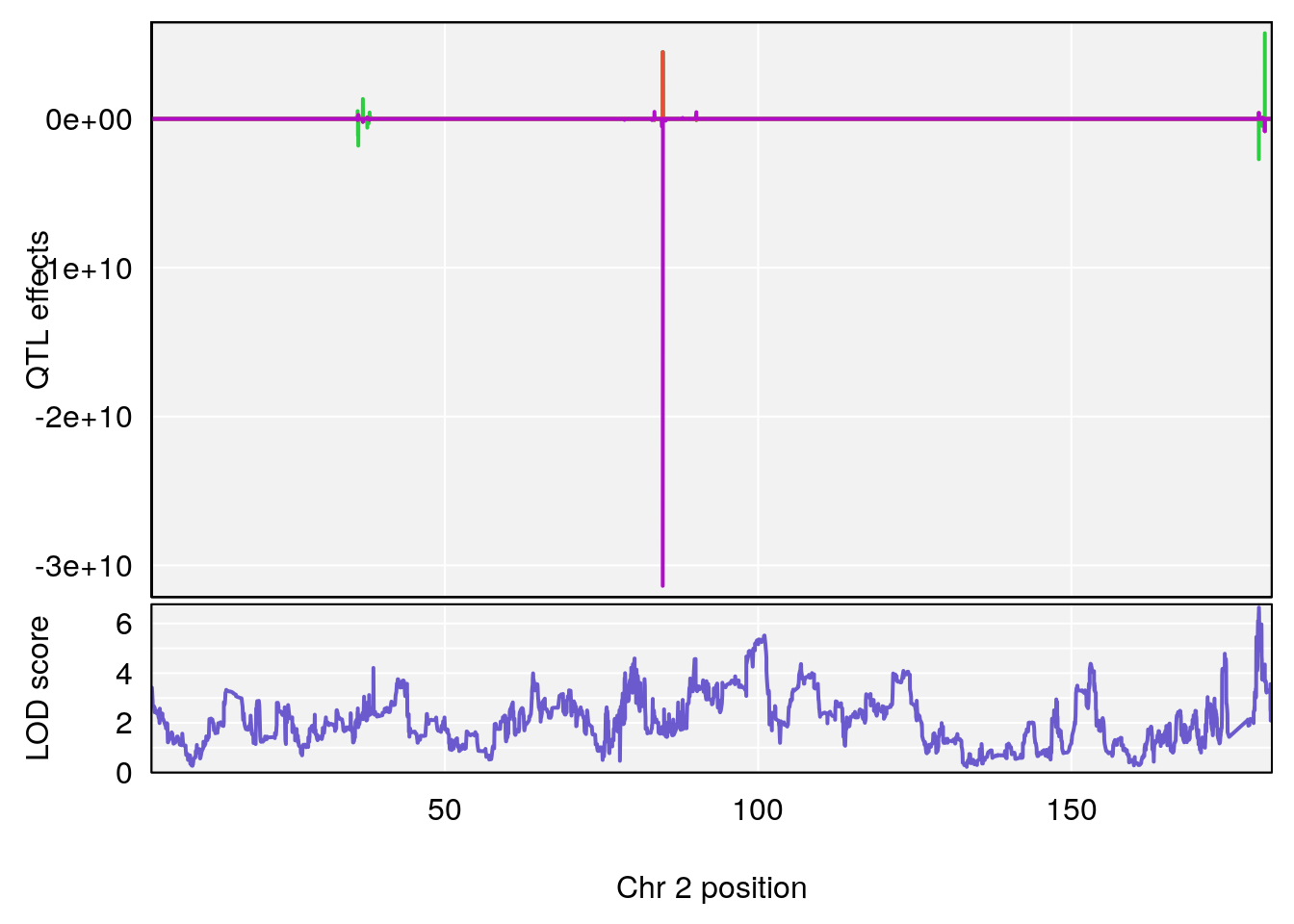
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
# [1] 2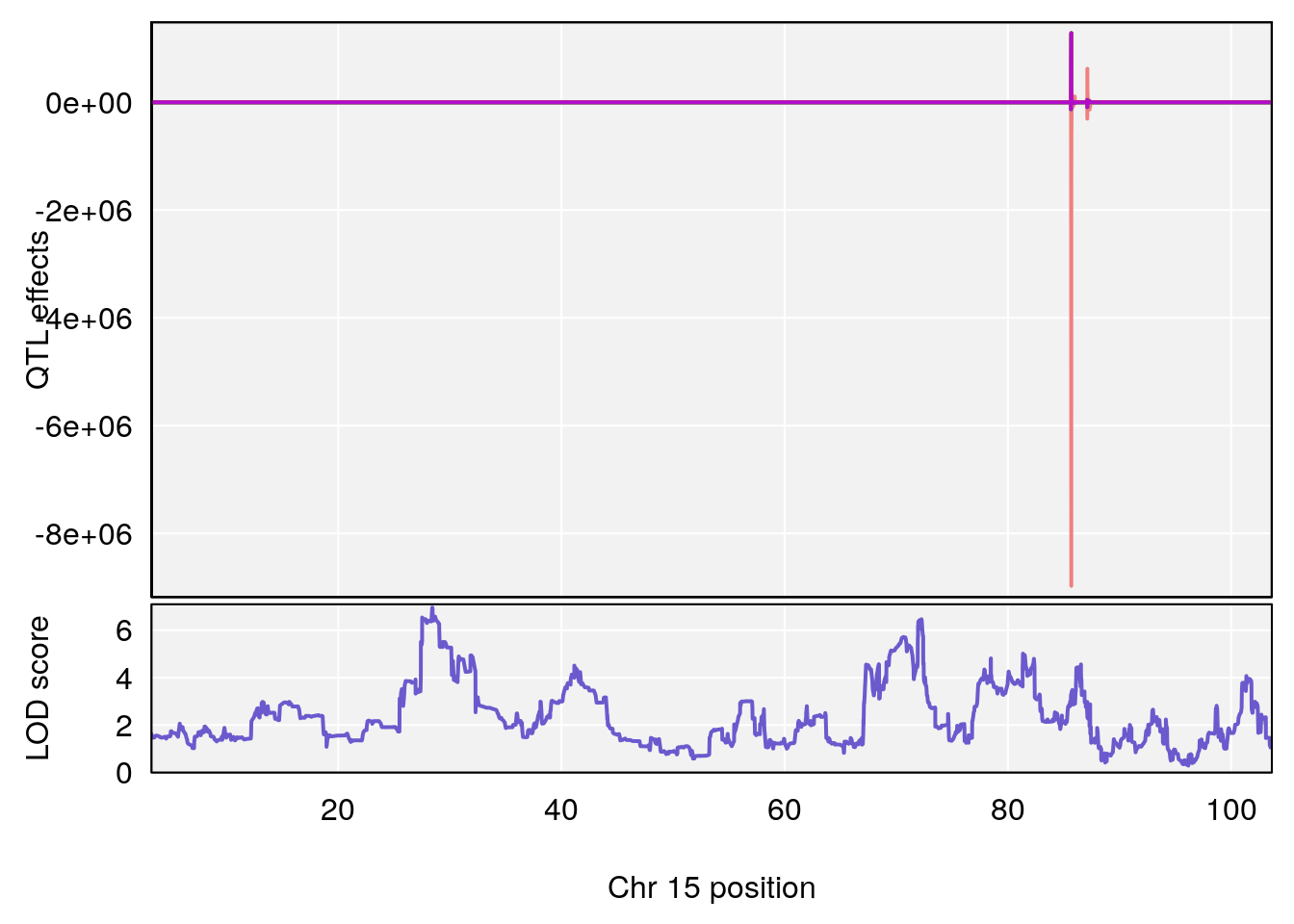
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
# [1] 3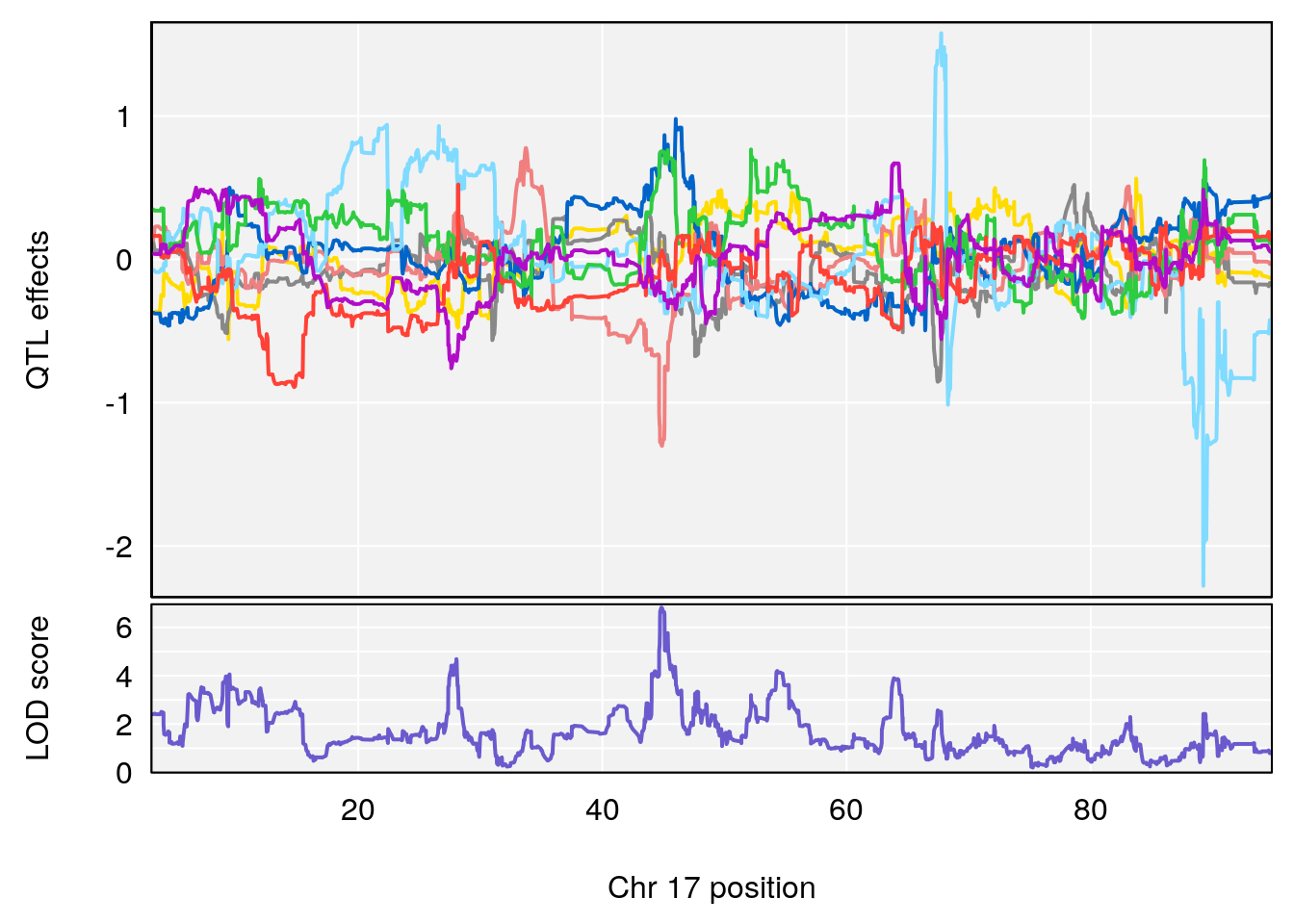
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
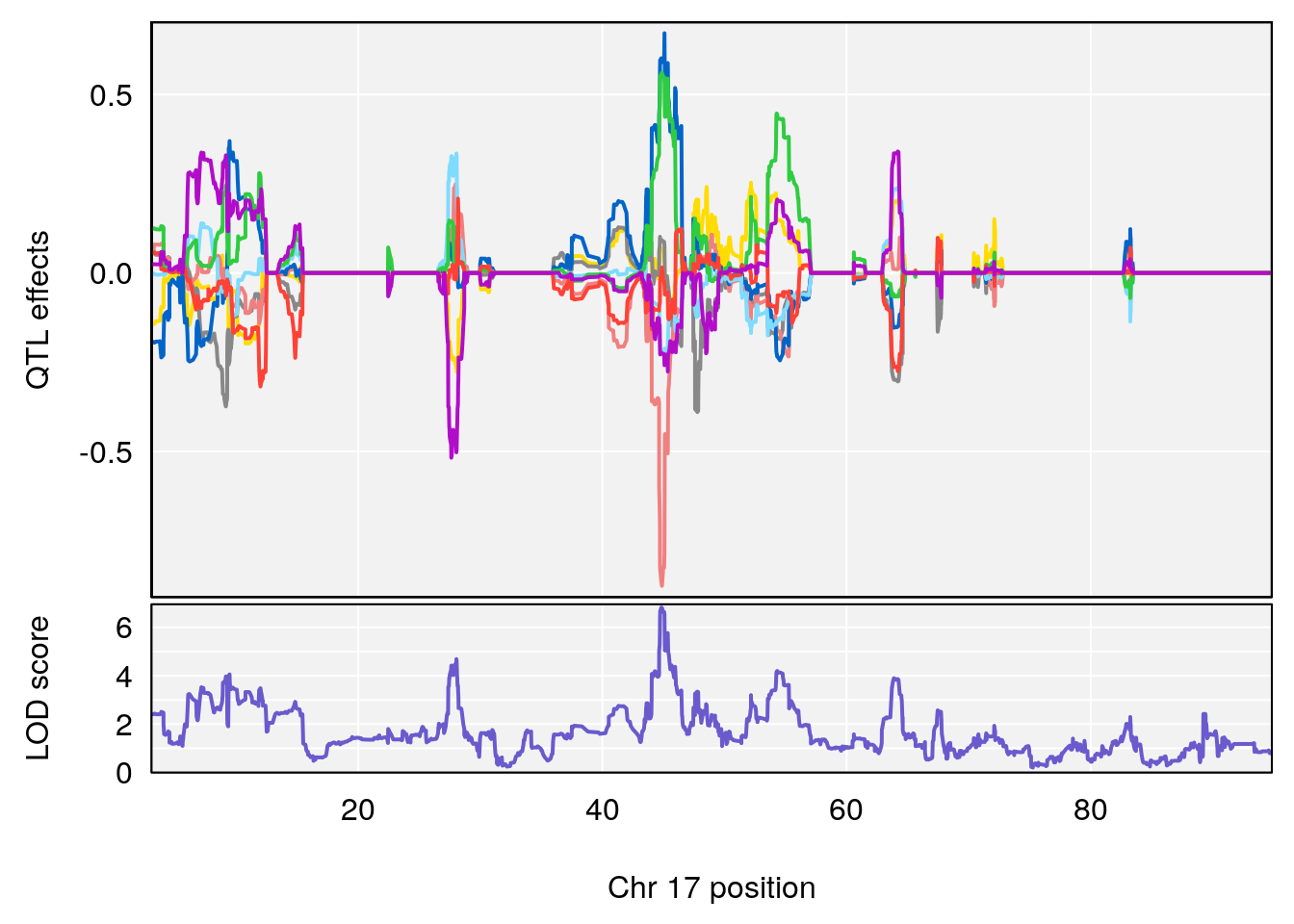
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
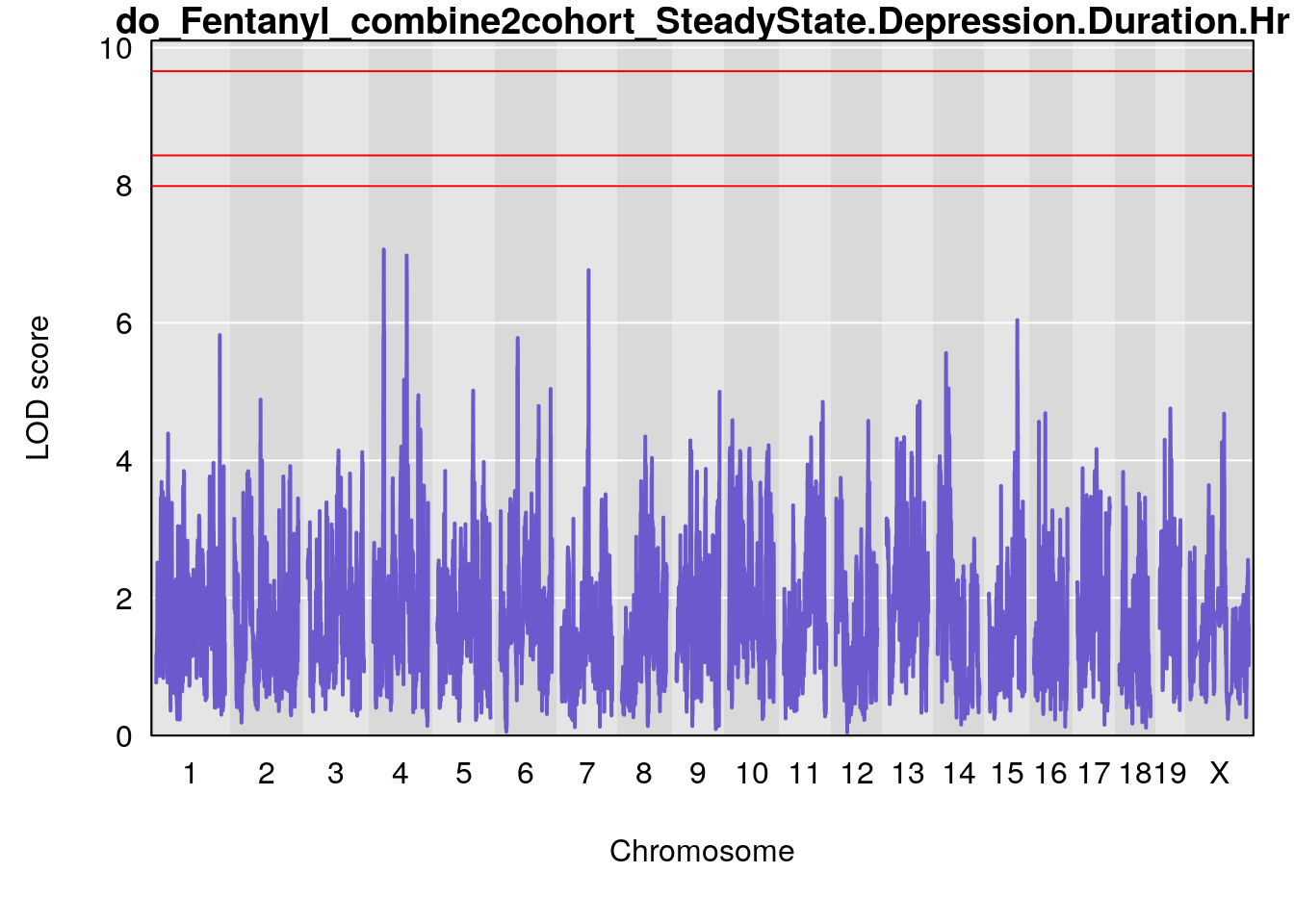
# [1] "SteadyState.Depression.Duration.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
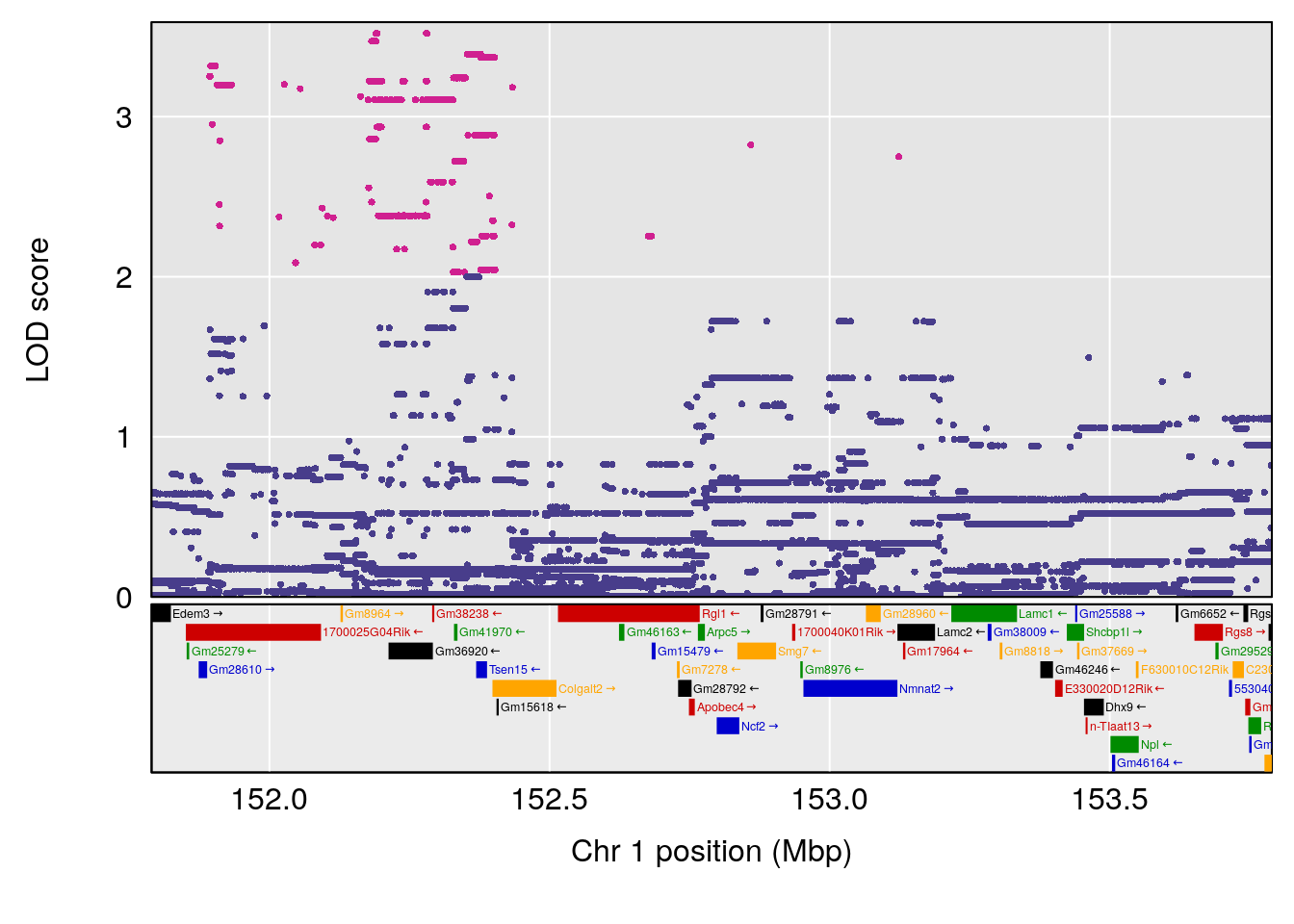
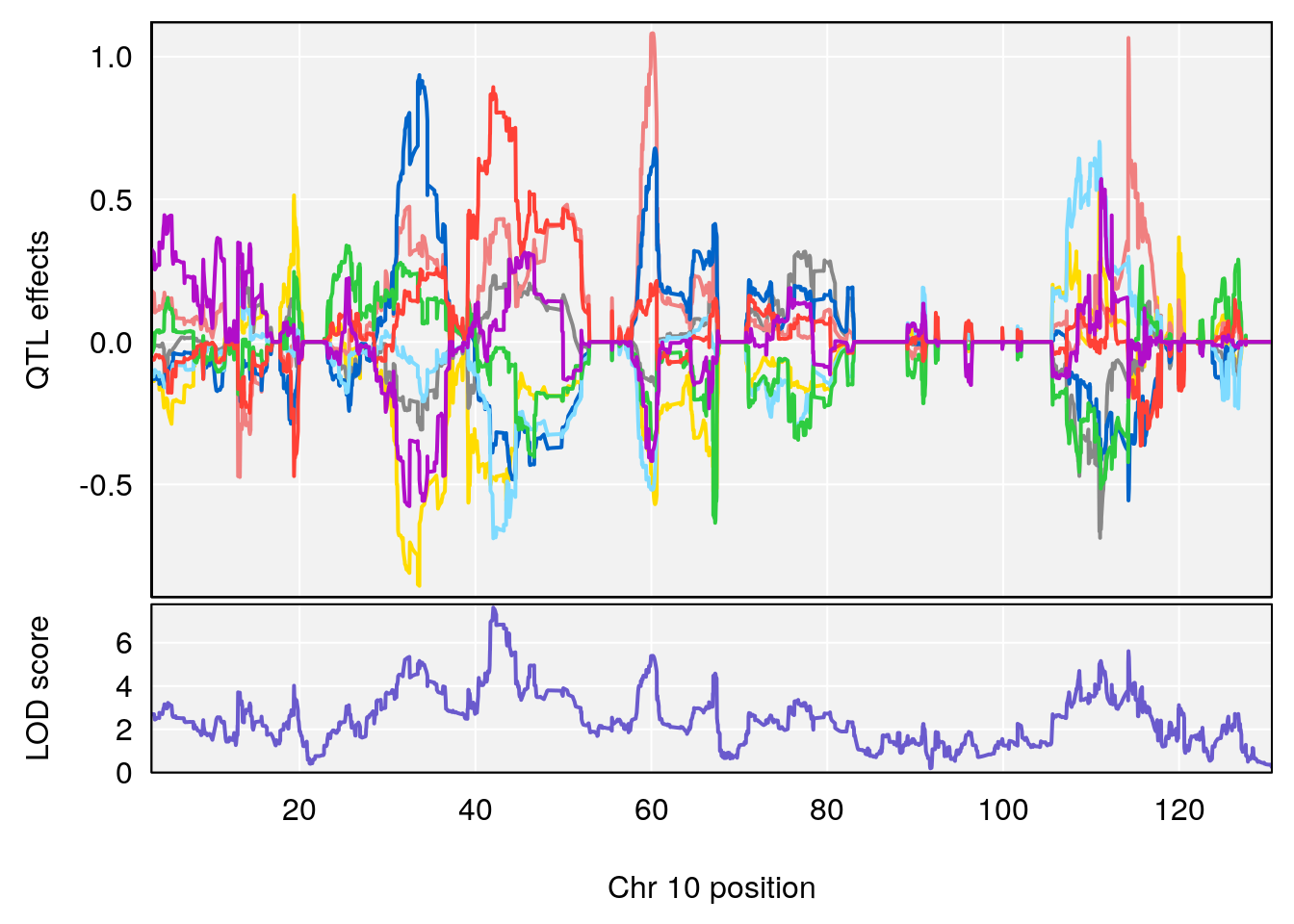
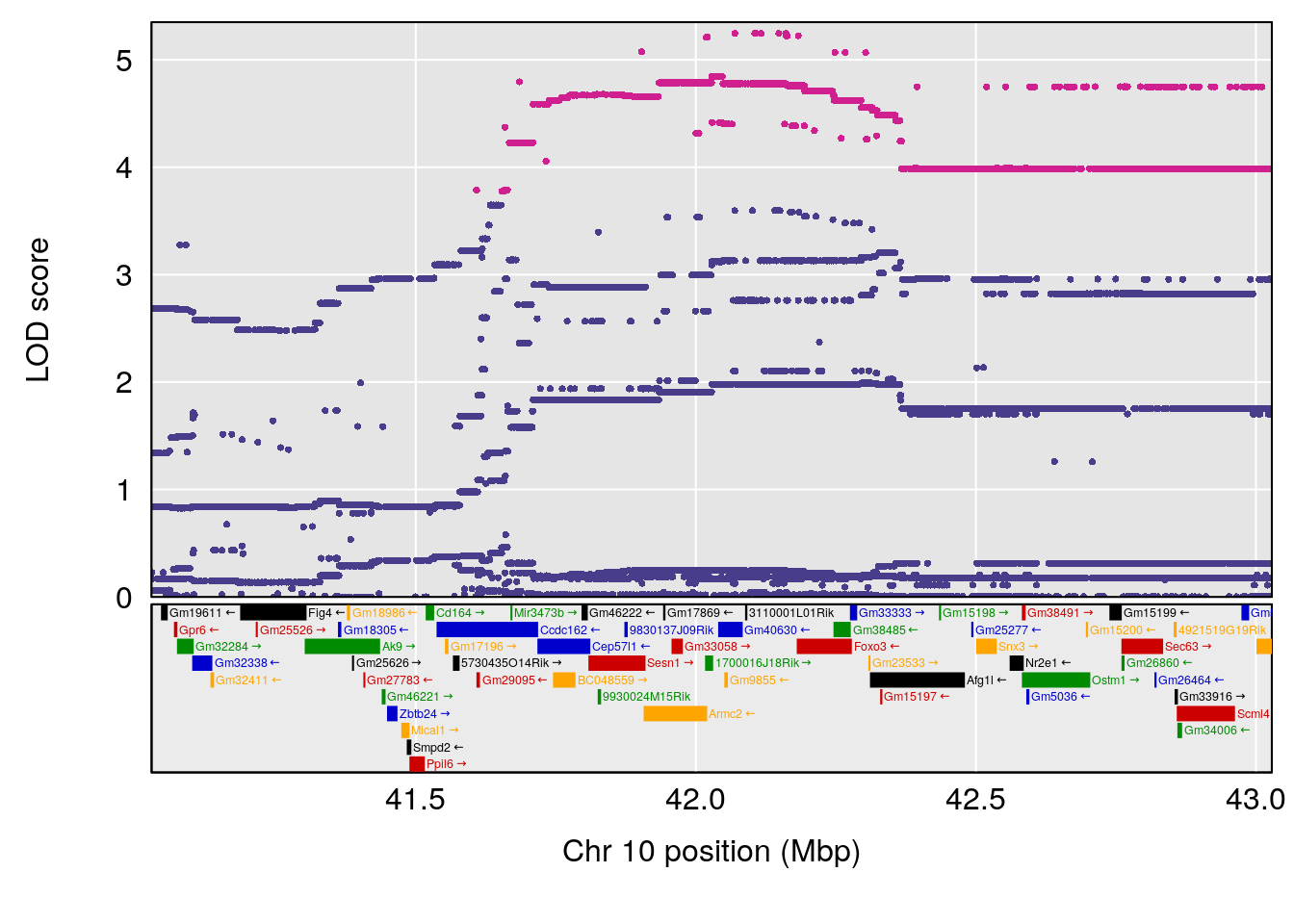
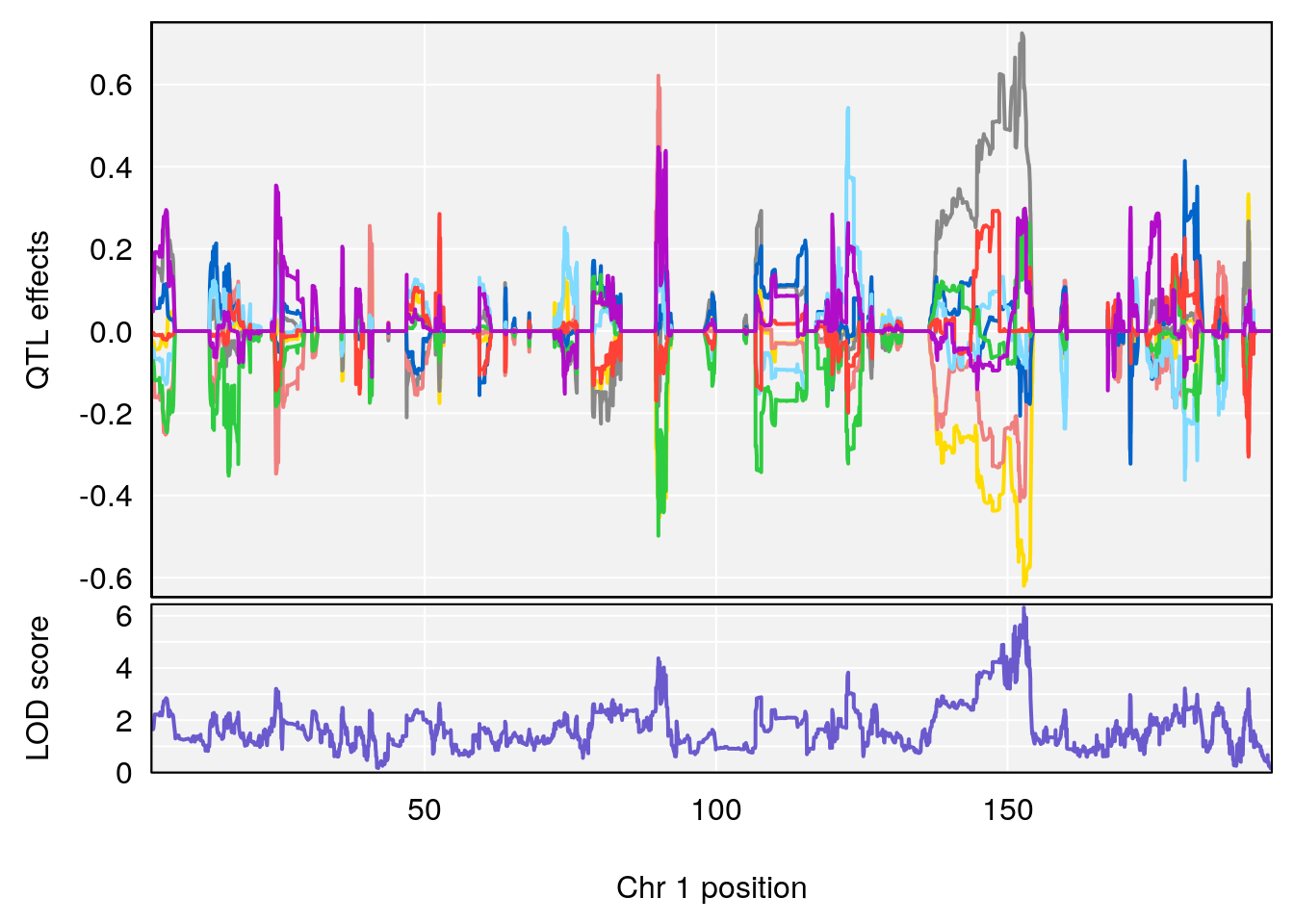
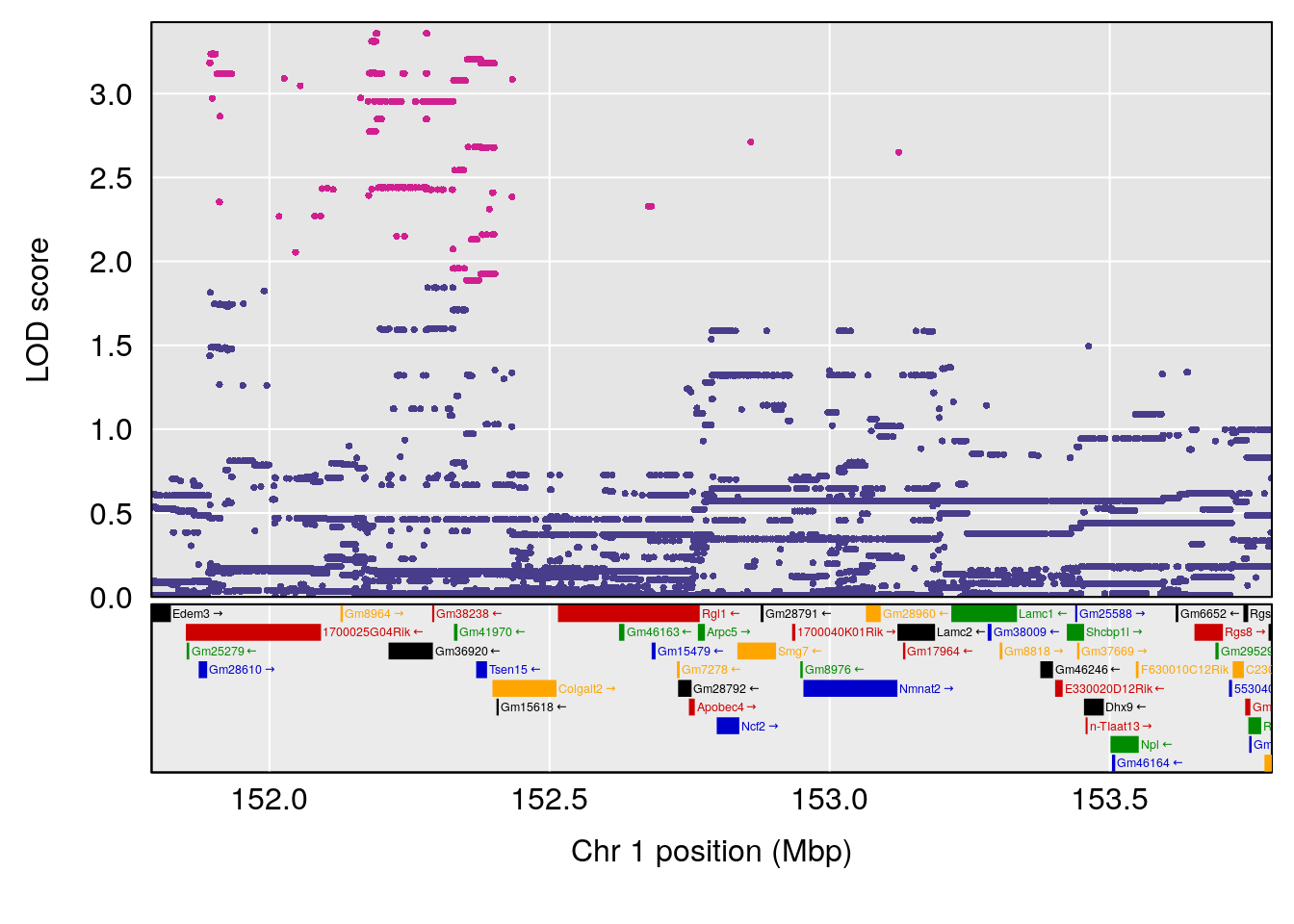
# 1 1 pheno1 1 152.78955 6.378383 151.00100 153.36692
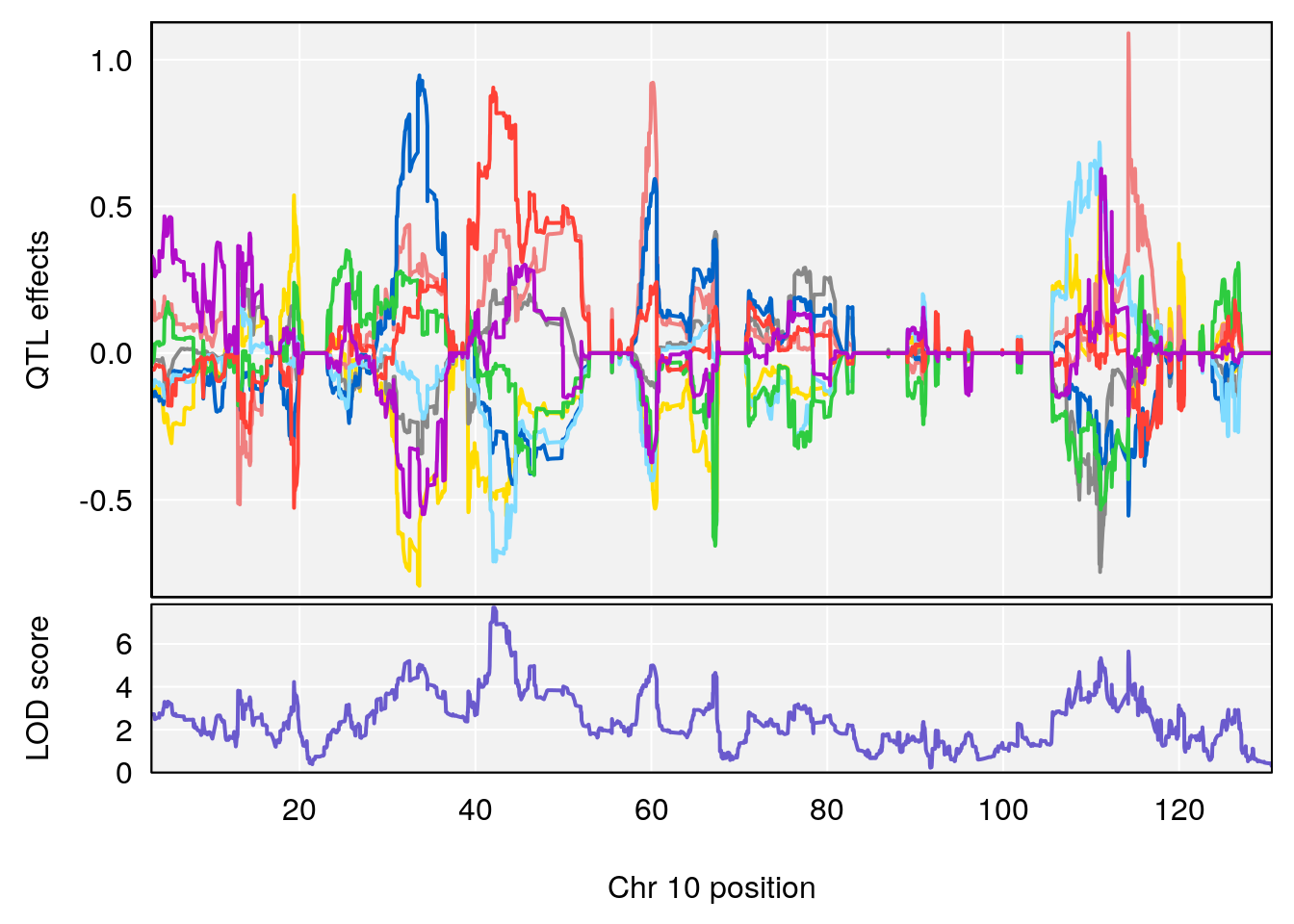
# 2 1 pheno1 10 42.02827 7.628133 41.66399 43.86722
# [1] 1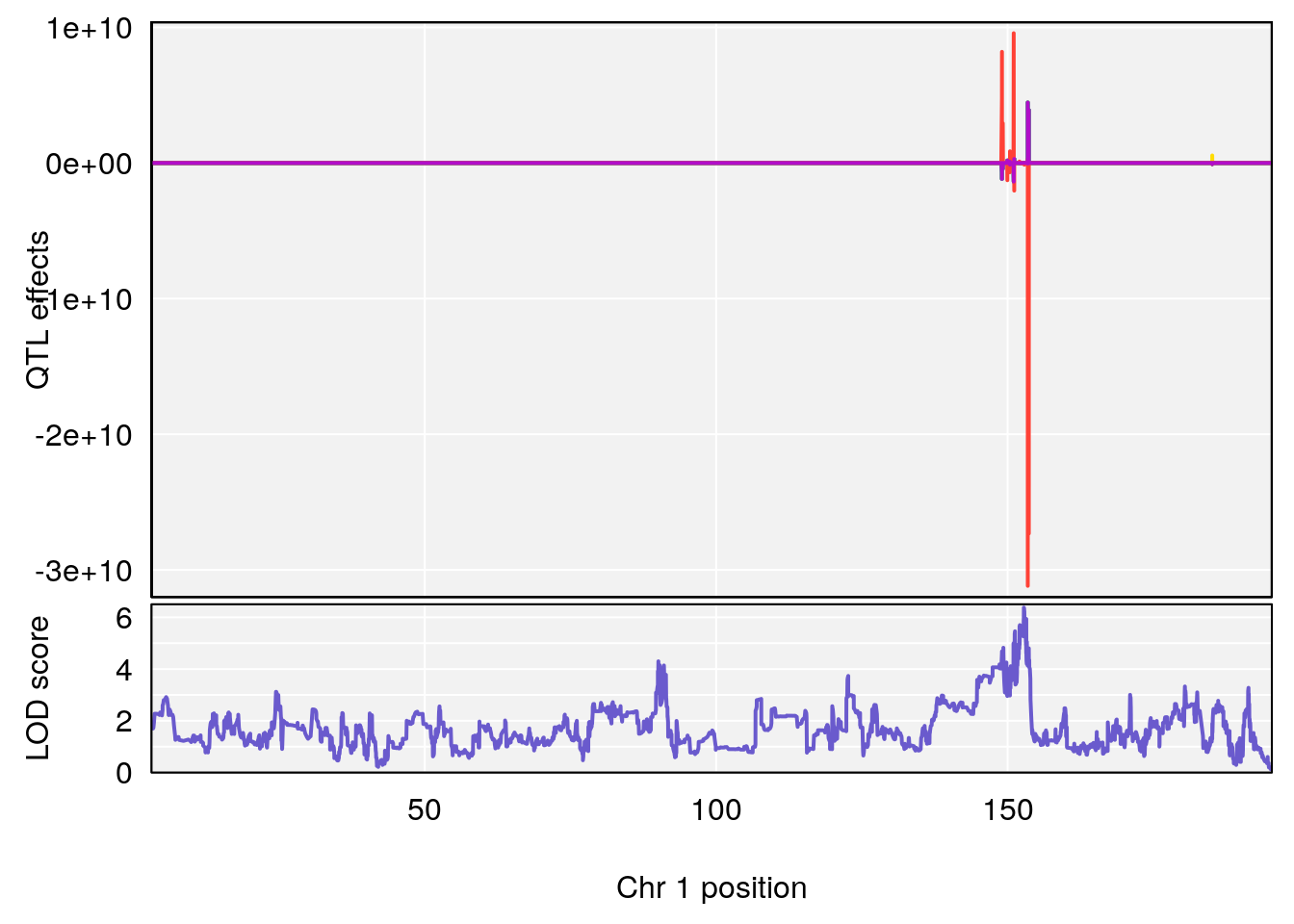
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
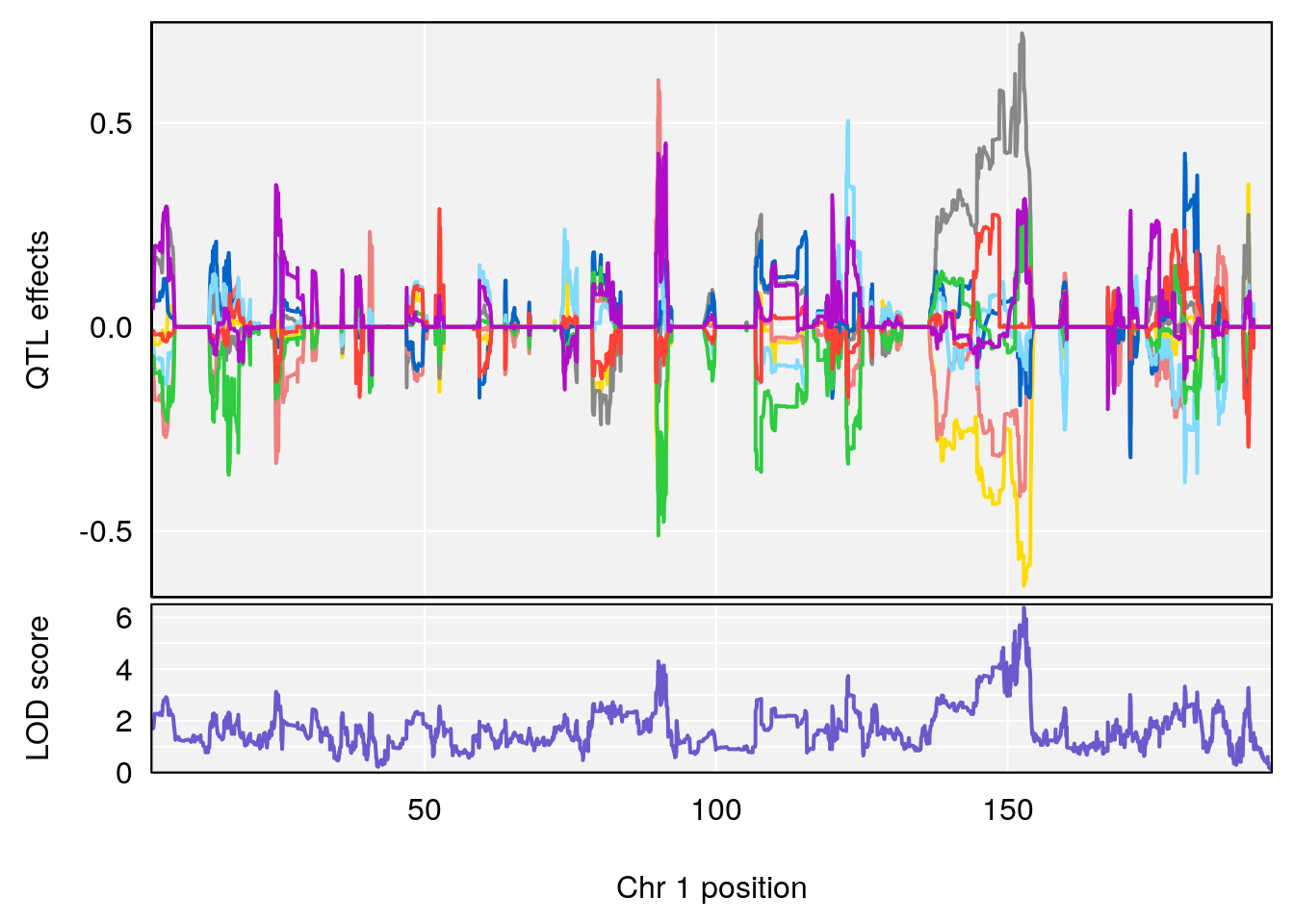
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
# [1] 2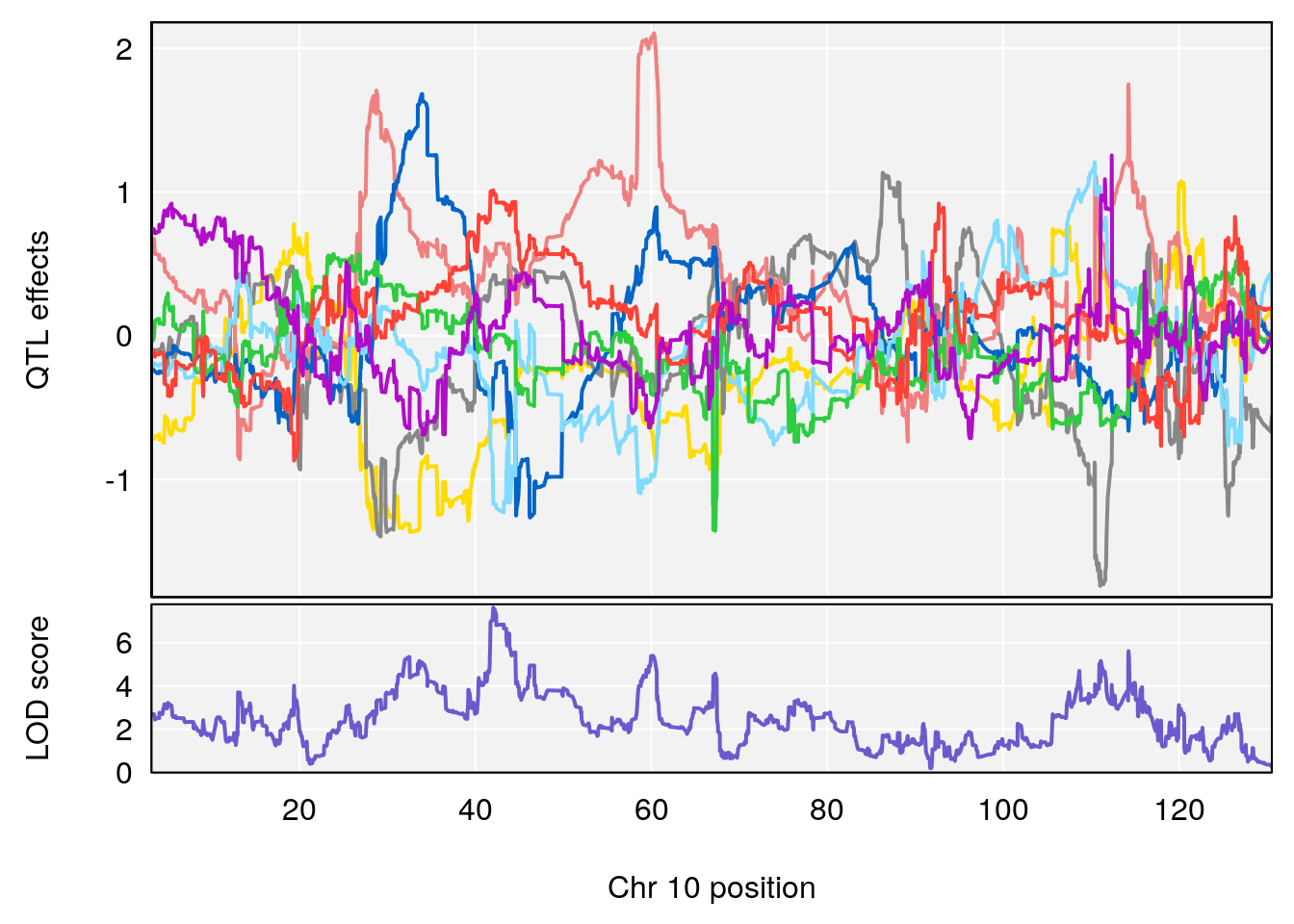
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
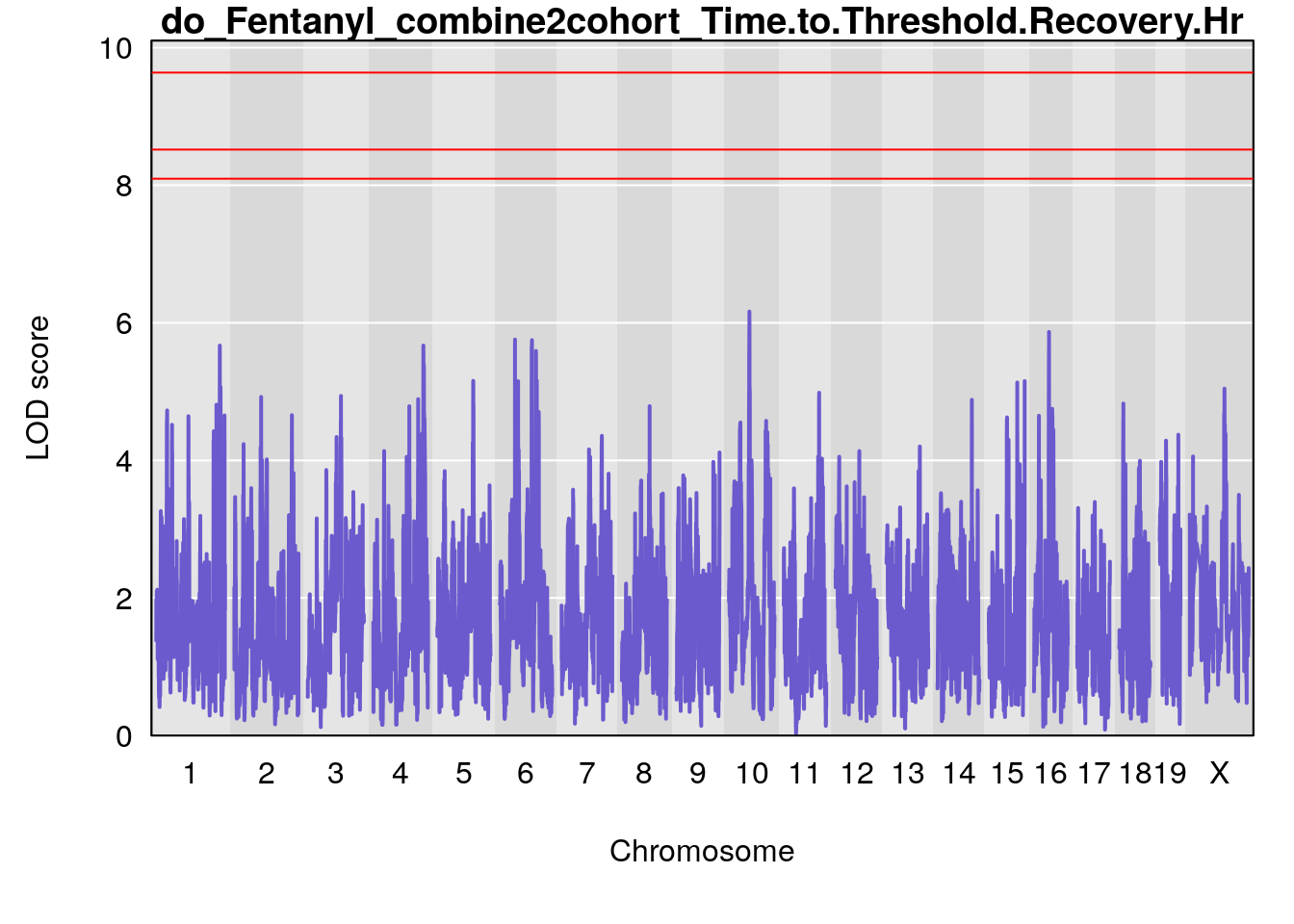
# [1] "Time.to.Threshold.Recovery.Hr"
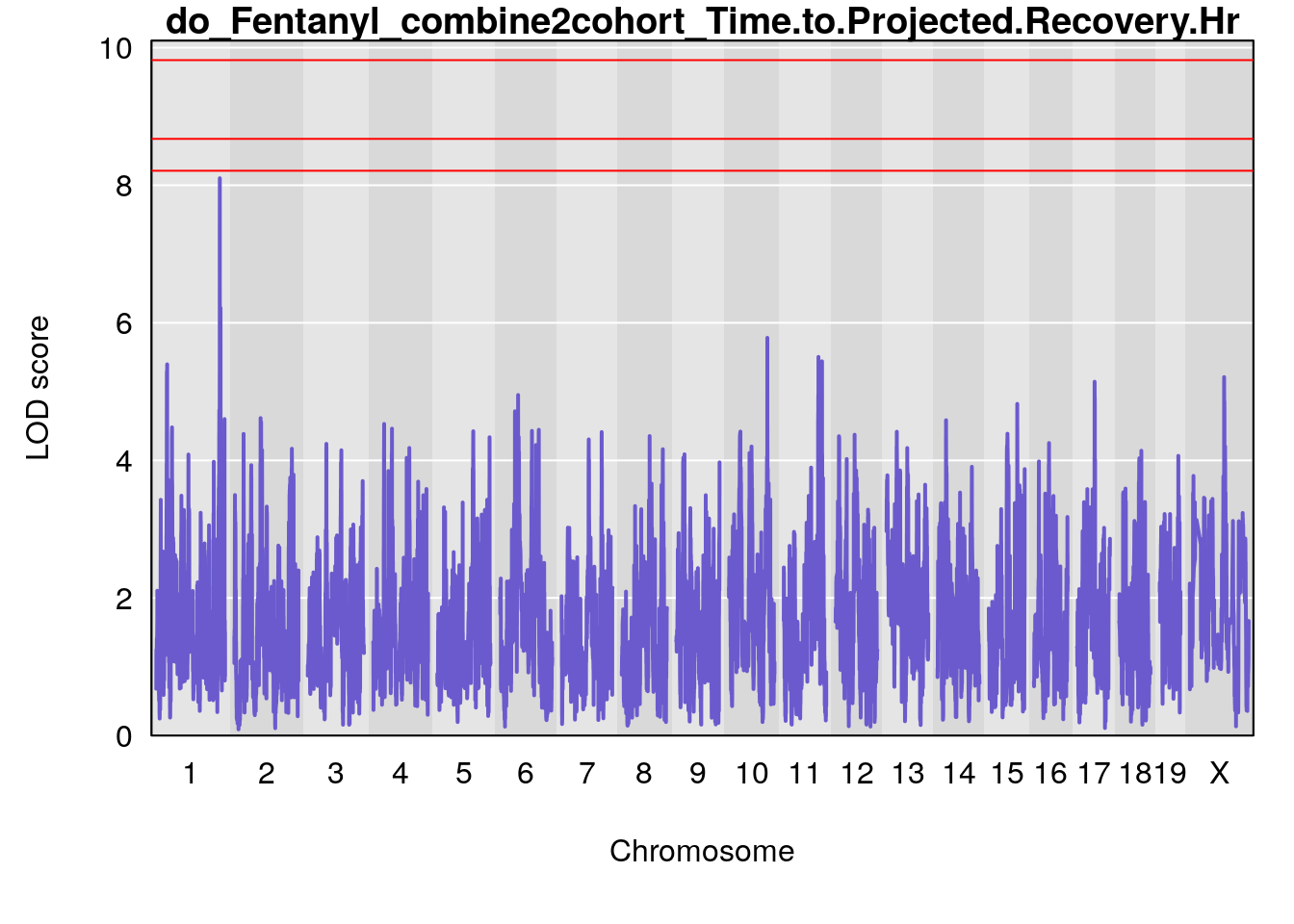
# [1] "Time.to.Projected.Recovery.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
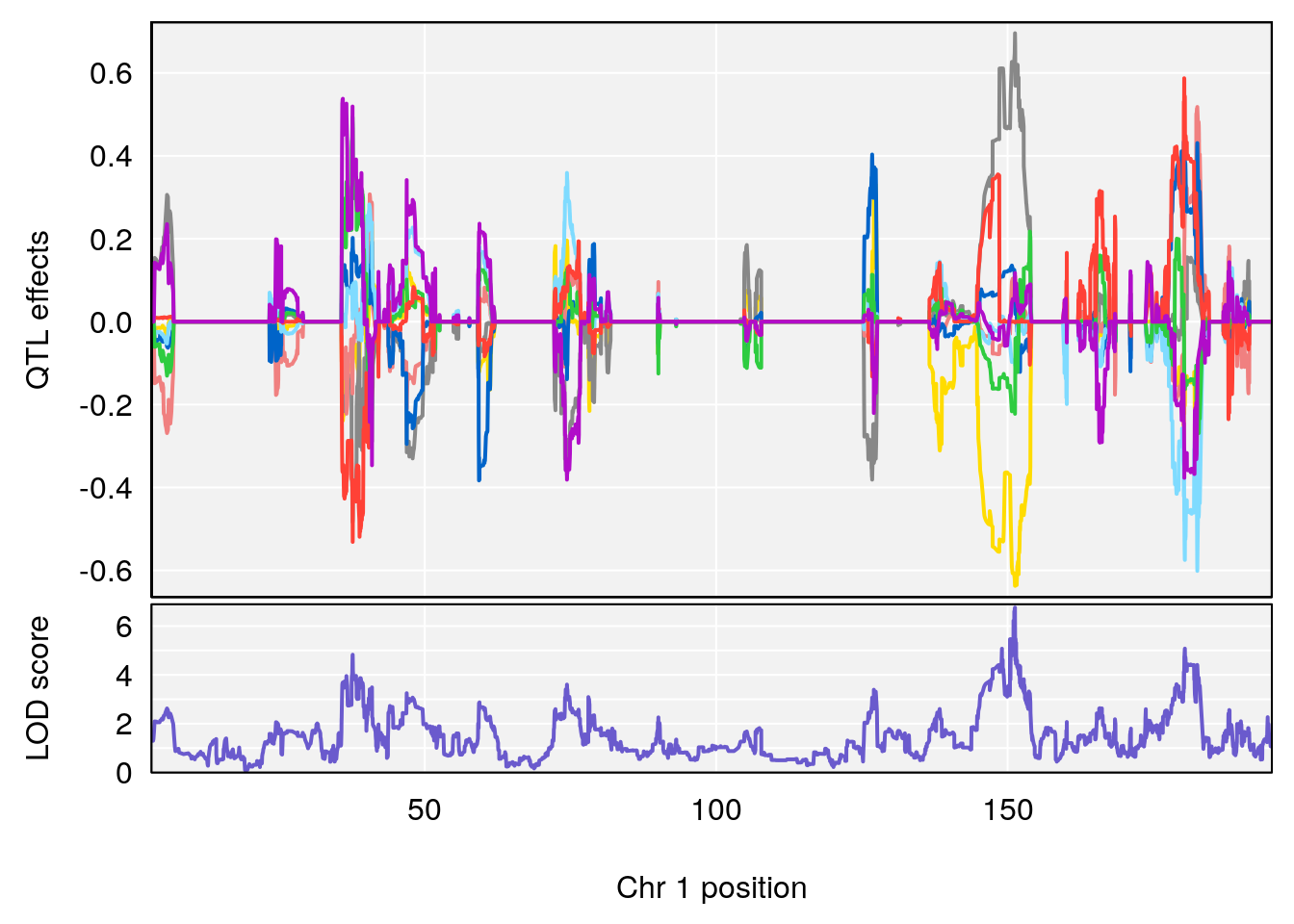
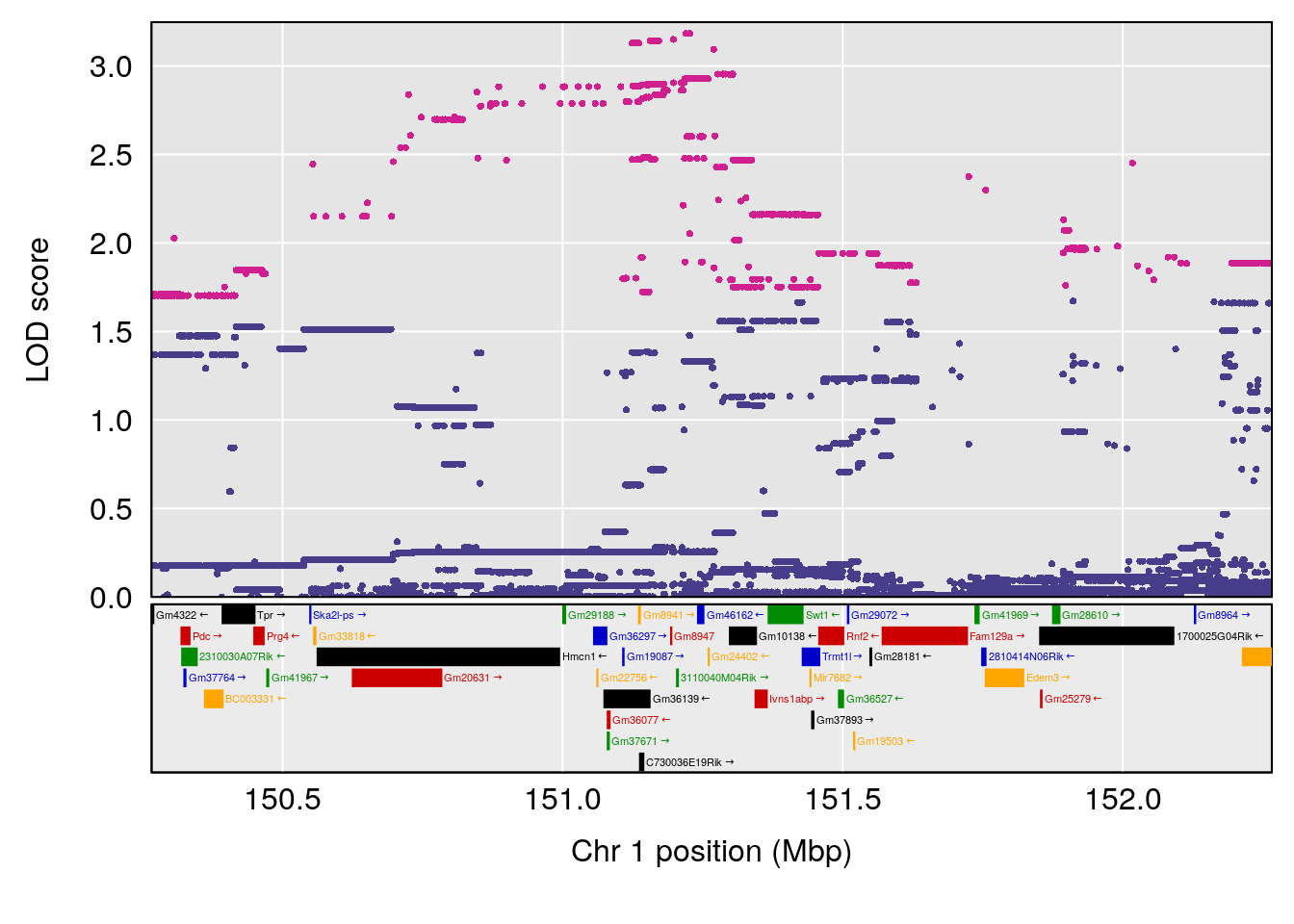
# 1 1 pheno1 1 151.2661 6.756996 150.4074 151.4769
# [1] 1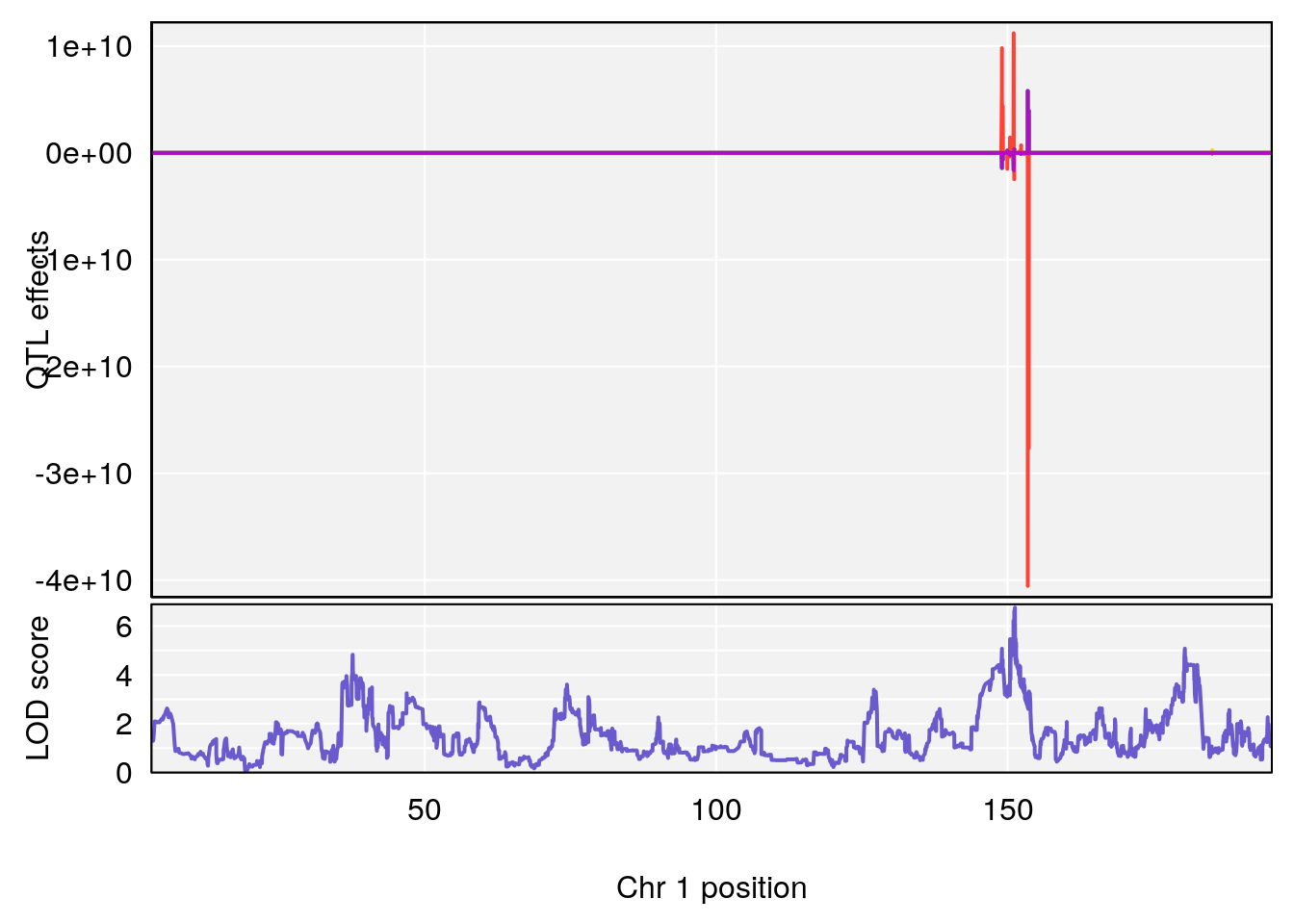
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
# [1] "Start.of.Recovery.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 152.78955 6.310882 149.02638 153.36692
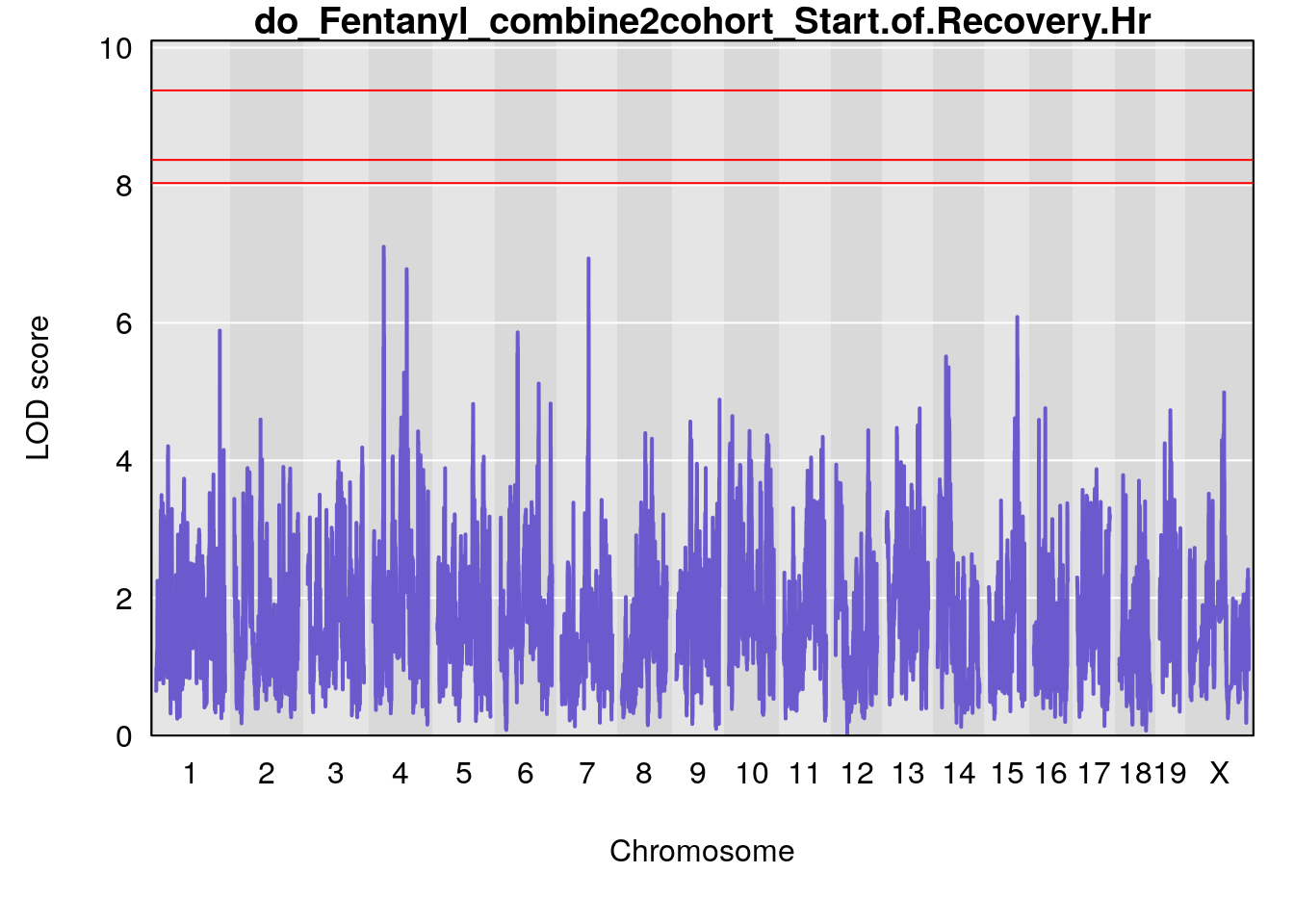
# 2 1 pheno1 10 42.16135 7.694886 41.66399 43.86722
# 3 1 pheno1 15 93.60493 6.059735 88.73612 93.88737
# [1] 1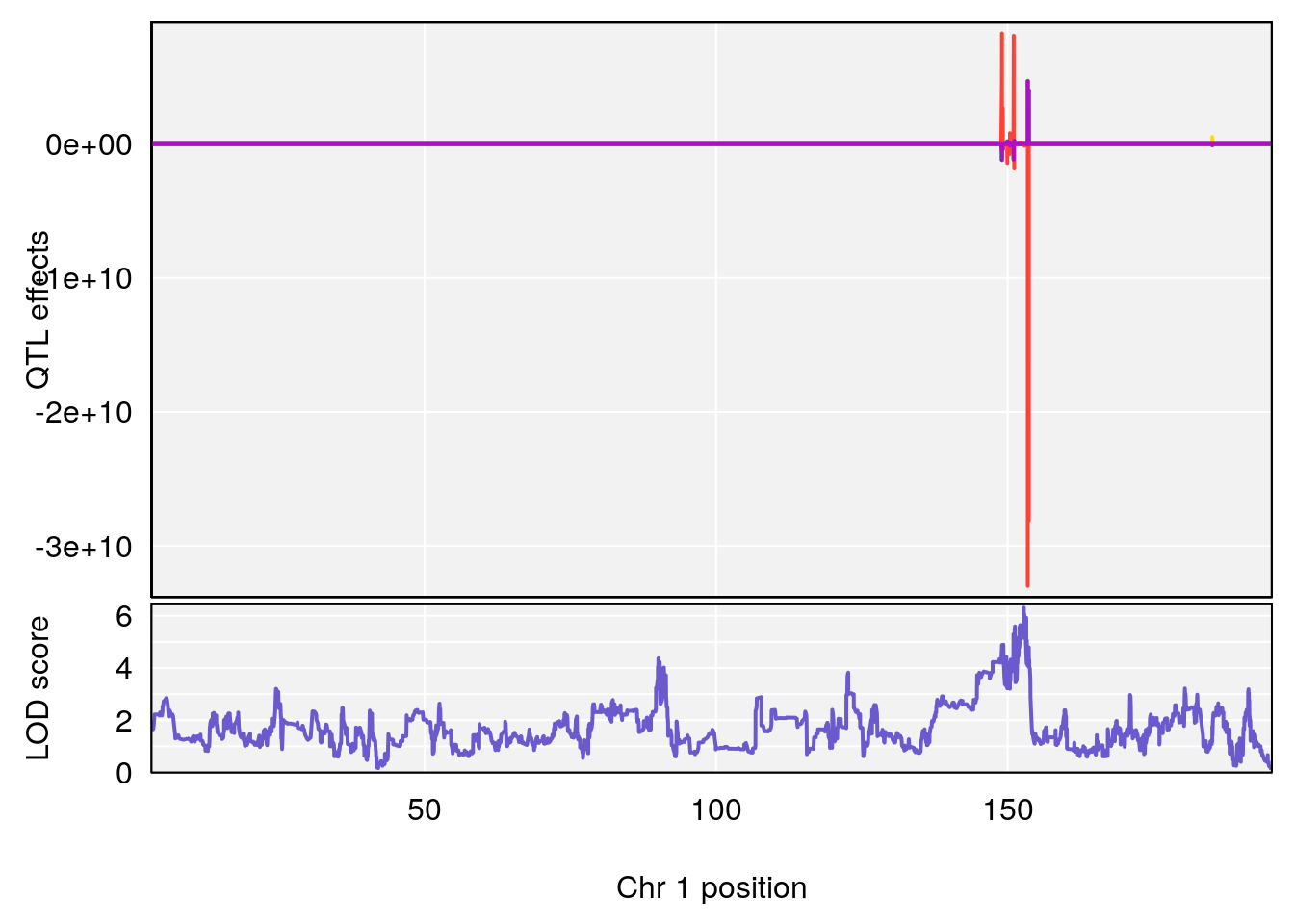
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
# [1] 2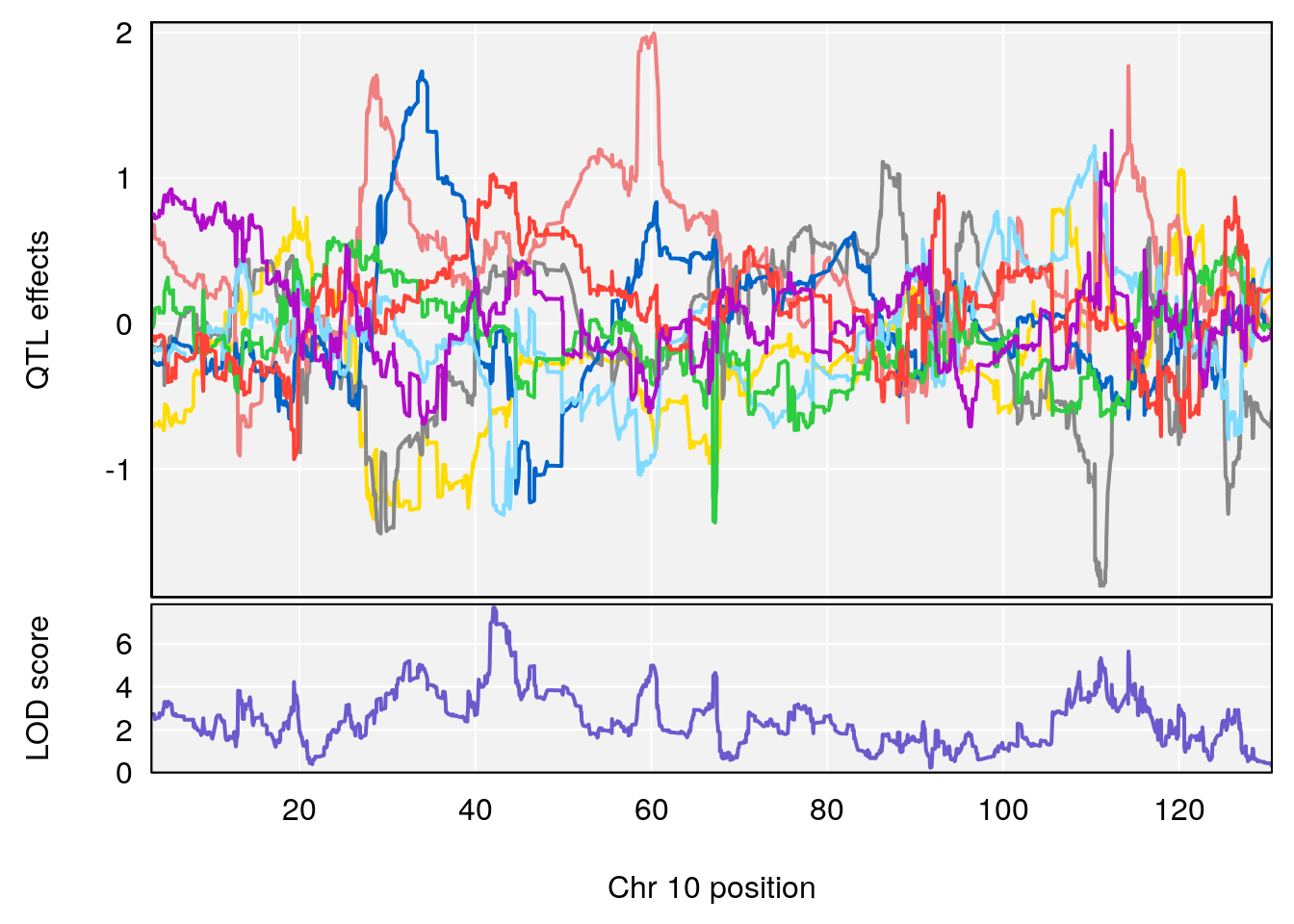
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
# [1] 3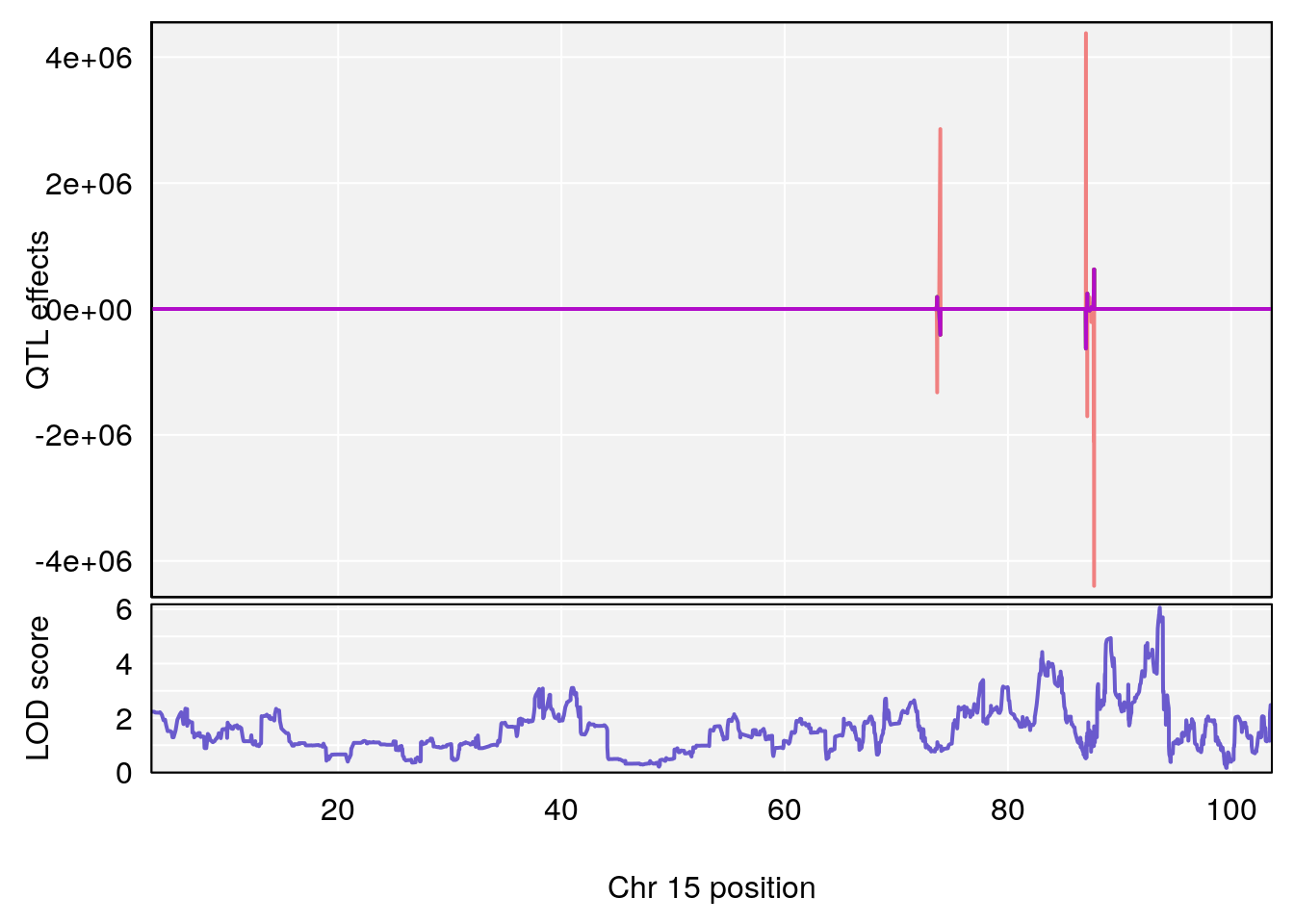
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
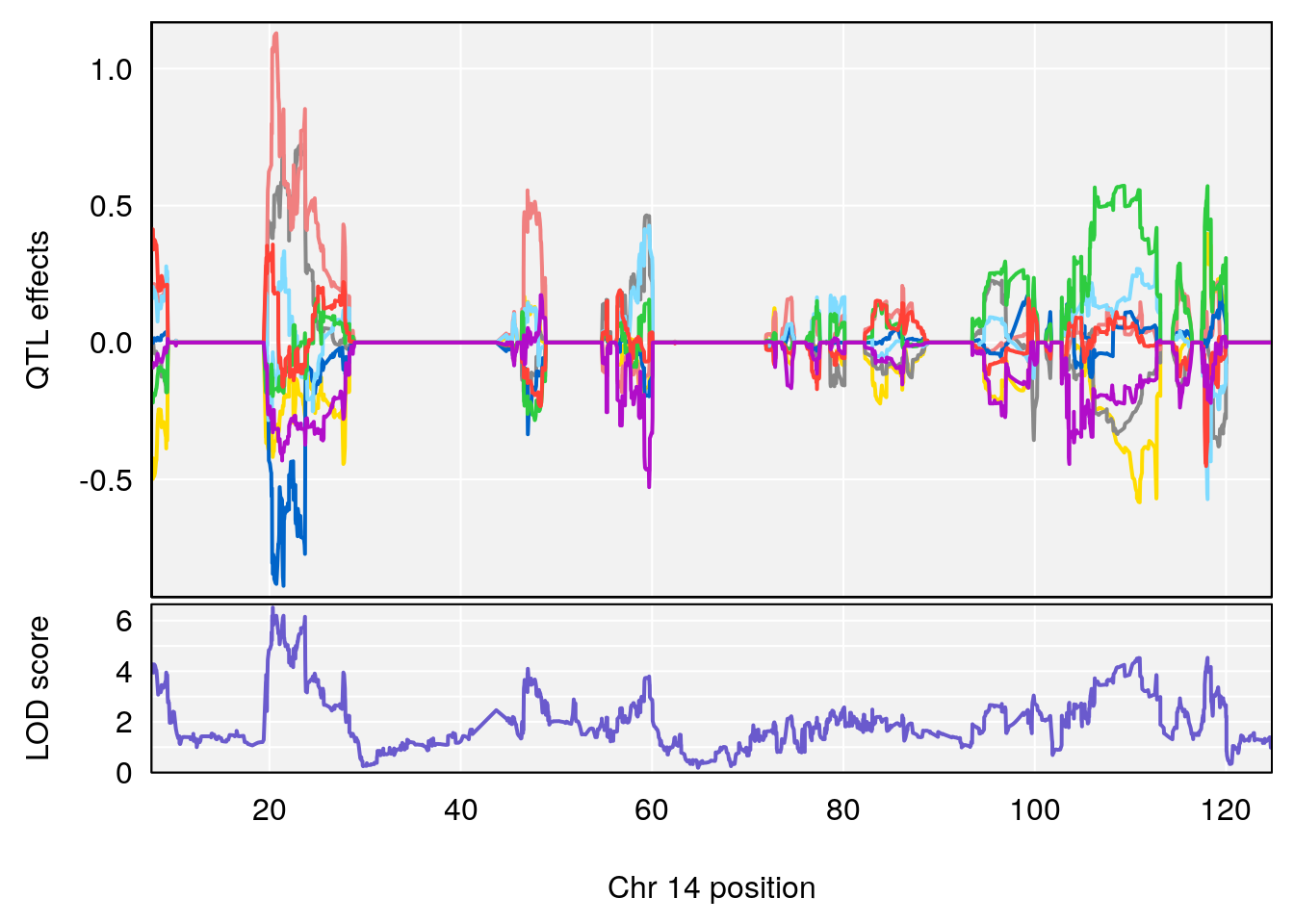
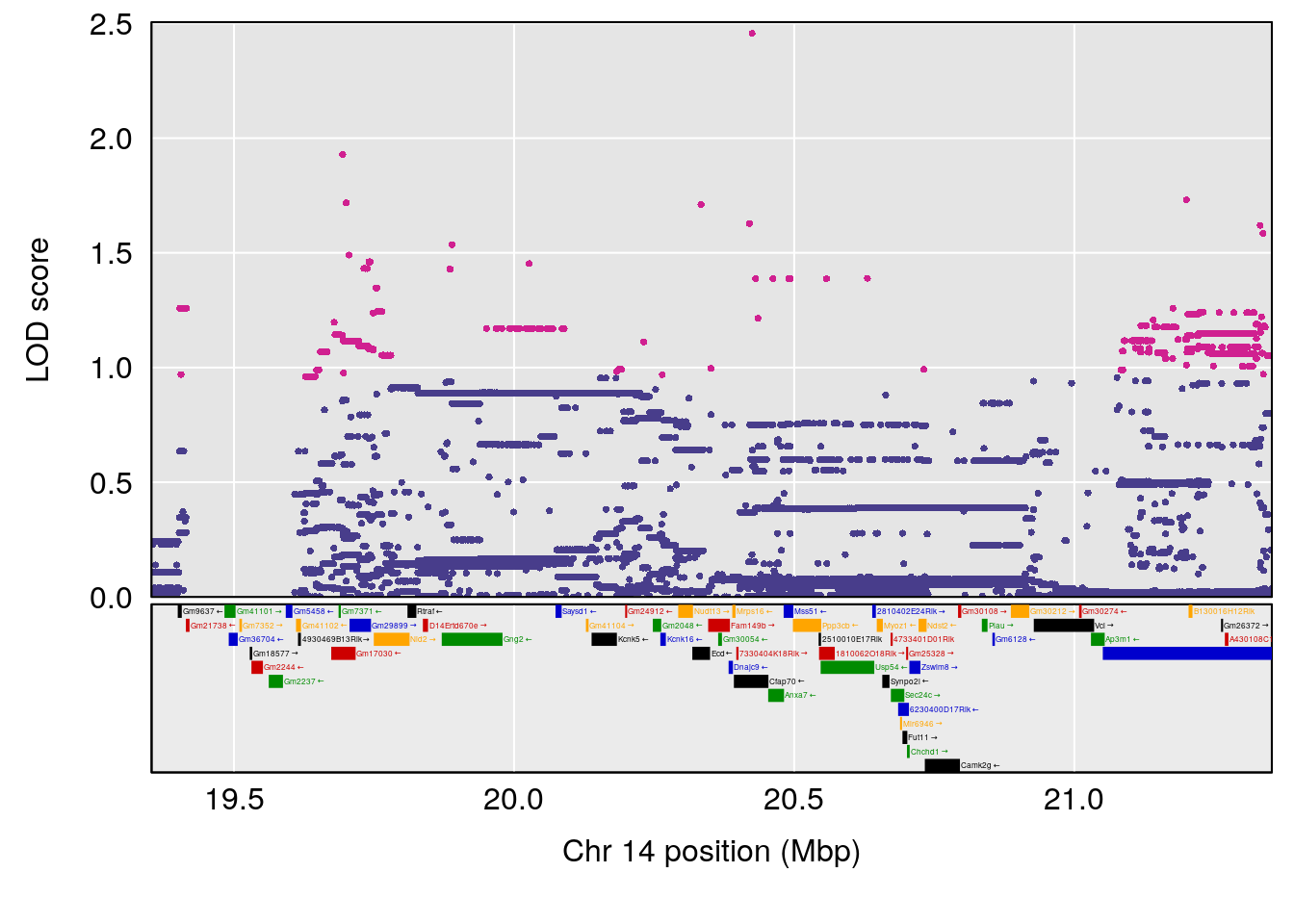
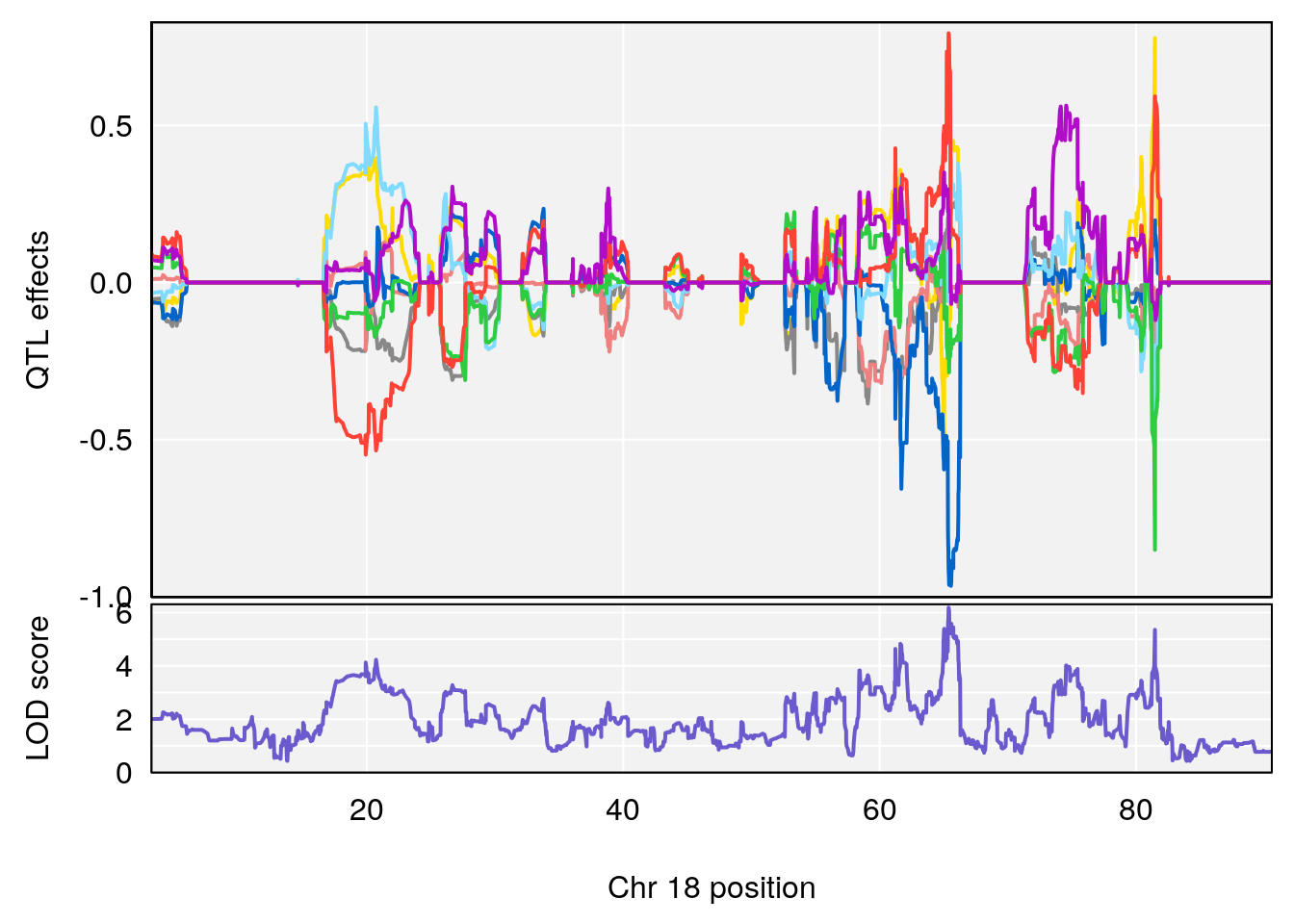
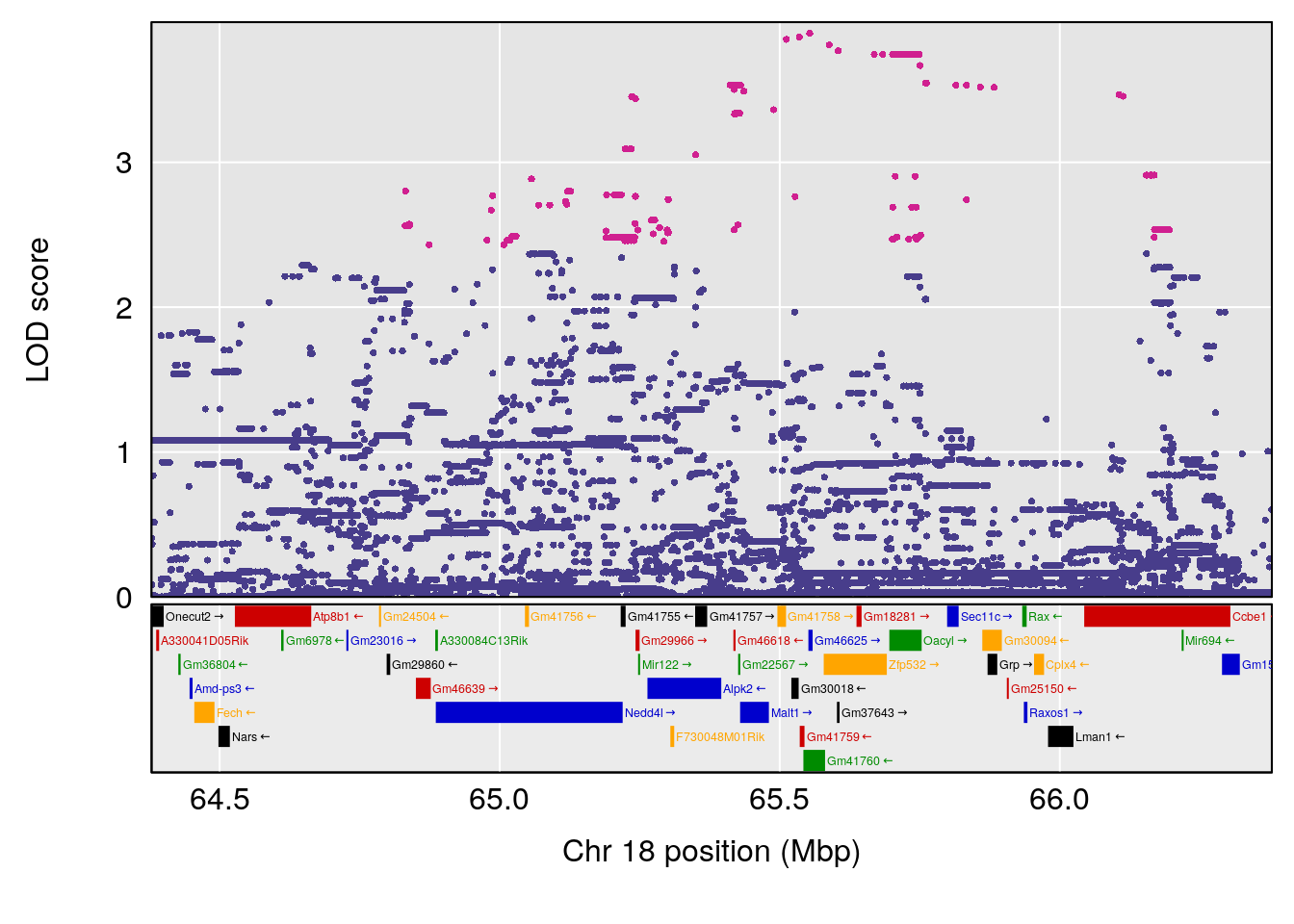
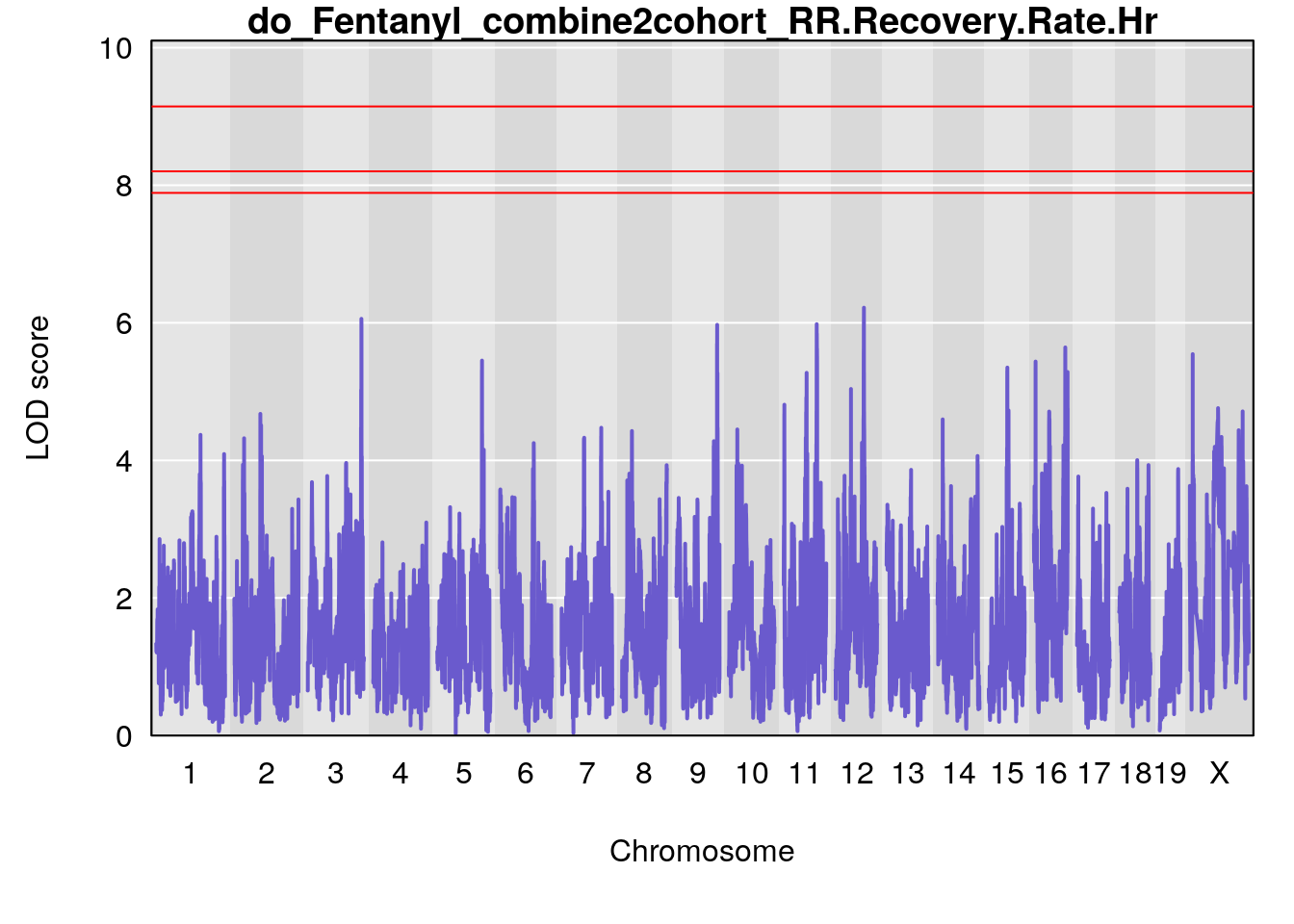
# [1] "RR.Recovery.Rate.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 14 20.35257 6.513019 20.15234 23.74698
# 2 1 pheno1 18 65.37854 6.187039 61.59291 81.48908
# [1] 1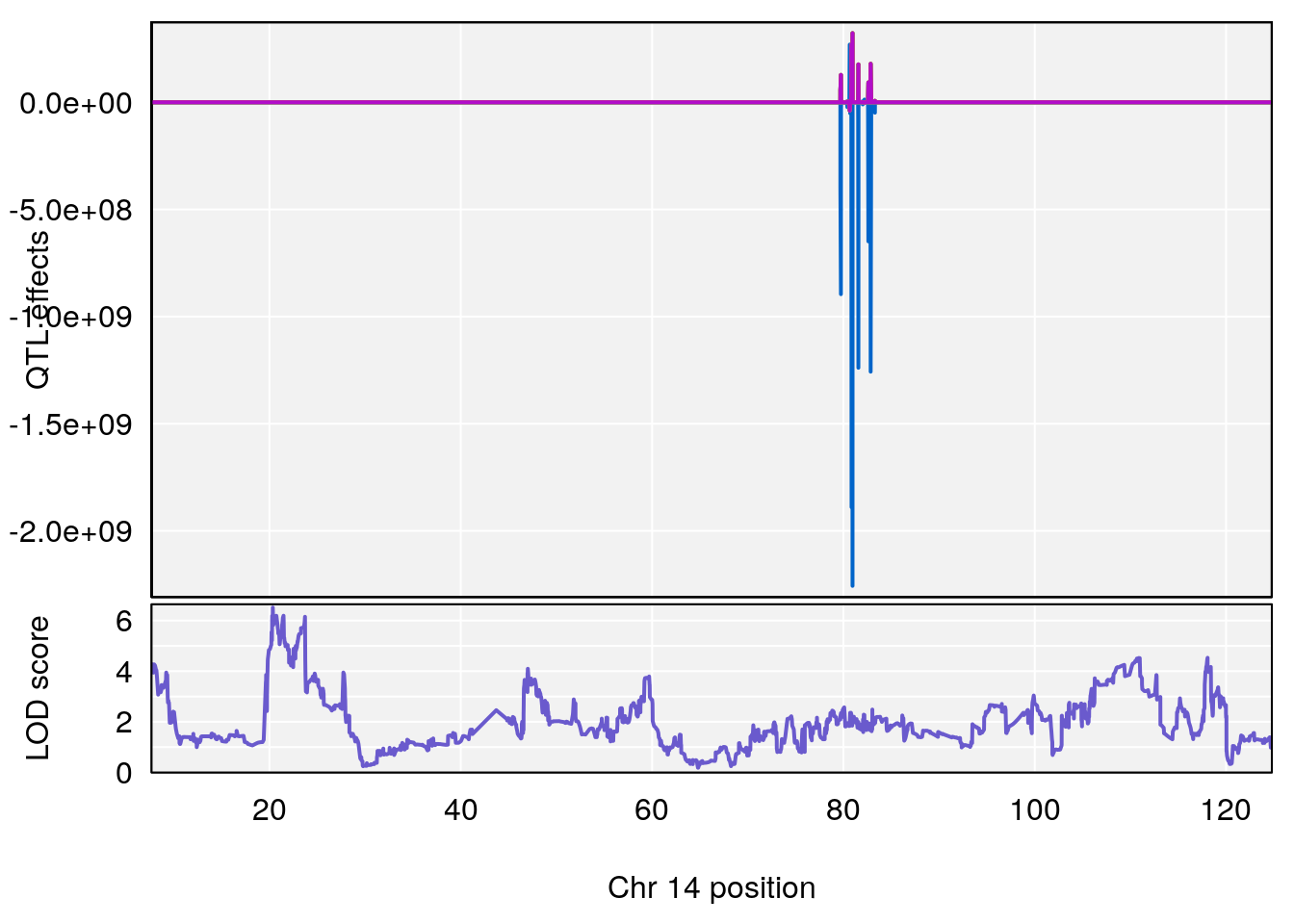
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
# [1] 2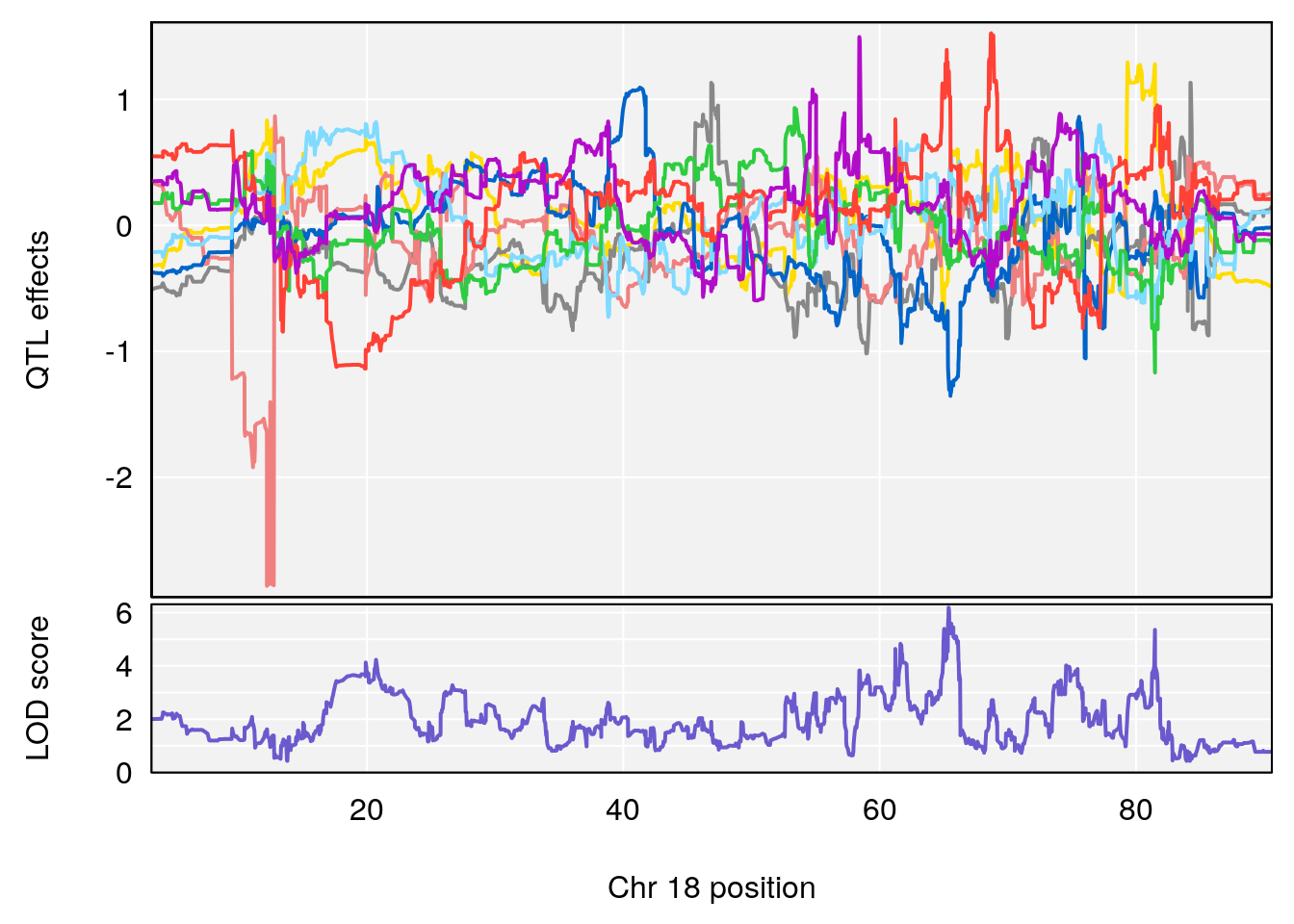
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
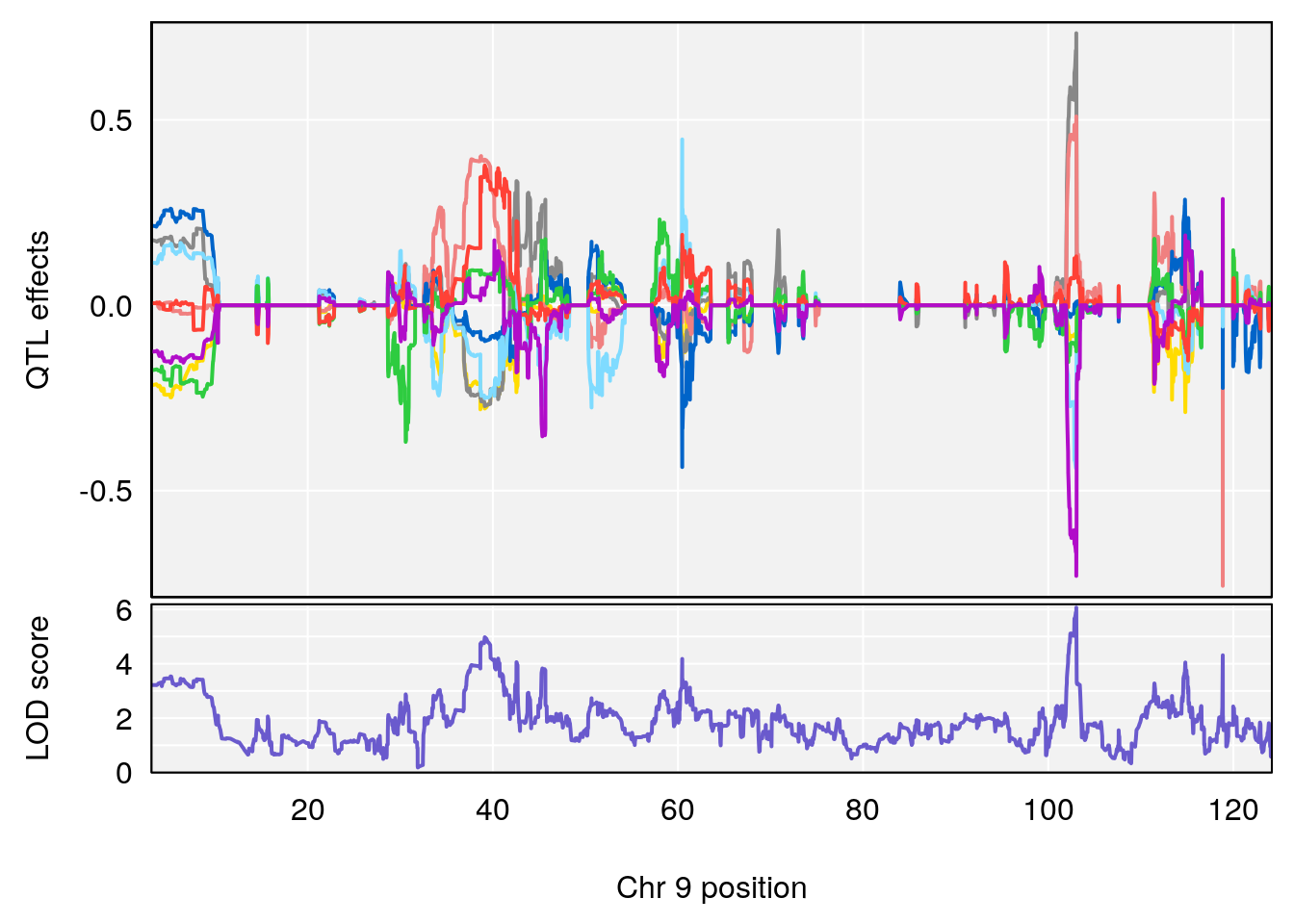
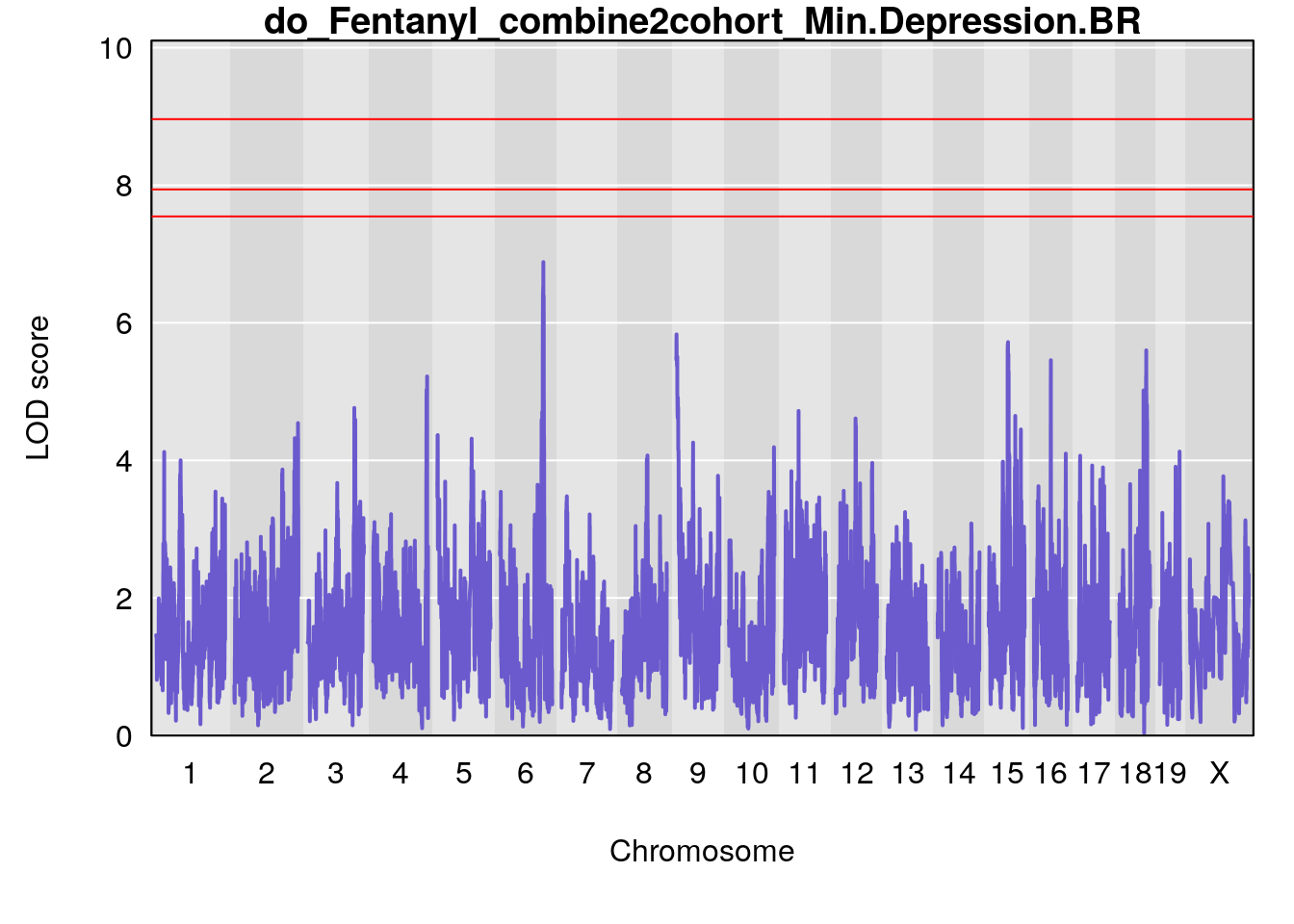
# [1] "Min.Depression.BR"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 9 103.0304 6.065324 38.64793 103.0599
# [1] 1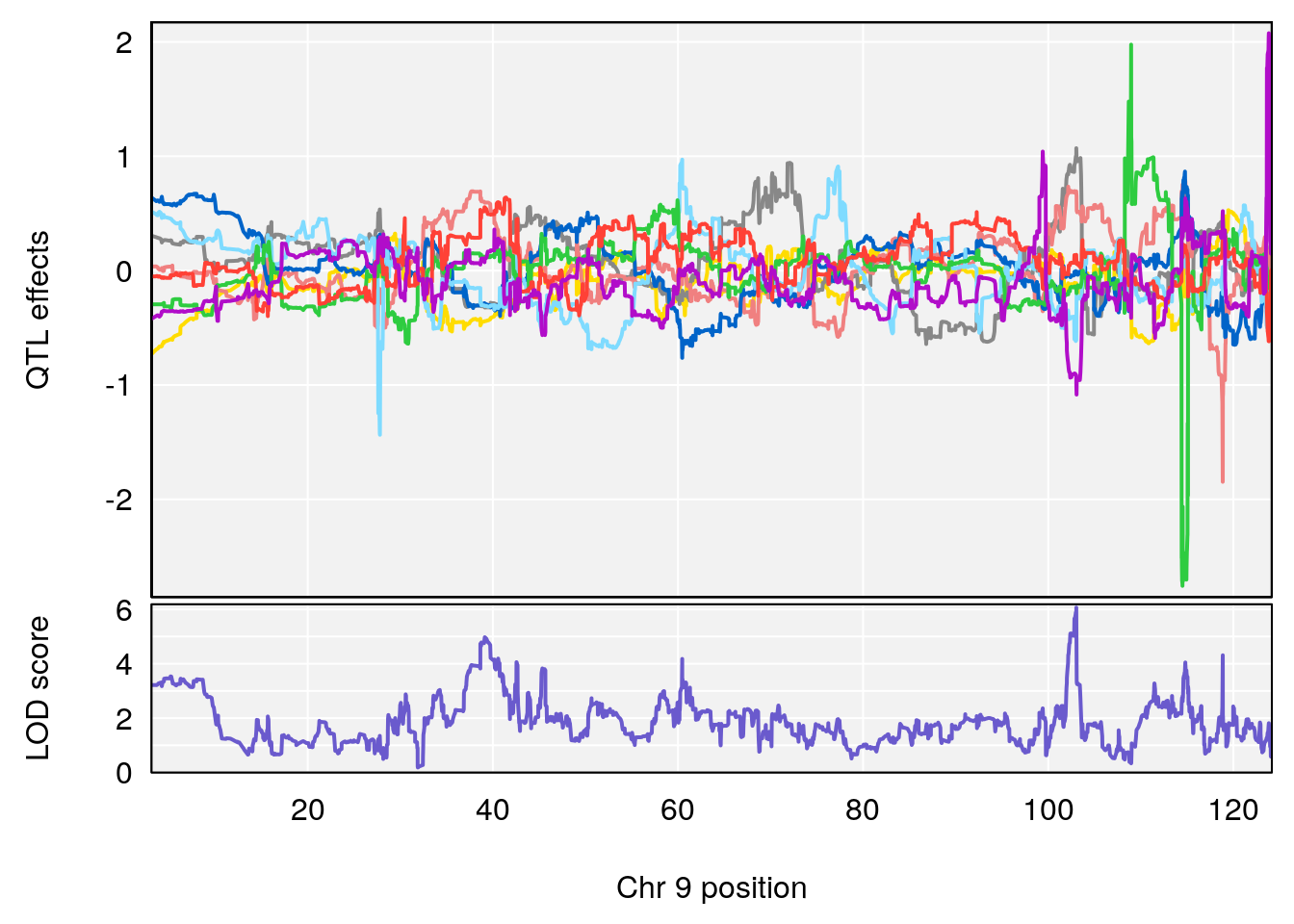
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
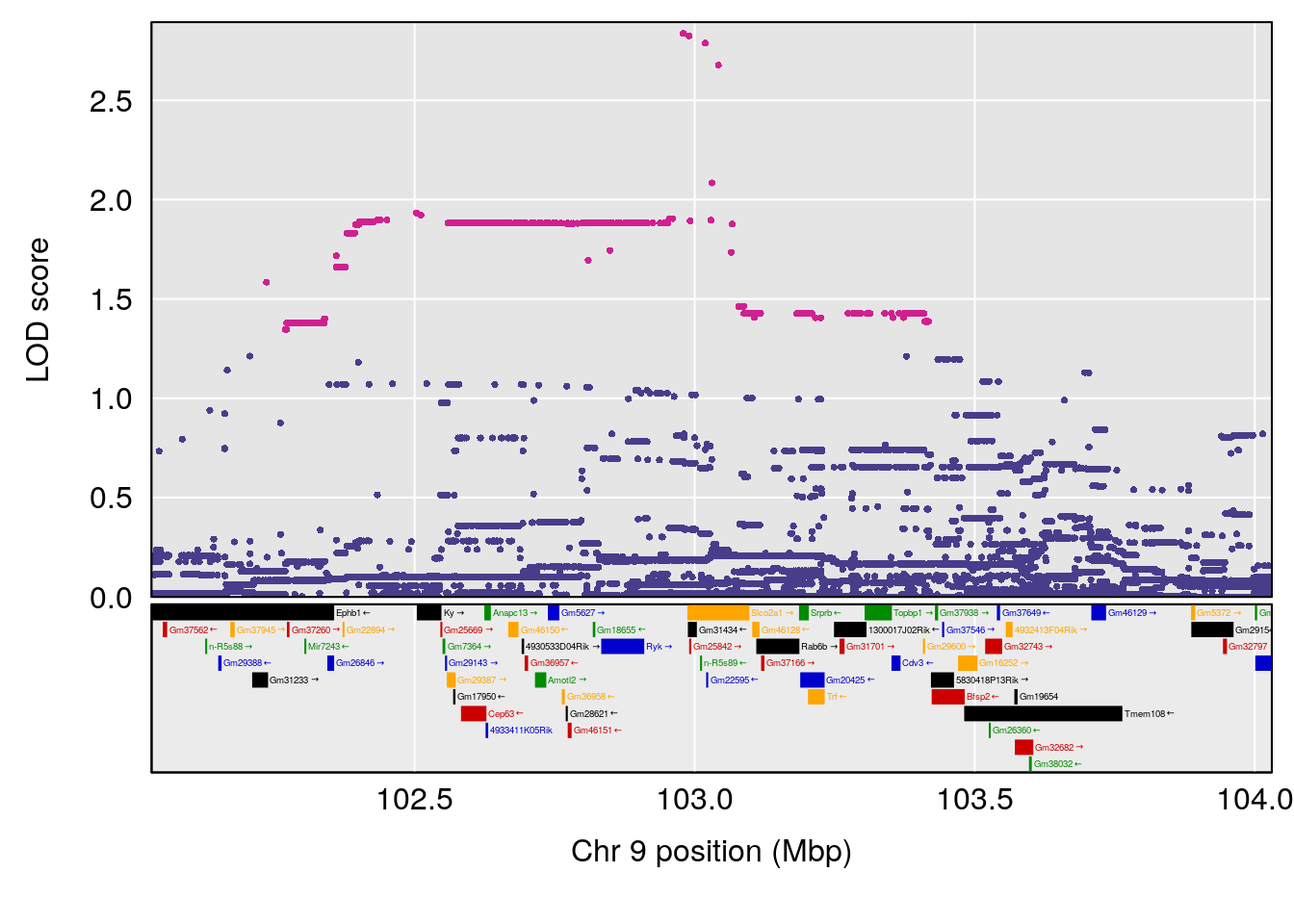
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
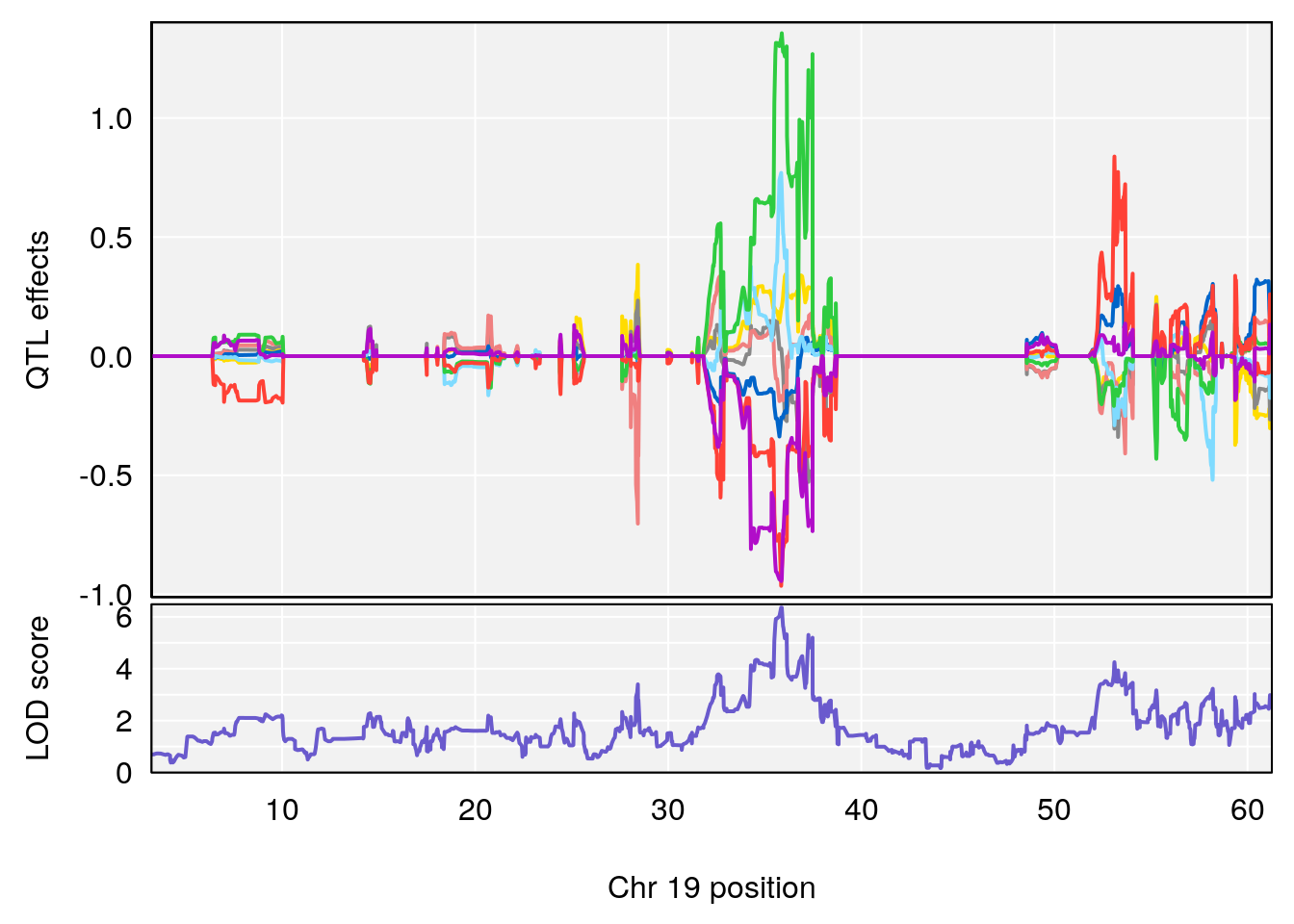
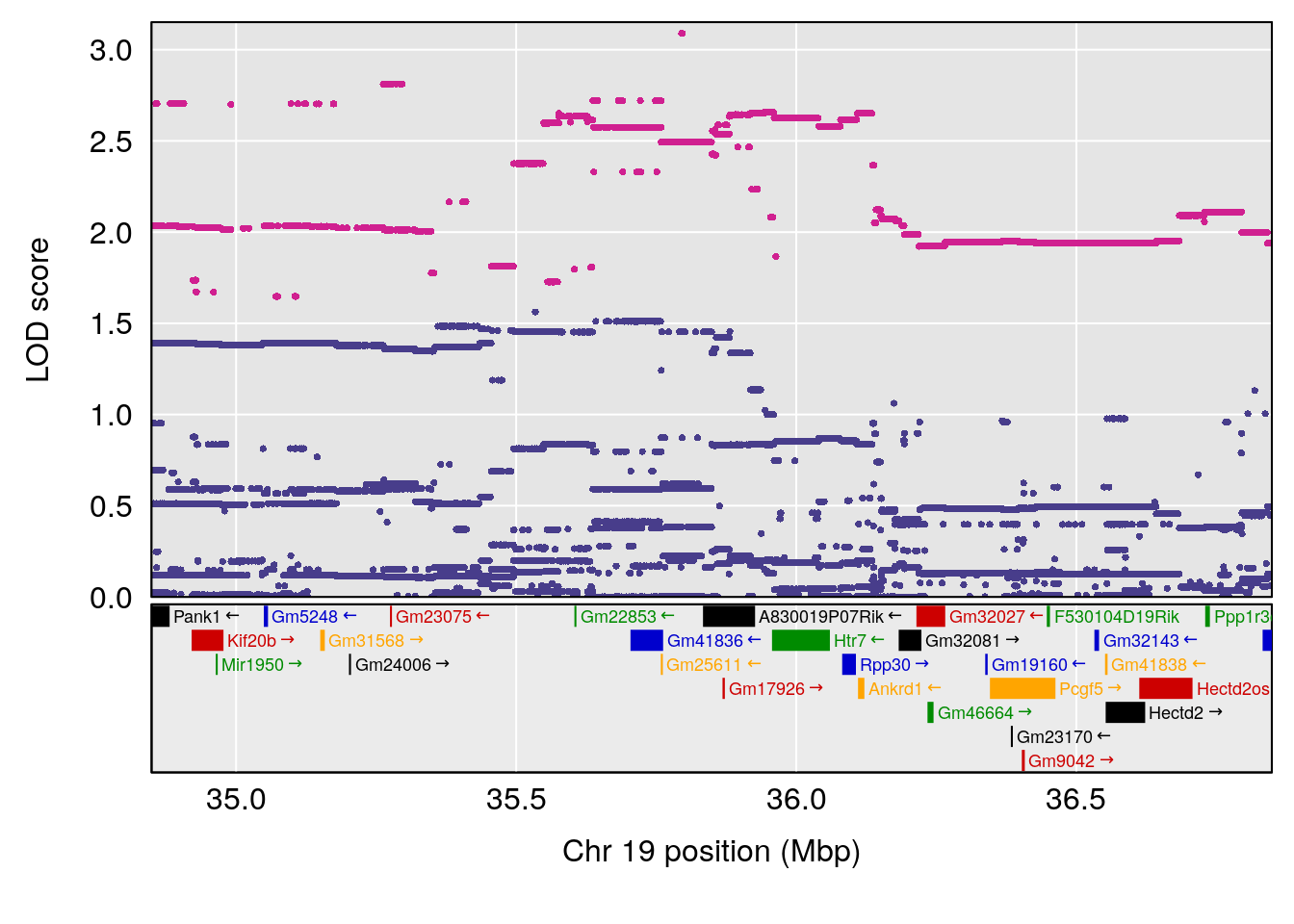
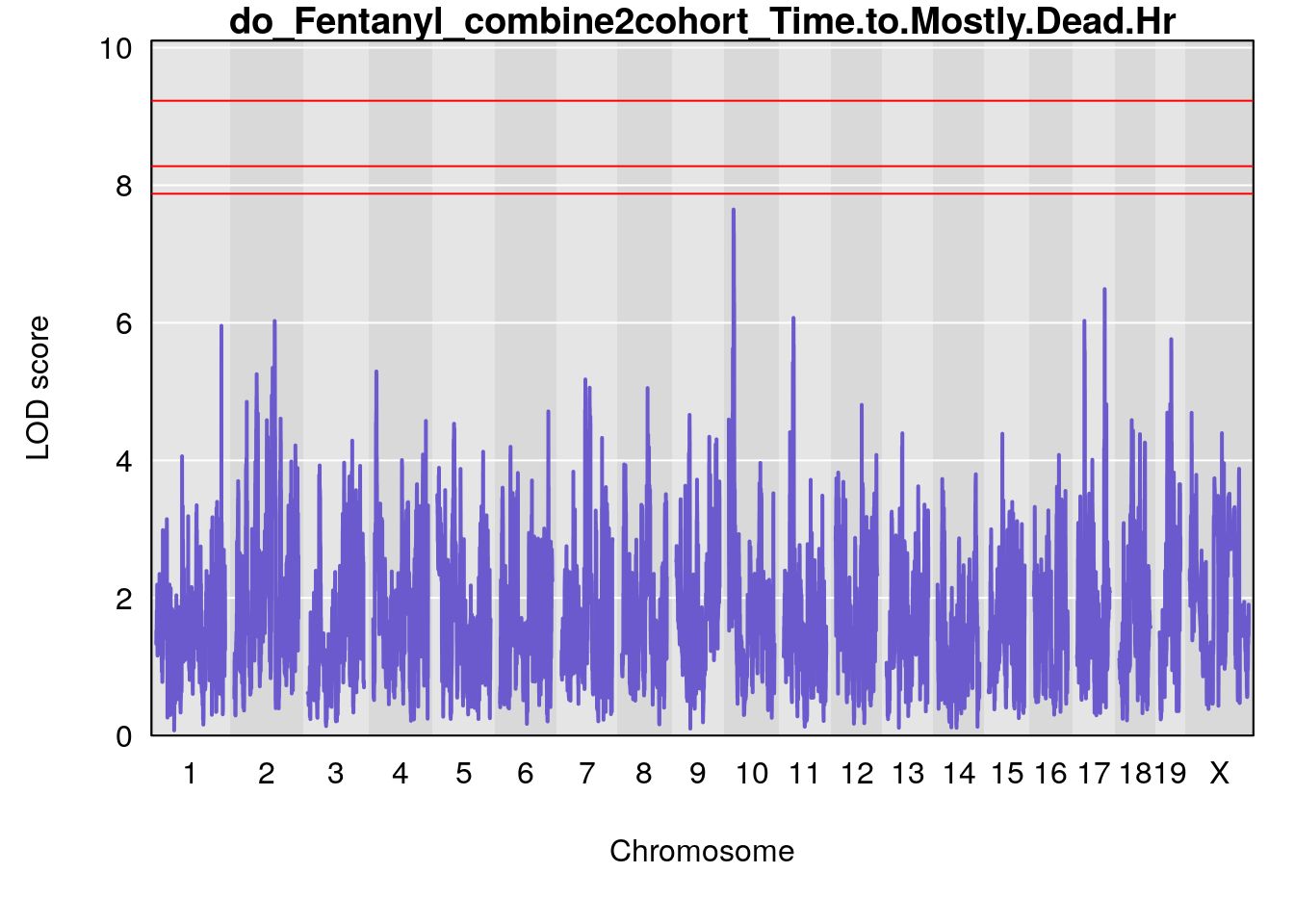
# [1] "Time.to.Mostly.Dead.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 19 35.84901 6.359785 35.4551 37.47958
# [1] 1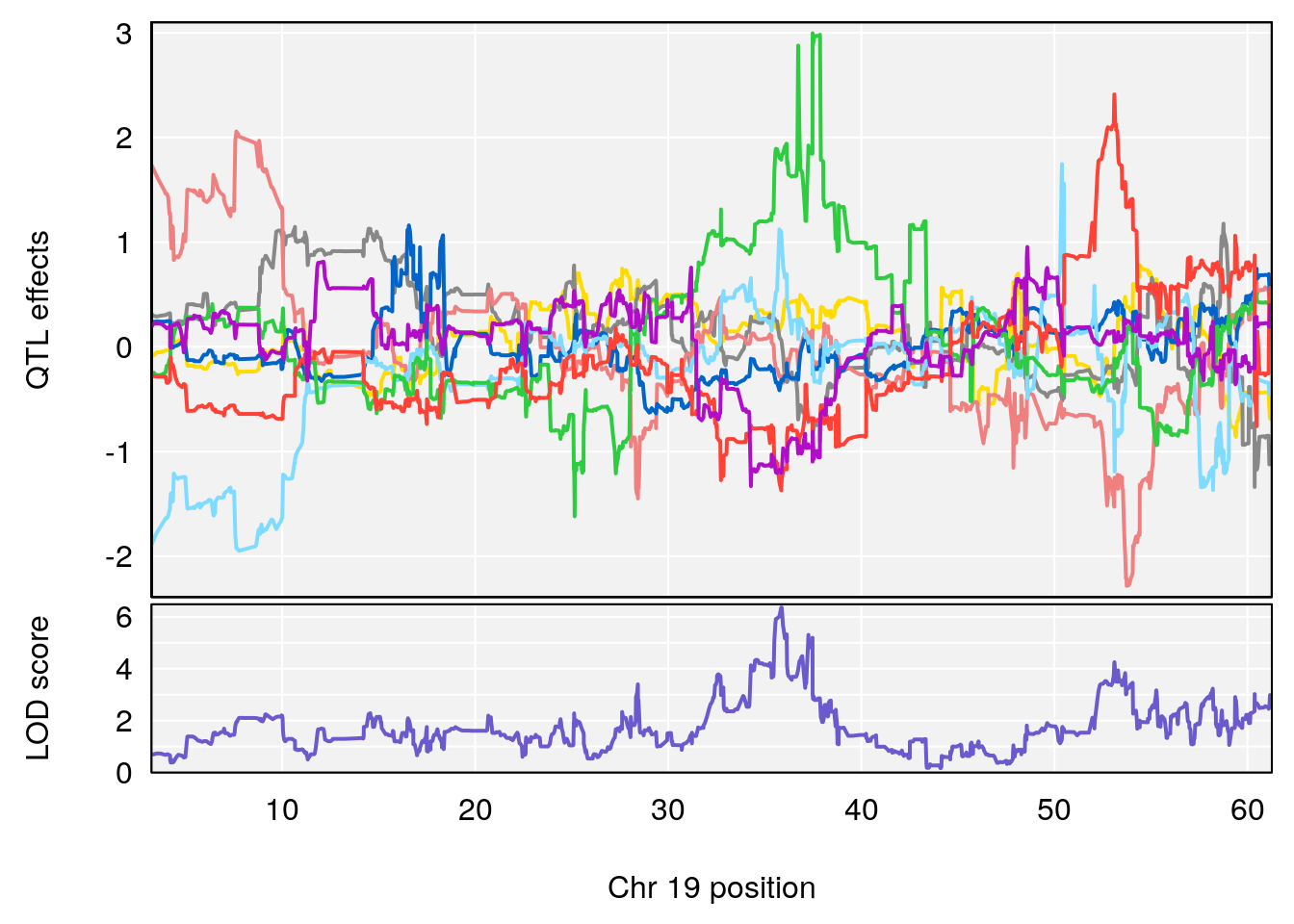
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
# [1] "Time.to.Recovery.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
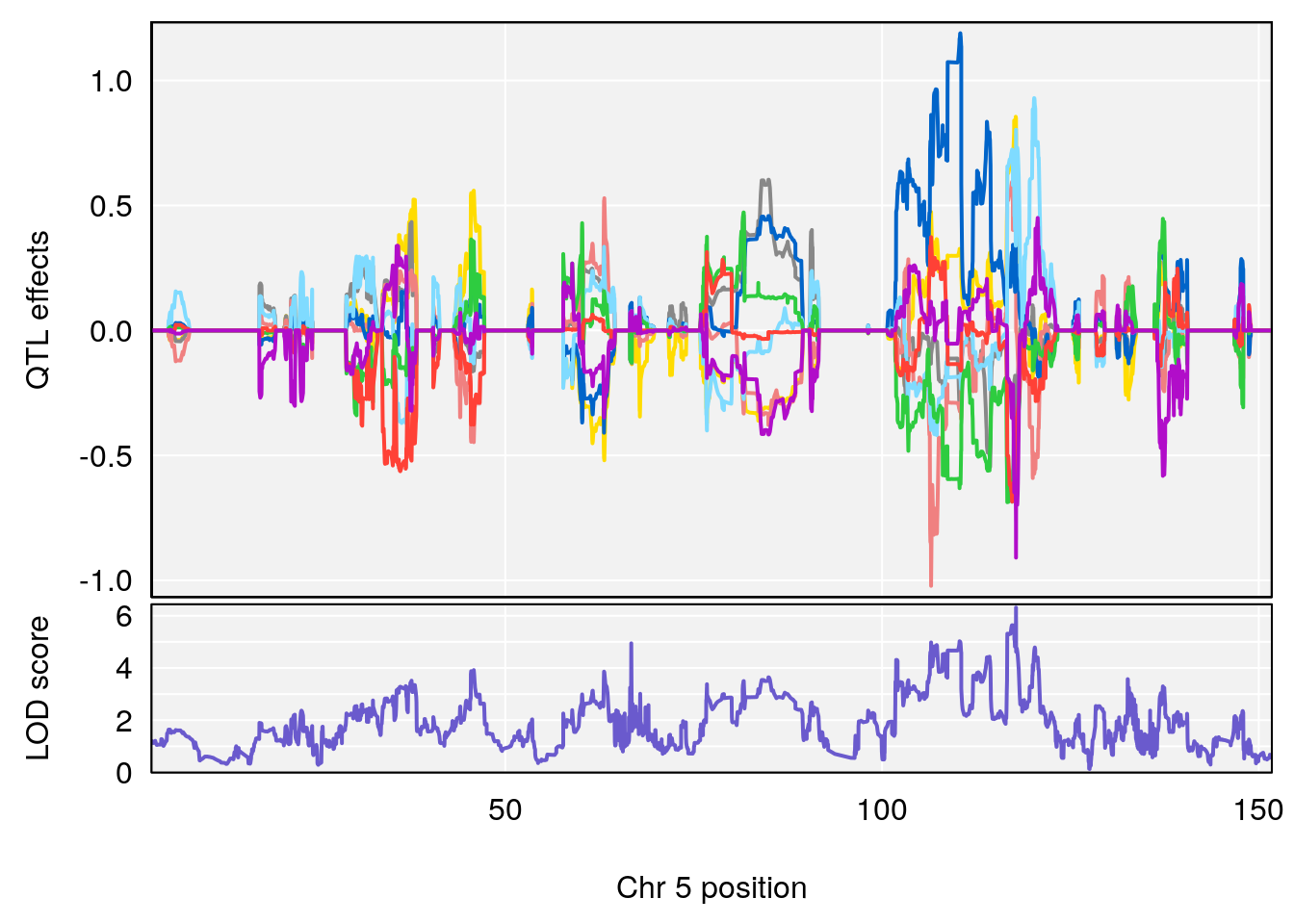
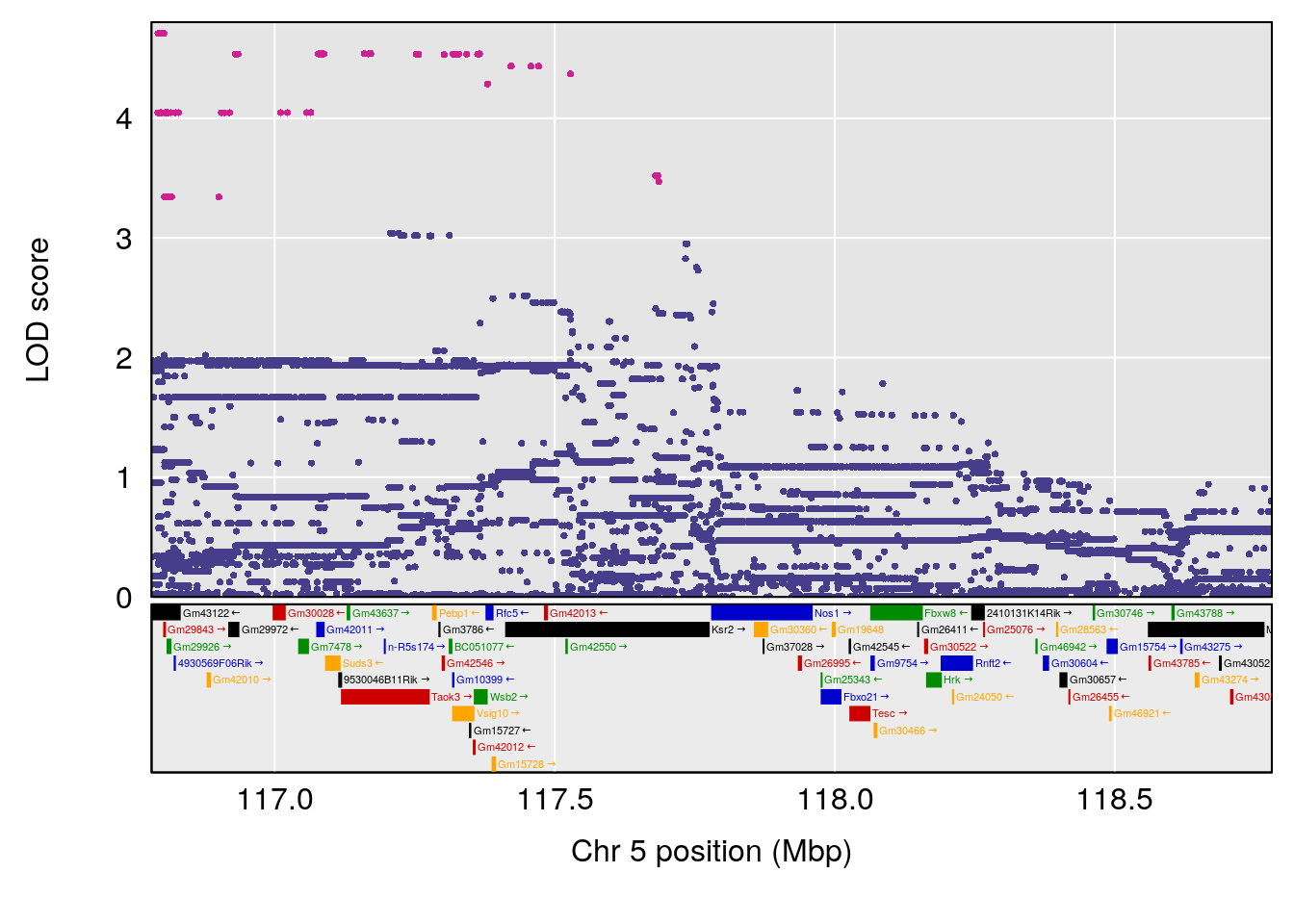
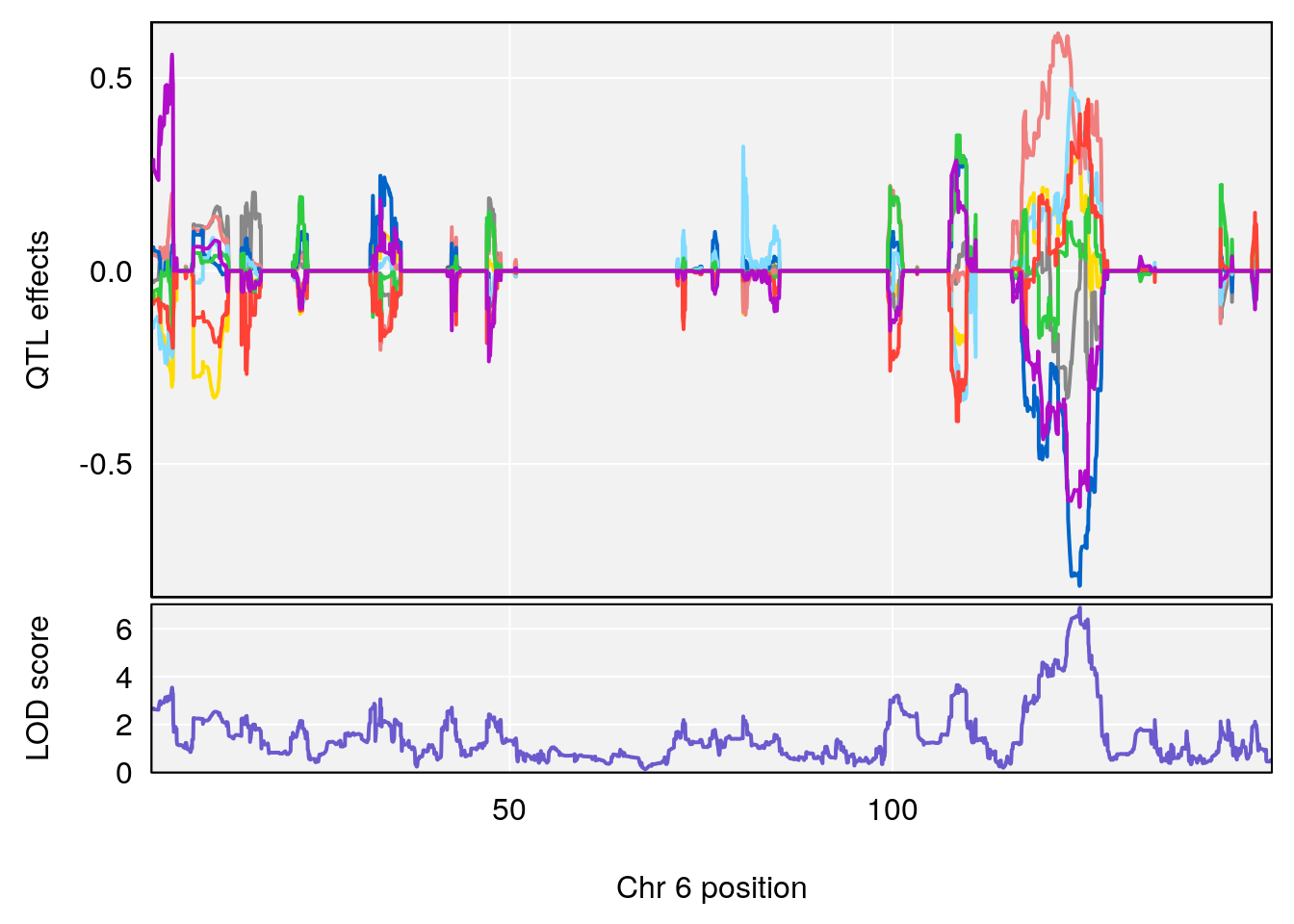
# 1 1 pheno1 5 117.7803 6.310941 66.67504 117.79601
# 2 1 pheno1 6 53.5762 6.096846 53.51503 53.91867
# [1] 1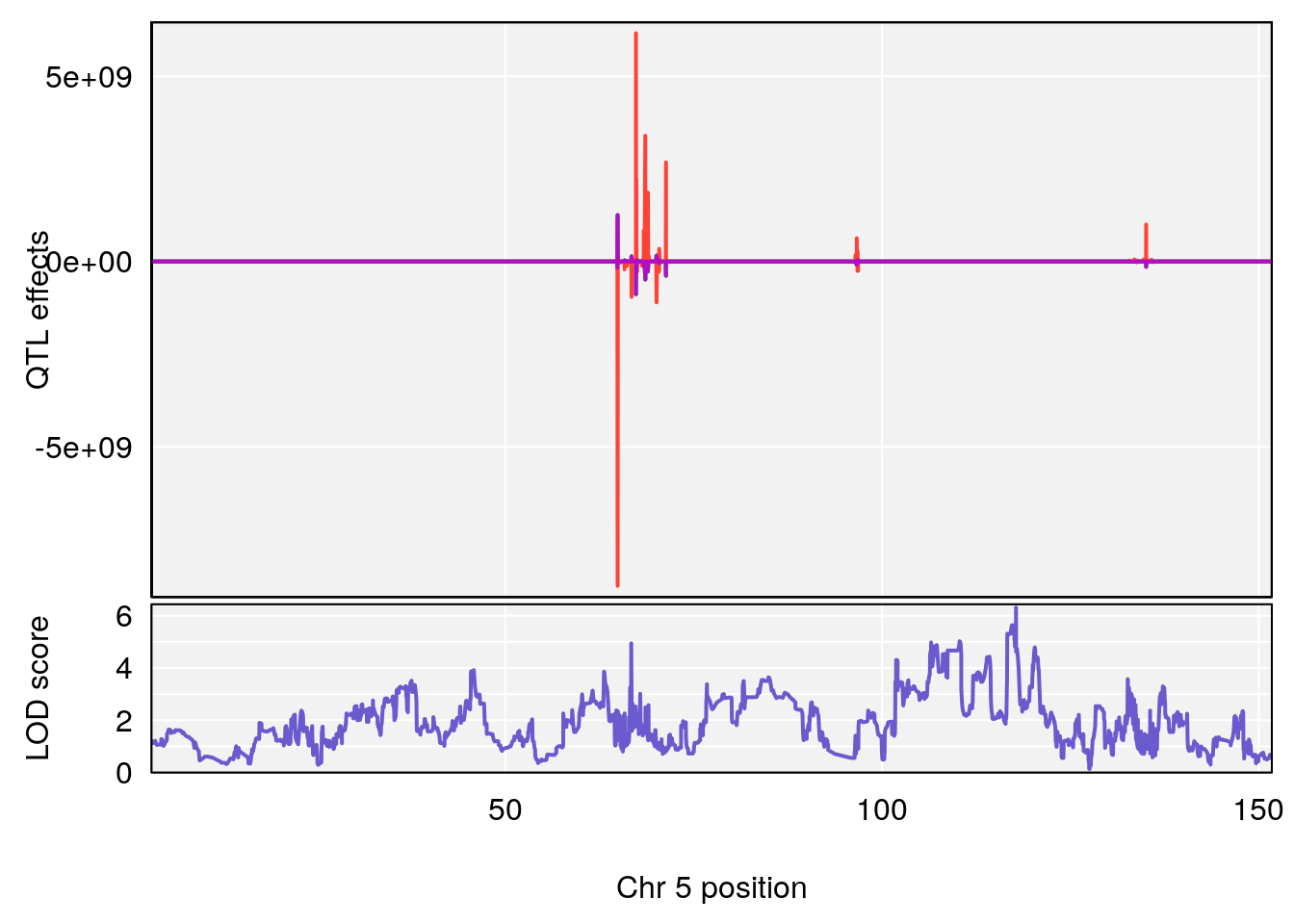
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
# [1] 2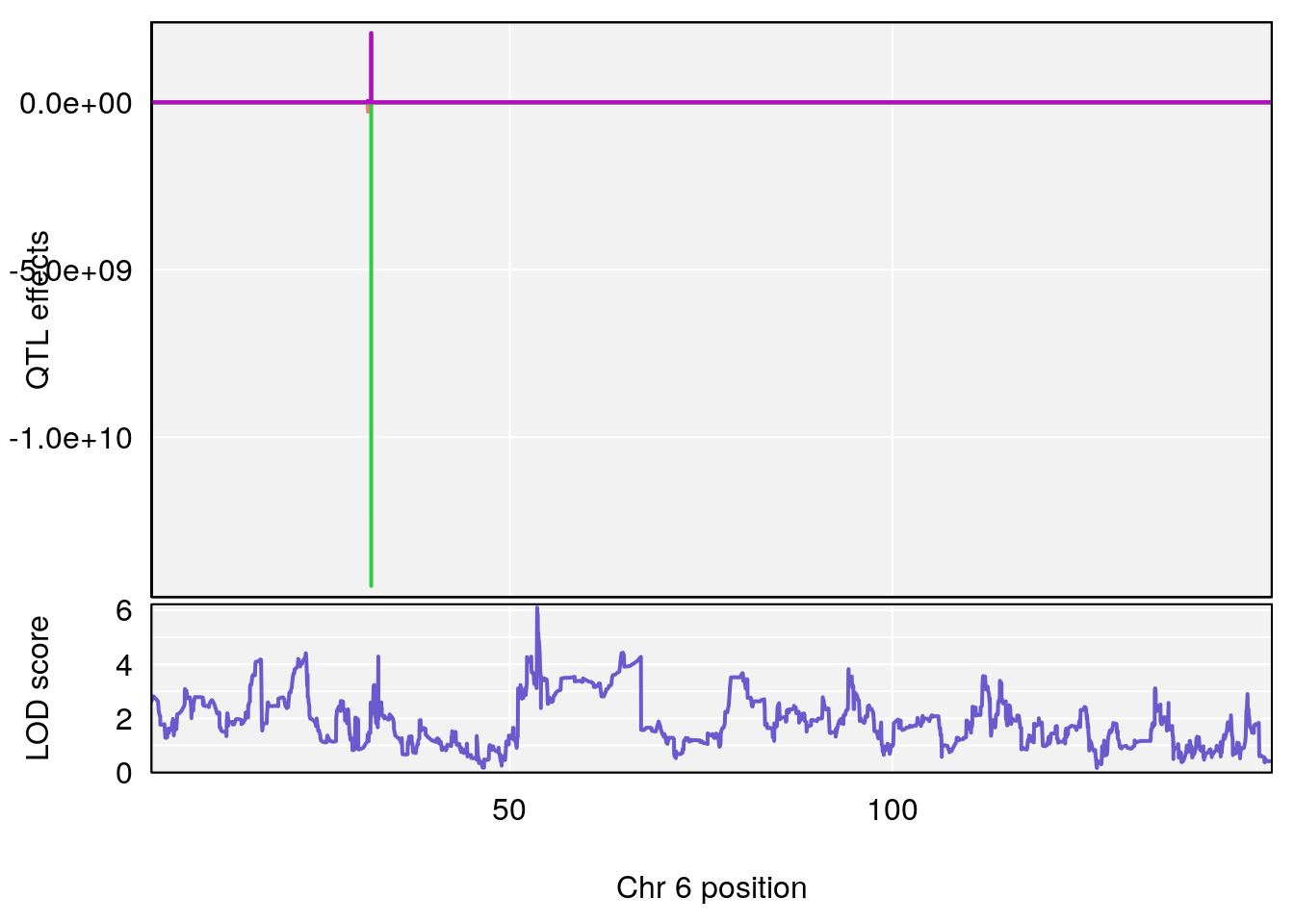
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
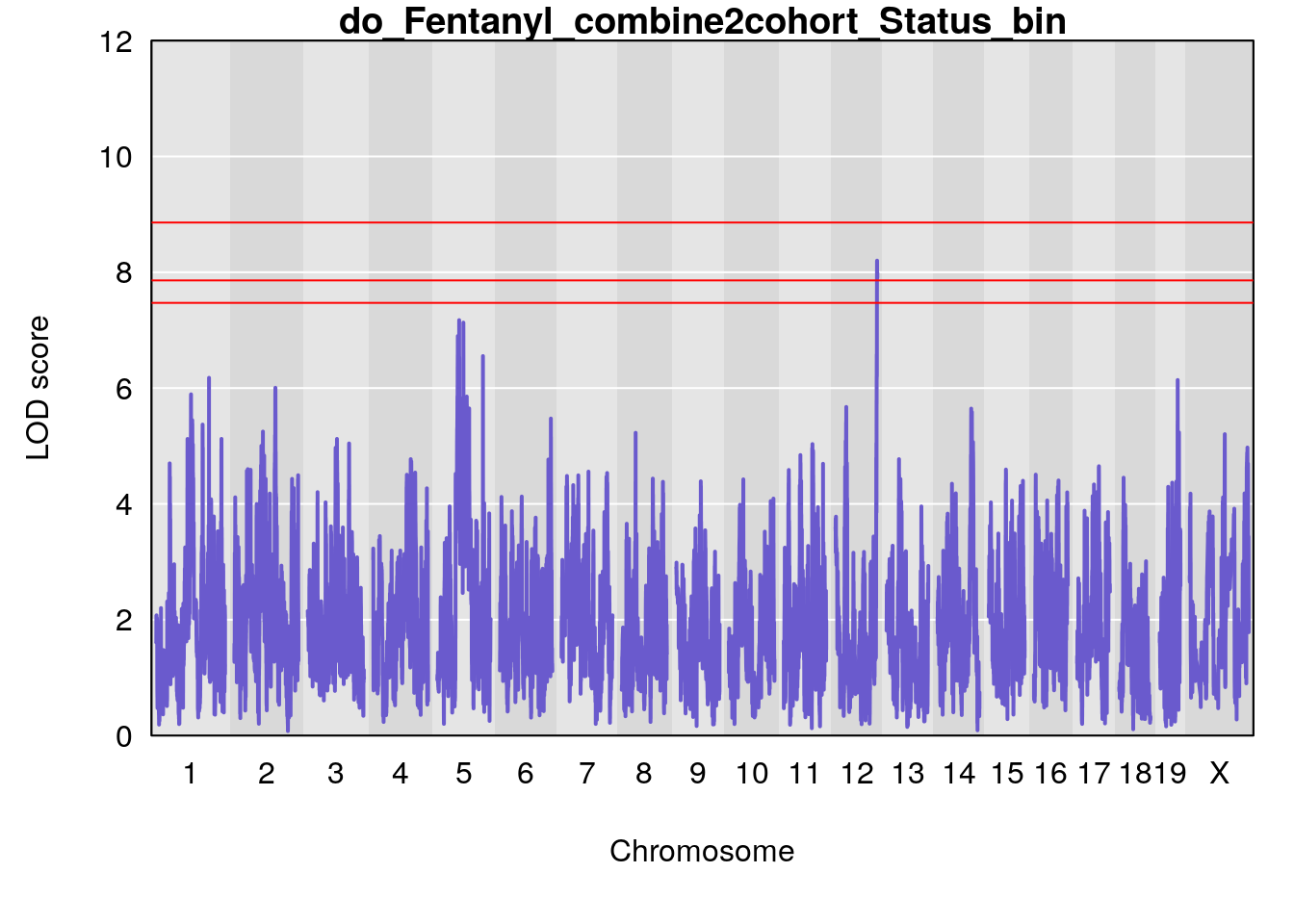
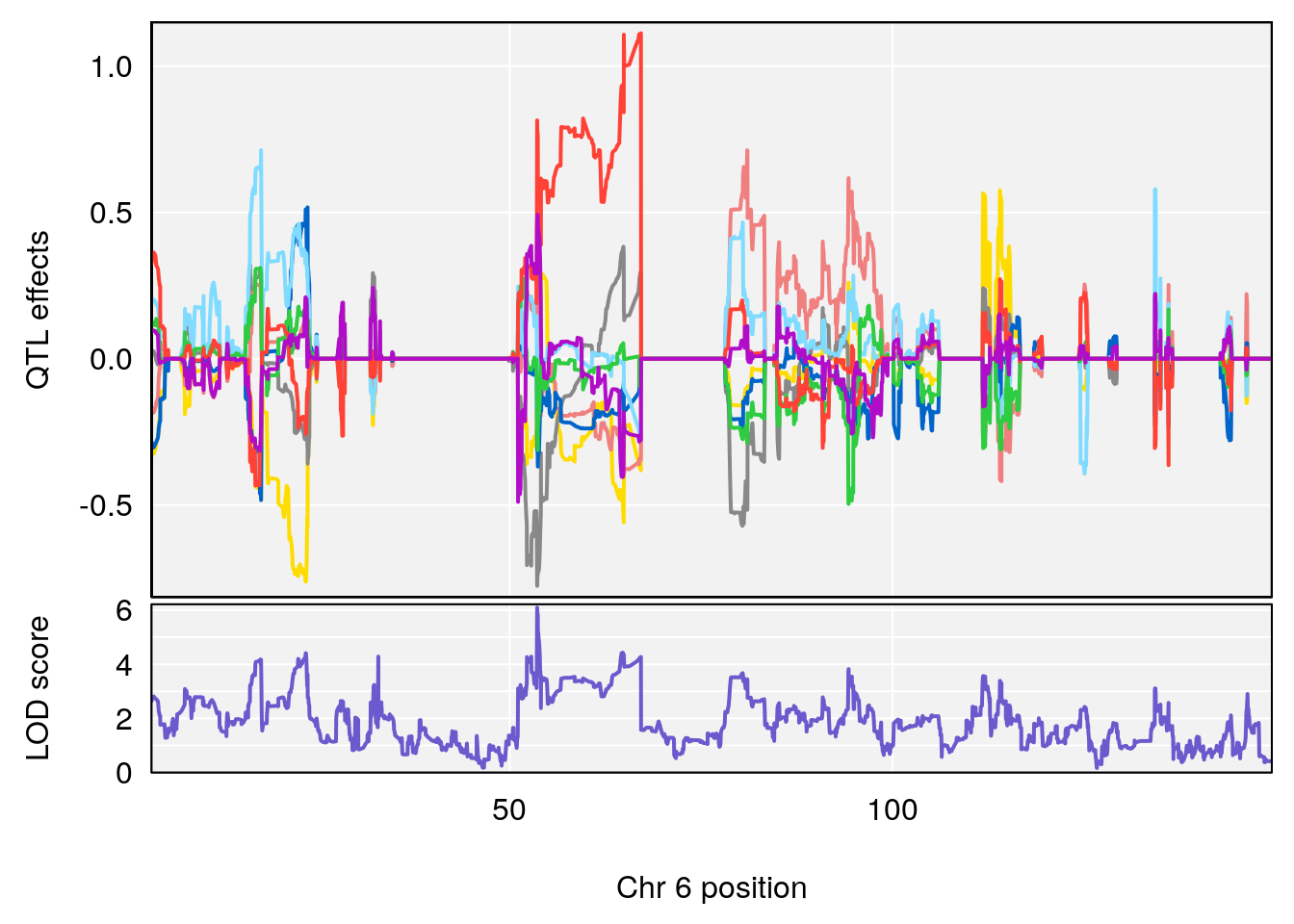
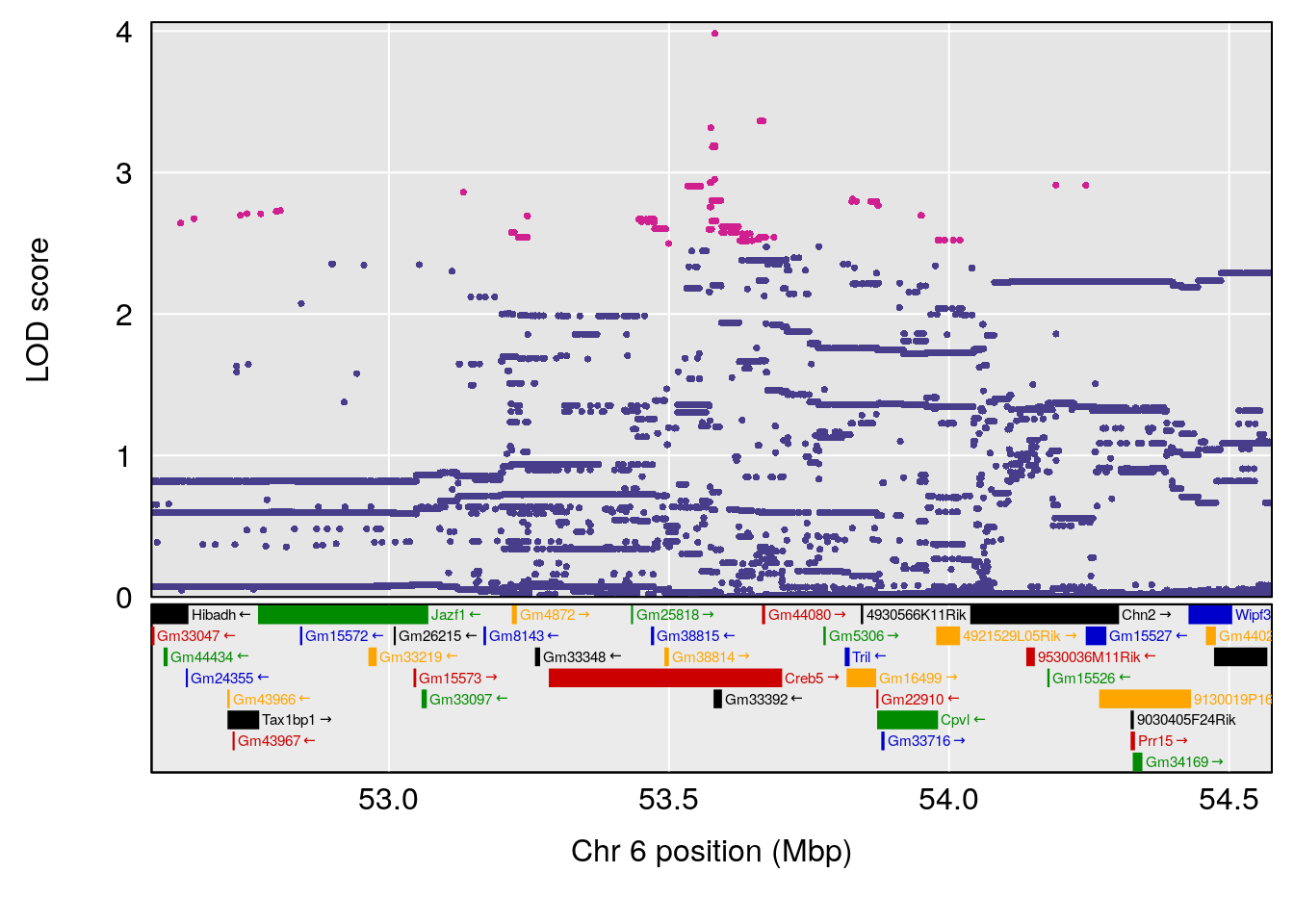
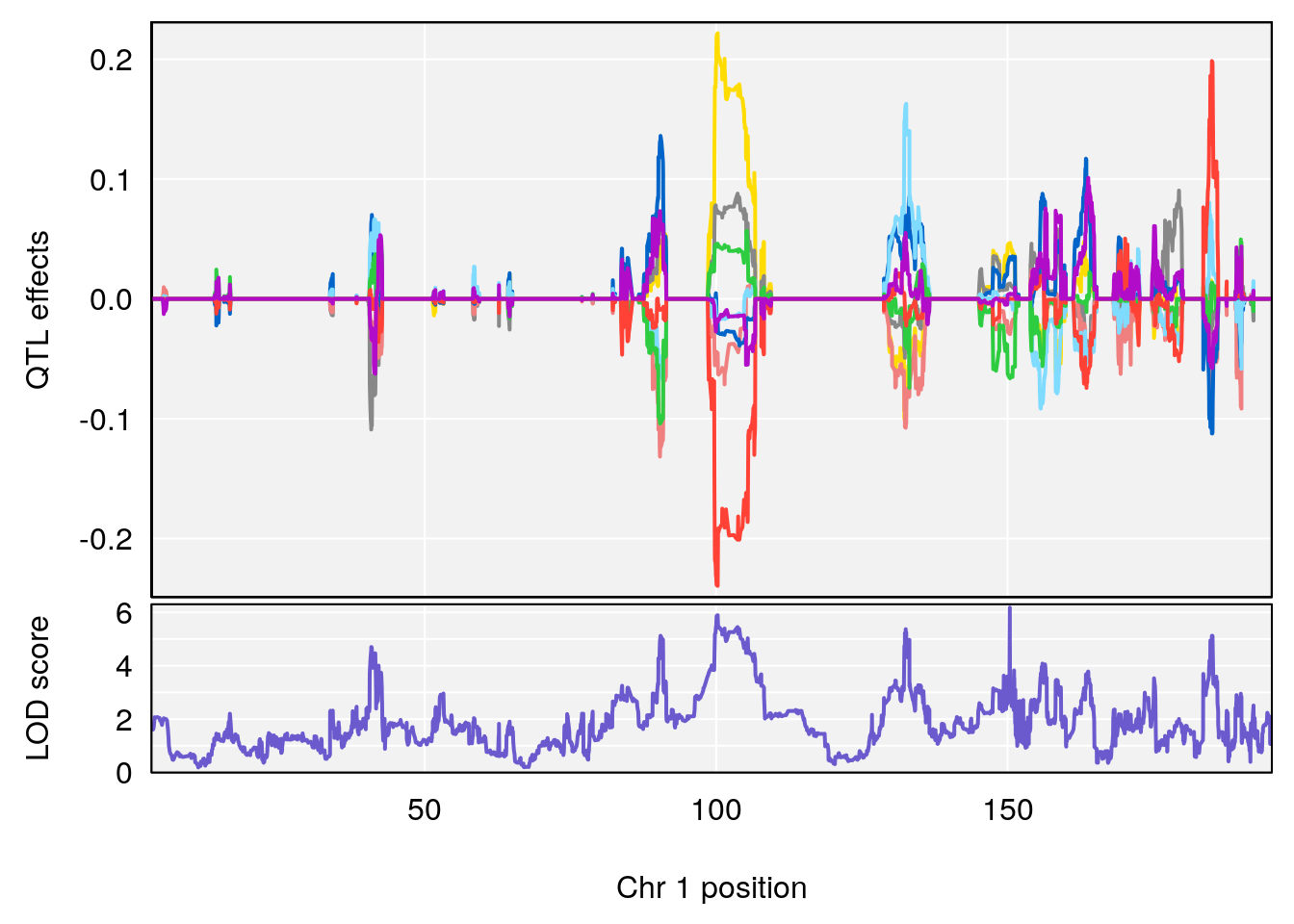
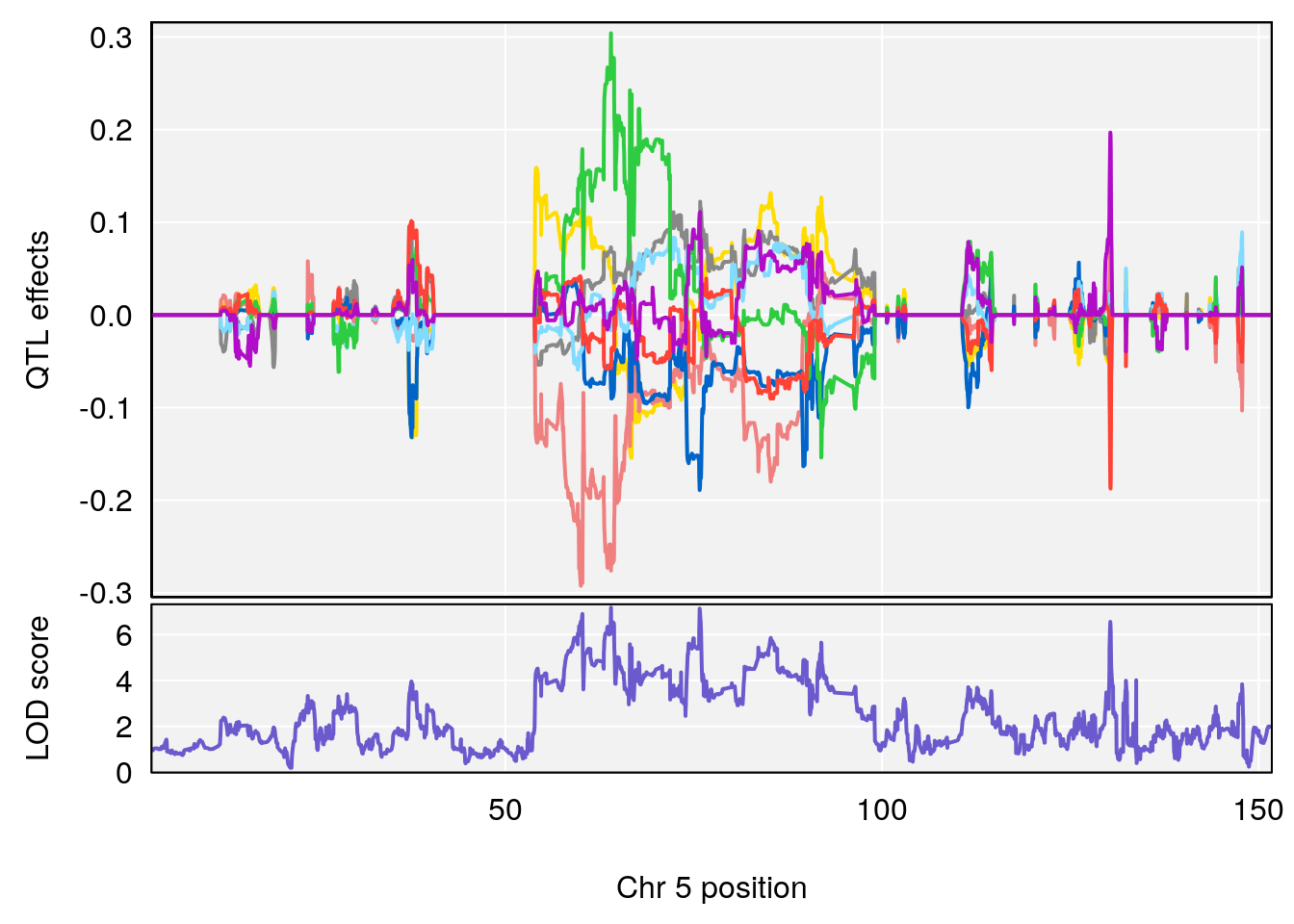
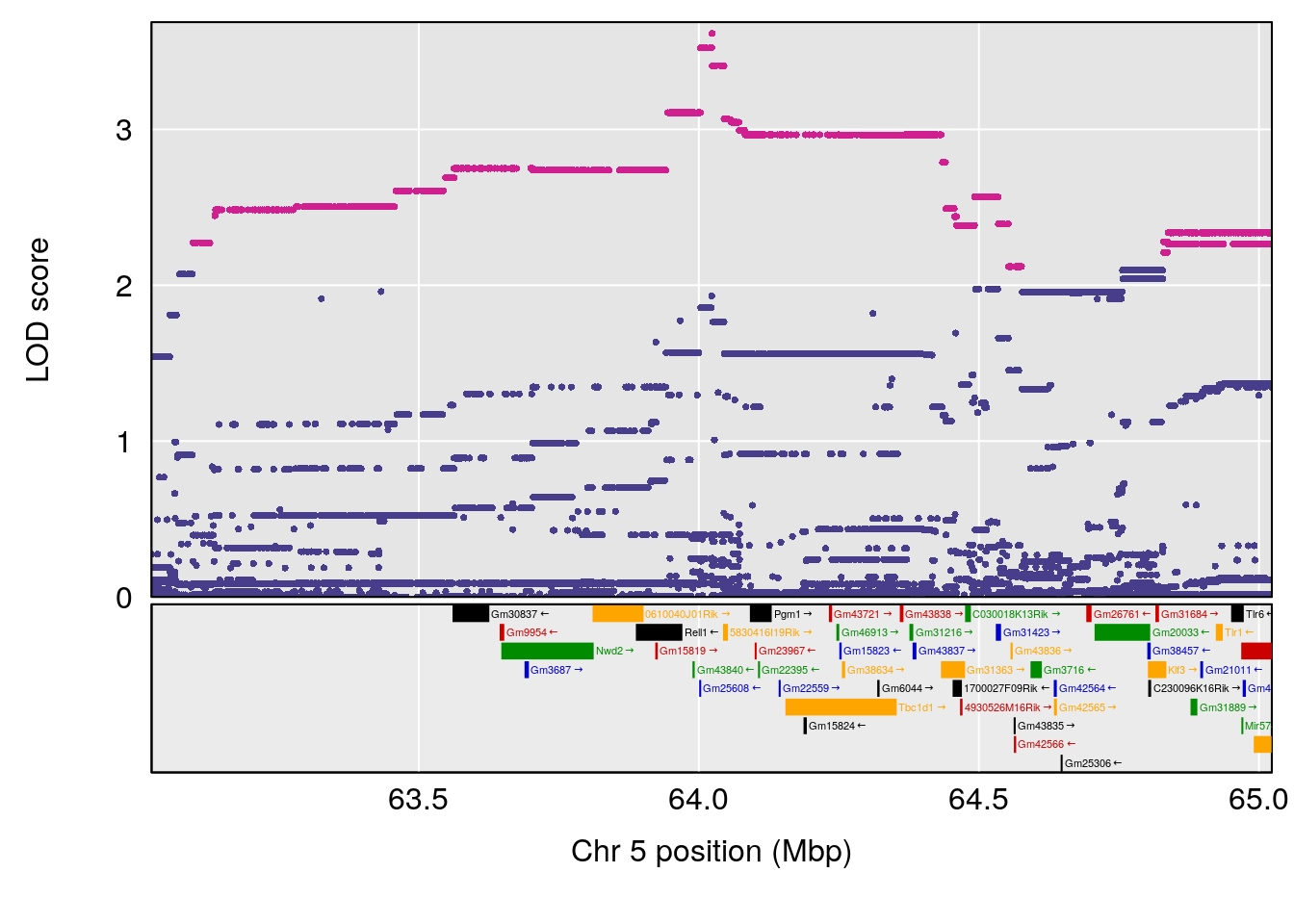
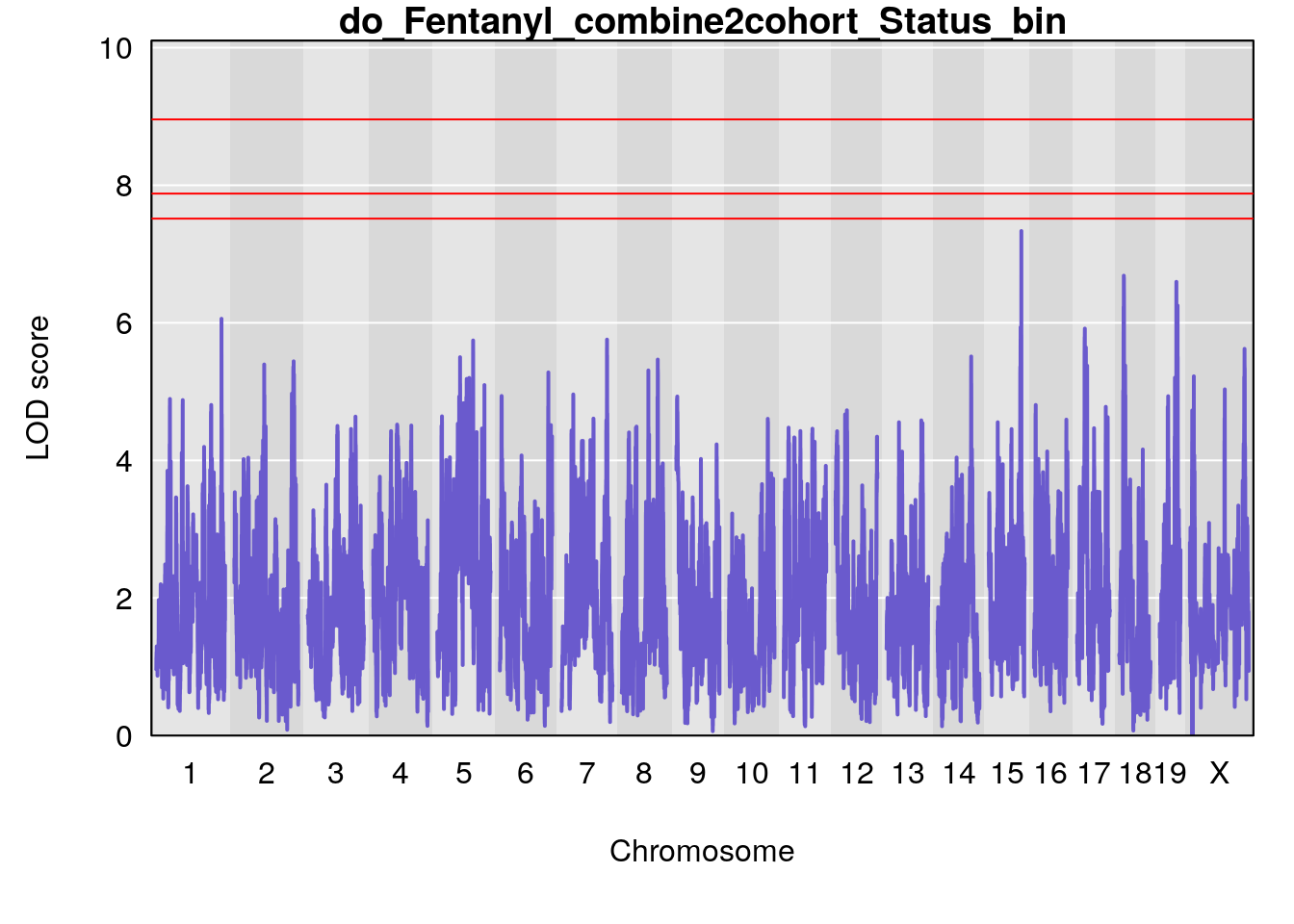
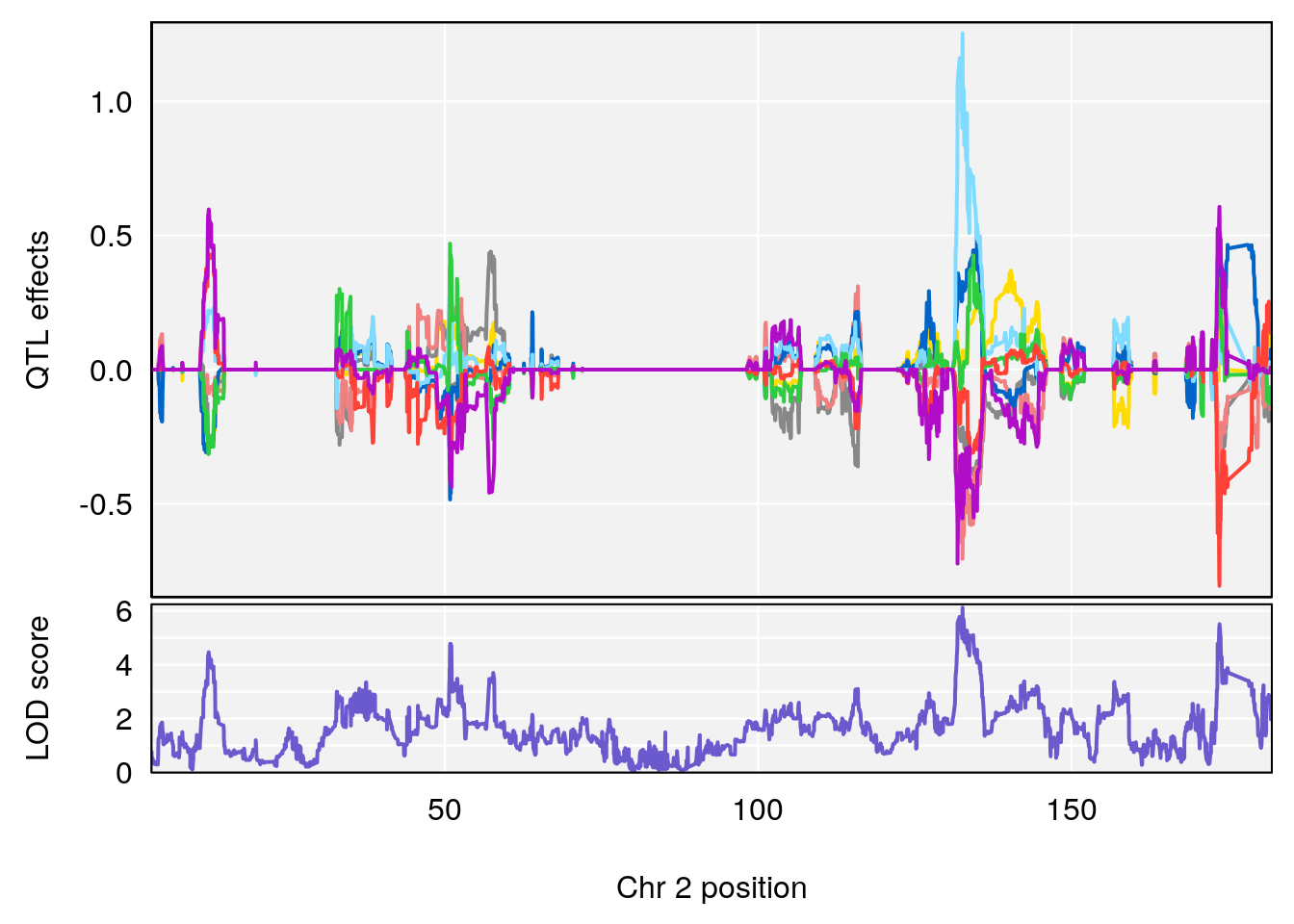
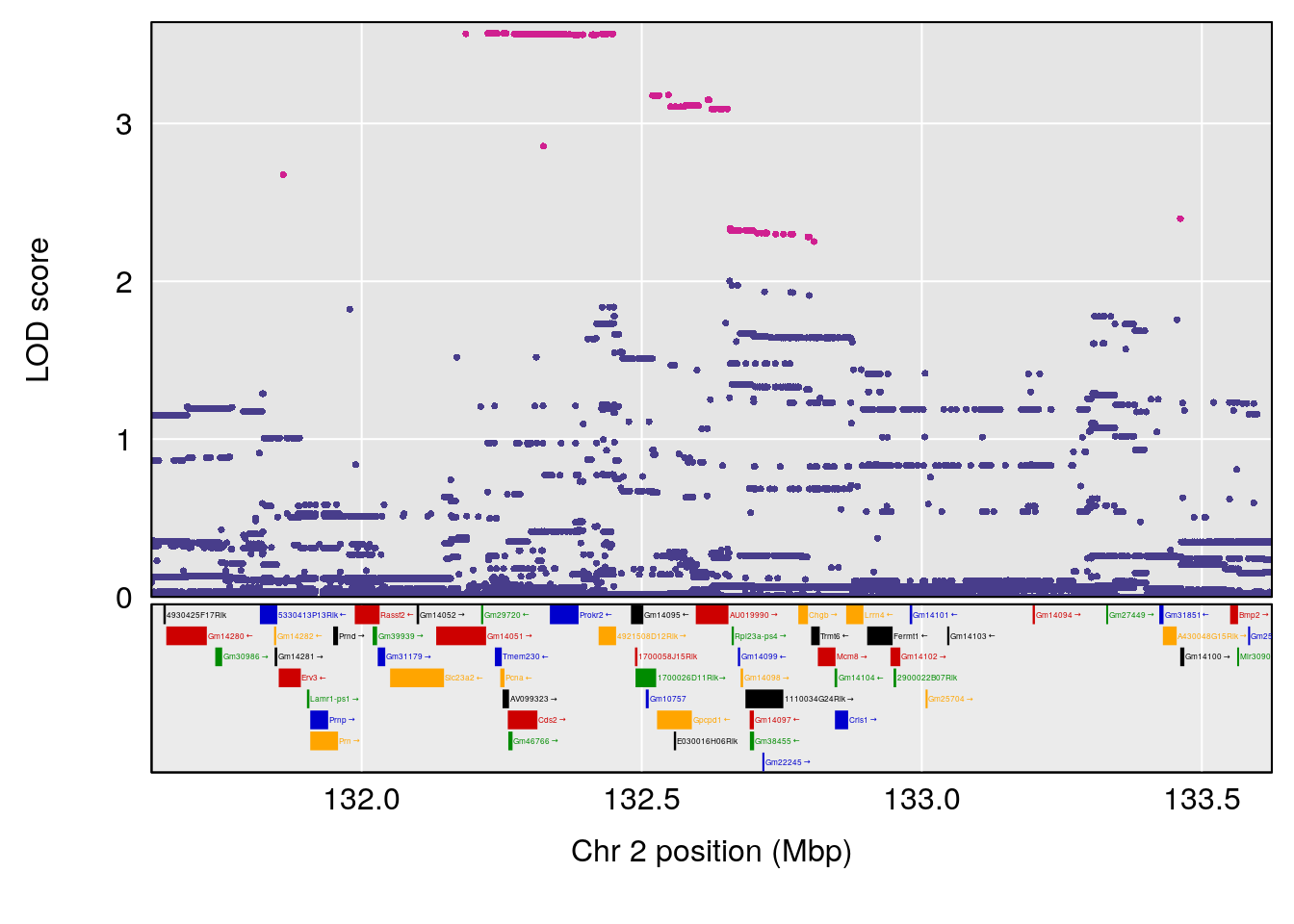
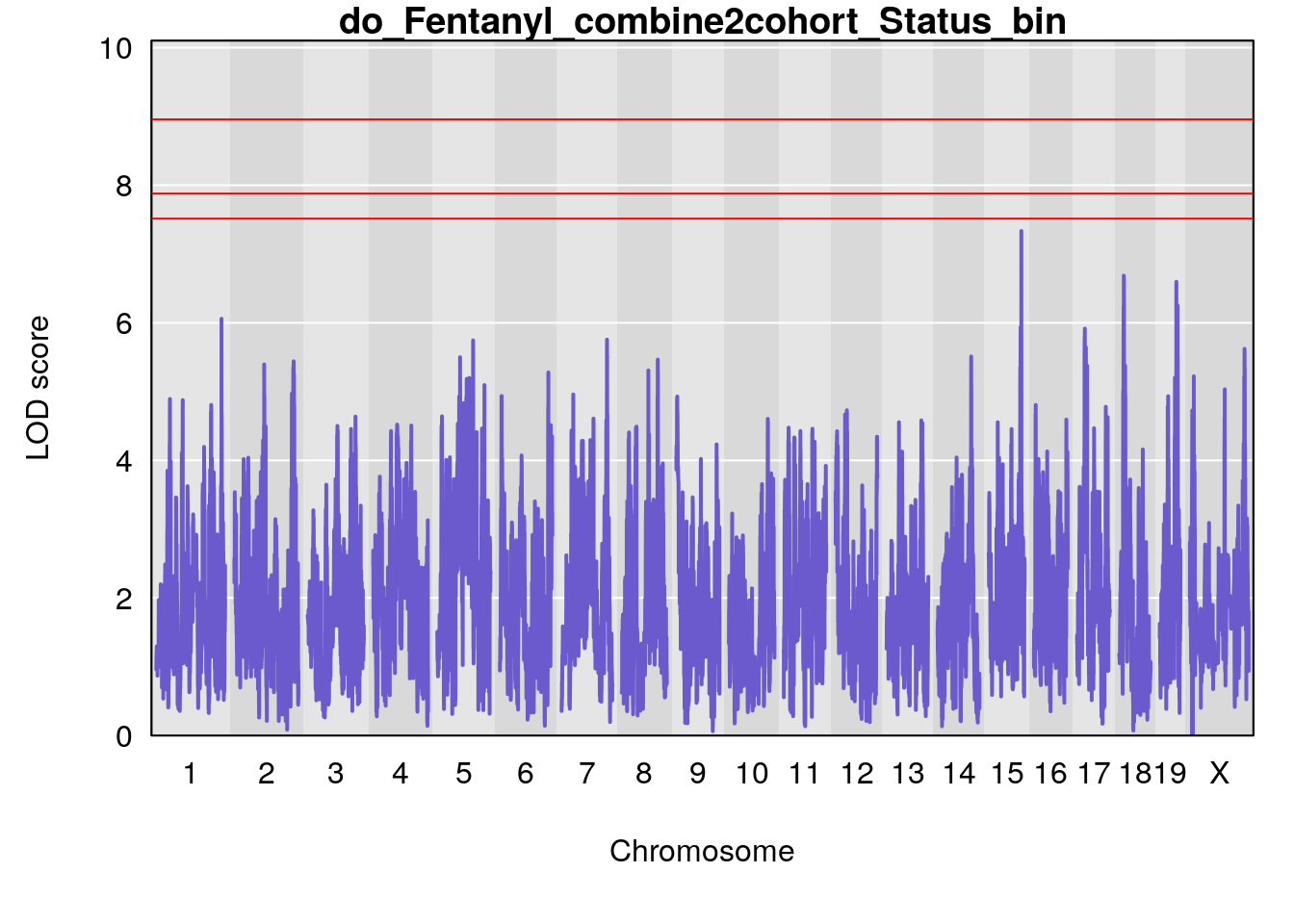
# [1] "Status_bin"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 150.38336 6.177322 40.80906 185.20043
# 2 1 pheno1 2 117.58205 6.005846 37.99636 118.56768
# 3 1 pheno1 5 64.02294 7.173261 59.01797 130.37092
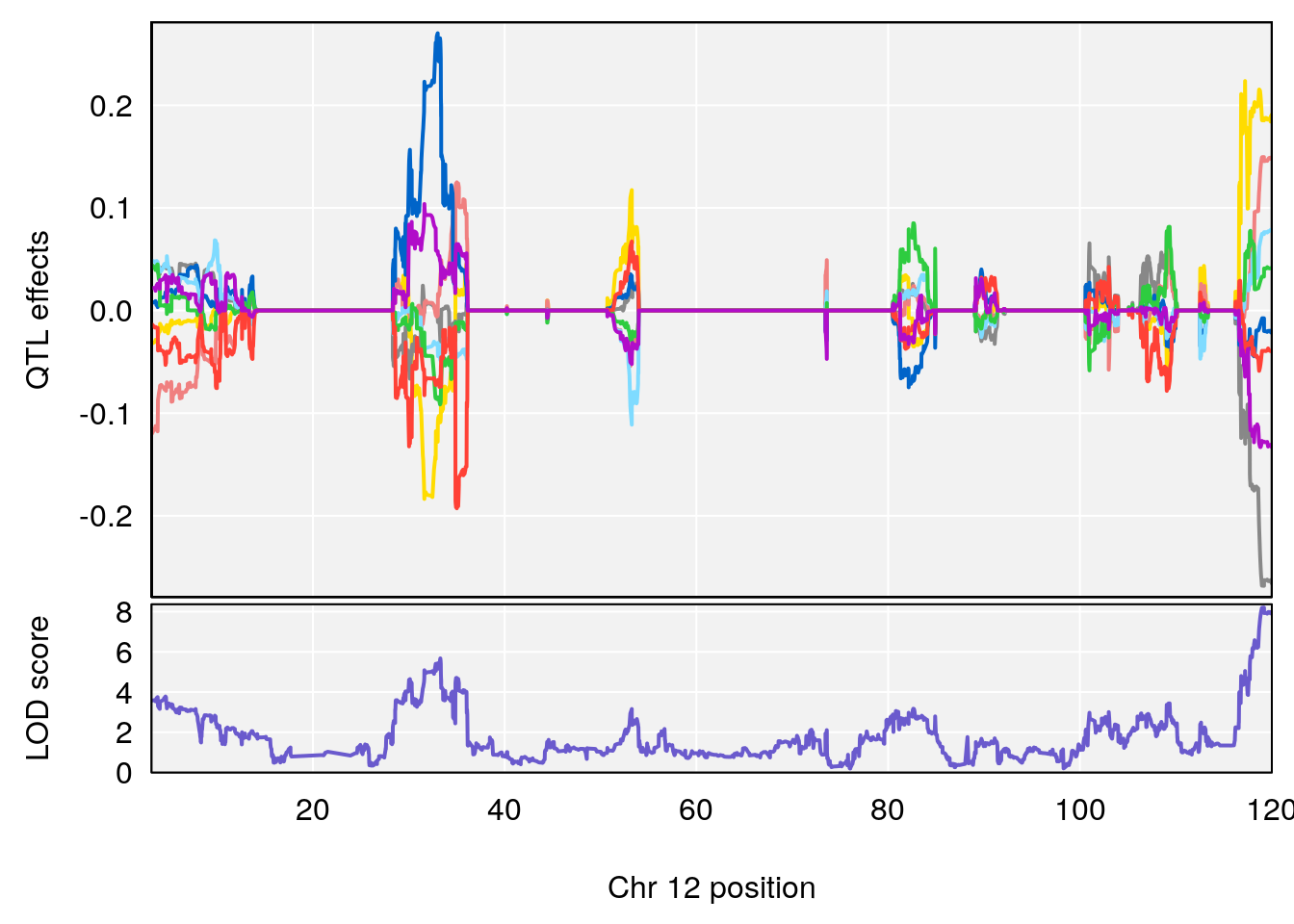
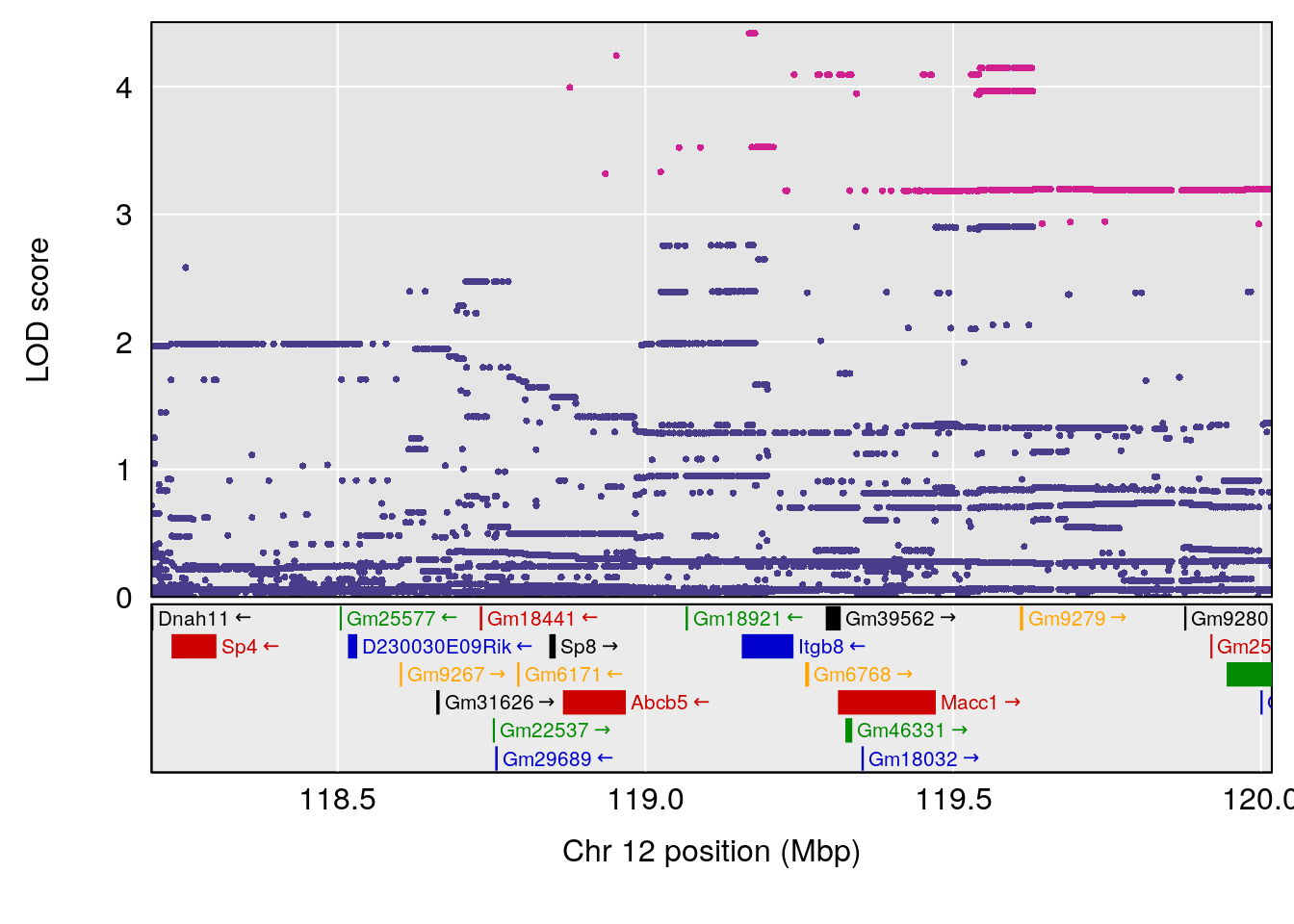
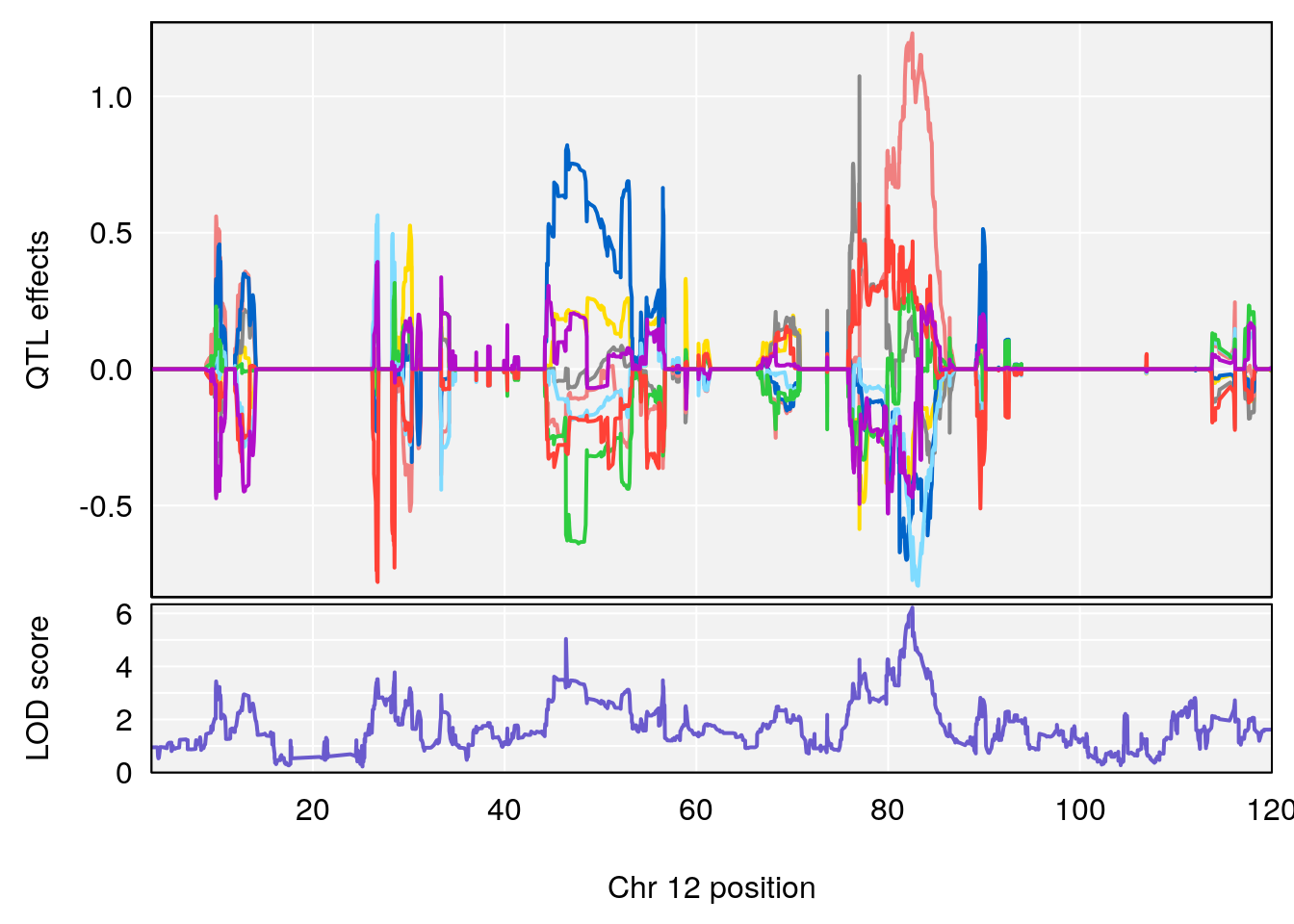
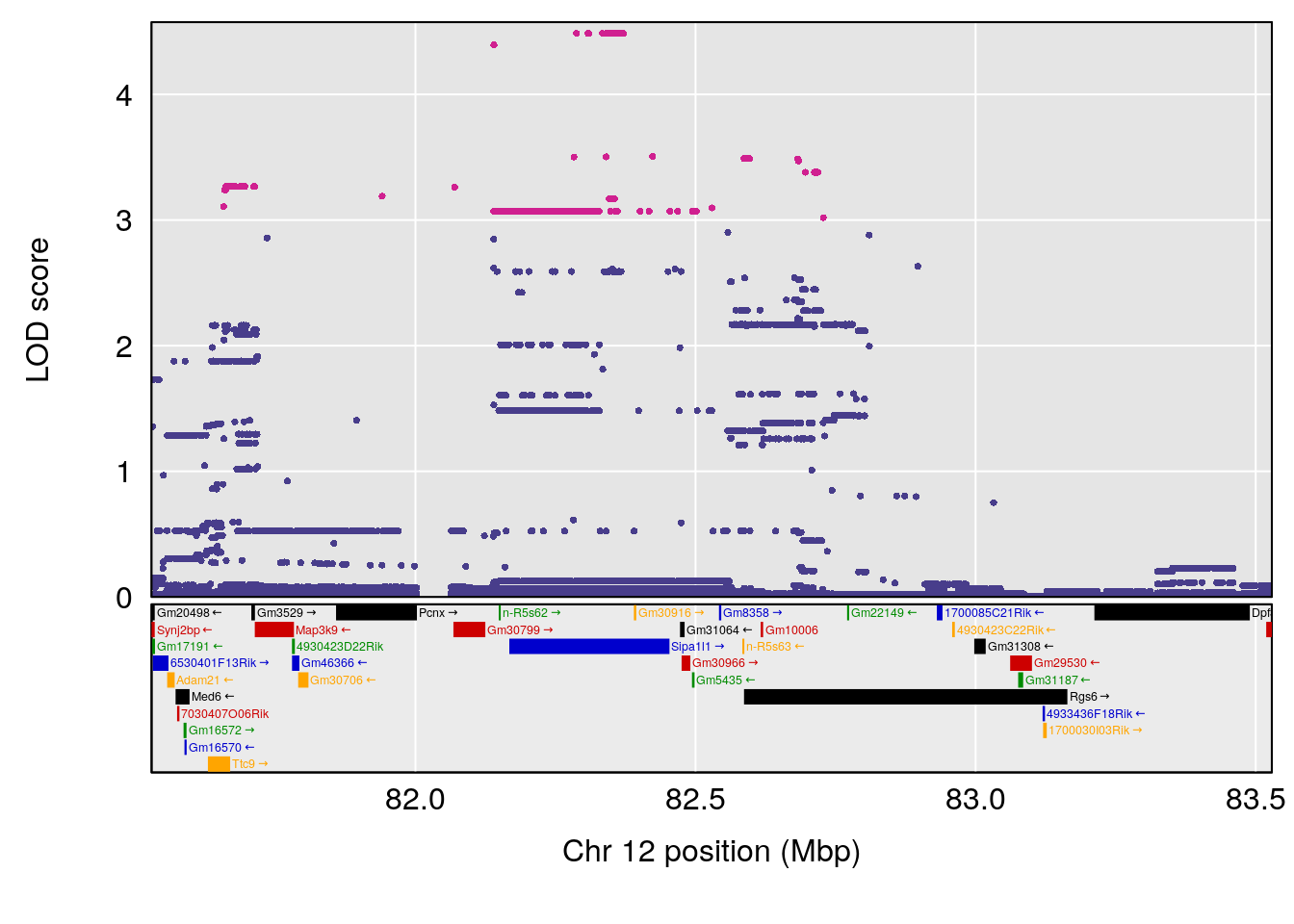
# 4 1 pheno1 12 119.19756 8.201177 118.59974 120.01743
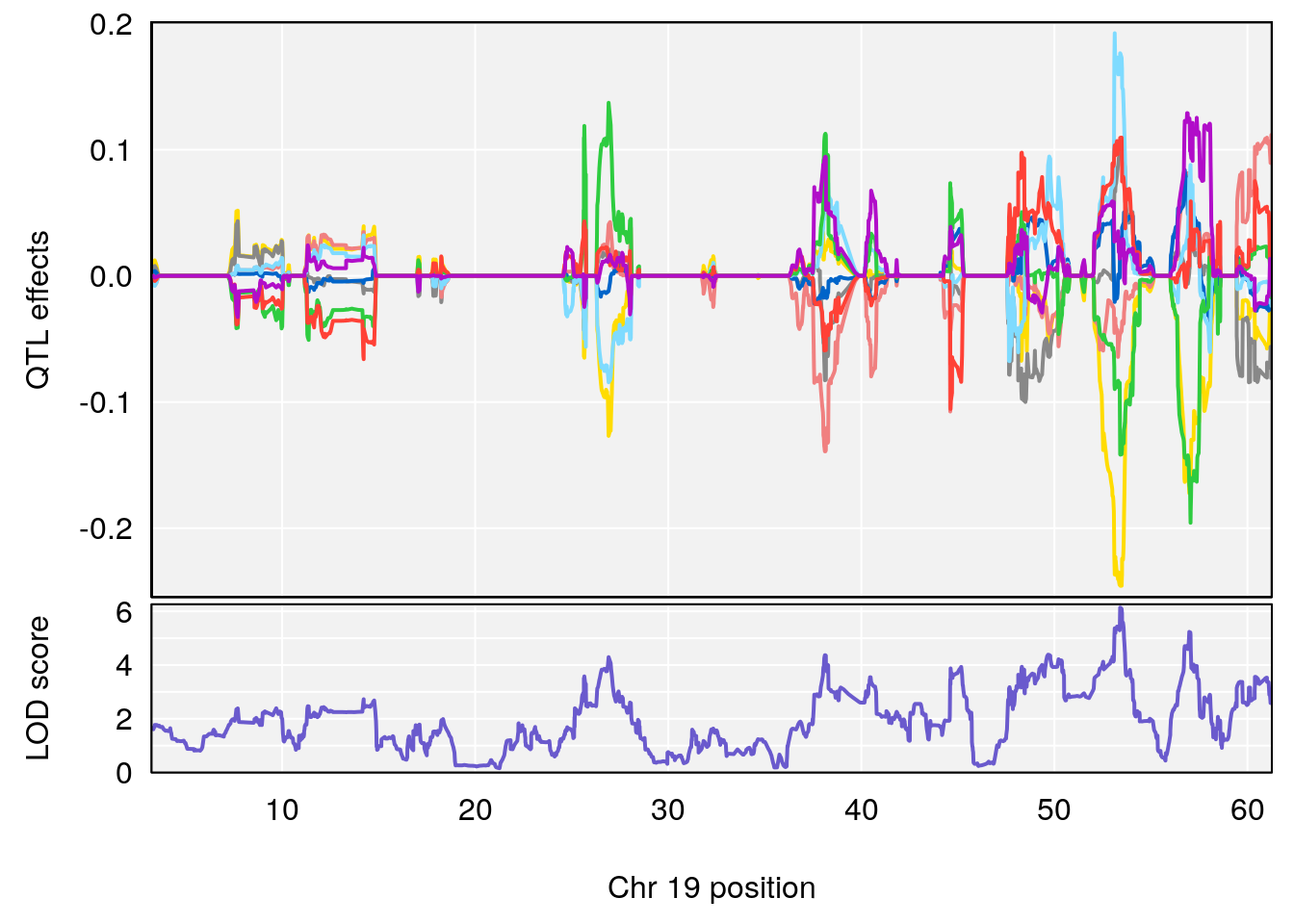
# 5 1 pheno1 19 53.40556 6.139452 53.08070 57.06435
# [1] 1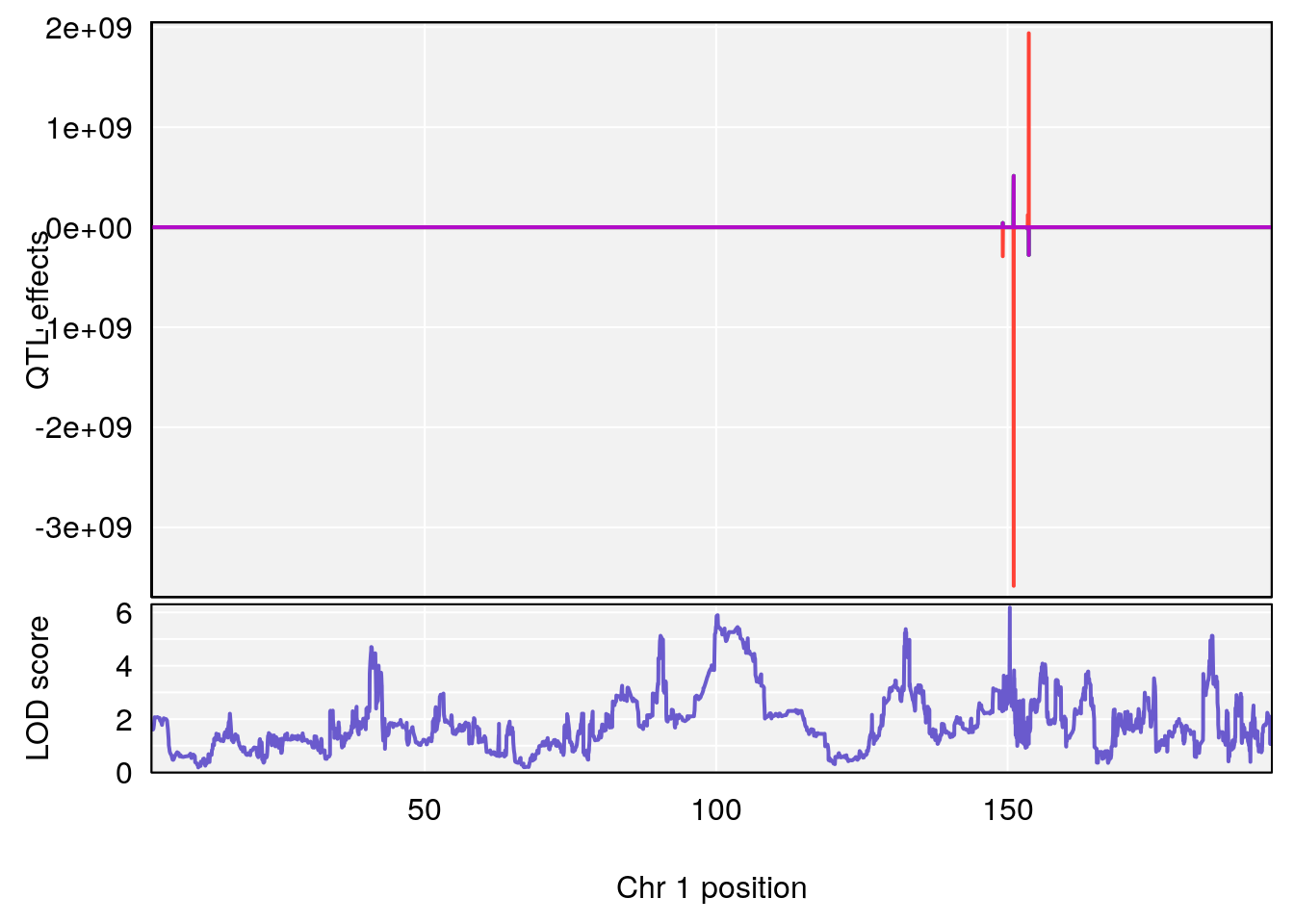
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
# [1] 2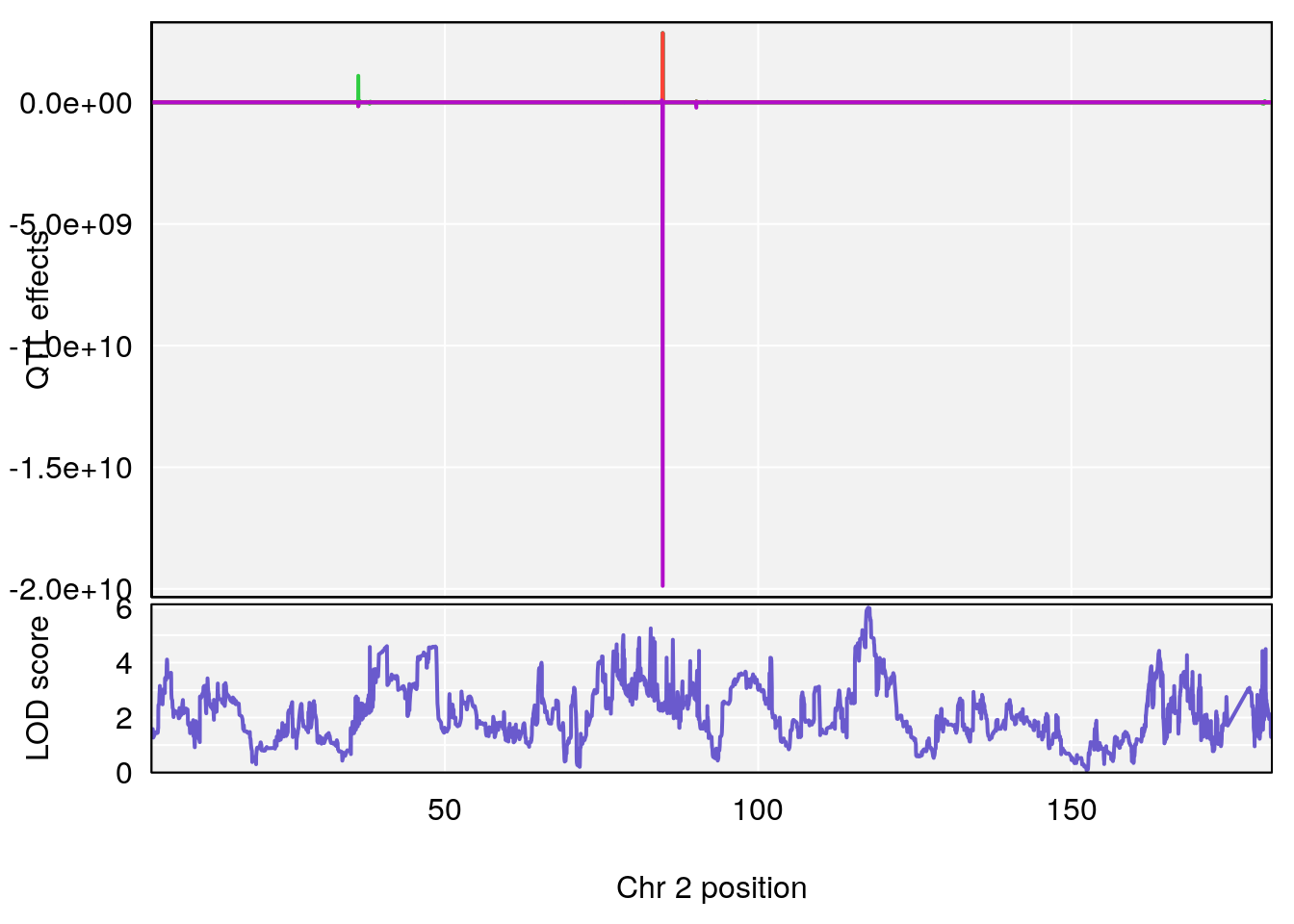
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
# [1] 3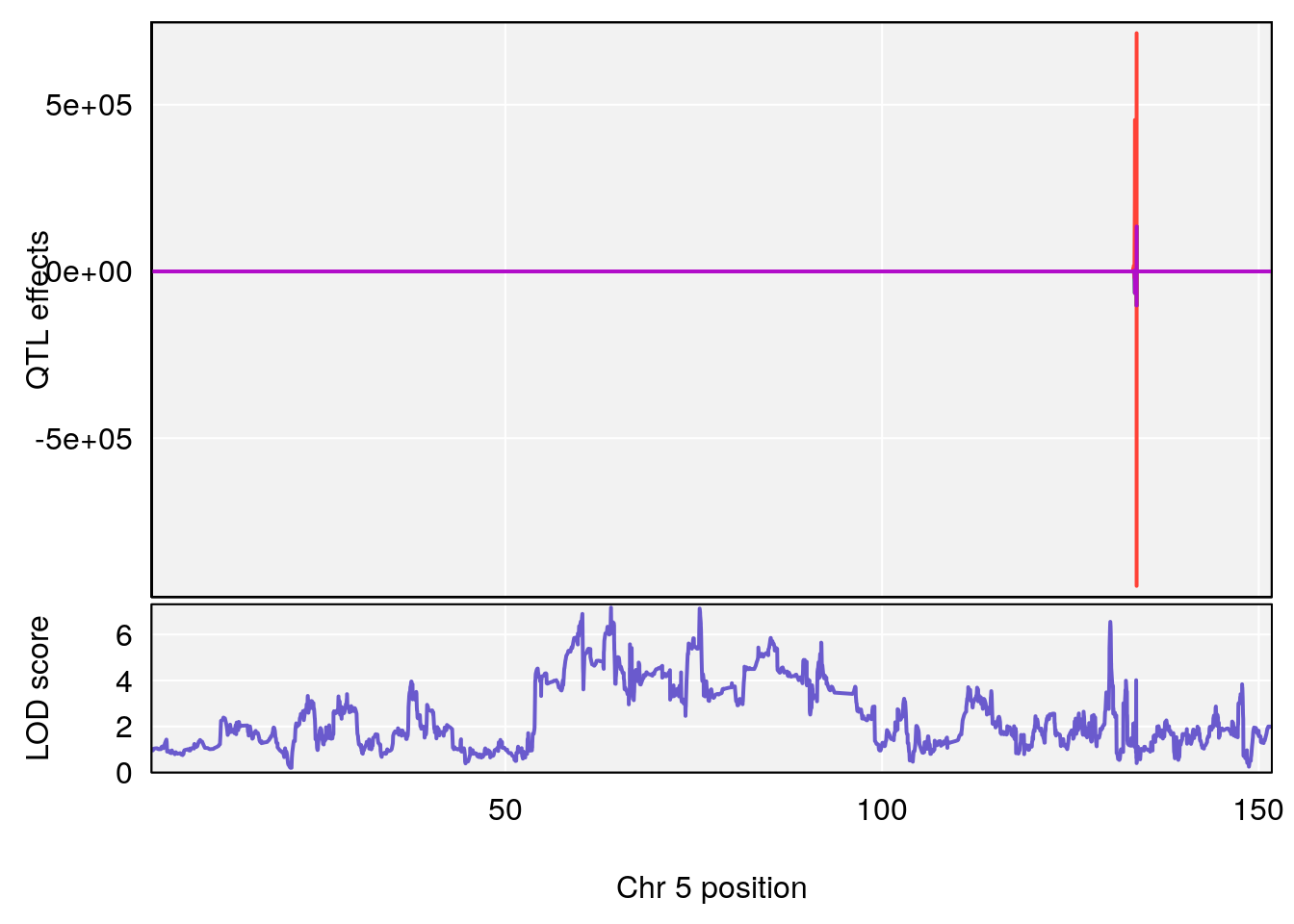
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
# [1] 4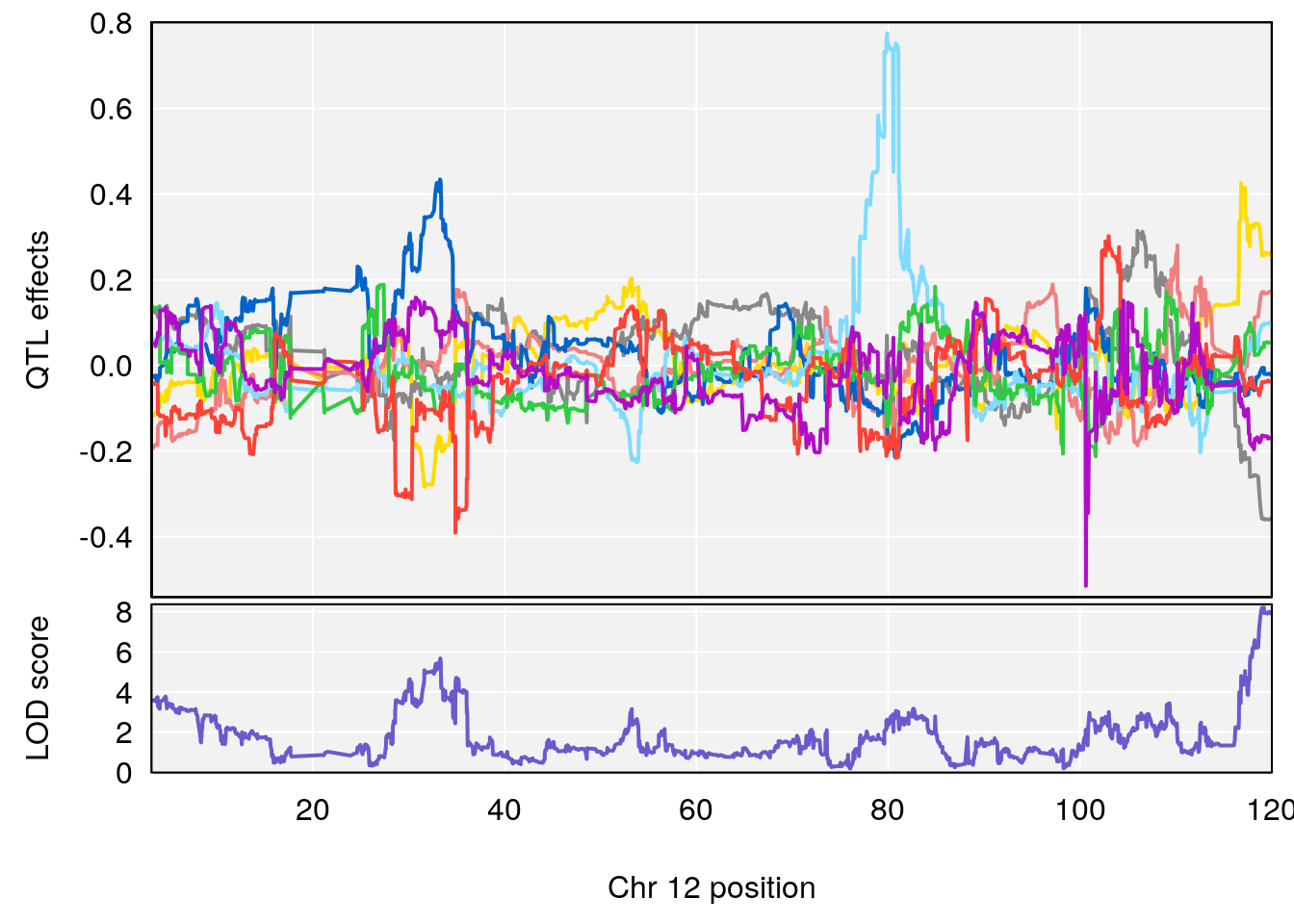
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
# [1] 5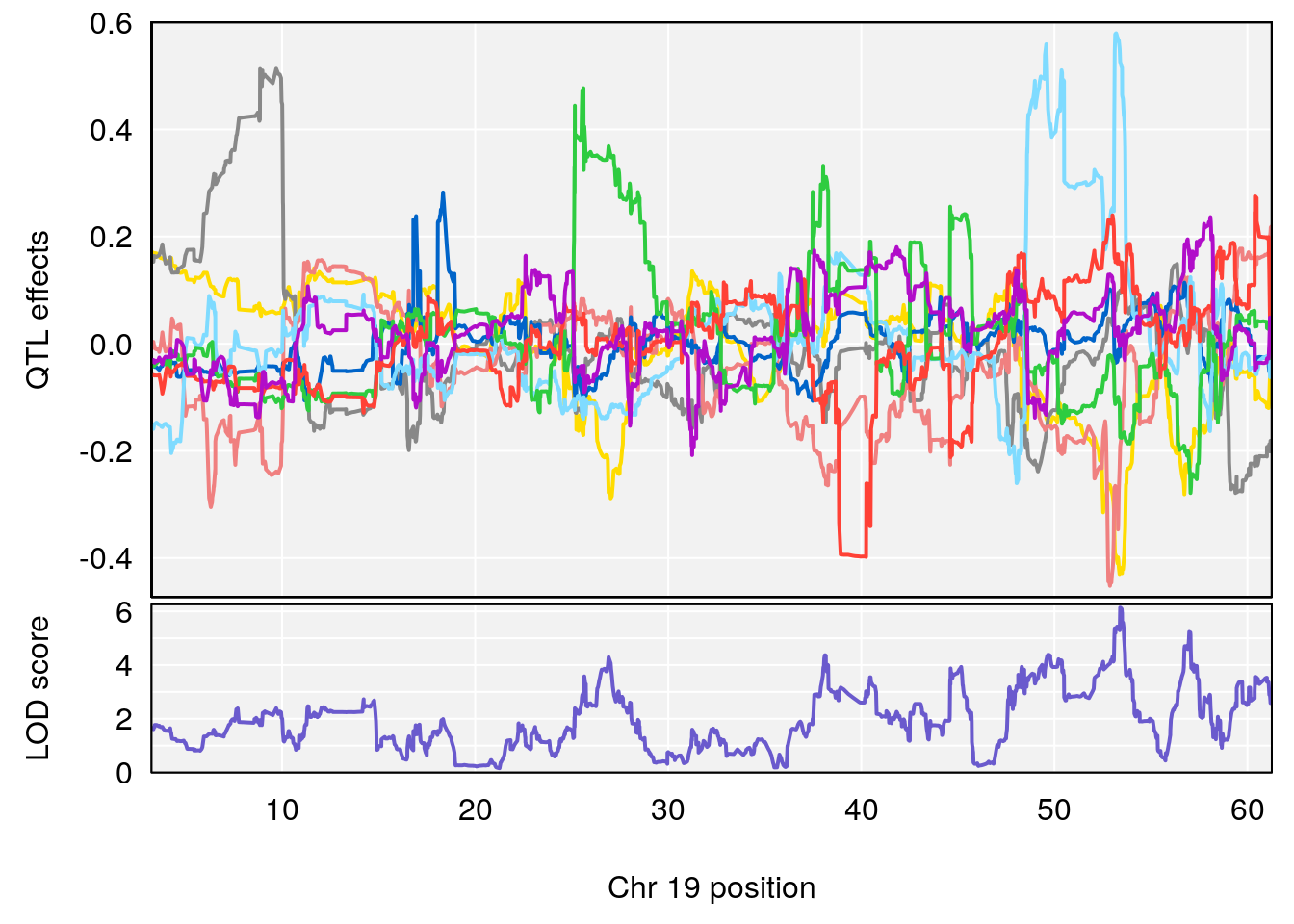
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
| Version | Author | Date |
|---|---|---|
| 0971ef2 | xhyuo | 2022-07-05 |
#save peaks coeff plot
for(i in names(qtl.out)){
print(i)
peaks <- find_peaks(qtl.out[[i]], map=pmap, threshold=6, drop=1.5)
if(nrow(peaks) > 0){
fname <- paste("output/do_Fentanyl_combine2cohort_", str_replace_all(i, "[[:punct:]]", "") ,"_coefplot.pdf",sep="")
pdf(file = fname, width = 16, height =8)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.out[[i]], bgcolor="gray95", legend=NULL)
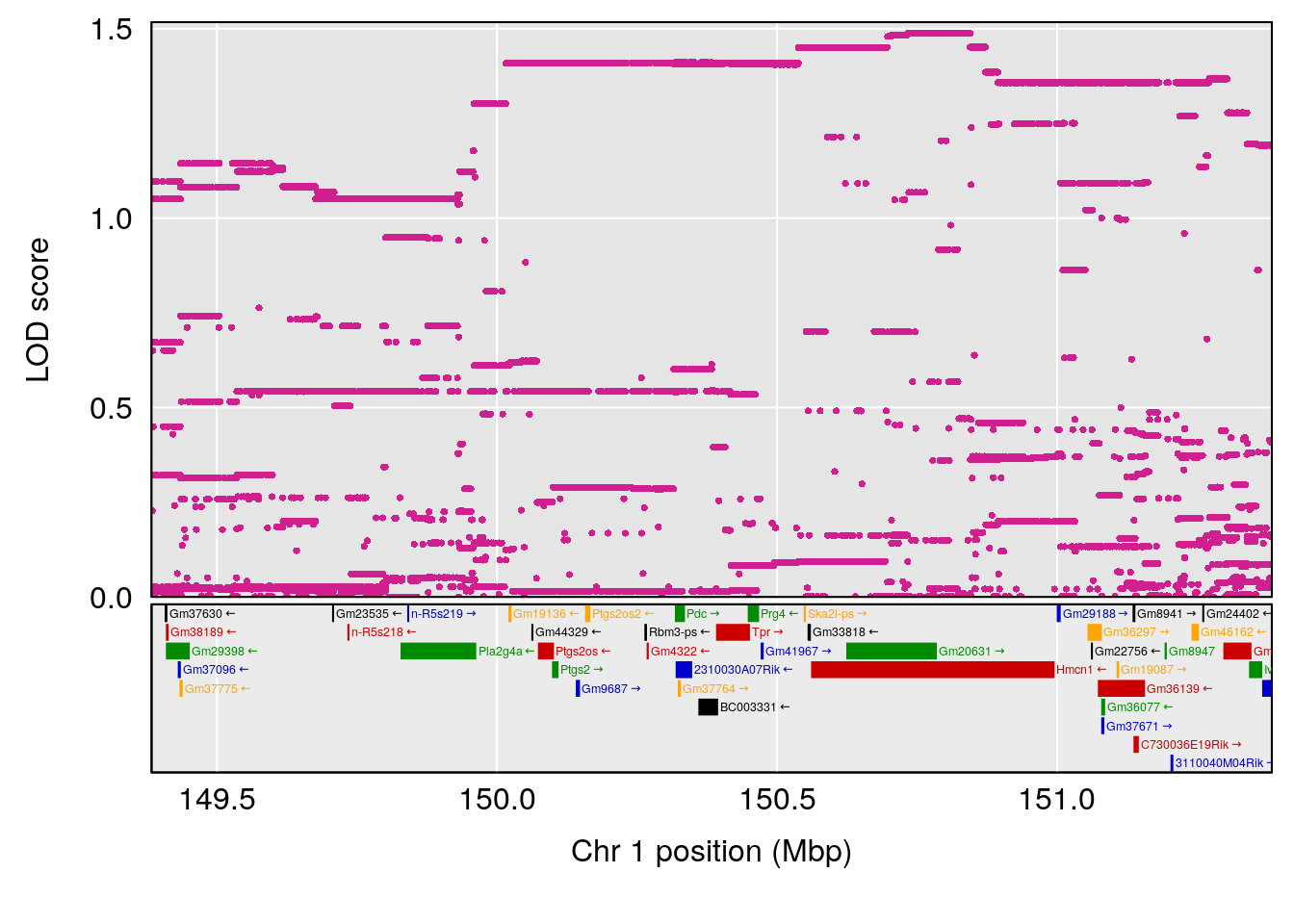
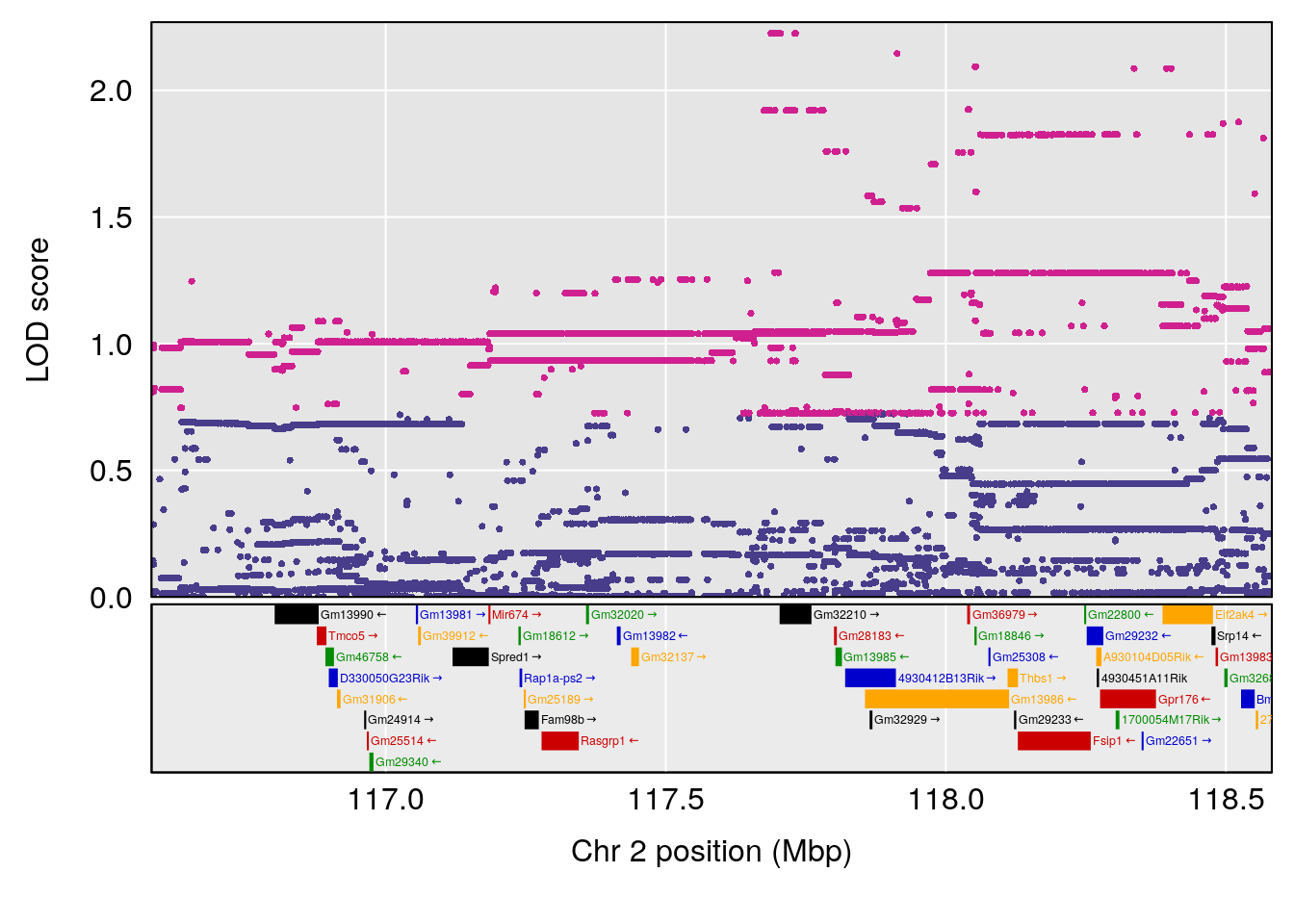
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
dev.off()
}
}
# [1] "Time.to.Dead.Hr"
# [1] 1
# [1] 2
# [1] 3
# [1] "RR.Depression.Rate.Hr"
# [1] "Time.to.Steady.RR.Depression.Hr"
# [1] "Min.Depression.RR"
# [1] 1
# [1] 2
# [1] "Mean.Depression.BR"
# [1] 1
# [1] 2
# [1] 3
# [1] "SteadyState.Depression.Duration.Hr"
# [1] 1
# [1] 2
# [1] "Time.to.Threshold.Recovery.Hr"
# [1] "Time.to.Projected.Recovery.Hr"
# [1] 1
# [1] "Start.of.Recovery.Hr"
# [1] 1
# [1] 2
# [1] 3
# [1] "RR.Recovery.Rate.Hr"
# [1] 1
# [1] 2
# [1] "Min.Depression.BR"
# [1] 1
# [1] "Time.to.Mostly.Dead.Hr"
# [1] 1
# [1] "Time.to.Recovery.Hr"
# [1] 1
# [1] 2
# [1] "Status_bin"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] 5
#save peaks coeff blup plot
for(i in names(qtl.out)){
print(i)
peaks <- find_peaks(qtl.out[[i]], map=pmap, threshold=6, drop=1.5)
if(nrow(peaks) > 0) {
fname <- paste("output/do_Fentanyl_combine2cohort_", str_replace_all(i, "[[:punct:]]", "") ,"_coefplot_blup.pdf",sep="")
pdf(file = fname, width = 16, height =8)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.out[[i]], bgcolor="gray95", legend=NULL)
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
dev.off()
}
}
# [1] "Time.to.Dead.Hr"
# [1] 1
# [1] 2
# [1] 3
# [1] "RR.Depression.Rate.Hr"
# [1] "Time.to.Steady.RR.Depression.Hr"
# [1] "Min.Depression.RR"
# [1] 1
# [1] 2
# [1] "Mean.Depression.BR"
# [1] 1
# [1] 2
# [1] 3
# [1] "SteadyState.Depression.Duration.Hr"
# [1] 1
# [1] 2
# [1] "Time.to.Threshold.Recovery.Hr"
# [1] "Time.to.Projected.Recovery.Hr"
# [1] 1
# [1] "Start.of.Recovery.Hr"
# [1] 1
# [1] 2
# [1] 3
# [1] "RR.Recovery.Rate.Hr"
# [1] 1
# [1] 2
# [1] "Min.Depression.BR"
# [1] 1
# [1] "Time.to.Mostly.Dead.Hr"
# [1] 1
# [1] "Time.to.Recovery.Hr"
# [1] 1
# [1] 2
# [1] "Status_bin"
# [1] 1
# [1] 2
# [1] 3
# [1] 4
# [1] 5heritability
#plot heritability by qtl2 array --------------
herit <- data.frame(Phenotype = names(unlist(qtl.hsq)),
Heritability = round(unlist(qtl.hsq),3))
herit <- herit %>%
arrange(desc(Heritability))
herit$Phenotype <- factor(herit$Phenotype, levels = herit$Phenotype)
#histgram
p2 <- ggplot(data=herit, aes(x=Phenotype, y=Heritability)) + #, fill=Domain, color = Domain)) +
geom_bar(stat="identity", fill = "blue", color = "blue", show.legend = FALSE) +
scale_y_continuous(breaks=seq(0.0, 1.0, 0.1)) +
geom_text(aes(label = Heritability, y = Heritability + 0.005), position = position_dodge(0.9),vjust = 0) +
theme(axis.text.x = element_text(angle = 90, hjust = 1)) +
ggtitle("DO do_Fentanyl_combine2cohort Heritability by qtl2 array")
p2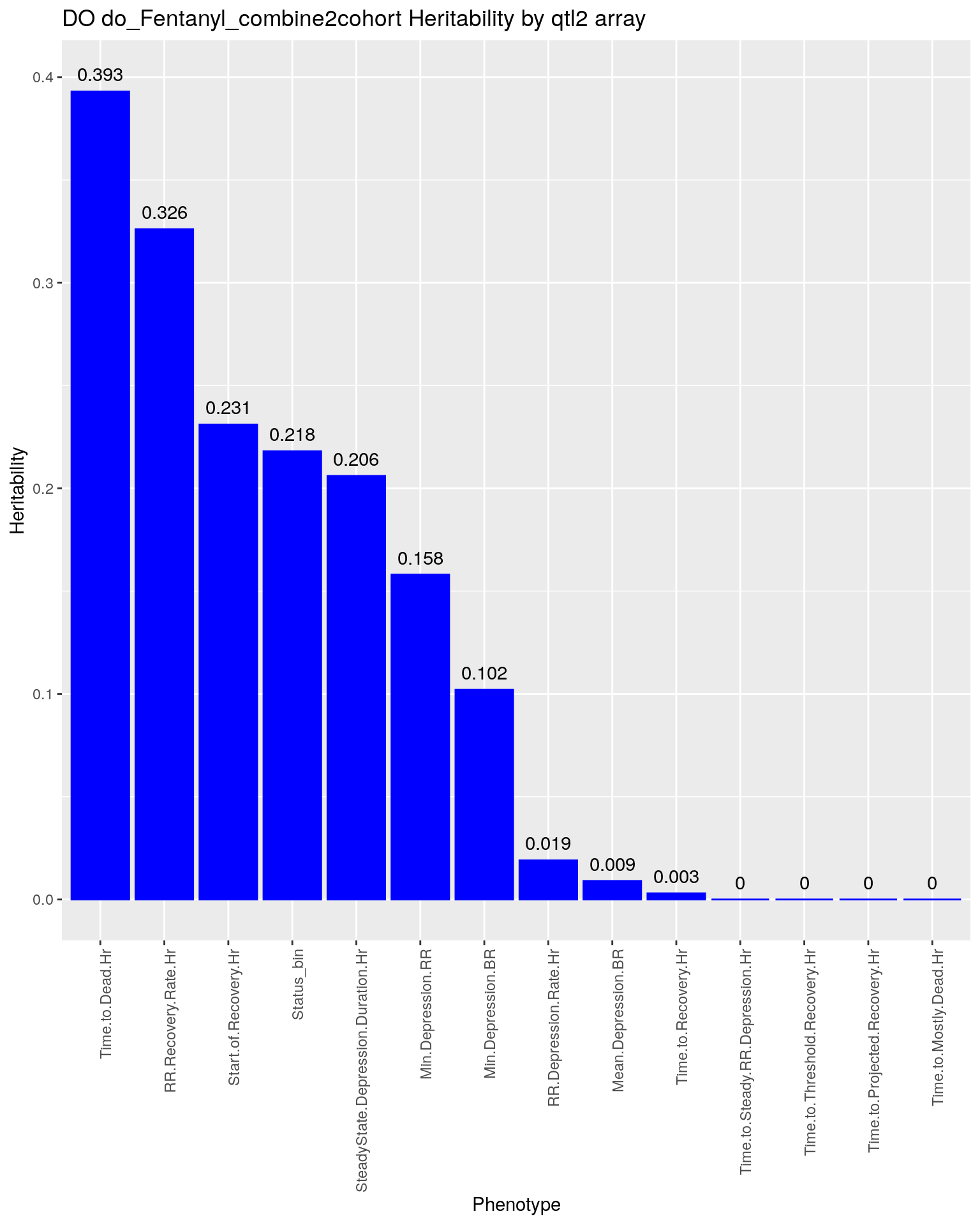
#plot heritability by GCTA
h <- read.csv("data/FinalReport/GCTA/DO_Fentanyl_combining2Cohort/DO_Fentanyl_combining2Cohort_heritability_by_GCTA.csv", header = TRUE)
h <- h %>%
arrange(desc(Heritability))
h$Phenotype <- factor(h$Phenotype, levels = h$Phenotype)
h$Heritability <- round(h$Heritability,2)
#histgram
pdf(file = paste0("data/FinalReport/GCTA/DO_Fentanyl_combining2Cohort/DO_Fentanyl_combining2Cohort","_heritability_by_GCTA.pdf"), height = 10, width = 10)
p3<-ggplot(data=h, aes(x=Phenotype, y=Heritability)) + #, fill=Domain, color = Domain)) +
geom_bar(stat="identity", fill = "blue", color = "blue", show.legend = FALSE) +
scale_y_continuous(breaks=seq(0.0, 1.0, 0.1)) +
geom_text(aes(label = Heritability, y = Heritability + 0.005), position = position_dodge(0.9),vjust = 0) +
theme(axis.text.x = element_text(angle = 90, hjust = 1)) +
ggtitle("Heritability by GCTA")
p3
dev.off()
# png
# 2
# png
# 2
p3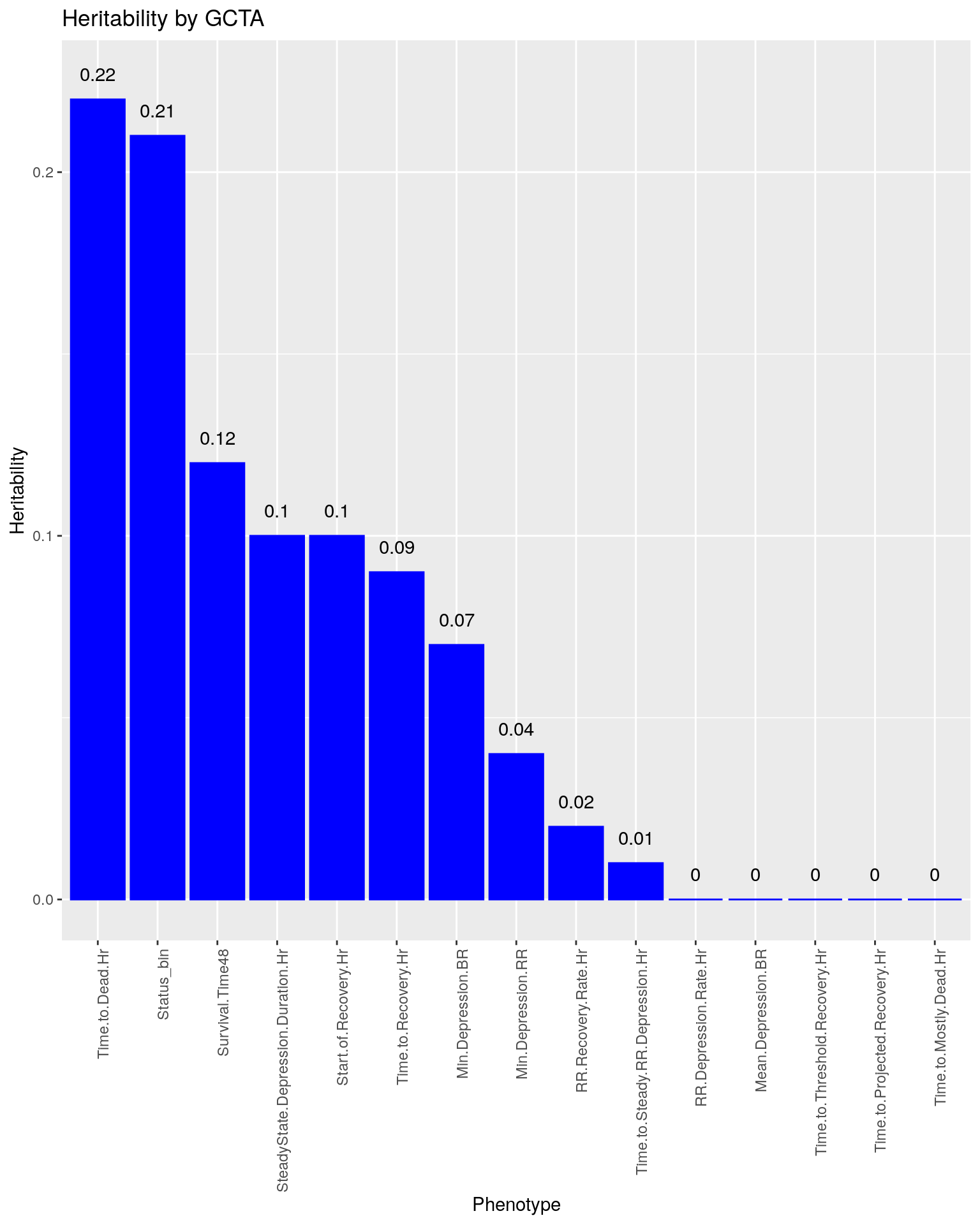
Plot qtl mapping on array male
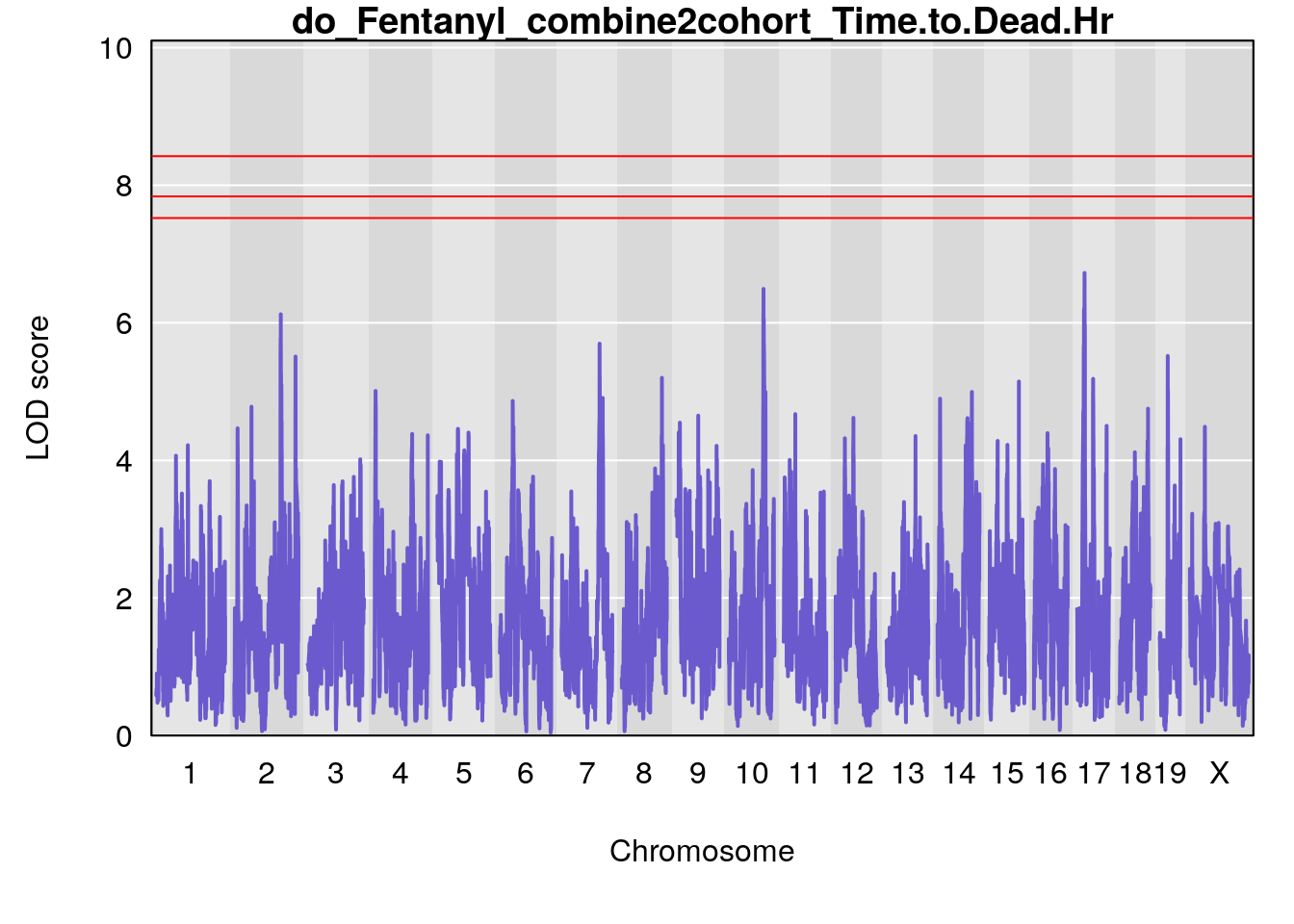
load("output/Fentanyl/qtl.fentanyl.combine2cohort.out.male.RData")
#pmap and gmap
chr.names <- c("1","2","3","4","5","6","7","8","9","10","11","12","13","14","15","16","17","18","19", "X")
gmap <- list()
pmap <- list()
for (chr in chr.names){
gmap[[chr]] <- gm$gmap[[chr]]
pmap[[chr]] <- gm$pmap[[chr]]
}
attr(gmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
attr(pmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
#genome-wide plot
for(i in names(qtl.out)){
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.out[[i]]) # overall maximum LOD score
plot(qtl.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, max(sapply(qtl.out, maxlod)) +2))
abline(h=summary(qtl.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("do_Fentanyl_combine2cohort_",i))
}
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
#peaks coeff plot
for(i in names(qtl.out)){
print(i)
peaks <- find_peaks(qtl.out[[i]], map=pmap, threshold=6, drop=1.5)
if(nrow(peaks) > 0) {
print(peaks)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.out[[i]], bgcolor="gray95", legend=NULL)
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.out[[i]], bgcolor="gray95", legend=NULL)
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
}
}
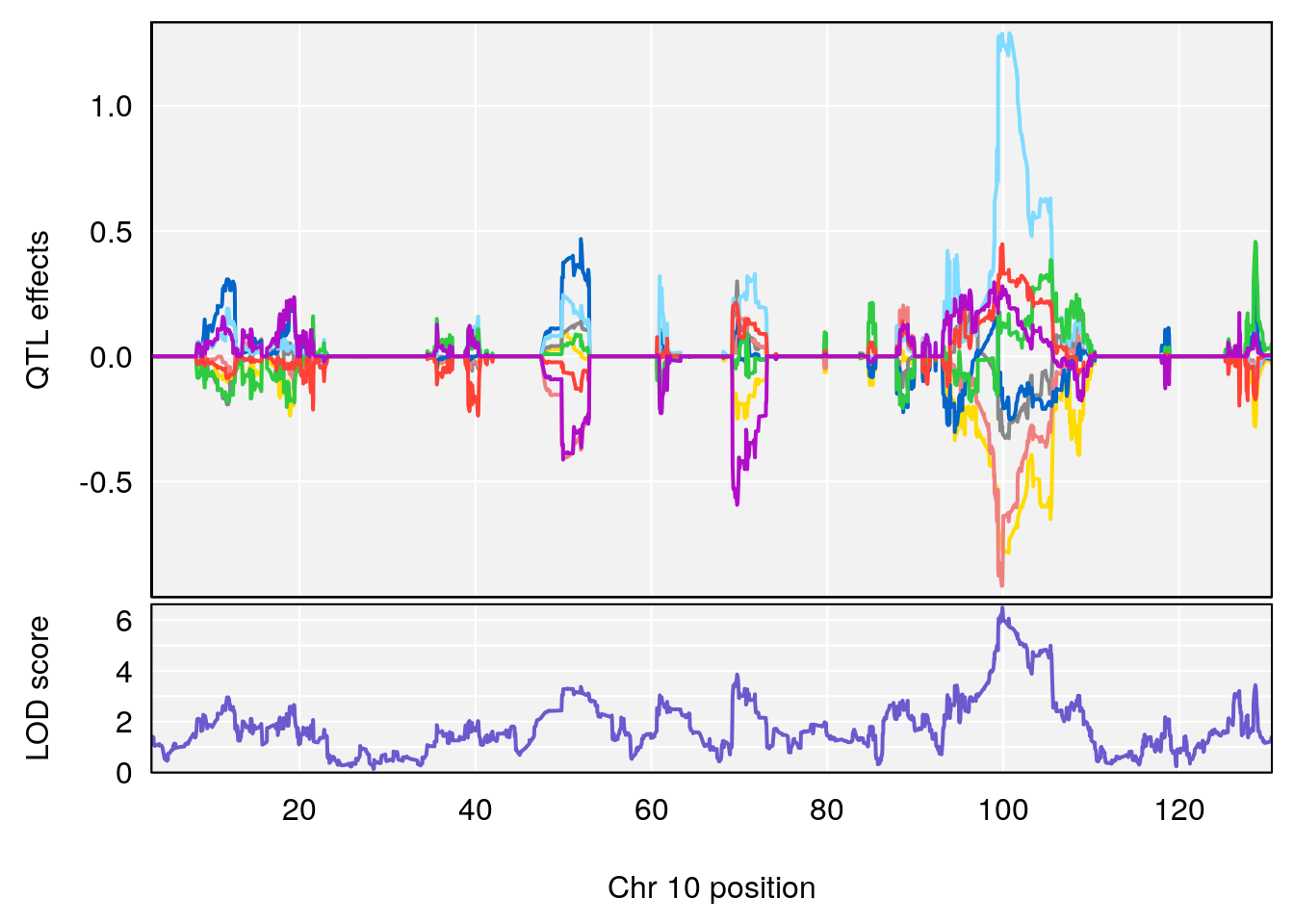
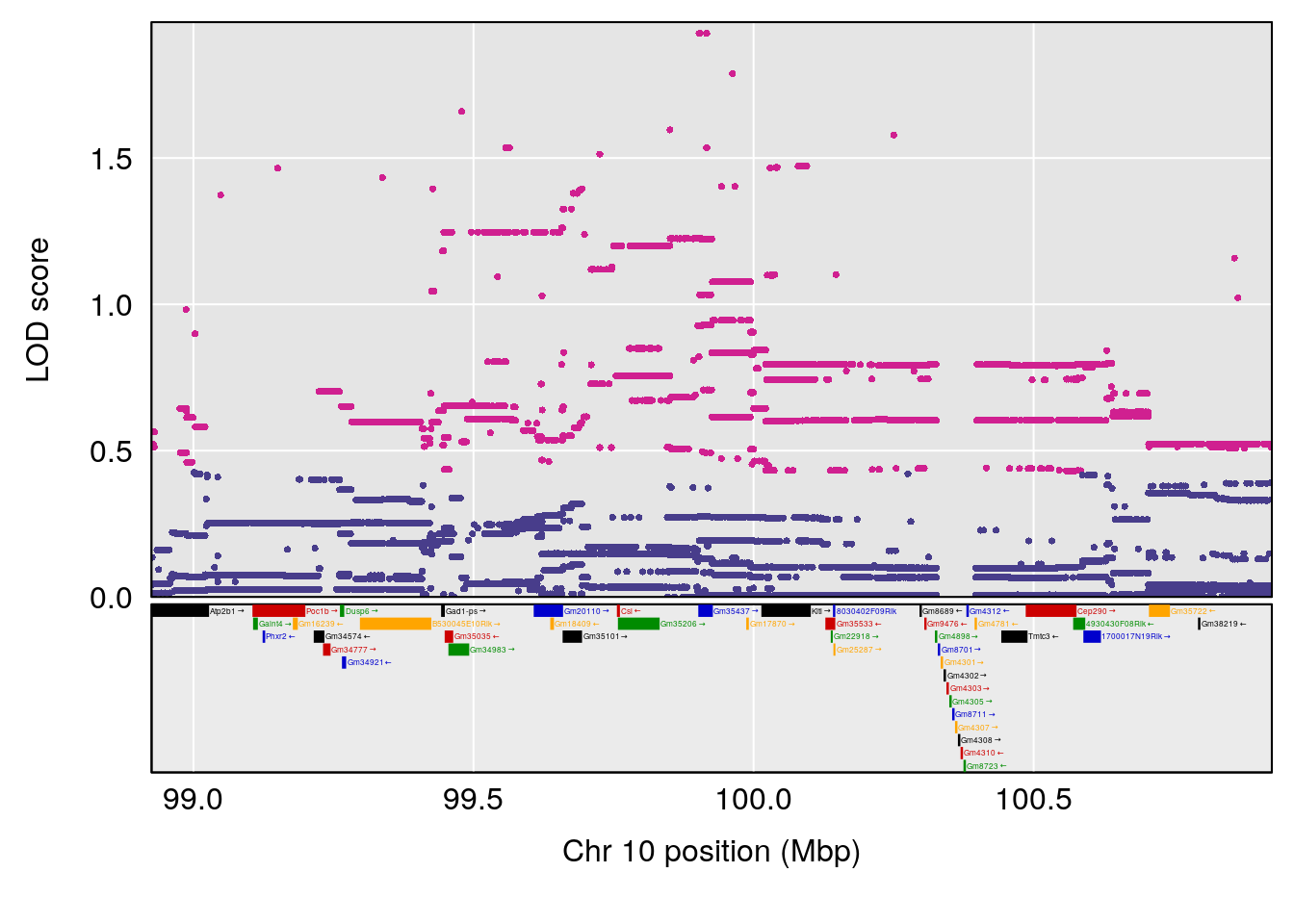
# [1] "Time.to.Dead.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 2 132.62489 6.124096 50.81340 173.86901
# 2 1 pheno1 10 99.92521 6.492863 99.28182 105.40417
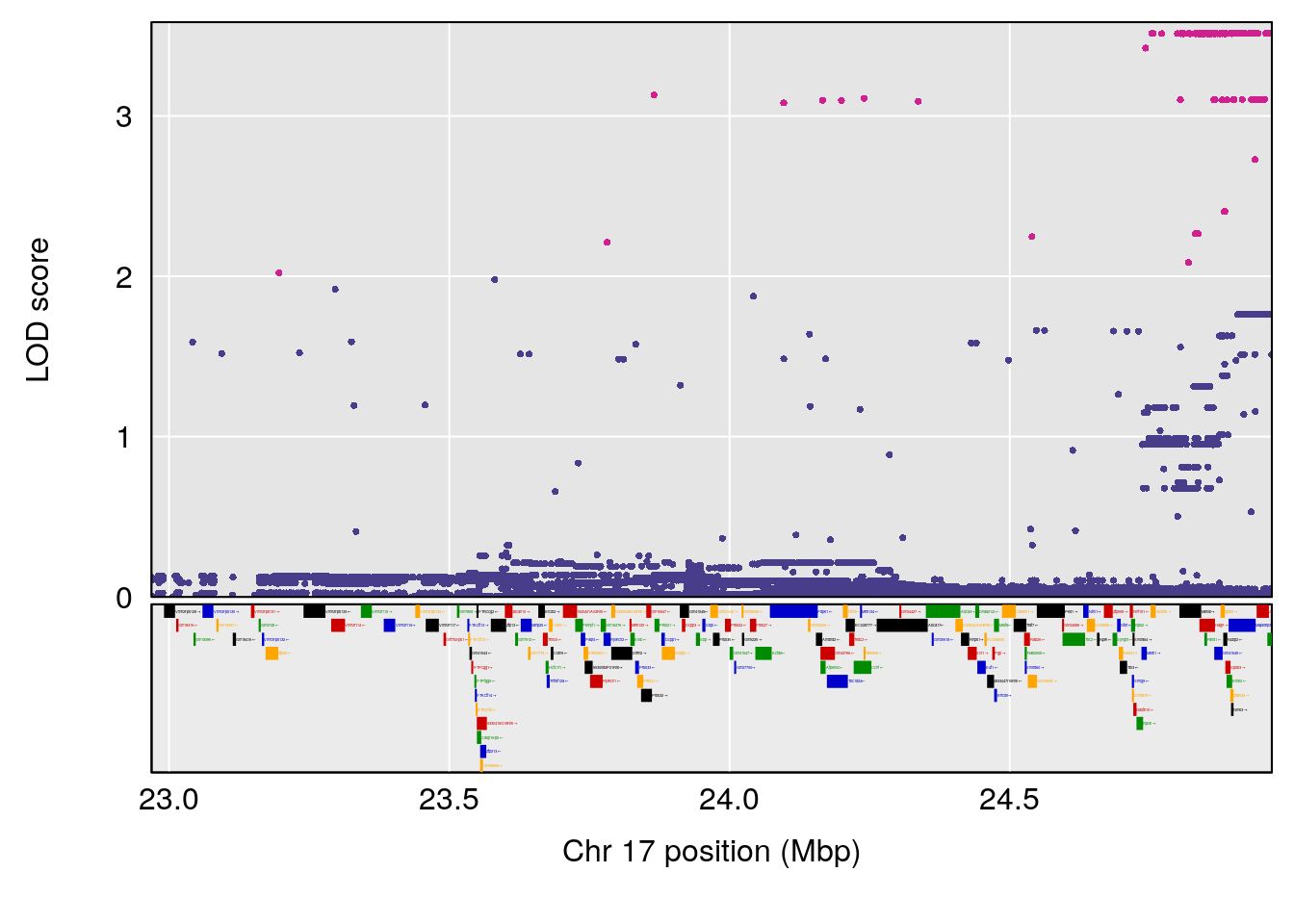
# 3 1 pheno1 17 23.96788 6.725145 21.59514 25.08344
# [1] 1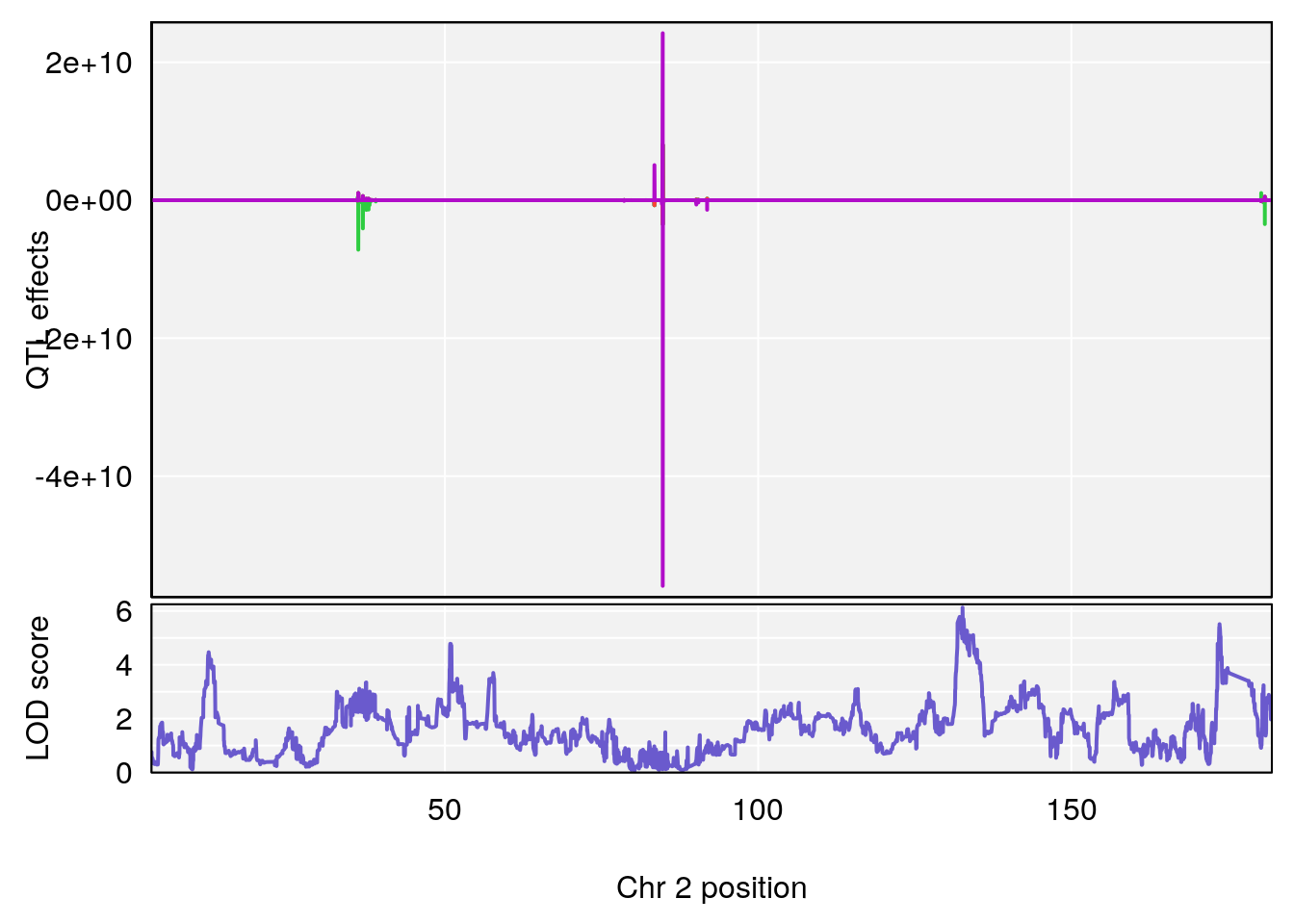
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2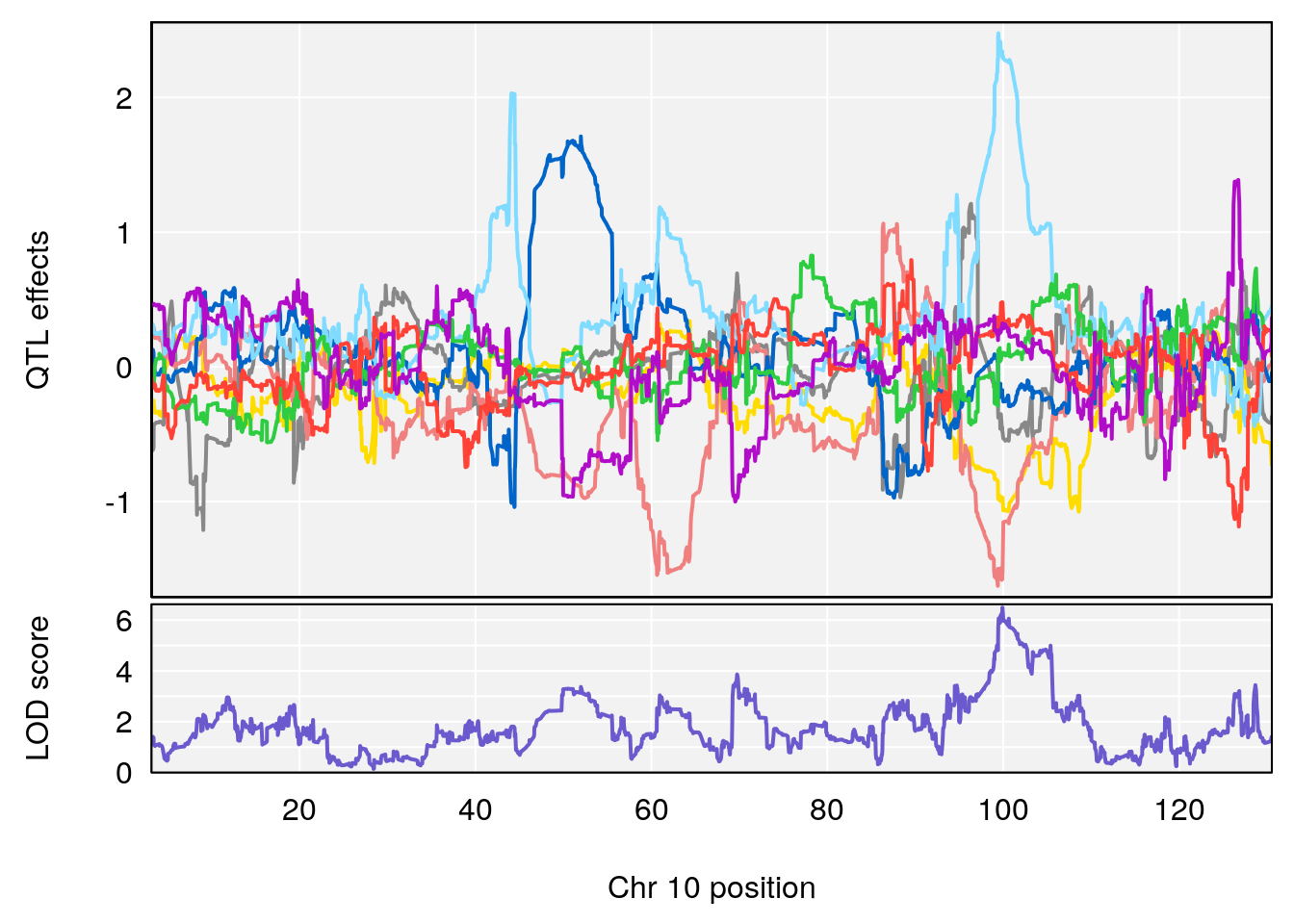
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 3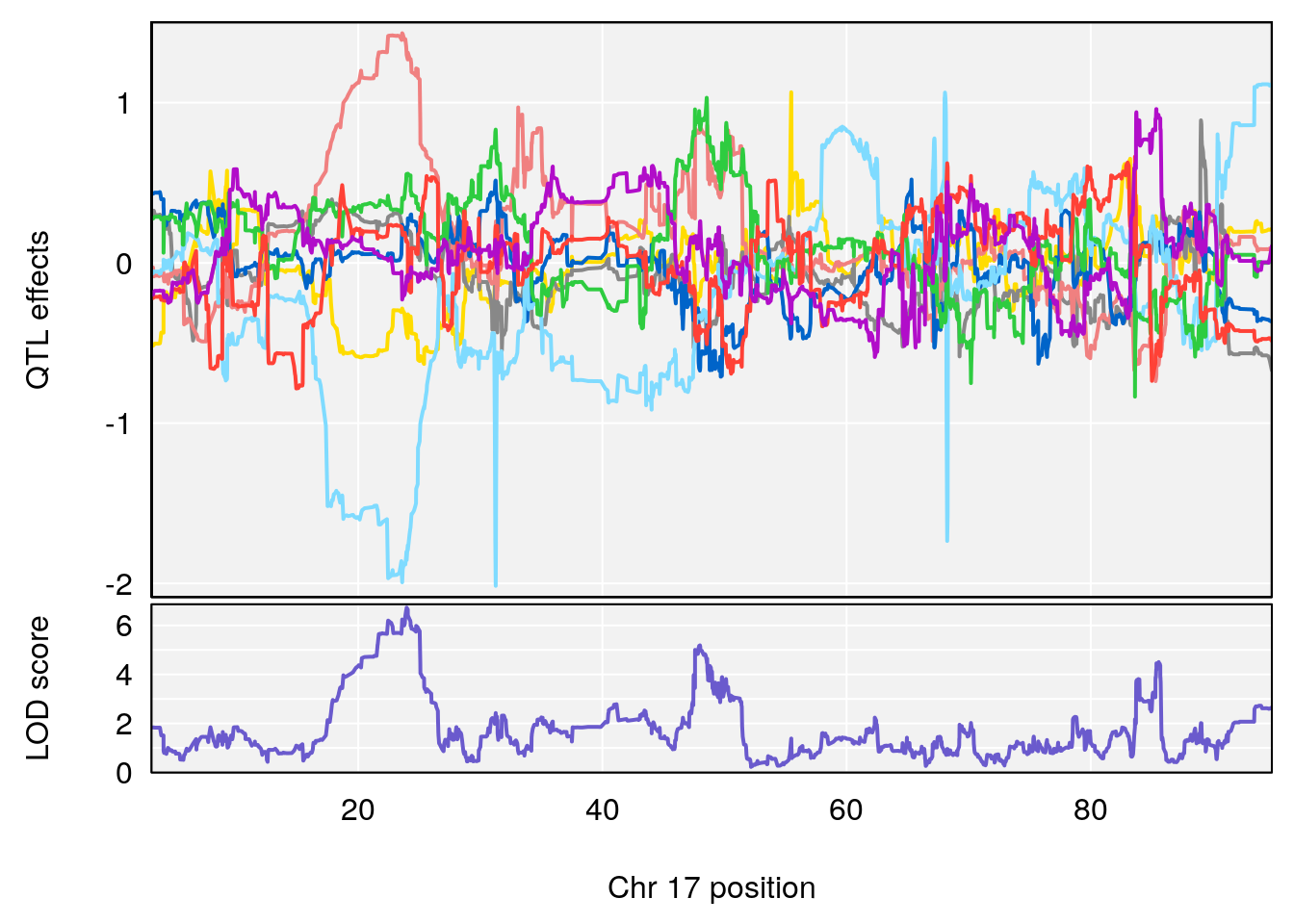
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
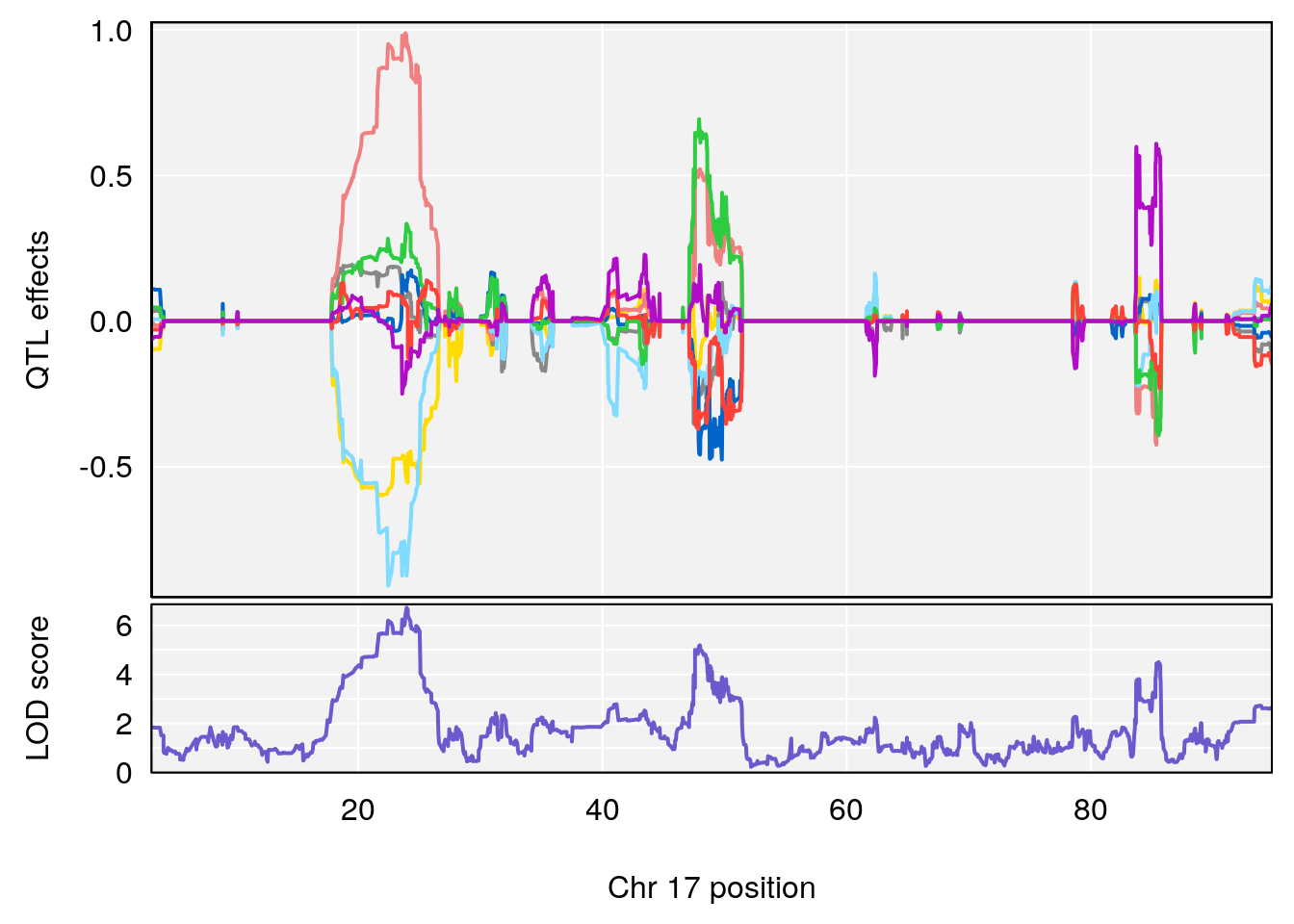
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "RR.Depression.Rate.Hr"
# [1] "Time.to.Steady.RR.Depression.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
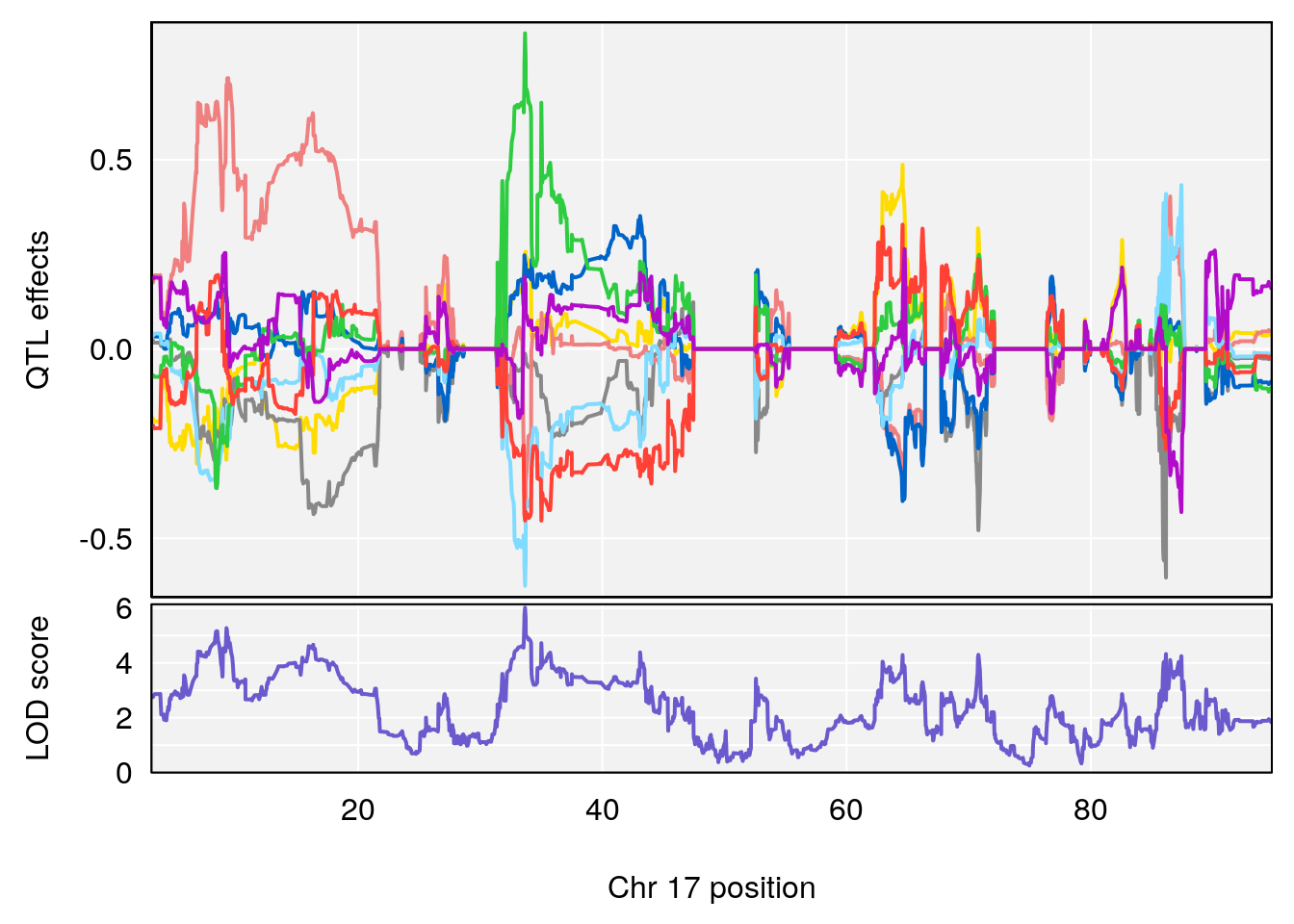
# 1 1 pheno1 17 33.66364 6.021735 7.725897 35.02144
# [1] 1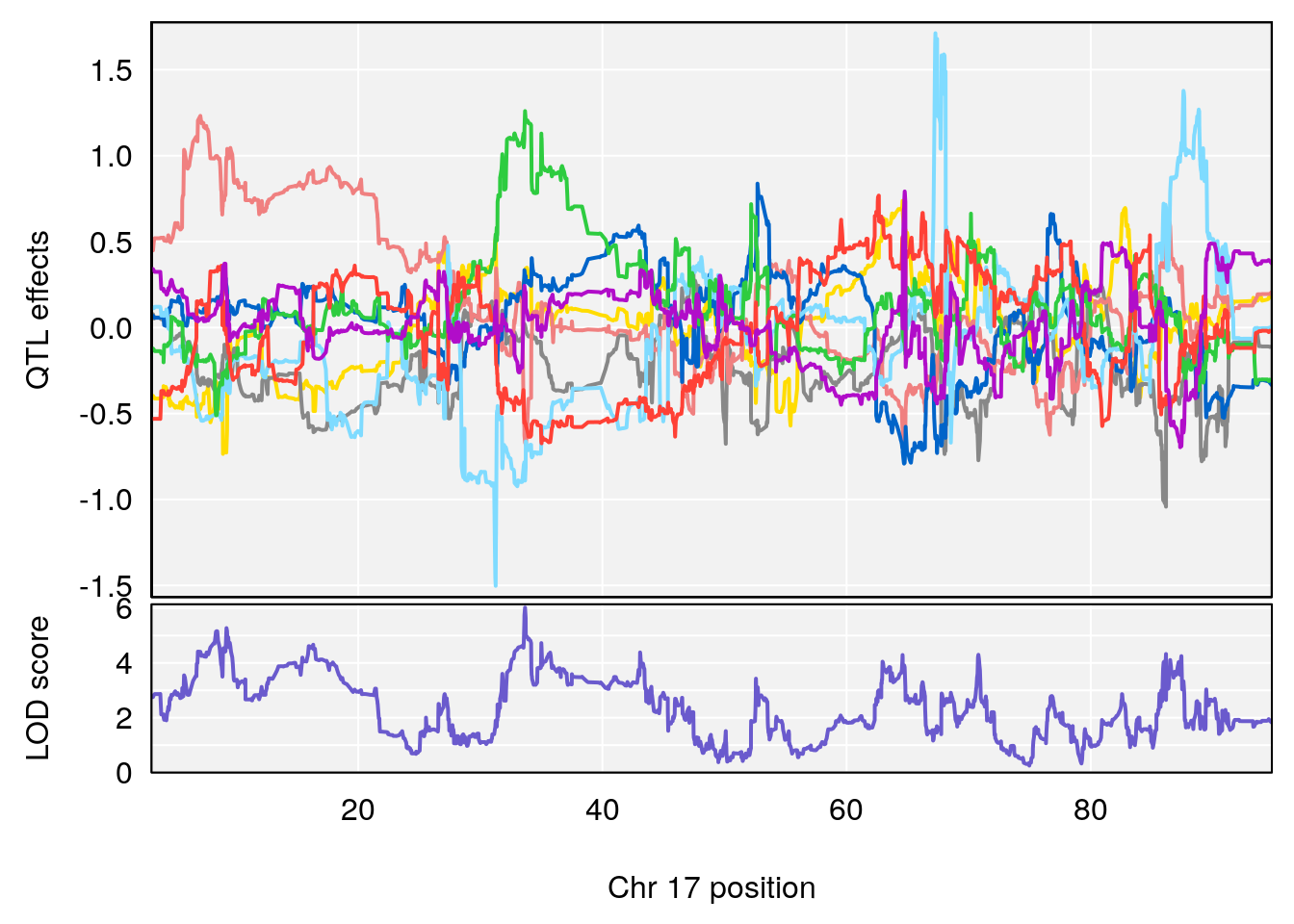
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
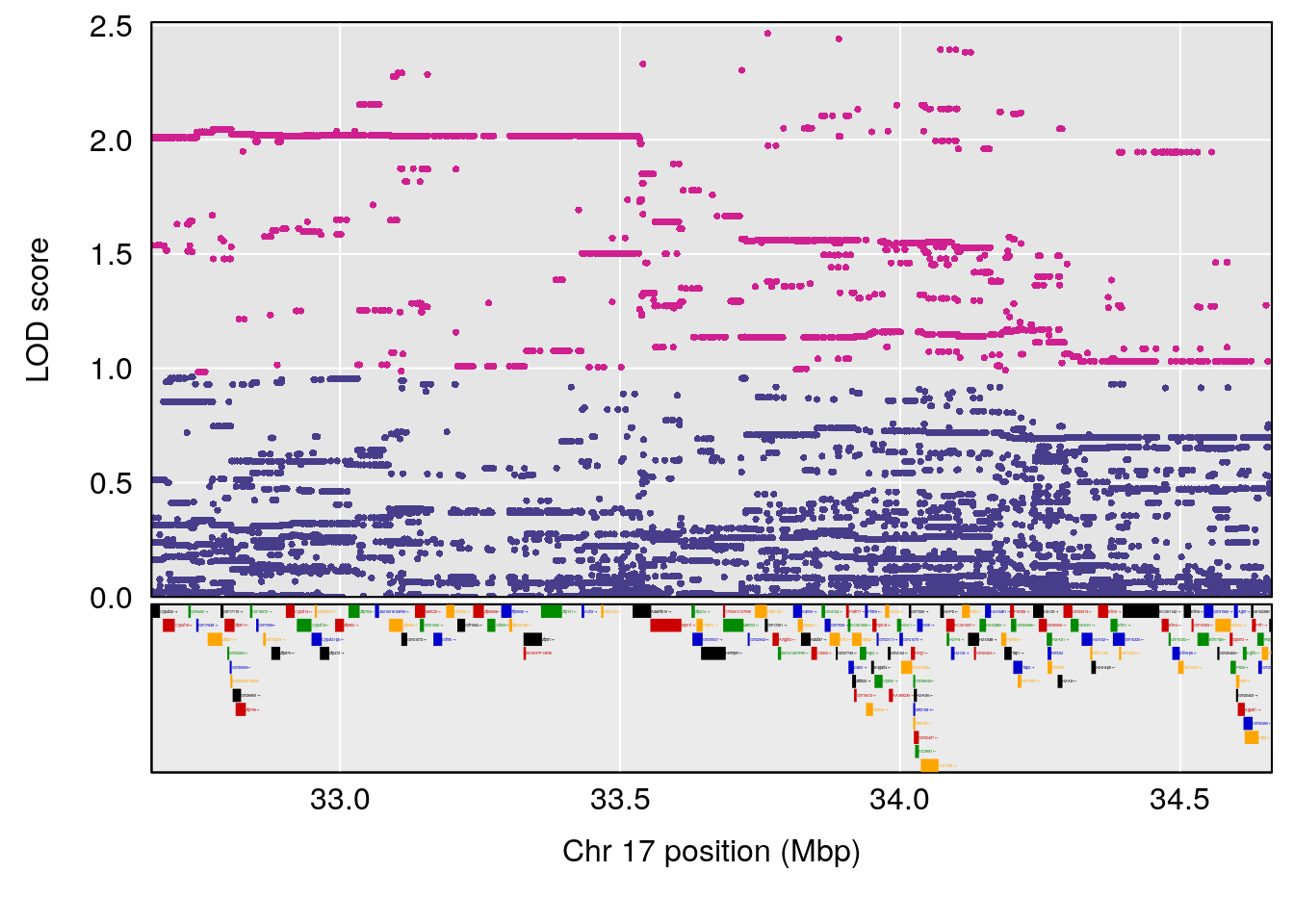
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "Min.Depression.RR"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 14 101.798 6.961191 100.3058 103.3399
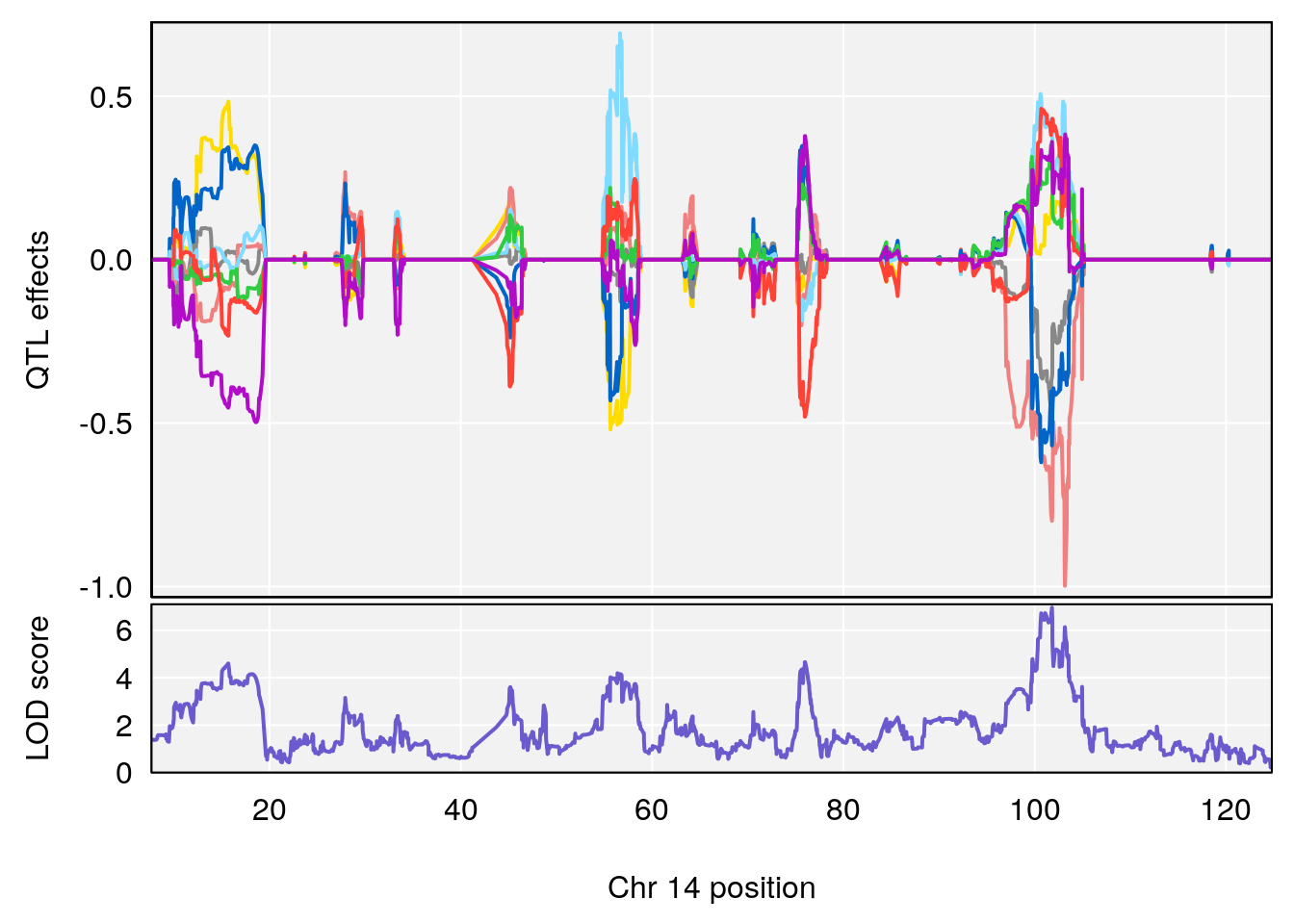
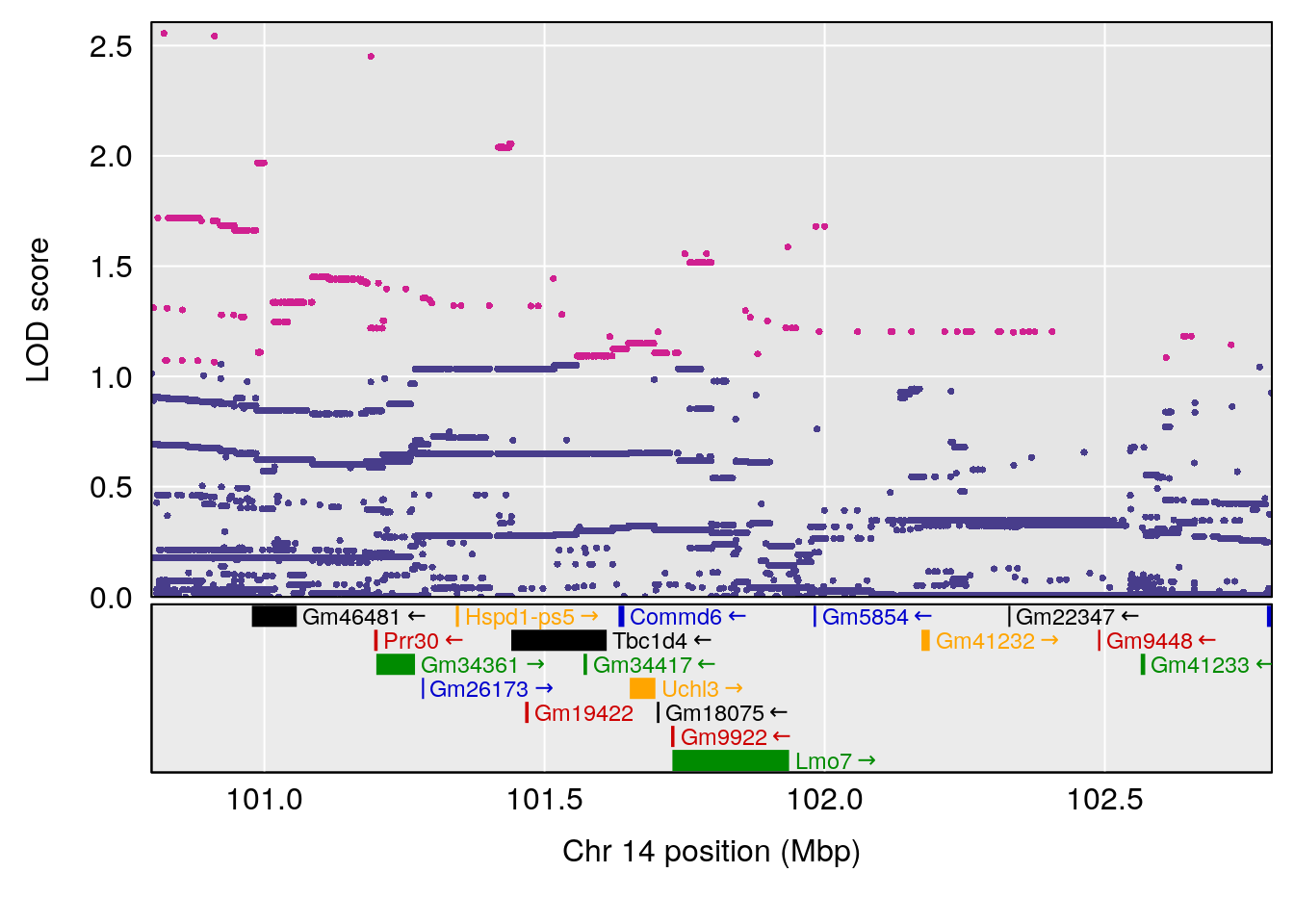
# [1] 1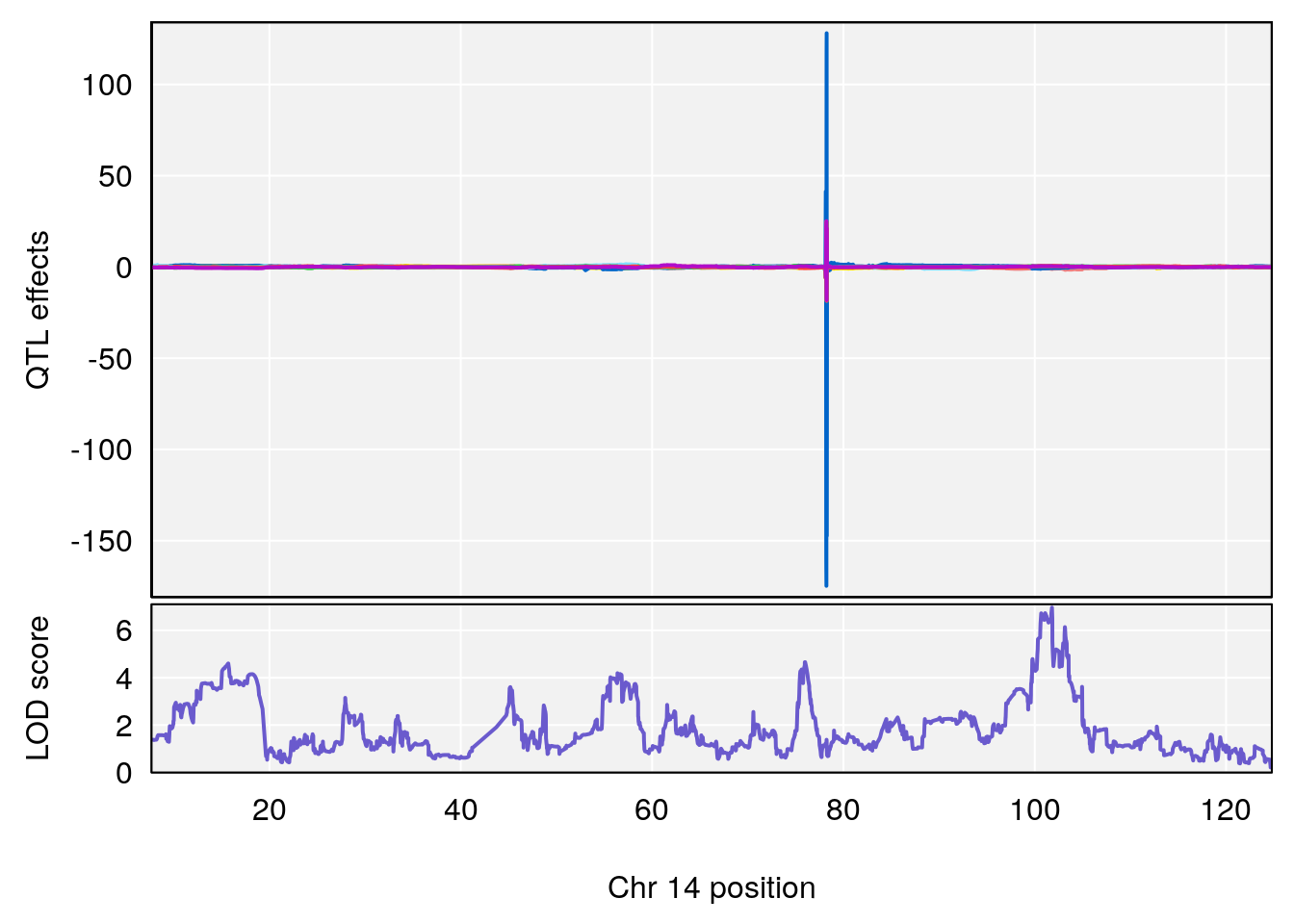
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "Mean.Depression.BR"
# [1] "SteadyState.Depression.Duration.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
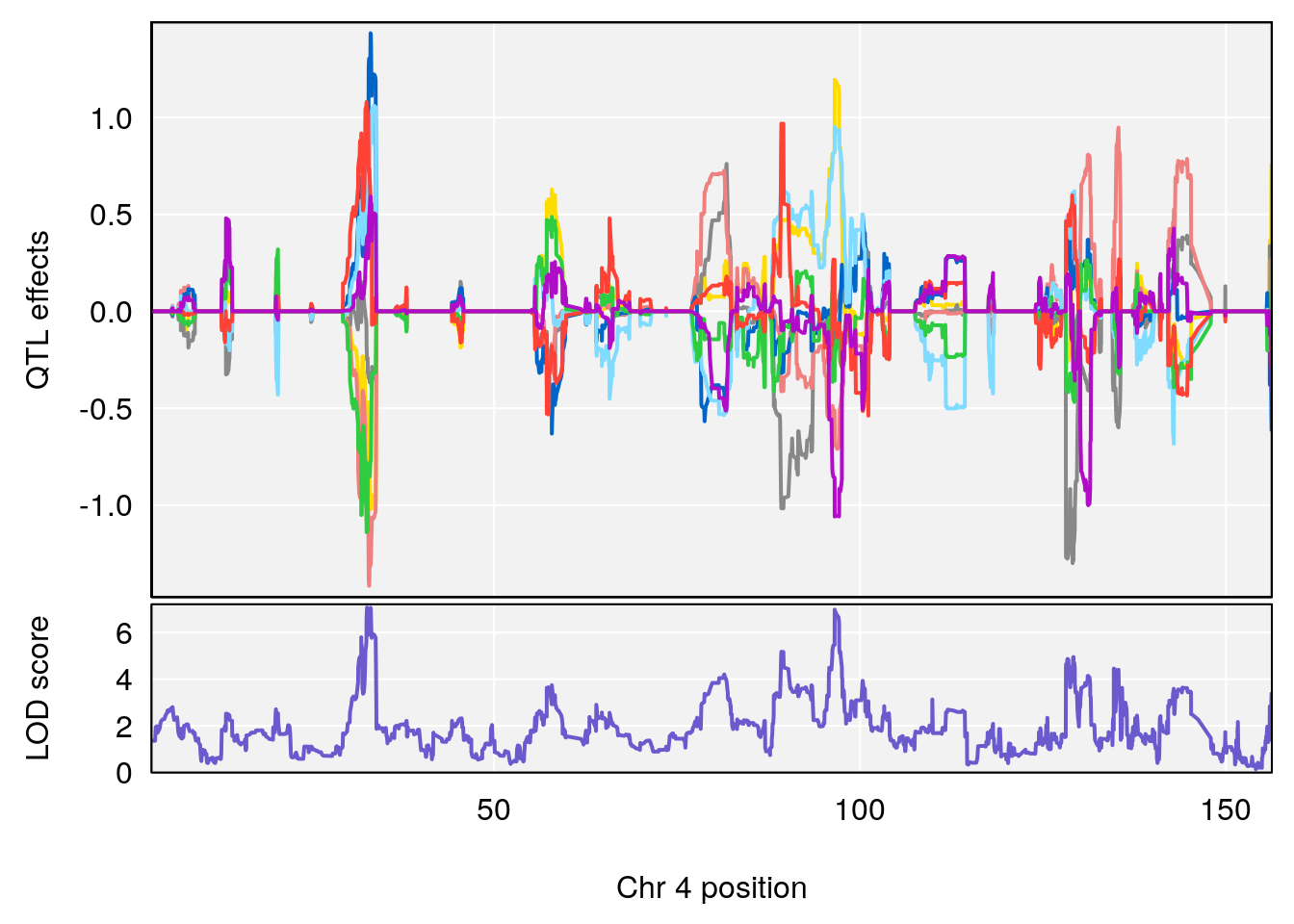
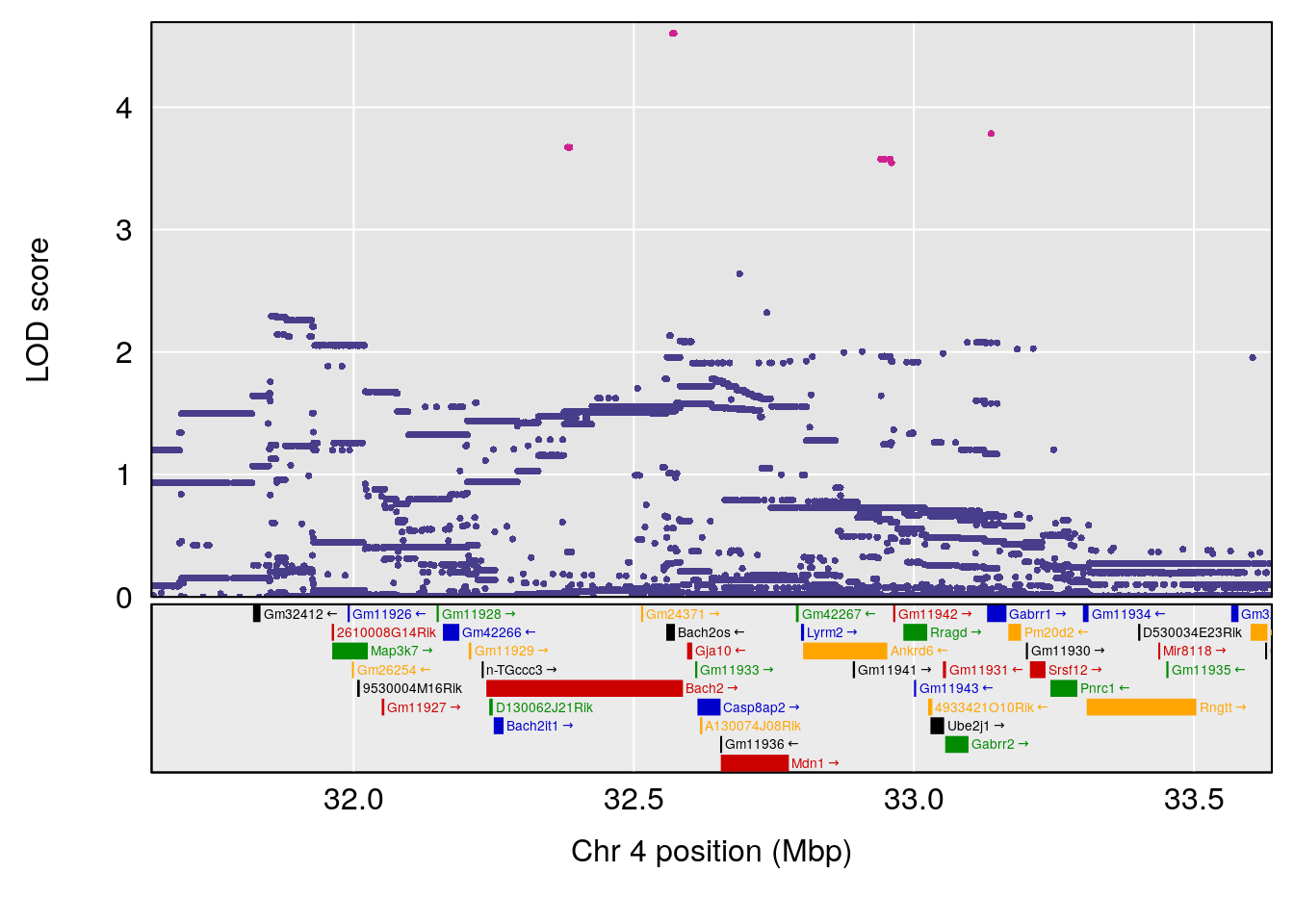
# 1 1 pheno1 4 32.63908 7.066593 31.85079 97.16594
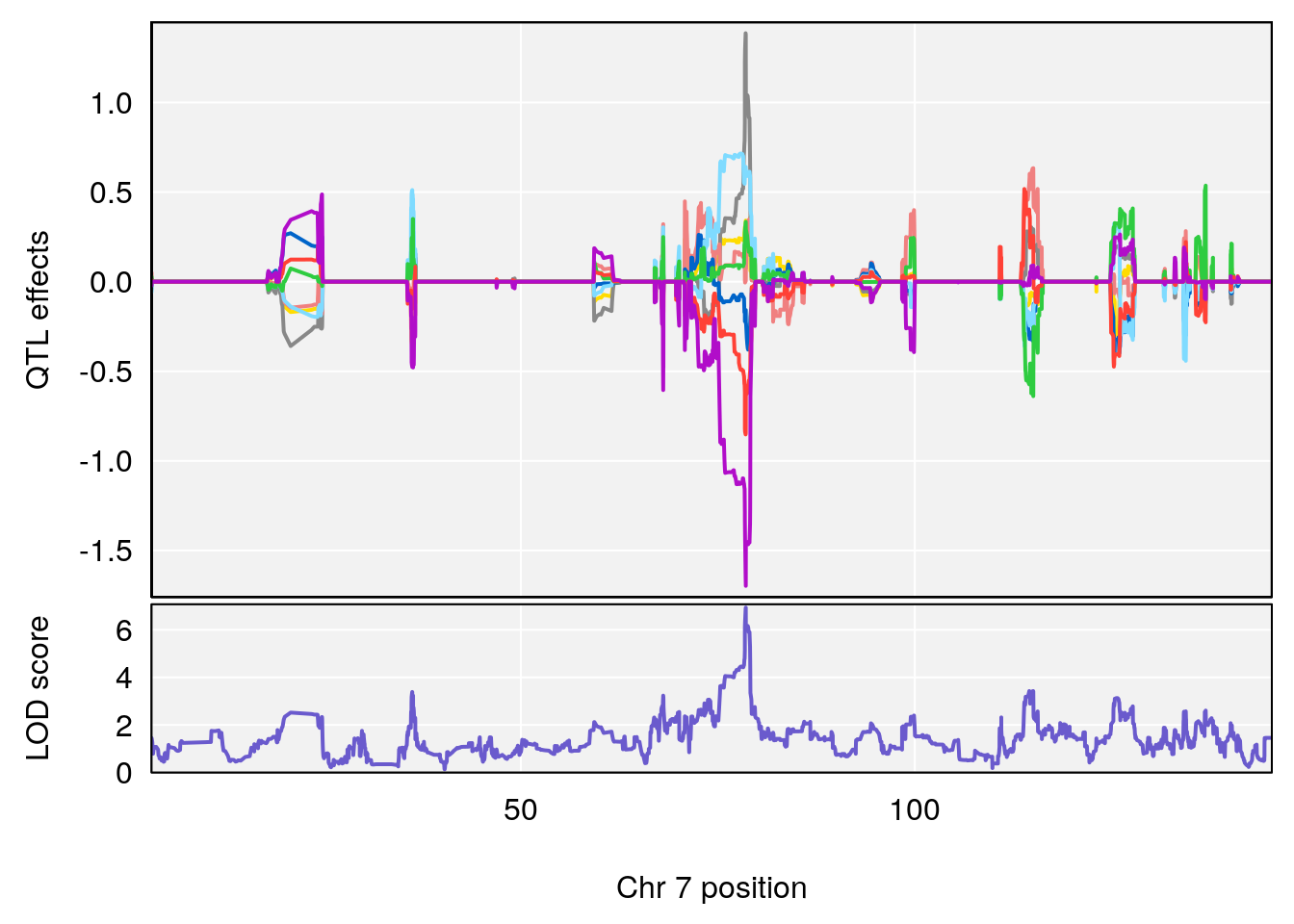
# 2 1 pheno1 7 78.55130 6.765506 78.43609 79.07406
# 3 1 pheno1 15 82.95976 6.039949 82.61311 84.16285
# [1] 1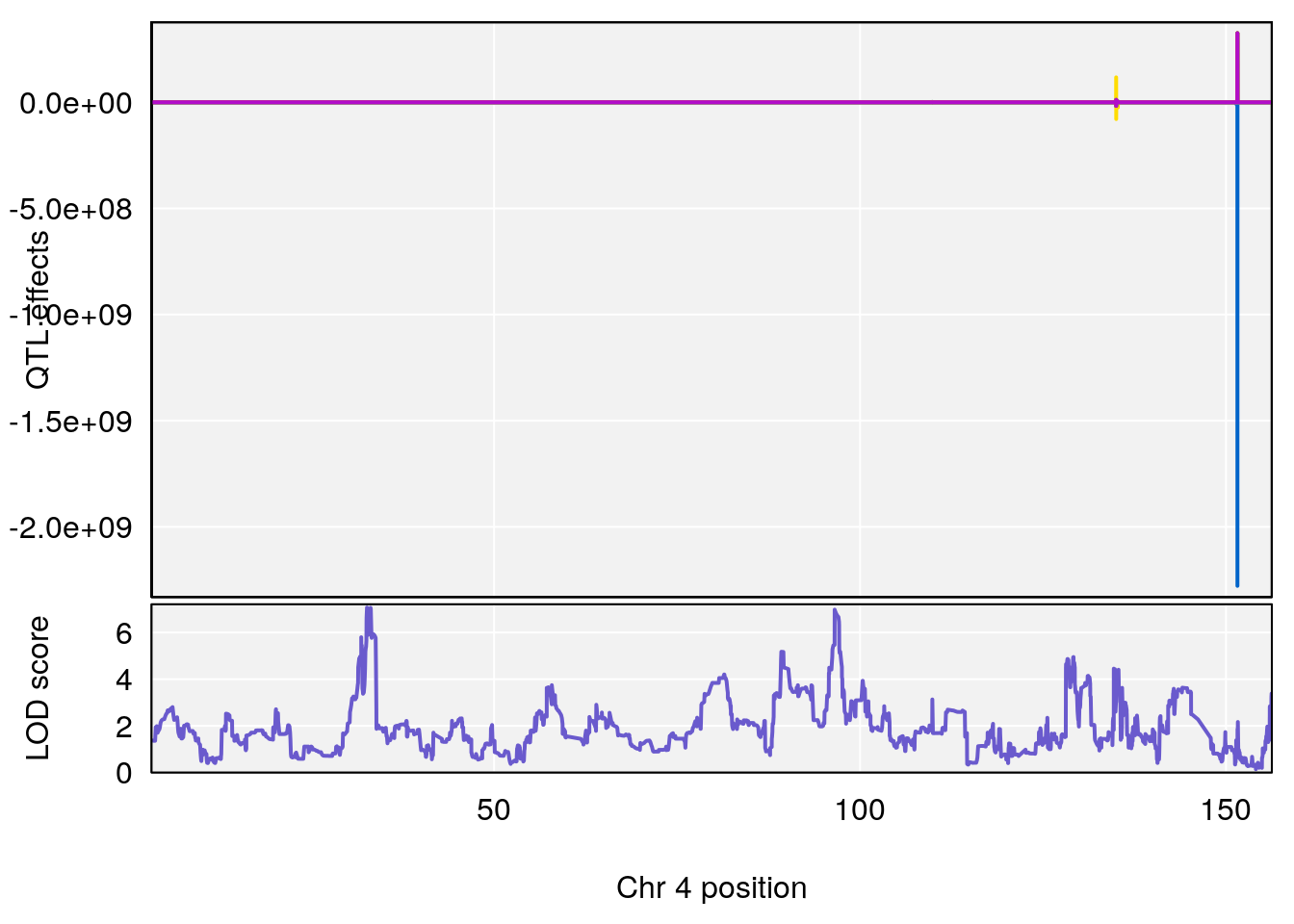
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2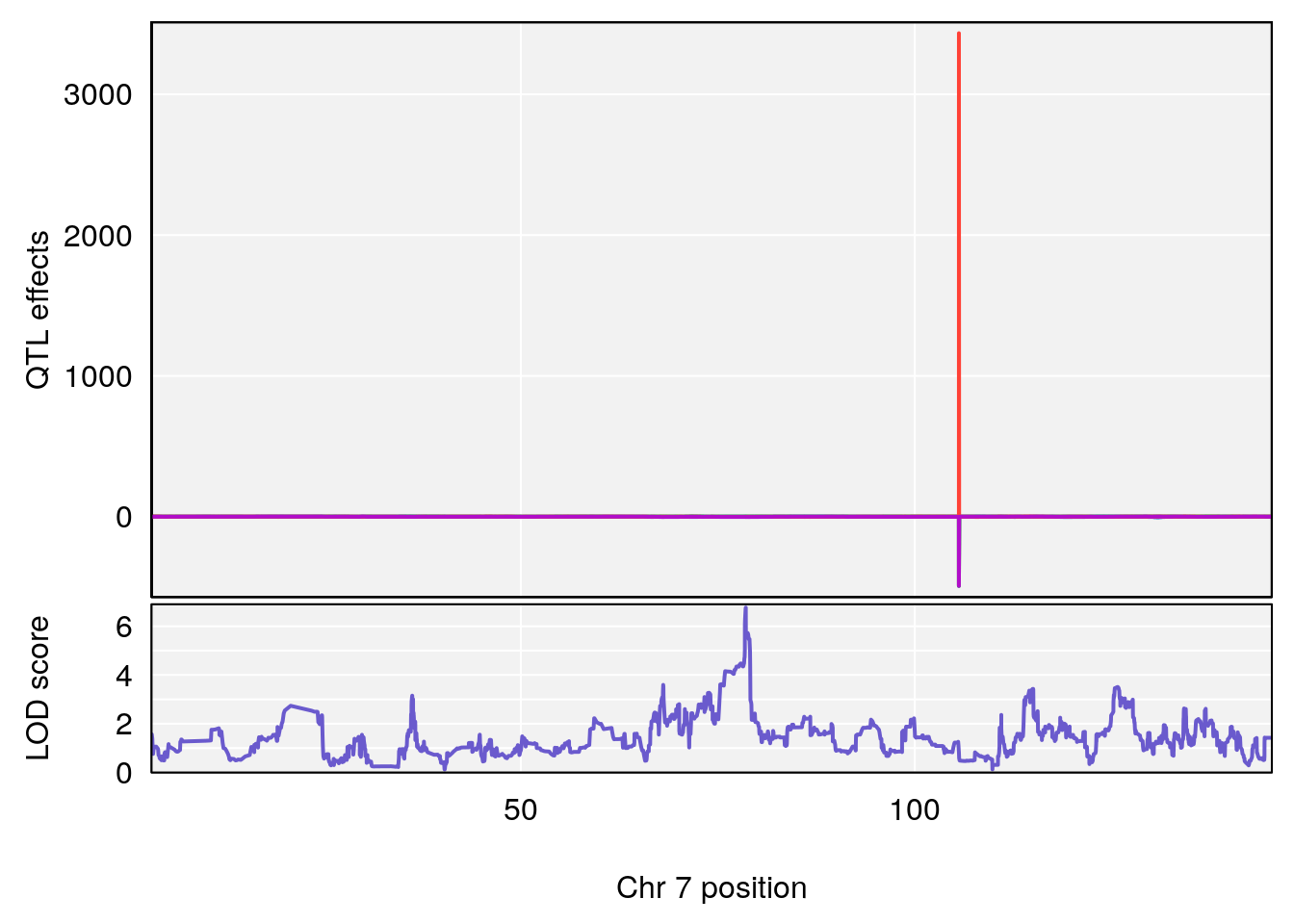
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
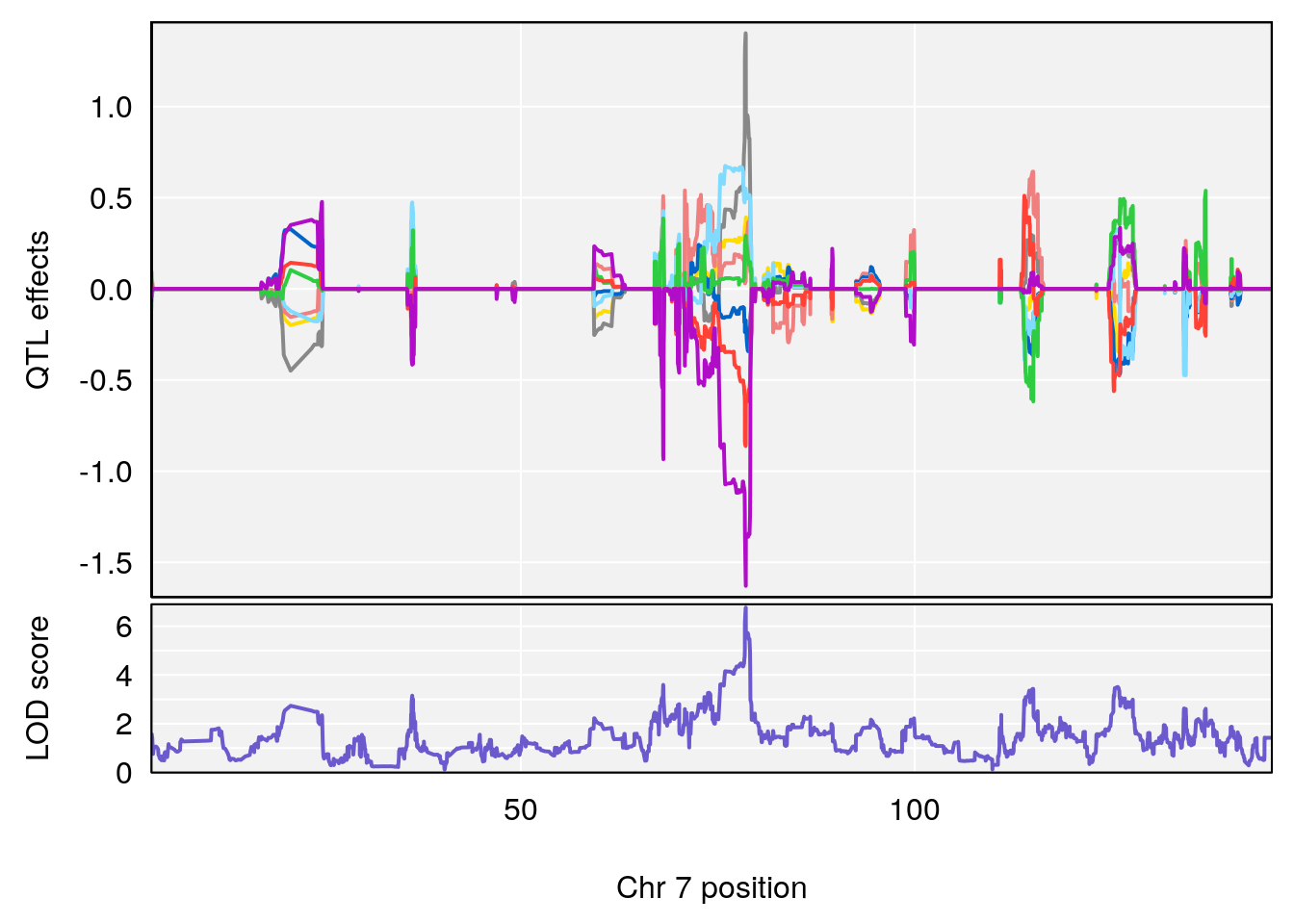
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
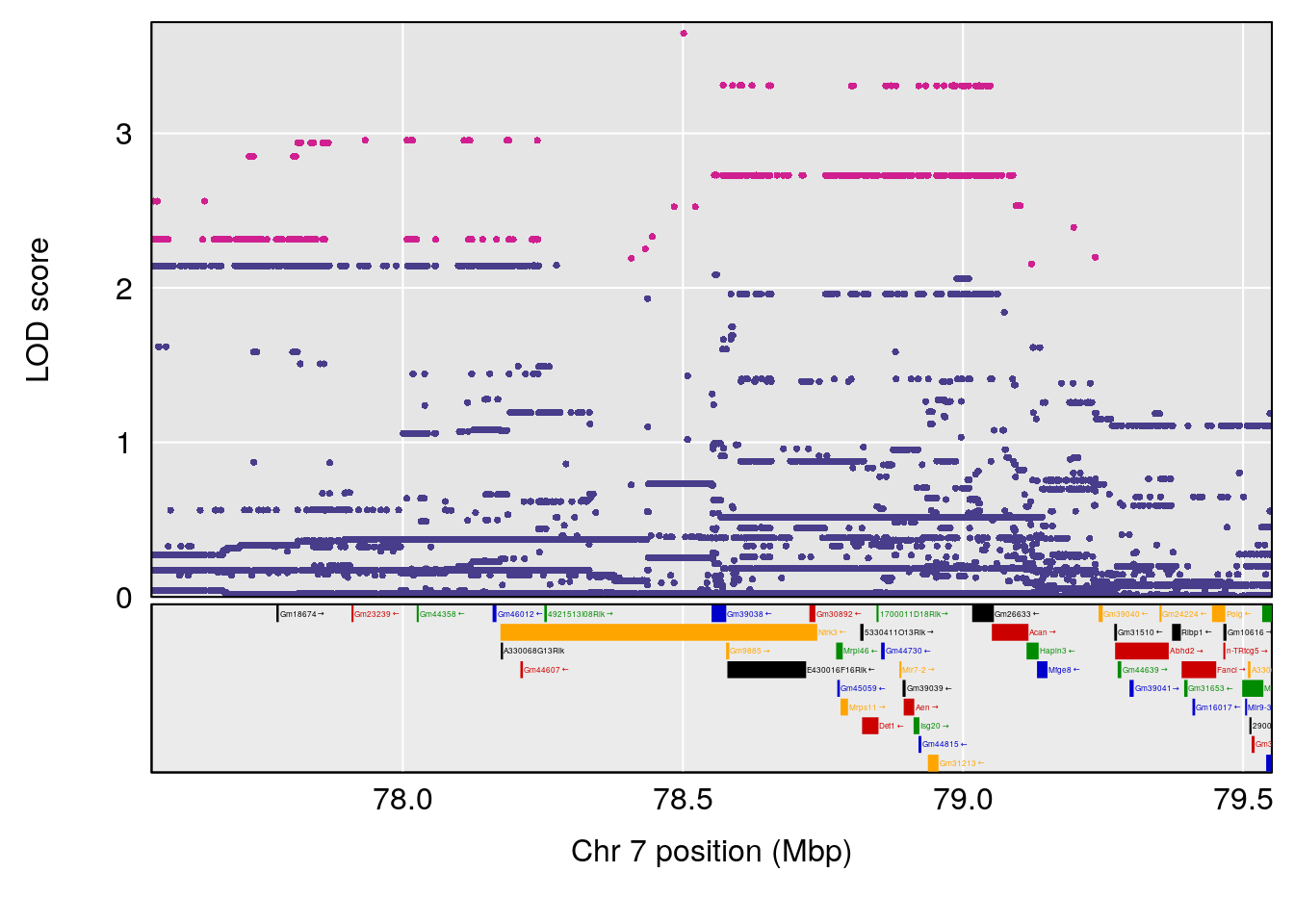
# [1] 3
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
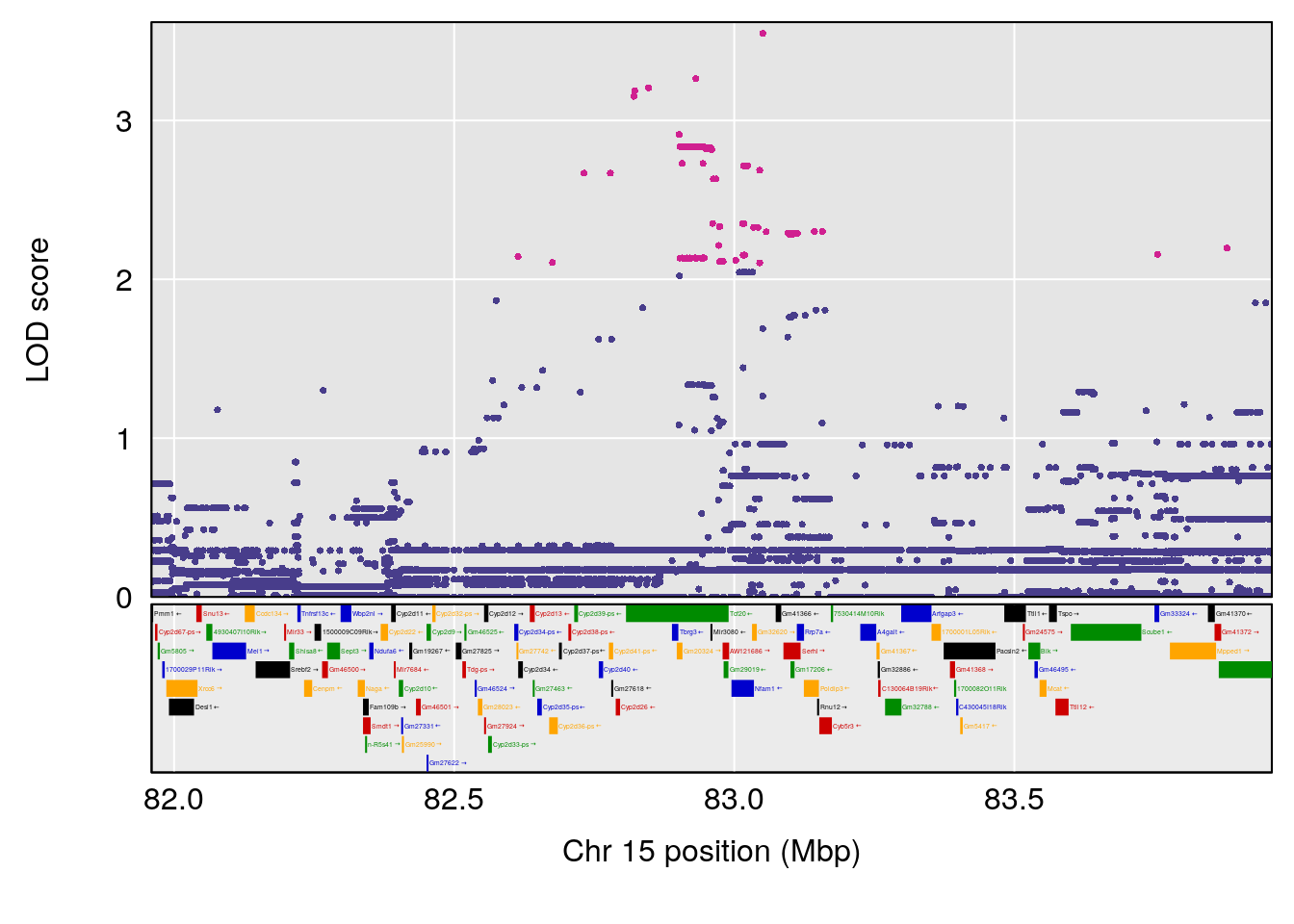
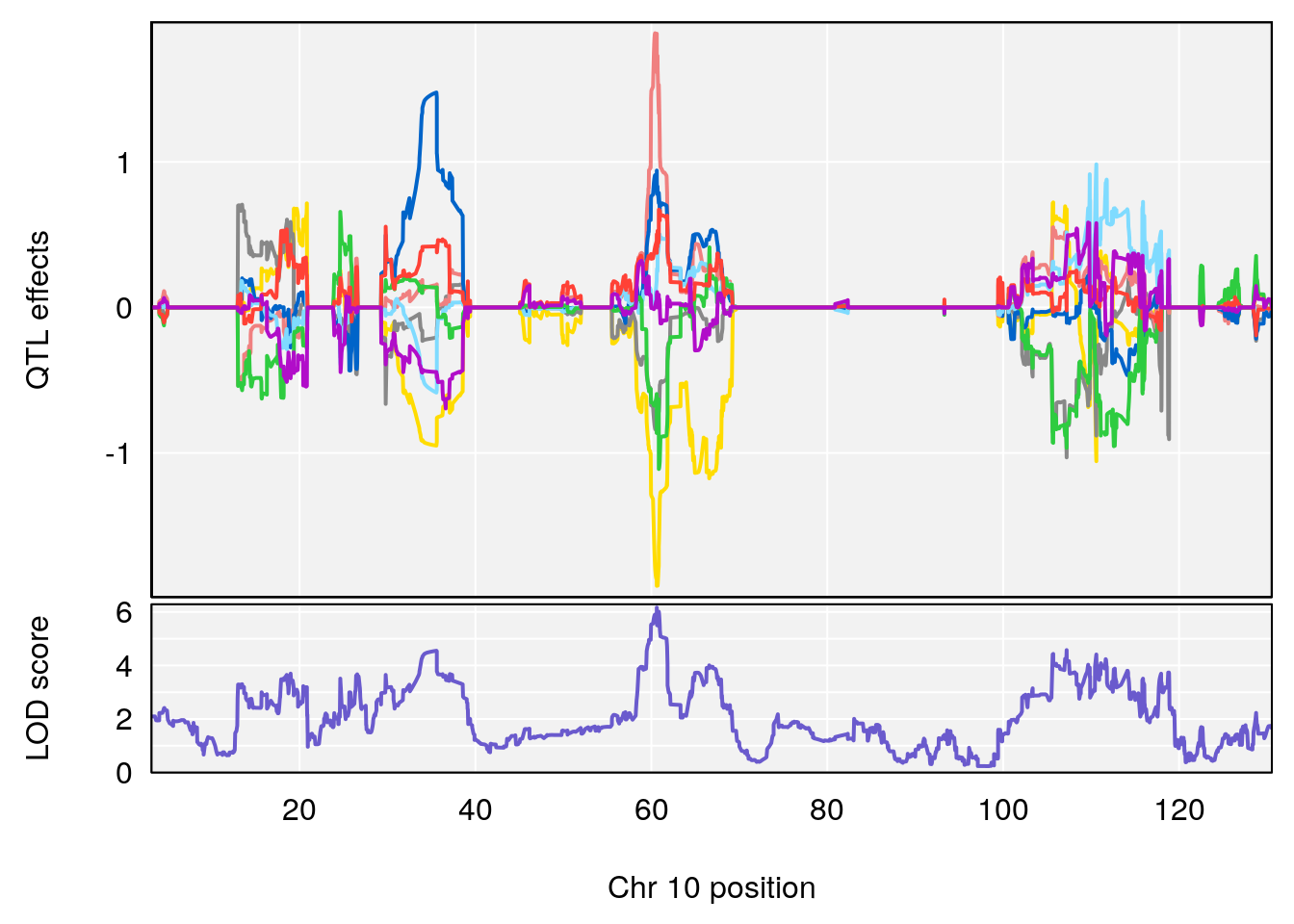
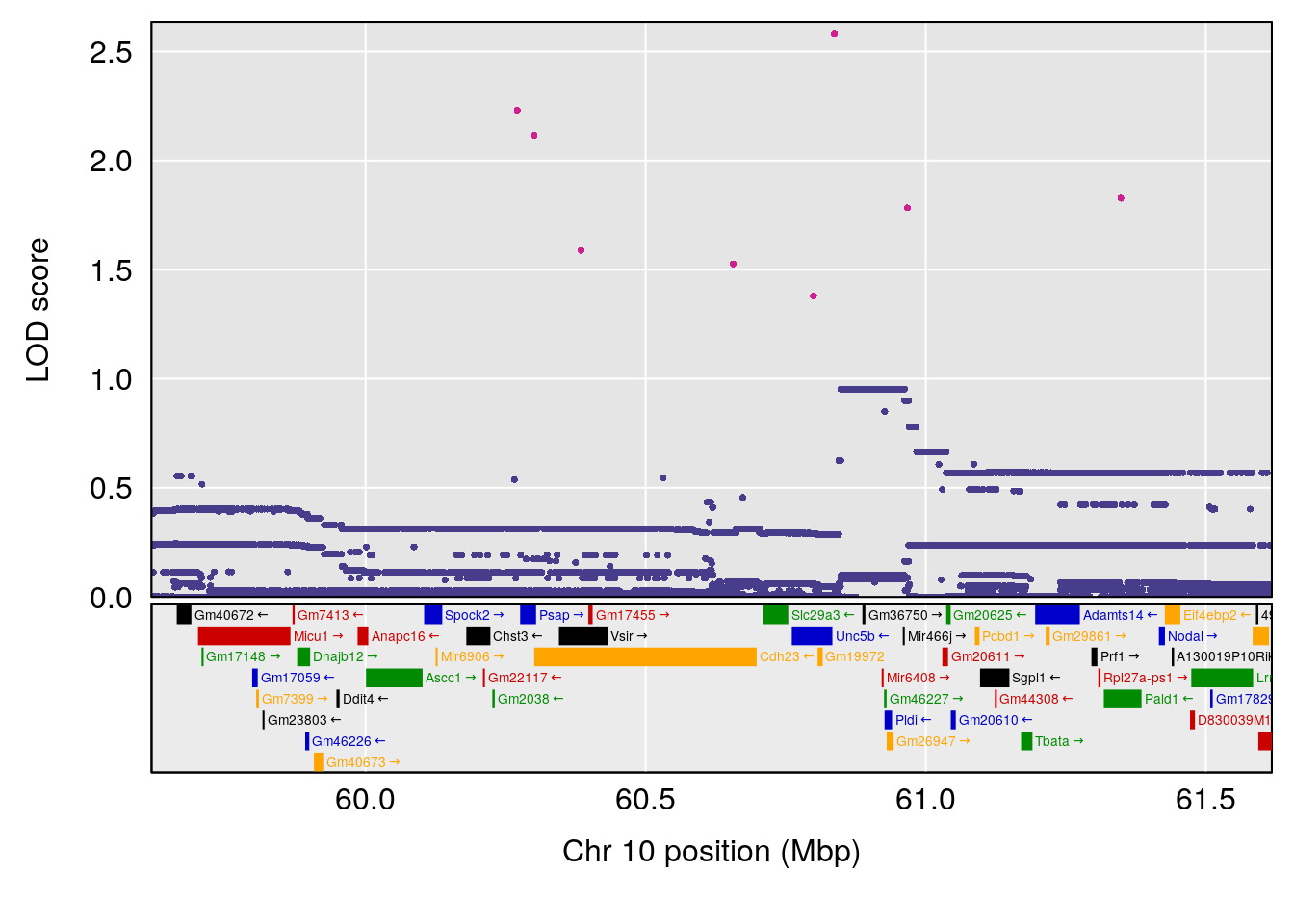
# [1] "Time.to.Threshold.Recovery.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 10 60.61789 6.163288 59.50257 61.79358
# [1] 1
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "Time.to.Projected.Recovery.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
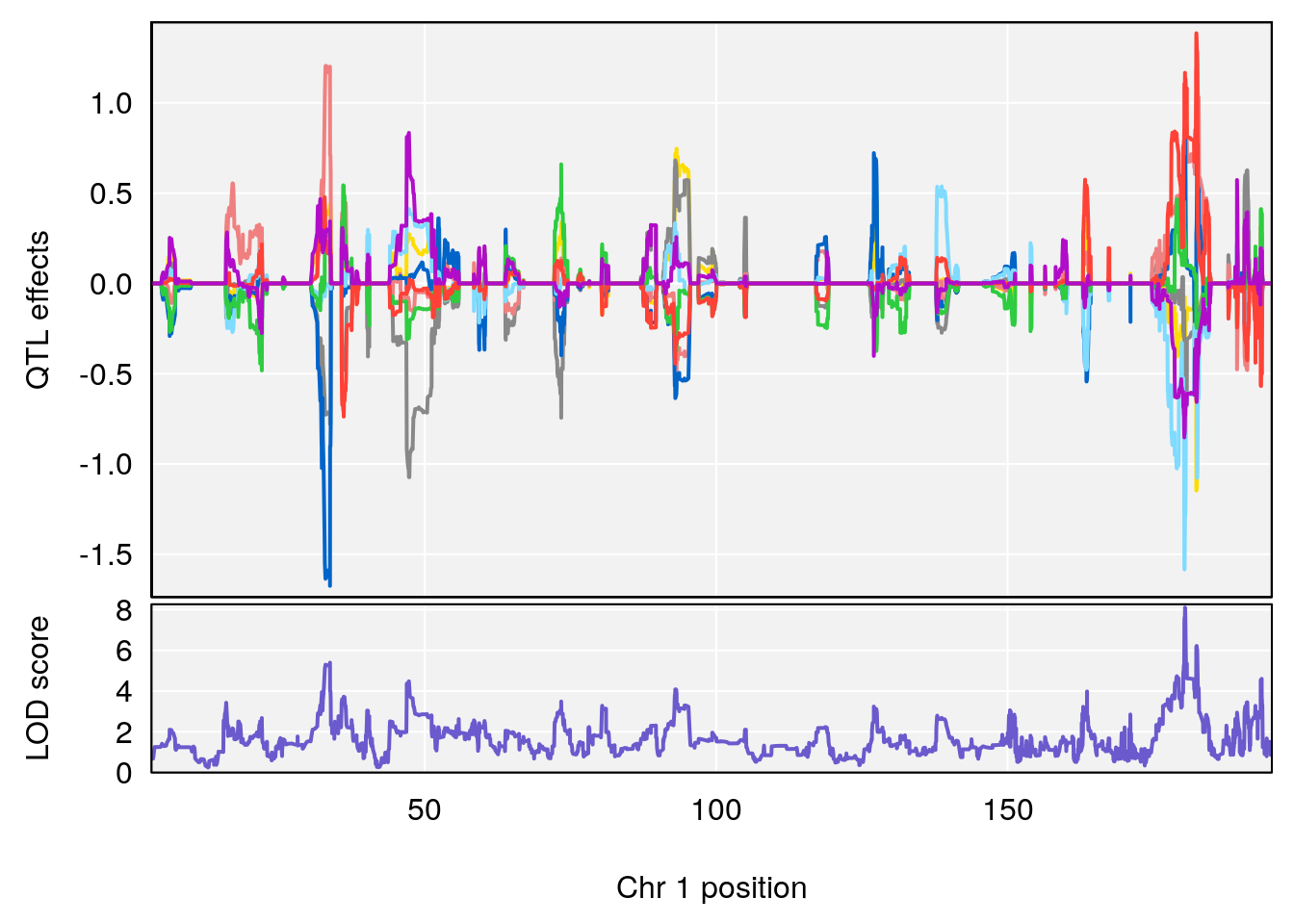
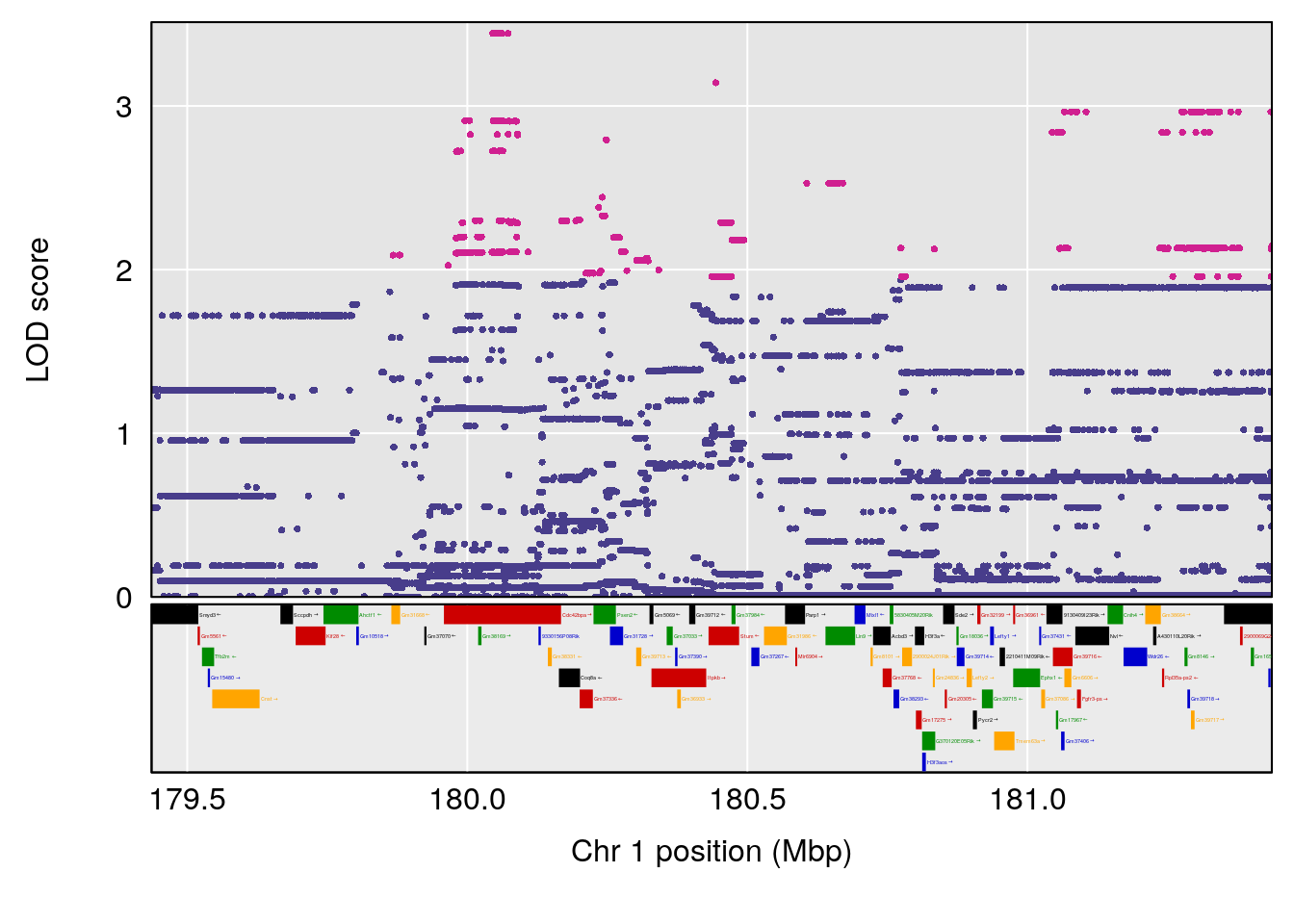
# 1 1 pheno1 1 180.4362 8.101813 180.3185 180.5471
# [1] 1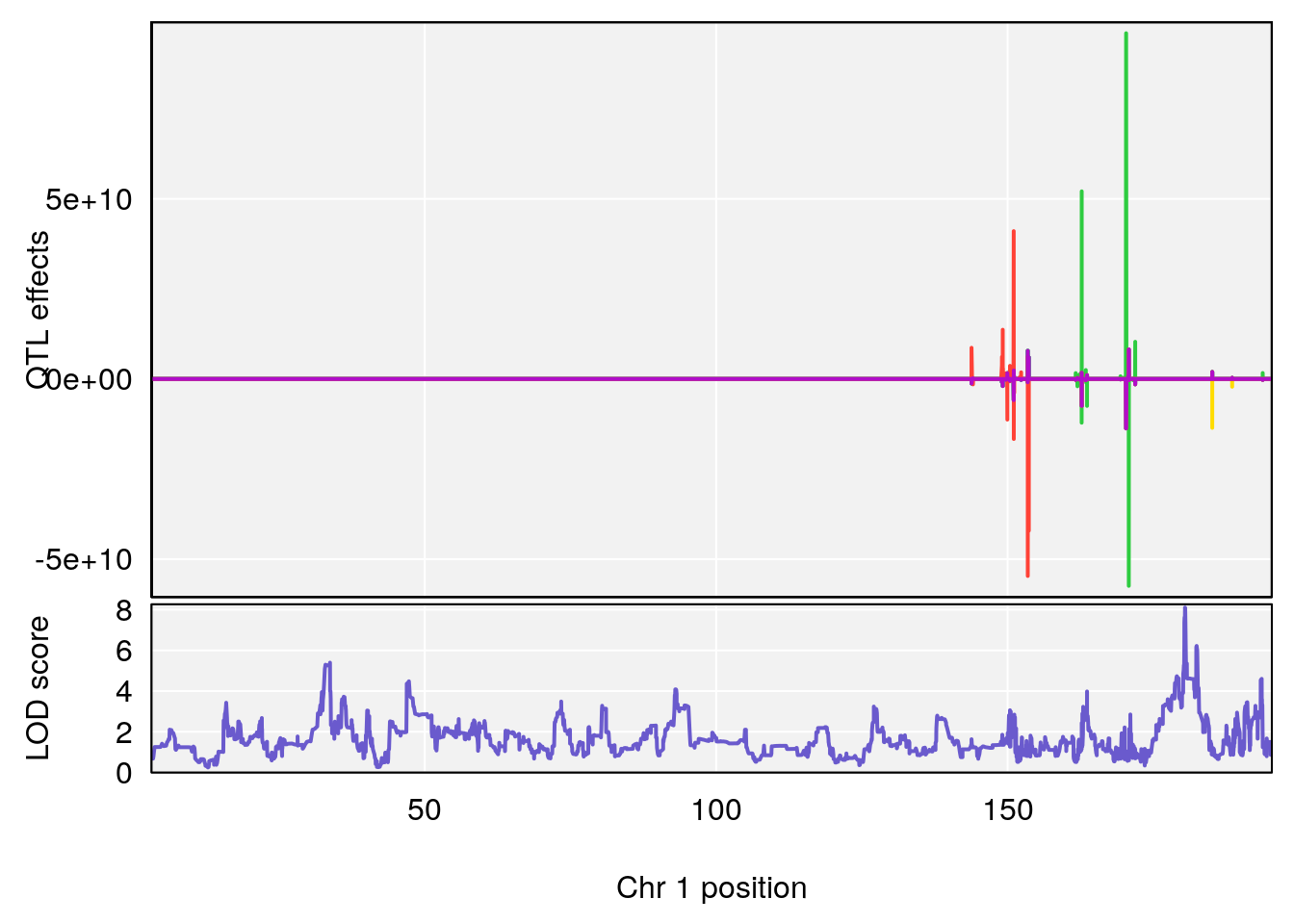
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
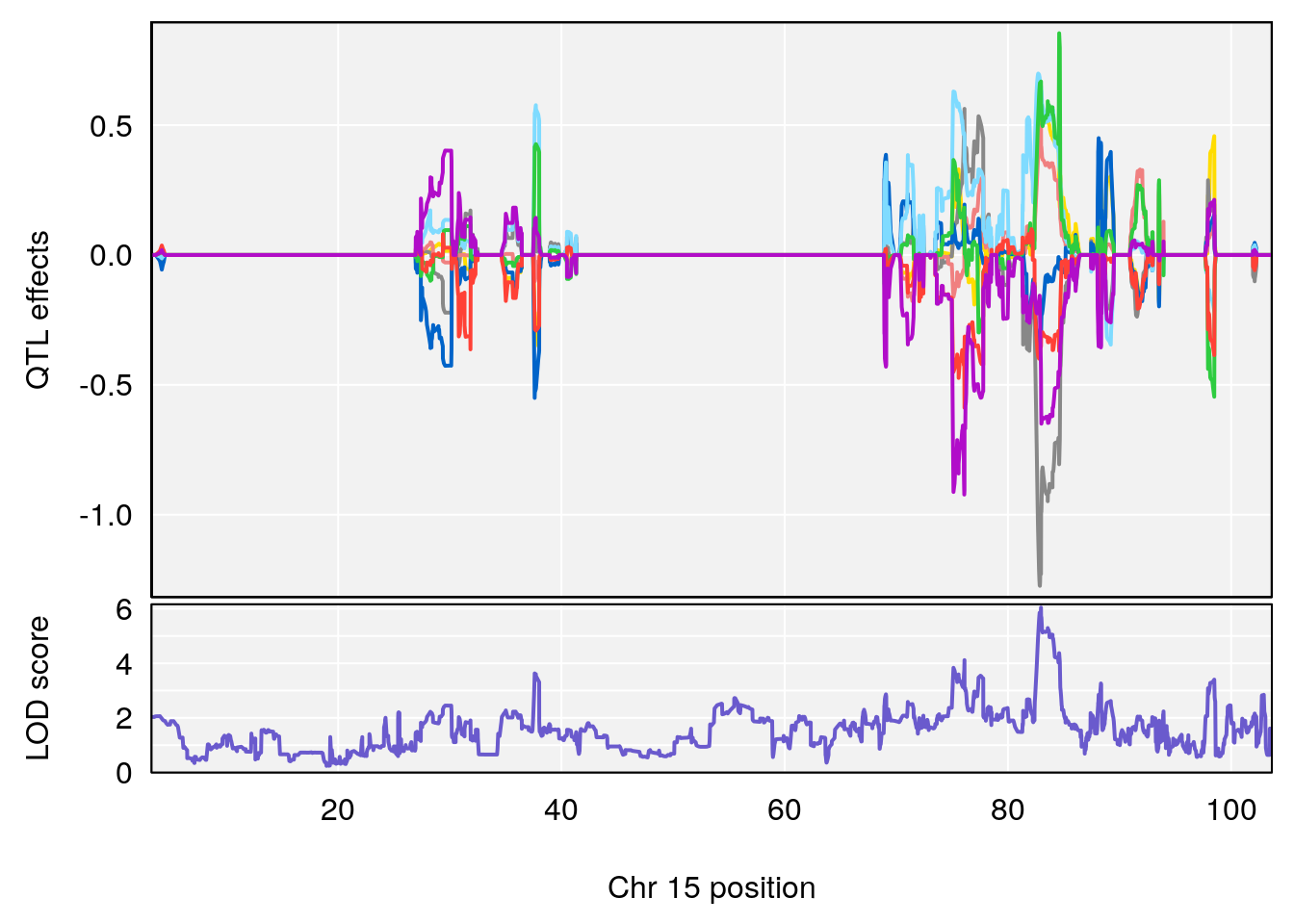
# [1] "Start.of.Recovery.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
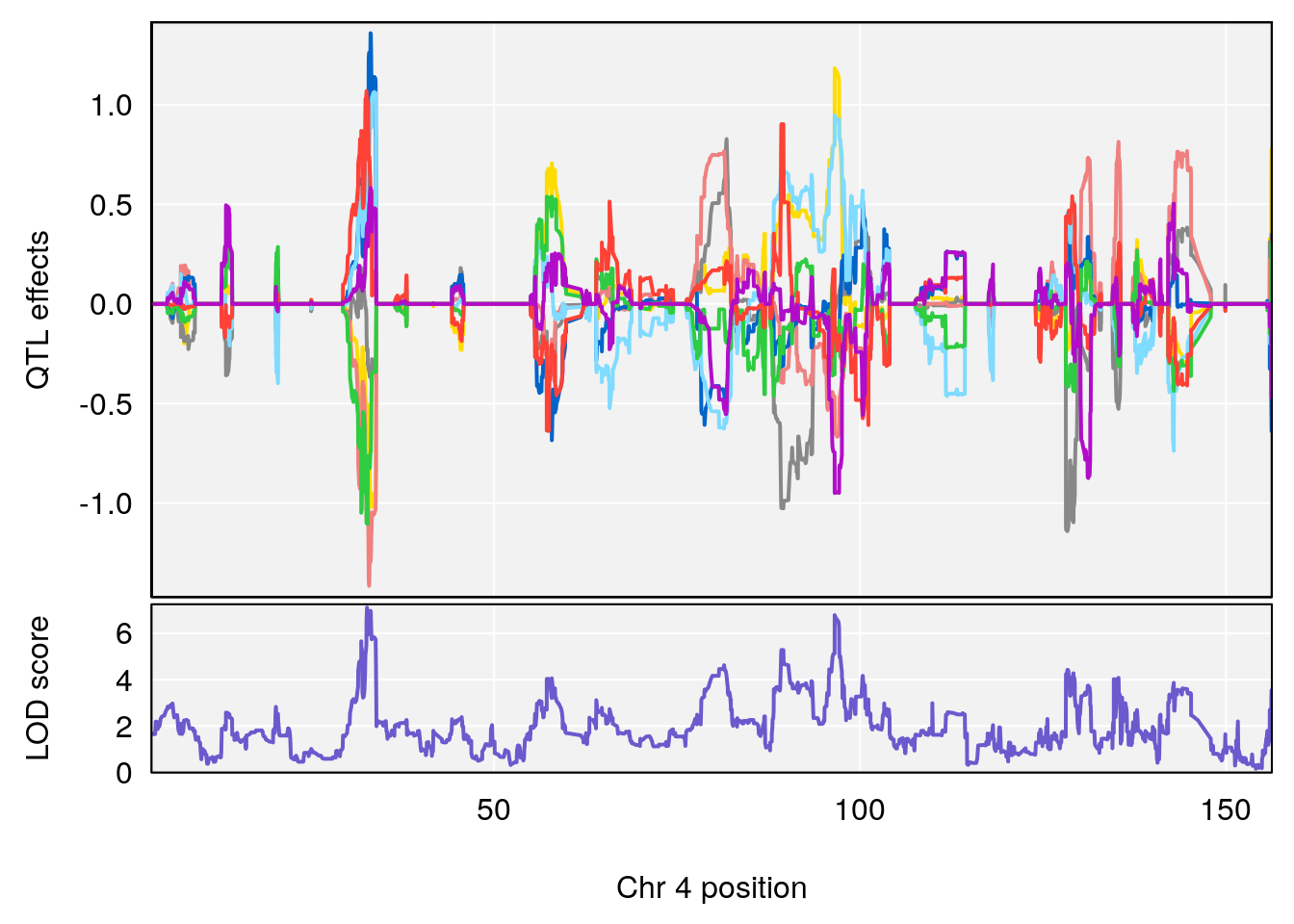
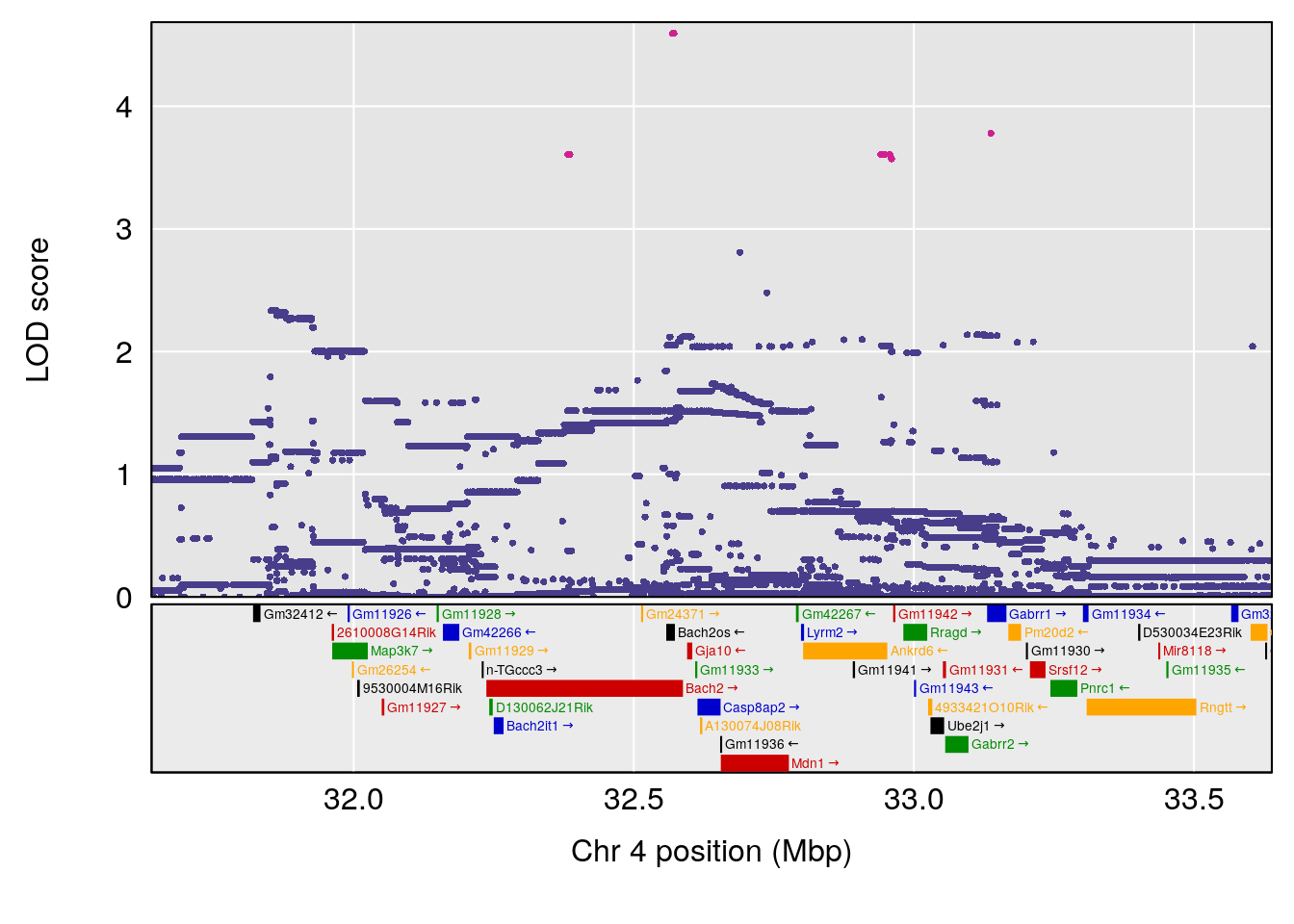
# 1 1 pheno1 4 32.63908 7.105498 31.85079 97.15470
# 2 1 pheno1 7 78.55130 6.935599 78.43609 79.07697
# 3 1 pheno1 15 82.95976 6.085487 76.08766 84.17742
# [1] 1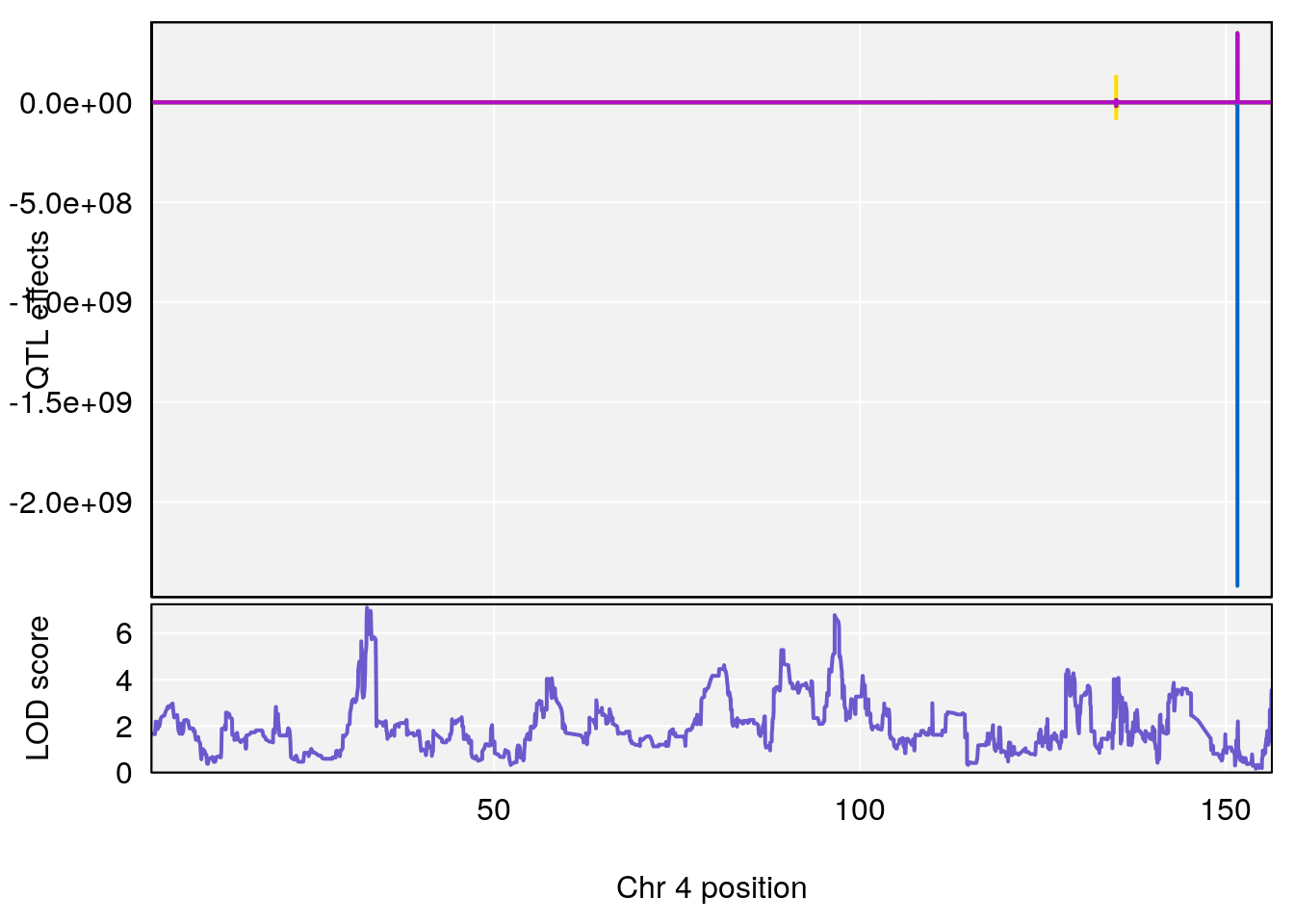
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
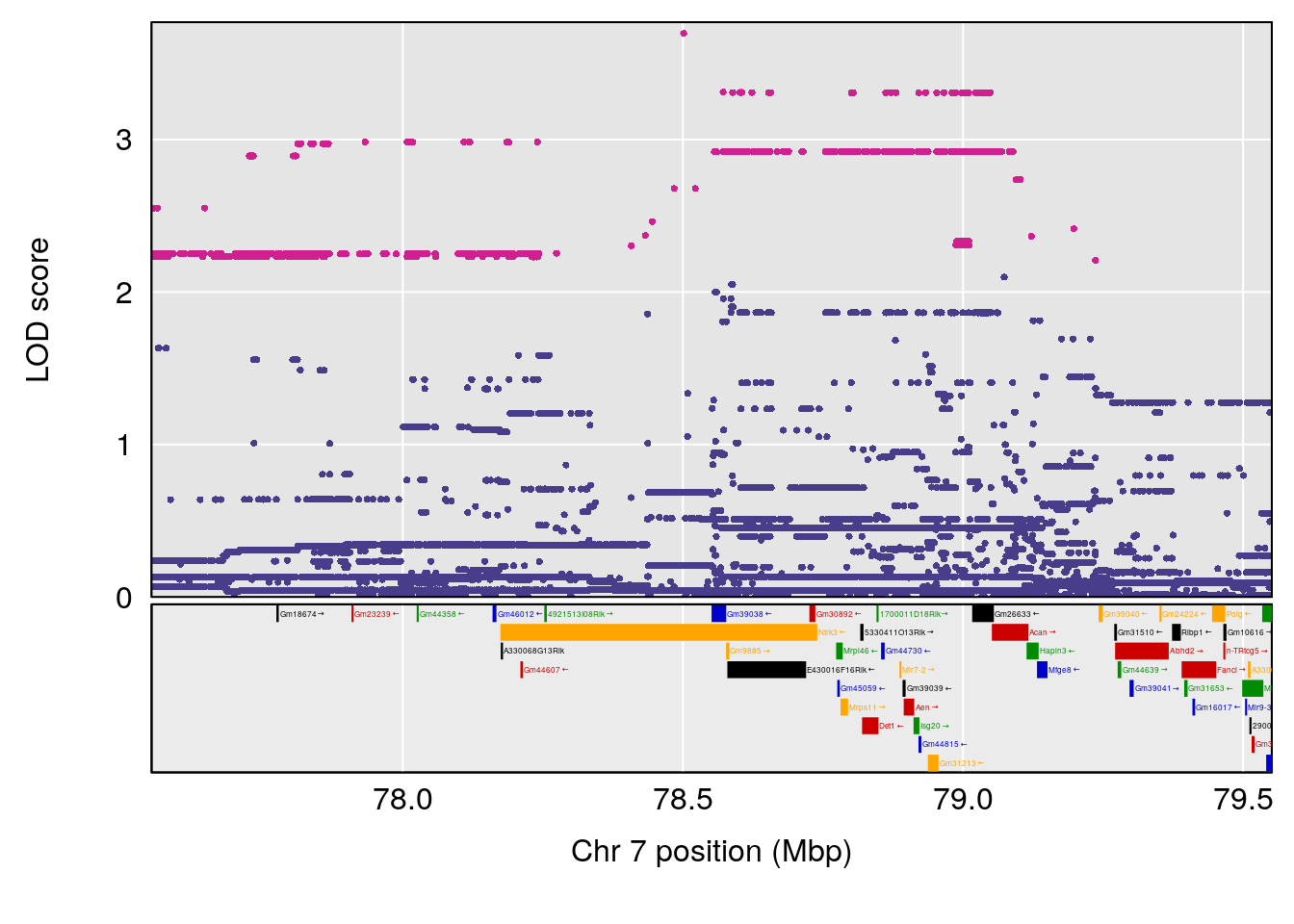
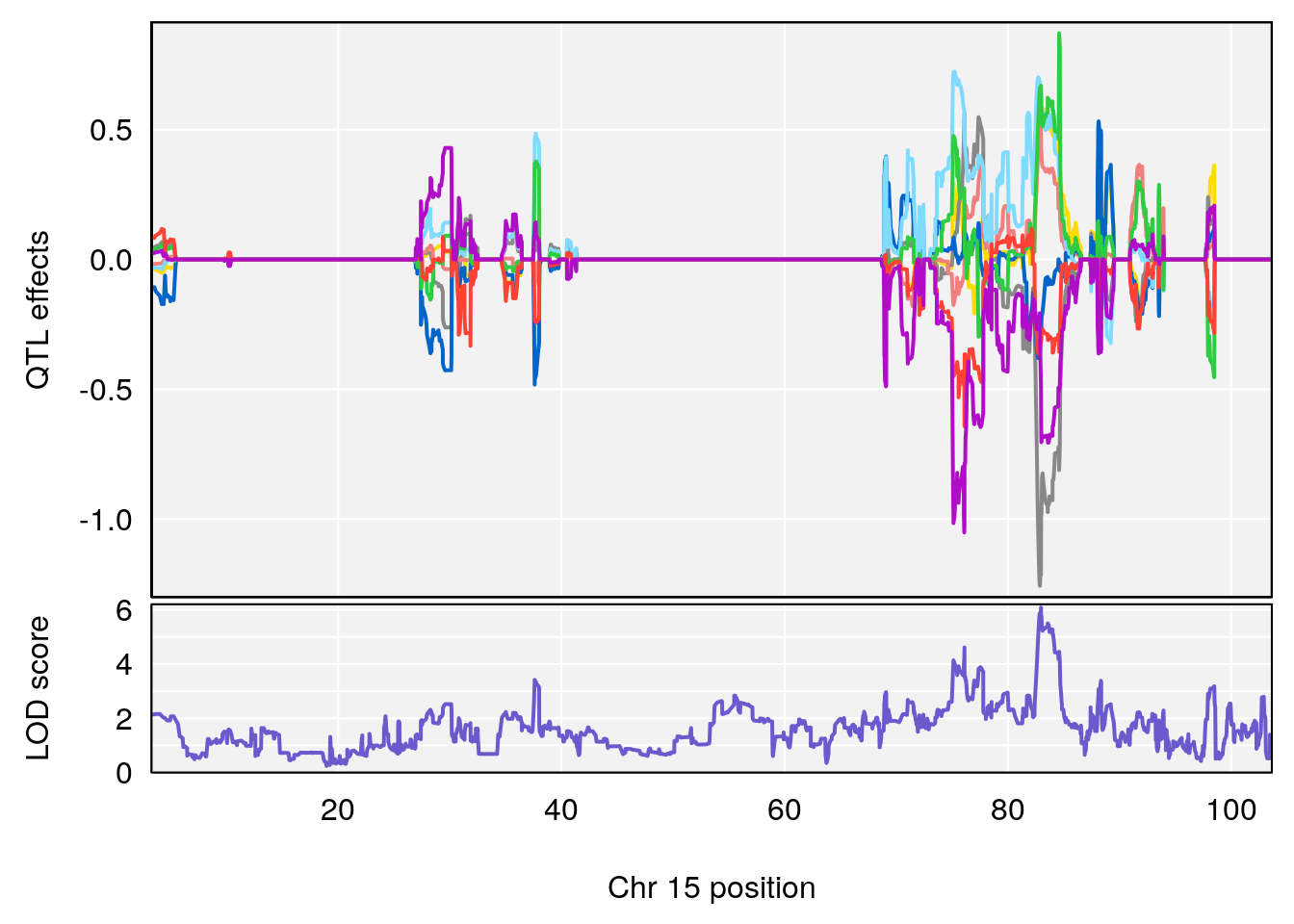
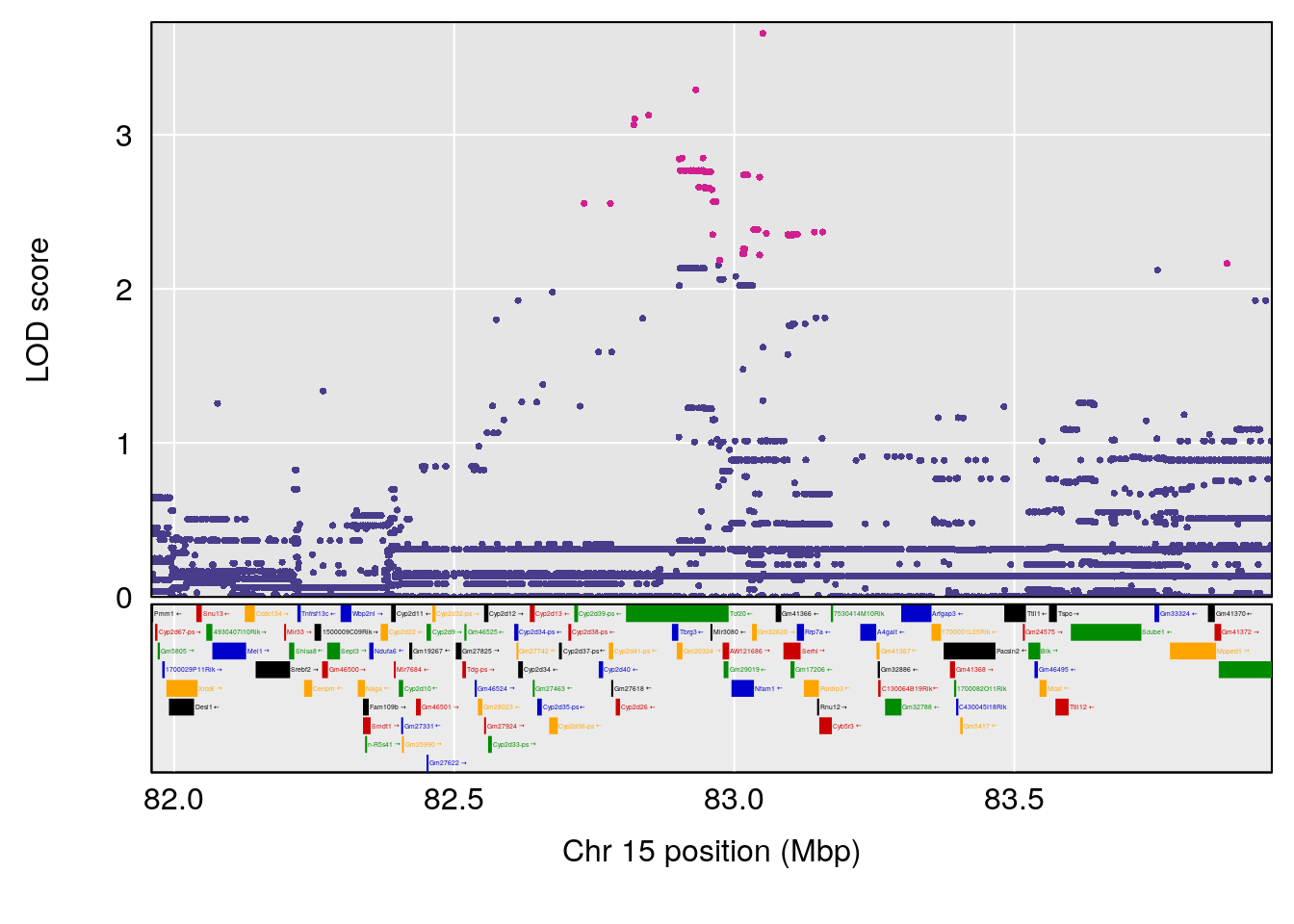
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 3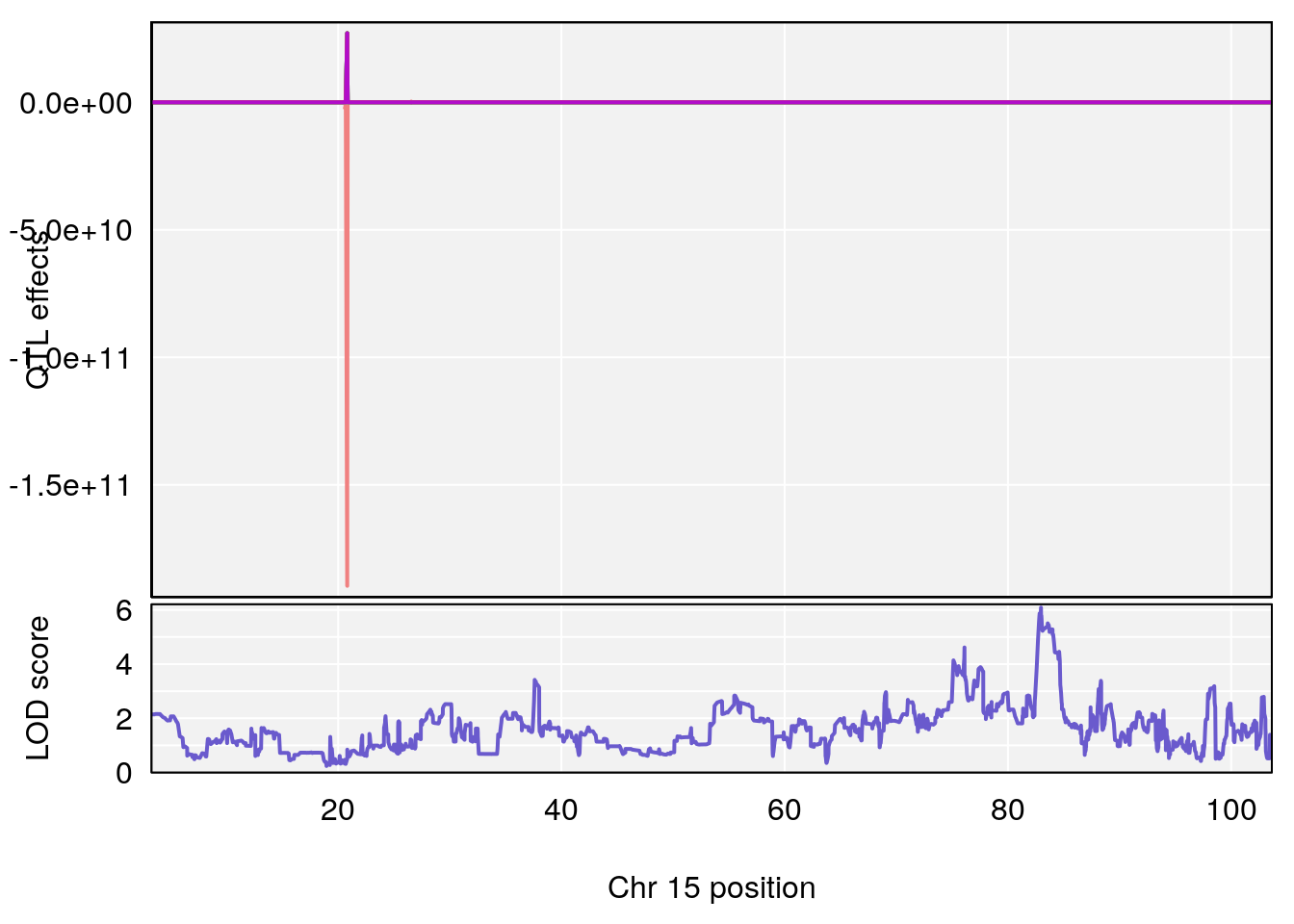
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "RR.Recovery.Rate.Hr"
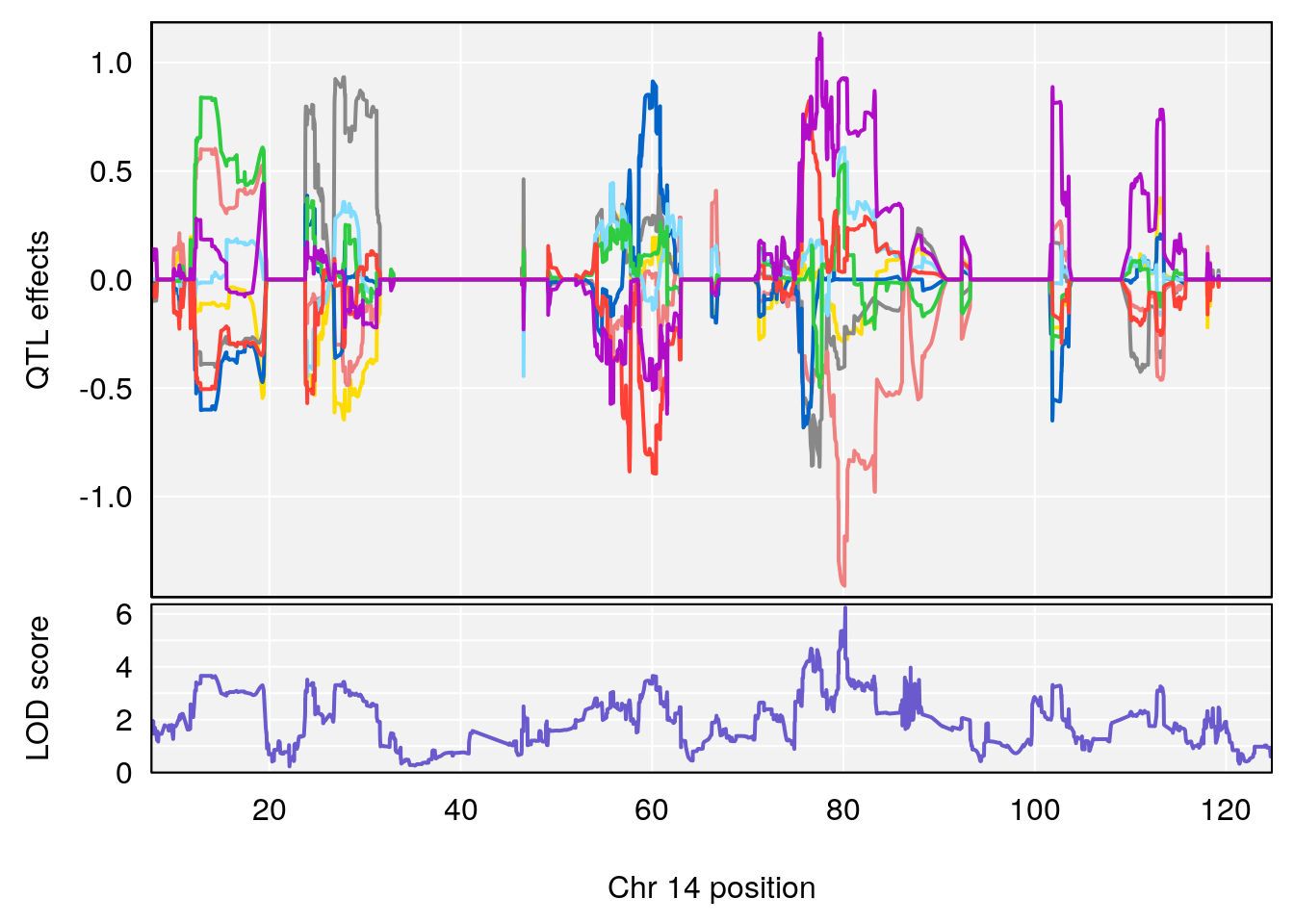
# lodindex lodcolumn chr pos lod ci_lo ci_hi
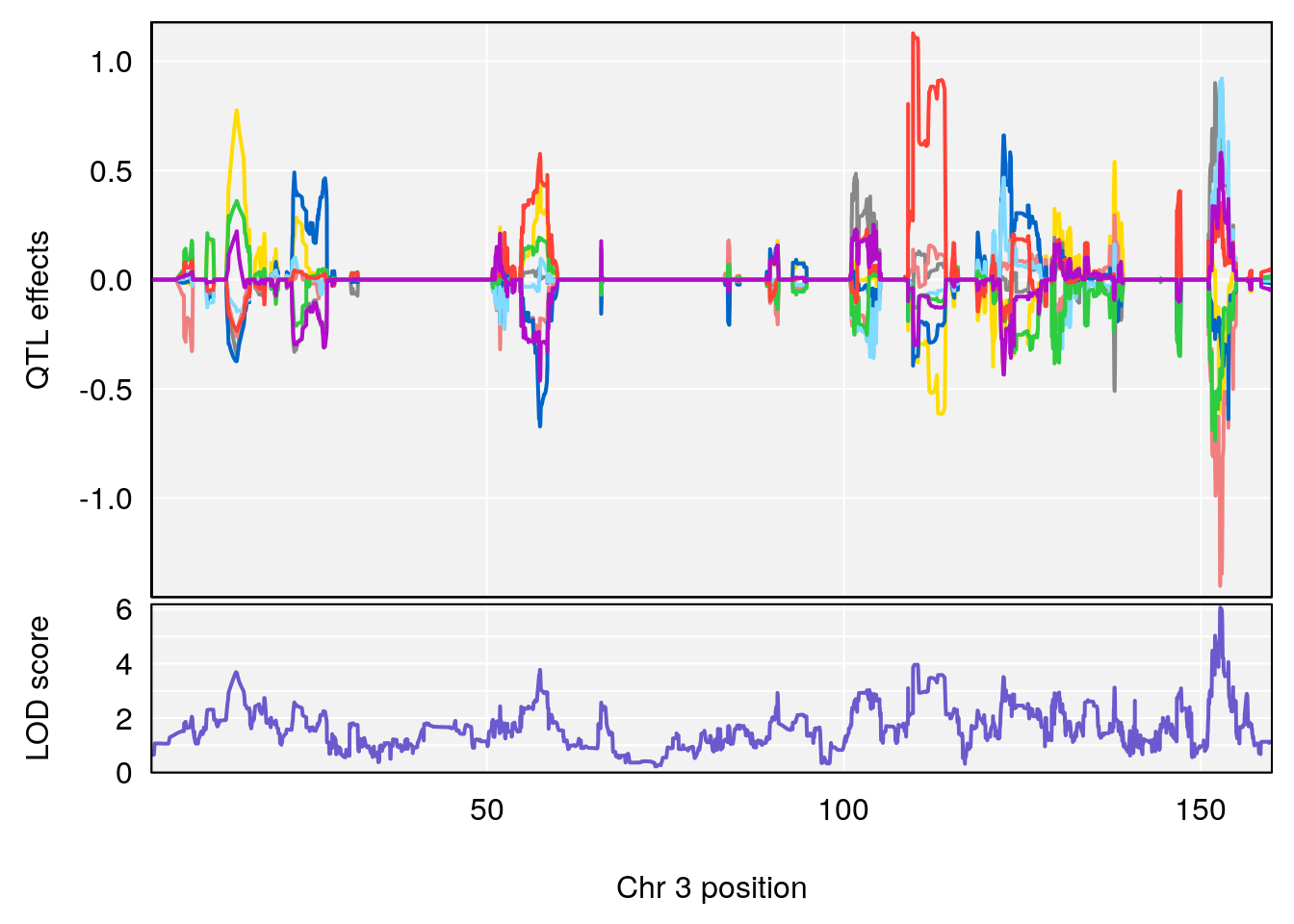
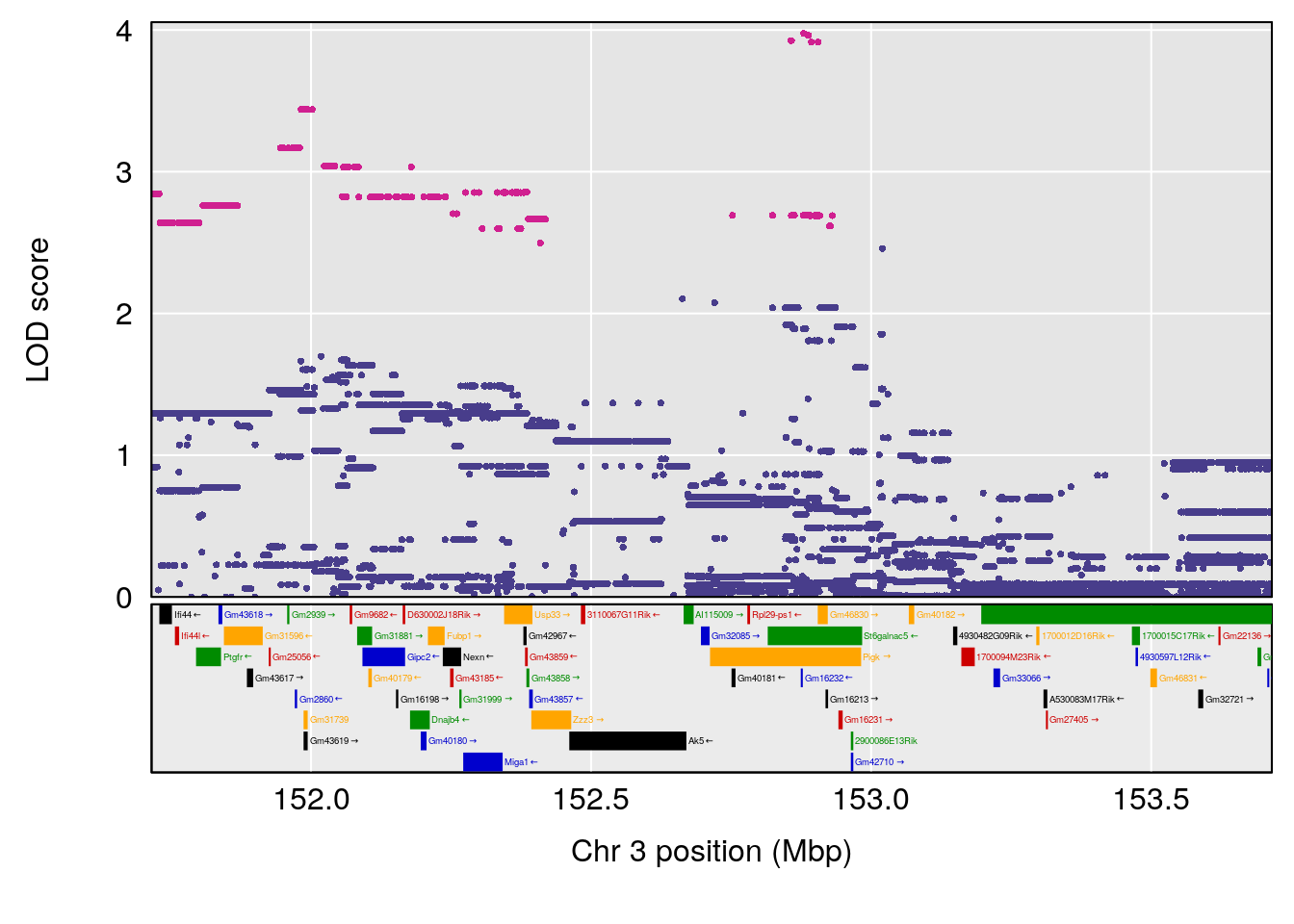
# 1 1 pheno1 3 152.71526 6.057558 151.92564 153.04206
# 2 1 pheno1 12 82.52881 6.219487 46.37269 82.84506
# [1] 1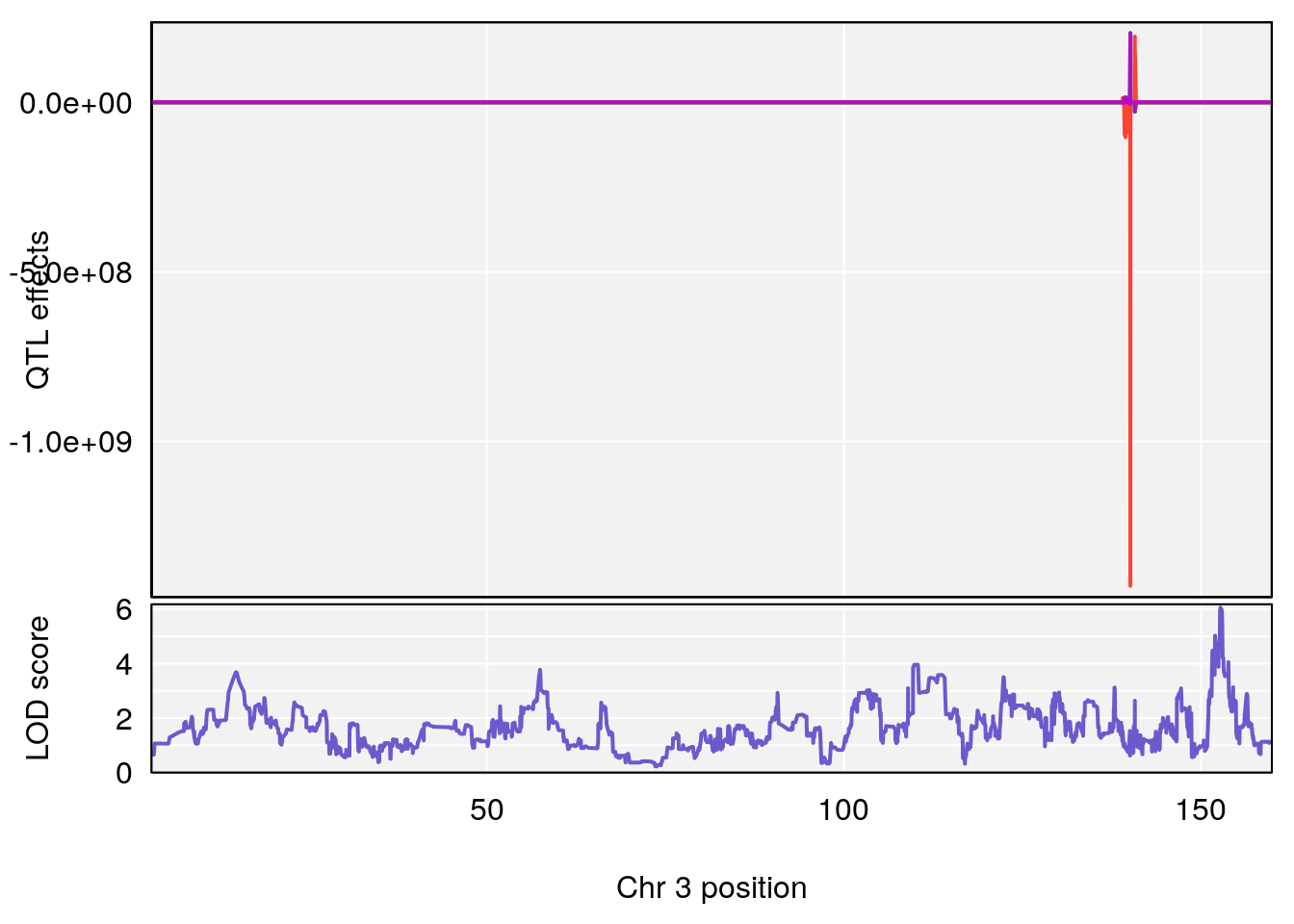
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2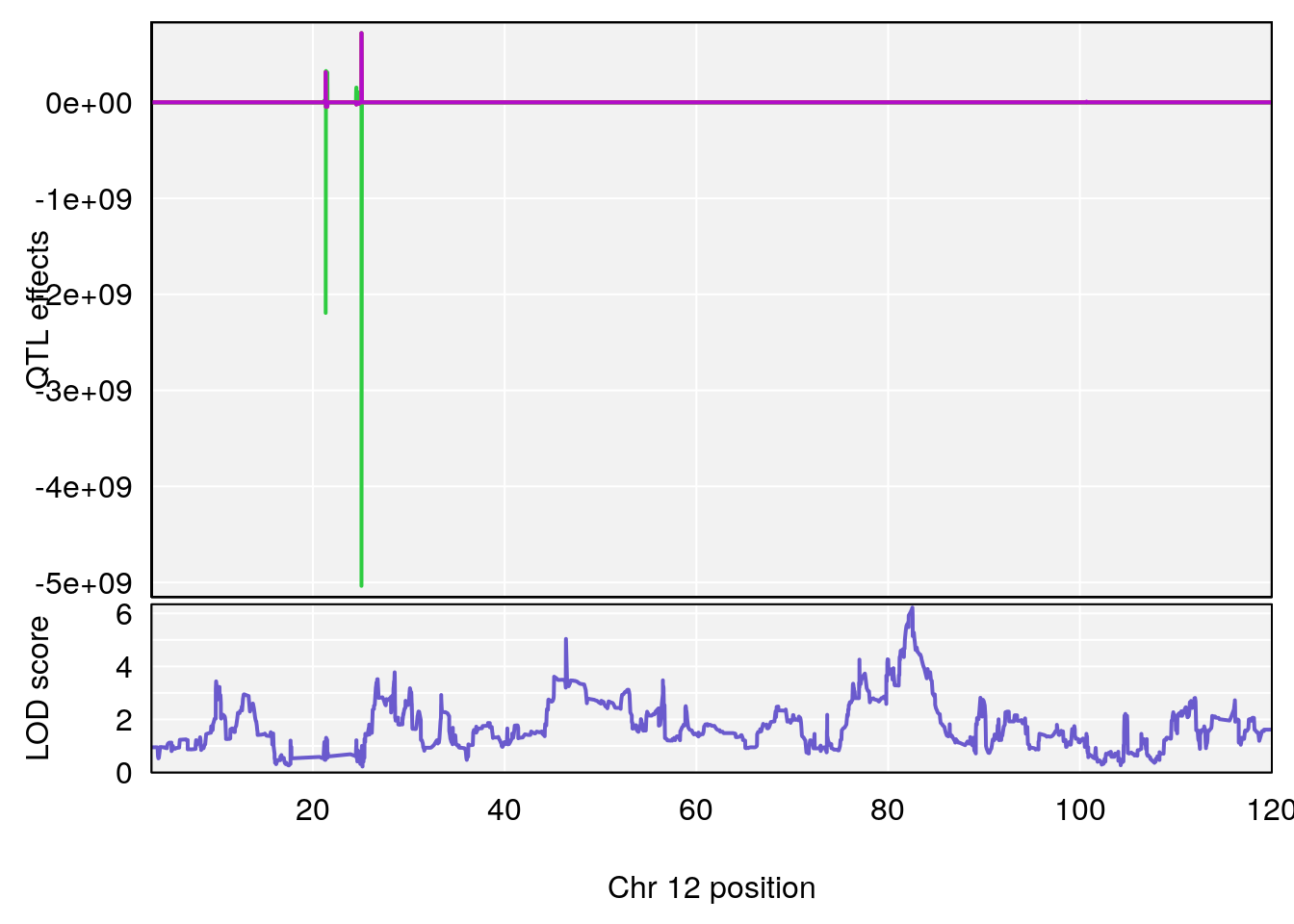
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "Min.Depression.BR"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 6 124.4987 6.882637 122.7691 125.5986
# [1] 1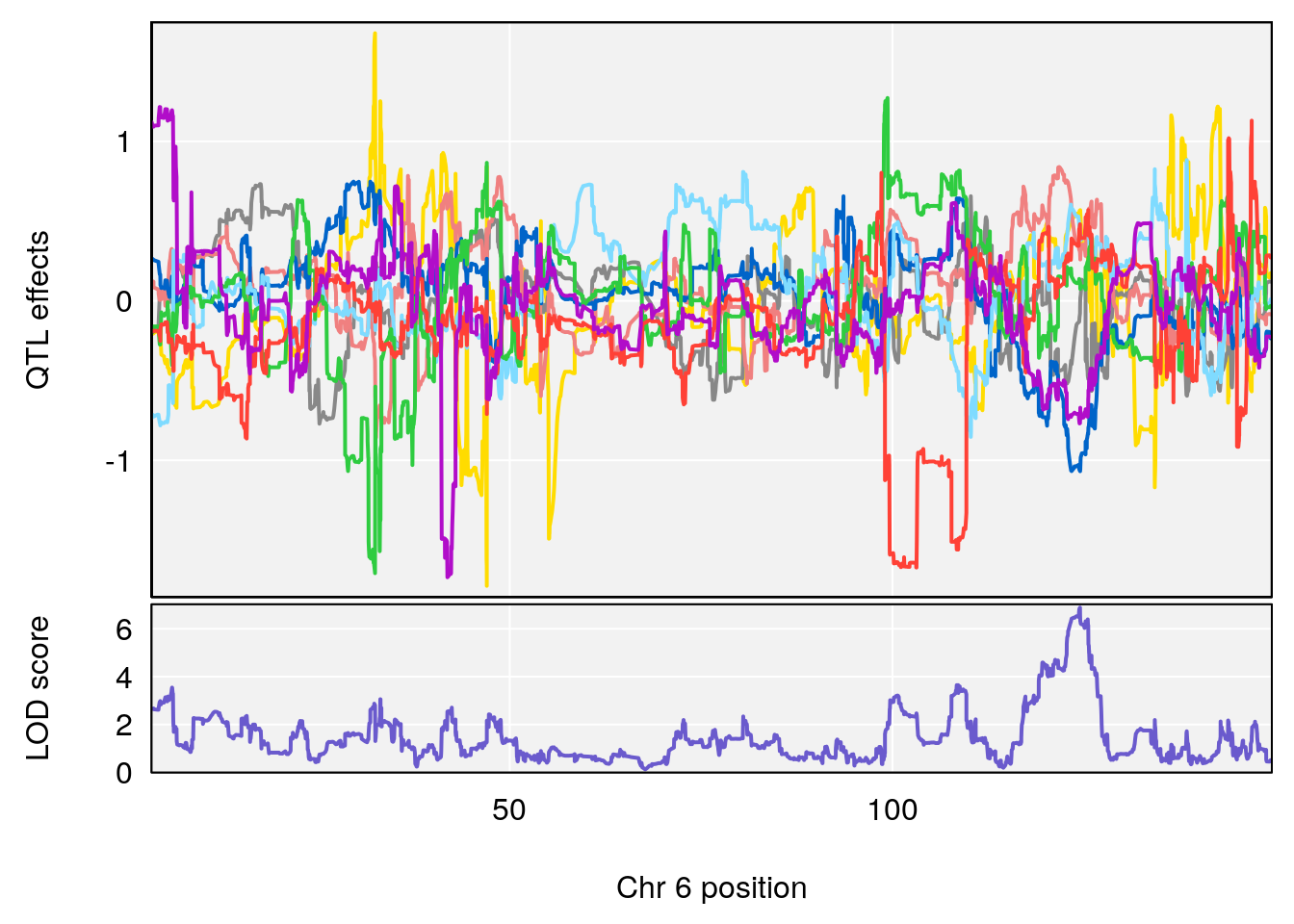
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
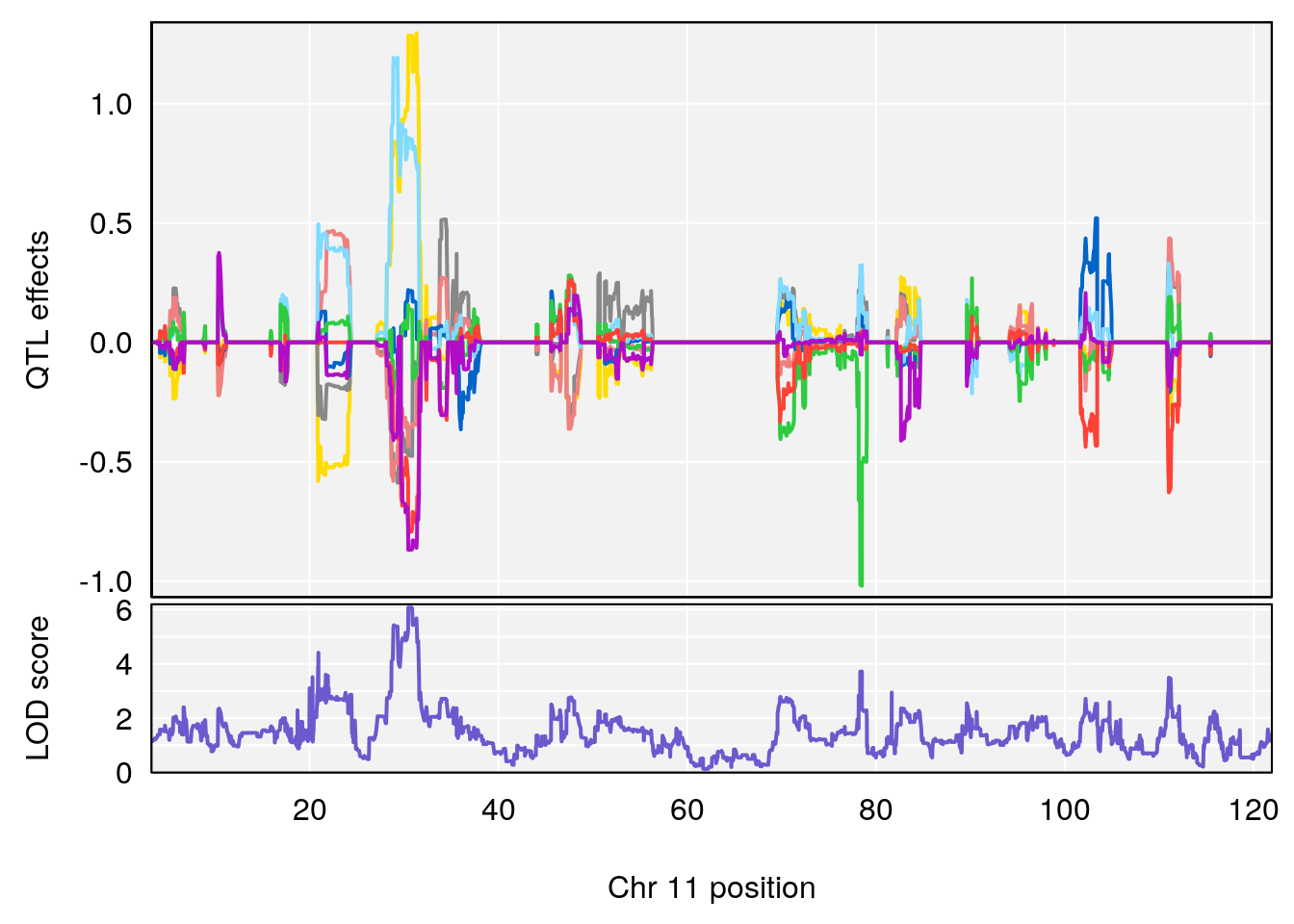
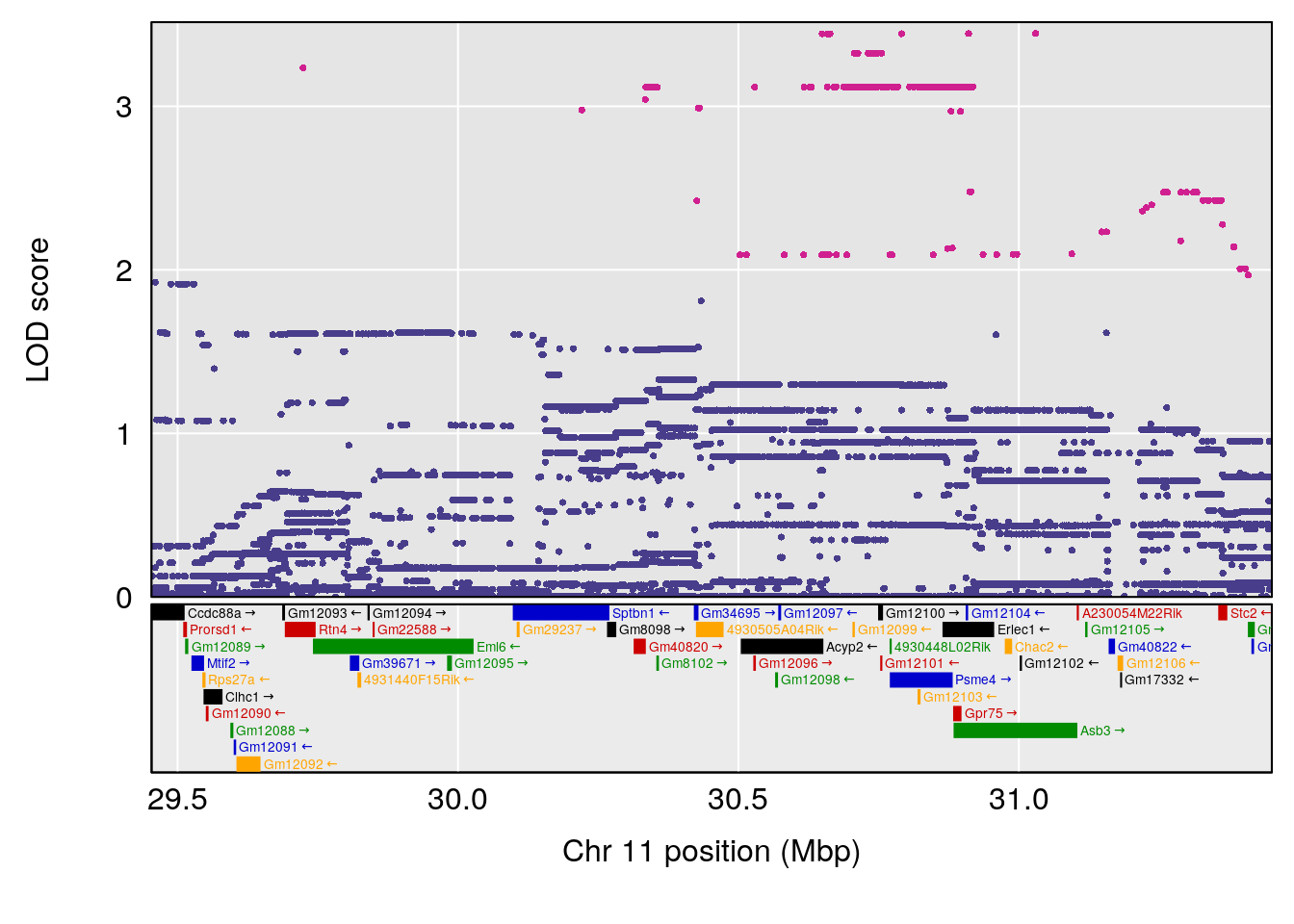
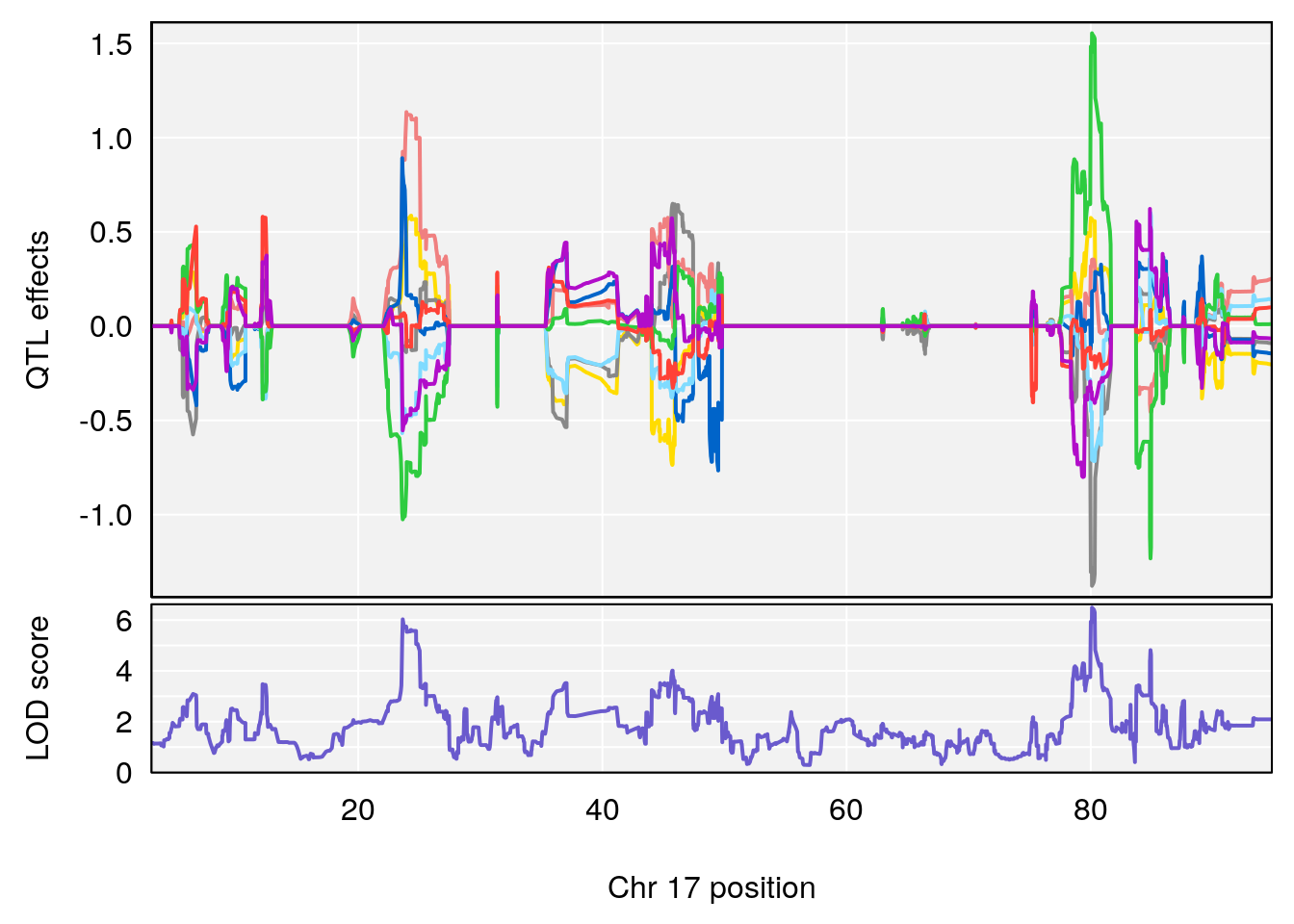
# [1] "Time.to.Mostly.Dead.Hr"
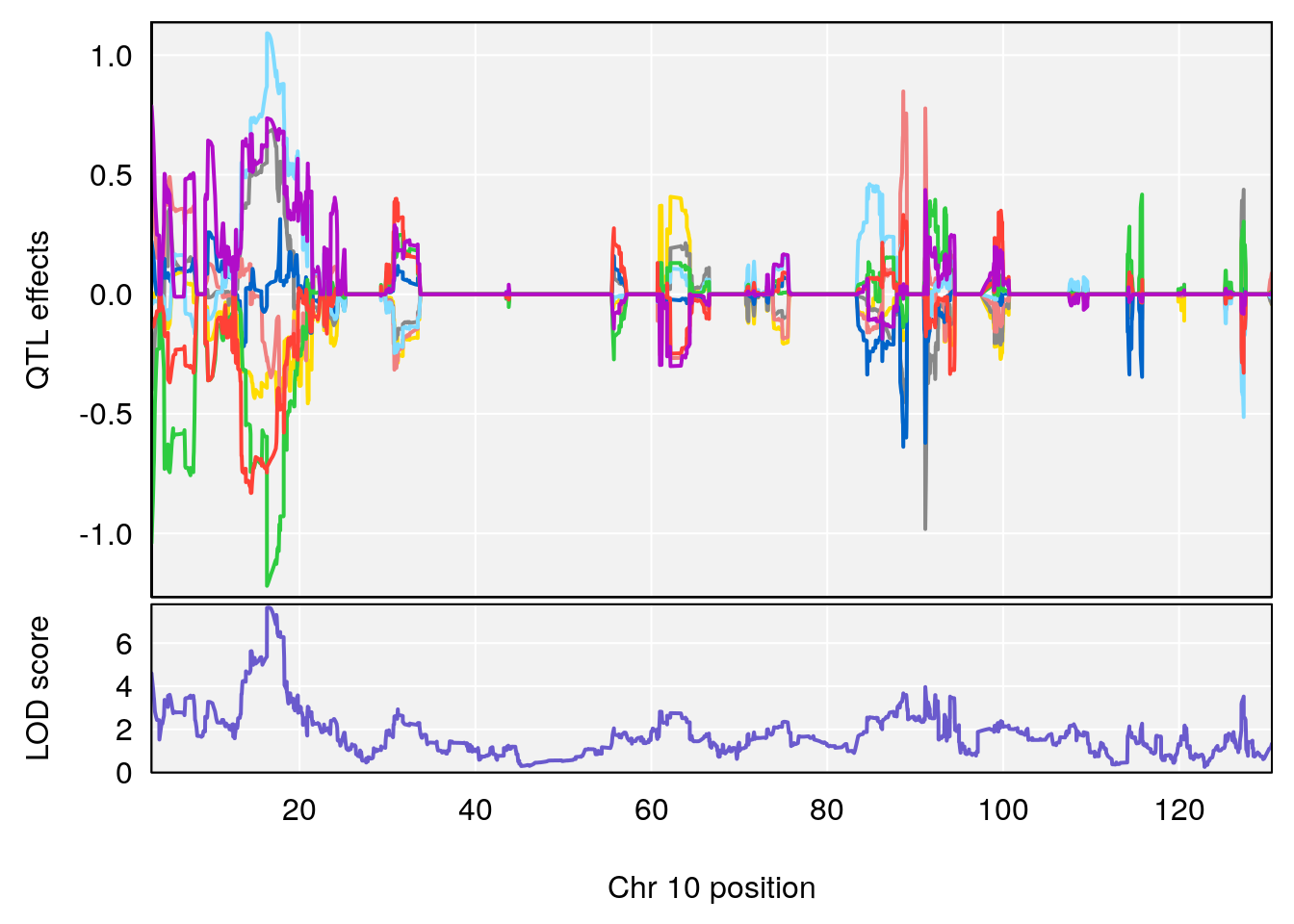
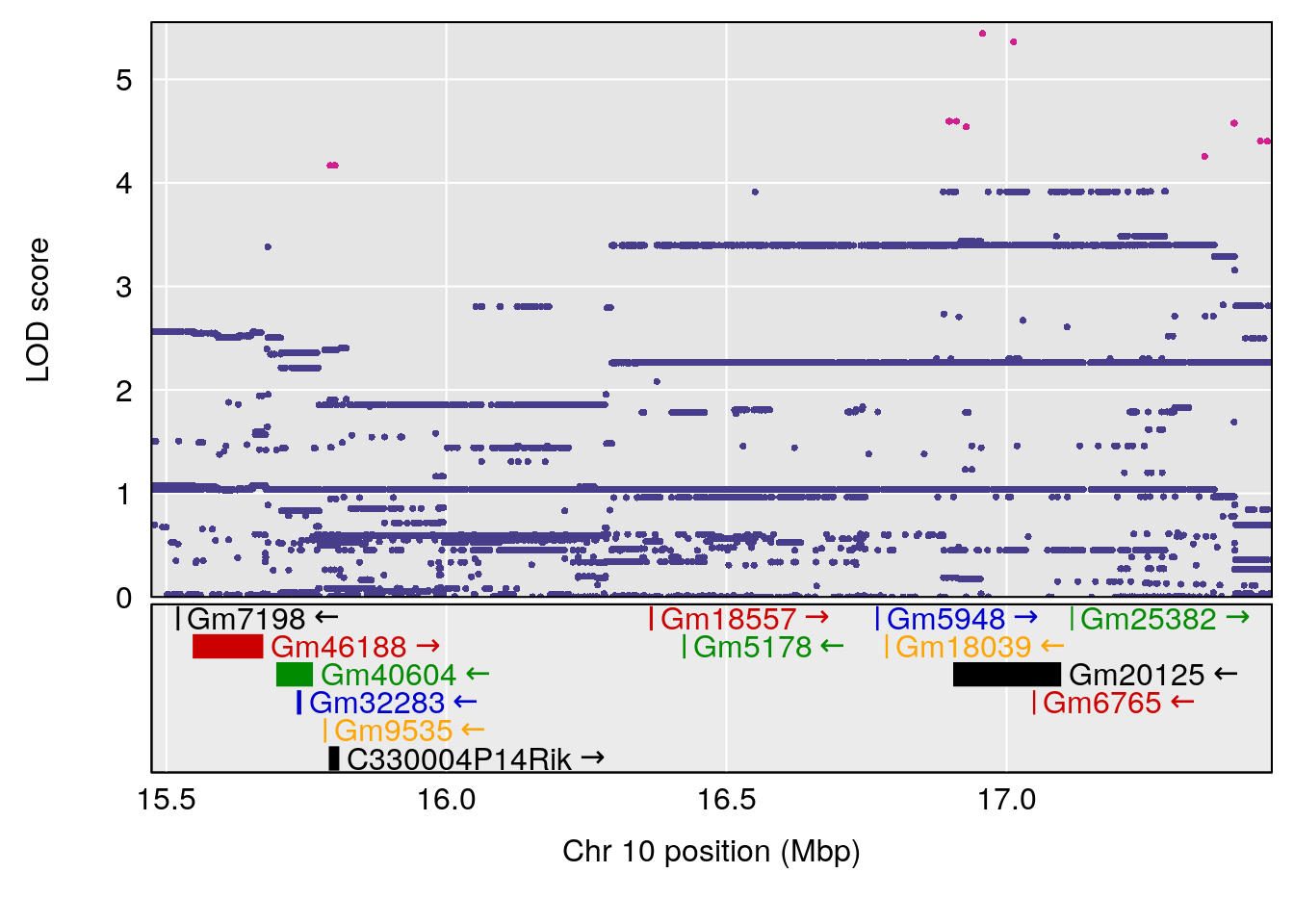
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 2 115.56002 6.027351 37.81483 132.19814
# 2 1 pheno1 10 16.47363 7.647309 16.28585 18.20019
# 3 1 pheno1 11 30.45329 6.071616 28.81595 31.56268
# 4 1 pheno1 17 80.09690 6.490398 23.55316 80.36008
# [1] 1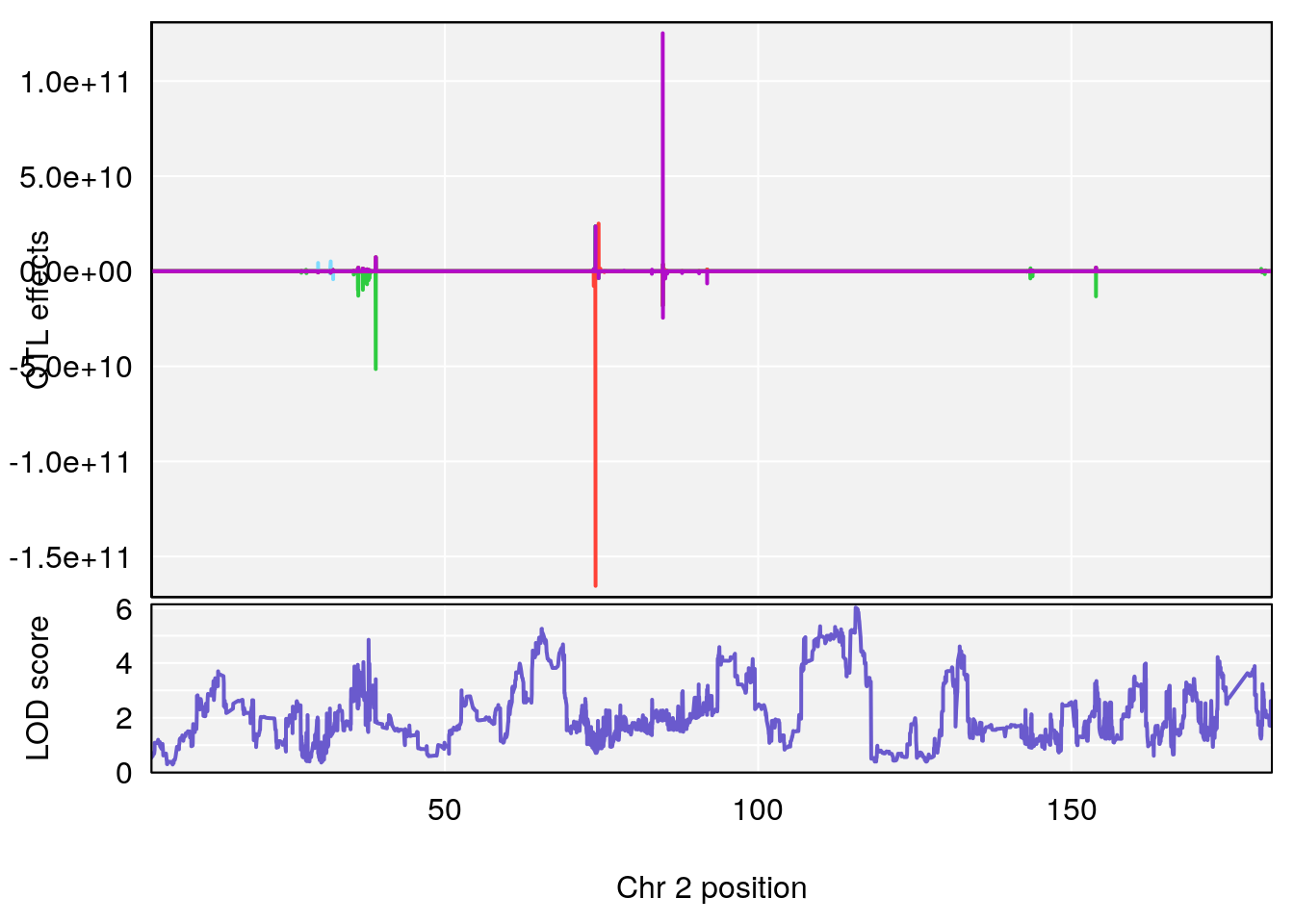
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2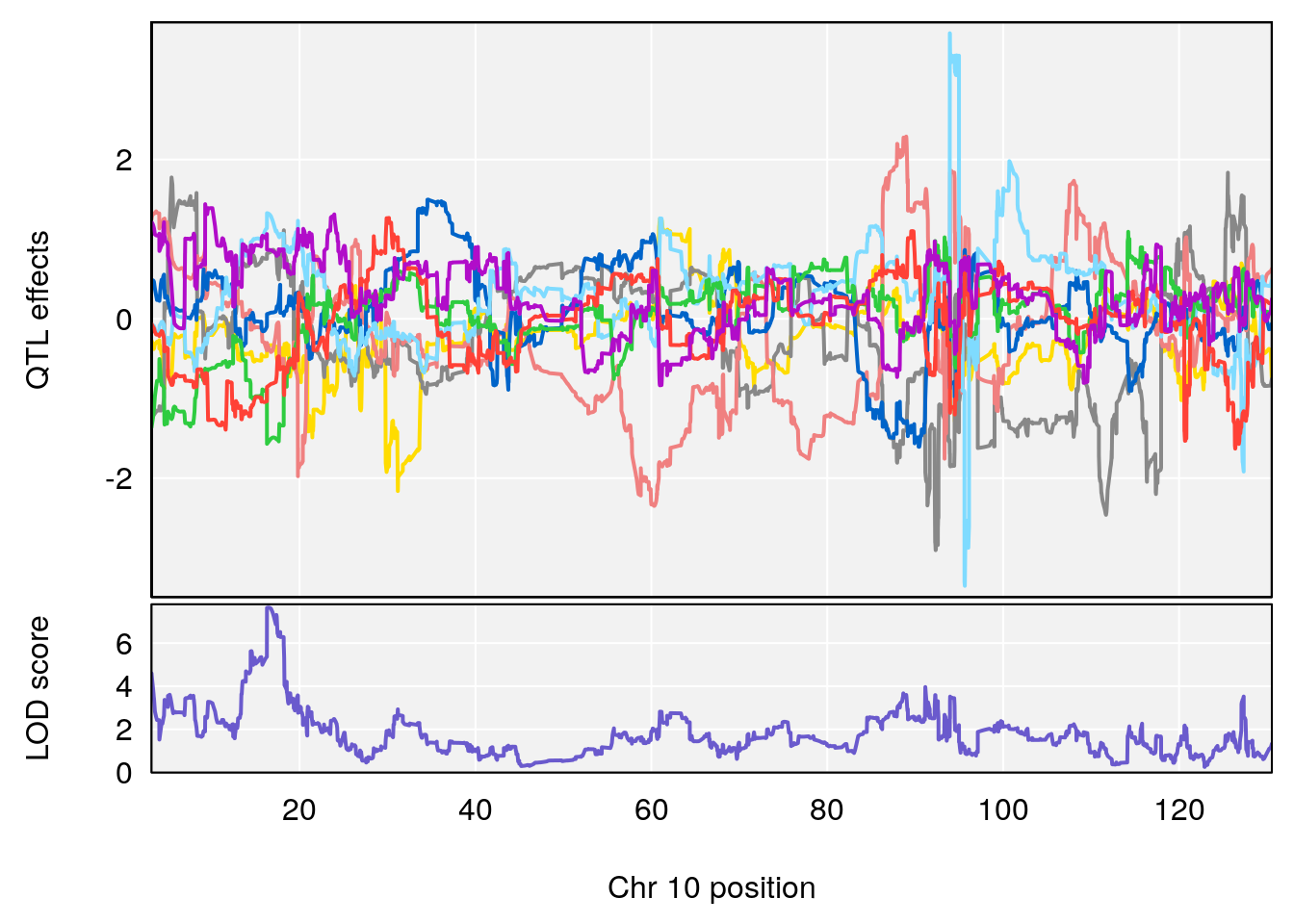
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 3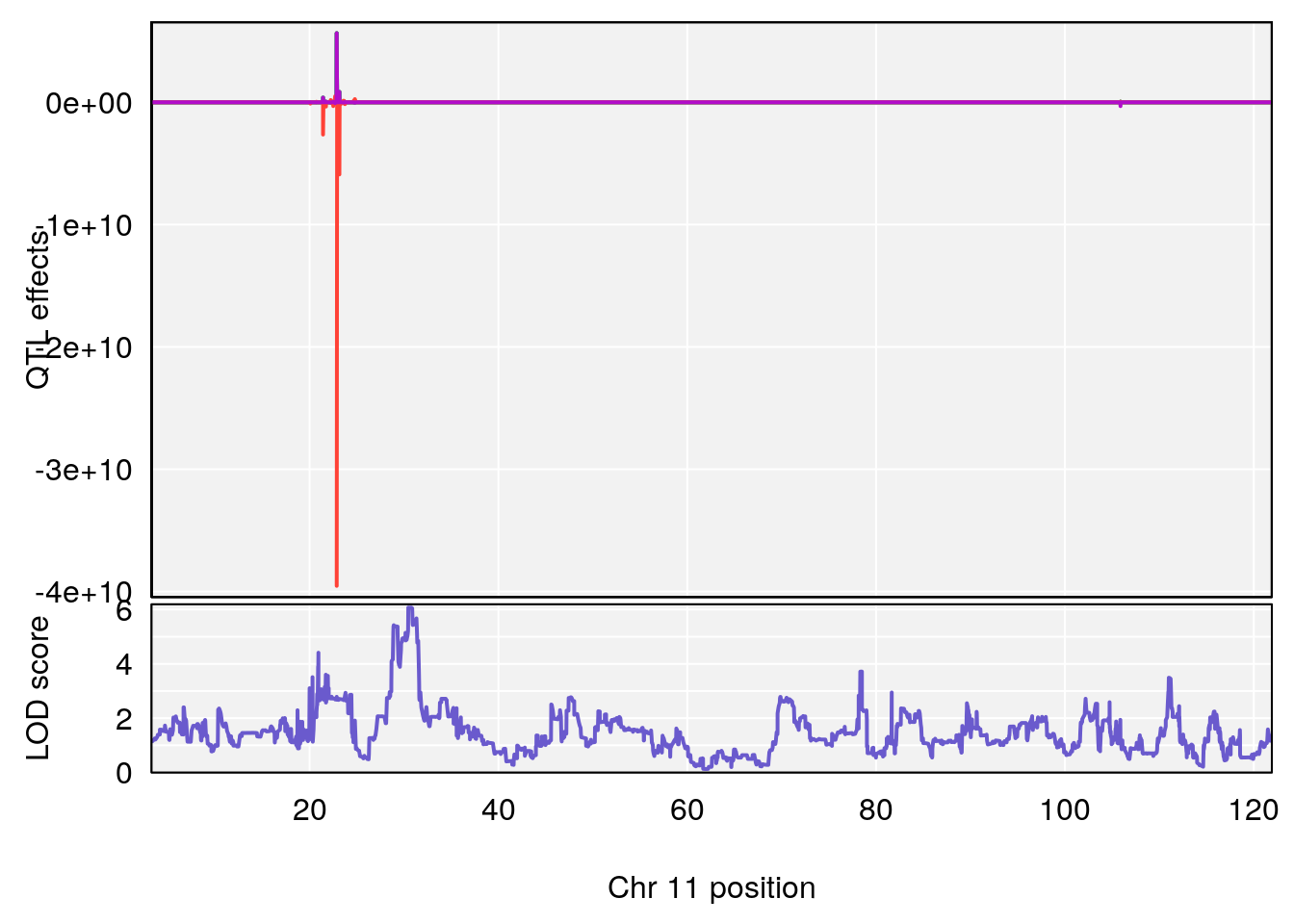
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 4
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
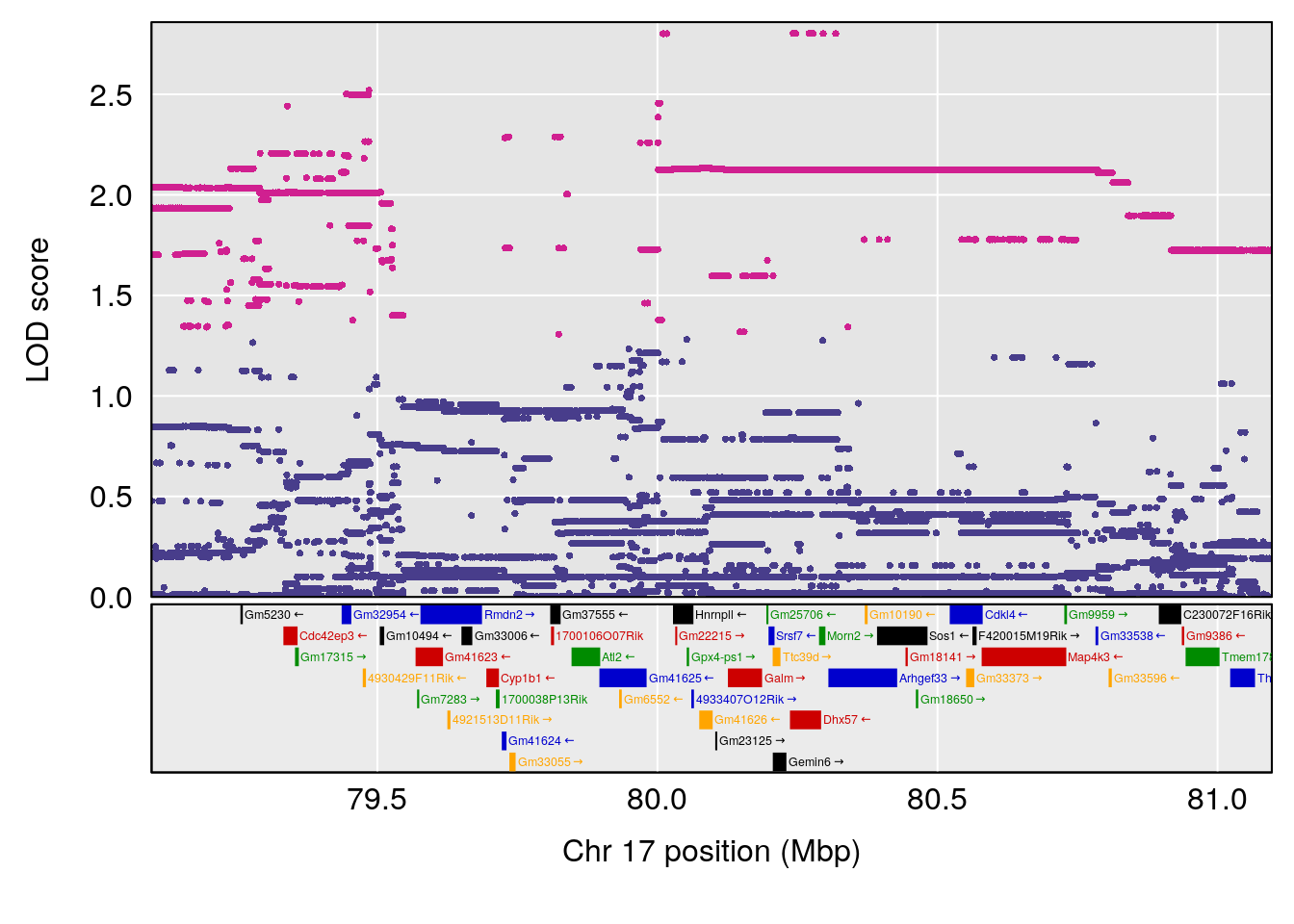
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
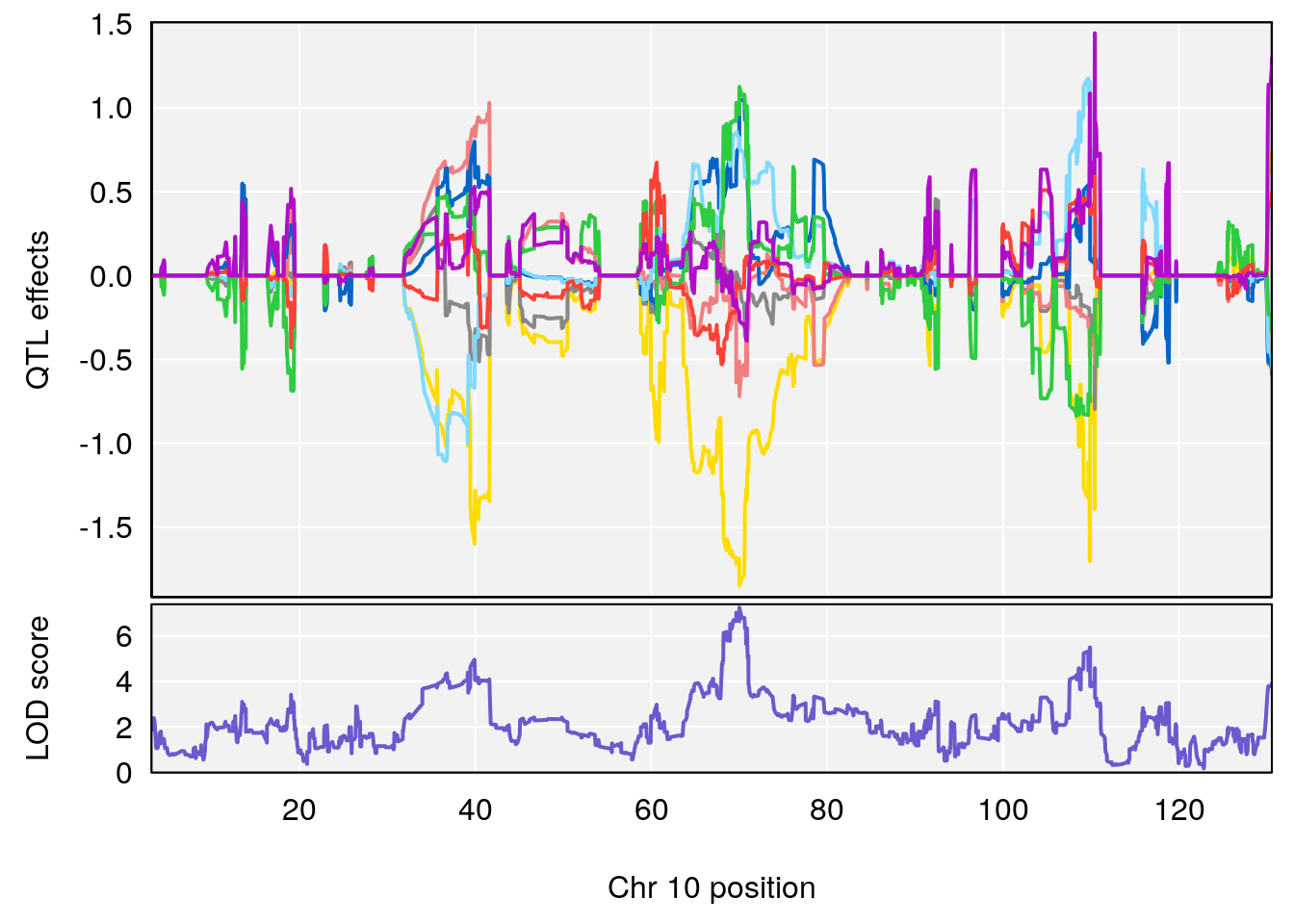
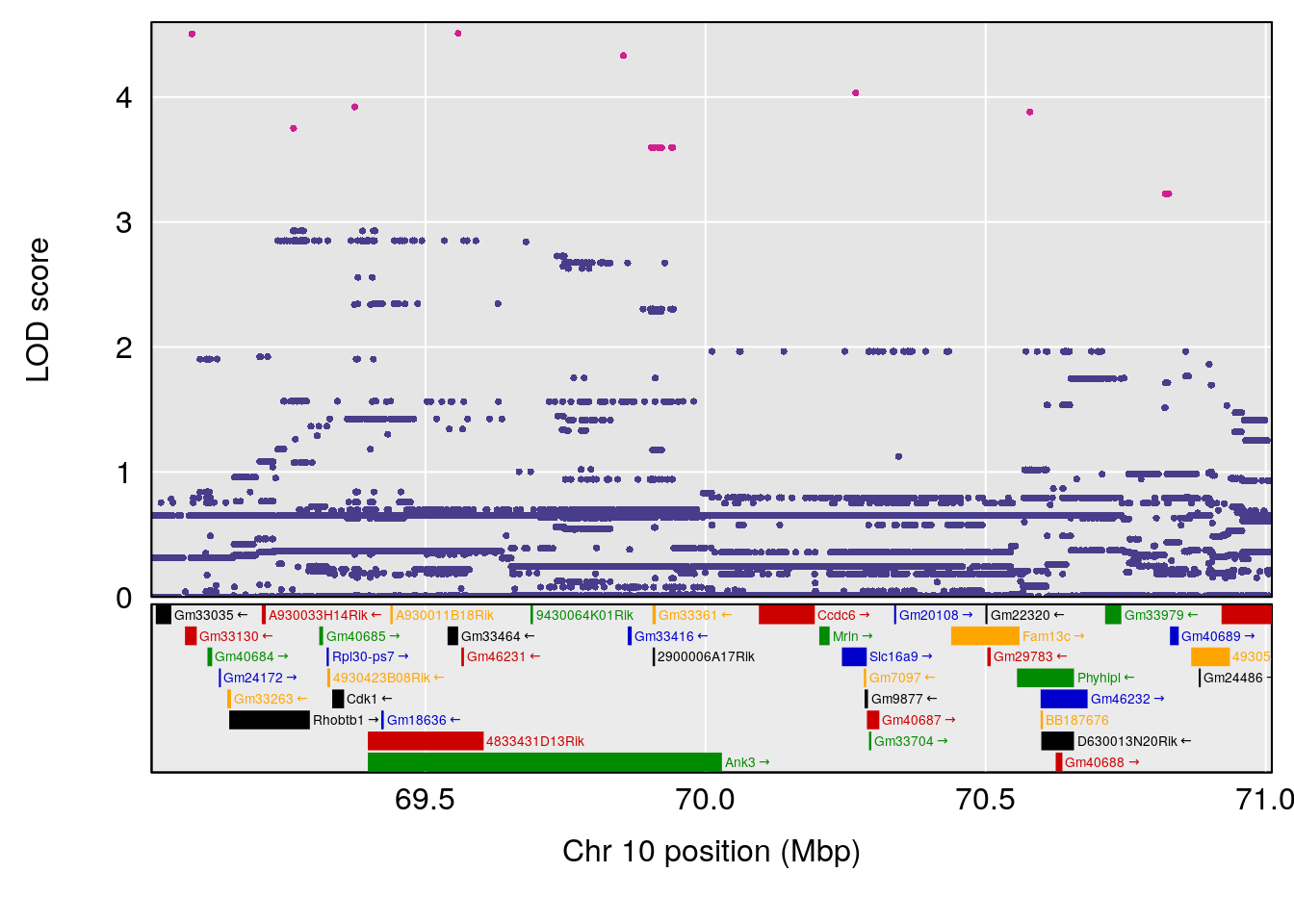
# [1] "Time.to.Recovery.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 3 148.59948 6.440788 53.60570 148.61250
# 2 1 pheno1 4 22.26230 6.109573 17.41576 137.08866
# 3 1 pheno1 10 70.01089 7.242967 68.18103 70.90480
# 4 1 pheno1 14 80.19357 6.242259 79.67940 80.21217
# 5 1 pheno1 18 23.93757 6.888692 22.73513 24.98189
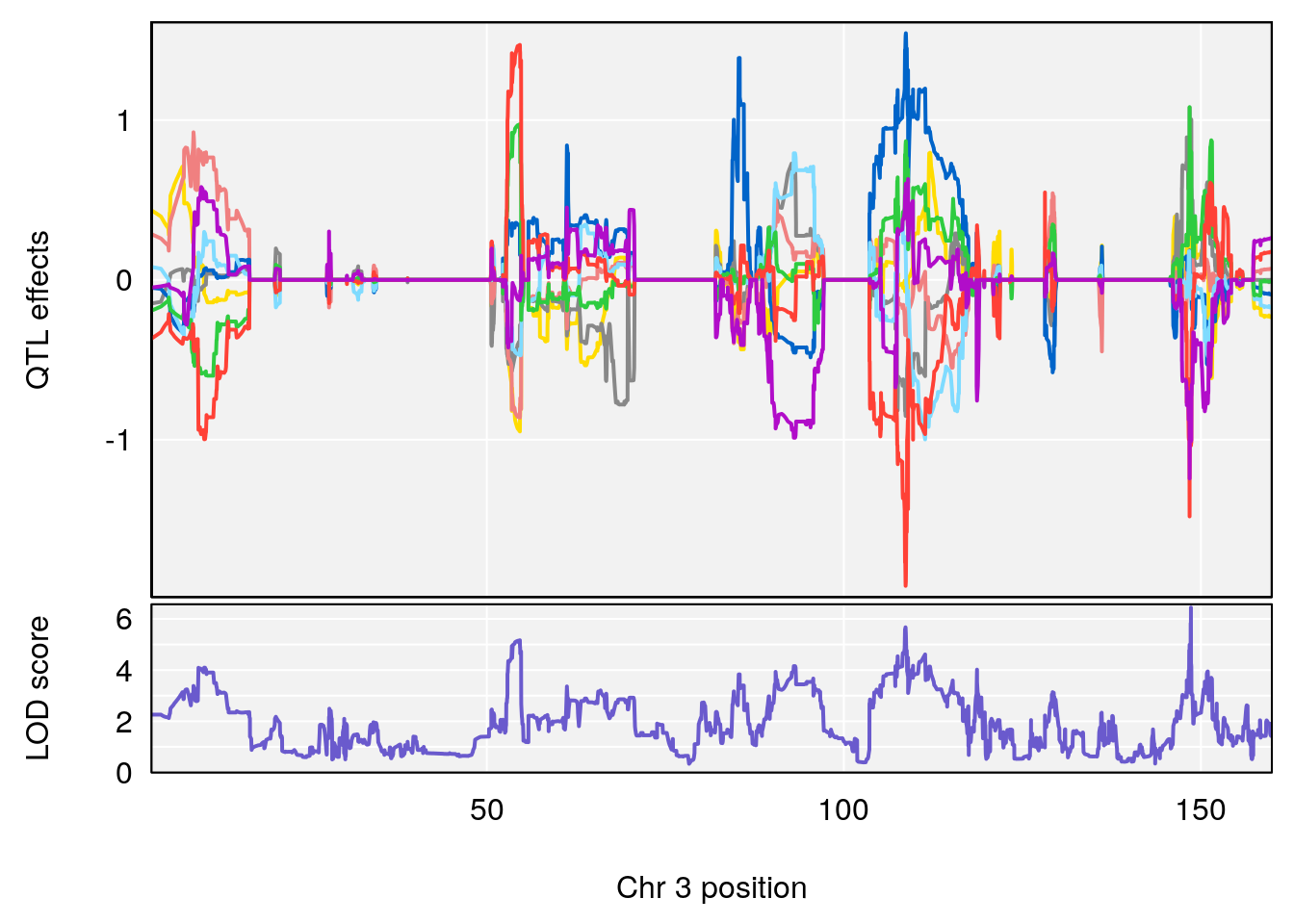
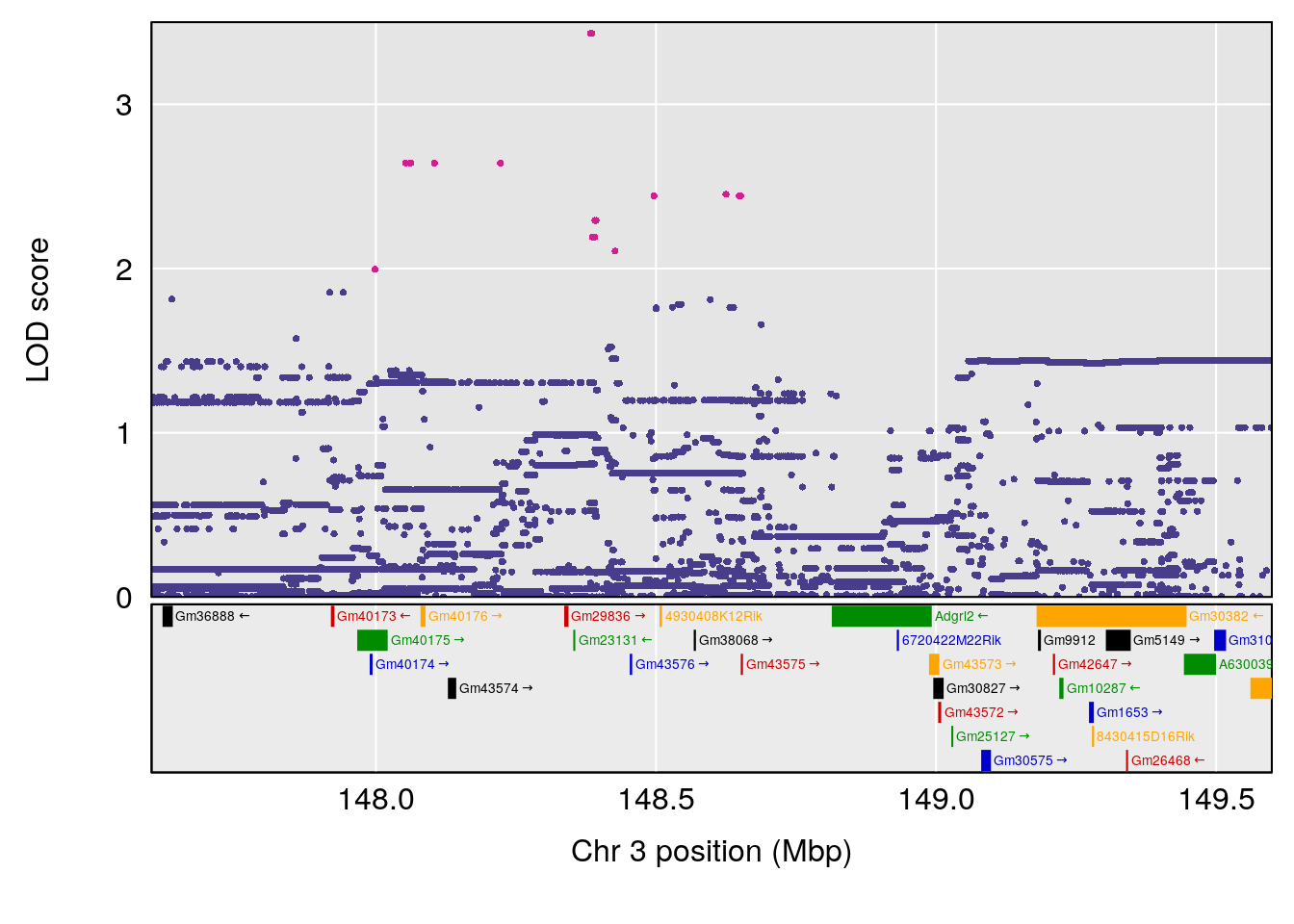
# [1] 1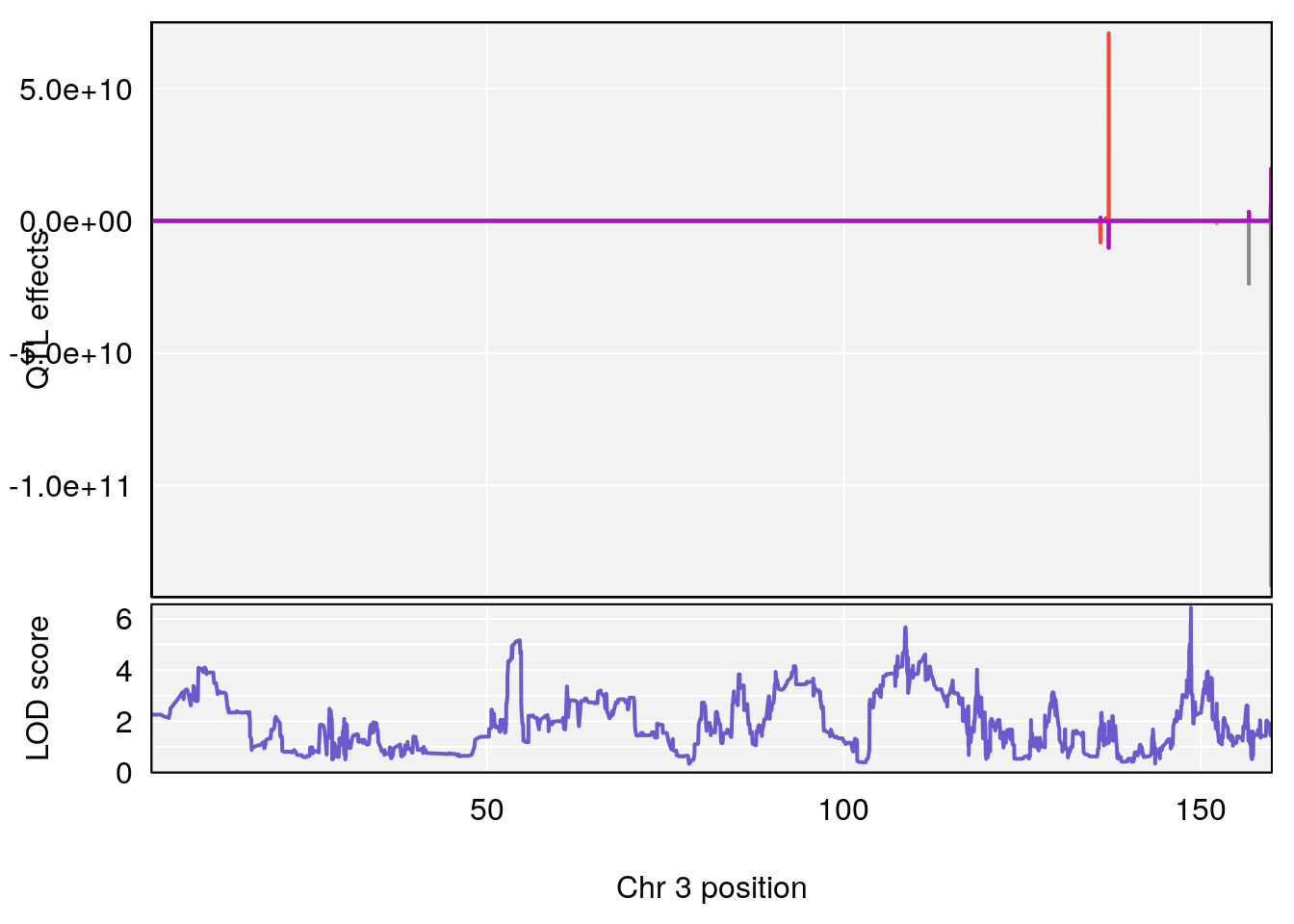
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
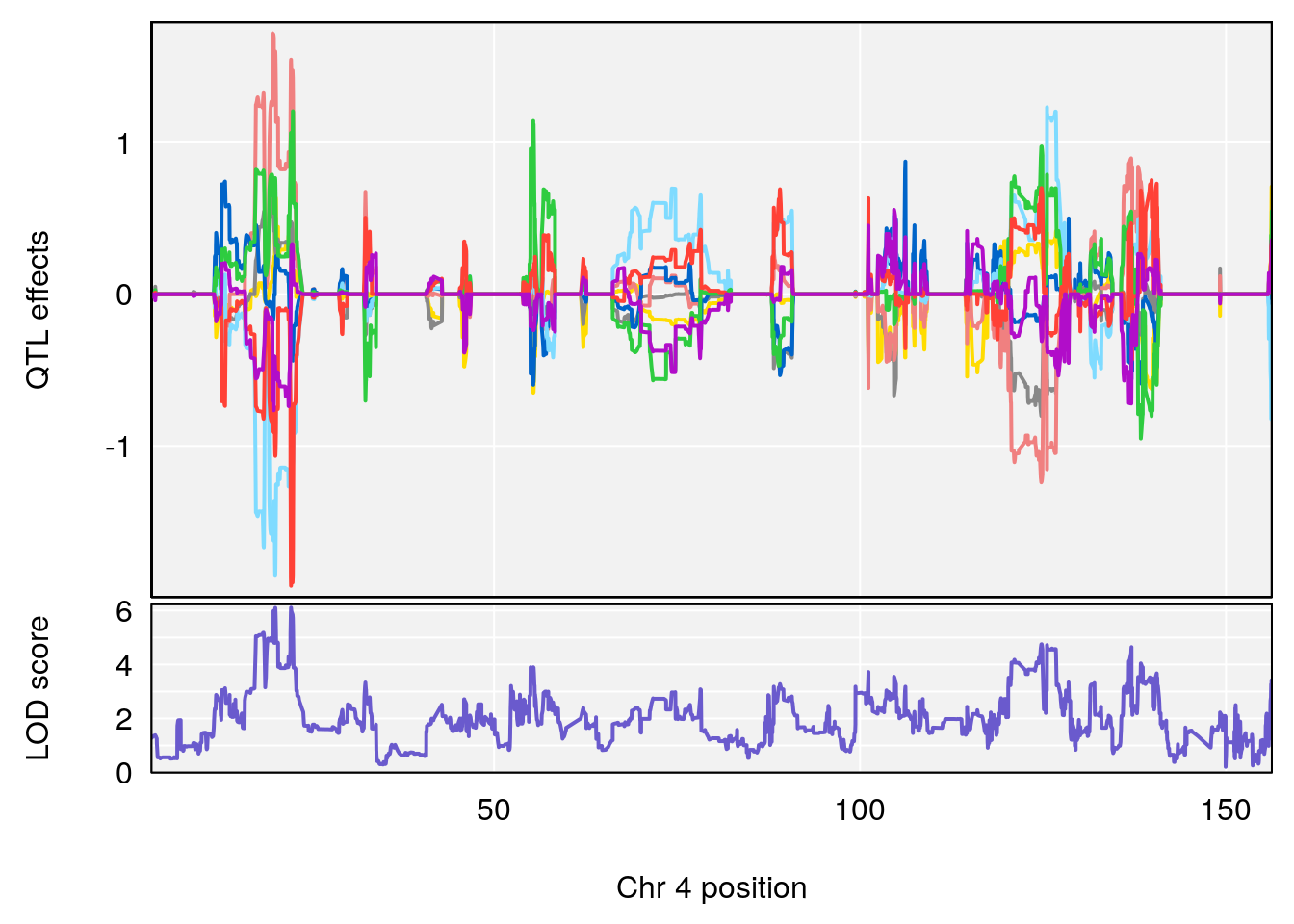
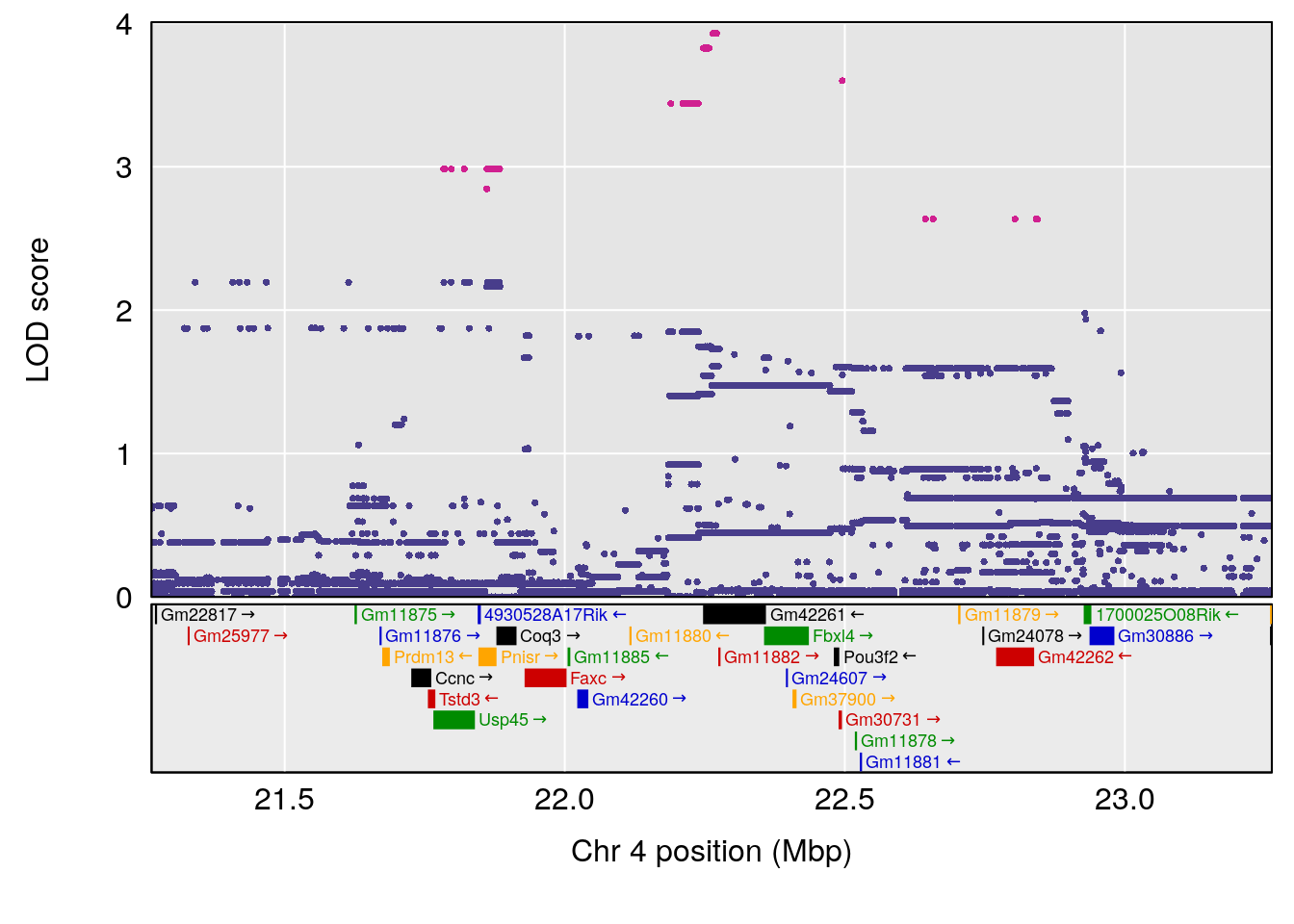
# [1] 2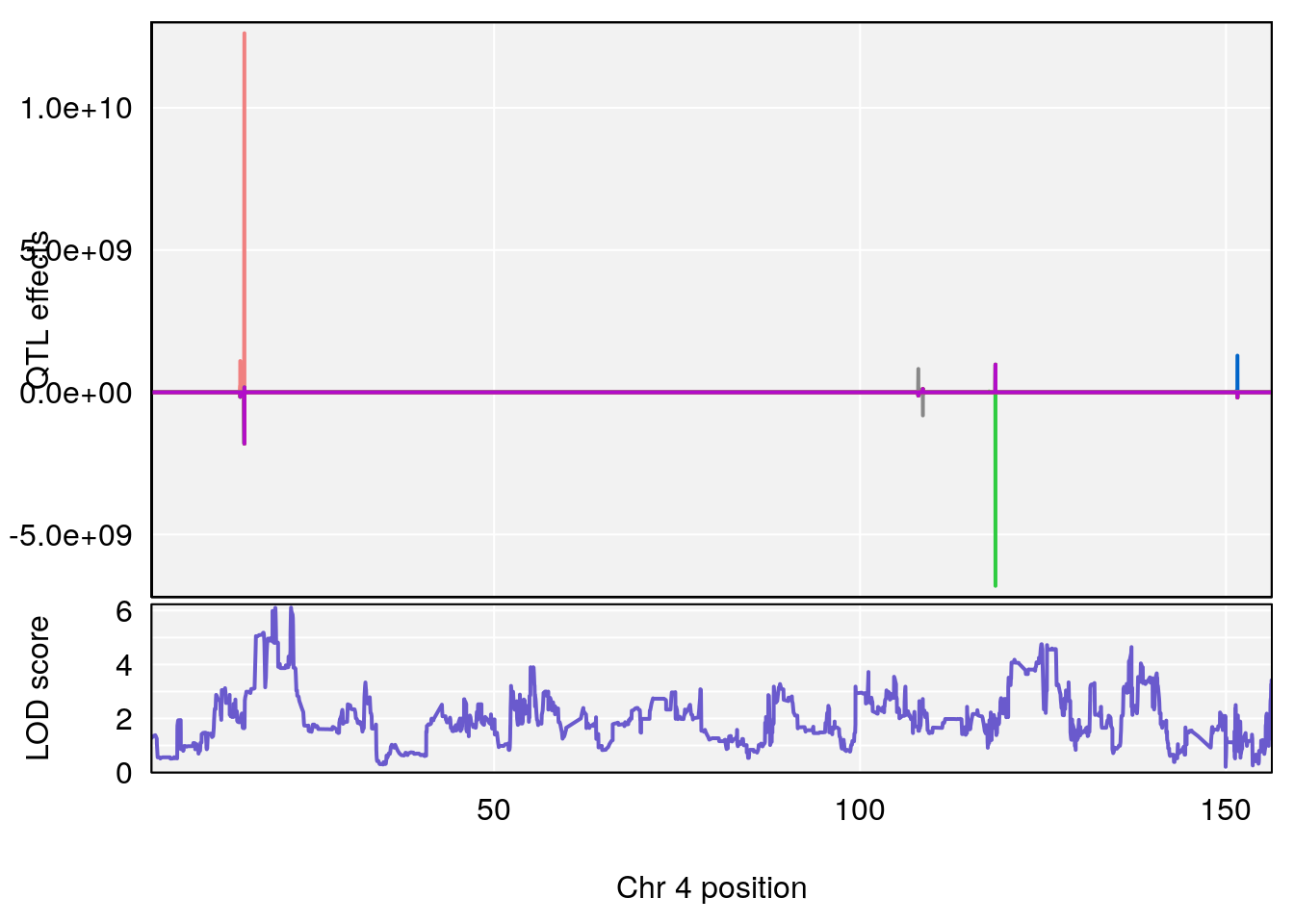
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 3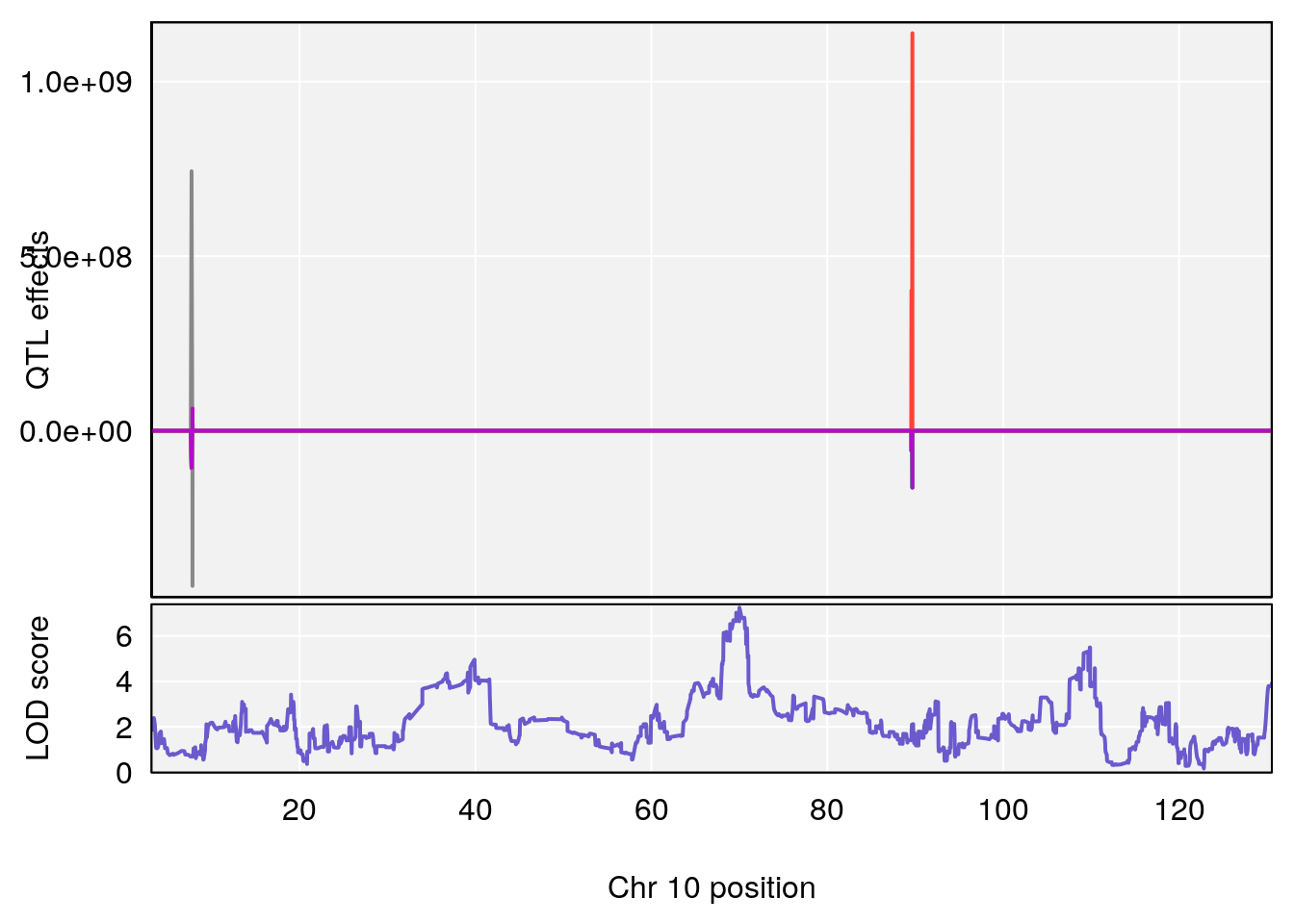
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 4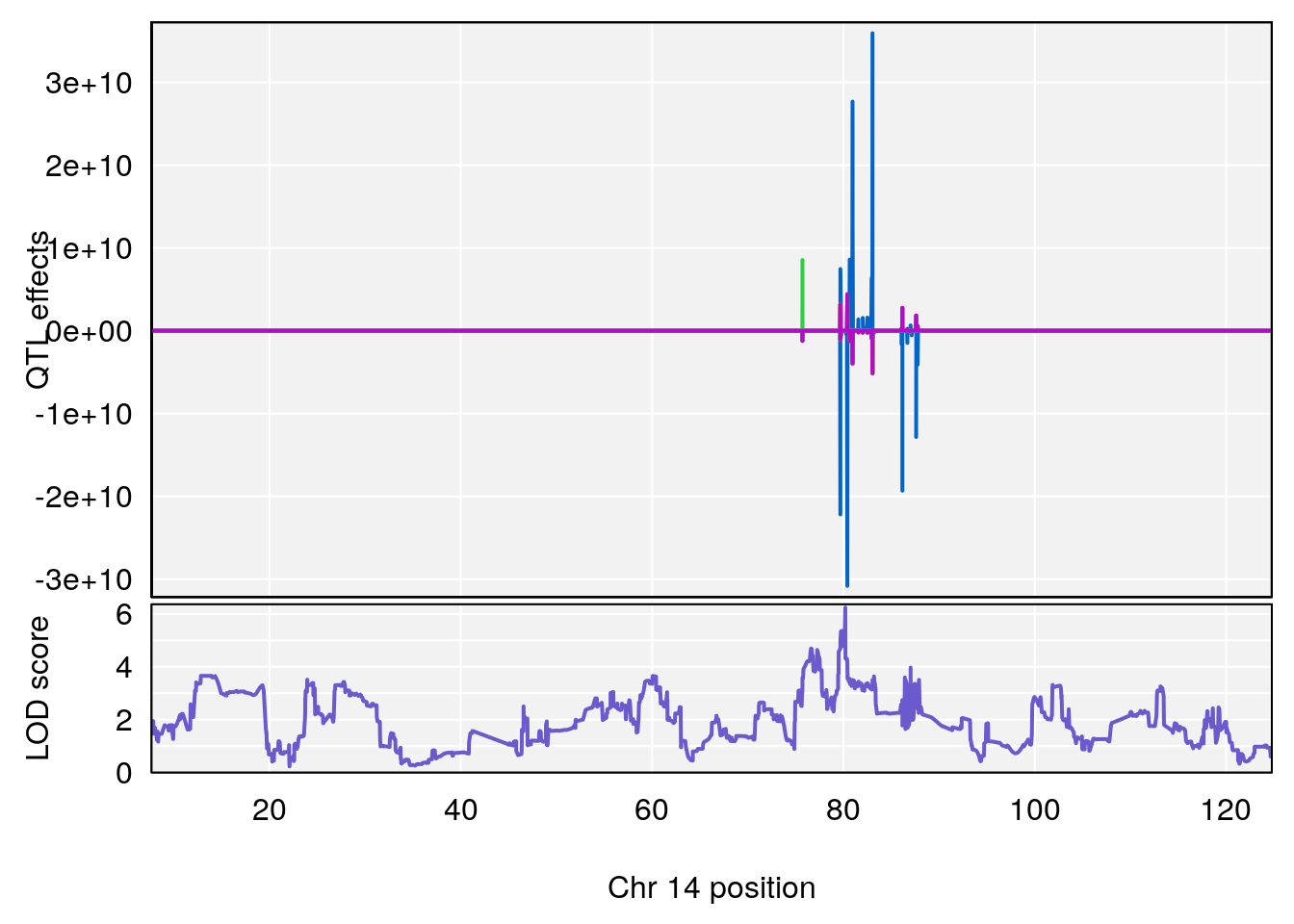
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
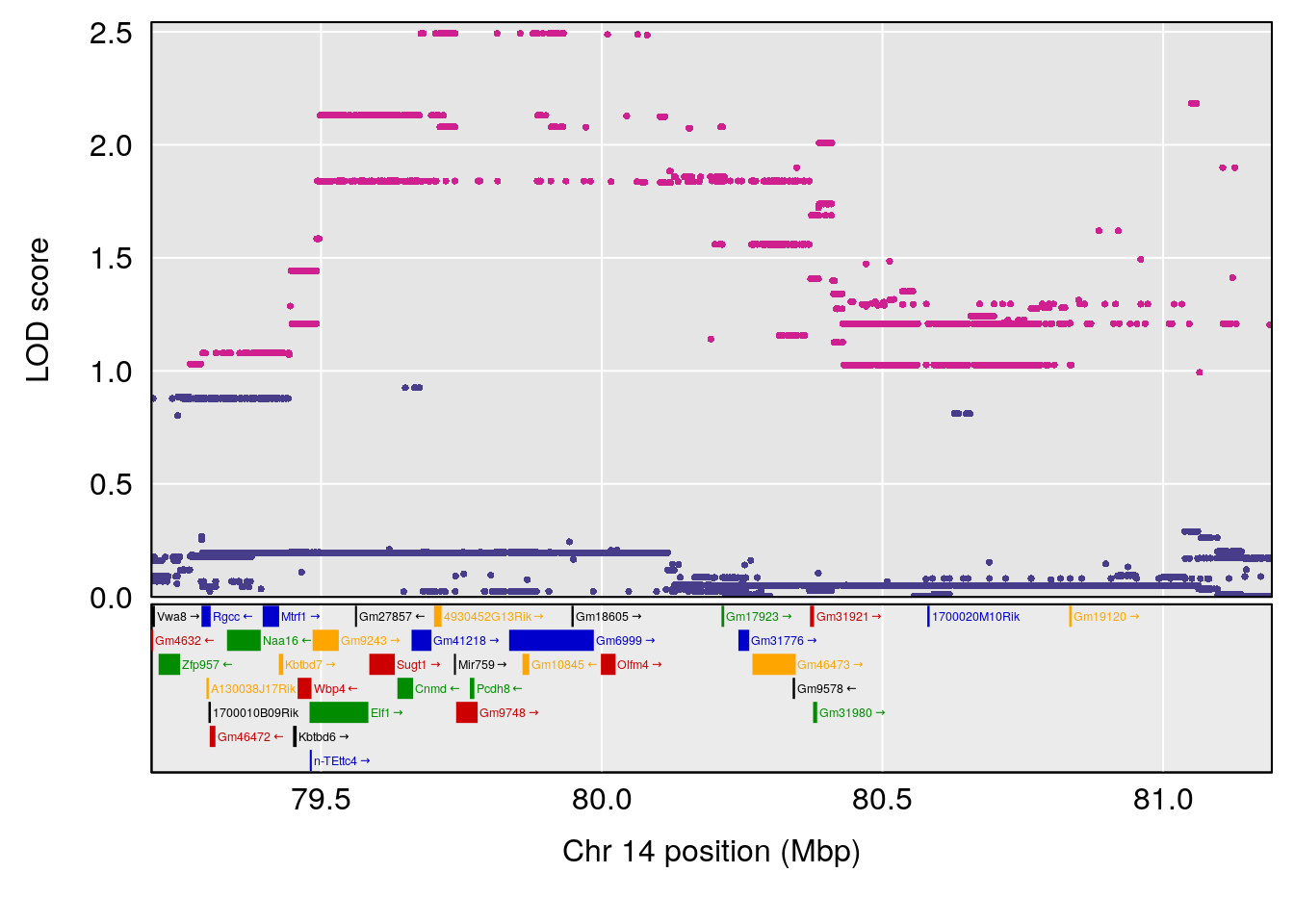
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 5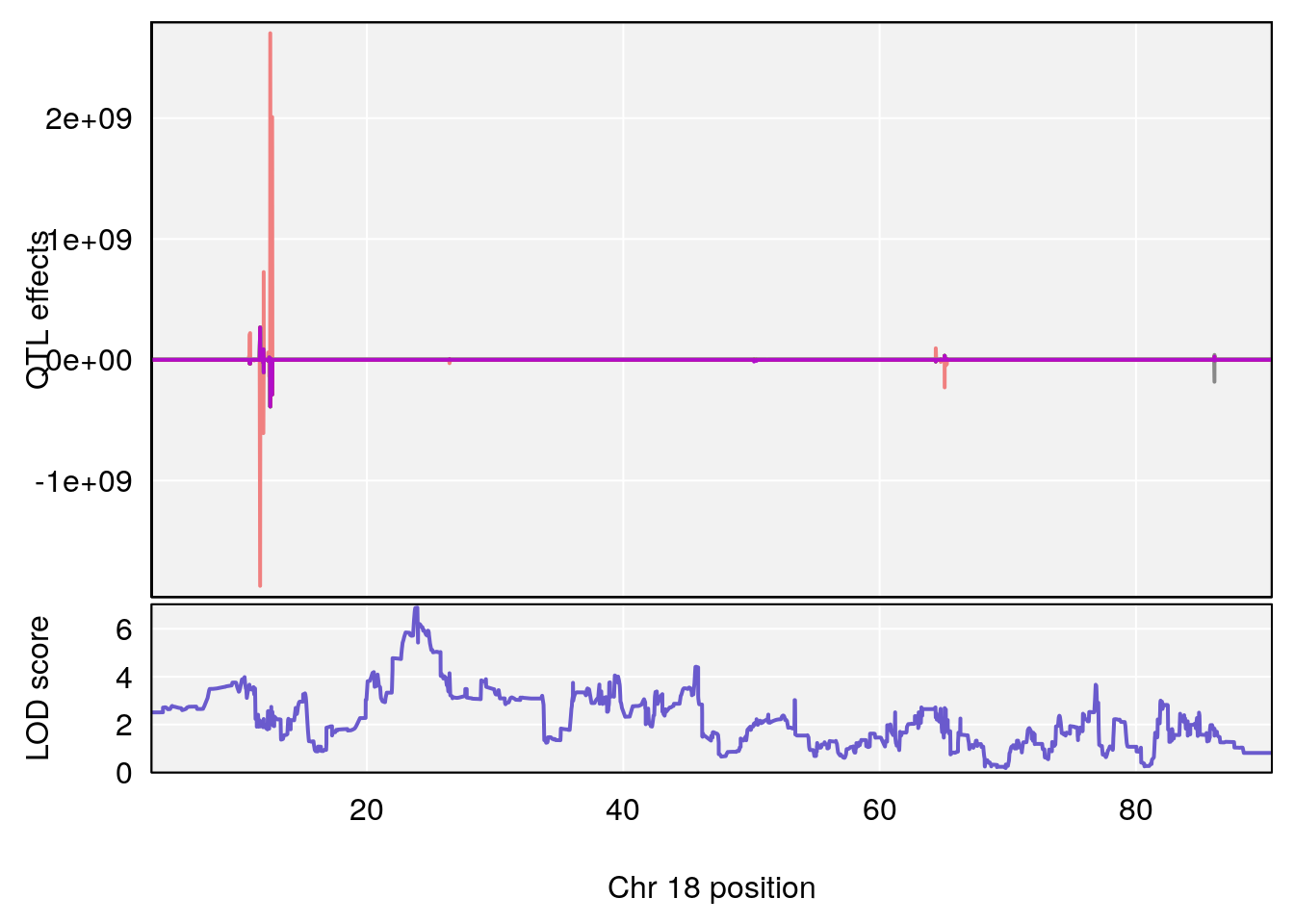
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
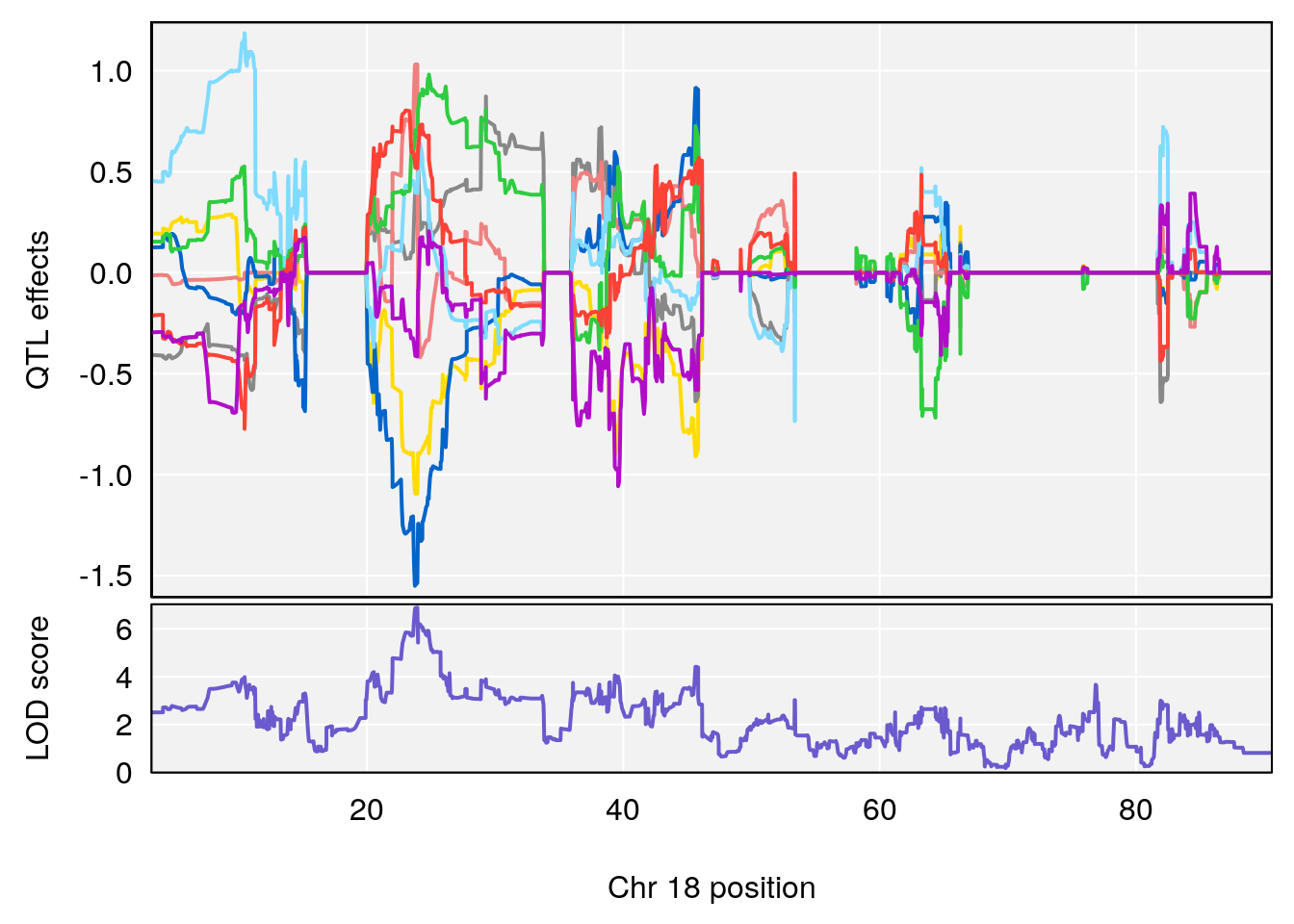
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
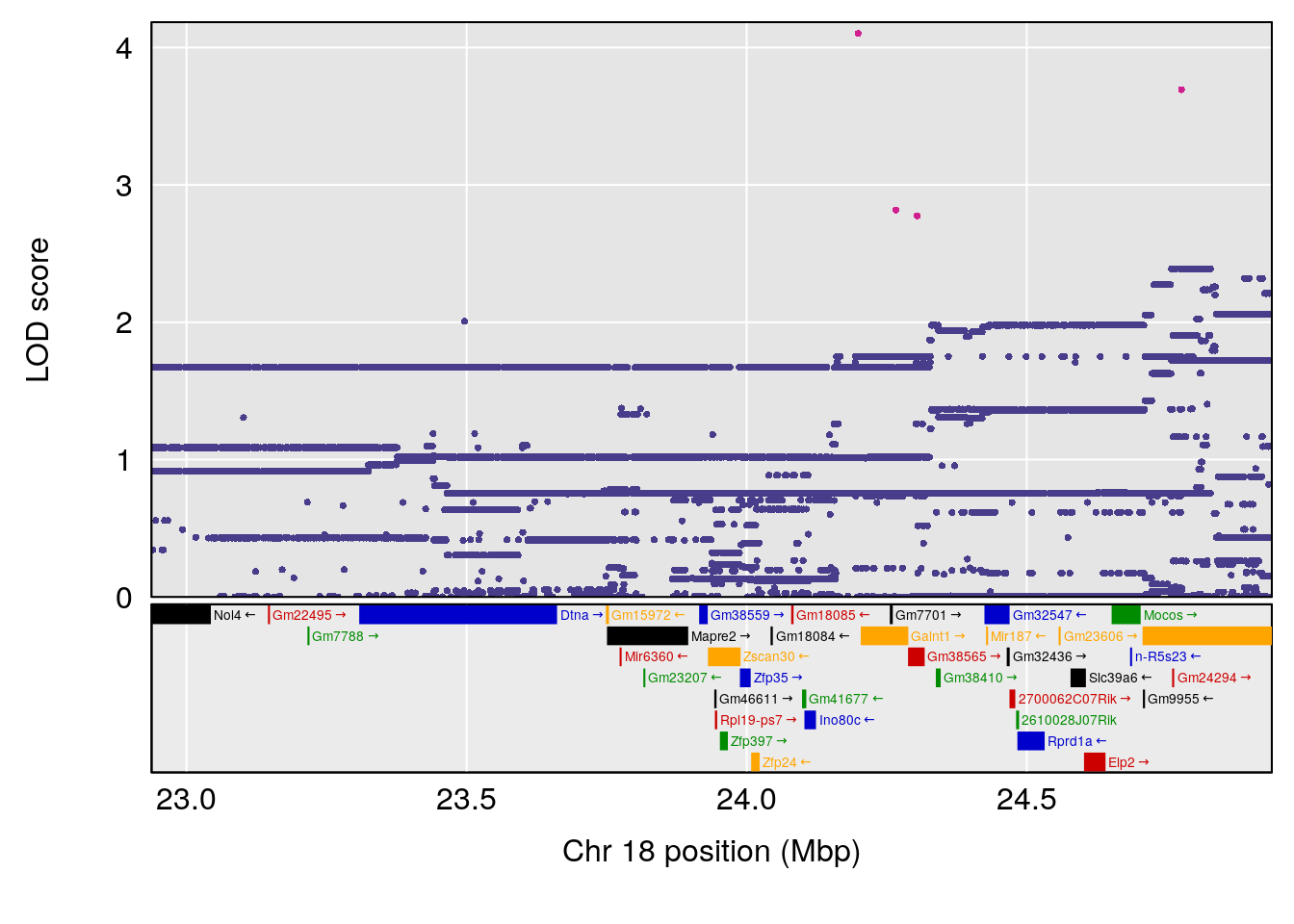
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
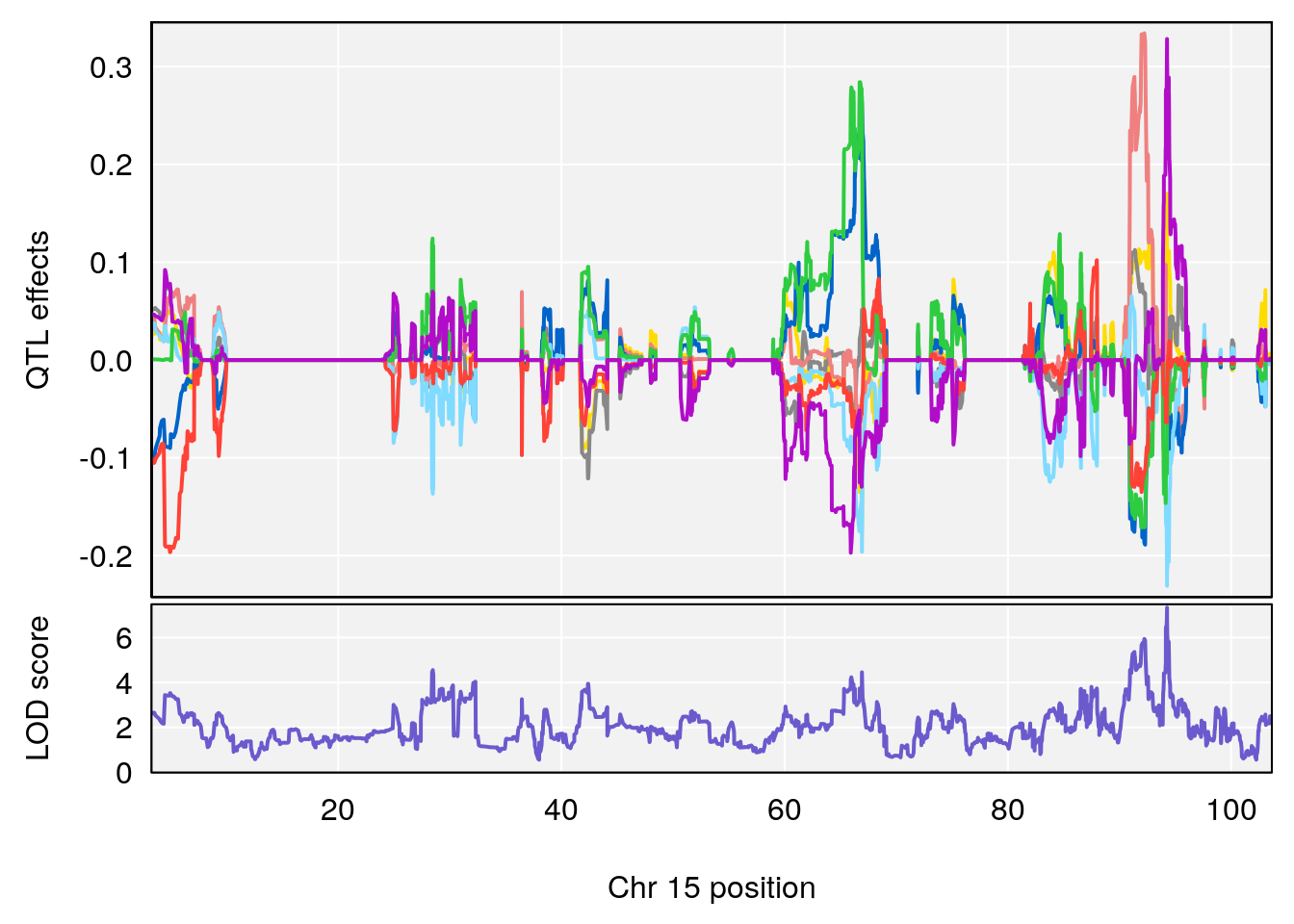
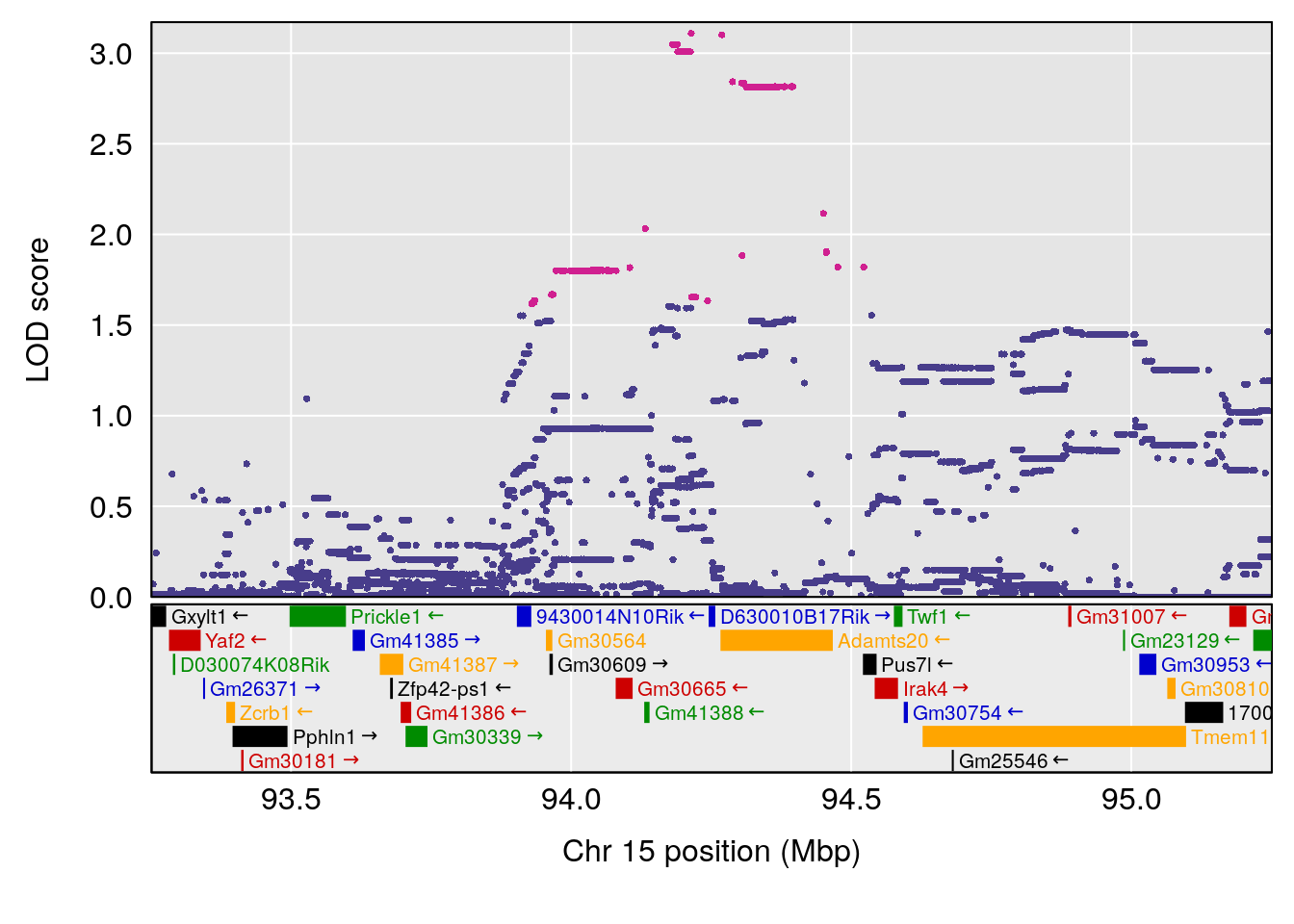
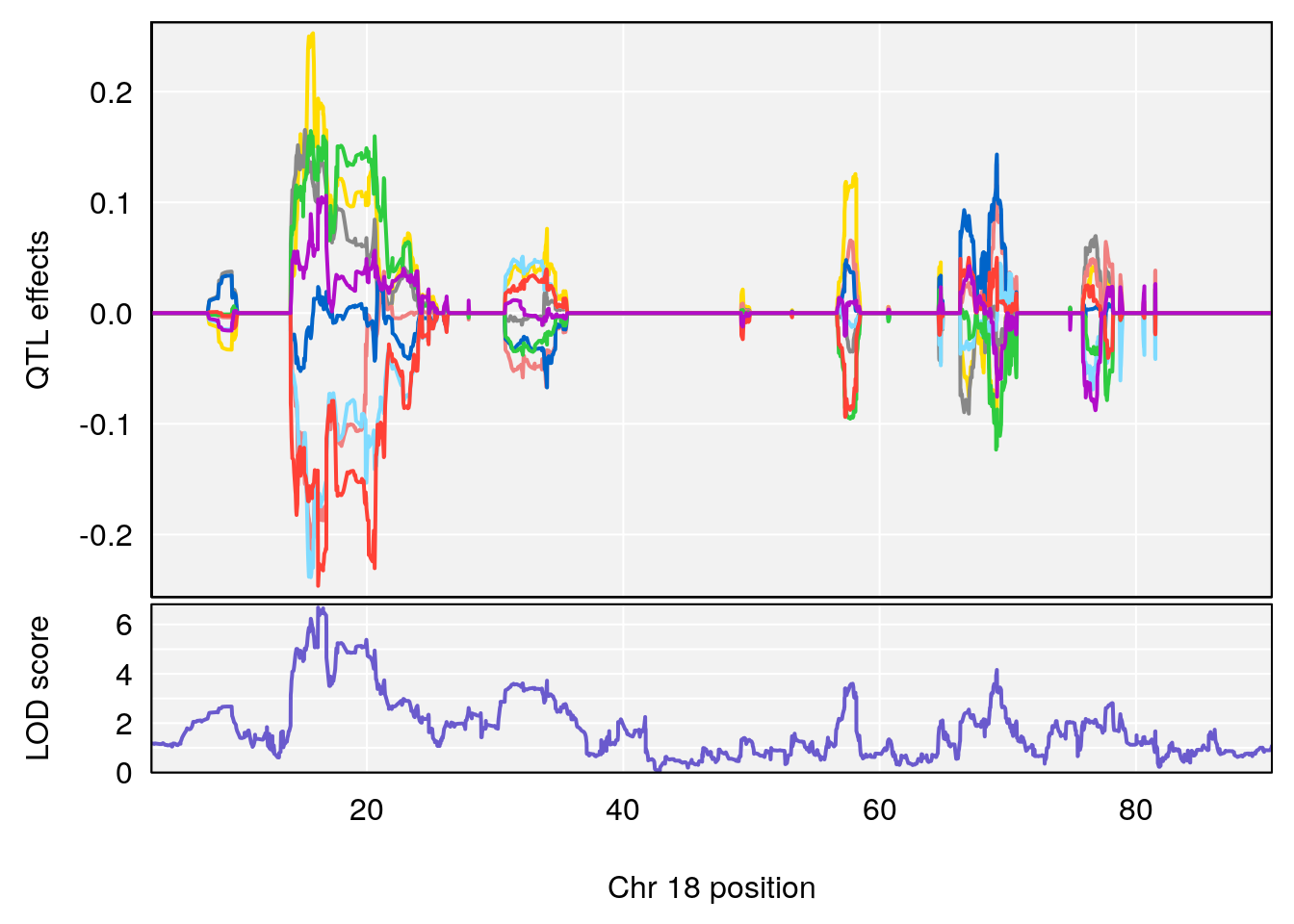
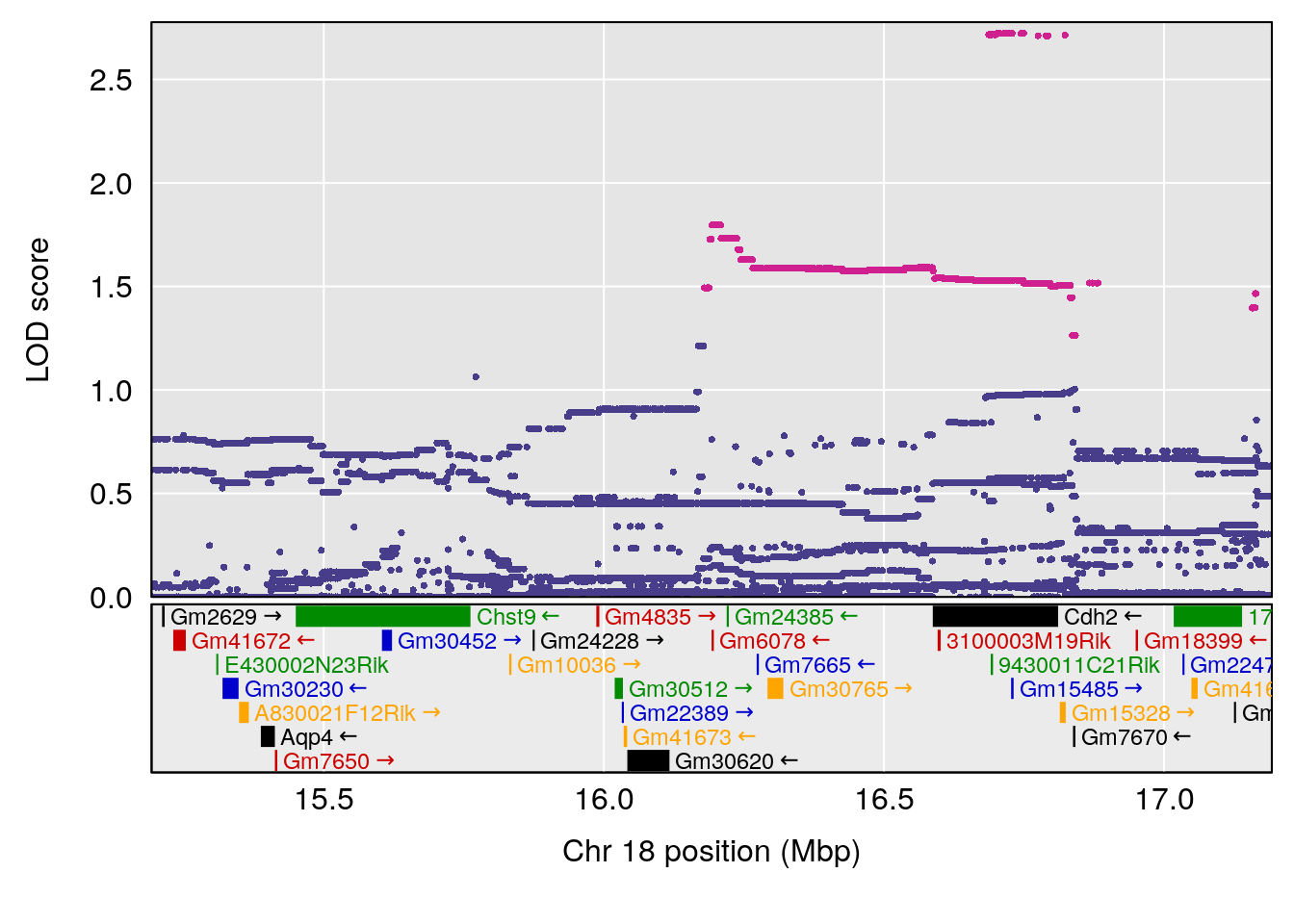
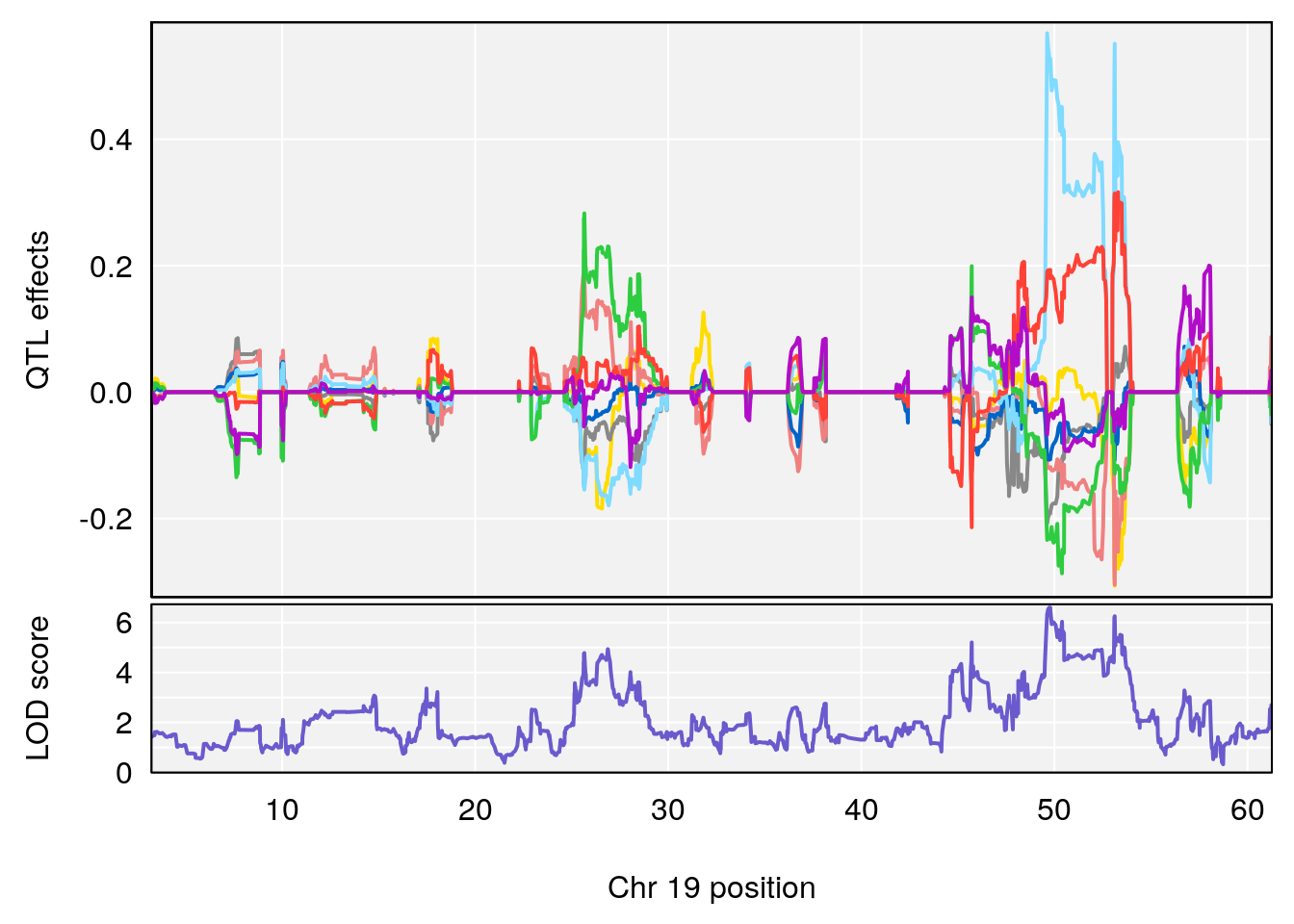
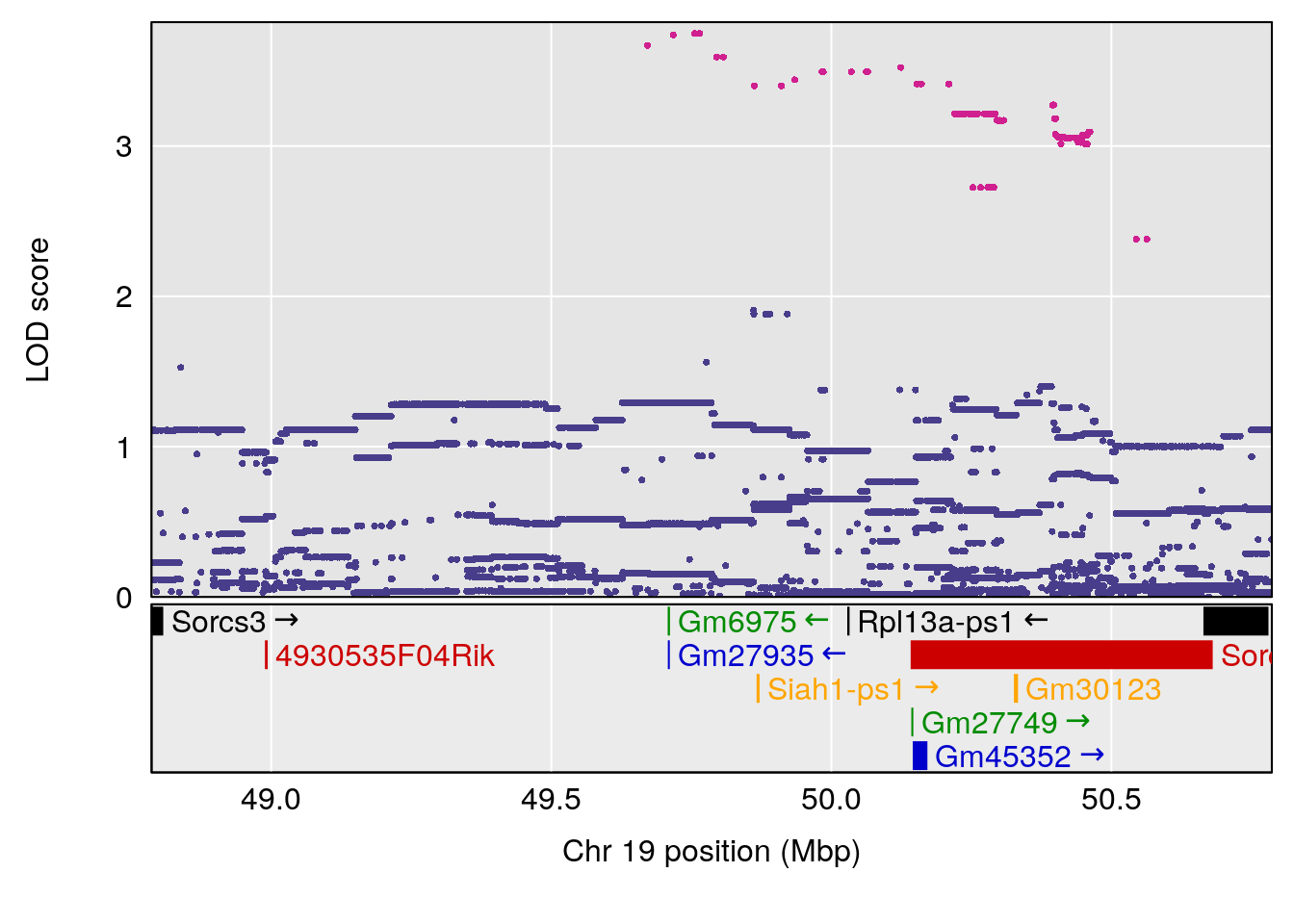
# [1] "Status_bin"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
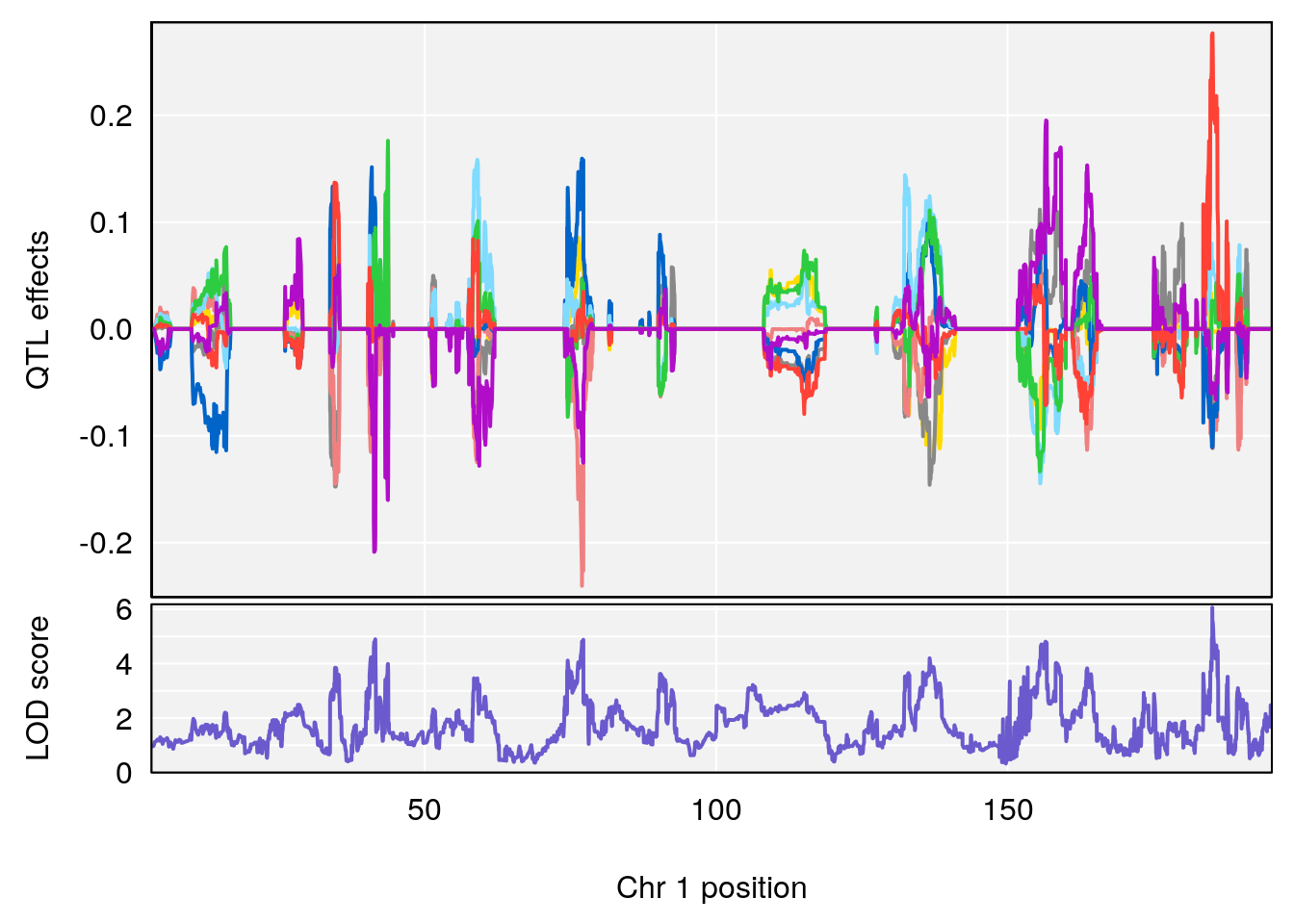
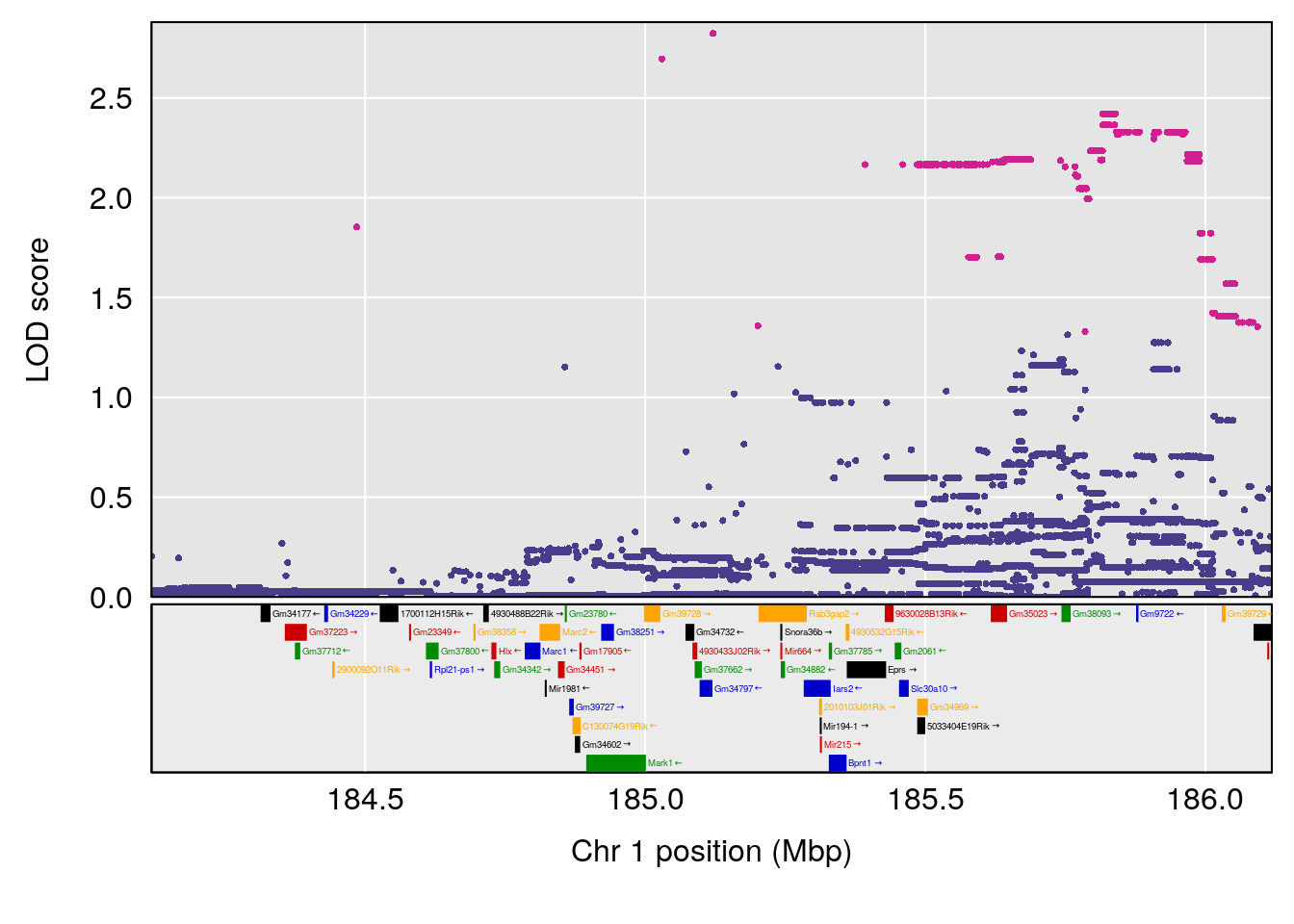
# 1 1 pheno1 1 185.11874 6.058648 41.25093 185.84088
# 2 1 pheno1 15 94.25093 7.334412 92.19023 94.31234
# 3 1 pheno1 18 16.19240 6.685013 15.31898 20.05685
# 4 1 pheno1 19 49.78637 6.595691 45.70921 53.50149
# [1] 1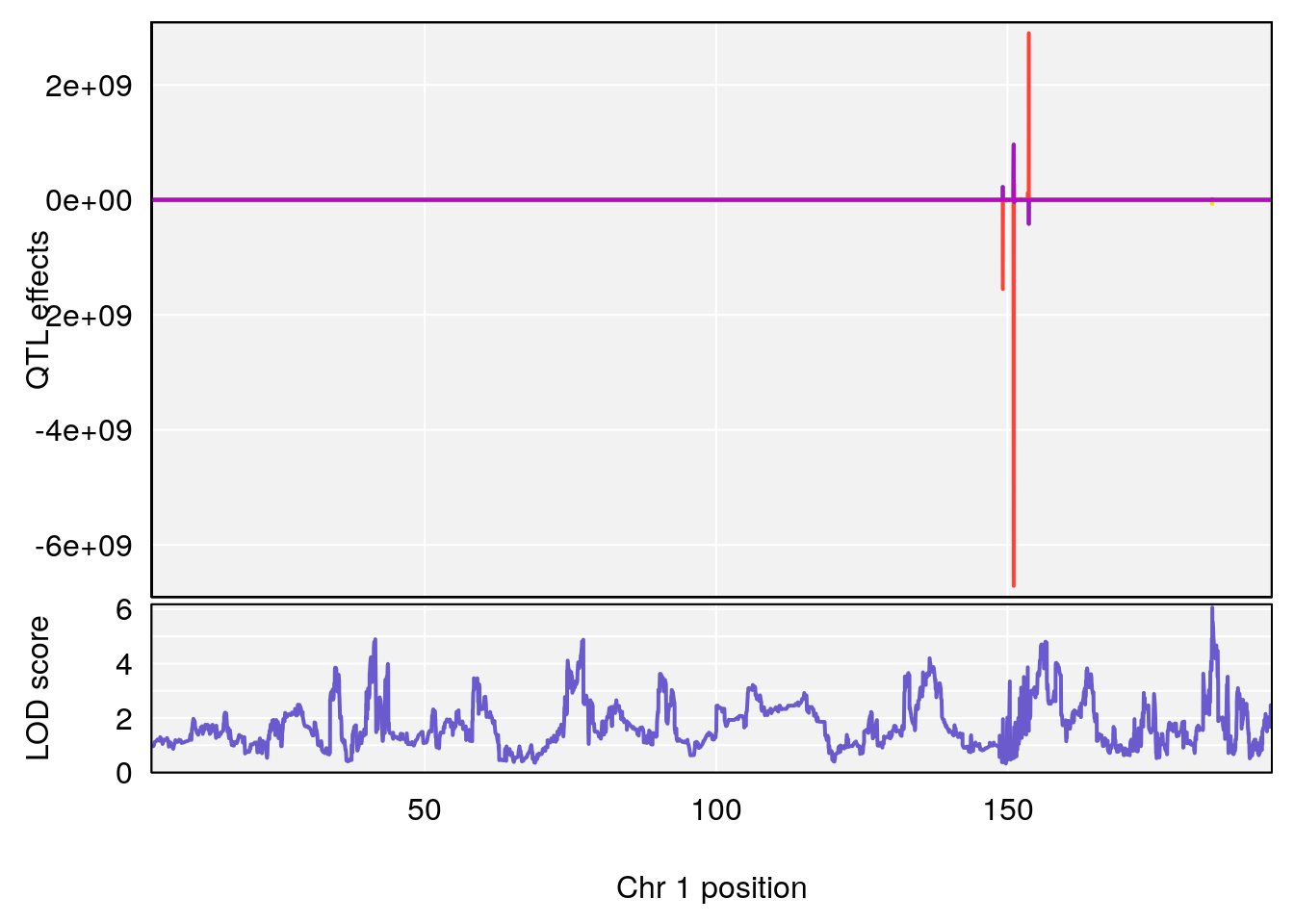
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2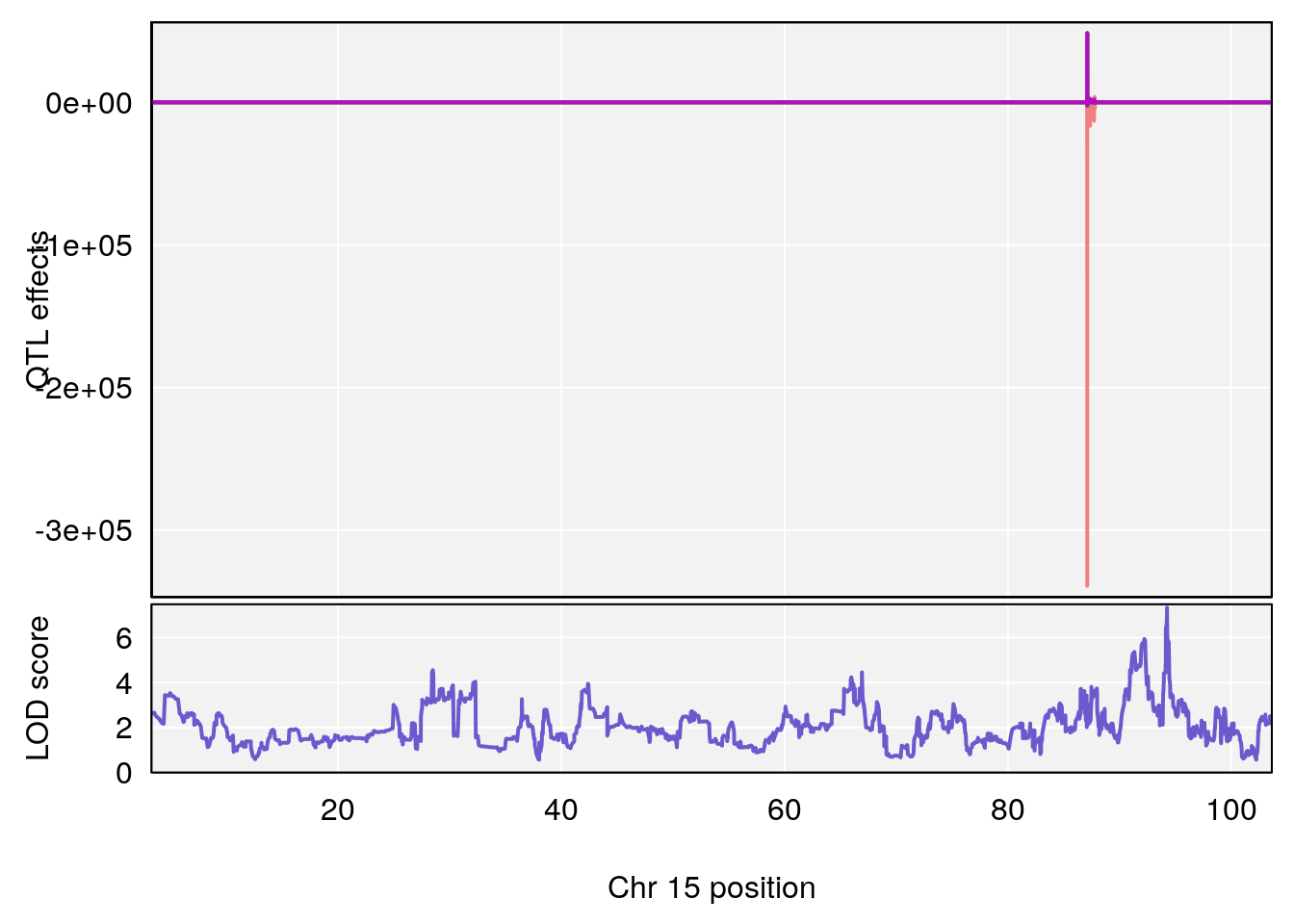
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 3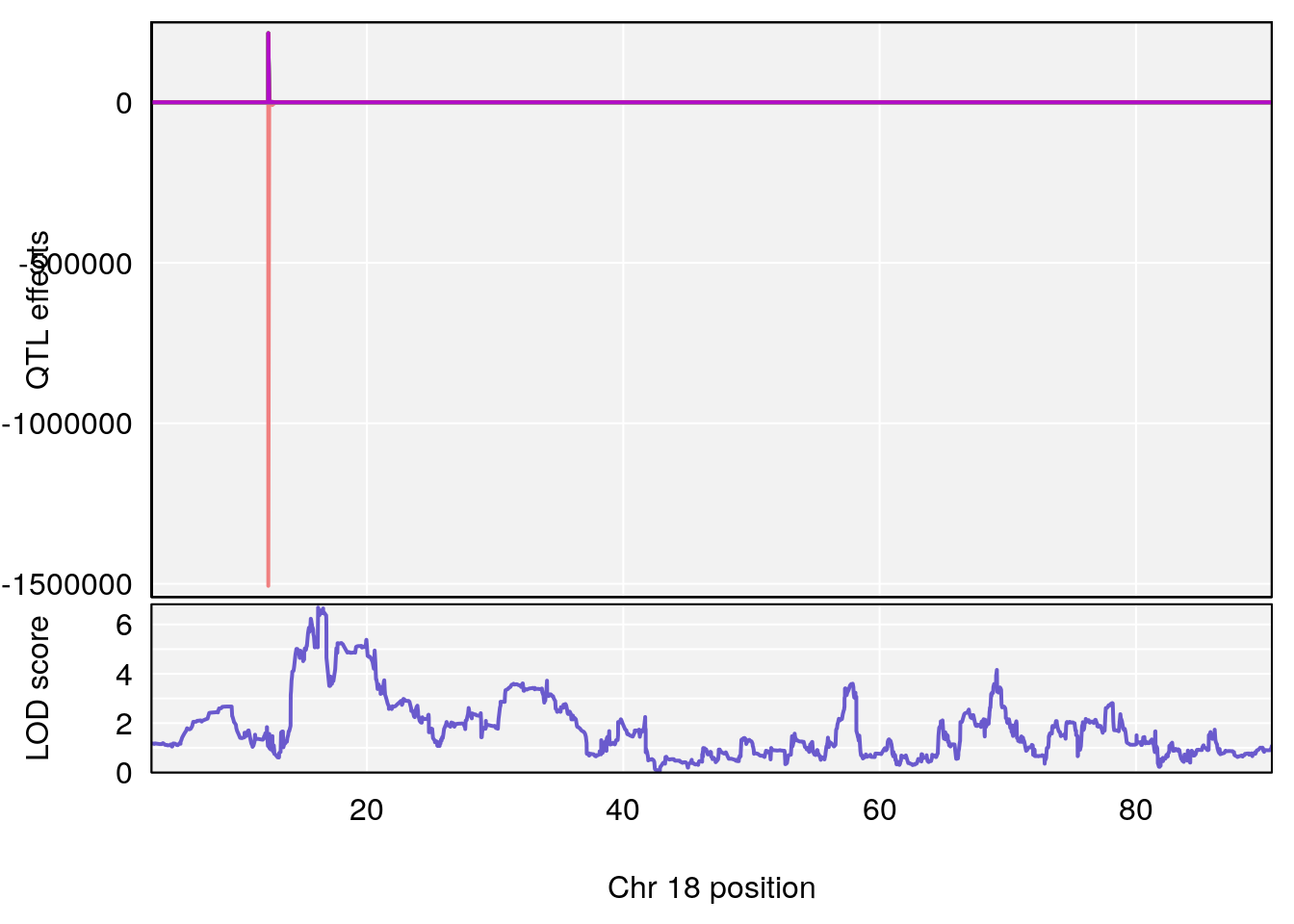
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 4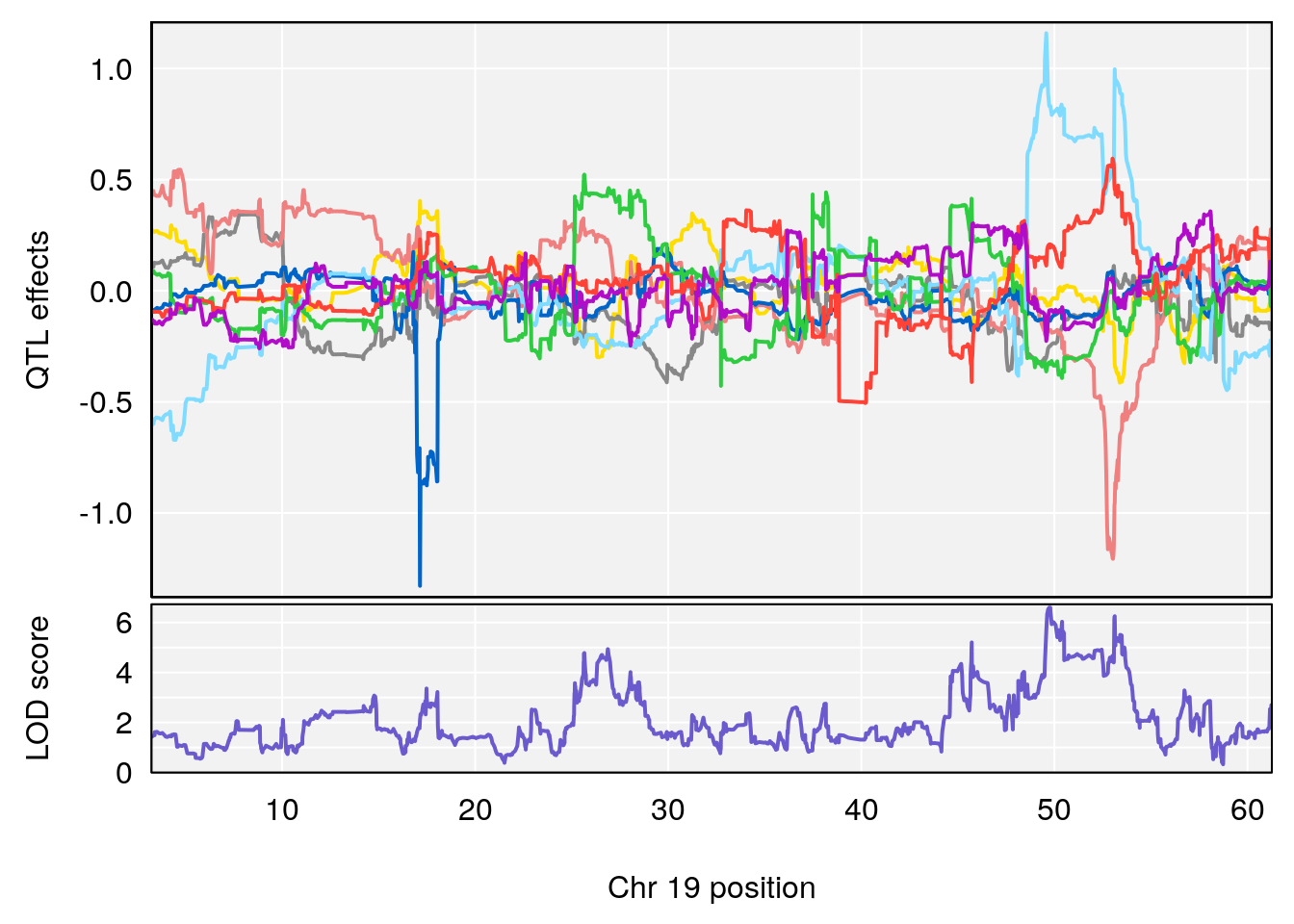
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
Plot qtl mapping on array female
load("output/Fentanyl/qtl.fentanyl.combine2cohort.out.male.RData")
#pmap and gmap
chr.names <- c("1","2","3","4","5","6","7","8","9","10","11","12","13","14","15","16","17","18","19", "X")
gmap <- list()
pmap <- list()
for (chr in chr.names){
gmap[[chr]] <- gm$gmap[[chr]]
pmap[[chr]] <- gm$pmap[[chr]]
}
attr(gmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
attr(pmap, "is_x_chr") <- structure(c(rep(FALSE,19),TRUE), names=1:20)
#genome-wide plot
for(i in names(qtl.out)){
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(qtl.out[[i]]) # overall maximum LOD score
plot(qtl.out[[i]], map=pmap, lodcolumn=1, col="slateblue", ylim=c(0, max(sapply(qtl.out, maxlod)) +2))
abline(h=summary(qtl.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[1]], col="red")
abline(h=summary(qtl.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[2]], col="red")
abline(h=summary(qtl.operm[[i]], alpha=c(0.10, 0.05, 0.01))[[3]], col="red")
title(main = paste0("do_Fentanyl_combine2cohort_",i))
}
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
#peaks coeff plot
for(i in names(qtl.out)){
print(i)
peaks <- find_peaks(qtl.out[[i]], map=pmap, threshold=6, drop=1.5)
if(nrow(peaks) > 0) {
print(peaks)
for(p in 1:dim(peaks)[1]) {
print(p)
chr <-peaks[p,3]
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
plot_coefCC(coef_c1[[i]][[p]], pmap[chr], scan1_output=qtl.out[[i]], bgcolor="gray95", legend=NULL)
plot_coefCC(coef_c2[[i]][[p]], pmap[chr], scan1_output=qtl.out[[i]], bgcolor="gray95", legend=NULL)
plot(out_snps[[i]][[p]]$lod, out_snps[[i]][[p]]$snpinfo, drop_hilit=1.5, genes=out_genes[[i]][[p]])
}
}
}
# [1] "Time.to.Dead.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 2 132.62489 6.124096 50.81340 173.86901
# 2 1 pheno1 10 99.92521 6.492863 99.28182 105.40417
# 3 1 pheno1 17 23.96788 6.725145 21.59514 25.08344
# [1] 1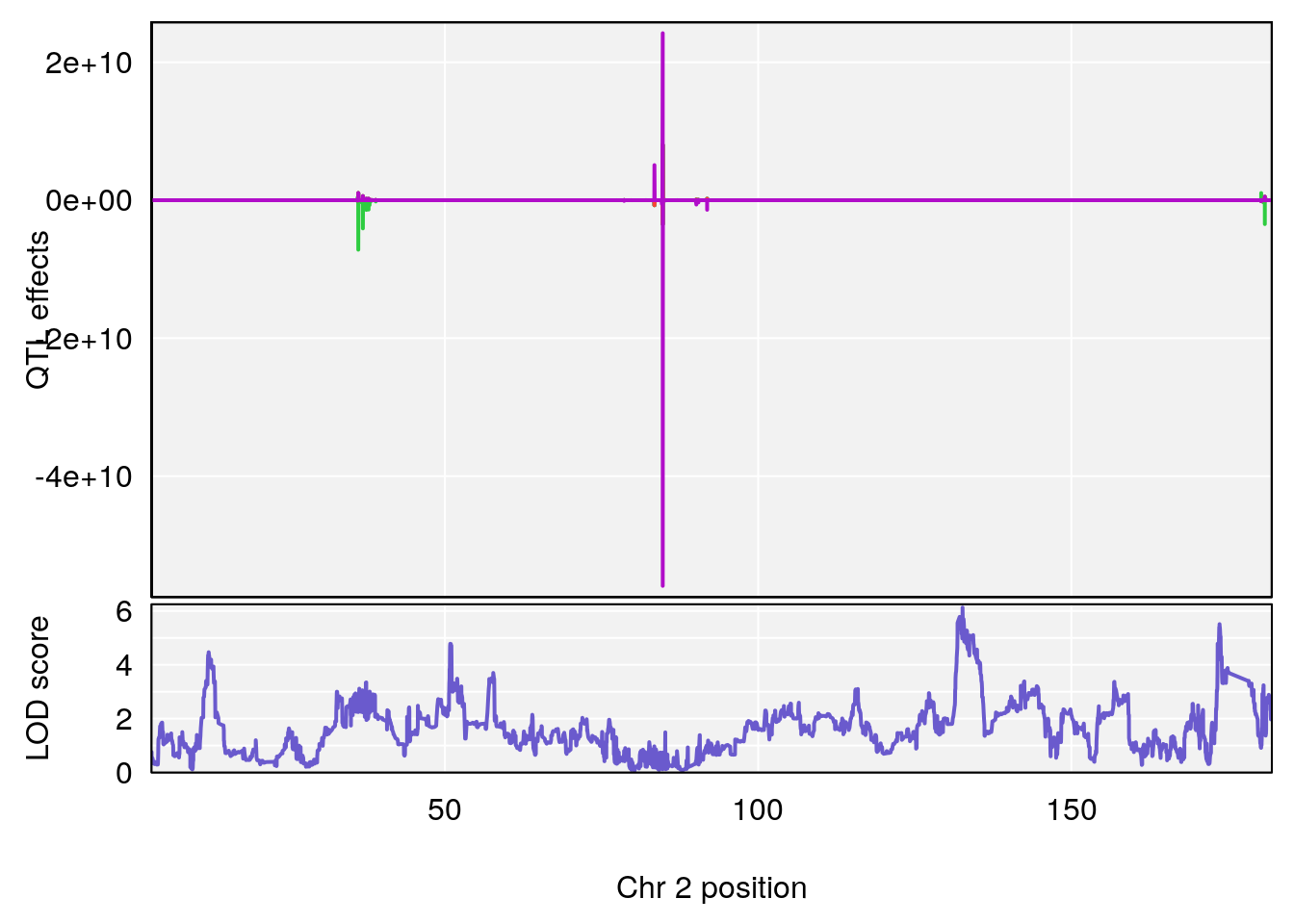
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
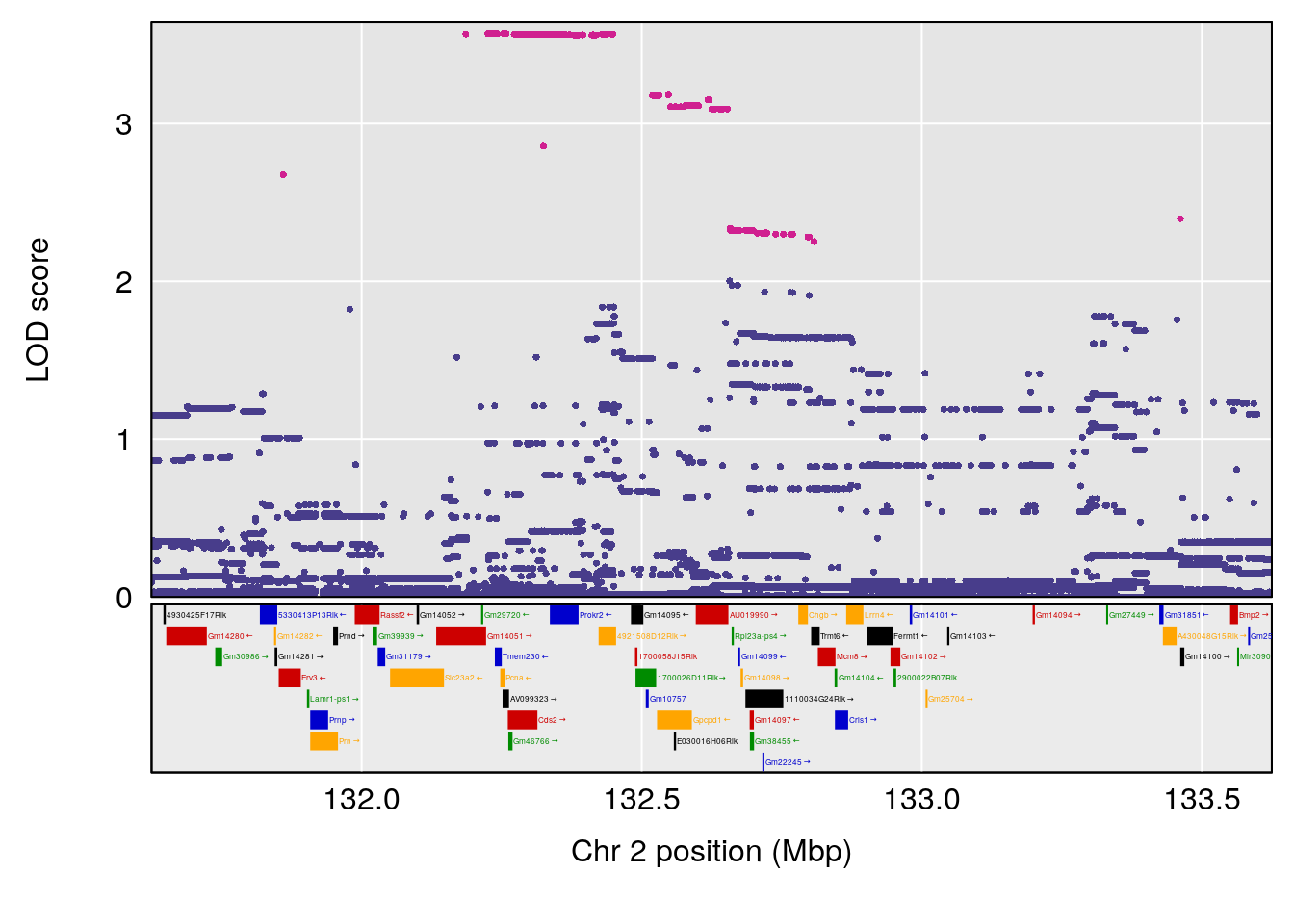
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2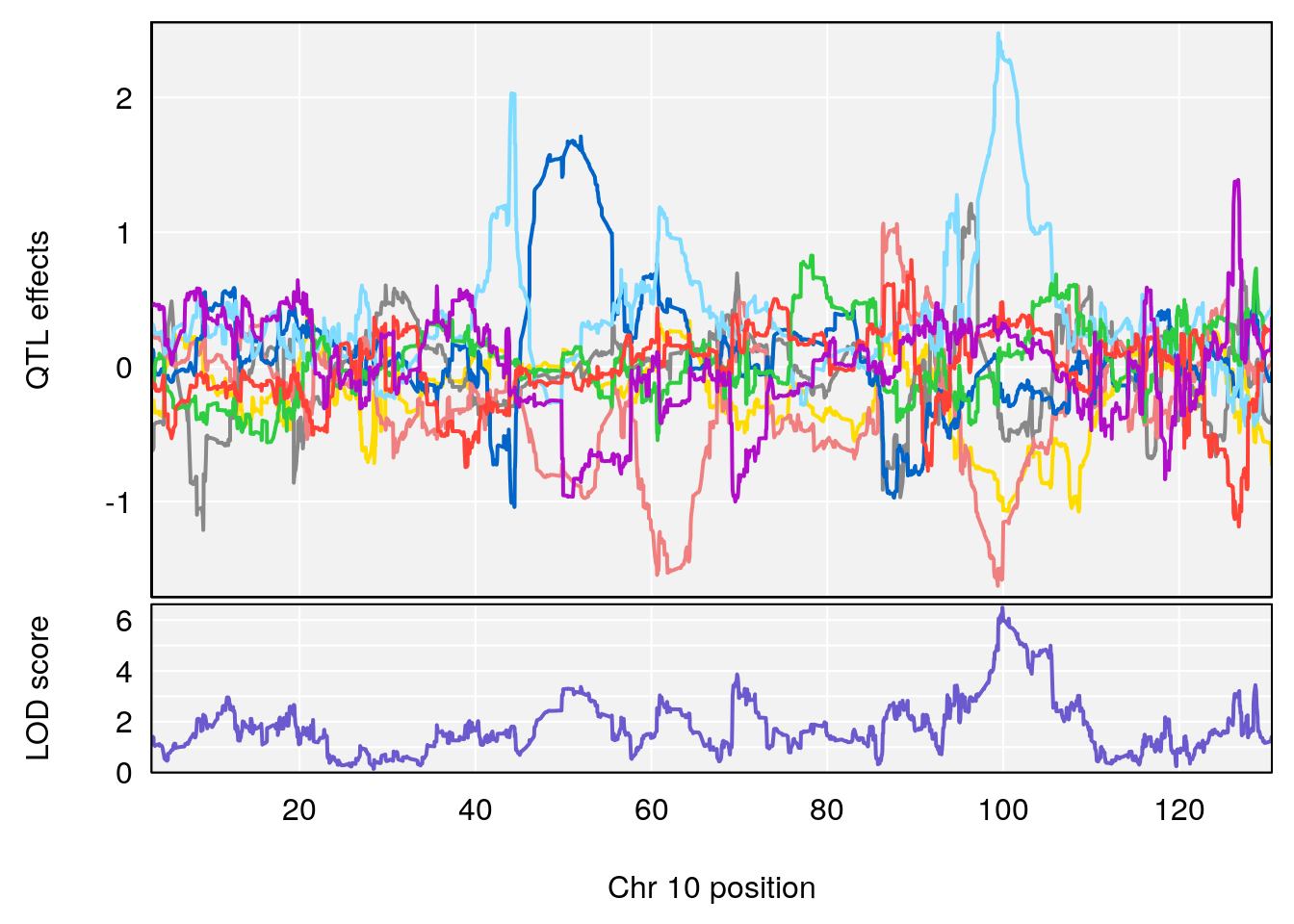
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
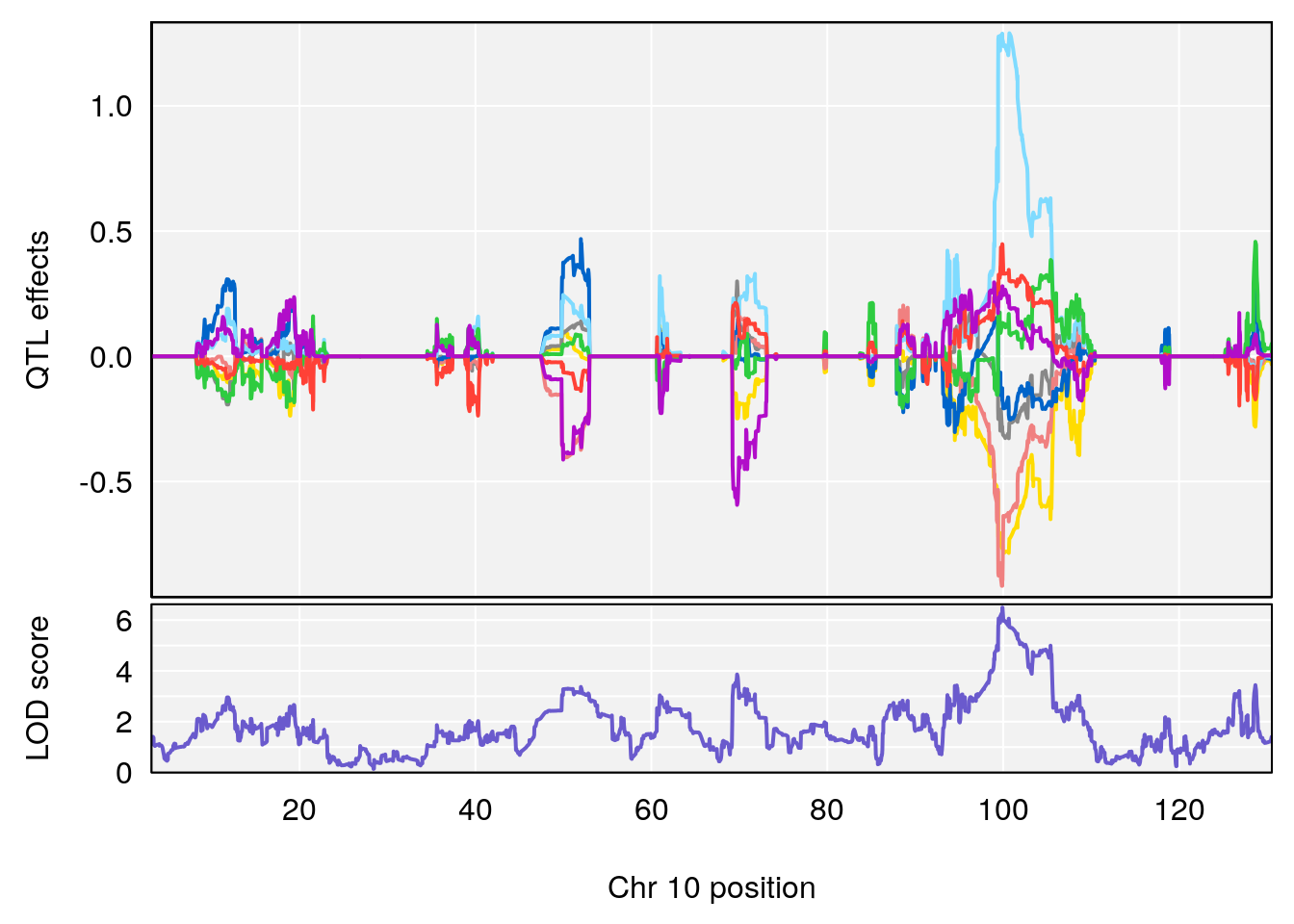
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
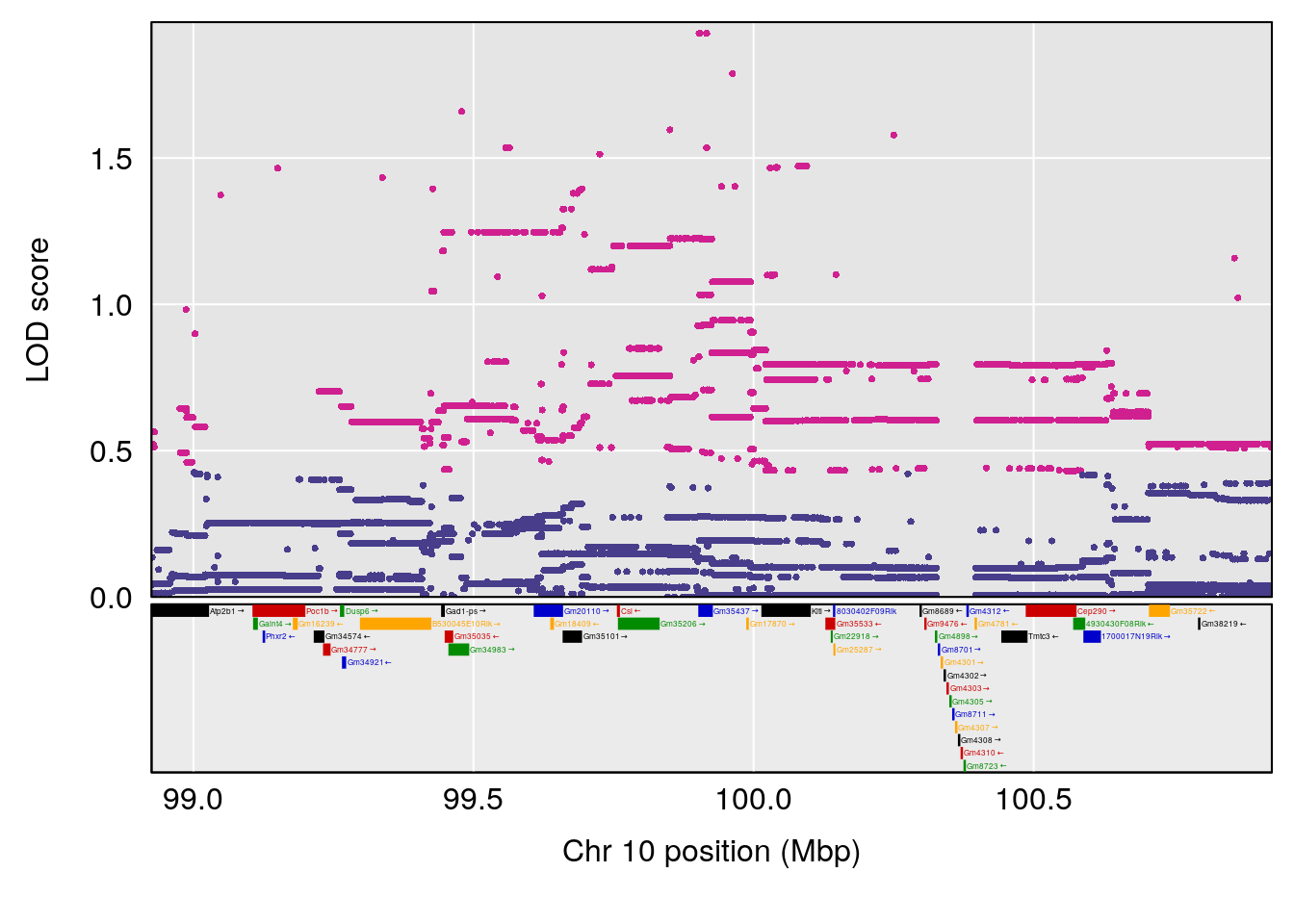
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
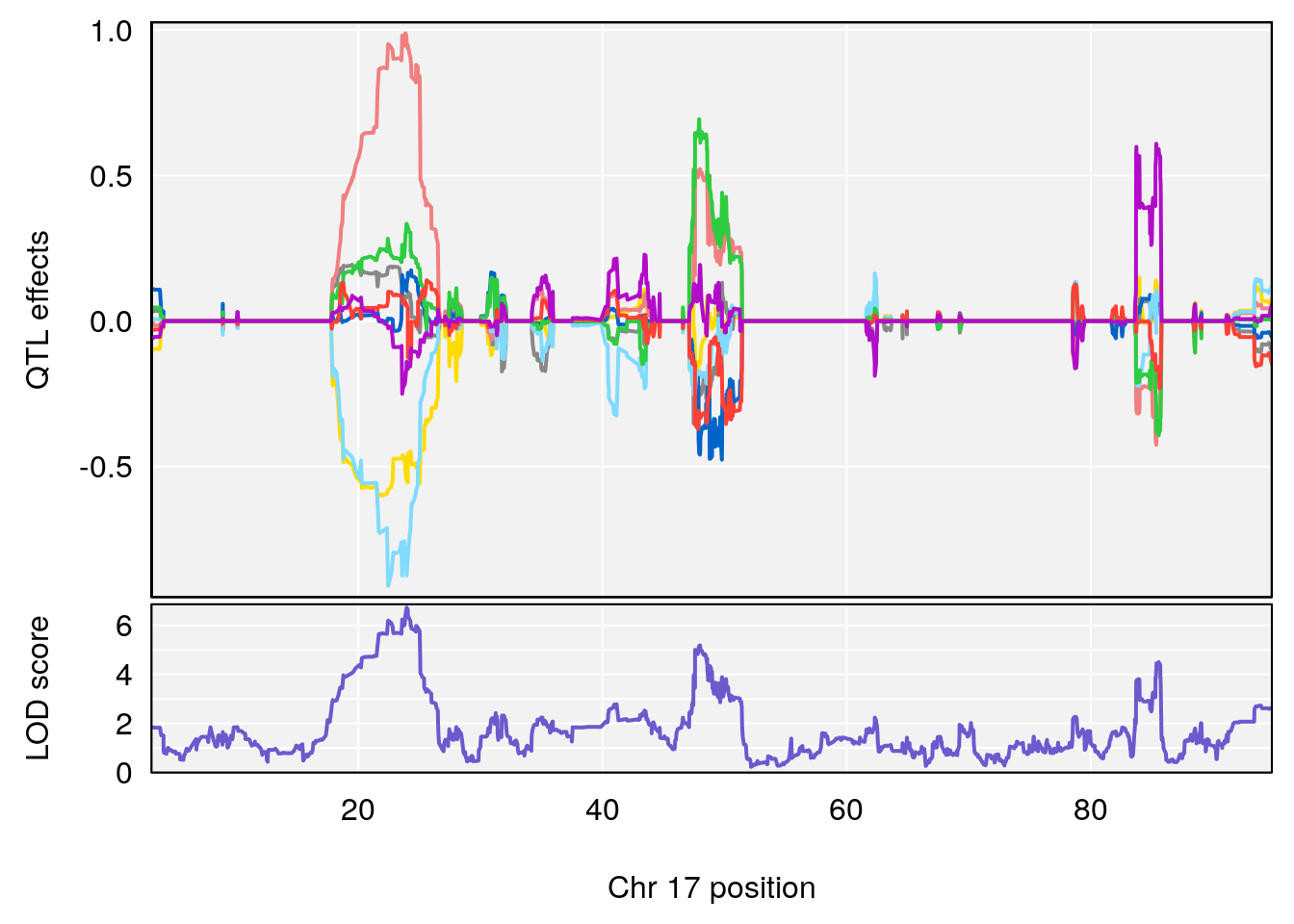
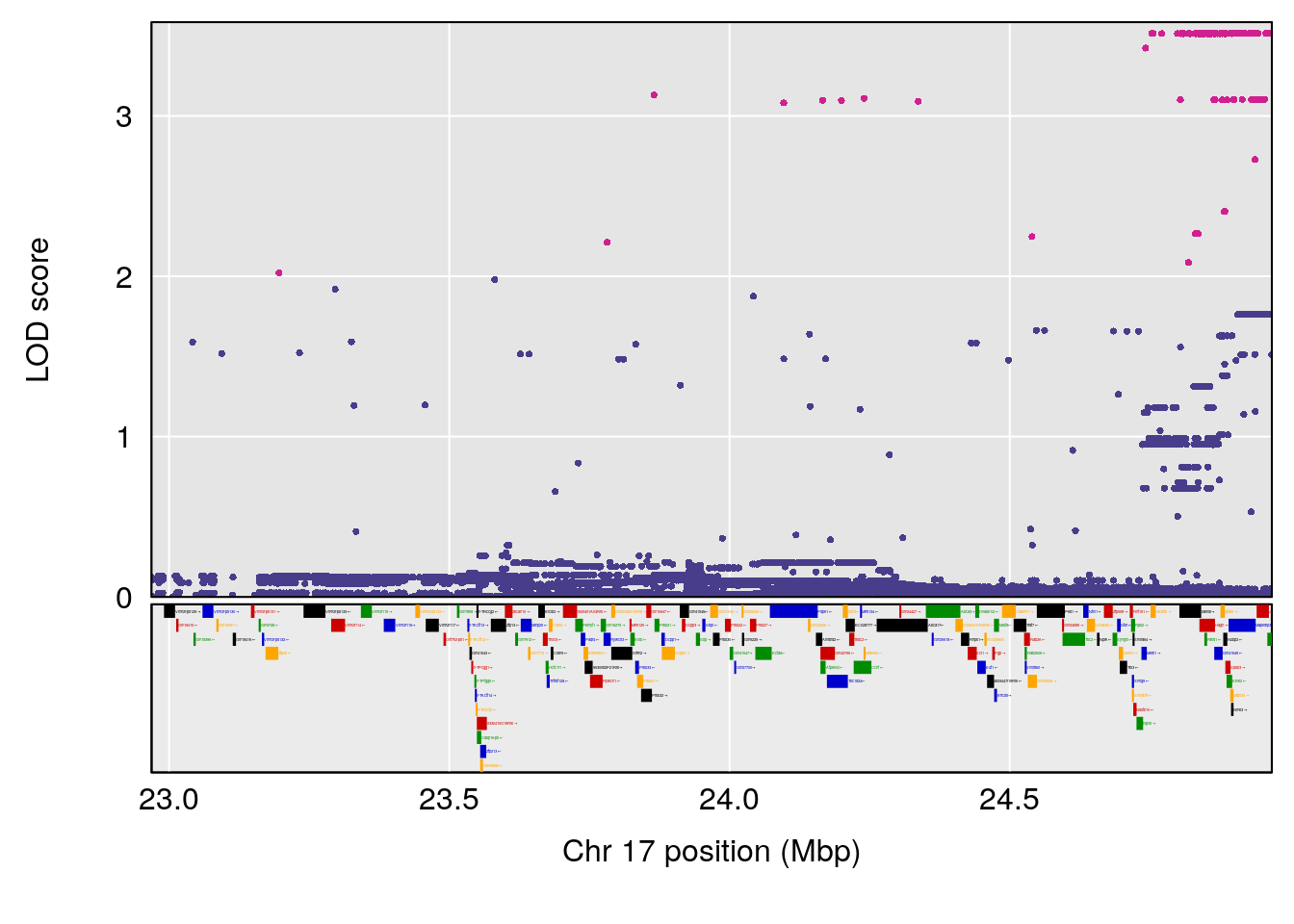
# [1] 3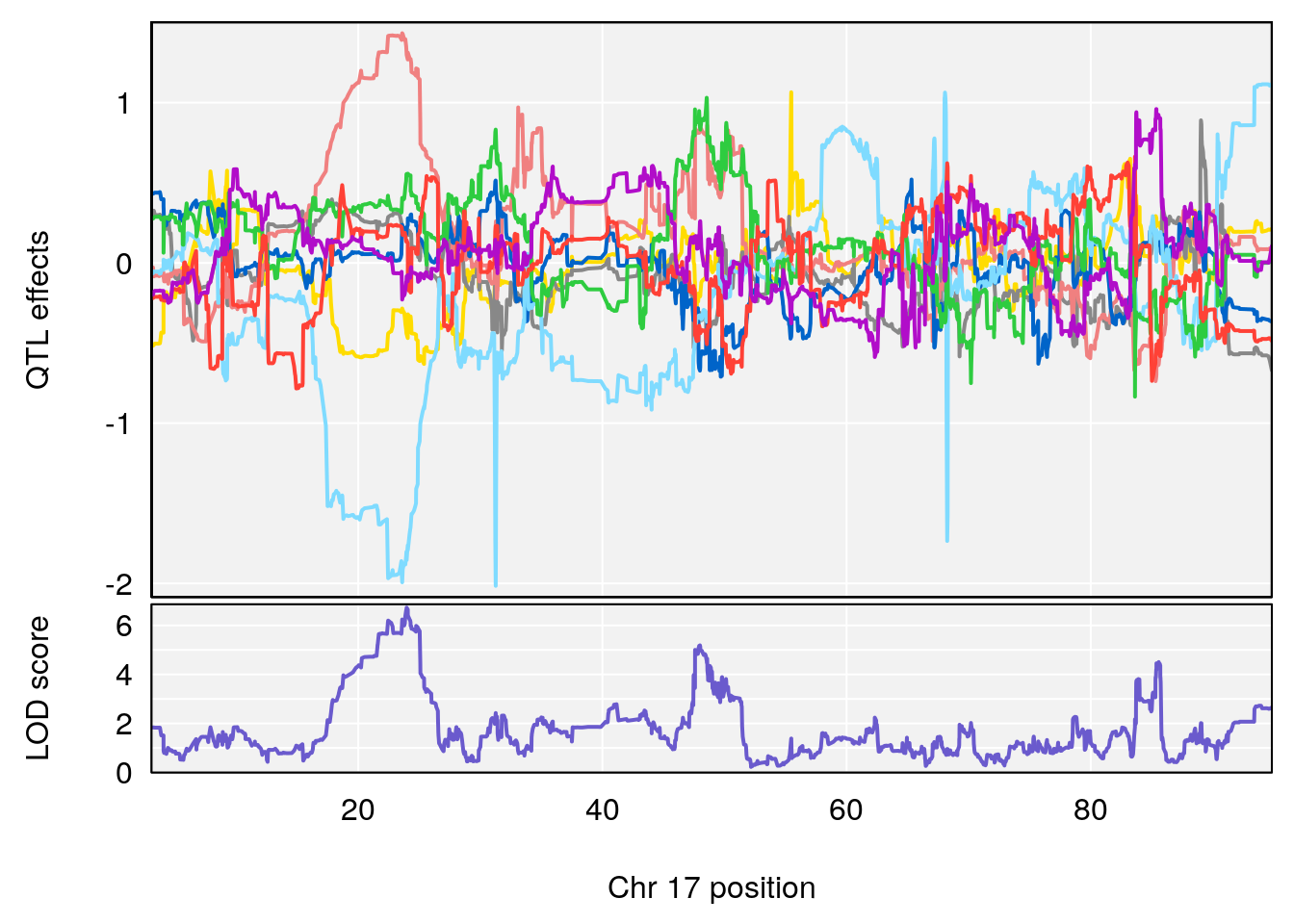
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "RR.Depression.Rate.Hr"
# [1] "Time.to.Steady.RR.Depression.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 17 33.66364 6.021735 7.725897 35.02144
# [1] 1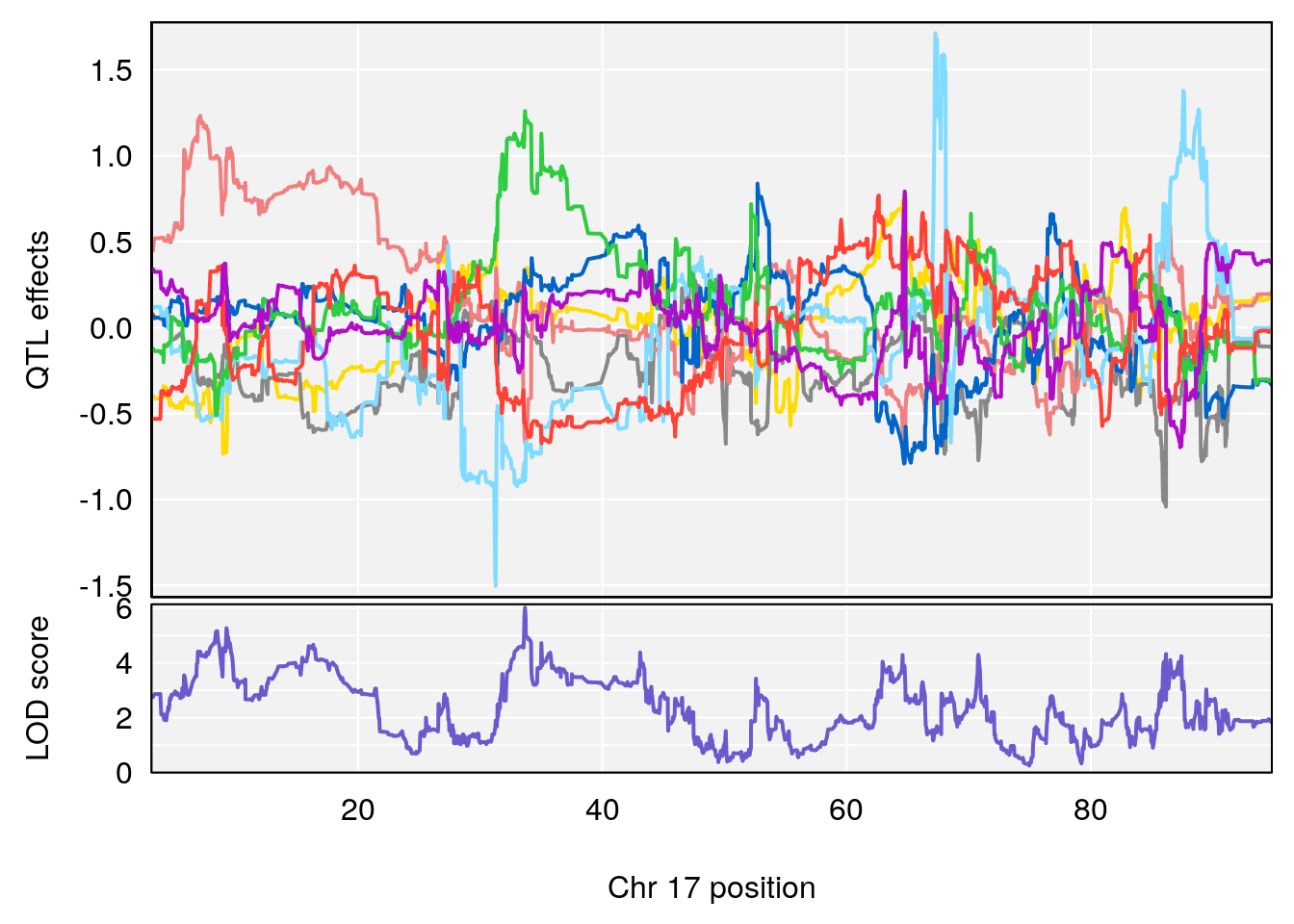
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
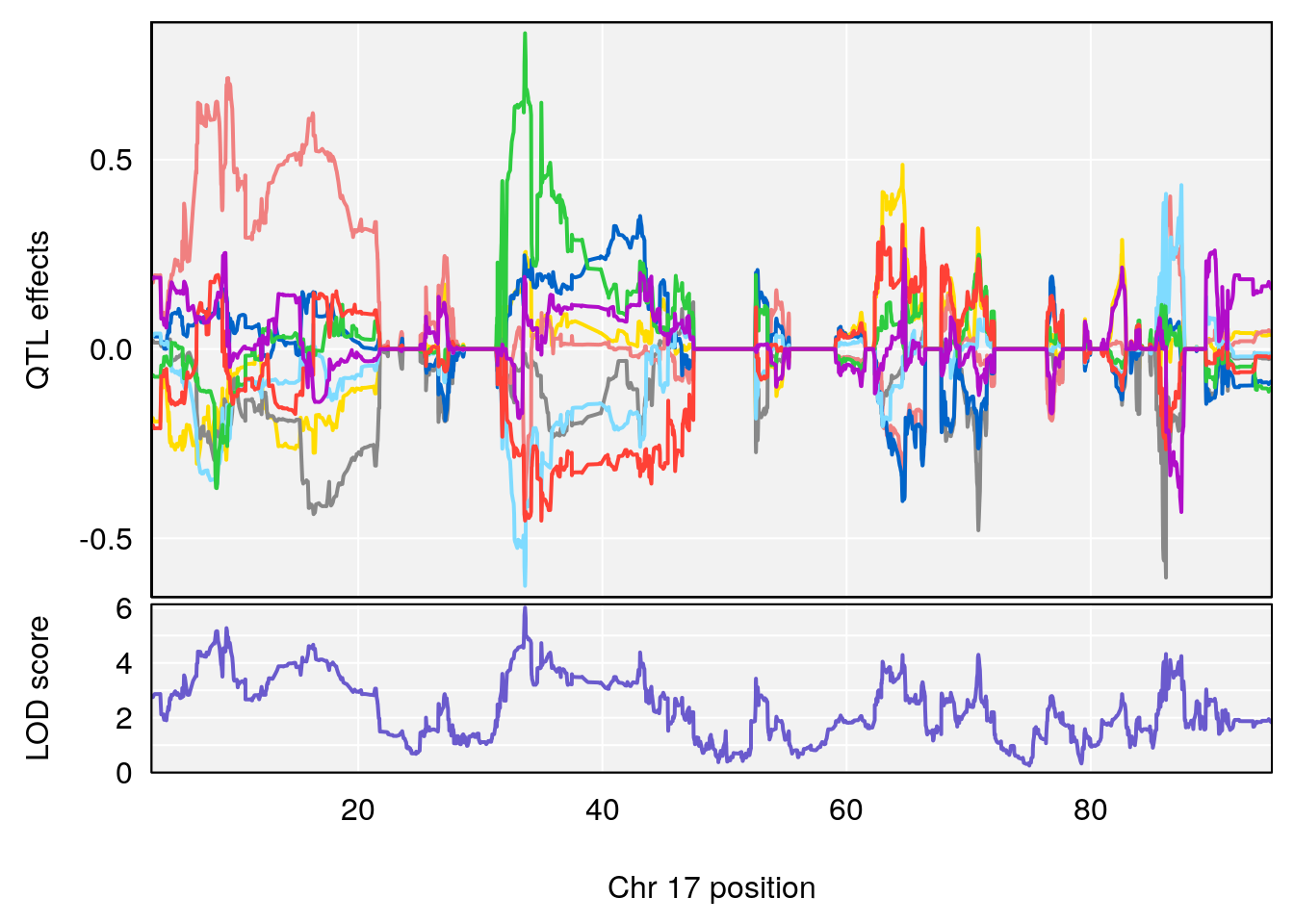
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
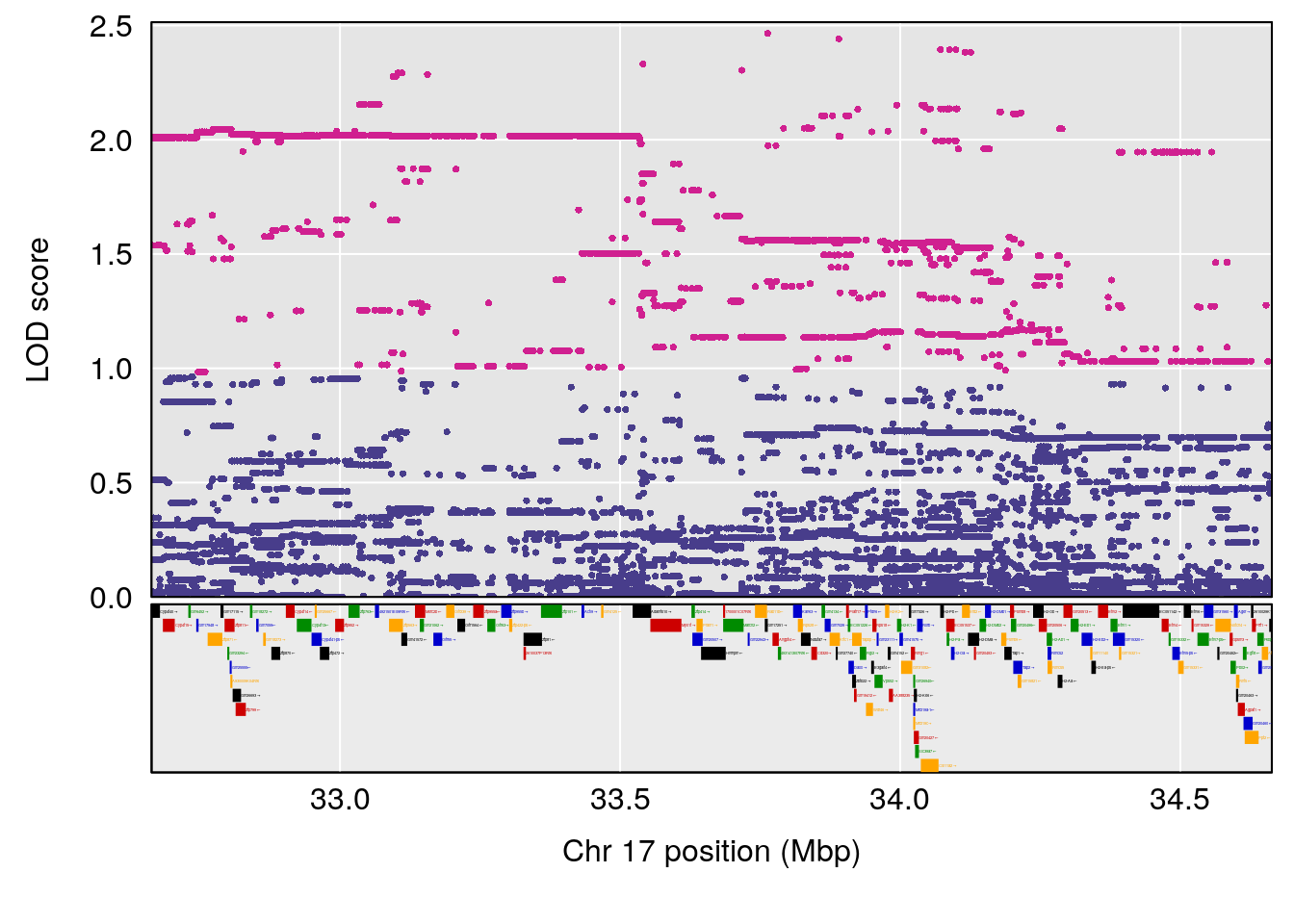
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "Min.Depression.RR"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 14 101.798 6.961191 100.3058 103.3399
# [1] 1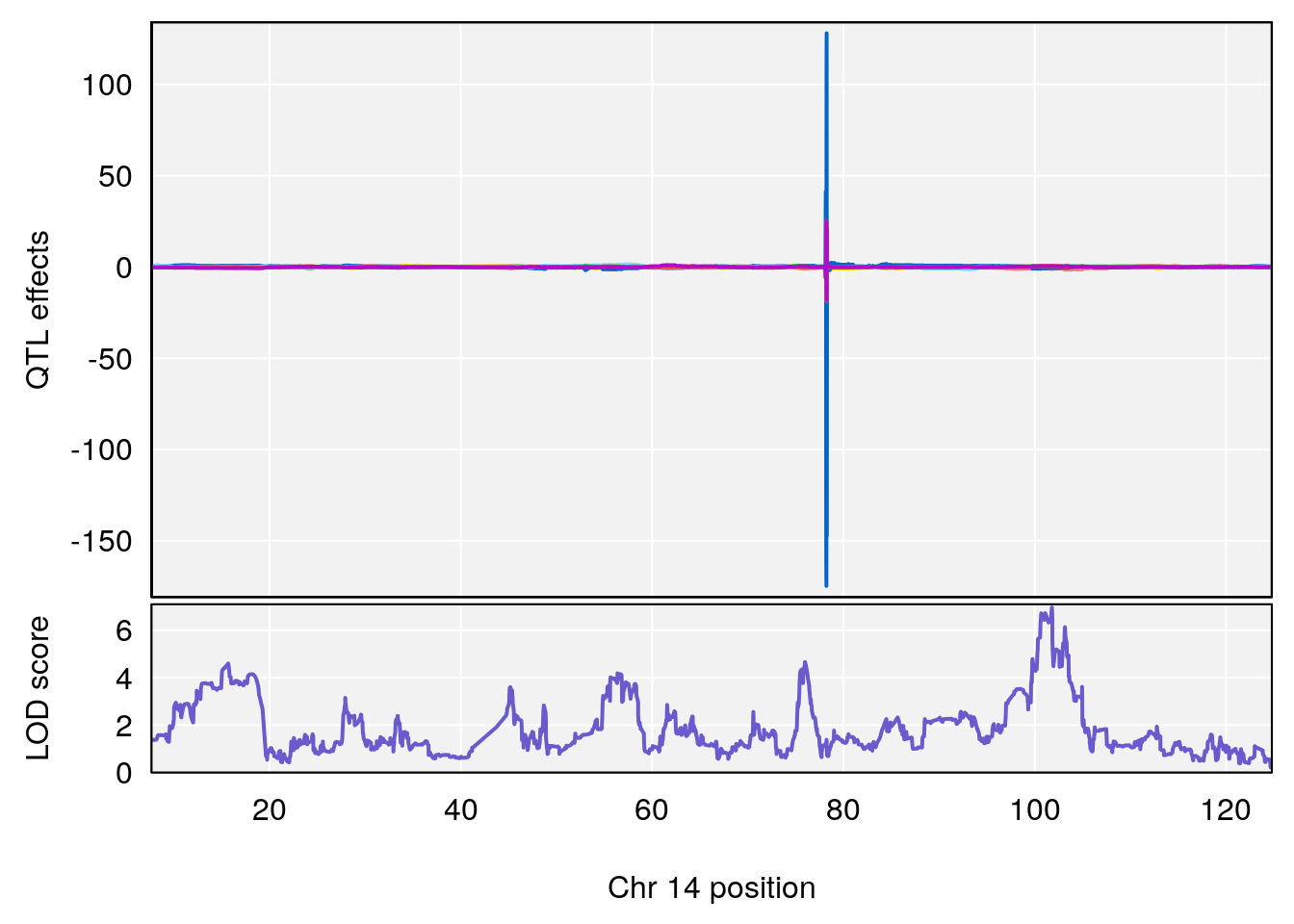
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
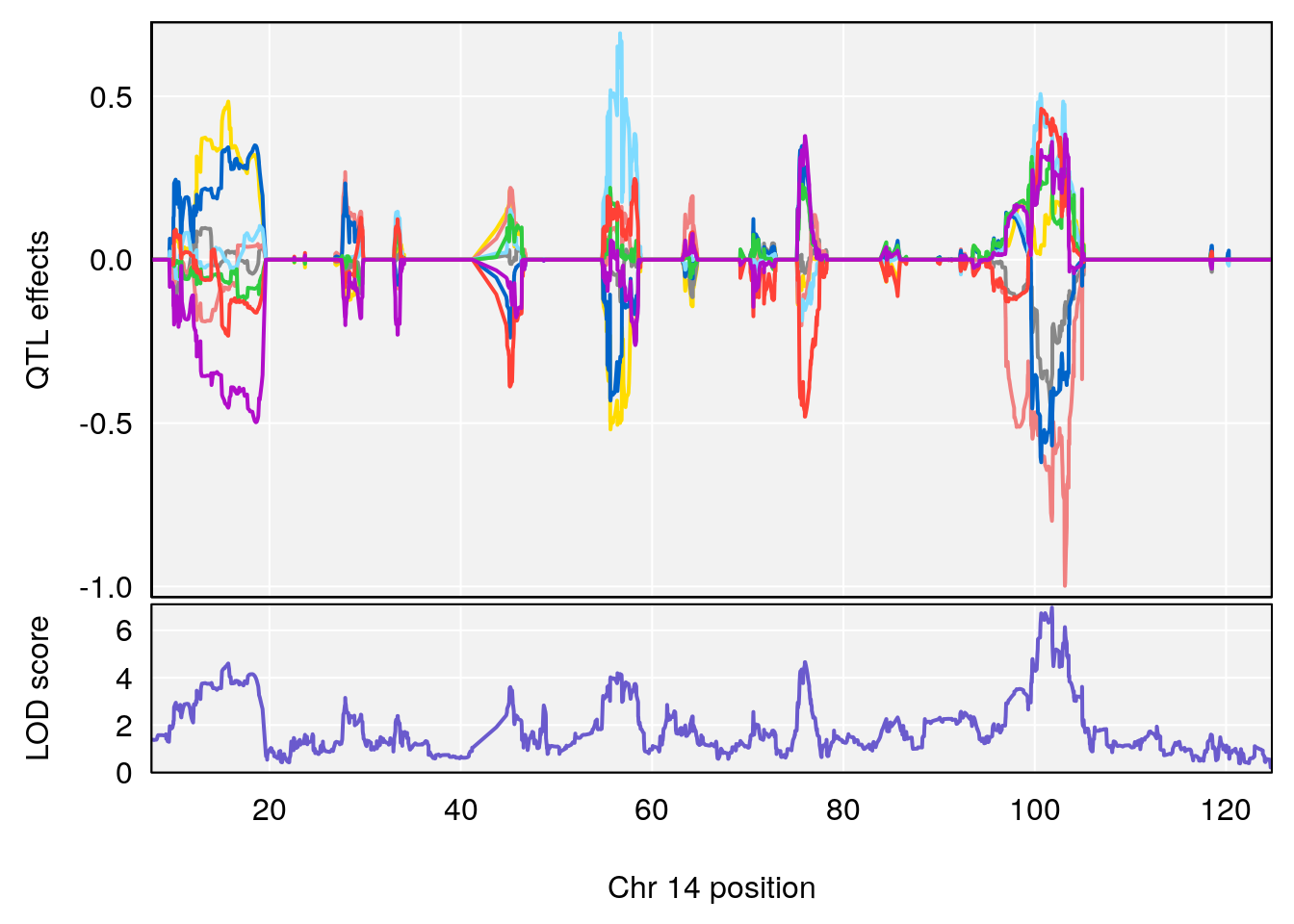
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
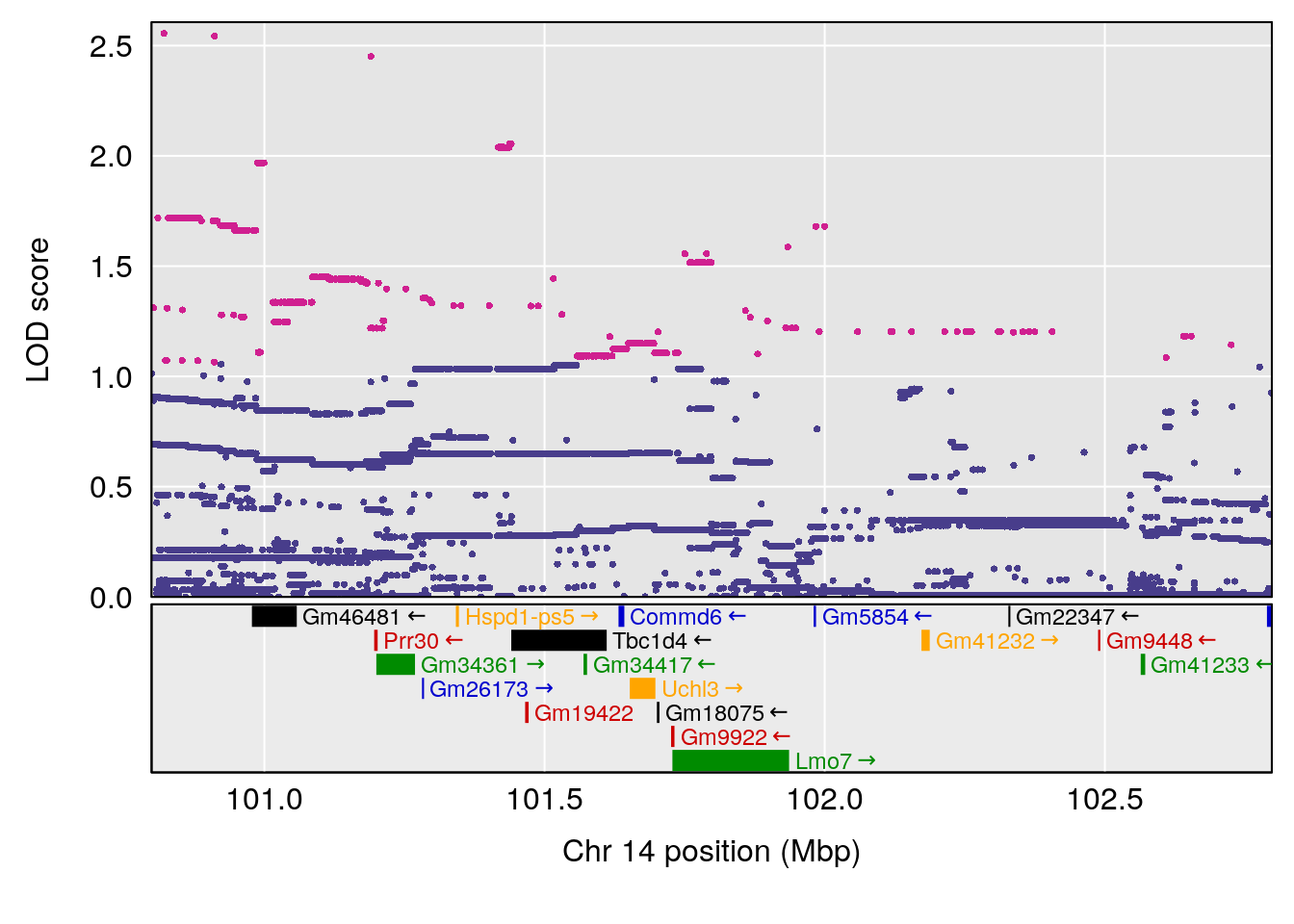
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "Mean.Depression.BR"
# [1] "SteadyState.Depression.Duration.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
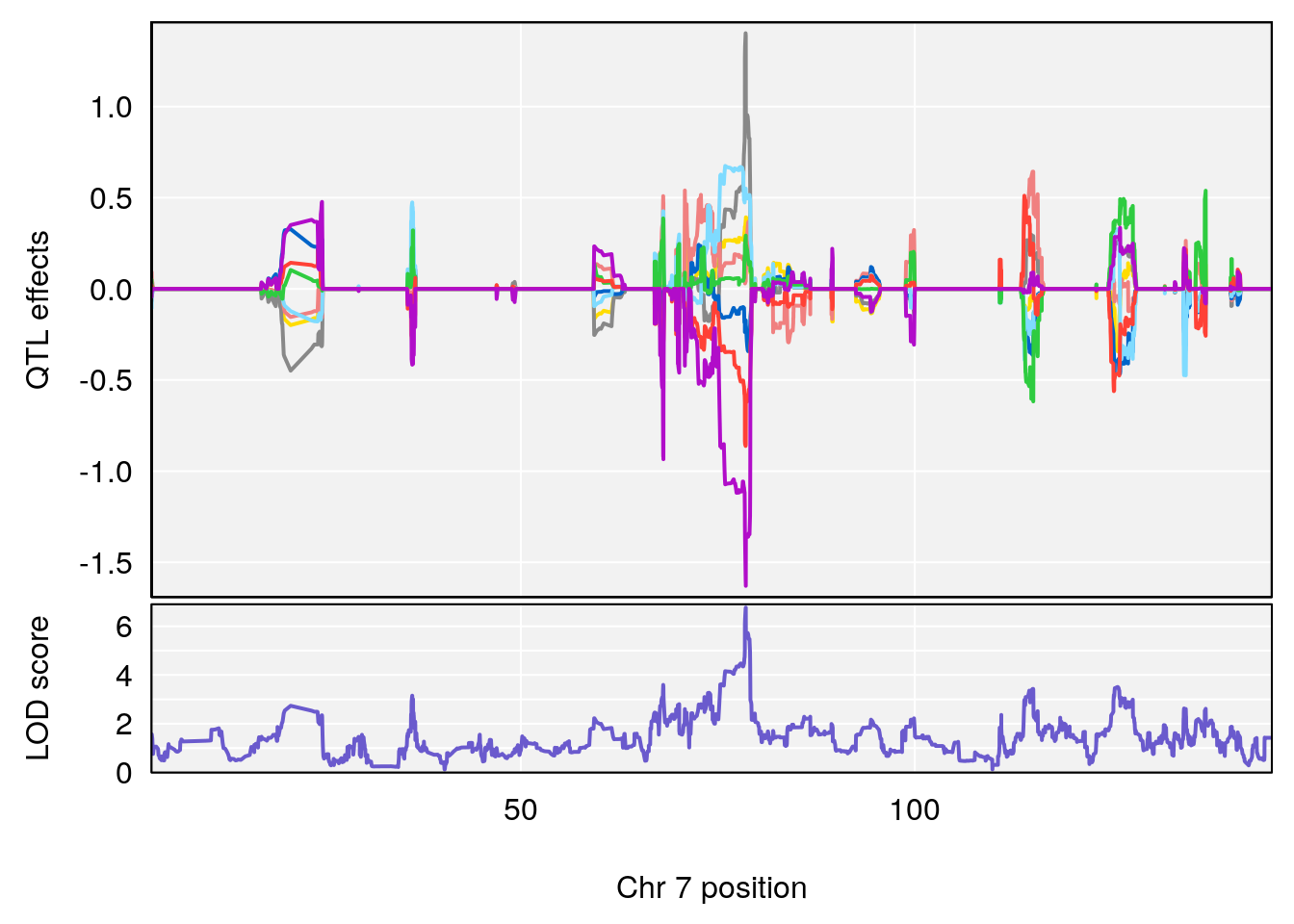
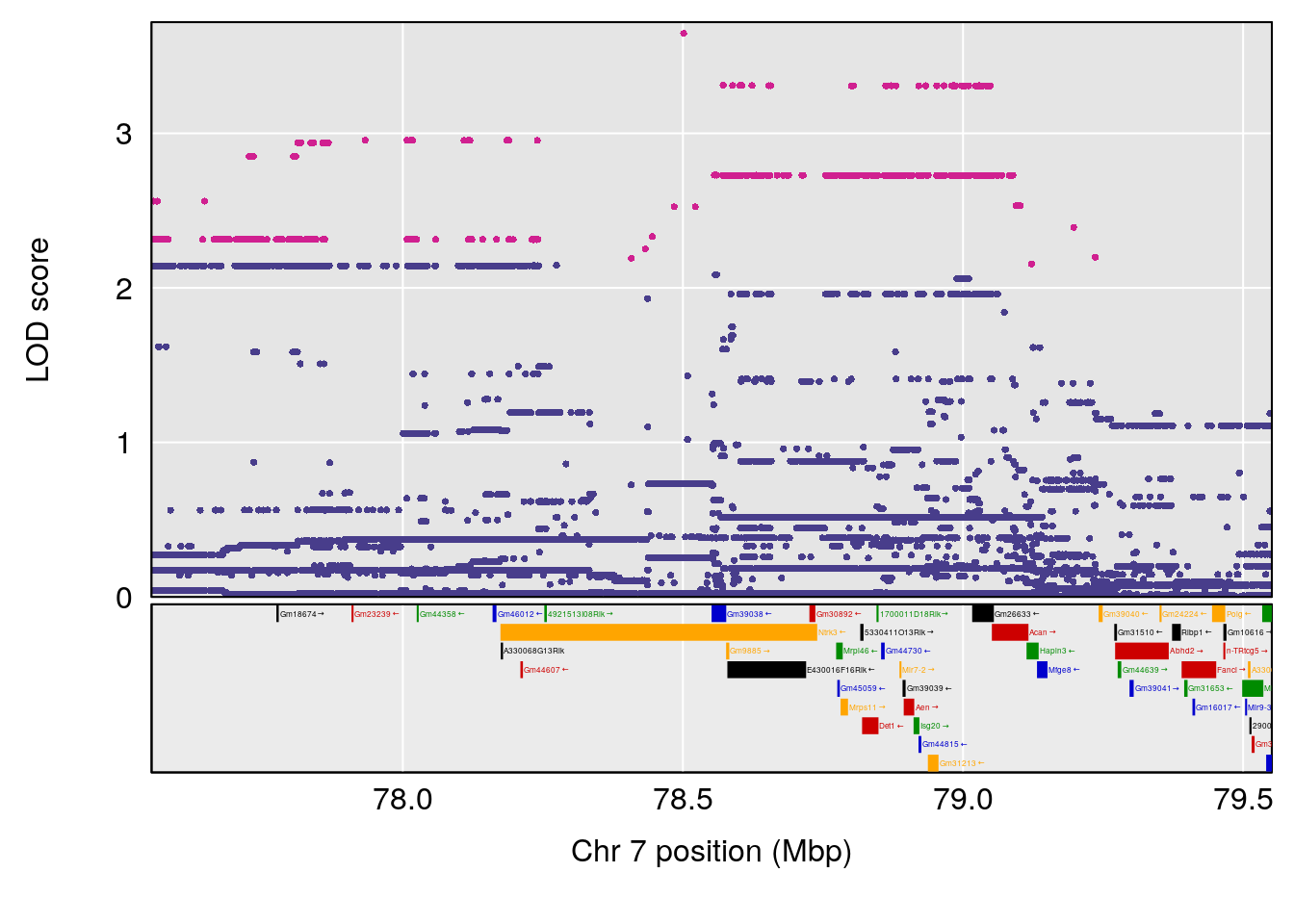
# 1 1 pheno1 4 32.63908 7.066593 31.85079 97.16594
# 2 1 pheno1 7 78.55130 6.765506 78.43609 79.07406
# 3 1 pheno1 15 82.95976 6.039949 82.61311 84.16285
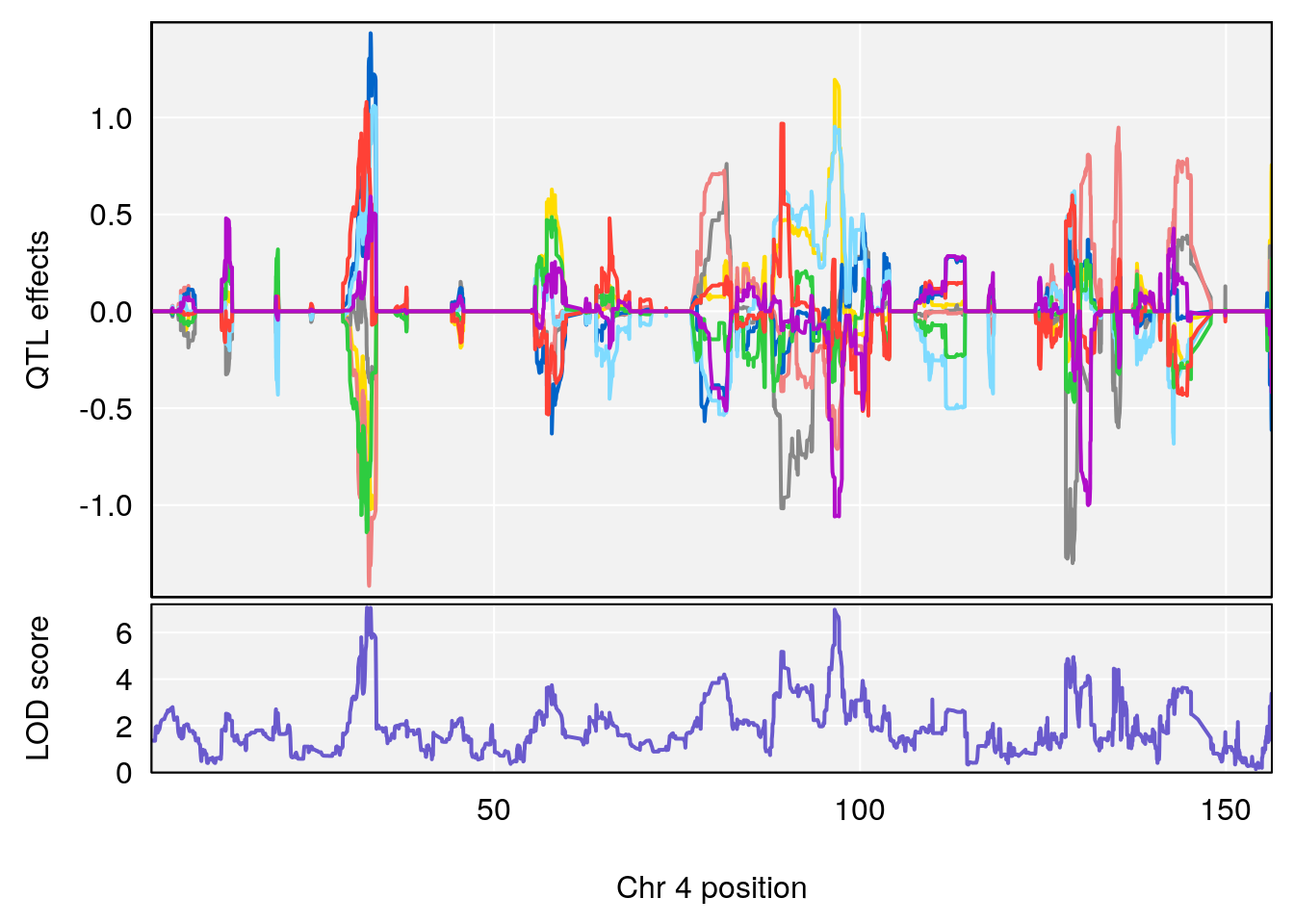
# [1] 1
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
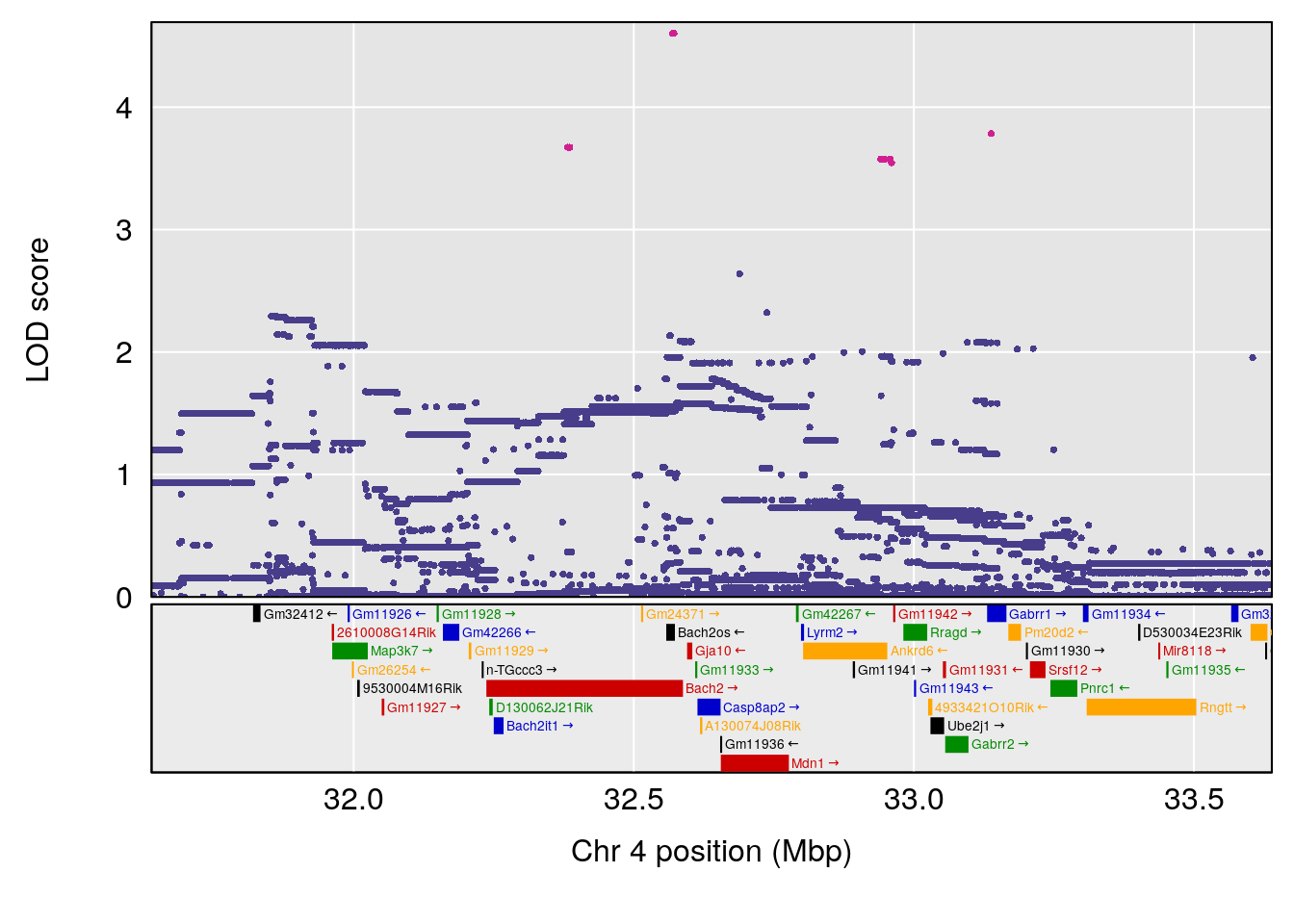
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2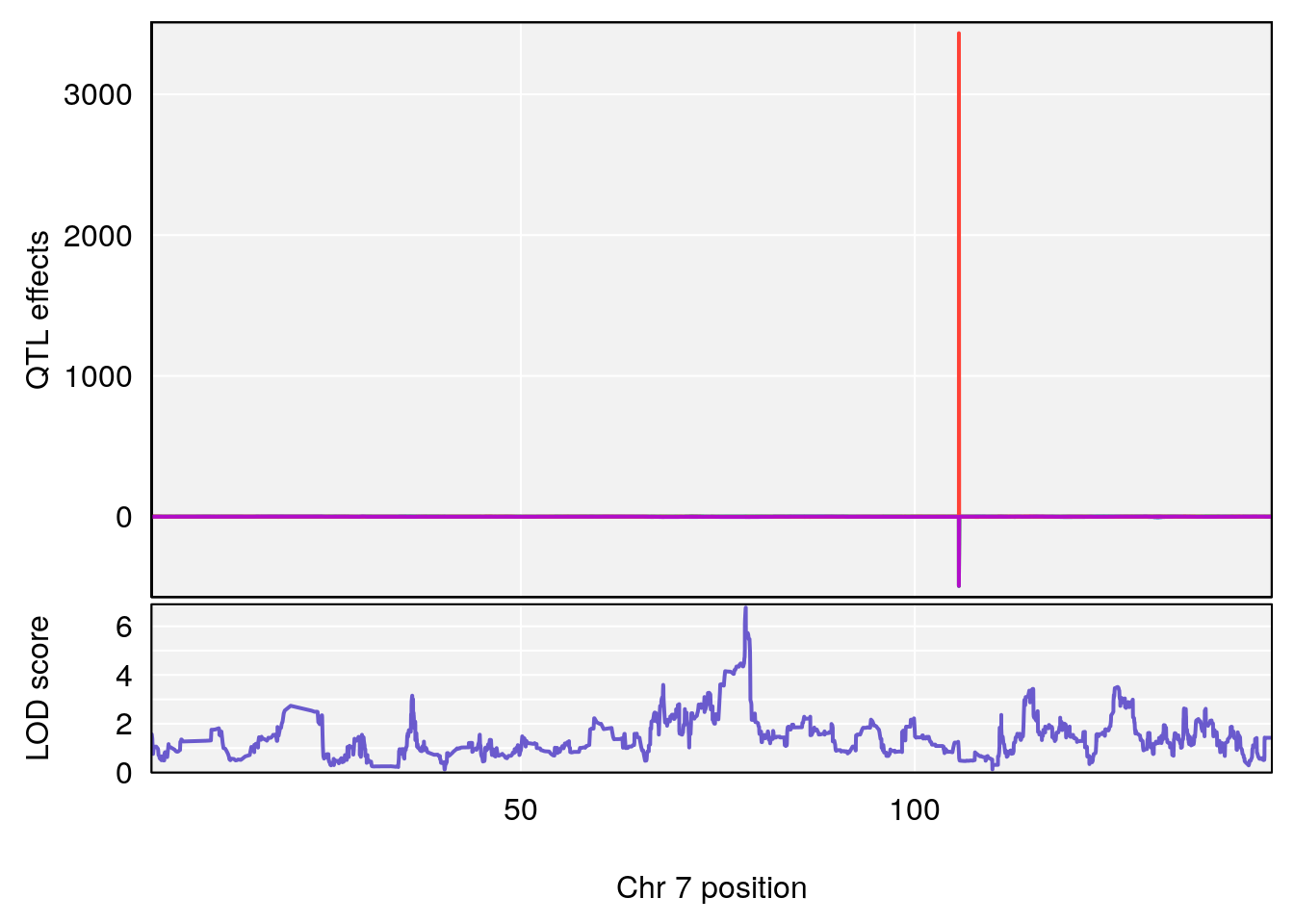
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 3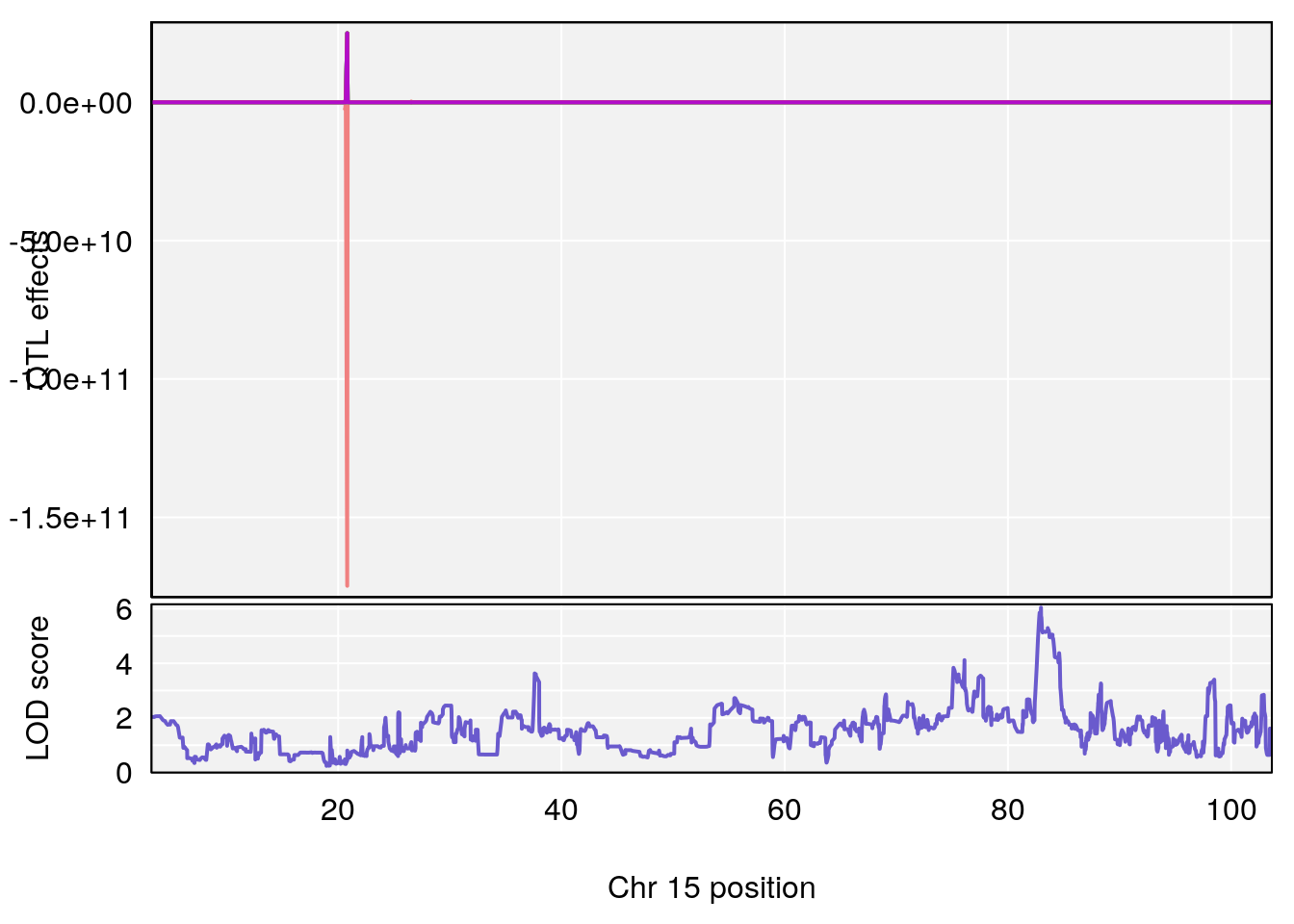
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
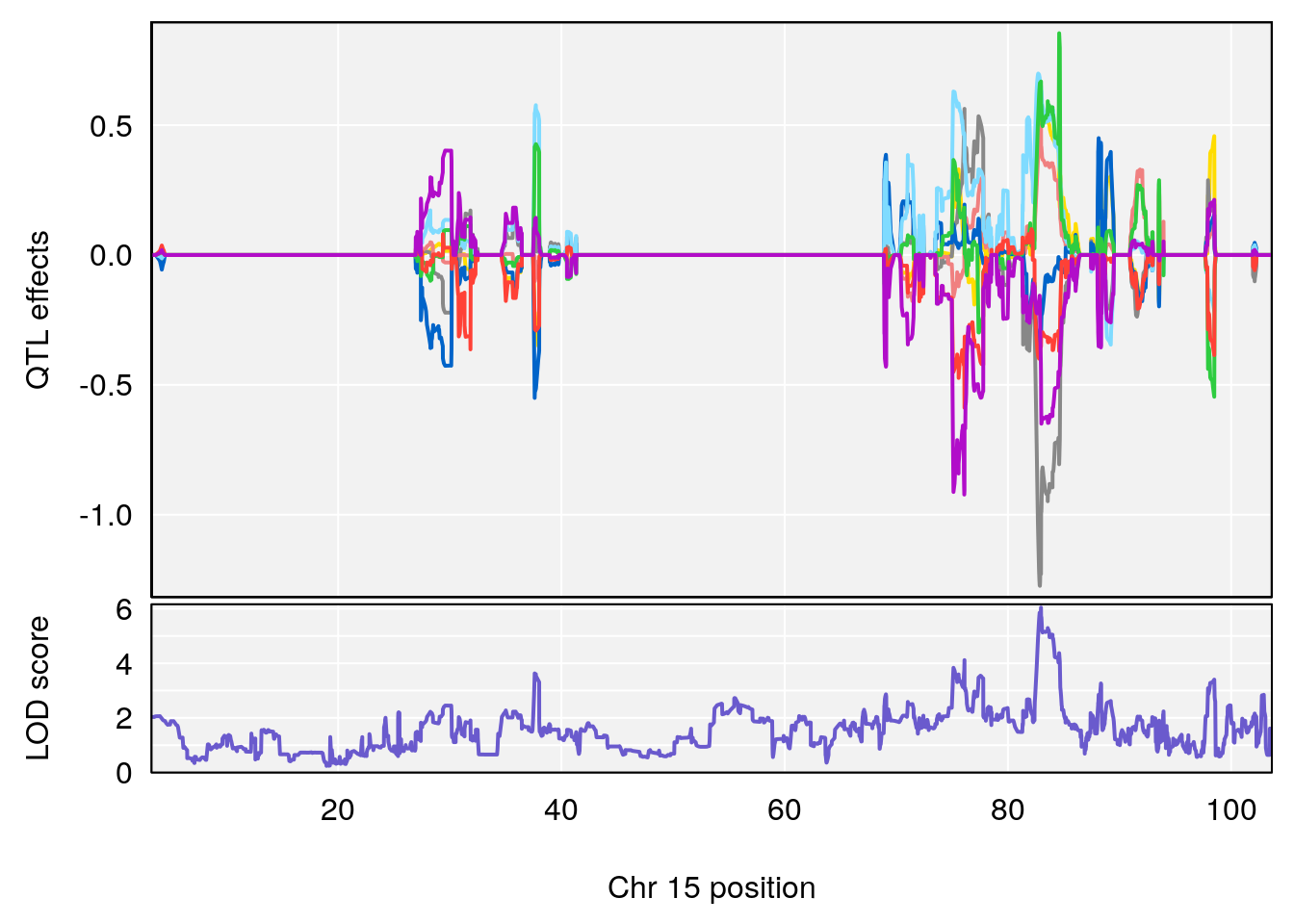
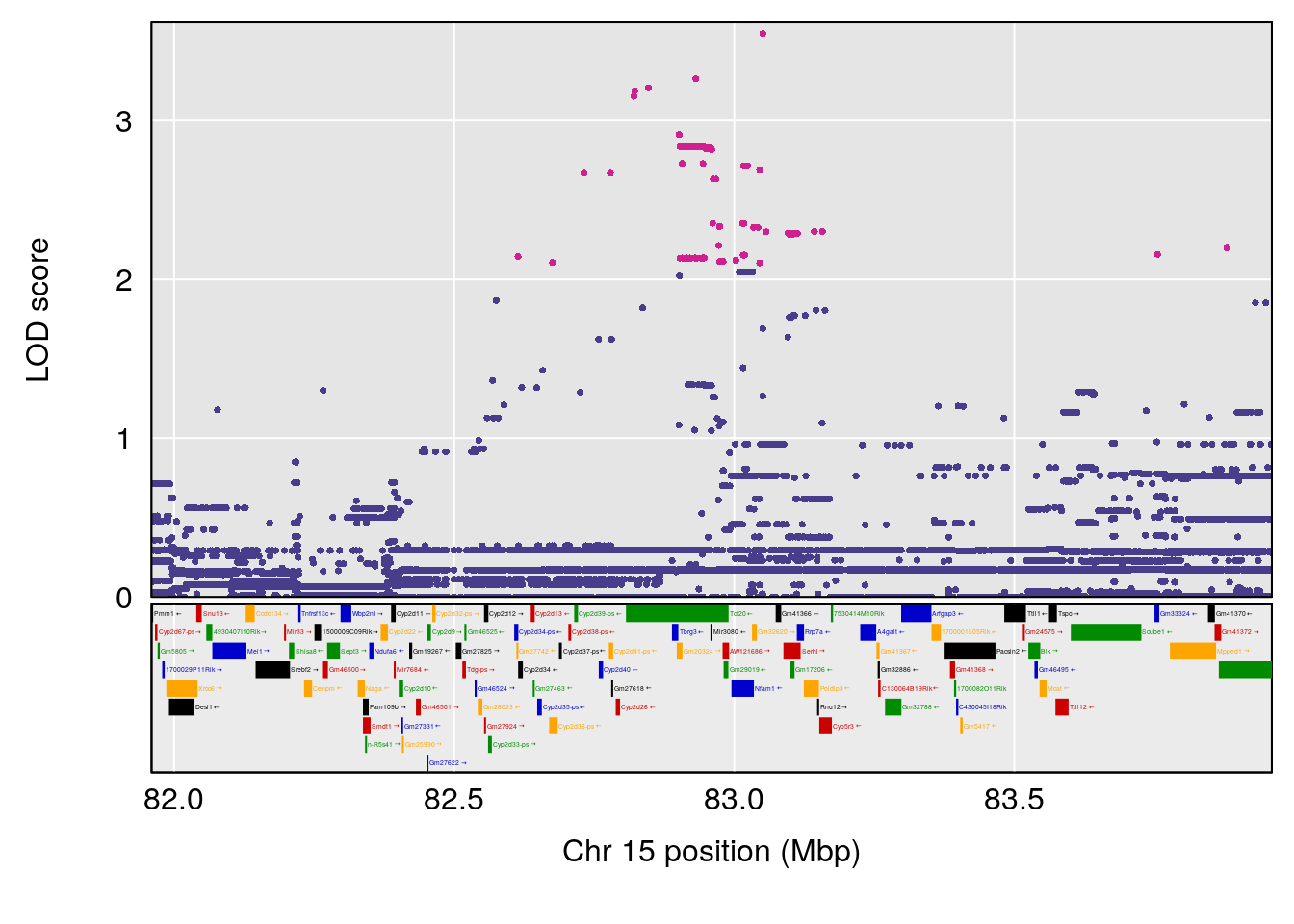
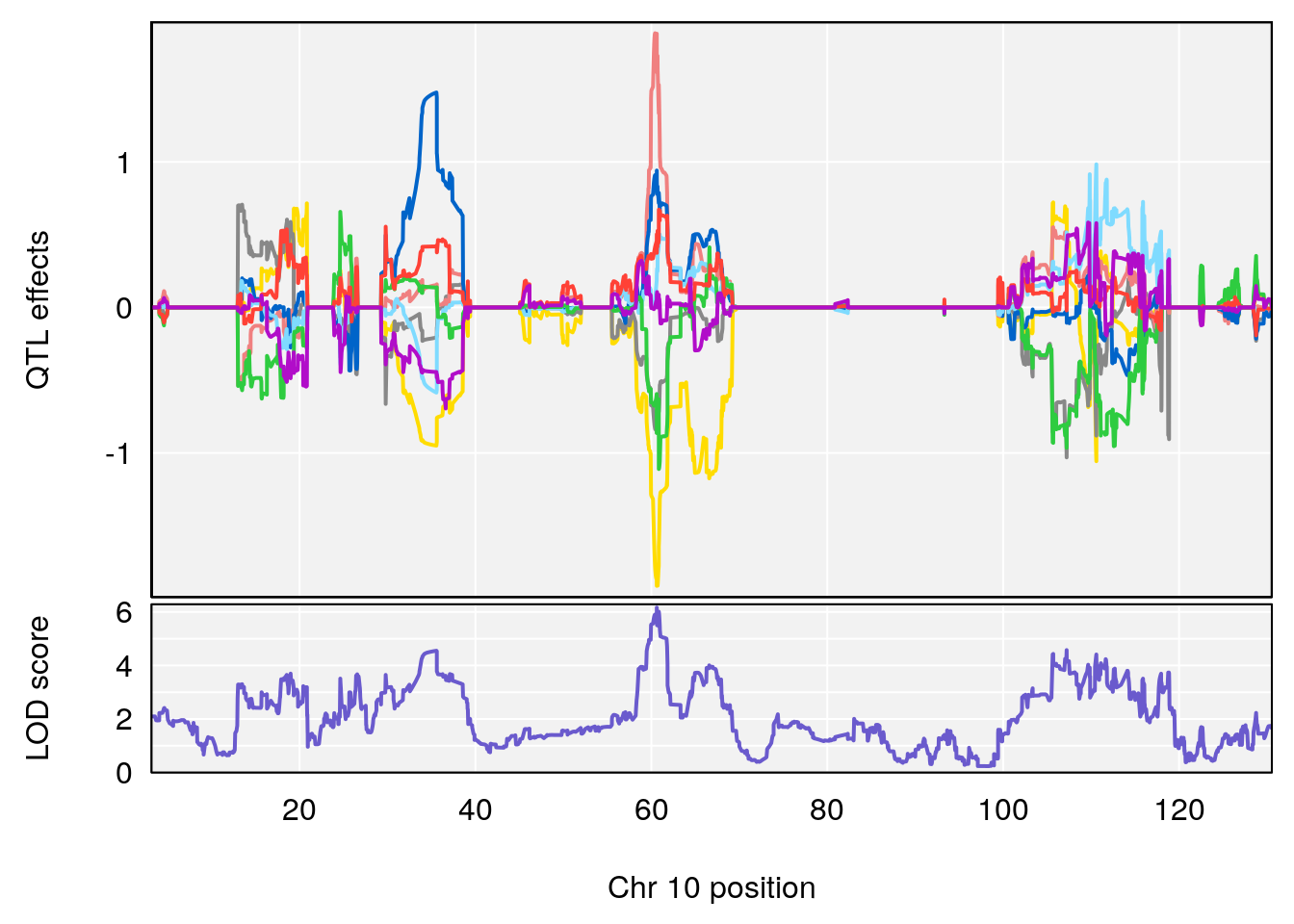
# [1] "Time.to.Threshold.Recovery.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 10 60.61789 6.163288 59.50257 61.79358
# [1] 1
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
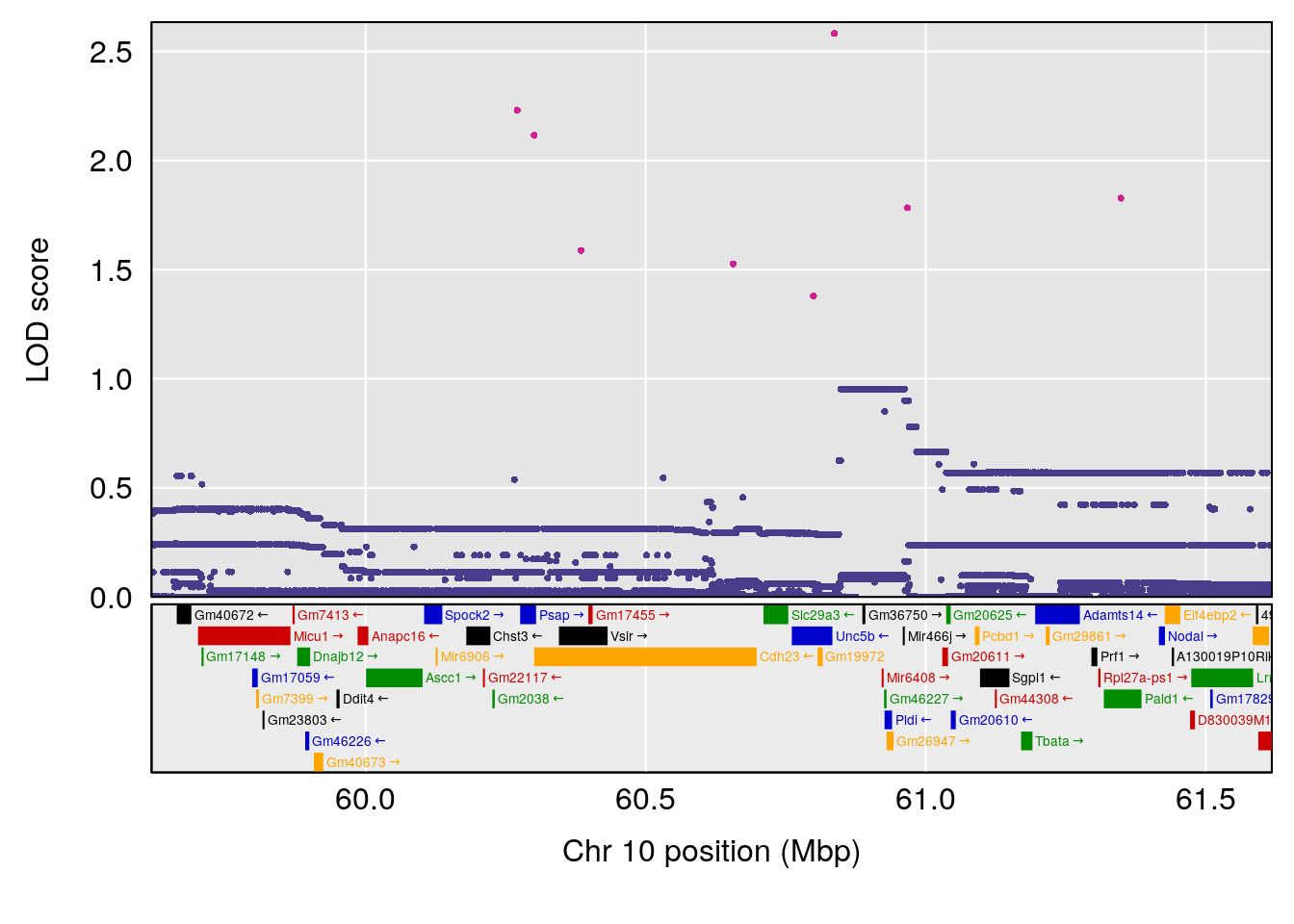
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "Time.to.Projected.Recovery.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 180.4362 8.101813 180.3185 180.5471
# [1] 1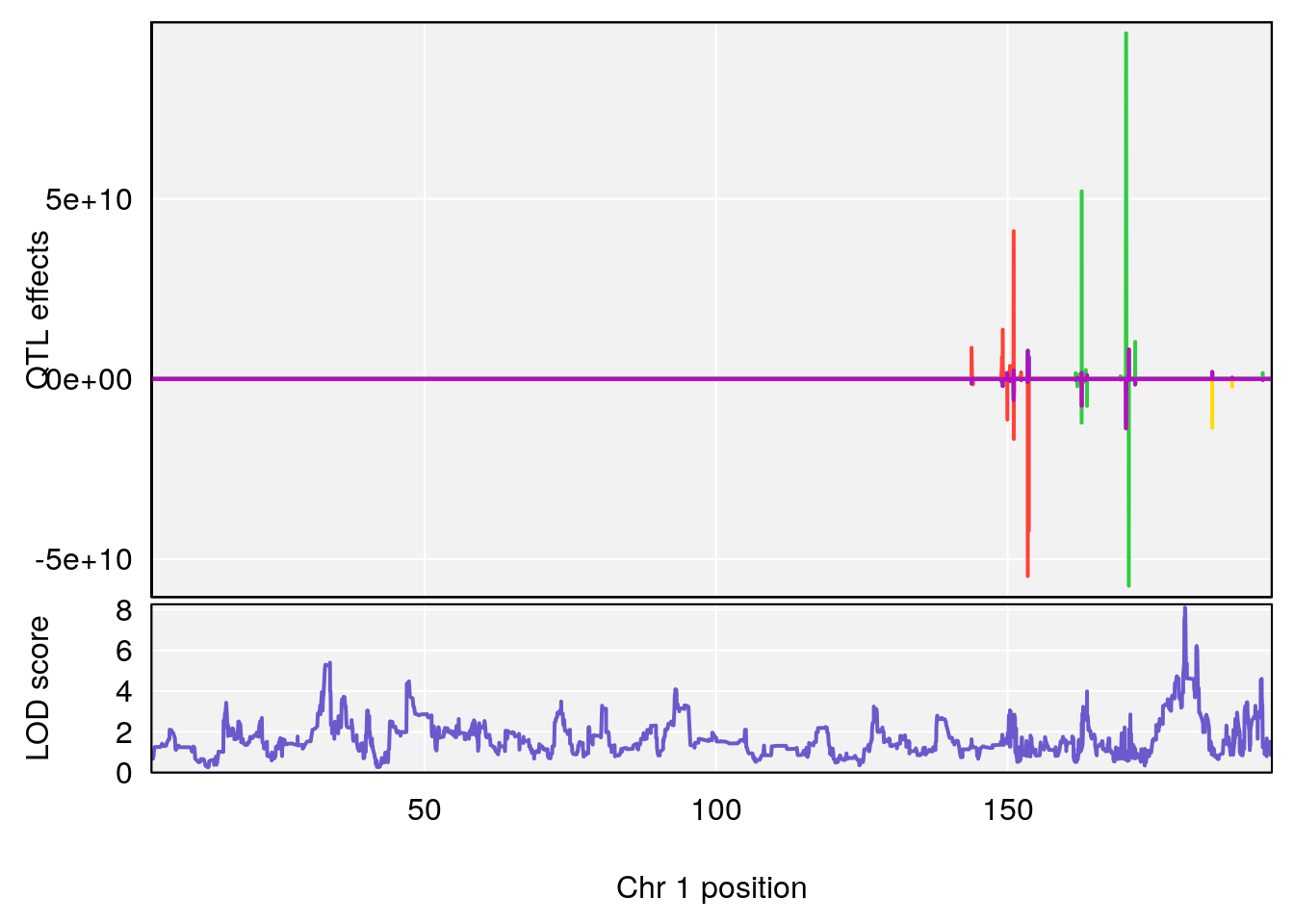
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
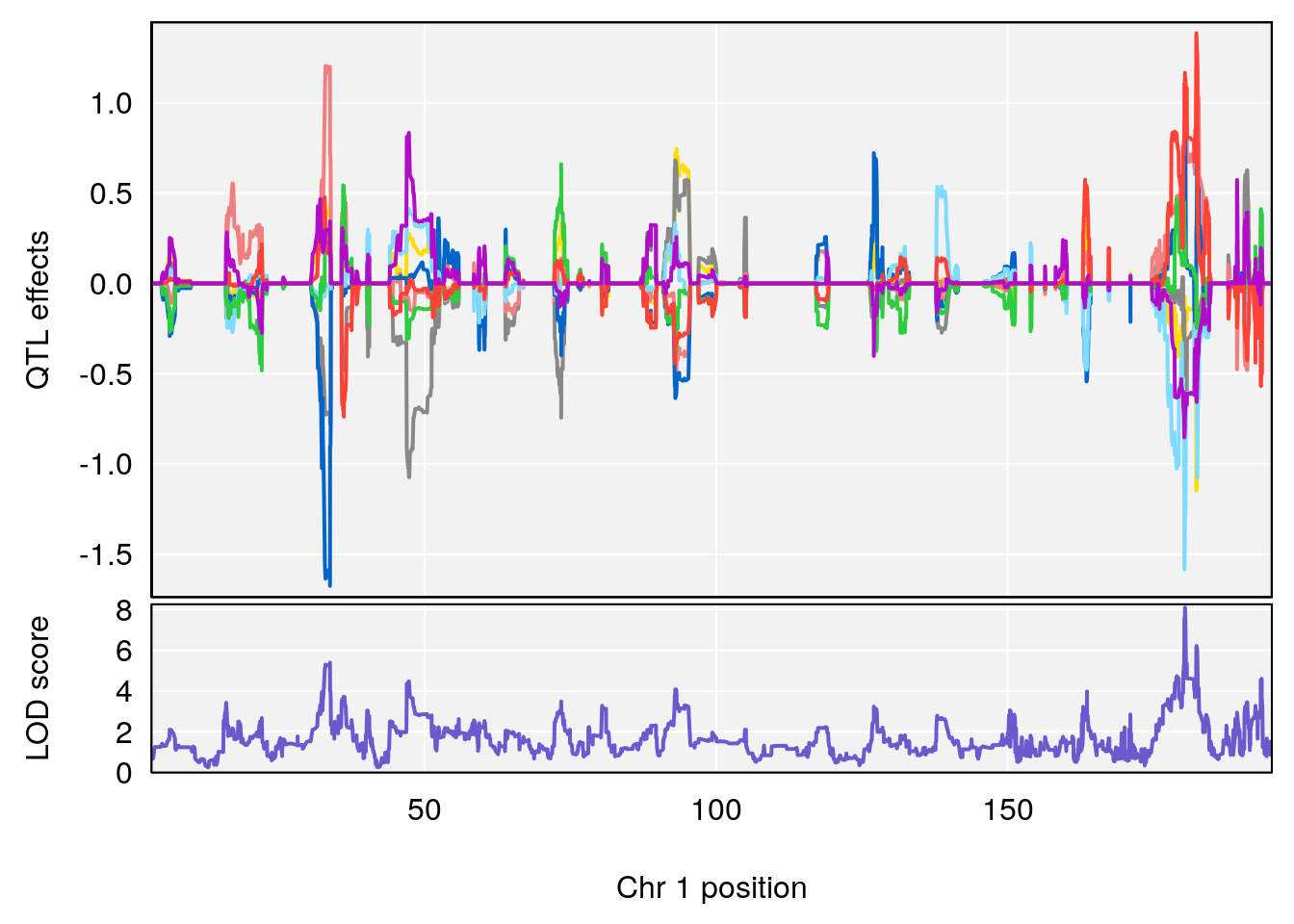
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
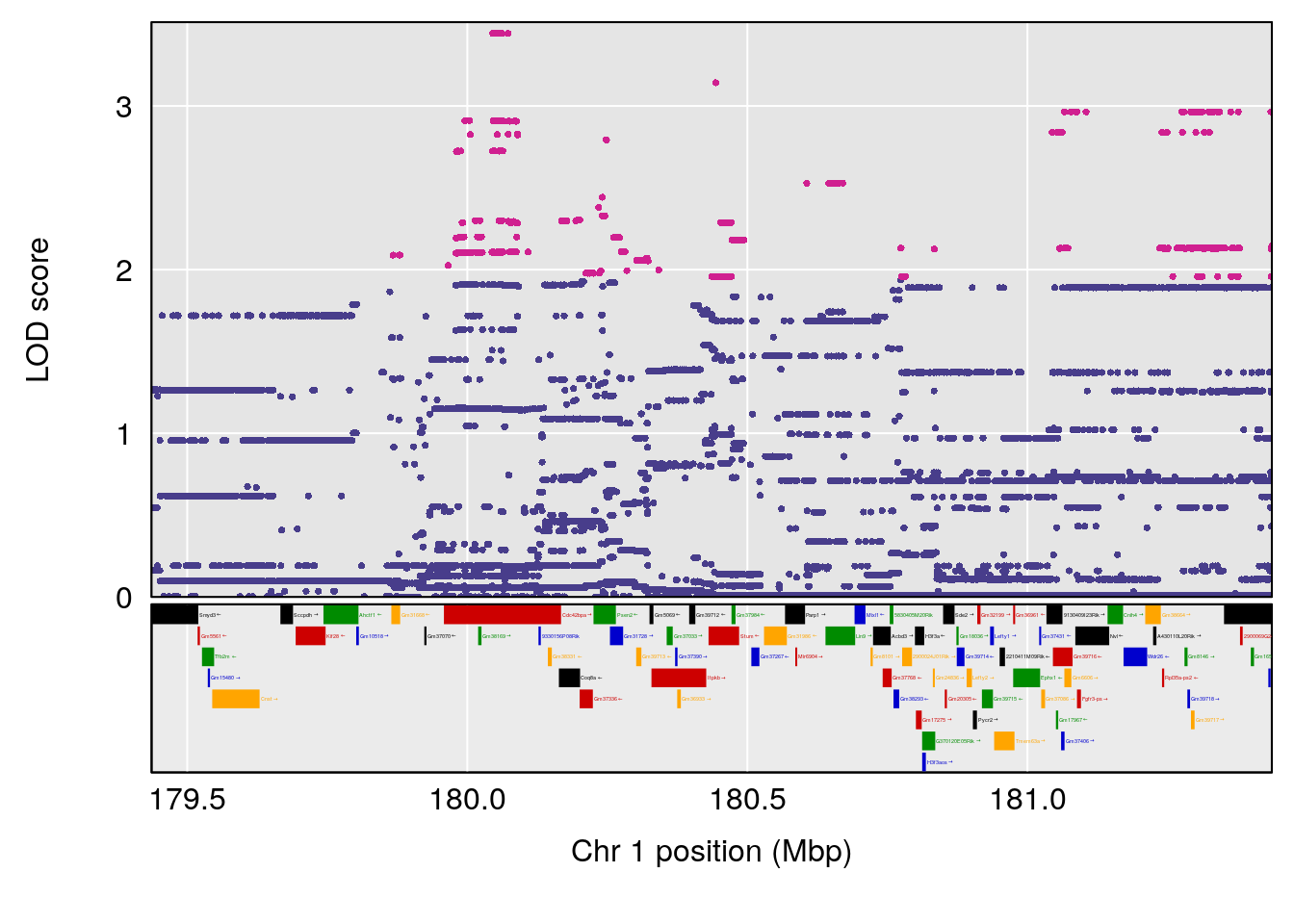
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "Start.of.Recovery.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 4 32.63908 7.105498 31.85079 97.15470
# 2 1 pheno1 7 78.55130 6.935599 78.43609 79.07697
# 3 1 pheno1 15 82.95976 6.085487 76.08766 84.17742
# [1] 1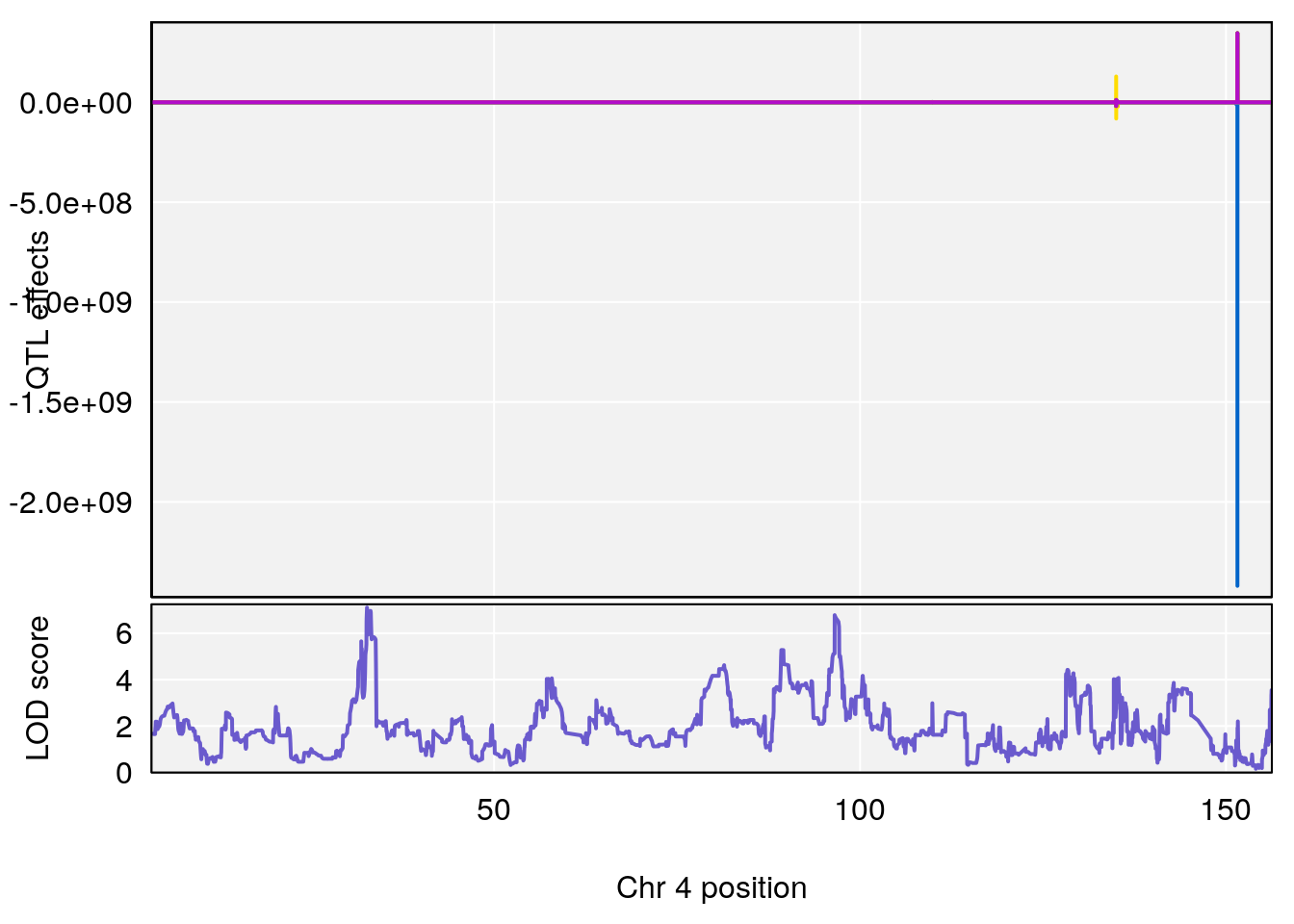
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
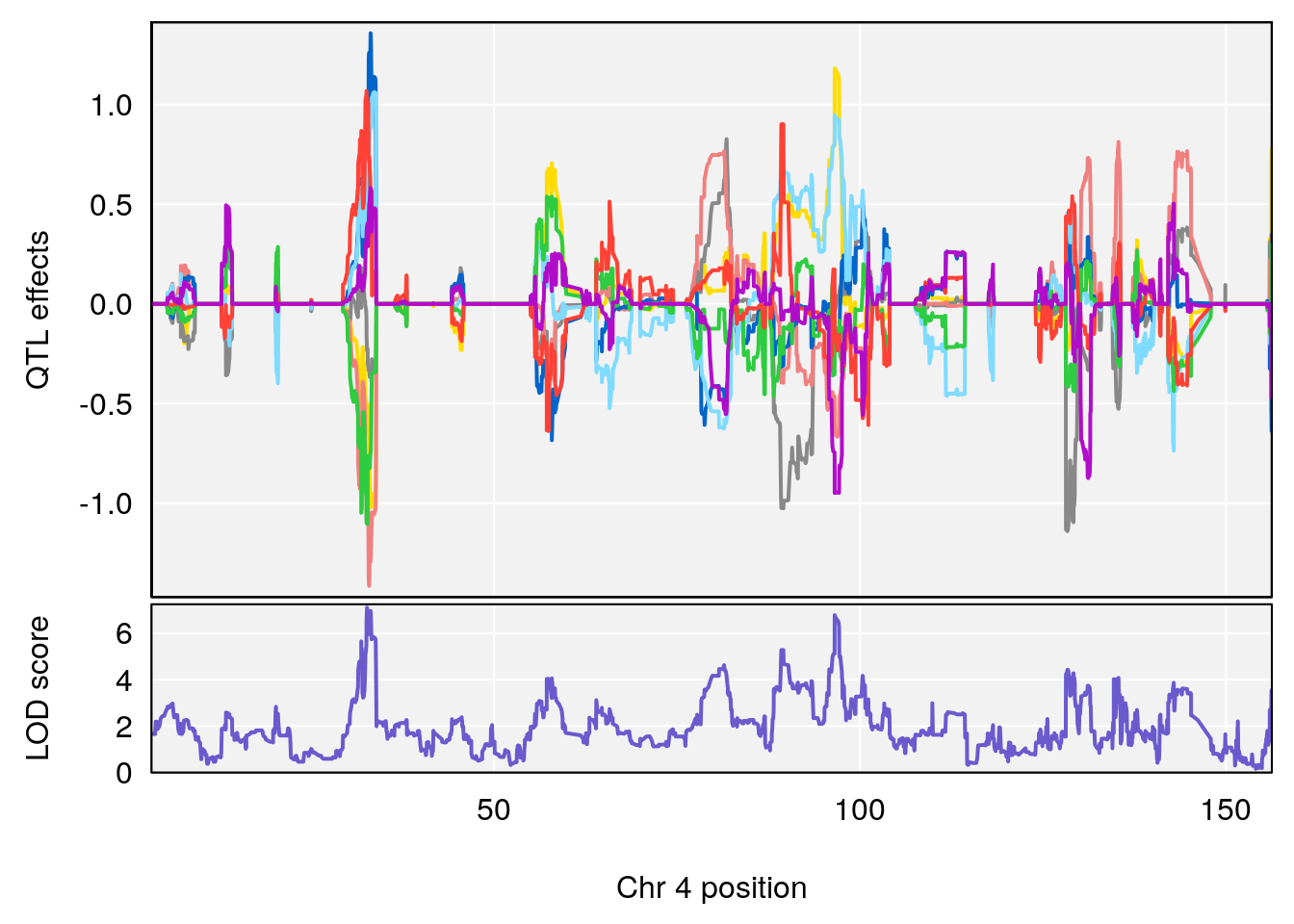
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
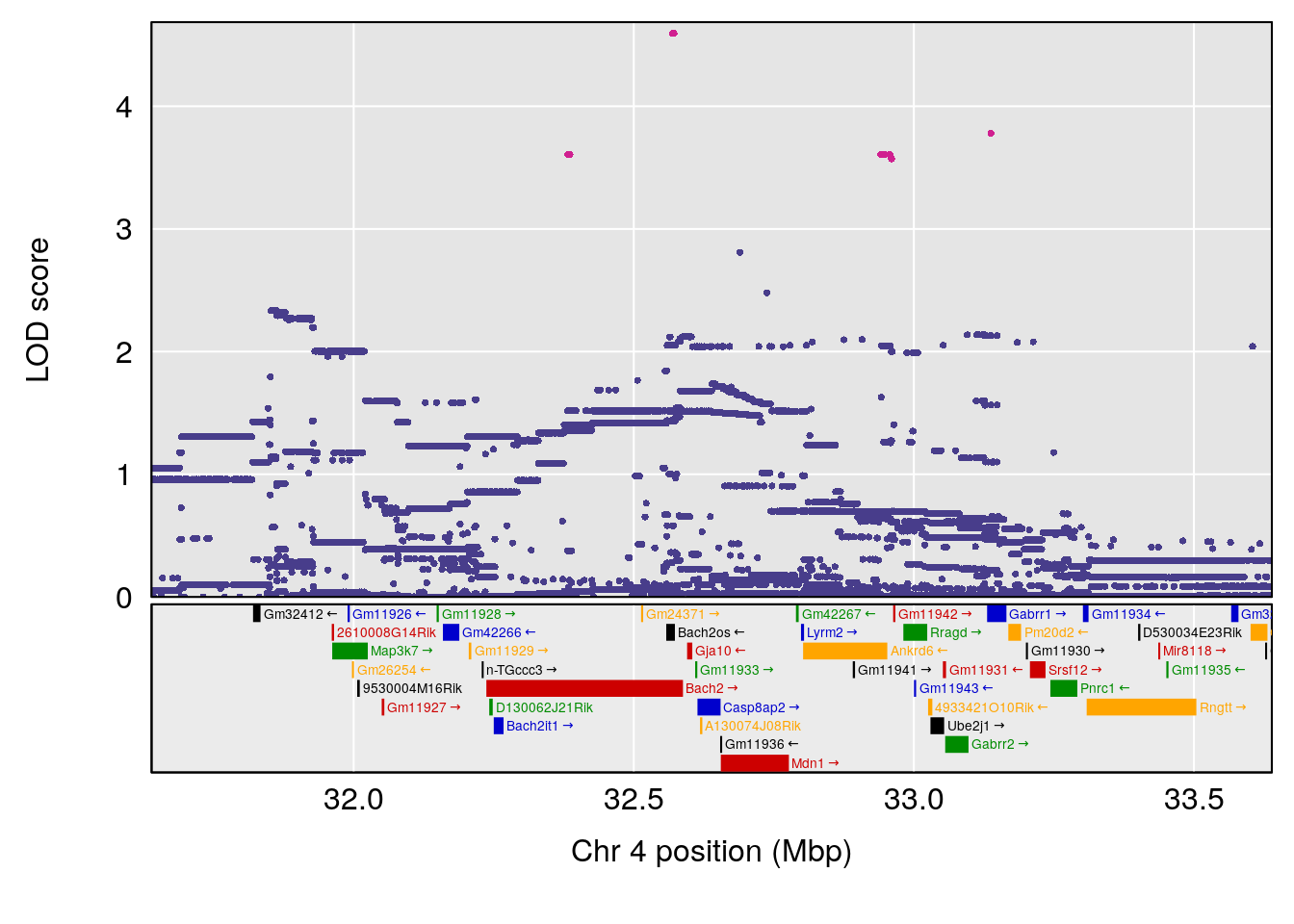
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2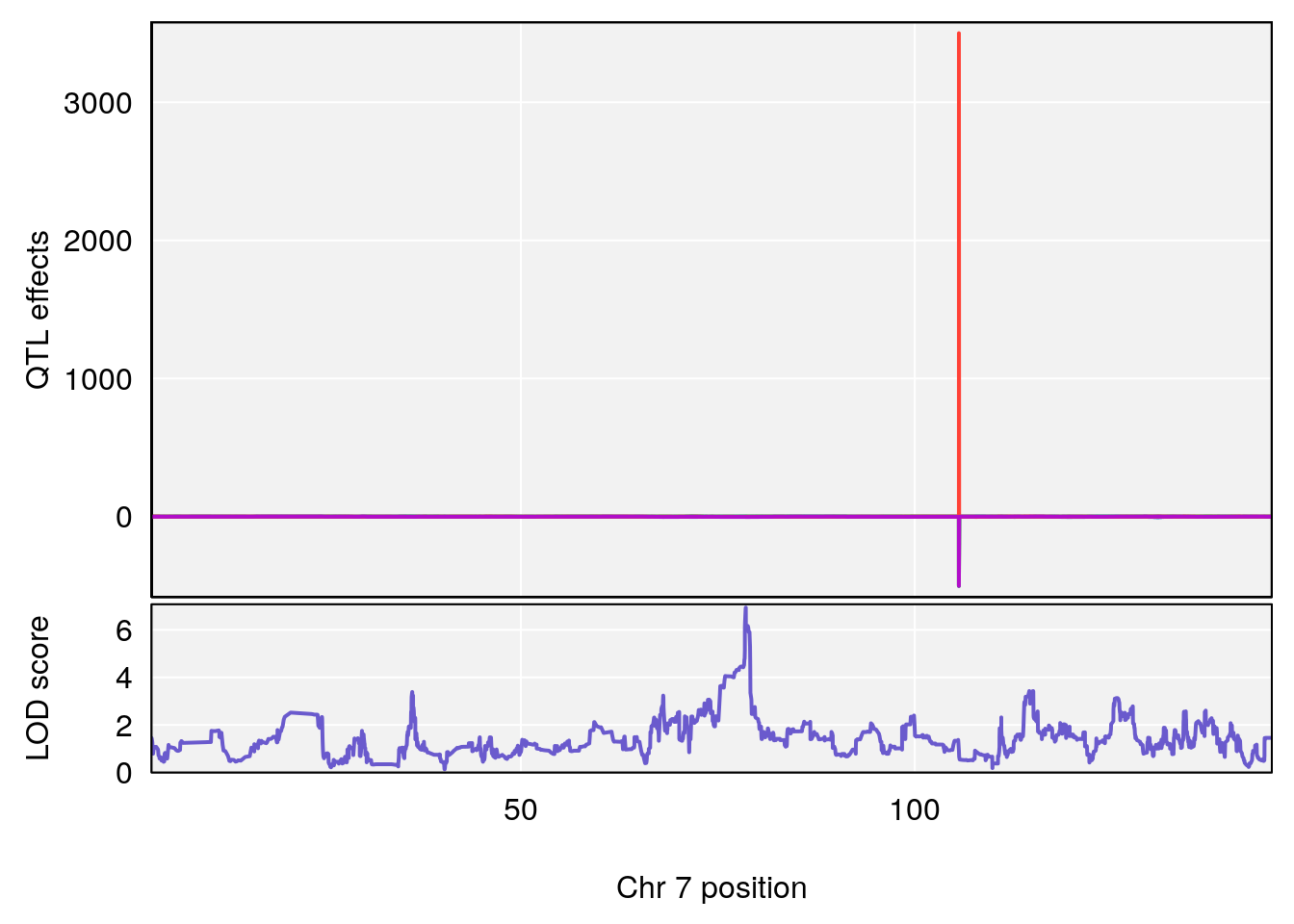
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
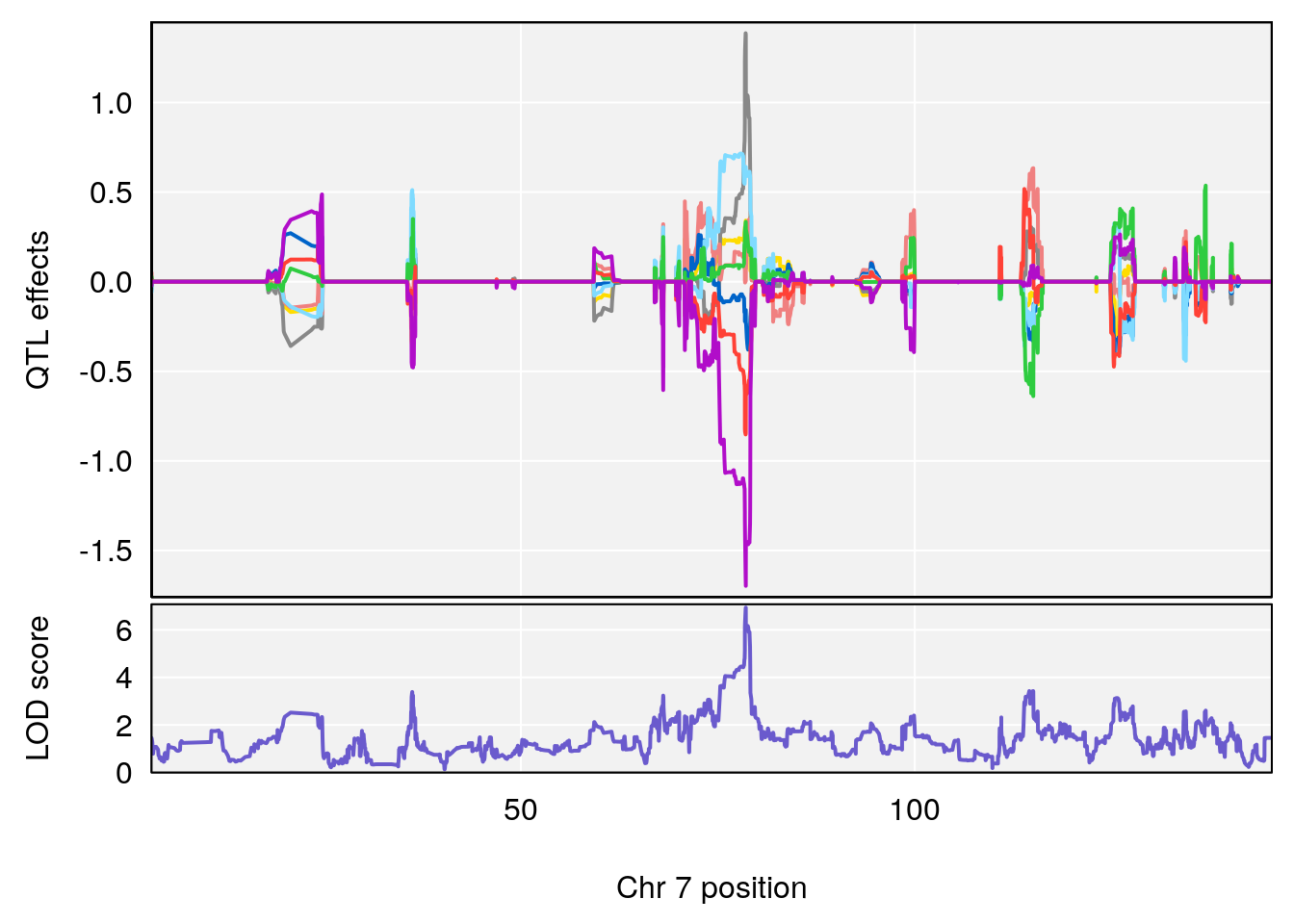
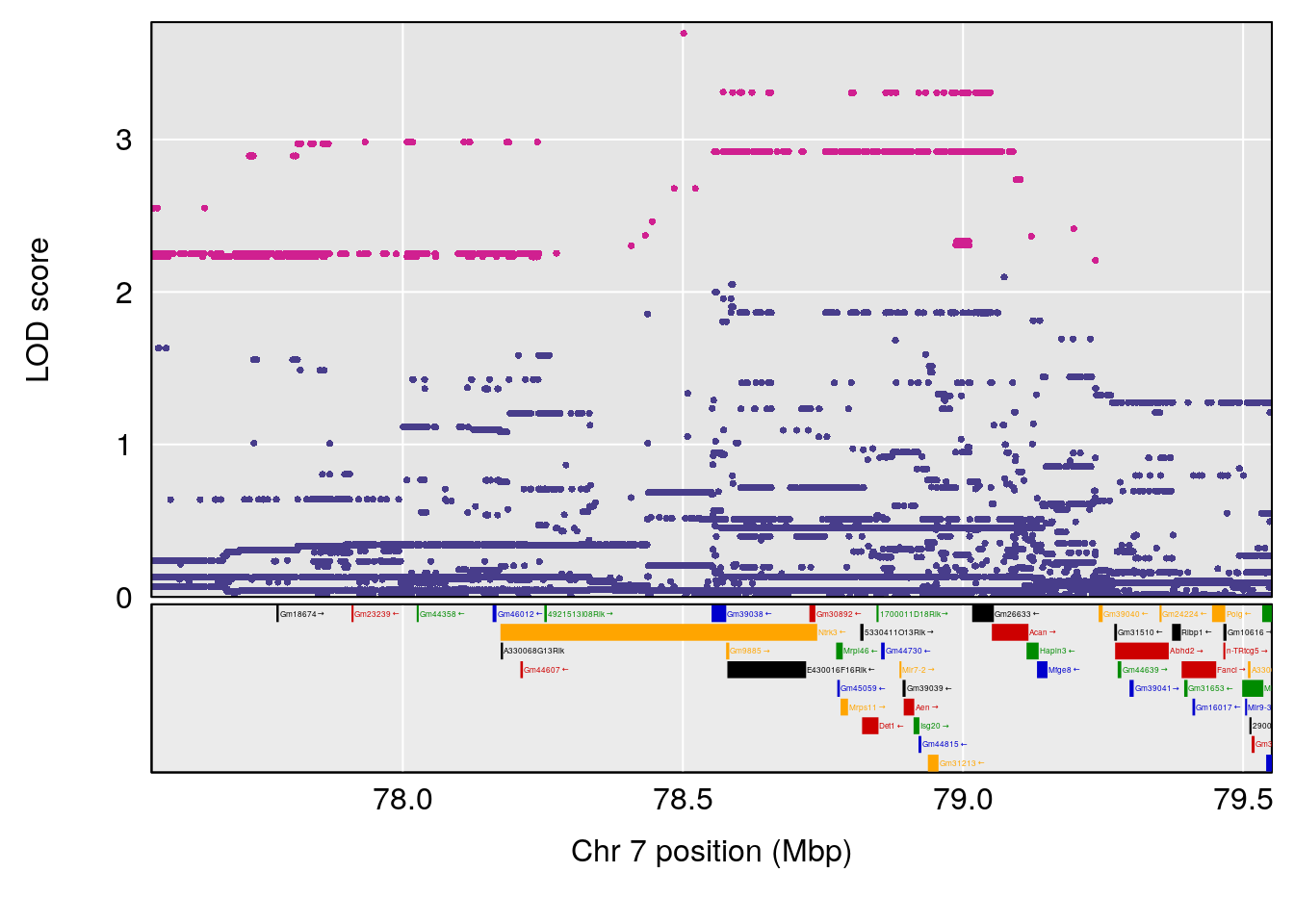
# [1] 3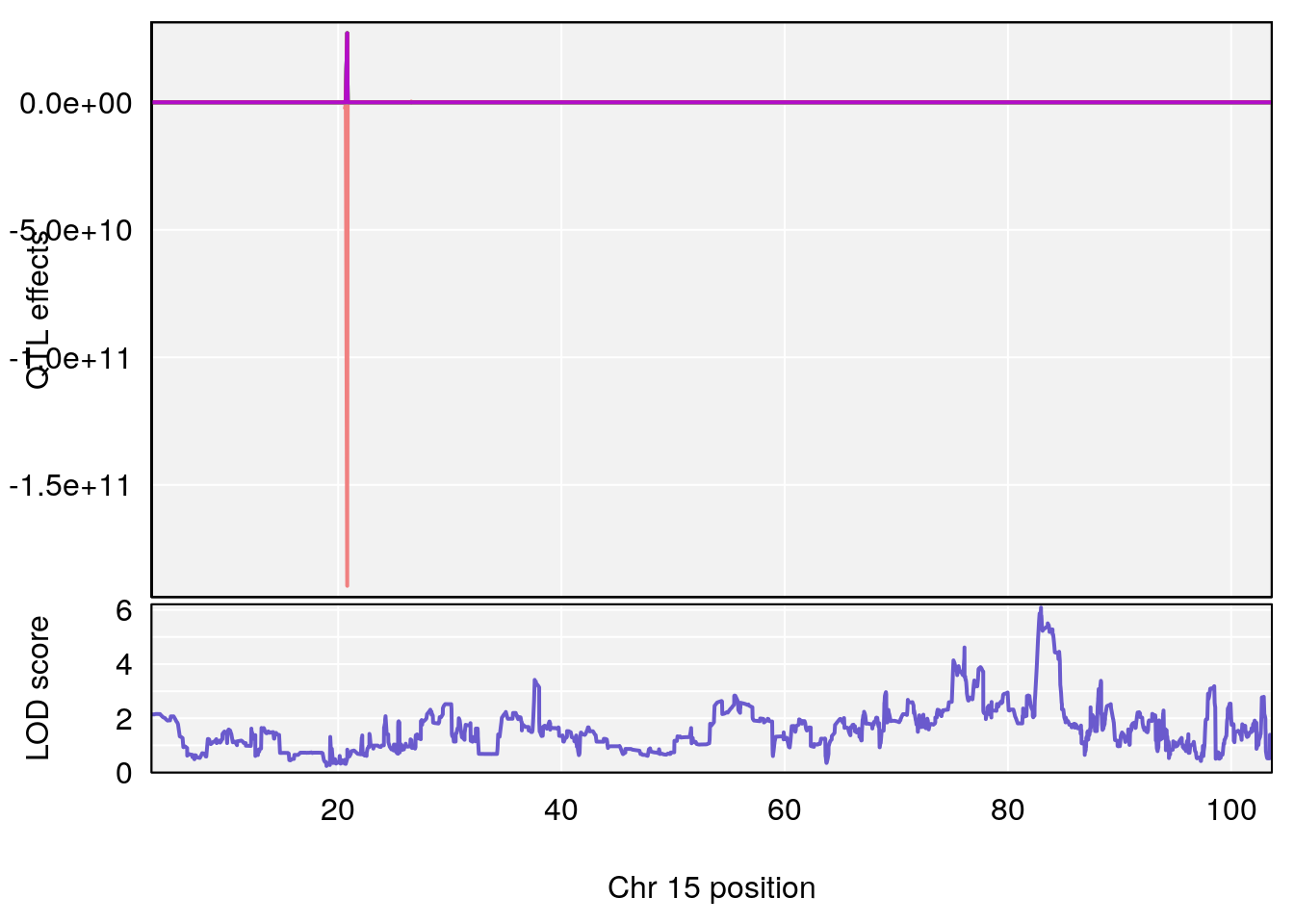
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
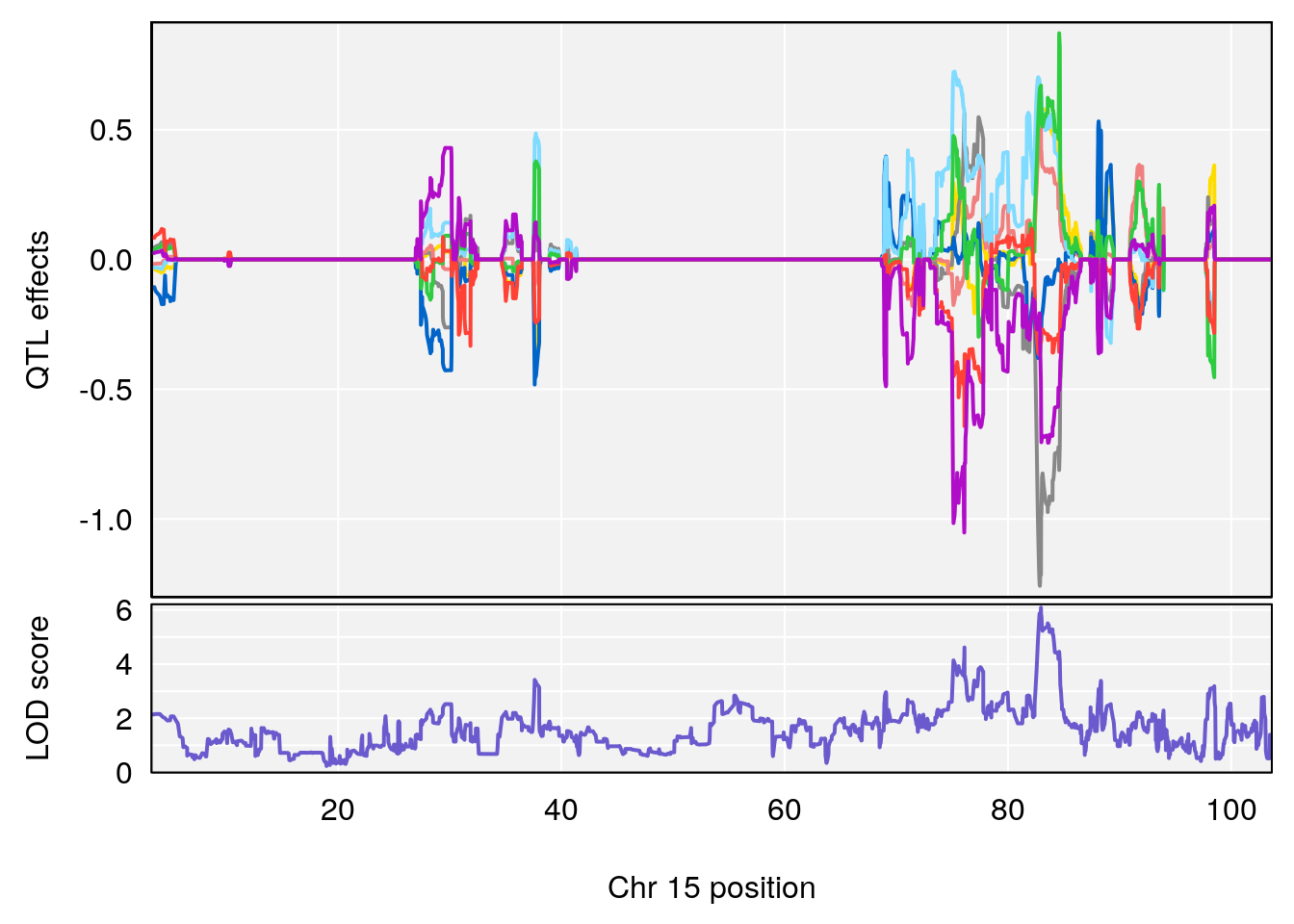
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
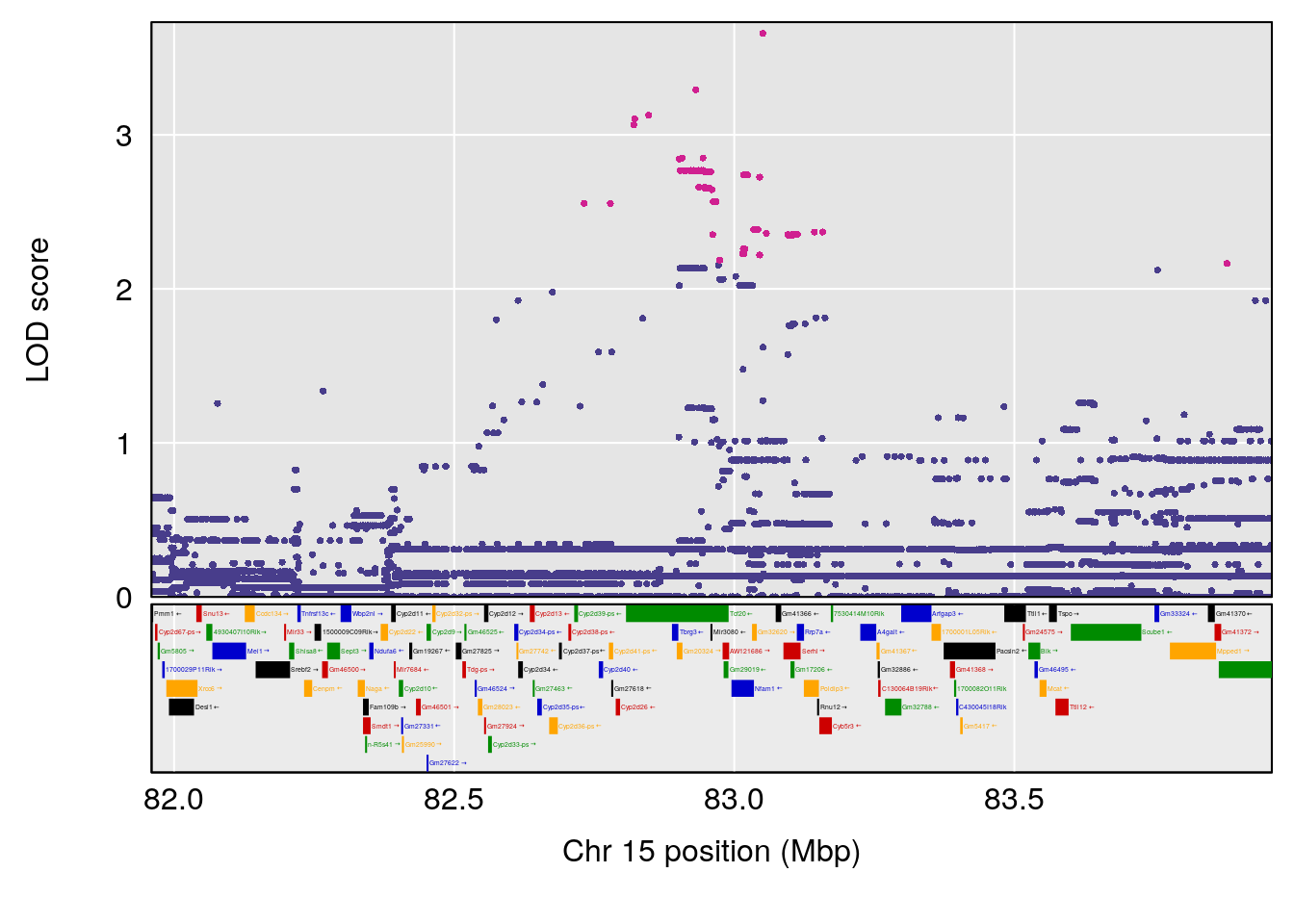
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "RR.Recovery.Rate.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 3 152.71526 6.057558 151.92564 153.04206
# 2 1 pheno1 12 82.52881 6.219487 46.37269 82.84506
# [1] 1
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
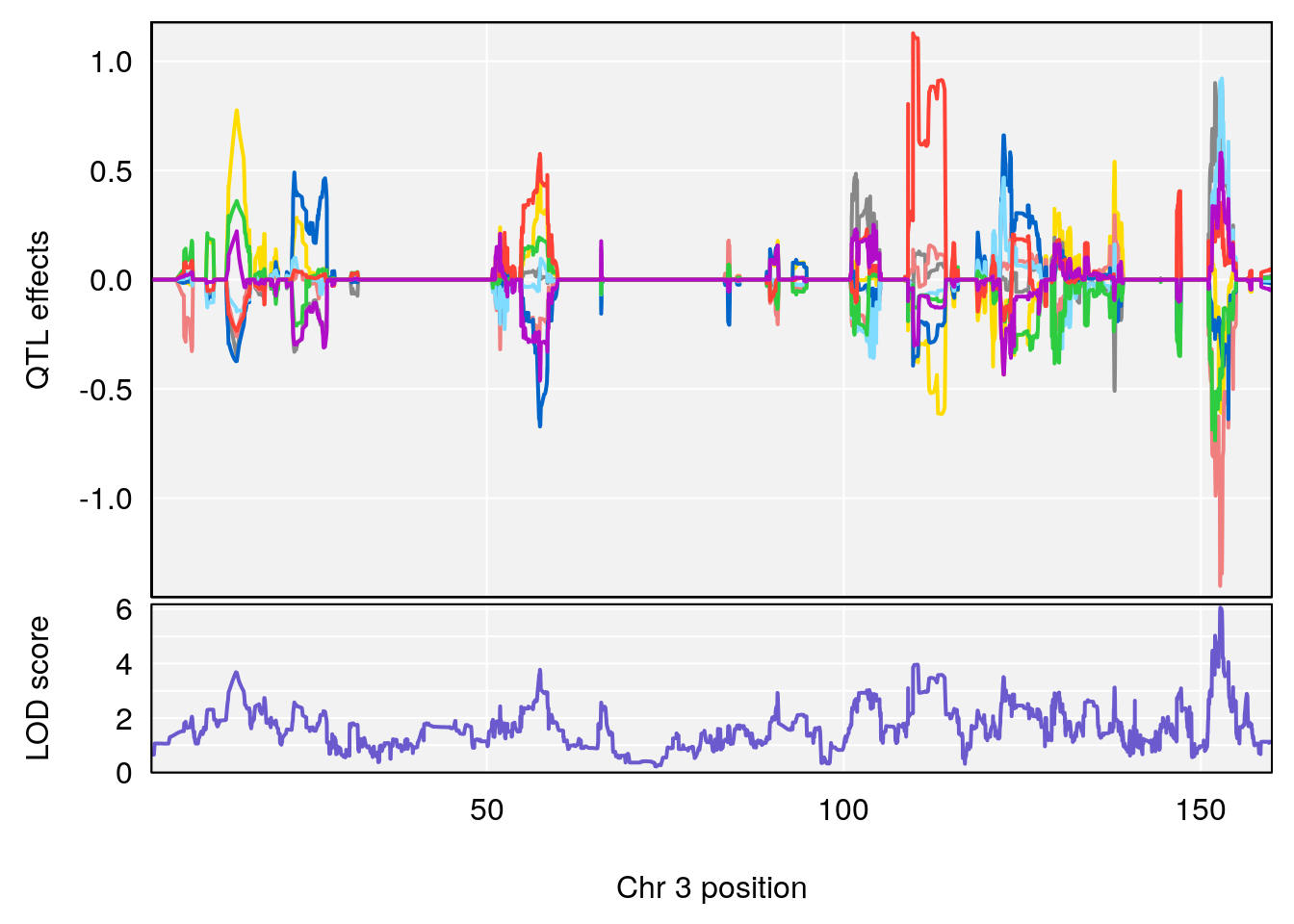
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
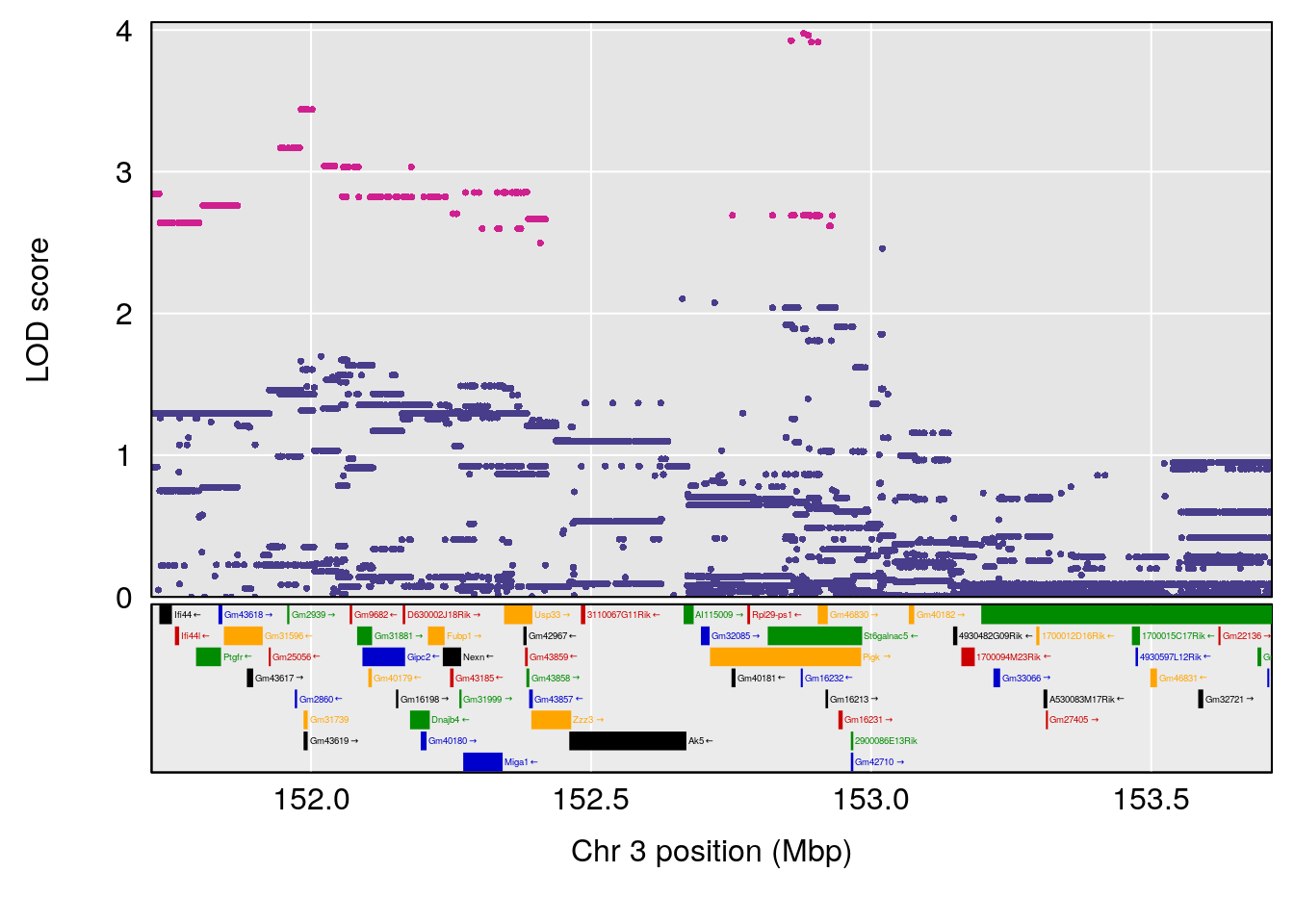
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2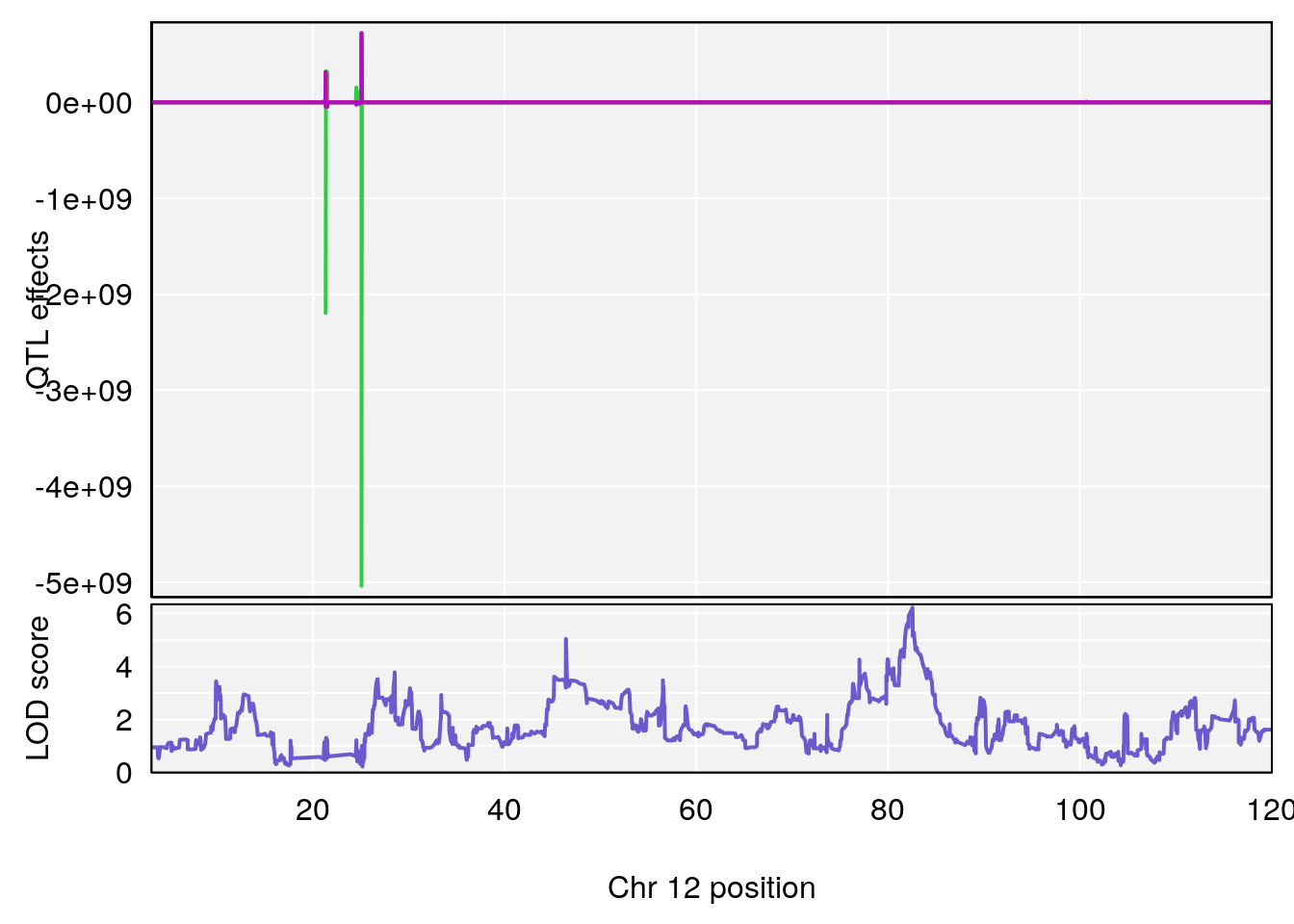
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
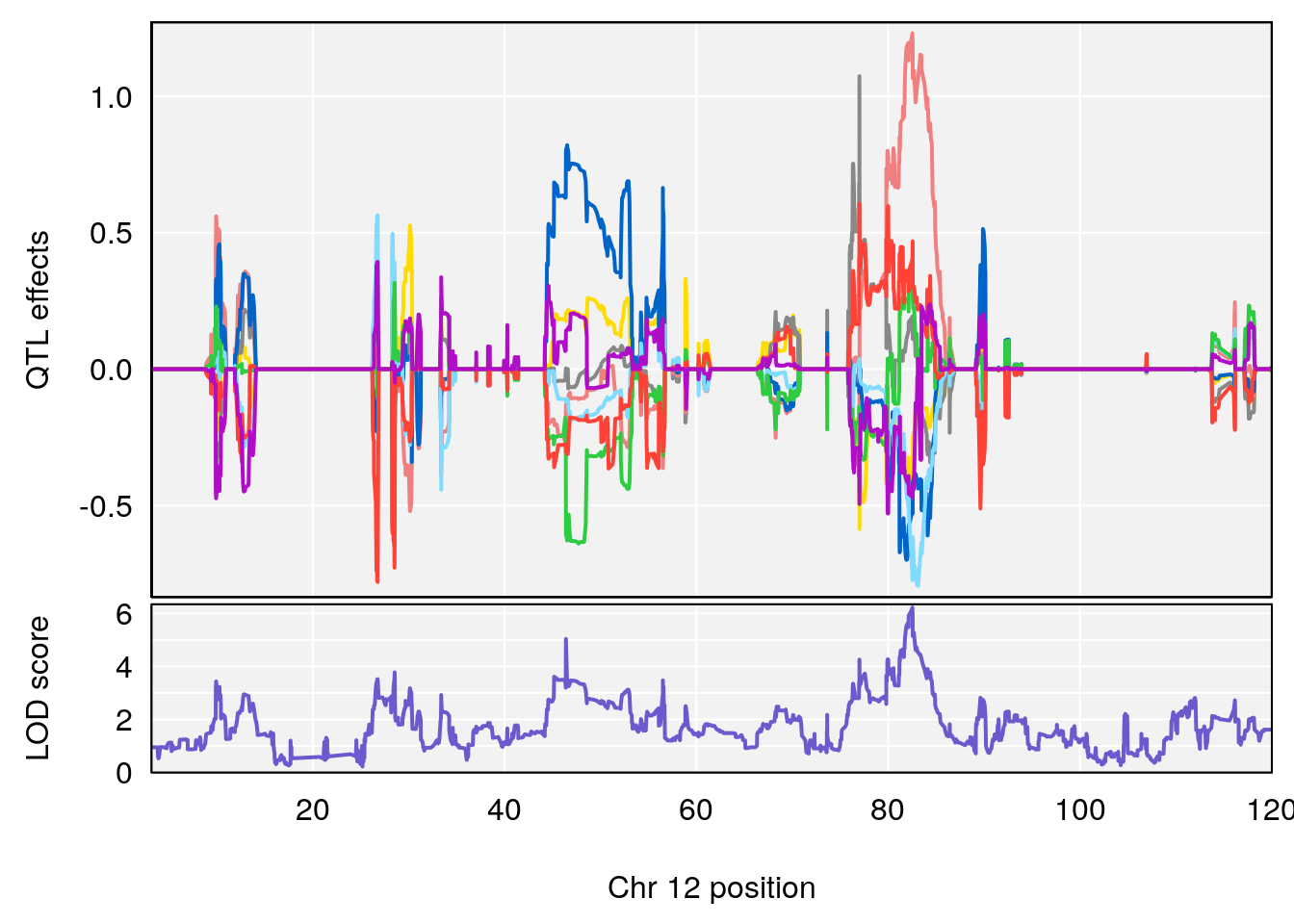
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
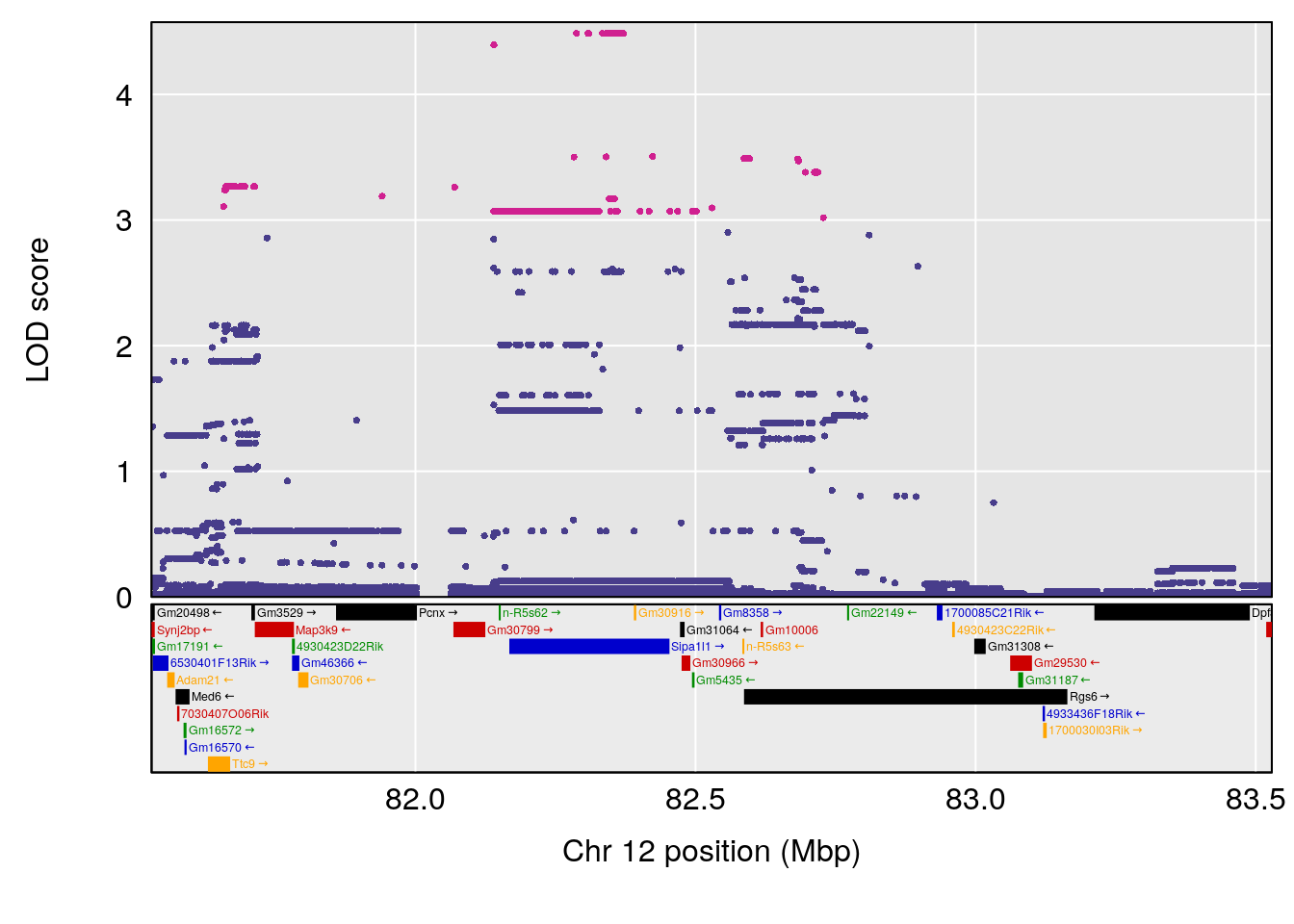
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "Min.Depression.BR"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 6 124.4987 6.882637 122.7691 125.5986
# [1] 1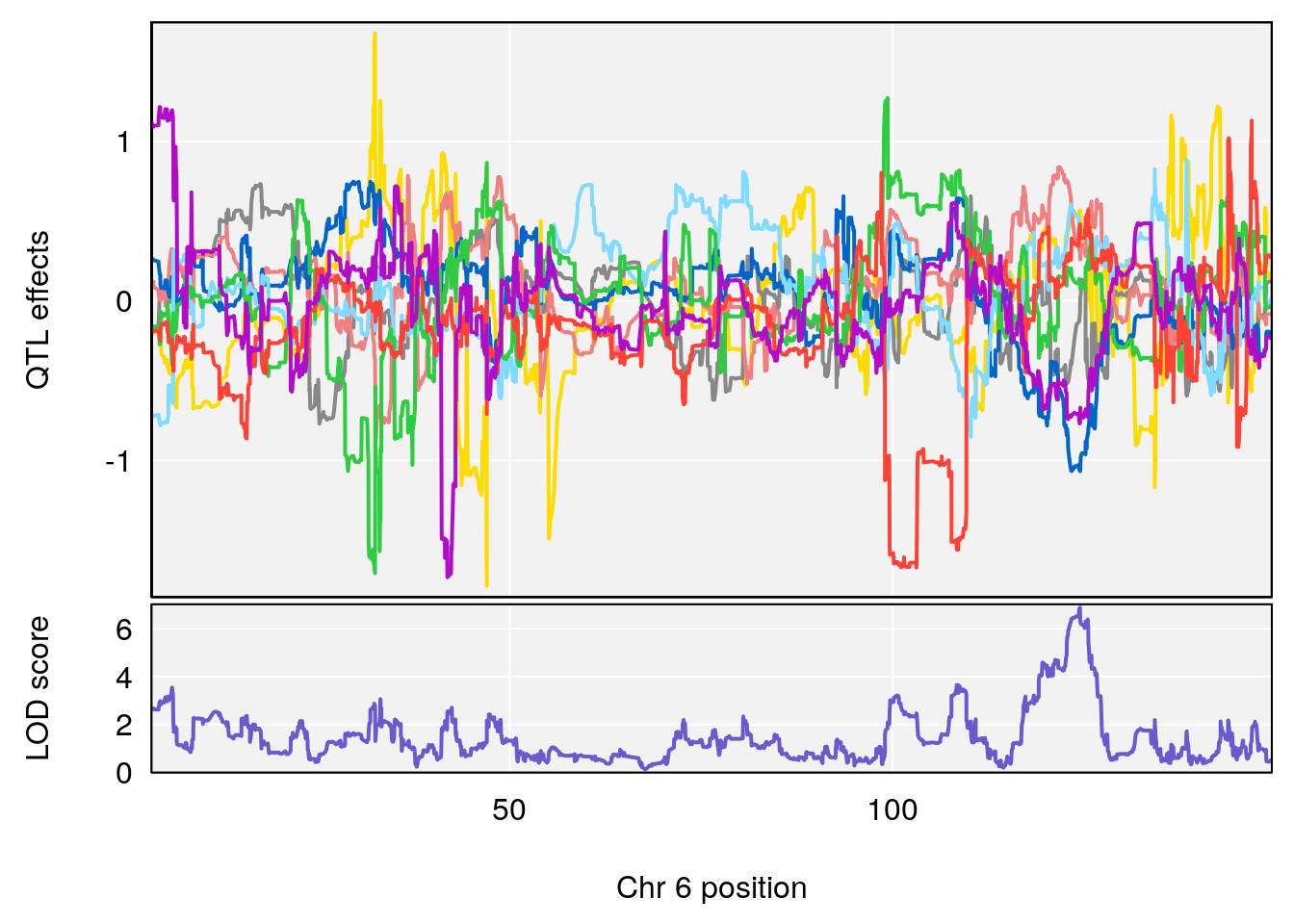
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
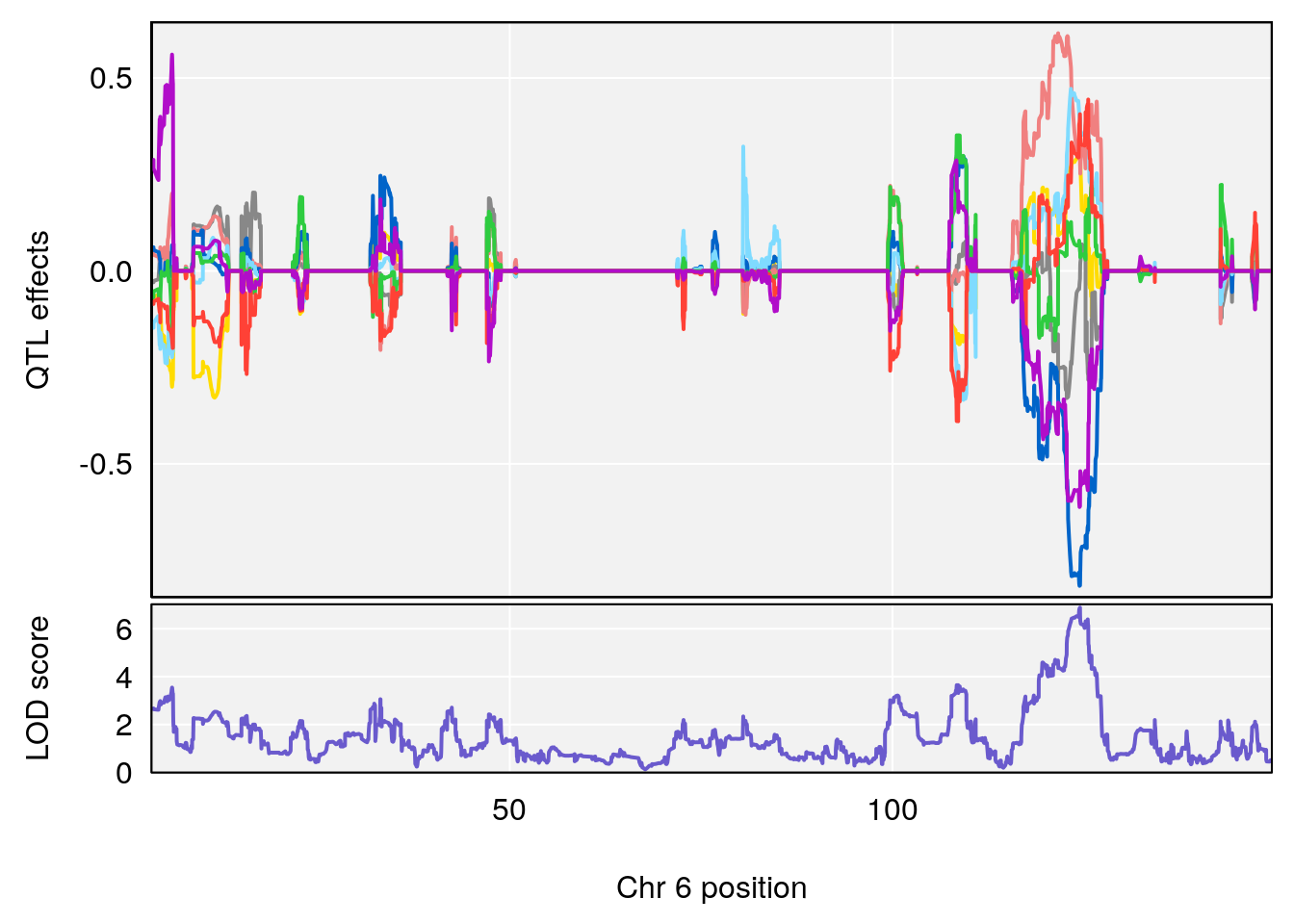
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
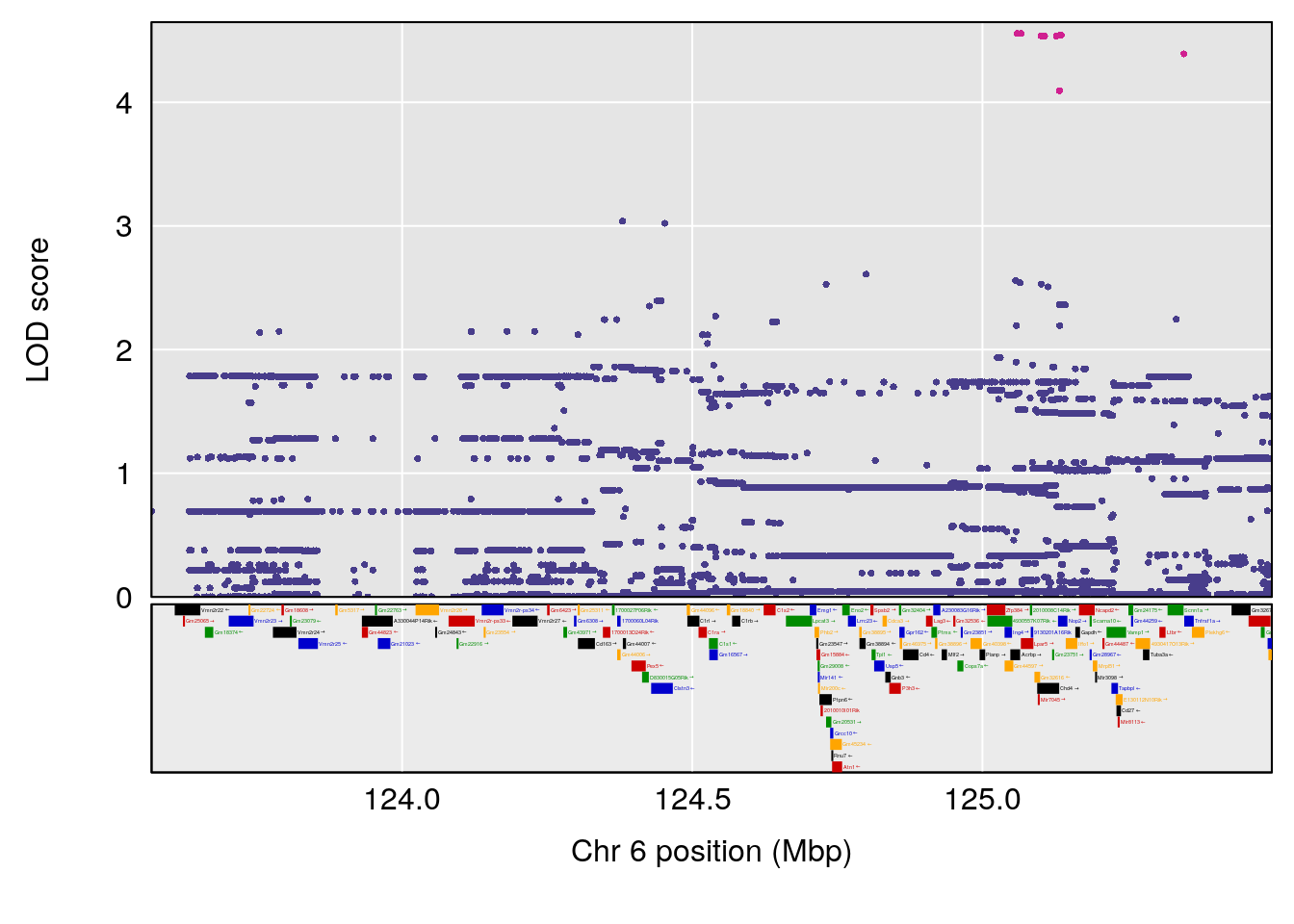
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "Time.to.Mostly.Dead.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 2 115.56002 6.027351 37.81483 132.19814
# 2 1 pheno1 10 16.47363 7.647309 16.28585 18.20019
# 3 1 pheno1 11 30.45329 6.071616 28.81595 31.56268
# 4 1 pheno1 17 80.09690 6.490398 23.55316 80.36008
# [1] 1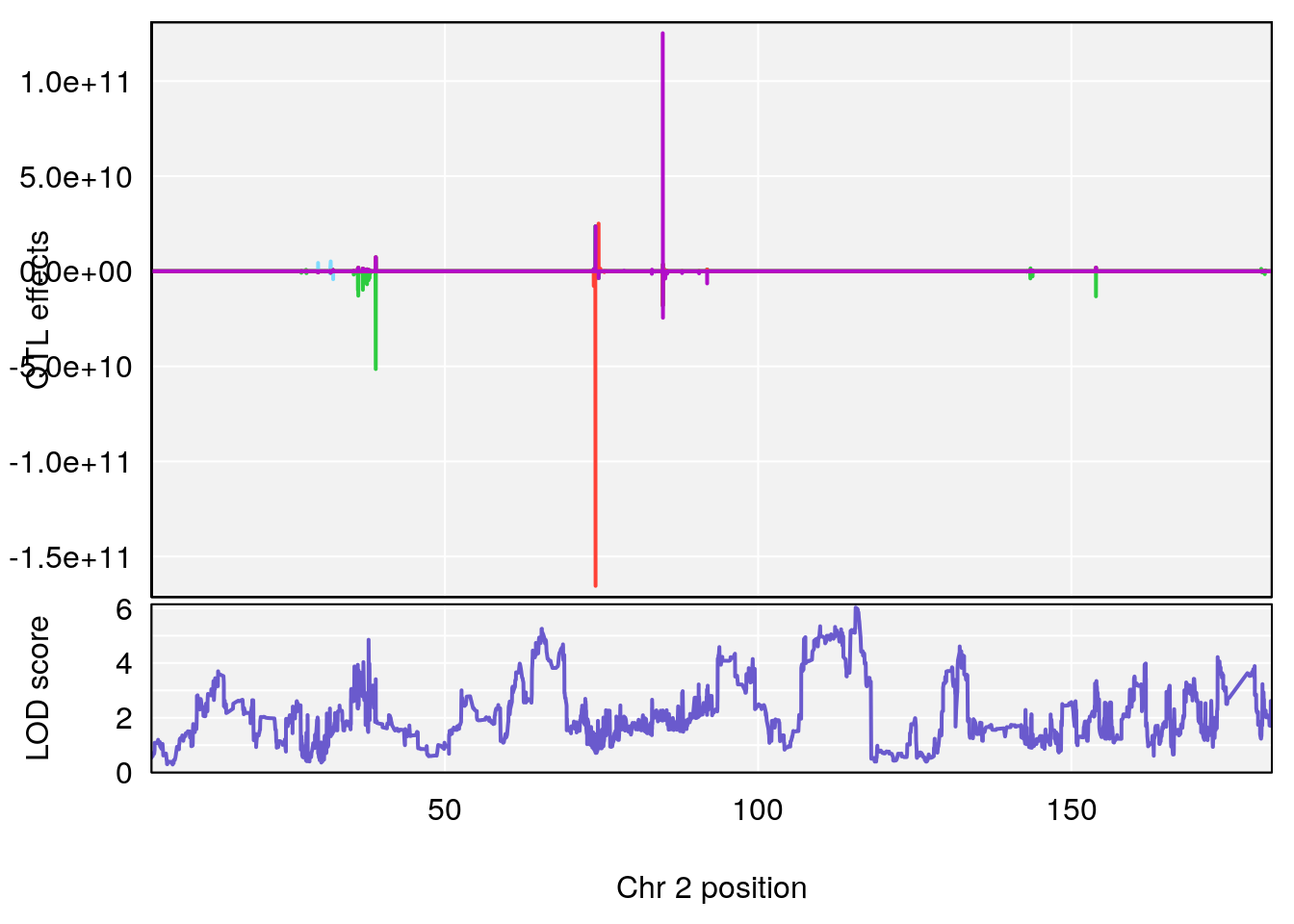
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
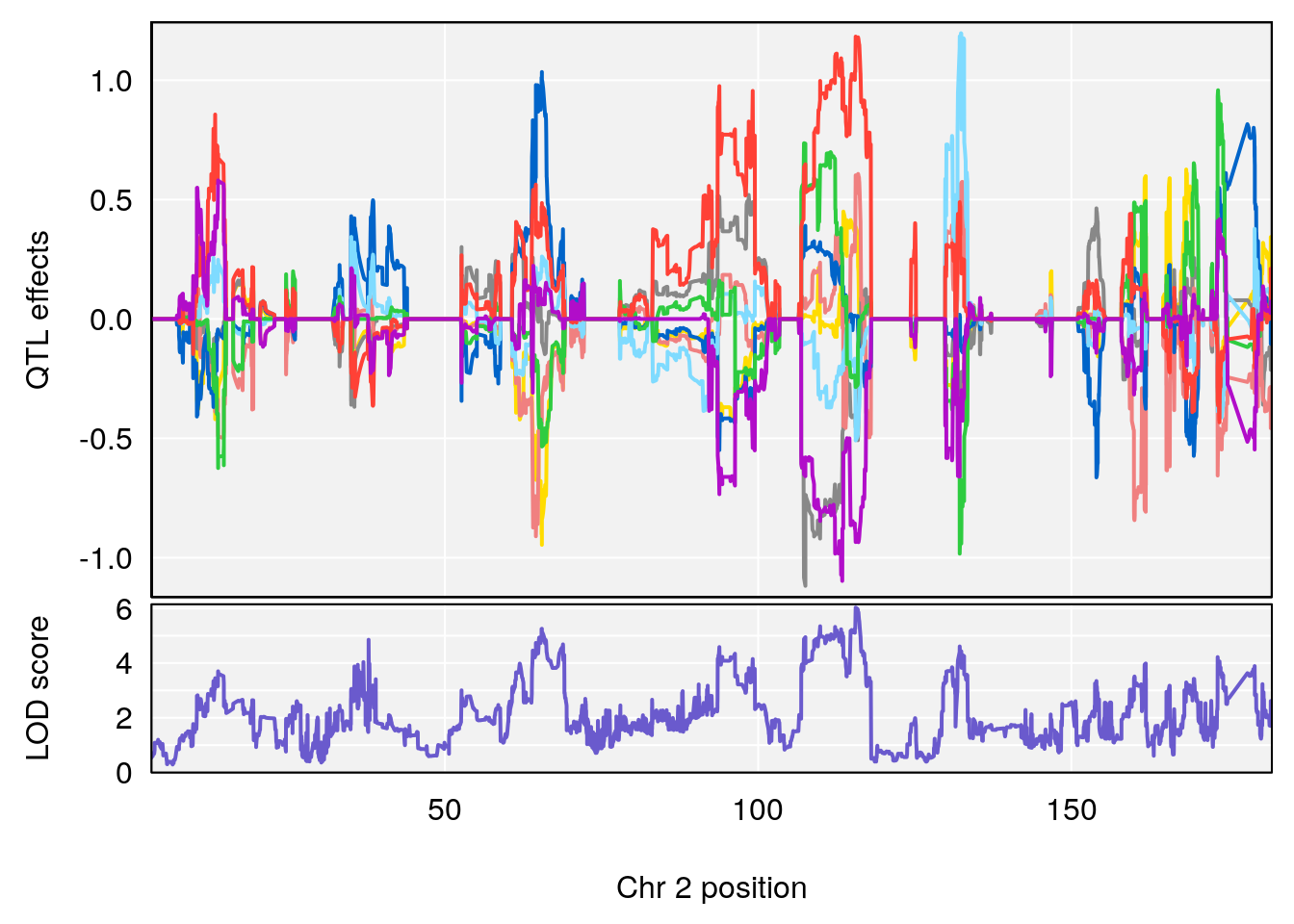
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
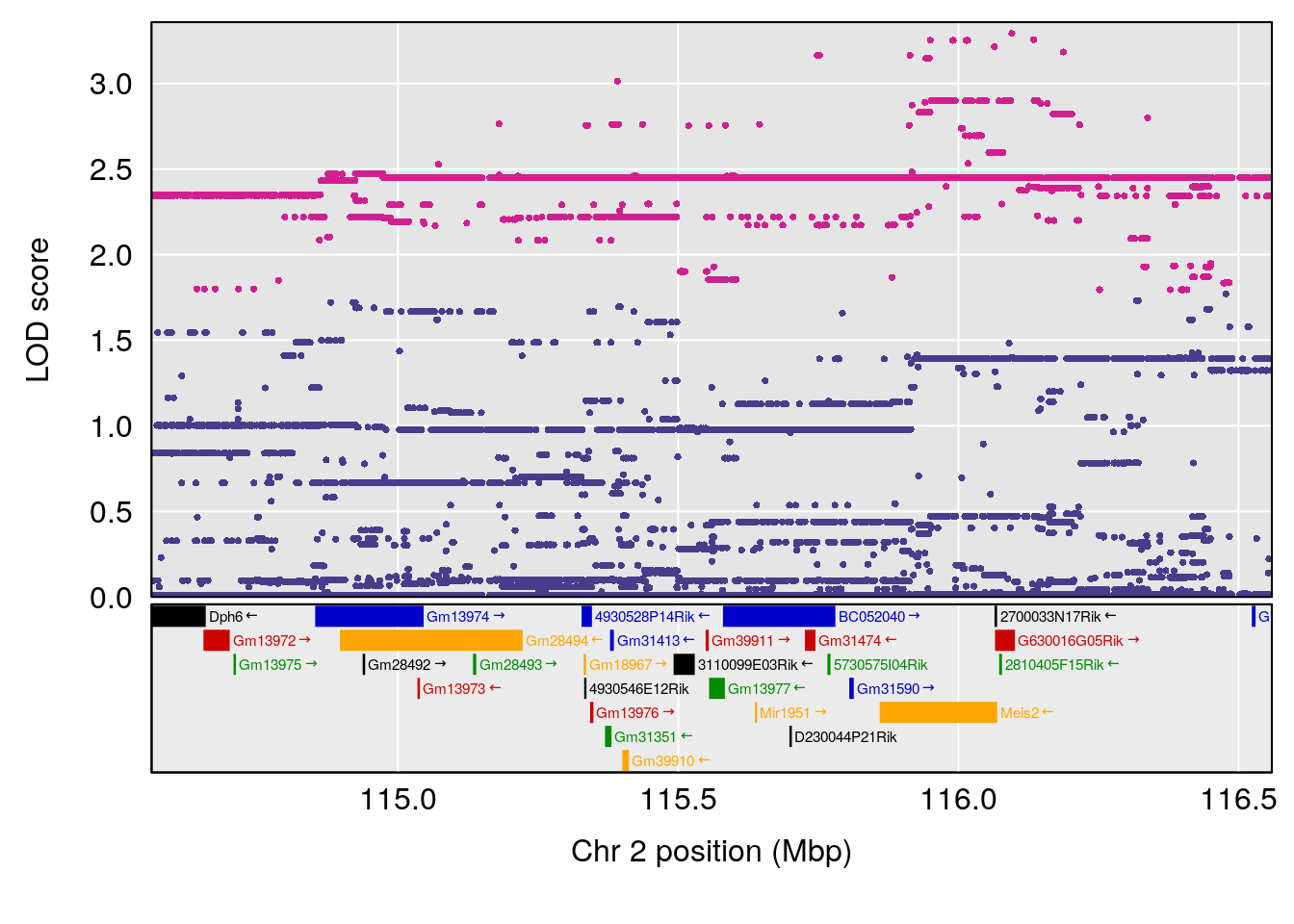
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2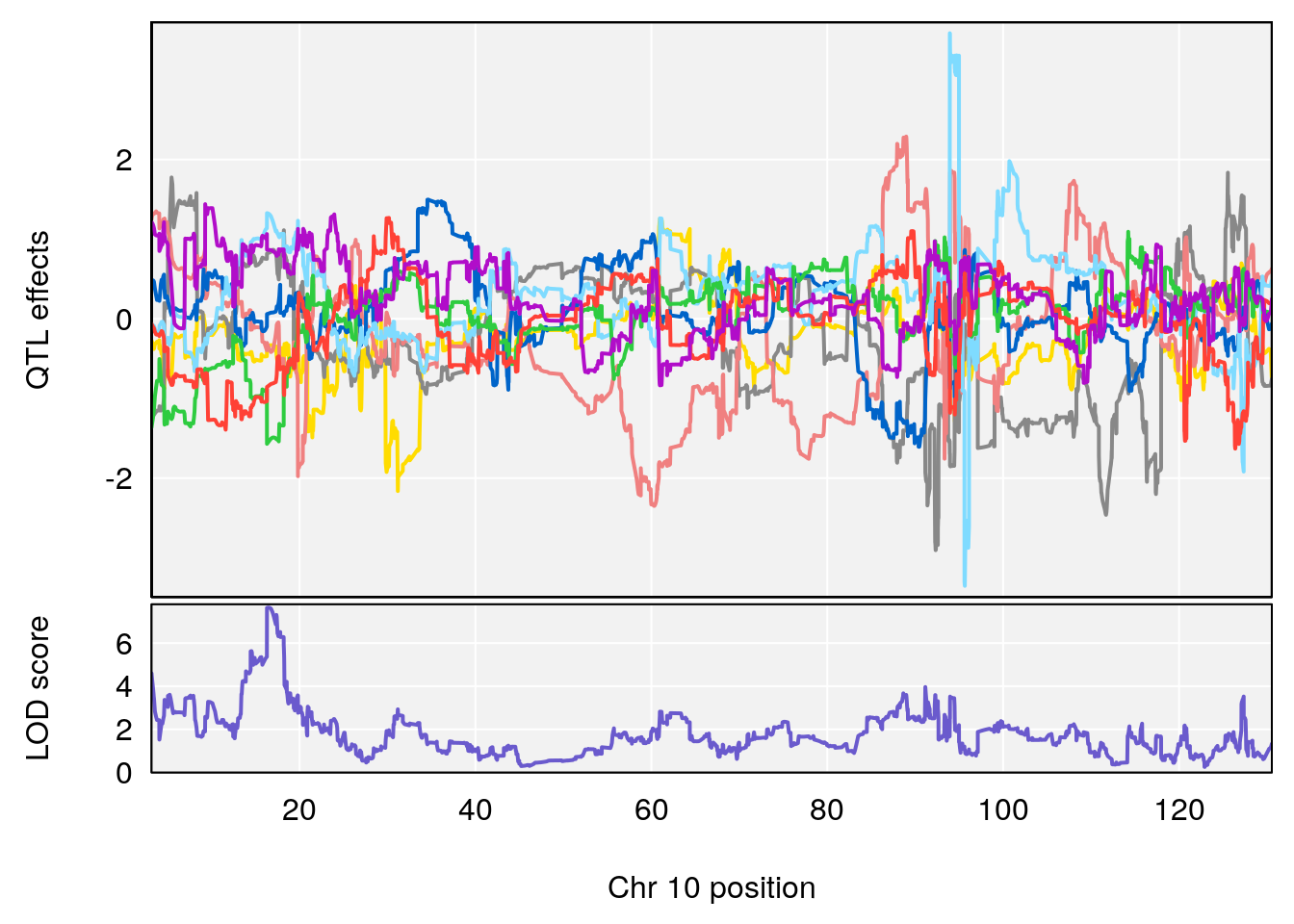
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
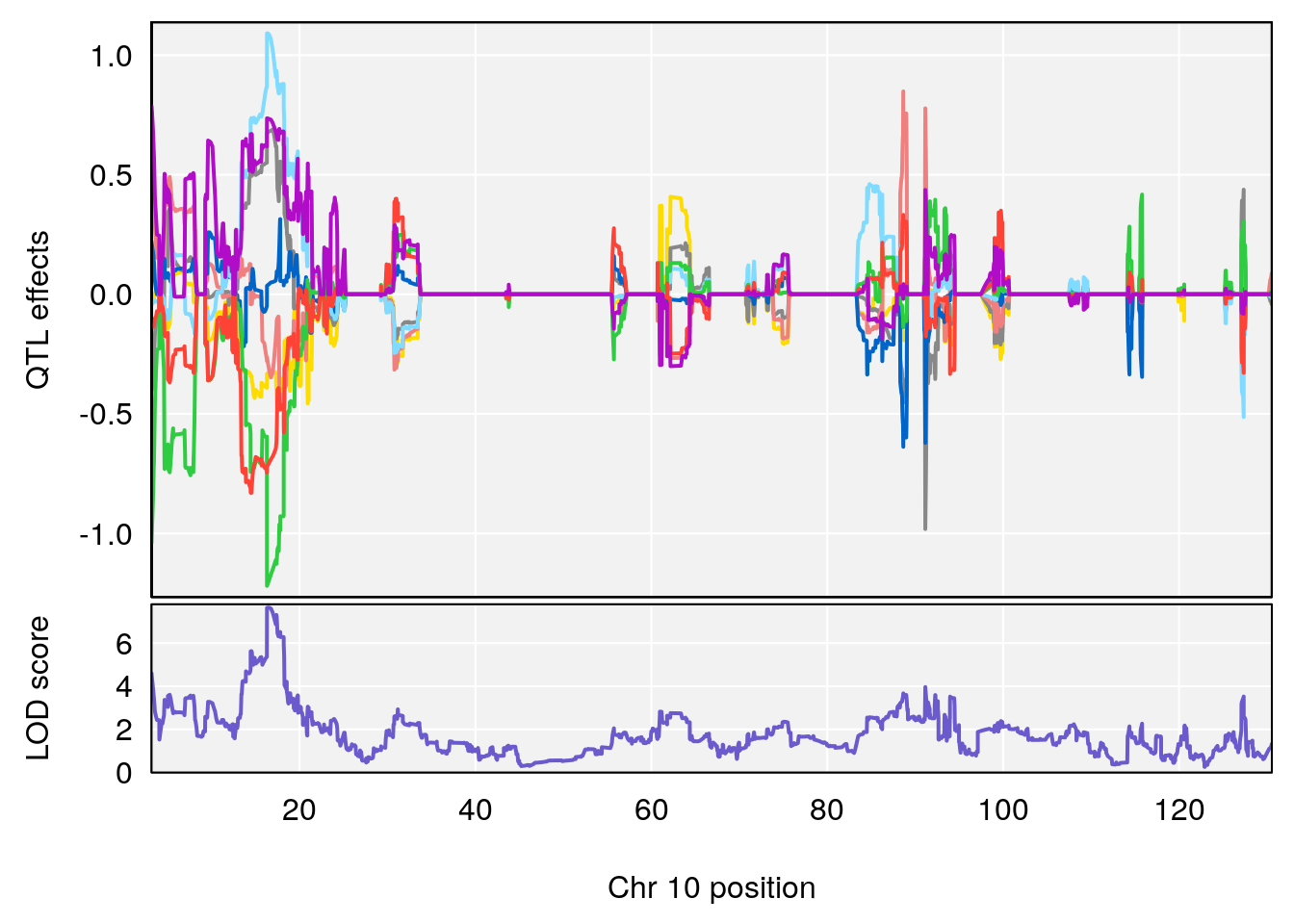
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
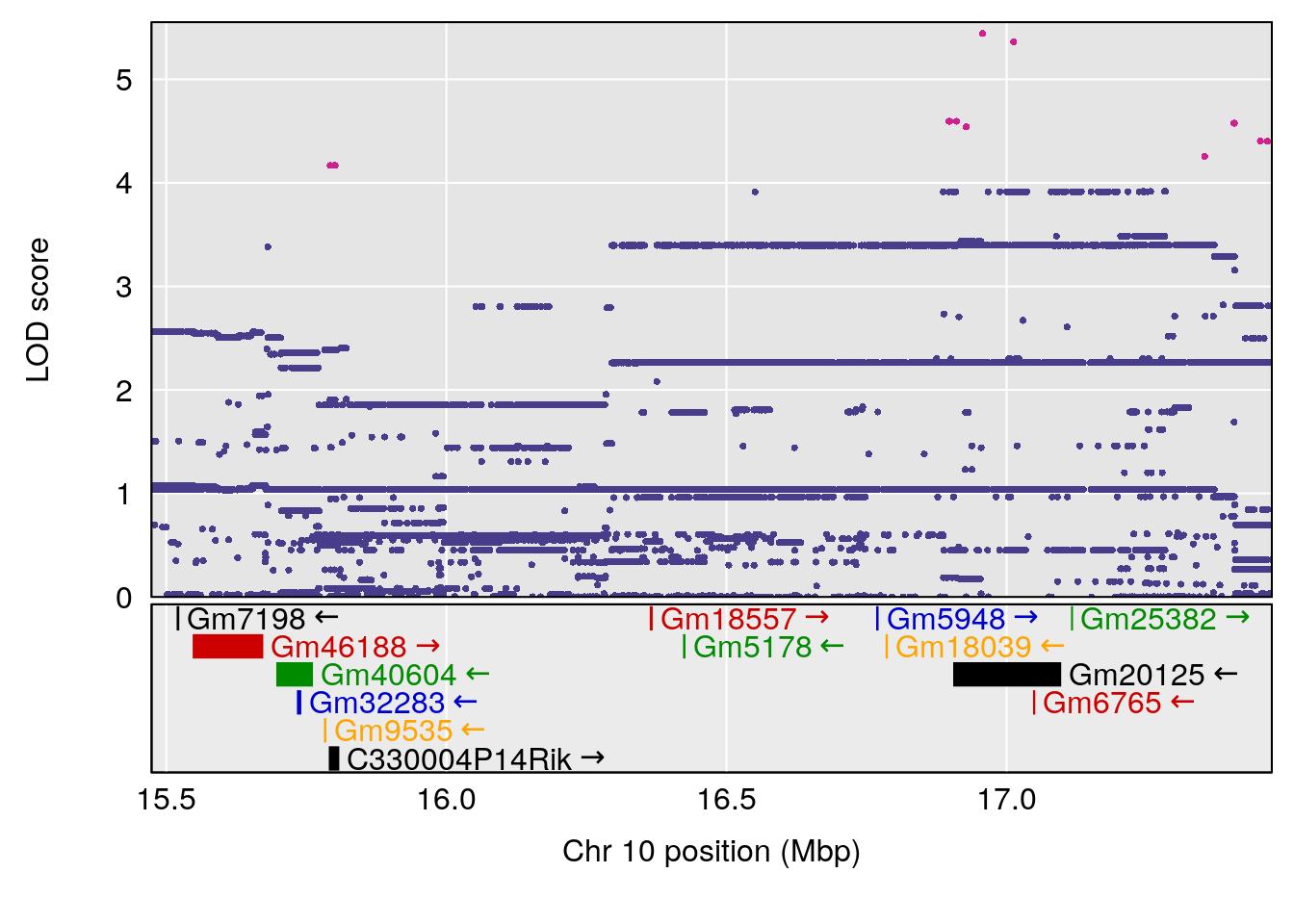
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 3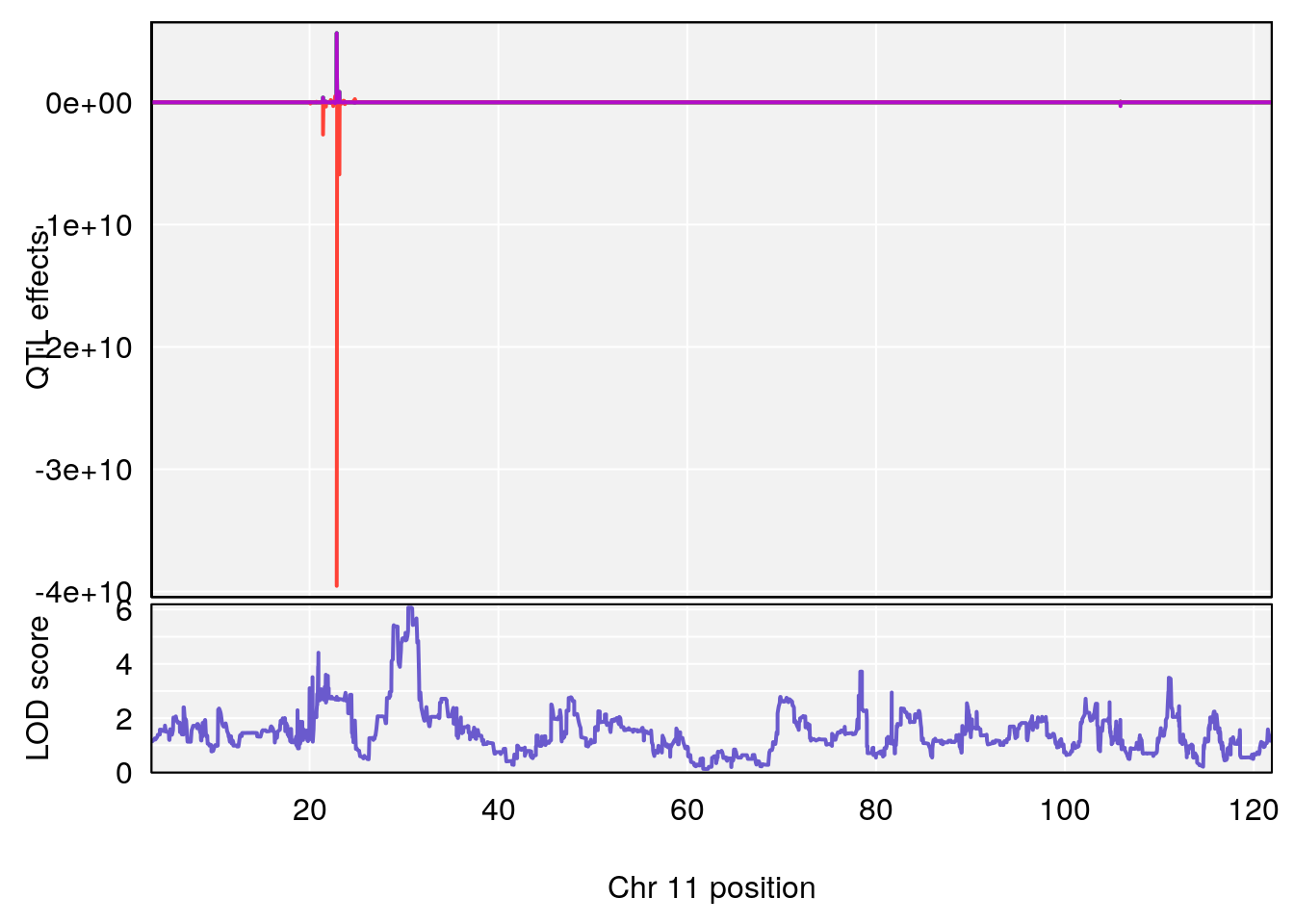
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
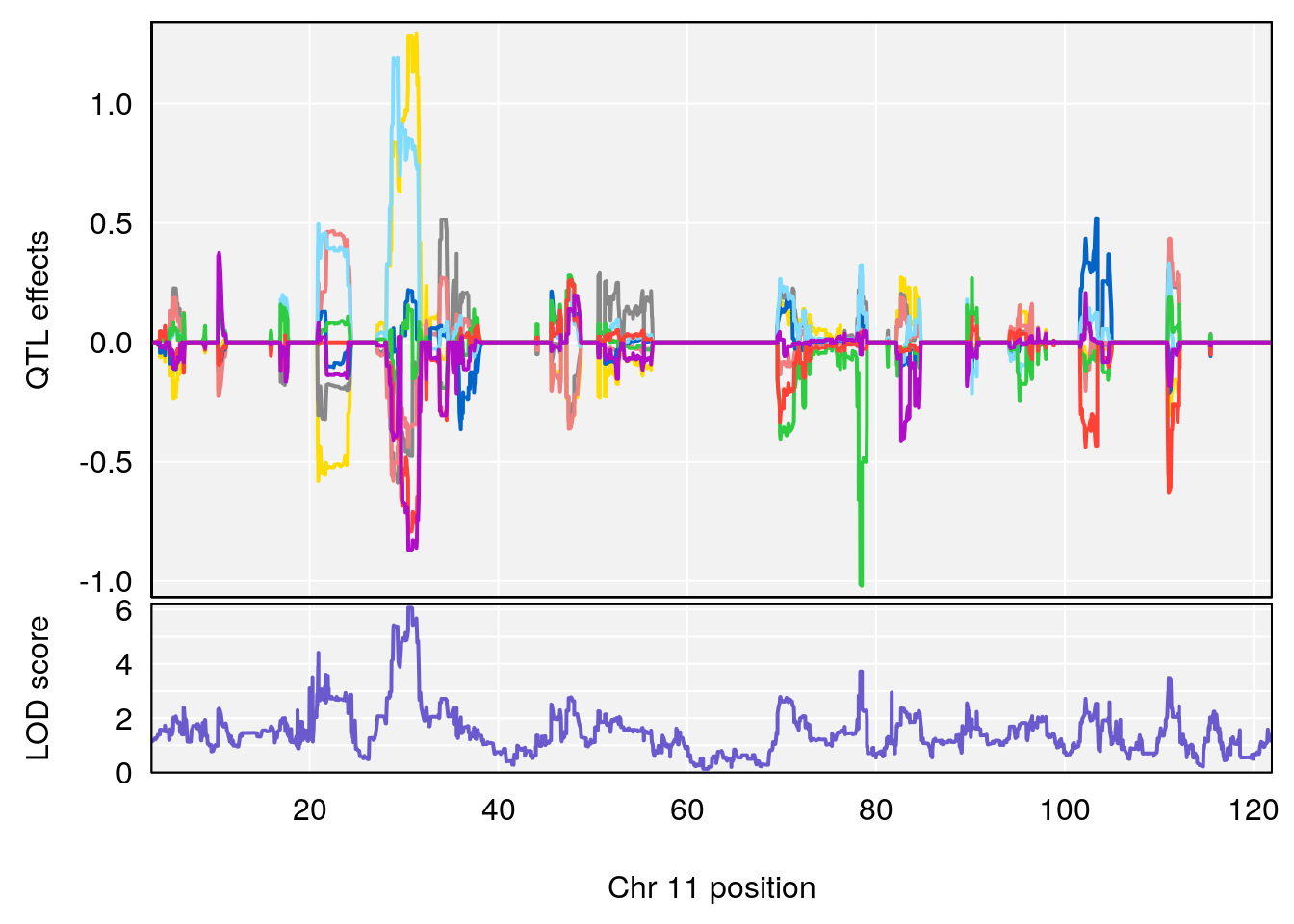
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
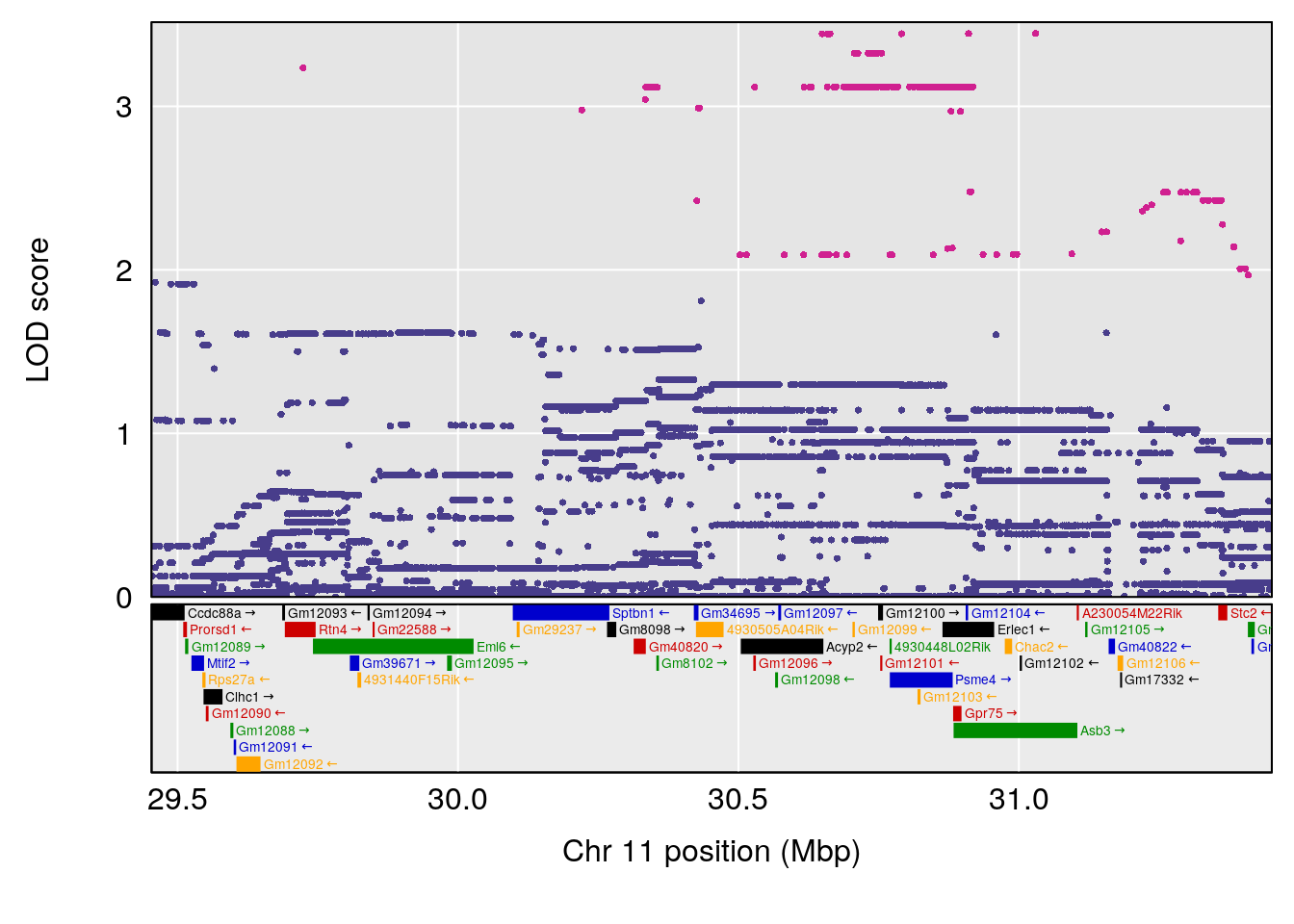
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 4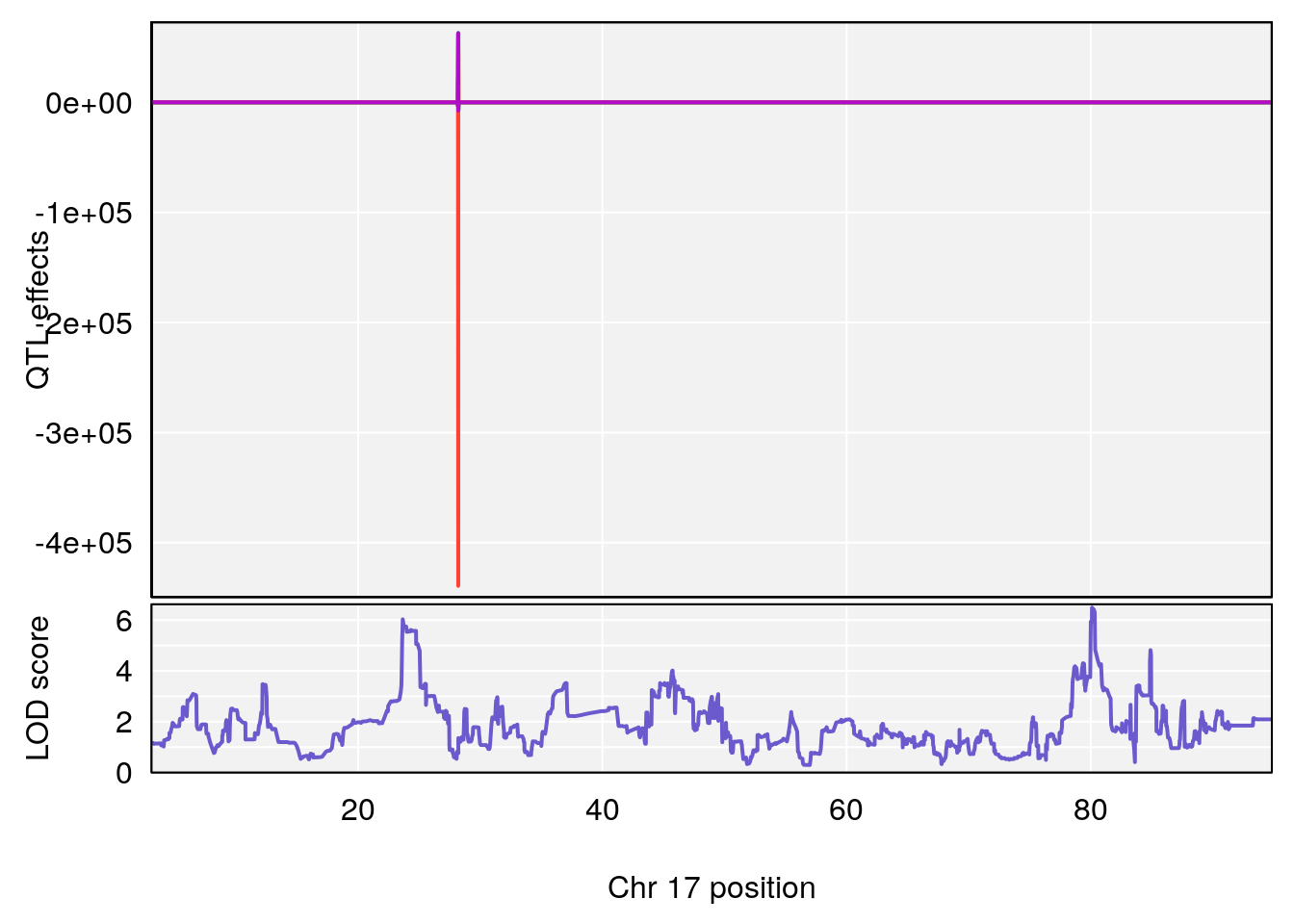
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
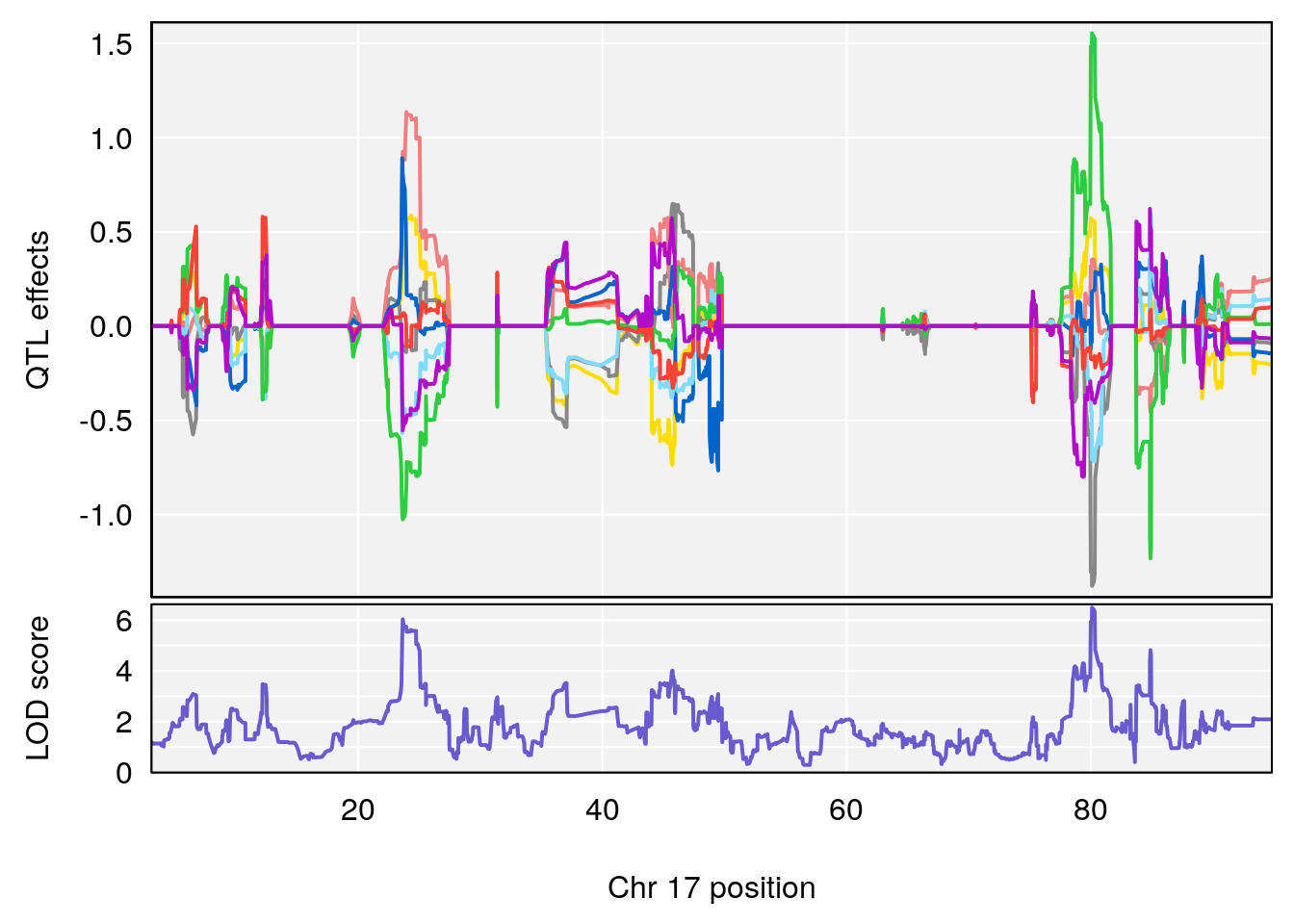
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
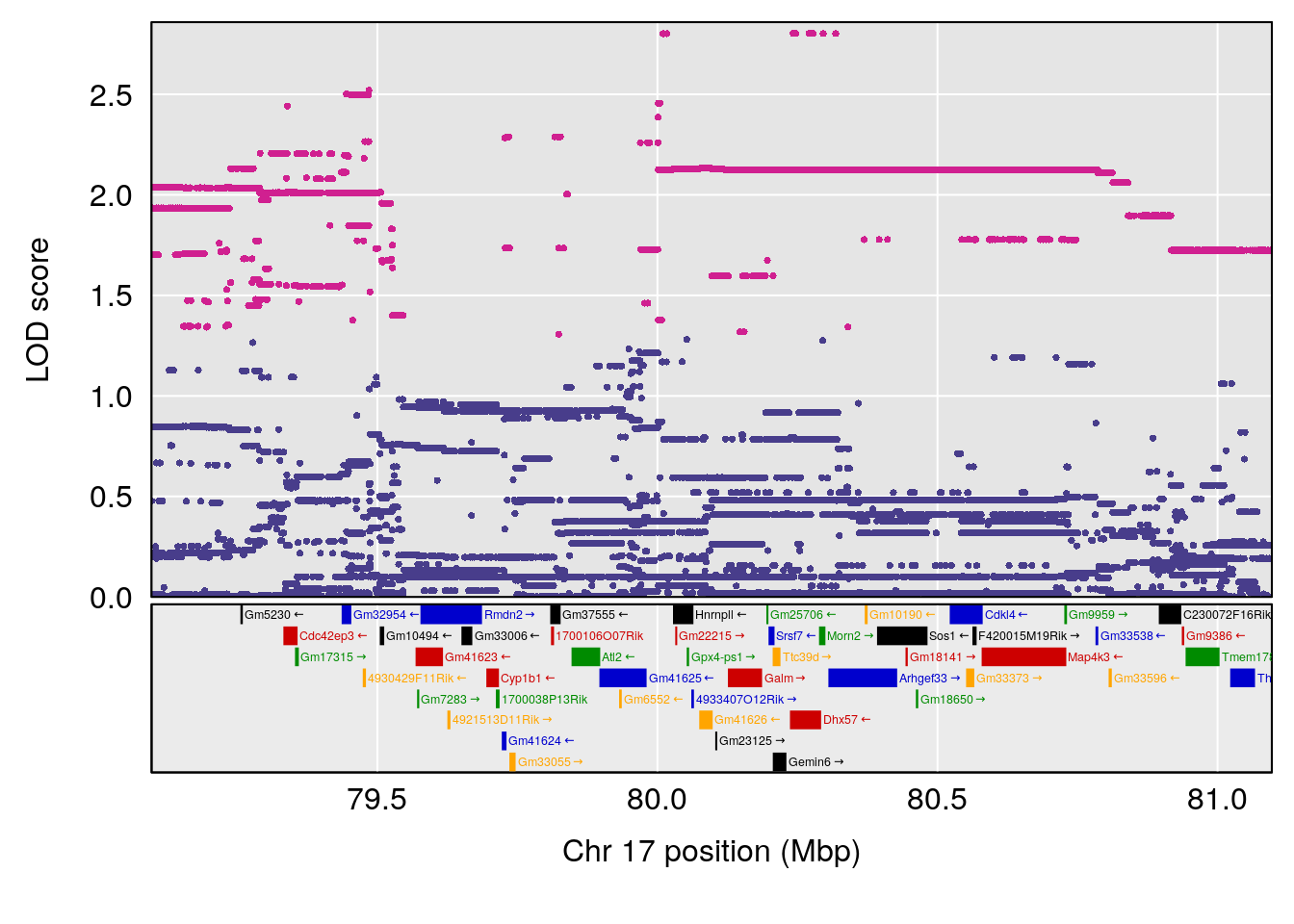
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "Time.to.Recovery.Hr"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 3 148.59948 6.440788 53.60570 148.61250
# 2 1 pheno1 4 22.26230 6.109573 17.41576 137.08866
# 3 1 pheno1 10 70.01089 7.242967 68.18103 70.90480
# 4 1 pheno1 14 80.19357 6.242259 79.67940 80.21217
# 5 1 pheno1 18 23.93757 6.888692 22.73513 24.98189
# [1] 1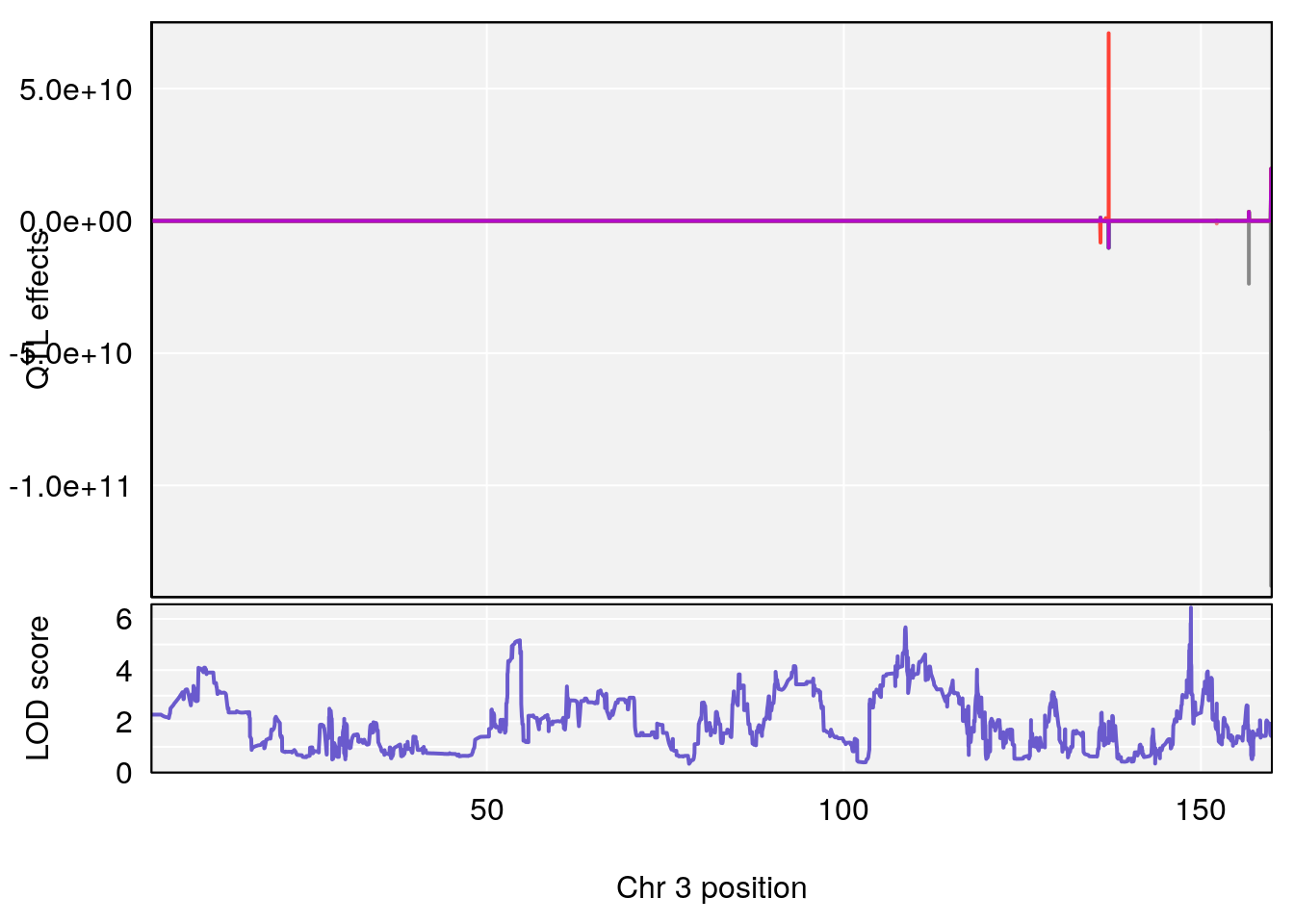
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
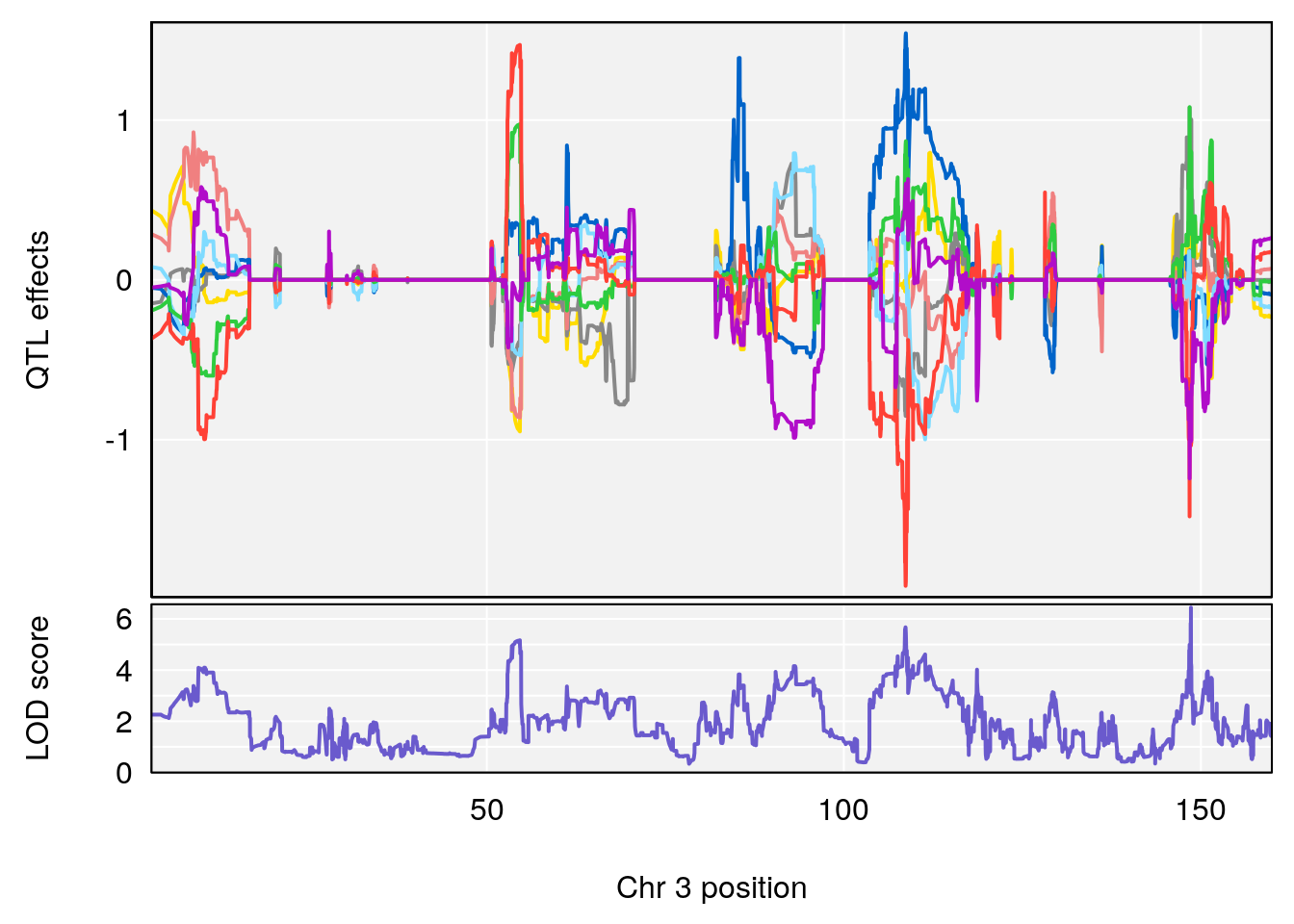
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
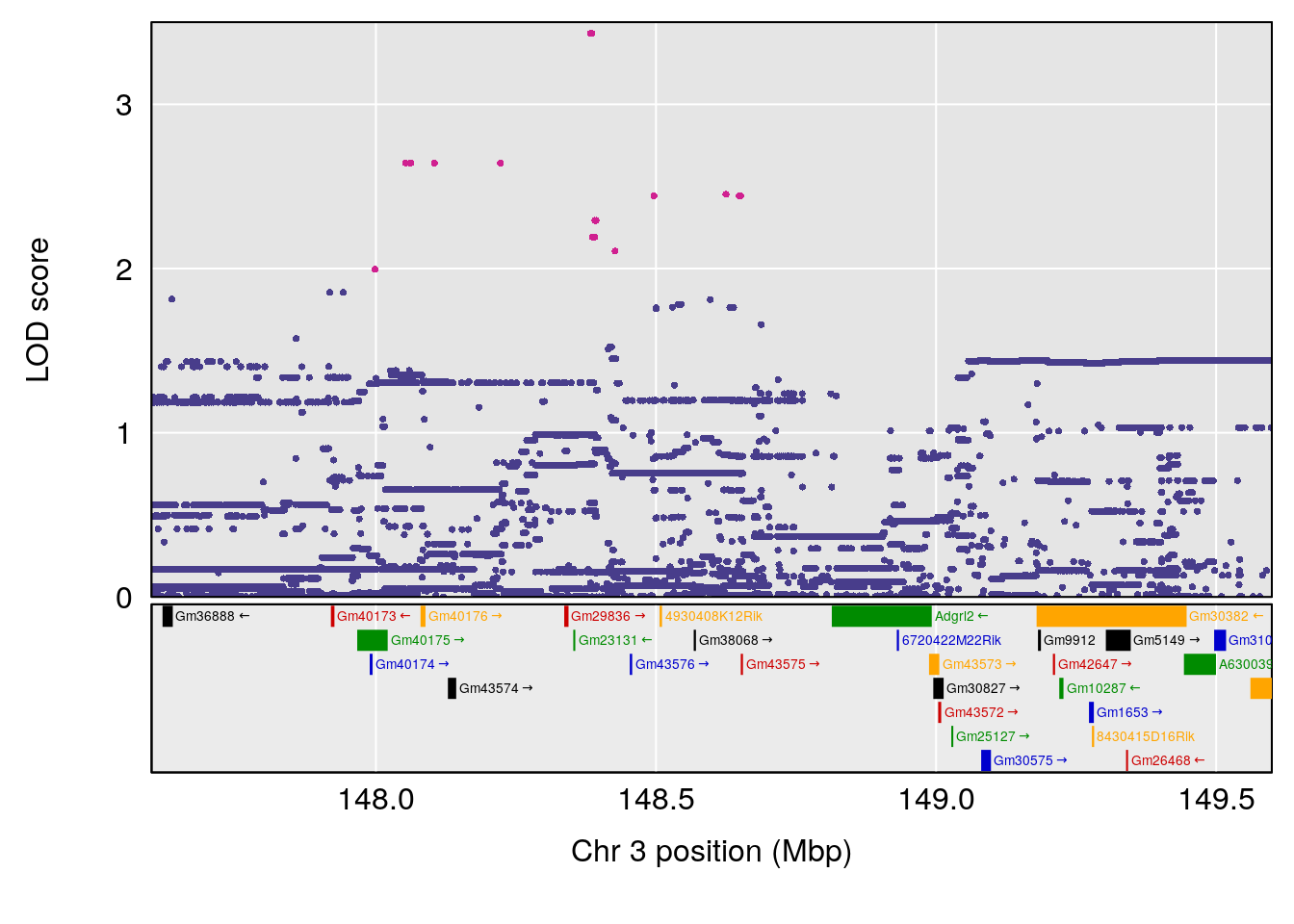
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
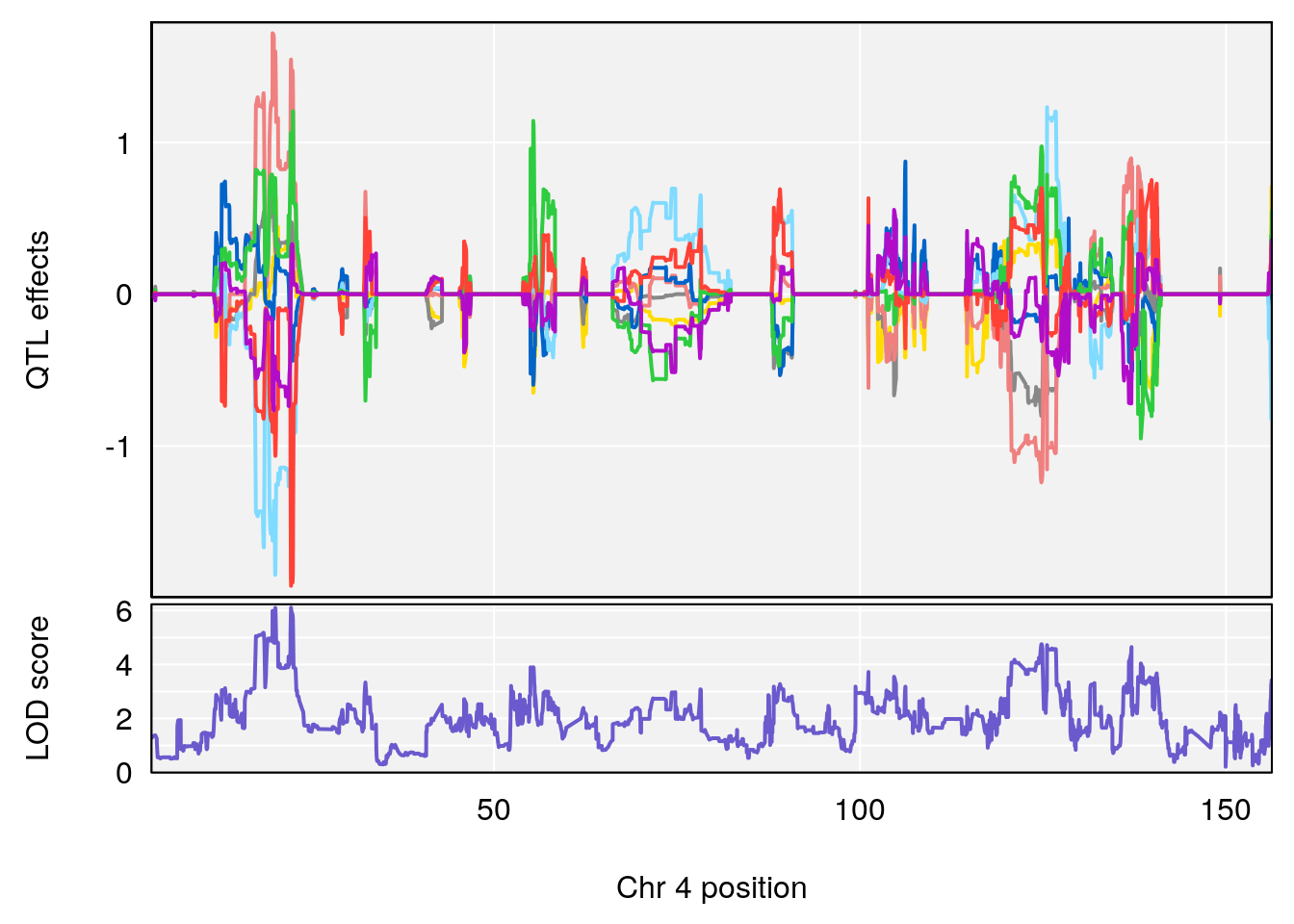
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
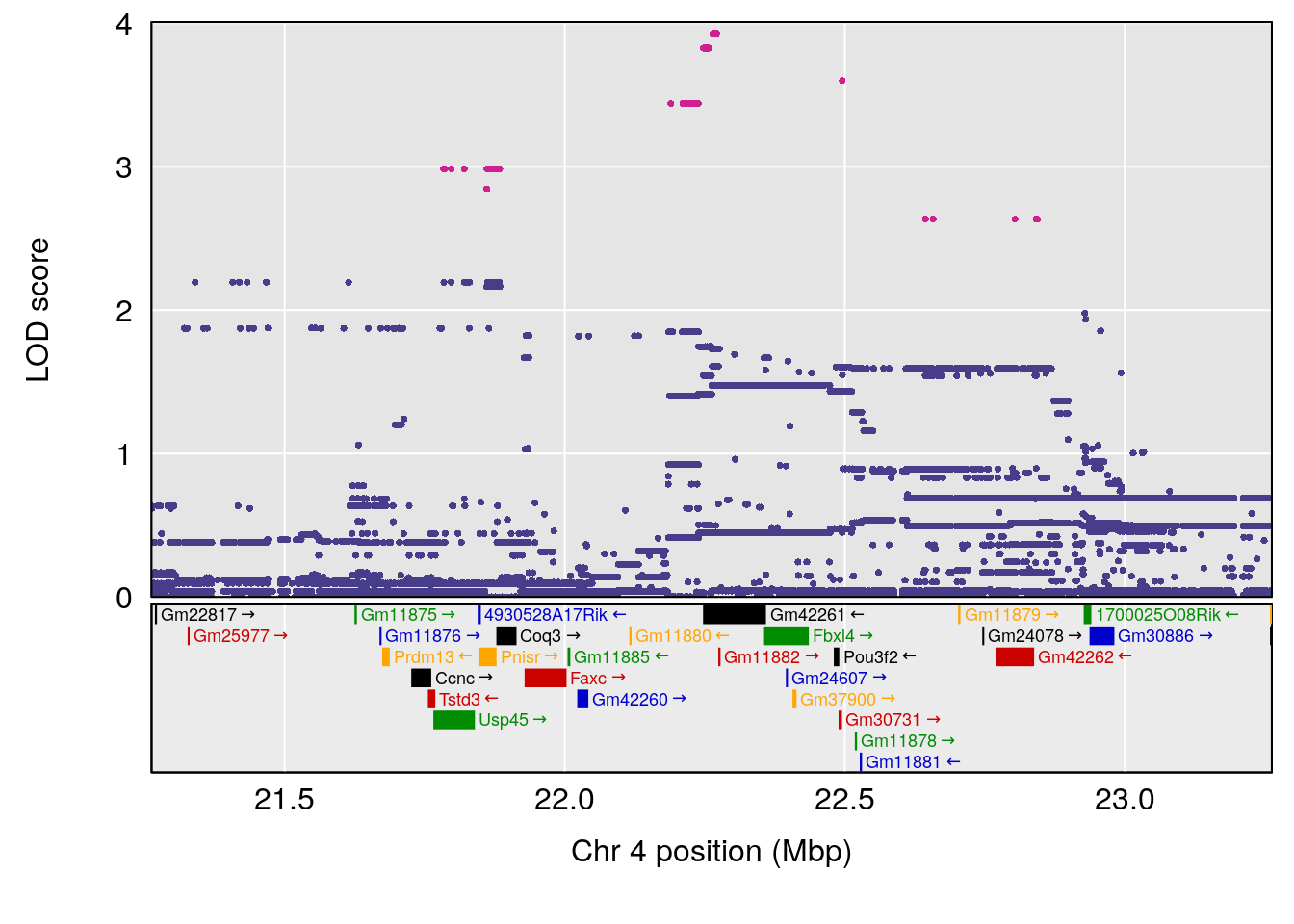
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 3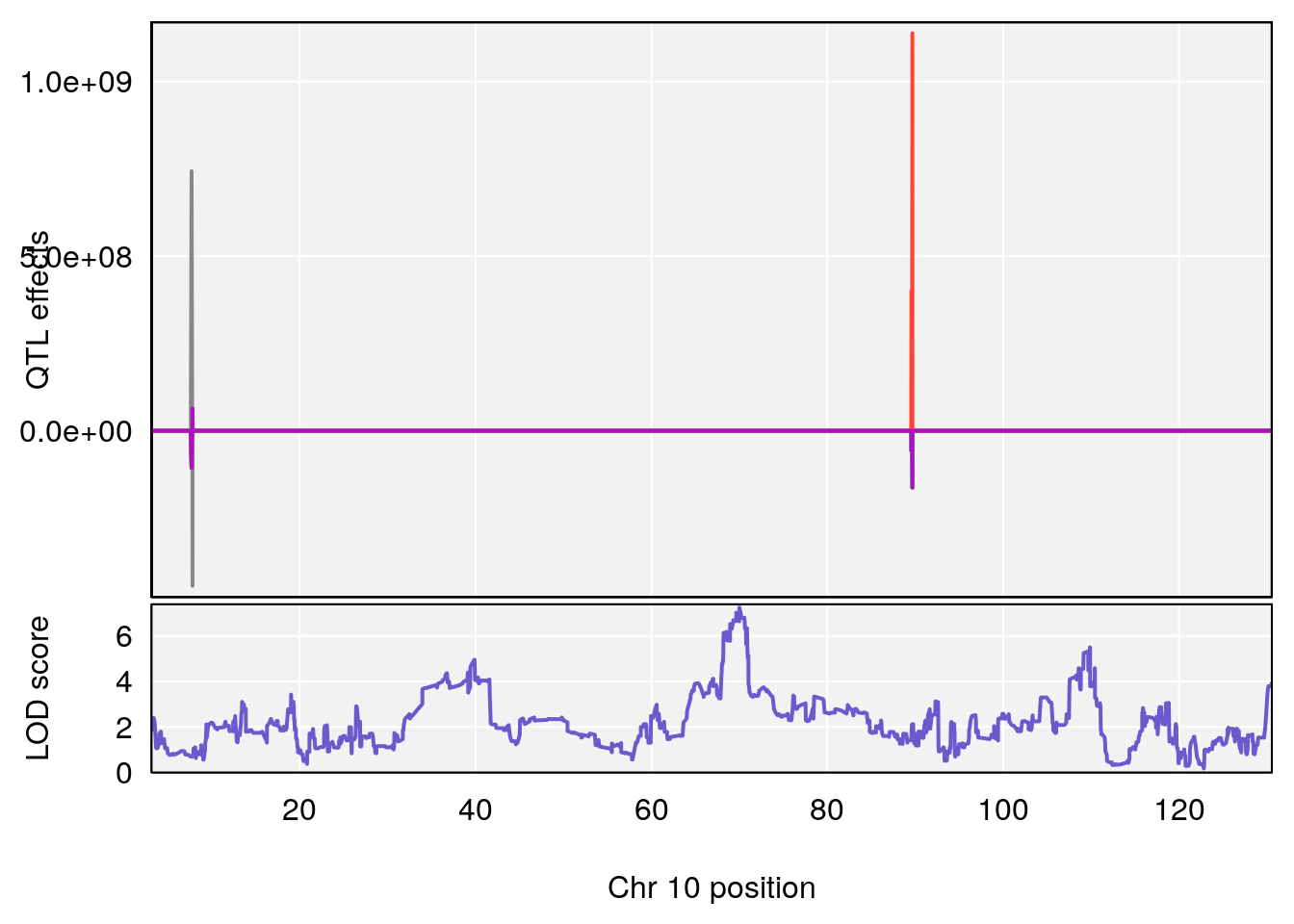
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
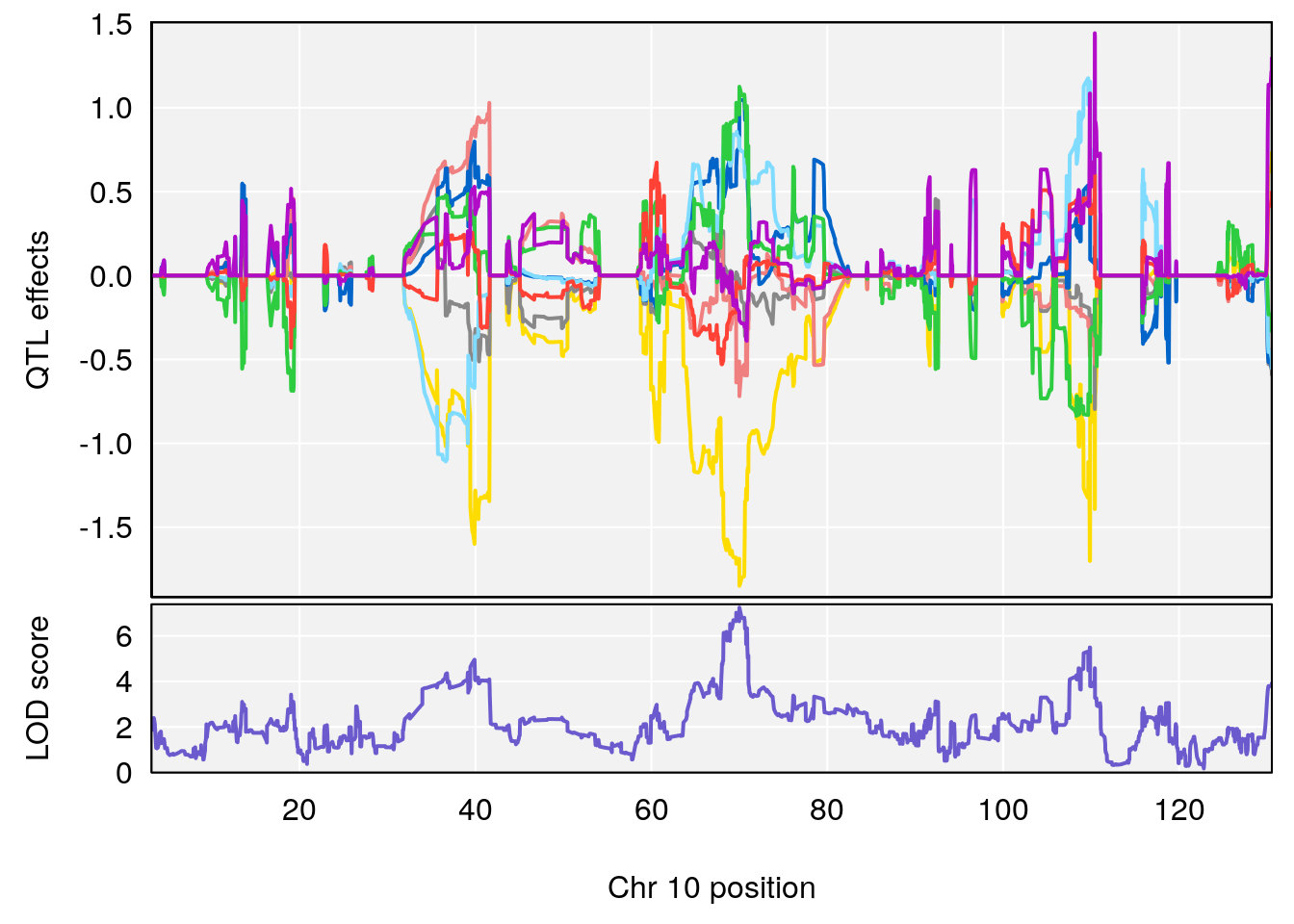
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
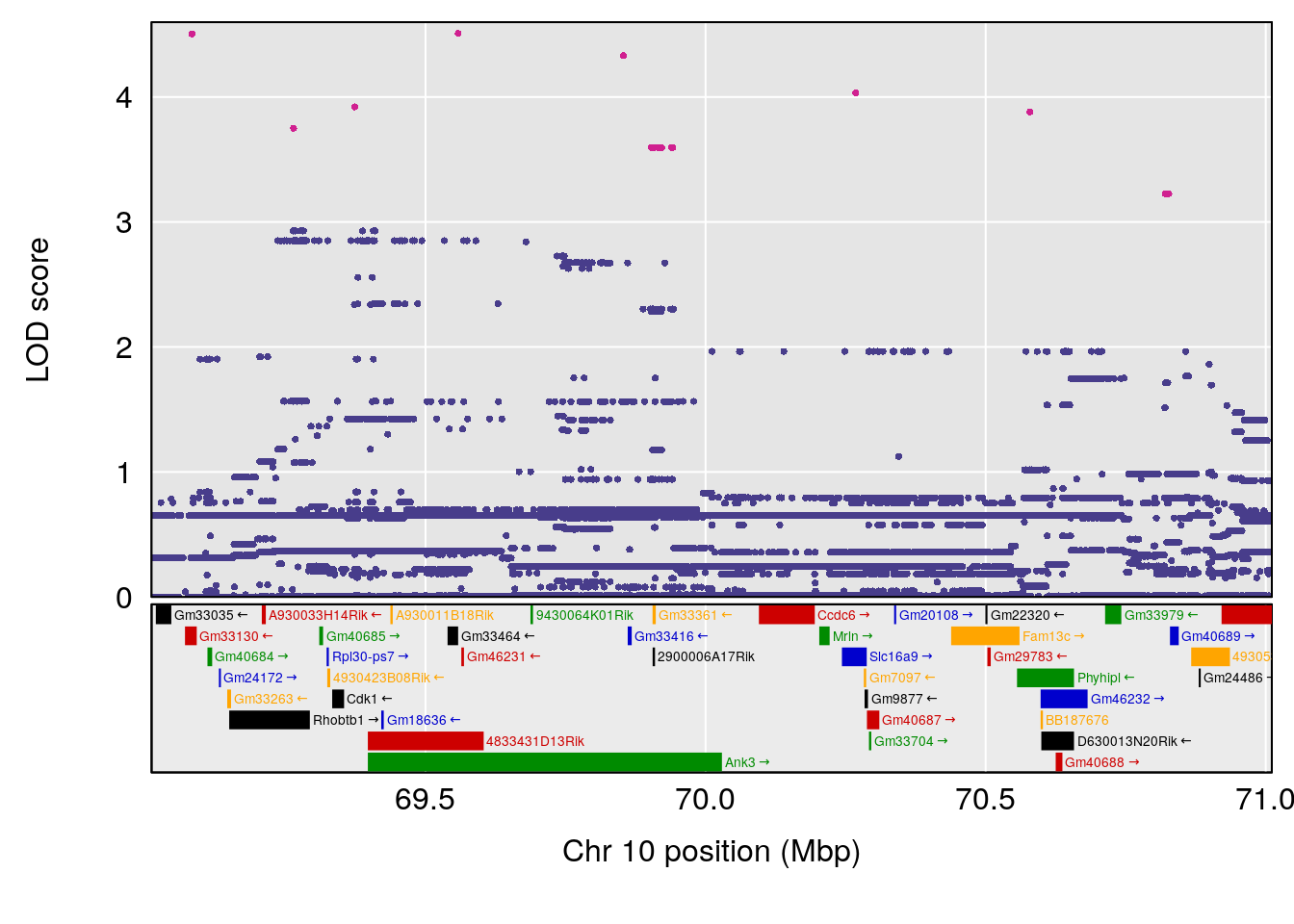
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 4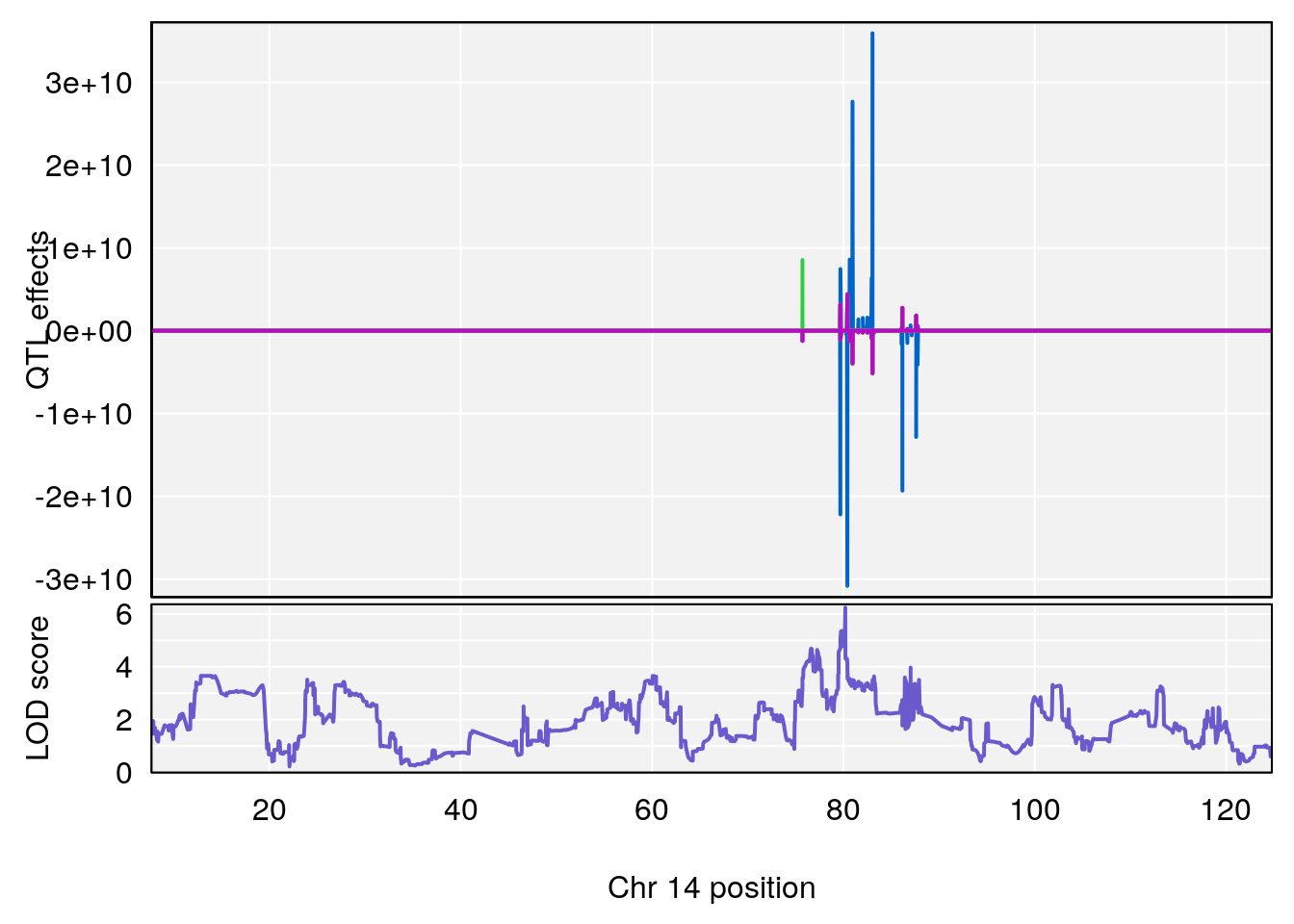
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
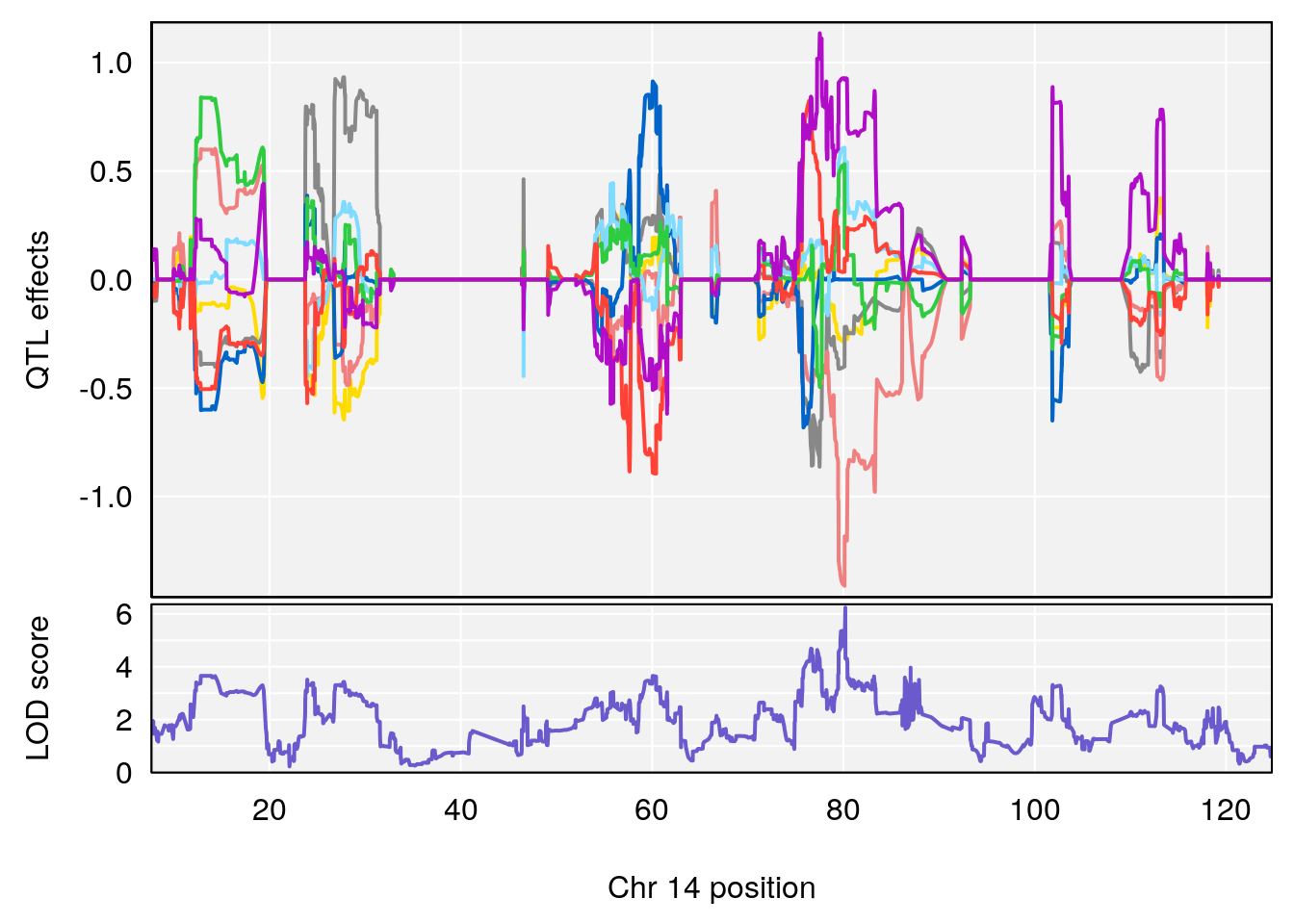
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
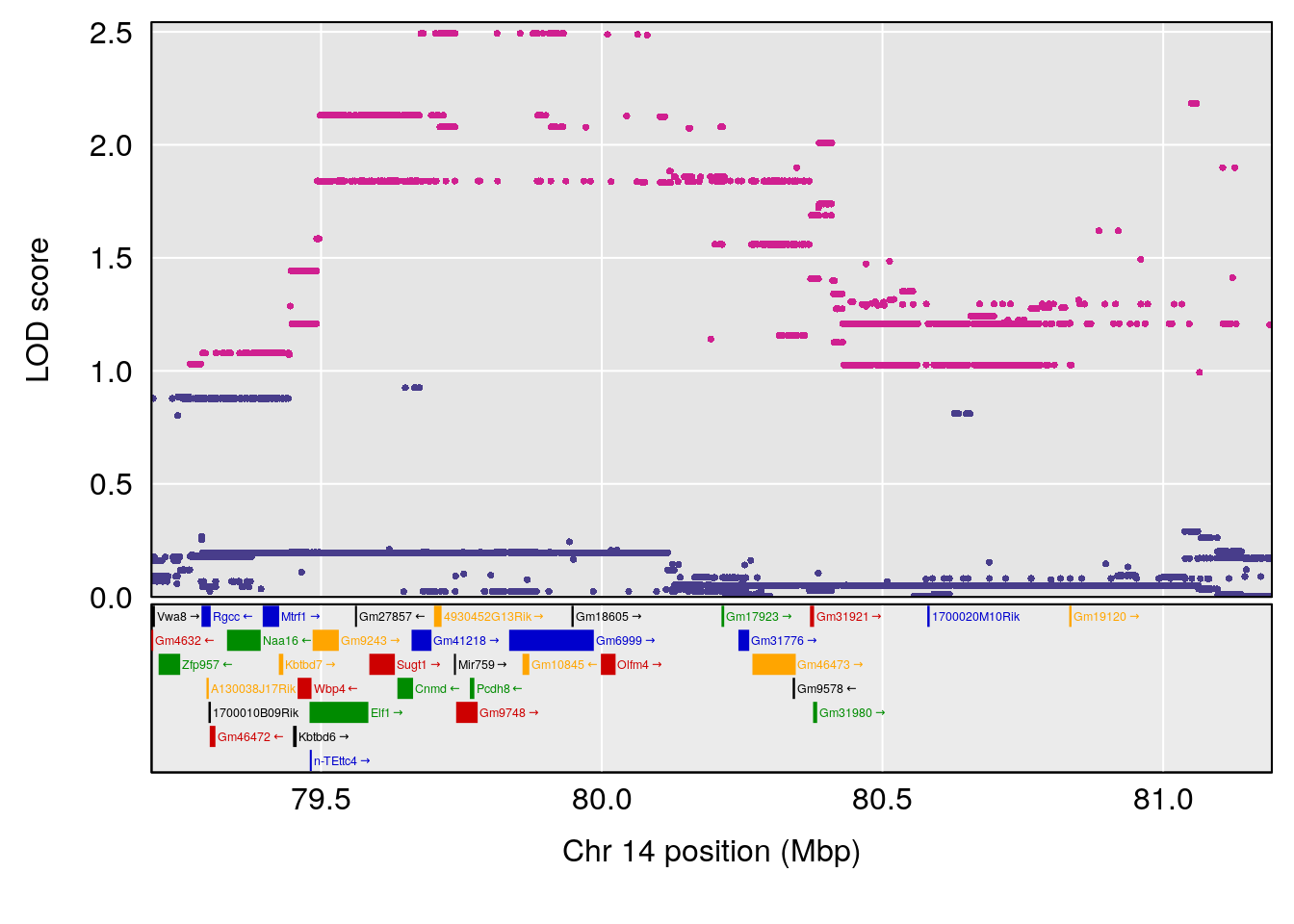
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 5
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
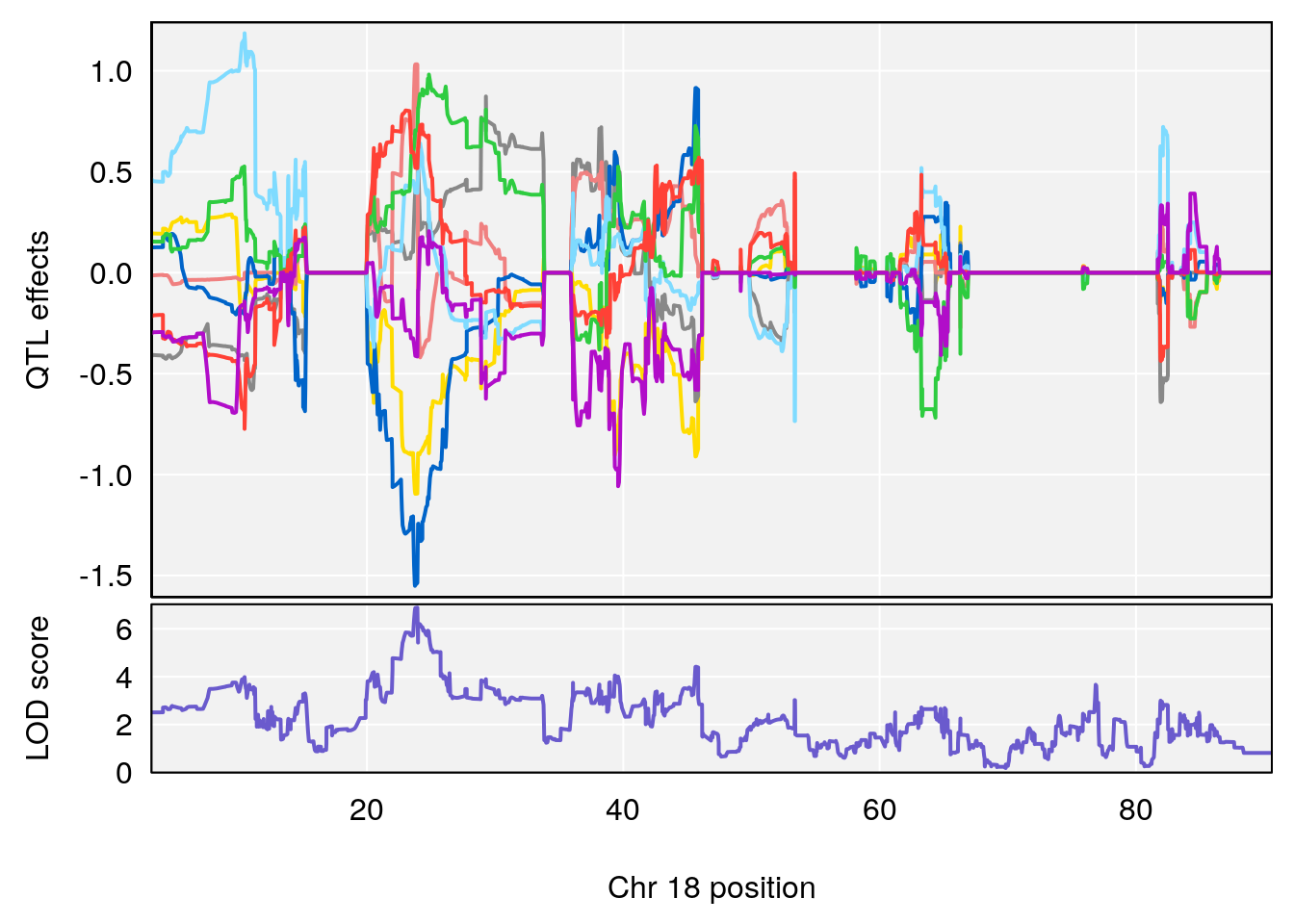
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
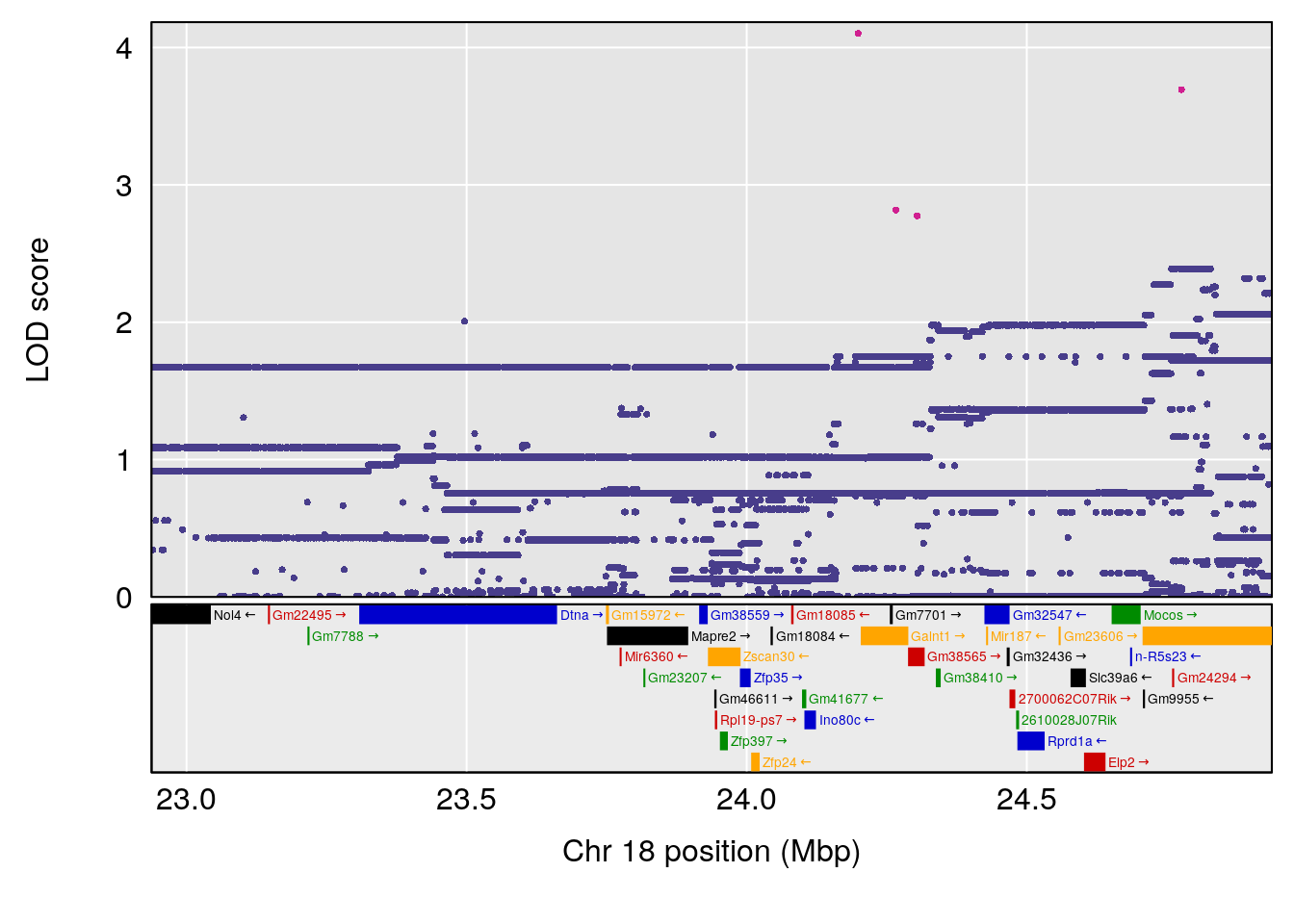
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] "Status_bin"
# lodindex lodcolumn chr pos lod ci_lo ci_hi
# 1 1 pheno1 1 185.11874 6.058648 41.25093 185.84088
# 2 1 pheno1 15 94.25093 7.334412 92.19023 94.31234
# 3 1 pheno1 18 16.19240 6.685013 15.31898 20.05685
# 4 1 pheno1 19 49.78637 6.595691 45.70921 53.50149
# [1] 1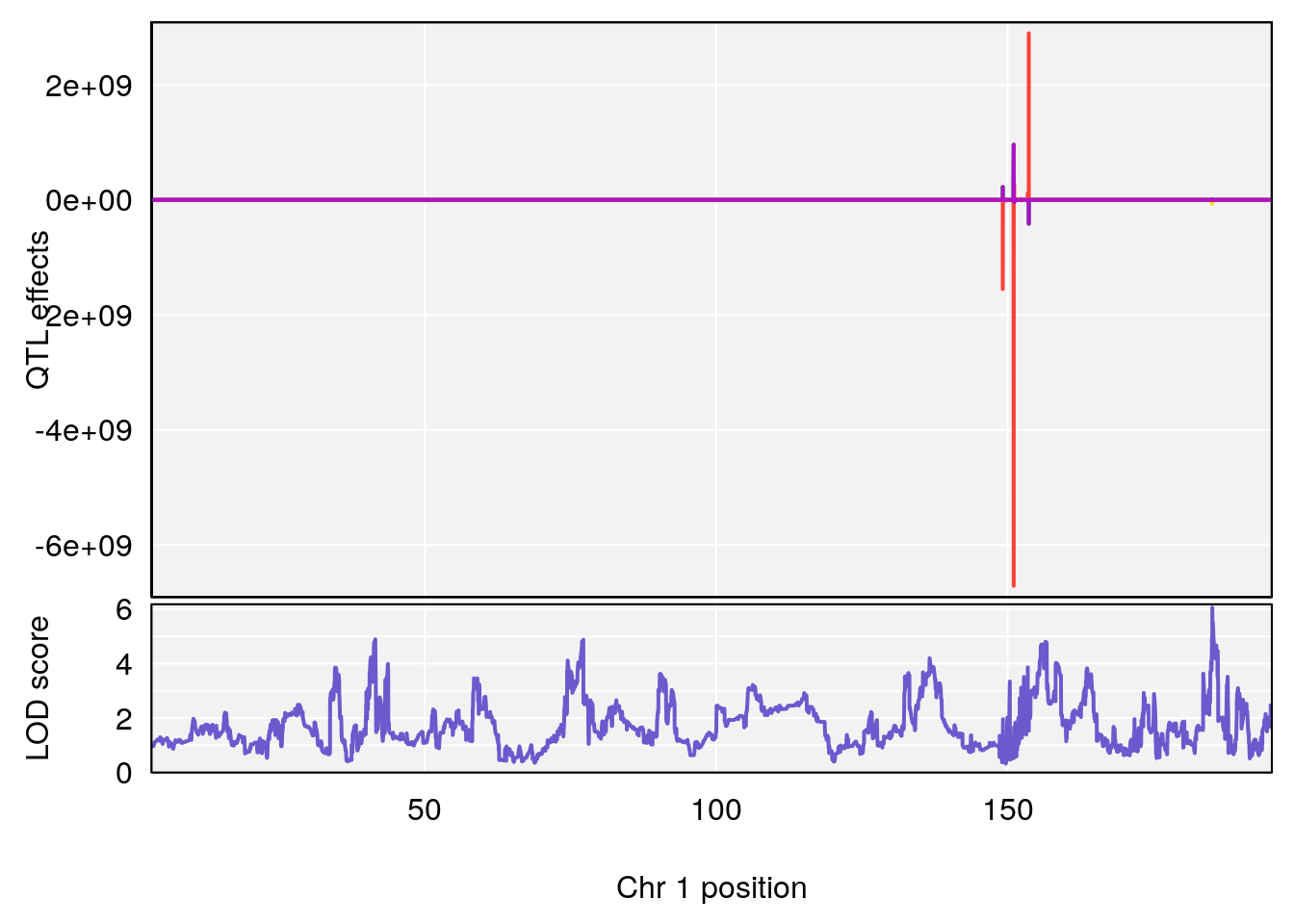
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
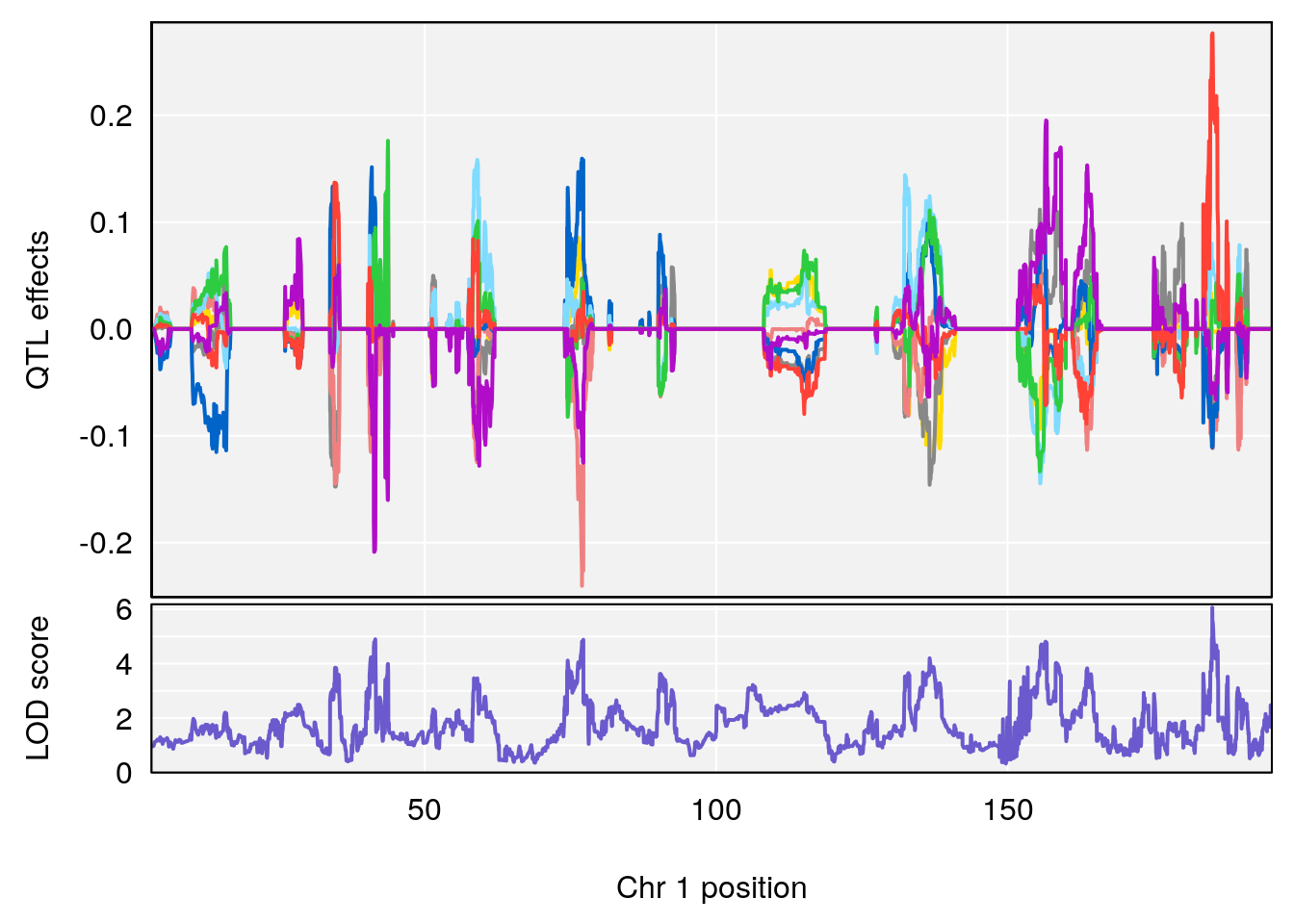
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
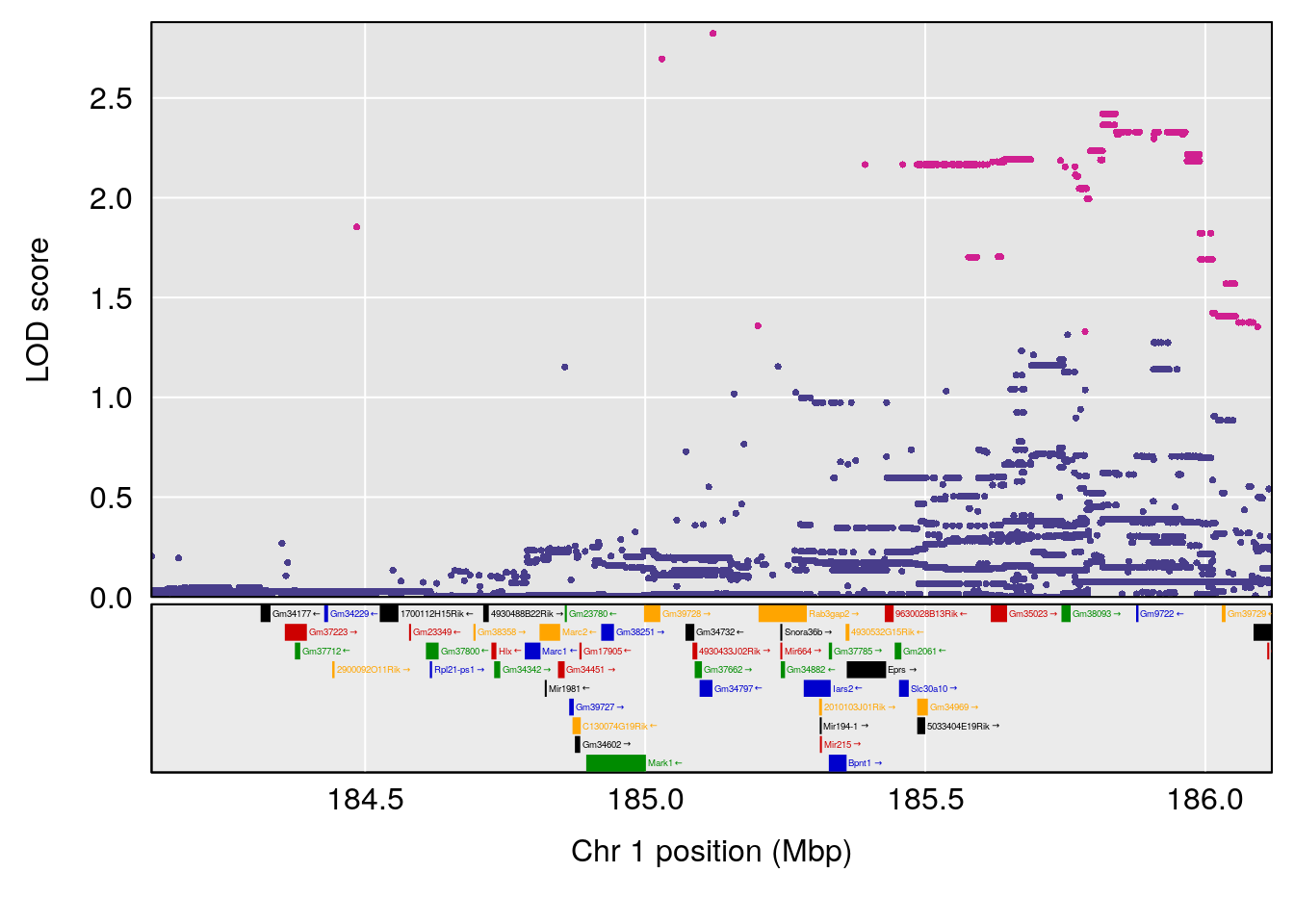
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 2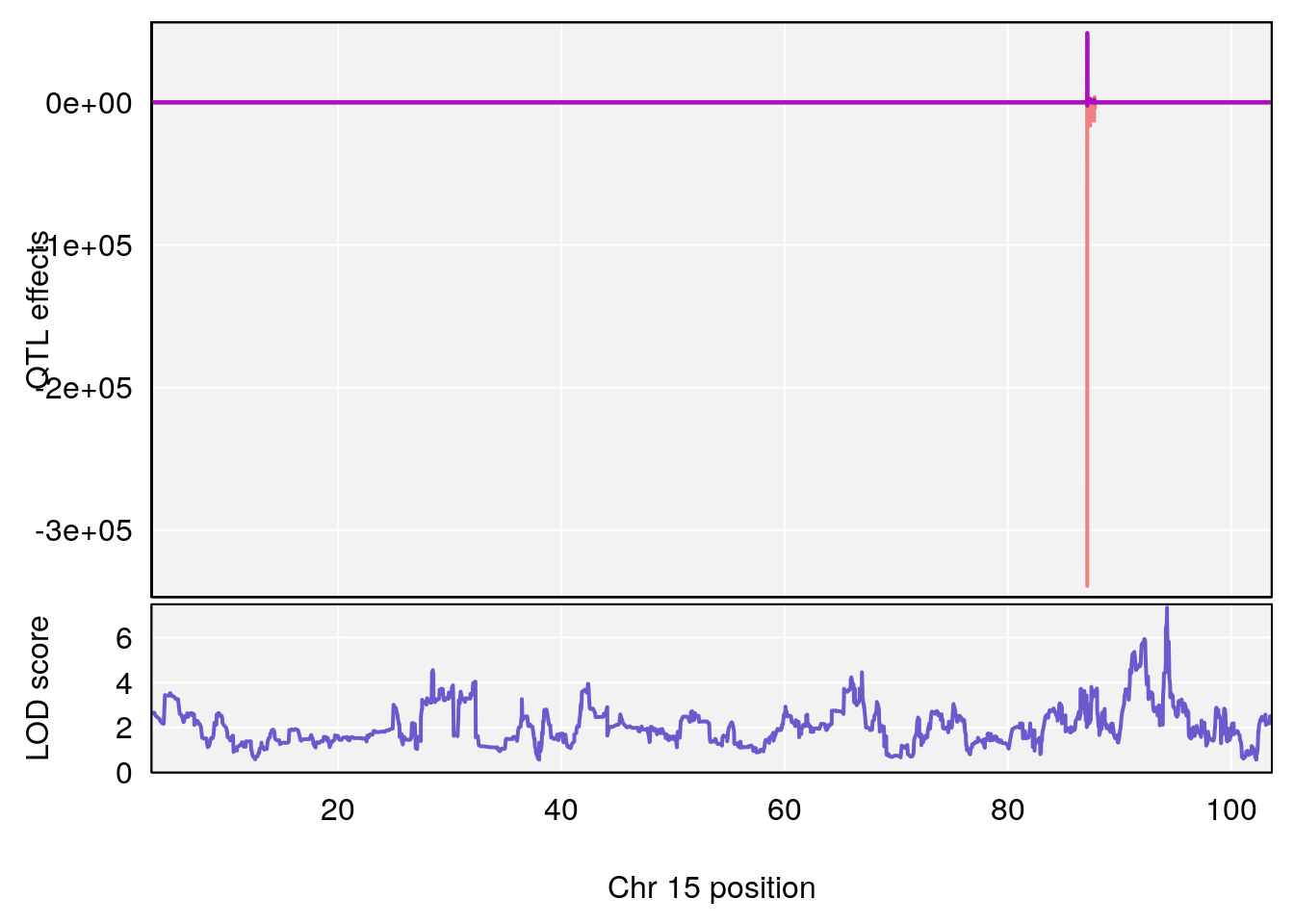
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
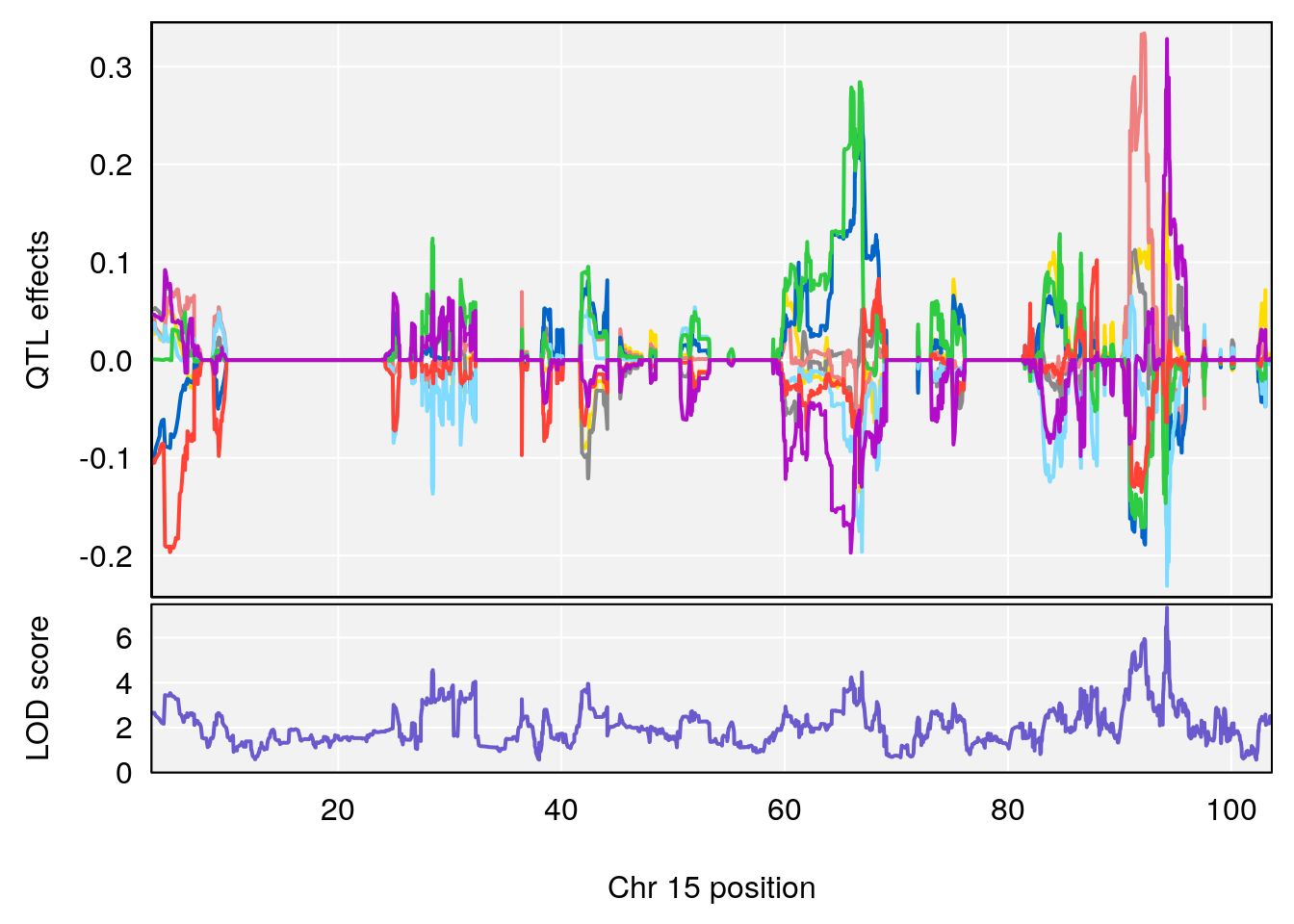
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
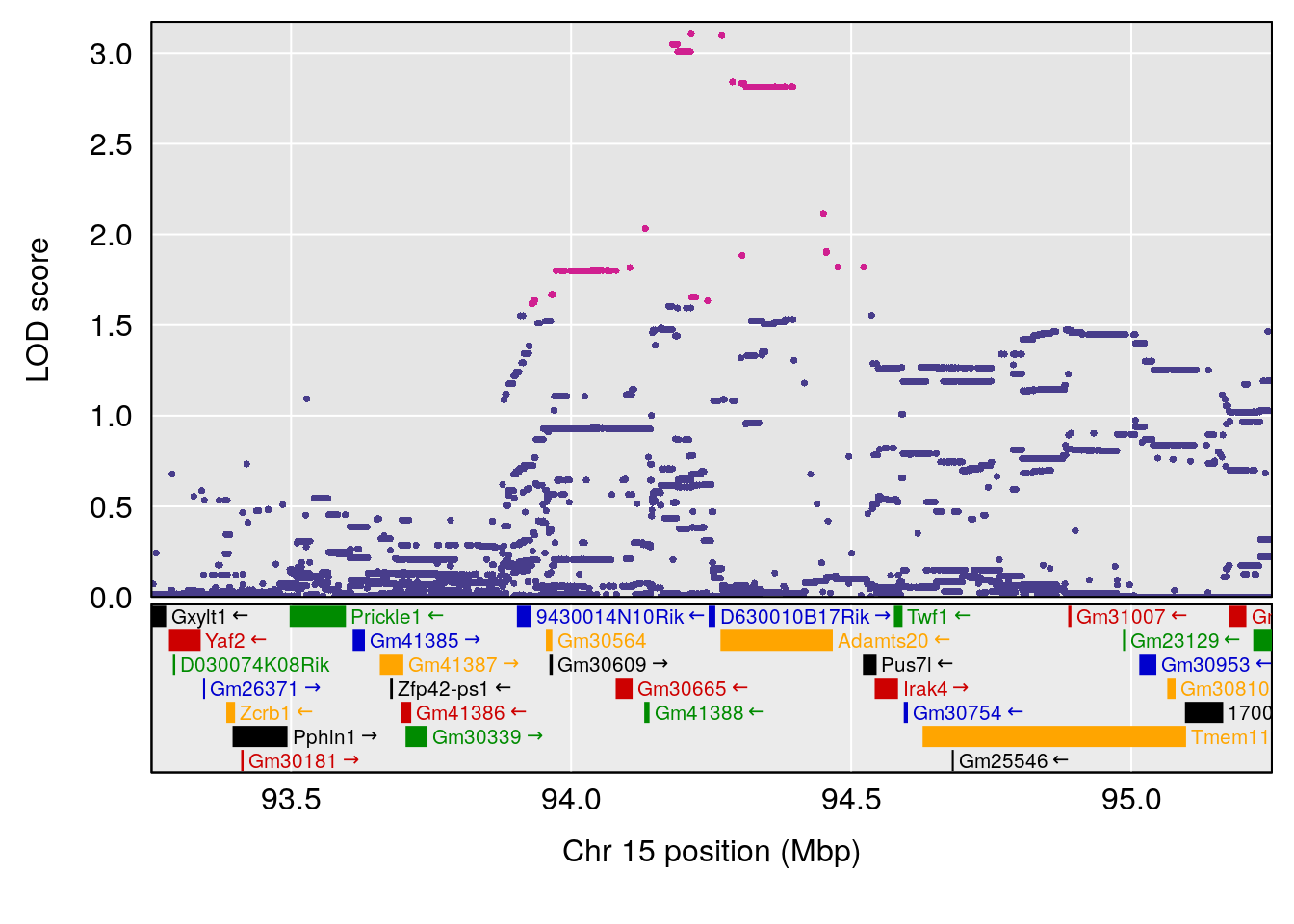
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 3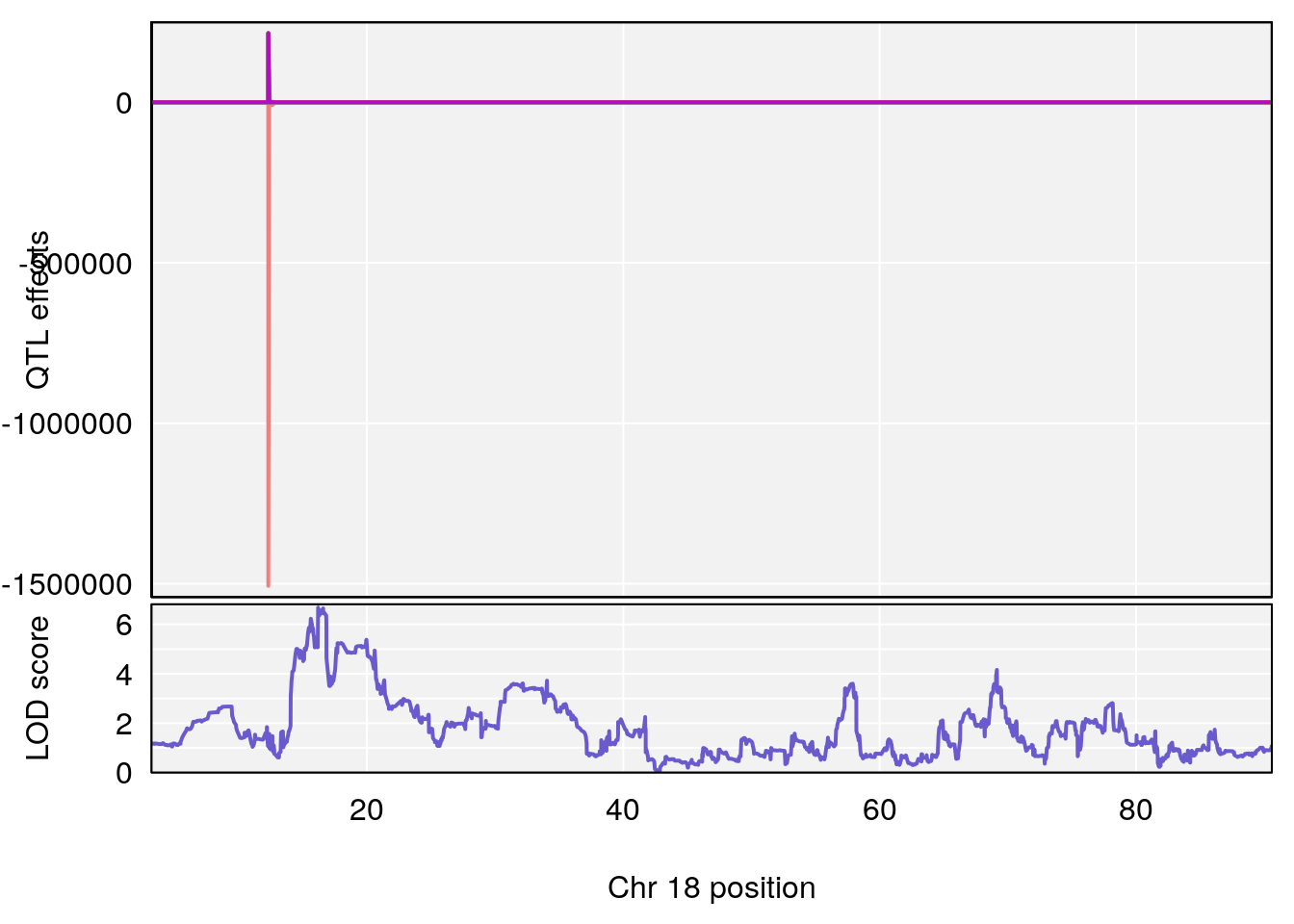
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
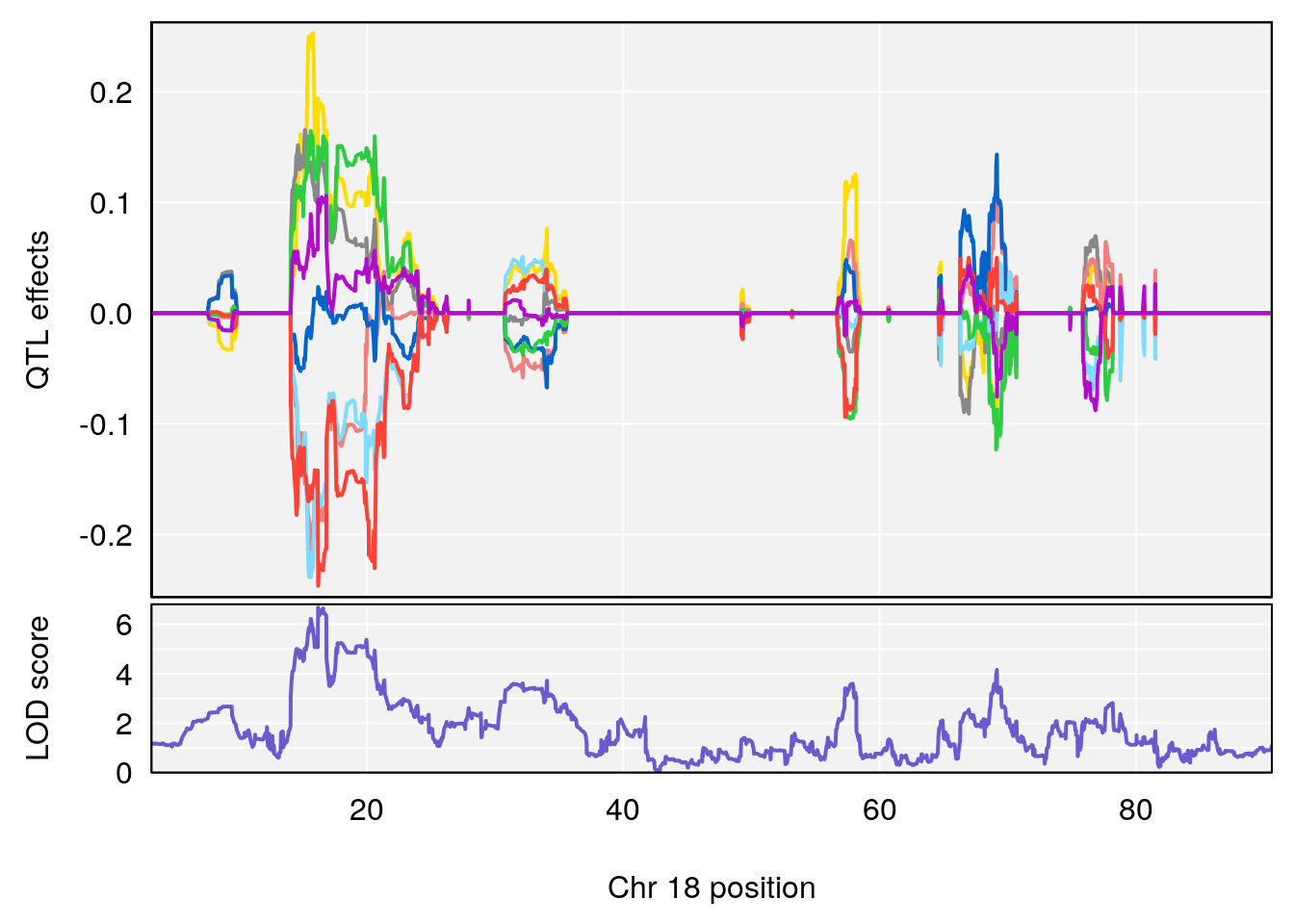
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
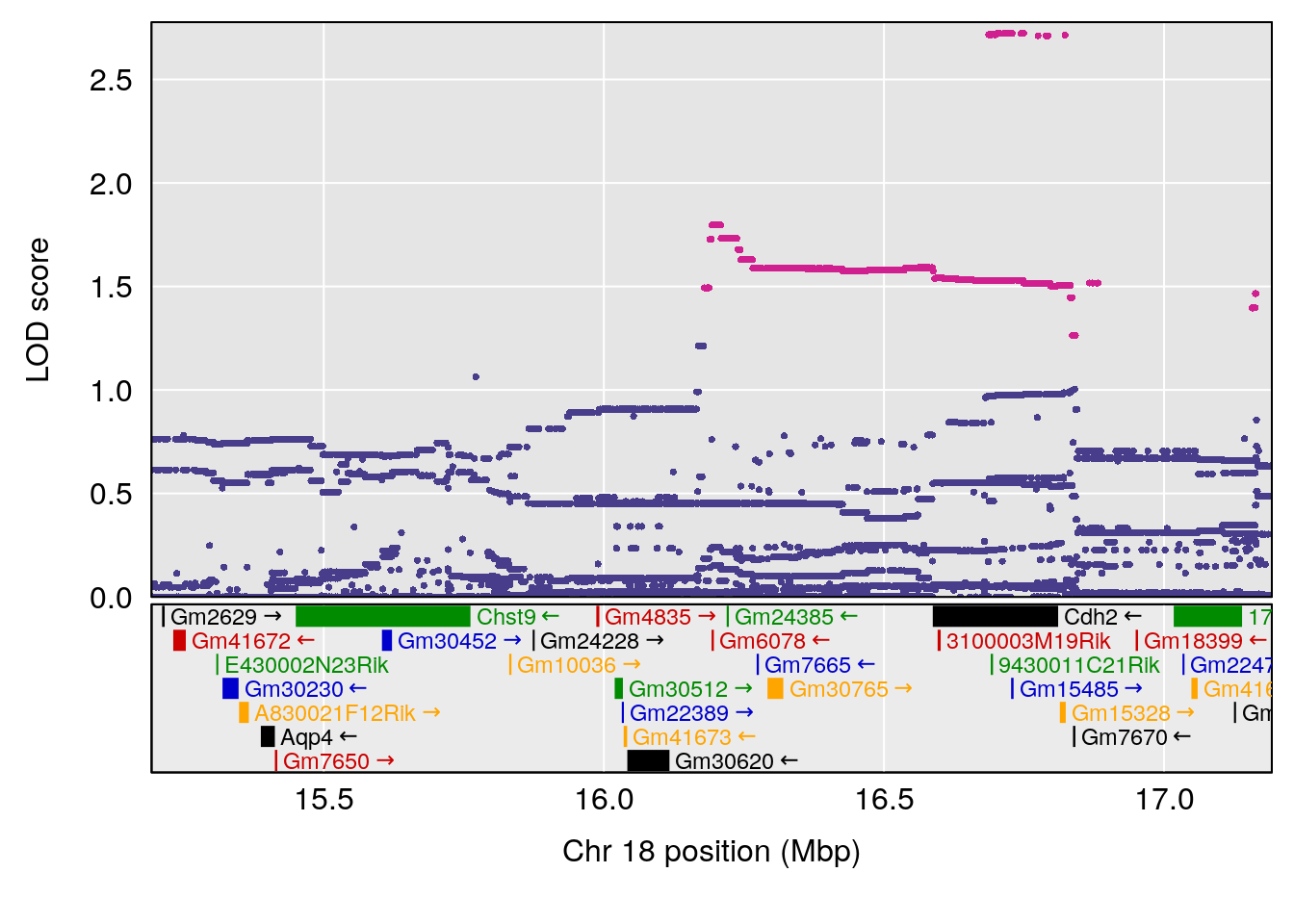
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
# [1] 4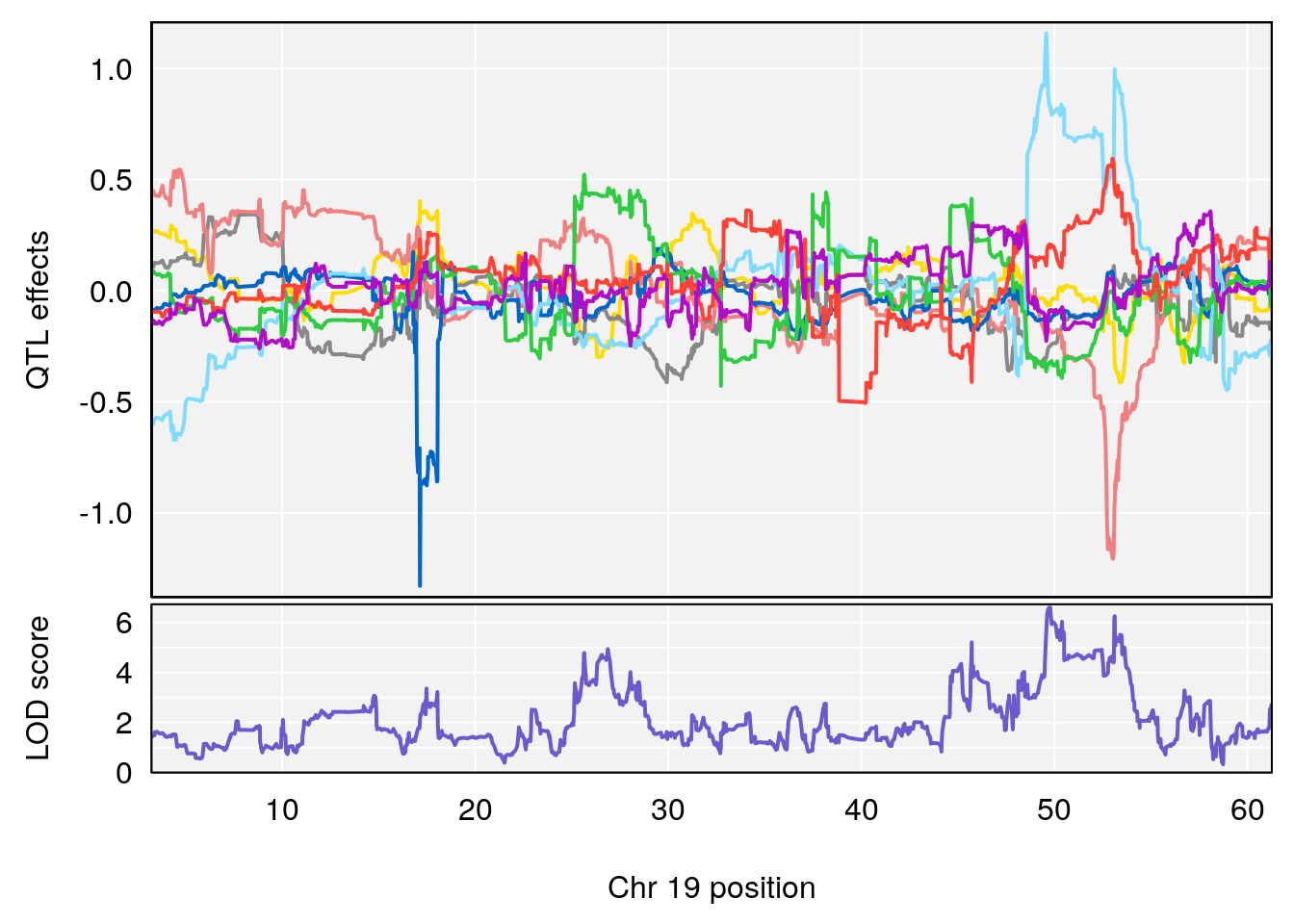
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
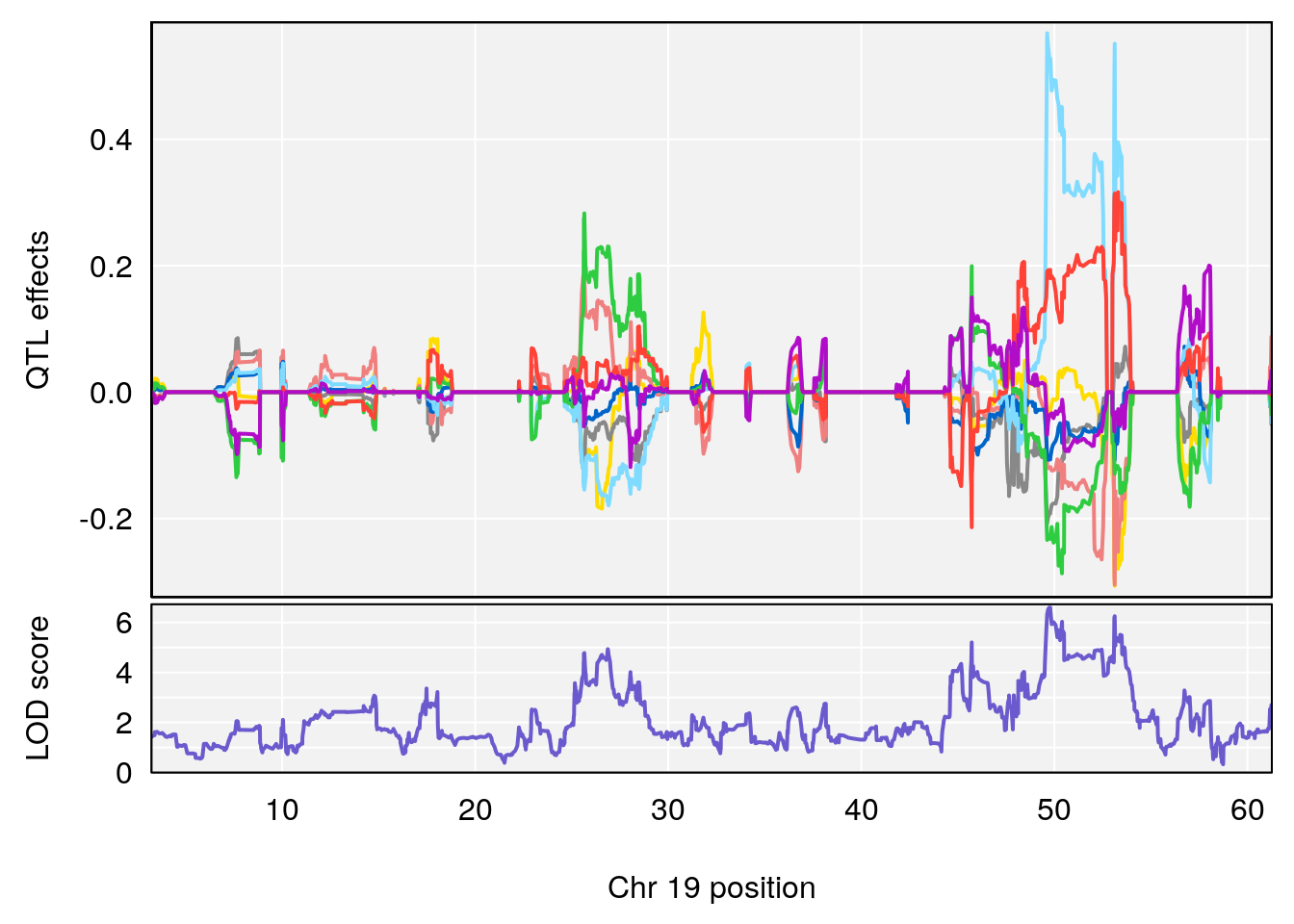
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
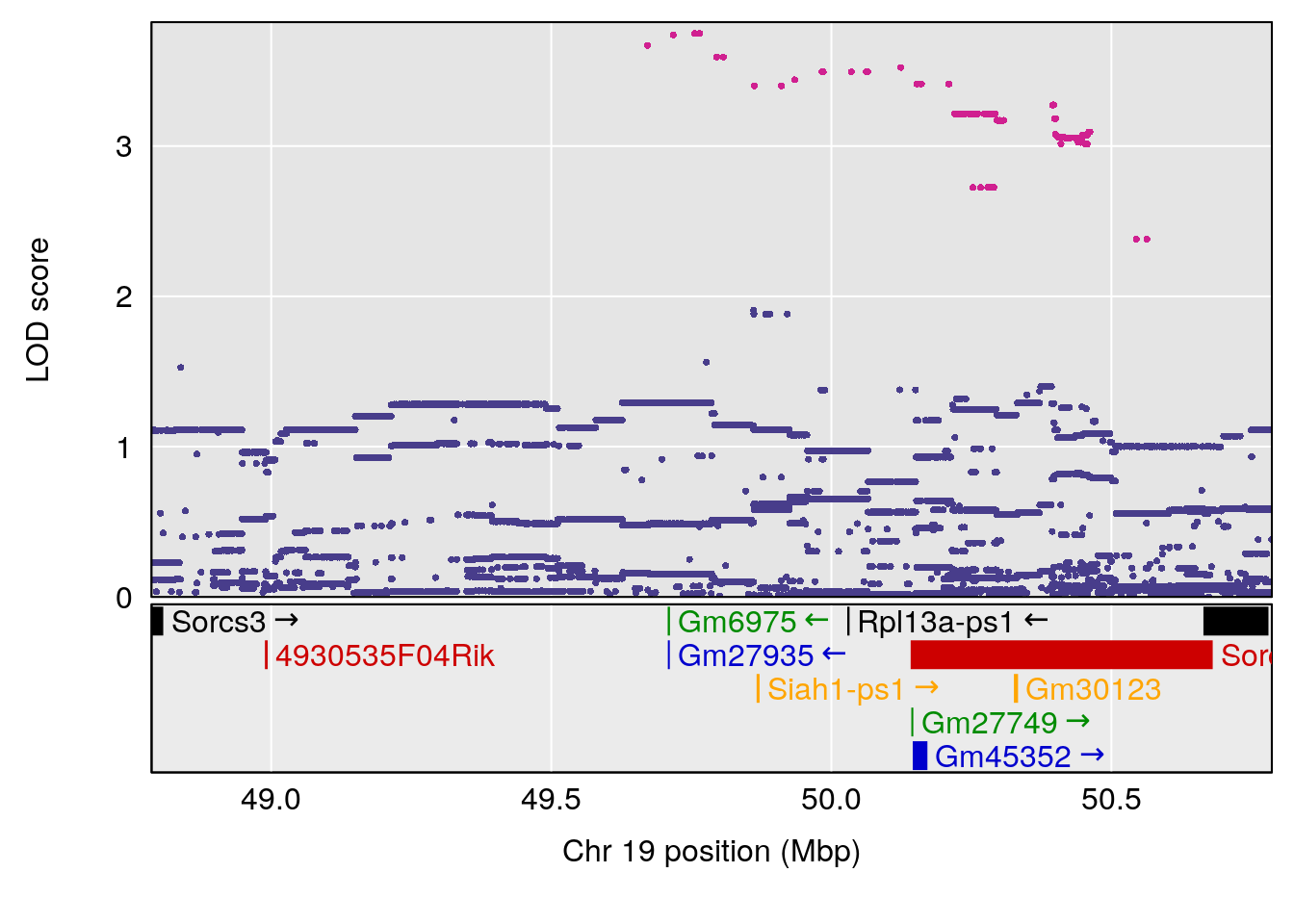
| Version | Author | Date |
|---|---|---|
| 8611541 | xhyuo | 2023-02-21 |
sessionInfo()
# R version 4.0.3 (2020-10-10)
# Platform: x86_64-pc-linux-gnu (64-bit)
# Running under: Ubuntu 20.04.2 LTS
#
# Matrix products: default
# BLAS/LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.8.so
#
# locale:
# [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
# [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
# [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=C
# [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
# [9] LC_ADDRESS=C LC_TELEPHONE=C
# [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
#
# attached base packages:
# [1] parallel stats graphics grDevices utils datasets methods
# [8] base
#
# other attached packages:
# [1] forcats_0.5.1 stringr_1.4.0 dplyr_1.0.4 purrr_0.3.4
# [5] readr_1.4.0 tidyr_1.1.2 tibble_3.0.6 tidyverse_1.3.0
# [9] openxlsx_4.2.3 abind_1.4-5 regress_1.3-21 survival_3.2-7
# [13] qtl2_0.24 GGally_2.1.0 gridExtra_2.3 ggplot2_3.3.3
# [17] workflowr_1.6.2
#
# loaded via a namespace (and not attached):
# [1] httr_1.4.2 bit64_4.0.5 jsonlite_1.7.2 splines_4.0.3
# [5] modelr_0.1.8 assertthat_0.2.1 highr_0.8 blob_1.2.1
# [9] cellranger_1.1.0 yaml_2.2.1 pillar_1.4.7 RSQLite_2.2.3
# [13] backports_1.2.1 lattice_0.20-41 glue_1.4.2 digest_0.6.27
# [17] RColorBrewer_1.1-2 promises_1.2.0.1 rvest_0.3.6 colorspace_2.0-0
# [21] htmltools_0.5.1.1 httpuv_1.5.5 Matrix_1.3-2 plyr_1.8.6
# [25] pkgconfig_2.0.3 broom_0.7.4 haven_2.3.1 scales_1.1.1
# [29] whisker_0.4 later_1.1.0.1 git2r_0.28.0 farver_2.0.3
# [33] generics_0.1.0 ellipsis_0.3.1 cachem_1.0.4 withr_2.4.1
# [37] cli_2.3.0 magrittr_2.0.1 crayon_1.4.1 readxl_1.3.1
# [41] memoise_2.0.0 evaluate_0.14 fs_1.5.0 xml2_1.3.2
# [45] tools_4.0.3 data.table_1.13.6 hms_1.0.0 lifecycle_1.0.0
# [49] reprex_1.0.0 munsell_0.5.0 zip_2.1.1 compiler_4.0.3
# [53] rlang_1.0.2 grid_4.0.3 rstudioapi_0.13 labeling_0.4.2
# [57] rmarkdown_2.6 gtable_0.3.0 DBI_1.1.1 reshape_0.8.8
# [61] R6_2.5.0 lubridate_1.7.9.2 knitr_1.31 fastmap_1.1.0
# [65] bit_4.0.4 rprojroot_2.0.2 stringi_1.5.3 Rcpp_1.0.6
# [69] vctrs_0.3.6 dbplyr_2.1.0 tidyselect_1.1.0 xfun_0.21This R Markdown site was created with workflowr