metabolomics_mouse_fecal
Hao He
2023-07-18
Last updated: 2023-07-18
Checks: 7 0
Knit directory: DO_Opioid/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200504) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 8566571. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/Picture1.png
Ignored: ieugwasr_oauth/
Untracked files:
Untracked: Rplot_rz.png
Untracked: TIMBR.test.prop.bm.MN.RET.RData
Untracked: TIMBR.test.random.RData
Untracked: TIMBR.test.rz.transformed_TVb_ml.RData
Untracked: analysis/DDO_morphine1_second_set_69k.stdout
Untracked: analysis/DO_Fentanyl.R
Untracked: analysis/DO_Fentanyl.err
Untracked: analysis/DO_Fentanyl.out
Untracked: analysis/DO_Fentanyl.sh
Untracked: analysis/DO_Fentanyl_69k.R
Untracked: analysis/DO_Fentanyl_69k.err
Untracked: analysis/DO_Fentanyl_69k.out
Untracked: analysis/DO_Fentanyl_69k.sh
Untracked: analysis/DO_Fentanyl_Cohort2_GCTA_herit.R
Untracked: analysis/DO_Fentanyl_Cohort2_gemma.R
Untracked: analysis/DO_Fentanyl_Cohort2_mapping.R
Untracked: analysis/DO_Fentanyl_Cohort2_mapping.err
Untracked: analysis/DO_Fentanyl_Cohort2_mapping.out
Untracked: analysis/DO_Fentanyl_Cohort2_mapping.sh
Untracked: analysis/DO_Fentanyl_GCTA_herit.R
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.R
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.err
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.out
Untracked: analysis/DO_Fentanyl_alternate_metrics_69k.sh
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.R
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.err
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.out
Untracked: analysis/DO_Fentanyl_alternate_metrics_array.sh
Untracked: analysis/DO_Fentanyl_array.R
Untracked: analysis/DO_Fentanyl_array.err
Untracked: analysis/DO_Fentanyl_array.out
Untracked: analysis/DO_Fentanyl_array.sh
Untracked: analysis/DO_Fentanyl_combining2Cohort_GCTA_herit.R
Untracked: analysis/DO_Fentanyl_combining2Cohort_gemma.R
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping.R
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping.err
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping.out
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping.sh
Untracked: analysis/DO_Fentanyl_combining2Cohort_mapping_CoxPH.R
Untracked: analysis/DO_Fentanyl_finalreport_to_plink.sh
Untracked: analysis/DO_Fentanyl_gemma.R
Untracked: analysis/DO_Fentanyl_gemma.err
Untracked: analysis/DO_Fentanyl_gemma.out
Untracked: analysis/DO_Fentanyl_gemma.sh
Untracked: analysis/DO_morphine1.R
Untracked: analysis/DO_morphine1.Rout
Untracked: analysis/DO_morphine1.sh
Untracked: analysis/DO_morphine1.stderr
Untracked: analysis/DO_morphine1.stdout
Untracked: analysis/DO_morphine1_SNP.R
Untracked: analysis/DO_morphine1_SNP.Rout
Untracked: analysis/DO_morphine1_SNP.sh
Untracked: analysis/DO_morphine1_SNP.stderr
Untracked: analysis/DO_morphine1_SNP.stdout
Untracked: analysis/DO_morphine1_combined.R
Untracked: analysis/DO_morphine1_combined.Rout
Untracked: analysis/DO_morphine1_combined.sh
Untracked: analysis/DO_morphine1_combined.stderr
Untracked: analysis/DO_morphine1_combined.stdout
Untracked: analysis/DO_morphine1_combined_69k.R
Untracked: analysis/DO_morphine1_combined_69k.Rout
Untracked: analysis/DO_morphine1_combined_69k.sh
Untracked: analysis/DO_morphine1_combined_69k.stderr
Untracked: analysis/DO_morphine1_combined_69k.stdout
Untracked: analysis/DO_morphine1_combined_69k_m2.R
Untracked: analysis/DO_morphine1_combined_69k_m2.Rout
Untracked: analysis/DO_morphine1_combined_69k_m2.sh
Untracked: analysis/DO_morphine1_combined_69k_m2.stderr
Untracked: analysis/DO_morphine1_combined_69k_m2.stdout
Untracked: analysis/DO_morphine1_combined_weight_DOB.R
Untracked: analysis/DO_morphine1_combined_weight_DOB.Rout
Untracked: analysis/DO_morphine1_combined_weight_DOB.err
Untracked: analysis/DO_morphine1_combined_weight_DOB.out
Untracked: analysis/DO_morphine1_combined_weight_DOB.sh
Untracked: analysis/DO_morphine1_combined_weight_DOB.stderr
Untracked: analysis/DO_morphine1_combined_weight_DOB.stdout
Untracked: analysis/DO_morphine1_combined_weight_age.R
Untracked: analysis/DO_morphine1_combined_weight_age.err
Untracked: analysis/DO_morphine1_combined_weight_age.out
Untracked: analysis/DO_morphine1_combined_weight_age.sh
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.R
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.err
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.out
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT.sh
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.R
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.err
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.out
Untracked: analysis/DO_morphine1_combined_weight_age_GAMMT_chr19.sh
Untracked: analysis/DO_morphine1_cph.R
Untracked: analysis/DO_morphine1_cph.Rout
Untracked: analysis/DO_morphine1_cph.sh
Untracked: analysis/DO_morphine1_second_set.R
Untracked: analysis/DO_morphine1_second_set.Rout
Untracked: analysis/DO_morphine1_second_set.sh
Untracked: analysis/DO_morphine1_second_set.stderr
Untracked: analysis/DO_morphine1_second_set.stdout
Untracked: analysis/DO_morphine1_second_set_69k.R
Untracked: analysis/DO_morphine1_second_set_69k.Rout
Untracked: analysis/DO_morphine1_second_set_69k.sh
Untracked: analysis/DO_morphine1_second_set_69k.stderr
Untracked: analysis/DO_morphine1_second_set_SNP.R
Untracked: analysis/DO_morphine1_second_set_SNP.Rout
Untracked: analysis/DO_morphine1_second_set_SNP.sh
Untracked: analysis/DO_morphine1_second_set_SNP.stderr
Untracked: analysis/DO_morphine1_second_set_SNP.stdout
Untracked: analysis/DO_morphine1_second_set_weight_DOB.R
Untracked: analysis/DO_morphine1_second_set_weight_DOB.Rout
Untracked: analysis/DO_morphine1_second_set_weight_DOB.err
Untracked: analysis/DO_morphine1_second_set_weight_DOB.out
Untracked: analysis/DO_morphine1_second_set_weight_DOB.sh
Untracked: analysis/DO_morphine1_second_set_weight_DOB.stderr
Untracked: analysis/DO_morphine1_second_set_weight_DOB.stdout
Untracked: analysis/DO_morphine1_second_set_weight_age.R
Untracked: analysis/DO_morphine1_second_set_weight_age.Rout
Untracked: analysis/DO_morphine1_second_set_weight_age.err
Untracked: analysis/DO_morphine1_second_set_weight_age.out
Untracked: analysis/DO_morphine1_second_set_weight_age.sh
Untracked: analysis/DO_morphine1_second_set_weight_age.stderr
Untracked: analysis/DO_morphine1_second_set_weight_age.stdout
Untracked: analysis/DO_morphine1_weight_DOB.R
Untracked: analysis/DO_morphine1_weight_DOB.sh
Untracked: analysis/DO_morphine1_weight_age.R
Untracked: analysis/DO_morphine1_weight_age.sh
Untracked: analysis/DO_morphine_gemma.R
Untracked: analysis/DO_morphine_gemma.err
Untracked: analysis/DO_morphine_gemma.out
Untracked: analysis/DO_morphine_gemma.sh
Untracked: analysis/DO_morphine_gemma_firstmin.R
Untracked: analysis/DO_morphine_gemma_firstmin.err
Untracked: analysis/DO_morphine_gemma_firstmin.out
Untracked: analysis/DO_morphine_gemma_firstmin.sh
Untracked: analysis/DO_morphine_gemma_withpermu.R
Untracked: analysis/DO_morphine_gemma_withpermu.err
Untracked: analysis/DO_morphine_gemma_withpermu.out
Untracked: analysis/DO_morphine_gemma_withpermu.sh
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.R
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.err
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.out
Untracked: analysis/DO_morphine_gemma_withpermu_firstbatch_min.depression.sh
Untracked: analysis/Plot_DO_morphine1_SNP.R
Untracked: analysis/Plot_DO_morphine1_SNP.Rout
Untracked: analysis/Plot_DO_morphine1_SNP.sh
Untracked: analysis/Plot_DO_morphine1_SNP.stderr
Untracked: analysis/Plot_DO_morphine1_SNP.stdout
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.R
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.Rout
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.sh
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.stderr
Untracked: analysis/Plot_DO_morphine1_second_set_SNP.stdout
Untracked: analysis/download_GSE100356_sra.sh
Untracked: analysis/fentanyl_2cohorts_coxph.R
Untracked: analysis/fentanyl_2cohorts_coxph.err
Untracked: analysis/fentanyl_2cohorts_coxph.out
Untracked: analysis/fentanyl_2cohorts_coxph.sh
Untracked: analysis/fentanyl_scanone.cph.R
Untracked: analysis/fentanyl_scanone.cph.err
Untracked: analysis/fentanyl_scanone.cph.out
Untracked: analysis/fentanyl_scanone.cph.sh
Untracked: analysis/geo_rnaseq.R
Untracked: analysis/heritability_first_second_batch.R
Untracked: analysis/morphine_fentanyl_survival_time.R
Untracked: analysis/nf-rnaseq-b6.R
Untracked: analysis/plot_fentanyl_2cohorts_coxph.R
Untracked: analysis/scripts/
Untracked: analysis/tibmr.R
Untracked: analysis/timbr_demo.R
Untracked: analysis/workflow_proc.R
Untracked: analysis/workflow_proc.sh
Untracked: analysis/workflow_proc.stderr
Untracked: analysis/workflow_proc.stdout
Untracked: analysis/x.R
Untracked: code/PLINKtoCSVR.R
Untracked: code/cfw/
Untracked: code/gemma_plot.R
Untracked: code/process.sanger.snp.R
Untracked: code/reconst_utils.R
Untracked: data/69k_grid_pgmap.RData
Untracked: data/CC_SARS-1/
Untracked: data/Composite Post Kevins Program Group 2 Fentanyl Prepped for Hao.xlsx
Untracked: data/DO_WBP_Data_JAB_to_map.xlsx
Untracked: data/Fentanyl_alternate_metrics.xlsx
Untracked: data/FinalReport/
Untracked: data/GM/
Untracked: data/GM_covar.csv
Untracked: data/GM_covar_07092018_morphine.csv
Untracked: data/Jackson_Lab_Bubier_MURGIGV01/
Untracked: data/MPD_Upload_October.csv
Untracked: data/MPD_Upload_October_updated_sex.csv
Untracked: data/Master Fentanyl DO Study Sheet.xlsx
Untracked: data/MasterMorphine Second Set DO w DOB2.xlsx
Untracked: data/MasterMorphine Second Set DO.xlsx
Untracked: data/Morphine CC DO mice Updated with Published inbred strains.csv
Untracked: data/Morphine_CC_DO_mice_Updated_with_Published_inbred_strains.csv
Untracked: data/cc_variants.sqlite
Untracked: data/combined/
Untracked: data/fentanyl/
Untracked: data/fentanyl2/
Untracked: data/fentanyl_1_2/
Untracked: data/fentanyl_2cohorts_coxph_data.Rdata
Untracked: data/first/
Untracked: data/founder_geno.csv
Untracked: data/genetic_map.csv
Untracked: data/gm.json
Untracked: data/gwas.sh
Untracked: data/marker_grid_0.02cM_plus.txt
Untracked: data/metabolomics_mouse_fecal/
Untracked: data/mouse_genes_mgi.sqlite
Untracked: data/pheno.csv
Untracked: data/pheno_qtl2.csv
Untracked: data/pheno_qtl2_07092018_morphine.csv
Untracked: data/pheno_qtl2_w_dob.csv
Untracked: data/physical_map.csv
Untracked: data/rnaseq/
Untracked: data/sample_geno.csv
Untracked: data/second/
Untracked: figure/
Untracked: glimma-plots/
Untracked: output/DO_Fentanyl_Cohort2_MinDepressionRR_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_MinDepressionRR_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_RRDepressionRateHrSLOPE_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_RRRecoveryRateHrSLOPE_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_RRRecoveryRateHrSLOPE_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_StartofRecoveryHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_StartofRecoveryHr_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_Statusbin_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_Statusbin_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_SteadyStateDepressionDurationHrINTERVAL_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoDead(Hr)_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoDeadHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoDeadHr_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoProjectedRecoveryHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoProjectedRecoveryHr_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoSteadyRRDepression(Hr)_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoSteadyRRDepressionHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoSteadyRRDepressionHr_coefplot_blup.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoThresholdRecoveryHr_coefplot.pdf
Untracked: output/DO_Fentanyl_Cohort2_TimetoThresholdRecoveryHr_coefplot_blup.pdf
Untracked: output/DO_morphine_Min.depression.png
Untracked: output/DO_morphine_Min.depression22222_violin_chr5.pdf
Untracked: output/DO_morphine_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_Min.depression_coefplot_blup_chr5.png
Untracked: output/DO_morphine_Min.depression_coefplot_blup_chrX.png
Untracked: output/DO_morphine_Min.depression_coefplot_chr5.png
Untracked: output/DO_morphine_Min.depression_coefplot_chrX.png
Untracked: output/DO_morphine_Min.depression_peak_genes_chr5.png
Untracked: output/DO_morphine_Min.depression_violin_chr5.png
Untracked: output/DO_morphine_Recovery.Time.png
Untracked: output/DO_morphine_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr11.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr4.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr7.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_blup_chr9.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr11.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr4.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr7.png
Untracked: output/DO_morphine_Recovery.Time_coefplot_chr9.png
Untracked: output/DO_morphine_Status_bin.png
Untracked: output/DO_morphine_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_Survival.Time.png
Untracked: output/DO_morphine_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_Survival.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_Survival.Time_coefplot_blup_chr17.png
Untracked: output/DO_morphine_Survival.Time_coefplot_blup_chr8.png
Untracked: output/DO_morphine_Survival.Time_coefplot_chr17.png
Untracked: output/DO_morphine_Survival.Time_coefplot_chr8.png
Untracked: output/DO_morphine_combine_batch_peak_violin.pdf
Untracked: output/DO_morphine_combined_69k_m2_Min.depression.png
Untracked: output/DO_morphine_combined_69k_m2_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_combined_69k_m2_Recovery.Time.png
Untracked: output/DO_morphine_combined_69k_m2_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_combined_69k_m2_Status_bin.png
Untracked: output/DO_morphine_combined_69k_m2_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_combined_69k_m2_Survival.Time.png
Untracked: output/DO_morphine_combined_69k_m2_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_combined_69k_m2_Survival.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_coxph_24hrs_kinship_QTL.png
Untracked: output/DO_morphine_cphout.RData
Untracked: output/DO_morphine_first_batch_peak_in_second_batch_violin.pdf
Untracked: output/DO_morphine_first_batch_peak_in_second_batch_violin_sidebyside.pdf
Untracked: output/DO_morphine_first_batch_peak_violin.pdf
Untracked: output/DO_morphine_operm.cph.RData
Untracked: output/DO_morphine_second_batch_on_first_batch_peak_violin.pdf
Untracked: output/DO_morphine_second_batch_peak_ch6surv_on_first_batchviolin.pdf
Untracked: output/DO_morphine_second_batch_peak_ch6surv_on_first_batchviolin2.pdf
Untracked: output/DO_morphine_second_batch_peak_in_first_batch_violin.pdf
Untracked: output/DO_morphine_second_batch_peak_in_first_batch_violin_sidebyside.pdf
Untracked: output/DO_morphine_second_batch_peak_violin.pdf
Untracked: output/DO_morphine_secondbatch_69k_Min.depression.png
Untracked: output/DO_morphine_secondbatch_69k_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_69k_Recovery.Time.png
Untracked: output/DO_morphine_secondbatch_69k_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_69k_Status_bin.png
Untracked: output/DO_morphine_secondbatch_69k_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_69k_Survival.Time.png
Untracked: output/DO_morphine_secondbatch_69k_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_69k_Survival.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Min.depression.png
Untracked: output/DO_morphine_secondbatch_Min.depression_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Min.depression_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Recovery.Time.png
Untracked: output/DO_morphine_secondbatch_Recovery.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Recovery.Time_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Status_bin.png
Untracked: output/DO_morphine_secondbatch_Status_bin_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Status_bin_coefplot_blup.pdf
Untracked: output/DO_morphine_secondbatch_Survival.Time.png
Untracked: output/DO_morphine_secondbatch_Survival.Time_coefplot.pdf
Untracked: output/DO_morphine_secondbatch_Survival.Time_coefplot_blup.pdf
Untracked: output/Fentanyl/
Untracked: output/KPNA3.pdf
Untracked: output/SSC4D.pdf
Untracked: output/TIMBR.test.RData
Untracked: output/apr_69kchr_combined.RData
Untracked: output/apr_69kchr_k_loco_combined.rds
Untracked: output/apr_69kchr_second_set.RData
Untracked: output/combine_batch_variation.RData
Untracked: output/combined_gm.RData
Untracked: output/combined_gm.k_loco.rds
Untracked: output/combined_gm.k_overall.rds
Untracked: output/combined_gm.probs_8state.rds
Untracked: output/coxph/
Untracked: output/do.morphine.RData
Untracked: output/do.morphine.k_loco.rds
Untracked: output/do.morphine.probs_36state.rds
Untracked: output/do.morphine.probs_8state.rds
Untracked: output/do_Fentanyl_combine2cohort_MeanDepressionBR_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_MeanDepressionBR_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_MinDepressionBR_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_MinDepressionBR_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_MinDepressionRR_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_MinDepressionRR_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_RRRecoveryRateHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_RRRecoveryRateHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_StartofRecoveryHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_StartofRecoveryHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_Statusbin_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_Statusbin_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_SteadyStateDepressionDurationHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_SteadyStateDepressionDurationHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_SurvivalTime_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_SurvivalTime_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoDeadHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoDeadHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoMostlyDeadHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoMostlyDeadHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoProjectedRecoveryHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoProjectedRecoveryHr_coefplot_blup.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoRecoveryHr_coefplot.pdf
Untracked: output/do_Fentanyl_combine2cohort_TimetoRecoveryHr_coefplot_blup.pdf
Untracked: output/first_batch_variation.RData
Untracked: output/first_second_survival_peak_chr.xlsx
Untracked: output/hsq_1_first_batch_herit_qtl2.RData
Untracked: output/hsq_2_second_batch_herit_qtl2.RData
Untracked: output/morphine_fentanyl_survival_time.pdf
Untracked: output/old_temp/
Untracked: output/out_1_operm.RData
Untracked: output/pr_69kchr_combined.RData
Untracked: output/pr_69kchr_second_set.RData
Untracked: output/qtl.morphine.69k.out.combined.RData
Untracked: output/qtl.morphine.69k.out.combined_m2.RData
Untracked: output/qtl.morphine.69k.out.second_set.RData
Untracked: output/qtl.morphine.operm.RData
Untracked: output/qtl.morphine.out.RData
Untracked: output/qtl.morphine.out.combined_gm.RData
Untracked: output/qtl.morphine.out.combined_gm.female.RData
Untracked: output/qtl.morphine.out.combined_gm.male.RData
Untracked: output/qtl.morphine.out.combined_weight_DOB.RData
Untracked: output/qtl.morphine.out.combined_weight_age.RData
Untracked: output/qtl.morphine.out.female.RData
Untracked: output/qtl.morphine.out.male.RData
Untracked: output/qtl.morphine.out.second_set.RData
Untracked: output/qtl.morphine.out.second_set.female.RData
Untracked: output/qtl.morphine.out.second_set.male.RData
Untracked: output/qtl.morphine.out.second_set.weight_DOB.RData
Untracked: output/qtl.morphine.out.second_set.weight_age.RData
Untracked: output/qtl.morphine.out.weight_DOB.RData
Untracked: output/qtl.morphine.out.weight_age.RData
Untracked: output/qtl.morphine1.snpout.RData
Untracked: output/qtl.morphine2.snpout.RData
Untracked: output/second_batch_pheno.csv
Untracked: output/second_batch_variation.RData
Untracked: output/second_set_apr_69kchr_k_loco.rds
Untracked: output/second_set_gm.RData
Untracked: output/second_set_gm.k_loco.rds
Untracked: output/second_set_gm.probs_36state.rds
Untracked: output/second_set_gm.probs_8state.rds
Untracked: output/topSNP_chr5_mindepression.csv
Untracked: output/zoompeak_Min.depression_9.pdf
Untracked: output/zoompeak_Recovery.Time_16.pdf
Untracked: output/zoompeak_Status_bin_11.pdf
Untracked: output/zoompeak_Survival.Time_1.pdf
Untracked: output/zoompeak_fentanyl_Survival.Time_2.pdf
Untracked: sra-tools_v2.10.7.sif
Unstaged changes:
Modified: .gitignore
Modified: _workflowr.yml
Modified: analysis/marker_violin.Rmd
Modified: output/CC_SARS_Chr16_QTL_interval.pdf
Modified: output/CC_SARS_Chr16_plotGeno.pdf
Modified: output/CC_SARS_Chr16_plotGeno.png
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/metabolomics_mouse_fecal.Rmd) and HTML (docs/metabolomics_mouse_fecal.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 8566571 | xhyuo | 2023-07-18 | qtl snp/gene plot |
| html | ff42a20 | xhyuo | 2023-07-17 | Build site. |
| Rmd | 5ab28b1 | xhyuo | 2023-07-17 | qtl plot |
| html | 44a7ef8 | xhyuo | 2023-07-12 | Build site. |
| Rmd | 0fe14ed | xhyuo | 2023-07-12 | qtl |
| html | 7473070 | xhyuo | 2023-07-03 | Build site. |
| Rmd | e84fb30 | xhyuo | 2023-07-03 | h2_se |
| html | 75ea822 | xhyuo | 2023-06-25 | Build site. |
| Rmd | f2c07ec | xhyuo | 2023-06-25 | h2 |
Last update: 2023-07-18
Loading libraries
library(ggplot2)
library(gridExtra)
library(GGally)
library(parallel)
library(qtl)
library(parallel)
library(survival)
library(regress)
library(abind)
library(tidyverse)
library(broman)
library(qtl2)
library(qtlcharts)
library(DT)
library(biomaRt)
rz.transform <- function(y) {
rankY=rank(y, ties.method="average", na.last="keep")
rzT=qnorm(rankY/(length(na.exclude(rankY))+1))
return(rzT)
}
# estimated SE of estimated heritability
# Visscher & Goddard (2015), https://doi.org/10.1534/genetics.114.171017
#
# lambda_i = eigenvalues of kinship matrix
# a = sum[ (lam_i-1)^2 / (1 + h^2(lam_i - 1))^2 ]
# b = sum[ (lam_i - 1) / (1 + h^2(lam_i - 1)) ]
# N = sample size
#
# var(h_hat^2) = 2 / (a - b^2/N)
se_herit <-
function(pheno, kinship)
{
h2 <- as.numeric(est_herit(pheno, kinship))
d <- decomp_kinship(kinship)
lam <- d$values
a <- sapply(h2, function(hsq) sum( (lam-1)^2 / (1 + hsq*(lam-1))^2 ) )
b <- sapply(h2, function(hsq) sum( (lam-1) / (1 + hsq*(lam-1)) ) )
n <- length(lam)
setNames( sqrt(2 / (a - b^2/n)), colnames(pheno) )
}Heritability and QTL
# Read phenotype data -----------------------------------------------------
sampleinfo <- readxl::read_excel(path = "data/metabolomics_mouse_fecal/Untargeted metabolomics_74 Mouse Fecal_YanjiaoZhou_06122023.xlsx", sheet = 1,
range = "A5:C79") %>%
dplyr::rename(., lab_ID = 1) %>%
dplyr::filter(!(lab_ID %in% c(11, 67))) %>%
dplyr::mutate(lab_ID = as.character(lab_ID))
#first dataset
rawdata1 = readxl::read_excel(path = "data/metabolomics_mouse_fecal/Untargeted metabolomics_74 Mouse Fecal_YanjiaoZhou_06122023.xlsx", sheet = 2) %>%
dplyr::mutate(Index = paste0("Dat1_", as.character(1:nrow(.))), .before = 1) %>%
dplyr::select(1:3, Fecal_1:Fecal_74)
#second dataset
rawdata2 = readxl::read_excel(path = "data/metabolomics_mouse_fecal/Bile acids_74 Fecal_YanjiaoZhou_07072023.xlsx", sheet = 2) %>%
dplyr::rename(CV = 2, Name = Sample) %>%
dplyr::filter(!is.na(CV)) %>%
dplyr::mutate(Index = paste0("Dat2_", as.character(1:nrow(.))), .before = 1) %>%
dplyr::select(1:2, starts_with("Fecal_")) %>%
dplyr::mutate(Formula = "", .after = Name)
#rbind two dataset
all.equal(colnames(rawdata1), colnames(rawdata2))
# [1] TRUE
rawdata <- rbind(rawdata1, rawdata2)
#make it longer format by t()
rawdata_long <- rawdata %>%
dplyr::select(-1:-3) %>%
t() %>%
as.data.frame() %>%
rownames_to_column(var = "lab_ID") %>%
dplyr::mutate(lab_ID = str_remove_all(lab_ID, "Fecal_")) %>%
dplyr::mutate(across(V1:last_col(), rz.transform))
colnames(rawdata_long)[-1] = rawdata$Index
#left_join
do.pheno <- left_join(sampleinfo[, 1:2], rawdata_long) %>%
as.data.frame()
# Read genotype data -----------------------------------------------------------
load("/projects/csna/csna_workflow/data/Jackson_Lab_12_batches/gm_DO3173_qc_newid.RData")#gm_after_qc
overlap.id = intersect(ind_ids(gm_after_qc), as.character(do.pheno$ID))
#subset
gm = gm_after_qc[overlap.id, ]
do.pheno = do.pheno[do.pheno$ID %in% overlap.id, ]
do.pheno = do.pheno[match(ind_ids(gm), do.pheno$ID) , ]
#add sex into do.pheno
load("/projects/csna/csna_workflow/data/Jackson_Lab_12_batches/qc_info.RData")
do.pheno <- left_join(do.pheno, qc_info[, c(1,3)], by = c("ID" = "name")) %>%
dplyr::relocate(sex, .after = "ID")
rownames(do.pheno) = do.pheno$ID
all.equal(ind_ids(gm), do.pheno$ID)
# [1] TRUE
if(FALSE){
#genotype probability for 68 subjects
pr = calc_genoprob(gm, cores = 20, quiet = FALSE)
saveRDS(pr, file = "data/metabolomics_mouse_fecal/pr.rds")
apr <- genoprob_to_alleleprob(pr, cores = 20)
saveRDS(apr, file = "data/metabolomics_mouse_fecal/apr.rds")
#Calculating a kinship matrix
k <- calc_kinship(pr, "overall", use_allele_probs = FALSE, quiet = TRUE, cores = 20)
saveRDS(k, file = "data/metabolomics_mouse_fecal/k.rds")
k_loco <- calc_kinship(apr, "loco", cores = 20)
saveRDS(k_loco, file = "data/metabolomics_mouse_fecal/k_loco.rds")
#addcovar
options(na.action='na.pass')
addcovar = model.matrix(~sex, data = do.pheno[, c("sex"), drop=F])[,-1, drop=F]
colnames(addcovar) <- c("sex")
rownames(addcovar) <- rownames(do.pheno)
#herit
hsq_se <- map_dfr(colnames(do.pheno)[-1:-3], function(x){
phe = do.pheno[,x]
names(phe) <- do.pheno$ID
h2 = est_herit(pheno = phe,
kinship = k,
addcovar = addcovar,
cores = 20)
d <- decomp_kinship(k)
lam <- d$values
a <- sapply(h2, function(hsq) sum( (lam-1)^2 / (1 + hsq*(lam-1))^2 ) )
b <- sapply(h2, function(hsq) sum( (lam-1) / (1 + hsq*(lam-1)) ) )
n <- length(lam)
se <- sqrt(2 / (a - b^2/n))
data.frame(h2 = h2, se = se)
})
hsq.df <- data.frame(Index = colnames(do.pheno)[-1:-3],
hsq_se) %>%
dplyr::left_join(rawdata[, 1:3])
save(hsq.df, file = "data/metabolomics_mouse_fecal/hsq.out")
#qtl
out <- scan1(genoprobs = apr,
pheno = do.pheno[, -1:-3],
addcovar = addcovar,
kinship = k_loco,
cores = 20)
save(out, file = "data/metabolomics_mouse_fecal/qtl.out")
#permutation
operm <- scan1perm(genoprobs = apr,
pheno = do.pheno[, 4, drop=F],
addcovar = addcovar,
kinship = k_loco,
cores = 20,
n_perm = 1000)
save(operm, file = "data/metabolomics_mouse_fecal/operm.out")
#5% significance thresholds
cutoff = summary(operm, alpha=c(0.05))
cutoff
# LOD thresholds (1000 permutations)
# Dat1_1
# 0.05 8.25
#peaks
peaks <- find_peaks(out, gm$gmap, threshold = cutoff, drop = 1.5)
save(peaks, file = "data/metabolomics_mouse_fecal/peaks.out")
}
load("data/metabolomics_mouse_fecal/hsq.out")
load("data/metabolomics_mouse_fecal/peaks.out")
#histogram raw data
for(i in c(233, 460, 3408, 710, 3806, 3805)){
#histogram
dat = data.frame(pheno = t(rawdata[i, 4:77]))
p <- ggplot(data=dat, aes(dat$pheno)) +
geom_histogram() +
ylab("Number of DO mice") + xlab(paste0("raw ", rawdata[rawdata$Index ==paste0("Dat1_", i), "Name"]," ",
rawdata[rawdata$Index ==paste0("Dat1_", i), "Formula"]))
print(p)
}
# Warning: Use of `dat$pheno` is discouraged. Use `pheno` instead.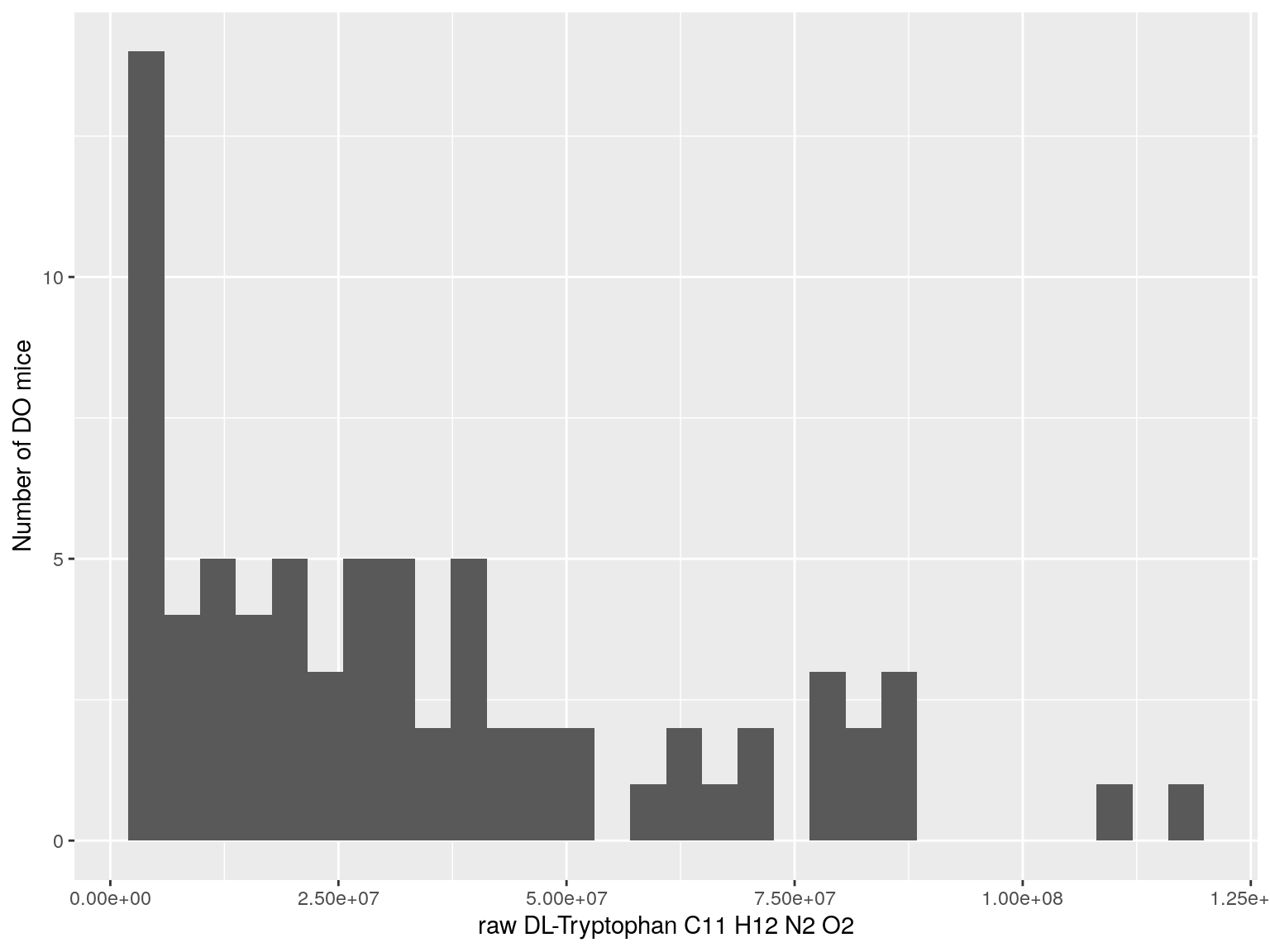
| Version | Author | Date |
|---|---|---|
| 44a7ef8 | xhyuo | 2023-07-12 |
# Warning: Use of `dat$pheno` is discouraged. Use `pheno` instead.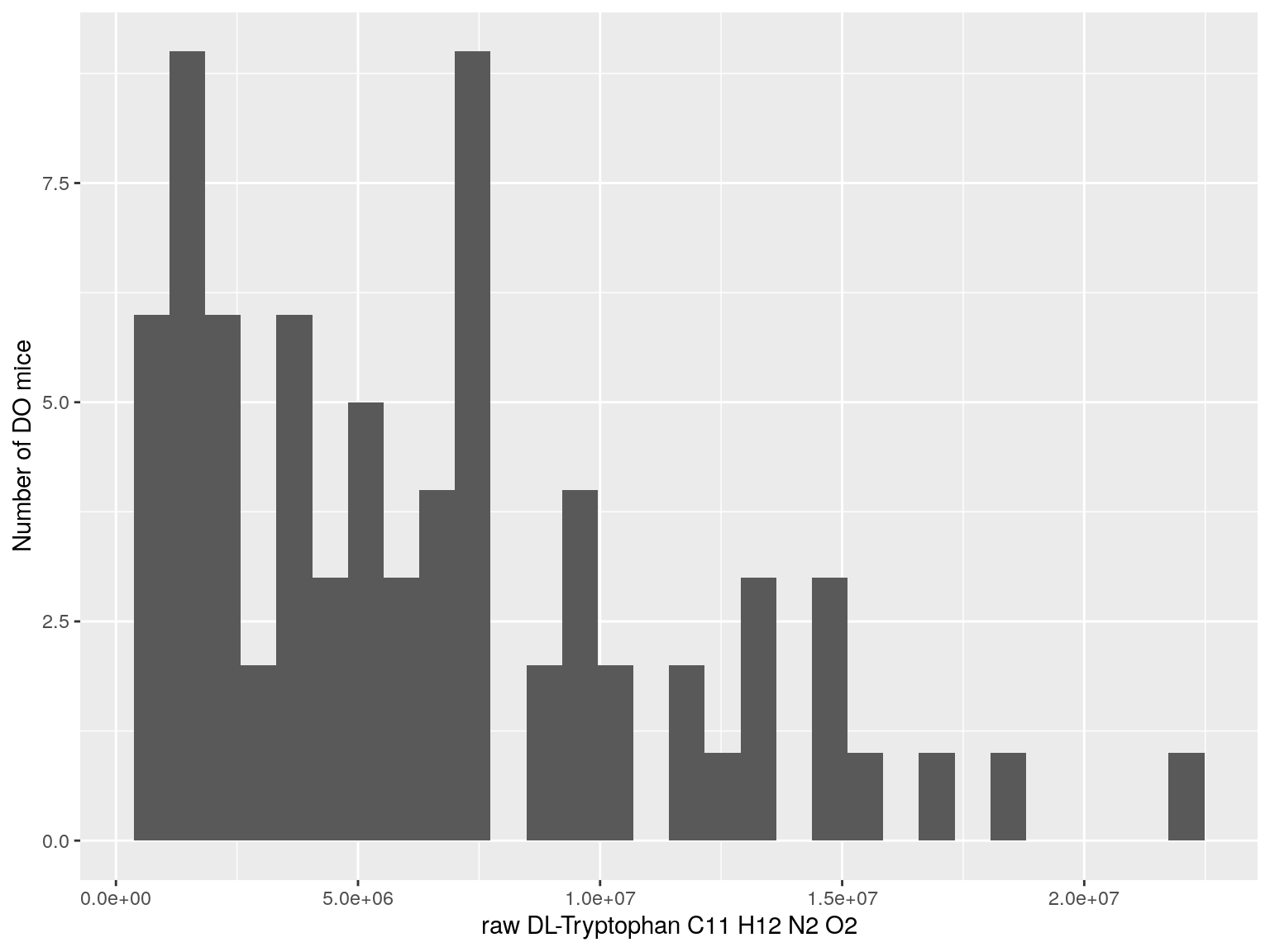
| Version | Author | Date |
|---|---|---|
| 44a7ef8 | xhyuo | 2023-07-12 |
# Warning: Use of `dat$pheno` is discouraged. Use `pheno` instead.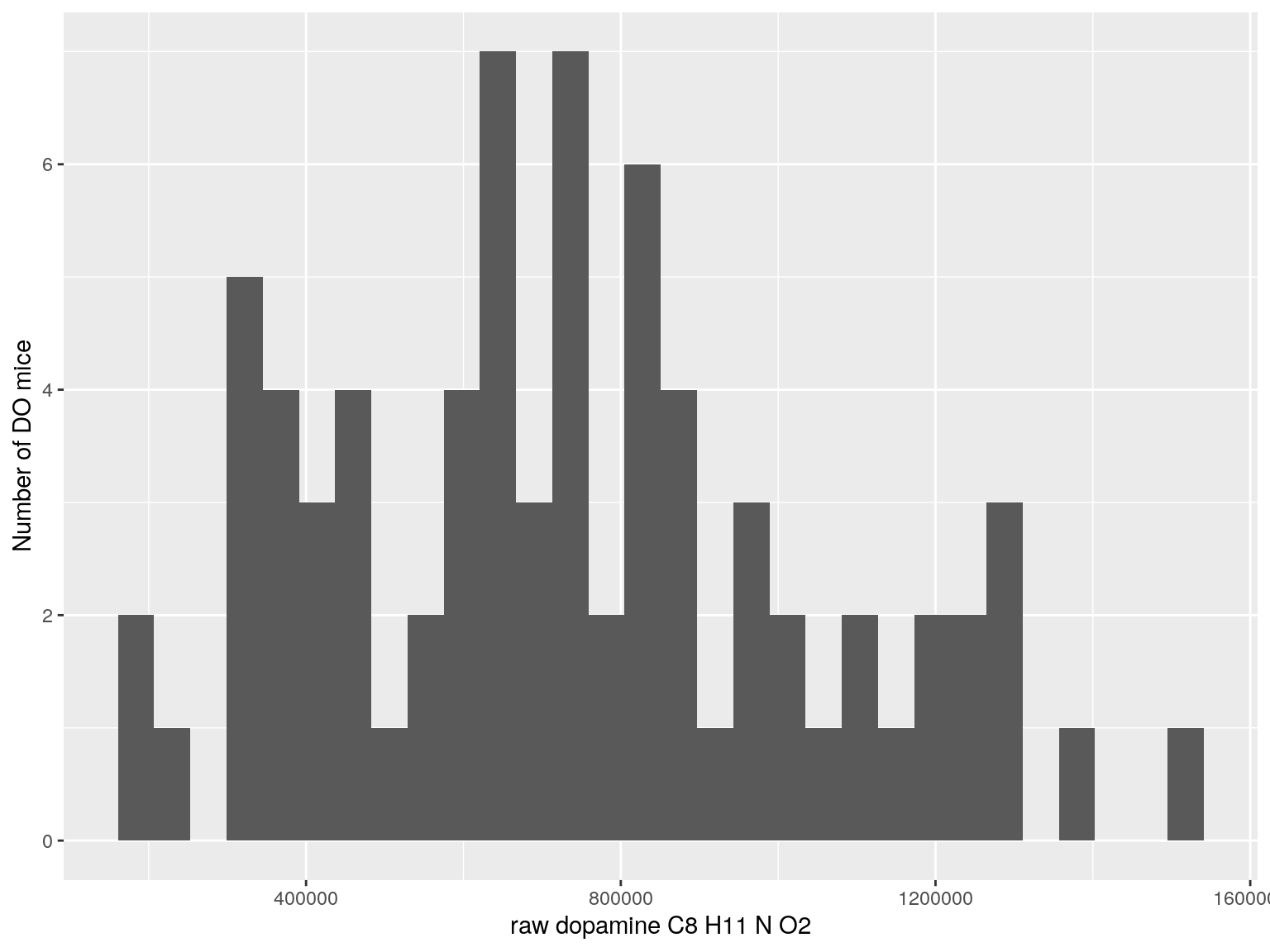
| Version | Author | Date |
|---|---|---|
| 44a7ef8 | xhyuo | 2023-07-12 |
# Warning: Use of `dat$pheno` is discouraged. Use `pheno` instead.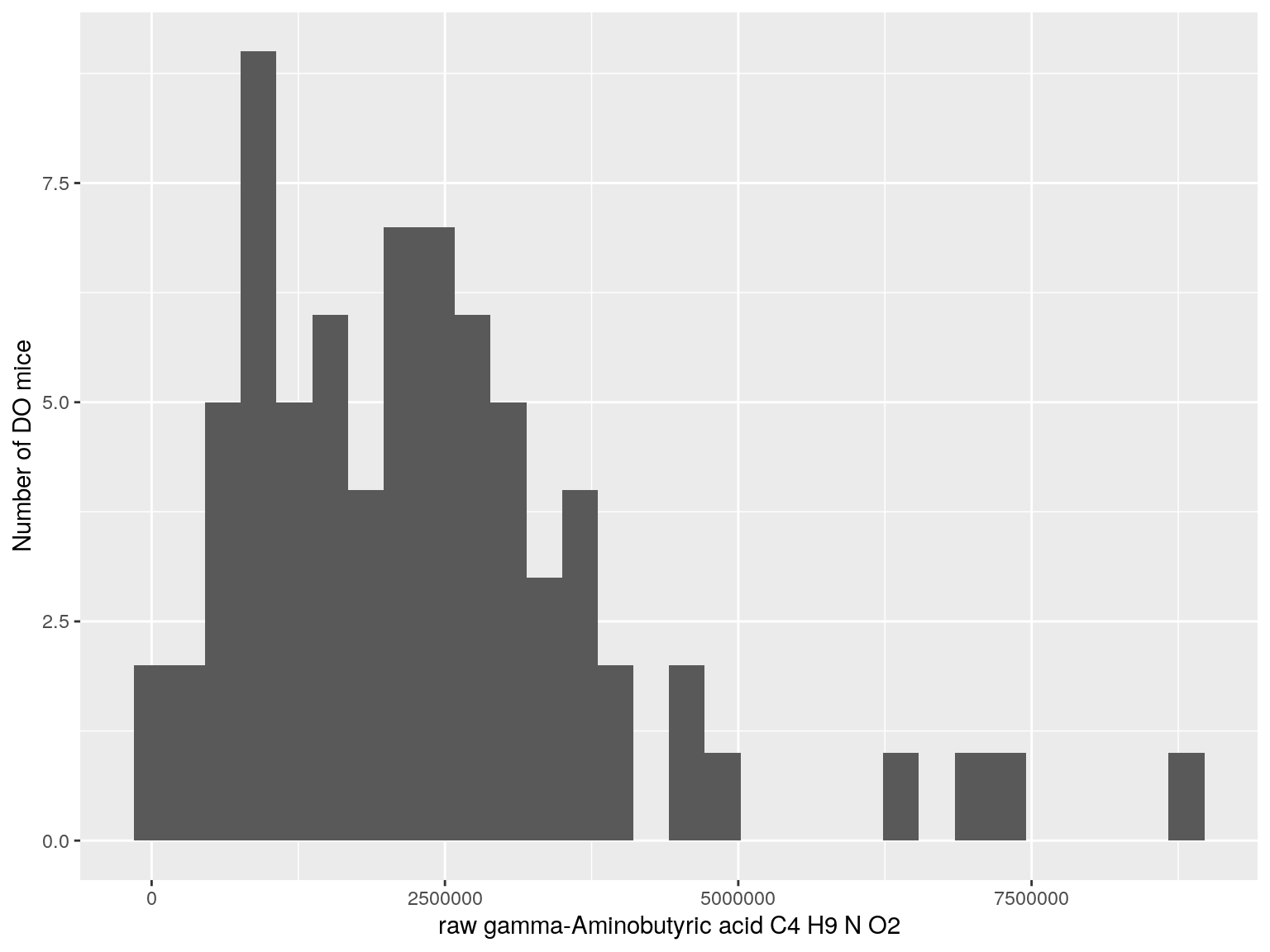
| Version | Author | Date |
|---|---|---|
| 44a7ef8 | xhyuo | 2023-07-12 |
# Warning: Use of `dat$pheno` is discouraged. Use `pheno` instead.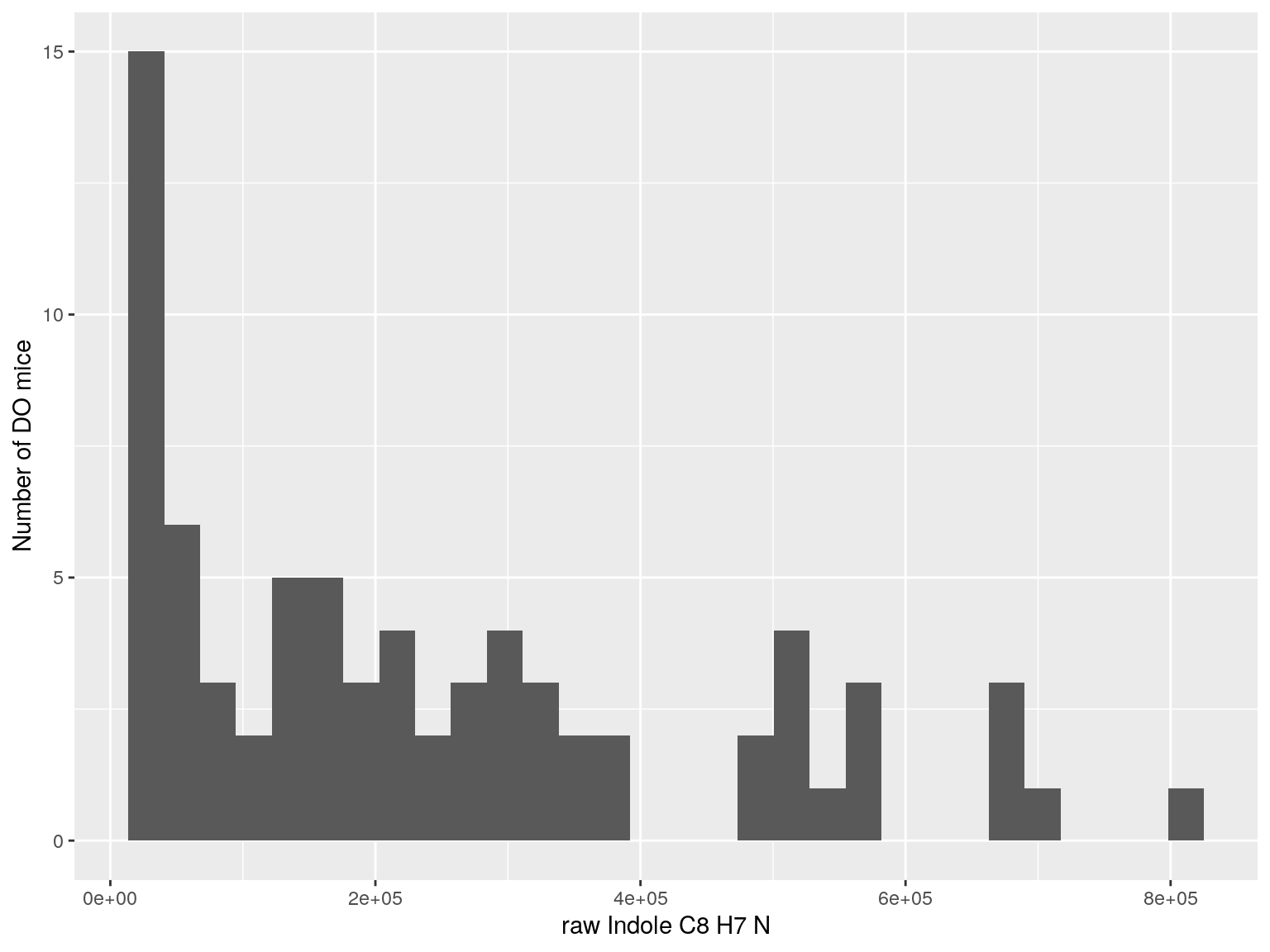
| Version | Author | Date |
|---|---|---|
| 44a7ef8 | xhyuo | 2023-07-12 |
# Warning: Use of `dat$pheno` is discouraged. Use `pheno` instead.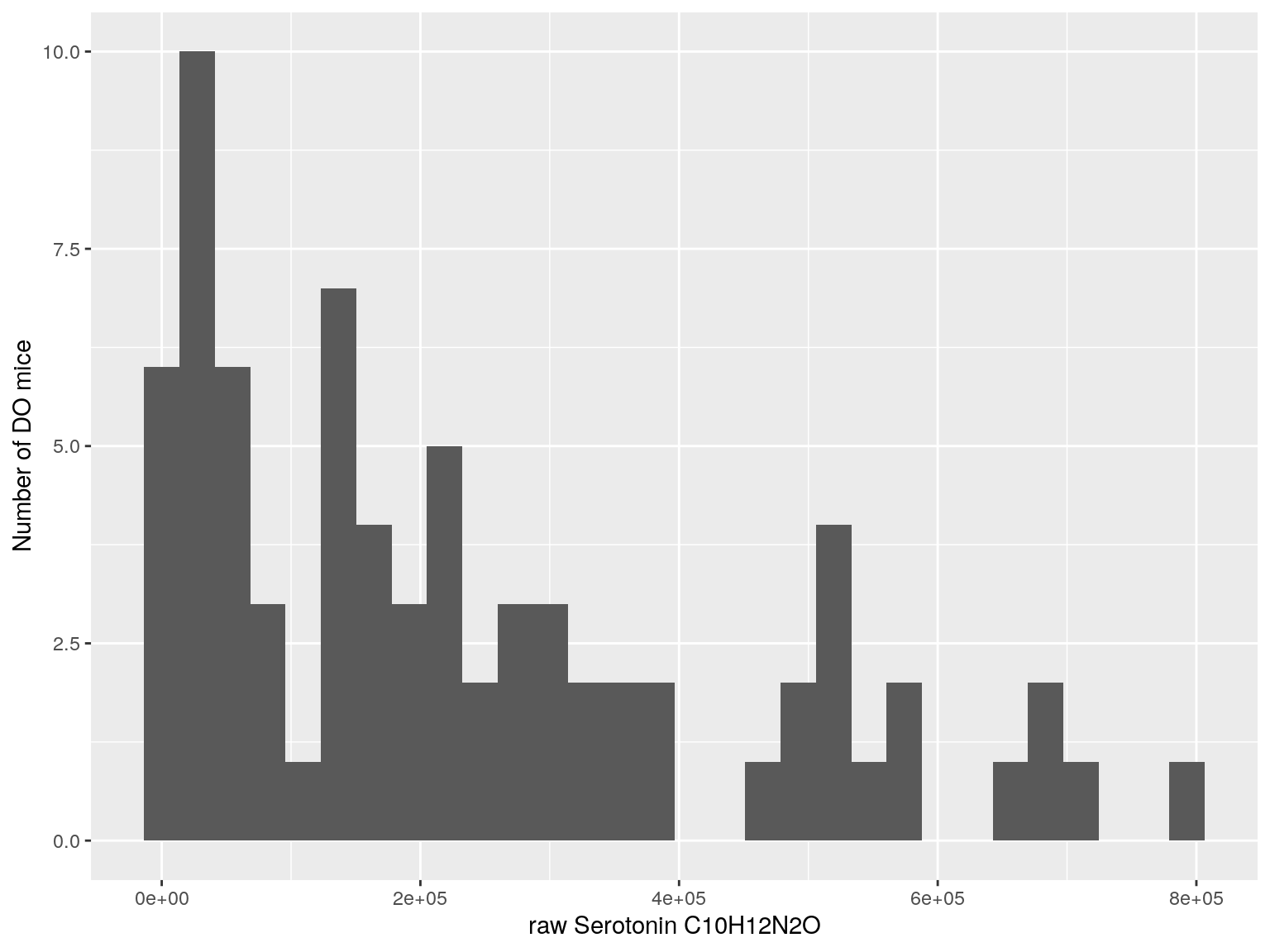
| Version | Author | Date |
|---|---|---|
| 44a7ef8 | xhyuo | 2023-07-12 |
#histogram rankZ data
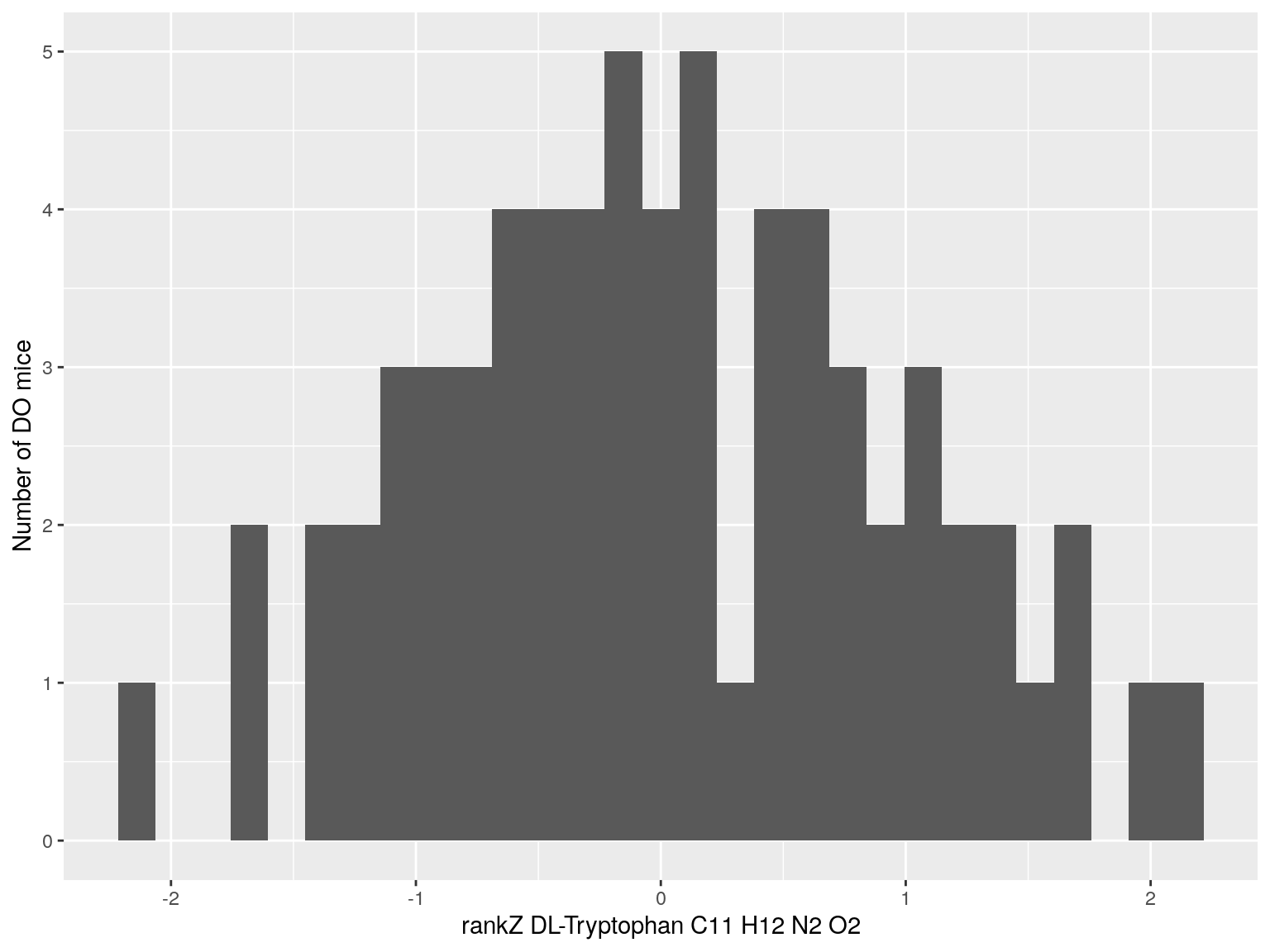
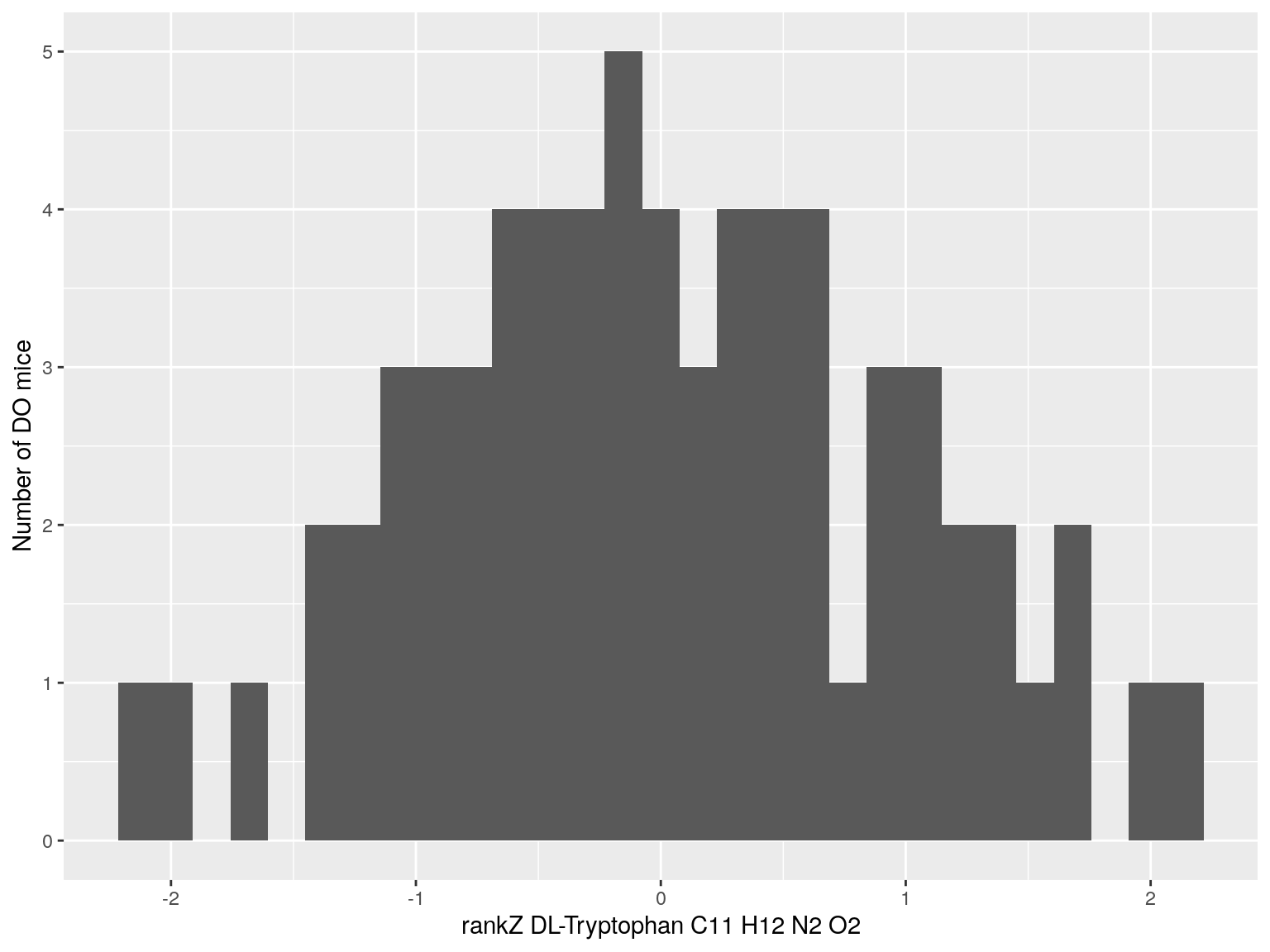
for(i in c(233, 460, 3408, 710, 3806, 3805)){
p <- ggplot(data=do.pheno, aes(do.pheno[, paste0("Dat1_", i)])) +
geom_histogram() +
ylab("Number of DO mice") + xlab(paste0("rankZ ", hsq.df[hsq.df$Index ==paste0("Dat1_", i), "Name"]," ",
hsq.df[hsq.df$Index ==paste0("Dat1_", i), "Formula"]))
print(p)
}
| Version | Author | Date |
|---|---|---|
| 44a7ef8 | xhyuo | 2023-07-12 |
| Version | Author | Date |
|---|---|---|
| 44a7ef8 | xhyuo | 2023-07-12 |
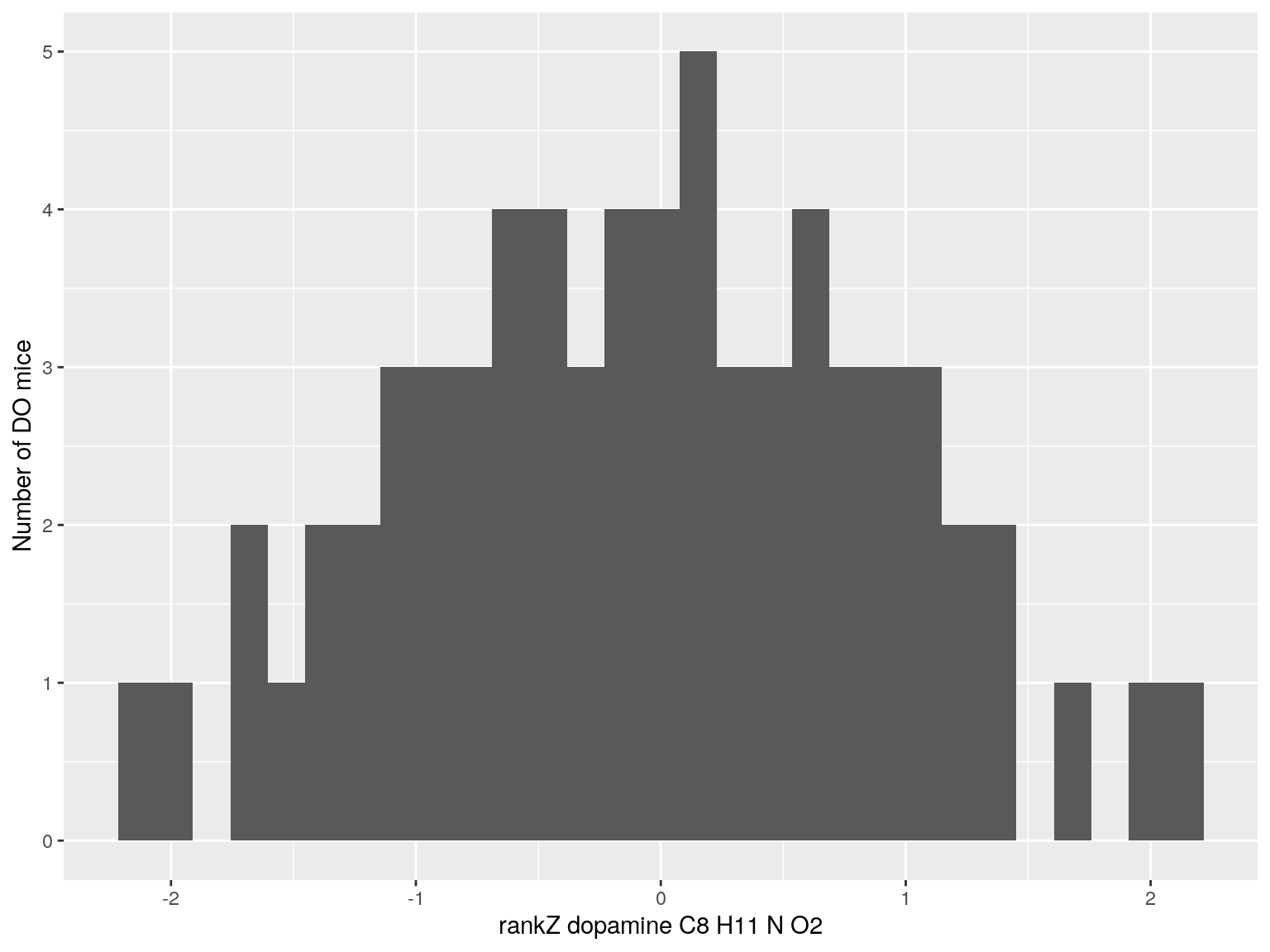
| Version | Author | Date |
|---|---|---|
| 44a7ef8 | xhyuo | 2023-07-12 |
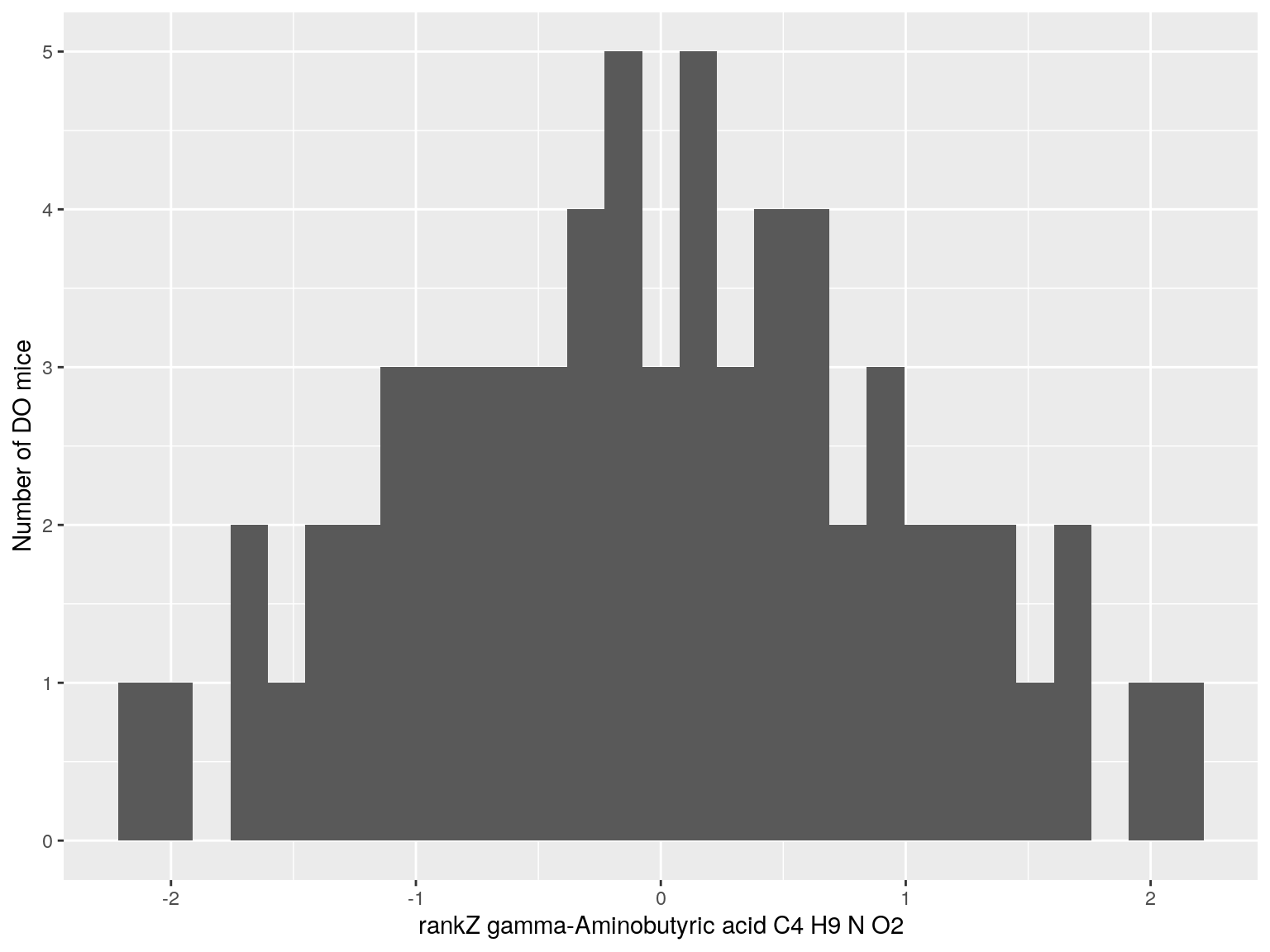
| Version | Author | Date |
|---|---|---|
| 44a7ef8 | xhyuo | 2023-07-12 |
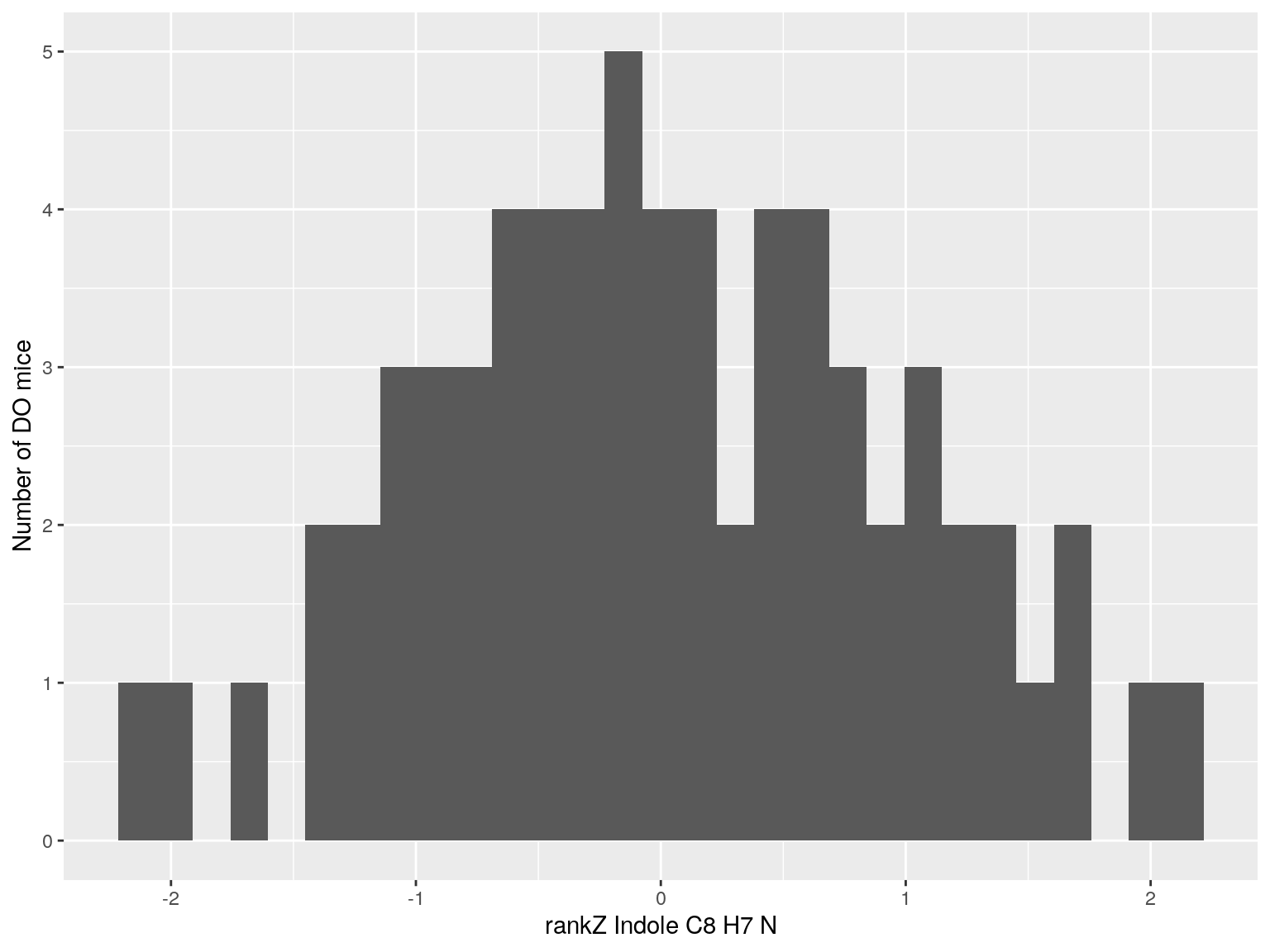
| Version | Author | Date |
|---|---|---|
| 44a7ef8 | xhyuo | 2023-07-12 |
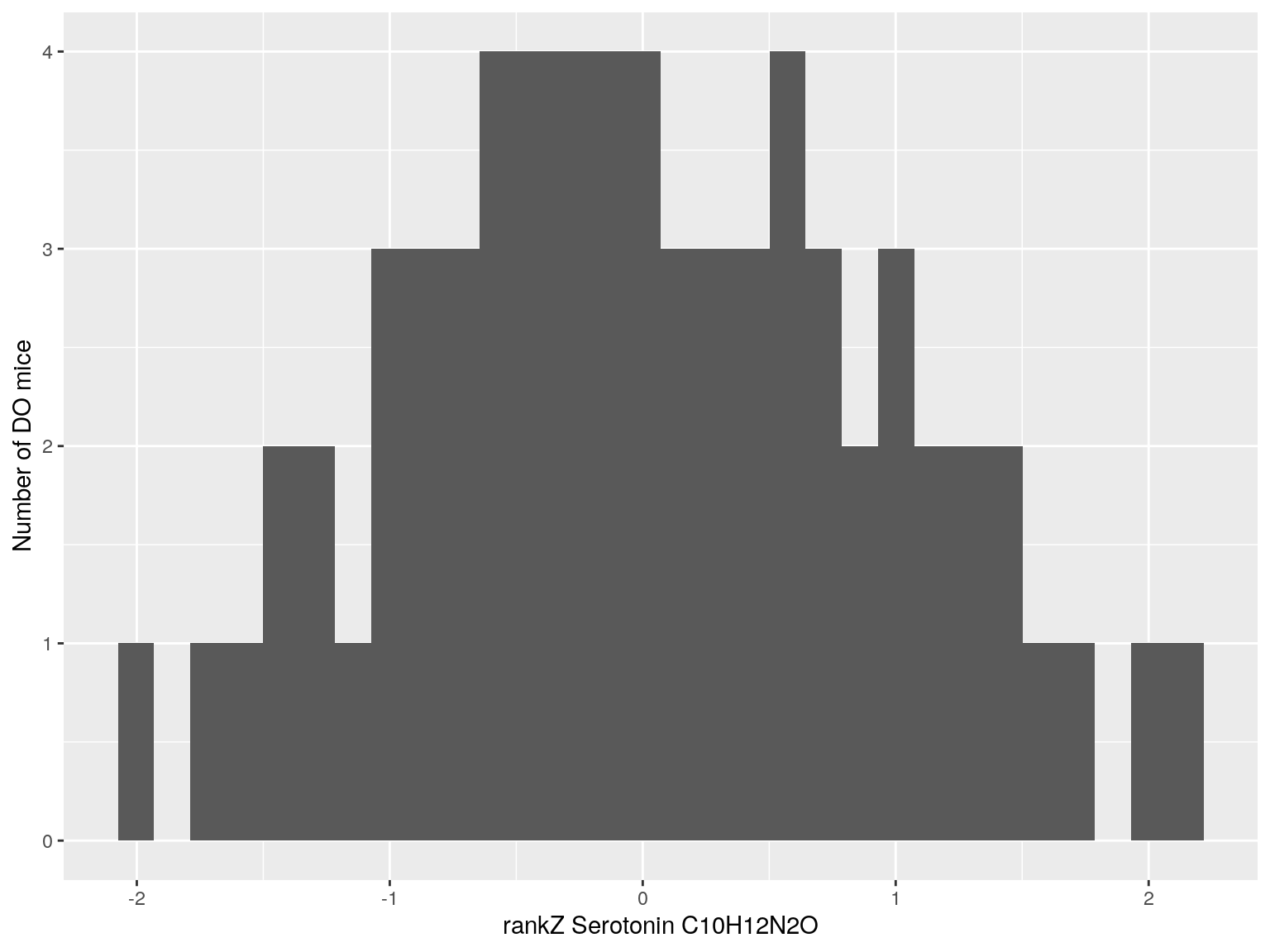
| Version | Author | Date |
|---|---|---|
| 44a7ef8 | xhyuo | 2023-07-12 |
Heritability Results
#display hsq.df
DT::datatable(hsq.df[, c(1,4,5,2,3)],
filter = list(position = 'top', clear = FALSE),
extensions = 'Buttons',
options = list(dom = 'Blfrtip',
buttons = c('csv', 'excel'),
lengthMenu = list(c(10,25,50,-1),
c(10,25,50,"All")),
pageLength = 40,
scrollY = "300px",
scrollX = "40px"),
caption = htmltools::tags$caption(style = 'caption-side: top; text-align: left; color:black; font-size:200% ;','Heritability'))QTL Results
load("data/metabolomics_mouse_fecal/qtl.out")
load("data/metabolomics_mouse_fecal/operm.out")
# overall maximum
max.dt <- data.frame(lodcolumn = colnames(out)) %>%
dplyr::mutate(max.lod = map_dbl(lodcolumn, function(x){
maxlod(out[, x])
})) %>%
dplyr::mutate(pvalue = map_dbl(max.lod, function(x){
sum(operm[,1] > x)/1000
})) %>%
dplyr::mutate(fdr = p.adjust(pvalue, method = "fdr"))
#display QTL peaks
peaks_dat <- left_join(peaks, hsq.df[, c(1,4,5,2,3)], by = c("lodcolumn" = "Index")) %>%
inner_join(max.dt, c("lodcolumn" = "lodcolumn", "lod" = "max.lod")) %>%
dplyr::relocate(pvalue, .after = lod) %>%
dplyr::relocate(fdr, .after = pvalue)
index = peaks_dat[peaks_dat$fdr <= 0.05, "lodcolumn"]
query_variants <- create_variant_query_func("data/cc_variants.sqlite")
query_genes <- create_gene_query_func("data/mouse_genes_mgi.sqlite")
apr <- readRDS("data/metabolomics_mouse_fecal/apr.rds")
k_loco <- readRDS("data/metabolomics_mouse_fecal/k_loco.rds")
#addcovar
options(na.action='na.pass')
addcovar = model.matrix(~sex, data = do.pheno[, c("sex"), drop=F])[,-1, drop=F]
colnames(addcovar) <- c("sex")
rownames(addcovar) <- rownames(do.pheno)
cutoff = 8.25
if(FALSE){
#peaks
#peak coeff and SNP
coef_c1 <- list()
coef_c2 <- list()
out_snps <- list()
out_genes <- list()
for(j in index) {
chr <- find_markerpos(gm, names(which.max(out[,j])))$chr
phe <- do.pheno[, j]
names(phe) <- rownames(do.pheno)
#peak coeff
coef_c1[[j]] <- scan1coef(genoprobs=apr[,chr],
pheno=phe,
kinship=k_loco[[chr]],
addcovar=addcovar)
#peak blup
coef_c2[[j]] <- scan1blup(genoprobs=apr[,chr],
pheno=phe,
kinship=k_loco[[chr]],
addcovar=addcovar,
cores = 20)
#peak SNPs
peak_Mbp <- find_markerpos(gm, names(which.max(out[,j])))$pmap
variants <- query_variants(chr, peak_Mbp - 1, peak_Mbp + 1)
out_snps[[j]] <- scan1snps(genoprobs = apr[,chr],
map = gm$pmap,
pheno = phe,
kinship = k_loco[[chr]],
addcovar = addcovar,
query_func=query_variants,
chr=chr,
start=peak_Mbp - 1,
end=peak_Mbp + 1,
keep_all_snps=TRUE)
out_genes[[j]] <- query_genes(chr, peak_Mbp - 1, peak_Mbp + 1)
}
save(coef_c1, coef_c2,
out_snps, out_genes, file = "data/metabolomics_mouse_fecal/index_out_coef_snp_gene.RData")
}
load("data/metabolomics_mouse_fecal/index_out_coef_snp_gene.RData")
#genome-wide plot
for(j in index){
par(mar=c(5.1, 4.1, 1.1, 1.1))
ymx <- maxlod(out[, j]) # overall maximum LOD score
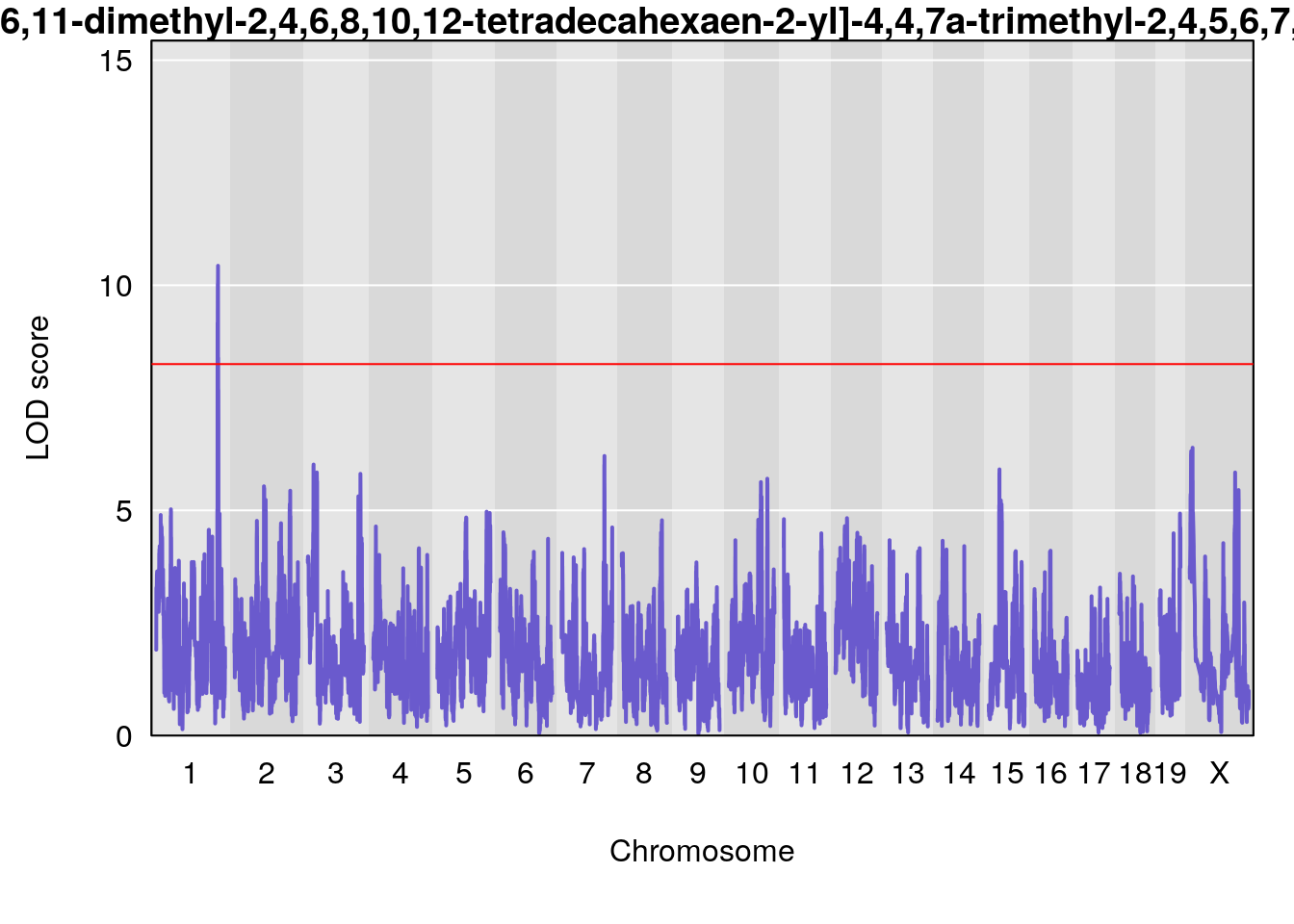
plot(out, map=gm$pmap, lodcolumn=j, col="slateblue", ylim=c(0, ymx+5))
abline(h=8.25, col="red")
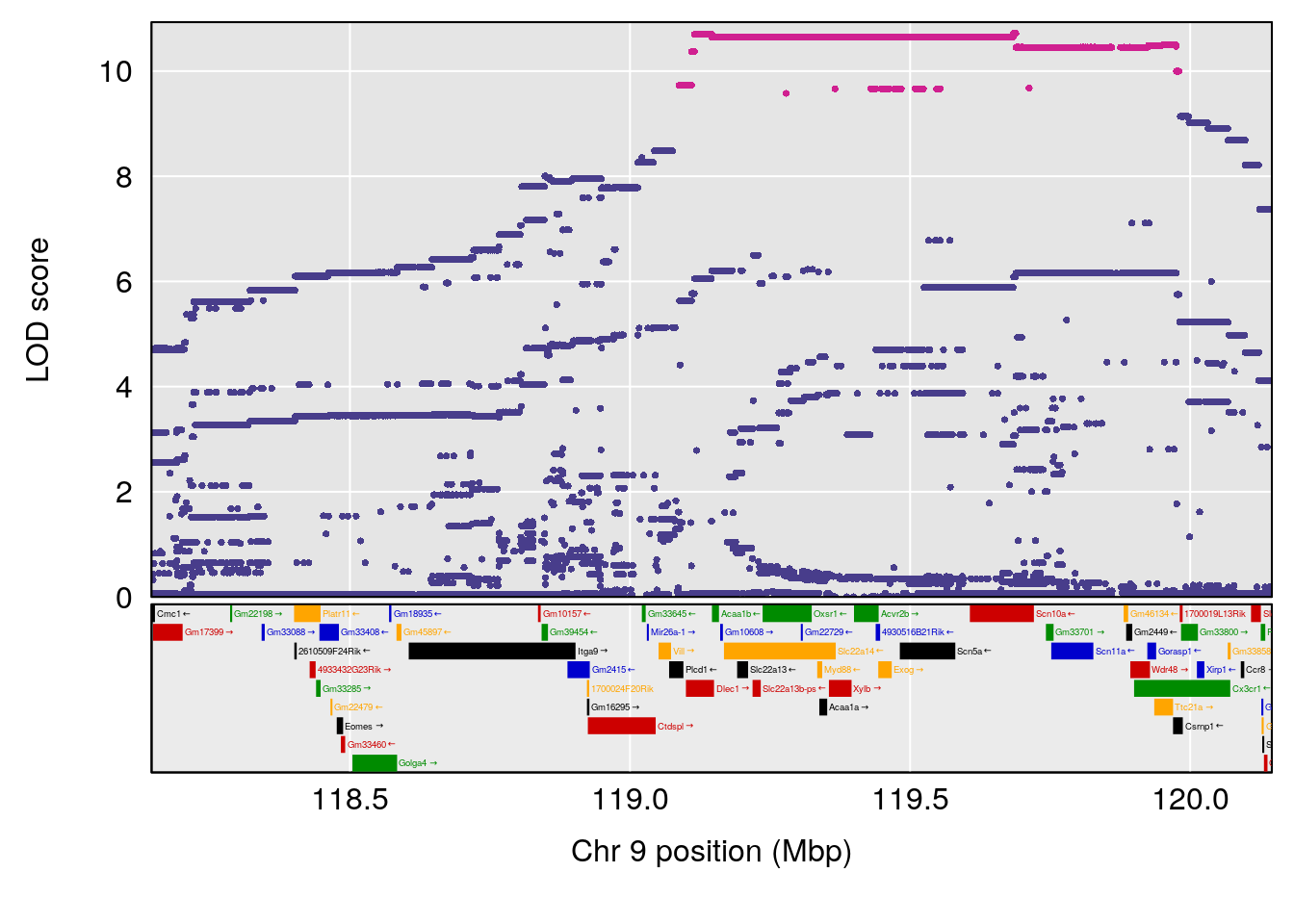
title(main = paste0(peaks_dat[peaks_dat$lodcolumn == j, "Name"],
peaks_dat[peaks_dat$lodcolumn == j, "Formula"]))
print(j)
chr <- find_markerpos(gm, names(which.max(out[,j])))$chr
print(peaks_dat[peaks_dat$lodcolumn == j, ])
#coeff plot
par(mar=c(4.1, 4.1, 0.6, 0.6))
#plot_coefCC(coef_c1[[j]], gm$pmap[chr], scan1_output=subset(out, gm$pmap, lodcolumn=j), bgcolor="gray95", legend=NULL)
plot_coefCC(coef_c2[[j]], gm$pmap[chr], scan1_output=subset(out, gm$pmap, lodcolumn=j), bgcolor="gray95", legend=NULL)
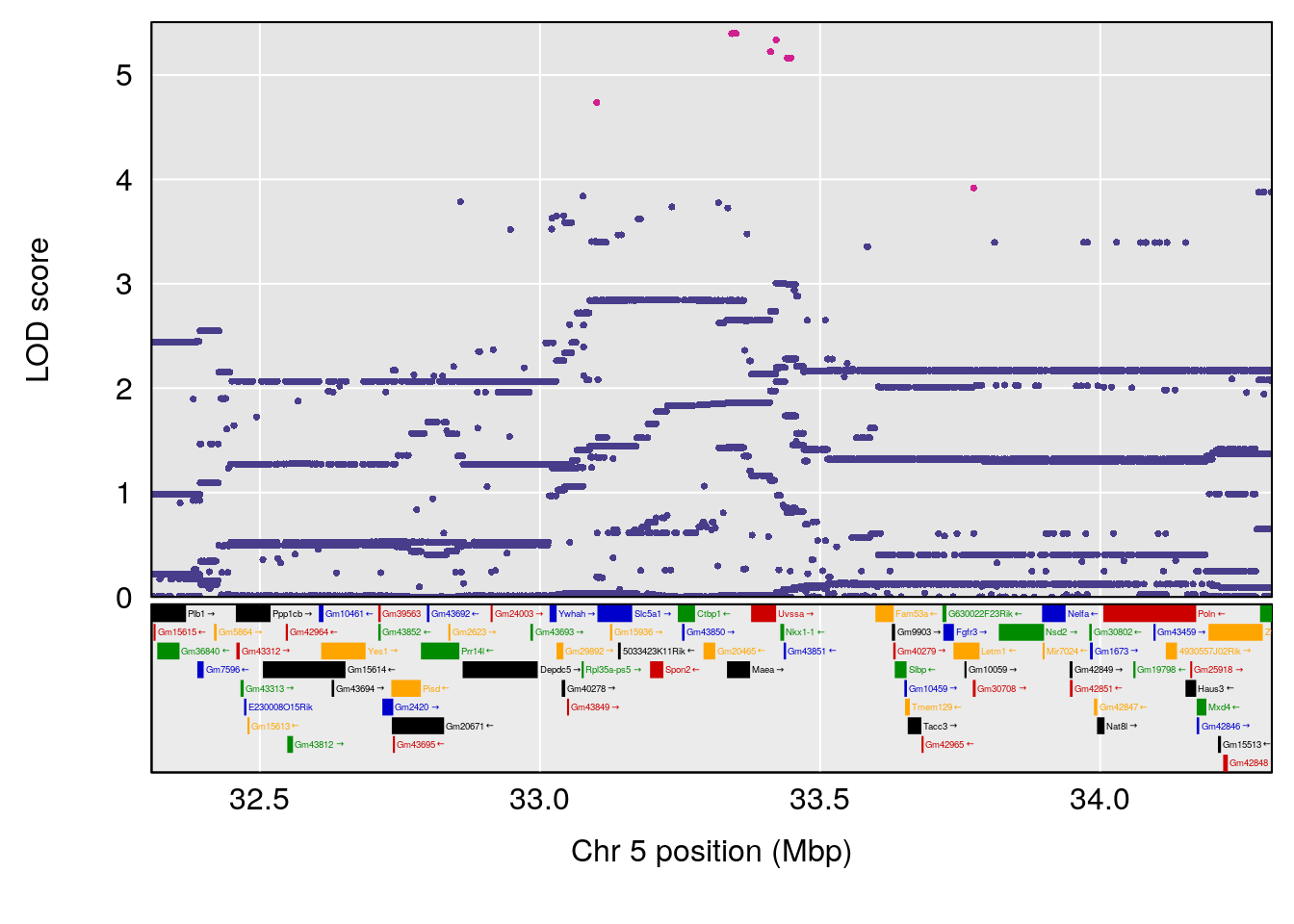
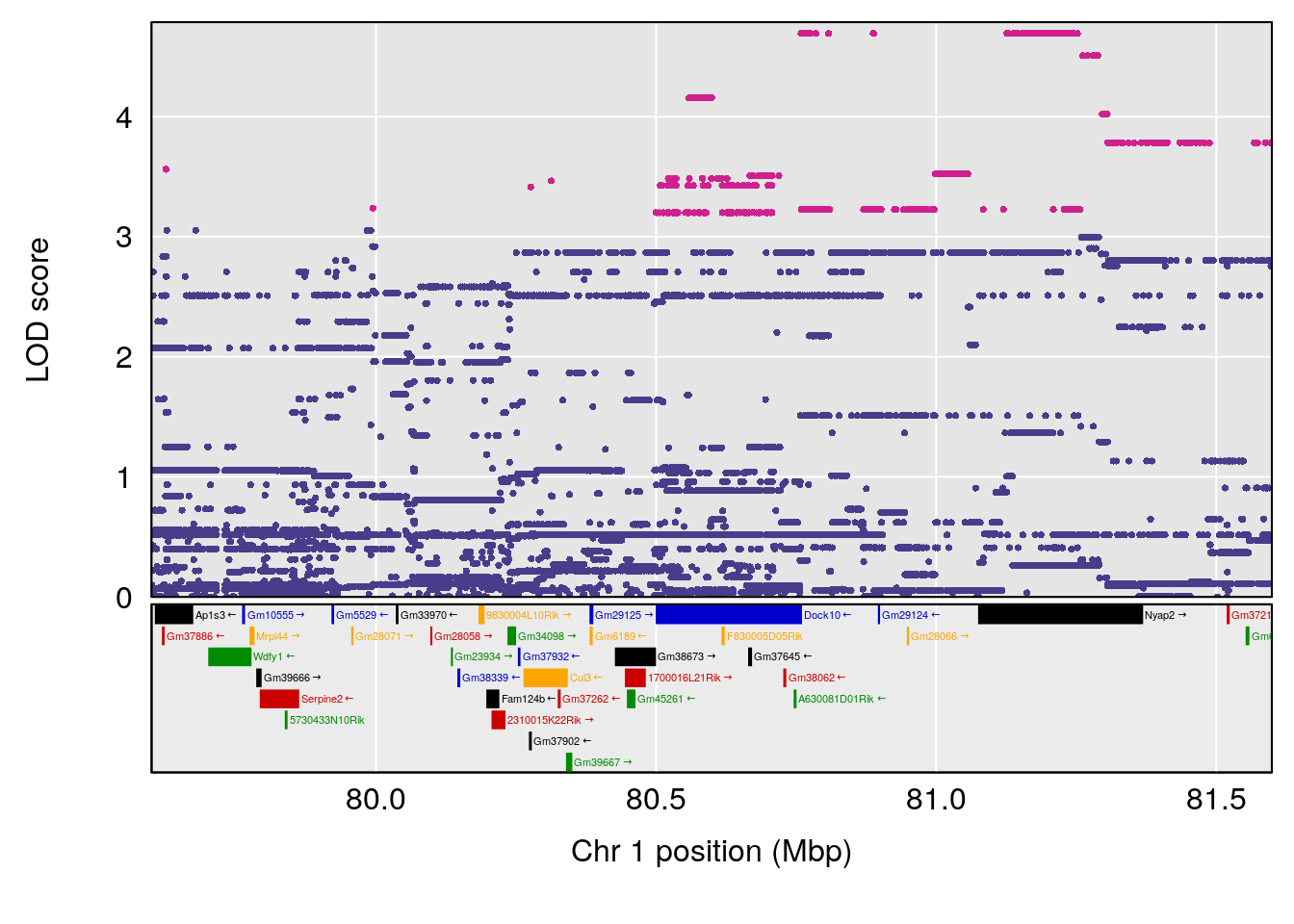
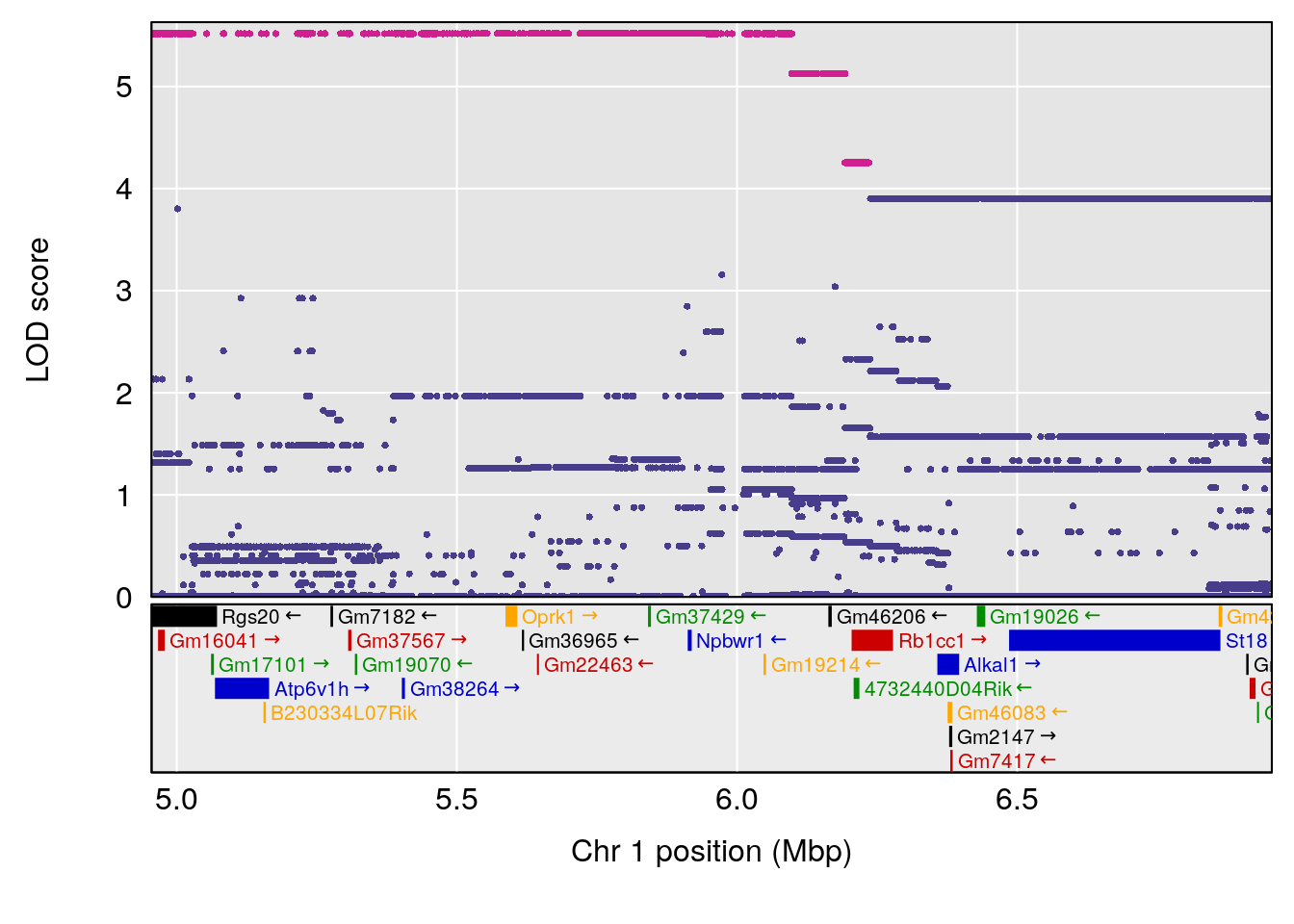
plot(out_snps[[j]]$lod, out_snps[[j]]$snpinfo, drop_hilit=1.5, genes=out_genes[[j]])
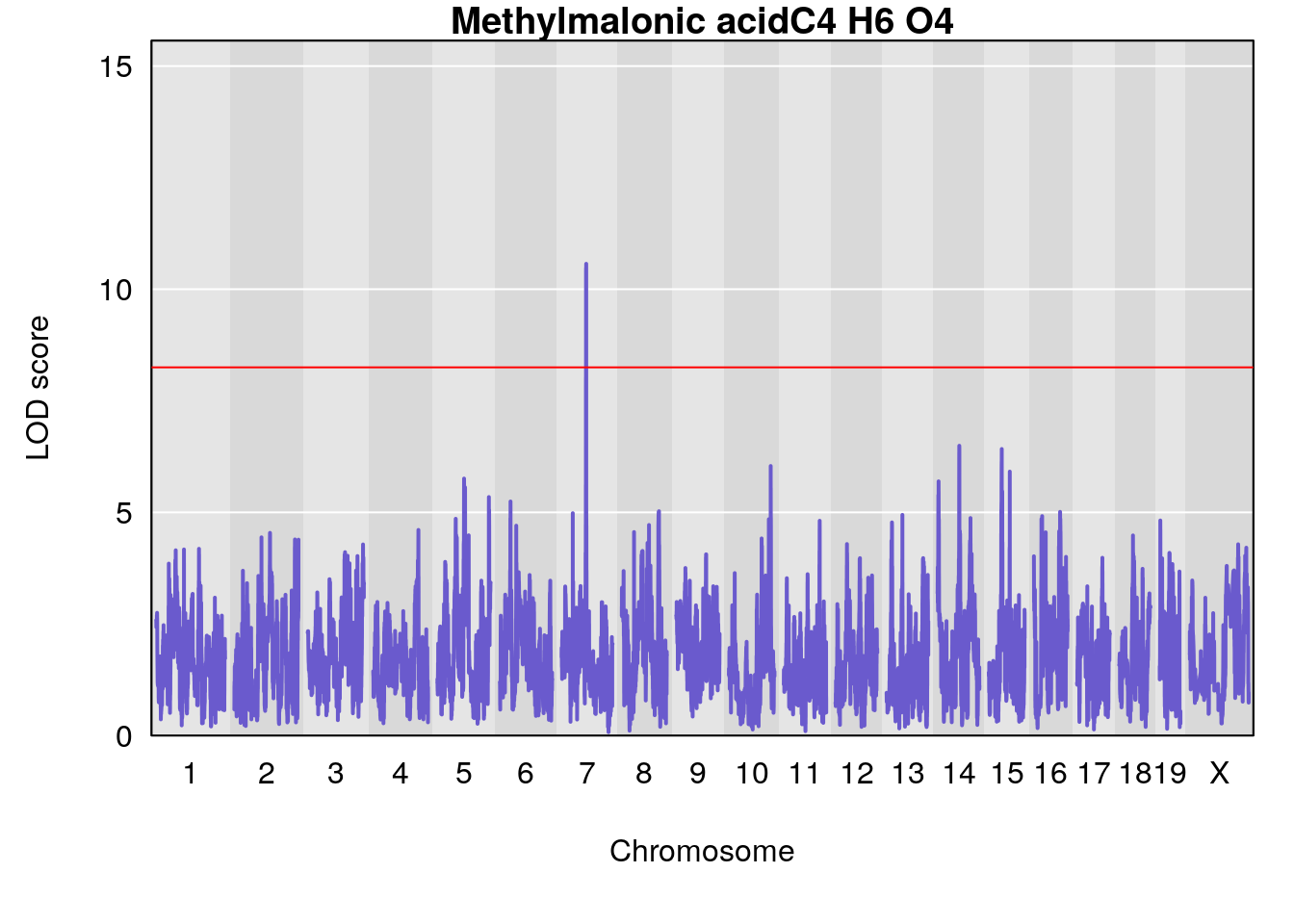
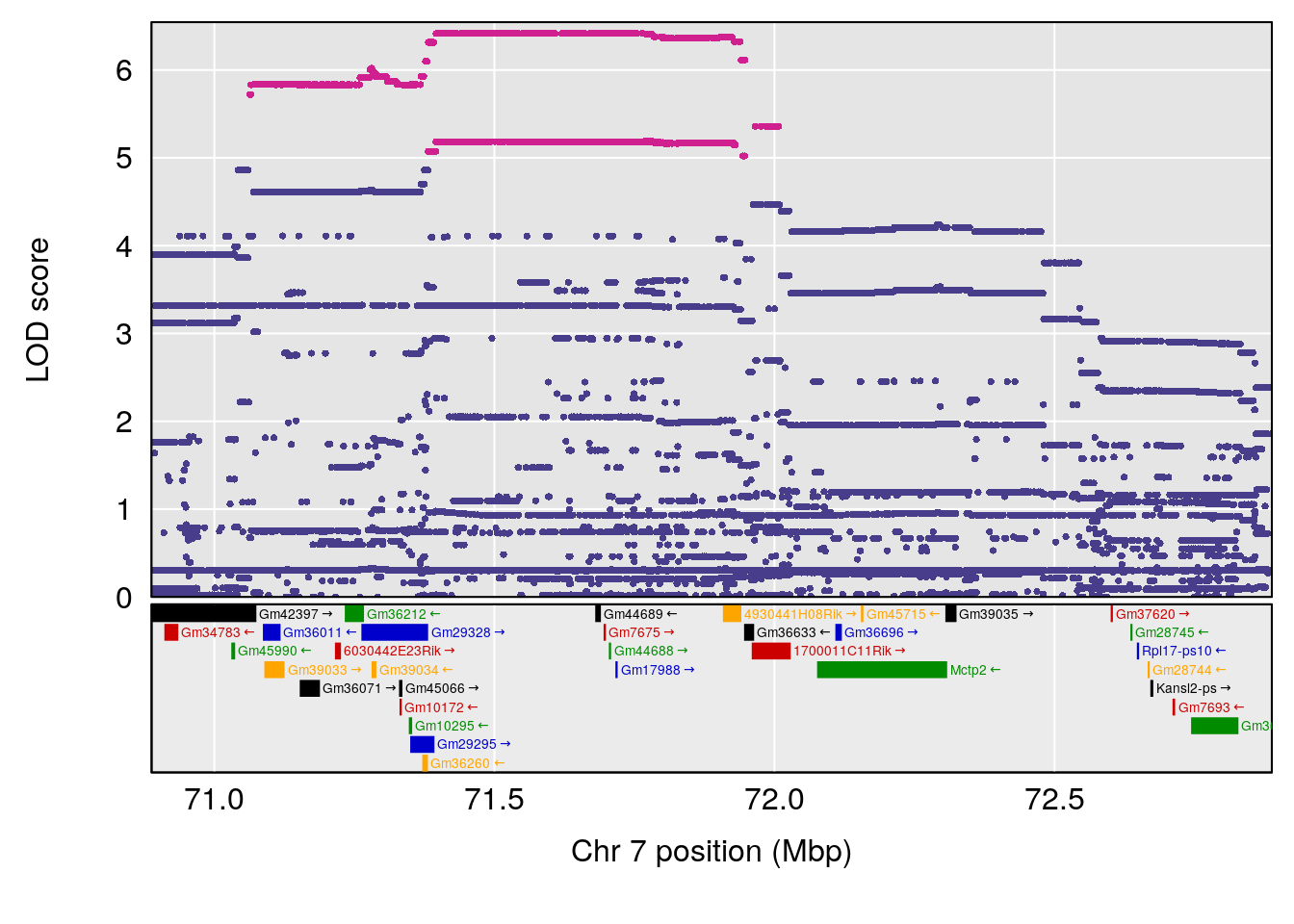
}
| Version | Author | Date |
|---|---|---|
| ff42a20 | xhyuo | 2023-07-17 |
# [1] "Dat1_159"
# lodindex lodcolumn chr pos lod pvalue fdr ci_lo ci_hi
# 13 159 Dat1_159 7 41.146 10.56996 0 0 40.713 41.214
# Name Formula h2 se
# 13 Methylmalonic acid C4 H6 O4 0.07132245 0.478351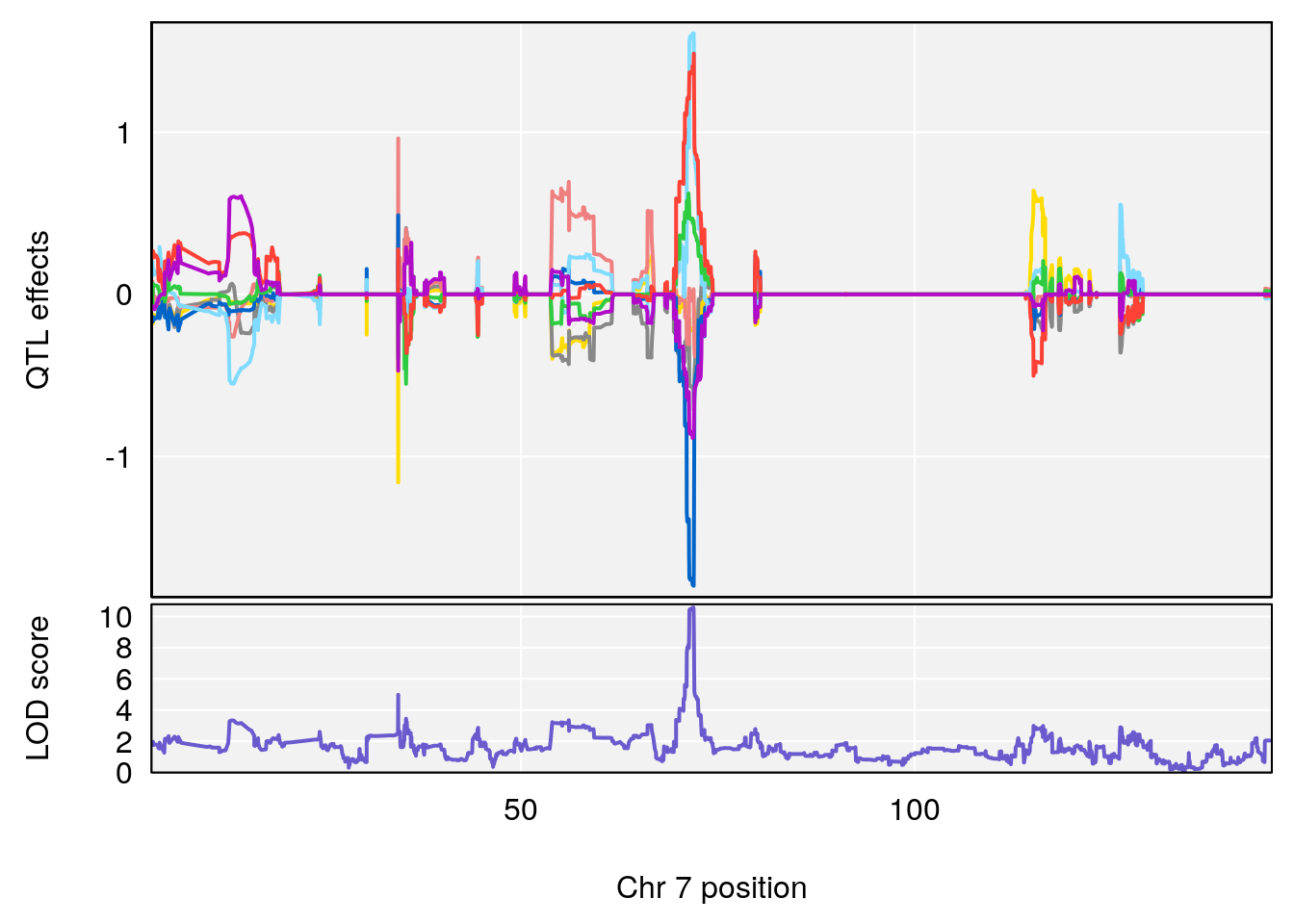
| Version | Author | Date |
|---|---|---|
| ff42a20 | xhyuo | 2023-07-17 |
| Version | Author | Date |
|---|---|---|
| ff42a20 | xhyuo | 2023-07-17 |
| Version | Author | Date |
|---|---|---|
| ff42a20 | xhyuo | 2023-07-17 |
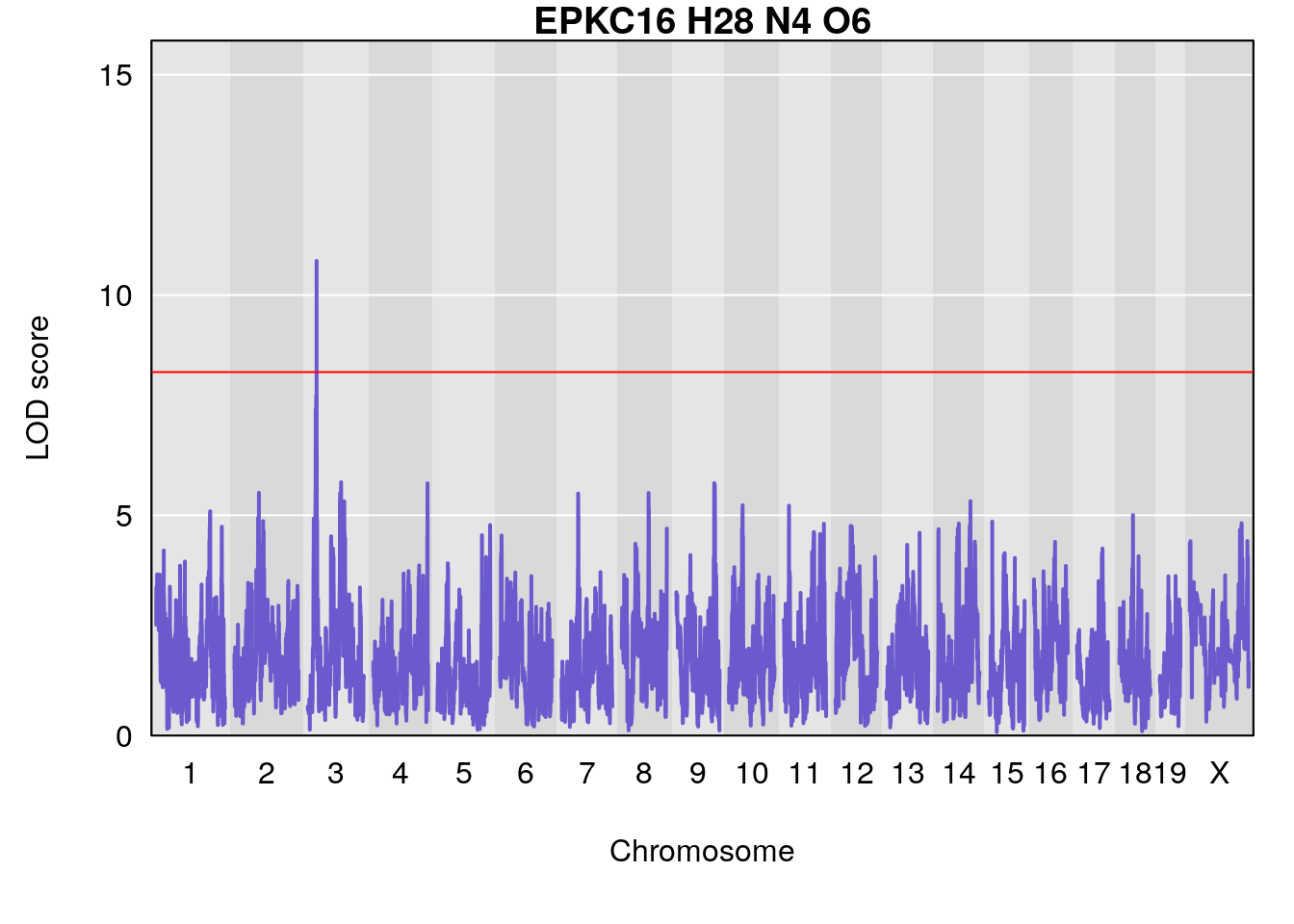
# [1] "Dat1_358"
# lodindex lodcolumn chr pos lod pvalue fdr ci_lo ci_hi Name
# 29 358 Dat1_358 3 11.064 10.7755 0 0 11.059 11.173 EPK
# Formula h2 se
# 29 C16 H28 N4 O6 0.8829885 0.6020464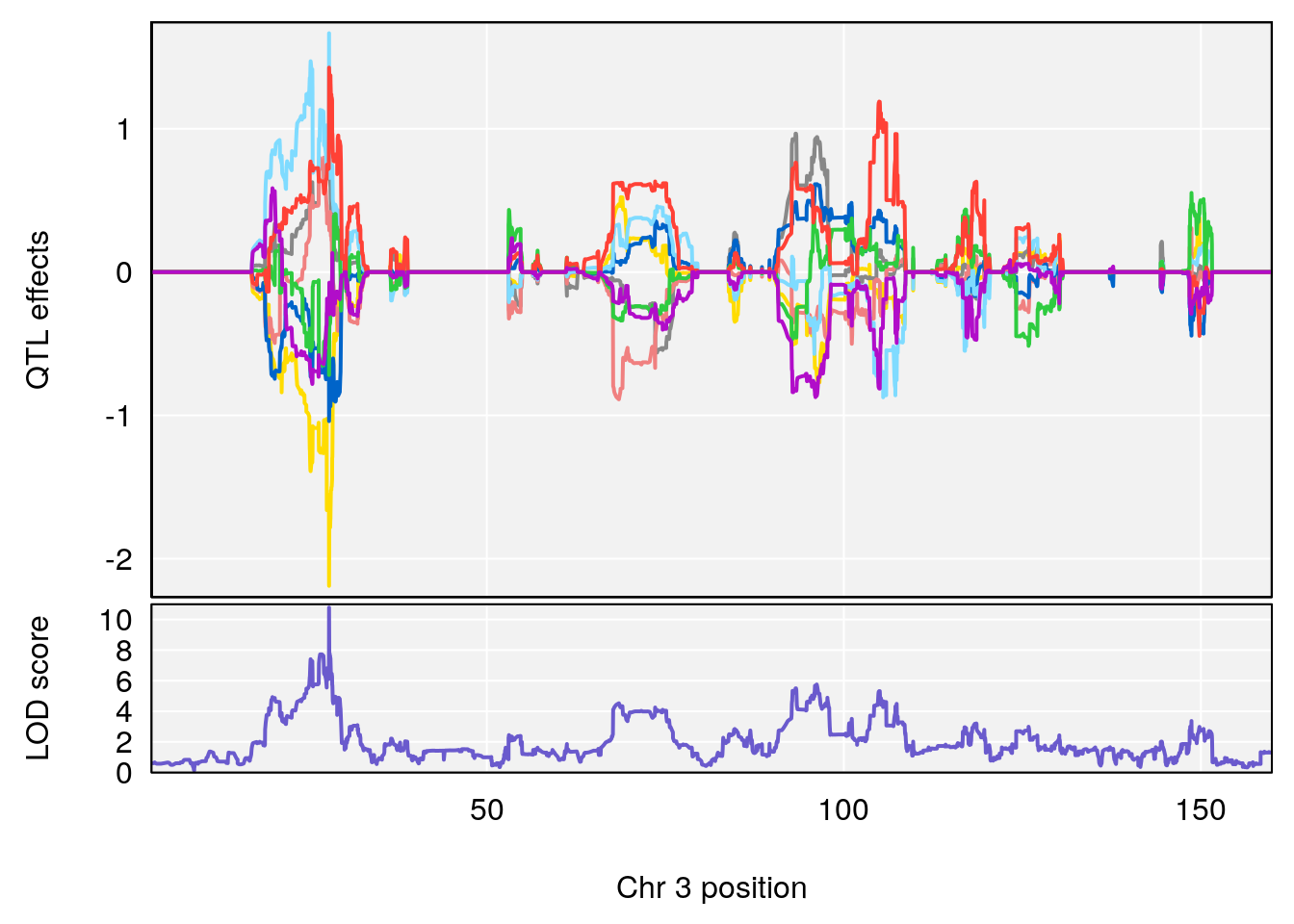
| Version | Author | Date |
|---|---|---|
| ff42a20 | xhyuo | 2023-07-17 |
| Version | Author | Date |
|---|---|---|
| ff42a20 | xhyuo | 2023-07-17 |
| Version | Author | Date |
|---|---|---|
| ff42a20 | xhyuo | 2023-07-17 |
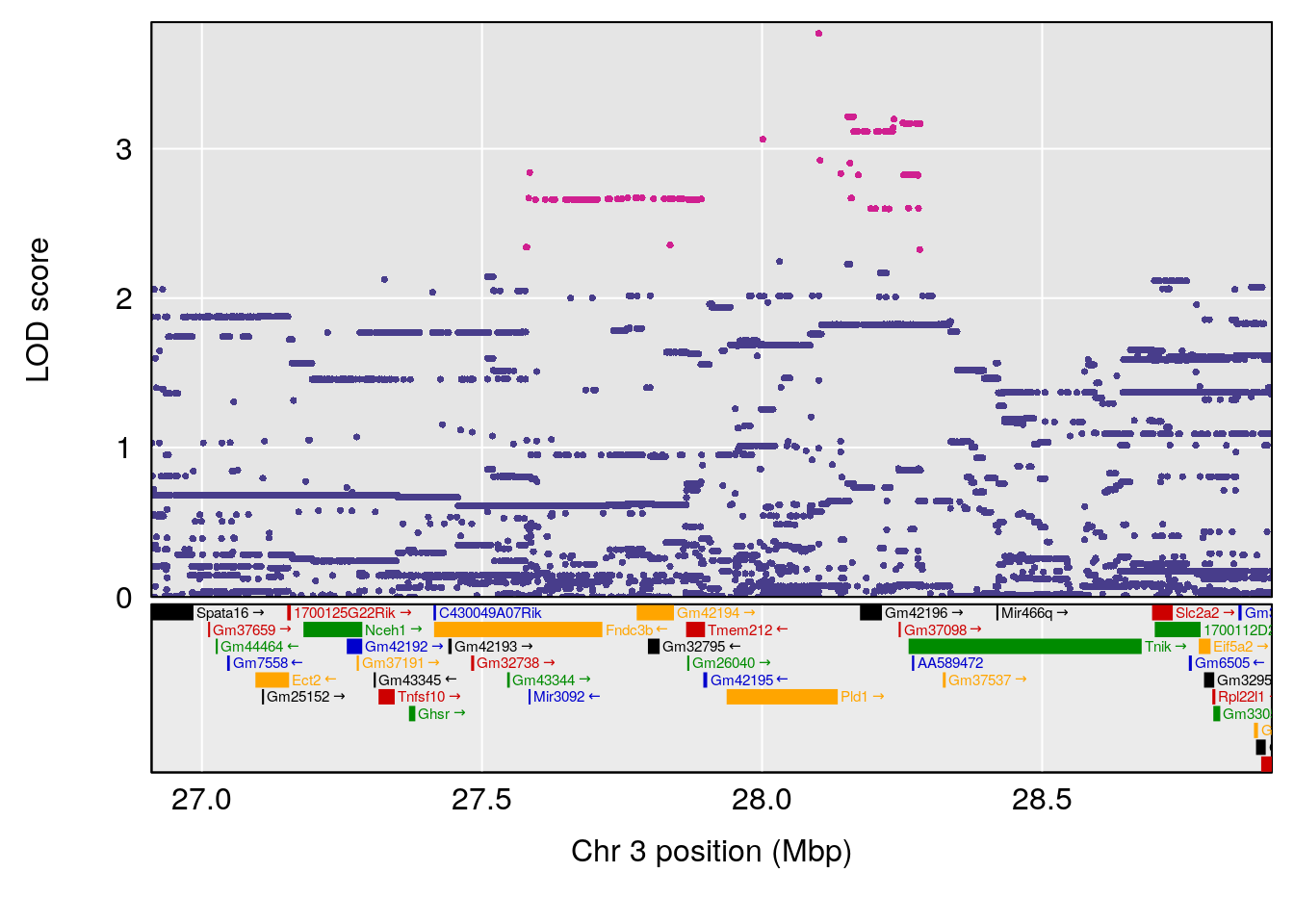
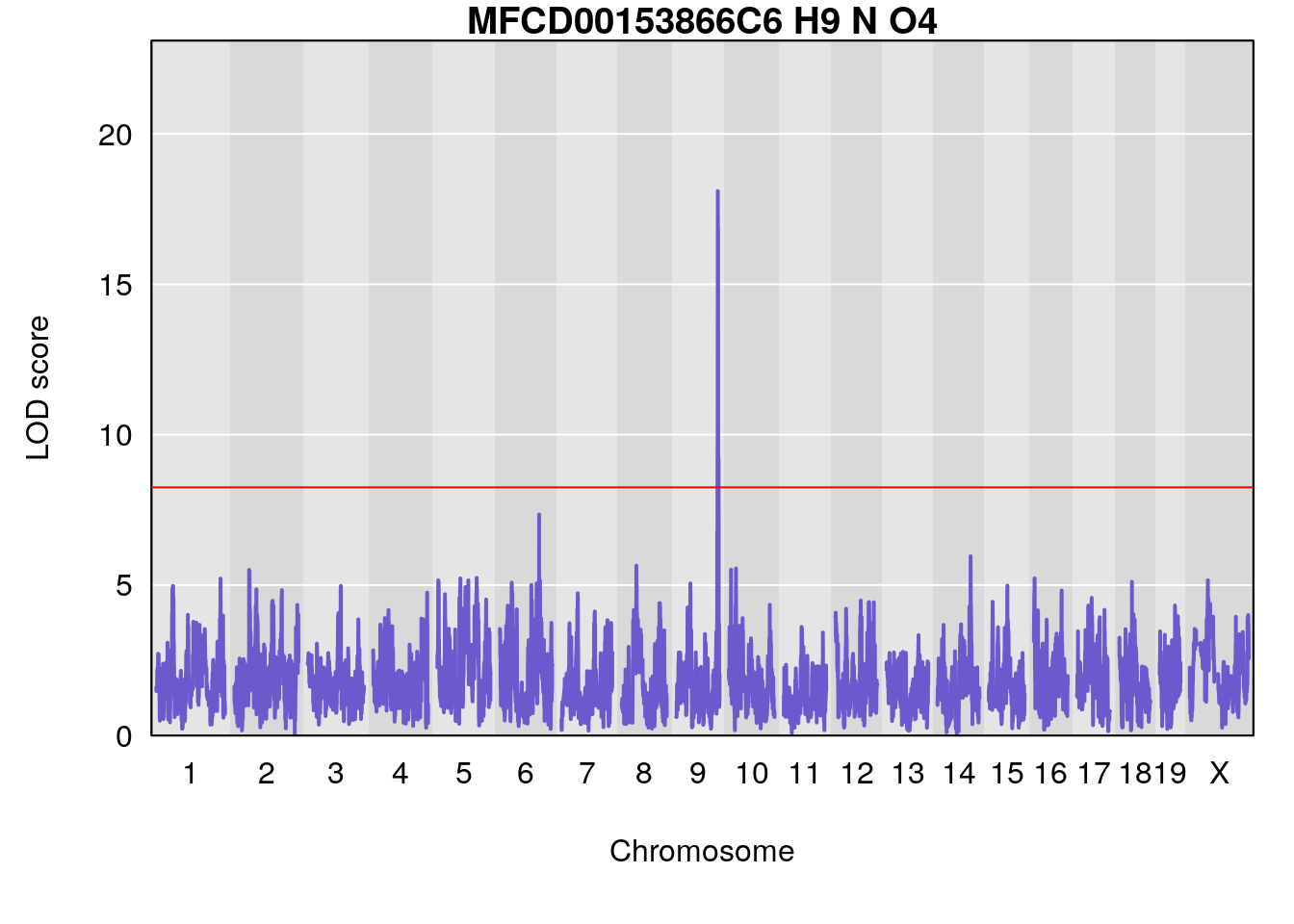
# [1] "Dat1_377"
# lodindex lodcolumn chr pos lod pvalue fdr ci_lo ci_hi Name
# 33 377 Dat1_377 9 71.322 18.09949 0 0 71.32 71.403 MFCD00153866
# Formula h2 se
# 33 C6 H9 N O4 1 0.5731433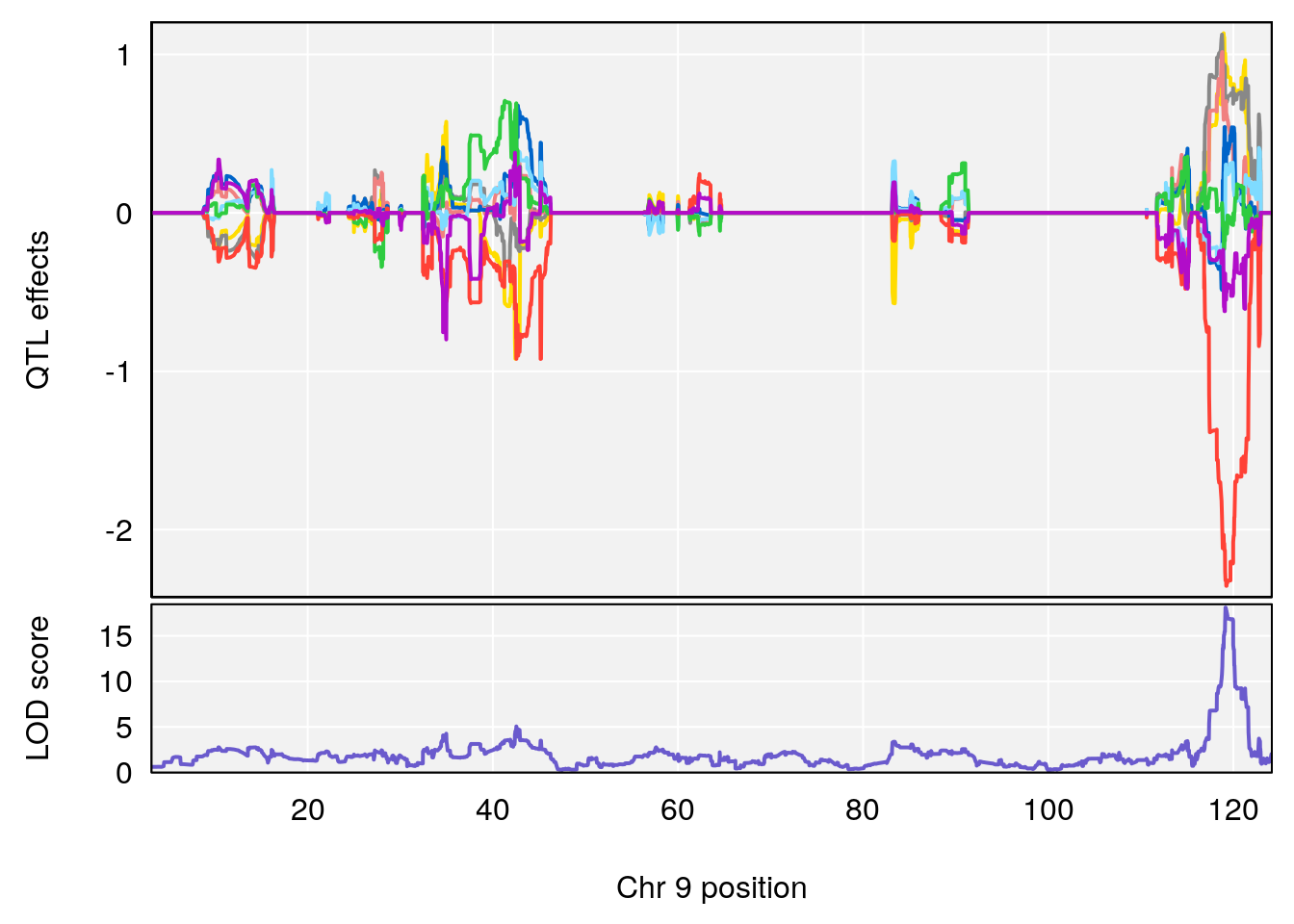
| Version | Author | Date |
|---|---|---|
| ff42a20 | xhyuo | 2023-07-17 |
| Version | Author | Date |
|---|---|---|
| ff42a20 | xhyuo | 2023-07-17 |
| Version | Author | Date |
|---|---|---|
| ff42a20 | xhyuo | 2023-07-17 |
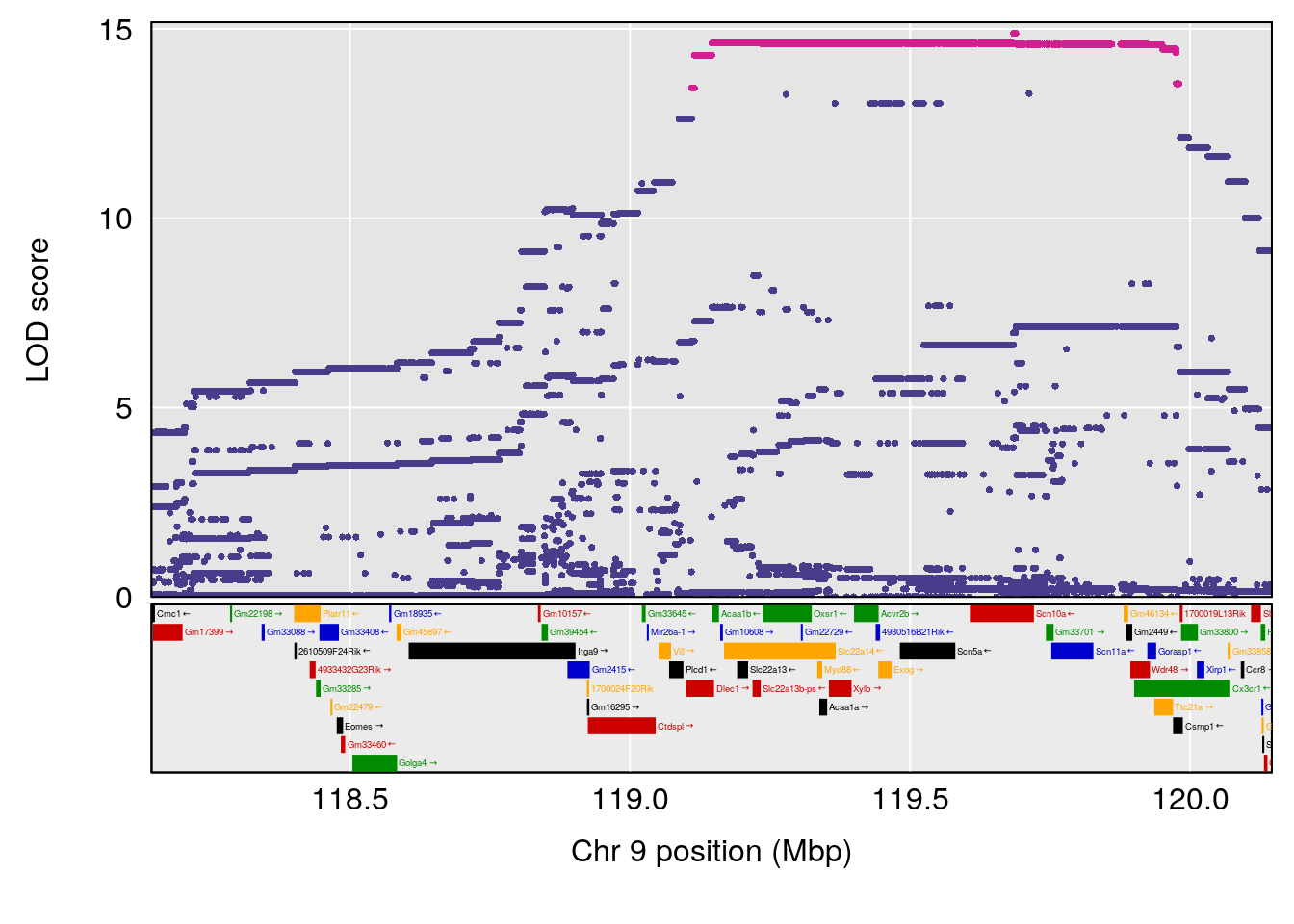
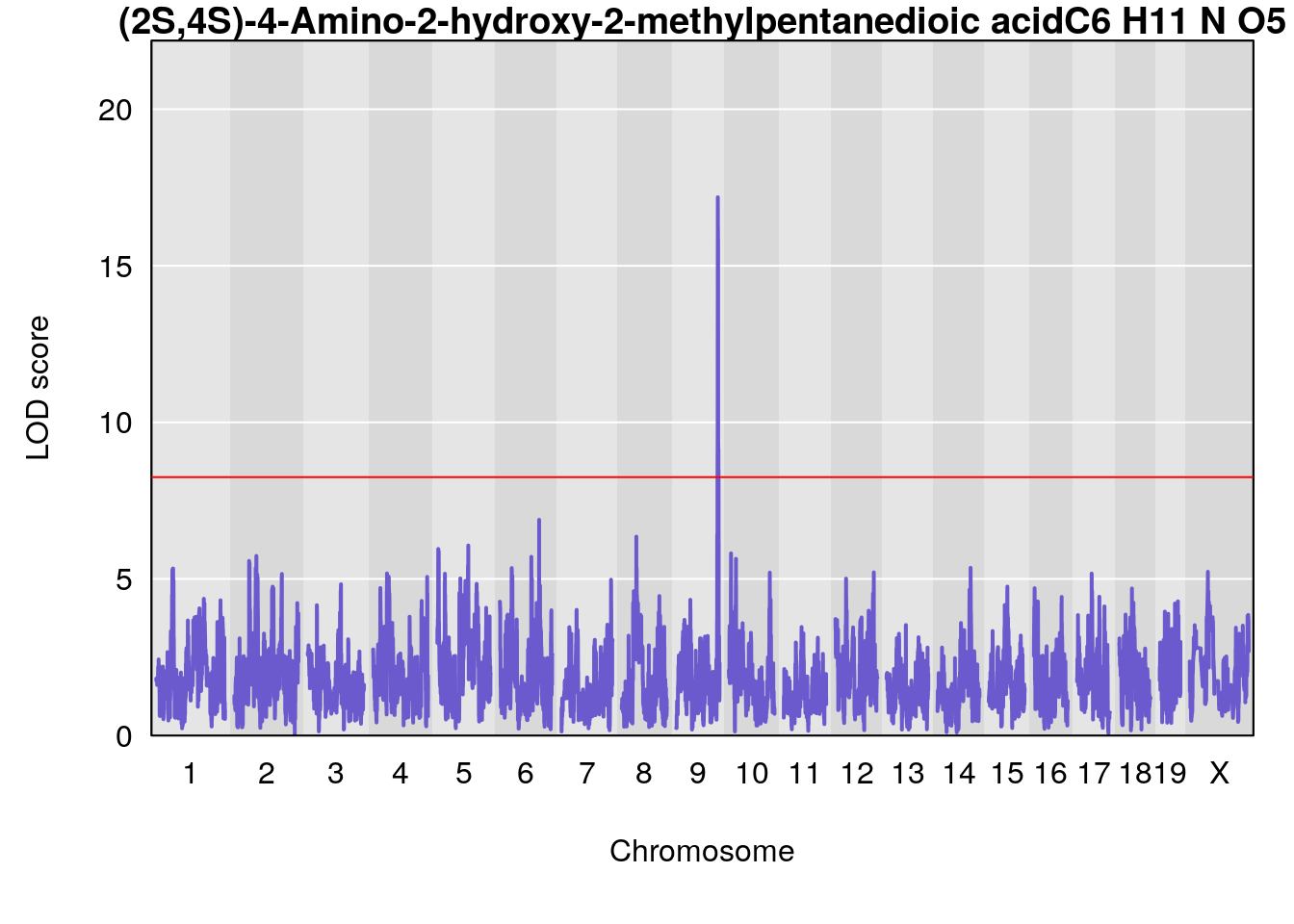
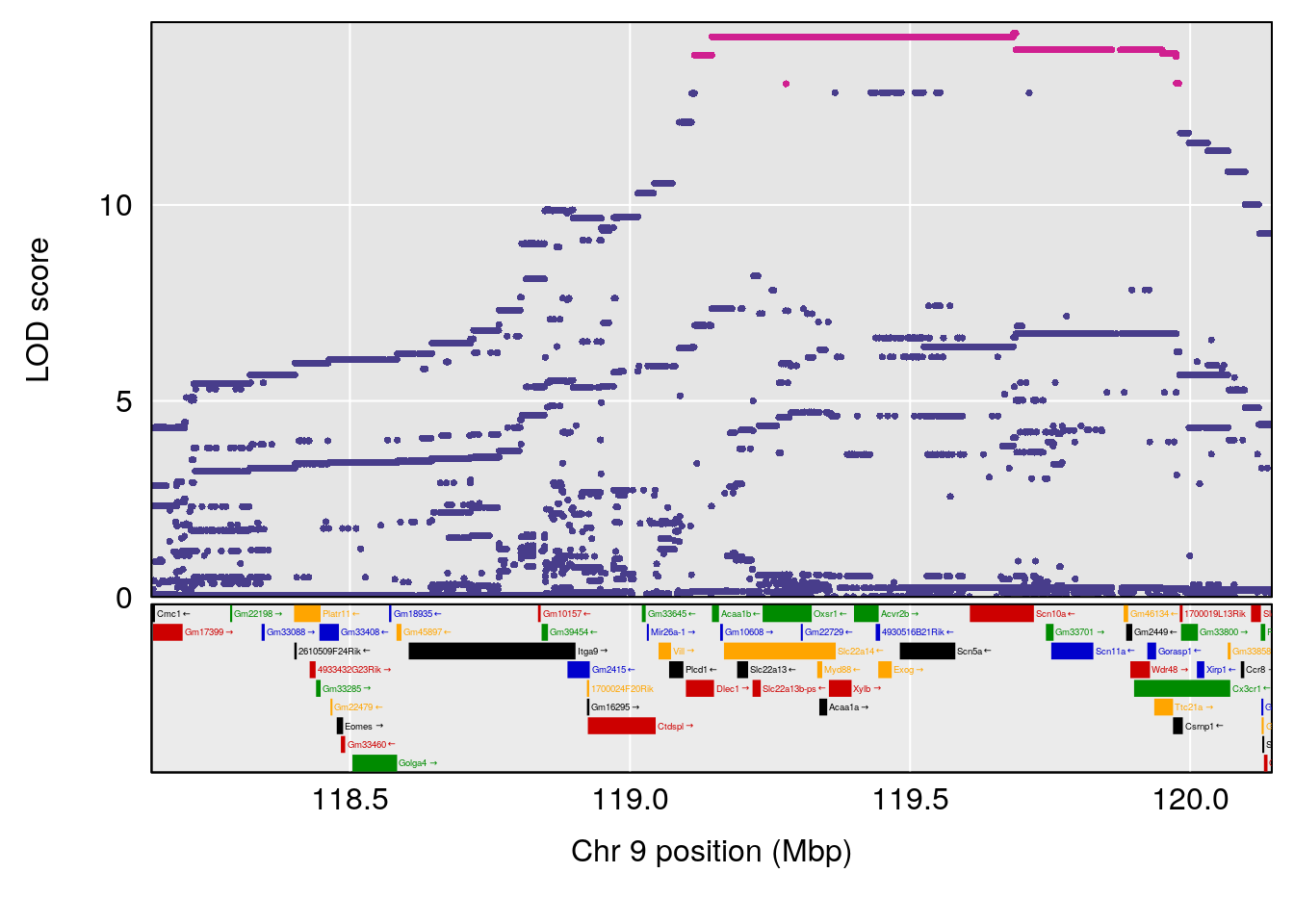
# [1] "Dat1_378"
# lodindex lodcolumn chr pos lod pvalue fdr ci_lo ci_hi
# 34 378 Dat1_378 9 71.322 17.18862 0 0 71.321 71.402
# Name Formula h2 se
# 34 (2S,4S)-4-Amino-2-hydroxy-2-methylpentanedioic acid C6 H11 N O5 1 0.5731433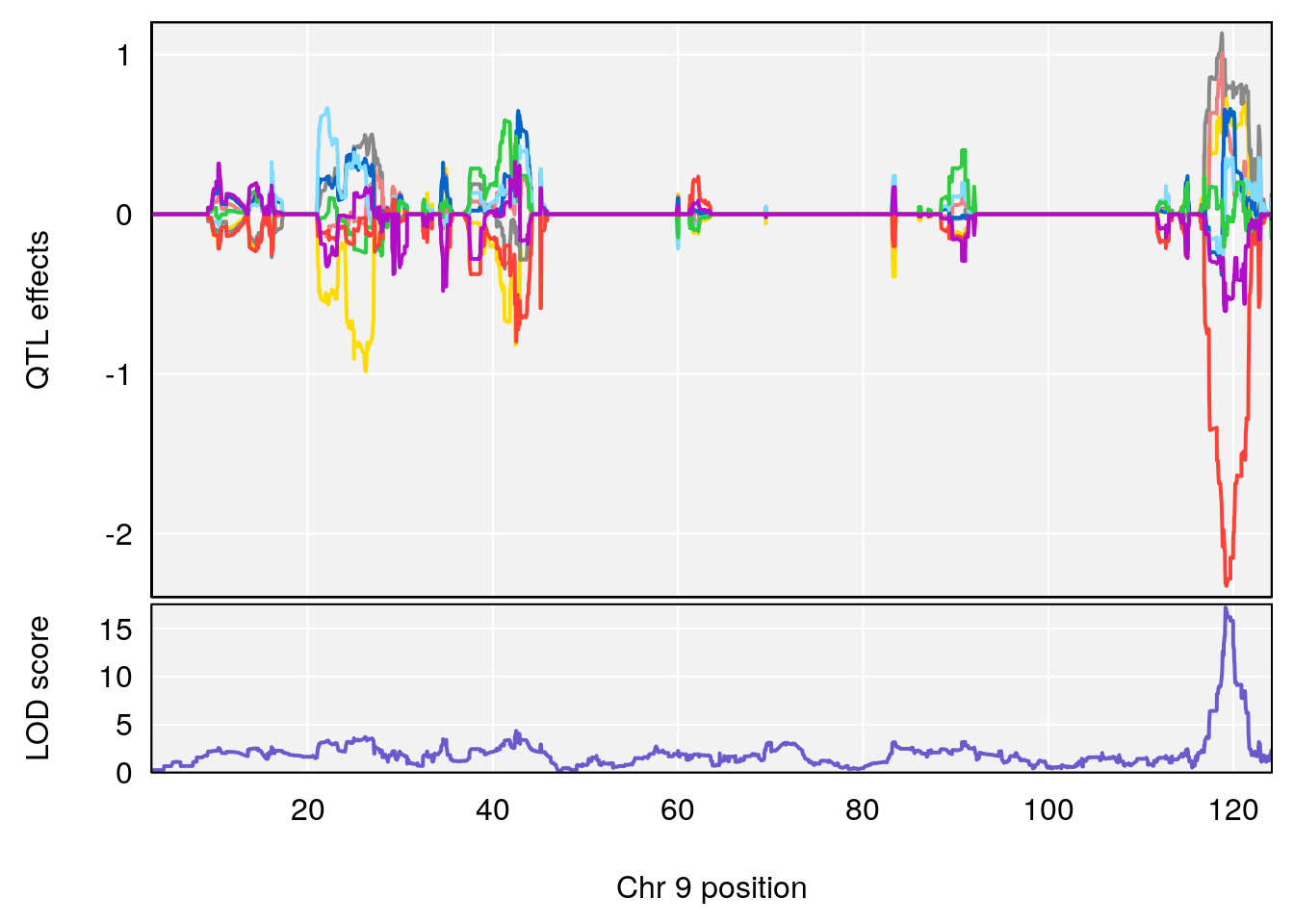
| Version | Author | Date |
|---|---|---|
| ff42a20 | xhyuo | 2023-07-17 |
| Version | Author | Date |
|---|---|---|
| ff42a20 | xhyuo | 2023-07-17 |
| Version | Author | Date |
|---|---|---|
| ff42a20 | xhyuo | 2023-07-17 |
# [1] "Dat1_536"
# lodindex lodcolumn chr pos lod pvalue fdr ci_lo ci_hi
# 47 536 Dat1_536 5 17.535 10.53676 0 0 17.497 17.652
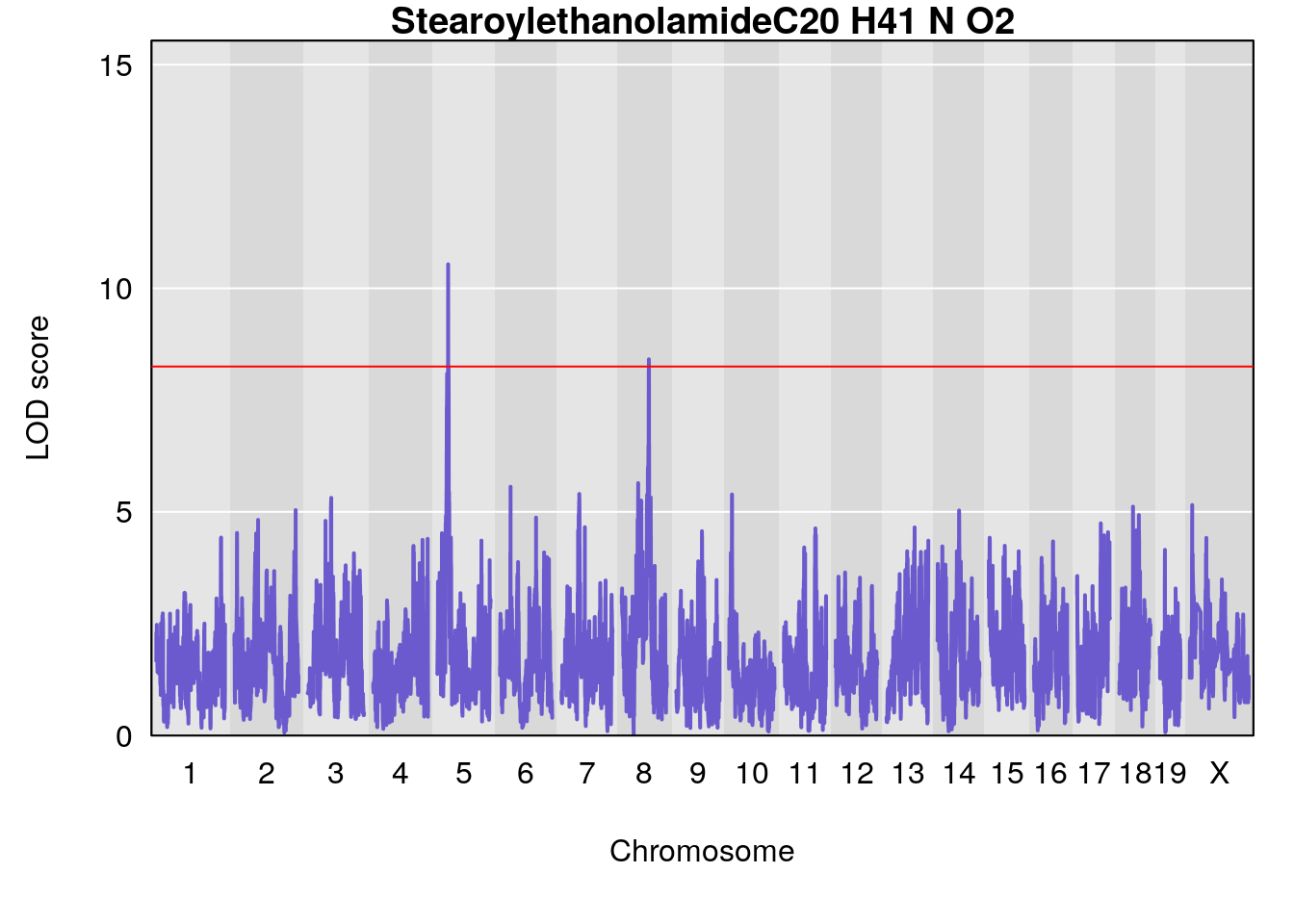
# Name Formula h2 se
# 47 Stearoylethanolamide C20 H41 N O2 0 0.4230553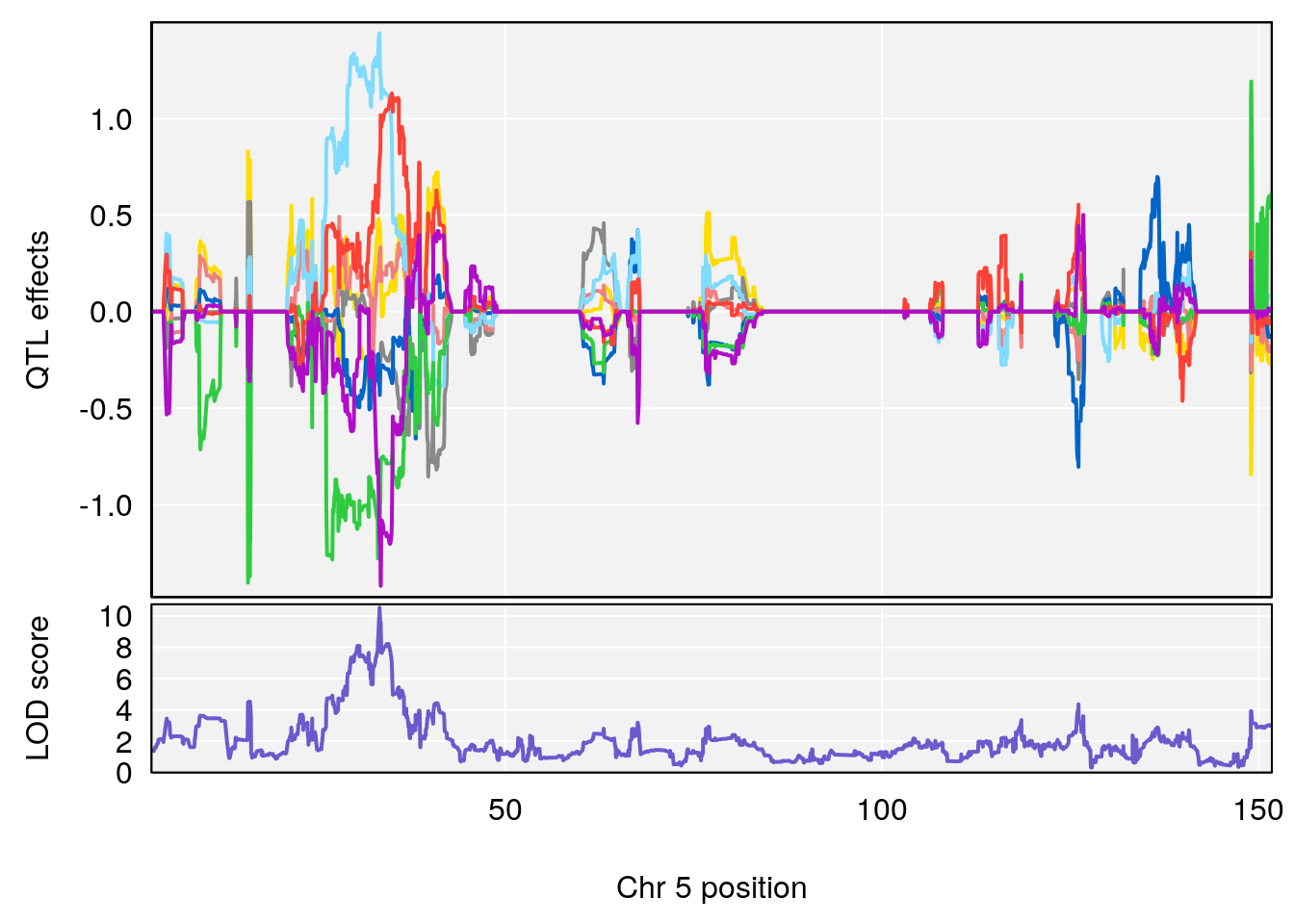
| Version | Author | Date |
|---|---|---|
| ff42a20 | xhyuo | 2023-07-17 |
| Version | Author | Date |
|---|---|---|
| ff42a20 | xhyuo | 2023-07-17 |
| Version | Author | Date |
|---|---|---|
| ff42a20 | xhyuo | 2023-07-17 |
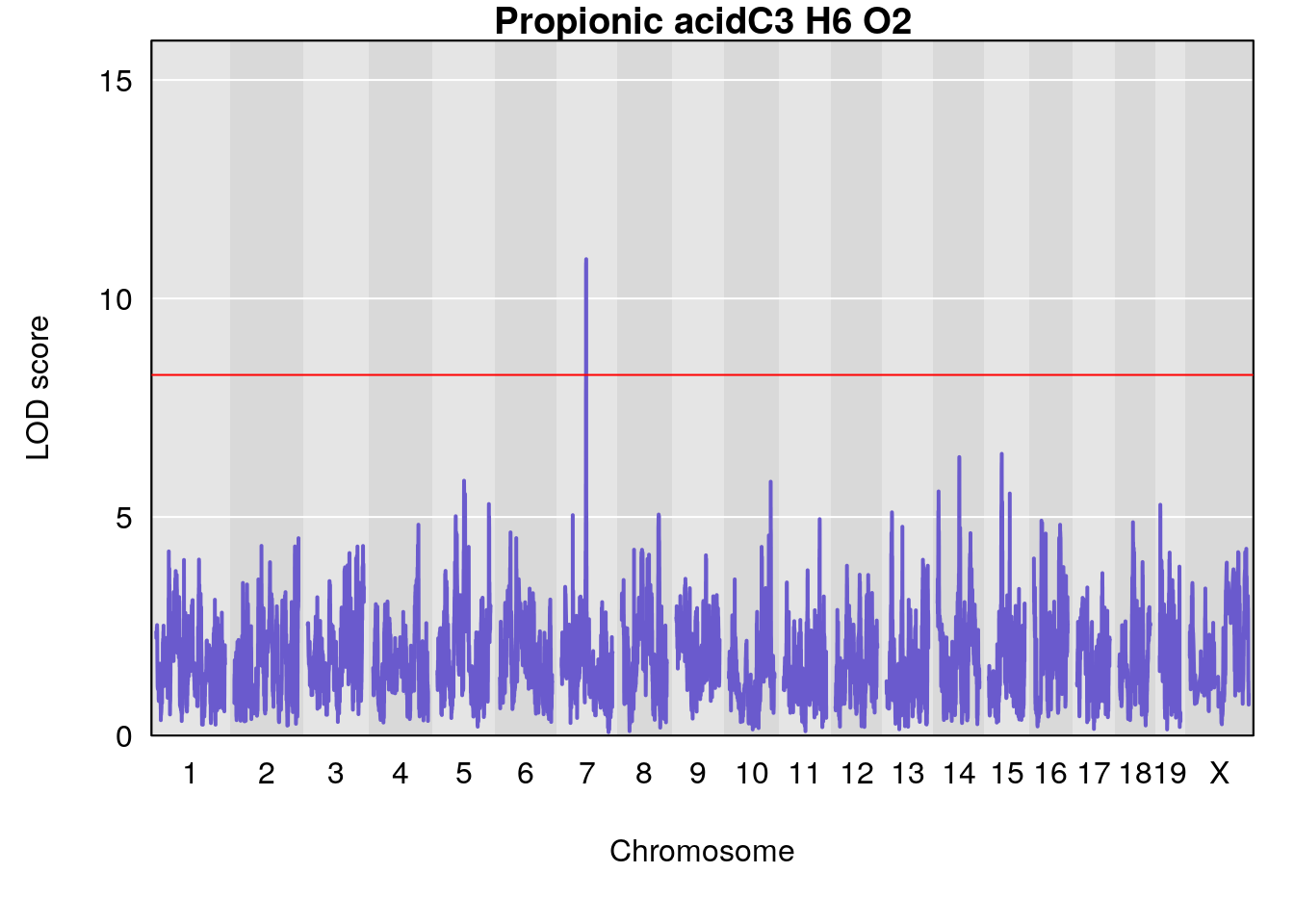
# [1] "Dat1_673"
# lodindex lodcolumn chr pos lod pvalue fdr ci_lo ci_hi
# 59 673 Dat1_673 7 41.153 10.89921 0 0 40.713 41.214
# Name Formula h2 se
# 59 Propionic acid C3 H6 O2 0 0.4230553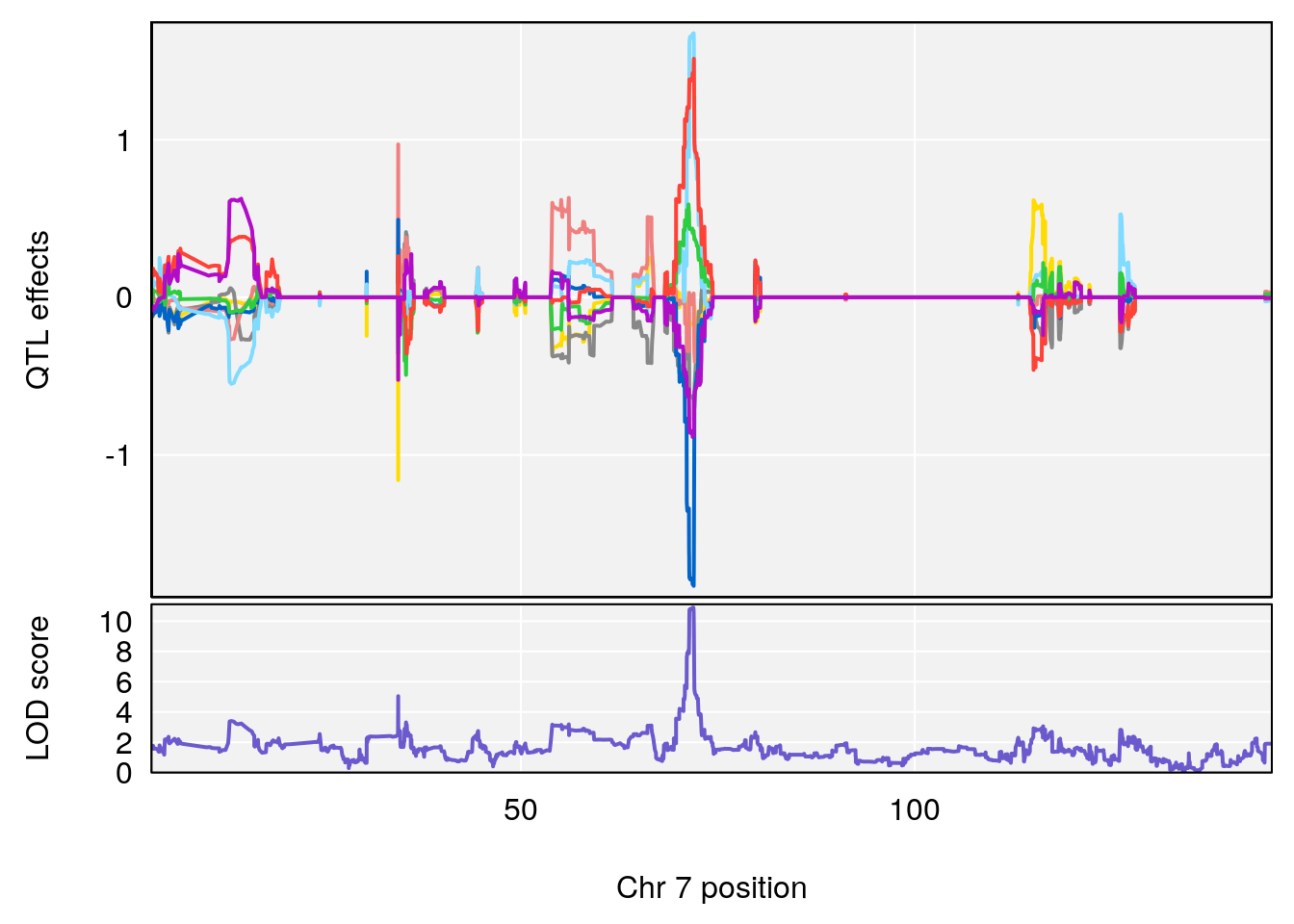
| Version | Author | Date |
|---|---|---|
| ff42a20 | xhyuo | 2023-07-17 |
| Version | Author | Date |
|---|---|---|
| ff42a20 | xhyuo | 2023-07-17 |
| Version | Author | Date |
|---|---|---|
| ff42a20 | xhyuo | 2023-07-17 |
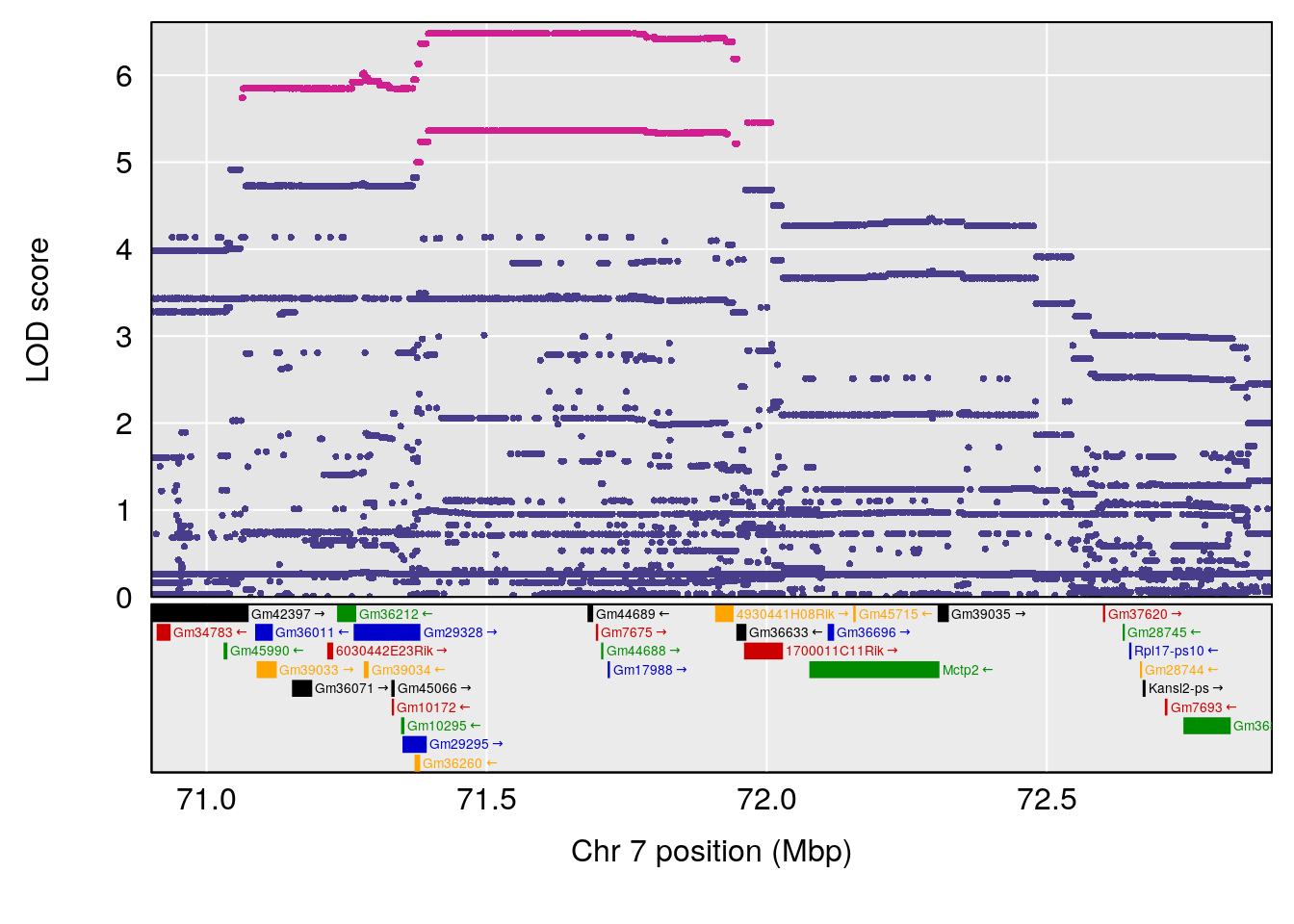
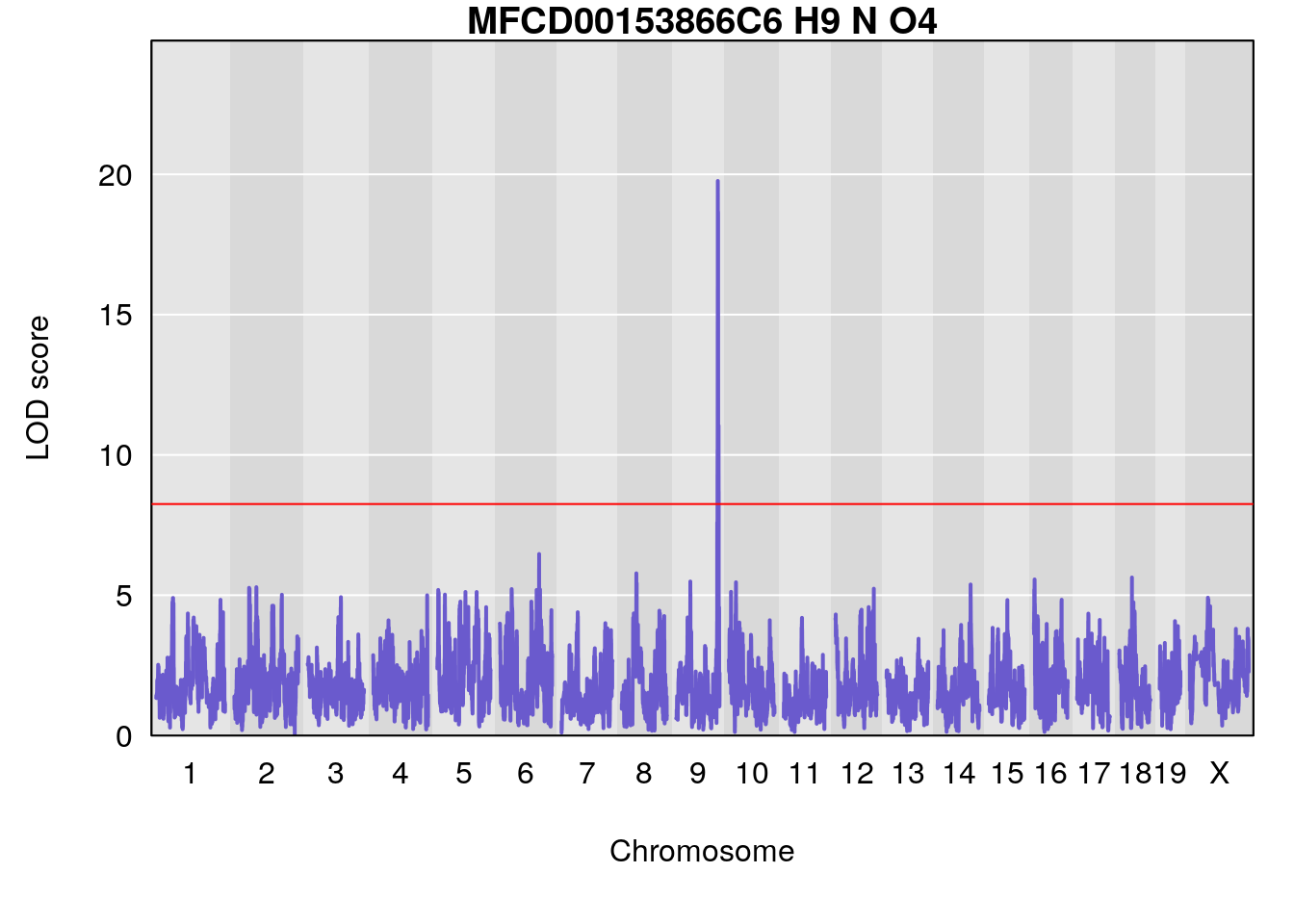
# [1] "Dat1_769"
# lodindex lodcolumn chr pos lod pvalue fdr ci_lo ci_hi Name
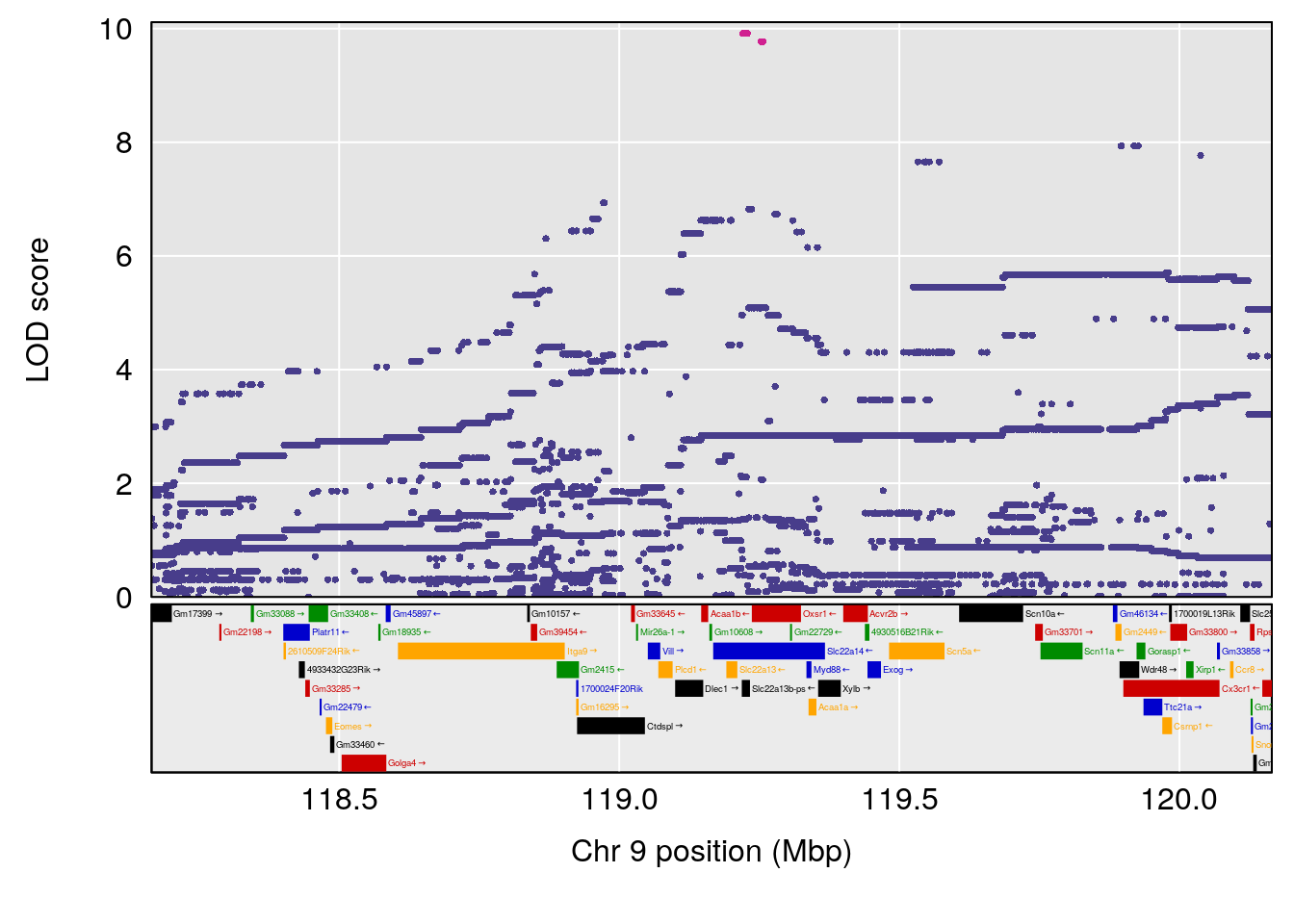
# 62 769 Dat1_769 9 71.322 19.76486 0 0 71.32 71.403 MFCD00153866
# Formula h2 se
# 62 C6 H9 N O4 1 0.5731433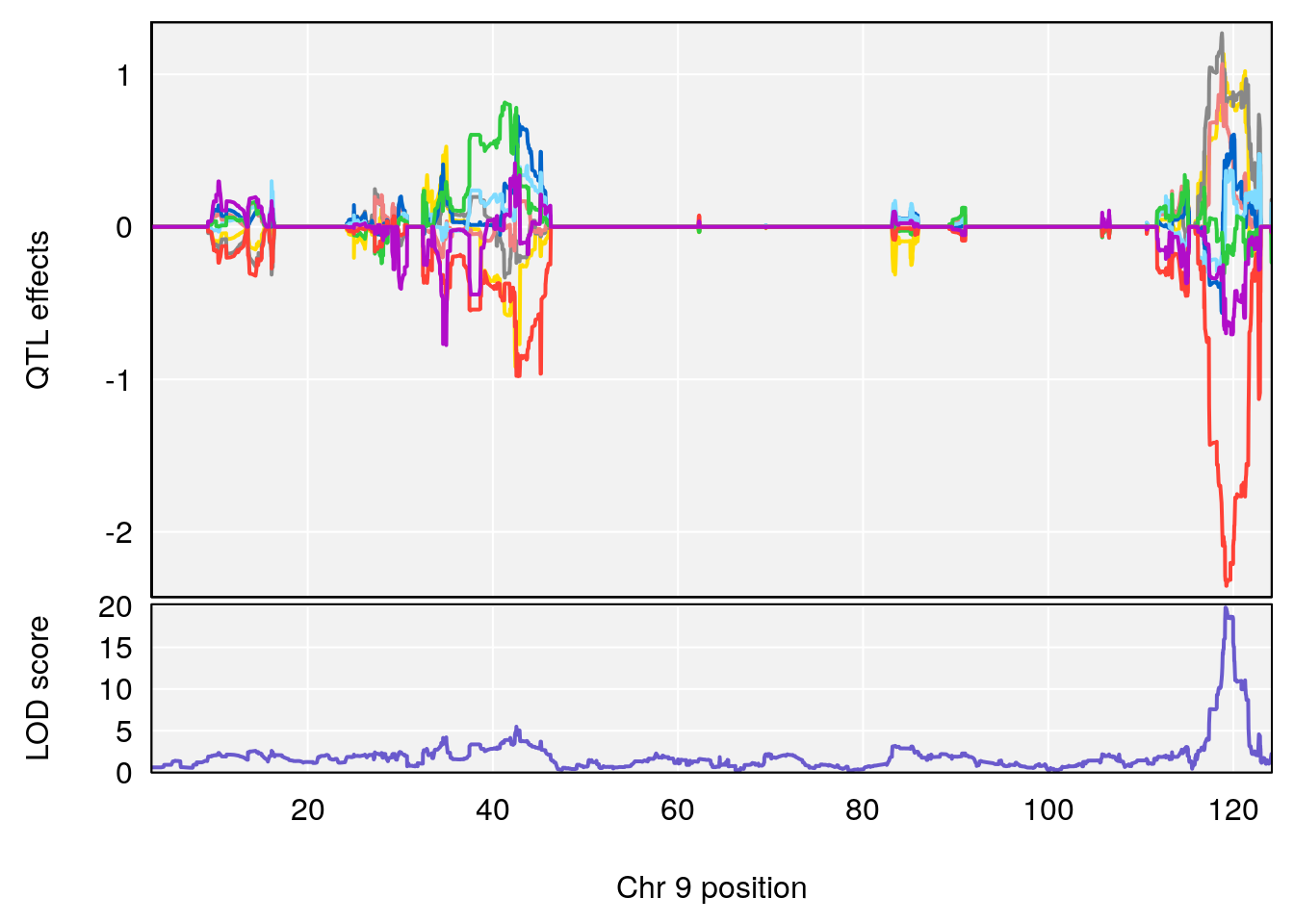
| Version | Author | Date |
|---|---|---|
| ff42a20 | xhyuo | 2023-07-17 |
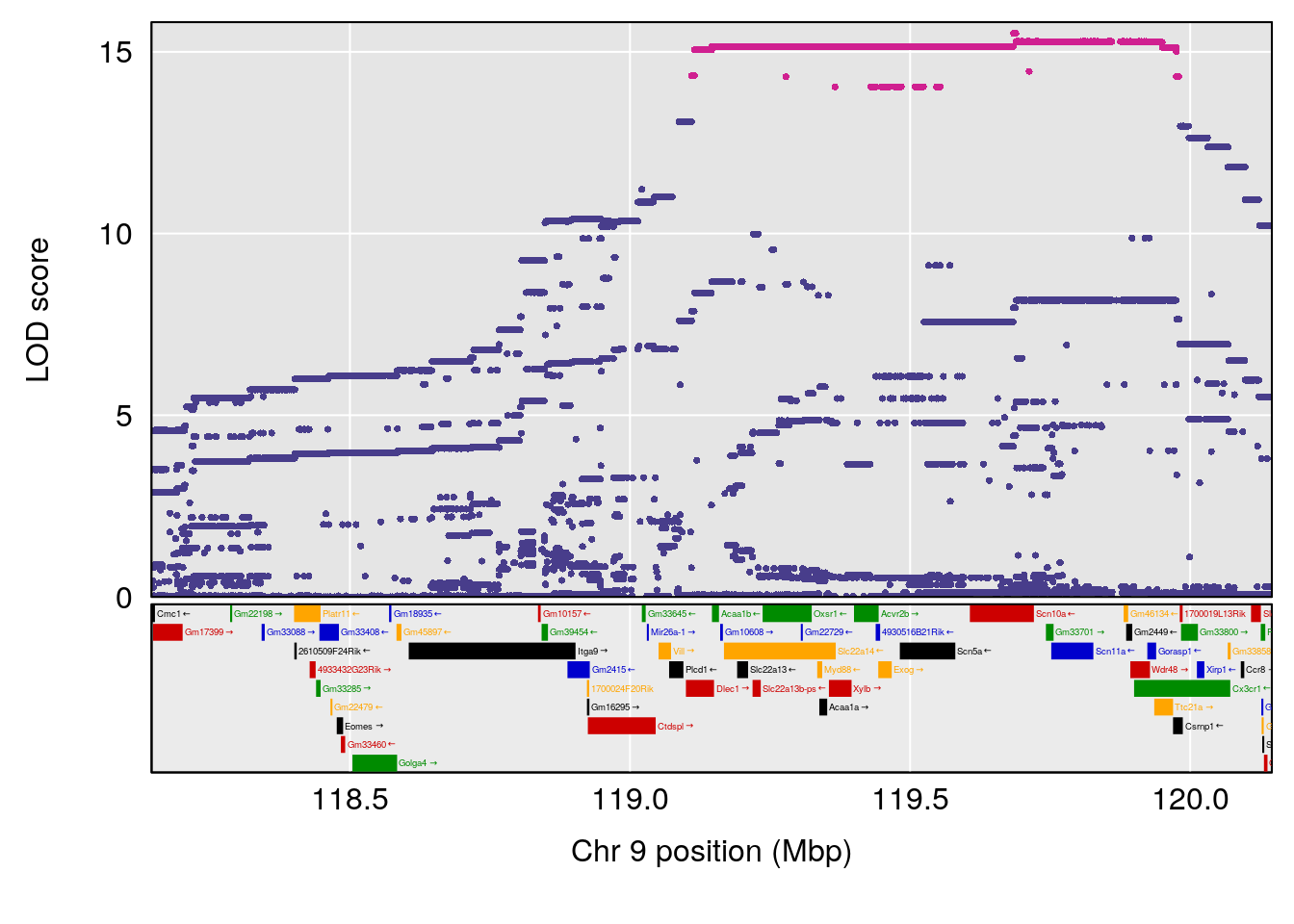
| Version | Author | Date |
|---|---|---|
| ff42a20 | xhyuo | 2023-07-17 |
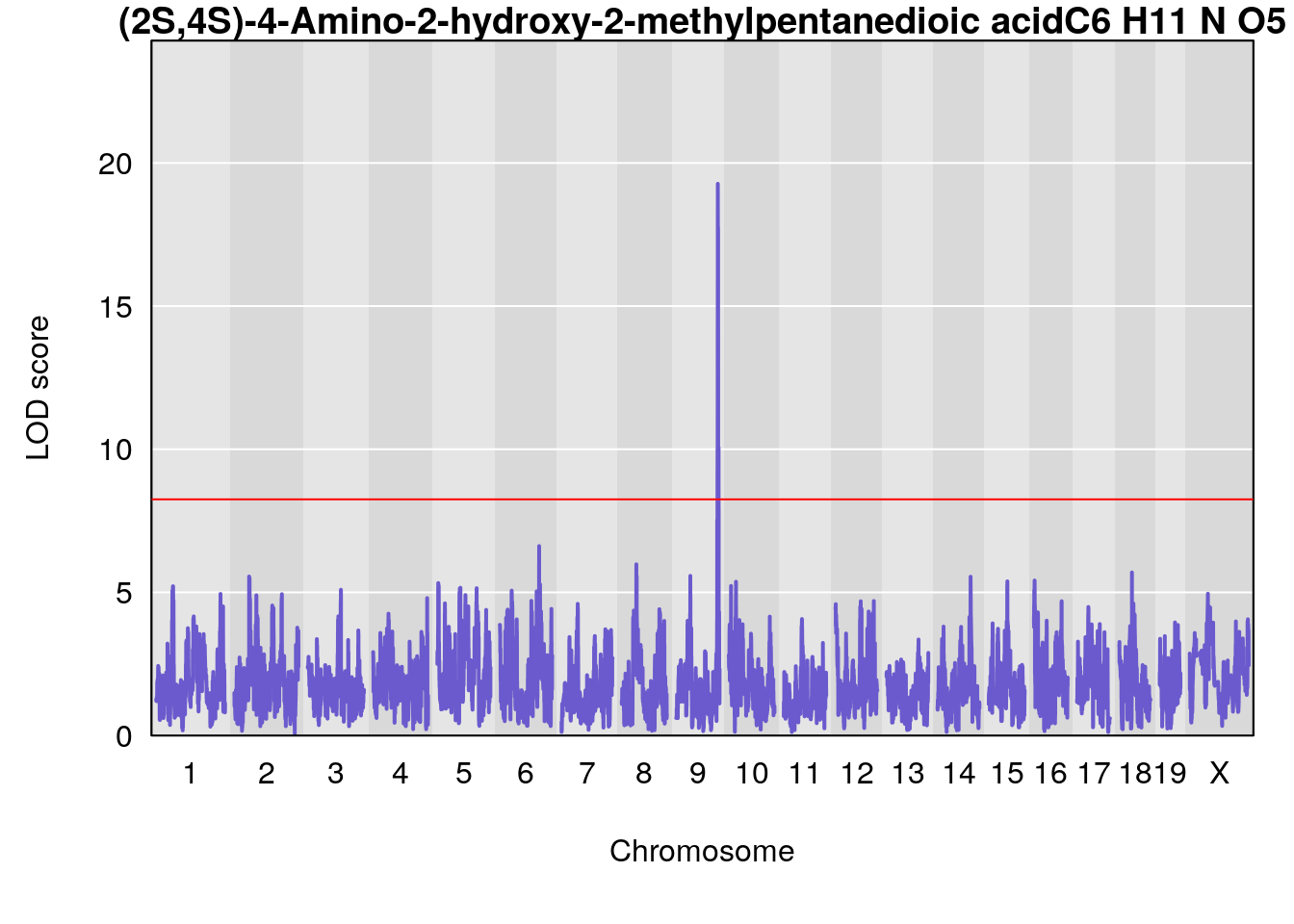
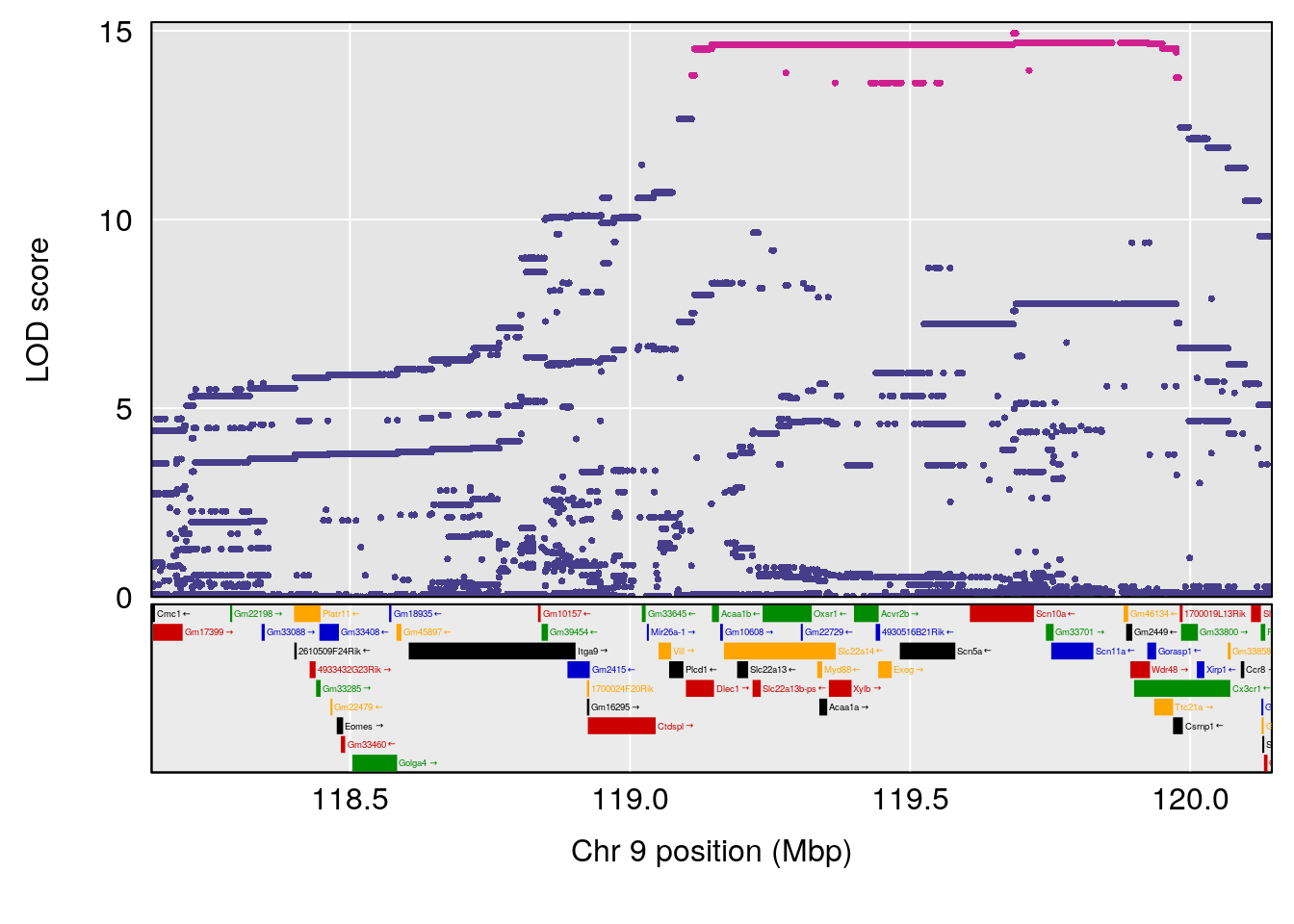
# [1] "Dat1_770"
# lodindex lodcolumn chr pos lod pvalue fdr ci_lo ci_hi
# 63 770 Dat1_770 9 71.322 19.27253 0 0 71.32 71.329
# Name Formula h2 se
# 63 (2S,4S)-4-Amino-2-hydroxy-2-methylpentanedioic acid C6 H11 N O5 1 0.5731433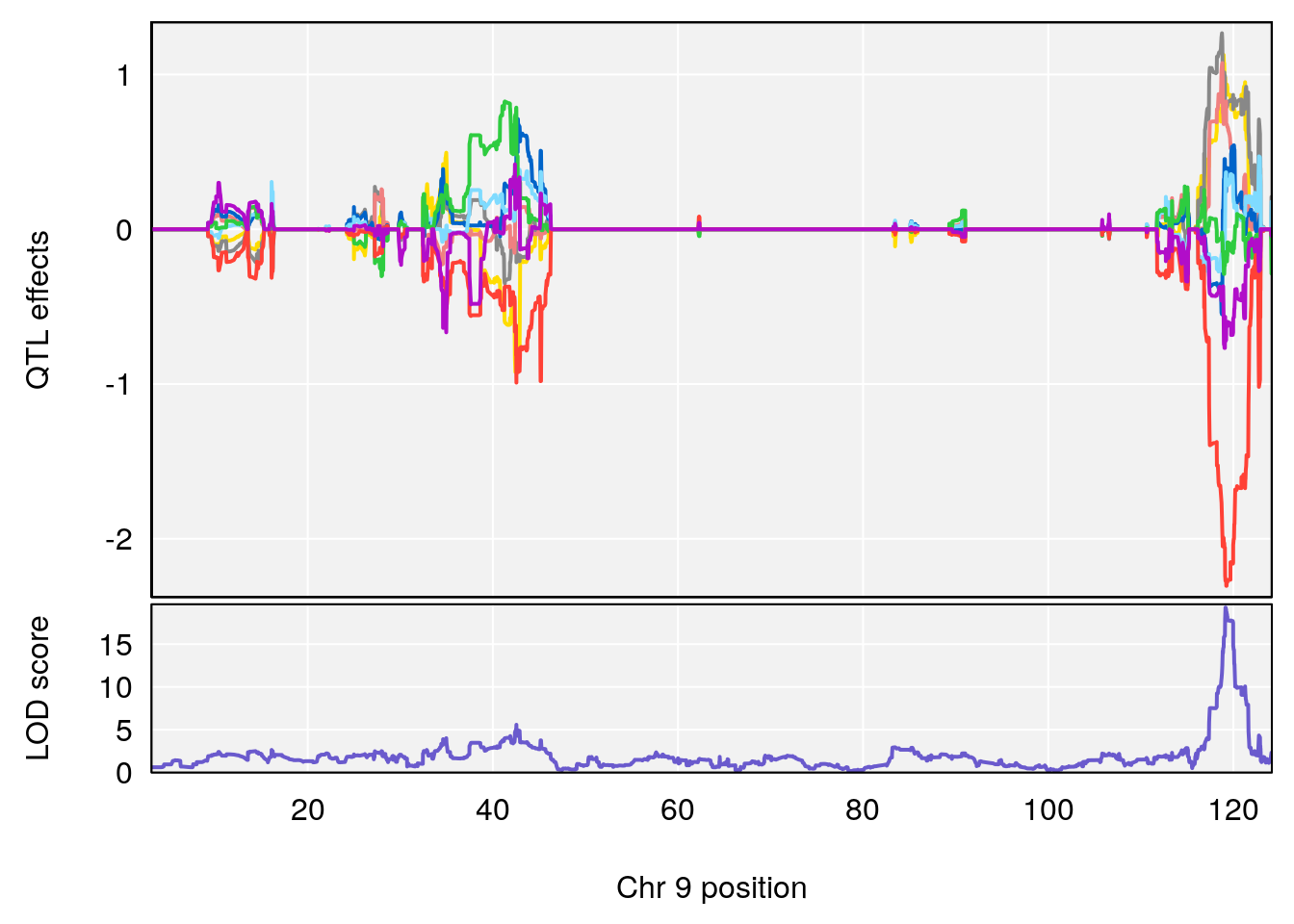
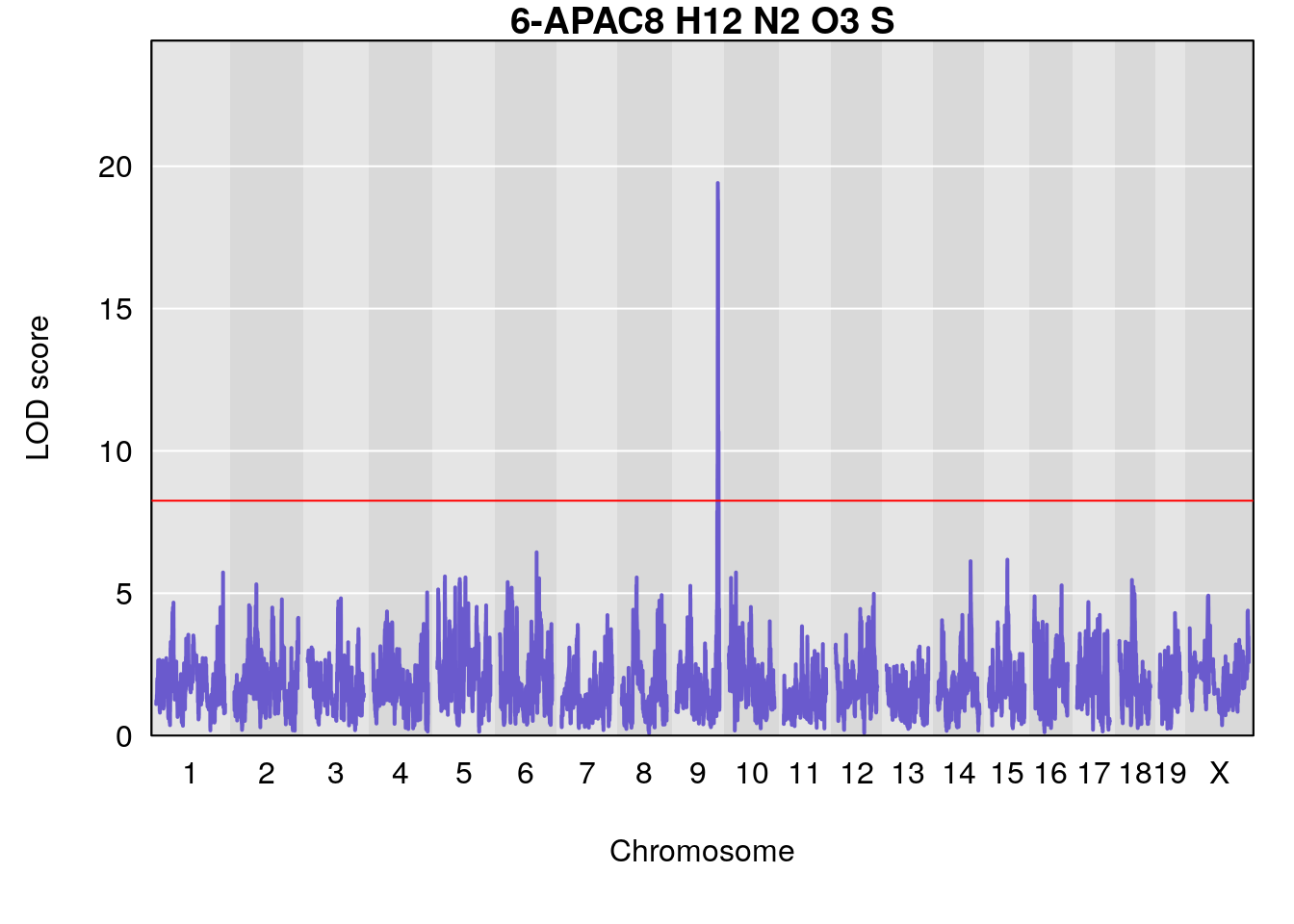
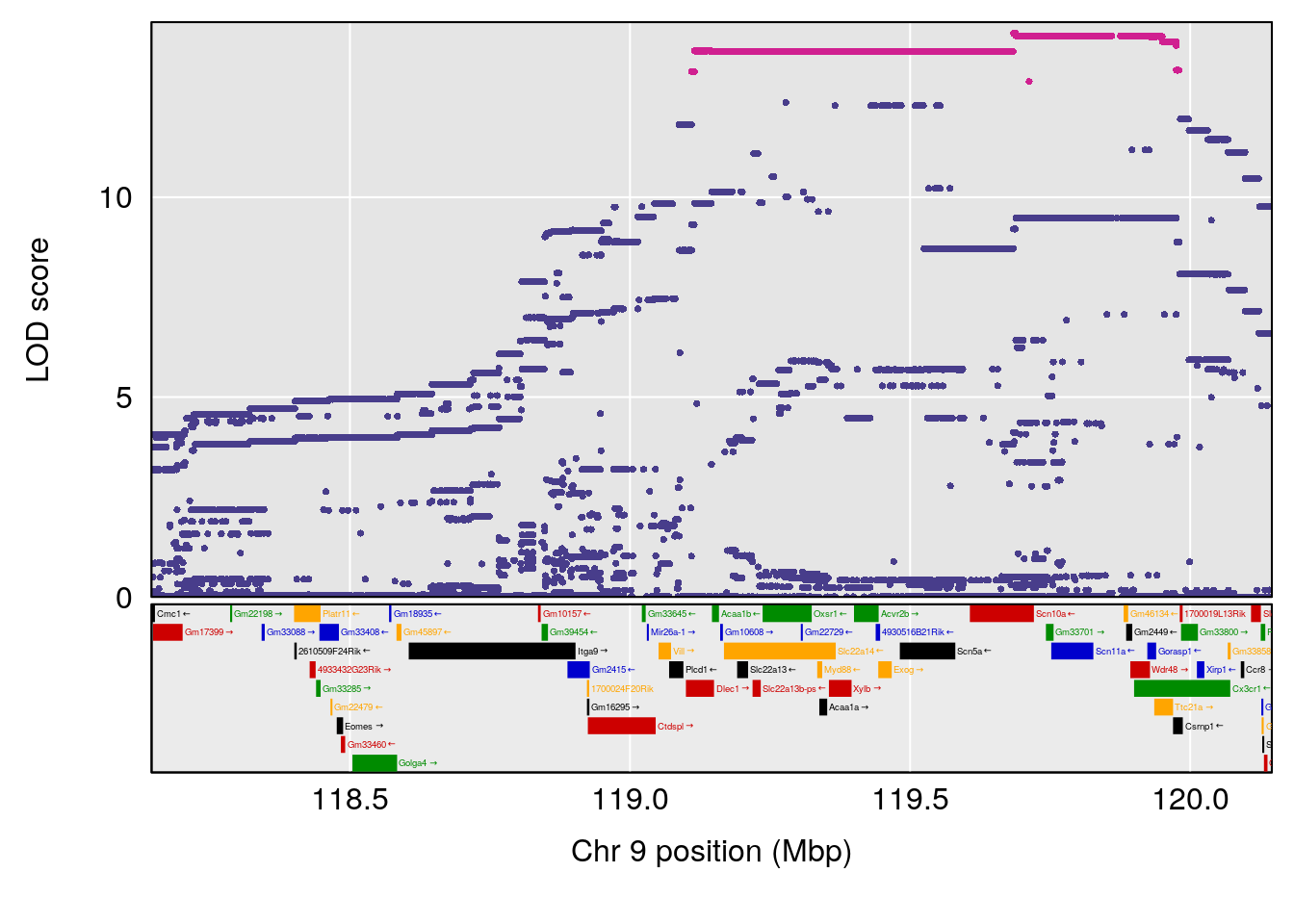
# [1] "Dat1_776"
# lodindex lodcolumn chr pos lod pvalue fdr ci_lo ci_hi Name
# 65 776 Dat1_776 9 71.322 19.4123 0 0 71.32 71.403 6-APA
# Formula h2 se
# 65 C8 H12 N2 O3 S 1 0.5731433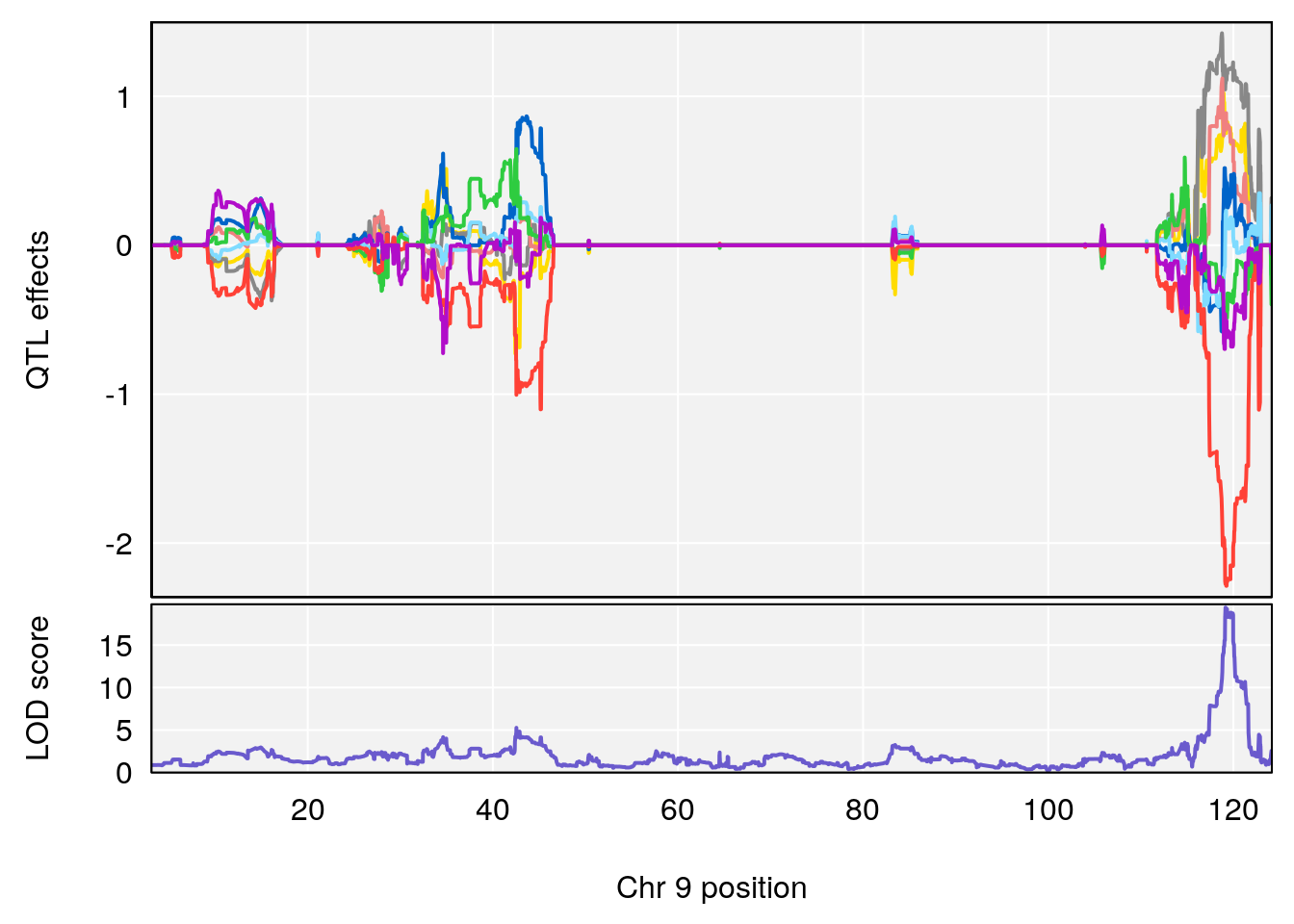
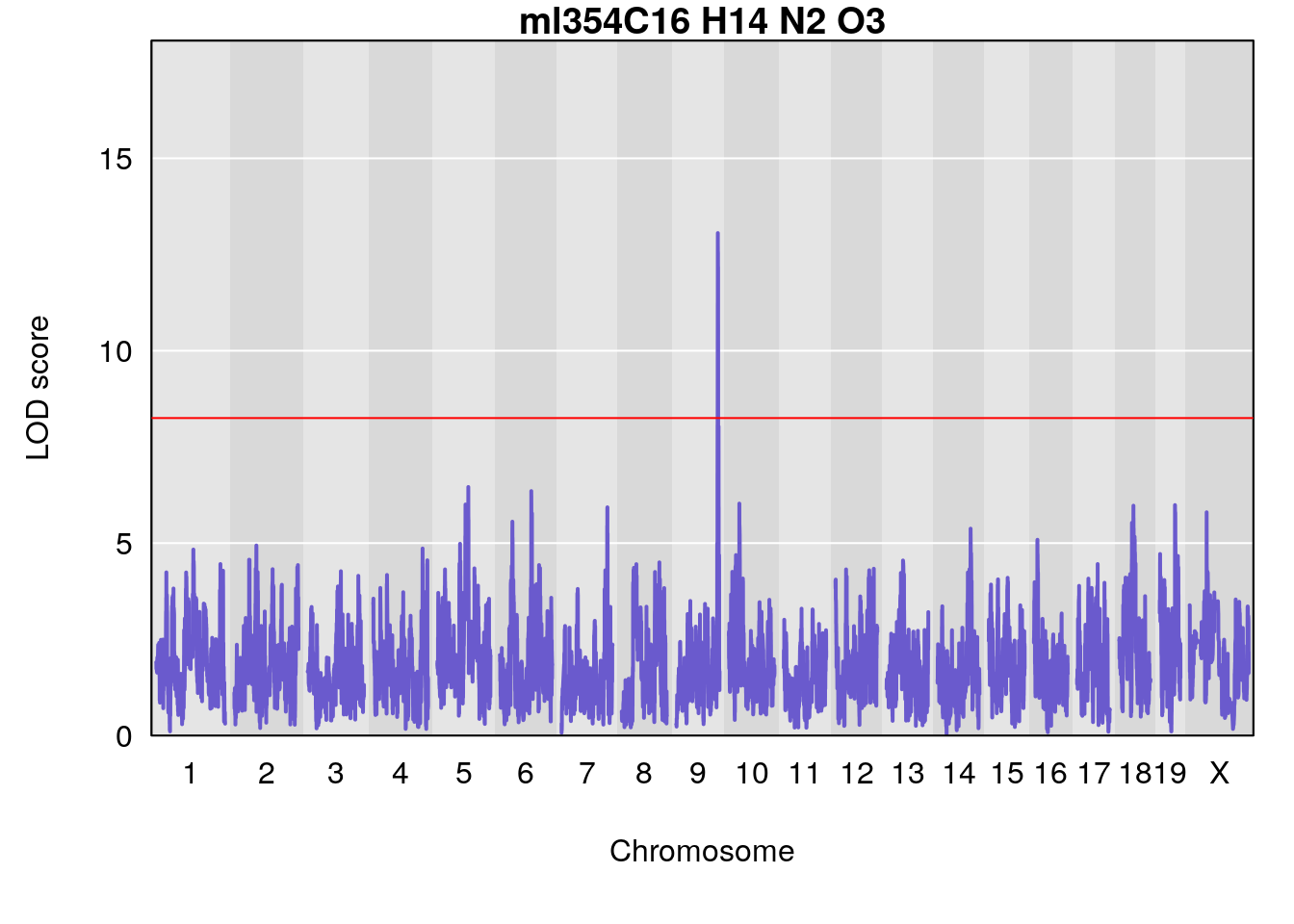
# [1] "Dat1_1099"
# lodindex lodcolumn chr pos lod pvalue fdr ci_lo ci_hi Name
# 94 1099 Dat1_1099 9 71.324 13.0579 0 0 71.32 71.403 ml354
# Formula h2 se
# 94 C16 H14 N2 O3 1 0.5731433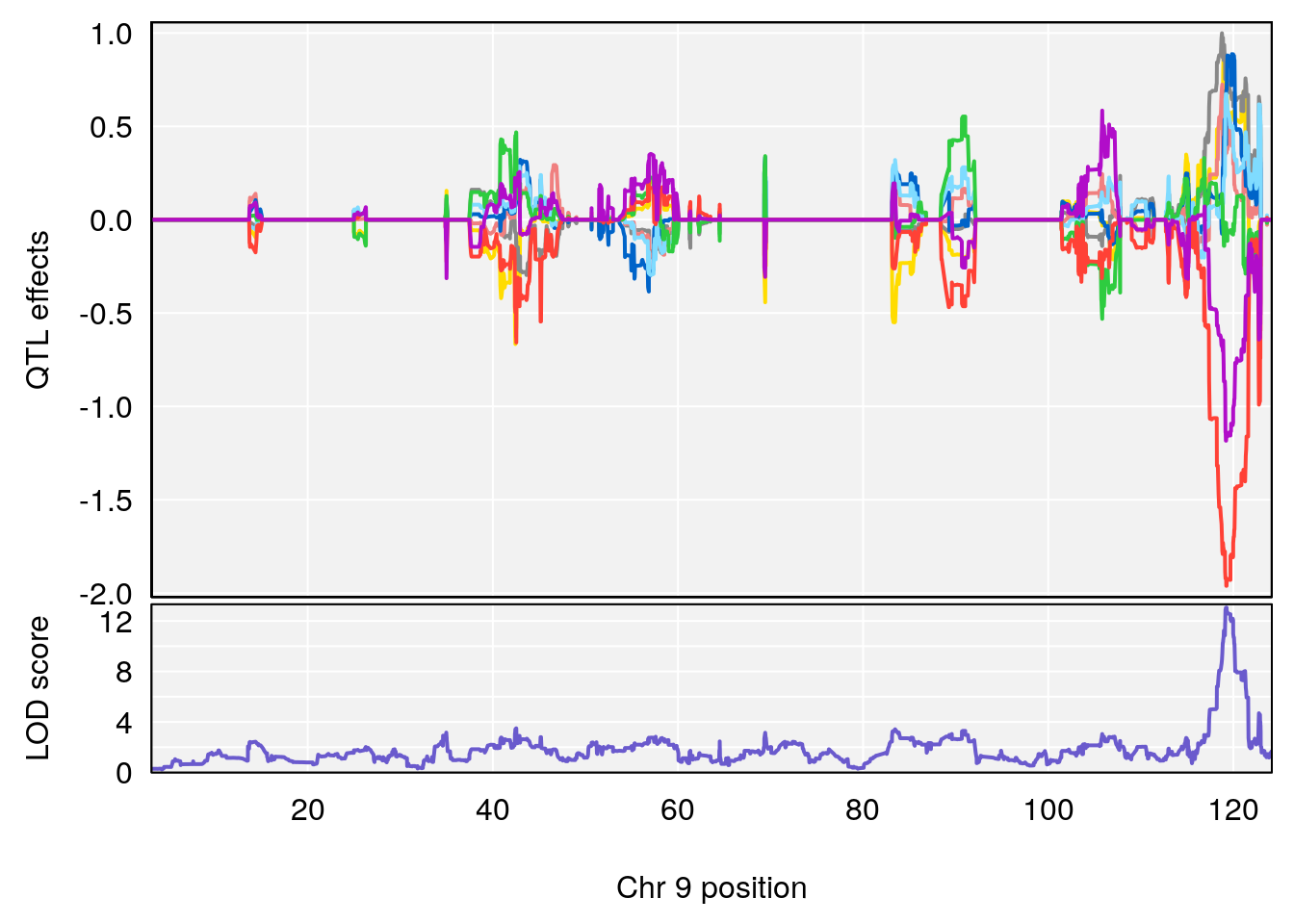
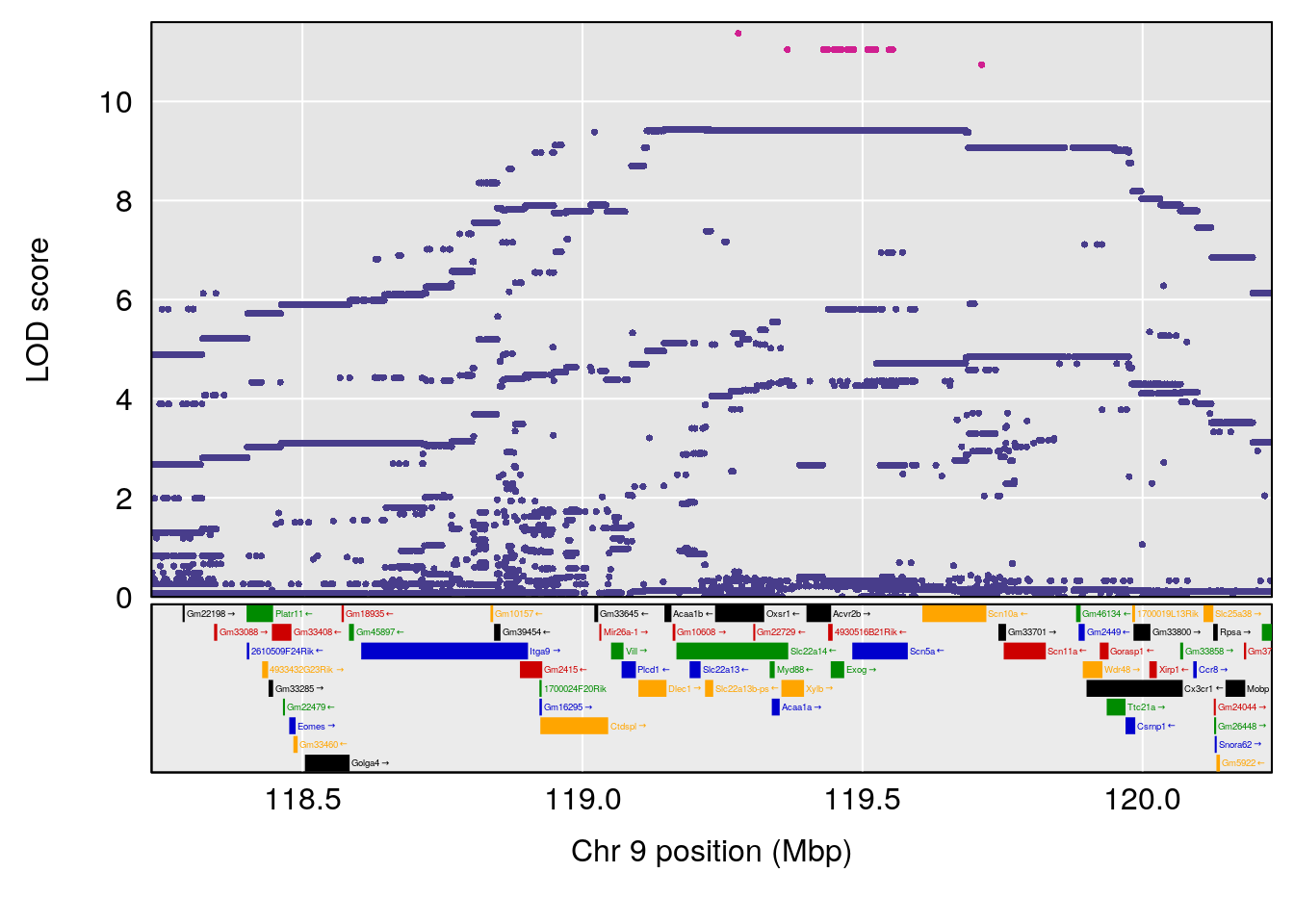
# [1] "Dat1_1993"
# lodindex lodcolumn chr pos lod pvalue fdr ci_lo ci_hi
# 154 1993 Dat1_1993 9 71.322 12.91449 0 0 71.32 71.327
# Name Formula h2 se
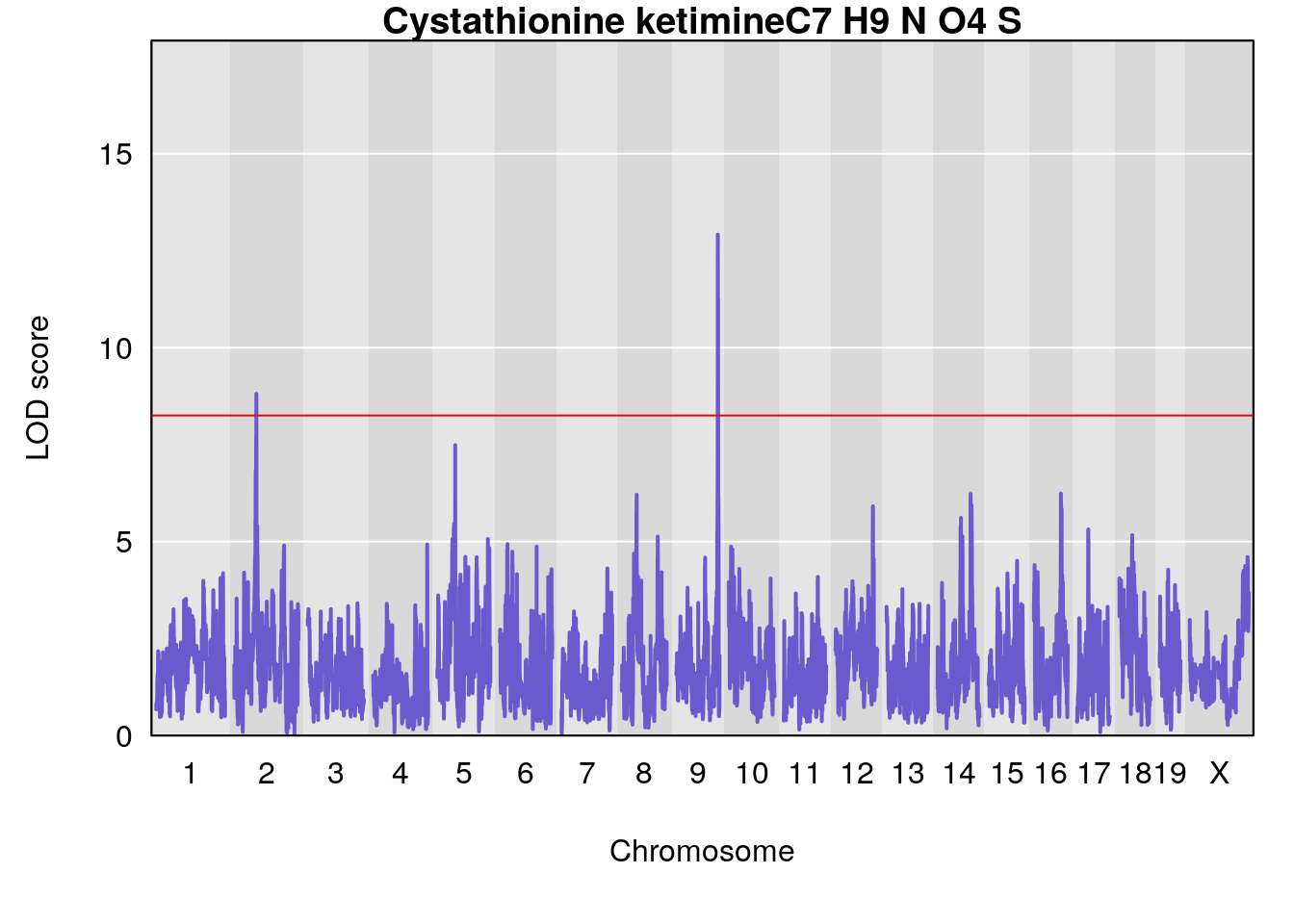
# 154 Cystathionine ketimine C7 H9 N O4 S 1 0.5731433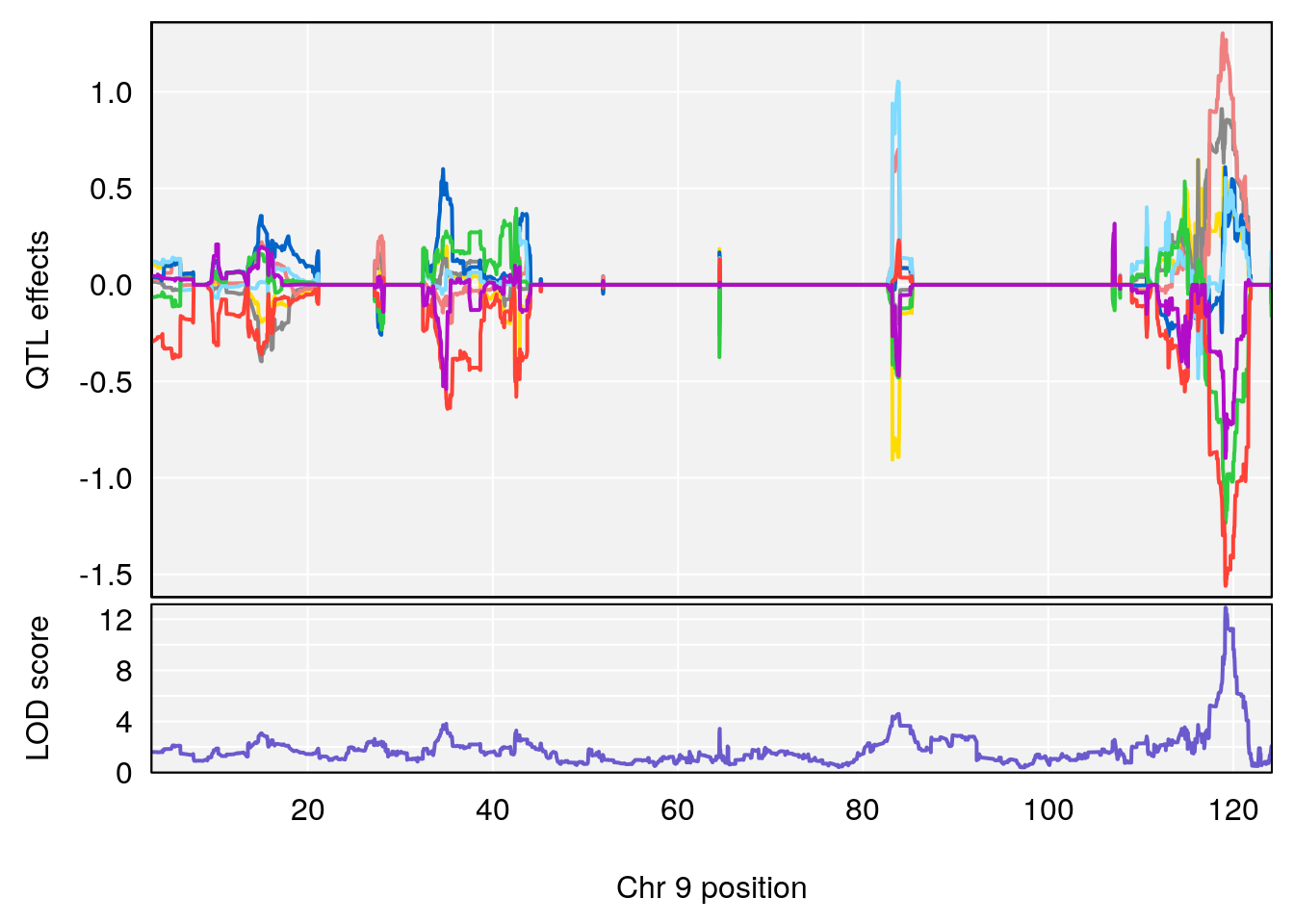
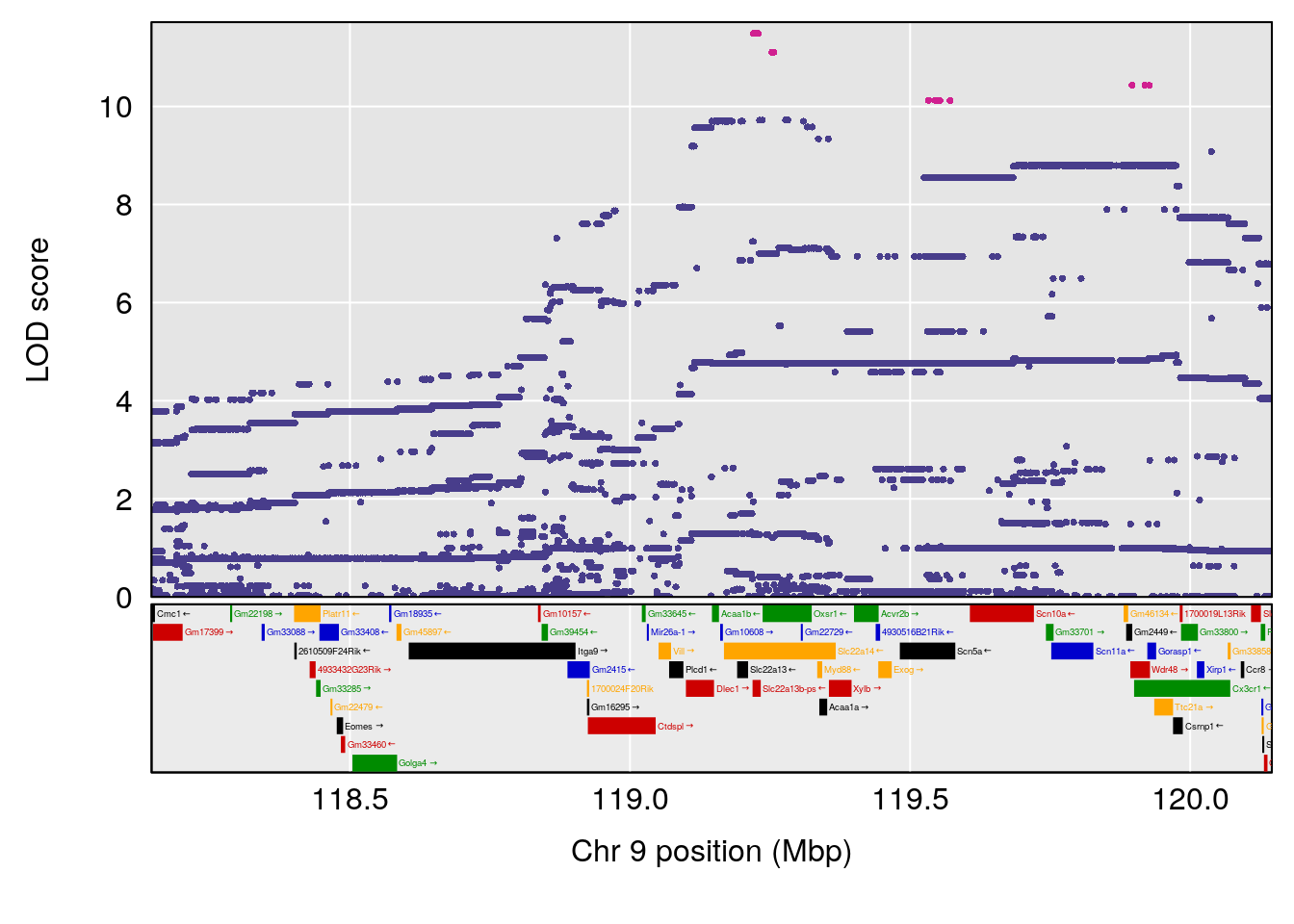
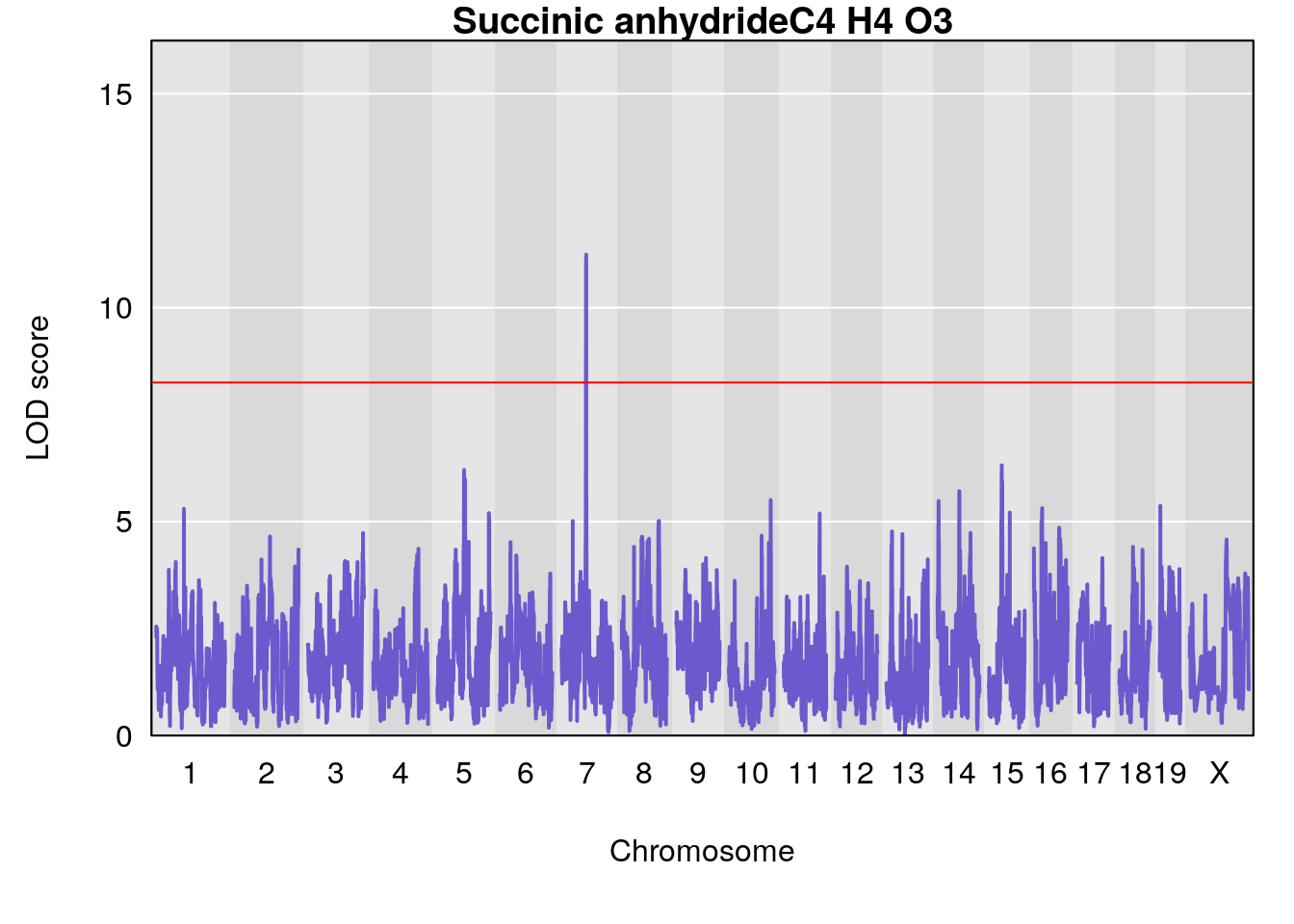
# [1] "Dat1_2005"
# lodindex lodcolumn chr pos lod pvalue fdr ci_lo ci_hi
# 156 2005 Dat1_2005 7 41.113 11.23992 0 0 40.7 41.214
# Name Formula h2 se
# 156 Succinic anhydride C4 H4 O3 0.129272 0.5160077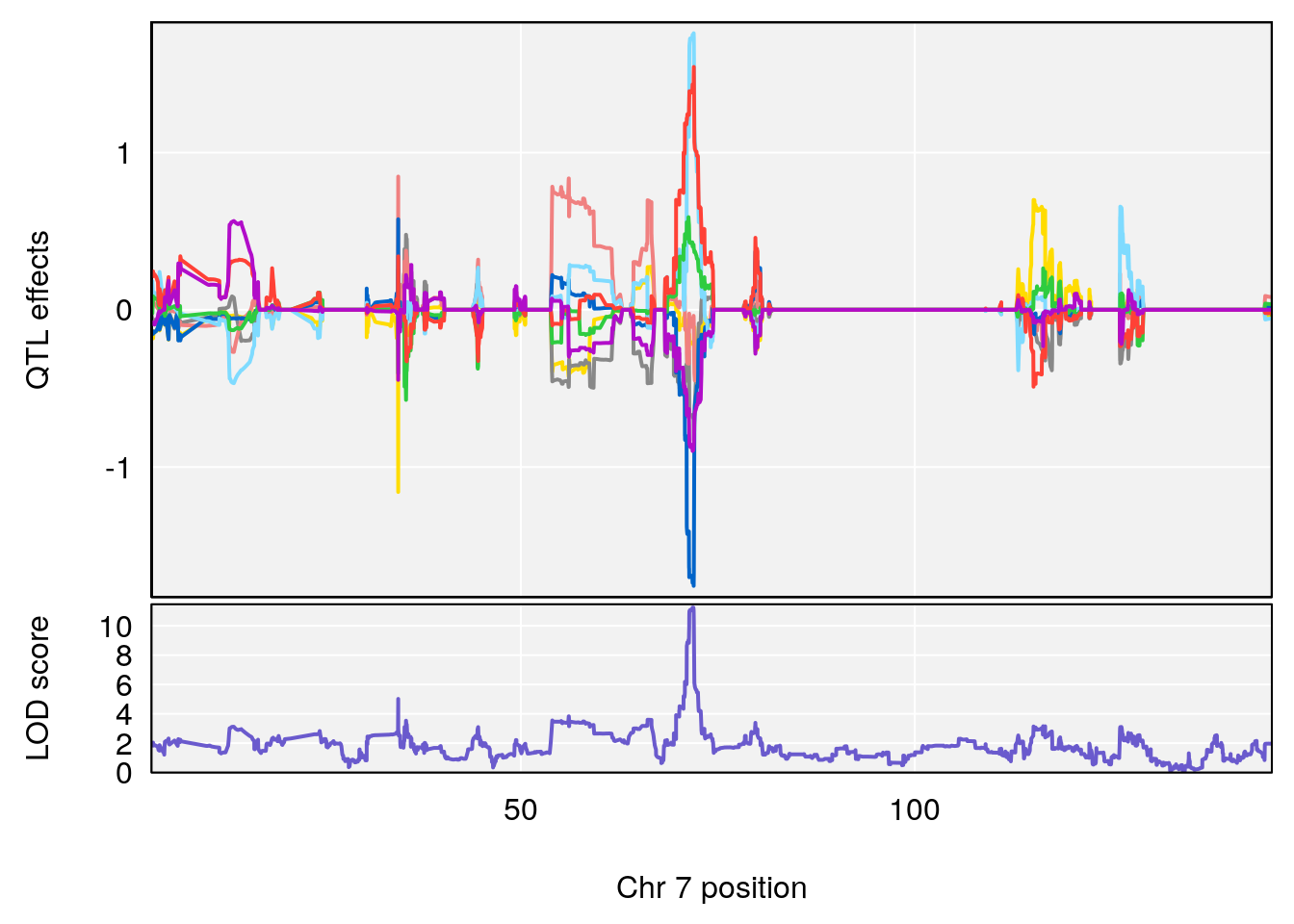
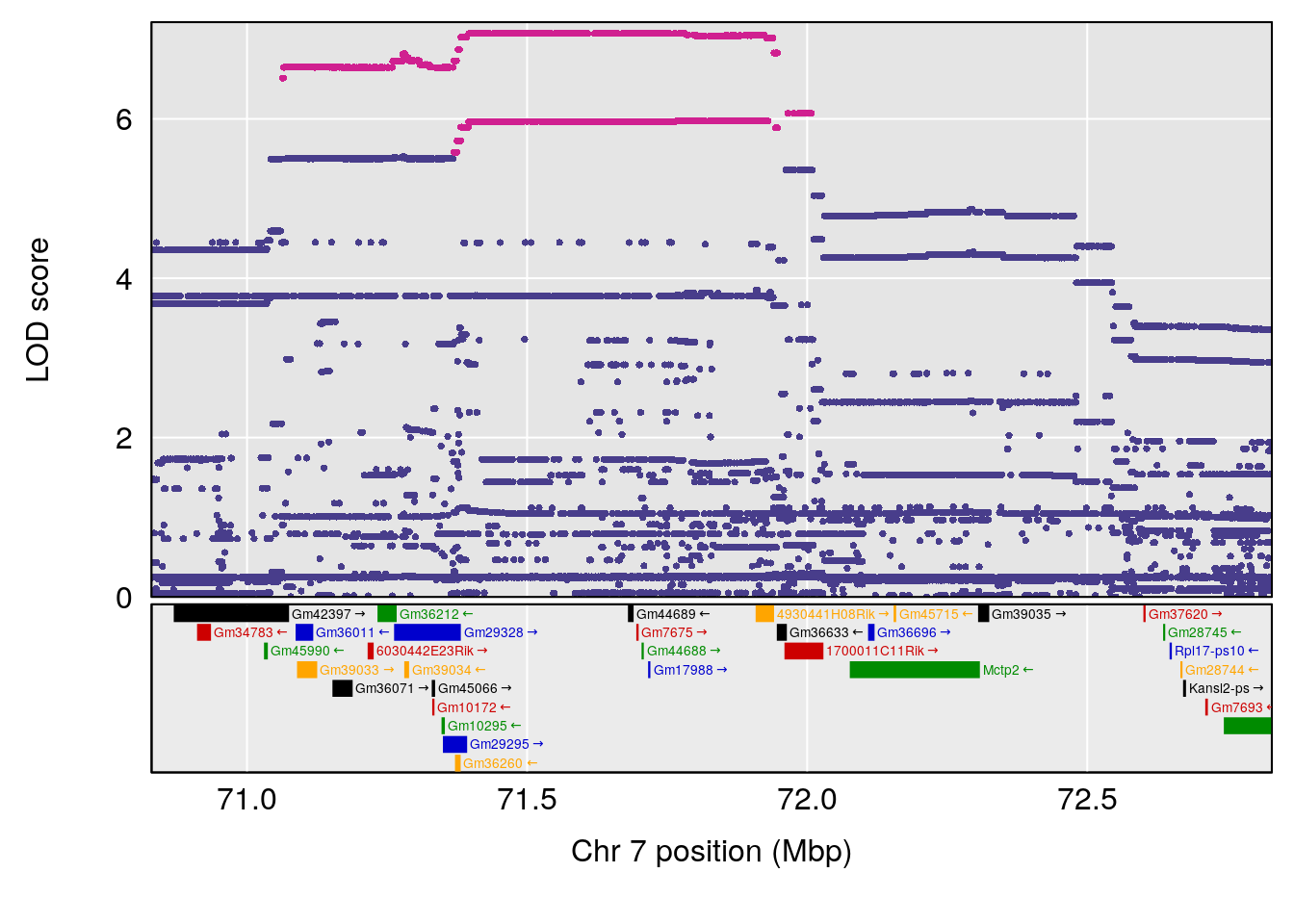
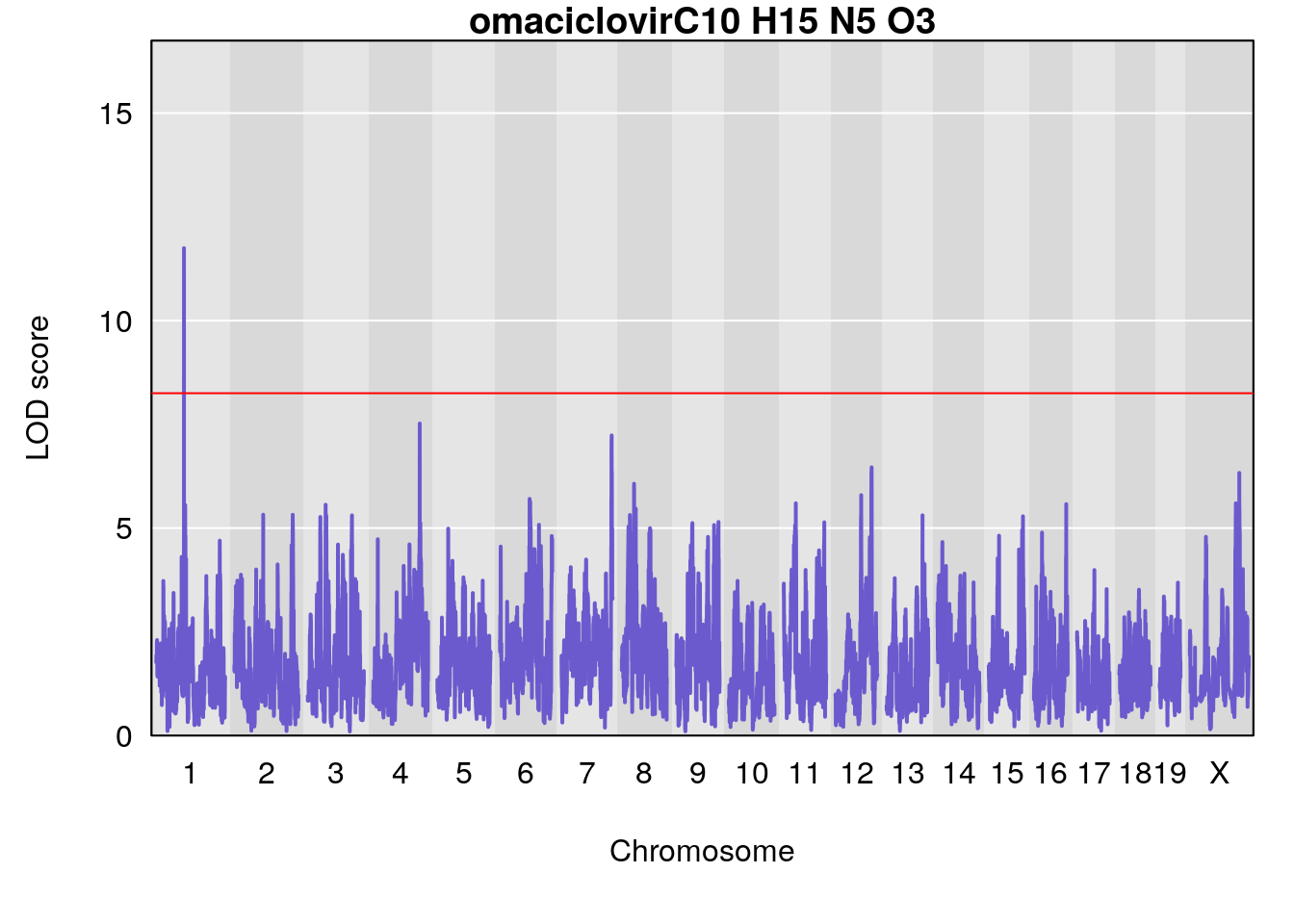
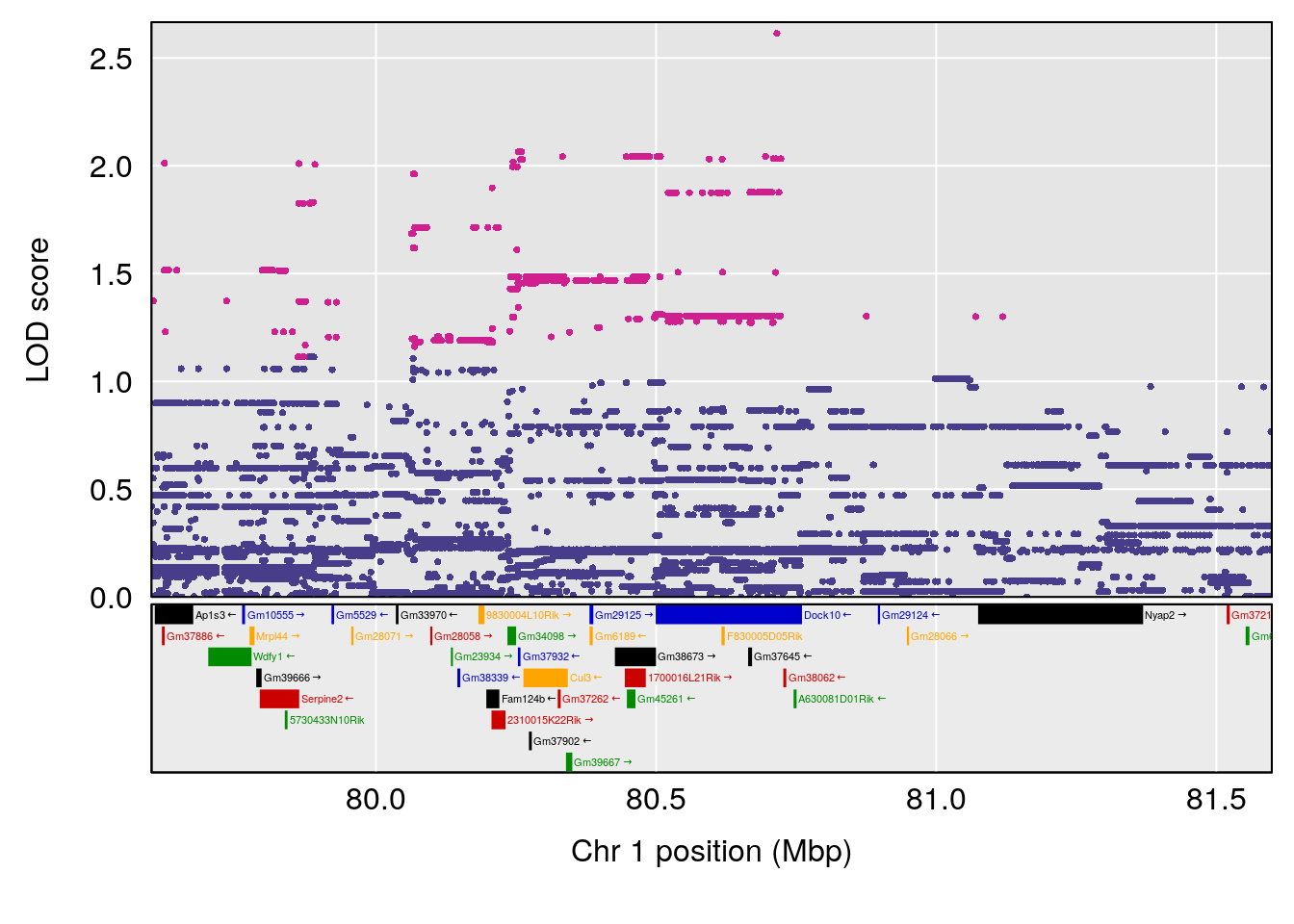
# [1] "Dat1_2487"
# lodindex lodcolumn chr pos lod pvalue fdr ci_lo ci_hi Name
# 196 2487 Dat1_2487 1 41.479 11.74653 0 0 41.451 41.494 omaciclovir
# Formula h2 se
# 196 C10 H15 N5 O3 1 0.5731433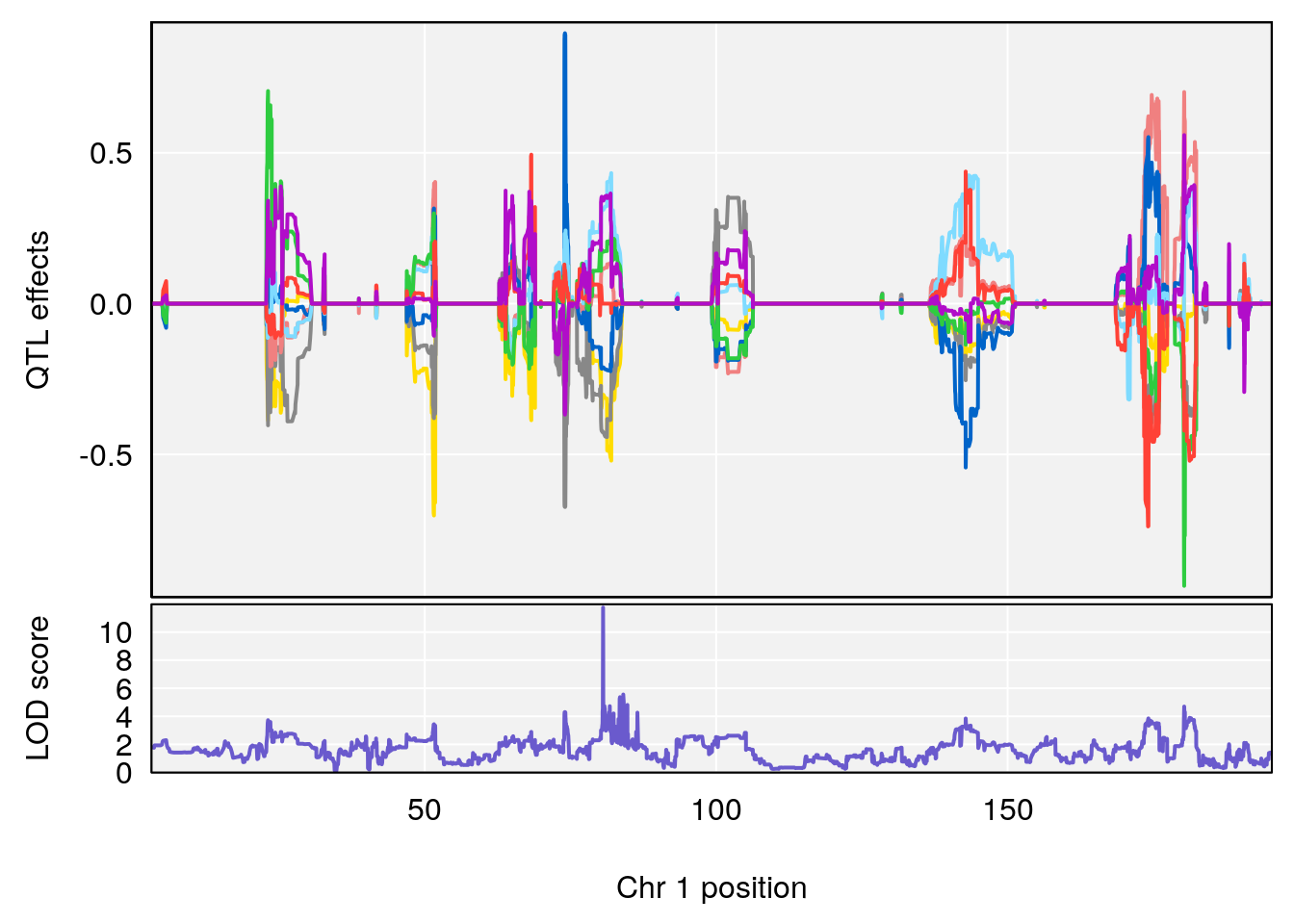
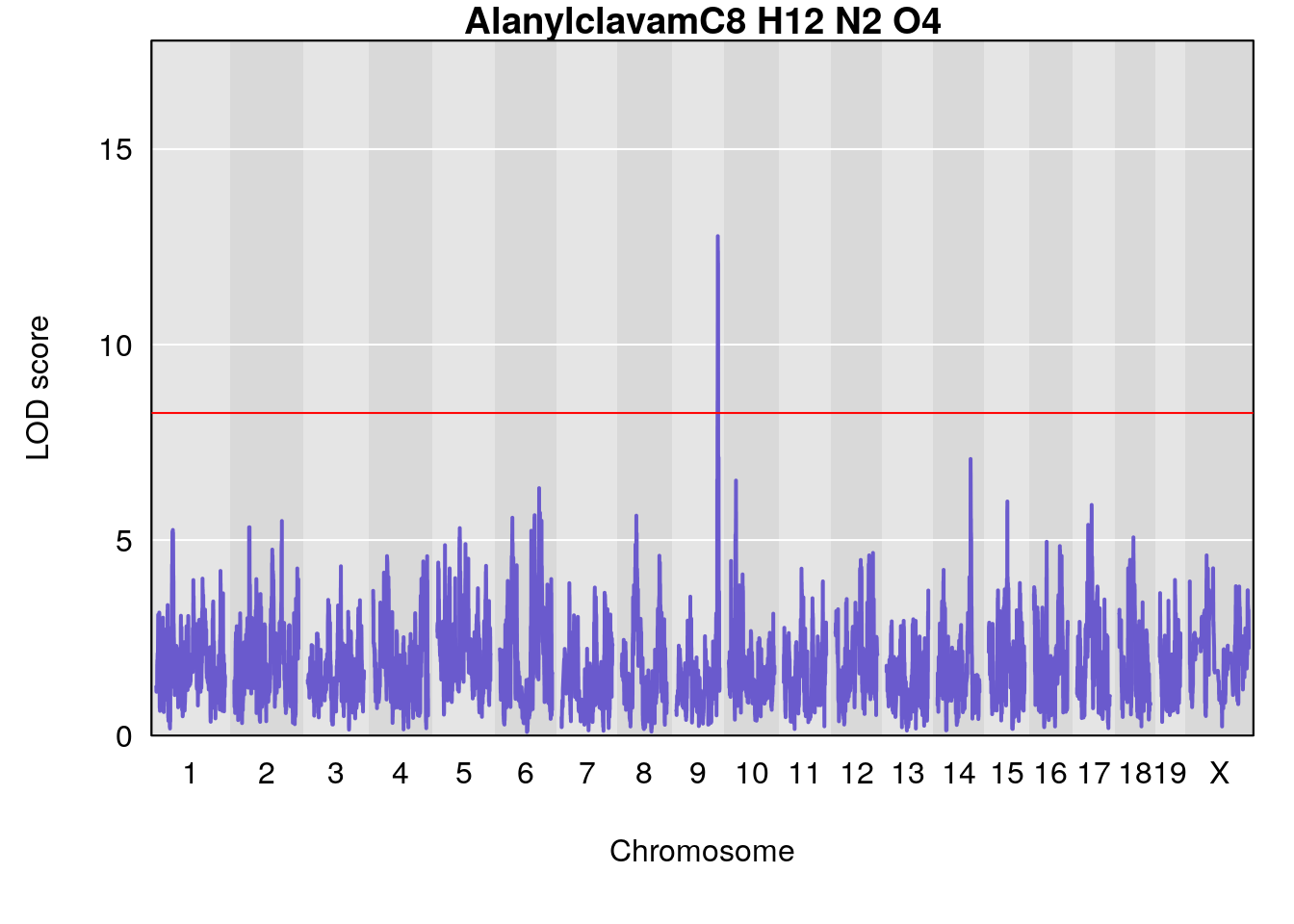
# [1] "Dat1_3046"
# lodindex lodcolumn chr pos lod pvalue fdr ci_lo ci_hi Name
# 227 3046 Dat1_3046 9 71.322 12.77517 0 0 71.32 71.403 Alanylclavam
# Formula h2 se
# 227 C8 H12 N2 O4 1 0.5731433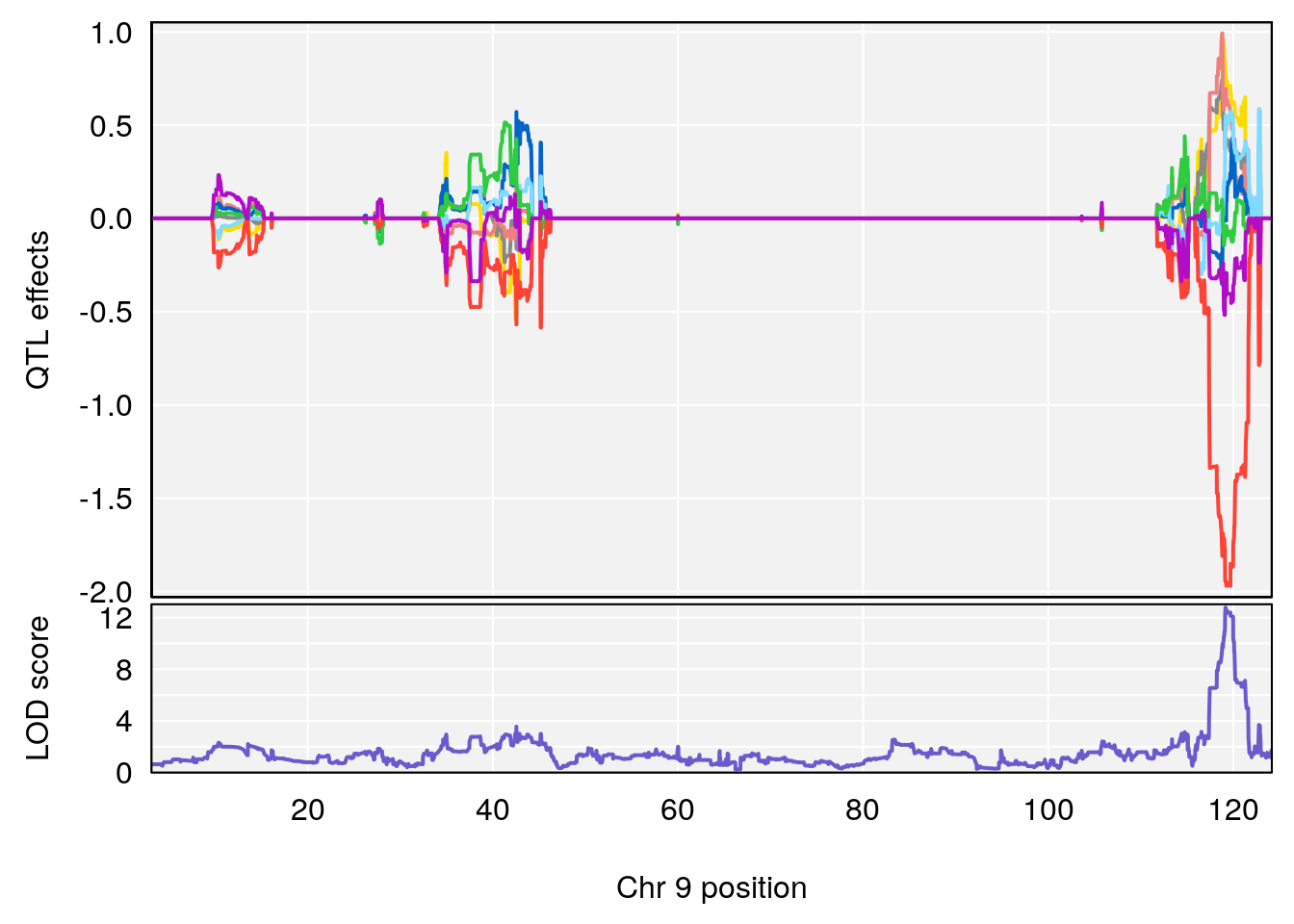
# [1] "Dat1_3099"
# lodindex lodcolumn chr pos lod pvalue fdr ci_lo ci_hi
# 229 3099 Dat1_3099 1 81.375 10.43377 0 0 80.938 81.407
# Name
# 229 2-[(2Z,4E,6E,8E,10E,12E)-14-Hydroxy-6,11-dimethyl-2,4,6,8,10,12-tetradecahexaen-2-yl]-4,4,7a-trimethyl-2,4,5,6,7,7a-hexahydro-1-benzofuran-6-ol
# Formula h2 se
# 229 C27 H38 O3 1 0.5731433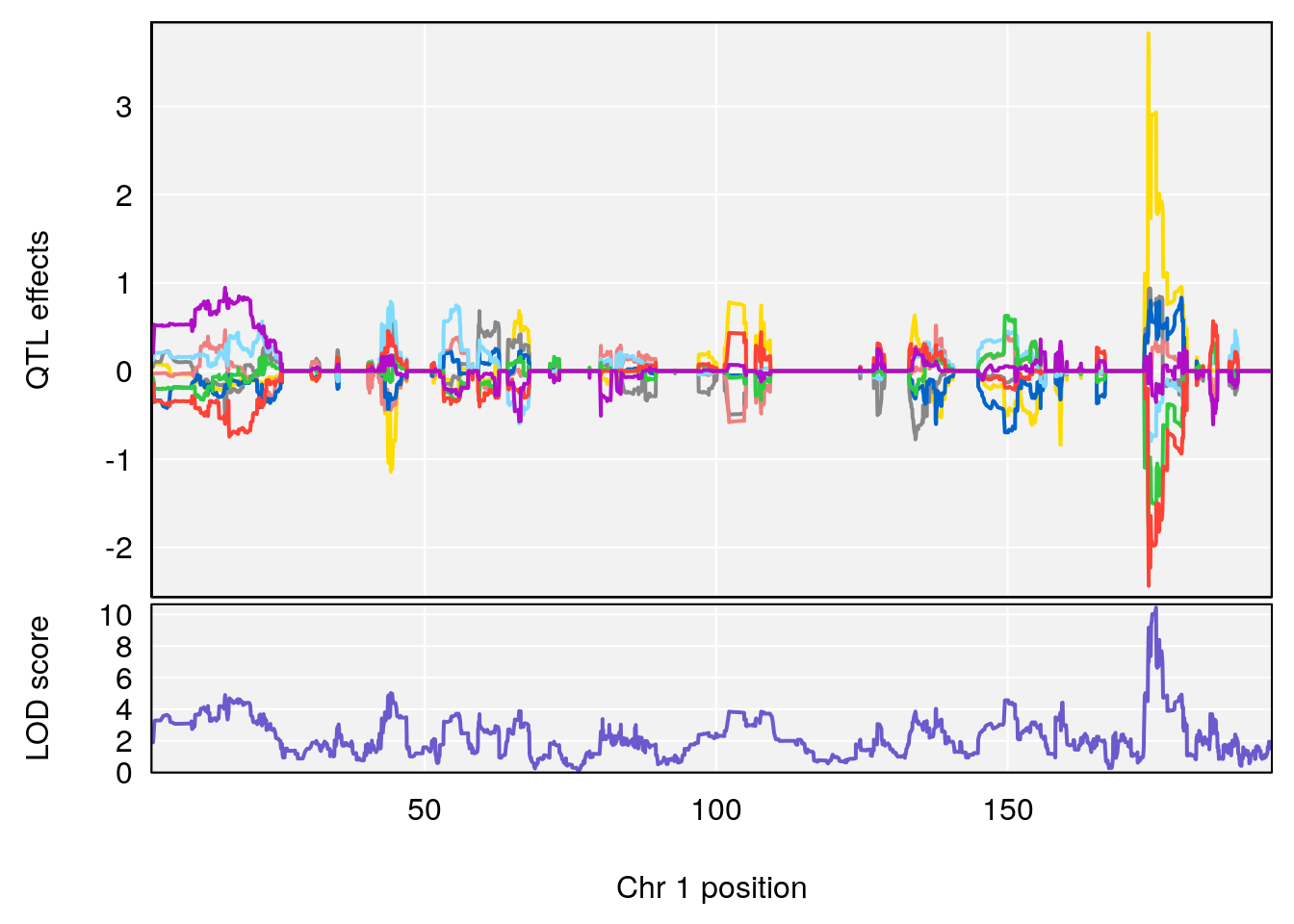
# [1] "Dat1_3271"
# lodindex lodcolumn chr pos lod pvalue fdr ci_lo ci_hi Name
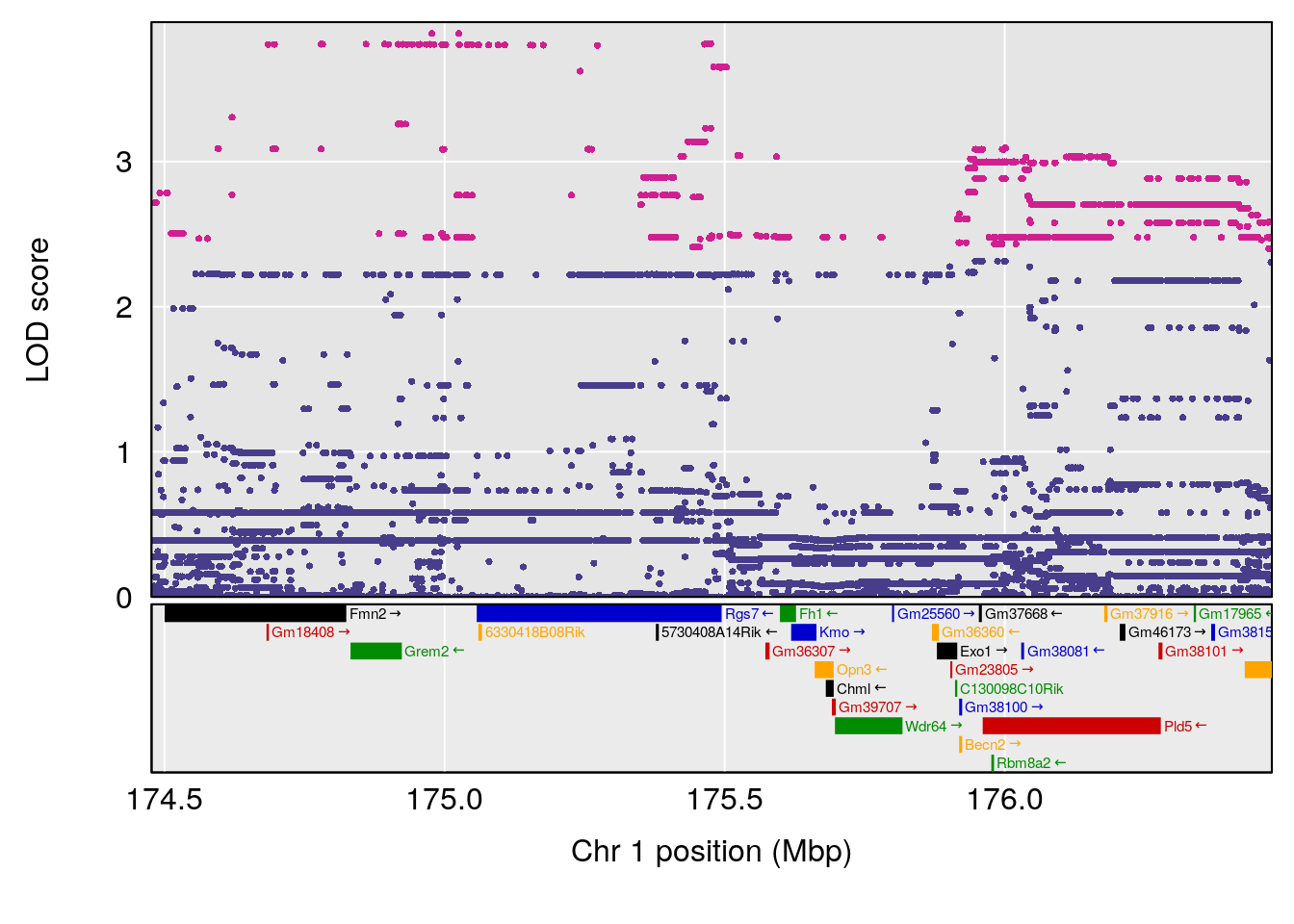
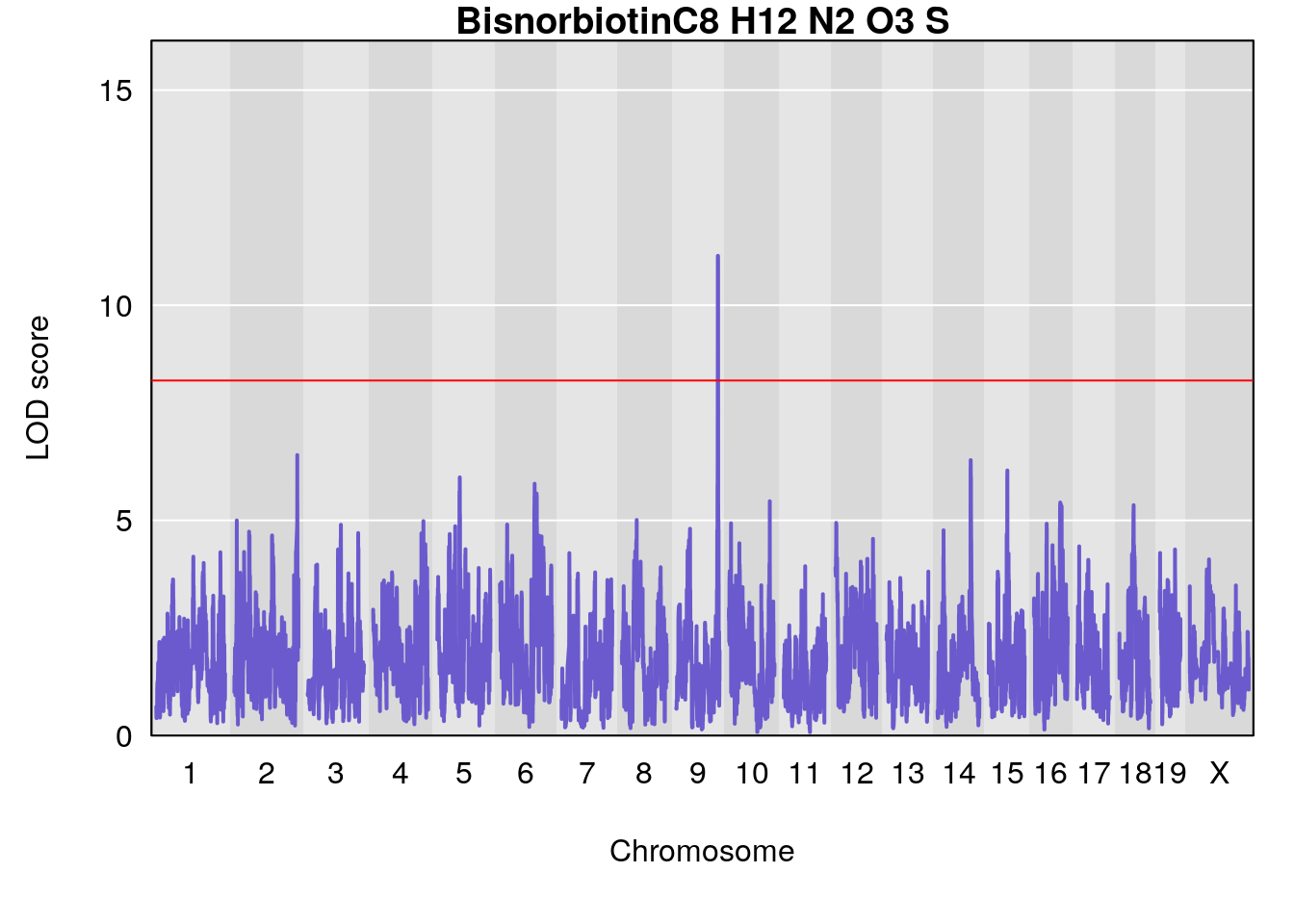
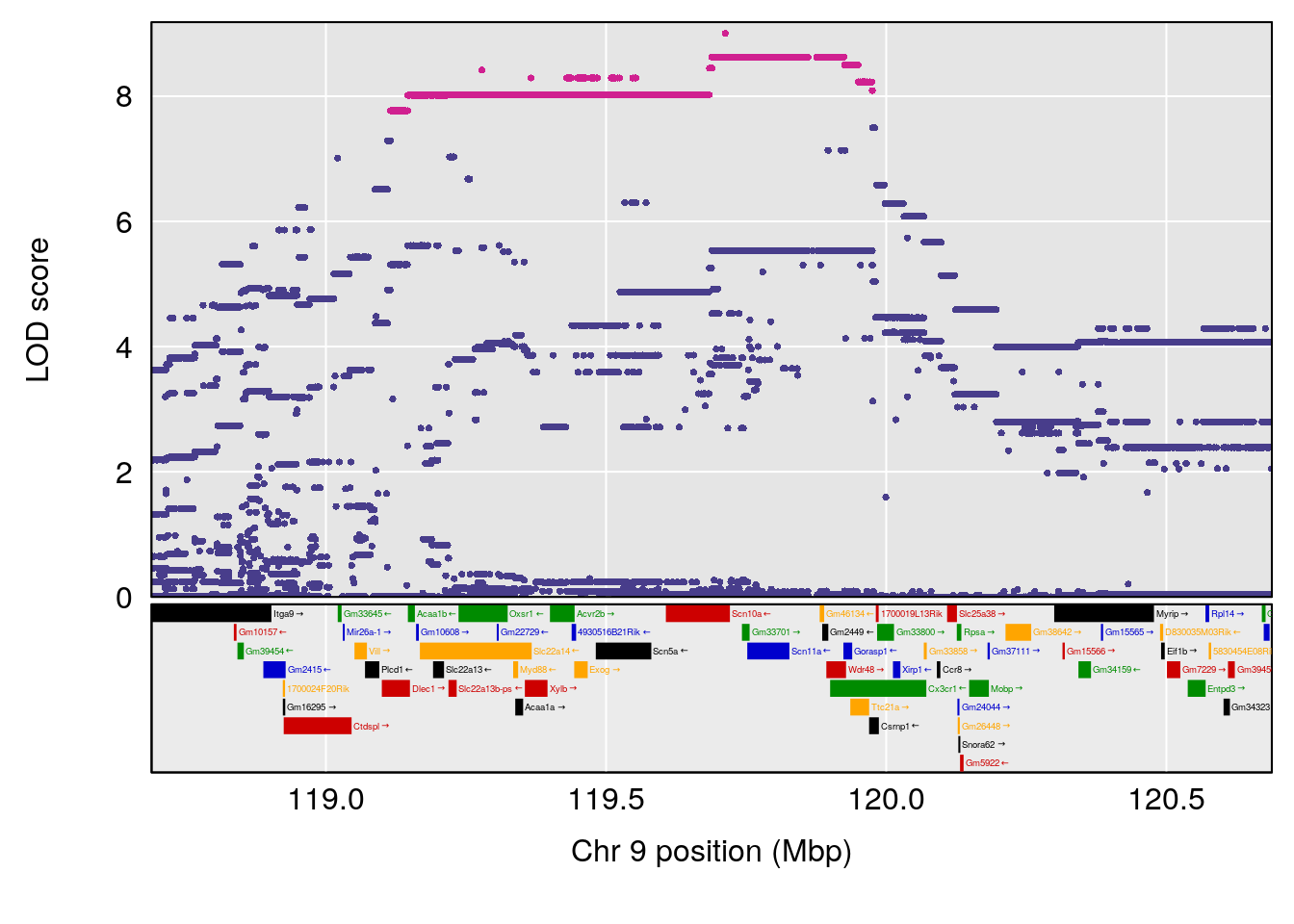
# 238 3271 Dat1_3271 9 71.342 11.14863 0 0 71.32 71.403 Bisnorbiotin
# Formula h2 se
# 238 C8 H12 N2 O3 S 1 0.5731433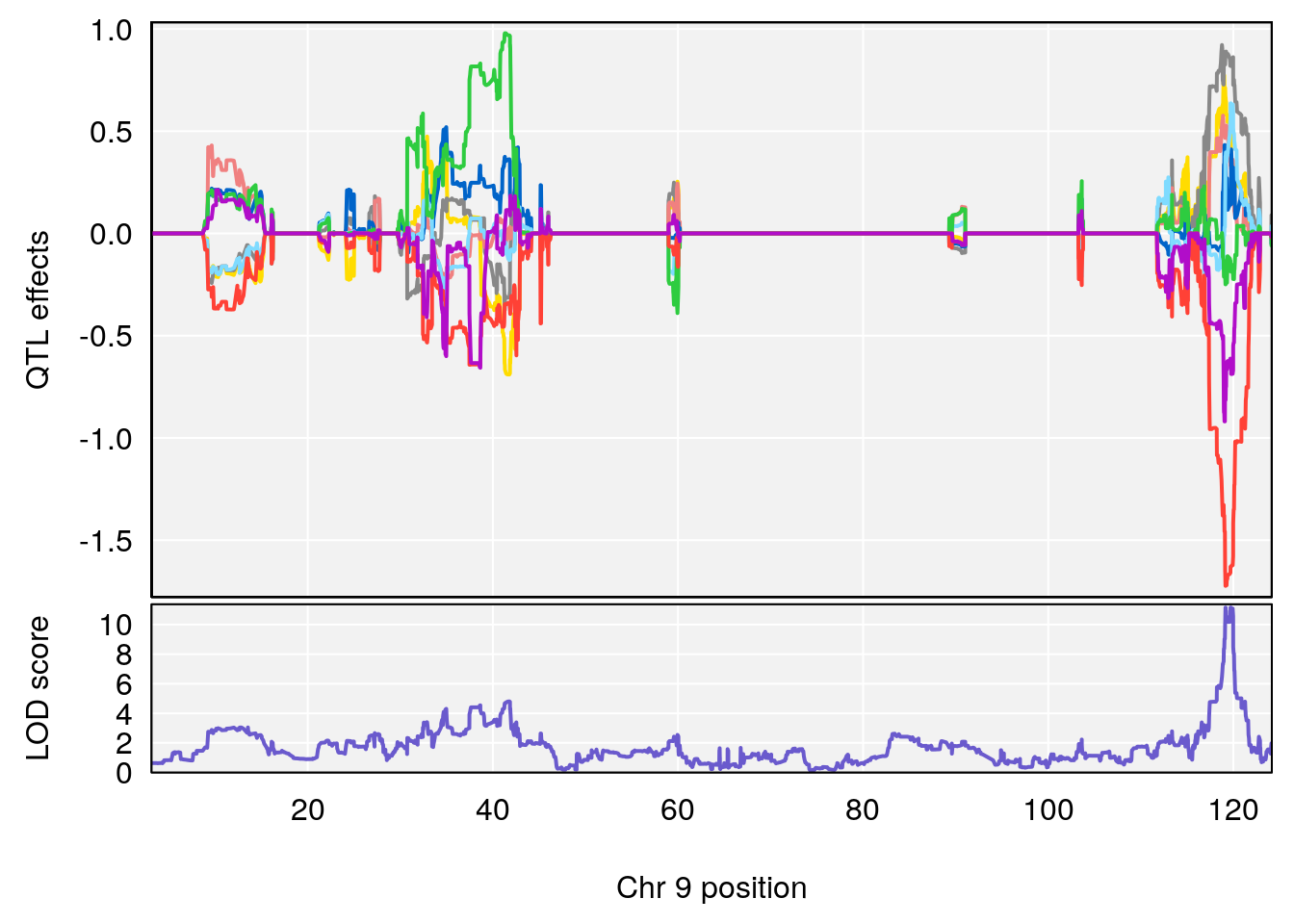
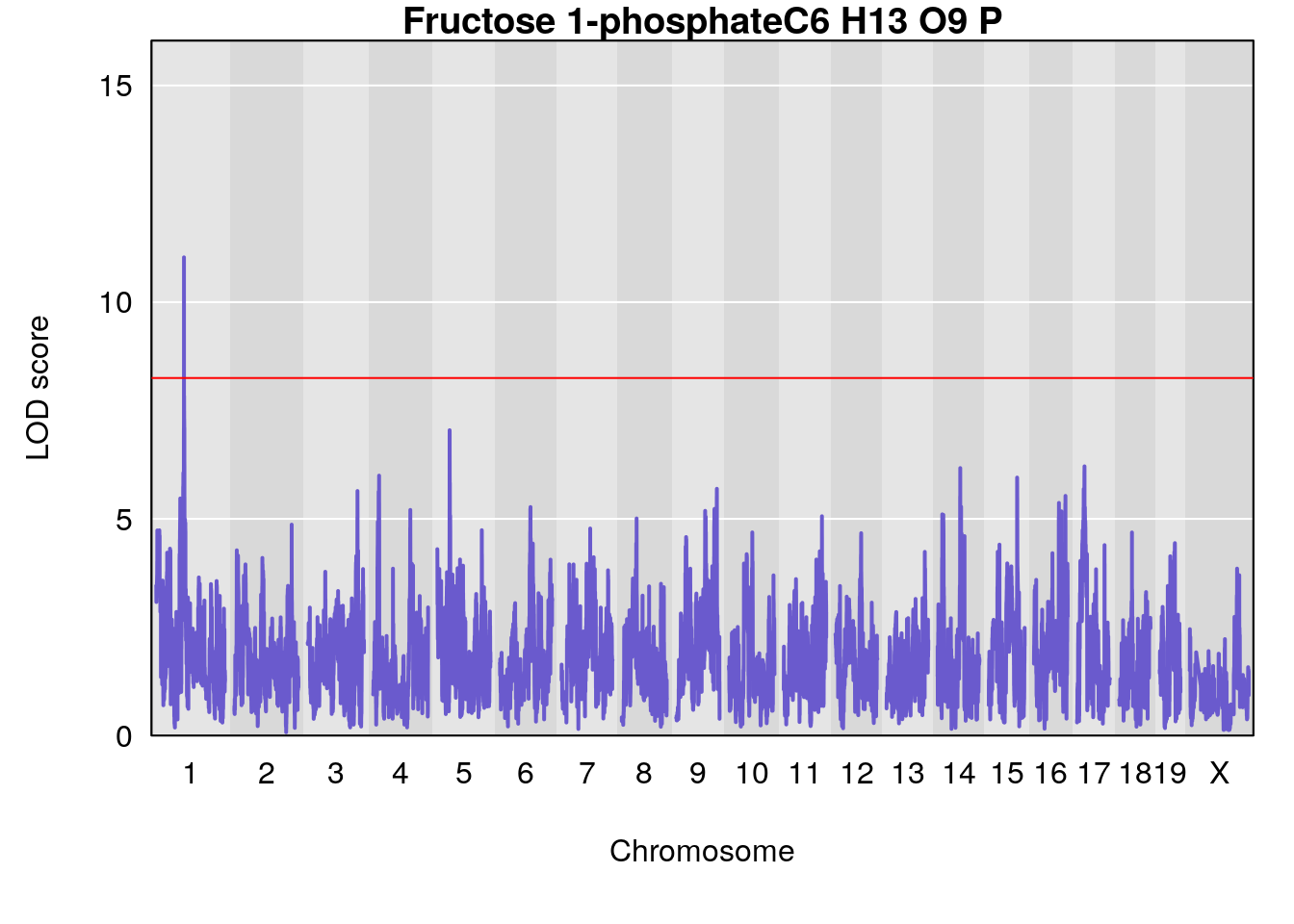
# [1] "Dat1_3325"
# lodindex lodcolumn chr pos lod pvalue fdr ci_lo ci_hi
# 240 3325 Dat1_3325 1 41.479 11.03427 0 0 41.451 41.494
# Name Formula h2 se
# 240 Fructose 1-phosphate C6 H13 O9 P 1 0.5731433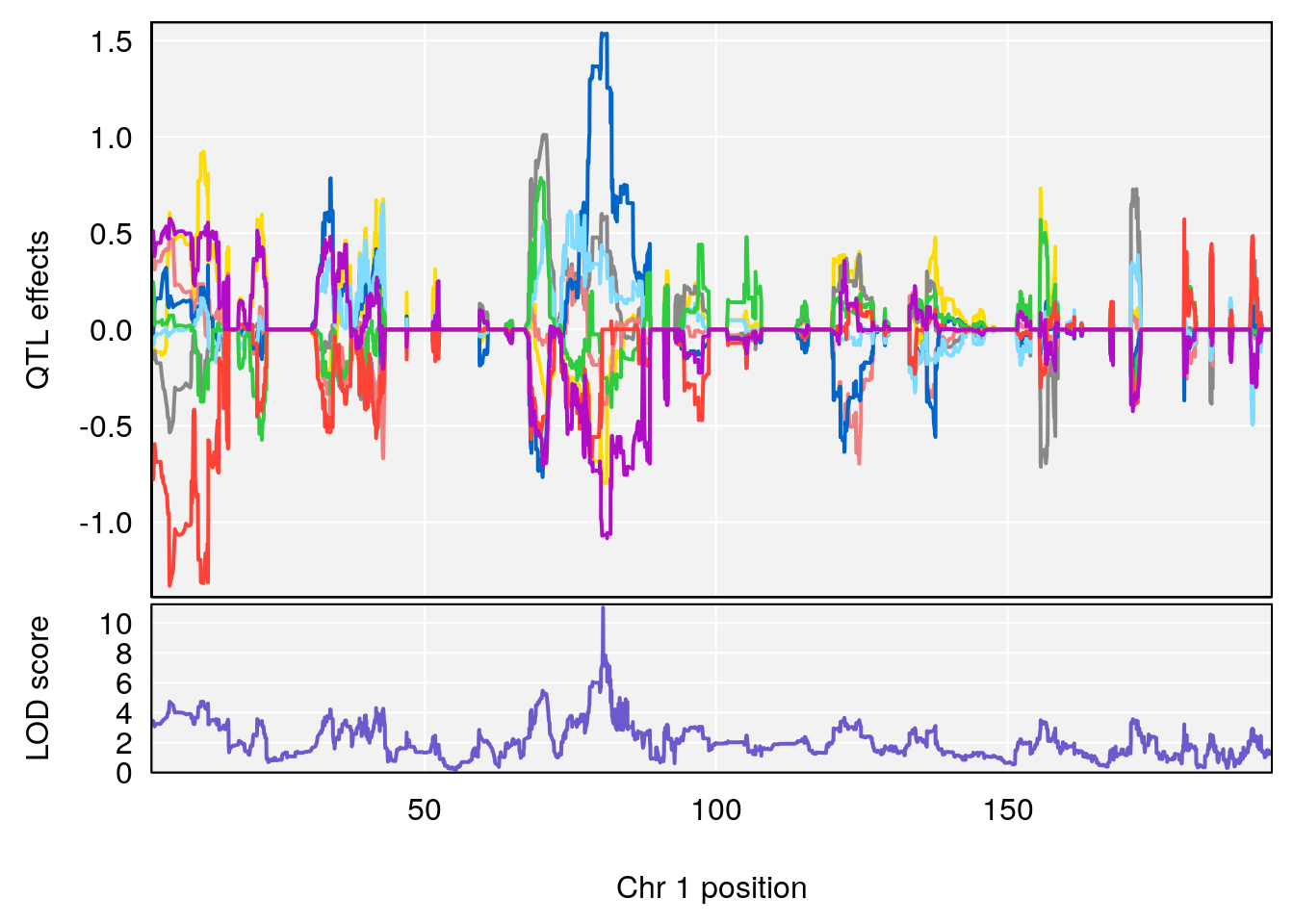
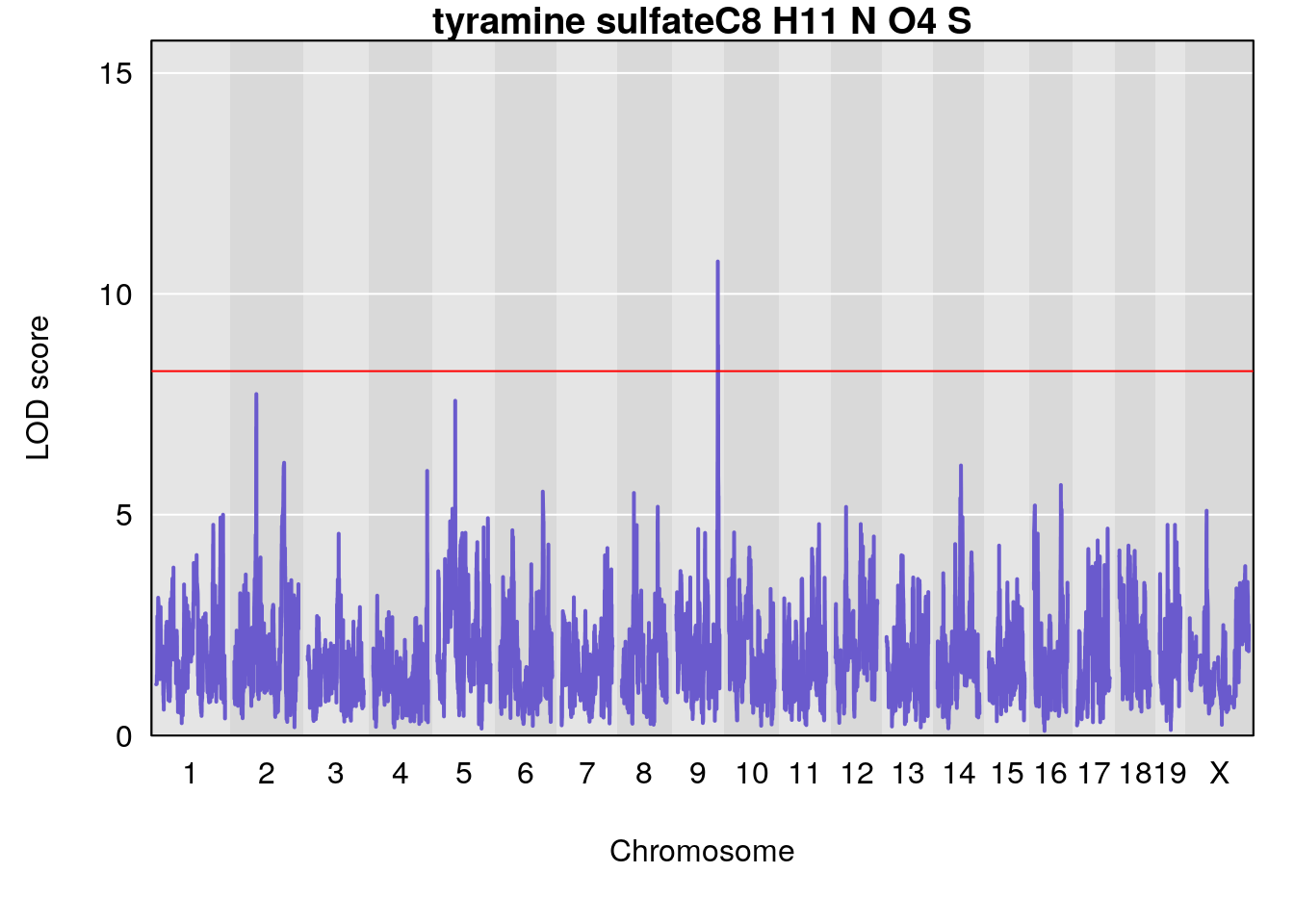
# [1] "Dat1_3439"
# lodindex lodcolumn chr pos lod pvalue fdr ci_lo ci_hi
# 248 3439 Dat1_3439 9 71.322 10.73324 0 0 71.32 71.327
# Name Formula h2 se
# 248 tyramine sulfate C8 H11 N O4 S 1 0.5731433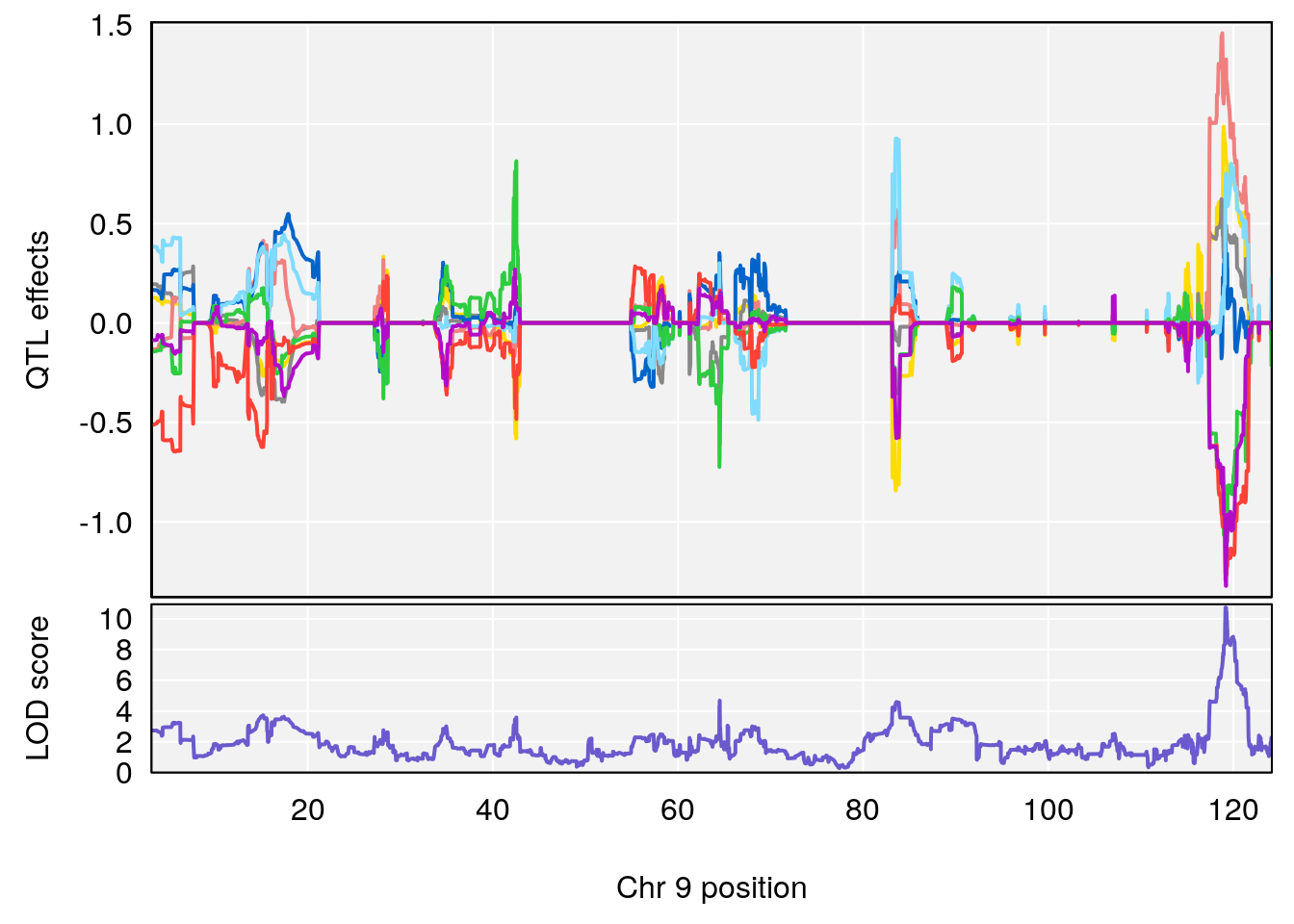
# [1] "Dat1_4031"
# lodindex lodcolumn chr pos lod pvalue fdr ci_lo ci_hi
# 290 4031 Dat1_4031 17 51.763 10.50568 0 0 51.458 52.617
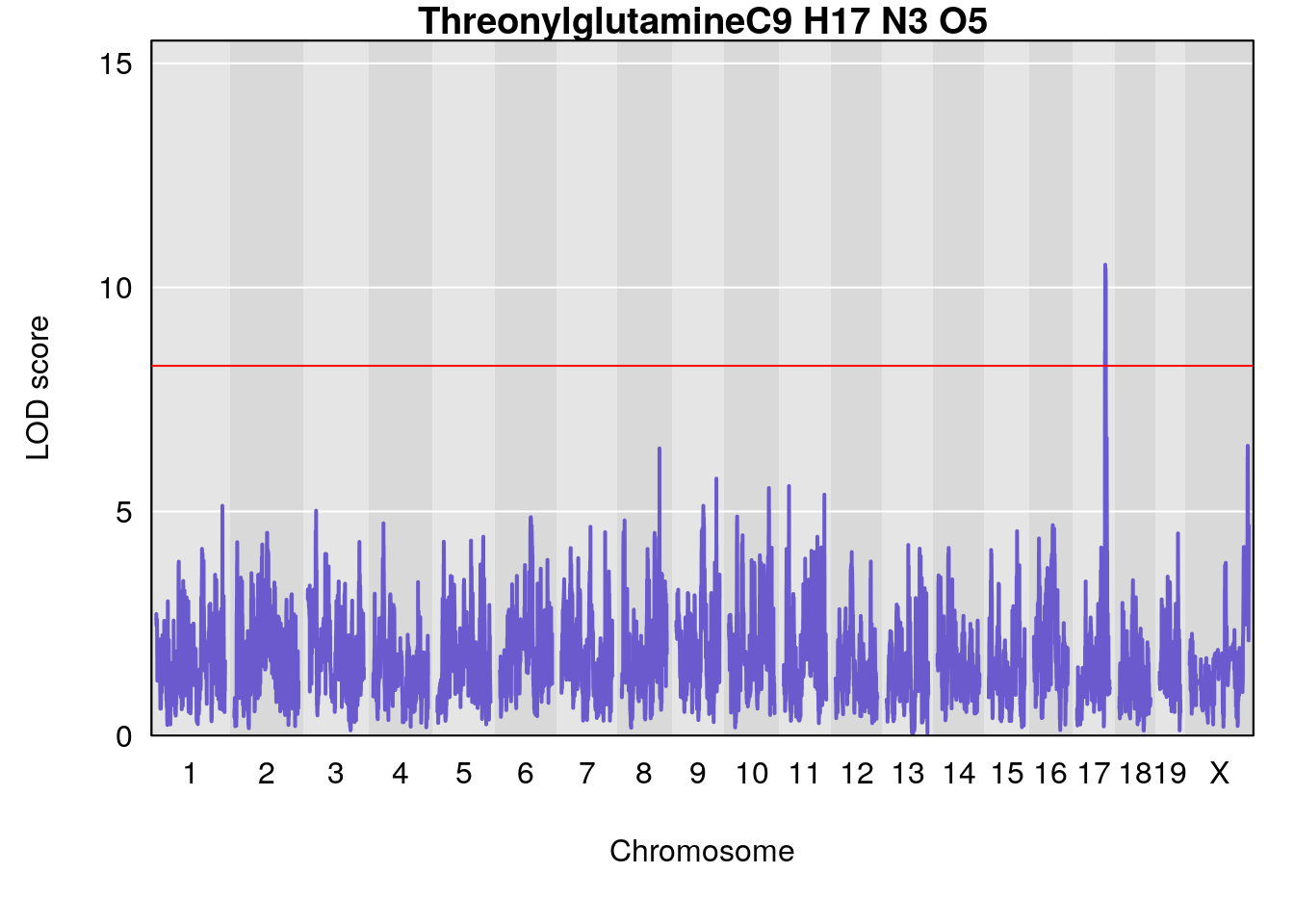
# Name Formula h2 se
# 290 Threonylglutamine C9 H17 N3 O5 1 0.5731433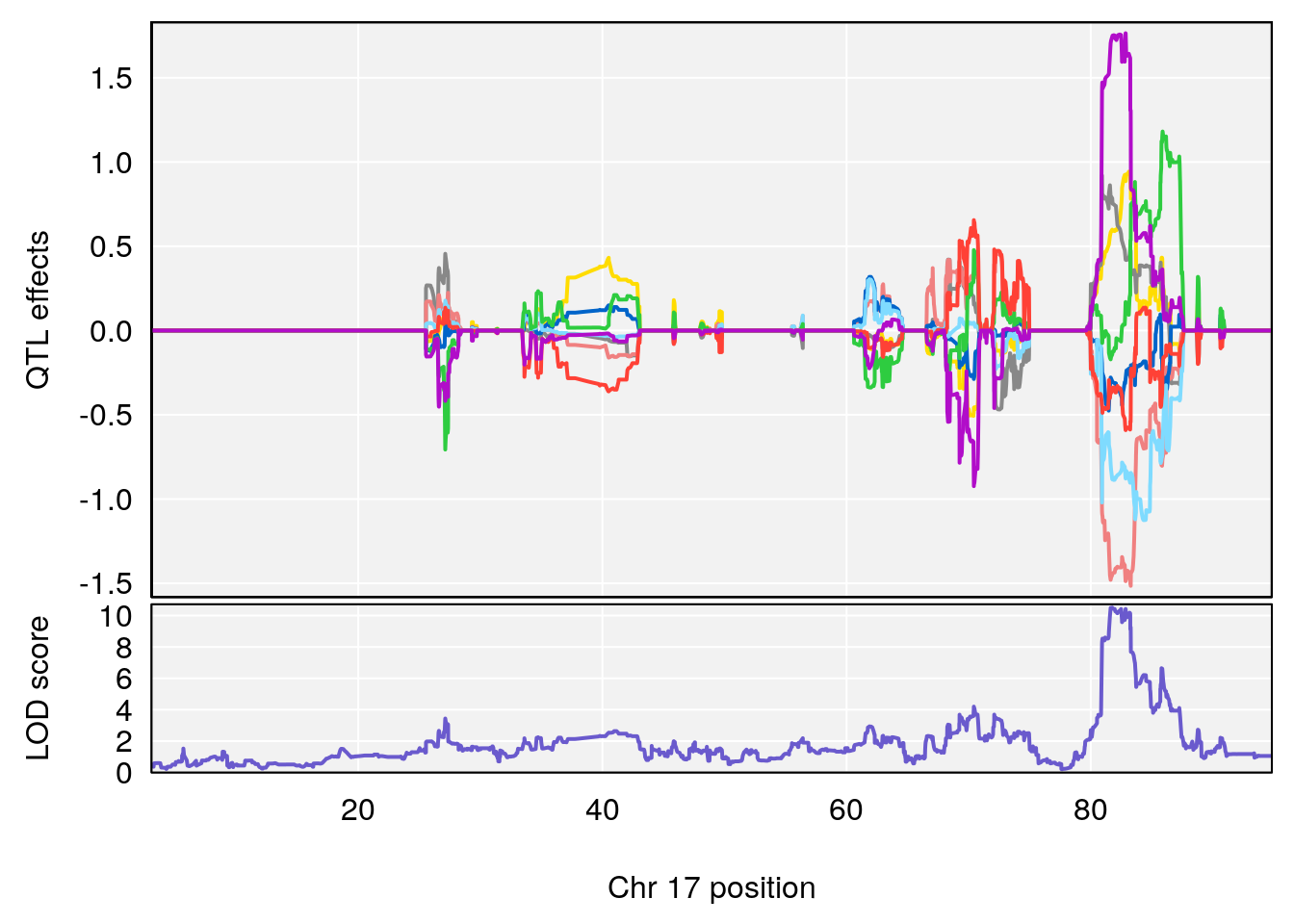
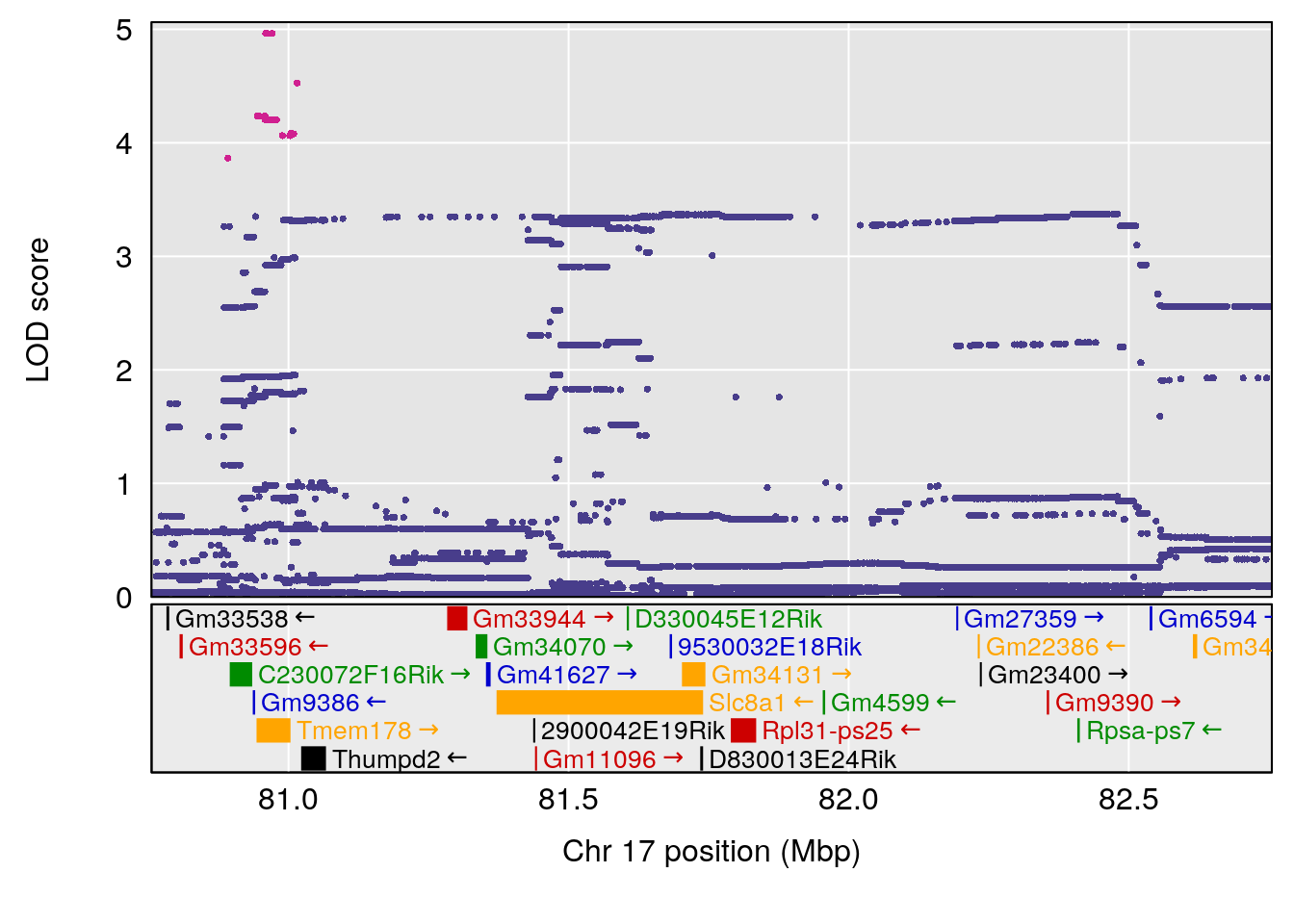
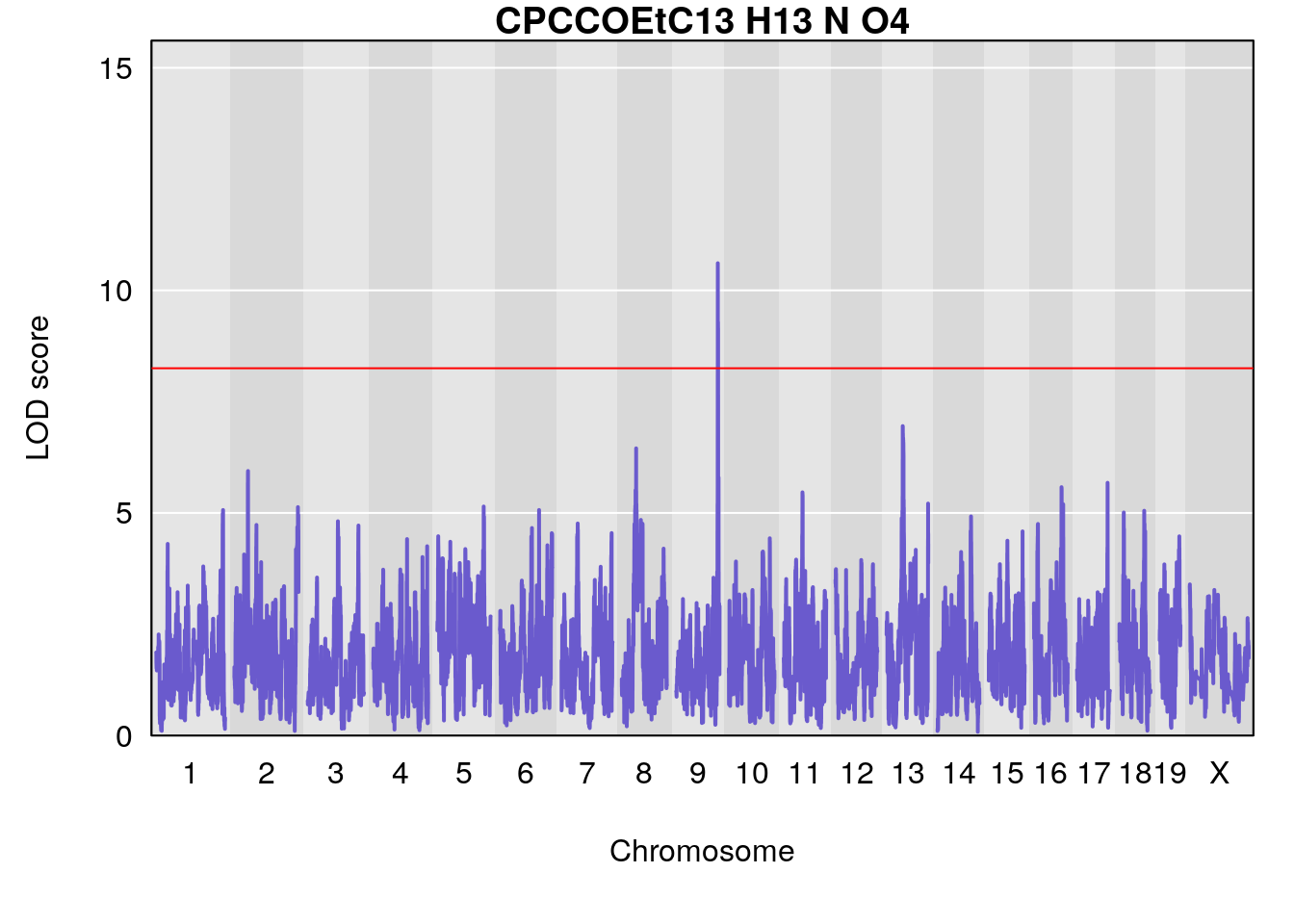
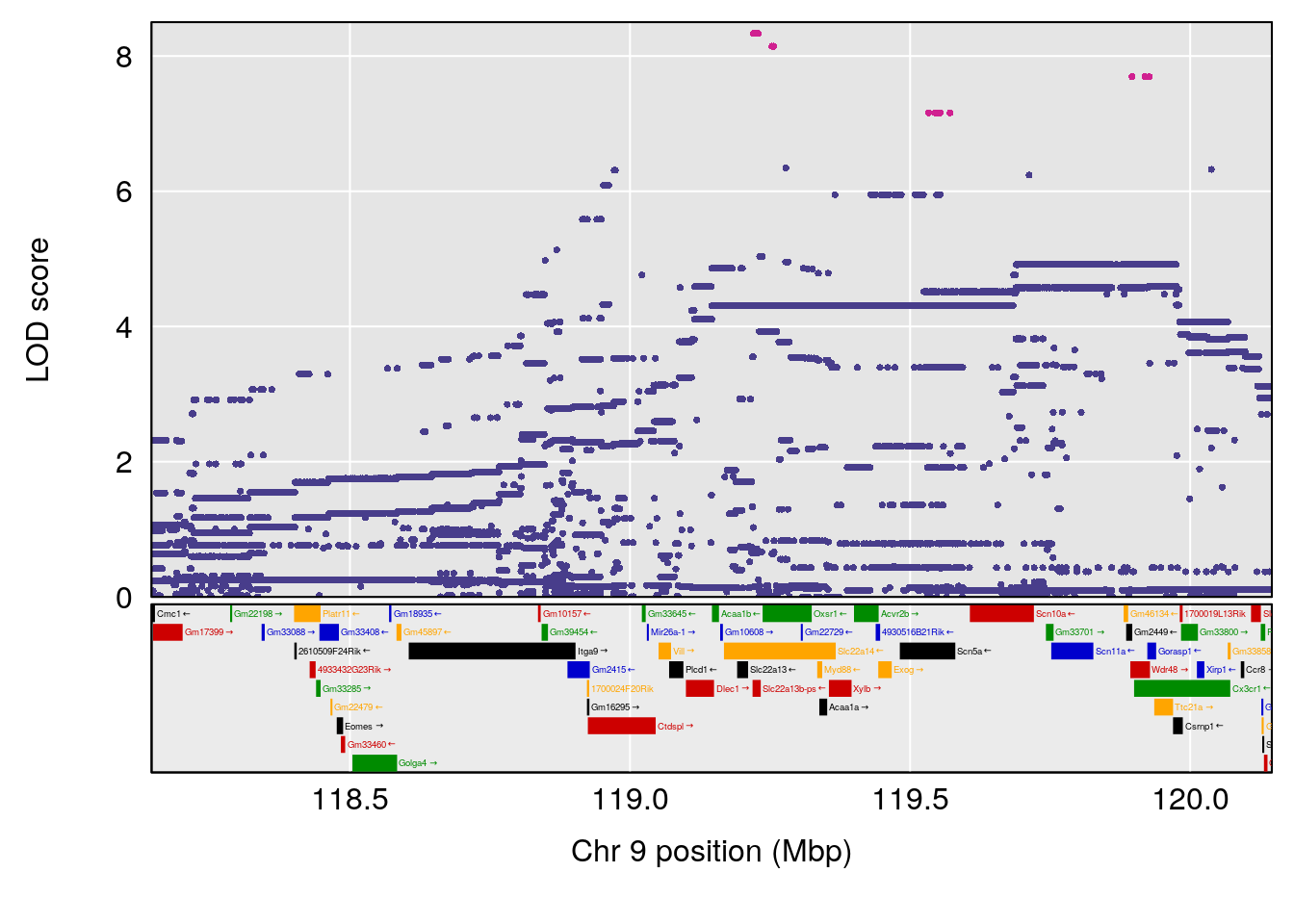
# [1] "Dat1_4112"
# lodindex lodcolumn chr pos lod pvalue fdr ci_lo ci_hi Name
# 295 4112 Dat1_4112 9 71.322 10.61034 0 0 71.32 71.403 CPCCOEt
# Formula h2 se
# 295 C13 H13 N O4 0.2104277 0.558635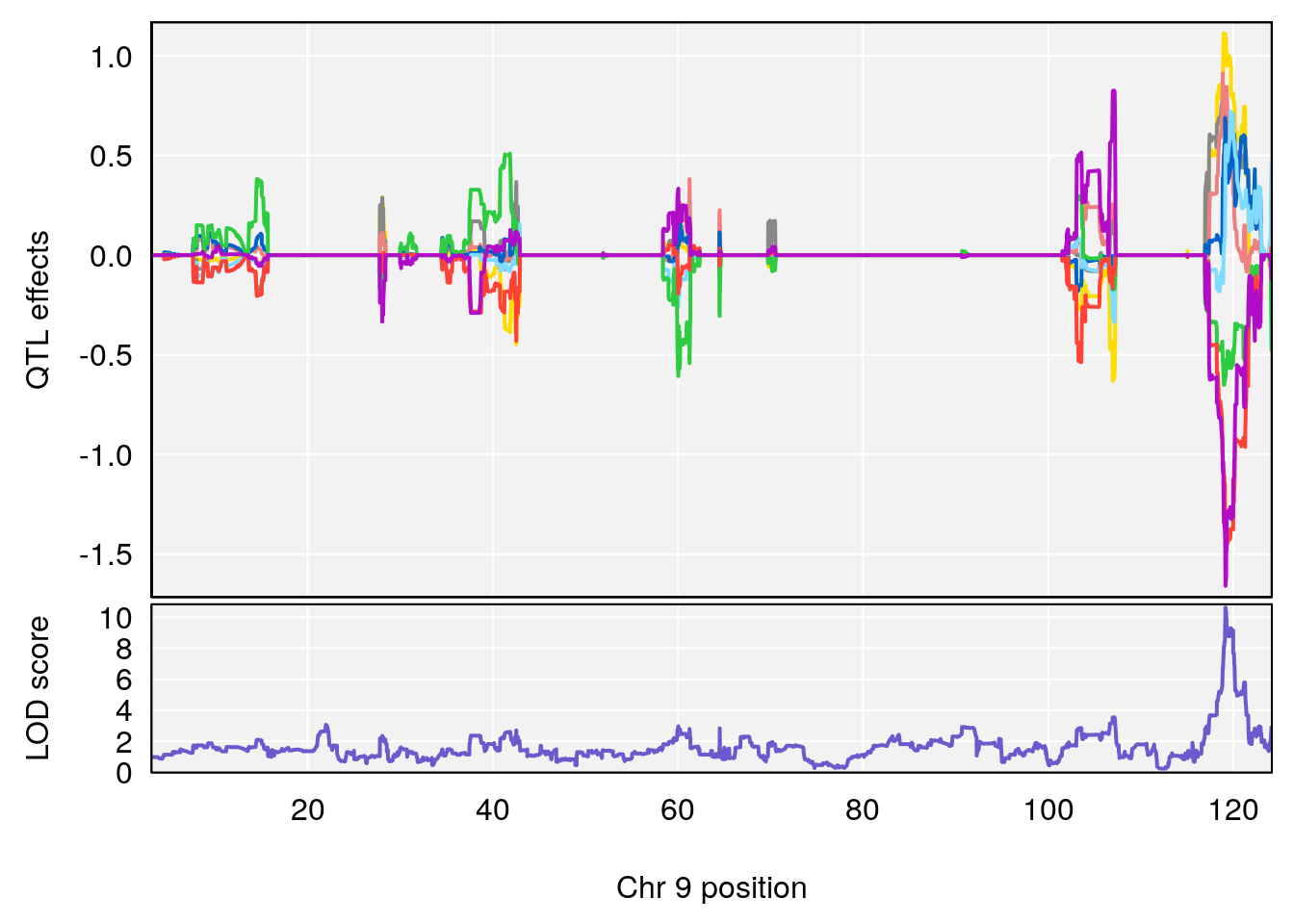
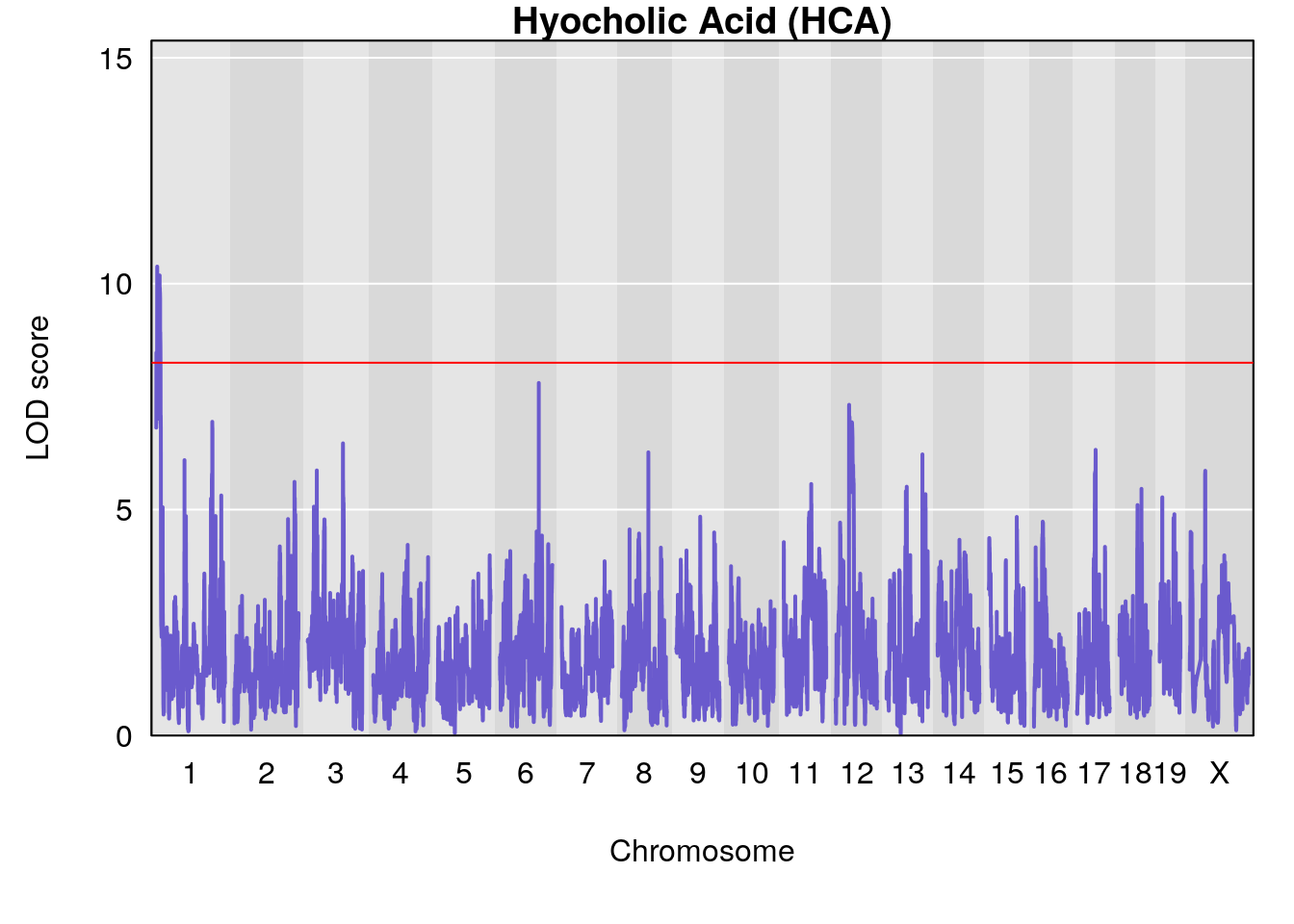
# [1] "Dat2_30"
# lodindex lodcolumn chr pos lod pvalue fdr ci_lo ci_hi
# 341 4744 Dat2_30 1 1.893 10.37835 0 0 1.879 4.439
# Name Formula h2 se
# 341 Hyocholic Acid (HCA) 0.1034566 0.5000092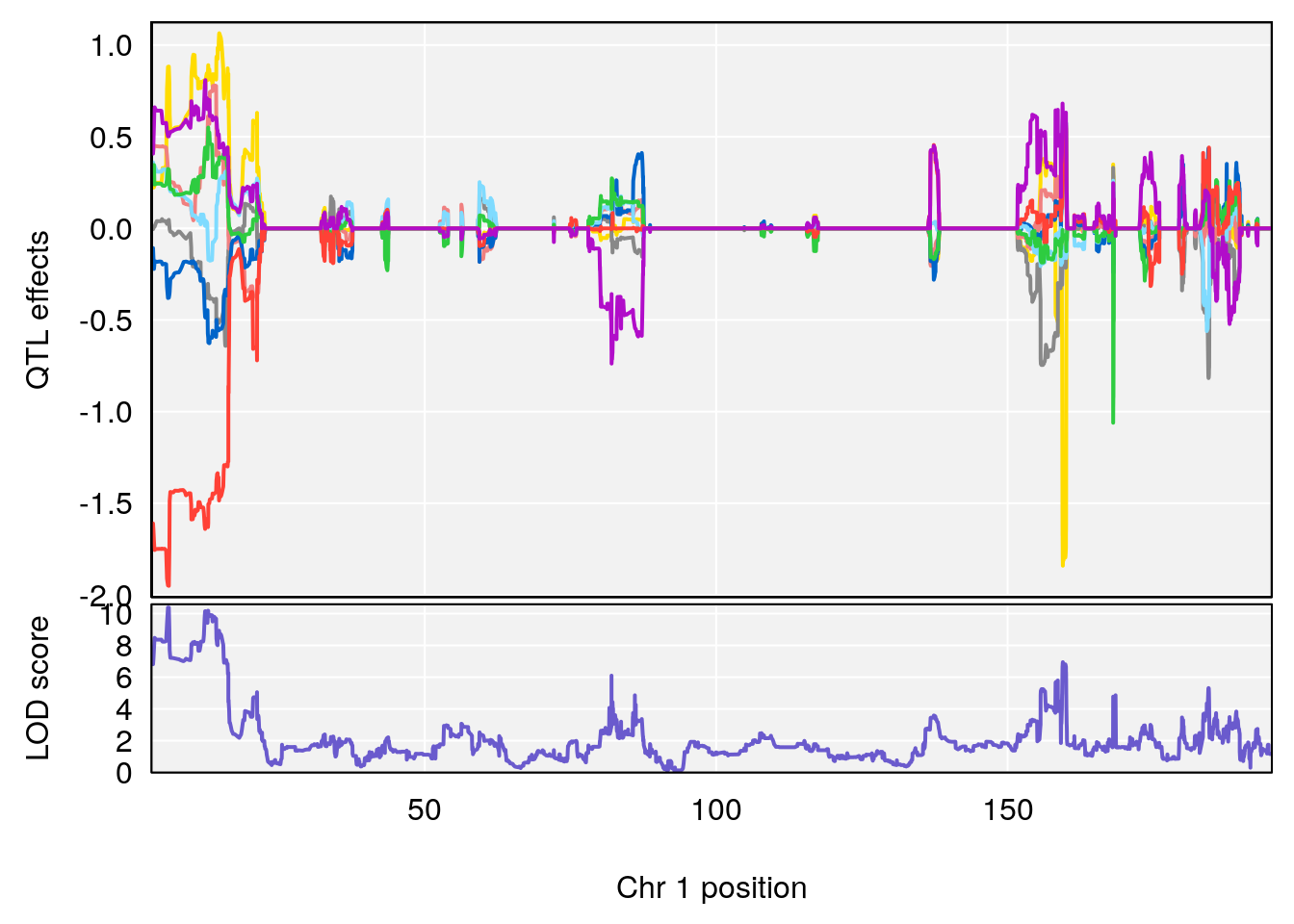
#QTL peaks
DT::datatable(peaks_dat,
filter = list(position = 'top', clear = FALSE),
extensions = 'Buttons',
options = list(dom = 'Blfrtip',
buttons = c('csv', 'excel'),
lengthMenu = list(c(10,25,50,-1),
c(10,25,50,"All")),
pageLength = 40,
scrollY = "300px",
scrollX = "40px"),
caption = htmltools::tags$caption(style = 'caption-side: top; text-align: left; color:black; font-size:200% ;','QTL Peaks'))
sessionInfo()
# R version 4.0.3 (2020-10-10)
# Platform: x86_64-pc-linux-gnu (64-bit)
# Running under: Ubuntu 20.04.2 LTS
#
# Matrix products: default
# BLAS/LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.8.so
#
# locale:
# [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
# [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
# [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=C
# [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
# [9] LC_ADDRESS=C LC_TELEPHONE=C
# [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
#
# attached base packages:
# [1] parallel stats graphics grDevices utils datasets methods
# [8] base
#
# other attached packages:
# [1] biomaRt_2.46.3 DT_0.17 qtlcharts_0.12-10 qtl2_0.24
# [5] broman_0.72-4 forcats_0.5.1 stringr_1.4.0 dplyr_1.0.4
# [9] purrr_0.3.4 readr_1.4.0 tidyr_1.1.2 tibble_3.0.6
# [13] tidyverse_1.3.0 abind_1.4-5 regress_1.3-21 survival_3.2-7
# [17] qtl_1.47-9 GGally_2.1.0 gridExtra_2.3 ggplot2_3.3.3
# [21] workflowr_1.6.2
#
# loaded via a namespace (and not attached):
# [1] fs_1.5.0 lubridate_1.7.9.2 bit64_4.0.5
# [4] progress_1.2.2 RColorBrewer_1.1-2 httr_1.4.2
# [7] rprojroot_2.0.2 tools_4.0.3 backports_1.2.1
# [10] R6_2.5.0 BiocGenerics_0.36.1 DBI_1.1.1
# [13] colorspace_2.0-0 withr_2.4.1 prettyunits_1.1.1
# [16] tidyselect_1.1.0 rematch_1.0.1 curl_4.3
# [19] bit_4.0.4 compiler_4.0.3 git2r_0.28.0
# [22] Biobase_2.50.0 cli_2.3.0 rvest_0.3.6
# [25] xml2_1.3.2 labeling_0.4.2 scales_1.1.1
# [28] askpass_1.1 rappdirs_0.3.3 digest_0.6.27
# [31] rmarkdown_2.6 pkgconfig_2.0.3 htmltools_0.5.1.1
# [34] highr_0.8 dbplyr_2.1.0 fastmap_1.1.0
# [37] htmlwidgets_1.5.3 rlang_1.0.2 readxl_1.3.1
# [40] rstudioapi_0.13 RSQLite_2.2.3 farver_2.0.3
# [43] generics_0.1.0 jsonlite_1.7.2 crosstalk_1.1.1
# [46] magrittr_2.0.1 Matrix_1.3-2 S4Vectors_0.28.1
# [49] Rcpp_1.0.6 munsell_0.5.0 lifecycle_1.0.0
# [52] stringi_1.5.3 whisker_0.4 yaml_2.2.1
# [55] BiocFileCache_1.14.0 plyr_1.8.6 grid_4.0.3
# [58] blob_1.2.1 promises_1.2.0.1 crayon_1.4.1
# [61] lattice_0.20-41 haven_2.3.1 splines_4.0.3
# [64] hms_1.0.0 knitr_1.31 pillar_1.4.7
# [67] stats4_4.0.3 reprex_1.0.0 XML_3.99-0.5
# [70] glue_1.4.2 evaluate_0.14 data.table_1.13.6
# [73] modelr_0.1.8 vctrs_0.3.6 httpuv_1.5.5
# [76] cellranger_1.1.0 openssl_1.4.3 gtable_0.3.0
# [79] reshape_0.8.8 assertthat_0.2.1 cachem_1.0.4
# [82] xfun_0.21 broom_0.7.4 later_1.1.0.1
# [85] IRanges_2.24.1 AnnotationDbi_1.52.0 memoise_2.0.0
# [88] ellipsis_0.3.1This R Markdown site was created with workflowr